├── .editorconfig
├── .gitattributes
├── .github
├── ISSUE_TEMPLATE
│ ├── bug_report.md
│ ├── custom.md
│ └── feature_request.md
└── workflows
│ └── ci.yml
├── .gitignore
├── .pre-commit-config.yaml
├── CHANGELOG.md
├── CONTRIBUTING.md
├── LICENSE
├── README.md
├── data
├── README.md
└── _scripts
│ ├── 1_download_all.sh
│ ├── 2_prepare.py
│ ├── cosmx_convert.py
│ ├── cosmx_download.sh
│ ├── merscope_convert.py
│ ├── merscope_download.sh
│ ├── stereoseq_convert.py
│ ├── stereoseq_download.sh
│ ├── xenium_convert.py
│ └── xenium_download.sh
├── docs
├── api
│ ├── Novae.md
│ ├── dataloader.md
│ ├── metrics.md
│ ├── modules.md
│ ├── plot.md
│ └── utils.md
├── assets
│ ├── Figure1.png
│ ├── banner.png
│ ├── logo_favicon.png
│ ├── logo_small_black.png
│ └── logo_white.png
├── cite_us.md
├── faq.md
├── getting_started.md
├── index.md
├── javascripts
│ └── mathjax.js
└── tutorials
│ ├── LEE_AGING_CEREBELLUM_UP.json
│ ├── hallmarks_pathways.json
│ ├── input_modes.md
│ ├── main_usage.ipynb
│ ├── mouse_hallmarks.json
│ ├── proteins.ipynb
│ └── resolutions.ipynb
├── mkdocs.yml
├── novae
├── __init__.py
├── _constants.py
├── _logging.py
├── _settings.py
├── data
│ ├── __init__.py
│ ├── convert.py
│ ├── datamodule.py
│ └── dataset.py
├── model.py
├── module
│ ├── __init__.py
│ ├── aggregate.py
│ ├── augment.py
│ ├── embed.py
│ ├── encode.py
│ └── swav.py
├── monitor
│ ├── __init__.py
│ ├── callback.py
│ ├── eval.py
│ └── log.py
├── plot
│ ├── __init__.py
│ ├── _bar.py
│ ├── _graph.py
│ ├── _heatmap.py
│ ├── _spatial.py
│ └── _utils.py
└── utils
│ ├── __init__.py
│ ├── _build.py
│ ├── _correct.py
│ ├── _data.py
│ ├── _mode.py
│ ├── _preprocess.py
│ ├── _utils.py
│ └── _validate.py
├── poetry.lock
├── pyproject.toml
├── scripts
├── README.md
├── __init__.py
├── config.py
├── config
│ ├── README.md
│ ├── _example.yaml
│ ├── all_16.yaml
│ ├── all_17.yaml
│ ├── all_brain.yaml
│ ├── all_human.yaml
│ ├── all_human2.yaml
│ ├── all_mouse.yaml
│ ├── all_new.yaml
│ ├── all_ruche.yaml
│ ├── all_spot.yaml
│ ├── brain.yaml
│ ├── brain2.yaml
│ ├── breast.yaml
│ ├── breast_zs.yaml
│ ├── breast_zs2.yaml
│ ├── colon.yaml
│ ├── colon_retrain.yaml
│ ├── colon_zs.yaml
│ ├── colon_zs2.yaml
│ ├── local_tests.yaml
│ ├── lymph_node.yaml
│ ├── missing.yaml
│ ├── ovarian.yaml
│ ├── revision.yaml
│ ├── revision_tests.yaml
│ ├── toy_cpu_seed0.yaml
│ └── toy_missing.yaml
├── missing_domain.py
├── revision
│ ├── cpu.sh
│ ├── heterogeneous.py
│ ├── heterogeneous_start.py
│ ├── mgc.py
│ ├── missing_domains.py
│ ├── perturbations.py
│ └── seg_robustness.py
├── ruche
│ ├── README.md
│ ├── agent.sh
│ ├── convert.sh
│ ├── cpu.sh
│ ├── debug_cpu.sh
│ ├── debug_gpu.sh
│ ├── download.sh
│ ├── gpu.sh
│ ├── prepare.sh
│ └── test.sh
├── sweep
│ ├── README.md
│ ├── cpu.yaml
│ ├── debug.yaml
│ ├── gpu.yaml
│ ├── gpu_ruche.yaml
│ ├── lung.yaml
│ ├── revision.yaml
│ └── toy.yaml
├── toy_missing_domain.py
├── train.py
└── utils.py
├── setup.py
└── tests
├── __init__.py
├── _utils.py
├── test_correction.py
├── test_dataset.py
├── test_metrics.py
├── test_misc.py
├── test_model.py
├── test_plots.py
└── test_utils.py
/.editorconfig:
--------------------------------------------------------------------------------
1 | max_line_length = 120
2 |
3 | [*.json]
4 | indent_style = space
5 | indent_size = 4
6 |
7 | [*.yaml]
8 | indent_size = 2
9 |
--------------------------------------------------------------------------------
/.gitattributes:
--------------------------------------------------------------------------------
1 | # Ignoring notebooks for language statistics on Github
2 | .ipynb -linguist-detectable
3 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/bug_report.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Bug report
3 | about: Create a report to help us improve
4 | title: "[Bug]"
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | ## Description
11 | A clear and concise description of what the bug is.
12 |
13 | ## Reproducing the issue
14 | Steps to reproduce the behavior.
15 |
16 | ## Expected behavior
17 | A clear and concise description of what you expected to happen.
18 |
19 | ## System
20 | - OS: [e.g. Linux]
21 | - Python version [e.g. 3.10.7]
22 |
23 | Dependencies versions
24 |
25 | Paste here what 'pip list' gives you.
26 |
27 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/custom.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Custom issue template
3 | about: Describe this issue template's purpose here.
4 | title: ''
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/feature_request.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Feature request
3 | about: Suggest an idea for this project
4 | title: "[Feature]"
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Is your feature request related to a problem? Please describe.**
11 | A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
12 |
13 | **Describe the solution you'd like**
14 | A clear and concise description of what you want to happen.
15 |
16 | **Describe alternatives you've considered**
17 | A clear and concise description of any alternative solutions or features you've considered.
18 |
19 | **Additional context**
20 | Add any other context or screenshots about the feature request here.
21 |
--------------------------------------------------------------------------------
/.github/workflows/ci.yml:
--------------------------------------------------------------------------------
1 | name: test_deploy_publish
2 | on:
3 | push:
4 | tags:
5 | - v*
6 | pull_request:
7 | branches: [main]
8 |
9 | jobs:
10 | pre-commit:
11 | runs-on: ubuntu-latest
12 | steps:
13 | - uses: actions/checkout@v3
14 | - uses: actions/setup-python@v3
15 | - uses: pre-commit/action@v3.0.1
16 |
17 | build:
18 | needs: [pre-commit]
19 | runs-on: ubuntu-latest
20 | strategy:
21 | matrix:
22 | python-version: ["3.10", "3.11", "3.12"]
23 | steps:
24 | - uses: actions/checkout@v4
25 | - uses: actions/setup-python@v5
26 | with:
27 | python-version: ${{ matrix.python-version }}
28 | cache: "pip"
29 | - run: pip install '.[dev]'
30 |
31 | - name: Run tests
32 | run: pytest --cov
33 |
34 | - name: Deploy doc
35 | if: matrix.python-version == '3.10' && contains(github.ref, 'tags')
36 | run: mkdocs gh-deploy --force
37 |
38 | - name: Upload results to Codecov
39 | if: matrix.python-version == '3.10'
40 | uses: codecov/codecov-action@v4
41 | with:
42 | token: ${{ secrets.CODECOV_TOKEN }}
43 |
44 | publish:
45 | needs: [build]
46 | if: contains(github.ref, 'tags')
47 | runs-on: ubuntu-latest
48 | steps:
49 | - uses: actions/checkout@v3
50 | - name: Build and publish to pypi
51 | uses: JRubics/poetry-publish@v1.17
52 | with:
53 | python_version: "3.10"
54 | pypi_token: ${{ secrets.PYPI_TOKEN }}
55 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # OS related
2 | .DS_Store
3 |
4 | # Ruche related
5 | jobs/
6 |
7 | # Tests related
8 | tests/*
9 | !tests/*.py
10 |
11 | # IDE related
12 | .vscode/*
13 | !.vscode/settings.json
14 | !.vscode/tasks.json
15 | !.vscode/launch.json
16 | !.vscode/extensions.json
17 | *.code-workspace
18 | **/.vscode
19 |
20 | # Documentation
21 | site
22 |
23 | # Logs
24 | outputs
25 | multirun
26 | lightning_logs
27 | novae_*
28 | checkpoints
29 |
30 | # Data files
31 | data/*
32 | !data/_scripts
33 | !data/README.md
34 | !data/*.py
35 | !data/*.sh
36 |
37 | # Results files
38 | results/*
39 |
40 | # Jupyter Notebook
41 | .ipynb_checkpoints
42 | *.ipynb
43 | !docs/tutorials/*.ipynb
44 |
45 | # Misc
46 | logs/
47 | test.h5ad
48 | test.ckpt
49 | wandb/
50 | .env
51 | .autoenv
52 | !**/.gitkeep
53 | exploration
54 |
55 | # pyenv
56 | .python-version
57 |
58 | # Byte-compiled / optimized / DLL files
59 | __pycache__/
60 | *.py[cod]
61 | *$py.class
62 |
63 | # C extensions
64 | *.so
65 |
66 | # Distribution / packaging
67 | .Python
68 | build/
69 | develop-eggs/
70 | dist/
71 | downloads/
72 | eggs/
73 | .eggs/
74 | lib/
75 | lib64/
76 | parts/
77 | sdist/
78 | var/
79 | wheels/
80 | pip-wheel-metadata/
81 | share/python-wheels/
82 | *.egg-info/
83 | .installed.cfg
84 | *.egg
85 | MANIFEST
86 |
87 | # PyInstaller
88 | # Usually these files are written by a python script from a template
89 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
90 | *.manifest
91 | *.spec
92 |
93 | # Installer logs
94 | pip-log.txt
95 | pip-delete-this-directory.txt
96 |
97 | # Unit test / coverage reports
98 | htmlcov/
99 | .tox/
100 | .nox/
101 | .coverage
102 | .coverage.*
103 | .cache
104 | nosetests.xml
105 | coverage.xml
106 | *.cover
107 | *.py,cover
108 | .hypothesis/
109 | .pytest_cache/
110 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | fail_fast: false
2 | default_language_version:
3 | python: python3
4 | default_stages:
5 | - pre-commit
6 | - pre-push
7 | minimum_pre_commit_version: 2.16.0
8 | repos:
9 | - repo: https://github.com/astral-sh/ruff-pre-commit
10 | rev: v0.11.5
11 | hooks:
12 | - id: ruff
13 | - id: ruff-format
14 | - repo: https://github.com/pre-commit/pre-commit-hooks
15 | rev: v4.5.0
16 | hooks:
17 | - id: trailing-whitespace
18 | - id: end-of-file-fixer
19 | - id: check-yaml
20 | - id: debug-statements
21 |
--------------------------------------------------------------------------------
/CHANGELOG.md:
--------------------------------------------------------------------------------
1 | ## [0.2.4] - 2025-03-26
2 |
3 | Hotfix (#18)
4 |
5 | ## [0.2.3] - 2025-03-21
6 |
7 | ### Added
8 | - New Visium-HD and Visium tutorials
9 | - Infer default plot size for plots (median neighbors distrance)
10 | - Can filter by technology in `load_dataset`
11 |
12 | ### Fixed
13 | - Fix `model.plot_prototype_covariance` and `model.plot_prototype_weights`
14 | - Edge case: ensure that even negative max-weights are above the prototype threshold
15 |
16 | ### Changed
17 | - `novae.utils.spatial_neighbors` can be now called via `novae.spatial_neighbors`
18 | - Store distances after `spatial_neighbors` instead of `1` when `coord_type="grid"`
19 |
20 | ## [0.2.2] - 2024-12-17
21 |
22 | ### Added
23 | - `load_dataset`: add `custom_filter` and `dry_run` arguments
24 | - added `min_prototypes_ratio` argument in `fine_tune` to run `init_slide_queue`
25 | - Added tutorials for proteins data + minor docs improvements
26 |
27 | ### Fixed
28 | - Ensure reset clustering if multiple zero-shot (#9)
29 |
30 | ### Changed
31 | - Removed the docs formatting (better for autocompletion)
32 | - Reorder parameters in Novae `__init__` (sorted by importance)
33 |
34 | ## [0.2.1] - 2024-12-04
35 |
36 | ### Added
37 | - `novae.utils.quantile_scaling` for proteins expression
38 |
39 | ### Fixed
40 | - Fix autocompletion using `__new__` to trick hugging_face inheritance
41 |
42 |
43 | ## [0.2.0] - 2024-12-03
44 |
45 | ### Added
46 |
47 | - `novae.plot.connectivities(...)` to show the cells neighbors
48 | - `novae.settings.auto_processing = False` to enforce using your own preprocessing
49 | - Tutorials update (more plots and more details)
50 |
51 | ### Fixed
52 |
53 | - Issue with `library_id` in `novae.plot.domains` (#8)
54 | - Set `pandas>=2.0.0` in the dependencies (#5)
55 |
56 | ### Breaking changes
57 |
58 | - `novae.utils.spatial_neighbors` must always be run, to force the user having more control on it
59 | - For multi-slide mode, the `slide_key` argument should now be used in `novae.utils.spatial_neighbors` (and only there)
60 | - Drop python 3.9 support (because dropped in `anndata`)
61 |
62 | ## [0.1.0] - 2024-09-11
63 |
64 | First official `novae` release. Preprint coming soon.
65 |
--------------------------------------------------------------------------------
/CONTRIBUTING.md:
--------------------------------------------------------------------------------
1 | # Contributing to *novae*
2 |
3 | Contributions are welcome as we aim to continue improving `novae`. For instance, you can contribute by:
4 |
5 | - Opening an issue
6 | - Discussing the current state of the code
7 | - Making a Pull Request (PR)
8 |
9 | If you want to open a PR, follow the following instructions.
10 |
11 | ## Making a Pull Request (PR)
12 |
13 | To add some new code to **novae**, you should:
14 |
15 | 1. Fork the repository
16 | 2. Install `novae` in editable mode with the `dev` extra (see below)
17 | 3. Create your personal branch from `main`
18 | 4. Implement your changes according to the 'Coding guidelines' below
19 | 5. Create a pull request on the `main` branch of the original repository. Add explanations about your developed features, and wait for discussion and validation of your pull request
20 |
21 | ## Installing `novae` in editable mode
22 |
23 | When contributing, installing `novae` in editable mode is recommended. We also recommend installing the `dev` extra.
24 |
25 | For that, choose between `pip` and `poetry` as below:
26 |
27 | ```sh
28 | git clone https://github.com/MICS-Lab/novae.git
29 | cd novae
30 |
31 | pip install -e '.[dev]' # pip installation
32 | poetry install -E dev # poetry installation
33 | ```
34 |
35 | ## Coding guidelines
36 |
37 | ### Styling and formatting
38 |
39 | We use [`pre-commit`](https://pre-commit.com/) to run code quality controls before the commits. This will run `ruff` and others minor checks.
40 |
41 |
42 | You can set it up at the root of the repository like this:
43 | ```sh
44 | pre-commit install
45 | ```
46 |
47 | Then, it will run the pre-commit automatically before each commit.
48 |
49 | You can also run the pre-commit manually:
50 | ```sh
51 | pre-commit run --all-files
52 | ```
53 |
54 | Apart from this, we recommend to follow the standard styling conventions:
55 | - Follow the [PEP8](https://peps.python.org/pep-0008/) style guide.
56 | - Provide meaningful names to all your variables and functions.
57 | - Provide type hints to your function inputs/outputs.
58 | - Add docstrings in the Google style.
59 | - Try as much as possible to follow the same coding style as the rest of the repository.
60 |
61 | ### Testing
62 |
63 | When create a pull request, tests are run automatically. But you can also run the tests yourself before making the PR. For that, run `pytest` at the root of the repository. You can also add new tests in the `./tests` directory.
64 |
65 | To check the coverage of the tests:
66 |
67 | ```sh
68 | coverage run -m pytest
69 | coverage report # command line report
70 | coverage html # html report
71 | ```
72 |
73 | ### Documentation
74 |
75 | You can update the documentation in the `./docs` directory. Refer to the [mkdocs-material documentation](https://squidfunk.github.io/mkdocs-material/) for more help.
76 |
77 | To serve the documentation locally:
78 |
79 | ```sh
80 | mkdocs serve
81 | ```
82 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | BSD 3-Clause License
2 |
3 | Copyright (c) 2024, Quentin Blampey
4 | All rights reserved.
5 |
6 | Redistribution and use in source and binary forms, with or without
7 | modification, are permitted provided that the following conditions are met:
8 |
9 | 1. Redistributions of source code must retain the above copyright notice, this
10 | list of conditions and the following disclaimer.
11 |
12 | 2. Redistributions in binary form must reproduce the above copyright notice,
13 | this list of conditions and the following disclaimer in the documentation
14 | and/or other materials provided with the distribution.
15 |
16 | 3. Neither the name of the copyright holder nor the names of its
17 | contributors may be used to endorse or promote products derived from
18 | this software without specific prior written permission.
19 |
20 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
21 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
22 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
23 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
24 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
25 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
26 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
27 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
28 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
29 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
30 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |  3 |
3 |
4 |
5 |
6 |
7 | [](https://pypi.org/project/novae)
8 | [](https://pepy.tech/project/novae)
9 | [](https://mics-lab.github.io/novae)
10 | 
11 | [](https://github.com/MICS-Lab/novae/blob/main/LICENSE)
12 | [](https://codecov.io/gh/MICS-Lab/novae)
13 | [](https://github.com/astral-sh/ruff)
14 |
15 |
16 |
17 |
18 | 💫 Graph-based foundation model for spatial transcriptomics data
19 |
20 |
21 | Novae is a deep learning model for spatial domain assignments of spatial transcriptomics data (at both single-cell or spot resolution). It works across multiple gene panels, tissues, and technologies. Novae offers several additional features, including: (i) native batch-effect correction, (ii) analysis of spatially variable genes and pathways, and (iii) architecture analysis of tissue slides.
22 |
23 | ## Documentation
24 |
25 | Check [Novae's documentation](https://mics-lab.github.io/novae/) to get started. It contains installation explanations, API details, and tutorials.
26 |
27 | ## Overview
28 |
29 |
30 | 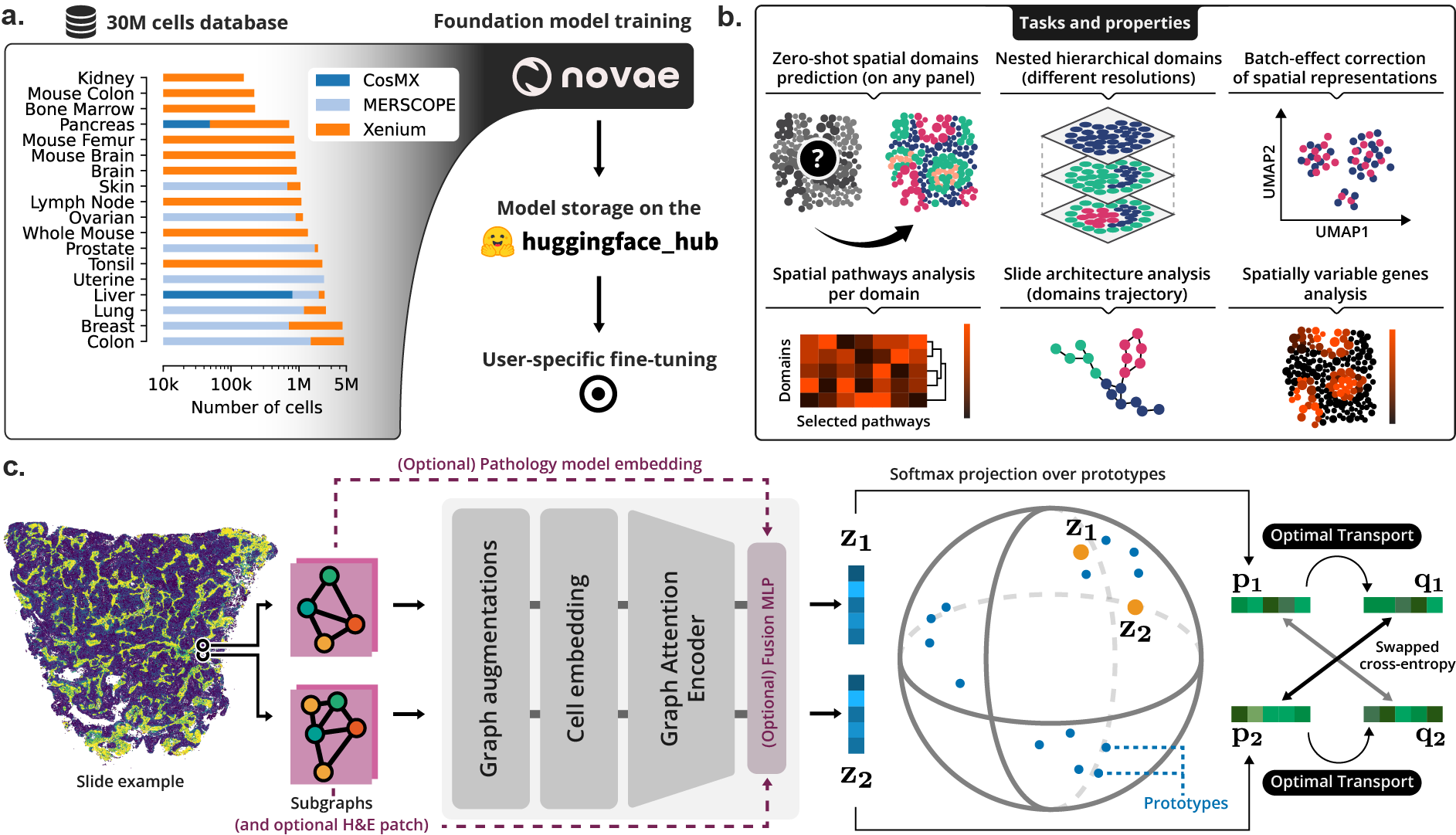 31 |
31 |
32 |
33 | > **(a)** Novae was trained on a large dataset, and is shared on [Hugging Face Hub](https://huggingface.co/collections/MICS-Lab/novae-669cdf1754729d168a69f6bd). **(b)** Illustration of the main tasks and properties of Novae. **(c)** Illustration of the method behind Novae (self-supervision on graphs, adapted from [SwAV](https://arxiv.org/abs/2006.09882)).
34 |
35 | ## Installation
36 |
37 | ### PyPI
38 |
39 | `novae` can be installed via `PyPI` on all OS, for `python>=3.10`.
40 |
41 | ```
42 | pip install novae
43 | ```
44 |
45 | ### Editable mode
46 |
47 | To install `novae` in editable mode (e.g., to contribute), clone the repository and choose among the options below.
48 |
49 | ```sh
50 | pip install -e . # pip, minimal dependencies
51 | pip install -e '.[dev]' # pip, all extras
52 | poetry install # poetry, minimal dependencies
53 | poetry install --all-extras # poetry, all extras
54 | ```
55 |
56 | ## Usage
57 |
58 | Here is a minimal usage example. For more details, refer to the [documentation](https://mics-lab.github.io/novae/).
59 |
60 | ```python
61 | import novae
62 |
63 | model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
64 |
65 | model.compute_representations(adata, zero_shot=True)
66 | model.assign_domains(adata)
67 | ```
68 |
69 | ## Cite us
70 |
71 | You can cite our [preprint](https://www.biorxiv.org/content/10.1101/2024.09.09.612009v1) as below:
72 |
73 | ```txt
74 | @article{blampeyNovae2024,
75 | title = {Novae: A Graph-Based Foundation Model for Spatial Transcriptomics Data},
76 | author = {Blampey, Quentin and Benkirane, Hakim and Bercovici, Nadege and Andre, Fabrice and Cournede, Paul-Henry},
77 | year = {2024},
78 | pages = {2024.09.09.612009},
79 | publisher = {bioRxiv},
80 | doi = {10.1101/2024.09.09.612009},
81 | }
82 | ```
83 |
--------------------------------------------------------------------------------
/data/README.md:
--------------------------------------------------------------------------------
1 | # Public datasets
2 |
3 | We detail below how to download public spatial transcriptomics datasets.
4 |
5 | ## Option 1: Hugging Face Hub
6 |
7 | We store our dataset on [Hugging Face Hub](https://huggingface.co/datasets/MICS-Lab/novae).
8 | To automatically download these slides, you can use the [`novae.utils.load_dataset`](https://mics-lab.github.io/novae/api/utils/#novae.utils.load_dataset) function.
9 |
10 | NB: not all slides are uploaded on Hugging Face yet, but we are progressively adding new slides. To get the full dataset right now, use the "Option 2" below.
11 |
12 | ## Option 2: Download
13 |
14 | For consistency, all the scripts below need to be executed at the root of the `data` directory (i.e., `novae/data`).
15 |
16 | ### MERSCOPE (18 samples)
17 |
18 | Requirements: the `gsutil` command line should be installed (see [here](https://cloud.google.com/storage/docs/gsutil_install)) and a Python environment with `scanpy`.
19 |
20 | ```sh
21 | # download all MERSCOPE datasets
22 | sh _scripts/merscope_download.sh
23 |
24 | # convert all datasets to h5ad files
25 | python _scripts/merscope_convert.py
26 | ```
27 |
28 | ### Xenium (20+ samples)
29 |
30 | Requirements: a Python environment with `spatialdata-io` installed.
31 |
32 | ```sh
33 | # download all Xenium datasets
34 | sh _scripts/xenium_download.sh
35 |
36 | # convert all datasets to h5ad files
37 | python _scripts/xenium_convert.py
38 | ```
39 |
40 | ### CosMX (3 samples)
41 |
42 | Requirements: a Python environment with `scanpy` installed.
43 |
44 | ```sh
45 | # download all CosMX datasets
46 | sh _scripts/cosmx_download.sh
47 |
48 | # convert all datasets to h5ad files
49 | python _scripts/cosmx_convert.py
50 | ```
51 |
52 | ### All datasets
53 |
54 | All above datasets can be downloaded using a single command line. Make sure you have all the requirements listed above.
55 |
56 | ```sh
57 | sh _scripts/1_download_all.sh
58 | ```
59 |
60 | ### Preprocess and prepare for training
61 |
62 | The script bellow will copy all `adata.h5ad` files into a single directory, compute UMAPs, and minor preprocessing. See the `argparse` helper of this script for more details.
63 |
64 | ```sh
65 | python _scripts/2_prepare.py
66 | ```
67 |
68 | ### Usage
69 |
70 | These datasets can be used during training (see the `scripts` directory at the root of the `novae` repository).
71 |
72 | ## Notes
73 | - Missing technologies: CosMX, Curio Seeker, Resolve
74 | - Public institute datasets with [STOmics DB](https://db.cngb.org/stomics/)
75 | - Some Xenium datasets are available outside of the main "10X Datasets" page:
76 | - https://www.10xgenomics.com/products/visium-hd-spatial-gene-expression/dataset-human-crc
77 |
--------------------------------------------------------------------------------
/data/_scripts/1_download_all.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | # download all MERSCOPE datasets
4 | sh merscope_download.sh
5 |
6 | # convert all datasets to h5ad files
7 | python merscope_convert.py
8 |
9 | # download all Xenium datasets
10 | sh xenium_download.sh
11 |
12 | # convert all datasets to h5ad files
13 | python xenium_convert.py
14 |
15 | # download all CosMX datasets
16 | sh cosmx_download.sh
17 |
18 | # convert all datasets to h5ad files
19 | python cosmx_convert.py
20 |
--------------------------------------------------------------------------------
/data/_scripts/2_prepare.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import anndata
5 | import scanpy as sc
6 | from anndata import AnnData
7 |
8 | import novae
9 |
10 | MIN_CELLS = 50
11 | DELAUNAY_RADIUS = 100
12 |
13 |
14 | def preprocess(adata: AnnData, compute_umap: bool = False):
15 | sc.pp.filter_genes(adata, min_cells=MIN_CELLS)
16 | adata.layers["counts"] = adata.X.copy()
17 | sc.pp.normalize_total(adata)
18 | sc.pp.log1p(adata)
19 |
20 | if compute_umap:
21 | sc.pp.neighbors(adata)
22 | sc.tl.umap(adata)
23 |
24 | novae.utils.spatial_neighbors(adata, radius=[0, DELAUNAY_RADIUS])
25 |
26 |
27 | def main(args):
28 | data_path: Path = Path(args.path).absolute()
29 | out_dir = data_path / args.name
30 |
31 | if not out_dir.exists():
32 | out_dir.mkdir()
33 |
34 | for dataset in args.datasets:
35 | dataset_dir: Path = data_path / dataset
36 | for file in dataset_dir.glob("**/adata.h5ad"):
37 | print("Reading file", file)
38 |
39 | adata = anndata.read_h5ad(file)
40 | adata.obs["technology"] = dataset
41 |
42 | if "slide_id" not in adata.obs:
43 | print(" (no slide_id in obs, skipping)")
44 | continue
45 |
46 | out_file = out_dir / f"{adata.obs['slide_id'].iloc[0]}.h5ad"
47 |
48 | if out_file.exists() and not args.overwrite:
49 | print(" (already exists)")
50 | continue
51 |
52 | preprocess(adata, compute_umap=args.umap)
53 | adata.write_h5ad(out_file)
54 |
55 |
56 | if __name__ == "__main__":
57 | parser = argparse.ArgumentParser()
58 | parser.add_argument(
59 | "-p",
60 | "--path",
61 | type=str,
62 | default=".",
63 | help="Path to spatial directory",
64 | )
65 | parser.add_argument(
66 | "-n",
67 | "--name",
68 | type=str,
69 | default="all",
70 | help="Name of the resulting data directory",
71 | )
72 | parser.add_argument(
73 | "-d",
74 | "--datasets",
75 | nargs="+",
76 | default=["xenium", "merscope", "cosmx"],
77 | help="List of dataset names to concatenate",
78 | )
79 | parser.add_argument(
80 | "-o",
81 | "--overwrite",
82 | action="store_true",
83 | help="Overwrite existing output files",
84 | )
85 | parser.add_argument(
86 | "-u",
87 | "--umap",
88 | action="store_true",
89 | help="Whether to compute the UMAP embedding",
90 | )
91 |
92 | args = parser.parse_args()
93 | main(args)
94 |
--------------------------------------------------------------------------------
/data/_scripts/cosmx_convert.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import anndata
5 | import pandas as pd
6 | from scipy.sparse import csr_matrix
7 |
8 |
9 | def convert_to_h5ad(dataset_dir: Path):
10 | print(f"Reading {dataset_dir}")
11 | res_path = dataset_dir / "adata.h5ad"
12 |
13 | if res_path.exists():
14 | print(f"File {res_path} already existing.")
15 | return
16 |
17 | slide_id = f"cosmx_{dataset_dir.name}"
18 |
19 | counts_files = list(dataset_dir.glob("*exprMat_file.csv"))
20 | metadata_files = list(dataset_dir.glob("*metadata_file.csv"))
21 |
22 | if len(counts_files) != 1 or len(metadata_files) != 1:
23 | print(f"Did not found both exprMat and metadata csv inside {dataset_dir}. Skipping this directory.")
24 | return
25 |
26 | data = pd.read_csv(counts_files[0], index_col=[0, 1])
27 | obs = pd.read_csv(metadata_files[0], index_col=[0, 1])
28 |
29 | data.index = data.index.map(lambda x: f"{x[0]}-{x[1]}")
30 | obs.index = obs.index.map(lambda x: f"{x[0]}-{x[1]}")
31 |
32 | if len(data) != len(obs):
33 | cell_ids = list(set(data.index) & set(obs.index))
34 | data = data.loc[cell_ids]
35 | obs = obs.loc[cell_ids]
36 |
37 | obs.index = obs.index.astype(str) + f"_{slide_id}"
38 | data.index = obs.index
39 |
40 | is_gene = ~data.columns.str.lower().str.contains("SystemControl")
41 |
42 | adata = anndata.AnnData(data.loc[:, is_gene], obs=obs)
43 |
44 | adata.obsm["spatial"] = adata.obs[["CenterX_global_px", "CenterY_global_px"]].values * 0.120280945
45 | adata.obs["slide_id"] = pd.Series(slide_id, index=adata.obs_names, dtype="category")
46 |
47 | adata.X = csr_matrix(adata.X)
48 | adata.write_h5ad(res_path)
49 |
50 | print(f"Created file at path {res_path}")
51 |
52 |
53 | def main(args):
54 | path = Path(args.path).absolute() / "cosmx"
55 |
56 | print(f"Reading all datasets inside {path}")
57 |
58 | for dataset_dir in path.iterdir():
59 | if dataset_dir.is_dir():
60 | convert_to_h5ad(dataset_dir)
61 |
62 |
63 | if __name__ == "__main__":
64 | parser = argparse.ArgumentParser()
65 | parser.add_argument(
66 | "-p",
67 | "--path",
68 | type=str,
69 | default=".",
70 | help="Path to spatial directory (containing the 'cosmx' directory)",
71 | )
72 |
73 | main(parser.parse_args())
74 |
--------------------------------------------------------------------------------
/data/_scripts/cosmx_download.sh:
--------------------------------------------------------------------------------
1 | # Pancreas
2 | PANCREAS_FLAT_FILES="https://smi-public.objects.liquidweb.services/cosmx-wtx/Pancreas-CosMx-WTx-FlatFiles.zip"
3 | PANCREAS_OUTPUT_ZIP="cosmx/pancreas/Pancreas-CosMx-WTx-FlatFiles.zip"
4 |
5 | mkdir -p cosmx/pancreas
6 |
7 | if [ -f $PANCREAS_OUTPUT_ZIP ]; then
8 | echo "File $PANCREAS_OUTPUT_ZIP already exists."
9 | else
10 | echo "Downloading $PANCREAS_FLAT_FILES to $PANCREAS_OUTPUT_ZIP"
11 | curl $PANCREAS_FLAT_FILES -o $PANCREAS_OUTPUT_ZIP
12 | unzip $PANCREAS_OUTPUT_ZIP -d cosmx/pancreas
13 | fi
14 |
15 | # Normal Liver
16 | mkdir -p cosmx/normal_liver
17 | METADATA_FILE="https://nanostring.app.box.com/index.php?rm=box_download_shared_file&shared_name=id16si2dckxqqpilexl2zg90leo57grn&file_id=f_1392279064291"
18 | METADATA_OUTPUT="cosmx/normal_liver/metadata_file.csv"
19 | if [ -f $METADATA_OUTPUT ]; then
20 | echo "File $METADATA_OUTPUT already exists."
21 | else
22 | echo "Downloading $METADATA_FILE to $METADATA_OUTPUT"
23 | curl -L $METADATA_FILE -o $METADATA_OUTPUT
24 | fi
25 | COUNT_FILE="https://nanostring.app.box.com/index.php?rm=box_download_shared_file&shared_name=id16si2dckxqqpilexl2zg90leo57grn&file_id=f_1392318918584"
26 | COUNT_OUTPUT="cosmx/normal_liver/exprMat_file.csv"
27 | if [ -f $COUNT_OUTPUT ]; then
28 | echo "File $COUNT_OUTPUT already exists."
29 | else
30 | echo "Downloading $COUNT_FILE to $COUNT_OUTPUT"
31 | curl -L $COUNT_FILE -o $COUNT_OUTPUT
32 | fi
33 |
34 | # Cancer Liver
35 | mkdir -p cosmx/cancer_liver
36 | METADATA_FILE="https://nanostring.app.box.com/index.php?rm=box_download_shared_file&shared_name=id16si2dckxqqpilexl2zg90leo57grn&file_id=f_1392293795557"
37 | METADATA_OUTPUT="cosmx/cancer_liver/metadata_file.csv"
38 | if [ -f $METADATA_OUTPUT ]; then
39 | echo "File $METADATA_OUTPUT already exists."
40 | else
41 | echo "Downloading $METADATA_FILE to $METADATA_OUTPUT"
42 | curl -L $METADATA_FILE -o $METADATA_OUTPUT
43 | fi
44 | COUNT_FILE="https://nanostring.app.box.com/index.php?rm=box_download_shared_file&shared_name=id16si2dckxqqpilexl2zg90leo57grn&file_id=f_1392441469377"
45 | COUNT_OUTPUT="cosmx/cancer_liver/exprMat_file.csv"
46 | if [ -f $COUNT_OUTPUT ]; then
47 | echo "File $COUNT_OUTPUT already exists."
48 | else
49 | echo "Downloading $COUNT_FILE to $COUNT_OUTPUT"
50 | curl -L $COUNT_FILE -o $COUNT_OUTPUT
51 | fi
52 |
--------------------------------------------------------------------------------
/data/_scripts/merscope_convert.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import anndata
5 | import numpy as np
6 | import pandas as pd
7 | from scipy.sparse import csr_matrix
8 |
9 |
10 | def convert_to_h5ad(dataset_dir: Path):
11 | res_path = dataset_dir / "adata.h5ad"
12 |
13 | if res_path.exists():
14 | print(f"File {res_path} already existing.")
15 | return
16 |
17 | region = "region_0"
18 | slide_id = f"{dataset_dir.name}_{region}"
19 |
20 | data_dir = dataset_dir / "cell_by_gene.csv"
21 | obs_dir = dataset_dir / "cell_metadata.csv"
22 |
23 | if not data_dir.exists() or not obs_dir.exists():
24 | print(f"Did not found both csv inside {dataset_dir}. Skipping this directory.")
25 | return
26 |
27 | data = pd.read_csv(data_dir, index_col=0, dtype={"cell": str})
28 | obs = pd.read_csv(obs_dir, index_col=0, dtype={"EntityID": str})
29 |
30 | obs.index = obs.index.astype(str) + f"_{slide_id}"
31 | data.index = data.index.astype(str) + f"_{slide_id}"
32 | obs = obs.loc[data.index]
33 |
34 | is_gene = ~data.columns.str.lower().str.contains("blank")
35 |
36 | adata = anndata.AnnData(data.loc[:, is_gene], dtype=np.uint16, obs=obs)
37 |
38 | adata.obsm["spatial"] = adata.obs[["center_x", "center_y"]].values
39 | adata.obs["region"] = pd.Series(region, index=adata.obs_names, dtype="category")
40 | adata.obs["slide_id"] = pd.Series(slide_id, index=adata.obs_names, dtype="category")
41 |
42 | adata.X = csr_matrix(adata.X)
43 | adata.write_h5ad(res_path)
44 |
45 | print(f"Created file at path {res_path}")
46 |
47 |
48 | def main(args):
49 | path = Path(args.path).absolute() / "merscope"
50 |
51 | print(f"Reading all datasets inside {path}")
52 |
53 | for dataset_dir in path.iterdir():
54 | if dataset_dir.is_dir():
55 | convert_to_h5ad(dataset_dir)
56 |
57 |
58 | if __name__ == "__main__":
59 | parser = argparse.ArgumentParser()
60 | parser.add_argument(
61 | "-p",
62 | "--path",
63 | type=str,
64 | default=".",
65 | help="Path to spatial directory (containing the 'merscope' directory)",
66 | )
67 |
68 | main(parser.parse_args())
69 |
--------------------------------------------------------------------------------
/data/_scripts/merscope_download.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | BUCKET_NAME="vz-ffpe-showcase"
4 | OUTPUT_DIR="./merscope"
5 |
6 | for BUCKET_DIR in $(gsutil ls -d gs://$BUCKET_NAME/*); do
7 | DATASET_NAME=$(basename $BUCKET_DIR)
8 | OUTPUT_DATASET_DIR=$OUTPUT_DIR/$DATASET_NAME
9 |
10 | mkdir -p $OUTPUT_DATASET_DIR
11 |
12 | for BUCKET_FILE in ${BUCKET_DIR}{cell_by_gene,cell_metadata}.csv; do
13 | FILE_NAME=$(basename $BUCKET_FILE)
14 |
15 | if [ -f $OUTPUT_DATASET_DIR/$FILE_NAME ]; then
16 | echo "File $FILE_NAME already exists in $OUTPUT_DATASET_DIR"
17 | else
18 | echo "Copying $BUCKET_FILE to $OUTPUT_DATASET_DIR"
19 | gsutil cp "$BUCKET_FILE" $OUTPUT_DATASET_DIR
20 | echo "Copied successfully"
21 | fi
22 | done
23 | done
24 |
--------------------------------------------------------------------------------
/data/_scripts/stereoseq_convert.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import anndata
5 | import pandas as pd
6 | from scipy.sparse import csr_matrix
7 |
8 |
9 | def convert_to_h5ad(dataset_dir: Path):
10 | res_path = dataset_dir / "adata.h5ad"
11 |
12 | if res_path.exists():
13 | print(f"File {res_path} already existing.")
14 | return
15 |
16 | slide_id = f"stereoseq_{dataset_dir.name}"
17 |
18 | h5ad_files = dataset_dir.glob(".h5ad")
19 |
20 | if len(h5ad_files) != 1:
21 | print(f"Found {len(h5ad_files)} h5ad file inside {dataset_dir}. Skipping this directory.")
22 | return
23 |
24 | adata = anndata.read_h5ad(h5ad_files[0])
25 | adata.X = adata.layers["raw_counts"]
26 | del adata.layers["raw_counts"]
27 |
28 | adata.obsm["spatial"] = adata.obsm["spatial"].astype(float).values
29 | adata.obs["slide_id"] = pd.Series(slide_id, index=adata.obs_names, dtype="category")
30 |
31 | adata.X = csr_matrix(adata.X)
32 | adata.write_h5ad(res_path)
33 |
34 | print(f"Created file at path {res_path}")
35 |
36 |
37 | def main(args):
38 | path = Path(args.path).absolute() / "stereoseq"
39 |
40 | print(f"Reading all datasets inside {path}")
41 |
42 | for dataset_dir in path.iterdir():
43 | if dataset_dir.is_dir():
44 | convert_to_h5ad(dataset_dir)
45 |
46 |
47 | if __name__ == "__main__":
48 | parser = argparse.ArgumentParser()
49 | parser.add_argument(
50 | "-p",
51 | "--path",
52 | type=str,
53 | default=".",
54 | help="Path to spatial directory (containing the 'stereoseq' directory)",
55 | )
56 |
57 | main(parser.parse_args())
58 |
--------------------------------------------------------------------------------
/data/_scripts/stereoseq_download.sh:
--------------------------------------------------------------------------------
1 | H5AD_REMOTE_PATHS=(\
2 | "https://ftp.cngb.org/pub/SciRAID/stomics/STDS0000062/stomics/FP200000498TL_D2_stereoseq.h5ad"\
3 | "https://ftp.cngb.org/pub/SciRAID/stomics/STDS0000062/stomics/FP200000498TL_E4_stereoseq.h5ad"\
4 | "https://ftp.cngb.org/pub/SciRAID/stomics/STDS0000062/stomics/FP200000498TL_E5_stereoseq.h5ad"\
5 | )
6 |
7 | OUTPUT_DIR="stereoseq"
8 | mkdir -p $OUTPUT_DIR
9 |
10 | for H5AD_REMOTE_PATH in "${H5AD_REMOTE_PATHS[@]}"
11 | do
12 | DATASET_NAME=$(basename $H5AD_REMOTE_PATH)
13 | OUTPUT_DATASET_DIR=${OUTPUT_DIR}/${DATASET_NAME%.h5ad}
14 | OUTPUT_DATASET=$OUTPUT_DATASET_DIR/${DATASET_NAME}
15 |
16 | mkdir -p $OUTPUT_DATASET_DIR
17 |
18 | if [ -f $OUTPUT_DATASET ]; then
19 | echo "File $OUTPUT_DATASET_DIR already exists"
20 | else
21 | echo "Downloading $H5AD_REMOTE_PATH to $OUTPUT_DATASET"
22 | # curl $H5AD_REMOTE_PATH -o $OUTPUT_DATASET
23 | echo "Successfully downloaded"
24 | fi
25 | done
26 |
--------------------------------------------------------------------------------
/data/_scripts/xenium_convert.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import anndata
5 | import pandas as pd
6 | from spatialdata_io.readers.xenium import _get_tables_and_circles
7 |
8 |
9 | def convert_to_h5ad(dataset_dir: Path):
10 | res_path = dataset_dir / "adata.h5ad"
11 |
12 | if res_path.exists():
13 | print(f"File {res_path} already existing.")
14 | return
15 |
16 | adata: anndata.AnnData = _get_tables_and_circles(dataset_dir, False, {"region": "region_0"})
17 | adata.obs["cell_id"] = adata.obs["cell_id"].apply(lambda x: x if (isinstance(x, (str, int))) else x.decode("utf-8"))
18 |
19 | slide_id = dataset_dir.name
20 | adata.obs.index = adata.obs["cell_id"].astype(str).values + f"_{slide_id}"
21 |
22 | adata.obs["slide_id"] = pd.Series(slide_id, index=adata.obs_names, dtype="category")
23 |
24 | adata.write_h5ad(res_path)
25 |
26 | print(f"Created file at path {res_path}")
27 |
28 |
29 | def main(args):
30 | path = Path(args.path).absolute() / "xenium"
31 |
32 | print(f"Reading all datasets inside {path}")
33 |
34 | for dataset_dir in path.iterdir():
35 | if dataset_dir.is_dir():
36 | print(f"In {dataset_dir}")
37 | try:
38 | convert_to_h5ad(dataset_dir)
39 | except:
40 | print(f"Failed to convert {dataset_dir}")
41 |
42 |
43 | if __name__ == "__main__":
44 | parser = argparse.ArgumentParser()
45 | parser.add_argument(
46 | "-p",
47 | "--path",
48 | type=str,
49 | default=".",
50 | help="Path to spatial directory (containing the 'xenium' directory)",
51 | )
52 |
53 | main(parser.parse_args())
54 |
--------------------------------------------------------------------------------
/docs/api/Novae.md:
--------------------------------------------------------------------------------
1 | ::: novae.Novae
2 |
--------------------------------------------------------------------------------
/docs/api/dataloader.md:
--------------------------------------------------------------------------------
1 | ::: novae.data.AnnDataTorch
2 |
3 | ::: novae.data.NovaeDataset
4 |
5 | ::: novae.data.NovaeDatamodule
6 |
--------------------------------------------------------------------------------
/docs/api/metrics.md:
--------------------------------------------------------------------------------
1 | ::: novae.monitor.jensen_shannon_divergence
2 |
3 | ::: novae.monitor.fide_score
4 |
5 | ::: novae.monitor.mean_fide_score
6 |
--------------------------------------------------------------------------------
/docs/api/modules.md:
--------------------------------------------------------------------------------
1 | ::: novae.module.AttentionAggregation
2 |
3 | ::: novae.module.CellEmbedder
4 |
5 | ::: novae.module.GraphAugmentation
6 |
7 | ::: novae.module.GraphEncoder
8 |
9 | ::: novae.module.SwavHead
10 |
--------------------------------------------------------------------------------
/docs/api/plot.md:
--------------------------------------------------------------------------------
1 | ::: novae.plot.domains
2 |
3 | ::: novae.plot.domains_proportions
4 |
5 | ::: novae.plot.connectivities
6 |
7 | ::: novae.plot.pathway_scores
8 |
9 | ::: novae.plot.paga
10 |
11 | ::: novae.plot.spatially_variable_genes
12 |
--------------------------------------------------------------------------------
/docs/api/utils.md:
--------------------------------------------------------------------------------
1 | ::: novae.spatial_neighbors
2 |
3 | ::: novae.batch_effect_correction
4 |
5 | ::: novae.utils.quantile_scaling
6 |
7 | ::: novae.utils.prepare_adatas
8 |
9 | ::: novae.utils.load_dataset
10 |
11 | ::: novae.utils.toy_dataset
12 |
--------------------------------------------------------------------------------
/docs/assets/Figure1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/docs/assets/Figure1.png
--------------------------------------------------------------------------------
/docs/assets/banner.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/docs/assets/banner.png
--------------------------------------------------------------------------------
/docs/assets/logo_favicon.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/docs/assets/logo_favicon.png
--------------------------------------------------------------------------------
/docs/assets/logo_small_black.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/docs/assets/logo_small_black.png
--------------------------------------------------------------------------------
/docs/assets/logo_white.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/docs/assets/logo_white.png
--------------------------------------------------------------------------------
/docs/cite_us.md:
--------------------------------------------------------------------------------
1 | You can cite our [preprint](https://www.biorxiv.org/content/10.1101/2024.09.09.612009v1) as below:
2 |
3 | ```txt
4 | @article{blampeyNovae2024,
5 | title = {Novae: A Graph-Based Foundation Model for Spatial Transcriptomics Data},
6 | author = {Blampey, Quentin and Benkirane, Hakim and Bercovici, Nadege and Andre, Fabrice and Cournede, Paul-Henry},
7 | year = {2024},
8 | pages = {2024.09.09.612009},
9 | publisher = {bioRxiv},
10 | doi = {10.1101/2024.09.09.612009},
11 | }
12 | ```
13 |
14 | This library has been developed by Quentin Blampey, PhD student in biomathematics and deep learning. The following institutions funded this work:
15 |
16 | - Lab of Mathematics and Computer Science (MICS), **CentraleSupélec** (Engineering School, Paris-Saclay University).
17 | - PRISM center, **Gustave Roussy Institute** (Cancer campus, Paris-Saclay University).
18 |
--------------------------------------------------------------------------------
/docs/faq.md:
--------------------------------------------------------------------------------
1 | # Frequently asked questions
2 |
3 | ### How to use the GPU?
4 |
5 | Using a GPU may significantly speed up Novae's training or inference.
6 |
7 | If you have a valid GPU for PyTorch, you can set the `accelerator` argument (e.g., one of `["cpu", "gpu", "tpu", "hpu", "mps", "auto"]`) in the following methods: [model.fit()](../api/Novae/#novae.Novae.fit), [model.fine_tune()](../api/Novae/#novae.Novae.fine_tune), [model.compute_representations()](../api/Novae/#novae.Novae.compute_representations).
8 |
9 | When using a GPU, we also highly recommend setting multiple workers to speed up the dataset `__getitem__`. For that, you'll need to set the `num_workers` argument in the previous methods, according to the number of CPUs available (`num_workers=8` is usually a good value).
10 |
11 | For more details, refer to the API of the [PyTorch Lightning Trainer](https://lightning.ai/docs/pytorch/stable/common/trainer.html#trainer-class-api) and to the API of the [PyTorch DataLoader](https://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader).
12 |
13 | ### How to load a pretrained model?
14 |
15 | We highly recommend loading a pre-trained Novae model instead of re-training from scratch. For that, choose an available Novae model name on [our HuggingFace collection](https://huggingface.co/collections/MICS-Lab/novae-669cdf1754729d168a69f6bd), and provide this name to the [model.save_pretrained()](../api/Novae/#novae.Novae.save_pretrained) method:
16 |
17 | ```python
18 | from novae import Novae
19 |
20 | model = Novae.from_pretrained("MICS-Lab/novae-human-0") # or any valid model name
21 | ```
22 |
23 | ### How to avoid overcorrecting?
24 |
25 | By default, Novae corrects the batch-effect to get shared spatial domains across slides.
26 | The batch information is used only during training (`fit` or `fine_tune`), which should prevent Novae from overcorrecting in `zero_shot` mode.
27 |
28 | If not using the `zero_shot` mode, you can provide the `min_prototypes_ratio` parameter to control batch effect correction: either (i) in the `fine_tune` method itself, or (ii) during the model initialization (if retraining a model from scratch).
29 |
30 | For instance, if `min_prototypes_ratio=0.5`, Novae expects each slide to contain at least 50% of the prototypes (each prototype can be interpreted as an "elementary spatial domain"). Therefore, the lower `min_prototypes_ratio`, the lower the batch-effect correction. Conversely, if `min_prototypes_ratio=1`, all prototypes are expected to be found in all slides (this doesn't mean the proportions will be the same overall slides, though).
31 |
32 | ### How do I save my own model?
33 |
34 | If you have trained or fine-tuned your own Novae model, you can save it for later use. For that, use the [model.save_pretrained()](../api/Novae/#novae.Novae.save_pretrained) method as below:
35 |
36 | ```python
37 | model.save_pretrained(save_directory="./my-model-directory")
38 | ```
39 |
40 | Then, you can load this model back via the [model.from_pretrained()](../api/Novae/#novae.Novae.from_pretrained) method:
41 |
42 | ```python
43 | from novae import Novae
44 |
45 | model = Novae.from_pretrained("./my-model-directory")
46 | ```
47 |
48 | ### How to turn lazy loading on or off?
49 |
50 | By default, lazy loading is used only on large datasets. To enforce a specific behavior, you can do the following:
51 |
52 | ```python
53 | # never use lazy loading
54 | novae.settings.disable_lazy_loading()
55 |
56 | # always use lazy loading
57 | novae.settings.enable_lazy_loading()
58 |
59 | # use lazy loading only for AnnData objects with 1M+ cells
60 | novae.settings.enable_lazy_loading(n_obs_threshold=1_000_000)
61 | ```
62 |
63 | ### How to update the logging level?
64 |
65 | The logging level can be updated as below:
66 |
67 | ```python
68 | import logging
69 | from novae import log
70 |
71 | log.setLevel(logging.ERROR) # or any other level, e.g. logging.DEBUG
72 | ```
73 |
74 | ### How to disable auto-preprocessing
75 |
76 | By default, Novae automatically run data preprocessing for you. If you don't want that, you can run the line below.
77 |
78 | ```python
79 | novae.settings.auto_preprocessing = False
80 | ```
81 |
82 | ### How long does it take to use Novae?
83 |
84 | The `pip` installation of Novae usually takes less than a minute on a standard laptop. The inference time depends on the number of cells, but typically takes 5-20 minutes on a CPUs, or 30sec to 2 minutes on a GPU (expect it to be roughly 10x times faster on a GPU).
85 |
86 | ### How to contribute?
87 |
88 | If you want to contribute, check our [contributing guide](https://github.com/MICS-Lab/novae/blob/main/CONTRIBUTING.md).
89 |
90 | ### How to resolve any other issue?
91 |
92 | If you have any bugs/questions/suggestions, don't hesitate to [open a new issue](https://github.com/MICS-Lab/novae/issues).
93 |
--------------------------------------------------------------------------------
/docs/getting_started.md:
--------------------------------------------------------------------------------
1 | ## Installation
2 |
3 | Novae can be installed on every OS via `pip` or [`poetry`](https://python-poetry.org/docs/), on any Python version from `3.10` to `3.12` (included). By default, we recommend using `python==3.10`.
4 |
5 | !!! note "Advice (optional)"
6 |
7 | We advise creating a new environment via a package manager, except if you use Poetry, which will automatically create the environment.
8 |
9 | For instance, you can create a new `conda` environment:
10 |
11 | ```bash
12 | conda create --name novae python=3.10
13 | conda activate novae
14 | ```
15 |
16 | Choose one of the following, depending on your needs. It should take at most a few minutes.
17 |
18 | === "From PyPI"
19 |
20 | ``` bash
21 | pip install novae
22 | ```
23 |
24 | === "pip (editable mode)"
25 |
26 | ``` bash
27 | git clone https://github.com/MICS-Lab/novae.git
28 | cd novae
29 |
30 | pip install -e . # no extra
31 | pip install -e '.[dev]' # all extras
32 | ```
33 |
34 | === "Poetry (editable mode)"
35 |
36 | ``` bash
37 | git clone https://github.com/MICS-Lab/novae.git
38 | cd novae
39 |
40 | poetry install --all-extras
41 | ```
42 |
43 | ## Next steps
44 |
45 | - We recommend to start with our [first tutorial](../tutorials/main_usage).
46 | - You can also read the [API](../api/Novae).
47 | - If you have questions, please check our [FAQ](../faq) or open an issue on the [GitHub repository](https://github.com/MICS-Lab/novae).
48 | - If you want to contribute, check our [contributing guide](https://github.com/MICS-Lab/novae/blob/main/CONTRIBUTING.md).
49 |
--------------------------------------------------------------------------------
/docs/index.md:
--------------------------------------------------------------------------------
1 |
2 |  3 |
3 |
4 |
5 |
6 | 💫 Graph-based foundation model for spatial transcriptomics data
7 |
8 |
9 | Novae is a deep learning model for spatial domain assignments of spatial transcriptomics data (at both single-cell or spot resolution). It works across multiple gene panels, tissues, and technologies. Novae offers several additional features, including: (i) native batch-effect correction, (ii) analysis of spatially variable genes and pathways, and (iii) architecture analysis of tissue slides.
10 |
11 | ## Overview
12 |
13 |
14 |  15 |
15 |
16 |
17 | > **(a)** Novae was trained on a large dataset, and is shared on [Hugging Face Hub](https://huggingface.co/collections/MICS-Lab/novae-669cdf1754729d168a69f6bd). **(b)** Illustration of the main tasks and properties of Novae. **(c)** Illustration of the method behind Novae (self-supervision on graphs, adapted from [SwAV](https://arxiv.org/abs/2006.09882)).
18 |
19 |
20 | ## Why using Novae
21 |
22 | - It is already pretrained on a large dataset (pan human/mouse tissues, brain, ...). Therefore, you can compute spatial domains in a zero-shot manner (i.e., without fine-tuning).
23 | - It has been developed to find consistent domains across many slides. This also works if you have different technologies (e.g., MERSCOPE/Xenium) and multiple gene panels.
24 | - You can natively correct batch-effect, without using external tools.
25 | - After inference, the spatial domain assignment is super fast, allowing you to try multiple resolutions easily.
26 | - It supports many downstream tasks, all included inside one framework.
27 |
--------------------------------------------------------------------------------
/docs/javascripts/mathjax.js:
--------------------------------------------------------------------------------
1 | window.MathJax = {
2 | tex: {
3 | inlineMath: [["\\(", "\\)"]],
4 | displayMath: [["\\[", "\\]"]],
5 | processEscapes: true,
6 | processEnvironments: true
7 | },
8 | options: {
9 | ignoreHtmlClass: ".*|",
10 | processHtmlClass: "arithmatex"
11 | }
12 | };
13 |
14 | document$.subscribe(() => {
15 | MathJax.typesetPromise()
16 | })
17 |
--------------------------------------------------------------------------------
/docs/tutorials/LEE_AGING_CEREBELLUM_UP.json:
--------------------------------------------------------------------------------
1 | {
2 | "LEE_AGING_CEREBELLUM_UP" : {"systematicName":"MM1023","pmid":"10888876","exactSource":"Table 5S","geneSymbols":["Acadvl","Agt","Amh","Apc","Apoe","Axl","B2m","Bcl2a1a","Bdnf","C1qa","C1qb","C1qc","C4b","Capn2","Ccl21b","Cd24a","Cd68","Cdk4","Cst7","Ctsd","Ctsh","Ctss","Ctsz","Dnajb2","Efs","Eif2b5","Eprs1","Eps15","F2","Fcrl2","Fos","Gbp3","Gck","Gfap","Gnb2","Gng11","H2-Ab1","Hexb","Hmox1","Hnrnph3","Hoxa4","Hoxd12","Hspa8","Iars1","Ifi27","Ifit1","Ighm","Impa1","Irf7","Irgm1","Itgb5","Lgals3","Lgals3bp","Lmnb1","Mnat1","Mpeg1","Myh8","Nfya","Nos3","Notch1","Nr4a1","Or2c1","Pglyrp1","Pigf","Psg-ps1","Ptbp2","Ptpro","Rhog","Rpsa","Selplg","Sez6","Sipa1l2","Slc11a1","Slc7a3","Snta1","Spp1","Tbc1d1","Tbx6","Tgfbr3","Thbs2","Trappc5","Trim30a","Tyms","Ube2h","Wdfy3","Zfp40"],"msigdbURL":"https://www.gsea-msigdb.org/gsea/msigdb/mouse/geneset/LEE_AGING_CEREBELLUM_UP","externalDetailsURL":[],"filteredBySimilarity":[],"externalNamesForSimilarTerms":[],"collection":"M2:CGP"}
3 | }
4 |
--------------------------------------------------------------------------------
/docs/tutorials/input_modes.md:
--------------------------------------------------------------------------------
1 | Depending on your data and preferences, you can use 4 types of inputs.
2 | Specifically, it depends on whether (i) you have one or multiple slides and (ii) you prefer to concatenate your data.
3 |
4 | !!! info
5 | In all cases, the data structure is [AnnData](https://anndata.readthedocs.io/en/latest/). We may support MuData in the future.
6 |
7 | ## 1. One slide mode
8 |
9 | This case is the easiest one. You simply have one `AnnData` object corresponding to one slide.
10 |
11 | You can follow the first section of the [main usage tutorial](../main_usage).
12 |
13 | ## 2. Multiple slides, one AnnData object
14 |
15 | If you have multiple slides with the same gene panel, you can concatenate them into one `AnnData` object. In that case, make sure you keep a column in `adata.obs` that denotes which cell corresponds to which slide.
16 |
17 | Then, remind this column, and pass it to [`novae.spatial_neighbors`](../../api/utils/#novae.spatial_neighbors).
18 |
19 | !!! example
20 | For instance, you can do:
21 | ```python
22 | novae.spatial_neighbors(adata, slide_key="my-slide-id-column")
23 | ```
24 |
25 | ## 3. Multiple slides, one AnnData object per slide
26 |
27 | If you have multiple slides, you may prefer to keep one `AnnData` object for each slide. This is also convenient if you have different gene panels and can't concatenate your data.
28 |
29 | That case is pretty easy, since most functions and methods of Novae also support a **list of `AnnData` objects** as inputs. Therefore, simply pass a list of `AnnData` object, as below:
30 |
31 | !!! example
32 | ```python
33 | adatas = [adata_1, adata_2, ...]
34 |
35 | novae.spatial_neighbors(adatas)
36 |
37 | model.compute_representations(adatas, zero_shot=True)
38 | ```
39 |
40 | ## 4. Multiple slides, multiple slides per AnnData object
41 |
42 | If you have multiple slides and multiple panels, instead of the above option, you could have one `AnnData` object per panel, and multiple slides inside each `AnnData` object. In that case, make sure you keep a column in `adata.obs` that denotes which cell corresponds to which slide.
43 |
44 | Then, remind this column, and pass it to [`novae.spatial_neighbors`](../../api/utils/#novae.spatial_neighbors). The other functions don't need this argument.
45 |
46 | !!! example
47 | For instance, you can do:
48 | ```python
49 | adatas = [adata_1, adata_2, ...]
50 |
51 | novae.spatial_neighbors(adatas, slide_key="my-slide-id-column")
52 |
53 | model.compute_representations(adatas, zero_shot=True)
54 | ```
55 |
--------------------------------------------------------------------------------
/mkdocs.yml:
--------------------------------------------------------------------------------
1 | site_name: Novae
2 | repo_name: MICS-Lab/novae
3 | repo_url: https://github.com/MICS-Lab/novae
4 | copyright: Copyright © 2024 Quentin Blampey
5 | theme:
6 | name: material
7 | logo: assets/logo_small_black.png
8 | favicon: assets/logo_favicon.png
9 | palette:
10 | scheme: slate
11 | primary: white
12 | nav:
13 | - Home: index.md
14 | - Getting started: getting_started.md
15 | - Tutorials:
16 | - Main usage: tutorials/main_usage.ipynb
17 | - Different input modes: tutorials/input_modes.md
18 | - Usage on proteins: tutorials/proteins.ipynb
19 | - Spot/bin technologies: tutorials/resolutions.ipynb
20 | - API:
21 | - Novae model: api/Novae.md
22 | - Utils: api/utils.md
23 | - Plotting: api/plot.md
24 | - Advanced:
25 | - Metrics: api/metrics.md
26 | - Modules: api/modules.md
27 | - Dataloader: api/dataloader.md
28 | - FAQ: faq.md
29 | - Cite us: cite_us.md
30 |
31 | plugins:
32 | - search
33 | - mkdocstrings:
34 | handlers:
35 | python:
36 | options:
37 | show_root_heading: true
38 | heading_level: 3
39 | - mkdocs-jupyter:
40 | include_source: True
41 | markdown_extensions:
42 | - admonition
43 | - attr_list
44 | - md_in_html
45 | - pymdownx.details
46 | - pymdownx.highlight:
47 | anchor_linenums: true
48 | - pymdownx.inlinehilite
49 | - pymdownx.snippets
50 | - pymdownx.superfences
51 | - pymdownx.arithmatex:
52 | generic: true
53 | - pymdownx.tabbed:
54 | alternate_style: true
55 | extra_javascript:
56 | - javascripts/mathjax.js
57 | - https://polyfill.io/v3/polyfill.min.js?features=es6
58 | - https://cdn.jsdelivr.net/npm/mathjax@3/es5/tex-mml-chtml.js
59 |
--------------------------------------------------------------------------------
/novae/__init__.py:
--------------------------------------------------------------------------------
1 | import importlib.metadata
2 | import logging
3 |
4 | __version__ = importlib.metadata.version("novae")
5 |
6 | from ._logging import configure_logger
7 | from ._settings import settings
8 | from .model import Novae
9 | from . import utils
10 | from . import data
11 | from . import monitor
12 | from . import plot
13 | from .utils import spatial_neighbors, batch_effect_correction
14 |
15 | log = logging.getLogger("novae")
16 | configure_logger(log)
17 |
--------------------------------------------------------------------------------
/novae/_constants.py:
--------------------------------------------------------------------------------
1 | class Keys:
2 | # obs keys
3 | LEAVES: str = "novae_leaves"
4 | DOMAINS_PREFIX: str = "novae_domains_"
5 | IS_VALID_OBS: str = "neighborhood_valid"
6 | SLIDE_ID: str = "novae_sid"
7 |
8 | # obsm keys
9 | REPR: str = "novae_latent"
10 | REPR_CORRECTED: str = "novae_latent_corrected"
11 |
12 | # obsp keys
13 | ADJ: str = "spatial_distances"
14 | ADJ_LOCAL: str = "spatial_distances_local"
15 | ADJ_PAIR: str = "spatial_distances_pair"
16 |
17 | # var keys
18 | VAR_MEAN: str = "mean"
19 | VAR_STD: str = "std"
20 | IS_KNOWN_GENE: str = "in_vocabulary"
21 | HIGHLY_VARIABLE: str = "highly_variable"
22 | USE_GENE: str = "novae_use_gene"
23 |
24 | # layer keys
25 | COUNTS_LAYER: str = "counts"
26 |
27 | # misc keys
28 | UNS_TISSUE: str = "novae_tissue"
29 | ADATA_INDEX: str = "adata_index"
30 | N_BATCHES: str = "n_batches"
31 | NOVAE_VERSION: str = "novae_version"
32 |
33 |
34 | class Nums:
35 | # training constants
36 | EPS: float = 1e-8
37 | MIN_DATASET_LENGTH: int = 50_000
38 | MAX_DATASET_LENGTH_RATIO: float = 0.02
39 | DEFAULT_SAMPLE_CELLS: int = 100_000
40 | WARMUP_EPOCHS: int = 1
41 |
42 | # distances constants and thresholds (in microns)
43 | CELLS_CHARACTERISTIC_DISTANCE: int = 20 # characteristic distance between two cells, in microns
44 | MAX_MEAN_DISTANCE_RATIO: float = 8
45 |
46 | # genes constants
47 | N_HVG_THRESHOLD: int = 500
48 | MIN_GENES_FOR_HVG: int = 100
49 | MIN_GENES: int = 20
50 |
51 | # swav head constants
52 | SWAV_EPSILON: float = 0.05
53 | SINKHORN_ITERATIONS: int = 3
54 | QUEUE_SIZE: int = 2
55 | QUEUE_WEIGHT_THRESHOLD_RATIO: float = 0.99
56 |
57 | # misc nums
58 | MEAN_NGH_TH_WARNING: float = 3.5
59 | N_OBS_THRESHOLD: int = 2_000_000 # above this number, lazy loading is used
60 | RATIO_VALID_CELLS_TH: float = 0.7
61 |
--------------------------------------------------------------------------------
/novae/_logging.py:
--------------------------------------------------------------------------------
1 | import logging
2 |
3 | log = logging.getLogger(__name__)
4 |
5 |
6 | class ColorFormatter(logging.Formatter):
7 | grey = "\x1b[38;20m"
8 | blue = "\x1b[36;20m"
9 | yellow = "\x1b[33;20m"
10 | red = "\x1b[31;20m"
11 | bold_red = "\x1b[31;1m"
12 | reset = "\x1b[0m"
13 |
14 | prefix = "[%(levelname)s] (%(name)s)"
15 | suffix = "%(message)s"
16 |
17 | FORMATS = {
18 | logging.DEBUG: f"{grey}{prefix}{reset} {suffix}",
19 | logging.INFO: f"{blue}{prefix}{reset} {suffix}",
20 | logging.WARNING: f"{yellow}{prefix}{reset} {suffix}",
21 | logging.ERROR: f"{red}{prefix}{reset} {suffix}",
22 | logging.CRITICAL: f"{bold_red}{prefix}{reset} {suffix}",
23 | }
24 |
25 | def format(self, record):
26 | log_fmt = self.FORMATS.get(record.levelno)
27 | formatter = logging.Formatter(log_fmt)
28 | return formatter.format(record)
29 |
30 |
31 | def configure_logger(log: logging.Logger):
32 | log.setLevel(logging.INFO)
33 |
34 | consoleHandler = logging.StreamHandler()
35 | consoleHandler.setFormatter(ColorFormatter())
36 |
37 | log.addHandler(consoleHandler)
38 | log.propagate = False
39 |
--------------------------------------------------------------------------------
/novae/_settings.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 | from ._constants import Nums
4 |
5 |
6 | class Settings:
7 | # misc settings
8 | auto_preprocessing: bool = True
9 |
10 | def disable_lazy_loading(self):
11 | """Disable lazy loading of subgraphs in the NovaeDataset."""
12 | Nums.N_OBS_THRESHOLD = np.inf
13 |
14 | def enable_lazy_loading(self, n_obs_threshold: int = 0):
15 | """Enable lazy loading of subgraphs in the NovaeDataset.
16 |
17 | Args:
18 | n_obs_threshold: Lazy loading is used above this number of cells in an AnnData object.
19 | """
20 | Nums.N_OBS_THRESHOLD = n_obs_threshold
21 |
22 | @property
23 | def warmup_epochs(self):
24 | return Nums.WARMUP_EPOCHS
25 |
26 | @warmup_epochs.setter
27 | def warmup_epochs(self, value: int):
28 | Nums.WARMUP_EPOCHS = value
29 |
30 |
31 | settings = Settings()
32 |
--------------------------------------------------------------------------------
/novae/data/__init__.py:
--------------------------------------------------------------------------------
1 | from .convert import AnnDataTorch
2 | from .dataset import NovaeDataset
3 | from .datamodule import NovaeDatamodule
4 |
--------------------------------------------------------------------------------
/novae/data/convert.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import torch
3 | from anndata import AnnData
4 | from sklearn.preprocessing import LabelEncoder
5 | from torch import Tensor
6 |
7 | from .._constants import Keys, Nums
8 | from ..module import CellEmbedder
9 | from ..utils import sparse_std
10 |
11 |
12 | class AnnDataTorch:
13 | tensors: list[Tensor] | None
14 | genes_indices_list: list[Tensor]
15 |
16 | def __init__(self, adatas: list[AnnData], cell_embedder: CellEmbedder):
17 | """Converting AnnData objects to PyTorch tensors.
18 |
19 | Args:
20 | adatas: A list of `AnnData` objects.
21 | cell_embedder: A [novae.module.CellEmbedder][] object.
22 | """
23 | super().__init__()
24 | self.adatas = adatas
25 | self.cell_embedder = cell_embedder
26 |
27 | self.genes_indices_list = [self._adata_to_genes_indices(adata) for adata in self.adatas]
28 | self.tensors = None
29 |
30 | self.means, self.stds, self.label_encoder = self._compute_means_stds()
31 |
32 | # Tensors are loaded in memory for low numbers of cells
33 | if sum(adata.n_obs for adata in self.adatas) < Nums.N_OBS_THRESHOLD:
34 | self.tensors = [self.to_tensor(adata) for adata in self.adatas]
35 |
36 | def _adata_to_genes_indices(self, adata: AnnData) -> Tensor:
37 | return self.cell_embedder.genes_to_indices(adata.var_names[self._keep_var(adata)])[None, :]
38 |
39 | def _keep_var(self, adata: AnnData) -> AnnData:
40 | return adata.var[Keys.USE_GENE]
41 |
42 | def _compute_means_stds(self) -> tuple[Tensor, Tensor, LabelEncoder]:
43 | means, stds = {}, {}
44 |
45 | for adata in self.adatas:
46 | slide_ids = adata.obs[Keys.SLIDE_ID]
47 | for slide_id in slide_ids.cat.categories:
48 | adata_slide = adata[adata.obs[Keys.SLIDE_ID] == slide_id, self._keep_var(adata)]
49 |
50 | mean = adata_slide.X.mean(0)
51 | mean = mean.A1 if isinstance(mean, np.matrix) else mean
52 | means[slide_id] = mean.astype(np.float32)
53 |
54 | std = adata_slide.X.std(0) if isinstance(adata_slide.X, np.ndarray) else sparse_std(adata_slide.X, 0).A1
55 | stds[slide_id] = std.astype(np.float32)
56 |
57 | label_encoder = LabelEncoder()
58 | label_encoder.fit(list(means.keys()))
59 |
60 | means = [torch.tensor(means[slide_id]) for slide_id in label_encoder.classes_]
61 | stds = [torch.tensor(stds[slide_id]) for slide_id in label_encoder.classes_]
62 |
63 | return means, stds, label_encoder
64 |
65 | def to_tensor(self, adata: AnnData) -> Tensor:
66 | """Get the normalized gene expressions of the cells in the dataset.
67 | Only the genes of interest are kept (known genes and highly variable).
68 |
69 | Args:
70 | adata: An `AnnData` object.
71 |
72 | Returns:
73 | A `Tensor` containing the normalized gene expresions.
74 | """
75 | adata = adata[:, self._keep_var(adata)]
76 |
77 | if len(np.unique(adata.obs[Keys.SLIDE_ID])) == 1:
78 | slide_id_index = self.label_encoder.transform([adata.obs.iloc[0][Keys.SLIDE_ID]])[0]
79 | mean, std = self.means[slide_id_index], self.stds[slide_id_index]
80 | else:
81 | slide_id_indices = self.label_encoder.transform(adata.obs[Keys.SLIDE_ID])
82 | mean = torch.stack([self.means[i] for i in slide_id_indices]) # TODO: avoid stack (only if not fast enough)
83 | std = torch.stack([self.stds[i] for i in slide_id_indices])
84 |
85 | X = adata.X if isinstance(adata.X, np.ndarray) else adata.X.toarray()
86 | X = torch.tensor(X, dtype=torch.float32)
87 | X = (X - mean) / (std + Nums.EPS)
88 |
89 | return X
90 |
91 | def __getitem__(self, item: tuple[int, slice]) -> tuple[Tensor, Tensor]:
92 | """Get the expression values for a subset of cells (corresponding to a subgraph).
93 |
94 | Args:
95 | item: A `tuple` containing the index of the `AnnData` object and the indices of the cells in the neighborhoods.
96 |
97 | Returns:
98 | A `Tensor` of normalized gene expressions and a `Tensor` of gene indices.
99 | """
100 | adata_index, obs_indices = item
101 |
102 | if self.tensors is not None:
103 | return self.tensors[adata_index][obs_indices], self.genes_indices_list[adata_index]
104 |
105 | adata = self.adatas[adata_index]
106 | adata_view = adata[obs_indices]
107 |
108 | return self.to_tensor(adata_view), self.genes_indices_list[adata_index]
109 |
--------------------------------------------------------------------------------
/novae/data/datamodule.py:
--------------------------------------------------------------------------------
1 | import lightning as L
2 | from anndata import AnnData
3 | from torch_geometric.loader import DataLoader
4 |
5 | from ..module import CellEmbedder
6 | from . import NovaeDataset
7 |
8 |
9 | class NovaeDatamodule(L.LightningDataModule):
10 | """
11 | Datamodule used for training and inference. Small wrapper around the [novae.data.NovaeDataset][]
12 | """
13 |
14 | def __init__(
15 | self,
16 | adatas: list[AnnData],
17 | cell_embedder: CellEmbedder,
18 | batch_size: int,
19 | n_hops_local: int,
20 | n_hops_view: int,
21 | num_workers: int = 0,
22 | sample_cells: int | None = None,
23 | ) -> None:
24 | super().__init__()
25 | self.dataset = NovaeDataset(
26 | adatas,
27 | cell_embedder=cell_embedder,
28 | batch_size=batch_size,
29 | n_hops_local=n_hops_local,
30 | n_hops_view=n_hops_view,
31 | sample_cells=sample_cells,
32 | )
33 | self.batch_size = batch_size
34 | self.num_workers = num_workers
35 |
36 | def train_dataloader(self) -> DataLoader:
37 | """Get a Pytorch dataloader for prediction.
38 |
39 | Returns:
40 | The training dataloader.
41 | """
42 | self.dataset.training = True
43 | return DataLoader(
44 | self.dataset,
45 | batch_size=self.batch_size,
46 | shuffle=False,
47 | drop_last=True,
48 | num_workers=self.num_workers,
49 | )
50 |
51 | def predict_dataloader(self) -> DataLoader:
52 | """Get a Pytorch dataloader for prediction or inference.
53 |
54 | Returns:
55 | The prediction dataloader.
56 | """
57 | self.dataset.training = False
58 | return DataLoader(
59 | self.dataset,
60 | batch_size=self.batch_size,
61 | shuffle=False,
62 | drop_last=False,
63 | num_workers=self.num_workers,
64 | )
65 |
--------------------------------------------------------------------------------
/novae/module/__init__.py:
--------------------------------------------------------------------------------
1 | from .aggregate import AttentionAggregation
2 | from .embed import CellEmbedder
3 | from .augment import GraphAugmentation
4 | from .encode import GraphEncoder
5 | from .swav import SwavHead
6 |
--------------------------------------------------------------------------------
/novae/module/aggregate.py:
--------------------------------------------------------------------------------
1 | import lightning as L
2 | from torch import Tensor, nn
3 | from torch_geometric.nn.aggr import Aggregation
4 | from torch_geometric.nn.inits import reset
5 | from torch_geometric.utils import softmax
6 |
7 |
8 | class AttentionAggregation(Aggregation, L.LightningModule):
9 | """Aggregate the node embeddings using attention."""
10 |
11 | def __init__(self, output_size: int):
12 | """
13 |
14 | Args:
15 | output_size: Size of the representations, i.e. the encoder outputs (`O` in the article).
16 | """
17 | super().__init__()
18 | self.attention_aggregation = ProjectionLayers(output_size) # for backward compatibility when loading models
19 |
20 | def forward(
21 | self,
22 | x: Tensor,
23 | index: Tensor | None = None,
24 | ptr: None = None,
25 | dim_size: None = None,
26 | dim: int = -2,
27 | ) -> Tensor:
28 | """Performs attention aggragation.
29 |
30 | Args:
31 | x: The nodes embeddings representing `B` total graphs.
32 | index: The Pytorch Geometric index used to know to which graph each node belongs.
33 |
34 | Returns:
35 | A tensor of shape `(B, O)` of graph embeddings.
36 | """
37 | gate = self.attention_aggregation.gate_nn(x)
38 | x = self.attention_aggregation.nn(x)
39 |
40 | gate = softmax(gate, index, dim=dim)
41 |
42 | return self.reduce(gate * x, index, dim=dim)
43 |
44 | def reset_parameters(self):
45 | reset(self.attention_aggregation.gate_nn)
46 | reset(self.attention_aggregation.nn)
47 |
48 | def __repr__(self) -> str:
49 | return f"{self.__class__.__name__}(gate_nn={self.attention_aggregation.gate_nn}, nn={self.attention_aggregation.nn})"
50 |
51 |
52 | class ProjectionLayers(L.LightningModule):
53 | """
54 | Small class for backward compatibility when loading models
55 | Contains the projection layers used for the attention aggregation
56 | """
57 |
58 | def __init__(self, output_size):
59 | super().__init__()
60 | self.gate_nn = nn.Linear(output_size, 1)
61 | self.nn = nn.Linear(output_size, output_size)
62 |
--------------------------------------------------------------------------------
/novae/module/augment.py:
--------------------------------------------------------------------------------
1 | import lightning as L
2 | import torch
3 | from torch.distributions import Exponential
4 | from torch_geometric.data import Batch
5 |
6 |
7 | class GraphAugmentation(L.LightningModule):
8 | """Perform graph augmentation for Novae. It adds noise to the data and keeps a subset of the genes."""
9 |
10 | def __init__(
11 | self,
12 | panel_subset_size: float,
13 | background_noise_lambda: float,
14 | sensitivity_noise_std: float,
15 | ):
16 | """
17 |
18 | Args:
19 | panel_subset_size: Ratio of genes kept from the panel during augmentation.
20 | background_noise_lambda: Parameter of the exponential distribution for the noise augmentation.
21 | sensitivity_noise_std: Standard deviation for the multiplicative for for the noise augmentation.
22 | """
23 | super().__init__()
24 | self.panel_subset_size = panel_subset_size
25 | self.background_noise_lambda = background_noise_lambda
26 | self.sensitivity_noise_std = sensitivity_noise_std
27 |
28 | self.background_noise_distribution = Exponential(torch.tensor(float(background_noise_lambda)))
29 |
30 | def noise(self, data: Batch):
31 | """Add noise (inplace) to the data as detailed in the article.
32 |
33 | Args:
34 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs.
35 | """
36 | sample_shape = (data.batch_size, data.x.shape[1])
37 |
38 | additions = self.background_noise_distribution.sample(sample_shape=sample_shape).to(self.device)
39 | gaussian_noise = torch.randn(sample_shape, device=self.device)
40 | factors = (1 + gaussian_noise * self.sensitivity_noise_std).clip(0, 2)
41 |
42 | for i in range(data.batch_size):
43 | start, stop = data.ptr[i], data.ptr[i + 1]
44 | data.x[start:stop] = data.x[start:stop] * factors[i] + additions[i]
45 |
46 | def panel_subset(self, data: Batch):
47 | """
48 | Keep a ratio of `panel_subset_size` of the input genes (inplace operation).
49 |
50 | Args:
51 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs.
52 | """
53 | n_total = len(data.genes_indices[0])
54 | n_subset = int(n_total * self.panel_subset_size)
55 |
56 | gene_subset_indices = torch.randperm(n_total)[:n_subset]
57 |
58 | data.x = data.x[:, gene_subset_indices]
59 | data.genes_indices = data.genes_indices[:, gene_subset_indices]
60 |
61 | def forward(self, data: Batch) -> Batch:
62 | """Perform data augmentation (`noise` and `panel_subset`).
63 |
64 | Args:
65 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs.
66 |
67 | Returns:
68 | The augmented `Data` object
69 | """
70 | self.panel_subset(data)
71 | self.noise(data)
72 | return data
73 |
--------------------------------------------------------------------------------
/novae/module/embed.py:
--------------------------------------------------------------------------------
1 | import json
2 | import logging
3 | from pathlib import Path
4 |
5 | import lightning as L
6 | import numpy as np
7 | import pandas as pd
8 | import torch
9 | import torch.nn.functional as F
10 | from anndata import AnnData
11 | from scipy.sparse import issparse
12 | from sklearn.decomposition import PCA
13 | from sklearn.neighbors import KDTree
14 | from torch import nn
15 | from torch_geometric.data import Data
16 |
17 | from .. import utils
18 | from .._constants import Keys
19 |
20 | log = logging.getLogger(__name__)
21 |
22 |
23 | class CellEmbedder(L.LightningModule):
24 | """Convert a cell into an embedding using a gene embedding matrix."""
25 |
26 | def __init__(
27 | self,
28 | gene_names: list[str] | dict[str, int],
29 | embedding_size: int | None,
30 | embedding: torch.Tensor | None = None,
31 | ) -> None:
32 | """
33 |
34 | Args:
35 | gene_names: Name of the genes to be used in the embedding, or dictionnary of index to name.
36 | embedding_size: Size of the embeddings of the genes (`E` in the article). Optional if `embedding` is provided.
37 | embedding: Optional pre-trained embedding matrix. If provided, `embedding_size` shouldn't be provided.
38 | """
39 | super().__init__()
40 | assert (embedding_size is None) ^ (embedding is None), "Either embedding_size or embedding must be provided"
41 |
42 | if isinstance(gene_names, dict):
43 | self.gene_to_index = {gene.lower(): index for gene, index in gene_names.items()}
44 | self.gene_names = sorted(self.gene_to_index, key=self.gene_to_index.get)
45 | _check_gene_to_index(self.gene_to_index)
46 | else:
47 | self.gene_names = [gene.lower() for gene in gene_names]
48 | self.gene_to_index = {gene: i for i, gene in enumerate(self.gene_names)}
49 |
50 | self.voc_size = len(self.gene_names)
51 |
52 | if embedding is None:
53 | self.embedding_size = embedding_size
54 | self.embedding = nn.Embedding(self.voc_size, embedding_size)
55 | else:
56 | self.embedding_size = embedding.size(1)
57 | self.embedding = nn.Embedding.from_pretrained(embedding)
58 |
59 | self.linear = nn.Linear(self.embedding_size, self.embedding_size)
60 | self._init_linear()
61 |
62 | @torch.no_grad()
63 | def _init_linear(self):
64 | self.linear.weight.data.copy_(torch.eye(self.embedding_size))
65 | self.linear.bias.data.zero_()
66 |
67 | @classmethod
68 | def from_scgpt_embedding(cls, scgpt_model_dir: str) -> "CellEmbedder":
69 | """Initialize the CellEmbedder from a scGPT pretrained model directory.
70 |
71 | Args:
72 | scgpt_model_dir: Path to a directory containing a scGPT checkpoint, i.e. a `vocab.json` and a `best_model.pt` file.
73 |
74 | Returns:
75 | A CellEmbedder instance.
76 | """
77 | scgpt_model_dir = Path(scgpt_model_dir)
78 |
79 | vocab_file = scgpt_model_dir / "vocab.json"
80 |
81 | with open(vocab_file) as file:
82 | gene_to_index: dict[str, int] = json.load(file)
83 |
84 | checkpoint = torch.load(scgpt_model_dir / "best_model.pt", map_location=torch.device("cpu"))
85 | embedding = checkpoint["encoder.embedding.weight"]
86 |
87 | return cls(gene_to_index, None, embedding=embedding)
88 |
89 | def genes_to_indices(self, gene_names: pd.Index | list[str], as_torch: bool = True) -> torch.Tensor | np.ndarray:
90 | """Convert gene names to their corresponding indices.
91 |
92 | Args:
93 | gene_names: Names of the gene names to convert.
94 | as_torch: Whether to return a `torch` tensor or a `numpy` array.
95 |
96 | Returns:
97 | A tensor or array of gene indices.
98 | """
99 | indices = [self.gene_to_index[gene] for gene in utils.lower_var_names(gene_names)]
100 |
101 | if as_torch:
102 | return torch.tensor(indices, dtype=torch.long)
103 |
104 | return np.array(indices, dtype=np.int16)
105 |
106 | def forward(self, data: Data) -> Data:
107 | """Embed the input data.
108 |
109 | Args:
110 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs. The number of node features is variable.
111 |
112 | Returns:
113 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs. Each node now has a size of `E`.
114 | """
115 | genes_embeddings = self.embedding(data.genes_indices[0])
116 | genes_embeddings = self.linear(genes_embeddings)
117 | genes_embeddings = F.normalize(genes_embeddings, dim=0, p=2)
118 |
119 | data.x = data.x @ genes_embeddings
120 | return data
121 |
122 | def pca_init(self, adatas: list[AnnData] | None):
123 | """Initialize the Noave embeddings with PCA components.
124 |
125 | Args:
126 | adatas: A list of `AnnData` objects to use for PCA initialization.
127 | """
128 | if adatas is None:
129 | return
130 |
131 | adatas = [adata[:, adata.var[Keys.USE_GENE]] for adata in adatas]
132 |
133 | adata = max(adatas, key=lambda adata: adata.n_vars)
134 |
135 | if adata.X.shape[1] <= self.embedding_size:
136 | log.warning(
137 | f"PCA with {self.embedding_size} components can not be run on shape {adata.X.shape}.\nTo use PCA initialization, set a lower `embedding_size` (<{adata.X.shape[1]}) in novae.Novae()."
138 | )
139 | return
140 |
141 | X = adata.X.toarray() if issparse(adata.X) else adata.X
142 |
143 | log.info("Running PCA embedding initialization")
144 |
145 | pca = PCA(n_components=self.embedding_size)
146 | pca.fit(X.astype(np.float32))
147 |
148 | indices = self.genes_to_indices(adata.var_names)
149 | self.embedding.weight.data[indices] = torch.tensor(pca.components_.T)
150 |
151 | known_var_names = utils.lower_var_names(adata.var_names)
152 |
153 | for other_adata in adatas:
154 | other_var_names = utils.lower_var_names(other_adata.var_names)
155 | where_in = np.isin(other_var_names, known_var_names)
156 |
157 | if where_in.all():
158 | continue
159 |
160 | X = other_adata[:, where_in].X.toarray().T
161 | Y = other_adata[:, ~where_in].X.toarray().T
162 |
163 | tree = KDTree(X)
164 | _, ind = tree.query(Y, k=1)
165 | neighbor_indices = self.genes_to_indices(other_adata[:, where_in].var_names[ind[:, 0]])
166 |
167 | indices = self.genes_to_indices(other_adata[:, ~where_in].var_names)
168 | self.embedding.weight.data[indices] = self.embedding.weight.data[neighbor_indices].clone()
169 |
170 |
171 | def _check_gene_to_index(gene_to_index: dict[str, int]):
172 | values = list(set(gene_to_index.values()))
173 |
174 | assert len(values) == len(gene_to_index), "gene_to_index should be a dictionnary with unique values"
175 |
176 | assert min(values) == 0 and max(values) == len(values) - 1, (
177 | "gene_to_index should be a dictionnary with continuous indices starting from 0"
178 | )
179 |
--------------------------------------------------------------------------------
/novae/module/encode.py:
--------------------------------------------------------------------------------
1 | import lightning as L
2 | from torch import Tensor
3 | from torch_geometric.data import Batch
4 | from torch_geometric.nn.models import GAT
5 |
6 | from . import AttentionAggregation
7 |
8 |
9 | class GraphEncoder(L.LightningModule):
10 | """Graph encoder of Novae. It uses a graph attention network."""
11 |
12 | def __init__(
13 | self,
14 | embedding_size: int,
15 | hidden_size: int,
16 | num_layers: int,
17 | output_size: int,
18 | heads: int,
19 | ) -> None:
20 | """
21 | Args:
22 | embedding_size: Size of the embeddings of the genes (`E` in the article).
23 | hidden_size: The size of the hidden layers in the GAT.
24 | num_layers: The number of layers in the GAT.
25 | output_size: Size of the representations, i.e. the encoder outputs (`O` in the article).
26 | heads: The number of attention heads in the GAT.
27 | """
28 | super().__init__()
29 | self.gnn = GAT(
30 | embedding_size,
31 | hidden_channels=hidden_size,
32 | num_layers=num_layers,
33 | out_channels=output_size,
34 | edge_dim=1,
35 | v2=True,
36 | heads=heads,
37 | act="ELU",
38 | )
39 |
40 | self.node_aggregation = AttentionAggregation(output_size)
41 |
42 | def forward(self, data: Batch) -> Tensor:
43 | """Encode the input data.
44 |
45 | Args:
46 | data: A Pytorch Geometric `Data` object representing a batch of `B` graphs.
47 |
48 | Returns:
49 | A tensor of shape `(B, O)` containing the encoded graphs.
50 | """
51 | out = self.gnn(x=data.x, edge_index=data.edge_index, edge_attr=data.edge_attr)
52 | return self.node_aggregation(out, index=data.batch)
53 |
--------------------------------------------------------------------------------
/novae/monitor/__init__.py:
--------------------------------------------------------------------------------
1 | from .eval import (
2 | jensen_shannon_divergence,
3 | mean_fide_score,
4 | fide_score,
5 | entropy,
6 | heuristic,
7 | mean_normalized_entropy,
8 | )
9 |
--------------------------------------------------------------------------------
/novae/monitor/callback.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import numpy as np
3 | import scanpy as sc
4 | import seaborn as sns
5 | from anndata import AnnData
6 | from lightning import Trainer
7 | from lightning.pytorch.callbacks import Callback
8 |
9 | from .._constants import Keys
10 | from ..model import Novae
11 | from .eval import heuristic, mean_fide_score
12 | from .log import log_plt_figure, save_pdf_figure
13 |
14 |
15 | class LogProtoCovCallback(Callback):
16 | def on_train_epoch_end(self, trainer: Trainer, model: Novae) -> None:
17 | C = model.swav_head.prototypes.data.numpy(force=True)
18 |
19 | plt.figure(figsize=(10, 10))
20 | sns.clustermap(np.cov(C))
21 | log_plt_figure("prototypes_covariance")
22 |
23 |
24 | class LogTissuePrototypeWeights(Callback):
25 | def on_train_epoch_end(self, trainer: Trainer, model: Novae) -> None:
26 | if model.swav_head.queue is None:
27 | return
28 |
29 | model.plot_prototype_weights()
30 | save_pdf_figure(f"tissue_prototype_weights_e{model.current_epoch}")
31 | log_plt_figure("tissue_prototype_weights")
32 |
33 |
34 | class ValidationCallback(Callback):
35 | def __init__(
36 | self,
37 | adatas: list[AnnData] | None,
38 | accelerator: str = "cpu",
39 | num_workers: int = 0,
40 | slide_name_key: str = "slide_id",
41 | k: int = 7,
42 | ):
43 | assert adatas is None or len(adatas) == 1, "ValidationCallback only supports single slide mode for now"
44 | self.adata = adatas[0] if adatas is not None else None
45 | self.accelerator = accelerator
46 | self.num_workers = num_workers
47 | self.slide_name_key = slide_name_key
48 | self.k = k
49 |
50 | self._max_heuristic = 0.0
51 |
52 | def on_train_epoch_end(self, trainer: Trainer, model: Novae):
53 | if self.adata is None:
54 | return
55 |
56 | model.mode.trained = True # trick to avoid assert error in compute_representations
57 |
58 | model.compute_representations(
59 | self.adata, accelerator=self.accelerator, num_workers=self.num_workers, zero_shot=True
60 | )
61 | model.swav_head.hierarchical_clustering()
62 |
63 | obs_key = model.assign_domains(self.adata, n_domains=self.k)
64 |
65 | plt.figure()
66 | sc.pl.spatial(self.adata, color=obs_key, spot_size=20, img_key=None, show=False)
67 | slide_name_key = self.slide_name_key if self.slide_name_key in self.adata.obs else Keys.SLIDE_ID
68 | log_plt_figure(f"val_{self.k}_{self.adata.obs[slide_name_key].iloc[0]}")
69 |
70 | fide = mean_fide_score(self.adata, obs_key=obs_key, n_classes=self.k)
71 | model.log("metrics/val_mean_fide_score", fide)
72 |
73 | heuristic_ = heuristic(self.adata, obs_key=obs_key, n_classes=self.k)
74 | model.log("metrics/val_heuristic", heuristic_)
75 |
76 | self._max_heuristic = max(self._max_heuristic, heuristic_)
77 | model.log("metrics/val_max_heuristic", self._max_heuristic)
78 |
79 | model.mode.zero_shot = False
80 |
--------------------------------------------------------------------------------
/novae/monitor/eval.py:
--------------------------------------------------------------------------------
1 | import logging
2 |
3 | import numpy as np
4 | from anndata import AnnData
5 | from sklearn import metrics
6 |
7 | from .._constants import Keys, Nums
8 |
9 | log = logging.getLogger(__name__)
10 |
11 |
12 | def mean_fide_score(
13 | adatas: AnnData | list[AnnData], obs_key: str, slide_key: str | None = None, n_classes: int | None = None
14 | ) -> float:
15 | """Mean FIDE score over all slides. A low score indicates a great domain continuity.
16 |
17 | Args:
18 | adatas: An `AnnData` object, or a list of `AnnData` objects.
19 | obs_key: Key of `adata.obs` containing the domains annotation.
20 | slide_key: Optional key of `adata.obs` containing the ID of each slide. Not needed if each `adata` is a slide.
21 | n_classes: Optional number of classes. This can be useful if not all classes are predicted, for a fair comparision.
22 |
23 | Returns:
24 | The FIDE score averaged for all slides.
25 | """
26 | return np.mean([
27 | fide_score(adata, obs_key, n_classes=n_classes)
28 | for adata in _iter_uid(adatas, slide_key=slide_key, obs_key=obs_key)
29 | ])
30 |
31 |
32 | def fide_score(adata: AnnData, obs_key: str, n_classes: int | None = None) -> float:
33 | """F1-score of intra-domain edges (FIDE). A high score indicates a great domain continuity.
34 |
35 | Note:
36 | The F1-score is computed for every class, then all F1-scores are averaged. If some classes
37 | are not predicted, the `n_classes` argument allows to pad with zeros before averaging the F1-scores.
38 |
39 | Args:
40 | adata: An `AnnData` object
41 | obs_key: Key of `adata.obs` containing the domains annotation.
42 | n_classes: Optional number of classes. This can be useful if not all classes are predicted, for a fair comparision.
43 |
44 | Returns:
45 | The FIDE score.
46 | """
47 | i_left, i_right = adata.obsp[Keys.ADJ].nonzero()
48 | classes_left, classes_right = adata.obs.iloc[i_left][obs_key].values, adata.obs.iloc[i_right][obs_key].values
49 |
50 | where_valid = ~classes_left.isna() & ~classes_right.isna()
51 | classes_left, classes_right = classes_left[where_valid], classes_right[where_valid]
52 |
53 | f1_scores = metrics.f1_score(classes_left, classes_right, average=None)
54 |
55 | if n_classes is None:
56 | return f1_scores.mean()
57 |
58 | assert n_classes >= len(f1_scores), f"Expected {n_classes:=}, but found {len(f1_scores)}, which is greater"
59 |
60 | return np.pad(f1_scores, (0, n_classes - len(f1_scores))).mean()
61 |
62 |
63 | def jensen_shannon_divergence(adatas: AnnData | list[AnnData], obs_key: str, slide_key: str | None = None) -> float:
64 | """Jensen-Shannon divergence (JSD) over all slides
65 |
66 | Args:
67 | adatas: One or a list of AnnData object(s)
68 | obs_key: Key of `adata.obs` containing the domains annotation.
69 | slide_key: Optional key of `adata.obs` containing the ID of each slide. Not needed if each `adata` is a slide.
70 |
71 | Returns:
72 | The Jensen-Shannon divergence score for all slides
73 | """
74 | all_categories = set()
75 | for adata in _iter_uid(adatas, slide_key=slide_key, obs_key=obs_key):
76 | all_categories.update(adata.obs[obs_key].cat.categories)
77 | all_categories = sorted(all_categories)
78 |
79 | distributions = []
80 | for adata in _iter_uid(adatas, slide_key=slide_key, obs_key=obs_key):
81 | value_counts = adata.obs[obs_key].value_counts(sort=False)
82 | distribution = np.zeros(len(all_categories))
83 |
84 | for i, category in enumerate(all_categories):
85 | if category in value_counts:
86 | distribution[i] = value_counts[category]
87 |
88 | distributions.append(distribution)
89 |
90 | return _jensen_shannon_divergence(np.array(distributions))
91 |
92 |
93 | def _jensen_shannon_divergence(distributions: np.ndarray) -> float:
94 | """Compute the Jensen-Shannon divergence (JSD) for a multiple probability distributions.
95 |
96 | The lower the score, the better distribution of clusters among the different batches.
97 |
98 | Args:
99 | distributions: An array of shape (B, C), where B is the number of batches, and C is the number of clusters. For each batch, it contains the percentage of each cluster among cells.
100 |
101 | Returns:
102 | A float corresponding to the JSD
103 | """
104 | distributions = distributions / distributions.sum(1)[:, None]
105 | mean_distribution = np.mean(distributions, 0)
106 |
107 | return entropy(mean_distribution) - np.mean([entropy(dist) for dist in distributions])
108 |
109 |
110 | def entropy(distribution: np.ndarray) -> float:
111 | """Shannon entropy
112 |
113 | Args:
114 | distribution: An array of probabilities (should sum to one)
115 |
116 | Returns:
117 | The Shannon entropy
118 | """
119 | return -(distribution * np.log2(distribution + Nums.EPS)).sum()
120 |
121 |
122 | def mean_normalized_entropy(
123 | adatas: AnnData | list[AnnData], n_classes: int, obs_key: str, slide_key: str | None = None
124 | ) -> float:
125 | return np.mean([
126 | _mean_normalized_entropy(adata, obs_key, n_classes=n_classes)
127 | for adata in _iter_uid(adatas, slide_key=slide_key, obs_key=obs_key)
128 | ])
129 |
130 |
131 | def _mean_normalized_entropy(adata: AnnData, obs_key: str, n_classes: int) -> float:
132 | distribution = adata.obs[obs_key].value_counts(normalize=True).values
133 | distribution = np.pad(distribution, (0, n_classes - len(distribution)), mode="constant")
134 | entropy_ = entropy(distribution)
135 |
136 | return entropy_ / np.log2(n_classes)
137 |
138 |
139 | def heuristic(adata: AnnData | list[AnnData], obs_key: str, n_classes: int, slide_key: str | None = None) -> float:
140 | """Heuristic score to evaluate the quality of the clustering.
141 |
142 | Args:
143 | adata: An `AnnData` object
144 | obs_key: The key in `adata.obs` that contains the domains.
145 | n_classes: The number of classes.
146 | slide_key: The key in `adata.obs` that contains the slide id.
147 |
148 | Returns:
149 | The heuristic score.
150 | """
151 | return np.mean([
152 | _heuristic(adata, obs_key, n_classes) for adata in _iter_uid(adata, slide_key=slide_key, obs_key=obs_key)
153 | ])
154 |
155 |
156 | def _heuristic(adata: AnnData, obs_key: str, n_classes: int) -> float:
157 | fide_ = fide_score(adata, obs_key, n_classes=n_classes)
158 |
159 | distribution = adata.obs[obs_key].value_counts(normalize=True).values
160 | distribution = np.pad(distribution, (0, n_classes - len(distribution)), mode="constant")
161 | entropy_ = entropy(distribution)
162 |
163 | return fide_ * entropy_ / np.log2(n_classes)
164 |
165 |
166 | def _iter_uid(adatas: AnnData | list[AnnData], slide_key: str | None = None, obs_key: str | None = None):
167 | """Iterate over all slides, and make sure `adata.obs[obs_key]` is categorical.
168 |
169 | Args:
170 | adatas: One or a list of AnnData object(s).
171 | slide_key: The key in `adata.obs` that contains the slide id.
172 | obs_key: The key in `adata.obs` that contains the domain id.
173 |
174 | Yields:
175 | One `AnnData` per slide.
176 | """
177 | if isinstance(adatas, AnnData):
178 | adatas = [adatas]
179 |
180 | if obs_key is not None:
181 | categories = set.union(*[set(adata.obs[obs_key].astype("category").cat.categories) for adata in adatas])
182 | for adata in adatas:
183 | adata.obs[obs_key] = adata.obs[obs_key].astype("category").cat.set_categories(categories)
184 |
185 | for adata in adatas:
186 | if slide_key is None:
187 | yield adata
188 | continue
189 |
190 | for slide_id in adata.obs[slide_key].unique():
191 | adata_yield = adata[adata.obs[slide_key] == slide_id]
192 |
193 | yield adata_yield
194 |
--------------------------------------------------------------------------------
/novae/monitor/log.py:
--------------------------------------------------------------------------------
1 | import io
2 | from pathlib import Path
3 |
4 | import matplotlib.pyplot as plt
5 | import wandb
6 | from PIL import Image
7 |
8 | from ..utils import repository_root
9 |
10 |
11 | def log_plt_figure(name: str, dpi: int = 300) -> None:
12 | img_buf = io.BytesIO()
13 | plt.savefig(img_buf, format="png", bbox_inches="tight", dpi=dpi)
14 | wandb.log({name: wandb.Image(Image.open(img_buf))})
15 | plt.close()
16 |
17 |
18 | def save_pdf_figure(name: str):
19 | plt.savefig(wandb_results_dir() / f"{name}.pdf", format="pdf", bbox_inches="tight")
20 |
21 |
22 | def wandb_results_dir() -> Path:
23 | res_dir: Path = repository_root() / "data" / "results" / wandb.run.name
24 | res_dir.mkdir(parents=True, exist_ok=True)
25 | return res_dir
26 |
--------------------------------------------------------------------------------
/novae/plot/__init__.py:
--------------------------------------------------------------------------------
1 | from ._graph import _domains_hierarchy, paga, connectivities
2 | from ._spatial import domains, spatially_variable_genes
3 | from ._heatmap import _weights_clustermap, pathway_scores
4 | from ._bar import domains_proportions
5 |
--------------------------------------------------------------------------------
/novae/plot/_bar.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import pandas as pd
3 | import seaborn as sns
4 | from anndata import AnnData
5 |
6 | from .. import utils
7 | from ._utils import get_categorical_color_palette
8 |
9 |
10 | def domains_proportions(
11 | adata: AnnData | list[AnnData],
12 | obs_key: str | None = None,
13 | slide_name_key: str | None = None,
14 | figsize: tuple[int, int] = (2, 5),
15 | show: bool = True,
16 | ):
17 | """Show the proportion of each domain in the slide(s).
18 |
19 | Args:
20 | adata: One `AnnData` object, or a list of `AnnData` objects.
21 | obs_key: The key in `adata.obs` that contains the Novae domains. By default, the last available domain key is shown.
22 | figsize: Matplotlib figure size.
23 | show: Whether to show the plot.
24 | """
25 | adatas = [adata] if isinstance(adata, AnnData) else adata
26 | slide_name_key = utils.check_slide_name_key(adatas, slide_name_key)
27 | obs_key = utils.check_available_domains_key(adatas, obs_key)
28 |
29 | all_domains, colors = get_categorical_color_palette(adatas, obs_key)
30 |
31 | names, series = [], []
32 | for adata_slide in utils.iter_slides(adatas):
33 | names.append(adata_slide.obs[slide_name_key].iloc[0])
34 | series.append(adata_slide.obs[obs_key].value_counts(normalize=True))
35 |
36 | df = pd.concat(series, axis=1)
37 | df.columns = names
38 |
39 | df.T.plot(kind="bar", stacked=True, figsize=figsize, color=dict(zip(all_domains, colors)))
40 | sns.despine(offset=10, trim=True)
41 | plt.legend(bbox_to_anchor=(1.04, 0.5), loc="center left", borderaxespad=0, frameon=False)
42 | plt.ylabel("Proportion")
43 | plt.xticks(rotation=90)
44 |
45 | if show:
46 | plt.show()
47 |
--------------------------------------------------------------------------------
/novae/plot/_graph.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import numpy as np
3 | import scanpy as sc
4 | import seaborn as sns
5 | from anndata import AnnData
6 | from matplotlib.collections import LineCollection
7 | from scipy.cluster.hierarchy import dendrogram
8 | from sklearn.cluster import AgglomerativeClustering
9 |
10 | from .. import utils
11 | from .._constants import Keys
12 | from ._utils import _subplots_per_slide, get_categorical_color_palette
13 |
14 |
15 | def _leaves_count(clustering: AgglomerativeClustering) -> np.ndarray:
16 | counts = np.zeros(clustering.children_.shape[0])
17 | n_samples = len(clustering.labels_)
18 | for i, merge in enumerate(clustering.children_):
19 | current_count = 0
20 | for child_idx in merge:
21 | if child_idx < n_samples:
22 | current_count += 1 # leaf node
23 | else:
24 | current_count += counts[child_idx - n_samples]
25 | counts[i] = current_count
26 | return counts
27 |
28 |
29 | def _domains_hierarchy(
30 | clustering: AgglomerativeClustering,
31 | max_level: int = 10,
32 | hline_level: int | list[int] | None = None,
33 | leaf_font_size: int = 10,
34 | **kwargs,
35 | ) -> None:
36 | assert max_level > 1
37 |
38 | size = clustering.children_.shape[0]
39 | original_ymax = max_level + 1
40 | original_ticks = np.arange(1, original_ymax)
41 | height = original_ymax + np.arange(size) - size
42 |
43 | if hline_level is not None:
44 | hline_level = [hline_level] if isinstance(hline_level, int) else hline_level
45 | for level in hline_level:
46 | plt.hlines(original_ymax - level, 0, 1e5, colors="r", linestyles="dashed")
47 |
48 | linkage_matrix = np.column_stack([clustering.children_, height.clip(0), _leaves_count(clustering)]).astype(float)
49 |
50 | ddata = dendrogram(
51 | linkage_matrix,
52 | color_threshold=-1,
53 | leaf_font_size=leaf_font_size,
54 | p=max_level + 1,
55 | truncate_mode="lastp",
56 | above_threshold_color="#ccc",
57 | **kwargs,
58 | )
59 |
60 | for i, d in zip(ddata["icoord"][::-1], ddata["dcoord"][::-1]):
61 | x, y = 0.5 * sum(i[1:3]), d[1]
62 | plt.plot(x, y, "ko")
63 | plt.annotate(
64 | f"D{2 * size - max_level + int(y)}",
65 | (x, y),
66 | xytext=(0, -8),
67 | textcoords="offset points",
68 | va="top",
69 | ha="center",
70 | )
71 |
72 | plt.yticks(original_ticks, original_ymax - original_ticks)
73 |
74 | plt.xlabel(None)
75 | plt.ylabel("Domains level")
76 | plt.title("Domains hierarchy")
77 | plt.xlabel("Number of domains in node (or prototype ID if no parenthesis)")
78 | sns.despine(offset=10, trim=True, bottom=True)
79 |
80 |
81 | def paga(adata: AnnData, obs_key: str | None = None, show: bool = True, **paga_plot_kwargs: int):
82 | """Plot a PAGA graph.
83 |
84 | Info:
85 | Currently, this function only supports one slide per call.
86 |
87 | Args:
88 | adata: An AnnData object.
89 | obs_key: Name of the key from `adata.obs` containing the Novae domains. By default, the last available domain key is shown.
90 | show: Whether to show the plot.
91 | **paga_plot_kwargs: Additional arguments for `sc.pl.paga`.
92 | """
93 | assert isinstance(adata, AnnData), f"For now, only AnnData objects are supported, received {type(adata)}"
94 |
95 | obs_key = utils.check_available_domains_key([adata], obs_key)
96 |
97 | get_categorical_color_palette([adata], obs_key)
98 |
99 | adata_clean = adata[~adata.obs[obs_key].isna()]
100 |
101 | if "paga" not in adata.uns or adata.uns["paga"]["groups"] != obs_key:
102 | sc.pp.neighbors(adata_clean, use_rep=Keys.REPR)
103 | sc.tl.paga(adata_clean, groups=obs_key)
104 |
105 | adata.uns["paga"] = adata_clean.uns["paga"]
106 | adata.uns[f"{obs_key}_sizes"] = adata_clean.uns[f"{obs_key}_sizes"]
107 |
108 | sc.pl.paga(adata_clean, title=f"PAGA graph ({obs_key})", show=False, **paga_plot_kwargs)
109 | sns.despine(offset=10, trim=True, bottom=True)
110 |
111 | if show:
112 | plt.show()
113 |
114 |
115 | def connectivities(
116 | adata: AnnData,
117 | ngh_threshold: int | None = 2,
118 | cell_size: int = 2,
119 | ncols: int = 4,
120 | fig_size_per_slide: tuple[int, int] = (5, 5),
121 | linewidths: float = 0.1,
122 | line_color: str = "#333",
123 | cmap: str = "rocket",
124 | color_isolated_cells: str = "orangered",
125 | show: bool = True,
126 | ):
127 | """Show the graph of the spatial connectivities between cells. By default,
128 | the cells which have a number of neighbors inferior to `ngh_threshold` are shown
129 | in red. If `ngh_threshold` is `None`, the cells are colored by the number of neighbors.
130 |
131 | !!! info "Quality control"
132 | This plot is useful to check the quality of the spatial connectivities obtained via [novae.spatial_neighbors][].
133 | Make sure few cells (e.g., less than 5%) have a number of neighbors below `ngh_threshold`.
134 | If too many cells are isolated, you may want to increase the `radius` parameter in [novae.spatial_neighbors][].
135 | Conversely, if there are some less that are really **far from each other**, but still connected, so may want to decrease the `radius` parameter to **disconnect** them.
136 |
137 | Args:
138 | adata: An AnnData object.
139 | ngh_threshold: Only cells with a number of neighbors below this threshold are shown (with color `color_isolated_cells`). If `None`, cells are colored by the number of neighbors.
140 | cell_size: Size of the dots for each cell. By default, it uses the median distance between neighbor cells.
141 | ncols: Number of columns to be shown.
142 | fig_size_per_slide: Size of the figure for each slide.
143 | linewidths: Width of the lines/edges connecting the cells.
144 | line_color: Color of the lines/edges.
145 | cmap: Name of the colormap to use for the number of neighbors.
146 | color_isolated_cells: Color for the cells with a number of neighbors below `ngh_threshold` (if not `None`).
147 | show: Whether to show the plot.
148 | """
149 | adatas = [adata] if isinstance(adata, AnnData) else adata
150 |
151 | fig, axes = _subplots_per_slide(adatas, ncols, fig_size_per_slide)
152 |
153 | for i, adata in enumerate(utils.iter_slides(adatas)):
154 | ax = axes[i // ncols, i % ncols]
155 |
156 | utils.check_has_spatial_adjancency(adata)
157 |
158 | X, A = adata.obsm["spatial"], adata.obsp[Keys.ADJ]
159 |
160 | ax.invert_yaxis()
161 | ax.axes.set_aspect("equal")
162 |
163 | rows, cols = A.nonzero()
164 | mask = rows < cols
165 | rows, cols = rows[mask], cols[mask]
166 | edge_segments = np.stack([X[rows], X[cols]], axis=1)
167 | edges = LineCollection(edge_segments, color=line_color, linewidths=linewidths, zorder=1)
168 | ax.add_collection(edges)
169 |
170 | n_neighbors = (A > 0).sum(1).A1
171 |
172 | if ngh_threshold is None:
173 | _ = ax.scatter(X[:, 0], X[:, 1], c=n_neighbors, s=cell_size, zorder=2, cmap=cmap)
174 | plt.colorbar(_, ax=ax)
175 | else:
176 | isolated_cells = n_neighbors < ngh_threshold
177 | ax.scatter(X[isolated_cells, 0], X[isolated_cells, 1], color=color_isolated_cells, s=cell_size, zorder=2)
178 |
179 | ax.set_title(adata.obs[Keys.SLIDE_ID].iloc[0])
180 |
181 | [fig.delaxes(ax) for ax in axes.flatten() if not ax.has_data()] # remove unused subplots
182 |
183 | title = "Node connectivities" + (f" (threshold={ngh_threshold} neighbors)" if ngh_threshold is not None else "")
184 |
185 | if i == 0:
186 | axes[0, 0].set_title(title)
187 | else:
188 | fig.suptitle(title, fontsize=14)
189 |
190 | if show:
191 | plt.show()
192 |
--------------------------------------------------------------------------------
/novae/plot/_heatmap.py:
--------------------------------------------------------------------------------
1 | import json
2 | import logging
3 |
4 | import matplotlib.patches as mpatches
5 | import matplotlib.pyplot as plt
6 | import numpy as np
7 | import pandas as pd
8 | import scanpy as sc
9 | import seaborn as sns
10 | from anndata import AnnData
11 |
12 | from .. import utils
13 | from .._constants import Keys
14 |
15 | log = logging.getLogger(__name__)
16 |
17 |
18 | def _weights_clustermap(
19 | weights: np.ndarray,
20 | adatas: list[AnnData] | None,
21 | slide_ids: list[str],
22 | show_yticklabels: bool = False,
23 | show_tissue_legend: bool = True,
24 | figsize: tuple[int] = (6, 4),
25 | vmin: float = 0,
26 | vmax: float = 1,
27 | **kwargs: int,
28 | ) -> None:
29 | row_colors = None
30 | if show_tissue_legend and adatas is not None and all(Keys.UNS_TISSUE in adata.uns for adata in adatas):
31 | tissues = list({adata.uns[Keys.UNS_TISSUE] for adata in adatas})
32 | tissue_colors = {tissue: sns.color_palette("tab20")[i] for i, tissue in enumerate(tissues)}
33 |
34 | row_colors = []
35 | for slide_id in slide_ids:
36 | for adata in adatas:
37 | if adata.obs[Keys.SLIDE_ID].iloc[0] == slide_id:
38 | row_colors.append(tissue_colors[adata.uns[Keys.UNS_TISSUE]])
39 | break
40 | else:
41 | row_colors.append("gray")
42 | log.info(f"Using {row_colors=}")
43 |
44 | sns.clustermap(
45 | weights,
46 | yticklabels=slide_ids if show_yticklabels else False,
47 | xticklabels=False,
48 | vmin=vmin,
49 | vmax=vmax,
50 | figsize=figsize,
51 | row_colors=row_colors,
52 | **kwargs,
53 | )
54 |
55 | if show_tissue_legend and row_colors is not None:
56 | handles = [mpatches.Patch(facecolor=color, label=tissue) for tissue, color in tissue_colors.items()]
57 | ax = plt.gcf().axes[3]
58 | ax.legend(handles=handles, bbox_to_anchor=(1.04, 0.5), loc="center left", borderaxespad=0, frameon=False)
59 |
60 |
61 | TEMP_KEY = "_temp"
62 |
63 |
64 | def pathway_scores(
65 | adata: AnnData,
66 | pathways: dict[str, list[str]] | str,
67 | obs_key: str | None = None,
68 | pathway_name: str | None = None,
69 | slide_name_key: str | None = None,
70 | return_df: bool = False,
71 | figsize: tuple[int, int] = (10, 5),
72 | min_pathway_size: int = 4,
73 | show: bool = True,
74 | **kwargs: int,
75 | ) -> pd.DataFrame | None:
76 | """Show a heatmap of either (i) the score of multiple pathways for each domain, or (ii) one pathway score for each domain and for each slide.
77 | To use the latter case, provide `pathway_name`, or make sure to have only one pathway in `pathways`.
78 |
79 | Info:
80 | Currently, this function only supports one AnnData object per call.
81 |
82 | Args:
83 | adata: An `AnnData` object.
84 | pathways: Either a dictionary of pathways (keys are pathway names, values are lists of gene names), or a path to a [GSEA](https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) JSON file.
85 | obs_key: Key in `adata.obs` that contains the domains. By default, it will use the last available Novae domain key.
86 | pathway_name: If `None`, all pathways will be shown (first mode). If not `None`, this specific pathway will be shown, for all domains and all slides (second mode).
87 | slide_name_key: Key of `adata.obs` that contains the slide names. By default, uses the Novae unique slide ID.
88 | return_df: Whether to return the DataFrame.
89 | figsize: Matplotlib figure size.
90 | min_pathway_size: Minimum number of known genes in the pathway to be considered.
91 | show: Whether to show the plot.
92 |
93 | Returns:
94 | A DataFrame of scores per domain if `return_df` is True.
95 | """
96 | assert isinstance(adata, AnnData), f"For now, only one AnnData object is supported, received {type(adata)}"
97 |
98 | obs_key = utils.check_available_domains_key([adata], obs_key)
99 |
100 | if isinstance(pathways, str):
101 | pathways = _load_gsea_json(pathways)
102 | log.info(f"Loaded {len(pathways)} pathway(s)")
103 |
104 | if len(pathways) == 1:
105 | pathway_name = next(iter(pathways.keys()))
106 |
107 | if pathway_name is not None:
108 | gene_names = pathways[pathway_name]
109 | is_valid = _get_pathway_score(adata, gene_names, min_pathway_size)
110 | assert is_valid, f"Pathway '{pathway_name}' has less than {min_pathway_size} genes in the dataset."
111 | else:
112 | scores = {}
113 |
114 | for key, gene_names in pathways.items():
115 | is_valid = _get_pathway_score(adata, gene_names, min_pathway_size)
116 | if is_valid:
117 | scores[key] = adata.obs[TEMP_KEY]
118 |
119 | if pathway_name is not None:
120 | log.info(f"Plot mode: {pathway_name} score per domain per slide")
121 |
122 | slide_name_key = utils.check_slide_name_key(adata, slide_name_key)
123 |
124 | df = adata.obs.groupby([obs_key, slide_name_key], observed=True)[TEMP_KEY].mean().unstack()
125 | df.columns.name = slide_name_key
126 |
127 | assert len(df) > 1, f"Found {len(df)} valid slide. Minimum 2 required."
128 | else:
129 | log.info(f"Plot mode: {len(scores)} pathways scores per domain")
130 |
131 | assert len(scores) > 1, f"Found {len(scores)} valid pathway. Minimum 2 required."
132 |
133 | df = pd.DataFrame(scores)
134 | df[obs_key] = adata.obs[obs_key]
135 | df = df.groupby(obs_key, observed=True).mean()
136 | df.columns.name = "Pathways"
137 |
138 | del adata.obs[TEMP_KEY]
139 |
140 | df = df.fillna(0)
141 |
142 | g = sns.clustermap(df, figsize=figsize, **kwargs)
143 | plt.setp(g.ax_heatmap.yaxis.get_majorticklabels(), rotation=0)
144 |
145 | if show:
146 | plt.show()
147 |
148 | if return_df:
149 | return df
150 |
151 |
152 | def _get_pathway_score(adata: AnnData, gene_names: list[str], min_pathway_size: int) -> bool:
153 | lower_var_names = adata.var_names.str.lower()
154 |
155 | gene_names = np.array([gene_name.lower() for gene_name in gene_names])
156 | gene_names = adata.var_names[np.isin(lower_var_names, gene_names)]
157 |
158 | if len(gene_names) >= min_pathway_size:
159 | sc.tl.score_genes(adata, gene_names, score_name=TEMP_KEY)
160 | return True
161 | return False
162 |
163 |
164 | def _load_gsea_json(path: str) -> dict[str, list[str]]:
165 | with open(path) as f:
166 | content: dict = json.load(f)
167 | assert all("geneSymbols" in value for value in content.values()), (
168 | "Missing 'geneSymbols' key in JSON file. Expected a valid GSEA JSON file."
169 | )
170 | return {key: value["geneSymbols"] for key, value in content.items()}
171 |
--------------------------------------------------------------------------------
/novae/plot/_spatial.py:
--------------------------------------------------------------------------------
1 | import logging
2 |
3 | import matplotlib.pyplot as plt
4 | import numpy as np
5 | import pandas as pd
6 | import scanpy as sc
7 | import seaborn as sns
8 | from anndata import AnnData
9 | from matplotlib.lines import Line2D
10 | from scanpy._utils import sanitize_anndata
11 |
12 | from .. import utils
13 | from .._constants import Keys
14 | from ._utils import (
15 | _get_default_cell_size,
16 | _subplots_per_slide,
17 | get_categorical_color_palette,
18 | )
19 |
20 | log = logging.getLogger(__name__)
21 |
22 |
23 | def domains(
24 | adata: AnnData | list[AnnData],
25 | obs_key: str | None = None,
26 | slide_name_key: str | None = None,
27 | cell_size: int | None = None,
28 | ncols: int = 4,
29 | fig_size_per_slide: tuple[int, int] = (5, 5),
30 | na_color: str = "#ccc",
31 | show: bool = True,
32 | library_id: str | None = None,
33 | **kwargs: int,
34 | ):
35 | """Show the Novae spatial domains for all slides in the `AnnData` object.
36 |
37 | Info:
38 | Make sure you have already your Novae domains assigned to the `AnnData` object. You can use `model.assign_domains(...)` to do so.
39 |
40 | Args:
41 | adata: An `AnnData` object, or a list of `AnnData` objects.
42 | obs_key: Name of the key from `adata.obs` containing the Novae domains. By default, the last available domain key is shown.
43 | slide_name_key: Key of `adata.obs` that contains the slide names. By default, uses the Novae unique slide ID.
44 | cell_size: Size of the cells or spots. By default, it uses the median distance between neighbor cells.
45 | ncols: Number of columns to be shown.
46 | fig_size_per_slide: Size of the figure for each slide.
47 | na_color: Color for cells that does not belong to any domain (i.e. cells with a too small neighborhood).
48 | show: Whether to show the plot.
49 | library_id: `library_id` argument for `sc.pl.spatial`.
50 | **kwargs: Additional arguments for `sc.pl.spatial`.
51 | """
52 | if obs_key is not None:
53 | assert str(obs_key).startswith(Keys.DOMAINS_PREFIX), f"Received {obs_key=}, which is not a valid Novae obs_key"
54 |
55 | adatas = adata if isinstance(adata, list) else [adata]
56 | slide_name_key = utils.check_slide_name_key(adatas, slide_name_key)
57 | obs_key = utils.check_available_domains_key(adatas, obs_key)
58 |
59 | for adata in adatas:
60 | sanitize_anndata(adata)
61 |
62 | all_domains, colors = get_categorical_color_palette(adatas, obs_key)
63 | cell_size = cell_size or _get_default_cell_size(adata)
64 |
65 | fig, axes = _subplots_per_slide(adatas, ncols, fig_size_per_slide)
66 |
67 | for i, adata in enumerate(utils.iter_slides(adatas)):
68 | ax = axes[i // ncols, i % ncols]
69 | slide_name = adata.obs[slide_name_key].iloc[0]
70 | assert len(np.unique(adata.obs[slide_name_key])) == 1
71 |
72 | sc.pl.spatial(
73 | adata,
74 | spot_size=cell_size,
75 | color=obs_key,
76 | ax=ax,
77 | show=False,
78 | library_id=library_id,
79 | **kwargs,
80 | )
81 | sns.despine(ax=ax, offset=10, trim=True)
82 | ax.get_legend().remove()
83 | ax.set_title(slide_name)
84 |
85 | [fig.delaxes(ax) for ax in axes.flatten() if not ax.has_data()] # remove unused subplots
86 |
87 | title = f"Novae domains ({obs_key})"
88 |
89 | if i == 0:
90 | axes[0, 0].set_title(title)
91 | else:
92 | fig.suptitle(title, fontsize=14, y=1.15)
93 |
94 | handles = [
95 | Line2D([0], [0], marker="o", color="w", markerfacecolor=color, markersize=8, linestyle="None")
96 | for color in [*colors, na_color]
97 | ]
98 | fig.legend(
99 | handles,
100 | [*all_domains, "NA"],
101 | loc="upper center" if i > 1 else "center left",
102 | bbox_to_anchor=(0.5, 1.1) if i > 1 else (1.04, 0.5),
103 | borderaxespad=0,
104 | frameon=False,
105 | ncol=len(colors) // (3 if i > 1 else 10) + 1,
106 | )
107 |
108 | if show:
109 | plt.show()
110 |
111 |
112 | def spatially_variable_genes(
113 | adata: AnnData,
114 | obs_key: str | None = None,
115 | top_k: int = 5,
116 | cell_size: int | None = None,
117 | min_positive_ratio: float = 0.05,
118 | return_list: bool = False,
119 | show: bool = True,
120 | **kwargs: int,
121 | ) -> None | list[str]:
122 | """Plot the most spatially variable genes (SVG) for a given `AnnData` object.
123 |
124 | !!! info
125 | Currently, this function only supports one slide per call.
126 |
127 | Args:
128 | adata: An `AnnData` object corresponding to one slide.
129 | obs_key: Key in `adata.obs` that contains the domains. By default, it will use the last available Novae domain key.
130 | top_k: Number of SVG to be shown.
131 | cell_size: Size of the cells or spots (`spot_size` argument of `sc.pl.spatial`). By default, it uses the median distance between neighbor cells.
132 | min_positive_ratio: Genes whose "ratio of cells expressing it" is lower than this threshold are not considered.
133 | return_list: Whether to return the list of SVG instead of plotting them.
134 | show: Whether to show the plot.
135 | **kwargs: Additional arguments for `sc.pl.spatial`.
136 |
137 | Returns:
138 | A list of SVG names if `return_list` is `True`.
139 | """
140 | assert isinstance(adata, AnnData), f"Received adata of type {type(adata)}. Currently only AnnData is supported."
141 |
142 | obs_key = utils.check_available_domains_key([adata], obs_key)
143 |
144 | sc.tl.rank_genes_groups(adata, groupby=obs_key)
145 | df = pd.concat(
146 | [
147 | sc.get.rank_genes_groups_df(adata, domain).set_index("names")["logfoldchanges"]
148 | for domain in adata.obs[obs_key].cat.categories
149 | ],
150 | axis=1,
151 | )
152 |
153 | where = (adata.X > 0).mean(0) > min_positive_ratio
154 | valid_vars = adata.var_names[where.A1 if isinstance(where, np.matrix) else where]
155 | assert len(valid_vars) >= top_k, (
156 | f"Only {len(valid_vars)} genes are available. Please decrease `top_k` or `min_positive_ratio`."
157 | )
158 |
159 | svg = df.std(1).loc[valid_vars].sort_values(ascending=False).head(top_k).index
160 |
161 | if return_list:
162 | return svg.tolist()
163 |
164 | sc.pl.spatial(adata, color=svg, spot_size=cell_size or _get_default_cell_size(adata), show=show, **kwargs)
165 |
--------------------------------------------------------------------------------
/novae/plot/_utils.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import numpy as np
3 | import seaborn as sns
4 | from anndata import AnnData
5 |
6 | from .._constants import Keys
7 |
8 |
9 | def get_categorical_color_palette(adatas: list[AnnData], obs_key: str) -> tuple[list[str], list[str]]:
10 | key_added = f"{obs_key}_colors"
11 |
12 | all_domains = sorted(set.union(*[set(adata.obs[obs_key].cat.categories) for adata in adatas]))
13 |
14 | n_colors = len(all_domains)
15 | colors = list(sns.color_palette("tab10" if n_colors <= 10 else "tab20", n_colors=n_colors).as_hex())
16 | for adata in adatas:
17 | adata.obs[obs_key] = adata.obs[obs_key].cat.set_categories(all_domains)
18 | adata.uns[key_added] = colors
19 |
20 | return all_domains, colors
21 |
22 |
23 | def _subplots_per_slide(
24 | adatas: list[AnnData], ncols: int, fig_size_per_slide: tuple[int, int]
25 | ) -> tuple[plt.Figure, np.ndarray]:
26 | n_slides = sum(len(adata.obs[Keys.SLIDE_ID].cat.categories) for adata in adatas)
27 | ncols = n_slides if n_slides < ncols else ncols
28 | nrows = (n_slides + ncols - 1) // ncols
29 |
30 | fig, axes = plt.subplots(
31 | nrows, ncols, figsize=(ncols * fig_size_per_slide[0], nrows * fig_size_per_slide[1]), squeeze=False
32 | )
33 |
34 | return fig, axes
35 |
36 |
37 | def _get_default_cell_size(adata: AnnData | list[AnnData]) -> float:
38 | if isinstance(adata, list):
39 | adata = max(adata, key=lambda adata: adata.n_obs)
40 |
41 | assert Keys.ADJ in adata.obsp, (
42 | f"Expected {Keys.ADJ} in adata.obsp. Please run `novae.spatial_neighbors(...)` first."
43 | )
44 |
45 | return np.median(adata.obsp[Keys.ADJ].data)
46 |
--------------------------------------------------------------------------------
/novae/utils/__init__.py:
--------------------------------------------------------------------------------
1 | from ._utils import (
2 | repository_root,
3 | tqdm,
4 | fill_invalid_indices,
5 | lower_var_names,
6 | wandb_log_dir,
7 | pretty_num_parameters,
8 | pretty_model_repr,
9 | parse_device_args,
10 | valid_indices,
11 | unique_leaves_indices,
12 | unique_obs,
13 | sparse_std,
14 | iter_slides,
15 | )
16 | from ._build import spatial_neighbors
17 | from ._validate import check_available_domains_key, prepare_adatas, check_has_spatial_adjancency, check_slide_name_key
18 | from ._data import load_local_dataset, load_wandb_artifact, toy_dataset, load_dataset
19 | from ._mode import Mode
20 | from ._correct import batch_effect_correction
21 | from ._preprocess import quantile_scaling
22 |
--------------------------------------------------------------------------------
/novae/utils/_correct.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 | from anndata import AnnData
4 |
5 | from .._constants import Keys
6 |
7 |
8 | def _slides_indices(adatas: list[AnnData], only_valid_obs: bool = True) -> tuple[list[int], list[np.ndarray]]:
9 | adata_indices, slides_obs_indices = [], []
10 |
11 | for i, adata in enumerate(adatas):
12 | slide_ids = adata.obs[Keys.SLIDE_ID].cat.categories
13 |
14 | for slide_id in slide_ids:
15 | condition = adata.obs[Keys.SLIDE_ID] == slide_id
16 | if only_valid_obs:
17 | condition = condition & adata.obs[Keys.IS_VALID_OBS]
18 |
19 | adata_indices.append(i)
20 | slides_obs_indices.append(np.where(condition)[0])
21 |
22 | return adata_indices, slides_obs_indices
23 |
24 |
25 | def _domains_counts(adata: AnnData, i: int, obs_key: str) -> pd.DataFrame:
26 | df = adata.obs[[Keys.SLIDE_ID, obs_key]].groupby(Keys.SLIDE_ID, observed=False)[obs_key].value_counts().unstack()
27 | df[Keys.ADATA_INDEX] = i
28 | return df
29 |
30 |
31 | def _domains_counts_per_slide(adatas: list[AnnData], obs_key: str) -> pd.DataFrame:
32 | return pd.concat([_domains_counts(adata, i, obs_key) for i, adata in enumerate(adatas)], axis=0)
33 |
34 |
35 | def batch_effect_correction(adatas: list[AnnData], obs_key: str) -> None:
36 | for adata in adatas:
37 | assert obs_key in adata.obs, f"Did not found `adata.obs['{obs_key}']`"
38 | assert Keys.REPR in adata.obsm, (
39 | f"Did not found `adata.obsm['{Keys.REPR}']`. Please run `model.compute_representations(...)` first"
40 | )
41 |
42 | adata_indices, slides_obs_indices = _slides_indices(adatas)
43 |
44 | domains_counts_per_slide = _domains_counts_per_slide(adatas, obs_key)
45 | domains = domains_counts_per_slide.columns[:-1]
46 | ref_slide_ids: pd.Series = domains_counts_per_slide[domains].idxmax(axis=0)
47 |
48 | def _centroid_reference(domain: str, slide_id: str, obs_key: str):
49 | adata_ref_index: int = domains_counts_per_slide[Keys.ADATA_INDEX].loc[slide_id]
50 | adata_ref = adatas[adata_ref_index]
51 | where = (adata_ref.obs[Keys.SLIDE_ID] == slide_id) & (adata_ref.obs[obs_key] == domain)
52 | return adata_ref.obsm[Keys.REPR][where].mean(0)
53 |
54 | centroids_reference = pd.DataFrame({
55 | domain: _centroid_reference(domain, slide_id, obs_key) for domain, slide_id in ref_slide_ids.items()
56 | })
57 |
58 | for adata in adatas:
59 | adata.obsm[Keys.REPR_CORRECTED] = adata.obsm[Keys.REPR].copy()
60 |
61 | for adata_index, obs_indices in zip(adata_indices, slides_obs_indices):
62 | adata = adatas[adata_index]
63 |
64 | for domain in domains:
65 | if adata.obs[Keys.SLIDE_ID].iloc[obs_indices[0]] == ref_slide_ids.loc[domain]:
66 | continue # reference for this domain
67 |
68 | indices_domain = obs_indices[adata.obs.iloc[obs_indices][obs_key] == domain]
69 | if len(indices_domain) == 0:
70 | continue
71 |
72 | centroid_reference = centroids_reference[domain].values
73 | centroid = adata.obsm[Keys.REPR][indices_domain].mean(0)
74 |
75 | adata.obsm[Keys.REPR_CORRECTED][indices_domain] += centroid_reference - centroid
76 |
--------------------------------------------------------------------------------
/novae/utils/_mode.py:
--------------------------------------------------------------------------------
1 | class Mode:
2 | """Novae mode class, used to store states variables related to training and inference."""
3 |
4 | zero_shot_clustering_attrs: list[str] = ["_clustering_zero", "_clusters_levels_zero"]
5 | normal_clustering_attrs: list[str] = ["_clustering", "_clusters_levels"]
6 | all_clustering_attrs: list[str] = normal_clustering_attrs + zero_shot_clustering_attrs
7 |
8 | def __init__(self):
9 | self.zero_shot = False
10 | self.trained = False
11 | self.pretrained = False
12 |
13 | def __repr__(self) -> str:
14 | return f"Mode({dict(self.__dict__.items())})"
15 |
16 | ### Mode modifiers
17 |
18 | def from_pretrained(self):
19 | self.zero_shot = False
20 | self.trained = True
21 | self.pretrained = True
22 |
23 | def fine_tune(self):
24 | assert self.pretrained, "Fine-tuning requires a pretrained model."
25 | self.zero_shot = False
26 |
27 | def fit(self):
28 | self.zero_shot = False
29 | self.trained = False
30 |
31 | ### Mode-specific attributes
32 |
33 | @property
34 | def clustering_attr(self):
35 | return "_clustering_zero" if self.zero_shot else "_clustering"
36 |
37 | @property
38 | def clusters_levels_attr(self):
39 | return "_clusters_levels_zero" if self.zero_shot else "_clusters_levels"
40 |
--------------------------------------------------------------------------------
/novae/utils/_preprocess.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 | from anndata import AnnData
4 |
5 | from .._constants import Nums
6 | from . import iter_slides
7 | from ._validate import _check_has_slide_id
8 |
9 |
10 | def quantile_scaling(
11 | adata: AnnData | list[AnnData],
12 | multiplier: float = 5,
13 | quantile: float = 0.2,
14 | per_slide: bool = True,
15 | ) -> pd.DataFrame:
16 | """Preprocess fluorescence data from `adata.X` using quantiles of expression.
17 | For each column `X`, we compute `asinh(X / 5*Q(0.2, X))`, and store them back.
18 |
19 | Args:
20 | adata: An `AnnData` object, or a list of `AnnData` objects.
21 | multiplier: The multiplier for the quantile.
22 | quantile: The quantile to compute.
23 | per_slide: Whether to compute the quantile per slide. If `False`, the quantile is computed for each `AnnData` object.
24 | """
25 | _check_has_slide_id(adata)
26 |
27 | if isinstance(adata, list):
28 | for adata_ in adata:
29 | quantile_scaling(adata_, multiplier, quantile, per_slide=per_slide)
30 | return
31 |
32 | if not per_slide:
33 | return _quantile_scaling(adata, multiplier, quantile)
34 |
35 | for adata_ in iter_slides(adata):
36 | _quantile_scaling(adata_, multiplier, quantile)
37 |
38 |
39 | def _quantile_scaling(adata: AnnData, multiplier: float, quantile: float):
40 | df = adata.to_df()
41 |
42 | divider = multiplier * np.quantile(df, quantile, axis=0)
43 | divider[divider == 0] = df.max(axis=0)[divider == 0] + Nums.EPS
44 |
45 | adata.X = np.arcsinh(df / divider)
46 |
--------------------------------------------------------------------------------
/novae/utils/_utils.py:
--------------------------------------------------------------------------------
1 | import importlib
2 | import logging
3 | from pathlib import Path
4 |
5 | import numpy as np
6 | import pandas as pd
7 | import torch
8 | from anndata import AnnData
9 | from lightning.pytorch.trainer.connectors.accelerator_connector import (
10 | _AcceleratorConnector,
11 | )
12 | from scipy.sparse import csr_matrix
13 | from torch import Tensor
14 |
15 | from .._constants import Keys
16 |
17 | log = logging.getLogger(__name__)
18 |
19 |
20 | def unique_obs(adata: AnnData | list[AnnData], obs_key: str) -> set:
21 | if isinstance(adata, list):
22 | return set.union(*[unique_obs(adata_, obs_key) for adata_ in adata])
23 | return set(adata.obs[obs_key].dropna().unique())
24 |
25 |
26 | def unique_leaves_indices(adata: AnnData | list[AnnData]) -> set:
27 | leaves = unique_obs(adata, Keys.LEAVES)
28 | return np.array([int(x[1:]) for x in leaves])
29 |
30 |
31 | def valid_indices(adata: AnnData) -> np.ndarray:
32 | return np.where(adata.obs[Keys.IS_VALID_OBS])[0]
33 |
34 |
35 | def lower_var_names(var_names: pd.Index | list[str]) -> pd.Index | list[str]:
36 | if isinstance(var_names, pd.Index):
37 | return var_names.str.lower()
38 | return [name.lower() for name in var_names]
39 |
40 |
41 | def sparse_std(a: csr_matrix, axis=None) -> np.matrix:
42 | a_squared = a.multiply(a)
43 | return np.sqrt(a_squared.mean(axis) - np.square(a.mean(axis)))
44 |
45 |
46 | def fill_invalid_indices(
47 | out: np.ndarray | Tensor,
48 | n_obs: int,
49 | valid_indices: list[int],
50 | fill_value: float | str = np.nan,
51 | dtype: object = None,
52 | ) -> np.ndarray:
53 | if isinstance(out, Tensor):
54 | out = out.numpy(force=True)
55 |
56 | dtype = np.float32 if dtype is None else dtype
57 |
58 | if isinstance(fill_value, str):
59 | dtype = object
60 |
61 | res = np.full((n_obs, *out.shape[1:]), fill_value, dtype=dtype)
62 | res[valid_indices] = out
63 | return res
64 |
65 |
66 | def parse_device_args(accelerator: str = "cpu") -> torch.device:
67 | """Updated from scvi-tools"""
68 | connector = _AcceleratorConnector(accelerator=accelerator)
69 | _accelerator = connector._accelerator_flag
70 | _devices = connector._devices_flag
71 |
72 | if _accelerator == "cpu":
73 | return torch.device("cpu")
74 |
75 | if isinstance(_devices, list):
76 | device_idx = _devices[0]
77 | elif isinstance(_devices, str) and "," in _devices:
78 | device_idx = _devices.split(",")[0]
79 | else:
80 | device_idx = _devices
81 |
82 | return torch.device(f"{_accelerator}:{device_idx}")
83 |
84 |
85 | def repository_root() -> Path:
86 | """Get the path to the root of the repository (dev-mode users only)
87 |
88 | Returns:
89 | `novae` repository path
90 | """
91 | path = Path(__file__).parents[2]
92 |
93 | if path.name != "novae":
94 | log.warning(f"Trying to get the novae repository path, but it seems it was not installed in dev mode: {path}")
95 |
96 | return path
97 |
98 |
99 | def wandb_log_dir() -> Path:
100 | return repository_root() / "wandb"
101 |
102 |
103 | def tqdm(*args, desc="DataLoader", **kwargs):
104 | # check if ipywidgets is installed before importing tqdm.auto
105 | # to ensure it won't fail and a progress bar is displayed
106 | if importlib.util.find_spec("ipywidgets") is not None:
107 | from tqdm.auto import tqdm as _tqdm
108 | else:
109 | from tqdm import tqdm as _tqdm
110 |
111 | return _tqdm(*args, desc=desc, **kwargs)
112 |
113 |
114 | def pretty_num_parameters(model: torch.nn.Module) -> str:
115 | n_params = sum(p.numel() for p in model.parameters())
116 |
117 | if n_params < 1_000_000:
118 | return f"{n_params / 1_000:.1f}K"
119 |
120 | return f"{n_params / 1_000_000:.1f}M"
121 |
122 |
123 | def pretty_model_repr(info_dict: dict[str, str], model_name: str = "Novae") -> str:
124 | rows = [f"{model_name} model"] + [f"{k}: {v}" for k, v in info_dict.items()]
125 | return "\n ├── ".join(rows[:-1]) + "\n └── " + rows[-1]
126 |
127 |
128 | def iter_slides(adatas: AnnData | list[AnnData]):
129 | """Iterate over all slides.
130 |
131 | Args:
132 | adatas: One or a list of AnnData object(s).
133 |
134 | Yields:
135 | One `AnnData` per slide.
136 | """
137 | if isinstance(adatas, AnnData):
138 | adatas = [adatas]
139 |
140 | for adata in adatas:
141 | slide_ids = adata.obs[Keys.SLIDE_ID].unique()
142 |
143 | if len(slide_ids) == 1:
144 | yield adata
145 | continue
146 |
147 | for slide_id in slide_ids:
148 | yield adata[adata.obs[Keys.SLIDE_ID] == slide_id]
149 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.poetry]
2 | name = "novae"
3 | version = "0.2.4"
4 | description = "Graph-based foundation model for spatial transcriptomics data"
5 | documentation = "https://mics-lab.github.io/novae/"
6 | homepage = "https://mics-lab.github.io/novae/"
7 | repository = "https://github.com/MICS-Lab/novae"
8 | authors = ["Quentin Blampey "]
9 | packages = [{ include = "novae" }]
10 | license = "BSD-3-Clause"
11 | readme = "README.md"
12 | classifiers = [
13 | "License :: OSI Approved :: BSD License",
14 | "Operating System :: MacOS :: MacOS X",
15 | "Operating System :: POSIX :: Linux",
16 | "Operating System :: Microsoft :: Windows",
17 | "Programming Language :: Python :: 3",
18 | "Topic :: Scientific/Engineering",
19 | ]
20 |
21 | [tool.poetry.dependencies]
22 | python = ">=3.10,<3.13"
23 | scanpy = ">=1.9.8"
24 | lightning = ">=2.2.1"
25 | torch = ">=2.2.1"
26 | torch-geometric = ">=2.5.2"
27 | huggingface-hub = ">=0.24.0"
28 | safetensors = ">=0.4.3"
29 | pandas = ">=2.0.0"
30 |
31 | ruff = { version = ">=0.11.4", optional = true }
32 | mypy = { version = ">=1.15.0", optional = true }
33 | pre-commit = { version = ">=3.8.0", optional = true }
34 | pytest = { version = ">=7.1.3", optional = true }
35 | pytest-cov = { version = ">=5.0.0", optional = true }
36 | wandb = { version = ">=0.17.2", optional = true }
37 | pyyaml = { version = ">=6.0.1", optional = true }
38 | pydantic = { version = ">=2.8.2", optional = true }
39 | ipykernel = { version = ">=6.22.0", optional = true }
40 | ipywidgets = { version = ">=8.1.2", optional = true }
41 | mkdocs-material = { version = ">=8.5.6", optional = true }
42 | mkdocs-jupyter = { version = ">=0.21.0", optional = true }
43 | mkdocstrings = { version = ">=0.19.0", optional = true }
44 | mkdocstrings-python = { version = ">=0.7.1", optional = true }
45 |
46 | [tool.poetry.extras]
47 | dev = [
48 | "ruff",
49 | "mypy",
50 | "pre-commit",
51 | "pytest",
52 | "pytest-cov",
53 | "wandb",
54 | "pyyaml",
55 | "pydantic",
56 | "ipykernel",
57 | "ipywidgets",
58 | "mkdocs-material",
59 | "mkdocs-jupyter",
60 | "mkdocstrings",
61 | "mkdocstrings-python",
62 | ]
63 |
64 | [build-system]
65 | requires = ["poetry-core>=1.0.0"]
66 | build-backend = "poetry.core.masonry.api"
67 |
68 | [tool.mypy]
69 | files = ["novae"]
70 | no_implicit_optional = true
71 | check_untyped_defs = true
72 | warn_return_any = true
73 | warn_unused_ignores = true
74 | show_error_codes = true
75 | ignore_missing_imports = true
76 |
77 | [tool.pytest.ini_options]
78 | testpaths = ["tests"]
79 | python_files = "test_*.py"
80 |
81 |
82 | [tool.ruff]
83 | target-version = "py39"
84 | line-length = 120
85 | fix = true
86 |
87 | [tool.ruff.lint]
88 | select = [
89 | # flake8-2020
90 | "YTT",
91 | # flake8-bandit
92 | "S",
93 | # flake8-bugbear
94 | "B",
95 | # flake8-builtins
96 | "A",
97 | # flake8-comprehensions
98 | "C4",
99 | # flake8-debugger
100 | "T10",
101 | # flake8-simplify
102 | "SIM",
103 | # isort
104 | "I",
105 | # mccabe
106 | "C90",
107 | # pycodestyle
108 | "E",
109 | "W",
110 | # pyflakes
111 | "F",
112 | # pygrep-hooks
113 | "PGH",
114 | # pyupgrade
115 | "UP",
116 | # ruff
117 | "RUF",
118 | # tryceratops
119 | "TRY",
120 | ]
121 | ignore = [
122 | # LineTooLong
123 | "E501",
124 | # DoNotAssignLambda
125 | "E731",
126 | # DoNotUseAssert
127 | "S101",
128 | "TRY003",
129 | "RUF012",
130 | "B904",
131 | "E722",
132 | ]
133 |
134 | [tool.ruff.lint.per-file-ignores]
135 | "tests/*" = ["S101"]
136 | "__init__.py" = ["F401", "I001"]
137 | "*.ipynb" = ["F401"]
138 |
139 | [tool.ruff.format]
140 | preview = true
141 |
142 | [tool.coverage.report]
143 | skip_empty = true
144 |
145 | [tool.coverage.run]
146 | source = ["novae"]
147 | omit = ["**/test_*.py", "novae/monitor/log.py", "novae/monitor/callback.py"]
148 |
--------------------------------------------------------------------------------
/scripts/README.md:
--------------------------------------------------------------------------------
1 | # Pre-training scripts
2 |
3 | These scripts are used to pretrain and monitor Novae with Weight & Biases. To see the actualy source code of Novae, refer to the `novae` directory.
4 |
5 | ## Setup
6 |
7 | For monitoring, `novae` must be installed with the `dev` extra, for instance via pip:
8 |
9 | ```sh
10 | pip install -e '.[dev]'
11 | ```
12 |
13 | ## Usage
14 |
15 | In the `data` directory, make sure to have `.h5ad` files. You can use the downloading scripts to get public data.
16 | The corresponding `AnnData` object should contain raw counts, or preprocessed with `normalize_total` and `log1p`.
17 |
18 | ### Normal training
19 |
20 | Choose a config inside the `config` directory.
21 |
22 | ```sh
23 | python -m scripts.train --config .yaml
24 | ```
25 |
26 | ### Sweep training
27 |
28 | Choose a sweep config inside the `sweep` directory.
29 |
30 | Inside the `scripts` directory, initialize the sweep with:
31 | ```sh
32 | wandb sweep --project novae sweep/.yaml
33 | ```
34 |
35 | Run the sweep with:
36 | ```sh
37 | wandb agent --count 1
38 | ```
39 |
40 | ### Slurm usage
41 |
42 | ⚠️ Warning: the scripts below are specific to one cluster (Flamingo, Gustave Roussy). You'll need to update the `.sh` scripts according to your cluster.
43 |
44 | In the `slurm` directory:
45 | - `train.sh` / `train_cpu.sh` for training
46 | - `download.sh` to download public data
47 | - `sbatch agent.sh SWEEP_ID COUNT` to run agents (where SWEEP_ID comes from `wandb sweep --project novae sweep/.yaml`)
48 |
49 | E.g., on ruche:
50 | ```sh
51 | module load anaconda3/2024.06/gcc-13.2.0 && source activate novae
52 | wandb sweep --project novae sweep/gpu_ruche.yaml
53 |
54 | cd ruche
55 | sbatch agent.sh SWEEP_ID
56 | ```
57 |
--------------------------------------------------------------------------------
/scripts/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/scripts/__init__.py
--------------------------------------------------------------------------------
/scripts/config.py:
--------------------------------------------------------------------------------
1 | from pydantic import BaseModel
2 |
3 |
4 | class DataConfig(BaseModel):
5 | train_dataset: str = "all"
6 | val_dataset: str | None = None
7 |
8 | files_black_list: list[str] = []
9 |
10 |
11 | class PostTrainingConfig(BaseModel):
12 | n_domains: list[int] = [7, 10]
13 | log_umap: bool = False
14 | log_metrics: bool = False
15 | log_domains: bool = False
16 | save_h5ad: bool = False
17 | delete_X: bool = False
18 |
19 |
20 | class Config(BaseModel):
21 | project: str = "novae"
22 | wandb_artefact: str | None = None
23 | zero_shot: bool = False
24 | sweep: bool = False
25 | seed: int = 0
26 |
27 | data: DataConfig = DataConfig()
28 | post_training: PostTrainingConfig = PostTrainingConfig()
29 |
30 | model_kwargs: dict = {}
31 | fit_kwargs: dict = {}
32 | wandb_init_kwargs: dict = {}
33 |
--------------------------------------------------------------------------------
/scripts/config/README.md:
--------------------------------------------------------------------------------
1 | # Config
2 |
3 | These `.yaml` files are used when training with Weight & Biases with the `train.py` script.
4 |
5 | ## Description
6 |
7 | This is a minimal YAML config used to explain its structure. The dataset names are relative to the `data` directory. If a directory, loads every `.h5ad` files inside it. Can also be a file, or a file pattern.
8 |
9 | ```yaml
10 | data:
11 | train_dataset: merscope # training dataset name
12 | val_dataset: xenium # eval dataset name
13 |
14 | model_kwargs: # Novae model kwargs
15 | heads: 4
16 |
17 | fit_kwargs: # Trainer kwargs (from Lightning)
18 | max_epochs: 3
19 | log_every_n_steps: 10
20 | accelerator: "cpu"
21 |
22 | wandb_init_kwargs: # wandb.init kwargs
23 | mode: "online"
24 | ```
25 |
--------------------------------------------------------------------------------
/scripts/config/_example.yaml:
--------------------------------------------------------------------------------
1 | project: novae
2 | wandb_artefact: novae/novae/xxx
3 |
4 | post_training:
5 | n_domains: [7, 10]
6 | log_umap: true
7 | save_h5ad: true
8 |
9 | data:
10 | train_dataset: all
11 | val_dataset: false
12 |
13 | model_kwargs:
14 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
15 | n_hops_view: 3
16 | heads: 16
17 | hidden_size: 128
18 |
19 | fit_kwargs:
20 | max_epochs: 30
21 | lr: 0.0001
22 | accelerator: gpu
23 | num_workers: 8
24 |
25 | wandb_init_kwargs:
26 | disabled: true
27 |
--------------------------------------------------------------------------------
/scripts/config/all_16.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: all
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 32
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 256
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.15
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0002
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 5
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_17.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: all
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 64
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.15
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0002
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 5
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20]
26 | log_metrics: true
27 | save_h5ad: false
28 | log_umap: false
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_brain.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: /gpfs/workdir/shared/prime/spatial/brain
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_brain
7 | n_hops_view: 3
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 512
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.4
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0001
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20, 25]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_human.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: /gpfs/workdir/shared/prime/spatial/human
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 512
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.5
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0001
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20, 25]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_human2.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: /gpfs/workdir/shared/prime/spatial/human
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 2
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 512
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.75
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0001
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20, 25]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_mouse.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: /gpfs/workdir/shared/prime/spatial/mouse
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 512
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.4
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0001
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20, 25]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_new.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: all
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 16
9 | hidden_size: 128
10 | temperature: 0.1
11 | num_prototypes: 512
12 | background_noise_lambda: 5
13 | panel_subset_size: 0.8
14 | min_prototypes_ratio: 0.15
15 |
16 | fit_kwargs:
17 | max_epochs: 30
18 | lr: 0.0001
19 | accelerator: "gpu"
20 | num_workers: 8
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [15, 20, 25]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/all_ruche.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: all
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 3
8 | heads: 16
9 | num_prototypes: 512
10 | panel_subset_size: 0.8
11 | temperature: 0.1
12 |
13 | fit_kwargs:
14 | max_epochs: 50
15 | accelerator: "gpu"
16 | num_workers: 8
17 | patience: 3
18 | min_delta: 0.05
19 |
--------------------------------------------------------------------------------
/scripts/config/all_spot.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: all
3 | val_dataset: igr/202305031337_hBreast-slide-B-4h-photobleach_VMSC09302
4 |
5 | model_kwargs:
6 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
7 | n_hops_view: 2
8 | heads: 8
9 | hidden_size: 128
10 | temperature: 0.1
11 | n_hops_local: 1
12 | num_prototypes: 256
13 | background_noise_lambda: 5
14 | panel_subset_size: 0.8
15 | min_prototypes_ratio: 0.15
16 |
17 | fit_kwargs:
18 | max_epochs: 30
19 | lr: 0.0002
20 | accelerator: "gpu"
21 | num_workers: 8
22 | patience: 5
23 | min_delta: 0.025
24 |
25 | post_training:
26 | n_domains: [15, 20]
27 | log_metrics: true
28 | save_h5ad: true
29 | log_umap: true
30 | log_domains: true
31 | delete_X: true
32 |
--------------------------------------------------------------------------------
/scripts/config/brain.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: brain
6 |
7 | fit_kwargs:
8 | max_epochs: 10
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 |
--------------------------------------------------------------------------------
/scripts/config/brain2.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: brain2
6 |
7 | fit_kwargs:
8 | max_epochs: 10
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 |
--------------------------------------------------------------------------------
/scripts/config/breast.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: breast
6 |
7 | fit_kwargs:
8 | max_epochs: 10
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 |
--------------------------------------------------------------------------------
/scripts/config/breast_zs.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 | zero_shot: true
4 |
5 | data:
6 | train_dataset: breast
7 |
8 | post_training:
9 | n_domains: [7, 10, 15]
10 | log_metrics: true
11 | save_h5ad: true
12 | log_umap: true
13 | log_domains: true
14 |
--------------------------------------------------------------------------------
/scripts/config/breast_zs2.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y4f53mpp:v30
3 | zero_shot: true
4 |
5 | data:
6 | train_dataset: breast
7 |
8 | post_training:
9 | n_domains: [7, 10, 15]
10 | log_metrics: true
11 | save_h5ad: true
12 | log_umap: true
13 | log_domains: true
14 |
--------------------------------------------------------------------------------
/scripts/config/colon.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: colon
6 |
7 | fit_kwargs:
8 | max_epochs: 15
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 | delete_X: true
21 |
--------------------------------------------------------------------------------
/scripts/config/colon_retrain.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 |
3 | model_kwargs:
4 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
5 | n_hops_view: 3
6 | heads: 16
7 | hidden_size: 128
8 | temperature: 0.1
9 | num_prototypes: 512
10 | background_noise_lambda: 5
11 | panel_subset_size: 0.8
12 |
13 | data:
14 | train_dataset: colon
15 |
16 | fit_kwargs:
17 | max_epochs: 20
18 | accelerator: "gpu"
19 | num_workers: 8
20 | patience: 3
21 | min_delta: 0.05
22 |
23 | post_training:
24 | n_domains: [7, 10, 15]
25 | log_metrics: true
26 | save_h5ad: true
27 | log_umap: true
28 | log_domains: true
29 | delete_X: true
30 |
--------------------------------------------------------------------------------
/scripts/config/colon_zs.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 | zero_shot: true
4 |
5 | data:
6 | train_dataset: colon
7 |
8 | post_training:
9 | n_domains: [7, 10, 15]
10 | log_metrics: true
11 | save_h5ad: true
12 | log_umap: true
13 | log_domains: true
14 | delete_X: true
15 |
--------------------------------------------------------------------------------
/scripts/config/colon_zs2.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y4f53mpp:v30
3 | zero_shot: true
4 |
5 | data:
6 | train_dataset: colon
7 |
8 | post_training:
9 | n_domains: [7, 10, 15]
10 | log_metrics: true
11 | save_h5ad: true
12 | log_umap: true
13 | log_domains: true
14 | delete_X: true
15 |
--------------------------------------------------------------------------------
/scripts/config/local_tests.yaml:
--------------------------------------------------------------------------------
1 | project: novae_tests
2 |
3 | data:
4 | train_dataset: toy_train
5 | val_dataset: toy_val
6 |
7 | model_kwargs:
8 | embedding_size: 50
9 |
10 | fit_kwargs:
11 | max_epochs: 2
12 | accelerator: "cpu"
13 |
14 | post_training:
15 | n_domains: [2, 3]
16 | log_umap: true
17 | log_domains: true
18 |
--------------------------------------------------------------------------------
/scripts/config/lymph_node.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: lymph_node
6 |
7 | fit_kwargs:
8 | max_epochs: 10
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 |
--------------------------------------------------------------------------------
/scripts/config/missing.yaml:
--------------------------------------------------------------------------------
1 | project: novae
2 | seed: 0
3 |
4 | data:
5 | train_dataset: toy_ari/v2
6 |
7 | model_kwargs:
8 | n_hops_view: 2
9 | n_hops_local: 2
10 |
11 | fit_kwargs:
12 | max_epochs: 40
13 | accelerator: "gpu"
14 | num_workers: 4
15 |
--------------------------------------------------------------------------------
/scripts/config/ovarian.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | wandb_artefact: novae/novae/model-y8i2da7y:v30
3 |
4 | data:
5 | train_dataset: ovarian
6 |
7 | fit_kwargs:
8 | max_epochs: 10
9 | accelerator: "gpu"
10 | num_workers: 8
11 | patience: 3
12 | min_delta: 0.05
13 |
14 | post_training:
15 | n_domains: [7, 10, 15]
16 | log_metrics: true
17 | save_h5ad: true
18 | log_umap: true
19 | log_domains: true
20 |
--------------------------------------------------------------------------------
/scripts/config/revision.yaml:
--------------------------------------------------------------------------------
1 | data:
2 | train_dataset: /gpfs/workdir/blampeyq/novae/data/_hyper
3 |
4 | model_kwargs:
5 | scgpt_model_dir: /gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human
6 | n_hops_view: 2
7 | background_noise_lambda: 5
8 | panel_subset_size: 0.8
9 | min_prototypes_ratio: 0.6
10 |
11 | fit_kwargs:
12 | max_epochs: 40
13 | accelerator: "gpu"
14 | num_workers: 8
15 | patience: 5
16 | min_delta: 0.015
17 |
18 | post_training:
19 | n_domains: [8, 10, 12]
20 | log_metrics: true
21 | log_domains: true
22 |
--------------------------------------------------------------------------------
/scripts/config/revision_tests.yaml:
--------------------------------------------------------------------------------
1 | project: novae_tests
2 |
3 | data:
4 | train_dataset: toy_train
5 |
6 | model_kwargs:
7 | n_hops_view: 2
8 | background_noise_lambda: 5
9 | panel_subset_size: 0.8
10 | min_prototypes_ratio: 0.6
11 |
12 | fit_kwargs:
13 | max_epochs: 1
14 | accelerator: "cpu"
15 | patience: 4
16 | min_delta: 0.025
17 |
18 | post_training:
19 | n_domains: [8, 10, 12]
20 | log_metrics: true
21 | log_domains: true
22 |
--------------------------------------------------------------------------------
/scripts/config/toy_cpu_seed0.yaml:
--------------------------------------------------------------------------------
1 | project: novae_eval
2 | seed: 0
3 |
4 | data:
5 | train_dataset: toy_ari/v2
6 |
7 | model_kwargs:
8 | n_hops_view: 1
9 | n_hops_local: 1
10 | heads: 16
11 | hidden_size: 128
12 | temperature: 0.1
13 | num_prototypes: 256
14 | background_noise_lambda: 5
15 | panel_subset_size: 0.8
16 | min_prototypes_ratio: 1
17 |
18 | fit_kwargs:
19 | max_epochs: 30
20 | lr: 0.0005
21 | patience: 6
22 | min_delta: 0.025
23 |
24 | post_training:
25 | n_domains: [7, 10, 15]
26 | log_metrics: true
27 | save_h5ad: true
28 | log_umap: true
29 | log_domains: true
30 | delete_X: true
31 |
--------------------------------------------------------------------------------
/scripts/config/toy_missing.yaml:
--------------------------------------------------------------------------------
1 | project: novae
2 | seed: 0
3 |
4 | data:
5 | train_dataset: toy_ari/v2
6 |
7 | model_kwargs:
8 | n_hops_view: 1
9 | n_hops_local: 1
10 |
11 | fit_kwargs:
12 | max_epochs: 30
13 | accelerator: "gpu"
14 | num_workers: 4
15 |
--------------------------------------------------------------------------------
/scripts/missing_domain.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | from pathlib import Path
3 |
4 | import scanpy as sc
5 | import wandb
6 |
7 | import novae
8 | from novae.monitor.log import log_plt_figure
9 |
10 | from .utils import init_wandb_logger, read_config
11 |
12 |
13 | def main(args: argparse.Namespace) -> None:
14 | config = read_config(args)
15 |
16 | path = Path("/gpfs/workdir/blampeyq/novae/data/_lung_robustness")
17 | adata1_split = sc.read_h5ad(path / "v1_split.h5ad")
18 | adata2_full = sc.read_h5ad(path / "v2_full.h5ad")
19 | adatas = [adata1_split, adata2_full]
20 |
21 | logger = init_wandb_logger(config)
22 |
23 | model = novae.Novae(adatas, **config.model_kwargs)
24 | model.fit(logger=logger, **config.fit_kwargs)
25 |
26 | model.compute_representations(adatas)
27 | obs_key = model.assign_domains(adatas, level=7)
28 |
29 | novae.plot.domains(adatas, obs_key=obs_key, show=False)
30 | log_plt_figure(f"domains_{obs_key}")
31 |
32 | adata2_split = adata2_full[adata2_full.obsm["spatial"][:, 0] < 5000].copy()
33 | jsd = novae.monitor.jensen_shannon_divergence([adata1_split, adata2_split], obs_key=obs_key)
34 |
35 | wandb.log({"metrics/jsd": jsd})
36 |
37 |
38 | if __name__ == "__main__":
39 | parser = argparse.ArgumentParser()
40 | parser.add_argument(
41 | "-c",
42 | "--config",
43 | type=str,
44 | required=True,
45 | help="Fullname of the YAML config to be used for training (see under the `config` directory)",
46 | )
47 | parser.add_argument(
48 | "-s",
49 | "--sweep",
50 | nargs="?",
51 | default=False,
52 | const=True,
53 | help="Whether it is a sweep or not",
54 | )
55 |
56 | main(parser.parse_args())
57 |
--------------------------------------------------------------------------------
/scripts/revision/cpu.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --mem=80G
5 | #SBATCH --cpus-per-task=8
6 | #SBATCH --partition=cpu_long
7 |
8 | module purge
9 | module load anaconda3/2022.10/gcc-11.2.0 && source activate novae
10 |
11 | cd /gpfs/workdir/blampeyq/novae/scripts/revision
12 |
13 | # Execute training
14 | python -u $1
15 |
--------------------------------------------------------------------------------
/scripts/revision/heterogeneous.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import scanpy as sc
3 |
4 | import novae
5 | from novae._constants import Nums
6 |
7 | Nums.QUEUE_WEIGHT_THRESHOLD_RATIO = 0.9999999
8 | Nums.WARMUP_EPOCHS = 4
9 |
10 | suffix = "_constants_fit_all9"
11 |
12 | dir_name = "/gpfs/workdir/blampeyq/novae/data/_heterogeneous"
13 |
14 | adatas = [
15 | sc.read_h5ad(f"{dir_name}/Xenium_V1_Human_Colon_Cancer_P2_CRC_Add_on_FFPE_outs.h5ad"),
16 | # sc.read_h5ad(f"{dir_name}/Xenium_V1_Human_Brain_GBM_FFPE_outs.h5ad"),
17 | sc.read_h5ad(f"{dir_name}/Xenium_V1_hLymphNode_nondiseased_section_outs.h5ad"),
18 | ]
19 |
20 | adatas[0].uns["novae_tissue"] = "colon"
21 | # adatas[1].uns["novae_tissue"] = "brain"
22 | adatas[1].uns["novae_tissue"] = "lymph_node"
23 |
24 | novae.utils.spatial_neighbors(adatas, radius=80)
25 |
26 | model = novae.Novae(
27 | adatas,
28 | scgpt_model_dir="/gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human",
29 | min_prototypes_ratio=0.3,
30 | heads=16,
31 | hidden_size=128,
32 | temperature=0.1,
33 | num_prototypes=1024,
34 | background_noise_lambda=5,
35 | panel_subset_size=0.8,
36 | # num_prototypes=512,
37 | # temperature=0.5,
38 | )
39 | model.fit(max_epochs=30)
40 | model.compute_representations()
41 |
42 | # model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
43 | # model.fine_tune(adatas, min_prototypes_ratio=0.25, reference="all")
44 | # model.compute_representations(adatas)
45 |
46 | for level in range(7, 15):
47 | model.assign_domains(level=level)
48 |
49 | model.plot_prototype_weights()
50 | plt.savefig(f"{dir_name}/prototype_weights{suffix}.pdf", bbox_inches="tight")
51 | model.plot_prototype_weights(assign_zeros=False)
52 | plt.savefig(f"{dir_name}/prototype_weights{suffix}_nz.pdf", bbox_inches="tight")
53 |
54 | for i, adata in enumerate(adatas):
55 | del adata.X
56 | for key in list(adata.layers.keys()):
57 | del adata.layers[key]
58 | adata.write_h5ad(f"{dir_name}/{i}_res{suffix}.h5ad")
59 |
--------------------------------------------------------------------------------
/scripts/revision/heterogeneous_start.py:
--------------------------------------------------------------------------------
1 | import matplotlib.pyplot as plt
2 | import scanpy as sc
3 |
4 | import novae
5 | from novae._constants import Nums
6 |
7 | Nums.QUEUE_WEIGHT_THRESHOLD_RATIO = 0.99
8 |
9 | suffix = ""
10 |
11 | dir_name = "/gpfs/workdir/blampeyq/novae/data/_heterogeneous"
12 |
13 | adatas = [
14 | sc.read_h5ad(f"{dir_name}/Xenium_V1_Human_Colon_Cancer_P2_CRC_Add_on_FFPE_outs.h5ad"),
15 | # sc.read_h5ad(f"{dir_name}/Xenium_V1_Human_Brain_GBM_FFPE_outs.h5ad"),
16 | sc.read_h5ad(f"{dir_name}/Xenium_V1_hLymphNode_nondiseased_section_outs.h5ad"),
17 | ]
18 |
19 | adatas[0].uns["novae_tissue"] = "colon"
20 | # adatas[1].uns["novae_tissue"] = "brain"
21 | adatas[1].uns["novae_tissue"] = "lymph_node"
22 |
23 | for adata in adatas:
24 | adata.obs["novae_tissue"] = adata.uns["novae_tissue"]
25 |
26 | novae.utils.spatial_neighbors(adatas, radius=80)
27 |
28 | # model = novae.Novae(adatas)
29 | # model.mode.trained = True
30 | # model.compute_representations(adatas)
31 | # model.assign_domains(adatas)
32 |
33 | # adata = sc.concat(adatas, join="inner")
34 | # adata = sc.pp.subsample(adata, n_obs=100_000, copy=True)
35 | # sc.pp.neighbors(adata, use_rep="novae_latent")
36 | # sc.tl.umap(adata)
37 | # sc.pl.umap(adata, color=["novae_domains_7", "novae_tissue"])
38 | # plt.savefig(f"{dir_name}/umap_start{suffix}.png", bbox_inches="tight")
39 |
40 | model = novae.Novae(
41 | adatas,
42 | scgpt_model_dir="/gpfs/workdir/blampeyq/checkpoints/scgpt/scGPT_human",
43 | )
44 | model.mode.trained = True
45 | model.compute_representations(adatas)
46 | model.assign_domains(adatas)
47 |
48 | adata = sc.concat(adatas, join="inner")
49 | adata = sc.pp.subsample(adata, n_obs=100_000, copy=True)
50 | sc.pp.neighbors(adata, use_rep="novae_latent")
51 | sc.tl.umap(adata)
52 | sc.pl.umap(adata, color=["novae_domains_7", "novae_tissue"])
53 | plt.savefig(f"{dir_name}/umap_start_scgpt{suffix}.png", bbox_inches="tight")
54 |
--------------------------------------------------------------------------------
/scripts/revision/mgc.py:
--------------------------------------------------------------------------------
1 | import anndata
2 | import matplotlib.pyplot as plt
3 |
4 | import novae
5 |
6 | adata = anndata.read_h5ad("/gpfs/workdir/blampeyq/novae/data/_mgc/MGC_merged_adata_clean_graph.h5ad")
7 |
8 | novae.utils.quantile_scaling(adata)
9 |
10 | model = novae.Novae(adata, embedding_size=62) # 63 proteins
11 |
12 | model.fit()
13 |
14 | model.compute_representations()
15 |
16 | model.plot_domains_hierarchy(max_level=16)
17 | plt.savefig("/gpfs/workdir/blampeyq/novae/data/_mgc/domains_hierarchy.pdf", bbox_inches="tight")
18 |
19 | for level in range(7, 15):
20 | model.assign_domains(level=level)
21 |
22 | adata.write_h5ad("/gpfs/workdir/blampeyq/novae/data/_mgc/MGC_merged_adata_clean_graph_domains.h5ad")
23 |
--------------------------------------------------------------------------------
/scripts/revision/missing_domains.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import anndata
4 | import matplotlib.pyplot as plt
5 | import scanpy as sc
6 |
7 | import novae
8 | from novae._constants import Nums
9 |
10 | Nums.WARMUP_EPOCHS = 1
11 | # Nums.SWAV_EPSILON = 0.01
12 |
13 | suffix = "_numproto_5"
14 |
15 | path = Path("/gpfs/workdir/blampeyq/novae/data/_lung_robustness")
16 |
17 | adata1_split = sc.read_h5ad(path / "v1_split.h5ad")
18 | adata2_full = sc.read_h5ad(path / "v2_full.h5ad")
19 | # adata2_split = sc.read_h5ad(path / "v2_split.h5ad")
20 |
21 | adata_join = anndata.concat([adata1_split, adata2_full], join="inner")
22 | # adatas = [adata1_split, adata2_full]
23 |
24 | novae.spatial_neighbors(adata_join, slide_key="slide_id", radius=80)
25 |
26 | # shared_genes = adata1_split.var_names.intersection(adata2_full.var_names)
27 | # adata1_split = adata1_split[:, shared_genes].copy()
28 | # adata2_full = adata2_full[:, shared_genes].copy()
29 | # adatas = [adata1_split, adata2_full]
30 |
31 | model = novae.Novae(
32 | adata_join,
33 | num_prototypes=2000,
34 | heads=8,
35 | hidden_size=128,
36 | min_prototypes_ratio=0.5,
37 | )
38 | model.fit()
39 | model.compute_representations()
40 |
41 | # model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
42 | # model.fine_tune(adatas, min_prototypes_ratio=0.5, reference="largest")
43 | # model.compute_representations(adatas)
44 |
45 | obs_key = model.assign_domains(adata_join, resolution=1)
46 | for res in [0.3, 0.35, 0.4, 0.45, 0.5]:
47 | obs_key = model.assign_domains(adata_join, resolution=res)
48 | obs_key = model.assign_domains(adata_join, level=7)
49 |
50 | model.plot_prototype_weights()
51 | plt.savefig(path / f"prototype_weights{suffix}.pdf", bbox_inches="tight")
52 |
53 | # model.umap_prototypes()
54 | # plt.savefig(path / f"umap_prototypes{suffix}.png", bbox_inches="tight")
55 |
56 | adatas = [adata_join[adata_join.obs["novae_sid"] == ID] for ID in adata_join.obs["novae_sid"].unique()]
57 | names = ["v1_split", "v2_full"]
58 |
59 | ### Save
60 |
61 | for adata, name in zip(adatas, names):
62 | adata.obs[f"{obs_key}_split_ft"] = adata.obs[obs_key]
63 | del adata.X
64 | for key in list(adata.layers.keys()):
65 | del adata.layers[key]
66 | adata.write_h5ad(path / f"{name}_res2{suffix}.h5ad")
67 |
--------------------------------------------------------------------------------
/scripts/revision/perturbations.py:
--------------------------------------------------------------------------------
1 | import anndata
2 | import torch
3 | from torch_geometric.utils import scatter, softmax
4 |
5 | import novae
6 | from novae._constants import Nums
7 |
8 | adatas = [
9 | anndata.read_h5ad("/gpfs/workdir/blampeyq/novae/data/_perturbation/HumanBreastCancerPatient1_region_0.h5ad"),
10 | anndata.read_h5ad(
11 | "/gpfs/workdir/blampeyq/novae/data/_perturbation/Xenium_V1_Human_Colon_Cancer_P2_CRC_Add_on_FFPE_outs.h5ad"
12 | ),
13 | ]
14 |
15 | model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
16 |
17 | novae.utils.spatial_neighbors(adatas, radius=80)
18 |
19 | # Zero-shot
20 | print("Zero-shot")
21 | model.compute_representations(adatas, zero_shot=True)
22 |
23 | for level in range(7, 15):
24 | model.assign_domains(adatas, level=level)
25 |
26 | for adata in adatas:
27 | adata.obsm["novae_latent_normal"] = adata.obsm["novae_latent"]
28 |
29 | # Attention heterogeneity
30 | print("Attention heterogeneity")
31 | for adata in adatas:
32 | model._datamodule = model._init_datamodule(adata)
33 |
34 | with torch.no_grad():
35 | _entropies = torch.tensor([], dtype=torch.float32)
36 | gat = model.encoder.gnn
37 |
38 | for data_batch in model.datamodule.predict_dataloader():
39 | averaged_attentions_list = []
40 |
41 | data = data_batch["main"]
42 | data = model._embed_pyg_data(data)
43 |
44 | x = data.x
45 |
46 | for i, conv in enumerate(gat.convs):
47 | x, (index, attentions) = conv(
48 | x, data.edge_index, edge_attr=data.edge_attr, return_attention_weights=True
49 | )
50 | averaged_attentions = scatter(attentions.mean(1), index[0], dim_size=len(data.x), reduce="mean")
51 | averaged_attentions_list.append(averaged_attentions)
52 | if i < gat.num_layers - 1:
53 | x = gat.act(x)
54 |
55 | attention_scores = torch.stack(averaged_attentions_list).mean(0)
56 | attention_scores = softmax(attention_scores / 0.01, data.batch, dim=0)
57 | attention_entropy = scatter(-attention_scores * (attention_scores + Nums.EPS).log2(), index=data.batch)
58 | _entropies = torch.cat([_entropies, attention_entropy])
59 |
60 | adata.obs["attention_entropies"] = 0.0
61 | adata.obs.loc[adata.obs["neighborhood_valid"], "attention_entropies"] = _entropies.numpy(force=True)
62 |
63 | # Shuffle nodes
64 | print("Shuffle nodes")
65 | novae.settings.shuffle_nodes = True
66 | model.compute_representations(adatas)
67 |
68 | for adata in adatas:
69 | adata.obs["rs_shuffle"] = novae.utils.get_relative_sensitivity(adata, "novae_latent_normal", "novae_latent")
70 | novae.settings.shuffle_nodes = False
71 |
72 | # Edge length drop
73 | print("Edge length drop")
74 | for adata in adatas:
75 | adata.obsp["spatial_distances"].data[:] = 0.01
76 |
77 | model.compute_representations(adatas)
78 |
79 | for adata in adatas:
80 | adata.obs["rs_edge_length"] = novae.utils.get_relative_sensitivity(adata, "novae_latent_normal", "novae_latent")
81 |
82 | # Saving results
83 |
84 | for adata in adatas:
85 | del adata.X
86 | for key in list(adata.layers.keys()):
87 | del adata.layers[key]
88 |
89 | adatas[0].write_h5ad(
90 | "/gpfs/workdir/blampeyq/novae/data/_perturbation/HumanBreastCancerPatient1_region_0_perturbed.h5ad"
91 | )
92 | adatas[1].write_h5ad(
93 | "/gpfs/workdir/blampeyq/novae/data/_perturbation/Xenium_V1_Human_Colon_Cancer_P2_CRC_Add_on_FFPE_outs_perturbed.h5ad"
94 | )
95 |
--------------------------------------------------------------------------------
/scripts/revision/seg_robustness.py:
--------------------------------------------------------------------------------
1 | import anndata
2 |
3 | import novae
4 |
5 | adatas = [
6 | anndata.read_h5ad("/gpfs/workdir/blampeyq/novae/data/_colon_seg/adata_graph.h5ad"),
7 | anndata.read_h5ad("/gpfs/workdir/blampeyq/novae/data/_colon_seg/adata_default_graph.h5ad"),
8 | ]
9 |
10 | suffix = "_2"
11 |
12 | model = novae.Novae(
13 | adatas,
14 | num_prototypes=3000,
15 | heads=8,
16 | hidden_size=128,
17 | min_prototypes_ratio=1,
18 | )
19 | model.fit()
20 | model.compute_representations()
21 |
22 | # model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
23 | # model.fine_tune(adatas)
24 | # model.compute_representations(adatas)
25 |
26 | for level in range(7, 15):
27 | model.assign_domains(adatas, level=level)
28 |
29 | adatas[0].write_h5ad(f"/gpfs/workdir/blampeyq/novae/data/_colon_seg/adata_graph_domains{suffix}.h5ad")
30 | adatas[1].write_h5ad(f"/gpfs/workdir/blampeyq/novae/data/_colon_seg/adata_default_graph_domains{suffix}.h5ad")
31 |
--------------------------------------------------------------------------------
/scripts/ruche/README.md:
--------------------------------------------------------------------------------
1 | # Scripts for training (rûche HPC)
2 |
--------------------------------------------------------------------------------
/scripts/ruche/agent.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --time=16:00:00
5 | #SBATCH --partition=gpu
6 | #SBATCH --mem=160G
7 | #SBATCH --cpus-per-task=8
8 | #SBATCH --gres=gpu:1
9 |
10 | module purge
11 | module load anaconda3/2022.10/gcc-11.2.0
12 | source activate novae-gpu
13 |
14 | cd /gpfs/workdir/blampeyq/novae
15 |
16 | # Get config
17 | SWEEP_ID=${1}
18 | AGENT_COUNT=${2:-1}
19 | echo "Running $AGENT_COUNT sequential agent(s) for SWEEP_ID=$SWEEP_ID"
20 |
21 | WANDB__SERVICE_WAIT=300
22 | export WANDB__SERVICE_WAIT
23 |
24 | # Run one agent
25 | wandb agent $SWEEP_ID --count $AGENT_COUNT
26 |
--------------------------------------------------------------------------------
/scripts/ruche/convert.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --mem=40G
5 | #SBATCH --cpus-per-task=8
6 | #SBATCH --partition=cpu_med
7 |
8 | module purge
9 | module load anaconda3/2022.10/gcc-11.2.0 && source activate spatial
10 |
11 | cd /gpfs/workdir/blampeyq/novae/data
12 |
13 | python -u xenium_convert.py
14 |
--------------------------------------------------------------------------------
/scripts/ruche/cpu.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --mem=80G
5 | #SBATCH --cpus-per-task=8
6 | #SBATCH --partition=cpu_long
7 |
8 | module purge
9 | module load anaconda3/2022.10/gcc-11.2.0 && source activate novae
10 |
11 | cd /gpfs/workdir/blampeyq/novae
12 |
13 | # Get config
14 | DEFAULT_CONFIG=swav_cpu_0.yaml
15 | CONFIG=${1:-$DEFAULT_CONFIG}
16 | echo Running with CONFIG=$CONFIG
17 |
18 | WANDB__SERVICE_WAIT=300
19 | export WANDB__SERVICE_WAIT
20 |
21 | # Execute training
22 | python -u -m scripts.train --config $CONFIG
23 |
--------------------------------------------------------------------------------
/scripts/ruche/debug_cpu.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --time=00:30:00
5 | #SBATCH --partition=cpu_short
6 | #SBATCH --mem=20G
7 | #SBATCH --cpus-per-task=8
8 |
9 | module purge
10 | module load anaconda3/2022.10/gcc-11.2.0
11 | source activate novae
12 |
13 | cd /gpfs/workdir/blampeyq/novae/scripts
14 |
15 | # Get config
16 | DEFAULT_CONFIG=debug_cpu.yaml
17 | CONFIG=${1:-$DEFAULT_CONFIG}
18 | echo Running with CONFIG=$CONFIG
19 |
20 | WANDB__SERVICE_WAIT=300
21 | export WANDB__SERVICE_WAIT
22 |
23 | # Execute training
24 | CUDA_LAUNCH_BLOCKING=1 python -u train.py --config $CONFIG
25 |
--------------------------------------------------------------------------------
/scripts/ruche/debug_gpu.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --time=00:30:00
5 | #SBATCH --partition=gpu
6 | #SBATCH --mem=32gb
7 | #SBATCH --cpus-per-task=8
8 | #SBATCH --gres=gpu:1
9 |
10 | module purge
11 | module load anaconda3/2022.10/gcc-11.2.0
12 | source activate novae-gpu
13 |
14 | cd /gpfs/workdir/blampeyq/novae/scripts
15 |
16 | # Get config
17 | DEFAULT_CONFIG=debug_gpu.yaml
18 | CONFIG=${1:-$DEFAULT_CONFIG}
19 | echo Running with CONFIG=$CONFIG
20 |
21 | WANDB__SERVICE_WAIT=300
22 | export WANDB__SERVICE_WAIT
23 |
24 | # Execute training
25 | CUDA_LAUNCH_BLOCKING=1 python -u train.py --config $CONFIG
26 |
--------------------------------------------------------------------------------
/scripts/ruche/download.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --mem=16G
5 | #SBATCH --time=01:00:00
6 | #SBATCH --cpus-per-task=1
7 | #SBATCH --partition=cpu_med
8 |
9 | module purge
10 | module load anaconda3/2022.10/gcc-11.2.0
11 | source activate novae
12 |
13 | cd /gpfs/workdir/blampeyq/novae/data
14 |
15 | # download all MERSCOPE datasets
16 | sh _scripts/merscope_download.sh
17 |
18 | # convert all datasets to h5ad files
19 | python _scripts/merscope_convert.py
20 |
--------------------------------------------------------------------------------
/scripts/ruche/gpu.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --time=24:00:00
5 | #SBATCH --partition=gpu
6 | #SBATCH --mem=300G
7 | #SBATCH --cpus-per-task=8
8 | #SBATCH --gres=gpu:1
9 |
10 | module purge
11 | module load anaconda3/2022.10/gcc-11.2.0
12 | source activate novae-gpu
13 |
14 | cd /gpfs/workdir/blampeyq/novae
15 |
16 | # Get config
17 | DEFAULT_CONFIG=all_6.yaml
18 | CONFIG=${1:-$DEFAULT_CONFIG}
19 | echo Running with CONFIG=$CONFIG
20 |
21 | WANDB__SERVICE_WAIT=300
22 | export WANDB__SERVICE_WAIT
23 |
24 | # Execute training
25 | python -u -m scripts.train --config $CONFIG
26 |
--------------------------------------------------------------------------------
/scripts/ruche/prepare.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --time=16:00:00
5 | #SBATCH --partition=gpu
6 | #SBATCH --mem=160G
7 | #SBATCH --cpus-per-task=8
8 | #SBATCH --gres=gpu:1
9 |
10 | module purge
11 | module load anaconda3/2022.10/gcc-11.2.0 && source activate novae
12 |
13 | cd /gpfs/workdir/blampeyq/novae/data
14 |
15 | python -u _scripts/2_prepare.py -n all3 -d merscope -u
16 |
--------------------------------------------------------------------------------
/scripts/ruche/test.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=novae
3 | #SBATCH --output=/gpfs/workdir/blampeyq/.jobs_outputs/%j
4 | #SBATCH --mem=16G
5 | #SBATCH --cpus-per-task=4
6 | #SBATCH --partition=cpu_short
7 |
8 | module purge
9 | cd /gpfs/workdir/blampeyq/novae/data
10 |
11 |
12 | OUTPUT_DIR="./xenium"
13 |
14 | mkdir -p $OUTPUT_DIR
15 |
16 | /usr/bin/unzip -v
17 |
--------------------------------------------------------------------------------
/scripts/sweep/README.md:
--------------------------------------------------------------------------------
1 | ## Wandb sweep configuration
2 |
3 | This is the structure of a wandb config file:
4 |
5 | ```yaml
6 | method: random # search strategy (grid/random/bayes)
7 |
8 | metric:
9 | goal: minimize
10 | name: train/loss_epoch # the metric to be optimized
11 |
12 | parameters: # parameters to hyperoptimize (kwargs arguments of Novae)
13 | heads:
14 | values: [1, 4, 8]
15 | lr:
16 | min: 0.0001
17 | max: 0.01
18 |
19 | command:
20 | - python
21 | - train.py
22 | - --config
23 | - swav_cpu_0.yaml # name of the config under the scripts/config directory
24 | - --sweep
25 | ```
26 |
--------------------------------------------------------------------------------
/scripts/sweep/cpu.yaml:
--------------------------------------------------------------------------------
1 | method: random
2 | metric:
3 | goal: maximize
4 | name: metrics/val_mean_fide_score
5 | parameters:
6 | embedding_size:
7 | values: [32, 64, 128]
8 | heads:
9 | values: [8, 16]
10 | num_prototypes:
11 | values: [1024, 2048]
12 | batch_size:
13 | values: [512, 1024, 2048]
14 | output_size:
15 | values: [64, 128, 256]
16 | num_layers:
17 | values: [8, 12, 16]
18 | hidden_size:
19 | values: [64, 128, 256]
20 | panel_subset_size:
21 | min: 0.1
22 | max: 0.4
23 | gene_expression_dropout:
24 | min: 0.1
25 | max: 0.3
26 | background_noise_lambda:
27 | min: 2
28 | max: 8
29 | sensitivity_noise_std:
30 | min: 0.01
31 | max: 0.1
32 | lr:
33 | min: 0.0001
34 | max: 0.001
35 |
36 | command:
37 | - python
38 | - -m
39 | - scripts.train
40 | - --config
41 | - swav_cpu_0.yaml
42 | - --sweep
43 |
--------------------------------------------------------------------------------
/scripts/sweep/debug.yaml:
--------------------------------------------------------------------------------
1 | method: random
2 | metric:
3 | goal: minimize
4 | name: train/loss_epoch
5 | parameters:
6 | heads:
7 | values: [1, 4, 8]
8 | lr:
9 | min: 0.0001
10 | max: 0.01
11 |
12 | command:
13 | - python
14 | - -m
15 | - scripts.train
16 | - --config
17 | - debug_online.yaml
18 | - --sweep
19 |
--------------------------------------------------------------------------------
/scripts/sweep/gpu.yaml:
--------------------------------------------------------------------------------
1 | method: random
2 | metric:
3 | goal: maximize
4 | name: metrics/val_mean_fide_score
5 | parameters:
6 | n_hops_view:
7 | values: [2, 3]
8 | heads:
9 | values: [8, 16]
10 | num_prototypes:
11 | values: [256, 512, 1024]
12 | output_size:
13 | values: [128, 256, 512]
14 | num_layers:
15 | values: [8, 12, 16]
16 | hidden_size:
17 | values: [128, 256, 512]
18 | panel_subset_size:
19 | min: 0.4
20 | max: 0.9
21 | background_noise_lambda:
22 | min: 2
23 | max: 8
24 | sensitivity_noise_std:
25 | min: 0.01
26 | max: 0.1
27 | lr:
28 | min: 0.0001
29 | max: 0.001
30 | temperature:
31 | min: 0.05
32 | max: 0.2
33 |
34 | command:
35 | - python
36 | - -m
37 | - scripts.train
38 | - --config
39 | - all.yaml
40 | - --sweep
41 |
--------------------------------------------------------------------------------
/scripts/sweep/gpu_ruche.yaml:
--------------------------------------------------------------------------------
1 | method: random
2 | metric:
3 | goal: maximize
4 | name: metrics/val_heuristic
5 | parameters:
6 | output_size:
7 | values: [64, 128]
8 | num_layers:
9 | values: [10, 14, 18]
10 | hidden_size:
11 | values: [64, 128]
12 | background_noise_lambda:
13 | min: 3
14 | max: 5
15 | sensitivity_noise_std:
16 | min: 0.08
17 | max: 0.16
18 | lr:
19 | values: [0.00005, 0.0001, 0.0002, 0.0004]
20 |
21 | command:
22 | - python
23 | - -m
24 | - scripts.train
25 | - --config
26 | - all_ruche.yaml
27 | - --sweep
28 |
--------------------------------------------------------------------------------
/scripts/sweep/lung.yaml:
--------------------------------------------------------------------------------
1 | method: bayes
2 | metric:
3 | goal: minimize
4 | name: metrics/jsd
5 | parameters:
6 | num_prototypes:
7 | min: 1000
8 | max: 8000
9 | temperature:
10 | min: 0.05
11 | max: 0.20
12 | lr:
13 | min: 0.00005
14 | max: 0.001
15 | min_delta:
16 | values: [ 0.001, 0.005, 0.01, 0.05, 0.1 ]
17 | SWAV_EPSILON:
18 | min: 0.005
19 | max: 0.05
20 | QUEUE_SIZE:
21 | values: [ 2, 4, 8 ]
22 | WARMUP_EPOCHS:
23 | values: [ 0, 2, 4, 8 ]
24 |
25 | command:
26 | - python
27 | - -m
28 | - scripts.missing_domain
29 | - --config
30 | - missing.yaml
31 | - --sweep
32 |
--------------------------------------------------------------------------------
/scripts/sweep/revision.yaml:
--------------------------------------------------------------------------------
1 | method: random
2 | metric:
3 | goal: maximize
4 | name: metrics/train_heuristic_8_domains
5 | parameters:
6 | heads:
7 | values: [2, 4, 8, 16]
8 | num_layers:
9 | values: [2, 6, 9, 12, 15]
10 | num_prototypes:
11 | values: [16, 64, 128, 256, 512, 1024]
12 | batch_size:
13 | values: [64, 128, 256, 512, 1024]
14 | temperature:
15 | values: [0.025, 0.05, 0.1, 0.2, 0.4]
16 | lr:
17 | values: [0.00005, 0.0001, 0.0002, 0.0005, 0.001]
18 |
19 | command:
20 | - python
21 | - -m
22 | - scripts.train
23 | - --config
24 | - revision.yaml
25 | - --sweep
26 |
--------------------------------------------------------------------------------
/scripts/sweep/toy.yaml:
--------------------------------------------------------------------------------
1 | method: bayes
2 | metric:
3 | goal: maximize
4 | name: metrics/score
5 | parameters:
6 | num_prototypes:
7 | values: [ 1_000, 3_000, 8_000 ]
8 | temperature:
9 | min: 0.1
10 | max: 0.2
11 | lr:
12 | values: [ 0.0001, 0.0005, 0.001 ]
13 | min_delta:
14 | values: [ 0.001, 0.005, 0.01, 0.05, 0.1 ]
15 | SWAV_EPSILON:
16 | min: 0.01
17 | max: 0.05
18 |
19 | command:
20 | - python
21 | - -m
22 | - scripts.toy_missing_domain
23 | - --config
24 | - toy_missing.yaml
25 | - --sweep
26 |
--------------------------------------------------------------------------------
/scripts/toy_missing_domain.py:
--------------------------------------------------------------------------------
1 | import argparse
2 |
3 | import wandb
4 | from sklearn.metrics.cluster import adjusted_rand_score
5 |
6 | import novae
7 | from novae.monitor.log import log_plt_figure
8 |
9 | from .utils import init_wandb_logger, read_config
10 |
11 |
12 | def main(args: argparse.Namespace) -> None:
13 | config = read_config(args)
14 |
15 | adatas = novae.utils.toy_dataset(n_panels=2, xmax=2_000, n_domains=7, n_vars=300)
16 |
17 | adatas[1] = adatas[1][adatas[1].obs["domain"] != "domain_6", :].copy()
18 | novae.spatial_neighbors(adatas)
19 |
20 | logger = init_wandb_logger(config)
21 |
22 | model = novae.Novae(adatas, **config.model_kwargs)
23 | model.fit(logger=logger, **config.fit_kwargs)
24 |
25 | model.compute_representations(adatas)
26 | obs_key = model.assign_domains(adatas, level=7)
27 |
28 | novae.plot.domains(adatas, obs_key=obs_key, show=False)
29 | log_plt_figure(f"domains_{obs_key}")
30 |
31 | ari = adjusted_rand_score(adatas[0].obs[obs_key], adatas[0].obs["domain"])
32 |
33 | adatas[0] = adatas[0][adatas[0].obs["domain"] != "domain_6", :].copy()
34 | accuracy = (adatas[0].obs[obs_key].values.astype(str) == adatas[1].obs[obs_key].values.astype(str)).mean()
35 |
36 | wandb.log({"metrics/score": ari * accuracy, "metrics/ari": ari, "metrics/accuracy": accuracy})
37 |
38 |
39 | if __name__ == "__main__":
40 | parser = argparse.ArgumentParser()
41 | parser.add_argument(
42 | "-c",
43 | "--config",
44 | type=str,
45 | required=True,
46 | help="Fullname of the YAML config to be used for training (see under the `config` directory)",
47 | )
48 | parser.add_argument(
49 | "-s",
50 | "--sweep",
51 | nargs="?",
52 | default=False,
53 | const=True,
54 | help="Whether it is a sweep or not",
55 | )
56 |
57 | main(parser.parse_args())
58 |
--------------------------------------------------------------------------------
/scripts/train.py:
--------------------------------------------------------------------------------
1 | """
2 | Novae model training with Weight & Biases monitoring
3 | This is **not** the actual Novae source code. Instead, see the `novae` directory
4 | """
5 |
6 | import argparse
7 |
8 | import novae
9 |
10 | from .utils import get_callbacks, init_wandb_logger, post_training, read_config
11 |
12 |
13 | def main(args: argparse.Namespace) -> None:
14 | config = read_config(args)
15 |
16 | adatas = novae.utils.load_local_dataset(config.data.train_dataset, files_black_list=config.data.files_black_list)
17 | adatas_val = novae.utils.load_local_dataset(config.data.val_dataset) if config.data.val_dataset else None
18 |
19 | logger = init_wandb_logger(config)
20 | callbacks = get_callbacks(config, adatas_val)
21 |
22 | if config.wandb_artefact is not None:
23 | model = novae.Novae._load_wandb_artifact(config.wandb_artefact)
24 |
25 | if not config.zero_shot:
26 | model.fine_tune(adatas, logger=logger, callbacks=callbacks, **config.fit_kwargs)
27 | else:
28 | model = novae.Novae(adatas, **config.model_kwargs)
29 | model.fit(logger=logger, callbacks=callbacks, **config.fit_kwargs)
30 |
31 | post_training(model, adatas, config)
32 |
33 |
34 | if __name__ == "__main__":
35 | parser = argparse.ArgumentParser()
36 | parser.add_argument(
37 | "-c",
38 | "--config",
39 | type=str,
40 | required=True,
41 | help="Fullname of the YAML config to be used for training (see under the `config` directory)",
42 | )
43 | parser.add_argument(
44 | "-s",
45 | "--sweep",
46 | nargs="?",
47 | default=False,
48 | const=True,
49 | help="Whether it is a sweep or not",
50 | )
51 |
52 | main(parser.parse_args())
53 |
--------------------------------------------------------------------------------
/scripts/utils.py:
--------------------------------------------------------------------------------
1 | import argparse
2 |
3 | import lightning as L
4 | import numpy as np
5 | import pandas as pd
6 | import scanpy as sc
7 | import wandb
8 | import yaml
9 | from anndata import AnnData
10 | from lightning.pytorch.callbacks import ModelCheckpoint
11 | from lightning.pytorch.loggers import WandbLogger
12 |
13 | import novae
14 | from novae import log
15 | from novae._constants import Keys, Nums
16 | from novae.monitor import (
17 | jensen_shannon_divergence,
18 | mean_fide_score,
19 | mean_normalized_entropy,
20 | )
21 | from novae.monitor.callback import (
22 | LogProtoCovCallback,
23 | LogTissuePrototypeWeights,
24 | ValidationCallback,
25 | )
26 | from novae.monitor.log import log_plt_figure, wandb_results_dir
27 |
28 | from .config import Config
29 |
30 |
31 | def init_wandb_logger(config: Config) -> WandbLogger:
32 | wandb.init(project=config.project, **config.wandb_init_kwargs)
33 |
34 | if config.sweep:
35 | sweep_config = dict(wandb.config)
36 |
37 | fit_kwargs_args = ["lr", "patience", "min_delta"]
38 |
39 | for arg in fit_kwargs_args:
40 | if arg in sweep_config:
41 | config.fit_kwargs[arg] = sweep_config[arg]
42 | del sweep_config[arg]
43 | for key, value in sweep_config.items():
44 | if hasattr(Nums, key):
45 | log.info(f"Nums.{key} = {value}")
46 | setattr(Nums, key, value)
47 | else:
48 | config.model_kwargs[key] = value
49 |
50 | log.info(f"Full config:\n{config.model_dump()}")
51 |
52 | assert "slide_key" not in config.model_kwargs, "For now, please provide one adata per file."
53 |
54 | wandb_logger = WandbLogger(
55 | save_dir=novae.utils.wandb_log_dir(),
56 | log_model="all",
57 | project=config.project,
58 | )
59 |
60 | config_flat = pd.json_normalize(config.model_dump(), sep=".").to_dict(orient="records")[0]
61 | wandb_logger.experiment.config.update(config_flat)
62 |
63 | return wandb_logger
64 |
65 |
66 | def _get_hardware_kwargs(config: Config) -> dict:
67 | return {
68 | "num_workers": config.fit_kwargs.get("num_workers", 0),
69 | "accelerator": config.fit_kwargs.get("accelerator", "cpu"),
70 | }
71 |
72 |
73 | def get_callbacks(config: Config, adatas_val: list[AnnData] | None) -> list[L.Callback] | None:
74 | if config.wandb_init_kwargs.get("mode") == "disabled":
75 | return None
76 |
77 | validation_callback = [ValidationCallback(adatas_val, **_get_hardware_kwargs(config))] if adatas_val else []
78 |
79 | if config.sweep:
80 | return validation_callback
81 |
82 | return [
83 | *validation_callback,
84 | ModelCheckpoint(monitor="metrics/val_heuristic", mode="max", save_last=True, save_top_k=1),
85 | LogProtoCovCallback(),
86 | LogTissuePrototypeWeights(),
87 | ]
88 |
89 |
90 | def read_config(args: argparse.Namespace) -> Config:
91 | with open(novae.utils.repository_root() / "scripts" / "config" / args.config) as f:
92 | config = yaml.safe_load(f)
93 | config = Config(**config, sweep=args.sweep)
94 |
95 | log.info(f"Using {config.seed}")
96 | L.seed_everything(config.seed)
97 |
98 | return config
99 |
100 |
101 | def post_training(model: novae.Novae, adatas: list[AnnData], config: Config): # noqa: C901
102 | wandb.log({"num_parameters": sum(p.numel() for p in model.parameters())})
103 |
104 | keys_repr = ["log_umap", "log_metrics", "log_domains"]
105 | if any(getattr(config.post_training, key) for key in keys_repr):
106 | model.compute_representations(adatas, **_get_hardware_kwargs(config), zero_shot=config.zero_shot)
107 | for n_domains in config.post_training.n_domains:
108 | model.assign_domains(adatas, n_domains=n_domains)
109 |
110 | if config.post_training.log_domains:
111 | for n_domains in config.post_training.n_domains:
112 | obs_key = model.assign_domains(adatas, n_domains=n_domains)
113 | novae.plot.domains(adatas, obs_key, show=False)
114 | log_plt_figure(f"domains_{n_domains=}")
115 |
116 | if config.post_training.log_metrics:
117 | for n_domains in config.post_training.n_domains:
118 | obs_key = model.assign_domains(adatas, n_domains=n_domains)
119 | jsd = jensen_shannon_divergence(adatas, obs_key)
120 | fide = mean_fide_score(adatas, obs_key, n_classes=n_domains)
121 | mne = mean_normalized_entropy(adatas, n_classes=n_domains, obs_key=obs_key)
122 | log.info(f"[{n_domains=}] JSD: {jsd}, FIDE: {fide}, MNE: {mne}")
123 | wandb.log({
124 | f"metrics/jsd_{n_domains}_domains": jsd,
125 | f"metrics/fid_{n_domains}_domainse": fide,
126 | f"metrics/mne_{n_domains}_domains": mne,
127 | f"metrics/train_heuristic_{n_domains}_domains": fide * mne,
128 | })
129 |
130 | if config.post_training.log_umap:
131 | _log_umap(model, adatas, config)
132 |
133 | if config.post_training.save_h5ad:
134 | for adata in adatas:
135 | if config.post_training.delete_X:
136 | del adata.X
137 | if "counts" in adata.layers:
138 | del adata.layers["counts"]
139 | _save_h5ad(adata)
140 |
141 |
142 | def _log_umap(model: novae.Novae, adatas: list[AnnData], config: Config, n_obs_th: int = 500_000):
143 | for adata in adatas:
144 | if "novae_tissue" in adata.uns:
145 | adata.obs["tissue"] = adata.uns["novae_tissue"]
146 |
147 | for n_domains in config.post_training.n_domains:
148 | obs_key = model.assign_domains(adatas, n_domains=n_domains)
149 | model.batch_effect_correction(adatas, obs_key=obs_key)
150 |
151 | latent_conc = np.concatenate([adata.obsm[Keys.REPR_CORRECTED] for adata in adatas], axis=0)
152 | obs_conc = pd.concat([adata.obs for adata in adatas], axis=0, join="inner")
153 | adata_conc = AnnData(obsm={Keys.REPR_CORRECTED: latent_conc}, obs=obs_conc)
154 |
155 | if "cell_id" in adata_conc.obs:
156 | del adata_conc.obs["cell_id"] # can't be saved for some reasons
157 | _save_h5ad(adata_conc, "adata_conc")
158 |
159 | if adata_conc.n_obs > n_obs_th:
160 | sc.pp.subsample(adata_conc, n_obs=n_obs_th)
161 |
162 | sc.pp.neighbors(adata_conc, use_rep=Keys.REPR_CORRECTED)
163 | sc.tl.umap(adata_conc)
164 |
165 | colors = [obs_key]
166 | for key in ["tissue", "technology"]:
167 | if key in adata_conc.obs:
168 | colors.append(key)
169 |
170 | sc.pl.umap(adata_conc, color=colors, show=False)
171 | log_plt_figure(f"umap_{n_domains=}")
172 |
173 |
174 | def _save_h5ad(adata: AnnData, stem: str | None = None):
175 | if stem is None:
176 | stem = adata.obs["slide_id"].iloc[0] if "slide_id" in adata.obs else str(id(adata))
177 |
178 | out_path = wandb_results_dir() / f"{stem}.h5ad"
179 | log.info(f"Writing adata file to {out_path}: {adata}")
180 | adata.write_h5ad(out_path)
181 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | from setuptools import setup
2 |
3 | if __name__ == "__main__":
4 | setup()
5 |
--------------------------------------------------------------------------------
/tests/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/MICS-Lab/novae/b641a0bd1947759317c9e7ab4d997bb7a4a00932/tests/__init__.py
--------------------------------------------------------------------------------
/tests/_utils.py:
--------------------------------------------------------------------------------
1 | import anndata
2 | import numpy as np
3 | import pandas as pd
4 | from anndata import AnnData
5 |
6 | import novae
7 |
8 | domains = [
9 | ["D1", "D2", "D3", "D4", "D5"],
10 | ["D1", "D1", "D2", "D2", "D3"],
11 | ["D1", "D1", "D1", "D2", np.nan],
12 | ]
13 |
14 | spatial_coords = np.array([
15 | [[0, 0], [0, 1], [0, 2], [3, 3], [1, 1]],
16 | [[0, 0], [0, 1], [0, 2], [3, 3], [1, 1]],
17 | [[0, 0], [0, 1], [0, 2], [3, 3], [1, 1]],
18 | ])
19 |
20 |
21 | def _get_adata(i: int):
22 | values = domains[i]
23 | adata = AnnData(obs=pd.DataFrame({"domain": values}, index=[str(i) for i in range(len(values))]))
24 | adata.obs["slide_key"] = f"slide_{i}"
25 | adata.obsm["spatial"] = spatial_coords[i]
26 | novae.utils.spatial_neighbors(adata, radius=[0, 1.5])
27 | return adata
28 |
29 |
30 | adatas = [_get_adata(i) for i in range(len(domains))]
31 |
32 | adata = adatas[0]
33 |
34 | adata_concat = anndata.concat(adatas)
35 |
36 | # o
37 | #
38 | # o-o
39 | #
40 | # o-o-o
41 | spatial_coords2 = np.array(
42 | [[0, 0], [0, 1], [0, 2], [2, 0], [2, 1], [4, 0]],
43 | )
44 |
45 | adata_line = AnnData(obs=pd.DataFrame(index=[str(i) for i in range(len(spatial_coords2))]))
46 | adata_line.obsm["spatial"] = spatial_coords2
47 | novae.utils.spatial_neighbors(adata_line, radius=1.5)
48 |
--------------------------------------------------------------------------------
/tests/test_correction.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 | from anndata import AnnData
4 |
5 | import novae
6 | from novae._constants import Keys
7 |
8 | obs1 = pd.DataFrame(
9 | {
10 | Keys.SLIDE_ID: ["a", "a", "b", "b", "a", "b", "b"],
11 | "domains_key": ["N1", "N1", "N1", "N1", "N3", "N1", "N2"],
12 | Keys.IS_VALID_OBS: [True, True, True, True, True, True, True],
13 | },
14 | index=[f"{i}_1" for i in range(7)],
15 | )
16 |
17 | latent1 = np.array([
18 | [1, 2],
19 | [3, 4],
20 | [5, 6],
21 | [7, 8],
22 | [9, 10],
23 | [12, 13],
24 | [-2, -2],
25 | ]).astype(np.float32)
26 |
27 | expected1 = np.array([
28 | [7, 8],
29 | [9, 10],
30 | [5, 6],
31 | [7, 8],
32 | [9, 10],
33 | [12, 13],
34 | [-2 + 35, -2 + 34],
35 | ]).astype(np.float32)
36 |
37 | adata1 = AnnData(obs=obs1, obsm={Keys.REPR: latent1})
38 |
39 | obs2 = pd.DataFrame(
40 | {
41 | Keys.SLIDE_ID: ["c", "c", "c", "c", "c"],
42 | "domains_key": ["N2", "N1", np.nan, "N2", "N2"],
43 | Keys.IS_VALID_OBS: [True, True, False, True, True],
44 | },
45 | index=[f"{i}_2" for i in range(5)],
46 | )
47 |
48 | latent2 = np.array([
49 | [-1, -3],
50 | [0, -10],
51 | [0, 0],
52 | [0, -1],
53 | [100, 100],
54 | ]).astype(np.float32)
55 |
56 | expected2 = np.array([
57 | [-1, -3],
58 | [0 + 8, -10 + 19],
59 | [0, 0],
60 | [0, -1],
61 | [100, 100],
62 | ]).astype(np.float32)
63 |
64 | adata2 = AnnData(obs=obs2, obsm={Keys.REPR: latent2})
65 |
66 | adatas = [adata1, adata2]
67 | for adata in adatas:
68 | for key in [Keys.SLIDE_ID, "domains_key"]:
69 | adata.obs[key] = adata.obs[key].astype("category")
70 |
71 |
72 | def test_batch_effect_correction():
73 | novae.utils.batch_effect_correction(adatas, "domains_key")
74 | assert (adata1.obsm[Keys.REPR_CORRECTED] == expected1).all()
75 | assert (adata2.obsm[Keys.REPR_CORRECTED] == expected2).all()
76 |
--------------------------------------------------------------------------------
/tests/test_dataset.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pytest
3 |
4 | import novae
5 | from novae._constants import Keys
6 |
7 | N_PANELS = 2
8 | N_SLIDES_PER_PANEL = 3
9 |
10 |
11 | adatas = novae.utils.toy_dataset(n_panels=N_PANELS, n_slides_per_panel=N_SLIDES_PER_PANEL)
12 |
13 | single_adata = adatas[0]
14 |
15 |
16 | def test_raise_invalid_slide_id():
17 | with pytest.raises(AssertionError):
18 | novae.utils.spatial_neighbors(adatas, slide_key="key_not_in_obs")
19 |
20 |
21 | def test_single_panel():
22 | novae.utils.spatial_neighbors(single_adata)
23 | model = novae.Novae(single_adata)
24 | model._datamodule = model._init_datamodule()
25 |
26 | assert len(model.dataset.slides_metadata) == 1
27 | assert model.dataset.obs_ilocs is not None
28 |
29 |
30 | def test_single_panel_slide_key():
31 | novae.utils.spatial_neighbors(single_adata, slide_key="slide_key")
32 | model = novae.Novae(single_adata)
33 | model._datamodule = model._init_datamodule()
34 |
35 | assert len(model.dataset.slides_metadata) == N_SLIDES_PER_PANEL
36 | assert model.dataset.obs_ilocs is not None
37 |
38 | _ensure_batch_same_slide(model)
39 |
40 |
41 | def test_multi_panel():
42 | novae.utils.spatial_neighbors(adatas)
43 | model = novae.Novae(adatas)
44 | model._datamodule = model._init_datamodule()
45 |
46 | assert len(model.dataset.slides_metadata) == N_PANELS
47 | assert model.dataset.obs_ilocs is None
48 |
49 | _ensure_batch_same_slide(model)
50 |
51 |
52 | def test_multi_panel_slide_key():
53 | novae.utils.spatial_neighbors(adatas, slide_key="slide_key")
54 | model = novae.Novae(adatas)
55 | model._datamodule = model._init_datamodule()
56 |
57 | assert len(model.dataset.slides_metadata) == N_PANELS * N_SLIDES_PER_PANEL
58 | assert model.dataset.obs_ilocs is None
59 |
60 | _ensure_batch_same_slide(model)
61 |
62 |
63 | def _ensure_batch_same_slide(model: novae.Novae):
64 | n_obs_dataset = model.dataset.shuffled_obs_ilocs.shape[0]
65 | assert n_obs_dataset % model.hparams.batch_size == 0
66 |
67 | for batch_index in range(n_obs_dataset // model.hparams.batch_size):
68 | sub_obs_ilocs = model.dataset.shuffled_obs_ilocs[
69 | batch_index * model.hparams.batch_size : (batch_index + 1) * model.hparams.batch_size
70 | ]
71 | unique_adata_indices = np.unique(sub_obs_ilocs[:, 0])
72 | assert len(unique_adata_indices) == 1
73 | slide_ids = adatas[unique_adata_indices[0]].obs[Keys.SLIDE_ID].iloc[sub_obs_ilocs[:, 1]]
74 | assert len(np.unique(slide_ids)) == 1
75 |
--------------------------------------------------------------------------------
/tests/test_metrics.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from pytest import approx
3 |
4 | import novae
5 | from novae.monitor.eval import _jensen_shannon_divergence, entropy
6 |
7 | from ._utils import adata_concat, adatas
8 |
9 | true_distributions = np.array([
10 | [0.2] * 5,
11 | [0.4, 0.4, 0.2, 0, 0],
12 | [0.75, 0.25, 0, 0, 0],
13 | ])
14 |
15 | true_jsd = _jensen_shannon_divergence(true_distributions)
16 |
17 |
18 | def test_heuristic():
19 | novae.monitor.heuristic(adatas, "domain", n_classes=5)
20 |
21 |
22 | def test_jensen_shannon_divergence():
23 | jsd = novae.monitor.jensen_shannon_divergence(adatas, "domain")
24 |
25 | assert jsd == approx(true_jsd)
26 |
27 |
28 | def test_jensen_shannon_divergence_concat():
29 | jsd = novae.monitor.jensen_shannon_divergence(adata_concat, "domain", slide_key="slide_key")
30 |
31 | assert jsd == approx(true_jsd)
32 |
33 |
34 | def test_jensen_shannon_divergence_manual():
35 | assert _jensen_shannon_divergence(np.ones((1, 5))) == approx(0.0)
36 | assert _jensen_shannon_divergence(np.ones((2, 5))) == approx(0.0)
37 |
38 | distribution = np.array([
39 | [0.3, 0.2, 0.5],
40 | [0.1, 0.1, 0.8],
41 | [0.2, 0.3, 0.5],
42 | [0, 0, 1],
43 | ])
44 |
45 | means = np.array([0.15, 0.15, 0.7])
46 |
47 | entropy_means = entropy(means)
48 |
49 | assert entropy_means == approx(1.18, rel=1e-2)
50 |
51 | jsd_manual = entropy_means - 0.25 * sum(entropy(d) for d in distribution)
52 |
53 | jsd = _jensen_shannon_divergence(distribution)
54 |
55 | assert jsd == approx(jsd_manual)
56 |
57 |
58 | def test_fide_score():
59 | fide = novae.monitor.fide_score(adatas[0], "domain", n_classes=5)
60 |
61 | assert fide == 0
62 |
63 | fide = novae.monitor.fide_score(adatas[1], "domain", n_classes=3)
64 |
65 | assert fide == approx(0.4 / 3)
66 |
67 | fide = novae.monitor.fide_score(adatas[1], "domain", n_classes=5)
68 |
69 | assert fide == approx(0.4 / 5)
70 |
71 | fide = novae.monitor.fide_score(adatas[2], "domain", n_classes=1)
72 |
73 | assert fide == approx(1)
74 |
75 | fide = novae.monitor.fide_score(adatas[2], "domain", n_classes=5)
76 |
77 | assert fide == approx(1 / 5)
78 |
--------------------------------------------------------------------------------
/tests/test_misc.py:
--------------------------------------------------------------------------------
1 | import novae
2 | from novae._constants import Nums
3 |
4 | adata = novae.utils.toy_dataset(xmax=200)[0]
5 |
6 |
7 | def test_settings():
8 | novae.settings.disable_lazy_loading()
9 |
10 | novae.utils.spatial_neighbors(adata)
11 | model = novae.Novae(adata)
12 | model._datamodule = model._init_datamodule()
13 | assert model.dataset.anndata_torch.tensors is not None
14 |
15 | novae.settings.enable_lazy_loading(n_obs_threshold=100)
16 | novae.utils.spatial_neighbors(adata)
17 | model = novae.Novae(adata)
18 | model._datamodule = model._init_datamodule()
19 | assert model.dataset.anndata_torch.tensors is None
20 |
21 | novae.settings.warmup_epochs = 2
22 | assert novae.settings.warmup_epochs == Nums.WARMUP_EPOCHS
23 |
24 |
25 | def test_repr():
26 | novae.utils.spatial_neighbors(adata)
27 | model = novae.Novae(adata)
28 |
29 | repr(model)
30 | repr(model.mode)
31 |
--------------------------------------------------------------------------------
/tests/test_model.py:
--------------------------------------------------------------------------------
1 | import json
2 |
3 | import anndata
4 | import numpy as np
5 | import pandas as pd
6 | import pytest
7 | import torch
8 |
9 | import novae
10 | from novae._constants import Keys
11 | from novae.utils._data import GENE_NAMES_SUBSET
12 |
13 | adatas = novae.utils.toy_dataset(
14 | n_panels=2,
15 | n_slides_per_panel=2,
16 | xmax=100,
17 | n_domains=2,
18 | compute_spatial_neighbors=True,
19 | )
20 | adata = adatas[0]
21 |
22 |
23 | def _generate_fake_scgpt_inputs():
24 | gene_names = GENE_NAMES_SUBSET[:100]
25 | indices = [7, 4, 3, 0, 1, 2, 9, 5, 6, 8, *list(range(10, 100))]
26 |
27 | vocab = dict(zip(gene_names, indices))
28 |
29 | with open("tests/vocab.json", "w") as f:
30 | json.dump(vocab, f, indent=4)
31 |
32 | torch.save({"encoder.embedding.weight": torch.randn(len(vocab), 16)}, "tests/best_model.pt")
33 |
34 |
35 | _generate_fake_scgpt_inputs()
36 |
37 |
38 | # def test_load_wandb_artifact():
39 | # novae.Novae._load_wandb_artifact("novae/novae/model-4i8e9g2v:v17")
40 |
41 |
42 | def test_load_huggingface_model():
43 | model = novae.Novae.from_pretrained("MICS-Lab/novae-test")
44 |
45 | assert model.cell_embedder.embedding.weight.requires_grad is False
46 |
47 |
48 | def test_train():
49 | adatas = novae.utils.toy_dataset(
50 | n_panels=2,
51 | n_slides_per_panel=2,
52 | xmax=60,
53 | n_domains=2,
54 | )
55 |
56 | novae.utils.spatial_neighbors(adatas)
57 | model = novae.Novae(adatas, num_prototypes=10)
58 |
59 | with pytest.raises(AssertionError): # should raise an error because the model has not been trained
60 | model.compute_representations()
61 |
62 | model.fit(max_epochs=3)
63 | model.compute_representations()
64 | model.compute_representations(num_workers=2)
65 |
66 | # obs_key = model.assign_domains(n_domains=2)
67 | obs_key = model.assign_domains(level=2)
68 |
69 | model.batch_effect_correction()
70 |
71 | adatas[0].obs.iloc[0][obs_key] = np.nan
72 |
73 | novae.monitor.mean_fide_score(adatas, obs_key=obs_key)
74 | novae.monitor.jensen_shannon_divergence(adatas, obs_key=obs_key)
75 |
76 | novae.monitor.mean_fide_score(adatas, obs_key=obs_key, slide_key="slide_key")
77 | novae.monitor.jensen_shannon_divergence(adatas, obs_key=obs_key, slide_key="slide_key")
78 |
79 | adatas[0].write_h5ad("tests/test.h5ad") # ensures the output can be saved
80 |
81 | model.compute_representations(adatas, zero_shot=True)
82 |
83 | with pytest.raises(AssertionError):
84 | model.fine_tune(adatas, max_epochs=1)
85 |
86 | model.mode.pretrained = True
87 |
88 | model.fine_tune(adatas, max_epochs=1)
89 |
90 |
91 | @pytest.mark.parametrize("slide_key", [None, "slide_key"])
92 | def test_representation_single_panel(slide_key: str | None):
93 | adata = novae.utils.toy_dataset(
94 | n_panels=1,
95 | n_slides_per_panel=2,
96 | xmax=100,
97 | n_domains=2,
98 | )[0]
99 |
100 | novae.utils.spatial_neighbors(adata, slide_key=slide_key)
101 |
102 | model = novae.Novae(adata)
103 | model._datamodule = model._init_datamodule()
104 | model.mode.trained = True
105 |
106 | model.compute_representations()
107 |
108 | domains = adata.obs[Keys.LEAVES].copy()
109 |
110 | model.compute_representations()
111 |
112 | assert domains.equals(adata.obs[Keys.LEAVES])
113 |
114 | novae.utils.spatial_neighbors(adata, slide_key=slide_key)
115 | model.compute_representations([adata])
116 |
117 | assert domains.equals(adata.obs[Keys.LEAVES])
118 |
119 | if slide_key is not None:
120 | sids = adata.obs[slide_key].unique()
121 | adatas = [adata[adata.obs[slide_key] == sid] for sid in sids]
122 |
123 | novae.utils.spatial_neighbors(adatas)
124 | model.compute_representations(adatas)
125 |
126 | adata_concat = anndata.concat(adatas)
127 |
128 | assert domains.equals(adata_concat.obs[Keys.LEAVES].loc[domains.index])
129 |
130 | novae.utils.spatial_neighbors(adatas, slide_key=slide_key)
131 | model.compute_representations(adatas)
132 |
133 | adata_concat = anndata.concat(adatas)
134 |
135 | assert domains.equals(adata_concat.obs[Keys.LEAVES].loc[domains.index])
136 |
137 |
138 | @pytest.mark.parametrize("slide_key", [None, "slide_key"])
139 | def test_representation_multi_panel(slide_key: str | None):
140 | adatas = novae.utils.toy_dataset(
141 | n_panels=3,
142 | n_slides_per_panel=2,
143 | xmax=100,
144 | n_domains=2,
145 | )
146 |
147 | novae.utils.spatial_neighbors(adatas, slide_key=slide_key)
148 | model = novae.Novae(adatas)
149 | model._datamodule = model._init_datamodule()
150 | model.mode.trained = True
151 |
152 | model.compute_representations()
153 |
154 | domains_series = pd.concat([adata.obs[Keys.LEAVES].copy() for adata in adatas])
155 |
156 | novae.utils.spatial_neighbors(adatas, slide_key=slide_key)
157 | model.compute_representations(adatas)
158 |
159 | domains_series2 = pd.concat([adata.obs[Keys.LEAVES].copy() for adata in adatas])
160 |
161 | assert domains_series.equals(domains_series2.loc[domains_series.index])
162 |
163 | adata_split = [
164 | adata[adata.obs[Keys.SLIDE_ID] == sid].copy() for adata in adatas for sid in adata.obs[Keys.SLIDE_ID].unique()
165 | ]
166 |
167 | model.compute_representations(adata_split)
168 |
169 | domains_series2 = pd.concat([adata.obs[Keys.LEAVES] for adata in adata_split])
170 |
171 | assert domains_series.equals(domains_series2.loc[domains_series.index])
172 |
173 |
174 | @pytest.mark.parametrize("slide_key", [None, "slide_key"])
175 | @pytest.mark.parametrize("scgpt_model_dir", [None, "tests"])
176 | def test_saved_model_identical(slide_key: str | None, scgpt_model_dir: str | None):
177 | adata = novae.utils.toy_dataset(
178 | n_panels=1,
179 | n_slides_per_panel=2,
180 | xmax=100,
181 | n_domains=2,
182 | )[0]
183 |
184 | # using weird parameters
185 | novae.utils.spatial_neighbors(adata, slide_key=slide_key)
186 | model = novae.Novae(
187 | adata,
188 | embedding_size=67,
189 | output_size=78,
190 | n_hops_local=4,
191 | n_hops_view=3,
192 | heads=2,
193 | hidden_size=46,
194 | num_layers=3,
195 | batch_size=345,
196 | temperature=0.13,
197 | num_prototypes=212,
198 | panel_subset_size=0.62,
199 | background_noise_lambda=7.7,
200 | sensitivity_noise_std=0.042,
201 | scgpt_model_dir=scgpt_model_dir,
202 | )
203 |
204 | assert model.cell_embedder.embedding.weight.requires_grad is (scgpt_model_dir is None)
205 |
206 | model._datamodule = model._init_datamodule()
207 | model.mode.trained = True
208 |
209 | model.compute_representations()
210 | model.assign_domains()
211 |
212 | domains = adata.obs[Keys.LEAVES].copy()
213 | representations = adata.obsm[Keys.REPR].copy()
214 |
215 | model.save_pretrained("tests/test_model")
216 |
217 | new_model = novae.Novae.from_pretrained("tests/test_model")
218 |
219 | novae.utils.spatial_neighbors(adata, slide_key=slide_key)
220 | new_model.compute_representations(adata)
221 | new_model.assign_domains(adata)
222 |
223 | assert (adata.obsm[Keys.REPR] == representations).all()
224 | assert domains.equals(adata.obs[Keys.LEAVES])
225 |
226 | for name, param in model.named_parameters():
227 | assert torch.equal(param, new_model.state_dict()[name])
228 |
229 |
230 | def test_safetensors_parameters_names():
231 | from huggingface_hub import hf_hub_download
232 | from safetensors import safe_open
233 |
234 | local_file = hf_hub_download(repo_id="MICS-Lab/novae-human-0", filename="model.safetensors")
235 | with safe_open(local_file, framework="pt", device="cpu") as f:
236 | pretrained_model_names = f.keys()
237 |
238 | model = novae.Novae(adata)
239 |
240 | actual_names = [name for name, _ in model.named_parameters()]
241 |
242 | assert set(pretrained_model_names) == set(actual_names)
243 |
244 |
245 | def test_reset_clusters_zero_shot():
246 | adata = novae.utils.toy_dataset()[0]
247 |
248 | novae.utils.spatial_neighbors(adata)
249 |
250 | model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
251 |
252 | model.compute_representations(adata, zero_shot=True)
253 | clusters_levels = model.swav_head.clusters_levels.copy()
254 |
255 | adata = adata[:550].copy()
256 |
257 | model.compute_representations(adata, zero_shot=True)
258 |
259 | assert not (model.swav_head.clusters_levels == clusters_levels).all()
260 |
--------------------------------------------------------------------------------
/tests/test_plots.py:
--------------------------------------------------------------------------------
1 | import pytest
2 |
3 | import novae
4 | from novae._constants import Keys
5 |
6 | adatas = novae.utils.toy_dataset(n_panels=2, xmax=200)
7 | novae.utils.spatial_neighbors(adatas)
8 |
9 | model = novae.Novae(adatas)
10 |
11 | model.mode.trained = True
12 | model.compute_representations()
13 | model.assign_domains()
14 |
15 |
16 | @pytest.mark.parametrize("hline_level", [None, 3, [2, 4]])
17 | def test_plot_domains_hierarchy(hline_level: int | list[int] | None):
18 | model.plot_domains_hierarchy(hline_level=hline_level)
19 |
20 |
21 | def test_plot_prototype_weights():
22 | model.plot_prototype_weights()
23 |
24 | adatas[0].uns[Keys.UNS_TISSUE] = "breast"
25 | adatas[1].uns[Keys.UNS_TISSUE] = "lung"
26 |
27 | model.plot_prototype_weights()
28 |
29 |
30 | def test_plot_domains():
31 | novae.plot.domains(adatas, show=False)
32 |
33 |
34 | def test_plot_connectivities():
35 | novae.plot.connectivities(adatas, ngh_threshold=2, show=False)
36 | novae.plot.connectivities(adatas, ngh_threshold=None, show=False)
37 |
--------------------------------------------------------------------------------
/tests/test_utils.py:
--------------------------------------------------------------------------------
1 | import anndata
2 | import numpy as np
3 | import pandas as pd
4 | import pytest
5 | from anndata import AnnData
6 |
7 | import novae
8 | from novae._constants import Keys
9 | from novae.data.dataset import _to_adjacency_local, _to_adjacency_view
10 | from novae.utils._build import _set_unique_slide_ids
11 |
12 | from ._utils import adata, adata_concat, adata_line
13 |
14 | true_connectivities = np.array([
15 | [0, 1, 0, 0, 1],
16 | [1, 0, 1, 0, 1],
17 | [0, 1, 0, 0, 1],
18 | [0, 0, 0, 0, 0],
19 | [1, 1, 1, 0, 0],
20 | ])
21 |
22 |
23 | def test_build():
24 | connectivities = adata.obsp["spatial_connectivities"]
25 |
26 | assert connectivities.shape[0] == adata.n_obs
27 |
28 | assert (connectivities.todense() == true_connectivities).all()
29 |
30 |
31 | def test_set_unique_slide_ids():
32 | adatas = novae.utils.toy_dataset(
33 | xmax=200,
34 | n_panels=2,
35 | n_slides_per_panel=1,
36 | n_vars=30,
37 | slide_ids_unique=False,
38 | )
39 |
40 | _set_unique_slide_ids(adatas, slide_key="slide_key")
41 |
42 | assert adatas[0].obs[Keys.SLIDE_ID].iloc[0] == f"{id(adatas[0])}_slide_0"
43 |
44 | adatas = novae.utils.toy_dataset(
45 | xmax=200,
46 | n_panels=2,
47 | n_slides_per_panel=1,
48 | n_vars=30,
49 | slide_ids_unique=True,
50 | )
51 |
52 | _set_unique_slide_ids(adatas, slide_key="slide_key")
53 |
54 | assert adatas[0].obs[Keys.SLIDE_ID].iloc[0] == "slide_0_0"
55 |
56 |
57 | def test_build_slide_key():
58 | adata_ = adata_concat.copy()
59 | novae.utils.spatial_neighbors(adata_, radius=1.5, slide_key="slide_key")
60 |
61 | true_connectivities = np.array([
62 | [0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
63 | [1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
64 | [0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
65 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
66 | [1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
67 | [0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0],
68 | [0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0],
69 | [0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0],
70 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
71 | [0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0],
72 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1],
73 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1],
74 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1],
75 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
76 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0],
77 | ])
78 |
79 | assert (adata_.obsp["spatial_connectivities"].todense() == true_connectivities).all()
80 |
81 |
82 | def test_build_slide_key_disjoint_indices():
83 | adata = novae.utils.toy_dataset(
84 | n_panels=1,
85 | n_slides_per_panel=1,
86 | xmax=100,
87 | n_domains=2,
88 | )[0]
89 |
90 | adata2 = adata.copy()
91 | adata2.obs["slide_key"] = "slide_key2"
92 | adata2.obs_names = adata2.obs_names + "_2"
93 |
94 | novae.utils.spatial_neighbors(adata)
95 | novae.utils.spatial_neighbors(adata2)
96 |
97 | n1, n2 = len(adata.obsp["spatial_connectivities"].data), len(adata2.obsp["spatial_connectivities"].data)
98 | assert n1 == n2
99 |
100 | adata_concat = anndata.concat([adata, adata2], axis=0).copy()
101 |
102 | novae.utils.spatial_neighbors(adata_concat, slide_key="slide_key")
103 |
104 | assert len(adata_concat.obsp["spatial_connectivities"].data) == n1 + n2
105 |
106 |
107 | def test_build_technology():
108 | adata_cosmx = adata.copy()
109 | adata_cosmx.obs[["CenterX_global_px", "CenterY_global_px"]] = adata_cosmx.obsm["spatial"]
110 | del adata_cosmx.obsm["spatial"]
111 | novae.utils.spatial_neighbors(adata_cosmx, technology="cosmx")
112 |
113 | del adata_cosmx.obs["CenterY_global_px"]
114 |
115 | # one column is missing in obs
116 | with pytest.raises(AssertionError):
117 | novae.utils.spatial_neighbors(adata_cosmx, technology="cosmx")
118 |
119 |
120 | def test_invalid_build():
121 | adata_invalid = anndata.AnnData(obs=pd.DataFrame(index=["0", "1", "2"]))
122 |
123 | with pytest.raises(AssertionError):
124 | novae.utils.spatial_neighbors(adata_invalid, radius=[0, 1.5])
125 |
126 | adata_invalid.obsm["spatial"] = np.array([[0, 0, 0], [0, 1, 2], [0, 2, 4]])
127 |
128 | with pytest.raises(AssertionError):
129 | novae.utils.spatial_neighbors(adata_invalid, radius=[0, 1.5])
130 |
131 | with pytest.raises(AssertionError):
132 | novae.utils.spatial_neighbors(adata_invalid, radius=2, technology="unknown")
133 |
134 | with pytest.raises(AssertionError):
135 | novae.utils.spatial_neighbors(adata_invalid, radius=1, technology="cosmx", pixel_size=0.1)
136 |
137 |
138 | def test_to_adjacency_local():
139 | adjancency_local = _to_adjacency_local(adata.obsp["spatial_connectivities"], 1)
140 |
141 | assert (
142 | (adjancency_local.todense() > 0)
143 | == np.array([
144 | [True, True, False, False, True],
145 | [True, True, True, False, True],
146 | [False, True, True, False, True],
147 | [False, False, False, True, False],
148 | [True, True, True, False, True],
149 | ])
150 | ).all()
151 |
152 | adjancency_local = _to_adjacency_local(adata.obsp["spatial_connectivities"], 2)
153 |
154 | assert (
155 | (adjancency_local.todense() > 0)
156 | == np.array([
157 | [True, True, True, False, True],
158 | [True, True, True, False, True],
159 | [True, True, True, False, True],
160 | [False, False, False, False, False], # unconnected node => no local connections with n_hop >= 2
161 | [True, True, True, False, True],
162 | ])
163 | ).all()
164 |
165 | adjancency_local = _to_adjacency_local(adata_line.obsp["spatial_connectivities"], 1)
166 |
167 | assert (
168 | (adjancency_local.todense() > 0)
169 | == np.array([
170 | [True, True, False, False, False, False],
171 | [True, True, True, False, False, False],
172 | [False, True, True, False, False, False],
173 | [False, False, False, True, True, False],
174 | [False, False, False, True, True, False],
175 | [False, False, False, False, False, True],
176 | ])
177 | ).all()
178 |
179 | adjancency_local = _to_adjacency_local(adata_line.obsp["spatial_connectivities"], 2)
180 |
181 | assert (
182 | (adjancency_local.todense() > 0)
183 | == np.array([
184 | [True, True, True, False, False, False],
185 | [True, True, True, False, False, False],
186 | [True, True, True, False, False, False],
187 | [False, False, False, True, True, False],
188 | [False, False, False, True, True, False],
189 | [False, False, False, False, False, False], # unconnected node => no local connections with n_hop >= 2
190 | ])
191 | ).all()
192 |
193 |
194 | def test_to_adjacency_view():
195 | adjancency_view = _to_adjacency_view(adata.obsp["spatial_connectivities"], 2)
196 |
197 | assert (
198 | (adjancency_view.todense() > 0)
199 | == np.array([
200 | [False, False, True, False, False],
201 | [False, False, False, False, False],
202 | [True, False, False, False, False],
203 | [False, False, False, False, False],
204 | [False, False, False, False, False],
205 | ])
206 | ).all()
207 |
208 | adjancency_view = _to_adjacency_view(adata.obsp["spatial_connectivities"], 3)
209 |
210 | assert adjancency_view.sum() == 0
211 |
212 | adjancency_view = _to_adjacency_view(adata_line.obsp["spatial_connectivities"], 1)
213 |
214 | assert (
215 | (adjancency_view.todense() > 0)
216 | == np.array([
217 | [False, True, False, False, False, False],
218 | [True, False, True, False, False, False],
219 | [False, True, False, False, False, False],
220 | [False, False, False, False, True, False],
221 | [False, False, False, True, False, False],
222 | [False, False, False, False, False, False],
223 | ])
224 | ).all()
225 |
226 | adjancency_view = _to_adjacency_view(adata_line.obsp["spatial_connectivities"], 2)
227 |
228 | assert (
229 | (adjancency_view.todense() > 0)
230 | == np.array([
231 | [False, False, True, False, False, False],
232 | [False, False, False, False, False, False],
233 | [True, False, False, False, False, False],
234 | [False, False, False, False, False, False],
235 | [False, False, False, False, False, False],
236 | [False, False, False, False, False, False],
237 | ])
238 | ).all()
239 |
240 | adjancency_view = _to_adjacency_view(adata_line.obsp["spatial_connectivities"], 3)
241 |
242 | assert adjancency_view.sum() == 0
243 |
244 |
245 | def test_check_slide_name_key():
246 | obs = pd.DataFrame({Keys.SLIDE_ID: ["slide1", "slide2"], "name": ["sample1", "sample2"]})
247 | adata = AnnData(obs=obs)
248 |
249 | assert novae.utils.check_slide_name_key(adata, None) == Keys.SLIDE_ID
250 | novae.utils.check_slide_name_key(adata, "name")
251 |
252 | obs = pd.DataFrame({Keys.SLIDE_ID: ["slide1", "slide2", "slide2"], "name": ["sample1", "sample2", "sample3"]})
253 | adata2 = AnnData(obs=obs)
254 |
255 | assert novae.utils.check_slide_name_key(adata2, None) == Keys.SLIDE_ID
256 | with pytest.raises(AssertionError):
257 | novae.utils.check_slide_name_key(adata2, "name")
258 |
259 | del adata2.obs[Keys.SLIDE_ID]
260 | with pytest.raises(AssertionError):
261 | novae.utils.check_slide_name_key(adata2, None)
262 |
--------------------------------------------------------------------------------
 3 |
3 |  3 |
3 |  31 |
31 |  3 |
3 |  15 |
15 |