├── .gitattributes
├── .gitignore
├── .idea
├── JOS-3.iml
├── inspectionProfiles
│ └── profiles_settings.xml
├── misc.xml
├── modules.xml
├── vcs.xml
└── workspace.xml
├── LICENSE
├── README.md
├── example
├── ex_result.png
├── example.png
├── example.py
├── example2.png
└── example_v2.py
├── requirements.txt
├── setup.py
├── src
└── jos3
│ ├── __init__.py
│ ├── comfmod.py
│ ├── construction.py
│ ├── jos3.py
│ ├── matrix.py
│ ├── params.py
│ └── thermoregulation.py
└── test.py
/.gitattributes:
--------------------------------------------------------------------------------
1 | # Auto detect text files and perform LF normalization
2 | * text=auto
3 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | *.pyc
2 | *.pypirc
3 | dist/
4 | build/
5 | jos3.egg-info/
6 | .eggs/
7 | *.csv
8 | test.py
--------------------------------------------------------------------------------
/.idea/JOS-3.iml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
--------------------------------------------------------------------------------
/.idea/inspectionProfiles/profiles_settings.xml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
--------------------------------------------------------------------------------
/.idea/misc.xml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
--------------------------------------------------------------------------------
/.idea/modules.xml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
--------------------------------------------------------------------------------
/.idea/vcs.xml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
--------------------------------------------------------------------------------
/.idea/workspace.xml:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
16 |
17 |
18 |
19 |
22 |
23 |
24 |
25 |
26 |
27 |
28 |
29 |
30 |
31 |
32 |
33 |
34 |
35 |
36 |
37 |
38 |
39 | {
40 | "keyToString": {
41 | "RunOnceActivity.OpenProjectViewOnStart": "true",
42 | "RunOnceActivity.ShowReadmeOnStart": "true",
43 | "last_opened_file_path": "C:/Users/monyo/PycharmProjects/TanabeLab/JOS-3/example",
44 | "settings.editor.selected.configurable": "com.jetbrains.python.configuration.PyActiveSdkModuleConfigurable"
45 | },
46 | "keyToStringList": {
47 | "ChangesTree.GroupingKeys": [
48 | ]
49 | }
50 | }
51 |
52 |
53 |
54 |
55 |
56 |
57 |
58 |
59 |
60 |
61 |
62 |
63 |
64 |
65 |
66 |
67 |
68 |
69 |
70 |
71 |
72 |
73 |
74 |
75 |
76 |
77 |
78 | 1678143595738
79 |
80 |
81 | 1678143595738
82 |
83 |
84 | 1678143797996
85 |
86 |
87 |
88 | 1678143797996
89 |
90 |
91 | 1678143830453
92 |
93 |
94 |
95 | 1678143830453
96 |
97 |
98 | 1678224357603
99 |
100 |
101 |
102 | 1678224357603
103 |
104 |
105 | 1678225149594
106 |
107 |
108 |
109 | 1678225149594
110 |
111 |
112 | 1678225197165
113 |
114 |
115 |
116 | 1678225197165
117 |
118 |
119 | 1678226035820
120 |
121 |
122 |
123 | 1678226035820
124 |
125 |
126 | 1678226288756
127 |
128 |
129 |
130 | 1678226288756
131 |
132 |
133 | 1678226556553
134 |
135 |
136 |
137 | 1678226556553
138 |
139 |
140 | 1678226596283
141 |
142 |
143 |
144 | 1678226596283
145 |
146 |
147 | 1678226660566
148 |
149 |
150 |
151 | 1678226660566
152 |
153 |
154 | 1678226783289
155 |
156 |
157 |
158 | 1678226783289
159 |
160 |
161 | 1678227559471
162 |
163 |
164 |
165 | 1678227559471
166 |
167 |
168 | 1678227998095
169 |
170 |
171 |
172 | 1678227998095
173 |
174 |
175 | 1678228273682
176 |
177 |
178 |
179 | 1678228273682
180 |
181 |
182 | 1678228708068
183 |
184 |
185 |
186 | 1678228708068
187 |
188 |
189 | 1678230498803
190 |
191 |
192 |
193 | 1678230498803
194 |
195 |
196 | 1678231146618
197 |
198 |
199 |
200 | 1678231146618
201 |
202 |
203 | 1678231180771
204 |
205 |
206 |
207 | 1678231180771
208 |
209 |
210 | 1678231746139
211 |
212 |
213 |
214 | 1678231746139
215 |
216 |
217 | 1678234000768
218 |
219 |
220 |
221 | 1678234000768
222 |
223 |
224 | 1678234045721
225 |
226 |
227 |
228 | 1678234045721
229 |
230 |
231 | 1678234064009
232 |
233 |
234 |
235 | 1678234064009
236 |
237 |
238 | 1678234914780
239 |
240 |
241 |
242 | 1678234914780
243 |
244 |
245 | 1678234928161
246 |
247 |
248 |
249 | 1678234928161
250 |
251 |
252 | 1678235357027
253 |
254 |
255 |
256 | 1678235357027
257 |
258 |
259 | 1678236823721
260 |
261 |
262 |
263 | 1678236823721
264 |
265 |
266 | 1678240477814
267 |
268 |
269 |
270 | 1678240477814
271 |
272 |
273 | 1678298678622
274 |
275 |
276 |
277 | 1678298678622
278 |
279 |
280 | 1678298819359
281 |
282 |
283 |
284 | 1678298819359
285 |
286 |
287 | 1678299430977
288 |
289 |
290 |
291 | 1678299430977
292 |
293 |
294 | 1678299719036
295 |
296 |
297 |
298 | 1678299719036
299 |
300 |
301 | 1678299874056
302 |
303 |
304 |
305 | 1678299874056
306 |
307 |
308 | 1678301447612
309 |
310 |
311 |
312 | 1678301447612
313 |
314 |
315 | 1678312798359
316 |
317 |
318 |
319 | 1678312798359
320 |
321 |
322 | 1678313421894
323 |
324 |
325 |
326 | 1678313421894
327 |
328 |
329 | 1678314065902
330 |
331 |
332 |
333 | 1678314065902
334 |
335 |
336 | 1678314243602
337 |
338 |
339 |
340 | 1678314243602
341 |
342 |
343 | 1678314317808
344 |
345 |
346 |
347 | 1678314317808
348 |
349 |
350 | 1678314444417
351 |
352 |
353 |
354 | 1678314444417
355 |
356 |
357 | 1678314538336
358 |
359 |
360 |
361 | 1678314538336
362 |
363 |
364 | 1678315090912
365 |
366 |
367 |
368 | 1678315090912
369 |
370 |
371 | 1678315405231
372 |
373 |
374 |
375 | 1678315405231
376 |
377 |
378 | 1678315431312
379 |
380 |
381 |
382 | 1678315431312
383 |
384 |
385 |
386 |
387 |
388 |
389 |
408 |
409 |
410 |
411 |
412 |
413 |
414 |
415 |
416 |
417 |
418 |
419 |
420 |
421 |
422 |
423 |
424 |
425 |
426 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2020 Yoshito Takahashi
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Joint system thermoregulation model (JOS-3)
2 |
3 | [Joint system thermoregulation model (JOS-3)](https://www.sciencedirect.com/science/article/pii/S0378778820333612)
4 | is a numerical model to simulate human thermal physiology such as skin temperature, core temperature,
5 | sweating rate, and so on at 17 local body parts as well as the whole body.
6 |
7 | This model was developed at [Shin-ichi Tanabe Laboratory, Waseda University](https://www.tanabe.arch.waseda.ac.jp/en/)
8 | and was derived from [65 Multi-Node model](https://doi.org/10.1016/S0378-7788(02)00014-2)
9 | and [JOS-2 model](https://doi.org/10.1016/j.buildenv.2013.04.013).
10 |
11 | Please cite us if you use this package and describe which version you used:
12 | Y. Takahashi, A. Nomoto, S. Yoda, R. Hisayama, M. Ogata, Y. Ozeki, S. Tanabe,
13 | Thermoregulation Model JOS-3 with New Open Source Code, Energy & Buildings (2020),
14 | doi: https://doi.org/10.1016/j.enbuild.2020.110575
15 |
16 | # Note
17 |
18 | Please also check [pythermalcomfort](https://github.com/CenterForTheBuiltEnvironment/pythermalcomfort) :
19 | F. Tartarini, S. Schiavon, pythermalcomfort: A Python package for thermal comfort research, SoftwareX (2020),
20 | doi: https://doi.org/10.1016/j.softx.2020.100578.
21 |
22 | ## Mnshiv[i] in Equation (41)
23 | Mnshiv[i] in Equation (41) is not utilized in the python code.
24 | This is because non-shivering thermogenesis does not involve activity and may not lead to an increase in blood flow.
25 | The current code, line 854 in thermoregulation.py is below.
26 | ```python
27 | for i, bn in enumerate(BODY_NAMES):
28 | # If the segment has a muscle layer, muscle blood flow increases.
29 | if not IDICT[bn]["muscle"] is None:
30 | bf_ms[i] += (mwork[i] + mshiv[i])/1.163
31 | # In other segments, core blood flow increase, instead of muscle blood flow.
32 | else:
33 | bf_cr[i] += (mwork[i] + mshiv[i])/1.163
34 | ```
35 |
36 | ## Fix in version 0.5.0
37 | * AVA blood flow function was corrected from;
38 | ```python
39 | sig_ava_hand = 0.265 * (err_bcr + 0.43) + 0.953 * (err_msk + 0.1905) + 0.9126
40 | sig_ava_foot = 0.265 * (err_bcr - 0.97) + 0.953 * (err_msk - 0.0095) + 0.9126
41 | ```
42 | to;
43 | ```python
44 | sig_ava_hand = 0.265 * (err_msk + 0.43) + 0.953 * (err_bcr + 0.1905) + 0.9126
45 | sig_ava_foot = 0.265 * (err_msk - 0.997) + 0.953 * (err_bcr + 0.0095) + 0.9126
46 | ```
47 |
48 | ## Fix in version 0.4.0
49 | * Pelvis capacity was corrected from 13.834 to 4.488.
50 |
51 |
52 | # Requirement
53 |
54 | * python3
55 | * numpy
56 |
57 | # Documentation
58 | https://pythermalcomfort.readthedocs.io/
59 |
60 | # Installation
61 |
62 | You can install the model with:
63 | ```bash
64 | pip install jos3
65 | ```
66 |
67 | If you have not installed numpy in your environment, please do so with:
68 |
69 | ```bash
70 | pip install numpy
71 | ```
72 |
73 | # Example
74 |
75 | ## Step 0: Import packages
76 |
77 | ```python
78 | import jos3
79 | import numpy as np
80 | import pandas as pd
81 | import matplotlib.pyplot as plt
82 | ```
83 |
84 | ## Step 1: Build model and set body built
85 |
86 | As a first step, you need to build a model and set a body built that you want to simulate.
87 |
88 | ### Parameters for JOS3 class:
89 |
90 | * height (float, optional) : Body height [m]. The default is 1.72.
91 | * weight (float, optional) : Body weight [kg]. The default is 74.43.
92 | * fat (float, optional) : Fat percentage [%]. The default is 15.
93 | * age (int, optional) : Age [years]. The default is 20.
94 | * sex (str, optional) : Sex ("male" or "female"). The default is "male".
95 | * ci (float, optional) : Cardiac index [L/min/m2]. The default is 2.6432.
96 | * bmr_equation (str, optional) : BMR equation. The default is "harris-benedict".
97 | * To use the equation for Japanese, type "japanese".
98 | * bsa_equation (str, optional) : BSA equation. The default is "dubois".
99 | * You can choose "dubois", "fujimoto", "kruazumi", "takahira".
100 | * ex_output (list/int, optional) : Extra output. The default is "None",
101 | which outputs only important parameters such as local skin temperatures or core temperature.
102 | * Set the parameters as the list format.
103 | (for example, if you want to see the data of ) ["BFsk", "BFcr", "Tar"].
104 | * If you want to see the all outputs, set ex_output to "all".
105 |
106 | ### Example code to built a model and set body buit
107 |
108 | ```python
109 | model = jos3.JOS3(height=1.7,
110 | weight=60,
111 | fat=20,
112 | age=30,
113 | sex="male",
114 | bmr_equation="japanese",
115 | bsa_equation="fujimoto",
116 | ex_output="all"
117 | )
118 | ```
119 |
120 | ## Step 2: Set environmental conditions
121 |
122 | Next, you need to set thermal environmental conditions that you want to simulate.
123 |
124 | If you want to simulate non-uniform thermal environment,
125 | use numpy.ndarray (or list-like data) and input the data separately to local bodies.
126 | You can also input a clothing insulation value for each body part individually as well as for the whole body.
127 |
128 | If you want to simulate transient thermal environment,
129 | alternate between entering environmental information and executing the simulate() method.
130 | After the simulate() method is executed, the environment input values are inherited,
131 | so you only need to enter the input parameters that you want to change.
132 |
133 | ### Environmental parameters
134 |
135 | Input parameters of environmental conditions are set as the Setter format.
136 |
137 | If you set the different conditions in each body parts, set them as a list-type object.
138 |
139 | List-type input must be 17 lengths and means the input of "Head", "Neck", "Chest",
140 | "Back", "Pelvis", "Left-Shoulder", "Left-Arm", "Left-Hand", "Right-Shoulder", "Right-Arm",
141 | "Right-Hand", "Left-Thigh", "Left-Leg", "Left-Foot", "Right-Thigh", "Right-Leg" and "Right-Foot".
142 |
143 | * Ta (float or list) : Air temperature [oC].
144 | * Tr (float or list) : Mean radiant temperature [oC].
145 | * To (float or list) : Operative temperature [oC].
146 | This parameter can be input only when air temperature and mean radiant temperature are equal.
147 | * Va (float or list) : Air velocity [m/s].
148 | * RH (float or list) : Relative humidity [%].
149 | * Icl (float or list) : Clothing insulation [clo].
150 | * [Reference for clothing insulation for the whole body](https://pythermalcomfort.readthedocs.io/en/latest/reference/pythermalcomfort.html#clothing-insulation-of-typical-ensembles-clo)
151 | * Reference for local clothing insulation: [A.Nomoto et al. (2019)](https://onlinelibrary.wiley.com/doi/full/10.1002/2475-8876.12124)
152 | * PAR (float) Physical activity ratio [-]. The default is 1.2.
153 | * This equals the ratio of metabolic rate to basal metabolic rate.
154 | * PAR is for calculation metabolic rate considering personal characteristics such as gender or age.
155 | * If you want to input a specific value of metabolic rate like 58.2 W/m2, check the basal metabolic rate
156 | for the simulated people using Getter (there is an example at the bottom of this document),
157 | and set PAR such that the metabolic rate is 58.2 W/m2.
158 | * PAR of sitting quietly is 1.2.
159 | * posture (str) : posture [-]. The default is "standing".
160 | * choose posture from "standing", "sitting" or "lying".
161 | * This parameter affects convective and radiant heat transfer coefficients for local body parts
162 |
163 | ### Example code to simulate non-uniform and transient conditions
164 | ```python
165 | # Set the first condition
166 | model.Ta = 28 # Air temperature [oC]
167 | model.Tr = 30 # Mean radiant temperature [oC]
168 | model.RH = 40 # Relative humidity [%]
169 | model.Va = 0.2 # Air velocity [m/s]
170 | model.PAR = 1.2 # Physical activity ratio [-], assuming a sitting position
171 | model.posture = 'sitting' # Posture [-], assuming a sitting position
172 | model.Icl = np.array([ # Clothing insulation [clo]
173 | 0.00, # Head
174 | 0.00, # Neck
175 | 1.14, # Chest
176 | 0.84, # Back
177 | 1.04, # Pelvis
178 | 0.84, # Left-Shoulder
179 | 0.42, # Left-Arm
180 | 0.00, # Left-Hand
181 | 0.84, # Right-Shoulder
182 | 0.42, # Right-Arm
183 | 0.00, # Right-Hand
184 | 0.58, # Left-Thigh
185 | 0.62, # Left-Leg
186 | 0.82, # Left-Foot
187 | 0.58, # Right-Thigh
188 | 0.62, # Right-Leg
189 | 0.82, # Right-Foot
190 | ])
191 | # Execute JOS-3 model
192 | model.simulate(times=30, # Number of loops of a simulation
193 | dtime=60, # Time delta [sec]. The default is 60.
194 | ) # Exposure time = 30 [loops] * 60 [sec] = 30 [min]
195 |
196 | # Set the next condition (You only need to change the parameters that you want to change)
197 | model.To = 20 # Change operative temperature
198 | model.Va = np.array([ # Air velocity [m/s], assuming to use a desk fan
199 | 0.2, # Head
200 | 0.4, # Neck
201 | 0.4, # Chest
202 | 0.1, # Back
203 | 0.1, # Pelvis
204 | 0.4, # Left-Shoulder
205 | 0.4, # Left-Arm
206 | 0.4, # Left-Hand
207 | 0.4, # Right-Shoulder
208 | 0.4, # Right-Arm
209 | 0.4, # Right-Hand
210 | 0.1, # Left-Thigh
211 | 0.1, # Left-Leg
212 | 0.1, # Left-Foot
213 | 0.1, # Right-Thigh
214 | 0.1, # Right-Leg
215 | 0.1, # Right-Foot
216 | ])
217 | # Execute JOS-3 model
218 | model.simulate(times=60, # Number of loops of a simulation
219 | dtime=60, # Time delta [sec]. The default is 60.
220 | ) # Additional exposure time = 60 [loops] * 60 [sec] = 60 [min]
221 |
222 | # Set the next condition (You only need to change the parameters that you want to change)
223 | model.Ta = 30 # Change air temperature [oC]
224 | model.Tr = 35 # Change mean radiant temperature [oC]
225 | # Execute JOS-3 model
226 | model.simulate(times=30, # Number of loops of a simulation
227 | dtime=60, # Time delta [sec]. The default is 60.
228 | ) # Additional exposure time = 30 [loops] * 60 [sec] = 30 [min]
229 | ```
230 |
231 | ## Step 3: Show and output results
232 |
233 | As explained above, output parameters can be added arbitrarily by setting ex_output in list format when creating JOS objects.
234 | The output parameters are suffixed with "Head," "Neck," "Chest," etc. for each body part.
235 |
236 | ### Defalt output parameters
237 |
238 | * CO : Cardiac output (the sum of the whole blood flow) [L/h]
239 | * CycleTime: The counts of executing one cycle calculation [-]
240 | * Met : Total heat production of the whole body [W]
241 | * ModTime : Simulation times [sec]
242 | * RES : Heat loss by the respiration [W]
243 | * THLsk : Heat loss from the skin of the body part [W]
244 | * Tcr : Core temperature of the body part [oC]
245 | * Tsk : Skin temperature of the body part [oC]
246 | * TskMean : Mean skin temperature of the body [oC]
247 | * Wet : Local skin wettedness of the body part [-]
248 | * WetMean : Mean skin wettedness of the body [-]
249 | * Wle : Weight loss rate by the evaporation and respiration of the whole body [g/sec]
250 | * dt : Time delta of the model [sec]
251 |
252 | ### Show results
253 | ```python
254 | # Show the results
255 | df = pd.DataFrame(model.dict_results()) # Make pandas.DataFrame
256 | df.TskMean.plot() # Plot time series of mean skin temperature.
257 | plt.show('example.png') # Show the plot
258 | ```
259 |
260 | 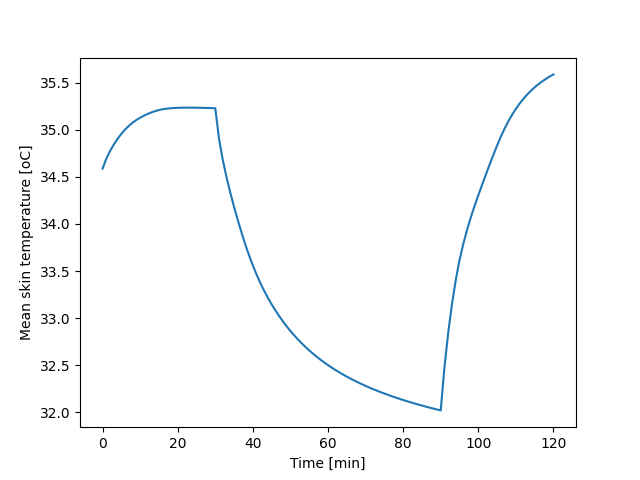
261 |
262 | ### Export results as csv file
263 | ```python
264 | # Exporte the results as csv
265 | model.to_csv('example.csv')
266 | ```
267 |
268 | ### Extra output parameters
269 |
270 | * Age : Age [years]
271 | * BFava_foot: AVA blood flow rate of one foot [L/h]
272 | * BFava_hand: AVA blood flow rate of one hand [L/h]
273 | * BFcr : Core blood flow rate of the body part [L/h]
274 | * BFfat : Fat blood flow rate of the body part [L/h]
275 | * BFms : Muscle blood flow rate of the body part [L/h]
276 | * BFsk : Skin blood flow rate of the body part [L/h]
277 | * BSA : Body surface area of the body part [m2]
278 | * Emax : Maximum evaporative heat loss at the skin of th body part [W]
279 | * Esk : Evaporative heat loss at the skin of the body part [W]
280 | * Esweat : Evaporative heat loss at the skin by only sweating of the body part [W]
281 | * Fat : Body fat rate [%]
282 | * Height : Body heigh [m]
283 | * Icl : Clothing insulation value of the body part [clo]
284 | * LHLsk : Latent heat loss at the skin of the body part [W]
285 | * Mbasecr : Core heat production by basal metaborism of th body part [W]
286 | * Mbasefat: Fat heat production by basal metaborism of th body part [W]
287 | * Mbasems : Muscle heat production by basal metaborism of th body part [W]
288 | * Mbasesk : Skin heat production by basal metaborism of th body part [W]
289 | * Mnst : Core heat production by non-shivering of the body part [W]
290 | * Mshiv : Core or muscle heat production by shivering of th body part [W]
291 | * Mwork : Core or muscle heat production by work of the body part [W]
292 | * Name : Name of the model [-]
293 | * PAR : Physical activity ratio [-]
294 | * Qcr : Core total heat production of the body part [W]
295 | * Qfat : Fat total heat production of the body part [W]
296 | * Qms : Muscle total heat production of the body part [W]
297 | * Qsk : Skin total heat production of the body part [W]
298 | * RESlh : Latent heat loss by respiration of the body part [W]
299 | * RESsh : Sensible heat loss by respiration of the body part [W]
300 | * RH : Relative humidity of the body part [%]
301 | * Ret : Total evaporative heat resistance of the body part [m2.kPa/W]
302 | * Rt : Total heat resistance of the body part [m2.K/W]
303 | * SHLsk : Sensible heat loss at the skin of the body part [W]
304 | * Setptcr : Set point skin temperatre of the body part [oC]
305 | * Setptsk : Set point core temperatre of the body part [oC]
306 | * Sex : Male or female [-]
307 | * Ta : Air temperature of the body part [oC]
308 | * Tar : Arterial temperature of the body part [oC]
309 | * Tcb : Central blood temperature [oC]
310 | * Tfat : Fat temperature of the body part [oC]
311 | * Tms : Muscle temperature as the body part [oC]
312 | * To : Operative temperature of the body part [oC]
313 | * Tr : Mean radiant temperature of the body part [oC]
314 | * Tsve : Superfical vein temperature of the body part [oC]
315 | * Tve : Vein temperature of the body part [oC]
316 | * Va : Air velocity of the body part [m/s]
317 | * Weight : Body weight [kg]
318 |
319 | ### Example code to check the output parameters
320 | ```python
321 | # Show the documentaion of the output parameters
322 | print(jos3.show_outparam_docs())
323 | ```
324 |
325 | ## Getter
326 | JOS3 has some useful getters to check the current parameters.
327 |
328 | ### Getter parameters
329 |
330 | * BSA (numpy.ndarray (17,)) : Body surface areas by local body segments [m2].
331 | * Rt (numpy.ndarray (17,)) : Dry heat resistances between the skin and ambience areas by local body segments [K.m2/W].
332 | * Ret (numpy.ndarray (17,)) : Wet (Evaporative) heat resistances between the skin and ambience areas by local body segments [Pa.m2/W].
333 | * Wet (numpy.ndarray (17,)) : Skin wettedness on local body segments [-].
334 | * WetMean (float) : Mean skin wettedness of the whole body [-].
335 | * TskMean (float) : Mean skin temperature of the whole body [oC].
336 | * Tsk (numpy.ndarray (17,)) : Skin temperatures by the local body segments [oC].
337 | * Tcr (numpy.ndarray (17,)) : Core temperatures by the local body segments [oC].
338 | * Tcb (numpy.ndarray (1,)) : Central blood pool temperatures [oC].
339 | * Tar (numpy.ndarray (17,)) : Arterial temperatures by the local body segments [oC].
340 | * Tve (numpy.ndarray (17,)) : Vein temperatures by the local body segments [oC].
341 | * Tsve (numpy.ndarray (12,)) : Superfical vein temperatures by the local body segments [oC].
342 | * Tms (numpy.ndarray (2,)) : Muscle temperatures of Head and Pelvis [oC].
343 | * Tfat (numpy.ndarray (2,)) : Fat temperatures of Head and Pelvis [oC].
344 | * BMR (float) : Basal metabolic rate [W/m2].
345 |
346 | ### Example code
347 |
348 | ```python
349 | # Check basal metabolic rate [W/m2] using Getters
350 | model.BMR
351 | ```
352 |
353 | # Contact
354 |
355 | * Yoshito Takahashi (takahashiyoshito64@gmail.com)
356 | * Akihisa Nomoto (monyo323232@gmail.com)
357 |
358 | # License
359 |
360 | jos3 is under [MIT license](https://en.wikipedia.org/wiki/MIT_License).
361 |
--------------------------------------------------------------------------------
/example/ex_result.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/TanabeLab/JOS-3/3c74ee2af2f79aa360093cc517e38bf465ec8c5b/example/ex_result.png
--------------------------------------------------------------------------------
/example/example.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/TanabeLab/JOS-3/3c74ee2af2f79aa360093cc517e38bf465ec8c5b/example/example.png
--------------------------------------------------------------------------------
/example/example.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import jos3
3 |
4 | model = jos3.JOS3(height=1.7, weight=60, age=30) # Builds a model
5 |
6 | # Set the first condition
7 | model.To = 28 # Operative temperature [oC]
8 | model.RH = 40 # Relative humidity [%]
9 | model.Va = 0.2 # Air velocity [m/s]
10 | model.PAR = 1.2 # Physical activity ratio [-]
11 | model.simulate(60) # Exposre time = 60 [min]
12 |
13 | # Set the next condition
14 | model.To = 20 # Changes only operative temperature
15 | model.simulate(60) # Additional exposre time = 60 [min]
16 |
17 | # Show the results
18 | import pandas as pd
19 | df = pd.DataFrame(model.dict_results()) # Make pandas.DataFrame
20 | df.TskMean.plot() # Show the graph of mean skin temp.
21 |
22 | # Exporting the results as csv
23 | model.to_csv()
24 |
25 | # Show the documentaion of the output parameters
26 | print(jos3.show_outparam_docs())
27 |
28 | # Check basal metabolic rate [W/m2] using Getters
29 | model.BMR
--------------------------------------------------------------------------------
/example/example2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/TanabeLab/JOS-3/3c74ee2af2f79aa360093cc517e38bf465ec8c5b/example/example2.png
--------------------------------------------------------------------------------
/example/example_v2.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import jos3
3 | import numpy as np
4 | import pandas as pd
5 | import matplotlib.pyplot as plt
6 |
7 | """
8 | 1. Builds a model and set a body built
9 |
10 | As a first step, you need to build a model and set a body built that you want to simulate.
11 |
12 | The following are the parameters for JOS3 class.
13 |
14 | Parameters
15 | -------
16 | height : float, optional
17 | Body height [m]. The default is 1.72.
18 | weight : float, optional
19 | Body weight [kg]. The default is 74.43.
20 | fat : float, optional
21 | Fat percentage [%]. The default is 15.
22 | age : int, optional
23 | Age [years]. The default is 20.
24 | sex : str, optional
25 | Sex ("male" or "female"). The default is "male".
26 | ci : float, optional
27 | Cardiac index [L/min/m2]. The default is 2.6432.
28 | bmr_equation : str, optional
29 | Choose a BMR equation. The default is "harris-benedict".
30 | To use the equation for Japanese, enter "japanese".
31 | bsa_equation : str, optional
32 | Choose a BSA equation.
33 | You can choose "dubois", "fujimoto", "kruazumi", "takahira".
34 | The default is "dubois".
35 | ex_output : None, list or "all", optional
36 | If you want to get extra output parameters, set the parameters as the list format like ["BFsk", "BFcr", "Tar"].
37 | If ex_output is "all", all parameters are output.
38 | The default is None, which outputs only important parameters such as local skin temperatures.
39 | """
40 |
41 | model = jos3.JOS3(height=1.7,
42 | weight=60,
43 | fat=20,
44 | age=30,
45 | sex="male",
46 | bmr_equation="japanese",
47 | bsa_equation="fujimoto",
48 | ex_output="all",
49 | )
50 |
51 | """
52 | 2. Set environmental conditions
53 |
54 | Next, you need to set thermal environmental conditions that you want to simulate.
55 |
56 | If you want to simulate non-uniform thermal environment, use numpy.ndarray and input the data separately to local bodies.
57 | You can also input a clothing insulation value for each body part individually as well as for the whole body.
58 |
59 | If you want to simulate transient thermal environment,
60 | alternate between entering environmental information and executing the simulate() method.
61 | After the simulate() method is executed, the environment input values are inherited,
62 | so you only need to enter the input parameters that you want to change.
63 |
64 | The following are the parameters for JOS-3 class.
65 |
66 | Setter & Getter
67 | -------
68 | Input parameters of environmental conditions are set as the Setter format.
69 | If you set the different conditons in each body parts, set the list.
70 | List input must be 17 lengths and means the input of "Head", "Neck", "Chest",
71 | "Back", "Pelvis", "LShoulder", "LArm", "LHand", "RShoulder", "RArm",
72 | "RHand", "LThigh", "LLeg", "LFoot", "RThigh", "RLeg" and "RFoot".
73 |
74 | Ta : float or list
75 | Air temperature [oC].
76 | Tr : float or list
77 | Mean radiant temperature [oC].
78 | To : float or list
79 | Operative temperature [oC].
80 | This parameter can be input only when air temperature and mean radiant temperature are equal.
81 | Va : float or list
82 | Air velocity [m/s].
83 | RH : float or list
84 | Relative humidity [%].
85 | Icl : float or list
86 | Clothing insulation [clo].
87 | PAR : float
88 | Physical activity ratio [-].
89 | This equals the ratio of metabolic rate to basal metabolic rate.
90 | PAR of sitting quietly is 1.2.
91 | The default is 1.2.
92 | posture : str
93 | Choose a posture from "standing", "sitting" or "lying".
94 | This parameter affects convective and radiant heat transfer coefficients for local body parts
95 | The default is "standing".
96 | bodytemp : numpy.ndarray (85,)
97 | All segment temperatures of JOS-3
98 |
99 | Getter
100 | -------
101 | JOS3 has some useful getters to check the current parameters.
102 |
103 | BSA : numpy.ndarray (17,)
104 | Body surface areas by local body segments [m2].
105 | Rt : numpy.ndarray (17,)
106 | Dry heat resistances between the skin and ambience areas by local body segments [K.m2/W].
107 | Ret : numpy.ndarray (17,)
108 | Wet (Evaporative) heat resistances between the skin and ambience areas by local body segments [Pa.m2/W].
109 | Wet : numpy.ndarray (17,)
110 | Skin wettedness on local body segments [-].
111 | WetMean : float
112 | Mean skin wettedness of the whole body [-].
113 | TskMean : float
114 | Mean skin temperature of the whole body [oC].
115 | Tsk : numpy.ndarray (17,)
116 | Skin temperatures by the local body segments [oC].
117 | Tcr : numpy.ndarray (17,)
118 | Skin temperatures by the local body segments [oC].
119 | Tcb : numpy.ndarray (1,)
120 | Core temperatures by the local body segments [oC].
121 | Tar : numpy.ndarray (17,)
122 | Arterial temperatures by the local body segments [oC].
123 | Tve : numpy.ndarray (17,)
124 | Vein temperatures by the local body segments [oC].
125 | Tsve : numpy.ndarray (12,)
126 | Superfical vein temperatures by the local body segments [oC].
127 | Tms : numpy.ndarray (2,)
128 | Muscle temperatures of Head and Pelvis [oC].
129 | Tfat : numpy.ndarray (2,)
130 | Fat temperatures of Head and Pelvis [oC].
131 | BMR : float
132 | Basal metabolic rate [W/m2].
133 | """
134 |
135 | # Set the first condition
136 | model.Ta = 28 # Air temperature [oC]
137 | model.Tr = 30 # Mean radiant temperature [oC]
138 | model.RH = 40 # Relative humidity [%]
139 | model.Va = 0.2 # Air velocity [m/s]
140 | model.PAR = 1.2 # Physical activity ratio [-], assuming a sitting position
141 | model.posture = 'sitting' # Posture [-], assuming a sitting position
142 | model.Icl = np.array([ # Clothing insulation [clo]
143 | 0.00, # Head
144 | 0.00, # Neck
145 | 1.14, # Chest
146 | 0.84, # Back
147 | 1.04, # Pelvis
148 | 0.84, # Left-Shoulder
149 | 0.42, # Left-Arm
150 | 0.00, # Left-Hand
151 | 0.84, # Right-Shoulder
152 | 0.42, # Right-Arm
153 | 0.00, # Right-Hand
154 | 0.58, # Left-Thigh
155 | 0.62, # Left-Leg
156 | 0.82, # Left-Foot
157 | 0.58, # Right-Thigh
158 | 0.62, # Right-Leg
159 | 0.82, # Right-Foot
160 | ])
161 | # Execute JOS-3 model
162 | model.simulate(times=30, # Number of loops of a simulation
163 | dtime=60, # Time delta [sec]. The default is 60.
164 | ) # Exposure time = 30 [loops] * 60 [sec] = 30 [min]
165 |
166 | # Set the next condition (You only need to change the parameters that you want to change)
167 | model.To = 20 # Change operative temperature
168 | model.Va = np.array([ # Air velocity [m/s], assuming to use a desk fan
169 | 0.2, # Head
170 | 0.4, # Neck
171 | 0.4, # Chest
172 | 0.1, # Back
173 | 0.1, # Pelvis
174 | 0.4, # Left-Shoulder
175 | 0.4, # Left-Arm
176 | 0.4, # Left-Hand
177 | 0.4, # Right-Shoulder
178 | 0.4, # Right-Arm
179 | 0.4, # Right-Hand
180 | 0.1, # Left-Thigh
181 | 0.1, # Left-Leg
182 | 0.1, # Left-Foot
183 | 0.1, # Right-Thigh
184 | 0.1, # Right-Leg
185 | 0.1, # Right-Foot
186 | ])
187 | # Execute JOS-3 model
188 | model.simulate(times=60, # Number of loops of a simulation
189 | dtime=60, # Time delta [sec]. The default is 60.
190 | ) # Additional exposure time = 60 [loops] * 60 [sec] = 60 [min]
191 |
192 | # Set the next condition (You only need to change the parameters that you want to change)
193 | model.Ta = 30 # Change air temperature [oC]
194 | model.Tr = 35 # Change mean radiant temperature [oC]
195 | # Execute JOS-3 model
196 | model.simulate(times=30, # Number of loops of a simulation
197 | dtime=60, # Time delta [sec]. The default is 60.
198 | ) # Additional exposure time = 30 [loops] * 60 [sec] = 30 [min]
199 |
200 | # Show the results
201 | df = pd.DataFrame(model.dict_results()) # Make pandas.DataFrame
202 | df.TskMean.plot() # Plot time series of mean skin temperature.
203 | plt.ylabel('Mean skin temperature [oC]') # Set y-label as 'Mean skin temperature [oC]'
204 | plt.xlabel('Time [min]') # Set x-label as 'Time [min]'
205 | plt.savefig('example.png') # Save plot at the same directory
206 | plt.show() # Show the plot
207 |
208 | # Exporting the results as csv
209 | model.to_csv('example.csv')
210 |
211 | # Show the documentaion of the output parameters
212 | print(jos3.show_outparam_docs())
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | numpy
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | # -*- coding:utf-8 -*-
2 | from __future__ import absolute_import
3 | from __future__ import unicode_literals
4 | import os
5 | from setuptools import setup, find_packages
6 |
7 |
8 | try:
9 | with open('README.md') as f:
10 | readme = f.read()
11 | except IOError:
12 | readme = ''
13 |
14 | # version
15 | here = os.path.dirname(os.path.abspath(__file__))
16 | initpath = os.path.join(here, "src", 'jos3', '__init__.py')
17 |
18 | for line in open(initpath):
19 | if "version" in line:
20 | line = line.split('=')[1].strip().replace('"', "").replace("'", "")
21 | version = line
22 |
23 | def _requires_from_file(filename):
24 | return open(filename).read().splitlines()
25 |
26 | setup(
27 | name="jos3",
28 | version=version,
29 | url='https://github.com/TanabeLab/JOS-3',
30 | author='Yoshito Takahashi',
31 | author_email='takahashiyoshito64@gmail.com',
32 | description='Joint-thermoregulation system, JOS-3',
33 | long_description=readme,
34 | long_description_content_type='text/markdown',
35 | packages=find_packages("src"),
36 | package_dir={'': "src"},
37 | # install_requires=_requires_from_file('requirements.txt'),
38 | # setup_requires=["numpy"],
39 | # tests_requires=["numpy", "pandas", "matplotlib"],
40 | license="MIT",
41 | classifiers=[
42 | 'Operating System :: OS Independent',
43 | 'Intended Audience :: Science/Research',
44 | 'Intended Audience :: Education',
45 | 'Programming Language :: Python :: 3',
46 | 'Programming Language :: Python :: 3.6',
47 | 'Programming Language :: Python :: 3.7',
48 | 'Programming Language :: Python :: 3.8',
49 | 'Programming Language :: Python :: 3.9',
50 | 'Programming Language :: Python :: Implementation :: CPython',
51 | 'License :: OSI Approved :: MIT License',
52 | ],)
53 |
--------------------------------------------------------------------------------
/src/jos3/__init__.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | from jos3.jos3 import *
3 | __version__ = '0.5.0'
--------------------------------------------------------------------------------
/src/jos3/comfmod.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import math
3 |
4 | def pmv(ta, tr, va, rh, met, clo, wmet=0):
5 | """
6 | Get PMV value based on the 2017 ASHRAE Handbook—Fundamentals, Chapter 9:
7 | Thermal Comfort, Equations 63 - 68.
8 |
9 | Parameters
10 | ----------
11 | ta : float, optional
12 | Air temperature [oC]
13 | tr : float, optional
14 | Mean radiant temperature [oC]
15 | va : float, optional
16 | Air velocity [m/s]
17 | rh : float, optional
18 | Relative humidity [%]
19 | met : float, optional
20 | Metabolic rate [met]
21 | clo : float, optional
22 | Clothing insulation [clo]
23 | wmet : float, optional
24 | External work [met], optional. The default is 0.
25 |
26 | Returns
27 | -------
28 | PMV value
29 | """
30 |
31 | met *= 58.15 # chage unit [met] to [W/m2]
32 | wmet *= 58.15 # chage unit [met] to [W/m2]
33 | mw = met - wmet # heat production [W/m2]
34 |
35 | if clo < 0.5: fcl = 1 + 0.2*clo # clothing area factor [-]
36 | else: fcl = 1.05 + 0.1*clo
37 |
38 | antoine = lambda x: math.e**(16.6536-(4030.183/(x+235))) # antoine's formula
39 | pa = antoine(ta) * rh/100 # vapor pressure [kPa]
40 | rcl = 0.155 * clo # clothing thermal resistance [K.m2/W]
41 | hcf = 12.1 * va**0.5 # forced convective heat transfer coefficience
42 |
43 | hc = hcf # initial convective heat transfer coefficience
44 | tcl = (34 + ta) / 2 # initial clothing temp.
45 |
46 | # Cal. clothing temp. by iterative calculation method
47 | for i in range(100):
48 | # clothing temp. [oC]
49 | tcliter = 35.7 - 0.028 * mw \
50 | - rcl * (39.6 * 10**(-9) * fcl * ((tcl+273)**4 - (tr+273)**4) \
51 | + fcl * hc * (tcl - ta)) # Eq.68
52 | # new clothin temp. [oC]
53 | tcl = (tcliter + tcl) / 2
54 |
55 | hcn = 2.38 * abs(tcl - ta)**0.25 # natural convective heat transfer coefficience
56 |
57 | # select forced or natural convection
58 | if hcn > hcf: hc = hcf
59 | else: hc = hcf
60 |
61 | # terminate iterative calculation
62 | if abs(tcliter - tcl) < 0.0001:
63 | break
64 |
65 | # tcl = 35.7 - 0.0275 * mw \
66 | # - rcl * (mw - 3.05 * (5.73 - 0.007 * mw - pa) \
67 | # - 0.42 * (mw - 58.15) - 0.0173 * met * (5.87 - pa) \
68 | # + 0.0014 * met * (34 - ta)) # Eq.64
69 |
70 | # Heat loss of human body
71 | rad = 3.96 * (10**(-8)) * fcl * ((tcl+273)**4 - (tr+273)**4) # by radiation
72 | conv = fcl * hc * (tcl - ta) # by convction
73 | diff = 3.05 * (5.73 - 0.007 * mw - pa) # by insensive perspiration
74 | sweat = max(0, 0.42 * (mw - 58.15)) # by sweating
75 | res = 0.0173 * met * (5.87 - pa) + 0.0014 * met * (34 - ta) # by repiration
76 | load = mw - rad - conv - diff - sweat - res
77 |
78 | pmv_value = (0.303 * math.exp(-0.036 * met) + 0.028) * load # Eq.63
79 |
80 | return pmv_value
81 |
82 | def preferred_temp(va=0.1, rh=50, met=1, clo=0):
83 | """
84 | Calculate operative temperature [oC] at PMV=0.
85 |
86 | Parameters
87 | ----------
88 | va : float, optional
89 | Air velocity [m/s]. The default is 0.1.
90 | rh : float, optional
91 | Relative humidity [%]. The default is 50.
92 | met : float, optional
93 | Metabolic rate [met]. The default is 1.
94 | clo : float, optional
95 | Clothing insulation [clo]. The default is 0.
96 |
97 | Returns
98 | -------
99 | to : float
100 | Operative temperature [oC].
101 | """
102 |
103 | to = 28 # initial temp
104 | for i in range(100):
105 | vpmv = pmv(to, to, va, rh, met, clo)
106 | if abs(vpmv) < 0.001: break
107 | else: to = to - vpmv/3
108 | return to
--------------------------------------------------------------------------------
/src/jos3/construction.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import numpy as np
3 |
4 | # Import from relative path
5 | try:
6 | from .matrix import NUM_NODES, IDICT, BODY_NAMES
7 | # Import from absolute path
8 | # These codes are for debugging
9 | except ImportError:
10 | from jos3.matrix import NUM_NODES, IDICT, BODY_NAMES
11 |
12 |
13 | _BSAst = np.array([
14 | 0.110, 0.029, 0.175, 0.161, 0.221,
15 | 0.096, 0.063, 0.050, 0.096, 0.063, 0.050,
16 | 0.209, 0.112, 0.056, 0.209, 0.112, 0.056,])
17 |
18 | dubois = lambda height, weight: 0.2025 * (height ** 0.725) * (weight ** 0.425)
19 | takahira = lambda height, weight: 0.2042 * (height ** 0.725) * (weight ** 0.425)
20 | fujimoto = lambda height, weight: 0.1882 * (height ** 0.663) * (weight ** 0.444)
21 | kurazumi = lambda height, weight: 0.2440 * (height ** 0.693) * (weight ** 0.383)
22 |
23 | def body_surface_area(height=1.72, weight=74.43, equation="dubois",):
24 | """
25 | Calculate body surface area (BSA) [m2].
26 |
27 | Parameters
28 | ----------
29 | height : float, optional
30 | Body height [m]. The default is 1.72.
31 | weight : float, optional
32 | Body weight [kg]. The default is 74.43.
33 | equation : str, optional
34 | The equation name (str) of bsa calculation. Choose a name from "dubois",
35 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
36 |
37 | Returns
38 | -------
39 | bsa : float
40 | Body surface area (BSA) [m2].
41 | """
42 |
43 | if equation == "dubois":
44 | bsa = dubois(height, weight)
45 | elif equation == "takahira":
46 | bsa = takahira(height, weight)
47 | elif equation == "fujimoto":
48 | bsa = fujimoto(height, weight)
49 | elif equation == "kurazumi":
50 | bsa = kurazumi(height, weight)
51 |
52 | return bsa
53 |
54 |
55 | def bsa_rate(height=1.72, weight=74.43, equation="dubois",):
56 | """
57 | Calculate the rate of BSA to standard body.
58 |
59 | Parameters
60 | ----------
61 | height : float, optional
62 | Body height [m]. The default is 1.72.
63 | weight : float, optional
64 | Body weight [kg]. The default is 74.43.
65 | equation : str, optional
66 | The equation name (str) of bsa calculation. Choose a name from "dubois",
67 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
68 |
69 | Returns
70 | -------
71 | bsa_rate : float
72 | The ratio of BSA to the standard body [-].
73 | """
74 | bsa_all = body_surface_area(height, weight, equation,)
75 | bsa_rate = bsa_all/_BSAst.sum() # The BSA ratio to the standard body (1.87m2)
76 | return bsa_rate
77 |
78 |
79 | def localbsa(height=1.72, weight=74.43, equation="dubois",):
80 | """
81 | Calculate local body surface area (BSA) [m2].
82 |
83 | The local body surface area has been derived from 65MN.
84 | The head have been devided to head and neck based on Smith's model.
85 | Head = 0.1396*0.1117/0.1414 (65MN_Head * Smith_Head / Smith_Head+Neck)
86 | Neck = 0.1396*0.0297/0.1414 (65MN_Head * Smith_Neck / Smith_Head+Neck)
87 |
88 | Parameters
89 | ----------
90 | height : float, optional
91 | Body height [m]. The default is 1.72.
92 | weight : float, optional
93 | Body weight [kg]. The default is 74.43.
94 | equation : str, optional
95 | The equation name (str) of bsa calculation. Choose a name from "dubois",
96 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
97 |
98 | Returns
99 | -------
100 | localbsa : ndarray(17,)
101 | Local body surface area (BSA) [m2].
102 | bsa_rate : float
103 | The ratio of BSA to the standard body [-].
104 |
105 | """
106 | _bsa_rate = bsa_rate(height, weight, equation,) # The BSA ratio to the standard body (1.87m2)
107 | bsa = _BSAst * _bsa_rate
108 | return bsa
109 |
110 |
111 | def weight_rate(weight=74.43,):
112 | """
113 | Calculate the ratio of the body weitht to the standard body (74.43 kg).
114 |
115 | The standard values of local body weights are as below.
116 | weight_local = np.array([
117 | 3.18, 0.84, 12.4, 11.03, 17.57,
118 | 2.16, 1.37, 0.34, 2.16, 1.37, 0.34,
119 | 7.01, 3.34, 0.48, 7.01, 3.34, 0.48])
120 | The data have been derived from 65MN.
121 | The weight of neck is extracted from the weight of 65MN's head based on
122 | the local body surface area of Smith's model.
123 |
124 | Parameters
125 | ----------
126 | weight : float, optional
127 | The body weight [kg]. The default is 74.43.
128 |
129 | Returns
130 | -------
131 | weight_rate : float
132 | The ratio of the body weight to the standard body (74.43 kg).
133 | weight_rate = weight / 74.43
134 | """
135 | rate = weight / 74.43
136 | return rate
137 |
138 |
139 | def bfb_rate(height=1.72, weight=74.43, equation="dubois", age=20, ci=2.59,):
140 | """
141 | Calculate the ratio of basal blood flow (BFB) of the standard body (290 L/h).
142 |
143 | Parameters
144 | ----------
145 | height : float, optional
146 | Body height [m]. The default is 1.72.
147 | weight : float, optional
148 | Body weight [kg]. The default is 74.43.
149 | equation : str, optional
150 | The equation name (str) of bsa calculation. Choose a name from "dubois",
151 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
152 | age : float, optional
153 | Age [years]. The default is 20.
154 | ci : float, optional
155 | Cardiac index [L/min/㎡]. The default is 2.59.
156 |
157 | Returns
158 | -------
159 | bfb_rate : float
160 | Basal blood flow rate.
161 | """
162 |
163 | ci *= 60 # Change unit [L/min/㎡] to [L/h/㎡]
164 |
165 | # Decrease of BFB by aging
166 | if age < 50:
167 | ci *= 1
168 | elif age < 60:
169 | ci *= 0.85
170 | elif age < 70:
171 | ci *= 0.75
172 | else: # age >= 70
173 | ci *= 0.7
174 |
175 | bfb_all = ci * bsa_rate(height, weight, equation) * _BSAst.sum() # [L/h]
176 | _bfb_rate = bfb_all / 290
177 | return _bfb_rate
178 |
179 |
180 | def conductance(height=1.72, weight=74.43, equation="dubois", fat=15,):
181 | """
182 | Calculate thermal conductance between layers [W/K].
183 |
184 | Parameters
185 | ----------
186 | height : float, optional
187 | Body height [m]. The default is 1.72.
188 | weight : float, optional
189 | Body weight [kg]. The default is 74.43.
190 | equation : str, optional
191 | The equation name (str) of bsa calculation. Choose a name from "dubois",
192 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
193 | fat : float, optional
194 | Body fat rate [%]. The default is 15.
195 |
196 | Returns
197 | -------
198 | conductance : numpy.ndarray
199 | Thermal conductance between layers [W/K].
200 | The shape is (NUM_NODES, NUM_NODES).
201 | """
202 |
203 | if fat < 12.5:
204 | cdt_cr_sk = np.array([
205 | 1.341, 0.930, 1.879, 1.729, 2.370,
206 | 1.557, 1.018, 2.210, 1.557, 1.018, 2.210,
207 | 2.565, 1.378, 3.404, 2.565, 1.378, 3.404
208 | ])
209 | elif fat < 17.5:
210 | cdt_cr_sk = np.array([
211 | 1.311, 0.909, 1.785, 1.643, 2.251,
212 | 1.501, 0.982, 2.183, 1.501, 0.982, 2.183,
213 | 2.468, 1.326, 3.370, 2.468, 1.326, 3.370
214 | ])
215 | elif fat < 22.5:
216 | cdt_cr_sk = np.array([
217 | 1.282, 0.889, 1.698, 1.563, 2.142,
218 | 1.448, 0.947, 2.156, 1.448, 0.947, 2.156,
219 | 2.375, 1.276, 3.337, 2.375, 1.276, 3.337
220 | ])
221 | elif fat < 27.5:
222 | cdt_cr_sk = np.array([
223 | 1.255, 0.870, 1.618, 1.488, 2.040,
224 | 1.396, 0.913, 2.130, 1.396, 0.913, 2.130,
225 | 2.285, 1.227, 3.304, 2.285, 1.227, 3.304
226 | ])
227 | else: #fat >= 27.5
228 | cdt_cr_sk = np.array([
229 | 1.227, 0.852, 1.542, 1.419, 1.945,
230 | 1.346, 0.880, 1.945, 1.346, 0.880, 1.945,
231 | 2.198, 1.181, 3.271, 2.198, 1.181, 3.271
232 | ])
233 |
234 | cdt_cr_ms = np.zeros(17) # core to muscle [W/K]
235 | cdt_ms_fat = np.zeros(17) # muscle to fat [W/K]
236 | cdt_fat_sk = np.zeros(17) # fat to skin [W/K]
237 |
238 | # Head and Pelvis consists of 65MN's conductances
239 | cdt_cr_ms[0] = 1.601 # Head
240 | cdt_ms_fat[0] = 13.222
241 | cdt_fat_sk[0] = 16.008
242 | cdt_cr_ms[4] = 3.0813 # Pelvis
243 | cdt_ms_fat[4] = 10.3738
244 | cdt_fat_sk[4] = 41.4954
245 |
246 | # vessel to core
247 | # The shape is a cylinder.
248 | # It is assumed that the inner is vascular radius, 2.5mm and the outer is
249 | # stolwijk's core radius.
250 | # The heat transer coefficient of the core is assumed as the Michel's
251 | # counter-flow model 0.66816 [W/(m・K)].
252 | cdt_ves_cr = np.array([
253 | 0, 0, 0, 0, 0,
254 | 0.586, 0.383, 1.534, 0.586, 0.383, 1.534,
255 | 0.810, 0.435, 1.816, 0.810, 0.435, 1.816,])
256 | #superficial vein to skin
257 | cdt_sfv_sk = np.array([
258 | 0, 0, 0, 0, 0,
259 | 57.735, 37.768, 16.634, 57.735, 37.768, 16.634,
260 | 102.012, 54.784, 24.277, 102.012, 54.784, 24.277,])
261 |
262 | # art to vein (counter-flow) [W/K]
263 | # The data has been derived Mitchell's model.
264 | # THe values = 15.869 [W/(m・K)] * the segment lenght [m]
265 | cdt_art_vein = np.array([
266 | 0, 0, 0, 0, 0,
267 | 0.537, 0.351, 0.762, 0.537, 0.351, 0.762,
268 | 0.826, 0.444, 0.992, 0.826, 0.444, 0.992
269 | ])
270 |
271 | # Changes values by body size based on the standard body.
272 | wr = weight_rate(weight)
273 | bsar = bsa_rate(height, weight, equation)
274 | # Head, Neck (Sphere shape)
275 | cdt_cr_sk[:2] *= wr/bsar
276 | cdt_cr_ms[:2] *= wr/bsar
277 | cdt_ms_fat[:2] *= wr/bsar
278 | cdt_fat_sk[:2] *= wr/bsar
279 | cdt_ves_cr[:2] *= wr/bsar
280 | cdt_sfv_sk[:2] *= wr/bsar
281 | cdt_art_vein[:2] *= wr/bsar
282 | # Others (Cylinder shape)
283 | cdt_cr_sk[2:] *= bsar**2/wr
284 | cdt_cr_ms[2:] *= bsar**2/wr
285 | cdt_ms_fat[2:] *= bsar**2/wr
286 | cdt_fat_sk[2:] *= bsar**2/wr
287 | cdt_ves_cr[2:] *= bsar**2/wr
288 | cdt_sfv_sk[2:] *= bsar**2/wr
289 | cdt_art_vein[2:] *= bsar**2/wr
290 |
291 | cdt_whole = np.zeros((NUM_NODES, NUM_NODES))
292 | for i, bn in enumerate(BODY_NAMES):
293 | # Dictionary of indecies in each body segment
294 | # key = layer name, value = index of matrix
295 | indexof = IDICT[bn]
296 |
297 | # Common
298 | cdt_whole[indexof["artery"], indexof["vein"]] = cdt_art_vein[i] # art to vein
299 | cdt_whole[indexof["artery"], indexof["core"]] = cdt_ves_cr[i] # art to cr
300 | cdt_whole[indexof["vein"], indexof["core"]] = cdt_ves_cr[i] # vein to cr
301 |
302 | # Only limbs
303 | if i >= 5:
304 | cdt_whole[indexof["sfvein"], indexof["skin"]] = cdt_sfv_sk[i] # sfv to sk
305 |
306 | # If the segment has a muscle or fat layer
307 | if not indexof["muscle"] is None: # or not indexof["fat"] is None
308 | cdt_whole[indexof["core"], indexof["muscle"]] = cdt_cr_ms[i] # cr to ms
309 | cdt_whole[indexof["muscle"], indexof["fat"]] = cdt_ms_fat[i] # ms to fat
310 | cdt_whole[indexof["fat"], indexof["skin"]] = cdt_fat_sk[i] # fat to sk
311 |
312 | else:
313 | cdt_whole[indexof["core"], indexof["skin"]] = cdt_cr_sk[i] # cr to sk
314 |
315 | # Creates a symmetrical matrix
316 | cdt_whole = cdt_whole + cdt_whole.T
317 |
318 | return cdt_whole.copy()
319 |
320 |
321 | def capacity(height=1.72, weight=74.43, equation="dubois", age=20, ci=2.59):
322 | """
323 | Calculate the thermal capacity [J/K].
324 |
325 | The values of vascular and central blood capacity have been derived from
326 | Yokoyama's model.
327 | The specific heat of blood is assumed as 1.0 [kcal/L.K].
328 |
329 | Parameters
330 | ----------
331 | height : float, optional
332 | Body height [m]. The default is 1.72.
333 | weight : float, optional
334 | Body weight [kg]. The default is 74.43.
335 | equation : str, optional
336 | The equation name (str) of bsa calculation. Choose a name from "dubois",
337 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
338 | age : float, optional
339 | Age [years]. The default is 20.
340 | ci : float, optional
341 | Cardiac index [L/min/㎡]. The default is 2.59.
342 |
343 | Returns
344 | -------
345 | capacity : numpy.ndarray.
346 | Thermal capacity [W/K].
347 | The shape is (NUM_NODES).
348 | """
349 | # artery [Wh/K]
350 | cap_art = np.array([
351 | 0.096, 0.025, 0.12, 0.111, 0.265,
352 | 0.0186, 0.0091, 0.0044, 0.0186, 0.0091, 0.0044,
353 | 0.0813, 0.04, 0.0103, 0.0813, 0.04, 0.0103,])
354 |
355 | # vein [Wh/K]
356 | cap_vein = np.array([
357 | 0.321, 0.085, 0.424, 0.39, 0.832,
358 | 0.046, 0.024, 0.01, 0.046, 0.024, 0.01,
359 | 0.207, 0.1, 0.024, 0.207, 0.1, 0.024,])
360 |
361 | # superficial vein [Wh/K]
362 | cap_sfv = np.array([
363 | 0, 0, 0, 0, 0,
364 | 0.025, 0.015, 0.011, 0.025, 0.015, 0.011,
365 | 0.074, 0.05, 0.021, 0.074, 0.05, 0.021,])
366 |
367 | # central blood [Wh/K]
368 | cap_cb = 1.999
369 |
370 | # core [Wh/K]
371 | cap_cr = np.array([
372 | 1.7229, 0.564, 10.2975, 9.3935, 4.488,
373 | 1.6994, 1.1209, 0.1536, 1.6994, 1.1209, 0.1536,
374 | 5.3117, 2.867, 0.2097, 5.3117, 2.867, 0.2097,])
375 |
376 | # muscle [Wh/K]
377 | cap_ms = np.array([
378 | 0.305, 0.0, 0.0, 0.0, 7.409,
379 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
380 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
381 |
382 | # fat [Wh/K]
383 | cap_fat = np.array([
384 | 0.203, 0.0, 0.0, 0.0, 1.947,
385 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
386 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
387 |
388 | # skin [Wh/K]
389 | cap_sk = np.array([
390 | 0.1885, 0.058, 0.441, 0.406, 0.556,
391 | 0.126, 0.084, 0.088, 0.126, 0.084, 0.088,
392 | 0.334, 0.169, 0.107, 0.334, 0.169, 0.107,])
393 |
394 | # Changes the values based on the standard body
395 | bfbr = bfb_rate(height, weight, equation, age, ci)
396 | wr = weight_rate(weight)
397 | cap_art *= bfbr
398 | cap_vein *= bfbr
399 | cap_sfv *= bfbr
400 | cap_cb *= bfbr
401 | cap_cr *= wr
402 | cap_ms *= wr
403 | cap_fat *= wr
404 | cap_sk *= wr
405 |
406 | cap_whole = np.zeros(NUM_NODES)
407 | cap_whole[0] = cap_cb
408 |
409 | for i, bn in enumerate(BODY_NAMES):
410 | # Dictionary of indecies in each body segment
411 | # key = layer name, value = index of matrix
412 | indexof = IDICT[bn]
413 |
414 | # Common
415 | cap_whole[indexof["artery"]] = cap_art[i]
416 | cap_whole[indexof["vein"]] = cap_vein[i]
417 | cap_whole[indexof["core"]] = cap_cr[i]
418 | cap_whole[indexof["skin"]] = cap_sk[i]
419 |
420 | # Only limbs
421 | if i >= 5:
422 | cap_whole[indexof["sfvein"]] = cap_sfv[i]
423 |
424 | # If the segment has a muscle or fat layer
425 | if not indexof["muscle"] is None: # or not indexof["fat"] is None
426 | cap_whole[indexof["muscle"]] = cap_ms[i]
427 | cap_whole[indexof["fat"]] = cap_fat[i]
428 |
429 | cap_whole *= 3600 # Changes unit [Wh/K] to [J/K]
430 | return cap_whole
--------------------------------------------------------------------------------
/src/jos3/jos3.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import csv
3 | import datetime as dt
4 | import os
5 |
6 | import numpy as np
7 | # Import from relative path
8 | try:
9 | from . import thermoregulation as threg
10 | from . import matrix
11 | from .matrix import NUM_NODES, INDEX, VINDEX, BODY_NAMES, remove_bodyname
12 | from .comfmod import preferred_temp
13 | from . import construction as cons
14 | from .construction import _BSAst
15 | from .params import ALL_OUT_PARAMS, show_outparam_docs
16 | # Import from absolute path
17 | # These codes are for debugging
18 | except ImportError:
19 | from jos3 import thermoregulation as threg
20 | from jos3 import matrix
21 | from jos3.matrix import NUM_NODES, INDEX, VINDEX, BODY_NAMES, remove_bodyname
22 | from jos3.comfmod import preferred_temp
23 | from jos3 import construction as cons
24 | from jos3.construction import _BSAst
25 | from jos3.params import ALL_OUT_PARAMS, show_outparam_docs
26 |
27 |
28 | class JOS3():
29 | """
30 | JOS-3 is a numeric model to simulate a human thermoregulation.
31 | You can see all the system of the model from following journal.
32 |
33 | Y.Takahashi, A.Nomoto, S.Yoda, R.Hisayama, M.Ogata, Y.Ozeki, S.Tanabe,
34 | Thermoregulation Model JOS-3 with New Open Source Code, Energy & Buildings (2020),
35 | doi: https://doi.org/10.1016/j.enbuild.2020.110575
36 |
37 |
38 | Parameters
39 | -------
40 | height : float, optional
41 | Body height [m]. The default is 1.72.
42 | weight : float, optional
43 | Body weight [kg]. The default is 74.43.
44 | fat : float, optional
45 | Fat percentage [%]. The default is 15.
46 | age : int, optional
47 | Age [years]. The default is 20.
48 | sex : str, optional
49 | Sex ("male" or "female"). The default is "male".
50 | ci : float, optional
51 | Cardiac index [L/min/m2]. The default is 2.6432.
52 | bmr_equation : str, optional
53 | Choose a BMR equation. The default is "harris-benedict".
54 | To use the equation for Japanese, enter "japanese".
55 | bsa_equation : str, optional
56 | Choose a BSA equation.
57 | You can choose "dubois", "fujimoto", "kruazumi", "takahira".
58 | The default is "dubois".
59 | ex_output : None, list or "all", optional
60 | If you want to get extra output parameters, set the parameters as the list format like ["BFsk", "BFcr", "Tar"].
61 | If ex_output is "all", all parameters are output.
62 | The default is None, which outputs only important parameters such as local skin temperatures.
63 |
64 |
65 | Setter & Getter
66 | -------
67 | Input parameters of environmental conditions are set as the Setter format.
68 | If you set the different conditons in each body parts, set the list.
69 | List input must be 17 lengths and means the input of "Head", "Neck", "Chest",
70 | "Back", "Pelvis", "LShoulder", "LArm", "LHand", "RShoulder", "RArm",
71 | "RHand", "LThigh", "LLeg", "LFoot", "RThigh", "RLeg" and "RFoot".
72 |
73 | Ta : float or list
74 | Air temperature [oC].
75 | Tr : float or list
76 | Mean radiant temperature [oC].
77 | To : float or list
78 | Operative temperature [oC].
79 | Va : float or list
80 | Air velocity [m/s].
81 | RH : float or list
82 | Relative humidity [%].
83 | Icl : float or list
84 | Clothing insulation [clo].
85 | PAR : float
86 | Physical activity ratio [-].
87 | This equals the ratio of metaboric rate to basal metablic rate.
88 | PAR of sitting quietly is 1.2.
89 | posture : str
90 | Choose a posture from standing, sitting or lying.
91 | bodytemp : numpy.ndarray (85,)
92 | All segment temperatures of JOS-3
93 |
94 | Getter
95 | -------
96 | JOS3 has some useful getters to check the current parameters.
97 |
98 | BSA : numpy.ndarray (17,)
99 | Body surface areas by local body segments [m2].
100 | Rt : numpy.ndarray (17,)
101 | Dry heat resistances between the skin and ambience areas by local body segments [K.m2/W].
102 | Ret : numpy.ndarray (17,)
103 | Wet (Evaporative) heat resistances between the skin and ambience areas by local body segments [Pa.m2/W].
104 | Wet : numpy.ndarray (17,)
105 | Skin wettedness on local body segments [-].
106 | WetMean : float
107 | Mean skin wettedness of the whole body [-].
108 | TskMean : float
109 | Mean skin temperature of the whole body [oC].
110 | Tsk : numpy.ndarray (17,)
111 | Skin temperatures by the local body segments [oC].
112 | Tcr : numpy.ndarray (17,)
113 | Skin temperatures by the local body segments [oC].
114 | Tcb : numpy.ndarray (1,)
115 | Core temperatures by the local body segments [oC].
116 | Tar : numpy.ndarray (17,)
117 | Arterial temperatures by the local body segments [oC].
118 | Tve : numpy.ndarray (17,)
119 | Vein temperatures by the local body segments [oC].
120 | Tsve : numpy.ndarray (12,)
121 | Superfical vein temperatures by the local body segments [oC].
122 | Tms : numpy.ndarray (2,)
123 | Muscle temperatures of Head and Pelvis [oC].
124 | Tfat : numpy.ndarray (2,)
125 | Fat temperatures of Head and Pelvis [oC].
126 | BMR : float
127 | Basal metabolic rate [W/m2].
128 |
129 |
130 | Examples
131 | -------
132 |
133 | Make a model:
134 |
135 | >>> import jos3
136 | >>> model = jos3.JOS3(height=1.7, weight=60, age=30)
137 |
138 | Set the first phase:
139 |
140 | >>> model.To = 28 # Operative temperature [oC]
141 | >>> model.RH = 40 # Relative humidity [%]
142 | >>> model.Va = 0.2 # Air velocity [m/s]
143 | >>> model.PAR = 1.2 # Physical activity ratio [-]
144 | >>> model.posture = "sitting" # Set the posture
145 | >>> model.simulate(60) # Exposre time = 60 [min]
146 |
147 | Set the next phase:
148 |
149 | >>> model.To = 20 # Change only operative temperature
150 | >>> model.simulate(60) # Additional exposre time = 60 [min]
151 |
152 |
153 | Show the results:
154 |
155 | >>> import pandas as pd
156 | >>> df = pd.DataFrame(model.dict_results()) # Make pandas.DataFrame
157 | >>> df.TskMean.plot() # Show the graph of mean skin temp.
158 |
159 | Exporting the results as csv:
160 |
161 | >>> model.to_csv(folder="C:/Users/takahashi/Desktop")
162 |
163 | Show the documentaion of the output parameters:
164 |
165 | >>> print(jos3.show_outparam_docs())
166 |
167 | Check basal metabolic rate [W/m2] using Getters:
168 |
169 | >>> model.BMR
170 | """
171 |
172 |
173 | def __init__(
174 | self,
175 | height=1.72,
176 | weight=74.43,
177 | fat=15,
178 | age=20,
179 | sex="male",
180 | ci=2.59,
181 | bmr_equation="harris-benedict",
182 | bsa_equation="dubois",
183 | ex_output=None,
184 | ):
185 |
186 | self._height = height
187 | self._weight = weight
188 | self._fat = fat
189 | self._sex = sex
190 | self._age = age
191 | self._ci = ci
192 | self._bmr_equation = bmr_equation
193 | self._bsa_equation = bsa_equation
194 | self._ex_output = ex_output
195 |
196 | # Body surface area [m2]
197 | self._bsa_rate = cons.bsa_rate(height, weight, bsa_equation,)
198 | # Body surface area rate [-]
199 | self._bsa = cons.localbsa(height, weight, bsa_equation,)
200 | # Basal blood flow rate [-]
201 | self._bfb_rate = cons.bfb_rate(height, weight, bsa_equation, age, ci)
202 | # Thermal conductance [W/K]
203 | self._cdt = cons.conductance(height, weight, bsa_equation, fat,)
204 | # Thermal capacity [J/K]
205 | self._cap = cons.capacity(height, weight, bsa_equation, age, ci)
206 |
207 | # Set point temp [oC]
208 | self.setpt_cr = np.ones(17)*37 # core
209 | self.setpt_sk = np.ones(17)*34 # skin
210 |
211 | # Initial body temp [oC]
212 | self._bodytemp = np.ones(NUM_NODES) * 36
213 |
214 | # Default values of input condition
215 | self._ta = np.ones(17)*28.8
216 | self._tr = np.ones(17)*28.8
217 | self._rh = np.ones(17)*50
218 | self._va = np.ones(17)*0.1
219 | self._clo = np.zeros(17)
220 | self._iclo = np.ones(17) * 0.45
221 | self._par = 1.25 # Physical activity ratio
222 | self._posture = "standing"
223 | self._hc = None
224 | self._hr = None
225 | self.ex_q = np.zeros(NUM_NODES)
226 | self._t = dt.timedelta(0) # Elapsed time
227 | self._cycle = 0 # Cycle time
228 | self.model_name = "JOS3"
229 | self.options = {
230 | "nonshivering_thermogenesis": True,
231 | "cold_acclimated": False,
232 | "shivering_threshold": False,
233 | "limit_dshiv/dt": False,
234 | "bat_positive": False,
235 | "ava_zero": False,
236 | "shivering": False,}
237 | threg.PRE_SHIV = 0 # reset
238 | self._history = []
239 | self._t = dt.timedelta(0) # Elapsed time
240 | self._cycle = 0 # Cycle time
241 | self._atmospheric_pressure = 101.33 # kPa. Used to fix the hc, he values
242 |
243 | # Reset setpoint temperature
244 | dictout = self._reset_setpt()
245 | self._history.append(dictout) # Save the last model parameters
246 |
247 |
248 | def _reset_setpt(self):
249 | """
250 | Reset setpoint temperature by steady state calculation.
251 | Be careful, input parameters (Ta, Tr, RH, Va, Icl, PAR) and body
252 | tempertures are also resetted.
253 |
254 | Returns
255 | -------
256 | Parameters of JOS-3 : dict
257 | """
258 | # Set operative temperature under PMV=0 environment
259 | # PAR = 1.25
260 | # 1 met = 58.15 W/m2
261 | met = self.BMR * 1.25 / 58.15 # [met]
262 | self.To = preferred_temp(met=met)
263 | self.RH = 50
264 | self.Va = 0.1
265 | self.Icl = 0
266 | self.PAR = 1.25
267 |
268 | # Steady-calculation
269 | self.options["ava_zero"] = True
270 | for t in range(10):
271 | dictout = self._run(dtime=60000, passive=True)
272 |
273 | # Set new setpoint temperatures
274 | self.setpt_cr = self.Tcr
275 | self.setpt_sk = self.Tsk
276 | self.options["ava_zero"] = False
277 |
278 | return dictout
279 |
280 |
281 | def simulate(self, times, dtime=60, output=True):
282 | """
283 | Execute JOS-3 model.
284 |
285 | Parameters
286 | ----------
287 | times : int
288 | Number of loops of a simulation
289 | dtime : int or float, optional
290 | Time delta [sec]. The default is 60.
291 | output : bool, optional
292 | If you don't record paramters, set False. The default is True.
293 |
294 | Returns
295 | -------
296 | None.
297 |
298 | """
299 | for t in range(times):
300 | self._t += dt.timedelta(0, dtime)
301 | self._cycle += 1
302 | dictdata = self._run(dtime=dtime, output=output)
303 | if output:
304 | # self.history.append(dictdata)
305 | self._history.append(dictdata)
306 |
307 |
308 | def _run(self, dtime=60, passive=False, output=True):
309 | """
310 | Run a model for a once and get model parameters.
311 |
312 | Parameters
313 | ----------
314 | dtime : int or float, optional
315 | Time delta [sec]. The default is 60.
316 | passive : bool, optional
317 | If you run a passive model, set True. The default is False.
318 | output : bool, optional
319 | If you don't need paramters, set False. The default is True.
320 |
321 | Returns
322 | -------
323 | dictout : dictionary
324 | Output parameters.
325 |
326 | """
327 | tcr = self.Tcr
328 | tsk = self.Tsk
329 |
330 | # Convective and radiative heat transfer coefficient [W/K.m2]
331 | hc = threg.fixed_hc(threg.conv_coef(self._posture, self._va, self._ta, tsk,), self._va)
332 | hr = threg.fixed_hr(threg.rad_coef(self._posture,))
333 | # Manual setting
334 | if self._hc is not None:
335 | hc = self._hc
336 | if self._hr is not None:
337 | hr = self._hr
338 |
339 |
340 | # Operarive temp. [oC], heat and evaporative heat resistance [m2.K/W], [m2.kPa/W]
341 | to = threg.operative_temp(self._ta, self._tr, hc, hr,)
342 | r_t = threg.dry_r(hc, hr, self._clo, pt=self._atmospheric_pressure)
343 | r_et = threg.wet_r(hc, self._clo, iclo=self._iclo, pt=self._atmospheric_pressure)
344 |
345 | #------------------------------------------------------------------
346 | # Thermoregulation
347 | #------------------------------------------------------------------
348 | # Setpoint temperature of thermoregulation
349 | if passive:
350 | setpt_cr = tcr.copy()
351 | setpt_sk = tsk.copy()
352 | else:
353 | setpt_cr = self.setpt_cr.copy()
354 | setpt_sk = self.setpt_sk.copy()
355 | # Difference between setpoint and body temperatures
356 | err_cr = tcr - setpt_cr
357 | err_sk = tsk - setpt_sk
358 |
359 | # Skinwettedness [-], Esk, Emax, Esw [W]
360 | wet, e_sk, e_max, e_sweat = threg.evaporation(
361 | err_cr, err_sk, tsk,
362 | self._ta, self._rh, r_et,
363 | self._height, self._weight, self._bsa_equation, self._age)
364 |
365 | # Skin blood flow, basal skin blood flow [L/h]
366 | bf_sk = threg.skin_bloodflow(err_cr, err_sk,
367 | self._height, self._weight, self._bsa_equation, self._age, self._ci)
368 |
369 | # Hand, Foot AVA blood flow [L/h]
370 | bf_ava_hand, bf_ava_foot = threg.ava_bloodflow(err_cr, err_sk,
371 | self._height, self._weight, self._bsa_equation, self._age, self._ci)
372 | if self.options["ava_zero"] and passive:
373 | bf_ava_hand = 0
374 | bf_ava_foot = 0
375 |
376 | # Thermogenesis by shivering [W]
377 | mshiv = threg.shivering(
378 | err_cr, err_sk, tcr, tsk,
379 | self._height, self._weight, self._bsa_equation, self._age, self._sex, dtime,
380 | self.options,)
381 |
382 | # Thermogenesis by non-shivering [W]
383 | if self.options["nonshivering_thermogenesis"]:
384 | mnst = threg.nonshivering(err_cr, err_sk,
385 | self._height, self._weight, self._bsa_equation, self._age,
386 | self.options["cold_acclimated"], self.options["bat_positive"])
387 | else: # not consider NST
388 | mnst = np.zeros(17)
389 |
390 | #------------------------------------------------------------------
391 | # Thermogenesis
392 | #------------------------------------------------------------------
393 | # Basal thermogenesis [W]
394 | mbase = threg.local_mbase(
395 | self._height, self._weight, self._age, self._sex,
396 | self._bmr_equation,)
397 | mbase_all = sum([m.sum() for m in mbase])
398 |
399 | # Thermogenesis by work [W]
400 | mwork = threg.local_mwork(mbase_all, self._par)
401 |
402 | # Sum of thermogenesis in core, muscle, fat, skin [W]
403 | qcr, qms, qfat, qsk = threg.sum_m(mbase, mwork, mshiv, mnst,)
404 | qall = qcr.sum() + qms.sum() + qfat.sum() + qsk.sum()
405 |
406 | #------------------------------------------------------------------
407 | # Other
408 | #------------------------------------------------------------------
409 | # Blood flow in core, muscle, fat [L/h]
410 | bf_cr, bf_ms, bf_fat = threg.crmsfat_bloodflow(mwork, mshiv,
411 | self._height, self._weight, self._bsa_equation, self._age, self._ci)
412 |
413 | # Heat loss by respiratory
414 | p_a = threg.antoine(self._ta)*self._rh/100
415 | res_sh, res_lh = threg.resp_heatloss(self._ta[0], p_a[0], qall)
416 |
417 | # Sensible heat loss [W]
418 | shlsk = (tsk - to) / r_t * self._bsa
419 |
420 | # Cardiac output [L/h]
421 | co = threg.sum_bf(
422 | bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot)
423 |
424 | # Weight loss rate by evaporation [g/sec]
425 | wlesk = (e_sweat + 0.06*e_max) / 2418
426 | wleres = res_lh / 2418
427 |
428 | #------------------------------------------------------------------
429 | # Matrix
430 | #------------------------------------------------------------------
431 | # Matrix A
432 | # (83, 83,) ndarray
433 | bf_art, bf_vein = matrix.vessel_bloodflow(
434 | bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot
435 | )
436 | bf_local = matrix.localarr(
437 | bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot
438 | )
439 | bf_whole = matrix.wholebody(
440 | bf_art, bf_vein, bf_ava_hand, bf_ava_foot
441 | )
442 | arr_bf = np.zeros((NUM_NODES,NUM_NODES))
443 | arr_bf += bf_local
444 | arr_bf += bf_whole
445 |
446 | arr_bf /= self._cap.reshape((NUM_NODES,1)) # Change unit [W/K] to [/sec]
447 | arr_bf *= dtime # Change unit [/sec] to [-]
448 |
449 | arr_cdt = self._cdt.copy()
450 | arr_cdt /= self._cap.reshape((NUM_NODES,1)) # Change unit [W/K] to [/sec]
451 | arr_cdt *= dtime # Change unit [/sec] to [-]
452 |
453 | arrB = np.zeros(NUM_NODES)
454 | arrB[INDEX["skin"]] += 1/r_t*self._bsa
455 | arrB /= self._cap # Change unit [W/K] to [/sec]
456 | arrB *= dtime # Change unit [/sec] to [-]
457 |
458 | arrA_tria = -(arr_cdt + arr_bf)
459 |
460 | arrA_dia = arr_cdt + arr_bf
461 | arrA_dia = arrA_dia.sum(axis=1) + arrB
462 | arrA_dia = np.diag(arrA_dia)
463 | arrA_dia += np.eye(NUM_NODES)
464 |
465 | arrA = arrA_tria + arrA_dia
466 | arrA_inv = np.linalg.inv(arrA)
467 |
468 | # Matrix Q [W] / [J/K] * [sec] = [-]
469 | # Thermogensis
470 | arrQ = np.zeros(NUM_NODES)
471 | arrQ[INDEX["core"]] += qcr
472 | arrQ[INDEX["muscle"]] += qms[VINDEX["muscle"]]
473 | arrQ[INDEX["fat"]] += qfat[VINDEX["fat"]]
474 | arrQ[INDEX["skin"]] += qsk
475 |
476 | # Respiratory [W]
477 | arrQ[INDEX["core"][2]] -= res_sh + res_lh #Chest core
478 |
479 | # Sweating [W]
480 | arrQ[INDEX["skin"]] -= e_sk
481 |
482 | # Extra heat gain [W]
483 | arrQ += self.ex_q.copy()
484 |
485 | arrQ /= self._cap # Change unit [W]/[J/K] to [K/sec]
486 | arrQ *= dtime # Change unit [K/sec] to [K]
487 |
488 | # Boundary batrix [℃]
489 | arr_to = np.zeros(NUM_NODES)
490 | arr_to[INDEX["skin"]] += to
491 |
492 | # all
493 | arr = self._bodytemp + arrB * arr_to + arrQ
494 |
495 | #------------------------------------------------------------------
496 | # New body temp. [oC]
497 | #------------------------------------------------------------------
498 | self._bodytemp = np.dot(arrA_inv, arr)
499 |
500 | #------------------------------------------------------------------
501 | # Output paramters
502 | #------------------------------------------------------------------
503 | dictout = {}
504 | if output: # Default output
505 | dictout["CycleTime"] = self._cycle
506 | dictout["ModTime"] = self._t
507 | dictout["dt"] = dtime
508 | dictout["TskMean"] = self.TskMean
509 | dictout["Tsk"] = self.Tsk
510 | dictout["Tcr"] = self.Tcr
511 | dictout["WetMean"] = np.average(wet, weights=_BSAst)
512 | dictout["Wet"] = wet
513 | dictout["Wle"] = (wlesk.sum() + wleres)
514 | dictout["CO"] = co
515 | dictout["Met"] = qall
516 | dictout["RES"] = res_sh + res_lh
517 | dictout["THLsk"] = shlsk + e_sk
518 |
519 |
520 | detailout = {}
521 | if self._ex_output and output:
522 | detailout["Name"] = self.model_name
523 | detailout["Height"] = self._height
524 | detailout["Weight"] = self._weight
525 | detailout["BSA"] = self._bsa
526 | detailout["Fat"] = self._fat
527 | detailout["Sex"] = self._sex
528 | detailout["Age"] = self._age
529 | detailout["Setptcr"] = setpt_cr
530 | detailout["Setptsk"] = setpt_sk
531 | detailout["Tcb"] = self.Tcb
532 | detailout["Tar"] = self.Tar
533 | detailout["Tve"] = self.Tve
534 | detailout["Tsve"] = self.Tsve
535 | detailout["Tms"] = self.Tms
536 | detailout["Tfat"] = self.Tfat
537 | detailout["To"] = to
538 | detailout["Rt"] = r_t
539 | detailout["Ret"] = r_et
540 | detailout["Ta"] = self._ta.copy()
541 | detailout["Tr"] = self._tr.copy()
542 | detailout["RH"] = self._rh.copy()
543 | detailout["Va"] = self._va.copy()
544 | detailout["PAR"] = self._par
545 | detailout["Icl"] = self._clo.copy()
546 | detailout["Esk"] = e_sk
547 | detailout["Emax"] = e_max
548 | detailout["Esweat"] = e_sweat
549 | detailout["BFcr"] = bf_cr
550 | detailout["BFms"] = bf_ms[VINDEX["muscle"]]
551 | detailout["BFfat"] = bf_fat[VINDEX["fat"]]
552 | detailout["BFsk"] = bf_sk
553 | detailout["BFava_hand"] = bf_ava_hand
554 | detailout["BFava_foot"] = bf_ava_foot
555 | detailout["Mbasecr"] = mbase[0]
556 | detailout["Mbasems"] = mbase[1][VINDEX["muscle"]]
557 | detailout["Mbasefat"] = mbase[2][VINDEX["fat"]]
558 | detailout["Mbasesk"] = mbase[3]
559 | detailout["Mwork"] = mwork
560 | detailout["Mshiv"] = mshiv
561 | detailout["Mnst"] = mnst
562 | detailout["Qcr"] = qcr
563 | detailout["Qms"] = qms[VINDEX["muscle"]]
564 | detailout["Qfat"] = qfat[VINDEX["fat"]]
565 | detailout["Qsk"] = qsk
566 | dictout["SHLsk"] = shlsk
567 | dictout["LHLsk"] = e_sk
568 | dictout["RESsh"] = res_sh
569 | dictout["RESlh"] = res_lh
570 |
571 |
572 | if self._ex_output == "all":
573 | dictout.update(detailout)
574 | elif isinstance(self._ex_output, list): # if ex_out type is list
575 | outkeys = detailout.keys()
576 | for key in self._ex_output:
577 | if key in outkeys:
578 | dictout[key] = detailout[key]
579 | return dictout
580 |
581 |
582 | def dict_results(self):
583 | """
584 | Get results as pandas.DataFrame format.
585 |
586 | Returns
587 | -------
588 | Dictionaly of the results
589 | """
590 | if not self._history:
591 | print("The model has no data.")
592 | return None
593 |
594 | def check_word_contain(word, *args):
595 | """
596 | Check if word contains *args.
597 | """
598 | boolfilter = False
599 | for arg in args:
600 | if arg in word:
601 | boolfilter = True
602 | return boolfilter
603 |
604 | # Set column titles
605 | # If the values are iter, add the body names as suffix words.
606 | # If the values are not iter and the single value data, convert it to iter.
607 | key2keys = {} # Column keys
608 | for key, value in self._history[0].items():
609 | try:
610 | length = len(value)
611 | if isinstance(value, str):
612 | keys = [key] # str is iter. Convert to list without suffix
613 | elif check_word_contain(key, "sve", "sfv", "superficialvein"):
614 | keys = [key+BODY_NAMES[i] for i in VINDEX["sfvein"]]
615 | elif check_word_contain(key, "ms", "muscle"):
616 | keys = [key+BODY_NAMES[i] for i in VINDEX["muscle"]]
617 | elif check_word_contain(key, "fat"):
618 | keys = [key+BODY_NAMES[i] for i in VINDEX["fat"]]
619 | elif length == 17: # if data contains 17 values
620 | keys = [key+bn for bn in BODY_NAMES]
621 | else:
622 | keys = [key+BODY_NAMES[i] for i in range(length)]
623 | except TypeError: # if the value is not iter.
624 | keys= [key] # convert to iter
625 | key2keys.update({key: keys})
626 |
627 | data = []
628 | for i, dictout in enumerate(self._history):
629 | row = {}
630 | for key, value in dictout.items():
631 | keys = key2keys[key]
632 | if len(keys) == 1:
633 | values = [value] # make list if value is not iter
634 | else:
635 | values = value
636 | row.update(dict(zip(keys, values)))
637 | data.append(row)
638 |
639 | outdict = dict(zip(data[0].keys(), [[] for i in range(len(data[0].keys()))]))
640 | for row in data:
641 | for k in data[0].keys():
642 | outdict[k].append(row[k])
643 | return outdict
644 |
645 |
646 | def to_csv(self, path=None, folder=None, unit=True, meanig=True):
647 | """
648 | Export results as csv format.
649 |
650 | Parameters
651 | ----------
652 | path : str, optional
653 | Output path. If you don't use the default file name, set a name.
654 | The default is None.
655 | folder : str, optional
656 | Output folder. If you use the default file name with the current time,
657 | set a only folder path.
658 | The default is None.
659 | unit : bool, optional
660 | Write units in csv file. The default is True.
661 | meaning : bool, optional
662 | Write meanings of the parameters in csv file. The default is True.
663 |
664 |
665 | Examples
666 | ----------
667 | >>> import jos3
668 | >>> model = jos3.JOS3()
669 | >>> model.simulate(60)
670 | >>> model.to_csv(folder="C:/Users/takahashi/desktop")
671 | """
672 |
673 | if path is None:
674 | nowtime = dt.datetime.now().strftime("%Y%m%d-%H%M%S")
675 | path = "{}_{}.csv".format(self.model_name, nowtime)
676 | if folder:
677 | os.makedirs(folder, exist_ok=True)
678 | path = folder + os.sep + path
679 | elif not ((path[-4:] == ".csv") or (path[-4:] == ".txt")):
680 | path += ".csv"
681 | dictout = self.dict_results()
682 |
683 | columns = [k for k in dictout.keys()]
684 | units = []
685 | meanigs = []
686 | for col in columns:
687 | param, rbn = remove_bodyname(col)
688 | if param in ALL_OUT_PARAMS:
689 | u = ALL_OUT_PARAMS[param]["unit"]
690 | units.append(u)
691 |
692 | m = ALL_OUT_PARAMS[param]["meaning"]
693 | if rbn:
694 | meanigs.append(m.replace("body part", rbn))
695 | else:
696 | meanigs.append(m)
697 | else:
698 | units.append("")
699 | meanigs.append("")
700 |
701 | with open(path, "wt", newline="") as f:
702 | writer = csv.writer(f)
703 | writer.writerow(list(columns))
704 | if unit: writer.writerow(units)
705 | if meanig: writer.writerow(meanigs)
706 | for i in range(len(dictout["CycleTime"])):
707 | row = []
708 | for k in columns:
709 | row.append(dictout[k][i])
710 | writer.writerow(row)
711 |
712 |
713 | #--------------------------------------------------------------------------
714 | # Setter
715 | #--------------------------------------------------------------------------
716 | def _set_ex_q(self, tissue, value):
717 | """
718 | Set extra heat gain by tissue name.
719 |
720 | Parameters
721 | ----------
722 | tissue : str
723 | Tissue name. "core", "skin", or "artery".... If you set value to
724 | Head muscle and other segment's core, set "all_muscle".
725 | value : int, float, array
726 | Heat gain [W]
727 |
728 | Returns
729 | -------
730 | array
731 | Extra heat gain of model.
732 | """
733 | self.ex_q[INDEX[tissue]] = value
734 | return self.ex_q
735 |
736 |
737 | #--------------------------------------------------------------------------
738 | # Setter & getter
739 | #--------------------------------------------------------------------------
740 |
741 | @property
742 | def Ta(self):
743 | """
744 | Getter
745 |
746 | Returns
747 | -------
748 | Ta : numpy.ndarray (17,)
749 | Air temperature [oC].
750 | """
751 | return self._ta
752 | @Ta.setter
753 | def Ta(self, inp):
754 | self._ta = _to17array(inp)
755 |
756 | @property
757 | def Tr(self):
758 | """
759 | Getter
760 |
761 | Returns
762 | -------
763 | Tr : numpy.ndarray (17,)
764 | Mean radiant temperature [oC].
765 | """
766 | return self._tr
767 | @Tr.setter
768 | def Tr(self, inp):
769 | self._tr = _to17array(inp)
770 |
771 | @property
772 | def To(self):
773 | """
774 | Getter
775 |
776 | Returns
777 | -------
778 | To : numpy.ndarray (17,)
779 | Operative temperature [oC].
780 | """
781 | hc = threg.fixed_hc(threg.conv_coef(self._posture, self._va, self._ta, self.Tsk,), self._va)
782 | hr = threg.fixed_hr(threg.rad_coef(self._posture,))
783 | to = threg.operative_temp(self._ta, self._tr, hc, hr,)
784 | return to
785 | @To.setter
786 | def To(self, inp):
787 | self._ta = _to17array(inp)
788 | self._tr = _to17array(inp)
789 |
790 | @property
791 | def RH(self):
792 | """
793 | Getter
794 |
795 | Returns
796 | -------
797 | RH : numpy.ndarray (17,)
798 | Relative humidity [%].
799 | """
800 | return self._rh
801 | @RH.setter
802 | def RH(self, inp):
803 | self._rh = _to17array(inp)
804 |
805 | @property
806 | def Va(self):
807 | """
808 | Getter
809 |
810 | Returns
811 | -------
812 | Va : numpy.ndarray (17,)
813 | Air velocity [m/s].
814 | """
815 | return self._va
816 | @Va.setter
817 | def Va(self, inp):
818 | self._va = _to17array(inp)
819 |
820 | @property
821 | def posture(self):
822 | """
823 | Getter
824 |
825 | Returns
826 | -------
827 | posture : str
828 | Current JOS3 posture.
829 | """
830 | return self._posture
831 | @posture.setter
832 | def posture(self, inp):
833 | if inp == 0:
834 | self._posture = "standing"
835 | elif inp == 1:
836 | self._posture = "sitting"
837 | elif inp == 2:
838 | self._posture = "lying"
839 | elif type(inp) == str:
840 | if inp.lower() == "standing":
841 | self._posture = "standing"
842 | elif inp.lower() in ["sitting", "sedentary"]:
843 | self._posture = "sitting"
844 | elif inp.lower() in ["lying", "supine"]:
845 | self._posture = "lying"
846 | else:
847 | self._posture = "standing"
848 | print('posture must be 0="standing", 1="sitting" or 2="lying".')
849 | print('posture was set "standing".')
850 |
851 | @property
852 | def Icl(self):
853 | """
854 | Getter
855 |
856 | Returns
857 | -------
858 | Icl : numpy.ndarray (17,)
859 | Clothing insulation [clo].
860 | """
861 | return self._clo
862 | @Icl.setter
863 | def Icl(self, inp):
864 | self._clo = _to17array(inp)
865 |
866 | @property
867 | def PAR(self):
868 | """
869 | Getter
870 |
871 | Returns
872 | -------
873 | PAR : float
874 | Physical activity ratio [-].
875 | This equals the ratio of metaboric rate to basal metablic rate.
876 | PAR of sitting quietly is 1.2.
877 | """
878 | return self._par
879 | @PAR.setter
880 | def PAR(self, inp):
881 | self._par = inp
882 |
883 | @property
884 | def bodytemp(self):
885 | """
886 | Getter
887 |
888 | Returns
889 | -------
890 | bodytemp : numpy.ndarray (85,)
891 | All segment temperatures of JOS-3
892 | """
893 | return self._bodytemp
894 | @bodytemp.setter
895 | def bodytemp(self, inp):
896 | self._bodytemp = inp.copy()
897 |
898 | #--------------------------------------------------------------------------
899 | # Getter
900 | #--------------------------------------------------------------------------
901 |
902 | @property
903 | def BSA(self):
904 | """
905 | Getter
906 |
907 | Returns
908 | -------
909 | BSA : numpy.ndarray (17,)
910 | Body surface areas by local body segments [m2].
911 | """
912 | return self._bsa.copy()
913 |
914 | @property
915 | def Rt(self):
916 | """
917 | Getter
918 |
919 | Returns
920 | -------
921 | Rt : numpy.ndarray (17,)
922 | Dry heat resistances between the skin and ambience areas by local body segments [K.m2/W].
923 | """
924 | hc = threg.fixed_hc(threg.conv_coef(self._posture, self._va, self._ta, self.Tsk,), self._va)
925 | hr = threg.fixed_hr(threg.rad_coef(self._posture,))
926 | return threg.dry_r(hc, hr, self._clo)
927 |
928 | @property
929 | def Ret(self):
930 | """
931 | Getter
932 |
933 | Returns
934 | -------
935 | Ret : numpy.ndarray (17,)
936 | Wet (Evaporative) heat resistances between the skin and ambience areas by local body segments [Pa.m2/W].
937 | """
938 | hc = threg.fixed_hc(threg.conv_coef(self._posture, self._va, self._ta, self.Tsk,), self._va)
939 | return threg.wet_r(hc, self._clo, self._iclo)
940 |
941 | @property
942 | def Wet(self):
943 | """
944 | Getter
945 |
946 | Returns
947 | -------
948 | Wet : numpy.ndarray (17,)
949 | Skin wettedness on local body segments [-].
950 | """
951 | err_cr = self.Tcr - self.setpt_cr
952 | err_sk = self.Tsk - self.setpt_sk
953 | wet, *_ = threg.evaporation(err_cr, err_sk,
954 | self._ta, self._rh, self.Ret, self._bsa_rate, self._age)
955 | return wet
956 |
957 | @property
958 | def WetMean(self):
959 | """
960 | Getter
961 |
962 | Returns
963 | -------
964 | WetMean : float
965 | Mean skin wettedness of the whole body [-].
966 | """
967 | wet = self.Wet
968 | return np.average(wet, weights=_BSAst)
969 |
970 |
971 |
972 | @property
973 | def TskMean(self):
974 | """
975 | Getter
976 |
977 | Returns
978 | -------
979 | TskMean : float
980 | Mean skin temperature of the whole body [oC].
981 | """
982 | return np.average(self._bodytemp[INDEX["skin"]], weights=_BSAst)
983 |
984 | @property
985 | def Tsk(self):
986 | """
987 | Getter
988 |
989 | Returns
990 | -------
991 | Tsk : numpy.ndarray (17,)
992 | Skin temperatures by the local body segments [oC].
993 | """
994 | return self._bodytemp[INDEX["skin"]].copy()
995 |
996 | @property
997 | def Tcr(self):
998 | """
999 | Getter
1000 |
1001 | Returns
1002 | -------
1003 | Tcr : numpy.ndarray (17,)
1004 | Skin temperatures by the local body segments [oC].
1005 | """
1006 | return self._bodytemp[INDEX["core"]].copy()
1007 |
1008 | @property
1009 | def Tcb(self):
1010 | """
1011 | Getter
1012 |
1013 | Returns
1014 | -------
1015 | Tcb : numpy.ndarray (1,)
1016 | Core temperatures by the local body segments [oC].
1017 | """
1018 | return self._bodytemp[0].copy()
1019 |
1020 | @property
1021 | def Tar(self):
1022 | """
1023 | Getter
1024 |
1025 | Returns
1026 | -------
1027 | Tar : numpy.ndarray (17,)
1028 | Arterial temperatures by the local body segments [oC].
1029 | """
1030 | return self._bodytemp[INDEX["artery"]].copy()
1031 |
1032 | @property
1033 | def Tve(self):
1034 | """
1035 | Getter
1036 |
1037 | Returns
1038 | -------
1039 | Tve : numpy.ndarray (17,)
1040 | Vein temperatures by the local body segments [oC].
1041 | """
1042 | return self._bodytemp[INDEX["vein"]].copy()
1043 |
1044 | @property
1045 | def Tsve(self):
1046 | """
1047 | Getter
1048 |
1049 | Returns
1050 | -------
1051 | Tsve : numpy.ndarray (12,)
1052 | Superfical vein temperatures by the local body segments [oC].
1053 | """
1054 | return self._bodytemp[INDEX["sfvein"]].copy()
1055 |
1056 | @property
1057 | def Tms(self):
1058 | """
1059 | Getter
1060 |
1061 | Returns
1062 | -------
1063 | Tms : numpy.ndarray (2,)
1064 | Muscle temperatures of Head and Pelvis [oC].
1065 | """
1066 | return self._bodytemp[INDEX["muscle"]].copy()
1067 |
1068 | @property
1069 | def Tfat(self):
1070 | """
1071 | Getter
1072 |
1073 | Returns
1074 | -------
1075 | Tfat : numpy.ndarray (2,)
1076 | Fat temperatures of Head and Pelvis [oC].
1077 | """
1078 | return self._bodytemp[INDEX["fat"]].copy()
1079 |
1080 | @property
1081 | def bodyname(self):
1082 | """
1083 | Getter
1084 |
1085 | Returns
1086 | -------
1087 | bodyname : list
1088 | JOS3 body names,
1089 | "Head", "Neck", "Chest", "Back", "Pelvis",
1090 | "LShoulder", "LArm", "LHand",
1091 | "RShoulder", "RArm", "RHand",
1092 | "LThigh", "LLeg", "LHand",
1093 | "RThigh", "RLeg" and "RHand".
1094 | """
1095 | body = [
1096 | "Head", "Neck", "Chest", "Back", "Pelvis",
1097 | "LShoulder", "LArm", "LHand",
1098 | "RShoulder", "RArm", "RHand",
1099 | "LThigh", "LLeg", "LHand",
1100 | "RThigh", "RLeg", "RHand",]
1101 | return body
1102 |
1103 | @property
1104 | def results(self):
1105 | return self.dict_results()
1106 |
1107 | @property
1108 | def BMR(self):
1109 | """
1110 | Getter
1111 |
1112 | Returns
1113 | -------
1114 | BMR : float
1115 | Basal metabolic rate [W/m2].
1116 | """
1117 | bmr = threg.basal_met(
1118 | self._height, self._weight, self._age,
1119 | self._sex, self._bmr_equation,)
1120 | return bmr / self.BSA.sum()
1121 |
1122 |
1123 | def _to17array(inp):
1124 | """
1125 | Make ndarray (17,).
1126 |
1127 | Parameters
1128 | ----------
1129 | inp : int, float, ndarray, list
1130 | Number you make as 17array.
1131 |
1132 | Returns
1133 | -------
1134 | ndarray
1135 | """
1136 | try:
1137 | if len(inp) == 17:
1138 | array = np.array(inp)
1139 | else:
1140 | first_item = inp[0]
1141 | array = np.ones(17)*first_item
1142 | except:
1143 | array = np.ones(17)*inp
1144 | return array.copy()
1145 |
1146 | if __name__ == "__main__":
1147 | import jos3
--------------------------------------------------------------------------------
/src/jos3/matrix.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import numpy as np
3 |
4 | def sub2whole(subarr_list):
5 | ishape = 0
6 | jshape = 0
7 | for subarr in subarr_list:
8 | ishape += subarr.shape[0]
9 | jshape += subarr.shape[1]
10 |
11 | wholearr = np.zeros((ishape, jshape))
12 | i = 0
13 | j = 0
14 | for subarr in subarr_list:
15 | iend = i + subarr.shape[0]
16 | jend = j + subarr.shape[1]
17 | wholearr[i:iend, j:jend] = subarr.copy()
18 | i += subarr.shape[0]
19 | j += subarr.shape[1]
20 |
21 | return wholearr
22 |
23 |
24 | BODY_NAMES = [
25 | "Head", "Neck", "Chest", "Back", "Pelvis",
26 | "LShoulder", "LArm", "LHand",
27 | "RShoulder", "RArm", "RHand",
28 | "LThigh", "LLeg", "LFoot",
29 | "RThigh", "RLeg", "RFoot"]
30 | LAYER_NAMES = ["artery", "vein", "sfvein", "core", "muscle", "fat", "skin"]
31 |
32 | def index_order():
33 | """
34 | Defines the index's order of the matrix
35 | Returns
36 | -------
37 | indexdict : nested dictionary
38 | keys are BODY_NAMES and LAYER_NAMES
39 | """
40 | # Defines exsisting layers as 1 or None
41 | indexdict = {}
42 |
43 | for key in ["Head", "Pelvis"]:
44 | indexdict[key] = {
45 | "artery": 1, "vein": 1, "sfvein": None,
46 | "core": 1, "muscle": 1, "fat": 1, "skin": 1}
47 |
48 | for key in ["Neck", "Chest", "Back"]:
49 | indexdict[key] = {
50 | "artery": 1, "vein": 1, "sfvein": None,
51 | "core": 1, "muscle": None, "fat": None, "skin": 1}
52 |
53 | for key in BODY_NAMES[5:]: # limb segments
54 | indexdict[key] = {
55 | "artery": 1, "vein": 1, "sfvein": 1,
56 | "core": 1, "muscle": None, "fat": None, "skin": 1}
57 |
58 | # Sets ordered indices in the matrix
59 | indexdict["CB"] = 0
60 | order_count = 1
61 | for bn in BODY_NAMES:
62 | for ln in LAYER_NAMES:
63 | if not indexdict[bn][ln] is None:
64 | indexdict[bn][ln] = order_count
65 | order_count += 1
66 |
67 | return indexdict, order_count
68 | IDICT, NUM_NODES = index_order()
69 |
70 |
71 | def index_bylayer(layer):
72 | """
73 | Get indices of the matrix by the layer name.
74 | Parameters
75 | ----------
76 | layer : str
77 | Layer name of jos.
78 | ex) artery, vein, sfvein, core, muscle, fat or skin.
79 | Returns
80 | -------
81 | indices of the matrix : list
82 | """
83 |
84 | # Gets indices by the layer name
85 | outindex = []
86 | for bn in BODY_NAMES:
87 | for ln in LAYER_NAMES:
88 | if (layer.lower() == ln) and IDICT[bn][ln]:
89 | outindex.append(IDICT[bn][ln])
90 | return outindex
91 |
92 |
93 | def validindex_bylayer(layer):
94 | """
95 | Get indices of the matrix by the layer name.
96 | Parameters
97 | ----------
98 | layer : str
99 | Layer name of jos.
100 | ex) artery, vein, sfvein, core, muscle, fat or skin.
101 | Returns
102 | -------
103 | indices of the matrix : list
104 | """
105 |
106 | # Gets valid indices of the layer name
107 | outindex = []
108 | for i, bn in enumerate(BODY_NAMES):
109 | if IDICT[bn][layer]:
110 | outindex.append(i)
111 | return outindex
112 |
113 | # Constant parameters of the matrix' indicies
114 | INDEX = {}
115 | VINDEX = {}
116 | for key in LAYER_NAMES:
117 | INDEX[key] = index_bylayer(key)
118 | VINDEX[key] = validindex_bylayer(key)
119 |
120 |
121 | def localarr(bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot):
122 | """
123 | Create matrix to calculate heat exchage by blood flow in each segment [W/K]
124 | 1.067 [Wh/(L・K)] * Bloodflow [L/h] = [W/K]
125 | """
126 | bf_local = np.zeros((NUM_NODES, NUM_NODES))
127 | for i, bn in enumerate(BODY_NAMES):
128 | # Dictionary of indecies in each body segment
129 | # key = layer name, value = index of matrix
130 | indexof = IDICT[bn]
131 |
132 | # Common
133 | bf_local[indexof["core"], indexof["artery"]] = 1.067*bf_cr[i] # art to cr
134 | bf_local[indexof["skin"], indexof["artery"]] = 1.067*bf_sk[i] # art to sk
135 | bf_local[indexof["vein"], indexof["core"]] = 1.067*bf_cr[i] # vein to cr
136 | bf_local[indexof["vein"], indexof["skin"]] = 1.067*bf_sk[i] # vein to sk
137 |
138 | # If the segment has a muslce or fat layer
139 | if not indexof["muscle"] is None:
140 | bf_local[indexof["muscle"], indexof["artery"]] = 1.067*bf_ms[i] # art to ms
141 | bf_local[indexof["vein"], indexof["muscle"]] = 1.067*bf_ms[i] # vein to ms
142 | if not indexof["fat"] is None:
143 | bf_local[indexof["fat"], indexof["artery"]] = 1.067*bf_fat[i] # art to fat
144 | bf_local[indexof["vein"], indexof["fat"]] = 1.067*bf_fat[i] # vein to fat
145 |
146 | # Only hand
147 | if i == 7 or i == 10:
148 | bf_local[indexof["sfvein"], indexof["artery"]] = 1.067*bf_ava_hand # art to sfvein
149 | # Only foot
150 | if i == 13 or i == 16:
151 | bf_local[indexof["sfvein"], indexof["artery"]] = 1.067*bf_ava_foot # art to sfvein
152 |
153 | return bf_local
154 |
155 |
156 | def vessel_bloodflow(bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot):
157 | """
158 | Get artery and vein blood flow rate [l/h]
159 | """
160 | xbf = bf_cr + bf_ms + bf_fat + bf_sk
161 |
162 | bf_art = np.zeros(17)
163 | bf_vein = np.zeros(17)
164 |
165 | #Head
166 | bf_art[0] = xbf[0]
167 | bf_vein[0] = xbf[0]
168 |
169 | #Neck (+Head)
170 | bf_art[1] = xbf[1] + xbf[0]
171 | bf_vein[1] = xbf[1] + xbf[0]
172 |
173 | #Chest
174 | bf_art[2] = xbf[2]
175 | bf_vein[2] = xbf[2]
176 |
177 | #Back
178 | bf_art[3] = xbf[3]
179 | bf_vein[3] = xbf[3]
180 |
181 | #Pelvis (+Thighs, Legs, Feet, AVA_Feet)
182 | bf_art[4] = xbf[4] + xbf[11:17].sum() + 2*bf_ava_foot
183 | bf_vein[4] = xbf[4] + xbf[11:17].sum() + 2*bf_ava_foot
184 |
185 | #L.Shoulder (+Arm, Hand, (arteryのみAVA_Hand))
186 | bf_art[5] = xbf[5:8].sum() + bf_ava_hand
187 | bf_vein[5] = xbf[5:8].sum()
188 |
189 | #L.Arm (+Hand)
190 | bf_art[6] = xbf[6:8].sum() + bf_ava_hand
191 | bf_vein[6] = xbf[6:8].sum()
192 |
193 | #L.Hand
194 | bf_art[7] = xbf[7] + bf_ava_hand
195 | bf_vein[7] = xbf[7]
196 |
197 | #R.Shoulder (+Arm, Hand, (arteryのみAVA_Hand))
198 | bf_art[8] = xbf[8:11].sum() + bf_ava_hand
199 | bf_vein[8] = xbf[8:11].sum()
200 |
201 | #R.Arm (+Hand)
202 | bf_art[9] = xbf[9:11].sum() + bf_ava_hand
203 | bf_vein[9] = xbf[9:11].sum()
204 |
205 | #R.Hand
206 | bf_art[10] = xbf[10] + bf_ava_hand
207 | bf_vein[10] = xbf[10]
208 |
209 | #L.Thigh (+Leg, Foot, (arteryのみAVA_Foot))
210 | bf_art[11] = xbf[11:14].sum() + bf_ava_foot
211 | bf_vein[11] = xbf[11:14].sum()
212 |
213 | #L.Leg (+Foot)
214 | bf_art[12] = xbf[12:14].sum() + bf_ava_foot
215 | bf_vein[12] = xbf[12:14].sum()
216 |
217 | #L.Foot
218 | bf_art[13] = xbf[13] + bf_ava_foot
219 | bf_vein[13] = xbf[13]
220 |
221 | #R.Thigh (+Leg, Foot, (arteryのみAVA_Foot))
222 | bf_art[14] = xbf[14:17].sum() + bf_ava_foot
223 | bf_vein[14] = xbf[14:17].sum()
224 |
225 | #R.Leg (+Foot)
226 | bf_art[15] = xbf[15:17].sum() + bf_ava_foot
227 | bf_vein[15] = xbf[15:17].sum()
228 |
229 | #R.Foot
230 | bf_art[16] = xbf[16] + bf_ava_foot
231 | bf_vein[16] = xbf[16]
232 |
233 | return bf_art, bf_vein
234 |
235 |
236 | def wholebody(bf_art, bf_vein, bf_ava_hand, bf_ava_foot):
237 | """
238 | Create matrix to calculate heat exchage by blood flow between segments [W/K]
239 | """
240 |
241 | def flow(up, down, bloodflow):
242 | arr = np.zeros((NUM_NODES, NUM_NODES))
243 | # Coefficient = 1.067 [Wh/L.K]
244 | arr[down,up] = 1.067*bloodflow # Change unit [L/h] to [W/K]
245 | return arr
246 |
247 | arr83 = np.zeros((NUM_NODES, NUM_NODES))
248 | # Matrix offsets of segments
249 | CB = IDICT["CB"]
250 | Head = IDICT["Head"]["artery"]
251 | Neck = IDICT["Neck"]["artery"]
252 | Chest = IDICT["Chest"]["artery"]
253 | Back = IDICT["Back"]["artery"]
254 | Pelvis = IDICT["Pelvis"]["artery"]
255 | LShoulder = IDICT["LShoulder"]["artery"]
256 | LArm = IDICT["LArm"]["artery"]
257 | LHand = IDICT["LHand"]["artery"]
258 | RShoulder = IDICT["RShoulder"]["artery"]
259 | RArm = IDICT["RArm"]["artery"]
260 | RHand = IDICT["RHand"]["artery"]
261 | LThigh = IDICT["LThigh"]["artery"]
262 | LLeg = IDICT["LLeg"]["artery"]
263 | LFoot = IDICT["LFoot"]["artery"]
264 | RThigh = IDICT["RThigh"]["artery"]
265 | RLeg = IDICT["RLeg"]["artery"]
266 | RFoot = IDICT["RFoot"]["artery"]
267 |
268 |
269 | arr83 += flow(CB, Neck, bf_art[1]) #CB to Neck.art
270 | arr83 += flow(Neck, Head, bf_art[0]) #Neck.art to Head.art
271 | arr83 += flow(Head+1, Neck+1, bf_vein[0]) #Head.vein to Neck.vein
272 | arr83 += flow(Neck+1, CB, bf_vein[1]) #Neck.vein to CB
273 |
274 | arr83 += flow(CB, Chest, bf_art[2]) #CB to Chest.art
275 | arr83 += flow(Chest+1, CB, bf_vein[2]) #Chest.vein to CB
276 |
277 | arr83 += flow(CB, Back, bf_art[3]) #CB to Back.art
278 | arr83 += flow(Back+1, CB, bf_vein[3]) #Back.vein to CB
279 |
280 | arr83 += flow(CB, Pelvis, bf_art[4]) #CB to Pelvis.art
281 | arr83 += flow(Pelvis+1, CB, bf_vein[4]) #Pelvis.vein to CB
282 |
283 | arr83 += flow(CB, LShoulder, bf_art[5]) #CB to LShoulder.art
284 | arr83 += flow(LShoulder, LArm, bf_art[6]) #LShoulder.art to LArm.art
285 | arr83 += flow(LArm, LHand, bf_art[7]) #LArm.art to LHand.art
286 | arr83 += flow(LHand+1, LArm+1, bf_vein[7]) #LHand.vein to LArm.vein
287 | arr83 += flow(LArm+1, LShoulder+1, bf_vein[6]) #LArm.vein to LShoulder.vein
288 | arr83 += flow(LShoulder+1, CB, bf_vein[5]) #LShoulder.vein to CB
289 | arr83 += flow(LHand+2, LArm+2, bf_ava_hand) #LHand.sfvein to LArm.sfvein
290 | arr83 += flow(LArm+2, LShoulder+2, bf_ava_hand) #LArm.sfvein to LShoulder.sfvein
291 | arr83 += flow(LShoulder+2, CB, bf_ava_hand) #LShoulder.sfvein to CB
292 |
293 | arr83 += flow(CB, RShoulder, bf_art[8]) #CB to RShoulder.art
294 | arr83 += flow(RShoulder, RArm, bf_art[9]) #RShoulder.art to RArm.art
295 | arr83 += flow(RArm, RHand, bf_art[10]) #RArm.art to RHand.art
296 | arr83 += flow(RHand+1, RArm+1, bf_vein[10]) #RHand.vein to RArm.vein
297 | arr83 += flow(RArm+1, RShoulder+1, bf_vein[9]) #RArm.vein to RShoulder.vein
298 | arr83 += flow(RShoulder+1, CB, bf_vein[8]) #RShoulder.vein to CB
299 | arr83 += flow(RHand+2, RArm+2, bf_ava_hand) #RHand.sfvein to RArm.sfvein
300 | arr83 += flow(RArm+2, RShoulder+2, bf_ava_hand) #RArm.sfvein to RShoulder.sfvein
301 | arr83 += flow(RShoulder+2, CB, bf_ava_hand) #RShoulder.sfvein to CB
302 |
303 | arr83 += flow(Pelvis, LThigh, bf_art[11]) #Pelvis to LThigh.art
304 | arr83 += flow(LThigh, LLeg, bf_art[12]) #LThigh.art to LLeg.art
305 | arr83 += flow(LLeg, LFoot, bf_art[13]) #LLeg.art to LFoot.art
306 | arr83 += flow(LFoot+1, LLeg+1, bf_vein[13]) #LFoot.vein to LLeg.vein
307 | arr83 += flow(LLeg+1, LThigh+1, bf_vein[12]) #LLeg.vein to LThigh.vein
308 | arr83 += flow(LThigh+1, Pelvis+1, bf_vein[11]) #LThigh.vein to Pelvis
309 | arr83 += flow(LFoot+2, LLeg+2, bf_ava_foot) #LFoot.sfvein to LLeg.sfvein
310 | arr83 += flow(LLeg+2, LThigh+2, bf_ava_foot) #LLeg.sfvein to LThigh.sfvein
311 | arr83 += flow(LThigh+2, Pelvis+1, bf_ava_foot) #LThigh.vein to Pelvis
312 |
313 | arr83 += flow(Pelvis, RThigh, bf_art[14]) #Pelvis to RThigh.art
314 | arr83 += flow(RThigh, RLeg, bf_art[15]) #RThigh.art to RLeg.art
315 | arr83 += flow(RLeg, RFoot, bf_art[16]) #RLeg.art to RFoot.art
316 | arr83 += flow(RFoot+1, RLeg+1, bf_vein[16]) #RFoot.vein to RLeg.vein
317 | arr83 += flow(RLeg+1, RThigh+1, bf_vein[15]) #RLeg.vein to RThigh.vein
318 | arr83 += flow(RThigh+1, Pelvis+1, bf_vein[14]) #RThigh.vein to Pelvis
319 | arr83 += flow(RFoot+2, RLeg+2, bf_ava_foot) #RFoot.sfvein to RLeg.sfvein
320 | arr83 += flow(RLeg+2, RThigh+2, bf_ava_foot) #RLeg.sfvein to RThigh.sfvein
321 | arr83 += flow(RThigh+2, Pelvis+1, bf_ava_foot) #RThigh.vein to Pelvis
322 |
323 | return arr83
324 |
325 |
326 | def remove_bodyname(text):
327 | """
328 | Removing the body name from the parameter name.
329 |
330 | Parameters
331 | ----------
332 | text : str
333 | Parameter name
334 |
335 | Returns
336 | -------
337 | rtext : str
338 | Parameter name removed the body name.
339 | removed : str
340 | The removed body name
341 |
342 | """
343 |
344 | rtext = text
345 | removed = None
346 | for bn in BODY_NAMES:
347 | if bn in text:
348 | rtext = rtext.replace(bn, "")
349 | removed = bn
350 | break
351 | return rtext, removed
--------------------------------------------------------------------------------
/src/jos3/params.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import textwrap
4 |
5 | ALL_OUT_PARAMS = {
6 | 'Age': {'ex_output': True,
7 | 'meaning': 'Age',
8 | 'suffix': None,
9 | 'unit': 'years'},
10 |
11 | 'BFava_foot': {'ex_output': True,
12 | 'meaning': 'AVA blood flow rate of one foot',
13 | 'suffix': None,
14 | 'unit': 'L/h'},
15 |
16 | 'BFava_hand': {'ex_output': True,
17 | 'meaning': 'AVA blood flow rate of one hand',
18 | 'suffix': None,
19 | 'unit': 'L/h'},
20 |
21 | 'BFcr': {'ex_output': True,
22 | 'meaning': 'Core blood flow rate of the body part',
23 | 'suffix': 'Body name',
24 | 'unit': 'L/h'},
25 |
26 | 'BFfat': {'ex_output': True,
27 | 'meaning': 'Fat blood flow rate of the body part',
28 | 'suffix': 'Body name',
29 | 'unit': 'L/h'},
30 |
31 | 'BFms': {'ex_output': True,
32 | 'meaning': 'Muscle blood flow rate of the body part',
33 | 'suffix': 'Body name',
34 | 'unit': 'L/h'},
35 |
36 | 'BFsk': {'ex_output': True,
37 | 'meaning': 'Skin blood flow rate of the body part',
38 | 'suffix': 'Body name',
39 | 'unit': 'L/h'},
40 |
41 | 'BSA': {'ex_output': True,
42 | 'meaning': 'Body surface area of the body part',
43 | 'suffix': 'Body name',
44 | 'unit': 'm2'},
45 |
46 | 'CO': {'ex_output': False,
47 | 'meaning': 'Cardiac output (the sum of the whole blood flow)',
48 | 'suffix': None,
49 | 'unit': 'L/h'},
50 |
51 | 'CycleTime': {'ex_output': False,
52 | 'meaning': 'The counts of executing one cycle calculation',
53 | 'suffix': None,
54 | 'unit': '-'},
55 |
56 | 'Emax': {'ex_output': True,
57 | 'meaning': 'Maximum evaporative heat loss at the skin of th body '
58 | 'part',
59 | 'suffix': 'Body name',
60 | 'unit': 'W'},
61 |
62 | 'Esk': {'ex_output': True,
63 | 'meaning': 'Evaporative heat loss at the skin of the body part',
64 | 'suffix': 'Body name',
65 | 'unit': 'W'},
66 |
67 | 'Esweat': {'ex_output': True,
68 | 'meaning': 'Evaporative heat loss at the skin by only sweating of '
69 | 'the body part',
70 | 'suffix': 'Body name',
71 | 'unit': 'W'},
72 |
73 | 'Fat': {'ex_output': True,
74 | 'meaning': 'Body fat rate',
75 | 'suffix': None,
76 | 'unit': '%'},
77 |
78 | 'Height': {'ex_output': True,
79 | 'meaning': 'Body heigh',
80 | 'suffix': None,
81 | 'unit': 'm'},
82 |
83 | 'Icl': {'ex_output': True,
84 | 'meaning': 'Clothing insulation value of the body part',
85 | 'suffix': 'Body name',
86 | 'unit': 'clo'},
87 |
88 | 'LHLsk': {'ex_output': True,
89 | 'meaning': 'Latent heat loss at the skin of the body part',
90 | 'suffix': 'Body name',
91 | 'unit': 'W'},
92 |
93 | 'Mbasecr': {'ex_output': True,
94 | 'meaning': 'Core heat production by basal metaborism of th body '
95 | 'part',
96 | 'suffix': 'Body name',
97 | 'unit': 'W'},
98 |
99 | 'Mbasefat': {'ex_output': True,
100 | 'meaning': 'Fat heat production by basal metaborism of th body '
101 | 'part',
102 | 'suffix': 'Body name',
103 | 'unit': 'W'},
104 |
105 | 'Mbasems': {'ex_output': True,
106 | 'meaning': 'Muscle heat production by basal metaborism of th body '
107 | 'part',
108 | 'suffix': 'Body name',
109 | 'unit': 'W'},
110 |
111 | 'Mbasesk': {'ex_output': True,
112 | 'meaning': 'Skin heat production by basal metaborism of th body '
113 | 'part',
114 | 'suffix': 'Body name',
115 | 'unit': 'W'},
116 |
117 | 'Met': {'ex_output': False,
118 | 'meaning': 'Total heat production of the whole body',
119 | 'suffix': None,
120 | 'unit': 'W'},
121 |
122 | 'Mnst': {'ex_output': True,
123 | 'meaning': 'Core heat production by non-shivering of the body part',
124 | 'suffix': 'Body name',
125 | 'unit': 'W'},
126 |
127 | 'ModTime': {'ex_output': False,
128 | 'meaning': 'Simulation times',
129 | 'suffix': None,
130 | 'unit': 'sec'},
131 |
132 | 'Mshiv': {'ex_output': True,
133 | 'meaning': 'Core or muscle heat production by shivering of th body '
134 | 'part',
135 | 'suffix': 'Body name',
136 | 'unit': 'W'},
137 |
138 | 'Mwork': {'ex_output': True,
139 | 'meaning': 'Core or muscle heat production by work of the body part',
140 | 'suffix': 'Body name',
141 | 'unit': 'W'},
142 |
143 | 'Name': {'ex_output': True,
144 | 'meaning': 'Name of the model',
145 | 'suffix': None,
146 | 'unit': '-'},
147 |
148 | 'PAR': {'ex_output': True,
149 | 'meaning': 'Physical activity ratio',
150 | 'suffix': None,
151 | 'unit': '-'},
152 |
153 | 'Qcr': {'ex_output': True,
154 | 'meaning': 'Core total heat production of the body part',
155 | 'suffix': 'Body name',
156 | 'unit': 'W'},
157 |
158 | 'Qfat': {'ex_output': True,
159 | 'meaning': 'Fat total heat production of the body part',
160 | 'suffix': 'Body name',
161 | 'unit': 'W'},
162 |
163 | 'Qms': {'ex_output': True,
164 | 'meaning': 'Muscle total heat production of the body part',
165 | 'suffix': 'Body name',
166 | 'unit': 'W'},
167 |
168 | 'Qsk': {'ex_output': True,
169 | 'meaning': 'Skin total heat production of the body part',
170 | 'suffix': 'Body name',
171 | 'unit': 'W'},
172 |
173 | 'RES': {'ex_output': False,
174 | 'meaning': 'Heat loss by the respiration',
175 | 'suffix': None,
176 | 'unit': 'W'},
177 |
178 | 'RESlh': {'ex_output': True,
179 | 'meaning': 'Latent heat loss by respiration of the body part',
180 | 'suffix': 'Body name',
181 | 'unit': 'W'},
182 | 'RESsh': {'ex_output': True,
183 | 'meaning': 'Sensible heat loss by respiration of the body part',
184 | 'suffix': 'Body name',
185 | 'unit': 'W'},
186 |
187 | 'RH': {'ex_output': True,
188 | 'meaning': 'Relative humidity of the body part',
189 | 'suffix': 'Body name',
190 | 'unit': '%'},
191 |
192 | 'Ret': {'ex_output': True,
193 | 'meaning': 'Total evaporative heat resistance of the body part',
194 | 'suffix': 'Body name',
195 | 'unit': 'm2.kPa/W'},
196 |
197 | 'Rt': {'ex_output': True,
198 | 'meaning': 'Total heat resistance of the body part',
199 | 'suffix': 'Body name',
200 | 'unit': 'm2.K/W'},
201 |
202 | 'SHLsk': {'ex_output': True,
203 | 'meaning': 'Sensible heat loss at the skin of the body part',
204 | 'suffix': 'Body name',
205 | 'unit': 'W'},
206 |
207 | 'Setptcr': {'ex_output': True,
208 | 'meaning': 'Set point skin temperatre of the body part',
209 | 'suffix': 'Body name',
210 | 'unit': 'oC'},
211 |
212 | 'Setptsk': {'ex_output': True,
213 | 'meaning': 'Set point core temperatre of the body part',
214 | 'suffix': 'Body name',
215 | 'unit': 'oC'},
216 |
217 | 'Sex': {'ex_output': True,
218 | 'meaning': 'Male or female',
219 | 'suffix': None,
220 | 'unit': '-'},
221 |
222 | 'THLsk': {'ex_output': False,
223 | 'meaning': 'Heat loss from the skin of the body part',
224 | 'suffix': 'Body name',
225 | 'unit': 'W'},
226 |
227 | 'Ta': {'ex_output': True,
228 | 'meaning': 'Air temperature of the body part',
229 | 'suffix': 'Body name',
230 | 'unit': 'oC'},
231 |
232 | 'Tar': {'ex_output': True,
233 | 'meaning': 'Arterial temperature of the body part',
234 | 'suffix': 'Body name',
235 | 'unit': 'oC'},
236 |
237 | 'Tcb': {'ex_output': True,
238 | 'meaning': 'Central blood temperature',
239 | 'suffix': None,
240 | 'unit': 'oC'},
241 |
242 | 'Tcr': {'ex_output': False,
243 | 'meaning': 'Core temperature of the body part',
244 | 'suffix': 'Body name',
245 | 'unit': 'oC'},
246 |
247 | 'Tfat': {'ex_output': True,
248 | 'meaning': 'Fat temperature of the body part',
249 | 'suffix': 'Body name',
250 | 'unit': 'oC'},
251 |
252 | 'Tms': {'ex_output': True,
253 | 'meaning': 'Muscle temperature as the body part',
254 | 'suffix': 'Body name',
255 | 'unit': 'oC'},
256 |
257 | 'To': {'ex_output': True,
258 | 'meaning': 'Operative temperature of the body part',
259 | 'suffix': 'Body name',
260 | 'unit': 'oC'},
261 |
262 | 'Tr': {'ex_output': True,
263 | 'meaning': 'Mean radiant temperature of the body part',
264 | 'suffix': 'Body name',
265 | 'unit': 'oC'},
266 |
267 | 'Tsk': {'ex_output': False,
268 | 'meaning': 'Skin temperature of the body part',
269 | 'suffix': 'Body name',
270 | 'unit': 'oC'},
271 |
272 | 'TskMean': {'ex_output': False,
273 | 'meaning': 'Mean skin temperature of the body',
274 | 'suffix': None,
275 | 'unit': 'oC'},
276 |
277 | 'Tsve': {'ex_output': True,
278 | 'meaning': 'Superfical vein temperature of the body part',
279 | 'suffix': 'Body name',
280 | 'unit': 'oC'},
281 |
282 | 'Tve': {'ex_output': True,
283 | 'meaning': 'Vein temperature of the body part',
284 | 'suffix': 'Body name',
285 | 'unit': 'oC'},
286 |
287 | 'Va': {'ex_output': True,
288 | 'meaning': 'Air velocity of the body part',
289 | 'suffix': 'Body name',
290 | 'unit': 'm/s'},
291 |
292 | 'Weight': {'ex_output': True,
293 | 'meaning': 'Body weight',
294 | 'suffix': None,
295 | 'unit': 'kg'},
296 |
297 | 'Wet': {'ex_output': False,
298 | 'meaning': 'Local skin wettedness of the body part',
299 | 'suffix': 'Body name',
300 | 'unit': '-'},
301 |
302 | 'WetMean': {'ex_output': False,
303 | 'meaning': 'Mean skin wettedness of the body',
304 | 'suffix': None,
305 | 'unit': '-'},
306 |
307 | 'Wle': {'ex_output': False,
308 | 'meaning': 'Weight loss rate by the evaporation and respiration of '
309 | 'the whole body',
310 | 'suffix': None,
311 | 'unit': 'g/sec'},
312 |
313 | 'dt': {'ex_output': False,
314 | 'meaning': 'Time delta of the model',
315 | 'suffix': None,
316 | 'unit': 'sec'}}
317 |
318 |
319 | def show_outparam_docs():
320 | """
321 | Show the documentation of the output parameters.
322 |
323 | Returns
324 | -------
325 | docstirng : str
326 | Text of the documentation of the output parameters
327 |
328 | """
329 |
330 |
331 | outparams = textwrap.dedent("""
332 | Output parameters
333 | -------
334 | """)
335 |
336 | exoutparams = textwrap.dedent("""
337 | Extra output parameters
338 | -------
339 | """)
340 |
341 | sortkeys = list(ALL_OUT_PARAMS.keys())
342 | sortkeys.sort()
343 | for key in sortkeys:
344 | value = ALL_OUT_PARAMS[key]
345 |
346 | line = "{}: {} [{}]".format(key.ljust(8), value["meaning"], value["unit"])
347 |
348 | if value["ex_output"]:
349 | exoutparams += line + "\n"
350 | else:
351 | outparams += line + "\n"
352 |
353 | docs = outparams + "\n" + exoutparams
354 | docs = textwrap.indent(docs.strip(), " ")
355 |
356 | return docs
357 |
358 | if __name__ == "__main__":
359 | show_outparam_docs()
--------------------------------------------------------------------------------
/src/jos3/thermoregulation.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import numpy as np
3 | import math
4 |
5 | # Import from relative path
6 | try:
7 | from .matrix import NUM_NODES, IDICT, BODY_NAMES
8 | from . import construction as cons
9 | # Import from absolute path
10 | # These codes are for debugging
11 | except ImportError:
12 | from jos3.matrix import NUM_NODES, IDICT, BODY_NAMES
13 | from jos3 import construction as cons
14 |
15 |
16 | _BSAst = np.array([
17 | 0.110, 0.029, 0.175, 0.161, 0.221,
18 | 0.096, 0.063, 0.050, 0.096, 0.063, 0.050,
19 | 0.209, 0.112, 0.056, 0.209, 0.112, 0.056,])
20 |
21 |
22 | def conv_coef(posture="standing", va=0.1, ta=28.8, tsk=34.0,):
23 | """
24 | Calculate convective heat transfer coefficient (hc) [W/K.m2]
25 |
26 | Parameters
27 | ----------
28 | posture : str, optional
29 | Select posture from standing, sitting or lying.
30 | The default is "standing".
31 | va : float or iter, optional
32 | Air velocity [m/s]. If iter is input, its length should be 17.
33 | The default is 0.1.
34 | ta : float or iter, optional
35 | Air temperature [oC]. If iter is input, its length should be 17.
36 | The default is 28.8.
37 | tsk : float or iter, optional
38 | Skin temperature [oC]. If iter is input, its length should be 17.
39 | The default is 34.0.
40 |
41 | Returns
42 | -------
43 | hc : numpy.ndarray
44 | Convective heat transfer coefficient (hc) [W/K.m2].
45 |
46 | """
47 | # Natural convection
48 | if posture.lower() == "standing":
49 | # Ichihara et al., 1997, https://doi.org/10.3130/aija.62.45_5
50 | hc_natural = np.array([
51 | 4.48, 4.48, 2.97, 2.91, 2.85,
52 | 3.61, 3.55, 3.67, 3.61, 3.55, 3.67,
53 | 2.80, 2.04, 2.04, 2.80, 2.04, 2.04,])
54 | elif posture.lower() in ["sitting", "sedentary"]:
55 | # Ichihara et al., 1997, https://doi.org/10.3130/aija.62.45_5
56 | hc_natural = np.array([
57 | 4.75, 4.75, 3.12, 2.48, 1.84,
58 | 3.76, 3.62, 2.06, 3.76, 3.62, 2.06,
59 | 2.98, 2.98, 2.62, 2.98, 2.98, 2.62,])
60 |
61 | elif posture.lower() in ["lying", "supine"]:
62 | # Kurazumi et al., 2008, https://doi.org/10.20718/jjpa.13.1_17
63 | # The values are applied under cold environment.
64 | hc_a = np.array([
65 | 1.105, 1.105, 1.211, 1.211, 1.211,
66 | 0.913, 2.081, 2.178, 0.913, 2.081, 2.178,
67 | 0.945, 0.385, 0.200, 0.945, 0.385, 0.200,])
68 | hc_b = np.array([
69 | 0.345, 0.345, 0.046, 0.046, 0.046,
70 | 0.373, 0.850, 0.297, 0.373, 0.850, 0.297,
71 | 0.447, 0.580, 0.966, 0.447, 0.580, 0.966,])
72 | hc_natural = hc_a * (abs(ta - tsk) ** hc_b)
73 |
74 | # Forced convection
75 | # Ichihara et al., 1997, https://doi.org/10.3130/aija.62.45_5
76 | hc_a = np.array([
77 | 15.0, 15.0, 11.0, 17.0, 13.0,
78 | 17.0, 17.0, 20.0, 17.0, 17.0, 20.0,
79 | 14.0, 15.8, 15.1, 14.0, 15.8, 15.1,])
80 | hc_b = np.array([
81 | 0.62, 0.62, 0.67, 0.49, 0.60,
82 | 0.59, 0.61, 0.60, 0.59, 0.61, 0.60,
83 | 0.61, 0.74, 0.62, 0.61, 0.74, 0.62,])
84 | hc_forced = hc_a * (va ** hc_b)
85 |
86 | # Select natural or forced hc.
87 | # If local va is under 0.2 m/s, the hc valuse is natural.
88 | hc = np.where(va<0.2, hc_natural, hc_forced) # hc [W/K.m2)]
89 |
90 | return hc
91 |
92 |
93 | def rad_coef(posture="standing"):
94 | """
95 | Calculate radiative heat transfer coefficient (hr) [W/K.m2]
96 |
97 | Parameters
98 | ----------
99 | posture : str, optional
100 | Select posture from standing, sitting or lying.
101 | The default is "standing".
102 |
103 | Returns
104 | -------
105 | hc : numpy.ndarray
106 | Radiative heat transfer coefficient (hr) [W/K.m2].
107 |
108 | """
109 |
110 |
111 | if posture.lower() == "standing":
112 | # Ichihara et al., 1997, https://doi.org/10.3130/aija.62.45_5
113 | hr = np.array([
114 | 4.89, 4.89, 4.32, 4.09, 4.32,
115 | 4.55, 4.43, 4.21, 4.55, 4.43, 4.21,
116 | 4.77, 5.34, 6.14, 4.77, 5.34, 6.14,])
117 | elif posture.lower() in ["sitting", "sedentary"]:
118 | # Ichihara et al., 1997, https://doi.org/10.3130/aija.62.45_5
119 | hr = np.array([
120 | 4.96, 4.96, 3.99, 4.64, 4.21,
121 | 4.96, 4.21, 4.74, 4.96, 4.21, 4.74,
122 | 4.10, 4.74, 6.36, 4.10, 4.74, 6.36,])
123 | elif posture.lower() in ["lying", "supine"]:
124 | # Kurazumi et al., 2008, https://doi.org/10.20718/jjpa.13.1_17
125 | hr = np.array([
126 | 5.475, 5.475, 3.463, 3.463, 3.463,
127 | 4.249, 4.835, 4.119, 4.249, 4.835, 4.119,
128 | 4.440, 5.547, 6.085, 4.440, 5.547, 6.085,])
129 | return hr
130 |
131 |
132 | def fixed_hc(hc, va):
133 | """
134 | Fixes hc values to fit tow-node-model's values.
135 | """
136 | mean_hc = np.average(hc, weights=_BSAst)
137 | mean_va = np.average(va, weights=_BSAst)
138 | mean_hc_whole = max(3, 8.600001*(mean_va**0.53))
139 | _fixed_hc = hc * mean_hc_whole/mean_hc
140 | return _fixed_hc
141 |
142 |
143 | def fixed_hr(hr):
144 | """
145 | Fixes hr values to fit tow-node-model's values.
146 | """
147 | mean_hr = np.average(hr, weights=_BSAst)
148 | _fixed_hr = hr * 4.7/mean_hr
149 | return _fixed_hr
150 |
151 | def operative_temp(ta, tr, hc, hr):
152 | to = (hc*ta + hr*tr) / (hc + hr)
153 | return to
154 |
155 |
156 | def clo_area_factor(clo):
157 | fcl = np.where(clo<0.5, clo*0.2+1, clo*0.1+1.05)
158 | return fcl
159 |
160 |
161 | def dry_r(hc, hr, clo, pt=101.33):
162 | """
163 | Calculate total sensible thermal resistance.
164 |
165 | Parameters
166 | ----------
167 | hc : float or array
168 | Convective heat transfer coefficient (hc) [W/K.m2].
169 | hr : float or array
170 | Radiative heat transfer coefficient (hr) [W/K.m2].
171 | clo : float or array
172 | Clothing insulation [clo].
173 | pt : float
174 | Local atmospheric pressure [kPa].
175 | Corrected hc (hcc) is calculated as follows:
176 | hcc = hc * ((pt / 101.33) ** 0.55)
177 |
178 | Returns
179 | -------
180 | rt : float or array
181 | Total sensible thermal resistance between skin and ambient.
182 | """
183 | fcl = clo_area_factor(clo)
184 | hcc = hc * ((pt / 101.33) ** 0.55)
185 | r_a = 1/(hcc+hr)
186 | r_cl = 0.155*clo
187 | r_t = r_a/fcl + r_cl
188 | return r_t
189 |
190 |
191 | def wet_r(hc, clo, iclo=0.45, lewis_rate=16.5, pt=101.33):
192 | """
193 | Calculate total evaporative thermal resistance.
194 |
195 | Parameters
196 | ----------
197 | hc : float or array
198 | Convective heat transfer coefficient (hc) [W/K.m2].
199 | clo : float or array
200 | Clothing insulation [clo].
201 | iclo : float, or array, optional
202 | Clothin vapor permeation efficiency [-]. The default is 0.45.
203 | lewis_rate : float, optional
204 | Lewis rate [K/kPa]. The default is 16.5.
205 | pt : float
206 | Local atmospheric pressure [kPa].
207 | Corrected he (hec) is calculated as follows:
208 | hec = he * ((101.33 / pt) ** 0.45)
209 |
210 | Returns
211 | -------
212 | ret : float or array
213 | Total evaporative thermal resistance.
214 |
215 | """
216 | fcl = clo_area_factor(clo)
217 | r_cl = 0.155 * clo
218 | he = hc * lewis_rate
219 | hec = he * ((101.33 / pt) ** 0.45)
220 | r_ea = 1 / hec
221 | r_ecl = r_cl / (lewis_rate * iclo)
222 | r_et = r_ea / fcl + r_ecl
223 | return r_et
224 |
225 |
226 | def heat_resistances(
227 | ta=np.ones(17)*28.8,
228 | tr=np.ones(17)*28.8,
229 | va=np.ones(17)*0.1,
230 | tsk=np.ones(17)*34,
231 | clo=np.zeros(17),
232 | posture="standing",
233 | iclo=np.ones(17)*0.45,
234 | options={},):
235 |
236 | hc = fixed_hc(conv_coef(posture, va, ta, tsk,))
237 | hr = fixed_hr(rad_coef(posture,))
238 | to = operative_temp(ta, tr, hc, hr,)
239 | fcl = clo_area_factor(clo,)
240 | r_t, r_a, r_cl = dry_r(hc, hr, clo)
241 | r_et, r_ea, r_ecl = wet_r(hc, clo, iclo)
242 |
243 | return to, r_t, r_et, r_a, r_cl, r_ea, r_ecl, fcl
244 |
245 |
246 | def error_signals(err_cr=0, err_sk=0):
247 | """
248 | Calculate WRMS and CLDS signals of thermoregulation
249 |
250 | Parameters
251 | ----------
252 | err_cr, err_sk : float or array, optional
253 | Difference between setpoint and body temperatures.
254 | The default is 0.
255 |
256 | Returns
257 | -------
258 | wrms, clds : array
259 | WRMS and CLDS signals.
260 | """
261 |
262 | # SKINR
263 | receptor = np.array([
264 | 0.0549, 0.0146, 0.1492, 0.1321, 0.2122,
265 | 0.0227, 0.0117, 0.0923, 0.0227, 0.0117, 0.0923,
266 | 0.0501, 0.0251, 0.0167, 0.0501, 0.0251, 0.0167,])
267 |
268 | # wrms signal
269 | wrm = np.maximum(err_sk, 0)
270 | wrm *= receptor
271 | wrms = wrm.sum()
272 | # clds signal
273 | cld = np.minimum(err_sk, 0)
274 | cld *= -receptor
275 | clds = cld.sum()
276 |
277 | return wrms, clds
278 |
279 |
280 | # Antoine equation [kPa]
281 | antoine = lambda x: math.e**(16.6536-(4030.183/(x+235)))
282 | # Tetens equation [kPa]
283 | tetens = lambda x: 0.61078*10**(7.5*x/(x+237.3))
284 |
285 |
286 | def evaporation(err_cr, err_sk, tsk, ta, rh, ret,
287 | height=1.72, weight=74.43, equation="dubois", age=20):
288 | """
289 | Calculate evaporative heat loss.
290 |
291 | Parameters
292 | ----------
293 | err_cr, err_sk : array
294 | Difference between setpoint and body temperatures [oC].
295 | tsk : array
296 | Skin temperatures [oC].
297 | ta : array
298 | Air temperatures at local body segments [oC].
299 | rh : array
300 | Relative humidity at local body segments [%].
301 | ret : array
302 | Total evaporative thermal resistances [m2.K/W].
303 | height : float, optional
304 | Body height [m]. The default is 1.72.
305 | weight : float, optional
306 | Body weight [kg]. The default is 74.43.
307 | equation : str, optional
308 | The equation name (str) of bsa calculation. Choose a name from "dubois",
309 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
310 | age : float, optional
311 | Age [years]. The default is 20.
312 |
313 | Returns
314 | -------
315 | wet : array
316 | Local skin wettedness [-].
317 | e_sk : array
318 | Evaporative heat loss at the skin by sweating and diffuse [W].
319 | e_max : array
320 | Maximum evaporative heat loss at the skin [W].
321 | e_sweat : TYPE
322 | Evaporative heat loss at the skin by only sweating [W].
323 |
324 | """
325 |
326 | wrms, clds = error_signals(err_cr, err_sk,) # Thermoregulation signals
327 | bsar = cons.bsa_rate(height, weight, equation,) # BSA rate
328 | bsa = _BSAst * bsar # BSA
329 | p_a = antoine(ta)*rh/100 # Saturated vapor pressure of ambient [kPa]
330 | p_sk_s = antoine(tsk) # Saturated vapor pressure at the skin [kPa]
331 |
332 | e_max = (p_sk_s - p_a) / ret * bsa # Maximum evaporative heat loss
333 |
334 | # SKINS
335 | skin_sweat = np.array([
336 | 0.064, 0.017, 0.146, 0.129, 0.206,
337 | 0.051, 0.026, 0.0155, 0.051, 0.026, 0.0155,
338 | 0.073, 0.036, 0.0175, 0.073, 0.036, 0.0175,])
339 |
340 | sig_sweat = (371.2*err_cr[0]) + (33.64*(wrms-clds))
341 | sig_sweat = max(sig_sweat, 0)
342 | sig_sweat *= bsar
343 |
344 | # Signal decrement by aging
345 | if age < 60:
346 | sd_sweat = np.ones(17)
347 | else: #age >= 60
348 | sd_sweat = np.array([
349 | 0.69, 0.69, 0.59, 0.52, 0.40,
350 | 0.75, 0.75, 0.75, 0.75, 0.75, 0.75,
351 | 0.40, 0.40, 0.40, 0.40, 0.40, 0.40,])
352 |
353 | e_sweat = skin_sweat * sig_sweat * sd_sweat * 2**((err_sk)/10)
354 | wet = 0.06 + 0.94*(e_sweat/e_max)
355 | wet = np.minimum(wet, 1) # Wettedness' upper limit
356 | e_sk = wet * e_max
357 | e_sweat = (wet - 0.06) / 0.94 * e_max # Effective sweating
358 | return wet, e_sk, e_max, e_sweat
359 |
360 |
361 | def skin_bloodflow(err_cr, err_sk,
362 | height=1.72, weight=74.43, equation="dubois", age=20, ci=2.59,):
363 | """
364 | Calculate skin blood flow rate (BFsk) [L/h].
365 |
366 | Parameters
367 | ----------
368 | err_cr, err_sk : array
369 | Difference between setpoint and body temperatures [oC].
370 | height : float, optional
371 | Body height [m]. The default is 1.72.
372 | weight : float, optional
373 | Body weight [kg]. The default is 74.43.
374 | equation : str, optional
375 | The equation name (str) of bsa calculation. Choose a name from "dubois",
376 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
377 | age : float, optional
378 | Age [years]. The default is 20.
379 | ci : float, optional
380 | Cardiac index [L/min/㎡]. The default is 2.59.
381 |
382 | Returns
383 | -------
384 | BFsk : array
385 | Skin blood flow rate [L/h].
386 |
387 | """
388 |
389 | wrms, clds = error_signals(err_cr, err_sk)
390 |
391 | # BFBsk
392 | bfb_sk = np.array([
393 | 1.754, 0.325, 1.967, 1.475, 2.272,
394 | 0.91, 0.508, 1.114, 0.91, 0.508, 1.114,
395 | 1.456, 0.651, 0.934, 1.456, 0.651, 0.934,])
396 | # SKIND
397 | skin_dilat = np.array([
398 | 0.0692, 0.0992, 0.0580, 0.0679, 0.0707,
399 | 0.0400, 0.0373, 0.0632, 0.0400, 0.0373, 0.0632,
400 | 0.0736, 0.0411, 0.0623, 0.0736, 0.0411, 0.0623,])
401 | # SKINC
402 | skin_stric = np.array([
403 | 0.0213, 0.0213, 0.0638, 0.0638, 0.0638,
404 | 0.0213, 0.0213, 0.1489, 0.0213, 0.0213, 0.1489,
405 | 0.0213, 0.0213, 0.1489, 0.0213, 0.0213, 0.1489,])
406 |
407 | sig_dilat = (100.5*err_cr[0]) + (6.4*(wrms-clds))
408 | sig_stric = (-10.8*err_cr[0]) + (-10.8*(wrms-clds))
409 | sig_dilat = max(sig_dilat, 0)
410 | sig_stric = max(sig_stric, 0)
411 |
412 | # Signal decrement by aging
413 | if age < 60:
414 | sd_dilat = np.ones(17)
415 | sd_stric = np.ones(17)
416 | else: #age >= 60
417 | sd_dilat = np.array([
418 | 0.91, 0.91, 0.47, 0.47, 0.31,
419 | 0.47, 0.47, 0.47, 0.47, 0.47, 0.47,
420 | 0.31, 0.31, 0.31, 0.31, 0.31, 0.31,
421 | ])
422 | sd_stric = np.ones(17)
423 |
424 | #皮膚血流量 [L/h]
425 | bf_sk = (1 + skin_dilat * sd_dilat * sig_dilat) / \
426 | (1 + skin_stric * sd_stric * sig_stric) * bfb_sk * 2**(err_sk/6)
427 |
428 | bfbr = cons.bfb_rate(height, weight, equation, age, ci,)
429 | bf_sk *= bfbr
430 | return bf_sk
431 |
432 |
433 | def ava_bloodflow(err_cr, err_sk,
434 | height=1.72, weight=74.43, equation="dubois", age=20, ci=2.59,):
435 | """
436 | Calculate areteriovenous anastmoses (AVA) blood flow rate [L/h] based on
437 | Takemori's model, 1995.
438 |
439 | Parameters
440 | ----------
441 | err_cr, err_sk : array
442 | Difference between setpoint and body temperatures [oC].
443 | height : float, optional
444 | Body height [m]. The default is 1.72.
445 | weight : float, optional
446 | Body weight [kg]. The default is 74.43.
447 | equation : str, optional
448 | The equation name (str) of bsa calculation. Choose a name from "dubois",
449 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
450 | age : float, optional
451 | Age [years]. The default is 20.
452 | ci : float, optional
453 | Cardiac index [L/min/m2]. The default is 2.59.
454 |
455 | Returns
456 | -------
457 | BFava_hand, BFava_foot : array
458 | AVA blood flow rate at hand and foot [L/h].
459 |
460 | """
461 | # Cal. mean error body core temp.
462 | cap_bcr = [10.2975, 9.3935, 13.834] # Thermal capacity at Chest, Back and Pelvis
463 | err_bcr = np.average(err_cr[2:5], weights=cap_bcr)
464 |
465 | # Cal. mean error skin temp.
466 | bsa = _BSAst
467 | err_msk = np.average(err_sk, weights=bsa)
468 |
469 | # Openbess of AVA [-]
470 | sig_ava_hand = 0.265 * (err_msk + 0.43) + 0.953 * (err_bcr + 0.1905) + 0.9126
471 | sig_ava_foot = 0.265 * (err_msk - 0.997) + 0.953 * (err_bcr + 0.0095) + 0.9126
472 |
473 | sig_ava_hand = min(sig_ava_hand, 1)
474 | sig_ava_hand = max(sig_ava_hand, 0)
475 | sig_ava_foot = min(sig_ava_foot, 1)
476 | sig_ava_foot = max(sig_ava_foot, 0)
477 |
478 | bfbr = bfbr = cons.bfb_rate(height, weight, equation, age, ci,)
479 | # AVA blood flow rate [L/h]
480 | bf_ava_hand = 1.71 * bfbr * sig_ava_hand # Hand
481 | bf_ava_foot = 2.16 * bfbr * sig_ava_foot # Foot
482 | return bf_ava_hand, bf_ava_foot
483 |
484 |
485 | def basal_met(height=1.72, weight=74.43, age=20,
486 | sex="male", equation="harris-benedict"):
487 | """
488 | Calculate basal metabolic rate [W].
489 |
490 | Parameters
491 | ----------
492 | height : float, optional
493 | Body height [m]. The default is 1.72.
494 | weight : float, optional
495 | Body weight [kg]. The default is 74.43.
496 | age : float, optional
497 | Age [years]. The default is 20.
498 | sex : str, optional
499 | Choose male or female. The default is "male".
500 | equation : str, optional
501 | Choose harris-benedict or ganpule. The default is "harris-benedict".
502 |
503 | Returns
504 | -------
505 | BMR : float
506 | Basal metabolic rate [W].
507 |
508 | """
509 |
510 | if equation=="harris-benedict":
511 | if sex=="male":
512 | bmr = 88.362 + 13.397*weight + 500.3*height - 5.677*age
513 | else:
514 | bmr = 447.593 + 9.247*weight + 479.9*height - 4.330*age
515 |
516 | elif equation=="harris-benedict_origin":
517 | if sex=="male":
518 | bmr = 66.4730 + 13.7516*weight + 500.33*height - 6.7550*age
519 | else:
520 | bmr = 655.0955 + 9.5634*weight + 184.96*height - 4.6756*age
521 |
522 | elif equation=="japanese" or equation=="ganpule":
523 | # Ganpule et al., 2007, https://doi.org/10.1038/sj.ejcn.1602645
524 | if sex=="male":
525 | bmr = 0.0481*weight + 2.34*height - 0.0138*age - 0.4235
526 | else:
527 | bmr = 0.0481*weight + 2.34*height - 0.0138*age - 0.9708
528 | bmr *= 1000 / 4.186
529 |
530 | bmr *= 0.048 # [kcal/day] to [W]
531 |
532 | return bmr
533 |
534 |
535 | def local_mbase(height=1.72, weight=74.43, age=20,
536 | sex="male", equation="harris-benedict"):
537 | """
538 | Calculate local basal metabolic rate [W].
539 |
540 | Parameters
541 | ----------
542 | height : float, optional
543 | Body height [m]. The default is 1.72.
544 | weight : float, optional
545 | Body weight [kg]. The default is 74.43.
546 | age : float, optional
547 | Age [years]. The default is 20.
548 | sex : str, optional
549 | Choose male or female. The default is "male".
550 | equation : str, optional
551 | Choose harris-benedict or ganpule. The default is "harris-benedict".
552 |
553 | Returns
554 | -------
555 | mbase : array
556 | Local basal metabolic rate (Mbase) [W].
557 |
558 | """
559 |
560 | mbase_all = basal_met(height, weight, age, sex, equation)
561 | # Distribution coefficient of basal metabolic rate
562 | mbf_cr = np.array([
563 | 0.19551, 0.00324, 0.28689, 0.25677, 0.09509,
564 | 0.01435, 0.00409, 0.00106, 0.01435, 0.00409, 0.00106,
565 | 0.01557, 0.00422, 0.00250, 0.01557, 0.00422, 0.00250,])
566 | mbf_ms = np.array([
567 | 0.00252, 0.0, 0.0, 0.0, 0.04804,
568 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
569 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
570 | mbf_fat = np.array([
571 | 0.00127, 0.0, 0.0, 0.0, 0.00950,
572 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
573 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
574 | mbf_sk = np.array([
575 | 0.00152, 0.00033, 0.00211, 0.00187, 0.00300,
576 | 0.00059, 0.00031, 0.00059, 0.00059, 0.00031, 0.00059,
577 | 0.00144, 0.00027, 0.00118, 0.00144, 0.00027, 0.00118,])
578 |
579 | mbase_cr = mbf_cr * mbase_all
580 | mbase_ms = mbf_ms * mbase_all
581 | mbase_fat = mbf_fat * mbase_all
582 | mbase_sk = mbf_sk * mbase_all
583 | return mbase_cr, mbase_ms, mbase_fat, mbase_sk
584 |
585 |
586 | def local_mwork(bmr, par):
587 | """
588 | Calculate local metabolic rate by work [W]
589 |
590 | Parameters
591 | ----------
592 | bmr : float
593 | Basal metbolic rate [W].
594 | par : float
595 | Physical activity ratio [-].
596 |
597 | Returns
598 | -------
599 | Mwork : array
600 | Local metabolic rate by work [W].
601 |
602 | """
603 | mwork_all = (par-1) * bmr
604 | mwf = np.array([
605 | 0, 0, 0.091, 0.08, 0.129,
606 | 0.0262, 0.0139, 0.005, 0.0262, 0.0139, 0.005,
607 | 0.2010, 0.0990, 0.005, 0.2010, 0.0990, 0.005])
608 | mwork = mwork_all * mwf
609 | return mwork
610 |
611 |
612 | PRE_SHIV = 0
613 | def shivering(err_cr, err_sk, tcr, tsk,
614 | height=1.72, weight=74.43, equation="dubois", age=20, sex="male", dtime=60,
615 | options={}):
616 | """
617 | Calculate local metabolic rate by shivering [W].
618 |
619 | Parameters
620 | ----------
621 | err_cr, err_sk : array
622 | Difference between setpoint and body temperatures [oC].
623 | tcr, tsk : array
624 | Core and skin temperatures [oC].
625 | height : float, optional
626 | Body height [m]. The default is 1.72.
627 | weight : float, optional
628 | Body weight [kg]. The default is 74.43.
629 | equation : str, optional
630 | The equation name (str) of bsa calculation. Choose a name from "dubois",
631 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
632 | age : float, optional
633 | Age [years]. The default is 20.
634 | sex : str, optional
635 | Choose male or female. The default is "male".
636 | dtime : float, optional
637 | Interval of analysis time. The default is 60.
638 |
639 | Returns
640 | -------
641 | Mshiv : array
642 | Local metabolic rate by shivering [W].
643 |
644 | """
645 | wrms, clds = error_signals(err_cr, err_sk,)
646 | shivf = np.array([
647 | 0.0339, 0.0436, 0.27394, 0.24102, 0.38754,
648 | 0.00243, 0.00137, 0.0002, 0.00243, 0.00137, 0.0002,
649 | 0.0039, 0.00175, 0.00035, 0.0039, 0.00175, 0.00035,])
650 | sig_shiv = 24.36 * clds * (-err_cr[0])
651 | sig_shiv = max(sig_shiv, 0)
652 |
653 | if options:
654 | if options["shivering_threshold"]:
655 | # Asaka, 2016
656 | # Threshold of starting shivering
657 | tskm = np.average(tsk, weights=_BSAst) # Mean skin temp.
658 | if tskm < 31:
659 | thres = 36.6
660 | else:
661 | if sex == "male":
662 | thres = -0.2436 * tskm + 44.10
663 | else: # sex == "female":
664 | thres = -0.2250 * tskm + 43.05
665 | # Second threshold of starting shivering
666 | if thres < tcr[0]:
667 | sig_shiv = 0
668 |
669 | global PRE_SHIV # Previous shivering thermogenesis [W]
670 | if options:
671 | if options["limit_dshiv/dt"]:
672 | # Asaka, 2016
673 | # dshiv < 0.0077 [W/s]
674 | dshiv = sig_shiv - PRE_SHIV
675 | if options["limit_dshiv/dt"] is True: # default is 0.0077 [W/s]
676 | limit_dshiv = 0.0077 * dtime
677 | else:
678 | limit_dshiv = options["limit_dshiv/dt"] * dtime
679 | if dshiv > limit_dshiv:
680 | sig_shiv = limit_dshiv + PRE_SHIV
681 | elif dshiv < -limit_dshiv:
682 | sig_shiv = -limit_dshiv + PRE_SHIV
683 | PRE_SHIV = sig_shiv
684 |
685 | # Signal sd_shiv by aging
686 | if age < 30:
687 | sd_shiv = np.ones(17)
688 | elif age < 40:
689 | sd_shiv = np.ones(17) * 0.97514
690 | elif age < 50:
691 | sd_shiv = np.ones(17) * 0.95028
692 | elif age < 60:
693 | sd_shiv = np.ones(17) * 0.92818
694 | elif age < 70:
695 | sd_shiv = np.ones(17) * 0.90055
696 | elif age < 80:
697 | sd_shiv = np.ones(17) * 0.86188
698 | else: #age >= 80
699 | sd_shiv = np.ones(17) * 0.82597
700 |
701 | bsar = cons.bsa_rate(height, weight, equation)
702 | mshiv = shivf * bsar * sd_shiv * sig_shiv
703 | return mshiv
704 |
705 | shivering
706 | def nonshivering(err_cr, err_sk,
707 | height=1.72, weight=74.43, equation="dubois", age=20,
708 | coldacclimation=False, batpositive=True,
709 | options={},):
710 | """
711 | Calculate local metabolic rate by non-shivering [W]
712 |
713 | Parameters
714 | ----------
715 | err_cr, err_sk : array
716 | Difference between setpoint and body temperatures [oC].
717 | height : float, optional
718 | Body height [m]. The default is 1.72.
719 | weight : float, optional
720 | Body weight [kg]. The default is 74.43.
721 | equation : str, optional
722 | The equation name (str) of bsa calculation. Choose a name from "dubois",
723 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
724 | age : float, optional
725 | Age [years]. The default is 20.
726 | coldacclimation : bool, optional
727 | Whether the subject acclimates cold enviroment or not.
728 | The default is False.
729 | batpositive : bool, optional
730 | Whether BAT ativity is positive or not.
731 | The default is True.
732 |
733 | Returns
734 | -------
735 | Mnst : array
736 | Local metabolic rate by non-shivering [W].
737 |
738 | """
739 | # NST (Non-Shivering Thermogenesis) model, Asaka, 2016
740 | wrms, clds = error_signals(err_cr, err_sk, )
741 |
742 | bmi = weight / height**2
743 |
744 | # BAT: brown adipose tissue [SUV]
745 | bat = 10**(-0.10502 * bmi + 2.7708)
746 |
747 | # age factor
748 | if age < 30:
749 | bat *= 1.61
750 | elif age < 40:
751 | bat *= 1.00
752 | else: # age >= 40
753 | bat *= 0.80
754 |
755 | if coldacclimation:
756 | bat += 3.46
757 |
758 | if not batpositive:
759 | # incidence age factor: T.Yoneshiro 2011
760 | if age < 30:
761 | bat *= 44/83
762 | elif age < 40:
763 | bat *= 15/38
764 | elif age < 50:
765 | bat *= 7/26
766 | elif age < 50:
767 | bat *= 1/8
768 | else: # age > 60
769 | bat *= 0
770 |
771 | # NST limit
772 | thres = ((1.80 * bat + 2.43) + 5.62) # [W]
773 |
774 | sig_nst = 2.8 * clds # [W]
775 | sig_nst = min(sig_nst, thres)
776 |
777 | mnstf = np.array([
778 | 0.000, 0.190, 0.000, 0.190, 0.190,
779 | 0.215, 0.000, 0.000, 0.215, 0.000, 0.000,
780 | 0.000, 0.000, 0.000, 0.000, 0.000, 0.000,])
781 | bsar = cons.bsa_rate(height, weight, equation)
782 | mnst = bsar * mnstf * sig_nst
783 | return mnst
784 |
785 |
786 | def sum_m(mbase, mwork, mshiv, mnst):
787 | qcr = mbase[0].copy()
788 | qms = mbase[1].copy()
789 | qfat = mbase[2].copy()
790 | qsk = mbase[3].copy()
791 |
792 | for i, bn in enumerate(BODY_NAMES):
793 | # If the segment has a muscle layer, muscle heat production increases by the activity.
794 | if not IDICT[bn]["muscle"] is None:
795 | qms[i] += mwork[i] + mshiv[i]
796 | # In other segments, core heat production increase, instead of muscle.
797 | else:
798 | qcr[i] += mwork[i] + mshiv[i]
799 | qcr += mnst # Non-shivering thermogenesis occurs in core layers
800 | return qcr, qms, qfat, qsk
801 |
802 |
803 | def crmsfat_bloodflow(mwork, mshiv,
804 | height=1.72, weight=74.43, equation="dubois", age=20, ci=2.59,):
805 | """
806 | Calculate core, muslce and fat blood flow rate [L/h].
807 |
808 | Parameters
809 | ----------
810 | mwork : array
811 | Metablic rate by work [W].
812 | mshiv : array
813 | Metablic rate by shivering [W].
814 | height : float, optional
815 | Body height [m]. The default is 1.72.
816 | weight : float, optional
817 | Body weight [kg]. The default is 74.43.
818 | equation : str, optional
819 | The equation name (str) of bsa calculation. Choose a name from "dubois",
820 | "takahira", "fujimoto", or "kurazumi". The default is "dubois".
821 | age : float, optional
822 | Age [years]. The default is 20.
823 | ci : float, optional
824 | Cardiac index [L/min/㎡]. The default is 2.59.
825 |
826 | Returns
827 | -------
828 | BFcr, BFms, BFfat : array
829 | Core, muslce and fat blood flow rate [L/h].
830 |
831 | """
832 | # Basal blood flow rate [L/h]
833 | # core, CBFB
834 | bfb_cr = np.array([
835 | 35.251, 15.240, 89.214, 87.663, 18.686,
836 | 1.808, 0.940, 0.217, 1.808, 0.940, 0.217,
837 | 1.406, 0.164, 0.080, 1.406, 0.164, 0.080,])
838 | # muscle, MSBFB
839 | bfb_ms = np.array([
840 | 0.682, 0.0, 0.0, 0.0, 12.614,
841 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
842 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
843 | # fat, FTBFB
844 | bfb_fat = np.array([
845 | 0.265, 0.0, 0.0, 0.0, 2.219,
846 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
847 | 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,])
848 |
849 | bfbr = cons.bfb_rate(height, weight, equation, age, ci)
850 | bf_cr = bfb_cr * bfbr
851 | bf_ms = bfb_ms * bfbr
852 | bf_fat = bfb_fat * bfbr
853 |
854 | for i, bn in enumerate(BODY_NAMES):
855 | # If the segment has a muscle layer, muscle blood flow increases.
856 | if not IDICT[bn]["muscle"] is None:
857 | bf_ms[i] += (mwork[i] + mshiv[i])/1.163
858 | # In other segments, core blood flow increase, instead of muscle blood flow.
859 | else:
860 | bf_cr[i] += (mwork[i] + mshiv[i])/1.163
861 | return bf_cr, bf_ms, bf_fat
862 |
863 |
864 | def sum_bf(bf_cr, bf_ms, bf_fat, bf_sk, bf_ava_hand, bf_ava_foot):
865 | co = 0
866 | co += bf_cr.sum()
867 | co += bf_ms.sum()
868 | co += bf_fat.sum()
869 | co += bf_sk.sum()
870 | co += 2*bf_ava_hand
871 | co += 2*bf_ava_foot
872 | return co
873 |
874 |
875 | def resp_heatloss(t, p, met):
876 | res_sh = 0.0014 * met * (34 - t) #顕熱
877 | res_lh = 0.0173 * met * (5.87 - p) #潜熱
878 | return res_sh, res_lh
879 |
880 |
881 | def get_lts(ta):
882 | return 2.418*1000
--------------------------------------------------------------------------------
/test.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | import sys
3 | import os
4 | sys.path.append(os.path.dirname(__file__) + "/src")
5 |
6 | import jos3
7 | from jos3 import JOS3
8 |
9 | if __name__ == "__main__":
10 | mod = JOS3(ex_output="all")
11 | mod.PAR = 2
12 | mod.To = 40
13 | mod.RH = 70
14 | mod.Va = 2
15 | mod.Icl = 0.6
16 | mod.simulate(60)
17 |
18 | d = mod.dict_results()
19 | # for k in d.keys():
20 | # print(jos3.remove_bodyname(k))
21 | mod.to_csv(folder="E:/desktop")
22 |
23 |
24 | mod = JOS3()
25 | print("\nNeutral")
26 | print("TcrHead: {:.3f} [oC]".format(mod.Tcr[0]))
27 | print("TskMean: {:.3f} [oC]".format(mod.TskMean))
28 |
29 | mod = JOS3()
30 | mod.PAR = 2
31 | mod.To = 40
32 | mod.RH = 70
33 | mod.Va = 2
34 | mod.Icl = 0.6
35 | mod.simulate(60)
36 | print("\nAfter Hot Exposure")
37 | print("TcrHead: {:.3f} [oC]".format(mod.Tcr[0]))
38 | print("TskMean: {:.3f} [oC]".format(mod.TskMean))
39 |
40 | mod = JOS3()
41 | mod.PAR = 1.2
42 | mod.To = 10
43 | mod.RH = 20
44 | mod.Va = 3
45 | mod.Icl = 0.1
46 | mod.simulate(60)
47 | print("\nAfter Cold Exposure")
48 | print("TcrHead: {:.3f} [oC]".format(mod.Tcr[0]))
49 | print("TskMean: {:.3f} [oC]".format(mod.TskMean))
50 |
51 | # Measure calculation time
52 | import time
53 | stime = time.time()
54 | mod = JOS3()
55 | mod.To = 30
56 | mod.simulate(60)
57 | mod.To = 20
58 | mod.simulate(60)
59 | mod.To = 40
60 | mod.simulate(60)
61 | mod.To = 10
62 | mod.simulate(60)
63 | etime = time.time()
64 | print("Default output")
65 | print("Calculation time {:.2f} [sec]".format(etime-stime))
66 |
67 | stime = time.time()
68 | mod = JOS3(ex_output=["BFsk", "BFcr", "Emax"])
69 | mod.To = 30
70 | mod.simulate(60)
71 | mod.To = 20
72 | mod.simulate(60)
73 | mod.To = 40
74 | mod.simulate(60)
75 | mod.To = 10
76 | mod.simulate(60)
77 | etime = time.time()
78 | print("Extra output")
79 | print("Calculation time {:.2f} [sec]".format(etime-stime))
80 |
81 | stime = time.time()
82 | mod = JOS3(ex_output="all")
83 | mod.To = 30
84 | mod.simulate(60)
85 | mod.To = 20
86 | mod.simulate(60)
87 | mod.To = 40
88 | mod.simulate(60)
89 | mod.To = 10
90 | mod.simulate(60)
91 | etime = time.time()
92 | print("All output")
93 | print("Calculation time {:.2f} [sec]".format(etime-stime))
--------------------------------------------------------------------------------