├── 3D reconstruction
├── vec.m
├── soft.m
├── linear_gradient_b.m
├── power_iters.m
├── read_tiff_stack.m
├── tv3d_iso_Haar.m
├── MiniscopeFISTA.m
├── Miniscope_shift_varying_recon.m
└── Miniscope_3d_shift_varying_main.m
├── LICENSE
├── README.md
└── Adaptive Stitching with Nanoscribe
├── TipSlicerUpdated.py
└── Adaptive-Stitching-Nanoscribe.ipynb
/3D reconstruction/vec.m:
--------------------------------------------------------------------------------
1 | function y = vec(x)
2 | y=x(:);
--------------------------------------------------------------------------------
/3D reconstruction/soft.m:
--------------------------------------------------------------------------------
1 | function y = soft(x,T)
2 |
3 | y = sign(x).*max(abs(x) - T, 0);
4 | %y = y./(y+T) .* x;
5 |
6 |
7 |
--------------------------------------------------------------------------------
/3D reconstruction/linear_gradient_b.m:
--------------------------------------------------------------------------------
1 | function [f, g] = linear_gradient_b(x,A,At,b)
2 | % Generalized gradient step for ||Ax-b|| cost function
3 | % Takes in variable, x, linear operator, A, and adjoint, At. Does not
4 | % require passing in Atb
5 | %
6 | % outputs:
7 | % g: Gradient. Does not need to be a vector.
8 | % f: Cost function value
9 |
10 | u = A(x);
11 | g = At(u-b);
12 | f = norm(u(:)-b(:));
--------------------------------------------------------------------------------
/3D reconstruction/power_iters.m:
--------------------------------------------------------------------------------
1 | function mu_out = power_iters(A,sz)
2 | %A = @(x)Aadj_3d(A3d(x));
3 | %sz = [Ny, Nx, Nz];
4 | bk = gpuArray(single(randn(sz)));
5 | k = 0;
6 | maxiter = 100;
7 | mu = gpuArray(zeros(1));
8 | resid = 1e10;
9 | while resid > .0001 && k<=maxiter
10 | k = k+1;
11 | Abk = A(bk);
12 | mu(k) = transpose(conj(bk(:)))*Abk(:)/(norm(bk(:))^2);
13 | if k >= 2
14 | resid = abs(mu(k) - mu(k-1));
15 | else
16 | resid = 1e10;
17 | end
18 | bknorm = norm(bk(:));
19 | bk = Abk/bknorm;
20 | fprintf('iter %i \t Eigenvalue %.6f \t residual %.6f \n',k,mu,resid)
21 | end
22 | mu_out = mu(end);
--------------------------------------------------------------------------------
/3D reconstruction/read_tiff_stack.m:
--------------------------------------------------------------------------------
1 | function stack = read_tiff_stack(path,varargin)
2 | %stack = read_tiff_stack(path,downsample_ratio,list_of_images)
3 | %list_of_images is a vector of indices to read out. if empty, read all.
4 |
5 |
6 |
7 | info = imfinfo(path);
8 | num_images_in = numel(info);
9 |
10 | if nargin>1
11 | ds = varargin{1};
12 | if nargin > 2
13 | k_list = varargin{2};
14 | else
15 | k_list = 1:num_images_in;
16 | end
17 | else
18 | ds = 1;
19 | end
20 | num_planes = length(k_list);
21 | if info(1).SamplesPerPixel == 1
22 |
23 | stack = zeros(info(1).Height/ds, info(1).Width/ds,num_planes);
24 | else
25 | stack = zeros(info(1).Height/ds, info(1).Width/ds,info(1).SamplesPerPixel,num_planes);
26 | end
27 | n = 0;
28 | for k = k_list
29 | n = n+1;
30 | if info(1).SamplesPerPixel == 1
31 | stack(:,:,n) = imresize(imread(path, k, 'Info', info),1/ds,'box');
32 | else
33 | stack(:,:,:,n) = imresize(imread(path, k, 'Info', info),1/ds,'box');
34 | end
35 |
36 | end
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | BSD 3-Clause License
2 |
3 | Copyright (c) 2020, Waller Lab
4 | All rights reserved.
5 |
6 | Redistribution and use in source and binary forms, with or without
7 | modification, are permitted provided that the following conditions are met:
8 |
9 | 1. Redistributions of source code must retain the above copyright notice, this
10 | list of conditions and the following disclaimer.
11 |
12 | 2. Redistributions in binary form must reproduce the above copyright notice,
13 | this list of conditions and the following disclaimer in the documentation
14 | and/or other materials provided with the distribution.
15 |
16 | 3. Neither the name of the copyright holder nor the names of its
17 | contributors may be used to endorse or promote products derived from
18 | this software without specific prior written permission.
19 |
20 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
21 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
22 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
23 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
24 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
25 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
26 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
27 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
28 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
29 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
30 |
--------------------------------------------------------------------------------
/3D reconstruction/tv3d_iso_Haar.m:
--------------------------------------------------------------------------------
1 | function y = tv3d_iso_Haar(x, tau, alpha)
2 |
3 | % Private functions here
4 | % circshift does circular shifting
5 | % indexing: x(5:10), 1 indexed. Use x(5:4:end-6) to index in strides of 4
6 | % to the 6th-to-last element
7 | D = 3;
8 | gamma = 1; %step size
9 | thresh = sqrt(2) * 2 * D * tau * gamma;
10 | y = zeros(size(x), 'like', x);
11 | for axis = 1 : 3
12 | if axis == 3
13 | t_scale = alpha;
14 | else
15 | t_scale = 1;
16 | end
17 | y = y + iht3(ht3(x, axis, false, thresh*t_scale), axis, false);
18 | y = y + iht3(ht3(x, axis, true, thresh*t_scale), axis, true);
19 | end
20 | y = y / (2 * D);
21 | return
22 |
23 | function w = ht3(x, ax, shift, thresh)
24 | s = size(x);
25 | w = zeros(s, 'like', x);
26 | C = 1 / sqrt(2);
27 | if shift
28 | x = circshift(x, -1, ax);
29 | end
30 | m = floor(s(ax) / 2);
31 | if ax == 1
32 | w(1:m, :, :) = C * (x(2:2:end, :, :) + x(1:2:end, :, :)); % use diff or circhisft?
33 | w((m + 1):end, :, :) = hs_soft(C * (x(2:2:end, :, :) - x(1:2:end, :, :)), thresh);
34 | %w((m + 1):end, :, :) = hs_soft(w((m + 1):end, :, :), thresh);
35 | elseif ax == 2
36 | w(:, 1:m, :) = C * (x(:, 2:2:end, :) + x(:, 1:2:end, :));
37 | w(:, (m + 1):end, :) = hs_soft(C * (x(:, 2:2:end, :) - x(:, 1:2:end, :)), thresh);
38 | %w(:, (m + 1):end, :) = hs_soft(w(:, (m + 1):end, :), thresh);
39 | else
40 | w(:, :, 1:m) = C * (x(:, :, 2:2:end) + x(:, :, 1:2:end));
41 | w(:, :, (m + 1):end) = hs_soft(C * (x(:, :, 2:2:end) - x(:, :, 1:2:end)), thresh);
42 | %w(:, :, (m + 1):end) = hs_soft(w(:, :, (m + 1):end), thresh);
43 | end
44 | return
45 |
46 | function y = iht3(w, ax, shift)
47 | s = size(w);
48 | y = zeros(s, 'like', w);
49 | C = 1 / sqrt(2);
50 | m = floor(s(ax) / 2);
51 | if ax == 1
52 | y(1:2:end, :, :) = C * (w(1:m, :, :) - w((m + 1):end, :, :));
53 | y(2:2:end, :, :) = C * (w(1:m, :, :) + w((m + 1):end, :, :));
54 | elseif ax == 2
55 | y(:, 1:2:end, :) = C * (w(:, 1:m, :) - w(:, (m + 1):end, :));
56 | y(:, 2:2:end, :) = C * (w(:, 1:m, :) + w(:, (m + 1):end, :));
57 | else
58 | y(:, :, 1:2:end) = C * (w(:, :, 1:m) - w(:, :, (m + 1):end));
59 | y(:, :, 2:2:end) = C * (w(:, :, 1:m) + w(:, :, (m + 1):end));
60 | end
61 | if shift
62 | y = circshift(y, 1, ax);
63 | end
64 | return
65 |
66 | function threshed = hs_soft(x,tau)
67 |
68 | threshed = max(abs(x)-tau,0);
69 | threshed = threshed.*sign(x);
70 | return
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | 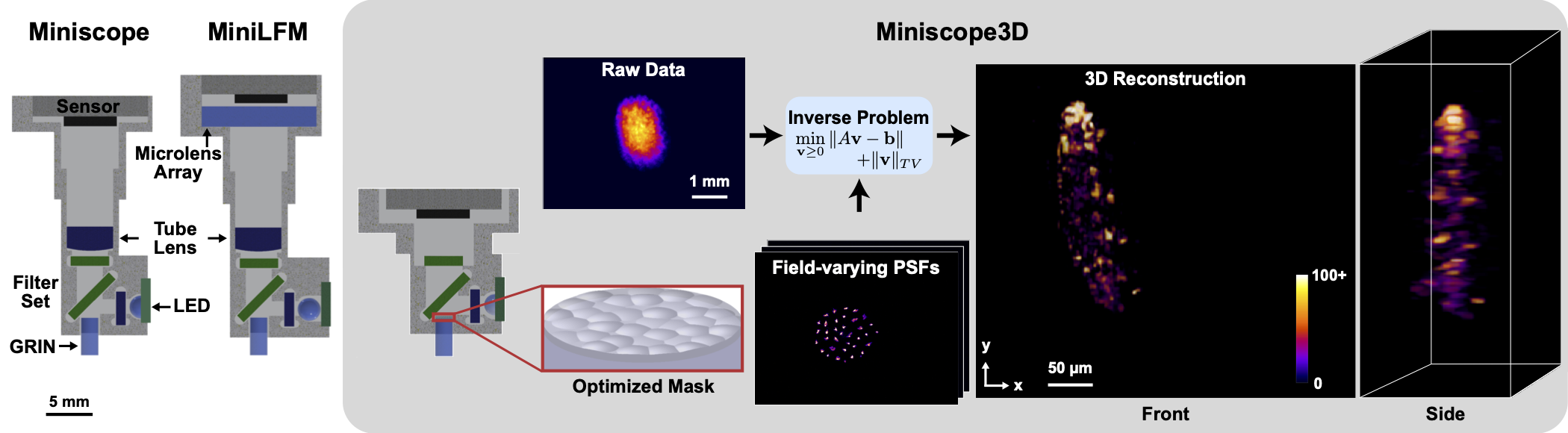
2 |
3 | # [Miniscope3D](https://waller-lab.github.io/Miniscope3D/)
4 |
5 |
6 |
7 | ## Paper
8 | [Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy](https://www.nature.com/articles/s41377-020-00403-7)
9 |
10 | Please cite the following paper when using this code or data:
11 |
12 |
13 | ```
14 | Yanny, Kyrollos, et al. "Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy." Light: Science & Applications 9.1 (2020): 1-13.
15 |
16 | @article{yanny2020miniscope3d,
17 | title={Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy},
18 | author={Yanny, Kyrollos and Antipa, Nick and Liberti, William and Dehaeck, Sam and Monakhova, Kristina and Liu, Fanglin Linda and Shen, Konlin and Ng, Ren and Waller, Laura},
19 | journal={Light: Science \& Applications},
20 | volume={9},
21 | number={1},
22 | pages={1--13},
23 | year={2020},
24 | publisher={Nature Publishing Group}
25 | }
26 |
27 | ```
28 |
29 |
30 | ## Contents
31 |
32 | 1. [Data](#Data)
33 | 2. [Setup](#Setup)
34 | 3. [Description](#Description)
35 |
36 | ## Data
37 | Sample data and PSFs (needed to run the code) can be found [here](https://drive.google.com/drive/folders/1E9Rt8edPrkn80qfUbDhEnLme98MPtQ2w?usp=sharing)
38 |
39 | This includes the following files:
40 | * calibration.mat - includes the calibratated point spread function, filter function, and wavelength list
41 | * four sample raw measurements
42 |
43 |
44 | ## Setup
45 | Clone this project using:
46 | ```
47 | git clone https://github.com/Waller-Lab/Miniscope3D.git
48 | ```
49 | ### 1) 3D Reconstruction
50 | First, download the sample data folder mentioned above. This contains PSF components and weights needed for the shift-varying forward model as well as waterbear raw data.
51 |
52 | Then, open the Miniscope_3d_shift_varying_main.m file in the 3D Reconstruction folder. This file is the main file that runs the reconstruction. You first need to edit the psf_path variable to the folder path where you downloaded the weights and components (SVD_2_5um_PSF_5um_1_ds4_components.mat and SVD_2_5um_PSF_5um_1_ds4_weights.mat).
53 |
54 | Then, run the file, a UI will show up asking you to choose a measurement file and background file. The measurement file is the data you want to deconvolve (e.g. waterbear raw data) and the background is a background image captured before taking the data. This background is subtracted from the data file (to account for back reflections)
55 |
56 | ### 2) Adaptive Stitching with Nanoscribe
57 | This code performs adaptive stitching for randomly spaced microlenses that are multifocal and have aberrations added to them. The adaptive stitching is needed for the microlenses to be correctly 3D printed with Nanoscribe. The code places the stitching artifacts at the boundaries of the microlenses and thus maximizes optical quality.
58 |
59 | The demonstration here will be for the 36 multifocal microlenses with astigmatism and tilt added. This is the phase mask used in the paper. The code also provides the necessary .gwl files needed for the print-job on Nanoscribe
60 |
61 |
62 |
63 | ## Description
64 | This repository contains code for the Miniscope3D paper. The code includes: 1) 3D Reconstruction with Shif-Varying model (matlab). This code reconstructs 3D volumes from a single 2D image. 2) Adaptive Stitching with Nanoscribe (python). This code performs the adaptive stitching needed to 3D print the phase mask with Nanoscribe. We might be adding Microlens phase mask optimization (python) in the future.
65 |
66 |
--------------------------------------------------------------------------------
/3D reconstruction/MiniscopeFISTA.m:
--------------------------------------------------------------------------------
1 | function [out,varargout] = MiniscopeFISTA(GradErrHandle,ProxFunc,xk,options)
2 |
3 | % Out = MiniscopeFISTA(GradErrHanle,ProxHandle,AxyTxy0,measurement,options)
4 | %
5 | % GradErrHandle: handle for function that computes error and gradient at
6 | % each step
7 | %
8 | % ProxFunc: handle for function that does projection step
9 | %
10 | % xk : initial guess for volume
11 | %
12 | % options : structure with optimization options.
13 | %
14 | % Nick Antipa and Kyrollos Yanny 2020
15 |
16 | if ~isfield(options,'convTol')
17 | options.convTol = 1e-9;
18 | end
19 | if ~isfield(options,'residTol')
20 | options.residTol = 1e-2;
21 | end
22 | if ~isfield(options,'xsize')
23 | options.xsize = size(A,2);
24 | end
25 | if ~isfield(options,'momentum')
26 | options.momentum = 'nesterov';
27 | end
28 | if ~isfield(options,'disp_figs')
29 | options.disp_figs = 0;
30 | end
31 | if ~isfield(options,'restarting')
32 | options.restarting = 0;
33 | end
34 | if ~isfield(options,'print_interval')
35 | options.print_interval = 1;
36 | end
37 | if ~isfield(options,'color_map')
38 | options.color_map = 'parula';
39 | end
40 | if ~isfield(options,'save_progress')
41 | options.save_progress = 0;
42 | end
43 | if ~isfield(options,'restart_interval')
44 | options.restart_interval = 0;
45 | end
46 | if ~isfield(options,'disp_crop')
47 | options.disp_crop = @(x)x;
48 | end
49 | if ~isfield(options,'disp_prctl')
50 | options.disp_prctl = 99.999;
51 | end
52 | if options.save_progress
53 | if ~isfield(options,'save_progress')
54 | options.progress_file = 'prox_progress.avi';
55 | end
56 | if exist(options.progress_file,'file')
57 | overwrite_mov = input('video file exists. Overwrite? y to overwrite, n to abort.');
58 | if strcmpi(overwrite_mov,'n')
59 | new_mov_name = input('input new name (no extension): ');
60 | options.progress_file = [new_mov_name,'.avi'];
61 | end
62 | end
63 | options.vidObj = VideoWriter(options.progress_file);
64 | open(options.vidObj);
65 | end
66 |
67 | % Initialize
68 | step_num = 0;
69 | fun_val = zeros([options.maxIter,1],'like',xk);
70 | tk = ones(1,'like',xk);
71 | yk = xk;
72 | f = 1e12*ones(1,'like',xk);
73 | tic
74 | while (step_num < options.maxIter) && (f>options.residTol)
75 |
76 | step_num = step_num+1;
77 | [f_kp1, g] = GradErrHandle(yk);
78 | fun_val(step_num) = f_kp1;
79 | [x_kp1, norm_x] = ProxFunc(yk-options.stepsize*g);
80 | fun_val(step_num) = fun_val(step_num)+norm_x; %To do: this should be tau*norm_x?
81 |
82 | % Compute momentum terms
83 | t_kp1 = (1+sqrt(1+4*tk^2))/2;
84 | beta_kp1 = (tk-1)/t_kp1;
85 | restart = (yk(:)-x_kp1(:))'*vec(x_kp1 - xk);
86 | yk = x_kp1+beta_kp1*(x_kp1 - xk);
87 |
88 | if step_num == 1
89 | if options.known_input
90 | fprintf('Iteration \t objective \t ||x|| \t momentum \t MSE \t PSNR\n');
91 | else
92 | fprintf('Iter\t ||Ax-b|| \t ||x|| \t Obj \t sparsity \t momentum \t elapsed time\n');
93 | end
94 | end
95 | if restart<0 && mod(step_num,options.restart_interval)==0
96 | fprintf('reached momentum reset interval\n')
97 | restart = Inf;
98 | end
99 |
100 | if ~mod(step_num,options.print_interval)
101 |
102 | if options.known_input
103 | fprintf('%i\t %6.4e\t %6.4e\t %.3f\t %6.4e\t %.2f dB\n',...
104 | step_num,f,norm_x,tk,...
105 | norm(options.xin(:) - yk(:)),...
106 | psnr(gather(yk),options.xin,255));
107 | else
108 | telapse = toc;
109 | fprintf('%i\t%6.4e\t%6.4e\t%6.4e\t%6.4e\t%.3f\t%.4f\n',...
110 | step_num,f,norm_x,fun_val(step_num),...
111 | nnz(x_kp1)/numel(x_kp1)*100,tk,telapse)
112 | end
113 | tic
114 | end
115 |
116 | if ~mod(step_num,options.disp_fig_interval)
117 | if options.disp_figs
118 | draw_figures(yk,options);
119 | end
120 |
121 | if options.save_progress
122 |
123 | frame = getframe(options.fighandle);
124 | writeVideo(options.vidObj,frame);
125 |
126 | end
127 | end
128 |
129 | if restart>0 && options.restarting
130 | tk = 1;
131 | fprintf('restarting momentum \n')
132 | yk = x_kp1;
133 | else
134 | tk = t_kp1;
135 | end
136 | xk = x_kp1;
137 | f = f_kp1;
138 | if abs(restart)=options.maxIter
151 | fprintf('Reached max number of iterations. Stopping. \n');
152 | end
153 |
154 | out = yk;
155 |
156 | if nargout>1
157 | varargout{1} = fun_val;
158 | end
159 |
160 | draw_figures(out,options)
161 |
162 | if options.save_progress
163 | close(options.vidObj);
164 | end
165 |
166 | return
167 |
168 |
169 | function draw_figures(xk,options)
170 | set(0,'CurrentFigure',options.fighandle)
171 | if numel(options.xsize)==2
172 | imagesc(options.disp_crop(xk))
173 | axis image
174 | colorbar
175 | colormap(options.color_map);
176 | %caxis(gather([prctile(xk(:),.1) prctile(xk(:),90)]))
177 | elseif numel(options.xsize)==3
178 | xk = gather(xk);
179 | set(0,'CurrentFigure',options.fighandle)
180 | subplot(1,3,1)
181 |
182 | im1 = squeeze(max(xk,[],3));
183 | imagesc(im1);
184 | hold on
185 | axis image
186 | colormap parula
187 | %colorbar
188 | caxis([0 prctile(im1(:),options.disp_prctl)])
189 | set(gca,'fontSize',6)
190 | axis off
191 | hold off
192 | set(0,'CurrentFigure',options.fighandle)
193 | subplot(1,3,2)
194 | im2 = squeeze(max(xk,[],1));
195 | imagesc(im2);
196 | hold on
197 | %axis image
198 | colormap parula
199 | %colorbar
200 | set(gca,'fontSize',8)
201 | caxis([0 prctile(im2(:),options.disp_prctl)])
202 | axis off
203 | hold off
204 | drawnow
205 | set(0,'CurrentFigure',options.fighandle)
206 | subplot(1,3,3)
207 | im3 = squeeze(max(xk,[],2));
208 | imagesc(im3);
209 | hold on
210 | %axis image
211 | colormap parula
212 | colorbar
213 | set(gca,'fontSize',8)
214 | caxis([0 prctile(im3(:),options.disp_prctl)]);
215 | axis off
216 | hold off
217 | elseif numel(options.xsize)==1
218 | plot(xk)
219 | end
220 | drawnow
221 |
222 |
--------------------------------------------------------------------------------
/3D reconstruction/Miniscope_shift_varying_recon.m:
--------------------------------------------------------------------------------
1 | % Read in PSF
2 | % This is designed to work with measurements in a folder, then in a
3 | % parallel folder, save the recons (i.e. measurements../recons/)
4 |
5 |
6 |
7 | %psf_path = 'D:\Kyrollos\RandoscopeNanoscribe\RandoscopeNanoscribe\Miniscope3D\psf_svd_12comps_23z_240xy_20190619';
8 | %psf_path = 'D:\Antipa\Randoscopev2_PSFs\Data_8_21_2019\SVD_2_5um_PSF_20um_1';
9 |
10 | psf_path = 'D:\Antipa\Randoscopev2_PSFs\20190912_recalibration\SVD_2p5_um_PSF_5um_1_green_channel';
11 | %psf_path = 'T:\Antipa\Randoscopev2_PSFs\20190912_recalibration\SVD_2p5_um_PSF_5um_1_green_channel';
12 | comps_path = [psf_path,'\SVD_2_5um_PSF_5um_1_ds2_components_green_SubAvg.mat'];
13 | weights_path = [psf_path,'\SVD_2_5um_PSF_5um_1_ds2_weights_interp_green_SubAvg.mat'];
14 |
15 | %%
16 | %comps_path = [psf_path,'/SVD_2_5um_PSF_5um_1_ds2_components_green_NoFro.mat'];
17 | %weights_path = [psf_path,'/SVD_2_5um_PSF_5um_1_ds2_weights_interp_green_NoFro.mat'];
18 | fprintf('loading components\n')
19 |
20 | h_in = load(comps_path);
21 | fprintf('done.\nLoading weights\n')
22 | weights_in = load(weights_path);
23 | fprintf('done loading PSF data\n')
24 | %%
25 |
26 | %Get names of files/paths
27 | [meas_name,data_path,~] = uigetfile('*.*','Select measurement','T:\Randoscope\RandoscopeV2_data');
28 | dots = strfind(meas_name,'.');
29 | fext = meas_name(dots(end):end);
30 | if strcmpi(fext,'.tif')
31 | [bg_name, bg_path,~] = uigetfile('*.*',['Select background for ',meas_name],fullfile([data_path,'../']));
32 | else
33 | bg_name = 'NONE';
34 | bg_path = 'NONE';
35 | end
36 |
37 | meas_path = [data_path,meas_name];
38 | bg_path = [bg_path,bg_name];
39 |
40 | if strcmpi(fext,'.tif')
41 | ome = strfind(meas_name,'.ome');
42 | bg_ome = strfind(bg_name,'.ome');
43 | meta_name = [meas_name(1:ome-1),'_metadata.txt'];
44 | bg_meta_name = [bg_name(1:bg_ome-1),'_metadata.txt'];
45 |
46 | % Inline function to open a metadata file, convert to characters then
47 | % decode (it's in json format).
48 | parse_json = @(x)jsondecode(transpose(fread(fopen(x),'*char')));
49 |
50 | % Construct full system paths to images
51 | meta_path = [data_path,meta_name];
52 | bg_meta_path = [bg_path,bg_meta_name];
53 |
54 |
55 | % Get json data from measurement and background images
56 |
57 | file_info = parse_json(meta_path);
58 | bg_info = parse_json(bg_meta_path);
59 |
60 | params.data_format = file_info.FrameKey_0_0_0.x50890959_DataFormat;
61 | params.bg_format = bg_info.FrameKey_0_0_0.x50890959_DataFormat;
62 | params.ds_raw = file_info.FrameKey_0_0_0.Binning;
63 | params.ds_bg = bg_info.FrameKey_0_0_0.Binning;
64 | elseif strcmpi(fext,'.mat')
65 | params.data_format = 'mat';
66 | params.bg_format = 'NONE';
67 | params.ds_raw = 4;
68 | mat_var_name = 'vid_bgrm_ds';
69 | meta_path = 'NONE';
70 | file_info='NONE';
71 | bg_info='NONE';
72 | end
73 |
74 |
75 |
76 |
77 | params.demosaic = contains(lower(params.data_format),'raw');
78 | params.demosaic_bg = contains(lower(params.bg_format),'raw');
79 | params.bg_name = bg_name;
80 | params.meas_path = meas_path;
81 | params.meta_path = meta_path;
82 | params.bg_path = bg_path;
83 | params.meas_info = file_info;
84 | params.bg_info = bg_info;
85 | %%
86 | %Waterbear_20190905\waterbear_big_lastone_20_3_30ms';
87 | %bg_path = 'D:\Randoscope\RandoscopeV2_data\Waterbear_20190905\waterbear_big_lastone_bck_20_3_30ms_1';
88 | %= 'waterbear_big_lastone_3_MMStack_Default.ome.tif';
89 | %bg_name = 'waterbear_big_lastone_bck_20_3_30ms_1_MMStack_Default.ome.tif';
90 |
91 |
92 |
93 |
94 |
95 | %for zd = 9
96 |
97 | %data_path = 'Z:\kyrollos\RandoscopeNanoscribe\Nanoscribe_pdms\Data_8_21_2019\real_res_target_10um_1'; %<--folder where the measurements are
98 | %bg_path = 'Z:\kyrollos\RandoscopeNanoscribe\Nanoscribe_pdms\Data_8_21_2019\bck_real_res_target_10um_1';
99 |
100 |
101 | params.data_tiff_format = 'time'; %Use 'time' if tiff stacks are at the same location over time, use 'z' if they are z stacks'
102 | params.tiff_color = 2; %use 'rgb' or 'mono'. Use number (1,2,3) for r,g, or b only
103 | params.meas_depth = 82; %If using 3D tiff or list of files, which slice was processed?
104 |
105 | params.ds_z = 1; %z downsampling ratio
106 | params.meas_bias = 0;
107 |
108 | params.ds = 4; % Global downsampling ratio (i.e.final image-to-sensor ratio)
109 | params.ds_psf = 2; %PSf downsample ratio (how much to further downsample -- if preprocessing included downsampling, use 1)
110 | params.ds_meas = params.ds/params.ds_raw; % How much to further downsample measurement?
111 | params.z_range = 1:44; %Must be even number!! Range of z slices to be solved for. If this is a scalar, 2D. Use this for subsampling z also (e.g. 1:4:... to do every 4th image)
112 | params.rank = 12;
113 | useGpu = 1; %cannot fit ds=2 on gpu unless we limit z range!!!!
114 | params.psf_norm = 'fro'; %Use max, slice, fro, or none
115 |
116 | %meas_name = ['real_res_target_10um_1_MMStack_Img_',num2str(params.meas_depth),'_000_000.ome.tif']; %<--- name of measurement
117 | %bg_name = ['bck_real_res_target_10um_1_MMStack_Img_',num2str(params.meas_depth),'_000_000.ome.tif'];
118 |
119 |
120 |
121 |
122 |
123 | % Make sure h and weights are in order y,x,z,rank
124 | fprintf('permuting PSF data\n')
125 | h = permute(h_in.comps_out(:,:,1:params.rank,params.z_range),[1,2,4,3]);
126 | weights = permute(weights_in.weights_out(:,:,1:params.rank,params.z_range),[1,2,4,3]);
127 | fprintf('Done permuting. Resampling PSF\n');
128 |
129 | %clear h_in;
130 | %clear weights_in;
131 | h = single(imresize(squeeze(h),1/params.ds_psf,'box'));
132 | weights = single(imresize(squeeze(weights),1/params.ds_psf,'box'));
133 |
134 | % Normalize weights to have maximum sum through rank of 1
135 | weights_norm = max(sum(weights(size(weights,1)/2,size(weights,2)/2,:,:),4),[],3);
136 | weights = weights/weights_norm;
137 | fprintf('Done. PSF ready!\n')
138 | %clear h_permute;
139 | %clear weights_permute;
140 |
141 | %%
142 | switch lower(params.psf_norm)
143 | case('max')
144 | h = h/max(h(:));
145 | case('none')
146 | case('fro')
147 | h = h/norm(vec(h));
148 | case('slice')
149 | for sl = 1:Nz
150 | slice_norm = norm(h(:,:,sl,1),'fro');
151 | for cp = 1:Nr
152 | h(:,:,sl,cp) = h(:,:,sl,cp)/slice_norm;
153 | end
154 | end
155 | end
156 |
157 | H = fft2(ifftshift(ifftshift(h,1),2));
158 | Hconj = conj(H);
159 | if useGpu
160 | H = gpuArray(H);
161 | Hconj = gpuArray(Hconj);
162 | weights = gpuArray(weights);
163 | end
164 |
165 |
166 | % Read in data
167 |
168 | %%
169 | init_style = 'loaded'; %Use 'loaded' to load initialization, 'zeros' to start from scratch. Admm will run 2D deconv, then replicate result to all time points
170 | im_tag = 'rank1_soft_of_TV_TEST';
171 | if strcmpi(params.data_format,'mat')
172 | data_in = load(meas_path,mat_var_name);
173 | data_in = data_in.(mat_var_name);
174 | end
175 |
176 |
177 | for meas_slice = 1:33
178 |
179 | params.meas_slice = meas_slice; %Slices to load from tiff stack. If 'all' used, it will average.
180 | %params.meas_slice = 'all';
181 | if ~strcmpi(params.data_format,'mat')
182 | switch lower(params.data_tiff_format)
183 | case('z')
184 | data_raw = double(read_tiff_stack(meas_path,params.ds_meas,params.meas_depth));
185 | bg_in = double(read_tiff_stack(bg_path,params.ds_meas,params.meas_depth));
186 | case('time')
187 | bg_raw = read_tiff_stack(bg_path,1);
188 | if strcmpi(params.meas_slice,'all')
189 | data_raw = mean(double(read_tiff_stack(meas_path,1)),4); %Average out the time variable
190 | if params.demosaic
191 | data_demos = imresize(double(demosaic(uint16(data_in),'grbg')),params.ds_raw/params.ds,'box');
192 | else
193 | data_demos = imresize(data_raw,params.ds_raw/params.ds,'box');
194 | end
195 | else
196 | data_in = read_tiff_stack(meas_path,1,params.meas_slice);
197 |
198 |
199 | if params.demosaic
200 | data_demos = imresize(double(demosaic(uint16(data_in),'grbg')),params.ds_raw/params.ds,'box');
201 | %grbg
202 | else
203 | data_demos = imresize(mean(data_in,4),params.ds_raw/params.ds,'box');
204 | end
205 | end
206 | if params.demosaic_bg
207 | bg_in = imresize(double(demosaic(uint16(mean(bg_raw,3)),'grbg')),params.ds_bg/params.ds,'box');
208 | else
209 | bg_in = imresize(mean(double(bg_raw),4),params.ds_bg/params.ds,'box');
210 | end
211 |
212 | % data_raw = data_raw(:,:,:,1);
213 |
214 | end
215 |
216 | if strcmpi(params.tiff_color,'rgb')
217 | data = mean(data_demos,3);
218 | bg = mean(bg_in,3); %Average out color. Change to (:,:,color) to select one channel
219 | elseif isnumeric(params.tiff_color)
220 | data = data_demos(:,:,params.tiff_color);
221 | bg = bg_in(:,:,params.tiff_color);
222 | end
223 | else
224 | data = data_in(:,:,meas_slice);
225 | bg = 0;
226 | end
227 |
228 |
229 | data = data - bg - params.meas_bias;
230 | b = data/max(data(:));
231 |
232 |
233 | % data_r = data_in(:,:,1);
234 | % data_g = data_in(:,:,2);
235 | % data_b = data_in(:,:,3);
236 |
237 |
238 | %Nx = size(h,2);
239 | %Ny = size(h,1);
240 | if numel(size(h)) == 3
241 | [Ny, Nx, Nr] = size(h);
242 | Nz = 1;
243 | else
244 | [Ny, Nx, Nz, Nr] = size(h);
245 | end
246 |
247 | %define crop and pad operators to handle 2D fft convolution
248 | pad2d = @(x)padarray(x,[size(h,1)/2,size(h,2)/2],0,'both');
249 | ccL = size(h,2)/2+1;
250 | ccU = 3*size(h,2)/2;
251 | rcL = size(h,1)/2+1;
252 | rcU = 3*size(h,1)/2;
253 |
254 | %cc = gpuArray((size(h,2)/2+1):(3*size(h,2)/2));
255 | %rc = gpuArray((size(h,1)/2+1):(3*size(h,1)/2));
256 | crop2d = @(x)x(rcL:rcU,ccL:ccU);
257 |
258 |
259 |
260 |
261 |
262 |
263 |
264 | if strcmpi(init_style, 'zeros')
265 | xinit = zeros(Ny, Nx, Nz);
266 | elseif strcmpi(init_style,'loaded')
267 | if ~exist('xinit')
268 | xinit = zeros(Ny,Nx,Nz);
269 | else
270 | xinit = xhat_out(:,:,:);
271 | end
272 | elseif strcmpi(init_style,'admm')
273 | xinit_2d = gpuArray(single(zeros(Ny, Nx, 3)));
274 |
275 | for n = 1:3
276 | xinit_2d(:,:,n) = admm2d_solver(gpuArray(single(b(:,:,n))), gpuArray(single(h(:,:,n))),[],.001);
277 |
278 | imagesc(2*xinit_2d/max(xinit_2d(:)))
279 | end
280 | end
281 |
282 |
283 |
284 |
285 |
286 |
287 |
288 | options.color_map = 'parula';
289 |
290 |
291 |
292 | options.convTol = 15e-4;
293 |
294 | %options.xsize = [256,256];
295 | options.maxIter = 2000;
296 | options.residTol = 5e-5;
297 | options.momentum = 'nesterov';
298 | options.disp_figs = 1;
299 | options.disp_fig_interval = 40; %display image this often
300 | if Nz == 1
301 | options.xsize = [Ny, Nx];
302 | else
303 | options.xsize=[Ny, Nx, Nz];
304 | end
305 | options.print_interval = 20;
306 |
307 | figure(2)
308 | clf
309 | imagesc(b)
310 | axis image
311 |
312 | h1 = figure(1);
313 | clf
314 | options.fighandle = h1;
315 | nocrop = @(x)x;
316 | options.known_input = 0;
317 |
318 |
319 |
320 |
321 |
322 | large = 0;
323 | if Nz > 1
324 | if large == 0
325 | A = @(x)A_svd_3d(x, weights,H);
326 |
327 | Aadj = @(y)A_adj_svd_3d(y, weights, Hconj);
328 | else
329 | weights=gpuArray(weights);
330 | H = gpuArray(H);
331 | Hconj = gpuArray(Hconj);
332 | b = gpuArray(single(b));
333 | A = @(x)A_svd_3d_large(x,weights,H);
334 | Aadj = @(y)A_adj_svd_3d_large(y, weights, Hconj);
335 | end
336 | elseif Nz == 1
337 | A = @(x)A_svd(H, weights, x, nocrop);
338 | Aadj = @(y)A_adj_svd(Hconj,weights,y,nocrop);
339 | end
340 |
341 |
342 |
343 |
344 | %options.stepsize = .1e-3; for ds=4
345 | if params.ds == 4
346 | if strcmpi(params.psf_norm ,'fro')
347 | if Nz == 18
348 | options.stepsize = 3e-3;
349 | elseif Nz == 12
350 | options.stepsize = .4e-2;
351 | fprintf('foo\n')
352 | elseif Nz == 14
353 | options.stepsize = 4e-3;
354 | elseif Nz == 20
355 | if params.rank == 12
356 | options.stepsize = 3e-3;
357 | elseif params.rank == 8
358 | options.stepsize = 1e-3;
359 | elseif params.rank == 18
360 | options.stepsize = 4e-3;
361 |
362 |
363 | end
364 | elseif Nz>20
365 | options.stepsize = .014; %015 is nice?
366 | end
367 |
368 | else
369 | options.stepsize = 3e-6;
370 | end
371 |
372 | elseif params.ds == 2
373 | options.stepsize = 0.7e-3;
374 | end
375 | %.3e-4 is good for waterbears
376 | params.tau1 = options.stepsize*.3e-3; %was 0.5e-7 %.000005 works pretty well for v1 camera, .0002 for v2
377 | params.tau_soft = options.stepsize * 3e-3;
378 | tau_iso = (.25e-4);
379 | params.z_tv_weight = 1; %z weighting in anisotropic TV
380 | tau2 = .001 %Auxilliary
381 | TVnorm3d = @(x)sum(sum(sum(abs(x))));
382 |
383 |
384 | if useGpu
385 |
386 | grad_handle = @(x)linear_gradient_b(x, A, Aadj, gpuArray(single(b)));
387 |
388 | params.tau1 = gpuArray(params.tau1);
389 | params.tau_soft = gpuArray(params.tau_soft);
390 | tau_iso = gpuArray(tau_iso);
391 | params.z_tv_weight = gpuArray(params.z_tv_weight);
392 | options.stepsize = gpuArray(options.stepsize);
393 |
394 | else
395 | if ~large
396 | grad_handle = @(x)linear_gradient_b(x, A, Aadj, single(b));
397 | else
398 | grad_handle = @(x)linear_gradient_large(x,A,Aadj,gpuArray(single(b)));
399 | end
400 |
401 | end
402 |
403 | %Prox
404 | %prox_handle = @(x)deal(x.*(x>=0), abs(sum(sum(sum(x(x<0))))));
405 |
406 |
407 |
408 | %prox_handle = @(x)deal(1/3*(x.*(x>=0) + soft(x, tau2) + tv3dApproxHaar(x, params.tau1)), TVnorm3d(x));
409 |
410 |
411 |
412 |
413 | if ~strcmpi(params.data_format,'mat')
414 | if Nz>1
415 | prox_handle = @(x)deal(1/2*(max(x,0) + (tv3d_iso_Haar((x), params.tau1, params.z_tv_weight))), params.tau1*TVnorm3d(x));
416 | elseif Nz == 1
417 | prox_handle = @(x)deal(.5*tv2d_aniso_haar(x,params.tau1*options.stepsize) + ...
418 | .5*max(x,0), params.tau1*options.stepsize*TVnorm(x));
419 | end
420 | else
421 | % prox_handle = @(x)deal(.5*(soft(x,params.tau_soft) +...
422 | % tv3d_iso_Haar(x, params.tau1, params.z_tv_weight)), ...
423 | % params.tau1*TVnorm3d(x));
424 | prox_handle = @(x)deal(soft(tv3d_iso_Haar(x, params.tau1, params.z_tv_weight),params.tau_soft), ...
425 | params.tau1*TVnorm3d(x));
426 | %prox_handle=@(x)deal(soft(x,params.tau_soft),params.tau_soft*sum(abs(vec(x))));
427 | end
428 | TVpars.epsilon = 1e-7;
429 | TVpars.MAXITER = 100;
430 | TVpars.alpha = .3;
431 | %prox_handle = @(x)deal(hsvid_TV3DFista(x, tau_iso, 0, 10, TVpars) , hsvid_TVnorm3d(x));
432 |
433 | if strcmpi(init_style, 'zeros')
434 | xinit = zeros(Ny, Nx, Nz);
435 |
436 | end
437 |
438 | if useGpu
439 |
440 | TVpars.epsilon = gpuArray(TVpars.epsilon);
441 | TVpars.MAXITER = gpuArray(TVpars.MAXITER);
442 | TVpars.alpha = gpuArray(TVpars.alpha);
443 | xinit = gpuArray(single(xinit));
444 | success = false;
445 | while success == false %This shouldn't be necessary, but it deals with restarting when GPU runs OOM
446 | try
447 | [xhat, f2] = MiniscopeFISTA(grad_handle,prox_handle,xinit,options);
448 | success = true;
449 | catch
450 | success = false;
451 | end
452 | end
453 |
454 | else
455 | if large
456 | xinit = gpuArray(xinit);
457 | end
458 | [xhat, f2] = MiniscopeFISTA(grad_handle,prox_handle,xinit,options);
459 | end
460 |
461 |
462 |
463 |
464 |
465 |
466 |
467 | datestamp = datetime;
468 | tiff_string = sprintf('%03d',meas_slice);
469 | date_string = datestr(datestamp,'yyyy-mmm-dd_HHMMSS');
470 | save_str = ['../recons/',date_string,'_',meas_name(1:end-4),'_',im_tag,'_',tiff_string];
471 | full_path = fullfile(data_path,save_str);
472 | mkdir(full_path);
473 |
474 |
475 |
476 | imout = gather(xhat/prctile(xhat(:),100*(numel(xhat)-10)/numel(xhat))); %Saturate only 10 pixels
477 | xhat_out = gather(xhat);
478 |
479 | params.tau1 = gather(params.tau1);
480 | params.tau_soft = gather(params.tau_soft);
481 | imbase = meas_name(1:end-4);
482 | mkdir([full_path, '/png/']);
483 | filebase = [full_path, '/png/', imbase];
484 | f_out = gather(f2);
485 | out_names = {};
486 | for n= 1:size(imout,3)
487 | out_names{n} = [filebase,'_',sprintf('Z_%.3i_T_',params.z_range(n)),...
488 | tiff_string,'_',im_tag,'.png'];
489 | imwrite(imout(:,:,n),out_names{n});
490 | fprintf('writing image %i of %i\n',n,size(xhat,3))
491 | end
492 |
493 | fprintf('zipping...\n')
494 | zip([full_path, '/png/', imbase],out_names)
495 | fprintf('done zipping\n')
496 |
497 | fprintf('writing .mat\n')
498 | options.fighandle = []
499 |
500 | save([full_path,'/',meas_name(1:end-4),'_',date_string,'_',im_tag,'_',tiff_string,'.mat'], 'tau_iso','TVpars','xhat_out', 'options', 'comps_path','weights_path', 'b','params')
501 | fprintf('done writing .mat\n')
502 | % gpuDevice(1)
503 | clear xhat
504 | clear f2
505 |
506 |
507 | if params.ds == 2
508 | gpuDevice(1)
509 | end
510 | end
511 | %end
512 |
513 | %%
514 | % imagesc(brain_recon.xhat(:,:,10))
515 | % axis image
--------------------------------------------------------------------------------
/3D reconstruction/Miniscope_3d_shift_varying_main.m:
--------------------------------------------------------------------------------
1 | % Read in PSF
2 | % This is designed to work with measurements in a folder, then in a
3 | % parallel folder, save the recons (i.e. measurements../recons/)
4 |
5 |
6 |
7 | %psf_path = 'D:\Kyrollos\RandoscopeNanoscribe\RandoscopeNanoscribe\Miniscope3D\psf_svd_12comps_23z_240xy_20190619';
8 | %psf_path = 'D:\Antipa\Randoscopev2_PSFs\Data_8_21_2019\SVD_2_5um_PSF_20um_1';
9 |
10 | psf_path = 'D:\Antipa\Randoscopev2_PSFs\20190912_recalibration\SVD_2p5_um_PSF_5um_1_green_channel';
11 | %psf_path = 'T:\Antipa\Randoscopev2_PSFs\20190912_recalibration\SVD_2p5_um_PSF_5um_1_green_channel';
12 | comps_path = [psf_path,'\SVD_2_5um_PSF_5um_1_ds2_components_green_SubAvg.mat'];
13 | weights_path = [psf_path,'\SVD_2_5um_PSF_5um_1_ds2_weights_interp_green_SubAvg.mat'];
14 |
15 | %%
16 | %comps_path = [psf_path,'/SVD_2_5um_PSF_5um_1_ds2_components_green_NoFro.mat'];
17 | %weights_path = [psf_path,'/SVD_2_5um_PSF_5um_1_ds2_weights_interp_green_NoFro.mat'];
18 | fprintf('loading components\n')
19 |
20 | h_in = load(comps_path);
21 | fprintf('done.\nLoading weights\n')
22 | weights_in = load(weights_path);
23 | fprintf('done loading PSF data\n')
24 |
25 |
26 | %%
27 |
28 | %Get names of files/paths
29 | [meas_name,data_path,~] = uigetfile('*.*','Select measurement','T:\Randoscope\RandoscopeV2_data');
30 | dots = strfind(meas_name,'.');
31 | fext = meas_name(dots(end):end);
32 | if strcmpi(fext,'.tif')
33 | [bg_name, bg_path,~] = uigetfile('*.*',['Select background for ',meas_name],fullfile([data_path,'../']));
34 | else
35 | bg_name = 'NONE';
36 | bg_path = 'NONE';
37 | end
38 |
39 |
40 |
41 |
42 |
43 |
44 | meas_path = [data_path,meas_name];
45 | bg_path = [bg_path,bg_name];
46 |
47 | if strcmpi(fext,'.tif')
48 | ome = strfind(meas_name,'.ome');
49 | bg_ome = strfind(bg_name,'.ome');
50 | meta_name = [meas_name(1:ome-1),'_metadata.txt'];
51 | bg_meta_name = [bg_name(1:bg_ome-1),'_metadata.txt'];
52 |
53 | % Inline function to open a metadata file, convert to characters then
54 | % decode (it's in json format).
55 | parse_json = @(x)jsondecode(transpose(fread(fopen(x),'*char')));
56 |
57 | % Construct full system paths to images
58 | meta_path = [data_path,meta_name];
59 | bg_meta_path = [bg_path,bg_meta_name];
60 |
61 |
62 | % Get json data from measurement and background images
63 |
64 | file_info = parse_json(meta_path);
65 | bg_info = parse_json(bg_meta_path);
66 |
67 | params.data_format = file_info.FrameKey_0_0_0.x50890959_DataFormat;
68 | params.bg_format = bg_info.FrameKey_0_0_0.x50890959_DataFormat;
69 | params.ds_raw = file_info.FrameKey_0_0_0.Binning;
70 | params.ds_bg = bg_info.FrameKey_0_0_0.Binning;
71 | elseif strcmpi(fext,'.mat')
72 | params.data_format = 'mat';
73 | params.bg_format = 'NONE';
74 | params.ds_raw = 4;
75 | mat_var_name = 'vid_bgrm_ds';
76 | meta_path = 'NONE';
77 | file_info='NONE';
78 | bg_info='NONE';
79 |
80 | end
81 |
82 |
83 |
84 |
85 | params.demosaic = contains(lower(params.data_format),'raw');
86 | params.demosaic_bg = contains(lower(params.bg_format),'raw');
87 | params.bg_name = bg_name;
88 | params.meas_path = meas_path;
89 | params.meta_path = meta_path;
90 | params.bg_path = bg_path;
91 | params.meas_info = file_info;
92 | params.bg_info = bg_info;
93 | %%
94 | %Waterbear_20190905\waterbear_big_lastone_20_3_30ms';
95 | %bg_path = 'D:\Randoscope\RandoscopeV2_data\Waterbear_20190905\waterbear_big_lastone_bck_20_3_30ms_1';
96 | %= 'waterbear_big_lastone_3_MMStack_Default.ome.tif';
97 | %bg_name = 'waterbear_big_lastone_bck_20_3_30ms_1_MMStack_Default.ome.tif';
98 |
99 |
100 |
101 |
102 |
103 | %for zd = 9
104 |
105 | %data_path = 'Z:\kyrollos\RandoscopeNanoscribe\Nanoscribe_pdms\Data_8_21_2019\real_res_target_10um_1'; %<--folder where the measurements are
106 | %bg_path = 'Z:\kyrollos\RandoscopeNanoscribe\Nanoscribe_pdms\Data_8_21_2019\bck_real_res_target_10um_1';
107 |
108 |
109 | params.data_tiff_format = 'time'; %Use 'time' if tiff stacks are at the same location over time, use 'z' if they are z stacks'
110 | params.tiff_color = 2; %use 'rgb' or 'mono'. Use number (1,2,3) for r,g, or b only
111 | params.meas_depth = 82; %If using 3D tiff or list of files, which slice was processed?
112 |
113 | params.ds_z = 1; %z downsampling ratio
114 | params.meas_bias = 0;
115 |
116 | params.ds = 4; % Global downsampling ratio (i.e.final image-to-sensor ratio)
117 | params.ds_psf = 2; %PSf downsample ratio (how much to further downsample -- if preprocessing included downsampling, use 1)
118 | params.ds_meas = params.ds/params.ds_raw; % How much to further downsample measurement?
119 | params.z_range = 1:44; %Must be even number!! Range of z slices to be solved for. If this is a scalar, 2D. Use this for subsampling z also (e.g. 1:4:... to do every 4th image)
120 | params.rank = 12;
121 | useGpu = 1; %cannot fit ds=2 on gpu unless we limit z range!!!!
122 | params.psf_norm = 'fro'; %Use max, slice, fro, or none
123 |
124 | %meas_name = ['real_res_target_10um_1_MMStack_Img_',num2str(params.meas_depth),'_000_000.ome.tif']; %<--- name of measurement
125 | %bg_name = ['bck_real_res_target_10um_1_MMStack_Img_',num2str(params.meas_depth),'_000_000.ome.tif'];
126 |
127 |
128 |
129 |
130 |
131 | % Make sure h and weights are in order y,x,z,rank
132 | fprintf('permuting PSF data\n')
133 | h = permute(h_in.comps_out(:,:,1:params.rank,params.z_range),[1,2,4,3]);
134 | weights = permute(weights_in.weights_out(:,:,1:params.rank,params.z_range),[1,2,4,3]);
135 | fprintf('Done permuting. Resampling PSF\n');
136 |
137 | %clear h_in;
138 | %clear weights_in;
139 | h = single(imresize(squeeze(h),1/params.ds_psf,'box'));
140 | weights = single(imresize(squeeze(weights),1/params.ds_psf,'box'));
141 |
142 | % Normalize weights to have maximum sum through rank of 1
143 | weights_norm = max(sum(weights(size(weights,1)/2,size(weights,2)/2,:,:),4),[],3);
144 | weights = weights/weights_norm;
145 | fprintf('Done. PSF ready!\n')
146 | %clear h_permute;
147 | %clear weights_permute;
148 |
149 | %%
150 | switch lower(params.psf_norm)
151 | case('max')
152 | h = h/max(h(:));
153 | case('none')
154 | case('fro')
155 | h = h/norm(vec(h));
156 | case('slice')
157 | for sl = 1:Nz
158 | slice_norm = norm(h(:,:,sl,1),'fro');
159 | for cp = 1:Nr
160 | h(:,:,sl,cp) = h(:,:,sl,cp)/slice_norm;
161 | end
162 | end
163 | end
164 |
165 | H = fft2(ifftshift(ifftshift(h,1),2));
166 | Hconj = conj(H);
167 | if useGpu
168 | H = gpuArray(H);
169 | Hconj = gpuArray(Hconj);
170 | weights = gpuArray(weights);
171 | end
172 |
173 |
174 | % Read in data
175 |
176 | %%
177 | init_style = 'zeros'; %Use 'loaded' to load initialization, 'zeros' to start from scratch. Admm will run 2D deconv, then replicate result to all time points
178 | im_tag = 'rank1_soft_of_TV_TEST';
179 | if strcmpi(params.data_format,'mat')
180 | data_in = load(meas_path,mat_var_name);
181 | data_in = data_in.(mat_var_name);
182 | end
183 |
184 |
185 | for meas_slice = 1:33
186 |
187 | params.meas_slice = meas_slice; %Slices to load from tiff stack. If 'all' used, it will average.
188 | %params.meas_slice = 'all';
189 | if ~strcmpi(params.data_format,'mat')
190 | switch lower(params.data_tiff_format)
191 | case('z')
192 | data_raw = double(read_tiff_stack(meas_path,params.ds_meas,params.meas_depth));
193 | bg_in = double(read_tiff_stack(bg_path,params.ds_meas,params.meas_depth));
194 | case('time')
195 | bg_raw = read_tiff_stack(bg_path,1);
196 | if strcmpi(params.meas_slice,'all')
197 | data_raw = mean(double(read_tiff_stack(meas_path,1)),4); %Average out the time variable
198 | if params.demosaic
199 | data_demos = imresize(double(demosaic(uint16(data_in),'grbg')),params.ds_raw/params.ds,'box');
200 | else
201 | data_demos = imresize(data_raw,params.ds_raw/params.ds,'box');
202 | end
203 | else
204 | data_in = read_tiff_stack(meas_path,1,params.meas_slice);
205 |
206 |
207 | if params.demosaic
208 | data_demos = imresize(double(demosaic(uint16(data_in),'grbg')),params.ds_raw/params.ds,'box');
209 | %grbg
210 | else
211 | data_demos = imresize(mean(data_in,4),params.ds_raw/params.ds,'box');
212 | end
213 | end
214 | if params.demosaic_bg
215 | bg_in = imresize(double(demosaic(uint16(mean(bg_raw,3)),'grbg')),params.ds_bg/params.ds,'box');
216 | else
217 | bg_in = imresize(mean(double(bg_raw),4),params.ds_bg/params.ds,'box');
218 | end
219 |
220 | % data_raw = data_raw(:,:,:,1);
221 |
222 | end
223 |
224 | if strcmpi(params.tiff_color,'rgb')
225 | data = mean(data_demos,3);
226 | bg = mean(bg_in,3); %Average out color. Change to (:,:,color) to select one channel
227 | elseif isnumeric(params.tiff_color)
228 | data = data_demos(:,:,params.tiff_color);
229 | bg = bg_in(:,:,params.tiff_color);
230 | end
231 | else
232 | data = data_in(:,:,meas_slice);
233 | bg = 0;
234 | end
235 |
236 |
237 | data = data - bg - params.meas_bias;
238 | b = data/max(data(:));
239 |
240 |
241 | % data_r = data_in(:,:,1);
242 | % data_g = data_in(:,:,2);
243 | % data_b = data_in(:,:,3);
244 |
245 |
246 | %Nx = size(h,2);
247 | %Ny = size(h,1);
248 | if numel(size(h)) == 3
249 | [Ny, Nx, Nr] = size(h);
250 | Nz = 1;
251 | else
252 | [Ny, Nx, Nz, Nr] = size(h);
253 | end
254 |
255 | %define crop and pad operators to handle 2D fft convolution
256 | pad2d = @(x)padarray(x,[size(h,1)/2,size(h,2)/2],0,'both');
257 | ccL = size(h,2)/2+1;
258 | ccU = 3*size(h,2)/2;
259 | rcL = size(h,1)/2+1;
260 | rcU = 3*size(h,1)/2;
261 |
262 | %cc = gpuArray((size(h,2)/2+1):(3*size(h,2)/2));
263 | %rc = gpuArray((size(h,1)/2+1):(3*size(h,1)/2));
264 | crop2d = @(x)x(rcL:rcU,ccL:ccU);
265 |
266 |
267 |
268 |
269 |
270 |
271 |
272 | if strcmpi(init_style, 'zeros')
273 | xinit = zeros(Ny, Nx, Nz);
274 | elseif strcmpi(init_style,'loaded')
275 | if ~exist('xinit')
276 | xinit = zeros(Ny,Nx,Nz);
277 | else
278 | xinit = xhat_out(:,:,:);
279 | end
280 | elseif strcmpi(init_style,'admm')
281 | xinit_2d = gpuArray(single(zeros(Ny, Nx, 3)));
282 |
283 | for n = 1:3
284 | xinit_2d(:,:,n) = admm2d_solver(gpuArray(single(b(:,:,n))), gpuArray(single(h(:,:,n))),[],.001);
285 |
286 | imagesc(2*xinit_2d/max(xinit_2d(:)))
287 | end
288 | end
289 |
290 |
291 |
292 |
293 |
294 |
295 |
296 | options.color_map = 'parula';
297 |
298 |
299 |
300 | options.convTol = 15e-4;
301 |
302 | %options.xsize = [256,256];
303 | options.maxIter = 2000;
304 | options.residTol = 5e-5;
305 | options.momentum = 'nesterov';
306 | options.disp_figs = 1;
307 | options.disp_fig_interval = 40; %display image this often
308 | if Nz == 1
309 | options.xsize = [Ny, Nx];
310 | else
311 | options.xsize=[Ny, Nx, Nz];
312 | end
313 | options.print_interval = 20;
314 |
315 | figure(2)
316 | clf
317 | imagesc(b)
318 | axis image
319 |
320 | h1 = figure(1);
321 | clf
322 | options.fighandle = h1;
323 | nocrop = @(x)x;
324 | options.known_input = 0;
325 |
326 |
327 |
328 |
329 |
330 | large = 0;
331 | if Nz > 1
332 | if large == 0
333 | A = @(x)A_svd_3d(x, weights,H);
334 |
335 | Aadj = @(y)A_adj_svd_3d(y, weights, Hconj);
336 | else
337 | weights=gpuArray(weights);
338 | H = gpuArray(H);
339 | Hconj = gpuArray(Hconj);
340 | b = gpuArray(single(b));
341 | A = @(x)A_svd_3d_large(x,weights,H);
342 | Aadj = @(y)A_adj_svd_3d_large(y, weights, Hconj);
343 | end
344 | elseif Nz == 1
345 | A = @(x)A_svd(H, weights, x, nocrop);
346 | Aadj = @(y)A_adj_svd(Hconj,weights,y,nocrop);
347 | end
348 |
349 |
350 |
351 |
352 | %options.stepsize = .1e-3; for ds=4
353 | if params.ds == 4
354 | if strcmpi(params.psf_norm ,'fro')
355 | if Nz == 18
356 | options.stepsize = 3e-3;
357 | elseif Nz == 12
358 | options.stepsize = .4e-2;
359 | fprintf('foo\n')
360 | elseif Nz == 14
361 | options.stepsize = 4e-3;
362 | elseif Nz == 20
363 | if params.rank == 12
364 | options.stepsize = 3e-3;
365 | elseif params.rank == 8
366 | options.stepsize = 1e-3;
367 | elseif params.rank == 18
368 | options.stepsize = 4e-3;
369 |
370 |
371 | end
372 | elseif Nz>20
373 | options.stepsize = .014; %015 is nice?
374 | end
375 |
376 | else
377 | options.stepsize = 3e-6;
378 | end
379 |
380 | elseif params.ds == 2
381 | options.stepsize = 0.7e-3;
382 | end
383 | %.3e-4 is good for waterbears
384 | params.tau1 = options.stepsize*.3e-3; %was 0.5e-7 %.000005 works pretty well for v1 camera, .0002 for v2
385 | params.tau_soft = options.stepsize * 3e-3;
386 | tau_iso = (.25e-4);
387 | params.z_tv_weight = 1; %z weighting in anisotropic TV
388 | tau2 = .001 %Auxilliary
389 | TVnorm3d = @(x)sum(sum(sum(abs(x))));
390 |
391 |
392 | if useGpu
393 |
394 | grad_handle = @(x)linear_gradient_b(x, A, Aadj, gpuArray(single(b)));
395 |
396 | params.tau1 = gpuArray(params.tau1);
397 | params.tau_soft = gpuArray(params.tau_soft);
398 | tau_iso = gpuArray(tau_iso);
399 | params.z_tv_weight = gpuArray(params.z_tv_weight);
400 | options.stepsize = gpuArray(options.stepsize);
401 |
402 | else
403 | if ~large
404 | grad_handle = @(x)linear_gradient_b(x, A, Aadj, single(b));
405 | else
406 | grad_handle = @(x)linear_gradient_large(x,A,Aadj,gpuArray(single(b)));
407 | end
408 |
409 | end
410 |
411 | %Prox
412 | %prox_handle = @(x)deal(x.*(x>=0), abs(sum(sum(sum(x(x<0))))));
413 |
414 |
415 |
416 | %prox_handle = @(x)deal(1/3*(x.*(x>=0) + soft(x, tau2) + tv3dApproxHaar(x, params.tau1)), TVnorm3d(x));
417 |
418 |
419 |
420 |
421 | if ~strcmpi(params.data_format,'mat')
422 | if Nz>1
423 | prox_handle = @(x)deal(1/2*(max(x,0) + (tv3d_iso_Haar((x), params.tau1, params.z_tv_weight))), params.tau1*TVnorm3d(x));
424 | elseif Nz == 1
425 | prox_handle = @(x)deal(.5*tv2d_aniso_haar(x,params.tau1*options.stepsize) + ...
426 | .5*max(x,0), params.tau1*options.stepsize*TVnorm(x));
427 | end
428 | else
429 | % prox_handle = @(x)deal(.5*(soft(x,params.tau_soft) +...
430 | % tv3d_iso_Haar(x, params.tau1, params.z_tv_weight)), ...
431 | % params.tau1*TVnorm3d(x));
432 | prox_handle = @(x)deal(soft(tv3d_iso_Haar(x, params.tau1, params.z_tv_weight),params.tau_soft), ...
433 | params.tau1*TVnorm3d(x));
434 | %prox_handle=@(x)deal(soft(x,params.tau_soft),params.tau_soft*sum(abs(vec(x))));
435 | end
436 | TVpars.epsilon = 1e-7;
437 | TVpars.MAXITER = 100;
438 | TVpars.alpha = .3;
439 | %prox_handle = @(x)deal(hsvid_TV3DFista(x, tau_iso, 0, 10, TVpars) , hsvid_TVnorm3d(x));
440 |
441 | if strcmpi(init_style, 'zeros')
442 | xinit = zeros(Ny, Nx, Nz);
443 |
444 | end
445 |
446 | if useGpu
447 |
448 | TVpars.epsilon = gpuArray(TVpars.epsilon);
449 | TVpars.MAXITER = gpuArray(TVpars.MAXITER);
450 | TVpars.alpha = gpuArray(TVpars.alpha);
451 | xinit = gpuArray(single(xinit));
452 | success = false;
453 | while success == false %This shouldn't be necessary, but it deals with restarting when GPU runs OOM
454 | try
455 | [xhat, f2] = MiniscopeFISTA(grad_handle,prox_handle,xinit,options);
456 | success = true;
457 | catch
458 | success = false;
459 | end
460 | end
461 |
462 | else
463 | if large
464 | xinit = gpuArray(xinit);
465 | end
466 | [xhat, f2] = MiniscopeFISTA(grad_handle,prox_handle,xinit,options);
467 | end
468 |
469 |
470 |
471 |
472 |
473 |
474 |
475 | datestamp = datetime;
476 | tiff_string = sprintf('%03d',meas_slice);
477 | date_string = datestr(datestamp,'yyyy-mmm-dd_HHMMSS');
478 | save_str = ['../recons/',date_string,'_',meas_name(1:end-4),'_',im_tag,'_',tiff_string];
479 | full_path = fullfile(data_path,save_str);
480 | mkdir(full_path);
481 |
482 |
483 |
484 | imout = gather(xhat/prctile(xhat(:),100*(numel(xhat)-10)/numel(xhat))); %Saturate only 10 pixels
485 | xhat_out = gather(xhat);
486 |
487 | params.tau1 = gather(params.tau1);
488 | params.tau_soft = gather(params.tau_soft);
489 | imbase = meas_name(1:end-4);
490 | mkdir([full_path, '/png/']);
491 | filebase = [full_path, '/png/', imbase];
492 | f_out = gather(f2);
493 | out_names = {};
494 | for n= 1:size(imout,3)
495 | out_names{n} = [filebase,'_',sprintf('Z_%.3i_T_',params.z_range(n)),...
496 | tiff_string,'_',im_tag,'.png'];
497 | imwrite(imout(:,:,n),out_names{n});
498 | fprintf('writing image %i of %i\n',n,size(xhat,3))
499 | end
500 |
501 | fprintf('zipping...\n')
502 | zip([full_path, '/png/', imbase],out_names)
503 | fprintf('done zipping\n')
504 |
505 | fprintf('writing .mat\n')
506 | options.fighandle = []
507 |
508 | save([full_path,'/',meas_name(1:end-4),'_',date_string,'_',im_tag,'_',tiff_string,'.mat'], 'tau_iso','TVpars','xhat_out', 'options', 'comps_path','weights_path', 'b','params')
509 | fprintf('done writing .mat\n')
510 | % gpuDevice(1)
511 | clear xhat
512 | clear f2
513 |
514 |
515 | if params.ds == 2
516 | gpuDevice(1)
517 | end
518 | end
519 | %end
520 |
521 | %%
522 | % imagesc(brain_recon.xhat(:,:,10))
523 | % axis image
--------------------------------------------------------------------------------
/Adaptive Stitching with Nanoscribe/TipSlicerUpdated.py:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env python
2 | import numpy as np
3 | import numpy.random as npRand
4 | import matplotlib.pyplot as plt
5 | import matplotlib.colors as colcol
6 | import matplotlib.cm as cmx
7 | import cv2
8 | import tables
9 | import scipy.ndimage as im
10 | from scipy.spatial import Voronoi
11 | import os
12 | import glob
13 | import re
14 | import scipy.signal as signal
15 |
16 | def squareSlice(X,Y,Z,extraParams):
17 | amp,period,heightOff=extraParams
18 | ResMat=(amp*np.cos(X*2.0*np.pi/period)+amp*np.cos(Y*2.0*np.pi/period)+amp+heightOff)>Z
19 | return ResMat
20 |
21 | #Z=sin(X)+sin(-X/2+sqrt(3)/2*Y) + sin(-X/2-sqrt(3)/2*Y)
22 | # minimum Z = heightOff
23 | # max Z = heightOff + 4.5*amp
24 | def hexagonalSlice(X,Y,Z,extraParams):
25 | amp,period,heightOff=extraParams
26 | ResMat=(amp*np.cos(X*2.0*np.pi/period)+amp*np.cos((-0.5*X+np.sqrt(3.0)/2.0*Y)*2.0*np.pi/period)+amp*np.cos((-X/2.0-np.sqrt(3.0)/2.0*Y)*2.0*np.pi/period)+1.5*amp+heightOff)>Z
27 | return ResMat
28 |
29 | def polarSineSlice(X,Y,Z,extraParams):
30 | xc,yc,period,amp,offset=extraParams
31 | D=np.sqrt((X-xc)**2+(Y-yc)**2)*2.0*np.pi/period
32 | ResMat=amp*np.cos(D)+offset>Z
33 | return ResMat
34 |
35 | # height falls off as the sum of the distances to the two focal points of an ellipse
36 | # smallAxis determines the scale and is the distance from centre to edge - perpendicular to separating line,
37 | # maxHeight scales the height
38 | def ellipticalConeSlice(X,Y,Z,extraParams):
39 | centreX,centreY,sepDist,sepAngle,smallAxis,maxHeight=extraParams
40 | centre=np.array([centreX,centreY])
41 | cosang=np.array([np.cos(np.deg2rad(sepAngle)),np.sin(np.deg2rad(sepAngle))])
42 | f1=centre+sepDist/2.0 * cosang
43 | f2=centre-sepDist/2.0 * cosang
44 | D=np.sqrt((f1[0]-X)**2+(f1[1]-Y)**2)+np.sqrt((f2[0]-X)**2+(f2[1]-Y)**2)
45 | heightOff=sepDist/np.cos(np.arctan(2*smallAxis/sepDist))
46 | ResMat=maxHeight*(heightOff-D)/(heightOff-sepDist)>Z
47 | return ResMat
48 |
49 | # will create a X,Y & Z matrix based on the given stlMesh
50 | def getMeshXYZ(stlMesh,hashing,slicing):
51 | # now determine the extent of the figure, in order to create X,Y and Z axes
52 | xext=stlMesh[:,:,0].max()-stlMesh[:,:,0].min()
53 | yext=stlMesh[:,:,1].max()-stlMesh[:,:,1].min()
54 | zext=stlMesh[:,:,2].max()-stlMesh[:,:,2].min()
55 | maxd=max(xext,yext)
56 | maxd=max(maxd,zext)
57 |
58 | Xmins=stlMesh[:,:,0].min(axis=1)
59 | XMI=Xmins.min()
60 | Xmaxs=stlMesh[:,:,0].max(axis=1)
61 | XMA=Xmaxs.max()
62 | Ymins=stlMesh[:,:,1].min(axis=1)
63 | YMI=Ymins.min()
64 | Ymaxs=stlMesh[:,:,1].max(axis=1)
65 | YMA=Ymaxs.max()
66 | Zmins=stlMesh[:,:,2].min(axis=1)
67 | ZMI=Zmins.min()
68 | Zmaxs=stlMesh[:,:,2].max(axis=1)
69 | ZMA=Zmaxs.max()
70 |
71 | offsetXY=5*hashing
72 | offsetZ=2*slicing
73 |
74 | # should set the range to match the hashing resolution! otherwise discrepancy with grid used later
75 | # assuming the grid goes through zero => each border should be a multiple of het hashing
76 | nrxmin=np.int(np.round((XMI-offsetXY)/hashing))
77 | nrxmax=np.int(np.round((XMA+offsetXY)/hashing))+1
78 | nrymin=np.int(np.round((YMI-offsetXY)/hashing))
79 | nrymax=np.int(np.round((YMA+offsetXY)/hashing))+1
80 |
81 | X,Y=np.meshgrid(np.arange(nrxmin*hashing,nrxmax*hashing,hashing),np.arange(nrymin*hashing,nrymax*hashing,hashing))
82 | Z=np.arange(ZMI,ZMA+offsetZ,slicing) # should start from zero
83 |
84 | # print('Maximum extent: {} with {} slices'.format(maxd,len(Z)))
85 | if (maxd<1):
86 | print('Warning, max extent is <1: {}'.format(maxd))
87 |
88 | return X,Y,Z
89 |
90 | def loadStl(stlName,mulDimension=1.,centred=True,zToZero=True,flipZ=False):
91 | from stl import mesh
92 | mm = mesh.Mesh.from_file(stlName)
93 | mm2=mm.vectors*mulDimension
94 |
95 | if (centred):
96 | xCentre=(mm2[:,:,0].min()+mm2[:,:,0].max())/2.0
97 | yCentre=(mm2[:,:,1].min()+mm2[:,:,1].max())/2.0
98 | mm2[:,:,0]=mm2[:,:,0]-xCentre
99 | mm2[:,:,1]=mm2[:,:,1]-yCentre
100 | if (flipZ):
101 | mm2[:,:,2]=mm2[:,:,2].max()-mm2[:,:,2]
102 | if (zToZero):
103 | mm2[:,:,2]=mm2[:,:,2]-mm2[:,:,2].min()
104 |
105 | Zmins=mm2[:,:,2].min(axis=1)
106 | Zmaxs=mm2[:,:,2].max(axis=1)
107 |
108 | return mm2,Zmins,Zmaxs
109 |
110 |
111 | # complete slicing into a hdf5 file
112 | # first load the stl with loadStl, this also gives the Zmins and Zmaxs values
113 | # when giving X & Y smaller than total size STL => not possible to get a good infill => use inFillMaster (hdf5-stack) with smaller resolution possibly
114 | def stlToStack(mesh,h5name,X,Y,Z,Zmins,Zmaxs,inFillMaster='',writingMask=np.ones((1,1)),epsilon=0.001,onlyShell=False):
115 | zslices=len(Z)
116 | height=X.shape[0]
117 | width=X.shape[1]
118 |
119 | if (writingMask.shape[0]==1):
120 | writingMask=np.ones(X.shape)
121 |
122 |
123 | compFilt=tables.Filters(complevel=3)
124 | with tables.open_file(h5name,filters=compFilt,mode="w") as h5file:
125 | Array3D=h5file.create_carray("/",'data',tables.BoolAtom(),(zslices,height,width),filters=compFilt)
126 | ArrayX=h5file.create_carray("/",'X',tables.Float32Atom(),(height,width))
127 | ArrayY=h5file.create_carray("/",'Y',tables.Float32Atom(),(height,width))
128 | ArrayZ=h5file.create_carray("/",'Z',tables.Float32Atom(),(zslices,))
129 | ArrayX[:]=X.copy()
130 | ArrayY[:]=Y.copy()
131 | ArrayZ[:]=Z.copy()
132 | h5file.root.data.attrs.GalvoCentre=[X[0].mean(),Y[:,0].mean(),Z.min()]
133 | h5file.root.data.attrs.PiezoCentre=[X[0].min(),Y[:,0].min(),Z.min()]
134 |
135 | if (inFillMaster!=''):
136 | with tables.open_file(inFillMaster,mode='r') as infill:
137 | Xglob=infill.root.X[:]
138 | Yglob=infill.root.Y[:]
139 | Zglob=infill.root.Z[:]
140 | Mglob=infill.root.data
141 |
142 | doCarving=False
143 | dXg=Xglob[0,1]-Xglob[0,0]
144 | dX=X[0,1]-X[0,0]
145 | dZg=Zglob[1]-Zglob[0]
146 | dZ=Z[1]-Z[0]
147 |
148 | if ((dX==dXg) and (dZ==dZg)):
149 | doCarving=True
150 |
151 | for i in range(len(Z)):
152 |
153 | if (doCarving):
154 | zind=np.searchsorted(Zglob,Z[i]) # check if it shouldn't be -1
155 | if (zind>=len(Zglob)):
156 | zind=len(Zglob)-1
157 | tempS=carvePiece(X,Y,Xglob,Yglob,Mglob[zind])
158 | Array3D[i,:,:]=tempS*writingMask
159 | else:
160 | # print(i)
161 | try:
162 | tempS=singleSliceStl(mesh,Zmins,Zmaxs,Z[i],X,Y,epsilon,onlyShell=True)
163 | zind=np.searchsorted(Zglob,Z[i])
164 | if (zind>=len(Zglob)):
165 | zind=len(Zglob)-1
166 | G=Mglob[zind][:]
167 | Ger=im.binary_erosion(G,iterations=3) # perhaps keep on eroding until number connected components changes? Or extract skeleton? => should reduce in-fill time?
168 | nrLabs,Labs=cv2.connectedComponents(Ger.astype(np.uint8))
169 | # print('First erosion: ',nrLabs)
170 | oldNrLabs=nrLabs
171 | if (nrLabs>1):
172 | while (oldNrLabs==nrLabs):
173 | Ger=im.binary_erosion(Ger,iterations=3)
174 | nrLabs,Labs=cv2.connectedComponents(Ger.astype(np.uint8))
175 | # print('Next erosion: ',nrLabs)
176 | tempS=fillInFromTemplate(tempS,X,Y,Ger,Xglob,Yglob)
177 | Array3D[i,:,:]=tempS*writingMask
178 | except:
179 | print('Problem with slice {} at position {}'.format(i,[X[0].mean(),Y[:,0].mean(),Z.min()]))
180 | # now just take the slice below the current one.
181 | if (i>0):
182 | Array3D[i,:,:]=Array3D[i-1,:,:]
183 | else:
184 | for i in range(len(Z)):
185 | # print(i)
186 | #try:
187 | Array3D[i,:,:]=writingMask*singleSliceStl(mesh,Zmins,Zmaxs,Z[i],X,Y,epsilon,onlyShell=onlyShell)
188 | #except:
189 | # now just take the slice below the current one. I could perhaps take the average of above vs below
190 | # print('Problem with slice {}'.format(i))
191 | #Array3D[i,:,:]=singleSliceStl(mm2,mm.normals*mulDimension,Zmins,Zmaxs,Z[i-1],X,Y,epsilon) # Not a perfect solution, but better than nothing!!
192 | return
193 |
194 |
195 | # will start with the simplest mask and complexify when needed (=> to function pointer probably)
196 | def applyStackMask(stackName,multMatrix):
197 | with tables.open_file(stackName,mode='a') as H:
198 | D=H.root.data
199 | for i in range(len(D)):
200 | D[i,:multMatrix.shape[0],:multMatrix.shape[1]]=D[i,:multMatrix.shape[0],:multMatrix.shape[1]]*multMatrix
201 | return
202 |
203 |
204 | # give as input already the loaded stl file with the mesh and the normals separately
205 | def singleSliceStl(mesh,Zmins,Zmaxs,Zslice,X,Y,epsilon=0.001,onlyShell=False,debug=False):
206 |
207 | lines=[]
208 | gI=np.argwhere(np.logical_and((Zmaxs>=Zslice),(Zmins<=Zslice)))[:,0]
209 | triangs=mesh[gI]
210 |
211 | T=np.zeros((X.shape[0],X.shape[1]),dtype=np.uint8) # this will be the filled in image
212 |
213 | for triang in triangs:
214 | dists=triang[:,2]-Zslice
215 | copoints=np.argwhere(np.abs(dists)1)):
218 | # print('In copoints situation',copoints.shape)
219 | # if (len(copoints)==2):
220 | tArray=triang[copoints[:,0]]
221 |
222 | if (len(copoints)!=2):
223 | tArray=np.row_stack((tArray,triang[copoints[0,0]]))
224 | #print(tArray)
225 |
226 | lines.append(tArray)
227 |
228 | if ((len(tArray)<3)):
229 | pp=triang[copoints[:,0]]
230 |
231 | else: # for in-plane triangles, add the centre point as a seed point
232 | vv=tArray[:3,:2]
233 | yy=convertToPixels(X,Y,vv)
234 | cv2.fillConvexPoly(T,yy.reshape(-1,1,2),255)
235 |
236 | else:
237 | ps=[]
238 | if (len(copoints)==1):
239 | poi=triang[copoints[:,0]]
240 | ps.append([poi[0,0],poi[0,1],Zslice])
241 |
242 |
243 | if (dists[0]*dists[1]<0):
244 | # print('Case 1')
245 | x1,y1,z1=triang[0,:]
246 | x2,y2,z2=triang[1,:]
247 | ts=(Zslice-z1)/(z2-z1)
248 | xs=(x2-x1)*ts+x1
249 | ys=(y2-y1)*ts+y1
250 | ps.append([xs,ys,Zslice])
251 |
252 | if (dists[1]*dists[2]<0):
253 | # print('Case 2')
254 | x1,y1,z1=triang[1,:]
255 | x2,y2,z2=triang[2,:]
256 | ts=(Zslice-z1)/(z2-z1)
257 | xs=(x2-x1)*ts+x1
258 | ys=(y2-y1)*ts+y1
259 | ps.append([xs,ys,Zslice])
260 |
261 | if (dists[0]*dists[2]<0):
262 | # print('Case 3')
263 | x1,y1,z1=triang[0,:]
264 | x2,y2,z2=triang[2,:]
265 | ts=(Zslice-z1)/(z2-z1)
266 | xs=(x2-x1)*ts+x1
267 | ys=(y2-y1)*ts+y1
268 | ps.append([xs,ys,Zslice])
269 |
270 | if (len(ps)==0):
271 | print('Freaky!!! Not case 1, 2 or 3')
272 |
273 | if (len(ps)>0):
274 | pp=np.asarray(ps)
275 | # print(pp)
276 | lines.append(pp)
277 |
278 |
279 |
280 | li=[(convertToPixels(X,Y,lines[ind][:,:2])).reshape(-1,1,2) for ind in range(len(lines))]
281 | li3=[ np.round(litem).astype(np.int32) for litem in li]
282 | res=cv2.polylines(T,li3,False,255,lineType=8)
283 |
284 | if (onlyShell):
285 | if (debug):
286 | return T,gI,lines
287 | else:
288 | return T
289 |
290 | if (T.sum()!=0):
291 | T=colourStlShell(T)
292 | return T
293 |
294 | def colourStlShell(TT):
295 | nrLabs,Labs=cv2.connectedComponents(TT)
296 | im,cnts,hier = cv2.findContours(TT, cv2.RETR_TREE, cv2.CHAIN_APPROX_NONE)
297 | hier=hier[0]
298 |
299 | bigTree=[]
300 | gen=np.argwhere(hier[:,3]==-1)[:,0]
301 |
302 | thisGen,nextGen=checkGeneration(gen,hier,cnts,Labs)
303 | bigTree.append(thisGen)
304 | while (len(nextGen)>0):
305 | thisGen,nextGen=checkGeneration(nextGen,hier,cnts,Labs)
306 | bigTree.append(thisGen)
307 |
308 | N=np.zeros((TT.shape))
309 | genNr=0
310 | for gen in bigTree:
311 | if (genNr%2==0): # should be white
312 | # print('Should paint white: {}'.format(gen))
313 | for gg in gen:
314 | cv2.drawContours(N,cnts,gg,1,-1)
315 | genNr=genNr+1
316 | else:
317 | # print('Should paint black: {}'.format(gen))
318 | for gg in gen:
319 | cv2.drawContours(N,cnts,gg,0,-1)
320 | for gg in gen:
321 | cv2.drawContours(N,cnts,gg,1,1)
322 | genNr=genNr+1
323 | return N
324 |
325 | def checkGeneration(gen,hier,cnts,Labs):
326 | thisGen=gen
327 | nextGen=[]
328 |
329 | findex=0
330 | while (findexXglob.shape[1]-1):
387 | endOffX=carveX[1]-Xglob.shape[1]+1
388 | carveX[1]=Xglob.shape[1]-1
389 | #print(carveX,startOffX,endOffX)
390 |
391 |
392 | carveY=ncoord[:,1]
393 | startOffY=0
394 | endOffY=0
395 | if (carveY[0]<0):
396 | startOffY=-carveY[0]
397 | carveY[0]=0
398 | if (carveY[1]>Xglob.shape[0]-1):
399 | endOffY=carveY[1]-Xglob.shape[0]+1
400 | carveY[1]=Xglob.shape[0]-1
401 | #print(carveY,startOffY,endOffY)
402 |
403 | M=np.zeros(Xloc.shape)
404 | M[startOffY:M.shape[0]-endOffY,startOffX:M.shape[1]-endOffX]=Mslice[carveY[0]:carveY[1]+1,carveX[0]:carveX[1]+1]
405 | return M
406 |
407 |
408 | def fillInFromTemplate(I,Xloc,Yloc,Template,X,Y):
409 | Mask=np.zeros((I.shape[0]+2,I.shape[1]+2),dtype=np.uint8)
410 | Mask[1:-1,1:-1]=I[:]
411 | TestIm=np.zeros(I.shape,np.uint8)
412 | gI=np.where(Template>0)
413 | Xnon=X[gI]
414 | Ynon=Y[gI]
415 | ggI=np.where(np.logical_and(YnonYloc.min(),np.logical_and(Xnon>Xloc.min(),XnontesBig[:,:-1])
480 | edgeXEnd=(tesBig[:,1:]0):
544 | startInd=max(0,sliceInd-amShells)
545 | stopInd=min(len(sliceIndList),sliceInd+amShells+1)
546 | extInds=sliceIndList[startInd:stopInd]
547 | Rstack=np.zeros((len(extInds),D.shape[1],D.shape[2]),dtype=bool)
548 | for jj in np.arange(len(extInds)):
549 | Rstack[jj]=D[extInds[jj]][:]
550 |
551 | extraStart=0
552 | extend=False
553 | if (sliceIndlen(sliceIndList)-1):
557 | extend=True
558 | if (extend):
559 | #print(startInd,stopInd,extraStart,extraStart+len(Rstack),len(Rstack))
560 | Rstack2=np.zeros((2*amShells+1,Rstack.shape[1],Rstack.shape[2]),dtype=bool)
561 | Rstack2[extraStart:extraStart+len(Rstack)]=Rstack
562 | Rstack=Rstack2
563 |
564 | RR=Rstack[amShells]
565 | if (RR.sum()==0):
566 | return sliceList,sliceCodes,sliceZList,sliceDist
567 |
568 | Xr=X.copy()
569 | Yr=Y.copy()
570 | erosionKernel=disk(hatchStep)
571 |
572 | # First do in-plane shells
573 | for i in range(amShells):
574 | if (RR.sum()>0):
575 | if (len(contApprox)==1):
576 | gSlice=getShell(RR.astype(np.uint8),Xr,Yr,zslice,contApprox[0],random=randomShell)
577 | else:
578 | if (i==0):
579 | gSlice=getShell(RR.astype(np.uint8),Xr,Yr,zslice,contApprox[0],random=randomShell)
580 | else:
581 | gSlice=getShell(RR.astype(np.uint8),Xr,Yr,zslice,contApprox[1],random=randomShell)
582 | sliceList.append(gSlice)
583 | sliceCodes.append(1) # 1 is for shell
584 | sliceZList.append(zslice)
585 | nDist=0
586 | for gArr in gSlice:
587 | nDist=nDist+np.sum(np.sqrt(np.sum((gArr[1:]-gArr[:-1])**2,axis=1)))
588 | sliceDist.append(nDist)
589 | if ((i==0) and (divider>1)): # should reduce resolution at this stage if necessary
590 | Rstack=Rstack[:,::divider,::divider]
591 | RR=Rstack[amShells]
592 | Xr=X[::divider,::divider]
593 | Yr=Y[::divider,::divider]
594 | #RR=im.binary_erosion(RR,iterations=iterations)
595 | RR=im.binary_erosion(RR,structure=erosionKernel)
596 | if (RR.sum()==0):
597 | return sliceList,sliceCodes,sliceZList,sliceDist
598 |
599 | # now check if some floors/ceilings need to be printed.
600 | # Rstack=Rstack[:,::iterations,::iterations]
601 | # RR=RR[::iterations,::iterations]
602 | # Xr=Xr[::iterations,::iterations]
603 | # Yr=Yr[::iterations,::iterations]
604 |
605 | rotAngle=-1
606 | if (len(np.atleast_1d(atAngles))>1):
607 | rotAngle=atAngles[sliceInd%len(atAngles)]
608 | else:
609 | if (atAngles==-1):
610 | rotAngle=np.random.randint(0,360)
611 | else:
612 | rotAngle=atAngles
613 |
614 |
615 | if (doTopBottom): # if you are doing shell + solid => better to switch of top-bottom.
616 | inds=np.arange(len(Rstack))
617 | inds=np.delete(inds,amShells)
618 | Cmin=Rstack[inds].min(axis=0)
619 |
620 |
621 | FillIn=np.logical_and(RR,np.logical_not(Cmin))
622 | if (FillIn.sum()>0):
623 | # now hatch these ceilings/floors without coarsening..
624 | gHatch,gDist=getHatching(FillIn.astype(np.uint8),Xr,Yr,zslice,angle=rotAngle,subdiv=hatchStep,crossed=True)
625 | sliceList.append(gHatch)
626 | sliceCodes.append(3) # 3 is for solid (straight lines with overlap), this is used for bottom/top hatching as well.
627 | sliceZList.append(zslice)
628 | sliceDist.append(gDist)
629 |
630 | RR=np.logical_and(RR,np.logical_not(FillIn))
631 |
632 | else: # no shells, only do scaffolds => Solid mode...
633 | Xr=X.copy()
634 | Yr=Y.copy()
635 | rotAngle=-1
636 | if (len(np.atleast_1d(atAngles))>1):
637 | rotAngle=atAngles[slicInd%len(atAngles)]
638 | else:
639 | if (atAngles==-1):
640 | rotAngle=np.random.randint(0,360)
641 | else:
642 | rotAngle=atAngles
643 |
644 | #print('Will try to hatch')
645 | # Finally, hatch in a coarse way the remaining material => Scaffolding
646 | if (scaffoldStep>0):
647 | if (RR.sum()>0):
648 | gHatch,gDist=getHatching(RR.astype(np.uint8),Xr,Yr,zslice,angle=rotAngle,subdiv=scaffoldStep)
649 | if (len(gHatch)>0):
650 | sliceList.append(gHatch)
651 | sliceCodes.append(2) # 2 is for scaffolding (straight lines with no hatching, doubles slicing)
652 | sliceZList.append(zslice)
653 | sliceDist.append(gDist)
654 |
655 | return sliceList,sliceCodes,sliceZList,sliceDist
656 |
657 | #~ def getFullShellScaffold(h5name,amShells,scaffoldJump,atAngles=[0,90,180,270],contApprox=['SIMPLE'],divider=1,iterations=1,randomShell=True,doTopBottom=True):
658 | #~ h5=tables.open_file(h5name,mode='r')
659 |
660 | #~ X=h5.root.X[:]
661 | #~ Y=h5.root.Y[:]
662 | #~ Z=h5.root.Z[:]
663 | #~ D=h5.root.data
664 |
665 | #~ allSliceList=[]
666 | #~ allSliceCodes=[]
667 | #~ allSliceZList=[]
668 | #~ allSliceDistList=[]
669 | #~ for slicInd in range(0,len(Z),1):
670 | #~ # for slicInd in range(0,65,1):
671 | #~ sList,sCodes,sZs,newDists=getSliceOfShellScaffold(D,X,Y,Z,slicInd,amShells,scaffoldJump,atAngles,contApprox,divider,iterations,randomShell,doTopBottom)
672 | #~ if (len(sCodes)==0):
673 | #~ continue
674 | #~ for j in range(len(sList)):
675 | #~ allSliceList.append(sList[j])
676 | #~ allSliceCodes.append(sCodes[j])
677 | #~ allSliceZList.append(sZs[j])
678 | #~ # print(slicInd,j,len(sList),len(newDists),sum(newDists))
679 | #~ allSliceDistList.append(newDists[j])
680 | #~ h5.close()
681 | #~ return allSliceList,allSliceCodes,allSliceZList,allSliceDistList
682 |
683 | # amShells gives the amount of contours to include
684 | # scaffoldStep gives the amount of lines in the stack to skip while making the solid infill => total dist = hatching of stack * scaffoldStep
685 | # hatchStep is similar to the above, but now for the distance between contours and the distance for the top and bottom 'shells'
686 | # codeSpeeds is typically a list of three speeds for the three line-types: contour, top/bottom shell, scaffold or inner infill
687 | # codeIntens is a list giving the intensities to go with the above speeds
688 | # atAngles: between each slicing layer, the view will be rotated by the next angle to get better uniformity
689 | # sliceIndexList: when empty all slices in the stack are done, otherwise only the indices in this list are used. => Variable slicing distance!
690 | # contApprox: contour approximation technique => see opencv documentation
691 | # divider: reduce image stack size after first contour extraction to speed up execution; not really used anymore
692 | # randomShell: whether to start at a random position along the contour
693 | # doTopBottom: when writing contours or shells, should he also make upper/lower shells? Switch off when in solid mode
694 | # writeHeader: make own gwl header or avoid when writing one of the blocks in a big stitch
695 | # writeColourH5: can also create a colour-coded hdf5-file for visualisation of the instructions
696 | #
697 | # returns the lengths written per coded line-type: combined with the codeSpeeds, this allows for an easy time-estimation for the structure
698 |
699 | # a simple Zscaler function giving 1 as an intensity multiplier independent of input height
700 | def noBaseReflection(zheight):
701 | return 1.0
702 | def stackToGwl(stackName,gwlName,amShells,scaffoldStep,hatchStep,atAngles=[0,90,180,270],codeSpeeds=[10000,10000,10000],codeIntens=[80,80,80],sliceIndexList=[],contApprox=['L1'],divider=1,randomShell=True,doTopBottom=True,writeHeader=True,writeColourH5=False,interfacePos=-1,ZscalerFunc=noBaseReflection):
703 | with tables.open_file(stackName,mode='r') as h5:
704 | X=h5.root.X[:]
705 | Y=h5.root.Y[:]
706 | Z=h5.root.Z[:]
707 | D=h5.root.data
708 | GC=h5.root.data.attrs.GalvoCentre
709 |
710 | allSliceList=[]
711 | allSliceCodes=[]
712 | allSliceZList=[]
713 | allSliceDistList=[]
714 | if (len(sliceIndexList)==0):
715 | sliceIndexList=range(0,len(Z))
716 |
717 | for slicInd in np.arange(len(sliceIndexList)):
718 | sList,sCodes,sZs,newDists=getSliceOfShellScaffold(D,X,Y,Z,sliceIndexList,slicInd,amShells,scaffoldStep,hatchStep,atAngles,contApprox,divider,randomShell,doTopBottom)
719 | if (len(sCodes)==0):
720 | continue
721 | for j in range(len(sList)):
722 | allSliceList.append(sList[j])
723 | allSliceCodes.append(sCodes[j])
724 | allSliceZList.append(sZs[j])
725 | # print(slicInd,j,len(sList),len(newDists),sum(newDists))
726 | allSliceDistList.append(newDists[j])
727 |
728 | writeGwlFile(gwlName,allSliceList,allSliceCodes,allSliceZList,codeSpeeds,codeIntens,stagePosition=GC,writeHeader=writeHeader,interfacePos=interfacePos,debug=True,ZscalerFunc=ZscalerFunc)
729 | if (writeColourH5):
730 | Z2=Z[sliceIndexList]
731 | writeH5ColourGwl(gwlName[:-4]+'_col.h5',allSliceList,allSliceCodes,allSliceZList,X,Y,Z2)
732 | return getCodeLengths(allSliceDistList,allSliceCodes)
733 |
734 | def getCodeLengths(allSliceDistList,allSliceCodes):
735 | if (len(allSliceCodes)!=0):
736 | amCodes=max(np.unique(allSliceCodes))
737 | codeLengths=np.zeros(amCodes)
738 | try:
739 | for i in range(len(allSliceDistList)):
740 | codeLengths[allSliceCodes[i]-1]=codeLengths[allSliceCodes[i]-1]+allSliceDistList[i]
741 | except:
742 | print('Problem with codelength calculation!')
743 | print(allSliceDistList)
744 | print(allSliceCodes)
745 | return codeLengths
746 | else:
747 | return [0]
748 |
749 |
750 |
751 | def writeGwlHeader(F,interfacePos,CodeSpeeds,CodeIntensities):
752 | F.write('GalvoScanMode\r')
753 | F.write('ContinuousMode\r\r')
754 | # F.write('ConnectPointsOff\r') # not used for GT in continuous mode
755 | F.write('InvertZAxis 1\r') # DiLL configuration
756 | F.write('% DefocusFactor 0.6\r') # for air objective
757 | F.write('TiltCorrectionOff\r') # not available with GT
758 | F.write('StageVelocity 200\r')
759 | F.write('PowerScaling 1\r')
760 | F.write('GalvoAcceleration 2\r\r')
761 |
762 | F.write('var $folderNumber = 1\r\r')
763 |
764 | F.write('% 1 is for shell, 2 for interior and 3 for top/bottom\r')
765 | for j in range(len(CodeSpeeds)):
766 | F.write('var $Speed{} = {}\r'.format(j+1,CodeSpeeds[j]))
767 | F.write('var $Power{} = {}\r'.format(j+1,CodeIntensities[j]))
768 | # F.write('local $block = 0\r')
769 |
770 |
771 | F.write('\r')
772 | F.write('DebugModeOn\r')
773 | F.write('TimeStampOn\r')
774 |
775 | F.write ('var $perSliceDebug = 0\r')
776 | F.write ('var $perBlockDebug = 1\r')
777 | F.write ('var $writePics = 1\r\r')
778 |
779 | F.write('XOffset 0\r') # Will use offsets later so initialise here
780 | F.write('YOffset 0\r')
781 | F.write('ZOffset 0\r\r') # Can add 3.5 here for a proper interface positioning
782 |
783 | F.write('var $doTiltCorrection = 0 % When >0, a correction for the interface location will be used instead of FindInterfaceAt\r')
784 | F.write('var $tiltdX=0 % The coefficient a in the fitted plane formula: Z=ax+by+c\r')
785 | F.write('var $tiltdY=0 % The coefficent b in the above formula\r')
786 | F.write('var $tiltInterface=3 % The equivalent FindInterfaceAt location\r')
787 | F.write('var $tiltMargin=20 % This is the amount of +- correction that will be allowed\r\r')
788 | F.write('if $doTiltCorrection>0 \r')
789 | F.write(' local $tiltInt=$tiltMargin+$tiltInterface\r')
790 | F.write(' FindInterfaceAt $tiltInt\r')
791 | F.write(' ZDrivePosition\r')
792 | F.write(' AddZOffset $tiltMargin\r')
793 | F.write('end\r\r')
794 |
795 |
796 |
797 | F.write('var $interfacePos = {} % if >=0 will be done for each block => clashes with TiltCorrection!!\r'.format(interfacePos))
798 | F.write('var $piezoInterf = 10 % piezo will be moved here to do the FindInterface\r')
799 | F.write('var $piezoWrite = 150 % afterwards it will come back here to write the block\r\r')
800 |
801 | F.write('var $currX = 0\r') # This will be used for relative positioning of different stitching blocks or arrays
802 | F.write('var $currY = 0\r')
803 | F.write('var $currZ = 0\r\r')
804 |
805 | F.write('% Write out the static parameters to the log file\r')
806 | F.write('ShowParameter\r')
807 | F.write('MessageOut "Speed1 %d Speed2 %d Speed3 %d" #($Speed1,$Speed2,$Speed3)\r')
808 | F.write('MessageOut "Power1 %d Power2 %d Power3 %d" #($Power1,$Power2,$Power3)\r')
809 | F.write('MessageOut "tiltMargin %d tiltInterface %d interfacePos %d" #($tiltMargin,$tiltInterface,$interfacePos)\r\r')
810 | F.write('% ----------- End of header -----------\r\r')
811 | return
812 |
813 |
814 |
815 | # Writes to GWL file
816 | # The adaptation of intensity on Z-position will be handled here.
817 | # stagePosition is used for stitching multiple blocks. Will move stage to this position.
818 | # In the header the current location is initialised to (0,0,0) and will then move stage to given location.
819 | # X,Y and ZOffsets are next incremented to this stagePosition, so as to centre the coordinate system for Galvowriting.
820 | # so make sure that all coordinates are reachable when the coordinates are centered on this stagePosition
821 | # interfacePos <0 implies that FindInterfaceAt is not called for this block, if >=0 it is called with the given value
822 | def writeGwlFile(filename,allSliceList,allSliceCodes,allSliceZList,CodeSpeeds=[10000,10000,10000],CodeIntensities=[80,80,80],stagePosition=[0,0,0],interfacePos=-1,ZscalerFunc=noBaseReflection,writeHeader=True,debug=True):
823 | with open(filename,mode="w") as F:
824 | if (writeHeader):
825 | writeGwlHeader(F,interfacePos,CodeSpeeds,CodeIntensities)
826 |
827 |
828 | # first handle relative movement of stage if necessary
829 | # also adapt X,Y and ZOffset parameters to have properly centered Galvo-coordinates for Nanoscribe
830 | # even if inside the gwl blocks, the original coordinate system is maintained.
831 | F.write('NewStructure\r')
832 | F.write('% The following commands are used for relative positioning.\r')
833 | F.write('% If you change them for manual positioning, do not forget to change (remove) the AddX(Y or Z)Offset values as well\r')
834 | F.write('local $locX = {:.0f}\r'.format(stagePosition[0])) # target values
835 | F.write('local $locY = {:.0f}\r'.format(stagePosition[1]))
836 | F.write('local $locZ = {:.0f}\r\r'.format(stagePosition[2]))
837 |
838 | F.write('local $xmov = $locX-$currX\r')
839 | F.write('MoveStageX $xmov\r')
840 | F.write('AddXOffset -$xmov\r')
841 | F.write('local $ymov = $locY-$currY\r')
842 | F.write('MoveStageY $ymov\r')
843 | F.write('AddYOffset -$ymov\r\r')
844 |
845 | F.write('local $zmov = $locZ-$currZ\r')
846 | # F.write('AddZDrivePosition $zmov\r')
847 | F.write('AddZOffset -$zmov\r\r')
848 |
849 | F.write('% Now find the interface if $interfacePos >=0\r')
850 | F.write('if $interfacePos >= 0 \r')
851 | F.write(' PiezoGotoY $piezoInterf \r')
852 | F.write(' FindInterfaceAt $interfacePos \r')
853 | F.write(' PiezoGotoY $piezoWrite \r')
854 | F.write(' ZDrivePosition\r')
855 | F.write(' set $currZ=0 % after a findinterface I am at zero again.\r')
856 | F.write('end\r\r')
857 |
858 | F.write('local $zmov = $locZ-$currZ\r')
859 | F.write('AddZDrivePosition $zmov\r')
860 | # F.write('AddZOffset -$zmov\r\r')
861 |
862 | F.write('set $currX=$locX\r')
863 | F.write('set $currY=$locY\r')
864 | F.write('set $currZ=$locZ\r\r')
865 |
866 |
867 |
868 | F.write('% Now handle the interface tilt correction => clashes with FindInterfaceAt\r')
869 | F.write('local $tiltCorr = 0\r')
870 | F.write('if $doTiltCorrection >0 \r')
871 | F.write(' set $tiltCorr = $tiltdX*$currX+$tiltdY*$currY\r')
872 | F.write(' AddZOffset $tiltCorr\r')
873 | F.write('end\r\r')
874 |
875 | F.write('% Now write out all the relevant numbers to the log file \r')
876 | F.write('MessageOut "tiltCorr %_2f " #($tiltCorr)\r')
877 | F.write('MessageOut "locX %_2f locY %_2f locZ %_2f" #($locX,$locY,$locZ)\r')
878 |
879 |
880 | Zscaler=1.0
881 | pointCounter=0
882 | lineCounter=0
883 | oldCode=-1
884 | oldZ=-1
885 | zCount=len(np.unique(allSliceZList))
886 | zIndex=0 # a counter to allow following the slice progress
887 | writeSliceInfo=False
888 | for i in np.arange(len(allSliceList)):
889 | sliceArray=allSliceList[i]
890 | zz=allSliceZList[i]
891 | cod=allSliceCodes[i] # can modify speed etc based on this code
892 | if ((cod!=oldCode) or (zz!=oldZ)):
893 | if (debug and (zz!=oldZ)):
894 | # first write old slice info
895 | F.write('if $perSliceDebug>0 \r')
896 | F.write(' MessageOut "Finished slice {0:05d} out of {1:d} for block %05d" #($folderNumber)\r'.format(zIndex,zCount))
897 | F.write(' if $writePics>0 \r')
898 | F.write(' Wait 0.1\r')
899 | filenam='PicPerSlice/Pic_Block_%05d_{0:05d}.tif'.format(zIndex) # I could also add the timestamp in the name or location (in case of stitching)
900 | F.write(' CapturePhoto "{}" #($folderNumber)\r'.format(filenam))
901 | F.write(' end\r')
902 |
903 |
904 | zIndex=zIndex+1
905 | F.write(' MessageOut "Started slice {0:05d} out of {1:d} for block %05d" #($folderNumber)\r'.format(zIndex,zCount))
906 | F.write('end\r\r')
907 |
908 | F.write('ScanSpeed $Speed{}\r'.format(cod))
909 | F.write('LaserPower $Power{}\r'.format(cod))
910 | Zscaler=ZscalerFunc(zz)
911 | F.write('MultLaserPower {0:.2f}\r\r'.format(Zscaler))
912 |
913 |
914 | oldCode=cod
915 | oldZ=zz
916 |
917 | for j in np.arange(len(sliceArray)):
918 | for k in np.arange(len(sliceArray[j])):
919 | if (sliceArray[j].shape[1]==3):
920 | F.write('{0:.2f} {1:.2f} {2:.2f}\r'.format(sliceArray[j][k,0],sliceArray[j][k,1],sliceArray[j][k,2]))
921 | elif (sliceArray[j].shape[1]==4):
922 | F.write('{0:.2f} {1:.2f} {2:.2f} {3:.2f}\r'.format(sliceArray[j][k,0],sliceArray[j][k,1],sliceArray[j][k,2],sliceArray[j][k,3]))
923 | pointCounter=pointCounter+1
924 | F.write("write\r\r")
925 | lineCounter=lineCounter+1
926 |
927 | if (debug):
928 | F.write('if $perSliceDebug>0 \r')
929 | F.write('MessageOut "Finished slice {0:05d} out of {1:d} for block %05d" #($folderNumber)\r'.format(zIndex,zCount))
930 | F.write(' if $writePics>0 \r')
931 | F.write(' Wait 0.1\r')
932 | filenam='PicPerSlice/Pic_Block_%05d_{0:05d}.tif'.format(zIndex) # I could also add the timestamp in the name or location (in case of stitching)
933 | F.write(' CapturePhoto "{}" #($folderNumber)\r'.format(filenam))
934 | F.write(' end\r')
935 | F.write('end\r\r')
936 |
937 | if (writeHeader): # if you don't want a header, than it is likely that you are not writing a single block => save the log only once!
938 | messfile='MyLogfile Block%d' # should make this name more unique in case of stitching but cannot make new folder
939 | if (debug):
940 | F.write('SaveMessages "{}" #($folderNumber)\r'.format(messfile))
941 |
942 | F.write('% Now remove the interface tilt correction again, if applied\r')
943 | F.write('if $doTiltCorrection >0 \r')
944 | F.write(' AddZOffset -$tiltCorr\r')
945 | F.write('end\r\r')
946 |
947 | return pointCounter,lineCounter
948 |
949 | # Writes an empty GWL file, used for interface tilt determination
950 | def writeEmptyGwlFile(filename,stagePosition=[0,0,0],interfacePos=0,writeHeader=True,debug=True):
951 | with open(filename,mode="w") as F:
952 | if (writeHeader):
953 | writeGwlHeader(F,interfacePos,[0],[0])
954 |
955 |
956 | # first handle relative movement of stage if necessary
957 | # also adapt X,Y and ZOffset parameters to have properly centered Galvo-coordinates for Nanoscribe
958 | # even if inside the gwl blocks, the original coordinate system is maintained.
959 | F.write('% The following commands are used for relative positioning.\r')
960 | F.write('% If you change them for manual positioning, do not forget to change (remove) the AddX(Y or Z)Offset values as well\r')
961 | F.write('local $locX = {:.0f}\r'.format(stagePosition[0])) # target values
962 | F.write('local $locY = {:.0f}\r'.format(stagePosition[1]))
963 | F.write('local $locZ = {:.0f}\r\r'.format(stagePosition[2]))
964 |
965 | F.write('local $xmov = $locX-$currX\r')
966 | F.write('MoveStageX $xmov\r')
967 | F.write('AddXOffset -$xmov\r')
968 | F.write('local $ymov = $locY-$currY\r')
969 | F.write('MoveStageY $ymov\r')
970 | F.write('AddYOffset -$ymov\r')
971 | F.write('local $zmov = $locZ-$currZ\r')
972 | F.write('AddZDrivePosition $zmov\r')
973 | F.write('AddZOffset -$zmov\r')
974 |
975 | F.write('set $currX=$locX\r')
976 | F.write('set $currY=$locY\r')
977 | F.write('set $currZ=$locZ\r\r')
978 |
979 | # handle using FindInterfaceAt
980 | F.write('if $interfacePos >= 0 \r')
981 | F.write(' PiezoGotoY $piezoInterf \r')
982 | F.write(' FindInterfaceAt $interfacePos \r')
983 | F.write(' PiezoGotoY $piezoWrite \r')
984 | F.write(' ZDrivePosition\r')
985 | F.write('end\r\r')
986 |
987 |
988 | return
989 |
990 | def writeGwlForRegex(outGwlName,filePattern,codeSpeeds=[10000,10000,10000],codeIntensities=[80,80,80],interfacePos=-1,debug=True):
991 | fp=os.path.split(filePattern)
992 | if (fp[0]==''):
993 | fnames,fnumbs=findFiles(filePattern)
994 | else:
995 | fnames,fnumbs=findFiles(fp[1],fp[0])
996 | with open(outGwlName,mode="w") as F:
997 | writeGwlHeader(F,interfacePos,codeSpeeds,codeIntensities)
998 |
999 | blockCounter=0
1000 | for ff in fnames:
1001 | F.write('if $perBlockDebug>0 \r')
1002 | F.write(' MessageOut "Starting block {0:d} out of {1:d}"\r'.format(blockCounter,len(fnames)))
1003 | F.write('end \r')
1004 | #F.write('include {}\r'.format(os.path.split(ff)[1])) # perhaps use relpath first and replace / with \
1005 | tempPath=os.path.relpath(ff)
1006 | startP=tempPath.find('/')
1007 | F.write('include {}\r'.format(tempPath[startP+1:].replace('/','\\')))
1008 |
1009 | F.write('if $perBlockDebug>0 \r')
1010 | F.write(' MessageOut "Finished block {0:d} out of {1:d}"\r'.format(blockCounter,len(fnames)))
1011 | F.write(' if $writePics>0 \r')
1012 | filenam='PicPerBlock/Block_%05d.tif'
1013 | F.write(' Wait 1\r')
1014 | F.write(' CapturePhoto "{}" #($folderNumber)\r'.format(filenam))
1015 | F.write(' end\r')
1016 |
1017 | F.write('end \r\r\r')
1018 |
1019 | F.write('set $folderNumber=$folderNumber+1\r')
1020 | blockCounter=blockCounter+1
1021 |
1022 | if (debug):
1023 | F.write('SaveMessages "MyLogfile"\r')
1024 |
1025 | return
1026 |
1027 |
1028 |
1029 |
1030 |
1031 |
1032 | # other good colourmaps 'viridis', 'magma','plasma','inferno', 'coolwarm', 'rainbow','spectral'
1033 | def writePointCloud(allSliceList,allSliceCodes,filename,divider=10,colorName='coolwarm',goodCodes=[0,1,2,3,4]):
1034 | #just modified the input parameters
1035 | pointCounter=0
1036 | pointList=np.zeros((0,3))
1037 | pointList=[]
1038 | for i in np.arange(len(allSliceList)):
1039 | cod=allSliceCodes[i] # can modify speed etc based on this code
1040 | if (cod not in goodCodes):
1041 | continue
1042 | sliceArray=allSliceList[i]
1043 | for contour in sliceArray:
1044 | for pp in contour:
1045 | if (np.random.randint(divider)==0):
1046 | # pointList=np.append(pointList,contour,axis=0)
1047 | pointList.append(pp)
1048 |
1049 | Pts=np.asarray(pointList)
1050 |
1051 | Hs=np.unique(np.asarray(Pts[:,2]))
1052 |
1053 | amPoints=len(Pts)
1054 | F=open(filename,mode='w')
1055 | F.write('# .PCD v.7 - Point Cloud Data file format\n')
1056 | F.write('VERSION .7\n')
1057 | F.write('FIELDS x y z rgb\n')
1058 | F.write('SIZE 4 4 4 4\n')
1059 | F.write('TYPE F F F I\n')
1060 | F.write('COUNT 1 1 1 1\n')
1061 | F.write('WIDTH {}\n'.format(amPoints))
1062 | F.write('HEIGHT 1\n')
1063 | F.write('VIEWPOINT 0 0 0 1 0 0 0\n')
1064 | F.write('POINTS {}\n'.format(amPoints))
1065 | F.write('DATA ascii\n')
1066 |
1067 | mappie = plt.get_cmap(colorName)
1068 | cNorm=colcol.Normalize(vmin=Hs.min(),vmax=Hs.max())
1069 | scalarMap = cmx.ScalarMappable(norm=cNorm, cmap=mappie)
1070 | cc=scalarMap.to_rgba(Hs)
1071 | rgb = (((cc[:,0]*255).astype(int) << 16) + ((cc[:,1]*255).astype(int) << 8) + (cc[:,2]*255).astype(int))
1072 |
1073 |
1074 | for pp in Pts:
1075 | gi=np.searchsorted(Hs,pp[2]) # only 1 height per slice
1076 | col=rgb[gi]
1077 | F.write('{:.4f} {:.4f} {:.4f} {:}\n'.format(pp[0],pp[1],pp[2],col))
1078 |
1079 | F.close()
1080 | return amPoints
1081 |
1082 | # other good colourmaps 'viridis','magma','plasma','inferno','rainbow','spectral'
1083 | def writePointCloud2(allSliceList,allSliceCodes,allSliceZList,filename,divider=10,colorName='coolwarm'):
1084 | zmin=np.min(allSliceZList)
1085 | zmax=np.max(allSliceZList)
1086 |
1087 |
1088 | F=open(filename,mode='w')
1089 | F.write('# .PCD v.7 - Point Cloud Data file format\n')
1090 | F.write('VERSION .7\n')
1091 | F.write('FIELDS x y z rgb\n')
1092 | F.write('SIZE 4 4 4 4\n')
1093 | F.write('TYPE F F F I\n')
1094 | F.write('COUNT 1 1 1 1\n')
1095 | F.write('WIDTH ')
1096 | writeLoc1=F.tell()
1097 | filler=' ' # this should be larger than amount of points
1098 | F.write(filler+'\n')
1099 | F.write('HEIGHT 1\n')
1100 | F.write('VIEWPOINT 0 0 0 1 0 0 0\n')
1101 | F.write('POINTS ')
1102 | writeLoc2=F.tell()
1103 | F.write(filler+'\n')
1104 | F.write('DATA ascii\n')
1105 |
1106 | Hs=np.arange(zmin,zmax,0.1)
1107 | mappie = plt.get_cmap(colorName)
1108 | cNorm=colcol.Normalize(vmin=Hs.min(),vmax=Hs.max())
1109 | scalarMap = cmx.ScalarMappable(norm=cNorm, cmap=mappie)
1110 | cc=scalarMap.to_rgba(Hs)
1111 | rgb = (((cc[:,0]*255).astype(int) << 16) + ((cc[:,1]*255).astype(int) << 8) + (cc[:,2]*255).astype(int))
1112 |
1113 | pointCounter=0
1114 | for i in np.arange(len(allSliceList)):
1115 | cod=allSliceCodes[i] # can modify speed etc based on this code
1116 | if (cod!=2):
1117 | continue
1118 | sliceArray=allSliceList[i]
1119 | gi=np.searchsorted(Hs,sliceArray[0][0,2]) # only 1 height per slice
1120 | col=rgb[gi]
1121 | for contour in sliceArray:
1122 | for pp in contour:
1123 | if (np.random.randint(divider)==0):
1124 | F.write('{:.4f} {:.4f} {:.4f} {:}\n'.format(pp[0],pp[1],pp[2],col))
1125 | pointCounter=pointCounter+1
1126 |
1127 | F.close()
1128 |
1129 | amPoints='{}'.format(pointCounter)
1130 | neededSpaces=len(filler)-len(amPoints)
1131 | F=open(filename,mode='r+')
1132 |
1133 | F.seek(writeLoc1)
1134 | F.write(amPoints)
1135 | for i in np.arange(neededSpaces):
1136 | F.write(' ')
1137 | F.write('\n')
1138 |
1139 | F.seek(writeLoc2)
1140 | F.write(amPoints)
1141 | for i in np.arange(neededSpaces):
1142 | F.write(' ')
1143 | F.write('\n')
1144 | F.close()
1145 |
1146 | return pointCounter
1147 |
1148 | # X, Y & Z are ranges with their values. Will make a 2d matrix of X & Y myself
1149 | def writeEmptyStack(h5name,X,Y,Z,compression=True):
1150 | zslices=len(Z)
1151 | Xm,Ym=np.meshgrid(X,Y)
1152 | height=Xm.shape[0]
1153 | width=Xm.shape[1]
1154 |
1155 | compFilt=None
1156 | if compression:
1157 | compFilt=tables.Filters(complevel=3)
1158 |
1159 | with tables.open_file(h5name,filters=compFilt,mode="w") as h5file:
1160 | #Array3D=h5file.createCArray("/",'data',tables.UInt8Atom(),(amount,height,width),filters=compFilt)
1161 | Array3D=h5file.create_carray("/",'data',tables.BoolAtom(),(zslices,height,width),filters=compFilt)
1162 | ArrayX=h5file.create_carray("/",'X',tables.Float32Atom(),(height,width))
1163 | ArrayY=h5file.create_carray("/",'Y',tables.Float32Atom(),(height,width))
1164 | ArrayZ=h5file.create_carray("/",'Z',tables.Float32Atom(),(zslices,))
1165 | ArrayX[:]=Xm.copy()
1166 | ArrayY[:]=Ym.copy()
1167 | ArrayZ[:]=Z.copy()
1168 |
1169 | return
1170 |
1171 | def formulaToStack(h5name,X,Y,Z,sliceFormula,extraParams=[],writingMask=np.ones((1,1)),compression=True):
1172 | zslices=len(Z)
1173 | height=X.shape[0]
1174 | width=X.shape[1]
1175 |
1176 | if (writingMask.shape[0]==1):
1177 | writingMask=np.ones(X.shape)
1178 |
1179 | compFilt=None
1180 | if compression:
1181 | compFilt=tables.Filters(complevel=3)
1182 |
1183 | with tables.open_file(h5name,filters=compFilt,mode="w") as h5file:
1184 | #Array3D=h5file.createCArray("/",'data',tables.UInt8Atom(),(amount,height,width),filters=compFilt)
1185 | Array3D=h5file.create_carray("/",'data',tables.BoolAtom(),(zslices,height,width),filters=compFilt)
1186 | ArrayX=h5file.create_carray("/",'X',tables.Float32Atom(),(height,width))
1187 | ArrayY=h5file.create_carray("/",'Y',tables.Float32Atom(),(height,width))
1188 | ArrayZ=h5file.create_carray("/",'Z',tables.Float32Atom(),(zslices,))
1189 | ArrayX[:]=X.copy()
1190 | ArrayY[:]=Y.copy()
1191 | ArrayZ[:]=Z.copy()
1192 | for i in range(len(Z)):
1193 | Array3D[i,:,:]=writingMask*sliceFormula(X,Y,Z[i],extraParams)
1194 | # Still add GalvoCentre and PiezoCentre attributes
1195 | h5file.root.data.attrs.GalvoCentre=[X[0].mean(),Y[:,0].mean(),Z.min()]
1196 | h5file.root.data.attrs.PiezoCentre=[X[0].min(),Y[:,0].min(),Z.min()]
1197 | return
1198 |
1199 | def formulaToStack2(h5name,X,Y,Z,sliceFormula,maskFormula,extraParams,compression=True):
1200 | zslices=len(Z)
1201 | height=X.shape[0]
1202 | width=X.shape[1]
1203 |
1204 | compFilt=None
1205 | if compression:
1206 | compFilt=tables.Filters(complevel=3)
1207 |
1208 | with tables.open_file(h5name,filters=compFilt,mode="w") as h5file:
1209 | #Array3D=h5file.createCArray("/",'data',tables.UInt8Atom(),(amount,height,width),filters=compFilt)
1210 | Array3D=h5file.create_carray("/",'data',tables.BoolAtom(),(zslices,height,width),filters=compFilt)
1211 | ArrayX=h5file.create_carray("/",'X',tables.Float32Atom(),(height,width))
1212 | ArrayY=h5file.create_carray("/",'Y',tables.Float32Atom(),(height,width))
1213 | ArrayZ=h5file.create_carray("/",'Z',tables.Float32Atom(),(zslices,))
1214 | ArrayX[:]=X.copy()
1215 | ArrayY[:]=Y.copy()
1216 | ArrayZ[:]=Z.copy()
1217 | for i in range(len(Z)):
1218 | mask=maskFormula(X,Y,Z[i],extraParams)
1219 | Array3D[i,:,:]=mask*sliceFormula(X,Y,Z[i],extraParams)
1220 | # Still add GalvoCentre and PiezoCentre attributes
1221 | h5file.root.data.attrs.GalvoCentre=[X[0].mean(),Y[:,0].mean(),Z.min()]
1222 | h5file.root.data.attrs.PiezoCentre=[X[0].min(),Y[:,0].min(),Z.min()]
1223 | return
1224 |
1225 | def writeH5ColourGwl(h5name,allSliceList,allSliceCodes,allSliceZList,X,Y,Z,compression=True,zprecision=0.05):
1226 | zslices=len(Z)
1227 | height,width=X.shape
1228 | hashing=X[0,1]-X[0,0]
1229 | compFilt=None
1230 | if compression:
1231 | compFilt=tables.Filters(complevel=3)
1232 |
1233 | with tables.open_file(h5name,filters=compFilt,mode="w") as h5file:
1234 |
1235 | Array3D=h5file.create_carray("/",'data',tables.UInt8Atom(),(zslices,height,width*3),filters=compFilt)
1236 | ArrayX=h5file.create_carray("/",'X',tables.Float32Atom(),(height,width))
1237 | ArrayY=h5file.create_carray("/",'Y',tables.Float32Atom(),(height,width))
1238 | ArrayZ=h5file.create_carray("/",'Z',tables.Float32Atom(),(zslices,))
1239 | ArrayX[:]=X.copy()
1240 | ArrayY[:]=Y.copy()
1241 | ArrayZ[:]=Z.copy()
1242 | h5file.root.data.set_attr('PIXFORMAT',b'RGB8') # so it can be visualised by gigaviewer
1243 |
1244 | currZ=-1
1245 | T=np.zeros((X.shape[0],X.shape[1],3),dtype=np.uint8)
1246 | pos=0
1247 | colours=[(255,0,0),(0,255,0),(0,0,255),(120,120,120)]
1248 | for i in range(len(allSliceZList)):
1249 | if (np.abs(allSliceZList[i]-currZ)>zprecision): # first time at this height => do initialisation
1250 | pos=np.searchsorted(Z,allSliceZList[i]-zprecision)
1251 | #print(i,allSliceZList[i],currZ,pos)
1252 | Array3D[pos,:,:]=T.reshape((T.shape[0],T.shape[1]*3)) # now write old T (for that one is finished now), pos 0 will be overwritten but ok
1253 | T=np.zeros((X.shape[0],X.shape[1],3),dtype=np.uint8) # make new T ready for next layers
1254 | currZ=allSliceZList[i]
1255 |
1256 | # NEED to accumulate all images on a given height (add colours not replace)!
1257 |
1258 | profs=allSliceList[i]
1259 | li=[(convertToPixels(X,Y,profs[ind][:,:2])).reshape(-1,1,2) for ind in range(len(profs))]
1260 | #li=[profs[ind][:,:2].reshape(-1,1,2) for ind in range(len(profs))]
1261 |
1262 | #li2=(li-X.min())/hashing # Only correct when X.min() = Y.min() => should substract differently for X and Y
1263 | li3=[ np.round(litem).astype(np.int32) for litem in li]
1264 | # li=[((profs[ind][:,:2]-X.min())/(X.max()-X.min())*X.shape[0]).astype(np.int32).reshape(-1,1,2) for ind in range(len(profs))]
1265 | res=cv2.polylines(T,li3,False,colours[allSliceCodes[i]-1])
1266 | Array3D[pos,:,:]=T.reshape((T.shape[0],T.shape[1]*3)) # don't forget last slice
1267 |
1268 | def disk(radius, dtype=np.uint8):
1269 | """Generates a flat, disk-shaped structuring element.
1270 | A pixel is within the neighborhood if the euclidean distance between
1271 | it and the origin is no greater than radius.
1272 | Parameters
1273 | ----------
1274 | radius : int
1275 | The radius of the disk-shaped structuring element.
1276 | Other Parameters
1277 | ----------------
1278 | dtype : data-type
1279 | The data type of the structuring element.
1280 | Returns
1281 | -------
1282 | selem : ndarray
1283 | The structuring element where elements of the neighborhood
1284 | are 1 and 0 otherwise.
1285 | """
1286 | L = np.arange(-radius, radius + 1)
1287 | X, Y = np.meshgrid(L, L)
1288 | return np.array((X ** 2 + Y ** 2) <= radius ** 2, dtype=dtype)
1289 |
1290 |
1291 | ### Typical regex would be "drops0ts(\d+).dat", placing the brackets will make the sort based on captured integer
1292 | def findFiles(regex,dirName=os.curdir,isFloat=False):
1293 | filenames=glob.glob(os.path.join(os.path.abspath(dirName),"*"))
1294 | if len(filenames) == 0:
1295 | print("No pictures found")
1296 | return (0,None)
1297 | # two steps, first get a list of those filenames matching the given regex
1298 | newList=[]
1299 | numList=[]
1300 | for gfile in filenames:
1301 | mat=re.search(regex,gfile)
1302 | if (mat):
1303 | newList.append(gfile)
1304 | if (len(mat.groups())>0):
1305 | if (isFloat):
1306 | nr=float(mat.groups()[-1])
1307 | else:
1308 | nr=int(mat.groups()[-1])
1309 | numList.append(nr)
1310 |
1311 | # now sort the files according to their number. Always fitting regex \d+.dat to extract number for each file
1312 | if (len(numList)>0):
1313 | sI=np.argsort(numList)
1314 | sortF= [newList[i] for i in sI]
1315 | else:
1316 | newList.sort()
1317 | sortF=newList
1318 |
1319 | return sortF,np.sort(numList)
1320 |
1321 | def getSquareStitchCentres(T,writeRad,X,Y):
1322 | dX=X[0,1]-X[0,0]
1323 | dist_pix=int(round(2*writeRad/dX))
1324 | xx=np.arange(X.min(),X.max(),2*writeRad)
1325 | yy=np.arange(Y.min(),Y.max(),2*writeRad)
1326 | NX,NY=np.meshgrid(xx,yy)
1327 | pp=np.column_stack((NX.ravel(),NY.ravel()))
1328 | #NC_pix=FormulaSlicer.convertToPixels(X,Y,NC)
1329 |
1330 | #pp=calcHexCenters(amY,amX,writeRad)
1331 | # now only keep those centres which have some work to do
1332 | regions, vertices = makeVoronoiPolygons(pp)
1333 | goodRegions=[]
1334 | goodPoints=[]
1335 | for i in np.arange(len(pp)):
1336 | polygon = vertices[regions[i]]
1337 | MM=makeVoronoiMask(polygon,X,Y)
1338 | if ((MM*T).sum()!=0):
1339 | goodPoints.append(pp[i])
1340 | goodRegions.append(regions[i])
1341 |
1342 | hexPoints=np.asarray(goodPoints)
1343 | return hexPoints
1344 |
1345 | def getHexStitchCentres(T,writeRad,X,Y):
1346 | amX=int(round((X.max()-X.min())*1.2/(2*writeRad)))
1347 | amY=int(round((Y.max()-Y.min())*1.4/(2*writeRad)))
1348 | pp=calcHexCenters(amY,amX,writeRad)
1349 | # now only keep those centres which have some work to do
1350 | regions, vertices = makeVoronoiPolygons(pp)
1351 | goodRegions=[]
1352 | goodPoints=[]
1353 | for i in np.arange(len(pp)):
1354 | polygon = vertices[regions[i]]
1355 | MM=makeVoronoiMask(polygon,X,Y)
1356 | if ((MM*T).sum()!=0):
1357 | goodPoints.append(pp[i])
1358 | goodRegions.append(regions[i])
1359 |
1360 | hexPoints=np.asarray(goodPoints)
1361 | return hexPoints
1362 |
1363 | # this will return the coords of the circle centers in a tesselation
1364 | def calcHexCenters(nrows,ncols,R):
1365 | offx=R*np.sqrt(3)
1366 | offy=1.5*R
1367 | offExtra=np.sqrt(3)*R/2.0
1368 |
1369 | coordList=[]
1370 |
1371 | # draw circumscribing circles
1372 | for i in np.arange(ncols):
1373 | for j in np.arange(nrows):
1374 | if np.logical_and(j%2==1,i==0):
1375 | cx=i*offx-j%2*offExtra
1376 | cy=j*offy
1377 | coordList.append((cx,cy))
1378 | cx=i*offx+j%2*offExtra
1379 | cy=j*offy
1380 | coordList.append((cx,cy))
1381 |
1382 | coords=np.asarray(coordList)
1383 | centerx=(coords[:,0].max()+coords[:,0].min())/2.0
1384 | centery=(coords[:,1].max()+coords[:,1].min())/2.0
1385 | centCoord=coords-[centerx,centery]
1386 |
1387 | return centCoord
1388 |
1389 | def getOptimalStitchCentres(T,VoteRadius,WriteRadius,X,Y):
1390 | dPix=X[0,1]-X[0,0]
1391 | VoteRad_pix=int(round(VoteRadius/dPix))
1392 | WriteRad_pix=int(round(WriteRadius/dPix))
1393 |
1394 | In=T.copy()
1395 | centList=[]
1396 | while(In.sum()!=0):
1397 | Vote=getVoteFullCircle(In,VoteRad_pix)#,oldT,oldVote)
1398 | cp,Out=getVoteCentre(In,Vote,WriteRad_pix)
1399 | diff=(In-Out).sum()
1400 | if (diff==0): # something went wrong with differential voting => go back to absolute voting
1401 | print('Something went wrong with voting')
1402 | In=Out.copy()
1403 | centList.append(cp)
1404 | Cents=np.asarray(centList)
1405 | Xc=X[Cents[:,1],Cents[:,0]]
1406 | Yc=Y[Cents[:,1],Cents[:,0]]
1407 | points=np.column_stack((Xc,Yc)).astype(np.float64)
1408 | return points
1409 |
1410 | def getOptimalStitchCentres_Bulk(T,VoteRadius,WriteRadius,X,Y):
1411 | dPix=X[0,1]-X[0,0]
1412 | VoteRad_pix=int(round(VoteRadius/dPix))
1413 | WriteRad_pix=int(round(WriteRadius/dPix))
1414 |
1415 | In=T.copy()
1416 | centList=[]
1417 | while(In.sum()!=0):
1418 | Vote=getVoteFullCircle_Bulk(In,VoteRad_pix)#,oldT,oldVote)
1419 | cp,Out=getVoteCentre(In,Vote,WriteRad_pix)
1420 | diff=(In-Out).sum()
1421 | if (diff==0): # something went wrong with differential voting => go back to absolute voting
1422 | print('Something went wrong with voting')
1423 | In=Out.copy()
1424 | centList.append(cp)
1425 | Cents=np.asarray(centList)
1426 | Xc=X[Cents[:,1],Cents[:,0]]
1427 | Yc=Y[Cents[:,1],Cents[:,0]]
1428 | points=np.column_stack((Xc,Yc)).astype(np.float64)
1429 | return points
1430 |
1431 | def getVoteFullCircle(T,VoteRad):
1432 | Disc=disk(VoteRad)
1433 | # E=cv2.Canny(T.astype(np.uint8),1,1)
1434 | Sx=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,1,0)
1435 | Sy=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,0,1)
1436 | SS,Sang=cv2.cartToPolar(Sx,Sy)
1437 | E=np.where(SS>1,1,0)
1438 | RE=signal.fftconvolve(E.astype(np.uint8),Disc,'same')
1439 | return RE/Disc.sum()
1440 |
1441 | # apply gaussian filter on disk. But make sure that you are not voting outside of original disk
1442 | # => should probably rescale the disk. => make smaller disk in large enough matrix and apply
1443 | # gaussian on that.
1444 | def getVoteFullCircle_gaus(T,VoteRad,gaus):
1445 | Disc=disk(VoteRad)
1446 | # E=cv2.Canny(T.astype(np.uint8),1,1)
1447 | Sx=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,1,0)
1448 | Sy=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,0,1)
1449 | SS,Sang=cv2.cartToPolar(Sx,Sy)
1450 | E=np.where(SS>1,1,0)
1451 | RE=signal.fftconvolve(E.astype(np.uint8),Disc,'same')
1452 | return RE/Disc.sum()
1453 |
1454 | def getVoteFullCircle_Bulk(T,VoteRad):
1455 | Disc=disk(VoteRad)
1456 | # E=cv2.Canny(T.astype(np.uint8),1,1)
1457 | #~ Sx=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,1,0)
1458 | #~ Sy=cv2.Sobel(T.astype(np.uint8),cv2.CV_64F,0,1)
1459 | #~ SS,Sang=cv2.cartToPolar(Sx,Sy)
1460 | #~ E=np.where(SS>1,1,0)
1461 | RE=signal.fftconvolve(T.astype(np.uint8),Disc,'same')
1462 | return RE/Disc.sum()
1463 |
1464 |
1465 | # This will cast votes in a circle of VoteRadius around all contourpoints
1466 | def getVoteFullCircle_old(T2,VoteRad,T=np.zeros((1,1)),Vote=np.zeros((1,1))):
1467 | if (T.sum()==0):
1468 | P=np.zeros(T2.shape,dtype=np.uint8)
1469 | R=cv2.findContours(T2.astype(np.uint8),cv2.RETR_LIST,cv2.CHAIN_APPROX_NONE)
1470 | Res=np.zeros(T2.shape,dtype=np.int32)
1471 | for CC in R[1]:
1472 | CC=CC.reshape((-1,2))
1473 |
1474 | for cc in CC:
1475 | P[:]=0
1476 | # cc=CC[150]
1477 | P=cv2.circle(P,(cc[0],cc[1]),VoteRad,1,-1)
1478 | #Pb=cv2.GaussianBlur(P,(kernel,kernel),0)
1479 | Res=Res+P
1480 | return Res
1481 | else:
1482 | try:
1483 | Tdif=T-T2
1484 | Cdif=cv2.findContours((Tdif).astype(np.uint8),cv2.RETR_LIST,cv2.CHAIN_APPROX_NONE)
1485 | Cold=cv2.findContours((T).astype(np.uint8),cv2.RETR_LIST,cv2.CHAIN_APPROX_NONE)
1486 |
1487 |
1488 | AllC_old=Cold[1][0].reshape((-1,2))
1489 | for nC in Cold[1][1:]:
1490 | AllC_old=np.row_stack((AllC_old,nC.reshape((-1,2))))
1491 | #print(AllC_old.shape)
1492 |
1493 | #print(T2.sum(),len(Cdif[1]),Tdif.sum())
1494 | AllC_dif=Cdif[1][0].reshape((-1,2))
1495 | for nC in Cdif[1][1:]:
1496 | AllC_dif=np.row_stack((AllC_dif,nC.reshape((-1,2))))
1497 |
1498 |
1499 | P=np.zeros(T.shape,dtype=np.uint8)
1500 | Sub=np.zeros(T.shape,dtype=np.int32)
1501 | Add=np.zeros(T.shape,dtype=np.int32)
1502 | for difE in AllC_dif:
1503 | dis=np.abs(AllC_old-difE)
1504 | if (len(np.argwhere(dis[:,0]+dis[:,1]<0.5))>0):
1505 | P[:]=0
1506 | P=cv2.circle(P,(difE[0],difE[1]),VoteRad,1,-1)
1507 | Sub=Sub+P
1508 | else:
1509 | P[:]=0
1510 | P=cv2.circle(P,(difE[0],difE[1]),VoteRad,1,-1)
1511 | Add=Add+P
1512 |
1513 | NewVote=Vote.astype(np.int32)+Add-Sub
1514 | except: # something went wrong with the differential voting system => use classic voting!
1515 | NewVote=getVoteFullCircle(T2,VoteRad)
1516 | return NewVote
1517 |
1518 | # This will take the original matrix + the vote => find the elected
1519 | # centre point and substract a zone with WriteRadius from the material matrix T => return T2
1520 | def getVoteCentre(T,Vote,WriteRad):
1521 | mm=np.argmax(Vote*T)
1522 | #mm=np.argmax(Vote) # While sometimes better, it can get stuck (happened for free-standing)
1523 | xmax=mm%Vote.shape[1]
1524 | ymax=np.int(mm/Vote.shape[1])
1525 | R2=np.zeros(T.shape,dtype=np.float)
1526 | R2=cv2.circle(R2,(xmax,ymax),WriteRad,1,-1)
1527 | T2=np.where(T-R2>0,1,0)
1528 | return np.array([xmax,ymax]),T2
1529 |
1530 |
1531 | def makeVoronoiPolygons(points):
1532 | vor = Voronoi(points)
1533 | regions, vertices = voronoi_finite_polygons_2d(vor)
1534 | return regions,vertices
1535 |
1536 | def voronoi_finite_polygons_2d(vor, radius=None):
1537 | """
1538 | Reconstruct infinite voronoi regions in a 2D diagram to finite
1539 | regions.
1540 |
1541 | Parameters
1542 | ----------
1543 | vor : Voronoi
1544 | Input diagram
1545 | radius : float, optional
1546 | Distance to 'points at infinity'.
1547 |
1548 | Returns
1549 | -------
1550 | regions : list of tuples
1551 | Indices of vertices in each revised Voronoi regions.
1552 | vertices : list of tuples
1553 | Coordinates for revised Voronoi vertices. Same as coordinates
1554 | of input vertices, with 'points at infinity' appended to the
1555 | end.
1556 |
1557 | """
1558 |
1559 | if vor.points.shape[1] != 2:
1560 | raise ValueError("Requires 2D input")
1561 |
1562 | new_regions = []
1563 | new_vertices = vor.vertices.tolist()
1564 |
1565 | center = vor.points.mean(axis=0)
1566 | if radius is None:
1567 | radius = vor.points.ptp().max()*2
1568 |
1569 | # Construct a map containing all ridges for a given point
1570 | all_ridges = {}
1571 | for (p1, p2), (v1, v2) in zip(vor.ridge_points, vor.ridge_vertices):
1572 | all_ridges.setdefault(p1, []).append((p2, v1, v2))
1573 | all_ridges.setdefault(p2, []).append((p1, v1, v2))
1574 |
1575 | # Reconstruct infinite regions
1576 | for p1, region in enumerate(vor.point_region):
1577 | vertices = vor.regions[region]
1578 |
1579 | if all([v >= 0 for v in vertices]):
1580 | # finite region
1581 | new_regions.append(vertices)
1582 | continue
1583 |
1584 | # reconstruct a non-finite region
1585 | ridges = all_ridges[p1]
1586 | new_region = [v for v in vertices if v >= 0]
1587 |
1588 | for p2, v1, v2 in ridges:
1589 | if v2 < 0:
1590 | v1, v2 = v2, v1
1591 | if v1 >= 0:
1592 | # finite ridge: already in the region
1593 | continue
1594 |
1595 | # Compute the missing endpoint of an infinite ridge
1596 |
1597 | t = vor.points[p2] - vor.points[p1] # tangent
1598 | t /= np.linalg.norm(t)
1599 | n = np.array([-t[1], t[0]]) # normal
1600 |
1601 | midpoint = vor.points[[p1, p2]].mean(axis=0)
1602 | direction = np.sign(np.dot(midpoint - center, n)) * n
1603 | far_point = vor.vertices[v2] + direction * radius
1604 |
1605 | new_region.append(len(new_vertices))
1606 | new_vertices.append(far_point.tolist())
1607 |
1608 | # sort region counterclockwise
1609 | vs = np.asarray([new_vertices[v] for v in new_region])
1610 | c = vs.mean(axis=0)
1611 | angles = np.arctan2(vs[:,1] - c[1], vs[:,0] - c[0])
1612 | new_region = np.array(new_region)[np.argsort(angles)]
1613 |
1614 | # finish
1615 | new_regions.append(new_region.tolist())
1616 |
1617 | return new_regions, np.asarray(new_vertices)
1618 |
1619 | def convertToPixels(X,Y,realList):
1620 | X_start = X[0,0]
1621 | X_delta = X[0,1]-X[0,0]
1622 | Y_start = Y[0,0]
1623 | Y_delta = Y[1,0]-Y[0,0]
1624 |
1625 | if (realList.shape[1]==2):
1626 | CoordX_pix=(np.round((realList[:,0]-X_start)/X_delta)).astype(np.int)
1627 | CoordY_pix=(np.round((realList[:,1]-Y_start)/Y_delta)).astype(np.int)
1628 | coords_pix=np.column_stack((CoordX_pix,CoordY_pix))
1629 | return coords_pix
1630 | elif (realList.shape[1]==1):
1631 | return (np.round((realList)/X_delta)).astype(np.int) # can only assume X_delta and Y_delta are the same
1632 |
1633 |
1634 | def makeVoronoiMask(polygon,X,Y):
1635 | M=np.zeros(X.shape,dtype=np.uint8)
1636 | # sssx=[np.argmin(np.abs(xx-ppp[0])) for ppp in polygon]
1637 | # sssy=[np.argmin(np.abs(yy-ppp[1])) for ppp in polygon]
1638 | # kj=np.column_stack((sssx,sssy)) # or other order!!
1639 | kj=convertToPixels(X,Y,polygon)
1640 | M=cv2.fillPoly(M,[kj.astype(np.int32)],1,100)
1641 | return M
1642 |
1643 | def makeCircularMask(centre,radius,xx,yy):
1644 | M=np.zeros((len(yy),len(xx)),dtype=np.uint8)
1645 | kj=convertToPixels(X,Y,np.asarray(centre))
1646 | dpix=xx[1]-xx[0]
1647 | rad_pix=int(round(radius/dpix))
1648 | M=cv2.circle(M,(kj[0],kj[1]),rad_pix,1,-1)
1649 | return M
1650 |
1651 | def getInfill(points,writeRadius,T,X,Y):
1652 | regions, vertices = makeVoronoiPolygons(points)
1653 | myL=[]
1654 | for i in np.arange(len(points)):
1655 | polygon = vertices[regions[i]]
1656 | MM=makeVoronoiMask(polygon,X,Y)
1657 | myMaterial=(MM*T).sum()
1658 | myL.append(myMaterial)
1659 |
1660 | return np.asarray(myL)
1661 |
1662 | def writeTSPfile(filename,pp):
1663 | import scipy.spatial.distance as dist
1664 | DD=dist.pdist(pp,'cityblock')
1665 |
1666 |
1667 | F=open(filename,mode='w')
1668 | F.write('NAME: {}\n'.format(filename[:-4]))
1669 | F.write('TYPE: TSP\n')
1670 | F.write('COMMENT: Nanoscribe Blocks in Ham Path\n')
1671 | F.write('DIMENSION: {}\n'.format(len(pp)+1))
1672 | F.write('EDGE_WEIGHT_TYPE: EXPLICIT\n')
1673 | F.write('EDGE_WEIGHT_FORMAT: UPPER_ROW\n')
1674 | F.write('DISPLAY_DATA_TYPE: NO_DISPLAY\n')
1675 | F.write('EDGE_WEIGHT_SECTION\n')
1676 |
1677 | pcounter=0
1678 | st=''
1679 | for cc in np.arange(len(pp)): # start with a zero line to transform into ham path problem
1680 | st=st+'0 '
1681 | F.write(st+'\n')
1682 |
1683 | for c in np.arange(len(pp)-1,0,-1):
1684 | st=''
1685 | for i in np.arange(c):
1686 | st=st+'{:d} '.format(int(DD[pcounter]))
1687 | pcounter=pcounter+1
1688 | F.write(st+'\n')
1689 |
1690 |
1691 | F.write('EOF')
1692 |
1693 | F.close()
1694 | return
1695 |
1696 | def readTSPfile(filename):
1697 | Sol=open(filename,mode='r').readlines()
1698 | OrdList=[]
1699 | for line in Sol[1:]:
1700 | els=line.split()
1701 | for el in els:
1702 | OrdList.append(int(el))
1703 | OO=np.asarray(OrdList)
1704 | if (OO[0]==0):
1705 | OO=OO[1:]-1 # cut away the fictitious first point
1706 | else:
1707 | print('First point is not the fictitious one: investigate')
1708 | return OO
1709 |
1710 | def TSPnearest(pp):
1711 | import scipy.spatial.distance as dist
1712 | DD=dist.pdist(pp,'cityblock')
1713 | Dmat=dist.squareform(DD)
1714 | order=(np.argsort(Dmat,axis=1))[:,1:] # cut out the first point as it is always the diagonal point
1715 |
1716 | pathList=[]
1717 | distList=[]
1718 |
1719 | for stp in np.arange(len(pp)):
1720 | visList=[stp]
1721 | dist=0
1722 | currP=stp
1723 | while (len(visList)0,1,0)
1765 | goodLabs=np.unique(RemMask*LL)[1:]
1766 |
1767 | T2=np.where(T-RemMask>0,1,0)
1768 | return np.array([xmax,ymax]),T2,goodLabs,crossing
1769 |
1770 | def makeFreeMaterialMatrix(Tloc,seeds):
1771 | TestIm=np.zeros(Tloc.shape,np.uint8)
1772 | Mask=np.zeros((Tloc.shape[0]+2,Tloc.shape[1]+2),dtype=np.uint8)
1773 | Mask[1:-1,1:-1]=Tloc[:]
1774 | Mask=1-Mask
1775 | for seed in seeds:
1776 | rr,TestIm,Mask,rect=cv2.floodFill(TestIm,Mask,(seed[0].astype(np.int32),seed[1].astype(np.int32)),1)
1777 | return TestIm
1778 |
1779 | def getOptimalFreestandingStitchCentres(T,VoteRadius,WriteRadius,X,Y,closing=0):
1780 | dPix=X[0,1]-X[0,0]
1781 | VoteRad_pix=int(round(VoteRadius/dPix))
1782 | WriteRad_pix=int(round(WriteRadius/dPix))
1783 |
1784 | L,n=im.label(T,np.ones((3,3)))
1785 | posLabels=[]
1786 | for i in np.arange(1,n+1):
1787 | Lcoords=np.argwhere(L==i)
1788 | meanLabel=Lcoords.mean(axis=0)
1789 | posLabels.append([meanLabel[1],meanLabel[0],i])
1790 | Pos=np.round(np.asarray(posLabels)).astype(np.int)
1791 | #print(Pos.shape,X.shape)
1792 | realPos=np.column_stack((X[0][Pos[:,0]],Y[:,0][Pos[:,1]]))
1793 | if (len(Pos)==0):
1794 | print('Big problem with algorithm, check debugoutput')
1795 | return L,posLabels,T
1796 |
1797 |
1798 | In=T.copy()
1799 | if (closing==0):
1800 | Voters=In.copy()
1801 | else:
1802 | #Voters=im.binary_closing(In,iterations=closing).astype(np.uint8)
1803 | Voters=im.binary_dilation(In,iterations=closing).astype(np.uint8)
1804 |
1805 | centList=[]
1806 | labPosList=[]
1807 | labsList=[]
1808 | #VoteList=[]
1809 | while(In.sum()!=0):
1810 | Vote=getVoteFullCircle(Voters,VoteRad_pix)
1811 | # VoteList.append(Vote)
1812 | cp,Out,goodLabs,crossing=getVoteCentre_freestanding(In,L,Vote,WriteRad_pix,True)
1813 | diff=(In-Out).sum()
1814 | if (diff==0): # something went wrong with differential voting => go back to absolute voting
1815 | print('Something wrong with voting, will do plan B')
1816 | TempIn=np.where(L==crossing[0],1,0)
1817 | Vote=getVoteFullCircle(TempIn,VoteRad_pix)
1818 | cp,Out,goodLabs,crossing=getVoteCentre_freestanding(In,L,Vote,WriteRad_pix,True)
1819 | diff=(In-Out).sum()
1820 | if (diff==0):
1821 | print('Do not know what to do, will abort')
1822 | return In,Voters,Vote
1823 | break
1824 | In=Out.copy()
1825 | if (closing==0):
1826 | Voters=In.copy()
1827 | else:
1828 | Voters=im.binary_closing(In,iterations=closing).astype(np.uint8)
1829 | centList.append(cp)
1830 | labPosList.append(realPos[goodLabs-1])
1831 | labsList.append(goodLabs)
1832 | Cents=np.asarray(centList)
1833 | Xc=X[Cents[:,1],Cents[:,0]]
1834 | Yc=Y[Cents[:,1],Cents[:,0]]
1835 | points=np.column_stack((Xc,Yc)).astype(np.float64)
1836 | return points,labPosList,labsList#,VoteList,Voters
1837 |
--------------------------------------------------------------------------------
/Adaptive Stitching with Nanoscribe/Adaptive-Stitching-Nanoscribe.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "# Adaptive Stitching\n",
8 | "This code performs adaptive stitching for randomly spaced microlenses that are multifocal and have aberrations added to them. The adaptive stitching is needed for the microlenses to be correctly 3D printed with Nanoscribe. The code places the stitching artifacts at the boundaries of the microlenses and thus maximizes optical quality. \n",
9 | "\n",
10 | "The demonstration here will be for the 36 multifocal microlenses with astigmatism and tilt added. This was used in the paper: **Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy**"
11 | ]
12 | },
13 | {
14 | "cell_type": "code",
15 | "execution_count": 1,
16 | "metadata": {},
17 | "outputs": [
18 | {
19 | "name": "stdout",
20 | "output_type": "stream",
21 | "text": [
22 | "Using matplotlib backend: agg\n",
23 | "Populating the interactive namespace from numpy and matplotlib\n"
24 | ]
25 | }
26 | ],
27 | "source": [
28 | "%pylab\n",
29 | "%matplotlib inline\n",
30 | "import matplotlib.pyplot as plt\n",
31 | "import TipSlicerUpdated\n",
32 | "import random\n",
33 | "import matplotlib.pyplot as plt\n",
34 | "import numpy as np\n",
35 | "import os\n",
36 | "import scipy.ndimage.morphology as im"
37 | ]
38 | },
39 | {
40 | "cell_type": "code",
41 | "execution_count": 2,
42 | "metadata": {},
43 | "outputs": [],
44 | "source": [
45 | "# this is the internal potential calculation for a single lens extracted for reuse in the masking def\n",
46 | "def AberPotential(X,Y,z,params):\n",
47 | " xpos,ypos,rlist,r_lenslet,height,aperR,indexPoint,Z=params\n",
48 | " Xn = X/1800.\n",
49 | " Yn = Y/1800.\n",
50 | " n=indexPoint\n",
51 | " Xnzern=Xn- xpos[n]/1800.\n",
52 | " Ynzern=Yn- ypos[n]/1800.\n",
53 | " r = np.sqrt(np.square(Xnzern) + np.square(Ynzern))\n",
54 | " Z2 = Z[n,0] * 2.*Xnzern\n",
55 | " Z3 = Z[n,1] * 2.*Ynzern\n",
56 | " Z5 = Z[n,2] * 2.*np.sqrt(6.)*Xnzern*Ynzern\n",
57 | " Z6 = Z[n,3] * np.sqrt(6.)*(Xnzern**2-Ynzern**2)\n",
58 | " #Z12 = Z[n,4] * np.sqrt(10.)*(Xnzern**2-Ynzern**2)*(4*r**2-3)\n",
59 | " #Z13 = Z[n,5] * 2*np.sqrt(10.)*Xnzern*Ynzern*(4*r**2-3)\n",
60 | " #Z23 = Z[n,6] * 2*np.sqrt(14.)*Xnzern*Ynzern*(15*r**4-20*r**2+6)\n",
61 | " #Z24 = Z[n,7] * np.sqrt(14.)*(Xnzern**2-Ynzern**2)*(15*r**4-20*r**2+6)\n",
62 | " ZW = Z5+Z6+Z2+Z3#+Z12+Z13+Z23+Z24\n",
63 | " sph1 = ZW+np.real(np.sqrt(0j+rlist[n]**2 - (X-xpos[n])**2 - (Y-ypos[n])**2))-np.real(np.sqrt(0j+rlist[n]**2-r_lenslet**2))-(z-height)\n",
64 | " return sph1\n",
65 | "\n",
66 | "\n",
67 | "def sliceFormulaAber(X,Y,z,params):\n",
68 | " xpos,ypos,rlist,r_lenslet,height,aperR,indexPoint,aber=params\n",
69 | " aper=np.sqrt(X**2+Y**2) <= aperR\n",
70 | " Z=aber\n",
71 | " if z<=height: \n",
72 | " return aper\n",
73 | " else: \n",
74 | " T = np.zeros(len(X))\n",
75 | " inds=range(len(xpos))\n",
76 | " if (indexPoint>-1):\n",
77 | " inds=[indexPoint]\n",
78 | " for n in inds:\n",
79 | " sph1=AberPotential(X,Y,z,[xpos,ypos,rlist,r_lenslet,height,aperR,n,aber])\n",
80 | " T = np.maximum(T,sph1)\n",
81 | " T=(T>0)\n",
82 | " return (T)*aper\n",
83 | " \n",
84 | "def maskFormulaAber(X,Y,z,params):\n",
85 | " xpos,ypos,rlist,r_lenslet,height,aperR,indexPoint,aber=params\n",
86 | " aper=np.sqrt(X**2+Y**2) <= aperR\n",
87 | " if (z<=height):\n",
88 | " z=height \n",
89 | " DD=np.zeros((len(xpos),X.shape[0],X.shape[1]))\n",
90 | " for i in np.arange(len(DD)):\n",
91 | " DD[i]=-AberPotential(X,Y,z,[xpos,ypos,rlist,r_lenslet,height,aperR,i,aber])*aper\n",
92 | " Div=np.argmin(DD,axis=0)\n",
93 | " if (indexPoint==-1):\n",
94 | " return Div\n",
95 | " else:\n",
96 | " return np.where(Div==indexPoint,1,0)*aper\n",
97 | "\n",
98 | "# extract the centres from the structure itself\n",
99 | "def getCentresAber(X,Y,z,params):\n",
100 | " xpos,ypos,rlist,r_lenslet,height,aperR,indexPoint,aber=params\n",
101 | " aper=np.sqrt(X**2+Y**2) <= aperR\n",
102 | " centList=[]\n",
103 | " for i in np.arange(len(xpos)):\n",
104 | " DD=-AberPotential(X,Y,z,[xpos,ypos,rlist,r_lenslet,height,aperR,i,aber])\n",
105 | " xc,yc=np.unravel_index(np.argmin(DD*aper),DD.shape)\n",
106 | " centList.append([yc,xc])\n",
107 | " Cents=np.asarray(centList)\n",
108 | " return Cents"
109 | ]
110 | },
111 | {
112 | "cell_type": "markdown",
113 | "metadata": {},
114 | "source": [
115 | "# Define microlens array parameters"
116 | ]
117 | },
118 | {
119 | "cell_type": "code",
120 | "execution_count": 3,
121 | "metadata": {},
122 | "outputs": [],
123 | "source": [
124 | "#all units are in microns\n",
125 | "r_lenslet=200 #average diameter of each microlens\n",
126 | "samples = (1800,1800) #how many samples\n",
127 | "xgrng = np.array((-900, 900)) #physical dimension of the microlens array in microns (1.8 mm here)\n",
128 | "xgrng2 = np.array((-1800, 1800)) #in microns\n",
129 | "ygrng=xgrng\n",
130 | "ygrng2=xgrng2\n",
131 | "\n",
132 | "\n",
133 | "yg = np.linspace(ygrng[0], ygrng[1], samples[0])\n",
134 | "xg=yg\n",
135 | "px=xg[1] - xg[0]\n",
136 | "py=px\n",
137 | "X,Y= np.meshgrid(xg,yg)\n",
138 | "\n",
139 | "yg2 = np.linspace(ygrng2[0], ygrng2[1], samples[0])\n",
140 | "xg2=yg2\n",
141 | "px2=xg2[1] - xg2[0]\n",
142 | "py2=px2\n",
143 | "X2,Y2= np.meshgrid(xg2,yg2)\n",
144 | "\n",
145 | "Xn = X2/np.max(X2)\n",
146 | "Yn = Y2/np.max(Y2)\n",
147 | "\n",
148 | "# xposition list in mm\n",
149 | "xpos=np.array([ 0.43641585, 0.29636857, 0.21274264, 0.6394599 , 0.65087426,\n",
150 | " -0.49792665, -0.11775845, 0.59266114, -0.1886654 , -0.45093933,\n",
151 | " -0.06002499, 0.49039292, 0.18374896, 0.32612935, 0.5355058 ,\n",
152 | " -0.07172677, 0.75601995, -0.27811506, -0.24367642, -0.22418435,\n",
153 | " 0.22841486, -0.67916805, -0.6805813 , -0.00778827, 0.8232382 ,\n",
154 | " -0.30767533, 0.19155966, 0.46077853, 0.10799856, -0.23324046,\n",
155 | " -0.4479409 , 0.13673823, -0.42627445, -0.02470369, -0.46301848,\n",
156 | " -0.6288782 ], dtype=float32)\n",
157 | "# yposition list in mm\n",
158 | "ypos=np.array([-0.0961623 , -0.590606 , 0.07416802, 0.08558229, -0.2063948 ,\n",
159 | " 0.22398773, -0.66743815, -0.4089036 , -0.48776114, -0.01421136,\n",
160 | " 0.48614654, 0.47054946, -0.29256105, -0.4106429 , -0.5639799 ,\n",
161 | " 0.6772651 , 0.26205382, 0.5754221 , 0.10239594, -0.2096889 ,\n",
162 | " 0.47864714, -0.39620727, 0.18052123, 0.17819166, 0.07138762,\n",
163 | " -0.68207985, -0.7970757 , 0.18449856, -0.4582133 , 0.37643924,\n",
164 | " 0.50502807, 0.66083544, -0.18756227, -0.07771606, -0.5855127 ,\n",
165 | " -0.00837374], dtype=float32)\n",
166 | "#radius of curvature in mm\n",
167 | "rlist=np.array([ 6.306523 , 6.453186 , 16.322765 , 7.4996486, 12.613046 ,\n",
168 | " 5.780979 , 12.064652 , 6.032326 , 17.342937 , 6.6068335,\n",
169 | " 7.1150513, 19.8205 , 8.408697 , 11.09948 , 7.9282 ,\n",
170 | " 9.91025 , 8.161383 , 13.87435 , 7.3022895, 11.561958 ,\n",
171 | " 14.604579 , 9.249567 , 9.568518 , 6.767976 , 5.903979 ,\n",
172 | " 7.707972 , 18.499134 , 10.672577 , 6.937175 , 6.1663775,\n",
173 | " 15.415944 , 10.277296 , 8.671469 , 8.951194 , 5.663 ,\n",
174 | " 13.213667 ],dtype=float32) \n",
175 | "\n",
176 | "#zernike list in mm\n",
177 | "zernlist=np.array([[-3.03727644e-03, 3.45289009e-03, -8.50316486e-04,\n",
178 | " -9.94477305e-04],\n",
179 | " [ 2.08902825e-03, -5.25590964e-03, -5.57003869e-03,\n",
180 | " 1.66087621e-03],\n",
181 | " [-1.54656521e-03, -7.22034927e-03, -1.47716681e-04,\n",
182 | " 1.33255846e-03],\n",
183 | " [-6.46650698e-03, -1.09545095e-03, -1.48787047e-03,\n",
184 | " 2.08820638e-04],\n",
185 | " [ 4.51811217e-03, -1.51357264e-03, 1.89111437e-04,\n",
186 | " 3.22496460e-04],\n",
187 | " [-1.23638997e-03, 1.33150117e-03, -5.25358878e-03,\n",
188 | " 9.76655632e-03],\n",
189 | " [ 4.36959649e-03, -7.47654727e-03, 3.64801125e-03,\n",
190 | " -1.86670839e-03],\n",
191 | " [-1.29632989e-03, -8.81011318e-03, -2.17094715e-03,\n",
192 | " 5.12582343e-03],\n",
193 | " [-3.18442448e-03, -7.02016230e-04, -7.80638075e-04,\n",
194 | " 8.95439996e-04],\n",
195 | " [ 1.33037404e-03, -4.07017302e-03, 1.83174980e-03,\n",
196 | " 4.63185087e-03],\n",
197 | " [ 6.37175108e-04, 3.62162525e-03, -1.38857099e-03,\n",
198 | " -1.69407239e-03],\n",
199 | " [ 4.18911048e-04, 2.99920258e-03, -6.04745280e-03,\n",
200 | " -2.02366011e-03],\n",
201 | " [ 7.84681237e-04, 2.57954700e-03, 8.30374379e-03,\n",
202 | " -5.63086197e-03],\n",
203 | " [-9.27667075e-04, 7.60378689e-03, 5.11155883e-03,\n",
204 | " -2.85442872e-03],\n",
205 | " [ 3.69066279e-03, 1.44687016e-03, 3.64636071e-03,\n",
206 | " -1.35618821e-03],\n",
207 | " [ 3.47679667e-03, 9.66886338e-03, 6.05431778e-05,\n",
208 | " 2.10903143e-03],\n",
209 | " [-7.06169615e-03, 1.05272122e-02, -2.37144646e-03,\n",
210 | " 2.14290898e-03],\n",
211 | " [-3.13660316e-03, 7.06096133e-03, -8.40417400e-04,\n",
212 | " -2.18926976e-03],\n",
213 | " [ 7.11004948e-03, 2.00309485e-04, 1.43178422e-02,\n",
214 | " 9.40302107e-03],\n",
215 | " [ 5.73336380e-03, 4.81330231e-03, -6.11291872e-03,\n",
216 | " -5.15368767e-03],\n",
217 | " [-2.86504888e-04, -1.82941454e-04, 8.74243851e-04,\n",
218 | " 1.06786657e-03],\n",
219 | " [-4.43635369e-03, 3.36597041e-05, -3.35925579e-04,\n",
220 | " -2.17877914e-06],\n",
221 | " [-4.91749262e-03, 5.81631484e-03, 6.35021715e-04,\n",
222 | " 1.07655076e-04],\n",
223 | " [-7.96477543e-04, 1.53168791e-03, 7.15573411e-03,\n",
224 | " -7.31880311e-03],\n",
225 | " [ 5.98072400e-03, 1.42488685e-02, -8.81085114e-04,\n",
226 | " -4.13347734e-03],\n",
227 | " [-4.52623982e-03, -2.22414453e-03, 2.17066554e-05,\n",
228 | " -1.89099193e-03],\n",
229 | " [-3.83377355e-03, 6.57709083e-04, 6.44097291e-03,\n",
230 | " 1.02351094e-03],\n",
231 | " [-3.02775938e-04, 2.83448049e-03, 1.72653550e-03,\n",
232 | " -2.10265117e-03],\n",
233 | " [ 1.72412605e-03, -8.58071493e-04, 6.27913105e-04,\n",
234 | " -4.08340804e-03],\n",
235 | " [ 2.68175127e-03, -1.63576938e-03, -9.31847002e-03,\n",
236 | " -1.93845030e-04],\n",
237 | " [-5.83293755e-03, 1.95736508e-03, -2.00207127e-04,\n",
238 | " -1.04814884e-03],\n",
239 | " [ 3.55686457e-03, 7.30788289e-03, -9.41661303e-04,\n",
240 | " 4.80500096e-03],\n",
241 | " [ 6.76476862e-04, -2.14314481e-04, -7.66097510e-04,\n",
242 | " -2.71079480e-03],\n",
243 | " [-3.65834520e-03, 7.38649350e-03, 1.80285275e-02,\n",
244 | " -1.13024702e-02],\n",
245 | " [-7.70746637e-03, -1.72380230e-03, 5.40864607e-03,\n",
246 | " -1.24156917e-03],\n",
247 | " [-4.90240427e-03, -6.24271634e-04, 2.45964224e-03,\n",
248 | " -1.91784347e-04]],dtype=float32)\n",
249 | " \n",
250 | "\n",
251 | "\n",
252 | "Nlenslets=36 #number of microlenses\n",
253 | "#convert from mm to microns\n",
254 | "xpos=np.reshape(xpos,(36))*1000\n",
255 | "ypos=np.reshape(ypos,(36))*1000\n",
256 | "rlist=np.reshape(rlist,(36))*1000\n",
257 | "zernlist=zernlist*1000\n",
258 | "\n",
259 | "height=50 #how much of support material to be printed under the phase mask (in microns)\n",
260 | "aperR=900 #radius of phase mask (collection of microlenses)"
261 | ]
262 | },
263 | {
264 | "cell_type": "code",
265 | "execution_count": 4,
266 | "metadata": {},
267 | "outputs": [],
268 | "source": [
269 | "X3,Y3=np.meshgrid(np.arange(-1000,1000),np.arange(-1000,1000))"
270 | ]
271 | },
272 | {
273 | "cell_type": "markdown",
274 | "metadata": {},
275 | "source": [
276 | "# Let's visualize our microlens array after adaptive stitching"
277 | ]
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": 8,
282 | "metadata": {},
283 | "outputs": [
284 | {
285 | "data": {
286 | "text/plain": [
287 | ""
288 | ]
289 | },
290 | "execution_count": 8,
291 | "metadata": {},
292 | "output_type": "execute_result"
293 | },
294 | {
295 | "data": {
296 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAQgAAAD8CAYAAACLgjpEAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjMuMiwgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy8vihELAAAACXBIWXMAAAsTAAALEwEAmpwYAAAuUElEQVR4nO3deZgc9X3n8fe3qq+5D82MbiEhRoAwxgiBccAkGMzlA+MQB9tr45gN6/iIY3vXC5tnH3s33l07uzEbP8k6wYHYzjoGmzhrknjtAD7YAAIkDiEQQiOha3SN5j77qPruH1UtWqPpme6uljTH9/U880xPdXV1ldTzmd/vV1W/r6gqxhgzFedM74AxZvaygDDGFGUBYYwpygLCGFOUBYQxpigLCGNMUac9IETkBhHZISJdInLX6X5/Y0zp5HReByEiLvAa8E7gAPAs8EFVfeW07YQxpmSnuwVxGdClqrtVNQM8ANx8mvfBGFOi2Gl+v+XA/oKfDwBvLVxBRO4E7gSoq6u75Lzzzjt9e2fMArRly5Zjqto+1XOnOyBmpKr3AvcCbNy4UTdv3nyG98iY+U1E9hZ77nR3MbqBlQU/rwiXGWNmodMdEM8CnSKyRkQSwG3Aw6d5H4wxJTqtXQxVzYnIp4GfAS5wv6q+fDr3wRhTutM+BqGqPwF+crrf1xhTPruS0hhTlAWEMaYoCwhjTFEWEMaYoiwgjDFFWUAYY4qygDDGFGUBYYwpygLCGFOUBYQxpigLCGNMURYQxpiiLCCMMUVZQBhjirKAMMYUVXFAiMhKEfmFiLwiIi+LyGfD5a0i8oiI7Ay/t4TLRUS+EdbD2CoiG6p1EMaYUyNKCyIHfEFV1wOXA58SkfXAXcBjqtoJPBb+DHAj0Bl+3Ql8M8J7G2NOg4oDQlUPqepz4eNhYDvBtPY3A98JV/sO8L7w8c3AdzWwCWgWkaWVvr8x5tSryhiEiKwGLgaeBhar6qHwqcPA4vDxVDUxllfj/Y0xp0bkgBCReuDvgD9Q1aHC5zSo61dWbT8RuVNENovI5p6enqi7Z4yJIFJAiEicIBy+p6o/ChcfyXcdwu9Hw+Ul1cRQ1XtVdaOqbmxvn7LYjzHmNIlyFkOA+4Dtqvr1gqceBm4PH98O/Lhg+UfDsxmXA4MFXRFjzCwUZdr7K4CPAC+JyAvhsv8AfBX4gYjcAewFPhA+9xPgJqALGAN+J8J7G2NOg4oDQlX/BZAiT18zxfoKfKrS9zPGnH52JaUxpigLCGNMURYQxpiiLCCMMUVZQBhjirKAMMYUZQFhjCnKAsIYU5QFhDGmKAsIY0xRFhDGmKIsIIwxRVlAGGOKsoAwxhRlAWGMKcoCwhhTVDUmrXVF5HkR+cfw5zUi8nRYIOdBEUmEy5Phz13h86ujvrcx5tSqRgviswQ1MfK+BtyjqucA/cAd4fI7gP5w+T3hesaYWSzqrNYrgHcBfxX+LMA7gIfCVSYXzskX1HkIuCZc3xgzS0VtQfxP4IuAH/68CBhQ1Vz4c2FxnOOFc8LnB8P1T2B1MYyZPaJMe/9u4Kiqbqni/lhdDGNmkajT3r9XRG4CUkAj8KcENTdjYSuhsDhOvnDOARGJAU1Ab4T3N8acYlGK996tqitUdTVwG/BzVf0w8Avg1nC1yYVz8gV1bg3XL6ssnzHm9DoV10H8e+DzItJFMMZwX7j8PmBRuPzzwF2n4L2NMVUUpYtxnKr+Evhl+Hg3cNkU60wAv1WN9zPGnB52JaWpmmuv+ArXXvGVM70bpoqq0oIwC0Opv/zXXvEVcISdH05wxVteO+n5v738W9XeNXOKWECYKUVuCfjKoi0uExfGSLm5E5760KbfPWl1C43ZyQLCHHfT478PwI5nVrNGgIjnmJr2ZHh9oJXzFx2dcd3C0LCwmD0sIBa4fCgUajy/l8wvmkgMZCJt2x3P0b+7FUoIiLxjE3Ws+8rXWfNQP7zezc8G74+0DyYaC4gFaqpgyFtcP8KB89tY8lS0gEChscvB3yg4Mn1zxFdh0641nP3XsGbzNvA8AK6vDy6d+dnId6Z7uTlFLCAWmOmCIc8RZXh9hsXPCOJF62c0dHsMZVM0J8aLrjOWi7P90U7Ov38vfl//lOtYUJwZFhALRCnBUGjZij4yLW0kj6UjvW+yP0v3YBPN7VMHxECmhsPfW82aH76Mn8nOuD0LitPLAmKee/f/+wy+ln9XfVNygoOrY3REDAgn4zPYXwdT3Hc3kKnhyHdW0/7QNjTsUpTKguL0sAul5rHuA0t5c1P3zCtOwRFlZLUPUWfsUEX64ictHsvFOfy91bQ99MZ4QyXyQWFODQuIeai3ezm93ctJicPvtDzF4tRwRdtxlo+h8YgfEYXEwInb8FXY/mgnHT98JVI45F3xm/+D9XffE3k75mQWEPNMb/fyE35udRw+0v4kKXfm/v1k7c0jZBpO/utfrsQwJ3RzNu1aw9n370VLGHOYicRijLcFH2MLieqzgJhHJodD3kWJEa5etGPGU42TNSTSjC9yI++XU3C29NhEHWu+LUXPVpRLGhvI1r8RPhYS1WUBMU8UC4e899RvZ2nNYFnbjDk+E23VnTZ096ZVJJ49+f6MSnlLW/EnNXIsJKrHAmKOy483zKRBHH6zdTMx8Wdct1C6JeL11gJeCmJOcD3E6n8Yq8q4Q97oilqmOkmz/u57LCiqIOqs1s0i8pCIvCoi20XkbSLSKiKPiMjO8HtLuK6IyDfCuhhbRWRDdQ5h4SolGAptSA5wUfOBsl6Tq9PKzmQIZJsS7HlPLefdsoNVNf2cXX+MgXNrK9hYkbdIJhldPP1H2EIimqgtiD8Ffqqq5wEXEdTHuAt4TFU7gcd4Y+aoG4HO8OtO4JsR33tBKzccAFyE9zY+T22stEuoh9Ip6vc4wZmMMkJCYw5HLquDLxzl1hueYF19cC9GvZum42N70AvWlr3vU2prOWH8oRgLicpFmdW6CbiKcEo5Vc2o6gAn1r+YXBfjuxrYRDC57dJK338hqyQc8s6O5bikad+M6x0camTon5bS0pUlWxfDq4mBM8Mvo8DE4iRdH3e49iObuKJ990kDo29pPkDXH8RwmhorPoa89KoWtMQxVAuJykRpQawBeoC/Dkvv/ZWI1AGLVfVQuM5hYHH4+HhdjFBhzYzjrC7G9KKEQ9719duKtiJ8FV7Zt5TY37XS3JU9fsu3l3TI1sfQRJHWhCP0XlhL8jOH+PDFz1AzzWnVD6zfwq5PnI3EKr+QV2Ixhs5KTL+OD04W3AmIj8BFn7aQKFeUS61jwAbgM6r6tIj8KZMmolVVFSnv3Jqq3gvcC7Bx40ab9foUWBHLcX7DYbb0rzq+zFfhwGATuc0tLH/Zw03nTnqdukK2Noab8XEnPPCD/x6NOey/toa33/Aii5NDM76/I8q1N23h2V0baP27rVDB5ObS1kq65Y2kEgXxwB2H+KgSH/OJjStO1kc8RZTI81ssRFFaEAeAA6r6dPjzQwSBcSTfdQi/5ycDyNfFyCusmWFKcLh7WVW24yJc0/AyMfFJezFeO9zO679YTcP3Gul4LoubnuZMh4StiYY4ftLFq43R9cE473nPUyWFQ15zfIyVv7uT7KXnVnQME2sWoW7QQkj1KY2ve7S+kqV1R5qG/RlSvTliYx5OVhGf4+Fw9bVfrej9FqoodTEOA/tFJP8/fA3wCifWv5hcF+Oj4dmMy4HBgq6ImUHnD/+IB4fWV217y90Rtm5dTf8DK1jyQIrFm7PER0s/BaoO5Gpd9l+b4INv20TSObnFMZPzGo7Q/4UR5Kzyuk0SizHeHqd+v0/r9ixNu4JAcNN+Sa0EC4nSRT2L8RngeyKyFXgL8F+BrwLvFJGdwLXhzwA/AXYDXcC3gE9GfO8FY+0D/wXfc/jLHVfyZLq1Ktt8cPASlv0S6rtzOJkK294CetZ42VdoFrpx5XZevbsBp7Wl9LdtaaLmaJbaIzO0dqZhIVGaSAGhqi+EdTTfrKrvU9V+Ve1V1WtUtVNVr1XVvnBdVdVPqepaVb1QVTdX5xDmt7UP/Jfjj9MTcb786nvZkU1G2mYW5a9+cTWx8cp+ufLUETpaS+9WFHPbhZvZ8bmVSG3NjOtKIk52eXVC0kJiZnYl5RwzMFTL53Z8gN25yseX//fghSx9Ivq+eAmhvWY08nYcUW65+mn23nkukpjm5jARWL4EL2XTmJwu9i89ixW2Hgod62/gi7tu5RvnPMgyt7zmfY+v3Puj61kyUv6YwWR+XGicZiq5ciSdHO/8zWd4vP9SOjYVtEpcwauJk6t1ydW5xIdylHm1+LSuvvar/OJRqwJZjAXELFUsHPL2HW3lztyHuefcB+mMlXbbtIdy6/P/msXPVudeCD8GSad691UAjKwEL9F04sL82UyFhm5I9kUPt0IWEsVZF2MOO9rXyO9v/yDPZRpKWv/zB26k4W+bIk9Em+clhBo34szXBR7vXktNjwSBUPiVJzDRXN5l3yYaC4hZaKbWQ6HegXo+/9IH+PHoarwi5/g8lH/bfR2v/fl64iPV+4sfm1BGc9EGTPMGsrWMbm2d8TRlrlbI1kefo2IyG7CcmgXEPDA2luRrL13P13quZFRP7KBnUd7/yr9i+z1vItVX3e6A+JDV6nyEfvraelLHZm4aqEMwiU2VWxFOtooDG/OIBcQsU07roVAu6/LjHW/mE7t/i4Oe0Of7HPZgfy7Ohrb9HL4xS39nHC9Zvf9yJ6sMZKLfvv1c/0qS22Y+xZmXrRcyjdUbPnMyHvG+MW44/+6qbXO+sEHKeURVSHsxev0aUvLGwOWHWp/itqueZuDKWh48dhmPP3kBi5+GxHB5LYrgXgyHTIOQbhayjYqO10LTzK+dLO3HeHlwKdu7llO3K05snJJbBerAWIdDfFSQXOXjKaKKM+ERGxwHz1oQUxGt4EaZ02Xjxo26efPCuZ6q0tZDXjKV5e4Lfkpn4vC062XV5Z8G38KDv/o1ljxJ8XGJ8L6L8UUO4+1CukXxa3208NRqzOfCzgMlTa+fVZe9Y6280L0c3VlPzRE5fhWnH5eSb90GQKGm16fuYGUT34qvxIbTOGOZ4zed5f10+3+raJtzlYhsUdWNUz1nLYh5QkS5/qztrI3PXCg3Lh7va97Cu977At/9tSv4l7+/mLZtueNnN/KhMLZUSC/y8VPTtDRyDi/tWkHsHI/1jScHU2EoeLvrqT0i1E5oOBj5xi+meEHLoOSxBYGJVofYeIxkf+mnPUUVJ+3hDqWRbHVPl85H1oKYJaK2HlYv7uU/rvmHE7oWperxGvj8lg/Q+NM6JtqEsaU+fm2ZTe6kxxXn7mJVTR8DuVp2DbXRdaid2N4UqWOCezwUivMTQrljnk4WGvfliI1O310SVZyMjzuaQcZnPjW7kFoR1oKY5xLJLB9Z/lRF4QDQ7g5z/6Xf5hOJf8XIkfrKdiLt8sSOtTw5ci41B10Sw9B0fHygtD9C4hFMQFvGGQo/DsMrXBr3Ku7EyaEmvuJkvCAYMrmTuhNmehYQc5yIcs1Zr7EucSTSdlyUmBNxoC7tUnPQJdlf2S+h+IqoTDlL9XS8pDC0Kha0JMa9YDsZHyeTw5nIQa66p3cXEjvNOQtE6V4sWzTI+1u24ES8QSGDy0QmehWtXF2Ev9BKxfdZeClhrCNGvHeM+NERYn2jOCPpisPBTnkGLCDmsHgix0dWbqLWiVaBG8BXB8+LfvVRrl7RmSa3nXZHqHhquNiEImnrRlRT1LoYnxORl0Vkm4h8X0RSIrJGRJ4O6188KCKJcN1k+HNX+PzqqhzBAiWivH3VLi5IVmfWvgmN42WjX8LsJ3386eeSnZZoOH/kTBScXDD/ZKpXqe/2qOuOHpTmRBWPQYjIcuD3gfWqOi4iPwBuA24C7lHVB0TkL4A7CGpg3AH0q+o5InIb8DXgtyMfwRzX+cM/QiT4i6tFOt/5eX/FUVzXJxnPsbRxiA8seiZy1yJvwKvFr0JAIJCtC2aSrkjYzTjpbEYYCIkhpe5QhuThUaSnDx0ewZ9Ig+8hsRiyYhmajJBQBW44/+4FdTZjKlEHKWNAjYhkgVrgEPAO4EPh898BvkwQEDeHjyGY4PbPRER0Np9nPQ3+9rK/osdrIKMuB7MteJMadSnJsiQ+QByPDneEJidLs+PgiNCVdfGqdFPC4VwTVKGLAeDVKtIzxS95qfLdDAnCIjGoNO2eILHzIN6xXjSXY6pY1FwOv6cXWbYYHOs9V0PFAaGq3SLyP4B9wDjwz8AWYEBV81egFNa+OF4XQ1VzIjIILAKOFW5XRO4kqLzFqlWrmO8uSSaAfNN4bIa1k+FXoNkZodevzt2UeybamPK3rlyi1O2H9i1DDJzXQKah/NARVRxPqD3q0/RSL/7ufWg6TSmXNfmjo7hDI9AcvTCPiVZZq4WgVbAGWAbUATdE3SFVvTec53Jje3t71M3Na21u9W573j3SRtnnF/NEIeZT1zHK297UxehKcPYdZdGv9rNo2ziJkRLHFULuBCx9rIf6h57F274TTZc3tuD39tlVklUSpYtxLfC6qvYAiMiPgCsISurFwlZEYe2LfF2MAyISI7jFpzfC+y94KYnxh7vfz4G+5hOWx2IeHQ0jtNeMcHbtMVanjrE83kezM4aLnjRu4atD91AFf3FFkaRP58ojfHD5M1ya2ktKPH508z5+/uClcLCH+Kv7WbQnhbe0ldEVtUw0O/iJ4lnkeND+TB/e9p3l709Iczl0cAjaqjO57UIWJSD2AZeLSC1BF+MaYDPwC+BW4AFOrotxO/BU+PzPF/r4g3+4M9LrD+TS9D60grYjJ/cN0tLAfgf2uevIpYRMozDermSWZVm9qodrFu/g4to9LHJGGfBrGRyqK+/NXWXJij4+e/bP2ZA6gFtwbvLd9S/x1++9ntV/EZRO1IkJnNcP0rDHobE2hd9UR7a1llydSy7loK6AKrFxpebQKN4rr0X6dwHw+geJNTWi8WjDbAt9oDLKGMTTIvIQ8ByQA54nKJn3T8ADIvKVcNl94UvuA/5GRLqAPoIzHiaClzJLSAxNnbH5UnR4iptRkkPQcAB43iUrS/jH1FJ+2OYw3OnRclY/3lgZH4Wkx7vetI3fa/8lKTn5QqSE+PzGzc+x54GlMFAwAa366OgYMjpG4iAE57+dE58fGaUqfzd8Dx0ahkWl19swJ4sUr6r6JeBLkxbvBi6bYt0J4LeivJ850bbxFbgVFL0Rhfi4Et/v0bgfcslWnLeCn5p5lFJSHv/mkse5pfGFE1oNk32i/Zf8zjs+x+IfzVA3Y9IMWFrFeRn84RGc5sbgjIbYRJaVsHNBc9hrox1VmQLezSjJ3pk/CqWGA0Cd5EjcchSJl3n5tlYvIJyGeiSbQ9IZJJMF3yaFKZcFhEEUao7O0BJxlVsufL6kcMj7Uuc/MrG+vLqb1eI0NCA1NcFl175CzkMyWSSdAc8rraK46+C1RJ9Sby6zuzkNEFyhKGkHTU7xV1aU1auP8vFFT5QcDgBnx/vY8644575Yxo5I9L9ZkkziNE1xVsZXQBFVEAkGMCddUKU1CcZW1tN3boyRc3LUtkevHDaXWUDMYUtTQ+wWKr65qZCbUWJjDlOV/XTrc9y95idTDkhOu02U9/36M2z/X2edOFg5DXEdtLJpLQKOi9vaMv2Yw/GgyILjoIk46eVNHN2QZPTCCRZ39LLI9VgUYTfmC+tizGEX1BzAi1fp8uhkMCekZOWNr1zwlarJ0OzOdJXn1D7UuomBSxaX/oIoF3+J4C5qhViJf/fCrsdoZys9vz/GousOsmppH0nX5o/Is4A4Q6JeAwHw5mQ32broAaECQ2c5eCk/aI3kv/zga6y7nk+/8qGKLutudjIcvjFbctdBIly34DY3I6ky99F1OHilS2tt8RqjVz367yrep7nOAmIOOyeujC2OHhDjixwmFk8zwq9wbHcrn3r5wxWFxG0XPQtNJU5l57rglN+KcBsbkfoyL/YCMitaqT1voOzXLRQWEGeIs6TyS4nz6p0U6fOjVdf24sLwalBnhoEMhd7XW/j0Kx9i2C/v1OVvNm9m8OKO0lYWwUmUt32nrg6ZalByxhcKhy+vpWWa1gPA49f+9/K3PU9YQMxx71//An6F3XYVGF7pkKsv8fqAsCXx73bfyqiW3hVodjIcvqL0lk453QQnlcJpaS55/UJam2LsomgBO99ZQMxxn1z0/xhdWllCZBocxpeWefGQwq7ty/jjgzeUNRfFxZd0IckSf/FjsZLWdVIpnEWtFV8lOXRBK0vaByt67UJhAXEGjfgT9HtjHMqNMOiPM+JPMOZnyKqHV+IVhVmETAO44z5ORpES52P0XRhaI/jxCs6R+vDE1nV8f+DSkl/y0SVPkl1V+u37Tm3ttL/4x8Oh0olhHOHoBod41Jm85zm7DuIMumHbh/BUyORcEjEPVxRHlNp4hvp4mo7UCMuSA6xJ9nBe4hArY1lanBRxccmqxyf2/zpb//JCVm0dRDxFRcAV/LiLJhy8uIOfdIKydpMmkh3rcMk0V/7LIRnhO8+/jbGLEuT8E39Jk06OlvgoTe44HbEh2t0hVsX66XtTLR2lDr24DrGzVpLreKPwp1cbJ9McI1Pn0Pr0EUhXfsGE19pIbN1wxa9fKCwgzqD+0TcqWhdOOd9LcHnvdoLrB0TAcXxS8RwddSOc09DDjzdfzDl/k6Vt5I0msqhCTnFzPowH/7mFoeEnXbykQ6bRZWSVUtYsLlPQrMOjB9Yx3STWIkrC9ahPpBnshMXJZEkTwOiydrruTnLFmt044ZVgjihxxyPruzz5fy5i9fcPVFx0d+CCBtoaotUSWQisizEHqILnOYxOJHi9t5V/eOVC1j7gERuZuYScqCI5H3c8S3xggtSRMeoOpnGqMOFSvD4zbTgE+y6kczF6x+qoP7efIzedNfM1ES1NvPrZeq4751Xq3Aw1bpYaN0vSyeGgJJ0cF7xrB71XLGPGHZiK69B7oeBEDMiFwAJiLupJEhuqfIp3dzRD8w4qn2IO0JhSUzNzQBWKuT5j1w8zvuGsoutIKsVrn+jgxjdvm3ZbHckRmu/Yz/BblpYdEn5THe6akbJes1DNGBAicr+IHBWRbQXLWkXkERHZGX5vCZeLiHwjrH2xVUQ2FLzm9nD9nSJy+6k5nLll+y1fruh1NUec45W4K9XUNUZ8OMJFVjUesQouSa5LZdhzq0w9kYvrsu+2s7j6ypeOdyumc07DMbzfO8bo+iVlhcTI2kbaGku7CWshXwMBpbUgvs3Jk9HeBTymqp3AY+HPADcCneHXnQTT3SMirQQTy7yVYDKZL+VDxZSvpid609gdy9L8GpW1IgRSJXQvimldMsiBd3ec1NUYevsazn/fDpJl9H8uXtSN/wfHGLisxO6GI/Se70avQ7pAzBgQqvo4wRRxhW4mqHlB+P19Bcu/q4FNBBPYLgWuBx5R1T5V7QceoQozYC9Evu+QHKzOh7tp1zixsfJ/yzXhU5Msr3tRyBHIXj5MrnPZ8WX+yiXkPt5LR7L8pv+bWw/S9Mn9HHnnCogVuSbEdfBb6hnasIzxlTbjdakqHYNYrKqHwseHgfztesdrX4TydTGKLT+JiNwpIptFZHNPT0+Fuzd/eZ4QH61OQLijGer3lv+6WEMWd6ZLs2dQX5Nm77tqg2pYqRSv/et6Nrbvn/mFRZxd38t5H9/Oro8tx8/f9+EIWptiYt1iDl+3nL03t3L0Eof4gMvhgYZI+79QRD7NqaoqUr3hYFW9l2DyWzZu3GjDzKdYbBxWnHsU1/EZSSfJeQ7j6TjZTAw/7ULGQTw5XlRH40ptbXVqYMbPH2LobasZWeZy9SVbI2+vOT7OO256jsfOW0f7D5eTrXUYWS7kajX8UxiWMPTA21NPb6fHorrKbmNfKCoNiCMislRVD4VdiKPh8nzti7x8XYxu4DcmLf9lhe89r2y/5cuc//dfPiPvrTGHvusm+PSK509Y7quDh5D24wzmajiSbuDQWBOHBhvJ5RwSserMl+A6Pt0353DiGca9ODVulJliAg7KBUsPsfXytbhpKDabjpOF8V2NDHb6NNVMXUh0oQ9QQuVdjHyNCzi59sVHw7MZlwODYVfkZ8B1ItISDk5eFy4zZ9DAefV8YP2Wk5Y74hMXj3p3guXJfjY07uNdS17i4+ue4v2dL5KMRe/D+wpjY8H9Fn7W4bnuFYx60YvuZtXh+Z1n4aZnHltx08LQrmYGx1OR33e+KuU05/cJit2cKyIHROQO4KvAO0VkJ0GFra+Gq/+EYNr7LuBbwCcBVLUP+CPg2fDrP4fLTJlcV8nVRr98xU+4DL1vhI5EaVPBQRAcHYkhNrTtx414FmA8ncDLvDGgmBlLsPnQStJ+tF7v8z0riB8t/Xbx2HgQEn1jNTOvvADJbC5utXHjRt28efOZ3o3TopxuRu0/NtL2fLS7EHs2NvGez/yKplj5fXBfHV4YXsH23iUVvXc65zIyVAP+yX/lm1tHuHTJPuIVzOc/kK3hqWfOIzZa/pkZPw6JtUO0NwTXRyyk7oWIbFHVjVM9Z1dSzkHjEWeR8mri+O/tqygcIGhJXNjQTXtd+ackMzmX0eHUlOEAMNBfx3NHV5LV8j+amw+srCgcIBiTyOxs5GBvE36EK0znGwuIOWh8sY/GKv+vO3J5HbeufiHSPsTF49LWvWWNR2RyLiMjKdSbZt9V6O2tLzskDo43kttb4rR2RTgesKeW/d02n3WeBcQsUc5l19KRJttU/tyQANnmFEvfs5daN/qpypbYKG9edJBSznIfD4dcCR85FXqPNZQcEj7Ci/tW4FZ+7dZx4sPej/376BuaJywg5qBkKsPA2vJH/FWE/dcmuabj1arty1A2xdCWNgZeWcSx7ib6h2oZz8TJeQ75uWsmsjFGhksMhwK9xxrYfGTmgct9oy043XYm4lSw+SBmkXKuieh/k0/r9iTuSCaYB6IE4yvquPLal4iXWQCnmNfGlvDCw+tp2RcMKKq4qBvDS9SQrRPS9ZBu99GWTMV3jvb3NvC0t4pLl+yf8joJH2H7/iVU4cwrADvv/nx1NjRPWEDMMSJQm8xwza89x9FLGti85yxiu1LUHoSaPiU+4r1xp6cI2XqHiSaH8Q4h9tZ+Lqg/WJX96E638POfbDgeDhDMPyM5xclBfEyhB7JHhN7LXCRR+WnRkYFansqtZl17D054diOnLsOZJMdG6nAOWuvhVLHTnLNQsVZEzPU5t+0ob2/posk98XblrMYY85MMejVMhNPSu+LTEhul1smQkByeOgz70X+ZjmUbePCfr6TthdI+O8cuErzF0QcIknuSpMJrdkWDIMIPzurk6qJ/jhdq62G605zWgpgDXNdnacMwb2/vYkWiD4eT/xrHJUeTmzspOE4gQb3McmajnmzES/H9n19B+4ul/0KmeoSRjoonnwaCWbXiwxAbP/l9EwOQq8FG1E4B+yedhfJnNFzXZ0XzIO9dvY0PLn+GVYljU4ZDqRx84lJ5Z33ES3Hfk1fR/lx501kmBxTKHKA8iSe4U98yQWxcSQxLpCLGC7X1MBNrQcxS53cc4S1NB4q2GCqVcrJMeOVVrgIY85Lc9+RVLP4Xh3IvcoxNKDLuQDzCceQc3EzxBEj2KV5S8FKzt8s8F1kLYpb60RXfjNximIqDT0rKu2sy7ce578Vfo+MJt+xwgODagthotI+aO+Iw3ckX8SDVo0iu/H6MtR6Ks4CYxT627qlTst1dEx1sGjibrM5ckSvtx7n3hStpeySFE2EezEovgc5LDDozdiHcNNQcZdogmczCYXrWxVhgBr06/m7rBqQvzpYlq3jr6j1c2NBN0jm5VXFCOOSiNd2LjR+UQn0hXuJNp7ExJdUjTLQFM2+baKwFMctVsxXh4/A3ey7DORZHPEG7a3hq03n8xear+PHBi9gz0Ubaj+OrU9VwgOCve8Vn1Cec4LqKEsVHlJoj4GSmH7i01sPMrAUxB3xs3VN8+7W3Rd7OE4Od9O5chBRc1Sie4B5LsP/YMvbFl+I35KhrGWd0MEXHLxJVCQegorGLvESfS7knX2LjSu0hmGgLp5yb1MOxcChNpXUx/ruIvBrWvvh7EWkueO7usC7GDhG5vmD5DeGyLhG5C1OWqC2JY7lGHt12PpItPhYgWcHtizOxq5HkvmTJhYBPJfWEVE/lt3DXHFFSvRIMXoaHY+FQukrrYjwCvElV3wy8BtwNICLrgduAC8LX/C8RcUXEBf6coG7GeuCD4bqmDJWGhI/D917fiNtX+ulNr0ZJN1WvBzq6Url47T7Wrz7IqmW9tCwaJlWfxk14MM0M2c5wjPhI5UElPiQGlLqDSmJILBzKNGMXQ1UfF5HVk5b9c8GPm4Bbw8c3Aw+oahp4XUS6CArlAHSp6m4AEXkgXPeVaLtvSvHEYCcDu1rLun5SBcY7IDEkkc5eAAytdvjdd/+M9anu48s8dchqjGE/RU+uge50C/tGW9g/1MLgSIpcJoZ6Qu1hpyotGScTnAY15anGGMTHgQfDx8sJAiOvsP7F5LoYb51qYyJyJ0FVLlatWlWF3Ztfyh2P6PPqeXTb+bjTdC2K8WqUiUVC7dHKf7FGVji8+7eePCEcILhPxJUMKSdDe2woeL4JvKUOw34Ne9Nt/Kqnk6NbqvcZ2HKftR7KFakNKSJ/COSA71Vnd4K6GKq6UVU3tre3V2uz80qpXQ0fhwf3XYLbX9nfgaAVoeSSlY0BjLc5XPPbz3BVQ+nzT7ji0+yOclHtXj616ucMv238pAHGSlg4VKbigBCRjwHvBj6sb9wSOl1djKmWmwqVEhIvjK7iyGvtkap4+3EYXS747tTbUAF1wIsLuZSQaRDG2xxGljsMbUjzzqbpq3RPxxWfL274GSMroyWEhUPlKgoIEbkB+CLwXlUtnPn0YeA2EUmKyBqCIr7PEEx13ykia0QkQTCQ+XC0XTfThcSYn+QfXrkwuBYgomyDMrJCGFnunPi1wmFkpcPwSoeRFcLoMmG8Q8g0gZcCBuMMe9Gmk18e7+Pcd+zCL//2EcDCIaoZ255hXYzfANpE5ABBle67gSTwiAT38G5S1U+o6ssi8gOCwccc8ClV9cLtfJqgWI4L3K+qL5+C41lw8iExeVzi4SMXIUcqm7dyMvFAY+CV2VOJjwj7s62sd6M1Fj+y9Cm+eP4aWreWHnYWDNVhE8bMI/mQ2Jdp474nrsIdmflei5mIgjsuweQsZVKBS2/cxm+3PxN5Px46dilb//pNOCXMO2PhUB6ri7FAfGzdU/g4/GD3xVUJBwD88m5+KiQKLx+rrLjOZO9ufZHBzpn/mFk4VJcFxDzz8XVPMPp6U9W253jlTQ4zWV9PI14FRXAmSzkZztu4l+luQLVwqD67F2Meev2zXwDg7D/5eqTtBJPQRhvkdPtiDHi1LIqVX4VrslsWP8/X29dQe/jExLJgOHWsBTGP7f5CxF8crbx7kRcbE3alF0fbSGhJfIDx80+8b9zC4dSyFsQ8lw+JSloT4kfrXkDQAnl+YCWX1e2KtqHQZWv30PX4uTx3rwXD6WABsUBUEhQR5rd9YxsKO4+2v3HBfUQ/eNtfQvQ7302JLCAWmFKDQhQcrzpVrieO1TDhJ0iVco6yiPec/VJV9sWUxwJigZoxKBSqNV9ufMDlcK6J1Ymesl9rwXBmWUAscIUDmSeERRWvn3MnhJdHl5ccEBYKs4cFhDmuMCzO+erXIw9Q5okPL/Yu410tLxRdx0JhdrKAMFPquuvkswTnfemeird36Egz3loHV3wLgznEAsKU7NX/9Llpn88HSPH17q7yHplTzQLCVM1MAWLmHruS0hhTlAWEMaaoiupiFDz3BRFREWkLfxYR+UZY+2KriGwoWPd2EdkZft1e3cMwxpwKldbFQERWAtcB+woW30gwzVwnwczU3wzXbSWYieqtBNPgf0lEWqLsuDHm1JsxIFT1caBviqfuIZiXsvBs+c3AdzWwCWgWkaXA9cAjqtqnqv0EhXdOCh1jzOxS6aS1NwPdqvripKeWc3L9i+XTLDfGzGJln+YUkVrgPxB0L6rOCucYM3tU0oJYC6wBXhSRPQQ1Lp4TkSVUoS6GFc4xZvYoOyBU9SVV7VDV1aq6mqC7sEFVDxPUuvhoeDbjcmBQVQ8RTHd/nYi0hIOT14XLjDGzWCmnOb8PPAWcKyIHROSOaVb/CbAb6AK+BXwSQFX7gD8iKKDzLPCfw2XGmFnM6mIYs8BZXQxjTEUsIIwxRVlAGGOKsoAwxhRlAWGMKcoCwhhTlAWEMaYoCwhjTFEWEMaYoiwgjDFFWUAYY4qygDDGFGUBYYwpygLCGFOUBYQxpqiK62KIyGdE5FUReVlE/rhg+d1hXYwdInJ9wfIbwmVdInJXdQ/DGHMqlDJp7beBPwO+m18gIlcTTHF/kaqmRaQjXL4euA24AFgGPCoi68KX/TnwToIp6p4VkYdV9ZVqHYgxpvpmDAhVfVxEVk9a/HvAV1U1Ha5zNFx+M/BAuPx1EekiKJQD0KWquwFE5IFwXQsIY2axSscg1gFvF5GnReRXInJpuNzqYhgzj5RdF6Pgda3A5cClwA9E5Oxq7JDVxTBm9qi0BXEA+FFYYu8ZwAfasLoYxswrlQbE/wGuBggHIRPAMYK6GLeJSFJE1hAU8X2GYKr7ThFZIyIJgoHMhyPuuzHmFJuxixHWxfgNoE1EDhBU6b4fuD889ZkBbtdg/vyXReQHBIOPOeBTquqF2/k0QbEcF7hfVV8+BcdjjKkiq4thzAJndTGMMRWxgDDGFGUBYYwpygLCGFOUBYQxpigLCGNMURYQxpiiLCCMMUVZQBhjirKAMMYUZQFhjCnKAsIYU5QFhDGmKAsIY0xRFhDGmKIsIIwxRVlAGGOKsoAwxhRlAWGMKWpWz0kpIj3AKMGM2QtFG3a8891sO+azVHXKGhOzOiAARGRzsQk15yM73vlvLh2zdTGMMUVZQBhjipoLAXHvmd6B08yOd/6bM8c868cgjDFnzlxoQRhjzhALCGNMUbM2IETkBhHZISJdInLXmd6fahGRPSLykoi8ICKbw2WtIvKIiOwMv7eEy0VEvhH+G2wVkQ1ndu9LIyL3i8jRsLhzflnZxygit4fr7xSR28/EsZSiyPF+WUS6w//nF0TkpoLn7g6Pd4eIXF+wfPZ95lV11n0RVADfBZwNJIAXgfVner+qdGx7gLZJy/4YuCt8fBfwtfDxTcD/BQS4HHj6TO9/icd4FbAB2FbpMQKtwO7we0v4uOVMH1sZx/tl4N9Ose768POcBNaEn3N3tn7mZ2sL4jKgS1V3q2oGeAC4+Qzv06l0M/Cd8PF3gPcVLP+uBjYBzSKy9AzsX1lU9XGgb9Lico/xeuARVe1T1X7gEeCGU77zFShyvMXcDDygqmlVfR3oIvi8z8rP/GwNiOXA/oKfD4TL5gMF/llEtojIneGyxap6KHx8GFgcPp5P/w7lHuN8OPZPh92m+/NdKubY8c7WgJjPrlTVDcCNwKdE5KrCJzVoh87rc88L4RiBbwJrgbcAh4A/OaN7U6HZGhDdwMqCn1eEy+Y8Ve0Ovx8F/p6gaXkk33UIvx8NV59P/w7lHuOcPnZVPaKqnqr6wLcI/p9hjh3vbA2IZ4FOEVkjIgngNuDhM7xPkYlInYg05B8D1wHbCI4tP0p/O/Dj8PHDwEfDkf7LgcGCZvpcU+4x/gy4TkRawub5deGyOWHSWNEtBP/PEBzvbSKSFJE1QCfwDLP1M3+mR0mnGRm+CXiNYGT3D8/0/lTpmM4mGJ1+EXg5f1zAIuAxYCfwKNAaLhfgz8N/g5eAjWf6GEo8zu8TNKuzBH3pOyo5RuDjBIN4XcDvnOnjKvN4/yY8nq0Ev+hLC9b/w/B4dwA3FiyfdZ95u9TaGFPUbO1iGGNmAQsIY0xRFhDGmKIsIIwxRVlAGGOKsoAwxhRlAWGMKer/A/cHrPyJeGhgAAAAAElFTkSuQmCC\n",
297 | "text/plain": [
298 | ""
299 | ]
300 | },
301 | "metadata": {
302 | "needs_background": "light"
303 | },
304 | "output_type": "display_data"
305 | }
306 | ],
307 | "source": [
308 | "z=50 # this is for visualization only, this the height in microns where you want to see the slice\n",
309 | "\n",
310 | "lens=-1 # putting this to a number between 0 and Nlenslets will show you what each block does\n",
311 | " # setting it to -1 will show all lenses..\n",
312 | "clf()\n",
313 | "Back=sliceFormulaAber(X2,Y2,z,[xpos,ypos,rlist,r_lenslet,height,aperR,-1,zernlist])\n",
314 | "T=sliceFormulaAber(X2,Y2,z,[xpos,ypos,rlist,r_lenslet,height,aperR,lens,zernlist])\n",
315 | "Mask=maskFormulaAber(X2,Y2,z,[xpos,ypos,rlist,r_lenslet,height,aperR,lens,zernlist])\n",
316 | "Res=T*(Mask)\n",
317 | "Back=np.where(Back==0,NaN,Back)\n",
318 | "imshow(Back+Res)\n",
319 | "\n"
320 | ]
321 | },
322 | {
323 | "cell_type": "markdown",
324 | "metadata": {},
325 | "source": [
326 | "## Now get the Centres of the lenses (most negative dist) as xpos, ypos not ideal"
327 | ]
328 | },
329 | {
330 | "cell_type": "code",
331 | "execution_count": 9,
332 | "metadata": {},
333 | "outputs": [],
334 | "source": [
335 | "# Note that there might be a small dependency on the height, taken 53 here but this can be changed..\n",
336 | "Cents=getCentresAber(X2,Y2,51,[xpos,ypos,rlist,r_lenslet,height,aperR,-1,zernlist])\n",
337 | "CCentsX=X2[0,Cents[:,0]]\n",
338 | "CCentsY=Y2[Cents[:,1],0]"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": 10,
344 | "metadata": {},
345 | "outputs": [
346 | {
347 | "data": {
348 | "text/plain": [
349 | "(-810.9505280711508, 939.0216787103948, -876.3868816008894, 860.3779877709837)"
350 | ]
351 | },
352 | "execution_count": 10,
353 | "metadata": {},
354 | "output_type": "execute_result"
355 | },
356 | {
357 | "data": {
358 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAQsAAAD4CAYAAAD7JMNRAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjMuMiwgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy8vihELAAAACXBIWXMAAAsTAAALEwEAmpwYAAAWyUlEQVR4nO3df7AdZX3H8fe3icSKAoFEmvLDGxTtBJ1qCJTWHzWA/IjWOK3D4DgFkSljipYfbdNQ5kYH/jHRFmV0sChY6VBj/FUdBiuBXJ3pdBJIMPwIELkJiMQAQRA7tYVGv/3jeU6y9+ace/ee3T3n2d3Pa2bn7nnO5u7uPbnf++yzz36/5u6IiEznt4Z9ACJSDwoWIpKLgoWI5KJgISK5KFiISC6zh30AecybN89HRkaGfRgirbB169Zn3X3+5PZaBIuRkRG2bNky7MMQaQUz+0m3dl2GiEguChYikouChYjkUkqwMLMrzGy7mT1oZl81s5eb2UIz22xm42b2NTM7JG47J74ej++PlHEMIlKtwsHCzI4B/gpY4u5vBGYB5wNrgOvc/XXA88DF8Z9cDDwf26+L24lI4sq6DJkN/LaZzQZeAewBTge+Ed//CvC+uL48via+f4aZWUnHIWVbuxbGxia2jY2FdmmVwsHC3XcDnwaeIASJF4CtwC/cfV/c7EngmLh+DPDT+G/3xe2Pmvx9zewSM9tiZlv27t1b9DClX6ecAueddyBgjI2F16ecMtzjkoEr4zJkLqG3sBD4XeBQ4Jyi39fdb3T3Je6+ZP78g+aHyKAsXQrr14cAsXp1+Lp+fWiXVinjMuRM4DF33+vu/wd8C3grcES8LAE4Ftgd13cDxwHE9w8Hfl7CcUhVli6FFSvg2mvDVwWKViojWDwBnGZmr4hjD2cADwFjwPvjNhcC34nr342vie9vdGXgSdvYGNxwA4yOhq+TxzCkFcoYs9hMGKi8F3ggfs8bgb8DrjSzccKYxE3xn9wEHBXbrwRWFT0GqVBnjGL9erjmmgOXJAoYrWN1+KO+ZMkS17MhQ7J2bRjMzF56jI3BPffAypX1249My8y2uvuSye2awSlTW7ny4DGKpUvL/wXWXZfk1eKpU2mB7F2XFSvC2IjuuiRFPQtJh+66JE3BQtKhuy5JU7CQNOiuS/IULFLS5ucw7rln4hhFZwzjnnuGe1yyn4JFStp8R2BQd12kb7obkhLdEZCEqWeRGt0RkEQpWKRGdwQkUQoWKdEdAUmYgkVKdEdAEqYHyURkAj1IJiKFKFiISC4KFiKSS1lFho4ws2+Y2SNm9rCZ/aGZHWlmG8zs0fh1btzWzOz6WGTofjNbXMYxiEi1yupZfBb4d3f/PeD3gYcJ6fLucvcTgbs4kD7vXODEuFwC3FDSMYhIhcooBXA48A5ijk13f8ndf8HEYkKTiwzd4sEmQhbwBUWPQ0SqVUbPYiGwF/iymf3IzL5kZocCR7v7nrjNU8DRcX1/kaEoW4BoPxUZEklLGcFiNrAYuMHd3wL8N5MydsdU/zOa0KEiQyJpKSNYPAk8GUsCQCgLsBh4unN5Eb8+E9/fX2QoyhYgEpFElVE35Cngp2b2htjUKTKULSY0ucjQBfGuyGnAC5nLFRFJVFn5LD4G3GpmhwC7gIsIgWi9mV0M/AQ4L257O7AMGAd+FbcVSZdqmgAlBQt33wYcNJec0MuYvK0Dl5ax34HQfxTpZDDrPOSXfTq4RTSDczptTnUngSrJAwoW09N/FAFlMEPBIp+2/Udpc5bxXpTBTMEil7b9R9Gl10TKYBa4e/LLySef7EOzcaP7vHnha7fXTdU5z9HRdpzvVNasOfj8N24M7Q0EbPEuv4fKlDWdNt8NWb06XHqNjoa/qNIKvTJlKVhId52ut+qXtI7S6kl+KV2ja7A1GQoWcrCUsoxrsDUZugyR9OmSaKB0GSL11bZ5LolSsJD0tW2eS6IULCRtKQ22tpyChaQtpcHWltMAp4hMoAFOESmktGBhZrNidu/b4uuFZrY5FhP6WsyihZnNia/H4/sjZR2DiFSnzJ7FZYTiQh1rgOvc/XXA88DFsf1i4PnYfl3cTkQSV1b5wmOBdwNfiq8NOJ2Q6RsOLjLUKT70DeCMuL2IJKysnsVngJXAb+Lro4BfuPu++DpbSGh/kaH4/gtx+wlUZEgkLWWUL3wP8Iy7by3hePZzFRkSSUoZPYu3Au81s8eBdYTLj88Saph2sodnCwntLzIU3z8c+HkJx9FeejKz/mrwGZZRZOgqdz/W3UeA84GN7v5BYAx4f9xscpGhTvGh98ft05/skTI9mVl/dfgMu6XP6ncB3gncFtdPAO4mFBP6OjAntr88vh6P758w3fcdalq9ulAavPpL5DOkR1q9oefXzLMoWOQ0Oho+0tHRYR+J9CuBz7BXsNAMzqbQk5n1l/hnqGDRBHoys/5q8BkqWDSBnsysvxp8hnrqVEQm0FOnIlKIgoWI5KJgISK5KFiISC4KFiKSi4KFiOSiYCEiudQ/WNTg0V6RJqh/sBj0o70KTtJS9Q8WnWmx550Hq1cfmF9fVT3MOuQdEKlA/YMFDLZw7qCDkwyeeo9dNSNYDPrRXlX1bjb1HrvrluRiJgshn+YY8BCwHbgsth8JbAAejV/nxnYDridkyrofWDzdPqZMftPJLtTJKjT5dRUSyWgkFWrxZ0xVmbKABZ1feOBVwI+BRcBaYFVsXwWsievLgO/FoHEasHm6fUwZLNasOfiD3LgxtFdhGMEpBYP+OacggaxVw1BZsDjoG4bEvO8CdgAL/EBA2RHX/wn4QGb7/dv1WpJKq9fGXxr39gVJ9SyqDRbACPAEcBihyFCn3TqvgduAt2XeuwtY0uV7XQJsAbYcf/zxlf+AJIe2/AK1LTBO0itYlFkY+ZXAN4HL3f2X2ffiAcwoy46ryFB62jKwW4OsVbmVeWenWwSZ6QK8DPg+cGWmrZmXIW3Wlp5Fk/TRS6KqnkUsanwT8LC7/2PmrWwxoclFhi6w4DTgBXffU/Q4pGI1SCgrXZQ4L6is8oV/DpxuZtvisgz4JPAuM3sUODO+Brgd2EW4dfpF4C9LOAapWpO65m1T0uWjEvaKNF2nV7hiRZi0OE3PQgl7RdqoxMtHBQuRJivx8lGXIU21dm14liHb3RwbC/9JVq4c3nFJ8nQZ0jZ6GEpKNnvYByAVyd4yyzmwJTIV9SyarC0zLmUgFCyabNB5PiRoaPIcBYum0ozL4WnoeJGCRVNpxuXwNDT1om6dilRl9eowXjQ6Gnp3NaFbpyKD1MDxIgULkbI1dLxIwUKkbA0dL9KYhYhMoDELESlEwUJ6a+jkIunP0IKFmZ1jZjvMbNzMVg3rOGQKDZ1cJP0ZSrAws1nA54FzCQWJPmBmi4ZxLDKFhk4ukv4Mq2dxKjDu7rvc/SVgHbB8SMciU9HDaBINK1gcA/w08/rJ2LafmV1iZlvMbMvevXvzf2ddZ5ergZOLpD/JDnB6v0WGdJ1dnoZOLpL+DCtY7CZUX+84NrYVl8J1dlN6Nw2dXCR96lZ5qOqFkKFrF7AQOAS4Dzip1/Z9VSQbZgXsltfKlHqj6lqnMwxQ+4CPEkoePgysd/ftpe1g2NfZKfRu6qgpPbKm6hZBUltm1LNI6a/6MHs3dZTSZ9dipNSzqFQq19nD7t3U0TB6ZOrN5NctgqS21K6Kuv5CFjPIHpk+q4PQmp5FClLp3dTRoHtkGl/Kr1sESW2pXc9C+jPMv/IaX9oP9SwkecPqkWl8KRclv5F2y85SXbr04NctpOQ3It1ofCk39SxEZAL1LESkEAULEclFwaLpNENRSqJg0XTK7yElmT3sA5CKZWcorlgR5hG0+Lag9E89izZQHk0pgYJFG2iGopRAwaJKKQwuKo+mlKRQsDCzT5nZI2Z2v5l928yOyLx3VSwgtMPMzs60t6e4UAqDi5qhKGXp9nRZ3gU4C5gd19cAa+L6IkJezTmEPJs7gVlx2QmcwIHcm4um20+tnzrtPDk5Otr6PAlSD1Tx1Km73+EhnybAJkKWbggFg9a5+4vu/hgwTigs1L7iQhpclIYoc8ziw8D34nqvIkLTFhfq6LvIUGo0uCgNMW2wMLM7zezBLsvyzDZXA/uAW8s6MO+3yFBKNLgo/UphcHySaYOFu5/p7m/ssnwHwMw+BLwH+GC83oHeRYSqKy6UIg0uSr9SGByfrNtARt4FOAd4CJg/qf0kJg5w7iIMbs6ouFBnqfUAp0i/hjQ4To8BzqLTvT8XA8IGMwPY5O4fcfftZrY+BpJ9wKXu/msAM+sUF5oF3OxlFhcSaZLs4Pjo6NAHx5X8RtKydm3oamd/McbGwqXbypXDO65h6Fx6DPiZHiW/kXpI8Vp9GBIcHFewkLSojkeQ4OC4LkMkTatXH7hWv+aaYR9Nq+gyROpDE9mSpGAhaUnwWl0CBQtJS9Fr9QRnPjaFgoWkZeXKgwczly7Nf9tUd1Mqoxyc0izKOVoZ9SykeZQWoBIKFtI8uptSCQULaRbdTamMgoU0S4IzH5tCMzhFZALN4BSRQhQsRCQXBQsRyaWUYGFmf21mbmbz4mszs+tjIaH7zWxxZtsLzezRuFxYxv41xVekeoWDhZkdRyg29ESm+VzgxLhcAtwQtz0S+DjwB4QaIh83s7lFj0FTfEWqV0bP4jpgJZC9rbIcuCXm/9wEHGFmC4CzgQ3u/py7Pw9sICT9LUYJU0QqV7TW6XJgt7vfN+mtwRcZ0hRfkUpN+yCZmd0J/E6Xt64G/p5wCVI6d78RuBHCPItp/8HkKb5LlypgiJRo2mDh7md2azezNxHqf9wXywAcC9xrZqcydZGhd05q/0Efxz1RdopvJ0joUkSkVH1fhrj7A+7+ancfcfcRwiXFYnd/CvgucEG8K3Ia8IK77yHUCznLzObGgc2zYlsxmuIrUrmq8lncDiwjVE//FXARgLs/Z2bXAp3f4mvc/bnCe+uWGEWXISKlKm1SVuxhPBvX3d0vdffXuvub3H1LZrub3f11cflyWfuXGmjLfJiGnqdmcMrgtGU+TFPPs1sB1NQWFUZukCEV+x24Gp8nPQojq2chg9WW+TANPE8FCxmstqS8a+B5KljUUV0H0NqS8q6h56lgUUd1HUBry3yYhp6n0urVVSdAqDaGlExp9ZqmgQNokjYFi7pq4ACapE3Boo4aOoAmaVOwqKOGDqDVXl3vUuWkYFFHRSuNSzXqepcqJ1VRFylLwyu4q2chUqYG36VSsBApU4PvUilYiJSl4Xepyqgb8jEze8TMtpvZ2kz7VbHI0A4zOzvTfk5sGzezVUX3P3ANH/GWApp+l6rbc+t5F2ApcCcwJ75+dfy6CLgPmENI6rsTmBWXncAJwCFxm0XT7SepfBadPAWd/ASTX4vUHD3yWRS9G7IC+KS7vxgDzzOxfTmwLrY/ZmbjhApkAOPuvgvAzNbFbR8qeByD0/ARb5Feil6GvB54u5ltNrMfmlnnhvLgiwwNUoNHvEV6mTZYmNmdZvZgl2U5YZ7GkcBpwN8C6y0WESnK3W909yXuvmT+/PllfMvyNHjEu9Y0nlSpvosMAZjZCuBb8TrnbjP7DTCP3kWGmKK9HlTQKF2dGZSdzyL7WUlhRS9D/o0wyImZvZ4waPksocjQ+WY2x8wWEqqp302oF3KimS00s0OA8+O29dH0Ee86U4HsShUd4LwZuNnMHgReAi6MvYztZraeMHC5D7jU3X8NYGYfJVQhmwXc7O7bCx7DYKmgUdqy40mjo/pcSqRMWdIsyiBWmDJlSfM1fAblsClYSHNoPKlSugwRkQl0GSIihShYpECTiaQGFCxS0PB0bNIMSquXAj2cJjWgnkUq9HCaJE7BIhV6OE0Sp2CRAk0mkhpQsEiBJhNJDWhSlohMoElZko/mfEgPChYykeZ8SA+aZyETac6H9KCehRxMcz6ki0LBwszebGabzGxbzMR9amw3M7s+FhK638wWZ/7NhWb2aFwuLHoCUoEq5nxoLKT+uhUTybsAdwDnxvVlwA8y698DjJD5e3NsPxLYFb/Ojetzp9tPUkWGmq6qIkoqzlQb9CgyVPQyxIHD4vrhwM/i+nLglrjvTcARZrYAOBvY4O7PufvzwAbgnILHIGWqas6HkunWXtEBzsuB75vZpwmXNH8U20spMgRcAnD88ccXPEzJrcqExEqmW2tFiwytAK5w9+OAK4CbyjowT7nIkPRHz7/UWtEiQ7cAl8WXXwe+FNd7FRnaDbxzUvsPch+t1JeKM9Ve0TGLnwF/HNdPBx6N698FLoh3RU4DXnD3PYR6IWeZ2VwzmwucFduk6fT8S+0VHbP4C+CzZjYb+F/iGANwO+GOyDjwK+AiAHd/zsyuJVQmA7jG3Z8reAxSByrOVHuFgoW7/wdwcpd2By7t8W9uJlQyE5Ea0QxOEclFwUJEclGwEJFcFCxEJJdaZMoys73AT6bYZB7w7IAOJ6V9D3v/bd33sPdf9b5f4+4HzYSsRbCYjplt8S5pwJq+72Hvv637Hvb+h7VvXYaISC4KFiKSS1OCxY0t3few99/WfQ97/0PZdyPGLESkek3pWYhIxRQsRCSX2gULM/taTBC8zcweN7NtsX3EzP4n894XMv/mZDN7ICYQvt7MrM99f8LMdmf2sSzz3lXx++8ws7Mz7efEtnEzW1XgvD9lZo/EBMjfNrMjBnXeXY6llHOaZh/HmdmYmT1kZtvN7LLYPuPPoM/9Px5/dtvMbEtsO9LMNsRk0xtimoUpE1T3sd83ZM5tm5n90swuH9R5T6lbYs66LMA/AKvj+gjwYI/t7iYkDjZCIuFz+9zfJ4C/6dK+CLgPmAMsBHYCs+KyEzgBOCRus6jPfZ8FzI7ra4A1gzrvSd+ztHOaZj8LgMVx/VXAj+PPeUafQYH9Pw7Mm9S2FlgV11dlPoOuCapL+lk/BbxmUOc91VK7nkVH/Ct5HvDVabZbABzm7ps8/HRvAd5X8uEsB9a5+4vu/hghj8epcRl3913u/hKwLm47Y+5+h7vviy83EbKM9VTheZd2TlNx9z3ufm9c/y/gYXrka416fQZlWg58Ja5/hQM/z14Jqos6A9jp7lPNXh7EeQM1vAzJeDvwtLs/mmlbaGY/MrMfmtnbY9sxhMTAHT2TBOf00djVvLnTDaWEBMUz9GHCX7KOQZx3R1Xn1JOZjQBvATbHppl8Bv1y4A4z22oheTTA0R4yvkH4i390RfvuOJ+JfwwHcd49JRksbOokwR0fYOIPcg9wvLu/BbgS+FczO4wZmmbfNwCvBd4c9/cP/Z5jH/vubHM1sA+4NTaVct6pMrNXAt8ELnf3X1LxZ5DxNndfDJwLXGpm78i+GXtrlc07MLNDgPcSctvC4M67pyRrnfoUSYIBLKTx+1MyWbrc/UXgxbi+1cx2Aq8nJAnOdtk7yYP72nfmGL4I3BZf9kpQzBTtM963mX0IeA9wRvzPWtp5z8BU51oqM3sZIVDc6u7fAnD3pzPv5/0MZszdd8evz5jZtwld+6fNbIG774mXGc9Use/oXODezvkO6rynVMVASNULoTDRDye1zScO7BAG33YDR8bXkwf6lvW53wWZ9SsI14oAJzFxkGkXYXBqdlxfyIHBwJMKnPNDwPxBn/ek/ZV2TtPsxwjjLJ8p8hn0ue9DgVdl1v8z/vw/xcQBzrVx/d1MHOC8u4TzXwdcNMjznvaYqvimVS/APwMfmdT2Z8B2YBtwL/AnmfeWAA8SRoo/R5y52sd+/wV4ALifkME8+wFeHb//DjJ3HQgj5T+O711d4JzHCdem2+LyhUGdd5djKeWcptnH2wjd/Psz57ysn8+gj32fEH8B74s/26tj+1HAXYQs9ndyICgb8Pm47weAJQXP/VDg58DhRf7vlb1oureI5JLkAKeIpEfBQkRyUbAQkVwULEQkFwULEclFwUJEclGwEJFc/h+4hcakr5+7GwAAAABJRU5ErkJggg==\n",
359 | "text/plain": [
360 | ""
361 | ]
362 | },
363 | "metadata": {
364 | "needs_background": "light"
365 | },
366 | "output_type": "display_data"
367 | }
368 | ],
369 | "source": [
370 | "clf()\n",
371 | "plot(CCentsX,CCentsY,'rx')\n",
372 | "plt.axis('image')"
373 | ]
374 | },
375 | {
376 | "cell_type": "markdown",
377 | "metadata": {},
378 | "source": [
379 | "# Now start with Nanoscribe file"
380 | ]
381 | },
382 | {
383 | "cell_type": "code",
384 | "execution_count": 11,
385 | "metadata": {},
386 | "outputs": [],
387 | "source": [
388 | "optPoints=np.column_stack((CCentsX,CCentsY))"
389 | ]
390 | },
391 | {
392 | "cell_type": "code",
393 | "execution_count": 14,
394 | "metadata": {},
395 | "outputs": [
396 | {
397 | "name": "stdout",
398 | "output_type": "stream",
399 | "text": [
400 | "11402.334630350195\n"
401 | ]
402 | },
403 | {
404 | "data": {
405 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAX8AAAD4CAYAAAAEhuazAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjMuMiwgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy8vihELAAAACXBIWXMAAAsTAAALEwEAmpwYAABRBklEQVR4nO2dd3hUVfrHP2fSey+kFxIgdAi9iNIRCerq2law4e7adddd167rrmXXtrrr+rOhq2JDQAQRG70FCL0FSK+k9zJzfn/MTQgaaqbP+TzPPLn33Du5b+5Mvufc97znfYWUEoVCoVA4FzprG6BQKBQKy6PEX6FQKJwQJf4KhULhhCjxVygUCidEib9CoVA4Ia7WNuBcCA0NlQkJCdY2Q6FQKOyK7du3n5BShnV3zC7EPyEhgczMTGuboVAoFHaFECL3dMeU20ehUCicECX+CoVC4YQo8VcoFAonxCTiL4S4TwixTwixVwjxsRDCUwiRKITYIoTIFkJ8IoRw18710PazteMJprBBoVAoFOdOj8VfCBEN3A2kSykHAC7ANcBzwEtSyt5AFXCL9pZbgCqt/SXtPIVCoVBYEFO5fVwBLyGEK+ANFAOXAJ9rxxcCc7XtDG0f7fhkIYQwkR0KhUKhOAd6LP5SykLgH0AeRtGvAbYD1VLKdu20AiBa244G8rX3tmvnh/z89wohFgghMoUQmeXl5T01U6FQKBRdMIXbJwjjaD4RiAJ8gBk9/b1SyjellOlSyvSwsG7XKCgUF0RJTTP/XXOUpVmF7MyrorKhFZXaXOFsmGKR1xTguJSyHEAIsRgYBwQKIVy10X0MUKidXwjEAgWamygAqDCBHQrFOfHR1jxe/f7IKW1+Hq7EBnsTH+JNXIg3ccHexAf7EB/iTa8AT1xdVGCcwrEwhfjnAaOFEN5AEzAZyAR+BH4FLALmAUu185dp+5u04z9INexSWJCCykZ6BXiy8OaR5FU0klvZSF5FA7mVjRwqqeO7A6W06U9+JV11guggL2OHEGLsFDo6iLhgb3w87GKhvEJxCj3+1koptwghPgd2AO3ATuBN4GtgkRDir1rb29pb3gY+EEJkA5UYI4MUCouRX9VIXLA3qRF+pEb4/eK43iApqW0mt6KBvIpG8io7OohGduUXUdvcfsr5ob4exicGrTOIDzG+YoO9CfP1QMUzKGwRkwxZpJSPA4//rPkYMLKbc5uBq0xxXYXiQiioamJscuhpj7voBNGBXkQHejE2+ZfHaxrbyK1sIFfrGIxPDw1sOVbBkqxCuj7Heru7nNIpxAV7ExfiQ3ywN9FBXrgpd5LCSqjnVYVT0dKup6S2mdhgrwv+HQHebgzyDmRQTOAvjjW36SmsbjJ2CJorKb+ykeMnGlhzuJyWdkPnuToBUYFeWqfg84unBz9Ptwu2UaE4G0r8FU5FcXUzUkJMkLdZfr+nmwvJYb4kh/n+4pjBICmrazG6kSoatJ/Gp4dV+0qobGg95fz7p6Zy9+QUs9ipUCjxVzgVBVVNAMQEXfjI/0LR6QSRAZ5EBngyMjH4F8drm9vIqzA+Kbz2YzbLdxcp8VeYDeVwVDgV+VWNAMQGm2fk3xP8Pd0YEB3AzIG9mD0oisOl9ZTXtVjbLIWDosRf4VQUVDXiqhNE+HlY25QzMibZuOh98zG1BEZhHpT4K5yKgqomegXa/qKtAVH++Hm4svGoEn+FebDt/wCFVThR30J1Y+vZT7RD8isbiTXTZK8pcXXRMSopmE1HT1jbFIWDosRf0cmhkjoe+HQXY/7+Pb/+72b0BsdbeF1Q1WSVyd4LYXRSCDkVjRRVN1nbFIUDosTfyZFSsjH7BPPe2cr0l9eyYk8xF6WGcai0ji93Fp79F9gRzW16yupazBbmaWo6FqJtUq4fhRlQoZ5OSrvewIq9Jby59ih7C2sJ9XXngamp3DA6nkBvNzJe38BLqw9z2eBeeLi6WNtck1CojaB7ssDLkvSN9CPI242NRyu4cniMtc1ROBhK/J2MhpZ2Ps3M5+31xymoaiIp1Ie/XzGQy4dG4+l2UuQfnN6XG97ewoeb87h5fKIVLTYdJ2P87WPkr9MJxiSHsPlYBVJKlSNIYVKU+DsJZXXNLNyYw/8251HT1EZ6fBCPzU5jSr8IdLpfisr4lFDG9Q7htR+zuXpELL4OkLkyv9IY428vPn+AMUkhrNhTQl5lI/EhPtY2R+FA2P9/tOKMZJfV89a6YyzeUUibwcD0tEhum5jE8Pigs773wel9yXh9A2+tO8a9U1ItYK15Kahqws1FEOHnaW1Tzpkxmt9/49EKJf4Kk6LE3wGRUrItp4o31x7luwNleLjquCo9hlsnJJEYeu4CMjg2kJkDIvm/tcf4zeh4Qnxte2HU2SioaiQ60KvbJx1bJTnMh3A/DzYereDakXHWNkfhQCjxdyD0Bsm3+0r479pjZOVXE+Ttxt2TU7hxTDyhFyjcD0zrw6p9Jbz+41EeuyzNxBZblvyqJrvx93cghNHvvyFb+f0VpkWJvwPQ1Krn8+35vLX+OLkVxkIlT2f051fDY/Fy71mkTu9wX64aHsv/Nudy8/gEuxPPrhRWNTI1LcLaZpw3Y5NDWJpVRHZZPSndFJ9RKC4Ek8T5CyEChRCfCyEOCiEOCCHGCCGChRCrhRBHtJ9B2rlCCPGqECJbCLFbCDHMFDY4IxX1Lby0+jDjnvuBR5fuI9DbnX9fP4wf/zCJ34xJ6LHwd3DPlBQQ8PJ3R85+so3S1KrnRH2rXXZeY7v4/RUKU2GqRV6vAN9IKfsCg4EDwJ+B76WUKcD32j7ATCBFey0A/mMiG5yGnBMNPPzlHsY++wOvfH+EYXGBfLJgNEt+P5ZZA3vhYmKfdlSgF/PGxLN4RwGHS+tM+rstRUGV/UX6dBAb7E10oJda7KUwKT12+wghAoCJwHwAKWUr0CqEyAAmaactBH4C/gRkAO9rRds3a08NvaSUxT21xdHZkVfFm2uOsWp/CW46HZcPjea2iYn0Dje/K+D3k3qzaGs+/1h1iDdvTDf79UyNvcX4/5yxySF8u78Ug0Ha1YS1wnYxhc8/ESgH3hVCDAa2A/cAEV0EvQTocLZGA/ld3l+gtZ0i/kKIBRifDIiLc94oB4NB8v3BMt5ce5RtOVX4e7ryu4uSmT82gXB/y4UsBvm4s2BiEv9cfZgdeVUMizt7qKgt0THyj7XDkT/A2N4hfLa9gP3FtQyIDrC2OQoHwBRuH1dgGPAfKeVQoIGTLh4AtFH+eWUJk1K+KaVMl1Kmh4WFmcBM+6K5Tc/HW/OY8tIabns/k6LqZh6bncamhybz4Iy+FhX+Dm4en0iorzvPrTyIlPaV9C2/qgl3V90FRz1ZmzFJKs+PwrSYQvwLgAIp5RZt/3OMnUGpEKIXgPazTDteCMR2eX+M1qYAqhtbee2HI4x/7kceWrwHb3cXXr12KGv+OImbxyfiY8WVtj4ertx1SQpbjley9oh9pRouqGokJsi+Yvy7EhngSVKoD5tUcReFieixkkgpS4QQ+UKIPlLKQ8BkYL/2mgc8q/1cqr1lGXCnEGIRMAqoUf5+Y+qBt9cf55Nt+TS16bkoNYzbJyYxJjnEpmK7rx0Zx/+tO8bz3xxkQu9QuxHT/Er7i/H/OWOSQ1iys5A2vQE3Gy9Go7B9TDWMvAv4UAjhDhwDbsL4VPGpEOIWIBe4Wjt3BTALyAYatXOdlj0FNfx37VFW7ClGJwRzhkSxYGISfSP9rW1at7i76nhgWir3fbKLr/cUc9ngKGubdE4UVDUyMMa+feVjk0P5cEseewpr7G7ORWF7mET8pZRZQHchIJO7OVcCd5jiuvaKlJKfDpXz5tpjbDpWgZ+HK7dNSGL+uAR6Bdj+hOScwdH8d80x/vntIWYMiLT5UWh9SztVjW12UcHrTIxOCgaMfn8l/oqeolb4WpB2vYEvdxbyf+uOcbi0nkh/T/4yqy/XjIzD39PN2uadMy46wR+n9+GWhZl8mpnP9aPirW3SGbHnGP+uhPh60DfSj01HK7jj4t7WNqfHVDa0kl1Wz/ET9QyJDaJPpFq9bEmU+FuQV3/I5tXvj9A30o8Xrx7M7EFRuLva9qj5dFzSN5z0+CBe+e4IVwyNMdlqYnNQUNkR42/f4g9Gv/9HW/JoadfbRZEdKSXl9S1kl9ZzpKyeI2V1HCmtJ7usnoqGk3WiXXSCeWMSuHdqil0NhOwZJf4WorlNz/825zK5bzhvzUu3qUncC0EIwZ9m9uWqNzbx3sYcfjcp2domnZaTI3/7dvuAMb//uxtyyMqrZlRSiLXN6URKSXFNs1HgS+s4Wl7PEU3wa5raOs/z83QlJdyXKf0iSInwJTncl5hAL97bmMO7G4/z1e4iHp7Vj4whUXb/P2LrKPG3EMt3F1PZ0MrN4xMd5ks9IiGYS/qG85+fsrluZBwB3rY5YsuvasLTTUeor7u1Tekxo5JC0Aljnh9riL/BICmsbuocwRtH8/UcLaunvqW987wgbzdSIvy4dFAvUsJ9SQn3IyXCl3A/j26//89cPpCr02N5dOle7v0ki4+25vF0xgDlCjIjSvwtgJSShRtzSAn3ZWyy7YzWTMEfp/dh1qvreGPtUf40o6+1zTkFKSXLdxezZGchSaG+DtHpBni5MSA6gE1HK7hvqvmu0643kFfZyJEyo4vmSGkd2eXG7eY2Q+d5YX4epIT7cuWwaHpH+GlC73tBtR8Gxwby5e/H8cm2fJ5fdZBZr67jprEJ3DMlBT/lCjI5SvwtwI68avYU1vD03AEOIUBd6dfLn4zBUby74TjzxyYQYYWVx92RX9nIY0v38uOhcgZGB/DCVYOsbZLJGJMUwjsbjtPUqu/xXEtru4HcigbNXWP0yWeX1XOsvIFW/UmRjwrwpHeEH9ePCjEKfIQvvcP8TP6056ITXDcqjhkDInlh1UHe3nCcZbuKePjSfswZrFxBpkSJvwVYuDEHP09XrhgabW1TzML9U/uwfHcxr35/hGcuH2hVW9r1Bt7dkMOLqw8jBDw6O415Y+JxtfFw1PNhTHII/117jMzcSiaknFvqk+Y2PcfKGzrFvUPocysaaTcYU3UIAbFB3qSE+3JRnzBSwv3oHe5LcpiPxUfewT7u/P2KQVydHstjS/dxz6IsFm3N56mM/qqmgYlQ4m9mymqbWbGnmBvHJFg1NYM5iQvx5rpRcXy4Je+8S0Wakt0F1Ty0eA/7imqZ3Decp+YOIDrQ/iN8fs6IhGBcdYKNRyt+If4NLe2dk63ZHT/L6sirbETTeFx0gvgQb3qH+TJjQGQXkfe1uaitoXFBLLljHB9vzeOFVYeY+co6bhmfyN2TUxz2/8lSqLtnZj7ckodeSm4cY9ux8D3lzkt681lmAS+uPsy/rh1q0WvXt7Tzz28PsXBjDqG+Hvz7+mHMHBDpsC4CHw9XBscG8sOBMhJDfIyTr9povrC6qfM8NxdBYqgP/aMCyBgSTUqEceI1IdTbLsJEO3DRCW4YHc/MAZE8981B/rv2GEuzinhkdj8uHdjLYT9ncyPsITtjenq6zMzMtLYZ501ru4Fxz/3AgCh/3r1ppLXNMTv/WHWI137MZvld4y2Wdnj1/lIeX7qX4tpmrh8Vx4Mz+jpFnPhLqw/zyvfGymoerjqSw3w1cfeltxZZExfsbfOrry+E7blVPLpkL/uLaxnXO4Qn5wygd7ivtc2ySYQQ26WU3RbgUOJvRpZmFXLPoizeu2kEk/qEW9scs1Pb3MbE539kSGwg75m5syutbeaJZftYubeEPhF+/O2KgQyPd56UB42t7WzPrSIu2JuYIG+TV2+zdfQGyYdbcnlh1SGa2/TcMj6Juyf3xttdOTO6cibxd7xhgQ3x3sYcEkN9mHiOk3L2jr+nG7+flMxPh8rZbKbUwwaD5INNOUz55xq+P1jGH6f34au7xjuV8AN4u7syISWM+BAfpxN+MLqCbhyTwI9/mETGkGjeWHOUKf9cw8o9xXZXa8JaKPE3E7sLqtmZV82NY+LtJu2xKbhxTAKR/p48/43pC74cLKnlyjc28ujSfQyKDeDbeydyx8W97TZFhqLnhPp68I+rBvP5b8fg7+XG7z7cwY3vbOVYeb21TbN51H+NmXhvYw4+7i78aniMtU2xKJ5uLtw7JYUdedV8d6Ds7G84B5rb9Dz/zUFmv7qenBMNvHj1YP53yygSrBRVpLA90hOCWX7XeB6/LI2svGpmvLyOF1YdpKlVb23TbBYl/mbgRH0Ly3cVc+XwGKdcmfir4TEkhfrwwqqD6A09G/2vP3KC6S+v5d8/HSVjSDTfPzCJK4bFqAgPxS9wddFx07hEvv/DRcwe1IvXfzzKlBfX8M3eEuUK6gYl/mZg0dY8WvUGbhyTYG1TrIKri44/TO/D4dJ6luy8sAqdFfUt3PdJFje8vQWdEHx06yj+efVggn3sPz+PwryE+3ny4q+H8MmC0fh6uPLb/23npve2kXOiwdqm2RQmE38hhIsQYqcQYrm2nyiE2CKEyBZCfKJV+UII4aHtZ2vHE0xlgy3Qpjfwv815TEgJderws5kDIhkYHcCLqw/T0n7uj95SSj7NzGfyi2tYvruIuy7pzcp7JjC2d6gZrVU4IqOSQlh+93genZ1GZk4V015ay4vfHlKuIA1TjvzvAQ502X8OeElK2RuoAm7R2m8BqrT2l7TzHIZv95VSUtvMPCcd9XcghOBPM/pSWN3ER1vyzuk9x8rrufb/NvPg57vpHebL13dP4IFpffB0s58FSQrbws1Fxy3jE/nhgYuYOTCSV3/IZupLa1i9v9Taplkdk4i/ECIGuBR4S9sXwCXA59opC4G52naGto92fLJwIAfuwo05xAZ7cXFfx4/rPxvjU0IZmxzCaz9kn5Lu9+e0tht49fsjzHhlHfuKannm8gF8evsYUlUOF4WJCPf35JVrhvLxbaPxcnPhtvczueW9beRVNFrbNKthqpH/y8CDQEcawBCgWkrZ8R9fAHRkNYsG8gG04zXa+acghFgghMgUQmSWl5ebyEzzsr+olq05ldw4OsEpY6+748EZfaloaOXtdce7Pb4tp5JZr67jxdWHmZoWwff3X8T1o5wrPFZhOcYkh7Dingk8PKsfm49VMOWlNbz83WGa25zPFdRj8RdCzAbKpJTbTWBPJ1LKN6WU6VLK9LAw+1gktXBjDl5uLlydHmttU2yGIbGBzOgfyf+tO0ZFfUtne01jGw8t3s1Vb2yiqVXPO/PTef26YYTbSEpohePi5qLjtolJfP/AJKb3j+Tl744w7aW1/HDQuVxBphj5jwPmCCFygEUY3T2vAIFCiI611jFAR9hHIRALoB0PAMyzHNSCVDW0siSrkLlDo222opW1+MP0VBpb2/n3T0eRUvLVriImv7iGT7blc+v4RL69byKX9I2wtpkKJyMywJN/XTuUj24dhZuL4Ob3Mrl1YSb5lc7hCuqx+EspH5JSxkgpE4BrgB+klNcDPwK/0k6bByzVtpdp+2jHf5AOEIT7SWY+Le0G5o117OydF0LvcD9+NTyGDzblcuM7W7nr4530CvBk2Z3jeWR2mkrNq7AqY3uHsvKeiTw0sy8bj55gyotrePX7Iw7vCjJnnP+fgPuFENkYffpva+1vAyFa+/3An81og0XQGyQfbMpldFIwfSP9rW2OTXLvlFQQxoyMj1zajy9/P9ZimT8VirPh7qrj9ouS+f6Bi5iSFsGLqw8z4+W1rD1sH/ONF4LK6mkCVu0r4fYPtvPGDcOYMaCXtc2xWfYX1RLs405kgPLrK2yb9UdO8NiyvRwrb2D+2AT+PLOvXYYcq6yeZmbhxhyiAjyZ0k/5rc9EWpS/En6FXTA+JZQVd0/gpnEJvLcxh4zXNnCopM7aZpkUJf495HBpHRuPVnCDg9WJVSicHU83Fx6/rD/v3TSCioZWLnttPe9tOO4weYKUWvWQhRtzcHfVcc2IOGubolAozMCkPuF8c+8ExiWH8MRX+7n5vW2c6BK2bK8o8e8BNU1tLN5RSMbgKJVwTKFwYEJ9PXhn/gienNOfDUcrmPHyWn48ZJqU5dZCiX8P+Cwzn6Y2PfPGJljbFIVCYWaEEMwbm8BXd44nxMeDm97dxhPL9tltSKgS/wvEYJB8sDmX9PggFbKoUDgRfSL9WHrnOOaPNU4Gz319A4dL7W8yWIn/BfLT4TJyKxrVqF+hcEI83Vx4Yk5/3p0/ghP1LVz2r/W8vynHriaDlfhfIO9tzCXC34MZAyKtbYpCobASF/cNZ+U9ExmTHMJjS/dx68LMU3JY2TJK/C+Ao+X1rD1czvWj4nFT4Z0KhVMT5ufBu/NH8PhlaazLPsH0l9exxg5WBivlugA+2JSLu4uOa0eq8E6FQmGcDL5pXCLL7hxHsI8b897ZylNf7T+vKnaWRon/eVLf0s7n2wu4dFAvwvw8rG2OQqGwIfpG+rPszvHMGxPPOxuOM/f1jRyx0clgJf7nyeIdBdS3tKuJXoVC0S2ebi48mTGAd+anU1bbzOx/reeDzbk2NxmsxP88kFKycGMOg2MDGRIbaG1zFAqFDXNJ3whW3juB0UkhPLpkL7e9v92mJoMdOpF6TWMbs19bh5uLDncXHW4uOtxchHHf9Wf7Hcddf7avtbm76Nh8rJKj5Q2M6x3Ckp2FJ9/veubff/J3a8d1OlWmUKFwAsL9PHl3/gje3ZjDcysPMuOVdbx49WAmpFi/OqFDp3SubW7jiaX7aNUbaNMbaNNL2vQGWtt/tt9xvP1n+3qJ3mCe++OqEyc7B62j8PFw5Y0bhtE7XBUuVygcjf1FtdyzaCdHyuq5dXwif5zRBw9X86aJPlNKZ4cWf1OgNxg7hKPl9Vz66np+nR7LnZf0/mXn0d5NZ6J1KKfsa+d0bJ/siAwsySriupFxPDGnv1X+VoXCnOgNEgFO/dTb3Kbnma8P8MHmXNJ6+fPqtUPMOtg7k/j32O0jhIgF3gciAAm8KaV8RQgRDHwCJAA5wNVSyiohhMBY43cW0AjMl1Lu6Kkd5sJFJ3DRubBkZyGuOsH901KJMFOR8YYWPV/tKuLhS/up9QMKh+PxZXtZvb+Ul64ewtjeodY2xyp4urnw9NwBXJQaxoNf7Gb2v9bzyKVpXD8qDqM0Wg5TKEw78ICUMg0YDdwhhEjDWJ7xeyllCvA9J8s1zgRStNcC4D8msMGsNLa288m2fGYMiDSb8ANcPjSaioZWhy4dp3BefjhQRmltC9e/vYXnvzlIm95gbZOsxpS0CL65ZwIjEoJ5ZMleFnywncqGVovaYIoC7sUdI3cpZR1wAIgGMoCF2mkLgbnadgbwvjSyGQgUQth07cMlO4uobW5nvpnDOy/qE0awjzuLdxSa9ToKhaUpqWmmqKaZP07vwzUj4vj3T0e56o1N5FU0Wts0qxHu78nCm0byyKX9WHOonBkvr2X9kRMWu75JfQtCiARgKLAFiJBSFmuHSjC6hcDYMeR3eVuB1vbz37VACJEphMgsL7feSLgjvLN/lD/D44PMei03Fx1zBkex+kApNU1tZr2WwkhuRQNvrTvGN3tLrG2KQ7MzrwqAsckh/P2Kgfz7+mEcK69n1qvrWJrlvIMdnU5w64QkvrxjLH6ertzw9hb+tuIAre3mfyoymfgLIXyBL4B7pZS1XY9J46zyec0sSynflFKmSynTw8KsFxa1+Vglh0rrmDcmwSI+uSuGRdPabmDFnuKzn6w4b6SU7Cmo4Z/fHmL6S2u56IWf+OvXB/jv2qPWNs2h2ZFXhbuLjrQofwBmDezFinsm0DfSj3sWZfGHz3bR0NJuZSutR/+oAJbfNYHrR8Xx5tpjXP7vDWSX1Zv1miYRfyGEG0bh/1BKuVhrLu1w52g/O8reFAKxXd4eo7XZJAs35hDk7cacIVEWud7A6ACSw3z4Url+TEab3sD6Iyd4bOlexj77A5e9tp7Xf8wm0NuNRy7th5uLIK2Xv7XNdGh25lUzINr/lNDGmCBvFi0Yzd2TU1i8o4DZ/1rP3sIaK1ppXbzcXXjm8oG8+ZvhFFU3Mftf6/hoS57ZVgb3WPy16J23gQNSyhe7HFoGzNO25wFLu7TfKIyMBmq6uIdsisLqJr7dX8KvR8Th6WbeeNwOhBBcMSyGrTmV5Fc6rz+0p9S3tPP17mLuWbSTYU+v5oa3t/BpZj4DowP4x1WDyXxkKp/cPoZJfcJo00u1YtuMtLYb2F1Yw7C4X7pNXV103D81lY9uG01zm57L/72Bt9Ydw2Cm9TX2wLT+kXz5+3H4erjxly/38PeVB81yHVOs8B0H/AbYI4TI0tr+AjwLfCqEuAXIBa7Wjq3AGOaZjTHU8yYT2GAW/rc5F4AbRls2e+fcodH849tDfLmzkLsnp1j02vZMWW0z3x0o49v9JWzMrqBVbyDYx50Z/SOZ1j+S8b1D8XI/tRPfmVcNwNC4QMsb7CQcKK6ltd3A0G7Ev4PRSSGsvGcCf/piN3/9+gDrjpzgH1cNdqrkiR1PqEuzCvl2fymNrXqiAjxJjTDPOoAei7+Ucj1wOmf45G7Ol8AdPb2uuWlu07Noax5T0yKICfK26LWjA70YnRjC4h0F3HVJb4vH/9oT2WX1fLu/hNX7SzuFPC7YmxvHxDOtfyTD44NwOcOioqz8avw8XEkK9bWQxc7HDm2yd1h84BnPC/R2540bhvPhljyeXr6fmVoqhImp1k+FYC6klOzIq2LJziK+3lNMZUMrAV5uZAyJZu6QKEYkBJttUZxD5/bpCct2FVHV2Ga17J1XDIvmj5/vZkdetdmjjOwJg0GyM7+a1ftL+XZ/CcfKGwAYFBPAA1NTmdY/ktQI33PuMHcVVDMoNsCpV52amx151fQK8KRXgNdZzxVCcMPoeEYkBHPXxzu48Z2t3D4xiQem9cHd1XEWPh4urWNpViFLs4ooqGrC003HlH4RZAyJ5qLUMIv8rUr8u6EjvLNPhB9jkkKsYsPMgb14dOlevtxZ4PTi39ymZ9PRCr7dX8p3B0opr2vBVScYkxzC/LEJTOkXQVTg2YWlu997sLiO2y9KMoPVig525lWdt1utT6Qfy+4cz1+/3s9/1x5j07EKXrlmKImhPuYx0gIUVTexbFcRS7OKOFBci07A+JQw7tcGLb4elpVjJf7dsD23in1FtTxz+QCruVx8PVyZ3j+Sr3YV8+jsNLMngLI1ahrb+PGQ0X+/5lA5Da16fNxdmNQ3nGlpEUzqE06Al1uPrrG3sIZ2g2RwTKBpjFb8grK6Zgqqmi5ogaSnmwt/nTuQ8b3D+NMXu5n96jqenjuAK4bFmN5QM1Hd2MqKPSUszSpka04lUsKQ2ECeuCyNSwdFWXVOQ4l/N7y3MQd/T1cuH/qLtWcW5fKh0SzNKuLHg+VOUSi+qLqJ1ftLWb2/lM3HKmg3SML8PMgYGs3UtAjGJoeYtBPMyq8GYIia7DUbO3KrAc442Xs2ZgyIZFBMAPd+ksX9n+5i3ZETPJXRHz/PnnX+5qKpVc/3B0tZsrOINYfLaNNLksJ8uG9KKhlDoogPsY2nFyX+P6O0tplv9pYwf2wC3u7WvT3je4cS5ufB4h0FDi3+Ukp+8/ZW1mcbl7Ynh/lw28QkpqZFMCQm0Gz++Kz8aqIDvQj3M1++JmdnZ34Vbi6C/lE9W0cRFejFx7eN5vUfs3n5u8PsyKvi1WuGMthGQnTb9QY2HK1gaVYhq/aW0NCqJ8Lfg/ljE8gYEk3/KH+bC9xQ4v8zPtyci15KbhyTYG1TcHXRkTE4ioWbcqhqaCXIx93aJpkFKWHTsQouSg3j0dlp9A63TORNVn41g2MDLHItZ2VnbjX9owJMsk7GRSe4e3IKY5NDuGdRFlf+ZyN/mN6HBROSrDJhL6UkK7+apVlFLN9dxIn6Vvw8XZk9KIqMIVGMSgo5Y6SZtVHi34WWdj0fbc3jkj7hxIVYNrzzdFwxLIa31h9n+e4ifmMDHZI50OkEQd5uRAV6WUz4T9S3UFDVxI1j4i1yPWekTW9gd2E114007T1OTwhmxd0TeOjL3Ty78iAbsk/wz6sGE27GjLtdOVpez9KdhSzdVURuRSPurjom9w0nY0g0k/qEWWxBaE9R4t+FFXuKOVHfalPF2dOi/Okb6cfinYUOK/4AwT7uVDZYrr7prg5/f6xzR1KZk4PFdTS3GcyygC7A243XrxvGom35PPnVPma+so5/XDWYi/uGm/xaYMxKunx3EUuyCtlbaIzUGZscyh0X92bGgEj8bXT+4Uwo8e/CextzSQrzYbyNFZq4Ylg0f1txkGPl9SSFOeZiJKP4Wy6feVZ+NS46wYBoldPHXJxc3GWeDlYIwbUj40iPD+Kuj3dy03vbuHlcIg/N6muSYkg1TW18s7eYpVlFbDpWgZTG9SSPXNqPOYOjLPakYS6U+Gtk5VezK7+aJ+f0t7kFPxlDonl25UG+3FnIA9P6WNscsxDs486hkjqLXS8rv5rUCD+rT+o7Mjvyqojw9yAqwLwimRLhx5I7xvHsyoO8s+E4nm46HpzR94J+V3Obnh8PlrEkq5AfD5bTqjeQEOLN3ZekMGdIFMkONPhS33yNhRtz8PVw5crhthdDHOHvybjeoXy5s5D7pqTaXOdkCiw58jcYJLvyq7l0kGUytTorO/OqGRYXZJEoF083F56Y05+Glnb+u/YYswb2YkD0uU3m6w2SzccqWLKzkG/2llDX0k6orwfXj45j7pBoBsUE2FykjilQ4g+U17WwfHcR14+Kt/gqu3PlimHR3PfJLrblVDLKSquOzUmwjwfVTW3oDdLsERLHKxqobW5niIr0MRsn6lvIq2y0eFLERy5N46fD5Tz4+W6W3jnutO4fKSV7CmtYmlXEV7uKKKtr6VxYOXdoFGOSQnB18Dratql0FubjrXm06aVNR35M7x+Jt/tevtxZ6JDiH+LjjpRQ1dhKqK95Vz1maQng1GSv+diRq/n7e7C460II8Hbjr3MHcPsH2/nvmqPcecmpWXGPn2hgaVYhy7KKOHaiAXcXHZP6hJExJJrJ/cLtJlLHFDi9+LfpDXy4JZeJqWE2PZnq7e7KzAG9+Hp3MU/M6e9wX9JgbQ1DZYMFxD+/Gh93F4uFlTojO/OrcXMR5+x6MSXT+0cye1AvXv0+m+n9IwnwdmP5rmKWZhWyq6AGIWBUYjALJiYxc0AvArztL1LHFDi9+H+zt4TS2hb+foXtjvo7uGJYNF/sKOC7A6XMdjB/dYgm/hX1rSerPZuJXQXVDIwJsOkFOPbOjtwq0nr5W22Q8sfpfVi+u5ipL63tbEvr5c9fZvXlssFR55Rh1NFxevFfuDGH+BBvJqWaJz7YlIxOCqFXgCeLdxQ6nPgH+54c+ZuT5jY9B4pruWW8yuRpLtr1BnYX1PDrEbFnP9mEtLTr+elQOcuyivjuQGlnu5ebC8vuHEeKmYqi2CtWE38hxAzgFcAFeEtK+aylbdhbWENmbhWPXNrPLiJoXHSCjCHR/N+6Y5TXtThUlaOTbh/zLvTaV1SryjaamYMldTS16c0W398Vg0Gy5XglS7MKWbGnmNrmdkJ83LlmRCxzhkTz7x+z2XD0hEni/h0Nq4i/EMIFeB2YChQA24QQy6SU+y1px/ubcvByc+GqdMuOUHrCFcOieWPNUb7aVcTN4xOtbY7JCPLW3D5mHvl3rOyNt5H0HY7ITm1x11AzdbBSSvYX17I0q4hlWUWU1Dbj7e7C9P6RzBkSxfjeoZ1i/8zlA5n64hr+vHg3H9062i4GeZbCWiP/kUC2lPIYgBBiEZABWEz8qxpaWZpVxK+Gx/Q4L7wlSY3wY0C0P4t3FjiU+Lu56AjwcjO726e0rhmAma+sI9TXnZRwP/pE+pES4UtqhB+p4X5OOwFoKnbmVRPm50FMkGn96nkVjSzbVciSrCKyy+px1QkuSg3jL5f2Y0q/8G4X7EUGePLwpf348+I9fLwtj+tH2f7cnqWwlvhHA/ld9guAUV1PEEIsABYAxMWZPlZ40bZ8WtoNNpXH51y5YmgMTy3fz+HSOrMVd7YGIT7uZh/53z81lbHJoRwpreNwaR2HS+v5LDOfhlZ95zkR/h7GjiDCj1StU0iJ8LPZNSC2xo68KobFBZpkYdSJ+ha+3m2M1NmhheiOSAjir3MHcOnAXueU6fbXI2L5ancRf19xkIv7hF9Q1TdHxGa/zVLKN4E3AdLT06Upf3e73sD/NucyNjnELsVzzpAonllxgMU7CvnzzAtbxm6LBPu4U1lvXvH3cHXhotQwLupSFNxgkBTVNHGktJ5DnZ1CHR9uyaW5zdB5XnSg1ymdQZ8IP3qH++Ll7lhhtz2hor6FnIpGrhl54QO2hpZ2vt1fwpKdRazPPoHeIOkb6ceDM/owZ3AUMUHn57ITQvDsFYOY9tJa/vLlHt6dP8IhV+yeL9YS/0Kgq6M9RmuzCBuOVlBY3cSjs9MsdUmTEurrwUWpYSzNKuSP0/s4TMhikI87+ZWNFr+uTieICfImJsj7lKyQeoOkoKqRQyV1HCmr53BpHYdK6tiQXUGr3tgpCAFxwd6khBufEvpE+pES7kdSmI/DrcU4Fzqqo53v4q7WdgPrjpSzJKuI1ftLaG4zEB3oxYKJSWQMiaJvZM8S8MUGe/PgjD48+dV+vtxZaFelIM2FtcR/G5AihEjEKPrXANdZ6uLHy+sBSE+w3xWelw+N5oeDZWw+VsE4G8tCeqGE+Lh3ioct4KITxIf4EB/iw7T+J9vb9QZyKxs5XGJ0G3U8Kfx0qIx2g/EhVScgIdSHVK1TSI00upESQ30cOvJkR14VrjrBwHNY3GUwSDJzqzojdaoa2wj0duPKYTHMHRrN8Lggk07QzhuTwPLdxTz51X7Gp4Q6fQU3q4i/lLJdCHEnsApjqOc7Usp9lrp+WV0LrjpBsLf9VsaamhaBn4crX+wocBjxD/Zxp6qhFSmlTT+Wu7roSA7zJTnMl5kDT7a3ths4fqKBw6V1HCmt63Qhfbu/BK1PwM1FkBjq0+k2So3wJSXCj/hgb4fIJbMjt5p+vfzP6Ao7WHIyUqewuglPNx1T0yKZOySKCSlhuLua5z7odILnrhzErFfX8fjSffznhuFmuY69YDWfv5RyBbDCGtcuq2sh1NfDrsO+PN1cmDkwkhV7SiySDM0SBPu4026Q1Da122XEjburjj6RxuihrjS36TlaXt85p3CktI49BTV8vbv4lPcmh/l2zimkap1DTJCX3XxP9QbJroJqruomM25BVSPLdhkF/2BJHS46wYSUUP4wPZVpaZH4WGgyvXe4L/dOSeH5bw6xck8xMwf2ssh1bRGbnfA1J2V1LUT42/8CqbHJoXyaWcDBklr6R9l/hsoQ345Y/xa7FP/T4enmQv+ogF98Ro2t7WSX1Z8yp7DteCVLs4o6z7l8aDQv/XqIhS2+MA6V1NHYenJxV1VDK1/vMUbqbMvpSPQWyFMZ/Zk1sJfZczidjgUTklixp5hHl+5jTHIIgXbsAegJzin+tc3nHTFgiwzX/sm251Y5hPgH+xjFoLKhlaSws5zsAHi7uzIoJpBBMYGntNc1t3GkrJ4nlu2zaIGbntJRuauwuolb3tvGmsPltBskvcN9+cO0VOYMjraJ2tiuLjqev3Iwc15bz1PL9/Pi1UOsbZJVcErxL69rscjSc3MTE+RFhL8HmTlV3OgA9X07k7tZsJyjLeLn6cawuCCSQn3YrgmqPdCxevr5bw4R6e/JzeMTyRgSRVovf5ubw0mL8ud3k5L51w/ZXDYoymy1f20ZpxP/Nr2BioZWwh0gL44QgvT4YLbn2o9AnImuaZ0Vxk6grrnd2macM1PSIvB0c2HWwF6MSgy2+bmKOy/pzTd7S/jLl3v49r6J+NlhEfaeYP/hBefJiXpj4jBHCfMaHh9EYXUTxTVN1jalxyjxPxU/T1fqm9uR0qRrHM3G9P6RPD13AGOSQ2xe+MG44O/5Xw2itLaZZ1cetLY5FsfpxL+stkP87X/kDyfXKmTm2P/o39PNBR93F2NOfwW+nq60G+Qpq4wVpmVoXBA3j0vkwy15bDpaYW1zLIrTiX9prTGxV7gDRPsAxphqNxfHcf34ups9rbO90OGGqGtus7Iljs0D0/oQH+LNnxfvpqlLjidHx+nEv6zOsdw+bi46hsQGkplbaW1TTEKwt/mTu9kL/p7GKblaO/L72yNe7i48e8Ugcisa+ee3h6xtjsVwSvEXAkJ9HSe2Nz0hiAPFdTS02L9IBPu4U9WoxB+MPn+Aegf4XG2dMckhXD8qjnc2HO8MWXV0nE78y+uaCfFxd4il9B0Mjw9Cb5A2lRfnQgn28TB7Zk97wddDuX0syZ9n9iXS35MHP99NS7vju38cRwHPkbLaFsIcxOXTwbD4IIRwjEnfEF+j28deIlzMScfI357CPe0ZP083nrliINll9bz2Q7a1zTE7zif+dS0OE+nTgb+nG30i/BzC7x/s405Lu4FGJ5p4Ox0nxV+N/C3FxX3CuWJYNP/56Sj7imqsbY5ZcbpFXmV1zfTrZX8FXM7G8PgglmYV2XSSt3a9gRP1rZTWNhtfdS2U1pzcLqttpqDKuF6htrnNYsm+bJWT0T5q5G9JHpudxtrDJ3jw890suWOcw6bgdqr/Lr1BcqK+1WEifboyPD6ID7fkcaikjrSonhW+OF8MBklVYyultS2U1jVTVttMSc3J7dLaFkprmzlR39KZ2rgDnYAwPw8i/T2JDfYmPSGI3mG+RPo73md0vnSUjVTib1kCvd15OqM/v/twB2+uPcYdF/e2tklmwanEv7KhFb1BOkyMf1fS44MB2J5baTLxl1JS19LeKeAlNc2aoLecHL3XtlBW10yb/pc++hAfd8L9PYnw9yCtlz8R/h5EBHgS4edJhNYe4uths08q1sZFJ/Bxd1HibwVmDuzFrIGRvPL9Eab3j6R3uK+1TTI5TiX+ZXXaAi8H8/kDxAZ7EebnQWZuFb85hyRvTa16yuo0Ua/tGKGfHKV3bDe1/dL37ufp2ineoxKDNUH3IMLfs1Psw/w88HB1vjKGpsaY30f5/K3Bk3MGsPHoGh78fBef/Xasww1SeiT+QogXgMuAVuAocJOUslo79hBwC6AH7pZSrtLaZwCvYKzg9ZaU8tme2HA+dKR2cLRoH+hI8hZEZk4VRdVNXQS95ZQReonmY+9u4ZCHq45IbWQ+MCaQKZ2C7qGJvVHYvd2dasxgVfw8XdXI30qE+Xnw2Ow07v90Fws35nDz+ERrm2RSevpfvBp4SCvL+BzwEPAnIUQaxrq8/YEo4DshRKr2nteBqUABsE0IsUxKub+HdpwTjjzyB0hPCGbl3hLGPvvDKe2uOkG4n9Hlkhzmy9jkEG2EbhTzSG3E7u/panOpd50dX09XtcjLilw+NJplu4p4YdUhpvSLsIl6BKaiR+Ivpfy2y+5m4FfadgawSErZAhwXQmQDI7Vj2VLKYwBCiEXauZYR/86Rv2OK/6+Gx9CuN+Dv5aYJunHEHuztbhdZFhW/xM/TjRq14tlqCCH42+UDmfbSWv68eDcf3jrKYQZIpnx+vxn4RNuOxtgZdFCgtQHk/6x9VHe/TAixAFgAEBcXZxIDy+paCPByw9PNMX3RAV5u3H5RsrXNUJgQP09XCiobrW2GUxMV6MVDs/ry8Jd7WbQtn2tHmkaPrM1ZA1iFEN8JIfZ288rocs7DQDvwoakMk1K+KaVMl1Kmh4WZpqZfWV2zw7p8FI6Jv6erSuxmA1w7Io7RScH87esDDlE7A85B/KWUU6SUA7p5LQUQQswHZgPXy5Nr8guB2C6/JkZrO127RSira3HIME+F4+Lr4Up9i4r2sTY6neC5KwfRZjDwyJd7HSL9SI+WrmmROw8Cc6SUXZ9NlwHXCCE8hBCJQAqwFdgGpAghEoUQ7hgnhZf1xIbzoay2xSEXeCkcFz9PN5rbDLTpVUEXaxMf4sPkfhF8f7CMPAdwxfXU5/8a4AGs1iZBNkspfyul3CeE+BTjRG47cIeUUg8ghLgTWIUx1PMdKeW+HtpwTkgpKVcjf4Wd0TW5W0eZS4V1+GpXEV/vLuZXw2OIC7b/qJ+eRvucdt2zlPIZ4Jlu2lcAK3py3QuhpqmNVr1BjfwVdkXXal5K/K3Hrvxq/vDZLkYkBPHM5QMcIuLHMTMWdcPJCl5q5K+wH1R+H+tTXNPEbe9nEubnwRs3DHeYletOI/6dtXuV+CvsiI5Sjpk5lQ5Rqc3eaGxt57b3M2loaefteSMI8XUc/XCadfodC7zCVbZIhR0RF+KNp5uOJ77az9NfH2BAdACjEoMZmRDMiIRgArzdrG2iw2IwSP7w2S72FdXy9rx0+kQ6Vip45xF/5fZR2CExQd5kPjKV7blVbD1ewbbjVby3IYc31x5DCOgT4cfIxGDjKyFYDW5MyMvfHWbFnhIentWPS/pGWNsck+NE4t+Mj7uL0xcIUdgfvh6uXJQaxkWpxsWOzW16duVXs/V4JVtzKvl8ewHvb8oFIDHUh5EJwZ0dQkyQl0NMTlqapVmFvPpDNlenx3DrBMdK6NaB0yihcYGXGhUp7B9PNxdGJYUwKikEgDa9gX1FtWw9XsHW41V8s6+ETzKNWVR6BXh2dgSjEoNJDvNVncFZ2JlXxR8/383IxGD+Onegw94vpxH/8toWh03opnBu3Fx0DIkNZEhsIAsmGn3Vh8vq2Hq8ki3HK9l4tIKlWUWAscDOiC5PBv16+TtcnvqeUFTdxIIPthPhb4zscXd13JgYpxH/srpmBkQHWNsMhcLs6HSCvpH+9I3058YxCUgpyaloZJvWGWzNqeCbfSUA+Hm4MjwhiAkpYcwfm+DUHUFjazu3LsykqVXPh7eOcvh1FU4k/iq1g8I5EUKQGOpDYqgPV48wptYqqm5iW46xM1h3pJyfDpUzqU8YyWGOV67wXDAYJPd/souDJbW8PX8EqRGOFdnTHY77TNOF+pZ2Glv1RKjUDgoFYExTnDEkmr9dPpC7Lk4BjJXcnJUXVx/mm30lPHxpGhf3Cbe2ORbBKT7tzgVeSvwVil/QUSnMx0nLcy7ZWchrP2ZzzYhYbh6XYG1zLIZTiH/nAi/l9lEofkFjqyb+ThgGvSOvige/2M2oxGCeynCMnD3ninOIv4PX7lUoekJ9ix43F+HQkS3dUVjdxIL3t9MrwNPhI3u6wym6+vI6NfJXKE5HY2s73k7m8mloMUb2tLTpWbRgFEEOHtnTHU7xiZfVteDuqsPfyyn+XIXivGho0XdmD3UGDAbJfZ9kcaiklnfmj6B3uONH9nSHUzznlNUaa/c6kz9PoThXGlra8XZ3jDTF58I/vj3Et/tLeXR2GpOcJLKnO0wi/kKIB4QQUggRqu0LIcSrQohsIcRuIcSwLufOE0Ic0V7zTHH9s2GM8Vf+foWiOxpa251msnfxjgL+/dNRrh0Zx/yxCdY2x6r0+BMXQsQC04C8Ls0zMdbtTQFGAf8BRgkhgoHHgXRAAtuFEMuklFU9teNMlNW10NtJF68oFGejsVWPj4fjj/y351bx5y/2MDopmKcy+ju9J8AUI/+XMBZx71rOPgN4XxrZDAQKIXoB04HVUspKTfBXAzNMYMMZqW9uJyu/mp8OlZn7UgqF3dHQ0u7wMf4FVY3c/kEmUYGe/Of64bi5OIXH+4z06A4IITKAQinlrp8digbyu+wXaG2na+/udy8QQmQKITLLy8t7YiavXjsUb3cX5r+7jbs+3tkZ+qlQKBzf7dMZ2dNu4K15I5wysqc7zir+QojvhBB7u3llAH8BHjOHYVLKN6WU6VLK9LCwsB79rpGJway8dwL3TUll1d4SJv9zDf/bnIvBIM/+ZoXCwWlo0TvshK/BILlnURZHyup5/bph9A5X7t8Ozir+UsopUsoBP38Bx4BEYJcQIgeIAXYIISKBQiC2y6+J0dpO1252PFxduGdKCivvncCAqAAeWbKXK9/YyMGSWktcXqGwWRpa2h021PP5VYf47kApj81OY2JqzwaRjsYFu32klHuklOFSygQpZQJGF84wKWUJsAy4UYv6GQ3USCmLgVXANCFEkBAiCONE8aqe/xnnTnKYLx/dNop/XjWY3IpGLn11PX9feaBzibtC4Uy06w20tBsccpHX59sLeGPNUa4fFceNY+KtbY7NYa5PfAUwC8gGGoGbAKSUlUKIp4Ft2nlPSSkrzWTDaRFCcOXwGC7pG87fVx7gv2uO8fXuYp7OGMDFfZ037lfhfDS06gEcLtonM6eSvyzew9jkEJ6YoyJ7usNkU97aE8AJbVtKKe+QUiZLKQdKKTO7nPeOlLK39nrXVNe/EIJ83Hn+V4P5ZMFoPFx13PTeNu74cEdnFlCFwtFxxKRu+ZWN3P7BdqKDvPj39cNUZM9pUHcFGJUUwop7JvDA1FRWHyhlyj/X8MGmHPRqQljh4DS0GEf+jjLhW69F9rTpDbw1L51AbxXZczocp7vvIR6uLtw1OYXZg6N4ZMkeHl26j893FPK3ywfQP0qVf1ScGwaDpKG1nbpm46u2uY265jbqmo0FhaamRRDqazurzRu0XP6OMOFrMEjuXbST7PJ63rtphNNWJTtX7P8TNzGJoT7875ZRLM0q4unl+5nz2gZuGZ/IvVNSHHJSTHESKSXNbQbqmtuobW4/5Wddczu1TW2aqJ96vGt7fUs7Z3pgrGxo5Y6Le1vujzoLDZrbxxG+2xuPVvDdgTIeubQfE1JUZM/ZsP9P3AwIIZg7NJpJfcJ47puDvLnWOCH8VEZ/JveLsLZ5itPQ2m7oFOquo+7ajv1TxPvn5xnb2/RndvXpBPh5uuHn6Yq/9jMmyBt/T1f8vYz7J49p53m54aoTzP7XelxtrEB6o+b2cYSR/4q9xXi7u3DDaBXZcy7Y/yduRgK93fn7FYO4YlgMf1m8h1sWZjJzQCSPX9afyABVG8CU6A2S+pbuR9edwt3S/Wi747zmNsNZr+Pr4XqKQIf6upMY6oO/l2unWPt5uhnFvIt4d7T7uLtcUORIx6pybxsT2c6Rv51H++gNklV7S7ikbziebvb9t1gK2/om2igjEoL5+u4J/N+6Y7z6/RHWHTnBH6al8psxCbjY2EjO2hRVN1Fc0/QzgT7V9921vUO8O+rIngkPV90pQuzv6Up0oNdJge4UdrdfjML9Pd3w9XS12ufVMcL2tjFh6pjwtffcPluPV1LR0MrMAb2sbYrdYN+fuAVxd9Vxx8W9mT2oF48s2csTX+1n8c5C/nb5QAZEqwlhgG/3lfDb/23v1uftqhOnjqI93EgI9f6FC8Xf063bUbifp5tdl9lrtNF4+o4JX1uz63xZubcYTzcdk/ooX/+5osT/PIkP8eH9m0eybFcRTy8/wJzX1nPTuETun5rqULHS50tpbTN/+mI3aVH+/HF6X03IO8TbDU83nVMvtGm00YlVR5jwNRgk3+wtYVJquFP/D54v6k5dAEIIMoZEMyk1nOdWHeTt9cdZuaeYJzMGMDXN+SaEDQbJHz7bRXObgVeuGapC7LrBVlfSNrbq8XJzsWv35Y68KsrqWpg5MNLaptgV9vscbQMEeLvxt8sH8sXvxuDn6cZt72ey4P1MiqqbrG2aRXlnw3HWHTnBY5elKeE/DY2ae8XLzbbGW/Ut7TbXIZ0vK/aU4O6q4xKVmuW8UOJvAobHB7P87vH8aUZf1h4pZ+qLa3hn/XGnWCG8v6iW5785xLS0CK4ZEXv2NzgpNjvyb2m3e5fPyr3FTEwJw8/Tzdrm2BVK/E2Em4uO301K5tt7LyI9IZinlu9n7usb2FNQY23TzEZzm557Fu0k0NuNZ68c5NQ+/bPRZKO+9YZWvV37yXcVVFNc08ws5fI5b+z3U7dR4kK8ee+mEXy9p5gnv9pPxuvrmTc2gQem9XGIhTRd+fuKAxwpq+eDW0YSrKojnRFbHfkbSzjalk1nQm+QZJfVs7ugmt0FNazPPoGbi1CLLy8Ax1IjG0EIwexBUUxICeOFVQd5b2MOK/eU8GRGf6b3d4wRyo8Hy1i4KZdbxyeqpfTnQGNLO0KAp6ttCW1Dq55AL9t0l0gpya1oZHdhDbvzjWK/t6imM2zW18OVAdH+3HlxbwJs9G+wZZT4m5EALzf+Ondg5wrh2z/YzpR+ETyZ0Z/oQC9rm3fBlNe18MfPd9E30o8/zuhjbXPsggYtqkZnY1E1DS3tRAdaf7W6lJKS2mZ25dewp9Ao9LsLaqhpagOM62z6R/lzdXosA6MDGBwbQFKor83dT3tCib8FGBYXxFd3jeed9cd5+bsjTH1xDfdPTWX+2ARc7SzXuJSSBz/fRV1zOx/dNhoPGxvJ2iqNrXqb8/eD9SZ8Kxta2VVQze78GqMLp7CG8roWAFx0gj4RfswaGMmgmEAGRgfQJ9JP5eU3MT3+1IUQdwF3AHrgaynlg1r7Q8AtWvvdUspVWvsM4BXABXhLSvlsT22wB9xcdNx+UTKzBvbisaV7+evXB1i8o5C/XzGQwbGB1jbvnHl/Uy4/HirnqYz+pEb4Wdscu6Gx1TZDKhta9Wafi6prbmNPYY02mjeO6guqjOHQQkBSqA8TeocyKCaAQbGBpPXyV/l5LECPPnUhxMVABjBYStkihAjX2tOAa4D+QBTwnRAiVXvb68BUjDV/twkhlkkp9/fEDnsiNtibd+aPYOXeEp5Yto+5/97AvDEJPDAt1eZD1Q6X1vHMigNc3CeM36jMiedFQ4vtjfyllDS0tJu0kEtzm559RbWdIr+roJpj5Q2dx2ODvRgcE8hvRsczKCaQAdH+Nv+9d1R6+m38HfCslLIFQEpZprVnAIu09uNCiGxgpHYsW0p5DEAIsUg712nEH4wTwrMG9mJ8Sij/XHWIhZtyWLm3mDsvSWHukCib/GdobtNz98c78fd05flfDVZhnedJY6vtRdW06g20G+QFh3q26Q0cKqnrHNHvKqjhcGld5/qWcD8PBsUEMndItHFUHxOoosJsiJ6KfyowQQjxDNAM/EFKuQ2IBjZ3Oa9AawPI/1n7qO5+sRBiAbAAIC4urodm2ib+nm48mTGAy4fF8PjSvTy6ZC9/X3GAOYOjuG5UHINiAq1tYicvrDrEwZI63p0/gjA/26lEZS80tOrx97StkX9jZ0bPs3dKeoPkWHn9KUK/v7iW1nZjGu0ALzcGxQQwuW8yA2MCGBwTqNKe2zhn/TYKIb4DuotPfFh7fzAwGhgBfCqESDKFYVLKN4E3AdLT0x16qeyQ2ECW3DGOXQU1fLQll6VZRSzals+AaH+uGxnPnCFRVl0jsPZwOW+vP868MfFcrJbQXxBNre308rctMexIo/3zGgNSSvIrm9hVUM2ewhp25Vezt7Cmc62Ct7sLA6IDmDfG6LoZFBNAXLC3ehq0M86qKFLKKac7JoT4HbBYSimBrUIIAxAKFAJd1/rHaG2cod2pEUIwJDaQIbGBPDI7jSU7C/loSx5/+XIPz3y9n4yh0Vw3Ms7i6aMrG1p54LNdpIT78tCsfha9tiPR0KK3uYIpHfHyTa16vt1XYhzVF9awp6CaqkYtxNJFR78of64cHsOgmEAGxwSQFOZr14ngFEZ6OpxcAlwM/KhN6LoDJ4BlwEdCiBcxTvimAFsBAaQIIRIxiv41wHU9tMHh8Pd048YxCfxmdDw78qr5aEseX2wv4KMteQyOCeC6UXFcNjjK7BOIUkr+9MVuahrbWHjTSBWB0QOMPn8bc/toKSceX7YPMIZYpkb4MS0tkkGxRtdNaoSfXddRUJyenn4b3wHeEULsBVqBedpTwD4hxKcYJ3LbgTuklHoAIcSdwCqMoZ7vSCn39dAGh0UIwfD4IIbHB/HY7DQW7zR2AH/6Yg9PLz/A3KFRXDcynrQof7Nc/+Ot+azeX8ojl/Yz2zWchYZWvUmjakxBv17+3D4xiQh/TwbHBpDWKwAvG7NRYT6EUattm/T0dJmZmWltM2wCKSXbc6v4aEsey/cU09puYEhsoPFpYFCUyf55s8vqmf2vdYxICGbhTSPVSsoe0K430Pvhldw3JZV7pqRY2xyFEyGE2C6lTO/umHqeszOEEKQnBPPir4ew9S+TeXR2GnXNbTz4+W5G/u07Hl+6l0MldT26Rmu7gXs/2YmXmwv/uGqwEv4e0thmm0ndFM6NbTkhFedFoLc7t4xP5OZxCWw9XslHW/P4eGs+CzflMjw+iOtGxnHpoF7n7at/cfVh9hbW8t/fDCfCxiJU7JHO4u025vNXODfq2+gACCEYlRTCqKQQHr+slS+2F/Dx1jwe+GwXTy3fzxXDjJFCKeeQjmHj0RP8d+1Rrh0Z5zAZSK1NR51cNfJX2BJK/B2MYB93bpuYxK0TEtl0rIKPtuTxv825vLshhxEJQVw3Ko6ZA7p/GqhubOX+T3aRGOLDo7NVWKep6Bj5e6loKYUNocTfQRFCMDY5lLHJoZyob+Fz7Wngvk928eRX+7lyWAzXjoyjd7ix5q6Ukr98uYcT9S18+ftxykVhQk6O/NU9VdgO6tvoBIT6evDbi5JZMCGp82lg4cYc3l5/nFGJwVw3Ko76lnZW7CnhTzP6MjDGsgvJHJ2mLitjFQpbQYm/E6HTCcb1DmVc71DK61r4bHs+H2/N455FWQCMTgpmwUSTZOdQdEGN/BW2iPo2Oilhfh78flJvfjsxmfXZJ/jxUBm3T0xWy/bNgPL5K2wRJf5Ojk4nmJgaxsRUVYfXXKiRv8IWUYu8FAoz06h8/gobRIm/QmFmGlvbcdEJPFSCNIUNob6NCoWZMZZwdFH57hU2hRJ/hcLMNLaatk6uQmEKlPgrFGamoVVvc7n8FQr1jVQ4LI8t3Yu/pxuRAZ70CvAkMsCTqAAvAr3dLOqCaWq1vSpeCoUSf4VD0q438P2BMkpqm9EbTq1Z4eGq6+wMegV40auzc/DqbA/xcTdZB9HQ0q7SZShsjh59I4UQQ4A3AE+MFbt+L6XcKoz/Na8As4BGYL6Ucof2nnnAI9qv+KuUcmFPbFAousPVRceGP1+C3iA5Ud9CcU0zJTVNFNc0d75KaprYllNJaW0zbfpTOwh3Fx2RnR3EyU6i61NEqI/HOdU6aGzVE+rrbq4/VaG4IHo6HHkeeFJKuVIIMUvbnwTMxFi3NwUYBfwHGCWECAYeB9IBCWwXQiyTUlb10A6FoltcdIIIf09jXYLYwG7PMRgkJxpaKOnoGKqbKK5t7tzfmVfNypoSWvWGU97n5mL83R1PDVGndA7GziLU14OG1nbi3L0t8NcqFOdOT8VfAh3FXQOAIm07A3hfq+e7WQgRKITohbFjWC2lrAQQQqwGZgAf99AOheKC0ekE4X6ehPt5Miim+3MMBkllY+vJDkJ7iijRtncXVLNqXzOt7ad2EK46gV5KhsUFWeAvUSjOnZ6K/73AKiHEPzBGDo3V2qOB/C7nFWhtp2v/BUKIBcACgLi4uB6aqVD0DJ1OEOrrQaivBwOiu896KqWkqrGN4pomSmqaKdJcS6W1LVydHmthixWKM3NW8RdCfAd0V9LpYWAycJ+U8gshxNXA28AUUxgmpXwTeBOMBdxN8TsVCnMihCDYx51gH3f6R6m02Arb5qziL6U8rZgLId4H7tF2PwPe0rYLga5DnRitrRCj66dr+0/nbK1CoVAoTEJPF3kVARdp25cAR7TtZcCNwshooEZKWQysAqYJIYKEEEHANK1NoVAoFBakpz7/24BXhBCuQDOajx5YgTHMMxtjqOdNAFLKSiHE08A27bynOiZ/FQqFQmE5eiT+Usr1wPBu2iVwx2ne8w7wTk+uq1AoFIqeoXL7KBQKhROixF+hUCicECX+CoVC4YQo8VcoFAonRBjnZm0bIUQ5kKvthgInrGjO6VB2nR+2ahfYrm3KrvPDVu0Cy9kWL6UM6+6AXYh/V4QQmVLKdGvb8XOUXeeHrdoFtmubsuv8sFW7wDZsU24fhUKhcEKU+CsUCoUTYo/i/6a1DTgNyq7zw1btAtu1Tdl1ftiqXWADttmdz1+hUCgUPcceR/4KhUKh6CFK/BUKhcIJsVnxF0IMEUJsFkJkCSEyhRAjtXYhhHhVCJEthNgthBjW5T3zhBBHtNc8M9p2lxDioBBinxDi+S7tD2l2HRJCTO/SPkNryxZC/NlcdnW53gNCCCmECNX2rXrPhBAvaPdrtxDiSyFEYJdjNnHPrHXNLteOFUL8KITYr32v7tHag4UQq7XPZ7WWCv2Mn6mZ7HMRQuwUQizX9hOFEFu0638ihHDX2j20/WzteIKZ7QoUQnyufb8OCCHG2MI9E0Lcp32Oe4UQHwshPG3lnnUipbTJF/AtMFPbngX81GV7JSCA0cAWrT0YOKb9DNK2g8xg18XAd4CHth+u/UwDdgEeQCJwFHDRXkeBJMBdOyfNjPctFmONhFwg1Ebu2TTAVdt+DnjOlu6ZZovFr/mz6/cChmnbfsBh7f48D/xZa/9zl3vX7WdqRvvuBz4Clmv7nwLXaNtvAL/Ttn8PvKFtXwN8Yma7FgK3atvuQKC17xnG0rTHAa8u92q+rdyzjpfNjvw5h+LwUsrNQEdx+OloxeGllFVAR3F4U/M74FkpZQuAlLKsi12LpJQtUsrjGGsZjNRe2VLKY1LKVmCRdq65eAl4EOP968Cq90xK+a2Usl3b3YyxgluHXbZwz7DSNTuRUhZLKXdo23XAAYwikoFR4NB+ztW2T/eZmhwhRAxwKVqlPiGEwFi86fPT2NVh7+fAZO18c9gVAEzEWD4WKWWrlLIaG7hnGNPlewljrRNvoBgbuGddsWXxvxd4QQiRD/wDeEhr73Fx+B6SCkzQHs/WCCFG2IhdCCEygEIp5a6fHbK6bV24GePoy9bsssY1u0V77B8KbAEipLEKHkAJEKFtW9LelzEOKAzafghQ3aVD73rtTru04zXa+eYgESgH3tVcUm8JIXyw8j2TUhZi1Kw8jKJfA2zHNu5ZJz2t5NUjhJWKw/fQLleMbpLRwAjgUyFEkiXsOgfb/oLRxWJxzmSXlHKpds7DQDvwoSVtsyeEEL7AF8C9UsrargNAKaUUQlg0NlsIMRsok1JuF0JMsuS1zwFXYBhwl5RyixDiFYxunk6sdM+CMI7mE4FqjPXNzeGF6BFWFX9po8Xhz2LX74DF0uig2yqEMGBM0nQ6uzhDu8lsE0IMxPhl26UJRgywQxgnyq16zzT75gOzgcnaveMMdnGGdnNxJlssghDCDaPwfyilXKw1lwohekkpizUXRYeb0VL2jgPmCCFmAZ4YXbGvYHSZuGoj1a7X7rCrQHN5BAAVZrALjKPnAinlFm3/c4zib+17NgU4LqUsBxBCLMZ4H23hnp3EEhMLF/LC6POcpG1PBrZr25dy6qTNVq09GOMkS5D2Og4Em8Gu32KsPQxGF1C+Zkt/Tp28PIZxEtFV207k5ERifwvcvxxOTvha+57NAPYDYT9rt5l7Zq3Pqcv1BfA+8PLP2l/g1MnL58/0mZrZxkmcnPD9jFMnL3+vbd/BqZOXn5rZpnVAH237Ce1+WfWeAaOAfRh9/QKjP/8uW7lnnXZa4iIXeAPHY/ST7cLo+xyutQvgdYyRGXuA9C7vuRnjpGE2cJOZ7HIH/gfsBXYAl3Q59rBm1yG0SCWtfRbG6I2jGN0glrh/OZwUf2vfs2yMnWSW9nrDRu+Zxa/Z5drjMU7S7+5yn2Zh9P1+DxzBGGUWfLbP1Iw2TuKk+CcBW7XP9jNORr95avvZ2vEkM9s0BMjU7tsSjIMYq98z4EngoKYTH2Ac4NjEPet4qfQOCoVC4YTYcrSPQqFQKMyEEn+FQqFwQpT4KxQKhROixF+hUCicECX+CoVC4YQo8VcoFAonRIm/QqFQOCH/DyHqNQwjF0eXAAAAAElFTkSuQmCC\n",
406 | "text/plain": [
407 | ""
408 | ]
409 | },
410 | "metadata": {
411 | "needs_background": "light"
412 | },
413 | "output_type": "display_data"
414 | }
415 | ],
416 | "source": [
417 | "# let's optimize the path for printing\n",
418 | "Ord,NNd=TipSlicerUpdated.TSPnearest(optPoints)\n",
419 | "plot(optPoints[Ord][:,0],optPoints[Ord][:,1],'-')\n",
420 | "print(NNd)"
421 | ]
422 | },
423 | {
424 | "cell_type": "markdown",
425 | "metadata": {},
426 | "source": [
427 | "### First check different FOV + Blocks to see if FOV big enough"
428 | ]
429 | },
430 | {
431 | "cell_type": "code",
432 | "execution_count": 15,
433 | "metadata": {},
434 | "outputs": [],
435 | "source": [
436 | "i=0\n",
437 | "Fov=400 # This is important! How big of a fov in microns you are trying to print before the stage moves (can get away with up to 400)\n",
438 | "hatching=0.2 \n",
439 | "am=np.int(np.round(Fov/hatching))\n",
440 | "z=50"
441 | ]
442 | },
443 | {
444 | "cell_type": "code",
445 | "execution_count": 18,
446 | "metadata": {},
447 | "outputs": [
448 | {
449 | "name": "stdout",
450 | "output_type": "stream",
451 | "text": [
452 | "Working on 22\n"
453 | ]
454 | }
455 | ],
456 | "source": [
457 | "xc=optPoints[Ord[i],0]\n",
458 | "yc=optPoints[Ord[i],1]\n",
459 | "\n",
460 | "xx=np.linspace(xc-Fov/2.0,xc+Fov/2.0,am)\n",
461 | "yy=np.linspace(yc-Fov/2.0,yc+Fov/2.0,am)\n",
462 | "Xloc,Yloc=np.meshgrid(xx,yy)\n",
463 | "print('Working on {}'.format(Ord[i]))\n",
464 | "\n",
465 | "#Back=sliceFormulaAber(Xloc,Yloc,z,[xpos,ypos,rlist,r_lenslet,height,aperR,-1,zernlist])\n",
466 | "T=sliceFormulaAber(Xloc,Yloc,z,[xpos,ypos,rlist,r_lenslet,height,aperR,Ord[i],zernlist])\n",
467 | "Mask=maskFormulaAber(Xloc,Yloc,z,[xpos,ypos,rlist,r_lenslet,height,aperR,Ord[i],zernlist])\n",
468 | "Res=T*(Mask)\n",
469 | "#Back=np.where(Back==0,NaN,Back)\n",
470 | "#imshow(Back+Res)\n",
471 | "#print(i)\n",
472 | "#i=i+1"
473 | ]
474 | },
475 | {
476 | "cell_type": "code",
477 | "execution_count": 22,
478 | "metadata": {},
479 | "outputs": [],
480 | "source": [
481 | "# this name will define the sub-folder that will be written to\n",
482 | "rootName='MicroAberStitch_36lenses_pdms_01_500fov_corrected'\n"
483 | ]
484 | },
485 | {
486 | "cell_type": "markdown",
487 | "metadata": {},
488 | "source": [
489 | "# Let's write the files need for the Nanoscribe printer\n",
490 | "This will take some time to run"
491 | ]
492 | },
493 | {
494 | "cell_type": "code",
495 | "execution_count": 23,
496 | "metadata": {},
497 | "outputs": [
498 | {
499 | "name": "stdout",
500 | "output_type": "stream",
501 | "text": [
502 | "Writing block 0\n",
503 | "Writing block 1\n",
504 | "Writing block 2\n",
505 | "Writing block 3\n",
506 | "Writing block 4\n",
507 | "Writing block 5\n",
508 | "Writing block 6\n",
509 | "Writing block 7\n",
510 | "Writing block 8\n",
511 | "Writing block 9\n",
512 | "Writing block 10\n",
513 | "Writing block 11\n",
514 | "Writing block 12\n",
515 | "Writing block 13\n",
516 | "Writing block 14\n",
517 | "Writing block 15\n",
518 | "Writing block 16\n",
519 | "Writing block 17\n",
520 | "Writing block 18\n",
521 | "Writing block 19\n",
522 | "Writing block 20\n",
523 | "Writing block 21\n",
524 | "Writing block 22\n",
525 | "Writing block 23\n",
526 | "Writing block 24\n",
527 | "Writing block 25\n",
528 | "Writing block 26\n",
529 | "Writing block 27\n",
530 | "Writing block 28\n",
531 | "Writing block 29\n",
532 | "Writing block 30\n",
533 | "Writing block 31\n",
534 | "Writing block 32\n",
535 | "Writing block 33\n",
536 | "Writing block 34\n",
537 | "Writing block 35\n"
538 | ]
539 | }
540 | ],
541 | "source": [
542 | "#%%%time\n",
543 | "# STEP 1: Write out the H5 stacks, intermediate format that is equivalent to a sliced STL\n",
544 | "# => only changes if you change parameters here (slicing,hashing,writing height)\n",
545 | "# => for other variations (number of shells etc..) you don't have to rerun this\n",
546 | "\n",
547 | "FovZ=54.120638 # should be maximum size of total element, used for slice determination\n",
548 | "height=50\n",
549 | "roughSlicing=2\n",
550 | "fineSlicing=0.1\n",
551 | "zzbase=np.arange(0,height,roughSlicing) # speed up writing of base\n",
552 | "zzlenses=np.arange(height,FovZ,fineSlicing)\n",
553 | "zz=np.append(zzbase,zzlenses)\n",
554 | "\n",
555 | "\n",
556 | "Fov=500 # 600 is quite a lot, but using the 25x, could be possible. \n",
557 | " # diameter of writing circle, using the Galvo and the 25x, 400µm is definitely writeable, \n",
558 | " # it just implies the voxel might get deformed by optical aberrations of the objective\n",
559 | " # => you can expect some quality decrease but no doubt still better than having a stitching line..\n",
560 | "\n",
561 | "hatching=0.2 #should be 0.2\n",
562 | "am=np.int(np.round(Fov/hatching)) \n",
563 | "\n",
564 | "\n",
565 | "\n",
566 | "#overlapXY_multiplier=3 # this is how much the different stitching blocks would overlap (this x hatching). \n",
567 | "# the masks are not overlapping for the moment, if we want this, needs to be implemented in masking function..\n",
568 | "\n",
569 | "if (not os.path.exists(rootName)):\n",
570 | " os.mkdir(rootName)\n",
571 | "if (not os.path.exists(rootName+'/h5data')):\n",
572 | " os.mkdir(rootName+'/h5data')\n",
573 | "if (not os.path.exists(rootName+'/data')):\n",
574 | " os.mkdir(rootName+'/data')\n",
575 | "genBlockName=rootName+'/h5data/'+rootName+'_Block_{:05d}.h5'\n",
576 | "genGwlName=rootName+'/data/'+rootName+'_Block_{:05d}.gwl'\n",
577 | "genRegex=rootName+'/data/.+gwl'\n",
578 | "fullGwlName=rootName+'/'+rootName+'.gwl'\n",
579 | "\n",
580 | "for i in np.arange(len(Ord)):\n",
581 | "#for i in np.arange(5):\n",
582 | " # now make coordinate systems\n",
583 | " print('Writing block {}'.format(i))\n",
584 | " xc=optPoints[Ord[i],0]\n",
585 | " yc=optPoints[Ord[i],1]\n",
586 | "\n",
587 | " xx=np.linspace(xc-Fov/2.0,xc+Fov/2.0,am)\n",
588 | " yy=np.linspace(yc-Fov/2.0,yc+Fov/2.0,am)\n",
589 | " Xloc,Yloc=np.meshgrid(xx,yy)\n",
590 | " \n",
591 | " # make specific mask\n",
592 | "# MM=maskFormula(Xloc,Yloc,z,maskParams)\n",
593 | " # if you need overlap, do binary_dilation on the mask\n",
594 | "# MM=im.binary_dilation(MM,iterations=overlapXY_multiplier)\n",
595 | " \n",
596 | " # with these coordinate systems, apply formulaToStack\n",
597 | " blockName=genBlockName.format(i)\n",
598 | " \n",
599 | " extras=[xpos,ypos,rlist,r_lenslet,height,aperR,Ord[i],zernlist]\n",
600 | " Mask=maskFormulaAber(Xloc,Yloc,50.1,[xpos,ypos,rlist,r_lenslet,height,aperR,Ord[i],zernlist])\n",
601 | " \n",
602 | " # if you need overlap between the masks, apply some dilation here..\n",
603 | " Mask=im.binary_dilation(Mask,iterations=3)\n",
604 | " \n",
605 | " TipSlicerUpdated.formulaToStack(blockName,Xloc,Yloc,zz,sliceFormulaAber,writingMask=Mask,extraParams=extras)\n",
606 | " \n",
607 | " "
608 | ]
609 | },
610 | {
611 | "cell_type": "code",
612 | "execution_count": 24,
613 | "metadata": {},
614 | "outputs": [
615 | {
616 | "name": "stdout",
617 | "output_type": "stream",
618 | "text": [
619 | "Writing block 0\n",
620 | "Writing block 1\n",
621 | "Writing block 2\n",
622 | "Writing block 3\n",
623 | "Writing block 4\n",
624 | "Writing block 5\n",
625 | "Writing block 6\n",
626 | "Writing block 7\n",
627 | "Writing block 8\n",
628 | "Writing block 9\n",
629 | "Writing block 10\n",
630 | "Writing block 11\n",
631 | "Writing block 12\n",
632 | "Writing block 13\n",
633 | "Writing block 14\n",
634 | "Writing block 15\n",
635 | "Writing block 16\n",
636 | "Writing block 17\n",
637 | "Writing block 18\n",
638 | "Writing block 19\n",
639 | "Writing block 20\n",
640 | "Writing block 21\n",
641 | "Writing block 22\n",
642 | "Writing block 23\n",
643 | "Writing block 24\n",
644 | "Writing block 25\n",
645 | "Writing block 26\n",
646 | "Writing block 27\n",
647 | "Writing block 28\n",
648 | "Writing block 29\n",
649 | "Writing block 30\n",
650 | "Writing block 31\n",
651 | "Writing block 32\n",
652 | "Writing block 33\n",
653 | "Writing block 34\n",
654 | "Writing block 35\n"
655 | ]
656 | }
657 | ],
658 | "source": [
659 | "#%%%time\n",
660 | "# STEP 2: Write out GWL files based on H5 files from above\n",
661 | "\n",
662 | "amShells=5\n",
663 | "ScaffStep=2 # for solid infill => 2*hatching is spacing here\n",
664 | "hatchStep=1 # for contour distance and top-bottom infill, not used if amShells==1 and doTopBottom=False\n",
665 | "\n",
666 | "\n",
667 | "# the definition below will be used to increase the writing laser power when the slicing is larger\n",
668 | "# below the base height, the intensity will be ? times larger, above it the nominal one will be used.\n",
669 | "def intensityMultiplier(z):\n",
670 | " if (z
