├── .gitignore
├── .vscode
└── settings.json
├── LICENSE
├── README.md
├── README.rst
├── complexCGR.pdf
├── complexcgr
├── __init__.py
├── cgr.py
├── complexcgr.py
├── complexfcgr.py
├── fcgr.py
├── fcgr_kmc.py
├── fcgr_samples.py
└── icgr.py
├── img
├── ACG-complexCGR.png
├── ACG.jpg
├── ACG_16bits.jpg
├── CGA.jpg
├── CGAN.jpg
├── SAMD00003784.jpeg
├── SAMD00032020.jpeg
├── SAMD00111209.jpeg
├── SAMD00111306.jpeg
├── complexcgr-readme.png
├── logo-complexCGR-nb.png
├── logo-complexCGR-v2-nb.png
├── logo-complexCGR-v2.png
├── logo-complexCGR-v3-nb.png
├── logo-complexCGR-v3.png
└── logo-complexCGR.png
├── poetry.lock
├── pyproject.toml
└── tests
├── __init__.py
├── test_cgr.py
├── test_complexcgr.py
├── test_complexfcgr.py
├── test_complexfcgr_savefig.py
├── test_fcgr.py
└── test_icgr.py
/.gitignore:
--------------------------------------------------------------------------------
1 | # img/
2 | # Byte-compiled / optimized / DLL files
3 | __pycache__/
4 | *.py[cod]
5 | *$py.class
6 | env.yml
7 | # C extensions
8 | *.so
9 |
10 | # Distribution / packaging
11 | .Python
12 | build/
13 | develop-eggs/
14 | dist/
15 | downloads/
16 | eggs/
17 | .eggs/
18 | lib/
19 | lib64/
20 | parts/
21 | sdist/
22 | var/
23 | wheels/
24 | pip-wheel-metadata/
25 | share/python-wheels/
26 | *.egg-info/
27 | .installed.cfg
28 | *.egg
29 | MANIFEST
30 |
31 | # PyInstaller
32 | # Usually these files are written by a python script from a template
33 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
34 | *.manifest
35 | *.spec
36 |
37 | # Installer logs
38 | pip-log.txt
39 | pip-delete-this-directory.txt
40 |
41 | # Unit test / coverage reports
42 | htmlcov/
43 | .tox/
44 | .nox/
45 | .coverage
46 | .coverage.*
47 | .cache
48 | nosetests.xml
49 | coverage.xml
50 | *.cover
51 | *.py,cover
52 | .hypothesis/
53 | .pytest_cache/
54 |
55 | # Translations
56 | *.mo
57 | *.pot
58 |
59 | # Django stuff:
60 | *.log
61 | local_settings.py
62 | db.sqlite3
63 | db.sqlite3-journal
64 |
65 | # Flask stuff:
66 | instance/
67 | .webassets-cache
68 |
69 | # Scrapy stuff:
70 | .scrapy
71 |
72 | # Sphinx documentation
73 | docs/_build/
74 |
75 | # PyBuilder
76 | target/
77 |
78 | # Jupyter Notebook
79 | .ipynb_checkpoints
80 |
81 | # IPython
82 | profile_default/
83 | ipython_config.py
84 |
85 | # pyenv
86 | .python-version
87 |
88 | # pipenv
89 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
90 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
91 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
92 | # install all needed dependencies.
93 | #Pipfile.lock
94 |
95 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
96 | __pypackages__/
97 |
98 | # Celery stuff
99 | celerybeat-schedule
100 | celerybeat.pid
101 |
102 | # SageMath parsed files
103 | *.sage.py
104 |
105 | # Environments
106 | .env
107 | .venv
108 | env/
109 | venv/
110 | ENV/
111 | env.bak/
112 | venv.bak/
113 |
114 | # Spyder project settings
115 | .spyderproject
116 | .spyproject
117 |
118 | # Rope project settings
119 | .ropeproject
120 |
121 | # mkdocs documentation
122 | /site
123 |
124 | # mypy
125 | .mypy_cache/
126 | .dmypy.json
127 | dmypy.json
128 |
129 | # Pyre type checker
130 | .pyre/
131 |
--------------------------------------------------------------------------------
/.vscode/settings.json:
--------------------------------------------------------------------------------
1 | {
2 | "workbench.colorCustomizations": {
3 | "activityBar.activeBackground": "#65c89b",
4 | "activityBar.activeBorder": "#945bc4",
5 | "activityBar.background": "#65c89b",
6 | "activityBar.foreground": "#15202b",
7 | "activityBar.inactiveForeground": "#15202b99",
8 | "activityBarBadge.background": "#945bc4",
9 | "activityBarBadge.foreground": "#e7e7e7",
10 | "sash.hoverBorder": "#65c89b",
11 | "statusBar.background": "#42b883",
12 | "statusBar.foreground": "#15202b",
13 | "statusBarItem.hoverBackground": "#359268",
14 | "statusBarItem.remoteBackground": "#42b883",
15 | "statusBarItem.remoteForeground": "#15202b",
16 | "titleBar.activeBackground": "#42b883",
17 | "titleBar.activeForeground": "#15202b",
18 | "titleBar.inactiveBackground": "#42b88399",
19 | "titleBar.inactiveForeground": "#15202b99",
20 | "commandCenter.border": "#15202b99"
21 | },
22 | "peacock.color": "#42b883"
23 | }
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2021 Koke
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |  2 |
3 | # `complexcgr`
4 | This library have classes around the *Chaos Game Representation* for DNA sequence
5 |
6 | **The FCGR helps to visualize a k-mer distribution** The `FCGR` of a sequence is an image showing the distribution of the $k$-mers
7 | given a chosen $k$. The frequencies of all $k$-mers are distributed in the position of a matrix of $2^k \times 2^k$,
8 | which considers all the possible $k$-mers: $4^k$.
9 |
10 | The position that a $k$-mer uses in the matrix depends on the encoding given by the `CGR`.
11 |
12 | Some examples of [bacterial assemblies](https://zenodo.org/records/4602622) ([see reference](https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001421)) are shown below.
13 | The name of the species and the `sample_id` is in the title of each image ([see an example with the first image](https://www.ebi.ac.uk/ena/browser/view/SAMEA2658585)). These images were
14 | created using the 6-mers of each assembly and the class `FCGR` of this library.
15 |
16 | |  |
17 | |:--:|
18 | |10 different species of bacteria represented by their FCGR (6-mers)| -->
19 |
20 | ## Installation
21 | [pypi](https://pypi.org/project/complexcgr/)
22 | ___
23 | ```shell
24 | pip install complexcgr
25 | ```
26 |
27 | to update to the latest version
28 | ```shell
29 | pip install complexcgr --upgrade
30 | ```
31 |
32 | ## How to use
33 | ___
34 | ### 1. `CGR` Chaos Game Representation of DNA
35 | ```python
36 | from complexcgr import CGR
37 |
38 | # Instantiate class CGR
39 | cgr = CGR()
40 |
41 | # encode a sequence
42 | cgr.encode("ACGT")
43 | # > CGRCoords(N=4, x=0.1875, y=-0.5625)
44 |
45 | # recover a sequence from CGR coordinates
46 | cgr.decode(N=4,x=0.1875,y=-0.5625)
47 | # > "ACGT"
48 | ```
49 |
50 | ### 2. `FCGR` Frequency Matrix of Chaos Game Representation of DNA
51 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
52 | will not be considered for the calculation of the frequency matrix CGR
53 | ```python
54 | import random; random.seed(42)
55 | from complexcgr import FCGR
56 |
57 | # set the k-mer
58 | fcgr = FCGR(k=8) # (256x256) array
59 |
60 | # Generate a random sequence without T's
61 | seq = "".join(random.choice("ACG") for _ in range(300_000))
62 | chaos = fcgr(seq) # an array with the frequencies of each k-mer
63 | fcgr.plot(chaos)
64 | ```
65 | |  |
66 | |:--:|
67 | |FCGR representation for a sequence without T's|
68 |
69 |
70 | You can save the image with
71 | ```python
72 | fcgr.save_img(chaos, path="img/ACG.jpg")
73 | ```
74 | *Formats allowed are defined by PIL.*
75 |
76 | You can also generate the image in 16 (or more bits), to avoid losing information of k-mer frequencies
77 | ```python
78 | # Generate image in 16-bits (default is 8-bits)
79 | fcgr = FCGR(k=8, bits=16) # (256x256) array. When using plot() it will be rescaled to [0,65535] colors
80 | ```
81 |
82 |
83 | ```python
84 | # Generate a random sequence without T's and lots of N's
85 | seq = "".join(random.choice("ACGN") for _ in range(300_000))
86 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
87 | fcgr.plot(chaos)
88 | ```
89 |
90 |
91 | ||
92 | |:--:|
93 | |FCGR representation for a sequence without T's and lots of N's|
94 |
95 |
96 |
97 | ### 3. `iCGR` integer Chaos Game Representation of DNA
98 | ```python
99 | from complexcgr import iCGR
100 |
101 | # Instantiate class CGR
102 | icgr = iCGR()
103 |
104 | # encode a sequence
105 | icgr.encode("ACGT")
106 | # > CGRCoords(N=4, x=3, y=-9)
107 |
108 | # recover a sequence from CGR coordinates
109 | icgr.decode(N=4,x=3,y=-9)
110 | # > "ACGT"
111 | ```
112 |
113 | ### 4. `ComplexCGR` Complex Chaos Game Representation of DNA (ComplexCGR)
114 |
115 | ```python
116 | from complexcgr import ComplexCGR
117 |
118 | # Instantiate class CGR
119 | ccgr = ComplexCGR()
120 |
121 | # encode a sequence
122 | ccgr.encode("ACGT")
123 | # > CGRCoords(k=228,N=4)
124 |
125 | # recover a sequence from ComplexCGR coordinates
126 | ccgr.decode(k=228,N=4)
127 | # > "ACGT"
128 |
129 | ```
130 |
131 | ### 5. `ComplexFCGR` Frequency Matrix of Complex Chaos Game Representation of DNA
132 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
133 | will not be considered for the calculation of the frequency matrix CGR
134 | ```python
135 | import random; random.seed(42)
136 | from complexcgr import FCGR
137 |
138 | # set the k-mer desired
139 | cfcgr = ComplexFCGR(k=8) # 8-mers
140 |
141 | # Generate a random sequence without T's
142 | seq = "".join(random.choice("ACG") for _ in range(300_000))
143 | fig = cfcgr(seq)
144 |
145 | ```
146 | |  |
147 | |:--:|
148 | |ComplexFCGR representation for a sequence without T's|
149 |
150 |
151 | You can save the image with
152 | ```python
153 | cfcgr.save(fig, path="img/ACG-ComplexCGR.png")
154 | ```
155 | *Currently the plot must be saved as png*
156 |
157 | ___
158 | ## Advice for Real applications
159 |
160 | **Count k-mers** could be the bottleneck for large sequences (> 100000 bp).
161 | Note that the class `FCGR` (and `ComplexCGR`) has implemented a naive approach to count k-mers, this is intended since in practice state-of-the-art tools like KMC or Jellyfish are used to count k-mers very efficiently.
162 |
163 | We provide the class `FCGRKmc`, that receives as input the file generated by the following pipeline using [KMC3](https://github.com/refresh-bio/KMC)
164 |
165 | Make sure to have `kmc` installed. One recommended way is to create a conda environment and [install it there](https://anaconda.org/bioconda/kmc)
166 |
167 | ```bash
168 | kmer_size=6
169 | input="path/to/sequence.fa"
170 | output="path/to/count-kmers.txt"
171 |
172 | mkdir -p tmp-kmc
173 | kmc -v -k$kmer_size -m4 -sm -ci0 -cs100000 -b -t4 -fa $input $input "tmp-kmc"
174 | kmc_tools -t4 -v transform $input dump $output
175 | rm -r $input.kmc_pre $input.kmc_suf
176 | ```
177 | the output file `path/to/count-kmers.txt` can be used with `FCGRKmc`
178 |
179 | ```python
180 | from complexcgr import FCGRKmc
181 |
182 | kmer = 6
183 | fcgr = FCGRKmc(kmer)
184 |
185 | arr = fcgr("path/to/count-kmers.txt") # k-mer counts ordered in a matrix of 2^k x 2^k
186 |
187 |
188 | # to visualize the distribution of k-mers.
189 | # Frequencies are scaled between [min, max] values.
190 | # White color corresponds to the minimum value of frequency
191 | # Black color corresponds to the maximum value of frequency
192 | fcgr.plot(arr)
193 |
194 | # Save it with numpy
195 | import numpy as np
196 | np.save("path_save/fcgr.npy",arr)
197 | ```
198 |
199 | ___
200 | # Videos
201 |
202 | **CGR encoding**
203 |
204 | [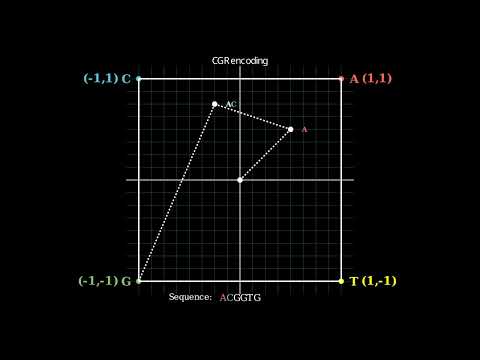](https://youtu.be/HU15ge0fkOY)
205 |
206 | **CGR encoding of all k-mers**
207 |
208 | [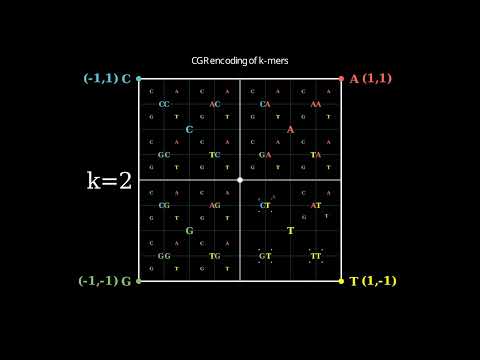](https://youtu.be/oYLT11Q9n5M)
209 |
210 | **ComplexCGR encoding**
211 |
212 | [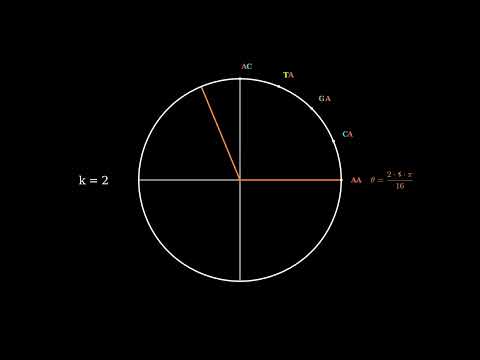](https://youtu.be/xKyXplS5KFk)
213 |
214 | **ComplexCGR and Symmetry**
215 |
216 | [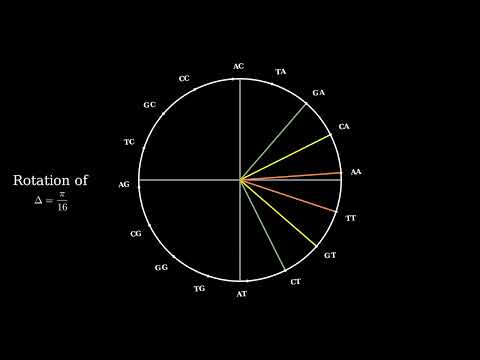](https://www.youtube.com/YmcxVId4_4w)
217 |
218 |
219 | # Functionalities/TODO list
220 | ___
221 | > version 0.8.0:
222 | A list of available classes and functionalities are listed below:
223 |
224 |
225 | **Encoders**
226 | The encoders are functions that map a sequence $s \in \{A,C,G,T\}$ to a point in the plane.
227 | `CGR`, `iCGR`, and `ComplexCGR`.
228 |
229 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
230 | - [x] encode a sequence.
231 | - [x] recover a sequence from a CGR encoding.
232 |
233 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
234 |
235 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
236 | - [x] encode a sequence.
237 | - [x] recover a sequence from a CGR encoding.
238 |
239 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
240 | - [x] encode a sequence
241 | - [x] recover a sequence from an iCGR encoding
242 |
243 | `ComplexCGR`: encodes a DNA sequence in 2 integers $(k,N)$.
244 | - [x] encode a sequence
245 | - [x] recover a sequence from a ComplexCGR encoding
246 | - [x] plot sequence of ComplexCGR encodings
247 |
248 | **Image for distribution of k-mers**
249 |
250 | - [x] `FCGR` Frequency Matrix CGR: representation as an image for k-mer representativity, based on CGR.
251 | - [x] generates FCGR from an arbitrary n-long sequence.
252 | - [x] plot FCGR.
253 | - [x] save FCGR generated.
254 | - [x] save FCGR in different bits.
255 | - [x] `FCGRKmc` Same as `FCGR` but receives as input the file with k-mer counts generated with [KMC](https://github.com/refresh-bio/KMC)
256 | - [x] `ComplexFCGR`: Frequency ComplexCGR: representation as an image (circle) for k-mer representativity, based on ComplexCGR.
257 | - [x] generates ComplexFCGR from an arbitrary n-long sequence.
258 | - [x] plot ComplexFCGR.
259 | - [x] save ComplexFCGR generated.
260 |
261 | - [ ] `PercentileFCGR`:
262 | - [A universal DNA signature for the Tree of Life. EcoEvoRxiv](https://doi.org/10.32942/X24891)
263 | - [ ] `SpacedFCGR`: Create FCGR from spaced-mers
264 | - [Spaced seeds improve k-mer-based metagenomic classification, Bioninformatics](https://academic.oup.com/bioinformatics/article/31/22/3584/240663)
265 | - [Effects of spaced k-mers on alignment-free genotyping](https://academic.oup.com/bioinformatics/article/39/Supplement_1/i213/7210439)
266 |
267 | # Author
268 |
269 | `complexcgr` is developed by [Jorge Avila Cartes](https://github.com/jorgeavilacartes/)
270 |
271 |
272 | # Related publications
273 |
274 | - [Accurate and fast clade assignment via deep learning and frequency chaos game representation](https://doi.org/10.1093/gigascience/giac119)
275 | - [PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases](https://doi.org/10.1101/2025.03.19.644115)
--------------------------------------------------------------------------------
/README.rst:
--------------------------------------------------------------------------------
1 | # complexCGR
2 |
3 | A complex representation for DNA, that preserves some properties of the CGR from the 'Chaos Game Representation for gene structure' by Jeffrey in 1990.
4 |
5 | Each nucleotide will be assigned to one quadrant (I,II,III and IV). This distribution will be the same for the CGR and complexCGR implementations (I:A,II:C,III:G,IV:T)
--------------------------------------------------------------------------------
/complexCGR.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/complexCGR.pdf
--------------------------------------------------------------------------------
/complexcgr/__init__.py:
--------------------------------------------------------------------------------
1 | __version__ ="0.8.0"
2 |
3 | # encodings
4 | from .cgr import CGR
5 | from .icgr import iCGR
6 | from .complexcgr import ComplexCGR
7 |
8 | # from k-mers
9 | from .fcgr import FCGR
10 | from .fcgr_kmc import FCGRKmc
11 | from .complexfcgr import ComplexFCGR
--------------------------------------------------------------------------------
/complexcgr/cgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from typing import Dict, Optional
3 | from collections import namedtuple
4 |
5 | # coordinates for x+iy
6 | Coord = namedtuple("Coord", ["x","y"])
7 |
8 | # coordinates for a CGR encoding
9 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
10 |
11 | # coordinates for each nucleotide in the 2d-plane
12 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
13 |
14 | class CGR:
15 | "Chaos Game Representation for DNA"
16 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
17 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
18 | self.cgr_coords = CGRCoords(0,0,0)
19 |
20 | def nucleotide_by_coords(self,x,y):
21 | "Get nucleotide by coordinates (x,y)"
22 | # filter nucleotide by coordinates
23 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
24 |
25 | return list(filtered.keys())[0]
26 |
27 | def forward(self, nucleotide: str):
28 | "Compute next CGR coordinates"
29 | x = (self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x)/2
30 | y = (self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y)/2
31 |
32 | # update cgr_coords
33 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y)
34 |
35 | def backward(self,):
36 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
37 | # get current nucleotide based on coordinates
38 | n_x,n_y = self.coords_current_nucleotide()
39 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
40 |
41 | # update coordinates to the previous one
42 | x = 2*self.cgr_coords.x - n_x
43 | y = 2*self.cgr_coords.y - n_y
44 |
45 | # update cgr_coords
46 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
47 |
48 | return nucleotide
49 |
50 | def coords_current_nucleotide(self,):
51 | x = 1 if self.cgr_coords.x>0 else -1
52 | y = 1 if self.cgr_coords.y>0 else -1
53 | return x,y
54 |
55 | def encode(self, sequence: str):
56 | "From DNA sequence to CGR"
57 | # reset starting position to (0,0,0)
58 | self.reset_coords()
59 | for nucleotide in sequence:
60 | self.forward(nucleotide)
61 | return self.cgr_coords
62 |
63 | def reset_coords(self,):
64 | self.cgr_coords = CGRCoords(0,0,0)
65 |
66 | def decode(self, N:int, x:int, y:int)->str:
67 | "From CGR to DNA sequence"
68 | self.cgr_coords = CGRCoords(N,x,y)
69 |
70 | # decoded sequence
71 | sequence = []
72 |
73 | # Recover the entire genome
74 | while self.cgr_coords.N>0:
75 | nucleotide = self.backward()
76 | sequence.append(nucleotide)
77 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/complexcgr/complexcgr.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List
2 | from collections import namedtuple
3 |
4 | # coordinates for x+iy
5 | Coord = namedtuple("Coord", ["x","y"])
6 |
7 | # coordinates for a CGR encoding
8 | CGRCoords = namedtuple("CGRCoord", ["k","N"])
9 |
10 | class ComplexCGR:
11 | "Complex Chaos Game Representation of DNA"
12 | def __init__(self, nucleotide_order: List[str] = ["A","C","G","T"]):
13 | self.nucleotide_order = nucleotide_order
14 | self.cgr_coords = CGRCoords(0,0) # complexCGR coordinates
15 |
16 | def id(self, nucleotide):
17 | return self.nucleotide_order.index(nucleotide)
18 |
19 | def forward(self, nucleotide: str):
20 | "compute next complexCGR coordinates"
21 | k = self.id(nucleotide)*4**(self.cgr_coords.N) + self.cgr_coords.k
22 |
23 | # update cgr_coords
24 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N+1)
25 |
26 | def backward(self,):
27 | "compute last complexCGR coordinates"
28 | nucleotide = self.current_nucleotide()
29 |
30 | # compute previous k
31 | k = self.cgr_coords.k - self.id(nucleotide)*4**(self.cgr_coords.N-1)
32 |
33 | # update cgr_coords
34 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N-1)
35 |
36 | return nucleotide
37 |
38 | def encode(self, sequence: str):
39 | "From DNA to complexCGR"
40 | self.reset_coords()
41 | for nucleotide in sequence:
42 | self.forward(nucleotide)
43 | return self.cgr_coords
44 |
45 | def decode(self, k: int, N: int):
46 | "From complexCGR to DNA"
47 | self.cgr_coords = CGRCoords(k,N)
48 |
49 | # decoded sequence
50 | sequence = []
51 |
52 | # Recover the entire genome
53 | while self.cgr_coords.N>0:
54 | nucleotide = self.backward()
55 | sequence.append(nucleotide)
56 | return "".join(sequence[::-1])
57 |
58 | def current_nucleotide(self,):
59 | "Get current nucleotide based on k and N"
60 | k,N = self.cgr_coords.k, self.cgr_coords.N
61 | alpha = k/4**N
62 | if alpha <0.25:
63 | return self.nucleotide_order[0]
64 | elif alpha <0.5:
65 | return self.nucleotide_order[1]
66 | elif alpha < 0.75:
67 | return self.nucleotide_order[2]
68 | else:
69 | return self.nucleotide_order[3]
70 |
71 | def reset_coords(self,):
72 | self.cgr_coords = CGRCoords(0,0)
73 |
--------------------------------------------------------------------------------
/complexcgr/complexfcgr.py:
--------------------------------------------------------------------------------
1 | from . import ComplexCGR
2 | import matplotlib.pyplot as plt
3 | from itertools import product
4 | from collections import defaultdict
5 | from tqdm import tqdm
6 | from PIL import Image
7 | import numpy as np
8 |
9 |
10 | class ComplexFCGR(ComplexCGR):
11 | """Circular density plot based on CGR"""
12 |
13 | def __init__(self, k: int):
14 | super().__init__()
15 | self.k = k # k-mer representation
16 | self.kmers = product("ACGT", repeat=self.k) # all kmers of length k
17 | self.freq_kmer = None # dict to save representativity of each kmer
18 | self.probabilities = None # dict to save probabilities/density for each kmer
19 | self.fig = None # to save matlotlib figure and then save as image
20 |
21 | def __call__(self, sequence: str, w=1):
22 | self.count_kmers(sequence)
23 | self.kmer_probabilities(sequence)
24 | self.plot(w)
25 |
26 | def count_kmers(self, sequence: str):
27 | self.freq_kmer = defaultdict(int)
28 | # representativity of kmers
29 | len_seq = len(sequence)
30 | for j,_ in enumerate(sequence):
31 | if j+self.k <= len_seq:
32 | subseq = sequence[j:j+self.k]
33 | if "N" not in subseq:
34 | self.freq_kmer[subseq] +=1
35 |
36 | def kmer_probabilities(self, sequence: str):
37 | self.probabilities = defaultdict(float)
38 | N=len(sequence)
39 | for key, value in self.freq_kmer.items():
40 | self.probabilities[key] = float(value) / (N - self.k + 1)
41 |
42 | def plot(self, w: int = 1):
43 | "Given a FCGR, plot it in grayscale"
44 | ax = plt.subplot(111, polar=True)
45 | center, bottom, width, height = self.compute_input_plot()
46 |
47 | # scale width by 'w'
48 | width = [w*_ for _ in width]
49 | print("generating plot")
50 | ax.bar(x=center, # center of the angle
51 | width=width, # width of the angle
52 | bottom=bottom, # lowest value
53 | height=height, # highest value
54 | )
55 |
56 | ax.axes.get_xaxis().set_visible(False)
57 | ax.axes.get_yaxis().set_visible(False)
58 |
59 | return ax.figure
60 |
61 | def save(self, ccgr, path: str):
62 | "save complexFCGR as image"
63 | # get figure
64 | fig = self.plot()
65 |
66 | # transform figure to Image
67 | img = self.fig2img(fig)
68 |
69 | # save image
70 | img.save(path)
71 | plt.close(fig)
72 |
73 | @staticmethod
74 | def fig2img(fig):
75 | "Convert a Matplotlib figure to a PIL Image and return it"
76 | #https://stackoverflow.com/questions/57316491/how-to-convert-matplotlib-figure-to-pil-image-object-without-saving-image
77 | import io
78 | buf = io.BytesIO()
79 | fig.savefig(buf)
80 | buf.seek(0)
81 | img = Image.open(buf)
82 | return img
83 |
84 |
85 | def compute_input_plot(self,):
86 | "Compute input for plot CFCGR"
87 | delta = 2*np.pi/4**self.k # angle between consecutive roots
88 |
89 | center = []
90 | width = []
91 | height = []
92 | bottom = []
93 |
94 | for kmer in tqdm(self.probabilities):
95 | h = self.probabilities.get(kmer,0.0) # height (density) of the bar in the circle
96 | theta = 2*self.encode(kmer).k*np.pi/4**self.k # angle of k-esim root
97 | c = theta + delta/2 # center of the angle
98 | center.append(c)
99 | bottom.append(0)
100 | width.append(50*delta)
101 | height.append(h)
102 |
103 | return center, bottom, width, height
--------------------------------------------------------------------------------
/complexcgr/fcgr.py:
--------------------------------------------------------------------------------
1 | from . import CGR
2 | from PIL import Image
3 | from itertools import product
4 | from collections import defaultdict
5 | import numpy as np
6 |
7 | NUC_COMPLEMENT = {n:c for n,c in zip ("ACGT","TGCA")}
8 |
9 | class FCGR(CGR):
10 | """Frequency matrix CGR
11 | an (2**k x 2**k) 2D representation will be created for a
12 | n-long sequence.
13 | - k represents the k-mer.
14 | - 2**k x 2**k = 4**k the total number of k-mers (sequences of length k)
15 | """
16 |
17 | def __init__(self, k: int, use_canonical_kmers: bool = False ,bits: int = 8):
18 | super().__init__()
19 | self.k = k # k-mer representation
20 | self.use_canonical_kmers = use_canonical_kmers
21 | self.kmers = list("".join(kmer) for kmer in product("ACGT", repeat=self.k))
22 |
23 | if use_canonical_kmers is True:
24 | self.kmer2pixel = self._kmer2pixel_canonical_kmers()
25 | else:
26 | self.kmer2pixel = self.kmer2pixel_position()
27 |
28 | self.bits = bits
29 | self.max_color = 2**bits-1
30 |
31 | def __call__(self, sequence: str):
32 | "Given a DNA sequence, returns an array with his FCGR"
33 | self.count_kmers(sequence)
34 |
35 | # Create an empty array to save the FCGR values
36 | array_size = int(2**self.k)
37 | fcgr = np.zeros((array_size,array_size))
38 |
39 | # Assign frequency to each box in the matrix
40 | for kmer, freq in self.freq_kmer.items():
41 | pos_x, pos_y = self.kmer2pixel[kmer]
42 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
43 | return fcgr
44 |
45 | def count_kmer(self, kmer):
46 | if "N" not in kmer:
47 | self.freq_kmer[kmer] += 1
48 |
49 | def count_kmers(self, sequence: str):
50 | self.freq_kmer = defaultdict(int)
51 | # representativity of kmers
52 | last_j = len(sequence) - self.k + 1
53 | kmers = (sequence[i:(i+self.k)] for i in range(last_j))
54 | # count kmers in a dictionary
55 | list(self.count_kmer(kmer) for kmer in kmers)
56 |
57 | def pixel_position(self, kmer: str):
58 | "Get pixel position in the FCGR matrix for a k-mer"
59 |
60 | coords = self.encode(kmer)
61 | N,x,y = coords.N, coords.x, coords.y
62 |
63 | # Coordinates from [-1,1]² to [1,2**k]²
64 | np_coords = np.array([(x + 1)/2, (y + 1)/2]) # move coordinates from [-1,1]² to [0,1]²
65 | np_coords *= 2**self.k # rescale coordinates from [0,1]² to [0,2**k]²
66 | x,y = np.ceil(np_coords) # round to upper integer
67 |
68 | # Turn coordinates (cx,cy) into pixel (px,py) position
69 | # px = 2**k-cy+1, py = cx
70 | return 2**self.k-int(y)+1, int(x)
71 |
72 | def kmer2pixel_position(self,):
73 | kmer2pixel = dict()
74 | for kmer in self.kmers:
75 | kmer2pixel[kmer] = self.pixel_position(kmer)
76 | return kmer2pixel
77 |
78 | def plot(self, fcgr):
79 | "Given a FCGR, plot it in grayscale"
80 | img_pil = self.array2img(fcgr)
81 | return img_pil
82 |
83 | def save_img(self, fcgr, path: str):
84 | "Save image in grayscale for the FCGR provided as input"
85 | img_pil = self.array2img(fcgr)
86 | img_pil.save(path)
87 |
88 | def array2img(self, array):
89 | "Array to PIL image"
90 | m, M = array.min(), array.max()
91 | # rescale to [0,1]
92 | img_rescaled = (array - m) / (M-m)
93 |
94 | # invert colors black->white
95 | img_array = np.ceil(self.max_color - img_rescaled*self.max_color)
96 | dtype = eval(f"np.int{self.bits}")
97 | img_array = np.array(img_array, dtype=dtype)
98 |
99 | # convert to Image
100 | img_pil = Image.fromarray(img_array,'L')
101 | return img_pil
102 |
103 | # # --------------- canonical k-mers ---------------- # #
104 | @staticmethod
105 | def reverse_complement(kmer: str):
106 | rev_kmer = list(kmer)[::-1]
107 | return "".join([NUC_COMPLEMENT[n] for n in rev_kmer])
108 |
109 | def get_canonical_kmer(self, kmer: str):
110 | rev_complement = self.reverse_complement(kmer)
111 | return kmer if kmer < rev_complement else rev_complement
112 |
113 | def _kmer2pixel_canonical_kmers(self,):
114 | # change pixel position of non-canonical kmers
115 | kmer2pixel = dict()
116 | for kmer in self.kmers:
117 | canonical_kmer = self.get_canonical_kmer(kmer)
118 | if kmer != canonical_kmer:
119 | kmer2pixel[kmer] = kmer2pixel[canonical_kmer]
120 | return kmer2pixel
--------------------------------------------------------------------------------
/complexcgr/fcgr_kmc.py:
--------------------------------------------------------------------------------
1 | from . import FCGR
2 |
3 | import gzip
4 | import numpy as np
5 |
6 | class FCGRKmc(FCGR):
7 | """
8 | Create FCGR with the option of using canonical kmers from KMC output
9 | """
10 | def __init__(self, k: int, use_canonical_kmers: bool=False):
11 | super().__init__(k, use_canonical_kmers)
12 | self.k = k # k-mer representation
13 | self.use_canonical_kmers = use_canonical_kmers
14 |
15 | def __call__(self, path_kmc_output):
16 | "Given a path to a kmc output file, return the FCGR using canonical kmers as an array"
17 | # Create an empty array to save the FCGR values
18 | array_size = int(2**self.k)
19 | fcgr = np.zeros((array_size,array_size))
20 |
21 | if str(path_kmc_output).endswith(".txt"):
22 | with open(path_kmc_output) as fp:
23 | for line in fp:
24 | kmer, freq = line.split("\t")
25 | pos_x, pos_y = self.kmer2pixel[kmer]
26 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
27 | else:
28 | with gzip.open(path_kmc_output,'rt') as f:
29 | for line in f:
30 | line = line.strip()
31 | kmer, freq = line.split()
32 | pos_x, pos_y = self.kmer2pixel[kmer]
33 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
34 |
35 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/fcgr_samples.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from Bio import SeqIO
3 | from collections import defaultdict
4 | from pathlib import Path
5 | from tqdm import tqdm
6 | from typing import List, Union
7 | from complexcgr import FCGR
8 |
9 | # input for FCGR Samples
10 | _path_fastq = Union[str, Path] # path can be a string or a Path instance
11 | _fastq = Union[_path_fastq,List[_path_fastq]] # either a single path or a list of paths to fastq files
12 |
13 | class FCGRSamples(FCGR):
14 |
15 | def __init__(self, k: int, bits: int = 8):
16 | super().__init__(k, bits)
17 |
18 | def __call__(self, path_fastq: _fastq, consider_quality: bool = False):
19 | "Given a (list) of fastq files, return the FCGR matrix"
20 |
21 | ## ---- New for reads ----
22 |
23 | # for each call, we start to count the frequencies
24 | self.freq_kmer = defaultdict(int)
25 | if consider_quality is True:
26 | self.qual_kmer = defaultdict(int)
27 |
28 | # transform to list to iterate
29 | path_fastq = path_fastq if type(path_fastq) is list else [path_fastq]
30 |
31 | # For each file, count kmers on their reads
32 | for path in path_fastq:
33 | fastq = self.load_fastq(path)
34 | n_reads = self.count_reads(path)
35 |
36 | # count kmers in each read
37 | for sample in tqdm(fastq,total=n_reads,desc=f"Counting kmers on {str(Path(path).stem)}"):
38 | read = sample.seq
39 | if consider_quality is True:
40 | qual = sample.letter_annotations["phred_quality"]
41 | self.count_kmers_qualities(str(read),qual)
42 | else:
43 | self.count_kmers(str(read))
44 |
45 | ## ---- End new for reads ----
46 |
47 | # Create an empty array to save the FCGR values
48 | if consider_quality is False:
49 | array_size = int(2**self.k)
50 | fcgr = np.zeros((array_size,array_size))
51 | else:
52 | array_size = int(2**self.k)
53 | fcgr = np.zeros((array_size,array_size,2))
54 |
55 | # Assign frequency to each box in the matrix
56 | for kmer, freq in self.freq_kmer.items():
57 | pos_x, pos_y = self.kmer2pixel[kmer]
58 |
59 | if consider_quality is True:
60 | fcgr[int(pos_x)-1,int(pos_y)-1,0] = freq
61 | fcgr[int(pos_x)-1,int(pos_y)-1,1] = self.qual_kmer[kmer]
62 | else:
63 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
64 |
65 | return fcgr if consider_quality is False else self.rescale_fcgr_qualities(fcgr)
66 |
67 |
68 | def load_fastq(self, path):
69 | "Load a fastq file"
70 | fastq = SeqIO.parse(str(path), "fastq")
71 | return fastq
72 |
73 | def count_kmers(self, read: str):
74 | # representativity of kmers
75 | last_j = len(read) - self.k + 1
76 | kmers = (read[i:(i+self.k)] for i in range(last_j))
77 | # count kmers in a dictionary
78 | list(self.count_kmer(kmer) for kmer in kmers)
79 |
80 | @staticmethod
81 | def count_reads(path):
82 | "Count reads in a file to define progress bar"
83 | n_reads = 0
84 | with open(str(path)) as fp:
85 | for line in fp:
86 | if line.startswith("@"):
87 | n_reads += 1
88 | return n_reads
89 |
90 | def count_kmer(self, kmer):
91 | if "N" not in kmer:
92 | self.freq_kmer[kmer] += 1
93 |
94 |
95 | def count_kmer_quality(self, kmer, qmer):
96 | """Count kmers and qualities
97 | For quality of a kmer, the mean of qualities for each nucleotide in the kmer will be saved"""
98 | if "N" not in kmer:
99 | self.freq_kmer[kmer] += 1
100 | self.qual_kmer[kmer] += qmer.mean()
101 |
102 | def count_kmers_qualities(self,read,qual):
103 | "Count kmers and qualities"
104 |
105 | # representativity of kmers
106 | last_j = len(read) - self.k + 1
107 | kmers = (read[i:(i+self.k)] for i in range(last_j))
108 | qmers = (np.array(qual[i:(i+self.k)]) for i in range(last_j)) # qualities for each kmer
109 |
110 | # count kmers and qualities in a dictionary
111 | list(self.count_kmer_quality(kmer,qmer) for (kmer,qmer) in zip(kmers,qmers))
112 |
113 | @staticmethod
114 | def rescale_fcgr_qualities(fcgr):
115 | "Divide each cumulate quality by the freq of its kmer"
116 | freqs, quals = fcgr[:,:,0], fcgr[:,:,1]
117 | fcgr[:,:,1] = np.divide(quals, freqs, out = np.zeros_like(quals), where = quals!=0)
118 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/icgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from itertools import product
3 | from tqdm import tqdm
4 | from typing import Dict, Optional
5 | from collections import defaultdict, namedtuple
6 | import numpy as np
7 |
8 | # coordinates for x+iy
9 | Coord = namedtuple("Coord", ["x","y"])
10 |
11 | # coordinates for a CGR encoding
12 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
13 |
14 | # coordinates for each nucleotide in the 2d-plane
15 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
16 |
17 | class iCGR:
18 | "integer Chaos Game Representation for DNA"
19 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
20 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
21 | self.cgr_coords = CGRCoords(0,0,0)
22 |
23 | def nucleotide_by_coords(self,x,y):
24 | "Get nucleotide by coordinates (x,y)"
25 | # filter nucleotide by coordinates
26 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
27 |
28 | return list(filtered.keys())[0]
29 |
30 | def forward(self, nucleotide: str):
31 | "Compute next CGR coordinates"
32 | # current length
33 | N = self.cgr_coords.N
34 |
35 | # compute next coordinates
36 | x = self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x * 2**N
37 | y = self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y * 2**N
38 |
39 | # update cgr_coords: iCGR starts in the corner of the first nucleotide
40 |
41 | if N>0:
42 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y,)
43 | else:
44 | self.cgr_coords = CGRCoords(1,
45 | self.nucleotide_coords.get(nucleotide).x,
46 | self.nucleotide_coords.get(nucleotide).y,
47 | )
48 |
49 | def backward(self,):
50 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
51 | # get current nucleotide based on coordinates
52 | n_x,n_y = self.coords_current_nucleotide()
53 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
54 |
55 | # current length
56 | N = self.cgr_coords.N
57 |
58 | # update coordinates to the previous one
59 | x = self.cgr_coords.x - self.nucleotide_coords.get(nucleotide).x * 2**(N-1)
60 | y = self.cgr_coords.y - self.nucleotide_coords.get(nucleotide).y * 2**(N-1)
61 |
62 | # update cgr_coords
63 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
64 |
65 | return nucleotide
66 |
67 | def coords_current_nucleotide(self,):
68 | x = 1 if self.cgr_coords.x>0 else -1
69 | y = 1 if self.cgr_coords.y>0 else -1
70 | return x,y
71 |
72 | def encode(self, sequence: str):
73 | "From DNA sequence to CGR"
74 | # reset starting position to (0,0,0)
75 | self.reset_coords()
76 | for nucleotide in sequence:
77 | self.forward(nucleotide)
78 | return self.cgr_coords
79 |
80 | def reset_coords(self,):
81 | self.cgr_coords = CGRCoords(0,0,0)
82 |
83 | def decode(self, N:int, x:int, y:int)->str:
84 | "From CGR to DNA sequence"
85 | self.cgr_coords = CGRCoords(N,x,y)
86 |
87 | # decoded sequence
88 | sequence = []
89 |
90 | # Recover the entire genome
91 | while self.cgr_coords.N>0:
92 | nucleotide = self.backward()
93 | sequence.append(nucleotide)
94 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/img/ACG-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG-complexCGR.png
--------------------------------------------------------------------------------
/img/ACG.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG.jpg
--------------------------------------------------------------------------------
/img/ACG_16bits.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG_16bits.jpg
--------------------------------------------------------------------------------
/img/CGA.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGA.jpg
--------------------------------------------------------------------------------
/img/CGAN.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGAN.jpg
--------------------------------------------------------------------------------
/img/SAMD00003784.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00003784.jpeg
--------------------------------------------------------------------------------
/img/SAMD00032020.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00032020.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111209.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111209.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111306.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111306.jpeg
--------------------------------------------------------------------------------
/img/complexcgr-readme.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/complexcgr-readme.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3.png
--------------------------------------------------------------------------------
/img/logo-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR.png
--------------------------------------------------------------------------------
/poetry.lock:
--------------------------------------------------------------------------------
1 | # This file is automatically @generated by Poetry 1.7.1 and should not be changed by hand.
2 |
3 | [[package]]
4 | name = "atomicwrites"
5 | version = "1.4.1"
6 | description = "Atomic file writes."
7 | optional = false
8 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
9 | files = [
10 | {file = "atomicwrites-1.4.1.tar.gz", hash = "sha256:81b2c9071a49367a7f770170e5eec8cb66567cfbbc8c73d20ce5ca4a8d71cf11"},
11 | ]
12 |
13 | [[package]]
14 | name = "attrs"
15 | version = "23.2.0"
16 | description = "Classes Without Boilerplate"
17 | optional = false
18 | python-versions = ">=3.7"
19 | files = [
20 | {file = "attrs-23.2.0-py3-none-any.whl", hash = "sha256:99b87a485a5820b23b879f04c2305b44b951b502fd64be915879d77a7e8fc6f1"},
21 | {file = "attrs-23.2.0.tar.gz", hash = "sha256:935dc3b529c262f6cf76e50877d35a4bd3c1de194fd41f47a2b7ae8f19971f30"},
22 | ]
23 |
24 | [package.extras]
25 | cov = ["attrs[tests]", "coverage[toml] (>=5.3)"]
26 | dev = ["attrs[tests]", "pre-commit"]
27 | docs = ["furo", "myst-parser", "sphinx", "sphinx-notfound-page", "sphinxcontrib-towncrier", "towncrier", "zope-interface"]
28 | tests = ["attrs[tests-no-zope]", "zope-interface"]
29 | tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
30 | tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
31 |
32 | [[package]]
33 | name = "biopython"
34 | version = "1.82"
35 | description = "Freely available tools for computational molecular biology."
36 | optional = false
37 | python-versions = ">=3.8"
38 | files = [
39 | {file = "biopython-1.82-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:713be5e1e8571ea151864544dfcd2637eaf98c67a6e47b69781d325feb02f6b9"},
40 | {file = "biopython-1.82-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5bc908e84278bf61ac901a0c1b695c85ce49491957e0ab07e1e2afc216882525"},
41 | {file = "biopython-1.82-cp310-cp310-win32.whl", hash = "sha256:f53aff30f01ac9a8aa1fd42dab347a96ee5312945098dfa38f9a5125f6e913fd"},
42 | {file = "biopython-1.82-cp310-cp310-win_amd64.whl", hash = "sha256:0f3f685a2d85348b8c84c6eebe58a74cef7e1158ac52ae5aa91789436907f0e1"},
43 | {file = "biopython-1.82-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c9bc080b7dc6cd6acbf2576766136e8e54acbcc6b7a29ad8075cb5a73f4cfb97"},
44 | {file = "biopython-1.82-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c7802b5033edaf34bd2fbd96b87f96999a6808caa0064c91087c7f1340624898"},
45 | {file = "biopython-1.82-cp311-cp311-win32.whl", hash = "sha256:ec35893b32458f6fa0c98d5f02d801a5fe76961ac66473455f51c0575562c594"},
46 | {file = "biopython-1.82-cp311-cp311-win_amd64.whl", hash = "sha256:40297d418bb142a7ea1a47faedc24bbe21b0ba455f3805373264470e5cb6ec5c"},
47 | {file = "biopython-1.82-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:96f77bd6540683c8958e4ebf55b58e8caa26d20db72891915d06670b783e9cdb"},
48 | {file = "biopython-1.82-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:49b829c2276a0711106663af632c7e203695a5cf082d488d6e10b8aca5ec63e4"},
49 | {file = "biopython-1.82-cp312-cp312-win32.whl", hash = "sha256:54a9b200d4960ed2a4ec85faac2124ffa278afa58d22e07e2d15ca7509219ea1"},
50 | {file = "biopython-1.82-cp312-cp312-win_amd64.whl", hash = "sha256:762547037038d42c9ac98877fc32d4aeba798077129df7790cb178256171a4e5"},
51 | {file = "biopython-1.82-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:cb013256742d31e24fd5093d55511f58bc6a93b8ac723d2ca3b33c7904f1ca14"},
52 | {file = "biopython-1.82-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:16829760fab60ba17141b7a81494fed837c6f29ea28cdcfb3070764c8e8f2ff1"},
53 | {file = "biopython-1.82-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c6af82894dd4f736d737fdea62008a19966251ba39aef39a0ce8b41321305ad5"},
54 | {file = "biopython-1.82-cp38-cp38-win32.whl", hash = "sha256:a1ff583b2f8b314a4c2d612aad4f4b73827a5f741b391f3a834b0c192a4f1527"},
55 | {file = "biopython-1.82-cp38-cp38-win_amd64.whl", hash = "sha256:0198f23776d9cb9d514a797e81a024f486adac973b252a2f4696995a5a163745"},
56 | {file = "biopython-1.82-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c88e9f87d1b07958e7e1e25ff20efced4796daf650e5290692b5c3ff9aebf9d3"},
57 | {file = "biopython-1.82-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8e2533db9275c3e1438151260844e7683f91308da320df02a4c28fb7053ea3a9"},
58 | {file = "biopython-1.82-cp39-cp39-win32.whl", hash = "sha256:80f48d220b92e747300281602f52e98ec6c537ddd4921b580d9a0368bae05ff5"},
59 | {file = "biopython-1.82-cp39-cp39-win_amd64.whl", hash = "sha256:73d619611bd7cf9d2ad8b3217b08d3796acfabb765b0771ad2ff7ee2e46a59b0"},
60 | {file = "biopython-1.82.tar.gz", hash = "sha256:a9b10d959ae88a9744a91c6ce3601f4c86e7ec41679bc93c29f679218f6167bb"},
61 | ]
62 |
63 | [package.dependencies]
64 | numpy = "*"
65 |
66 | [[package]]
67 | name = "colorama"
68 | version = "0.4.6"
69 | description = "Cross-platform colored terminal text."
70 | optional = false
71 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,!=3.3.*,!=3.4.*,!=3.5.*,!=3.6.*,>=2.7"
72 | files = [
73 | {file = "colorama-0.4.6-py2.py3-none-any.whl", hash = "sha256:4f1d9991f5acc0ca119f9d443620b77f9d6b33703e51011c16baf57afb285fc6"},
74 | {file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
75 | ]
76 |
77 | [[package]]
78 | name = "contourpy"
79 | version = "1.2.0"
80 | description = "Python library for calculating contours of 2D quadrilateral grids"
81 | optional = false
82 | python-versions = ">=3.9"
83 | files = [
84 | {file = "contourpy-1.2.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:0274c1cb63625972c0c007ab14dd9ba9e199c36ae1a231ce45d725cbcbfd10a8"},
85 | {file = "contourpy-1.2.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ab459a1cbbf18e8698399c595a01f6dcc5c138220ca3ea9e7e6126232d102bb4"},
86 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6fdd887f17c2f4572ce548461e4f96396681212d858cae7bd52ba3310bc6f00f"},
87 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:5d16edfc3fc09968e09ddffada434b3bf989bf4911535e04eada58469873e28e"},
88 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1c203f617abc0dde5792beb586f827021069fb6d403d7f4d5c2b543d87edceb9"},
89 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b69303ceb2e4d4f146bf82fda78891ef7bcd80c41bf16bfca3d0d7eb545448aa"},
90 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:884c3f9d42d7218304bc74a8a7693d172685c84bd7ab2bab1ee567b769696df9"},
91 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4a1b1208102be6e851f20066bf0e7a96b7d48a07c9b0cfe6d0d4545c2f6cadab"},
92 | {file = "contourpy-1.2.0-cp310-cp310-win32.whl", hash = "sha256:34b9071c040d6fe45d9826cbbe3727d20d83f1b6110d219b83eb0e2a01d79488"},
93 | {file = "contourpy-1.2.0-cp310-cp310-win_amd64.whl", hash = "sha256:bd2f1ae63998da104f16a8b788f685e55d65760cd1929518fd94cd682bf03e41"},
94 | {file = "contourpy-1.2.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:dd10c26b4eadae44783c45ad6655220426f971c61d9b239e6f7b16d5cdaaa727"},
95 | {file = "contourpy-1.2.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:5c6b28956b7b232ae801406e529ad7b350d3f09a4fde958dfdf3c0520cdde0dd"},
96 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ebeac59e9e1eb4b84940d076d9f9a6cec0064e241818bcb6e32124cc5c3e377a"},
97 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:139d8d2e1c1dd52d78682f505e980f592ba53c9f73bd6be102233e358b401063"},
98 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1e9dc350fb4c58adc64df3e0703ab076f60aac06e67d48b3848c23647ae4310e"},
99 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:18fc2b4ed8e4a8fe849d18dce4bd3c7ea637758c6343a1f2bae1e9bd4c9f4686"},
100 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:16a7380e943a6d52472096cb7ad5264ecee36ed60888e2a3d3814991a0107286"},
101 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8d8faf05be5ec8e02a4d86f616fc2a0322ff4a4ce26c0f09d9f7fb5330a35c95"},
102 | {file = "contourpy-1.2.0-cp311-cp311-win32.whl", hash = "sha256:67b7f17679fa62ec82b7e3e611c43a016b887bd64fb933b3ae8638583006c6d6"},
103 | {file = "contourpy-1.2.0-cp311-cp311-win_amd64.whl", hash = "sha256:99ad97258985328b4f207a5e777c1b44a83bfe7cf1f87b99f9c11d4ee477c4de"},
104 | {file = "contourpy-1.2.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:575bcaf957a25d1194903a10bc9f316c136c19f24e0985a2b9b5608bdf5dbfe0"},
105 | {file = "contourpy-1.2.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:9e6c93b5b2dbcedad20a2f18ec22cae47da0d705d454308063421a3b290d9ea4"},
106 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:464b423bc2a009088f19bdf1f232299e8b6917963e2b7e1d277da5041f33a779"},
107 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:68ce4788b7d93e47f84edd3f1f95acdcd142ae60bc0e5493bfd120683d2d4316"},
108 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3d7d1f8871998cdff5d2ff6a087e5e1780139abe2838e85b0b46b7ae6cc25399"},
109 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6e739530c662a8d6d42c37c2ed52a6f0932c2d4a3e8c1f90692ad0ce1274abe0"},
110 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:247b9d16535acaa766d03037d8e8fb20866d054d3c7fbf6fd1f993f11fc60ca0"},
111 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:461e3ae84cd90b30f8d533f07d87c00379644205b1d33a5ea03381edc4b69431"},
112 | {file = "contourpy-1.2.0-cp312-cp312-win32.whl", hash = "sha256:1c2559d6cffc94890b0529ea7eeecc20d6fadc1539273aa27faf503eb4656d8f"},
113 | {file = "contourpy-1.2.0-cp312-cp312-win_amd64.whl", hash = "sha256:491b1917afdd8638a05b611a56d46587d5a632cabead889a5440f7c638bc6ed9"},
114 | {file = "contourpy-1.2.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5fd1810973a375ca0e097dee059c407913ba35723b111df75671a1976efa04bc"},
115 | {file = "contourpy-1.2.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:999c71939aad2780f003979b25ac5b8f2df651dac7b38fb8ce6c46ba5abe6ae9"},
116 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b7caf9b241464c404613512d5594a6e2ff0cc9cb5615c9475cc1d9b514218ae8"},
117 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:266270c6f6608340f6c9836a0fb9b367be61dde0c9a9a18d5ece97774105ff3e"},
118 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:dbd50d0a0539ae2e96e537553aff6d02c10ed165ef40c65b0e27e744a0f10af8"},
119 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:11f8d2554e52f459918f7b8e6aa20ec2a3bce35ce95c1f0ef4ba36fbda306df5"},
120 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:ce96dd400486e80ac7d195b2d800b03e3e6a787e2a522bfb83755938465a819e"},
121 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6d3364b999c62f539cd403f8123ae426da946e142312a514162adb2addd8d808"},
122 | {file = "contourpy-1.2.0-cp39-cp39-win32.whl", hash = "sha256:1c88dfb9e0c77612febebb6ac69d44a8d81e3dc60f993215425b62c1161353f4"},
123 | {file = "contourpy-1.2.0-cp39-cp39-win_amd64.whl", hash = "sha256:78e6ad33cf2e2e80c5dfaaa0beec3d61face0fb650557100ee36db808bfa6843"},
124 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:be16975d94c320432657ad2402f6760990cb640c161ae6da1363051805fa8108"},
125 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b95a225d4948b26a28c08307a60ac00fb8671b14f2047fc5476613252a129776"},
126 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:0d7e03c0f9a4f90dc18d4e77e9ef4ec7b7bbb437f7f675be8e530d65ae6ef956"},
127 | {file = "contourpy-1.2.0.tar.gz", hash = "sha256:171f311cb758de7da13fc53af221ae47a5877be5a0843a9fe150818c51ed276a"},
128 | ]
129 |
130 | [package.dependencies]
131 | numpy = ">=1.20,<2.0"
132 |
133 | [package.extras]
134 | bokeh = ["bokeh", "selenium"]
135 | docs = ["furo", "sphinx (>=7.2)", "sphinx-copybutton"]
136 | mypy = ["contourpy[bokeh,docs]", "docutils-stubs", "mypy (==1.6.1)", "types-Pillow"]
137 | test = ["Pillow", "contourpy[test-no-images]", "matplotlib"]
138 | test-no-images = ["pytest", "pytest-cov", "pytest-xdist", "wurlitzer"]
139 |
140 | [[package]]
141 | name = "cycler"
142 | version = "0.12.1"
143 | description = "Composable style cycles"
144 | optional = false
145 | python-versions = ">=3.8"
146 | files = [
147 | {file = "cycler-0.12.1-py3-none-any.whl", hash = "sha256:85cef7cff222d8644161529808465972e51340599459b8ac3ccbac5a854e0d30"},
148 | {file = "cycler-0.12.1.tar.gz", hash = "sha256:88bb128f02ba341da8ef447245a9e138fae777f6a23943da4540077d3601eb1c"},
149 | ]
150 |
151 | [package.extras]
152 | docs = ["ipython", "matplotlib", "numpydoc", "sphinx"]
153 | tests = ["pytest", "pytest-cov", "pytest-xdist"]
154 |
155 | [[package]]
156 | name = "fonttools"
157 | version = "4.47.0"
158 | description = "Tools to manipulate font files"
159 | optional = false
160 | python-versions = ">=3.8"
161 | files = [
162 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:2d2404107626f97a221dc1a65b05396d2bb2ce38e435f64f26ed2369f68675d9"},
163 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:c01f409be619a9a0f5590389e37ccb58b47264939f0e8d58bfa1f3ba07d22671"},
164 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d986b66ff722ef675b7ee22fbe5947a41f60a61a4da15579d5e276d897fbc7fa"},
165 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e8acf6dd0434b211b3bd30d572d9e019831aae17a54016629fa8224783b22df8"},
166 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:495369c660e0c27233e3c572269cbe520f7f4978be675f990f4005937337d391"},
167 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c59227d7ba5b232281c26ae04fac2c73a79ad0e236bca5c44aae904a18f14faf"},
168 | {file = "fonttools-4.47.0-cp310-cp310-win32.whl", hash = "sha256:59a6c8b71a245800e923cb684a2dc0eac19c56493e2f896218fcf2571ed28984"},
169 | {file = "fonttools-4.47.0-cp310-cp310-win_amd64.whl", hash = "sha256:52c82df66201f3a90db438d9d7b337c7c98139de598d0728fb99dab9fd0495ca"},
170 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:854421e328d47d70aa5abceacbe8eef231961b162c71cbe7ff3f47e235e2e5c5"},
171 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:511482df31cfea9f697930f61520f6541185fa5eeba2fa760fe72e8eee5af88b"},
172 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ce0e2c88c8c985b7b9a7efcd06511fb0a1fe3ddd9a6cd2895ef1dbf9059719d7"},
173 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e7a0a8848726956e9d9fb18c977a279013daadf0cbb6725d2015a6dd57527992"},
174 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e869da810ae35afb3019baa0d0306cdbab4760a54909c89ad8904fa629991812"},
175 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:dd23848f877c3754f53a4903fb7a593ed100924f9b4bff7d5a4e2e8a7001ae11"},
176 | {file = "fonttools-4.47.0-cp311-cp311-win32.whl", hash = "sha256:bf1810635c00f7c45d93085611c995fc130009cec5abdc35b327156aa191f982"},
177 | {file = "fonttools-4.47.0-cp311-cp311-win_amd64.whl", hash = "sha256:61df4dee5d38ab65b26da8efd62d859a1eef7a34dcbc331299a28e24d04c59a7"},
178 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:e3f4d61f3a8195eac784f1d0c16c0a3105382c1b9a74d99ac4ba421da39a8826"},
179 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:174995f7b057e799355b393e97f4f93ef1f2197cbfa945e988d49b2a09ecbce8"},
180 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea592e6a09b71cb7a7661dd93ac0b877a6228e2d677ebacbad0a4d118494c86d"},
181 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:40bdbe90b33897d9cc4a39f8e415b0fcdeae4c40a99374b8a4982f127ff5c767"},
182 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:843509ae9b93db5aaf1a6302085e30bddc1111d31e11d724584818f5b698f500"},
183 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:9acfa1cdc479e0dde528b61423855913d949a7f7fe09e276228298fef4589540"},
184 | {file = "fonttools-4.47.0-cp312-cp312-win32.whl", hash = "sha256:66c92ec7f95fd9732550ebedefcd190a8d81beaa97e89d523a0d17198a8bda4d"},
185 | {file = "fonttools-4.47.0-cp312-cp312-win_amd64.whl", hash = "sha256:e8fa20748de55d0021f83754b371432dca0439e02847962fc4c42a0e444c2d78"},
186 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c75e19971209fbbce891ebfd1b10c37320a5a28e8d438861c21d35305aedb81c"},
187 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e79f1a3970d25f692bbb8c8c2637e621a66c0d60c109ab48d4a160f50856deff"},
188 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:562681188c62c024fe2c611b32e08b8de2afa00c0c4e72bed47c47c318e16d5c"},
189 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a77a60315c33393b2bd29d538d1ef026060a63d3a49a9233b779261bad9c3f71"},
190 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:b4fabb8cc9422efae1a925160083fdcbab8fdc96a8483441eb7457235df625bd"},
191 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2a78dba8c2a1e9d53a0fb5382979f024200dc86adc46a56cbb668a2249862fda"},
192 | {file = "fonttools-4.47.0-cp38-cp38-win32.whl", hash = "sha256:e6b968543fde4119231c12c2a953dcf83349590ca631ba8216a8edf9cd4d36a9"},
193 | {file = "fonttools-4.47.0-cp38-cp38-win_amd64.whl", hash = "sha256:4a9a51745c0439516d947480d4d884fa18bd1458e05b829e482b9269afa655bc"},
194 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:62d8ddb058b8e87018e5dc26f3258e2c30daad4c87262dfeb0e2617dd84750e6"},
195 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5dde0eab40faaa5476133123f6a622a1cc3ac9b7af45d65690870620323308b4"},
196 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f4da089f6dfdb822293bde576916492cd708c37c2501c3651adde39804630538"},
197 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:253bb46bab970e8aae254cebf2ae3db98a4ef6bd034707aa68a239027d2b198d"},
198 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:1193fb090061efa2f9e2d8d743ae9850c77b66746a3b32792324cdce65784154"},
199 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:084511482dd265bce6dca24c509894062f0117e4e6869384d853f46c0e6d43be"},
200 | {file = "fonttools-4.47.0-cp39-cp39-win32.whl", hash = "sha256:97620c4af36e4c849e52661492e31dc36916df12571cb900d16960ab8e92a980"},
201 | {file = "fonttools-4.47.0-cp39-cp39-win_amd64.whl", hash = "sha256:e77bdf52185bdaf63d39f3e1ac3212e6cfa3ab07d509b94557a8902ce9c13c82"},
202 | {file = "fonttools-4.47.0-py3-none-any.whl", hash = "sha256:d6477ba902dd2d7adda7f0fd3bfaeb92885d45993c9e1928c9f28fc3961415f7"},
203 | {file = "fonttools-4.47.0.tar.gz", hash = "sha256:ec13a10715eef0e031858c1c23bfaee6cba02b97558e4a7bfa089dba4a8c2ebf"},
204 | ]
205 |
206 | [package.extras]
207 | all = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "fs (>=2.2.0,<3)", "lxml (>=4.0,<5)", "lz4 (>=1.7.4.2)", "matplotlib", "munkres", "pycairo", "scipy", "skia-pathops (>=0.5.0)", "sympy", "uharfbuzz (>=0.23.0)", "unicodedata2 (>=15.1.0)", "xattr", "zopfli (>=0.1.4)"]
208 | graphite = ["lz4 (>=1.7.4.2)"]
209 | interpolatable = ["munkres", "pycairo", "scipy"]
210 | lxml = ["lxml (>=4.0,<5)"]
211 | pathops = ["skia-pathops (>=0.5.0)"]
212 | plot = ["matplotlib"]
213 | repacker = ["uharfbuzz (>=0.23.0)"]
214 | symfont = ["sympy"]
215 | type1 = ["xattr"]
216 | ufo = ["fs (>=2.2.0,<3)"]
217 | unicode = ["unicodedata2 (>=15.1.0)"]
218 | woff = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "zopfli (>=0.1.4)"]
219 |
220 | [[package]]
221 | name = "importlib-resources"
222 | version = "6.1.1"
223 | description = "Read resources from Python packages"
224 | optional = false
225 | python-versions = ">=3.8"
226 | files = [
227 | {file = "importlib_resources-6.1.1-py3-none-any.whl", hash = "sha256:e8bf90d8213b486f428c9c39714b920041cb02c184686a3dee24905aaa8105d6"},
228 | {file = "importlib_resources-6.1.1.tar.gz", hash = "sha256:3893a00122eafde6894c59914446a512f728a0c1a45f9bb9b63721b6bacf0b4a"},
229 | ]
230 |
231 | [package.dependencies]
232 | zipp = {version = ">=3.1.0", markers = "python_version < \"3.10\""}
233 |
234 | [package.extras]
235 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
236 | testing = ["pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy (>=0.9.1)", "pytest-ruff", "zipp (>=3.17)"]
237 |
238 | [[package]]
239 | name = "iniconfig"
240 | version = "2.0.0"
241 | description = "brain-dead simple config-ini parsing"
242 | optional = false
243 | python-versions = ">=3.7"
244 | files = [
245 | {file = "iniconfig-2.0.0-py3-none-any.whl", hash = "sha256:b6a85871a79d2e3b22d2d1b94ac2824226a63c6b741c88f7ae975f18b6778374"},
246 | {file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
247 | ]
248 |
249 | [[package]]
250 | name = "kiwisolver"
251 | version = "1.4.5"
252 | description = "A fast implementation of the Cassowary constraint solver"
253 | optional = false
254 | python-versions = ">=3.7"
255 | files = [
256 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:05703cf211d585109fcd72207a31bb170a0f22144d68298dc5e61b3c946518af"},
257 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:146d14bebb7f1dc4d5fbf74f8a6cb15ac42baadee8912eb84ac0b3b2a3dc6ac3"},
258 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ef7afcd2d281494c0a9101d5c571970708ad911d028137cd558f02b851c08b4"},
259 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9eaa8b117dc8337728e834b9c6e2611f10c79e38f65157c4c38e9400286f5cb1"},
260 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ec20916e7b4cbfb1f12380e46486ec4bcbaa91a9c448b97023fde0d5bbf9e4ff"},
261 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:39b42c68602539407884cf70d6a480a469b93b81b7701378ba5e2328660c847a"},
262 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:aa12042de0171fad672b6c59df69106d20d5596e4f87b5e8f76df757a7c399aa"},
263 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:2a40773c71d7ccdd3798f6489aaac9eee213d566850a9533f8d26332d626b82c"},
264 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:19df6e621f6d8b4b9c4d45f40a66839294ff2bb235e64d2178f7522d9170ac5b"},
265 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:83d78376d0d4fd884e2c114d0621624b73d2aba4e2788182d286309ebdeed770"},
266 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:e391b1f0a8a5a10ab3b9bb6afcfd74f2175f24f8975fb87ecae700d1503cdee0"},
267 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:852542f9481f4a62dbb5dd99e8ab7aedfeb8fb6342349a181d4036877410f525"},
268 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:59edc41b24031bc25108e210c0def6f6c2191210492a972d585a06ff246bb79b"},
269 | {file = "kiwisolver-1.4.5-cp310-cp310-win32.whl", hash = "sha256:a6aa6315319a052b4ee378aa171959c898a6183f15c1e541821c5c59beaa0238"},
270 | {file = "kiwisolver-1.4.5-cp310-cp310-win_amd64.whl", hash = "sha256:d0ef46024e6a3d79c01ff13801cb19d0cad7fd859b15037aec74315540acc276"},
271 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:11863aa14a51fd6ec28688d76f1735f8f69ab1fabf388851a595d0721af042f5"},
272 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:8ab3919a9997ab7ef2fbbed0cc99bb28d3c13e6d4b1ad36e97e482558a91be90"},
273 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fcc700eadbbccbf6bc1bcb9dbe0786b4b1cb91ca0dcda336eef5c2beed37b797"},
274 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:dfdd7c0b105af050eb3d64997809dc21da247cf44e63dc73ff0fd20b96be55a9"},
275 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:76c6a5964640638cdeaa0c359382e5703e9293030fe730018ca06bc2010c4437"},
276 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:bbea0db94288e29afcc4c28afbf3a7ccaf2d7e027489c449cf7e8f83c6346eb9"},
277 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ceec1a6bc6cab1d6ff5d06592a91a692f90ec7505d6463a88a52cc0eb58545da"},
278 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:040c1aebeda72197ef477a906782b5ab0d387642e93bda547336b8957c61022e"},
279 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f91de7223d4c7b793867797bacd1ee53bfe7359bd70d27b7b58a04efbb9436c8"},
280 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:faae4860798c31530dd184046a900e652c95513796ef51a12bc086710c2eec4d"},
281 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:b0157420efcb803e71d1b28e2c287518b8808b7cf1ab8af36718fd0a2c453eb0"},
282 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:06f54715b7737c2fecdbf140d1afb11a33d59508a47bf11bb38ecf21dc9ab79f"},
283 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:fdb7adb641a0d13bdcd4ef48e062363d8a9ad4a182ac7647ec88f695e719ae9f"},
284 | {file = "kiwisolver-1.4.5-cp311-cp311-win32.whl", hash = "sha256:bb86433b1cfe686da83ce32a9d3a8dd308e85c76b60896d58f082136f10bffac"},
285 | {file = "kiwisolver-1.4.5-cp311-cp311-win_amd64.whl", hash = "sha256:6c08e1312a9cf1074d17b17728d3dfce2a5125b2d791527f33ffbe805200a355"},
286 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:32d5cf40c4f7c7b3ca500f8985eb3fb3a7dfc023215e876f207956b5ea26632a"},
287 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:f846c260f483d1fd217fe5ed7c173fb109efa6b1fc8381c8b7552c5781756192"},
288 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5ff5cf3571589b6d13bfbfd6bcd7a3f659e42f96b5fd1c4830c4cf21d4f5ef45"},
289 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7269d9e5f1084a653d575c7ec012ff57f0c042258bf5db0954bf551c158466e7"},
290 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:da802a19d6e15dffe4b0c24b38b3af68e6c1a68e6e1d8f30148c83864f3881db"},
291 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:3aba7311af82e335dd1e36ffff68aaca609ca6290c2cb6d821a39aa075d8e3ff"},
292 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:763773d53f07244148ccac5b084da5adb90bfaee39c197554f01b286cf869228"},
293 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2270953c0d8cdab5d422bee7d2007f043473f9d2999631c86a223c9db56cbd16"},
294 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:d099e745a512f7e3bbe7249ca835f4d357c586d78d79ae8f1dcd4d8adeb9bda9"},
295 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:74db36e14a7d1ce0986fa104f7d5637aea5c82ca6326ed0ec5694280942d1162"},
296 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:7e5bab140c309cb3a6ce373a9e71eb7e4873c70c2dda01df6820474f9889d6d4"},
297 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:0f114aa76dc1b8f636d077979c0ac22e7cd8f3493abbab152f20eb8d3cda71f3"},
298 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:88a2df29d4724b9237fc0c6eaf2a1adae0cdc0b3e9f4d8e7dc54b16812d2d81a"},
299 | {file = "kiwisolver-1.4.5-cp312-cp312-win32.whl", hash = "sha256:72d40b33e834371fd330fb1472ca19d9b8327acb79a5821d4008391db8e29f20"},
300 | {file = "kiwisolver-1.4.5-cp312-cp312-win_amd64.whl", hash = "sha256:2c5674c4e74d939b9d91dda0fae10597ac7521768fec9e399c70a1f27e2ea2d9"},
301 | {file = "kiwisolver-1.4.5-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:3a2b053a0ab7a3960c98725cfb0bf5b48ba82f64ec95fe06f1d06c99b552e130"},
302 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3cd32d6c13807e5c66a7cbb79f90b553642f296ae4518a60d8d76243b0ad2898"},
303 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:59ec7b7c7e1a61061850d53aaf8e93db63dce0c936db1fda2658b70e4a1be709"},
304 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da4cfb373035def307905d05041c1d06d8936452fe89d464743ae7fb8371078b"},
305 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:2400873bccc260b6ae184b2b8a4fec0e4082d30648eadb7c3d9a13405d861e89"},
306 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:1b04139c4236a0f3aff534479b58f6f849a8b351e1314826c2d230849ed48985"},
307 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:4e66e81a5779b65ac21764c295087de82235597a2293d18d943f8e9e32746265"},
308 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:7931d8f1f67c4be9ba1dd9c451fb0eeca1a25b89e4d3f89e828fe12a519b782a"},
309 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:b3f7e75f3015df442238cca659f8baa5f42ce2a8582727981cbfa15fee0ee205"},
310 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:bbf1d63eef84b2e8c89011b7f2235b1e0bf7dacc11cac9431fc6468e99ac77fb"},
311 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:4c380469bd3f970ef677bf2bcba2b6b0b4d5c75e7a020fb863ef75084efad66f"},
312 | {file = "kiwisolver-1.4.5-cp37-cp37m-win32.whl", hash = "sha256:9408acf3270c4b6baad483865191e3e582b638b1654a007c62e3efe96f09a9a3"},
313 | {file = "kiwisolver-1.4.5-cp37-cp37m-win_amd64.whl", hash = "sha256:5b94529f9b2591b7af5f3e0e730a4e0a41ea174af35a4fd067775f9bdfeee01a"},
314 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:11c7de8f692fc99816e8ac50d1d1aef4f75126eefc33ac79aac02c099fd3db71"},
315 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:53abb58632235cd154176ced1ae8f0d29a6657aa1aa9decf50b899b755bc2b93"},
316 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:88b9f257ca61b838b6f8094a62418421f87ac2a1069f7e896c36a7d86b5d4c29"},
317 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3195782b26fc03aa9c6913d5bad5aeb864bdc372924c093b0f1cebad603dd712"},
318 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:fc579bf0f502e54926519451b920e875f433aceb4624a3646b3252b5caa9e0b6"},
319 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5a580c91d686376f0f7c295357595c5a026e6cbc3d77b7c36e290201e7c11ecb"},
320 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:cfe6ab8da05c01ba6fbea630377b5da2cd9bcbc6338510116b01c1bc939a2c18"},
321 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:d2e5a98f0ec99beb3c10e13b387f8db39106d53993f498b295f0c914328b1333"},
322 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a51a263952b1429e429ff236d2f5a21c5125437861baeed77f5e1cc2d2c7c6da"},
323 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:3edd2fa14e68c9be82c5b16689e8d63d89fe927e56debd6e1dbce7a26a17f81b"},
324 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:74d1b44c6cfc897df648cc9fdaa09bc3e7679926e6f96df05775d4fb3946571c"},
325 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:76d9289ed3f7501012e05abb8358bbb129149dbd173f1f57a1bf1c22d19ab7cc"},
326 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:92dea1ffe3714fa8eb6a314d2b3c773208d865a0e0d35e713ec54eea08a66250"},
327 | {file = "kiwisolver-1.4.5-cp38-cp38-win32.whl", hash = "sha256:5c90ae8c8d32e472be041e76f9d2f2dbff4d0b0be8bd4041770eddb18cf49a4e"},
328 | {file = "kiwisolver-1.4.5-cp38-cp38-win_amd64.whl", hash = "sha256:c7940c1dc63eb37a67721b10d703247552416f719c4188c54e04334321351ced"},
329 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:9407b6a5f0d675e8a827ad8742e1d6b49d9c1a1da5d952a67d50ef5f4170b18d"},
330 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:15568384086b6df3c65353820a4473575dbad192e35010f622c6ce3eebd57af9"},
331 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:0dc9db8e79f0036e8173c466d21ef18e1befc02de8bf8aa8dc0813a6dc8a7046"},
332 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:cdc8a402aaee9a798b50d8b827d7ecf75edc5fb35ea0f91f213ff927c15f4ff0"},
333 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6c3bd3cde54cafb87d74d8db50b909705c62b17c2099b8f2e25b461882e544ff"},
334 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:955e8513d07a283056b1396e9a57ceddbd272d9252c14f154d450d227606eb54"},
335 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:346f5343b9e3f00b8db8ba359350eb124b98c99efd0b408728ac6ebf38173958"},
336 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b9098e0049e88c6a24ff64545cdfc50807818ba6c1b739cae221bbbcbc58aad3"},
337 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:00bd361b903dc4bbf4eb165f24d1acbee754fce22ded24c3d56eec268658a5cf"},
338 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:7b8b454bac16428b22560d0a1cf0a09875339cab69df61d7805bf48919415901"},
339 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:f1d072c2eb0ad60d4c183f3fb44ac6f73fb7a8f16a2694a91f988275cbf352f9"},
340 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:31a82d498054cac9f6d0b53d02bb85811185bcb477d4b60144f915f3b3126342"},
341 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6512cb89e334e4700febbffaaa52761b65b4f5a3cf33f960213d5656cea36a77"},
342 | {file = "kiwisolver-1.4.5-cp39-cp39-win32.whl", hash = "sha256:9db8ea4c388fdb0f780fe91346fd438657ea602d58348753d9fb265ce1bca67f"},
343 | {file = "kiwisolver-1.4.5-cp39-cp39-win_amd64.whl", hash = "sha256:59415f46a37f7f2efeec758353dd2eae1b07640d8ca0f0c42548ec4125492635"},
344 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:5c7b3b3a728dc6faf3fc372ef24f21d1e3cee2ac3e9596691d746e5a536de920"},
345 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:620ced262a86244e2be10a676b646f29c34537d0d9cc8eb26c08f53d98013390"},
346 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:378a214a1e3bbf5ac4a8708304318b4f890da88c9e6a07699c4ae7174c09a68d"},
347 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aaf7be1207676ac608a50cd08f102f6742dbfc70e8d60c4db1c6897f62f71523"},
348 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:ba55dce0a9b8ff59495ddd050a0225d58bd0983d09f87cfe2b6aec4f2c1234e4"},
349 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:fd32ea360bcbb92d28933fc05ed09bffcb1704ba3fc7942e81db0fd4f81a7892"},
350 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:5e7139af55d1688f8b960ee9ad5adafc4ac17c1c473fe07133ac092310d76544"},
351 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:dced8146011d2bc2e883f9bd68618b8247387f4bbec46d7392b3c3b032640126"},
352 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c9bf3325c47b11b2e51bca0824ea217c7cd84491d8ac4eefd1e409705ef092bd"},
353 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5794cf59533bc3f1b1c821f7206a3617999db9fbefc345360aafe2e067514929"},
354 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e368f200bbc2e4f905b8e71eb38b3c04333bddaa6a2464a6355487b02bb7fb09"},
355 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:e5d706eba36b4c4d5bc6c6377bb6568098765e990cfc21ee16d13963fab7b3e7"},
356 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:85267bd1aa8880a9c88a8cb71e18d3d64d2751a790e6ca6c27b8ccc724bcd5ad"},
357 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:210ef2c3a1f03272649aff1ef992df2e724748918c4bc2d5a90352849eb40bea"},
358 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:11d011a7574eb3b82bcc9c1a1d35c1d7075677fdd15de527d91b46bd35e935ee"},
359 | {file = "kiwisolver-1.4.5.tar.gz", hash = "sha256:e57e563a57fb22a142da34f38acc2fc1a5c864bc29ca1517a88abc963e60d6ec"},
360 | ]
361 |
362 | [[package]]
363 | name = "matplotlib"
364 | version = "3.8.2"
365 | description = "Python plotting package"
366 | optional = false
367 | python-versions = ">=3.9"
368 | files = [
369 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_10_12_x86_64.whl", hash = "sha256:09796f89fb71a0c0e1e2f4bdaf63fb2cefc84446bb963ecdeb40dfee7dfa98c7"},
370 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6f9c6976748a25e8b9be51ea028df49b8e561eed7809146da7a47dbecebab367"},
371 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b78e4f2cedf303869b782071b55fdde5987fda3038e9d09e58c91cc261b5ad18"},
372 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e208f46cf6576a7624195aa047cb344a7f802e113bb1a06cfd4bee431de5e31"},
373 | {file = "matplotlib-3.8.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:46a569130ff53798ea5f50afce7406e91fdc471ca1e0e26ba976a8c734c9427a"},
374 | {file = "matplotlib-3.8.2-cp310-cp310-win_amd64.whl", hash = "sha256:830f00640c965c5b7f6bc32f0d4ce0c36dfe0379f7dd65b07a00c801713ec40a"},
375 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_10_12_x86_64.whl", hash = "sha256:d86593ccf546223eb75a39b44c32788e6f6440d13cfc4750c1c15d0fcb850b63"},
376 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:9a5430836811b7652991939012f43d2808a2db9b64ee240387e8c43e2e5578c8"},
377 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b9576723858a78751d5aacd2497b8aef29ffea6d1c95981505877f7ac28215c6"},
378 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5ba9cbd8ac6cf422f3102622b20f8552d601bf8837e49a3afed188d560152788"},
379 | {file = "matplotlib-3.8.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:03f9d160a29e0b65c0790bb07f4f45d6a181b1ac33eb1bb0dd225986450148f0"},
380 | {file = "matplotlib-3.8.2-cp311-cp311-win_amd64.whl", hash = "sha256:3773002da767f0a9323ba1a9b9b5d00d6257dbd2a93107233167cfb581f64717"},
381 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_10_12_x86_64.whl", hash = "sha256:4c318c1e95e2f5926fba326f68177dee364aa791d6df022ceb91b8221bd0a627"},
382 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:091275d18d942cf1ee9609c830a1bc36610607d8223b1b981c37d5c9fc3e46a4"},
383 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b0f3b8ea0e99e233a4bcc44590f01604840d833c280ebb8fe5554fd3e6cfe8d"},
384 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d7b1704a530395aaf73912be741c04d181f82ca78084fbd80bc737be04848331"},
385 | {file = "matplotlib-3.8.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:533b0e3b0c6768eef8cbe4b583731ce25a91ab54a22f830db2b031e83cca9213"},
386 | {file = "matplotlib-3.8.2-cp312-cp312-win_amd64.whl", hash = "sha256:0f4fc5d72b75e2c18e55eb32292659cf731d9d5b312a6eb036506304f4675630"},
387 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_10_12_x86_64.whl", hash = "sha256:deaed9ad4da0b1aea77fe0aa0cebb9ef611c70b3177be936a95e5d01fa05094f"},
388 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:172f4d0fbac3383d39164c6caafd3255ce6fa58f08fc392513a0b1d3b89c4f89"},
389 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c7d36c2209d9136cd8e02fab1c0ddc185ce79bc914c45054a9f514e44c787917"},
390 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5864bdd7da445e4e5e011b199bb67168cdad10b501750367c496420f2ad00843"},
391 | {file = "matplotlib-3.8.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:ef8345b48e95cee45ff25192ed1f4857273117917a4dcd48e3905619bcd9c9b8"},
392 | {file = "matplotlib-3.8.2-cp39-cp39-win_amd64.whl", hash = "sha256:7c48d9e221b637c017232e3760ed30b4e8d5dfd081daf327e829bf2a72c731b4"},
393 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-macosx_10_12_x86_64.whl", hash = "sha256:aa11b3c6928a1e496c1a79917d51d4cd5d04f8a2e75f21df4949eeefdf697f4b"},
394 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1095fecf99eeb7384dabad4bf44b965f929a5f6079654b681193edf7169ec20"},
395 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:bddfb1db89bfaa855912261c805bd0e10218923cc262b9159a49c29a7a1c1afa"},
396 | {file = "matplotlib-3.8.2.tar.gz", hash = "sha256:01a978b871b881ee76017152f1f1a0cbf6bd5f7b8ff8c96df0df1bd57d8755a1"},
397 | ]
398 |
399 | [package.dependencies]
400 | contourpy = ">=1.0.1"
401 | cycler = ">=0.10"
402 | fonttools = ">=4.22.0"
403 | importlib-resources = {version = ">=3.2.0", markers = "python_version < \"3.10\""}
404 | kiwisolver = ">=1.3.1"
405 | numpy = ">=1.21,<2"
406 | packaging = ">=20.0"
407 | pillow = ">=8"
408 | pyparsing = ">=2.3.1"
409 | python-dateutil = ">=2.7"
410 |
411 | [[package]]
412 | name = "numpy"
413 | version = "1.26.2"
414 | description = "Fundamental package for array computing in Python"
415 | optional = false
416 | python-versions = ">=3.9"
417 | files = [
418 | {file = "numpy-1.26.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:3703fc9258a4a122d17043e57b35e5ef1c5a5837c3db8be396c82e04c1cf9b0f"},
419 | {file = "numpy-1.26.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:cc392fdcbd21d4be6ae1bb4475a03ce3b025cd49a9be5345d76d7585aea69440"},
420 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:36340109af8da8805d8851ef1d74761b3b88e81a9bd80b290bbfed61bd2b4f75"},
421 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:bcc008217145b3d77abd3e4d5ef586e3bdfba8fe17940769f8aa09b99e856c00"},
422 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:3ced40d4e9e18242f70dd02d739e44698df3dcb010d31f495ff00a31ef6014fe"},
423 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b272d4cecc32c9e19911891446b72e986157e6a1809b7b56518b4f3755267523"},
424 | {file = "numpy-1.26.2-cp310-cp310-win32.whl", hash = "sha256:22f8fc02fdbc829e7a8c578dd8d2e15a9074b630d4da29cda483337e300e3ee9"},
425 | {file = "numpy-1.26.2-cp310-cp310-win_amd64.whl", hash = "sha256:26c9d33f8e8b846d5a65dd068c14e04018d05533b348d9eaeef6c1bd787f9919"},
426 | {file = "numpy-1.26.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:b96e7b9c624ef3ae2ae0e04fa9b460f6b9f17ad8b4bec6d7756510f1f6c0c841"},
427 | {file = "numpy-1.26.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:aa18428111fb9a591d7a9cc1b48150097ba6a7e8299fb56bdf574df650e7d1f1"},
428 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:06fa1ed84aa60ea6ef9f91ba57b5ed963c3729534e6e54055fc151fad0423f0a"},
429 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:96ca5482c3dbdd051bcd1fce8034603d6ebfc125a7bd59f55b40d8f5d246832b"},
430 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:854ab91a2906ef29dc3925a064fcd365c7b4da743f84b123002f6139bcb3f8a7"},
431 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:f43740ab089277d403aa07567be138fc2a89d4d9892d113b76153e0e412409f8"},
432 | {file = "numpy-1.26.2-cp311-cp311-win32.whl", hash = "sha256:a2bbc29fcb1771cd7b7425f98b05307776a6baf43035d3b80c4b0f29e9545186"},
433 | {file = "numpy-1.26.2-cp311-cp311-win_amd64.whl", hash = "sha256:2b3fca8a5b00184828d12b073af4d0fc5fdd94b1632c2477526f6bd7842d700d"},
434 | {file = "numpy-1.26.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:a4cd6ed4a339c21f1d1b0fdf13426cb3b284555c27ac2f156dfdaaa7e16bfab0"},
435 | {file = "numpy-1.26.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5d5244aabd6ed7f312268b9247be47343a654ebea52a60f002dc70c769048e75"},
436 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a3cdb4d9c70e6b8c0814239ead47da00934666f668426fc6e94cce869e13fd7"},
437 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:aa317b2325f7aa0a9471663e6093c210cb2ae9c0ad824732b307d2c51983d5b6"},
438 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:174a8880739c16c925799c018f3f55b8130c1f7c8e75ab0a6fa9d41cab092fd6"},
439 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f79b231bf5c16b1f39c7f4875e1ded36abee1591e98742b05d8a0fb55d8a3eec"},
440 | {file = "numpy-1.26.2-cp312-cp312-win32.whl", hash = "sha256:4a06263321dfd3598cacb252f51e521a8cb4b6df471bb12a7ee5cbab20ea9167"},
441 | {file = "numpy-1.26.2-cp312-cp312-win_amd64.whl", hash = "sha256:b04f5dc6b3efdaab541f7857351aac359e6ae3c126e2edb376929bd3b7f92d7e"},
442 | {file = "numpy-1.26.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:4eb8df4bf8d3d90d091e0146f6c28492b0be84da3e409ebef54349f71ed271ef"},
443 | {file = "numpy-1.26.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:1a13860fdcd95de7cf58bd6f8bc5a5ef81c0b0625eb2c9a783948847abbef2c2"},
444 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:64308ebc366a8ed63fd0bf426b6a9468060962f1a4339ab1074c228fa6ade8e3"},
445 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:baf8aab04a2c0e859da118f0b38617e5ee65d75b83795055fb66c0d5e9e9b818"},
446 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d73a3abcac238250091b11caef9ad12413dab01669511779bc9b29261dd50210"},
447 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:b361d369fc7e5e1714cf827b731ca32bff8d411212fccd29ad98ad622449cc36"},
448 | {file = "numpy-1.26.2-cp39-cp39-win32.whl", hash = "sha256:bd3f0091e845164a20bd5a326860c840fe2af79fa12e0469a12768a3ec578d80"},
449 | {file = "numpy-1.26.2-cp39-cp39-win_amd64.whl", hash = "sha256:2beef57fb031dcc0dc8fa4fe297a742027b954949cabb52a2a376c144e5e6060"},
450 | {file = "numpy-1.26.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:1cc3d5029a30fb5f06704ad6b23b35e11309491c999838c31f124fee32107c79"},
451 | {file = "numpy-1.26.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:94cc3c222bb9fb5a12e334d0479b97bb2df446fbe622b470928f5284ffca3f8d"},
452 | {file = "numpy-1.26.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:fe6b44fb8fcdf7eda4ef4461b97b3f63c466b27ab151bec2366db8b197387841"},
453 | {file = "numpy-1.26.2.tar.gz", hash = "sha256:f65738447676ab5777f11e6bbbdb8ce11b785e105f690bc45966574816b6d3ea"},
454 | ]
455 |
456 | [[package]]
457 | name = "packaging"
458 | version = "23.2"
459 | description = "Core utilities for Python packages"
460 | optional = false
461 | python-versions = ">=3.7"
462 | files = [
463 | {file = "packaging-23.2-py3-none-any.whl", hash = "sha256:8c491190033a9af7e1d931d0b5dacc2ef47509b34dd0de67ed209b5203fc88c7"},
464 | {file = "packaging-23.2.tar.gz", hash = "sha256:048fb0e9405036518eaaf48a55953c750c11e1a1b68e0dd1a9d62ed0c092cfc5"},
465 | ]
466 |
467 | [[package]]
468 | name = "pillow"
469 | version = "10.1.0"
470 | description = "Python Imaging Library (Fork)"
471 | optional = false
472 | python-versions = ">=3.8"
473 | files = [
474 | {file = "Pillow-10.1.0-cp310-cp310-macosx_10_10_x86_64.whl", hash = "sha256:1ab05f3db77e98f93964697c8efc49c7954b08dd61cff526b7f2531a22410106"},
475 | {file = "Pillow-10.1.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6932a7652464746fcb484f7fc3618e6503d2066d853f68a4bd97193a3996e273"},
476 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a5f63b5a68daedc54c7c3464508d8c12075e56dcfbd42f8c1bf40169061ae666"},
477 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c0949b55eb607898e28eaccb525ab104b2d86542a85c74baf3a6dc24002edec2"},
478 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:ae88931f93214777c7a3aa0a8f92a683f83ecde27f65a45f95f22d289a69e593"},
479 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:b0eb01ca85b2361b09480784a7931fc648ed8b7836f01fb9241141b968feb1db"},
480 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:d27b5997bdd2eb9fb199982bb7eb6164db0426904020dc38c10203187ae2ff2f"},
481 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:7df5608bc38bd37ef585ae9c38c9cd46d7c81498f086915b0f97255ea60c2818"},
482 | {file = "Pillow-10.1.0-cp310-cp310-win_amd64.whl", hash = "sha256:41f67248d92a5e0a2076d3517d8d4b1e41a97e2df10eb8f93106c89107f38b57"},
483 | {file = "Pillow-10.1.0-cp311-cp311-macosx_10_10_x86_64.whl", hash = "sha256:1fb29c07478e6c06a46b867e43b0bcdb241b44cc52be9bc25ce5944eed4648e7"},
484 | {file = "Pillow-10.1.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:2cdc65a46e74514ce742c2013cd4a2d12e8553e3a2563c64879f7c7e4d28bce7"},
485 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50d08cd0a2ecd2a8657bd3d82c71efd5a58edb04d9308185d66c3a5a5bed9610"},
486 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:062a1610e3bc258bff2328ec43f34244fcec972ee0717200cb1425214fe5b839"},
487 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:61f1a9d247317fa08a308daaa8ee7b3f760ab1809ca2da14ecc88ae4257d6172"},
488 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:a646e48de237d860c36e0db37ecaecaa3619e6f3e9d5319e527ccbc8151df061"},
489 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:47e5bf85b80abc03be7455c95b6d6e4896a62f6541c1f2ce77a7d2bb832af262"},
490 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:a92386125e9ee90381c3369f57a2a50fa9e6aa8b1cf1d9c4b200d41a7dd8e992"},
491 | {file = "Pillow-10.1.0-cp311-cp311-win_amd64.whl", hash = "sha256:0f7c276c05a9767e877a0b4c5050c8bee6a6d960d7f0c11ebda6b99746068c2a"},
492 | {file = "Pillow-10.1.0-cp312-cp312-macosx_10_10_x86_64.whl", hash = "sha256:a89b8312d51715b510a4fe9fc13686283f376cfd5abca8cd1c65e4c76e21081b"},
493 | {file = "Pillow-10.1.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:00f438bb841382b15d7deb9a05cc946ee0f2c352653c7aa659e75e592f6fa17d"},
494 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3d929a19f5469b3f4df33a3df2983db070ebb2088a1e145e18facbc28cae5b27"},
495 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9a92109192b360634a4489c0c756364c0c3a2992906752165ecb50544c251312"},
496 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:0248f86b3ea061e67817c47ecbe82c23f9dd5d5226200eb9090b3873d3ca32de"},
497 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:9882a7451c680c12f232a422730f986a1fcd808da0fd428f08b671237237d651"},
498 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:1c3ac5423c8c1da5928aa12c6e258921956757d976405e9467c5f39d1d577a4b"},
499 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:806abdd8249ba3953c33742506fe414880bad78ac25cc9a9b1c6ae97bedd573f"},
500 | {file = "Pillow-10.1.0-cp312-cp312-win_amd64.whl", hash = "sha256:eaed6977fa73408b7b8a24e8b14e59e1668cfc0f4c40193ea7ced8e210adf996"},
501 | {file = "Pillow-10.1.0-cp38-cp38-macosx_10_10_x86_64.whl", hash = "sha256:fe1e26e1ffc38be097f0ba1d0d07fcade2bcfd1d023cda5b29935ae8052bd793"},
502 | {file = "Pillow-10.1.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:7a7e3daa202beb61821c06d2517428e8e7c1aab08943e92ec9e5755c2fc9ba5e"},
503 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:24fadc71218ad2b8ffe437b54876c9382b4a29e030a05a9879f615091f42ffc2"},
504 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fa1d323703cfdac2036af05191b969b910d8f115cf53093125e4058f62012c9a"},
505 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:912e3812a1dbbc834da2b32299b124b5ddcb664ed354916fd1ed6f193f0e2d01"},
506 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:7dbaa3c7de82ef37e7708521be41db5565004258ca76945ad74a8e998c30af8d"},
507 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:9d7bc666bd8c5a4225e7ac71f2f9d12466ec555e89092728ea0f5c0c2422ea80"},
508 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:baada14941c83079bf84c037e2d8b7506ce201e92e3d2fa0d1303507a8538212"},
509 | {file = "Pillow-10.1.0-cp38-cp38-win_amd64.whl", hash = "sha256:2ef6721c97894a7aa77723740a09547197533146fba8355e86d6d9a4a1056b14"},
510 | {file = "Pillow-10.1.0-cp39-cp39-macosx_10_10_x86_64.whl", hash = "sha256:0a026c188be3b443916179f5d04548092e253beb0c3e2ee0a4e2cdad72f66099"},
511 | {file = "Pillow-10.1.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:04f6f6149f266a100374ca3cc368b67fb27c4af9f1cc8cb6306d849dcdf12616"},
512 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bb40c011447712d2e19cc261c82655f75f32cb724788df315ed992a4d65696bb"},
513 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1a8413794b4ad9719346cd9306118450b7b00d9a15846451549314a58ac42219"},
514 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:c9aeea7b63edb7884b031a35305629a7593272b54f429a9869a4f63a1bf04c34"},
515 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:b4005fee46ed9be0b8fb42be0c20e79411533d1fd58edabebc0dd24626882cfd"},
516 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0152565c6aa6ebbfb1e5d8624140a440f2b99bf7afaafbdbf6430426497f28"},
517 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:d921bc90b1defa55c9917ca6b6b71430e4286fc9e44c55ead78ca1a9f9eba5f2"},
518 | {file = "Pillow-10.1.0-cp39-cp39-win_amd64.whl", hash = "sha256:cfe96560c6ce2f4c07d6647af2d0f3c54cc33289894ebd88cfbb3bcd5391e256"},
519 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-macosx_10_10_x86_64.whl", hash = "sha256:937bdc5a7f5343d1c97dc98149a0be7eb9704e937fe3dc7140e229ae4fc572a7"},
520 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b1c25762197144e211efb5f4e8ad656f36c8d214d390585d1d21281f46d556ba"},
521 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:afc8eef765d948543a4775f00b7b8c079b3321d6b675dde0d02afa2ee23000b4"},
522 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:883f216eac8712b83a63f41b76ddfb7b2afab1b74abbb413c5df6680f071a6b9"},
523 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-macosx_10_10_x86_64.whl", hash = "sha256:b920e4d028f6442bea9a75b7491c063f0b9a3972520731ed26c83e254302eb1e"},
524 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1c41d960babf951e01a49c9746f92c5a7e0d939d1652d7ba30f6b3090f27e412"},
525 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:1fafabe50a6977ac70dfe829b2d5735fd54e190ab55259ec8aea4aaea412fa0b"},
526 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:3b834f4b16173e5b92ab6566f0473bfb09f939ba14b23b8da1f54fa63e4b623f"},
527 | {file = "Pillow-10.1.0.tar.gz", hash = "sha256:e6bf8de6c36ed96c86ea3b6e1d5273c53f46ef518a062464cd7ef5dd2cf92e38"},
528 | ]
529 |
530 | [package.extras]
531 | docs = ["furo", "olefile", "sphinx (>=2.4)", "sphinx-copybutton", "sphinx-inline-tabs", "sphinx-removed-in", "sphinxext-opengraph"]
532 | tests = ["check-manifest", "coverage", "defusedxml", "markdown2", "olefile", "packaging", "pyroma", "pytest", "pytest-cov", "pytest-timeout"]
533 |
534 | [[package]]
535 | name = "pluggy"
536 | version = "1.3.0"
537 | description = "plugin and hook calling mechanisms for python"
538 | optional = false
539 | python-versions = ">=3.8"
540 | files = [
541 | {file = "pluggy-1.3.0-py3-none-any.whl", hash = "sha256:d89c696a773f8bd377d18e5ecda92b7a3793cbe66c87060a6fb58c7b6e1061f7"},
542 | {file = "pluggy-1.3.0.tar.gz", hash = "sha256:cf61ae8f126ac6f7c451172cf30e3e43d3ca77615509771b3a984a0730651e12"},
543 | ]
544 |
545 | [package.extras]
546 | dev = ["pre-commit", "tox"]
547 | testing = ["pytest", "pytest-benchmark"]
548 |
549 | [[package]]
550 | name = "py"
551 | version = "1.11.0"
552 | description = "library with cross-python path, ini-parsing, io, code, log facilities"
553 | optional = false
554 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*"
555 | files = [
556 | {file = "py-1.11.0-py2.py3-none-any.whl", hash = "sha256:607c53218732647dff4acdfcd50cb62615cedf612e72d1724fb1a0cc6405b378"},
557 | {file = "py-1.11.0.tar.gz", hash = "sha256:51c75c4126074b472f746a24399ad32f6053d1b34b68d2fa41e558e6f4a98719"},
558 | ]
559 |
560 | [[package]]
561 | name = "pyparsing"
562 | version = "3.1.1"
563 | description = "pyparsing module - Classes and methods to define and execute parsing grammars"
564 | optional = false
565 | python-versions = ">=3.6.8"
566 | files = [
567 | {file = "pyparsing-3.1.1-py3-none-any.whl", hash = "sha256:32c7c0b711493c72ff18a981d24f28aaf9c1fb7ed5e9667c9e84e3db623bdbfb"},
568 | {file = "pyparsing-3.1.1.tar.gz", hash = "sha256:ede28a1a32462f5a9705e07aea48001a08f7cf81a021585011deba701581a0db"},
569 | ]
570 |
571 | [package.extras]
572 | diagrams = ["jinja2", "railroad-diagrams"]
573 |
574 | [[package]]
575 | name = "pytest"
576 | version = "6.2.5"
577 | description = "pytest: simple powerful testing with Python"
578 | optional = false
579 | python-versions = ">=3.6"
580 | files = [
581 | {file = "pytest-6.2.5-py3-none-any.whl", hash = "sha256:7310f8d27bc79ced999e760ca304d69f6ba6c6649c0b60fb0e04a4a77cacc134"},
582 | {file = "pytest-6.2.5.tar.gz", hash = "sha256:131b36680866a76e6781d13f101efb86cf674ebb9762eb70d3082b6f29889e89"},
583 | ]
584 |
585 | [package.dependencies]
586 | atomicwrites = {version = ">=1.0", markers = "sys_platform == \"win32\""}
587 | attrs = ">=19.2.0"
588 | colorama = {version = "*", markers = "sys_platform == \"win32\""}
589 | iniconfig = "*"
590 | packaging = "*"
591 | pluggy = ">=0.12,<2.0"
592 | py = ">=1.8.2"
593 | toml = "*"
594 |
595 | [package.extras]
596 | testing = ["argcomplete", "hypothesis (>=3.56)", "mock", "nose", "requests", "xmlschema"]
597 |
598 | [[package]]
599 | name = "python-dateutil"
600 | version = "2.8.2"
601 | description = "Extensions to the standard Python datetime module"
602 | optional = false
603 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,>=2.7"
604 | files = [
605 | {file = "python-dateutil-2.8.2.tar.gz", hash = "sha256:0123cacc1627ae19ddf3c27a5de5bd67ee4586fbdd6440d9748f8abb483d3e86"},
606 | {file = "python_dateutil-2.8.2-py2.py3-none-any.whl", hash = "sha256:961d03dc3453ebbc59dbdea9e4e11c5651520a876d0f4db161e8674aae935da9"},
607 | ]
608 |
609 | [package.dependencies]
610 | six = ">=1.5"
611 |
612 | [[package]]
613 | name = "six"
614 | version = "1.16.0"
615 | description = "Python 2 and 3 compatibility utilities"
616 | optional = false
617 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*"
618 | files = [
619 | {file = "six-1.16.0-py2.py3-none-any.whl", hash = "sha256:8abb2f1d86890a2dfb989f9a77cfcfd3e47c2a354b01111771326f8aa26e0254"},
620 | {file = "six-1.16.0.tar.gz", hash = "sha256:1e61c37477a1626458e36f7b1d82aa5c9b094fa4802892072e49de9c60c4c926"},
621 | ]
622 |
623 | [[package]]
624 | name = "toml"
625 | version = "0.10.2"
626 | description = "Python Library for Tom's Obvious, Minimal Language"
627 | optional = false
628 | python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
629 | files = [
630 | {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
631 | {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
632 | ]
633 |
634 | [[package]]
635 | name = "tqdm"
636 | version = "4.66.1"
637 | description = "Fast, Extensible Progress Meter"
638 | optional = false
639 | python-versions = ">=3.7"
640 | files = [
641 | {file = "tqdm-4.66.1-py3-none-any.whl", hash = "sha256:d302b3c5b53d47bce91fea46679d9c3c6508cf6332229aa1e7d8653723793386"},
642 | {file = "tqdm-4.66.1.tar.gz", hash = "sha256:d88e651f9db8d8551a62556d3cff9e3034274ca5d66e93197cf2490e2dcb69c7"},
643 | ]
644 |
645 | [package.dependencies]
646 | colorama = {version = "*", markers = "platform_system == \"Windows\""}
647 |
648 | [package.extras]
649 | dev = ["pytest (>=6)", "pytest-cov", "pytest-timeout", "pytest-xdist"]
650 | notebook = ["ipywidgets (>=6)"]
651 | slack = ["slack-sdk"]
652 | telegram = ["requests"]
653 |
654 | [[package]]
655 | name = "zipp"
656 | version = "3.17.0"
657 | description = "Backport of pathlib-compatible object wrapper for zip files"
658 | optional = false
659 | python-versions = ">=3.8"
660 | files = [
661 | {file = "zipp-3.17.0-py3-none-any.whl", hash = "sha256:0e923e726174922dce09c53c59ad483ff7bbb8e572e00c7f7c46b88556409f31"},
662 | {file = "zipp-3.17.0.tar.gz", hash = "sha256:84e64a1c28cf7e91ed2078bb8cc8c259cb19b76942096c8d7b84947690cabaf0"},
663 | ]
664 |
665 | [package.extras]
666 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
667 | testing = ["big-O", "jaraco.functools", "jaraco.itertools", "more-itertools", "pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-ignore-flaky", "pytest-mypy (>=0.9.1)", "pytest-ruff"]
668 |
669 | [metadata]
670 | lock-version = "2.0"
671 | python-versions = "^3.9"
672 | content-hash = "5e3a8b71c1d10601d5e0c2e8e72d1ea2cd79f51cbe48d1f7512dd3fc591030c2"
673 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.poetry]
2 | name = "complexcgr"
3 | version = "0.8.0"
4 | description = "complex Chaos Game Representation for DNA"
5 | authors = ["Jorge Avila "]
6 | maintainers = ["Jorge Avila "]
7 | repository = "https://github.com/AlgoLab/complexCGR"
8 | license = "MIT"
9 |
10 | [tool.poetry.dependencies]
11 | python = "^3.9"
12 | tqdm = "^4.61.2"
13 | matplotlib = "^3.4.2"
14 | Pillow = "^10.0"
15 | numpy = "^1.22.3"
16 | biopython = "^1.79"
17 |
18 | [tool.poetry.dev-dependencies]
19 | pytest = "^6.2.4"
20 |
21 | [build-system]
22 | requires = ["poetry-core>=1.0.0"]
23 | build-backend = "poetry.core.masonry.api"
24 |
--------------------------------------------------------------------------------
/tests/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/tests/__init__.py
--------------------------------------------------------------------------------
/tests/test_cgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import CGR
2 |
3 | def test_cgr():
4 | "Encoding for each nucleotide"
5 | cgr = CGR()
6 |
7 | cgr.encode("A")
8 | assert cgr.cgr_coords.N == 1
9 | assert cgr.cgr_coords.x == 0.5
10 | assert cgr.cgr_coords.y == 0.5
11 |
12 | cgr.encode("C")
13 | assert cgr.cgr_coords.N == 1
14 | assert cgr.cgr_coords.x == -0.5
15 | assert cgr.cgr_coords.y == 0.5
16 |
17 | cgr.encode("G")
18 | assert cgr.cgr_coords.N == 1
19 | assert cgr.cgr_coords.x == -0.5
20 | assert cgr.cgr_coords.y == -0.5
21 |
22 | cgr.encode("T")
23 | assert cgr.cgr_coords.N == 1
24 | assert cgr.cgr_coords.x == 0.5
25 | assert cgr.cgr_coords.y == -0.5
26 |
27 | def test_cgr_decode():
28 | "Decoding each nucleotide"
29 | cgr = CGR()
30 |
31 | assert cgr.decode(N=1,x=0.5,y=0.5) == "A"
32 | assert cgr.decode(N=1,x=-0.5,y=0.5) == "C"
33 | assert cgr.decode(N=1,x=-0.5,y=-0.5) == "G"
34 | assert cgr.decode(N=1,x=0.5,y=-0.5) == "T"
--------------------------------------------------------------------------------
/tests/test_complexcgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import (
2 | __version__,
3 | ComplexCGR,
4 | )
5 |
6 | def test_version():
7 | assert __version__ == '0.8.0'
8 |
9 | def test_complexCGR():
10 | ccgr = ComplexCGR()
11 |
12 | # nucleotide A
13 | encode = ccgr.encode("A")
14 | assert encode.k == 0
15 | assert encode.N == 1
16 |
17 |
18 | # nucleotide C
19 | encode = ccgr.encode("C")
20 | assert encode.k == 1
21 | assert encode.N == 1
22 |
23 |
24 | # nucleotide G
25 | encode = ccgr.encode("G")
26 | assert encode.k == 2
27 | assert encode.N == 1
28 |
29 |
30 | # nucleotide T
31 | encode = ccgr.encode("T")
32 | assert encode.k == 3
33 | assert encode.N == 1
34 |
35 | def test_complexCGR_decode():
36 | ccgr = ComplexCGR()
37 |
38 | # nucleotide A
39 | decode = ccgr.decode(k=0,N=1)
40 | assert decode == "A"
41 |
42 |
43 | # nucleotide C
44 | decode = ccgr.decode(k=1,N=1)
45 | assert decode == "C"
46 |
47 |
48 | # nucleotide G
49 | decode = ccgr.decode(k=2,N=1)
50 | assert decode == "G"
51 |
52 |
53 | # nucleotide T
54 | decode = ccgr.decode(k=3,N=1)
55 | assert decode == "T"
56 |
57 | def test_encode_decode_compatibility():
58 | ccgr = ComplexCGR()
59 |
60 | seq = "ACGT"
61 | encode = ccgr.encode(seq)
62 | decode = ccgr.decode(encode.k, encode.N)
63 | assert seq == decode
--------------------------------------------------------------------------------
/tests/test_complexfcgr.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/tests/test_complexfcgr.py
--------------------------------------------------------------------------------
/tests/test_complexfcgr_savefig.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | import random
3 | from complexcgr import ComplexFCGR
4 |
5 | BASEPATH = Path().parent.resolve()
6 | def test_savefig():
7 | fcgr = ComplexFCGR(k=8)
8 | seq = "".join(random.choice("ACG") for _ in range(300_000))
9 | fig = fcgr(seq)
10 | fcgr.save(fig, path=BASEPATH.joinpath("img/ACG-complexCGR.png"))
11 |
--------------------------------------------------------------------------------
/tests/test_fcgr.py:
--------------------------------------------------------------------------------
1 | import random
2 | from complexcgr import FCGR
3 |
4 | def test_fcgr():
5 | "frecuency matrix CGR"
6 | fcgr = FCGR(k=1)
7 | fcgr.count_kmers("ACGT")
8 |
9 | assert fcgr.freq_kmer.get("A") == 1
10 | assert fcgr.freq_kmer.get("C") == 1
11 | assert fcgr.freq_kmer.get("G") == 1
12 | assert fcgr.freq_kmer.get("T") == 1
13 |
14 | def test_savefig():
15 | fcgr = FCGR(k=8)
16 | # Generate a random sequence without T's
17 | seq = "".join(random.choice("ACG") for _ in range(30_000))
18 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
19 | fcgr.save_img(chaos, path="img/ACG.jpg")
20 |
21 | def test_savefig_16bits():
22 | fcgr = FCGR(k=8, bits=16)
23 | # Generate a random sequence without T's
24 | seq = "".join(random.choice("ACG") for _ in range(30_000))
25 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
26 | fcgr.save_img(chaos, path="img/ACG_16bits.jpg")
27 |
28 |
29 |
30 |
31 |
--------------------------------------------------------------------------------
/tests/test_icgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import iCGR
2 |
3 | def test_cgr():
4 | "Encoding for each nucleotide"
5 | cgr = iCGR()
6 |
7 | cgr.encode("A")
8 | assert cgr.cgr_coords.N == 1
9 | assert cgr.cgr_coords.x == 1
10 | assert cgr.cgr_coords.y == 1
11 |
12 | cgr.encode("C")
13 | assert cgr.cgr_coords.N == 1
14 | assert cgr.cgr_coords.x == -1
15 | assert cgr.cgr_coords.y == 1
16 |
17 | cgr.encode("G")
18 | assert cgr.cgr_coords.N == 1
19 | assert cgr.cgr_coords.x == -1
20 | assert cgr.cgr_coords.y == -1
21 |
22 | cgr.encode("T")
23 | assert cgr.cgr_coords.N == 1
24 | assert cgr.cgr_coords.x == 1
25 | assert cgr.cgr_coords.y == -1
26 |
27 |
28 | def test_cgr_decode():
29 | "Decoding each nucleotide"
30 | cgr = iCGR()
31 |
32 | assert cgr.decode(N=1,x=1,y=1) == "A"
33 | assert cgr.decode(N=1,x=-1,y=1) == "C"
34 | assert cgr.decode(N=1,x=-1,y=-1) == "G"
35 | assert cgr.decode(N=1,x=1,y=-1) == "T"
36 | assert cgr.decode(N=4,x=3,y=-9) == "ACGT"
--------------------------------------------------------------------------------
2 |
3 | # `complexcgr`
4 | This library have classes around the *Chaos Game Representation* for DNA sequence
5 |
6 | **The FCGR helps to visualize a k-mer distribution** The `FCGR` of a sequence is an image showing the distribution of the $k$-mers
7 | given a chosen $k$. The frequencies of all $k$-mers are distributed in the position of a matrix of $2^k \times 2^k$,
8 | which considers all the possible $k$-mers: $4^k$.
9 |
10 | The position that a $k$-mer uses in the matrix depends on the encoding given by the `CGR`.
11 |
12 | Some examples of [bacterial assemblies](https://zenodo.org/records/4602622) ([see reference](https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001421)) are shown below.
13 | The name of the species and the `sample_id` is in the title of each image ([see an example with the first image](https://www.ebi.ac.uk/ena/browser/view/SAMEA2658585)). These images were
14 | created using the 6-mers of each assembly and the class `FCGR` of this library.
15 |
16 | |  |
17 | |:--:|
18 | |10 different species of bacteria represented by their FCGR (6-mers)| -->
19 |
20 | ## Installation
21 | [pypi](https://pypi.org/project/complexcgr/)
22 | ___
23 | ```shell
24 | pip install complexcgr
25 | ```
26 |
27 | to update to the latest version
28 | ```shell
29 | pip install complexcgr --upgrade
30 | ```
31 |
32 | ## How to use
33 | ___
34 | ### 1. `CGR` Chaos Game Representation of DNA
35 | ```python
36 | from complexcgr import CGR
37 |
38 | # Instantiate class CGR
39 | cgr = CGR()
40 |
41 | # encode a sequence
42 | cgr.encode("ACGT")
43 | # > CGRCoords(N=4, x=0.1875, y=-0.5625)
44 |
45 | # recover a sequence from CGR coordinates
46 | cgr.decode(N=4,x=0.1875,y=-0.5625)
47 | # > "ACGT"
48 | ```
49 |
50 | ### 2. `FCGR` Frequency Matrix of Chaos Game Representation of DNA
51 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
52 | will not be considered for the calculation of the frequency matrix CGR
53 | ```python
54 | import random; random.seed(42)
55 | from complexcgr import FCGR
56 |
57 | # set the k-mer
58 | fcgr = FCGR(k=8) # (256x256) array
59 |
60 | # Generate a random sequence without T's
61 | seq = "".join(random.choice("ACG") for _ in range(300_000))
62 | chaos = fcgr(seq) # an array with the frequencies of each k-mer
63 | fcgr.plot(chaos)
64 | ```
65 | |  |
66 | |:--:|
67 | |FCGR representation for a sequence without T's|
68 |
69 |
70 | You can save the image with
71 | ```python
72 | fcgr.save_img(chaos, path="img/ACG.jpg")
73 | ```
74 | *Formats allowed are defined by PIL.*
75 |
76 | You can also generate the image in 16 (or more bits), to avoid losing information of k-mer frequencies
77 | ```python
78 | # Generate image in 16-bits (default is 8-bits)
79 | fcgr = FCGR(k=8, bits=16) # (256x256) array. When using plot() it will be rescaled to [0,65535] colors
80 | ```
81 |
82 |
83 | ```python
84 | # Generate a random sequence without T's and lots of N's
85 | seq = "".join(random.choice("ACGN") for _ in range(300_000))
86 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
87 | fcgr.plot(chaos)
88 | ```
89 |
90 |
91 | ||
92 | |:--:|
93 | |FCGR representation for a sequence without T's and lots of N's|
94 |
95 |
96 |
97 | ### 3. `iCGR` integer Chaos Game Representation of DNA
98 | ```python
99 | from complexcgr import iCGR
100 |
101 | # Instantiate class CGR
102 | icgr = iCGR()
103 |
104 | # encode a sequence
105 | icgr.encode("ACGT")
106 | # > CGRCoords(N=4, x=3, y=-9)
107 |
108 | # recover a sequence from CGR coordinates
109 | icgr.decode(N=4,x=3,y=-9)
110 | # > "ACGT"
111 | ```
112 |
113 | ### 4. `ComplexCGR` Complex Chaos Game Representation of DNA (ComplexCGR)
114 |
115 | ```python
116 | from complexcgr import ComplexCGR
117 |
118 | # Instantiate class CGR
119 | ccgr = ComplexCGR()
120 |
121 | # encode a sequence
122 | ccgr.encode("ACGT")
123 | # > CGRCoords(k=228,N=4)
124 |
125 | # recover a sequence from ComplexCGR coordinates
126 | ccgr.decode(k=228,N=4)
127 | # > "ACGT"
128 |
129 | ```
130 |
131 | ### 5. `ComplexFCGR` Frequency Matrix of Complex Chaos Game Representation of DNA
132 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
133 | will not be considered for the calculation of the frequency matrix CGR
134 | ```python
135 | import random; random.seed(42)
136 | from complexcgr import FCGR
137 |
138 | # set the k-mer desired
139 | cfcgr = ComplexFCGR(k=8) # 8-mers
140 |
141 | # Generate a random sequence without T's
142 | seq = "".join(random.choice("ACG") for _ in range(300_000))
143 | fig = cfcgr(seq)
144 |
145 | ```
146 | |  |
147 | |:--:|
148 | |ComplexFCGR representation for a sequence without T's|
149 |
150 |
151 | You can save the image with
152 | ```python
153 | cfcgr.save(fig, path="img/ACG-ComplexCGR.png")
154 | ```
155 | *Currently the plot must be saved as png*
156 |
157 | ___
158 | ## Advice for Real applications
159 |
160 | **Count k-mers** could be the bottleneck for large sequences (> 100000 bp).
161 | Note that the class `FCGR` (and `ComplexCGR`) has implemented a naive approach to count k-mers, this is intended since in practice state-of-the-art tools like KMC or Jellyfish are used to count k-mers very efficiently.
162 |
163 | We provide the class `FCGRKmc`, that receives as input the file generated by the following pipeline using [KMC3](https://github.com/refresh-bio/KMC)
164 |
165 | Make sure to have `kmc` installed. One recommended way is to create a conda environment and [install it there](https://anaconda.org/bioconda/kmc)
166 |
167 | ```bash
168 | kmer_size=6
169 | input="path/to/sequence.fa"
170 | output="path/to/count-kmers.txt"
171 |
172 | mkdir -p tmp-kmc
173 | kmc -v -k$kmer_size -m4 -sm -ci0 -cs100000 -b -t4 -fa $input $input "tmp-kmc"
174 | kmc_tools -t4 -v transform $input dump $output
175 | rm -r $input.kmc_pre $input.kmc_suf
176 | ```
177 | the output file `path/to/count-kmers.txt` can be used with `FCGRKmc`
178 |
179 | ```python
180 | from complexcgr import FCGRKmc
181 |
182 | kmer = 6
183 | fcgr = FCGRKmc(kmer)
184 |
185 | arr = fcgr("path/to/count-kmers.txt") # k-mer counts ordered in a matrix of 2^k x 2^k
186 |
187 |
188 | # to visualize the distribution of k-mers.
189 | # Frequencies are scaled between [min, max] values.
190 | # White color corresponds to the minimum value of frequency
191 | # Black color corresponds to the maximum value of frequency
192 | fcgr.plot(arr)
193 |
194 | # Save it with numpy
195 | import numpy as np
196 | np.save("path_save/fcgr.npy",arr)
197 | ```
198 |
199 | ___
200 | # Videos
201 |
202 | **CGR encoding**
203 |
204 | [](https://youtu.be/HU15ge0fkOY)
205 |
206 | **CGR encoding of all k-mers**
207 |
208 | [](https://youtu.be/oYLT11Q9n5M)
209 |
210 | **ComplexCGR encoding**
211 |
212 | [](https://youtu.be/xKyXplS5KFk)
213 |
214 | **ComplexCGR and Symmetry**
215 |
216 | [](https://www.youtube.com/YmcxVId4_4w)
217 |
218 |
219 | # Functionalities/TODO list
220 | ___
221 | > version 0.8.0:
222 | A list of available classes and functionalities are listed below:
223 |
224 |
225 | **Encoders**
226 | The encoders are functions that map a sequence $s \in \{A,C,G,T\}$ to a point in the plane.
227 | `CGR`, `iCGR`, and `ComplexCGR`.
228 |
229 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
230 | - [x] encode a sequence.
231 | - [x] recover a sequence from a CGR encoding.
232 |
233 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
234 |
235 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
236 | - [x] encode a sequence.
237 | - [x] recover a sequence from a CGR encoding.
238 |
239 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
240 | - [x] encode a sequence
241 | - [x] recover a sequence from an iCGR encoding
242 |
243 | `ComplexCGR`: encodes a DNA sequence in 2 integers $(k,N)$.
244 | - [x] encode a sequence
245 | - [x] recover a sequence from a ComplexCGR encoding
246 | - [x] plot sequence of ComplexCGR encodings
247 |
248 | **Image for distribution of k-mers**
249 |
250 | - [x] `FCGR` Frequency Matrix CGR: representation as an image for k-mer representativity, based on CGR.
251 | - [x] generates FCGR from an arbitrary n-long sequence.
252 | - [x] plot FCGR.
253 | - [x] save FCGR generated.
254 | - [x] save FCGR in different bits.
255 | - [x] `FCGRKmc` Same as `FCGR` but receives as input the file with k-mer counts generated with [KMC](https://github.com/refresh-bio/KMC)
256 | - [x] `ComplexFCGR`: Frequency ComplexCGR: representation as an image (circle) for k-mer representativity, based on ComplexCGR.
257 | - [x] generates ComplexFCGR from an arbitrary n-long sequence.
258 | - [x] plot ComplexFCGR.
259 | - [x] save ComplexFCGR generated.
260 |
261 | - [ ] `PercentileFCGR`:
262 | - [A universal DNA signature for the Tree of Life. EcoEvoRxiv](https://doi.org/10.32942/X24891)
263 | - [ ] `SpacedFCGR`: Create FCGR from spaced-mers
264 | - [Spaced seeds improve k-mer-based metagenomic classification, Bioninformatics](https://academic.oup.com/bioinformatics/article/31/22/3584/240663)
265 | - [Effects of spaced k-mers on alignment-free genotyping](https://academic.oup.com/bioinformatics/article/39/Supplement_1/i213/7210439)
266 |
267 | # Author
268 |
269 | `complexcgr` is developed by [Jorge Avila Cartes](https://github.com/jorgeavilacartes/)
270 |
271 |
272 | # Related publications
273 |
274 | - [Accurate and fast clade assignment via deep learning and frequency chaos game representation](https://doi.org/10.1093/gigascience/giac119)
275 | - [PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases](https://doi.org/10.1101/2025.03.19.644115)
--------------------------------------------------------------------------------
/README.rst:
--------------------------------------------------------------------------------
1 | # complexCGR
2 |
3 | A complex representation for DNA, that preserves some properties of the CGR from the 'Chaos Game Representation for gene structure' by Jeffrey in 1990.
4 |
5 | Each nucleotide will be assigned to one quadrant (I,II,III and IV). This distribution will be the same for the CGR and complexCGR implementations (I:A,II:C,III:G,IV:T)
--------------------------------------------------------------------------------
/complexCGR.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/complexCGR.pdf
--------------------------------------------------------------------------------
/complexcgr/__init__.py:
--------------------------------------------------------------------------------
1 | __version__ ="0.8.0"
2 |
3 | # encodings
4 | from .cgr import CGR
5 | from .icgr import iCGR
6 | from .complexcgr import ComplexCGR
7 |
8 | # from k-mers
9 | from .fcgr import FCGR
10 | from .fcgr_kmc import FCGRKmc
11 | from .complexfcgr import ComplexFCGR
--------------------------------------------------------------------------------
/complexcgr/cgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from typing import Dict, Optional
3 | from collections import namedtuple
4 |
5 | # coordinates for x+iy
6 | Coord = namedtuple("Coord", ["x","y"])
7 |
8 | # coordinates for a CGR encoding
9 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
10 |
11 | # coordinates for each nucleotide in the 2d-plane
12 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
13 |
14 | class CGR:
15 | "Chaos Game Representation for DNA"
16 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
17 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
18 | self.cgr_coords = CGRCoords(0,0,0)
19 |
20 | def nucleotide_by_coords(self,x,y):
21 | "Get nucleotide by coordinates (x,y)"
22 | # filter nucleotide by coordinates
23 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
24 |
25 | return list(filtered.keys())[0]
26 |
27 | def forward(self, nucleotide: str):
28 | "Compute next CGR coordinates"
29 | x = (self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x)/2
30 | y = (self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y)/2
31 |
32 | # update cgr_coords
33 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y)
34 |
35 | def backward(self,):
36 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
37 | # get current nucleotide based on coordinates
38 | n_x,n_y = self.coords_current_nucleotide()
39 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
40 |
41 | # update coordinates to the previous one
42 | x = 2*self.cgr_coords.x - n_x
43 | y = 2*self.cgr_coords.y - n_y
44 |
45 | # update cgr_coords
46 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
47 |
48 | return nucleotide
49 |
50 | def coords_current_nucleotide(self,):
51 | x = 1 if self.cgr_coords.x>0 else -1
52 | y = 1 if self.cgr_coords.y>0 else -1
53 | return x,y
54 |
55 | def encode(self, sequence: str):
56 | "From DNA sequence to CGR"
57 | # reset starting position to (0,0,0)
58 | self.reset_coords()
59 | for nucleotide in sequence:
60 | self.forward(nucleotide)
61 | return self.cgr_coords
62 |
63 | def reset_coords(self,):
64 | self.cgr_coords = CGRCoords(0,0,0)
65 |
66 | def decode(self, N:int, x:int, y:int)->str:
67 | "From CGR to DNA sequence"
68 | self.cgr_coords = CGRCoords(N,x,y)
69 |
70 | # decoded sequence
71 | sequence = []
72 |
73 | # Recover the entire genome
74 | while self.cgr_coords.N>0:
75 | nucleotide = self.backward()
76 | sequence.append(nucleotide)
77 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/complexcgr/complexcgr.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List
2 | from collections import namedtuple
3 |
4 | # coordinates for x+iy
5 | Coord = namedtuple("Coord", ["x","y"])
6 |
7 | # coordinates for a CGR encoding
8 | CGRCoords = namedtuple("CGRCoord", ["k","N"])
9 |
10 | class ComplexCGR:
11 | "Complex Chaos Game Representation of DNA"
12 | def __init__(self, nucleotide_order: List[str] = ["A","C","G","T"]):
13 | self.nucleotide_order = nucleotide_order
14 | self.cgr_coords = CGRCoords(0,0) # complexCGR coordinates
15 |
16 | def id(self, nucleotide):
17 | return self.nucleotide_order.index(nucleotide)
18 |
19 | def forward(self, nucleotide: str):
20 | "compute next complexCGR coordinates"
21 | k = self.id(nucleotide)*4**(self.cgr_coords.N) + self.cgr_coords.k
22 |
23 | # update cgr_coords
24 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N+1)
25 |
26 | def backward(self,):
27 | "compute last complexCGR coordinates"
28 | nucleotide = self.current_nucleotide()
29 |
30 | # compute previous k
31 | k = self.cgr_coords.k - self.id(nucleotide)*4**(self.cgr_coords.N-1)
32 |
33 | # update cgr_coords
34 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N-1)
35 |
36 | return nucleotide
37 |
38 | def encode(self, sequence: str):
39 | "From DNA to complexCGR"
40 | self.reset_coords()
41 | for nucleotide in sequence:
42 | self.forward(nucleotide)
43 | return self.cgr_coords
44 |
45 | def decode(self, k: int, N: int):
46 | "From complexCGR to DNA"
47 | self.cgr_coords = CGRCoords(k,N)
48 |
49 | # decoded sequence
50 | sequence = []
51 |
52 | # Recover the entire genome
53 | while self.cgr_coords.N>0:
54 | nucleotide = self.backward()
55 | sequence.append(nucleotide)
56 | return "".join(sequence[::-1])
57 |
58 | def current_nucleotide(self,):
59 | "Get current nucleotide based on k and N"
60 | k,N = self.cgr_coords.k, self.cgr_coords.N
61 | alpha = k/4**N
62 | if alpha <0.25:
63 | return self.nucleotide_order[0]
64 | elif alpha <0.5:
65 | return self.nucleotide_order[1]
66 | elif alpha < 0.75:
67 | return self.nucleotide_order[2]
68 | else:
69 | return self.nucleotide_order[3]
70 |
71 | def reset_coords(self,):
72 | self.cgr_coords = CGRCoords(0,0)
73 |
--------------------------------------------------------------------------------
/complexcgr/complexfcgr.py:
--------------------------------------------------------------------------------
1 | from . import ComplexCGR
2 | import matplotlib.pyplot as plt
3 | from itertools import product
4 | from collections import defaultdict
5 | from tqdm import tqdm
6 | from PIL import Image
7 | import numpy as np
8 |
9 |
10 | class ComplexFCGR(ComplexCGR):
11 | """Circular density plot based on CGR"""
12 |
13 | def __init__(self, k: int):
14 | super().__init__()
15 | self.k = k # k-mer representation
16 | self.kmers = product("ACGT", repeat=self.k) # all kmers of length k
17 | self.freq_kmer = None # dict to save representativity of each kmer
18 | self.probabilities = None # dict to save probabilities/density for each kmer
19 | self.fig = None # to save matlotlib figure and then save as image
20 |
21 | def __call__(self, sequence: str, w=1):
22 | self.count_kmers(sequence)
23 | self.kmer_probabilities(sequence)
24 | self.plot(w)
25 |
26 | def count_kmers(self, sequence: str):
27 | self.freq_kmer = defaultdict(int)
28 | # representativity of kmers
29 | len_seq = len(sequence)
30 | for j,_ in enumerate(sequence):
31 | if j+self.k <= len_seq:
32 | subseq = sequence[j:j+self.k]
33 | if "N" not in subseq:
34 | self.freq_kmer[subseq] +=1
35 |
36 | def kmer_probabilities(self, sequence: str):
37 | self.probabilities = defaultdict(float)
38 | N=len(sequence)
39 | for key, value in self.freq_kmer.items():
40 | self.probabilities[key] = float(value) / (N - self.k + 1)
41 |
42 | def plot(self, w: int = 1):
43 | "Given a FCGR, plot it in grayscale"
44 | ax = plt.subplot(111, polar=True)
45 | center, bottom, width, height = self.compute_input_plot()
46 |
47 | # scale width by 'w'
48 | width = [w*_ for _ in width]
49 | print("generating plot")
50 | ax.bar(x=center, # center of the angle
51 | width=width, # width of the angle
52 | bottom=bottom, # lowest value
53 | height=height, # highest value
54 | )
55 |
56 | ax.axes.get_xaxis().set_visible(False)
57 | ax.axes.get_yaxis().set_visible(False)
58 |
59 | return ax.figure
60 |
61 | def save(self, ccgr, path: str):
62 | "save complexFCGR as image"
63 | # get figure
64 | fig = self.plot()
65 |
66 | # transform figure to Image
67 | img = self.fig2img(fig)
68 |
69 | # save image
70 | img.save(path)
71 | plt.close(fig)
72 |
73 | @staticmethod
74 | def fig2img(fig):
75 | "Convert a Matplotlib figure to a PIL Image and return it"
76 | #https://stackoverflow.com/questions/57316491/how-to-convert-matplotlib-figure-to-pil-image-object-without-saving-image
77 | import io
78 | buf = io.BytesIO()
79 | fig.savefig(buf)
80 | buf.seek(0)
81 | img = Image.open(buf)
82 | return img
83 |
84 |
85 | def compute_input_plot(self,):
86 | "Compute input for plot CFCGR"
87 | delta = 2*np.pi/4**self.k # angle between consecutive roots
88 |
89 | center = []
90 | width = []
91 | height = []
92 | bottom = []
93 |
94 | for kmer in tqdm(self.probabilities):
95 | h = self.probabilities.get(kmer,0.0) # height (density) of the bar in the circle
96 | theta = 2*self.encode(kmer).k*np.pi/4**self.k # angle of k-esim root
97 | c = theta + delta/2 # center of the angle
98 | center.append(c)
99 | bottom.append(0)
100 | width.append(50*delta)
101 | height.append(h)
102 |
103 | return center, bottom, width, height
--------------------------------------------------------------------------------
/complexcgr/fcgr.py:
--------------------------------------------------------------------------------
1 | from . import CGR
2 | from PIL import Image
3 | from itertools import product
4 | from collections import defaultdict
5 | import numpy as np
6 |
7 | NUC_COMPLEMENT = {n:c for n,c in zip ("ACGT","TGCA")}
8 |
9 | class FCGR(CGR):
10 | """Frequency matrix CGR
11 | an (2**k x 2**k) 2D representation will be created for a
12 | n-long sequence.
13 | - k represents the k-mer.
14 | - 2**k x 2**k = 4**k the total number of k-mers (sequences of length k)
15 | """
16 |
17 | def __init__(self, k: int, use_canonical_kmers: bool = False ,bits: int = 8):
18 | super().__init__()
19 | self.k = k # k-mer representation
20 | self.use_canonical_kmers = use_canonical_kmers
21 | self.kmers = list("".join(kmer) for kmer in product("ACGT", repeat=self.k))
22 |
23 | if use_canonical_kmers is True:
24 | self.kmer2pixel = self._kmer2pixel_canonical_kmers()
25 | else:
26 | self.kmer2pixel = self.kmer2pixel_position()
27 |
28 | self.bits = bits
29 | self.max_color = 2**bits-1
30 |
31 | def __call__(self, sequence: str):
32 | "Given a DNA sequence, returns an array with his FCGR"
33 | self.count_kmers(sequence)
34 |
35 | # Create an empty array to save the FCGR values
36 | array_size = int(2**self.k)
37 | fcgr = np.zeros((array_size,array_size))
38 |
39 | # Assign frequency to each box in the matrix
40 | for kmer, freq in self.freq_kmer.items():
41 | pos_x, pos_y = self.kmer2pixel[kmer]
42 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
43 | return fcgr
44 |
45 | def count_kmer(self, kmer):
46 | if "N" not in kmer:
47 | self.freq_kmer[kmer] += 1
48 |
49 | def count_kmers(self, sequence: str):
50 | self.freq_kmer = defaultdict(int)
51 | # representativity of kmers
52 | last_j = len(sequence) - self.k + 1
53 | kmers = (sequence[i:(i+self.k)] for i in range(last_j))
54 | # count kmers in a dictionary
55 | list(self.count_kmer(kmer) for kmer in kmers)
56 |
57 | def pixel_position(self, kmer: str):
58 | "Get pixel position in the FCGR matrix for a k-mer"
59 |
60 | coords = self.encode(kmer)
61 | N,x,y = coords.N, coords.x, coords.y
62 |
63 | # Coordinates from [-1,1]² to [1,2**k]²
64 | np_coords = np.array([(x + 1)/2, (y + 1)/2]) # move coordinates from [-1,1]² to [0,1]²
65 | np_coords *= 2**self.k # rescale coordinates from [0,1]² to [0,2**k]²
66 | x,y = np.ceil(np_coords) # round to upper integer
67 |
68 | # Turn coordinates (cx,cy) into pixel (px,py) position
69 | # px = 2**k-cy+1, py = cx
70 | return 2**self.k-int(y)+1, int(x)
71 |
72 | def kmer2pixel_position(self,):
73 | kmer2pixel = dict()
74 | for kmer in self.kmers:
75 | kmer2pixel[kmer] = self.pixel_position(kmer)
76 | return kmer2pixel
77 |
78 | def plot(self, fcgr):
79 | "Given a FCGR, plot it in grayscale"
80 | img_pil = self.array2img(fcgr)
81 | return img_pil
82 |
83 | def save_img(self, fcgr, path: str):
84 | "Save image in grayscale for the FCGR provided as input"
85 | img_pil = self.array2img(fcgr)
86 | img_pil.save(path)
87 |
88 | def array2img(self, array):
89 | "Array to PIL image"
90 | m, M = array.min(), array.max()
91 | # rescale to [0,1]
92 | img_rescaled = (array - m) / (M-m)
93 |
94 | # invert colors black->white
95 | img_array = np.ceil(self.max_color - img_rescaled*self.max_color)
96 | dtype = eval(f"np.int{self.bits}")
97 | img_array = np.array(img_array, dtype=dtype)
98 |
99 | # convert to Image
100 | img_pil = Image.fromarray(img_array,'L')
101 | return img_pil
102 |
103 | # # --------------- canonical k-mers ---------------- # #
104 | @staticmethod
105 | def reverse_complement(kmer: str):
106 | rev_kmer = list(kmer)[::-1]
107 | return "".join([NUC_COMPLEMENT[n] for n in rev_kmer])
108 |
109 | def get_canonical_kmer(self, kmer: str):
110 | rev_complement = self.reverse_complement(kmer)
111 | return kmer if kmer < rev_complement else rev_complement
112 |
113 | def _kmer2pixel_canonical_kmers(self,):
114 | # change pixel position of non-canonical kmers
115 | kmer2pixel = dict()
116 | for kmer in self.kmers:
117 | canonical_kmer = self.get_canonical_kmer(kmer)
118 | if kmer != canonical_kmer:
119 | kmer2pixel[kmer] = kmer2pixel[canonical_kmer]
120 | return kmer2pixel
--------------------------------------------------------------------------------
/complexcgr/fcgr_kmc.py:
--------------------------------------------------------------------------------
1 | from . import FCGR
2 |
3 | import gzip
4 | import numpy as np
5 |
6 | class FCGRKmc(FCGR):
7 | """
8 | Create FCGR with the option of using canonical kmers from KMC output
9 | """
10 | def __init__(self, k: int, use_canonical_kmers: bool=False):
11 | super().__init__(k, use_canonical_kmers)
12 | self.k = k # k-mer representation
13 | self.use_canonical_kmers = use_canonical_kmers
14 |
15 | def __call__(self, path_kmc_output):
16 | "Given a path to a kmc output file, return the FCGR using canonical kmers as an array"
17 | # Create an empty array to save the FCGR values
18 | array_size = int(2**self.k)
19 | fcgr = np.zeros((array_size,array_size))
20 |
21 | if str(path_kmc_output).endswith(".txt"):
22 | with open(path_kmc_output) as fp:
23 | for line in fp:
24 | kmer, freq = line.split("\t")
25 | pos_x, pos_y = self.kmer2pixel[kmer]
26 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
27 | else:
28 | with gzip.open(path_kmc_output,'rt') as f:
29 | for line in f:
30 | line = line.strip()
31 | kmer, freq = line.split()
32 | pos_x, pos_y = self.kmer2pixel[kmer]
33 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
34 |
35 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/fcgr_samples.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from Bio import SeqIO
3 | from collections import defaultdict
4 | from pathlib import Path
5 | from tqdm import tqdm
6 | from typing import List, Union
7 | from complexcgr import FCGR
8 |
9 | # input for FCGR Samples
10 | _path_fastq = Union[str, Path] # path can be a string or a Path instance
11 | _fastq = Union[_path_fastq,List[_path_fastq]] # either a single path or a list of paths to fastq files
12 |
13 | class FCGRSamples(FCGR):
14 |
15 | def __init__(self, k: int, bits: int = 8):
16 | super().__init__(k, bits)
17 |
18 | def __call__(self, path_fastq: _fastq, consider_quality: bool = False):
19 | "Given a (list) of fastq files, return the FCGR matrix"
20 |
21 | ## ---- New for reads ----
22 |
23 | # for each call, we start to count the frequencies
24 | self.freq_kmer = defaultdict(int)
25 | if consider_quality is True:
26 | self.qual_kmer = defaultdict(int)
27 |
28 | # transform to list to iterate
29 | path_fastq = path_fastq if type(path_fastq) is list else [path_fastq]
30 |
31 | # For each file, count kmers on their reads
32 | for path in path_fastq:
33 | fastq = self.load_fastq(path)
34 | n_reads = self.count_reads(path)
35 |
36 | # count kmers in each read
37 | for sample in tqdm(fastq,total=n_reads,desc=f"Counting kmers on {str(Path(path).stem)}"):
38 | read = sample.seq
39 | if consider_quality is True:
40 | qual = sample.letter_annotations["phred_quality"]
41 | self.count_kmers_qualities(str(read),qual)
42 | else:
43 | self.count_kmers(str(read))
44 |
45 | ## ---- End new for reads ----
46 |
47 | # Create an empty array to save the FCGR values
48 | if consider_quality is False:
49 | array_size = int(2**self.k)
50 | fcgr = np.zeros((array_size,array_size))
51 | else:
52 | array_size = int(2**self.k)
53 | fcgr = np.zeros((array_size,array_size,2))
54 |
55 | # Assign frequency to each box in the matrix
56 | for kmer, freq in self.freq_kmer.items():
57 | pos_x, pos_y = self.kmer2pixel[kmer]
58 |
59 | if consider_quality is True:
60 | fcgr[int(pos_x)-1,int(pos_y)-1,0] = freq
61 | fcgr[int(pos_x)-1,int(pos_y)-1,1] = self.qual_kmer[kmer]
62 | else:
63 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
64 |
65 | return fcgr if consider_quality is False else self.rescale_fcgr_qualities(fcgr)
66 |
67 |
68 | def load_fastq(self, path):
69 | "Load a fastq file"
70 | fastq = SeqIO.parse(str(path), "fastq")
71 | return fastq
72 |
73 | def count_kmers(self, read: str):
74 | # representativity of kmers
75 | last_j = len(read) - self.k + 1
76 | kmers = (read[i:(i+self.k)] for i in range(last_j))
77 | # count kmers in a dictionary
78 | list(self.count_kmer(kmer) for kmer in kmers)
79 |
80 | @staticmethod
81 | def count_reads(path):
82 | "Count reads in a file to define progress bar"
83 | n_reads = 0
84 | with open(str(path)) as fp:
85 | for line in fp:
86 | if line.startswith("@"):
87 | n_reads += 1
88 | return n_reads
89 |
90 | def count_kmer(self, kmer):
91 | if "N" not in kmer:
92 | self.freq_kmer[kmer] += 1
93 |
94 |
95 | def count_kmer_quality(self, kmer, qmer):
96 | """Count kmers and qualities
97 | For quality of a kmer, the mean of qualities for each nucleotide in the kmer will be saved"""
98 | if "N" not in kmer:
99 | self.freq_kmer[kmer] += 1
100 | self.qual_kmer[kmer] += qmer.mean()
101 |
102 | def count_kmers_qualities(self,read,qual):
103 | "Count kmers and qualities"
104 |
105 | # representativity of kmers
106 | last_j = len(read) - self.k + 1
107 | kmers = (read[i:(i+self.k)] for i in range(last_j))
108 | qmers = (np.array(qual[i:(i+self.k)]) for i in range(last_j)) # qualities for each kmer
109 |
110 | # count kmers and qualities in a dictionary
111 | list(self.count_kmer_quality(kmer,qmer) for (kmer,qmer) in zip(kmers,qmers))
112 |
113 | @staticmethod
114 | def rescale_fcgr_qualities(fcgr):
115 | "Divide each cumulate quality by the freq of its kmer"
116 | freqs, quals = fcgr[:,:,0], fcgr[:,:,1]
117 | fcgr[:,:,1] = np.divide(quals, freqs, out = np.zeros_like(quals), where = quals!=0)
118 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/icgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from itertools import product
3 | from tqdm import tqdm
4 | from typing import Dict, Optional
5 | from collections import defaultdict, namedtuple
6 | import numpy as np
7 |
8 | # coordinates for x+iy
9 | Coord = namedtuple("Coord", ["x","y"])
10 |
11 | # coordinates for a CGR encoding
12 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
13 |
14 | # coordinates for each nucleotide in the 2d-plane
15 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
16 |
17 | class iCGR:
18 | "integer Chaos Game Representation for DNA"
19 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
20 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
21 | self.cgr_coords = CGRCoords(0,0,0)
22 |
23 | def nucleotide_by_coords(self,x,y):
24 | "Get nucleotide by coordinates (x,y)"
25 | # filter nucleotide by coordinates
26 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
27 |
28 | return list(filtered.keys())[0]
29 |
30 | def forward(self, nucleotide: str):
31 | "Compute next CGR coordinates"
32 | # current length
33 | N = self.cgr_coords.N
34 |
35 | # compute next coordinates
36 | x = self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x * 2**N
37 | y = self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y * 2**N
38 |
39 | # update cgr_coords: iCGR starts in the corner of the first nucleotide
40 |
41 | if N>0:
42 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y,)
43 | else:
44 | self.cgr_coords = CGRCoords(1,
45 | self.nucleotide_coords.get(nucleotide).x,
46 | self.nucleotide_coords.get(nucleotide).y,
47 | )
48 |
49 | def backward(self,):
50 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
51 | # get current nucleotide based on coordinates
52 | n_x,n_y = self.coords_current_nucleotide()
53 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
54 |
55 | # current length
56 | N = self.cgr_coords.N
57 |
58 | # update coordinates to the previous one
59 | x = self.cgr_coords.x - self.nucleotide_coords.get(nucleotide).x * 2**(N-1)
60 | y = self.cgr_coords.y - self.nucleotide_coords.get(nucleotide).y * 2**(N-1)
61 |
62 | # update cgr_coords
63 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
64 |
65 | return nucleotide
66 |
67 | def coords_current_nucleotide(self,):
68 | x = 1 if self.cgr_coords.x>0 else -1
69 | y = 1 if self.cgr_coords.y>0 else -1
70 | return x,y
71 |
72 | def encode(self, sequence: str):
73 | "From DNA sequence to CGR"
74 | # reset starting position to (0,0,0)
75 | self.reset_coords()
76 | for nucleotide in sequence:
77 | self.forward(nucleotide)
78 | return self.cgr_coords
79 |
80 | def reset_coords(self,):
81 | self.cgr_coords = CGRCoords(0,0,0)
82 |
83 | def decode(self, N:int, x:int, y:int)->str:
84 | "From CGR to DNA sequence"
85 | self.cgr_coords = CGRCoords(N,x,y)
86 |
87 | # decoded sequence
88 | sequence = []
89 |
90 | # Recover the entire genome
91 | while self.cgr_coords.N>0:
92 | nucleotide = self.backward()
93 | sequence.append(nucleotide)
94 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/img/ACG-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG-complexCGR.png
--------------------------------------------------------------------------------
/img/ACG.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG.jpg
--------------------------------------------------------------------------------
/img/ACG_16bits.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG_16bits.jpg
--------------------------------------------------------------------------------
/img/CGA.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGA.jpg
--------------------------------------------------------------------------------
/img/CGAN.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGAN.jpg
--------------------------------------------------------------------------------
/img/SAMD00003784.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00003784.jpeg
--------------------------------------------------------------------------------
/img/SAMD00032020.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00032020.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111209.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111209.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111306.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111306.jpeg
--------------------------------------------------------------------------------
/img/complexcgr-readme.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/complexcgr-readme.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3.png
--------------------------------------------------------------------------------
/img/logo-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR.png
--------------------------------------------------------------------------------
/poetry.lock:
--------------------------------------------------------------------------------
1 | # This file is automatically @generated by Poetry 1.7.1 and should not be changed by hand.
2 |
3 | [[package]]
4 | name = "atomicwrites"
5 | version = "1.4.1"
6 | description = "Atomic file writes."
7 | optional = false
8 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
9 | files = [
10 | {file = "atomicwrites-1.4.1.tar.gz", hash = "sha256:81b2c9071a49367a7f770170e5eec8cb66567cfbbc8c73d20ce5ca4a8d71cf11"},
11 | ]
12 |
13 | [[package]]
14 | name = "attrs"
15 | version = "23.2.0"
16 | description = "Classes Without Boilerplate"
17 | optional = false
18 | python-versions = ">=3.7"
19 | files = [
20 | {file = "attrs-23.2.0-py3-none-any.whl", hash = "sha256:99b87a485a5820b23b879f04c2305b44b951b502fd64be915879d77a7e8fc6f1"},
21 | {file = "attrs-23.2.0.tar.gz", hash = "sha256:935dc3b529c262f6cf76e50877d35a4bd3c1de194fd41f47a2b7ae8f19971f30"},
22 | ]
23 |
24 | [package.extras]
25 | cov = ["attrs[tests]", "coverage[toml] (>=5.3)"]
26 | dev = ["attrs[tests]", "pre-commit"]
27 | docs = ["furo", "myst-parser", "sphinx", "sphinx-notfound-page", "sphinxcontrib-towncrier", "towncrier", "zope-interface"]
28 | tests = ["attrs[tests-no-zope]", "zope-interface"]
29 | tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
30 | tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
31 |
32 | [[package]]
33 | name = "biopython"
34 | version = "1.82"
35 | description = "Freely available tools for computational molecular biology."
36 | optional = false
37 | python-versions = ">=3.8"
38 | files = [
39 | {file = "biopython-1.82-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:713be5e1e8571ea151864544dfcd2637eaf98c67a6e47b69781d325feb02f6b9"},
40 | {file = "biopython-1.82-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5bc908e84278bf61ac901a0c1b695c85ce49491957e0ab07e1e2afc216882525"},
41 | {file = "biopython-1.82-cp310-cp310-win32.whl", hash = "sha256:f53aff30f01ac9a8aa1fd42dab347a96ee5312945098dfa38f9a5125f6e913fd"},
42 | {file = "biopython-1.82-cp310-cp310-win_amd64.whl", hash = "sha256:0f3f685a2d85348b8c84c6eebe58a74cef7e1158ac52ae5aa91789436907f0e1"},
43 | {file = "biopython-1.82-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c9bc080b7dc6cd6acbf2576766136e8e54acbcc6b7a29ad8075cb5a73f4cfb97"},
44 | {file = "biopython-1.82-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c7802b5033edaf34bd2fbd96b87f96999a6808caa0064c91087c7f1340624898"},
45 | {file = "biopython-1.82-cp311-cp311-win32.whl", hash = "sha256:ec35893b32458f6fa0c98d5f02d801a5fe76961ac66473455f51c0575562c594"},
46 | {file = "biopython-1.82-cp311-cp311-win_amd64.whl", hash = "sha256:40297d418bb142a7ea1a47faedc24bbe21b0ba455f3805373264470e5cb6ec5c"},
47 | {file = "biopython-1.82-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:96f77bd6540683c8958e4ebf55b58e8caa26d20db72891915d06670b783e9cdb"},
48 | {file = "biopython-1.82-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:49b829c2276a0711106663af632c7e203695a5cf082d488d6e10b8aca5ec63e4"},
49 | {file = "biopython-1.82-cp312-cp312-win32.whl", hash = "sha256:54a9b200d4960ed2a4ec85faac2124ffa278afa58d22e07e2d15ca7509219ea1"},
50 | {file = "biopython-1.82-cp312-cp312-win_amd64.whl", hash = "sha256:762547037038d42c9ac98877fc32d4aeba798077129df7790cb178256171a4e5"},
51 | {file = "biopython-1.82-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:cb013256742d31e24fd5093d55511f58bc6a93b8ac723d2ca3b33c7904f1ca14"},
52 | {file = "biopython-1.82-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:16829760fab60ba17141b7a81494fed837c6f29ea28cdcfb3070764c8e8f2ff1"},
53 | {file = "biopython-1.82-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c6af82894dd4f736d737fdea62008a19966251ba39aef39a0ce8b41321305ad5"},
54 | {file = "biopython-1.82-cp38-cp38-win32.whl", hash = "sha256:a1ff583b2f8b314a4c2d612aad4f4b73827a5f741b391f3a834b0c192a4f1527"},
55 | {file = "biopython-1.82-cp38-cp38-win_amd64.whl", hash = "sha256:0198f23776d9cb9d514a797e81a024f486adac973b252a2f4696995a5a163745"},
56 | {file = "biopython-1.82-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c88e9f87d1b07958e7e1e25ff20efced4796daf650e5290692b5c3ff9aebf9d3"},
57 | {file = "biopython-1.82-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8e2533db9275c3e1438151260844e7683f91308da320df02a4c28fb7053ea3a9"},
58 | {file = "biopython-1.82-cp39-cp39-win32.whl", hash = "sha256:80f48d220b92e747300281602f52e98ec6c537ddd4921b580d9a0368bae05ff5"},
59 | {file = "biopython-1.82-cp39-cp39-win_amd64.whl", hash = "sha256:73d619611bd7cf9d2ad8b3217b08d3796acfabb765b0771ad2ff7ee2e46a59b0"},
60 | {file = "biopython-1.82.tar.gz", hash = "sha256:a9b10d959ae88a9744a91c6ce3601f4c86e7ec41679bc93c29f679218f6167bb"},
61 | ]
62 |
63 | [package.dependencies]
64 | numpy = "*"
65 |
66 | [[package]]
67 | name = "colorama"
68 | version = "0.4.6"
69 | description = "Cross-platform colored terminal text."
70 | optional = false
71 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,!=3.3.*,!=3.4.*,!=3.5.*,!=3.6.*,>=2.7"
72 | files = [
73 | {file = "colorama-0.4.6-py2.py3-none-any.whl", hash = "sha256:4f1d9991f5acc0ca119f9d443620b77f9d6b33703e51011c16baf57afb285fc6"},
74 | {file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
75 | ]
76 |
77 | [[package]]
78 | name = "contourpy"
79 | version = "1.2.0"
80 | description = "Python library for calculating contours of 2D quadrilateral grids"
81 | optional = false
82 | python-versions = ">=3.9"
83 | files = [
84 | {file = "contourpy-1.2.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:0274c1cb63625972c0c007ab14dd9ba9e199c36ae1a231ce45d725cbcbfd10a8"},
85 | {file = "contourpy-1.2.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ab459a1cbbf18e8698399c595a01f6dcc5c138220ca3ea9e7e6126232d102bb4"},
86 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6fdd887f17c2f4572ce548461e4f96396681212d858cae7bd52ba3310bc6f00f"},
87 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:5d16edfc3fc09968e09ddffada434b3bf989bf4911535e04eada58469873e28e"},
88 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1c203f617abc0dde5792beb586f827021069fb6d403d7f4d5c2b543d87edceb9"},
89 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b69303ceb2e4d4f146bf82fda78891ef7bcd80c41bf16bfca3d0d7eb545448aa"},
90 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:884c3f9d42d7218304bc74a8a7693d172685c84bd7ab2bab1ee567b769696df9"},
91 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4a1b1208102be6e851f20066bf0e7a96b7d48a07c9b0cfe6d0d4545c2f6cadab"},
92 | {file = "contourpy-1.2.0-cp310-cp310-win32.whl", hash = "sha256:34b9071c040d6fe45d9826cbbe3727d20d83f1b6110d219b83eb0e2a01d79488"},
93 | {file = "contourpy-1.2.0-cp310-cp310-win_amd64.whl", hash = "sha256:bd2f1ae63998da104f16a8b788f685e55d65760cd1929518fd94cd682bf03e41"},
94 | {file = "contourpy-1.2.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:dd10c26b4eadae44783c45ad6655220426f971c61d9b239e6f7b16d5cdaaa727"},
95 | {file = "contourpy-1.2.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:5c6b28956b7b232ae801406e529ad7b350d3f09a4fde958dfdf3c0520cdde0dd"},
96 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ebeac59e9e1eb4b84940d076d9f9a6cec0064e241818bcb6e32124cc5c3e377a"},
97 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:139d8d2e1c1dd52d78682f505e980f592ba53c9f73bd6be102233e358b401063"},
98 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1e9dc350fb4c58adc64df3e0703ab076f60aac06e67d48b3848c23647ae4310e"},
99 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:18fc2b4ed8e4a8fe849d18dce4bd3c7ea637758c6343a1f2bae1e9bd4c9f4686"},
100 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:16a7380e943a6d52472096cb7ad5264ecee36ed60888e2a3d3814991a0107286"},
101 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8d8faf05be5ec8e02a4d86f616fc2a0322ff4a4ce26c0f09d9f7fb5330a35c95"},
102 | {file = "contourpy-1.2.0-cp311-cp311-win32.whl", hash = "sha256:67b7f17679fa62ec82b7e3e611c43a016b887bd64fb933b3ae8638583006c6d6"},
103 | {file = "contourpy-1.2.0-cp311-cp311-win_amd64.whl", hash = "sha256:99ad97258985328b4f207a5e777c1b44a83bfe7cf1f87b99f9c11d4ee477c4de"},
104 | {file = "contourpy-1.2.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:575bcaf957a25d1194903a10bc9f316c136c19f24e0985a2b9b5608bdf5dbfe0"},
105 | {file = "contourpy-1.2.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:9e6c93b5b2dbcedad20a2f18ec22cae47da0d705d454308063421a3b290d9ea4"},
106 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:464b423bc2a009088f19bdf1f232299e8b6917963e2b7e1d277da5041f33a779"},
107 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:68ce4788b7d93e47f84edd3f1f95acdcd142ae60bc0e5493bfd120683d2d4316"},
108 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3d7d1f8871998cdff5d2ff6a087e5e1780139abe2838e85b0b46b7ae6cc25399"},
109 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6e739530c662a8d6d42c37c2ed52a6f0932c2d4a3e8c1f90692ad0ce1274abe0"},
110 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:247b9d16535acaa766d03037d8e8fb20866d054d3c7fbf6fd1f993f11fc60ca0"},
111 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:461e3ae84cd90b30f8d533f07d87c00379644205b1d33a5ea03381edc4b69431"},
112 | {file = "contourpy-1.2.0-cp312-cp312-win32.whl", hash = "sha256:1c2559d6cffc94890b0529ea7eeecc20d6fadc1539273aa27faf503eb4656d8f"},
113 | {file = "contourpy-1.2.0-cp312-cp312-win_amd64.whl", hash = "sha256:491b1917afdd8638a05b611a56d46587d5a632cabead889a5440f7c638bc6ed9"},
114 | {file = "contourpy-1.2.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5fd1810973a375ca0e097dee059c407913ba35723b111df75671a1976efa04bc"},
115 | {file = "contourpy-1.2.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:999c71939aad2780f003979b25ac5b8f2df651dac7b38fb8ce6c46ba5abe6ae9"},
116 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b7caf9b241464c404613512d5594a6e2ff0cc9cb5615c9475cc1d9b514218ae8"},
117 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:266270c6f6608340f6c9836a0fb9b367be61dde0c9a9a18d5ece97774105ff3e"},
118 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:dbd50d0a0539ae2e96e537553aff6d02c10ed165ef40c65b0e27e744a0f10af8"},
119 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:11f8d2554e52f459918f7b8e6aa20ec2a3bce35ce95c1f0ef4ba36fbda306df5"},
120 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:ce96dd400486e80ac7d195b2d800b03e3e6a787e2a522bfb83755938465a819e"},
121 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6d3364b999c62f539cd403f8123ae426da946e142312a514162adb2addd8d808"},
122 | {file = "contourpy-1.2.0-cp39-cp39-win32.whl", hash = "sha256:1c88dfb9e0c77612febebb6ac69d44a8d81e3dc60f993215425b62c1161353f4"},
123 | {file = "contourpy-1.2.0-cp39-cp39-win_amd64.whl", hash = "sha256:78e6ad33cf2e2e80c5dfaaa0beec3d61face0fb650557100ee36db808bfa6843"},
124 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:be16975d94c320432657ad2402f6760990cb640c161ae6da1363051805fa8108"},
125 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b95a225d4948b26a28c08307a60ac00fb8671b14f2047fc5476613252a129776"},
126 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:0d7e03c0f9a4f90dc18d4e77e9ef4ec7b7bbb437f7f675be8e530d65ae6ef956"},
127 | {file = "contourpy-1.2.0.tar.gz", hash = "sha256:171f311cb758de7da13fc53af221ae47a5877be5a0843a9fe150818c51ed276a"},
128 | ]
129 |
130 | [package.dependencies]
131 | numpy = ">=1.20,<2.0"
132 |
133 | [package.extras]
134 | bokeh = ["bokeh", "selenium"]
135 | docs = ["furo", "sphinx (>=7.2)", "sphinx-copybutton"]
136 | mypy = ["contourpy[bokeh,docs]", "docutils-stubs", "mypy (==1.6.1)", "types-Pillow"]
137 | test = ["Pillow", "contourpy[test-no-images]", "matplotlib"]
138 | test-no-images = ["pytest", "pytest-cov", "pytest-xdist", "wurlitzer"]
139 |
140 | [[package]]
141 | name = "cycler"
142 | version = "0.12.1"
143 | description = "Composable style cycles"
144 | optional = false
145 | python-versions = ">=3.8"
146 | files = [
147 | {file = "cycler-0.12.1-py3-none-any.whl", hash = "sha256:85cef7cff222d8644161529808465972e51340599459b8ac3ccbac5a854e0d30"},
148 | {file = "cycler-0.12.1.tar.gz", hash = "sha256:88bb128f02ba341da8ef447245a9e138fae777f6a23943da4540077d3601eb1c"},
149 | ]
150 |
151 | [package.extras]
152 | docs = ["ipython", "matplotlib", "numpydoc", "sphinx"]
153 | tests = ["pytest", "pytest-cov", "pytest-xdist"]
154 |
155 | [[package]]
156 | name = "fonttools"
157 | version = "4.47.0"
158 | description = "Tools to manipulate font files"
159 | optional = false
160 | python-versions = ">=3.8"
161 | files = [
162 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:2d2404107626f97a221dc1a65b05396d2bb2ce38e435f64f26ed2369f68675d9"},
163 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:c01f409be619a9a0f5590389e37ccb58b47264939f0e8d58bfa1f3ba07d22671"},
164 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d986b66ff722ef675b7ee22fbe5947a41f60a61a4da15579d5e276d897fbc7fa"},
165 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e8acf6dd0434b211b3bd30d572d9e019831aae17a54016629fa8224783b22df8"},
166 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:495369c660e0c27233e3c572269cbe520f7f4978be675f990f4005937337d391"},
167 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c59227d7ba5b232281c26ae04fac2c73a79ad0e236bca5c44aae904a18f14faf"},
168 | {file = "fonttools-4.47.0-cp310-cp310-win32.whl", hash = "sha256:59a6c8b71a245800e923cb684a2dc0eac19c56493e2f896218fcf2571ed28984"},
169 | {file = "fonttools-4.47.0-cp310-cp310-win_amd64.whl", hash = "sha256:52c82df66201f3a90db438d9d7b337c7c98139de598d0728fb99dab9fd0495ca"},
170 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:854421e328d47d70aa5abceacbe8eef231961b162c71cbe7ff3f47e235e2e5c5"},
171 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:511482df31cfea9f697930f61520f6541185fa5eeba2fa760fe72e8eee5af88b"},
172 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ce0e2c88c8c985b7b9a7efcd06511fb0a1fe3ddd9a6cd2895ef1dbf9059719d7"},
173 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e7a0a8848726956e9d9fb18c977a279013daadf0cbb6725d2015a6dd57527992"},
174 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e869da810ae35afb3019baa0d0306cdbab4760a54909c89ad8904fa629991812"},
175 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:dd23848f877c3754f53a4903fb7a593ed100924f9b4bff7d5a4e2e8a7001ae11"},
176 | {file = "fonttools-4.47.0-cp311-cp311-win32.whl", hash = "sha256:bf1810635c00f7c45d93085611c995fc130009cec5abdc35b327156aa191f982"},
177 | {file = "fonttools-4.47.0-cp311-cp311-win_amd64.whl", hash = "sha256:61df4dee5d38ab65b26da8efd62d859a1eef7a34dcbc331299a28e24d04c59a7"},
178 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:e3f4d61f3a8195eac784f1d0c16c0a3105382c1b9a74d99ac4ba421da39a8826"},
179 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:174995f7b057e799355b393e97f4f93ef1f2197cbfa945e988d49b2a09ecbce8"},
180 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea592e6a09b71cb7a7661dd93ac0b877a6228e2d677ebacbad0a4d118494c86d"},
181 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:40bdbe90b33897d9cc4a39f8e415b0fcdeae4c40a99374b8a4982f127ff5c767"},
182 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:843509ae9b93db5aaf1a6302085e30bddc1111d31e11d724584818f5b698f500"},
183 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:9acfa1cdc479e0dde528b61423855913d949a7f7fe09e276228298fef4589540"},
184 | {file = "fonttools-4.47.0-cp312-cp312-win32.whl", hash = "sha256:66c92ec7f95fd9732550ebedefcd190a8d81beaa97e89d523a0d17198a8bda4d"},
185 | {file = "fonttools-4.47.0-cp312-cp312-win_amd64.whl", hash = "sha256:e8fa20748de55d0021f83754b371432dca0439e02847962fc4c42a0e444c2d78"},
186 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c75e19971209fbbce891ebfd1b10c37320a5a28e8d438861c21d35305aedb81c"},
187 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e79f1a3970d25f692bbb8c8c2637e621a66c0d60c109ab48d4a160f50856deff"},
188 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:562681188c62c024fe2c611b32e08b8de2afa00c0c4e72bed47c47c318e16d5c"},
189 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a77a60315c33393b2bd29d538d1ef026060a63d3a49a9233b779261bad9c3f71"},
190 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:b4fabb8cc9422efae1a925160083fdcbab8fdc96a8483441eb7457235df625bd"},
191 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2a78dba8c2a1e9d53a0fb5382979f024200dc86adc46a56cbb668a2249862fda"},
192 | {file = "fonttools-4.47.0-cp38-cp38-win32.whl", hash = "sha256:e6b968543fde4119231c12c2a953dcf83349590ca631ba8216a8edf9cd4d36a9"},
193 | {file = "fonttools-4.47.0-cp38-cp38-win_amd64.whl", hash = "sha256:4a9a51745c0439516d947480d4d884fa18bd1458e05b829e482b9269afa655bc"},
194 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:62d8ddb058b8e87018e5dc26f3258e2c30daad4c87262dfeb0e2617dd84750e6"},
195 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5dde0eab40faaa5476133123f6a622a1cc3ac9b7af45d65690870620323308b4"},
196 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f4da089f6dfdb822293bde576916492cd708c37c2501c3651adde39804630538"},
197 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:253bb46bab970e8aae254cebf2ae3db98a4ef6bd034707aa68a239027d2b198d"},
198 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:1193fb090061efa2f9e2d8d743ae9850c77b66746a3b32792324cdce65784154"},
199 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:084511482dd265bce6dca24c509894062f0117e4e6869384d853f46c0e6d43be"},
200 | {file = "fonttools-4.47.0-cp39-cp39-win32.whl", hash = "sha256:97620c4af36e4c849e52661492e31dc36916df12571cb900d16960ab8e92a980"},
201 | {file = "fonttools-4.47.0-cp39-cp39-win_amd64.whl", hash = "sha256:e77bdf52185bdaf63d39f3e1ac3212e6cfa3ab07d509b94557a8902ce9c13c82"},
202 | {file = "fonttools-4.47.0-py3-none-any.whl", hash = "sha256:d6477ba902dd2d7adda7f0fd3bfaeb92885d45993c9e1928c9f28fc3961415f7"},
203 | {file = "fonttools-4.47.0.tar.gz", hash = "sha256:ec13a10715eef0e031858c1c23bfaee6cba02b97558e4a7bfa089dba4a8c2ebf"},
204 | ]
205 |
206 | [package.extras]
207 | all = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "fs (>=2.2.0,<3)", "lxml (>=4.0,<5)", "lz4 (>=1.7.4.2)", "matplotlib", "munkres", "pycairo", "scipy", "skia-pathops (>=0.5.0)", "sympy", "uharfbuzz (>=0.23.0)", "unicodedata2 (>=15.1.0)", "xattr", "zopfli (>=0.1.4)"]
208 | graphite = ["lz4 (>=1.7.4.2)"]
209 | interpolatable = ["munkres", "pycairo", "scipy"]
210 | lxml = ["lxml (>=4.0,<5)"]
211 | pathops = ["skia-pathops (>=0.5.0)"]
212 | plot = ["matplotlib"]
213 | repacker = ["uharfbuzz (>=0.23.0)"]
214 | symfont = ["sympy"]
215 | type1 = ["xattr"]
216 | ufo = ["fs (>=2.2.0,<3)"]
217 | unicode = ["unicodedata2 (>=15.1.0)"]
218 | woff = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "zopfli (>=0.1.4)"]
219 |
220 | [[package]]
221 | name = "importlib-resources"
222 | version = "6.1.1"
223 | description = "Read resources from Python packages"
224 | optional = false
225 | python-versions = ">=3.8"
226 | files = [
227 | {file = "importlib_resources-6.1.1-py3-none-any.whl", hash = "sha256:e8bf90d8213b486f428c9c39714b920041cb02c184686a3dee24905aaa8105d6"},
228 | {file = "importlib_resources-6.1.1.tar.gz", hash = "sha256:3893a00122eafde6894c59914446a512f728a0c1a45f9bb9b63721b6bacf0b4a"},
229 | ]
230 |
231 | [package.dependencies]
232 | zipp = {version = ">=3.1.0", markers = "python_version < \"3.10\""}
233 |
234 | [package.extras]
235 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
236 | testing = ["pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy (>=0.9.1)", "pytest-ruff", "zipp (>=3.17)"]
237 |
238 | [[package]]
239 | name = "iniconfig"
240 | version = "2.0.0"
241 | description = "brain-dead simple config-ini parsing"
242 | optional = false
243 | python-versions = ">=3.7"
244 | files = [
245 | {file = "iniconfig-2.0.0-py3-none-any.whl", hash = "sha256:b6a85871a79d2e3b22d2d1b94ac2824226a63c6b741c88f7ae975f18b6778374"},
246 | {file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
247 | ]
248 |
249 | [[package]]
250 | name = "kiwisolver"
251 | version = "1.4.5"
252 | description = "A fast implementation of the Cassowary constraint solver"
253 | optional = false
254 | python-versions = ">=3.7"
255 | files = [
256 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:05703cf211d585109fcd72207a31bb170a0f22144d68298dc5e61b3c946518af"},
257 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:146d14bebb7f1dc4d5fbf74f8a6cb15ac42baadee8912eb84ac0b3b2a3dc6ac3"},
258 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ef7afcd2d281494c0a9101d5c571970708ad911d028137cd558f02b851c08b4"},
259 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9eaa8b117dc8337728e834b9c6e2611f10c79e38f65157c4c38e9400286f5cb1"},
260 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ec20916e7b4cbfb1f12380e46486ec4bcbaa91a9c448b97023fde0d5bbf9e4ff"},
261 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:39b42c68602539407884cf70d6a480a469b93b81b7701378ba5e2328660c847a"},
262 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:aa12042de0171fad672b6c59df69106d20d5596e4f87b5e8f76df757a7c399aa"},
263 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:2a40773c71d7ccdd3798f6489aaac9eee213d566850a9533f8d26332d626b82c"},
264 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:19df6e621f6d8b4b9c4d45f40a66839294ff2bb235e64d2178f7522d9170ac5b"},
265 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:83d78376d0d4fd884e2c114d0621624b73d2aba4e2788182d286309ebdeed770"},
266 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:e391b1f0a8a5a10ab3b9bb6afcfd74f2175f24f8975fb87ecae700d1503cdee0"},
267 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:852542f9481f4a62dbb5dd99e8ab7aedfeb8fb6342349a181d4036877410f525"},
268 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:59edc41b24031bc25108e210c0def6f6c2191210492a972d585a06ff246bb79b"},
269 | {file = "kiwisolver-1.4.5-cp310-cp310-win32.whl", hash = "sha256:a6aa6315319a052b4ee378aa171959c898a6183f15c1e541821c5c59beaa0238"},
270 | {file = "kiwisolver-1.4.5-cp310-cp310-win_amd64.whl", hash = "sha256:d0ef46024e6a3d79c01ff13801cb19d0cad7fd859b15037aec74315540acc276"},
271 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:11863aa14a51fd6ec28688d76f1735f8f69ab1fabf388851a595d0721af042f5"},
272 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:8ab3919a9997ab7ef2fbbed0cc99bb28d3c13e6d4b1ad36e97e482558a91be90"},
273 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fcc700eadbbccbf6bc1bcb9dbe0786b4b1cb91ca0dcda336eef5c2beed37b797"},
274 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:dfdd7c0b105af050eb3d64997809dc21da247cf44e63dc73ff0fd20b96be55a9"},
275 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:76c6a5964640638cdeaa0c359382e5703e9293030fe730018ca06bc2010c4437"},
276 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:bbea0db94288e29afcc4c28afbf3a7ccaf2d7e027489c449cf7e8f83c6346eb9"},
277 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ceec1a6bc6cab1d6ff5d06592a91a692f90ec7505d6463a88a52cc0eb58545da"},
278 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:040c1aebeda72197ef477a906782b5ab0d387642e93bda547336b8957c61022e"},
279 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f91de7223d4c7b793867797bacd1ee53bfe7359bd70d27b7b58a04efbb9436c8"},
280 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:faae4860798c31530dd184046a900e652c95513796ef51a12bc086710c2eec4d"},
281 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:b0157420efcb803e71d1b28e2c287518b8808b7cf1ab8af36718fd0a2c453eb0"},
282 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:06f54715b7737c2fecdbf140d1afb11a33d59508a47bf11bb38ecf21dc9ab79f"},
283 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:fdb7adb641a0d13bdcd4ef48e062363d8a9ad4a182ac7647ec88f695e719ae9f"},
284 | {file = "kiwisolver-1.4.5-cp311-cp311-win32.whl", hash = "sha256:bb86433b1cfe686da83ce32a9d3a8dd308e85c76b60896d58f082136f10bffac"},
285 | {file = "kiwisolver-1.4.5-cp311-cp311-win_amd64.whl", hash = "sha256:6c08e1312a9cf1074d17b17728d3dfce2a5125b2d791527f33ffbe805200a355"},
286 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:32d5cf40c4f7c7b3ca500f8985eb3fb3a7dfc023215e876f207956b5ea26632a"},
287 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:f846c260f483d1fd217fe5ed7c173fb109efa6b1fc8381c8b7552c5781756192"},
288 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5ff5cf3571589b6d13bfbfd6bcd7a3f659e42f96b5fd1c4830c4cf21d4f5ef45"},
289 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7269d9e5f1084a653d575c7ec012ff57f0c042258bf5db0954bf551c158466e7"},
290 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:da802a19d6e15dffe4b0c24b38b3af68e6c1a68e6e1d8f30148c83864f3881db"},
291 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:3aba7311af82e335dd1e36ffff68aaca609ca6290c2cb6d821a39aa075d8e3ff"},
292 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:763773d53f07244148ccac5b084da5adb90bfaee39c197554f01b286cf869228"},
293 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2270953c0d8cdab5d422bee7d2007f043473f9d2999631c86a223c9db56cbd16"},
294 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:d099e745a512f7e3bbe7249ca835f4d357c586d78d79ae8f1dcd4d8adeb9bda9"},
295 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:74db36e14a7d1ce0986fa104f7d5637aea5c82ca6326ed0ec5694280942d1162"},
296 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:7e5bab140c309cb3a6ce373a9e71eb7e4873c70c2dda01df6820474f9889d6d4"},
297 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:0f114aa76dc1b8f636d077979c0ac22e7cd8f3493abbab152f20eb8d3cda71f3"},
298 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:88a2df29d4724b9237fc0c6eaf2a1adae0cdc0b3e9f4d8e7dc54b16812d2d81a"},
299 | {file = "kiwisolver-1.4.5-cp312-cp312-win32.whl", hash = "sha256:72d40b33e834371fd330fb1472ca19d9b8327acb79a5821d4008391db8e29f20"},
300 | {file = "kiwisolver-1.4.5-cp312-cp312-win_amd64.whl", hash = "sha256:2c5674c4e74d939b9d91dda0fae10597ac7521768fec9e399c70a1f27e2ea2d9"},
301 | {file = "kiwisolver-1.4.5-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:3a2b053a0ab7a3960c98725cfb0bf5b48ba82f64ec95fe06f1d06c99b552e130"},
302 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3cd32d6c13807e5c66a7cbb79f90b553642f296ae4518a60d8d76243b0ad2898"},
303 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:59ec7b7c7e1a61061850d53aaf8e93db63dce0c936db1fda2658b70e4a1be709"},
304 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da4cfb373035def307905d05041c1d06d8936452fe89d464743ae7fb8371078b"},
305 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:2400873bccc260b6ae184b2b8a4fec0e4082d30648eadb7c3d9a13405d861e89"},
306 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:1b04139c4236a0f3aff534479b58f6f849a8b351e1314826c2d230849ed48985"},
307 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:4e66e81a5779b65ac21764c295087de82235597a2293d18d943f8e9e32746265"},
308 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:7931d8f1f67c4be9ba1dd9c451fb0eeca1a25b89e4d3f89e828fe12a519b782a"},
309 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:b3f7e75f3015df442238cca659f8baa5f42ce2a8582727981cbfa15fee0ee205"},
310 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:bbf1d63eef84b2e8c89011b7f2235b1e0bf7dacc11cac9431fc6468e99ac77fb"},
311 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:4c380469bd3f970ef677bf2bcba2b6b0b4d5c75e7a020fb863ef75084efad66f"},
312 | {file = "kiwisolver-1.4.5-cp37-cp37m-win32.whl", hash = "sha256:9408acf3270c4b6baad483865191e3e582b638b1654a007c62e3efe96f09a9a3"},
313 | {file = "kiwisolver-1.4.5-cp37-cp37m-win_amd64.whl", hash = "sha256:5b94529f9b2591b7af5f3e0e730a4e0a41ea174af35a4fd067775f9bdfeee01a"},
314 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:11c7de8f692fc99816e8ac50d1d1aef4f75126eefc33ac79aac02c099fd3db71"},
315 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:53abb58632235cd154176ced1ae8f0d29a6657aa1aa9decf50b899b755bc2b93"},
316 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:88b9f257ca61b838b6f8094a62418421f87ac2a1069f7e896c36a7d86b5d4c29"},
317 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3195782b26fc03aa9c6913d5bad5aeb864bdc372924c093b0f1cebad603dd712"},
318 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:fc579bf0f502e54926519451b920e875f433aceb4624a3646b3252b5caa9e0b6"},
319 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5a580c91d686376f0f7c295357595c5a026e6cbc3d77b7c36e290201e7c11ecb"},
320 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:cfe6ab8da05c01ba6fbea630377b5da2cd9bcbc6338510116b01c1bc939a2c18"},
321 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:d2e5a98f0ec99beb3c10e13b387f8db39106d53993f498b295f0c914328b1333"},
322 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a51a263952b1429e429ff236d2f5a21c5125437861baeed77f5e1cc2d2c7c6da"},
323 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:3edd2fa14e68c9be82c5b16689e8d63d89fe927e56debd6e1dbce7a26a17f81b"},
324 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:74d1b44c6cfc897df648cc9fdaa09bc3e7679926e6f96df05775d4fb3946571c"},
325 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:76d9289ed3f7501012e05abb8358bbb129149dbd173f1f57a1bf1c22d19ab7cc"},
326 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:92dea1ffe3714fa8eb6a314d2b3c773208d865a0e0d35e713ec54eea08a66250"},
327 | {file = "kiwisolver-1.4.5-cp38-cp38-win32.whl", hash = "sha256:5c90ae8c8d32e472be041e76f9d2f2dbff4d0b0be8bd4041770eddb18cf49a4e"},
328 | {file = "kiwisolver-1.4.5-cp38-cp38-win_amd64.whl", hash = "sha256:c7940c1dc63eb37a67721b10d703247552416f719c4188c54e04334321351ced"},
329 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:9407b6a5f0d675e8a827ad8742e1d6b49d9c1a1da5d952a67d50ef5f4170b18d"},
330 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:15568384086b6df3c65353820a4473575dbad192e35010f622c6ce3eebd57af9"},
331 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:0dc9db8e79f0036e8173c466d21ef18e1befc02de8bf8aa8dc0813a6dc8a7046"},
332 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:cdc8a402aaee9a798b50d8b827d7ecf75edc5fb35ea0f91f213ff927c15f4ff0"},
333 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6c3bd3cde54cafb87d74d8db50b909705c62b17c2099b8f2e25b461882e544ff"},
334 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:955e8513d07a283056b1396e9a57ceddbd272d9252c14f154d450d227606eb54"},
335 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:346f5343b9e3f00b8db8ba359350eb124b98c99efd0b408728ac6ebf38173958"},
336 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b9098e0049e88c6a24ff64545cdfc50807818ba6c1b739cae221bbbcbc58aad3"},
337 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:00bd361b903dc4bbf4eb165f24d1acbee754fce22ded24c3d56eec268658a5cf"},
338 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:7b8b454bac16428b22560d0a1cf0a09875339cab69df61d7805bf48919415901"},
339 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:f1d072c2eb0ad60d4c183f3fb44ac6f73fb7a8f16a2694a91f988275cbf352f9"},
340 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:31a82d498054cac9f6d0b53d02bb85811185bcb477d4b60144f915f3b3126342"},
341 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6512cb89e334e4700febbffaaa52761b65b4f5a3cf33f960213d5656cea36a77"},
342 | {file = "kiwisolver-1.4.5-cp39-cp39-win32.whl", hash = "sha256:9db8ea4c388fdb0f780fe91346fd438657ea602d58348753d9fb265ce1bca67f"},
343 | {file = "kiwisolver-1.4.5-cp39-cp39-win_amd64.whl", hash = "sha256:59415f46a37f7f2efeec758353dd2eae1b07640d8ca0f0c42548ec4125492635"},
344 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:5c7b3b3a728dc6faf3fc372ef24f21d1e3cee2ac3e9596691d746e5a536de920"},
345 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:620ced262a86244e2be10a676b646f29c34537d0d9cc8eb26c08f53d98013390"},
346 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:378a214a1e3bbf5ac4a8708304318b4f890da88c9e6a07699c4ae7174c09a68d"},
347 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aaf7be1207676ac608a50cd08f102f6742dbfc70e8d60c4db1c6897f62f71523"},
348 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:ba55dce0a9b8ff59495ddd050a0225d58bd0983d09f87cfe2b6aec4f2c1234e4"},
349 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:fd32ea360bcbb92d28933fc05ed09bffcb1704ba3fc7942e81db0fd4f81a7892"},
350 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:5e7139af55d1688f8b960ee9ad5adafc4ac17c1c473fe07133ac092310d76544"},
351 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:dced8146011d2bc2e883f9bd68618b8247387f4bbec46d7392b3c3b032640126"},
352 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c9bf3325c47b11b2e51bca0824ea217c7cd84491d8ac4eefd1e409705ef092bd"},
353 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5794cf59533bc3f1b1c821f7206a3617999db9fbefc345360aafe2e067514929"},
354 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e368f200bbc2e4f905b8e71eb38b3c04333bddaa6a2464a6355487b02bb7fb09"},
355 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:e5d706eba36b4c4d5bc6c6377bb6568098765e990cfc21ee16d13963fab7b3e7"},
356 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:85267bd1aa8880a9c88a8cb71e18d3d64d2751a790e6ca6c27b8ccc724bcd5ad"},
357 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:210ef2c3a1f03272649aff1ef992df2e724748918c4bc2d5a90352849eb40bea"},
358 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:11d011a7574eb3b82bcc9c1a1d35c1d7075677fdd15de527d91b46bd35e935ee"},
359 | {file = "kiwisolver-1.4.5.tar.gz", hash = "sha256:e57e563a57fb22a142da34f38acc2fc1a5c864bc29ca1517a88abc963e60d6ec"},
360 | ]
361 |
362 | [[package]]
363 | name = "matplotlib"
364 | version = "3.8.2"
365 | description = "Python plotting package"
366 | optional = false
367 | python-versions = ">=3.9"
368 | files = [
369 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_10_12_x86_64.whl", hash = "sha256:09796f89fb71a0c0e1e2f4bdaf63fb2cefc84446bb963ecdeb40dfee7dfa98c7"},
370 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6f9c6976748a25e8b9be51ea028df49b8e561eed7809146da7a47dbecebab367"},
371 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b78e4f2cedf303869b782071b55fdde5987fda3038e9d09e58c91cc261b5ad18"},
372 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e208f46cf6576a7624195aa047cb344a7f802e113bb1a06cfd4bee431de5e31"},
373 | {file = "matplotlib-3.8.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:46a569130ff53798ea5f50afce7406e91fdc471ca1e0e26ba976a8c734c9427a"},
374 | {file = "matplotlib-3.8.2-cp310-cp310-win_amd64.whl", hash = "sha256:830f00640c965c5b7f6bc32f0d4ce0c36dfe0379f7dd65b07a00c801713ec40a"},
375 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_10_12_x86_64.whl", hash = "sha256:d86593ccf546223eb75a39b44c32788e6f6440d13cfc4750c1c15d0fcb850b63"},
376 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:9a5430836811b7652991939012f43d2808a2db9b64ee240387e8c43e2e5578c8"},
377 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b9576723858a78751d5aacd2497b8aef29ffea6d1c95981505877f7ac28215c6"},
378 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5ba9cbd8ac6cf422f3102622b20f8552d601bf8837e49a3afed188d560152788"},
379 | {file = "matplotlib-3.8.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:03f9d160a29e0b65c0790bb07f4f45d6a181b1ac33eb1bb0dd225986450148f0"},
380 | {file = "matplotlib-3.8.2-cp311-cp311-win_amd64.whl", hash = "sha256:3773002da767f0a9323ba1a9b9b5d00d6257dbd2a93107233167cfb581f64717"},
381 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_10_12_x86_64.whl", hash = "sha256:4c318c1e95e2f5926fba326f68177dee364aa791d6df022ceb91b8221bd0a627"},
382 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:091275d18d942cf1ee9609c830a1bc36610607d8223b1b981c37d5c9fc3e46a4"},
383 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b0f3b8ea0e99e233a4bcc44590f01604840d833c280ebb8fe5554fd3e6cfe8d"},
384 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d7b1704a530395aaf73912be741c04d181f82ca78084fbd80bc737be04848331"},
385 | {file = "matplotlib-3.8.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:533b0e3b0c6768eef8cbe4b583731ce25a91ab54a22f830db2b031e83cca9213"},
386 | {file = "matplotlib-3.8.2-cp312-cp312-win_amd64.whl", hash = "sha256:0f4fc5d72b75e2c18e55eb32292659cf731d9d5b312a6eb036506304f4675630"},
387 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_10_12_x86_64.whl", hash = "sha256:deaed9ad4da0b1aea77fe0aa0cebb9ef611c70b3177be936a95e5d01fa05094f"},
388 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:172f4d0fbac3383d39164c6caafd3255ce6fa58f08fc392513a0b1d3b89c4f89"},
389 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c7d36c2209d9136cd8e02fab1c0ddc185ce79bc914c45054a9f514e44c787917"},
390 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5864bdd7da445e4e5e011b199bb67168cdad10b501750367c496420f2ad00843"},
391 | {file = "matplotlib-3.8.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:ef8345b48e95cee45ff25192ed1f4857273117917a4dcd48e3905619bcd9c9b8"},
392 | {file = "matplotlib-3.8.2-cp39-cp39-win_amd64.whl", hash = "sha256:7c48d9e221b637c017232e3760ed30b4e8d5dfd081daf327e829bf2a72c731b4"},
393 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-macosx_10_12_x86_64.whl", hash = "sha256:aa11b3c6928a1e496c1a79917d51d4cd5d04f8a2e75f21df4949eeefdf697f4b"},
394 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1095fecf99eeb7384dabad4bf44b965f929a5f6079654b681193edf7169ec20"},
395 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:bddfb1db89bfaa855912261c805bd0e10218923cc262b9159a49c29a7a1c1afa"},
396 | {file = "matplotlib-3.8.2.tar.gz", hash = "sha256:01a978b871b881ee76017152f1f1a0cbf6bd5f7b8ff8c96df0df1bd57d8755a1"},
397 | ]
398 |
399 | [package.dependencies]
400 | contourpy = ">=1.0.1"
401 | cycler = ">=0.10"
402 | fonttools = ">=4.22.0"
403 | importlib-resources = {version = ">=3.2.0", markers = "python_version < \"3.10\""}
404 | kiwisolver = ">=1.3.1"
405 | numpy = ">=1.21,<2"
406 | packaging = ">=20.0"
407 | pillow = ">=8"
408 | pyparsing = ">=2.3.1"
409 | python-dateutil = ">=2.7"
410 |
411 | [[package]]
412 | name = "numpy"
413 | version = "1.26.2"
414 | description = "Fundamental package for array computing in Python"
415 | optional = false
416 | python-versions = ">=3.9"
417 | files = [
418 | {file = "numpy-1.26.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:3703fc9258a4a122d17043e57b35e5ef1c5a5837c3db8be396c82e04c1cf9b0f"},
419 | {file = "numpy-1.26.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:cc392fdcbd21d4be6ae1bb4475a03ce3b025cd49a9be5345d76d7585aea69440"},
420 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:36340109af8da8805d8851ef1d74761b3b88e81a9bd80b290bbfed61bd2b4f75"},
421 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:bcc008217145b3d77abd3e4d5ef586e3bdfba8fe17940769f8aa09b99e856c00"},
422 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:3ced40d4e9e18242f70dd02d739e44698df3dcb010d31f495ff00a31ef6014fe"},
423 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b272d4cecc32c9e19911891446b72e986157e6a1809b7b56518b4f3755267523"},
424 | {file = "numpy-1.26.2-cp310-cp310-win32.whl", hash = "sha256:22f8fc02fdbc829e7a8c578dd8d2e15a9074b630d4da29cda483337e300e3ee9"},
425 | {file = "numpy-1.26.2-cp310-cp310-win_amd64.whl", hash = "sha256:26c9d33f8e8b846d5a65dd068c14e04018d05533b348d9eaeef6c1bd787f9919"},
426 | {file = "numpy-1.26.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:b96e7b9c624ef3ae2ae0e04fa9b460f6b9f17ad8b4bec6d7756510f1f6c0c841"},
427 | {file = "numpy-1.26.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:aa18428111fb9a591d7a9cc1b48150097ba6a7e8299fb56bdf574df650e7d1f1"},
428 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:06fa1ed84aa60ea6ef9f91ba57b5ed963c3729534e6e54055fc151fad0423f0a"},
429 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:96ca5482c3dbdd051bcd1fce8034603d6ebfc125a7bd59f55b40d8f5d246832b"},
430 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:854ab91a2906ef29dc3925a064fcd365c7b4da743f84b123002f6139bcb3f8a7"},
431 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:f43740ab089277d403aa07567be138fc2a89d4d9892d113b76153e0e412409f8"},
432 | {file = "numpy-1.26.2-cp311-cp311-win32.whl", hash = "sha256:a2bbc29fcb1771cd7b7425f98b05307776a6baf43035d3b80c4b0f29e9545186"},
433 | {file = "numpy-1.26.2-cp311-cp311-win_amd64.whl", hash = "sha256:2b3fca8a5b00184828d12b073af4d0fc5fdd94b1632c2477526f6bd7842d700d"},
434 | {file = "numpy-1.26.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:a4cd6ed4a339c21f1d1b0fdf13426cb3b284555c27ac2f156dfdaaa7e16bfab0"},
435 | {file = "numpy-1.26.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5d5244aabd6ed7f312268b9247be47343a654ebea52a60f002dc70c769048e75"},
436 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a3cdb4d9c70e6b8c0814239ead47da00934666f668426fc6e94cce869e13fd7"},
437 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:aa317b2325f7aa0a9471663e6093c210cb2ae9c0ad824732b307d2c51983d5b6"},
438 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:174a8880739c16c925799c018f3f55b8130c1f7c8e75ab0a6fa9d41cab092fd6"},
439 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f79b231bf5c16b1f39c7f4875e1ded36abee1591e98742b05d8a0fb55d8a3eec"},
440 | {file = "numpy-1.26.2-cp312-cp312-win32.whl", hash = "sha256:4a06263321dfd3598cacb252f51e521a8cb4b6df471bb12a7ee5cbab20ea9167"},
441 | {file = "numpy-1.26.2-cp312-cp312-win_amd64.whl", hash = "sha256:b04f5dc6b3efdaab541f7857351aac359e6ae3c126e2edb376929bd3b7f92d7e"},
442 | {file = "numpy-1.26.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:4eb8df4bf8d3d90d091e0146f6c28492b0be84da3e409ebef54349f71ed271ef"},
443 | {file = "numpy-1.26.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:1a13860fdcd95de7cf58bd6f8bc5a5ef81c0b0625eb2c9a783948847abbef2c2"},
444 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:64308ebc366a8ed63fd0bf426b6a9468060962f1a4339ab1074c228fa6ade8e3"},
445 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:baf8aab04a2c0e859da118f0b38617e5ee65d75b83795055fb66c0d5e9e9b818"},
446 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d73a3abcac238250091b11caef9ad12413dab01669511779bc9b29261dd50210"},
447 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:b361d369fc7e5e1714cf827b731ca32bff8d411212fccd29ad98ad622449cc36"},
448 | {file = "numpy-1.26.2-cp39-cp39-win32.whl", hash = "sha256:bd3f0091e845164a20bd5a326860c840fe2af79fa12e0469a12768a3ec578d80"},
449 | {file = "numpy-1.26.2-cp39-cp39-win_amd64.whl", hash = "sha256:2beef57fb031dcc0dc8fa4fe297a742027b954949cabb52a2a376c144e5e6060"},
450 | {file = "numpy-1.26.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:1cc3d5029a30fb5f06704ad6b23b35e11309491c999838c31f124fee32107c79"},
451 | {file = "numpy-1.26.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:94cc3c222bb9fb5a12e334d0479b97bb2df446fbe622b470928f5284ffca3f8d"},
452 | {file = "numpy-1.26.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:fe6b44fb8fcdf7eda4ef4461b97b3f63c466b27ab151bec2366db8b197387841"},
453 | {file = "numpy-1.26.2.tar.gz", hash = "sha256:f65738447676ab5777f11e6bbbdb8ce11b785e105f690bc45966574816b6d3ea"},
454 | ]
455 |
456 | [[package]]
457 | name = "packaging"
458 | version = "23.2"
459 | description = "Core utilities for Python packages"
460 | optional = false
461 | python-versions = ">=3.7"
462 | files = [
463 | {file = "packaging-23.2-py3-none-any.whl", hash = "sha256:8c491190033a9af7e1d931d0b5dacc2ef47509b34dd0de67ed209b5203fc88c7"},
464 | {file = "packaging-23.2.tar.gz", hash = "sha256:048fb0e9405036518eaaf48a55953c750c11e1a1b68e0dd1a9d62ed0c092cfc5"},
465 | ]
466 |
467 | [[package]]
468 | name = "pillow"
469 | version = "10.1.0"
470 | description = "Python Imaging Library (Fork)"
471 | optional = false
472 | python-versions = ">=3.8"
473 | files = [
474 | {file = "Pillow-10.1.0-cp310-cp310-macosx_10_10_x86_64.whl", hash = "sha256:1ab05f3db77e98f93964697c8efc49c7954b08dd61cff526b7f2531a22410106"},
475 | {file = "Pillow-10.1.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6932a7652464746fcb484f7fc3618e6503d2066d853f68a4bd97193a3996e273"},
476 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a5f63b5a68daedc54c7c3464508d8c12075e56dcfbd42f8c1bf40169061ae666"},
477 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c0949b55eb607898e28eaccb525ab104b2d86542a85c74baf3a6dc24002edec2"},
478 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:ae88931f93214777c7a3aa0a8f92a683f83ecde27f65a45f95f22d289a69e593"},
479 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:b0eb01ca85b2361b09480784a7931fc648ed8b7836f01fb9241141b968feb1db"},
480 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:d27b5997bdd2eb9fb199982bb7eb6164db0426904020dc38c10203187ae2ff2f"},
481 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:7df5608bc38bd37ef585ae9c38c9cd46d7c81498f086915b0f97255ea60c2818"},
482 | {file = "Pillow-10.1.0-cp310-cp310-win_amd64.whl", hash = "sha256:41f67248d92a5e0a2076d3517d8d4b1e41a97e2df10eb8f93106c89107f38b57"},
483 | {file = "Pillow-10.1.0-cp311-cp311-macosx_10_10_x86_64.whl", hash = "sha256:1fb29c07478e6c06a46b867e43b0bcdb241b44cc52be9bc25ce5944eed4648e7"},
484 | {file = "Pillow-10.1.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:2cdc65a46e74514ce742c2013cd4a2d12e8553e3a2563c64879f7c7e4d28bce7"},
485 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50d08cd0a2ecd2a8657bd3d82c71efd5a58edb04d9308185d66c3a5a5bed9610"},
486 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:062a1610e3bc258bff2328ec43f34244fcec972ee0717200cb1425214fe5b839"},
487 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:61f1a9d247317fa08a308daaa8ee7b3f760ab1809ca2da14ecc88ae4257d6172"},
488 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:a646e48de237d860c36e0db37ecaecaa3619e6f3e9d5319e527ccbc8151df061"},
489 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:47e5bf85b80abc03be7455c95b6d6e4896a62f6541c1f2ce77a7d2bb832af262"},
490 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:a92386125e9ee90381c3369f57a2a50fa9e6aa8b1cf1d9c4b200d41a7dd8e992"},
491 | {file = "Pillow-10.1.0-cp311-cp311-win_amd64.whl", hash = "sha256:0f7c276c05a9767e877a0b4c5050c8bee6a6d960d7f0c11ebda6b99746068c2a"},
492 | {file = "Pillow-10.1.0-cp312-cp312-macosx_10_10_x86_64.whl", hash = "sha256:a89b8312d51715b510a4fe9fc13686283f376cfd5abca8cd1c65e4c76e21081b"},
493 | {file = "Pillow-10.1.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:00f438bb841382b15d7deb9a05cc946ee0f2c352653c7aa659e75e592f6fa17d"},
494 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3d929a19f5469b3f4df33a3df2983db070ebb2088a1e145e18facbc28cae5b27"},
495 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9a92109192b360634a4489c0c756364c0c3a2992906752165ecb50544c251312"},
496 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:0248f86b3ea061e67817c47ecbe82c23f9dd5d5226200eb9090b3873d3ca32de"},
497 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:9882a7451c680c12f232a422730f986a1fcd808da0fd428f08b671237237d651"},
498 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:1c3ac5423c8c1da5928aa12c6e258921956757d976405e9467c5f39d1d577a4b"},
499 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:806abdd8249ba3953c33742506fe414880bad78ac25cc9a9b1c6ae97bedd573f"},
500 | {file = "Pillow-10.1.0-cp312-cp312-win_amd64.whl", hash = "sha256:eaed6977fa73408b7b8a24e8b14e59e1668cfc0f4c40193ea7ced8e210adf996"},
501 | {file = "Pillow-10.1.0-cp38-cp38-macosx_10_10_x86_64.whl", hash = "sha256:fe1e26e1ffc38be097f0ba1d0d07fcade2bcfd1d023cda5b29935ae8052bd793"},
502 | {file = "Pillow-10.1.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:7a7e3daa202beb61821c06d2517428e8e7c1aab08943e92ec9e5755c2fc9ba5e"},
503 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:24fadc71218ad2b8ffe437b54876c9382b4a29e030a05a9879f615091f42ffc2"},
504 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fa1d323703cfdac2036af05191b969b910d8f115cf53093125e4058f62012c9a"},
505 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:912e3812a1dbbc834da2b32299b124b5ddcb664ed354916fd1ed6f193f0e2d01"},
506 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:7dbaa3c7de82ef37e7708521be41db5565004258ca76945ad74a8e998c30af8d"},
507 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:9d7bc666bd8c5a4225e7ac71f2f9d12466ec555e89092728ea0f5c0c2422ea80"},
508 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:baada14941c83079bf84c037e2d8b7506ce201e92e3d2fa0d1303507a8538212"},
509 | {file = "Pillow-10.1.0-cp38-cp38-win_amd64.whl", hash = "sha256:2ef6721c97894a7aa77723740a09547197533146fba8355e86d6d9a4a1056b14"},
510 | {file = "Pillow-10.1.0-cp39-cp39-macosx_10_10_x86_64.whl", hash = "sha256:0a026c188be3b443916179f5d04548092e253beb0c3e2ee0a4e2cdad72f66099"},
511 | {file = "Pillow-10.1.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:04f6f6149f266a100374ca3cc368b67fb27c4af9f1cc8cb6306d849dcdf12616"},
512 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bb40c011447712d2e19cc261c82655f75f32cb724788df315ed992a4d65696bb"},
513 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1a8413794b4ad9719346cd9306118450b7b00d9a15846451549314a58ac42219"},
514 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:c9aeea7b63edb7884b031a35305629a7593272b54f429a9869a4f63a1bf04c34"},
515 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:b4005fee46ed9be0b8fb42be0c20e79411533d1fd58edabebc0dd24626882cfd"},
516 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0152565c6aa6ebbfb1e5d8624140a440f2b99bf7afaafbdbf6430426497f28"},
517 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:d921bc90b1defa55c9917ca6b6b71430e4286fc9e44c55ead78ca1a9f9eba5f2"},
518 | {file = "Pillow-10.1.0-cp39-cp39-win_amd64.whl", hash = "sha256:cfe96560c6ce2f4c07d6647af2d0f3c54cc33289894ebd88cfbb3bcd5391e256"},
519 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-macosx_10_10_x86_64.whl", hash = "sha256:937bdc5a7f5343d1c97dc98149a0be7eb9704e937fe3dc7140e229ae4fc572a7"},
520 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b1c25762197144e211efb5f4e8ad656f36c8d214d390585d1d21281f46d556ba"},
521 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:afc8eef765d948543a4775f00b7b8c079b3321d6b675dde0d02afa2ee23000b4"},
522 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:883f216eac8712b83a63f41b76ddfb7b2afab1b74abbb413c5df6680f071a6b9"},
523 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-macosx_10_10_x86_64.whl", hash = "sha256:b920e4d028f6442bea9a75b7491c063f0b9a3972520731ed26c83e254302eb1e"},
524 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1c41d960babf951e01a49c9746f92c5a7e0d939d1652d7ba30f6b3090f27e412"},
525 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:1fafabe50a6977ac70dfe829b2d5735fd54e190ab55259ec8aea4aaea412fa0b"},
526 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:3b834f4b16173e5b92ab6566f0473bfb09f939ba14b23b8da1f54fa63e4b623f"},
527 | {file = "Pillow-10.1.0.tar.gz", hash = "sha256:e6bf8de6c36ed96c86ea3b6e1d5273c53f46ef518a062464cd7ef5dd2cf92e38"},
528 | ]
529 |
530 | [package.extras]
531 | docs = ["furo", "olefile", "sphinx (>=2.4)", "sphinx-copybutton", "sphinx-inline-tabs", "sphinx-removed-in", "sphinxext-opengraph"]
532 | tests = ["check-manifest", "coverage", "defusedxml", "markdown2", "olefile", "packaging", "pyroma", "pytest", "pytest-cov", "pytest-timeout"]
533 |
534 | [[package]]
535 | name = "pluggy"
536 | version = "1.3.0"
537 | description = "plugin and hook calling mechanisms for python"
538 | optional = false
539 | python-versions = ">=3.8"
540 | files = [
541 | {file = "pluggy-1.3.0-py3-none-any.whl", hash = "sha256:d89c696a773f8bd377d18e5ecda92b7a3793cbe66c87060a6fb58c7b6e1061f7"},
542 | {file = "pluggy-1.3.0.tar.gz", hash = "sha256:cf61ae8f126ac6f7c451172cf30e3e43d3ca77615509771b3a984a0730651e12"},
543 | ]
544 |
545 | [package.extras]
546 | dev = ["pre-commit", "tox"]
547 | testing = ["pytest", "pytest-benchmark"]
548 |
549 | [[package]]
550 | name = "py"
551 | version = "1.11.0"
552 | description = "library with cross-python path, ini-parsing, io, code, log facilities"
553 | optional = false
554 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*"
555 | files = [
556 | {file = "py-1.11.0-py2.py3-none-any.whl", hash = "sha256:607c53218732647dff4acdfcd50cb62615cedf612e72d1724fb1a0cc6405b378"},
557 | {file = "py-1.11.0.tar.gz", hash = "sha256:51c75c4126074b472f746a24399ad32f6053d1b34b68d2fa41e558e6f4a98719"},
558 | ]
559 |
560 | [[package]]
561 | name = "pyparsing"
562 | version = "3.1.1"
563 | description = "pyparsing module - Classes and methods to define and execute parsing grammars"
564 | optional = false
565 | python-versions = ">=3.6.8"
566 | files = [
567 | {file = "pyparsing-3.1.1-py3-none-any.whl", hash = "sha256:32c7c0b711493c72ff18a981d24f28aaf9c1fb7ed5e9667c9e84e3db623bdbfb"},
568 | {file = "pyparsing-3.1.1.tar.gz", hash = "sha256:ede28a1a32462f5a9705e07aea48001a08f7cf81a021585011deba701581a0db"},
569 | ]
570 |
571 | [package.extras]
572 | diagrams = ["jinja2", "railroad-diagrams"]
573 |
574 | [[package]]
575 | name = "pytest"
576 | version = "6.2.5"
577 | description = "pytest: simple powerful testing with Python"
578 | optional = false
579 | python-versions = ">=3.6"
580 | files = [
581 | {file = "pytest-6.2.5-py3-none-any.whl", hash = "sha256:7310f8d27bc79ced999e760ca304d69f6ba6c6649c0b60fb0e04a4a77cacc134"},
582 | {file = "pytest-6.2.5.tar.gz", hash = "sha256:131b36680866a76e6781d13f101efb86cf674ebb9762eb70d3082b6f29889e89"},
583 | ]
584 |
585 | [package.dependencies]
586 | atomicwrites = {version = ">=1.0", markers = "sys_platform == \"win32\""}
587 | attrs = ">=19.2.0"
588 | colorama = {version = "*", markers = "sys_platform == \"win32\""}
589 | iniconfig = "*"
590 | packaging = "*"
591 | pluggy = ">=0.12,<2.0"
592 | py = ">=1.8.2"
593 | toml = "*"
594 |
595 | [package.extras]
596 | testing = ["argcomplete", "hypothesis (>=3.56)", "mock", "nose", "requests", "xmlschema"]
597 |
598 | [[package]]
599 | name = "python-dateutil"
600 | version = "2.8.2"
601 | description = "Extensions to the standard Python datetime module"
602 | optional = false
603 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,>=2.7"
604 | files = [
605 | {file = "python-dateutil-2.8.2.tar.gz", hash = "sha256:0123cacc1627ae19ddf3c27a5de5bd67ee4586fbdd6440d9748f8abb483d3e86"},
606 | {file = "python_dateutil-2.8.2-py2.py3-none-any.whl", hash = "sha256:961d03dc3453ebbc59dbdea9e4e11c5651520a876d0f4db161e8674aae935da9"},
607 | ]
608 |
609 | [package.dependencies]
610 | six = ">=1.5"
611 |
612 | [[package]]
613 | name = "six"
614 | version = "1.16.0"
615 | description = "Python 2 and 3 compatibility utilities"
616 | optional = false
617 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*"
618 | files = [
619 | {file = "six-1.16.0-py2.py3-none-any.whl", hash = "sha256:8abb2f1d86890a2dfb989f9a77cfcfd3e47c2a354b01111771326f8aa26e0254"},
620 | {file = "six-1.16.0.tar.gz", hash = "sha256:1e61c37477a1626458e36f7b1d82aa5c9b094fa4802892072e49de9c60c4c926"},
621 | ]
622 |
623 | [[package]]
624 | name = "toml"
625 | version = "0.10.2"
626 | description = "Python Library for Tom's Obvious, Minimal Language"
627 | optional = false
628 | python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
629 | files = [
630 | {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
631 | {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
632 | ]
633 |
634 | [[package]]
635 | name = "tqdm"
636 | version = "4.66.1"
637 | description = "Fast, Extensible Progress Meter"
638 | optional = false
639 | python-versions = ">=3.7"
640 | files = [
641 | {file = "tqdm-4.66.1-py3-none-any.whl", hash = "sha256:d302b3c5b53d47bce91fea46679d9c3c6508cf6332229aa1e7d8653723793386"},
642 | {file = "tqdm-4.66.1.tar.gz", hash = "sha256:d88e651f9db8d8551a62556d3cff9e3034274ca5d66e93197cf2490e2dcb69c7"},
643 | ]
644 |
645 | [package.dependencies]
646 | colorama = {version = "*", markers = "platform_system == \"Windows\""}
647 |
648 | [package.extras]
649 | dev = ["pytest (>=6)", "pytest-cov", "pytest-timeout", "pytest-xdist"]
650 | notebook = ["ipywidgets (>=6)"]
651 | slack = ["slack-sdk"]
652 | telegram = ["requests"]
653 |
654 | [[package]]
655 | name = "zipp"
656 | version = "3.17.0"
657 | description = "Backport of pathlib-compatible object wrapper for zip files"
658 | optional = false
659 | python-versions = ">=3.8"
660 | files = [
661 | {file = "zipp-3.17.0-py3-none-any.whl", hash = "sha256:0e923e726174922dce09c53c59ad483ff7bbb8e572e00c7f7c46b88556409f31"},
662 | {file = "zipp-3.17.0.tar.gz", hash = "sha256:84e64a1c28cf7e91ed2078bb8cc8c259cb19b76942096c8d7b84947690cabaf0"},
663 | ]
664 |
665 | [package.extras]
666 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
667 | testing = ["big-O", "jaraco.functools", "jaraco.itertools", "more-itertools", "pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-ignore-flaky", "pytest-mypy (>=0.9.1)", "pytest-ruff"]
668 |
669 | [metadata]
670 | lock-version = "2.0"
671 | python-versions = "^3.9"
672 | content-hash = "5e3a8b71c1d10601d5e0c2e8e72d1ea2cd79f51cbe48d1f7512dd3fc591030c2"
673 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.poetry]
2 | name = "complexcgr"
3 | version = "0.8.0"
4 | description = "complex Chaos Game Representation for DNA"
5 | authors = ["Jorge Avila "]
6 | maintainers = ["Jorge Avila "]
7 | repository = "https://github.com/AlgoLab/complexCGR"
8 | license = "MIT"
9 |
10 | [tool.poetry.dependencies]
11 | python = "^3.9"
12 | tqdm = "^4.61.2"
13 | matplotlib = "^3.4.2"
14 | Pillow = "^10.0"
15 | numpy = "^1.22.3"
16 | biopython = "^1.79"
17 |
18 | [tool.poetry.dev-dependencies]
19 | pytest = "^6.2.4"
20 |
21 | [build-system]
22 | requires = ["poetry-core>=1.0.0"]
23 | build-backend = "poetry.core.masonry.api"
24 |
--------------------------------------------------------------------------------
/tests/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/tests/__init__.py
--------------------------------------------------------------------------------
/tests/test_cgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import CGR
2 |
3 | def test_cgr():
4 | "Encoding for each nucleotide"
5 | cgr = CGR()
6 |
7 | cgr.encode("A")
8 | assert cgr.cgr_coords.N == 1
9 | assert cgr.cgr_coords.x == 0.5
10 | assert cgr.cgr_coords.y == 0.5
11 |
12 | cgr.encode("C")
13 | assert cgr.cgr_coords.N == 1
14 | assert cgr.cgr_coords.x == -0.5
15 | assert cgr.cgr_coords.y == 0.5
16 |
17 | cgr.encode("G")
18 | assert cgr.cgr_coords.N == 1
19 | assert cgr.cgr_coords.x == -0.5
20 | assert cgr.cgr_coords.y == -0.5
21 |
22 | cgr.encode("T")
23 | assert cgr.cgr_coords.N == 1
24 | assert cgr.cgr_coords.x == 0.5
25 | assert cgr.cgr_coords.y == -0.5
26 |
27 | def test_cgr_decode():
28 | "Decoding each nucleotide"
29 | cgr = CGR()
30 |
31 | assert cgr.decode(N=1,x=0.5,y=0.5) == "A"
32 | assert cgr.decode(N=1,x=-0.5,y=0.5) == "C"
33 | assert cgr.decode(N=1,x=-0.5,y=-0.5) == "G"
34 | assert cgr.decode(N=1,x=0.5,y=-0.5) == "T"
--------------------------------------------------------------------------------
/tests/test_complexcgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import (
2 | __version__,
3 | ComplexCGR,
4 | )
5 |
6 | def test_version():
7 | assert __version__ == '0.8.0'
8 |
9 | def test_complexCGR():
10 | ccgr = ComplexCGR()
11 |
12 | # nucleotide A
13 | encode = ccgr.encode("A")
14 | assert encode.k == 0
15 | assert encode.N == 1
16 |
17 |
18 | # nucleotide C
19 | encode = ccgr.encode("C")
20 | assert encode.k == 1
21 | assert encode.N == 1
22 |
23 |
24 | # nucleotide G
25 | encode = ccgr.encode("G")
26 | assert encode.k == 2
27 | assert encode.N == 1
28 |
29 |
30 | # nucleotide T
31 | encode = ccgr.encode("T")
32 | assert encode.k == 3
33 | assert encode.N == 1
34 |
35 | def test_complexCGR_decode():
36 | ccgr = ComplexCGR()
37 |
38 | # nucleotide A
39 | decode = ccgr.decode(k=0,N=1)
40 | assert decode == "A"
41 |
42 |
43 | # nucleotide C
44 | decode = ccgr.decode(k=1,N=1)
45 | assert decode == "C"
46 |
47 |
48 | # nucleotide G
49 | decode = ccgr.decode(k=2,N=1)
50 | assert decode == "G"
51 |
52 |
53 | # nucleotide T
54 | decode = ccgr.decode(k=3,N=1)
55 | assert decode == "T"
56 |
57 | def test_encode_decode_compatibility():
58 | ccgr = ComplexCGR()
59 |
60 | seq = "ACGT"
61 | encode = ccgr.encode(seq)
62 | decode = ccgr.decode(encode.k, encode.N)
63 | assert seq == decode
--------------------------------------------------------------------------------
/tests/test_complexfcgr.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/tests/test_complexfcgr.py
--------------------------------------------------------------------------------
/tests/test_complexfcgr_savefig.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | import random
3 | from complexcgr import ComplexFCGR
4 |
5 | BASEPATH = Path().parent.resolve()
6 | def test_savefig():
7 | fcgr = ComplexFCGR(k=8)
8 | seq = "".join(random.choice("ACG") for _ in range(300_000))
9 | fig = fcgr(seq)
10 | fcgr.save(fig, path=BASEPATH.joinpath("img/ACG-complexCGR.png"))
11 |
--------------------------------------------------------------------------------
/tests/test_fcgr.py:
--------------------------------------------------------------------------------
1 | import random
2 | from complexcgr import FCGR
3 |
4 | def test_fcgr():
5 | "frecuency matrix CGR"
6 | fcgr = FCGR(k=1)
7 | fcgr.count_kmers("ACGT")
8 |
9 | assert fcgr.freq_kmer.get("A") == 1
10 | assert fcgr.freq_kmer.get("C") == 1
11 | assert fcgr.freq_kmer.get("G") == 1
12 | assert fcgr.freq_kmer.get("T") == 1
13 |
14 | def test_savefig():
15 | fcgr = FCGR(k=8)
16 | # Generate a random sequence without T's
17 | seq = "".join(random.choice("ACG") for _ in range(30_000))
18 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
19 | fcgr.save_img(chaos, path="img/ACG.jpg")
20 |
21 | def test_savefig_16bits():
22 | fcgr = FCGR(k=8, bits=16)
23 | # Generate a random sequence without T's
24 | seq = "".join(random.choice("ACG") for _ in range(30_000))
25 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
26 | fcgr.save_img(chaos, path="img/ACG_16bits.jpg")
27 |
28 |
29 |
30 |
31 |
--------------------------------------------------------------------------------
/tests/test_icgr.py:
--------------------------------------------------------------------------------
1 | from complexcgr import iCGR
2 |
3 | def test_cgr():
4 | "Encoding for each nucleotide"
5 | cgr = iCGR()
6 |
7 | cgr.encode("A")
8 | assert cgr.cgr_coords.N == 1
9 | assert cgr.cgr_coords.x == 1
10 | assert cgr.cgr_coords.y == 1
11 |
12 | cgr.encode("C")
13 | assert cgr.cgr_coords.N == 1
14 | assert cgr.cgr_coords.x == -1
15 | assert cgr.cgr_coords.y == 1
16 |
17 | cgr.encode("G")
18 | assert cgr.cgr_coords.N == 1
19 | assert cgr.cgr_coords.x == -1
20 | assert cgr.cgr_coords.y == -1
21 |
22 | cgr.encode("T")
23 | assert cgr.cgr_coords.N == 1
24 | assert cgr.cgr_coords.x == 1
25 | assert cgr.cgr_coords.y == -1
26 |
27 |
28 | def test_cgr_decode():
29 | "Decoding each nucleotide"
30 | cgr = iCGR()
31 |
32 | assert cgr.decode(N=1,x=1,y=1) == "A"
33 | assert cgr.decode(N=1,x=-1,y=1) == "C"
34 | assert cgr.decode(N=1,x=-1,y=-1) == "G"
35 | assert cgr.decode(N=1,x=1,y=-1) == "T"
36 | assert cgr.decode(N=4,x=3,y=-9) == "ACGT"
--------------------------------------------------------------------------------
 2 |
3 | # `complexcgr`
4 | This library have classes around the *Chaos Game Representation* for DNA sequence
5 |
6 | **The FCGR helps to visualize a k-mer distribution** The `FCGR` of a sequence is an image showing the distribution of the $k$-mers
7 | given a chosen $k$. The frequencies of all $k$-mers are distributed in the position of a matrix of $2^k \times 2^k$,
8 | which considers all the possible $k$-mers: $4^k$.
9 |
10 | The position that a $k$-mer uses in the matrix depends on the encoding given by the `CGR`.
11 |
12 | Some examples of [bacterial assemblies](https://zenodo.org/records/4602622) ([see reference](https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001421)) are shown below.
13 | The name of the species and the `sample_id` is in the title of each image ([see an example with the first image](https://www.ebi.ac.uk/ena/browser/view/SAMEA2658585)). These images were
14 | created using the 6-mers of each assembly and the class `FCGR` of this library.
15 |
16 | |  |
17 | |:--:|
18 | |10 different species of bacteria represented by their FCGR (6-mers)| -->
19 |
20 | ## Installation
21 | [pypi](https://pypi.org/project/complexcgr/)
22 | ___
23 | ```shell
24 | pip install complexcgr
25 | ```
26 |
27 | to update to the latest version
28 | ```shell
29 | pip install complexcgr --upgrade
30 | ```
31 |
32 | ## How to use
33 | ___
34 | ### 1. `CGR` Chaos Game Representation of DNA
35 | ```python
36 | from complexcgr import CGR
37 |
38 | # Instantiate class CGR
39 | cgr = CGR()
40 |
41 | # encode a sequence
42 | cgr.encode("ACGT")
43 | # > CGRCoords(N=4, x=0.1875, y=-0.5625)
44 |
45 | # recover a sequence from CGR coordinates
46 | cgr.decode(N=4,x=0.1875,y=-0.5625)
47 | # > "ACGT"
48 | ```
49 |
50 | ### 2. `FCGR` Frequency Matrix of Chaos Game Representation of DNA
51 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
52 | will not be considered for the calculation of the frequency matrix CGR
53 | ```python
54 | import random; random.seed(42)
55 | from complexcgr import FCGR
56 |
57 | # set the k-mer
58 | fcgr = FCGR(k=8) # (256x256) array
59 |
60 | # Generate a random sequence without T's
61 | seq = "".join(random.choice("ACG") for _ in range(300_000))
62 | chaos = fcgr(seq) # an array with the frequencies of each k-mer
63 | fcgr.plot(chaos)
64 | ```
65 | |  |
66 | |:--:|
67 | |FCGR representation for a sequence without T's|
68 |
69 |
70 | You can save the image with
71 | ```python
72 | fcgr.save_img(chaos, path="img/ACG.jpg")
73 | ```
74 | *Formats allowed are defined by PIL.*
75 |
76 | You can also generate the image in 16 (or more bits), to avoid losing information of k-mer frequencies
77 | ```python
78 | # Generate image in 16-bits (default is 8-bits)
79 | fcgr = FCGR(k=8, bits=16) # (256x256) array. When using plot() it will be rescaled to [0,65535] colors
80 | ```
81 |
82 |
83 | ```python
84 | # Generate a random sequence without T's and lots of N's
85 | seq = "".join(random.choice("ACGN") for _ in range(300_000))
86 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
87 | fcgr.plot(chaos)
88 | ```
89 |
90 |
91 | ||
92 | |:--:|
93 | |FCGR representation for a sequence without T's and lots of N's|
94 |
95 |
96 |
97 | ### 3. `iCGR` integer Chaos Game Representation of DNA
98 | ```python
99 | from complexcgr import iCGR
100 |
101 | # Instantiate class CGR
102 | icgr = iCGR()
103 |
104 | # encode a sequence
105 | icgr.encode("ACGT")
106 | # > CGRCoords(N=4, x=3, y=-9)
107 |
108 | # recover a sequence from CGR coordinates
109 | icgr.decode(N=4,x=3,y=-9)
110 | # > "ACGT"
111 | ```
112 |
113 | ### 4. `ComplexCGR` Complex Chaos Game Representation of DNA (ComplexCGR)
114 |
115 | ```python
116 | from complexcgr import ComplexCGR
117 |
118 | # Instantiate class CGR
119 | ccgr = ComplexCGR()
120 |
121 | # encode a sequence
122 | ccgr.encode("ACGT")
123 | # > CGRCoords(k=228,N=4)
124 |
125 | # recover a sequence from ComplexCGR coordinates
126 | ccgr.decode(k=228,N=4)
127 | # > "ACGT"
128 |
129 | ```
130 |
131 | ### 5. `ComplexFCGR` Frequency Matrix of Complex Chaos Game Representation of DNA
132 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
133 | will not be considered for the calculation of the frequency matrix CGR
134 | ```python
135 | import random; random.seed(42)
136 | from complexcgr import FCGR
137 |
138 | # set the k-mer desired
139 | cfcgr = ComplexFCGR(k=8) # 8-mers
140 |
141 | # Generate a random sequence without T's
142 | seq = "".join(random.choice("ACG") for _ in range(300_000))
143 | fig = cfcgr(seq)
144 |
145 | ```
146 | |  |
147 | |:--:|
148 | |ComplexFCGR representation for a sequence without T's|
149 |
150 |
151 | You can save the image with
152 | ```python
153 | cfcgr.save(fig, path="img/ACG-ComplexCGR.png")
154 | ```
155 | *Currently the plot must be saved as png*
156 |
157 | ___
158 | ## Advice for Real applications
159 |
160 | **Count k-mers** could be the bottleneck for large sequences (> 100000 bp).
161 | Note that the class `FCGR` (and `ComplexCGR`) has implemented a naive approach to count k-mers, this is intended since in practice state-of-the-art tools like KMC or Jellyfish are used to count k-mers very efficiently.
162 |
163 | We provide the class `FCGRKmc`, that receives as input the file generated by the following pipeline using [KMC3](https://github.com/refresh-bio/KMC)
164 |
165 | Make sure to have `kmc` installed. One recommended way is to create a conda environment and [install it there](https://anaconda.org/bioconda/kmc)
166 |
167 | ```bash
168 | kmer_size=6
169 | input="path/to/sequence.fa"
170 | output="path/to/count-kmers.txt"
171 |
172 | mkdir -p tmp-kmc
173 | kmc -v -k$kmer_size -m4 -sm -ci0 -cs100000 -b -t4 -fa $input $input "tmp-kmc"
174 | kmc_tools -t4 -v transform $input dump $output
175 | rm -r $input.kmc_pre $input.kmc_suf
176 | ```
177 | the output file `path/to/count-kmers.txt` can be used with `FCGRKmc`
178 |
179 | ```python
180 | from complexcgr import FCGRKmc
181 |
182 | kmer = 6
183 | fcgr = FCGRKmc(kmer)
184 |
185 | arr = fcgr("path/to/count-kmers.txt") # k-mer counts ordered in a matrix of 2^k x 2^k
186 |
187 |
188 | # to visualize the distribution of k-mers.
189 | # Frequencies are scaled between [min, max] values.
190 | # White color corresponds to the minimum value of frequency
191 | # Black color corresponds to the maximum value of frequency
192 | fcgr.plot(arr)
193 |
194 | # Save it with numpy
195 | import numpy as np
196 | np.save("path_save/fcgr.npy",arr)
197 | ```
198 |
199 | ___
200 | # Videos
201 |
202 | **CGR encoding**
203 |
204 | [](https://youtu.be/HU15ge0fkOY)
205 |
206 | **CGR encoding of all k-mers**
207 |
208 | [](https://youtu.be/oYLT11Q9n5M)
209 |
210 | **ComplexCGR encoding**
211 |
212 | [](https://youtu.be/xKyXplS5KFk)
213 |
214 | **ComplexCGR and Symmetry**
215 |
216 | [](https://www.youtube.com/YmcxVId4_4w)
217 |
218 |
219 | # Functionalities/TODO list
220 | ___
221 | > version 0.8.0:
222 | A list of available classes and functionalities are listed below:
223 |
224 |
225 | **Encoders**
226 | The encoders are functions that map a sequence $s \in \{A,C,G,T\}$ to a point in the plane.
227 | `CGR`, `iCGR`, and `ComplexCGR`.
228 |
229 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
230 | - [x] encode a sequence.
231 | - [x] recover a sequence from a CGR encoding.
232 |
233 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
234 |
235 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
236 | - [x] encode a sequence.
237 | - [x] recover a sequence from a CGR encoding.
238 |
239 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
240 | - [x] encode a sequence
241 | - [x] recover a sequence from an iCGR encoding
242 |
243 | `ComplexCGR`: encodes a DNA sequence in 2 integers $(k,N)$.
244 | - [x] encode a sequence
245 | - [x] recover a sequence from a ComplexCGR encoding
246 | - [x] plot sequence of ComplexCGR encodings
247 |
248 | **Image for distribution of k-mers**
249 |
250 | - [x] `FCGR` Frequency Matrix CGR: representation as an image for k-mer representativity, based on CGR.
251 | - [x] generates FCGR from an arbitrary n-long sequence.
252 | - [x] plot FCGR.
253 | - [x] save FCGR generated.
254 | - [x] save FCGR in different bits.
255 | - [x] `FCGRKmc` Same as `FCGR` but receives as input the file with k-mer counts generated with [KMC](https://github.com/refresh-bio/KMC)
256 | - [x] `ComplexFCGR`: Frequency ComplexCGR: representation as an image (circle) for k-mer representativity, based on ComplexCGR.
257 | - [x] generates ComplexFCGR from an arbitrary n-long sequence.
258 | - [x] plot ComplexFCGR.
259 | - [x] save ComplexFCGR generated.
260 |
261 | - [ ] `PercentileFCGR`:
262 | - [A universal DNA signature for the Tree of Life. EcoEvoRxiv](https://doi.org/10.32942/X24891)
263 | - [ ] `SpacedFCGR`: Create FCGR from spaced-mers
264 | - [Spaced seeds improve k-mer-based metagenomic classification, Bioninformatics](https://academic.oup.com/bioinformatics/article/31/22/3584/240663)
265 | - [Effects of spaced k-mers on alignment-free genotyping](https://academic.oup.com/bioinformatics/article/39/Supplement_1/i213/7210439)
266 |
267 | # Author
268 |
269 | `complexcgr` is developed by [Jorge Avila Cartes](https://github.com/jorgeavilacartes/)
270 |
271 |
272 | # Related publications
273 |
274 | - [Accurate and fast clade assignment via deep learning and frequency chaos game representation](https://doi.org/10.1093/gigascience/giac119)
275 | - [PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases](https://doi.org/10.1101/2025.03.19.644115)
--------------------------------------------------------------------------------
/README.rst:
--------------------------------------------------------------------------------
1 | # complexCGR
2 |
3 | A complex representation for DNA, that preserves some properties of the CGR from the 'Chaos Game Representation for gene structure' by Jeffrey in 1990.
4 |
5 | Each nucleotide will be assigned to one quadrant (I,II,III and IV). This distribution will be the same for the CGR and complexCGR implementations (I:A,II:C,III:G,IV:T)
--------------------------------------------------------------------------------
/complexCGR.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/complexCGR.pdf
--------------------------------------------------------------------------------
/complexcgr/__init__.py:
--------------------------------------------------------------------------------
1 | __version__ ="0.8.0"
2 |
3 | # encodings
4 | from .cgr import CGR
5 | from .icgr import iCGR
6 | from .complexcgr import ComplexCGR
7 |
8 | # from k-mers
9 | from .fcgr import FCGR
10 | from .fcgr_kmc import FCGRKmc
11 | from .complexfcgr import ComplexFCGR
--------------------------------------------------------------------------------
/complexcgr/cgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from typing import Dict, Optional
3 | from collections import namedtuple
4 |
5 | # coordinates for x+iy
6 | Coord = namedtuple("Coord", ["x","y"])
7 |
8 | # coordinates for a CGR encoding
9 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
10 |
11 | # coordinates for each nucleotide in the 2d-plane
12 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
13 |
14 | class CGR:
15 | "Chaos Game Representation for DNA"
16 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
17 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
18 | self.cgr_coords = CGRCoords(0,0,0)
19 |
20 | def nucleotide_by_coords(self,x,y):
21 | "Get nucleotide by coordinates (x,y)"
22 | # filter nucleotide by coordinates
23 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
24 |
25 | return list(filtered.keys())[0]
26 |
27 | def forward(self, nucleotide: str):
28 | "Compute next CGR coordinates"
29 | x = (self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x)/2
30 | y = (self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y)/2
31 |
32 | # update cgr_coords
33 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y)
34 |
35 | def backward(self,):
36 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
37 | # get current nucleotide based on coordinates
38 | n_x,n_y = self.coords_current_nucleotide()
39 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
40 |
41 | # update coordinates to the previous one
42 | x = 2*self.cgr_coords.x - n_x
43 | y = 2*self.cgr_coords.y - n_y
44 |
45 | # update cgr_coords
46 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
47 |
48 | return nucleotide
49 |
50 | def coords_current_nucleotide(self,):
51 | x = 1 if self.cgr_coords.x>0 else -1
52 | y = 1 if self.cgr_coords.y>0 else -1
53 | return x,y
54 |
55 | def encode(self, sequence: str):
56 | "From DNA sequence to CGR"
57 | # reset starting position to (0,0,0)
58 | self.reset_coords()
59 | for nucleotide in sequence:
60 | self.forward(nucleotide)
61 | return self.cgr_coords
62 |
63 | def reset_coords(self,):
64 | self.cgr_coords = CGRCoords(0,0,0)
65 |
66 | def decode(self, N:int, x:int, y:int)->str:
67 | "From CGR to DNA sequence"
68 | self.cgr_coords = CGRCoords(N,x,y)
69 |
70 | # decoded sequence
71 | sequence = []
72 |
73 | # Recover the entire genome
74 | while self.cgr_coords.N>0:
75 | nucleotide = self.backward()
76 | sequence.append(nucleotide)
77 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/complexcgr/complexcgr.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List
2 | from collections import namedtuple
3 |
4 | # coordinates for x+iy
5 | Coord = namedtuple("Coord", ["x","y"])
6 |
7 | # coordinates for a CGR encoding
8 | CGRCoords = namedtuple("CGRCoord", ["k","N"])
9 |
10 | class ComplexCGR:
11 | "Complex Chaos Game Representation of DNA"
12 | def __init__(self, nucleotide_order: List[str] = ["A","C","G","T"]):
13 | self.nucleotide_order = nucleotide_order
14 | self.cgr_coords = CGRCoords(0,0) # complexCGR coordinates
15 |
16 | def id(self, nucleotide):
17 | return self.nucleotide_order.index(nucleotide)
18 |
19 | def forward(self, nucleotide: str):
20 | "compute next complexCGR coordinates"
21 | k = self.id(nucleotide)*4**(self.cgr_coords.N) + self.cgr_coords.k
22 |
23 | # update cgr_coords
24 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N+1)
25 |
26 | def backward(self,):
27 | "compute last complexCGR coordinates"
28 | nucleotide = self.current_nucleotide()
29 |
30 | # compute previous k
31 | k = self.cgr_coords.k - self.id(nucleotide)*4**(self.cgr_coords.N-1)
32 |
33 | # update cgr_coords
34 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N-1)
35 |
36 | return nucleotide
37 |
38 | def encode(self, sequence: str):
39 | "From DNA to complexCGR"
40 | self.reset_coords()
41 | for nucleotide in sequence:
42 | self.forward(nucleotide)
43 | return self.cgr_coords
44 |
45 | def decode(self, k: int, N: int):
46 | "From complexCGR to DNA"
47 | self.cgr_coords = CGRCoords(k,N)
48 |
49 | # decoded sequence
50 | sequence = []
51 |
52 | # Recover the entire genome
53 | while self.cgr_coords.N>0:
54 | nucleotide = self.backward()
55 | sequence.append(nucleotide)
56 | return "".join(sequence[::-1])
57 |
58 | def current_nucleotide(self,):
59 | "Get current nucleotide based on k and N"
60 | k,N = self.cgr_coords.k, self.cgr_coords.N
61 | alpha = k/4**N
62 | if alpha <0.25:
63 | return self.nucleotide_order[0]
64 | elif alpha <0.5:
65 | return self.nucleotide_order[1]
66 | elif alpha < 0.75:
67 | return self.nucleotide_order[2]
68 | else:
69 | return self.nucleotide_order[3]
70 |
71 | def reset_coords(self,):
72 | self.cgr_coords = CGRCoords(0,0)
73 |
--------------------------------------------------------------------------------
/complexcgr/complexfcgr.py:
--------------------------------------------------------------------------------
1 | from . import ComplexCGR
2 | import matplotlib.pyplot as plt
3 | from itertools import product
4 | from collections import defaultdict
5 | from tqdm import tqdm
6 | from PIL import Image
7 | import numpy as np
8 |
9 |
10 | class ComplexFCGR(ComplexCGR):
11 | """Circular density plot based on CGR"""
12 |
13 | def __init__(self, k: int):
14 | super().__init__()
15 | self.k = k # k-mer representation
16 | self.kmers = product("ACGT", repeat=self.k) # all kmers of length k
17 | self.freq_kmer = None # dict to save representativity of each kmer
18 | self.probabilities = None # dict to save probabilities/density for each kmer
19 | self.fig = None # to save matlotlib figure and then save as image
20 |
21 | def __call__(self, sequence: str, w=1):
22 | self.count_kmers(sequence)
23 | self.kmer_probabilities(sequence)
24 | self.plot(w)
25 |
26 | def count_kmers(self, sequence: str):
27 | self.freq_kmer = defaultdict(int)
28 | # representativity of kmers
29 | len_seq = len(sequence)
30 | for j,_ in enumerate(sequence):
31 | if j+self.k <= len_seq:
32 | subseq = sequence[j:j+self.k]
33 | if "N" not in subseq:
34 | self.freq_kmer[subseq] +=1
35 |
36 | def kmer_probabilities(self, sequence: str):
37 | self.probabilities = defaultdict(float)
38 | N=len(sequence)
39 | for key, value in self.freq_kmer.items():
40 | self.probabilities[key] = float(value) / (N - self.k + 1)
41 |
42 | def plot(self, w: int = 1):
43 | "Given a FCGR, plot it in grayscale"
44 | ax = plt.subplot(111, polar=True)
45 | center, bottom, width, height = self.compute_input_plot()
46 |
47 | # scale width by 'w'
48 | width = [w*_ for _ in width]
49 | print("generating plot")
50 | ax.bar(x=center, # center of the angle
51 | width=width, # width of the angle
52 | bottom=bottom, # lowest value
53 | height=height, # highest value
54 | )
55 |
56 | ax.axes.get_xaxis().set_visible(False)
57 | ax.axes.get_yaxis().set_visible(False)
58 |
59 | return ax.figure
60 |
61 | def save(self, ccgr, path: str):
62 | "save complexFCGR as image"
63 | # get figure
64 | fig = self.plot()
65 |
66 | # transform figure to Image
67 | img = self.fig2img(fig)
68 |
69 | # save image
70 | img.save(path)
71 | plt.close(fig)
72 |
73 | @staticmethod
74 | def fig2img(fig):
75 | "Convert a Matplotlib figure to a PIL Image and return it"
76 | #https://stackoverflow.com/questions/57316491/how-to-convert-matplotlib-figure-to-pil-image-object-without-saving-image
77 | import io
78 | buf = io.BytesIO()
79 | fig.savefig(buf)
80 | buf.seek(0)
81 | img = Image.open(buf)
82 | return img
83 |
84 |
85 | def compute_input_plot(self,):
86 | "Compute input for plot CFCGR"
87 | delta = 2*np.pi/4**self.k # angle between consecutive roots
88 |
89 | center = []
90 | width = []
91 | height = []
92 | bottom = []
93 |
94 | for kmer in tqdm(self.probabilities):
95 | h = self.probabilities.get(kmer,0.0) # height (density) of the bar in the circle
96 | theta = 2*self.encode(kmer).k*np.pi/4**self.k # angle of k-esim root
97 | c = theta + delta/2 # center of the angle
98 | center.append(c)
99 | bottom.append(0)
100 | width.append(50*delta)
101 | height.append(h)
102 |
103 | return center, bottom, width, height
--------------------------------------------------------------------------------
/complexcgr/fcgr.py:
--------------------------------------------------------------------------------
1 | from . import CGR
2 | from PIL import Image
3 | from itertools import product
4 | from collections import defaultdict
5 | import numpy as np
6 |
7 | NUC_COMPLEMENT = {n:c for n,c in zip ("ACGT","TGCA")}
8 |
9 | class FCGR(CGR):
10 | """Frequency matrix CGR
11 | an (2**k x 2**k) 2D representation will be created for a
12 | n-long sequence.
13 | - k represents the k-mer.
14 | - 2**k x 2**k = 4**k the total number of k-mers (sequences of length k)
15 | """
16 |
17 | def __init__(self, k: int, use_canonical_kmers: bool = False ,bits: int = 8):
18 | super().__init__()
19 | self.k = k # k-mer representation
20 | self.use_canonical_kmers = use_canonical_kmers
21 | self.kmers = list("".join(kmer) for kmer in product("ACGT", repeat=self.k))
22 |
23 | if use_canonical_kmers is True:
24 | self.kmer2pixel = self._kmer2pixel_canonical_kmers()
25 | else:
26 | self.kmer2pixel = self.kmer2pixel_position()
27 |
28 | self.bits = bits
29 | self.max_color = 2**bits-1
30 |
31 | def __call__(self, sequence: str):
32 | "Given a DNA sequence, returns an array with his FCGR"
33 | self.count_kmers(sequence)
34 |
35 | # Create an empty array to save the FCGR values
36 | array_size = int(2**self.k)
37 | fcgr = np.zeros((array_size,array_size))
38 |
39 | # Assign frequency to each box in the matrix
40 | for kmer, freq in self.freq_kmer.items():
41 | pos_x, pos_y = self.kmer2pixel[kmer]
42 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
43 | return fcgr
44 |
45 | def count_kmer(self, kmer):
46 | if "N" not in kmer:
47 | self.freq_kmer[kmer] += 1
48 |
49 | def count_kmers(self, sequence: str):
50 | self.freq_kmer = defaultdict(int)
51 | # representativity of kmers
52 | last_j = len(sequence) - self.k + 1
53 | kmers = (sequence[i:(i+self.k)] for i in range(last_j))
54 | # count kmers in a dictionary
55 | list(self.count_kmer(kmer) for kmer in kmers)
56 |
57 | def pixel_position(self, kmer: str):
58 | "Get pixel position in the FCGR matrix for a k-mer"
59 |
60 | coords = self.encode(kmer)
61 | N,x,y = coords.N, coords.x, coords.y
62 |
63 | # Coordinates from [-1,1]² to [1,2**k]²
64 | np_coords = np.array([(x + 1)/2, (y + 1)/2]) # move coordinates from [-1,1]² to [0,1]²
65 | np_coords *= 2**self.k # rescale coordinates from [0,1]² to [0,2**k]²
66 | x,y = np.ceil(np_coords) # round to upper integer
67 |
68 | # Turn coordinates (cx,cy) into pixel (px,py) position
69 | # px = 2**k-cy+1, py = cx
70 | return 2**self.k-int(y)+1, int(x)
71 |
72 | def kmer2pixel_position(self,):
73 | kmer2pixel = dict()
74 | for kmer in self.kmers:
75 | kmer2pixel[kmer] = self.pixel_position(kmer)
76 | return kmer2pixel
77 |
78 | def plot(self, fcgr):
79 | "Given a FCGR, plot it in grayscale"
80 | img_pil = self.array2img(fcgr)
81 | return img_pil
82 |
83 | def save_img(self, fcgr, path: str):
84 | "Save image in grayscale for the FCGR provided as input"
85 | img_pil = self.array2img(fcgr)
86 | img_pil.save(path)
87 |
88 | def array2img(self, array):
89 | "Array to PIL image"
90 | m, M = array.min(), array.max()
91 | # rescale to [0,1]
92 | img_rescaled = (array - m) / (M-m)
93 |
94 | # invert colors black->white
95 | img_array = np.ceil(self.max_color - img_rescaled*self.max_color)
96 | dtype = eval(f"np.int{self.bits}")
97 | img_array = np.array(img_array, dtype=dtype)
98 |
99 | # convert to Image
100 | img_pil = Image.fromarray(img_array,'L')
101 | return img_pil
102 |
103 | # # --------------- canonical k-mers ---------------- # #
104 | @staticmethod
105 | def reverse_complement(kmer: str):
106 | rev_kmer = list(kmer)[::-1]
107 | return "".join([NUC_COMPLEMENT[n] for n in rev_kmer])
108 |
109 | def get_canonical_kmer(self, kmer: str):
110 | rev_complement = self.reverse_complement(kmer)
111 | return kmer if kmer < rev_complement else rev_complement
112 |
113 | def _kmer2pixel_canonical_kmers(self,):
114 | # change pixel position of non-canonical kmers
115 | kmer2pixel = dict()
116 | for kmer in self.kmers:
117 | canonical_kmer = self.get_canonical_kmer(kmer)
118 | if kmer != canonical_kmer:
119 | kmer2pixel[kmer] = kmer2pixel[canonical_kmer]
120 | return kmer2pixel
--------------------------------------------------------------------------------
/complexcgr/fcgr_kmc.py:
--------------------------------------------------------------------------------
1 | from . import FCGR
2 |
3 | import gzip
4 | import numpy as np
5 |
6 | class FCGRKmc(FCGR):
7 | """
8 | Create FCGR with the option of using canonical kmers from KMC output
9 | """
10 | def __init__(self, k: int, use_canonical_kmers: bool=False):
11 | super().__init__(k, use_canonical_kmers)
12 | self.k = k # k-mer representation
13 | self.use_canonical_kmers = use_canonical_kmers
14 |
15 | def __call__(self, path_kmc_output):
16 | "Given a path to a kmc output file, return the FCGR using canonical kmers as an array"
17 | # Create an empty array to save the FCGR values
18 | array_size = int(2**self.k)
19 | fcgr = np.zeros((array_size,array_size))
20 |
21 | if str(path_kmc_output).endswith(".txt"):
22 | with open(path_kmc_output) as fp:
23 | for line in fp:
24 | kmer, freq = line.split("\t")
25 | pos_x, pos_y = self.kmer2pixel[kmer]
26 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
27 | else:
28 | with gzip.open(path_kmc_output,'rt') as f:
29 | for line in f:
30 | line = line.strip()
31 | kmer, freq = line.split()
32 | pos_x, pos_y = self.kmer2pixel[kmer]
33 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
34 |
35 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/fcgr_samples.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from Bio import SeqIO
3 | from collections import defaultdict
4 | from pathlib import Path
5 | from tqdm import tqdm
6 | from typing import List, Union
7 | from complexcgr import FCGR
8 |
9 | # input for FCGR Samples
10 | _path_fastq = Union[str, Path] # path can be a string or a Path instance
11 | _fastq = Union[_path_fastq,List[_path_fastq]] # either a single path or a list of paths to fastq files
12 |
13 | class FCGRSamples(FCGR):
14 |
15 | def __init__(self, k: int, bits: int = 8):
16 | super().__init__(k, bits)
17 |
18 | def __call__(self, path_fastq: _fastq, consider_quality: bool = False):
19 | "Given a (list) of fastq files, return the FCGR matrix"
20 |
21 | ## ---- New for reads ----
22 |
23 | # for each call, we start to count the frequencies
24 | self.freq_kmer = defaultdict(int)
25 | if consider_quality is True:
26 | self.qual_kmer = defaultdict(int)
27 |
28 | # transform to list to iterate
29 | path_fastq = path_fastq if type(path_fastq) is list else [path_fastq]
30 |
31 | # For each file, count kmers on their reads
32 | for path in path_fastq:
33 | fastq = self.load_fastq(path)
34 | n_reads = self.count_reads(path)
35 |
36 | # count kmers in each read
37 | for sample in tqdm(fastq,total=n_reads,desc=f"Counting kmers on {str(Path(path).stem)}"):
38 | read = sample.seq
39 | if consider_quality is True:
40 | qual = sample.letter_annotations["phred_quality"]
41 | self.count_kmers_qualities(str(read),qual)
42 | else:
43 | self.count_kmers(str(read))
44 |
45 | ## ---- End new for reads ----
46 |
47 | # Create an empty array to save the FCGR values
48 | if consider_quality is False:
49 | array_size = int(2**self.k)
50 | fcgr = np.zeros((array_size,array_size))
51 | else:
52 | array_size = int(2**self.k)
53 | fcgr = np.zeros((array_size,array_size,2))
54 |
55 | # Assign frequency to each box in the matrix
56 | for kmer, freq in self.freq_kmer.items():
57 | pos_x, pos_y = self.kmer2pixel[kmer]
58 |
59 | if consider_quality is True:
60 | fcgr[int(pos_x)-1,int(pos_y)-1,0] = freq
61 | fcgr[int(pos_x)-1,int(pos_y)-1,1] = self.qual_kmer[kmer]
62 | else:
63 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
64 |
65 | return fcgr if consider_quality is False else self.rescale_fcgr_qualities(fcgr)
66 |
67 |
68 | def load_fastq(self, path):
69 | "Load a fastq file"
70 | fastq = SeqIO.parse(str(path), "fastq")
71 | return fastq
72 |
73 | def count_kmers(self, read: str):
74 | # representativity of kmers
75 | last_j = len(read) - self.k + 1
76 | kmers = (read[i:(i+self.k)] for i in range(last_j))
77 | # count kmers in a dictionary
78 | list(self.count_kmer(kmer) for kmer in kmers)
79 |
80 | @staticmethod
81 | def count_reads(path):
82 | "Count reads in a file to define progress bar"
83 | n_reads = 0
84 | with open(str(path)) as fp:
85 | for line in fp:
86 | if line.startswith("@"):
87 | n_reads += 1
88 | return n_reads
89 |
90 | def count_kmer(self, kmer):
91 | if "N" not in kmer:
92 | self.freq_kmer[kmer] += 1
93 |
94 |
95 | def count_kmer_quality(self, kmer, qmer):
96 | """Count kmers and qualities
97 | For quality of a kmer, the mean of qualities for each nucleotide in the kmer will be saved"""
98 | if "N" not in kmer:
99 | self.freq_kmer[kmer] += 1
100 | self.qual_kmer[kmer] += qmer.mean()
101 |
102 | def count_kmers_qualities(self,read,qual):
103 | "Count kmers and qualities"
104 |
105 | # representativity of kmers
106 | last_j = len(read) - self.k + 1
107 | kmers = (read[i:(i+self.k)] for i in range(last_j))
108 | qmers = (np.array(qual[i:(i+self.k)]) for i in range(last_j)) # qualities for each kmer
109 |
110 | # count kmers and qualities in a dictionary
111 | list(self.count_kmer_quality(kmer,qmer) for (kmer,qmer) in zip(kmers,qmers))
112 |
113 | @staticmethod
114 | def rescale_fcgr_qualities(fcgr):
115 | "Divide each cumulate quality by the freq of its kmer"
116 | freqs, quals = fcgr[:,:,0], fcgr[:,:,1]
117 | fcgr[:,:,1] = np.divide(quals, freqs, out = np.zeros_like(quals), where = quals!=0)
118 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/icgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from itertools import product
3 | from tqdm import tqdm
4 | from typing import Dict, Optional
5 | from collections import defaultdict, namedtuple
6 | import numpy as np
7 |
8 | # coordinates for x+iy
9 | Coord = namedtuple("Coord", ["x","y"])
10 |
11 | # coordinates for a CGR encoding
12 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
13 |
14 | # coordinates for each nucleotide in the 2d-plane
15 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
16 |
17 | class iCGR:
18 | "integer Chaos Game Representation for DNA"
19 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
20 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
21 | self.cgr_coords = CGRCoords(0,0,0)
22 |
23 | def nucleotide_by_coords(self,x,y):
24 | "Get nucleotide by coordinates (x,y)"
25 | # filter nucleotide by coordinates
26 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
27 |
28 | return list(filtered.keys())[0]
29 |
30 | def forward(self, nucleotide: str):
31 | "Compute next CGR coordinates"
32 | # current length
33 | N = self.cgr_coords.N
34 |
35 | # compute next coordinates
36 | x = self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x * 2**N
37 | y = self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y * 2**N
38 |
39 | # update cgr_coords: iCGR starts in the corner of the first nucleotide
40 |
41 | if N>0:
42 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y,)
43 | else:
44 | self.cgr_coords = CGRCoords(1,
45 | self.nucleotide_coords.get(nucleotide).x,
46 | self.nucleotide_coords.get(nucleotide).y,
47 | )
48 |
49 | def backward(self,):
50 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
51 | # get current nucleotide based on coordinates
52 | n_x,n_y = self.coords_current_nucleotide()
53 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
54 |
55 | # current length
56 | N = self.cgr_coords.N
57 |
58 | # update coordinates to the previous one
59 | x = self.cgr_coords.x - self.nucleotide_coords.get(nucleotide).x * 2**(N-1)
60 | y = self.cgr_coords.y - self.nucleotide_coords.get(nucleotide).y * 2**(N-1)
61 |
62 | # update cgr_coords
63 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
64 |
65 | return nucleotide
66 |
67 | def coords_current_nucleotide(self,):
68 | x = 1 if self.cgr_coords.x>0 else -1
69 | y = 1 if self.cgr_coords.y>0 else -1
70 | return x,y
71 |
72 | def encode(self, sequence: str):
73 | "From DNA sequence to CGR"
74 | # reset starting position to (0,0,0)
75 | self.reset_coords()
76 | for nucleotide in sequence:
77 | self.forward(nucleotide)
78 | return self.cgr_coords
79 |
80 | def reset_coords(self,):
81 | self.cgr_coords = CGRCoords(0,0,0)
82 |
83 | def decode(self, N:int, x:int, y:int)->str:
84 | "From CGR to DNA sequence"
85 | self.cgr_coords = CGRCoords(N,x,y)
86 |
87 | # decoded sequence
88 | sequence = []
89 |
90 | # Recover the entire genome
91 | while self.cgr_coords.N>0:
92 | nucleotide = self.backward()
93 | sequence.append(nucleotide)
94 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/img/ACG-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG-complexCGR.png
--------------------------------------------------------------------------------
/img/ACG.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG.jpg
--------------------------------------------------------------------------------
/img/ACG_16bits.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG_16bits.jpg
--------------------------------------------------------------------------------
/img/CGA.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGA.jpg
--------------------------------------------------------------------------------
/img/CGAN.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGAN.jpg
--------------------------------------------------------------------------------
/img/SAMD00003784.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00003784.jpeg
--------------------------------------------------------------------------------
/img/SAMD00032020.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00032020.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111209.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111209.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111306.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111306.jpeg
--------------------------------------------------------------------------------
/img/complexcgr-readme.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/complexcgr-readme.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3.png
--------------------------------------------------------------------------------
/img/logo-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR.png
--------------------------------------------------------------------------------
/poetry.lock:
--------------------------------------------------------------------------------
1 | # This file is automatically @generated by Poetry 1.7.1 and should not be changed by hand.
2 |
3 | [[package]]
4 | name = "atomicwrites"
5 | version = "1.4.1"
6 | description = "Atomic file writes."
7 | optional = false
8 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
9 | files = [
10 | {file = "atomicwrites-1.4.1.tar.gz", hash = "sha256:81b2c9071a49367a7f770170e5eec8cb66567cfbbc8c73d20ce5ca4a8d71cf11"},
11 | ]
12 |
13 | [[package]]
14 | name = "attrs"
15 | version = "23.2.0"
16 | description = "Classes Without Boilerplate"
17 | optional = false
18 | python-versions = ">=3.7"
19 | files = [
20 | {file = "attrs-23.2.0-py3-none-any.whl", hash = "sha256:99b87a485a5820b23b879f04c2305b44b951b502fd64be915879d77a7e8fc6f1"},
21 | {file = "attrs-23.2.0.tar.gz", hash = "sha256:935dc3b529c262f6cf76e50877d35a4bd3c1de194fd41f47a2b7ae8f19971f30"},
22 | ]
23 |
24 | [package.extras]
25 | cov = ["attrs[tests]", "coverage[toml] (>=5.3)"]
26 | dev = ["attrs[tests]", "pre-commit"]
27 | docs = ["furo", "myst-parser", "sphinx", "sphinx-notfound-page", "sphinxcontrib-towncrier", "towncrier", "zope-interface"]
28 | tests = ["attrs[tests-no-zope]", "zope-interface"]
29 | tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
30 | tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
31 |
32 | [[package]]
33 | name = "biopython"
34 | version = "1.82"
35 | description = "Freely available tools for computational molecular biology."
36 | optional = false
37 | python-versions = ">=3.8"
38 | files = [
39 | {file = "biopython-1.82-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:713be5e1e8571ea151864544dfcd2637eaf98c67a6e47b69781d325feb02f6b9"},
40 | {file = "biopython-1.82-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5bc908e84278bf61ac901a0c1b695c85ce49491957e0ab07e1e2afc216882525"},
41 | {file = "biopython-1.82-cp310-cp310-win32.whl", hash = "sha256:f53aff30f01ac9a8aa1fd42dab347a96ee5312945098dfa38f9a5125f6e913fd"},
42 | {file = "biopython-1.82-cp310-cp310-win_amd64.whl", hash = "sha256:0f3f685a2d85348b8c84c6eebe58a74cef7e1158ac52ae5aa91789436907f0e1"},
43 | {file = "biopython-1.82-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c9bc080b7dc6cd6acbf2576766136e8e54acbcc6b7a29ad8075cb5a73f4cfb97"},
44 | {file = "biopython-1.82-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c7802b5033edaf34bd2fbd96b87f96999a6808caa0064c91087c7f1340624898"},
45 | {file = "biopython-1.82-cp311-cp311-win32.whl", hash = "sha256:ec35893b32458f6fa0c98d5f02d801a5fe76961ac66473455f51c0575562c594"},
46 | {file = "biopython-1.82-cp311-cp311-win_amd64.whl", hash = "sha256:40297d418bb142a7ea1a47faedc24bbe21b0ba455f3805373264470e5cb6ec5c"},
47 | {file = "biopython-1.82-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:96f77bd6540683c8958e4ebf55b58e8caa26d20db72891915d06670b783e9cdb"},
48 | {file = "biopython-1.82-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:49b829c2276a0711106663af632c7e203695a5cf082d488d6e10b8aca5ec63e4"},
49 | {file = "biopython-1.82-cp312-cp312-win32.whl", hash = "sha256:54a9b200d4960ed2a4ec85faac2124ffa278afa58d22e07e2d15ca7509219ea1"},
50 | {file = "biopython-1.82-cp312-cp312-win_amd64.whl", hash = "sha256:762547037038d42c9ac98877fc32d4aeba798077129df7790cb178256171a4e5"},
51 | {file = "biopython-1.82-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:cb013256742d31e24fd5093d55511f58bc6a93b8ac723d2ca3b33c7904f1ca14"},
52 | {file = "biopython-1.82-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:16829760fab60ba17141b7a81494fed837c6f29ea28cdcfb3070764c8e8f2ff1"},
53 | {file = "biopython-1.82-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c6af82894dd4f736d737fdea62008a19966251ba39aef39a0ce8b41321305ad5"},
54 | {file = "biopython-1.82-cp38-cp38-win32.whl", hash = "sha256:a1ff583b2f8b314a4c2d612aad4f4b73827a5f741b391f3a834b0c192a4f1527"},
55 | {file = "biopython-1.82-cp38-cp38-win_amd64.whl", hash = "sha256:0198f23776d9cb9d514a797e81a024f486adac973b252a2f4696995a5a163745"},
56 | {file = "biopython-1.82-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c88e9f87d1b07958e7e1e25ff20efced4796daf650e5290692b5c3ff9aebf9d3"},
57 | {file = "biopython-1.82-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8e2533db9275c3e1438151260844e7683f91308da320df02a4c28fb7053ea3a9"},
58 | {file = "biopython-1.82-cp39-cp39-win32.whl", hash = "sha256:80f48d220b92e747300281602f52e98ec6c537ddd4921b580d9a0368bae05ff5"},
59 | {file = "biopython-1.82-cp39-cp39-win_amd64.whl", hash = "sha256:73d619611bd7cf9d2ad8b3217b08d3796acfabb765b0771ad2ff7ee2e46a59b0"},
60 | {file = "biopython-1.82.tar.gz", hash = "sha256:a9b10d959ae88a9744a91c6ce3601f4c86e7ec41679bc93c29f679218f6167bb"},
61 | ]
62 |
63 | [package.dependencies]
64 | numpy = "*"
65 |
66 | [[package]]
67 | name = "colorama"
68 | version = "0.4.6"
69 | description = "Cross-platform colored terminal text."
70 | optional = false
71 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,!=3.3.*,!=3.4.*,!=3.5.*,!=3.6.*,>=2.7"
72 | files = [
73 | {file = "colorama-0.4.6-py2.py3-none-any.whl", hash = "sha256:4f1d9991f5acc0ca119f9d443620b77f9d6b33703e51011c16baf57afb285fc6"},
74 | {file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
75 | ]
76 |
77 | [[package]]
78 | name = "contourpy"
79 | version = "1.2.0"
80 | description = "Python library for calculating contours of 2D quadrilateral grids"
81 | optional = false
82 | python-versions = ">=3.9"
83 | files = [
84 | {file = "contourpy-1.2.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:0274c1cb63625972c0c007ab14dd9ba9e199c36ae1a231ce45d725cbcbfd10a8"},
85 | {file = "contourpy-1.2.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ab459a1cbbf18e8698399c595a01f6dcc5c138220ca3ea9e7e6126232d102bb4"},
86 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6fdd887f17c2f4572ce548461e4f96396681212d858cae7bd52ba3310bc6f00f"},
87 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:5d16edfc3fc09968e09ddffada434b3bf989bf4911535e04eada58469873e28e"},
88 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1c203f617abc0dde5792beb586f827021069fb6d403d7f4d5c2b543d87edceb9"},
89 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b69303ceb2e4d4f146bf82fda78891ef7bcd80c41bf16bfca3d0d7eb545448aa"},
90 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:884c3f9d42d7218304bc74a8a7693d172685c84bd7ab2bab1ee567b769696df9"},
91 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4a1b1208102be6e851f20066bf0e7a96b7d48a07c9b0cfe6d0d4545c2f6cadab"},
92 | {file = "contourpy-1.2.0-cp310-cp310-win32.whl", hash = "sha256:34b9071c040d6fe45d9826cbbe3727d20d83f1b6110d219b83eb0e2a01d79488"},
93 | {file = "contourpy-1.2.0-cp310-cp310-win_amd64.whl", hash = "sha256:bd2f1ae63998da104f16a8b788f685e55d65760cd1929518fd94cd682bf03e41"},
94 | {file = "contourpy-1.2.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:dd10c26b4eadae44783c45ad6655220426f971c61d9b239e6f7b16d5cdaaa727"},
95 | {file = "contourpy-1.2.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:5c6b28956b7b232ae801406e529ad7b350d3f09a4fde958dfdf3c0520cdde0dd"},
96 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ebeac59e9e1eb4b84940d076d9f9a6cec0064e241818bcb6e32124cc5c3e377a"},
97 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:139d8d2e1c1dd52d78682f505e980f592ba53c9f73bd6be102233e358b401063"},
98 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1e9dc350fb4c58adc64df3e0703ab076f60aac06e67d48b3848c23647ae4310e"},
99 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:18fc2b4ed8e4a8fe849d18dce4bd3c7ea637758c6343a1f2bae1e9bd4c9f4686"},
100 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:16a7380e943a6d52472096cb7ad5264ecee36ed60888e2a3d3814991a0107286"},
101 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8d8faf05be5ec8e02a4d86f616fc2a0322ff4a4ce26c0f09d9f7fb5330a35c95"},
102 | {file = "contourpy-1.2.0-cp311-cp311-win32.whl", hash = "sha256:67b7f17679fa62ec82b7e3e611c43a016b887bd64fb933b3ae8638583006c6d6"},
103 | {file = "contourpy-1.2.0-cp311-cp311-win_amd64.whl", hash = "sha256:99ad97258985328b4f207a5e777c1b44a83bfe7cf1f87b99f9c11d4ee477c4de"},
104 | {file = "contourpy-1.2.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:575bcaf957a25d1194903a10bc9f316c136c19f24e0985a2b9b5608bdf5dbfe0"},
105 | {file = "contourpy-1.2.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:9e6c93b5b2dbcedad20a2f18ec22cae47da0d705d454308063421a3b290d9ea4"},
106 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:464b423bc2a009088f19bdf1f232299e8b6917963e2b7e1d277da5041f33a779"},
107 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:68ce4788b7d93e47f84edd3f1f95acdcd142ae60bc0e5493bfd120683d2d4316"},
108 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3d7d1f8871998cdff5d2ff6a087e5e1780139abe2838e85b0b46b7ae6cc25399"},
109 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6e739530c662a8d6d42c37c2ed52a6f0932c2d4a3e8c1f90692ad0ce1274abe0"},
110 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:247b9d16535acaa766d03037d8e8fb20866d054d3c7fbf6fd1f993f11fc60ca0"},
111 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:461e3ae84cd90b30f8d533f07d87c00379644205b1d33a5ea03381edc4b69431"},
112 | {file = "contourpy-1.2.0-cp312-cp312-win32.whl", hash = "sha256:1c2559d6cffc94890b0529ea7eeecc20d6fadc1539273aa27faf503eb4656d8f"},
113 | {file = "contourpy-1.2.0-cp312-cp312-win_amd64.whl", hash = "sha256:491b1917afdd8638a05b611a56d46587d5a632cabead889a5440f7c638bc6ed9"},
114 | {file = "contourpy-1.2.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5fd1810973a375ca0e097dee059c407913ba35723b111df75671a1976efa04bc"},
115 | {file = "contourpy-1.2.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:999c71939aad2780f003979b25ac5b8f2df651dac7b38fb8ce6c46ba5abe6ae9"},
116 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b7caf9b241464c404613512d5594a6e2ff0cc9cb5615c9475cc1d9b514218ae8"},
117 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:266270c6f6608340f6c9836a0fb9b367be61dde0c9a9a18d5ece97774105ff3e"},
118 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:dbd50d0a0539ae2e96e537553aff6d02c10ed165ef40c65b0e27e744a0f10af8"},
119 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:11f8d2554e52f459918f7b8e6aa20ec2a3bce35ce95c1f0ef4ba36fbda306df5"},
120 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:ce96dd400486e80ac7d195b2d800b03e3e6a787e2a522bfb83755938465a819e"},
121 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6d3364b999c62f539cd403f8123ae426da946e142312a514162adb2addd8d808"},
122 | {file = "contourpy-1.2.0-cp39-cp39-win32.whl", hash = "sha256:1c88dfb9e0c77612febebb6ac69d44a8d81e3dc60f993215425b62c1161353f4"},
123 | {file = "contourpy-1.2.0-cp39-cp39-win_amd64.whl", hash = "sha256:78e6ad33cf2e2e80c5dfaaa0beec3d61face0fb650557100ee36db808bfa6843"},
124 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:be16975d94c320432657ad2402f6760990cb640c161ae6da1363051805fa8108"},
125 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b95a225d4948b26a28c08307a60ac00fb8671b14f2047fc5476613252a129776"},
126 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:0d7e03c0f9a4f90dc18d4e77e9ef4ec7b7bbb437f7f675be8e530d65ae6ef956"},
127 | {file = "contourpy-1.2.0.tar.gz", hash = "sha256:171f311cb758de7da13fc53af221ae47a5877be5a0843a9fe150818c51ed276a"},
128 | ]
129 |
130 | [package.dependencies]
131 | numpy = ">=1.20,<2.0"
132 |
133 | [package.extras]
134 | bokeh = ["bokeh", "selenium"]
135 | docs = ["furo", "sphinx (>=7.2)", "sphinx-copybutton"]
136 | mypy = ["contourpy[bokeh,docs]", "docutils-stubs", "mypy (==1.6.1)", "types-Pillow"]
137 | test = ["Pillow", "contourpy[test-no-images]", "matplotlib"]
138 | test-no-images = ["pytest", "pytest-cov", "pytest-xdist", "wurlitzer"]
139 |
140 | [[package]]
141 | name = "cycler"
142 | version = "0.12.1"
143 | description = "Composable style cycles"
144 | optional = false
145 | python-versions = ">=3.8"
146 | files = [
147 | {file = "cycler-0.12.1-py3-none-any.whl", hash = "sha256:85cef7cff222d8644161529808465972e51340599459b8ac3ccbac5a854e0d30"},
148 | {file = "cycler-0.12.1.tar.gz", hash = "sha256:88bb128f02ba341da8ef447245a9e138fae777f6a23943da4540077d3601eb1c"},
149 | ]
150 |
151 | [package.extras]
152 | docs = ["ipython", "matplotlib", "numpydoc", "sphinx"]
153 | tests = ["pytest", "pytest-cov", "pytest-xdist"]
154 |
155 | [[package]]
156 | name = "fonttools"
157 | version = "4.47.0"
158 | description = "Tools to manipulate font files"
159 | optional = false
160 | python-versions = ">=3.8"
161 | files = [
162 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:2d2404107626f97a221dc1a65b05396d2bb2ce38e435f64f26ed2369f68675d9"},
163 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:c01f409be619a9a0f5590389e37ccb58b47264939f0e8d58bfa1f3ba07d22671"},
164 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d986b66ff722ef675b7ee22fbe5947a41f60a61a4da15579d5e276d897fbc7fa"},
165 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e8acf6dd0434b211b3bd30d572d9e019831aae17a54016629fa8224783b22df8"},
166 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:495369c660e0c27233e3c572269cbe520f7f4978be675f990f4005937337d391"},
167 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c59227d7ba5b232281c26ae04fac2c73a79ad0e236bca5c44aae904a18f14faf"},
168 | {file = "fonttools-4.47.0-cp310-cp310-win32.whl", hash = "sha256:59a6c8b71a245800e923cb684a2dc0eac19c56493e2f896218fcf2571ed28984"},
169 | {file = "fonttools-4.47.0-cp310-cp310-win_amd64.whl", hash = "sha256:52c82df66201f3a90db438d9d7b337c7c98139de598d0728fb99dab9fd0495ca"},
170 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:854421e328d47d70aa5abceacbe8eef231961b162c71cbe7ff3f47e235e2e5c5"},
171 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:511482df31cfea9f697930f61520f6541185fa5eeba2fa760fe72e8eee5af88b"},
172 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ce0e2c88c8c985b7b9a7efcd06511fb0a1fe3ddd9a6cd2895ef1dbf9059719d7"},
173 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e7a0a8848726956e9d9fb18c977a279013daadf0cbb6725d2015a6dd57527992"},
174 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e869da810ae35afb3019baa0d0306cdbab4760a54909c89ad8904fa629991812"},
175 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:dd23848f877c3754f53a4903fb7a593ed100924f9b4bff7d5a4e2e8a7001ae11"},
176 | {file = "fonttools-4.47.0-cp311-cp311-win32.whl", hash = "sha256:bf1810635c00f7c45d93085611c995fc130009cec5abdc35b327156aa191f982"},
177 | {file = "fonttools-4.47.0-cp311-cp311-win_amd64.whl", hash = "sha256:61df4dee5d38ab65b26da8efd62d859a1eef7a34dcbc331299a28e24d04c59a7"},
178 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:e3f4d61f3a8195eac784f1d0c16c0a3105382c1b9a74d99ac4ba421da39a8826"},
179 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:174995f7b057e799355b393e97f4f93ef1f2197cbfa945e988d49b2a09ecbce8"},
180 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea592e6a09b71cb7a7661dd93ac0b877a6228e2d677ebacbad0a4d118494c86d"},
181 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:40bdbe90b33897d9cc4a39f8e415b0fcdeae4c40a99374b8a4982f127ff5c767"},
182 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:843509ae9b93db5aaf1a6302085e30bddc1111d31e11d724584818f5b698f500"},
183 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:9acfa1cdc479e0dde528b61423855913d949a7f7fe09e276228298fef4589540"},
184 | {file = "fonttools-4.47.0-cp312-cp312-win32.whl", hash = "sha256:66c92ec7f95fd9732550ebedefcd190a8d81beaa97e89d523a0d17198a8bda4d"},
185 | {file = "fonttools-4.47.0-cp312-cp312-win_amd64.whl", hash = "sha256:e8fa20748de55d0021f83754b371432dca0439e02847962fc4c42a0e444c2d78"},
186 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c75e19971209fbbce891ebfd1b10c37320a5a28e8d438861c21d35305aedb81c"},
187 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e79f1a3970d25f692bbb8c8c2637e621a66c0d60c109ab48d4a160f50856deff"},
188 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:562681188c62c024fe2c611b32e08b8de2afa00c0c4e72bed47c47c318e16d5c"},
189 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a77a60315c33393b2bd29d538d1ef026060a63d3a49a9233b779261bad9c3f71"},
190 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:b4fabb8cc9422efae1a925160083fdcbab8fdc96a8483441eb7457235df625bd"},
191 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2a78dba8c2a1e9d53a0fb5382979f024200dc86adc46a56cbb668a2249862fda"},
192 | {file = "fonttools-4.47.0-cp38-cp38-win32.whl", hash = "sha256:e6b968543fde4119231c12c2a953dcf83349590ca631ba8216a8edf9cd4d36a9"},
193 | {file = "fonttools-4.47.0-cp38-cp38-win_amd64.whl", hash = "sha256:4a9a51745c0439516d947480d4d884fa18bd1458e05b829e482b9269afa655bc"},
194 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:62d8ddb058b8e87018e5dc26f3258e2c30daad4c87262dfeb0e2617dd84750e6"},
195 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5dde0eab40faaa5476133123f6a622a1cc3ac9b7af45d65690870620323308b4"},
196 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f4da089f6dfdb822293bde576916492cd708c37c2501c3651adde39804630538"},
197 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:253bb46bab970e8aae254cebf2ae3db98a4ef6bd034707aa68a239027d2b198d"},
198 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:1193fb090061efa2f9e2d8d743ae9850c77b66746a3b32792324cdce65784154"},
199 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:084511482dd265bce6dca24c509894062f0117e4e6869384d853f46c0e6d43be"},
200 | {file = "fonttools-4.47.0-cp39-cp39-win32.whl", hash = "sha256:97620c4af36e4c849e52661492e31dc36916df12571cb900d16960ab8e92a980"},
201 | {file = "fonttools-4.47.0-cp39-cp39-win_amd64.whl", hash = "sha256:e77bdf52185bdaf63d39f3e1ac3212e6cfa3ab07d509b94557a8902ce9c13c82"},
202 | {file = "fonttools-4.47.0-py3-none-any.whl", hash = "sha256:d6477ba902dd2d7adda7f0fd3bfaeb92885d45993c9e1928c9f28fc3961415f7"},
203 | {file = "fonttools-4.47.0.tar.gz", hash = "sha256:ec13a10715eef0e031858c1c23bfaee6cba02b97558e4a7bfa089dba4a8c2ebf"},
204 | ]
205 |
206 | [package.extras]
207 | all = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "fs (>=2.2.0,<3)", "lxml (>=4.0,<5)", "lz4 (>=1.7.4.2)", "matplotlib", "munkres", "pycairo", "scipy", "skia-pathops (>=0.5.0)", "sympy", "uharfbuzz (>=0.23.0)", "unicodedata2 (>=15.1.0)", "xattr", "zopfli (>=0.1.4)"]
208 | graphite = ["lz4 (>=1.7.4.2)"]
209 | interpolatable = ["munkres", "pycairo", "scipy"]
210 | lxml = ["lxml (>=4.0,<5)"]
211 | pathops = ["skia-pathops (>=0.5.0)"]
212 | plot = ["matplotlib"]
213 | repacker = ["uharfbuzz (>=0.23.0)"]
214 | symfont = ["sympy"]
215 | type1 = ["xattr"]
216 | ufo = ["fs (>=2.2.0,<3)"]
217 | unicode = ["unicodedata2 (>=15.1.0)"]
218 | woff = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "zopfli (>=0.1.4)"]
219 |
220 | [[package]]
221 | name = "importlib-resources"
222 | version = "6.1.1"
223 | description = "Read resources from Python packages"
224 | optional = false
225 | python-versions = ">=3.8"
226 | files = [
227 | {file = "importlib_resources-6.1.1-py3-none-any.whl", hash = "sha256:e8bf90d8213b486f428c9c39714b920041cb02c184686a3dee24905aaa8105d6"},
228 | {file = "importlib_resources-6.1.1.tar.gz", hash = "sha256:3893a00122eafde6894c59914446a512f728a0c1a45f9bb9b63721b6bacf0b4a"},
229 | ]
230 |
231 | [package.dependencies]
232 | zipp = {version = ">=3.1.0", markers = "python_version < \"3.10\""}
233 |
234 | [package.extras]
235 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
236 | testing = ["pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy (>=0.9.1)", "pytest-ruff", "zipp (>=3.17)"]
237 |
238 | [[package]]
239 | name = "iniconfig"
240 | version = "2.0.0"
241 | description = "brain-dead simple config-ini parsing"
242 | optional = false
243 | python-versions = ">=3.7"
244 | files = [
245 | {file = "iniconfig-2.0.0-py3-none-any.whl", hash = "sha256:b6a85871a79d2e3b22d2d1b94ac2824226a63c6b741c88f7ae975f18b6778374"},
246 | {file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
247 | ]
248 |
249 | [[package]]
250 | name = "kiwisolver"
251 | version = "1.4.5"
252 | description = "A fast implementation of the Cassowary constraint solver"
253 | optional = false
254 | python-versions = ">=3.7"
255 | files = [
256 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:05703cf211d585109fcd72207a31bb170a0f22144d68298dc5e61b3c946518af"},
257 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:146d14bebb7f1dc4d5fbf74f8a6cb15ac42baadee8912eb84ac0b3b2a3dc6ac3"},
258 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ef7afcd2d281494c0a9101d5c571970708ad911d028137cd558f02b851c08b4"},
259 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9eaa8b117dc8337728e834b9c6e2611f10c79e38f65157c4c38e9400286f5cb1"},
260 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ec20916e7b4cbfb1f12380e46486ec4bcbaa91a9c448b97023fde0d5bbf9e4ff"},
261 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:39b42c68602539407884cf70d6a480a469b93b81b7701378ba5e2328660c847a"},
262 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:aa12042de0171fad672b6c59df69106d20d5596e4f87b5e8f76df757a7c399aa"},
263 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:2a40773c71d7ccdd3798f6489aaac9eee213d566850a9533f8d26332d626b82c"},
264 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:19df6e621f6d8b4b9c4d45f40a66839294ff2bb235e64d2178f7522d9170ac5b"},
265 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:83d78376d0d4fd884e2c114d0621624b73d2aba4e2788182d286309ebdeed770"},
266 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:e391b1f0a8a5a10ab3b9bb6afcfd74f2175f24f8975fb87ecae700d1503cdee0"},
267 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:852542f9481f4a62dbb5dd99e8ab7aedfeb8fb6342349a181d4036877410f525"},
268 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:59edc41b24031bc25108e210c0def6f6c2191210492a972d585a06ff246bb79b"},
269 | {file = "kiwisolver-1.4.5-cp310-cp310-win32.whl", hash = "sha256:a6aa6315319a052b4ee378aa171959c898a6183f15c1e541821c5c59beaa0238"},
270 | {file = "kiwisolver-1.4.5-cp310-cp310-win_amd64.whl", hash = "sha256:d0ef46024e6a3d79c01ff13801cb19d0cad7fd859b15037aec74315540acc276"},
271 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:11863aa14a51fd6ec28688d76f1735f8f69ab1fabf388851a595d0721af042f5"},
272 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:8ab3919a9997ab7ef2fbbed0cc99bb28d3c13e6d4b1ad36e97e482558a91be90"},
273 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fcc700eadbbccbf6bc1bcb9dbe0786b4b1cb91ca0dcda336eef5c2beed37b797"},
274 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:dfdd7c0b105af050eb3d64997809dc21da247cf44e63dc73ff0fd20b96be55a9"},
275 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:76c6a5964640638cdeaa0c359382e5703e9293030fe730018ca06bc2010c4437"},
276 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:bbea0db94288e29afcc4c28afbf3a7ccaf2d7e027489c449cf7e8f83c6346eb9"},
277 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ceec1a6bc6cab1d6ff5d06592a91a692f90ec7505d6463a88a52cc0eb58545da"},
278 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:040c1aebeda72197ef477a906782b5ab0d387642e93bda547336b8957c61022e"},
279 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f91de7223d4c7b793867797bacd1ee53bfe7359bd70d27b7b58a04efbb9436c8"},
280 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:faae4860798c31530dd184046a900e652c95513796ef51a12bc086710c2eec4d"},
281 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:b0157420efcb803e71d1b28e2c287518b8808b7cf1ab8af36718fd0a2c453eb0"},
282 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:06f54715b7737c2fecdbf140d1afb11a33d59508a47bf11bb38ecf21dc9ab79f"},
283 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:fdb7adb641a0d13bdcd4ef48e062363d8a9ad4a182ac7647ec88f695e719ae9f"},
284 | {file = "kiwisolver-1.4.5-cp311-cp311-win32.whl", hash = "sha256:bb86433b1cfe686da83ce32a9d3a8dd308e85c76b60896d58f082136f10bffac"},
285 | {file = "kiwisolver-1.4.5-cp311-cp311-win_amd64.whl", hash = "sha256:6c08e1312a9cf1074d17b17728d3dfce2a5125b2d791527f33ffbe805200a355"},
286 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:32d5cf40c4f7c7b3ca500f8985eb3fb3a7dfc023215e876f207956b5ea26632a"},
287 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:f846c260f483d1fd217fe5ed7c173fb109efa6b1fc8381c8b7552c5781756192"},
288 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5ff5cf3571589b6d13bfbfd6bcd7a3f659e42f96b5fd1c4830c4cf21d4f5ef45"},
289 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7269d9e5f1084a653d575c7ec012ff57f0c042258bf5db0954bf551c158466e7"},
290 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:da802a19d6e15dffe4b0c24b38b3af68e6c1a68e6e1d8f30148c83864f3881db"},
291 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:3aba7311af82e335dd1e36ffff68aaca609ca6290c2cb6d821a39aa075d8e3ff"},
292 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:763773d53f07244148ccac5b084da5adb90bfaee39c197554f01b286cf869228"},
293 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2270953c0d8cdab5d422bee7d2007f043473f9d2999631c86a223c9db56cbd16"},
294 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:d099e745a512f7e3bbe7249ca835f4d357c586d78d79ae8f1dcd4d8adeb9bda9"},
295 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:74db36e14a7d1ce0986fa104f7d5637aea5c82ca6326ed0ec5694280942d1162"},
296 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:7e5bab140c309cb3a6ce373a9e71eb7e4873c70c2dda01df6820474f9889d6d4"},
297 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:0f114aa76dc1b8f636d077979c0ac22e7cd8f3493abbab152f20eb8d3cda71f3"},
298 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:88a2df29d4724b9237fc0c6eaf2a1adae0cdc0b3e9f4d8e7dc54b16812d2d81a"},
299 | {file = "kiwisolver-1.4.5-cp312-cp312-win32.whl", hash = "sha256:72d40b33e834371fd330fb1472ca19d9b8327acb79a5821d4008391db8e29f20"},
300 | {file = "kiwisolver-1.4.5-cp312-cp312-win_amd64.whl", hash = "sha256:2c5674c4e74d939b9d91dda0fae10597ac7521768fec9e399c70a1f27e2ea2d9"},
301 | {file = "kiwisolver-1.4.5-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:3a2b053a0ab7a3960c98725cfb0bf5b48ba82f64ec95fe06f1d06c99b552e130"},
302 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3cd32d6c13807e5c66a7cbb79f90b553642f296ae4518a60d8d76243b0ad2898"},
303 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:59ec7b7c7e1a61061850d53aaf8e93db63dce0c936db1fda2658b70e4a1be709"},
304 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da4cfb373035def307905d05041c1d06d8936452fe89d464743ae7fb8371078b"},
305 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:2400873bccc260b6ae184b2b8a4fec0e4082d30648eadb7c3d9a13405d861e89"},
306 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:1b04139c4236a0f3aff534479b58f6f849a8b351e1314826c2d230849ed48985"},
307 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:4e66e81a5779b65ac21764c295087de82235597a2293d18d943f8e9e32746265"},
308 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:7931d8f1f67c4be9ba1dd9c451fb0eeca1a25b89e4d3f89e828fe12a519b782a"},
309 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:b3f7e75f3015df442238cca659f8baa5f42ce2a8582727981cbfa15fee0ee205"},
310 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:bbf1d63eef84b2e8c89011b7f2235b1e0bf7dacc11cac9431fc6468e99ac77fb"},
311 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:4c380469bd3f970ef677bf2bcba2b6b0b4d5c75e7a020fb863ef75084efad66f"},
312 | {file = "kiwisolver-1.4.5-cp37-cp37m-win32.whl", hash = "sha256:9408acf3270c4b6baad483865191e3e582b638b1654a007c62e3efe96f09a9a3"},
313 | {file = "kiwisolver-1.4.5-cp37-cp37m-win_amd64.whl", hash = "sha256:5b94529f9b2591b7af5f3e0e730a4e0a41ea174af35a4fd067775f9bdfeee01a"},
314 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:11c7de8f692fc99816e8ac50d1d1aef4f75126eefc33ac79aac02c099fd3db71"},
315 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:53abb58632235cd154176ced1ae8f0d29a6657aa1aa9decf50b899b755bc2b93"},
316 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:88b9f257ca61b838b6f8094a62418421f87ac2a1069f7e896c36a7d86b5d4c29"},
317 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3195782b26fc03aa9c6913d5bad5aeb864bdc372924c093b0f1cebad603dd712"},
318 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:fc579bf0f502e54926519451b920e875f433aceb4624a3646b3252b5caa9e0b6"},
319 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5a580c91d686376f0f7c295357595c5a026e6cbc3d77b7c36e290201e7c11ecb"},
320 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:cfe6ab8da05c01ba6fbea630377b5da2cd9bcbc6338510116b01c1bc939a2c18"},
321 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:d2e5a98f0ec99beb3c10e13b387f8db39106d53993f498b295f0c914328b1333"},
322 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a51a263952b1429e429ff236d2f5a21c5125437861baeed77f5e1cc2d2c7c6da"},
323 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:3edd2fa14e68c9be82c5b16689e8d63d89fe927e56debd6e1dbce7a26a17f81b"},
324 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:74d1b44c6cfc897df648cc9fdaa09bc3e7679926e6f96df05775d4fb3946571c"},
325 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:76d9289ed3f7501012e05abb8358bbb129149dbd173f1f57a1bf1c22d19ab7cc"},
326 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:92dea1ffe3714fa8eb6a314d2b3c773208d865a0e0d35e713ec54eea08a66250"},
327 | {file = "kiwisolver-1.4.5-cp38-cp38-win32.whl", hash = "sha256:5c90ae8c8d32e472be041e76f9d2f2dbff4d0b0be8bd4041770eddb18cf49a4e"},
328 | {file = "kiwisolver-1.4.5-cp38-cp38-win_amd64.whl", hash = "sha256:c7940c1dc63eb37a67721b10d703247552416f719c4188c54e04334321351ced"},
329 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:9407b6a5f0d675e8a827ad8742e1d6b49d9c1a1da5d952a67d50ef5f4170b18d"},
330 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:15568384086b6df3c65353820a4473575dbad192e35010f622c6ce3eebd57af9"},
331 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:0dc9db8e79f0036e8173c466d21ef18e1befc02de8bf8aa8dc0813a6dc8a7046"},
332 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:cdc8a402aaee9a798b50d8b827d7ecf75edc5fb35ea0f91f213ff927c15f4ff0"},
333 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6c3bd3cde54cafb87d74d8db50b909705c62b17c2099b8f2e25b461882e544ff"},
334 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:955e8513d07a283056b1396e9a57ceddbd272d9252c14f154d450d227606eb54"},
335 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:346f5343b9e3f00b8db8ba359350eb124b98c99efd0b408728ac6ebf38173958"},
336 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b9098e0049e88c6a24ff64545cdfc50807818ba6c1b739cae221bbbcbc58aad3"},
337 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:00bd361b903dc4bbf4eb165f24d1acbee754fce22ded24c3d56eec268658a5cf"},
338 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:7b8b454bac16428b22560d0a1cf0a09875339cab69df61d7805bf48919415901"},
339 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:f1d072c2eb0ad60d4c183f3fb44ac6f73fb7a8f16a2694a91f988275cbf352f9"},
340 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:31a82d498054cac9f6d0b53d02bb85811185bcb477d4b60144f915f3b3126342"},
341 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6512cb89e334e4700febbffaaa52761b65b4f5a3cf33f960213d5656cea36a77"},
342 | {file = "kiwisolver-1.4.5-cp39-cp39-win32.whl", hash = "sha256:9db8ea4c388fdb0f780fe91346fd438657ea602d58348753d9fb265ce1bca67f"},
343 | {file = "kiwisolver-1.4.5-cp39-cp39-win_amd64.whl", hash = "sha256:59415f46a37f7f2efeec758353dd2eae1b07640d8ca0f0c42548ec4125492635"},
344 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:5c7b3b3a728dc6faf3fc372ef24f21d1e3cee2ac3e9596691d746e5a536de920"},
345 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:620ced262a86244e2be10a676b646f29c34537d0d9cc8eb26c08f53d98013390"},
346 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:378a214a1e3bbf5ac4a8708304318b4f890da88c9e6a07699c4ae7174c09a68d"},
347 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aaf7be1207676ac608a50cd08f102f6742dbfc70e8d60c4db1c6897f62f71523"},
348 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:ba55dce0a9b8ff59495ddd050a0225d58bd0983d09f87cfe2b6aec4f2c1234e4"},
349 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:fd32ea360bcbb92d28933fc05ed09bffcb1704ba3fc7942e81db0fd4f81a7892"},
350 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:5e7139af55d1688f8b960ee9ad5adafc4ac17c1c473fe07133ac092310d76544"},
351 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:dced8146011d2bc2e883f9bd68618b8247387f4bbec46d7392b3c3b032640126"},
352 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c9bf3325c47b11b2e51bca0824ea217c7cd84491d8ac4eefd1e409705ef092bd"},
353 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5794cf59533bc3f1b1c821f7206a3617999db9fbefc345360aafe2e067514929"},
354 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e368f200bbc2e4f905b8e71eb38b3c04333bddaa6a2464a6355487b02bb7fb09"},
355 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:e5d706eba36b4c4d5bc6c6377bb6568098765e990cfc21ee16d13963fab7b3e7"},
356 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:85267bd1aa8880a9c88a8cb71e18d3d64d2751a790e6ca6c27b8ccc724bcd5ad"},
357 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:210ef2c3a1f03272649aff1ef992df2e724748918c4bc2d5a90352849eb40bea"},
358 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:11d011a7574eb3b82bcc9c1a1d35c1d7075677fdd15de527d91b46bd35e935ee"},
359 | {file = "kiwisolver-1.4.5.tar.gz", hash = "sha256:e57e563a57fb22a142da34f38acc2fc1a5c864bc29ca1517a88abc963e60d6ec"},
360 | ]
361 |
362 | [[package]]
363 | name = "matplotlib"
364 | version = "3.8.2"
365 | description = "Python plotting package"
366 | optional = false
367 | python-versions = ">=3.9"
368 | files = [
369 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_10_12_x86_64.whl", hash = "sha256:09796f89fb71a0c0e1e2f4bdaf63fb2cefc84446bb963ecdeb40dfee7dfa98c7"},
370 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6f9c6976748a25e8b9be51ea028df49b8e561eed7809146da7a47dbecebab367"},
371 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b78e4f2cedf303869b782071b55fdde5987fda3038e9d09e58c91cc261b5ad18"},
372 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e208f46cf6576a7624195aa047cb344a7f802e113bb1a06cfd4bee431de5e31"},
373 | {file = "matplotlib-3.8.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:46a569130ff53798ea5f50afce7406e91fdc471ca1e0e26ba976a8c734c9427a"},
374 | {file = "matplotlib-3.8.2-cp310-cp310-win_amd64.whl", hash = "sha256:830f00640c965c5b7f6bc32f0d4ce0c36dfe0379f7dd65b07a00c801713ec40a"},
375 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_10_12_x86_64.whl", hash = "sha256:d86593ccf546223eb75a39b44c32788e6f6440d13cfc4750c1c15d0fcb850b63"},
376 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:9a5430836811b7652991939012f43d2808a2db9b64ee240387e8c43e2e5578c8"},
377 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b9576723858a78751d5aacd2497b8aef29ffea6d1c95981505877f7ac28215c6"},
378 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5ba9cbd8ac6cf422f3102622b20f8552d601bf8837e49a3afed188d560152788"},
379 | {file = "matplotlib-3.8.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:03f9d160a29e0b65c0790bb07f4f45d6a181b1ac33eb1bb0dd225986450148f0"},
380 | {file = "matplotlib-3.8.2-cp311-cp311-win_amd64.whl", hash = "sha256:3773002da767f0a9323ba1a9b9b5d00d6257dbd2a93107233167cfb581f64717"},
381 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_10_12_x86_64.whl", hash = "sha256:4c318c1e95e2f5926fba326f68177dee364aa791d6df022ceb91b8221bd0a627"},
382 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:091275d18d942cf1ee9609c830a1bc36610607d8223b1b981c37d5c9fc3e46a4"},
383 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b0f3b8ea0e99e233a4bcc44590f01604840d833c280ebb8fe5554fd3e6cfe8d"},
384 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d7b1704a530395aaf73912be741c04d181f82ca78084fbd80bc737be04848331"},
385 | {file = "matplotlib-3.8.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:533b0e3b0c6768eef8cbe4b583731ce25a91ab54a22f830db2b031e83cca9213"},
386 | {file = "matplotlib-3.8.2-cp312-cp312-win_amd64.whl", hash = "sha256:0f4fc5d72b75e2c18e55eb32292659cf731d9d5b312a6eb036506304f4675630"},
387 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_10_12_x86_64.whl", hash = "sha256:deaed9ad4da0b1aea77fe0aa0cebb9ef611c70b3177be936a95e5d01fa05094f"},
388 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:172f4d0fbac3383d39164c6caafd3255ce6fa58f08fc392513a0b1d3b89c4f89"},
389 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c7d36c2209d9136cd8e02fab1c0ddc185ce79bc914c45054a9f514e44c787917"},
390 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5864bdd7da445e4e5e011b199bb67168cdad10b501750367c496420f2ad00843"},
391 | {file = "matplotlib-3.8.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:ef8345b48e95cee45ff25192ed1f4857273117917a4dcd48e3905619bcd9c9b8"},
392 | {file = "matplotlib-3.8.2-cp39-cp39-win_amd64.whl", hash = "sha256:7c48d9e221b637c017232e3760ed30b4e8d5dfd081daf327e829bf2a72c731b4"},
393 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-macosx_10_12_x86_64.whl", hash = "sha256:aa11b3c6928a1e496c1a79917d51d4cd5d04f8a2e75f21df4949eeefdf697f4b"},
394 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1095fecf99eeb7384dabad4bf44b965f929a5f6079654b681193edf7169ec20"},
395 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:bddfb1db89bfaa855912261c805bd0e10218923cc262b9159a49c29a7a1c1afa"},
396 | {file = "matplotlib-3.8.2.tar.gz", hash = "sha256:01a978b871b881ee76017152f1f1a0cbf6bd5f7b8ff8c96df0df1bd57d8755a1"},
397 | ]
398 |
399 | [package.dependencies]
400 | contourpy = ">=1.0.1"
401 | cycler = ">=0.10"
402 | fonttools = ">=4.22.0"
403 | importlib-resources = {version = ">=3.2.0", markers = "python_version < \"3.10\""}
404 | kiwisolver = ">=1.3.1"
405 | numpy = ">=1.21,<2"
406 | packaging = ">=20.0"
407 | pillow = ">=8"
408 | pyparsing = ">=2.3.1"
409 | python-dateutil = ">=2.7"
410 |
411 | [[package]]
412 | name = "numpy"
413 | version = "1.26.2"
414 | description = "Fundamental package for array computing in Python"
415 | optional = false
416 | python-versions = ">=3.9"
417 | files = [
418 | {file = "numpy-1.26.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:3703fc9258a4a122d17043e57b35e5ef1c5a5837c3db8be396c82e04c1cf9b0f"},
419 | {file = "numpy-1.26.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:cc392fdcbd21d4be6ae1bb4475a03ce3b025cd49a9be5345d76d7585aea69440"},
420 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:36340109af8da8805d8851ef1d74761b3b88e81a9bd80b290bbfed61bd2b4f75"},
421 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:bcc008217145b3d77abd3e4d5ef586e3bdfba8fe17940769f8aa09b99e856c00"},
422 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:3ced40d4e9e18242f70dd02d739e44698df3dcb010d31f495ff00a31ef6014fe"},
423 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b272d4cecc32c9e19911891446b72e986157e6a1809b7b56518b4f3755267523"},
424 | {file = "numpy-1.26.2-cp310-cp310-win32.whl", hash = "sha256:22f8fc02fdbc829e7a8c578dd8d2e15a9074b630d4da29cda483337e300e3ee9"},
425 | {file = "numpy-1.26.2-cp310-cp310-win_amd64.whl", hash = "sha256:26c9d33f8e8b846d5a65dd068c14e04018d05533b348d9eaeef6c1bd787f9919"},
426 | {file = "numpy-1.26.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:b96e7b9c624ef3ae2ae0e04fa9b460f6b9f17ad8b4bec6d7756510f1f6c0c841"},
427 | {file = "numpy-1.26.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:aa18428111fb9a591d7a9cc1b48150097ba6a7e8299fb56bdf574df650e7d1f1"},
428 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:06fa1ed84aa60ea6ef9f91ba57b5ed963c3729534e6e54055fc151fad0423f0a"},
429 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:96ca5482c3dbdd051bcd1fce8034603d6ebfc125a7bd59f55b40d8f5d246832b"},
430 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:854ab91a2906ef29dc3925a064fcd365c7b4da743f84b123002f6139bcb3f8a7"},
431 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:f43740ab089277d403aa07567be138fc2a89d4d9892d113b76153e0e412409f8"},
432 | {file = "numpy-1.26.2-cp311-cp311-win32.whl", hash = "sha256:a2bbc29fcb1771cd7b7425f98b05307776a6baf43035d3b80c4b0f29e9545186"},
433 | {file = "numpy-1.26.2-cp311-cp311-win_amd64.whl", hash = "sha256:2b3fca8a5b00184828d12b073af4d0fc5fdd94b1632c2477526f6bd7842d700d"},
434 | {file = "numpy-1.26.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:a4cd6ed4a339c21f1d1b0fdf13426cb3b284555c27ac2f156dfdaaa7e16bfab0"},
435 | {file = "numpy-1.26.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5d5244aabd6ed7f312268b9247be47343a654ebea52a60f002dc70c769048e75"},
436 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a3cdb4d9c70e6b8c0814239ead47da00934666f668426fc6e94cce869e13fd7"},
437 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:aa317b2325f7aa0a9471663e6093c210cb2ae9c0ad824732b307d2c51983d5b6"},
438 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:174a8880739c16c925799c018f3f55b8130c1f7c8e75ab0a6fa9d41cab092fd6"},
439 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f79b231bf5c16b1f39c7f4875e1ded36abee1591e98742b05d8a0fb55d8a3eec"},
440 | {file = "numpy-1.26.2-cp312-cp312-win32.whl", hash = "sha256:4a06263321dfd3598cacb252f51e521a8cb4b6df471bb12a7ee5cbab20ea9167"},
441 | {file = "numpy-1.26.2-cp312-cp312-win_amd64.whl", hash = "sha256:b04f5dc6b3efdaab541f7857351aac359e6ae3c126e2edb376929bd3b7f92d7e"},
442 | {file = "numpy-1.26.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:4eb8df4bf8d3d90d091e0146f6c28492b0be84da3e409ebef54349f71ed271ef"},
443 | {file = "numpy-1.26.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:1a13860fdcd95de7cf58bd6f8bc5a5ef81c0b0625eb2c9a783948847abbef2c2"},
444 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:64308ebc366a8ed63fd0bf426b6a9468060962f1a4339ab1074c228fa6ade8e3"},
445 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:baf8aab04a2c0e859da118f0b38617e5ee65d75b83795055fb66c0d5e9e9b818"},
446 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d73a3abcac238250091b11caef9ad12413dab01669511779bc9b29261dd50210"},
447 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:b361d369fc7e5e1714cf827b731ca32bff8d411212fccd29ad98ad622449cc36"},
448 | {file = "numpy-1.26.2-cp39-cp39-win32.whl", hash = "sha256:bd3f0091e845164a20bd5a326860c840fe2af79fa12e0469a12768a3ec578d80"},
449 | {file = "numpy-1.26.2-cp39-cp39-win_amd64.whl", hash = "sha256:2beef57fb031dcc0dc8fa4fe297a742027b954949cabb52a2a376c144e5e6060"},
450 | {file = "numpy-1.26.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:1cc3d5029a30fb5f06704ad6b23b35e11309491c999838c31f124fee32107c79"},
451 | {file = "numpy-1.26.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:94cc3c222bb9fb5a12e334d0479b97bb2df446fbe622b470928f5284ffca3f8d"},
452 | {file = "numpy-1.26.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:fe6b44fb8fcdf7eda4ef4461b97b3f63c466b27ab151bec2366db8b197387841"},
453 | {file = "numpy-1.26.2.tar.gz", hash = "sha256:f65738447676ab5777f11e6bbbdb8ce11b785e105f690bc45966574816b6d3ea"},
454 | ]
455 |
456 | [[package]]
457 | name = "packaging"
458 | version = "23.2"
459 | description = "Core utilities for Python packages"
460 | optional = false
461 | python-versions = ">=3.7"
462 | files = [
463 | {file = "packaging-23.2-py3-none-any.whl", hash = "sha256:8c491190033a9af7e1d931d0b5dacc2ef47509b34dd0de67ed209b5203fc88c7"},
464 | {file = "packaging-23.2.tar.gz", hash = "sha256:048fb0e9405036518eaaf48a55953c750c11e1a1b68e0dd1a9d62ed0c092cfc5"},
465 | ]
466 |
467 | [[package]]
468 | name = "pillow"
469 | version = "10.1.0"
470 | description = "Python Imaging Library (Fork)"
471 | optional = false
472 | python-versions = ">=3.8"
473 | files = [
474 | {file = "Pillow-10.1.0-cp310-cp310-macosx_10_10_x86_64.whl", hash = "sha256:1ab05f3db77e98f93964697c8efc49c7954b08dd61cff526b7f2531a22410106"},
475 | {file = "Pillow-10.1.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6932a7652464746fcb484f7fc3618e6503d2066d853f68a4bd97193a3996e273"},
476 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a5f63b5a68daedc54c7c3464508d8c12075e56dcfbd42f8c1bf40169061ae666"},
477 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c0949b55eb607898e28eaccb525ab104b2d86542a85c74baf3a6dc24002edec2"},
478 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:ae88931f93214777c7a3aa0a8f92a683f83ecde27f65a45f95f22d289a69e593"},
479 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:b0eb01ca85b2361b09480784a7931fc648ed8b7836f01fb9241141b968feb1db"},
480 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:d27b5997bdd2eb9fb199982bb7eb6164db0426904020dc38c10203187ae2ff2f"},
481 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:7df5608bc38bd37ef585ae9c38c9cd46d7c81498f086915b0f97255ea60c2818"},
482 | {file = "Pillow-10.1.0-cp310-cp310-win_amd64.whl", hash = "sha256:41f67248d92a5e0a2076d3517d8d4b1e41a97e2df10eb8f93106c89107f38b57"},
483 | {file = "Pillow-10.1.0-cp311-cp311-macosx_10_10_x86_64.whl", hash = "sha256:1fb29c07478e6c06a46b867e43b0bcdb241b44cc52be9bc25ce5944eed4648e7"},
484 | {file = "Pillow-10.1.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:2cdc65a46e74514ce742c2013cd4a2d12e8553e3a2563c64879f7c7e4d28bce7"},
485 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50d08cd0a2ecd2a8657bd3d82c71efd5a58edb04d9308185d66c3a5a5bed9610"},
486 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:062a1610e3bc258bff2328ec43f34244fcec972ee0717200cb1425214fe5b839"},
487 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:61f1a9d247317fa08a308daaa8ee7b3f760ab1809ca2da14ecc88ae4257d6172"},
488 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:a646e48de237d860c36e0db37ecaecaa3619e6f3e9d5319e527ccbc8151df061"},
489 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:47e5bf85b80abc03be7455c95b6d6e4896a62f6541c1f2ce77a7d2bb832af262"},
490 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:a92386125e9ee90381c3369f57a2a50fa9e6aa8b1cf1d9c4b200d41a7dd8e992"},
491 | {file = "Pillow-10.1.0-cp311-cp311-win_amd64.whl", hash = "sha256:0f7c276c05a9767e877a0b4c5050c8bee6a6d960d7f0c11ebda6b99746068c2a"},
492 | {file = "Pillow-10.1.0-cp312-cp312-macosx_10_10_x86_64.whl", hash = "sha256:a89b8312d51715b510a4fe9fc13686283f376cfd5abca8cd1c65e4c76e21081b"},
493 | {file = "Pillow-10.1.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:00f438bb841382b15d7deb9a05cc946ee0f2c352653c7aa659e75e592f6fa17d"},
494 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3d929a19f5469b3f4df33a3df2983db070ebb2088a1e145e18facbc28cae5b27"},
495 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9a92109192b360634a4489c0c756364c0c3a2992906752165ecb50544c251312"},
496 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:0248f86b3ea061e67817c47ecbe82c23f9dd5d5226200eb9090b3873d3ca32de"},
497 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:9882a7451c680c12f232a422730f986a1fcd808da0fd428f08b671237237d651"},
498 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:1c3ac5423c8c1da5928aa12c6e258921956757d976405e9467c5f39d1d577a4b"},
499 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:806abdd8249ba3953c33742506fe414880bad78ac25cc9a9b1c6ae97bedd573f"},
500 | {file = "Pillow-10.1.0-cp312-cp312-win_amd64.whl", hash = "sha256:eaed6977fa73408b7b8a24e8b14e59e1668cfc0f4c40193ea7ced8e210adf996"},
501 | {file = "Pillow-10.1.0-cp38-cp38-macosx_10_10_x86_64.whl", hash = "sha256:fe1e26e1ffc38be097f0ba1d0d07fcade2bcfd1d023cda5b29935ae8052bd793"},
502 | {file = "Pillow-10.1.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:7a7e3daa202beb61821c06d2517428e8e7c1aab08943e92ec9e5755c2fc9ba5e"},
503 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:24fadc71218ad2b8ffe437b54876c9382b4a29e030a05a9879f615091f42ffc2"},
504 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fa1d323703cfdac2036af05191b969b910d8f115cf53093125e4058f62012c9a"},
505 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:912e3812a1dbbc834da2b32299b124b5ddcb664ed354916fd1ed6f193f0e2d01"},
506 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:7dbaa3c7de82ef37e7708521be41db5565004258ca76945ad74a8e998c30af8d"},
507 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:9d7bc666bd8c5a4225e7ac71f2f9d12466ec555e89092728ea0f5c0c2422ea80"},
508 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:baada14941c83079bf84c037e2d8b7506ce201e92e3d2fa0d1303507a8538212"},
509 | {file = "Pillow-10.1.0-cp38-cp38-win_amd64.whl", hash = "sha256:2ef6721c97894a7aa77723740a09547197533146fba8355e86d6d9a4a1056b14"},
510 | {file = "Pillow-10.1.0-cp39-cp39-macosx_10_10_x86_64.whl", hash = "sha256:0a026c188be3b443916179f5d04548092e253beb0c3e2ee0a4e2cdad72f66099"},
511 | {file = "Pillow-10.1.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:04f6f6149f266a100374ca3cc368b67fb27c4af9f1cc8cb6306d849dcdf12616"},
512 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bb40c011447712d2e19cc261c82655f75f32cb724788df315ed992a4d65696bb"},
513 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1a8413794b4ad9719346cd9306118450b7b00d9a15846451549314a58ac42219"},
514 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:c9aeea7b63edb7884b031a35305629a7593272b54f429a9869a4f63a1bf04c34"},
515 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:b4005fee46ed9be0b8fb42be0c20e79411533d1fd58edabebc0dd24626882cfd"},
516 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0152565c6aa6ebbfb1e5d8624140a440f2b99bf7afaafbdbf6430426497f28"},
517 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:d921bc90b1defa55c9917ca6b6b71430e4286fc9e44c55ead78ca1a9f9eba5f2"},
518 | {file = "Pillow-10.1.0-cp39-cp39-win_amd64.whl", hash = "sha256:cfe96560c6ce2f4c07d6647af2d0f3c54cc33289894ebd88cfbb3bcd5391e256"},
519 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-macosx_10_10_x86_64.whl", hash = "sha256:937bdc5a7f5343d1c97dc98149a0be7eb9704e937fe3dc7140e229ae4fc572a7"},
520 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b1c25762197144e211efb5f4e8ad656f36c8d214d390585d1d21281f46d556ba"},
521 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:afc8eef765d948543a4775f00b7b8c079b3321d6b675dde0d02afa2ee23000b4"},
522 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:883f216eac8712b83a63f41b76ddfb7b2afab1b74abbb413c5df6680f071a6b9"},
523 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-macosx_10_10_x86_64.whl", hash = "sha256:b920e4d028f6442bea9a75b7491c063f0b9a3972520731ed26c83e254302eb1e"},
524 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1c41d960babf951e01a49c9746f92c5a7e0d939d1652d7ba30f6b3090f27e412"},
525 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:1fafabe50a6977ac70dfe829b2d5735fd54e190ab55259ec8aea4aaea412fa0b"},
526 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:3b834f4b16173e5b92ab6566f0473bfb09f939ba14b23b8da1f54fa63e4b623f"},
527 | {file = "Pillow-10.1.0.tar.gz", hash = "sha256:e6bf8de6c36ed96c86ea3b6e1d5273c53f46ef518a062464cd7ef5dd2cf92e38"},
528 | ]
529 |
530 | [package.extras]
531 | docs = ["furo", "olefile", "sphinx (>=2.4)", "sphinx-copybutton", "sphinx-inline-tabs", "sphinx-removed-in", "sphinxext-opengraph"]
532 | tests = ["check-manifest", "coverage", "defusedxml", "markdown2", "olefile", "packaging", "pyroma", "pytest", "pytest-cov", "pytest-timeout"]
533 |
534 | [[package]]
535 | name = "pluggy"
536 | version = "1.3.0"
537 | description = "plugin and hook calling mechanisms for python"
538 | optional = false
539 | python-versions = ">=3.8"
540 | files = [
541 | {file = "pluggy-1.3.0-py3-none-any.whl", hash = "sha256:d89c696a773f8bd377d18e5ecda92b7a3793cbe66c87060a6fb58c7b6e1061f7"},
542 | {file = "pluggy-1.3.0.tar.gz", hash = "sha256:cf61ae8f126ac6f7c451172cf30e3e43d3ca77615509771b3a984a0730651e12"},
543 | ]
544 |
545 | [package.extras]
546 | dev = ["pre-commit", "tox"]
547 | testing = ["pytest", "pytest-benchmark"]
548 |
549 | [[package]]
550 | name = "py"
551 | version = "1.11.0"
552 | description = "library with cross-python path, ini-parsing, io, code, log facilities"
553 | optional = false
554 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*"
555 | files = [
556 | {file = "py-1.11.0-py2.py3-none-any.whl", hash = "sha256:607c53218732647dff4acdfcd50cb62615cedf612e72d1724fb1a0cc6405b378"},
557 | {file = "py-1.11.0.tar.gz", hash = "sha256:51c75c4126074b472f746a24399ad32f6053d1b34b68d2fa41e558e6f4a98719"},
558 | ]
559 |
560 | [[package]]
561 | name = "pyparsing"
562 | version = "3.1.1"
563 | description = "pyparsing module - Classes and methods to define and execute parsing grammars"
564 | optional = false
565 | python-versions = ">=3.6.8"
566 | files = [
567 | {file = "pyparsing-3.1.1-py3-none-any.whl", hash = "sha256:32c7c0b711493c72ff18a981d24f28aaf9c1fb7ed5e9667c9e84e3db623bdbfb"},
568 | {file = "pyparsing-3.1.1.tar.gz", hash = "sha256:ede28a1a32462f5a9705e07aea48001a08f7cf81a021585011deba701581a0db"},
569 | ]
570 |
571 | [package.extras]
572 | diagrams = ["jinja2", "railroad-diagrams"]
573 |
574 | [[package]]
575 | name = "pytest"
576 | version = "6.2.5"
577 | description = "pytest: simple powerful testing with Python"
578 | optional = false
579 | python-versions = ">=3.6"
580 | files = [
581 | {file = "pytest-6.2.5-py3-none-any.whl", hash = "sha256:7310f8d27bc79ced999e760ca304d69f6ba6c6649c0b60fb0e04a4a77cacc134"},
582 | {file = "pytest-6.2.5.tar.gz", hash = "sha256:131b36680866a76e6781d13f101efb86cf674ebb9762eb70d3082b6f29889e89"},
583 | ]
584 |
585 | [package.dependencies]
586 | atomicwrites = {version = ">=1.0", markers = "sys_platform == \"win32\""}
587 | attrs = ">=19.2.0"
588 | colorama = {version = "*", markers = "sys_platform == \"win32\""}
589 | iniconfig = "*"
590 | packaging = "*"
591 | pluggy = ">=0.12,<2.0"
592 | py = ">=1.8.2"
593 | toml = "*"
594 |
595 | [package.extras]
596 | testing = ["argcomplete", "hypothesis (>=3.56)", "mock", "nose", "requests", "xmlschema"]
597 |
598 | [[package]]
599 | name = "python-dateutil"
600 | version = "2.8.2"
601 | description = "Extensions to the standard Python datetime module"
602 | optional = false
603 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,>=2.7"
604 | files = [
605 | {file = "python-dateutil-2.8.2.tar.gz", hash = "sha256:0123cacc1627ae19ddf3c27a5de5bd67ee4586fbdd6440d9748f8abb483d3e86"},
606 | {file = "python_dateutil-2.8.2-py2.py3-none-any.whl", hash = "sha256:961d03dc3453ebbc59dbdea9e4e11c5651520a876d0f4db161e8674aae935da9"},
607 | ]
608 |
609 | [package.dependencies]
610 | six = ">=1.5"
611 |
612 | [[package]]
613 | name = "six"
614 | version = "1.16.0"
615 | description = "Python 2 and 3 compatibility utilities"
616 | optional = false
617 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*"
618 | files = [
619 | {file = "six-1.16.0-py2.py3-none-any.whl", hash = "sha256:8abb2f1d86890a2dfb989f9a77cfcfd3e47c2a354b01111771326f8aa26e0254"},
620 | {file = "six-1.16.0.tar.gz", hash = "sha256:1e61c37477a1626458e36f7b1d82aa5c9b094fa4802892072e49de9c60c4c926"},
621 | ]
622 |
623 | [[package]]
624 | name = "toml"
625 | version = "0.10.2"
626 | description = "Python Library for Tom's Obvious, Minimal Language"
627 | optional = false
628 | python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
629 | files = [
630 | {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
631 | {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
632 | ]
633 |
634 | [[package]]
635 | name = "tqdm"
636 | version = "4.66.1"
637 | description = "Fast, Extensible Progress Meter"
638 | optional = false
639 | python-versions = ">=3.7"
640 | files = [
641 | {file = "tqdm-4.66.1-py3-none-any.whl", hash = "sha256:d302b3c5b53d47bce91fea46679d9c3c6508cf6332229aa1e7d8653723793386"},
642 | {file = "tqdm-4.66.1.tar.gz", hash = "sha256:d88e651f9db8d8551a62556d3cff9e3034274ca5d66e93197cf2490e2dcb69c7"},
643 | ]
644 |
645 | [package.dependencies]
646 | colorama = {version = "*", markers = "platform_system == \"Windows\""}
647 |
648 | [package.extras]
649 | dev = ["pytest (>=6)", "pytest-cov", "pytest-timeout", "pytest-xdist"]
650 | notebook = ["ipywidgets (>=6)"]
651 | slack = ["slack-sdk"]
652 | telegram = ["requests"]
653 |
654 | [[package]]
655 | name = "zipp"
656 | version = "3.17.0"
657 | description = "Backport of pathlib-compatible object wrapper for zip files"
658 | optional = false
659 | python-versions = ">=3.8"
660 | files = [
661 | {file = "zipp-3.17.0-py3-none-any.whl", hash = "sha256:0e923e726174922dce09c53c59ad483ff7bbb8e572e00c7f7c46b88556409f31"},
662 | {file = "zipp-3.17.0.tar.gz", hash = "sha256:84e64a1c28cf7e91ed2078bb8cc8c259cb19b76942096c8d7b84947690cabaf0"},
663 | ]
664 |
665 | [package.extras]
666 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
667 | testing = ["big-O", "jaraco.functools", "jaraco.itertools", "more-itertools", "pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-ignore-flaky", "pytest-mypy (>=0.9.1)", "pytest-ruff"]
668 |
669 | [metadata]
670 | lock-version = "2.0"
671 | python-versions = "^3.9"
672 | content-hash = "5e3a8b71c1d10601d5e0c2e8e72d1ea2cd79f51cbe48d1f7512dd3fc591030c2"
673 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.poetry]
2 | name = "complexcgr"
3 | version = "0.8.0"
4 | description = "complex Chaos Game Representation for DNA"
5 | authors = ["Jorge Avila
2 |
3 | # `complexcgr`
4 | This library have classes around the *Chaos Game Representation* for DNA sequence
5 |
6 | **The FCGR helps to visualize a k-mer distribution** The `FCGR` of a sequence is an image showing the distribution of the $k$-mers
7 | given a chosen $k$. The frequencies of all $k$-mers are distributed in the position of a matrix of $2^k \times 2^k$,
8 | which considers all the possible $k$-mers: $4^k$.
9 |
10 | The position that a $k$-mer uses in the matrix depends on the encoding given by the `CGR`.
11 |
12 | Some examples of [bacterial assemblies](https://zenodo.org/records/4602622) ([see reference](https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001421)) are shown below.
13 | The name of the species and the `sample_id` is in the title of each image ([see an example with the first image](https://www.ebi.ac.uk/ena/browser/view/SAMEA2658585)). These images were
14 | created using the 6-mers of each assembly and the class `FCGR` of this library.
15 |
16 | |  |
17 | |:--:|
18 | |10 different species of bacteria represented by their FCGR (6-mers)| -->
19 |
20 | ## Installation
21 | [pypi](https://pypi.org/project/complexcgr/)
22 | ___
23 | ```shell
24 | pip install complexcgr
25 | ```
26 |
27 | to update to the latest version
28 | ```shell
29 | pip install complexcgr --upgrade
30 | ```
31 |
32 | ## How to use
33 | ___
34 | ### 1. `CGR` Chaos Game Representation of DNA
35 | ```python
36 | from complexcgr import CGR
37 |
38 | # Instantiate class CGR
39 | cgr = CGR()
40 |
41 | # encode a sequence
42 | cgr.encode("ACGT")
43 | # > CGRCoords(N=4, x=0.1875, y=-0.5625)
44 |
45 | # recover a sequence from CGR coordinates
46 | cgr.decode(N=4,x=0.1875,y=-0.5625)
47 | # > "ACGT"
48 | ```
49 |
50 | ### 2. `FCGR` Frequency Matrix of Chaos Game Representation of DNA
51 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
52 | will not be considered for the calculation of the frequency matrix CGR
53 | ```python
54 | import random; random.seed(42)
55 | from complexcgr import FCGR
56 |
57 | # set the k-mer
58 | fcgr = FCGR(k=8) # (256x256) array
59 |
60 | # Generate a random sequence without T's
61 | seq = "".join(random.choice("ACG") for _ in range(300_000))
62 | chaos = fcgr(seq) # an array with the frequencies of each k-mer
63 | fcgr.plot(chaos)
64 | ```
65 | |  |
66 | |:--:|
67 | |FCGR representation for a sequence without T's|
68 |
69 |
70 | You can save the image with
71 | ```python
72 | fcgr.save_img(chaos, path="img/ACG.jpg")
73 | ```
74 | *Formats allowed are defined by PIL.*
75 |
76 | You can also generate the image in 16 (or more bits), to avoid losing information of k-mer frequencies
77 | ```python
78 | # Generate image in 16-bits (default is 8-bits)
79 | fcgr = FCGR(k=8, bits=16) # (256x256) array. When using plot() it will be rescaled to [0,65535] colors
80 | ```
81 |
82 |
83 | ```python
84 | # Generate a random sequence without T's and lots of N's
85 | seq = "".join(random.choice("ACGN") for _ in range(300_000))
86 | chaos = fcgr(seq) # an array with the probabilities of each k-mer
87 | fcgr.plot(chaos)
88 | ```
89 |
90 |
91 | ||
92 | |:--:|
93 | |FCGR representation for a sequence without T's and lots of N's|
94 |
95 |
96 |
97 | ### 3. `iCGR` integer Chaos Game Representation of DNA
98 | ```python
99 | from complexcgr import iCGR
100 |
101 | # Instantiate class CGR
102 | icgr = iCGR()
103 |
104 | # encode a sequence
105 | icgr.encode("ACGT")
106 | # > CGRCoords(N=4, x=3, y=-9)
107 |
108 | # recover a sequence from CGR coordinates
109 | icgr.decode(N=4,x=3,y=-9)
110 | # > "ACGT"
111 | ```
112 |
113 | ### 4. `ComplexCGR` Complex Chaos Game Representation of DNA (ComplexCGR)
114 |
115 | ```python
116 | from complexcgr import ComplexCGR
117 |
118 | # Instantiate class CGR
119 | ccgr = ComplexCGR()
120 |
121 | # encode a sequence
122 | ccgr.encode("ACGT")
123 | # > CGRCoords(k=228,N=4)
124 |
125 | # recover a sequence from ComplexCGR coordinates
126 | ccgr.decode(k=228,N=4)
127 | # > "ACGT"
128 |
129 | ```
130 |
131 | ### 5. `ComplexFCGR` Frequency Matrix of Complex Chaos Game Representation of DNA
132 | Input for FCGR only accept sequences in $\{A,C,G,T,N\}$, but all $k$-mers that contains an $N$
133 | will not be considered for the calculation of the frequency matrix CGR
134 | ```python
135 | import random; random.seed(42)
136 | from complexcgr import FCGR
137 |
138 | # set the k-mer desired
139 | cfcgr = ComplexFCGR(k=8) # 8-mers
140 |
141 | # Generate a random sequence without T's
142 | seq = "".join(random.choice("ACG") for _ in range(300_000))
143 | fig = cfcgr(seq)
144 |
145 | ```
146 | |  |
147 | |:--:|
148 | |ComplexFCGR representation for a sequence without T's|
149 |
150 |
151 | You can save the image with
152 | ```python
153 | cfcgr.save(fig, path="img/ACG-ComplexCGR.png")
154 | ```
155 | *Currently the plot must be saved as png*
156 |
157 | ___
158 | ## Advice for Real applications
159 |
160 | **Count k-mers** could be the bottleneck for large sequences (> 100000 bp).
161 | Note that the class `FCGR` (and `ComplexCGR`) has implemented a naive approach to count k-mers, this is intended since in practice state-of-the-art tools like KMC or Jellyfish are used to count k-mers very efficiently.
162 |
163 | We provide the class `FCGRKmc`, that receives as input the file generated by the following pipeline using [KMC3](https://github.com/refresh-bio/KMC)
164 |
165 | Make sure to have `kmc` installed. One recommended way is to create a conda environment and [install it there](https://anaconda.org/bioconda/kmc)
166 |
167 | ```bash
168 | kmer_size=6
169 | input="path/to/sequence.fa"
170 | output="path/to/count-kmers.txt"
171 |
172 | mkdir -p tmp-kmc
173 | kmc -v -k$kmer_size -m4 -sm -ci0 -cs100000 -b -t4 -fa $input $input "tmp-kmc"
174 | kmc_tools -t4 -v transform $input dump $output
175 | rm -r $input.kmc_pre $input.kmc_suf
176 | ```
177 | the output file `path/to/count-kmers.txt` can be used with `FCGRKmc`
178 |
179 | ```python
180 | from complexcgr import FCGRKmc
181 |
182 | kmer = 6
183 | fcgr = FCGRKmc(kmer)
184 |
185 | arr = fcgr("path/to/count-kmers.txt") # k-mer counts ordered in a matrix of 2^k x 2^k
186 |
187 |
188 | # to visualize the distribution of k-mers.
189 | # Frequencies are scaled between [min, max] values.
190 | # White color corresponds to the minimum value of frequency
191 | # Black color corresponds to the maximum value of frequency
192 | fcgr.plot(arr)
193 |
194 | # Save it with numpy
195 | import numpy as np
196 | np.save("path_save/fcgr.npy",arr)
197 | ```
198 |
199 | ___
200 | # Videos
201 |
202 | **CGR encoding**
203 |
204 | [](https://youtu.be/HU15ge0fkOY)
205 |
206 | **CGR encoding of all k-mers**
207 |
208 | [](https://youtu.be/oYLT11Q9n5M)
209 |
210 | **ComplexCGR encoding**
211 |
212 | [](https://youtu.be/xKyXplS5KFk)
213 |
214 | **ComplexCGR and Symmetry**
215 |
216 | [](https://www.youtube.com/YmcxVId4_4w)
217 |
218 |
219 | # Functionalities/TODO list
220 | ___
221 | > version 0.8.0:
222 | A list of available classes and functionalities are listed below:
223 |
224 |
225 | **Encoders**
226 | The encoders are functions that map a sequence $s \in \{A,C,G,T\}$ to a point in the plane.
227 | `CGR`, `iCGR`, and `ComplexCGR`.
228 |
229 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
230 | - [x] encode a sequence.
231 | - [x] recover a sequence from a CGR encoding.
232 |
233 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
234 |
235 | `CGR` Chaos Game Representation: encodes a DNA sequence in 3 numbers $(N,x,y)$
236 | - [x] encode a sequence.
237 | - [x] recover a sequence from a CGR encoding.
238 |
239 | `iCGR` integer CGR: encodes a DNA sequence in 3 integers $(N,x,y)$.
240 | - [x] encode a sequence
241 | - [x] recover a sequence from an iCGR encoding
242 |
243 | `ComplexCGR`: encodes a DNA sequence in 2 integers $(k,N)$.
244 | - [x] encode a sequence
245 | - [x] recover a sequence from a ComplexCGR encoding
246 | - [x] plot sequence of ComplexCGR encodings
247 |
248 | **Image for distribution of k-mers**
249 |
250 | - [x] `FCGR` Frequency Matrix CGR: representation as an image for k-mer representativity, based on CGR.
251 | - [x] generates FCGR from an arbitrary n-long sequence.
252 | - [x] plot FCGR.
253 | - [x] save FCGR generated.
254 | - [x] save FCGR in different bits.
255 | - [x] `FCGRKmc` Same as `FCGR` but receives as input the file with k-mer counts generated with [KMC](https://github.com/refresh-bio/KMC)
256 | - [x] `ComplexFCGR`: Frequency ComplexCGR: representation as an image (circle) for k-mer representativity, based on ComplexCGR.
257 | - [x] generates ComplexFCGR from an arbitrary n-long sequence.
258 | - [x] plot ComplexFCGR.
259 | - [x] save ComplexFCGR generated.
260 |
261 | - [ ] `PercentileFCGR`:
262 | - [A universal DNA signature for the Tree of Life. EcoEvoRxiv](https://doi.org/10.32942/X24891)
263 | - [ ] `SpacedFCGR`: Create FCGR from spaced-mers
264 | - [Spaced seeds improve k-mer-based metagenomic classification, Bioninformatics](https://academic.oup.com/bioinformatics/article/31/22/3584/240663)
265 | - [Effects of spaced k-mers on alignment-free genotyping](https://academic.oup.com/bioinformatics/article/39/Supplement_1/i213/7210439)
266 |
267 | # Author
268 |
269 | `complexcgr` is developed by [Jorge Avila Cartes](https://github.com/jorgeavilacartes/)
270 |
271 |
272 | # Related publications
273 |
274 | - [Accurate and fast clade assignment via deep learning and frequency chaos game representation](https://doi.org/10.1093/gigascience/giac119)
275 | - [PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases](https://doi.org/10.1101/2025.03.19.644115)
--------------------------------------------------------------------------------
/README.rst:
--------------------------------------------------------------------------------
1 | # complexCGR
2 |
3 | A complex representation for DNA, that preserves some properties of the CGR from the 'Chaos Game Representation for gene structure' by Jeffrey in 1990.
4 |
5 | Each nucleotide will be assigned to one quadrant (I,II,III and IV). This distribution will be the same for the CGR and complexCGR implementations (I:A,II:C,III:G,IV:T)
--------------------------------------------------------------------------------
/complexCGR.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/complexCGR.pdf
--------------------------------------------------------------------------------
/complexcgr/__init__.py:
--------------------------------------------------------------------------------
1 | __version__ ="0.8.0"
2 |
3 | # encodings
4 | from .cgr import CGR
5 | from .icgr import iCGR
6 | from .complexcgr import ComplexCGR
7 |
8 | # from k-mers
9 | from .fcgr import FCGR
10 | from .fcgr_kmc import FCGRKmc
11 | from .complexfcgr import ComplexFCGR
--------------------------------------------------------------------------------
/complexcgr/cgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from typing import Dict, Optional
3 | from collections import namedtuple
4 |
5 | # coordinates for x+iy
6 | Coord = namedtuple("Coord", ["x","y"])
7 |
8 | # coordinates for a CGR encoding
9 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
10 |
11 | # coordinates for each nucleotide in the 2d-plane
12 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
13 |
14 | class CGR:
15 | "Chaos Game Representation for DNA"
16 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
17 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
18 | self.cgr_coords = CGRCoords(0,0,0)
19 |
20 | def nucleotide_by_coords(self,x,y):
21 | "Get nucleotide by coordinates (x,y)"
22 | # filter nucleotide by coordinates
23 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
24 |
25 | return list(filtered.keys())[0]
26 |
27 | def forward(self, nucleotide: str):
28 | "Compute next CGR coordinates"
29 | x = (self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x)/2
30 | y = (self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y)/2
31 |
32 | # update cgr_coords
33 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y)
34 |
35 | def backward(self,):
36 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
37 | # get current nucleotide based on coordinates
38 | n_x,n_y = self.coords_current_nucleotide()
39 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
40 |
41 | # update coordinates to the previous one
42 | x = 2*self.cgr_coords.x - n_x
43 | y = 2*self.cgr_coords.y - n_y
44 |
45 | # update cgr_coords
46 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
47 |
48 | return nucleotide
49 |
50 | def coords_current_nucleotide(self,):
51 | x = 1 if self.cgr_coords.x>0 else -1
52 | y = 1 if self.cgr_coords.y>0 else -1
53 | return x,y
54 |
55 | def encode(self, sequence: str):
56 | "From DNA sequence to CGR"
57 | # reset starting position to (0,0,0)
58 | self.reset_coords()
59 | for nucleotide in sequence:
60 | self.forward(nucleotide)
61 | return self.cgr_coords
62 |
63 | def reset_coords(self,):
64 | self.cgr_coords = CGRCoords(0,0,0)
65 |
66 | def decode(self, N:int, x:int, y:int)->str:
67 | "From CGR to DNA sequence"
68 | self.cgr_coords = CGRCoords(N,x,y)
69 |
70 | # decoded sequence
71 | sequence = []
72 |
73 | # Recover the entire genome
74 | while self.cgr_coords.N>0:
75 | nucleotide = self.backward()
76 | sequence.append(nucleotide)
77 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/complexcgr/complexcgr.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List
2 | from collections import namedtuple
3 |
4 | # coordinates for x+iy
5 | Coord = namedtuple("Coord", ["x","y"])
6 |
7 | # coordinates for a CGR encoding
8 | CGRCoords = namedtuple("CGRCoord", ["k","N"])
9 |
10 | class ComplexCGR:
11 | "Complex Chaos Game Representation of DNA"
12 | def __init__(self, nucleotide_order: List[str] = ["A","C","G","T"]):
13 | self.nucleotide_order = nucleotide_order
14 | self.cgr_coords = CGRCoords(0,0) # complexCGR coordinates
15 |
16 | def id(self, nucleotide):
17 | return self.nucleotide_order.index(nucleotide)
18 |
19 | def forward(self, nucleotide: str):
20 | "compute next complexCGR coordinates"
21 | k = self.id(nucleotide)*4**(self.cgr_coords.N) + self.cgr_coords.k
22 |
23 | # update cgr_coords
24 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N+1)
25 |
26 | def backward(self,):
27 | "compute last complexCGR coordinates"
28 | nucleotide = self.current_nucleotide()
29 |
30 | # compute previous k
31 | k = self.cgr_coords.k - self.id(nucleotide)*4**(self.cgr_coords.N-1)
32 |
33 | # update cgr_coords
34 | self.cgr_coords = CGRCoords(k,self.cgr_coords.N-1)
35 |
36 | return nucleotide
37 |
38 | def encode(self, sequence: str):
39 | "From DNA to complexCGR"
40 | self.reset_coords()
41 | for nucleotide in sequence:
42 | self.forward(nucleotide)
43 | return self.cgr_coords
44 |
45 | def decode(self, k: int, N: int):
46 | "From complexCGR to DNA"
47 | self.cgr_coords = CGRCoords(k,N)
48 |
49 | # decoded sequence
50 | sequence = []
51 |
52 | # Recover the entire genome
53 | while self.cgr_coords.N>0:
54 | nucleotide = self.backward()
55 | sequence.append(nucleotide)
56 | return "".join(sequence[::-1])
57 |
58 | def current_nucleotide(self,):
59 | "Get current nucleotide based on k and N"
60 | k,N = self.cgr_coords.k, self.cgr_coords.N
61 | alpha = k/4**N
62 | if alpha <0.25:
63 | return self.nucleotide_order[0]
64 | elif alpha <0.5:
65 | return self.nucleotide_order[1]
66 | elif alpha < 0.75:
67 | return self.nucleotide_order[2]
68 | else:
69 | return self.nucleotide_order[3]
70 |
71 | def reset_coords(self,):
72 | self.cgr_coords = CGRCoords(0,0)
73 |
--------------------------------------------------------------------------------
/complexcgr/complexfcgr.py:
--------------------------------------------------------------------------------
1 | from . import ComplexCGR
2 | import matplotlib.pyplot as plt
3 | from itertools import product
4 | from collections import defaultdict
5 | from tqdm import tqdm
6 | from PIL import Image
7 | import numpy as np
8 |
9 |
10 | class ComplexFCGR(ComplexCGR):
11 | """Circular density plot based on CGR"""
12 |
13 | def __init__(self, k: int):
14 | super().__init__()
15 | self.k = k # k-mer representation
16 | self.kmers = product("ACGT", repeat=self.k) # all kmers of length k
17 | self.freq_kmer = None # dict to save representativity of each kmer
18 | self.probabilities = None # dict to save probabilities/density for each kmer
19 | self.fig = None # to save matlotlib figure and then save as image
20 |
21 | def __call__(self, sequence: str, w=1):
22 | self.count_kmers(sequence)
23 | self.kmer_probabilities(sequence)
24 | self.plot(w)
25 |
26 | def count_kmers(self, sequence: str):
27 | self.freq_kmer = defaultdict(int)
28 | # representativity of kmers
29 | len_seq = len(sequence)
30 | for j,_ in enumerate(sequence):
31 | if j+self.k <= len_seq:
32 | subseq = sequence[j:j+self.k]
33 | if "N" not in subseq:
34 | self.freq_kmer[subseq] +=1
35 |
36 | def kmer_probabilities(self, sequence: str):
37 | self.probabilities = defaultdict(float)
38 | N=len(sequence)
39 | for key, value in self.freq_kmer.items():
40 | self.probabilities[key] = float(value) / (N - self.k + 1)
41 |
42 | def plot(self, w: int = 1):
43 | "Given a FCGR, plot it in grayscale"
44 | ax = plt.subplot(111, polar=True)
45 | center, bottom, width, height = self.compute_input_plot()
46 |
47 | # scale width by 'w'
48 | width = [w*_ for _ in width]
49 | print("generating plot")
50 | ax.bar(x=center, # center of the angle
51 | width=width, # width of the angle
52 | bottom=bottom, # lowest value
53 | height=height, # highest value
54 | )
55 |
56 | ax.axes.get_xaxis().set_visible(False)
57 | ax.axes.get_yaxis().set_visible(False)
58 |
59 | return ax.figure
60 |
61 | def save(self, ccgr, path: str):
62 | "save complexFCGR as image"
63 | # get figure
64 | fig = self.plot()
65 |
66 | # transform figure to Image
67 | img = self.fig2img(fig)
68 |
69 | # save image
70 | img.save(path)
71 | plt.close(fig)
72 |
73 | @staticmethod
74 | def fig2img(fig):
75 | "Convert a Matplotlib figure to a PIL Image and return it"
76 | #https://stackoverflow.com/questions/57316491/how-to-convert-matplotlib-figure-to-pil-image-object-without-saving-image
77 | import io
78 | buf = io.BytesIO()
79 | fig.savefig(buf)
80 | buf.seek(0)
81 | img = Image.open(buf)
82 | return img
83 |
84 |
85 | def compute_input_plot(self,):
86 | "Compute input for plot CFCGR"
87 | delta = 2*np.pi/4**self.k # angle between consecutive roots
88 |
89 | center = []
90 | width = []
91 | height = []
92 | bottom = []
93 |
94 | for kmer in tqdm(self.probabilities):
95 | h = self.probabilities.get(kmer,0.0) # height (density) of the bar in the circle
96 | theta = 2*self.encode(kmer).k*np.pi/4**self.k # angle of k-esim root
97 | c = theta + delta/2 # center of the angle
98 | center.append(c)
99 | bottom.append(0)
100 | width.append(50*delta)
101 | height.append(h)
102 |
103 | return center, bottom, width, height
--------------------------------------------------------------------------------
/complexcgr/fcgr.py:
--------------------------------------------------------------------------------
1 | from . import CGR
2 | from PIL import Image
3 | from itertools import product
4 | from collections import defaultdict
5 | import numpy as np
6 |
7 | NUC_COMPLEMENT = {n:c for n,c in zip ("ACGT","TGCA")}
8 |
9 | class FCGR(CGR):
10 | """Frequency matrix CGR
11 | an (2**k x 2**k) 2D representation will be created for a
12 | n-long sequence.
13 | - k represents the k-mer.
14 | - 2**k x 2**k = 4**k the total number of k-mers (sequences of length k)
15 | """
16 |
17 | def __init__(self, k: int, use_canonical_kmers: bool = False ,bits: int = 8):
18 | super().__init__()
19 | self.k = k # k-mer representation
20 | self.use_canonical_kmers = use_canonical_kmers
21 | self.kmers = list("".join(kmer) for kmer in product("ACGT", repeat=self.k))
22 |
23 | if use_canonical_kmers is True:
24 | self.kmer2pixel = self._kmer2pixel_canonical_kmers()
25 | else:
26 | self.kmer2pixel = self.kmer2pixel_position()
27 |
28 | self.bits = bits
29 | self.max_color = 2**bits-1
30 |
31 | def __call__(self, sequence: str):
32 | "Given a DNA sequence, returns an array with his FCGR"
33 | self.count_kmers(sequence)
34 |
35 | # Create an empty array to save the FCGR values
36 | array_size = int(2**self.k)
37 | fcgr = np.zeros((array_size,array_size))
38 |
39 | # Assign frequency to each box in the matrix
40 | for kmer, freq in self.freq_kmer.items():
41 | pos_x, pos_y = self.kmer2pixel[kmer]
42 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
43 | return fcgr
44 |
45 | def count_kmer(self, kmer):
46 | if "N" not in kmer:
47 | self.freq_kmer[kmer] += 1
48 |
49 | def count_kmers(self, sequence: str):
50 | self.freq_kmer = defaultdict(int)
51 | # representativity of kmers
52 | last_j = len(sequence) - self.k + 1
53 | kmers = (sequence[i:(i+self.k)] for i in range(last_j))
54 | # count kmers in a dictionary
55 | list(self.count_kmer(kmer) for kmer in kmers)
56 |
57 | def pixel_position(self, kmer: str):
58 | "Get pixel position in the FCGR matrix for a k-mer"
59 |
60 | coords = self.encode(kmer)
61 | N,x,y = coords.N, coords.x, coords.y
62 |
63 | # Coordinates from [-1,1]² to [1,2**k]²
64 | np_coords = np.array([(x + 1)/2, (y + 1)/2]) # move coordinates from [-1,1]² to [0,1]²
65 | np_coords *= 2**self.k # rescale coordinates from [0,1]² to [0,2**k]²
66 | x,y = np.ceil(np_coords) # round to upper integer
67 |
68 | # Turn coordinates (cx,cy) into pixel (px,py) position
69 | # px = 2**k-cy+1, py = cx
70 | return 2**self.k-int(y)+1, int(x)
71 |
72 | def kmer2pixel_position(self,):
73 | kmer2pixel = dict()
74 | for kmer in self.kmers:
75 | kmer2pixel[kmer] = self.pixel_position(kmer)
76 | return kmer2pixel
77 |
78 | def plot(self, fcgr):
79 | "Given a FCGR, plot it in grayscale"
80 | img_pil = self.array2img(fcgr)
81 | return img_pil
82 |
83 | def save_img(self, fcgr, path: str):
84 | "Save image in grayscale for the FCGR provided as input"
85 | img_pil = self.array2img(fcgr)
86 | img_pil.save(path)
87 |
88 | def array2img(self, array):
89 | "Array to PIL image"
90 | m, M = array.min(), array.max()
91 | # rescale to [0,1]
92 | img_rescaled = (array - m) / (M-m)
93 |
94 | # invert colors black->white
95 | img_array = np.ceil(self.max_color - img_rescaled*self.max_color)
96 | dtype = eval(f"np.int{self.bits}")
97 | img_array = np.array(img_array, dtype=dtype)
98 |
99 | # convert to Image
100 | img_pil = Image.fromarray(img_array,'L')
101 | return img_pil
102 |
103 | # # --------------- canonical k-mers ---------------- # #
104 | @staticmethod
105 | def reverse_complement(kmer: str):
106 | rev_kmer = list(kmer)[::-1]
107 | return "".join([NUC_COMPLEMENT[n] for n in rev_kmer])
108 |
109 | def get_canonical_kmer(self, kmer: str):
110 | rev_complement = self.reverse_complement(kmer)
111 | return kmer if kmer < rev_complement else rev_complement
112 |
113 | def _kmer2pixel_canonical_kmers(self,):
114 | # change pixel position of non-canonical kmers
115 | kmer2pixel = dict()
116 | for kmer in self.kmers:
117 | canonical_kmer = self.get_canonical_kmer(kmer)
118 | if kmer != canonical_kmer:
119 | kmer2pixel[kmer] = kmer2pixel[canonical_kmer]
120 | return kmer2pixel
--------------------------------------------------------------------------------
/complexcgr/fcgr_kmc.py:
--------------------------------------------------------------------------------
1 | from . import FCGR
2 |
3 | import gzip
4 | import numpy as np
5 |
6 | class FCGRKmc(FCGR):
7 | """
8 | Create FCGR with the option of using canonical kmers from KMC output
9 | """
10 | def __init__(self, k: int, use_canonical_kmers: bool=False):
11 | super().__init__(k, use_canonical_kmers)
12 | self.k = k # k-mer representation
13 | self.use_canonical_kmers = use_canonical_kmers
14 |
15 | def __call__(self, path_kmc_output):
16 | "Given a path to a kmc output file, return the FCGR using canonical kmers as an array"
17 | # Create an empty array to save the FCGR values
18 | array_size = int(2**self.k)
19 | fcgr = np.zeros((array_size,array_size))
20 |
21 | if str(path_kmc_output).endswith(".txt"):
22 | with open(path_kmc_output) as fp:
23 | for line in fp:
24 | kmer, freq = line.split("\t")
25 | pos_x, pos_y = self.kmer2pixel[kmer]
26 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
27 | else:
28 | with gzip.open(path_kmc_output,'rt') as f:
29 | for line in f:
30 | line = line.strip()
31 | kmer, freq = line.split()
32 | pos_x, pos_y = self.kmer2pixel[kmer]
33 | fcgr[int(pos_x)-1,int(pos_y)-1] += int(freq)
34 |
35 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/fcgr_samples.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from Bio import SeqIO
3 | from collections import defaultdict
4 | from pathlib import Path
5 | from tqdm import tqdm
6 | from typing import List, Union
7 | from complexcgr import FCGR
8 |
9 | # input for FCGR Samples
10 | _path_fastq = Union[str, Path] # path can be a string or a Path instance
11 | _fastq = Union[_path_fastq,List[_path_fastq]] # either a single path or a list of paths to fastq files
12 |
13 | class FCGRSamples(FCGR):
14 |
15 | def __init__(self, k: int, bits: int = 8):
16 | super().__init__(k, bits)
17 |
18 | def __call__(self, path_fastq: _fastq, consider_quality: bool = False):
19 | "Given a (list) of fastq files, return the FCGR matrix"
20 |
21 | ## ---- New for reads ----
22 |
23 | # for each call, we start to count the frequencies
24 | self.freq_kmer = defaultdict(int)
25 | if consider_quality is True:
26 | self.qual_kmer = defaultdict(int)
27 |
28 | # transform to list to iterate
29 | path_fastq = path_fastq if type(path_fastq) is list else [path_fastq]
30 |
31 | # For each file, count kmers on their reads
32 | for path in path_fastq:
33 | fastq = self.load_fastq(path)
34 | n_reads = self.count_reads(path)
35 |
36 | # count kmers in each read
37 | for sample in tqdm(fastq,total=n_reads,desc=f"Counting kmers on {str(Path(path).stem)}"):
38 | read = sample.seq
39 | if consider_quality is True:
40 | qual = sample.letter_annotations["phred_quality"]
41 | self.count_kmers_qualities(str(read),qual)
42 | else:
43 | self.count_kmers(str(read))
44 |
45 | ## ---- End new for reads ----
46 |
47 | # Create an empty array to save the FCGR values
48 | if consider_quality is False:
49 | array_size = int(2**self.k)
50 | fcgr = np.zeros((array_size,array_size))
51 | else:
52 | array_size = int(2**self.k)
53 | fcgr = np.zeros((array_size,array_size,2))
54 |
55 | # Assign frequency to each box in the matrix
56 | for kmer, freq in self.freq_kmer.items():
57 | pos_x, pos_y = self.kmer2pixel[kmer]
58 |
59 | if consider_quality is True:
60 | fcgr[int(pos_x)-1,int(pos_y)-1,0] = freq
61 | fcgr[int(pos_x)-1,int(pos_y)-1,1] = self.qual_kmer[kmer]
62 | else:
63 | fcgr[int(pos_x)-1,int(pos_y)-1] = freq
64 |
65 | return fcgr if consider_quality is False else self.rescale_fcgr_qualities(fcgr)
66 |
67 |
68 | def load_fastq(self, path):
69 | "Load a fastq file"
70 | fastq = SeqIO.parse(str(path), "fastq")
71 | return fastq
72 |
73 | def count_kmers(self, read: str):
74 | # representativity of kmers
75 | last_j = len(read) - self.k + 1
76 | kmers = (read[i:(i+self.k)] for i in range(last_j))
77 | # count kmers in a dictionary
78 | list(self.count_kmer(kmer) for kmer in kmers)
79 |
80 | @staticmethod
81 | def count_reads(path):
82 | "Count reads in a file to define progress bar"
83 | n_reads = 0
84 | with open(str(path)) as fp:
85 | for line in fp:
86 | if line.startswith("@"):
87 | n_reads += 1
88 | return n_reads
89 |
90 | def count_kmer(self, kmer):
91 | if "N" not in kmer:
92 | self.freq_kmer[kmer] += 1
93 |
94 |
95 | def count_kmer_quality(self, kmer, qmer):
96 | """Count kmers and qualities
97 | For quality of a kmer, the mean of qualities for each nucleotide in the kmer will be saved"""
98 | if "N" not in kmer:
99 | self.freq_kmer[kmer] += 1
100 | self.qual_kmer[kmer] += qmer.mean()
101 |
102 | def count_kmers_qualities(self,read,qual):
103 | "Count kmers and qualities"
104 |
105 | # representativity of kmers
106 | last_j = len(read) - self.k + 1
107 | kmers = (read[i:(i+self.k)] for i in range(last_j))
108 | qmers = (np.array(qual[i:(i+self.k)]) for i in range(last_j)) # qualities for each kmer
109 |
110 | # count kmers and qualities in a dictionary
111 | list(self.count_kmer_quality(kmer,qmer) for (kmer,qmer) in zip(kmers,qmers))
112 |
113 | @staticmethod
114 | def rescale_fcgr_qualities(fcgr):
115 | "Divide each cumulate quality by the freq of its kmer"
116 | freqs, quals = fcgr[:,:,0], fcgr[:,:,1]
117 | fcgr[:,:,1] = np.divide(quals, freqs, out = np.zeros_like(quals), where = quals!=0)
118 | return fcgr
--------------------------------------------------------------------------------
/complexcgr/icgr.py:
--------------------------------------------------------------------------------
1 | "From original work: CGR for gene structure"
2 | from itertools import product
3 | from tqdm import tqdm
4 | from typing import Dict, Optional
5 | from collections import defaultdict, namedtuple
6 | import numpy as np
7 |
8 | # coordinates for x+iy
9 | Coord = namedtuple("Coord", ["x","y"])
10 |
11 | # coordinates for a CGR encoding
12 | CGRCoords = namedtuple("CGRCoords", ["N","x","y"])
13 |
14 | # coordinates for each nucleotide in the 2d-plane
15 | DEFAULT_COORDS = dict(A=Coord(1,1),C=Coord(-1,1),G=Coord(-1,-1),T=Coord(1,-1))
16 |
17 | class iCGR:
18 | "integer Chaos Game Representation for DNA"
19 | def __init__(self, coords: Optional[Dict[chr,tuple]]=None):
20 | self.nucleotide_coords = DEFAULT_COORDS if coords is None else coords
21 | self.cgr_coords = CGRCoords(0,0,0)
22 |
23 | def nucleotide_by_coords(self,x,y):
24 | "Get nucleotide by coordinates (x,y)"
25 | # filter nucleotide by coordinates
26 | filtered = dict(filter(lambda item: item[1] == Coord(x,y), self.nucleotide_coords.items()))
27 |
28 | return list(filtered.keys())[0]
29 |
30 | def forward(self, nucleotide: str):
31 | "Compute next CGR coordinates"
32 | # current length
33 | N = self.cgr_coords.N
34 |
35 | # compute next coordinates
36 | x = self.cgr_coords.x + self.nucleotide_coords.get(nucleotide).x * 2**N
37 | y = self.cgr_coords.y + self.nucleotide_coords.get(nucleotide).y * 2**N
38 |
39 | # update cgr_coords: iCGR starts in the corner of the first nucleotide
40 |
41 | if N>0:
42 | self.cgr_coords = CGRCoords(self.cgr_coords.N+1,x,y,)
43 | else:
44 | self.cgr_coords = CGRCoords(1,
45 | self.nucleotide_coords.get(nucleotide).x,
46 | self.nucleotide_coords.get(nucleotide).y,
47 | )
48 |
49 | def backward(self,):
50 | "Compute last CGR coordinates. Current nucleotide can be inferred from (x,y)"
51 | # get current nucleotide based on coordinates
52 | n_x,n_y = self.coords_current_nucleotide()
53 | nucleotide = self.nucleotide_by_coords(n_x,n_y)
54 |
55 | # current length
56 | N = self.cgr_coords.N
57 |
58 | # update coordinates to the previous one
59 | x = self.cgr_coords.x - self.nucleotide_coords.get(nucleotide).x * 2**(N-1)
60 | y = self.cgr_coords.y - self.nucleotide_coords.get(nucleotide).y * 2**(N-1)
61 |
62 | # update cgr_coords
63 | self.cgr_coords = CGRCoords(self.cgr_coords.N-1,x,y)
64 |
65 | return nucleotide
66 |
67 | def coords_current_nucleotide(self,):
68 | x = 1 if self.cgr_coords.x>0 else -1
69 | y = 1 if self.cgr_coords.y>0 else -1
70 | return x,y
71 |
72 | def encode(self, sequence: str):
73 | "From DNA sequence to CGR"
74 | # reset starting position to (0,0,0)
75 | self.reset_coords()
76 | for nucleotide in sequence:
77 | self.forward(nucleotide)
78 | return self.cgr_coords
79 |
80 | def reset_coords(self,):
81 | self.cgr_coords = CGRCoords(0,0,0)
82 |
83 | def decode(self, N:int, x:int, y:int)->str:
84 | "From CGR to DNA sequence"
85 | self.cgr_coords = CGRCoords(N,x,y)
86 |
87 | # decoded sequence
88 | sequence = []
89 |
90 | # Recover the entire genome
91 | while self.cgr_coords.N>0:
92 | nucleotide = self.backward()
93 | sequence.append(nucleotide)
94 | return "".join(sequence[::-1])
--------------------------------------------------------------------------------
/img/ACG-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG-complexCGR.png
--------------------------------------------------------------------------------
/img/ACG.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG.jpg
--------------------------------------------------------------------------------
/img/ACG_16bits.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/ACG_16bits.jpg
--------------------------------------------------------------------------------
/img/CGA.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGA.jpg
--------------------------------------------------------------------------------
/img/CGAN.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/CGAN.jpg
--------------------------------------------------------------------------------
/img/SAMD00003784.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00003784.jpeg
--------------------------------------------------------------------------------
/img/SAMD00032020.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00032020.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111209.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111209.jpeg
--------------------------------------------------------------------------------
/img/SAMD00111306.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/SAMD00111306.jpeg
--------------------------------------------------------------------------------
/img/complexcgr-readme.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/complexcgr-readme.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v2.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3-nb.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3-nb.png
--------------------------------------------------------------------------------
/img/logo-complexCGR-v3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR-v3.png
--------------------------------------------------------------------------------
/img/logo-complexCGR.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/AlgoLab/complexCGR/930a3804af8d43f81db3a383390ad56d5afaa72d/img/logo-complexCGR.png
--------------------------------------------------------------------------------
/poetry.lock:
--------------------------------------------------------------------------------
1 | # This file is automatically @generated by Poetry 1.7.1 and should not be changed by hand.
2 |
3 | [[package]]
4 | name = "atomicwrites"
5 | version = "1.4.1"
6 | description = "Atomic file writes."
7 | optional = false
8 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*"
9 | files = [
10 | {file = "atomicwrites-1.4.1.tar.gz", hash = "sha256:81b2c9071a49367a7f770170e5eec8cb66567cfbbc8c73d20ce5ca4a8d71cf11"},
11 | ]
12 |
13 | [[package]]
14 | name = "attrs"
15 | version = "23.2.0"
16 | description = "Classes Without Boilerplate"
17 | optional = false
18 | python-versions = ">=3.7"
19 | files = [
20 | {file = "attrs-23.2.0-py3-none-any.whl", hash = "sha256:99b87a485a5820b23b879f04c2305b44b951b502fd64be915879d77a7e8fc6f1"},
21 | {file = "attrs-23.2.0.tar.gz", hash = "sha256:935dc3b529c262f6cf76e50877d35a4bd3c1de194fd41f47a2b7ae8f19971f30"},
22 | ]
23 |
24 | [package.extras]
25 | cov = ["attrs[tests]", "coverage[toml] (>=5.3)"]
26 | dev = ["attrs[tests]", "pre-commit"]
27 | docs = ["furo", "myst-parser", "sphinx", "sphinx-notfound-page", "sphinxcontrib-towncrier", "towncrier", "zope-interface"]
28 | tests = ["attrs[tests-no-zope]", "zope-interface"]
29 | tests-mypy = ["mypy (>=1.6)", "pytest-mypy-plugins"]
30 | tests-no-zope = ["attrs[tests-mypy]", "cloudpickle", "hypothesis", "pympler", "pytest (>=4.3.0)", "pytest-xdist[psutil]"]
31 |
32 | [[package]]
33 | name = "biopython"
34 | version = "1.82"
35 | description = "Freely available tools for computational molecular biology."
36 | optional = false
37 | python-versions = ">=3.8"
38 | files = [
39 | {file = "biopython-1.82-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:713be5e1e8571ea151864544dfcd2637eaf98c67a6e47b69781d325feb02f6b9"},
40 | {file = "biopython-1.82-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5bc908e84278bf61ac901a0c1b695c85ce49491957e0ab07e1e2afc216882525"},
41 | {file = "biopython-1.82-cp310-cp310-win32.whl", hash = "sha256:f53aff30f01ac9a8aa1fd42dab347a96ee5312945098dfa38f9a5125f6e913fd"},
42 | {file = "biopython-1.82-cp310-cp310-win_amd64.whl", hash = "sha256:0f3f685a2d85348b8c84c6eebe58a74cef7e1158ac52ae5aa91789436907f0e1"},
43 | {file = "biopython-1.82-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:c9bc080b7dc6cd6acbf2576766136e8e54acbcc6b7a29ad8075cb5a73f4cfb97"},
44 | {file = "biopython-1.82-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c7802b5033edaf34bd2fbd96b87f96999a6808caa0064c91087c7f1340624898"},
45 | {file = "biopython-1.82-cp311-cp311-win32.whl", hash = "sha256:ec35893b32458f6fa0c98d5f02d801a5fe76961ac66473455f51c0575562c594"},
46 | {file = "biopython-1.82-cp311-cp311-win_amd64.whl", hash = "sha256:40297d418bb142a7ea1a47faedc24bbe21b0ba455f3805373264470e5cb6ec5c"},
47 | {file = "biopython-1.82-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:96f77bd6540683c8958e4ebf55b58e8caa26d20db72891915d06670b783e9cdb"},
48 | {file = "biopython-1.82-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:49b829c2276a0711106663af632c7e203695a5cf082d488d6e10b8aca5ec63e4"},
49 | {file = "biopython-1.82-cp312-cp312-win32.whl", hash = "sha256:54a9b200d4960ed2a4ec85faac2124ffa278afa58d22e07e2d15ca7509219ea1"},
50 | {file = "biopython-1.82-cp312-cp312-win_amd64.whl", hash = "sha256:762547037038d42c9ac98877fc32d4aeba798077129df7790cb178256171a4e5"},
51 | {file = "biopython-1.82-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:cb013256742d31e24fd5093d55511f58bc6a93b8ac723d2ca3b33c7904f1ca14"},
52 | {file = "biopython-1.82-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:16829760fab60ba17141b7a81494fed837c6f29ea28cdcfb3070764c8e8f2ff1"},
53 | {file = "biopython-1.82-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:c6af82894dd4f736d737fdea62008a19966251ba39aef39a0ce8b41321305ad5"},
54 | {file = "biopython-1.82-cp38-cp38-win32.whl", hash = "sha256:a1ff583b2f8b314a4c2d612aad4f4b73827a5f741b391f3a834b0c192a4f1527"},
55 | {file = "biopython-1.82-cp38-cp38-win_amd64.whl", hash = "sha256:0198f23776d9cb9d514a797e81a024f486adac973b252a2f4696995a5a163745"},
56 | {file = "biopython-1.82-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:c88e9f87d1b07958e7e1e25ff20efced4796daf650e5290692b5c3ff9aebf9d3"},
57 | {file = "biopython-1.82-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:8e2533db9275c3e1438151260844e7683f91308da320df02a4c28fb7053ea3a9"},
58 | {file = "biopython-1.82-cp39-cp39-win32.whl", hash = "sha256:80f48d220b92e747300281602f52e98ec6c537ddd4921b580d9a0368bae05ff5"},
59 | {file = "biopython-1.82-cp39-cp39-win_amd64.whl", hash = "sha256:73d619611bd7cf9d2ad8b3217b08d3796acfabb765b0771ad2ff7ee2e46a59b0"},
60 | {file = "biopython-1.82.tar.gz", hash = "sha256:a9b10d959ae88a9744a91c6ce3601f4c86e7ec41679bc93c29f679218f6167bb"},
61 | ]
62 |
63 | [package.dependencies]
64 | numpy = "*"
65 |
66 | [[package]]
67 | name = "colorama"
68 | version = "0.4.6"
69 | description = "Cross-platform colored terminal text."
70 | optional = false
71 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,!=3.3.*,!=3.4.*,!=3.5.*,!=3.6.*,>=2.7"
72 | files = [
73 | {file = "colorama-0.4.6-py2.py3-none-any.whl", hash = "sha256:4f1d9991f5acc0ca119f9d443620b77f9d6b33703e51011c16baf57afb285fc6"},
74 | {file = "colorama-0.4.6.tar.gz", hash = "sha256:08695f5cb7ed6e0531a20572697297273c47b8cae5a63ffc6d6ed5c201be6e44"},
75 | ]
76 |
77 | [[package]]
78 | name = "contourpy"
79 | version = "1.2.0"
80 | description = "Python library for calculating contours of 2D quadrilateral grids"
81 | optional = false
82 | python-versions = ">=3.9"
83 | files = [
84 | {file = "contourpy-1.2.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:0274c1cb63625972c0c007ab14dd9ba9e199c36ae1a231ce45d725cbcbfd10a8"},
85 | {file = "contourpy-1.2.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:ab459a1cbbf18e8698399c595a01f6dcc5c138220ca3ea9e7e6126232d102bb4"},
86 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6fdd887f17c2f4572ce548461e4f96396681212d858cae7bd52ba3310bc6f00f"},
87 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:5d16edfc3fc09968e09ddffada434b3bf989bf4911535e04eada58469873e28e"},
88 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1c203f617abc0dde5792beb586f827021069fb6d403d7f4d5c2b543d87edceb9"},
89 | {file = "contourpy-1.2.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b69303ceb2e4d4f146bf82fda78891ef7bcd80c41bf16bfca3d0d7eb545448aa"},
90 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:884c3f9d42d7218304bc74a8a7693d172685c84bd7ab2bab1ee567b769696df9"},
91 | {file = "contourpy-1.2.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:4a1b1208102be6e851f20066bf0e7a96b7d48a07c9b0cfe6d0d4545c2f6cadab"},
92 | {file = "contourpy-1.2.0-cp310-cp310-win32.whl", hash = "sha256:34b9071c040d6fe45d9826cbbe3727d20d83f1b6110d219b83eb0e2a01d79488"},
93 | {file = "contourpy-1.2.0-cp310-cp310-win_amd64.whl", hash = "sha256:bd2f1ae63998da104f16a8b788f685e55d65760cd1929518fd94cd682bf03e41"},
94 | {file = "contourpy-1.2.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:dd10c26b4eadae44783c45ad6655220426f971c61d9b239e6f7b16d5cdaaa727"},
95 | {file = "contourpy-1.2.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:5c6b28956b7b232ae801406e529ad7b350d3f09a4fde958dfdf3c0520cdde0dd"},
96 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ebeac59e9e1eb4b84940d076d9f9a6cec0064e241818bcb6e32124cc5c3e377a"},
97 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:139d8d2e1c1dd52d78682f505e980f592ba53c9f73bd6be102233e358b401063"},
98 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:1e9dc350fb4c58adc64df3e0703ab076f60aac06e67d48b3848c23647ae4310e"},
99 | {file = "contourpy-1.2.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:18fc2b4ed8e4a8fe849d18dce4bd3c7ea637758c6343a1f2bae1e9bd4c9f4686"},
100 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:16a7380e943a6d52472096cb7ad5264ecee36ed60888e2a3d3814991a0107286"},
101 | {file = "contourpy-1.2.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:8d8faf05be5ec8e02a4d86f616fc2a0322ff4a4ce26c0f09d9f7fb5330a35c95"},
102 | {file = "contourpy-1.2.0-cp311-cp311-win32.whl", hash = "sha256:67b7f17679fa62ec82b7e3e611c43a016b887bd64fb933b3ae8638583006c6d6"},
103 | {file = "contourpy-1.2.0-cp311-cp311-win_amd64.whl", hash = "sha256:99ad97258985328b4f207a5e777c1b44a83bfe7cf1f87b99f9c11d4ee477c4de"},
104 | {file = "contourpy-1.2.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:575bcaf957a25d1194903a10bc9f316c136c19f24e0985a2b9b5608bdf5dbfe0"},
105 | {file = "contourpy-1.2.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:9e6c93b5b2dbcedad20a2f18ec22cae47da0d705d454308063421a3b290d9ea4"},
106 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:464b423bc2a009088f19bdf1f232299e8b6917963e2b7e1d277da5041f33a779"},
107 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:68ce4788b7d93e47f84edd3f1f95acdcd142ae60bc0e5493bfd120683d2d4316"},
108 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:3d7d1f8871998cdff5d2ff6a087e5e1780139abe2838e85b0b46b7ae6cc25399"},
109 | {file = "contourpy-1.2.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:6e739530c662a8d6d42c37c2ed52a6f0932c2d4a3e8c1f90692ad0ce1274abe0"},
110 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:247b9d16535acaa766d03037d8e8fb20866d054d3c7fbf6fd1f993f11fc60ca0"},
111 | {file = "contourpy-1.2.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:461e3ae84cd90b30f8d533f07d87c00379644205b1d33a5ea03381edc4b69431"},
112 | {file = "contourpy-1.2.0-cp312-cp312-win32.whl", hash = "sha256:1c2559d6cffc94890b0529ea7eeecc20d6fadc1539273aa27faf503eb4656d8f"},
113 | {file = "contourpy-1.2.0-cp312-cp312-win_amd64.whl", hash = "sha256:491b1917afdd8638a05b611a56d46587d5a632cabead889a5440f7c638bc6ed9"},
114 | {file = "contourpy-1.2.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5fd1810973a375ca0e097dee059c407913ba35723b111df75671a1976efa04bc"},
115 | {file = "contourpy-1.2.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:999c71939aad2780f003979b25ac5b8f2df651dac7b38fb8ce6c46ba5abe6ae9"},
116 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b7caf9b241464c404613512d5594a6e2ff0cc9cb5615c9475cc1d9b514218ae8"},
117 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:266270c6f6608340f6c9836a0fb9b367be61dde0c9a9a18d5ece97774105ff3e"},
118 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:dbd50d0a0539ae2e96e537553aff6d02c10ed165ef40c65b0e27e744a0f10af8"},
119 | {file = "contourpy-1.2.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:11f8d2554e52f459918f7b8e6aa20ec2a3bce35ce95c1f0ef4ba36fbda306df5"},
120 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:ce96dd400486e80ac7d195b2d800b03e3e6a787e2a522bfb83755938465a819e"},
121 | {file = "contourpy-1.2.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6d3364b999c62f539cd403f8123ae426da946e142312a514162adb2addd8d808"},
122 | {file = "contourpy-1.2.0-cp39-cp39-win32.whl", hash = "sha256:1c88dfb9e0c77612febebb6ac69d44a8d81e3dc60f993215425b62c1161353f4"},
123 | {file = "contourpy-1.2.0-cp39-cp39-win_amd64.whl", hash = "sha256:78e6ad33cf2e2e80c5dfaaa0beec3d61face0fb650557100ee36db808bfa6843"},
124 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:be16975d94c320432657ad2402f6760990cb640c161ae6da1363051805fa8108"},
125 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b95a225d4948b26a28c08307a60ac00fb8671b14f2047fc5476613252a129776"},
126 | {file = "contourpy-1.2.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:0d7e03c0f9a4f90dc18d4e77e9ef4ec7b7bbb437f7f675be8e530d65ae6ef956"},
127 | {file = "contourpy-1.2.0.tar.gz", hash = "sha256:171f311cb758de7da13fc53af221ae47a5877be5a0843a9fe150818c51ed276a"},
128 | ]
129 |
130 | [package.dependencies]
131 | numpy = ">=1.20,<2.0"
132 |
133 | [package.extras]
134 | bokeh = ["bokeh", "selenium"]
135 | docs = ["furo", "sphinx (>=7.2)", "sphinx-copybutton"]
136 | mypy = ["contourpy[bokeh,docs]", "docutils-stubs", "mypy (==1.6.1)", "types-Pillow"]
137 | test = ["Pillow", "contourpy[test-no-images]", "matplotlib"]
138 | test-no-images = ["pytest", "pytest-cov", "pytest-xdist", "wurlitzer"]
139 |
140 | [[package]]
141 | name = "cycler"
142 | version = "0.12.1"
143 | description = "Composable style cycles"
144 | optional = false
145 | python-versions = ">=3.8"
146 | files = [
147 | {file = "cycler-0.12.1-py3-none-any.whl", hash = "sha256:85cef7cff222d8644161529808465972e51340599459b8ac3ccbac5a854e0d30"},
148 | {file = "cycler-0.12.1.tar.gz", hash = "sha256:88bb128f02ba341da8ef447245a9e138fae777f6a23943da4540077d3601eb1c"},
149 | ]
150 |
151 | [package.extras]
152 | docs = ["ipython", "matplotlib", "numpydoc", "sphinx"]
153 | tests = ["pytest", "pytest-cov", "pytest-xdist"]
154 |
155 | [[package]]
156 | name = "fonttools"
157 | version = "4.47.0"
158 | description = "Tools to manipulate font files"
159 | optional = false
160 | python-versions = ">=3.8"
161 | files = [
162 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:2d2404107626f97a221dc1a65b05396d2bb2ce38e435f64f26ed2369f68675d9"},
163 | {file = "fonttools-4.47.0-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:c01f409be619a9a0f5590389e37ccb58b47264939f0e8d58bfa1f3ba07d22671"},
164 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:d986b66ff722ef675b7ee22fbe5947a41f60a61a4da15579d5e276d897fbc7fa"},
165 | {file = "fonttools-4.47.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e8acf6dd0434b211b3bd30d572d9e019831aae17a54016629fa8224783b22df8"},
166 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:495369c660e0c27233e3c572269cbe520f7f4978be675f990f4005937337d391"},
167 | {file = "fonttools-4.47.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:c59227d7ba5b232281c26ae04fac2c73a79ad0e236bca5c44aae904a18f14faf"},
168 | {file = "fonttools-4.47.0-cp310-cp310-win32.whl", hash = "sha256:59a6c8b71a245800e923cb684a2dc0eac19c56493e2f896218fcf2571ed28984"},
169 | {file = "fonttools-4.47.0-cp310-cp310-win_amd64.whl", hash = "sha256:52c82df66201f3a90db438d9d7b337c7c98139de598d0728fb99dab9fd0495ca"},
170 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:854421e328d47d70aa5abceacbe8eef231961b162c71cbe7ff3f47e235e2e5c5"},
171 | {file = "fonttools-4.47.0-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:511482df31cfea9f697930f61520f6541185fa5eeba2fa760fe72e8eee5af88b"},
172 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ce0e2c88c8c985b7b9a7efcd06511fb0a1fe3ddd9a6cd2895ef1dbf9059719d7"},
173 | {file = "fonttools-4.47.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:e7a0a8848726956e9d9fb18c977a279013daadf0cbb6725d2015a6dd57527992"},
174 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:e869da810ae35afb3019baa0d0306cdbab4760a54909c89ad8904fa629991812"},
175 | {file = "fonttools-4.47.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:dd23848f877c3754f53a4903fb7a593ed100924f9b4bff7d5a4e2e8a7001ae11"},
176 | {file = "fonttools-4.47.0-cp311-cp311-win32.whl", hash = "sha256:bf1810635c00f7c45d93085611c995fc130009cec5abdc35b327156aa191f982"},
177 | {file = "fonttools-4.47.0-cp311-cp311-win_amd64.whl", hash = "sha256:61df4dee5d38ab65b26da8efd62d859a1eef7a34dcbc331299a28e24d04c59a7"},
178 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:e3f4d61f3a8195eac784f1d0c16c0a3105382c1b9a74d99ac4ba421da39a8826"},
179 | {file = "fonttools-4.47.0-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:174995f7b057e799355b393e97f4f93ef1f2197cbfa945e988d49b2a09ecbce8"},
180 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:ea592e6a09b71cb7a7661dd93ac0b877a6228e2d677ebacbad0a4d118494c86d"},
181 | {file = "fonttools-4.47.0-cp312-cp312-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:40bdbe90b33897d9cc4a39f8e415b0fcdeae4c40a99374b8a4982f127ff5c767"},
182 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:843509ae9b93db5aaf1a6302085e30bddc1111d31e11d724584818f5b698f500"},
183 | {file = "fonttools-4.47.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:9acfa1cdc479e0dde528b61423855913d949a7f7fe09e276228298fef4589540"},
184 | {file = "fonttools-4.47.0-cp312-cp312-win32.whl", hash = "sha256:66c92ec7f95fd9732550ebedefcd190a8d81beaa97e89d523a0d17198a8bda4d"},
185 | {file = "fonttools-4.47.0-cp312-cp312-win_amd64.whl", hash = "sha256:e8fa20748de55d0021f83754b371432dca0439e02847962fc4c42a0e444c2d78"},
186 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:c75e19971209fbbce891ebfd1b10c37320a5a28e8d438861c21d35305aedb81c"},
187 | {file = "fonttools-4.47.0-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:e79f1a3970d25f692bbb8c8c2637e621a66c0d60c109ab48d4a160f50856deff"},
188 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:562681188c62c024fe2c611b32e08b8de2afa00c0c4e72bed47c47c318e16d5c"},
189 | {file = "fonttools-4.47.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:a77a60315c33393b2bd29d538d1ef026060a63d3a49a9233b779261bad9c3f71"},
190 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:b4fabb8cc9422efae1a925160083fdcbab8fdc96a8483441eb7457235df625bd"},
191 | {file = "fonttools-4.47.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:2a78dba8c2a1e9d53a0fb5382979f024200dc86adc46a56cbb668a2249862fda"},
192 | {file = "fonttools-4.47.0-cp38-cp38-win32.whl", hash = "sha256:e6b968543fde4119231c12c2a953dcf83349590ca631ba8216a8edf9cd4d36a9"},
193 | {file = "fonttools-4.47.0-cp38-cp38-win_amd64.whl", hash = "sha256:4a9a51745c0439516d947480d4d884fa18bd1458e05b829e482b9269afa655bc"},
194 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:62d8ddb058b8e87018e5dc26f3258e2c30daad4c87262dfeb0e2617dd84750e6"},
195 | {file = "fonttools-4.47.0-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:5dde0eab40faaa5476133123f6a622a1cc3ac9b7af45d65690870620323308b4"},
196 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:f4da089f6dfdb822293bde576916492cd708c37c2501c3651adde39804630538"},
197 | {file = "fonttools-4.47.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:253bb46bab970e8aae254cebf2ae3db98a4ef6bd034707aa68a239027d2b198d"},
198 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:1193fb090061efa2f9e2d8d743ae9850c77b66746a3b32792324cdce65784154"},
199 | {file = "fonttools-4.47.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:084511482dd265bce6dca24c509894062f0117e4e6869384d853f46c0e6d43be"},
200 | {file = "fonttools-4.47.0-cp39-cp39-win32.whl", hash = "sha256:97620c4af36e4c849e52661492e31dc36916df12571cb900d16960ab8e92a980"},
201 | {file = "fonttools-4.47.0-cp39-cp39-win_amd64.whl", hash = "sha256:e77bdf52185bdaf63d39f3e1ac3212e6cfa3ab07d509b94557a8902ce9c13c82"},
202 | {file = "fonttools-4.47.0-py3-none-any.whl", hash = "sha256:d6477ba902dd2d7adda7f0fd3bfaeb92885d45993c9e1928c9f28fc3961415f7"},
203 | {file = "fonttools-4.47.0.tar.gz", hash = "sha256:ec13a10715eef0e031858c1c23bfaee6cba02b97558e4a7bfa089dba4a8c2ebf"},
204 | ]
205 |
206 | [package.extras]
207 | all = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "fs (>=2.2.0,<3)", "lxml (>=4.0,<5)", "lz4 (>=1.7.4.2)", "matplotlib", "munkres", "pycairo", "scipy", "skia-pathops (>=0.5.0)", "sympy", "uharfbuzz (>=0.23.0)", "unicodedata2 (>=15.1.0)", "xattr", "zopfli (>=0.1.4)"]
208 | graphite = ["lz4 (>=1.7.4.2)"]
209 | interpolatable = ["munkres", "pycairo", "scipy"]
210 | lxml = ["lxml (>=4.0,<5)"]
211 | pathops = ["skia-pathops (>=0.5.0)"]
212 | plot = ["matplotlib"]
213 | repacker = ["uharfbuzz (>=0.23.0)"]
214 | symfont = ["sympy"]
215 | type1 = ["xattr"]
216 | ufo = ["fs (>=2.2.0,<3)"]
217 | unicode = ["unicodedata2 (>=15.1.0)"]
218 | woff = ["brotli (>=1.0.1)", "brotlicffi (>=0.8.0)", "zopfli (>=0.1.4)"]
219 |
220 | [[package]]
221 | name = "importlib-resources"
222 | version = "6.1.1"
223 | description = "Read resources from Python packages"
224 | optional = false
225 | python-versions = ">=3.8"
226 | files = [
227 | {file = "importlib_resources-6.1.1-py3-none-any.whl", hash = "sha256:e8bf90d8213b486f428c9c39714b920041cb02c184686a3dee24905aaa8105d6"},
228 | {file = "importlib_resources-6.1.1.tar.gz", hash = "sha256:3893a00122eafde6894c59914446a512f728a0c1a45f9bb9b63721b6bacf0b4a"},
229 | ]
230 |
231 | [package.dependencies]
232 | zipp = {version = ">=3.1.0", markers = "python_version < \"3.10\""}
233 |
234 | [package.extras]
235 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
236 | testing = ["pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-mypy (>=0.9.1)", "pytest-ruff", "zipp (>=3.17)"]
237 |
238 | [[package]]
239 | name = "iniconfig"
240 | version = "2.0.0"
241 | description = "brain-dead simple config-ini parsing"
242 | optional = false
243 | python-versions = ">=3.7"
244 | files = [
245 | {file = "iniconfig-2.0.0-py3-none-any.whl", hash = "sha256:b6a85871a79d2e3b22d2d1b94ac2824226a63c6b741c88f7ae975f18b6778374"},
246 | {file = "iniconfig-2.0.0.tar.gz", hash = "sha256:2d91e135bf72d31a410b17c16da610a82cb55f6b0477d1a902134b24a455b8b3"},
247 | ]
248 |
249 | [[package]]
250 | name = "kiwisolver"
251 | version = "1.4.5"
252 | description = "A fast implementation of the Cassowary constraint solver"
253 | optional = false
254 | python-versions = ">=3.7"
255 | files = [
256 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_universal2.whl", hash = "sha256:05703cf211d585109fcd72207a31bb170a0f22144d68298dc5e61b3c946518af"},
257 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:146d14bebb7f1dc4d5fbf74f8a6cb15ac42baadee8912eb84ac0b3b2a3dc6ac3"},
258 | {file = "kiwisolver-1.4.5-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6ef7afcd2d281494c0a9101d5c571970708ad911d028137cd558f02b851c08b4"},
259 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:9eaa8b117dc8337728e834b9c6e2611f10c79e38f65157c4c38e9400286f5cb1"},
260 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:ec20916e7b4cbfb1f12380e46486ec4bcbaa91a9c448b97023fde0d5bbf9e4ff"},
261 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:39b42c68602539407884cf70d6a480a469b93b81b7701378ba5e2328660c847a"},
262 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:aa12042de0171fad672b6c59df69106d20d5596e4f87b5e8f76df757a7c399aa"},
263 | {file = "kiwisolver-1.4.5-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:2a40773c71d7ccdd3798f6489aaac9eee213d566850a9533f8d26332d626b82c"},
264 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:19df6e621f6d8b4b9c4d45f40a66839294ff2bb235e64d2178f7522d9170ac5b"},
265 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_i686.whl", hash = "sha256:83d78376d0d4fd884e2c114d0621624b73d2aba4e2788182d286309ebdeed770"},
266 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_ppc64le.whl", hash = "sha256:e391b1f0a8a5a10ab3b9bb6afcfd74f2175f24f8975fb87ecae700d1503cdee0"},
267 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_s390x.whl", hash = "sha256:852542f9481f4a62dbb5dd99e8ab7aedfeb8fb6342349a181d4036877410f525"},
268 | {file = "kiwisolver-1.4.5-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:59edc41b24031bc25108e210c0def6f6c2191210492a972d585a06ff246bb79b"},
269 | {file = "kiwisolver-1.4.5-cp310-cp310-win32.whl", hash = "sha256:a6aa6315319a052b4ee378aa171959c898a6183f15c1e541821c5c59beaa0238"},
270 | {file = "kiwisolver-1.4.5-cp310-cp310-win_amd64.whl", hash = "sha256:d0ef46024e6a3d79c01ff13801cb19d0cad7fd859b15037aec74315540acc276"},
271 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_universal2.whl", hash = "sha256:11863aa14a51fd6ec28688d76f1735f8f69ab1fabf388851a595d0721af042f5"},
272 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:8ab3919a9997ab7ef2fbbed0cc99bb28d3c13e6d4b1ad36e97e482558a91be90"},
273 | {file = "kiwisolver-1.4.5-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:fcc700eadbbccbf6bc1bcb9dbe0786b4b1cb91ca0dcda336eef5c2beed37b797"},
274 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:dfdd7c0b105af050eb3d64997809dc21da247cf44e63dc73ff0fd20b96be55a9"},
275 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:76c6a5964640638cdeaa0c359382e5703e9293030fe730018ca06bc2010c4437"},
276 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:bbea0db94288e29afcc4c28afbf3a7ccaf2d7e027489c449cf7e8f83c6346eb9"},
277 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:ceec1a6bc6cab1d6ff5d06592a91a692f90ec7505d6463a88a52cc0eb58545da"},
278 | {file = "kiwisolver-1.4.5-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:040c1aebeda72197ef477a906782b5ab0d387642e93bda547336b8957c61022e"},
279 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:f91de7223d4c7b793867797bacd1ee53bfe7359bd70d27b7b58a04efbb9436c8"},
280 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_i686.whl", hash = "sha256:faae4860798c31530dd184046a900e652c95513796ef51a12bc086710c2eec4d"},
281 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_ppc64le.whl", hash = "sha256:b0157420efcb803e71d1b28e2c287518b8808b7cf1ab8af36718fd0a2c453eb0"},
282 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_s390x.whl", hash = "sha256:06f54715b7737c2fecdbf140d1afb11a33d59508a47bf11bb38ecf21dc9ab79f"},
283 | {file = "kiwisolver-1.4.5-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:fdb7adb641a0d13bdcd4ef48e062363d8a9ad4a182ac7647ec88f695e719ae9f"},
284 | {file = "kiwisolver-1.4.5-cp311-cp311-win32.whl", hash = "sha256:bb86433b1cfe686da83ce32a9d3a8dd308e85c76b60896d58f082136f10bffac"},
285 | {file = "kiwisolver-1.4.5-cp311-cp311-win_amd64.whl", hash = "sha256:6c08e1312a9cf1074d17b17728d3dfce2a5125b2d791527f33ffbe805200a355"},
286 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_universal2.whl", hash = "sha256:32d5cf40c4f7c7b3ca500f8985eb3fb3a7dfc023215e876f207956b5ea26632a"},
287 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:f846c260f483d1fd217fe5ed7c173fb109efa6b1fc8381c8b7552c5781756192"},
288 | {file = "kiwisolver-1.4.5-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5ff5cf3571589b6d13bfbfd6bcd7a3f659e42f96b5fd1c4830c4cf21d4f5ef45"},
289 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:7269d9e5f1084a653d575c7ec012ff57f0c042258bf5db0954bf551c158466e7"},
290 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:da802a19d6e15dffe4b0c24b38b3af68e6c1a68e6e1d8f30148c83864f3881db"},
291 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:3aba7311af82e335dd1e36ffff68aaca609ca6290c2cb6d821a39aa075d8e3ff"},
292 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:763773d53f07244148ccac5b084da5adb90bfaee39c197554f01b286cf869228"},
293 | {file = "kiwisolver-1.4.5-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:2270953c0d8cdab5d422bee7d2007f043473f9d2999631c86a223c9db56cbd16"},
294 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:d099e745a512f7e3bbe7249ca835f4d357c586d78d79ae8f1dcd4d8adeb9bda9"},
295 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_i686.whl", hash = "sha256:74db36e14a7d1ce0986fa104f7d5637aea5c82ca6326ed0ec5694280942d1162"},
296 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_ppc64le.whl", hash = "sha256:7e5bab140c309cb3a6ce373a9e71eb7e4873c70c2dda01df6820474f9889d6d4"},
297 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_s390x.whl", hash = "sha256:0f114aa76dc1b8f636d077979c0ac22e7cd8f3493abbab152f20eb8d3cda71f3"},
298 | {file = "kiwisolver-1.4.5-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:88a2df29d4724b9237fc0c6eaf2a1adae0cdc0b3e9f4d8e7dc54b16812d2d81a"},
299 | {file = "kiwisolver-1.4.5-cp312-cp312-win32.whl", hash = "sha256:72d40b33e834371fd330fb1472ca19d9b8327acb79a5821d4008391db8e29f20"},
300 | {file = "kiwisolver-1.4.5-cp312-cp312-win_amd64.whl", hash = "sha256:2c5674c4e74d939b9d91dda0fae10597ac7521768fec9e399c70a1f27e2ea2d9"},
301 | {file = "kiwisolver-1.4.5-cp37-cp37m-macosx_10_9_x86_64.whl", hash = "sha256:3a2b053a0ab7a3960c98725cfb0bf5b48ba82f64ec95fe06f1d06c99b552e130"},
302 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3cd32d6c13807e5c66a7cbb79f90b553642f296ae4518a60d8d76243b0ad2898"},
303 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:59ec7b7c7e1a61061850d53aaf8e93db63dce0c936db1fda2658b70e4a1be709"},
304 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:da4cfb373035def307905d05041c1d06d8936452fe89d464743ae7fb8371078b"},
305 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:2400873bccc260b6ae184b2b8a4fec0e4082d30648eadb7c3d9a13405d861e89"},
306 | {file = "kiwisolver-1.4.5-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:1b04139c4236a0f3aff534479b58f6f849a8b351e1314826c2d230849ed48985"},
307 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_aarch64.whl", hash = "sha256:4e66e81a5779b65ac21764c295087de82235597a2293d18d943f8e9e32746265"},
308 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_i686.whl", hash = "sha256:7931d8f1f67c4be9ba1dd9c451fb0eeca1a25b89e4d3f89e828fe12a519b782a"},
309 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_ppc64le.whl", hash = "sha256:b3f7e75f3015df442238cca659f8baa5f42ce2a8582727981cbfa15fee0ee205"},
310 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_s390x.whl", hash = "sha256:bbf1d63eef84b2e8c89011b7f2235b1e0bf7dacc11cac9431fc6468e99ac77fb"},
311 | {file = "kiwisolver-1.4.5-cp37-cp37m-musllinux_1_1_x86_64.whl", hash = "sha256:4c380469bd3f970ef677bf2bcba2b6b0b4d5c75e7a020fb863ef75084efad66f"},
312 | {file = "kiwisolver-1.4.5-cp37-cp37m-win32.whl", hash = "sha256:9408acf3270c4b6baad483865191e3e582b638b1654a007c62e3efe96f09a9a3"},
313 | {file = "kiwisolver-1.4.5-cp37-cp37m-win_amd64.whl", hash = "sha256:5b94529f9b2591b7af5f3e0e730a4e0a41ea174af35a4fd067775f9bdfeee01a"},
314 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_universal2.whl", hash = "sha256:11c7de8f692fc99816e8ac50d1d1aef4f75126eefc33ac79aac02c099fd3db71"},
315 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_10_9_x86_64.whl", hash = "sha256:53abb58632235cd154176ced1ae8f0d29a6657aa1aa9decf50b899b755bc2b93"},
316 | {file = "kiwisolver-1.4.5-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:88b9f257ca61b838b6f8094a62418421f87ac2a1069f7e896c36a7d86b5d4c29"},
317 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3195782b26fc03aa9c6913d5bad5aeb864bdc372924c093b0f1cebad603dd712"},
318 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:fc579bf0f502e54926519451b920e875f433aceb4624a3646b3252b5caa9e0b6"},
319 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:5a580c91d686376f0f7c295357595c5a026e6cbc3d77b7c36e290201e7c11ecb"},
320 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_i686.manylinux1_i686.whl", hash = "sha256:cfe6ab8da05c01ba6fbea630377b5da2cd9bcbc6338510116b01c1bc939a2c18"},
321 | {file = "kiwisolver-1.4.5-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.whl", hash = "sha256:d2e5a98f0ec99beb3c10e13b387f8db39106d53993f498b295f0c914328b1333"},
322 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:a51a263952b1429e429ff236d2f5a21c5125437861baeed77f5e1cc2d2c7c6da"},
323 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_i686.whl", hash = "sha256:3edd2fa14e68c9be82c5b16689e8d63d89fe927e56debd6e1dbce7a26a17f81b"},
324 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_ppc64le.whl", hash = "sha256:74d1b44c6cfc897df648cc9fdaa09bc3e7679926e6f96df05775d4fb3946571c"},
325 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_s390x.whl", hash = "sha256:76d9289ed3f7501012e05abb8358bbb129149dbd173f1f57a1bf1c22d19ab7cc"},
326 | {file = "kiwisolver-1.4.5-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:92dea1ffe3714fa8eb6a314d2b3c773208d865a0e0d35e713ec54eea08a66250"},
327 | {file = "kiwisolver-1.4.5-cp38-cp38-win32.whl", hash = "sha256:5c90ae8c8d32e472be041e76f9d2f2dbff4d0b0be8bd4041770eddb18cf49a4e"},
328 | {file = "kiwisolver-1.4.5-cp38-cp38-win_amd64.whl", hash = "sha256:c7940c1dc63eb37a67721b10d703247552416f719c4188c54e04334321351ced"},
329 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_universal2.whl", hash = "sha256:9407b6a5f0d675e8a827ad8742e1d6b49d9c1a1da5d952a67d50ef5f4170b18d"},
330 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:15568384086b6df3c65353820a4473575dbad192e35010f622c6ce3eebd57af9"},
331 | {file = "kiwisolver-1.4.5-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:0dc9db8e79f0036e8173c466d21ef18e1befc02de8bf8aa8dc0813a6dc8a7046"},
332 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:cdc8a402aaee9a798b50d8b827d7ecf75edc5fb35ea0f91f213ff927c15f4ff0"},
333 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:6c3bd3cde54cafb87d74d8db50b909705c62b17c2099b8f2e25b461882e544ff"},
334 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:955e8513d07a283056b1396e9a57ceddbd272d9252c14f154d450d227606eb54"},
335 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl", hash = "sha256:346f5343b9e3f00b8db8ba359350eb124b98c99efd0b408728ac6ebf38173958"},
336 | {file = "kiwisolver-1.4.5-cp39-cp39-manylinux_2_17_s390x.manylinux2014_s390x.whl", hash = "sha256:b9098e0049e88c6a24ff64545cdfc50807818ba6c1b739cae221bbbcbc58aad3"},
337 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:00bd361b903dc4bbf4eb165f24d1acbee754fce22ded24c3d56eec268658a5cf"},
338 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_i686.whl", hash = "sha256:7b8b454bac16428b22560d0a1cf0a09875339cab69df61d7805bf48919415901"},
339 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_ppc64le.whl", hash = "sha256:f1d072c2eb0ad60d4c183f3fb44ac6f73fb7a8f16a2694a91f988275cbf352f9"},
340 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_s390x.whl", hash = "sha256:31a82d498054cac9f6d0b53d02bb85811185bcb477d4b60144f915f3b3126342"},
341 | {file = "kiwisolver-1.4.5-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:6512cb89e334e4700febbffaaa52761b65b4f5a3cf33f960213d5656cea36a77"},
342 | {file = "kiwisolver-1.4.5-cp39-cp39-win32.whl", hash = "sha256:9db8ea4c388fdb0f780fe91346fd438657ea602d58348753d9fb265ce1bca67f"},
343 | {file = "kiwisolver-1.4.5-cp39-cp39-win_amd64.whl", hash = "sha256:59415f46a37f7f2efeec758353dd2eae1b07640d8ca0f0c42548ec4125492635"},
344 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-macosx_10_9_x86_64.whl", hash = "sha256:5c7b3b3a728dc6faf3fc372ef24f21d1e3cee2ac3e9596691d746e5a536de920"},
345 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:620ced262a86244e2be10a676b646f29c34537d0d9cc8eb26c08f53d98013390"},
346 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:378a214a1e3bbf5ac4a8708304318b4f890da88c9e6a07699c4ae7174c09a68d"},
347 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:aaf7be1207676ac608a50cd08f102f6742dbfc70e8d60c4db1c6897f62f71523"},
348 | {file = "kiwisolver-1.4.5-pp37-pypy37_pp73-win_amd64.whl", hash = "sha256:ba55dce0a9b8ff59495ddd050a0225d58bd0983d09f87cfe2b6aec4f2c1234e4"},
349 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-macosx_10_9_x86_64.whl", hash = "sha256:fd32ea360bcbb92d28933fc05ed09bffcb1704ba3fc7942e81db0fd4f81a7892"},
350 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_i686.manylinux2010_i686.whl", hash = "sha256:5e7139af55d1688f8b960ee9ad5adafc4ac17c1c473fe07133ac092310d76544"},
351 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_12_x86_64.manylinux2010_x86_64.whl", hash = "sha256:dced8146011d2bc2e883f9bd68618b8247387f4bbec46d7392b3c3b032640126"},
352 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c9bf3325c47b11b2e51bca0824ea217c7cd84491d8ac4eefd1e409705ef092bd"},
353 | {file = "kiwisolver-1.4.5-pp38-pypy38_pp73-win_amd64.whl", hash = "sha256:5794cf59533bc3f1b1c821f7206a3617999db9fbefc345360aafe2e067514929"},
354 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:e368f200bbc2e4f905b8e71eb38b3c04333bddaa6a2464a6355487b02bb7fb09"},
355 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_12_i686.manylinux2010_i686.manylinux_2_17_i686.manylinux2014_i686.whl", hash = "sha256:e5d706eba36b4c4d5bc6c6377bb6568098765e990cfc21ee16d13963fab7b3e7"},
356 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:85267bd1aa8880a9c88a8cb71e18d3d64d2751a790e6ca6c27b8ccc724bcd5ad"},
357 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:210ef2c3a1f03272649aff1ef992df2e724748918c4bc2d5a90352849eb40bea"},
358 | {file = "kiwisolver-1.4.5-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:11d011a7574eb3b82bcc9c1a1d35c1d7075677fdd15de527d91b46bd35e935ee"},
359 | {file = "kiwisolver-1.4.5.tar.gz", hash = "sha256:e57e563a57fb22a142da34f38acc2fc1a5c864bc29ca1517a88abc963e60d6ec"},
360 | ]
361 |
362 | [[package]]
363 | name = "matplotlib"
364 | version = "3.8.2"
365 | description = "Python plotting package"
366 | optional = false
367 | python-versions = ">=3.9"
368 | files = [
369 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_10_12_x86_64.whl", hash = "sha256:09796f89fb71a0c0e1e2f4bdaf63fb2cefc84446bb963ecdeb40dfee7dfa98c7"},
370 | {file = "matplotlib-3.8.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6f9c6976748a25e8b9be51ea028df49b8e561eed7809146da7a47dbecebab367"},
371 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b78e4f2cedf303869b782071b55fdde5987fda3038e9d09e58c91cc261b5ad18"},
372 | {file = "matplotlib-3.8.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:4e208f46cf6576a7624195aa047cb344a7f802e113bb1a06cfd4bee431de5e31"},
373 | {file = "matplotlib-3.8.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:46a569130ff53798ea5f50afce7406e91fdc471ca1e0e26ba976a8c734c9427a"},
374 | {file = "matplotlib-3.8.2-cp310-cp310-win_amd64.whl", hash = "sha256:830f00640c965c5b7f6bc32f0d4ce0c36dfe0379f7dd65b07a00c801713ec40a"},
375 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_10_12_x86_64.whl", hash = "sha256:d86593ccf546223eb75a39b44c32788e6f6440d13cfc4750c1c15d0fcb850b63"},
376 | {file = "matplotlib-3.8.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:9a5430836811b7652991939012f43d2808a2db9b64ee240387e8c43e2e5578c8"},
377 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:b9576723858a78751d5aacd2497b8aef29ffea6d1c95981505877f7ac28215c6"},
378 | {file = "matplotlib-3.8.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5ba9cbd8ac6cf422f3102622b20f8552d601bf8837e49a3afed188d560152788"},
379 | {file = "matplotlib-3.8.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:03f9d160a29e0b65c0790bb07f4f45d6a181b1ac33eb1bb0dd225986450148f0"},
380 | {file = "matplotlib-3.8.2-cp311-cp311-win_amd64.whl", hash = "sha256:3773002da767f0a9323ba1a9b9b5d00d6257dbd2a93107233167cfb581f64717"},
381 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_10_12_x86_64.whl", hash = "sha256:4c318c1e95e2f5926fba326f68177dee364aa791d6df022ceb91b8221bd0a627"},
382 | {file = "matplotlib-3.8.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:091275d18d942cf1ee9609c830a1bc36610607d8223b1b981c37d5c9fc3e46a4"},
383 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:1b0f3b8ea0e99e233a4bcc44590f01604840d833c280ebb8fe5554fd3e6cfe8d"},
384 | {file = "matplotlib-3.8.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d7b1704a530395aaf73912be741c04d181f82ca78084fbd80bc737be04848331"},
385 | {file = "matplotlib-3.8.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:533b0e3b0c6768eef8cbe4b583731ce25a91ab54a22f830db2b031e83cca9213"},
386 | {file = "matplotlib-3.8.2-cp312-cp312-win_amd64.whl", hash = "sha256:0f4fc5d72b75e2c18e55eb32292659cf731d9d5b312a6eb036506304f4675630"},
387 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_10_12_x86_64.whl", hash = "sha256:deaed9ad4da0b1aea77fe0aa0cebb9ef611c70b3177be936a95e5d01fa05094f"},
388 | {file = "matplotlib-3.8.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:172f4d0fbac3383d39164c6caafd3255ce6fa58f08fc392513a0b1d3b89c4f89"},
389 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:c7d36c2209d9136cd8e02fab1c0ddc185ce79bc914c45054a9f514e44c787917"},
390 | {file = "matplotlib-3.8.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:5864bdd7da445e4e5e011b199bb67168cdad10b501750367c496420f2ad00843"},
391 | {file = "matplotlib-3.8.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:ef8345b48e95cee45ff25192ed1f4857273117917a4dcd48e3905619bcd9c9b8"},
392 | {file = "matplotlib-3.8.2-cp39-cp39-win_amd64.whl", hash = "sha256:7c48d9e221b637c017232e3760ed30b4e8d5dfd081daf327e829bf2a72c731b4"},
393 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-macosx_10_12_x86_64.whl", hash = "sha256:aa11b3c6928a1e496c1a79917d51d4cd5d04f8a2e75f21df4949eeefdf697f4b"},
394 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:d1095fecf99eeb7384dabad4bf44b965f929a5f6079654b681193edf7169ec20"},
395 | {file = "matplotlib-3.8.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:bddfb1db89bfaa855912261c805bd0e10218923cc262b9159a49c29a7a1c1afa"},
396 | {file = "matplotlib-3.8.2.tar.gz", hash = "sha256:01a978b871b881ee76017152f1f1a0cbf6bd5f7b8ff8c96df0df1bd57d8755a1"},
397 | ]
398 |
399 | [package.dependencies]
400 | contourpy = ">=1.0.1"
401 | cycler = ">=0.10"
402 | fonttools = ">=4.22.0"
403 | importlib-resources = {version = ">=3.2.0", markers = "python_version < \"3.10\""}
404 | kiwisolver = ">=1.3.1"
405 | numpy = ">=1.21,<2"
406 | packaging = ">=20.0"
407 | pillow = ">=8"
408 | pyparsing = ">=2.3.1"
409 | python-dateutil = ">=2.7"
410 |
411 | [[package]]
412 | name = "numpy"
413 | version = "1.26.2"
414 | description = "Fundamental package for array computing in Python"
415 | optional = false
416 | python-versions = ">=3.9"
417 | files = [
418 | {file = "numpy-1.26.2-cp310-cp310-macosx_10_9_x86_64.whl", hash = "sha256:3703fc9258a4a122d17043e57b35e5ef1c5a5837c3db8be396c82e04c1cf9b0f"},
419 | {file = "numpy-1.26.2-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:cc392fdcbd21d4be6ae1bb4475a03ce3b025cd49a9be5345d76d7585aea69440"},
420 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:36340109af8da8805d8851ef1d74761b3b88e81a9bd80b290bbfed61bd2b4f75"},
421 | {file = "numpy-1.26.2-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:bcc008217145b3d77abd3e4d5ef586e3bdfba8fe17940769f8aa09b99e856c00"},
422 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:3ced40d4e9e18242f70dd02d739e44698df3dcb010d31f495ff00a31ef6014fe"},
423 | {file = "numpy-1.26.2-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:b272d4cecc32c9e19911891446b72e986157e6a1809b7b56518b4f3755267523"},
424 | {file = "numpy-1.26.2-cp310-cp310-win32.whl", hash = "sha256:22f8fc02fdbc829e7a8c578dd8d2e15a9074b630d4da29cda483337e300e3ee9"},
425 | {file = "numpy-1.26.2-cp310-cp310-win_amd64.whl", hash = "sha256:26c9d33f8e8b846d5a65dd068c14e04018d05533b348d9eaeef6c1bd787f9919"},
426 | {file = "numpy-1.26.2-cp311-cp311-macosx_10_9_x86_64.whl", hash = "sha256:b96e7b9c624ef3ae2ae0e04fa9b460f6b9f17ad8b4bec6d7756510f1f6c0c841"},
427 | {file = "numpy-1.26.2-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:aa18428111fb9a591d7a9cc1b48150097ba6a7e8299fb56bdf574df650e7d1f1"},
428 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:06fa1ed84aa60ea6ef9f91ba57b5ed963c3729534e6e54055fc151fad0423f0a"},
429 | {file = "numpy-1.26.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:96ca5482c3dbdd051bcd1fce8034603d6ebfc125a7bd59f55b40d8f5d246832b"},
430 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:854ab91a2906ef29dc3925a064fcd365c7b4da743f84b123002f6139bcb3f8a7"},
431 | {file = "numpy-1.26.2-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:f43740ab089277d403aa07567be138fc2a89d4d9892d113b76153e0e412409f8"},
432 | {file = "numpy-1.26.2-cp311-cp311-win32.whl", hash = "sha256:a2bbc29fcb1771cd7b7425f98b05307776a6baf43035d3b80c4b0f29e9545186"},
433 | {file = "numpy-1.26.2-cp311-cp311-win_amd64.whl", hash = "sha256:2b3fca8a5b00184828d12b073af4d0fc5fdd94b1632c2477526f6bd7842d700d"},
434 | {file = "numpy-1.26.2-cp312-cp312-macosx_10_9_x86_64.whl", hash = "sha256:a4cd6ed4a339c21f1d1b0fdf13426cb3b284555c27ac2f156dfdaaa7e16bfab0"},
435 | {file = "numpy-1.26.2-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:5d5244aabd6ed7f312268b9247be47343a654ebea52a60f002dc70c769048e75"},
436 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:6a3cdb4d9c70e6b8c0814239ead47da00934666f668426fc6e94cce869e13fd7"},
437 | {file = "numpy-1.26.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:aa317b2325f7aa0a9471663e6093c210cb2ae9c0ad824732b307d2c51983d5b6"},
438 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:174a8880739c16c925799c018f3f55b8130c1f7c8e75ab0a6fa9d41cab092fd6"},
439 | {file = "numpy-1.26.2-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:f79b231bf5c16b1f39c7f4875e1ded36abee1591e98742b05d8a0fb55d8a3eec"},
440 | {file = "numpy-1.26.2-cp312-cp312-win32.whl", hash = "sha256:4a06263321dfd3598cacb252f51e521a8cb4b6df471bb12a7ee5cbab20ea9167"},
441 | {file = "numpy-1.26.2-cp312-cp312-win_amd64.whl", hash = "sha256:b04f5dc6b3efdaab541f7857351aac359e6ae3c126e2edb376929bd3b7f92d7e"},
442 | {file = "numpy-1.26.2-cp39-cp39-macosx_10_9_x86_64.whl", hash = "sha256:4eb8df4bf8d3d90d091e0146f6c28492b0be84da3e409ebef54349f71ed271ef"},
443 | {file = "numpy-1.26.2-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:1a13860fdcd95de7cf58bd6f8bc5a5ef81c0b0625eb2c9a783948847abbef2c2"},
444 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:64308ebc366a8ed63fd0bf426b6a9468060962f1a4339ab1074c228fa6ade8e3"},
445 | {file = "numpy-1.26.2-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:baf8aab04a2c0e859da118f0b38617e5ee65d75b83795055fb66c0d5e9e9b818"},
446 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:d73a3abcac238250091b11caef9ad12413dab01669511779bc9b29261dd50210"},
447 | {file = "numpy-1.26.2-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:b361d369fc7e5e1714cf827b731ca32bff8d411212fccd29ad98ad622449cc36"},
448 | {file = "numpy-1.26.2-cp39-cp39-win32.whl", hash = "sha256:bd3f0091e845164a20bd5a326860c840fe2af79fa12e0469a12768a3ec578d80"},
449 | {file = "numpy-1.26.2-cp39-cp39-win_amd64.whl", hash = "sha256:2beef57fb031dcc0dc8fa4fe297a742027b954949cabb52a2a376c144e5e6060"},
450 | {file = "numpy-1.26.2-pp39-pypy39_pp73-macosx_10_9_x86_64.whl", hash = "sha256:1cc3d5029a30fb5f06704ad6b23b35e11309491c999838c31f124fee32107c79"},
451 | {file = "numpy-1.26.2-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:94cc3c222bb9fb5a12e334d0479b97bb2df446fbe622b470928f5284ffca3f8d"},
452 | {file = "numpy-1.26.2-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:fe6b44fb8fcdf7eda4ef4461b97b3f63c466b27ab151bec2366db8b197387841"},
453 | {file = "numpy-1.26.2.tar.gz", hash = "sha256:f65738447676ab5777f11e6bbbdb8ce11b785e105f690bc45966574816b6d3ea"},
454 | ]
455 |
456 | [[package]]
457 | name = "packaging"
458 | version = "23.2"
459 | description = "Core utilities for Python packages"
460 | optional = false
461 | python-versions = ">=3.7"
462 | files = [
463 | {file = "packaging-23.2-py3-none-any.whl", hash = "sha256:8c491190033a9af7e1d931d0b5dacc2ef47509b34dd0de67ed209b5203fc88c7"},
464 | {file = "packaging-23.2.tar.gz", hash = "sha256:048fb0e9405036518eaaf48a55953c750c11e1a1b68e0dd1a9d62ed0c092cfc5"},
465 | ]
466 |
467 | [[package]]
468 | name = "pillow"
469 | version = "10.1.0"
470 | description = "Python Imaging Library (Fork)"
471 | optional = false
472 | python-versions = ">=3.8"
473 | files = [
474 | {file = "Pillow-10.1.0-cp310-cp310-macosx_10_10_x86_64.whl", hash = "sha256:1ab05f3db77e98f93964697c8efc49c7954b08dd61cff526b7f2531a22410106"},
475 | {file = "Pillow-10.1.0-cp310-cp310-macosx_11_0_arm64.whl", hash = "sha256:6932a7652464746fcb484f7fc3618e6503d2066d853f68a4bd97193a3996e273"},
476 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:a5f63b5a68daedc54c7c3464508d8c12075e56dcfbd42f8c1bf40169061ae666"},
477 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:c0949b55eb607898e28eaccb525ab104b2d86542a85c74baf3a6dc24002edec2"},
478 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_aarch64.whl", hash = "sha256:ae88931f93214777c7a3aa0a8f92a683f83ecde27f65a45f95f22d289a69e593"},
479 | {file = "Pillow-10.1.0-cp310-cp310-manylinux_2_28_x86_64.whl", hash = "sha256:b0eb01ca85b2361b09480784a7931fc648ed8b7836f01fb9241141b968feb1db"},
480 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_aarch64.whl", hash = "sha256:d27b5997bdd2eb9fb199982bb7eb6164db0426904020dc38c10203187ae2ff2f"},
481 | {file = "Pillow-10.1.0-cp310-cp310-musllinux_1_1_x86_64.whl", hash = "sha256:7df5608bc38bd37ef585ae9c38c9cd46d7c81498f086915b0f97255ea60c2818"},
482 | {file = "Pillow-10.1.0-cp310-cp310-win_amd64.whl", hash = "sha256:41f67248d92a5e0a2076d3517d8d4b1e41a97e2df10eb8f93106c89107f38b57"},
483 | {file = "Pillow-10.1.0-cp311-cp311-macosx_10_10_x86_64.whl", hash = "sha256:1fb29c07478e6c06a46b867e43b0bcdb241b44cc52be9bc25ce5944eed4648e7"},
484 | {file = "Pillow-10.1.0-cp311-cp311-macosx_11_0_arm64.whl", hash = "sha256:2cdc65a46e74514ce742c2013cd4a2d12e8553e3a2563c64879f7c7e4d28bce7"},
485 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:50d08cd0a2ecd2a8657bd3d82c71efd5a58edb04d9308185d66c3a5a5bed9610"},
486 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:062a1610e3bc258bff2328ec43f34244fcec972ee0717200cb1425214fe5b839"},
487 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_aarch64.whl", hash = "sha256:61f1a9d247317fa08a308daaa8ee7b3f760ab1809ca2da14ecc88ae4257d6172"},
488 | {file = "Pillow-10.1.0-cp311-cp311-manylinux_2_28_x86_64.whl", hash = "sha256:a646e48de237d860c36e0db37ecaecaa3619e6f3e9d5319e527ccbc8151df061"},
489 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_aarch64.whl", hash = "sha256:47e5bf85b80abc03be7455c95b6d6e4896a62f6541c1f2ce77a7d2bb832af262"},
490 | {file = "Pillow-10.1.0-cp311-cp311-musllinux_1_1_x86_64.whl", hash = "sha256:a92386125e9ee90381c3369f57a2a50fa9e6aa8b1cf1d9c4b200d41a7dd8e992"},
491 | {file = "Pillow-10.1.0-cp311-cp311-win_amd64.whl", hash = "sha256:0f7c276c05a9767e877a0b4c5050c8bee6a6d960d7f0c11ebda6b99746068c2a"},
492 | {file = "Pillow-10.1.0-cp312-cp312-macosx_10_10_x86_64.whl", hash = "sha256:a89b8312d51715b510a4fe9fc13686283f376cfd5abca8cd1c65e4c76e21081b"},
493 | {file = "Pillow-10.1.0-cp312-cp312-macosx_11_0_arm64.whl", hash = "sha256:00f438bb841382b15d7deb9a05cc946ee0f2c352653c7aa659e75e592f6fa17d"},
494 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:3d929a19f5469b3f4df33a3df2983db070ebb2088a1e145e18facbc28cae5b27"},
495 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:9a92109192b360634a4489c0c756364c0c3a2992906752165ecb50544c251312"},
496 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_aarch64.whl", hash = "sha256:0248f86b3ea061e67817c47ecbe82c23f9dd5d5226200eb9090b3873d3ca32de"},
497 | {file = "Pillow-10.1.0-cp312-cp312-manylinux_2_28_x86_64.whl", hash = "sha256:9882a7451c680c12f232a422730f986a1fcd808da0fd428f08b671237237d651"},
498 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_aarch64.whl", hash = "sha256:1c3ac5423c8c1da5928aa12c6e258921956757d976405e9467c5f39d1d577a4b"},
499 | {file = "Pillow-10.1.0-cp312-cp312-musllinux_1_1_x86_64.whl", hash = "sha256:806abdd8249ba3953c33742506fe414880bad78ac25cc9a9b1c6ae97bedd573f"},
500 | {file = "Pillow-10.1.0-cp312-cp312-win_amd64.whl", hash = "sha256:eaed6977fa73408b7b8a24e8b14e59e1668cfc0f4c40193ea7ced8e210adf996"},
501 | {file = "Pillow-10.1.0-cp38-cp38-macosx_10_10_x86_64.whl", hash = "sha256:fe1e26e1ffc38be097f0ba1d0d07fcade2bcfd1d023cda5b29935ae8052bd793"},
502 | {file = "Pillow-10.1.0-cp38-cp38-macosx_11_0_arm64.whl", hash = "sha256:7a7e3daa202beb61821c06d2517428e8e7c1aab08943e92ec9e5755c2fc9ba5e"},
503 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:24fadc71218ad2b8ffe437b54876c9382b4a29e030a05a9879f615091f42ffc2"},
504 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:fa1d323703cfdac2036af05191b969b910d8f115cf53093125e4058f62012c9a"},
505 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_aarch64.whl", hash = "sha256:912e3812a1dbbc834da2b32299b124b5ddcb664ed354916fd1ed6f193f0e2d01"},
506 | {file = "Pillow-10.1.0-cp38-cp38-manylinux_2_28_x86_64.whl", hash = "sha256:7dbaa3c7de82ef37e7708521be41db5565004258ca76945ad74a8e998c30af8d"},
507 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_aarch64.whl", hash = "sha256:9d7bc666bd8c5a4225e7ac71f2f9d12466ec555e89092728ea0f5c0c2422ea80"},
508 | {file = "Pillow-10.1.0-cp38-cp38-musllinux_1_1_x86_64.whl", hash = "sha256:baada14941c83079bf84c037e2d8b7506ce201e92e3d2fa0d1303507a8538212"},
509 | {file = "Pillow-10.1.0-cp38-cp38-win_amd64.whl", hash = "sha256:2ef6721c97894a7aa77723740a09547197533146fba8355e86d6d9a4a1056b14"},
510 | {file = "Pillow-10.1.0-cp39-cp39-macosx_10_10_x86_64.whl", hash = "sha256:0a026c188be3b443916179f5d04548092e253beb0c3e2ee0a4e2cdad72f66099"},
511 | {file = "Pillow-10.1.0-cp39-cp39-macosx_11_0_arm64.whl", hash = "sha256:04f6f6149f266a100374ca3cc368b67fb27c4af9f1cc8cb6306d849dcdf12616"},
512 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl", hash = "sha256:bb40c011447712d2e19cc261c82655f75f32cb724788df315ed992a4d65696bb"},
513 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1a8413794b4ad9719346cd9306118450b7b00d9a15846451549314a58ac42219"},
514 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_aarch64.whl", hash = "sha256:c9aeea7b63edb7884b031a35305629a7593272b54f429a9869a4f63a1bf04c34"},
515 | {file = "Pillow-10.1.0-cp39-cp39-manylinux_2_28_x86_64.whl", hash = "sha256:b4005fee46ed9be0b8fb42be0c20e79411533d1fd58edabebc0dd24626882cfd"},
516 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_aarch64.whl", hash = "sha256:4d0152565c6aa6ebbfb1e5d8624140a440f2b99bf7afaafbdbf6430426497f28"},
517 | {file = "Pillow-10.1.0-cp39-cp39-musllinux_1_1_x86_64.whl", hash = "sha256:d921bc90b1defa55c9917ca6b6b71430e4286fc9e44c55ead78ca1a9f9eba5f2"},
518 | {file = "Pillow-10.1.0-cp39-cp39-win_amd64.whl", hash = "sha256:cfe96560c6ce2f4c07d6647af2d0f3c54cc33289894ebd88cfbb3bcd5391e256"},
519 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-macosx_10_10_x86_64.whl", hash = "sha256:937bdc5a7f5343d1c97dc98149a0be7eb9704e937fe3dc7140e229ae4fc572a7"},
520 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:b1c25762197144e211efb5f4e8ad656f36c8d214d390585d1d21281f46d556ba"},
521 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:afc8eef765d948543a4775f00b7b8c079b3321d6b675dde0d02afa2ee23000b4"},
522 | {file = "Pillow-10.1.0-pp310-pypy310_pp73-win_amd64.whl", hash = "sha256:883f216eac8712b83a63f41b76ddfb7b2afab1b74abbb413c5df6680f071a6b9"},
523 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-macosx_10_10_x86_64.whl", hash = "sha256:b920e4d028f6442bea9a75b7491c063f0b9a3972520731ed26c83e254302eb1e"},
524 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl", hash = "sha256:1c41d960babf951e01a49c9746f92c5a7e0d939d1652d7ba30f6b3090f27e412"},
525 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-manylinux_2_28_x86_64.whl", hash = "sha256:1fafabe50a6977ac70dfe829b2d5735fd54e190ab55259ec8aea4aaea412fa0b"},
526 | {file = "Pillow-10.1.0-pp39-pypy39_pp73-win_amd64.whl", hash = "sha256:3b834f4b16173e5b92ab6566f0473bfb09f939ba14b23b8da1f54fa63e4b623f"},
527 | {file = "Pillow-10.1.0.tar.gz", hash = "sha256:e6bf8de6c36ed96c86ea3b6e1d5273c53f46ef518a062464cd7ef5dd2cf92e38"},
528 | ]
529 |
530 | [package.extras]
531 | docs = ["furo", "olefile", "sphinx (>=2.4)", "sphinx-copybutton", "sphinx-inline-tabs", "sphinx-removed-in", "sphinxext-opengraph"]
532 | tests = ["check-manifest", "coverage", "defusedxml", "markdown2", "olefile", "packaging", "pyroma", "pytest", "pytest-cov", "pytest-timeout"]
533 |
534 | [[package]]
535 | name = "pluggy"
536 | version = "1.3.0"
537 | description = "plugin and hook calling mechanisms for python"
538 | optional = false
539 | python-versions = ">=3.8"
540 | files = [
541 | {file = "pluggy-1.3.0-py3-none-any.whl", hash = "sha256:d89c696a773f8bd377d18e5ecda92b7a3793cbe66c87060a6fb58c7b6e1061f7"},
542 | {file = "pluggy-1.3.0.tar.gz", hash = "sha256:cf61ae8f126ac6f7c451172cf30e3e43d3ca77615509771b3a984a0730651e12"},
543 | ]
544 |
545 | [package.extras]
546 | dev = ["pre-commit", "tox"]
547 | testing = ["pytest", "pytest-benchmark"]
548 |
549 | [[package]]
550 | name = "py"
551 | version = "1.11.0"
552 | description = "library with cross-python path, ini-parsing, io, code, log facilities"
553 | optional = false
554 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*, !=3.3.*, !=3.4.*"
555 | files = [
556 | {file = "py-1.11.0-py2.py3-none-any.whl", hash = "sha256:607c53218732647dff4acdfcd50cb62615cedf612e72d1724fb1a0cc6405b378"},
557 | {file = "py-1.11.0.tar.gz", hash = "sha256:51c75c4126074b472f746a24399ad32f6053d1b34b68d2fa41e558e6f4a98719"},
558 | ]
559 |
560 | [[package]]
561 | name = "pyparsing"
562 | version = "3.1.1"
563 | description = "pyparsing module - Classes and methods to define and execute parsing grammars"
564 | optional = false
565 | python-versions = ">=3.6.8"
566 | files = [
567 | {file = "pyparsing-3.1.1-py3-none-any.whl", hash = "sha256:32c7c0b711493c72ff18a981d24f28aaf9c1fb7ed5e9667c9e84e3db623bdbfb"},
568 | {file = "pyparsing-3.1.1.tar.gz", hash = "sha256:ede28a1a32462f5a9705e07aea48001a08f7cf81a021585011deba701581a0db"},
569 | ]
570 |
571 | [package.extras]
572 | diagrams = ["jinja2", "railroad-diagrams"]
573 |
574 | [[package]]
575 | name = "pytest"
576 | version = "6.2.5"
577 | description = "pytest: simple powerful testing with Python"
578 | optional = false
579 | python-versions = ">=3.6"
580 | files = [
581 | {file = "pytest-6.2.5-py3-none-any.whl", hash = "sha256:7310f8d27bc79ced999e760ca304d69f6ba6c6649c0b60fb0e04a4a77cacc134"},
582 | {file = "pytest-6.2.5.tar.gz", hash = "sha256:131b36680866a76e6781d13f101efb86cf674ebb9762eb70d3082b6f29889e89"},
583 | ]
584 |
585 | [package.dependencies]
586 | atomicwrites = {version = ">=1.0", markers = "sys_platform == \"win32\""}
587 | attrs = ">=19.2.0"
588 | colorama = {version = "*", markers = "sys_platform == \"win32\""}
589 | iniconfig = "*"
590 | packaging = "*"
591 | pluggy = ">=0.12,<2.0"
592 | py = ">=1.8.2"
593 | toml = "*"
594 |
595 | [package.extras]
596 | testing = ["argcomplete", "hypothesis (>=3.56)", "mock", "nose", "requests", "xmlschema"]
597 |
598 | [[package]]
599 | name = "python-dateutil"
600 | version = "2.8.2"
601 | description = "Extensions to the standard Python datetime module"
602 | optional = false
603 | python-versions = "!=3.0.*,!=3.1.*,!=3.2.*,>=2.7"
604 | files = [
605 | {file = "python-dateutil-2.8.2.tar.gz", hash = "sha256:0123cacc1627ae19ddf3c27a5de5bd67ee4586fbdd6440d9748f8abb483d3e86"},
606 | {file = "python_dateutil-2.8.2-py2.py3-none-any.whl", hash = "sha256:961d03dc3453ebbc59dbdea9e4e11c5651520a876d0f4db161e8674aae935da9"},
607 | ]
608 |
609 | [package.dependencies]
610 | six = ">=1.5"
611 |
612 | [[package]]
613 | name = "six"
614 | version = "1.16.0"
615 | description = "Python 2 and 3 compatibility utilities"
616 | optional = false
617 | python-versions = ">=2.7, !=3.0.*, !=3.1.*, !=3.2.*"
618 | files = [
619 | {file = "six-1.16.0-py2.py3-none-any.whl", hash = "sha256:8abb2f1d86890a2dfb989f9a77cfcfd3e47c2a354b01111771326f8aa26e0254"},
620 | {file = "six-1.16.0.tar.gz", hash = "sha256:1e61c37477a1626458e36f7b1d82aa5c9b094fa4802892072e49de9c60c4c926"},
621 | ]
622 |
623 | [[package]]
624 | name = "toml"
625 | version = "0.10.2"
626 | description = "Python Library for Tom's Obvious, Minimal Language"
627 | optional = false
628 | python-versions = ">=2.6, !=3.0.*, !=3.1.*, !=3.2.*"
629 | files = [
630 | {file = "toml-0.10.2-py2.py3-none-any.whl", hash = "sha256:806143ae5bfb6a3c6e736a764057db0e6a0e05e338b5630894a5f779cabb4f9b"},
631 | {file = "toml-0.10.2.tar.gz", hash = "sha256:b3bda1d108d5dd99f4a20d24d9c348e91c4db7ab1b749200bded2f839ccbe68f"},
632 | ]
633 |
634 | [[package]]
635 | name = "tqdm"
636 | version = "4.66.1"
637 | description = "Fast, Extensible Progress Meter"
638 | optional = false
639 | python-versions = ">=3.7"
640 | files = [
641 | {file = "tqdm-4.66.1-py3-none-any.whl", hash = "sha256:d302b3c5b53d47bce91fea46679d9c3c6508cf6332229aa1e7d8653723793386"},
642 | {file = "tqdm-4.66.1.tar.gz", hash = "sha256:d88e651f9db8d8551a62556d3cff9e3034274ca5d66e93197cf2490e2dcb69c7"},
643 | ]
644 |
645 | [package.dependencies]
646 | colorama = {version = "*", markers = "platform_system == \"Windows\""}
647 |
648 | [package.extras]
649 | dev = ["pytest (>=6)", "pytest-cov", "pytest-timeout", "pytest-xdist"]
650 | notebook = ["ipywidgets (>=6)"]
651 | slack = ["slack-sdk"]
652 | telegram = ["requests"]
653 |
654 | [[package]]
655 | name = "zipp"
656 | version = "3.17.0"
657 | description = "Backport of pathlib-compatible object wrapper for zip files"
658 | optional = false
659 | python-versions = ">=3.8"
660 | files = [
661 | {file = "zipp-3.17.0-py3-none-any.whl", hash = "sha256:0e923e726174922dce09c53c59ad483ff7bbb8e572e00c7f7c46b88556409f31"},
662 | {file = "zipp-3.17.0.tar.gz", hash = "sha256:84e64a1c28cf7e91ed2078bb8cc8c259cb19b76942096c8d7b84947690cabaf0"},
663 | ]
664 |
665 | [package.extras]
666 | docs = ["furo", "jaraco.packaging (>=9.3)", "jaraco.tidelift (>=1.4)", "rst.linker (>=1.9)", "sphinx (<7.2.5)", "sphinx (>=3.5)", "sphinx-lint"]
667 | testing = ["big-O", "jaraco.functools", "jaraco.itertools", "more-itertools", "pytest (>=6)", "pytest-black (>=0.3.7)", "pytest-checkdocs (>=2.4)", "pytest-cov", "pytest-enabler (>=2.2)", "pytest-ignore-flaky", "pytest-mypy (>=0.9.1)", "pytest-ruff"]
668 |
669 | [metadata]
670 | lock-version = "2.0"
671 | python-versions = "^3.9"
672 | content-hash = "5e3a8b71c1d10601d5e0c2e8e72d1ea2cd79f51cbe48d1f7512dd3fc591030c2"
673 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.poetry]
2 | name = "complexcgr"
3 | version = "0.8.0"
4 | description = "complex Chaos Game Representation for DNA"
5 | authors = ["Jorge Avila