├── .github
└── ISSUE_TEMPLATE
│ └── summer-2020-user-question.md
├── .gitignore
├── DLCcourse.md
├── DeepLabCut-WhatNextQuestionMark.pdf
├── JAX-TutorialOct2022.pdf
├── README.md
├── SICB-tutorialJan2022_shared.pdf
├── demos_colab
├── DeepLabCut_ParentingMouse_Colab_VideoAnalysis.ipynb
└── short-parenting.mp4
├── part1-labeling.pdf
├── part2-network.pdf
├── part3-analysis.pdf
└── runtraining_andevaluation.py
/.github/ISSUE_TEMPLATE/summer-2020-user-question.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Summer 2020 user question
3 | about: template for users to ask questions about the summer 2020 content.
4 | title: ''
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | 1: Please search the issues and if related post there. Also please consider posting on the Image Forum: https://forum.image.sc/tags/deeplabcut
11 |
12 | 2. Here are the community guidelines from the main code base: https://github.com/AlexEMG/DeepLabCut#community-support-developers--help
13 |
14 | We as that on https://github.com/AlexEMG/DeepLabCut you only issue bug reports; **all** questions should go on the Image Forum: https://forum.image.sc/tags/deeplabcut.
15 |
16 | This repo's "Issues" is to start discussions for students of the course!
17 | ______________________________________________________________________________
18 |
19 | DISCUSSION QUESTION:
20 |
21 | - What is your question?
22 |
23 |
24 |
25 | - What resources have you considered? (list papers, videos, blog searches, etc).
26 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | *.egg-info/
24 | .installed.cfg
25 | *.egg
26 | MANIFEST
27 |
28 | # PyInstaller
29 | # Usually these files are written by a python script from a template
30 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
31 | *.manifest
32 | *.spec
33 |
34 | # Installer logs
35 | pip-log.txt
36 | pip-delete-this-directory.txt
37 |
38 | # Unit test / coverage reports
39 | htmlcov/
40 | .tox/
41 | .coverage
42 | .coverage.*
43 | .cache
44 | nosetests.xml
45 | coverage.xml
46 | *.cover
47 | .hypothesis/
48 | .pytest_cache/
49 |
50 | # Translations
51 | *.mo
52 | *.pot
53 |
54 | # Django stuff:
55 | *.log
56 | local_settings.py
57 | db.sqlite3
58 |
59 | # Flask stuff:
60 | instance/
61 | .webassets-cache
62 |
63 | # Scrapy stuff:
64 | .scrapy
65 |
66 | # Sphinx documentation
67 | docs/_build/
68 |
69 | # PyBuilder
70 | target/
71 |
72 | # Jupyter Notebook
73 | .ipynb_checkpoints
74 |
75 | # pyenv
76 | .python-version
77 |
78 | # celery beat schedule file
79 | celerybeat-schedule
80 |
81 | # SageMath parsed files
82 | *.sage.py
83 |
84 | # Environments
85 | .env
86 | .venv
87 | env/
88 | venv/
89 | ENV/
90 | env.bak/
91 | venv.bak/
92 |
93 | # Spyder project settings
94 | .spyderproject
95 | .spyproject
96 |
97 | # Rope project settings
98 | .ropeproject
99 |
100 | # mkdocs documentation
101 | /site
102 |
103 | # mypy
104 | .mypy_cache/
105 |
--------------------------------------------------------------------------------
/DLCcourse.md:
--------------------------------------------------------------------------------
1 | ## Got Behavior? Get Poses ...  2 |
3 | This document is an outline of resources for a course for those wanting to learn to use Python and DeepLabCut (while responsibly isolating due to COVID-19!). We expect it to take *roughly* 1-2 weeks to get through if you do it rigorously. To get the basics, it should take 1-2 days.
4 |
5 | [CLICK HERE to launch the interactive graphic to get started!](https://view.genial.ly/5fb40a49f8a0ef13943d4e5e/horizontal-infographic-review-learning-to-use-deeplabcut) (mini preview below) Or, jump in below!
6 |
7 |
2 |
3 | This document is an outline of resources for a course for those wanting to learn to use Python and DeepLabCut (while responsibly isolating due to COVID-19!). We expect it to take *roughly* 1-2 weeks to get through if you do it rigorously. To get the basics, it should take 1-2 days.
4 |
5 | [CLICK HERE to launch the interactive graphic to get started!](https://view.genial.ly/5fb40a49f8a0ef13943d4e5e/horizontal-infographic-review-learning-to-use-deeplabcut) (mini preview below) Or, jump in below!
6 |
7 |
8 |  9 |
9 |
10 |
11 | You can also chat with one another on Gitter or Twitter:
12 | [](https://gitter.im/DeepLabCut/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
13 | [](https://twitter.com/DeepLabCut)
14 |
15 |
16 | ## Installation:
17 |
18 | You need: Anaconda for python3 and DeepLabCut installed (CPU version)
19 | - you should have a [CPU version of DeepLabCut installed on your laptop](https://github.com/AlexEMG/DeepLabCut/blob/master/conda-environments/README.md). We will assume you don't all have GPUs at home, so we will
20 | utilize cloud-computing resources for those steps.
21 |
22 | - **WATCH:** overview of conda: [Python Tutorial: Anaconda - Installation and Using Conda](https://www.youtube.com/watch?v=YJC6ldI3hWk)
23 |
24 | - **ACTION:** [Install DeepLabCut](https://github.com/AlexEMG/DeepLabCut/blob/master/docs/installation.md)
25 |
26 | ## Outline:
27 |
28 | ### **The basics of computing in Python, terminal, and overview of DLC:**
29 |
30 | - **Learning:** Using the program terminal / cmd on your computer: [Video Tutorial!](https://www.youtube.com/watch?v=5XgBd6rjuDQ)
31 |
32 | - **Learning:** although minimal to no Python coding is required (i.e. you could use the DLC GUI to run the full program without it), here are some resources you may want to check out. [Software Carpentry: Programming with Python](https://swcarpentry.github.io/python-novice-inflammation/)
33 |
34 | - **Learning:** learning and teaching signal processing, and overview from Prof. Demba Ba [talk at JupyterCon](https://www.youtube.com/watch?v=ywz-LLYwkQQ)
35 |
36 | - **Learning:** Watch a talk from Alexander Mathis (a lead DeepLabCut developer) [talk about DeepLabCut!](https://www.youtube.com/watch?v=ZjWPHM0sL4E)
37 |
38 | - **DEMO:** Can I DEMO DEEPLABCUT (DLC) quickly?
39 | - Yes: [you can click through this DEMO notebook](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_DEMO_mouse_openfield.ipynb)
40 | - AND follow along with me: [Video Tutorial!](https://www.youtube.com/watch?v=DRT-Cq2vdWs)
41 |
42 |
43 | - **WATCH:** How do you know DLC is installed properly? (i.e. how to use our test script!) [Video Tutorial!](https://youtu.be/IOWtKn3l33s)
44 |
45 |
46 | 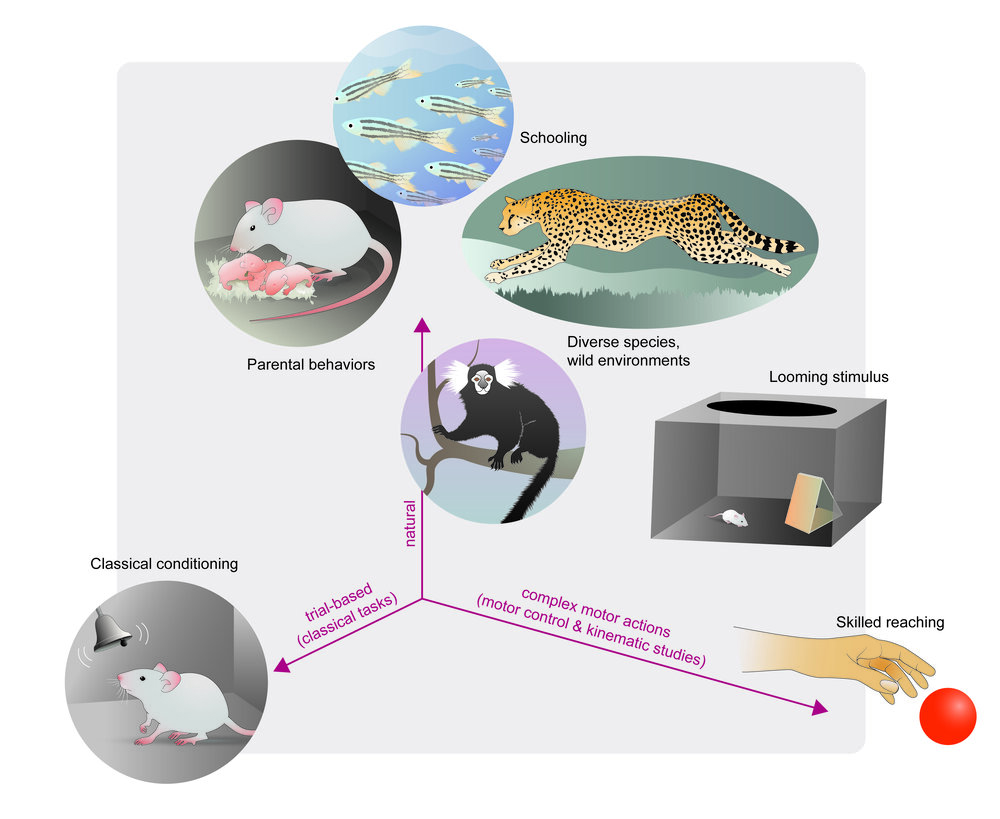 47 |
48 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
49 |
50 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
51 | https://arxiv.org/abs/2009.00564
52 |
53 | - **WATCH:** There are a lot of docs... where to begin: [Video Tutorial!](https://www.youtube.com/watch?v=A9qZidI7tL8)
54 |
55 | ### **Module 1: getting started on data**
56 |
57 | **What you need:** any videos where you can see the animals/objects, etc.
58 | You can use our demo videos, grab some from the internet, or use whatever older data you have. Any camera, color/monochrome, etc will work. Find diverse videos, and label what you want to track well :)
59 | - IF YOU ARE PART OF THE COURSE: you will be contributing to the DLC Model Zoo :smile:
60 |
61 | :purple_heart: **NOTE:** if you want to contribute back to community-science, please get in touch with us as we have a LOT of data we want to label to be able to share back with everyone; So, if you want to help sign up here (labeling can be on data we provide or possibly yours): https://forms.gle/KRtdKKYB57ZkaBwH7 :purple_heart:
62 |
63 | - **Slides:** [Overview of starting new projects](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part1-labeling.pdf)
64 | - **READ ME PLEASE:** [DeepLabCut, the science](https://rdcu.be/4Rep)
65 | - **READ ME PLEASE:** [DeepLabCut, the user guide](https://rdcu.be/bHpHN)
66 | - **WATCH:** Video tutorial 1: [using the Project Manager GUI](https://www.youtube.com/watch?v=KcXogR-p5Ak)
67 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
68 | - **WATCH:** Video tutorial 2: [using the Project Manager GUI for multi-animal pose estimation](https://www.youtube.com/watch?v=Kp-stcTm77g)
69 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
70 | - **WATCH:** Video tutorial 3: [using ipython/pythonw (more functions!)](https://www.youtube.com/watch?v=7xwOhUcIGio)
71 | - multi-animal DLC: [labeling](https://www.youtube.com/watch?v=Kp-stcTm77g)
72 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
73 |
74 | - **June 5th RECAP: AFTER LABELING (ACTION/WATCH):**
75 | - IF YOU LABELED FOR THE MODEL ZOO, [please upload your labeled data here!!](https://docs.google.com/forms/d/e/1FAIpQLSf0z6CWihGOxBUiALpN-ms4hr42xHNPAbvfeI3WxZRbEk9Reg/viewform)
76 | - Once you label on your laptop and you want to train on the cloud, please upload your project folder to google drive, and then use this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb) for single animal projects/model zoo, etc; and this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_maDLC_TrainNetwork_VideoAnalysis.ipynb) if you have a multi-animal project to create a training set, train, and start evaluating.
77 | - [VIDEO on using COLAB with your data](https://www.youtube.com/watch?v=qJGs8nxx80A)
78 |
79 |
80 | ### **Module 2: Neural Networks**
81 |
82 | - **Slides:** [Overview of creating training and test data, and training networks](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part2-network.pdf)
83 | - **READ ME PLEASE:** [What are convolutional neural networks?](https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53)
84 |
85 | - **READ ME PLEASE:** Here is a new paper from us describing challenges in robust pose estimation, why PRE-TRAINING really matters - which was our major scientific contribution to low-data input pose-estimation - and it describes new networks that are available to you. [Pretraining boosts out-of-domain robustness for pose estimation](https://paperswithcode.com/paper/pretraining-boosts-out-of-domain-robustness)
86 |
87 | - **MORE DETAILS:** ImageNet: check out the original paper and dataset: http://www.image-net.org/ (link to [ppt from Dr. Fei-Fei Li](http://www.image-net.org/papers/ImageNet_2010.ppt))
88 |
89 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
90 | https://arxiv.org/abs/2009.00564
91 |
92 |
47 |
48 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
49 |
50 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
51 | https://arxiv.org/abs/2009.00564
52 |
53 | - **WATCH:** There are a lot of docs... where to begin: [Video Tutorial!](https://www.youtube.com/watch?v=A9qZidI7tL8)
54 |
55 | ### **Module 1: getting started on data**
56 |
57 | **What you need:** any videos where you can see the animals/objects, etc.
58 | You can use our demo videos, grab some from the internet, or use whatever older data you have. Any camera, color/monochrome, etc will work. Find diverse videos, and label what you want to track well :)
59 | - IF YOU ARE PART OF THE COURSE: you will be contributing to the DLC Model Zoo :smile:
60 |
61 | :purple_heart: **NOTE:** if you want to contribute back to community-science, please get in touch with us as we have a LOT of data we want to label to be able to share back with everyone; So, if you want to help sign up here (labeling can be on data we provide or possibly yours): https://forms.gle/KRtdKKYB57ZkaBwH7 :purple_heart:
62 |
63 | - **Slides:** [Overview of starting new projects](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part1-labeling.pdf)
64 | - **READ ME PLEASE:** [DeepLabCut, the science](https://rdcu.be/4Rep)
65 | - **READ ME PLEASE:** [DeepLabCut, the user guide](https://rdcu.be/bHpHN)
66 | - **WATCH:** Video tutorial 1: [using the Project Manager GUI](https://www.youtube.com/watch?v=KcXogR-p5Ak)
67 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
68 | - **WATCH:** Video tutorial 2: [using the Project Manager GUI for multi-animal pose estimation](https://www.youtube.com/watch?v=Kp-stcTm77g)
69 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
70 | - **WATCH:** Video tutorial 3: [using ipython/pythonw (more functions!)](https://www.youtube.com/watch?v=7xwOhUcIGio)
71 | - multi-animal DLC: [labeling](https://www.youtube.com/watch?v=Kp-stcTm77g)
72 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
73 |
74 | - **June 5th RECAP: AFTER LABELING (ACTION/WATCH):**
75 | - IF YOU LABELED FOR THE MODEL ZOO, [please upload your labeled data here!!](https://docs.google.com/forms/d/e/1FAIpQLSf0z6CWihGOxBUiALpN-ms4hr42xHNPAbvfeI3WxZRbEk9Reg/viewform)
76 | - Once you label on your laptop and you want to train on the cloud, please upload your project folder to google drive, and then use this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb) for single animal projects/model zoo, etc; and this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_maDLC_TrainNetwork_VideoAnalysis.ipynb) if you have a multi-animal project to create a training set, train, and start evaluating.
77 | - [VIDEO on using COLAB with your data](https://www.youtube.com/watch?v=qJGs8nxx80A)
78 |
79 |
80 | ### **Module 2: Neural Networks**
81 |
82 | - **Slides:** [Overview of creating training and test data, and training networks](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part2-network.pdf)
83 | - **READ ME PLEASE:** [What are convolutional neural networks?](https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53)
84 |
85 | - **READ ME PLEASE:** Here is a new paper from us describing challenges in robust pose estimation, why PRE-TRAINING really matters - which was our major scientific contribution to low-data input pose-estimation - and it describes new networks that are available to you. [Pretraining boosts out-of-domain robustness for pose estimation](https://paperswithcode.com/paper/pretraining-boosts-out-of-domain-robustness)
86 |
87 | - **MORE DETAILS:** ImageNet: check out the original paper and dataset: http://www.image-net.org/ (link to [ppt from Dr. Fei-Fei Li](http://www.image-net.org/papers/ImageNet_2010.ppt))
88 |
89 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
90 | https://arxiv.org/abs/2009.00564
91 |
92 | 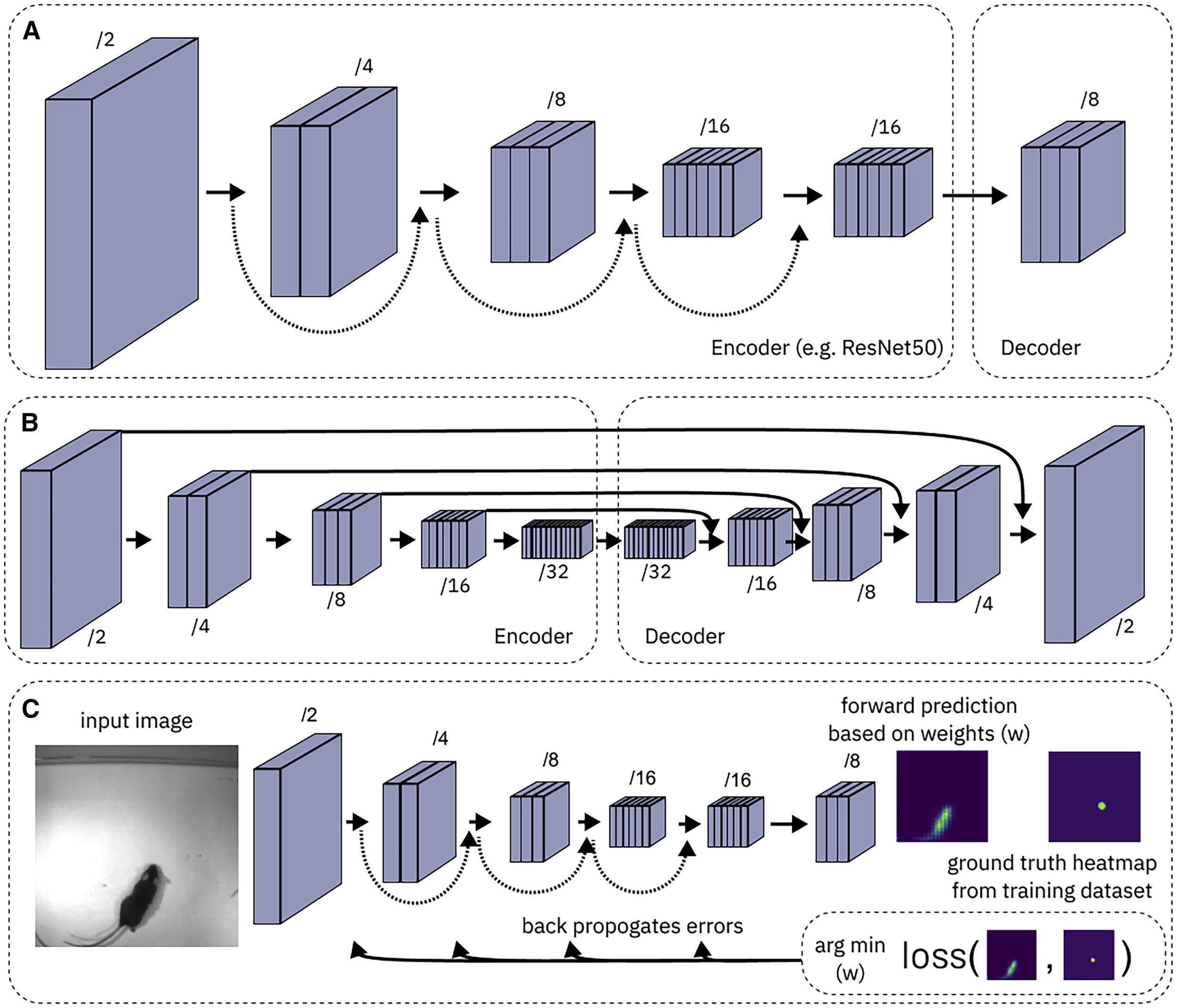 93 |
94 | Before you create a training/test set, please read/watch:
95 | - **More information:** [Which types neural networks are available, and what should I use?](https://github.com/AlexEMG/DeepLabCut/wiki/What-neural-network-should-I-use%3F)
96 | - **WATCH:** Video tutorial 1: [How to test different networks in a controlled way](https://www.youtube.com/watch?v=WXCVr6xAcCA)
97 | - Now, decide what model(s) you want to test.
98 | - IF you want to train on your CPU, then run the step `create_training_dataset`, in the GUI etc. on your own computer.
99 | - IF you want to use GPUs on google colab, [**(1)** watch this FIRST/follow along here!](https://www.youtube.com/watch?v=qJGs8nxx80A) **(2)** move your whole project folder to Google Drive, and then [**use this notebook**](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb)
100 |
101 | **MODULE 2 webinar**: https://youtu.be/ILsuC4icBU0
102 |
103 |
104 | ### **Module 3: Evalution of network performance**
105 |
106 | - **Slides** [Evalute your network](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part3-analysis.pdf)
107 | - **WATCH:** [Evaluate the network in ipython](https://www.youtube.com/watch?v=bgfnz1wtlpo)
108 | - why evaluation matters; how to benchmark; analyzing a video and using scoremaps, conf. readouts, etc.
109 |
110 | ### **Module 4: Scaling your analysis to many new videos**
111 |
112 | Once you have good networks, you can deploy them. You can create "cron jobs" to run a timed analysis script, for example. We run this daily on new videos collected in the lab. Check out a simple script to get started, and read more below:
113 |
114 | - [Analyzing videos in batches, over many folders, setting up automated data processing](https://github.com/DeepLabCut/DLCutils/tree/master/SCALE_YOUR_ANALYSIS)
115 |
116 | - How to automate your analysis in the lab: [datajoint.io](https://datajoint.io), Cron Jobs: [schedule your code runs](https://www.ostechnix.com/a-beginners-guide-to-cron-jobs/)
117 | ### **Module 5: Got Poses? Now what ...**
118 |
119 | Pose estimation took away the painful part of digitizing your data, but now what? There is a rich set of tools out there to help you create your own custom analysis, or use others (and edit them to your needs). Check out more below:
120 |
121 | - [Helper code and packages for use on DLC outputs](https://github.com/DeepLabCut/DLCutils)
122 |
123 | - Create your own machine learning classifiers: https://scikit-learn.org/stable/
124 |
125 | - **REVIEW PAPER:** [Toward a Science of Computational Ethology](https://www.sciencedirect.com/science/article/pii/S0896627314007934)
126 |
127 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
128 |
129 | - **REVIEW PAPER:** [Big behavior: challenges and opportunities in a new era of deep behavior profiling](https://www.nature.com/articles/s41386-020-0751-7)
130 |
131 | - **READ**: [Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning](https://www.pnas.org/content/112/38/E5351)
132 |
133 |
134 |
135 |
136 |
137 | *compiled and edited by Mackenzie Mathis*
138 |
--------------------------------------------------------------------------------
/DeepLabCut-WhatNextQuestionMark.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/DeepLabCut-WhatNextQuestionMark.pdf
--------------------------------------------------------------------------------
/JAX-TutorialOct2022.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/JAX-TutorialOct2022.pdf
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | [](https://twitter.com/DeepLabCut)
2 |
3 |
93 |
94 | Before you create a training/test set, please read/watch:
95 | - **More information:** [Which types neural networks are available, and what should I use?](https://github.com/AlexEMG/DeepLabCut/wiki/What-neural-network-should-I-use%3F)
96 | - **WATCH:** Video tutorial 1: [How to test different networks in a controlled way](https://www.youtube.com/watch?v=WXCVr6xAcCA)
97 | - Now, decide what model(s) you want to test.
98 | - IF you want to train on your CPU, then run the step `create_training_dataset`, in the GUI etc. on your own computer.
99 | - IF you want to use GPUs on google colab, [**(1)** watch this FIRST/follow along here!](https://www.youtube.com/watch?v=qJGs8nxx80A) **(2)** move your whole project folder to Google Drive, and then [**use this notebook**](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb)
100 |
101 | **MODULE 2 webinar**: https://youtu.be/ILsuC4icBU0
102 |
103 |
104 | ### **Module 3: Evalution of network performance**
105 |
106 | - **Slides** [Evalute your network](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part3-analysis.pdf)
107 | - **WATCH:** [Evaluate the network in ipython](https://www.youtube.com/watch?v=bgfnz1wtlpo)
108 | - why evaluation matters; how to benchmark; analyzing a video and using scoremaps, conf. readouts, etc.
109 |
110 | ### **Module 4: Scaling your analysis to many new videos**
111 |
112 | Once you have good networks, you can deploy them. You can create "cron jobs" to run a timed analysis script, for example. We run this daily on new videos collected in the lab. Check out a simple script to get started, and read more below:
113 |
114 | - [Analyzing videos in batches, over many folders, setting up automated data processing](https://github.com/DeepLabCut/DLCutils/tree/master/SCALE_YOUR_ANALYSIS)
115 |
116 | - How to automate your analysis in the lab: [datajoint.io](https://datajoint.io), Cron Jobs: [schedule your code runs](https://www.ostechnix.com/a-beginners-guide-to-cron-jobs/)
117 | ### **Module 5: Got Poses? Now what ...**
118 |
119 | Pose estimation took away the painful part of digitizing your data, but now what? There is a rich set of tools out there to help you create your own custom analysis, or use others (and edit them to your needs). Check out more below:
120 |
121 | - [Helper code and packages for use on DLC outputs](https://github.com/DeepLabCut/DLCutils)
122 |
123 | - Create your own machine learning classifiers: https://scikit-learn.org/stable/
124 |
125 | - **REVIEW PAPER:** [Toward a Science of Computational Ethology](https://www.sciencedirect.com/science/article/pii/S0896627314007934)
126 |
127 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
128 |
129 | - **REVIEW PAPER:** [Big behavior: challenges and opportunities in a new era of deep behavior profiling](https://www.nature.com/articles/s41386-020-0751-7)
130 |
131 | - **READ**: [Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning](https://www.pnas.org/content/112/38/E5351)
132 |
133 |
134 |
135 |
136 |
137 | *compiled and edited by Mackenzie Mathis*
138 |
--------------------------------------------------------------------------------
/DeepLabCut-WhatNextQuestionMark.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/DeepLabCut-WhatNextQuestionMark.pdf
--------------------------------------------------------------------------------
/JAX-TutorialOct2022.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/JAX-TutorialOct2022.pdf
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | [](https://twitter.com/DeepLabCut)
2 |
3 |
4 |  5 |
5 |
6 |
7 |
8 | **Due to COVID-19 we have released a free [self-paced online course!](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/DLCcourse.md)**
9 |
10 | [CLICK HERE to lauch the interactive graphic to get started!](https://view.genial.ly/5fb40a49f8a0ef13943d4e5e/horizontal-infographic-review-learning-to-use-deeplabcut) (mini preview below)
11 |
12 |
13 |  14 |
14 |
15 |
16 | # Recent Workshop slides:
17 |
18 | - October, 2022; [Slides from the quick DeepLabCut demo](JAX-TutorialOct2022.pdf) at the [SHORT COURSE ON THE APPLICATION OF MACHINE LEARNING FOR AUTOMATED QUANTIFICATION OF BEHAVIOR at JAX Laboratory](https://www.jax.org/education-and-learning/education-calendar/2022/october/short-course-on-the-application-of-machine-learning-for-automated-quantific)
19 | - January, 2022, [Slides for Society of Integrative and Comparative Biology Workshop on DeepLabCut](SICB-tutorialJan2022_shared.pdf)
20 |
21 | # DeepLabCut 2020 Workshop Materials:
22 |
23 | Notice that details on installation, usage and examples can be found on the standard repository: https://github.com/DeepLabCut/DeepLabCut
24 |
25 | Here, we share selected materials from our workshops for using DeepLabCut:
26 |
27 | [Demo session 1: project creation and labeling](part1-labeling.pdf)
28 |
29 | [Demo session 2: network selection training and evaluation](part2-network.pdf)
30 |
31 | [Demo session 3: analysis and active learning](part3-analysis.pdf)
32 |
33 | [Outlook slides on analysis avenues with postural data](DeepLabCut-WhatNextQuestionMark.pdf)
34 |
35 | [Example script for automating analysis with deeplabcut](runtraining_andevaluation.py)
36 |
37 | [User guide for DeepLabCut published in Nature Protcols](https://www.nature.com/articles/s41596-019-0176-0)
38 |
39 | And please check the [GitHub documentation](https://github.com/AlexEMG/DeepLabCut/blob/master/docs/UseOverviewGuide.md) for more features and updates!
40 |
41 | [YouTube Tutorial Videos](https://www.youtube.com/channel/UC2HEbWpC_1v6i9RnDMy-dfA)
42 |
43 | # Interactive Demos
44 |
45 | You can also use Google Colaboratory to run the demo data (without any installation):
46 | [](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_DEMO_mouse_openfield.ipynb)
47 |
48 | Notice that more demos can be found in the examples folder of the [DeepLabCut github repo](https://github.com/AlexEMG/DeepLabCut/tree/master/examples).
49 |
50 | # Past and present workshop schedule
51 |
52 | - October 2022: Workshop on DeepLabCut at the the [SHORT COURSE ON THE APPLICATION OF MACHINE LEARNING FOR AUTOMATED QUANTIFICATION OF BEHAVIOR at JAX Laboratory](https://www.jax.org/education-and-learning/education-calendar/2022/october/short-course-on-the-application-of-machine-learning-for-automated-quantific); slides available [here](JAX-TutorialOct2022.pdf)
53 | - January 2022: Workshop on DeepLabCut at the Society of Integrative and Comparative Biology Workshop in Phoenix, USA (slides available [here](SICB-tutorialJan2022_shared.pdf))
54 | - October 2021: International Neural Regeneration Symposium Workshop 1: Machine & Deep Learning Models in Nantong, China
55 | - June 2021: Virtual visit to Marine Biological Laboratory, Woods Hole, MA as part of [The Neural Systems and Behavior Course in Woods Hole](https://www.mbl.edu/nsb/) -- see: https://twitter.com/NSB_MBL/status/1401548173798264837
56 | - December, 2020: [DLC-live](https://github.com/DeepLabCut/DeepLabCut-live) was published (one of the topics the hackathon contributed to!)
57 | - March 5-6, 2020: First DLC Hackathon: focus on hardware integration. Many exciting developments will appear -- Stay tuned!
58 | - Jan 16-17, 2020: Second Annual Workshop at the Rowland Institute at Harvard!
59 | - July 2019: [Neural Data Science Course at Cold Spring Harbor Laboratory](https://meetings.cshl.edu/courses.aspx?course=c-neudata&year=19)
60 | - June 2019: Marine Biological Laboratory, Woods Hole, MA as part of [The Neural Systems and Behavior Course in Woods Hole](https://www.mbl.edu/nsb/). The ribbon fin tracking below is from that course. See http://www.mousemotorlab.org/deeplabcutdetails for more details.
61 | - June 29-July 6, 2019: CVSS 2019: Black Forest, Germany (http://orga.cvss.cc/)
62 | - Jan 28-29 2019: [Workshop at the Rowland Institute at Harvard](https://www2.rowland.harvard.edu/news/adaptive-motor-control-workshop)
63 | - Jan 7 2019: Marine Biological Laboratory, Woods Hole, MA (locally organized by Dr. Yisrael Schnytzer)
64 | - Dec 13-14 2018: Nencki Institute of Experimental Biology, Warsaw (locally organized by Mateusz Kostecki)
65 | - Dec 10-11 2018: Ludwig-Maximilians-University, Munich (locally organized by Kate Stynik)
66 |
67 |
68 |
69 | 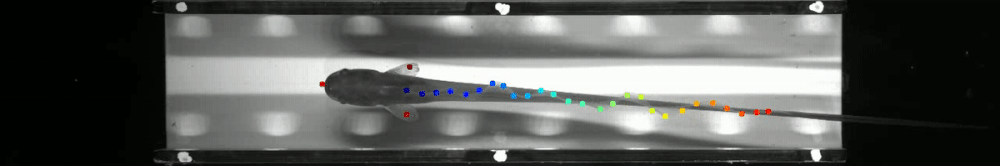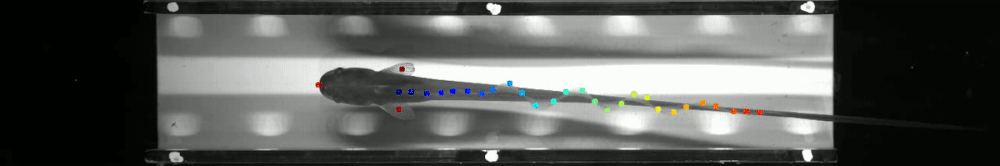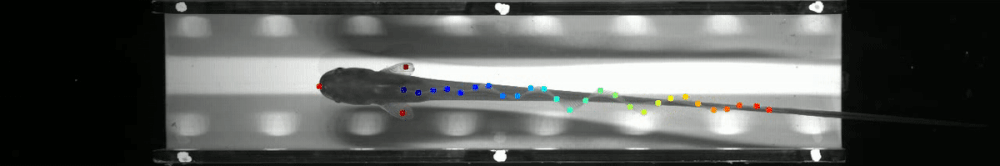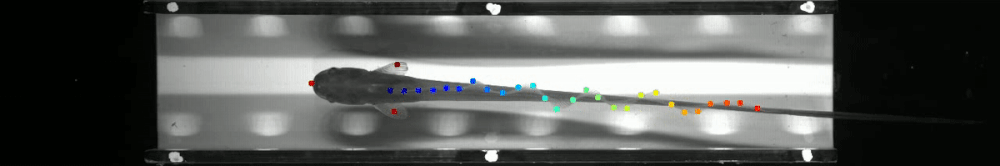 70 |
70 |
71 |
--------------------------------------------------------------------------------
/SICB-tutorialJan2022_shared.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/SICB-tutorialJan2022_shared.pdf
--------------------------------------------------------------------------------
/demos_colab/short-parenting.mp4:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/demos_colab/short-parenting.mp4
--------------------------------------------------------------------------------
/part1-labeling.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/part1-labeling.pdf
--------------------------------------------------------------------------------
/part2-network.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/part2-network.pdf
--------------------------------------------------------------------------------
/part3-analysis.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/part3-analysis.pdf
--------------------------------------------------------------------------------
/runtraining_andevaluation.py:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env python3
2 | # -*- coding: utf-8 -*-
3 | """
4 | Created on Sat Nov 17 14:12:43 2018
5 |
6 | An example script to automate analysis on 3 different GPUs for different projects. Feel free to adapt this to your needs!
7 |
8 | @author: alex
9 |
10 | First start container:
11 | python3 runtraining_andevaluation.py 1 (2 or 3)
12 |
13 | """
14 |
15 | import subprocess, sys
16 | import numpy as np
17 | import itertools
18 | import os
19 |
20 | import deeplabcut
21 |
22 | Maxiter=int(1.5*10**5)
23 |
24 | model=int(sys.argv[1])
25 |
26 | Projects=[['project1-phoenix-2019-01-28'],['ComplexWheelD3-12-Fumi-2019-01-28', 'maze-ariel-2019-01-28'], ['TBI-BvA-2019-01-28','group-eli-2019-01-28']]
27 |
28 | shuffle=1
29 |
30 | prefix='/home/alex/DLC-workshopRowland'
31 |
32 | for project in Projects[model]:
33 | projectpath=os.path.join(prefix,project)
34 | config=os.path.join(projectpath,'config.yaml')
35 |
36 | cfg=deeplabcut.auxiliaryfunctions.read_config(config)
37 | previous_path=cfg['project_path']
38 |
39 | cfg['project_path']=projectpath
40 | deeplabcut.auxiliaryfunctions.write_config(config,cfg)
41 |
42 | print("This is the name of the script: ", sys.argv[0])
43 | print("Shuffle: ", shuffle)
44 | print("config: ", config)
45 |
46 | deeplabcut.create_training_dataset(config, Shuffles=[shuffle],windows2linux=True)
47 |
48 | deeplabcut.train_network(config, shuffle=shuffle, max_snapshots_to_keep=5, maxiters=Maxiter)
49 | print("Evaluating...")
50 | deeplabcut.evaluate_network(config, Shuffles=[shuffle],plotting=True)
51 |
52 | print("Analyzing videos..., switching to last snapshot...")
53 | #cfg=deeplabcut.auxiliaryfunctions.read_config(config)
54 | #cfg['snapshotindex']=-1
55 | #deeplabcut.auxiliaryfunctions.write_config(config,cfg)
56 |
57 | for vtype in ['.mp4','.m4v','.mpg']:
58 | try:
59 | deeplabcut.analyze_videos(config,[str(os.path.join(projectpath,'videos'))],shuffle=shuffle,videotype=vtype,save_as_csv=True)
60 | except:
61 | pass
62 |
63 | print("DONE WITH ", project," resetting to original path")
64 | cfg['project_path']=previous_path
65 | deeplabcut.auxiliaryfunctions.write_config(config,cfg)
66 |
--------------------------------------------------------------------------------
 9 |
9 |  2 |
3 | This document is an outline of resources for a course for those wanting to learn to use Python and DeepLabCut (while responsibly isolating due to COVID-19!). We expect it to take *roughly* 1-2 weeks to get through if you do it rigorously. To get the basics, it should take 1-2 days.
4 |
5 | [CLICK HERE to launch the interactive graphic to get started!](https://view.genial.ly/5fb40a49f8a0ef13943d4e5e/horizontal-infographic-review-learning-to-use-deeplabcut) (mini preview below) Or, jump in below!
6 |
7 |
2 |
3 | This document is an outline of resources for a course for those wanting to learn to use Python and DeepLabCut (while responsibly isolating due to COVID-19!). We expect it to take *roughly* 1-2 weeks to get through if you do it rigorously. To get the basics, it should take 1-2 days.
4 |
5 | [CLICK HERE to launch the interactive graphic to get started!](https://view.genial.ly/5fb40a49f8a0ef13943d4e5e/horizontal-infographic-review-learning-to-use-deeplabcut) (mini preview below) Or, jump in below!
6 |
7 |  9 |
9 |  47 |
48 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
49 |
50 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
51 | https://arxiv.org/abs/2009.00564
52 |
53 | - **WATCH:** There are a lot of docs... where to begin: [Video Tutorial!](https://www.youtube.com/watch?v=A9qZidI7tL8)
54 |
55 | ### **Module 1: getting started on data**
56 |
57 | **What you need:** any videos where you can see the animals/objects, etc.
58 | You can use our demo videos, grab some from the internet, or use whatever older data you have. Any camera, color/monochrome, etc will work. Find diverse videos, and label what you want to track well :)
59 | - IF YOU ARE PART OF THE COURSE: you will be contributing to the DLC Model Zoo :smile:
60 |
61 | :purple_heart: **NOTE:** if you want to contribute back to community-science, please get in touch with us as we have a LOT of data we want to label to be able to share back with everyone; So, if you want to help sign up here (labeling can be on data we provide or possibly yours): https://forms.gle/KRtdKKYB57ZkaBwH7 :purple_heart:
62 |
63 | - **Slides:** [Overview of starting new projects](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part1-labeling.pdf)
64 | - **READ ME PLEASE:** [DeepLabCut, the science](https://rdcu.be/4Rep)
65 | - **READ ME PLEASE:** [DeepLabCut, the user guide](https://rdcu.be/bHpHN)
66 | - **WATCH:** Video tutorial 1: [using the Project Manager GUI](https://www.youtube.com/watch?v=KcXogR-p5Ak)
67 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
68 | - **WATCH:** Video tutorial 2: [using the Project Manager GUI for multi-animal pose estimation](https://www.youtube.com/watch?v=Kp-stcTm77g)
69 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
70 | - **WATCH:** Video tutorial 3: [using ipython/pythonw (more functions!)](https://www.youtube.com/watch?v=7xwOhUcIGio)
71 | - multi-animal DLC: [labeling](https://www.youtube.com/watch?v=Kp-stcTm77g)
72 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
73 |
74 | - **June 5th RECAP: AFTER LABELING (ACTION/WATCH):**
75 | - IF YOU LABELED FOR THE MODEL ZOO, [please upload your labeled data here!!](https://docs.google.com/forms/d/e/1FAIpQLSf0z6CWihGOxBUiALpN-ms4hr42xHNPAbvfeI3WxZRbEk9Reg/viewform)
76 | - Once you label on your laptop and you want to train on the cloud, please upload your project folder to google drive, and then use this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb) for single animal projects/model zoo, etc; and this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_maDLC_TrainNetwork_VideoAnalysis.ipynb) if you have a multi-animal project to create a training set, train, and start evaluating.
77 | - [VIDEO on using COLAB with your data](https://www.youtube.com/watch?v=qJGs8nxx80A)
78 |
79 |
80 | ### **Module 2: Neural Networks**
81 |
82 | - **Slides:** [Overview of creating training and test data, and training networks](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part2-network.pdf)
83 | - **READ ME PLEASE:** [What are convolutional neural networks?](https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53)
84 |
85 | - **READ ME PLEASE:** Here is a new paper from us describing challenges in robust pose estimation, why PRE-TRAINING really matters - which was our major scientific contribution to low-data input pose-estimation - and it describes new networks that are available to you. [Pretraining boosts out-of-domain robustness for pose estimation](https://paperswithcode.com/paper/pretraining-boosts-out-of-domain-robustness)
86 |
87 | - **MORE DETAILS:** ImageNet: check out the original paper and dataset: http://www.image-net.org/ (link to [ppt from Dr. Fei-Fei Li](http://www.image-net.org/papers/ImageNet_2010.ppt))
88 |
89 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
90 | https://arxiv.org/abs/2009.00564
91 |
92 |
47 |
48 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
49 |
50 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
51 | https://arxiv.org/abs/2009.00564
52 |
53 | - **WATCH:** There are a lot of docs... where to begin: [Video Tutorial!](https://www.youtube.com/watch?v=A9qZidI7tL8)
54 |
55 | ### **Module 1: getting started on data**
56 |
57 | **What you need:** any videos where you can see the animals/objects, etc.
58 | You can use our demo videos, grab some from the internet, or use whatever older data you have. Any camera, color/monochrome, etc will work. Find diverse videos, and label what you want to track well :)
59 | - IF YOU ARE PART OF THE COURSE: you will be contributing to the DLC Model Zoo :smile:
60 |
61 | :purple_heart: **NOTE:** if you want to contribute back to community-science, please get in touch with us as we have a LOT of data we want to label to be able to share back with everyone; So, if you want to help sign up here (labeling can be on data we provide or possibly yours): https://forms.gle/KRtdKKYB57ZkaBwH7 :purple_heart:
62 |
63 | - **Slides:** [Overview of starting new projects](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part1-labeling.pdf)
64 | - **READ ME PLEASE:** [DeepLabCut, the science](https://rdcu.be/4Rep)
65 | - **READ ME PLEASE:** [DeepLabCut, the user guide](https://rdcu.be/bHpHN)
66 | - **WATCH:** Video tutorial 1: [using the Project Manager GUI](https://www.youtube.com/watch?v=KcXogR-p5Ak)
67 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
68 | - **WATCH:** Video tutorial 2: [using the Project Manager GUI for multi-animal pose estimation](https://www.youtube.com/watch?v=Kp-stcTm77g)
69 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
70 | - **WATCH:** Video tutorial 3: [using ipython/pythonw (more functions!)](https://www.youtube.com/watch?v=7xwOhUcIGio)
71 | - multi-animal DLC: [labeling](https://www.youtube.com/watch?v=Kp-stcTm77g)
72 | - Please go from project creation (use >1 video!) to labeling your data, and then check the labels!
73 |
74 | - **June 5th RECAP: AFTER LABELING (ACTION/WATCH):**
75 | - IF YOU LABELED FOR THE MODEL ZOO, [please upload your labeled data here!!](https://docs.google.com/forms/d/e/1FAIpQLSf0z6CWihGOxBUiALpN-ms4hr42xHNPAbvfeI3WxZRbEk9Reg/viewform)
76 | - Once you label on your laptop and you want to train on the cloud, please upload your project folder to google drive, and then use this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb) for single animal projects/model zoo, etc; and this [COLAB NOTEBOOK](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_maDLC_TrainNetwork_VideoAnalysis.ipynb) if you have a multi-animal project to create a training set, train, and start evaluating.
77 | - [VIDEO on using COLAB with your data](https://www.youtube.com/watch?v=qJGs8nxx80A)
78 |
79 |
80 | ### **Module 2: Neural Networks**
81 |
82 | - **Slides:** [Overview of creating training and test data, and training networks](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part2-network.pdf)
83 | - **READ ME PLEASE:** [What are convolutional neural networks?](https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53)
84 |
85 | - **READ ME PLEASE:** Here is a new paper from us describing challenges in robust pose estimation, why PRE-TRAINING really matters - which was our major scientific contribution to low-data input pose-estimation - and it describes new networks that are available to you. [Pretraining boosts out-of-domain robustness for pose estimation](https://paperswithcode.com/paper/pretraining-boosts-out-of-domain-robustness)
86 |
87 | - **MORE DETAILS:** ImageNet: check out the original paper and dataset: http://www.image-net.org/ (link to [ppt from Dr. Fei-Fei Li](http://www.image-net.org/papers/ImageNet_2010.ppt))
88 |
89 | - **NEW! REVIEW PAPER:** A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
90 | https://arxiv.org/abs/2009.00564
91 |
92 |  93 |
94 | Before you create a training/test set, please read/watch:
95 | - **More information:** [Which types neural networks are available, and what should I use?](https://github.com/AlexEMG/DeepLabCut/wiki/What-neural-network-should-I-use%3F)
96 | - **WATCH:** Video tutorial 1: [How to test different networks in a controlled way](https://www.youtube.com/watch?v=WXCVr6xAcCA)
97 | - Now, decide what model(s) you want to test.
98 | - IF you want to train on your CPU, then run the step `create_training_dataset`, in the GUI etc. on your own computer.
99 | - IF you want to use GPUs on google colab, [**(1)** watch this FIRST/follow along here!](https://www.youtube.com/watch?v=qJGs8nxx80A) **(2)** move your whole project folder to Google Drive, and then [**use this notebook**](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb)
100 |
101 | **MODULE 2 webinar**: https://youtu.be/ILsuC4icBU0
102 |
103 |
104 | ### **Module 3: Evalution of network performance**
105 |
106 | - **Slides** [Evalute your network](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part3-analysis.pdf)
107 | - **WATCH:** [Evaluate the network in ipython](https://www.youtube.com/watch?v=bgfnz1wtlpo)
108 | - why evaluation matters; how to benchmark; analyzing a video and using scoremaps, conf. readouts, etc.
109 |
110 | ### **Module 4: Scaling your analysis to many new videos**
111 |
112 | Once you have good networks, you can deploy them. You can create "cron jobs" to run a timed analysis script, for example. We run this daily on new videos collected in the lab. Check out a simple script to get started, and read more below:
113 |
114 | - [Analyzing videos in batches, over many folders, setting up automated data processing](https://github.com/DeepLabCut/DLCutils/tree/master/SCALE_YOUR_ANALYSIS)
115 |
116 | - How to automate your analysis in the lab: [datajoint.io](https://datajoint.io), Cron Jobs: [schedule your code runs](https://www.ostechnix.com/a-beginners-guide-to-cron-jobs/)
117 | ### **Module 5: Got Poses? Now what ...**
118 |
119 | Pose estimation took away the painful part of digitizing your data, but now what? There is a rich set of tools out there to help you create your own custom analysis, or use others (and edit them to your needs). Check out more below:
120 |
121 | - [Helper code and packages for use on DLC outputs](https://github.com/DeepLabCut/DLCutils)
122 |
123 | - Create your own machine learning classifiers: https://scikit-learn.org/stable/
124 |
125 | - **REVIEW PAPER:** [Toward a Science of Computational Ethology](https://www.sciencedirect.com/science/article/pii/S0896627314007934)
126 |
127 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
128 |
129 | - **REVIEW PAPER:** [Big behavior: challenges and opportunities in a new era of deep behavior profiling](https://www.nature.com/articles/s41386-020-0751-7)
130 |
131 | - **READ**: [Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning](https://www.pnas.org/content/112/38/E5351)
132 |
133 |
134 |
135 |
136 |
137 | *compiled and edited by Mackenzie Mathis*
138 |
--------------------------------------------------------------------------------
/DeepLabCut-WhatNextQuestionMark.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/DeepLabCut-WhatNextQuestionMark.pdf
--------------------------------------------------------------------------------
/JAX-TutorialOct2022.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/JAX-TutorialOct2022.pdf
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | [](https://twitter.com/DeepLabCut)
2 |
3 |
93 |
94 | Before you create a training/test set, please read/watch:
95 | - **More information:** [Which types neural networks are available, and what should I use?](https://github.com/AlexEMG/DeepLabCut/wiki/What-neural-network-should-I-use%3F)
96 | - **WATCH:** Video tutorial 1: [How to test different networks in a controlled way](https://www.youtube.com/watch?v=WXCVr6xAcCA)
97 | - Now, decide what model(s) you want to test.
98 | - IF you want to train on your CPU, then run the step `create_training_dataset`, in the GUI etc. on your own computer.
99 | - IF you want to use GPUs on google colab, [**(1)** watch this FIRST/follow along here!](https://www.youtube.com/watch?v=qJGs8nxx80A) **(2)** move your whole project folder to Google Drive, and then [**use this notebook**](https://github.com/DeepLabCut/DeepLabCut/blob/master/examples/COLAB/COLAB_YOURDATA_TrainNetwork_VideoAnalysis.ipynb)
100 |
101 | **MODULE 2 webinar**: https://youtu.be/ILsuC4icBU0
102 |
103 |
104 | ### **Module 3: Evalution of network performance**
105 |
106 | - **Slides** [Evalute your network](https://github.com/DeepLabCut/DeepLabCut-Workshop-Materials/blob/master/part3-analysis.pdf)
107 | - **WATCH:** [Evaluate the network in ipython](https://www.youtube.com/watch?v=bgfnz1wtlpo)
108 | - why evaluation matters; how to benchmark; analyzing a video and using scoremaps, conf. readouts, etc.
109 |
110 | ### **Module 4: Scaling your analysis to many new videos**
111 |
112 | Once you have good networks, you can deploy them. You can create "cron jobs" to run a timed analysis script, for example. We run this daily on new videos collected in the lab. Check out a simple script to get started, and read more below:
113 |
114 | - [Analyzing videos in batches, over many folders, setting up automated data processing](https://github.com/DeepLabCut/DLCutils/tree/master/SCALE_YOUR_ANALYSIS)
115 |
116 | - How to automate your analysis in the lab: [datajoint.io](https://datajoint.io), Cron Jobs: [schedule your code runs](https://www.ostechnix.com/a-beginners-guide-to-cron-jobs/)
117 | ### **Module 5: Got Poses? Now what ...**
118 |
119 | Pose estimation took away the painful part of digitizing your data, but now what? There is a rich set of tools out there to help you create your own custom analysis, or use others (and edit them to your needs). Check out more below:
120 |
121 | - [Helper code and packages for use on DLC outputs](https://github.com/DeepLabCut/DLCutils)
122 |
123 | - Create your own machine learning classifiers: https://scikit-learn.org/stable/
124 |
125 | - **REVIEW PAPER:** [Toward a Science of Computational Ethology](https://www.sciencedirect.com/science/article/pii/S0896627314007934)
126 |
127 | - **REVIEW PAPER:** The state of animal pose estimation w/ deep learning i.e. "Deep learning tools for the measurement of animal behavior in neuroscience" [arXiv](https://arxiv.org/abs/1909.13868) & [published version](https://www.sciencedirect.com/science/article/pii/S0959438819301151)
128 |
129 | - **REVIEW PAPER:** [Big behavior: challenges and opportunities in a new era of deep behavior profiling](https://www.nature.com/articles/s41386-020-0751-7)
130 |
131 | - **READ**: [Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning](https://www.pnas.org/content/112/38/E5351)
132 |
133 |
134 |
135 |
136 |
137 | *compiled and edited by Mackenzie Mathis*
138 |
--------------------------------------------------------------------------------
/DeepLabCut-WhatNextQuestionMark.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/DeepLabCut-WhatNextQuestionMark.pdf
--------------------------------------------------------------------------------
/JAX-TutorialOct2022.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/DeepLabCut/DeepLabCut-Workshop-Materials/39d29ab39274eb2ca116816c1d545a01e4185c3d/JAX-TutorialOct2022.pdf
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | [](https://twitter.com/DeepLabCut)
2 |
3 |  5 |
5 |  14 |
14 |  70 |
70 |