

Prediction of Drug-likeness using Graph Convolutional Attention Network.
4 |
A deep learning-based process related to the screening of SARS-CoV2 3CL inhibitors.
4 |
5 |
6 | ## process
7 |
8 | 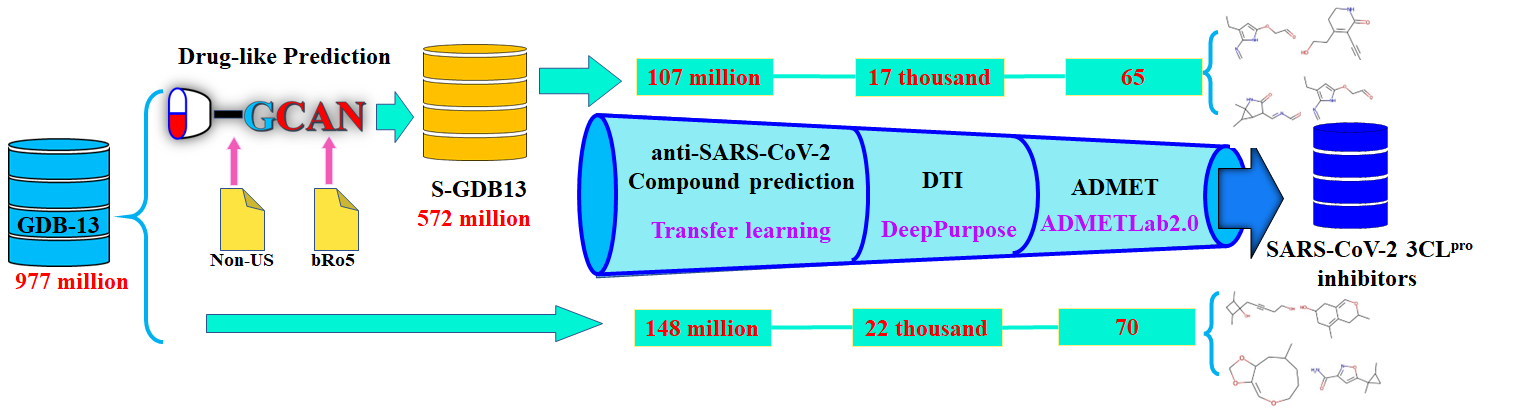
9 |
10 | Coronavirus disease 2019 (COVID-19) is a highly infectious disease caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). It is urgent to find potential antiviral drugs against SARS-CoV-2 in a short time. Deep learning-based virtual screening is one of the approaches that can rapidly search against large molecular libraries. Here, SARS-CoV-2 3C-like protease (SARS-CoV-2 3CLpro) was chosen as the target. As shown in Figure bellow, the utility of D-GCAN is evaluated by comparing the screening results on the GDB-13 and S-GDB13 databases. The process was carried out with the help of the transfer learning method (Wang et al., 2021), DeepPurpose (Huang et al., 2020), and ADMETLab2.0 (Xiong et al., 2021).
11 |
12 | These databases were firstly screened by using a transfer learning method (COVIDVS) proposed by Wang et al (Wang et al., 2021), which was reported for screening inhibitors against SARS-CoV-2. The model was trained on the dataset containing inhibitors against HCoV-OC43, SARS-CoV and MERS-CoV. All of these viruses as well as SARS-CoV-2 belong to β-coronaviruses. They have high consistency in essential functional proteins (Wu et al., 2020; Shen et al.; Pillaiyar et al., 2020). Then, the trained model was fine-tuned by the transfer learning approach with the dataset containing drugs against SARS-CoV-2. In this way, 107 million drug-like molecules were screened out. Then, drug-target interaction prediction (DTI) was carried out based on DeepPurpose (Huang et al., 2020), which provided pretrained model for the interaction prediction between drugs and SARS-CoV-2 3CLpro target. The interaction binding score was evaluated by the dissociation equilibrium constant (Kd). After this step, 17 thousand molecules with high affinity were obtained. Finally, ADMET properties were widely chosen and used for screening SARS-CoV-2 inhibitors (Gajjar et al., 2021; Roy et al., 2021; Dhameliya et al., 2022). These properties were calculated by using ADMETLab2.0 (Xiong et al., 2021), and 65 candidates with good properties were selected.
13 |
14 | ## COVIDVS
15 |
16 | COVIDVS models are Chemprop models trained with anti-beta-coronavirus actives/inactives collected from published papers and fine-tuned with anti-SARS-CoV-2 actives/inactives.
17 |
18 |
19 |
20 | ## DeepPurpose
21 |
22 | DeepPurpose has provied the pretrained model by predicting the interaction between a target (SARS-CoV2 3CL Protease) and a list of repurposing drugs from a curated drug library of 81 antiviral drugs. The Binding Score is the Kd values. Results aggregated from five pretrained model on BindingDB dataset.
23 |
24 |
25 |
26 | ## AMETLab2.0
27 |
28 | Undesirable pharmacokinetics and toxicity of candidate compounds are the main reasons for the failure of drug development, and it has been widely recognized that absorption, distribution, metabolism, excretion and toxicity (ADMET) of chemicals should be evaluated as early as possible. ADMETlab 2.0 is an enhanced version of the widely used [ADMETlab](http://admet.scbdd.com/) for systematical evaluation of ADMET properties, as well as some physicochemical properties and medicinal chemistry friendliness. With significant updates to functional modules, predictive models, explanations, and the user interface, ADMETlab 2.0 has greater capacity to assist medicinal chemists in accelerating the drug research and development process.
29 |
30 |
31 |
32 | ## Acknowledgement
33 |
34 | Dhameliya,T.M. *et al.* (2022) Systematic virtual screening in search of SARS CoV-2 inhibitors against spike glycoprotein: pharmacophore screening, molecular docking, ADMET analysis and MD simulations. *Mol Divers*.
35 |
36 | Gajjar,N.D. *et al.* (2021) In search of RdRp and Mpro inhibitors against SARS CoV-2: Molecular docking, molecular dynamic simulations and ADMET analysis. *Journal of Molecular Structure*, **1239**, 130488.
37 |
38 | Huang,K. *et al.* (2020) DeepPurpose: a deep learning library for drug–target interaction prediction. *Bioinformatics*, **36**, 5545–5547.
39 |
40 | Pillaiyar,T. *et al.* (2020) Recent discovery and development of inhibitors targeting coronaviruses. *Drug Discovery Today*, **25**, 668–688.
41 |
42 | Roy,R. *et al.* (2021) Finding potent inhibitors against SARS-CoV-2 main protease through virtual screening, ADMET, and molecular dynamics simulation studies. *Journal of Biomolecular Structure and Dynamics*, **0**, 1–13.
43 |
44 | Shen,L. *et al.* High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. *Journal of Virology*, **93**, e00023-19.
45 |
46 | Wang,S. *et al.* (2021) A transferable deep learning approach to fast screen potential antiviral drugs against SARS-CoV-2. *Briefings in Bioinformatics*.
47 |
48 | Wu,F. *et al.* (2020) A new coronavirus associated with human respiratory disease in China. *Nature*, **579**, 265–269.
49 |
50 | Xiong,G. *et al.* (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. *Nucleic Acids Research*, **49**, W5–W14.
51 |
52 |
53 |
54 |
--------------------------------------------------------------------------------
/Discussion/preprocess.py:
--------------------------------------------------------------------------------
1 | from collections import defaultdict
2 |
3 | import numpy as np
4 |
5 | from rdkit import Chem
6 |
7 | import torch
8 | atom_dict = defaultdict(lambda: len(atom_dict))
9 | bond_dict = defaultdict(lambda: len(bond_dict))
10 | fingerprint_dict = defaultdict(lambda: len(fingerprint_dict))
11 | edge_dict = defaultdict(lambda: len(edge_dict))
12 | radius=1
13 | if torch.cuda.is_available():
14 | device = torch.device('cuda')
15 | print('The code uses a GPU!')
16 | else:
17 | device = torch.device('cpu')
18 | print('The code uses a CPU...')
19 | def create_atoms(mol, atom_dict):
20 | """Transform the atom types in a molecule (e.g., H, C, and O)
21 | into the indices (e.g., H=0, C=1, and O=2).
22 | Note that each atom index considers the aromaticity.
23 | """
24 | atoms = [a.GetSymbol() for a in mol.GetAtoms()]
25 | for a in mol.GetAromaticAtoms():
26 | i = a.GetIdx()
27 | atoms[i] = (atoms[i], 'aromatic')
28 | atoms = [atom_dict[a] for a in atoms]
29 | return np.array(atoms)
30 |
31 |
32 | def create_ijbonddict(mol, bond_dict):
33 | """Create a dictionary, in which each key is a node ID
34 | and each value is the tuples of its neighboring node
35 | and chemical bond (e.g., single and double) IDs.
36 |
37 | """
38 | i_jbond_dict = defaultdict(lambda: [])
39 | for b in mol.GetBonds():
40 | i, j = b.GetBeginAtomIdx(), b.GetEndAtomIdx()
41 | bond = bond_dict[str(b.GetBondType())]
42 | i_jbond_dict[i].append((j, bond))
43 | i_jbond_dict[j].append((i, bond))
44 | return i_jbond_dict
45 |
46 |
47 | def extract_fingerprints(radius, atoms, i_jbond_dict,
48 | fingerprint_dict, edge_dict):
49 | """Extract the fingerprints from a molecular graph
50 | based on Weisfeiler-Lehman algorithm.
51 |
52 | """
53 |
54 | if (len(atoms) == 1) or (radius == 0):
55 | nodes = [fingerprint_dict[a] for a in atoms]
56 |
57 | else:
58 | nodes = atoms
59 | i_jedge_dict = i_jbond_dict
60 |
61 | for _ in range(radius):
62 |
63 | """Update each node ID considering its neighboring nodes and edges.
64 | The updated node IDs are the fingerprint IDs.。
65 | """
66 | nodes_ = []
67 | for i, j_edge in i_jedge_dict.items():
68 | neighbors = [(nodes[j], edge) for j, edge in j_edge]
69 | fingerprint = (nodes[i], tuple(sorted(neighbors)))
70 | nodes_.append(fingerprint_dict[fingerprint])
71 |
72 | """Also update each edge ID considering
73 | its two nodes on both sides.
74 | """
75 | i_jedge_dict_ = defaultdict(lambda: [])
76 | for i, j_edge in i_jedge_dict.items():
77 | for j, edge in j_edge:
78 | both_side = tuple(sorted((nodes[i], nodes[j])))
79 | edge = edge_dict[(both_side, edge)]
80 | i_jedge_dict_[i].append((j, edge))
81 |
82 | nodes = nodes_
83 | i_jedge_dict = i_jedge_dict_
84 |
85 | return np.array(nodes)
86 |
87 |
88 | def split_dataset(dataset, ratio):
89 | """Shuffle and split a dataset.洗牌和拆分数据集"""
90 | np.random.seed(1234) # fix the seed for shuffle为洗牌修正种子.
91 | np.random.shuffle(dataset)
92 | n = int(ratio * len(dataset))
93 | return dataset[:n], dataset[n:]
94 |
95 |
96 | def create_dataset(filename,path,dataname):
97 | dir_dataset = path+dataname
98 | print(filename)
99 | """Load a dataset."""
100 | with open(dir_dataset + filename, 'r') as f:
101 | smiles_property = f.readline().strip().split()
102 | data_original = f.read().strip().split('\n')
103 |
104 | """Exclude the data contains '.' in its smiles.排除含.的数据"""
105 | data_original = [data for data in data_original
106 | if '.' not in data.split()[0]]
107 | dataset = []
108 | for data in data_original:
109 |
110 | smiles, property = data.strip().split()
111 |
112 | """Create each data with the above defined functions."""
113 | mol = Chem.AddHs(Chem.MolFromSmiles(smiles))
114 | atoms = create_atoms(mol, atom_dict)
115 | molecular_size = len(atoms)
116 | i_jbond_dict = create_ijbonddict(mol, bond_dict)
117 | fingerprints = extract_fingerprints(radius, atoms, i_jbond_dict,
118 | fingerprint_dict, edge_dict)
119 | adjacency = Chem.GetAdjacencyMatrix(mol)
120 |
121 | """Transform the above each data of numpy
122 | to pytorch tensor on a device (i.e., CPU or GPU).
123 | """
124 | fingerprints = torch.LongTensor(fingerprints).to(device)
125 | adjacency = torch.FloatTensor(adjacency).to(device)
126 | property = torch.FloatTensor([int(property)]).to(device)
127 |
128 | dataset.append((smiles,fingerprints, adjacency, molecular_size, property))
129 |
130 | return dataset
131 |
132 | dataset_train = create_dataset('data_train.txt')
133 | dataset_train, dataset_dev = split_dataset(dataset_train, 0.9)
134 | dataset_test = create_dataset('data_test.txt')
135 |
136 | N_fingerprints = len(fingerprint_dict)
137 |
138 | return dataset_train, dataset_dev, dataset_test, N_fingerprints
139 |
--------------------------------------------------------------------------------
/Test/test.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 1,
6 | "id": "65b363bc",
7 | "metadata": {},
8 | "outputs": [],
9 | "source": [
10 | "import predict"
11 | ]
12 | },
13 | {
14 | "cell_type": "code",
15 | "execution_count": 3,
16 | "id": "4cc4f418",
17 | "metadata": {},
18 | "outputs": [
19 | {
20 | "name": "stdout",
21 | "output_type": "stream",
22 | "text": [
23 | "The code uses a GPU!\n",
24 | "../dataset/data_test.txt\n",
25 | "bacc_dev: 0.5119539230602043\n",
26 | "pre_dev: 0.5080213903743316\n",
27 | "rec_dev: 0.8837209302325582\n",
28 | "f1_dev: 0.6451612903225807\n",
29 | "mcc_dev: 0.03575604067764825\n",
30 | "sp_dev: 0.14018691588785046\n",
31 | "q__dev: 0.5454545454545454\n",
32 | "acc_dev: 0.5128205128205128\n"
33 | ]
34 | }
35 | ],
36 | "source": [
37 | "test1 = predict.predict('../dataset/data_test.txt',property=True)#Drugs from FDA"
38 | ]
39 | },
40 | {
41 | "cell_type": "code",
42 | "execution_count": 2,
43 | "id": "8f0eba5e",
44 | "metadata": {},
45 | "outputs": [
46 | {
47 | "name": "stdout",
48 | "output_type": "stream",
49 | "text": [
50 | "The code uses a GPU!\n",
51 | "../dataset/world_wide.txt\n",
52 | "bacc_dev: 0.46604215456674475\n",
53 | "pre_dev: 0.47987043035631655\n",
54 | "rec_dev: 0.8095238095238095\n",
55 | "f1_dev: 0.6025566531086578\n",
56 | "mcc_dev: -0.09345868862125822\n",
57 | "sp_dev: 0.12256049960967993\n",
58 | "q__dev: 0.3915211970074813\n",
59 | "acc_dev: 0.46604215456674475\n"
60 | ]
61 | }
62 | ],
63 | "source": [
64 | "test2 = predict.predict('../dataset/world_wide.txt',property=True)#Drugs from non-US"
65 | ]
66 | },
67 | {
68 | "cell_type": "code",
69 | "execution_count": 4,
70 | "id": "75c3a192",
71 | "metadata": {},
72 | "outputs": [
73 | {
74 | "name": "stdout",
75 | "output_type": "stream",
76 | "text": [
77 | "The code uses a GPU!\n",
78 | "../dataset/beyondRo5.txt\n",
79 | "1\n",
80 | "1\n",
81 | "1\n",
82 | "1\n",
83 | "1\n",
84 | "1\n",
85 | "1\n",
86 | "1\n",
87 | "1\n",
88 | "1\n",
89 | "1\n",
90 | "1\n",
91 | "1\n",
92 | "1\n",
93 | "1\n",
94 | "1\n",
95 | "1\n",
96 | "1\n",
97 | "1\n",
98 | "1\n",
99 | "1\n",
100 | "1\n",
101 | "1\n",
102 | "1\n",
103 | "1\n",
104 | "1\n",
105 | "1\n",
106 | "1\n",
107 | "1\n",
108 | "1\n",
109 | "1\n",
110 | "1\n",
111 | "1\n",
112 | "1\n",
113 | "1\n",
114 | "1\n",
115 | "1\n",
116 | "1\n",
117 | "1\n",
118 | "1\n",
119 | "1\n",
120 | "1\n",
121 | "1\n",
122 | "1\n",
123 | "1\n",
124 | "1\n",
125 | "1\n",
126 | "1\n",
127 | "1\n",
128 | "1\n",
129 | "1\n",
130 | "1\n",
131 | "1\n",
132 | "1\n",
133 | "1\n",
134 | "1\n",
135 | "1\n",
136 | "1\n",
137 | "1\n",
138 | "1\n",
139 | "1\n",
140 | "1\n",
141 | "1\n",
142 | "1\n",
143 | "1\n",
144 | "1\n",
145 | "1\n",
146 | "1\n",
147 | "1\n",
148 | "1\n",
149 | "1\n",
150 | "1\n",
151 | "1\n",
152 | "1\n",
153 | "1\n",
154 | "1\n",
155 | "1\n",
156 | "1\n",
157 | "1\n",
158 | "1\n",
159 | "1\n",
160 | "1\n",
161 | "1\n",
162 | "1\n",
163 | "1\n",
164 | "1\n",
165 | "1\n",
166 | "1\n",
167 | "1\n",
168 | "1\n",
169 | "1\n",
170 | "1\n",
171 | "1\n",
172 | "1\n",
173 | "1\n",
174 | "1\n",
175 | "1\n",
176 | "1\n",
177 | "1\n",
178 | "1\n",
179 | "1\n",
180 | "1\n",
181 | "1\n",
182 | "1\n",
183 | "1\n",
184 | "1\n",
185 | "1\n",
186 | "1\n",
187 | "1\n",
188 | "1\n",
189 | "1\n",
190 | "1\n",
191 | "1\n",
192 | "1\n",
193 | "1\n",
194 | "1\n",
195 | "1\n",
196 | "1\n",
197 | "1\n",
198 | "1\n",
199 | "1\n",
200 | "1\n",
201 | "1\n",
202 | "1\n",
203 | "1\n",
204 | "1\n",
205 | "1\n",
206 | "1\n",
207 | "1\n",
208 | "1\n",
209 | "1\n",
210 | "1\n",
211 | "1\n",
212 | "1\n",
213 | "1\n",
214 | "1\n"
215 | ]
216 | }
217 | ],
218 | "source": [
219 | "test3 = predict.predict('../dataset/beyondRo5.txt',property=False)#Drugs beyond Ro5"
220 | ]
221 | },
222 | {
223 | "cell_type": "code",
224 | "execution_count": null,
225 | "id": "147a96e5",
226 | "metadata": {},
227 | "outputs": [],
228 | "source": []
229 | }
230 | ],
231 | "metadata": {
232 | "kernelspec": {
233 | "display_name": "Python 3",
234 | "language": "python",
235 | "name": "python3"
236 | },
237 | "language_info": {
238 | "codemirror_mode": {
239 | "name": "ipython",
240 | "version": 3
241 | },
242 | "file_extension": ".py",
243 | "mimetype": "text/x-python",
244 | "name": "python",
245 | "nbconvert_exporter": "python",
246 | "pygments_lexer": "ipython3",
247 | "version": "3.8.8"
248 | }
249 | },
250 | "nbformat": 4,
251 | "nbformat_minor": 5
252 | }
253 |
--------------------------------------------------------------------------------
/DGCAN/train.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Wed Apr 27 20:09:31 2022
4 |
5 | @author:Jinyu-Sun
6 | """
7 |
8 | import timeit
9 | import sys
10 | import numpy as np
11 | import math
12 | import torch
13 | import torch.nn as nn
14 | import torch.nn.functional as F
15 | import torch.optim as optim
16 | import pickle
17 | from sklearn.metrics import roc_auc_score, roc_curve,auc
18 | from sklearn.metrics import confusion_matrix
19 | import preprocess as pp
20 | import pandas as pd
21 | import matplotlib.pyplot as plt
22 | from DGCAN import MolecularGraphNeuralNetwork,Trainer,Tester
23 |
24 | def metrics(cnf_matrix):
25 | '''Evaluation Metrics'''
26 | tn = cnf_matrix[0, 0]
27 | tp = cnf_matrix[1, 1]
28 | fn = cnf_matrix[1, 0]
29 | fp = cnf_matrix[0, 1]
30 |
31 | bacc = ((tp / (tp + fn)) + (tn / (tn + fp))) / 2 # balance accurance
32 | pre = tp / (tp + fp) # precision/q+
33 | rec = tp / (tp + fn) # recall/se
34 | sp = tn / (tn + fp)
35 | q_ = tn / (tn + fn)
36 | f1 = 2 * pre * rec / (pre + rec) # f1score
37 | mcc = ((tp * tn) - (fp * fn)) / math.sqrt(
38 | (tp + fp) * (tp + fn) * (tn + fp) * (tn + fn)) # Matthews correlation coefficient
39 | acc = (tp + tn) / (tp + fp + fn + tn) # accurancy
40 |
41 | print('bacc:', bacc)
42 | print('pre:', pre)
43 | print('rec:', rec)
44 | print('f1:', f1)
45 | print('mcc:', mcc)
46 | print('sp:', sp)
47 | print('q_:', q_)
48 | print('acc:', acc)
49 |
50 |
51 | def train (test_name, radius, dim, layer_hidden, layer_output, dropout, batch_train,
52 | batch_test, lr, lr_decay, decay_interval, iteration, N , dataset_train):
53 | '''
54 |
55 | Parameters
56 | ----------
57 | data_test='../dataset/data_test.txt', #test set
58 | radius = 1, #hops of radius subgraph: 1, 2
59 | dim = 64, #dimension of graph convolution layers
60 | layer_hidden = 4, #Number of graph convolution layers
61 | layer_output = 10, #Number of dense layers
62 | dropout = 0.45, #drop out rate :0-1
63 | batch_train = 8, # batch of training set
64 | batch_test = 8, #batch of test set
65 | lr =3e-4, #learning rate: 1e-5,1e-4,3e-4, 5e-4, 1e-3, 3e-3,5e-3
66 | lr_decay = 0.85, #Learning rate decay:0.5, 0.75, 0.85, 0.9
67 | decay_interval = 25,#Number of iterations for learning rate decay:10,25,30,50
68 | iteration = 140, #Number of iterations
69 | N = 5000, #length of embedding: 2000,3000,5000,7000
70 | dataset_train='../dataset/data_train.txt') #training set
71 |
72 | Returns
73 | -------
74 | res_test : results
75 | Predicting results.
76 |
77 | '''
78 | dataset_test = test_name
79 | (radius, dim, layer_hidden, layer_output,
80 | batch_train, batch_test, decay_interval,
81 | iteration, dropout) = map(int, [radius, dim, layer_hidden, layer_output,
82 | batch_train, batch_test,
83 | decay_interval, iteration, dropout])
84 | lr, lr_decay = map(float, [lr, lr_decay])
85 | if torch.cuda.is_available():
86 | device = torch.device('cuda')
87 | print('The code uses a GPU!')
88 | else:
89 | device = torch.device('cpu')
90 | print('The code uses a CPU...')
91 |
92 | lr, lr_decay = map(float, [lr, lr_decay])
93 |
94 | print('-' * 100)
95 | print('Just a moment......')
96 | print('-' * 100)
97 | path = ''
98 | dataname = ''
99 |
100 | dataset_train= pp.create_dataset(dataset_train,path,dataname)
101 | #dataset_train,dataset_test = pp.split_dataset(dataset_train,0.9)
102 | #dataset_test= pp.create_dataset(dataset_dev,path,dataname)
103 | dataset_test= pp.create_dataset(dataset_test,path,dataname)
104 | np.random.seed(0)
105 | np.random.shuffle(dataset_train)
106 | print('The preprocess has finished!')
107 | print('-' * 100)
108 |
109 | print('Creating a model.')
110 | torch.manual_seed(0)
111 | model = MolecularGraphNeuralNetwork(
112 | N, dim, layer_hidden, layer_output, dropout).to(device)
113 | trainer = Trainer(model,lr,batch_train)
114 | tester = Tester(model,batch_test)
115 | print('# of model parameters:',
116 | sum([np.prod(p.size()) for p in model.parameters()]))
117 | print('-' * 100)
118 | file_result = path + '../DGCAN/results/AUC' + '.txt'
119 | # file_result = '../output/result--' + setting + '.txt'
120 | result = 'Epoch\tTime(sec)\tLoss_train\tLoss_test\tAUC_train\tAUC_test'
121 | file_test_result = path + 'test_prediction' + '.txt'
122 | file_predictions = path + 'train_prediction' + '.txt'
123 | file_model = '../DGCAN/model/model' + '.pth'
124 | with open(file_result, 'w') as f:
125 | f.write(result + '\n')
126 |
127 | print('Start training.')
128 | print('The result is saved in the output directory every epoch!')
129 |
130 | np.random.seed(0)
131 |
132 | start = timeit.default_timer()
133 |
134 | for epoch in range(iteration):
135 | epoch += 1
136 | if epoch % decay_interval == 0:
137 | trainer.optimizer.param_groups[0]['lr'] *= lr_decay
138 | # [‘amsgrad’, ‘params’, ‘lr’, ‘betas’, ‘weight_decay’, ‘eps’]
139 | prediction_train, loss_train, train_res = trainer.train(dataset_train)
140 | prediction_test, loss_test, test_res = tester.test_classifier(dataset_test)
141 |
142 | time = timeit.default_timer() - start
143 |
144 | if epoch == 1:
145 | minutes = time * iteration / 60
146 | hours = int(minutes / 60)
147 | minutes = int(minutes - 60 * hours)
148 | print('The training will finish in about',
149 | hours, 'hours', minutes, 'minutes.')
150 | print('-' * 100)
151 | print(result)
152 |
153 | result = '\t'.join(map(str, [epoch, time, loss_train, loss_test, prediction_train, prediction_test]))
154 | tester.save_result(result, file_result)
155 | tester.save_model(model, file_model)
156 | print(result)
157 | model.eval()
158 | prediction_test, loss_test, test_res = tester.test_classifier(dataset_test)
159 | res_test = test_res.T
160 |
161 | cnf_matrix = confusion_matrix(res_test[:, 0], res_test[:, 1])
162 | fpr, tpr, thresholds = roc_curve(res_test[:, 0], res_test[:, 1])
163 | AUC = auc(fpr, tpr)
164 | print('auc:',AUC)
165 | metrics(cnf_matrix)
166 | return res_test
--------------------------------------------------------------------------------
/screening/DTI.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "c40219cd",
6 | "metadata": {},
7 | "source": [
8 | "# Drug Target Interaction Prediction by using DeepPurpose"
9 | ]
10 | },
11 | {
12 | "cell_type": "markdown",
13 | "id": "23b43a6f",
14 | "metadata": {},
15 | "source": [
16 | "DeepPurpose has provied the convinient way for DTI prediction especially for SARS_CoV2_Protease. "
17 | ]
18 | },
19 | {
20 | "cell_type": "markdown",
21 | "id": "38ab44af",
22 | "metadata": {},
23 | "source": [
24 | "## Installation"
25 | ]
26 | },
27 | {
28 | "cell_type": "code",
29 | "execution_count": null,
30 | "id": "1ffbb4b3",
31 | "metadata": {},
32 | "outputs": [],
33 | "source": [
34 | "pip\n",
35 | "conda create -n DeepPurpose python=3.6\n",
36 | "conda activate DeepPurpose\n",
37 | "conda install -c conda-forge rdkit\n",
38 | "conda install -c conda-forge notebook\n",
39 | "pip install git+https://github.com/bp-kelley/descriptastorus \n",
40 | "pip install DeepPurpose\n",
41 | "\n",
42 | "or Build from Source\n",
43 | "\n",
44 | "git clone https://github.com/kexinhuang12345/DeepPurpose.git ## Download code repository\n",
45 | "cd DeepPurpose ## Change directory to DeepPurpose\n",
46 | "conda env create -f environment.yml ## Build virtual environment with all packages installed using conda\n",
47 | "conda activate DeepPurpose ## Activate conda environment (use \"source activate DeepPurpose\" for anaconda 4.4 or earlier) \n",
48 | "jupyter notebook ## open the jupyter notebook with the conda env\n",
49 | "\n",
50 | "## run our code, e.g. click a file in the DEMO folder\n",
51 | "... ...\n",
52 | "\n",
53 | "conda deactivate ## when done, exit conda environment "
54 | ]
55 | },
56 | {
57 | "cell_type": "markdown",
58 | "id": "26e590fe",
59 | "metadata": {},
60 | "source": [
61 | "## Run"
62 | ]
63 | },
64 | {
65 | "cell_type": "code",
66 | "execution_count": null,
67 | "id": "55207f2c",
68 | "metadata": {},
69 | "outputs": [],
70 | "source": [
71 | "import os\n",
72 | "os.chdir('../')\n",
73 | "from DeepPurpose import utils\n",
74 | "from DeepPurpose import DTI as models\n",
75 | "X_drug, X_target, y = process_BindingDB(download_BindingDB(SAVE_PATH),\n",
76 | " y = 'Kd', \n",
77 | " binary = False, \n",
78 | " convert_to_log = True)\n",
79 | "\n",
80 | "# Type in the encoding names for drug/protein.\n",
81 | "drug_encoding, target_encoding = 'MPNN', 'CNN'\n",
82 | "\n",
83 | "# Data processing, here we select cold protein split setup.\n",
84 | "train, val, test = data_process(X_drug, X_target, y, \n",
85 | " drug_encoding, target_encoding, \n",
86 | " split_method='cold_protein', \n",
87 | " frac=[0.7,0.1,0.2])\n",
88 | "\n",
89 | "# Generate new model using default parameters; also allow model tuning via input parameters.\n",
90 | "config = generate_config(drug_encoding, target_encoding, transformer_n_layer_target = 8)\n",
91 | "net = models.model_initialize(**config)\n",
92 | "\n",
93 | "# Train the new model.\n",
94 | "# Detailed output including a tidy table storing validation loss, metrics, AUC curves figures and etc. are stored in the ./result folder.\n",
95 | "net.train(train, val, test)\n",
96 | "\n",
97 | "# or simply load pretrained model from a model directory path or reproduced model name such as DeepDTA\n",
98 | "net = models.model_pretrained(MODEL_PATH_DIR or MODEL_NAME)\n",
99 | "\n",
100 | "X_repurpose, drug_name, drug_cid = load_broad_repurposing_hub(SAVE_PATH)\n",
101 | "target, target_name = load_SARS_CoV2_Protease_3CL()\n",
102 | "\n",
103 | "_ = models.virtual_screening(smiles, target, net, drug_name, target_name)"
104 | ]
105 | },
106 | {
107 | "cell_type": "markdown",
108 | "id": "bab0c86d",
109 | "metadata": {},
110 | "source": [
111 | "## Results"
112 | ]
113 | },
114 | {
115 | "cell_type": "code",
116 | "execution_count": null,
117 | "id": "1136255f",
118 | "metadata": {},
119 | "outputs": [],
120 | "source": [
121 | "+-------+-----------+------------------------+---------------+\n",
122 | "| Rank | Drug Name | Target Name | Binding Score |\n",
123 | "+-------+-----------+------------------------+---------------+\n",
124 | "| 1 | Drug 4565 | SARS-CoV2 3CL Protease | 8.96 |\n",
125 | "| 2 | Drug 4570 | SARS-CoV2 3CL Protease | 12.42 |\n",
126 | "| 3 | Drug 3690 | SARS-CoV2 3CL Protease | 12.86 |\n",
127 | "| 4 | Drug 3068 | SARS-CoV2 3CL Protease | 13.36 |\n",
128 | "| 5 | Drug 8387 | SARS-CoV2 3CL Protease | 13.47 |\n",
129 | "| 6 | Drug 5176 | SARS-CoV2 3CL Protease | 14.47 |\n",
130 | "| 7 | Drug 438 | SARS-CoV2 3CL Protease | 14.67 |\n",
131 | "| 8 | Drug 4507 | SARS-CoV2 3CL Protease | 16.11 |\n",
132 | "```\n",
133 | "```\n",
134 | "| 9978 | Drug 1377 | SARS-CoV2 3CL Protease | 460788.11 |\n",
135 | "| 9979 | Drug 3768 | SARS-CoV2 3CL Protease | 479737.13 |\n",
136 | "| 9980 | Drug 5106 | SARS-CoV2 3CL Protease | 485684.14 |\n",
137 | "| 9981 | Drug 3765 | SARS-CoV2 3CL Protease | 505994.35 |\n",
138 | "| 9982 | Drug 2207 | SARS-CoV2 3CL Protease | 510293.39 |\n",
139 | "| 9983 | Drug 1161 | SARS-CoV2 3CL Protease | 525921.93 |\n",
140 | "| 9984 | Drug 2477 | SARS-CoV2 3CL Protease | 533613.12 |\n",
141 | "| 9985 | Drug 3320 | SARS-CoV2 3CL Protease | 538902.46 |\n",
142 | "| 9986 | Drug 3783 | SARS-CoV2 3CL Protease | 542639.17 |\n",
143 | "| 9987 | Drug 4834 | SARS-CoV2 3CL Protease | 603510.00 |\n",
144 | "| 9988 | Drug 9653 | SARS-CoV2 3CL Protease | 611796.89 |\n",
145 | "| 9989 | Drug 6606 | SARS-CoV2 3CL Protease | 671138.31 |\n",
146 | "| 9990 | Drug 160 | SARS-CoV2 3CL Protease | 697775.04 |\n",
147 | "| 9991 | Drug 3851 | SARS-CoV2 3CL Protease | 792134.96 |\n",
148 | "| 9992 | Drug 5208 | SARS-CoV2 3CL Protease | 832708.75 |\n",
149 | "| 9993 | Drug 2786 | SARS-CoV2 3CL Protease | 905739.10 |\n",
150 | "| 9994 | Drug 6612 | SARS-CoV2 3CL Protease | 968825.66 |\n",
151 | "| 9995 | Drug 6609 | SARS-CoV2 3CL Protease | 1088788.87 |\n",
152 | "| 9996 | Drug 801 | SARS-CoV2 3CL Protease | 1186364.21 |\n",
153 | "| 9997 | Drug 3844 | SARS-CoV2 3CL Protease | 1199274.11 |\n",
154 | "| 9998 | Drug 3842 | SARS-CoV2 3CL Protease | 1559694.06 |\n",
155 | "| 9999 | Drug 4486 | SARS-CoV2 3CL Protease | 1619297.87 |\n",
156 | "| 10000 | Drug 800 | SARS-CoV2 3CL Protease | 1623061.65 |\n",
157 | "+-------+-----------+------------------------+---------------+"
158 | ]
159 | },
160 | {
161 | "cell_type": "markdown",
162 | "id": "83ff4364",
163 | "metadata": {},
164 | "source": [
165 | "## Acknowledgement"
166 | ]
167 | },
168 | {
169 | "cell_type": "code",
170 | "execution_count": null,
171 | "id": "ab0dd49f",
172 | "metadata": {},
173 | "outputs": [],
174 | "source": [
175 | "This project incorporates code from the following repo:\n",
176 | " \n",
177 | " https://github.com/kexinhuang12345/DeepPurpose\n",
178 | " "
179 | ]
180 | }
181 | ],
182 | "metadata": {
183 | "kernelspec": {
184 | "display_name": "Python 3 (ipykernel)",
185 | "language": "python",
186 | "name": "python3"
187 | },
188 | "language_info": {
189 | "codemirror_mode": {
190 | "name": "ipython",
191 | "version": 3
192 | },
193 | "file_extension": ".py",
194 | "mimetype": "text/x-python",
195 | "name": "python",
196 | "nbconvert_exporter": "python",
197 | "pygments_lexer": "ipython3",
198 | "version": "3.8.8"
199 | }
200 | },
201 | "nbformat": 4,
202 | "nbformat_minor": 5
203 | }
204 |
--------------------------------------------------------------------------------
/screening/Dataset/finetunev1.csv:
--------------------------------------------------------------------------------
1 | smiles,new_label
2 | CCN(CC)CCCC(C)Nc1ccnc2cc(Cl)ccc12,1
3 | Cc1cccc(C)c1OCC(=O)NC(Cc1ccccc1)C(O)CC(Cc1ccccc1)NC(=O)C(C(C)C)N1CCCNC1=O,1
4 | O=C(Nc1ccc([N+](=O)[O-])cc1Cl)c1cc(Cl)ccc1O,1
5 | CN(C)C(=O)C(CCN1CCC(O)(c2ccc(Cl)cc2)CC1)(c1ccccc1)c1ccccc1,1
6 | CC1OC(OC2CC(O)C3(CO)C4C(O)CC5(C)C(C6=CC(=O)OC6)CCC5(O)C4CCC3(O)C2)C(O)C(O)C1O,1
7 | CCN(CC)CCOc1ccc(C(O)(Cc2ccc(Cl)cc2)c2ccc(C)cc2)cc1,1
8 | COc1ccc2cc1Oc1ccc(cc1)CC1c3cc(c(OC)cc3CCN1C)Oc1c(OC)c(OC)cc3c1C(C2)N(C)CC3,1
9 | CCN(CC)Cc1cc(Nc2ccnc3cc(Cl)ccc23)ccc1O,1
10 | O=C1NCN(c2ccccc2)C12CCN(CCCC(c1ccc(F)cc1)c1ccc(F)cc1)CC2,1
11 | Cc1ccc(NC(=O)c2ccc(CN3CCN(C)CC3)cc2)cc1Nc1nccc(-c2cccnc2)n1,1
12 | CCN(CC)CCOc1ccc2c(c1)C(=O)c1cc(OCCN(CC)CC)ccc1-2,1
13 | COc1cc2c3cc1Oc1c(OC)c(OC)cc4c1C(Cc1ccc(O)c(c1)Oc1ccc(cc1)CC3N(C)CC2)N(C)CC4,1
14 | OC(c1cc(C(F)(F)F)nc2c(C(F)(F)F)cccc12)C1CCCCN1,1
15 | OCCN1CCN(CCCN2c3ccccc3Sc3ccc(C(F)(F)F)cc32)CC1,1
16 | O=C(Nc1cc(Cl)cc(Cl)c1O)c1c(O)c(Cl)cc(Cl)c1Cl,1
17 | CCN(CCO)CCCC(C)Nc1ccnc2cc(Cl)ccc12,1
18 | CC(CN1c2ccccc2Sc2ccccc21)N(C)C,1
19 | CCSc1ccc2c(c1)N(CCCN1CCN(C)CC1)c1ccccc1S2,1
20 | CC1OC(OC2C(O)CC(OC3C(O)CC(OC4CCC5(C)C(CCC6C5CC(O)C5(C)C(C7=CC(=O)OC7)CCC65O)C4)OC3C)OC2C)CC(O)C1O,1
21 | CN(C)CCCN1c2ccccc2CCc2ccc(Cl)cc21,1
22 | CN(C)CCCN1c2ccccc2Sc2ccc(Cl)cc21,1
23 | CCN(CC)CCOc1ccc(C(=C(Cl)c2ccccc2)c2ccccc2)cc1,1
24 | CCC1OC(=O)C(C)C(OC2CC(C)(OC)C(O)C(C)O2)C(C)C(OC2OC(C)CC(N(C)C)C2O)C(C)(O)CC(C)CN(C)C(C)C(O)C1(C)O,1
25 | COc1ncnc(NS(=O)(=O)c2ccc(N)cc2)c1OC,1
26 | COc1ccc2nc(S(=O)Cc3ncc(C)c(OC)c3C)[nH]c2c1,1
27 | CN(C)CCOc1ccc(C(=C(CCCl)c2ccccc2)c2ccccc2)cc1,1
28 | CSc1ccc2c(c1)N(CCC1CCCCN1C)c1ccccc1S2,1
29 | CC=CCC(C)C(O)C1C(=O)NC(CC)C(=O)N(C)CC(=O)N(C)C(CC(C)C)C(=O)NC(C(C)C)C(=O)N(C)C(CC(C)C)C(=O)NC(C)C(=O)NC(C)C(=O)N(C)C(CC(C)C)C(=O)N(C)C(CC(C)C)C(=O)N(C)C(C(C)C)C(=O)N1C,1
30 | CC(C)=CCCC1(C)C=Cc2c(O)c3c(c(CC=C(C)C)c2O1)OC12C(=CC4CC1C(C)(C)OC2(CC=C(C)C(=O)O)C4O)C3=O,1
31 | C=CC1CN2CCC1CC2C(O)c1ccnc2ccc(OC)cc12,1
32 | CCCCCC(=O)OC1(C(C)=O)CCC2C3CCC4=CC(=O)CCC4(C)C3CCC21C,1
33 | COc1ccc2cc1Oc1ccc(cc1)CC1c3c(cc4c(c3Oc3cc5c(cc3OC)CCN(C)C5C2)OCO4)CCN1C,1
34 | CCC(=C(c1ccccc1)c1ccc(OCCN(C)C)cc1)c1ccccc1,1
35 | OC1(c2ccc(Cl)c(C(F)(F)F)c2)CCN(CCCC(c2ccc(F)cc2)c2ccc(F)cc2)CC1,1
36 | Clc1ccc(Cn2c(CN3CCCC3)nc3ccccc32)cc1,1
37 | C1CCC(C(CC2CCCCN2)C2CCCCC2)CC1,1
38 | Oc1c(Cl)cc(Cl)c(Cl)c1Cc1c(O)c(Cl)cc(Cl)c1Cl,1
39 | CCC(C(=O)O)C1CCC(C)C(C(C)C(O)C(C)C(=O)C(CC)C2OC3(C=CC(O)C4(CCC(C)(C5CCC(O)(CC)C(C)O5)O4)O3)C(C)CC2C)O1,1
40 | Cc1cc(Nc2ncc(Cl)c(Nc3ccccc3S(=O)(=O)C(C)C)n2)c(OC(C)C)cc1C1CCNCC1,1
41 | CC1(C)C=Cc2c(c3c(c4c(=O)c(-c5ccc(O)cc5)coc24)OC(C)(C)CC3)O1,1
42 | C=CC(=O)Nc1cc(Nc2nccc(-c3cn(C)c4ccccc34)n2)c(OC)cc1N(C)CCN(C)C,1
43 | Cc1c(-c2ccc(O)cc2)n(Cc2ccc(OCCN3CCCCCC3)cc2)c2ccc(O)cc12,1
44 | CCCCCCOC(C)c1cccc(-c2csc(NC(=O)c3cc(Cl)c(C=C(C)C(=O)O)c(Cl)c3)n2)c1OC,1
45 | CC(C)=CCc1c2c(c3occ(-c4ccc(O)cc4)c(=O)c3c1O)C=CC(C)(C)O2,1
46 | CCCCc1oc2ccc(NS(C)(=O)=O)cc2c1C(=O)c1ccc(OCCCN(CCCC)CCCC)cc1,1
47 | CC(C)C(=O)OCC(=O)C12OC(C3CCCCC3)OC1CC1C3CCC4=CC(=O)C=CC4(C)C3C(O)CC12C,1
48 | CC1(C)C=Cc2c(c3c(c4c(=O)c(-c5ccc(O)c(O)c5)coc24)OC(C)(C)CC3)O1,1
49 | CCCCCOc1ccc(-c2ccc(-c3ccc(C(=O)NC4CC(O)C(O)NC(=O)C5C(O)C(C)CN5C(=O)C(C(C)O)NC(=O)C(C(O)C(O)c5ccc(O)cc5)NC(=O)C5CC(O)CN5C(=O)C(C(C)O)NC4=O)cc3)cc2)cc1,1
50 | CC(C)(C)c1cc(C(C)(C)C)c(NC(=O)c2c[nH]c3ccccc3c2=O)cc1O,1
51 | CCC(=C(c1ccc(OCCN(C)C)cc1)c1cccc(O)c1)c1ccccc1,1
52 | CCN1CCN(Cc2ccc(Nc3ncc(F)c(-c4cc(F)c5nc(C)n(C(C)C)c5c4)n3)nc2)CC1,1
53 | CCc1nc(C(N)=O)c(Nc2ccc(N3CCC(N4CCN(C)CC4)CC3)c(OC)c2)nc1NC1CCOCC1,1
54 | CC(C)(C)c1ccc(C(=O)CCCN2CCC(OC(c3ccccc3)c3ccccc3)CC2)cc1,1
55 | c1ccc2c(c1)Sc1ccccc1N2CC1CN2CCC1CC2,1
56 | CC1=NN(c2ccc(C)c(C)c2)C(=O)C1=NNc1cccc(-c2cccc(C(=O)O)c2)c1O,1
57 | COC(=O)NC(C(=O)NC(Cc1ccccc1)C(O)CN(Cc1ccc(-c2ccccn2)cc1)NC(=O)C(NC(=O)OC)C(C)(C)C)C(C)(C)C,1
58 | CN1C2CCC1CC(OC(c1ccccc1)c1ccccc1)C2,1
59 | CC(C)N1CCN(c2ccc(OCC3COC(Cn4cncn4)(c4ccc(Cl)cc4Cl)O3)cc2)CC1,1
60 | C=CCOc1ccccc1OCC(O)CNC(C)C,1
61 | CCCCCC(O)C=CC1C(O)CC(=O)C1CCCCCCC(=O)O,1
62 | CC1CCOC2Cn3cc(C(=O)NCc4ccc(F)cc4F)c(=O)c(O)c3C(=O)N12,1
63 | OCCN1CCN(CCCN2c3ccccc3C=Cc3ccccc32)CC1,1
64 | CCOC(=O)c1c(CSc2ccccc2)n(C)c2cc(Br)c(O)c(CN(C)C)c12,1
65 | CC(C)c1nc(CN(C)C(=O)NC(C(=O)NC(Cc2ccccc2)CC(O)C(Cc2ccccc2)NC(=O)OCc2cncs2)C(C)C)cs1,1
66 | Cc1c(O)cccc1C(=O)NC(CSc1ccccc1)C(O)CN1CC2CCCCC2CC1C(=O)NC(C)(C)C,1
67 | CC(C)(C)NC(=O)C1CC2CCCCC2CN1CC(O)C(Cc1ccccc1)NC(=O)C(CC(N)=O)NC(=O)c1ccc2ccccc2n1,1
68 | CCCC1(CCc2ccccc2)CC(O)=C(C(CC)c2cccc(NS(=O)(=O)c3ccc(C(F)(F)F)cn3)c2)C(=O)O1,1
69 | CC(C)CN(CC(O)C(Cc1ccccc1)NC(=O)OC1CCOC1)S(=O)(=O)c1ccc(N)cc1,1
70 | CC(C)CN(CC(O)C(Cc1ccccc1)NC(=O)OC1COC2OCCC12)S(=O)(=O)c1ccc(N)cc1,1
71 | CC(C)(C)NC(=O)C1CN(Cc2cccnc2)CCN1CC(O)CC(Cc1ccccc1)C(=O)NC1c2ccccc2CC1O,1
72 | Nc1ccn(C2OC(CO)C(O)C2(F)F)c(=O)n1,0
73 | CCC1CN2CCc3cc(OC)c(OC)cc3C2CC1CC1NCCc2cc(OC)c(OC)cc21,0
74 | CN(C)CCCN1c2ccccc2Sc2ccccc21,0
75 | CC(=O)OC1CC(OC2C(O)CC(OC3C(O)CC(OC4CCC5(C)C(CCC6C5CC(O)C5(C)C(C7=CC(=O)OC7)CCC65O)C4)OC3C)OC2C)OC(C)C1OC1OC(CO)C(O)C(O)C1O,0
76 | COc1ccc(CC2NCC(O)C2OC(C)=O)cc1,0
77 | Nc1ccc(N=Nc2ccccc2)c(N)n1,0
78 | CC(C)NCC(O)COc1cccc2ccccc12,0
79 | CN(C)CCCSC(=N)N,0
80 | CN1CCCC1Cc1c[nH]c2ccc(CCS(=O)(=O)c3ccccc3)cc12,0
81 | NC1CONC1=O,0
82 | Nc1c2c(nc3ccccc13)CCCC2,0
83 | CC(=O)C1CCC2C3CC=C4CC(O)CCC4(C)C3CCC12C,0
84 | O=C1CC2(CCCC2)CC(=O)N1CCCCN1CCN(c2ncccn2)CC1,0
85 | CCN(CC)CCCC(C)Nc1c2ccc(Cl)cc2nc2ccc(OC)cc12,0
86 | CCCCOc1ccc(C(=O)CCN2CCCCC2)cc1,0
87 | O=C(CCCN1CCC2(CC1)C(=O)NCN2c1ccccc1)c1ccc(F)cc1,0
88 | CC(C(O)c1ccc(O)cc1)N1CCC(Cc2ccccc2)CC1,0
89 | Cn1nnnc1SCC1=C(C(=O)[O-])N2C(=O)C(NC(=O)C(O)c3ccccc3)C2SC1,0
90 | CCOc1ccc2nc(S(N)(=O)=O)sc2c1,0
91 | CC=Cc1ccc(OC)cc1,0
92 | CC(CCc1ccccc1)NCC(O)c1ccc(O)c(C(N)=O)c1,0
93 | CC1COc2c(N3CCN(C)CC3)c(F)cc3c(=O)c(C(=O)O)cn1c23,0
94 | CC(C)(N)Cc1ccccc1,0
95 | CC12CCC3C(=CCc4cc(O)ccc43)C1CCC2=O,0
96 | COc1cc2c(cc1OC)C(=O)C(CC1CCN(Cc3ccccc3)CC1)C2,0
97 | CC(CCc1ccccc1)NC(C)C(O)c1ccc(O)cc1,0
98 | C=C1CC2C(CCC3(C)C(=O)CCC23)C2(C)C=CC(=O)C=C12,0
99 | NC(CCC(=O)NC(CSSCC(NC(=O)CCC(N)C(=O)O)C(=O)NCC(=O)O)C(=O)NCC(=O)O)C(=O)O,0
100 | CC(C=CC1=C(C)CCCC1(C)C)=CC=CC(C)=CC(=O)O,0
101 | CC12CCC3C(CCC4CC(O)CCC43C)C1CCC2=O,0
102 | Cc1ccc(C(=O)C(C)CN2CCCCC2)cc1,0
103 | CN(C)CCN(Cc1ccccc1)c1ccccn1,0
104 | COc1ccccc1OCC(O)CN1CCN(CC(=O)Nc2c(C)cccc2C)CC1,0
105 | COc1cc(N)c(Cl)cc1C(=O)NC1CCN(Cc2ccccc2)CC1,0
106 | CC1(C)OC2CC3C4CCC5=CC(=O)C=CC5(C)C4C(O)CC3(C)C2(C(=O)CO)O1,0
107 | CCn1cc(C(=O)O)c(=O)c2cc(F)c(N3CCNCC3)nc21,0
108 | Nc1c(Br)cc(Br)cc1CNC1CCC(O)CC1,0
109 | CC12CCC3C(CCC4=C(O)C(=O)CCC43C)C1CCC2=O,0
110 | NCCc1ccc(O)c(O)c1,0
111 | CC(C)(C)NCC(O)c1ccc(O)c(CO)c1,0
112 | CCCN1CC(CSC)CC2c3cccc4[nH]cc(c34)CC21,0
113 | CN(C)CCOC(=O)C(c1ccccc1)C1(O)CCCC1,0
114 | c1ccc2c(c1)OCC(C1=NCCN1)O2,0
115 | CCCc1cc(C(N)=S)ccn1,0
116 | CN(C)CCOC(c1ccccc1)c1ccccc1,0
117 | O=C1c2c(O)cccc2Cc2cccc(O)c21,0
118 | CN(C)C(=O)COC(=O)Cc1ccc(OC(=O)c2ccc(N=C(N)N)cc2)cc1,0
119 | NC(=O)c1nc(F)c[nH]c1=O,0
120 | COC1C(OC(C)=O)CC(=O)OC(C)CC=CC=CC(OC2CCC(N(C)C)C(C)O2)C(C)CC(CC=O)C1OC1OC(C)C(OC2CC(C)(O)C(O)C(C)O2)C(N(C)C)C1O,0
121 | CC12CC(=O)C3C(CCC4CC(O)CCC43C)C1CCC2C(=O)CO,0
122 | COC(c1ccccc1)(c1ccccc1)C(Oc1nc(C)cc(C)n1)C(=O)O,0
123 | CC1CCC2C(C)C(O)OC3OC4(C)CCC1C32OO4,0
124 | CCCCOc1cc(C(=O)OCCN(CC)CC)ccc1N,0
125 | c1ccc(CNCCNCc2ccccc2)cc1,0
126 | CC(=C(CCOC(=O)c1ccccc1)SSC(CCOC(=O)c1ccccc1)=C(C)N(C=O)Cc1cnc(C)nc1N)N(C=O)Cc1cnc(C)nc1N,0
127 | CC(C)=CC(C)=NNc1nncc2ccccc12,0
128 | CCOc1nc2cccc(C(=O)O)c2n1Cc1ccc(-c2ccccc2-c2nn[nH]n2)cc1,0
129 | COc1ccc(CC2c3cc(OC)c(OC)cc3CC[N+]2(C)CCC(=O)OCCCCCOC(=O)CC[N+]2(C)CCc3cc(OC)c(OC)cc3C2Cc2ccc(OC)c(OC)c2)cc1OC,0
130 | CC(=O)Oc1ccc(C(=C2CCCCC2)c2ccc(OC(C)=O)cc2)cc1,0
131 | CNCC(O)c1ccc(OC(=O)C(C)(C)C)c(OC(=O)C(C)(C)C)c1,0
132 | CCc1ccc(C(=O)C(C)CN2CCCCC2)cc1,0
133 | Oc1ccc(C2CNCCc3c2cc(O)c(O)c3Cl)cc1,0
134 | COc1ccc(CC(C)NCC(O)c2ccc(O)c(NC=O)c2)cc1,0
135 | CCC(=O)OC(OP(=O)(CCCCc1ccccc1)CC(=O)N1CC(C2CCCCC2)CC1C(=O)O)C(C)C,0
136 | CC(=C(CCO)SSCC1CCCO1)N(C=O)Cc1cnc(C)nc1N,0
137 | CC(C)C(=O)c1c(C(C)C)nn2ccccc12,0
138 | CCCNC(C)(C)COC(=O)c1ccccc1,0
139 | CCC1(c2cccc(O)c2)CCCCN(C)C1,0
140 | CN1CCN2c3ncccc3Cc3ccccc3C2C1,0
141 | CCCCC(C)(O)CC=CC1C(O)CC(=O)C1CCCCCCC(=O)OC,0
142 | COc1ccc2cc(CCC(C)=O)ccc2c1,0
143 | C=C1CCC2(O)C3Cc4ccc(O)c5c4C2(CCN3CC2CC2)C1O5,0
144 | Cc1cc2c(s1)Nc1ccccc1N=C2N1CCN(C)CC1,0
145 | CCCc1nc(C(C)(C)O)c(C(=O)O)n1Cc1ccc(-c2ccccc2-c2nn[nH]n2)cc1,0
146 | Cc1nccn1CC1CCc2c(c3ccccc3n2C)C1=O,0
147 | CC12CC(=CO)C(=O)CC1CCC1C2CCC2(C)C1CCC2(C)O,0
148 | O=C1Nc2ccccc2C1(c1ccc(O)cc1)c1ccc(O)cc1,0
149 | CCOC(=O)OC1(C(=O)COC(=O)CC)CCC2C3CCC4=CC(=O)C=CC4(C)C3C(O)CC21C,0
150 | CC1CN(c2c(F)c(N)c3c(=O)c(C(=O)O)cn(C4CC4)c3c2F)CC(C)N1,0
151 | Cc1ccc(O)c(C(CCN(C(C)C)C(C)C)c2ccccc2)c1,0
152 | CNCc1cc(-c2ccccc2F)n(S(=O)(=O)c2cccnc2)c1,0
153 | O=P(O)(O)C(O)(Cn1ccnc1)P(=O)(O)O,0
154 | O=C1OC2(c3cc(Br)c([O-])cc3Oc3c2cc(Br)c([O-])c3[Hg])c2ccccc21,0
155 | CC12C=CC(=O)C=C1CCC1C2CCC2(C)C1CCC2(C)O,0
156 |
--------------------------------------------------------------------------------
/Discussion/GPC.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Tue Apr 27 22:04:01 2021
4 |
5 | @author: BM109X32G-10GPU-02
6 | """
7 |
8 | # -*- coding: utf-8 -*-
9 | """
10 | Created on Sun Nov 15 13:46:29 2020
11 |
12 | @author: de''
13 | """
14 |
15 | # -*- coding: utf-8 -*-
16 | """
17 | Created on Sun Nov 15 10:40:57 2020
18 |
19 | @author: de''
20 | """
21 |
22 | from sklearn.datasets import make_blobs
23 | import json
24 | import numpy as np
25 | import math
26 | from tqdm import tqdm
27 | from scipy import sparse
28 | from sklearn.metrics import roc_auc_score,roc_curve,auc
29 | from sklearn.metrics import confusion_matrix
30 | from sklearn.gaussian_process.kernels import RBF
31 | import pandas as pd
32 | import matplotlib.pyplot as plt
33 | from rdkit import Chem
34 | from sklearn.gaussian_process import GaussianProcessClassifier as GPC
35 | from sklearn.ensemble import RandomForestClassifier
36 | from sklearn.model_selection import train_test_split
37 | from sklearn.preprocessing import MinMaxScaler
38 | from sklearn.neural_network import MLPClassifier
39 | from sklearn.svm import SVC
40 | from tensorflow.keras.models import Model, load_model
41 | from tensorflow.keras.layers import Dense, Input, Flatten, Conv1D, MaxPooling1D, concatenate
42 | from tensorflow.keras import metrics, optimizers
43 | from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
44 |
45 | def split_smiles(smiles, kekuleSmiles=True):
46 | try:
47 | mol = Chem.MolFromSmiles(smiles)
48 | smiles = Chem.MolToSmiles(mol, kekuleSmiles=kekuleSmiles)
49 | except:
50 | pass
51 | splitted_smiles = []
52 | for j, k in enumerate(smiles):
53 | if len(smiles) == 1:
54 | return [smiles]
55 | if j == 0:
56 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
57 | splitted_smiles.append(k + smiles[j + 1])
58 | else:
59 | splitted_smiles.append(k)
60 | elif j != 0 and j < len(smiles) - 1:
61 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
62 | splitted_smiles.append(k + smiles[j + 1])
63 | elif k.islower() and smiles[j - 1].isupper() and k != "c":
64 | pass
65 | else:
66 | splitted_smiles.append(k)
67 |
68 | elif j == len(smiles) - 1:
69 | if k.islower() and smiles[j - 1].isupper() and k != "c":
70 | pass
71 | else:
72 | splitted_smiles.append(k)
73 | return splitted_smiles
74 |
75 | def get_maxlen(all_smiles, kekuleSmiles=True):
76 | maxlen = 0
77 | for smi in tqdm(all_smiles):
78 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
79 | if spt is None:
80 | continue
81 | maxlen = max(maxlen, len(spt))
82 | return maxlen
83 | def get_dict(all_smiles, save_path, kekuleSmiles=True):
84 | words = [' ']
85 | for smi in tqdm(all_smiles):
86 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
87 | if spt is None:
88 | continue
89 | for w in spt:
90 | if w in words:
91 | continue
92 | else:
93 | words.append(w)
94 | with open(save_path, 'w') as js:
95 | json.dump(words, js)
96 | return words
97 |
98 | def one_hot_coding(smi, words, kekuleSmiles=True, max_len=1000):
99 | coord_j = []
100 | coord_k = []
101 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
102 | if spt is None:
103 | return None
104 | for j,w in enumerate(spt):
105 | if j >= max_len:
106 | break
107 | try:
108 | k = words.index(w)
109 | except:
110 | continue
111 | coord_j.append(j)
112 | coord_k.append(k)
113 | data = np.repeat(1, len(coord_j))
114 | output = sparse.csr_matrix((data, (coord_j, coord_k)), shape=(max_len, len(words)))
115 | return output
116 |
117 | if __name__ == "__main__":
118 | data_train= pd.read_csv('E:/code/drug/drugnn/data_train.csv')
119 | data_test=pd.read_csv('E:/code/drug/drugnn/worddrug.csv')
120 | inchis = list(data_train['SMILES'])
121 | rts = list(data_train['type'])
122 |
123 | smiles, targets = [], []
124 | for i, inc in enumerate(tqdm(inchis)):
125 | mol = Chem.MolFromSmiles(inc)
126 | if mol is None:
127 | continue
128 | else:
129 | smi = Chem.MolToSmiles(mol)
130 | smiles.append(smi)
131 | targets.append(rts[i])
132 |
133 | words = get_dict(smiles, save_path='E:\code\FingerID Reference\drug-likeness/dict.json')
134 |
135 | features = []
136 | for i, smi in enumerate(tqdm(smiles)):
137 | xi = one_hot_coding(smi, words, max_len=600)
138 | if xi is not None:
139 | features.append(xi.todense())
140 | features = np.asarray(features)
141 | targets = np.asarray(targets)
142 | X_train=features
143 | Y_train=targets
144 |
145 |
146 | # physical_devices = tf.config.experimental.list_physical_devices('CPU')
147 | # assert len(physical_devices) > 0, "Not enough GPU hardware devices available"
148 | # tf.config.experimental.set_memory_growth(physical_devices[0], True)
149 |
150 |
151 |
152 | inchis = list(data_test['SMILES'])
153 | rts = list(data_test['type'])
154 |
155 | smiles, targets = [], []
156 | for i, inc in enumerate(tqdm(inchis)):
157 | mol = Chem.MolFromSmiles(inc)
158 | if mol is None:
159 | continue
160 | else:
161 | smi = Chem.MolToSmiles(mol)
162 | smiles.append(smi)
163 | targets.append(rts[i])
164 |
165 | # words = get_dict(smiles, save_path='D:/工作文件/work.Data/CNN/dict.json')
166 |
167 | features = []
168 | for i, smi in enumerate(tqdm(smiles)):
169 | xi = one_hot_coding(smi, words, max_len=600)
170 | if xi is not None:
171 | features.append(xi.todense())
172 | features = np.asarray(features)
173 | targets = np.asarray(targets)
174 | X_test=features
175 | Y_test=targets
176 |
177 | # kernel = 1.0 * RBF(0.8)
178 | #model = RandomForestClassifier(n_estimators=10,max_features='auto', max_depth=None,min_samples_split=2, bootstrap=True)
179 | model = GPC( random_state=111)
180 |
181 | # earlyStopping = EarlyStopping(monitor='val_loss', patience=0.05, verbose=0, mode='min')
182 | #mcp_save = ModelCheckpoint('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5', save_best_only=True, monitor='accuracy', mode='auto')
183 | # reduce_lr_loss = ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=3, verbose=1, epsilon=1e-4, mode='min')
184 | from tensorflow.keras import backend as K
185 | X_train = K.cast_to_floatx(X_train).reshape((np.size(X_train,0),np.size(X_train,1)*np.size(X_train,2)))
186 |
187 | Y_train = K.cast_to_floatx(Y_train)
188 |
189 | # X_train,Y_train = make_blobs(n_samples=300, n_features=n_features, centers=6)
190 | model.fit(X_train, Y_train)
191 |
192 |
193 | # model = load_model('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5')
194 | Y_predict = model.predict(K.cast_to_floatx(X_test).reshape((np.size(X_test,0),np.size(X_test,1)*np.size(X_test,2))))
195 | #Y_predict = model.predict(X_test)#训练数据
196 | x = list(Y_test)
197 | y = list(Y_predict)
198 | from pandas.core.frame import DataFrame
199 | x=DataFrame(x)
200 | y=DataFrame(y)
201 | # X= pd.concat([x,y], axis=1)
202 | #X.to_csv('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/molecularGNN_smiles-master/0825/single-CNN-seed444.csv')
203 | Y_predict = [1 if i >0.4 else 0 for i in Y_predict]
204 |
205 | cnf_matrix=confusion_matrix(Y_test, Y_predict)
206 | cnf_matrix
207 |
208 | tn = cnf_matrix[0,0]
209 | tp = cnf_matrix[1,1]
210 | fn = cnf_matrix[1,0]
211 | fp = cnf_matrix[0,1]
212 |
213 | bacc = ((tp/(tp+fn))+(tn/(tn+fp)))/2#balance accurance
214 | pre = tp/(tp+fp)#precision/q+
215 | rec = tp/(tp+fn)#recall/se
216 | sp=tn/(tn+fp)
217 | q_=tn/(tn+fn)
218 | f1 = 2*pre*rec/(pre+rec)#f1score
219 | mcc = ((tp*tn) - (fp*fn))/math.sqrt((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))#Matthews correlation coefficient
220 | acc=(tp+tn)/(tp+fp+fn+tn)#accurancy
221 | fpr, tpr, thresholds =roc_curve(Y_test, Y_predict)
222 | AUC = auc(fpr, tpr)
223 | print('bacc:',bacc)
224 | print('pre:',pre)
225 | print('rec:',rec)
226 | print('f1:',f1)
227 | print('mcc:',mcc)
228 | print('sp:',sp)
229 | print('q_:',q_)
230 | print('acc:',acc)
231 | print('auc:',AUC)
232 |
--------------------------------------------------------------------------------
/Discussion/svc.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Sun Nov 15 13:46:29 2020
4 |
5 | @author: de''
6 | """
7 |
8 | # -*- coding: utf-8 -*-
9 | """
10 | Created on Sun Nov 15 10:40:57 2020
11 |
12 | @author: de''
13 | """
14 |
15 | from sklearn.datasets import make_blobs
16 | import json
17 | import numpy as np
18 | import math
19 | from tqdm import tqdm
20 | from scipy import sparse
21 | from sklearn.metrics import roc_auc_score,roc_curve,auc
22 | from sklearn.metrics import confusion_matrix
23 |
24 | import pandas as pd
25 | import matplotlib.pyplot as plt
26 | from rdkit import Chem
27 |
28 | from sklearn.ensemble import RandomForestClassifier
29 | from sklearn.model_selection import train_test_split
30 | from sklearn.preprocessing import MinMaxScaler
31 | from sklearn.neural_network import MLPClassifier
32 | from sklearn.svm import SVC
33 | from tensorflow.keras.models import Model, load_model
34 | from tensorflow.keras.layers import Dense, Input, Flatten, Conv1D, MaxPooling1D, concatenate
35 | from tensorflow.keras import metrics, optimizers
36 | from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
37 |
38 | def split_smiles(smiles, kekuleSmiles=True):
39 | try:
40 | mol = Chem.MolFromSmiles(smiles)
41 | smiles = Chem.MolToSmiles(mol, kekuleSmiles=kekuleSmiles)
42 | except:
43 | pass
44 | splitted_smiles = []
45 | for j, k in enumerate(smiles):
46 | if len(smiles) == 1:
47 | return [smiles]
48 | if j == 0:
49 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
50 | splitted_smiles.append(k + smiles[j + 1])
51 | else:
52 | splitted_smiles.append(k)
53 | elif j != 0 and j < len(smiles) - 1:
54 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
55 | splitted_smiles.append(k + smiles[j + 1])
56 | elif k.islower() and smiles[j - 1].isupper() and k != "c":
57 | pass
58 | else:
59 | splitted_smiles.append(k)
60 |
61 | elif j == len(smiles) - 1:
62 | if k.islower() and smiles[j - 1].isupper() and k != "c":
63 | pass
64 | else:
65 | splitted_smiles.append(k)
66 | return splitted_smiles
67 |
68 | def get_maxlen(all_smiles, kekuleSmiles=True):
69 | maxlen = 0
70 | for smi in tqdm(all_smiles):
71 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
72 | if spt is None:
73 | continue
74 | maxlen = max(maxlen, len(spt))
75 | return maxlen
76 | def get_dict(all_smiles, save_path, kekuleSmiles=True):
77 | words = [' ']
78 | for smi in tqdm(all_smiles):
79 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
80 | if spt is None:
81 | continue
82 | for w in spt:
83 | if w in words:
84 | continue
85 | else:
86 | words.append(w)
87 | with open(save_path, 'w') as js:
88 | json.dump(words, js)
89 | return words
90 |
91 | def one_hot_coding(smi, words, kekuleSmiles=True, max_len=1000):
92 | coord_j = []
93 | coord_k = []
94 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
95 | if spt is None:
96 | return None
97 | for j,w in enumerate(spt):
98 | if j >= max_len:
99 | break
100 | try:
101 | k = words.index(w)
102 | except:
103 | continue
104 | coord_j.append(j)
105 | coord_k.append(k)
106 | data = np.repeat(1, len(coord_j))
107 | output = sparse.csr_matrix((data, (coord_j, coord_k)), shape=(max_len, len(words)))
108 | return output
109 |
110 | if __name__ == "__main__":
111 |
112 |
113 | data_train= pd.read_csv('E:/code/drug/drugnn/data_train.csv')
114 | data_test=pd.read_csv('E:/code/drug/drugnn/bro5.csv')
115 | inchis = list(data_train['SMILES'])
116 | rts = list(data_train['type'])
117 |
118 | smiles, targets = [], []
119 | for i, inc in enumerate(tqdm(inchis)):
120 | mol = Chem.MolFromSmiles(inc)
121 | if mol is None:
122 | continue
123 | else:
124 | smi = Chem.MolToSmiles(mol)
125 | smiles.append(smi)
126 | targets.append(rts[i])
127 |
128 | words = get_dict(smiles, save_path='E:\code\FingerID Reference\drug-likeness/dict.json')
129 |
130 | features = []
131 | for i, smi in enumerate(tqdm(smiles)):
132 | xi = one_hot_coding(smi, words, max_len=2000)
133 | if xi is not None:
134 | features.append(xi.todense())
135 | features = np.asarray(features)
136 | targets = np.asarray(targets)
137 | X_train=features
138 | Y_train=targets
139 |
140 |
141 | # physical_devices = tf.config.experimental.list_physical_devices('CPU')
142 | # assert len(physical_devices) > 0, "Not enough GPU hardware devices available"
143 | # tf.config.experimental.set_memory_growth(physical_devices[0], True)
144 |
145 |
146 |
147 | inchis = list(data_test['SMILES'])
148 | rts = list(data_test['type'])
149 |
150 | smiles, targets = [], []
151 | for i, inc in enumerate(tqdm(inchis)):
152 | mol = Chem.MolFromSmiles(inc)
153 | if mol is None:
154 | continue
155 | else:
156 | smi = Chem.MolToSmiles(mol)
157 | smiles.append(smi)
158 | targets.append(rts[i])
159 |
160 | # words = get_dict(smiles, save_path='D:/工作文件/work.Data/CNN/dict.json')
161 |
162 | features = []
163 | for i, smi in enumerate(tqdm(smiles)):
164 | xi = one_hot_coding(smi, words, max_len=2000)
165 | if xi is not None:
166 | features.append(xi.todense())
167 | features = np.asarray(features)
168 | targets = np.asarray(targets)
169 | X_test=features
170 | Y_test=targets
171 |
172 |
173 | #model = RandomForestClassifier(n_estimators=10,max_features='auto', max_depth=None,min_samples_split=2, bootstrap=True)
174 | #model = MLPClassifier(rangdom_state=1,max_iter=300)
175 | model = SVC(C=500, kernel='rbf', gamma='auto',
176 | coef0=0.0, shrinking=True,probability=False, tol=0.0001, cache_size=200, class_weight=None, verbose=False, max_iter=- 1, decision_function_shape='ovr', break_ties=False, random_state=None)
177 |
178 | # earlyStopping = EarlyStopping(monitor='val_loss', patience=0.05, verbose=0, mode='min')
179 | #mcp_save = ModelCheckpoint('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5', save_best_only=True, monitor='accuracy', mode='auto')
180 | # reduce_lr_loss = ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=3, verbose=1, epsilon=1e-4, mode='min')
181 | from tensorflow.keras import backend as K
182 | X_train = K.cast_to_floatx(X_train).reshape((np.size(X_train,0),np.size(X_train,1)*np.size(X_train,2)))
183 |
184 | Y_train = K.cast_to_floatx(Y_train)
185 |
186 | # X_train,Y_train = make_blobs(n_samples=300, n_features=n_features, centers=6)
187 | model.fit(X_train, Y_train)
188 |
189 |
190 | # model = load_model('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5')
191 | Y_predict = model.predict(K.cast_to_floatx(X_test).reshape((np.size(X_test,0),np.size(X_test,1)*np.size(X_test,2))))
192 | #Y_predict = model.predict(X_test)#训练数据
193 | x = list(Y_test)
194 | y = list(Y_predict)
195 | from pandas.core.frame import DataFrame
196 | x=DataFrame(x)
197 | y=DataFrame(y)
198 | # X= pd.concat([x,y], axis=1)
199 | #X.to_csv('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/molecularGNN_smiles-master/0825/single-CNN-seed444.csv')
200 | Y_predict = [1 if i >0.5 else 0 for i in Y_predict]
201 |
202 | cnf_matrix=confusion_matrix(Y_test, Y_predict)
203 | cnf_matrix
204 |
205 | tn = cnf_matrix[0,0]
206 | tp = cnf_matrix[1,1]
207 | fn = cnf_matrix[1,0]
208 | fp = cnf_matrix[0,1]
209 |
210 | bacc = ((tp/(tp+fn))+(tn/(tn+fp)))/2#balance accurance
211 | pre = tp/(tp+fp)#precision/q+
212 | rec = tp/(tp+fn)#recall/se
213 | sp=tn/(tn+fp)
214 | q_=tn/(tn+fn)
215 | f1 = 2*pre*rec/(pre+rec)#f1score
216 | mcc = ((tp*tn) - (fp*fn))/math.sqrt((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))#Matthews correlation coefficient
217 | acc=(tp+tn)/(tp+fp+fn+tn)#accurancy
218 | fpr, tpr, thresholds =roc_curve(Y_test, Y_predict)
219 | AUC = auc(fpr, tpr)
220 | print('bacc:',bacc)
221 | print('pre:',pre)
222 | print('rec:',rec)
223 | print('f1:',f1)
224 | print('mcc:',mcc)
225 | print('sp:',sp)
226 | print('q_:',q_)
227 | print('acc:',acc)
228 | print('auc:',AUC)
--------------------------------------------------------------------------------
/DGCAN/preprocess.py:
--------------------------------------------------------------------------------
1 |
2 | # -*- coding: utf-8 -*-
3 | """
4 | Created on Wed Apr 27 20:09:31 2022
5 |
6 | @author:Jinyu-Sun
7 | """
8 |
9 | from collections import defaultdict
10 | import numpy as np
11 | from rdkit import Chem
12 | import torch
13 |
14 | device = torch.device('cuda')
15 |
16 | atom_dict = defaultdict(lambda: len(atom_dict))
17 | bond_dict = defaultdict(lambda: len(bond_dict))
18 | fingerprint_dict = defaultdict(lambda: len(fingerprint_dict))
19 | edge_dict = defaultdict(lambda: len(edge_dict))
20 | radius=1
21 | def create_atoms(mol, atom_dict):
22 | """Transform the atom types in a molecule (e.g., H, C, and O)
23 | into the indices (e.g., H=0, C=1, and O=2).
24 | Note that each atom index considers the aromaticity.
25 | """
26 | atoms = [a.GetSymbol() for a in mol.GetAtoms()]
27 | for a in mol.GetAromaticAtoms():
28 | i = a.GetIdx()
29 | atoms[i] = (atoms[i], 'aromatic')
30 | atoms = [atom_dict[a] for a in atoms]
31 | return np.array(atoms)
32 |
33 |
34 | def create_ijbonddict(mol, bond_dict):
35 | """Create a dictionary, in which each key is a node ID
36 | and each value is the tuples of its neighboring node
37 | and chemical bond (e.g., single and double) IDs.
38 |
39 | """
40 | i_jbond_dict = defaultdict(lambda: [])
41 | for b in mol.GetBonds():
42 | i, j = b.GetBeginAtomIdx(), b.GetEndAtomIdx()
43 | bond = bond_dict[str(b.GetBondType())]
44 | i_jbond_dict[i].append((j, bond))
45 | i_jbond_dict[j].append((i, bond))
46 | return i_jbond_dict
47 |
48 |
49 | def extract_fingerprints(radius, atoms, i_jbond_dict,
50 | fingerprint_dict, edge_dict):
51 | """Extract the fingerprints from a molecular graph
52 | based on Weisfeiler-Lehman algorithm.
53 | """
54 |
55 | if (len(atoms) == 1) or (radius == 0):
56 | nodes = [fingerprint_dict[a] for a in atoms]
57 |
58 | else:
59 | nodes = atoms
60 | i_jedge_dict = i_jbond_dict
61 |
62 | for _ in range(radius):
63 |
64 | """Update each node ID considering its neighboring nodes and edges.
65 | The updated node IDs are the fingerprint IDs.。
66 | """
67 | nodes_ = []
68 | for i, j_edge in i_jedge_dict.items():
69 | neighbors = [(nodes[j], edge) for j, edge in j_edge]
70 | fingerprint = (nodes[i], tuple(sorted(neighbors)))
71 | nodes_.append(fingerprint_dict[fingerprint])
72 |
73 | """Also update each edge ID considering
74 | its two nodes on both sides.
75 | """
76 | i_jedge_dict_ = defaultdict(lambda: [])
77 | for i, j_edge in i_jedge_dict.items():
78 | for j, edge in j_edge:
79 | both_side = tuple(sorted((nodes[i], nodes[j])))
80 | edge = edge_dict[(both_side, edge)]
81 | i_jedge_dict_[i].append((j, edge))
82 |
83 | nodes = nodes_
84 | i_jedge_dict = i_jedge_dict_
85 |
86 | return np.array(nodes)
87 |
88 |
89 | def split_dataset(dataset, ratio):
90 | """Shuffle and split a dataset."""
91 | np.random.seed(1234) # fix the seed for shuffle
92 | # np.random.shuffle(dataset)
93 | n = int(ratio * len(dataset))
94 | return dataset[:n], dataset[n:]
95 | def create_testdataset(filename,path,dataname,property):
96 | dir_dataset = path+dataname
97 | print(filename)
98 | """Load a dataset."""
99 | if property== False:
100 | with open(dir_dataset + filename, 'r') as f:
101 | #smiles_property = f.readline().strip().split()
102 | data_original = f.read().strip().split()
103 | data_original = [data for data in data_original
104 | if '.' not in data.split()[0]]
105 | dataset = []
106 | for data in data_original:
107 | smiles = data
108 | try:
109 | """Create each data with the above defined functions."""
110 | mol = Chem.AddHs(Chem.MolFromSmiles(smiles))
111 | atoms = create_atoms(mol, atom_dict)
112 | molecular_size = len(atoms)
113 | i_jbond_dict = create_ijbonddict(mol, bond_dict)
114 | fingerprints = extract_fingerprints(radius, atoms, i_jbond_dict,
115 | fingerprint_dict, edge_dict)
116 | adjacency = Chem.GetAdjacencyMatrix(mol)
117 | """Transform the above each data of numpy
118 | to pytorch tensor on a device (i.e., CPU or GPU).
119 | """
120 | fingerprints = torch.LongTensor(fingerprints).to(device)
121 | adjacency = torch.FloatTensor(adjacency).to(device)
122 | proper = torch.LongTensor([int(0)]).to(device)
123 | dataset.append((smiles,fingerprints, adjacency, molecular_size,proper ))
124 | except:
125 | print(smiles)
126 | elif property== True:
127 | with open(dir_dataset + filename, 'r') as f:
128 | # smiles_property = f.readline().strip().split()
129 | data_original = f.read().strip().split('\n')
130 |
131 | data_original = [data for data in data_original
132 | if '.' not in data.split()[0]]
133 | dataset = []
134 | for data in data_original:
135 | smiles, proper = data.strip().split()
136 | try:
137 | """Create each data with the above defined functions."""
138 | mol = Chem.AddHs(Chem.MolFromSmiles(smiles))
139 | atoms = create_atoms(mol, atom_dict)
140 | molecular_size = len(atoms)
141 | i_jbond_dict = create_ijbonddict(mol, bond_dict)
142 | fingerprints = extract_fingerprints(radius, atoms, i_jbond_dict,
143 | fingerprint_dict, edge_dict)
144 | adjacency = Chem.GetAdjacencyMatrix(mol)
145 |
146 | """Transform the above each data of numpy
147 | to pytorch tensor on a device (i.e., CPU or GPU).
148 | """
149 | fingerprints = torch.LongTensor(fingerprints).to(device)
150 | adjacency = torch.FloatTensor(adjacency).to(device)
151 | proper = torch.LongTensor([int(proper)]).to(device)
152 | dataset.append((smiles,fingerprints, adjacency, molecular_size, proper))
153 | except:
154 | print(smiles+'is error')
155 | return dataset
156 |
157 | def create_dataset(filename,path,dataname):
158 | dir_dataset = path+dataname
159 | print(filename)
160 | """Load a dataset."""
161 | try:
162 | with open(dir_dataset + filename, 'r') as f:
163 | smiles_property = f.readline().strip().split()
164 | data_original = f.read().strip().split('\n')
165 | except:
166 | with open(dir_dataset + filename, 'r') as f:
167 | smiles_property = f.readline().strip().split()
168 | data_original = f.read().strip().split('\n')
169 |
170 | """Exclude the data contains '.' in its smiles.排除含.的数据"""
171 | data_original = [data for data in data_original
172 | if '.' not in data.split()[0]]
173 | dataset = []
174 | for data in data_original:
175 | smiles, property = data.strip().split()
176 | try:
177 | """Create each data with the above defined functions."""
178 | mol = Chem.AddHs(Chem.MolFromSmiles(smiles))
179 | atoms = create_atoms(mol, atom_dict)
180 | molecular_size = len(atoms)
181 | i_jbond_dict = create_ijbonddict(mol, bond_dict)
182 | fingerprints = extract_fingerprints(radius, atoms, i_jbond_dict,
183 | fingerprint_dict, edge_dict)
184 | adjacency = Chem.GetAdjacencyMatrix(mol)
185 | """
186 | Transform the above each data of numpy
187 | to pytorch tensor on a device (i.e., CPU or GPU).
188 | """
189 | fingerprints = torch.LongTensor(fingerprints).to(device)
190 | adjacency = torch.FloatTensor(adjacency).to(device)
191 | property = torch.LongTensor([int(property)]).to(device)
192 | dataset.append((smiles,fingerprints, adjacency, molecular_size, property))
193 | except:
194 | print(smiles)
195 | return dataset
196 |
197 |
198 |
--------------------------------------------------------------------------------
/Discussion/RF.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Tue Apr 27 22:08:23 2021
4 |
5 | @author:Jinyusun
6 | """
7 |
8 |
9 | from sklearn.datasets import make_blobs
10 | import json
11 | import numpy as np
12 | import math
13 | from tqdm import tqdm
14 | from scipy import sparse
15 | from sklearn.metrics import roc_auc_score,roc_curve,auc
16 | from sklearn.metrics import confusion_matrix
17 |
18 | import pandas as pd
19 | import matplotlib.pyplot as plt
20 | from rdkit import Chem
21 |
22 | from sklearn.ensemble import RandomForestClassifier
23 | from sklearn.model_selection import train_test_split
24 | from sklearn.preprocessing import MinMaxScaler
25 | from sklearn.neural_network import MLPClassifier
26 | from sklearn.svm import SVC
27 | from tensorflow.keras.models import Model, load_model
28 | from tensorflow.keras.layers import Dense, Input, Flatten, Conv1D, MaxPooling1D, concatenate
29 | from tensorflow.keras import metrics, optimizers
30 | from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
31 |
32 | def split_smiles(smiles, kekuleSmiles=True):
33 | try:
34 | mol = Chem.MolFromSmiles(smiles)
35 | smiles = Chem.MolToSmiles(mol, kekuleSmiles=kekuleSmiles)
36 | except:
37 | pass
38 | splitted_smiles = []
39 | for j, k in enumerate(smiles):
40 | if len(smiles) == 1:
41 | return [smiles]
42 | if j == 0:
43 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
44 | splitted_smiles.append(k + smiles[j + 1])
45 | else:
46 | splitted_smiles.append(k)
47 | elif j != 0 and j < len(smiles) - 1:
48 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
49 | splitted_smiles.append(k + smiles[j + 1])
50 | elif k.islower() and smiles[j - 1].isupper() and k != "c":
51 | pass
52 | else:
53 | splitted_smiles.append(k)
54 |
55 | elif j == len(smiles) - 1:
56 | if k.islower() and smiles[j - 1].isupper() and k != "c":
57 | pass
58 | else:
59 | splitted_smiles.append(k)
60 | return splitted_smiles
61 |
62 | def get_maxlen(all_smiles, kekuleSmiles=True):
63 | maxlen = 0
64 | for smi in tqdm(all_smiles):
65 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
66 | if spt is None:
67 | continue
68 | maxlen = max(maxlen, len(spt))
69 | return maxlen
70 | def get_dict(all_smiles, save_path, kekuleSmiles=True):
71 | words = [' ']
72 | for smi in tqdm(all_smiles):
73 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
74 | if spt is None:
75 | continue
76 | for w in spt:
77 | if w in words:
78 | continue
79 | else:
80 | words.append(w)
81 | with open(save_path, 'w') as js:
82 | json.dump(words, js)
83 | return words
84 |
85 | def one_hot_coding(smi, words, kekuleSmiles=True, max_len=1000):
86 | coord_j = []
87 | coord_k = []
88 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
89 | if spt is None:
90 | return None
91 | for j,w in enumerate(spt):
92 | if j >= max_len:
93 | break
94 | try:
95 | k = words.index(w)
96 | except:
97 | continue
98 | coord_j.append(j)
99 | coord_k.append(k)
100 | data = np.repeat(1, len(coord_j))
101 | output = sparse.csr_matrix((data, (coord_j, coord_k)), shape=(max_len, len(words)))
102 | return output
103 | def split_dataset(dataset, ratio):
104 | """Shuffle and split a dataset."""
105 | # np.random.seed(111) # fix the seed for shuffle.
106 | #np.random.shuffle(dataset)
107 | n = int(ratio * len(dataset))
108 | return dataset[:n], dataset[n:]
109 | def edit_dataset(drug,non_drug,task):
110 | # np.random.seed(111) # fix the seed for shuffle.
111 |
112 | # np.random.shuffle(non_drug)
113 | non_drug=non_drug[0:len(drug)]
114 |
115 |
116 | # np.random.shuffle(non_drug)
117 | # np.random.shuffle(drug)
118 | dataset_train_drug, dataset_test_drug = split_dataset(drug, 0.9)
119 | # dataset_train_drug,dataset_dev_drug = split_dataset(dataset_train_drug, 0.9)
120 | dataset_train_no, dataset_test_no = split_dataset(non_drug, 0.9)
121 | # dataset_train_no,dataset_dev_no = split_dataset(dataset_train_no, 0.9)
122 | dataset_train = pd.concat([dataset_train_drug,dataset_train_no], axis=0)
123 | dataset_test=pd.concat([ dataset_test_drug,dataset_test_no], axis=0)
124 | # dataset_dev = dataset_dev_drug+dataset_dev_no
125 | return dataset_train, dataset_test
126 | if __name__ == "__main__":
127 | data_train= pd.read_csv('E:/code/drug/drugnn/data_train.csv')
128 | data_test=pd.read_csv('E:/code/drug/drugnn/bro5.csv')
129 | inchis = list(data_train['SMILES'])
130 | rts = list(data_train['type'])
131 |
132 | smiles, targets = [], []
133 | for i, inc in enumerate(tqdm(inchis)):
134 | mol = Chem.MolFromSmiles(inc)
135 | if mol is None:
136 | continue
137 | else:
138 | smi = Chem.MolToSmiles(mol)
139 | smiles.append(smi)
140 | targets.append(rts[i])
141 |
142 | words = get_dict(smiles, save_path='E:\code\FingerID Reference\drug-likeness/dict.json')
143 |
144 | features = []
145 | for i, smi in enumerate(tqdm(smiles)):
146 | xi = one_hot_coding(smi, words, max_len=600)
147 | if xi is not None:
148 | features.append(xi.todense())
149 | features = np.asarray(features)
150 | targets = np.asarray(targets)
151 | X_train=features
152 | Y_train=targets
153 |
154 |
155 | # physical_devices = tf.config.experimental.list_physical_devices('CPU')
156 | # assert len(physical_devices) > 0, "Not enough GPU hardware devices available"
157 | # tf.config.experimental.set_memory_growth(physical_devices[0], True)
158 |
159 |
160 |
161 | inchis = list(data_test['SMILES'])
162 | rts = list(data_test['type'])
163 |
164 | smiles, targets = [], []
165 | for i, inc in enumerate(tqdm(inchis)):
166 | mol = Chem.MolFromSmiles(inc)
167 | if mol is None:
168 | continue
169 | else:
170 | smi = Chem.MolToSmiles(mol)
171 | smiles.append(smi)
172 | targets.append(rts[i])
173 |
174 | # words = get_dict(smiles, save_path='D:/工作文件/work.Data/CNN/dict.json')
175 |
176 | features = []
177 | for i, smi in enumerate(tqdm(smiles)):
178 | xi = one_hot_coding(smi, words, max_len=600)
179 | if xi is not None:
180 | features.append(xi.todense())
181 | features = np.asarray(features)
182 | targets = np.asarray(targets)
183 | X_test=features
184 | Y_test=targets
185 | n_features=10
186 |
187 | model = RandomForestClassifier(n_estimators=5,max_features='auto', max_depth=None,min_samples_split=5, bootstrap=True)
188 | #model = MLPClassifier(rangdom_state=1,max_iter=300)
189 | #model = SVC()
190 |
191 | # earlyStopping = EarlyStopping(monitor='val_loss', patience=0.05, verbose=0, mode='min')
192 | #mcp_save = ModelCheckpoint('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5', save_best_only=True, monitor='accuracy', mode='auto')
193 | # reduce_lr_loss = ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=3, verbose=1, epsilon=1e-4, mode='min')
194 | from tensorflow.keras import backend as K
195 | X_train = K.cast_to_floatx(X_train).reshape((np.size(X_train,0),np.size(X_train,1)*np.size(X_train,2)))
196 |
197 | Y_train = K.cast_to_floatx(Y_train)
198 |
199 | # X_train,Y_train = make_blobs(n_samples=300, n_features=n_features, centers=6)
200 | model.fit(X_train, Y_train)
201 |
202 |
203 | # model = load_model('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5')
204 | Y_predict = model.predict(K.cast_to_floatx(X_test).reshape((np.size(X_test,0),np.size(X_test,1)*np.size(X_test,2))))
205 | #Y_predict = model.predict(X_test)#训练数据
206 | x = list(Y_test)
207 | y = list(Y_predict)
208 | from pandas.core.frame import DataFrame
209 | x=DataFrame(x)
210 | y=DataFrame(y)
211 | # X= pd.concat([x,y], axis=1)
212 | #X.to_csv('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/molecularGNN_smiles-master/0825/single-CNN-seed444.csv')
213 | Y_predict = [1 if i >0.4 else 0 for i in Y_predict]
214 |
215 | cnf_matrix=confusion_matrix(Y_test, Y_predict)
216 | cnf_matrix
217 |
218 | tn = cnf_matrix[0,0]
219 | tp = cnf_matrix[1,1]
220 | fn = cnf_matrix[1,0]
221 | fp = cnf_matrix[0,1]
222 |
223 | bacc = ((tp/(tp+fn))+(tn/(tn+fp)))/2#balance accurance
224 | pre = tp/(tp+fp)#precision/q+
225 | rec = tp/(tp+fn)#recall/se
226 | sp=tn/(tn+fp)
227 | q_=tn/(tn+fn)
228 | f1 = 2*pre*rec/(pre+rec)#f1score
229 | mcc = ((tp*tn) - (fp*fn))/math.sqrt((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))#Matthews correlation coefficient
230 | acc=(tp+tn)/(tp+fp+fn+tn)#accurancy
231 | fpr, tpr, thresholds =roc_curve(Y_test, Y_predict)
232 | AUC = auc(fpr, tpr)
233 | print('bacc:',bacc)
234 | print('pre:',pre)
235 | print('rec:',rec)
236 | print('f1:',f1)
237 | print('mcc:',mcc)
238 | print('sp:',sp)
239 | print('q_:',q_)
240 | print('acc:',acc)
241 | print('auc:',AUC)
242 |
--------------------------------------------------------------------------------
/Discussion/CNN.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Sun Nov 15 10:40:57 2020
4 |
5 | @author: de''
6 | """
7 |
8 | import json
9 | import numpy as np
10 | import math
11 | from tqdm import tqdm
12 | from scipy import sparse
13 | from sklearn.metrics import roc_auc_score,roc_curve,auc
14 | from sklearn.metrics import confusion_matrix
15 |
16 | import pandas as pd
17 | import matplotlib.pyplot as plt
18 | from rdkit import Chem
19 |
20 | from sklearn.model_selection import train_test_split
21 | from sklearn.preprocessing import MinMaxScaler
22 | from sklearn.metrics import mean_absolute_error, r2_score,median_absolute_error
23 | from tensorflow.keras.models import Model, load_model
24 | from tensorflow.keras.layers import Dense, Input, Flatten, Conv1D, MaxPooling1D, concatenate
25 | from tensorflow.keras import metrics, optimizers
26 | from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
27 |
28 | def split_smiles(smiles, kekuleSmiles=True):
29 | try:
30 | mol = Chem.MolFromSmiles(smiles)
31 | smiles = Chem.MolToSmiles(mol, kekuleSmiles=kekuleSmiles)
32 | except:
33 | pass
34 | splitted_smiles = []
35 | for j, k in enumerate(smiles):
36 | if len(smiles) == 1:
37 | return [smiles]
38 | if j == 0:
39 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
40 | splitted_smiles.append(k + smiles[j + 1])
41 | else:
42 | splitted_smiles.append(k)
43 | elif j != 0 and j < len(smiles) - 1:
44 | if k.isupper() and smiles[j + 1].islower() and smiles[j + 1] != "c":
45 | splitted_smiles.append(k + smiles[j + 1])
46 | elif k.islower() and smiles[j - 1].isupper() and k != "c":
47 | pass
48 | else:
49 | splitted_smiles.append(k)
50 |
51 | elif j == len(smiles) - 1:
52 | if k.islower() and smiles[j - 1].isupper() and k != "c":
53 | pass
54 | else:
55 | splitted_smiles.append(k)
56 | return splitted_smiles
57 |
58 | def get_maxlen(all_smiles, kekuleSmiles=True):
59 | maxlen = 0
60 | for smi in tqdm(all_smiles):
61 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
62 | if spt is None:
63 | continue
64 | maxlen = max(maxlen, len(spt))
65 | return maxlen
66 | def get_dict(all_smiles, save_path, kekuleSmiles=True):
67 | words = [' ']
68 | for smi in tqdm(all_smiles):
69 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
70 | if spt is None:
71 | continue
72 | for w in spt:
73 | if w in words:
74 | continue
75 | else:
76 | words.append(w)
77 | with open(save_path, 'w') as js:
78 | json.dump(words, js)
79 | return words
80 |
81 | def one_hot_coding(smi, words, kekuleSmiles=True, max_len=1000):
82 | coord_j = []
83 | coord_k = []
84 | spt = split_smiles(smi, kekuleSmiles=kekuleSmiles)
85 | if spt is None:
86 | return None
87 | for j,w in enumerate(spt):
88 | if j >= max_len:
89 | break
90 | try:
91 | k = words.index(w)
92 | except:
93 | continue
94 | coord_j.append(j)
95 | coord_k.append(k)
96 | data = np.repeat(1, len(coord_j))
97 | output = sparse.csr_matrix((data, (coord_j, coord_k)), shape=(max_len, len(words)))
98 | return output
99 | def split_dataset(dataset, ratio):
100 | """Shuffle and split a dataset."""
101 | # np.random.seed(111) # fix the seed for shuffle.
102 | #np.random.shuffle(dataset)
103 | n = int(ratio * len(dataset))
104 | return dataset[:n], dataset[n:]
105 | def edit_dataset(drug,non_drug,task):
106 | # np.random.seed(111) # fix the seed for shuffle.
107 |
108 | # np.random.shuffle(non_drug)
109 | non_drug=non_drug[0:len(drug)]
110 |
111 |
112 | # np.random.shuffle(non_drug)
113 | # np.random.shuffle(drug)
114 | dataset_train_drug, dataset_test_drug = split_dataset(drug, 0.9)
115 | # dataset_train_drug,dataset_dev_drug = split_dataset(dataset_train_drug, 0.9)
116 | dataset_train_no, dataset_test_no = split_dataset(non_drug, 0.9)
117 | # dataset_train_no,dataset_dev_no = split_dataset(dataset_train_no, 0.9)
118 | dataset_train = pd.concat([dataset_train_drug,dataset_train_no], axis=0)
119 | dataset_test=pd.concat([ dataset_test_drug,dataset_test_no], axis=0)
120 | # dataset_dev = dataset_dev_drug+dataset_dev_no
121 | return dataset_train, dataset_test
122 | if __name__ == "__main__":
123 | data_train= pd.read_csv('E:/code/drug/drugnn/data_train.csv')
124 | data_test=pd.read_csv('E:/code/drug/drugnn/bro5.csv')
125 | inchis = list(data_train['SMILES'])
126 | rts = list(data_train['type'])
127 |
128 | smiles, targets = [], []
129 | for i, inc in enumerate(tqdm(inchis)):
130 | mol = Chem.MolFromSmiles(inc)
131 | if mol is None:
132 | continue
133 | else:
134 | smi = Chem.MolToSmiles(mol)
135 | smiles.append(smi)
136 | targets.append(rts[i])
137 |
138 | words = get_dict(smiles, save_path='E:\code\FingerID Reference\drug-likeness/dict.json')
139 |
140 | features = []
141 | for i, smi in enumerate(tqdm(smiles)):
142 | xi = one_hot_coding(smi, words, max_len=600)
143 | if xi is not None:
144 | features.append(xi.todense())
145 | features = np.asarray(features)
146 | targets = np.asarray(targets)
147 | X_train=features
148 | Y_train=targets
149 |
150 | import tensorflow as tf
151 | # physical_devices = tf.config.experimental.list_physical_devices('CPU')
152 | # assert len(physical_devices) > 0, "Not enough GPU hardware devices available"
153 | # tf.config.experimental.set_memory_growth(physical_devices[0], True)
154 |
155 |
156 |
157 | inchis = list(data_test['SMILES'])
158 | rts = list(data_test['type'])
159 |
160 | smiles, targets = [], []
161 | for i, inc in enumerate(tqdm(inchis)):
162 | mol = Chem.MolFromSmiles(inc)
163 | if mol is None:
164 | continue
165 | else:
166 | smi = Chem.MolToSmiles(mol)
167 | smiles.append(smi)
168 | targets.append(rts[i])
169 |
170 | # words = get_dict(smiles, save_path='D:/工作文件/work.Data/CNN/dict.json')
171 |
172 | features = []
173 | for i, smi in enumerate(tqdm(smiles)):
174 | xi = one_hot_coding(smi, words, max_len=600)
175 | if xi is not None:
176 | features.append(xi.todense())
177 | features = np.asarray(features)
178 | targets = np.asarray(targets)
179 | X_test=features
180 | Y_test=targets
181 | layer_in = Input(shape=(X_train.shape[1:3]), name="smile")
182 | layer_conv = layer_in
183 | for i in range(6):
184 | layer_conv = Conv1D(128, kernel_size=4, activation='relu', kernel_initializer='normal')(layer_conv)
185 | layer_conv = MaxPooling1D(pool_size=2)(layer_conv)
186 | layer_dense = Flatten()(layer_conv)
187 |
188 | for i in range(1):
189 | layer_dense = Dense(32, activation="relu", kernel_initializer='normal')(layer_dense)
190 | layer_output = Dense(1, activation="sigmoid", name="output")(layer_dense)
191 |
192 | # earlyStopping = EarlyStopping(monitor='val_loss', patience=0.05, verbose=0, mode='min')
193 | #mcp_save = ModelCheckpoint('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5', save_best_only=True, monitor='accuracy', mode='auto')
194 | # reduce_lr_loss = ReduceLROnPlateau(monitor='val_loss', factor=0.1, patience=3, verbose=1, epsilon=1e-4, mode='min')
195 |
196 | model = Model(layer_in, outputs = layer_output)
197 | opt = optimizers.Adam(lr=0.0005)
198 | model.compile(optimizer=opt, loss='binary_crossentropy', metrics=['accuracy'])
199 | from tensorflow.keras import backend as K #转换为张量
200 | X_train = K.cast_to_floatx(X_train)
201 | Y_train = K.cast_to_floatx(Y_train)
202 | history = model.fit(X_train, Y_train, epochs=12)
203 |
204 | # plot loss
205 | plt.plot(history.history['loss'])
206 | plt.plot(history.history['accuracy'])
207 | # plt.plot(history.history['val_loss'])
208 | # plt.plot(history.history['val_accuracy'])
209 | plt.ylabel('values')
210 | plt.xlabel('epoch')
211 | # plt.legend(['loss', 'mae', 'val_loss', 'val_mae'], loc='upper left')
212 | plt.show()
213 | # model = load_model('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/single_model.h5')
214 | Y_predict = model.predict(K.cast_to_floatx(X_test))
215 | #Y_predict = model.predict(X_test)#训练数据
216 | x = list(Y_test)
217 | y = list(Y_predict)
218 | from pandas.core.frame import DataFrame
219 | x=DataFrame(x)
220 | y=DataFrame(y)
221 | # X= pd.concat([x,y], axis=1)
222 | #X.to_csv('C:/Users/sunjinyu/Desktop/FingerID Reference/drug-likeness/CNN/molecularGNN_smiles-master/0825/single-CNN-seed444.csv')
223 | Y_predict = [1 if i >0.4 else 0 for i in Y_predict]
224 |
225 | cnf_matrix=confusion_matrix(Y_test, Y_predict)
226 | cnf_matrix
227 |
228 | tn = cnf_matrix[0,0]
229 | tp = cnf_matrix[1,1]
230 | fn = cnf_matrix[1,0]
231 | fp = cnf_matrix[0,1]
232 |
233 | bacc = ((tp/(tp+fn))+(tn/(tn+fp)))/2#balance accurance
234 | pre = tp/(tp+fp)#precision/q+
235 | rec = tp/(tp+fn)#recall/se
236 | sp=tn/(tn+fp)
237 | q_=tn/(tn+fn)
238 | f1 = 2*pre*rec/(pre+rec)#f1score
239 | mcc = ((tp*tn) - (fp*fn))/math.sqrt((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))#Matthews correlation coefficient
240 | acc=(tp+tn)/(tp+fp+fn+tn)#accurancy
241 | fpr, tpr, thresholds =roc_curve(Y_test, Y_predict)
242 | AUC = auc(fpr, tpr)
243 | print('bacc:',bacc)
244 | print('pre:',pre)
245 | print('rec:',rec)
246 | print('f1:',f1)
247 | print('mcc:',mcc)
248 | print('sp:',sp)
249 | print('q_:',q_)
250 | print('acc:',acc)
251 | print('auc:',AUC)
--------------------------------------------------------------------------------
/DGCAN/DGCAN.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | Created on Wed Apr 27 18:04:58 2022
4 |
5 | @author:Jinyu-Sun
6 | """
7 |
8 | #coding=utf-8
9 | import timeit
10 | import sys
11 | import numpy as np
12 | import math
13 | import torch
14 | import torch.nn as nn
15 | import torch.nn.functional as F
16 | import torch.optim as optim
17 | import pickle
18 | from sklearn.metrics import roc_auc_score, roc_curve
19 | from sklearn.metrics import confusion_matrix
20 | import preprocess as pp
21 | import pandas as pd
22 | import matplotlib.pyplot as plt
23 |
24 | if torch.cuda.is_available():
25 | device = torch.device('cuda')
26 |
27 | else:
28 | device = torch.device('cpu')
29 |
30 | torch.cuda.empty_cache()
31 | class GraphAttentionLayer(nn.Module):
32 | def __init__(self, in_features, out_features, dropout, alpha, concat=True):
33 | super(GraphAttentionLayer, self).__init__()
34 | self.dropout = dropout
35 | self.concat = concat
36 | self.in_features = in_features #dim of input feature
37 | self.out_features = out_features #dim of output feature
38 | self.alpha = alpha # negative_slope leakyrelu
39 | self.W = nn.Parameter(torch.zeros(size=(in_features, out_features)))
40 |
41 | self.a = nn.Parameter(torch.zeros(size=(2 * out_features, 1)))
42 | torch.nn.init.xavier_uniform_(self.W , gain=2.0)
43 | #torch.nn.init.kaiming_uniform_(self.a, a=0, mode='fan_in', nonlinearity='leaky_relu')
44 | torch.nn.init.xavier_uniform_(self.W , gain=1.9)
45 | self.leakyrelu = nn.LeakyReLU(self.alpha)
46 |
47 | def forward(self, input, adj):
48 | """
49 | input: input_feature [N, in_features] in_features indicates the number of elements of the input feature vector of the node
50 | adj: adjacency matrix of the graph dimension [N, N] non-zero is one, data structure basics
51 | """
52 | h = torch.mm(input, self.W) # [N, out_features]
53 | N = h.size()[0] #Number of nodes of the graph
54 | a_input = torch.cat([h.repeat(1, N).view(N * N, -1), h.repeat(N, 1)], dim=1).view(N, -1, 2 * self.out_features) # [N, N, 2*out_features]
55 | e = self.leakyrelu(torch.matmul(a_input, self.a).squeeze(2))
56 | zero_vec =-9e10 *torch.ones_like(e)
57 | attention = torch.where(adj > 0, e, zero_vec)
58 | # indicates that if the adjacency matrix element is greater than 0, then the two nodes are connected and the attention factor at that position is retained.
59 | # Otherwise it is necessary to mask and set to a very small value, the reason is that this minimum value will be disregarded during softmax.
60 | attention = F.softmax(attention, dim=1)
61 | attention = F.dropout(attention, self.dropout, training=self.training)
62 | h_prime = torch.matmul(attention, h)
63 | if self.concat:
64 | return F.elu(h_prime)
65 | else:
66 | return h_prime
67 |
68 |
69 | class GAT(nn.Module):
70 | def __init__(self, nfeat, nhid, dropout, alpha, nheads):
71 | super(GAT, self).__init__()
72 | """
73 | n_heads indicates that there are several GAL layers, which are finally stitched together, similar to self-attention
74 | to extract features from different subspaces.
75 | """
76 | self.dropout = dropout
77 | self.attentions = [GraphAttentionLayer(nfeat, nhid, dropout=dropout, alpha=alpha, concat=True) for _ in
78 | range(nheads)]
79 | for i, attention in enumerate(self.attentions):
80 | self.add_module('attention_{}'.format(i), attention)
81 |
82 | self.out_att = GraphAttentionLayer(nhid,56, dropout=dropout, alpha=alpha, concat=False)
83 | self.nheads=nheads
84 |
85 | def forward(self, x, adj):
86 | x = F.dropout(x, self.dropout, training=self.training)
87 | #x = torch.cat([att(x, adj) for att in self.attentions], dim=1)
88 |
89 | z=torch.zeros_like(self.attentions[1](x, adj))
90 | for att in self.attentions:
91 | z=torch.add(z, att(x, adj))
92 | x = z/self.nheads
93 | x = F.dropout(x, self.dropout, training=self.training)
94 | x = F.elu(self.out_att(x, adj))
95 | return F.softmax(x, dim=1)
96 |
97 | class MolecularGraphNeuralNetwork(nn.Module):
98 | def __init__(self, N_fingerprints, dim, layer_hidden, layer_output, dropout):
99 | super(MolecularGraphNeuralNetwork, self).__init__()
100 | self.layer_hidden=layer_hidden
101 | self.layer_output=layer_output
102 | self.embed_fingerprint = nn.Embedding(N_fingerprints, dim)
103 | self.W_fingerprint = nn.ModuleList([nn.Linear(dim, dim) for _ in range(layer_hidden)])
104 |
105 | self.W_output = nn.ModuleList([nn.Linear(56,56) for _ in range(layer_output)])
106 | self.W_property = nn.Linear(56, 2)
107 |
108 | self.dropout = dropout
109 | self.alpha = 0.25
110 | self.nheads = 2
111 | self.attentions = GAT(dim, dim, dropout, alpha=self.alpha, nheads=self.nheads).to(device)
112 |
113 | def pad(self, matrices, pad_value):
114 | """Pad the list of matrices
115 | with a pad_value (e.g., 0) for batch processing.
116 | For example, given a list of matrices [A, B, C],
117 | we obtain a new matrix [A00, 0B0, 00C],
118 | where 0 is the zero (i.e., pad value) matrix.
119 | """
120 | shapes = [m.shape for m in matrices]
121 | M, N = sum([s[0] for s in shapes]), sum([s[1] for s in shapes])
122 | zeros = torch.FloatTensor(np.zeros((M, N))).to(device)
123 | pad_matrices = pad_value + zeros
124 | i, j = 0, 0
125 | for k, matrix in enumerate(matrices):
126 | m, n = shapes[k]
127 | pad_matrices[i:i + m, j:j + n] = matrix
128 | i += m
129 | j += n
130 | return pad_matrices
131 |
132 | def update(self, matrix, vectors, layer):
133 | hidden_vectors = torch.relu(self.W_fingerprint[layer](vectors))
134 |
135 | return hidden_vectors + torch.matmul(matrix, hidden_vectors)
136 |
137 | def sum(self, vectors, axis):
138 | sum_vectors = [torch.sum(v, 0) for v in torch.split(vectors, axis)]
139 | return torch.stack(sum_vectors)
140 |
141 | def gnn(self, inputs):
142 | """Cat or pad each input data for batch processing."""
143 | Smiles, fingerprints, adjacencies, molecular_sizes = inputs
144 | fingerprints = torch.cat(fingerprints)
145 | adj = self.pad(adjacencies, 0)
146 | """GNN layer (update the fingerprint vectors)."""
147 | fingerprint_vectors = self.embed_fingerprint(fingerprints)
148 |
149 | for l in range(self.layer_hidden):
150 | hs = self.update(adj, fingerprint_vectors, l)
151 | fingerprint_vectors = F.normalize(hs, 2, 1)
152 | """Attention layer"""
153 | molecular_vectors = self.attentions(fingerprint_vectors, adj)
154 | """Molecular vector by sum or mean of the fingerprint vectors."""
155 | molecular_vectors = self.sum(molecular_vectors, molecular_sizes)
156 | return Smiles, molecular_vectors
157 |
158 | def mlp(self, vectors):
159 | """Regressor based on multilayer perceptron."""
160 | for l in range(self.layer_output):
161 |
162 | vectors = torch.relu(self.W_output[l](vectors))
163 | outputs = torch.sigmoid(self.W_property(vectors))
164 | return outputs
165 |
166 | def forward_classifier(self, data_batch, train):
167 |
168 | inputs = data_batch[:-1]
169 | correct_labels = torch.cat(data_batch[-1])
170 |
171 | if train:
172 | Smiles, molecular_vectors = self.gnn(inputs)
173 | predicted_scores = self.mlp(molecular_vectors)
174 | '''loss function'''
175 | loss = F.cross_entropy(predicted_scores, correct_labels)
176 | predicted_scores = predicted_scores.to('cpu').data.numpy()
177 | predicted_scores = [s[1] for s in predicted_scores]
178 | correct_labels = correct_labels.to('cpu').data.numpy()
179 | return Smiles,loss, predicted_scores, correct_labels
180 | else:
181 | with torch.no_grad():
182 | Smiles, molecular_vectors = self.gnn(inputs)
183 | predicted_scores = self.mlp(molecular_vectors)
184 | loss = F.cross_entropy(predicted_scores, correct_labels)
185 | predicted_scores = predicted_scores.to('cpu').data.numpy()
186 | predicted_scores = [s[1] for s in predicted_scores]
187 | correct_labels = correct_labels.to('cpu').data.numpy()
188 |
189 | return Smiles, loss, predicted_scores, correct_labels
190 |
191 |
192 | class Trainer(object):
193 | def __init__(self, model,lr,batch_train):
194 | self.model = model
195 | self.batch_train=batch_train
196 | self.lr=lr
197 | self.optimizer = optim.Adam(self.model.parameters(), lr=self.lr)
198 |
199 | def train(self, dataset):
200 | np.random.shuffle(dataset)
201 | N = len(dataset)
202 | loss_total = 0
203 | SMILES,P, C = '',[], []
204 | for i in range(0, N, self.batch_train):
205 | data_batch = list(zip(*dataset[i:i + self.batch_train]))
206 | Smiles,loss, predicted_scores, correct_labels = self.model.forward_classifier(data_batch, train=True)
207 | SMILES += ' '.join(Smiles) + ' '
208 | P.append(predicted_scores)
209 | C.append(correct_labels)
210 | self.optimizer.zero_grad()
211 | loss.backward()
212 | self.optimizer.step()
213 | loss_total += loss.item()
214 | tru = np.concatenate(C)
215 | pre = np.concatenate(P)
216 | AUC = roc_auc_score(tru, pre)
217 | SMILES = SMILES.strip().split()
218 | pred = [1 if i > 0.15 else 0 for i in pre]
219 | predictions = np.stack((tru, pred, pre))
220 | return AUC, loss_total, predictions
221 |
222 |
223 | class Tester(object):
224 | def __init__(self, model,batch_test):
225 | self.model = model
226 | self.batch_test=batch_test
227 | def test_classifier(self, dataset):
228 | N = len(dataset)
229 | loss_total = 0
230 | SMILES, P, C = '', [], []
231 | for i in range(0, N, self.batch_test):
232 | data_batch = list(zip(*dataset[i:i + self.batch_test]))
233 | (Smiles, loss, predicted_scores, correct_labels) = self.model.forward_classifier(
234 | data_batch, train=False)
235 | SMILES += ' '.join(Smiles) + ' '
236 | loss_total += loss.item()
237 | P.append(predicted_scores)
238 | C.append(correct_labels)
239 | SMILES = SMILES.strip().split()
240 | tru = np.concatenate(C)
241 | pre = np.concatenate(P)
242 | pred = [1 if i >0.15 else 0 for i in pre]
243 | #AUC = roc_auc_score(tru, pre)
244 | cnf_matrix=confusion_matrix(tru,pred)
245 | tn = cnf_matrix[0, 0]
246 | tp = cnf_matrix[1, 1]
247 | fn = cnf_matrix[1, 0]
248 | fp = cnf_matrix[0, 1]
249 | acc = (tp + tn) / (tp + fp + fn + tn)
250 | # Tru=map(str,np.concatenate(C))
251 | # Pre=map(str,np.concatenate(P))
252 | # predictions = '\n'.join(['\t'.join(x) for x in zip(SMILES, Tru, Pre)])
253 | predictions = np.stack((tru, pred, pre))
254 | return acc, loss_total, predictions
255 |
256 | def save_result(self, result, filename):
257 | with open(filename, 'a') as f:
258 | f.write(result + '\n')
259 |
260 | def save_predictions(self, predictions, filename):
261 | with open(filename, 'w') as f:
262 | f.write('Smiles\tCorrect\tPredict\n')
263 | f.write(predictions + '\n')
264 |
265 | def save_model(self, model, filename):
266 | torch.save(model.state_dict(), filename)
267 |
268 | def dump_dictionary(dictionary, filename):
269 | with open('../DGCAN/model'+filename, 'wb') as f:
270 | pickle.dump(dict(dictionary), f)
271 |
272 |
273 |
--------------------------------------------------------------------------------
/DGCAN/results/AUC.txt:
--------------------------------------------------------------------------------
1 | Epoch Time(sec) Loss_train Loss_test AUC_train AUC_test
2 | 1 7.2395853999999815 318.02376973629 33.23613902926445 0.6330387783115992 0.5
3 | 2 13.431995 275.59704649448395 28.36697283387184 0.7726390395642837 0.7757009345794392
4 | 3 19.53742679999999 258.0953543186188 26.692712754011154 0.8227923594659818 0.7827102803738317
5 | 4 25.411535499999985 244.29262351989746 25.99587020277977 0.8555440089003034 0.8247663551401869
6 | 5 31.270511900000002 235.61571648716927 26.206634640693665 0.8711811167445258 0.8271028037383178
7 | 6 37.1334444 235.10905063152313 24.74921104311943 0.8782201330617486 0.8504672897196262
8 | 7 43.15194819999999 230.60763642191887 24.482909947633743 0.8858224754798858 0.8644859813084113
9 | 8 49.21481349999999 225.9473716020584 25.168260991573334 0.894445889698231 0.8738317757009346
10 | 9 55.39354119999999 220.88472372293472 23.143073588609695 0.9038094737308211 0.8808411214953271
11 | 10 61.388609 220.29008296132088 23.10640263557434 0.9080117951159034 0.8925233644859814
12 | 11 67.3816218 220.04156962037086 23.67304638028145 0.905873895764607 0.8714953271028038
13 | 12 73.3649585 214.85031658411026 23.34747040271759 0.9159177330794608 0.8551401869158879
14 | 13 79.32801410000002 212.33444252610207 23.14932319521904 0.9178444716275156 0.8714953271028038
15 | 14 85.31854019999997 211.54040449857712 22.778073489665985 0.9219235005645715 0.8901869158878505
16 | 15 91.33987619999999 208.26400744915009 22.901916056871414 0.9267551530985012 0.8995327102803738
17 | 16 98.36570660000001 209.3945328295231 23.913705557584763 0.9246417461863752 0.8878504672897196
18 | 17 104.39726789999997 206.03158766031265 23.282782286405563 0.930114906901056 0.8901869158878505
19 | 18 110.4006412 207.53857171535492 22.225304275751114 0.9226543992295261 0.9018691588785047
20 | 19 116.81378369999999 204.79183167219162 23.475462794303894 0.926265719441185 0.8785046728971962
21 | 20 122.6911399 205.36031165719032 22.78501933813095 0.9291473863661524 0.8878504672897196
22 | 21 128.57552929999997 202.3321330845356 23.385528802871704 0.9302150906635376 0.8855140186915887
23 | 22 134.4078144 202.55410113930702 23.08760157227516 0.9293607611751943 0.8925233644859814
24 | 23 140.28498439999998 198.95897144079208 22.36356022953987 0.9377836694932141 0.8948598130841121
25 | 24 146.0898661 197.13710144162178 23.3654263317585 0.9351247869019417 0.8785046728971962
26 | 25 151.9243611 256.36723348498344 22.31619429588318 0.7880461675559591 0.8901869158878505
27 | 26 157.7956577 199.1333883702755 22.19395723938942 0.9381437221865521 0.9018691588785047
28 | 27 163.5824619 195.61116680502892 21.885735362768173 0.9357744592290832 0.8995327102803738
29 | 28 169.4125838 196.21020331978798 21.808892458677292 0.937718217946731 0.9065420560747663
30 | 29 175.33045549999997 196.93134278059006 22.267054110765457 0.9385652135408595 0.897196261682243
31 | 30 181.20051949999998 195.89555063843727 22.040870487689972 0.9386104622844113 0.8995327102803738
32 | 31 187.0336099 194.0237057507038 22.781775504350662 0.9417760754533178 0.8714953271028038
33 | 32 192.90557769999998 193.68072113394737 22.449314266443253 0.9423293001527663 0.8948598130841121
34 | 33 198.73298769999997 192.5338954925537 22.377066612243652 0.9452480516748955 0.8785046728971962
35 | 34 204.81813569999997 192.58278796076775 23.285291463136673 0.9402174153696283 0.8808411214953271
36 | 35 210.6708656 196.01435166597366 24.061037868261337 0.9359749651294087 0.8691588785046729
37 | 36 216.5086068 193.9636361002922 22.313345968723297 0.936061864857086 0.8901869158878505
38 | 37 222.3077349 192.51033294200897 22.285043627023697 0.9435566896185268 0.9042056074766355
39 | 38 228.07614589999997 188.01407945156097 22.830698162317276 0.94882975955897 0.8995327102803738
40 | 39 233.88910299999998 193.91294729709625 22.711496233940125 0.9402640478668054 0.8808411214953271
41 | 40 239.76910729999997 192.2110168337822 21.79123494029045 0.9453036785706378 0.9042056074766355
42 | 41 245.54325319999998 189.3926584124565 23.183754086494446 0.9464263178869528 0.8925233644859814
43 | 42 251.29193049999998 197.67854461073875 24.210958123207092 0.9356472922709057 0.8644859813084113
44 | 43 257.07465279999997 195.8016073703766 22.462971657514572 0.9351026468439347 0.897196261682243
45 | 44 262.8480727 191.97943636775017 23.224840223789215 0.940738813735692 0.8878504672897196
46 | 45 268.5777391 190.848837941885 22.82283341884613 0.9461044567936768 0.897196261682243
47 | 46 274.3567221 190.04618108272552 22.433310955762863 0.9440021199105542 0.897196261682243
48 | 47 280.1616897 190.5216095149517 22.426137387752533 0.9457633615250072 0.9042056074766355
49 | 48 285.9260854 185.92078268527985 22.22221177816391 0.9480603925432284 0.8995327102803738
50 | 49 291.66768609999997 187.782156676054 22.887968957424164 0.94423528239644 0.8901869158878505
51 | 50 297.4925078 187.28414443135262 21.483285009860992 0.9458896982310093 0.9088785046728972
52 | 51 303.2552989 185.18417713046074 21.38184556365013 0.9481489527752565 0.9205607476635514
53 | 52 309.0365314 182.16105404496193 24.673764526844025 0.9509743009276684 0.8714953271028038
54 | 53 314.8158657 188.7527618408203 23.513393253087997 0.9457229559191445 0.8878504672897196
55 | 54 320.5944295 185.709531635046 21.631520986557007 0.9463949066796555 0.9135514018691588
56 | 55 326.40547119999997 185.12931755185127 22.429152816534042 0.944120984346979 0.9018691588785047
57 | 56 332.1655878 182.88407680392265 21.58257967233658 0.9498509697345405 0.9135514018691588
58 | 57 337.97326309999994 182.04424741864204 21.475889027118683 0.9525705991099698 0.9182242990654206
59 | 58 343.78313679999997 182.934487760067 21.883195608854294 0.949658766355968 0.9088785046728972
60 | 59 349.55780849999996 184.17358297109604 21.290808767080307 0.9471534804171187 0.9182242990654206
61 | 60 355.38770680000005 181.42354640364647 21.597694754600525 0.949361674452587 0.9135514018691588
62 | 61 361.28093179999996 187.25566163659096 21.6785786151886 0.9452393340270551 0.9065420560747663
63 | 62 367.0356904 181.59250125288963 21.666670441627502 0.9521657127991676 0.9112149532710281
64 | 63 372.82640919999994 179.8839019536972 22.01644539833069 0.9538880709367459 0.9065420560747663
65 | 64 378.58320630000003 182.93770709633827 22.33838379383087 0.9484886642903003 0.8995327102803738
66 | 65 384.42532470000003 181.58496183156967 22.23741576075554 0.950153319901698 0.8995327102803738
67 | 66 390.227352 182.49673774838448 21.934344708919525 0.9507491642128103 0.9042056074766355
68 | 67 396.04737980000004 180.1727076768875 22.335491836071014 0.9534794484911551 0.9042056074766355
69 | 68 401.8080486 182.3468733727932 21.559545934200287 0.953210031660283 0.9158878504672897
70 | 69 407.56943720000004 177.6970148384571 21.813909739255905 0.9542144984169858 0.9158878504672897
71 | 70 413.3951455 179.4230616092682 21.47458705306053 0.9503143888236987 0.9182242990654206
72 | 71 419.26894319999997 187.4236896932125 21.997405976057053 0.9434714503952 0.9112149532710281
73 | 72 425.0120637 185.64457353949547 21.80859535932541 0.9493346912568912 0.9112149532710281
74 | 73 430.8028591 183.28448390960693 22.7678345143795 0.942662231275046 0.8995327102803738
75 | 74 436.5995785 181.35295176506042 21.99883532524109 0.9524445391546925 0.9088785046728972
76 | 75 442.4056693 180.42559936642647 22.20555028319359 0.9545189242145815 0.9065420560747663
77 | 76 448.17074230000003 177.4391260445118 21.684407979249954 0.9532330019704651 0.9088785046728972
78 | 77 453.95868340000004 187.34195244312286 22.080274641513824 0.9482340536232203 0.9042056074766355
79 | 78 459.8026899 184.663908213377 22.014076620340347 0.9470819403546837 0.9088785046728972
80 | 79 465.5365296 178.98830798268318 21.47413921356201 0.9541025527486882 0.9158878504672897
81 | 80 471.3356651 178.3373854458332 21.658688694238663 0.9525628500896672 0.9088785046728972
82 | 81 477.18991429999994 176.8597036600113 22.024581998586655 0.9535130737042532 0.9065420560747663
83 | 82 483.0233918 177.2030012011528 21.766023725271225 0.957143212965218 0.9135514018691588
84 | 83 488.7872165 176.38141465187073 21.79708757996559 0.9572966712422787 0.9112149532710281
85 | 84 494.52968880000003 174.46399101614952 21.843281388282776 0.956292066110213 0.9112149532710281
86 | 85 500.3899477 175.65917918086052 21.409487038850784 0.9559244027719352 0.9182242990654206
87 | 86 506.2031879 176.77976202964783 21.49503728747368 0.9538743717758542 0.9228971962616822
88 | 87 512.0002034 179.85141596198082 21.427332252264023 0.9514658102154229 0.9158878504672897
89 | 88 517.7809181 178.52282038331032 21.403560250997543 0.9539066132353267 0.9158878504672897
90 | 89 523.6232049 177.05544209480286 21.411171078681946 0.9535158412115041 0.9135514018691588
91 | 90 529.4495936 176.31908676028252 21.366370409727097 0.955243734363584 0.9228971962616822
92 | 91 535.3322216 176.20382365584373 21.893729746341705 0.9558952055704386 0.9088785046728972
93 | 92 541.1628488 175.1233125925064 22.482848435640335 0.9574447328802002 0.9042056074766355
94 | 93 547.003017 176.93210792541504 21.549375027418137 0.9527253027652932 0.9135514018691588
95 | 94 552.8402735 173.00296890735626 21.5932075381279 0.9579943598202227 0.9205607476635514
96 | 95 558.6800595 179.9282302260399 23.50808882713318 0.9537645017379945 0.8878504672897196
97 | 96 564.5009782 174.56020081043243 22.648311734199524 0.9556187315960767 0.8995327102803738
98 | 97 570.3044254 176.7171704173088 21.891200184822083 0.9524900646489693 0.9088785046728972
99 | 98 576.1318849 178.38612964749336 22.245244562625885 0.9540746009254544 0.9088785046728972
100 | 99 581.9812099 177.73075929284096 24.06598174571991 0.9527554685943277 0.8551401869158879
101 | 100 587.8207973 177.71725061535835 23.09346652030945 0.9563492151349435 0.8925233644859814
102 | 101 593.7006995 173.766254901886 22.383565932512283 0.9588301470099851 0.8995327102803738
103 | 102 599.5459518 173.67894527316093 21.84225881099701 0.9581018774769188 0.9112149532710281
104 | 103 605.2735841 175.92625331878662 22.260210156440735 0.956732791639914 0.9065420560747663
105 | 104 611.1396088 174.88757956027985 21.68474268913269 0.95813674806828 0.9158878504672897
106 | 105 616.9215246 176.1177335381508 22.008816480636597 0.9573001306263422 0.9065420560747663
107 | 106 622.727265 174.49301874637604 21.83292892575264 0.9588928310492174 0.9088785046728972
108 | 107 628.5301017 172.7093889117241 21.64242872595787 0.9596258053446101 0.9135514018691588
109 | 108 634.310622 177.16424638032913 21.55477637052536 0.9525133117098767 0.9158878504672897
110 | 109 640.0827119 178.4102607667446 21.33096119761467 0.9554701164567051 0.9158878504672897
111 | 110 645.8604367 174.0306807756424 21.91256058216095 0.9572584796422166 0.9135514018691588
112 | 111 651.6639703 174.55561447143555 21.62583690881729 0.9593071268846725 0.9158878504672897
113 | 112 657.5478062 172.49658674001694 22.11896824836731 0.9582462029800518 0.9112149532710281
114 | 113 663.3088726999999 173.48215851187706 21.262643307447433 0.9584646976775077 0.9205607476635514
115 | 114 669.1280201 174.29942700266838 21.171885669231415 0.9568379569154472 0.9205607476635514
116 | 115 674.9632922 171.870591878891 21.214154481887817 0.9590825436712644 0.9205607476635514
117 | 116 680.7173928 176.74994710087776 22.71759131550789 0.9544714614652291 0.8901869158878505
118 | 117 686.4742894999999 184.73798117041588 21.845041394233704 0.9432161478513075 0.9112149532710281
119 | 118 692.2357881 177.2356958091259 21.59891825914383 0.9571596796333606 0.9158878504672897
120 | 119 698.004266 176.63044354319572 21.46969723701477 0.9547083600859034 0.9158878504672897
121 | 120 703.704268 172.92364439368248 21.97585704922676 0.9586079161777404 0.9065420560747663
122 | 121 709.4959497 180.26964315772057 21.951832473278046 0.948673395399296 0.9088785046728972
123 | 122 715.3568411 173.81644931435585 21.459405571222305 0.9592527453671928 0.9205607476635514
124 | 123 721.1472172 176.33700492978096 21.565876573324203 0.9538397779352181 0.9205607476635514
125 | 124 726.9662815 172.69166892766953 21.18627032637596 0.9598193924768083 0.9205607476635514
126 | 125 732.7549476 171.38375091552734 21.528594940900803 0.9606258440897115 0.9158878504672897
127 | 126 738.5203703 171.18208953738213 22.116858184337616 0.9604675426749618 0.9088785046728972
128 | 127 744.3382166 171.14211875200272 22.178175538778305 0.9611412923151859 0.9112149532710281
129 | 128 750.1037539 169.92953234910965 21.887178242206573 0.9611555449775278 0.9112149532710281
130 | 129 755.8869365 174.96560329198837 22.6447791159153 0.9555907797728429 0.8995327102803738
131 | 130 761.6342917 178.70918104052544 22.300085812807083 0.9529475335975381 0.9042056074766355
132 | 131 767.4047745 174.0145247578621 22.113102048635483 0.9579599043549494 0.9088785046728972
133 | 132 773.1641292 172.59970355033875 21.501632899045944 0.9596043571634159 0.9182242990654206
134 | 133 779.0603237 172.19679167866707 22.107417851686478 0.9594648747979719 0.9088785046728972
135 | 134 784.8799782 173.3250037431717 23.72211918234825 0.9588380344056502 0.8761682242990654
136 | 135 790.6637747999999 173.15374860167503 22.544156223535538 0.9600561527221201 0.897196261682243
137 | 136 796.4695435 171.82129180431366 22.224039256572723 0.9606888048796689 0.9065420560747663
138 | 137 802.2469289 171.67953670024872 22.0196373462677 0.9615298503332077 0.9042056074766355
139 | 138 808.016359 172.37993958592415 22.44371086359024 0.9589339285318929 0.8995327102803738
140 | 139 813.7643964 174.99391075968742 21.463502824306488 0.9590761784045874 0.9158878504672897
141 | 140 819.5146855 174.17848363518715 21.192844033241272 0.959691256891093 0.9228971962616822
142 |
--------------------------------------------------------------------------------
/DGCAN/predict.py:
--------------------------------------------------------------------------------
1 |
2 |
3 | # -*- coding: utf-8 -*-
4 | """
5 | Created on Wed Apr 27 20:09:31 2022
6 |
7 | @author:Jinyu-Sun
8 | """
9 |
10 | import timeit
11 | import sys
12 | import numpy as np
13 | import math
14 | import torch
15 | import torch.nn as nn
16 | import torch.nn.functional as F
17 | import torch.optim as optim
18 | import pickle
19 | from sklearn.metrics import roc_auc_score, roc_curve
20 | from sklearn.metrics import confusion_matrix
21 | import preprocess as pp
22 | import pandas as pd
23 | import matplotlib.pyplot as plt
24 | torch.cuda.empty_cache()
25 | if torch.cuda.is_available():
26 | device = torch.device('cuda')
27 |
28 | else:
29 | device = torch.device('cpu')
30 |
31 | class GraphAttentionLayer(nn.Module):
32 | def __init__(self, in_features, out_features, dropout, alpha, concat=True):
33 | super(GraphAttentionLayer, self).__init__()
34 | self.dropout = dropout
35 | self.concat = concat
36 | self.in_features = in_features #dim of input feature
37 | self.out_features = out_features #dim of output feature
38 | self.alpha = alpha # negative_slope leakyrelu
39 | self.W = nn.Parameter(torch.zeros(size=(in_features, out_features)))
40 | self.a = nn.Parameter(torch.zeros(size=(2 * out_features, 1)))
41 | self.leakyrelu = nn.LeakyReLU(self.alpha)
42 |
43 | def forward(self, input, adj):
44 | """
45 | input: input_feature [N, in_features] in_features indicates the number of elements of the input feature vector of the node
46 | adj: adjacency matrix of the graph dimension [N, N] non-zero is one, data structure basics
47 | """
48 | h = torch.mm(input, self.W) # [N, out_features]
49 | N = h.size()[0] #Number of nodes of the graph
50 | a_input = torch.cat([h.repeat(1, N).view(N * N, -1), h.repeat(N, 1)], dim=1).view(N, -1, 2 * self.out_features) # [N, N, 2*out_features]
51 | e = self.leakyrelu(torch.matmul(a_input, self.a).squeeze(2))
52 | zero_vec =-9e10 *torch.ones_like(e)
53 | attention = torch.where(adj > 0, e, zero_vec)
54 | # indicates that if the adjacency matrix element is greater than 0, then the two nodes are connected and the attention factor at that position is retained.
55 | # Otherwise it is necessary to mask and set to a very small value, the reason is that this minimum value will be disregarded during softmax.
56 | attention = F.softmax(attention, dim=1)
57 | attention = F.dropout(attention, self.dropout, training=self.training)
58 | h_prime = torch.matmul(attention, h)
59 | if self.concat:
60 | return F.elu(h_prime)
61 | else:
62 | return h_prime
63 |
64 |
65 | class GAT(nn.Module):
66 | def __init__(self, nfeat, nhid, dropout, alpha, nheads):
67 | super(GAT, self).__init__()
68 | """
69 | n_heads indicates that there are several GAL layers, which are finally stitched together, similar to self-attention
70 | to extract features from different subspaces.
71 | """
72 | self.dropout = dropout
73 | self.attentions = [GraphAttentionLayer(nfeat, nhid, dropout=dropout, alpha=alpha, concat=True) for _ in
74 | range(nheads)]
75 | for i, attention in enumerate(self.attentions):
76 | self.add_module('attention_{}'.format(i), attention)
77 |
78 | self.out_att = GraphAttentionLayer(nhid,56, dropout=dropout, alpha=alpha, concat=False)
79 | self.nheads=nheads
80 |
81 | def forward(self, x, adj):
82 | x = F.dropout(x, self.dropout, training=self.training)
83 | #x = torch.cat([att(x, adj) for att in self.attentions], dim=1)
84 |
85 | z=torch.zeros_like(self.attentions[1](x, adj))
86 | for att in self.attentions:
87 | z=torch.add(z, att(x, adj))
88 | x = z/self.nheads
89 | x = F.dropout(x, self.dropout, training=self.training)
90 | x = F.elu(self.out_att(x, adj))
91 | return F.softmax(x, dim=1)
92 |
93 | class MolecularGraphNeuralNetwork(nn.Module):
94 | def __init__(self, N_fingerprints, dim, layer_hidden, layer_output, dropout):
95 | super(MolecularGraphNeuralNetwork, self).__init__()
96 | self.layer_hidden=layer_hidden
97 | self.layer_output=layer_output
98 | self.embed_fingerprint = nn.Embedding(N_fingerprints, dim)
99 | self.W_fingerprint = nn.ModuleList([nn.Linear(dim, dim) for _ in range(layer_hidden)])
100 |
101 | self.W_output = nn.ModuleList([nn.Linear(56,56) for _ in range(layer_output)])
102 | self.W_property = nn.Linear(56, 2)
103 |
104 | self.dropout = dropout
105 | self.alpha = 0.25
106 | self.nheads = 2
107 | self.attentions = GAT(dim, dim, dropout, alpha=self.alpha, nheads=self.nheads).to(device)
108 |
109 | def pad(self, matrices, pad_value):
110 | """Pad the list of matrices
111 | with a pad_value (e.g., 0) for batch processing.
112 | For example, given a list of matrices [A, B, C],
113 | we obtain a new matrix [A00, 0B0, 00C],
114 | where 0 is the zero (i.e., pad value) matrix.
115 | """
116 | shapes = [m.shape for m in matrices]
117 | M, N = sum([s[0] for s in shapes]), sum([s[1] for s in shapes])
118 | zeros = torch.FloatTensor(np.zeros((M, N))).to(device)
119 | pad_matrices = pad_value + zeros
120 | i, j = 0, 0
121 | for k, matrix in enumerate(matrices):
122 | m, n = shapes[k]
123 | pad_matrices[i:i + m, j:j + n] = matrix
124 | i += m
125 | j += n
126 | return pad_matrices
127 |
128 | def update(self, matrix, vectors, layer):
129 | hidden_vectors = torch.relu(self.W_fingerprint[layer](vectors))
130 |
131 | return hidden_vectors + torch.matmul(matrix, hidden_vectors)
132 |
133 | def sum(self, vectors, axis):
134 | sum_vectors = [torch.sum(v, 0) for v in torch.split(vectors, axis)]
135 | return torch.stack(sum_vectors)
136 |
137 | def gnn(self, inputs):
138 | """Cat or pad each input data for batch processing."""
139 | Smiles, fingerprints, adjacencies, molecular_sizes = inputs
140 | fingerprints = torch.cat(fingerprints)
141 | adj = self.pad(adjacencies, 0)
142 | """GNN layer (update the fingerprint vectors)."""
143 | fingerprint_vectors = self.embed_fingerprint(fingerprints)
144 |
145 | for l in range(self.layer_hidden):
146 | hs = self.update(adj, fingerprint_vectors, l)
147 | fingerprint_vectors = F.normalize(hs, 2, 1)
148 | """Attention layer"""
149 | molecular_vectors = self.attentions(fingerprint_vectors, adj)
150 | """Molecular vector by sum or mean of the fingerprint vectors."""
151 | molecular_vectors = self.sum(molecular_vectors, molecular_sizes)
152 | return Smiles, molecular_vectors
153 |
154 | def mlp(self, vectors):
155 | """Regressor based on multilayer perceptron."""
156 | for l in range(self.layer_output):
157 |
158 | vectors = torch.relu(self.W_output[l](vectors))
159 | outputs = torch.sigmoid(self.W_property(vectors))
160 | return outputs
161 |
162 | def forward_classifier(self, data_batch):
163 |
164 | inputs = data_batch[:-1]
165 | correct_labels = torch.cat(data_batch[-1])
166 |
167 |
168 | with torch.no_grad():
169 | Smiles, molecular_vectors = self.gnn(inputs)
170 | predicted_scores = self.mlp(molecular_vectors)
171 |
172 | predicted_scores = predicted_scores.to('cpu').data.numpy()
173 | predicted_scores = [s[1] for s in predicted_scores]
174 | correct_labels = correct_labels.to('cpu').data.numpy()
175 |
176 | return Smiles,predicted_scores, correct_labels
177 |
178 |
179 | class Tester(object):
180 | def __init__(self, model,batch_test):
181 | self.model = model
182 | self.batch_test=batch_test
183 | def test_classifier(self, dataset):
184 | N = len(dataset)
185 | SMILES, P, C = '', [], []
186 | for i in range(0, N, self.batch_test):
187 | data_batch = list(zip(*dataset[i:i + self.batch_test]))
188 | Smiles, predicted_scores, correct_labels = self.model.forward_classifier( data_batch)
189 | SMILES += ' '.join(Smiles) + ' '
190 |
191 | P.append(predicted_scores)
192 | C.append(correct_labels)
193 | SMILES = SMILES.strip().split()
194 | tru = np.concatenate(C)
195 | pre = np.concatenate(P)
196 | pred = [1 if i >0.15 else 0 for i in pre]
197 | #AUC = roc_auc_score(tru, pre)
198 | cnf_matrix=confusion_matrix(tru,pred)
199 | tn = cnf_matrix[0, 0]
200 | tp = cnf_matrix[1, 1]
201 | fn = cnf_matrix[1, 0]
202 | fp = cnf_matrix[0, 1]
203 | acc = (tp + tn) / (tp + fp + fn + tn)
204 | # Tru=map(str,np.concatenate(C))
205 | # Pre=map(str,np.concatenate(P))
206 | # predictions = '\n'.join(['\t'.join(x) for x in zip(SMILES, Tru, Pre)])
207 | predictions = np.stack((tru, pred, pre))
208 | return acc, predictions
209 |
210 | def save_result(self, result, filename):
211 | with open(filename, 'a') as f:
212 | f.write(result + '\n')
213 |
214 | def save_predictions(self, predictions, filename):
215 | with open(filename, 'w') as f:
216 | f.write('Smiles\tCorrect\tPredict\n')
217 | f.write(predictions + '\n')
218 |
219 | def dump_dictionary(dictionary, filename):
220 | with open('../DGCAN/model'+filename, 'wb') as f:
221 | pickle.dump(dict(dictionary), f)
222 | def metrics(cnd_matrix):
223 | '''Evaluation Metrics'''
224 |
225 | tn = cnd_matrix[0, 0]
226 | tp = cnd_matrix[1, 1]
227 | fn = cnd_matrix[1, 0]
228 | fp = cnd_matrix[0, 1]
229 |
230 | bacc = ((tp / (tp + fn)) + (tn / (tn + fp))) / 2 # balance accurance
231 | pre = tp / (tp + fp) # precision/q+
232 | rec = tp / (tp + fn) # recall/se
233 | sp = tn / (tn + fp)
234 | q_ = tn / (tn + fn)
235 | f1 = 2 * pre * rec / (pre + rec) # f1score
236 | mcc = ((tp * tn) - (fp * fn)) / math.sqrt(
237 | (tp + fp) * (tp + fn) * (tn + fp) * (tn + fn)) # Matthews correlation coefficient
238 | acc = (tp + tn) / (tp + fp + fn + tn) # accurancy
239 |
240 | print('bacc:', bacc)
241 | print('pre:', pre)
242 | print('rec:', rec)
243 | print('f1:', f1)
244 | print('mcc:', mcc)
245 | print('sp:', sp)
246 | print('q_:', q_)
247 | print('acc:', acc)
248 |
249 |
250 | def predict (test_name, property, radius, dim, layer_hidden, layer_output, dropout, batch_train,
251 | batch_test, lr, lr_decay, decay_interval, iteration, N):
252 | '''
253 |
254 | Parameters
255 | ----------
256 | data_test = '../dataset/data_test.txt', #test set
257 | radius = 1, #hops of radius subgraph: 1, 2
258 | dim = 52, #dimension of graph convolution layers
259 | layer_hidden = 4, #Number of graph convolution layers
260 | layer_output = 10, #Number of dense layers
261 | dropout = 0.45, #drop out rate :0-1
262 | batch_train = 8, # batch of training set
263 | batch_test = 8, #batch of test set
264 | lr =3e-4, #learning rate: 1e-5,1e-4,3e-4, 5e-4, 1e-3, 3e-3,5e-3
265 | lr_decay = 0.85, #Learning rate decay:0.5, 0.75, 0.85, 0.9
266 | decay_interval = 25,#Number of iterations for learning rate decay:10,25,30,50
267 | iteration = 140, #Number of iterations
268 | N = 5000, #length of embedding: 2000,3000,5000,7000
269 | dataset_train = ('../dataset/data_train.txt') #training set
270 |
271 | Returns
272 | -------
273 | res_dev
274 | Predicting results
275 |
276 | '''
277 | (radius, dim, layer_hidden, layer_output,
278 | batch_train, batch_test, decay_interval,
279 | iteration, dropout) = map(int, [radius, dim, layer_hidden, layer_output,
280 | batch_train, batch_test,
281 | decay_interval, iteration, dropout])
282 |
283 | lr, lr_decay = map(float, [lr, lr_decay])
284 | if torch.cuda.is_available():
285 | device = torch.device('cuda')
286 | print('The code uses a GPU!')
287 | else:
288 | device = torch.device('cpu')
289 | print('The code uses a CPU...')
290 |
291 | lr, lr_decay = map(float, [lr, lr_decay])
292 | path = ''
293 | dataname = ''
294 | torch.manual_seed(0)
295 | model = MolecularGraphNeuralNetwork(
296 | N, dim, layer_hidden, layer_output, dropout).to(device)
297 | model.load_state_dict(torch.load(r'model/model.pth'))
298 | model.eval()
299 | tester = Tester(model,batch_test)
300 | dataset_dev=pp.create_testdataset(test_name, path, dataname,property)
301 | np.random.seed(0)
302 | #np.random.shuffle(dataset_dev)
303 | prediction_dev, dev_res = tester.test_classifier(dataset_dev)
304 | if property == True:
305 | res_dev = dev_res.T
306 | cnd_matrix=confusion_matrix(res_dev[:,0], res_dev[:,1])
307 | cnd_matrix
308 | metrics(cnd_matrix)
309 | elif property == False:
310 | res_dev = dev_res.T[:,1]
311 |
312 | return res_dev
313 |
--------------------------------------------------------------------------------
/dataset/bRo5.txt:
--------------------------------------------------------------------------------
1 | CC1=NC=C(N=C1)C(=O)NCCC1=CC=C(C=C1)S(=O)(=O)NC(=O)NC1CCCCC1 1
2 | CN1CCN(CC1)C(=O)O[C@@H]1N(C(=O)C2=NC=CN=C12)C1=NC=C(Cl)C=C1 1
3 | [H][C@@]12CCO[C@]1([H])OC[C@@H]2OC(=O)N[C@@H](CC1=CC=CC=C1)[C@H](O)CN(CC(C)C)S(=O)(=O)C1=CC=C(N)C=C1 1
4 | COC1=C(OC)C=C2[C@@H](CN(C)CCCN3CCC4=CC(OC)=C(OC)C=C4CC3=O)CC2=C1 1
5 | [H][C@]12SC(C)(C)[C@@H](N1C(=O)[C@H]2NC(=O)[C@H](C(O)=O)C1=CSC=C1)C(O)=O 1
6 | CC[N+](C)(CC)CCOC(=O)C1C2=CC=CC=C2OC2=CC=CC=C12 1
7 | CC(C)O 1
8 | COC1=CC2=C(NC(=N2)[S@@](=O)CC2=NC=C(C)C(OC)=C2C)C=C1 1
9 | COC1=C(OC)C=C2C(N)=NC(=NC2=C1)N1CCN(CC1)C(=O)C1CCCO1 1
10 | CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=O 1
11 | CN(CCCCCCCCCCN(C)C(=O)OC1=CC=CC(=C1)[N+](C)(C)C)C(=O)OC1=CC=CC(=C1)[N+](C)(C)C 1
12 | NC[C@H](O)C1=CC(O)=C(O)C=C1 1
13 | C[N+]1(C)CCC(C1)OC(=O)C(O)(C1CCCC1)C1=CC=CC=C1 1
14 | CCC1(CCC(=O)NC1=O)C1=CC=CC=C1 1
15 | OCC(O)COC1=CC=C(Cl)C=C1 1
16 | O=[N+]([O-])c1cc([N+](=O)[O-])c(O)c([N+](=O)[O-])c1 1
17 | NC1=NC(=O)C2=C(N1)N(CCC(CO)CO)C=N2 1
18 | CC1=CC(CN2CCC(CC2)=C2C3=CC=C(Cl)C=C3CCC3=C2N=CC=C3)=CN=C1 1
19 | CSCCNC1=C2N=CN([C@@H]3O[C@H](COP(O)(=O)OP(O)(=O)C(Cl)(Cl)P(O)(O)=O)[C@@H](O)[C@H]3O)C2=NC(SCCC(F)(F)F)=N1 1
20 | CCC(C1=CC=CC=C1)C1=C(O)C2=C(OC1=O)C=CC=C2 1
21 | CS(=O)(=O)OCCCCOS(C)(=O)=O 1
22 | CN1[C@@H](CNC2=CC=C(C=C2)C(=O)N[C@@H](CCC(O)=O)C(O)=O)CNC2=C1C(=O)N=C(N)N2 1
23 | CC(C)(C)C(=O)OCOP(=O)(COCCN1C=NC2=C(N)N=CN=C12)OCOC(=O)C(C)(C)C 1
24 | CNC[C@H](O)C1=CC(O)=C(O)C=C1 1
25 | CC(C)(C)NCC(O)C1=CC(Cl)=C(N)C(Cl)=C1 1
26 | CCCCOC1=CC=C(OCCCN2CCOCC2)C=C1 1
27 | CC1(C)C[C@@H]1C(=O)N\C(=C/CCCCSC[C@H](N)C(O)=O)C(O)=O 1
28 | CC(C)[C@@H](C)\C=C\[C@@H](C)[C@@]1([H])CC[C@@]2([H])\C(CCC[C@]12C)=C\C=C1\C[C@@H](O)CCC1=C 1
29 | CN1C[C@@H](C2=CC(=CC=C2)S(=O)(=O)NCCOCCOCCNC(=O)NCCCCNC(=O)NCCOCCOCCNS(=O)(=O)C2=CC(=CC=C2)[C@@H]2CN(C)CC3=C2C=C(Cl)C=C3Cl)C2=C(C1)C(Cl)=CC(Cl)=C2 1
30 | CN1CCOC(C2=CC=CC=C2)C2=CC=CC=C2C1 1
31 | CC(C(O)=O)C1=CC(OC2=CC=CC=C2)=CC=C1 1
32 | [H][C@]12SC(C)(C)[C@@H](N1C(=O)[C@H]2NC(=O)[C@H](NC(=O)N1CCN(CC)C(=O)C1=O)C1=CC=CC=C1)C(O)=O 1
33 | CC(C)NCC(O)C1=CC(O)=C(O)C=C1 1
34 | [H][C@@]12CCCN1C(=O)[C@H](CC(C)C)N1C(=O)[C@](NC(=O)[C@H]3CN(C)[C@]4([H])CC5=CNC6=CC=CC(=C56)[C@@]4([H])C3)(O[C@@]21O)C(C)C 1
35 | [H][C@]12CC[C@]([H])(C[C@@H](C1)OC(=O)C(O)C1=CC=CC=C1)N2C 1
36 | NC(N)=N 1
37 | OC1=C(CC2=C(O)C(Cl)=CC(Cl)=C2Cl)C(Cl)=C(Cl)C=C1Cl 1
38 | C\C(N(CC1=CN=C(C)NC1=N)C=O)=C(\CCO)SSCC1CCCO1 1
39 | ONC(=O)\C=C\C1=CC=CC(=C1)S(=O)(=O)NC1=CC=CC=C1 1
40 | CN1C(C(=O)NC2=CC=CC=N2)=C(O)C2=C(C=C(Cl)S2)S1(=O)=O 1
41 | CCOC1=NC2=C(N1CC1=CC=C(C=C1)C1=CC=CC=C1C1=NOC(=O)N1)C(=CC=C2)C(=O)OCC1=C(C)OC(=O)O1 1
42 | C[N+]1=C(\C=N\O)C=CC=C1 1
43 | NC(=N)NC(N)=N 1
44 | CC(C)=C[C@@H]1[C@@H](C(=O)OC2CC(=O)C(CC=C)=C2C)C1(C)C 1
45 | CCCCN1CCCCC1C(=O)NC1=C(C)C=CC=C1C 1
46 | FC(F)(F)CNC(=O)C1(CCCCN2CCC(CC2)NC(=O)C2=C(C=CC=C2)C2=CC=C(C=C2)C(F)(F)F)C2=CC=CC=C2C2=CC=CC=C12 1
47 | NC1=CC=C(C=C1)C(O)=O 1
48 | [H][C@@]12C[C@@H](C)[C@](OC(=O)CC)(C(=O)SCF)[C@@]1(C)C[C@H](O)[C@@]1(F)[C@@]2([H])C[C@H](F)C2=CC(=O)C=C[C@]12C 1
49 | CCCCCOC1=CC=C(C=C1)C1=CC(=NO1)C1=CC=C(C=C1)C(=O)N[C@H]1C[C@@H](O)[C@@H](O)NC(=O)[C@@H]2[C@@H](O)[C@@H](C)CN2C(=O)[C@@H](NC(=O)[C@@H](NC(=O)[C@@H]2C[C@@H](O)CN2C(=O)[C@@H](NC1=O)[C@@H](C)O)[C@H](O)[C@@H](O)C1=CC(OS(O)(=O)=O)=C(O)C=C1)[C@H](O)CC(N)=O 1
50 | C=CC[C@@H](CCC)C(=O)O 1
51 | [H][C@@]12CC[C@](OC(=O)CCC)(C(=O)CO)[C@@]1(C)C[C@H](O)[C@@]1([H])[C@@]2([H])CCC2=CC(=O)CC[C@]12C 1
52 | OC(C(O)=O)C1=CC=CC=C1 1
53 | [H][C@@]12OC3=C(O)C=CC4=C3[C@@]11CCN(C)[C@]([H])(C4)[C@]1([H])C=C[C@@H]2O 1
54 | COC1=CC=CC(=C1)N1CCN(CC1)C1=NC2=C(C=CC=C2F)[C@H](CC(O)=O)N1C1=CC(=CC=C1OC)C(F)(F)F 1
55 | [H][C@]1(O)CO[C@]2([H])[C@]([H])(O)CO[C@]12[H] 1
56 | COc1cc2c(cc1OC)[C@@H]1C[C@H](O)[C@@H](CC(C)C)CN1CC2 1
57 | CC[C@@H](c1ccccc1)[C@H](c1ccc(OCCNC)cc1)c1ccc(O[C@@H]2O[C@H](C(=O)O)[C@@H](O)[C@@H](O)[C@@H]2O)cc1 1
58 | C[C@@H]1CNc2c(cccc2S(=O)(=O)N[C@@H](CCCNC(=N)N)C(=O)N2CC[C@@H](C)C[C@@H]2C(=O)O)C1 1
59 | O=C[C@H](O)[C@@H](O)[C@H](O)CO 1
60 | O=C1c2cccc(O)c2C(=O)c2c(O)cccc21 1
61 | C[C@]12CC[C@@H]3c4ccc(O)c(O)c4CC[C@H]3[C@@H]1CC[C@@H]2O 1
62 | CCNCC#CCOC(=O)[C@](O)(c1ccccc1)C1CCCCC1 1
63 | CCN(CC)CC(=O)Nc1c(C)ccc(O)c1C 1
64 | NC(=O)c1c[nH]c(=O)cn1 1
65 | C[C@H]1O[C@@H](n2cc(F)c(=O)[nH]c2=O)[C@H](O)[C@@H]1O 1
66 | C=C1CC[C@@]2(O)[C@H]3Cc4ccc(O)c5c4[C@@]2(CCN3CC2CC2)[C@H]1O5 1
67 | CCN(CC)C(=O)N1CC[N+](C)([O-])CC1 1
68 | COc1ccccc1O 1
69 | CCCCCCCCCCCC[N+](C)(C)CCOc1ccccc1 1
70 | C[N+]1([O-])CCN(C2=Nc3cc(Cl)ccc3Nc3ccccc32)CC1 1
71 | CCCCCc1cc(O)c2c(c1)OC(C)(C)[C@H]1CC[C@]3(C)O[C@@H]3[C@@H]21 1
72 | NC(=O)N1c2ccccc2[C@H](O)[C@@H](O)c2ccccc21 1
73 | CC(C)(O)CNc1nc(Nc2ccnc(C(F)(F)F)c2)nc(-c2cccc(C(F)(F)F)n2)n1 1
74 | CCCCCCCCCCC(=O)O[C@H]1CC[C@H]2[C@@H]3CCC4=CC(=O)CC[C@]4(C)[C@H]3CC[C@]12C 1
75 | O[Si](O)(O)O 1
76 | CN1C(=O)NC(=O)[C@@](C)(C2=C[C@@H](O)CCC2)C1=O 1
77 | C[C@@H]([C@H](O)c1ccccc1)N(C)C 1
78 | O=C1N[C@H](O)[C@@H](c2ccccc2)CO1 1
79 | Nc1nc2c(c(=O)[nH]1)N[C@@H](CNc1ccc(C(=O)N[C@H](CCC(=O)O)C(=O)O)cc1)CN2 1
80 | O=[N+]([O-])OCC(CO[N+](=O)[O-])(CO[N+](=O)[O-])CO[N+](=O)[O-] 1
81 | C[C@H](NC(=O)[C@@H](Cc1ccc2ccccc2c1)NC(=O)[C@@H](C)N)C(=O)N[C@@H](Cc1c[nH]c2ccccc12)C(=O)N[C@H](Cc1ccccc1)C(=O)N[C@@H](CCCCN)C(N)=O 1
82 | CC1=C(/C=C/C(C)=C/C=C/C(C)=C/C(=O)O)C(C)(C)CCC1=O 1
83 | OCCN1CCOCC1 1
84 | COC(=O)c1ccc(C)cc1O 1
85 | COc1cc(C[C@@](C)(O)C(=O)O)ccc1O 1
86 | CN1CCCN=C1/C=C/c1cccs1 1
87 | CCc1nc(C2CC2)c(C(N)=O)n1Cc1ccc2oc(-c3ccccc3NS(=O)(=O)C(F)(F)F)c(Br)c2c1 1
88 | NC[C@H](O)COc1ccccc1C(=O)CCc1ccccc1 1
89 | O=C(NC(=O)c1c(F)cccc1F)Nc1cc(Cl)c(OC(F)(F)[C@@H](F)C(F)(F)F)cc1Cl 1
90 | CC(C)C(=O)O[C@H](C)OC(=O)NCC1(CC(=O)O)CCCCC1 1
91 | CN(C(=O)CO)c1c(I)c(C(=O)NC[C@H](O)CO)c(I)c(C(=O)NC[C@H](O)CO)c1I 1
92 | C[C@]12C=CC(=O)C=C1CC[C@@H]1[C@@H]2CC[C@]2(C)C(=O)CC[C@@H]12 1
93 | CC(C)(C)[C@@H]1NC(=O)O[C@@H]2CCC[C@H]2OC/C=C/C(F)(F)c2nc3ccccc3nc2O[C@@H]2C[C@@H](C(=O)N[C@]3(C(=O)NS(=O)(=O)C4(C)CC4)C[C@H]3C(F)F)N(C2)C1=O 1
94 | CCN(CC)CCc1nc(-c2ccccc2)no1 1
95 | CN1C(=O)CC[C@H]1c1ccc[n+]([O-])c1 1
96 | CNCc1ccc(-c2[nH]c3cc(F)cc4c3c2CCNC4=O)cc1 1
97 | O=C(O[C@@H]1CN2CCC1CC2)c1ccccc1 1
98 | CC(C)[C@@H](C(=O)O)N(Cc1ccc(-c2ccccc2-c2nnn[nH]2)cc1)C(=O)CC[C@@H](C)O 1
99 | O=C1CN2Cc3c(ccc(Cl)c3Cl)NC2=N1 1
100 | CC#CC[C@@H](C)[C@H](O)/C=C/[C@@H]1[C@@H](O)C[C@@H]2C/C(=C/CCCC(=O)OCC(=O)c3ccccc3)C[C@H]21 1
101 | C[C@@H](c1cc2ccccc2s1)N(O[C@@H]1O[C@H](C(=O)O)[C@@H](O)[C@H](O)[C@H]1O)C(N)=O 1
102 | CCN(CC)CCOCCOC(=O)C(CC)(CC)c1ccccc1 1
103 | O=C(CSc1ccncc1)N[C@H]1C(=O)N2C(C(=O)O)=C(CO)CS[C@H]12 1
104 | COc1cc2c(cc1OC)[C@@H]1C[C@@H](O)[C@@H](CC(C)C)CN1CC2 1
105 | COc1ccc2[nH]c(C)c(CC(=O)O)c2c1 1
106 | CC1(C)S[C@@H]2[C@H](NC(=O)[C@H](C(=O)Oc3ccc4c(c3)CCC4)c3ccccc3)C(=O)N2[C@H]1C(=O)O 1
107 | [O-][n+]1ccccc1SSc1cccc[n+]1[O-] 1
108 | Cc1cc(OS(=O)(=O)O)c2ccccc2c1OS(=O)(=O)O 1
109 | CCCCCCCCCCCC(=O)OCN1C(=O)CCc2ccc(OCCCCN3CCN(c4cccc(Cl)c4Cl)CC3)cc21 1
110 | CCCCCCc1ccc(O)cc1O 1
111 | C[C@H](I)[C@H]1OC[C@@H](CO)O1 1
112 | CN1CCC[C@@H]1c1ccc[n+](C)c1 1
113 | C[C@H](CCC(=O)O)[C@H]1CC[C@H]2[C@@H]3C(=O)C[C@@H]4CC(=O)CC[C@]4(C)[C@H]3CC(=O)[C@@]21C 1
114 | CCC[C@@H](Cc1ccccc1)N1CCCC1 1
115 | CCCNC(=O)NS(=O)(=O)c1ccc(Cl)c(O)c1 1
116 | O=C(O)COc1ccc(C(=O)c2cccs2)c(Cl)c1Cl 1
117 | CNCCN1C(=O)[C@H](OC(C)=O)[C@@H](c2ccc(OC)cc2)Sc2ccccc21 1
118 | CCCCCCCCCC[N+](C)(C)CCCCCCCCCC 1
119 | O=C(O)CCCCCCNC1c2ccccc2CCc2ccccc21 1
120 | NC(=O)N1c2ccccc2C=Cc2cc(O)ccc21 1
121 | C/C(=C(/CCO)SSC[C@H]1CCCO1)N(C=O)Cc1cnc(C)nc1N 1
122 | O=C1NC(=O)[C@@H](N2C(=O)c3ccccc3C2=O)C[C@@H]1O 1
123 | CC(C)(C)NC[C@H](O)COc1cccc2c1SCCC2 1
124 | CN1C(=O)NC(=O)[C@](C)([C@]23CCCC[C@@H]2O3)C1=O 1
125 | N[C@H](CO)C(=O)NNCc1ccc(O)c(O)c1O 1
126 | OC[C@H]1O[C@](O)(CO[C@]2(CO)O[C@H](CO)[C@@H](O)[C@@H]2O)[C@@H](O)[C@@H]1O 1
127 | Nc1nccs1 1
128 | CCn1cc(C(=O)O)c(=O)c2cnc(N3CCCC3)nc21 1
129 | CN(Cc1nc2c(N)[nH]c(=N)[nH]c-2nc1=O)c1ccc(C(=O)N[C@@H](CCC(=O)O)C(=O)O)cc1 1
130 | C[C@]12C[C@H]3C[C@@](N)(C1)C[C@@](CO)(C3)C2 1
131 | CNCCC[C@](C#N)(c1ccc(OC)c(OC)c1)C(C)C 1
132 | CN1C(=O)[C@H](O[C@@H]2O[C@H](C(=O)O)[C@@H](O)[C@H](O)[C@H]2O)C[C@H]1c1cccnc1 1
133 | CCOc1c(N)c2c(c(OCC)c1OCC)[C@H]([C@@H]1c3c(cc4c(c3OC)OCO4)CCN1C)OC2=O 1
134 | O=C(c1ccc(O)cc1)c1ccc2n1CC[C@@H]2C(=O)O 1
135 | CN(c1nccc(=O)[nH]1)C1CCN(c2nc3ccccc3n2Cc2ccc(F)cc2)CC1 1
136 | CCCc1nnc(NC(=O)[C@@]2(C)Sc3ccccc3N(C)C2=O)s1 0
137 | COc1ccccc1OC(=O)Cn1nc(C)c([N+](=O)[O-])c1C 0
138 | Cc1ccc2c(c1)C(=O)NC[C@]1(CC[C@H](N(C)C(=O)c3ccno3)CC1)O2 0
139 | CC(=O)N[C@@H](C)C(=O)Nc1ccccc1N1CCCCC1 0
140 | CCn1/c(=N/C(=O)c2ccc([N+](=O)[O-])s2)sc2cc([N+](=O)[O-])ccc21 0
141 | O=C(Nc1cccc(N2CCCC2)c1)c1ccccc1 0
142 | O=C([C@H]1CCCO1)N1CCN(Cc2cn3cc(Cl)ccc3n2)CC1 0
143 | CN(C(=O)C[N@H+]1CC[C@H](C(=O)NC2CCCCC2)CC1)c1ccccc1 0
144 | O=C(CN1CCN(Cc2ccc(Cl)s2)CC1)Nc1cccc(S(=O)(=O)N2CCCCC2)c1 0
145 | COc1ccc2sc(N(Cc3ccccc3)C(=O)C34C[C@H]5C[C@@H](C3)C[C@@H](C4)C5)nc2c1 0
146 | COc1ccc(OC)c2sc(N(CCCN(C)C)C(=O)[C@H]3CC(=O)N(c4ccc(F)cc4)C3)nc12 0
147 | Cc1nc(-c2ccc(F)cc2)n(CC(=O)N(C)C2CCCCC2)c(=O)c1CCO 0
148 | Cn1c(C[N@@H+]2CC[C@H](CNc3ncccc3C#N)C2)nc2ccccc21 0
149 | CCCNC(=O)[C@H]1CCCN1C(=O)/C=C/c1ccc(SC(F)(F)F)cc1 0
150 | COc1cc(C(=O)OCC(=O)NC2CC2)ccc1OCCC(C)C 0
151 | NC(=O)[C@H]1CC(C(=O)Nc2ncn[n-]2)=NN1c1ccc(F)cc1 0
152 | Cn1c(=O)c2ccccc2n(CC(=O)NCc2ccccc2Cl)c1=O 0
153 | C[C@](O)(CNC(=O)c1cc2ccccc2o1)C1CC1 0
154 | COc1ccccc1C(=O)OCC(=O)N[C@@H](C)c1ccccc1 0
155 | COc1cccc(-c2nn(-c3ccc(C)cc3)cc2C(=O)N2CCC(N(C)C)CC2)c1 0
156 | CC(=O)Nc1ccc(/C(C)=N/O[C@@H](C)C(=O)Nc2ccc(F)c(F)c2)cc1 0
157 | COC(=O)C[C@@H]1CN(Cc2c(Cl)nc3sccn23)CCO1 0
158 | Cc1ccc(N2C[C@H](C(=O)N3CCC[C@H]([C@@H](C)O)C3)CCC2=O)cc1C 0
159 | CC[C@H](C)c1ccc(NC(=O)CSc2nnc(-c3cccnc3)n2N)cc1 0
160 | CC(=O)Nc1ccc(-c2nnc3n(Cc4ccc(C)cc4)c(=O)c4ccccc4n23)cc1 0
161 | CC(C)CNC(=O)C[C@H]1CSc2nc3c(cnn3-c3ccc(Cl)cc3)c(=O)n21 0
162 | O[C@]1(c2ccccc2)CC[N@H+](CCOc2cccc(Cl)c2)CC1 0
163 | CCc1nc2n(n1)CCC[C@@H]2NC(=O)N[C@@H]1CCC(C)(C)c2ccccc21 0
164 | C/C(=N\NC(=O)COc1ccccc1[N+](=O)[O-])c1ccc(-c2ccccc2)cc1 0
165 | O=C(CSc1ccc(F)cc1)N1CCN(c2nc3ccc(Br)cc3s2)CC1 0
166 | COCc1ccc(C[N@H+]2CCCC[C@@H]2CNC(=O)c2ccccc2)cc1 0
167 | Cc1cccc(Cl)c1OCc1cc(=O)n(C)c(=O)n1C 0
168 | CCOC(=O)CSc1nnc(-c2ccccc2[N+](=O)[O-])o1 0
169 | O=C(CC1=CCCCC1)NC[C@@H]1Cc2cc(F)cc(-c3cncnc3)c2O1 0
170 | CN(C)CCN(C(=O)c1ccc([N+](=O)[O-])cc1)c1nc2ccc(OC(F)(F)F)cc2s1 0
171 | CC(C)N(C(=O)/C=C/c1cncc(F)c1)C1CCC1 0
172 | Cc1cc(C)n2nc(C(=O)N/N=C/c3ccc(Cl)c(Cl)c3)nc2n1 0
173 | O=C(Nc1ccccc1F)c1nc(SCc2ccc(Cl)cc2)ncc1Br 0
174 | CCOC(=O)C1=C(c2ccccc2)N=c2s/c(=C\c3cc4c(cc3C)OCO4)c(=O)n2[C@H]1c1ccc(Cl)cc1 0
175 | COc1ccc(Br)c(C(=O)NCCCN2CCc3ccccc3C2)c1 0
176 | COc1ccc(S[C@@H](C)C(=O)Nc2ccc(CN3CCCC3)cc2)cc1 0
177 | CC(C)C[C@@H](N[C@@H](C)C(=O)Nc1ccc(S(N)(=O)=O)cc1)c1ccccc1 0
178 | C[C@H](NC(=O)NC[C@H](O)c1cccc(F)c1)c1ccc(F)cc1 0
179 | COc1ccc([C@@H](C)NCc2cccc(OC)c2OC)c(F)c1 0
180 | Cn1c(SCC(=O)N(c2ccccc2)[C@H]2CCS(=O)(=O)C2)nnc1C1CC1 0
181 | Cn1c(CSc2ncccn2)nnc1SCC(=O)O 0
182 | CC1=C(C#N)C(=O)N(C2CCCC2)C(=O)/C1=C/Nc1ccc([N+](=O)[O-])cc1C 0
183 | COc1cc([C@H]2Nc3ccc(OCCC(C)C)cc3[C@@H]3C=CC[C@@H]23)ccc1OC(C)=O 0
184 | CC(=O)NCC(=O)N1CCN(CC(=O)NC(C)C)CC1 0
185 | CCCc1ccc(OCC(=O)c2ccc3c(c2)N(CC(=O)NCc2cccnc2)C(=O)CO3)cc1 0
186 | COc1ccc(S(=O)(=O)N(C)c2ccc(C(=O)Nc3ccccc3SC)cc2)cc1OC 0
187 | Cc1cc([C@H]2CCC[N@@H+]2Cc2ccccc2Br)on1 0
188 | O=[N+]([O-])c1ccccc1-c1ccc(CN2CCC(n3cncn3)CC2)s1 0
189 | COc1cc([N+](=O)[O-])ccc1NC(=O)[C@@H]1CCCC[C@H]1C(=O)O 0
190 | Cc1ccc(OCc2cccc(C(=O)N(C3CCCC3)C3CC3)c2)cn1 0
191 | COc1ccc(CNC(=O)NCc2ccc(Cn3cnc4ccccc43)cc2)cc1 0
192 | CC1(C)CCc2cc(S(=O)(=O)NCc3csc(-c4cccs4)n3)ccc2O1 0
193 | CCn1c(SCC(=O)Nc2ccc(Br)cc2C(=O)O)nnc1C1CC1 0
194 | CC(=O)c1cccc(NC(=O)[C@H](N[C@H](C)c2ccc(F)cc2)c2ccccc2)c1 0
195 | C[C@@H](NC(=O)COc1ccc2c3c(c(=O)oc2c1)CCCC3)C(=O)NCC(=O)NCC(=O)O 0
196 | CC(C)(C)c1n[nH]c([C@H]2CN(C(=O)CCn3cncn3)CCO2)n1 0
197 | Cc1nc(C(C)(C)NC(=O)NCCOC[C@H]2CCCO2)sc1C 0
198 | O=C(O)c1ccc2c(c1)=N[C@@H](c1ccccc1Cl)N=2 0
199 | CN(CC(=O)N(C)c1cccc2ncccc12)C(=O)OC(C)(C)C 0
200 | C[C@@H]1COCC[N@H+]1Cc1ccc(-c2c(F)cccc2F)o1 0
201 | COc1ccc(NC(=O)c2nc(-c3ccco3)n(-c3cccc(C)c3)n2)cc1 0
202 | COc1ccc(N2CCN(Cc3cc(OC)c(O)c([N+](=O)[O-])c3)CC2)cc1 0
203 | COCCNC(=O)[C@@H]1CCCN([C@H]2CC[N@H+](Cc3cc(C)cc(C)c3)CC2)C1 0
204 | N#C[C@@H]1C(=O)Nc2nc(NCc3ccc4c(c3)OCCO4)[nH]c(=O)c2[C@@H]1/C=C/c1ccccc1 0
205 | Cc1ccc(C(C)C)cc1OCC(=O)Nc1ccc2c(c1)OCC(=O)N2 0
206 | CCCCOc1c(Cl)cc(C(=O)Nc2ccc(S(=O)(=O)N3C[C@H](C)O[C@@H](C)C3)cc2)cc1Cl 0
207 | Cc1nccc(CN2CCC[C@H](c3cc(=O)[nH]c(-c4ccncc4)n3)C2)n1 0
208 | COCc1ccc(CN[C@H]2CCCN(c3ccccc3F)C2=O)cc1 0
209 | Cc1ccc([C@H](NC(=O)CS(=O)(=O)Cc2cccc(Br)c2)C2CC2)cc1 0
210 | COc1cccc([C@@H]2[C@@H]3CCCC=C3[C@H](C#N)C(=N)C2(C#N)C#N)c1 0
211 | Cc1n[nH]c(C)c1[C@H]1CCCN1C(=O)COc1ccccc1 0
212 | O=C(NCc1cccs1)c1cccc(NC(=O)c2cc3sccn3c2)c1 0
213 | CC(C)[C@@H]1CCc2ccccc2N1C(=O)c1ccc2c(c1)C(=O)N(C)C2=O 0
214 | Cc1nc([N+](=O)[O-])cn1CC(=O)NCc1cccnc1 0
215 | CCN(CC(=O)Nc1nc(-c2ccc(OC)cc2)cn1-c1ccc(C(C)C)cc1)C(=O)c1ccc(C)cc1 0
216 | COc1ccc([C@@H]2c3c(n[nH]c3-c3ccc(Cl)cc3)C(=O)N2c2ccc(C)cc2)c(OC)c1 0
217 | O=C(Cn1nc2nc(Nc3ccccc3)ccn2c1=O)NCc1ccc(F)cc1F 0
218 | Cc1cccc(OCCCCNc2ccc(C)cc2C(=O)O)c1 0
219 | CC(C)n1cc(S(=O)(=O)N2CCC(n3cccc3)CC2)cn1 0
220 | O=C1CCc2ccc(C(=O)NCc3ccc(CC(=O)NC4CCCCC4)cc3)nc2N1 0
221 | CCOc1cccc(C(=O)NCC23CC4(CNC(=O)c5cccc(OCC)c5)C[C@H](C2)C[C@H](C3)C4)c1 0
222 | C[C@H](Oc1cccc(C=O)c1)C(=O)Nc1ccc(N2CCCCC2)cc1 0
223 | Cc1ccc(-c2[nH]ncc2C[N@H+](C)Cc2cc(C)on2)cc1 0
224 | Cc1cccc(Cl)c1NC(=O)[C@@H](C)c1ccc(Cl)s1 0
225 | Cc1nnc(NC(=O)CSc2nnc(C)c(O)n2)s1 0
226 | O=C(OCCN1CCCC1=O)[C@H]1C[C@H]1c1ccc(OC(F)(F)F)cc1 0
227 | Cc1ccccc1NC(=O)Cc1cc(-c2ccccc2)on1 0
228 | Cc1cccc(NCC(=O)N/N=C/c2ccc3c(c2)OCO3)c1 0
229 | Cc1ccc(C)n1N1C(=O)/C(=C/c2c(F)cccc2Cl)SC1=S 0
230 | COc1ccc(/N=C2\NC(=O)/C(=C\C=C\c3ccccc3)S2)cc1 0
231 | COc1cccc(NC2=NN=C(c3cc(C)n(-c4ncn[nH]4)c3C)CS2)c1 0
232 | Cc1nn(-c2ccccc2)c(C)c1CN(C)C(=O)Cc1ccc2c(c1)CCCC2 0
233 | Cc1cc(C)c([N+](=O)[O-])cc1NC(=O)N1CCC(c2ccn[nH]2)CC1 0
234 | COC(=O)c1cc(C2OCCO2)ccn1 0
235 | Cc1cccc(OCCCCn2c([C@@H](C)NC(=O)c3cccc(Br)c3)nc3ccccc32)c1 0
236 | CCc1cccc(CC)c1NC(=O)CN(c1ccc(F)cc1)S(C)(=O)=O 0
237 | O=C(NCc1ccc2c(c1)OCO2)c1ccc2c(=O)n(C[C@H]3CCCO3)c(=S)[nH]c2c1 0
238 | O=C(O)CNC(=O)CCCCCN1C(=O)/C(=C/C=C/c2ccccc2)SC1=S 0
239 | COc1cc(/C=C(\C#N)C(=O)Nc2cc(Cl)ccc2Cl)cc2c1OCCO2 0
240 | NC(=O)c1ccc(C[N@H+](CC(=O)N2CCC(C(=O)Nc3ccccc3)CC2)C2CC2)cc1 0
241 | COCCOc1ccc(C#N)cc1NC(=O)c1ccsc1 0
242 | Cn1c(=O)c2c(nc(CN3CCOCC3)n2Cc2ccccc2F)n(C)c1=O 0
243 | CC(=O)[C@H]1C(=O)C(=O)N(CCC(=O)O)[C@H]1c1ccc(C(C)C)cc1 0
244 | CCN1CCN(c2ccc(NC(=O)[C@@H](NC(=O)c3ccc(Cl)cc3)C(C)C)cc2C)CC1 0
245 | C#CCNC(=O)N1CCN(S(=O)(=O)c2ccc3c(c2)OCCO3)CC1 0
246 | O=C(c1ccc(Cl)cc1Cl)N1CCCN(c2ccc(C(F)(F)F)cn2)CC1 0
247 | CC(C)[C@H](CNC(=O)CCS(C)(=O)=O)N1CCc2ccccc2C1 0
248 | O=C(NCCNC(=O)c1cccnc1OCC(F)F)c1cccnc1 0
249 | CCc1cc(C(F)(F)F)n2nc([C@H]3CCCN3C(=O)[C@H]3CCS(=O)(=O)C3)cc2n1 0
250 | CCCN(CCC)CCCNC(=O)c1oc(=O)c2ccccc2c1-c1ccccc1 0
251 | CN(C(=O)c1cccc(-c2ccoc2)c1)c1ccc(F)c(F)c1 0
252 | Cc1cc(C)cc(NC(=S)N(CCCn2ccnc2)C[C@H]2CC=CCC2)c1 0
253 | CCc1ccc([N+](=O)[O-])cc1S(=O)(=O)NC(C)(C)CC 0
254 | Nc1cc(N2CCC[C@@H](c3[nH+]ccn3Cc3cscn3)C2)ncn1 0
255 | O=c1[nH]cc(-c2cc(Cl)cc(Cl)c2)c(=O)[nH]1 0
256 | O=C(O)c1nc(O)n(-c2cccc(Cl)c2)n1 0
257 | CC/N=C(/NCc1cc(C)on1)N(C)Cc1ccc(OC)cc1 0
258 | Fc1ccc(NCl)c(S)c1 0
259 | CN(CCCO)C(=O)OC(C)(C)C 0
260 | O=C1C[C@H](C(=O)Nc2ccccc2C(F)(F)F)c2c(nc(Nc3cc(Cl)cc(Cl)c3)[nH]c2=O)N1 0
--------------------------------------------------------------------------------
/Discussion/GNN.py:
--------------------------------------------------------------------------------
1 | import timeit
2 |
3 | import numpy as np
4 | import math
5 | import torch
6 | import torch.nn as nn
7 | import torch.nn.functional as F
8 | import torch.optim as optim
9 | import pickle
10 | from sklearn.metrics import roc_auc_score,roc_curve
11 | from sklearn.metrics import confusion_matrix
12 | import preprocess as pp
13 | import pandas as pd
14 | import matplotlib.pyplot as plt
15 |
16 |
17 | class MolecularGraphNeuralNetwork(nn.Module):
18 | def __init__(self, N_fingerprints, dim, layer_hidden, layer_output):
19 | super(MolecularGraphNeuralNetwork, self).__init__()
20 | self.embed_fingerprint = nn.Embedding(N_fingerprints, dim)
21 | self.W_fingerprint = nn.ModuleList([nn.Linear(dim, dim)
22 | for _ in range(layer_hidden)])
23 |
24 | self.W_output = nn.ModuleList([nn.Linear(dim, dim)
25 | for _ in range(layer_output)])
26 | self.W_property = nn.Linear(dim, 2)
27 |
28 |
29 | def pad(self, matrices, pad_value):
30 | """Pad the list of matrices
31 | with a pad_value (e.g., 0) for batch processing.
32 | For example, given a list of matrices [A, B, C],
33 | we obtain a new matrix [A00, 0B0, 00C],
34 | where 0 is the zero (i.e., pad value) matrix.
35 | """
36 | shapes = [m.shape for m in matrices]
37 | M, N = sum([s[0] for s in shapes]), sum([s[1] for s in shapes])
38 | zeros = torch.FloatTensor(np.zeros((M, N))).to(device)
39 | pad_matrices = pad_value + zeros
40 | i, j = 0, 0
41 | for k, matrix in enumerate(matrices):
42 | m, n = shapes[k]
43 | pad_matrices[i:i+m, j:j+n] = matrix
44 | i += m
45 | j += n
46 | return pad_matrices
47 |
48 | def update(self, matrix, vectors, layer):
49 | hidden_vectors = torch.relu(self.W_fingerprint[layer](vectors))
50 |
51 | return hidden_vectors + torch.matmul(matrix, hidden_vectors)
52 |
53 | def sum(self, vectors, axis):
54 | sum_vectors = [torch.sum(v, 0) for v in torch.split(vectors, axis)]
55 | return torch.stack(sum_vectors)
56 | def gnn(self, inputs):
57 |
58 | """Cat or pad each input data for batch processing."""
59 | Smiles,fingerprints, adjacencies, molecular_sizes = inputs
60 | fingerprints = torch.cat(fingerprints)
61 | adjacencies = self.pad(adjacencies, 0)
62 |
63 | """GNN layer (update the fingerprint vectors)."""
64 | fingerprint_vectors = self.embed_fingerprint(fingerprints)
65 | for l in range(layer_hidden):
66 | hs = self.update(adjacencies, fingerprint_vectors, l)
67 | fingerprint_vectors = F.normalize(hs, 2, 1) # normalize.
68 |
69 | """Molecular vector by sum or mean of the fingerprint vectors."""
70 | molecular_vectors = self.sum(fingerprint_vectors, molecular_sizes)
71 |
72 | return Smiles,molecular_vectors
73 |
74 | def mlp(self, vectors):
75 | """Classifier based on multilayer perceptron给予多层感知器的分类器."""
76 | for l in range(layer_output):
77 | vectors = torch.relu(self.W_output[l](vectors))
78 | outputs = torch.sigmoid(self.W_property(vectors))
79 | return outputs
80 |
81 |
82 | def forward_classifier(self, data_batch, train):
83 |
84 | inputs = data_batch[:-1]
85 | correct_labels = torch.cat(data_batch[-1])
86 |
87 | if train:
88 | Smiles,molecular_vectors = self.gnn(inputs)
89 |
90 | predicted_scores = self.mlp(molecular_vectors)
91 |
92 | loss = F.cross_entropy(predicted_scores, correct_labels.long())
93 | predicted_scores = predicted_scores.to('cpu').data.numpy()
94 | predicted_scores = [s[1] for s in predicted_scores]
95 |
96 |
97 | correct_labels = correct_labels.to('cpu').data.numpy()
98 | return loss,predicted_scores, correct_labels
99 | else:
100 | with torch.no_grad():
101 | Smiles,molecular_vectors = self.gnn(inputs)
102 | predicted_scores = self.mlp(molecular_vectors)
103 | loss = F.cross_entropy(predicted_scores, correct_labels.long())
104 | predicted_scores = predicted_scores.to('cpu').data.numpy()
105 | predicted_scores = [s[1] for s in predicted_scores]
106 | correct_labels = correct_labels.to('cpu').data.numpy()
107 |
108 | return Smiles,loss,predicted_scores, correct_labels
109 |
110 | class Trainer(object):
111 | def __init__(self, model):
112 | self.model = model
113 | self.optimizer = optim.Adam(self.model.parameters(), lr=lr)
114 |
115 | def train(self, dataset):
116 | np.random.shuffle(dataset)
117 | N = len(dataset)
118 | loss_total = 0
119 | P, C = [], []
120 | for i in range(0, N, batch_train):
121 | data_batch = list(zip(*dataset[i:i+batch_train]))
122 | loss,predicted_scores, correct_labels= self.model.forward_classifier(data_batch, train=True)
123 |
124 | P.append(predicted_scores)
125 | C.append(correct_labels)
126 | self.optimizer.zero_grad()
127 | loss.backward()
128 | self.optimizer.step()
129 | loss_total += loss.item()
130 | tru=np.concatenate(C)
131 | pre=np.concatenate(P)
132 | AUC = roc_auc_score(tru, pre)
133 | pred = [1 if i >0.4 else 0 for i in pre]
134 | predictions =np.stack((tru,pred,pre))
135 | return AUC, loss_total,predictions
136 |
137 |
138 | class Tester(object):
139 | def __init__(self, model):
140 | self.model = model
141 |
142 | def test_classifier(self, dataset):
143 | N = len(dataset)
144 | loss_total = 0
145 | SMILES,P, C ='', [], []
146 | for i in range(0, N, batch_test):
147 | data_batch = list(zip(*dataset[i:i+batch_test]))
148 | (Smiles,loss,predicted_scores,correct_labels) = self.model.forward_classifier(
149 | data_batch, train=False)
150 |
151 | SMILES += ' '.join(Smiles) + ' '
152 |
153 | loss_total += loss.item()
154 | P.append(predicted_scores)
155 | C.append(correct_labels)
156 | SMILES = SMILES.strip().split()
157 | tru=np.concatenate(C)
158 |
159 | pre=np.concatenate(P)
160 | AUC = roc_auc_score(tru, pre)
161 | pred = [1 if i >0.4 else 0 for i in pre]
162 | # Tru=map(str,np.concatenate(C))
163 | # Pre=map(str,np.concatenate(P))
164 | # predictions = '\n'.join(['\t'.join(x) for x in zip(SMILES, Tru, Pre)])
165 | predictions =np.stack((tru,pred,pre))
166 | return AUC, loss_total,predictions
167 | def save_result(self, result, filename):
168 | with open(filename, 'a') as f:
169 | f.write(result + '\n')
170 | def save_predictions(self, predictions, filename):
171 | with open(filename, 'w') as f:
172 | f.write('Smiles\tCorrect\tPredict\n')
173 | f.write(predictions + '\n')
174 | def save_model(self, model, filename):
175 | torch.save(model.state_dict(), filename)
176 | def split_dataset(dataset, ratio):
177 | """Shuffle and split a dataset."""
178 | np.random.seed(111) # fix the seed for shuffle.
179 | np.random.shuffle(dataset)
180 | n = int(ratio * len(dataset))
181 | return dataset[:n], dataset[n:]
182 | def edit_dataset(drug,non_drug,task):
183 | np.random.seed(111) # fix the seed for shuffle.
184 |
185 | if task =='balance':
186 | #np.random.shuffle(non_drug)
187 | non_drug=non_drug[0:len(drug)]
188 |
189 | else:
190 | np.random.shuffle(non_drug)
191 | np.random.shuffle(drug)
192 | dataset_train_drug, dataset_test_drug = split_dataset(drug, 0.9)
193 | # dataset_train_drug,dataset_dev_drug = split_dataset(dataset_train_drug, 0.9)
194 | dataset_train_no, dataset_test_no = split_dataset(non_drug, 0.9)
195 | # dataset_train_no,dataset_dev_no = split_dataset(dataset_train_no, 0.9)
196 | dataset_train = dataset_train_drug+dataset_train_no
197 | dataset_test= dataset_test_drug+dataset_test_no
198 | # dataset_dev = dataset_dev_drug+dataset_dev_no
199 | return dataset_train, dataset_test
200 |
201 | def dump_dictionary(dictionary, filename):
202 | with open(filename, 'wb') as f:
203 | pickle.dump(dict(dictionary), f)
204 | if __name__ == "__main__":
205 |
206 | radius=1
207 | dim=65
208 | layer_hidden=0
209 | layer_output=5
210 |
211 | batch_train=48
212 | batch_test=48
213 | lr=3e-4
214 | lr_decay=0.85
215 | decay_interval=10#下降间隔
216 | iteration=140
217 | N=5000
218 | (radius, dim, layer_hidden, layer_output,
219 | batch_train, batch_test, decay_interval,
220 | iteration) = map(int, [radius, dim, layer_hidden, layer_output,
221 | batch_train, batch_test,
222 | decay_interval, iteration])
223 | lr, lr_decay = map(float, [lr, lr_decay])
224 | if torch.cuda.is_available():
225 | device = torch.device('cuda')
226 | print('The code uses a GPU!')
227 | else:
228 | device = torch.device('cpu')
229 | print('The code uses a CPU...')
230 | print('-'*100)
231 |
232 | # print('Preprocessing the', dataset, 'dataset.')
233 | print('Just a moment......')
234 | print('-'*100)
235 | path='E:/code/drug/drugnn/'
236 | dataname=''
237 |
238 | dataset_train = pp.create_dataset('data_train.txt',path,dataname)
239 | dataset_test = pp.create_dataset('data_test.txt',path,dataname)
240 |
241 | #dataset_train, dataset_test = edit_dataset(dataset_drug, dataset_nondrug,'balance')
242 | #dataset_train, dataset_dev = split_dataset(dataset_train, 0.9)
243 | print('The preprocess has finished!')
244 | print('# of training data samples:', len(dataset_train))
245 | #print('# of development data samples:', len(dataset_dev))
246 | print('# of test data samples:', len(dataset_test))
247 | print('-'*100)
248 |
249 | print('Creating a model.')
250 | torch.manual_seed(111)
251 | model = MolecularGraphNeuralNetwork(
252 | N, dim, layer_hidden, layer_output).to(device)
253 | trainer = Trainer(model)
254 | tester = Tester(model)
255 | print('# of model parameters:',
256 | sum([np.prod(p.size()) for p in model.parameters()]))
257 | print('-'*100)
258 | file_result = path+'AUC'+'.txt'
259 | # file_result = '../output/result--' + setting + '.txt'
260 | result = 'Epoch\tTime(sec)\tLoss_train\tLoss_test\tAUC_train\tAUC_test'
261 | file_test_result = path+ 'test_prediction'+ '.txt'
262 | file_predictions = path+'train_prediction' +'.txt'
263 | file_model = path+'model'+'.h5'
264 | with open(file_result, 'w') as f:
265 | f.write(result + '\n')
266 |
267 | print('Start training.')
268 | print('The result is saved in the output directory every epoch!')
269 |
270 | np.random.seed(111)
271 |
272 | start = timeit.default_timer()
273 |
274 | for epoch in range(iteration):
275 |
276 | epoch += 1
277 | if epoch % decay_interval == 0:
278 | trainer.optimizer.param_groups[0]['lr'] *= lr_decay
279 | #[‘amsgrad’, ‘params’, ‘lr’, ‘betas’, ‘weight_decay’, ‘eps’]
280 | prediction_train,loss_train,train_res= trainer.train(dataset_train)
281 |
282 |
283 | #prediction_dev,dev_res = tester.test_classifier(dataset_dev)
284 | prediction_test,loss_test,test_res = tester.test_classifier(dataset_test)
285 |
286 |
287 | time = timeit.default_timer() - start
288 |
289 | if epoch == 1:
290 | minutes = time * iteration / 60
291 | hours = int(minutes / 60)
292 | minutes = int(minutes - 60 * hours)
293 | print('The training will finish in about',
294 | hours, 'hours', minutes, 'minutes.')
295 | print('-'*100)
296 | print(result)
297 |
298 | result = '\t'.join(map(str, [epoch, time, loss_train, loss_test,prediction_train,prediction_test]))
299 | tester.save_result(result, file_result)
300 |
301 | print(result)
302 |
303 |
304 |
305 | loss = pd.read_table(file_result)
306 | plt.plot(loss['Loss_train'], color='r',label='Loss of train set')
307 | plt.plot(loss['Loss_test'], color='y',label='Loss of train set')
308 | plt.plot(loss['AUC_train'], color='y',label='AUC of train set')
309 | plt.plot(loss['AUC_test'], color='b',label='AUC of test set')
310 | # plt.plot(loss['AUC_test'], color='y',label='AUC of test set')
311 | plt.ylabel('AUC')
312 | plt.xlabel('Epoch')
313 | plt.legend()
314 | plt.savefig(path+'loss.tif',dpi=300)
315 | plt.show()
316 | colors = ['#00CED1','#DC143C' ]
317 |
318 | target_names=np.array(['druglike','not-drug'])
319 | lw=2
320 | res_test = test_res.T
321 |
322 | for color,i,target_name in zip(colors,[1,0],target_names):
323 |
324 | plt.scatter((res_test[res_test[:,0]==i,0]),(res_test[res_test[:,0]==i,2]),color = color,alpha=.8,lw=lw,label=target_name)
325 | plt.legend(loc='best',shadow=False,scatterpoints=1)
326 | plt.title('the results of gnn classification')
327 | res_train = train_res.T
328 | cn_matrix=confusion_matrix(res_train[:,0], res_train[:,1])
329 | cn_matrix
330 |
331 | tn1 = cn_matrix[0,0]
332 | tp1 = cn_matrix[1,1]
333 | fn1 = cn_matrix[1,0]
334 | fp1 = cn_matrix[0,1]
335 |
336 |
337 | bacc_train = ((tp1/(tp1+fn1))+(tn1/(tn1+fp1)))/2#balance accurance
338 | pre_train = tp1/(tp1+fp1)#precision/q+
339 | rec_train = tp1/(tp1+fn1)#recall/se
340 | sp_train=tn1/(tn1+fp1)
341 | q__train=tn1/(tn1+fn1)
342 | f1_train = 2*pre_train*rec_train/(pre_train+rec_train)#f1score
343 | mcc_train = ((tp1*tn1) - (fp1*fn1))/math.sqrt((tp1+fp1)*(tp1+fn1)*(tn1+fp1)*(tn1+fn1))#Matthews correlation coefficient
344 | acc_train=(tp1+tn1)/(tp1+fp1+fn1+tn1)#accurancy
345 | fpr_train, tpr_train, thresholds_train =roc_curve(res_train[:,0],res_train[:,1])
346 | print('bacc_train:',bacc_train)
347 | print('pre_train:',pre_train)
348 | print('rec_train:',rec_train)
349 | print('f1_train:',f1_train)
350 | print('mcc_train:',mcc_train)
351 | print('sp_train:',sp_train)
352 | print('q__train:',q__train)
353 | print('acc_train:',acc_train)
354 |
355 |
356 | '''
357 | res_dev = dev_res.T
358 | cn_matrix=confusion_matrix(res_dev[:,0], res_dev[:,1])
359 | cn_matrix
360 |
361 | tn2 = cn_matrix[0,0]
362 | tp2 = cn_matrix[1,1]
363 | fn2 = cn_matrix[1,0]
364 | fp2 = cn_matrix[0,1]
365 |
366 |
367 | bacc_dev = ((tp2/(tp2+fn2))+(tn2/(tn2+fp2)))/2#balance accurance
368 | pre_dev= tp2/(tp2+fp2)#precision/q+
369 | rec_dev = tp2/(tp2+fn2)#recall/se
370 | sp_dev=tn2/(tn2+fp2)
371 | q__dev=tn2/(tn2+fn2)
372 | f1_dev = 2*pre_dev*rec_dev/(pre_dev+rec_dev)#f1score
373 | mcc_dev = ((tp2*tn2) - (fp2*fn2))/math.sqrt((tp2+fp2)*(tp2+fn2)*(tn2+fp2)*(tn2+fn2))#Matthews correlation coefficient
374 | acc_dev=(tp2+tn2)/(tp2+fp2+fn2+tn2)#accurancy
375 | fpr_dev, tpr_dev, thresholds_dev =roc_curve(res_dev[:,0],res_dev[:,1])
376 | print('bacc_dev:',bacc_dev)
377 | print('pre_dev:',pre_dev)
378 | print('rec_dev:',rec_dev)
379 | print('f1_dev:',f1_dev)
380 | print('mcc_dev:',mcc_dev)
381 | print('sp_dev:',sp_dev)
382 | print('q__dev:',q__dev)
383 | print('acc_dev:',acc_dev)
384 |
385 | '''
386 |
387 | cnf_matrix=confusion_matrix(res_test[:,0], res_test[:,1])
388 | cnf_matrix
389 |
390 | tn = cnf_matrix[0,0]
391 | tp = cnf_matrix[1,1]
392 | fn = cnf_matrix[1,0]
393 | fp = cnf_matrix[0,1]
394 |
395 | bacc = ((tp/(tp+fn))+(tn/(tn+fp)))/2#balance accurance
396 | pre = tp/(tp+fp)#precision/q+
397 | rec = tp/(tp+fn)#recall/se
398 | sp=tn/(tn+fp)
399 | q_=tn/(tn+fn)
400 | f1 = 2*pre*rec/(pre+rec)#f1score
401 | mcc = ((tp*tn) - (fp*fn))/math.sqrt((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))#Matthews correlation coefficient
402 | acc=(tp+tn)/(tp+fp+fn+tn)#accurancy
403 | fpr, tpr, thresholds =roc_curve(res_test[:,0], res_test[:,1])
404 | print('bacc:',bacc)
405 | print('pre:',pre)
406 | print('rec:',rec)
407 | print('f1:',f1)
408 | print('mcc:',mcc)
409 | print('sp:',sp)
410 | print('q_:',q_)
411 | print('acc:',acc)
412 | print('auc:',prediction_test)
413 |
414 |
--------------------------------------------------------------------------------
/dataset/withdrawn.txt:
--------------------------------------------------------------------------------
1 | O=C(Nc1ccc(Cl)c(Cl)c1)c1cc(Cl)cc(Cl)c1O 0
2 | CC(=O)NC1=CC=CC=C1 0
3 | CC(=O)NC1=C(C=CC(=C1)[As](=O)(O)O)O 0
4 | CC(=O)NC1=NC=C(N=N1)C=CC2=CC=C(O2)[N+](=O)[O-] 0
5 | CC(=O)OC1=CC=CC=C1C(=O)O 0
6 | CC1=CC(=C(C(=C1C=CC(=CC=CC(=CC(=O)O)C)C)C)C)OC 0
7 | C1=NC(=C2C(=N1)N(C=N2)C3C(C(C(O3)COP(=O)(O)O)O)O)N 0
8 | CC(C(=O)NC(C)C(=O)NC1C2C1CN(C2)C3=C(C=C4C(=O)C(=CN(C4=N3)C5=C(C=C(C=C5)F)F)C(=O)O)F)N 0
9 | C=CCOC1=C(C=C(C=C1)CC(=O)O)Cl 0
10 | CC(C)C(CC1=CC(=C(C=C1)OC)OCCCOC)CC(C(CC(C(C)C)C(=O)NCC(C)(C)C(=O)N)O)N 0
11 | CC1=C(N=CN1)CN2CCC3=C(C2=O)C4=CC=CC=C4N3C 0
12 | CCC(C(CC(C)N(C)C)(C1=CC=CC=C1)C2=CC=CC=C2)OC(=O)C 0
13 | CCCN(CCC)C(=O)CC1=C(N=C2N1C=C(C=C2)Cl)C3=CC=C(C=C3)Cl 0
14 | CCN(CC)C(C)C(=O)C1=CC=CC=C1 0
15 | C1CC2=CC=CC=C2C(C3=CC=CC=C31)NCCCCCCC(=O)O 0
16 | CCC1(CCC(=O)NC1=O)C2=CC=C(C=C2)N 0
17 | Cc1c(N(C)C)c(=O)n(-c2ccccc2)n1C 0
18 | CCC1(C(=O)NC(=O)NC1=O)CCC(C)C 0
19 | CC(C)CCOCC(CN1CCOCC1)OC(=O)C2=CC(=C(C(=C2)OC)OC)OC 0
20 | CC(CC1=CC=CC=C1)N 0
21 | C1=CC=C(C=C1)C(C#N)OC2C(C(C(C(O2)COC3C(C(C(C(O3)CO)O)O)O)O)O)O 0
22 | CC1CC2C(CCC3(C2CCC3(C(=O)C)OC(=O)C)C)C4(C1=CCCC4)C 0
23 | C1CN(CCN1CCOC(=O)C2=CC=CC=C2NC3=C4C=CC(=CC4=NC=C3)C(F)(F)F)C5=CC=CC(=C5)C(F)(F)F 0
24 | CC(C)C1(C(=O)NC(=O)NC1=O)CC=C 0
25 | COC1=CC=CC2=C3C(=C(C=C21)[N+](=O)[O-])C(=CC4=C3OCO4)C(=O)O 0
26 | COC1=CC=C(C=C1)CCN2CCC(CC2)NC3=NC4=CC=CC=C4N3CC5=CC=C(C=C5)F 0
27 | CC(=O)OCC1C(C(C(O1)N2C(=O)NC(=O)C=N2)OC(=O)C)OC(=O)C 0
28 | CCC1(C(=O)NC(=O)NC1=O)CC 0
29 | CCC(C)(C(=O)OCC)OC1=CC=C(C=C1)CC2=CC=C(C=C2)Cl 0
30 | C1=CC=C(C=C1)CN2C3=CC=CC=C3C(=N2)OCC(=O)O 0
31 | CC(CC1=CC(=CC=C1)C(F)(F)F)NCCOC(=O)C2=CC=CC=C2 0
32 | CC(C1=CC2=C(C=C1)OC(=N2)C3=CC=C(C=C3)Cl)C(=O)O 0
33 | CCC1=C(C2=CC=CC=C2O1)C(=O)C3=CC=C(C=C3)O 0
34 | CCC1=C(C2=CC=CC=C2O1)C(=O)C3=CC(=C(C(=C3)Br)O)Br 0
35 | CCC1=C(C2=CC=CC=C2O1)C(=O)C3=CC(=C(C(=C3)I)O)I 0
36 | CN(C)CCCOC1=NN(C2=CC=CC=C21)CC3=CC=CC=C3 0
37 | C1=CC=C(C=C1)CO 0
38 | CC(C)COCC(CN(CC1=CC=CC=C1)C2=CC=CC=C2)N3CCCC3 0
39 | CCC(=O)N1C2=CC=CC=C2N(C1=O)C3CCN(CC3)CCC(C#N)(C4=CC=CC=C4)C5=CC=CC=C5 0
40 | CC(CS(=O)(=O)C1=CC=C(C=C1)F)(C(=O)NC2=CC(=C(C=C2)C#N)C(F)(F)F)O 0
41 | C1=C(C=C(C(=C1SC2=C(C(=CC(=C2)Cl)Cl)O)O)Cl)Cl 0
42 | B(O)(O)O 0
43 | C1=CC(=C(C(=C1)C(=O)C2=CC=C(C=C2)Br)N)CC(=O)O 0
44 | CC(C)C(C(=O)NC(=O)N)Br 0
45 | CC1=NN=C2N1C3=C(C=C(S3)Br)C(=NC2)C4=CC=CC=C4Cl 0
46 | C1=CC2=C(C(=C(C=C2Br)Br)O)N=C1 0
47 | CCOC1=CC=C(C=C1)NC(=O)CC(C)O 0
48 | CC(C)(C)N1CCC(CC1)(C2=CC=CC=C2)C3=CC=CC=C3 0
49 | CCCCOC1=CC=C(C=C1)CC(=O)NO 0
50 | COC1=CC(=C(C(=C1)OC)C(=O)CCCN2CCCC2)OC 0
51 | CCCCN=C(N)N=C(N)N 0
52 | CCCCC(C(=O)N(C1=CC=CC=C1)NC2=CC=CC=C2)C(=O)O 0
53 | CCCC(=O)NC1=C(C=C(C(=C1I)C=C(CC)C(=O)O)I)I 0
54 | CC(C)(C)C(C)(C1CC23CCC1(C4C25CCN(C3CC6=C5C(=C(C=C6)O)O4)CC7CC7)OC)O 0
55 | CC(C(=O)C1=CC(=CC=C1)Cl)NC(C)(C)C 0
56 | CCCCOC(=O)C1=CC=C(C=C1)N 0
57 | CCN(CC(C)O)C1=NN=C(C=C1)NNC(=O)OCC 0
58 | CN1C2=C(C=C(C=C2)Cl)C(=NC(C1=O)OC(=O)N(C)C)C3=CC=CC=C3 0
59 | CCOc1nc2cccc(C(=O)O)c2n1Cc1ccc(-c2ccccc2-c2nnn[nH]2)cc1 0
60 | CC12CCC(=O)C=C1C=CC3C2CCC4(C3CCC45CCC(=O)O5)C 0
61 | CC1=C(C(CCC1=O)(C)C)C=CC(=CC=CC(=CC=CC=C(C)C=CC=C(C)C=CC2=C(C(=O)CCC2(C)C)C)C)C 0
62 | CCN(CC)CCOCCOC(=O)C1(c2ccccc2)CCCC1 0
63 | CN(C)CCOC(C1=CC=C(C=C1)Cl)C2=CC=CC=N2 0
64 | CCCC(C)(COC(=O)N)COC(=O)NC(C)C 0
65 | CCCCCCNC(=O)N1C=C(C(=O)NC1=O)F 0
66 | CC1=CC=C(C=C1)C2=CC(=NN2C3=CC=C(C=C3)S(=O)(=O)N)C(F)(F)F 0
67 | C1C(=C(N2C(S1)C(C2=O)NC(=O)CC3=CC=CS3)C(=O)[O-])C[N+]4=CC=CC=C4 0
68 | CC(C)C1=C(C(=C(C(=N1)C(C)C)COC)C2=CC=C(C=C2)F)C=CC(CC(CC(=O)O)O)O 0
69 | CC(CCC(=O)O)C1CCC2C1(CCC3C2C(CC4C3(CCC(C4)O)C)O)C 0
70 | C(C(Cl)(Cl)Cl)(O)O 0
71 | C1=CC(=CC=C1C(C(CO)NC(=O)C(Cl)Cl)O)[N+](=O)[O-] 0
72 | N=C(NCCCCCCNC(=N)NC(=N)Nc1ccc(Cl)cc1)NC(=N)Nc1ccc(Cl)cc1 0
73 | CC(=O)C1(CCC2C1(CCC3C2C=C(C4=CC(=O)CCC34C)Cl)C)O 0
74 | CC(=O)C1(CCC2C1(CCC3C2C=C(C4=CC(=O)CCC34C)Cl)C)OC(=O)C 0
75 | CN1C(S(=O)(=O)CCC1=O)C2=CC=C(C=C2)Cl 0
76 | C1=CC=C2C=C(C=CC2=C1)N(CCCl)CCCl 0
77 | C(Cl)(Cl)Cl 0
78 | CCN(CC)CCCC(C)NC1=C2C=CC(=CC2=NC=C1)Cl 0
79 | CC(C)(CC1=CC=C(C=C1)Cl)N 0
80 | C1C(C(OC2=CC(=CC(=C21)O)O)C3=CC(=C(C=C3)O)O)O 0
81 | C1=CC=C(C=C1)C2=NC3=CC=CC=C3C(=C2)C(=O)O 0
82 | COC1=CC(=CC(=C1OC)OC)C=CC(=O)N2CCN(CC2)CC(=O)N3CCCC3 0
83 | CC(C)(C(=O)O)OC1=CC=C(C=C1)C2CC2(Cl)Cl 0
84 | COC1CN(CCC1NC(=O)C2=CC(=C(C=C2OC)N)Cl)CCCOC3=CC=C(C=C3)F 0
85 | C1=CC2=C(C(=C(C=C2Cl)I)O)N=C1 0
86 | CC(CC1=CC=CC=C1)NCC2=CC=CC=C2Cl 0
87 | CC(CN(C)C)C(C)(CC1=CC=C(C=C1)Cl)O 0
88 | C1=CC(=CC=C1C(C2=CC=C(C=C2)Cl)C(Cl)(Cl)Cl)Cl 0
89 | CCOC(=O)C(C)(C)OC1=CC=C(C=C1)Cl 0
90 | CCOC(=O)NC(C)(C)CC1=CC=C(C=C1)Cl 0
91 | CN(C)CCCC1C2=CC=CC=C2NC3=C1C=C(C=C3)Cl 0
92 | CC1=C(C2=C(N1CC(=O)O)C=C(C=C2)OC)C(=O)C3=CC=C(C=C3)Cl 0
93 | C1=CC=C(C=C1)C2=NC(C(=O)NC3=C2C=C(C=C3)Cl)C(=O)O 0
94 | CN1CCN(CC1)C2=NC3=C(C=CC(=C3)Cl)NC4=CC=CC=C42 0
95 | [Co] 0
96 | CN1CCC23C4C1CC5=C2C(=C(C=C5)OC)OC3C(C=C4)O 0
97 | CCC(C1=CC=C(C=C1)OCCN(CC)CC)C(CC)C2=CC=C(C=C2)OCCN(CC)CC 0
98 | C1=CC=C2C(=C1)C=CC(=O)O2 0
99 | CC1CC(CC(C1)(C)C)OC(=O)C(C2=CC=CC=C2)O 0
100 | CCC1(C(=O)NC(=O)NC1=O)C2=CCCCC2 0
101 | CC(=O)OC1=CC=C(C=C1)C(=C2CCCCC2)C3=CC=C(C=C3)OC(=O)C 0
102 | CN1CCC(=C2C3=CC=CC=C3C=CC4=CC=CC=C42)CC1 0
103 | C1=CC2=C(C(=C1)O)C(=O)C3=C(C2=O)C=CC=C3O 0
104 | CC(CC1=CC=CC=C1)N 0
105 | CCNC(C)CC1=CC(=CC=C1)C(F)(F)F 0
106 | CCN(CC)CCOC1=CC2=C(C=C1)N=C(S2)N(C)C 0
107 | C1=CC(=CC=C1NC(=O)C2=C(C=CC(=C2)Br)O)Br 0
108 | CCN(CC)CCOC(=O)C1(CCCCC1)C2CCCCC2 0
109 | CC=C(C1=CC=C(C=C1)O)C(=CC)C2=CC=C(C=C2)O 0
110 | CCC(=C(CC)C1=CC=C(C=C1)O)C2=CC=C(C=C2)O 0
111 | CC(C)(COC(=O)C(C1=CC=CC=C1)(C2=CC=CC=C2)O)N(C)C 0
112 | C1CN(CCC1(C2=CC=CC=C2)C(=O)O)CCC(C#N)(C3=CC=CC=C3)C4=CC=CC=C4 0
113 | CC(C)[C@@]1(NC(=O)[C@@H]2C[C@@H]3c4cccc5[nH]cc(c45)C[C@H]3N(C)C2)O[C@@]2(O)[C@@H]3CCCN3C(=O)[C@H](Cc3ccccc3)N2C1=O 0
114 | CC(C)C[C@H]1C(=O)N2CCC[C@H]2[C@]2(O)O[C@](NC(=O)[C@@H]3C[C@@H]4c5cccc6[nH]cc(c56)C[C@H]4N(C)C3)(C(C)C)C(=O)N12 0
115 | CN1C[C@H](C(=O)N[C@]2(C)O[C@@]3(O)[C@@H]4CCCN4C(=O)[C@H](Cc4ccccc4)N3C2=O)C[C@@H]2c3cccc4[nH]cc(c34)C[C@H]21 0
116 | CN[C@@H]1[C@H](O[C@H]2[C@H](O[C@@H]3[C@@H](NC(=N)N)[C@H](O)[C@@H](NC(=N)N)[C@H](O)[C@H]3O)O[C@@H](C)[C@]2(O)CO)O[C@@H](CO)[C@H](O)[C@H]1O 0
117 | C1=C(OC(=C1)[N+](=O)[O-])C=CC2=CN=C(N=N2)N(CO)CO 0
118 | I[Sn](CC)(CC)I 0
119 | CC(CCC1=CC=CC=C1)NCC(C2=CC(=C(C=C2)O)C(=O)N)O 0
120 | CCC(C)CC(C)N 0
121 | C1=CC(=C(C=C1[N+](=O)[O-])[N+](=O)[O-])O 0
122 | CCCCCC(C=CC1C(CC(=O)C1CC=CCCCC(=O)O)O)O 0
123 | CC(CC1=CC=CC=C1)N2CCN(CC2)C(C)CC3=CC=CC=C3 0
124 | CN(C)CCOC(C1=CC=CC=C1)C2=CC=CC=C2 0
125 | CCOC(=O)C1(CCN(CC1)CCC(C#N)(C2=CC=CC=C2)C3=CC=CC=C3)C4=CC=CC=C4 0
126 | Cc1c(N(C)CS(=O)(=O)O)c(=O)n(-c2ccccc2)n1C 0
127 | CCN1C(=CC=CC=Cc2sc3ccccc3[n+]2CC)Sc2ccccc21 0
128 | CN(CCC1=CC=C(C=C1)NS(=O)(=O)C)CCOC2=CC=C(C=C2)NS(=O)(=O)C 0
129 | O=C(O[C@@H]1C[C@@H]2C[C@H]3C[C@H](C1)N2CC3=O)c1c[nH]c2ccccc12 0
130 | C5=C(C(OC3CC1N2C(CC(C1)C(C2)=O)C3)=O)C4=C(C=CC=C4)[NH]5 0
131 | C1CN(CCC1N2C3=C(C=C(C=C3)Cl)NC2=O)CCCN4C5=CC=CC=C5NC4=O 0
132 | CC1C2C(C3C(C(=O)C(=C(C3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O 0
133 | C2=C(C(C1=NC=CC=C1)(OCCN(C)C)C)C=CC=C2 0
134 | C1CN(CC=C1N2C3=CC=CC=C3NC2=O)CCCC(=O)C4=CC=C(C=C4)F 0
135 | CN1C2=C(C3=CC=CC=C3S1(=O)=O)OC(=O)N(C2=O)C4=CC=CC=N4 0
136 | C1=CC(=CC=C1S(=O)(=O)NC=NCCSCC2=CSC(=N2)N=C(N)N)Br 0
137 | CCC1CN2CCC3=CC(=C(C=C3C2CC1CC4C5=CC(=C(C=C5CCN4)OC)OC)OC)OC 0
138 | CN1CCCCC1CCC2=CC=CC=C2NC(=O)C3=CC=C(C=C3)OC 0
139 | CN[C@@H](C)[C@H](O)c1:c:c:c:c:c:1 0
140 | CNCC(C1=CC(=C(C=C1)O)O)O 0
141 | C=C1CC[C@H](O)C/C1=C/C=C1\CCC[C@@]2(C)[C@H]1CC[C@@H]2[C@H](C)/C=C/[C@H](C)C(C)C 0
142 | C(C(C(CO[N+](=O)[O-])O[N+](=O)[O-])O[N+](=O)[O-])O[N+](=O)[O-] 0
143 | CCC1C(C(C(C(=O)C(CC(C(C(C(C(C(=O)O1)C)OC2CC(C(C(O2)C)O)(C)OC)C)OC3C(C(CC(O3)C)N(C)C)O)(C)O)C)C)O)(C)O 0
144 | COC(=O)CCc1ccc(OCC(O)CNC(C)C)cc1 0
145 | CCO 0
146 | C#CC(O)(/C=C/Cl)CC 0
147 | CC12CCC3C(C1CCC2(C#C)O)CCC4=C3C=CC(=C4)O 0
148 | CCON=O 0
149 | C(CCl)Cl 0
150 | CCC1(CCC2C1(CCC3C2CCC4=CCCCC34)C)O 0
151 | CCNC1=NC2=C(C=C(C=C2)Cl)C(O1)(C)C3=CC=CC=C3 0
152 | CCNCC(C1=CC(=CC=C1)O)O 0
153 | CCOC(=O)C1=CN=CN1C(C)C2=CC=CC=C2 0
154 | CCOC(=O)C=C(C)C=CC=C(C)C=CC1=C(C(=C(C=C1C)OC)C)C 0
155 | C1=CC(=C(C(=C1C(=O)C2=CC(=C(C(=C2)O)O)O)O)O)O 0
156 | CCCCOCC(CN1C(=O)C(C(=O)NC1=O)(CC)C2=CC=CC=C2)OC(=O)N 0
157 | C1=CC=C(C=C1)C(COC(=O)N)COC(=O)N 0
158 | CCC(C1=CC=CC=C1)C(=O)OCCN2CCOC(C2C)C3=CC=CC=C3 0
159 | C1=CC=C(C(=C1)CC(=O)O)OC2=C(C=C(C=C2)Cl)Cl 0
160 | C1=CC(=CC=C1C2=NC(=CS2)CC(=O)O)Cl 0
161 | CC(CC1=CC=CC=C1)NCCN2C=NC3=C2C(=O)N(C(=O)N3C)C 0
162 | CCNC(C)CC1=CC(=CC=C1)C(F)(F)F 0
163 | CC(CC1=CC=C(C=C1)O)NCC(C2=CC(=CC(=C2)O)O)O 0
164 | CC(CC1=CC=CC=C1)NCCC#N 0
165 | O=C1NCC2(CCN(CCc3ccccc3)CC2)O1 0
166 | CC(=CCC1C(=O)N(N(C1=O)C2=CC=CC=C2)C3=CC=CC=C3)C 0
167 | C1CN(CCN1CC2=CC3=C(C=C2)OCO3)C(=O)COC4=CC=C(C=C4)Cl 0
168 | C1=CC=C(C(=C1)C(=O)OCC(CO)O)NC2=C3C=CC=C(C3=NC=C2)C(F)(F)F 0
169 | CN1C=C(C(=O)C2=C1C=C(C=C2)F)S(=O)C 0
170 | CS(=O)(=O)NC1=C(C=C2C(=C1)CCC2=O)OC3=C(C=C(C=C3)F)F 0
171 | COC(=O)NC1=NC2=C(N1)C=C(C=C2)C(=O)C3=CC=C(C=C3)F 0
172 | CC1CCC2=C3N1C=C(C(=O)C3=CC(=C2)F)C(=O)O 0
173 | CN1C(=O)CN=C(C2=C1C=CC(=C2)[N+](=O)[O-])C3=CC=CC=C3F 0
174 | CCOC(=O)Nc1ccc(NCc2ccc(F)cc2)nc1N 0
175 | CC1=CC2=C(C=C1)C(=NC(=O)N2C(C)C)C3=CC=C(C=C3)F 0
176 | C(C(F)(F)F)OCC(F)(F)F 0
177 | COCCCCC(=NOCCN)C1=CC=C(C=C1)C(F)(F)F 0
178 | CN(CC1=C(C=CC=C1Cl)NC(=O)C2=CC=CC=C2)CC(=O)N3CCOCC3 0
179 | C1COC(=O)N1N=CC2=CC=C(O2)[N+](=O)[O-] 0
180 | CC(C)C(CCCN(C)CCC1=CC(=C(C=C1)OC)OC)(C#N)C2=CC(=C(C(=C2)OC)OC)OC 0
181 | CC1CN(CCN1)C2=C(C=C3C(=C2OC)N(C=C(C3=O)C(=O)O)C4CC4)F 0
182 | CC1=CC(=C(C=C1)C)OCCCC(C)(C)C(=O)O 0
183 | CC(C(CN1C=NC=N1)(C2=C(C=C(C=C2)F)F)O)S(=O)(=O)C 0
184 | CC(C1CCC(C(O1)OC2C(CC(C(C2O)OC3C(C(C(CO3)(C)O)NC)O)N)N)N)NC 0
185 | C1=CC=C(C(=C1)C(=O)OCC(CO)O)NC2=C3C=CC(=CC3=NC=C2)Cl 0
186 | CCC1(CCC(=O)NC1=O)C2=CC=CC=C2 0
187 | CC1CN(CCN1)C2=C(C(=C3C(=C2)N(C=C(C3=O)C(=O)O)C4CC4)C)F 0
188 | C1CCCN(CCC1)CCN=C(N)N 0
189 | CCC1(C(=O)NC(=O)NC1=O)C2=CCCCCC2 0
190 | C1=C(C(=C(C(=C1Cl)Cl)CC2=C(C(=CC(=C2Cl)Cl)Cl)O)O)Cl 0
191 | CCC(C1=CC=C(C=C1)O)C(CC)C2=CC=C(C=C2)O 0
192 | CC1(C(=O)NC(=O)N(C1=O)C)C2=CCCCC2 0
193 | C1NC2=CC(=C(C=C2S(=O)(=O)N1)S(=O)(=O)N)Cl 0
194 | CN1CCC23C4C1CC5=C2C(=C(C=C5)O)OC3C(=O)CC4 0
195 | CC(C)CC1=CC=C(C=C1)CC(=O)O 0
196 | C1CNCCC1CCC2=CNC3=CC=CC=C32 0
197 | CC1=C(C2=C(N1C(=O)C3=CC=C(C=C3)Cl)C=CC(=C2)OC)CC(=O)O 0
198 | CC(C1=CC=C(C=C1)N2CC3=CC=CC=C3C2=O)C(=O)O 0
199 | C1CN(CCC1NC(=O)C2=CC=CC=C2)CCC3=CNC4=CC=CC=C43 0
200 | C/C=C(/C)C(=O)O[C@H]1C(C)=C[C@]23C(=O)[C@@H](C=C(CO)[C@@H](O)[C@]12O)[C@H]1[C@@H](C[C@H]3C)C1(C)C 0
201 | CC(=O)NCC1=C(C(=C(C(=C1I)C(=O)O)I)NC(=O)C)I 0
202 | CCOC(=O)CCCCCCCCC(C)C1=CC=CC=C1I 0
203 | CC(C)NNC(=O)C1=CC=NC=C1 0
204 | CC(C)Nc1ncccn1 0
205 | CC1=CC(=NO1)C(=O)NNCC2=CC=CC=C2 0
206 | CC(C)NCC(C1=CC(=C(C=C1)O)O)O 0
207 | CC1=C(C(CCC1)(C)C)C=CC(=CC=CC(=CC(=O)O)C)C 0
208 | CC1=CC(=NO1)NC(=O)C2=C(C3=CC=CC=C3S(=O)(=O)N2C)O 0
209 | CC(=O)N1CCN(CC1)C2=CC=C(C=C2)OCC3COC(O3)(CN4C=CN=C4)C5=C(C=C(C=C5)Cl)Cl 0
210 | CC(C1=CC(=CC=C1)C(=O)C2=CC=CC=C2)C(=O)O 0
211 | C1CN2C(=CC=C2C(=O)C3=CC=CC=C3)C1C(=O)O 0
212 | CS(=O)(=O)C1=CC(=CC2=C1N(C3=C2CCC3CC(=O)O)CC4=CC=C(C=C4)Cl)F 0
213 | C1=CC(=CC=C1C#N)C(C2=CC=C(C=C2)C#N)N3C=NC=N3 0
214 | CCC(C(CC(C)N(C)C)(C1=CC=CC=C1)C2=CC=CC=C2)OC(=O)C 0
215 | CC(CC1=CC=CC=C1)N 0
216 | C1CSC2=NC(CN21)C3=CC=CC=C3 0
217 | C1=CC(=C(C=C1C(CN)O)O)O 0
218 | C1(C(C(C(C(C1Cl)Cl)Cl)Cl)Cl)Cl 0
219 | CN(C)C(=O)C(CCN1CCC(CC1)(C2=CC=C(C=C2)Cl)O)(C3=CC=CC=C3)C4=CC=CC=C4 0
220 | CC1=CC(=C(C=C1)NC2=C(C=CC=C2Cl)F)CC(=O)O 0
221 | CC12CCC3C(C1CCC2(C#C)O)CCC4=CCCCC34 0
222 | C1CN2C(=N1)C3=CC=CC=C3C2(C4=CC=C(C=C4)Cl)O 0
223 | CC(C1=CC=CC=C1)NN 0
224 | CC1=CC(=CC=C1)CN2CCN(CC2)C(C3=CC=CC=C3)C4=CC=C(C=C4)Cl 0
225 | CN(C)CC(OC1=CC=CC=C1)OC2=CC=CC=C2 0
226 | CC(CC1=CC=CC=C1)NCCCCl 0
227 | CC1=CC2C(CCC3(C2CCC3(C(=O)C)OC(=O)C)C)C4(C1=CC(=O)CC4)C 0
228 | CCN(CC)CCCC(C)NC1=C2C=C(C=CC2=NC3=C1C=CC(=C3)Cl)OC 0
229 | CN1CCCC(C1)CN2C3=CC=CC=C3SC4=CC=CC=C42 0
230 | CC1=CC=CC=C1OCC(CO)O 0
231 | CCCC(C)(COC(=O)N)COC(=O)N 0
232 | O=C(Nc1ccccc1)c1cc(Br)cc(Br)c1O 0
233 | CC(CC1=CC=CC=C1)NC 0
234 | [C@H]23[C@@H]([C@@]1(C(=CC(=O)C=C1)CC2)C)CC[C@]4([C@H]3CC[C@]4(C)O)C 0
235 | CN(C)CCN(CC1=CC=CS1)C2=CC=CC=N2 0
236 | CC1=CC=CC=C1N2C(=NC3=CC=CC=C3C2=O)C 0
237 | CN1CCC2=CC(=C(C=C2C1CCC3=CC=C(C=C3)Cl)OC)OC 0
238 | COC(F)(F)C(Cl)Cl 0
239 | COC(=O)C(C1CCCCN1)C2=CC=CC=C2 0
240 | CCC1(C(=O)C(CNC1=O)C)CC 0
241 | CC1=CC(=C(C(=C1OC(=O)C)C)C)OCC(CNC(C)C)O 0
242 | CC(=O)NC1=C(C(=C(C(=C1I)C(=O)NC2C(C(C(OC2O)CO)O)O)I)N(C)C(=O)C)I 0
243 | CN1CCN2C(C1)C3=CC=CC=C3CC4=CC=CC=C42 0
244 | CC(C)C1C2=C(CCC1(CCN(C)CCCC3=NC4=CC=CC=C4N3)OC(=O)COC)C=C(C=C2)F 0
245 | CCCCN1CC(C(C(C1CO)O)O)O 0
246 | CC1=CC(=NN=C1NCCN2CCOCC2)C3=CC=CC=C3 0
247 | CN(C)C1C2CC3CC4=C(C=CC(=C4C(=C3C(=O)C2(C(=C(C1=O)C(=O)N)O)O)O)O)N(C)C 0
248 | CCOC(=NC1=C[N+](=NO1)N2CCOCC2)[O-] 0
249 | CC1=CC(=C(C=C1OC(=O)C)C(C)C)OCCN(C)C 0
250 | CC(C1=CC(=C(C=C1)Cl)Cl)N2C(=O)CC(=N2)N 0
251 | CCCCCCCCCC(=O)OC1CCC2C1(CCC3C2CCC4=CC(=O)CCC34)C 0
252 | CC12CCC3C(C1CCC2OC(=O)CCC4=CC=CC=C4)CCC5=CC(=O)CCC35 0
253 | CCC1=NN(C(=O)N1CCOC2=CC=CC=C2)CCCN3CCN(CC3)C4=CC(=CC=C4)Cl 0
254 | CC1=C2C(=NC=C1)N(C3=C(C=CC=N3)C(=O)N2)C4CC4 0
255 | C1=CC=C(C=C1)CNC(=O)CCNNC(=O)C2=CC=NC=C2 0
256 | CO[C@]12C[C@@H](COC(=O)c3cncc(Br)c3)CN(C)[C@@H]1Cc1cn(C)c3cccc2c13 0
257 | CC1=C(C(C(=C(N1)C)C(=O)OC)C2=CC=CC=C2[N+](=O)[O-])C(=O)OC 0
258 | C1=CC(=CC=C1C(=O)NN=CC2=CC=C(O2)[N+](=O)[O-])O 0
259 | CCN(CC)C(=O)C1=CN=CC=C1 0
260 | CS(=O)(=O)NC1=C(C=C(C=C1)[N+](=O)[O-])OC2=CC=CC=C2 0
261 | CN(C)CC1=CC=C(O1)CSCCNC(=C[N+](=O)[O-])NCC2=CC3=C(C=C2)OCO3 0
262 | CC1=NC(=CN1C2=CC=C(C=C2)[N+](=O)[O-])[N+](=O)[O-] 0
263 | C1=C(OC(=C1)[N+](=O)[O-])C=NNC(=O)N 0
264 | C1=C(OC(=C1)[N+](=O)[O-])C=CC(=NN=C(N)N)C=CC2=CC=C(O2)[N+](=O)[O-] 0
265 | C1=CC2=C(C=CC(=C2N=C1)O)[N+](=O)[O-] 0
266 | CN1CC(C2=C(C1)C(=CC=C2)N)C3=CC=CC=C3 0
267 | CN1CCC2=CC3=C(C(=C2C1C4C5=C(C(=C(C=C5)OC)OC)C(=O)O4)OC)OCO3 0
268 | CC1=C(C=CC2=C1OC(=O)C(=C2O)NC(=O)C3=CC(=C(C=C3)O)CC=C(C)C)OC4C(C(C(C(O4)(C)C)OC)OC(=O)N)O 0
269 | CC1=CN=C(C(=C1OC)C)CS(=O)C2=NC3=C(N2)C=C(C=C3)OC 0
270 | CC(C)NCC(C1=CC(=CC(=C1)O)O)O 0
271 | CCC(CC)(C1=CC=CC=C1)C(=O)OCCOCCN(CC)CC 0
272 | CCN(CC)CCC1=NC(=NO1)C2=CC=CC=C2 0
273 | CC(CN1C2=CC=CC=C2S(=O)(=O)C3=CC=CC=C31)CN(C)C 0
274 | COc1ccc2c3c1O[C@H]1C(=O)CC[C@@]4(O)[C@@H](C2)N(C)CC[C@]314 0
275 | CCCCC1C(=O)N(N(C1=O)C2=CC=C(C=C2)O)C3=CC=CC=C3 0
276 | CC(=O)OC1=CC=C(C=C1)C2(C3=CC=CC=C3NC2=O)C4=CC=C(C=C4)OC(=O)C 0
277 | C1=CC=C2C(=C1)C(C(=O)N2)(C3=CC=C(C=C3)O)C4=CC=C(C=C4)O 0
278 | CN(C)CC(=O)OC[C@@H](O)[C@@H](O)[C@H](O)[C@@H](O)C(=O)O 0
279 | C1C(C(C(C(C1N)OC2C(C(C(C(O2)CO)O)O)N)OC3C(C(C(O3)CO)OC4C(C(C(C(O4)CN)O)O)N)O)O)N 0
280 | CCC(=O)NS(=O)(=O)C1=CC=C(C=C1)C2=C(ON=C2C3=CC=CC=C3)C 0
281 | CN(CC#C)CC1=CC=CC=C1 0
282 | C1=CC=C(C=C1)C2C(=O)N=C(O2)N 0
283 | CCCC(C)C1(C(=O)NC(=O)NC1=O)CC 0
284 | O[C@@H]1CO[C@@H](O[C@@H]2CO[C@@H](O)[C@H](OS(O)(=O)=O)[C@H]2OS(O)(=O)=O)[C@H](OS(O)(=O)=O)[C@H]1OS(O)(=O)=O 0
285 | C1CCC2=NN=NN2CC1 0
286 | CCCN1CC(CC2C1CC3=CNC4=CC=CC2=C34)CSC 0
287 | C1CCC(CC1)C(CC2CCCCN2)C3CCCCC3 0
288 | CCOC1=CC=C(C=C1)NC(=O)C 0
289 | CC1=CC(=O)N(N1C)C2=CC=CC=C2 0
290 | C1=CC=C(C=C1)N=NC2=C(N=C(C=C2)N)N 0
291 | CC1C(OCCN1C)C2=CC=CC=C2 0
292 | C1=CC=C(C=C1)CCN=C(N)N=C(N)N 0
293 | C1=CC=C(C=C1)NNC(=O)N 0
294 | CC1C(OCCN1)C2=CC=CC=C2 0
295 | CCC1(C(=O)NC(=O)NC1=O)C2=CC=CC=C2 0
296 | C1=CC=C(C=C1)O 0
297 | C1=CC=C2C(=C1)C(=O)OC2(C3=CC=C(C=C3)O)C4=CC=C(C=C4)O 0
298 | CC(COC1=CC=CC=C1)NN 0
299 | CC(C)(CC1=CC=CC=C1)N 0
300 | CCCCC1C(=O)N(N(C1=O)C2=CC=CC=C2)C3=CC=CC=C3 0
301 | CNCC(C1=CC(=CC=C1)O)O 0
302 | CC(C(C1=CC=CC=C1)O)N 0
303 | CC(C(C1=CC=CC=C1)O)N 0
304 | [H][C@@](C)(N)[C@]([H])(O)C1=CC=CC=C1 0
305 | C1=CC=C(C(=C1)C(=O)NC2=CC=C(C=C2)S(=O)(=O)NC3=NC=CS3)C(=O)O 0
306 | CC(=NO)C1=CC=C(C=C1)OCC(=O)N2CCCCC2 0
307 | CCC1=CN=C(C=C1)CCOC2=CC=C(C=C2)CC3C(=O)NC(=O)S3 0
308 | C1CN(CCC1C(=O)N)CCCN2C3=CC=CC=C3SC4=C2C=C(C=C4)Cl 0
309 | CC[N+]1(CCCC(C1)OC(=O)C(C2=CC=CC=C2)(C3=CC=CC=C3)O)C 0
310 | C1CNCCN1 0
311 | C1CCNC(C1)C(C2=CC=CC=C2)(C3=CC=CC=C3)O 0
312 | CC(C1=CC(=C(C=C1)N2CC=CC2)Cl)C(=O)O 0
313 | C1C(O1)CCl 0
314 | CCCCCCC(CC=CCCCCCCCOC(=O)OCCOCC(COCCOC(=O)OCCCCCCCC=CCC(CCCCCC)O)OCCOC(=O)OCCCCCCCC=CCC(CCCCCC)O)O 0
315 | C=CN1CCCC1=O 0
316 | CC(C)NCC(COC1=CC=C(C=C1)NC(=O)C)O 0
317 | CCCNC1CCC2=C(C1)SC(=N2)N 0
318 | CC(CC1=CC=CC=C1)NCCC(C2=CC=CC=C2)C3=CC=CC=C3 0
319 | CC(C)(C)C1=CC(=CC(=C1O)C(C)(C)C)SC(C)(C)SC2=CC(=C(C(=C2)C(C)(C)C)O)C(C)(C)C 0
320 | CCCN(CCC)C(=O)C(CCC(=O)O)NC(=O)C1=CC=CC=C1 0
321 | CC(C)NCC(C1=CC2=CC=CC=C2C=C1)O 0
322 | CCCOC(=O)CC1=CC(=C(C=C1)OCC(=O)N(CC)CC)OC 0
323 | CC(C)C1=C(C(=CC=C1)C(C)C)O 0
324 | CCC(=O)OC(CC1=CC=CC=C1)(C2=CC=CC=C2)C(C)CN(C)C 0
325 | CC1=C(C(=O)N(N1C)C2=CC=CC=C2)C(C)C 0
326 | CC(CC1(C(=O)NC(=O)NC1=O)CC=C)O 0
327 | CC(C(C1=CC=CC=C1)O)NC 0
328 | C(C1(C(C=CNC1=O)=O)CC)C 0
329 | CC1=NC=C(C(=C1O)CO)CSSCC2=CN=C(C(=C2CO)O)C 0
330 | CCCC(C(=O)C1=CC=C(C=C1)C)N2CCCC2 0
331 | C1CC2CCCN2C1 0
332 | CNC(=C[N+](=O)[O-])NCCSCC1=CC=C(O1)CN(C)C 0
333 | CCC(=O)OC1C(CC2C1(CCC3C2CCC4C3(CC(C(C4)OC(=O)C)N5CCCCC5)C)C)[N+]6(CCCCC6)CC=C 0
334 | CCN1CCCC1CNC(=O)C2=C(C=CC(=C2OC)Br)OC 0
335 | COC(=O)[C@H]1[C@H]2C[C@@H]3c4[nH]c5cc(OC)ccc5c4CCN3C[C@H]2C[C@@H](OC(=O)c2cc(OC)c(OC)c(OC)c2)[C@@H]1OC 0
336 | [C@@H]2([N]1N=C(C(N)=O)N=C1)O[C@H](CO)[C@H]([C@H]2O)O 0
337 | CC1=C(N(N=C1C(=O)NN2CCCCC2)C3=C(C=C(C=C3)Cl)Cl)C4=CC=C(C=C4)Cl 0
338 | CS(=O)(=O)C1=CC=C(C=C1)C2=C(C(=O)OC2)C3=CC=CC=C3 0
339 | CN(CCOC1=CC=C(C=C1)CC2C(=O)NC(=O)S2)C3=CC=CC=N3 0
340 | CC1C2CCC3(C=CC(=O)C(=C3C2OC1=O)C)C 0
341 | CCCC(C)C1(C(=O)NC(=O)NC1=O)CC=C 0
342 | CC(CC1=CC=CC=C1)N(C)CC#C 0
343 | C1CN(CCC1C2=CN(C3=C2C=C(C=C3)Cl)C4=CC=C(C=C4)F)CCN5CCNC5=O 0
344 | CC(C)CC(C1(CCC1)C2=CC=C(C=C2)Cl)N(C)C 0
345 | CC1=CC2=C(C=C1CC(=O)C3=C(C=CS3)S(=O)(=O)NC4=C(C(=NO4)C)Cl)OCO2 0
346 | C1=C(C(=O)NC(=O)N1C2C(C(C(O2)CO)O)O)C=CBr 0
347 | CC(C)NCC(C1=CC=C(C=C1)NS(=O)(=O)C)O 0
348 | CC1CN(CC(N1)C)C2=C(C(=C3C(=C2F)N(C=C(C3=O)C(=O)O)C4CC4)N)F 0
349 | C1CCN2CC3CC(C2C1)CN4C3CCCC4 0
350 | C1CN2CC3=CCOC4CC(=O)N5C6C4C3CC2C61C7=CC=CC=C75 0
351 | C1=CC(=CC=C1N)S(=O)(=O)NC(=O)N 0
352 | CC(=CC(=O)NS(=O)(=O)C1=CC=C(C=C1)N)C 0
353 | COC1=NC(=NC(=C1)NS(=O)(=O)C2=CC=C(C=C2)N)OC 0
354 | CC1=CC(=NC(=N1)NS(=O)(=O)C2=CC=C(C=C2)N)C 0
355 | C1=CC(=CC=C1N)S(=O)(=O)N=C(N)N 0
356 | CC1=NN=C(S1)NS(=O)(=O)C2=CC=C(C=C2)N 0
357 | COC1=NN=C(C=C1)NS(=O)(=O)C2=CC=C(C=C2)N 0
358 | COC1=CN=C(N=C1)NS(=O)(=O)C2=CC=C(C=C2)N 0
359 | C1=CC(=CC=C1N)S(=O)(=O)N 0
360 | C1=CC(=CC=C1N)S(=O)(=O)NC2=NC=CS2 0
361 | CC1=CC(=NC(=N1)C)NS(=O)(=O)C2=CC=C(C=C2)N 0
362 | CCCCCCCCNC(C)C(C1=CC=C(C=C1)SC(C)C)O 0
363 | CC(C(C1=CC=C(C=C1)O)O)NC 0
364 | CC(C1=CC=C(C=C1)C(=O)C2=CC=CS2)C(=O)O 0
365 | CCCCC1(C(=O)N(N(C1=O)C2=CC=CC=C2)C3=CC=CC=C3)COC(=O)CCC(=O)O 0
366 | CCCCCN=C(N)NN=CC1=CNC2=C1C=C(C=C2)OC 0
367 | CCC[C@H](NC(=O)[C@@H]1[C@H]2CCC[C@H]2CN1C(=O)[C@@H](NC(=O)[C@@H](NC(=O)c3cnccn3)C4CCCCC4)C(C)(C)C)C(=O)C(=O)NC5CC5 0
368 | CC1CN(CCN1)C2=C(C=C3C(=C2)N(C=C(C3=O)C(=O)O)C4=C(C=C(C=C4)F)F)F 0
369 | CN1C2=C(C=C(C=C2)Cl)C(=NC(C1=O)O)C3=CC=CC=C3 0
370 | CC(C)N1CCN(CC1)C2=CC=C(C=C2)OCC3COC(O3)(CN4C=NC=N4)C5=C(C=C(C=C5)Cl)Cl 0
371 | CC(C)(C)C1=CC=C(C=C1)C(CCCN2CCC(CC2)C(C3=CC=CC=C3)(C4=CC=CC=C4)O)O 0
372 | CC(CC(C1=CC=CC=C1)C2=CC=CC=C2)NC(C)(C)C 0
373 | CCC(=O)OC1CCC2C1(CCC3C2CCC4=CC(=O)CCC34C)C 0
374 | C1=CC=C(C=C1)NC(=O)C2=C(C(=C(C(=C2Cl)Cl)Cl)Cl)O 0
375 | CC1(C2CC3C(C(=O)C(=C(C3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O 0
376 | CN1C(=O)CN=C(C2=C1C=CC(=C2)Cl)C3=CCCCC3 0
377 | C1CC(=O)NC(=O)C1N2C(=O)C3=CC=CC=C3C2=O 0
378 | CN1CCC(CC1)N(CC2=CC=CS2)C3=CC=CC=C3 0
379 | CC[C@H](C)[C@]1(CC)C(=NC(=S)NC1=O)[O-] 0
380 | CN1CCCCC1CCN2C3=CC=CC=C3SC4=C2C=C(C=C4)SC 0
381 | C1=CSC(=C1)C(=O)C2=C(C(=C(C=C2)OCC(=O)O)Cl)Cl 0
382 | CC1=CC(=C(C2=C1C=CC=N2)O)Br 0
383 | C1C(NCS1)C(=O)O 0
384 | CC1=C(C(=CC=C1)C)NC(=O)C(C)N 0
385 | CC1=CC=C(C=C1)C(=O)C2=CC(=C(C(=C2)O)O)[N+](=O)[O-] 0
386 | CN(CC(=O)O)C(=S)C1=CC=CC2=C1C=CC(=C2C(F)(F)F)OC 0
387 | [C@H]2(C1=CC(=CC=C1N([C@H](CC)C2)C(=O)OCC)C(F)(F)F)N(C(=O)OC)CC3=CC(=CC(=C3)C(F)(F)F)C(F)(F)F 0
388 | C1C(C1N)C2=CC=CC=C2 0
389 | C1CN(CCN1CCCN2C(=O)N3C=CC=CC3=N2)C4=CC(=CC=C4)Cl 0
390 | CC1=C(C(CCC1)(C)C)C=CC(=CC=CC(=CC(=O)O)C)C 0
391 | CC(=O)N1C2=CC=CC=C2C(C1=O)(C3=CC=C(C=C3)OC(=O)C)C4=CC=C(C=C4)OC(=O)C 0
392 | CC1=NN=C2N1C3=C(C=C(C=C3)Cl)C(=NC2)C4=CC=CC=C4Cl 0
393 | O=C(Nc1ccc(Br)cc1)c1cc(Br)cc(Br)c1O 0
394 | CC(Cl)(Cl)Cl 0
395 | CN(C)CCOC1=CC=C(C=C1)CNC(=O)C2=CC(=C(C(=C2)OC)OC)OC 0
396 | CCN(CC)CCOC1=CC=C(C=C1)C(CC2=CC=C(C=C2)Cl)(C3=CC=C(C=C3)C)O 0
397 | CC1=C(C2=C(CCC(O2)(C)COC3=CC=C(C=C3)CC4C(=O)NC(=O)S4)C(=C1O)C)C 0
398 | C1C2C(C2N)CN1C3=C(C=C4C(=O)C(=CN(C4=N3)C5=C(C=C(C=C5)F)F)C(=O)O)F 0
399 | C1=CC=C2C(=C1)C(=CN2)CC(C(=O)O)N 0
400 | CC(=O)O[C@]1(C(C)=O)CC[C@H]2[C@@H]3CCC4=CC(=O)CCC4=C3[C@@H](c3ccc(N(C)C)cc3)C[C@@]21C 0
401 | CCOC(=O)N 0
402 | CC1=C(C(=NO1)C2=CC=CC=C2)C3=CC=C(C=C3)S(=O)(=O)N 0
403 | COC1=CC(=CC(=C1OC)C(=O)NCC2CCCN2CC=C)S(=O)(=O)N 0
404 | CCC=C(C)C1(C(=O)NC(=O)NC1=O)CC 0
405 | CCC12CCCN3C1C4=C(CC3)C5=CC=CC=C5N4C(C2)(C(=O)OC)O 0
406 | C=CCl 0
407 | [C@]2(OC1=C(C(=C(C(=C1CC2)C)O)C)C)(CCC[C@@H](CCC[C@@H](CCCC(C)C)C)C)C 0
408 | CCOC(C(=O)C1=CC=C(C=C1)C2=CC=CC=C2)NC3=CC=C(C=C3)C(=O)O 0
409 | CCOC(=O)CNC(C1CCCCC1)C(=O)N2CCC2C(=O)NCC3=CC=C(C=C3)C(=NO)N 0
410 | CN(C)CC=C(C1=CC=C(C=C1)Br)C2=CN=CC=C2 0
411 | COC(CN1CCN(CC1)CC(C(C2=CC=CC=C2)OC)O)C3=CC=CC=C3 0
412 | CC1=C(N(C(=C1)CC(=O)O)C)C(=O)C2=CC=C(C=C2)Cl 0
413 | CN1CCN(CC1)C(=O)OC2C3=NC=CN=C3C(=O)N2C4=NC=C(C=C4)Cl 0
414 |
415 |
416 |
--------------------------------------------------------------------------------
/Tutorial.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "fa1cbc05",
6 | "metadata": {},
7 | "source": [
8 | "# D-GCAN Deep Dive"
9 | ]
10 | },
11 | {
12 | "cell_type": "markdown",
13 | "id": "d4a486bf",
14 | "metadata": {},
15 | "source": [
16 | "In this tutorial, we take a deep dive into D-GCAN and show how it builds a drug-likeness prediction model from scratch.\n",
17 | "\n",
18 | "Let's start!"
19 | ]
20 | },
21 | {
22 | "cell_type": "markdown",
23 | "id": "d7cacf2e",
24 | "metadata": {},
25 | "source": [
26 | "## Part I: Overview of D-GCAN and Data"
27 | ]
28 | },
29 | {
30 | "cell_type": "markdown",
31 | "id": "43a44a94",
32 | "metadata": {},
33 | "source": [
34 | "The drug-likeness has been widely used as a criterion to distinguish drug-like molecules from non-drugs. Developing reliable computational methods to predict drug-likeness of compounds is crucial to triage unpromising molecules and accelerate the drug discovery process.In this study, a deep learning method was developed to predict drug-likeness based on the graph convo-lutional attention network (D-GCAN) directly from molecular structures. The model combined the ad-vantages of graph convolution and attention mechanism. Results showed that the D-GCAN outper-formed other state-of-the-art models for drug-likeness prediction. Molecular graph was used as encoding method for drug-likeness prediction.\n",
35 | "\n",
36 | "A dataset with enough drugs and non-drugs is the prerequisite to train accurate deep neural network models for prediction of drug-likeness.In this study, D-GCAN model was trained on the dataset released by Beker, which consists of drug and non-drug sets (abbrevi-ated as: Drugs and Non-drugs). The Drugs set includes 2136 FDA small-molecule drugs assembled from Drugbank. The Non-drugs was chosen from ZINC15. Compounds with a maximum fingerprint-based Tanimoto similarity to drugs above 0.85 were removed, and standard binary classification was used to itera-tively refine the set of reliable negative set. Since the negative set is much larger than the positive set, it was randomly down-sampled to create a balanced dataset for model training. The dataset was randomly divided into training, validation, and test sets at ratio 8:1:1. In addition, two additional datasets, the non-US dataset and the bRo5 dataset, were used to test the performance of the model. The non-US dataset composes of 1281 word-wide drugs from Drugbank and an equal size of non-drugs from ZINC15. The bRo5 dataset includes 135 FDA and non-US drugs beyond Ro5 space (bRo5). The GDB-13 data-base was used to test the ability of D-GCAN in screening large-scale data. It consists of about 977 million drug-like small molecules according to Lipinski’s rule. All molecules contain up to 13 heavy atoms , and they were stored in the canonical SMILES. All the independent test datasets and validation dataset were not used in the training process.\n"
37 | ]
38 | },
39 | {
40 | "cell_type": "markdown",
41 | "id": "e597ea85",
42 | "metadata": {},
43 | "source": [
44 | "## Part II: To train the model"
45 | ]
46 | },
47 | {
48 | "cell_type": "code",
49 | "execution_count": 1,
50 | "id": "07e8ed86",
51 | "metadata": {},
52 | "outputs": [],
53 | "source": [
54 | "import train"
55 | ]
56 | },
57 | {
58 | "cell_type": "code",
59 | "execution_count": 2,
60 | "id": "dcca4e9f",
61 | "metadata": {},
62 | "outputs": [
63 | {
64 | "name": "stdout",
65 | "output_type": "stream",
66 | "text": [
67 | "The code uses a GPU!\n",
68 | "----------------------------------------------------------------------------------------------------\n",
69 | "Just a moment......\n",
70 | "----------------------------------------------------------------------------------------------------\n",
71 | "../dataset/data_train.txt\n",
72 | "../dataset/data_test.txt\n",
73 | "The preprocess has finished!\n",
74 | "# of training data samples: 3802\n",
75 | "# of test data samples: 428\n",
76 | "----------------------------------------------------------------------------------------------------\n",
77 | "Creating a model.\n",
78 | "# of model parameters: 311698\n",
79 | "----------------------------------------------------------------------------------------------------\n",
80 | "Start training.\n",
81 | "The result is saved in the output directory every epoch!\n",
82 | "The training will finish in about 0 hours 21 minutes.\n",
83 | "----------------------------------------------------------------------------------------------------\n",
84 | "Epoch\tTime(sec)\tLoss_train\tLoss_test\tAUC_train\tAUC_test\n",
85 | "1\t9.334350300000011\t318.02376973629\t33.23613902926445\t0.6330387783115992\t0.5116822429906542\n",
86 | "2\t16.232699300000007\t275.59704649448395\t28.36697283387184\t0.7726390395642837\t0.5233644859813084\n",
87 | "3\t22.533227600000004\t258.0953543186188\t26.692712754011154\t0.8227923594659818\t0.530373831775701\n",
88 | "4\t30.035969300000005\t244.29262351989746\t25.99587020277977\t0.8555440089003034\t0.5934579439252337\n",
89 | "5\t36.20996820000002\t235.61571648716927\t26.206634640693665\t0.8711811167445258\t0.544392523364486\n",
90 | "6\t42.15839070000001\t235.10905063152313\t24.74921104311943\t0.8782201330617486\t0.5771028037383178\n",
91 | "7\t48.091756000000004\t230.60763642191887\t24.482909947633743\t0.8858224754798858\t0.6308411214953271\n",
92 | "8\t53.9427063\t225.9473716020584\t25.168260991573334\t0.894445889698231\t0.5794392523364486\n",
93 | "9\t59.887297399999994\t220.88472372293472\t23.143073588609695\t0.9038094737308211\t0.6845794392523364\n",
94 | "10\t65.88160690000001\t220.29008296132088\t23.10640263557434\t0.9080117951159034\t0.6542056074766355\n",
95 | "11\t71.7950055\t220.04156962037086\t23.67304638028145\t0.905873895764607\t0.7289719626168224\n",
96 | "12\t77.81804110000002\t214.85031658411026\t23.34747040271759\t0.9159177330794608\t0.8200934579439252\n",
97 | "13\t83.67658890000001\t212.33444252610207\t23.14932319521904\t0.9178444716275156\t0.735981308411215\n",
98 | "14\t89.6064955\t211.54040449857712\t22.778073489665985\t0.9219235005645715\t0.7593457943925234\n",
99 | "15\t95.5943882\t208.26400744915009\t22.901916056871414\t0.9267551530985012\t0.7663551401869159\n",
100 | "16\t101.6206004\t209.3945328295231\t23.913705557584763\t0.9246417461863752\t0.6915887850467289\n",
101 | "17\t107.65320110000002\t206.03158766031265\t23.282782286405563\t0.930114906901056\t0.7313084112149533\n",
102 | "18\t113.61952920000002\t207.53857171535492\t22.225304275751114\t0.9226543992295261\t0.8037383177570093\n",
103 | "19\t120.54484979999998\t204.79183167219162\t23.475462794303894\t0.926265719441185\t0.735981308411215\n",
104 | "20\t126.48899880000002\t205.36031165719032\t22.78501933813095\t0.9291473863661524\t0.8107476635514018\n",
105 | "21\t132.3617932\t202.3321330845356\t23.385528802871704\t0.9302150906635376\t0.7897196261682243\n",
106 | "22\t138.31940950000003\t202.55410113930702\t23.08760157227516\t0.9293607611751943\t0.7686915887850467\n",
107 | "23\t144.17849070000003\t198.95897144079208\t22.36356022953987\t0.9377836694932141\t0.8294392523364486\n",
108 | "24\t150.04248400000003\t197.13710144162178\t23.3654263317585\t0.9351247869019417\t0.8084112149532711\n",
109 | "25\t155.89731129999998\t256.36723348498344\t22.31619429588318\t0.7880461675559591\t0.8411214953271028\n",
110 | "26\t161.73105990000002\t199.1333883702755\t22.19395723938942\t0.9381437221865521\t0.8200934579439252\n",
111 | "27\t167.57376660000003\t195.61116680502892\t21.885735362768173\t0.9357744592290832\t0.8621495327102804\n",
112 | "28\t173.4351409\t196.21020331978798\t21.808892458677292\t0.937718217946731\t0.8808411214953271\n",
113 | "29\t179.35259220000003\t196.93134278059006\t22.267054110765457\t0.9385652135408595\t0.8481308411214953\n",
114 | "30\t185.28482499999998\t195.89555063843727\t22.040870487689972\t0.9386104622844113\t0.8434579439252337\n",
115 | "31\t191.16947059999998\t194.0237057507038\t22.781775504350662\t0.9417760754533178\t0.8387850467289719\n",
116 | "32\t197.0987102\t193.68072113394737\t22.449314266443253\t0.9423293001527663\t0.8294392523364486\n",
117 | "33\t203.01129849999998\t192.5338954925537\t22.377066612243652\t0.9452480516748955\t0.8621495327102804\n",
118 | "34\t209.6004434\t192.58278796076775\t23.285291463136673\t0.9402174153696283\t0.8714953271028038\n",
119 | "35\t216.03077720000002\t196.01435166597366\t24.061037868261337\t0.9359749651294087\t0.8014018691588785\n",
120 | "36\t222.4218536\t193.9636361002922\t22.313345968723297\t0.936061864857086\t0.8434579439252337\n",
121 | "37\t228.7779303\t192.51033294200897\t22.285043627023697\t0.9435566896185268\t0.8317757009345794\n",
122 | "38\t235.1833634\t188.01407945156097\t22.830698162317276\t0.94882975955897\t0.8014018691588785\n",
123 | "39\t241.73452799999998\t193.91294729709625\t22.711496233940125\t0.9402640478668054\t0.8294392523364486\n",
124 | "40\t248.1350879\t192.2110168337822\t21.79123494029045\t0.9453036785706378\t0.8785046728971962\n",
125 | "41\t254.51809260000002\t189.3926584124565\t23.183754086494446\t0.9464263178869528\t0.822429906542056\n",
126 | "42\t260.67076799999995\t197.67854461073875\t24.210958123207092\t0.9356472922709057\t0.8714953271028038\n",
127 | "43\t266.8244926\t195.8016073703766\t22.462971657514572\t0.9351026468439347\t0.8107476635514018\n",
128 | "44\t273.0223882\t191.97943636775017\t23.224840223789215\t0.940738813735692\t0.8247663551401869\n",
129 | "45\t279.0850431\t190.848837941885\t22.82283341884613\t0.9461044567936768\t0.8060747663551402\n",
130 | "46\t285.1555958\t190.04618108272552\t22.433310955762863\t0.9440021199105542\t0.7967289719626168\n",
131 | "47\t291.1587971\t190.5216095149517\t22.426137387752533\t0.9457633615250072\t0.822429906542056\n",
132 | "48\t297.2169235\t185.92078268527985\t22.22221177816391\t0.9480603925432284\t0.8247663551401869\n",
133 | "49\t303.3877255\t187.782156676054\t22.887968957424164\t0.94423528239644\t0.8364485981308412\n",
134 | "50\t309.40409650000004\t187.28414443135262\t21.483285009860992\t0.9458896982310093\t0.8714953271028038\n",
135 | "51\t315.44450670000003\t185.18417713046074\t21.38184556365013\t0.9481489527752565\t0.8785046728971962\n",
136 | "52\t321.7301946\t182.16105404496193\t24.673764526844025\t0.9509743009276684\t0.7920560747663551\n",
137 | "53\t327.84551209999995\t188.7527618408203\t23.513393253087997\t0.9457229559191445\t0.8154205607476636\n",
138 | "54\t334.0279252\t185.709531635046\t21.631520986557007\t0.9463949066796555\t0.8808411214953271\n",
139 | "55\t340.16017209999995\t185.12931755185127\t22.429152816534042\t0.944120984346979\t0.8294392523364486\n",
140 | "56\t346.29319970000006\t182.88407680392265\t21.58257967233658\t0.9498509697345405\t0.8761682242990654\n",
141 | "57\t357.14253529999996\t182.04424741864204\t21.475889027118683\t0.9525705991099698\t0.8808411214953271\n",
142 | "58\t363.8143368\t182.934487760067\t21.883195608854294\t0.949658766355968\t0.8714953271028038\n",
143 | "59\t370.1603794\t184.17358297109604\t21.290808767080307\t0.9471534804171187\t0.883177570093458\n",
144 | "60\t376.4684089\t181.42354640364647\t21.597694754600525\t0.949361674452587\t0.9042056074766355\n",
145 | "61\t382.88365710000005\t187.25566163659096\t21.6785786151886\t0.9452393340270551\t0.866822429906542\n",
146 | "62\t389.3225355000001\t181.59250125288963\t21.666670441627502\t0.9521657127991676\t0.8598130841121495\n",
147 | "63\t396.0132287\t179.8839019536972\t22.01644539833069\t0.9538880709367459\t0.852803738317757\n",
148 | "64\t402.58914460000005\t182.93770709633827\t22.33838379383087\t0.9484886642903003\t0.8504672897196262\n",
149 | "65\t409.2413564000001\t181.58496183156967\t22.23741576075554\t0.950153319901698\t0.8785046728971962\n",
150 | "66\t415.7737618\t182.49673774838448\t21.934344708919525\t0.9507491642128103\t0.8785046728971962\n",
151 | "67\t422.29757770000003\t180.1727076768875\t22.335491836071014\t0.9534794484911551\t0.852803738317757\n",
152 | "68\t428.7057098\t182.3468733727932\t21.559545934200287\t0.953210031660283\t0.8855140186915887\n",
153 | "69\t435.1430776000001\t177.6970148384571\t21.813909739255905\t0.9542144984169858\t0.8785046728971962\n",
154 | "70\t441.59579740000004\t179.4230616092682\t21.47458705306053\t0.9503143888236987\t0.8925233644859814\n",
155 | "71\t447.9559312\t187.4236896932125\t21.997405976057053\t0.9434714503952\t0.8714953271028038\n",
156 | "72\t454.36015280000004\t185.64457353949547\t21.80859535932541\t0.9493346912568912\t0.8995327102803738\n",
157 | "73\t460.8502926\t183.28448390960693\t22.7678345143795\t0.942662231275046\t0.8504672897196262\n",
158 | "74\t467.7673221\t181.35295176506042\t21.99883532524109\t0.9524445391546925\t0.8317757009345794\n",
159 | "75\t474.20574880000004\t180.42559936642647\t22.20555028319359\t0.9545189242145815\t0.8621495327102804\n",
160 | "76\t480.7348668000001\t177.4391260445118\t21.684407979249954\t0.9532330019704651\t0.8878504672897196\n",
161 | "77\t488.22190610000007\t187.34195244312286\t22.080274641513824\t0.9482340536232203\t0.8738317757009346\n",
162 | "78\t498.7163915\t184.663908213377\t22.014076620340347\t0.9470819403546837\t0.8691588785046729\n"
163 | ]
164 | },
165 | {
166 | "name": "stdout",
167 | "output_type": "stream",
168 | "text": [
169 | "79\t507.72747260000006\t178.98830798268318\t21.47413921356201\t0.9541025527486882\t0.8598130841121495\n",
170 | "80\t516.8672275\t178.3373854458332\t21.658688694238663\t0.9525628500896672\t0.8901869158878505\n",
171 | "81\t526.2272582\t176.8597036600113\t22.024581998586655\t0.9535130737042532\t0.8644859813084113\n",
172 | "82\t537.2631751\t177.2030012011528\t21.766023725271225\t0.957143212965218\t0.8691588785046729\n",
173 | "83\t548.0884053000001\t176.38141465187073\t21.79708757996559\t0.9572966712422787\t0.8714953271028038\n",
174 | "84\t559.8119843000001\t174.46399101614952\t21.843281388282776\t0.956292066110213\t0.8574766355140186\n",
175 | "85\t569.1060591\t175.65917918086052\t21.409487038850784\t0.9559244027719352\t0.9018691588785047\n",
176 | "86\t576.5913633\t176.77976202964783\t21.49503728747368\t0.9538743717758542\t0.8785046728971962\n",
177 | "87\t584.3266782000001\t179.85141596198082\t21.427332252264023\t0.9514658102154229\t0.9018691588785047\n",
178 | "88\t595.5166383000001\t178.52282038331032\t21.403560250997543\t0.9539066132353267\t0.8644859813084113\n",
179 | "89\t605.9234084000001\t177.05544209480286\t21.411171078681946\t0.9535158412115041\t0.9042056074766355\n",
180 | "90\t613.9635365\t176.31908676028252\t21.366370409727097\t0.955243734363584\t0.8878504672897196\n",
181 | "91\t623.9597695\t176.20382365584373\t21.893729746341705\t0.9558952055704386\t0.8761682242990654\n",
182 | "92\t634.0385619\t175.1233125925064\t22.482848435640335\t0.9574447328802002\t0.8621495327102804\n",
183 | "93\t644.2752579\t176.93210792541504\t21.549375027418137\t0.9527253027652932\t0.8948598130841121\n",
184 | "94\t652.8900821000001\t173.00296890735626\t21.5932075381279\t0.9579943598202227\t0.8855140186915887\n",
185 | "95\t667.6088093000001\t179.9282302260399\t23.50808882713318\t0.9537645017379945\t0.8341121495327103\n",
186 | "96\t676.2838057\t174.56020081043243\t22.648311734199524\t0.9556187315960767\t0.8714953271028038\n",
187 | "97\t685.9453472\t176.7171704173088\t21.891200184822083\t0.9524900646489693\t0.897196261682243\n",
188 | "98\t694.2790274\t178.38612964749336\t22.245244562625885\t0.9540746009254544\t0.8808411214953271\n",
189 | "99\t702.7636145\t177.73075929284096\t24.06598174571991\t0.9527554685943277\t0.897196261682243\n",
190 | "100\t713.3440201000001\t177.71725061535835\t23.09346652030945\t0.9563492151349435\t0.8434579439252337\n",
191 | "101\t722.6864448\t173.766254901886\t22.383565932512283\t0.9588301470099851\t0.8785046728971962\n",
192 | "102\t731.0422685000001\t173.67894527316093\t21.84225881099701\t0.9581018774769188\t0.8855140186915887\n",
193 | "103\t739.0269975\t175.92625331878662\t22.260210156440735\t0.956732791639914\t0.8691588785046729\n",
194 | "104\t746.7896567\t174.88757956027985\t21.68474268913269\t0.95813674806828\t0.8901869158878505\n",
195 | "105\t754.1008847\t176.1177335381508\t22.008816480636597\t0.9573001306263422\t0.8785046728971962\n",
196 | "106\t761.2548227000001\t174.49301874637604\t21.83292892575264\t0.9588928310492174\t0.897196261682243\n",
197 | "107\t768.4392893\t172.7093889117241\t21.64242872595787\t0.9596258053446101\t0.8878504672897196\n",
198 | "108\t775.9572361\t177.16424638032913\t21.55477637052536\t0.9525133117098767\t0.9065420560747663\n",
199 | "109\t784.4066813000001\t178.4102607667446\t21.33096119761467\t0.9554701164567051\t0.9042056074766355\n",
200 | "110\t794.4927042\t174.0306807756424\t21.91256058216095\t0.9572584796422166\t0.8714953271028038\n",
201 | "111\t802.6228819\t174.55561447143555\t21.62583690881729\t0.9593071268846725\t0.9088785046728972\n",
202 | "112\t810.7530621000001\t172.49658674001694\t22.11896824836731\t0.9582462029800518\t0.8901869158878505\n",
203 | "113\t818.6795158\t173.48215851187706\t21.262643307447433\t0.9584646976775077\t0.897196261682243\n",
204 | "114\t827.9049124000001\t174.29942700266838\t21.171885669231415\t0.9568379569154472\t0.9088785046728972\n",
205 | "115\t837.2002482\t171.870591878891\t21.214154481887817\t0.9590825436712644\t0.9158878504672897\n",
206 | "116\t845.4472041\t176.74994710087776\t22.71759131550789\t0.9544714614652291\t0.8995327102803738\n",
207 | "117\t853.6806366000001\t184.73798117041588\t21.845041394233704\t0.9432161478513075\t0.8995327102803738\n",
208 | "118\t868.1707754\t177.2356958091259\t21.59891825914383\t0.9571596796333606\t0.9112149532710281\n",
209 | "119\t875.8412158\t176.63044354319572\t21.46969723701477\t0.9547083600859034\t0.9088785046728972\n",
210 | "120\t884.2734249\t172.92364439368248\t21.97585704922676\t0.9586079161777404\t0.9065420560747663\n",
211 | "121\t892.1201223\t180.26964315772057\t21.951832473278046\t0.948673395399296\t0.9018691588785047\n",
212 | "122\t899.5396582999999\t173.81644931435585\t21.459405571222305\t0.9592527453671928\t0.8925233644859814\n",
213 | "123\t906.7295165\t176.33700492978096\t21.565876573324203\t0.9538397779352181\t0.8761682242990654\n",
214 | "124\t914.7522263999999\t172.69166892766953\t21.18627032637596\t0.9598193924768083\t0.9112149532710281\n",
215 | "125\t923.6481627000001\t171.38375091552734\t21.528594940900803\t0.9606258440897115\t0.8995327102803738\n",
216 | "126\t932.1785648\t171.18208953738213\t22.116858184337616\t0.9604675426749618\t0.8785046728971962\n",
217 | "127\t939.5199154\t171.14211875200272\t22.178175538778305\t0.9611412923151859\t0.883177570093458\n",
218 | "128\t946.4239123\t169.92953234910965\t21.887178242206573\t0.9611555449775278\t0.8808411214953271\n",
219 | "129\t954.5059573000001\t174.96560329198837\t22.6447791159153\t0.9555907797728429\t0.8761682242990654\n",
220 | "130\t961.8763295000001\t178.70918104052544\t22.300085812807083\t0.9529475335975381\t0.8808411214953271\n",
221 | "131\t969.1789554\t174.0145247578621\t22.113102048635483\t0.9579599043549494\t0.8808411214953271\n",
222 | "132\t976.3019077000001\t172.59970355033875\t21.501632899045944\t0.9596043571634159\t0.9018691588785047\n",
223 | "133\t984.5187846000001\t172.19679167866707\t22.107417851686478\t0.9594648747979719\t0.8808411214953271\n",
224 | "134\t993.8344443000001\t173.3250037431717\t23.72211918234825\t0.9588380344056502\t0.8341121495327103\n",
225 | "135\t1002.1266977\t173.15374860167503\t22.544156223535538\t0.9600561527221201\t0.8738317757009346\n",
226 | "136\t1009.9465580000001\t171.82129180431366\t22.224039256572723\t0.9606888048796689\t0.8738317757009346\n",
227 | "137\t1018.5052494000001\t171.67953670024872\t22.0196373462677\t0.9615298503332077\t0.8901869158878505\n",
228 | "138\t1028.1396465\t172.37993958592415\t22.44371086359024\t0.9589339285318929\t0.8808411214953271\n",
229 | "139\t1037.3752773\t174.99391075968742\t21.463502824306488\t0.9590761784045874\t0.9018691588785047\n",
230 | "140\t1046.0815085\t174.17848363518715\t21.192844033241272\t0.959691256891093\t0.9042056074766355\n",
231 | "auc: 0.9042056074766356\n",
232 | "bacc: 0.9042056074766356\n",
233 | "pre: 0.9303482587064676\n",
234 | "rec: 0.8738317757009346\n",
235 | "f1: 0.9012048192771085\n",
236 | "mcc: 0.8099069874057296\n",
237 | "sp: 0.9345794392523364\n",
238 | "q_: 0.8810572687224669\n",
239 | "acc: 0.9042056074766355\n"
240 | ]
241 | }
242 | ],
243 | "source": [
244 | "\n",
245 | "\n",
246 | "tes = train.train('../dataset/data_test.txt', #test set \n",
247 | " radius = 1, #hops of radius subgraph: 1, 2 \n",
248 | " dim = 52, #dimension of graph convolution layers\n",
249 | " layer_hidden = 4, #Number of graph convolution layers\n",
250 | " layer_output = 10, #Number of dense layers\n",
251 | " dropout = 0.45, #drop out rate :0-1\n",
252 | " batch_train = 8, # batch of training set\n",
253 | " batch_test = 8, #batch of test set\n",
254 | " lr =3e-4, #learning rate: 1e-5,1e-4,3e-4, 5e-4, 1e-3, 3e-3,5e-3\n",
255 | " lr_decay = 0.85, #Learning rate decay:0.5, 0.75, 0.85, 0.9\n",
256 | " decay_interval = 25,#Number of iterations for learning rate decay:10,25,30,50\n",
257 | " iteration = 140, #Number of iterations \n",
258 | " N = 5000, #length of embedding: 2000,3000,5000,7000 \n",
259 | " dataset_train='../dataset/data_train.txt') #training set\n",
260 | "\n"
261 | ]
262 | },
263 | {
264 | "cell_type": "markdown",
265 | "id": "d4a50302",
266 | "metadata": {},
267 | "source": [
268 | "## Part III: To test the performance of the D-GCAN on independent model"
269 | ]
270 | },
271 | {
272 | "cell_type": "markdown",
273 | "id": "581857d7",
274 | "metadata": {},
275 | "source": [
276 | "We have provided the trained model. And it can be used directly as follow:\n",
277 | "\n",
278 | "We test the trained model on bRo5 dataset.\n",
279 | "\n"
280 | ]
281 | },
282 | {
283 | "cell_type": "code",
284 | "execution_count": null,
285 | "id": "3d3ee166",
286 | "metadata": {},
287 | "outputs": [],
288 | "source": []
289 | },
290 | {
291 | "cell_type": "code",
292 | "execution_count": 4,
293 | "id": "68722dcd",
294 | "metadata": {},
295 | "outputs": [],
296 | "source": [
297 | "import predict"
298 | ]
299 | },
300 | {
301 | "cell_type": "code",
302 | "execution_count": 5,
303 | "id": "97d307e5",
304 | "metadata": {},
305 | "outputs": [
306 | {
307 | "name": "stdout",
308 | "output_type": "stream",
309 | "text": [
310 | "The code uses a GPU!\n",
311 | "../dataset/bRo5.txt\n",
312 | "SMILESis error\n",
313 | "bacc: 0.9580740740740741\n",
314 | "pre: 0.9696969696969697\n",
315 | "rec: 0.9481481481481482\n",
316 | "f1: 0.9588014981273408\n",
317 | "mcc: 0.9155786319049269\n",
318 | "sp: 0.968\n",
319 | "q_: 0.9453125\n",
320 | "acc: 0.9576923076923077\n"
321 | ]
322 | }
323 | ],
324 | "source": [
325 | "test = predict.predict('../dataset/bRo5.txt',\n",
326 | " radius = 1,\n",
327 | " property = True, #True if drug-likeness is known \n",
328 | " dim = 52 ,\n",
329 | " layer_hidden = 4,\n",
330 | " layer_output = 10,\n",
331 | " dropout = 0.45,\n",
332 | " batch_train = 8,\n",
333 | " batch_test = 8,\n",
334 | " lr = 3e-4,\n",
335 | " lr_decay = 0.85,\n",
336 | " decay_interval = 25 ,\n",
337 | " iteration = 140,\n",
338 | " N = 5000)"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": null,
344 | "id": "5fe34a0c",
345 | "metadata": {},
346 | "outputs": [],
347 | "source": [
348 | "Feedbacks would also be appreciated and you can send me an email (jinyusun@csu.edu.cn)!"
349 | ]
350 | }
351 | ],
352 | "metadata": {
353 | "kernelspec": {
354 | "display_name": "Python 3 (ipykernel)",
355 | "language": "python",
356 | "name": "python3"
357 | },
358 | "language_info": {
359 | "codemirror_mode": {
360 | "name": "ipython",
361 | "version": 3
362 | },
363 | "file_extension": ".py",
364 | "mimetype": "text/x-python",
365 | "name": "python",
366 | "nbconvert_exporter": "python",
367 | "pygments_lexer": "ipython3",
368 | "version": "3.8.8"
369 | }
370 | },
371 | "nbformat": 4,
372 | "nbformat_minor": 5
373 | }
374 |
--------------------------------------------------------------------------------