├── COPYING
├── ChangeLog
├── Makefile
├── Readme.md
└── src
├── BloomFilter.cpp
├── BloomFilter.h
├── ReverseBloomFilter.cpp
├── ReverseBloomFilter.h
├── Rscripts
├── Q20Q30.R
├── base.R
└── quality.R
├── gc.cpp
├── gc.h
├── global_parameter.h
├── global_variable.cpp
├── global_variable.h
├── mGzip.cpp
├── mGzip.h
├── main.cpp
├── peFilterTmpOut.h
├── peprocess.cpp
├── peprocess.h
├── processHts.cpp
├── processHts.h
├── processStLFR.cpp
├── processStLFR.h
├── process_argv.cpp
├── process_argv.h
├── read_filter.cpp
├── read_filter.h
├── rmdup.cpp
├── rmdup.h
├── seprocess.cpp
├── seprocess.h
├── sequence.cpp
└── sequence.h
/COPYING:
--------------------------------------------------------------------------------
1 | GNU GENERAL PUBLIC LICENSE
2 |
3 | Version 3, 29 June 2007
4 |
5 | Copyright © 2007 Free Software Foundation, Inc.
6 |
7 | Everyone is permitted to copy and distribute verbatim copies of this license document, but changing it is not allowed.
8 |
9 | Preamble
10 |
11 | The GNU General Public License is a free, copyleft license for software and other kinds of works.
12 |
13 | The licenses for most software and other practical works are designed to take away your freedom to share and change the works. By contrast, the GNU General Public License is intended to guarantee your freedom to share and change all versions of a program--to make sure it remains free software for all its users. We, the Free Software Foundation, use the GNU General Public License for most of our software; it applies also to any other work released this way by its authors. You can apply it to your programs, too.
14 |
15 | When we speak of free software, we are referring to freedom, not price. Our General Public Licenses are designed to make sure that you have the freedom to distribute copies of free software (and charge for them if you wish), that you receive source code or can get it if you want it, that you can change the software or use pieces of it in new free programs, and that you know you can do these things.
16 |
17 | To protect your rights, we need to prevent others from denying you these rights or asking you to surrender the rights. Therefore, you have certain responsibilities if you distribute copies of the software, or if you modify it: responsibilities to respect the freedom of others.
18 |
19 | For example, if you distribute copies of such a program, whether gratis or for a fee, you must pass on to the recipients the same freedoms that you received. You must make sure that they, too, receive or can get the source code. And you must show them these terms so they know their rights.
20 |
21 | Developers that use the GNU GPL protect your rights with two steps: (1) assert copyright on the software, and (2) offer you this License giving you legal permission to copy, distribute and/or modify it.
22 |
23 | For the developers' and authors' protection, the GPL clearly explains that there is no warranty for this free software. For both users' and authors' sake, the GPL requires that modified versions be marked as changed, so that their problems will not be attributed erroneously to authors of previous versions.
24 |
25 | Some devices are designed to deny users access to install or run modified versions of the software inside them, although the manufacturer can do so. This is fundamentally incompatible with the aim of protecting users' freedom to change the software. The systematic pattern of such abuse occurs in the area of products for individuals to use, which is precisely where it is most unacceptable. Therefore, we have designed this version of the GPL to prohibit the practice for those products. If such problems arise substantially in other domains, we stand ready to extend this provision to those domains in future versions of the GPL, as needed to protect the freedom of users.
26 |
27 | Finally, every program is threatened constantly by software patents. States should not allow patents to restrict development and use of software on general-purpose computers, but in those that do, we wish to avoid the special danger that patents applied to a free program could make it effectively proprietary. To prevent this, the GPL assures that patents cannot be used to render the program non-free.
28 |
29 | The precise terms and conditions for copying, distribution and modification follow.
30 |
31 | TERMS AND CONDITIONS
32 |
33 | 0. Definitions.
34 |
35 | “This License” refers to version 3 of the GNU General Public License.
36 |
37 | “Copyright” also means copyright-like laws that apply to other kinds of works, such as semiconductor masks.
38 |
39 | “The Program” refers to any copyrightable work licensed under this License. Each licensee is addressed as “you”. “Licensees” and “recipients” may be individuals or organizations.
40 |
41 | To “modify” a work means to copy from or adapt all or part of the work in a fashion requiring copyright permission, other than the making of an exact copy. The resulting work is called a “modified version” of the earlier work or a work “based on” the earlier work.
42 |
43 | A “covered work” means either the unmodified Program or a work based on the Program.
44 |
45 | To “propagate” a work means to do anything with it that, without permission, would make you directly or secondarily liable for infringement under applicable copyright law, except executing it on a computer or modifying a private copy. Propagation includes copying, distribution (with or without modification), making available to the public, and in some countries other activities as well.
46 |
47 | To “convey” a work means any kind of propagation that enables other parties to make or receive copies. Mere interaction with a user through a computer network, with no transfer of a copy, is not conveying.
48 |
49 | An interactive user interface displays “Appropriate Legal Notices” to the extent that it includes a convenient and prominently visible feature that (1) displays an appropriate copyright notice, and (2) tells the user that there is no warranty for the work (except to the extent that warranties are provided), that licensees may convey the work under this License, and how to view a copy of this License. If the interface presents a list of user commands or options, such as a menu, a prominent item in the list meets this criterion.
50 |
51 | 1. Source Code.
52 |
53 | The “source code” for a work means the preferred form of the work for making modifications to it. “Object code” means any non-source form of a work.
54 |
55 | A “Standard Interface” means an interface that either is an official standard defined by a recognized standards body, or, in the case of interfaces specified for a particular programming language, one that is widely used among developers working in that language.
56 |
57 | The “System Libraries” of an executable work include anything, other than the work as a whole, that (a) is included in the normal form of packaging a Major Component, but which is not part of that Major Component, and (b) serves only to enable use of the work with that Major Component, or to implement a Standard Interface for which an implementation is available to the public in source code form. A “Major Component”, in this context, means a major essential component (kernel, window system, and so on) of the specific operating system (if any) on which the executable work runs, or a compiler used to produce the work, or an object code interpreter used to run it.
58 |
59 | The “Corresponding Source” for a work in object code form means all the source code needed to generate, install, and (for an executable work) run the object code and to modify the work, including scripts to control those activities. However, it does not include the work's System Libraries, or general-purpose tools or generally available free programs which are used unmodified in performing those activities but which are not part of the work. For example, Corresponding Source includes interface definition files associated with source files for the work, and the source code for shared libraries and dynamically linked subprograms that the work is specifically designed to require, such as by intimate data communication or control flow between those subprograms and other parts of the work.
60 |
61 | The Corresponding Source need not include anything that users can regenerate automatically from other parts of the Corresponding Source.
62 |
63 | The Corresponding Source for a work in source code form is that same work.
64 |
65 | 2. Basic Permissions.
66 |
67 | All rights granted under this License are granted for the term of copyright on the Program, and are irrevocable provided the stated conditions are met. This License explicitly affirms your unlimited permission to run the unmodified Program. The output from running a covered work is covered by this License only if the output, given its content, constitutes a covered work. This License acknowledges your rights of fair use or other equivalent, as provided by copyright law.
68 |
69 | You may make, run and propagate covered works that you do not convey, without conditions so long as your license otherwise remains in force. You may convey covered works to others for the sole purpose of having them make modifications exclusively for you, or provide you with facilities for running those works, provided that you comply with the terms of this License in conveying all material for which you do not control copyright. Those thus making or running the covered works for you must do so exclusively on your behalf, under your direction and control, on terms that prohibit them from making any copies of your copyrighted material outside their relationship with you.
70 |
71 | Conveying under any other circumstances is permitted solely under the conditions stated below. Sublicensing is not allowed; section 10 makes it unnecessary.
72 |
73 | 3. Protecting Users' Legal Rights From Anti-Circumvention Law.
74 |
75 | No covered work shall be deemed part of an effective technological measure under any applicable law fulfilling obligations under article 11 of the WIPO copyright treaty adopted on 20 December 1996, or similar laws prohibiting or restricting circumvention of such measures.
76 |
77 | When you convey a covered work, you waive any legal power to forbid circumvention of technological measures to the extent such circumvention is effected by exercising rights under this License with respect to the covered work, and you disclaim any intention to limit operation or modification of the work as a means of enforcing, against the work's users, your or third parties' legal rights to forbid circumvention of technological measures.
78 |
79 | 4. Conveying Verbatim Copies.
80 |
81 | You may convey verbatim copies of the Program's source code as you receive it, in any medium, provided that you conspicuously and appropriately publish on each copy an appropriate copyright notice; keep intact all notices stating that this License and any non-permissive terms added in accord with section 7 apply to the code; keep intact all notices of the absence of any warranty; and give all recipients a copy of this License along with the Program.
82 |
83 | You may charge any price or no price for each copy that you convey, and you may offer support or warranty protection for a fee.
84 |
85 | 5. Conveying Modified Source Versions.
86 |
87 | You may convey a work based on the Program, or the modifications to produce it from the Program, in the form of source code under the terms of section 4, provided that you also meet all of these conditions:
88 |
89 | a) The work must carry prominent notices stating that you modified it, and giving a relevant date.
90 | b) The work must carry prominent notices stating that it is released under this License and any conditions added under section 7. This requirement modifies the requirement in section 4 to “keep intact all notices”.
91 | c) You must license the entire work, as a whole, under this License to anyone who comes into possession of a copy. This License will therefore apply, along with any applicable section 7 additional terms, to the whole of the work, and all its parts, regardless of how they are packaged. This License gives no permission to license the work in any other way, but it does not invalidate such permission if you have separately received it.
92 | d) If the work has interactive user interfaces, each must display Appropriate Legal Notices; however, if the Program has interactive interfaces that do not display Appropriate Legal Notices, your work need not make them do so.
93 | A compilation of a covered work with other separate and independent works, which are not by their nature extensions of the covered work, and which are not combined with it such as to form a larger program, in or on a volume of a storage or distribution medium, is called an “aggregate” if the compilation and its resulting copyright are not used to limit the access or legal rights of the compilation's users beyond what the individual works permit. Inclusion of a covered work in an aggregate does not cause this License to apply to the other parts of the aggregate.
94 |
95 | 6. Conveying Non-Source Forms.
96 |
97 | You may convey a covered work in object code form under the terms of sections 4 and 5, provided that you also convey the machine-readable Corresponding Source under the terms of this License, in one of these ways:
98 |
99 | a) Convey the object code in, or embodied in, a physical product (including a physical distribution medium), accompanied by the Corresponding Source fixed on a durable physical medium customarily used for software interchange.
100 | b) Convey the object code in, or embodied in, a physical product (including a physical distribution medium), accompanied by a written offer, valid for at least three years and valid for as long as you offer spare parts or customer support for that product model, to give anyone who possesses the object code either (1) a copy of the Corresponding Source for all the software in the product that is covered by this License, on a durable physical medium customarily used for software interchange, for a price no more than your reasonable cost of physically performing this conveying of source, or (2) access to copy the Corresponding Source from a network server at no charge.
101 | c) Convey individual copies of the object code with a copy of the written offer to provide the Corresponding Source. This alternative is allowed only occasionally and noncommercially, and only if you received the object code with such an offer, in accord with subsection 6b.
102 | d) Convey the object code by offering access from a designated place (gratis or for a charge), and offer equivalent access to the Corresponding Source in the same way through the same place at no further charge. You need not require recipients to copy the Corresponding Source along with the object code. If the place to copy the object code is a network server, the Corresponding Source may be on a different server (operated by you or a third party) that supports equivalent copying facilities, provided you maintain clear directions next to the object code saying where to find the Corresponding Source. Regardless of what server hosts the Corresponding Source, you remain obligated to ensure that it is available for as long as needed to satisfy these requirements.
103 | e) Convey the object code using peer-to-peer transmission, provided you inform other peers where the object code and Corresponding Source of the work are being offered to the general public at no charge under subsection 6d.
104 | A separable portion of the object code, whose source code is excluded from the Corresponding Source as a System Library, need not be included in conveying the object code work.
105 |
106 | A “User Product” is either (1) a “consumer product”, which means any tangible personal property which is normally used for personal, family, or household purposes, or (2) anything designed or sold for incorporation into a dwelling. In determining whether a product is a consumer product, doubtful cases shall be resolved in favor of coverage. For a particular product received by a particular user, “normally used” refers to a typical or common use of that class of product, regardless of the status of the particular user or of the way in which the particular user actually uses, or expects or is expected to use, the product. A product is a consumer product regardless of whether the product has substantial commercial, industrial or non-consumer uses, unless such uses represent the only significant mode of use of the product.
107 |
108 | “Installation Information” for a User Product means any methods, procedures, authorization keys, or other information required to install and execute modified versions of a covered work in that User Product from a modified version of its Corresponding Source. The information must suffice to ensure that the continued functioning of the modified object code is in no case prevented or interfered with solely because modification has been made.
109 |
110 | If you convey an object code work under this section in, or with, or specifically for use in, a User Product, and the conveying occurs as part of a transaction in which the right of possession and use of the User Product is transferred to the recipient in perpetuity or for a fixed term (regardless of how the transaction is characterized), the Corresponding Source conveyed under this section must be accompanied by the Installation Information. But this requirement does not apply if neither you nor any third party retains the ability to install modified object code on the User Product (for example, the work has been installed in ROM).
111 |
112 | The requirement to provide Installation Information does not include a requirement to continue to provide support service, warranty, or updates for a work that has been modified or installed by the recipient, or for the User Product in which it has been modified or installed. Access to a network may be denied when the modification itself materially and adversely affects the operation of the network or violates the rules and protocols for communication across the network.
113 |
114 | Corresponding Source conveyed, and Installation Information provided, in accord with this section must be in a format that is publicly documented (and with an implementation available to the public in source code form), and must require no special password or key for unpacking, reading or copying.
115 |
116 | 7. Additional Terms.
117 |
118 | “Additional permissions” are terms that supplement the terms of this License by making exceptions from one or more of its conditions. Additional permissions that are applicable to the entire Program shall be treated as though they were included in this License, to the extent that they are valid under applicable law. If additional permissions apply only to part of the Program, that part may be used separately under those permissions, but the entire Program remains governed by this License without regard to the additional permissions.
119 |
120 | When you convey a copy of a covered work, you may at your option remove any additional permissions from that copy, or from any part of it. (Additional permissions may be written to require their own removal in certain cases when you modify the work.) You may place additional permissions on material, added by you to a covered work, for which you have or can give appropriate copyright permission.
121 |
122 | Notwithstanding any other provision of this License, for material you add to a covered work, you may (if authorized by the copyright holders of that material) supplement the terms of this License with terms:
123 |
124 | a) Disclaiming warranty or limiting liability differently from the terms of sections 15 and 16 of this License; or
125 | b) Requiring preservation of specified reasonable legal notices or author attributions in that material or in the Appropriate Legal Notices displayed by works containing it; or
126 | c) Prohibiting misrepresentation of the origin of that material, or requiring that modified versions of such material be marked in reasonable ways as different from the original version; or
127 | d) Limiting the use for publicity purposes of names of licensors or authors of the material; or
128 | e) Declining to grant rights under trademark law for use of some trade names, trademarks, or service marks; or
129 | f) Requiring indemnification of licensors and authors of that material by anyone who conveys the material (or modified versions of it) with contractual assumptions of liability to the recipient, for any liability that these contractual assumptions directly impose on those licensors and authors.
130 | All other non-permissive additional terms are considered “further restrictions” within the meaning of section 10. If the Program as you received it, or any part of it, contains a notice stating that it is governed by this License along with a term that is a further restriction, you may remove that term. If a license document contains a further restriction but permits relicensing or conveying under this License, you may add to a covered work material governed by the terms of that license document, provided that the further restriction does not survive such relicensing or conveying.
131 |
132 | If you add terms to a covered work in accord with this section, you must place, in the relevant source files, a statement of the additional terms that apply to those files, or a notice indicating where to find the applicable terms.
133 |
134 | Additional terms, permissive or non-permissive, may be stated in the form of a separately written license, or stated as exceptions; the above requirements apply either way.
135 |
136 | 8. Termination.
137 |
138 | You may not propagate or modify a covered work except as expressly provided under this License. Any attempt otherwise to propagate or modify it is void, and will automatically terminate your rights under this License (including any patent licenses granted under the third paragraph of section 11).
139 |
140 | However, if you cease all violation of this License, then your license from a particular copyright holder is reinstated (a) provisionally, unless and until the copyright holder explicitly and finally terminates your license, and (b) permanently, if the copyright holder fails to notify you of the violation by some reasonable means prior to 60 days after the cessation.
141 |
142 | Moreover, your license from a particular copyright holder is reinstated permanently if the copyright holder notifies you of the violation by some reasonable means, this is the first time you have received notice of violation of this License (for any work) from that copyright holder, and you cure the violation prior to 30 days after your receipt of the notice.
143 |
144 | Termination of your rights under this section does not terminate the licenses of parties who have received copies or rights from you under this License. If your rights have been terminated and not permanently reinstated, you do not qualify to receive new licenses for the same material under section 10.
145 |
146 | 9. Acceptance Not Required for Having Copies.
147 |
148 | You are not required to accept this License in order to receive or run a copy of the Program. Ancillary propagation of a covered work occurring solely as a consequence of using peer-to-peer transmission to receive a copy likewise does not require acceptance. However, nothing other than this License grants you permission to propagate or modify any covered work. These actions infringe copyright if you do not accept this License. Therefore, by modifying or propagating a covered work, you indicate your acceptance of this License to do so.
149 |
150 | 10. Automatic Licensing of Downstream Recipients.
151 |
152 | Each time you convey a covered work, the recipient automatically receives a license from the original licensors, to run, modify and propagate that work, subject to this License. You are not responsible for enforcing compliance by third parties with this License.
153 |
154 | An “entity transaction” is a transaction transferring control of an organization, or substantially all assets of one, or subdividing an organization, or merging organizations. If propagation of a covered work results from an entity transaction, each party to that transaction who receives a copy of the work also receives whatever licenses to the work the party's predecessor in interest had or could give under the previous paragraph, plus a right to possession of the Corresponding Source of the work from the predecessor in interest, if the predecessor has it or can get it with reasonable efforts.
155 |
156 | You may not impose any further restrictions on the exercise of the rights granted or affirmed under this License. For example, you may not impose a license fee, royalty, or other charge for exercise of rights granted under this License, and you may not initiate litigation (including a cross-claim or counterclaim in a lawsuit) alleging that any patent claim is infringed by making, using, selling, offering for sale, or importing the Program or any portion of it.
157 |
158 | 11. Patents.
159 |
160 | A “contributor” is a copyright holder who authorizes use under this License of the Program or a work on which the Program is based. The work thus licensed is called the contributor's “contributor version”.
161 |
162 | A contributor's “essential patent claims” are all patent claims owned or controlled by the contributor, whether already acquired or hereafter acquired, that would be infringed by some manner, permitted by this License, of making, using, or selling its contributor version, but do not include claims that would be infringed only as a consequence of further modification of the contributor version. For purposes of this definition, “control” includes the right to grant patent sublicenses in a manner consistent with the requirements of this License.
163 |
164 | Each contributor grants you a non-exclusive, worldwide, royalty-free patent license under the contributor's essential patent claims, to make, use, sell, offer for sale, import and otherwise run, modify and propagate the contents of its contributor version.
165 |
166 | In the following three paragraphs, a “patent license” is any express agreement or commitment, however denominated, not to enforce a patent (such as an express permission to practice a patent or covenant not to sue for patent infringement). To “grant” such a patent license to a party means to make such an agreement or commitment not to enforce a patent against the party.
167 |
168 | If you convey a covered work, knowingly relying on a patent license, and the Corresponding Source of the work is not available for anyone to copy, free of charge and under the terms of this License, through a publicly available network server or other readily accessible means, then you must either (1) cause the Corresponding Source to be so available, or (2) arrange to deprive yourself of the benefit of the patent license for this particular work, or (3) arrange, in a manner consistent with the requirements of this License, to extend the patent license to downstream recipients. “Knowingly relying” means you have actual knowledge that, but for the patent license, your conveying the covered work in a country, or your recipient's use of the covered work in a country, would infringe one or more identifiable patents in that country that you have reason to believe are valid.
169 |
170 | If, pursuant to or in connection with a single transaction or arrangement, you convey, or propagate by procuring conveyance of, a covered work, and grant a patent license to some of the parties receiving the covered work authorizing them to use, propagate, modify or convey a specific copy of the covered work, then the patent license you grant is automatically extended to all recipients of the covered work and works based on it.
171 |
172 | A patent license is “discriminatory” if it does not include within the scope of its coverage, prohibits the exercise of, or is conditioned on the non-exercise of one or more of the rights that are specifically granted under this License. You may not convey a covered work if you are a party to an arrangement with a third party that is in the business of distributing software, under which you make payment to the third party based on the extent of your activity of conveying the work, and under which the third party grants, to any of the parties who would receive the covered work from you, a discriminatory patent license (a) in connection with copies of the covered work conveyed by you (or copies made from those copies), or (b) primarily for and in connection with specific products or compilations that contain the covered work, unless you entered into that arrangement, or that patent license was granted, prior to 28 March 2007.
173 |
174 | Nothing in this License shall be construed as excluding or limiting any implied license or other defenses to infringement that may otherwise be available to you under applicable patent law.
175 |
176 | 12. No Surrender of Others' Freedom.
177 |
178 | If conditions are imposed on you (whether by court order, agreement or otherwise) that contradict the conditions of this License, they do not excuse you from the conditions of this License. If you cannot convey a covered work so as to satisfy simultaneously your obligations under this License and any other pertinent obligations, then as a consequence you may not convey it at all. For example, if you agree to terms that obligate you to collect a royalty for further conveying from those to whom you convey the Program, the only way you could satisfy both those terms and this License would be to refrain entirely from conveying the Program.
179 |
180 | 13. Use with the GNU Affero General Public License.
181 |
182 | Notwithstanding any other provision of this License, you have permission to link or combine any covered work with a work licensed under version 3 of the GNU Affero General Public License into a single combined work, and to convey the resulting work. The terms of this License will continue to apply to the part which is the covered work, but the special requirements of the GNU Affero General Public License, section 13, concerning interaction through a network will apply to the combination as such.
183 |
184 | 14. Revised Versions of this License.
185 |
186 | The Free Software Foundation may publish revised and/or new versions of the GNU General Public License from time to time. Such new versions will be similar in spirit to the present version, but may differ in detail to address new problems or concerns.
187 |
188 | Each version is given a distinguishing version number. If the Program specifies that a certain numbered version of the GNU General Public License “or any later version” applies to it, you have the option of following the terms and conditions either of that numbered version or of any later version published by the Free Software Foundation. If the Program does not specify a version number of the GNU General Public License, you may choose any version ever published by the Free Software Foundation.
189 |
190 | If the Program specifies that a proxy can decide which future versions of the GNU General Public License can be used, that proxy's public statement of acceptance of a version permanently authorizes you to choose that version for the Program.
191 |
192 | Later license versions may give you additional or different permissions. However, no additional obligations are imposed on any author or copyright holder as a result of your choosing to follow a later version.
193 |
194 | 15. Disclaimer of Warranty.
195 |
196 | THERE IS NO WARRANTY FOR THE PROGRAM, TO THE EXTENT PERMITTED BY APPLICABLE LAW. EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT HOLDERS AND/OR OTHER PARTIES PROVIDE THE PROGRAM “AS IS” WITHOUT WARRANTY OF ANY KIND, EITHER EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE ENTIRE RISK AS TO THE QUALITY AND PERFORMANCE OF THE PROGRAM IS WITH YOU. SHOULD THE PROGRAM PROVE DEFECTIVE, YOU ASSUME THE COST OF ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
197 |
198 | 16. Limitation of Liability.
199 |
200 | IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING WILL ANY COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MODIFIES AND/OR CONVEYS THE PROGRAM AS PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY GENERAL, SPECIAL, INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE USE OR INABILITY TO USE THE PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF DATA OR DATA BEING RENDERED INACCURATE OR LOSSES SUSTAINED BY YOU OR THIRD PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE WITH ANY OTHER PROGRAMS), EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGES.
201 |
202 | 17. Interpretation of Sections 15 and 16.
203 |
204 | If the disclaimer of warranty and limitation of liability provided above cannot be given local legal effect according to their terms, reviewing courts shall apply local law that most closely approximates an absolute waiver of all civil liability in connection with the Program, unless a warranty or assumption of liability accompanies a copy of the Program in return for a fee.
205 |
206 | END OF TERMS AND CONDITIONS
207 |
208 | How to Apply These Terms to Your New Programs
209 |
210 | If you develop a new program, and you want it to be of the greatest possible use to the public, the best way to achieve this is to make it free software which everyone can redistribute and change under these terms.
211 |
212 | To do so, attach the following notices to the program. It is safest to attach them to the start of each source file to most effectively state the exclusion of warranty; and each file should have at least the “copyright” line and a pointer to where the full notice is found.
213 |
214 |
215 | Copyright (C)
216 |
217 | This program is free software: you can redistribute it and/or modify
218 | it under the terms of the GNU General Public License as published by
219 | the Free Software Foundation, either version 3 of the License, or
220 | (at your option) any later version.
221 |
222 | This program is distributed in the hope that it will be useful,
223 | but WITHOUT ANY WARRANTY; without even the implied warranty of
224 | MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
225 | GNU General Public License for more details.
226 |
227 | You should have received a copy of the GNU General Public License
228 | along with this program. If not, see .
229 | Also add information on how to contact you by electronic and paper mail.
230 |
231 | If the program does terminal interaction, make it output a short notice like this when it starts in an interactive mode:
232 |
233 | Copyright (C)
234 | This program comes with ABSOLUTELY NO WARRANTY; for details type `show w'.
235 | This is free software, and you are welcome to redistribute it
236 | under certain conditions; type `show c' for details.
237 | The hypothetical commands `show w' and `show c' should show the appropriate parts of the General Public License. Of course, your program's commands might be different; for a GUI interface, you would use an “about box”.
238 |
239 | You should also get your employer (if you work as a programmer) or school, if any, to sign a “copyright disclaimer” for the program, if necessary. For more information on this, and how to apply and follow the GNU GPL, see .
240 |
241 | The GNU General Public License does not permit incorporating your program into proprietary programs. If your program is a subroutine library, you may consider it more useful to permit linking proprietary applications with the library. If this is what you want to do, use the GNU Lesser General Public License instead of this License. But first, please read .
242 |
--------------------------------------------------------------------------------
/ChangeLog:
--------------------------------------------------------------------------------
1 | 2012-12-19
2 | * Release 1.1.1
3 | * (FilterProcessor.cpp) Change the method of determining low mean-quality for PE reads
4 | From: Sum(Q)[read1,read2] / (len(read1)+len(read2)) < Threshold
5 | To: Sum(Q)[read1]/(len(read1) < Threshold || Sum(Q)[read2]/(len(read2) < Threshold
6 |
7 | 2012-12-21
8 | * (PeBuffer.cpp::getReads) Initialize result1 and result2
9 | * (PeBuffer.cpp::readTask) Initialize result at very beginning
10 |
11 | 2012-12-25
12 | * Release 1.1.2
13 | * Change deduplication module, replace BTree with std::map
14 | * (FilterProcessor) Use External sort and merge for low memory usage
15 |
16 | 2013-4-7
17 | * Release 1.2.0.2
18 | * sRNA module start outputing two files: small.txt, clean.txt
19 | * sRNA module, length distribution starts from 10
20 | * Change SRNACleanFA.cpp and SRNAProcessor.h
21 |
22 | 2013-4-9
23 | * Release 1.3.0
24 | * Tile-filter function added to FqBuffer and PeBuffer
25 | * Filter, FilterSRNA, FilterDGE modules support Tile-filter function
26 | * Filter, FilterSRNA, FilterDGE modules all use FqBuffer and PeBuffer
27 | * When filtering tile, FilterSRNA and FilterDGE will not mandatory output anymore, while DNA remains the same
28 |
29 | 2013-4-9
30 | * Release 1.4.0
31 | * FilterMeta added
32 |
33 | 2014-9-10
34 | * Release 1.4.2
35 | * Upper limit of length changed to 512 for FilterMeta
36 |
37 | 2014-9-10
38 | * Release 1.4.2.1
39 | * Upper limit of length changed to 1024 for Filter
40 |
41 | 2014-10-14
42 | * Release 1.4.2
43 | * Bug fixed when determing seq length of read2
44 | * Bug fixed when outputing seq length of read2 in FilterProcessor.cpp
45 |
46 | 2015-01-05
47 | * Release 1.5
48 | * Add --seqtype to process another name format
49 | * Add --polyatype for 2 methods of strip PolyA, 0 for AND, 1 for OR
50 | * Load adapterlist first and align adapter then
51 | * Filter index
52 |
53 | 2015-11-12
54 | * Release 1.5.3
55 | * Add –cutAdaptor which cut adapters at ends of reads. Only reads shorter then –BaseNum will be aborted。 -cut will be ignored when applied this param
56 | * Add –BaseNum which used with -cutAdapter
57 | * No more support of adapter list
58 |
59 | 2016-03-18
60 | * Fix bug of keeping raw gz data open when filtering tile
61 |
62 | 2016-04-25
63 | * Release 1.5.4
64 | * Fix bug of low data volume when filtering SE data
65 |
66 | 2016-06-30
67 | * Release 1.5.5
68 | * Add -3 to filter module, which accompanied with -d param to output duplicated reads while not counting them in QC stats
69 |
70 | 2016-08-22
71 | * Release 1.5.6
72 | * Add --fov which supporting filtering by FOV num in name of BGI-SEQ data, which can be used with --tile
73 |
74 | 2016-12-8
75 | * Release 1.6.0
76 | * Support compling for Windows
77 | * Support Streaming IO
78 |
79 | 2017-9-15
80 | * Release 1.6.2
81 | * Change CMakeList for automatic compling
82 | * Add Python script in assist of Streaming processing
83 | * Add plotting R-scripts for QC stats
84 |
85 | 2018-4-11
86 | * Release 1.6.3
87 | * Bug fixed of incompatibility between outType and Rmdup
88 | * Bug fixed when performing adapter cutting and length filtering
89 | * Add PolyX filtering function
90 |
91 | 2018-4-23
92 | * Release 1.6.4
93 | * Fullfil CMakeList
94 | * Add QC stats of trimming position
95 | * Optimize trimming algorithm
96 | * Fix most warning of compiling
97 |
98 | 2018-6-30
99 | * Release 1.6.5
100 | * Add low-Q filtering function to sRNA module
101 | * Bug fixed when filtering adapter
102 | * Add function of filtering contaminants
103 |
104 | 2018-8-17
105 | * Release 2.0 beta
106 | * Delete filterDGE module
107 | * Unify running logic of all modules, which mainly differ in adapter determination now.
108 | * Main pipeline includes counting raw data, filtering data and counting filtered data.
109 | * Merge params of modules. Default values mainly derive from filter module, so users should be cautious when using other modules.
110 | * filter and filterMeta modules support SE and PE, filtersRNA support SE only.
111 | * filter and filterMeta modules update adapter determination algorithms.
112 | * Have 2 running modes now: ssd and non-ssd. It is recommended to use ssd mode on high-speed IO storage(like SSD). Default to be non-ssd.
113 | * Remove deduplication function.
114 | * As thread number increases, running efficiency improves obviously. It costs 70 mins to process 1.3G reads (PE50) with 16 threads at non-ssd mode.
115 |
116 | 2018-12-1
117 | * Release 2.0.1
118 | * Bugs fixed
119 | * Change of split module for fastq files after filtering
120 | * Loose standard of determining adapters
121 |
122 | 2018-12-26
123 | * Release 2.0.2
124 | * Promise order consistent for multi-threading
125 | * Lower MEM and accelerate much
126 |
127 | 2019-1-10
128 | * Release 2.0.3
129 | * Bugs fixed of SE trimming 和 fov filtering
130 |
131 | 2019-2-1
132 | * Release 2.0.5
133 | * Add extraction function on filtered data.
134 | * Fix bugs of data loading
135 |
136 | 2019-3-31
137 | * Release 2.0.6
138 | * Fix the bug of unstead output
139 | * Optimize algorithms of global determination of contaminants
140 |
141 | 2019-5-17
142 | * Release 2.0.7
143 | * Fix bugs of running on MacOS and others
144 |
145 | 2019-6-28
146 | * Release 2.0.8
147 | * Update README to adapt to 2.X
148 | * Optimize the algorithm of filtering FOV
149 |
150 | 2019-7-24
151 | * Release 2.1.0
152 | * Change the output order to the same as input, which accelerate the split function
153 | * Add randomness of random extraction
154 |
155 | 2019-12-3
156 | * Release 2.1.1
157 | * Bugs fixed of SE trim、polyX and random extraction
158 |
159 | 2020-4-19
160 | * Release 2.1.2
161 | * Add filterHts module
162 |
163 | 2020-7-23
164 | * Release 2.1.3
165 | * Support adapter list as input
166 | * Add filterStLFR module
167 | * Fix bugs of adapter detection in filterHts module
168 |
169 | 2020-8-27
170 | * Release 2.1.4.beta.1
171 | * Bugs fixed
172 |
173 | 2020-11-27
174 | * Release 2.1.4.beta.3
175 | * add rmdup function by adopting reverse Bloom filter
176 |
177 | 2020-12-17
178 | * Release 2.1.5
179 | * add rmdup function by adopting reverse Bloom filter
180 |
181 | 2021-4-30
182 | * Release 2.1.6
183 | * Fix the bug of detecting adapter in the head of read
184 | * Increase the max threading num from 48 to 72
185 |
186 | 2022-1-16
187 | * Release 2.1.7
188 | * Fix the bug when totalReadsNum parameter is too large to do the extraction
189 |
190 | 2023-8-31
191 | * Release 2.1.8
192 | * Assign max value of Quality Score by parameter
193 | * Automatically set the max threading number by software
194 | * Max supported read length increased from 500 to 1000
195 |
196 | 2024-3-28
197 | * Release 2.1.9
198 | * Fix the bug of quality value getting out of bounds and some statistical values overflowing
199 | * Remove the obsolete description about parameter L
200 | * Add some description about usage of config file
201 |
--------------------------------------------------------------------------------
/Makefile:
--------------------------------------------------------------------------------
1 | cc := g++
2 | src := ./src
3 | obj := ./obj
4 |
5 | MIN_GCC_VERSION = 4.7
6 | GCC_VERSION := $(shell gcc -dumpversion)
7 | IS_GCC_ABOVE_MIN_VERSION := $(shell echo `gcc -dumpversion | cut -f1-2 -d.` \>= $(MIN_GCC_VERSION) | bc )
8 | ifeq "$(IS_GCC_ABOVE_MIN_VERSION)" "1"
9 | GCC_VERSION_STRING := "GCC version Passes, $(GCC_VERSION) >= $(MIN_GCC_VERSION)"
10 | else
11 | GCC_VERSION_STRING := "Warning: GCC version $(GCC_VERSION) is lower than $(MIN_GCC_VERSION)."
12 | endif
13 | $(info $(GCC_VERSION_STRING))
14 |
15 |
16 |
17 | all=SOAPnuke
18 | exe=SOAPnuke
19 | MIN_ZLIB_VERSION = 1.2.3.5
20 | #ZLIB_VERSION := $(shell readlink $(whereis libz) | sed 's/libz.so.\(.*\)$/\1/g')
21 | ZLIBLOCATION:=$(shell whereis libz)
22 | ZLIB_VERSION=$(shell readlink $(ZLIBLOCATION) | awk -F 'so.' '{print $$NF}')
23 | #$(info $(ZLIB_VERSION_STRING))
24 | IS_ZLIB_ABOVE_MIN_VERSION := $(shell expr "$(ZLIB_VERSION)" ">=" "$(MIN_ZLIB_VERSION)")
25 | ifeq "$(IS_ZLIB_ABOVE_MIN_VERSION)" "1"
26 | ZLIB_VERSION_STRING := "ZLIB version Passes, $(ZLIB_VERSION) >= $(MIN_ZLIB_VERSION)"
27 | else
28 | ZLIB_VERSION_STRING := "Warning: ZLIB version $(ZLIB_VERSION) is lower than $(MIN_ZLIB_VERSION)."
29 | endif
30 | $(info $(ZLIB_VERSION_STRING))
31 |
32 | #set USEHTS true if you want use filterHts module
33 | USEHTS=false
34 |
35 | source := $(wildcard ${src}/*.cpp)
36 | source := $(filter-out ./src/mGzip.cpp, $(source))
37 | LIBS := -lz -lpthread -labc
38 | LD_FLAGS := $(foreach librarydir,$(LIBRARY_DIRS),-L$(librarydir)) $(LIBS) $(LD_FLAGS)
39 | $(warning $(LD_FLAGS))
40 | DFLAG=-lz -lpthread
41 | CXXFLAGS=-std=c++11 -g -O3
42 | ifeq "$(USEHTS)" "true"
43 | CXXFLAGS+=-D _PROCESSHTS
44 | DFLAG+= -lhts
45 | else
46 | source := $(filter-out ./src/processHts.cpp, $(source))
47 | endif
48 | #$(warning $(source))
49 | objfile := $(patsubst %.cpp,${obj}/%.o,$(notdir ${source}))
50 | objfile:=$(filter-out processHts.o,$(objfile))
51 | $(exe):${objfile}
52 | $(cc) $(objfile) -o $@ $(DFLAG)
53 | ${obj}/%.o:${src}/%.cpp mk_dir
54 | $(cc) $(CXXFLAGS) -c $< -o $@
55 | mk_dir:
56 | @if test ! -d $(obj);\
57 | then\
58 | mkdir $(obj);\
59 | fi
60 | .PHONY:clean
61 | clean:
62 | rm -f obj/*.o
63 | rm -f SOAPnuke
64 |
--------------------------------------------------------------------------------
/Readme.md:
--------------------------------------------------------------------------------
1 |
2 |
3 | ## Introduction
4 |
5 | As a novel analysis tool developed for quality control and preprocessing of FASTQ and SAM/BAM data, SOAPnuke includes 5 modules for different usage scenarios, namely **filter**, **filterHts**, **filterStLFR**, **filtersRNA** and **filterMeta**.
6 |
7 | **filter**: Preprocess FASTQ files, include trimming (adapter, low quality end and etc.) if set, discarding (adapter, low quality, high N base ratio and etc.) and generating statistic report.
8 |
9 | **filterHts**: Preprocess BAM/CRAM files. The process procedure remains the same as filter module.
10 |
11 | (Note: Input BAM/CRAM files should be sorted by readID when it contains Paired-End data.)
12 |
13 | **filterStLFR**: Preprocess stLFR FASTQ files, added with a barcode-detection step at the beginning, and support FASTQ files list as input.
14 |
15 | **filtersRNA**: Preprocess sRNA FASTQ files. Since it is still under testing, please inform us if you encounter any bug.
16 |
17 | **filterMeta**: Preprocess Meta FASTQ files. Since it is still under testing, please inform us if you encounter any bug.
18 |
19 |
20 | ## PERFORMANCE
21 |
22 | SOAPnuke 2.X version shows an excellent performance compared with 1.X version. An great acceleration has been accomplished by refactoring the whole framework, optimizing multithreading and IO.
23 |
24 | This table presents a benchmark result on 628M Paired-End 150bp reads. As thread number increases, user time obviously decreases.
25 |
26 | |Software | ThreadNum | RunTime(min) | MaxMem(MB)|Parameter|
27 | | :-----| ----: | :----: | :----: | :----: |
28 | | SOAPnuke | 16| 35.7 |2270 | filter module
29 | | SOAPnuke | 8 | 48.4 |881 | filter module
30 | | SOAPnuke | 4 | 72.1 |275 | filter module
31 | | fastp | 8 | 62.0 |1004 |-A -w 8|
32 |
33 |
34 | ## Getting started
35 | #### Requirements
36 | gcc: 4.7 or higher
37 | zlib: 1.2.3.5 or higher
38 | htslib: 1.9 or higher
39 | pthread library
40 |
41 | #### Install
42 | git clone https://github.com/BGI-flexlab/SOAPnuke.git
43 | cd SOAPnuke
44 |
45 | // Considering rarely been used and complex compile dependency, we turn off filterHts module by default.
46 | // If you want to use filterHts module, please set USEHTS true in Makefile like this:
47 | // USEHTS=true
48 |
49 | make
50 |
51 |
52 | #### QuickStart
53 |
54 | All usages start with executable file **SOAPnuke**, and different modules are invoked with different sub-commands. Here are some usage examples:
55 | ```shell
56 | #filter:
57 |
58 | #QC the input fastq and extract 10M clean reads to the output files.
59 | echo "totalReadsNum=10000000" >config.txt
60 |
61 | SOAPnuke filter -1 test.r1.fq.gz -2 test.r2.fq.gz -C clean_1.fq.gz -D clean_2.fq.gz -o result -T 8 -c config.txt
62 |
63 |
64 | #filterHts:
65 |
66 | SOAPnuke filterHts --ref chr21.fa -1 input.bam -2 output.cram -o result
67 | SOAPnuke filterHts -1 input.bam -2 output.bam -o result
68 |
69 |
70 | #filterStLFR:
71 |
72 | filterStLFR -1 fq1.list -2 fq2.list -C clean1.gz -D clean2.gz -o result -T 8 -c config
73 | ```
74 |
75 | #### Detailed QC steps
76 |
77 | If set trim-related parameters(no trim if not set), do **trimming** first:
78 |
79 |
80 |
81 | **Read ID**
82 |
83 | If parameter “index” set in config file, remove index sequence from read ID.
84 |
85 | Once “index” is set, if seqType is 0(default value), read ID would be expected like:
86 |
87 | @FCD1PB1ACXX:4:1101:1799:2201#GAAGCACG/2,
88 |
89 | “#GAAGCACG” would be removed then.
90 |
91 | If seqType is 1, read ID would be expected like:
92 |
93 | @HISEQ:310:C5MH9ANXX:1:1101:3517:20432:N:0:TCGGTCAC,
94 |
95 | “:TCGGTCAC” would be removed then.
96 |
97 | **Read sequence and quality**
98 |
99 | First, the cutting length of all trimming type would be calculated, including hard trim, low quality end trim, adapter trim and tail-polyG trim. The longest cutting would be performed.
100 |
101 | - hard trim: directly remove a certain length sequence from head or tail on read sequence
102 | - low quality end trim: remove low quality base starting from end until quality higher than cutoff
103 | - adapter trim: when adapter was found, the base sequence and quality sequence would be trimmed from the start position which match adapter
104 | - tail-polyG trim: if polyG number is greater than cutoff, then these polyG sequence in tail would be trimmed
105 |
106 | Then do **filtering**:
107 |
108 | Note that the read pair would be both discarded both when any of which fails to pass QC.
109 |
110 | Priority(High to Low):
111 |
112 | - **Tile, may be used in some types of BGI data.**
113 |
114 |
115 | If you want to discard reads with certain tile ID, set the parameter like “1101-1104,1205”.
116 |
117 | - **Fov, may be used in data from zebra-platform.**
118 |
119 |
120 | If you want to discard reads with certain FOV ID, set the parameter like “C001R003,C003R004”.
121 |
122 | - **Minimal read length**
123 |
124 |
125 | Discard a read with sequence length shorter than the parameter.
126 |
127 | - **Maximal read length**
128 |
129 |
130 | Discard a read with sequence length longer than the parameter.
131 |
132 | - **N ratio**
133 |
134 |
135 | Discard a read with N base ratio not smaller than the parameter.
136 |
137 | - **High A ratio**
138 |
139 |
140 | Discard a read with A base ratio not smaller than the parameter.
141 |
142 | - **polyX number (X means any one base)**
143 |
144 |
145 | Discard a read with poly-X number not smaller than the parameter.
146 |
147 | - **Low quality base ratio**
148 |
149 |
150 | Discard a read with low-quality bases ratio not smaller than the parameter.
151 |
152 | - **Mean quality**
153 |
154 |
155 | Discard a read of which mean quality of sequence smaller than the parameter.
156 |
157 | - **Overlapped length if PE**
158 |
159 |
160 | Discard a read pair which is suspected to be overlapped longer then the parameter.
161 |
162 | - **Adapter**
163 |
164 |
165 | Discard a read which contains an adapter.
166 |
167 | ## Parameter
168 |
169 | ### Commonly used parameters
170 |
171 |
172 |
173 | #### filter module
174 |
175 | - -1 / --fq1
176 |
177 | fq1 file(required), .gz or normal text format are both supported
178 |
179 | - -2 / --fq2
180 |
181 | fq2 file(used when process PE data), format should be same as fq1 file, both are gz or both are normal text
182 |
183 | - -C / --cleanFq1
184 |
185 | reads which passed QC from fq1 file would output to this file
186 |
187 | - -D / --cleanFq2
188 |
189 | reads which passed QC from fq2 file would output to this file
190 |
191 | - -o / --out
192 |
193 | Output directory. Processed fq files and statistical results would be output to here
194 |
195 | - -f / --adapter1
196 |
197 | adapter sequence or list file of read1
198 |
199 | - -r / --adapter2
200 |
201 | adapter sequence or list file of read2
202 |
203 | - -J / --ada_trim
204 |
205 | trim read when find adapter, it’s a bool parameter, default is false which means discard the read when find adapter
206 |
207 | - -T / --thread
208 |
209 | threads number used in process, default value is 6
210 |
211 | - -c / --configFile
212 |
213 | config file which include uncommonly used parameters. Each line contains a parameter, e.g., for value needed parameter: adaMis=2, for bool parameter: contam_trim, which means set mode as discard when find contaminant sequence
214 |
215 | - -l / --lowQual
216 |
217 | low quality threshold, default value is 5
218 |
219 | - -q / --qualRate
220 |
221 | low quality rate threshold, default value is 0.5
222 |
223 | - -n / --nRate
224 |
225 | N rate threshold, default value is 0.05
226 |
227 | - -m / --mean
228 |
229 | low average quality threshold, if you want discard reads with low average quality, you can set a value. The software do NOT check this item by default
230 |
231 | - -p / --highA
232 |
233 | ratio of A threshold in a read, the software do NOT check this item by default
234 |
235 | - -g / --polyG_tail
236 |
237 | polyG number threshold in read tail, the software do NOT check this item by default
238 |
239 | - -X / --polyX
240 |
241 | polyX number threshold, the software do NOT check this item by default
242 |
243 | - -4 / --minReadLen
244 |
245 | read minimal length, default value is 30
246 |
247 | - -h / --help
248 |
249 | Show help information
250 |
251 | - -v / --version
252 |
253 | Show version information
254 |
255 |
256 |
257 |
258 | #### filterHts module
259 |
260 | Here we only present options different from **filter** module.
261 |
262 | - -E / --ref
263 |
264 | reference file(required when process cram format)
265 |
266 | - -1
267 |
268 | input bam/cram file(required)
269 |
270 | - -2
271 |
272 | output bam/cram file(required)
273 |
274 |
275 |
276 |
277 | #### filterStLFR module
278 |
279 | Here we only present options different from **filter** module.
280 |
281 | - -1 / --fq1
282 |
283 | Support FASTQ files list as input
284 |
285 | - -2 / --fq2
286 |
287 | Support FASTQ files list as input
288 |
289 |
290 |
291 |
292 | ### Uncommonly used parameters
293 |
294 | - ctMatchR
295 |
296 | Contaminant sequence shortest consistent matching ratio [default:0.2]
297 |
298 | - seqType
299 |
300 | Sequence fq name type, 0->old fastq name, 1->new fastq name [0]
301 |
302 | old fastq name: @FCD1PB1ACXX:4:1101:1799:2201#GAAGCACG/2
303 |
304 | new fastq name: @HISEQ:310:C5MH9ANXX:1:1101:3517:2043 2:N:0:TCGGTCAC
305 |
306 | - trimFq1
307 |
308 | trim fq1 file name(gz format) [optional]
309 |
310 | - trimFq2
311 |
312 | trim fq2 file name [optional]. If trim related parameters were set on, these output files would include the total reads which only do trimming. For example, if read A failed QC after trimming, it will still output to -R/-W, but not to -C/-D
313 |
314 | - tile
315 |
316 | tile number to ignore reads, such as [1101-1104,1205]
317 |
318 | - fov
319 |
320 | fov number to ignore reads (only for zebra-platform data), such as [C001R003,C003R004]
321 |
322 | - barcodeListPath
323 |
324 | barcode list of two columns:sequence and barcodeID
325 |
326 | - barcodeRegionStr
327 |
328 | barcode regions, such as: 101_10,117_10,145_10 or 101_10,117_10,133_10
329 |
330 | - notCutNoLFR
331 |
332 | do not cut sequence when fail found barcode
333 |
334 | - inputAsList
335 |
336 | input file list not a file
337 |
338 | - tenX
339 |
340 | output tenX format
341 |
342 | - outFileType
343 |
344 | output file format: fastq or fasta[default: fastq]
345 |
346 | - index
347 |
348 | remove index
349 |
350 | - totalReadsNum
351 |
352 | number/fraction of reads you want to keep in the output clean FASTQ file(cannot be assigned when -w is given). It will extract reads randomly through the total clean FASTQ file by default, you also can get the head reads for save time by add head suffix to the integer
353 |
354 | - trim
355 |
356 | trim some bp of the read's head and tail, they means: (PE type:read1's head and tail and read2's head and tail [0,0,0,0]; SE type:read head and tail [0,0])
357 |
358 | - trimBadHead
359 |
360 | Trim from head ends until meeting high-quality base or reach the length threshold, set (quality threshold,MaxLengthForTrim) [0,0]
361 |
362 | - trimBadTail
363 |
364 | Trim from tail ends until meeting high-quality base or reach the length threshold, set (quality threshold,MaxLengthForTrim) [0,0]
365 |
366 | - overlap
367 |
368 | filter the small insert size.Not filter until the value exceed 1 [-1]
369 |
370 | - mis
371 |
372 | the maximum mismatch ratio when find overlap between PE reads(depend on -O) [0.1]
373 |
374 | - patch
375 |
376 | reads number of a patch processed [400000]
377 |
378 | - qualSys
379 |
380 | quality system 1:64, 2:33 [default:2]
381 |
382 | - outQualSys
383 |
384 | out quality system 1:64, 2:33 [default:2]
385 |
386 | - maxReadLen
387 |
388 | read max length, default 49 for filtersRNA, the software do NOT check this item by default in other modules
389 |
390 | - cleanOutSplit
391 |
392 | max reads number in each output clean FASTQ file
393 |
394 | - pe_info
395 |
396 | Add /1, /2 at the end of FASTQ name. [default: not add]
397 |
398 | - baseConvert
399 |
400 | convert base when write data, example: TtoU , means convert base T to base U in the output
401 |
402 | - log
403 |
404 | log file output path
405 |
406 |
407 |
408 | ## Plotting
409 |
410 | The three scripts in src/Rscripts/ are used for plotting QC stats from SOAPnuke.
411 |
412 | **Q20Q30.R**
413 |
414 | USAGE:
415 |
416 | Rscript src/Rscripts/Q20Q30.R Distribution_of_Q20_Q30_bases_by_read_position_1.txt Distribution_of_Q20_Q30_bases_by_read_position_2.txt q2030.png
417 |
418 | 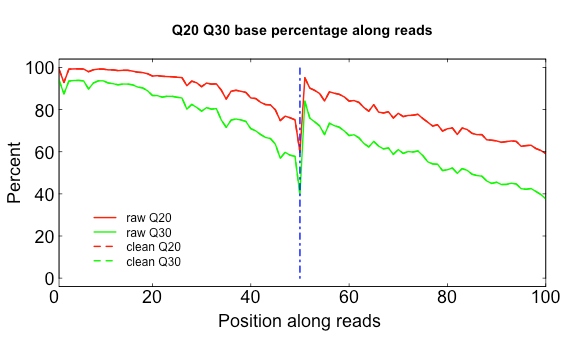
419 |
420 |
421 |
422 | **base.R**
423 |
424 | USAGE:
425 |
426 | Rscript src/Rscripts/base.R Base_distributions_by_read_position_1.txt Base_distributions_by_read_position_2.txt raw.png clean.png
427 |
428 | 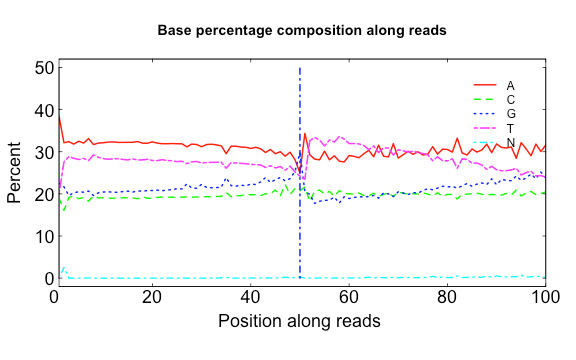
429 |
430 |
431 |
432 | **quality.R**
433 |
434 | Rscript src/Rscripts/quality.R Base_quality_value_distribution_by_read_position_1.txt Base_quality_value_distribution_by_read_position_2.txt rawQuality.png cleanQuality.png 0 0
435 |
436 | 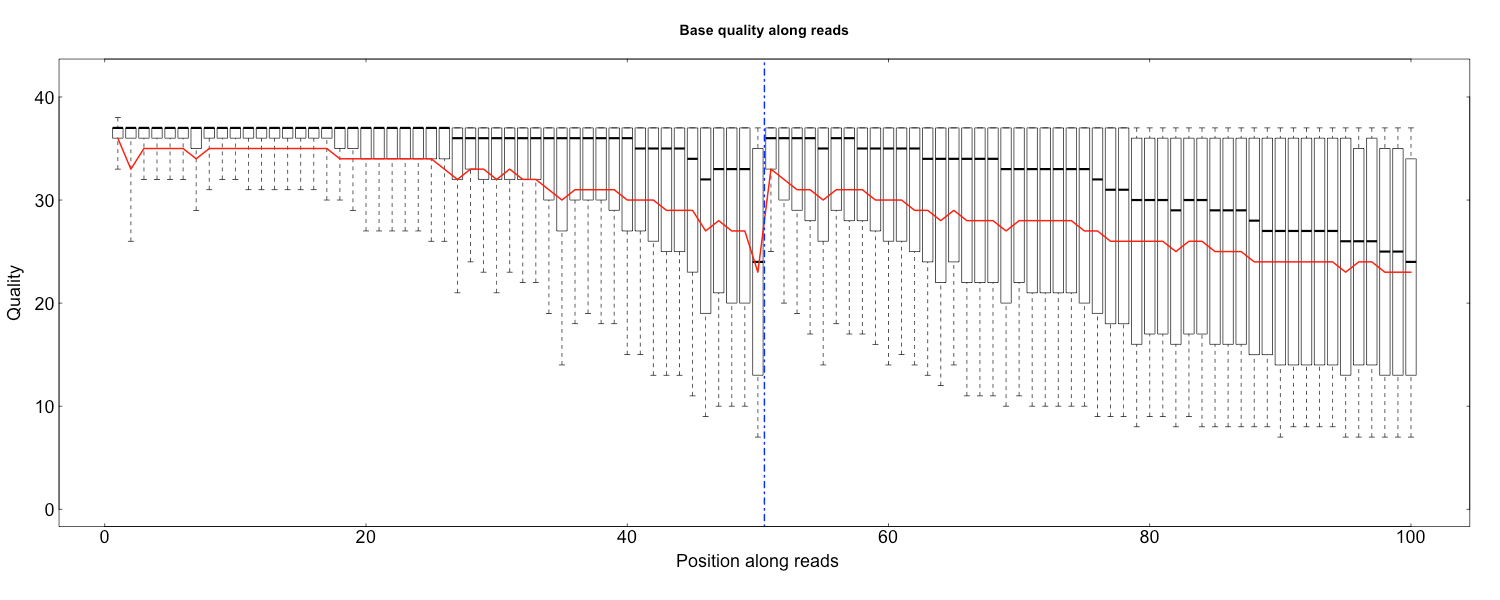
437 |
438 |
439 |
440 | ## Availability
441 |
442 | SOAPnuke is released under [GPLv3][1]. The latest source code is [freely
443 | available at github][2].
444 |
445 |
446 | ## Citing SOAPnuke
447 |
448 | - Chen Y, Chen Y, Shi C, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):1-6. doi:10.1093/gigascience/gix120 [PMID: [29220494][3]]
449 |
450 |
451 |
452 | [1]: http://en.wikipedia.org/wiki/GNU_General_Public_License
453 | [2]: https://github.com/BGI-flexlab/SOAPnuke
454 | [3]: http://www.ncbi.nlm.nih.gov/pubmed/29220494
455 |
456 |
457 |
--------------------------------------------------------------------------------
/src/BloomFilter.cpp:
--------------------------------------------------------------------------------
1 | //
2 | // Created by berry on 2020-10-09.
3 | //
4 |
5 | #include "BloomFilter.h"
6 | #include
7 |
8 | BloomFilter::BloomFilter(long long sampleSize, int multiple):multiple(20) {
9 | if(sampleSize==0){
10 | cerr<<"Error:no reads found in input file"<maxBfSize || realUseBitSize>maxBfSize){
16 | cerr<<"Error:reads number maybe is too large to do remove duplication"<> 27)) ^ str[i];
68 | }
69 | return hash;
70 | }
71 | case 2:{
72 | unsigned long hash = 0;
73 | for(int i = 0; i < str.length(); i++)
74 | {
75 | hash = str[i] + (hash << 6) + (hash << 16) - hash;
76 | }
77 | return hash;
78 | }
79 | case 3:{ // BKDRHash
80 | unsigned long seed = 131; // 31 131 1313 13131 131313 etc..
81 | unsigned long hash = 0;
82 | for(int i = 0; i < str.length(); i++)
83 | {
84 | hash = (hash * seed) + str[i];
85 | }
86 | return hash;
87 | }
88 | case 4:{
89 | unsigned long hash = 0;
90 | unsigned long x = 0;
91 | for(int i = 0; i < str.length(); i++)
92 | {
93 | hash = (hash << 4) + str[i];
94 | if((x = hash & 0xF0000000L) != 0)
95 | {
96 | hash ^= (x >> 24);
97 | }
98 | hash &= ~x;
99 | }
100 | return hash;
101 | }
102 | case 5:{
103 | unsigned long BitsInUnsignedInt = (unsigned long)(4 * 8);
104 | unsigned long ThreeQuarters = (unsigned long)((BitsInUnsignedInt * 3) / 4);
105 | unsigned long OneEighth = (unsigned long)(BitsInUnsignedInt / 8);
106 | unsigned long HighBits = (unsigned long)(0xFFFFFFFF) << (BitsInUnsignedInt - OneEighth);
107 | unsigned long hash = 0;
108 | unsigned long test = 0;
109 | for(int i = 0; i < str.length(); i++)
110 | {
111 | hash = (hash << OneEighth) + str[i];
112 | if((test = hash & HighBits) != 0)
113 | {
114 | hash = (( hash ^ (test >> ThreeQuarters)) & (~HighBits));
115 | }
116 | }
117 | return hash;
118 | }
119 | case 6:{
120 | unsigned long hash = 1315423911;
121 | for(int i = 0; i < str.length(); i++)
122 | {
123 | hash ^= ((hash << 5) + str[i] + (hash >> 2));
124 | }
125 | return hash;
126 | }
127 | case 7:{ //RSHash
128 | int b = 378551;
129 | int a = 63689;
130 | unsigned long hash = 0;
131 | for(int i = 0; i < str.length(); i++)
132 | {
133 | hash = hash * a + str[i];
134 | a = a * b;
135 | }
136 | return hash;
137 | }
138 | default:{
139 | cerr<<"Error:code error"<>smallIdx);
157 | }
158 |
159 | unsigned long BloomFilter::createHash2(string &str, int i) {
160 | string newStr=to_string(i)+str;
161 | return hash()(newStr);
162 | }
163 |
--------------------------------------------------------------------------------
/src/BloomFilter.h:
--------------------------------------------------------------------------------
1 | //
2 | // Created by berry on 2020-10-09.
3 | //
4 |
5 | #ifndef SOAPNUKE_BLOOMFILTER_H
6 | #define SOAPNUKE_BLOOMFILTER_H
7 |
8 | #include
9 | #include
10 | #include
11 | #include
12 | #include
13 | using namespace::std;
14 | #define maxBfSize 1024L*1024*1024*200
15 | class BloomFilter {
16 | private:
17 | int multiple;
18 | int hashNum;
19 | long realUseBitSize;
20 | long* indexs;
21 | char* arr;
22 | public:
23 | BloomFilter(long long sampleSize,int multiple);
24 | bool query(string& seq);
25 | void add();
26 | bool getPosStatus(long idx);
27 | void setPosStatus(long idx);
28 | static unsigned long createHash(string& seq,int i);
29 | float expectedFP();
30 | long realUseByteSize;
31 | ~BloomFilter();
32 |
33 | unsigned long createHash2(string &str, int i);
34 | };
35 |
36 |
37 | #endif //SOAPNUKE_BLOOMFILTER_H
38 |
--------------------------------------------------------------------------------
/src/ReverseBloomFilter.cpp:
--------------------------------------------------------------------------------
1 | //
2 | // Created by berry on 2020-10-19.
3 | //
4 |
5 | #include "ReverseBloomFilter.h"
6 |
7 | ReverseBloomFilter::ReverseBloomFilter(long readsNum, float multiple) {
8 | arrSize=readsNum*multiple;
9 | arr=new long[arrSize];
10 | }
11 |
12 | ReverseBloomFilter::ReverseBloomFilter(long readsNum, float multiple, long memSizeUsedInRmdup) {
13 | arrSize=readsNum*multiple;
14 | while (arrSize > maxRBfSize) {
15 | multiple -= 0.5;
16 | if (multiple < 1) {
17 | cerr << "Error:reads number maybe is too large to do remove duplication" << endl;
18 | exit(1);
19 | }
20 | }
21 | long actualMem=arrSize*8;
22 | long memG=actualMem/(1024*1024);
23 | if(actualMem>memSizeUsedInRmdup){
24 | arrSize=memSizeUsedInRmdup/8;
25 | cerr<<"Error:given memSize is small, maybe it should be at least "<()(a);
39 | while(curHash=arrSize){
56 | return -1;
57 | }else{
58 | arr[idx]=curHash;
59 | return 0;
60 | }
61 | }
62 |
--------------------------------------------------------------------------------
/src/ReverseBloomFilter.h:
--------------------------------------------------------------------------------
1 | //
2 | // Created by berry on 2020-10-19.
3 | //
4 |
5 | #ifndef SOAPNUKE_REVERSEBLOOMFILTER_H
6 | #define SOAPNUKE_REVERSEBLOOMFILTER_H
7 | #include
8 | #include
9 | #include
10 | #include
11 | #include
12 | using namespace ::std;
13 | #define maxRBfSize 1024L * 1024 * 1024 * 4
14 | #define minRBfSize 1024L * 1024 * 1000
15 | class ReverseBloomFilter
16 | {
17 | long idx;
18 | uint64_t curHash;
19 | long *arr;
20 |
21 | public:
22 | ReverseBloomFilter(long readsNum, float multiple);
23 | ReverseBloomFilter(long readsNum, float multiple, long memSizeUsedInRmdup);
24 | bool query(string &a);
25 | int add();
26 | long arrSize;
27 | };
28 |
29 | #endif // SOAPNUKE_REVERSEBLOOMFILTER_H
30 |
--------------------------------------------------------------------------------
/src/Rscripts/Q20Q30.R:
--------------------------------------------------------------------------------
1 | args <- commandArgs(TRUE)
2 | if(length(args) != 3){
3 | cat ("Usage:\n\tRscript Q20Q30.R