├── .github
├── ISSUE_TEMPLATE
│ ├── bug_report.md
│ └── feature_request.md
├── pull_request_template.md
└── workflows
│ └── premerge-py-min.yml
├── .gitignore
├── .pre-commit-config.yaml
├── LICENSE.txt
├── README.md
├── vista2d
├── README.md
├── configs
│ ├── hyper_parameters.yaml
│ ├── inference.json
│ └── metadata.json
├── datalists
│ ├── cellpose_datalist.json
│ ├── deepbacs_datalist.json
│ ├── group1_datalist.yaml
│ ├── kg_nuclei_datalist.json
│ ├── livecell_A172_datalist.json
│ ├── livecell_BT474_datalist.json
│ ├── livecell_BV2_datalist.json
│ ├── livecell_Huh7_datalist.json
│ ├── livecell_MCF7_datalist.json
│ ├── livecell_SHSY5Y_datalist.json
│ ├── livecell_SKOV3_datalist.json
│ ├── livecell_SkBr3_datalist.json
│ ├── nips_data_list.json
│ ├── omnipose_bact_f_dataset.json
│ ├── omnipose_bact_p_dataset.json
│ ├── omnipose_worm_dataset.json
│ ├── tissuenet_breast_datalist.json
│ ├── tissuenet_breast_imc_datalist.json
│ ├── tissuenet_breast_mibi_datalist.json
│ ├── tissuenet_breast_vectra_datalist.json
│ ├── tissuenet_gi_codex_datalist.json
│ ├── tissuenet_gi_datalist.json
│ ├── tissuenet_gi_mibi_datalist.json
│ ├── tissuenet_gi_mxif_datalist.json
│ ├── tissuenet_immune_cycif_datalist.json
│ ├── tissuenet_immune_datalist.json
│ ├── tissuenet_immune_mibi_datalist.json
│ ├── tissuenet_immune_vectra_datalist.json
│ ├── tissuenet_lung_cycif_datalist.json
│ ├── tissuenet_pancreas_codex_datalist.json
│ ├── tissuenet_pancreas_datalist.json
│ ├── tissuenet_pancreas_vectra_datalist.json
│ ├── tissuenet_skin_mibi_datalist.json
│ ├── yeaz_bf_dataset_list.json
│ ├── yeaz_dataset_list.json
│ └── yeaz_phc_dataset_list.json
├── docs
│ └── data_license.txt
├── download_preprocessor
│ ├── all_file_downloader.py
│ ├── cellpose_agreement.png
│ ├── cellpose_links.png
│ ├── data_tree.png
│ ├── generate_json.py
│ ├── kaggle_download.png
│ ├── omnipose_download.png
│ ├── process_data.py
│ ├── readme.md
│ ├── tissuenet_download.png
│ ├── tissuenet_login.png
│ └── urls.txt
├── large_files.yml
├── license.code
├── license.weights
├── scripts
│ ├── __init__.py
│ ├── cell_distributed_weighted_sampler.py
│ ├── cell_sam_wrapper.py
│ ├── components.py
│ ├── utils.py
│ └── workflow.py
├── setup.cfg
└── unit_tests
│ ├── test_vista2d.py
│ ├── test_vista2d_mgpu.py
│ └── utils.py
└── vista3d
├── LICENSE
├── NVIDIA OneWay Noncommercial License.txt
├── README.md
├── assets
└── imgs
│ ├── demo_gif.gif
│ ├── everything.gif
│ ├── finetune.png
│ ├── liver.gif
│ ├── model.png
│ ├── montage.png
│ ├── sam2.png
│ ├── scores.png
│ ├── unspecified.gif
│ ├── wholeBody.png
│ └── zeroshot.gif
├── configs
├── finetune
│ ├── infer_patch_auto_murine.yaml
│ ├── infer_patch_auto_word.yaml
│ ├── train_finetune_murine.yaml
│ └── train_finetune_word.yaml
├── infer.yaml
├── supported_eval
│ ├── infer_patch_auto.yaml
│ ├── infer_patch_autopoint.yaml
│ ├── infer_patch_point.yaml
│ └── infer_sam2_point.yaml
├── train
│ ├── hyper_parameters_stage1.yaml
│ ├── hyper_parameters_stage2.yaml
│ ├── hyper_parameters_stage3.yaml
│ └── hyper_parameters_stage4.yaml
└── zeroshot_eval
│ ├── infer_iter_point_adrenal.yaml
│ ├── infer_iter_point_hcc.yaml
│ ├── infer_iter_point_kits.yaml
│ └── infer_iter_point_murine.yaml
├── cvpr_workshop
├── Dockerfile
├── README.md
├── infer_cvpr.py
├── predict.sh
├── requirements.txt
├── train_cvpr.py
└── update_ckpt.py
├── data
├── README.md
├── __init__.py
├── dataset_weights.yaml
├── datasets.py
├── external
│ ├── Adrenal_Ki67_5_folds.json
│ ├── C4KC-KiTS_5_folds.json
│ ├── HCC-TACE-Seg_5_folds.json
│ ├── WORD.json
│ └── micro-ct-murine-native_5_folds.json
├── jsons
│ ├── AMOS22_5_folds.json
│ ├── AbdomenCT-1K_5_folds.json
│ ├── AeroPath_5_folds.json
│ ├── BTCV-Abdomen_5_folds.json
│ ├── BTCV-Cervix_5_folds.json
│ ├── CRLM-CT_5_folds.json
│ ├── CT-ORG_5_folds.json
│ ├── CTPelvic1K-CLINIC_5_folds.json
│ ├── Covid19_5_folds.json
│ ├── FLARE22_5_folds.json
│ ├── LIDC_5_folds.json
│ ├── Multi-organ-Abdominal-CT-btcv_5_folds.json
│ ├── Multi-organ-Abdominal-CT-tcia_5_folds.json
│ ├── NLST_5_folds.json
│ ├── Pancreas-CT_5_folds.json
│ ├── StonyBrook-CT_5_folds.json
│ ├── TCIA_Colon_5_folds.json
│ ├── Task03_5_folds.json
│ ├── Task06_5_folds.json
│ ├── Task07_5_folds.json
│ ├── Task08_5_folds.json
│ ├── Task09_5_folds.json
│ ├── Task10_5_folds.json
│ ├── TotalSegmentatorV2_5_folds.json
│ ├── VerSe_5_folds.json
│ ├── label_dict.json
│ └── label_mappings.json
└── make_datalists.py
├── requirements.txt
├── scripts
├── __init__.py
├── debugger.py
├── infer.py
├── slic_process_sam.py
├── sliding_window.py
├── train.py
├── train_finetune.py
├── utils
│ ├── sample_utils.py
│ ├── trans_utils.py
│ └── workflow_utils.py
└── validation
│ ├── build_vista3d_eval_only.py
│ ├── val_multigpu_autopoint_patch.py
│ ├── val_multigpu_point_iterative.py
│ ├── val_multigpu_point_patch.py
│ └── val_multigpu_sam2_point_iterative.py
├── tests
├── __init__.py
├── test_config.py
└── test_logger.py
└── vista3d
├── __init__.py
├── build_vista3d.py
└── modeling
├── __init__.py
├── class_head.py
├── point_head.py
├── sam_blocks.py
├── segresnetds.py
└── vista3d.py
/.github/ISSUE_TEMPLATE/bug_report.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Bug report
3 | about: Create a report to help us improve
4 | title: ''
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Describe the bug**
11 | A clear and concise description of what the bug is.
12 |
13 | **To Reproduce**
14 | Steps to reproduce the behavior:
15 | 1. Go to '...'
16 | 2. Install '....'
17 | 3. Run commands '....'
18 |
19 | **Expected behavior**
20 | A clear and concise description of what you expected to happen.
21 |
22 | **Screenshots**
23 | If applicable, add screenshots to help explain your problem.
24 |
25 | **Additional context**
26 | Add any other context about the problem here.
27 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/feature_request.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Feature request
3 | about: Suggest an idea for this project
4 | title: ''
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Is your feature request related to a problem? Please describe.**
11 | A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
12 |
13 | **Describe the solution you'd like**
14 | A clear and concise description of what you want to happen.
15 |
16 | **Describe alternatives you've considered**
17 | A clear and concise description of any alternative solutions or features you've considered.
18 |
19 | **Additional context**
20 | Add any other context or screenshots about the feature request here.
21 |
--------------------------------------------------------------------------------
/.github/pull_request_template.md:
--------------------------------------------------------------------------------
1 | Fixes # .
2 |
3 | ### Description
4 |
5 | A few sentences describing the changes proposed in this pull request.
6 |
7 | ### Types of changes
8 |
9 | - [x] Non-breaking change (fix or new feature that would not break existing functionality).
10 | - [ ] Breaking change (fix or new feature that would cause existing functionality to change).
11 | - [ ] New tests added to cover the changes.
12 | - [ ] In-line docstrings updated.
13 |
--------------------------------------------------------------------------------
/.github/workflows/premerge-py-min.yml:
--------------------------------------------------------------------------------
1 | name: premerge-min
2 |
3 | on:
4 | # quick tests for pull requests and the releasing branches
5 | push:
6 | branches:
7 | - vista3d
8 | - main
9 | pull_request:
10 |
11 | concurrency:
12 | # automatically cancel the previously triggered workflows when there's a newer version
13 | group: py-min-${{ github.event.pull_request.number || github.ref }}
14 | cancel-in-progress: true
15 |
16 | jobs:
17 | min-dep-py3: # min dependencies installed tests for different python
18 | runs-on: ubuntu-latest
19 | strategy:
20 | fail-fast: false
21 | matrix:
22 | python-version: ['3.9', '3.10', '3.11']
23 | timeout-minutes: 40
24 | steps:
25 | - uses: actions/checkout@v4
26 | - name: Set up Python ${{ matrix.python-version }}
27 | uses: actions/setup-python@v5
28 | with:
29 | python-version: ${{ matrix.python-version }}
30 | - name: Prepare pip wheel
31 | run: |
32 | which python
33 | python -m pip install --user --upgrade pip setuptools wheel
34 | - name: cache weekly timestamp
35 | id: pip-cache
36 | run: |
37 | echo "datew=$(date '+%Y-%V')" >> $GITHUB_OUTPUT
38 | echo "dir=$(pip cache dir)" >> $GITHUB_OUTPUT
39 | shell: bash
40 | - name: cache for pip
41 | uses: actions/cache@v4
42 | id: cache
43 | with:
44 | path: ${{ steps.pip-cache.outputs.dir }}
45 | key: ubuntu-latest-latest-pip-${{ steps.pip-cache.outputs.datew }}
46 | - name: Install the dependencies
47 | run: |
48 | python -m pip install torch --extra-index-url https://download.pytorch.org/whl/cpu
49 | python -m pip install "monai[all]"
50 | python -m pip list
51 | shell: bash
52 | - name: Run quick tests (CPU ${{ runner.os }})
53 | run: |
54 | python -c 'import torch; print(torch.__version__); print(torch.rand(5,3))'
55 | python -c "import monai; monai.config.print_config()"
56 | python -m unittest -v
57 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 | # Distribution / packaging
9 | .Python
10 | build/
11 | develop-eggs/
12 | dist/
13 | downloads/
14 | eggs/
15 | .eggs/
16 | # lib/
17 | lib64/
18 | parts/
19 | sdist/
20 | var/
21 | wheels/
22 | *.egg-info/
23 | .installed.cfg
24 | *.egg
25 | MANIFEST
26 | # PyInstaller
27 | # Usually these files are written by a python script from a template
28 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
29 | *.manifest
30 | *.spec
31 | # Installer logs
32 | pip-log.txt
33 | pip-delete-this-directory.txt
34 | # Unit test / coverage reports
35 | htmlcov/
36 | .tox/
37 | .coverage
38 | .coverage.*
39 | .coverage/
40 | .cache
41 | nosetests.xml
42 | coverage.xml
43 | *.cover
44 | .hypothesis/
45 | .pytest_cache/
46 | # Translations
47 | *.mo
48 | *.pot
49 | # Django stuff:
50 | *.log
51 | local_settings.py
52 | db.sqlite3
53 | # Flask stuff:
54 | instance/
55 | .webassets-cache
56 | # Scrapy stuff:
57 | .scrapy
58 | # Sphinx documentation
59 | docs/build/
60 | docs/source/_gen
61 | _build/
62 | # PyBuilder
63 | target/
64 | # Jupyter Notebook
65 | .ipynb_checkpoints
66 | # pyenv
67 | .python-version
68 | # celery beat schedule file
69 | celerybeat-schedule
70 | # SageMath parsed files
71 | *.sage.py
72 | # Environments
73 | .env
74 | .venv
75 | env/
76 | venv/
77 | ENV/
78 | env.bak/
79 | venv.bak/
80 | # Spyder project settings
81 | .spyderproject
82 | .spyproject
83 | # Rope project settings
84 | .ropeproject
85 | # mkdocs documentation
86 | /site
87 | # pytype cache
88 | .pytype/
89 | # mypy
90 | .mypy_cache/
91 | examples/scd_lvsegs.npz
92 | temp/
93 | .idea/
94 | # *~
95 | # Remove .pyre temporary config files
96 | .pyre
97 | .pyre_configuration
98 | # temporary editor files that should not be in git
99 | *.orig
100 | *.bak
101 | *.swp
102 | .DS_Store
103 | # VSCode
104 | .vscode/
105 | *.zip
106 | # profiling results
107 | *.prof
108 | runs
109 | *.gz
110 | *.pth
111 | *.pt
112 | lib/
113 | pip-wheel-metadata/
114 | share/python-wheels/
115 | .nox/
116 | *.py,cover
117 | db.sqlite3-journal
118 | docs/_build/
119 | # IPython
120 | profile_default/
121 | ipython_config.py
122 | # pipenv
123 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
124 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
125 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
126 | # install all needed dependencies.
127 | #Pipfile.lock
128 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
129 | __pypackages__/
130 | # Celery stuff
131 | celerybeat.pid
132 | .idea
133 | .dmypy.json
134 | dmypy.json
135 | # Pyre type checker
136 | .pyre/
137 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | default_language_version:
2 | python: python3
3 |
4 | ci:
5 | autofix_prs: true
6 | autoupdate_commit_msg: '[pre-commit.ci] pre-commit suggestions'
7 | autoupdate_schedule: quarterly
8 | # submodules: true
9 |
10 | repos:
11 | - repo: https://github.com/pre-commit/pre-commit-hooks
12 | rev: v5.0.0
13 | hooks:
14 | - id: end-of-file-fixer
15 | - id: trailing-whitespace
16 | - id: check-yaml
17 | - id: check-docstring-first
18 | - id: check-executables-have-shebangs
19 | - id: check-toml
20 | - id: check-case-conflict
21 | - id: check-added-large-files
22 | args: ['--maxkb=1024']
23 | - id: detect-private-key
24 | - id: forbid-new-submodules
25 | - id: pretty-format-json
26 | args: ['--autofix', '--no-sort-keys', '--indent=4']
27 | - id: end-of-file-fixer
28 | - id: mixed-line-ending
29 | - repo: https://github.com/psf/black
30 | rev: "24.10.0"
31 | hooks:
32 | - id: black
33 | - id: black-jupyter
34 | - repo: https://github.com/pycqa/isort
35 | rev: 5.13.2
36 | hooks:
37 | - id: isort
38 | args: ["--profile", "black"]
39 | - repo: https://github.com/astral-sh/ruff-pre-commit
40 | rev: v0.8.6
41 | hooks:
42 | - id: ruff
43 | args: ['--fix']
44 |
45 | - repo: https://github.com/asottile/yesqa
46 | rev: v1.5.0
47 | hooks:
48 | - id: yesqa
49 | name: Unused noqa
50 | additional_dependencies:
51 | - flake8>=3.8.1
52 | - flake8-bugbear
53 | - flake8-comprehensions
54 | - pep8-naming
55 | exclude: |
56 | (?x)^(
57 | .*/__init__.py|
58 | tests/utils.py
59 | )$
60 |
61 | # - repo: https://github.com/hadialqattan/pycln
62 | # rev: v2.1.3
63 | # hooks:
64 | # - id: pycln
65 | # args: [--config=pyproject.toml]
66 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
13 |
14 | # MONAI VISTA Repository
15 | This is the repository for VISTA3D and VISTA2D. For the older VISTA2.5d code, please checkout the vista2.5d branch
16 |
--------------------------------------------------------------------------------
/vista2d/README.md:
--------------------------------------------------------------------------------
1 |
13 |
14 | ## Overview
15 |
16 | The **VISTA2D** is a cell segmentation training and inference pipeline for cell imaging [[`Blog`](https://developer.nvidia.com/blog/advancing-cell-segmentation-and-morphology-analysis-with-nvidia-ai-foundation-model-vista-2d/)].
17 |
18 | A pretrained model was trained on collection of 15K public microscopy images. The data collection and training can be reproduced following the [tutorial](./download_preprocessor/). Alternatively, the model can be retrained on your own dataset. The pretrained vista2d model achieves good performance on diverse set of cell types, microscopy image modalities, and can be further finetuned if necessary. The codebase utilizes several components from other great works including [SegmentAnything](https://github.com/facebookresearch/segment-anything) and [Cellpose](https://www.cellpose.org/), which must be pip installed as dependencies. Vista2D codebase follows MONAI bundle format and its [specifications](https://docs.monai.io/en/stable/mb_specification.html).
19 |
20 |
21 |
22 |
23 | ### Model highlights
24 |
25 | - Robust deep learning algorithm based on transformers
26 | - Generalist model as compared to specialist models
27 | - Multiple dataset sources and file formats supported
28 | - Multiple modalities of imaging data collectively supported
29 | - Multi-GPU and multinode training support
30 |
31 |
32 | ### Generalization performance
33 |
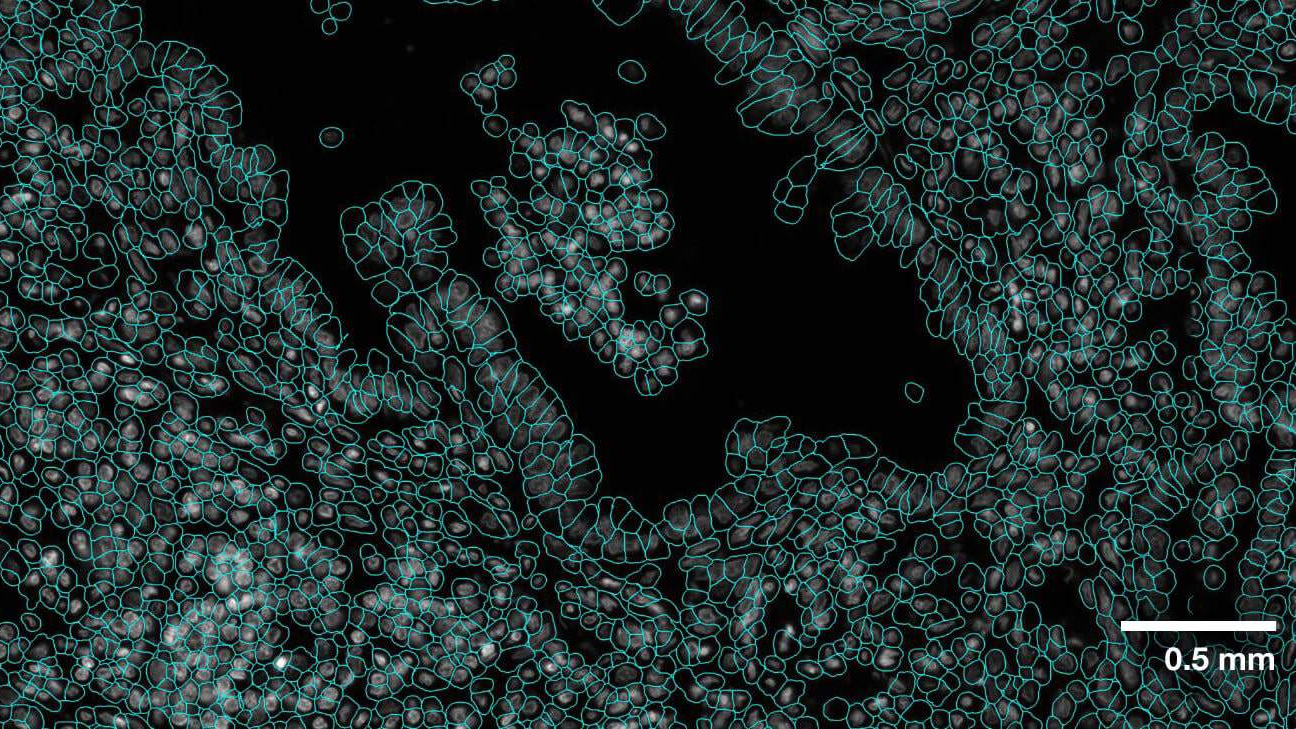
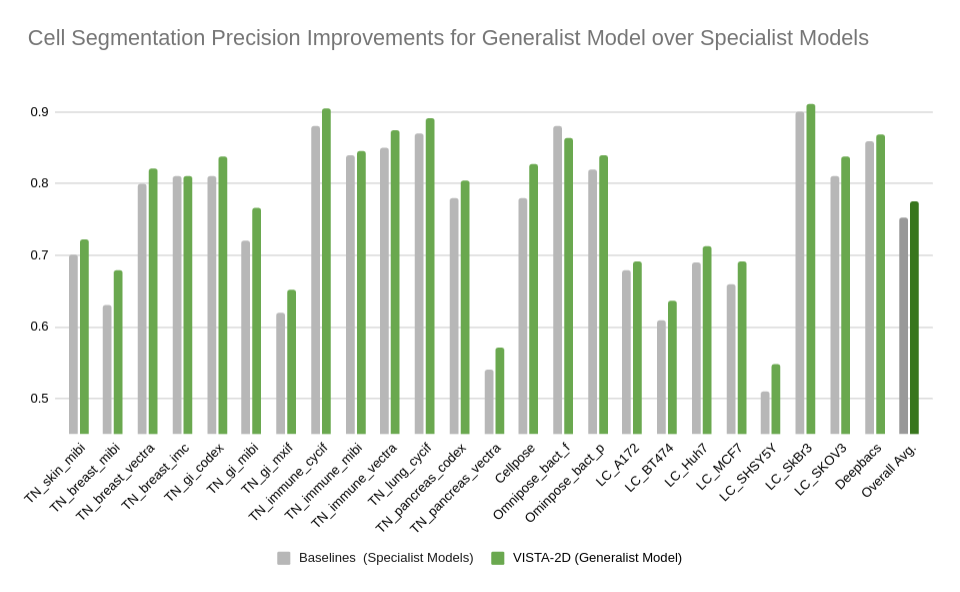
34 | Evaluation was performed for the VISTA2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and [more](./docs/data_license.txt). A total of ~15K annotated cell images were collected to train the generalist VISTA2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types. The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. Average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA2D model trained only on a particular dataset or a subset of data.
35 |
36 |
37 |
38 |
39 |
40 | ### Install dependencies
41 |
42 | ```
43 | pip install monai fire tifffile imagecodecs pillow fastremap
44 | pip install --no-deps cellpose natsort roifile
45 | pip install git+https://github.com/facebookresearch/segment-anything.git
46 | pip install mlflow psutil pynvml #optional for MLFlow support
47 | ```
48 |
49 | ### Execute training
50 | ```bash
51 | python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml
52 | ```
53 |
54 | #### Quick run with a few data points
55 | ```bash
56 | python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --quick True --train#trainer#max_epochs 3
57 | ```
58 |
59 | ### Execute multi-GPU training
60 | ```bash
61 | torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml
62 | ```
63 |
64 | ### Execute validation
65 | ```bash
66 | python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --pretrained_ckpt_name model.pt --mode eval
67 | ```
68 | (can append `--quick True` for quick demoing)
69 |
70 | ### Execute multi-GPU validation
71 | ```bash
72 | torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode eval
73 | ```
74 |
75 | ### Execute inference
76 | ```bash
77 | python -m monai.bundle run --config_file configs/inference.json
78 | ```
79 |
80 | ### Execute multi-GPU inference
81 | ```bash
82 | torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode infer --pretrained_ckpt_name model.pt
83 | ```
84 | (can append `--quick True` for quick demoing)
85 |
86 |
87 |
88 | #### Finetune starting from a trained checkpoint
89 | (we use a smaller learning rate, small number of epochs, and initialize from a checkpoint)
90 | ```bash
91 | python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --learning_rate=0.001 --train#trainer#max_epochs 20 --pretrained_ckpt_path /path/to/saved/model.pt
92 | ```
93 |
94 |
95 | #### Configuration options
96 |
97 | To disable the segmentation writing:
98 | ```
99 | --postprocessing []
100 | ```
101 |
102 | Load a checkpoint for validation or inference (relative path within results directory):
103 | ```
104 | --pretrained_ckpt_name "model.pt"
105 | ```
106 |
107 | Load a checkpoint for validation or inference (absolute path):
108 | ```
109 | --pretrained_ckpt_path "/path/to/another/location/model.pt"
110 | ```
111 |
112 | `--mode eval` or `--mode infer`will use the corresponding configurations from the `validate` or `infer`
113 | of the `configs/hyper_parameters.yaml`.
114 |
115 | By default the generated `model.pt` corresponds to the checkpoint at the best validation score,
116 | `model_final.pt` is the checkpoint after the latest training epoch.
117 |
118 |
119 | ### Development
120 |
121 | For development purposes it's possible to run the script directly (without monai bundle calls)
122 |
123 | ```bash
124 | python scripts/workflow.py --config_file configs/hyper_parameters.yaml ...
125 | torchrun --nproc_per_node=gpu -m scripts/workflow.py --config_file configs/hyper_parameters.yaml ..
126 | ```
127 |
128 | ### MLFlow support
129 |
130 | Enable MLFlow logging by specifying "mlflow_tracking_uri" (can be local or remote URL).
131 |
132 | ```bash
133 | python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mlflow_tracking_uri=http://127.0.0.1:8080
134 | ```
135 |
136 | Optionally use "--mlflow_run_name=.." to specify MLFlow experiment name, and "--mlflow_log_system_metrics=True/False" to enable logging of CPU/GPU resources (requires pip install psutil pynvml)
137 |
138 |
139 |
140 | ### Unit tests
141 |
142 | Test single GPU training:
143 | ```
144 | python unit_tests/test_vista2d.py
145 | ```
146 |
147 | Test multi-GPU training (may need to uncomment the `"--standalone"` in the `unit_tests/utils.py` file):
148 | ```
149 | python unit_tests/test_vista2d_mgpu.py
150 | ```
151 |
152 | ## Compute Requirements
153 | Min GPU memory requirements 16Gb.
154 |
155 |
156 | ## Contributing
157 | Vista2D codebase follows MONAI bundle format and its [specifications](https://docs.monai.io/en/stable/mb_specification.html).
158 | Make sure to run pre-commit before committing code changes to git
159 | ```bash
160 | pip install pre-commit
161 | python3 -m pre_commit run --all-files
162 | ```
163 |
164 |
165 | ## Community
166 |
167 | Join the conversation on Twitter [@ProjectMONAI](https://twitter.com/ProjectMONAI) or join
168 | our [Slack channel](https://projectmonai.slack.com/archives/C031QRE0M1C).
169 |
170 | Ask and answer questions on [MONAI VISTA's GitHub discussions tab](https://github.com/Project-MONAI/VISTA/discussions).
171 |
172 | ## License
173 |
174 | The codebase is under Apache 2.0 Licence. The model weight is released under CC-BY-NC-SA-4.0. For various public data licenses please see [data_license.txt](./docs/data_license.txt).
175 |
176 | ## Acknowledgement
177 | - [segment-anything](https://github.com/facebookresearch/segment-anything)
178 | - [Cellpose](https://www.cellpose.org/)
179 |
--------------------------------------------------------------------------------
/vista2d/configs/hyper_parameters.yaml:
--------------------------------------------------------------------------------
1 | imports:
2 | - $import os
3 |
4 | # seed: 28022024 # uncommend for deterministic results (but slower)
5 | seed: null
6 |
7 | bundle_root: "."
8 | ckpt_path: $os.path.join(@bundle_root, "models") # location to save checkpoints
9 | output_dir: $os.path.join(@bundle_root, "eval") # location to save events and logs
10 | log_output_file: $os.path.join(@output_dir, "vista_cell.log")
11 |
12 | mlflow_tracking_uri: null # enable mlflow logging, e.g. $@ckpt_path + '/mlruns/ or "http://127.0.0.1:8080" or a remote url
13 | mlflow_log_system_metrics: true # log system metrics to mlflow (requires: pip install psutil pynvml)
14 | mlflow_run_name: null # optional name of the current run
15 |
16 | ckpt_save: true # save checkpoints periodically
17 | amp: true
18 | amp_dtype: "float16" #float16 or bfloat16 (Ampere or newer)
19 | channels_last: true
20 | compile: false # complie the model for faster processing
21 |
22 | start_epoch: 0
23 | run_final_testing: true
24 | use_weighted_sampler: false # only applicable when using several dataset jsons for data_list_files
25 |
26 | pretrained_ckpt_name: null
27 | pretrained_ckpt_path: null
28 |
29 | # for commandline setting of a single dataset
30 | datalist: datalists/tissuenet_skin_mibi_datalist.json
31 | basedir: /data/tissuenet
32 | data_list_files:
33 | - {datalist: "@datalist", basedir: "@basedir"}
34 |
35 |
36 | fold: 0
37 | learning_rate: 0.01 # try 1.0e-4 if using AdamW
38 | quick: false # whether to use a small subset of data for quick testing
39 | roi_size: [256, 256]
40 |
41 | train:
42 | skip: false
43 | handlers: []
44 | trainer:
45 | num_warmup_epochs: 3
46 | max_epochs: 200
47 | num_epochs_per_saving: 1
48 | num_epochs_per_validation: null
49 | num_workers: 4
50 | batch_size: 1
51 | dataset:
52 | preprocessing:
53 | roi_size: "@roi_size"
54 | data:
55 | key: null # set to 'testing' to use this subset in periodic validations, instead of the the validation set
56 | data_list_files: "@data_list_files"
57 |
58 | dataset:
59 | data:
60 | key: "testing"
61 | data_list_files: "@data_list_files"
62 |
63 | validate:
64 | grouping: true
65 | evaluator:
66 | postprocessing: "@postprocessing"
67 | dataset:
68 | data: "@dataset#data"
69 | batch_size: 1
70 | num_workers: 4
71 | preprocessing: null
72 | postprocessing: null
73 | inferer: null
74 | handlers: null
75 | key_metric: null
76 |
77 | infer:
78 | evaluator:
79 | postprocessing: "@postprocessing"
80 | dataset:
81 | data: "@dataset#data"
82 |
83 |
84 | device: "$torch.device(('cuda:' + os.environ.get('LOCAL_RANK', '0')) if torch.cuda.is_available() else 'cpu')"
85 | network_def:
86 | _target_: scripts.cell_sam_wrapper.CellSamWrapper
87 | checkpoint: $os.path.join(@ckpt_path, "sam_vit_b_01ec64.pth")
88 | network: $@network_def.to(@device)
89 |
90 | loss_function:
91 | _target_: scripts.components.CellLoss
92 |
93 | key_metric:
94 | _target_: scripts.components.CellAcc
95 |
96 | # optimizer:
97 | # _target_: torch.optim.AdamW
98 | # params: $@network.parameters()
99 | # lr: "@learning_rate"
100 | # weight_decay: 1.0e-5
101 |
102 | optimizer:
103 | _target_: torch.optim.SGD
104 | params: $@network.parameters()
105 | momentum: 0.9
106 | lr: "@learning_rate"
107 | weight_decay: 1.0e-5
108 |
109 | lr_scheduler:
110 | _target_: monai.optimizers.lr_scheduler.WarmupCosineSchedule
111 | optimizer: "@optimizer"
112 | warmup_steps: "@train#trainer#num_warmup_epochs"
113 | warmup_multiplier: 0.1
114 | t_total: "@train#trainer#max_epochs"
115 |
116 | inferer:
117 | sliding_inferer:
118 | _target_: monai.inferers.SlidingWindowInfererAdapt
119 | roi_size: "@roi_size"
120 | sw_batch_size: 1
121 | overlap: 0.625
122 | mode: "gaussian"
123 | cache_roi_weight_map: true
124 | progress: false
125 |
126 | image_saver:
127 | _target_: scripts.components.SaveTiffd
128 | keys: "seg"
129 | output_dir: "@output_dir"
130 | nested_folder: false
131 |
132 | postprocessing:
133 | _target_: monai.transforms.Compose
134 | transforms:
135 | - "@image_saver"
136 |
--------------------------------------------------------------------------------
/vista2d/configs/inference.json:
--------------------------------------------------------------------------------
1 | {
2 | "imports": [
3 | "$import numpy as np"
4 | ],
5 | "bundle_root": ".",
6 | "ckpt_dir": "$@bundle_root + '/models'",
7 | "output_dir": "$@bundle_root + '/eval'",
8 | "output_ext": ".tif",
9 | "output_postfix": "trans",
10 | "roi_size": [
11 | 256,
12 | 256
13 | ],
14 | "input_dict": "${'image': '/cellpose_dataset/test/001_img.png'}",
15 | "device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
16 | "sam_ckpt_path": "$@ckpt_dir + '/sam_vit_b_01ec64.pth'",
17 | "pretrained_ckpt_path": "$@ckpt_dir + '/model.pt'",
18 | "image_key": "image",
19 | "channels_last": true,
20 | "use_amp": true,
21 | "amp_dtype": "$torch.float",
22 | "network_def": {
23 | "_target_": "scripts.cell_sam_wrapper.CellSamWrapper",
24 | "checkpoint": "@sam_ckpt_path"
25 | },
26 | "network": "$@network_def.to(@device)",
27 | "preprocessing_transforms": [
28 | {
29 | "_target_": "scripts.components.LoadTiffd",
30 | "keys": "@image_key"
31 | },
32 | {
33 | "_target_": "EnsureTyped",
34 | "keys": "@image_key",
35 | "data_type": "tensor",
36 | "dtype": "$torch.float"
37 | },
38 | {
39 | "_target_": "ScaleIntensityd",
40 | "keys": "@image_key",
41 | "minv": 0,
42 | "maxv": 1,
43 | "channel_wise": true
44 | },
45 | {
46 | "_target_": "ScaleIntensityRangePercentilesd",
47 | "keys": "image",
48 | "lower": 1,
49 | "upper": 99,
50 | "b_min": 0.0,
51 | "b_max": 1.0,
52 | "channel_wise": true,
53 | "clip": true
54 | }
55 | ],
56 | "preprocessing": {
57 | "_target_": "Compose",
58 | "transforms": "$@preprocessing_transforms "
59 | },
60 | "dataset": {
61 | "_target_": "Dataset",
62 | "data": "$[@input_dict]",
63 | "transform": "@preprocessing"
64 | },
65 | "dataloader": {
66 | "_target_": "ThreadDataLoader",
67 | "dataset": "@dataset",
68 | "batch_size": 1,
69 | "shuffle": false,
70 | "num_workers": 0
71 | },
72 | "inferer": {
73 | "_target_": "SlidingWindowInfererAdapt",
74 | "roi_size": "@roi_size",
75 | "sw_batch_size": 1,
76 | "overlap": 0.625,
77 | "mode": "gaussian",
78 | "cache_roi_weight_map": true,
79 | "progress": false
80 | },
81 | "postprocessing": {
82 | "_target_": "Compose",
83 | "transforms": [
84 | {
85 | "_target_": "ToDeviced",
86 | "keys": "pred",
87 | "device": "cpu"
88 | },
89 | {
90 | "_target_": "scripts.components.LogitsToLabelsd",
91 | "keys": "pred"
92 | },
93 | {
94 | "_target_": "scripts.components.SaveTiffExd",
95 | "keys": "pred",
96 | "output_dir": "@output_dir",
97 | "output_ext": "@output_ext",
98 | "output_postfix": "@output_postfix"
99 | }
100 | ]
101 | },
102 | "handlers": [
103 | {
104 | "_target_": "StatsHandler",
105 | "iteration_log": false
106 | }
107 | ],
108 | "checkpointloader": {
109 | "_target_": "CheckpointLoader",
110 | "load_path": "@pretrained_ckpt_path",
111 | "map_location": "cpu",
112 | "load_dict": {
113 | "state_dict": "@network"
114 | }
115 | },
116 | "evaluator": {
117 | "_target_": "SupervisedEvaluator",
118 | "device": "@device",
119 | "val_data_loader": "@dataloader",
120 | "network": "@network",
121 | "inferer": "@inferer",
122 | "postprocessing": "@postprocessing",
123 | "val_handlers": "@handlers",
124 | "amp": true
125 | },
126 | "initialize": [
127 | "$monai.utils.set_determinism(seed=123)",
128 | "$@checkpointloader(@evaluator)"
129 | ],
130 | "run": [
131 | "$@evaluator.run()"
132 | ]
133 | }
134 |
--------------------------------------------------------------------------------
/vista2d/configs/metadata.json:
--------------------------------------------------------------------------------
1 | {
2 | "schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
3 | "version": "0.2.0",
4 | "changelog": {
5 | "0.2.0": "support bundle inference json",
6 | "0.1.9": "finetuning checkpoint loading update",
7 | "0.1.8": "added manfests",
8 | "0.1.7": "prescaling to 0..1",
9 | "0.1.6": "data processing script instead of json generation, data licenses",
10 | "0.1.5": "improvements and mlflow support",
11 | "0.1.4": "add unit tests and use monai 1.3.1",
12 | "0.1.3": "update default optimizer",
13 | "0.1.2": "use_weighted_sampler defaults to false",
14 | "0.1.1": "remove unused code",
15 | "0.1.0": "update evaluator",
16 | "0.0.9": "do workflow_type and dataset data changes",
17 | "0.0.8": "let pretrained_ckpt_name refer to ckpt_path",
18 | "0.0.7": "update workflow input arg",
19 | "0.0.6": "update the transforms and model, partly verified the reimplementation",
20 | "0.0.5": "add inference logic and rename the entry point class from train.py to workflow.py",
21 | "0.0.4": "update loadimage and weighted sampler",
22 | "0.0.3": "fix final val error if override datalist",
23 | "0.0.2": "fix attr error",
24 | "0.0.1": "initialize the model package structure"
25 | },
26 | "monai_version": "1.3.1",
27 | "pytorch_version": "2.2.2",
28 | "numpy_version": "1.24.4",
29 | "einops": "0.7.0",
30 | "optional_packages_version": {
31 | "scikit-image": "0.22.0",
32 | "fastremap": "1.14.1",
33 | "cucim-cu12": "24.2.0",
34 | "gdown": "5.1.0",
35 | "fire": "0.6.0",
36 | "pyyaml": "6.0.1",
37 | "tensorboard": "2.16.2",
38 | "opencv-python": "4.7.0.68",
39 | "numba": "0.59.0",
40 | "torchvision": "0.17.2",
41 | "cellpose": "3.0.8",
42 | "segment_anything": "1.0",
43 | "mlflow": "2.13.1",
44 | "pynvml": "11.5.0"
45 | },
46 | "name": "VISTA-Cell",
47 | "task": "cell image segmentation",
48 | "description": "VISTA2D bundle for cell image analysis",
49 | "authors": "MONAI team",
50 | "copyright": "Copyright (c) MONAI Consortium",
51 | "data_type": "tiff",
52 | "image_classes": "1 channel data, intensity scaled to [0, 1]",
53 | "label_classes": "3-channel data",
54 | "pred_classes": "3 channels",
55 | "eval_metrics": {
56 | "mean_dice": 0.0
57 | },
58 | "intended_use": "This is an example, not to be used for diagnostic purposes",
59 | "references": [],
60 | "network_data_format": {
61 | "inputs": {
62 | "image": {
63 | "type": "image",
64 | "num_channels": 3,
65 | "spatial_shape": [
66 | 256,
67 | 256
68 | ],
69 | "format": "RGB",
70 | "value_range": [
71 | 0,
72 | 255
73 | ],
74 | "dtype": "float32",
75 | "is_patch_data": true,
76 | "channel_def": {
77 | "0": "image"
78 | }
79 | }
80 | },
81 | "outputs": {
82 | "pred": {
83 | "type": "image",
84 | "format": "segmentation",
85 | "num_channels": 3,
86 | "dtype": "float32",
87 | "value_range": [
88 | 0,
89 | 1
90 | ],
91 | "spatial_shape": [
92 | 256,
93 | 256
94 | ]
95 | }
96 | }
97 | }
98 | }
99 |
--------------------------------------------------------------------------------
/vista2d/datalists/group1_datalist.yaml:
--------------------------------------------------------------------------------
1 | - {datalist: datalists2/tissuenet_skin_mibi_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
2 | - {datalist: datalists2/tissuenet_breast_mibi_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
3 | - {datalist: datalists2/tissuenet_breast_vectra_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
4 | - {datalist: datalists2/tissuenet_breast_imc_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
5 | - {datalist: datalists2/tissuenet_gi_codex_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
6 | - {datalist: datalists2/tissuenet_gi_mibi_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
7 | - {datalist: datalists2/tissuenet_gi_mxif_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
8 | - {datalist: datalists2/tissuenet_immune_cycif_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
9 | - {datalist: datalists2/tissuenet_immune_mibi_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
10 | - {datalist: datalists2/tissuenet_immune_vectra_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
11 | - {datalist: datalists2/tissuenet_lung_cycif_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
12 | - {datalist: datalists2/tissuenet_pancreas_codex_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
13 | - {datalist: datalists2/tissuenet_pancreas_vectra_datalist.json, basedir: /data/vista2d_combined/tissuenet_dataset/tissuenet_1.0}
14 |
15 | - {datalist: datalists2/cellpose_datalist.json, basedir: /data/vista2d_combined/cellpose_dataset}
16 |
17 | - {datalist: datalists2/omnipose_bact_f_dataset.json, basedir: /data/vista2d_combined/omnipose_dataset}
18 | - {datalist: datalists2/omnipose_bact_p_dataset.json, basedir: /data/vista2d_combined/omnipose_dataset}
19 |
20 | - {datalist: datalists2/livecell_A172_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
21 | - {datalist: datalists2/livecell_BT474_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
22 | - {datalist: datalists2/livecell_Huh7_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
23 | - {datalist: datalists2/livecell_MCF7_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
24 | - {datalist: datalists2/livecell_SHSY5Y_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
25 | - {datalist: datalists2/livecell_SkBr3_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
26 | - {datalist: datalists2/livecell_SKOV3_datalist.json, basedir: /data/vista2d_combined/livecell_dataset/images}
27 |
28 | - {datalist: datalists2/deepbacs_datalist.json, basedir: /data/vista2d_combined/deepbacs_dataset}
29 | - {datalist: datalists2/nips_data_list.json, basedir: /data/vista2d_combined/nips_dataset}
30 | - {datalist: datalists2/kg_nuclei_datalist.json, basedir: /data/vista2d_combined/kaggle_dataset}
31 |
--------------------------------------------------------------------------------
/vista2d/datalists/yeaz_phc_dataset_list.json:
--------------------------------------------------------------------------------
1 | {
2 | "testing": [
3 | {
4 | "image": "gold-standard-PhC-plus-2/m_reexport1_crop_1_im.tif",
5 | "label": "gold-standard-PhC-plus-2/m_reexport1_crop_1_mask.tif"
6 | },
7 | {
8 | "image": "gold-standard-PhC-plus-2/d_reexport2_crop_3_im.tif",
9 | "label": "gold-standard-PhC-plus-2/d_reexport2_crop_3_mask.tif"
10 | },
11 | {
12 | "image": "gold-standard-PhC-plus-2/FOV4_20p_PhC_present_crop_1_im.tif",
13 | "label": "gold-standard-PhC-plus-2/FOV4_20p_PhC_present_crop_1_mask.tif"
14 | },
15 | {
16 | "image": "gold-standard-PhC-plus-2/d_reexport2_crop_2_im.tif",
17 | "label": "gold-standard-PhC-plus-2/d_reexport2_crop_2_mask.tif"
18 | },
19 | {
20 | "image": "gold-standard-PhC-plus-2/FOV10_20p_PhC_present_crop_1_im.tif",
21 | "label": "gold-standard-PhC-plus-2/FOV10_20p_PhC_present_crop_1_mask.tif"
22 | },
23 | {
24 | "image": "gold-standard-PhC-plus-2/d_reexport1_crop_3_im.tif",
25 | "label": "gold-standard-PhC-plus-2/d_reexport1_crop_3_mask.tif"
26 | },
27 | {
28 | "image": "gold-standard-PhC-plus-2/d_reexport1_crop_2_im.tif",
29 | "label": "gold-standard-PhC-plus-2/d_reexport1_crop_2_mask.tif"
30 | },
31 | {
32 | "image": "gold-standard-PhC-plus-2/d_reexport2_crop_1_im.tif",
33 | "label": "gold-standard-PhC-plus-2/d_reexport2_crop_1_mask.tif"
34 | }
35 | ],
36 | "training": [
37 | {
38 | "image": "gold-standard-PhC-plus-2/d_reexport1_crop_4_im.tif",
39 | "label": "gold-standard-PhC-plus-2/d_reexport1_crop_4_mask.tif",
40 | "fold": 0
41 | },
42 | {
43 | "image": "gold-standard-PhC-plus-2/FOV11_20p_PhC_present_crop_1_im.tif",
44 | "label": "gold-standard-PhC-plus-2/FOV11_20p_PhC_present_crop_1_mask.tif",
45 | "fold": 0
46 | },
47 | {
48 | "image": "gold-standard-PhC-plus-2/FOV13_20p_PhC_present_crop_1_im.tif",
49 | "label": "gold-standard-PhC-plus-2/FOV13_20p_PhC_present_crop_1_mask.tif",
50 | "fold": 0
51 | },
52 | {
53 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_2_im.tif",
54 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_2_mask.tif",
55 | "fold": 0
56 | },
57 | {
58 | "image": "gold-standard-PhC-plus-2/V11032020_p3_crop_1_im.tif",
59 | "label": "gold-standard-PhC-plus-2/V11032020_p3_crop_1_mask.tif",
60 | "fold": 0
61 | },
62 | {
63 | "image": "gold-standard-PhC-plus-2/FOV14_20p_PhC_present_crop_1_im.tif",
64 | "label": "gold-standard-PhC-plus-2/FOV14_20p_PhC_present_crop_1_mask.tif",
65 | "fold": 0
66 | },
67 | {
68 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_4_im.tif",
69 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_4_mask.tif",
70 | "fold": 0
71 | },
72 | {
73 | "image": "gold-standard-PhC-plus-2/FOV1_20p_PhC_present_crop_1_im.tif",
74 | "label": "gold-standard-PhC-plus-2/FOV1_20p_PhC_present_crop_1_mask.tif",
75 | "fold": 1
76 | },
77 | {
78 | "image": "gold-standard-PhC-plus-2/a_reexport3_crop_2_im.tif",
79 | "label": "gold-standard-PhC-plus-2/a_reexport3_crop_2_mask.tif",
80 | "fold": 1
81 | },
82 | {
83 | "image": "gold-standard-PhC-plus-2/v_clnnull_crop_1_im.tif",
84 | "label": "gold-standard-PhC-plus-2/v_clnnull_crop_1_mask.tif",
85 | "fold": 1
86 | },
87 | {
88 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_1_im.tif",
89 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_1_mask.tif",
90 | "fold": 1
91 | },
92 | {
93 | "image": "gold-standard-PhC-plus-2/FOV8_20p_PhC_present_crop_1_im.tif",
94 | "label": "gold-standard-PhC-plus-2/FOV8_20p_PhC_present_crop_1_mask.tif",
95 | "fold": 1
96 | },
97 | {
98 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_2_im.tif",
99 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_2_mask.tif",
100 | "fold": 1
101 | },
102 | {
103 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_3_im.tif",
104 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_3_mask.tif",
105 | "fold": 1
106 | },
107 | {
108 | "image": "gold-standard-PhC-plus-2/2020_3_24_AS20_1_AS18_Pos11_crop_2_im.tif",
109 | "label": "gold-standard-PhC-plus-2/2020_3_24_AS20_1_AS18_Pos11_crop_2_mask.tif",
110 | "fold": 2
111 | },
112 | {
113 | "image": "gold-standard-PhC-plus-2/2020_3_24_AS20_1_AS18_Pos11_crop_1_im.tif",
114 | "label": "gold-standard-PhC-plus-2/2020_3_24_AS20_1_AS18_Pos11_crop_1_mask.tif",
115 | "fold": 2
116 | },
117 | {
118 | "image": "gold-standard-PhC-plus-2/FOV10_20p_PhC_present_crop_2_im.tif",
119 | "label": "gold-standard-PhC-plus-2/FOV10_20p_PhC_present_crop_2_mask.tif",
120 | "fold": 2
121 | },
122 | {
123 | "image": "gold-standard-PhC-plus-2/FOV9_20p_PhC_present_crop_2_im.tif",
124 | "label": "gold-standard-PhC-plus-2/FOV9_20p_PhC_present_crop_2_mask.tif",
125 | "fold": 2
126 | },
127 | {
128 | "image": "gold-standard-PhC-plus-2/FOV5_20p_PhC_present_crop_1_im.tif",
129 | "label": "gold-standard-PhC-plus-2/FOV5_20p_PhC_present_crop_1_mask.tif",
130 | "fold": 2
131 | },
132 | {
133 | "image": "gold-standard-PhC-plus-2/FOV12_20p_PhC_present_crop_1_im.tif",
134 | "label": "gold-standard-PhC-plus-2/FOV12_20p_PhC_present_crop_1_mask.tif",
135 | "fold": 2
136 | },
137 | {
138 | "image": "gold-standard-PhC-plus-2/FOV9_20p_PhC_present_crop_1_im.tif",
139 | "label": "gold-standard-PhC-plus-2/FOV9_20p_PhC_present_crop_1_mask.tif",
140 | "fold": 2
141 | },
142 | {
143 | "image": "gold-standard-PhC-plus-2/FOV15_20p_PhC_present_crop_1_im.tif",
144 | "label": "gold-standard-PhC-plus-2/FOV15_20p_PhC_present_crop_1_mask.tif",
145 | "fold": 3
146 | },
147 | {
148 | "image": "gold-standard-PhC-plus-2/FOV7_20p_PhC_present_crop_1_im.tif",
149 | "label": "gold-standard-PhC-plus-2/FOV7_20p_PhC_present_crop_1_mask.tif",
150 | "fold": 3
151 | },
152 | {
153 | "image": "gold-standard-PhC-plus-2/FOV2_20p_PhC_present_crop_1_im.tif",
154 | "label": "gold-standard-PhC-plus-2/FOV2_20p_PhC_present_crop_1_mask.tif",
155 | "fold": 3
156 | },
157 | {
158 | "image": "gold-standard-PhC-plus-2/V11032020_p1_crop_1_im.tif",
159 | "label": "gold-standard-PhC-plus-2/V11032020_p1_crop_1_mask.tif",
160 | "fold": 3
161 | },
162 | {
163 | "image": "gold-standard-PhC-plus-2/v_cdc20null_crop_1_im.tif",
164 | "label": "gold-standard-PhC-plus-2/v_cdc20null_crop_1_mask.tif",
165 | "fold": 3
166 | },
167 | {
168 | "image": "gold-standard-PhC-plus-2/FOV3_20p_PhC_present_crop_1_im.tif",
169 | "label": "gold-standard-PhC-plus-2/FOV3_20p_PhC_present_crop_1_mask.tif",
170 | "fold": 3
171 | },
172 | {
173 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_1_im.tif",
174 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_16_crop_1_mask.tif",
175 | "fold": 3
176 | },
177 | {
178 | "image": "gold-standard-PhC-plus-2/d_reexport1_crop_1_im.tif",
179 | "label": "gold-standard-PhC-plus-2/d_reexport1_crop_1_mask.tif",
180 | "fold": 4
181 | },
182 | {
183 | "image": "gold-standard-PhC-plus-2/FOV6_20p_PhC_present_crop_1_im.tif",
184 | "label": "gold-standard-PhC-plus-2/FOV6_20p_PhC_present_crop_1_mask.tif",
185 | "fold": 4
186 | },
187 | {
188 | "image": "gold-standard-PhC-plus-2/V11032020_p2_crop_1_im.tif",
189 | "label": "gold-standard-PhC-plus-2/V11032020_p2_crop_1_mask.tif",
190 | "fold": 4
191 | },
192 | {
193 | "image": "gold-standard-PhC-plus-2/a_reexport3_crop_1_im.tif",
194 | "label": "gold-standard-PhC-plus-2/a_reexport3_crop_1_mask.tif",
195 | "fold": 4
196 | },
197 | {
198 | "image": "gold-standard-PhC-plus-2/a_reexport1_crop_1_im.tif",
199 | "label": "gold-standard-PhC-plus-2/a_reexport1_crop_1_mask.tif",
200 | "fold": 4
201 | },
202 | {
203 | "image": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_3_im.tif",
204 | "label": "gold-standard-PhC-plus-2/Ahmad_frame_19_crop_3_mask.tif",
205 | "fold": 4
206 | },
207 | {
208 | "image": "gold-standard-PhC-plus-2/a_reexport2_crop_1_im.tif",
209 | "label": "gold-standard-PhC-plus-2/a_reexport2_crop_1_mask.tif",
210 | "fold": 4
211 | }
212 | ]
213 | }
214 |
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/all_file_downloader.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import os
3 |
4 | import requests
5 | from tqdm import tqdm
6 |
7 |
8 | def download_files(url_dict, directory):
9 | if not os.path.exists(directory):
10 | os.makedirs(directory)
11 |
12 | for key, url in url_dict.items():
13 | if key == "nips_train.zip" or key == "nips_test.zip":
14 | if not os.path.exists(os.path.join(directory, "nips_dataset")):
15 | os.mkdir(os.path.join(directory, "nips_dataset"))
16 | base_dir = os.path.join(directory, "nips_dataset")
17 | elif key == "deepbacs.zip":
18 | if not os.path.exists(os.path.join(directory, "deepbacs_dataset")):
19 | os.mkdir(os.path.join(directory, "deepbacs_dataset"))
20 | base_dir = os.path.join(directory, "deepbacs_dataset")
21 | elif key == "livecell":
22 | if not os.path.exists(os.path.join(directory, "livecell_dataset")):
23 | os.mkdir(os.path.join(directory, "livecell_dataset"))
24 | base_dir = os.path.join(directory, "livecell_dataset")
25 | print(f"Downloading from {key}: {url}")
26 | os.system(url + base_dir)
27 | continue
28 |
29 | try:
30 | print(f"Downloading from {key}: {url}")
31 | response = requests.get(url, stream=True, allow_redirects=True)
32 | total_size = int(response.headers.get("content-length", 0))
33 |
34 | # Extract the filename from the URL or use the key as the filename
35 | filename = os.path.basename(key)
36 | file_path = os.path.join(base_dir, filename)
37 |
38 | # Write the content to a file in the specified directory with progress
39 | with open(file_path, "wb") as file, tqdm(
40 | desc=filename,
41 | total=total_size,
42 | unit="iB",
43 | unit_scale=True,
44 | unit_divisor=1024,
45 | ) as bar:
46 | for data in response.iter_content(chunk_size=1024):
47 | size = file.write(data)
48 | bar.update(size)
49 |
50 | print(f"Saved to {file_path}")
51 | except Exception as e:
52 | print(f"Failed to download from {key} ({url}). Reason: {str(e)}")
53 |
54 |
55 | def main():
56 | parser = argparse.ArgumentParser(description="Process some integers.")
57 | parser.add_argument(
58 | "--dir",
59 | type=str,
60 | help="Directory to download files to",
61 | default="/set/the/path",
62 | )
63 |
64 | args = parser.parse_args()
65 | directory = os.path.normpath(args.dir)
66 |
67 | url_dict = {
68 | "deepbacs.zip": "https://zenodo.org/records/5551009/files/DeepBacs_Data_Segmentation_StarDist_MIXED_dataset.zip?download=1",
69 | "nips_test.zip": "https://zenodo.org/records/10719375/files/Testing.zip?download=1",
70 | "nips_train.zip": "https://zenodo.org/records/10719375/files/Training-labeled.zip?download=1",
71 | "livecell": "wget --recursive --no-parent --cut-dirs=0 --timestamping -i urls.txt --directory-prefix=",
72 | # Add URLs with keys here
73 | }
74 | download_files(url_dict, directory)
75 |
76 |
77 | if __name__ == "__main__":
78 | main()
79 |
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/cellpose_agreement.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/cellpose_agreement.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/cellpose_links.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/cellpose_links.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/data_tree.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/data_tree.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/kaggle_download.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/kaggle_download.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/omnipose_download.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/omnipose_download.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/readme.md:
--------------------------------------------------------------------------------
1 | ## Tutorial: VISTA2D Model Creation
2 |
3 | This tutorial will guide the users to setting up all the datasets, running pre-processing, creation of organized json file lists which can be provided to VISTA-2D training pipeline.
4 | Some dataset need to be manually downloaded, others will be downloaded by a provided script. Please do not manually unzip any of the downloaded files, it will be automatically handled in the final step.
5 |
6 | ### List of Datasets
7 | 1.) [Cellpose](https://www.cellpose.org/dataset)
8 |
9 | 2.) [TissueNet](https://datasets.deepcell.org/login)
10 |
11 | 3.) [Kaggle Nuclei Segmentation](https://www.kaggle.com/c/data-science-bowl-2018/data)
12 |
13 | 4.) [Omnipose - OSF repository](https://osf.io/xmury/)
14 |
15 | 5.) [NIPS Cell Segmentation Challenge](https://neurips22-cellseg.grand-challenge.org/)
16 |
17 | 6.) [LiveCell](https://sartorius-research.github.io/LIVECell/)
18 |
19 | 7.) [Deepbacs](https://github.com/HenriquesLab/DeepBacs/wiki/Segmentation)
20 |
21 | Datasets 1-4 need to be manually downloaded, instructions to download them have been provided below.
22 |
23 | ### Manual Dataset Download Instructions
24 | #### 1.) Cellpose:
25 | The dataset can be downloaded from this [link](https://www.cellpose.org/dataset). Please see below screenshots to assist in downloading it
26 | 
27 | Please enter your email and accept terms and conditions to download the dataset.
28 |
29 | 
30 | Click on train.zip and test.zip to download both directories independently. They both need to be placed in a `cellpose_dataset` directory. The `cellpose_dataset` will have to be created by the user in the root data directory.
31 |
32 | #### 2.) TissueNet
33 | Login credentials have to be created at below provided link. Please see below screenshots for further assistance.
34 |
35 | 
36 | Please create an account at the provided [link](https://datasets.deepcell.org/login).
37 |
38 | 
39 | After logging in, the above page will be visible, please make sure that version 1.0 is selected for TissueNet before clicking on download button.
40 | All the downloaded files need to be placed in a `tissuenet_dataset` directory, this directory has to be created by the user.
41 |
42 | #### 3.) Kaggle Nuclei Segmentation
43 | Kaggle credentials are required in order to access this dataset at this [link](https://www.kaggle.com/c/data-science-bowl-2018/data), the user will have to register for the challenge to access and download the dataset.
44 | Please refer below screenshots for additional help.
45 |
46 | 
47 | The `Download All` button needs to be used so all files are downloaded, the files need to be placed in a directory created by the user `kaggle_dataset`.
48 |
49 | #### 4.) Omnipose
50 | The Omnipose dataset is hosted on an [OSF repository](https://osf.io/xmury/) and the dataset part needs to be downloaded from it. Please refer below screenshots for further assistance.
51 |
52 | 
53 | The `datasets` directory needs to be selected as highlighted in the screenshot, then `download as zip` needs to be pressed for downloading the dataset. The user will have to place all the files in
54 | a user created directory named `omnipose_dataset`.
55 |
56 | ### The remaining datasets will be downloaded by a python script.
57 | To run the script use the following example command `python all_file_downloader.py --download_path provide_the_same_root_data_path`
58 |
59 | After completion of downloading of all datasets, below is how the data root directory should look:
60 |
61 | 
62 |
63 | ### Process the downloaded data
64 | To execute VISTA-2D training pipeline, some datasets require label conversion. Please use the `root_data_path` as the input to the script, example command to execute the script is given below:
65 |
66 | `python generate_json.py --data_root provide_the_same_root_data_path`
67 |
68 | ### Generation of Json data lists (Optional)
69 | If one desires to generate JSON files from scratch, `generate_json.py` script performs both processing and creation of JSON files.
70 | To execute VISTA-2D training pipeline, some datasets require label conversion and then a json file list which the VISTA-2D training uses a format.
71 | Creating the json lists from the raw dataset sources, please use the `root_data_path` as the input to the script, example command to execute the script is given below:
72 |
73 | `python generate_json.py --data_root provide_the_same_root_data_path`
74 |
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/tissuenet_download.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/tissuenet_download.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/tissuenet_login.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/download_preprocessor/tissuenet_login.png
--------------------------------------------------------------------------------
/vista2d/download_preprocessor/urls.txt:
--------------------------------------------------------------------------------
1 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LICENSE
2 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/
3 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/
4 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell/
5 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/
6 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/
7 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/test.json
8 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/train.json
9 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/val.json
10 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/
11 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/test.json
12 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/train.json
13 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/val.json
14 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/
15 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/test.json
16 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/train.json
17 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/val.json
18 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/
19 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/test.json
20 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/train.json
21 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/val.json
22 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/
23 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/test.json
24 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/train.json
25 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/val.json
26 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/
27 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/test.json
28 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/train.json
29 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/val.json
30 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/
31 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/test.json
32 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/train.json
33 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/val.json
34 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/
35 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/test.json
36 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/train.json
37 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/val.json
38 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/images_per_celltype.zip
39 | http://livecell-dataset.s3.eu-central-1.amazonaws.com/README.md
40 |
--------------------------------------------------------------------------------
/vista2d/large_files.yml:
--------------------------------------------------------------------------------
1 | large_files:
2 | - path: "models/sam_vit_b_01ec64.pth"
3 | url: "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth"
4 |
--------------------------------------------------------------------------------
/vista2d/scripts/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista2d/scripts/__init__.py
--------------------------------------------------------------------------------
/vista2d/scripts/cell_distributed_weighted_sampler.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | # based on Pytorch DistributedSampler and WeightedRandomSampler combined
13 |
14 | import math
15 | from typing import Iterator, Optional, Sequence, TypeVar
16 |
17 | import torch
18 | import torch.distributed as dist
19 | from torch.utils.data import Dataset, Sampler
20 |
21 | __all__ = ["DistributedWeightedSampler"]
22 |
23 | T_co = TypeVar("T_co", covariant=True)
24 |
25 |
26 | class DistributedWeightedSampler(Sampler[T_co]):
27 | def __init__(
28 | self,
29 | dataset: Dataset,
30 | weights: Sequence[float],
31 | num_samples: int,
32 | num_replicas: Optional[int] = None,
33 | rank: Optional[int] = None,
34 | shuffle: bool = True,
35 | seed: int = 0,
36 | drop_last: bool = False,
37 | ) -> None:
38 | if (
39 | not isinstance(num_samples, int)
40 | or isinstance(num_samples, bool)
41 | or num_samples <= 0
42 | ):
43 | raise ValueError(
44 | f"num_samples should be a positive integer value, but got num_samples={num_samples}"
45 | )
46 |
47 | weights_tensor = torch.as_tensor(weights, dtype=torch.float)
48 | if len(weights_tensor.shape) != 1:
49 | raise ValueError(
50 | "weights should be a 1d sequence but given "
51 | f"weights have shape {tuple(weights_tensor.shape)}"
52 | )
53 |
54 | self.weights = weights_tensor
55 | self.num_samples = num_samples
56 |
57 | if num_replicas is None:
58 | if not dist.is_available():
59 | raise RuntimeError("Requires distributed package to be available")
60 | num_replicas = dist.get_world_size()
61 | if rank is None:
62 | if not dist.is_available():
63 | raise RuntimeError("Requires distributed package to be available")
64 | rank = dist.get_rank()
65 | if rank >= num_replicas or rank < 0:
66 | raise ValueError(
67 | f"Invalid rank {rank}, rank should be in the interval [0, {num_replicas - 1}]"
68 | )
69 | self.dataset = dataset
70 | self.num_replicas = num_replicas
71 | self.rank = rank
72 | self.epoch = 0
73 | self.drop_last = drop_last
74 | self.shuffle = shuffle
75 |
76 | if self.shuffle:
77 | self.num_samples = int(math.ceil(self.num_samples / self.num_replicas))
78 | else:
79 | # this is not used, as we always shuffle, the only reason to use this class
80 |
81 | # If the dataset length is evenly divisible by # of replicas, then there
82 | # is no need to drop any data, since the dataset will be split equally.
83 | if self.drop_last and len(self.dataset) % self.num_replicas != 0: # type: ignore[arg-type]
84 | # Split to nearest available length that is evenly divisible.
85 | # This is to ensure each rank receives the same amount of data when
86 | # using this Sampler.
87 | self.num_samples = math.ceil(
88 | (len(self.dataset) - self.num_replicas) / self.num_replicas # type: ignore[arg-type]
89 | )
90 | else:
91 | self.num_samples = math.ceil(len(self.dataset) / self.num_replicas) # type: ignore[arg-type]
92 |
93 | self.total_size = self.num_samples * self.num_replicas
94 | self.shuffle = shuffle
95 | self.seed = seed
96 |
97 | def __iter__(self) -> Iterator[T_co]:
98 | if self.shuffle:
99 | # deterministically shuffle based on epoch and seed
100 | g = torch.Generator()
101 | g.manual_seed(self.seed + self.epoch)
102 | indices = torch.multinomial(input=self.weights, num_samples=self.total_size, replacement=True, generator=g).tolist() # type: ignore[arg-type]
103 | else:
104 | # this is not used, as we always shuffle, the only reason to use this class
105 | indices = list(range(len(self.dataset))) # type: ignore[arg-type]
106 | if not self.drop_last:
107 | # add extra samples to make it evenly divisible
108 | padding_size = self.total_size - len(indices)
109 | if padding_size <= len(indices):
110 | indices += indices[:padding_size]
111 | else:
112 | indices += (indices * math.ceil(padding_size / len(indices)))[
113 | :padding_size
114 | ]
115 | else:
116 | # remove tail of data to make it evenly divisible.
117 | indices = indices[: self.total_size]
118 | assert len(indices) == self.total_size
119 |
120 | # subsample
121 | indices = indices[self.rank : self.total_size : self.num_replicas]
122 | assert len(indices) == self.num_samples
123 |
124 | return iter(indices)
125 |
126 | def __len__(self) -> int:
127 | return self.num_samples
128 |
129 | def set_epoch(self, epoch: int) -> None:
130 | self.epoch = epoch
131 |
--------------------------------------------------------------------------------
/vista2d/scripts/cell_sam_wrapper.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import torch
13 | from segment_anything.build_sam import build_sam_vit_b
14 | from torch import nn
15 | from torch.nn import functional as F
16 |
17 |
18 | class CellSamWrapper(torch.nn.Module):
19 | def __init__(
20 | self,
21 | auto_resize_inputs=True,
22 | network_resize_roi=[1024, 1024],
23 | checkpoint="sam_vit_b_01ec64.pth",
24 | return_features=False,
25 | *args,
26 | **kwargs,
27 | ) -> None:
28 | super().__init__(*args, **kwargs)
29 |
30 | print(
31 | f"CellSamWrapper auto_resize_inputs {auto_resize_inputs} network_resize_roi {network_resize_roi} checkpoint {checkpoint}"
32 | )
33 | self.network_resize_roi = network_resize_roi
34 | self.auto_resize_inputs = auto_resize_inputs
35 | self.return_features = return_features
36 |
37 | model = build_sam_vit_b(checkpoint=checkpoint)
38 |
39 | model.prompt_encoder = None
40 | model.mask_decoder = None
41 |
42 | model.mask_decoder = nn.Sequential(
43 | nn.BatchNorm2d(num_features=256),
44 | nn.ReLU(inplace=True),
45 | nn.ConvTranspose2d(
46 | 256,
47 | 128,
48 | kernel_size=3,
49 | stride=2,
50 | padding=1,
51 | output_padding=1,
52 | bias=False,
53 | ),
54 | nn.BatchNorm2d(num_features=128),
55 | nn.ReLU(inplace=True),
56 | nn.ConvTranspose2d(
57 | 128, 3, kernel_size=3, stride=2, padding=1, output_padding=1, bias=True
58 | ),
59 | )
60 |
61 | self.model = model
62 |
63 | def forward(self, x):
64 | # print("CellSamWrapper x0", x.shape)
65 | sh = x.shape[2:]
66 |
67 | if self.auto_resize_inputs:
68 | x = F.interpolate(x, size=self.network_resize_roi, mode="bilinear")
69 |
70 | # print("CellSamWrapper x1", x.shape)
71 | x = self.model.image_encoder(x) # shape: (1, 256, 64, 64)
72 | # print("CellSamWrapper image_embeddings", x.shape)
73 |
74 | if not self.return_features:
75 | x = self.model.mask_decoder(x)
76 | if self.auto_resize_inputs:
77 | x = F.interpolate(x, size=sh, mode="bilinear")
78 |
79 | # print("CellSamWrapper x final", x.shape)
80 | return x
81 |
--------------------------------------------------------------------------------
/vista2d/scripts/utils.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import logging
13 | import os
14 | import warnings
15 | from logging.config import fileConfig
16 | from pathlib import Path
17 |
18 | import numpy as np
19 | from monai.apps import get_logger
20 | from monai.apps.utils import DEFAULT_FMT
21 | from monai.bundle import ConfigParser
22 | from monai.utils import RankFilter, ensure_tuple
23 |
24 | logger = get_logger("VistaCell")
25 |
26 | np.set_printoptions(formatter={"float": "{: 0.3f}".format}, suppress=True)
27 | logging.getLogger("torch.nn.parallel.distributed").setLevel(logging.WARNING)
28 | warnings.filterwarnings(
29 | "ignore", message=".*Divide by zero.*"
30 | ) # intensity transform divide by zero warning
31 |
32 | LOGGING_CONFIG = {
33 | "version": 1,
34 | "disable_existing_loggers": False,
35 | "formatters": {"monai_default": {"format": DEFAULT_FMT}},
36 | "loggers": {
37 | "VistaCell": {
38 | "handlers": ["file", "console"],
39 | "level": "DEBUG",
40 | "propagate": False,
41 | },
42 | },
43 | "filters": {"rank_filter": {"()": RankFilter}},
44 | "handlers": {
45 | "file": {
46 | "class": "logging.FileHandler",
47 | "filename": "default.log",
48 | "mode": "a", # append or overwrite
49 | "level": "DEBUG",
50 | "formatter": "monai_default",
51 | "filters": ["rank_filter"],
52 | },
53 | "console": {
54 | "class": "logging.StreamHandler",
55 | "level": "INFO",
56 | "formatter": "monai_default",
57 | "filters": ["rank_filter"],
58 | },
59 | },
60 | }
61 |
62 |
63 | def parsing_bundle_config(config_file, logging_file=None, meta_file=None):
64 | if config_file is not None:
65 | _config_files = ensure_tuple(config_file)
66 | config_root_path = Path(_config_files[0]).parent
67 | for _config_file in _config_files:
68 | _config_file = Path(_config_file)
69 | if _config_file.parent != config_root_path:
70 | logger.warning(
71 | f"Not all config files are in '{config_root_path}'. If logging_file and meta_file are"
72 | f"not specified, '{config_root_path}' will be used as the default config root directory."
73 | )
74 | if not _config_file.is_file():
75 | raise FileNotFoundError(f"Cannot find the config file: {_config_file}.")
76 | else:
77 | config_root_path = Path("configs")
78 |
79 | logging_file = (
80 | str(config_root_path / "logging.conf") if logging_file is None else logging_file
81 | )
82 | if os.path.exists(logging_file):
83 | fileConfig(logging_file, disable_existing_loggers=False)
84 |
85 | parser = ConfigParser()
86 | parser.read_config(config_file)
87 | meta_file = (
88 | str(config_root_path / "metadata.json") if meta_file is None else meta_file
89 | )
90 | if isinstance(meta_file, str) and not os.path.exists(meta_file):
91 | logger.error(
92 | f"Cannot find the metadata config file: {meta_file}. "
93 | "Please see: https://docs.monai.io/en/stable/mb_specification.html"
94 | )
95 | else:
96 | parser.read_meta(f=meta_file)
97 |
98 | return parser
99 |
--------------------------------------------------------------------------------
/vista2d/setup.cfg:
--------------------------------------------------------------------------------
1 | [flake8]
2 | select = B,C,E,F,N,P,T4,W,B9
3 | max_line_length = 120
4 | # F403 'from module import *' used; unable to detect undefined names
5 | # F405 Name may be undefined, or defined from star import
6 | # E203 whitespace before ':' # incompatible with black style
7 | # E402 module level import not at top of file
8 | # E501 is not flexible enough, we're using B950 instead
9 | # E722 do not use bare 'except'
10 | # C408 ignored because we like the dict keyword argument syntax
11 | # W503 line break before binary operator
12 | ignore =
13 | E203,E302,E303,E305,E402,E501,E721,E722,E741,F403,F405,F821,F841,F999,C408,W291,W503,W504,B008,
14 | # N812 lowercase 'torch.nn.functional' imported as non lowercase 'F'
15 | N812,N818
16 | per_file_ignores = __init__.py: F401
17 | exclude = *.pyi,.git,.eggs,_version.py,versioneer.py,venv,.venv,_version.py
18 |
--------------------------------------------------------------------------------
/vista2d/unit_tests/test_vista2d.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import os
13 | import shutil
14 | import sys
15 | import tempfile
16 | import unittest
17 |
18 | import matplotlib.pyplot as plt
19 | import numpy as np
20 | from monai.bundle import create_workflow
21 | from parameterized import parameterized
22 | from utils import check_workflow
23 |
24 | TEST_CASE_TRAIN = [
25 | {
26 | "bundle_root": ".",
27 | "mode": "train",
28 | "train#trainer#max_epochs": 1,
29 | }
30 | ]
31 |
32 | TEST_CASE_INFER = [
33 | {
34 | "bundle_root": ".",
35 | "mode": "infer",
36 | }
37 | ]
38 |
39 |
40 | class TestVista2d(unittest.TestCase):
41 | def setUp(self):
42 | self.dataset_dir = tempfile.mkdtemp()
43 | self.tmp_output_dir = os.path.join(self.dataset_dir, "output")
44 | os.makedirs(self.tmp_output_dir, exist_ok=True)
45 | self.dataset_size = 5
46 | input_shape = (256, 256)
47 | for s in range(self.dataset_size):
48 | test_image = np.random.randint(low=0, high=2, size=input_shape).astype(
49 | np.int8
50 | )
51 | test_label = np.random.randint(low=0, high=2, size=input_shape).astype(
52 | np.int8
53 | )
54 | image_filename = os.path.join(self.dataset_dir, f"image_{s}.png")
55 | label_filename = os.path.join(self.dataset_dir, f"label_{s}.png")
56 | plt.imsave(image_filename, test_image, cmap="gray")
57 | plt.imsave(label_filename, test_label, cmap="gray")
58 |
59 | self.bundle_root = "."
60 | sys.path = [self.bundle_root] + sys.path
61 | from scripts.workflow import VistaCell
62 |
63 | self.workflow = VistaCell
64 |
65 | def tearDown(self):
66 | shutil.rmtree(self.dataset_dir)
67 |
68 | @parameterized.expand([TEST_CASE_INFER])
69 | def test_infer_config(self, override):
70 | # update override with dataset dir
71 | override["dataset#data"] = [
72 | {
73 | "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
74 | "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
75 | }
76 | for s in range(self.dataset_size)
77 | ]
78 | override["output_dir"] = self.tmp_output_dir
79 | workflow = create_workflow(
80 | workflow_name=self.workflow,
81 | config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
82 | meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
83 | **override,
84 | )
85 |
86 | # check_properties=False, need to add monai service properties later
87 | check_workflow(workflow, check_properties=False)
88 |
89 | expected_output_file = os.path.join(
90 | self.tmp_output_dir, f"image_{self.dataset_size-1}.tif"
91 | )
92 | self.assertTrue(os.path.isfile(expected_output_file))
93 |
94 | @parameterized.expand([TEST_CASE_TRAIN])
95 | def test_train_config(self, override):
96 | # update override with dataset dir
97 | override["train#dataset#data"] = [
98 | {
99 | "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
100 | "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
101 | }
102 | for s in range(self.dataset_size)
103 | ]
104 | override["dataset#data"] = override["train#dataset#data"]
105 |
106 | workflow = create_workflow(
107 | workflow_name=self.workflow,
108 | config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
109 | meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
110 | **override,
111 | )
112 |
113 | # check_properties=False, need to add monai service properties later
114 | check_workflow(workflow, check_properties=False)
115 |
116 | # follow up to use trained weights and test eval
117 | override["mode"] = "eval"
118 | override["pretrained_ckpt_name"] = "model.pt"
119 | workflow = create_workflow(
120 | workflow_name=self.workflow,
121 | config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
122 | meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
123 | **override,
124 | )
125 | check_workflow(workflow, check_properties=False)
126 |
127 |
128 | if __name__ == "__main__":
129 | unittest.main()
130 |
--------------------------------------------------------------------------------
/vista2d/unit_tests/test_vista2d_mgpu.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import os
13 | import shutil
14 | import sys
15 | import tempfile
16 | import unittest
17 |
18 | import matplotlib.pyplot as plt
19 | import numpy as np
20 | import torch
21 | from parameterized import parameterized
22 | from utils import export_config_and_run_mgpu_cmd
23 |
24 | TEST_CASE_TRAIN_MGPU = [
25 | {
26 | "bundle_root": ".",
27 | "workflow_type": "train",
28 | "train#trainer#max_epochs": 2,
29 | }
30 | ]
31 |
32 |
33 | class TestVista2d(unittest.TestCase):
34 | def setUp(self):
35 | self.dataset_dir = tempfile.mkdtemp()
36 | self.dataset_size = 5
37 | input_shape = (256, 256)
38 | for s in range(self.dataset_size):
39 | test_image = np.random.randint(low=0, high=2, size=input_shape).astype(
40 | np.int8
41 | )
42 | test_label = np.random.randint(low=0, high=2, size=input_shape).astype(
43 | np.int8
44 | )
45 | image_filename = os.path.join(self.dataset_dir, f"image_{s}.png")
46 | label_filename = os.path.join(self.dataset_dir, f"label_{s}.png")
47 | plt.imsave(image_filename, test_image, cmap="gray")
48 | plt.imsave(label_filename, test_label, cmap="gray")
49 |
50 | self.bundle_root = "."
51 | sys.path = [self.bundle_root] + sys.path
52 | from scripts.workflow import VistaCell
53 |
54 | self.workflow = VistaCell
55 |
56 | def tearDown(self):
57 | shutil.rmtree(self.dataset_dir)
58 |
59 | @parameterized.expand([TEST_CASE_TRAIN_MGPU])
60 | def test_train_mgpu_config(self, override):
61 | override["train#dataset#data"] = [
62 | {
63 | "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
64 | "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
65 | }
66 | for s in range(self.dataset_size)
67 | ]
68 | override["dataset#data"] = override["train#dataset#data"]
69 |
70 | output_path = os.path.join(self.bundle_root, "configs/train_override.json")
71 | n_gpu = torch.cuda.device_count()
72 | export_config_and_run_mgpu_cmd(
73 | workflow_name=self.workflow,
74 | bundle_root=self.bundle_root,
75 | config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
76 | meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
77 | override_dict=override,
78 | output_path=output_path,
79 | ngpu=n_gpu,
80 | )
81 |
82 |
83 | if __name__ == "__main__":
84 | unittest.main()
85 |
--------------------------------------------------------------------------------
/vista2d/unit_tests/utils.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 |
13 | import os
14 | import subprocess
15 |
16 | from monai.bundle import ConfigParser, create_workflow
17 |
18 |
19 | def export_overrided_config(config_file, override_dict, output_path):
20 | parser = ConfigParser()
21 | parser.read_config(config_file)
22 | parser.update(pairs=override_dict)
23 | ConfigParser.export_config_file(parser.config, output_path, indent=4)
24 |
25 |
26 | def produce_mgpu_cmd(config_file, meta_file, nnodes=1, nproc_per_node=2):
27 | cmd = [
28 | "torchrun",
29 | # "--standalone",
30 | f"--nnodes={nnodes}",
31 | f"--nproc_per_node={nproc_per_node}",
32 | "-m",
33 | "monai.bundle",
34 | "run_workflow",
35 | "scripts.workflow.VistaCell",
36 | "--config_file",
37 | config_file,
38 | "--meta_file",
39 | meta_file,
40 | ]
41 | return cmd
42 |
43 |
44 | def export_config_and_run_mgpu_cmd(
45 | config_file,
46 | meta_file,
47 | override_dict,
48 | output_path,
49 | workflow_name,
50 | bundle_root,
51 | workflow_type="train",
52 | nnode=1,
53 | ngpu=2,
54 | check_config=False,
55 | ):
56 | """
57 | step 1: override the config file and export it
58 | step 2: (optional) check the exported config file
59 | step 3: produce multi-gpu running command
60 | step 4: run produced command
61 | """
62 | export_overrided_config(
63 | config_file=config_file, override_dict=override_dict, output_path=output_path
64 | )
65 | if check_config is True:
66 | workflow = create_workflow(
67 | workflow_name=workflow_name,
68 | config_file=os.path.join(bundle_root, "configs/hyper_parameters.yaml"),
69 | meta_file=os.path.join(bundle_root, "configs/metadata.json"),
70 | **override_dict,
71 | )
72 | check_result = workflow.check_properties()
73 | if check_result is not None and len(check_result) > 0:
74 | raise ValueError(
75 | f"check properties for overrided mgpu configs failed: {check_result}"
76 | )
77 | cmd = produce_mgpu_cmd(
78 | config_file=output_path, meta_file=meta_file, nnodes=nnode, nproc_per_node=ngpu
79 | )
80 | env = os.environ.copy()
81 | # ensure customized library can be loaded in subprocess
82 | env["PYTHONPATH"] = override_dict.get("bundle_root", ".")
83 | subprocess.check_call(cmd, env=env)
84 |
85 |
86 | def check_workflow(workflow, check_properties: bool = False):
87 | if check_properties is True:
88 | check_result = workflow.check_properties()
89 | if check_result is not None and len(check_result) > 0:
90 | raise ValueError(f"check properties for workflow failed: {check_result}")
91 | workflow.run()
92 | workflow.finalize()
93 |
--------------------------------------------------------------------------------
/vista3d/NVIDIA OneWay Noncommercial License.txt:
--------------------------------------------------------------------------------
1 | NVIDIA License
2 |
3 | 1. Definitions
4 |
5 | “Licensor” means any person or entity that distributes its Work.
6 | “Work” means (a) the original work of authorship made available under this license, which may include software, documentation, or other files, and (b) any additions to or derivative works thereof that are made available under this license.
7 | The terms “reproduce,” “reproduction,” “derivative works,” and “distribution” have the meaning as provided under U.S. copyright law; provided, however, that for the purposes of this license, derivative works shall not include works that remain separable from, or merely link (or bind by name) to the interfaces of, the Work.
8 | Works are “made available” under this license by including in or with the Work either (a) a copyright notice referencing the applicability of this license to the Work, or (b) a copy of this license.
9 |
10 | 2. License Grant
11 |
12 | 2.1 Copyright Grant. Subject to the terms and conditions of this license, each Licensor grants to you a perpetual, worldwide, non-exclusive, royalty-free, copyright license to use, reproduce, prepare derivative works of, publicly display, publicly perform, sublicense and distribute its Work and any resulting derivative works in any form.
13 |
14 | 3. Limitations
15 |
16 | 3.1 Redistribution. You may reproduce or distribute the Work only if (a) you do so under this license, (b) you include a complete copy of this license with your distribution, and (c) you retain without modification any copyright, patent, trademark, or attribution notices that are present in the Work.
17 |
18 | 3.2 Derivative Works. You may specify that additional or different terms apply to the use, reproduction, and distribution of your derivative works of the Work (“Your Terms”) only if (a) Your Terms provide that the use limitation in Section 3.3 applies to your derivative works, and (b) you identify the specific derivative works that are subject to Your Terms. Notwithstanding Your Terms, this license (including the redistribution requirements in Section 3.1) will continue to apply to the Work itself.
19 |
20 | 3.3 Use Limitation. The Work and any derivative works thereof only may be used or intended for use non-commercially. Notwithstanding the foregoing, NVIDIA Corporation and its affiliates may use the Work and any derivative works commercially. As used herein, “non-commercially” means for research or evaluation purposes only.
21 |
22 | 3.4 Patent Claims. If you bring or threaten to bring a patent claim against any Licensor (including any claim, cross-claim or counterclaim in a lawsuit) to enforce any patents that you allege are infringed by any Work, then your rights under this license from such Licensor (including the grant in Section 2.1) will terminate immediately.
23 |
24 | 3.5 Trademarks. This license does not grant any rights to use any Licensor’s or its affiliates’ names, logos, or trademarks, except as necessary to reproduce the notices described in this license.
25 |
26 | 3.6 Termination. If you violate any term of this license, then your rights under this license (including the grant in Section 2.1) will terminate immediately.
27 |
28 | 4. Disclaimer of Warranty.
29 |
30 | THE WORK IS PROVIDED “AS IS” WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, EITHER EXPRESS OR IMPLIED, INCLUDING WARRANTIES OR CONDITIONS OF
31 | MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE, TITLE OR NON-INFRINGEMENT. YOU BEAR THE RISK OF UNDERTAKING ANY ACTIVITIES UNDER THIS LICENSE.
32 |
33 | 5. Limitation of Liability.
34 |
35 | EXCEPT AS PROHIBITED BY APPLICABLE LAW, IN NO EVENT AND UNDER NO LEGAL THEORY, WHETHER IN TORT (INCLUDING NEGLIGENCE), CONTRACT, OR OTHERWISE SHALL ANY LICENSOR BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES ARISING OUT OF OR RELATED TO THIS LICENSE, THE USE OR INABILITY TO USE THE WORK (INCLUDING BUT NOT LIMITED TO LOSS OF GOODWILL, BUSINESS INTERRUPTION, LOST PROFITS OR DATA, COMPUTER FAILURE OR MALFUNCTION, OR ANY OTHER DAMAGES OR LOSSES), EVEN IF THE LICENSOR HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGES.
36 |
--------------------------------------------------------------------------------
/vista3d/assets/imgs/demo_gif.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/demo_gif.gif
--------------------------------------------------------------------------------
/vista3d/assets/imgs/everything.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/everything.gif
--------------------------------------------------------------------------------
/vista3d/assets/imgs/finetune.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/finetune.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/liver.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/liver.gif
--------------------------------------------------------------------------------
/vista3d/assets/imgs/model.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/model.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/montage.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/montage.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/sam2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/sam2.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/scores.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/scores.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/unspecified.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/unspecified.gif
--------------------------------------------------------------------------------
/vista3d/assets/imgs/wholeBody.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/wholeBody.png
--------------------------------------------------------------------------------
/vista3d/assets/imgs/zeroshot.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/Project-MONAI/VISTA/8bb7572d163373ed73d136d2c0eacb152f79664c/vista3d/assets/imgs/zeroshot.gif
--------------------------------------------------------------------------------
/vista3d/configs/finetune/infer_patch_auto_murine.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | output_path: "$'/workspace/vista3d/work_dir_finetune_murine_' + str(@train_number)"
3 | ckpt: "$@output_path + '/model_fold0/best_metric_model.pt'"
4 | dataset_name: "murine"
5 | label_set: [0,1,2,3,4]
6 | mapped_label_set: [0,115,121,30,28]
7 | val_auto: true
8 | overlap: 0.625
9 | data_file_base_dir: '/data/micro-ct-murine/1_nativeCTdata_nifti/'
10 | data_list_file_path: './data/external/micro-ct-murine-native_5_folds.json'
11 | log_output_file: "$@output_path + '/test_set.log'"
12 | list_key: 'testing'
13 | five_fold: true
14 | fold: 0
15 | train_number: 89
16 | argmax_first: false
17 | input_channels: 1
18 | image_key: image
19 | label_key: label
20 | pixdim: [1,1,1]
21 | patch_size: [128, 128, 128]
22 | transforms_infer:
23 | _target_: Compose
24 | transforms:
25 | - _target_: LoadImaged
26 | ensure_channel_first: true
27 | image_only: true
28 | keys: ['@image_key','@label_key']
29 | - _target_: CopyItemsd
30 | names: 'label_gt'
31 | keys: '@label_key'
32 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
33 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
34 | - _target_: Orientationd
35 | axcodes: RAS
36 | keys: ['@image_key','@label_key']
37 | - _target_: CastToTyped
38 | dtype: [$torch.float32, $torch.uint8]
39 | keys: ['@image_key','@label_key']
40 | - _target_: EnsureTyped
41 | keys: ['@image_key','@label_key']
42 | track_meta: true
43 | model: "vista3d_segresnet_d"
44 |
--------------------------------------------------------------------------------
/vista3d/configs/finetune/infer_patch_auto_word.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | output_path: "$'/workspace/vista3d/work_dir_finetune_word_' + str(@train_number)"
3 | ckpt: "$@output_path + '/model_fold0/best_metric_model.pt'"
4 | dataset_name: "WORD"
5 | label_set: [0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16]
6 | mapped_label_set: [0,1,3,14,5,12,10,11,4,13,62,19,8,18,15,93,94]
7 | val_auto: true
8 | overlap: 0.625
9 | data_file_base_dir: '/data/WORD'
10 | data_list_file_path: './data/external/WORD.json'
11 | log_output_file: "$@output_path + '/test_set.log'"
12 | list_key: 'testing'
13 | five_fold: false
14 | fold: 0
15 | train_number: 100
16 | argmax_first: false
17 | input_channels: 1
18 | image_key: image
19 | label_key: label
20 | pixdim: [1,1,1]
21 | patch_size: [224, 224, 144]
22 | transforms_infer:
23 | _target_: Compose
24 | transforms:
25 | - _target_: LoadImaged
26 | ensure_channel_first: true
27 | image_only: true
28 | keys: ['@image_key','@label_key']
29 | - _target_: CopyItemsd
30 | names: 'label_gt'
31 | keys: '@label_key'
32 | - _target_: Spacingd
33 | keys: ["@image_key",'@label_key']
34 | pixdim: '@pixdim'
35 | mode: [bilinear,nearest]
36 | align_corners: [true, true]
37 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
38 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
39 | - _target_: Orientationd
40 | axcodes: RAS
41 | keys: ['@image_key','@label_key']
42 | - _target_: CastToTyped
43 | dtype: [$torch.float32, $torch.uint8]
44 | keys: ['@image_key','@label_key']
45 | - _target_: EnsureTyped
46 | keys: ['@image_key','@label_key']
47 | track_meta: true
48 | model: "vista3d_segresnet_d"
49 |
--------------------------------------------------------------------------------
/vista3d/configs/finetune/train_finetune_murine.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | train_number: 89
3 | comment: 'finetune on murine datasets.'
4 | bundle_root: $'./work_dir_ft_final_finetune_murine_' + str(@train_number)
5 | label_set: [0,1,2,3,4]
6 | mapped_label_set: [0,115,121,30,28]
7 | model: "vista3d_segresnet_d"
8 | use_folds: true
9 | data_file_base_dir: '/data/micro-ct-murine/1_nativeCTdata_nifti/'
10 | data_list_file_path: './data/external/micro-ct-murine-native_5_folds.json'
11 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
12 | drop_label_prob: 0
13 | drop_point_prob: 1
14 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
15 | fold: 0
16 | image_key: image
17 | input_channels: 1
18 | iter_num: 5
19 | label_key: label
20 | learning_rate: 0.00005
21 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/finetune_word.log'
22 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
23 | to_onehot_y: false}
24 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
25 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
26 | max_backprompt: null
27 | max_foreprompt: null
28 | ignore_labelset: false
29 | max_point: 3
30 | max_prompt: null
31 | num_epochs: 200
32 | freeze_epoch: 0
33 | freeze_head: 'point'
34 | save_last: false
35 | save_all: false
36 | num_epochs_per_validation: 5
37 | num_images_per_batch: 1
38 | num_patches_per_image: 2
39 | num_patches_per_iter: 1
40 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
41 | output_classes: 133
42 | overlap_ratio: 0.625
43 | patch_size: [224, 224, 144]
44 | random_seed: 0
45 | skip_iter_prob: 1
46 | transforms_train:
47 | _target_: Compose
48 | transforms:
49 | - _target_: LoadImaged
50 | ensure_channel_first: true
51 | image_only: true
52 | keys: ['@image_key', '@label_key']
53 | allow_missing_keys: true
54 | - _target_: CropForegroundd
55 | allow_smaller: true
56 | end_coord_key: null
57 | keys: ['@image_key', '@label_key']
58 | margin: 10
59 | source_key: '@image_key'
60 | start_coord_key: null
61 | allow_missing_keys: true
62 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
63 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
64 | - _target_: Orientationd
65 | axcodes: RAS
66 | keys: ['@image_key', '@label_key']
67 | allow_missing_keys: true
68 | - _target_: EnsureTyped
69 | keys: ['@image_key', '@label_key']
70 | allow_missing_keys: true
71 | track_meta: false
72 | - _target_: SpatialPadd
73 | keys: ['@image_key', '@label_key']
74 | allow_missing_keys: true
75 | mode: [constant, constant]
76 | spatial_size: '@patch_size'
77 | - _target_: RandCropByLabelClassesd
78 | keys:
79 | - '@image_key'
80 | - '@label_key'
81 | label_key: '@label_key'
82 | num_classes: 5
83 | num_samples: '@num_patches_per_image'
84 | spatial_size: '@patch_size'
85 | ratios: $tuple(float(i >= 0) for i in range(5))
86 | warn: false
87 | allow_missing_keys: true
88 | - _target_: RandZoomd
89 | keys:
90 | - '@image_key'
91 | - '@label_key'
92 | min_zoom: 0.8
93 | max_zoom: 1.2
94 | mode:
95 | - trilinear
96 | - nearest
97 | prob: 0.2
98 | allow_missing_keys: true
99 | - _target_: RandSimulateLowResolutiond
100 | keys:
101 | - '@image_key'
102 | zoom_range:
103 | - 0.3
104 | - 1
105 | prob: 0.2
106 | allow_missing_keys: true
107 | - _target_: RandGaussianSmoothd
108 | keys:
109 | - '@image_key'
110 | prob: 0.2
111 | sigma_x:
112 | - 0.5
113 | - 1
114 | sigma_y:
115 | - 0.5
116 | - 1
117 | sigma_z:
118 | - 0.5
119 | - 1
120 | - _target_: RandScaleIntensityd
121 | keys:
122 | - '@image_key'
123 | factors: 0.1
124 | prob: 0.2
125 | - _target_: RandShiftIntensityd
126 | keys:
127 | - '@image_key'
128 | offsets: 0.1
129 | prob: 0.2
130 | - _target_: RandGaussianNoised
131 | keys:
132 | - '@image_key'
133 | prob: 0.2
134 | mean: 0
135 | std: 0.2
136 | - _target_: CastToTyped
137 | dtype: [$torch.float32, $torch.int32]
138 | keys: ['@image_key', '@label_key']
139 | allow_missing_keys: true
140 |
141 | transforms_validate:
142 | _target_: Compose
143 | transforms:
144 | - _target_: LoadImaged
145 | ensure_channel_first: true
146 | image_only: true
147 | keys: ['@image_key', '@label_key']
148 | - _target_: CopyItemsd
149 | names: 'label_gt'
150 | keys: '@label_key'
151 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
152 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
153 | - _target_: Orientationd
154 | axcodes: RAS
155 | keys: ['@image_key', '@label_key']
156 | - _target_: CastToTyped
157 | dtype: [$torch.float32, $torch.uint8]
158 | keys: ['@image_key', '@label_key']
159 | - _target_: EnsureTyped
160 | keys: ['@image_key', '@label_key']
161 | track_meta: true
162 |
163 | transforms_infer: $@transforms_validate
164 |
--------------------------------------------------------------------------------
/vista3d/configs/finetune/train_finetune_word.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | train_number: 100
3 | bundle_root: $'./work_dir_finetune_word_' + str(@train_number)
4 | comment: 'finetune on WORD datasets.'
5 | label_set: [0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16]
6 | mapped_label_set: [0,1,3,14,5,12,10,11,4,13,62,19,8,18,15,93,94]
7 | model: "vista3d_segresnet_d"
8 | data_file_base_dir: '/data/WORD'
9 | data_list_file_path: './data/external/WORD.json'
10 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
11 | drop_label_prob: 0
12 | drop_point_prob: 1
13 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
14 | fold: 0
15 | image_key: image
16 | input_channels: 1
17 | iter_num: 5
18 | label_key: label
19 | learning_rate: 0.00005
20 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/finetune_word.log'
21 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
22 | to_onehot_y: false}
23 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
24 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
25 | max_backprompt: null
26 | max_foreprompt: null
27 | ignore_labelset: false
28 | max_point: 3
29 | max_prompt: null
30 | num_epochs: 200
31 | freeze_epoch: 0
32 | freeze_head: 'point'
33 | save_last: false
34 | save_all: false
35 | num_epochs_per_validation: 1
36 | num_images_per_batch: 1
37 | num_patches_per_image: 2
38 | num_patches_per_iter: 1
39 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
40 | output_classes: 133
41 | overlap_ratio: 0.625
42 | patch_size: [224, 224, 144]
43 | random_seed: 0
44 | resample_to_spacing: [1., 1., 1.]
45 | skip_iter_prob: 1
46 | transforms_train:
47 | _target_: Compose
48 | transforms:
49 | - _target_: LoadImaged
50 | ensure_channel_first: true
51 | image_only: true
52 | keys: ['@image_key', '@label_key']
53 | allow_missing_keys: true
54 | - _target_: CropForegroundd
55 | allow_smaller: true
56 | end_coord_key: null
57 | keys: ['@image_key', '@label_key']
58 | margin: 10
59 | source_key: '@image_key'
60 | start_coord_key: null
61 | allow_missing_keys: true
62 | - _target_: Spacingd
63 | keys: ["@image_key", "@label_key"]
64 | pixdim: '@resample_to_spacing'
65 | mode: [bilinear, nearest]
66 | align_corners: [true, true]
67 | allow_missing_keys: true
68 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
69 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
70 | - _target_: Orientationd
71 | axcodes: RAS
72 | keys: ['@image_key', '@label_key']
73 | allow_missing_keys: true
74 | - _target_: EnsureTyped
75 | keys: ['@image_key', '@label_key']
76 | allow_missing_keys: true
77 | track_meta: false
78 | - _target_: SpatialPadd
79 | keys: ['@image_key', '@label_key']
80 | allow_missing_keys: true
81 | mode: [constant, constant]
82 | spatial_size: '@patch_size'
83 | - _target_: RandCropByLabelClassesd
84 | keys:
85 | - '@image_key'
86 | - '@label_key'
87 | label_key: '@label_key'
88 | num_classes: 133
89 | num_samples: '@num_patches_per_image'
90 | spatial_size: '@patch_size'
91 | ratios: $tuple(float(i >= 0) for i in range(133))
92 | warn: false
93 | allow_missing_keys: true
94 | - _target_: RandZoomd
95 | keys:
96 | - '@image_key'
97 | - '@label_key'
98 | min_zoom: 0.8
99 | max_zoom: 1.2
100 | mode:
101 | - trilinear
102 | - nearest
103 | prob: 0.2
104 | allow_missing_keys: true
105 | - _target_: RandSimulateLowResolutiond
106 | keys:
107 | - '@image_key'
108 | zoom_range:
109 | - 0.3
110 | - 1
111 | prob: 0.2
112 | allow_missing_keys: true
113 | - _target_: RandGaussianSmoothd
114 | keys:
115 | - '@image_key'
116 | prob: 0.2
117 | sigma_x:
118 | - 0.5
119 | - 1
120 | sigma_y:
121 | - 0.5
122 | - 1
123 | sigma_z:
124 | - 0.5
125 | - 1
126 | - _target_: RandScaleIntensityd
127 | keys:
128 | - '@image_key'
129 | factors: 0.1
130 | prob: 0.2
131 | - _target_: RandShiftIntensityd
132 | keys:
133 | - '@image_key'
134 | offsets: 0.1
135 | prob: 0.2
136 | - _target_: RandGaussianNoised
137 | keys:
138 | - '@image_key'
139 | prob: 0.2
140 | mean: 0
141 | std: 0.2
142 | - _target_: CastToTyped
143 | dtype: [$torch.float32, $torch.int32]
144 | keys: ['@image_key', '@label_key']
145 | allow_missing_keys: true
146 |
147 | transforms_validate:
148 | _target_: Compose
149 | transforms:
150 | - _target_: LoadImaged
151 | ensure_channel_first: true
152 | image_only: true
153 | keys: ['@image_key', '@label_key']
154 | - _target_: CopyItemsd
155 | names: 'label_gt'
156 | keys: '@label_key'
157 | - _target_: Spacingd
158 | keys: ["@image_key", "@label_key"]
159 | pixdim: '@resample_to_spacing'
160 | mode: [bilinear, nearest]
161 | align_corners: [true, true]
162 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
163 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
164 | - _target_: Orientationd
165 | axcodes: RAS
166 | keys: ['@image_key', '@label_key']
167 | - _target_: CastToTyped
168 | dtype: [$torch.float32, $torch.uint8]
169 | keys: ['@image_key', '@label_key']
170 | - _target_: EnsureTyped
171 | keys: ['@image_key', '@label_key']
172 | track_meta: true
173 | transforms_infer: $@transforms_validate
174 |
--------------------------------------------------------------------------------
/vista3d/configs/infer.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | input_channels: 1
3 | patch_size: [128, 128, 128]
4 | bundle_root: './models'
5 | fold: 0
6 | infer: {ckpt_name: $@bundle_root + '/model.pt',
7 | output_path: $@bundle_root + '/prediction',
8 | log_output_file: $@bundle_root + '/inference.log'}
9 | resample_to_spacing: [1.5, 1.5, 1.5]
10 | model: "vista3d_segresnet_d"
11 | image_key: "image"
12 | transforms_infer:
13 | _target_: Compose
14 | transforms:
15 | - _target_: LoadImaged
16 | keys: "@image_key"
17 | image_only: True
18 | - _target_: EnsureChannelFirstd
19 | keys: "@image_key"
20 | - _target_: ScaleIntensityRanged
21 | a_max: 1053.678477684517
22 | a_min: -963.8247715525971
23 | b_max: 1.0
24 | b_min: 0.0
25 | clip: true
26 | keys: '@image_key'
27 | - _target_: Orientationd
28 | keys: "@image_key"
29 | axcodes: RAS
30 | - _target_: Spacingd
31 | keys: ["@image_key"]
32 | pixdim: "@resample_to_spacing"
33 | mode: [bilinear]
34 | align_corners: [true]
35 | - _target_: CastToTyped
36 | keys: "@image_key"
37 | dtype: "$torch.float32"
38 |
--------------------------------------------------------------------------------
/vista3d/configs/supported_eval/infer_patch_auto.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "TotalSegmentatorV2"
6 | label_set: null
7 | overlap: 0.625
8 | json_name: "$@dataset_name + '_5_folds.json'"
9 | data_file_base_dir: "$'/data/' + @dataset_name"
10 | data_list_file_path: "$'./data/jsons/' + @json_name"
11 | log_output_file: "$@output_path + '/validation_auto_' + @dataset_name + '.log'"
12 | list_key: 'testing'
13 | val_auto: true
14 | argmax_first: false
15 | fold: 0
16 | input_channels: 1
17 | image_key: image
18 | label_key: label
19 | patch_size: [128, 128, 128]
20 | transforms_infer:
21 | _target_: Compose
22 | transforms:
23 | - _target_: LoadImaged
24 | ensure_channel_first: true
25 | image_only: true
26 | keys: ['@image_key','@label_key']
27 | - _target_: CopyItemsd
28 | names: 'label_gt'
29 | keys: '@label_key'
30 | - _target_: CropForegroundd
31 | allow_smaller: true
32 | keys: ['@image_key', '@label_key']
33 | margin: 10
34 | source_key: '@image_key'
35 | - _target_: Spacingd
36 | keys: ["@image_key",'@label_key']
37 | pixdim: [1.5, 1.5, 1.5]
38 | mode: [bilinear,nearest]
39 | align_corners: [true, true]
40 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
41 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
42 | - _target_: Orientationd
43 | axcodes: RAS
44 | keys: ['@image_key','@label_key']
45 | - _target_: CastToTyped
46 | dtype: [$torch.float32, $torch.uint8]
47 | keys: ['@image_key','@label_key']
48 | - _target_: EnsureTyped
49 | keys: ['@image_key','@label_key']
50 | track_meta: true
51 | model: "vista3d_segresnet_d"
52 |
--------------------------------------------------------------------------------
/vista3d/configs/supported_eval/infer_patch_autopoint.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "TotalSegmentatorV2"
6 | label_set: null
7 | overlap: 0.625
8 | json_name: "$@dataset_name + '_5_folds.json'"
9 | data_file_base_dir: "$'/data/' + @dataset_name"
10 | data_list_file_path: "$'./data/jsons/' + @json_name"
11 | log_output_file: "$@output_path + '/validation_autopoint_patch_' + @dataset_name + '.log'"
12 | list_key: 'testing'
13 | save_metric: false
14 | argmax_first: false
15 | val_auto: false
16 | fold: 0
17 | input_channels: 1
18 | image_key: image
19 | label_key: label
20 | patch_size: [128, 128, 128]
21 | transforms_infer:
22 | _target_: Compose
23 | transforms:
24 | - _target_: LoadImaged
25 | ensure_channel_first: true
26 | image_only: true
27 | keys: ['@image_key','@label_key']
28 | - _target_: CopyItemsd
29 | names: 'label_gt'
30 | keys: '@label_key'
31 | - _target_: CropForegroundd
32 | allow_smaller: true
33 | keys: ['@image_key', '@label_key']
34 | margin: 10
35 | source_key: '@image_key'
36 | - _target_: Spacingd
37 | keys: ["@image_key",'@label_key']
38 | pixdim: [1.5, 1.5, 1.5]
39 | mode: [bilinear,nearest]
40 | align_corners: [true, true]
41 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
42 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
43 | - _target_: Orientationd
44 | axcodes: RAS
45 | keys: ['@image_key','@label_key']
46 | - _target_: CastToTyped
47 | dtype: [$torch.float32, $torch.uint8]
48 | keys: ['@image_key','@label_key']
49 | - _target_: EnsureTyped
50 | keys: ['@image_key','@label_key']
51 | track_meta: true
52 | model: "vista3d_segresnet_d"
53 |
--------------------------------------------------------------------------------
/vista3d/configs/supported_eval/infer_patch_point.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "TotalSegmentatorV2"
6 | label_set: null
7 | overlap: 0.625
8 | json_name: "$@dataset_name + '_5_folds.json'"
9 | data_file_base_dir: "$'/data/' + @dataset_name"
10 | data_list_file_path: "$'./data/jsons/' + @json_name"
11 | log_output_file: "$@output_path + '/validation_1point_' + @dataset_name + '.log'"
12 | list_key: 'testing'
13 | save_metric: false
14 | argmax_first: false
15 | val_auto: false
16 | fold: 0
17 | input_channels: 1
18 | image_key: image
19 | label_key: label
20 | patch_size: [128, 128, 128]
21 | transforms_infer:
22 | _target_: Compose
23 | transforms:
24 | - _target_: LoadImaged
25 | ensure_channel_first: true
26 | image_only: true
27 | keys: ['@image_key','@label_key']
28 | - _target_: CopyItemsd
29 | names: 'label_gt'
30 | keys: '@label_key'
31 | - _target_: CropForegroundd
32 | allow_smaller: true
33 | keys: ['@image_key', '@label_key']
34 | margin: 10
35 | source_key: '@image_key'
36 | - _target_: Spacingd
37 | keys: ["@image_key",'@label_key']
38 | pixdim: [1.5, 1.5, 1.5]
39 | mode: [bilinear,nearest]
40 | align_corners: [true, true]
41 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
42 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
43 | - _target_: Orientationd
44 | axcodes: RAS
45 | keys: ['@image_key','@label_key']
46 | - _target_: CastToTyped
47 | dtype: [$torch.float32, $torch.uint8]
48 | keys: ['@image_key','@label_key']
49 | - _target_: EnsureTyped
50 | keys: ['@image_key','@label_key']
51 | track_meta: true
52 | model: "vista3d_segresnet_d"
53 |
--------------------------------------------------------------------------------
/vista3d/configs/supported_eval/infer_sam2_point.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "./"
4 | ckpt: '/workspace/segment-anything-2/checkpoints/sam2_hiera_large.pt'
5 | model_cfg: 'sam2_hiera_l.yaml'
6 | dataset_name: "Task06"
7 | label_set: null
8 | json_name: "$@dataset_name + '_5_folds.json'"
9 | data_file_base_dir: "$'/data/' + @dataset_name"
10 | data_list_file_path: "$'./data/jsons/' + @json_name"
11 | log_output_file: "$@output_path + '/validation_auto_' + @dataset_name + '.log'"
12 | list_key: 'testing'
13 | saliency: true
14 | start_file: 0
15 | end_file: -1
16 | max_iter: 8
17 | argmax_first: false
18 | fold: 0
19 | image_key: image
20 | label_key: label
21 | transforms_infer:
22 | _target_: Compose
23 | transforms:
24 | - _target_: LoadImaged
25 | ensure_channel_first: true
26 | image_only: true
27 | keys: ['@image_key','@label_key']
28 | - _target_: CopyItemsd
29 | names: 'label_gt'
30 | keys: '@label_key'
31 | - {_target_: ScaleIntensityRanged, a_max: 1000, a_min: -1000,
32 | b_max: 255, b_min: 0, clip: true, keys: '@image_key'}
33 | - _target_: CastToTyped
34 | dtype: [$torch.float32, $torch.uint8]
35 | keys: ['@image_key','@label_key']
36 | - _target_: EnsureTyped
37 | keys: ['@image_key','@label_key']
38 | track_meta: true
39 | model: "vista3d_segresnet_d"
40 |
--------------------------------------------------------------------------------
/vista3d/configs/train/hyper_parameters_stage1.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | bundle_root: ./work_dir_stage1
3 | comments: "After training for several epoch, remove the unlabeled dataset from train_datasets."
4 | json_dir: ./data/jsons
5 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
6 | model: "vista3d_segresnet_d"
7 | weighted_sampling: false
8 | drop_label_prob: 1
9 | drop_point_prob: 0
10 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
11 | fold: 0
12 | image_key: image
13 | input_channels: 1
14 | iter_num: 5
15 | label_key: label
16 | label_sv_key: label_sv
17 | pseudo_label_key: pseudo_label
18 | learning_rate: 0.00002
19 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/training.log'
20 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
21 | to_onehot_y: false}
22 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
23 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
24 | max_backprompt: 0
25 | max_foreprompt: 4
26 | max_point: 3
27 | max_prompt: null
28 | num_epochs: 300
29 | freeze_epoch: 0
30 | freeze_head: 'auto'
31 | save_last: true
32 | save_all: false
33 | num_epochs_per_validation: 1
34 | num_images_per_batch: 1
35 | num_patches_per_image: 2
36 | num_patches_per_iter: 1
37 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
38 | output_classes: 133
39 | overlap_ratio: 0.5
40 | patch_size: [128, 128, 128]
41 | random_seed: 0
42 | resample_to_spacing: [1.5, 1.5, 1.5]
43 | skip_iter_prob: 0
44 | train_datasets: [CTPelvic1K-CLINIC, AbdomenCT-1K, AeroPath, AMOS22,
45 | BTCV-Abdomen, BTCV-Cervix, CT-ORG, FLARE22, Multi-organ-Abdominal-CT-btcv,
46 | Multi-organ-Abdominal-CT-tcia, Pancreas-CT, Task03, Task06, Task07,
47 | Task08, Task09, Task10, VerSe, CRLM-CT, TotalSegmentatorV2, NLST, LIDC, StonyBrook-CT, TCIA_Colon, Covid19]
48 | val_datasets: [TotalSegmentatorV2]
49 | transforms_train:
50 | _target_: Compose
51 | transforms:
52 | - _target_: LoadImaged
53 | ensure_channel_first: true
54 | image_only: true
55 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
56 | allow_missing_keys: true
57 | - _target_: CropForegroundd
58 | allow_smaller: true
59 | end_coord_key: null
60 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
61 | margin: 10
62 | source_key: '@image_key'
63 | start_coord_key: null
64 | allow_missing_keys: true
65 | - _target_: Spacingd
66 | keys: ["@image_key", "@label_key", '@label_sv_key', '@pseudo_label_key']
67 | pixdim: [1.5, 1.5, 1.5]
68 | mode: [bilinear, nearest, nearest, nearest]
69 | align_corners: [true, true, true, true]
70 | allow_missing_keys: true
71 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
72 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
73 | - _target_: Orientationd
74 | axcodes: RAS
75 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
76 | allow_missing_keys: true
77 | - _target_: EnsureTyped
78 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
79 | allow_missing_keys: true
80 | track_meta: false
81 | - _target_: SpatialPadd
82 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
83 | allow_missing_keys: true
84 | mode: [constant, constant, constant, constant]
85 | spatial_size: '@patch_size'
86 | - "Placeholder for dataset-specific transform"
87 | - _target_: CastToTyped
88 | dtype: [$torch.float32, $torch.int32, $torch.int32, $torch.int32]
89 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
90 | allow_missing_keys: true
91 | transforms_validate:
92 | _target_: Compose
93 | transforms:
94 | - _target_: LoadImaged
95 | ensure_channel_first: true
96 | image_only: true
97 | keys: ['@image_key', '@label_key']
98 | - _target_: CropForegroundd
99 | allow_smaller: true
100 | keys: ['@image_key', '@label_key']
101 | margin: 10
102 | source_key: '@image_key'
103 | - _target_: Spacingd
104 | keys: ["@image_key", "@label_key"]
105 | pixdim: [1.5, 1.5, 1.5]
106 | mode: [bilinear, nearest]
107 | align_corners: [true, true]
108 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
109 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
110 | - _target_: Orientationd
111 | axcodes: RAS
112 | keys: ['@image_key', '@label_key']
113 | - _target_: CastToTyped
114 | dtype: [$torch.float32, $torch.uint8]
115 | keys: ['@image_key', '@label_key']
116 | - _target_: EnsureTyped
117 | keys: ['@image_key', '@label_key']
118 | track_meta: true
119 | - "Placeholder for dataset-specific transform"
120 |
--------------------------------------------------------------------------------
/vista3d/configs/train/hyper_parameters_stage2.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | bundle_root: ./work_dir_stage2
3 | json_dir: ./data/jsons
4 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
5 | model: "vista3d_segresnet_d"
6 | weighted_sampling: false
7 | drop_label_prob: 1
8 | drop_point_prob: 0
9 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
10 | fold: 0
11 | image_key: image
12 | input_channels: 1
13 | iter_num: 5
14 | label_key: label
15 | label_sv_key: label_sv
16 | pseudo_label_key: pseudo_label
17 | learning_rate: 0.00002
18 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/training.log'
19 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
20 | to_onehot_y: false}
21 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
22 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
23 | max_backprompt: 0
24 | max_foreprompt: 4
25 | max_point: 3
26 | max_prompt: null
27 | num_epochs: 300
28 | freeze_epoch: 0
29 | freeze_head: 'auto'
30 | save_last: true
31 | save_all: true
32 | num_epochs_per_validation: 1
33 | num_images_per_batch: 1
34 | num_patches_per_image: 2
35 | num_patches_per_iter: 1
36 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
37 | output_classes: 133
38 | overlap_ratio: 0.5
39 | patch_size: [128, 128, 128]
40 | random_seed: 0
41 | resample_to_spacing: [1.5, 1.5, 1.5]
42 | skip_iter_prob: 0
43 | train_datasets: [CTPelvic1K-CLINIC, AbdomenCT-1K, AeroPath, AMOS22, BTCV-Abdomen,
44 | BTCV-Cervix, CT-ORG, FLARE22, Multi-organ-Abdominal-CT-btcv, Multi-organ-Abdominal-CT-tcia,
45 | Pancreas-CT, Task03, Task06, Task07, Task08, Task09, Task10, VerSe, CRLM-CT,
46 | TotalSegmentatorV2]
47 | val_datasets: ['CRLM-CT', 'AeroPath', 'Task03','Task06','Task07','Task08','Task10']
48 | transforms_train:
49 | _target_: Compose
50 | transforms:
51 | - _target_: LoadImaged
52 | ensure_channel_first: true
53 | image_only: true
54 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
55 | allow_missing_keys: true
56 | - _target_: CropForegroundd
57 | allow_smaller: true
58 | end_coord_key: null
59 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
60 | margin: 10
61 | source_key: '@image_key'
62 | start_coord_key: null
63 | allow_missing_keys: true
64 | - _target_: Spacingd
65 | keys: ["@image_key", "@label_key", '@label_sv_key', '@pseudo_label_key']
66 | pixdim: [1.5, 1.5, 1.5]
67 | mode: [bilinear, nearest, nearest, nearest]
68 | align_corners: [true, true, true, true]
69 | allow_missing_keys: true
70 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
71 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
72 | - _target_: Orientationd
73 | axcodes: RAS
74 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
75 | allow_missing_keys: true
76 | - _target_: EnsureTyped
77 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
78 | allow_missing_keys: true
79 | track_meta: false
80 | - _target_: SpatialPadd
81 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
82 | allow_missing_keys: true
83 | mode: [constant, constant, constant, constant]
84 | spatial_size: '@patch_size'
85 | - "Placeholder for dataset-specific transform"
86 | - _target_: CastToTyped
87 | dtype: [$torch.float32, $torch.int32, $torch.int32, $torch.int32]
88 | keys: ['@image_key', '@label_key', '@label_sv_key', '@pseudo_label_key']
89 | allow_missing_keys: true
90 | transforms_validate:
91 | _target_: Compose
92 | transforms:
93 | - _target_: LoadImaged
94 | ensure_channel_first: true
95 | image_only: true
96 | keys: ['@image_key', '@label_key']
97 | - _target_: CropForegroundd
98 | allow_smaller: true
99 | keys: ['@image_key', '@label_key']
100 | margin: 10
101 | source_key: '@image_key'
102 | - _target_: Spacingd
103 | keys: ["@image_key", "@label_key"]
104 | pixdim: [1.5, 1.5, 1.5]
105 | mode: [bilinear, nearest]
106 | align_corners: [true, true]
107 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
108 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
109 | - _target_: Orientationd
110 | axcodes: RAS
111 | keys: ['@image_key', '@label_key']
112 | - _target_: CastToTyped
113 | dtype: [$torch.float32, $torch.uint8]
114 | keys: ['@image_key', '@label_key']
115 | - _target_: EnsureTyped
116 | keys: ['@image_key', '@label_key']
117 | track_meta: true
118 | - "Placeholder for dataset-specific transform"
119 |
--------------------------------------------------------------------------------
/vista3d/configs/train/hyper_parameters_stage3.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | bundle_root: ./work_dir_stage3
3 | comments: "After training for several epoch, remove the unlabeled dataset from train_datasets."
4 | json_dir: ./data/jsons
5 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
6 | model: "vista3d_segresnet_d"
7 | weighted_sampling: false
8 | drop_label_prob: 0
9 | drop_point_prob: 1
10 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
11 | fold: 0
12 | image_key: image
13 | input_channels: 1
14 | iter_num: 5
15 | label_key: label
16 | label_sv_key: label_sv
17 | pseudo_label_key: pseudo_label
18 | learning_rate: 0.00002
19 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/training.log'
20 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
21 | to_onehot_y: false}
22 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
23 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
24 | max_backprompt: 4
25 | max_foreprompt: 32
26 | max_point: 3
27 | max_prompt: null
28 | num_epochs: 200
29 | freeze_epoch: 1000
30 | freeze_head: 'point'
31 | save_last: false
32 | save_all: false
33 | num_epochs_per_validation: 5
34 | num_images_per_batch: 1
35 | num_patches_per_image: 2
36 | num_patches_per_iter: 1
37 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
38 | output_classes: 133
39 | overlap_ratio: 0.5
40 | patch_size: [128, 128, 128]
41 | random_seed: 0
42 | resample_to_spacing: [1.5, 1.5, 1.5]
43 | skip_iter_prob: 1
44 | train_datasets: [CTPelvic1K-CLINIC, AbdomenCT-1K, AeroPath, AMOS22, BTCV-Abdomen,
45 | BTCV-Cervix, CT-ORG, FLARE22, Multi-organ-Abdominal-CT-btcv, Multi-organ-Abdominal-CT-tcia,
46 | Pancreas-CT, Task03, Task06, Task07, Task08, Task09, Task10, VerSe, CRLM-CT,
47 | TotalSegmentatorV2, NLST, LIDC, StonyBrook-CT, TCIA_Colon]
48 | val_datasets: ['TotalSegmentatorV2']
49 | transforms_train:
50 | _target_: Compose
51 | transforms:
52 | - _target_: LoadImaged
53 | ensure_channel_first: true
54 | image_only: true
55 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
56 | allow_missing_keys: true
57 | - _target_: DeleteItemsd
58 | keys: ['@label_sv_key']
59 | - _target_: CropForegroundd
60 | allow_smaller: true
61 | end_coord_key: null
62 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
63 | margin: 10
64 | source_key: '@image_key'
65 | start_coord_key: null
66 | allow_missing_keys: true
67 | - _target_: Spacingd
68 | keys: ["@image_key", "@label_key", '@pseudo_label_key']
69 | pixdim: [1.5, 1.5, 1.5]
70 | mode: [bilinear, nearest, nearest]
71 | align_corners: [true, true, true]
72 | allow_missing_keys: true
73 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
74 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
75 | - _target_: Orientationd
76 | axcodes: RAS
77 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
78 | allow_missing_keys: true
79 | - _target_: EnsureTyped
80 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
81 | allow_missing_keys: true
82 | track_meta: false
83 | - _target_: SpatialPadd
84 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
85 | allow_missing_keys: true
86 | mode: [constant, constant, constant]
87 | spatial_size: '@patch_size'
88 | - "Placeholder for dataset-specific transform"
89 | - _target_: CastToTyped
90 | dtype: [$torch.float32, $torch.int32, $torch.int32]
91 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
92 | allow_missing_keys: true
93 | transforms_validate:
94 | _target_: Compose
95 | transforms:
96 | - _target_: LoadImaged
97 | ensure_channel_first: true
98 | image_only: true
99 | keys: ['@image_key', '@label_key']
100 | - _target_: CropForegroundd
101 | allow_smaller: true
102 | keys: ['@image_key', '@label_key']
103 | margin: 10
104 | source_key: '@image_key'
105 | - _target_: Spacingd
106 | keys: ["@image_key", "@label_key"]
107 | pixdim: [1.5, 1.5, 1.5]
108 | mode: [bilinear, nearest]
109 | align_corners: [true, true]
110 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
111 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
112 | - _target_: Orientationd
113 | axcodes: RAS
114 | keys: ['@image_key', '@label_key']
115 | - _target_: CastToTyped
116 | dtype: [$torch.float32, $torch.uint8]
117 | keys: ['@image_key', '@label_key']
118 | - _target_: EnsureTyped
119 | keys: ['@image_key', '@label_key']
120 | track_meta: true
121 | - "Placeholder for dataset-specific transform"
122 |
--------------------------------------------------------------------------------
/vista3d/configs/train/hyper_parameters_stage4.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | bundle_root: ./work_dir_stage4
3 | json_dir: ./data/jsons
4 | balance_gt: true
5 | ckpt_path: $@bundle_root + '/model_fold' + str(@fold)
6 | model: "vista3d_segresnet_d"
7 | weighted_sampling: false
8 | drop_label_prob: 0
9 | drop_point_prob: 1
10 | finetune: {activate: true, exclude_vars: null, pretrained_ckpt_name: $'/workspace/vista3d/models/model.pt'}
11 | fold: 0
12 | image_key: image
13 | input_channels: 1
14 | iter_num: 5
15 | label_key: label
16 | label_sv_key: label_sv
17 | pseudo_label_key: pseudo_label
18 | learning_rate: 0.00002
19 | log_output_file: $@bundle_root + '/model_fold' + str(@fold) + '/training.log'
20 | loss: {_target_: DiceCELoss, include_background: false, sigmoid: true, smooth_dr: 1.0e-05, smooth_nr: 0, softmax: false, squared_pred: true,
21 | to_onehot_y: false}
22 | lr_scheduler: {_target_: monai.optimizers.WarmupCosineSchedule, optimizer: $@optimizer,
23 | t_total: $@num_epochs+1, warmup_multiplier: 0.1, warmup_steps: 0}
24 | max_backprompt: 4
25 | max_foreprompt: 32
26 | max_point: 3
27 | max_prompt: null
28 | num_epochs: 200
29 | freeze_epoch: 1000
30 | freeze_head: 'point'
31 | save_last: true
32 | save_all: true
33 | num_epochs_per_validation: 1
34 | num_images_per_batch: 1
35 | num_patches_per_image: 2
36 | num_patches_per_iter: 1
37 | optimizer: {_target_: torch.optim.AdamW, lr: '@learning_rate', weight_decay: 1.0e-05}
38 | output_classes: 133
39 | overlap_ratio: 0.5
40 | patch_size: [128, 128, 128]
41 | random_seed: 0
42 | resample_to_spacing: [1.5, 1.5, 1.5]
43 | skip_iter_prob: 1
44 | train_datasets: [CTPelvic1K-CLINIC, AbdomenCT-1K, AeroPath, AMOS22, BTCV-Abdomen,
45 | BTCV-Cervix, CT-ORG, FLARE22, Multi-organ-Abdominal-CT-btcv, Multi-organ-Abdominal-CT-tcia,
46 | Pancreas-CT, Task03, Task06, Task07, Task08, Task09, Task10, VerSe, CRLM-CT,
47 | TotalSegmentatorV2]
48 | val_datasets: ['CRLM-CT', 'AeroPath', 'Task03','Task06','Task07','Task08','Task10','Bone-NIH']
49 | transforms_train:
50 | _target_: Compose
51 | transforms:
52 | - _target_: LoadImaged
53 | ensure_channel_first: true
54 | image_only: true
55 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
56 | allow_missing_keys: true
57 | - _target_: DeleteItemsd
58 | keys: ['@label_sv_key']
59 | - _target_: CropForegroundd
60 | allow_smaller: true
61 | end_coord_key: null
62 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
63 | margin: 10
64 | source_key: '@image_key'
65 | start_coord_key: null
66 | allow_missing_keys: true
67 | - _target_: Spacingd
68 | keys: ["@image_key", "@label_key", '@pseudo_label_key']
69 | pixdim: [1.5, 1.5, 1.5]
70 | mode: [bilinear, nearest, nearest]
71 | align_corners: [true, true, true]
72 | allow_missing_keys: true
73 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
74 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
75 | - _target_: Orientationd
76 | axcodes: RAS
77 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
78 | allow_missing_keys: true
79 | - _target_: EnsureTyped
80 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
81 | allow_missing_keys: true
82 | track_meta: false
83 | - _target_: SpatialPadd
84 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
85 | allow_missing_keys: true
86 | mode: [constant, constant, constant]
87 | spatial_size: '@patch_size'
88 | - "Placeholder for dataset-specific transform"
89 | - _target_: CastToTyped
90 | dtype: [$torch.float32, $torch.int32, $torch.int32]
91 | keys: ['@image_key', '@label_key', '@pseudo_label_key']
92 | allow_missing_keys: true
93 | transforms_validate:
94 | _target_: Compose
95 | transforms:
96 | - _target_: LoadImaged
97 | ensure_channel_first: true
98 | image_only: true
99 | keys: ['@image_key', '@label_key']
100 | - _target_: CropForegroundd
101 | allow_smaller: true
102 | keys: ['@image_key', '@label_key']
103 | margin: 10
104 | source_key: '@image_key'
105 | - _target_: Spacingd
106 | keys: ["@image_key", "@label_key"]
107 | pixdim: [1.5, 1.5, 1.5]
108 | mode: [bilinear, nearest]
109 | align_corners: [true, true]
110 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
111 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
112 | - _target_: Orientationd
113 | axcodes: RAS
114 | keys: ['@image_key', '@label_key']
115 | - _target_: CastToTyped
116 | dtype: [$torch.float32, $torch.uint8]
117 | keys: ['@image_key', '@label_key']
118 | - _target_: EnsureTyped
119 | keys: ['@image_key', '@label_key']
120 | track_meta: true
121 | - "Placeholder for dataset-specific transform"
122 |
--------------------------------------------------------------------------------
/vista3d/configs/zeroshot_eval/infer_iter_point_adrenal.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "Adrenal_Ki67"
6 | label_set: [0,1]
7 | max_iter: 80
8 | overlap: 0.625
9 | json_name: "$@dataset_name + '_5_folds.json'"
10 | data_file_base_dir: "$'/data/' + @dataset_name"
11 | data_list_file_path: "$'./data/external/' + @json_name"
12 | log_output_file: $@output_path + '/inference_adrenal.log'
13 | list_key: 'all'
14 | fold: 0
15 | input_channels: 1
16 | image_key: image
17 | label_key: label
18 | patch_size: [128, 128, 128]
19 | transforms_infer:
20 | _target_: Compose
21 | transforms:
22 | - _target_: LoadImaged
23 | ensure_channel_first: true
24 | image_only: true
25 | keys: ['@image_key','@label_key']
26 | - _target_: CopyItemsd
27 | names: 'label_gt'
28 | keys: '@label_key'
29 | - _target_: Spacingd
30 | keys: ["@image_key",'@label_key']
31 | pixdim: [1.5, 1.5, 1.5]
32 | mode: [bilinear,nearest]
33 | align_corners: [true, true]
34 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
35 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
36 | - _target_: Orientationd
37 | axcodes: RAS
38 | keys: ['@image_key','@label_key']
39 | - _target_: CastToTyped
40 | dtype: [$torch.float32, $torch.uint8]
41 | keys: ['@image_key','@label_key']
42 | - _target_: EnsureTyped
43 | keys: ['@image_key','@label_key']
44 | track_meta: true
45 | model: "vista3d_segresnet_d"
46 |
--------------------------------------------------------------------------------
/vista3d/configs/zeroshot_eval/infer_iter_point_hcc.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "HCC-TACE-Seg"
6 | label_set: [0,2]
7 | max_iter: 80
8 | overlap: 0.625
9 | json_name: "$@dataset_name + '_5_folds.json'"
10 | data_file_base_dir: "$'/data/' + @dataset_name"
11 | data_list_file_path: "$'./data/external/' + @json_name"
12 | log_output_file: $@output_path + '/inference_hcc.log'
13 | list_key: 'all'
14 | fold: 0
15 | input_channels: 1
16 | image_key: image
17 | label_key: label
18 | patch_size: [128, 128, 128]
19 | transforms_infer:
20 | _target_: Compose
21 | transforms:
22 | - _target_: LoadImaged
23 | ensure_channel_first: true
24 | image_only: true
25 | keys: ['@image_key','@label_key']
26 | - _target_: CopyItemsd
27 | names: 'label_gt'
28 | keys: '@label_key'
29 | - _target_: Spacingd
30 | keys: ["@image_key",'@label_key']
31 | pixdim: [1.5, 1.5, 1.5]
32 | mode: [bilinear,nearest]
33 | align_corners: [true, true]
34 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
35 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
36 | - _target_: Orientationd
37 | axcodes: RAS
38 | keys: ['@image_key','@label_key']
39 | - _target_: CastToTyped
40 | dtype: [$torch.float32, $torch.uint8]
41 | keys: ['@image_key','@label_key']
42 | - _target_: EnsureTyped
43 | keys: ['@image_key','@label_key']
44 | track_meta: true
45 | model: "vista3d_segresnet_d"
46 |
--------------------------------------------------------------------------------
/vista3d/configs/zeroshot_eval/infer_iter_point_kits.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "C4KC-KiTS"
6 | label_set: [0,2]
7 | max_iter: 80
8 | overlap: 0.625
9 | json_name: "$@dataset_name + '_5_folds.json'"
10 | data_file_base_dir: "$'/data/' + @dataset_name + '/nifti'"
11 | data_list_file_path: "$'./data/external/' + @json_name"
12 | log_output_file: $@output_path + '/inference_kits.log'
13 | list_key: 'all'
14 | fold: 0
15 | input_channels: 1
16 | image_key: image
17 | label_key: label
18 | patch_size: [128, 128, 128]
19 | transforms_infer:
20 | _target_: Compose
21 | transforms:
22 | - _target_: LoadImaged
23 | ensure_channel_first: true
24 | image_only: true
25 | keys: ['@image_key','@label_key']
26 | - _target_: CopyItemsd
27 | names: 'label_gt'
28 | keys: '@label_key'
29 | - _target_: Spacingd
30 | keys: ["@image_key",'@label_key']
31 | pixdim: [1., 1., 1.]
32 | mode: [bilinear,nearest]
33 | align_corners: [true, true]
34 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
35 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
36 | - _target_: Orientationd
37 | axcodes: RAS
38 | keys: ['@image_key','@label_key']
39 | - _target_: CastToTyped
40 | dtype: [$torch.float32, $torch.uint8]
41 | keys: ['@image_key','@label_key']
42 | - _target_: EnsureTyped
43 | keys: ['@image_key','@label_key']
44 | track_meta: true
45 | model: "vista3d_segresnet_d"
46 |
--------------------------------------------------------------------------------
/vista3d/configs/zeroshot_eval/infer_iter_point_murine.yaml:
--------------------------------------------------------------------------------
1 | amp: true
2 | exps: models
3 | output_path: "$'/workspace/vista3d/' + @exps"
4 | ckpt: "$@output_path + '/model.pt'"
5 | dataset_name: "micro-ct-murine-native"
6 | label_set: [0,1,2,3,4]
7 | max_iter: 80
8 | overlap: 0.625
9 | json_name: "$@dataset_name + '_5_folds.json'"
10 | data_file_base_dir: '/data/micro-ct-murine/1_nativeCTdata_nifti'
11 | data_list_file_path: "$'./data/external/' + @json_name"
12 | log_output_file: $@output_path + '/inference_murine.log'
13 | list_key: 'all'
14 | fold: 0
15 | input_channels: 1
16 | image_key: image
17 | label_key: label
18 | patch_size: [128, 128, 128]
19 | transforms_infer:
20 | _target_: Compose
21 | transforms:
22 | - _target_: LoadImaged
23 | ensure_channel_first: true
24 | image_only: true
25 | keys: ['@image_key','@label_key']
26 | - _target_: CopyItemsd
27 | names: 'label_gt'
28 | keys: '@label_key'
29 | - {_target_: ScaleIntensityRanged, a_max: 1053.678477684517, a_min: -963.8247715525971,
30 | b_max: 1.0, b_min: 0.0, clip: true, keys: '@image_key'}
31 | - _target_: Orientationd
32 | axcodes: RAS
33 | keys: ['@image_key','@label_key']
34 | - _target_: CastToTyped
35 | dtype: [$torch.float32, $torch.uint8]
36 | keys: ['@image_key','@label_key']
37 | - _target_: EnsureTyped
38 | keys: ['@image_key','@label_key']
39 | track_meta: true
40 | model: "vista3d_segresnet_d"
41 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/Dockerfile:
--------------------------------------------------------------------------------
1 | # Use an appropriate base image with GPU support
2 | FROM nvidia/cuda:11.8.0-runtime-ubuntu22.04
3 | RUN apt-get update && apt-get install -y \
4 | python3 python3-pip && \
5 | rm -rf /var/lib/apt/lists/*
6 | # Set working directory
7 | WORKDIR /workspace
8 |
9 | # Copy inference script and requirements
10 | COPY infer_cvpr.py /workspace/infer.py
11 | COPY train_cvpr.py /workspace/train.py
12 | COPY update_ckpt.py /workspace/update_ckpt.py
13 | COPY Dockerfile /workspace/Dockerfile
14 | COPY requirements.txt /workspace/
15 | COPY model_epoch40.pth /workspace
16 | # Install Python dependencies
17 | RUN pip3 install -r requirements.txt
18 |
19 | # Copy the prediction script
20 | COPY predict.sh /workspace/predict.sh
21 | RUN chmod +x /workspace/predict.sh
22 |
23 | # Set default command
24 | CMD ["/bin/bash"]
25 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/README.md:
--------------------------------------------------------------------------------
1 |
13 |
14 | # Overview
15 | This repository is written for the "CVPR 2025: Foundation Models for Interactive 3D Biomedical Image Segmentation"([link](https://www.codabench.org/competitions/5263/)) challenge. It
16 | is based on MONAI 1.4. Many of the functions in the main VISTA3D repository are moved to MONAI 1.4 and this simplified folder will directly use components from MONAI.
17 |
18 | It is simplified to train interactive segmentation models across different modalities. The sophisticated transforms and recipes used for VISTA3D are removed. The finetuned VISTA3D checkpoint on the challenge subsets is available [here](https://drive.google.com/file/d/1r2KvHP_30nHR3LU7NJEdscVnlZ2hTtcd/view?usp=sharing)
19 |
20 | # Setup
21 | ```
22 | pip install -r requirements.txt
23 | ```
24 |
25 | # Training
26 | Download the challenge subsets finetuned [checkpoint](https://drive.google.com/file/d/1hQ8imaf4nNSg_43dYbPSJT0dr7JgAKWX/view?usp=sharing) or VISTA3D original [checkpoint]((https://drive.google.com/file/d/1DRYA2-AI-UJ23W1VbjqHsnHENGi0ShUl/view?usp=sharing)). Generate a json list that contains your traning data and update the json file path in the script.
27 | ```
28 | torchrun --nnodes=1 --nproc_per_node=8 train_cvpr.py
29 | ```
30 | The checkpoint saved by train_cvpr.py can be updated by `update_ckpt.py` to remove the additional `module` key due to multi-gpu training.
31 |
32 |
33 | # Inference
34 | You can directly download the [docker file](https://drive.google.com/file/d/1r2KvHP_30nHR3LU7NJEdscVnlZ2hTtcd/view?usp=sharing) for the challenge baseline.
35 | We provide a Dockerfile to satisfy the challenge format. For more details, refer to the [challenge website]((https://www.codabench.org/competitions/5263/)).
36 | ```
37 | docker build -t vista3d:latest .
38 | docker save -o vista3d.tar.gz vista3d:latest
39 | ```
40 | You can also directly run `predict.sh`. Download the finetuned checkpoint and modify the `--model=/your_downloaded_checkpoint`. Change `save_data=True` in `infer_cvpr.py` to save predictions to nifti files for visualization.
41 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/infer_cvpr.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import glob
3 |
4 | import monai
5 | import monai.transforms
6 | import nibabel as nib
7 | import numpy as np
8 | import torch
9 | from monai.apps.vista3d.inferer import point_based_window_inferer
10 | from monai.inferers import SlidingWindowInfererAdapt
11 | from monai.networks.nets.vista3d import vista3d132
12 | from monai.utils import optional_import
13 |
14 | tqdm, _ = optional_import("tqdm", name="tqdm")
15 | import os
16 |
17 | from train_cvpr import ROI_SIZE
18 |
19 | def convert_clicks(alldata):
20 | # indexes = list(alldata.keys())
21 | # data = [alldata[i] for i in indexes]
22 | data = alldata
23 | B = len(data) # Number of objects
24 | indexes = np.arange(1, B + 1).tolist()
25 | # Determine the maximum number of points across all objects
26 | max_N = max(len(obj["fg"]) + len(obj["bg"]) for obj in data)
27 |
28 | # Initialize padded arrays
29 | point_coords = np.zeros((B, max_N, 3), dtype=int)
30 | point_labels = np.full((B, max_N), -1, dtype=int)
31 |
32 | for i, obj in enumerate(data):
33 | points = []
34 | labels = []
35 |
36 | # Add foreground points
37 | for fg_point in obj["fg"]:
38 | points.append(fg_point)
39 | labels.append(1)
40 |
41 | # Add background points
42 | for bg_point in obj["bg"]:
43 | points.append(bg_point)
44 | labels.append(0)
45 |
46 | # Fill in the arrays
47 | point_coords[i, : len(points)] = points
48 | point_labels[i, : len(labels)] = labels
49 |
50 | return point_coords, point_labels, indexes
51 |
52 |
53 | if __name__ == "__main__":
54 | # set to true to save nifti files for visualization
55 | save_data = False
56 | point_inferer = True # use point based inferen
57 | parser = argparse.ArgumentParser()
58 | parser.add_argument("--test_img_path", type=str, default="./tests")
59 | parser.add_argument("--save_path", type=str, default="./outputs/")
60 | parser.add_argument("--model", type=str, default="checkpoints/model_final.pth")
61 | args = parser.parse_args()
62 | os.makedirs(args.save_path, exist_ok=True)
63 | # load model
64 | checkpoint_path = args.model
65 | model = vista3d132(in_channels=1)
66 | pretrained_ckpt = torch.load(checkpoint_path, map_location="cuda")
67 | model.load_state_dict(pretrained_ckpt, strict=True)
68 |
69 | # load data
70 | test_cases = glob.glob(os.path.join(args.test_img_path, "*.npz"))
71 | for img_path in test_cases:
72 | case_name = os.path.basename(img_path)
73 | print(case_name)
74 | img = np.load(img_path, allow_pickle=True)
75 | img_array = img["imgs"]
76 | spacing = img["spacing"]
77 | original_shape = img_array.shape
78 | affine = np.diag(spacing.tolist() + [1]) # 4x4 affine matrix
79 | if save_data:
80 | # Create a NIfTI image
81 | nifti_img = nib.Nifti1Image(img_array, affine)
82 | # Save the NIfTI file
83 | nib.save(nifti_img, img_path.replace(".npz", ".nii.gz"))
84 | nifti_img = nib.Nifti1Image(img["gts"], affine)
85 | # Save the NIfTI file
86 | nib.save(nifti_img, img_path.replace(".npz", "gts.nii.gz"))
87 | clicks = img.get("clicks", [{"fg": [[418, 138, 136]], "bg": []}])
88 | point_coords, point_labels, indexes = convert_clicks(clicks)
89 | # preprocess
90 | img_array = torch.from_numpy(img_array)
91 | img_array = img_array.unsqueeze(0)
92 | img_array = monai.transforms.ScaleIntensityRangePercentiles(
93 | lower=1, upper=99, b_min=0, b_max=1, clip=True
94 | )(img_array)
95 | img_array = img_array.unsqueeze(0) # add channel dim
96 | device = "cuda"
97 | # slidingwindow
98 | with torch.no_grad():
99 | if not point_inferer:
100 | model.NINF_VALUE = 0 # set to 0 in case sliding window is used.
101 | # directly using slidingwindow inferer is not optimal.
102 | val_outputs = (
103 | SlidingWindowInfererAdapt(
104 | roi_size=ROI_SIZE,
105 | sw_batch_size=1,
106 | with_coord=True,
107 | padding_mode="replicate",
108 | )(
109 | inputs=img_array.to(device),
110 | transpose=True,
111 | network=model.to(device),
112 | point_coords=torch.from_numpy(point_coords).to(device),
113 | point_labels=torch.from_numpy(point_labels).to(device),
114 | )[
115 | 0
116 | ]
117 | > 0
118 | )
119 | final_outputs = torch.zeros_like(val_outputs[0], dtype=torch.float32)
120 | for i, v in enumerate(val_outputs):
121 | final_outputs += indexes[i] * v
122 | else:
123 | # point based
124 | final_outputs = torch.zeros_like(img_array[0, 0], dtype=torch.float32)
125 | for i, v in enumerate(indexes):
126 | val_outputs = (
127 | point_based_window_inferer(
128 | inputs=img_array.to(device),
129 | roi_size=ROI_SIZE,
130 | transpose=True,
131 | with_coord=True,
132 | predictor=model.to(device),
133 | mode="gaussian",
134 | sw_device=device,
135 | device=device,
136 | center_only=True, # only crop the center
137 | point_coords=torch.from_numpy(point_coords[[i]]).to(device),
138 | point_labels=torch.from_numpy(point_labels[[i]]).to(device),
139 | )[0]
140 | > 0
141 | )
142 | final_outputs[val_outputs[0]] = v
143 | final_outputs = torch.nan_to_num(final_outputs)
144 | # save data
145 | if save_data:
146 | # Create a NIfTI image
147 | nifti_img = nib.Nifti1Image(
148 | final_outputs.to(torch.float32).data.cpu().numpy(), affine
149 | )
150 | # Save the NIfTI file
151 | nib.save(
152 | nifti_img,
153 | os.path.join(args.save_path, case_name.replace(".npz", ".nii.gz")),
154 | )
155 | np.savez_compressed(
156 | os.path.join(args.save_path, case_name),
157 | segs=final_outputs.to(torch.float32).data.cpu().numpy(),
158 | )
159 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/predict.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | # Run inference script with input/output folder paths
4 | python3 infer.py --test_img_path /workspace/inputs/ --save_path /workspace/outputs/ --model /workspace/model_final.pth
5 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/requirements.txt:
--------------------------------------------------------------------------------
1 | tensorboard
2 | matplotlib
3 | monai
4 | torchvision
5 | nibabel
6 | torch
7 | connected-components-3d
8 | pandas
9 | numpy
10 | scipy
11 | cupy-cuda12x
12 | cucim
13 | tqdm
14 |
--------------------------------------------------------------------------------
/vista3d/cvpr_workshop/update_ckpt.py:
--------------------------------------------------------------------------------
1 | import argparse
2 |
3 | import torch
4 |
5 |
6 | def remove_module_prefix(input_pth, output_pth):
7 | # Load the checkpoint
8 | checkpoint = torch.load(input_pth, map_location="cpu")["model"]
9 |
10 | # Modify the state_dict to remove 'module.' prefix
11 | new_state_dict = {}
12 | for key, value in checkpoint.items():
13 | if isinstance(value, dict) and "state_dict" in value:
14 | # If the checkpoint contains a 'state_dict' key (common in some saved models)
15 | new_state_dict = {
16 | k.replace("module.", ""): v for k, v in value["state_dict"].items()
17 | }
18 | value["state_dict"] = new_state_dict
19 | torch.save(value, output_pth)
20 | print(f"Updated weights saved to {output_pth}")
21 | return
22 | elif "module." in key:
23 | new_state_dict[key.replace("module.", "")] = value
24 | else:
25 | new_state_dict[key] = value
26 |
27 | # Save the modified weights
28 | torch.save(new_state_dict, output_pth)
29 | print(f"Updated weights saved to {output_pth}")
30 |
31 |
32 | if __name__ == "__main__":
33 | parser = argparse.ArgumentParser(
34 | description="Remove 'module.' prefix from PyTorch weights"
35 | )
36 | parser.add_argument("--input", required=True, help="Path to input .pth file")
37 | parser.add_argument(
38 | "--output", required=True, help="Path to save the modified .pth file"
39 | )
40 | args = parser.parse_args()
41 |
42 | remove_module_prefix(args.input, args.output)
43 |
--------------------------------------------------------------------------------
/vista3d/data/README.md:
--------------------------------------------------------------------------------
1 | #### Aggregating multiple datasets
2 |
3 | The training workflow requires one or multiple dataset JSON files to specifiy the image and segmentation pairs as well as dataset preprocessing transformations.
4 | Example files are located in the `data/jsons` folder.
5 |
6 | The JSON file has the following structure:
7 | ```python
8 | {
9 | "training": [
10 | {
11 | "image": "img1.nii.gz", # relative path to the primary image file
12 | "label": "label1.nii.gz", # optional relative path to the primary label file
13 | "pseudo_label": "p_label1.nii.gz", # optional relative path to the pseudo label file
14 | "pseudo_label_reliability": 1 # optional reliability score for pseudo label
15 | "label_sv": "label_sv1.nii.gz", # optional relative path to the supervoxel label file
16 | "fold": 0 # optional fold index for cross validation, fold 0 is used for training
17 | },
18 |
19 | ...
20 | ],
21 | "training_transform": [
22 | # a set of monai transform configuration for dataset-specific loading
23 | ],
24 | "original_label_dict": {"1": "liver", ...},
25 | "label_dict": {"1": "liver", ...}
26 | }
27 | ```
28 |
29 | During training, the JSON files will be consumed along with additional configurations, for example:
30 | ```py
31 | from data.datasets import get_datalist_with_dataset_name_and_transform
32 |
33 | train_files, _, dataset_specific_transforms, dataset_specific_transforms_val = \
34 | get_datalist_with_dataset_name_and_transform(
35 | datasets=train_datasets,
36 | fold_idx=fold,
37 | image_key=image_key,
38 | label_key=label_key,
39 | label_sv_key=label_sv_key,
40 | pseudo_label_key=pseudo_label_key,
41 | num_patches_per_image=parser.get_parsed_content("num_patches_per_image"),
42 | patch_size=parser.get_parsed_content("patch_size"),
43 | json_dir=json_dir)
44 | ```
45 |
46 | The following steps are necessary for creating a multi-dataset data loader for model training.
47 | Step 1 and 2 generate persistent JSON files based on the original dataset (the `image` and `label` pairs; without the additional pseudo label or supervoxel-based label), and only need to be run once when the JSON files don't exist.
48 |
49 | ##### 1. Generate data list JSON file
50 | ```
51 | python -m data.make_datalists
52 | ```
53 |
54 | This script reads image and label folders, lists all the nii.gz files,
55 | creates a JSON file in a format:
56 |
57 | ```json
58 | {
59 | "training": [
60 | {"image": "img0001.nii.gz", "label": "label0001.nii.gz", "fold": 0},
61 | {"image": "img0002.nii.gz", "label": "label0002.nii.gz", "fold": 2},
62 | ...
63 | ],
64 | "testing": [
65 | {"image": "img0003.nii.gz", "label": "label0003.nii.gz"},
66 | {"image": "img0004.nii.gz", "label": "label0004.nii.gz"},
67 | ...
68 | ]
69 | "original_label_dict": {"1": "liver", ...},
70 | "label_dict": {"1": "liver", ...}
71 | }
72 | ```
73 |
74 | This step includes a 5-fold cross validation splitting and
75 | some logic for 80-20 training/testing splitting. User need to modify the code in make_datalists.py for their own dataset. Meanwhile, the "training_transform" should manually added for each dataset.
76 |
77 | The `original_label_dict` corresponds to the original dataset label definitions.
78 | The `label_dict` modifies `original_label_dict` by simply rephrasing the terms.
79 | For example in Task06, `cancer` is renamed to `lung tumor`.
80 | The output of this step is multiple JSON files, each file corresponds
81 | to one dataset.
82 |
83 | ##### 2. Add label_dict.json and label_mapping.json
84 | Add new class indexes to `label_dict.json` and the local to global mapping to `label_mapping.json`.
85 |
86 | ## SupverVoxel Generation
87 | 1. Download the segment anything repo and download the ViT-H weights
88 | ```
89 | git clone https://github.com/facebookresearch/segment-anything.git
90 | mv segment-anything/segment_anything/ segment_anything/
91 | wget https://dl.fbaipublicfiles.com/segment_anything/sam_vit_h_4b8939.pth
92 | ```
93 | 2. Modify the code for supervoxel generation
94 | - Add this function to `predictor.py/SamPredictor`
95 | ```python
96 | @torch.no_grad()
97 | def get_feature_upsampled(self, input_image=None):
98 | if input_image is None:
99 | image_embeddings = self.model.mask_decoder.predict_masks_noprompt(self.features)
100 | else:
101 | image_embeddings = self.model.mask_decoder.predict_masks_noprompt(self.model.image_encoder(input_image))
102 | return image_embeddings
103 | ```
104 | - Add this function to `modeling/mask_decoder.py/MaskDecoder`
105 | ```python
106 | def predict_masks_noprompt(
107 | self,
108 | image_embeddings: torch.Tensor,
109 | ) -> Tuple[torch.Tensor, torch.Tensor]:
110 | """Predicts masks. See 'forward' for more details."""
111 | # Concatenate output tokens
112 |
113 | # Expand per-image data in batch direction to be per-mask
114 | src = image_embeddings
115 | # Upscale mask embeddings and predict masks using the mask tokens
116 | upscaled_embedding = self.output_upscaling(src)
117 |
118 | return upscaled_embedding
119 | ```
120 | 3. Run the supervoxel generation script. The processsing time is over 10 minutes, use `batch_infer` and multi-gpu for speed up.
121 | ```
122 | python -m scripts.slic_process_sam infer --image_file xxxx
123 | ```
124 |
--------------------------------------------------------------------------------
/vista3d/data/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
--------------------------------------------------------------------------------
/vista3d/data/dataset_weights.yaml:
--------------------------------------------------------------------------------
1 | # This is the weights for weighted sampling in stage2 and stage4
2 | {
3 | "CTPelvic1K-CLINIC": 1.3333333333333333,
4 | "AbdomenCT-1K": 0.15625,
5 | "AeroPath": 5.882352941176471,
6 | "AMOS22": 0.5208333333333334,
7 | "BTCV-Abdomen": 5.2631578947368425,
8 | "BTCV-Cervix": 5.2631578947368425,
9 | "CT-ORG": 1.1494252873563218,
10 | "FLARE22": 3.125,
11 | "Multi-organ-Abdominal-CT-btcv": 3.3333333333333335,
12 | "Multi-organ-Abdominal-CT-tcia": 3.7037037037037037,
13 | "Pancreas-CT": 1.9607843137254901,
14 | "Task03": 1.1904761904761905,
15 | "Task06": 2.5,
16 | "Task07": 0.5555555555555556,
17 | "Task08": 0.5181347150259067,
18 | "Task09": 3.8461538461538463,
19 | "Task10": 1.25,
20 | "VerSe": 0.41841004184100417,
21 | "Bone-NIH": 0.5291005291005291,
22 | "CRLM-CT": 0.7936507936507936,
23 | "TotalSegmentatorV2": 0.12755102040816327
24 | }
25 |
--------------------------------------------------------------------------------
/vista3d/data/datasets.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import os
13 | from pprint import pformat
14 |
15 | from monai import transforms
16 | from monai.apps import get_logger
17 | from monai.auto3dseg.utils import datafold_read
18 | from monai.bundle import ConfigParser
19 | from monai.utils import ensure_tuple, look_up_option
20 |
21 | # Define the root path to the orignal manual label files, supervoxels, and pseudolabels.
22 | all_base_dirs = {
23 | "AbdomenCT-1K": "/data/AbdomenCT-1K",
24 | "FLARE22": "/data/AbdomenCT-1K/FLARE22Train",
25 | "AMOS22": "/data/AMOS22",
26 | "BTCV-Abdomen": "/data/BTCV/Abdomen",
27 | "BTCV-Cervix": "/data/BTCV/Cervix",
28 | "CT-ORG": "/data/CT-ORG",
29 | "Multi-organ-Abdominal-CT-btcv": "/data/Multi-organ-Abdominal-CT/res_1.0mm_relabeled2",
30 | "Multi-organ-Abdominal-CT-tcia": "/data/Multi-organ-Abdominal-CT/res_1.0mm_relabeled2",
31 | "Pancreas-CT": "/data/Pancreas-CT",
32 | "Task06": "/data/Task06",
33 | "Task07": "/data/Task07",
34 | "Task08": "/data/Task08",
35 | "Task09": "/data/Task09",
36 | "Task10": "/data/Task10",
37 | "TotalSegmentator": "/data/TotalSegmentator",
38 | "TotalSegmentatorV2": "/data/TotalSegmentatorV2",
39 | "Task03": "/data/Task03",
40 | "Bone-NIH": "/data/Bone-NIH",
41 | "CRLM-CT": "/data/CRLM-CT/nifti",
42 | "VerSe": "/data/VerSe/",
43 | "AeroPath": "/data/AeroPath/",
44 | "CTPelvic1K-CLINIC": "/data/CTPelvic1K-CLINIC",
45 | "NLST": "/data/NLST",
46 | "LIDC": "/data/LIDC",
47 | "Covid19": "/data/Covid19",
48 | "TCIA_Colon": "/data/TCIA_Colon",
49 | "StonyBrook-CT": "/data/StonyBrook-CT",
50 | }
51 | # Notice the root path to supervoxel and pseudolabel has the same sub-folder structure as all_base_dirs
52 | # The path is generated by replacing json_base with supervoxel_base/pl_base in get_datalist_with_dataset_name
53 | json_base = "/data/"
54 | supervoxel_base = "/workspace_infer/supervoxel_sam/"
55 | pl_base = "/workspace_infer/V2_pseudo_12Feb2024/"
56 |
57 |
58 | cur_json_dir = os.path.join(os.path.dirname(__file__), "jsons")
59 | logger = get_logger(__name__)
60 |
61 |
62 | def get_json_files_k_folds(json_dir=None, base_dirs=None, k=5):
63 | """the json files are generated by data/make_datalists.py, stored at `json_dir`"""
64 | if json_dir is None:
65 | json_dir = cur_json_dir
66 | if base_dirs is None:
67 | base_dirs = all_base_dirs
68 | output_dict = {
69 | item: os.path.join(json_dir, f"{item}_{k}_folds.json") for item in base_dirs
70 | }
71 | logger.debug(pformat(output_dict))
72 | return output_dict
73 |
74 |
75 | def get_class_names(json_dir=None):
76 | """
77 | the list of class names, background is at 0
78 | """
79 | parser = ConfigParser.load_config_file(os.path.join(json_dir, "label_dict.json"))
80 | label_dict = dict(parser)
81 | label_dict["unspecified region"] = 0
82 | inv_label_dict = {v: k for k, v in label_dict.items()}
83 | label_list = []
84 | for i in range(len(label_dict)):
85 | label_list.append(inv_label_dict[i])
86 | return label_list
87 |
88 |
89 | def get_datalist_with_dataset_name(
90 | datasets=None, fold_idx=-1, key="training", json_dir=None, base_dirs=None
91 | ):
92 | """
93 | when `datasets` is None, it returns a list of all data from all datasets.
94 | when `datasets` is a list of dataset names, it returns a list of all data from the specified datasets.
95 |
96 | train_list's item format::
97 |
98 | {"image": image_file_path, "label": label_file_path, "dataset_name": dataset_name, "fold": fold_id}
99 |
100 | """
101 | if base_dirs is None:
102 | base_dirs = all_base_dirs # all_base_dirs is the broader set
103 | # get the list of training/validation files (absolute path)
104 | json_files = get_json_files_k_folds(json_dir=json_dir, base_dirs=base_dirs)
105 | if datasets is None:
106 | loading_dict = json_files.copy()
107 | else:
108 | loading_dict = {
109 | k: look_up_option(k, json_files) for k in ensure_tuple(datasets)
110 | }
111 | train_list, val_list = [], []
112 | for k, j in loading_dict.items():
113 | t, v = datafold_read(j, basedir=all_base_dirs[k], fold=fold_idx, key=key)
114 | for item in t:
115 | item["dataset_name"] = k
116 | if "label_sv" in item.keys():
117 | item["label_sv"] = item["label_sv"].replace(
118 | json_base, supervoxel_base, 1
119 | )
120 | if "pseudo_label" in item.keys():
121 | item["pseudo_label"] = item["pseudo_label"].replace(
122 | json_base, pl_base, 1
123 | )
124 | train_list += t
125 | for item in v:
126 | item["dataset_name"] = k
127 | if "label_sv" in item.keys():
128 | item["label_sv"] = item["label_sv"].replace(
129 | json_base, supervoxel_base, 1
130 | )
131 | if "pseudo_label" in item.keys():
132 | item["pseudo_label"] = item["pseudo_label"].replace(
133 | json_base, pl_base, 1
134 | )
135 |
136 | val_list += v
137 | logger.warning(
138 | f"data list from datasets={datasets} fold={fold_idx}: train={len(train_list)}, val={len(val_list)}"
139 | )
140 | return ensure_tuple(train_list), ensure_tuple(val_list)
141 |
142 |
143 | def get_datalist_with_dataset_name_and_transform(
144 | image_key,
145 | label_key,
146 | label_sv_key,
147 | pseudo_label_key,
148 | num_patches_per_image,
149 | patch_size,
150 | datasets=None,
151 | fold_idx=-1,
152 | key="training",
153 | json_dir=None,
154 | base_dirs=None,
155 | ):
156 | """
157 | when `datasets` is None, it returns a list of all data from all datasets.
158 | when `datasets` is a list of dataset names, it returns a list of all data from the specified datasets.
159 | Return file lists and specific transforms for each dataset.
160 |
161 | """
162 | if base_dirs is None:
163 | base_dirs = all_base_dirs # all_base_dirs is the broader set
164 | train_list, val_list = get_datalist_with_dataset_name(
165 | datasets=datasets,
166 | fold_idx=fold_idx,
167 | key=key,
168 | json_dir=json_dir,
169 | base_dirs=base_dirs,
170 | )
171 | # get the list of training/validation files (absolute path)
172 | json_files = get_json_files_k_folds(json_dir=json_dir, base_dirs=base_dirs)
173 | if datasets is None:
174 | loading_dict = json_files.copy()
175 | else:
176 | loading_dict = {
177 | k: look_up_option(k, json_files) for k in ensure_tuple(datasets)
178 | }
179 |
180 | dataset_transforms = {}
181 | dataset_transforms_val = {}
182 | for k, j in loading_dict.items():
183 | parser = ConfigParser()
184 | parser.read_config(j)
185 | # those parameters are required to initiate the transforms
186 | parser.update(
187 | pairs={
188 | "image_key": image_key,
189 | "label_key": label_key,
190 | "label_sv_key": label_sv_key,
191 | "pseudo_label_key": pseudo_label_key,
192 | "num_patches_per_image": num_patches_per_image,
193 | "patch_size": patch_size,
194 | }
195 | )
196 | transform = parser.get_parsed_content("training_transform")
197 | dataset_transforms[k] = transforms.Compose(transform)
198 | transform_val = parser.get_parsed_content("validation_transform", default=None)
199 | dataset_transforms_val[k] = (
200 | transforms.Compose(transform_val) if transform_val is not None else None
201 | )
202 | return (
203 | ensure_tuple(train_list),
204 | ensure_tuple(val_list),
205 | dataset_transforms,
206 | dataset_transforms_val,
207 | )
208 |
209 |
210 | def compute_dataset_weights(datalist, weight_path="./data/dataset_weights.yaml"):
211 | """based on class-wise weight, assign a weight to each training sample"""
212 | cfg = ConfigParser.load_config_file(weight_path)
213 | w = []
214 | for item in datalist:
215 | fg_w = cfg[item["dataset_name"]]
216 | w.append(fg_w)
217 | item["w"] = fg_w
218 | return w
219 |
220 |
221 | def calculate_dataset_weights(datalist):
222 | dataset_name = []
223 | dataset_counts = {}
224 | for item in datalist:
225 | dn = item["dataset_name"]
226 | if dn in dataset_name:
227 | dataset_counts[dn] += 1
228 | else:
229 | dataset_name.append(dn)
230 | dataset_counts[dn] = 1
231 | dataset_weights = {}
232 | non_tumor_count = 0
233 | tumor_count = 0
234 | for item in dataset_name:
235 | if item not in ["Task03", "Task06", "Task07", "Task08", "Task10", "Bone-NIH"]:
236 | non_tumor_count += dataset_counts[item]
237 | else:
238 | tumor_count += dataset_counts[item]
239 |
240 | for item in dataset_name:
241 | if item not in ["Task03", "Task06", "Task07", "Task08", "Task10", "Bone-NIH"]:
242 | dataset_weights[item] = 100 / dataset_counts[item] # non_tumor_count
243 | else:
244 | dataset_weights[item] = 100 / dataset_counts[item] # tumor_count
245 |
246 | dataset_prob = {}
247 | total_prob = 0
248 | for item in dataset_name:
249 | dataset_prob[item] = dataset_weights[item] * dataset_counts[item]
250 | total_prob += dataset_prob[item]
251 | for item in dataset_name:
252 | dataset_prob[item] /= total_prob
253 |
254 | import json
255 |
256 | with open("./dataset_counts.yaml", "w") as f:
257 | json.dump(dataset_counts, f, indent=4)
258 | with open("./dataset_weights.yaml", "w") as f:
259 | json.dump(dataset_weights, f, indent=4)
260 | with open("./dataset_prob.yaml", "w") as f:
261 | json.dump(dataset_prob, f, indent=4)
262 |
263 |
264 | if __name__ == "__main__":
265 | from monai.utils import optional_import
266 |
267 | fire, _ = optional_import("fire")
268 | fire.Fire()
269 |
--------------------------------------------------------------------------------
/vista3d/data/jsons/AeroPath_5_folds.json:
--------------------------------------------------------------------------------
1 | {
2 | "training": [
3 | {
4 | "image": "8/8_CT_HR.nii.gz",
5 | "pseudo_label": "8/8_CT_HR.nii.gz",
6 | "label": "8/8_CT_HR_label.nii.gz",
7 | "fold": 0,
8 | "pseudo_label_reliability": 0
9 | },
10 | {
11 | "image": "6/6_CT_HR.nii.gz",
12 | "pseudo_label": "6/6_CT_HR.nii.gz",
13 | "label": "6/6_CT_HR_label.nii.gz",
14 | "fold": 0,
15 | "pseudo_label_reliability": 1
16 | },
17 | {
18 | "image": "14/14_CT_HR.nii.gz",
19 | "pseudo_label": "14/14_CT_HR.nii.gz",
20 | "label": "14/14_CT_HR_label.nii.gz",
21 | "fold": 0,
22 | "pseudo_label_reliability": 1
23 | },
24 | {
25 | "image": "16/16_CT_HR.nii.gz",
26 | "pseudo_label": "16/16_CT_HR.nii.gz",
27 | "label": "16/16_CT_HR_label.nii.gz",
28 | "fold": 0,
29 | "pseudo_label_reliability": 0
30 | },
31 | {
32 | "image": "15/15_CT_HR.nii.gz",
33 | "pseudo_label": "15/15_CT_HR.nii.gz",
34 | "label": "15/15_CT_HR_label.nii.gz",
35 | "fold": 0,
36 | "pseudo_label_reliability": 0
37 | },
38 | {
39 | "image": "27/27_CT_HR.nii.gz",
40 | "pseudo_label": "27/27_CT_HR.nii.gz",
41 | "label": "27/27_CT_HR_label.nii.gz",
42 | "fold": 1,
43 | "pseudo_label_reliability": 0,
44 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/27_CT_HR/27_CT_HR_seg.nii.gz"
45 | },
46 | {
47 | "image": "20/20_CT_HR.nii.gz",
48 | "pseudo_label": "20/20_CT_HR.nii.gz",
49 | "label": "20/20_CT_HR_label.nii.gz",
50 | "fold": 1,
51 | "pseudo_label_reliability": 1,
52 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/20_CT_HR/20_CT_HR_seg.nii.gz"
53 | },
54 | {
55 | "image": "1/1_CT_HR.nii.gz",
56 | "pseudo_label": "1/1_CT_HR.nii.gz",
57 | "label": "1/1_CT_HR_label.nii.gz",
58 | "fold": 1,
59 | "pseudo_label_reliability": 0,
60 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/1_CT_HR/1_CT_HR_seg.nii.gz"
61 | },
62 | {
63 | "image": "18/18_CT_HR.nii.gz",
64 | "pseudo_label": "18/18_CT_HR.nii.gz",
65 | "label": "18/18_CT_HR_label.nii.gz",
66 | "fold": 1,
67 | "pseudo_label_reliability": 1,
68 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/18_CT_HR/18_CT_HR_seg.nii.gz"
69 | },
70 | {
71 | "image": "25/25_CT_HR.nii.gz",
72 | "pseudo_label": "25/25_CT_HR.nii.gz",
73 | "label": "25/25_CT_HR_label.nii.gz",
74 | "fold": 1,
75 | "pseudo_label_reliability": 0,
76 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/25_CT_HR/25_CT_HR_seg.nii.gz"
77 | },
78 | {
79 | "image": "13/13_CT_HR.nii.gz",
80 | "pseudo_label": "13/13_CT_HR.nii.gz",
81 | "label": "13/13_CT_HR_label.nii.gz",
82 | "fold": 2,
83 | "pseudo_label_reliability": 1,
84 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/13_CT_HR/13_CT_HR_seg.nii.gz"
85 | },
86 | {
87 | "image": "23/23_CT_HR.nii.gz",
88 | "pseudo_label": "23/23_CT_HR.nii.gz",
89 | "label": "23/23_CT_HR_label.nii.gz",
90 | "fold": 2,
91 | "pseudo_label_reliability": 0,
92 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/23_CT_HR/23_CT_HR_seg.nii.gz"
93 | },
94 | {
95 | "image": "5/5_CT_HR.nii.gz",
96 | "pseudo_label": "5/5_CT_HR.nii.gz",
97 | "label": "5/5_CT_HR_label.nii.gz",
98 | "fold": 2,
99 | "pseudo_label_reliability": 1,
100 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/5_CT_HR/5_CT_HR_seg.nii.gz"
101 | },
102 | {
103 | "image": "10/10_CT_HR.nii.gz",
104 | "pseudo_label": "10/10_CT_HR.nii.gz",
105 | "label": "10/10_CT_HR_label.nii.gz",
106 | "fold": 2,
107 | "pseudo_label_reliability": 0,
108 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/10_CT_HR/10_CT_HR_seg.nii.gz"
109 | },
110 | {
111 | "image": "17/17_CT_HR.nii.gz",
112 | "pseudo_label": "17/17_CT_HR.nii.gz",
113 | "label": "17/17_CT_HR_label.nii.gz",
114 | "fold": 3,
115 | "pseudo_label_reliability": 1,
116 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/17_CT_HR/17_CT_HR_seg.nii.gz"
117 | },
118 | {
119 | "image": "12/12_CT_HR.nii.gz",
120 | "pseudo_label": "12/12_CT_HR.nii.gz",
121 | "label": "12/12_CT_HR_label.nii.gz",
122 | "fold": 3,
123 | "pseudo_label_reliability": 0,
124 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/12_CT_HR/12_CT_HR_seg.nii.gz"
125 | },
126 | {
127 | "image": "3/3_CT_HR.nii.gz",
128 | "pseudo_label": "3/3_CT_HR.nii.gz",
129 | "label": "3/3_CT_HR_label.nii.gz",
130 | "fold": 3,
131 | "pseudo_label_reliability": 0,
132 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/3_CT_HR/3_CT_HR_seg.nii.gz"
133 | },
134 | {
135 | "image": "24/24_CT_HR.nii.gz",
136 | "pseudo_label": "24/24_CT_HR.nii.gz",
137 | "label": "24/24_CT_HR_label.nii.gz",
138 | "fold": 3,
139 | "pseudo_label_reliability": 0,
140 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/24_CT_HR/24_CT_HR_seg.nii.gz"
141 | },
142 | {
143 | "image": "9/9_CT_HR.nii.gz",
144 | "pseudo_label": "9/9_CT_HR.nii.gz",
145 | "label": "9/9_CT_HR_label.nii.gz",
146 | "fold": 4,
147 | "pseudo_label_reliability": 1,
148 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/9_CT_HR/9_CT_HR_seg.nii.gz"
149 | },
150 | {
151 | "image": "26/26_CT_HR.nii.gz",
152 | "pseudo_label": "26/26_CT_HR.nii.gz",
153 | "label": "26/26_CT_HR_label.nii.gz",
154 | "fold": 4,
155 | "pseudo_label_reliability": 0,
156 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/26_CT_HR/26_CT_HR_seg.nii.gz"
157 | },
158 | {
159 | "image": "11/11_CT_HR.nii.gz",
160 | "pseudo_label": "11/11_CT_HR.nii.gz",
161 | "label": "11/11_CT_HR_label.nii.gz",
162 | "fold": 4,
163 | "pseudo_label_reliability": 0,
164 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/11_CT_HR/11_CT_HR_seg.nii.gz"
165 | },
166 | {
167 | "image": "7/7_CT_HR.nii.gz",
168 | "pseudo_label": "7/7_CT_HR.nii.gz",
169 | "label": "7/7_CT_HR_label.nii.gz",
170 | "fold": 4,
171 | "pseudo_label_reliability": 0,
172 | "label_sv": "/workspace_infer/supervoxel_sam/AeroPath_100/7_CT_HR/7_CT_HR_seg.nii.gz"
173 | }
174 | ],
175 | "training_transform": [
176 | {
177 | "_target_": "RandCropByLabelClassesd",
178 | "keys": [
179 | "@image_key",
180 | "@label_key",
181 | "@pseudo_label_key",
182 | "@label_sv_key"
183 | ],
184 | "label_key": "@pseudo_label_key",
185 | "num_classes": 133,
186 | "num_samples": "@num_patches_per_image",
187 | "spatial_size": "@patch_size",
188 | "ratios": "$tuple(float(i >= 0) for i in range(133))",
189 | "warn": false,
190 | "allow_missing_keys": true
191 | },
192 | {
193 | "_target_": "RandZoomd",
194 | "keys": [
195 | "@image_key",
196 | "@label_key",
197 | "@pseudo_label_key",
198 | "@label_sv_key"
199 | ],
200 | "min_zoom": 0.8,
201 | "max_zoom": 1.2,
202 | "mode": [
203 | "trilinear",
204 | "nearest",
205 | "nearest",
206 | "nearest"
207 | ],
208 | "prob": 0.2,
209 | "allow_missing_keys": true
210 | },
211 | {
212 | "_target_": "RandSimulateLowResolutiond",
213 | "keys": [
214 | "@image_key"
215 | ],
216 | "zoom_range": [
217 | 0.3,
218 | 1
219 | ],
220 | "prob": 0.2,
221 | "allow_missing_keys": true
222 | },
223 | {
224 | "_target_": "RandGaussianSmoothd",
225 | "keys": [
226 | "@image_key"
227 | ],
228 | "prob": 0.2,
229 | "sigma_x": [
230 | 0.5,
231 | 1.0
232 | ],
233 | "sigma_y": [
234 | 0.5,
235 | 1.0
236 | ],
237 | "sigma_z": [
238 | 0.5,
239 | 1.0
240 | ]
241 | },
242 | {
243 | "_target_": "RandScaleIntensityd",
244 | "keys": [
245 | "@image_key"
246 | ],
247 | "factors": 0.1,
248 | "prob": 0.2
249 | },
250 | {
251 | "_target_": "RandShiftIntensityd",
252 | "keys": [
253 | "@image_key"
254 | ],
255 | "offsets": 0.1,
256 | "prob": 0.2
257 | },
258 | {
259 | "_target_": "RandGaussianNoised",
260 | "keys": [
261 | "@image_key"
262 | ],
263 | "prob": 0.2,
264 | "mean": 0.0,
265 | "std": 0.2
266 | }
267 | ],
268 | "label_dict": {
269 | "2": "airway"
270 | },
271 | "original_label_dict": {
272 | "1": "lung",
273 | "2": "airway"
274 | },
275 | "testing": [
276 | {
277 | "image": "19/19_CT_HR.nii.gz",
278 | "label": "19/19_CT_HR_label.nii.gz"
279 | },
280 | {
281 | "image": "2/2_CT_HR.nii.gz",
282 | "label": "2/2_CT_HR_label.nii.gz"
283 | },
284 | {
285 | "image": "22/22_CT_HR.nii.gz",
286 | "label": "22/22_CT_HR_label.nii.gz"
287 | },
288 | {
289 | "image": "21/21_CT_HR.nii.gz",
290 | "label": "21/21_CT_HR_label.nii.gz"
291 | },
292 | {
293 | "image": "4/4_CT_HR.nii.gz",
294 | "label": "4/4_CT_HR_label.nii.gz"
295 | }
296 | ]
297 | }
298 |
--------------------------------------------------------------------------------
/vista3d/data/jsons/label_dict.json:
--------------------------------------------------------------------------------
1 | {
2 | "liver": 1,
3 | "spleen": 3,
4 | "pancreas": 4,
5 | "right kidney": 5,
6 | "aorta": 6,
7 | "inferior vena cava": 7,
8 | "right adrenal gland": 8,
9 | "left adrenal gland": 9,
10 | "gallbladder": 10,
11 | "esophagus": 11,
12 | "stomach": 12,
13 | "duodenum": 13,
14 | "left kidney": 14,
15 | "bladder": 15,
16 | "portal vein and splenic vein": 17,
17 | "small bowel": 19,
18 | "brain": 22,
19 | "lung tumor": 23,
20 | "pancreatic tumor": 24,
21 | "hepatic vessel": 25,
22 | "hepatic tumor": 26,
23 | "colon cancer primaries": 27,
24 | "left lung upper lobe": 28,
25 | "left lung lower lobe": 29,
26 | "right lung upper lobe": 30,
27 | "right lung middle lobe": 31,
28 | "right lung lower lobe": 32,
29 | "vertebrae L5": 33,
30 | "vertebrae L4": 34,
31 | "vertebrae L3": 35,
32 | "vertebrae L2": 36,
33 | "vertebrae L1": 37,
34 | "vertebrae T12": 38,
35 | "vertebrae T11": 39,
36 | "vertebrae T10": 40,
37 | "vertebrae T9": 41,
38 | "vertebrae T8": 42,
39 | "vertebrae T7": 43,
40 | "vertebrae T6": 44,
41 | "vertebrae T5": 45,
42 | "vertebrae T4": 46,
43 | "vertebrae T3": 47,
44 | "vertebrae T2": 48,
45 | "vertebrae T1": 49,
46 | "vertebrae C7": 50,
47 | "vertebrae C6": 51,
48 | "vertebrae C5": 52,
49 | "vertebrae C4": 53,
50 | "vertebrae C3": 54,
51 | "vertebrae C2": 55,
52 | "vertebrae C1": 56,
53 | "trachea": 57,
54 | "left iliac artery": 58,
55 | "right iliac artery": 59,
56 | "left iliac vena": 60,
57 | "right iliac vena": 61,
58 | "colon": 62,
59 | "left rib 1": 63,
60 | "left rib 2": 64,
61 | "left rib 3": 65,
62 | "left rib 4": 66,
63 | "left rib 5": 67,
64 | "left rib 6": 68,
65 | "left rib 7": 69,
66 | "left rib 8": 70,
67 | "left rib 9": 71,
68 | "left rib 10": 72,
69 | "left rib 11": 73,
70 | "left rib 12": 74,
71 | "right rib 1": 75,
72 | "right rib 2": 76,
73 | "right rib 3": 77,
74 | "right rib 4": 78,
75 | "right rib 5": 79,
76 | "right rib 6": 80,
77 | "right rib 7": 81,
78 | "right rib 8": 82,
79 | "right rib 9": 83,
80 | "right rib 10": 84,
81 | "right rib 11": 85,
82 | "right rib 12": 86,
83 | "left humerus": 87,
84 | "right humerus": 88,
85 | "left scapula": 89,
86 | "right scapula": 90,
87 | "left clavicula": 91,
88 | "right clavicula": 92,
89 | "left femur": 93,
90 | "right femur": 94,
91 | "left hip": 95,
92 | "right hip": 96,
93 | "sacrum": 97,
94 | "left gluteus maximus": 98,
95 | "right gluteus maximus": 99,
96 | "left gluteus medius": 100,
97 | "right gluteus medius": 101,
98 | "left gluteus minimus": 102,
99 | "right gluteus minimus": 103,
100 | "left autochthon": 104,
101 | "right autochthon": 105,
102 | "left iliopsoas": 106,
103 | "right iliopsoas": 107,
104 | "left atrial appendage": 108,
105 | "brachiocephalic trunk": 109,
106 | "left brachiocephalic vein": 110,
107 | "right brachiocephalic vein": 111,
108 | "left common carotid artery": 112,
109 | "right common carotid artery": 113,
110 | "costal cartilages": 114,
111 | "heart": 115,
112 | "left kidney cyst": 116,
113 | "right kidney cyst": 117,
114 | "prostate": 118,
115 | "pulmonary vein": 119,
116 | "skull": 120,
117 | "spinal cord": 121,
118 | "sternum": 122,
119 | "left subclavian artery": 123,
120 | "right subclavian artery": 124,
121 | "superior vena cava": 125,
122 | "thyroid gland": 126,
123 | "vertebrae S1": 127,
124 | "bone lesion": 128,
125 | "airway": 132
126 | }
127 |
--------------------------------------------------------------------------------
/vista3d/requirements.txt:
--------------------------------------------------------------------------------
1 | fire==0.6.0
2 | matplotlib==3.8.3
3 | monai==1.3.2
4 | nibabel==5.2.1
5 | numpy==1.24.4
6 | Pillow==10.4.0
7 | PyYAML==6.0.2
8 | scipy==1.14.0
9 | scikit-image==0.24.0
10 | torch==2.0.1
11 | tqdm==4.66.2
12 | tensorboard==2.13.0
13 | einops==0.6.1
14 | ml-collections
15 | timm
16 | pytorch-ignite
17 | tensorboardX
18 |
--------------------------------------------------------------------------------
/vista3d/scripts/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
--------------------------------------------------------------------------------
/vista3d/scripts/debugger.py:
--------------------------------------------------------------------------------
1 | import copy
2 | from tkinter import Tk, filedialog, messagebox
3 |
4 | import fire
5 | import matplotlib.pyplot as plt
6 | import nibabel as nib
7 | import numpy as np
8 | from matplotlib.widgets import Button, TextBox
9 |
10 | from .infer import InferClass
11 | from .utils.workflow_utils import get_point_label
12 |
13 | inferer = InferClass(config_file=["./configs/infer.yaml"])
14 |
15 |
16 | class samm_visualizer:
17 | def __init__(self):
18 | self.clicked_points = []
19 | self.data = None
20 | self.mask_plot = None
21 | self.mask = None
22 | self.data_path = None
23 | self.circle_artists = []
24 | self.class_label = None
25 |
26 | def select_data_file(self):
27 | root = Tk()
28 | root.withdraw()
29 | file_path = filedialog.askopenfilename(
30 | title="Select Data File", initialfile=self.data_path
31 | )
32 | if not file_path:
33 | print("No file selected.")
34 | exit()
35 | # Load data from NIfTI file
36 | try:
37 | nifti_img = nib.load(file_path)
38 | data = nifti_img.get_fdata()
39 | if len(data.shape) == 4:
40 | data = data[..., 0] # Extract last element along the 4th dimension
41 | except FileNotFoundError:
42 | print("File not found.")
43 | exit()
44 | except nib.filebasedimages.ImageFileError:
45 | print("Invalid NIfTI file.")
46 | exit()
47 | self.data = data
48 | self.data_path = file_path
49 |
50 | def generate_mask(self):
51 | point = []
52 | point_label = []
53 | self.class_label = self.text_box.text
54 | if len(self.class_label) == 0:
55 | messagebox.showwarning(
56 | "Warning",
57 | "Label prompt is not specified. Assuming the point is for supported class. \
58 | For zero-shot, input random number > 132",
59 | )
60 | label_prompt = None
61 | prompt_class = None
62 | neg_id, pos_id = get_point_label(1)
63 | else:
64 | if self.class_label in [2, 20, 21]:
65 | messagebox.showwarning(
66 | "Warning",
67 | "Current debugger skip kidney (2), lung (20), and bone (21). Use their subclasses.",
68 | )
69 | return
70 | label_prompt = int(self.class_label)
71 | neg_id, pos_id = get_point_label(label_prompt)
72 | label_prompt = np.array([label_prompt])[np.newaxis, ...]
73 | prompt_class = copy.deepcopy(label_prompt)
74 | # if zero-shot
75 | if label_prompt is not None and label_prompt[0] > 132:
76 | label_prompt = None
77 | for p in self.clicked_points:
78 | point.append([p[1], p[0], p[2]])
79 | point_label.append(pos_id if p[3] == 1 else neg_id)
80 | if len(point) == 0:
81 | point = None
82 | point_label = None
83 | else:
84 | point = np.array(point)[np.newaxis, ...]
85 | point_label = np.array(point_label)[np.newaxis, ...]
86 | mask = inferer.infer(
87 | {"image": self.data_path},
88 | point,
89 | point_label,
90 | label_prompt,
91 | prompt_class,
92 | save_mask=True,
93 | point_start=self.point_start,
94 | )[0]
95 | nan_mask = np.isnan(mask)
96 | mask = mask.data.cpu().numpy() > 0.5
97 | mask = mask.astype(np.float32)
98 | mask[mask == 0] = np.nan
99 | if self.mask is None:
100 | self.mask = mask
101 | else:
102 | self.mask[~nan_mask] = mask[~nan_mask]
103 |
104 | def display_3d_slices(self):
105 | fig, ax = plt.subplots()
106 | assert self.data is not None, "Load data first."
107 | ax.volume = self.data
108 | ax.index = self.data.shape[2] // 2
109 | ax.imshow(self.data[:, :, ax.index], cmap="gray")
110 | ax.set_title(f"Slice {ax.index}")
111 | self.update_slice(ax)
112 | fig.canvas.mpl_connect("scroll_event", self.process_scroll)
113 | fig.canvas.mpl_connect("button_press_event", self.process_click)
114 | # Add numerical input box for slice index
115 | text_ax = plt.axes([0.45, 0.01, 0.2, 0.05]) # Position of the text box
116 | self.text_box = TextBox(text_ax, "Class prompt", initial=self.class_label)
117 | # Add a button
118 | button_ax = plt.axes([0.05, 0.01, 0.2, 0.05]) # Position of the button
119 | button = Button(button_ax, "Run")
120 |
121 | def on_button_click(event, ax=ax):
122 | # Define what happens when the button is clicked
123 | print("-- segmenting ---")
124 | self.generate_mask()
125 | print("-- done ---")
126 | print(
127 | "-- Note: Point only prompts will only do 128 cubic segmentation, a cropping artefact will be observed. ---"
128 | )
129 | print(
130 | "-- Note: Point without class will be treated as supported class, which has worse zero-shot ability. Try class > 132 to perform better zeroshot. ---"
131 | )
132 | print("-- Note: CTRL + Right Click will be adding negative points. ---")
133 | print(
134 | "-- Note: Click points on different foreground class will cause segmentation conflicts. Clear first. ---"
135 | )
136 | print(
137 | "-- Note: Click points not matching class prompts will also cause confusion. ---"
138 | )
139 |
140 | self.update_slice(ax)
141 | # self.point_start = len(self.clicked_points)
142 |
143 | button.on_clicked(on_button_click)
144 |
145 | button_ax_clear = plt.axes([0.75, 0.01, 0.2, 0.05]) # Position of the button
146 | button_clear = Button(button_ax_clear, "Clear")
147 |
148 | def on_button_click_clear(event, ax=ax):
149 | # Define what happens when the button is clicked
150 | inferer.clear_cache()
151 | # clear points
152 | self.clicked_points = []
153 | self.point_start = 0
154 | self.mask = None
155 | self.mask_plot.remove()
156 | self.mask_plot = None
157 | self.update_slice(ax)
158 |
159 | button_clear.on_clicked(on_button_click_clear)
160 |
161 | plt.show()
162 |
163 | def process_scroll(self, event):
164 | ax = event.inaxes
165 | try:
166 | if event.button == "up":
167 | self.previous_slice(ax)
168 | elif event.button == "down":
169 | self.next_slice(ax)
170 | except BaseException:
171 | pass
172 |
173 | def previous_slice(self, ax):
174 | if ax is None:
175 | return
176 | ax.index = (ax.index - 1) % ax.volume.shape[2]
177 | self.update_slice(ax)
178 |
179 | def next_slice(self, ax):
180 | if ax is None:
181 | return
182 | ax.index = (ax.index + 1) % ax.volume.shape[2]
183 | self.update_slice(ax)
184 |
185 | def update_slice(self, ax):
186 | # remove circles
187 | while len(self.circle_artists) > 0:
188 | ca = self.circle_artists.pop()
189 | ca.remove()
190 | # plot circles
191 | for x, y, z, label in self.clicked_points:
192 | if z == ax.index:
193 | color = "red" if (label == 1 or label == 3) else "blue"
194 | circle_artist = plt.Circle((x, y), 1, color=color, fill=False)
195 | self.circle_artists.append(circle_artist)
196 | ax.add_artist(circle_artist)
197 | ax.images[0].set_array(ax.volume[:, :, ax.index])
198 | if self.mask is not None and self.mask_plot is None:
199 | self.mask_plot = ax.imshow(
200 | np.zeros_like(self.mask[:, :, ax.index]) * np.nan,
201 | cmap="viridis",
202 | alpha=0.5,
203 | )
204 | if self.mask is not None and self.mask_plot is not None:
205 | self.mask_plot.set_data(self.mask[:, :, ax.index])
206 | self.mask_plot.set_visible(True)
207 | ax.set_title(f"Slice {ax.index}")
208 | ax.figure.canvas.draw()
209 |
210 | def process_click(self, event):
211 | try:
212 | ax = event.inaxes
213 | if ax is not None:
214 | x = int(event.xdata)
215 | y = int(event.ydata)
216 | z = ax.index
217 | print(f"Clicked coordinates: x={x}, y={y}, z={z}")
218 | if event.key == "control":
219 | point_label = 0
220 | else:
221 | point_label = 1
222 | self.clicked_points.append((x, y, z, point_label))
223 | self.update_slice(ax)
224 | except BaseException:
225 | pass
226 |
227 | def run(self):
228 | # File selection
229 | self.select_data_file()
230 | inferer.clear_cache()
231 | self.point_start = 0
232 | self.display_3d_slices()
233 |
234 |
235 | if __name__ == "__main__":
236 | from monai.utils import optional_import
237 |
238 | fire, _ = optional_import("fire")
239 | # using python -m interactive run
240 | fire.Fire(samm_visualizer)
241 |
--------------------------------------------------------------------------------
/vista3d/scripts/slic_process_sam.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import logging
13 | import os
14 | import sys
15 | import time
16 |
17 | import monai
18 | import numpy as np
19 | import torch
20 | import torch.distributed as dist
21 | from monai import transforms
22 | from monai.auto3dseg.utils import datafold_read
23 | from monai.data import partition_dataset
24 | from monai.utils import optional_import
25 | from segment_anything import SamPredictor, sam_model_registry
26 | from skimage.segmentation import slic
27 | from tqdm import tqdm
28 |
29 | from .train import CONFIG
30 | from .utils.trans_utils import dilate3d, erode3d
31 |
32 | rearrange, _ = optional_import("einops", name="rearrange")
33 | sys.path.insert(0, os.path.abspath(os.path.dirname(__file__)))
34 |
35 |
36 | def pad_to_divisible_by_16(image):
37 | # Get the dimensions of the input image
38 | depth, height, width = image.shape[-3:]
39 |
40 | # Calculate the padding required to make the dimensions divisible by 16
41 | pad_depth = (16 - (depth % 16)) % 16
42 | pad_height = (16 - (height % 16)) % 16
43 | pad_width = (16 - (width % 16)) % 16
44 |
45 | # Create a tuple with the padding values for each dimension
46 | padding = (0, pad_width, 0, pad_height, 0, pad_depth)
47 |
48 | # Pad the image
49 | padded_image = torch.nn.functional.pad(image, padding)
50 |
51 | return padded_image, padding
52 |
53 |
54 | class InferClass:
55 | def __init__(self):
56 | logging.basicConfig(stream=sys.stdout, level=logging.INFO)
57 | output_path = "./supervoxel_sam"
58 | if not os.path.exists(output_path):
59 | os.makedirs(output_path, exist_ok=True)
60 | self.amp = True
61 | CONFIG["handlers"]["file"]["filename"] = f"{output_path}/log.log"
62 | logging.config.dictConfig(CONFIG)
63 | self.device = torch.device("cuda:0")
64 | self.sam = sam_model_registry["vit_h"](checkpoint="sam_vit_h_4b8939.pth").to(
65 | self.device
66 | )
67 | self.model = SamPredictor(self.sam)
68 | return
69 |
70 | @torch.no_grad()
71 | @torch.cuda.amp.autocast()
72 | def infer(
73 | self,
74 | image_file="example/s1238.nii.gz",
75 | rank=0,
76 | output_dir="./supervoxel_sam/",
77 | data_root_dir=None,
78 | n_segments=400,
79 | ):
80 | """Infer a single image_file. If save_mask is true, save the argmax prediction to disk. If false,
81 | do not save and return the probability maps (usually used by autorunner emsembler).
82 | """
83 | pixel_mean = torch.Tensor([123.675, 116.28, 103.53]).view(1, 3, 1, 1)
84 | pixel_std = torch.Tensor([58.395, 57.12, 57.375]).view(1, 3, 1, 1)
85 | if not isinstance(image_file, list):
86 | image_file = [image_file]
87 |
88 | permute_pairs = [
89 | [(2, 0, 1), None],
90 | [(1, 0, 2), (0, 1, 3, 2)],
91 | [(0, 1, 2), (0, 3, 1, 2)],
92 | ]
93 | for file in image_file:
94 | if data_root_dir is not None:
95 | savefolder = os.path.join(
96 | output_dir,
97 | file.replace(data_root_dir, "").split("/")[0],
98 | file.replace(data_root_dir, "")
99 | .split("/")[1]
100 | .replace(".nii.gz", ""),
101 | )
102 | else:
103 | savefolder = os.path.join(
104 | output_dir, file.split("/")[-1].replace(".nii.gz", "")
105 | )
106 | if os.path.isdir(savefolder):
107 | print(f"{file} already exist. Skipped")
108 | continue
109 | try:
110 | batch_data = None
111 | batch_data = transforms.LoadImage(image_only=True)(file)
112 | orig_data = batch_data.clone()
113 | batch_data = transforms.ScaleIntensityRange(
114 | a_max=1000, a_min=-1000, b_max=255, b_min=0, clip=True
115 | )(batch_data)
116 | print(f"[{rank}] working on {file}")
117 | outputs = None
118 | torch.cuda.empty_cache()
119 | features_ = 0
120 | for views in permute_pairs:
121 | data = batch_data.permute(*views[0])
122 | features = []
123 | max_slice = 8
124 | for i in tqdm(range(int(np.ceil(data.shape[0] / max_slice)))):
125 | idx = (i * max_slice, min((i + 1) * max_slice, data.shape[0]))
126 | image = data[idx[0] : idx[1]]
127 | d, h, w = image.shape
128 | pad_h = 0 if h > w else w - h
129 | pad_w = 0 if w > h else h - w
130 | image = torch.nn.functional.pad(
131 | image, (0, pad_w, 0, pad_h, 0, 0)
132 | )
133 | image = monai.transforms.Resize(
134 | [d, 1024, 1024], mode="bilinear"
135 | )(image.unsqueeze(0)).squeeze(0)
136 | image = (
137 | torch.stack([image, image, image], -1)
138 | .permute(0, 3, 1, 2)
139 | .contiguous()
140 | )
141 | image = (image - pixel_mean) / pixel_std
142 | feature = self.model.get_feature_upsampled(
143 | image.to(f"cuda:{rank}")
144 | )
145 | feature = monai.transforms.Resize(
146 | [h + pad_h, w + pad_w, d], mode="bilinear"
147 | )(feature.permute(1, 2, 3, 0))[:, :h, :w]
148 | features.append(feature.cpu())
149 | features = torch.cat(features, -1)
150 | if views[1] is not None:
151 | features = features.permute(*views[1])
152 | features_ += features
153 | features = None
154 | start = time.time()
155 | outputs = slic(
156 | features_.numpy(),
157 | channel_axis=0,
158 | compactness=0.01,
159 | n_segments=n_segments,
160 | sigma=3,
161 | )
162 | features_ = None
163 | outputs = torch.from_numpy(outputs).cuda()
164 | print("slic took", time.time() - start)
165 | mask = monai.transforms.utils.get_largest_connected_component_mask(
166 | orig_data < -800, connectivity=None, num_components=1
167 | ).cuda()
168 | mask = dilate3d(mask, erosion=3)
169 | mask = erode3d(mask, erosion=3)
170 | outputs[mask.to(torch.bool)] = 0
171 | outputs = monai.data.MetaTensor(
172 | outputs, affine=batch_data.affine, meta=batch_data.meta
173 | )
174 | monai.transforms.SaveImage(
175 | output_dir=output_dir,
176 | output_postfix="seg",
177 | data_root_dir=data_root_dir,
178 | )(outputs.unsqueeze(0).cpu().to(torch.int16))
179 | except BaseException:
180 | print(f"{file} failed. Skipped.")
181 |
182 | @torch.no_grad()
183 | @torch.cuda.amp.autocast()
184 | def batch_infer(
185 | self,
186 | datalist=str,
187 | basedir=str,
188 | output_dir="./supervoxel_sam/",
189 | data_root_dir=None,
190 | n_segments=400,
191 | ):
192 | train_files, _ = datafold_read(datalist=datalist, basedir=basedir, fold=0)
193 | train_files = [_["image"] for _ in train_files]
194 | dist.init_process_group(backend="nccl", init_method="env://")
195 | world_size = dist.get_world_size()
196 | rank = dist.get_rank()
197 | # no need to wrap model with DistributedDataParallel
198 | self.model = SamPredictor(self.sam.to(f"cuda:{rank}"))
199 | infer_files = partition_dataset(
200 | data=train_files,
201 | shuffle=False,
202 | num_partitions=world_size,
203 | even_divisible=False,
204 | )[rank]
205 | self.infer(
206 | infer_files,
207 | rank=rank,
208 | output_dir=output_dir,
209 | data_root_dir=data_root_dir,
210 | n_segments=n_segments,
211 | )
212 |
213 |
214 | if __name__ == "__main__":
215 | from monai.utils import optional_import
216 |
217 | inferer = InferClass()
218 | fire, _ = optional_import("fire")
219 | fire.Fire(inferer)
220 |
--------------------------------------------------------------------------------
/vista3d/scripts/validation/build_vista3d_eval_only.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | from __future__ import annotations
13 |
14 | import copy
15 |
16 | import numpy as np
17 | import torch
18 | from monai.metrics import compute_dice
19 | from vista3d.modeling import (
20 | VISTA3D2,
21 | Class_Mapping_Classify,
22 | Point_Mapping_SAM,
23 | SegResNetDS2,
24 | )
25 |
26 | from ..utils.workflow_utils import get_next_points_auto_point
27 |
28 |
29 | class VISTA3D2_eval_only(VISTA3D2):
30 | @torch.no_grad()
31 | def point_head_iterative_trial(
32 | self,
33 | logits,
34 | labels,
35 | out,
36 | point_coords,
37 | point_labels,
38 | class_vector,
39 | prompt_class,
40 | n_trials=3,
41 | ):
42 | """The prompt class is the local label set while class vector is the mapped global label set"""
43 | logits_update = logits.detach().clone()
44 | for trial_idx in range(n_trials):
45 | if trial_idx == 0:
46 | point_coords, point_labels = get_next_points_auto_point(
47 | logits > 0, labels, prompt_class, class_vector, use_fg=True
48 | )
49 | else:
50 | point_coords, point_labels = get_next_points_auto_point(
51 | logits > 0, labels, prompt_class, class_vector, use_fg=False
52 | )
53 | mapping_index = ((point_labels != -1).sum(1) > 0).to(torch.bool)
54 | point_coords = point_coords[mapping_index]
55 | point_labels = point_labels[mapping_index]
56 | if (torch.sum(mapping_index) == 1 and mapping_index[0]) or torch.sum(
57 | mapping_index
58 | ) == 0:
59 | return logits
60 | if trial_idx == 0:
61 | best_dice = []
62 | for i in range(len(prompt_class)):
63 | dice = compute_dice(
64 | y_pred=(logits[[i]] > 0).to(labels.device),
65 | y=labels == prompt_class[i],
66 | ).item()
67 | if np.isnan(dice):
68 | best_dice.append(-(logits[[i]] > 0).sum())
69 | else:
70 | best_dice.append(dice)
71 |
72 | point_logits = self.point_head(
73 | out,
74 | point_coords,
75 | point_labels,
76 | class_vector=class_vector[mapping_index],
77 | )
78 |
79 | target_logits = self.connected_components_combine(
80 | logits, point_logits, point_coords, point_labels, mapping_index
81 | )
82 | combine_dice = []
83 | for i in range(len(prompt_class)):
84 | if mapping_index[i]:
85 | dice = compute_dice(
86 | y_pred=(target_logits[[i]] > 0).to(labels.device),
87 | y=(labels == prompt_class[i]),
88 | ).item()
89 | if np.isnan(dice):
90 | combine_dice.append(-(target_logits[[i]] > 0).sum())
91 | else:
92 | combine_dice.append(dice)
93 | else:
94 | combine_dice.append(-1)
95 | # check the dice for each label
96 | for i in range(len(prompt_class)):
97 | if prompt_class[i] == 0:
98 | continue
99 | if combine_dice[i] > best_dice[i]:
100 | # print(trial_idx, prompt_class[i], combine_dice[i], best_dice[i])
101 | logits_update[i] = copy.deepcopy(target_logits[i])
102 | best_dice[i] = copy.deepcopy(combine_dice[i])
103 |
104 | labels, target_logits, logits, best_dice, combine_dice = (
105 | None,
106 | None,
107 | None,
108 | None,
109 | None,
110 | )
111 | # force releasing memories that set to None
112 | torch.cuda.empty_cache()
113 | return logits_update
114 |
115 | def forward(
116 | self,
117 | input_images,
118 | point_coords=None,
119 | point_labels=None,
120 | class_vector=None,
121 | prompt_class=None,
122 | patch_coords=None,
123 | labels=None,
124 | label_set=None,
125 | prev_mask=None,
126 | radius=None,
127 | val_point_sampler=None,
128 | **kwargs,
129 | ):
130 | out, out_auto = self.image_encoder(
131 | input_images, with_point=True, with_label=True
132 | )
133 | input_images = None
134 | # force releasing memories that set to None
135 | torch.cuda.empty_cache()
136 | logits, _ = self.class_head(out_auto, class_vector)
137 | logits = self.point_head_iterative_trial(
138 | logits,
139 | labels[patch_coords],
140 | out,
141 | point_coords,
142 | point_labels,
143 | class_vector[0],
144 | prompt_class[0],
145 | n_trials=3,
146 | )
147 | return logits
148 |
149 |
150 | def build_vista3d_segresnet_decoder(
151 | encoder_embed_dim=48, in_channels=1, image_size=(96, 96, 96)
152 | ):
153 | segresnet = SegResNetDS2(
154 | in_channels=in_channels,
155 | blocks_down=(1, 2, 2, 4, 4),
156 | norm="instance",
157 | out_channels=encoder_embed_dim,
158 | init_filters=encoder_embed_dim,
159 | dsdepth=1,
160 | )
161 | point_head = Point_Mapping_SAM(feature_size=encoder_embed_dim, last_supported=132)
162 | class_head = Class_Mapping_Classify(
163 | n_classes=512, feature_size=encoder_embed_dim, use_mlp=True
164 | )
165 | vista = VISTA3D2_eval_only(
166 | image_encoder=segresnet,
167 | class_head=class_head,
168 | point_head=point_head,
169 | feature_size=encoder_embed_dim,
170 | )
171 | return vista
172 |
173 |
174 | vista_model_registry = {"vista3d_segresnet_d": build_vista3d_segresnet_decoder}
175 |
--------------------------------------------------------------------------------
/vista3d/tests/__init__.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
--------------------------------------------------------------------------------
/vista3d/tests/test_config.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import glob
13 | import os
14 | import unittest
15 |
16 | from monai.apps.utils import get_logger
17 | from monai.bundle import ConfigParser
18 |
19 |
20 | class TestConfig(unittest.TestCase):
21 | def test_vista3d_configs_parsing(self):
22 | config_dir = os.path.join(
23 | os.path.dirname(os.path.dirname(os.path.abspath(__file__))), "configs"
24 | )
25 | get_logger("TestConfig").info(config_dir)
26 |
27 | configs = glob.glob(os.path.join(config_dir, "**", "*.yaml"), recursive=True)
28 | for x in configs:
29 | parser = ConfigParser()
30 | parser.read_config(x)
31 | keys = sorted(parser.config.keys())
32 | # verify parser key fetching
33 | get_logger("TestConfig").info(
34 | f"{parser[keys[0]]}, {keys[0]}, {parser[keys[-1]]}, {keys[-1]}"
35 | )
36 |
37 |
38 | if __name__ == "__main__":
39 | unittest.main()
40 |
--------------------------------------------------------------------------------
/vista3d/tests/test_logger.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 | import logging
13 | import unittest
14 |
15 | from monai.apps.auto3dseg.auto_runner import logger
16 |
17 |
18 | class TestLogger(unittest.TestCase):
19 | def test_vista3d_logger(self):
20 | from scripts.train import CONFIG
21 |
22 | logging.config.dictConfig(CONFIG)
23 | logger.warning("check train logging format")
24 |
25 |
26 | if __name__ == "__main__":
27 | unittest.main()
28 |
--------------------------------------------------------------------------------
/vista3d/vista3d/__init__.py:
--------------------------------------------------------------------------------
1 | from .build_vista3d import vista_model_registry # noqa: F401
2 |
--------------------------------------------------------------------------------
/vista3d/vista3d/build_vista3d.py:
--------------------------------------------------------------------------------
1 | # Copyright (c) MONAI Consortium
2 | # Licensed under the Apache License, Version 2.0 (the "License");
3 | # you may not use this file except in compliance with the License.
4 | # You may obtain a copy of the License at
5 | # http://www.apache.org/licenses/LICENSE-2.0
6 | # Unless required by applicable law or agreed to in writing, software
7 | # distributed under the License is distributed on an "AS IS" BASIS,
8 | # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
9 | # See the License for the specific language governing permissions and
10 | # limitations under the License.
11 |
12 |
13 | from .modeling import VISTA3D2, Class_Mapping_Classify, Point_Mapping_SAM, SegResNetDS2
14 |
15 |
16 | def build_vista3d_segresnet_decoder(
17 | encoder_embed_dim=48, in_channels=1, image_size=(96, 96, 96)
18 | ):
19 | segresnet = SegResNetDS2(
20 | in_channels=in_channels,
21 | blocks_down=(1, 2, 2, 4, 4),
22 | norm="instance",
23 | out_channels=encoder_embed_dim,
24 | init_filters=encoder_embed_dim,
25 | dsdepth=1,
26 | )
27 | point_head = Point_Mapping_SAM(feature_size=encoder_embed_dim, last_supported=132)
28 | class_head = Class_Mapping_Classify(
29 | n_classes=512, feature_size=encoder_embed_dim, use_mlp=True
30 | )
31 | vista = VISTA3D2(
32 | image_encoder=segresnet,
33 | class_head=class_head,
34 | point_head=point_head,
35 | feature_size=encoder_embed_dim,
36 | )
37 | return vista
38 |
39 |
40 | vista_model_registry = {"vista3d_segresnet_d": build_vista3d_segresnet_decoder}
41 |
--------------------------------------------------------------------------------
/vista3d/vista3d/modeling/__init__.py:
--------------------------------------------------------------------------------
1 | from .class_head import Class_Mapping_Classify # noqa: F401
2 | from .point_head import Point_Mapping_SAM # noqa: F401
3 | from .segresnetds import SegResNetDS2 # noqa: F401
4 | from .vista3d import VISTA3D2 # noqa: F401
5 |
--------------------------------------------------------------------------------
/vista3d/vista3d/modeling/class_head.py:
--------------------------------------------------------------------------------
1 | import monai
2 | import torch
3 | import torch.nn as nn
4 |
5 |
6 | class Class_Mapping_Classify(nn.Module):
7 | def __init__(self, n_classes, feature_size, use_mlp=False):
8 | super().__init__()
9 | self.use_mlp = use_mlp
10 | if use_mlp:
11 | self.mlp = nn.Sequential(
12 | nn.Linear(feature_size, feature_size),
13 | nn.InstanceNorm1d(1),
14 | nn.GELU(),
15 | nn.Linear(feature_size, feature_size),
16 | )
17 | self.class_embeddings = nn.Embedding(n_classes, feature_size)
18 | self.image_post_mapping = nn.Sequential(
19 | monai.networks.blocks.UnetrBasicBlock(
20 | spatial_dims=3,
21 | in_channels=feature_size,
22 | out_channels=feature_size,
23 | kernel_size=3,
24 | stride=1,
25 | norm_name="instance",
26 | res_block=True,
27 | ),
28 | monai.networks.blocks.UnetrBasicBlock(
29 | spatial_dims=3,
30 | in_channels=feature_size,
31 | out_channels=feature_size,
32 | kernel_size=3,
33 | stride=1,
34 | norm_name="instance",
35 | res_block=True,
36 | ),
37 | )
38 |
39 | def forward(self, src, class_vector):
40 | b, c, h, w, d = src.shape
41 | src = self.image_post_mapping(src)
42 | class_embedding = self.class_embeddings(class_vector)
43 | if self.use_mlp:
44 | class_embedding = self.mlp(class_embedding)
45 | # [b,1,feat] @ [1,feat,dim], batch dimension become class_embedding batch dimension.
46 | masks = []
47 | for i in range(b):
48 | mask = (class_embedding @ src[[i]].view(1, c, h * w * d)).view(
49 | -1, 1, h, w, d
50 | )
51 | masks.append(mask)
52 | masks = torch.cat(masks, 1)
53 | return masks, class_embedding
54 |
--------------------------------------------------------------------------------
/vista3d/vista3d/modeling/point_head.py:
--------------------------------------------------------------------------------
1 | from __future__ import annotations
2 |
3 | import numpy as np
4 | import torch
5 | import torch.nn as nn
6 | from monai.utils import optional_import
7 |
8 | from .sam_blocks import MLP, PositionEmbeddingRandom, TwoWayTransformer
9 |
10 | rearrange, _ = optional_import("einops", name="rearrange")
11 |
12 |
13 | class Point_Mapping_SAM(nn.Module):
14 | def __init__(
15 | self,
16 | feature_size,
17 | max_prompt=32,
18 | num_add_mask_tokens=2,
19 | n_classes=512,
20 | last_supported=132,
21 | ):
22 | super().__init__()
23 | transformer_dim = feature_size
24 | self.max_prompt = max_prompt
25 | self.feat_downsample = nn.Sequential(
26 | nn.Conv3d(
27 | in_channels=feature_size,
28 | out_channels=feature_size,
29 | kernel_size=3,
30 | stride=2,
31 | padding=1,
32 | ),
33 | nn.InstanceNorm3d(feature_size),
34 | nn.GELU(),
35 | nn.Conv3d(
36 | in_channels=feature_size,
37 | out_channels=transformer_dim,
38 | kernel_size=3,
39 | stride=1,
40 | padding=1,
41 | ),
42 | nn.InstanceNorm3d(feature_size),
43 | )
44 |
45 | self.mask_downsample = nn.Conv3d(
46 | in_channels=2, out_channels=2, kernel_size=3, stride=2, padding=1
47 | )
48 |
49 | self.transformer = TwoWayTransformer(
50 | depth=2,
51 | embedding_dim=transformer_dim,
52 | mlp_dim=512,
53 | num_heads=4,
54 | )
55 | self.pe_layer = PositionEmbeddingRandom(transformer_dim // 2)
56 | self.point_embeddings = nn.ModuleList(
57 | [nn.Embedding(1, transformer_dim), nn.Embedding(1, transformer_dim)]
58 | )
59 | self.not_a_point_embed = nn.Embedding(1, transformer_dim)
60 | self.special_class_embed = nn.Embedding(1, transformer_dim)
61 | self.mask_tokens = nn.Embedding(1, transformer_dim)
62 |
63 | self.output_upscaling = nn.Sequential(
64 | nn.ConvTranspose3d(
65 | transformer_dim,
66 | transformer_dim,
67 | kernel_size=3,
68 | stride=2,
69 | padding=1,
70 | output_padding=1,
71 | ),
72 | nn.InstanceNorm3d(transformer_dim),
73 | nn.GELU(),
74 | nn.Conv3d(
75 | transformer_dim, transformer_dim, kernel_size=3, stride=1, padding=1
76 | ),
77 | )
78 |
79 | self.output_hypernetworks_mlps = MLP(
80 | transformer_dim, transformer_dim, transformer_dim, 3
81 | )
82 |
83 | ## MultiMask output
84 | self.num_add_mask_tokens = num_add_mask_tokens
85 | self.output_add_hypernetworks_mlps = nn.ModuleList(
86 | [
87 | MLP(transformer_dim, transformer_dim, transformer_dim, 3)
88 | for i in range(self.num_add_mask_tokens)
89 | ]
90 | )
91 | # class embedding
92 | self.n_classes = n_classes
93 | self.last_supported = last_supported
94 | self.class_embeddings = nn.Embedding(n_classes, feature_size)
95 | self.zeroshot_embed = nn.Embedding(1, transformer_dim)
96 | self.supported_embed = nn.Embedding(1, transformer_dim)
97 |
98 | def forward(self, out, point_coords, point_labels, class_vector=None):
99 | # downsample out
100 | out_low = self.feat_downsample(out)
101 | out_shape = out.shape[-3:]
102 | out = None
103 | torch.cuda.empty_cache()
104 | # embed points
105 | points = point_coords + 0.5 # Shift to center of pixel
106 | point_embedding = self.pe_layer.forward_with_coords(points, out_shape)
107 | point_embedding[point_labels == -1] = 0.0
108 | point_embedding[point_labels == -1] += self.not_a_point_embed.weight
109 | point_embedding[point_labels == 0] += self.point_embeddings[0].weight
110 | point_embedding[point_labels == 1] += self.point_embeddings[1].weight
111 | point_embedding[point_labels == 2] += (
112 | self.point_embeddings[0].weight + self.special_class_embed.weight
113 | )
114 | point_embedding[point_labels == 3] += (
115 | self.point_embeddings[1].weight + self.special_class_embed.weight
116 | )
117 | output_tokens = self.mask_tokens.weight
118 |
119 | output_tokens = output_tokens.unsqueeze(0).expand(
120 | point_embedding.size(0), -1, -1
121 | )
122 | if class_vector is None:
123 | tokens_all = torch.cat(
124 | (
125 | output_tokens,
126 | point_embedding,
127 | self.supported_embed.weight.unsqueeze(0).expand(
128 | point_embedding.size(0), -1, -1
129 | ),
130 | ),
131 | dim=1,
132 | )
133 | # tokens_all = torch.cat((output_tokens, point_embedding), dim=1)
134 | else:
135 | class_embeddings = []
136 | for i in class_vector:
137 | if i > self.last_supported:
138 | class_embeddings.append(self.zeroshot_embed.weight)
139 | else:
140 | class_embeddings.append(self.supported_embed.weight)
141 | class_embeddings = torch.stack(class_embeddings)
142 | tokens_all = torch.cat(
143 | (output_tokens, point_embedding, class_embeddings), dim=1
144 | )
145 | # cross attention
146 | masks = []
147 | max_prompt = self.max_prompt
148 | for i in range(int(np.ceil(tokens_all.shape[0] / max_prompt))):
149 | # remove variables in previous for loops to save peak memory for self.transformer
150 | src, upscaled_embedding, hyper_in = None, None, None
151 | torch.cuda.empty_cache()
152 | idx = (i * max_prompt, min((i + 1) * max_prompt, tokens_all.shape[0]))
153 | tokens = tokens_all[idx[0] : idx[1]]
154 | src = torch.repeat_interleave(out_low, tokens.shape[0], dim=0)
155 | pos_src = torch.repeat_interleave(
156 | self.pe_layer(out_low.shape[-3:]).unsqueeze(0), tokens.shape[0], dim=0
157 | )
158 | b, c, h, w, d = src.shape
159 | hs, src = self.transformer(src, pos_src, tokens)
160 | mask_tokens_out = hs[:, :1, :]
161 | hyper_in = self.output_hypernetworks_mlps(mask_tokens_out)
162 | src = src.transpose(1, 2).view(b, c, h, w, d)
163 | upscaled_embedding = self.output_upscaling(src)
164 | b, c, h, w, d = upscaled_embedding.shape
165 | masks.append(
166 | (hyper_in @ upscaled_embedding.view(b, c, h * w * d)).view(
167 | b, -1, h, w, d
168 | )
169 | )
170 | masks = torch.vstack(masks)
171 | return masks
172 |
--------------------------------------------------------------------------------