├── assets
├── intel.png
├── oatml.png
├── oxcs.png
└── turing.png
├── leaderboard
├── diabetic_retinopathy_diagnosis
│ ├── auc
│ │ ├── mfvi.csv
│ │ ├── random.csv
│ │ ├── deep_ensemble.csv
│ │ ├── deterministic.csv
│ │ ├── mc_dropout.csv
│ │ └── ensemble_mc_dropout.csv
│ ├── accuracy
│ │ ├── mfvi.csv
│ │ ├── random.csv
│ │ ├── deep_ensemble.csv
│ │ ├── deterministic.csv
│ │ ├── mc_dropout.csv
│ │ └── ensemble_mc_dropout.csv
│ └── README.md
├── aptos2019
│ ├── auc
│ │ ├── random.csv
│ │ ├── ensemble_mc_dropout.csv
│ │ ├── deterministic.csv
│ │ ├── mfvi.csv
│ │ ├── deep_ensemble.csv
│ │ └── mc_dropout.csv
│ └── accuracy
│ │ ├── deterministic.csv
│ │ ├── ensemble_mc_dropout.csv
│ │ ├── deep_ensemble.csv
│ │ ├── mc_dropout.csv
│ │ ├── random.csv
│ │ └── mfvi.csv
└── pretraining
│ ├── auc
│ ├── mc_dropout.csv
│ ├── random.csv
│ └── deterministic.csv
│ └── accuracy
│ ├── deterministic.csv
│ ├── mc_dropout.csv
│ └── random.csv
├── baselines
├── diabetic_retinopathy_diagnosis
│ ├── mfvi
│ │ ├── configs
│ │ │ ├── medium.cfg
│ │ │ └── realworld.cfg
│ │ ├── __init__.py
│ │ ├── main.py
│ │ └── model.py
│ ├── mc_dropout
│ │ ├── configs
│ │ │ ├── medium.cfg
│ │ │ └── realworld.cfg
│ │ ├── __init__.py
│ │ ├── main.py
│ │ └── model.py
│ ├── deep_ensembles
│ │ ├── __init__.py
│ │ ├── model.py
│ │ └── main.py
│ ├── deterministic
│ │ ├── __init__.py
│ │ ├── model.py
│ │ └── main.py
│ ├── ensemble_mc_dropout
│ │ ├── __init__.py
│ │ ├── model.py
│ │ └── main.py
│ └── README.md
└── __init__.py
├── bdlb
├── core
│ ├── __init__.py
│ ├── constants.py
│ ├── benchmark.py
│ ├── levels.py
│ ├── registered.py
│ ├── transforms.py
│ └── plotting.py
├── diabetic_retinopathy_diagnosis
│ ├── __init__.py
│ ├── tfds_adapter.py
│ └── benchmark.py
└── __init__.py
├── setup.py
├── .gitignore
├── README.md
└── LICENSE
/assets/intel.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/OATML/bdl-benchmarks/HEAD/assets/intel.png
--------------------------------------------------------------------------------
/assets/oatml.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/OATML/bdl-benchmarks/HEAD/assets/oatml.png
--------------------------------------------------------------------------------
/assets/oxcs.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/OATML/bdl-benchmarks/HEAD/assets/oxcs.png
--------------------------------------------------------------------------------
/assets/turing.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/OATML/bdl-benchmarks/HEAD/assets/turing.png
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/mfvi.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,86.6,0.9
3 | 0.6,85.4,1.2
4 | 0.7,84.0,1.0
5 | 0.8,83.0,0.9
6 | 0.9,81.8,0.8
7 | 1.0,82.1,1.3
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,81.8,1.2
3 | 0.6,82.1,1.1
4 | 0.7,82.0,1.3
5 | 0.8,81.8,1.1
6 | 0.9,82.1,1.0
7 | 1.0,82.0,0.9
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/mfvi.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,88.1,1.1
3 | 0.6,86.9,0.9
4 | 0.7,85.0,1.0
5 | 0.8,84.5,0.7
6 | 0.9,84.4,0.6
7 | 1.0,84.3,0.7
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,84.8,0.9
3 | 0.6,84.6,0.8
4 | 0.7,84.3,0.7
5 | 0.8,84.6,0.7
6 | 0.9,84.3,0.6
7 | 1.0,84.2,0.5
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/deep_ensemble.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,87.2,0.9
3 | 0.6,86.1,1.1
4 | 0.7,84.9,0.8
5 | 0.8,83.8,0.9
6 | 0.9,82.7,0.8
7 | 1.0,81.8,1.1
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,84.9,1.1
3 | 0.6,83.4,1.3
4 | 0.7,82.3,1.2
5 | 0.8,81.8,1.2
6 | 0.9,81.7,1.4
7 | 1.0,82.0,1.0
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,87.8,1.1
3 | 0.6,86.2,0.8
4 | 0.7,85.2,0.8
5 | 0.8,84.1,0.7
6 | 0.9,83.0,0.8
7 | 1.0,82.1,1.2
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/deep_ensemble.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,89.9,0.9
3 | 0.6,88.3,1.1
4 | 0.7,86.1,1.0
5 | 0.8,85.1,0.9
6 | 0.9,84.7,0.8
7 | 1.0,84.6,0.7
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,86.1,0.6
3 | 0.6,85.2,0.5
4 | 0.7,84.9,0.5
5 | 0.8,84.4,0.5
6 | 0.9,84.3,0.5
7 | 1.0,84.2,0.6
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,91.3,0.7
3 | 0.6,88.9,0.9
4 | 0.7,87.1,0.9
5 | 0.8,86.4,0.7
6 | 0.9,85.2,0.8
7 | 1.0,84.5,0.9
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/auc/ensemble_mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,88.1,1.2
3 | 0.6,86.7,1.0
4 | 0.7,85.4,1.0
5 | 0.8,84.4,0.6
6 | 0.9,83.2,0.8
7 | 1.0,82.5,1.1
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/accuracy/ensemble_mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,92.4,0.9
3 | 0.6,90.2,1.0
4 | 0.7,88.1,1.0
5 | 0.8,87.1,0.9
6 | 0.9,86.7,0.8
7 | 1.0,85.3,1.0
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mfvi/configs/medium.cfg:
--------------------------------------------------------------------------------
1 | --level=medium
2 | --output_dir=tmp/medium.mfvi
3 | --batch_size=64
4 | --num_epochs=50
5 | --num_base_filters=42
6 | --learning_rate=4e-4
7 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mfvi/configs/realworld.cfg:
--------------------------------------------------------------------------------
1 | --level=realworld
2 | --output_dir=tmp/realworld.mfvi
3 | --batch_size=64
4 | --num_epochs=50
5 | --num_base_filters=42
6 | --learning_rate=4e-4
7 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,74.1115282530884,1.2
3 | 0.6,73.98926352594685,1.1
4 | 0.7,74.31505347450982,1.3
5 | 0.8,73.63684036945445,1.1
6 | 0.9,73.70576881316478,1.0
7 | 1.0,74.44604600876292,0.9
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/auc/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,90.50087157323037,1.1
3 | 0.6,89.27465772369939,0.8
4 | 0.7,87.98128411710026,1.0
5 | 0.8,86.90757174461723,0.8
6 | 0.9,85.8656455334597,1.0
7 | 1.0,86.05092622000835,1.2
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/auc/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,84.69111482898622,1.2
3 | 0.6,84.80000240671389,1.1
4 | 0.7,84.74939574019838,1.3
5 | 0.8,84.64244653558231,1.1
6 | 0.9,84.86491640995733,1.0
7 | 1.0,84.75539927127676,0.9
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,77.5813901979666,1.2
3 | 0.6,76.49447682269734,1.1
4 | 0.7,77.70305205105299,1.1
5 | 0.8,76.14151490098507,1.1
6 | 0.9,76.72487320864245,1.1
7 | 1.0,76.5230841411558,1.2
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/auc/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,87.7653697027395,1.1
3 | 0.6,86.29581740991412,1.3
4 | 0.7,85.37533171580017,1.2
5 | 0.8,84.60957109993197,1.2
6 | 0.9,84.51952536111276,1.4
7 | 1.0,84.98894869973235,1.0
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/ensemble_mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,82.37751333962768,1.2
3 | 0.6,81.29736671796819,1.0
4 | 0.7,79.82695138171496,1.0
5 | 0.8,79.00445952639875,0.6
6 | 0.9,77.87282433308359,0.8
7 | 1.0,77.21169988706856,1.1
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/accuracy/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,89.81101313374596,0.6
3 | 0.6,88.20880358604539,0.5
4 | 0.7,88.67156209245978,0.5
5 | 0.8,88.3554739574559,0.5
6 | 0.9,88.26438144807636,0.5
7 | 1.0,87.48948402164251,0.6
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/accuracy/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,95.37552651460484,0.7
3 | 0.6,92.84161334475907,0.9
4 | 0.7,91.12201822951694,0.9
5 | 0.8,89.73503652410145,0.7
6 | 0.9,88.79844947088918,0.8
7 | 1.0,87.85851249188208,0.9
8 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mc_dropout/configs/medium.cfg:
--------------------------------------------------------------------------------
1 | --level=medium
2 | --output_dir=tmp/medium.mc_dropout
3 | --batch_size=64
4 | --num_epochs=50
5 | --num_base_filters=64
6 | --learning_rate=4e-4
7 | --dropout_rate=0.1
8 | --l2_reg=5e-5
9 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/ensemble_mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,87.1431711534417,1.3
3 | 0.6,84.93029656023879,1.4
4 | 0.7,82.88382528204772,1.4
5 | 0.8,81.23390412356164,1.3
6 | 0.9,80.87747459924013,1.2
7 | 1.0,79.40399028140253,1.4
8 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mc_dropout/configs/realworld.cfg:
--------------------------------------------------------------------------------
1 | --level=realworld
2 | --output_dir=tmp/realworld.mc_dropout
3 | --batch_size=64
4 | --num_epochs=50
5 | --num_base_filters=64
6 | --learning_rate=4e-4
7 | --dropout_rate=0.1
8 | --l2_reg=5e-5
9 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/deep_ensemble.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,84.33166109450175,0.7000000000000001
3 | 0.6,82.4987348870786,0.9
4 | 0.7,80.4561312734997,0.8
5 | 0.8,79.53716354380433,0.7000000000000001
6 | 0.9,79.30688328092553,0.6000000000000001
7 | 1.0,78.8957482417067,0.5
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,84.10168287156655,1.5
3 | 0.6,82.37704127549415,1.7000000000000002
4 | 0.7,79.93894164742707,1.7000000000000002
5 | 0.8,80.56345114826215,1.5
6 | 0.9,78.32015795734496,1.6
7 | 1.0,78.6132934251819,1.7000000000000002
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/deterministic.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,76.35435199554918,0.5
3 | 0.6,74.31748839691352,0.7
4 | 0.7,73.55963606499797,0.5999999999999999
5 | 0.8,71.63022802104602,0.5999999999999999
6 | 0.9,73.83468349205513,0.8
7 | 1.0,73.85692043415344,0.39999999999999997
8 |
--------------------------------------------------------------------------------
/leaderboard/pretraining/accuracy/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,87.84742483344976,0.6000000000000001
3 | 0.6,88.01581754189371,0.5
4 | 0.7,87.20753593919196,0.39999999999999997
5 | 0.8,87.61709745818298,0.39999999999999997
6 | 0.9,86.91129532409715,0.3
7 | 1.0,87.23000053924596,0.2
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/mfvi.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,77.85178917697054,0.09999999999999998
3 | 0.6,77.86896735637504,0.30000000000000004
4 | 0.7,75.55112560598127,0.0
5 | 0.8,75.2693398565609,0.09999999999999998
6 | 0.9,73.87362012532374,0.0
7 | 1.0,74.5137868570405,0.30000000000000004
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/random.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,76.91767480701789,0.3000000000000001
3 | 0.6,76.73097026546617,0.2
4 | 0.7,76.41935840750058,0.10000000000000003
5 | 0.8,75.93500461156489,0.10000000000000003
6 | 0.9,76.26363128215164,0.0
7 | 1.0,76.93718103862291,-0.09999999999999998
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/deep_ensemble.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,80.82331632570184,0.7000000000000001
3 | 0.6,80.61568433971902,0.9
4 | 0.7,78.56652659837609,0.6000000000000001
5 | 0.8,77.46110698253457,0.7000000000000001
6 | 0.9,77.00546441449661,0.6000000000000001
7 | 1.0,76.21950318328321,0.9
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/auc/mc_dropout.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,79.92363409730275,0.30000000000000004
3 | 0.6,78.50683527663989,0.0
4 | 0.7,76.86254965867806,0.19999999999999996
5 | 0.8,74.95801354030414,0.0
6 | 0.9,75.30579744841387,0.19999999999999996
7 | 1.0,74.0241101850024,0.3999999999999999
8 |
--------------------------------------------------------------------------------
/leaderboard/aptos2019/accuracy/mfvi.csv:
--------------------------------------------------------------------------------
1 | retained_data,mean,std
2 | 0.5,81.15193369214985,0.30000000000000004
3 | 0.6,80.18774373202086,0.09999999999999998
4 | 0.7,78.61797765336982,0.19999999999999996
5 | 0.8,76.99668588324471,-0.10000000000000009
6 | 0.9,77.59646926912359,-0.2
7 | 1.0,77.24944531248897,-0.10000000000000009
8 |
--------------------------------------------------------------------------------
/baselines/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/bdlb/core/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/bdlb/diabetic_retinopathy_diagnosis/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mfvi/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mc_dropout/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deep_ensembles/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deterministic/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/ensemble_mc_dropout/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
--------------------------------------------------------------------------------
/bdlb/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Global API."""
16 |
17 | from .core.registered import load
18 |
--------------------------------------------------------------------------------
/bdlb/core/constants.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Default values for some parameters of the API when no values are passed."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | import os
22 |
23 | # Root directory of the BDL Benchmarks module.
24 | BDLB_ROOT_DIR: str = os.path.abspath(
25 | os.path.join(os.path.dirname(__file__), os.path.pardir, os.path.pardir))
26 |
27 | # Directory where to store processed datasets.
28 | DATA_DIR: str = os.path.join(BDLB_ROOT_DIR, "data")
29 |
30 | # URL to hosted datasets
31 | DIABETIC_RETINOPATHY_DIAGNOSIS_URL_MEDIUM = "https://drive.google.com/uc?id=1WAvS-pQsVLxUJiClmKLnVNQkoKmRt2I_"
32 |
33 | # URL to hosted assets
34 | LEADERBOARD_DIR_URL: str = "https://drive.google.com/uc?id=1LQeAfqMQa4lot09qAuzWa3t6attmCeG-"
35 |
--------------------------------------------------------------------------------
/bdlb/core/benchmark.py:
--------------------------------------------------------------------------------
1 | # Copyright 2018 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Data structures and API for general benchmarks."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | import collections
22 |
23 | from .levels import Level
24 |
25 | # Abstract class for benchmark information.
26 | BenchmarkInfo = collections.namedtuple("BenchmarkInfo", [
27 | "description",
28 | "urls",
29 | "setup",
30 | "citation",
31 | ])
32 |
33 | # Container for train, validation and test sets.
34 | DataSplits = collections.namedtuple("DataSplits", [
35 | "train",
36 | "validation",
37 | "test",
38 | ])
39 |
40 |
41 | class Benchmark(object):
42 | """Abstract class for benchmark objects, specifying the core API."""
43 |
44 | def download_and_prepare(self):
45 | """Downloads and prepares necessary datasets for benchmark."""
46 | raise NotImplementedError()
47 |
48 | @property
49 | def info(self) -> BenchmarkInfo:
50 | """Text description of the benchmark."""
51 | raise NotImplementedError()
52 |
53 | @property
54 | def level(self) -> Level:
55 | """The downstream task level."""
56 | raise NotImplementedError()
57 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
16 | import os
17 |
18 | from setuptools import find_packages
19 | from setuptools import setup
20 |
21 | here = os.path.abspath(os.path.dirname(__file__))
22 |
23 | # Get the long description from the README file
24 | with open(os.path.join(here, "README.md"), encoding="utf-8") as f:
25 | long_description = f.read()
26 |
27 | setup(
28 | name="bdlb",
29 | version="0.0.2",
30 | description="BDL Benchmarks",
31 | long_description=long_description,
32 | long_description_content_type="text/markdown",
33 | url="https://github.com/oatml/bdl-benchmarks",
34 | author="Oxford Applied and Theoretical Machine Learning Group",
35 | author_email="oatml@googlegroups.com",
36 | license="Apache-2.0",
37 | packages=find_packages(),
38 | install_requires=[
39 | "numpy==1.18.5",
40 | "scipy==1.4.1",
41 | "pandas==1.0.4",
42 | "matplotlib==3.2.1",
43 | "seaborn==0.10.1",
44 | "scikit-learn==0.21.3",
45 | "kaggle==1.5.6",
46 | "opencv-python==4.2.0.34",
47 | "tensorflow-gpu==2.0.0-beta0",
48 | "tensorflow-probability==0.7.0",
49 | "tensorflow-datasets==1.1.0",
50 | ],
51 | classifiers=[
52 | "Development Status :: 3 - Alpha",
53 | "Intended Audience :: Researchers",
54 | "Topic :: Software Development :: Build Tools",
55 | "License :: OSI Approved :: Apache 2.0 License",
56 | "Programming Language :: Python :: 3.6",
57 | "Programming Language :: Python :: 3.7",
58 | ],
59 | )
60 |
--------------------------------------------------------------------------------
/bdlb/core/levels.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Downstream tasks levels."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | import enum
22 | from typing import Text
23 |
24 |
25 | class Level(enum.IntEnum):
26 | """Downstream task levels.
27 |
28 | TOY: Fewer examples and drastically reduced input dimensionality.
29 | This version is intended for sanity checks and debugging only.
30 | Training on a modern CPU should take five to ten minutes.
31 | MEDIUM: Fewer examples and reduced input dimensionality.
32 | This version is intended for prototyping before moving on to the
33 | real-world scale data. Training on a single modern GPU should take
34 | five or six hours.
35 | REALWORLD: The full dataset and input dimensionality. This version is
36 | intended for the evaluation of proposed techniques at a scale applicable
37 | to the real world. There are no guidelines for train time for the real-world
38 | version of the task, reflecting the fact that any improvement will translate to
39 | safer, more robust and reliable systems.
40 | """
41 |
42 | TOY = 0
43 | MEDIUM = 1
44 | REALWORLD = 2
45 |
46 | @classmethod
47 | def from_str(cls, strvalue: Text) -> "Level":
48 | """Parses a string value to ``Level``.
49 |
50 | Args:
51 | strvalue: `str`, the level in string format.
52 |
53 | Returns:
54 | The `IntEnum` ``Level`` object.
55 | """
56 | strvalue = strvalue.lower()
57 | if strvalue == "toy":
58 | return cls.TOY
59 | elif strvalue == "medium":
60 | return cls.MEDIUM
61 | elif strvalue == "realworld":
62 | return cls.REALWORLD
63 | else:
64 | raise ValueError(
65 | "Unrecognized level value '{}' provided.".format(strvalue))
66 |
--------------------------------------------------------------------------------
/leaderboard/diabetic_retinopathy_diagnosis/README.md:
--------------------------------------------------------------------------------
1 | # Diabetic Retinopathy Diagnosis
2 | The baseline results we evaluated on this benchmark are ranked below by AUC@50% data retained:
3 |
4 | | Method | AUC
(50% data retained) | Accuracy
(50% data retained) | AUC
(100% data retained) | Accuracy
(100% data retained) |
5 | | ------------------- | :-------------------------: | :-----------------------------: | :-------------------------: | :-------------------------------: |
6 | | Ensemble MC Dropout | 88.1 ± 1.2 | 92.4 ± 0.9 | 82.5 ± 1.1 | 85.3 ± 1.0 |
7 | | MC Dropout | 87.8 ± 1.1 | 91.3 ± 0.7 | 82.1 ± 0.9 | 84.5 ± 0.9 |
8 | | Deep Ensembles | 87.2 ± 0.9 | 89.9 ± 0.9 | 81.8 ± 1.1 | 84.6 ± 0.7 |
9 | | Mean-field VI | 86.6 ± 1.1 | 88.1 ± 1.1 | 82.1 ± 1.2 | 84.3 ± 0.7 |
10 | | Deterministic | 84.9 ± 1.1 | 86.1 ± 0.6 | 82.0 ± 1.0 | 84.2 ± 0.6 |
11 | | Random | 81.8 ± 1.2 | 84.8 ± 0.9 | 82.0 ± 0.9 | 84.2 ± 0.5 |
12 |
13 | These are also plotted below in an area under the receiver-operating characteristic curve (AUC) and binary accuracy for the different baselines. The methods that capture uncertainty score better when less data is retained, referring the least certain patients to expert doctors. The best scoring methods, _MC Dropout_, _mean-field variational inference_ and _Deep Ensembles_, estimate and use the predictive uncertainty. The deterministic deep model regularized by _standard dropout_ uses only aleatoric uncertainty and performs worse. Shading shows the standard error.
14 |
15 |
16 |  17 |
17 |  18 |
18 |  19 |
19 |
20 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # General
2 | .DS_Store
3 | .AppleDouble
4 | .LSOverride
5 | .vscode
6 |
7 | # Icon must end with two \r
8 | Icon
9 |
10 | # Thumbnails
11 | ._*
12 |
13 | # Files that might appear in the root of a volume
14 | .DocumentRevisions-V100
15 | .fseventsd
16 | .Spotlight-V100
17 | .TemporaryItems
18 | .Trashes
19 | .VolumeIcon.icns
20 | .com.apple.timemachine.donotpresent
21 |
22 | # Directories potentially created on remote AFP share
23 | .AppleDB
24 | .AppleDesktop
25 | Network Trash Folder
26 | Temporary Items
27 | .apdisk
28 |
29 | # Byte-compiled / optimized / DLL files
30 | __pycache__/
31 | *.py[cod]
32 | *$py.class
33 |
34 | # C extensions
35 | *.so
36 |
37 | # Distribution / packaging
38 | .Python
39 | build/
40 | develop-eggs/
41 | dist/
42 | downloads/
43 | eggs/
44 | .eggs/
45 | lib/
46 | lib64/
47 | parts/
48 | sdist/
49 | var/
50 | wheels/
51 | *.egg-info/
52 | .installed.cfg
53 | *.egg
54 | MANIFEST
55 |
56 | # PyInstaller
57 | # Usually these files are written by a python script from a template
58 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
59 | *.manifest
60 | *.spec
61 |
62 | # Installer logs
63 | pip-log.txt
64 | pip-delete-this-directory.txt
65 |
66 | # Unit test / coverage reports
67 | htmlcov/
68 | .tox/
69 | .coverage
70 | .coverage.*
71 | .cache
72 | nosetests.xml
73 | coverage.xml
74 | *.cover
75 | .hypothesis/
76 |
77 | # Translations
78 | *.mo
79 | *.pot
80 |
81 | # Django stuff:
82 | *.log
83 | .static_storage/
84 | .media/
85 | local_settings.py
86 |
87 | # Flask stuff:
88 | instance/
89 | .webassets-cache
90 |
91 | # Scrapy stuff:
92 | .scrapy
93 |
94 | # Sphinx documentation
95 | docs/_build/
96 |
97 | # PyBuilder
98 | target/
99 |
100 | # Jupyter Notebook
101 | .ipynb_checkpoints
102 |
103 | # pyenv
104 | .python-version
105 |
106 | # celery beat schedule file
107 | celerybeat-schedule
108 |
109 | # SageMath parsed files
110 | *.sage.py
111 |
112 | # Environments
113 | .env
114 | .venv
115 | env/
116 | venv/
117 | ENV/
118 | env.bak/
119 | venv.bak/
120 |
121 | # Spyder project settings
122 | .spyderproject
123 | .spyproject
124 |
125 | # Rope project settings
126 | .ropeproject
127 |
128 | # mkdocs documentation
129 | /site
130 |
131 | # mypy
132 | .mypy_cache/
133 |
134 | # temporary files
135 | tmp
136 |
137 | # direnv
138 | .envrc
139 |
140 | # Alpha release
141 | assets/*.graffle
142 | data
143 |
144 | # workstation files
145 | .style.yapf
146 | Makefile
147 | matplotlibrc
148 |
149 | # ckeckpoints
150 | baselines/*/*/checkpoints
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deterministic/model.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Uncertainty estimator for the deterministic deep model baseline."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 |
22 | def predict(x, model, num_samples, type="entropy"):
23 | """Simple sigmoid uncertainty estimator.
24 |
25 | Args:
26 | x: `numpy.ndarray`, datapoints from input space,
27 | with shape [B, H, W, 3], where B the batch size and
28 | H, W the input images height and width accordingly.
29 | model: `tensorflow.keras.Model`, a probabilistic model,
30 | which accepts input with shape [B, H, W, 3] and
31 | outputs sigmoid probability [0.0, 1.0], and also
32 | accepts boolean arguments `training=False` for
33 | disabling dropout at test time.
34 | type: (optional) `str`, type of uncertainty returns,
35 | one of {"entropy", "stddev"}.
36 |
37 | Returns:
38 | mean: `numpy.ndarray`, predictive mean, with shape [B].
39 | uncertainty: `numpy.ndarray`, ncertainty in prediction,
40 | with shape [B].
41 | """
42 | import numpy as np
43 | import scipy.stats
44 |
45 | # Get shapes of data
46 | B, _, _, _ = x.shape
47 |
48 | # Single forward pass from the deterministic model
49 | p = model(x, training=False)
50 |
51 | # Bernoulli output distribution
52 | dist = scipy.stats.bernoulli(p)
53 |
54 | # Predictive mean calculation
55 | mean = dist.mean()
56 |
57 | # Use predictive entropy for uncertainty
58 | if type == "entropy":
59 | uncertainty = dist.entropy()
60 | # Use predictive standard deviation for uncertainty
61 | elif type == "stddev":
62 | uncertainty = dist.std()
63 | else:
64 | raise ValueError(
65 | "Unrecognized type={} provided, use one of {'entropy', 'stddev'}".
66 | format(type))
67 |
68 | return mean, uncertainty
69 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deep_ensembles/model.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Uncertainty estimator for the Deep Ensemble baseline."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 |
22 | def predict(x, models, type="entropy"):
23 | """Deep Ensembles uncertainty estimator.
24 |
25 | Args:
26 | x: `numpy.ndarray`, datapoints from input space,
27 | with shape [B, H, W, 3], where B the batch size and
28 | H, W the input images height and width accordingly.
29 | models: `iterable` of `tensorflow.keras.Model`,
30 | a probabilistic model, which accepts input with
31 | shape [B, H, W, 3] and outputs sigmoid probability
32 | [0.0, 1.0], and also accepts boolean arguments
33 | `training=False` for disabling dropout at test time.
34 | type: (optional) `str`, type of uncertainty returns,
35 | one of {"entropy", "stddev"}.
36 |
37 | Returns:
38 | mean: `numpy.ndarray`, predictive mean, with shape [B].
39 | uncertainty: `numpy.ndarray`, ncertainty in prediction,
40 | with shape [B].
41 | """
42 | import numpy as np
43 | import scipy.stats
44 |
45 | # Get shapes of data
46 | B, _, _, _ = x.shape
47 |
48 | # Monte Carlo samples from different deterministic models
49 | mc_samples = np.asarray([model(x, training=False) for model in models
50 | ]).reshape(-1, B)
51 |
52 | # Bernoulli output distribution
53 | dist = scipy.stats.bernoulli(mc_samples.mean(axis=0))
54 |
55 | # Predictive mean calculation
56 | mean = dist.mean()
57 |
58 | # Use predictive entropy for uncertainty
59 | if type == "entropy":
60 | uncertainty = dist.entropy()
61 | # Use predictive standard deviation for uncertainty
62 | elif type == "stddev":
63 | uncertainty = dist.std()
64 | else:
65 | raise ValueError(
66 | "Unrecognized type={} provided, use one of {'entropy', 'stddev'}".
67 | format(type))
68 |
69 | return mean, uncertainty
70 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/ensemble_mc_dropout/model.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Uncertainty estimator for the Ensemble Monte Carlo Dropout baseline."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 |
22 | def predict(x, models, num_samples, type="entropy"):
23 | """Deep Ensembles uncertainty estimator.
24 |
25 | Args:
26 | x: `numpy.ndarray`, datapoints from input space,
27 | with shape [B, H, W, 3], where B the batch size and
28 | H, W the input images height and width accordingly.
29 | num_samples: `int`, number of Monte Carlo samples
30 | (i.e. forward passes from dropout) used for
31 | the calculation of predictive mean and uncertainty.
32 | type: (optional) `str`, type of uncertainty returns,
33 | one of {"entropy", "stddev"}.
34 | models: `iterable` of `tensorflow.keras.Model`,
35 | a probabilistic model, which accepts input with shape
36 | [B, H, W, 3] and outputs sigmoid probability [0.0, 1.0],
37 | and also accepts boolean arguments `training=True` for
38 | enabling dropout at test time.
39 |
40 | Returns:
41 | mean: `numpy.ndarray`, predictive mean, with shape [B].
42 | uncertainty: `numpy.ndarray`, ncertainty in prediction,
43 | with shape [B].
44 | """
45 | import numpy as np
46 | import scipy.stats
47 |
48 | # Get shapes of data
49 | B, _, _, _ = x.shape
50 |
51 | # Monte Carlo samples from different dropout mask at test time from different models
52 | mc_samples = np.asarray([
53 | model(x, training=True) for _ in range(num_samples) for model in models
54 | ]).reshape(-1, B)
55 |
56 | # Bernoulli output distribution
57 | dist = scipy.stats.bernoulli(mc_samples.mean(axis=0))
58 |
59 | # Predictive mean calculation
60 | mean = dist.mean()
61 |
62 | # Use predictive entropy for uncertainty
63 | if type == "entropy":
64 | uncertainty = dist.entropy()
65 | # Use predictive standard deviation for uncertainty
66 | elif type == "stddev":
67 | uncertainty = dist.std()
68 | else:
69 | raise ValueError(
70 | "Unrecognized type={} provided, use one of {'entropy', 'stddev'}".
71 | format(type))
72 |
73 | return mean, uncertainty
74 |
--------------------------------------------------------------------------------
/bdlb/core/registered.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Benchmarks registry handlers and definitions."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | from typing import Dict
22 | from typing import Optional
23 | from typing import Text
24 | from typing import Union
25 |
26 | from ..core.benchmark import Benchmark
27 | from ..core.levels import Level
28 | from ..diabetic_retinopathy_diagnosis.benchmark import \
29 | DiabeticRetinopathyDiagnosisBecnhmark
30 |
31 | # Internal registry containing
32 | _BENCHMARK_REGISTRY: Dict[Text, Benchmark] = {
33 | "diabetic_retinopathy_diagnosis": DiabeticRetinopathyDiagnosisBecnhmark
34 | }
35 |
36 |
37 | def load(
38 | benchmark: Text,
39 | level: Union[Text, Level] = "realworld",
40 | data_dir: Optional[Text] = None,
41 | download_and_prepare: bool = True,
42 | **dtask_kwargs,
43 | ) -> Benchmark:
44 | """Loads the named benchmark into a `bdlb.Benchmark`.
45 |
46 | Args:
47 | benchmark: `str`, the registerd name of `bdlb.Benchmark`.
48 | level: `bdlb.Level` or `str`, which level of the benchmark to load.
49 | data_dir: `str` (optional), directory to read/write data.
50 | Defaults to "~/.bdlb/data".
51 | download_and_prepare: (optional) `bool`, if the data is not available
52 | it downloads and preprocesses it.
53 | dtask_kwargs: key arguments for the benchmark contructor.
54 |
55 | Returns:
56 | A registered `bdlb.Benchmark` with `level` at `data_dir`.
57 |
58 | Raises:
59 | BenchmarkNotFoundError: if `name` is unrecognised.
60 | """
61 | if not benchmark in _BENCHMARK_REGISTRY:
62 | raise BenchmarkNotFoundError(benchmark)
63 | # Fetch benchmark object
64 | return _BENCHMARK_REGISTRY.get(benchmark)(
65 | level=level,
66 | data_dir=data_dir,
67 | download_and_prepare=download_and_prepare,
68 | **dtask_kwargs,
69 | )
70 |
71 |
72 | class BenchmarkNotFoundError(ValueError):
73 | """The requested `bdlb.Benchmark` was not found."""
74 |
75 | def __init__(self, name: Text):
76 | all_denchmarks_str = "\n\t- ".join([""] + list(_BENCHMARK_REGISTRY.keys()))
77 | error_str = (
78 | "Benchmark {name} not found. Available denchmarks: {benchmarks}\n",
79 | format(name=name, benchmarks=all_denchmarks_str))
80 | super(BenchmarkNotFoundError, self).__init__(error_str)
81 |
--------------------------------------------------------------------------------
/bdlb/core/transforms.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Data augmentation and transformations."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | from typing import Optional
22 | from typing import Sequence
23 | from typing import Tuple
24 |
25 | import numpy as np
26 | import tensorflow as tf
27 |
28 |
29 | class Transform(object):
30 | """Abstract transformation class."""
31 |
32 | def __call__(
33 | self,
34 | x: tf.Tensor,
35 | y: Optional[tf.Tensor] = None,
36 | ) -> Tuple[tf.Tensor, tf.Tensor]:
37 | raise NotImplementedError()
38 |
39 |
40 | class Compose(Transform):
41 | """Uber transformation, composing a list of transformations."""
42 |
43 | def __init__(self, transforms: Sequence[Transform]):
44 | """Constructs a composition of transformations.
45 |
46 | Args:
47 | transforms: `iterable`, sequence of transformations to be composed.
48 | """
49 | self.transforms = transforms
50 |
51 | def __call__(
52 | self,
53 | x: tf.Tensor,
54 | y: Optional[tf.Tensor] = None,
55 | ) -> Tuple[tf.Tensor, tf.Tensor]:
56 | """Returns a composite function of transformations.
57 |
58 | Args:
59 | x: `any`, raw data format.
60 | y: `optional`, raw data format.
61 |
62 | Returns:
63 | A composite function to be used with `tf.data.Dataset.map()`.
64 | """
65 | for f in self.transforms:
66 | x, y = f(x, y)
67 | return x, y
68 |
69 |
70 | class RandomAugment(Transform):
71 |
72 | def __init__(self, **config):
73 | """Constructs a tranformer from `config`.
74 |
75 | Args:
76 | **config: keyword arguments for
77 | `tensorflow.keras.preprocessing.image.ImageDataGenerator`
78 | """
79 | self.idg = tf.keras.preprocessing.image.ImageDataGenerator(**config)
80 |
81 | def __call__(self, x: tf.Tensor, y: tf.Tensor) -> Tuple[tf.Tensor, tf.Tensor]:
82 | """Returns a randomly augmented image and its label.
83 |
84 | Args:
85 | x: `tensorflow.Tensor`, an image, with shape [height, width, channels].
86 | y: `tensorflow.Tensor`, a target, with shape [].
87 |
88 |
89 | Returns:
90 | The processed tuple:
91 | * `x`: `tensorflow.Tensor`, the randomly augmented image,
92 | with shape [height, width, channels].
93 | * `y`: `tensorflow.Tensor`, the unchanged target, with shape [].
94 | """
95 | return tf.py_function(self._transform, inp=[x], Tout=tf.float32), y
96 |
97 | def _transform(self, x: tf.Tensor) -> tf.Tensor:
98 | """Helper function for `tensorflow.py_function`, will be removed when
99 | TensorFlow 2.0 is released."""
100 | return tf.cast(self.idg.random_transform(x.numpy()), tf.float32)
101 |
102 |
103 | class Resize(Transform):
104 |
105 | def __init__(self, target_height: int, target_width: int):
106 | """Constructs an image resizer.
107 |
108 | Args:
109 | target_height: `int`, number of pixels in height.
110 | target_width: `int`, number of pixels in width.
111 | """
112 | self.target_height = target_height
113 | self.target_width = target_width
114 |
115 | def __call__(self, x: tf.Tensor, y: tf.Tensor) -> Tuple[tf.Tensor, tf.Tensor]:
116 | """Returns a resized image."""
117 | return tf.image.resize(x, size=[self.target_height, self.target_width]), y
118 |
119 |
120 | class Normalize(Transform):
121 |
122 | def __init__(self, loc: np.ndarray, scale: np.ndarray):
123 | self.loc = loc
124 | self.scale = scale
125 |
126 | def __call__(self, x: tf.Tensor, y: tf.Tensor) -> Tuple[tf.Tensor, tf.Tensor]:
127 | return (x - self.loc) / self.scale, y
128 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deep_ensembles/main.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Script for training and evaluating Deep Ensemble baseline for Diabetic
16 | Retinopathy Diagnosis benchmark."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import functools
23 | import os
24 |
25 | import tensorflow as tf
26 | from absl import app
27 | from absl import flags
28 | from absl import logging
29 |
30 | import bdlb
31 | from baselines.diabetic_retinopathy_diagnosis.deep_ensembles.model import \
32 | predict
33 | from baselines.diabetic_retinopathy_diagnosis.mc_dropout.model import VGGDrop
34 | from bdlb.core import plotting
35 |
36 | tfk = tf.keras
37 |
38 | ##########################
39 | # Command line arguments #
40 | ##########################
41 | FLAGS = flags.FLAGS
42 | flags.DEFINE_spaceseplist(
43 | name="model_checkpoints",
44 | default=None,
45 | help="Paths to checkpoints of the models.",
46 | )
47 | flags.DEFINE_string(
48 | name="output_dir",
49 | default="/tmp",
50 | help="Path to store model, tensorboard and report outputs.",
51 | )
52 | flags.DEFINE_enum(
53 | name="level",
54 | default="medium",
55 | enum_values=["realworld", "medium"],
56 | help="Downstream task level, one of {'medium', 'realworld'}.",
57 | )
58 | flags.DEFINE_integer(
59 | name="batch_size",
60 | default=128,
61 | help="Batch size used for training.",
62 | )
63 | flags.DEFINE_integer(
64 | name="num_epochs",
65 | default=50,
66 | help="Number of epochs of training over the whole training set.",
67 | )
68 | flags.DEFINE_enum(
69 | name="uncertainty",
70 | default="entropy",
71 | enum_values=["stddev", "entropy"],
72 | help="Uncertainty type, one of those defined "
73 | "with `estimator` function.",
74 | )
75 | flags.DEFINE_integer(
76 | name="num_base_filters",
77 | default=32,
78 | help="Number of base filters in convolutional layers.",
79 | )
80 | flags.DEFINE_float(
81 | name="learning_rate",
82 | default=4e-4,
83 | help="ADAM optimizer learning rate.",
84 | )
85 | flags.DEFINE_float(
86 | name="dropout_rate",
87 | default=0.1,
88 | help="The rate of dropout, between [0.0, 1.0).",
89 | )
90 | flags.DEFINE_float(
91 | name="l2_reg",
92 | default=5e-5,
93 | help="The L2-regularization coefficient.",
94 | )
95 |
96 |
97 | def main(argv):
98 |
99 | print(argv)
100 | print(FLAGS)
101 |
102 | ##########################
103 | # Hyperparmeters & Model #
104 | ##########################
105 | input_shape = dict(medium=(256, 256, 3), realworld=(512, 512, 3))[FLAGS.level]
106 |
107 | hparams = dict(dropout_rate=FLAGS.dropout_rate,
108 | num_base_filters=FLAGS.num_base_filters,
109 | learning_rate=FLAGS.learning_rate,
110 | l2_reg=FLAGS.l2_reg,
111 | input_shape=input_shape)
112 | classifiers = list()
113 | for checkpoint in FLAGS.model_checkpoints:
114 | classifier = VGGDrop(**hparams)
115 | classifier.load_weights(checkpoint)
116 | classifier.summary()
117 | classifiers.append(classifier)
118 |
119 | #############
120 | # Load Task #
121 | #############
122 | dtask = bdlb.load(
123 | benchmark="diabetic_retinopathy_diagnosis",
124 | level=FLAGS.level,
125 | batch_size=FLAGS.batch_size,
126 | download_and_prepare=False, # do not download data from this script
127 | )
128 | _, _, ds_test = dtask.datasets
129 |
130 | ##############
131 | # Evaluation #
132 | ##############
133 | dtask.evaluate(functools.partial(predict,
134 | models=classifiers,
135 | type=FLAGS.uncertainty),
136 | dataset=ds_test,
137 | output_dir=FLAGS.output_dir)
138 |
139 |
140 | if __name__ == "__main__":
141 | app.run(main)
142 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/ensemble_mc_dropout/main.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Script for training and evaluating Ensemble Monte Carlo Dropout baseline for
16 | Diabetic Retinopathy Diagnosis benchmark."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import functools
23 | import os
24 |
25 | import tensorflow as tf
26 | from absl import app
27 | from absl import flags
28 | from absl import logging
29 |
30 | import bdlb
31 | from baselines.diabetic_retinopathy_diagnosis.ensemble_mc_dropout.model import \
32 | predict
33 | from baselines.diabetic_retinopathy_diagnosis.mc_dropout.model import VGGDrop
34 | from bdlb.core import plotting
35 |
36 | tfk = tf.keras
37 |
38 | ##########################
39 | # Command line arguments #
40 | ##########################
41 | FLAGS = flags.FLAGS
42 | flags.DEFINE_spaceseplist(
43 | name="model_checkpoints",
44 | default=None,
45 | help="Paths to checkpoints of the models.",
46 | )
47 | flags.DEFINE_string(
48 | name="output_dir",
49 | default="/tmp",
50 | help="Path to store model, tensorboard and report outputs.",
51 | )
52 | flags.DEFINE_enum(
53 | name="level",
54 | default="medium",
55 | enum_values=["realworld", "medium"],
56 | help="Downstream task level, one of {'medium', 'realworld'}.",
57 | )
58 | flags.DEFINE_integer(
59 | name="batch_size",
60 | default=128,
61 | help="Batch size used for training.",
62 | )
63 | flags.DEFINE_integer(
64 | name="num_epochs",
65 | default=50,
66 | help="Number of epochs of training over the whole training set.",
67 | )
68 | flags.DEFINE_integer(
69 | name="num_mc_samples",
70 | default=10,
71 | help="Number of Monte Carlo samples used for uncertainty estimation.",
72 | )
73 | flags.DEFINE_enum(
74 | name="uncertainty",
75 | default="entropy",
76 | enum_values=["stddev", "entropy"],
77 | help="Uncertainty type, one of those defined "
78 | "with `estimator` function.",
79 | )
80 | flags.DEFINE_integer(
81 | name="num_base_filters",
82 | default=32,
83 | help="Number of base filters in convolutional layers.",
84 | )
85 | flags.DEFINE_float(
86 | name="learning_rate",
87 | default=4e-4,
88 | help="ADAM optimizer learning rate.",
89 | )
90 | flags.DEFINE_float(
91 | name="dropout_rate",
92 | default=0.1,
93 | help="The rate of dropout, between [0.0, 1.0).",
94 | )
95 | flags.DEFINE_float(
96 | name="l2_reg",
97 | default=5e-5,
98 | help="The L2-regularization coefficient.",
99 | )
100 |
101 |
102 | def main(argv):
103 |
104 | print(argv)
105 | print(FLAGS)
106 |
107 | ##########################
108 | # Hyperparmeters & Model #
109 | ##########################
110 | input_shape = dict(medium=(256, 256, 3), realworld=(512, 512, 3))[FLAGS.level]
111 |
112 | hparams = dict(dropout_rate=FLAGS.dropout_rate,
113 | num_base_filters=FLAGS.num_base_filters,
114 | learning_rate=FLAGS.learning_rate,
115 | l2_reg=FLAGS.l2_reg,
116 | input_shape=input_shape)
117 | classifiers = list()
118 | for checkpoint in FLAGS.model_checkpoints:
119 | classifier = VGGDrop(**hparams)
120 | classifier.load_weights(checkpoint)
121 | classifier.summary()
122 | classifiers.append(classifier)
123 |
124 | #############
125 | # Load Task #
126 | #############
127 | dtask = bdlb.load(
128 | benchmark="diabetic_retinopathy_diagnosis",
129 | level=FLAGS.level,
130 | batch_size=FLAGS.batch_size,
131 | download_and_prepare=False, # do not download data from this script
132 | )
133 | _, _, ds_test = dtask.datasets
134 |
135 | ##############
136 | # Evaluation #
137 | ##############
138 | dtask.evaluate(functools.partial(predict,
139 | models=classifiers,
140 | num_samples=FLAGS.num_mc_samples,
141 | type=FLAGS.uncertainty),
142 | dataset=ds_test,
143 | output_dir=FLAGS.output_dir)
144 |
145 |
146 | if __name__ == "__main__":
147 | app.run(main)
148 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/deterministic/main.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Script for training and evaluating a deterministic baseline for Diabetic
16 | Retinopathy Diagnosis benchmark."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import functools
23 | import os
24 |
25 | import tensorflow as tf
26 | from absl import app
27 | from absl import flags
28 | from absl import logging
29 |
30 | import bdlb
31 | from baselines.diabetic_retinopathy_diagnosis.deterministic.model import \
32 | predict
33 | from baselines.diabetic_retinopathy_diagnosis.mc_dropout.model import VGGDrop
34 | from bdlb.core import plotting
35 |
36 | tfk = tf.keras
37 |
38 | ##########################
39 | # Command line arguments #
40 | ##########################
41 | FLAGS = flags.FLAGS
42 | flags.DEFINE_string(
43 | name="output_dir",
44 | default="/tmp",
45 | help="Path to store model, tensorboard and report outputs.",
46 | )
47 | flags.DEFINE_enum(
48 | name="level",
49 | default="medium",
50 | enum_values=["realworld", "medium"],
51 | help="Downstream task level, one of {'medium', 'realworld'}.",
52 | )
53 | flags.DEFINE_integer(
54 | name="batch_size",
55 | default=128,
56 | help="Batch size used for training.",
57 | )
58 | flags.DEFINE_integer(
59 | name="num_epochs",
60 | default=50,
61 | help="Number of epochs of training over the whole training set.",
62 | )
63 | flags.DEFINE_enum(
64 | name="uncertainty",
65 | default="entropy",
66 | enum_values=["stddev", "entropy"],
67 | help="Uncertainty type, one of those defined "
68 | "with `estimator` function.",
69 | )
70 | flags.DEFINE_integer(
71 | name="num_base_filters",
72 | default=32,
73 | help="Number of base filters in convolutional layers.",
74 | )
75 | flags.DEFINE_float(
76 | name="learning_rate",
77 | default=4e-4,
78 | help="ADAM optimizer learning rate.",
79 | )
80 | flags.DEFINE_float(
81 | name="dropout_rate",
82 | default=0.1,
83 | help="The rate of dropout, between [0.0, 1.0).",
84 | )
85 | flags.DEFINE_float(
86 | name="l2_reg",
87 | default=5e-5,
88 | help="The L2-regularization coefficient.",
89 | )
90 |
91 |
92 | def main(argv):
93 |
94 | print(argv)
95 | print(FLAGS)
96 |
97 | ##########################
98 | # Hyperparmeters & Model #
99 | ##########################
100 | input_shape = dict(medium=(256, 256, 3), realworld=(512, 512, 3))[FLAGS.level]
101 |
102 | hparams = dict(dropout_rate=FLAGS.dropout_rate,
103 | num_base_filters=FLAGS.num_base_filters,
104 | learning_rate=FLAGS.learning_rate,
105 | l2_reg=FLAGS.l2_reg,
106 | input_shape=input_shape)
107 | classifier = VGGDrop(**hparams)
108 | classifier.summary()
109 |
110 | #############
111 | # Load Task #
112 | #############
113 | dtask = bdlb.load(
114 | benchmark="diabetic_retinopathy_diagnosis",
115 | level=FLAGS.level,
116 | batch_size=FLAGS.batch_size,
117 | download_and_prepare=False, # do not download data from this script
118 | )
119 | ds_train, ds_validation, ds_test = dtask.datasets
120 |
121 | #################
122 | # Training Loop #
123 | #################
124 | history = classifier.fit(
125 | ds_train,
126 | epochs=FLAGS.num_epochs,
127 | validation_data=ds_validation,
128 | class_weight=dtask.class_weight(),
129 | callbacks=[
130 | tfk.callbacks.TensorBoard(
131 | log_dir=os.path.join(FLAGS.output_dir, "tensorboard"),
132 | update_freq="epoch",
133 | write_graph=True,
134 | histogram_freq=1,
135 | ),
136 | tfk.callbacks.ModelCheckpoint(
137 | filepath=os.path.join(
138 | FLAGS.output_dir,
139 | "checkpoints",
140 | "weights-{epoch}.ckpt",
141 | ),
142 | verbose=1,
143 | save_weights_only=True,
144 | )
145 | ],
146 | )
147 | plotting.tfk_history(history,
148 | output_dir=os.path.join(FLAGS.output_dir, "history"))
149 |
150 | ##############
151 | # Evaluation #
152 | ##############
153 | dtask.evaluate(functools.partial(predict,
154 | model=classifier,
155 | type=FLAGS.uncertainty),
156 | dataset=ds_test,
157 | output_dir=FLAGS.output_dir)
158 |
159 |
160 | if __name__ == "__main__":
161 | app.run(main)
162 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mfvi/main.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Script for training and evaluating Mean-Field Variational Inference baseline
16 | for Diabetic Retinopathy Diagnosis benchmark."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import functools
23 | import os
24 |
25 | import tensorflow as tf
26 | from absl import app
27 | from absl import flags
28 | from absl import logging
29 |
30 | import bdlb
31 | from bdlb.core import plotting
32 | from model import VGGFlipout
33 | from model import predict
34 |
35 | tfk = tf.keras
36 |
37 | ##########################
38 | # Command line arguments #
39 | ##########################
40 | FLAGS = flags.FLAGS
41 | flags.DEFINE_string(
42 | name="output_dir",
43 | default="/tmp",
44 | help="Path to store model, tensorboard and report outputs.",
45 | )
46 | flags.DEFINE_enum(
47 | name="level",
48 | default="medium",
49 | enum_values=["realworld", "medium"],
50 | help="Downstream task level, one of {'medium', 'realworld'}.",
51 | )
52 | flags.DEFINE_integer(
53 | name="batch_size",
54 | default=128,
55 | help="Batch size used for training.",
56 | )
57 | flags.DEFINE_integer(
58 | name="num_epochs",
59 | default=50,
60 | help="Number of epochs of training over the whole training set.",
61 | )
62 | flags.DEFINE_integer(
63 | name="num_mc_samples",
64 | default=10,
65 | help="Number of Monte Carlo samples used for uncertainty estimation.",
66 | )
67 | flags.DEFINE_enum(
68 | name="uncertainty",
69 | default="entropy",
70 | enum_values=["stddev", "entropy"],
71 | help="Uncertainty type, one of those defined "
72 | "with `estimator` function.",
73 | )
74 | flags.DEFINE_integer(
75 | name="num_base_filters",
76 | default=32,
77 | help="Number of base filters in convolutional layers.",
78 | )
79 | flags.DEFINE_float(
80 | name="learning_rate",
81 | default=4e-4,
82 | help="ADAM optimizer learning rate.",
83 | )
84 | flags.DEFINE_float(
85 | name="dropout_rate",
86 | default=0.1,
87 | help="The rate of dropout, between [0.0, 1.0).",
88 | )
89 | flags.DEFINE_float(
90 | name="l2_reg",
91 | default=5e-5,
92 | help="The L2-regularization coefficient.",
93 | )
94 |
95 |
96 | def main(argv):
97 |
98 | print(argv)
99 | print(FLAGS)
100 |
101 | ##########################
102 | # Hyperparmeters & Model #
103 | ##########################
104 | input_shape = dict(medium=(256, 256, 3), realworld=(512, 512, 3))[FLAGS.level]
105 |
106 | hparams = dict(num_base_filters=FLAGS.num_base_filters,
107 | learning_rate=FLAGS.learning_rate,
108 | input_shape=input_shape)
109 | classifier = VGGFlipout(**hparams)
110 | classifier.summary()
111 |
112 | #############
113 | # Load Task #
114 | #############
115 | dtask = bdlb.load(
116 | benchmark="diabetic_retinopathy_diagnosis",
117 | level=FLAGS.level,
118 | batch_size=FLAGS.batch_size,
119 | download_and_prepare=False, # do not download data from this script
120 | )
121 | ds_train, ds_validation, ds_test = dtask.datasets
122 |
123 | #################
124 | # Training Loop #
125 | #################
126 | history = classifier.fit(
127 | ds_train,
128 | epochs=FLAGS.num_epochs,
129 | validation_data=ds_validation,
130 | class_weight=dtask.class_weight(),
131 | callbacks=[
132 | tfk.callbacks.TensorBoard(
133 | log_dir=os.path.join(FLAGS.output_dir, "tensorboard"),
134 | update_freq="epoch",
135 | write_graph=True,
136 | histogram_freq=1,
137 | ),
138 | tfk.callbacks.ModelCheckpoint(
139 | filepath=os.path.join(
140 | FLAGS.output_dir,
141 | "checkpoints",
142 | "weights-{epoch}.ckpt",
143 | ),
144 | verbose=1,
145 | save_weights_only=True,
146 | )
147 | ],
148 | )
149 | plotting.tfk_history(history,
150 | output_dir=os.path.join(FLAGS.output_dir, "history"))
151 |
152 | ##############
153 | # Evaluation #
154 | ##############
155 | dtask.evaluate(functools.partial(predict,
156 | model=classifier,
157 | num_samples=FLAGS.num_mc_samples,
158 | type=FLAGS.uncertainty),
159 | dataset=ds_test,
160 | output_dir=FLAGS.output_dir)
161 |
162 |
163 | if __name__ == "__main__":
164 | app.run(main)
165 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mc_dropout/main.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Script for training and evaluating Monte Carlo baseline for Diabetic
16 | Retinopathy Diagnosis benchmark."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import functools
23 | import os
24 |
25 | import tensorflow as tf
26 | from absl import app
27 | from absl import flags
28 | from absl import logging
29 |
30 | import bdlb
31 | from baselines.diabetic_retinopathy_diagnosis.mc_dropout.model import VGGDrop
32 | from baselines.diabetic_retinopathy_diagnosis.mc_dropout.model import predict
33 | from bdlb.core import plotting
34 |
35 | tfk = tf.keras
36 |
37 | ##########################
38 | # Command line arguments #
39 | ##########################
40 | FLAGS = flags.FLAGS
41 | flags.DEFINE_string(
42 | name="output_dir",
43 | default="/tmp",
44 | help="Path to store model, tensorboard and report outputs.",
45 | )
46 | flags.DEFINE_enum(
47 | name="level",
48 | default="medium",
49 | enum_values=["realworld", "medium"],
50 | help="Downstream task level, one of {'medium', 'realworld'}.",
51 | )
52 | flags.DEFINE_integer(
53 | name="batch_size",
54 | default=128,
55 | help="Batch size used for training.",
56 | )
57 | flags.DEFINE_integer(
58 | name="num_epochs",
59 | default=50,
60 | help="Number of epochs of training over the whole training set.",
61 | )
62 | flags.DEFINE_integer(
63 | name="num_mc_samples",
64 | default=10,
65 | help="Number of Monte Carlo samples used for uncertainty estimation.",
66 | )
67 | flags.DEFINE_enum(
68 | name="uncertainty",
69 | default="entropy",
70 | enum_values=["stddev", "entropy"],
71 | help="Uncertainty type, one of those defined "
72 | "with `estimator` function.",
73 | )
74 | flags.DEFINE_integer(
75 | name="num_base_filters",
76 | default=32,
77 | help="Number of base filters in convolutional layers.",
78 | )
79 | flags.DEFINE_float(
80 | name="learning_rate",
81 | default=4e-4,
82 | help="ADAM optimizer learning rate.",
83 | )
84 | flags.DEFINE_float(
85 | name="dropout_rate",
86 | default=0.1,
87 | help="The rate of dropout, between [0.0, 1.0).",
88 | )
89 | flags.DEFINE_float(
90 | name="l2_reg",
91 | default=5e-5,
92 | help="The L2-regularization coefficient.",
93 | )
94 |
95 |
96 | def main(argv):
97 |
98 | print(argv)
99 | print(FLAGS)
100 |
101 | ##########################
102 | # Hyperparmeters & Model #
103 | ##########################
104 | input_shape = dict(medium=(256, 256, 3), realworld=(512, 512, 3))[FLAGS.level]
105 |
106 | hparams = dict(dropout_rate=FLAGS.dropout_rate,
107 | num_base_filters=FLAGS.num_base_filters,
108 | learning_rate=FLAGS.learning_rate,
109 | l2_reg=FLAGS.l2_reg,
110 | input_shape=input_shape)
111 | classifier = VGGDrop(**hparams)
112 | classifier.summary()
113 |
114 | #############
115 | # Load Task #
116 | #############
117 | dtask = bdlb.load(
118 | benchmark="diabetic_retinopathy_diagnosis",
119 | level=FLAGS.level,
120 | batch_size=FLAGS.batch_size,
121 | download_and_prepare=False, # do not download data from this script
122 | )
123 | ds_train, ds_validation, ds_test = dtask.datasets
124 |
125 | #################
126 | # Training Loop #
127 | #################
128 | history = classifier.fit(
129 | ds_train,
130 | epochs=FLAGS.num_epochs,
131 | validation_data=ds_validation,
132 | class_weight=dtask.class_weight(),
133 | callbacks=[
134 | tfk.callbacks.TensorBoard(
135 | log_dir=os.path.join(FLAGS.output_dir, "tensorboard"),
136 | update_freq="epoch",
137 | write_graph=True,
138 | histogram_freq=1,
139 | ),
140 | tfk.callbacks.ModelCheckpoint(
141 | filepath=os.path.join(
142 | FLAGS.output_dir,
143 | "checkpoints",
144 | "weights-{epoch}.ckpt",
145 | ),
146 | verbose=1,
147 | save_weights_only=True,
148 | )
149 | ],
150 | )
151 | plotting.tfk_history(history,
152 | output_dir=os.path.join(FLAGS.output_dir, "history"))
153 |
154 | ##############
155 | # Evaluation #
156 | ##############
157 | dtask.evaluate(functools.partial(predict,
158 | model=classifier,
159 | num_samples=FLAGS.num_mc_samples,
160 | type=FLAGS.uncertainty),
161 | dataset=ds_test,
162 | output_dir=FLAGS.output_dir)
163 |
164 |
165 | if __name__ == "__main__":
166 | app.run(main)
167 |
--------------------------------------------------------------------------------
/bdlb/core/plotting.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Helper functions for visualization."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | import os
22 | from typing import Dict
23 | from typing import Optional
24 | from typing import Text
25 |
26 | import matplotlib.pyplot as plt
27 | import pandas as pd
28 | import tensorflow as tf
29 |
30 | tfk = tf.keras
31 |
32 |
33 | def tfk_history(
34 | history: tfk.callbacks.History,
35 | output_dir: Optional[Text] = None,
36 | **ax_set_kwargs,
37 | ):
38 | """Visualization of `tensorflow.keras.callbacks.History`, similar to
39 | `TensorBoard`, in train and validation.
40 |
41 | Args:
42 | history: `tensorflow.keras.callbacks.History`, the logs of
43 | training a `tensorflow.keras.Model`.
44 | output_dir: (optional) `str`, the directory name to
45 | store the figures.
46 | """
47 | if not isinstance(history, tfk.callbacks.History):
48 | raise TypeError("`history` was expected to be of type "
49 | "`tensorflow.keras.callbacks.History`, "
50 | "but {} was provided.".format(type(history)))
51 | for metric in [k for k in history.history.keys() if not "val_" in k]:
52 | fig, ax = plt.subplots()
53 | ax.plot(history.history.get(metric), label="train")
54 | ax.plot(history.history.get("val_{}".format(metric)), label="validation")
55 | ax.set(title=metric, xlabel="epochs", **ax_set_kwargs)
56 | ax.legend()
57 | fig.tight_layout()
58 | if isinstance(output_dir, str):
59 | os.makedirs(output_dir, exist_ok=True)
60 | fig.savefig(
61 | os.path.join(output_dir, "{}.pdf".format(metric)),
62 | trasparent=True,
63 | dpi=300,
64 | format="pdf",
65 | )
66 | fig.show()
67 |
68 |
69 | def leaderboard(
70 | benchmark: Text,
71 | results: Optional[Dict[Text, pd.DataFrame]] = None,
72 | output_dir: Optional[Text] = None,
73 | leaderboard_dir: Optional[Text] = None,
74 | **ax_set_kwargs,
75 | ):
76 | """Generates a leaderboard for all metrics in `benchmark`, by appending the
77 | (optional) `results`.
78 |
79 | Args:
80 | benchmark: `str`, the registerd name of `bdlb.Benchmark`.
81 | results: (optional) `dict`, dictionary of `pandas.DataFrames`
82 | with the results from a new method to be plotted against
83 | the leaderboard.
84 | leaderboard_dir: (optional) `str`, the path to the parent
85 | directory with all the leaderboard results.
86 | output_dir: (optional) `str`, the directory name to
87 | store the figures.
88 | """
89 | from .constants import BDLB_ROOT_DIR
90 | COLORS = plt.rcParams['axes.prop_cycle'].by_key()['color']
91 | MARKERS = ["o", "D", "s", "8", "^", "*"]
92 |
93 | # The assumes path for stored baselines records
94 | leaderboard_dir = leaderboard_dir or os.path.join(BDLB_ROOT_DIR,
95 | "leaderboard")
96 | benchmark_dir = os.path.join(leaderboard_dir, benchmark)
97 | if not os.path.exists(benchmark_dir):
98 | ValueError("No leaderboard data found at {}".format(benchmark_dir))
99 |
100 | # Metrics for which values are stored
101 | metrics = [
102 | x for x in os.listdir(benchmark_dir)
103 | if os.path.isdir(os.path.join(benchmark_dir, x))
104 | ]
105 | for metric in metrics:
106 | fig, ax = plt.subplots()
107 | # Iterate over baselines
108 | baselines = [
109 | x for x in os.listdir(os.path.join(benchmark_dir, metric))
110 | if ".csv" in x
111 | ]
112 | for b, baseline in enumerate(baselines):
113 | baseline = baseline.replace(".csv", "")
114 | # Fetch results
115 | df = pd.read_csv(
116 | os.path.join(benchmark_dir, metric, "{}.csv".format(baseline)))
117 | # Parse columns
118 | retained_data = df["retained_data"]
119 | mean = df["mean"]
120 | std = df["std"]
121 | # Visualize mean with standard error
122 | ax.plot(
123 | retained_data,
124 | mean,
125 | label=baseline,
126 | color=COLORS[b % len(COLORS)],
127 | marker=MARKERS[b % len(MARKERS)],
128 | )
129 | ax.fill_between(
130 | retained_data,
131 | mean - std,
132 | mean + std,
133 | color=COLORS[b % len(COLORS)],
134 | alpha=0.25,
135 | )
136 | if results is not None:
137 | # Plot results from dictionary
138 | if metric in results:
139 | df = results[metric]
140 | baseline = df.name if hasattr(df, "name") else "new_method"
141 | # Parse columns
142 | retained_data = df["retained_data"]
143 | mean = df["mean"]
144 | std = df["std"]
145 | # Visualize mean with standard error
146 | ax.plot(
147 | retained_data,
148 | mean,
149 | label=baseline,
150 | color=COLORS[(b + 1) % len(COLORS)],
151 | marker=MARKERS[(b + 1) % len(MARKERS)],
152 | )
153 | ax.fill_between(
154 | retained_data,
155 | mean - std,
156 | mean + std,
157 | color=COLORS[(b + 1) % len(COLORS)],
158 | alpha=0.25,
159 | )
160 | ax.set(xlabel="retained data", ylabel=metric)
161 | ax.legend()
162 | fig.tight_layout()
163 | if isinstance(output_dir, str):
164 | os.makedirs(output_dir, exist_ok=True)
165 | fig.savefig(
166 | os.path.join(output_dir, "{}.pdf".format(metric)),
167 | trasparent=True,
168 | dpi=300,
169 | format="pdf",
170 | )
171 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Bayesian Deep Learning Benchmarks
2 |
3 | **This repository is no longer being updated.**
4 |
5 | Please refer to the [Diabetic Retinopathy Detection implementation in Google's 'uncertainty-baselines' repo](https://github.com/google/uncertainty-baselines/tree/master/baselines/diabetic_retinopathy_detection) for up-to-date baseline implementations.
6 |
7 | ## Overview
8 | In order to make real-world difference with **Bayesian Deep Learning** (BDL) tools, the tools must scale to real-world settings. And for that we, the research community, must be able to evaluate our inference tools (and iterate quickly) with real-world benchmark tasks. We should be able to do this without necessarily worrying about application-specific domain knowledge, like the expertise often required in medical applications for example. We require benchmarks to test for inference robustness, performance, and accuracy, in addition to cost and effort of development. These benchmarks should be at a variety of scales, ranging from toy `MNIST`-scale benchmarks for fast development cycles, to large data benchmarks which are truthful to real-world applications, capturing their constraints.
9 |
10 | Our BDL benchmarks should
11 | * provide a transparent, modular and consistent interface for the evaluation of deep probabilistic models on a variety of _downstream tasks_;
12 | * rely on expert-driven metrics of uncertainty quality (actual applications making use of BDL uncertainty in the real-world), but abstract away the expert-knowledge and eliminate the boilerplate steps necessary for running experiments on real-world datasets;
13 | * make it easy to compare the performance of new models against _well tuned baselines_, models that have been well-adopted by the machine learning community, under a fair and realistic setting (e.g., computational resources, model sizes, datasets);
14 | * provide reference implementations of baseline models (e.g., Monte Carlo Dropout Inference, Mean Field Variational Inference, Deep Ensembles), enabling rapid prototyping and easy development of new tools;
15 | * be independent of specific deep learning frameworks (e.g., not depend on `TensorFlow`, `PyTorch`, etc.), and integrate with the SciPy ecosystem (i.e., `NumPy`, `Pandas`, `Matplotlib`). Benchmarks are framework-agnostic, while baselines are framework-dependent.
16 |
17 | In this repo we strive to provide such well-needed benchmarks for the BDL community, and collect and maintain new baselines and benchmarks contributed by the community. **A colab notebook demonstrating the MNIST-like workflow of our benchmarks is [available here](notebooks/diabetic_retinopathy_diagnosis.ipynb)**.
18 |
19 | **We highly encourage you to contribute your models as new *baselines* for others to compete against, as well as contribute new *benchmarks* for others to evaluate their models on!**
20 |
21 | ## List of Benchmarks
22 |
23 | **Bayesian Deep Learning Benchmarks** (BDL Benchmarks or `bdlb` for short), is an open-source framework that aims to bridge the gap between the design of deep probabilistic machine learning models and their application to real-world problems. Our currently supported benchmarks are:
24 |
25 | - [x] [Diabetic Retinopathy Diagnosis](baselines/diabetic_retinopathy_diagnosis) (in [`alpha`](https://github.com/OATML/bdl-benchmarks/tree/alpha/), following [Leibig et al.](https://www.nature.com/articles/s41598-017-17876-z))
26 | - [x] [Deterministic](baselines/diabetic_retinopathy_diagnosis/deterministic)
27 | - [x] [Monte Carlo Dropout](baselines/diabetic_retinopathy_diagnosis/mc_dropout) (following [Gal and Ghahramani, 2015](https://arxiv.org/abs/1506.02142))
28 | - [x] [Mean-Field Variational Inference](baselines/diabetic_retinopathy_diagnosis/mfvi) (following [Peterson and Anderson, 1987](https://pdfs.semanticscholar.org/37fa/18c66b8130b9f9748d9c94472c5671fb5622.pdf), [Wen et al., 2018](https://arxiv.org/abs/1803.04386))
29 | - [x] [Deep Ensembles](baselines/diabetic_retinopathy_diagnosis/deep_ensembles) (following [Lakshminarayanan et al., 2016](https://arxiv.org/abs/1612.01474))
30 | - [x] [Ensemble MC Dropout](baselines/diabetic_retinopathy_diagnosis/deep_ensembles) (following [Smith and Gal, 2018](https://arxiv.org/abs/1803.08533))
31 |
32 | - [ ] Autonomous Vehicle's Scene Segmentation (in `pre-alpha`, following [Mukhoti et al.](https://arxiv.org/abs/1811.12709))
33 | - [ ] Galaxy Zoo (in `pre-alpha`, following [Walmsley et al.](https://arxiv.org/abs/1905.07424))
34 | - [ ] Fishyscapes (in `pre-alpha`, following [Blum et al.](https://arxiv.org/abs/1904.03215))
35 |
36 |
37 | ## Installation
38 |
39 | *BDL Benchmarks* is shipped as a PyPI package (Python3 compatible) installable as:
40 |
41 | ```
42 | pip3 install git+https://github.com/OATML/bdl-benchmarks.git
43 | ```
44 |
45 | The data downloading and preparation is benchmark-specific, and you can follow the relevant guides at `baselines//README.md` (e.g. [`baselines/diabetic_retinopathy_diagnosis/README.md`](baselines/diabetic_retinopathy_diagnosis/README.md)).
46 |
47 |
48 | ## Examples
49 |
50 | For example, the [Diabetic Retinopathy Diagnosis](baselines/diabetic_retinopathy_diagnosis) benchmark comes with several baselines, including MC Dropout, MFVI, Deep Ensembles, and more. These models are trained with images of blood vessels in the eye:
51 |
52 |
53 |  54 |
54 |
55 |
56 | The models try to predict diabetic retinopathy, and use their uncertainty for *prescreening* (sending patients the model is uncertain about to an expert for further examination). When you implement a new model, you can easily benchmark your model against existing baseline results provided in the repo, and generate plots using expert metrics (such as the AUC of retained data when referring 50% most uncertain patients to an expert):
57 |
58 |
59 |  60 |
60 |  61 |
61 |  62 |
62 |
63 |
64 | **You can even play with a [colab notebook](notebooks/diabetic_retinopathy_diagnosis.ipynb) to see the workflow of the benchmark**, and contribute your model for others to benchmark against.
65 |
66 |
67 | ## Cite as
68 |
69 | Please cite individual benchmarks when you use these, as well as the baselines you compare against. For the [Diabetic Retinopathy Diagnosis](baselines/diabetic_retinopathy_diagnosis) benchmark please see [here](baselines/diabetic_retinopathy_diagnosis#cite-as).
70 |
71 | ## Acknowledgements
72 |
73 | The repository is developed and maintained by the [Oxford Applied and Theoretical Machine Learning](http://oatml.cs.ox.ac.uk/) group, with sponsorship from:
74 |
75 |
76 |
77 |  |
78 |  |
79 |  |
80 |  |
81 |
82 |
83 |
84 | ## Contact Us
85 |
86 | Email us for questions at oatml@cs.ox.ac.uk, or submit any issues to improve the framework.
87 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mfvi/model.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Model definition of the VGGish network for Mean-Field Variational Inference
16 | baseline."""
17 |
18 | from __future__ import absolute_import
19 | from __future__ import division
20 | from __future__ import print_function
21 |
22 | import collections
23 | import functools
24 |
25 |
26 | def VGGFlipout(num_base_filters, learning_rate, input_shape):
27 | """VGG-like model with Flipout for diabetic retinopathy diagnosis.
28 |
29 | Args:
30 | num_base_filters: `int`, number of convolution filters in the
31 | first layer.
32 | learning_rate: `float`, ADAM optimizer learning rate.
33 | input_shape: `iterable`, the shape of the images in the input layer.
34 |
35 | Returns:
36 | A tensorflow.keras.Sequential VGG-like model with flipout.
37 | """
38 | import tensorflow as tf
39 | tfk = tf.keras
40 | tfkl = tfk.layers
41 | import tensorflow_probability as tfp
42 | tfpl = tfp.layers
43 | from bdlb.diabetic_retinopathy_diagnosis.benchmark import DiabeticRetinopathyDiagnosisBecnhmark
44 |
45 | # Feedforward neural network
46 | model = tfk.Sequential([
47 | tfkl.InputLayer(input_shape),
48 | # Block 1
49 | tfpl.Convolution2DFlipout(filters=num_base_filters,
50 | kernel_size=3,
51 | strides=(2, 2),
52 | padding="same"),
53 | tfkl.Activation("relu"),
54 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
55 | # Block 2
56 | tfpl.Convolution2DFlipout(filters=num_base_filters,
57 | kernel_size=3,
58 | strides=(1, 1),
59 | padding="same"),
60 | tfkl.Activation("relu"),
61 | tfpl.Convolution2DFlipout(filters=num_base_filters,
62 | kernel_size=3,
63 | strides=(1, 1),
64 | padding="same"),
65 | tfkl.Activation("relu"),
66 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
67 | # Block 3
68 | tfpl.Convolution2DFlipout(filters=num_base_filters * 2,
69 | kernel_size=3,
70 | strides=(1, 1),
71 | padding="same"),
72 | tfkl.Activation("relu"),
73 | tfpl.Convolution2DFlipout(filters=num_base_filters * 2,
74 | kernel_size=3,
75 | strides=(1, 1),
76 | padding="same"),
77 | tfkl.Activation("relu"),
78 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
79 | # Block 4
80 | tfpl.Convolution2DFlipout(filters=num_base_filters * 4,
81 | kernel_size=3,
82 | strides=(1, 1),
83 | padding="same"),
84 | tfkl.Activation("relu"),
85 | tfpl.Convolution2DFlipout(filters=num_base_filters * 4,
86 | kernel_size=3,
87 | strides=(1, 1),
88 | padding="same"),
89 | tfkl.Activation("relu"),
90 | tfpl.Convolution2DFlipout(filters=num_base_filters * 4,
91 | kernel_size=3,
92 | strides=(1, 1),
93 | padding="same"),
94 | tfkl.Activation("relu"),
95 | tfpl.Convolution2DFlipout(filters=num_base_filters * 4,
96 | kernel_size=3,
97 | strides=(1, 1),
98 | padding="same"),
99 | tfkl.Activation("relu"),
100 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
101 | # Block 5
102 | tfpl.Convolution2DFlipout(filters=num_base_filters * 8,
103 | kernel_size=3,
104 | strides=(1, 1),
105 | padding="same"),
106 | tfkl.Activation("relu"),
107 | tfpl.Convolution2DFlipout(filters=num_base_filters * 8,
108 | kernel_size=3,
109 | strides=(1, 1),
110 | padding="same"),
111 | tfkl.Activation("relu"),
112 | tfpl.Convolution2DFlipout(filters=num_base_filters * 8,
113 | kernel_size=3,
114 | strides=(1, 1),
115 | padding="same"),
116 | tfkl.Activation("relu"),
117 | tfpl.Convolution2DFlipout(filters=num_base_filters * 8,
118 | kernel_size=3,
119 | strides=(1, 1),
120 | padding="same"),
121 | tfkl.Activation("relu"),
122 | # Global poolings
123 | tfkl.Lambda(lambda x: tfk.backend.concatenate(
124 | [tfkl.GlobalAvgPool2D()
125 | (x), tfkl.GlobalMaxPool2D()(x)], axis=1)),
126 | # Fully-connected
127 | tfpl.DenseFlipout(1,),
128 | tfkl.Activation("sigmoid")
129 | ])
130 |
131 | model.compile(loss=DiabeticRetinopathyDiagnosisBecnhmark.loss(),
132 | optimizer=tfk.optimizers.Adam(learning_rate),
133 | metrics=DiabeticRetinopathyDiagnosisBecnhmark.metrics())
134 |
135 | return model
136 |
137 |
138 | def predict(x, model, num_samples, type="entropy"):
139 | """Monte Carlo dropout uncertainty estimator.
140 |
141 | Args:
142 | x: `numpy.ndarray`, datapoints from input space,
143 | with shape [B, H, W, 3], where B the batch size and
144 | H, W the input images height and width accordingly.
145 | model: `tensorflow.keras.Model`, a probabilistic model,
146 | which accepts input with shape [B, H, W, 3] and
147 | outputs sigmoid probability [0.0, 1.0].
148 | num_samples: `int`, number of Monte Carlo samples
149 | (i.e. forward passes from dropout) used for
150 | the calculation of predictive mean and uncertainty.
151 | type: (optional) `str`, type of uncertainty returns,
152 | one of {"entropy", "stddev"}.
153 |
154 | Returns:
155 | mean: `numpy.ndarray`, predictive mean, with shape [B].
156 | uncertainty: `numpy.ndarray`, ncertainty in prediction,
157 | with shape [B].
158 | """
159 | import numpy as np
160 | import scipy.stats
161 |
162 | # Get shapes of data
163 | B, _, _, _ = x.shape
164 |
165 | # Monte Carlo samples from different dropout mask at test time
166 | mc_samples = np.asarray([model(x) for _ in range(num_samples)]).reshape(-1, B)
167 |
168 | # Bernoulli output distribution
169 | dist = scipy.stats.bernoulli(mc_samples.mean(axis=0))
170 |
171 | # Predictive mean calculation
172 | mean = dist.mean()
173 |
174 | # Use predictive entropy for uncertainty
175 | if type == "entropy":
176 | uncertainty = dist.entropy()
177 | # Use predictive standard deviation for uncertainty
178 | elif type == "stddev":
179 | uncertainty = dist.std()
180 | else:
181 | raise ValueError(
182 | "Unrecognized type={} provided, use one of {'entropy', 'stddev'}".
183 | format(type))
184 |
185 | return mean, uncertainty
186 |
--------------------------------------------------------------------------------
/bdlb/diabetic_retinopathy_diagnosis/tfds_adapter.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 |
16 | import csv
17 | import io
18 | import os
19 | from typing import Sequence
20 |
21 | import numpy as np
22 | import tensorflow as tf
23 | import tensorflow_datasets as tfds
24 |
25 | os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
26 |
27 | cv2 = tfds.core.lazy_imports.cv2
28 |
29 |
30 | class DiabeticRetinopathyDiagnosisConfig(tfds.core.BuilderConfig):
31 | """BuilderConfig for DiabeticRetinopathyDiagnosis."""
32 |
33 | def __init__(
34 | self,
35 | target_height: int,
36 | target_width: int,
37 | crop: bool = False,
38 | scale: int = 500,
39 | **kwargs,
40 | ):
41 | """BuilderConfig for DiabeticRetinopathyDiagnosis.
42 |
43 | Args:
44 | target_height: `int`, number of pixels in height.
45 | target_width: `int`, number of pixels in width.
46 | scale: (optional) `int`, the radius of the neighborhood to apply
47 | Gaussian blur filtering.
48 | **kwargs: keyword arguments forward to super.
49 | """
50 | super(DiabeticRetinopathyDiagnosisConfig, self).__init__(**kwargs)
51 | self._target_height = target_height
52 | self._target_width = target_width
53 | self._scale = scale
54 |
55 | @property
56 | def target_height(self) -> int:
57 | return self._target_height
58 |
59 | @property
60 | def target_width(self) -> int:
61 | return self._target_width
62 |
63 | @property
64 | def scale(self) -> int:
65 | return self._scale
66 |
67 |
68 | class DiabeticRetinopathyDiagnosis(tfds.image.DiabeticRetinopathyDetection):
69 |
70 | BUILDER_CONFIGS: Sequence[DiabeticRetinopathyDiagnosisConfig] = [
71 | DiabeticRetinopathyDiagnosisConfig(
72 | name="medium",
73 | version="0.0.1",
74 | description="Images for Medium level.",
75 | target_height=256,
76 | target_width=256,
77 | ),
78 | DiabeticRetinopathyDiagnosisConfig(
79 | name="realworld",
80 | version="0.0.1",
81 | description="Images for RealWorld level.",
82 | target_height=512,
83 | target_width=512,
84 | ),
85 | ]
86 |
87 | def _info(self) -> tfds.core.DatasetInfo:
88 | return tfds.core.DatasetInfo(
89 | builder=self,
90 | description="A large set of high-resolution retina images taken under "
91 | "a variety of imaging conditions. "
92 | "Ehanced contrast and resized to {}x{}.".format(
93 | self.builder_config.target_height,
94 | self.builder_config.target_width),
95 | features=tfds.features.FeaturesDict({
96 | "name":
97 | tfds.features.Text(), # patient ID + eye. eg: "4_left".
98 | "image":
99 | tfds.features.Image(shape=(
100 | self.builder_config.target_height,
101 | self.builder_config.target_width,
102 | 3,
103 | )),
104 | # 0: (no DR)
105 | # 1: (with DR)
106 | "label":
107 | tfds.features.ClassLabel(num_classes=2),
108 | }),
109 | urls=["https://www.kaggle.com/c/diabetic-retinopathy-detection/data"],
110 | citation=tfds.image.diabetic_retinopathy_detection._CITATION,
111 | )
112 |
113 | def _generate_examples(self, images_dir_path, csv_path=None, csv_usage=None):
114 | """Yields Example instances from given CSV. Applies contrast enhancement as
115 | in https://github.com/btgraham/SparseConvNet/tree/kaggle_Diabetic_Retinopat
116 | hy_competition. Turns the multiclass (i.e. 5 classes) problem to binary
117 | classification according to
118 | https://www.nature.com/articles/s41598-017-17876-z.pdf.
119 |
120 | Args:
121 | images_dir_path: path to dir in which images are stored.
122 | csv_path: optional, path to csv file with two columns: name of image and
123 | label. If not provided, just scan image directory, don't set labels.
124 | csv_usage: optional, subset of examples from the csv file to use based on
125 | the "Usage" column from the csv.
126 | """
127 | if csv_path:
128 | with tf.io.gfile.GFile(csv_path) as csv_f:
129 | reader = csv.DictReader(csv_f)
130 | data = [(row["image"], int(row["level"]))
131 | for row in reader
132 | if csv_usage is None or row["Usage"] == csv_usage]
133 | else:
134 | data = [(fname[:-5], -1)
135 | for fname in tf.io.gfile.listdir(images_dir_path)
136 | if fname.endswith(".jpeg")]
137 | for name, label in data:
138 | record = {
139 | "name":
140 | name,
141 | "image":
142 | self._preprocess(
143 | tf.io.gfile.GFile("%s/%s.jpeg" % (images_dir_path, name),
144 | mode="rb"),
145 | target_height=self.builder_config.target_height,
146 | target_width=self.builder_config.target_width,
147 | ),
148 | "label":
149 | int(label > 1),
150 | }
151 |
152 | yield record
153 |
154 | @classmethod
155 | def _preprocess(
156 | cls,
157 | image_fobj,
158 | target_height: int,
159 | target_width: int,
160 | crop: bool = False,
161 | scale: int = 500,
162 | ) -> io.BytesIO:
163 | """Resize an image to have (roughly) the given number of target pixels.
164 |
165 | Args:
166 | image_fobj: File object containing the original image.
167 | target_height: `int`, number of pixels in height.
168 | target_width: `int`, number of pixels in width.
169 | crops: (optional) `bool`, if True crops the centre of the original
170 | image t the target size.
171 | scale: (optional) `int`, the radius of the neighborhood to apply
172 | Gaussian blur filtering.
173 |
174 | Returns:
175 | A file object.
176 | """
177 | # Decode image using OpenCV2.
178 | image = cv2.imdecode(np.fromstring(image_fobj.read(), dtype=np.uint8),
179 | flags=3)
180 | try:
181 | a = cls._get_radius(image, scale)
182 | b = np.zeros(a.shape)
183 | cv2.circle(img=b,
184 | center=(a.shape[1] // 2, a.shape[0] // 2),
185 | radius=int(scale * 0.9),
186 | color=(1, 1, 1),
187 | thickness=-1,
188 | lineType=8,
189 | shift=0)
190 | image = cv2.addWeighted(src1=a,
191 | alpha=4,
192 | src2=cv2.GaussianBlur(
193 | src=a, ksize=(0, 0), sigmaX=scale // 30),
194 | beta=-4,
195 | gamma=128) * b + 128 * (1 - b)
196 | except cv2.error:
197 | pass
198 | # Reshape image to target size
199 | image = cv2.resize(image, (target_height, target_width))
200 | # Encode the image with quality=72 and store it in a BytesIO object.
201 | _, buff = cv2.imencode(".jpg", image, [int(cv2.IMWRITE_JPEG_QUALITY), 72])
202 | return io.BytesIO(buff.tostring())
203 |
204 | @staticmethod
205 | def _get_radius(img: np.ndarray, scale: int) -> np.ndarray:
206 | """Returns radius of the circle to use.
207 |
208 | Args:
209 | img: `numpy.ndarray`, an image, with shape [height, width, 3].
210 | scale: `int`, the radius of the neighborhood.
211 |
212 | Returns:
213 | A resized image.
214 | """
215 | x = img[img.shape[0] // 2, ...].sum(axis=1)
216 | r = 0.5 * (x > x.mean() // 10).sum()
217 | s = scale * 1.0 / r
218 | return cv2.resize(img, (0, 0), fx=s, fy=s)
219 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/mc_dropout/model.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Model definition of the VGGish network for Monte Carlo Dropout baseline."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 |
22 | def VGGDrop(dropout_rate, num_base_filters, learning_rate, l2_reg, input_shape):
23 | """VGG-like model with dropout for diabetic retinopathy diagnosis.
24 |
25 | Args:
26 | dropout_rate: `float`, the rate of dropout, between [0.0, 1.0).
27 | num_base_filters: `int`, number of convolution filters in the
28 | first layer.
29 | learning_rate: `float`, ADAM optimizer learning rate.
30 | l2_reg: `float`, the L2-regularization coefficient.
31 | input_shape: `iterable`, the shape of the images in the input layer.
32 |
33 | Returns:
34 | A tensorflow.keras.Sequential VGG-like model with dropout.
35 | """
36 | import tensorflow as tf
37 | tfk = tf.keras
38 | tfkl = tfk.layers
39 | from bdlb.diabetic_retinopathy_diagnosis.benchmark import DiabeticRetinopathyDiagnosisBecnhmark
40 |
41 | # Feedforward neural network
42 | model = tfk.Sequential([
43 | tfkl.InputLayer(input_shape),
44 | # Block 1
45 | tfkl.Conv2D(filters=num_base_filters,
46 | kernel_size=3,
47 | strides=(2, 2),
48 | padding="same",
49 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
50 | tfkl.Activation("relu"),
51 | tfkl.Dropout(dropout_rate),
52 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
53 | # Block 2
54 | tfkl.Conv2D(filters=num_base_filters,
55 | kernel_size=3,
56 | strides=(1, 1),
57 | padding="same",
58 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
59 | tfkl.Activation("relu"),
60 | tfkl.Dropout(dropout_rate),
61 | tfkl.Conv2D(filters=num_base_filters,
62 | kernel_size=3,
63 | strides=(1, 1),
64 | padding="same",
65 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
66 | tfkl.Activation("relu"),
67 | tfkl.Dropout(dropout_rate),
68 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
69 | # Block 3
70 | tfkl.Conv2D(filters=num_base_filters * 2,
71 | kernel_size=3,
72 | strides=(1, 1),
73 | padding="same",
74 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
75 | tfkl.Activation("relu"),
76 | tfkl.Dropout(dropout_rate),
77 | tfkl.Conv2D(filters=num_base_filters * 2,
78 | kernel_size=3,
79 | strides=(1, 1),
80 | padding="same",
81 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
82 | tfkl.Activation("relu"),
83 | tfkl.Dropout(dropout_rate),
84 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
85 | # Block 4
86 | tfkl.Conv2D(filters=num_base_filters * 4,
87 | kernel_size=3,
88 | strides=(1, 1),
89 | padding="same",
90 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
91 | tfkl.Activation("relu"),
92 | tfkl.Dropout(dropout_rate),
93 | tfkl.Conv2D(filters=num_base_filters * 4,
94 | kernel_size=3,
95 | strides=(1, 1),
96 | padding="same",
97 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
98 | tfkl.Activation("relu"),
99 | tfkl.Dropout(dropout_rate),
100 | tfkl.Conv2D(filters=num_base_filters * 4,
101 | kernel_size=3,
102 | strides=(1, 1),
103 | padding="same",
104 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
105 | tfkl.Activation("relu"),
106 | tfkl.Dropout(dropout_rate),
107 | tfkl.Conv2D(filters=num_base_filters * 4,
108 | kernel_size=3,
109 | strides=(1, 1),
110 | padding="same",
111 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
112 | tfkl.Activation("relu"),
113 | tfkl.Dropout(dropout_rate),
114 | tfkl.MaxPooling2D(pool_size=3, strides=(2, 2), padding="same"),
115 | # Block 5
116 | tfkl.Conv2D(filters=num_base_filters * 8,
117 | kernel_size=3,
118 | strides=(1, 1),
119 | padding="same",
120 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
121 | tfkl.Activation("relu"),
122 | tfkl.Dropout(dropout_rate),
123 | tfkl.Conv2D(filters=num_base_filters * 8,
124 | kernel_size=3,
125 | strides=(1, 1),
126 | padding="same",
127 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
128 | tfkl.Activation("relu"),
129 | tfkl.Dropout(dropout_rate),
130 | tfkl.Conv2D(filters=num_base_filters * 8,
131 | kernel_size=3,
132 | strides=(1, 1),
133 | padding="same",
134 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
135 | tfkl.Activation("relu"),
136 | tfkl.Dropout(dropout_rate),
137 | tfkl.Conv2D(filters=num_base_filters * 8,
138 | kernel_size=3,
139 | strides=(1, 1),
140 | padding="same",
141 | kernel_regularizer=tfk.regularizers.l2(l2_reg)),
142 | tfkl.Activation("relu"),
143 | # Global poolings
144 | tfkl.Lambda(lambda x: tfk.backend.concatenate(
145 | [tfkl.GlobalAvgPool2D()
146 | (x), tfkl.GlobalMaxPool2D()(x)], axis=1)),
147 | # Fully-connected
148 | tfkl.Dense(1, kernel_regularizer=tfk.regularizers.l2(l2_reg)),
149 | tfkl.Activation("sigmoid")
150 | ])
151 |
152 | model.compile(loss=DiabeticRetinopathyDiagnosisBecnhmark.loss(),
153 | optimizer=tfk.optimizers.Adam(learning_rate),
154 | metrics=DiabeticRetinopathyDiagnosisBecnhmark.metrics())
155 |
156 | return model
157 |
158 |
159 | def predict(x, model, num_samples, type="entropy"):
160 | """Monte Carlo dropout uncertainty estimator.
161 |
162 | Args:
163 | x: `numpy.ndarray`, datapoints from input space,
164 | with shape [B, H, W, 3], where B the batch size and
165 | H, W the input images height and width accordingly.
166 | model: `tensorflow.keras.Model`, a probabilistic model,
167 | which accepts input with shape [B, H, W, 3] and
168 | outputs sigmoid probability [0.0, 1.0], and also
169 | accepts boolean arguments `training=True` for enabling
170 | dropout at test time.
171 | num_samples: `int`, number of Monte Carlo samples
172 | (i.e. forward passes from dropout) used for
173 | the calculation of predictive mean and uncertainty.
174 | type: (optional) `str`, type of uncertainty returns,
175 | one of {"entropy", "stddev"}.
176 |
177 | Returns:
178 | mean: `numpy.ndarray`, predictive mean, with shape [B].
179 | uncertainty: `numpy.ndarray`, ncertainty in prediction,
180 | with shape [B].
181 | """
182 | import numpy as np
183 | import scipy.stats
184 |
185 | # Get shapes of data
186 | B, _, _, _ = x.shape
187 |
188 | # Monte Carlo samples from different dropout mask at test time
189 | mc_samples = np.asarray([model(x, training=True) for _ in range(num_samples)
190 | ]).reshape(-1, B)
191 |
192 | # Bernoulli output distribution
193 | dist = scipy.stats.bernoulli(mc_samples.mean(axis=0))
194 |
195 | # Predictive mean calculation
196 | mean = dist.mean()
197 |
198 | # Use predictive entropy for uncertainty
199 | if type == "entropy":
200 | uncertainty = dist.entropy()
201 | # Use predictive standard deviation for uncertainty
202 | elif type == "stddev":
203 | uncertainty = dist.std()
204 | else:
205 | raise ValueError(
206 | "Unrecognized type={} provided, use one of {'entropy', 'stddev'}".
207 | format(type))
208 |
209 | return mean, uncertainty
210 |
--------------------------------------------------------------------------------
/baselines/diabetic_retinopathy_diagnosis/README.md:
--------------------------------------------------------------------------------
1 | # Diabetic Retinopathy Diagnosis
2 |
3 | Machine learning researchers often evaluate their predictions directly on the whole test set.
4 | But, in fact, in real-world settings we have additional choices available, like asking for more information when we are uncertain.
5 | Because of the importance of accurate diagnosis, it would be unreasonable _not_ to ask for further scans of the most ambiguous cases.
6 | Moreover, in this dataset, many images feature camera artefacts that distort results.
7 | In these cases, it is critically important that a model is able to tell when the information provided to it is not sufficiently reliable to classify the patient.
8 | Just like real medical professionals, any diagnostic algorithm should be able to flag cases that require more investigation by medical experts.
9 |
10 |
11 | 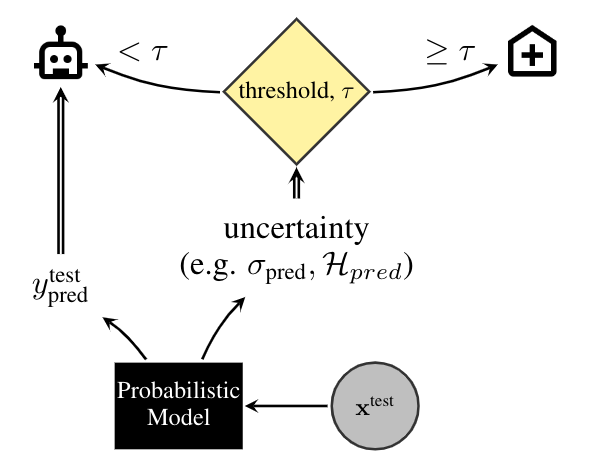 12 |
12 |
13 |
14 | This task is illustrated in the figure above, where a threshold, `τ`, is used to flag cases as certain and uncertain, with uncertain cases referred to an expert. Alternatively, the uncertainty estimates could be used to come up with a priority list, which could be matched to the available resources of a hospital, rather than waste diagnostic resources on patients for whom the diagnosis is clear cut.
15 |
16 | In order to simulate this process of referring the uncertain cases to experts and relying on the model's predictions for cases it is certain of, we assess the techniques by their diagnostic accuracy and area under receiver-operating-characteristic (ROC) curve, as a function of the
17 | referral rate. We expect the models with well-calibrated uncertainty to refer their least confident predictions to experts,
18 | improving their performance as the number of referrals increases.
19 |
20 | The accuracy of the binary classifier is defined as the ratio of correctly classified data-points over the size of the population.
21 | The receiver-operating-characteristic (ROC) curve illustrates
22 | the diagnostic ability of a binary classifier system as its discrimination threshold is varied.
23 | It is created by plotting the true positive rate (a.k.a. sensitivity) against the false positive rate (a.k.a. 1 - sensitivity).
24 | The quality of such a ROC curve can be summarized by its area under the curve (AUC), which varies between 0.5 (chance level) and 1.0 (best possible value).
25 |
26 |
27 |  28 |
28 |  29 |
29 |  30 |
30 |
31 |
32 | To get a better insight into the mechanics of these plots, below we show the relation between predictive uncertainty, e.g. entropy `H_{pred}` of MC Dropout (on y-axis), and maximum-likelihood, i.e. sigmoid probabilities `p(disease| image)` of a deterministic dropout model (on x-axis). In red are images classified incorrectly, and in green are images classified correctly. You can see that the model has higher *uncertainty* for the miss-classified images, whereas the softmax probabilities cannot distinguish red from green for low p (i.e. the plot is separable along the y-axis, but not the x-axis). Hence the uncertainty can be used as an indicator to drive referral.
33 |
34 |
35 | 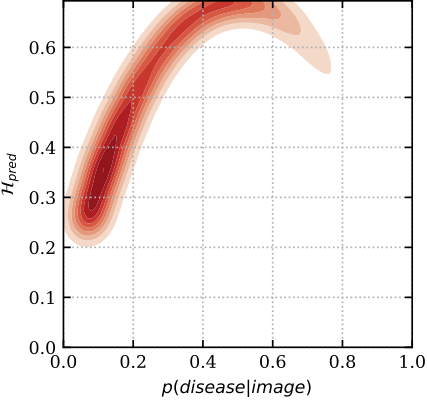 36 |
36 | 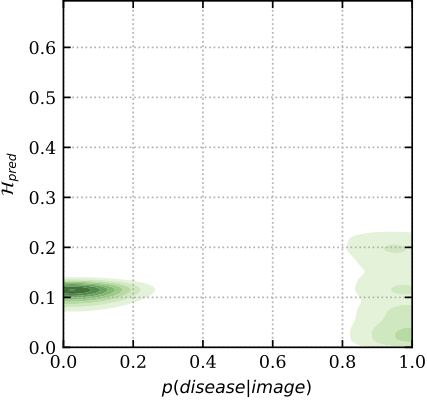 37 |
37 |
38 |
39 | ## Download and Prepare
40 |
41 | The raw data is licensed and hosted by [Kaggle](https://www.kaggle.com/c/diabetic-retinopathy-detection),
42 | hence you will need a Kaggle account to fetch it. The Kaggle Credentials can be found at
43 |
44 | ```
45 | https://www.kaggle.com//account -> "Create New API Key"
46 | ```
47 |
48 | After creating an API key you will need to accept the dataset license.
49 | Go to [the dateset page on Kaggle](https://www.kaggle.com/c/diabetic-retinopathy-detection/overview) and look
50 | for the button `I Understand and Accept` (make sure when reloading the page that the button does not pop up again).
51 |
52 | The [Kaggle command line interface](https://github.com/Kaggle/kaggle-api) is used for downloading the data, which
53 | assumes that the API token is stored at `~/.kaggle/kaggle.json`. Run the following commands to populate it:
54 |
55 | ```
56 | mkdir -p ~/.kaggle
57 | echo '{"username":"${KAGGLE_USERNAME}","key":"${KAGGLE_KEY}"}' > ~/.kaggle/kaggle.json
58 | chmod 600 ~/.kaggle/kaggle.json
59 | ```
60 |
61 | Download and prepare the data by running:
62 |
63 | ```
64 | python3 -u -c "from bdlb.diabetic_retinopathy_diagnosis.benchmark import DiabeticRetinopathyDiagnosisBecnhmark; DiabeticRetinopathyDiagnosisBecnhmark.download_and_prepare()"
65 | ```
66 |
67 | ## Run a Baseline
68 |
69 | Baseline we currently have implemented include:
70 | * [Deterministic](deterministic)
71 | * [Monte Carlo Dropout](mc_dropout) (following [Gal and Ghahramani](https://arxiv.org/abs/1506.02142))
72 | * [Mean-Field Variational Inference](mfvi) (following [Peterson and Anderson](https://pdfs.semanticscholar.org/37fa/18c66b8130b9f9748d9c94472c5671fb5622.pdf), [Wen et al., 2018](https://arxiv.org/abs/1803.04386))
73 | * [Deep Ensembles](deep_ensembles) (following [Lakshminarayanan et al.](https://arxiv.org/abs/1612.01474))
74 | * [Ensemble MC Dropout](deep_ensembles) (following [Smith and Gal](https://arxiv.org/abs/1803.08533))
75 |
76 |
77 | One executable script per baseline, `main.py`, is provided and can be used by running:
78 |
79 | ```
80 | python3 baselines/diabetic_retinopathy_diagnosis/mc_dropout/main.py \
81 | --level=medium \
82 | --dropout_rate=0.2 \
83 | --output_dir=tmp/medium.mc_dropout
84 | ```
85 |
86 | Or alternatively, use the `baselines/*/configs` for tuned hyperparameters per baseline:
87 |
88 | ```
89 | python3 baselines/diabetic_retinopathy_diagnosis/mc_dropout/main.py --flagfile=baselines/diabetic_retinopathy_diagnosis/mc_dropout/configs/medium.cfg
90 | ```
91 |
92 | ## Leaderboard
93 |
94 | The baseline results we evaluated on this benchmark are ranked below by AUC@50% data retained:
95 |
96 | | Method | AUC
(50% data retained) | Accuracy
(50% data retained) | AUC
(100% data retained) | Accuracy
(100% data retained) |
97 | | ------------------- | :-------------------------: | :-----------------------------: | :-------------------------: | :-------------------------------: |
98 | | Ensemble MC Dropout | 88.1 ± 1.2 | 92.4 ± 0.9 | 82.5 ± 1.1 | 85.3 ± 1.0 |
99 | | MC Dropout | 87.8 ± 1.1 | 91.3 ± 0.7 | 82.1 ± 0.9 | 84.5 ± 0.9 |
100 | | Deep Ensembles | 87.2 ± 0.9 | 89.9 ± 0.9 | 81.8 ± 1.1 | 84.6 ± 0.7 |
101 | | Mean-field VI | 86.6 ± 1.1 | 88.1 ± 1.1 | 82.1 ± 1.2 | 84.3 ± 0.7 |
102 | | Deterministic | 84.9 ± 1.1 | 86.1 ± 0.6 | 82.0 ± 1.0 | 84.2 ± 0.6 |
103 | | Random | 81.8 ± 1.2 | 84.8 ± 0.9 | 82.0 ± 0.9 | 84.2 ± 0.5 |
104 |
105 | ## Cite as
106 |
107 | > [**A Systematic Comparison of Bayesian Deep Learning Robustness in Diabetic Retinopathy Tasks**](https://arxiv.org/abs/1912.10481)
108 | > Angelos Filos, Sebastian Farquhar, Aidan N. Gomez, Tim G. J. Rudner, Zachary Kenton, Lewis Smith, Milad Alizadeh, Arnoud de Kroon & Yarin Gal
109 | > [Bayesian Deep Learning Workshop @ NeurIPS 2019](http://bayesiandeeplearning.org/) (BDL2019)
110 | > _arXiv 1912.10481_
111 |
112 | ```
113 | @article{filos2019systematic,
114 | title={A Systematic Comparison of Bayesian Deep Learning Robustness in Diabetic Retinopathy Tasks},
115 | author={Filos, Angelos and Farquhar, Sebastian and Gomez, Aidan N and Rudner, Tim GJ and Kenton, Zachary and Smith, Lewis and Alizadeh, Milad and de Kroon, Arnoud and Gal, Yarin},
116 | journal={arXiv preprint arXiv:1912.10481},
117 | year={2019}
118 | }
119 | ```
120 | Please cite individual baselines you compare to as well:
121 | - [Monte Carlo Dropout](mc_dropout) [[Gal and Ghahramani, 2015](https://arxiv.org/abs/1506.02142)]
122 | - [Mean-Field Variational Inference](mfvi) [[Peterson and Anderson, 1987](https://pdfs.semanticscholar.org/37fa/18c66b8130b9f9748d9c94472c5671fb5622.pdf); [Wen et al., 2018](https://arxiv.org/abs/1803.04386)]
123 | - [Deep Ensembles](deep_ensembles) [[Lakshminarayanan et al., 2016](https://arxiv.org/abs/1612.01474)]
124 | - [Ensemble MC Dropout](deep_ensembles) [[Smith and Gal, 2018](https://arxiv.org/abs/1803.08533)]
125 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | Copyright 2018, BDL Benchmark Authors. All rights reserved.
2 |
3 | Apache License
4 | Version 2.0, January 2004
5 | http://www.apache.org/licenses/
6 |
7 | TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
8 |
9 | 1. Definitions.
10 |
11 | "License" shall mean the terms and conditions for use, reproduction,
12 | and distribution as defined by Sections 1 through 9 of this document.
13 |
14 | "Licensor" shall mean the copyright owner or entity authorized by
15 | the copyright owner that is granting the License.
16 |
17 | "Legal Entity" shall mean the union of the acting entity and all
18 | other entities that control, are controlled by, or are under common
19 | control with that entity. For the purposes of this definition,
20 | "control" means (i) the power, direct or indirect, to cause the
21 | direction or management of such entity, whether by contract or
22 | otherwise, or (ii) ownership of fifty percent (50%) or more of the
23 | outstanding shares, or (iii) beneficial ownership of such entity.
24 |
25 | "You" (or "Your") shall mean an individual or Legal Entity
26 | exercising permissions granted by this License.
27 |
28 | "Source" form shall mean the preferred form for making modifications,
29 | including but not limited to software source code, documentation
30 | source, and configuration files.
31 |
32 | "Object" form shall mean any form resulting from mechanical
33 | transformation or translation of a Source form, including but
34 | not limited to compiled object code, generated documentation,
35 | and conversions to other media types.
36 |
37 | "Work" shall mean the work of authorship, whether in Source or
38 | Object form, made available under the License, as indicated by a
39 | copyright notice that is included in or attached to the work
40 | (an example is provided in the Appendix below).
41 |
42 | "Derivative Works" shall mean any work, whether in Source or Object
43 | form, that is based on (or derived from) the Work and for which the
44 | editorial revisions, annotations, elaborations, or other modifications
45 | represent, as a whole, an original work of authorship. For the purposes
46 | of this License, Derivative Works shall not include works that remain
47 | separable from, or merely link (or bind by name) to the interfaces of,
48 | the Work and Derivative Works thereof.
49 |
50 | "Contribution" shall mean any work of authorship, including
51 | the original version of the Work and any modifications or additions
52 | to that Work or Derivative Works thereof, that is intentionally
53 | submitted to Licensor for inclusion in the Work by the copyright owner
54 | or by an individual or Legal Entity authorized to submit on behalf of
55 | the copyright owner. For the purposes of this definition, "submitted"
56 | means any form of electronic, verbal, or written communication sent
57 | to the Licensor or its representatives, including but not limited to
58 | communication on electronic mailing lists, source code control systems,
59 | and issue tracking systems that are managed by, or on behalf of, the
60 | Licensor for the purpose of discussing and improving the Work, but
61 | excluding communication that is conspicuously marked or otherwise
62 | designated in writing by the copyright owner as "Not a Contribution."
63 |
64 | "Contributor" shall mean Licensor and any individual or Legal Entity
65 | on behalf of whom a Contribution has been received by Licensor and
66 | subsequently incorporated within the Work.
67 |
68 | 2. Grant of Copyright License. Subject to the terms and conditions of
69 | this License, each Contributor hereby grants to You a perpetual,
70 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
71 | copyright license to reproduce, prepare Derivative Works of,
72 | publicly display, publicly perform, sublicense, and distribute the
73 | Work and such Derivative Works in Source or Object form.
74 |
75 | 3. Grant of Patent License. Subject to the terms and conditions of
76 | this License, each Contributor hereby grants to You a perpetual,
77 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
78 | (except as stated in this section) patent license to make, have made,
79 | use, offer to sell, sell, import, and otherwise transfer the Work,
80 | where such license applies only to those patent claims licensable
81 | by such Contributor that are necessarily infringed by their
82 | Contribution(s) alone or by combination of their Contribution(s)
83 | with the Work to which such Contribution(s) was submitted. If You
84 | institute patent litigation against any entity (including a
85 | cross-claim or counterclaim in a lawsuit) alleging that the Work
86 | or a Contribution incorporated within the Work constitutes direct
87 | or contributory patent infringement, then any patent licenses
88 | granted to You under this License for that Work shall terminate
89 | as of the date such litigation is filed.
90 |
91 | 4. Redistribution. You may reproduce and distribute copies of the
92 | Work or Derivative Works thereof in any medium, with or without
93 | modifications, and in Source or Object form, provided that You
94 | meet the following conditions:
95 |
96 | (a) You must give any other recipients of the Work or
97 | Derivative Works a copy of this License; and
98 |
99 | (b) You must cause any modified files to carry prominent notices
100 | stating that You changed the files; and
101 |
102 | (c) You must retain, in the Source form of any Derivative Works
103 | that You distribute, all copyright, patent, trademark, and
104 | attribution notices from the Source form of the Work,
105 | excluding those notices that do not pertain to any part of
106 | the Derivative Works; and
107 |
108 | (d) If the Work includes a "NOTICE" text file as part of its
109 | distribution, then any Derivative Works that You distribute must
110 | include a readable copy of the attribution notices contained
111 | within such NOTICE file, excluding those notices that do not
112 | pertain to any part of the Derivative Works, in at least one
113 | of the following places: within a NOTICE text file distributed
114 | as part of the Derivative Works; within the Source form or
115 | documentation, if provided along with the Derivative Works; or,
116 | within a display generated by the Derivative Works, if and
117 | wherever such third-party notices normally appear. The contents
118 | of the NOTICE file are for informational purposes only and
119 | do not modify the License. You may add Your own attribution
120 | notices within Derivative Works that You distribute, alongside
121 | or as an addendum to the NOTICE text from the Work, provided
122 | that such additional attribution notices cannot be construed
123 | as modifying the License.
124 |
125 | You may add Your own copyright statement to Your modifications and
126 | may provide additional or different license terms and conditions
127 | for use, reproduction, or distribution of Your modifications, or
128 | for any such Derivative Works as a whole, provided Your use,
129 | reproduction, and distribution of the Work otherwise complies with
130 | the conditions stated in this License.
131 |
132 | 5. Submission of Contributions. Unless You explicitly state otherwise,
133 | any Contribution intentionally submitted for inclusion in the Work
134 | by You to the Licensor shall be under the terms and conditions of
135 | this License, without any additional terms or conditions.
136 | Notwithstanding the above, nothing herein shall supersede or modify
137 | the terms of any separate license agreement you may have executed
138 | with Licensor regarding such Contributions.
139 |
140 | 6. Trademarks. This License does not grant permission to use the trade
141 | names, trademarks, service marks, or product names of the Licensor,
142 | except as required for reasonable and customary use in describing the
143 | origin of the Work and reproducing the content of the NOTICE file.
144 |
145 | 7. Disclaimer of Warranty. Unless required by applicable law or
146 | agreed to in writing, Licensor provides the Work (and each
147 | Contributor provides its Contributions) on an "AS IS" BASIS,
148 | WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
149 | implied, including, without limitation, any warranties or conditions
150 | of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
151 | PARTICULAR PURPOSE. You are solely responsible for determining the
152 | appropriateness of using or redistributing the Work and assume any
153 | risks associated with Your exercise of permissions under this License.
154 |
155 | 8. Limitation of Liability. In no event and under no legal theory,
156 | whether in tort (including negligence), contract, or otherwise,
157 | unless required by applicable law (such as deliberate and grossly
158 | negligent acts) or agreed to in writing, shall any Contributor be
159 | liable to You for damages, including any direct, indirect, special,
160 | incidental, or consequential damages of any character arising as a
161 | result of this License or out of the use or inability to use the
162 | Work (including but not limited to damages for loss of goodwill,
163 | work stoppage, computer failure or malfunction, or any and all
164 | other commercial damages or losses), even if such Contributor
165 | has been advised of the possibility of such damages.
166 |
167 | 9. Accepting Warranty or Additional Liability. While redistributing
168 | the Work or Derivative Works thereof, You may choose to offer,
169 | and charge a fee for, acceptance of support, warranty, indemnity,
170 | or other liability obligations and/or rights consistent with this
171 | License. However, in accepting such obligations, You may act only
172 | on Your own behalf and on Your sole responsibility, not on behalf
173 | of any other Contributor, and only if You agree to indemnify,
174 | defend, and hold each Contributor harmless for any liability
175 | incurred by, or claims asserted against, such Contributor by reason
176 | of your accepting any such warranty or additional liability.
177 |
178 | END OF TERMS AND CONDITIONS
179 |
180 | APPENDIX: How to apply the Apache License to your work.
181 |
182 | To apply the Apache License to your work, attach the following
183 | boilerplate notice, with the fields enclosed by brackets "[]"
184 | replaced with your own identifying information. (Don't include
185 | the brackets!) The text should be enclosed in the appropriate
186 | comment syntax for the file format. We also recommend that a
187 | file or class name and description of purpose be included on the
188 | same "printed page" as the copyright notice for easier
189 | identification within third-party archives.
190 |
191 | Copyright 2018, Zac Kenton, Angelos Filos.
192 |
193 | Licensed under the Apache License, Version 2.0 (the "License");
194 | you may not use this file except in compliance with the License.
195 | You may obtain a copy of the License at
196 |
197 | http://www.apache.org/licenses/LICENSE-2.0
198 |
199 | Unless required by applicable law or agreed to in writing, software
200 | distributed under the License is distributed on an "AS IS" BASIS,
201 | WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
202 | See the License for the specific language governing permissions and
203 | limitations under the License.
--------------------------------------------------------------------------------
/bdlb/diabetic_retinopathy_diagnosis/benchmark.py:
--------------------------------------------------------------------------------
1 | # Copyright 2019 BDL Benchmarks Authors. All Rights Reserved.
2 | #
3 | # Licensed under the Apache License, Version 2.0 (the "License");
4 | # you may not use this file except in compliance with the License.
5 | # You may obtain a copy of the License at
6 | #
7 | # http://www.apache.org/licenses/LICENSE-2.0
8 | #
9 | # Unless required by applicable law or agreed to in writing, software
10 | # distributed under the License is distributed on an "AS IS" BASIS,
11 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
12 | # See the License for the specific language governing permissions and
13 | # limitations under the License.
14 | # ==============================================================================
15 | """Diabetic retinopathy diagnosis BDL Benchmark."""
16 |
17 | from __future__ import absolute_import
18 | from __future__ import division
19 | from __future__ import print_function
20 |
21 | import collections
22 | import os
23 | from typing import Callable
24 | from typing import Dict
25 | from typing import Optional
26 | from typing import Sequence
27 | from typing import Text
28 | from typing import Tuple
29 | from typing import Union
30 |
31 | import numpy as np
32 | import pandas as pd
33 | import tensorflow as tf
34 | from absl import logging
35 |
36 | from ..core import transforms
37 | from ..core.benchmark import Benchmark
38 | from ..core.benchmark import BenchmarkInfo
39 | from ..core.benchmark import DataSplits
40 | from ..core.constants import DATA_DIR
41 | from ..core.levels import Level
42 |
43 | tfk = tf.keras
44 |
45 | _DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR = os.path.join(
46 | DATA_DIR, "downloads", "manual", "diabetic_retinopathy_diagnosis")
47 |
48 |
49 | class DiabeticRetinopathyDiagnosisBecnhmark(Benchmark):
50 | """Diabetic retinopathy diagnosis benchmark class."""
51 |
52 | def __init__(
53 | self,
54 | level: Union[Text, Level],
55 | batch_size: int = 64,
56 | data_dir: Optional[Text] = None,
57 | download_and_prepare: bool = False,
58 | ):
59 | """Constructs a benchmark object.
60 |
61 | Args:
62 | level: `Level` or `str, downstream task level.
63 | batch_size: (optional) `int`, number of datapoints
64 | per mini-batch.
65 | data_dir: (optional) `str`, path to parent data directory.
66 | download_and_prepare: (optional) `bool`, if the data is not available
67 | it downloads and preprocesses it.
68 | """
69 | self.__level = level if isinstance(level, Level) else Level.from_str(level)
70 | try:
71 | self.__ds = self.load(level=level,
72 | batch_size=batch_size,
73 | data_dir=data_dir or DATA_DIR)
74 | except AssertionError:
75 | if not download_and_prepare:
76 | raise
77 | else:
78 | logging.info(
79 | "Data not found, `DiabeticRetinopathyDiagnosisBecnhmark.download_and_prepare()`"
80 | " is now running...")
81 | self.download_and_prepare()
82 |
83 | @classmethod
84 | def evaluate(
85 | cls,
86 | estimator: Callable[[np.ndarray], Tuple[np.ndarray, np.ndarray]],
87 | dataset: tf.data.Dataset,
88 | output_dir: Optional[Text] = None,
89 | name: Optional[Text] = None,
90 | ) -> Dict[Text, float]:
91 | """Evaluates an `estimator` on the `mode` benchmark dataset.
92 |
93 | Args:
94 | estimator: `lambda x: mu_x, uncertainty_x`, an uncertainty estimation
95 | function, which returns `mean_x` and predictive `uncertainty_x`.
96 | dataset: `tf.data.Dataset`, on which dataset to performance evaluation.
97 | output_dir: (optional) `str`, directory to save figures.
98 | name: (optional) `str`, the name of the method.
99 | """
100 | import inspect
101 | import tqdm
102 | import tensorflow_datasets as tfds
103 | from sklearn.metrics import roc_auc_score
104 | from sklearn.metrics import accuracy_score
105 | import matplotlib.pyplot as plt
106 |
107 | # Containers used for caching performance evaluation
108 | y_true = list()
109 | y_pred = list()
110 | y_uncertainty = list()
111 |
112 | # Convert to NumPy iterator if necessary
113 | ds = dataset if inspect.isgenerator(dataset) else tfds.as_numpy(dataset)
114 |
115 | for x, y in tqdm.tqdm(ds):
116 | # Sample from probabilistic model
117 | mean, uncertainty = estimator(x)
118 | # Cache predictions
119 | y_true.append(y)
120 | y_pred.append(mean)
121 | y_uncertainty.append(uncertainty)
122 |
123 | # Use vectorized NumPy containers
124 | y_true = np.concatenate(y_true).flatten()
125 | y_pred = np.concatenate(y_pred).flatten()
126 | y_uncertainty = np.concatenate(y_uncertainty).flatten()
127 | fractions = np.asarray([0.5, 0.6, 0.7, 0.8, 0.9, 1.0])
128 |
129 | # Metrics for evaluation
130 | metrics = zip(["accuracy", "auc"], cls.metrics())
131 |
132 | return {
133 | metric: cls._evaluate_metric(
134 | y_true,
135 | y_pred,
136 | y_uncertainty,
137 | fractions,
138 | lambda y_true, y_pred: metric_fn(y_true, y_pred).numpy(),
139 | name,
140 | ) for (metric, metric_fn) in metrics
141 | }
142 |
143 | @staticmethod
144 | def _evaluate_metric(
145 | y_true: np.ndarray,
146 | y_pred: np.ndarray,
147 | y_uncertainty: np.ndarray,
148 | fractions: Sequence[float],
149 | metric_fn: Callable[[np.ndarray, np.ndarray], float],
150 | name=None,
151 | ) -> pd.DataFrame:
152 | """Evaluate model predictive distribution on `metric_fn` at data retain
153 | `fractions`.
154 |
155 | Args:
156 | y_true: `numpy.ndarray`, the ground truth labels, with shape [N].
157 | y_pred: `numpy.ndarray`, the model predictions, with shape [N].
158 | y_uncertainty: `numpy.ndarray`, the model uncertainties,
159 | with shape [N].
160 | fractions: `iterable`, the percentages of data to retain for
161 | calculating `metric_fn`.
162 | metric_fn: `lambda(y_true, y_pred) -> float`, a metric
163 | function that provides a score given ground truths
164 | and predictions.
165 | name: (optional) `str`, the name of the method.
166 |
167 | Returns:
168 | A `pandas.DataFrame` with columns ["retained_data", "mean", "std"],
169 | that summarizes the scores at different data retained fractions.
170 | """
171 |
172 | N = y_true.shape[0]
173 |
174 | # Sorts indexes by ascending uncertainty
175 | I_uncertainties = np.argsort(y_uncertainty)
176 |
177 | # Score containers
178 | mean = np.empty_like(fractions)
179 | # TODO(filangel): do bootstrap sampling and estimate standard error
180 | std = np.zeros_like(fractions)
181 |
182 | for i, frac in enumerate(fractions):
183 | # Keep only the %-frac of lowest uncertainties
184 | I = np.zeros(N, dtype=bool)
185 | I[I_uncertainties[:int(N * frac)]] = True
186 | mean[i] = metric_fn(y_true[I], y_pred[I])
187 |
188 | # Store
189 | df = pd.DataFrame(dict(retained_data=fractions, mean=mean, std=std))
190 | df.name = name
191 |
192 | return df
193 |
194 | @property
195 | def datasets(self) -> tf.data.Dataset:
196 | """Pointer to the processed datasets."""
197 | return self.__ds
198 |
199 | @property
200 | def info(self) -> BenchmarkInfo:
201 | """Text description of the benchmark."""
202 | return BenchmarkInfo(description="", urls="", setup="", citation="")
203 |
204 | @property
205 | def level(self) -> Level:
206 | """The downstream task level."""
207 | return self.__level
208 |
209 | @staticmethod
210 | def loss() -> tfk.losses.Loss:
211 | """Loss used for training binary classifiers."""
212 | return tfk.losses.BinaryCrossentropy()
213 |
214 | @staticmethod
215 | def metrics() -> tfk.metrics.Metric:
216 | """Evaluation metrics used for monitoring training."""
217 | return [tfk.metrics.BinaryAccuracy(), tfk.metrics.AUC()]
218 |
219 | @staticmethod
220 | def class_weight() -> Sequence[float]:
221 | """Class weights used for rebalancing the dataset, by skewing the `loss`
222 | accordingly."""
223 | return [1.0, 4.0]
224 |
225 | @classmethod
226 | def load(
227 | cls,
228 | level: Union[Text, Level] = "realworld",
229 | batch_size: int = 64,
230 | data_dir: Optional[Text] = None,
231 | as_numpy: bool = False,
232 | ) -> DataSplits:
233 | """Loads the datasets for the benchmark.
234 |
235 | Args:
236 | level: `Level` or `str, downstream task level.
237 | batch_size: (optional) `int`, number of datapoints
238 | per mini-batch.
239 | data_dir: (optional) `str`, path to parent data directory.
240 | as_numpy: (optional) `bool`, if True returns python generators
241 | with `numpy.ndarray` outputs.
242 |
243 | Returns:
244 | A namedtuple with properties:
245 | * train: `tf.data.Dataset`, train dataset.
246 | * validation: `tf.data.Dataset`, validation dataset.
247 | * test: `tf.data.Dataset`, test dataset.
248 | """
249 | import tensorflow_datasets as tfds
250 | from .tfds_adapter import DiabeticRetinopathyDiagnosis
251 |
252 | # Fetch datasets
253 | try:
254 | ds_train, ds_validation, ds_test = DiabeticRetinopathyDiagnosis(
255 | data_dir=data_dir or DATA_DIR,
256 | config=level).as_dataset(split=["train", "validation", "test"],
257 | shuffle_files=True,
258 | batch_size=batch_size)
259 | except AssertionError as ae:
260 | raise AssertionError(
261 | str(ae) +
262 | " Run DiabeticRetinopathyDiagnosisBecnhmark.download_and_prepare()"
263 | " first and then retry.")
264 |
265 | # Parse task level
266 | level = level if isinstance(level, Level) else Level.from_str(level)
267 | # Dataset tranformations
268 | transforms_train, transforms_eval = cls._preprocessors()
269 | # Apply transformations
270 | ds_train = ds_train.map(transforms_train,
271 | num_parallel_calls=tf.data.experimental.AUTOTUNE)
272 | ds_validation = ds_validation.map(
273 | transforms_eval, num_parallel_calls=tf.data.experimental.AUTOTUNE)
274 | ds_test = ds_test.map(transforms_eval,
275 | num_parallel_calls=tf.data.experimental.AUTOTUNE)
276 |
277 | # Prefetches datasets to memory
278 | ds_train = ds_train.prefetch(tf.data.experimental.AUTOTUNE)
279 | ds_validation = ds_validation.prefetch(tf.data.experimental.AUTOTUNE)
280 | ds_test = ds_test.prefetch(tf.data.experimental.AUTOTUNE)
281 |
282 | if as_numpy:
283 | # Convert to NumPy iterators
284 | ds_train = tfds.as_numpy(ds_train)
285 | ds_validation = tfds.as_numpy(ds_validation)
286 | ds_test = tfds.as_numpy(ds_test)
287 |
288 | return DataSplits(ds_train, ds_validation, ds_test)
289 |
290 | @classmethod
291 | def download_and_prepare(cls, levels=None) -> None:
292 | """Downloads dataset from Kaggle, extracts zip files and processes it using
293 | `tensorflow_datasets`.
294 |
295 | Args:
296 | levels: (optional) `iterable` of `str`, specifies which
297 | levels from {'medium', 'realworld'} to prepare,
298 | if None it prepares all the levels.
299 |
300 | Raises:
301 | OSError: if `~/.kaggle/kaggle.json` is not set up.
302 | """
303 | # Disable GPU for data download, extraction and preparation
304 | import os
305 | os.environ["CUDA_VISIBLE_DEVICES"] = "-1"
306 | cls._download()
307 | # cls._extract()
308 | #cls._prepare(levels)
309 |
310 | @staticmethod
311 | def _download() -> None:
312 | """Downloads data from Kaggle using `tensorflow_datasets`.
313 |
314 | Raises:
315 | OSError: if `~/.kaggle/kaggle.json` is not set up.
316 | """
317 | import subprocess as sp
318 | import tensorflow_datasets as tfds
319 |
320 | # Append `/home/$USER/.local/bin` to path
321 | os.environ["PATH"] += ":/home/{}/.local/bin/".format(os.environ["USER"])
322 |
323 | # Download all files from Kaggle
324 | drd = tfds.download.kaggle.KaggleCompetitionDownloader(
325 | "diabetic-retinopathy-detection")
326 | try:
327 | for dfile in drd.competition_files:
328 | drd.download_file(dfile,
329 | output_dir=_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR)
330 | except sp.CalledProcessError as cpe:
331 | raise OSError(
332 | str(cpe) + "." +
333 | " Make sure you have ~/.kaggle/kaggle.json setup, fetched from the Kaggle website"
334 | " https://www.kaggle.com//account -> 'Create New API Key'."
335 | " Also accept the dataset license by going to"
336 | " https://www.kaggle.com/c/diabetic-retinopathy-detection/rules"
337 | " and look for the button 'I Understand and Accept' (make sure when reloading the"
338 | " page that the button does not pop up again).")
339 |
340 | @staticmethod
341 | def _extract() -> None:
342 | """Extracts zip files downloaded from Kaggle."""
343 | import glob
344 | import tqdm
345 | import zipfile
346 | import tempfile
347 |
348 | # Extract train and test original images
349 | for split in ["train", "test"]:
350 | # Extract ".zip.00*"" files to ""
351 | with tempfile.NamedTemporaryFile() as tmp:
352 | # Concatenate ".zip.00*" to ".zip"
353 | for fname in tqdm.tqdm(
354 | sorted(
355 | glob.glob(
356 | os.path.join(_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR,
357 | "{split}.zip.00*".format(split=split))))):
358 | # Unzip ".zip" to ""
359 | with open(fname, "rb") as ztmp:
360 | tmp.write(ztmp.read())
361 | with zipfile.ZipFile(tmp) as zfile:
362 | for image in tqdm.tqdm(iterable=zfile.namelist(),
363 | total=len(zfile.namelist())):
364 | zfile.extract(member=image,
365 | path=_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR)
366 | # Delete ".zip.00*" files
367 | for splitzip in os.listdir(_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR):
368 | if "{split}.zip.00".format(split=split) in splitzip:
369 | os.remove(
370 | os.path.join(_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR, splitzip))
371 |
372 | # Extract "sample.zip", "trainLabels.csv.zip"
373 | for fname in ["sample", "trainLabels.csv"]:
374 | zfname = os.path.join(_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR,
375 | "{fname}.zip".format(fname=fname))
376 | with zipfile.ZipFile(zfname) as zfile:
377 | zfile.extractall(_DIABETIC_RETINOPATHY_DIAGNOSIS_DATA_DIR)
378 | os.remove(zfname)
379 |
380 | @staticmethod
381 | def _prepare(levels=None) -> None:
382 | """Generates the TFRecord objects for medium and realworld experiments."""
383 | import multiprocessing
384 | from absl import logging
385 | from .tfds_adapter import DiabeticRetinopathyDiagnosis
386 | # Hangle each level individually
387 | for level in levels or ["medium", "realworld"]:
388 | dtask = DiabeticRetinopathyDiagnosis(data_dir=DATA_DIR, config=level)
389 | logging.debug("=== Preparing TFRecords for {} ===".format(level))
390 | dtask.download_and_prepare()
391 |
392 | @classmethod
393 | def _preprocessors(cls) -> Tuple[transforms.Transform, transforms.Transform]:
394 | """Applies transformations to the raw data."""
395 | import tensorflow_datasets as tfds
396 |
397 | # Transformation hyperparameters
398 | mean = np.asarray([0.42606387, 0.29752496, 0.21309826])
399 | stddev = np.asarray([0.27662534, 0.20280295, 0.1687619])
400 |
401 | class Parse(transforms.Transform):
402 | """Parses datapoints from raw `tf.data.Dataset`."""
403 |
404 | def __call__(self, x, y=None):
405 | """Returns `as_supervised` tuple."""
406 | return x["image"], x["label"]
407 |
408 | class CastX(transforms.Transform):
409 | """Casts image to `dtype`."""
410 |
411 | def __init__(self, dtype):
412 | """Constructs a type caster."""
413 | self.dtype = dtype
414 |
415 | def __call__(self, x, y):
416 | """Returns casted image (to `dtype`) and its (unchanged) label as
417 | tuple."""
418 | return tf.cast(x, self.dtype), y
419 |
420 | class To01X(transforms.Transform):
421 | """Rescales image to [min, max]=[0, 1]."""
422 |
423 | def __call__(self, x, y):
424 | """Returns rescaled image and its (unchanged) label as tuple."""
425 | return x / 255.0, y
426 |
427 | # Get augmentation schemes
428 | [augmentation_config,
429 | no_augmentation_config] = cls._ImageDataGenerator_config()
430 |
431 | # Transformations for train dataset
432 | transforms_train = transforms.Compose([
433 | Parse(),
434 | CastX(tf.float32),
435 | To01X(),
436 | transforms.Normalize(mean, stddev),
437 | # TODO(filangel): hangle batch with ImageDataGenerator
438 | # transforms.RandomAugment(**augmentation_config),
439 | ])
440 |
441 | # Transformations for validation/test dataset
442 | transforms_eval = transforms.Compose([
443 | Parse(),
444 | CastX(tf.float32),
445 | To01X(),
446 | transforms.Normalize(mean, stddev),
447 | # TODO(filangel): hangle batch with ImageDataGenerator
448 | # transforms.RandomAugment(**no_augmentation_config),
449 | ])
450 |
451 | return transforms_train, transforms_eval
452 |
453 | @staticmethod
454 | def _ImageDataGenerator_config():
455 | """Returns the configs for the
456 | `tensorflow.keras.preprocessing.image.ImageDataGenerator`, used for the
457 | random augmentation of the dataset, following the implementation of
458 | https://github.com/chleibig/disease-detection/blob/f3401b26aa9b832ff77afe93
459 | e3faa342f7d088e5/scripts/inspect_data_augmentation.py."""
460 | augmentation_config = dict(
461 | featurewise_center=False,
462 | samplewise_center=False,
463 | featurewise_std_normalization=False,
464 | samplewise_std_normalization=False,
465 | zca_whitening=False,
466 | rotation_range=180.0,
467 | width_shift_range=0.05,
468 | height_shift_range=0.05,
469 | shear_range=0.,
470 | zoom_range=0.10,
471 | channel_shift_range=0.,
472 | fill_mode="constant",
473 | cval=0.,
474 | horizontal_flip=True,
475 | vertical_flip=True,
476 | data_format="channels_last",
477 | )
478 | no_augmentation_config = dict(
479 | featurewise_center=False,
480 | samplewise_center=False,
481 | featurewise_std_normalization=False,
482 | samplewise_std_normalization=False,
483 | zca_whitening=False,
484 | rotation_range=0.0,
485 | width_shift_range=0.0,
486 | height_shift_range=0.0,
487 | shear_range=0.,
488 | zoom_range=0.0,
489 | channel_shift_range=0.,
490 | fill_mode="nearest",
491 | cval=0.,
492 | horizontal_flip=False,
493 | vertical_flip=False,
494 | data_format="channels_last",
495 | )
496 | return augmentation_config, no_augmentation_config
497 |
--------------------------------------------------------------------------------
 17 |
17 |  18 |
18 |  19 |
19 |  17 |
17 |  18 |
18 |  19 |
19 |  54 |
54 |  60 |
60 |  61 |
61 |  62 |
62 | 



 12 |
12 |  28 |
28 |  29 |
29 |  30 |
30 |  36 |
36 |  37 |
37 |