43 |
🧬 TF Inference on PBMC 3K
44 |
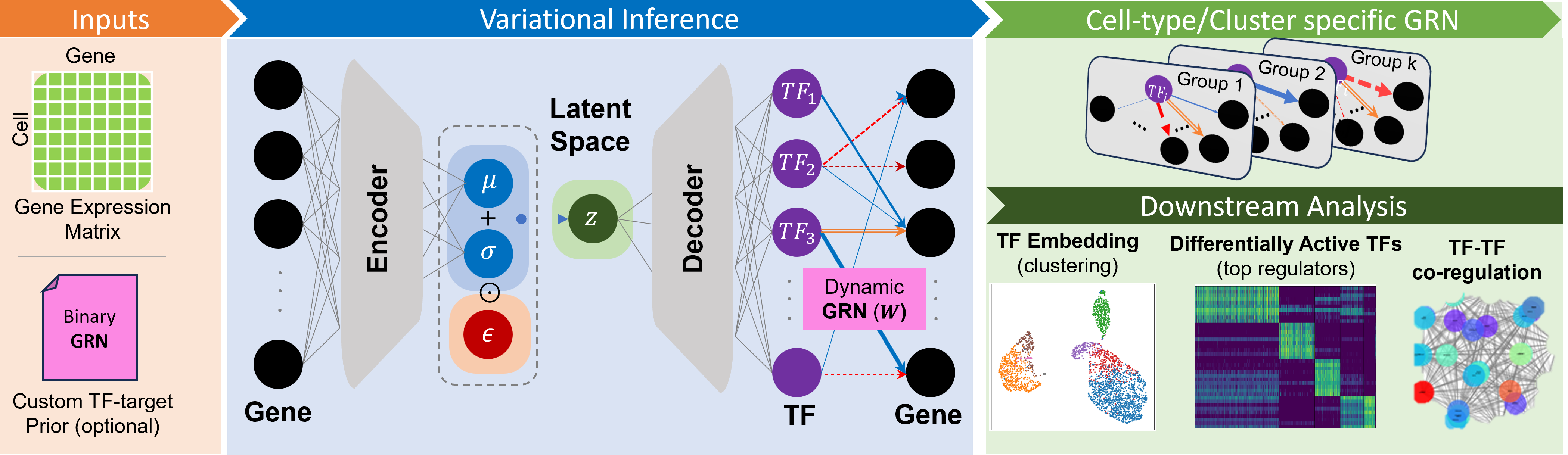
This tutorial walks you through the basic usage of scRegulate on a small PBMC 3K dataset.
45 |
49 |
52 |
📄 Reproducing Manuscript Results
53 |
This notebook replicates the preprocessing and analysis pipeline used in our paper.
54 |
58 |
 15 |
15 |