├── .gitignore
├── Evaluate_ADB_Project.ipynb
├── LICENSE
├── README.md
├── Report.pdf
├── Statistics IVF.xlsx
├── proposal.pdf
├── requirements.txt
├── src
├── .gitignore
├── Draft
│ ├── access.py
│ ├── access_wrong.py
│ ├── file.py
│ ├── gen.py
│ ├── genData.ipynb
│ ├── generate.py
│ └── test_data.py
├── IVF.py
├── Modules
│ ├── IVF.py
│ └── LSH.py
├── __init__.py
├── api.py
├── best_case_implementation.py
├── evaluation.py
├── main.py
├── notes.txt
├── pipeline.ipynb
├── utils.py
├── vec_db.py
└── worst_case_implementation.py
└── vector searching algorithms
├── LSH.ipynb
├── LSHHyperPlane.ipynb
├── Product Qunatization.ipynb
├── SplitBySign
├── Split_by_sign.ipynb
└── split_by_sign.py
└── clustering.ipynb
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 | *.vscode
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | share/python-wheels/
24 | *.egg-info/
25 | .installed.cfg
26 | *.egg
27 | MANIFEST
28 |
29 | # PyInstaller
30 | # Usually these files are written by a python script from a template
31 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
32 | *.manifest
33 | *.spec
34 |
35 | # Installer logs
36 | pip-log.txt
37 | pip-delete-this-directory.txt
38 |
39 | # Unit test / coverage reports
40 | htmlcov/
41 | .tox/
42 | .nox/
43 | .coverage
44 | .coverage.*
45 | .cache
46 | nosetests.xml
47 | coverage.xml
48 | *.cover
49 | *.py,cover
50 | .hypothesis/
51 | .pytest_cache/
52 | cover/
53 |
54 | # Translations
55 | *.mo
56 | *.pot
57 |
58 | # Django stuff:
59 | *.log
60 | local_settings.py
61 | db.sqlite3
62 | db.sqlite3-journal
63 |
64 | # Flask stuff:
65 | instance/
66 | .webassets-cache
67 |
68 | # Scrapy stuff:
69 | .scrapy
70 |
71 | # Sphinx documentation
72 | docs/_build/
73 |
74 | # PyBuilder
75 | .pybuilder/
76 | target/
77 |
78 | # Jupyter Notebook
79 | .ipynb_checkpoints
80 |
81 | # IPython
82 | profile_default/
83 | ipython_config.py
84 |
85 | # pyenv
86 | # For a library or package, you might want to ignore these files since the code is
87 | # intended to run in multiple environments; otherwise, check them in:
88 | # .python-version
89 |
90 | # pipenv
91 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
92 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
93 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
94 | # install all needed dependencies.
95 | #Pipfile.lock
96 |
97 | # poetry
98 | # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
99 | # This is especially recommended for binary packages to ensure reproducibility, and is more
100 | # commonly ignored for libraries.
101 | # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
102 | #poetry.lock
103 |
104 | # pdm
105 | # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
106 | #pdm.lock
107 | # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
108 | # in version control.
109 | # https://pdm.fming.dev/#use-with-ide
110 | .pdm.toml
111 |
112 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
113 | __pypackages__/
114 |
115 | # Celery stuff
116 | celerybeat-schedule
117 | celerybeat.pid
118 |
119 | # SageMath parsed files
120 | *.sage.py
121 |
122 | # Environments
123 | .env
124 | .venv
125 | env/
126 | venv/
127 | ENV/
128 | env.bak/
129 | venv.bak/
130 |
131 | # Spyder project settings
132 | .spyderproject
133 | .spyproject
134 |
135 | # Rope project settings
136 | .ropeproject
137 |
138 | # mkdocs documentation
139 | /site
140 |
141 | # mypy
142 | .mypy_cache/
143 | .dmypy.json
144 | dmypy.json
145 |
146 | # Pyre type checker
147 | .pyre/
148 |
149 | # pytype static type analyzer
150 | .pytype/
151 |
152 | # Cython debug symbols
153 | cython_debug/
154 |
155 | # PyCharm
156 | # JetBrains specific template is maintained in a separate JetBrains.gitignore that can
157 | # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
158 | # and can be added to the global gitignore or merged into this file. For a more nuclear
159 | # option (not recommended) you can uncomment the following to ignore the entire idea folder.
160 | #.idea/
161 |
162 | DataBase
163 | bucket_files
164 | .vscode
165 | modules/inverted_files
166 | ```
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2024 Ziad Sherif
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | IntelliQuery
2 |
3 | ## 📝 Table of Contents
4 |
5 | - [📝 Table of Contents](#-table-of-contents)
6 | - [📙 Overview ](#-overview-)
7 | - [Get Started ](#get-started-)
8 | - [Infernce Mode](#infernce-mode)
9 | - [Run Locally](#run-locally)
10 | - [Methods](#methods)
11 | - [Inverted File Inedex (IVF) ](#inverted-file-inedex-ivf-)
12 | - [Local Sensitive Hashing (LSH) ](#local-sensitive-hashing-lsh-)
13 | - [Product Qunatization (PQ) ](#product-qunatization-pq-)
14 | - [PQ-LSH ](#pq-lsh-)
15 | - [🕴 Contributors ](#-contributors-)
16 | - [📃 License ](#-license-)
17 |
18 | ## 📙 Overview
19 | Given the embedding of the search query we can efficent get the top matching k results form DB with 20M document.The objective of this project is to design and implement an indexing system for a
20 | semantic search database.
21 |
22 |
23 | ##  Get Started
24 | ### Infernce Mode
25 | ***Check Final Notebook***
26 | ```
27 | https://github.com/ZiadSheriif/IntelliQuery/blob/main/Evaluate_ADB_Project.ipynb
28 | ```
29 | ### Run Locally
30 |
31 | ***Clone Repo***
32 | ```
33 | git clone https://github.com/ZiadSheriif/IntelliQuery.git
34 | ```
35 | ***Install dependencies***
36 | ```
37 | pip install -r requirements.txt
38 | ```
39 | ***Run Indexer***
40 | ```
41 | $ python ./src/evaluation.py
42 | ```
43 |
44 |
45 | ##
Get Started
24 | ### Infernce Mode
25 | ***Check Final Notebook***
26 | ```
27 | https://github.com/ZiadSheriif/IntelliQuery/blob/main/Evaluate_ADB_Project.ipynb
28 | ```
29 | ### Run Locally
30 |
31 | ***Clone Repo***
32 | ```
33 | git clone https://github.com/ZiadSheriif/IntelliQuery.git
34 | ```
35 | ***Install dependencies***
36 | ```
37 | pip install -r requirements.txt
38 | ```
39 | ***Run Indexer***
40 | ```
41 | $ python ./src/evaluation.py
42 | ```
43 |
44 |
45 | ##  Methods
46 | ### Inverted File Inedex (IVF)
47 | This is out final Approach with Some Enhancements
48 | 1. Changed MiniBatchKMeans to regular KMeans
49 | 2. We calculate initial centroids with just the first chunk of data
50 | 3. Introduced parallel processing for different regions
51 |
Methods
46 | ### Inverted File Inedex (IVF)
47 | This is out final Approach with Some Enhancements
48 | 1. Changed MiniBatchKMeans to regular KMeans
49 | 2. We calculate initial centroids with just the first chunk of data
50 | 3. Introduced parallel processing for different regions
51 | 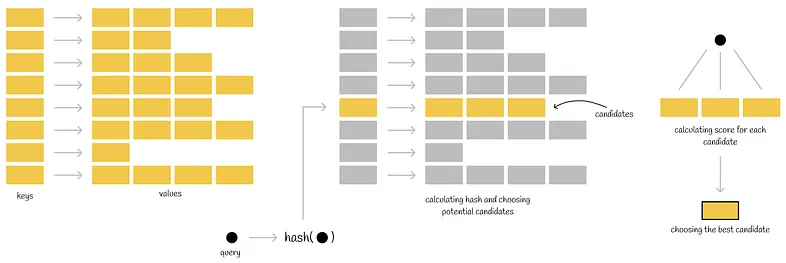 52 |
53 | ### Local Sensitive Hashing (LSH)
54 |
52 |
53 | ### Local Sensitive Hashing (LSH)
54 | 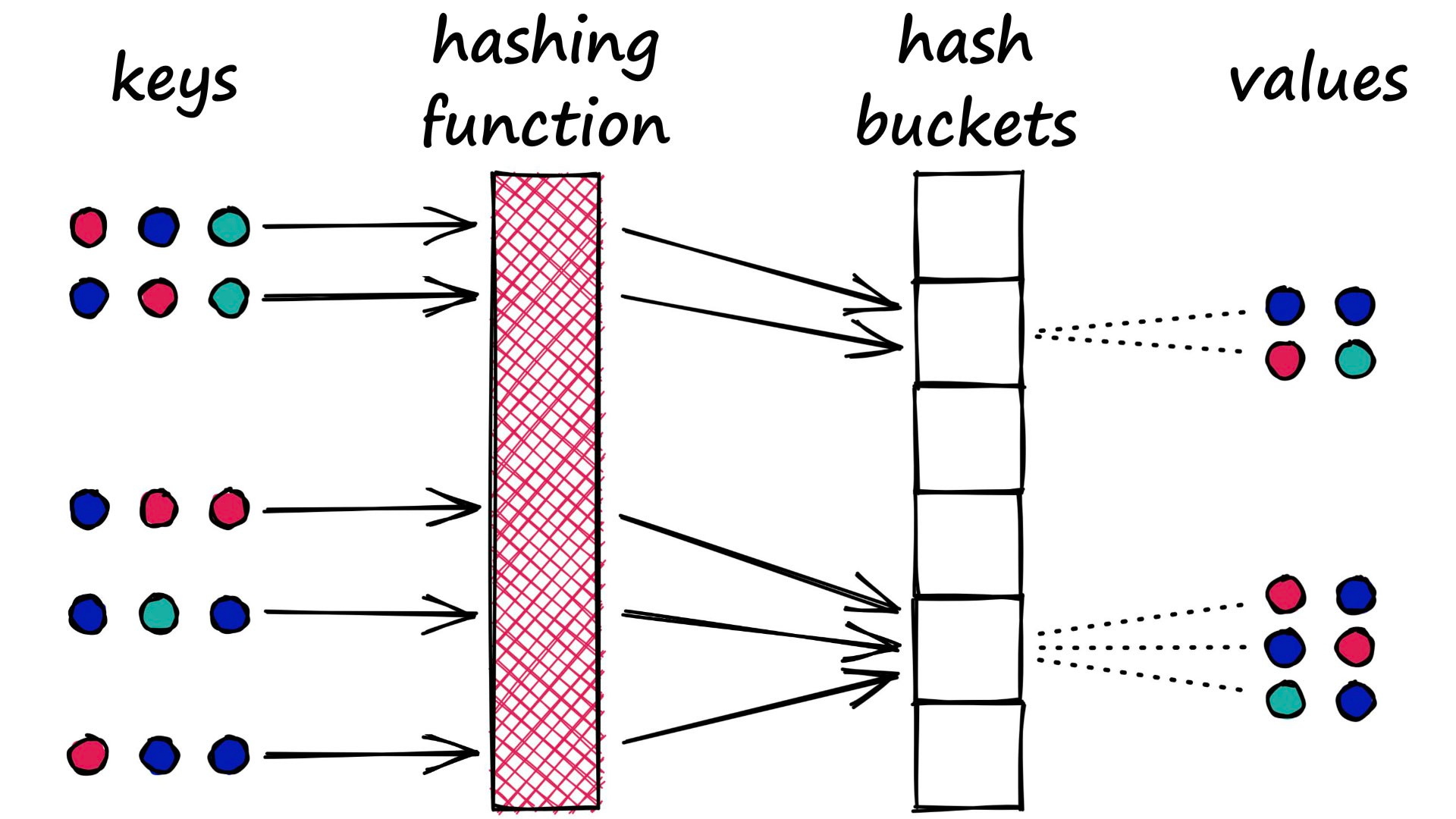 55 |
56 | ### Product Qunatization (PQ)
57 |
55 |
56 | ### Product Qunatization (PQ)
57 | 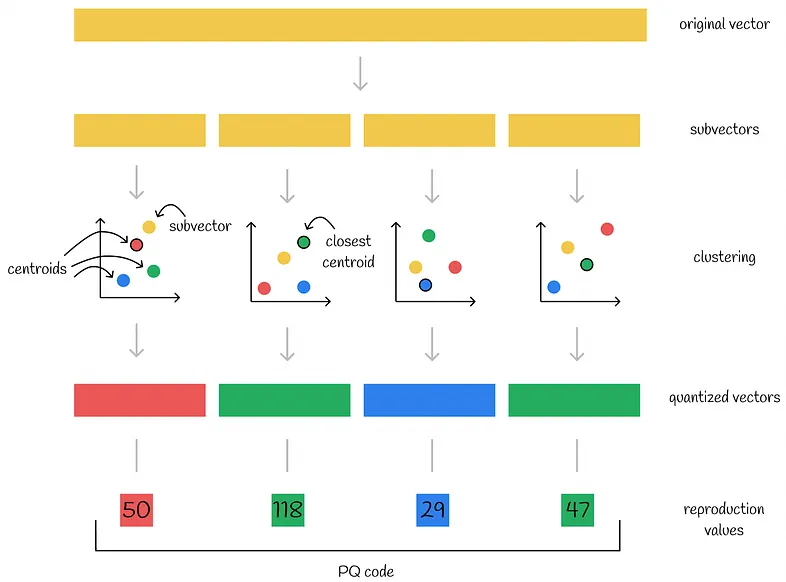 58 |
59 | ### PQ-LSH
60 | It Combines both LSH & PQ
61 |
62 | ## 🕴 Contributors
63 |
64 |
65 |
74 |
75 |
76 |
77 | ## 📃 License
78 |
79 | This software is licensed under MIT License, See [License](https://github.com/ZiadSheriif/sematic_search_DB/blob/main/LICENSE) for more information ©Ziad Sherif.
80 |
--------------------------------------------------------------------------------
/Report.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/Report.pdf
--------------------------------------------------------------------------------
/Statistics IVF.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/Statistics IVF.xlsx
--------------------------------------------------------------------------------
/proposal.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/proposal.pdf
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/requirements.txt
--------------------------------------------------------------------------------
/src/.gitignore:
--------------------------------------------------------------------------------
1 | Database/
2 | inverted_files/
3 |
4 | # Byte-compiled / optimized / DLL files
5 | __pycache__/
6 | *.py[cod]
7 | *$py.class
8 | *.csv
9 | *.bin
10 |
11 | DataBase
12 |
13 | # C extensions
14 | *.so
15 |
16 | # Distribution / packaging
17 | .Python
18 | build/
19 | develop-eggs/
20 | dist/
21 | downloads/
22 | eggs/
23 | .eggs/
24 | lib/
25 | lib64/
26 | parts/
27 | sdist/
28 | var/
29 | wheels/
30 | share/python-wheels/
31 | *.egg-info/
32 | .installed.cfg
33 | *.egg
34 | MANIFEST
35 |
36 | # PyInstaller
37 | # Usually these files are written by a python script from a template
38 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
39 | *.manifest
40 | *.spec
41 |
42 | # Installer logs
43 | pip-log.txt
44 | pip-delete-this-directory.txt
45 |
46 | # Unit test / coverage reports
47 | htmlcov/
48 | .tox/
49 | .nox/
50 | .coverage
51 | .coverage.*
52 | .cache
53 | nosetests.xml
54 | coverage.xml
55 | *.cover
56 | *.py,cover
57 | .hypothesis/
58 | .pytest_cache/
59 | cover/
60 |
61 | # Translations
62 | *.mo
63 | *.pot
64 |
65 | # Django stuff:
66 | *.log
67 | local_settings.py
68 | db.sqlite3
69 | db.sqlite3-journal
70 |
71 | # Flask stuff:
72 | instance/
73 | .webassets-cache
74 |
75 | # Scrapy stuff:
76 | .scrapy
77 |
78 | # Sphinx documentation
79 | docs/_build/
80 |
81 | # PyBuilder
82 | .pybuilder/
83 | target/
84 |
85 | # Jupyter Notebook
86 | .ipynb_checkpoints
87 |
88 | # IPython
89 | profile_default/
90 | ipython_config.py
91 |
92 | # pyenv
93 | # For a library or package, you might want to ignore these files since the code is
94 | # intended to run in multiple environments; otherwise, check them in:
95 | # .python-version
96 |
97 | # pipenv
98 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
99 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
100 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
101 | # install all needed dependencies.
102 | #Pipfile.lock
103 |
104 | # poetry
105 | # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
106 | # This is especially recommended for binary packages to ensure reproducibility, and is more
107 | # commonly ignored for libraries.
108 | # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
109 | #poetry.lock
110 |
111 | # pdm
112 | # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
113 | #pdm.lock
114 | # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
115 | # in version control.
116 | # https://pdm.fming.dev/#use-with-ide
117 | .pdm.toml
118 |
119 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
120 | __pypackages__/
121 |
122 | # Celery stuff
123 | celerybeat-schedule
124 | celerybeat.pid
125 |
126 | # SageMath parsed files
127 | *.sage.py
128 |
129 | # Environments
130 | .env
131 | .venv
132 | env/
133 | venv/
134 | ENV/
135 | env.bak/
136 | venv.bak/
137 |
138 | # Spyder project settings
139 | .spyderproject
140 | .spyproject
141 |
142 | # Rope project settings
143 | .ropeproject
144 |
145 | # mkdocs documentation
146 | /site
147 |
148 | # mypy
149 | .mypy_cache/
150 | .dmypy.json
151 | dmypy.json
152 |
153 | # Pyre type checker
154 | .pyre/
155 |
156 | # pytype static type analyzer
157 | .pytype/
158 |
159 | # Cython debug symbols
160 | cython_debug/
161 |
162 | # PyCharm
163 | # JetBrains specific template is maintained in a separate JetBrains.gitignore that can
164 | # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
165 | # and can be added to the global gitignore or merged into this file. For a more nuclear
166 | # option (not recommended) you can uncomment the following to ignore the entire idea folder.
167 | #.idea/
168 | bucket_files
169 | Modules/bucket_files
170 | DataBase
171 | .vscode
--------------------------------------------------------------------------------
/src/Draft/access.py:
--------------------------------------------------------------------------------
1 | import struct
2 |
3 | # Define the binary file name
4 | binary_file_name = 'records_with_index_name.bin'
5 |
6 | # Define the index of the element you want to access
7 | i = 9999 # Change this to the desired index
8 |
9 | # Calculate the position of the ith element based on record size
10 | record_size = struct.calcsize('I20s20s') # Size of packed data
11 | print(record_size)
12 | position = i * record_size
13 |
14 | # Get the address of the first block in the binary file
15 | binary_file_address = 0
16 | # with open(binary_file_name, 'rb') as file:
17 | # binary_file_address = file.tell()
18 | # print(binary_file_address)
19 |
20 | # Calculate the absolute position of the ith element
21 | absolute_position = binary_file_address + position
22 |
23 | # Open the binary file and seek to the absolute position of the ith element
24 | with open(binary_file_name, 'rb') as file:
25 | file.seek(absolute_position)
26 |
27 | # Read the packed data at the ith position
28 | packed_data = file.read(record_size)
29 |
30 | # Unpack the data
31 | index, name, phone = struct.unpack('I20s20s', packed_data)
32 | name = name.decode().strip('\0')
33 | phone = phone.decode().strip('\0')
34 |
35 | print(f'Index: {index}, Name: {name}, Phone: {phone}')
36 |
--------------------------------------------------------------------------------
/src/Draft/access_wrong.py:
--------------------------------------------------------------------------------
1 | # Open a file: file
2 | file = open('records_with_index_name_phone.bin',mode='r')
3 |
4 | # read all lines at once
5 | all_of_it = file.read()
6 |
7 | # close the file
8 | file.close()
--------------------------------------------------------------------------------
/src/Draft/file.py:
--------------------------------------------------------------------------------
1 | import csv
2 |
3 | # Generate 10,000 float values based on their index
4 | float_data = [float(i) for i in range(10000)]
5 |
6 | # Define the CSV file name
7 | csv_file_name = 'float_records_with_index.csv'
8 |
9 | # Write the float data with index to the CSV file
10 | with open(csv_file_name, 'w', newline='') as csvfile:
11 | writer = csv.writer(csvfile)
12 | writer.writerow(['Index', 'Value']) # Write a header row
13 | for index, value in enumerate(float_data):
14 | writer.writerow([index, value])
15 |
16 | # Get the address of the first block in the CSV file
17 | csv_file_address = None
18 | with open(csv_file_name, 'rb') as file:
19 | csv_file_address = file.tell()
20 |
21 | print(f'CSV file created: {csv_file_name}')
22 | print(f'Address of the first block in the CSV file: {csv_file_address}')
23 |
--------------------------------------------------------------------------------
/src/Draft/gen.py:
--------------------------------------------------------------------------------
1 | from worst_case_implementation import VecDBWorst
2 | import numpy as np
3 |
4 | # Function to generate random embeddings
5 | def generate_embeddings(num_records, embedding_dim):
6 | return [np.random.rand(embedding_dim).tolist() for _ in range(num_records)]
7 |
8 | # Create an instance of VecDB
9 | db = VecDBWorst()
10 |
11 | # Define parameters
12 | total_records = 10000 # 20 million records
13 | chunk_size = 10000 # Insert records in chunks of 10,000
14 |
15 | # Insert records in chunks
16 | for i in range(0, total_records, chunk_size):
17 | chunk_records = []
18 | for j in range(i + 1, i + chunk_size + 1):
19 | if j > total_records:
20 | break
21 | record = {"id": j, "embed": generate_embeddings(1, 70)[0]}
22 | # make this size of record to be fixed 1500 bytes
23 | # size_of_dummy_needed = 1500 - len(record["embed"])
24 |
25 | chunk_records.append(record)
26 |
27 | db.insert_records(chunk_records)
28 | print(f"Inserted {len(chunk_records)} records. Total records inserted: {j}")
29 |
30 | print("Insertion complete.")
31 |
--------------------------------------------------------------------------------
/src/Draft/generate.py:
--------------------------------------------------------------------------------
1 | import struct
2 |
3 | # Define the binary file name
4 | binary_file_name = 'records_with_index_name.bin'
5 |
6 | # Generate and write the records to the binary file
7 | with open(binary_file_name, 'wb') as file:
8 | for i in range(10000):
9 | # Generate example name and phone number (you can replace with your data source)
10 | name = f"Name-{i}"
11 | phone = f"Phone-{i}"
12 |
13 | # Ensure a fixed length for name and phone
14 | name = name.ljust(20, '\0') # 20 characters
15 | phone = phone.ljust(20, '\0') # 20 characters
16 |
17 | # Pack data into binary format (4 bytes for index, 20 bytes for name, and 20 bytes for phone)
18 | packed_data = struct.pack('I20s20s', i, name.encode(), phone.encode())
19 |
20 | # Write the packed data to the binary file
21 | file.write(packed_data)
--------------------------------------------------------------------------------
/src/Draft/test_data.py:
--------------------------------------------------------------------------------
1 | from datasets import load_dataset
2 |
3 | dataset = load_dataset("aadityaubhat/GPT-wiki-intro")
4 |
5 | print(dataset['train'][0])
--------------------------------------------------------------------------------
/src/IVF.py:
--------------------------------------------------------------------------------
1 | # import files
2 | from utils import *
3 | from sklearn.cluster import KMeans
4 |
5 | # improt libraries
6 | import heapq
7 | import numpy as np
8 |
9 |
10 | def IVF_index(file_path,K_means_metric,K_means_n_clusters,k_means_batch_size,k_means_max_iter,k_means_n_init,chunk_size,index_folder_path):
11 | '''
12 | file_path: path to the data .bin file
13 |
14 | K_means_metric: metric to be used in clustering cosine or euclidean' or TODO use SCANN idea ERORR Think of another way this isn't supported in kmeans
15 | K_means_n_clusters: No of Kmeans Clusters
16 | k_means_batch_size: kmeans batch size to be sampled at each iteration of fitting
17 | k_means_max_iter: max iteration by kmeans default [100] in sklearn
18 | k_means_n_init:The number of times the algorithm will be run with different centroid seeds.
19 |
20 | chunk_size: chunk_size: no of records to be processing together in while performing kmeans
21 |
22 | ivf_folder_path: Folder path to store regions of kmeans
23 | '''
24 | print("---IVF_index()----")
25 | # ############################################################### ################################# ###############################################################
26 | # ############################################################### Step(1):Clustering Data from file ###############################################################
27 | # ############################################################### ################################# ###############################################################
28 | kmeans = KMeans(n_clusters=K_means_n_clusters, max_iter=k_means_max_iter,n_init=k_means_n_init,random_state=42)
29 |
30 |
31 | # Use the first Chunck to only get teh centroids
32 | data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=0,chunk_size=1000000) #[{"id":,"embed":[]}]
33 | # TODO Remove this loop
34 | chunk_vectors=np.array([entry['embed'] for entry in data_chunk])
35 | kmeans.fit(chunk_vectors)
36 |
37 |
38 |
39 | # We need to Read Data from File chunk by chunk
40 | file_size = os.path.getsize(file_path)
41 | record_size=struct.calcsize(f"I{70}f")
42 | n_records=file_size/record_size
43 | no_chunks=math.ceil(n_records/chunk_size)
44 |
45 | # # Step(1) Getting centroids:
46 | # # Loop to get the Kmeans Centroids
47 | # for i in range(no_chunks):
48 | # data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=i*chunk_size,chunk_size=chunk_size) #[{"id":,"embed":[]}]
49 | # # TODO Remove this loop
50 | # chunk_vectors=np.array([entry['embed'] for entry in data_chunk])
51 | # kmeans.partial_fit(chunk_vectors)
52 |
53 | # Centroids
54 | K_means_centroids=kmeans.cluster_centers_
55 | # Saving Centroids #TODO Check precision of centroids after read and write in the file @Basma Elhoseny
56 | write_binary_file(file_path=index_folder_path+'/centroids.bin',data_to_write=K_means_centroids,format=f"{70}f")
57 |
58 | # ##################################################################
59 | # #TEST# Centroids are Written Correct #############################
60 | # ##################################################################
61 |
62 |
63 |
64 | # Step(2) Getting vectors of each regions
65 | for i in range(no_chunks):
66 | data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=i*chunk_size,chunk_size=chunk_size,dictionary_format=True) #[{109: np.array([70 dim])}]
67 |
68 | # Get Cluster for each one
69 | labels=kmeans.predict(list(data_chunk.values())) #Each vector corresponding centroid
70 |
71 |
72 | ids=np.array(list(data_chunk.keys()))
73 | vectors=np.array(list(data_chunk.values()))
74 | data_chunk=None #Clear Memory
75 |
76 | # Add vectors to their corresponding region
77 | for label in set(labels):
78 | region_ids=ids[labels==label] # get ids belonging to such region
79 | region_vectors=vectors[labels==label] # get vectors belonging to such region
80 | # Open file of this Region(cluster) Just Once for every Region :D
81 | with open(index_folder_path+f'/cluster{label}.bin', "ab") as fout:
82 | for i in range(len(region_ids)):
83 | #TODO Check whether store id of the vector @Basma Elhoseny
84 | data = struct.pack(f"I{70}f", region_ids[i],*region_vectors[i,:])
85 | fout.write(data)
86 |
87 |
88 |
89 | return
90 |
91 |
92 |
93 | def semantic_query_ivf(data_file_path, index_folder_path, query, top_k, n_regions):
94 | query = np.squeeze(np.array(query))
95 |
96 |
97 | # Read Centroids
98 | K_means_centroids = read_binary_file(index_folder_path + '/centroids.bin', f"70f")
99 |
100 |
101 | assert K_means_centroids.shape[0] > n_regions, "n_regions must be less than the number of regions"
102 |
103 |
104 | # Calculate distances to centroids
105 | distances = np.linalg.norm(K_means_centroids - query, axis=1)
106 | # Get indices of the nearest centroids
107 | nearest_regions = np.argsort(distances)[:n_regions]
108 |
109 |
110 | # Use a heap to keep track of the top k scores
111 | top_scores_heap = []
112 | for region in nearest_regions:
113 | records=read_binary_file_chunk(index_folder_path+f'/cluster{region}.bin', f'I{70}f', 0, chunk_size=100000000000,dictionary_format=True)
114 |
115 |
116 | # Vectorize cosine similarity calculation
117 | vectors = np.array([record for record in records.values()])
118 | dot_products = np.dot(vectors, query)
119 | norms = np.linalg.norm(vectors, axis=1) * np.linalg.norm(query)
120 | similarities = dot_products / norms
121 |
122 | # Process the scores and maintain a heap
123 | for score, id in zip(similarities, records.keys()):
124 | if len(top_scores_heap) < top_k:
125 | heapq.heappush(top_scores_heap, (score, id))
126 | else:
127 | heapq.heappushpop(top_scores_heap, (score, id))
128 |
129 | # Sort and get the top k scores
130 | top_scores_heap.sort(reverse=True)
131 | top_k_ids = [id for _, id in top_scores_heap]
132 |
133 | return top_k_ids
134 |
--------------------------------------------------------------------------------
/src/Modules/IVF.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | from sklearn.cluster import KMeans
4 | import time
5 | from scipy.spatial.distance import cosine
6 |
7 |
8 | class InvertedFileSystem:
9 | def __init__(self, n_clusters, data_dir):
10 | self.n_clusters = n_clusters
11 | self.data_dir = data_dir
12 | self.inverted_file_paths = [
13 | os.path.join(data_dir, f"inverted_file_{i}.npy") for i in range(n_clusters)
14 | ]
15 | self.centroids = None
16 |
17 | def build_index(self, data):
18 | # Cluster the data
19 | kmeans = KMeans(n_clusters=self.n_clusters, n_init=10)
20 | labels = kmeans.fit_predict(data)
21 | self.centroids = kmeans.cluster_centers_
22 |
23 | # Build inverted files
24 | inverted_files = [[] for _ in range(self.n_clusters)]
25 | for idx, label in enumerate(labels):
26 | inverted_files[label].append(idx)
27 |
28 | # Save inverted files to disk
29 | for i, inverted_file in enumerate(inverted_files):
30 | np.save(self.inverted_file_paths[i], inverted_file)
31 |
32 | def query(self, vector, top_k=5):
33 | # Assign vector to nearest cluster
34 | nearest_cluster = np.argmin(np.linalg.norm(self.centroids - vector, axis=1))
35 |
36 | # Load the corresponding inverted file from disk
37 | inverted_file = np.load(self.inverted_file_paths[nearest_cluster])
38 |
39 | # Search in the inverted file
40 | distances = [np.linalg.norm(vector - data[idx]) for idx in inverted_file]
41 | nearest_indices = np.argsort(distances)[:top_k]
42 |
43 | return [inverted_file[i] for i in nearest_indices]
44 |
45 |
46 | def brute_force_cosine_similarity(query_vector, data, top_k=5):
47 | # Calculate cosine similarities for each vector in the dataset
48 | similarities = [1 - cosine(query_vector, vector) for vector in data]
49 |
50 | # Get the indices of the top k most similar vectors
51 | nearest_indices = np.argsort(similarities)[-top_k:]
52 |
53 | # Return the indices and their cosine similarities
54 | return [idx for idx in reversed(nearest_indices)]
55 |
56 | def run_queries(n_queries, ivf, data, top_k=5):
57 | total_time_ivf = 0
58 | total_time_brute_force = 0
59 | total_score_ivf = 0
60 | ivf_results = []
61 | brute_force_results = []
62 |
63 | for _ in range(n_queries):
64 | query_vector = np.random.rand(70)
65 |
66 | start_time = time.time()
67 | ivf_result = ivf.query(query_vector, top_k)
68 | end_time = time.time()
69 | total_time_ivf += end_time - start_time
70 | ivf_results.append(ivf_result)
71 |
72 | start_time = time.time()
73 | brute_force_result = brute_force_cosine_similarity(query_vector, data, top_k)
74 | end_time = time.time()
75 | total_time_brute_force += end_time - start_time
76 | brute_force_results.append(brute_force_result)
77 |
78 | intersection = len(set(ivf_result).intersection(brute_force_result))
79 | total_score_ivf += intersection / top_k
80 |
81 | avg_time_ivf = total_time_ivf / n_queries
82 | avg_score_ivf = total_score_ivf / n_queries
83 | avg_time_brute_force = total_time_brute_force / n_queries
84 |
85 | print(f"IVF: Average time = {avg_time_ivf}, Average score = {avg_score_ivf}")
86 | print(f"Brute Force: Average time = {avg_time_brute_force}")
87 |
88 | # Calculate intersection of top k results
89 | intersection = set(ivf_result).intersection(brute_force_result)
90 | print(f"Intersection of top {top_k} results: {intersection}")
91 |
92 | # !testing IVF
93 | data_dir = "inverted_files"
94 | os.makedirs(data_dir, exist_ok=True)
95 | number_of_queries=10
96 | data_set=10000
97 |
98 | data = np.random.rand(data_set, 70)
99 | ivf = InvertedFileSystem(n_clusters=5, data_dir=data_dir)
100 | ivf.build_index(data)
101 |
102 | print("Dataset in k: ",data_set//1000)
103 | print("Number of Queries: ",number_of_queries)
104 |
105 | run_queries(number_of_queries, ivf, data)
106 |
107 |
108 | # # !testing IVF
109 | # data_dir = "inverted_files"
110 | # os.makedirs(data_dir, exist_ok=True)
111 |
112 | # data = np.random.rand(100000, 70)
113 | # ivf = InvertedFileSystem(n_clusters=3, data_dir=data_dir)
114 | # ivf.build_index(data)
115 |
116 | # query_vector = np.random.rand(70)
117 |
118 |
119 | # # brute force search
120 | # start_time = time.time()
121 | # brute_force_results = brute_force_cosine_similarity(query_vector, data, top_k=10)
122 | # brute_force_time = time.time() - start_time
123 | # print("Brute force top k: ", brute_force_results)
124 | # print("Brute force time: ", brute_force_time)
125 | # print("============================================")
126 | # # Timing IVF query
127 | # start_time = time.time()
128 | # top_k_results = ivf.query(query_vector, top_k=10)
129 | # ivf_time = time.time() - start_time
130 | # print("IVF top k: ", top_k_results)
131 | # print("IVF time: ", ivf_time)
132 |
133 |
134 | # # Get intersection
135 | # brute_force_set = set(brute_force_results)
136 | # ivf_set = set(top_k_results)
137 |
138 | # intersection = brute_force_set.intersection(ivf_set)
139 | # print("Intersection of Brute Force and IVF: ", intersection)
140 | # print("length of the intersection: ", len(intersection))
141 |
142 | # print("********************************************")
143 |
--------------------------------------------------------------------------------
/src/Modules/LSH.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import os
3 |
4 |
5 | from scipy.spatial.distance import cosine
6 | # from best_case_implementation import VecDBBest
7 |
8 |
9 | # TODO:
10 | # * 1) Build LSH function (indexing)
11 | # * 2) Build semantic query function (retrieval)
12 |
13 |
14 | def LSH_index(data, nbits, index_path, d=70):
15 | """
16 | Function to Build the LSH indexing
17 | data:[{'id':int,'embed':vector}]
18 | nbits: no of bits of the Buckets
19 | index_path:path of the Result to be saved
20 | d: vector dimension
21 | """
22 | # create nbits Random hyperplanes used for portioning
23 |
24 | plane_norms = np.random.rand(nbits, d) - 0.5
25 |
26 | #! for -1,1
27 | #? plane_norms = 2 * np.random.rand(nbits, d) - 1.0
28 |
29 | # If index Folder Doesn't Exist just Create it :D
30 | if not os.path.exists(index_path):
31 | os.makedirs(index_path)
32 |
33 |
34 | for item in data:
35 | vector = item["embed"]
36 | id = item["id"]
37 |
38 | # Dot Product with Random Planes
39 | data_dot_product = np.dot(vector, plane_norms.T)
40 |
41 | # Decision Making
42 | data_set_decision_hamming = (data_dot_product > 0) * 1
43 |
44 | # Bucket no. (Key)
45 | hash_str = "".join(data_set_decision_hamming.astype(str)) # 101001101
46 |
47 | # Add This vector to the bucket
48 | file_path = os.path.join(index_path, hash_str + ".txt")
49 |
50 | # Open File in Append Mode
51 | with open(file_path, "a") as file:
52 | file.write(str(id) + "\n")
53 |
54 | return plane_norms
55 |
56 | def get_top_k_hamming_distances(query, buckets, top_k):
57 | distances = []
58 | # Calculate Hamming distance for each bucket
59 | for bucket in buckets:
60 | hamming_distance = sum(bit1 != bit2 for bit1, bit2 in zip(query, bucket))
61 | distances.append((bucket, hamming_distance))
62 | # Sort distances and get the top K
63 | sorted_distances = sorted(distances, key=lambda x: x[1])
64 | top_k_distances = sorted_distances[:top_k]
65 | return top_k_distances
66 | def read_text_files_in_folder(folder_path):
67 | text_files_content = {}
68 |
69 | # Iterate over all files in the folder
70 | for filename in os.listdir(folder_path):
71 | file_path = os.path.join(folder_path, filename)
72 |
73 | # Check if the file is a text file
74 | if filename.endswith('.txt') and os.path.isfile(file_path):
75 | # Read the content of the text file
76 | with open(file_path, 'r', encoding='utf-8') as file:
77 | content = file.read()
78 | # Store content in the dictionary with the filename as the key
79 | text_files_content[filename] = content
80 |
81 | return text_files_content
82 |
83 |
84 |
85 |
86 | def semantic_query_lsh(query, plane_norms, index_path):
87 |
88 |
89 | """
90 | Function to Query the LSH indexing
91 | query:[] query vector
92 | plane_norms: [[]]
93 | index_path:path of the Index to be Search in
94 | """
95 | # Dot Product with Random Planes
96 | query_dot = np.dot(query, plane_norms.T)
97 |
98 | # Decision Making
99 | query_dot = (query_dot > 0) * 1
100 |
101 | query_dot = query_dot.squeeze()
102 | # Ensure query_dot is 1D for string conversion
103 | if query_dot.ndim == 0:
104 | query_dot = np.array([query_dot])
105 | # Bucket no. (Key)
106 | # hash_str = "".join(query_dot.astype(str)) # 101001101
107 | hash_str = "".join(map(str, query_dot.astype(int))) # Converts boolean array to int and then to string
108 |
109 | file_path = os.path.join(index_path, hash_str + ".txt")
110 | result = read_text_files_in_folder(index_path)
111 |
112 | list_buckets = []
113 | for filename, content in result.items():
114 | list_buckets.append(list(map(int, filename[:-4])))
115 | number_of_neighbours = 6
116 | min_hamming_buckets = get_top_k_hamming_distances(query_dot, list_buckets, number_of_neighbours)
117 | index_result =[]
118 | for (bucket, hamming_distance) in min_hamming_buckets:
119 | file_path = os.path.join(index_path, "".join(map(str,bucket)) + ".txt")
120 | try:
121 | list_1 = np.loadtxt(file_path, dtype=int)
122 | list_buckets = np.atleast_1d(list_1).tolist()
123 | index_result+=list_buckets
124 |
125 | except FileNotFoundError:
126 | # Handle the case where the file doesn't exist
127 | print(f"The file {file_path} doesn't exist. Setting index_result to a default value.")
128 | index_result = []
129 | return hash_str, np.array(index_result) # Bucket no
130 | # return index_result
131 |
132 |
133 |
134 |
--------------------------------------------------------------------------------
/src/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/src/__init__.py
--------------------------------------------------------------------------------
/src/api.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from worst_case_implementation import VecDBWorst
3 | from best_case_implementation import VecDBBest
4 | from typing import Dict, List, Annotated
5 |
6 | class DataApi:
7 | def __init__(self, file_path, worst = False, database_path="./DataBase",delete_db = True) -> None:
8 | self.file_path = file_path
9 | self.worst = worst
10 | if worst:
11 | self.db = VecDBWorst(self.file_path,delete_db)
12 | else:
13 | self.db = VecDBBest(self.file_path,database_path,delete_db)
14 | self.chunk_size = 10000
15 |
16 | # Function to generate random embeddings
17 | def __generate_embeddings(self,num_records, embedding_dim):
18 | return [np.random.rand(embedding_dim).tolist() for _ in range(num_records)]
19 |
20 |
21 | def generate_data_file(self,num_of_records):
22 | # Insert records in chunks
23 | for i in range(0, num_of_records, self.chunk_size):
24 | chunk_records = []
25 | for j in range(i + 1, i + self.chunk_size + 1):
26 | if j > num_of_records:
27 | break
28 | record = {"id": j, "embed": self.__generate_embeddings(1, 70)[0]}
29 | chunk_records.append(record)
30 |
31 | self.db.insert_records_binary(chunk_records)
32 | print(f"Inserted {len(chunk_records)} records. Total records inserted: {j}")
33 |
34 | print("Insertion complete.")
35 |
36 |
37 | def get_record_by_id(self,record_id):
38 | return self.db.read_record_by_id(record_id)
39 |
40 | def get_first_k_records(self,k):
41 | return self.db.get_top_k_records(k)
42 |
43 | def get_multiple_records_by_ids(self,record_ids):
44 | return self.db.read_multiple_records_by_id(record_ids)
45 |
46 | def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
47 | return self.db.insert_records_binary(rows)
48 |
49 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
50 | return self.db.insert_records(rows)
51 |
52 | def retrive(self, query:Annotated[List[float], 70], top_k = 5):
53 | return self.db.retrive(query,top_k)
54 |
55 |
--------------------------------------------------------------------------------
/src/best_case_implementation.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Annotated
2 | import numpy as np
3 | from utils import empty_folder
4 | from Modules.LSH import *
5 | import struct
6 | import time
7 |
8 | class VecDBBest:
9 | def __init__(self,file_path="./DataBase/data.bin", database_path = "./DataBase", new_db = True) -> None:
10 | '''
11 | Constructor
12 | '''
13 | self.file_path =file_path # Data File Path
14 | self.database_path= database_path # Path of the Folder to Create Indexes
15 |
16 | if new_db:
17 | # If New Data Base
18 | # Empty DataBase Folder
19 | empty_folder(self.database_path)
20 |
21 | # just open new file to delete the old one
22 | with open(self.file_path, "w") as fout:
23 | # if you need to add any head to the file
24 | pass
25 |
26 | def calculate_offset(self, record_id: int) -> int:
27 | # Calculate the offset for a given record ID
28 | record_size = struct.calcsize("I70f")
29 | return (record_id) * record_size
30 |
31 | def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
32 | with open(self.file_path, "ab") as fout: # Open the file in binary mode for appending
33 | for row in rows:
34 | id, embed = row["id"], row["embed"]
35 | # Pack the data into a binary format
36 | data = struct.pack(f"I{70}f", id, *embed)
37 | fout.write(data)
38 | self._build_index()
39 |
40 | def read_multiple_records_by_id(self, records_id: List[int]):
41 | record_size = struct.calcsize("I70f")
42 | records = {}
43 |
44 | with open(self.file_path, "rb") as fin:
45 | for i in range(len(records_id)):
46 | offset = self.calculate_offset(records_id[i])
47 | fin.seek(offset) # Move the file pointer to the calculated offset
48 | data = fin.read(record_size)
49 | if not data:
50 | records[records_id[i]] = None
51 | continue
52 |
53 | # Unpack the binary data into a dictionary
54 | unpacked_data = struct.unpack("I70f", data)
55 | id_value, floats = unpacked_data[0], unpacked_data[1:]
56 |
57 | # Create and return the record dictionary
58 | record = {"id": id_value, "embed": list(floats)}
59 | records[records_id[i]] = record
60 | return records
61 |
62 | def get_top_k_records(self,k):
63 | records = []
64 | record_size = struct.calcsize("I70f")
65 | with open(self.file_path,'rb') as fin:
66 | fin.seek(0)
67 | for i in range(k):
68 | data = fin.read(record_size)
69 | unpacked_data = struct.unpack("I70f", data)
70 | id_value, floats = unpacked_data[0], unpacked_data[1:]
71 |
72 | record = {"id": id_value, "embed": list(floats)}
73 | records.append(record)
74 | return records
75 |
76 | def _build_index(self,Level_1_nbits=5, Level_2_nbits=3, Level_3_nbits=3,Level_4_nbits=3)-> None:
77 |
78 | '''
79 | Build the Index
80 | '''
81 | top_k_records = 2000
82 |
83 | # measure the time
84 | start = time.time()
85 |
86 | # Layer 1 Indexing
87 | # TODO: Here we are reading the whole file: Change later
88 | level_1_in = self.get_top_k_records(top_k_records)
89 | level_1_planes = LSH_index(data=level_1_in, nbits=Level_1_nbits, index_path=self.database_path + "/Level1")
90 | np.save(self.database_path + "/Level1/"+'metadata.npy',level_1_planes)
91 | print("Layer 1 Finished")
92 | return
93 |
94 |
95 |
96 | # Layer 2 Indexing
97 | for file_name in os.listdir(self.database_path + "/Level1"):
98 | file_path = os.path.join(self.database_path + "/Level1", file_name)
99 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
100 | read_data_2 = np.loadtxt(file_path, dtype=int, ndmin=1)
101 | level_2_in = self.read_multiple_records_by_id(read_data_2)
102 | level_2_planes = LSH_index(data=level_2_in.values(), nbits=Level_2_nbits, index_path=self.database_path + "/Level2/" + file_name[:-4])
103 | np.save(self.database_path + "/Level2/" + file_name[:-4]+'/metadata.npy',level_2_planes)
104 | print("Layer 2 Finished")
105 | return
106 |
107 |
108 | # Layer 3 Indexing
109 | for folder_name in os.listdir(self.database_path + "/Level2"):
110 | folder_path = os.path.join(self.database_path + "/Level2", folder_name)
111 | for file_name in os.listdir(folder_path):
112 | file_path = os.path.join(folder_path, file_name)
113 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
114 | read_data_3 = np.loadtxt(file_path, dtype=int, ndmin=1)
115 | level_3_in = self.read_multiple_records_by_id(read_data_3)
116 | level_3_planes = LSH_index(data=level_3_in.values(), nbits=Level_3_nbits, index_path=self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4])
117 | np.save(self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4]+'/metadata.npy',level_3_planes)
118 | print("Layer 3 Finished")
119 |
120 | return
121 | # Layer 4 Indexing
122 | for folder_name in os.listdir(self.database_path + "/Level3"):
123 | folder_path = os.path.join(self.database_path + "/Level3", folder_name)
124 | for folder_name_2 in os.listdir(folder_path):
125 | folder_path_2 = os.path.join(folder_path, folder_name_2)
126 | for file_name in os.listdir(folder_path_2):
127 | file_path = os.path.join(folder_path_2, file_name)
128 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
129 | read_data_4 = np.loadtxt(file_path, dtype=int, ndmin=1)
130 | level_4_in = self.read_multiple_records_by_id(read_data_4)
131 | level_4_planes = LSH_index(data=level_4_in.values(), nbits=Level_4_nbits, index_path=self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4])
132 | np.save(self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4]+'/metadata.npy',level_4_planes)
133 | print("Layer 4 Finished")
134 |
135 |
136 | # measure the time
137 | end = time.time()
138 | print("Time taken by Indexing: ",end - start)
139 | def retrive(self, query:Annotated[List[float], 70],top_k = 5)-> [int]:
140 | '''
141 | Get the top_k vectors similar to the Query
142 |
143 | return: list of the top_k similar vectors Ordered by Cosine Similarity
144 | '''
145 |
146 | # Retrieve from Level 1
147 | level_1_planes = np.load(self.database_path + "/Level1"+'/metadata.npy')
148 | bucket_1,result = semantic_query_lsh(query, level_1_planes, self.database_path + "/Level1")
149 | print("length of first bucket",result.shape)
150 |
151 | if len(result) < top_k:
152 | print('level 1 smaller than top_k')
153 |
154 | # # Retrieve from Level 2
155 | # level_2_planes = np.load(self.database_path + "/Level2/"+bucket_1+'/metadata.npy')
156 | # bucket_2,result = semantic_query_lsh(query, level_2_planes, self.database_path + "/Level2/"+bucket_1)
157 | # print("length of second bucket",result.shape)
158 |
159 | # if len(result) < top_k:
160 | # print('level 2 smaller than top_k')
161 |
162 | # # Retrieve from Level 3
163 | # level_3_planes = np.load(self.database_path + "/Level3/"+bucket_1+'/'+bucket_2+'/metadata.npy')
164 | # bucket_3,result = semantic_query_lsh(query, level_3_planes, self.database_path + "/Level3/"+bucket_1+'/'+bucket_2)
165 | # print("length of third bucket",result.shape)
166 |
167 | # if len(result) < top_k:
168 | # print('level 3 smaller than top_k')
169 |
170 | # # Retrieve from Level 4
171 | # level_4_planes = np.load(self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3+'/metadata.npy')
172 | # bucket_4,result = semantic_query_lsh(query, level_4_planes, self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3)
173 | # print("length of fourth bucket",result.shape)
174 |
175 | # if len(result) < top_k:
176 | # print('level 4 smaller than top_k')
177 |
178 |
179 | # Retrieve from Data Base the Embeddings of the Vectors
180 | final_result= self.read_multiple_records_by_id(result)
181 |
182 | # Calculate the Cosine Similarity between the Query and the Vectors
183 | scores = []
184 | for row in final_result.values():
185 | id_value = row['id']
186 | embed_values = row['embed']
187 | score = self._cal_score(query, embed_values)
188 | scores.append((score, id_value))
189 | scores = sorted(scores, reverse=True)[:top_k]
190 | return [s[1] for s in scores]
191 |
192 |
193 |
194 |
195 | def _cal_score(self, vec1, vec2):
196 | dot_product = np.dot(vec1, vec2)
197 | norm_vec1 = np.linalg.norm(vec1)
198 | norm_vec2 = np.linalg.norm(vec2)
199 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
200 | return cosine_similarity
201 |
202 |
--------------------------------------------------------------------------------
/src/evaluation.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from worst_case_implementation import VecDBWorst
3 | from best_case_implementation import VecDBBest
4 | import argparse
5 | from utils import extract_embeds_array
6 | import pandas as pd
7 | from api import DataApi
8 | import os
9 | import time
10 | from dataclasses import dataclass
11 | from typing import List
12 |
13 | AVG_OVERX_ROWS = 1
14 |
15 | @dataclass
16 | class Result:
17 | run_time: float
18 | top_k: int
19 | db_ids: List[int]
20 | actual_ids: List[int]
21 |
22 | # def run_queries(db1,db2, np_rows, top_k, num_runs,delete=False):
23 | def run_queries(db, np_rows, top_k, num_runs, delete=False):
24 | results = []
25 | # results_worst = []
26 | # results_best = []
27 | for i in range(num_runs):
28 | if delete:
29 | query = np.random.random((1,70))
30 | np.save( "./DataBase/q"+str(i)+'.npy',query)
31 | else:
32 | query = np.load( "./DataBase/q"+str(i)+'.npy')
33 |
34 | tic = time.time()
35 | db_ids = db.retrive(query,top_k)
36 | toc = time.time()
37 | run_time= toc - tic
38 |
39 | actual_ids = np.argsort(np_rows.dot(query.T).T / (np.linalg.norm(np_rows, axis=1) * np.linalg.norm(query)), axis=1).squeeze().tolist()[::-1]
40 |
41 | toc = time.time()
42 | np_run_time = toc - tic
43 |
44 | results.append(Result(run_time,top_k,db_ids,actual_ids))
45 | return results
46 |
47 | def eval(results: List[Result]):
48 | # scores are negative. So getting 0 is the best score.
49 | scores = []
50 | run_time = []
51 | for res in results:
52 | run_time.append(res.run_time)
53 | # case for retireving number not equal to top_k, socre will be the lowest
54 | if len(set(res.db_ids)) != res.top_k or len(res.db_ids) != res.top_k:
55 | scores.append( -1 * len(res.actual_ids) * res.top_k)
56 | print('retrieving number not equal to top_k')
57 | continue
58 |
59 | score = 0
60 | for id in res.db_ids:
61 | try:
62 | ind = res.actual_ids.index(id)
63 | if ind > res.top_k * 3:
64 | # print("not in top top_k*3")
65 | score -= ind
66 | except:
67 | # print("not in ids")
68 | score -= len(res.actual_ids)
69 | scores.append(score)
70 |
71 | return sum(scores) / len(scores), sum(run_time) / len(run_time)
72 |
73 | def find_indices(list1, list2):
74 | """
75 | Find the indices of elements of list1 in list2.
76 |
77 | :param list1: The list containing elements whose indices are to be found.
78 | :param list2: The list in which to search for elements from list1.
79 | :return: A list of indices.
80 | """
81 | indices = []
82 | for element in list1:
83 | # Convert both to numpy arrays for consistent handling
84 | np_list2 = np.array(list2)
85 | # Find the index of element in list2
86 | found_indices = np.where(np_list2 == element)[0]
87 | if found_indices.size > 0:
88 | indices.append(found_indices[0])
89 |
90 | return indices
91 |
92 |
93 | def compare_results_print(worst_res,best_res,top_k):
94 | for i in range(len(worst_res)):
95 | actual_ids=worst_res[i].actual_ids
96 | db_ids_best=best_res[i].db_ids
97 | db_ids_worst=worst_res[i].db_ids
98 |
99 | run_time_worst=worst_res[i].run_time
100 | run_time_best=best_res[i].run_time
101 |
102 |
103 | print("=======================================")
104 | print("Best ids: ",db_ids_best)
105 | print("Actual ids: ",actual_ids[:top_k])
106 | print("Worst ids: ",db_ids_worst)
107 | print("Intersect: ",set(actual_ids[:top_k]).intersection(set(db_ids_best)))
108 | print("Intersection in top k indices in the best DB: ",find_indices(actual_ids[:top_k], db_ids_best))
109 |
110 | print("Time taken by Query (Best): ",run_time_best)
111 | print("Time taken by Query (Worst): ",run_time_worst)
112 | print("=======================================")
113 |
114 | if __name__ == "__main__":
115 | print("Hello Semantic LSH")
116 |
117 | number_of_records = 2000
118 | number_of_features = 70
119 | number_of_queries = 5
120 | top_k = 10
121 | print("******************************""")
122 | print("Number of records: ",number_of_records)
123 | print("Number of queries: ",number_of_queries)
124 | print("Top k: ",top_k)
125 | print("******************************""")
126 |
127 |
128 | folder_name = "DataBase"
129 | if not os.path.exists(folder_name):
130 | os.makedirs(folder_name)
131 |
132 | # Mode
133 | parser = argparse.ArgumentParser(description='Description of your script')
134 | parser.add_argument('-d','--delete', help='Description of the -d flag', action='store_true')
135 | args = parser.parse_args()
136 |
137 | # worst_db = VecDBWorst('./DataBase/data.csv',new_db=not args.delete)
138 | worst_api = DataApi('./DataBase/data_worst.csv',True,'./DataBase',args.delete)
139 | # best_db = VecDBBest('./DataBase/data.bin','./DataBase',new_db=not args.delete)
140 | best_api = DataApi('./DataBase/data.bin', False,'./DataBase',args.delete)
141 |
142 | if not args.delete:
143 | print("Reading")
144 | # records_np = pd.read_csv('./DataBase/data.csv',header=None)
145 | # rows_without_first_element = np.array([row[1:].tolist() for _, row in records_np.iterrows()])
146 | # records_np=rows_without_first_element
147 |
148 | records_database = np.array(best_api.get_first_k_records(number_of_records))
149 | records_np = extract_embeds_array(records_database)
150 | records_dict = records_database
151 | _len = len(records_np)
152 | else:
153 | # New
154 |
155 | # records_database = np.array(best_api.get_first_k_records(10000))
156 | print("Generating data files")
157 | records_np = np.random.random((number_of_records, number_of_features))
158 | # records_np = extract_embeds_array(records_database)
159 |
160 | records_dict = [{"id": i, "embed": list(row)} for i, row in enumerate(records_np)]
161 | # records_dict = records_database
162 | _len = len(records_np)
163 |
164 | worst_api.insert_records(records_dict)
165 | best_api.insert_records_binary(records_dict)
166 |
167 |
168 | # Worst
169 | res_worst = run_queries(worst_api, records_np, top_k, number_of_queries,args.delete)
170 | # Best
171 | res_best = run_queries(best_api, records_np, top_k, number_of_queries,False)
172 |
173 | compare_results_print(res_worst,res_best,top_k)
174 | print("Worst:",eval(res_worst))
175 | print("Best:",eval(res_best))
176 |
177 | # res = run_queries(best_api, records_np, 5, 3)
178 | # print("Best:",eval(res))
179 | # results_worst, results_best = run_queries(worst_api,best_api, records_np, top_k, number_of_queries)
180 | # print("Worst:",eval(results_worst))
181 | # print("Best:",eval(results_best))
182 |
183 | # records_np = np.concatenate([records_np, np.random.random((90000, 70))])
184 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
185 | # _len = len(records_np)
186 | # worst_db.insert_records(records_dict)
187 | # res = run_queries(worst_db, records_np, 5, 10)
188 | # print(eval(res))
189 |
190 | # records_np = np.concatenate([records_np, np.random.random((900000, 70))])
191 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
192 | # _len = len(records_np)
193 | # worst_db.insert_records(records_dict)
194 | # res = run_queries(worst_db, records_np, 5, 10)
195 | # eval(res)
196 |
197 | # records_np = np.concatenate([records_np, np.random.random((4000000, 70))])
198 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
199 | # _len = len(records_np)
200 | # db.insert_records(records_dict)
201 | # res = run_queries(db, records_np, 5, 10)
202 | # eval(res)
203 |
204 | # records_np = np.concatenate([records_np, np.random.random((5000000, 70))])
205 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
206 | # _len = len(records_np)
207 | # db.insert_records(records_dict)
208 | # res = run_queries(db, records_np, 5, 10)
209 | # eval(res)
210 |
211 | # records_np = np.concatenate([records_np, np.random.random((5000000, 70))])
212 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
213 | # _len = len(records_np)
214 | # db.insert_records(records_dict)
215 | # res = run_queries(db, records_np, 5, 10)
216 | # eval(res)
--------------------------------------------------------------------------------
/src/main.py:
--------------------------------------------------------------------------------
1 | from api import DataApi
2 |
3 |
4 | api_data = DataApi("test.bin")
5 |
6 | # api_data.generate_data_file(5000)
7 |
8 |
9 | records = api_data.get_multiple_records_by_ids([2, 1, 5, 8000])
10 | print(records[8000])
11 |
--------------------------------------------------------------------------------
/src/notes.txt:
--------------------------------------------------------------------------------
1 | 1- 20,000,000
2 | // ==>200
3 |
4 | 2- 100,000
5 | // ==>200
6 |
7 |
8 | 3- 5,000
9 | // ==>500
10 |
11 |
12 | 4- 10
13 |
14 | ============
15 |
16 | new query
17 | first level: 1-10
18 |
19 | second level: 1-500 ==> most load from ram <=200
20 |
21 |
22 | third level: 1-200
23 |
24 | fourth: level: 1-200
25 |
26 |
27 | final retreving: 910 record
28 |
29 | =============================
30 |
31 | n_probe =2
32 |
33 | new query
34 | first level: 2-10
35 | second level: 2-1000
36 | third level: 2-400
37 | fourth: level: 2-400
38 |
39 |
40 | ==================================
41 |
42 | 20,000,000 record
43 |
44 | 256 buckets ===> 256 file
45 |
46 |
47 |
48 |
49 |
50 |
--------------------------------------------------------------------------------
/src/pipeline.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": null,
6 | "metadata": {},
7 | "outputs": [],
8 | "source": [
9 | "# from Modules.LSH import semantic_query_lsh\n",
10 | "# from Modules.LSH import LSH\n",
11 | "\n",
12 | "\n",
13 | "# import numpy as np\n",
14 | "# import os"
15 | ]
16 | },
17 | {
18 | "cell_type": "code",
19 | "execution_count": null,
20 | "metadata": {},
21 | "outputs": [],
22 | "source": [
23 | "# file_path = \"./random_data.txt\"\n",
24 | "# read_data = np.loadtxt(file_path)\n",
25 | "# plane_norms = LSH(read_data, 8)\n",
26 | "# query=[read_data[0]]\n",
27 | "# folder_name = \"bucket_files\"\n",
28 | "# result = semantic_query_lsh(query, plane_norms,folder_name)"
29 | ]
30 | },
31 | {
32 | "cell_type": "code",
33 | "execution_count": 23,
34 | "metadata": {},
35 | "outputs": [
36 | {
37 | "name": "stdout",
38 | "output_type": "stream",
39 | "text": [
40 | "The autoreload extension is already loaded. To reload it, use:\n",
41 | " %reload_ext autoreload\n"
42 | ]
43 | }
44 | ],
45 | "source": [
46 | "%load_ext autoreload\n",
47 | "%autoreload 2\n",
48 | "\n",
49 | "\n",
50 | "from utils import *\n",
51 | "from Modules.LSH import*\n",
52 | "from api import *\n",
53 | "from evaluation import *\n",
54 | "from worst_case_implementation import VecDBWorst\n",
55 | "\n",
56 | "\n",
57 | "# datafile_path=\"../DataBase/random_data_10000.txt\""
58 | ]
59 | },
60 | {
61 | "cell_type": "code",
62 | "execution_count": 29,
63 | "metadata": {},
64 | "outputs": [
65 | {
66 | "name": "stdout",
67 | "output_type": "stream",
68 | "text": [
69 | "Inserted 10000 records. Total records inserted: 10000\n",
70 | "Insertion complete.\n"
71 | ]
72 | }
73 | ],
74 | "source": [
75 | "data_file='./DataBase/data.bin'\n",
76 | "Level_1_path='./DataBase/Level1'\n",
77 | "Level_2_path='./DataBase/Level2'\n",
78 | "Level_3_path='./DataBase/Level3'\n",
79 | "\n",
80 | "Level_1_nbits=8\n",
81 | "Level_2_nbits=3\n",
82 | "Level_3_nbits=3\n",
83 | "\n",
84 | "data_api = DataApi(data_file)\n",
85 | "data_api.generate_data_file(10000)"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": 33,
91 | "metadata": {},
92 | "outputs": [],
93 | "source": [
94 | "\n",
95 | "# Test LSH_index\n",
96 | "# Read Data From File\n",
97 | "read_data = data_api.get_top_k_records(10000)\n",
98 | "\n",
99 | "\n",
100 | "# Layer(1)\n",
101 | "level_1_in=read_data\n",
102 | "# TODO: Save Planes to be used in query Search\n",
103 | "level_1_planes=LSH_index(data=level_1_in, nbits=Level_1_nbits,index_path=Level_1_path)"
104 | ]
105 | },
106 | {
107 | "cell_type": "code",
108 | "execution_count": 34,
109 | "metadata": {},
110 | "outputs": [],
111 | "source": [
112 | "# Layer(2)\n",
113 | "# On Each Bucket Apply LSH\n",
114 | "\n",
115 | "# List all files in the directory\n",
116 | "files = os.listdir(Level_1_path)\n",
117 | "\n",
118 | "# TODO: Save Planes to be used in query Search\n",
119 | "level_2_planes={}\n",
120 | "\n",
121 | "# Loop over the files\n",
122 | "for file_name in files:\n",
123 | " file_path = os.path.join(Level_1_path, file_name)\n",
124 | " \n",
125 | " if os.path.isfile(file_path):\n",
126 | " # Read Data\n",
127 | " read_data_2 = np.loadtxt(file_path,dtype=int,ndmin=1)\n",
128 | "\n",
129 | " level_2_in=data_api.get_multiple_records_by_ids(read_data_2-1)\n",
130 | " # level_2_in = array_to_dictionary(values=vectors,keys=np.hstack(read_data_2))\n",
131 | "\n",
132 | " # # Apply LSH on this Bucket\n",
133 | " # level_2=arr[level_1]\n",
134 | " level_2_planes[file_name[:-4]]=LSH_index(data=level_2_in.values(), nbits=Level_2_nbits,index_path=Level_2_path+'/' + file_name[:-4])\n",
135 | "\n"
136 | ]
137 | },
138 | {
139 | "cell_type": "code",
140 | "execution_count": 9,
141 | "metadata": {},
142 | "outputs": [],
143 | "source": [
144 | "# Layer(3)\n",
145 | "# On Each Bucket Apply LSH\n",
146 | "\n",
147 | "# List all files in the directory\n",
148 | "folders = os.listdir(Level_2_path)\n",
149 | "\n",
150 | "# TODO: Save Planes to be used in query Search\n",
151 | "level_3_planes={}\n",
152 | "# file_3=folder{}\n",
153 | "# Loop over the folders\n",
154 | "for folder_name in folders:\n",
155 | " level_3_planes[folder_name]={}\n",
156 | " folder_path = os.path.join(Level_2_path, folder_name)\n",
157 | " files = os.listdir(folder_path)\n",
158 | " # Loop over the files\n",
159 | " for file_name in files:\n",
160 | " file_path = os.path.join(folder_path, file_name)\n",
161 | " \n",
162 | " if os.path.isfile(file_path):\n",
163 | " # Read Data\n",
164 | " read_data_3 = np.loadtxt(file_path,dtype=int,ndmin=1)\n",
165 | "\n",
166 | " level_3_in=data_api.get_multiple_records_by_ids(read_data_3)\n",
167 | "\n",
168 | " # # Apply LSH on this Bucket\n",
169 | " level_3_planes[folder_name][file_name[:-4]]=LSH_index(data=level_3_in.values(), nbits=Level_3_nbits,index_path=Level_3_path+'/'+folder_name+'/' + file_name[:-4])\n"
170 | ]
171 | },
172 | {
173 | "cell_type": "code",
174 | "execution_count": 22,
175 | "metadata": {},
176 | "outputs": [
177 | {
178 | "name": "stdout",
179 | "output_type": "stream",
180 | "text": [
181 | "bucket of level 1: 11110011\n",
182 | "=====================================\n",
183 | "bucket of level 2: 001\n",
184 | "=====================================\n",
185 | "bucket of level 3: 100\n",
186 | "Length of level 3 189\n",
187 | "Indices of level 3 [ 36 51 104 159 266 357 372 385 434 465 510 671 702 707\n",
188 | " 720 822 824 834 863 938 1034 1044 1165 1248 1264 1438 1505 1565\n",
189 | " 1613 1683 1712 1719 1771 1798 1812 1843 1953 2191 2238 2266 2330 2353\n",
190 | " 2594 2602 2624 2669 2730 2744 2825 2880 2894 2915 2942 2944 3080 3168\n",
191 | " 3286 3351 3490 3645 3648 3735 3798 3851 3859 3911 3986 4026 4030 4065\n",
192 | " 4121 4134 4187 4211 4232 4260 4391 4399 4476 4477 4489 4492 4545 4554\n",
193 | " 4591 4605 4660 4792 4905 4937 4953 4954 4970 4986 4987 5228 5249 5329\n",
194 | " 5398 5454 5471 5495 5584 5708 5712 5725 5744 5799 5899 5900 5908 5952\n",
195 | " 5987 6049 6072 6096 6144 6184 6209 6287 6344 6399 6479 6495 6536 6544\n",
196 | " 6662 6693 6848 6880 6915 6962 7080 7085 7187 7199 7213 7240 7390 7404\n",
197 | " 7417 7442 7531 7538 7554 7584 7625 7664 7708 7721 7765 7768 7808 7827\n",
198 | " 7955 8101 8170 8279 8284 8380 8444 8446 8454 8481 8552 8560 8565 8586\n",
199 | " 8676 8700 8761 8792 8912 8935 9007 9150 9336 9352 9354 9367 9586 9662\n",
200 | " 9745 9762 9794 9801 9859 9948 9972]\n",
201 | "=====================================\n",
202 | "target_vector [0.5522450804710388, 0.8917692303657532, 0.7913368344306946, 0.6000004410743713, 0.2616525888442993, 0.9615220427513123, 0.4808562695980072, 0.6019359827041626, 0.07978673279285431, 0.30365362763404846, 0.7390730381011963, 0.2133997678756714, 0.36366748809814453, 0.1835469752550125, 0.20069865882396698, 0.13891369104385376, 0.11978743225336075, 0.3913387358188629, 0.002954070921987295, 0.5194749236106873, 0.37845972180366516, 0.9680533409118652, 0.6960610747337341, 0.8805666565895081, 0.06497178226709366, 0.5662519335746765, 0.04004804417490959, 0.2919067144393921, 0.737677812576294, 0.10855083167552948, 0.3745698928833008, 0.37776005268096924, 0.9178327322006226, 0.7241680026054382, 0.12325477600097656, 0.3273957073688507, 0.9901415109634399, 0.4085298478603363, 0.6129018068313599, 0.1801413595676422, 0.9952824711799622, 0.3938077688217163, 0.913888692855835, 0.11249328404664993, 0.14214684069156647, 0.6679161787033081, 0.9495717287063599, 0.4362204968929291, 0.3122316896915436, 0.6952698230743408, 0.8448274731636047, 0.965186595916748, 0.35632771253585815, 0.9069381952285767, 0.42551901936531067, 0.9420151710510254, 0.022108066827058792, 0.6098361611366272, 0.897776186466217, 0.4446363151073456, 0.7102886438369751, 0.5624412894248962, 0.5420237183570862, 0.3291500210762024, 0.2226945161819458, 0.6429535150527954, 0.5322402119636536, 0.09856311231851578, 0.5489377379417419, 0.5590397715568542]\n"
203 | ]
204 | },
205 | {
206 | "ename": "TypeError",
207 | "evalue": "unhashable type: 'slice'",
208 | "output_type": "error",
209 | "traceback": [
210 | "\u001b[1;31m---------------------------------------------------------------------------\u001b[0m",
211 | "\u001b[1;31mTypeError\u001b[0m Traceback (most recent call last)",
212 | "\u001b[1;32md:\\Semantic-Search-Engine\\pipeline.ipynb Cell 8\u001b[0m line \u001b[0;36m2\n\u001b[0;32m 27\u001b[0m index_result_3\u001b[39m=\u001b[39mdata_api\u001b[39m.\u001b[39mget_multiple_records_by_ids(index_result_3)\n\u001b[0;32m 28\u001b[0m level3_res_vectors\u001b[39m=\u001b[39m[entry[\u001b[39m'\u001b[39m\u001b[39membed\u001b[39m\u001b[39m'\u001b[39m] \u001b[39mfor\u001b[39;00m entry \u001b[39min\u001b[39;00m index_result_3\u001b[39m.\u001b[39mvalues()]\n\u001b[1;32m---> 29\u001b[0m top_result,_\u001b[39m=\u001b[39mget_top_k_similar(query,index_result_3,\u001b[39m10\u001b[39;49m)\n\u001b[0;32m 30\u001b[0m \u001b[39m# print(\"Top k results: \",top_result[0])\u001b[39;00m\n\u001b[0;32m 31\u001b[0m \u001b[39m# print(\"=====================================\")\u001b[39;00m\n\u001b[0;32m 32\u001b[0m \u001b[39m# # get the intersection of the two lists level 2 and level3\u001b[39;00m\n\u001b[0;32m 33\u001b[0m \u001b[39m# count = sum(element in index_result_2 for element in index_result_3)\u001b[39;00m\n\u001b[0;32m 34\u001b[0m \u001b[39m# print(\"Intersection of the two layers: \",count)\u001b[39;00m\n",
213 | "File \u001b[1;32md:\\Semantic-Search-Engine\\Modules\\LSH.py:145\u001b[0m, in \u001b[0;36mget_top_k_similar\u001b[1;34m(target_vector, data, k)\u001b[0m\n\u001b[0;32m 143\u001b[0m \u001b[39m# Calculate cosine similarities using vectorized operations\u001b[39;00m\n\u001b[0;32m 144\u001b[0m \u001b[39mprint\u001b[39m(\u001b[39m\"\u001b[39m\u001b[39mtarget_vector\u001b[39m\u001b[39m\"\u001b[39m,target_vector)\n\u001b[1;32m--> 145\u001b[0m \u001b[39mprint\u001b[39m(\u001b[39m\"\u001b[39m\u001b[39mdata\u001b[39m\u001b[39m\"\u001b[39m,data[\u001b[39m1\u001b[39;49m:\u001b[39m5\u001b[39;49m])\n\u001b[0;32m 146\u001b[0m similarities \u001b[39m=\u001b[39m \u001b[39m1\u001b[39m \u001b[39m-\u001b[39m np\u001b[39m.\u001b[39marray([cosine(target_vector, vector) \u001b[39mfor\u001b[39;00m vector \u001b[39min\u001b[39;00m data])\n\u001b[0;32m 148\u001b[0m \u001b[39m# Find the indices of the top k most similar vectors\u001b[39;00m\n",
214 | "\u001b[1;31mTypeError\u001b[0m: unhashable type: 'slice'"
215 | ]
216 | }
217 | ],
218 | "source": [
219 | "# Query\n",
220 | "query=data_api.get_record_by_id(5)[5]['embed']\n",
221 | "# Layer (1)\n",
222 | "bucket_1,index_result_1 = semantic_query_lsh(query=query,plane_norms=level_1_planes,index_path=Level_1_path)\n",
223 | "print(\"bucket of level 1: \",bucket_1)\n",
224 | "# print(\"Length of level 1\",len(index_result_1))\n",
225 | "# print(\"Indices of level 1\",index_result_1)\n",
226 | "print(\"=====================================\")\n",
227 | "\n",
228 | "# Layer(2)\n",
229 | "bucket_2,index_result_2 = semantic_query_lsh(query=query,plane_norms=level_2_planes[bucket_1],index_path=Level_2_path+\"/\"+bucket_1)\n",
230 | "print(\"bucket of level 2: \",bucket_2)\n",
231 | "# print(\"Length of level 2\",len(index_result_2))\n",
232 | "# print(\"Indices of level 2\",index_result_2)\n",
233 | "print(\"=====================================\")\n",
234 | "\n",
235 | "# Layer(3)\n",
236 | "bucket_3,index_result_3 = semantic_query_lsh(query=query,plane_norms=level_3_planes[bucket_1][bucket_2],index_path=Level_3_path+\"/\"+bucket_1+'/'+bucket_2)\n",
237 | "print(\"bucket of level 3: \",bucket_3)\n",
238 | "print(\"Length of level 3\",len(index_result_3))\n",
239 | "print(\"Indices of level 3\",index_result_3)\n",
240 | "\n",
241 | "\n",
242 | "print(\"=====================================\")\n",
243 | "\n",
244 | "# get top 10 results from the last layer\n",
245 | "index_result_3=data_api.get_multiple_records_by_ids(index_result_3)\n",
246 | "level3_res_vectors=[entry['embed'] for entry in index_result_3.values()]\n",
247 | "top_result,_=get_top_k_similar(query,index_result_3,10)\n",
248 | "# print(\"Top k results: \",top_result[0])\n",
249 | "# print(\"=====================================\")\n",
250 | "# # get the intersection of the two lists level 2 and level3\n",
251 | "# count = sum(element in index_result_2 for element in index_result_3)\n",
252 | "# print(\"Intersection of the two layers: \",count)\n",
253 | "\n"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "np_rows = np.array([record['embed'] for record in read_data if 'embed' in record])\n",
263 | "# temp=[5, 966, 536, 1088, 5073, 5549]\n",
264 | "index_result_3_minus_one = [id - 1 for id in top_results[0]]\n",
265 | "res=run_queries(index_result_3_minus_one, np_rows, top_k=len(top_results), num_runs=1,query=np.array([query]))\n",
266 | "print(eval(res))"
267 | ]
268 | },
269 | {
270 | "cell_type": "code",
271 | "execution_count": null,
272 | "metadata": {},
273 | "outputs": [],
274 | "source": [
275 | "db = VecDBWorst()\n",
276 | "# records_np = np.random.random((10000, 70))\n",
277 | "records_np = np.array([record['embed'] for record in read_data if 'embed' in record])\n",
278 | "\n",
279 | "# records_dict = [{\"id\": i, \"embed\": list(row)} for i, row in enumerate(records_np)]\n",
280 | "records_dict=read_data\n",
281 | "\n",
282 | "# _len = len(records_np)\n",
283 | "db.insert_records(records_dict)\n",
284 | "db_ids=db.retrive(query, top_k=1)\n",
285 | "db_ids_minus_one = [id - 1 for id in db_ids]\n",
286 | "res = run_queries(db_ids_minus_one, records_np, 1, 1, np.array([query]))\n",
287 | "print(eval(res))\n"
288 | ]
289 | }
290 | ],
291 | "metadata": {
292 | "kernelspec": {
293 | "display_name": "Python 3",
294 | "language": "python",
295 | "name": "python3"
296 | },
297 | "language_info": {
298 | "codemirror_mode": {
299 | "name": "ipython",
300 | "version": 3
301 | },
302 | "file_extension": ".py",
303 | "mimetype": "text/x-python",
304 | "name": "python",
305 | "nbconvert_exporter": "python",
306 | "pygments_lexer": "ipython3",
307 | "version": "3.10.9"
308 | }
309 | },
310 | "nbformat": 4,
311 | "nbformat_minor": 2
312 | }
313 |
--------------------------------------------------------------------------------
/src/utils.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import shutil
3 | import os
4 | import math
5 | from typing import Dict, List, Annotated
6 | import struct
7 | import sys
8 |
9 |
10 | def save_20M_record(data):
11 | '''
12 | Given 20M record save them as required by the TA
13 | data: (20M,70)
14 | '''
15 |
16 | folder_name='./Data_TA'
17 | if not os.path.exists(folder_name):
18 | os.makedirs(folder_name)
19 |
20 | empty_folder(folder_name)
21 |
22 | files=['data_100K.bin',"data_1M.bin","data_5M.bin","data_10M.bin","data_15M.bin","data_20M.bin"]
23 | # files=["data_20M.bin"]
24 | limits=[10**5,10**6,5*10**6,10**7,15*10**6,2*10**7]
25 | # limits=[20*10**6]
26 | for i,file in enumerate(files):
27 | data_part=data[:limits[i]]
28 |
29 | # Append in Binary Mode
30 | with open(folder_name+'/'+file, "ab") as fout:
31 | for id,vector in enumerate(data_part):
32 | # Pack the data into a binary format
33 | unpacked_data = struct.pack(f"I{70}f", id, *vector)
34 | fout.write(unpacked_data)
35 |
36 | # # Test
37 | # print("len(data)",len(data))
38 | # # print(data[0])
39 | # print(data[-1])
40 | # read_data=read_binary_file_chunk('./Data_TA/data_100K.bin',f"I{70}f",start_index=0,chunk_size=1000000,dictionary_format=True)
41 | # print("len(read_data)",len(read_data))
42 | # # print(read_data[0])
43 | # print(read_data[10**5-1])
44 |
45 |
46 | # # Test
47 | # print("len(data)",len(data))
48 | # # print(data[0])
49 | # print(data[-1])
50 | # read_data=read_binary_file_chunk('./Data_TA/data_1M.bin',f"I{70}f",start_index=0,chunk_size=1000000,dictionary_format=True)
51 | # print("len(read_data)",len(read_data))
52 | # # print(read_data[0])
53 | # print(read_data[10**6-1])
54 |
55 | def read_binary_file(file_path,format):
56 | '''
57 | Read binary file from its format
58 | '''
59 | try:
60 | with open(file_path,"rb") as fin:
61 | file_size = os.path.getsize(file_path)
62 | record_size=struct.calcsize(format)
63 | n_records=file_size/record_size
64 | # print("n_records",n_records)
65 |
66 | fin.seek(0) #Move pointer to the beginning of the file
67 | data = fin.read(record_size * int(n_records))
68 | if not data:
69 | print("Empty File ",file_path,"🔴🔴")
70 | return None
71 | # Unpack the binary data
72 | data=np.frombuffer(data, dtype=np.dtype(format))
73 | return data
74 | except FileNotFoundError:
75 | print(f"The file '{file_path}' Not Found.")
76 |
77 | def write_binary_file(file_path,data_to_write,format):
78 | '''
79 | data_to_write: array of values with format as passed
80 | format: format of each element
81 | '''
82 | try:
83 | with open(file_path, "ab") as fout:
84 | # Pack the entire array into binary data
85 | binary_data = struct.pack(len(data_to_write)*format, *data_to_write.flatten())
86 | fout.write(binary_data)
87 | except FileNotFoundError:
88 | print(f"The file '{file_path}' could not be created.")
89 |
90 | def read_binary_file_chunk(file_path, record_format, start_index, chunk_size=10,dictionary_format=False):
91 | """
92 | This Function Reads Chunk from a binary File
93 | If remaining from file are < chunk size they are returned normally
94 |
95 | file_path:Path of the file to be read from
96 | record_format: format of the record ex:f"4I" 4 integers
97 | start_index: index of the record from which we start reading [0_indexed]
98 | chunk_size: no of records to be retrieved
99 |
100 | @return : None in case out of index of file
101 | the records
102 | """
103 |
104 | # Calculate record size
105 | record_size = struct.calcsize(record_format)
106 |
107 | # Open the binary file for reading
108 | with open(file_path, "rb") as fin:
109 | fin.seek(
110 | start_index * record_size
111 | ) # Move the file pointer to the calculated offset
112 |
113 | # Read a chunk of records
114 | # .read() moves the file pointer (cursor) forward by the number of bytes read.
115 | chunk_data = fin.read(record_size * (chunk_size))
116 | if len(chunk_data) == 0:
117 | print("Out Of File Index 🔥🔥")
118 | return None
119 |
120 | # file_size = os.path.getsize(file_path)
121 | # print("Current file position:", fin.tell())
122 | # print("File size:", file_size,"record_format",record_format,"record_size",record_size,"chunk_data len",len(chunk_data))
123 |
124 | if dictionary_format:
125 | records={}
126 | for i in range(0, len(chunk_data), record_size):
127 | #TODO Remove this loop @Basma Elhoseny
128 | unpacked_record = struct.unpack(record_format, chunk_data[i : i + record_size])
129 | id, vector = unpacked_record[0], unpacked_record[1:]
130 | records[id]=np.array(vector)
131 | return records
132 |
133 | # Unpack Data
134 | records = []
135 | for i in range(0, len(chunk_data), record_size):
136 | unpacked_record = struct.unpack(
137 | record_format, chunk_data[i : i + record_size]
138 | )
139 | id, vector = unpacked_record[0], unpacked_record[1:]
140 | record = {"id": id, "embed": list(vector)}
141 | records.append(record)
142 | return records
143 | def empty_folder(folder_path):
144 | """
145 | Function to Empty a folder given its path
146 | @param folder_path : path of the folder to be deleted
147 | """
148 | if not os.path.exists(folder_path):
149 | os.makedirs(folder_path)
150 | print("Created new ", folder_path, "successfully")
151 | return
152 |
153 | for filename in os.listdir(folder_path):
154 | file_path = os.path.join(folder_path, filename)
155 | try:
156 | if os.path.isfile(file_path):
157 | os.unlink(file_path)
158 | elif os.path.isdir(file_path):
159 | shutil.rmtree(file_path)
160 | except Exception as e:
161 | print(f"Error while deleting {file_path}: {e}")
162 | print("Deleted", folder_path, "successfully")
163 |
164 |
165 | def extract_embeds(dict):
166 | # {505: {'id': 505, 'embed': [0.8,....]}} --> [[0.8,....],[.......]]
167 | return [entry["embed"] for entry in dict.values()]
168 |
169 |
170 | def extract_embeds_array(arr):
171 | return np.array([entry["embed"] for entry in arr])
172 |
173 |
174 | def cal_score(vec1, vec2):
175 | dot_product = np.dot(vec1, vec2)
176 | norm_vec1 = np.linalg.norm(vec1)
177 | norm_vec2 = np.linalg.norm(vec2)
178 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
179 | return cosine_similarity
180 |
181 |
182 | def calculate_offset(record_id: int) -> int:
183 | # Calculate the offset for a given record ID
184 | record_size = struct.calcsize("I70f")
185 | return (record_id) * record_size
186 |
187 |
188 | def read_multiple_records_by_id(file_path, records_id: List[int],dictionary_format=False):
189 | record_size = struct.calcsize("I70f")
190 | records = {}
191 |
192 | records_dictionary={}
193 |
194 | with open(file_path, "rb") as fin:
195 | for i in range(len(records_id)):
196 | offset = calculate_offset(records_id[i])
197 | fin.seek(offset) # Move the file pointer to the calculated offset
198 | data = fin.read(record_size)
199 | if not data:
200 | records[records_id[i]] = None
201 | continue
202 |
203 | # Unpack the binary data into a dictionary
204 | unpacked_data = struct.unpack("I70f", data)
205 | id_value, floats = unpacked_data[0], unpacked_data[1:]
206 |
207 | if dictionary_format:
208 | records_dictionary[id_value]=list(floats)
209 | else:
210 | # Create and return the record dictionary
211 | record = {"id": id_value, "embed": list(floats)}
212 | records[records_id[i]] = record
213 |
214 | if dictionary_format: return records_dictionary
215 | return records
216 |
217 | # def generate_random(k=100):
218 | # # Sample data: k vectors with 70 features each
219 | # data = np.random.uniform(-1, 1, size=(k, 70))
220 |
221 | # # Write data to a text file
222 | # file_path = "../DataBase/random_data_"+str(k)+".txt"
223 | # np.savetxt(file_path, data)
224 |
225 | # # Read Data from File
226 | # # read_data = np.loadtxt(file_path)
227 |
228 |
229 | # def array_to_dictionary(values,keys=None):
230 | # '''
231 | # values: [array of values]
232 | # Keys: [array of Keys] optional if not passed the keys are indexed 0-N
233 | # '''
234 | # if(keys is None):
235 | # keys=range(0,len(values))
236 |
237 | # if(len(values)!=len(keys)):
238 | # print ("array_to_dictionary(): InCorrect Size of keys and values")
239 | # return None
240 |
241 | # dictionary_data = dict(zip(keys, values))
242 | # return dictionary_data
243 |
244 |
245 | # def get_vector_from_id(data_path,id):
246 | # '''
247 | # function to get the vector by its id [BADDDDDDDD Use Seek]
248 |
249 | # '''
250 | # read_data = np.loadtxt(data_path)
251 | # return read_data[id]
252 |
253 |
254 | # def check_dir(path):
255 | # if os.path.exists(path):
256 | # shutil.rmtree(path, ignore_errors=True, onerror=lambda func, path, exc: None)
257 | # os.makedirs(path)
258 |
259 |
260 | # Test generate_random()
261 | # generate_random(10000)
262 |
--------------------------------------------------------------------------------
/src/vec_db.py:
--------------------------------------------------------------------------------
1 |
2 | from typing import Dict, List, Annotated
3 | from utils import *
4 | import numpy as np
5 | import time
6 | import os
7 | # from Modules.LSH import LSH_index, semantic_query_lsh
8 | from IVF import IVF_index,semantic_query_ivf
9 |
10 | NUMBER_OF_RECORDS_BRUTE_FORCE = 20*10**3
11 |
12 | class VecDB:
13 | def __init__(self,file_path="./DataBase", new_db = True) -> None:
14 | '''
15 | Constructor
16 | '''
17 | self.file_path =file_path+'/data.bin' # Data File Path
18 | self.database_path= file_path # Path of the Folder to Create Indexes
19 | self.n_regions = None # Initialize n_regions
20 |
21 |
22 | if new_db:
23 | if not os.path.exists(self.database_path):
24 | os.makedirs(self.database_path)
25 |
26 | else:
27 | # If New DataBase Empty DataBase Folder
28 | empty_folder(self.database_path)
29 |
30 | # just open new file to delete the old one
31 | with open(self.file_path, "w") as fout:
32 | # if you need to add any head to the file
33 | pass
34 |
35 |
36 | self.level1=None
37 |
38 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
39 | # Append in Binary Mode
40 | with open(self.file_path, "ab") as fout:
41 | for row in rows:
42 | id, embed = row["id"], row["embed"]
43 | # Pack the data into a binary format
44 | data = struct.pack(f"I{70}f", id, *embed)
45 | fout.write(data)
46 | self._build_index()
47 |
48 |
49 | def _build_index(self,Level_1_nbits=5, Level_2_nbits=3, Level_3_nbits=3,Level_4_nbits=3)-> None:
50 |
51 | '''
52 | Build the Index
53 | '''
54 | file_size = os.path.getsize(self.file_path)
55 | record_size=struct.calcsize(f"I{70}f")
56 | n_records=file_size/record_size
57 | if(n_records==10*10**3):
58 | self.number_of_clusters=10
59 | elif(n_records==100*10**3):
60 | self.number_of_clusters=50
61 | elif(n_records==10**6):
62 | self.number_of_clusters=200
63 | elif(n_records==5*10**6):
64 | self.number_of_clusters=500
65 | elif(n_records==10*10**6):
66 | self.number_of_clusters=8000
67 |
68 | # if()
69 | # if(n_records==10**3):
70 | # self.number_of_clusters=50
71 | # if(n_records<=100000):
72 | # self.number_of_clusters=10
73 | # self.n_regions=5
74 | # else:
75 | # self.number_of_clusters=int(n_records/NUMBER_OF_RECORDS_BRUTE_FORCE)
76 | # self.number_of_clusters=NUMBER_OF_RECORDS_BRUTE_FORCE
77 | # self.n_regions=10
78 | print("Record Size: ",record_size)
79 | print("File Size: ",file_size)
80 | print("Building Index ..........")
81 | print("Number of records: ",n_records)
82 | print("Number of Clusters: ",self.number_of_clusters)
83 | # measure the time
84 | start = time.time()
85 |
86 | # Make Level1 Folder
87 | Level1_folder_path = self.database_path+'/Level1'
88 | if not os.path.exists(Level1_folder_path):
89 | os.makedirs(Level1_folder_path)
90 |
91 | # IVF Layer 1 Indexing
92 | chunk_size=100000
93 | print("chunk_size",chunk_size)
94 | IVF_index(file_path=self.file_path,K_means_metric='euclidean',K_means_n_clusters=self.number_of_clusters,k_means_batch_size=chunk_size,k_means_max_iter=100,k_means_n_init='auto',chunk_size=chunk_size,index_folder_path=Level1_folder_path)
95 |
96 |
97 | # # Layer 1 Indexing
98 | # # level_1_in = self.get_top_k_records(top_k_records)
99 | # level_1_planes = LSH_index(file_path=self.file_path, nbits=Level_1_nbits, chunk_size=1000,index_path=self.database_path + "/Level1")
100 | # np.save(self.database_path + "/Level1/"+'metadata.npy',level_1_planes)
101 | # print("Layer 1 Finished")
102 | # return
103 |
104 |
105 |
106 | # # Layer 2 Indexing
107 | # for file_name in os.listdir(self.database_path + "/Level1"):
108 | # file_path = os.path.join(self.database_path + "/Level1", file_name)
109 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
110 | # read_data_2 = np.loadtxt(file_path, dtype=int, ndmin=1)
111 | # level_2_in = self.read_multiple_records_by_id(read_data_2)
112 | # level_2_planes = LSH_index(data=level_2_in.values(), nbits=Level_2_nbits, index_path=self.database_path + "/Level2/" + file_name[:-4])
113 | # np.save(self.database_path + "/Level2/" + file_name[:-4]+'/metadata.npy',level_2_planes)
114 | # print("Layer 2 Finished")
115 | # return
116 |
117 |
118 | # # Layer 3 Indexing
119 | # for folder_name in os.listdir(self.database_path + "/Level2"):
120 | # folder_path = os.path.join(self.database_path + "/Level2", folder_name)
121 | # for file_name in os.listdir(folder_path):
122 | # file_path = os.path.join(folder_path, file_name)
123 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
124 | # read_data_3 = np.loadtxt(file_path, dtype=int, ndmin=1)
125 | # level_3_in = self.read_multiple_records_by_id(read_data_3)
126 | # level_3_planes = LSH_index(data=level_3_in.values(), nbits=Level_3_nbits, index_path=self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4])
127 | # np.save(self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4]+'/metadata.npy',level_3_planes)
128 | # print("Layer 3 Finished")
129 | # return
130 |
131 | # # Layer 4 Indexing
132 | # for folder_name in os.listdir(self.database_path + "/Level3"):
133 | # folder_path = os.path.join(self.database_path + "/Level3", folder_name)
134 | # for folder_name_2 in os.listdir(folder_path):
135 | # folder_path_2 = os.path.join(folder_path, folder_name_2)
136 | # for file_name in os.listdir(folder_path_2):
137 | # file_path = os.path.join(folder_path_2, file_name)
138 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
139 | # read_data_4 = np.loadtxt(file_path, dtype=int, ndmin=1)
140 | # level_4_in = self.read_multiple_records_by_id(read_data_4)
141 | # level_4_planes = LSH_index(data=level_4_in.values(), nbits=Level_4_nbits, index_path=self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4])
142 | # np.save(self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4]+'/metadata.npy',level_4_planes)
143 | # print("Layer 4 Finished")
144 |
145 |
146 | # measure the time
147 | end = time.time()
148 | print("Indexing Done ...... Time taken by Indexing: ",end - start)
149 | return
150 |
151 | def retrive(self, query:Annotated[List[float], 70],top_k = 5)-> [int]:
152 | '''
153 | Get the top_k vectors similar to the Query
154 |
155 | return: list of the top_k similar vectors Ordered by Cosine Similarity
156 | '''
157 | print(f"Retrieving top {top_k} ..........")
158 | Level1_folder_path = self.database_path+'/Level1'
159 |
160 | file_size = os.path.getsize(self.file_path)
161 | record_size=struct.calcsize(f"I{70}f")
162 | n_records=file_size/record_size
163 | if(n_records==10*10**3):
164 | n_probes=3
165 | elif(n_records==100*10**3):
166 | n_probes=10
167 | elif(n_records==10**6):
168 | n_probes=5
169 | elif(n_records==5*10**6):
170 | n_probes=15
171 | elif(n_records==10*10**6):
172 | n_probes=30
173 | elif(n_records==15*10**6):
174 | n_probes=256
175 | elif(n_records==20*10**6):
176 | n_probes=64
177 | # n_probes=0
178 | # if(n_records<=5*10**6):
179 | # n_probes=3
180 | # else:
181 | # n_probes=20
182 | final_result=semantic_query_ivf(data_file_path=self.file_path,index_folder_path=Level1_folder_path,query=query,top_k=top_k,n_regions=n_probes)
183 |
184 | return final_result
185 |
186 |
187 |
188 | # # Retrieve from Level 1
189 | # level_1_planes = np.load(self.database_path + "/Level1"+'/metadata.npy')
190 | # bucket_1,result = semantic_query_lsh(query, level_1_planes, self.database_path + "/Level1")
191 | # print("length of first bucket",result.shape)
192 |
193 | # if len(result) < top_k:
194 | # print('level 1 smaller than top_k')
195 |
196 | # # Retrieve from Level 2
197 | # level_2_planes = np.load(self.database_path + "/Level2/"+bucket_1+'/metadata.npy')
198 | # bucket_2,result = semantic_query_lsh(query, level_2_planes, self.database_path + "/Level2/"+bucket_1)
199 | # print("length of second bucket",result.shape)
200 |
201 | # if len(result) < top_k:
202 | # print('level 2 smaller than top_k')
203 |
204 | # # Retrieve from Level 3
205 | # level_3_planes = np.load(self.database_path + "/Level3/"+bucket_1+'/'+bucket_2+'/metadata.npy')
206 | # bucket_3,result = semantic_query_lsh(query, level_3_planes, self.database_path + "/Level3/"+bucket_1+'/'+bucket_2)
207 | # print("length of third bucket",result.shape)
208 |
209 | # if len(result) < top_k:
210 | # print('level 3 smaller than top_k')
211 |
212 | # # Retrieve from Level 4
213 | # level_4_planes = np.load(self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3+'/metadata.npy')
214 | # bucket_4,result = semantic_query_lsh(query, level_4_planes, self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3)
215 | # print("length of fourth bucket",result.shape)
216 |
217 | # if len(result) < top_k:
218 | # print('level 4 smaller than top_k')
219 |
220 |
221 | # # Retrieve from Data Base the Embeddings of the Vectors
222 | # final_result= read_multiple_records_by_id(self.file_path,result)
223 |
224 | # Calculate the Cosine Similarity between the Query and the Vectors
225 | # scores = []
226 | # for row in final_result.values():
227 | # id_value = row['id']
228 | # embed_values = row['embed']
229 | # score = self._cal_score(query, embed_values)
230 | # scores.append((score, id_value))
231 | # scores = sorted(scores, reverse=True)[:top_k]
232 | # return [s[1] for s in scores]
233 |
234 |
235 |
236 |
237 | def _cal_score(self, vec1, vec2):
238 | dot_product = np.dot(vec1, vec2)
239 | norm_vec1 = np.linalg.norm(vec1)
240 | norm_vec2 = np.linalg.norm(vec2)
241 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
242 | return cosine_similarity
--------------------------------------------------------------------------------
/src/worst_case_implementation.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Annotated
2 | import struct
3 | import numpy as np
4 |
5 | class VecDBWorst:
6 | def __init__(self, file_path = "saved_db.csv", new_db = True) -> None:
7 | self.file_path = file_path
8 | if new_db:
9 | # just open new file to delete the old one
10 | with open(self.file_path, "w") as fout:
11 | # if you need to add any head to the file
12 | pass
13 |
14 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
15 | with open(self.file_path, "a+") as fout:
16 | for row in rows:
17 | id, embed = row["id"], row["embed"]
18 | row_str = f"{id}," + ",".join([str(e) for e in embed])
19 | fout.write(f"{row_str}\n")
20 | self._build_index()
21 |
22 | # def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
23 | # with open(self.file_path, "ab") as fout: # Open the file in binary mode for appending
24 | # for row in rows:
25 | # id, embed = row["id"], row["embed"]
26 | # # Pack the data into a binary format
27 | # data = struct.pack(f"I{70}f", id, *embed)
28 | # fout.write(data)
29 | # self._build_index()
30 |
31 | # def calculate_offset(self, record_id: int) -> int:
32 | # # Calculate the offset for a given record ID
33 | # record_size = struct.calcsize("I70f")
34 | # return (record_id - 1) * record_size
35 |
36 | # def read_record_by_id(self, record_id: int) -> Dict[int, Annotated[List[float], 70]]:
37 | # record_size = struct.calcsize("I70f")
38 | # offset = self.calculate_offset(record_id)
39 |
40 | # with open(self.file_path, "rb") as fin:
41 | # fin.seek(offset) # Move the file pointer to the calculated offset
42 | # data = fin.read(record_size)
43 | # if not data:
44 | # return {} # Record not found
45 |
46 | # # Unpack the binary data into a dictionary
47 | # unpacked_data = struct.unpack("I70f", data)
48 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
49 |
50 | # # Create and return the record dictionary
51 | # record = {"id": id_value, "embed": list(floats)}
52 | # return {record_id: record}
53 |
54 | # def read_multiple_records_by_id(self, records_id: List[int]):
55 | # record_size = struct.calcsize("I70f")
56 | # records = {}
57 |
58 | # with open(self.file_path, "rb") as fin:
59 | # for i in range(len(records_id)):
60 | # offset = self.calculate_offset(records_id[i])
61 | # fin.seek(offset) # Move the file pointer to the calculated offset
62 | # data = fin.read(record_size)
63 | # if not data:
64 | # records[records_id[i]] = None
65 | # continue
66 |
67 | # # Unpack the binary data into a dictionary
68 | # unpacked_data = struct.unpack("I70f", data)
69 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
70 |
71 | # # Create and return the record dictionary
72 | # record = {"id": id_value, "embed": list(floats)}
73 | # records[records_id[i]] = record
74 | # return records
75 |
76 | # def get_top_k_records(self,k):
77 | # records = []
78 | # record_size = struct.calcsize("I70f")
79 | # with open(self.file_path,'rb') as fin:
80 | # fin.seek(0)
81 | # for i in range(k):
82 | # data = fin.read(record_size)
83 | # unpacked_data = struct.unpack("I70f", data)
84 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
85 |
86 | # record = {"id": id_value, "embed": list(floats)}
87 | # records.append(record)
88 | # return records
89 |
90 | def retrive(self, query: Annotated[List[float], 70], top_k = 5):
91 | scores = []
92 | with open(self.file_path, "r") as fin:
93 | for row in fin.readlines():
94 | row_splits = row.split(",")
95 | id = int(row_splits[0])
96 | embed = [float(e) for e in row_splits[1:]]
97 | score = self._cal_score(query, embed)
98 | scores.append((score, id))
99 | # here we assume that if two rows have the same score, return the lowest ID
100 | scores = sorted(scores, reverse=True)[:top_k]
101 | return [s[1] for s in scores]
102 |
103 | def _cal_score(self, vec1, vec2):
104 | dot_product = np.dot(vec1, vec2)
105 | norm_vec1 = np.linalg.norm(vec1)
106 | norm_vec2 = np.linalg.norm(vec2)
107 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
108 | return cosine_similarity
109 |
110 | def _build_index(self):
111 | pass
112 |
113 |
114 |
--------------------------------------------------------------------------------
/vector searching algorithms/LSHHyperPlane.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 19,
6 | "metadata": {},
7 | "outputs": [],
8 | "source": [
9 | "nbits = 3 # number of hyperplanes and binary vals to produce\n",
10 | "d = 70 # vector dimensions\n",
11 | "\n",
12 | "#! log2(8)"
13 | ]
14 | },
15 | {
16 | "cell_type": "code",
17 | "execution_count": 20,
18 | "metadata": {},
19 | "outputs": [
20 | {
21 | "data": {
22 | "text/plain": [
23 | "array([[-0.16225197, -0.1159841 , -0.0379947 , 0.47482455, -0.24329282,\n",
24 | " -0.17219515, 0.0889353 , -0.34083189, -0.0857109 , 0.19710836,\n",
25 | " -0.48189731, 0.04372259, -0.40512638, -0.49822556, -0.18287792,\n",
26 | " 0.31290614, -0.3228106 , -0.24989376, -0.30750153, 0.35378187,\n",
27 | " 0.32047542, -0.3405673 , 0.01704872, -0.29642438, -0.13589619,\n",
28 | " -0.28071414, -0.05584624, 0.09991783, -0.3077081 , 0.13647533,\n",
29 | " -0.4770819 , 0.38661425, 0.00783346, -0.3927221 , 0.24110202,\n",
30 | " 0.05990217, -0.06802645, 0.03282446, 0.30179191, 0.19156282,\n",
31 | " 0.1054492 , -0.00932327, 0.32185434, 0.49241347, 0.20733614,\n",
32 | " 0.18026476, -0.26101194, -0.3778801 , 0.4936665 , -0.41047846,\n",
33 | " 0.23792759, -0.23321223, 0.39830173, -0.13084787, -0.31116184,\n",
34 | " -0.00189201, 0.08791531, -0.14980787, 0.30558995, -0.00670045,\n",
35 | " 0.2243021 , -0.41023912, 0.18223908, -0.38580149, -0.11450846,\n",
36 | " -0.43534932, -0.30870211, 0.29227875, -0.15542413, 0.20264467],\n",
37 | " [ 0.14822377, -0.3276353 , 0.26702648, 0.03217607, -0.20768427,\n",
38 | " -0.46076215, -0.1540441 , -0.14252796, -0.21732578, -0.35428344,\n",
39 | " 0.2084557 , -0.18725843, -0.14300948, -0.16831679, -0.15468043,\n",
40 | " -0.07450581, 0.00233269, -0.43596823, 0.19815002, 0.19192439,\n",
41 | " 0.13743071, 0.1589349 , -0.37645398, -0.40863437, 0.18087113,\n",
42 | " 0.26399475, -0.20873789, 0.31885534, -0.47078825, 0.19251382,\n",
43 | " 0.39492556, 0.13612851, 0.05252917, 0.1066819 , -0.20602171,\n",
44 | " 0.019199 , -0.42388983, -0.29196827, -0.08782944, 0.38928403,\n",
45 | " -0.14570291, -0.04510013, -0.11215063, 0.08435185, 0.0814708 ,\n",
46 | " 0.30669655, 0.2207886 , 0.48463154, -0.07672077, 0.02202735,\n",
47 | " -0.43230734, -0.16302195, 0.42660854, 0.21663128, -0.37419712,\n",
48 | " -0.07194837, 0.38714646, -0.34736523, 0.17594598, -0.04035088,\n",
49 | " -0.40523731, -0.46061098, -0.18572369, 0.46951449, 0.04887038,\n",
50 | " -0.47345552, 0.21424037, 0.06851608, -0.44381182, 0.18315833],\n",
51 | " [-0.34032214, 0.20510388, -0.11437108, -0.25271871, -0.29298418,\n",
52 | " -0.39316298, -0.27235239, -0.02090773, 0.42378779, 0.36244202,\n",
53 | " -0.49353892, -0.37632702, 0.08773845, 0.23887437, -0.03724808,\n",
54 | " -0.23496996, 0.46748875, -0.2527981 , -0.39073735, 0.40062121,\n",
55 | " 0.10471371, -0.43568824, 0.14986386, 0.18503889, 0.37681242,\n",
56 | " -0.01243404, -0.39396771, 0.19966568, 0.08733691, 0.2509886 ,\n",
57 | " -0.30488297, 0.39487436, -0.38444297, -0.47168848, 0.40205414,\n",
58 | " -0.40537546, 0.03695501, -0.28056081, 0.42477745, 0.49012324,\n",
59 | " 0.29699303, -0.14461517, 0.37445295, -0.30211604, -0.39721614,\n",
60 | " 0.34692406, 0.11398823, 0.30746006, -0.22106426, -0.1443832 ,\n",
61 | " -0.07727599, 0.23807248, 0.32999453, 0.36904291, -0.01930504,\n",
62 | " -0.01021899, -0.25900161, 0.32297803, -0.08289675, 0.28510423,\n",
63 | " -0.35789496, -0.22445429, -0.43838493, -0.25795092, 0.18836288,\n",
64 | " 0.07583603, 0.24472323, 0.07588773, -0.46374612, 0.29371209]])"
65 | ]
66 | },
67 | "execution_count": 20,
68 | "metadata": {},
69 | "output_type": "execute_result"
70 | }
71 | ],
72 | "source": [
73 | "import numpy as np\n",
74 | "# create a set of 4 hyperplanes, with 2 dimensions\n",
75 | "plane_norms = np.random.rand(nbits, d) - .5\n",
76 | "plane_norms"
77 | ]
78 | },
79 | {
80 | "cell_type": "code",
81 | "execution_count": 21,
82 | "metadata": {},
83 | "outputs": [
84 | {
85 | "name": "stdout",
86 | "output_type": "stream",
87 | "text": [
88 | "[[0.24346879 0.20847865 0.09572784 0.55319155 0.07226476 0.87621505\n",
89 | " 0.58913737 0.46246208 0.3333929 0.96904595 0.89600564 0.59480015\n",
90 | " 0.77746555 0.54283576 0.67569602 0.23497124 0.07950847 0.31373656\n",
91 | " 0.14113477 0.408343 0.81078156 0.57261091 0.35708278 0.41438797\n",
92 | " 0.44477819 0.65966904 0.12290202 0.22656549 0.07834719 0.83130173\n",
93 | " 0.4011682 0.72348172 0.58621532 0.97192664 0.12448728 0.19814639\n",
94 | " 0.84652828 0.19786014 0.70758209 0.91906712 0.93421752 0.70310207\n",
95 | " 0.27726954 0.96761661 0.06944151 0.32237123 0.95214921 0.55453475\n",
96 | " 0.22046089 0.75767774 0.76303426 0.11226826 0.96778584 0.18364252\n",
97 | " 0.86959947 0.35425893 0.05710316 0.67524358 0.84838244 0.84003201\n",
98 | " 0.71003473 0.74628258 0.48265117 0.48526062 0.41682251 0.24458015\n",
99 | " 0.56040784 0.47847743 0.11747069 0.2841326 ]]\n",
100 | "[[0.08021692 0.11425865 0.66780691 0.64591999 0.72730137 0.37124926\n",
101 | " 0.90060496 0.29506503 0.14570761 0.05080957 0.84666837 0.89391903\n",
102 | " 0.24643966 0.44699371 0.64934757 0.67063548 0.75535129 0.75834603\n",
103 | " 0.36194954 0.35376051 0.86358269 0.67616135 0.0427738 0.70108469\n",
104 | " 0.27322976 0.30507254 0.39729602 0.43346906 0.20055928 0.0809176\n",
105 | " 0.05108797 0.16594553 0.60731689 0.25100448 0.60585635 0.18616766\n",
106 | " 0.35069704 0.88805346 0.07440514 0.91451748 0.14379966 0.30309109\n",
107 | " 0.6960067 0.5342824 0.90332074 0.95252648 0.74792365 0.09408956\n",
108 | " 0.44708244 0.75941983 0.49066048 0.6441386 0.63012995 0.98668287\n",
109 | " 0.60327176 0.62091147 0.72095104 0.59418854 0.84847164 0.18768005\n",
110 | " 0.50059785 0.83818945 0.58380742 0.79375409 0.38927237 0.30760971\n",
111 | " 0.90913216 0.75463438 0.32010021 0.71195468]]\n",
112 | "[array([[0.24346879, 0.20847865, 0.09572784, 0.55319155, 0.07226476,\n",
113 | " 0.87621505, 0.58913737, 0.46246208, 0.3333929 , 0.96904595,\n",
114 | " 0.89600564, 0.59480015, 0.77746555, 0.54283576, 0.67569602,\n",
115 | " 0.23497124, 0.07950847, 0.31373656, 0.14113477, 0.408343 ,\n",
116 | " 0.81078156, 0.57261091, 0.35708278, 0.41438797, 0.44477819,\n",
117 | " 0.65966904, 0.12290202, 0.22656549, 0.07834719, 0.83130173,\n",
118 | " 0.4011682 , 0.72348172, 0.58621532, 0.97192664, 0.12448728,\n",
119 | " 0.19814639, 0.84652828, 0.19786014, 0.70758209, 0.91906712,\n",
120 | " 0.93421752, 0.70310207, 0.27726954, 0.96761661, 0.06944151,\n",
121 | " 0.32237123, 0.95214921, 0.55453475, 0.22046089, 0.75767774,\n",
122 | " 0.76303426, 0.11226826, 0.96778584, 0.18364252, 0.86959947,\n",
123 | " 0.35425893, 0.05710316, 0.67524358, 0.84838244, 0.84003201,\n",
124 | " 0.71003473, 0.74628258, 0.48265117, 0.48526062, 0.41682251,\n",
125 | " 0.24458015, 0.56040784, 0.47847743, 0.11747069, 0.2841326 ]]), array([[0.08021692, 0.11425865, 0.66780691, 0.64591999, 0.72730137,\n",
126 | " 0.37124926, 0.90060496, 0.29506503, 0.14570761, 0.05080957,\n",
127 | " 0.84666837, 0.89391903, 0.24643966, 0.44699371, 0.64934757,\n",
128 | " 0.67063548, 0.75535129, 0.75834603, 0.36194954, 0.35376051,\n",
129 | " 0.86358269, 0.67616135, 0.0427738 , 0.70108469, 0.27322976,\n",
130 | " 0.30507254, 0.39729602, 0.43346906, 0.20055928, 0.0809176 ,\n",
131 | " 0.05108797, 0.16594553, 0.60731689, 0.25100448, 0.60585635,\n",
132 | " 0.18616766, 0.35069704, 0.88805346, 0.07440514, 0.91451748,\n",
133 | " 0.14379966, 0.30309109, 0.6960067 , 0.5342824 , 0.90332074,\n",
134 | " 0.95252648, 0.74792365, 0.09408956, 0.44708244, 0.75941983,\n",
135 | " 0.49066048, 0.6441386 , 0.63012995, 0.98668287, 0.60327176,\n",
136 | " 0.62091147, 0.72095104, 0.59418854, 0.84847164, 0.18768005,\n",
137 | " 0.50059785, 0.83818945, 0.58380742, 0.79375409, 0.38927237,\n",
138 | " 0.30760971, 0.90913216, 0.75463438, 0.32010021, 0.71195468]]), array([[0.77078444, 0.08654989, 0.53784898, 0.27799896, 0.8002419 ,\n",
139 | " 0.3718589 , 0.79333253, 0.93137511, 0.62069102, 0.24775348,\n",
140 | " 0.4272542 , 0.98292296, 0.8470418 , 0.52637634, 0.80102169,\n",
141 | " 0.9440776 , 0.68466439, 0.86571266, 0.27882657, 0.71765609,\n",
142 | " 0.75582515, 0.94039075, 0.20125464, 0.64221553, 0.83386962,\n",
143 | " 0.73863021, 0.93711781, 0.4794255 , 0.48793874, 0.86123148,\n",
144 | " 0.92146684, 0.81077912, 0.50759451, 0.18685755, 0.98628992,\n",
145 | " 0.97781323, 0.38057898, 0.87204991, 0.30697755, 0.49756831,\n",
146 | " 0.21495361, 0.38075046, 0.30418495, 0.02362606, 0.96563469,\n",
147 | " 0.80356302, 0.60805212, 0.30632618, 0.59294981, 0.08821319,\n",
148 | " 0.19220448, 0.17337163, 0.93316608, 0.46086827, 0.17464549,\n",
149 | " 0.16804619, 0.05238805, 0.95753146, 0.87498728, 0.5543621 ,\n",
150 | " 0.727997 , 0.54389248, 0.38181964, 0.47099822, 0.59929861,\n",
151 | " 0.35783896, 0.27050514, 0.98113893, 0.49137662, 0.05494532]])]\n"
152 | ]
153 | }
154 | ],
155 | "source": [
156 | "a=np.random.random((1, 70))\n",
157 | "b=np.random.random((1, 70))\n",
158 | "c=np.random.random((1, 70))\n",
159 | "# Repeat the array to generate a 1x70 array\n",
160 | "print(a)\n",
161 | "print(b)\n",
162 | "dataset=[a,b,c]\n",
163 | "print(dataset)\n"
164 | ]
165 | },
166 | {
167 | "cell_type": "code",
168 | "execution_count": 22,
169 | "metadata": {},
170 | "outputs": [
171 | {
172 | "data": {
173 | "text/plain": [
174 | "array([[-0.98219519, -1.00134517, 0.23751343]])"
175 | ]
176 | },
177 | "execution_count": 22,
178 | "metadata": {},
179 | "output_type": "execute_result"
180 | }
181 | ],
182 | "source": [
183 | "# calculate the dot product for each of these\n",
184 | "a_dot = np.dot(a, plane_norms.T)\n",
185 | "b_dot = np.dot(b, plane_norms.T)\n",
186 | "c_dot = np.dot(c, plane_norms.T)\n",
187 | "a_dot"
188 | ]
189 | },
190 | {
191 | "cell_type": "code",
192 | "execution_count": 23,
193 | "metadata": {},
194 | "outputs": [
195 | {
196 | "data": {
197 | "text/plain": [
198 | "array([[False, False, True]])"
199 | ]
200 | },
201 | "execution_count": 23,
202 | "metadata": {},
203 | "output_type": "execute_result"
204 | }
205 | ],
206 | "source": [
207 | " #! Dataset\n",
208 | "# we know that a positive dot product == +ve side of hyperplane\n",
209 | "# and negative dot product == -ve side of hyperplane\n",
210 | "a_dot = a_dot > 0\n",
211 | "b_dot = b_dot > 0\n",
212 | "c_dot = c_dot > 0\n",
213 | "a_dot"
214 | ]
215 | },
216 | {
217 | "cell_type": "code",
218 | "execution_count": 24,
219 | "metadata": {},
220 | "outputs": [
221 | {
222 | "data": {
223 | "text/plain": [
224 | "array([0, 0, 1])"
225 | ]
226 | },
227 | "execution_count": 24,
228 | "metadata": {},
229 | "output_type": "execute_result"
230 | }
231 | ],
232 | "source": [
233 | "# convert our boolean arrays to int arrays to make bucketing\n",
234 | "# easier (although is okay to use boolean for Hamming distance)\n",
235 | "a_dot = a_dot.astype(int)[0]\n",
236 | "b_dot = b_dot.astype(int)[0]\n",
237 | "c_dot = c_dot.astype(int)[0]\n",
238 | "a_dot"
239 | ]
240 | },
241 | {
242 | "cell_type": "code",
243 | "execution_count": 25,
244 | "metadata": {},
245 | "outputs": [
246 | {
247 | "name": "stdout",
248 | "output_type": "stream",
249 | "text": [
250 | "001\n",
251 | "000\n",
252 | "000\n",
253 | "{'001': [0], '000': [1, 2]}\n"
254 | ]
255 | }
256 | ],
257 | "source": [
258 | "vectors = [a_dot, b_dot, c_dot]\n",
259 | "buckets = {}\n",
260 | "i = 0\n",
261 | "\n",
262 | "for i in range(len(vectors)):\n",
263 | " # convert from array to string\n",
264 | " hash_str = ''.join(vectors[i].astype(str))\n",
265 | " print(hash_str)\n",
266 | " # create bucket if it doesn't exist\n",
267 | " if hash_str not in buckets.keys():\n",
268 | " buckets[hash_str] = []\n",
269 | " # add vector position to bucket\n",
270 | " buckets[hash_str].append(i)\n",
271 | "\n",
272 | "print(buckets)"
273 | ]
274 | },
275 | {
276 | "cell_type": "code",
277 | "execution_count": 26,
278 | "metadata": {},
279 | "outputs": [
280 | {
281 | "name": "stdout",
282 | "output_type": "stream",
283 | "text": [
284 | "[[0.98217599 0.0740474 0.60518903 0.61501807 0.37022865 0.59778156\n",
285 | " 0.37753566 0.44307262 0.00959564 0.43188369 0.77591898 0.40505507\n",
286 | " 0.40793131 0.30983161 0.20962565 0.47749091 0.64407774 0.07951662\n",
287 | " 0.21950209 0.5617165 0.27452938 0.73727983 0.10188798 0.15470781\n",
288 | " 0.9668675 0.63906846 0.76027852 0.59622264 0.90857345 0.49078738\n",
289 | " 0.17966669 0.29728534 0.76605938 0.89432476 0.17330878 0.91669595\n",
290 | " 0.327371 0.89819097 0.98084002 0.44954373 0.04567631 0.18179054\n",
291 | " 0.97601994 0.11805953 0.42048999 0.19162843 0.69235512 0.48898102\n",
292 | " 0.89062367 0.27993955 0.90024359 0.15118375 0.81479348 0.42161774\n",
293 | " 0.78538813 0.84025532 0.42421928 0.63838074 0.74416435 0.76683117\n",
294 | " 0.63133131 0.36330796 0.24528589 0.42388808 0.53415259 0.58918953\n",
295 | " 0.50997522 0.88050857 0.5938881 0.76094727]]\n",
296 | "Query belongs to bucket: [1, 2]\n"
297 | ]
298 | }
299 | ],
300 | "source": [
301 | " #! for tesing input query\n",
302 | "\n",
303 | "query=np.random.random((1, 70))\n",
304 | "print(query)\n",
305 | "\n",
306 | "query_dot = np.dot(query, plane_norms.T)\n",
307 | "query_dot = query_dot > 0\n",
308 | "query_dot = query_dot.astype(int)[0]\n",
309 | "\n",
310 | "# Convert the query array to a string\n",
311 | "query_hash_str = ''.join(query_dot.astype(str))\n",
312 | "\n",
313 | "# Check which bucket the query belongs to\n",
314 | "if query_hash_str in buckets.keys():\n",
315 | " bucket_containing_query = buckets[query_hash_str]\n",
316 | " print(\"Query belongs to bucket:\", bucket_containing_query)\n",
317 | "else:\n",
318 | " print(\"Query doesn't match any existing buckets\")\n",
319 | " print(0)\n",
320 | "\n"
321 | ]
322 | },
323 | {
324 | "cell_type": "code",
325 | "execution_count": 27,
326 | "metadata": {},
327 | "outputs": [
328 | {
329 | "name": "stdout",
330 | "output_type": "stream",
331 | "text": [
332 | "[[0.24346879 0.20847865 0.09572784 0.55319155 0.07226476 0.87621505\n",
333 | " 0.58913737 0.46246208 0.3333929 0.96904595 0.89600564 0.59480015\n",
334 | " 0.77746555 0.54283576 0.67569602 0.23497124 0.07950847 0.31373656\n",
335 | " 0.14113477 0.408343 0.81078156 0.57261091 0.35708278 0.41438797\n",
336 | " 0.44477819 0.65966904 0.12290202 0.22656549 0.07834719 0.83130173\n",
337 | " 0.4011682 0.72348172 0.58621532 0.97192664 0.12448728 0.19814639\n",
338 | " 0.84652828 0.19786014 0.70758209 0.91906712 0.93421752 0.70310207\n",
339 | " 0.27726954 0.96761661 0.06944151 0.32237123 0.95214921 0.55453475\n",
340 | " 0.22046089 0.75767774 0.76303426 0.11226826 0.96778584 0.18364252\n",
341 | " 0.86959947 0.35425893 0.05710316 0.67524358 0.84838244 0.84003201\n",
342 | " 0.71003473 0.74628258 0.48265117 0.48526062 0.41682251 0.24458015\n",
343 | " 0.56040784 0.47847743 0.11747069 0.2841326 ]]\n",
344 | "[[0.14634457 0.20457996 0.24741999 0.92396483 0.02756294 0.55294575\n",
345 | " 0.53786271 0.25648871 0.87692497 0.0504492 0.78309208 0.78032385\n",
346 | " 0.60510748 0.50301495 0.33663068 0.48482831 0.31022081 0.22420917\n",
347 | " 0.03656832 0.0576642 0.02176892 0.41431776 0.97839866 0.36627294\n",
348 | " 0.90726783 0.942912 0.10056186 0.27157551 0.35269652 0.65476284\n",
349 | " 0.3802201 0.10174655 0.59332797 0.41950798 0.51371538 0.66586081\n",
350 | " 0.86951989 0.30015139 0.39627816 0.90903098 0.71667395 0.27805884\n",
351 | " 0.43209455 0.17558953 0.55876165 0.44371013 0.54468845 0.95573234\n",
352 | " 0.8846391 0.69004526 0.78832344 0.97466743 0.01227129 0.26130364\n",
353 | " 0.46660631 0.64762417 0.39275623 0.7934695 0.29527433 0.66989054\n",
354 | " 0.73093381 0.26445753 0.17682595 0.24664441 0.41129241 0.83235075\n",
355 | " 0.72809304 0.64075518 0.98233083 0.85216486]]\n",
356 | "[[18.0160595]]\n",
357 | "0\n",
358 | "Query belongs to bucket: [0]\n"
359 | ]
360 | }
361 | ],
362 | "source": [
363 | "query=np.random.random((1, 70))\n",
364 | "# print(query)\n",
365 | "\n",
366 | "# Assuming 'query' is your query vector\n",
367 | "query_dot = np.dot(query, plane_norms.T)\n",
368 | "query_dot = query_dot > 0\n",
369 | "query_dot = query_dot.astype(int)[0]\n",
370 | "\n",
371 | "# Convert the query array to a string\n",
372 | "query_hash_str = ''.join(query_dot.astype(str))\n",
373 | "\n",
374 | "# Check which bucket the query belongs to\n",
375 | "if query_hash_str in buckets.keys():\n",
376 | " bucket_containing_query = buckets[query_hash_str]\n",
377 | " min_dist=100\n",
378 | " index=-1\n",
379 | " for vec in bucket_containing_query:\n",
380 | " print(dataset[vec])\n",
381 | " print(query)\n",
382 | " dot_res=np.dot(query,dataset[vec].T)\n",
383 | " print(dot_res)\n",
384 | " res = dot_res / (np.linalg.norm(query) * np.linalg.norm(dataset[vec].T))\n",
385 | " if res list2[j]:
28 | j += 1
29 | else:
30 | intersection.append(list1[i])
31 | i += 1
32 | j += 1
33 |
34 | return intersection
35 |
36 |
37 | def split_on_sign(data:[[float]],split_on)->int:
38 | '''
39 | @param data: Data to categorize
40 | @split_on: The max count to split on
41 |
42 | @return dictionary of the data splitted by sign +ve and -ve
43 | '''
44 | if(split_on is None or split_on>np.shape(data)[1]):
45 | #split on the whole size
46 | # split_on=np.shape(data)[1]
47 | split_on=10
48 |
49 | regions = []
50 | for col in data[:,:split_on].T: # Transpose the matrix to iterate over columns
51 | positive_region = (col >= 0)
52 | negative_region = (col < 0)
53 | regions.append(np.where(positive_region)[0])
54 | regions.append(np.where(negative_region)[0])
55 | return regions
56 |
57 | def search_on_sign(q:[float],regions:[[int]]):
58 | # O(m * n), where m is the average length of the input lists, and n is the number of input lists.
59 | # Check on sign of the feature
60 | intersect=None
61 | split_on=np.shape(regions)[0]//2
62 | for ind,feature in enumerate(q[:split_on]):
63 | if(ind==0):
64 | intersect=regions[0] if feature>=0 else regions[1]
65 | continue
66 | if(feature>=0):
67 | # positive
68 | intersect=sorted_list_intersection(intersect, regions[2*ind])
69 | else:
70 | #negative
71 | intersect=sorted_list_intersection(intersect, regions[2*ind+1])
72 | return intersect
--------------------------------------------------------------------------------
58 |
59 | ### PQ-LSH
60 | It Combines both LSH & PQ
61 |
62 | ## 🕴 Contributors
63 |
64 |
65 |
74 |
75 |
76 |
77 | ## 📃 License
78 |
79 | This software is licensed under MIT License, See [License](https://github.com/ZiadSheriif/sematic_search_DB/blob/main/LICENSE) for more information ©Ziad Sherif.
80 |
--------------------------------------------------------------------------------
/Report.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/Report.pdf
--------------------------------------------------------------------------------
/Statistics IVF.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/Statistics IVF.xlsx
--------------------------------------------------------------------------------
/proposal.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/proposal.pdf
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/requirements.txt
--------------------------------------------------------------------------------
/src/.gitignore:
--------------------------------------------------------------------------------
1 | Database/
2 | inverted_files/
3 |
4 | # Byte-compiled / optimized / DLL files
5 | __pycache__/
6 | *.py[cod]
7 | *$py.class
8 | *.csv
9 | *.bin
10 |
11 | DataBase
12 |
13 | # C extensions
14 | *.so
15 |
16 | # Distribution / packaging
17 | .Python
18 | build/
19 | develop-eggs/
20 | dist/
21 | downloads/
22 | eggs/
23 | .eggs/
24 | lib/
25 | lib64/
26 | parts/
27 | sdist/
28 | var/
29 | wheels/
30 | share/python-wheels/
31 | *.egg-info/
32 | .installed.cfg
33 | *.egg
34 | MANIFEST
35 |
36 | # PyInstaller
37 | # Usually these files are written by a python script from a template
38 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
39 | *.manifest
40 | *.spec
41 |
42 | # Installer logs
43 | pip-log.txt
44 | pip-delete-this-directory.txt
45 |
46 | # Unit test / coverage reports
47 | htmlcov/
48 | .tox/
49 | .nox/
50 | .coverage
51 | .coverage.*
52 | .cache
53 | nosetests.xml
54 | coverage.xml
55 | *.cover
56 | *.py,cover
57 | .hypothesis/
58 | .pytest_cache/
59 | cover/
60 |
61 | # Translations
62 | *.mo
63 | *.pot
64 |
65 | # Django stuff:
66 | *.log
67 | local_settings.py
68 | db.sqlite3
69 | db.sqlite3-journal
70 |
71 | # Flask stuff:
72 | instance/
73 | .webassets-cache
74 |
75 | # Scrapy stuff:
76 | .scrapy
77 |
78 | # Sphinx documentation
79 | docs/_build/
80 |
81 | # PyBuilder
82 | .pybuilder/
83 | target/
84 |
85 | # Jupyter Notebook
86 | .ipynb_checkpoints
87 |
88 | # IPython
89 | profile_default/
90 | ipython_config.py
91 |
92 | # pyenv
93 | # For a library or package, you might want to ignore these files since the code is
94 | # intended to run in multiple environments; otherwise, check them in:
95 | # .python-version
96 |
97 | # pipenv
98 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
99 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
100 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
101 | # install all needed dependencies.
102 | #Pipfile.lock
103 |
104 | # poetry
105 | # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
106 | # This is especially recommended for binary packages to ensure reproducibility, and is more
107 | # commonly ignored for libraries.
108 | # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
109 | #poetry.lock
110 |
111 | # pdm
112 | # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
113 | #pdm.lock
114 | # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
115 | # in version control.
116 | # https://pdm.fming.dev/#use-with-ide
117 | .pdm.toml
118 |
119 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
120 | __pypackages__/
121 |
122 | # Celery stuff
123 | celerybeat-schedule
124 | celerybeat.pid
125 |
126 | # SageMath parsed files
127 | *.sage.py
128 |
129 | # Environments
130 | .env
131 | .venv
132 | env/
133 | venv/
134 | ENV/
135 | env.bak/
136 | venv.bak/
137 |
138 | # Spyder project settings
139 | .spyderproject
140 | .spyproject
141 |
142 | # Rope project settings
143 | .ropeproject
144 |
145 | # mkdocs documentation
146 | /site
147 |
148 | # mypy
149 | .mypy_cache/
150 | .dmypy.json
151 | dmypy.json
152 |
153 | # Pyre type checker
154 | .pyre/
155 |
156 | # pytype static type analyzer
157 | .pytype/
158 |
159 | # Cython debug symbols
160 | cython_debug/
161 |
162 | # PyCharm
163 | # JetBrains specific template is maintained in a separate JetBrains.gitignore that can
164 | # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
165 | # and can be added to the global gitignore or merged into this file. For a more nuclear
166 | # option (not recommended) you can uncomment the following to ignore the entire idea folder.
167 | #.idea/
168 | bucket_files
169 | Modules/bucket_files
170 | DataBase
171 | .vscode
--------------------------------------------------------------------------------
/src/Draft/access.py:
--------------------------------------------------------------------------------
1 | import struct
2 |
3 | # Define the binary file name
4 | binary_file_name = 'records_with_index_name.bin'
5 |
6 | # Define the index of the element you want to access
7 | i = 9999 # Change this to the desired index
8 |
9 | # Calculate the position of the ith element based on record size
10 | record_size = struct.calcsize('I20s20s') # Size of packed data
11 | print(record_size)
12 | position = i * record_size
13 |
14 | # Get the address of the first block in the binary file
15 | binary_file_address = 0
16 | # with open(binary_file_name, 'rb') as file:
17 | # binary_file_address = file.tell()
18 | # print(binary_file_address)
19 |
20 | # Calculate the absolute position of the ith element
21 | absolute_position = binary_file_address + position
22 |
23 | # Open the binary file and seek to the absolute position of the ith element
24 | with open(binary_file_name, 'rb') as file:
25 | file.seek(absolute_position)
26 |
27 | # Read the packed data at the ith position
28 | packed_data = file.read(record_size)
29 |
30 | # Unpack the data
31 | index, name, phone = struct.unpack('I20s20s', packed_data)
32 | name = name.decode().strip('\0')
33 | phone = phone.decode().strip('\0')
34 |
35 | print(f'Index: {index}, Name: {name}, Phone: {phone}')
36 |
--------------------------------------------------------------------------------
/src/Draft/access_wrong.py:
--------------------------------------------------------------------------------
1 | # Open a file: file
2 | file = open('records_with_index_name_phone.bin',mode='r')
3 |
4 | # read all lines at once
5 | all_of_it = file.read()
6 |
7 | # close the file
8 | file.close()
--------------------------------------------------------------------------------
/src/Draft/file.py:
--------------------------------------------------------------------------------
1 | import csv
2 |
3 | # Generate 10,000 float values based on their index
4 | float_data = [float(i) for i in range(10000)]
5 |
6 | # Define the CSV file name
7 | csv_file_name = 'float_records_with_index.csv'
8 |
9 | # Write the float data with index to the CSV file
10 | with open(csv_file_name, 'w', newline='') as csvfile:
11 | writer = csv.writer(csvfile)
12 | writer.writerow(['Index', 'Value']) # Write a header row
13 | for index, value in enumerate(float_data):
14 | writer.writerow([index, value])
15 |
16 | # Get the address of the first block in the CSV file
17 | csv_file_address = None
18 | with open(csv_file_name, 'rb') as file:
19 | csv_file_address = file.tell()
20 |
21 | print(f'CSV file created: {csv_file_name}')
22 | print(f'Address of the first block in the CSV file: {csv_file_address}')
23 |
--------------------------------------------------------------------------------
/src/Draft/gen.py:
--------------------------------------------------------------------------------
1 | from worst_case_implementation import VecDBWorst
2 | import numpy as np
3 |
4 | # Function to generate random embeddings
5 | def generate_embeddings(num_records, embedding_dim):
6 | return [np.random.rand(embedding_dim).tolist() for _ in range(num_records)]
7 |
8 | # Create an instance of VecDB
9 | db = VecDBWorst()
10 |
11 | # Define parameters
12 | total_records = 10000 # 20 million records
13 | chunk_size = 10000 # Insert records in chunks of 10,000
14 |
15 | # Insert records in chunks
16 | for i in range(0, total_records, chunk_size):
17 | chunk_records = []
18 | for j in range(i + 1, i + chunk_size + 1):
19 | if j > total_records:
20 | break
21 | record = {"id": j, "embed": generate_embeddings(1, 70)[0]}
22 | # make this size of record to be fixed 1500 bytes
23 | # size_of_dummy_needed = 1500 - len(record["embed"])
24 |
25 | chunk_records.append(record)
26 |
27 | db.insert_records(chunk_records)
28 | print(f"Inserted {len(chunk_records)} records. Total records inserted: {j}")
29 |
30 | print("Insertion complete.")
31 |
--------------------------------------------------------------------------------
/src/Draft/generate.py:
--------------------------------------------------------------------------------
1 | import struct
2 |
3 | # Define the binary file name
4 | binary_file_name = 'records_with_index_name.bin'
5 |
6 | # Generate and write the records to the binary file
7 | with open(binary_file_name, 'wb') as file:
8 | for i in range(10000):
9 | # Generate example name and phone number (you can replace with your data source)
10 | name = f"Name-{i}"
11 | phone = f"Phone-{i}"
12 |
13 | # Ensure a fixed length for name and phone
14 | name = name.ljust(20, '\0') # 20 characters
15 | phone = phone.ljust(20, '\0') # 20 characters
16 |
17 | # Pack data into binary format (4 bytes for index, 20 bytes for name, and 20 bytes for phone)
18 | packed_data = struct.pack('I20s20s', i, name.encode(), phone.encode())
19 |
20 | # Write the packed data to the binary file
21 | file.write(packed_data)
--------------------------------------------------------------------------------
/src/Draft/test_data.py:
--------------------------------------------------------------------------------
1 | from datasets import load_dataset
2 |
3 | dataset = load_dataset("aadityaubhat/GPT-wiki-intro")
4 |
5 | print(dataset['train'][0])
--------------------------------------------------------------------------------
/src/IVF.py:
--------------------------------------------------------------------------------
1 | # import files
2 | from utils import *
3 | from sklearn.cluster import KMeans
4 |
5 | # improt libraries
6 | import heapq
7 | import numpy as np
8 |
9 |
10 | def IVF_index(file_path,K_means_metric,K_means_n_clusters,k_means_batch_size,k_means_max_iter,k_means_n_init,chunk_size,index_folder_path):
11 | '''
12 | file_path: path to the data .bin file
13 |
14 | K_means_metric: metric to be used in clustering cosine or euclidean' or TODO use SCANN idea ERORR Think of another way this isn't supported in kmeans
15 | K_means_n_clusters: No of Kmeans Clusters
16 | k_means_batch_size: kmeans batch size to be sampled at each iteration of fitting
17 | k_means_max_iter: max iteration by kmeans default [100] in sklearn
18 | k_means_n_init:The number of times the algorithm will be run with different centroid seeds.
19 |
20 | chunk_size: chunk_size: no of records to be processing together in while performing kmeans
21 |
22 | ivf_folder_path: Folder path to store regions of kmeans
23 | '''
24 | print("---IVF_index()----")
25 | # ############################################################### ################################# ###############################################################
26 | # ############################################################### Step(1):Clustering Data from file ###############################################################
27 | # ############################################################### ################################# ###############################################################
28 | kmeans = KMeans(n_clusters=K_means_n_clusters, max_iter=k_means_max_iter,n_init=k_means_n_init,random_state=42)
29 |
30 |
31 | # Use the first Chunck to only get teh centroids
32 | data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=0,chunk_size=1000000) #[{"id":,"embed":[]}]
33 | # TODO Remove this loop
34 | chunk_vectors=np.array([entry['embed'] for entry in data_chunk])
35 | kmeans.fit(chunk_vectors)
36 |
37 |
38 |
39 | # We need to Read Data from File chunk by chunk
40 | file_size = os.path.getsize(file_path)
41 | record_size=struct.calcsize(f"I{70}f")
42 | n_records=file_size/record_size
43 | no_chunks=math.ceil(n_records/chunk_size)
44 |
45 | # # Step(1) Getting centroids:
46 | # # Loop to get the Kmeans Centroids
47 | # for i in range(no_chunks):
48 | # data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=i*chunk_size,chunk_size=chunk_size) #[{"id":,"embed":[]}]
49 | # # TODO Remove this loop
50 | # chunk_vectors=np.array([entry['embed'] for entry in data_chunk])
51 | # kmeans.partial_fit(chunk_vectors)
52 |
53 | # Centroids
54 | K_means_centroids=kmeans.cluster_centers_
55 | # Saving Centroids #TODO Check precision of centroids after read and write in the file @Basma Elhoseny
56 | write_binary_file(file_path=index_folder_path+'/centroids.bin',data_to_write=K_means_centroids,format=f"{70}f")
57 |
58 | # ##################################################################
59 | # #TEST# Centroids are Written Correct #############################
60 | # ##################################################################
61 |
62 |
63 |
64 | # Step(2) Getting vectors of each regions
65 | for i in range(no_chunks):
66 | data_chunk=read_binary_file_chunk(file_path=file_path,record_format=f"I{70}f",start_index=i*chunk_size,chunk_size=chunk_size,dictionary_format=True) #[{109: np.array([70 dim])}]
67 |
68 | # Get Cluster for each one
69 | labels=kmeans.predict(list(data_chunk.values())) #Each vector corresponding centroid
70 |
71 |
72 | ids=np.array(list(data_chunk.keys()))
73 | vectors=np.array(list(data_chunk.values()))
74 | data_chunk=None #Clear Memory
75 |
76 | # Add vectors to their corresponding region
77 | for label in set(labels):
78 | region_ids=ids[labels==label] # get ids belonging to such region
79 | region_vectors=vectors[labels==label] # get vectors belonging to such region
80 | # Open file of this Region(cluster) Just Once for every Region :D
81 | with open(index_folder_path+f'/cluster{label}.bin', "ab") as fout:
82 | for i in range(len(region_ids)):
83 | #TODO Check whether store id of the vector @Basma Elhoseny
84 | data = struct.pack(f"I{70}f", region_ids[i],*region_vectors[i,:])
85 | fout.write(data)
86 |
87 |
88 |
89 | return
90 |
91 |
92 |
93 | def semantic_query_ivf(data_file_path, index_folder_path, query, top_k, n_regions):
94 | query = np.squeeze(np.array(query))
95 |
96 |
97 | # Read Centroids
98 | K_means_centroids = read_binary_file(index_folder_path + '/centroids.bin', f"70f")
99 |
100 |
101 | assert K_means_centroids.shape[0] > n_regions, "n_regions must be less than the number of regions"
102 |
103 |
104 | # Calculate distances to centroids
105 | distances = np.linalg.norm(K_means_centroids - query, axis=1)
106 | # Get indices of the nearest centroids
107 | nearest_regions = np.argsort(distances)[:n_regions]
108 |
109 |
110 | # Use a heap to keep track of the top k scores
111 | top_scores_heap = []
112 | for region in nearest_regions:
113 | records=read_binary_file_chunk(index_folder_path+f'/cluster{region}.bin', f'I{70}f', 0, chunk_size=100000000000,dictionary_format=True)
114 |
115 |
116 | # Vectorize cosine similarity calculation
117 | vectors = np.array([record for record in records.values()])
118 | dot_products = np.dot(vectors, query)
119 | norms = np.linalg.norm(vectors, axis=1) * np.linalg.norm(query)
120 | similarities = dot_products / norms
121 |
122 | # Process the scores and maintain a heap
123 | for score, id in zip(similarities, records.keys()):
124 | if len(top_scores_heap) < top_k:
125 | heapq.heappush(top_scores_heap, (score, id))
126 | else:
127 | heapq.heappushpop(top_scores_heap, (score, id))
128 |
129 | # Sort and get the top k scores
130 | top_scores_heap.sort(reverse=True)
131 | top_k_ids = [id for _, id in top_scores_heap]
132 |
133 | return top_k_ids
134 |
--------------------------------------------------------------------------------
/src/Modules/IVF.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | from sklearn.cluster import KMeans
4 | import time
5 | from scipy.spatial.distance import cosine
6 |
7 |
8 | class InvertedFileSystem:
9 | def __init__(self, n_clusters, data_dir):
10 | self.n_clusters = n_clusters
11 | self.data_dir = data_dir
12 | self.inverted_file_paths = [
13 | os.path.join(data_dir, f"inverted_file_{i}.npy") for i in range(n_clusters)
14 | ]
15 | self.centroids = None
16 |
17 | def build_index(self, data):
18 | # Cluster the data
19 | kmeans = KMeans(n_clusters=self.n_clusters, n_init=10)
20 | labels = kmeans.fit_predict(data)
21 | self.centroids = kmeans.cluster_centers_
22 |
23 | # Build inverted files
24 | inverted_files = [[] for _ in range(self.n_clusters)]
25 | for idx, label in enumerate(labels):
26 | inverted_files[label].append(idx)
27 |
28 | # Save inverted files to disk
29 | for i, inverted_file in enumerate(inverted_files):
30 | np.save(self.inverted_file_paths[i], inverted_file)
31 |
32 | def query(self, vector, top_k=5):
33 | # Assign vector to nearest cluster
34 | nearest_cluster = np.argmin(np.linalg.norm(self.centroids - vector, axis=1))
35 |
36 | # Load the corresponding inverted file from disk
37 | inverted_file = np.load(self.inverted_file_paths[nearest_cluster])
38 |
39 | # Search in the inverted file
40 | distances = [np.linalg.norm(vector - data[idx]) for idx in inverted_file]
41 | nearest_indices = np.argsort(distances)[:top_k]
42 |
43 | return [inverted_file[i] for i in nearest_indices]
44 |
45 |
46 | def brute_force_cosine_similarity(query_vector, data, top_k=5):
47 | # Calculate cosine similarities for each vector in the dataset
48 | similarities = [1 - cosine(query_vector, vector) for vector in data]
49 |
50 | # Get the indices of the top k most similar vectors
51 | nearest_indices = np.argsort(similarities)[-top_k:]
52 |
53 | # Return the indices and their cosine similarities
54 | return [idx for idx in reversed(nearest_indices)]
55 |
56 | def run_queries(n_queries, ivf, data, top_k=5):
57 | total_time_ivf = 0
58 | total_time_brute_force = 0
59 | total_score_ivf = 0
60 | ivf_results = []
61 | brute_force_results = []
62 |
63 | for _ in range(n_queries):
64 | query_vector = np.random.rand(70)
65 |
66 | start_time = time.time()
67 | ivf_result = ivf.query(query_vector, top_k)
68 | end_time = time.time()
69 | total_time_ivf += end_time - start_time
70 | ivf_results.append(ivf_result)
71 |
72 | start_time = time.time()
73 | brute_force_result = brute_force_cosine_similarity(query_vector, data, top_k)
74 | end_time = time.time()
75 | total_time_brute_force += end_time - start_time
76 | brute_force_results.append(brute_force_result)
77 |
78 | intersection = len(set(ivf_result).intersection(brute_force_result))
79 | total_score_ivf += intersection / top_k
80 |
81 | avg_time_ivf = total_time_ivf / n_queries
82 | avg_score_ivf = total_score_ivf / n_queries
83 | avg_time_brute_force = total_time_brute_force / n_queries
84 |
85 | print(f"IVF: Average time = {avg_time_ivf}, Average score = {avg_score_ivf}")
86 | print(f"Brute Force: Average time = {avg_time_brute_force}")
87 |
88 | # Calculate intersection of top k results
89 | intersection = set(ivf_result).intersection(brute_force_result)
90 | print(f"Intersection of top {top_k} results: {intersection}")
91 |
92 | # !testing IVF
93 | data_dir = "inverted_files"
94 | os.makedirs(data_dir, exist_ok=True)
95 | number_of_queries=10
96 | data_set=10000
97 |
98 | data = np.random.rand(data_set, 70)
99 | ivf = InvertedFileSystem(n_clusters=5, data_dir=data_dir)
100 | ivf.build_index(data)
101 |
102 | print("Dataset in k: ",data_set//1000)
103 | print("Number of Queries: ",number_of_queries)
104 |
105 | run_queries(number_of_queries, ivf, data)
106 |
107 |
108 | # # !testing IVF
109 | # data_dir = "inverted_files"
110 | # os.makedirs(data_dir, exist_ok=True)
111 |
112 | # data = np.random.rand(100000, 70)
113 | # ivf = InvertedFileSystem(n_clusters=3, data_dir=data_dir)
114 | # ivf.build_index(data)
115 |
116 | # query_vector = np.random.rand(70)
117 |
118 |
119 | # # brute force search
120 | # start_time = time.time()
121 | # brute_force_results = brute_force_cosine_similarity(query_vector, data, top_k=10)
122 | # brute_force_time = time.time() - start_time
123 | # print("Brute force top k: ", brute_force_results)
124 | # print("Brute force time: ", brute_force_time)
125 | # print("============================================")
126 | # # Timing IVF query
127 | # start_time = time.time()
128 | # top_k_results = ivf.query(query_vector, top_k=10)
129 | # ivf_time = time.time() - start_time
130 | # print("IVF top k: ", top_k_results)
131 | # print("IVF time: ", ivf_time)
132 |
133 |
134 | # # Get intersection
135 | # brute_force_set = set(brute_force_results)
136 | # ivf_set = set(top_k_results)
137 |
138 | # intersection = brute_force_set.intersection(ivf_set)
139 | # print("Intersection of Brute Force and IVF: ", intersection)
140 | # print("length of the intersection: ", len(intersection))
141 |
142 | # print("********************************************")
143 |
--------------------------------------------------------------------------------
/src/Modules/LSH.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import os
3 |
4 |
5 | from scipy.spatial.distance import cosine
6 | # from best_case_implementation import VecDBBest
7 |
8 |
9 | # TODO:
10 | # * 1) Build LSH function (indexing)
11 | # * 2) Build semantic query function (retrieval)
12 |
13 |
14 | def LSH_index(data, nbits, index_path, d=70):
15 | """
16 | Function to Build the LSH indexing
17 | data:[{'id':int,'embed':vector}]
18 | nbits: no of bits of the Buckets
19 | index_path:path of the Result to be saved
20 | d: vector dimension
21 | """
22 | # create nbits Random hyperplanes used for portioning
23 |
24 | plane_norms = np.random.rand(nbits, d) - 0.5
25 |
26 | #! for -1,1
27 | #? plane_norms = 2 * np.random.rand(nbits, d) - 1.0
28 |
29 | # If index Folder Doesn't Exist just Create it :D
30 | if not os.path.exists(index_path):
31 | os.makedirs(index_path)
32 |
33 |
34 | for item in data:
35 | vector = item["embed"]
36 | id = item["id"]
37 |
38 | # Dot Product with Random Planes
39 | data_dot_product = np.dot(vector, plane_norms.T)
40 |
41 | # Decision Making
42 | data_set_decision_hamming = (data_dot_product > 0) * 1
43 |
44 | # Bucket no. (Key)
45 | hash_str = "".join(data_set_decision_hamming.astype(str)) # 101001101
46 |
47 | # Add This vector to the bucket
48 | file_path = os.path.join(index_path, hash_str + ".txt")
49 |
50 | # Open File in Append Mode
51 | with open(file_path, "a") as file:
52 | file.write(str(id) + "\n")
53 |
54 | return plane_norms
55 |
56 | def get_top_k_hamming_distances(query, buckets, top_k):
57 | distances = []
58 | # Calculate Hamming distance for each bucket
59 | for bucket in buckets:
60 | hamming_distance = sum(bit1 != bit2 for bit1, bit2 in zip(query, bucket))
61 | distances.append((bucket, hamming_distance))
62 | # Sort distances and get the top K
63 | sorted_distances = sorted(distances, key=lambda x: x[1])
64 | top_k_distances = sorted_distances[:top_k]
65 | return top_k_distances
66 | def read_text_files_in_folder(folder_path):
67 | text_files_content = {}
68 |
69 | # Iterate over all files in the folder
70 | for filename in os.listdir(folder_path):
71 | file_path = os.path.join(folder_path, filename)
72 |
73 | # Check if the file is a text file
74 | if filename.endswith('.txt') and os.path.isfile(file_path):
75 | # Read the content of the text file
76 | with open(file_path, 'r', encoding='utf-8') as file:
77 | content = file.read()
78 | # Store content in the dictionary with the filename as the key
79 | text_files_content[filename] = content
80 |
81 | return text_files_content
82 |
83 |
84 |
85 |
86 | def semantic_query_lsh(query, plane_norms, index_path):
87 |
88 |
89 | """
90 | Function to Query the LSH indexing
91 | query:[] query vector
92 | plane_norms: [[]]
93 | index_path:path of the Index to be Search in
94 | """
95 | # Dot Product with Random Planes
96 | query_dot = np.dot(query, plane_norms.T)
97 |
98 | # Decision Making
99 | query_dot = (query_dot > 0) * 1
100 |
101 | query_dot = query_dot.squeeze()
102 | # Ensure query_dot is 1D for string conversion
103 | if query_dot.ndim == 0:
104 | query_dot = np.array([query_dot])
105 | # Bucket no. (Key)
106 | # hash_str = "".join(query_dot.astype(str)) # 101001101
107 | hash_str = "".join(map(str, query_dot.astype(int))) # Converts boolean array to int and then to string
108 |
109 | file_path = os.path.join(index_path, hash_str + ".txt")
110 | result = read_text_files_in_folder(index_path)
111 |
112 | list_buckets = []
113 | for filename, content in result.items():
114 | list_buckets.append(list(map(int, filename[:-4])))
115 | number_of_neighbours = 6
116 | min_hamming_buckets = get_top_k_hamming_distances(query_dot, list_buckets, number_of_neighbours)
117 | index_result =[]
118 | for (bucket, hamming_distance) in min_hamming_buckets:
119 | file_path = os.path.join(index_path, "".join(map(str,bucket)) + ".txt")
120 | try:
121 | list_1 = np.loadtxt(file_path, dtype=int)
122 | list_buckets = np.atleast_1d(list_1).tolist()
123 | index_result+=list_buckets
124 |
125 | except FileNotFoundError:
126 | # Handle the case where the file doesn't exist
127 | print(f"The file {file_path} doesn't exist. Setting index_result to a default value.")
128 | index_result = []
129 | return hash_str, np.array(index_result) # Bucket no
130 | # return index_result
131 |
132 |
133 |
134 |
--------------------------------------------------------------------------------
/src/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ZiadSheriif/IntelliQuery/e49a854ae66c9a22632927e4956bbf1f032cbcc9/src/__init__.py
--------------------------------------------------------------------------------
/src/api.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from worst_case_implementation import VecDBWorst
3 | from best_case_implementation import VecDBBest
4 | from typing import Dict, List, Annotated
5 |
6 | class DataApi:
7 | def __init__(self, file_path, worst = False, database_path="./DataBase",delete_db = True) -> None:
8 | self.file_path = file_path
9 | self.worst = worst
10 | if worst:
11 | self.db = VecDBWorst(self.file_path,delete_db)
12 | else:
13 | self.db = VecDBBest(self.file_path,database_path,delete_db)
14 | self.chunk_size = 10000
15 |
16 | # Function to generate random embeddings
17 | def __generate_embeddings(self,num_records, embedding_dim):
18 | return [np.random.rand(embedding_dim).tolist() for _ in range(num_records)]
19 |
20 |
21 | def generate_data_file(self,num_of_records):
22 | # Insert records in chunks
23 | for i in range(0, num_of_records, self.chunk_size):
24 | chunk_records = []
25 | for j in range(i + 1, i + self.chunk_size + 1):
26 | if j > num_of_records:
27 | break
28 | record = {"id": j, "embed": self.__generate_embeddings(1, 70)[0]}
29 | chunk_records.append(record)
30 |
31 | self.db.insert_records_binary(chunk_records)
32 | print(f"Inserted {len(chunk_records)} records. Total records inserted: {j}")
33 |
34 | print("Insertion complete.")
35 |
36 |
37 | def get_record_by_id(self,record_id):
38 | return self.db.read_record_by_id(record_id)
39 |
40 | def get_first_k_records(self,k):
41 | return self.db.get_top_k_records(k)
42 |
43 | def get_multiple_records_by_ids(self,record_ids):
44 | return self.db.read_multiple_records_by_id(record_ids)
45 |
46 | def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
47 | return self.db.insert_records_binary(rows)
48 |
49 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
50 | return self.db.insert_records(rows)
51 |
52 | def retrive(self, query:Annotated[List[float], 70], top_k = 5):
53 | return self.db.retrive(query,top_k)
54 |
55 |
--------------------------------------------------------------------------------
/src/best_case_implementation.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Annotated
2 | import numpy as np
3 | from utils import empty_folder
4 | from Modules.LSH import *
5 | import struct
6 | import time
7 |
8 | class VecDBBest:
9 | def __init__(self,file_path="./DataBase/data.bin", database_path = "./DataBase", new_db = True) -> None:
10 | '''
11 | Constructor
12 | '''
13 | self.file_path =file_path # Data File Path
14 | self.database_path= database_path # Path of the Folder to Create Indexes
15 |
16 | if new_db:
17 | # If New Data Base
18 | # Empty DataBase Folder
19 | empty_folder(self.database_path)
20 |
21 | # just open new file to delete the old one
22 | with open(self.file_path, "w") as fout:
23 | # if you need to add any head to the file
24 | pass
25 |
26 | def calculate_offset(self, record_id: int) -> int:
27 | # Calculate the offset for a given record ID
28 | record_size = struct.calcsize("I70f")
29 | return (record_id) * record_size
30 |
31 | def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
32 | with open(self.file_path, "ab") as fout: # Open the file in binary mode for appending
33 | for row in rows:
34 | id, embed = row["id"], row["embed"]
35 | # Pack the data into a binary format
36 | data = struct.pack(f"I{70}f", id, *embed)
37 | fout.write(data)
38 | self._build_index()
39 |
40 | def read_multiple_records_by_id(self, records_id: List[int]):
41 | record_size = struct.calcsize("I70f")
42 | records = {}
43 |
44 | with open(self.file_path, "rb") as fin:
45 | for i in range(len(records_id)):
46 | offset = self.calculate_offset(records_id[i])
47 | fin.seek(offset) # Move the file pointer to the calculated offset
48 | data = fin.read(record_size)
49 | if not data:
50 | records[records_id[i]] = None
51 | continue
52 |
53 | # Unpack the binary data into a dictionary
54 | unpacked_data = struct.unpack("I70f", data)
55 | id_value, floats = unpacked_data[0], unpacked_data[1:]
56 |
57 | # Create and return the record dictionary
58 | record = {"id": id_value, "embed": list(floats)}
59 | records[records_id[i]] = record
60 | return records
61 |
62 | def get_top_k_records(self,k):
63 | records = []
64 | record_size = struct.calcsize("I70f")
65 | with open(self.file_path,'rb') as fin:
66 | fin.seek(0)
67 | for i in range(k):
68 | data = fin.read(record_size)
69 | unpacked_data = struct.unpack("I70f", data)
70 | id_value, floats = unpacked_data[0], unpacked_data[1:]
71 |
72 | record = {"id": id_value, "embed": list(floats)}
73 | records.append(record)
74 | return records
75 |
76 | def _build_index(self,Level_1_nbits=5, Level_2_nbits=3, Level_3_nbits=3,Level_4_nbits=3)-> None:
77 |
78 | '''
79 | Build the Index
80 | '''
81 | top_k_records = 2000
82 |
83 | # measure the time
84 | start = time.time()
85 |
86 | # Layer 1 Indexing
87 | # TODO: Here we are reading the whole file: Change later
88 | level_1_in = self.get_top_k_records(top_k_records)
89 | level_1_planes = LSH_index(data=level_1_in, nbits=Level_1_nbits, index_path=self.database_path + "/Level1")
90 | np.save(self.database_path + "/Level1/"+'metadata.npy',level_1_planes)
91 | print("Layer 1 Finished")
92 | return
93 |
94 |
95 |
96 | # Layer 2 Indexing
97 | for file_name in os.listdir(self.database_path + "/Level1"):
98 | file_path = os.path.join(self.database_path + "/Level1", file_name)
99 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
100 | read_data_2 = np.loadtxt(file_path, dtype=int, ndmin=1)
101 | level_2_in = self.read_multiple_records_by_id(read_data_2)
102 | level_2_planes = LSH_index(data=level_2_in.values(), nbits=Level_2_nbits, index_path=self.database_path + "/Level2/" + file_name[:-4])
103 | np.save(self.database_path + "/Level2/" + file_name[:-4]+'/metadata.npy',level_2_planes)
104 | print("Layer 2 Finished")
105 | return
106 |
107 |
108 | # Layer 3 Indexing
109 | for folder_name in os.listdir(self.database_path + "/Level2"):
110 | folder_path = os.path.join(self.database_path + "/Level2", folder_name)
111 | for file_name in os.listdir(folder_path):
112 | file_path = os.path.join(folder_path, file_name)
113 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
114 | read_data_3 = np.loadtxt(file_path, dtype=int, ndmin=1)
115 | level_3_in = self.read_multiple_records_by_id(read_data_3)
116 | level_3_planes = LSH_index(data=level_3_in.values(), nbits=Level_3_nbits, index_path=self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4])
117 | np.save(self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4]+'/metadata.npy',level_3_planes)
118 | print("Layer 3 Finished")
119 |
120 | return
121 | # Layer 4 Indexing
122 | for folder_name in os.listdir(self.database_path + "/Level3"):
123 | folder_path = os.path.join(self.database_path + "/Level3", folder_name)
124 | for folder_name_2 in os.listdir(folder_path):
125 | folder_path_2 = os.path.join(folder_path, folder_name_2)
126 | for file_name in os.listdir(folder_path_2):
127 | file_path = os.path.join(folder_path_2, file_name)
128 | if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
129 | read_data_4 = np.loadtxt(file_path, dtype=int, ndmin=1)
130 | level_4_in = self.read_multiple_records_by_id(read_data_4)
131 | level_4_planes = LSH_index(data=level_4_in.values(), nbits=Level_4_nbits, index_path=self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4])
132 | np.save(self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4]+'/metadata.npy',level_4_planes)
133 | print("Layer 4 Finished")
134 |
135 |
136 | # measure the time
137 | end = time.time()
138 | print("Time taken by Indexing: ",end - start)
139 | def retrive(self, query:Annotated[List[float], 70],top_k = 5)-> [int]:
140 | '''
141 | Get the top_k vectors similar to the Query
142 |
143 | return: list of the top_k similar vectors Ordered by Cosine Similarity
144 | '''
145 |
146 | # Retrieve from Level 1
147 | level_1_planes = np.load(self.database_path + "/Level1"+'/metadata.npy')
148 | bucket_1,result = semantic_query_lsh(query, level_1_planes, self.database_path + "/Level1")
149 | print("length of first bucket",result.shape)
150 |
151 | if len(result) < top_k:
152 | print('level 1 smaller than top_k')
153 |
154 | # # Retrieve from Level 2
155 | # level_2_planes = np.load(self.database_path + "/Level2/"+bucket_1+'/metadata.npy')
156 | # bucket_2,result = semantic_query_lsh(query, level_2_planes, self.database_path + "/Level2/"+bucket_1)
157 | # print("length of second bucket",result.shape)
158 |
159 | # if len(result) < top_k:
160 | # print('level 2 smaller than top_k')
161 |
162 | # # Retrieve from Level 3
163 | # level_3_planes = np.load(self.database_path + "/Level3/"+bucket_1+'/'+bucket_2+'/metadata.npy')
164 | # bucket_3,result = semantic_query_lsh(query, level_3_planes, self.database_path + "/Level3/"+bucket_1+'/'+bucket_2)
165 | # print("length of third bucket",result.shape)
166 |
167 | # if len(result) < top_k:
168 | # print('level 3 smaller than top_k')
169 |
170 | # # Retrieve from Level 4
171 | # level_4_planes = np.load(self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3+'/metadata.npy')
172 | # bucket_4,result = semantic_query_lsh(query, level_4_planes, self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3)
173 | # print("length of fourth bucket",result.shape)
174 |
175 | # if len(result) < top_k:
176 | # print('level 4 smaller than top_k')
177 |
178 |
179 | # Retrieve from Data Base the Embeddings of the Vectors
180 | final_result= self.read_multiple_records_by_id(result)
181 |
182 | # Calculate the Cosine Similarity between the Query and the Vectors
183 | scores = []
184 | for row in final_result.values():
185 | id_value = row['id']
186 | embed_values = row['embed']
187 | score = self._cal_score(query, embed_values)
188 | scores.append((score, id_value))
189 | scores = sorted(scores, reverse=True)[:top_k]
190 | return [s[1] for s in scores]
191 |
192 |
193 |
194 |
195 | def _cal_score(self, vec1, vec2):
196 | dot_product = np.dot(vec1, vec2)
197 | norm_vec1 = np.linalg.norm(vec1)
198 | norm_vec2 = np.linalg.norm(vec2)
199 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
200 | return cosine_similarity
201 |
202 |
--------------------------------------------------------------------------------
/src/evaluation.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from worst_case_implementation import VecDBWorst
3 | from best_case_implementation import VecDBBest
4 | import argparse
5 | from utils import extract_embeds_array
6 | import pandas as pd
7 | from api import DataApi
8 | import os
9 | import time
10 | from dataclasses import dataclass
11 | from typing import List
12 |
13 | AVG_OVERX_ROWS = 1
14 |
15 | @dataclass
16 | class Result:
17 | run_time: float
18 | top_k: int
19 | db_ids: List[int]
20 | actual_ids: List[int]
21 |
22 | # def run_queries(db1,db2, np_rows, top_k, num_runs,delete=False):
23 | def run_queries(db, np_rows, top_k, num_runs, delete=False):
24 | results = []
25 | # results_worst = []
26 | # results_best = []
27 | for i in range(num_runs):
28 | if delete:
29 | query = np.random.random((1,70))
30 | np.save( "./DataBase/q"+str(i)+'.npy',query)
31 | else:
32 | query = np.load( "./DataBase/q"+str(i)+'.npy')
33 |
34 | tic = time.time()
35 | db_ids = db.retrive(query,top_k)
36 | toc = time.time()
37 | run_time= toc - tic
38 |
39 | actual_ids = np.argsort(np_rows.dot(query.T).T / (np.linalg.norm(np_rows, axis=1) * np.linalg.norm(query)), axis=1).squeeze().tolist()[::-1]
40 |
41 | toc = time.time()
42 | np_run_time = toc - tic
43 |
44 | results.append(Result(run_time,top_k,db_ids,actual_ids))
45 | return results
46 |
47 | def eval(results: List[Result]):
48 | # scores are negative. So getting 0 is the best score.
49 | scores = []
50 | run_time = []
51 | for res in results:
52 | run_time.append(res.run_time)
53 | # case for retireving number not equal to top_k, socre will be the lowest
54 | if len(set(res.db_ids)) != res.top_k or len(res.db_ids) != res.top_k:
55 | scores.append( -1 * len(res.actual_ids) * res.top_k)
56 | print('retrieving number not equal to top_k')
57 | continue
58 |
59 | score = 0
60 | for id in res.db_ids:
61 | try:
62 | ind = res.actual_ids.index(id)
63 | if ind > res.top_k * 3:
64 | # print("not in top top_k*3")
65 | score -= ind
66 | except:
67 | # print("not in ids")
68 | score -= len(res.actual_ids)
69 | scores.append(score)
70 |
71 | return sum(scores) / len(scores), sum(run_time) / len(run_time)
72 |
73 | def find_indices(list1, list2):
74 | """
75 | Find the indices of elements of list1 in list2.
76 |
77 | :param list1: The list containing elements whose indices are to be found.
78 | :param list2: The list in which to search for elements from list1.
79 | :return: A list of indices.
80 | """
81 | indices = []
82 | for element in list1:
83 | # Convert both to numpy arrays for consistent handling
84 | np_list2 = np.array(list2)
85 | # Find the index of element in list2
86 | found_indices = np.where(np_list2 == element)[0]
87 | if found_indices.size > 0:
88 | indices.append(found_indices[0])
89 |
90 | return indices
91 |
92 |
93 | def compare_results_print(worst_res,best_res,top_k):
94 | for i in range(len(worst_res)):
95 | actual_ids=worst_res[i].actual_ids
96 | db_ids_best=best_res[i].db_ids
97 | db_ids_worst=worst_res[i].db_ids
98 |
99 | run_time_worst=worst_res[i].run_time
100 | run_time_best=best_res[i].run_time
101 |
102 |
103 | print("=======================================")
104 | print("Best ids: ",db_ids_best)
105 | print("Actual ids: ",actual_ids[:top_k])
106 | print("Worst ids: ",db_ids_worst)
107 | print("Intersect: ",set(actual_ids[:top_k]).intersection(set(db_ids_best)))
108 | print("Intersection in top k indices in the best DB: ",find_indices(actual_ids[:top_k], db_ids_best))
109 |
110 | print("Time taken by Query (Best): ",run_time_best)
111 | print("Time taken by Query (Worst): ",run_time_worst)
112 | print("=======================================")
113 |
114 | if __name__ == "__main__":
115 | print("Hello Semantic LSH")
116 |
117 | number_of_records = 2000
118 | number_of_features = 70
119 | number_of_queries = 5
120 | top_k = 10
121 | print("******************************""")
122 | print("Number of records: ",number_of_records)
123 | print("Number of queries: ",number_of_queries)
124 | print("Top k: ",top_k)
125 | print("******************************""")
126 |
127 |
128 | folder_name = "DataBase"
129 | if not os.path.exists(folder_name):
130 | os.makedirs(folder_name)
131 |
132 | # Mode
133 | parser = argparse.ArgumentParser(description='Description of your script')
134 | parser.add_argument('-d','--delete', help='Description of the -d flag', action='store_true')
135 | args = parser.parse_args()
136 |
137 | # worst_db = VecDBWorst('./DataBase/data.csv',new_db=not args.delete)
138 | worst_api = DataApi('./DataBase/data_worst.csv',True,'./DataBase',args.delete)
139 | # best_db = VecDBBest('./DataBase/data.bin','./DataBase',new_db=not args.delete)
140 | best_api = DataApi('./DataBase/data.bin', False,'./DataBase',args.delete)
141 |
142 | if not args.delete:
143 | print("Reading")
144 | # records_np = pd.read_csv('./DataBase/data.csv',header=None)
145 | # rows_without_first_element = np.array([row[1:].tolist() for _, row in records_np.iterrows()])
146 | # records_np=rows_without_first_element
147 |
148 | records_database = np.array(best_api.get_first_k_records(number_of_records))
149 | records_np = extract_embeds_array(records_database)
150 | records_dict = records_database
151 | _len = len(records_np)
152 | else:
153 | # New
154 |
155 | # records_database = np.array(best_api.get_first_k_records(10000))
156 | print("Generating data files")
157 | records_np = np.random.random((number_of_records, number_of_features))
158 | # records_np = extract_embeds_array(records_database)
159 |
160 | records_dict = [{"id": i, "embed": list(row)} for i, row in enumerate(records_np)]
161 | # records_dict = records_database
162 | _len = len(records_np)
163 |
164 | worst_api.insert_records(records_dict)
165 | best_api.insert_records_binary(records_dict)
166 |
167 |
168 | # Worst
169 | res_worst = run_queries(worst_api, records_np, top_k, number_of_queries,args.delete)
170 | # Best
171 | res_best = run_queries(best_api, records_np, top_k, number_of_queries,False)
172 |
173 | compare_results_print(res_worst,res_best,top_k)
174 | print("Worst:",eval(res_worst))
175 | print("Best:",eval(res_best))
176 |
177 | # res = run_queries(best_api, records_np, 5, 3)
178 | # print("Best:",eval(res))
179 | # results_worst, results_best = run_queries(worst_api,best_api, records_np, top_k, number_of_queries)
180 | # print("Worst:",eval(results_worst))
181 | # print("Best:",eval(results_best))
182 |
183 | # records_np = np.concatenate([records_np, np.random.random((90000, 70))])
184 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
185 | # _len = len(records_np)
186 | # worst_db.insert_records(records_dict)
187 | # res = run_queries(worst_db, records_np, 5, 10)
188 | # print(eval(res))
189 |
190 | # records_np = np.concatenate([records_np, np.random.random((900000, 70))])
191 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
192 | # _len = len(records_np)
193 | # worst_db.insert_records(records_dict)
194 | # res = run_queries(worst_db, records_np, 5, 10)
195 | # eval(res)
196 |
197 | # records_np = np.concatenate([records_np, np.random.random((4000000, 70))])
198 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
199 | # _len = len(records_np)
200 | # db.insert_records(records_dict)
201 | # res = run_queries(db, records_np, 5, 10)
202 | # eval(res)
203 |
204 | # records_np = np.concatenate([records_np, np.random.random((5000000, 70))])
205 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
206 | # _len = len(records_np)
207 | # db.insert_records(records_dict)
208 | # res = run_queries(db, records_np, 5, 10)
209 | # eval(res)
210 |
211 | # records_np = np.concatenate([records_np, np.random.random((5000000, 70))])
212 | # records_dict = [{"id": i + _len, "embed": list(row)} for i, row in enumerate(records_np[_len:])]
213 | # _len = len(records_np)
214 | # db.insert_records(records_dict)
215 | # res = run_queries(db, records_np, 5, 10)
216 | # eval(res)
--------------------------------------------------------------------------------
/src/main.py:
--------------------------------------------------------------------------------
1 | from api import DataApi
2 |
3 |
4 | api_data = DataApi("test.bin")
5 |
6 | # api_data.generate_data_file(5000)
7 |
8 |
9 | records = api_data.get_multiple_records_by_ids([2, 1, 5, 8000])
10 | print(records[8000])
11 |
--------------------------------------------------------------------------------
/src/notes.txt:
--------------------------------------------------------------------------------
1 | 1- 20,000,000
2 | // ==>200
3 |
4 | 2- 100,000
5 | // ==>200
6 |
7 |
8 | 3- 5,000
9 | // ==>500
10 |
11 |
12 | 4- 10
13 |
14 | ============
15 |
16 | new query
17 | first level: 1-10
18 |
19 | second level: 1-500 ==> most load from ram <=200
20 |
21 |
22 | third level: 1-200
23 |
24 | fourth: level: 1-200
25 |
26 |
27 | final retreving: 910 record
28 |
29 | =============================
30 |
31 | n_probe =2
32 |
33 | new query
34 | first level: 2-10
35 | second level: 2-1000
36 | third level: 2-400
37 | fourth: level: 2-400
38 |
39 |
40 | ==================================
41 |
42 | 20,000,000 record
43 |
44 | 256 buckets ===> 256 file
45 |
46 |
47 |
48 |
49 |
50 |
--------------------------------------------------------------------------------
/src/pipeline.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": null,
6 | "metadata": {},
7 | "outputs": [],
8 | "source": [
9 | "# from Modules.LSH import semantic_query_lsh\n",
10 | "# from Modules.LSH import LSH\n",
11 | "\n",
12 | "\n",
13 | "# import numpy as np\n",
14 | "# import os"
15 | ]
16 | },
17 | {
18 | "cell_type": "code",
19 | "execution_count": null,
20 | "metadata": {},
21 | "outputs": [],
22 | "source": [
23 | "# file_path = \"./random_data.txt\"\n",
24 | "# read_data = np.loadtxt(file_path)\n",
25 | "# plane_norms = LSH(read_data, 8)\n",
26 | "# query=[read_data[0]]\n",
27 | "# folder_name = \"bucket_files\"\n",
28 | "# result = semantic_query_lsh(query, plane_norms,folder_name)"
29 | ]
30 | },
31 | {
32 | "cell_type": "code",
33 | "execution_count": 23,
34 | "metadata": {},
35 | "outputs": [
36 | {
37 | "name": "stdout",
38 | "output_type": "stream",
39 | "text": [
40 | "The autoreload extension is already loaded. To reload it, use:\n",
41 | " %reload_ext autoreload\n"
42 | ]
43 | }
44 | ],
45 | "source": [
46 | "%load_ext autoreload\n",
47 | "%autoreload 2\n",
48 | "\n",
49 | "\n",
50 | "from utils import *\n",
51 | "from Modules.LSH import*\n",
52 | "from api import *\n",
53 | "from evaluation import *\n",
54 | "from worst_case_implementation import VecDBWorst\n",
55 | "\n",
56 | "\n",
57 | "# datafile_path=\"../DataBase/random_data_10000.txt\""
58 | ]
59 | },
60 | {
61 | "cell_type": "code",
62 | "execution_count": 29,
63 | "metadata": {},
64 | "outputs": [
65 | {
66 | "name": "stdout",
67 | "output_type": "stream",
68 | "text": [
69 | "Inserted 10000 records. Total records inserted: 10000\n",
70 | "Insertion complete.\n"
71 | ]
72 | }
73 | ],
74 | "source": [
75 | "data_file='./DataBase/data.bin'\n",
76 | "Level_1_path='./DataBase/Level1'\n",
77 | "Level_2_path='./DataBase/Level2'\n",
78 | "Level_3_path='./DataBase/Level3'\n",
79 | "\n",
80 | "Level_1_nbits=8\n",
81 | "Level_2_nbits=3\n",
82 | "Level_3_nbits=3\n",
83 | "\n",
84 | "data_api = DataApi(data_file)\n",
85 | "data_api.generate_data_file(10000)"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": 33,
91 | "metadata": {},
92 | "outputs": [],
93 | "source": [
94 | "\n",
95 | "# Test LSH_index\n",
96 | "# Read Data From File\n",
97 | "read_data = data_api.get_top_k_records(10000)\n",
98 | "\n",
99 | "\n",
100 | "# Layer(1)\n",
101 | "level_1_in=read_data\n",
102 | "# TODO: Save Planes to be used in query Search\n",
103 | "level_1_planes=LSH_index(data=level_1_in, nbits=Level_1_nbits,index_path=Level_1_path)"
104 | ]
105 | },
106 | {
107 | "cell_type": "code",
108 | "execution_count": 34,
109 | "metadata": {},
110 | "outputs": [],
111 | "source": [
112 | "# Layer(2)\n",
113 | "# On Each Bucket Apply LSH\n",
114 | "\n",
115 | "# List all files in the directory\n",
116 | "files = os.listdir(Level_1_path)\n",
117 | "\n",
118 | "# TODO: Save Planes to be used in query Search\n",
119 | "level_2_planes={}\n",
120 | "\n",
121 | "# Loop over the files\n",
122 | "for file_name in files:\n",
123 | " file_path = os.path.join(Level_1_path, file_name)\n",
124 | " \n",
125 | " if os.path.isfile(file_path):\n",
126 | " # Read Data\n",
127 | " read_data_2 = np.loadtxt(file_path,dtype=int,ndmin=1)\n",
128 | "\n",
129 | " level_2_in=data_api.get_multiple_records_by_ids(read_data_2-1)\n",
130 | " # level_2_in = array_to_dictionary(values=vectors,keys=np.hstack(read_data_2))\n",
131 | "\n",
132 | " # # Apply LSH on this Bucket\n",
133 | " # level_2=arr[level_1]\n",
134 | " level_2_planes[file_name[:-4]]=LSH_index(data=level_2_in.values(), nbits=Level_2_nbits,index_path=Level_2_path+'/' + file_name[:-4])\n",
135 | "\n"
136 | ]
137 | },
138 | {
139 | "cell_type": "code",
140 | "execution_count": 9,
141 | "metadata": {},
142 | "outputs": [],
143 | "source": [
144 | "# Layer(3)\n",
145 | "# On Each Bucket Apply LSH\n",
146 | "\n",
147 | "# List all files in the directory\n",
148 | "folders = os.listdir(Level_2_path)\n",
149 | "\n",
150 | "# TODO: Save Planes to be used in query Search\n",
151 | "level_3_planes={}\n",
152 | "# file_3=folder{}\n",
153 | "# Loop over the folders\n",
154 | "for folder_name in folders:\n",
155 | " level_3_planes[folder_name]={}\n",
156 | " folder_path = os.path.join(Level_2_path, folder_name)\n",
157 | " files = os.listdir(folder_path)\n",
158 | " # Loop over the files\n",
159 | " for file_name in files:\n",
160 | " file_path = os.path.join(folder_path, file_name)\n",
161 | " \n",
162 | " if os.path.isfile(file_path):\n",
163 | " # Read Data\n",
164 | " read_data_3 = np.loadtxt(file_path,dtype=int,ndmin=1)\n",
165 | "\n",
166 | " level_3_in=data_api.get_multiple_records_by_ids(read_data_3)\n",
167 | "\n",
168 | " # # Apply LSH on this Bucket\n",
169 | " level_3_planes[folder_name][file_name[:-4]]=LSH_index(data=level_3_in.values(), nbits=Level_3_nbits,index_path=Level_3_path+'/'+folder_name+'/' + file_name[:-4])\n"
170 | ]
171 | },
172 | {
173 | "cell_type": "code",
174 | "execution_count": 22,
175 | "metadata": {},
176 | "outputs": [
177 | {
178 | "name": "stdout",
179 | "output_type": "stream",
180 | "text": [
181 | "bucket of level 1: 11110011\n",
182 | "=====================================\n",
183 | "bucket of level 2: 001\n",
184 | "=====================================\n",
185 | "bucket of level 3: 100\n",
186 | "Length of level 3 189\n",
187 | "Indices of level 3 [ 36 51 104 159 266 357 372 385 434 465 510 671 702 707\n",
188 | " 720 822 824 834 863 938 1034 1044 1165 1248 1264 1438 1505 1565\n",
189 | " 1613 1683 1712 1719 1771 1798 1812 1843 1953 2191 2238 2266 2330 2353\n",
190 | " 2594 2602 2624 2669 2730 2744 2825 2880 2894 2915 2942 2944 3080 3168\n",
191 | " 3286 3351 3490 3645 3648 3735 3798 3851 3859 3911 3986 4026 4030 4065\n",
192 | " 4121 4134 4187 4211 4232 4260 4391 4399 4476 4477 4489 4492 4545 4554\n",
193 | " 4591 4605 4660 4792 4905 4937 4953 4954 4970 4986 4987 5228 5249 5329\n",
194 | " 5398 5454 5471 5495 5584 5708 5712 5725 5744 5799 5899 5900 5908 5952\n",
195 | " 5987 6049 6072 6096 6144 6184 6209 6287 6344 6399 6479 6495 6536 6544\n",
196 | " 6662 6693 6848 6880 6915 6962 7080 7085 7187 7199 7213 7240 7390 7404\n",
197 | " 7417 7442 7531 7538 7554 7584 7625 7664 7708 7721 7765 7768 7808 7827\n",
198 | " 7955 8101 8170 8279 8284 8380 8444 8446 8454 8481 8552 8560 8565 8586\n",
199 | " 8676 8700 8761 8792 8912 8935 9007 9150 9336 9352 9354 9367 9586 9662\n",
200 | " 9745 9762 9794 9801 9859 9948 9972]\n",
201 | "=====================================\n",
202 | "target_vector [0.5522450804710388, 0.8917692303657532, 0.7913368344306946, 0.6000004410743713, 0.2616525888442993, 0.9615220427513123, 0.4808562695980072, 0.6019359827041626, 0.07978673279285431, 0.30365362763404846, 0.7390730381011963, 0.2133997678756714, 0.36366748809814453, 0.1835469752550125, 0.20069865882396698, 0.13891369104385376, 0.11978743225336075, 0.3913387358188629, 0.002954070921987295, 0.5194749236106873, 0.37845972180366516, 0.9680533409118652, 0.6960610747337341, 0.8805666565895081, 0.06497178226709366, 0.5662519335746765, 0.04004804417490959, 0.2919067144393921, 0.737677812576294, 0.10855083167552948, 0.3745698928833008, 0.37776005268096924, 0.9178327322006226, 0.7241680026054382, 0.12325477600097656, 0.3273957073688507, 0.9901415109634399, 0.4085298478603363, 0.6129018068313599, 0.1801413595676422, 0.9952824711799622, 0.3938077688217163, 0.913888692855835, 0.11249328404664993, 0.14214684069156647, 0.6679161787033081, 0.9495717287063599, 0.4362204968929291, 0.3122316896915436, 0.6952698230743408, 0.8448274731636047, 0.965186595916748, 0.35632771253585815, 0.9069381952285767, 0.42551901936531067, 0.9420151710510254, 0.022108066827058792, 0.6098361611366272, 0.897776186466217, 0.4446363151073456, 0.7102886438369751, 0.5624412894248962, 0.5420237183570862, 0.3291500210762024, 0.2226945161819458, 0.6429535150527954, 0.5322402119636536, 0.09856311231851578, 0.5489377379417419, 0.5590397715568542]\n"
203 | ]
204 | },
205 | {
206 | "ename": "TypeError",
207 | "evalue": "unhashable type: 'slice'",
208 | "output_type": "error",
209 | "traceback": [
210 | "\u001b[1;31m---------------------------------------------------------------------------\u001b[0m",
211 | "\u001b[1;31mTypeError\u001b[0m Traceback (most recent call last)",
212 | "\u001b[1;32md:\\Semantic-Search-Engine\\pipeline.ipynb Cell 8\u001b[0m line \u001b[0;36m2\n\u001b[0;32m 27\u001b[0m index_result_3\u001b[39m=\u001b[39mdata_api\u001b[39m.\u001b[39mget_multiple_records_by_ids(index_result_3)\n\u001b[0;32m 28\u001b[0m level3_res_vectors\u001b[39m=\u001b[39m[entry[\u001b[39m'\u001b[39m\u001b[39membed\u001b[39m\u001b[39m'\u001b[39m] \u001b[39mfor\u001b[39;00m entry \u001b[39min\u001b[39;00m index_result_3\u001b[39m.\u001b[39mvalues()]\n\u001b[1;32m---> 29\u001b[0m top_result,_\u001b[39m=\u001b[39mget_top_k_similar(query,index_result_3,\u001b[39m10\u001b[39;49m)\n\u001b[0;32m 30\u001b[0m \u001b[39m# print(\"Top k results: \",top_result[0])\u001b[39;00m\n\u001b[0;32m 31\u001b[0m \u001b[39m# print(\"=====================================\")\u001b[39;00m\n\u001b[0;32m 32\u001b[0m \u001b[39m# # get the intersection of the two lists level 2 and level3\u001b[39;00m\n\u001b[0;32m 33\u001b[0m \u001b[39m# count = sum(element in index_result_2 for element in index_result_3)\u001b[39;00m\n\u001b[0;32m 34\u001b[0m \u001b[39m# print(\"Intersection of the two layers: \",count)\u001b[39;00m\n",
213 | "File \u001b[1;32md:\\Semantic-Search-Engine\\Modules\\LSH.py:145\u001b[0m, in \u001b[0;36mget_top_k_similar\u001b[1;34m(target_vector, data, k)\u001b[0m\n\u001b[0;32m 143\u001b[0m \u001b[39m# Calculate cosine similarities using vectorized operations\u001b[39;00m\n\u001b[0;32m 144\u001b[0m \u001b[39mprint\u001b[39m(\u001b[39m\"\u001b[39m\u001b[39mtarget_vector\u001b[39m\u001b[39m\"\u001b[39m,target_vector)\n\u001b[1;32m--> 145\u001b[0m \u001b[39mprint\u001b[39m(\u001b[39m\"\u001b[39m\u001b[39mdata\u001b[39m\u001b[39m\"\u001b[39m,data[\u001b[39m1\u001b[39;49m:\u001b[39m5\u001b[39;49m])\n\u001b[0;32m 146\u001b[0m similarities \u001b[39m=\u001b[39m \u001b[39m1\u001b[39m \u001b[39m-\u001b[39m np\u001b[39m.\u001b[39marray([cosine(target_vector, vector) \u001b[39mfor\u001b[39;00m vector \u001b[39min\u001b[39;00m data])\n\u001b[0;32m 148\u001b[0m \u001b[39m# Find the indices of the top k most similar vectors\u001b[39;00m\n",
214 | "\u001b[1;31mTypeError\u001b[0m: unhashable type: 'slice'"
215 | ]
216 | }
217 | ],
218 | "source": [
219 | "# Query\n",
220 | "query=data_api.get_record_by_id(5)[5]['embed']\n",
221 | "# Layer (1)\n",
222 | "bucket_1,index_result_1 = semantic_query_lsh(query=query,plane_norms=level_1_planes,index_path=Level_1_path)\n",
223 | "print(\"bucket of level 1: \",bucket_1)\n",
224 | "# print(\"Length of level 1\",len(index_result_1))\n",
225 | "# print(\"Indices of level 1\",index_result_1)\n",
226 | "print(\"=====================================\")\n",
227 | "\n",
228 | "# Layer(2)\n",
229 | "bucket_2,index_result_2 = semantic_query_lsh(query=query,plane_norms=level_2_planes[bucket_1],index_path=Level_2_path+\"/\"+bucket_1)\n",
230 | "print(\"bucket of level 2: \",bucket_2)\n",
231 | "# print(\"Length of level 2\",len(index_result_2))\n",
232 | "# print(\"Indices of level 2\",index_result_2)\n",
233 | "print(\"=====================================\")\n",
234 | "\n",
235 | "# Layer(3)\n",
236 | "bucket_3,index_result_3 = semantic_query_lsh(query=query,plane_norms=level_3_planes[bucket_1][bucket_2],index_path=Level_3_path+\"/\"+bucket_1+'/'+bucket_2)\n",
237 | "print(\"bucket of level 3: \",bucket_3)\n",
238 | "print(\"Length of level 3\",len(index_result_3))\n",
239 | "print(\"Indices of level 3\",index_result_3)\n",
240 | "\n",
241 | "\n",
242 | "print(\"=====================================\")\n",
243 | "\n",
244 | "# get top 10 results from the last layer\n",
245 | "index_result_3=data_api.get_multiple_records_by_ids(index_result_3)\n",
246 | "level3_res_vectors=[entry['embed'] for entry in index_result_3.values()]\n",
247 | "top_result,_=get_top_k_similar(query,index_result_3,10)\n",
248 | "# print(\"Top k results: \",top_result[0])\n",
249 | "# print(\"=====================================\")\n",
250 | "# # get the intersection of the two lists level 2 and level3\n",
251 | "# count = sum(element in index_result_2 for element in index_result_3)\n",
252 | "# print(\"Intersection of the two layers: \",count)\n",
253 | "\n"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "np_rows = np.array([record['embed'] for record in read_data if 'embed' in record])\n",
263 | "# temp=[5, 966, 536, 1088, 5073, 5549]\n",
264 | "index_result_3_minus_one = [id - 1 for id in top_results[0]]\n",
265 | "res=run_queries(index_result_3_minus_one, np_rows, top_k=len(top_results), num_runs=1,query=np.array([query]))\n",
266 | "print(eval(res))"
267 | ]
268 | },
269 | {
270 | "cell_type": "code",
271 | "execution_count": null,
272 | "metadata": {},
273 | "outputs": [],
274 | "source": [
275 | "db = VecDBWorst()\n",
276 | "# records_np = np.random.random((10000, 70))\n",
277 | "records_np = np.array([record['embed'] for record in read_data if 'embed' in record])\n",
278 | "\n",
279 | "# records_dict = [{\"id\": i, \"embed\": list(row)} for i, row in enumerate(records_np)]\n",
280 | "records_dict=read_data\n",
281 | "\n",
282 | "# _len = len(records_np)\n",
283 | "db.insert_records(records_dict)\n",
284 | "db_ids=db.retrive(query, top_k=1)\n",
285 | "db_ids_minus_one = [id - 1 for id in db_ids]\n",
286 | "res = run_queries(db_ids_minus_one, records_np, 1, 1, np.array([query]))\n",
287 | "print(eval(res))\n"
288 | ]
289 | }
290 | ],
291 | "metadata": {
292 | "kernelspec": {
293 | "display_name": "Python 3",
294 | "language": "python",
295 | "name": "python3"
296 | },
297 | "language_info": {
298 | "codemirror_mode": {
299 | "name": "ipython",
300 | "version": 3
301 | },
302 | "file_extension": ".py",
303 | "mimetype": "text/x-python",
304 | "name": "python",
305 | "nbconvert_exporter": "python",
306 | "pygments_lexer": "ipython3",
307 | "version": "3.10.9"
308 | }
309 | },
310 | "nbformat": 4,
311 | "nbformat_minor": 2
312 | }
313 |
--------------------------------------------------------------------------------
/src/utils.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import shutil
3 | import os
4 | import math
5 | from typing import Dict, List, Annotated
6 | import struct
7 | import sys
8 |
9 |
10 | def save_20M_record(data):
11 | '''
12 | Given 20M record save them as required by the TA
13 | data: (20M,70)
14 | '''
15 |
16 | folder_name='./Data_TA'
17 | if not os.path.exists(folder_name):
18 | os.makedirs(folder_name)
19 |
20 | empty_folder(folder_name)
21 |
22 | files=['data_100K.bin',"data_1M.bin","data_5M.bin","data_10M.bin","data_15M.bin","data_20M.bin"]
23 | # files=["data_20M.bin"]
24 | limits=[10**5,10**6,5*10**6,10**7,15*10**6,2*10**7]
25 | # limits=[20*10**6]
26 | for i,file in enumerate(files):
27 | data_part=data[:limits[i]]
28 |
29 | # Append in Binary Mode
30 | with open(folder_name+'/'+file, "ab") as fout:
31 | for id,vector in enumerate(data_part):
32 | # Pack the data into a binary format
33 | unpacked_data = struct.pack(f"I{70}f", id, *vector)
34 | fout.write(unpacked_data)
35 |
36 | # # Test
37 | # print("len(data)",len(data))
38 | # # print(data[0])
39 | # print(data[-1])
40 | # read_data=read_binary_file_chunk('./Data_TA/data_100K.bin',f"I{70}f",start_index=0,chunk_size=1000000,dictionary_format=True)
41 | # print("len(read_data)",len(read_data))
42 | # # print(read_data[0])
43 | # print(read_data[10**5-1])
44 |
45 |
46 | # # Test
47 | # print("len(data)",len(data))
48 | # # print(data[0])
49 | # print(data[-1])
50 | # read_data=read_binary_file_chunk('./Data_TA/data_1M.bin',f"I{70}f",start_index=0,chunk_size=1000000,dictionary_format=True)
51 | # print("len(read_data)",len(read_data))
52 | # # print(read_data[0])
53 | # print(read_data[10**6-1])
54 |
55 | def read_binary_file(file_path,format):
56 | '''
57 | Read binary file from its format
58 | '''
59 | try:
60 | with open(file_path,"rb") as fin:
61 | file_size = os.path.getsize(file_path)
62 | record_size=struct.calcsize(format)
63 | n_records=file_size/record_size
64 | # print("n_records",n_records)
65 |
66 | fin.seek(0) #Move pointer to the beginning of the file
67 | data = fin.read(record_size * int(n_records))
68 | if not data:
69 | print("Empty File ",file_path,"🔴🔴")
70 | return None
71 | # Unpack the binary data
72 | data=np.frombuffer(data, dtype=np.dtype(format))
73 | return data
74 | except FileNotFoundError:
75 | print(f"The file '{file_path}' Not Found.")
76 |
77 | def write_binary_file(file_path,data_to_write,format):
78 | '''
79 | data_to_write: array of values with format as passed
80 | format: format of each element
81 | '''
82 | try:
83 | with open(file_path, "ab") as fout:
84 | # Pack the entire array into binary data
85 | binary_data = struct.pack(len(data_to_write)*format, *data_to_write.flatten())
86 | fout.write(binary_data)
87 | except FileNotFoundError:
88 | print(f"The file '{file_path}' could not be created.")
89 |
90 | def read_binary_file_chunk(file_path, record_format, start_index, chunk_size=10,dictionary_format=False):
91 | """
92 | This Function Reads Chunk from a binary File
93 | If remaining from file are < chunk size they are returned normally
94 |
95 | file_path:Path of the file to be read from
96 | record_format: format of the record ex:f"4I" 4 integers
97 | start_index: index of the record from which we start reading [0_indexed]
98 | chunk_size: no of records to be retrieved
99 |
100 | @return : None in case out of index of file
101 | the records
102 | """
103 |
104 | # Calculate record size
105 | record_size = struct.calcsize(record_format)
106 |
107 | # Open the binary file for reading
108 | with open(file_path, "rb") as fin:
109 | fin.seek(
110 | start_index * record_size
111 | ) # Move the file pointer to the calculated offset
112 |
113 | # Read a chunk of records
114 | # .read() moves the file pointer (cursor) forward by the number of bytes read.
115 | chunk_data = fin.read(record_size * (chunk_size))
116 | if len(chunk_data) == 0:
117 | print("Out Of File Index 🔥🔥")
118 | return None
119 |
120 | # file_size = os.path.getsize(file_path)
121 | # print("Current file position:", fin.tell())
122 | # print("File size:", file_size,"record_format",record_format,"record_size",record_size,"chunk_data len",len(chunk_data))
123 |
124 | if dictionary_format:
125 | records={}
126 | for i in range(0, len(chunk_data), record_size):
127 | #TODO Remove this loop @Basma Elhoseny
128 | unpacked_record = struct.unpack(record_format, chunk_data[i : i + record_size])
129 | id, vector = unpacked_record[0], unpacked_record[1:]
130 | records[id]=np.array(vector)
131 | return records
132 |
133 | # Unpack Data
134 | records = []
135 | for i in range(0, len(chunk_data), record_size):
136 | unpacked_record = struct.unpack(
137 | record_format, chunk_data[i : i + record_size]
138 | )
139 | id, vector = unpacked_record[0], unpacked_record[1:]
140 | record = {"id": id, "embed": list(vector)}
141 | records.append(record)
142 | return records
143 | def empty_folder(folder_path):
144 | """
145 | Function to Empty a folder given its path
146 | @param folder_path : path of the folder to be deleted
147 | """
148 | if not os.path.exists(folder_path):
149 | os.makedirs(folder_path)
150 | print("Created new ", folder_path, "successfully")
151 | return
152 |
153 | for filename in os.listdir(folder_path):
154 | file_path = os.path.join(folder_path, filename)
155 | try:
156 | if os.path.isfile(file_path):
157 | os.unlink(file_path)
158 | elif os.path.isdir(file_path):
159 | shutil.rmtree(file_path)
160 | except Exception as e:
161 | print(f"Error while deleting {file_path}: {e}")
162 | print("Deleted", folder_path, "successfully")
163 |
164 |
165 | def extract_embeds(dict):
166 | # {505: {'id': 505, 'embed': [0.8,....]}} --> [[0.8,....],[.......]]
167 | return [entry["embed"] for entry in dict.values()]
168 |
169 |
170 | def extract_embeds_array(arr):
171 | return np.array([entry["embed"] for entry in arr])
172 |
173 |
174 | def cal_score(vec1, vec2):
175 | dot_product = np.dot(vec1, vec2)
176 | norm_vec1 = np.linalg.norm(vec1)
177 | norm_vec2 = np.linalg.norm(vec2)
178 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
179 | return cosine_similarity
180 |
181 |
182 | def calculate_offset(record_id: int) -> int:
183 | # Calculate the offset for a given record ID
184 | record_size = struct.calcsize("I70f")
185 | return (record_id) * record_size
186 |
187 |
188 | def read_multiple_records_by_id(file_path, records_id: List[int],dictionary_format=False):
189 | record_size = struct.calcsize("I70f")
190 | records = {}
191 |
192 | records_dictionary={}
193 |
194 | with open(file_path, "rb") as fin:
195 | for i in range(len(records_id)):
196 | offset = calculate_offset(records_id[i])
197 | fin.seek(offset) # Move the file pointer to the calculated offset
198 | data = fin.read(record_size)
199 | if not data:
200 | records[records_id[i]] = None
201 | continue
202 |
203 | # Unpack the binary data into a dictionary
204 | unpacked_data = struct.unpack("I70f", data)
205 | id_value, floats = unpacked_data[0], unpacked_data[1:]
206 |
207 | if dictionary_format:
208 | records_dictionary[id_value]=list(floats)
209 | else:
210 | # Create and return the record dictionary
211 | record = {"id": id_value, "embed": list(floats)}
212 | records[records_id[i]] = record
213 |
214 | if dictionary_format: return records_dictionary
215 | return records
216 |
217 | # def generate_random(k=100):
218 | # # Sample data: k vectors with 70 features each
219 | # data = np.random.uniform(-1, 1, size=(k, 70))
220 |
221 | # # Write data to a text file
222 | # file_path = "../DataBase/random_data_"+str(k)+".txt"
223 | # np.savetxt(file_path, data)
224 |
225 | # # Read Data from File
226 | # # read_data = np.loadtxt(file_path)
227 |
228 |
229 | # def array_to_dictionary(values,keys=None):
230 | # '''
231 | # values: [array of values]
232 | # Keys: [array of Keys] optional if not passed the keys are indexed 0-N
233 | # '''
234 | # if(keys is None):
235 | # keys=range(0,len(values))
236 |
237 | # if(len(values)!=len(keys)):
238 | # print ("array_to_dictionary(): InCorrect Size of keys and values")
239 | # return None
240 |
241 | # dictionary_data = dict(zip(keys, values))
242 | # return dictionary_data
243 |
244 |
245 | # def get_vector_from_id(data_path,id):
246 | # '''
247 | # function to get the vector by its id [BADDDDDDDD Use Seek]
248 |
249 | # '''
250 | # read_data = np.loadtxt(data_path)
251 | # return read_data[id]
252 |
253 |
254 | # def check_dir(path):
255 | # if os.path.exists(path):
256 | # shutil.rmtree(path, ignore_errors=True, onerror=lambda func, path, exc: None)
257 | # os.makedirs(path)
258 |
259 |
260 | # Test generate_random()
261 | # generate_random(10000)
262 |
--------------------------------------------------------------------------------
/src/vec_db.py:
--------------------------------------------------------------------------------
1 |
2 | from typing import Dict, List, Annotated
3 | from utils import *
4 | import numpy as np
5 | import time
6 | import os
7 | # from Modules.LSH import LSH_index, semantic_query_lsh
8 | from IVF import IVF_index,semantic_query_ivf
9 |
10 | NUMBER_OF_RECORDS_BRUTE_FORCE = 20*10**3
11 |
12 | class VecDB:
13 | def __init__(self,file_path="./DataBase", new_db = True) -> None:
14 | '''
15 | Constructor
16 | '''
17 | self.file_path =file_path+'/data.bin' # Data File Path
18 | self.database_path= file_path # Path of the Folder to Create Indexes
19 | self.n_regions = None # Initialize n_regions
20 |
21 |
22 | if new_db:
23 | if not os.path.exists(self.database_path):
24 | os.makedirs(self.database_path)
25 |
26 | else:
27 | # If New DataBase Empty DataBase Folder
28 | empty_folder(self.database_path)
29 |
30 | # just open new file to delete the old one
31 | with open(self.file_path, "w") as fout:
32 | # if you need to add any head to the file
33 | pass
34 |
35 |
36 | self.level1=None
37 |
38 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
39 | # Append in Binary Mode
40 | with open(self.file_path, "ab") as fout:
41 | for row in rows:
42 | id, embed = row["id"], row["embed"]
43 | # Pack the data into a binary format
44 | data = struct.pack(f"I{70}f", id, *embed)
45 | fout.write(data)
46 | self._build_index()
47 |
48 |
49 | def _build_index(self,Level_1_nbits=5, Level_2_nbits=3, Level_3_nbits=3,Level_4_nbits=3)-> None:
50 |
51 | '''
52 | Build the Index
53 | '''
54 | file_size = os.path.getsize(self.file_path)
55 | record_size=struct.calcsize(f"I{70}f")
56 | n_records=file_size/record_size
57 | if(n_records==10*10**3):
58 | self.number_of_clusters=10
59 | elif(n_records==100*10**3):
60 | self.number_of_clusters=50
61 | elif(n_records==10**6):
62 | self.number_of_clusters=200
63 | elif(n_records==5*10**6):
64 | self.number_of_clusters=500
65 | elif(n_records==10*10**6):
66 | self.number_of_clusters=8000
67 |
68 | # if()
69 | # if(n_records==10**3):
70 | # self.number_of_clusters=50
71 | # if(n_records<=100000):
72 | # self.number_of_clusters=10
73 | # self.n_regions=5
74 | # else:
75 | # self.number_of_clusters=int(n_records/NUMBER_OF_RECORDS_BRUTE_FORCE)
76 | # self.number_of_clusters=NUMBER_OF_RECORDS_BRUTE_FORCE
77 | # self.n_regions=10
78 | print("Record Size: ",record_size)
79 | print("File Size: ",file_size)
80 | print("Building Index ..........")
81 | print("Number of records: ",n_records)
82 | print("Number of Clusters: ",self.number_of_clusters)
83 | # measure the time
84 | start = time.time()
85 |
86 | # Make Level1 Folder
87 | Level1_folder_path = self.database_path+'/Level1'
88 | if not os.path.exists(Level1_folder_path):
89 | os.makedirs(Level1_folder_path)
90 |
91 | # IVF Layer 1 Indexing
92 | chunk_size=100000
93 | print("chunk_size",chunk_size)
94 | IVF_index(file_path=self.file_path,K_means_metric='euclidean',K_means_n_clusters=self.number_of_clusters,k_means_batch_size=chunk_size,k_means_max_iter=100,k_means_n_init='auto',chunk_size=chunk_size,index_folder_path=Level1_folder_path)
95 |
96 |
97 | # # Layer 1 Indexing
98 | # # level_1_in = self.get_top_k_records(top_k_records)
99 | # level_1_planes = LSH_index(file_path=self.file_path, nbits=Level_1_nbits, chunk_size=1000,index_path=self.database_path + "/Level1")
100 | # np.save(self.database_path + "/Level1/"+'metadata.npy',level_1_planes)
101 | # print("Layer 1 Finished")
102 | # return
103 |
104 |
105 |
106 | # # Layer 2 Indexing
107 | # for file_name in os.listdir(self.database_path + "/Level1"):
108 | # file_path = os.path.join(self.database_path + "/Level1", file_name)
109 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
110 | # read_data_2 = np.loadtxt(file_path, dtype=int, ndmin=1)
111 | # level_2_in = self.read_multiple_records_by_id(read_data_2)
112 | # level_2_planes = LSH_index(data=level_2_in.values(), nbits=Level_2_nbits, index_path=self.database_path + "/Level2/" + file_name[:-4])
113 | # np.save(self.database_path + "/Level2/" + file_name[:-4]+'/metadata.npy',level_2_planes)
114 | # print("Layer 2 Finished")
115 | # return
116 |
117 |
118 | # # Layer 3 Indexing
119 | # for folder_name in os.listdir(self.database_path + "/Level2"):
120 | # folder_path = os.path.join(self.database_path + "/Level2", folder_name)
121 | # for file_name in os.listdir(folder_path):
122 | # file_path = os.path.join(folder_path, file_name)
123 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
124 | # read_data_3 = np.loadtxt(file_path, dtype=int, ndmin=1)
125 | # level_3_in = self.read_multiple_records_by_id(read_data_3)
126 | # level_3_planes = LSH_index(data=level_3_in.values(), nbits=Level_3_nbits, index_path=self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4])
127 | # np.save(self.database_path + "/Level3/" + folder_name + '/' + file_name[:-4]+'/metadata.npy',level_3_planes)
128 | # print("Layer 3 Finished")
129 | # return
130 |
131 | # # Layer 4 Indexing
132 | # for folder_name in os.listdir(self.database_path + "/Level3"):
133 | # folder_path = os.path.join(self.database_path + "/Level3", folder_name)
134 | # for folder_name_2 in os.listdir(folder_path):
135 | # folder_path_2 = os.path.join(folder_path, folder_name_2)
136 | # for file_name in os.listdir(folder_path_2):
137 | # file_path = os.path.join(folder_path_2, file_name)
138 | # if os.path.isfile(file_path) and file_name.lower().endswith(".txt"):
139 | # read_data_4 = np.loadtxt(file_path, dtype=int, ndmin=1)
140 | # level_4_in = self.read_multiple_records_by_id(read_data_4)
141 | # level_4_planes = LSH_index(data=level_4_in.values(), nbits=Level_4_nbits, index_path=self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4])
142 | # np.save(self.database_path + "/Level4/" + folder_name + '/' + folder_name_2 + '/' + file_name[:-4]+'/metadata.npy',level_4_planes)
143 | # print("Layer 4 Finished")
144 |
145 |
146 | # measure the time
147 | end = time.time()
148 | print("Indexing Done ...... Time taken by Indexing: ",end - start)
149 | return
150 |
151 | def retrive(self, query:Annotated[List[float], 70],top_k = 5)-> [int]:
152 | '''
153 | Get the top_k vectors similar to the Query
154 |
155 | return: list of the top_k similar vectors Ordered by Cosine Similarity
156 | '''
157 | print(f"Retrieving top {top_k} ..........")
158 | Level1_folder_path = self.database_path+'/Level1'
159 |
160 | file_size = os.path.getsize(self.file_path)
161 | record_size=struct.calcsize(f"I{70}f")
162 | n_records=file_size/record_size
163 | if(n_records==10*10**3):
164 | n_probes=3
165 | elif(n_records==100*10**3):
166 | n_probes=10
167 | elif(n_records==10**6):
168 | n_probes=5
169 | elif(n_records==5*10**6):
170 | n_probes=15
171 | elif(n_records==10*10**6):
172 | n_probes=30
173 | elif(n_records==15*10**6):
174 | n_probes=256
175 | elif(n_records==20*10**6):
176 | n_probes=64
177 | # n_probes=0
178 | # if(n_records<=5*10**6):
179 | # n_probes=3
180 | # else:
181 | # n_probes=20
182 | final_result=semantic_query_ivf(data_file_path=self.file_path,index_folder_path=Level1_folder_path,query=query,top_k=top_k,n_regions=n_probes)
183 |
184 | return final_result
185 |
186 |
187 |
188 | # # Retrieve from Level 1
189 | # level_1_planes = np.load(self.database_path + "/Level1"+'/metadata.npy')
190 | # bucket_1,result = semantic_query_lsh(query, level_1_planes, self.database_path + "/Level1")
191 | # print("length of first bucket",result.shape)
192 |
193 | # if len(result) < top_k:
194 | # print('level 1 smaller than top_k')
195 |
196 | # # Retrieve from Level 2
197 | # level_2_planes = np.load(self.database_path + "/Level2/"+bucket_1+'/metadata.npy')
198 | # bucket_2,result = semantic_query_lsh(query, level_2_planes, self.database_path + "/Level2/"+bucket_1)
199 | # print("length of second bucket",result.shape)
200 |
201 | # if len(result) < top_k:
202 | # print('level 2 smaller than top_k')
203 |
204 | # # Retrieve from Level 3
205 | # level_3_planes = np.load(self.database_path + "/Level3/"+bucket_1+'/'+bucket_2+'/metadata.npy')
206 | # bucket_3,result = semantic_query_lsh(query, level_3_planes, self.database_path + "/Level3/"+bucket_1+'/'+bucket_2)
207 | # print("length of third bucket",result.shape)
208 |
209 | # if len(result) < top_k:
210 | # print('level 3 smaller than top_k')
211 |
212 | # # Retrieve from Level 4
213 | # level_4_planes = np.load(self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3+'/metadata.npy')
214 | # bucket_4,result = semantic_query_lsh(query, level_4_planes, self.database_path + "/Level4/"+bucket_1+'/'+bucket_2+'/'+bucket_3)
215 | # print("length of fourth bucket",result.shape)
216 |
217 | # if len(result) < top_k:
218 | # print('level 4 smaller than top_k')
219 |
220 |
221 | # # Retrieve from Data Base the Embeddings of the Vectors
222 | # final_result= read_multiple_records_by_id(self.file_path,result)
223 |
224 | # Calculate the Cosine Similarity between the Query and the Vectors
225 | # scores = []
226 | # for row in final_result.values():
227 | # id_value = row['id']
228 | # embed_values = row['embed']
229 | # score = self._cal_score(query, embed_values)
230 | # scores.append((score, id_value))
231 | # scores = sorted(scores, reverse=True)[:top_k]
232 | # return [s[1] for s in scores]
233 |
234 |
235 |
236 |
237 | def _cal_score(self, vec1, vec2):
238 | dot_product = np.dot(vec1, vec2)
239 | norm_vec1 = np.linalg.norm(vec1)
240 | norm_vec2 = np.linalg.norm(vec2)
241 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
242 | return cosine_similarity
--------------------------------------------------------------------------------
/src/worst_case_implementation.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Annotated
2 | import struct
3 | import numpy as np
4 |
5 | class VecDBWorst:
6 | def __init__(self, file_path = "saved_db.csv", new_db = True) -> None:
7 | self.file_path = file_path
8 | if new_db:
9 | # just open new file to delete the old one
10 | with open(self.file_path, "w") as fout:
11 | # if you need to add any head to the file
12 | pass
13 |
14 | def insert_records(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
15 | with open(self.file_path, "a+") as fout:
16 | for row in rows:
17 | id, embed = row["id"], row["embed"]
18 | row_str = f"{id}," + ",".join([str(e) for e in embed])
19 | fout.write(f"{row_str}\n")
20 | self._build_index()
21 |
22 | # def insert_records_binary(self, rows: List[Dict[int, Annotated[List[float], 70]]]):
23 | # with open(self.file_path, "ab") as fout: # Open the file in binary mode for appending
24 | # for row in rows:
25 | # id, embed = row["id"], row["embed"]
26 | # # Pack the data into a binary format
27 | # data = struct.pack(f"I{70}f", id, *embed)
28 | # fout.write(data)
29 | # self._build_index()
30 |
31 | # def calculate_offset(self, record_id: int) -> int:
32 | # # Calculate the offset for a given record ID
33 | # record_size = struct.calcsize("I70f")
34 | # return (record_id - 1) * record_size
35 |
36 | # def read_record_by_id(self, record_id: int) -> Dict[int, Annotated[List[float], 70]]:
37 | # record_size = struct.calcsize("I70f")
38 | # offset = self.calculate_offset(record_id)
39 |
40 | # with open(self.file_path, "rb") as fin:
41 | # fin.seek(offset) # Move the file pointer to the calculated offset
42 | # data = fin.read(record_size)
43 | # if not data:
44 | # return {} # Record not found
45 |
46 | # # Unpack the binary data into a dictionary
47 | # unpacked_data = struct.unpack("I70f", data)
48 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
49 |
50 | # # Create and return the record dictionary
51 | # record = {"id": id_value, "embed": list(floats)}
52 | # return {record_id: record}
53 |
54 | # def read_multiple_records_by_id(self, records_id: List[int]):
55 | # record_size = struct.calcsize("I70f")
56 | # records = {}
57 |
58 | # with open(self.file_path, "rb") as fin:
59 | # for i in range(len(records_id)):
60 | # offset = self.calculate_offset(records_id[i])
61 | # fin.seek(offset) # Move the file pointer to the calculated offset
62 | # data = fin.read(record_size)
63 | # if not data:
64 | # records[records_id[i]] = None
65 | # continue
66 |
67 | # # Unpack the binary data into a dictionary
68 | # unpacked_data = struct.unpack("I70f", data)
69 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
70 |
71 | # # Create and return the record dictionary
72 | # record = {"id": id_value, "embed": list(floats)}
73 | # records[records_id[i]] = record
74 | # return records
75 |
76 | # def get_top_k_records(self,k):
77 | # records = []
78 | # record_size = struct.calcsize("I70f")
79 | # with open(self.file_path,'rb') as fin:
80 | # fin.seek(0)
81 | # for i in range(k):
82 | # data = fin.read(record_size)
83 | # unpacked_data = struct.unpack("I70f", data)
84 | # id_value, floats = unpacked_data[0], unpacked_data[1:]
85 |
86 | # record = {"id": id_value, "embed": list(floats)}
87 | # records.append(record)
88 | # return records
89 |
90 | def retrive(self, query: Annotated[List[float], 70], top_k = 5):
91 | scores = []
92 | with open(self.file_path, "r") as fin:
93 | for row in fin.readlines():
94 | row_splits = row.split(",")
95 | id = int(row_splits[0])
96 | embed = [float(e) for e in row_splits[1:]]
97 | score = self._cal_score(query, embed)
98 | scores.append((score, id))
99 | # here we assume that if two rows have the same score, return the lowest ID
100 | scores = sorted(scores, reverse=True)[:top_k]
101 | return [s[1] for s in scores]
102 |
103 | def _cal_score(self, vec1, vec2):
104 | dot_product = np.dot(vec1, vec2)
105 | norm_vec1 = np.linalg.norm(vec1)
106 | norm_vec2 = np.linalg.norm(vec2)
107 | cosine_similarity = dot_product / (norm_vec1 * norm_vec2)
108 | return cosine_similarity
109 |
110 | def _build_index(self):
111 | pass
112 |
113 |
114 |
--------------------------------------------------------------------------------
/vector searching algorithms/LSHHyperPlane.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 19,
6 | "metadata": {},
7 | "outputs": [],
8 | "source": [
9 | "nbits = 3 # number of hyperplanes and binary vals to produce\n",
10 | "d = 70 # vector dimensions\n",
11 | "\n",
12 | "#! log2(8)"
13 | ]
14 | },
15 | {
16 | "cell_type": "code",
17 | "execution_count": 20,
18 | "metadata": {},
19 | "outputs": [
20 | {
21 | "data": {
22 | "text/plain": [
23 | "array([[-0.16225197, -0.1159841 , -0.0379947 , 0.47482455, -0.24329282,\n",
24 | " -0.17219515, 0.0889353 , -0.34083189, -0.0857109 , 0.19710836,\n",
25 | " -0.48189731, 0.04372259, -0.40512638, -0.49822556, -0.18287792,\n",
26 | " 0.31290614, -0.3228106 , -0.24989376, -0.30750153, 0.35378187,\n",
27 | " 0.32047542, -0.3405673 , 0.01704872, -0.29642438, -0.13589619,\n",
28 | " -0.28071414, -0.05584624, 0.09991783, -0.3077081 , 0.13647533,\n",
29 | " -0.4770819 , 0.38661425, 0.00783346, -0.3927221 , 0.24110202,\n",
30 | " 0.05990217, -0.06802645, 0.03282446, 0.30179191, 0.19156282,\n",
31 | " 0.1054492 , -0.00932327, 0.32185434, 0.49241347, 0.20733614,\n",
32 | " 0.18026476, -0.26101194, -0.3778801 , 0.4936665 , -0.41047846,\n",
33 | " 0.23792759, -0.23321223, 0.39830173, -0.13084787, -0.31116184,\n",
34 | " -0.00189201, 0.08791531, -0.14980787, 0.30558995, -0.00670045,\n",
35 | " 0.2243021 , -0.41023912, 0.18223908, -0.38580149, -0.11450846,\n",
36 | " -0.43534932, -0.30870211, 0.29227875, -0.15542413, 0.20264467],\n",
37 | " [ 0.14822377, -0.3276353 , 0.26702648, 0.03217607, -0.20768427,\n",
38 | " -0.46076215, -0.1540441 , -0.14252796, -0.21732578, -0.35428344,\n",
39 | " 0.2084557 , -0.18725843, -0.14300948, -0.16831679, -0.15468043,\n",
40 | " -0.07450581, 0.00233269, -0.43596823, 0.19815002, 0.19192439,\n",
41 | " 0.13743071, 0.1589349 , -0.37645398, -0.40863437, 0.18087113,\n",
42 | " 0.26399475, -0.20873789, 0.31885534, -0.47078825, 0.19251382,\n",
43 | " 0.39492556, 0.13612851, 0.05252917, 0.1066819 , -0.20602171,\n",
44 | " 0.019199 , -0.42388983, -0.29196827, -0.08782944, 0.38928403,\n",
45 | " -0.14570291, -0.04510013, -0.11215063, 0.08435185, 0.0814708 ,\n",
46 | " 0.30669655, 0.2207886 , 0.48463154, -0.07672077, 0.02202735,\n",
47 | " -0.43230734, -0.16302195, 0.42660854, 0.21663128, -0.37419712,\n",
48 | " -0.07194837, 0.38714646, -0.34736523, 0.17594598, -0.04035088,\n",
49 | " -0.40523731, -0.46061098, -0.18572369, 0.46951449, 0.04887038,\n",
50 | " -0.47345552, 0.21424037, 0.06851608, -0.44381182, 0.18315833],\n",
51 | " [-0.34032214, 0.20510388, -0.11437108, -0.25271871, -0.29298418,\n",
52 | " -0.39316298, -0.27235239, -0.02090773, 0.42378779, 0.36244202,\n",
53 | " -0.49353892, -0.37632702, 0.08773845, 0.23887437, -0.03724808,\n",
54 | " -0.23496996, 0.46748875, -0.2527981 , -0.39073735, 0.40062121,\n",
55 | " 0.10471371, -0.43568824, 0.14986386, 0.18503889, 0.37681242,\n",
56 | " -0.01243404, -0.39396771, 0.19966568, 0.08733691, 0.2509886 ,\n",
57 | " -0.30488297, 0.39487436, -0.38444297, -0.47168848, 0.40205414,\n",
58 | " -0.40537546, 0.03695501, -0.28056081, 0.42477745, 0.49012324,\n",
59 | " 0.29699303, -0.14461517, 0.37445295, -0.30211604, -0.39721614,\n",
60 | " 0.34692406, 0.11398823, 0.30746006, -0.22106426, -0.1443832 ,\n",
61 | " -0.07727599, 0.23807248, 0.32999453, 0.36904291, -0.01930504,\n",
62 | " -0.01021899, -0.25900161, 0.32297803, -0.08289675, 0.28510423,\n",
63 | " -0.35789496, -0.22445429, -0.43838493, -0.25795092, 0.18836288,\n",
64 | " 0.07583603, 0.24472323, 0.07588773, -0.46374612, 0.29371209]])"
65 | ]
66 | },
67 | "execution_count": 20,
68 | "metadata": {},
69 | "output_type": "execute_result"
70 | }
71 | ],
72 | "source": [
73 | "import numpy as np\n",
74 | "# create a set of 4 hyperplanes, with 2 dimensions\n",
75 | "plane_norms = np.random.rand(nbits, d) - .5\n",
76 | "plane_norms"
77 | ]
78 | },
79 | {
80 | "cell_type": "code",
81 | "execution_count": 21,
82 | "metadata": {},
83 | "outputs": [
84 | {
85 | "name": "stdout",
86 | "output_type": "stream",
87 | "text": [
88 | "[[0.24346879 0.20847865 0.09572784 0.55319155 0.07226476 0.87621505\n",
89 | " 0.58913737 0.46246208 0.3333929 0.96904595 0.89600564 0.59480015\n",
90 | " 0.77746555 0.54283576 0.67569602 0.23497124 0.07950847 0.31373656\n",
91 | " 0.14113477 0.408343 0.81078156 0.57261091 0.35708278 0.41438797\n",
92 | " 0.44477819 0.65966904 0.12290202 0.22656549 0.07834719 0.83130173\n",
93 | " 0.4011682 0.72348172 0.58621532 0.97192664 0.12448728 0.19814639\n",
94 | " 0.84652828 0.19786014 0.70758209 0.91906712 0.93421752 0.70310207\n",
95 | " 0.27726954 0.96761661 0.06944151 0.32237123 0.95214921 0.55453475\n",
96 | " 0.22046089 0.75767774 0.76303426 0.11226826 0.96778584 0.18364252\n",
97 | " 0.86959947 0.35425893 0.05710316 0.67524358 0.84838244 0.84003201\n",
98 | " 0.71003473 0.74628258 0.48265117 0.48526062 0.41682251 0.24458015\n",
99 | " 0.56040784 0.47847743 0.11747069 0.2841326 ]]\n",
100 | "[[0.08021692 0.11425865 0.66780691 0.64591999 0.72730137 0.37124926\n",
101 | " 0.90060496 0.29506503 0.14570761 0.05080957 0.84666837 0.89391903\n",
102 | " 0.24643966 0.44699371 0.64934757 0.67063548 0.75535129 0.75834603\n",
103 | " 0.36194954 0.35376051 0.86358269 0.67616135 0.0427738 0.70108469\n",
104 | " 0.27322976 0.30507254 0.39729602 0.43346906 0.20055928 0.0809176\n",
105 | " 0.05108797 0.16594553 0.60731689 0.25100448 0.60585635 0.18616766\n",
106 | " 0.35069704 0.88805346 0.07440514 0.91451748 0.14379966 0.30309109\n",
107 | " 0.6960067 0.5342824 0.90332074 0.95252648 0.74792365 0.09408956\n",
108 | " 0.44708244 0.75941983 0.49066048 0.6441386 0.63012995 0.98668287\n",
109 | " 0.60327176 0.62091147 0.72095104 0.59418854 0.84847164 0.18768005\n",
110 | " 0.50059785 0.83818945 0.58380742 0.79375409 0.38927237 0.30760971\n",
111 | " 0.90913216 0.75463438 0.32010021 0.71195468]]\n",
112 | "[array([[0.24346879, 0.20847865, 0.09572784, 0.55319155, 0.07226476,\n",
113 | " 0.87621505, 0.58913737, 0.46246208, 0.3333929 , 0.96904595,\n",
114 | " 0.89600564, 0.59480015, 0.77746555, 0.54283576, 0.67569602,\n",
115 | " 0.23497124, 0.07950847, 0.31373656, 0.14113477, 0.408343 ,\n",
116 | " 0.81078156, 0.57261091, 0.35708278, 0.41438797, 0.44477819,\n",
117 | " 0.65966904, 0.12290202, 0.22656549, 0.07834719, 0.83130173,\n",
118 | " 0.4011682 , 0.72348172, 0.58621532, 0.97192664, 0.12448728,\n",
119 | " 0.19814639, 0.84652828, 0.19786014, 0.70758209, 0.91906712,\n",
120 | " 0.93421752, 0.70310207, 0.27726954, 0.96761661, 0.06944151,\n",
121 | " 0.32237123, 0.95214921, 0.55453475, 0.22046089, 0.75767774,\n",
122 | " 0.76303426, 0.11226826, 0.96778584, 0.18364252, 0.86959947,\n",
123 | " 0.35425893, 0.05710316, 0.67524358, 0.84838244, 0.84003201,\n",
124 | " 0.71003473, 0.74628258, 0.48265117, 0.48526062, 0.41682251,\n",
125 | " 0.24458015, 0.56040784, 0.47847743, 0.11747069, 0.2841326 ]]), array([[0.08021692, 0.11425865, 0.66780691, 0.64591999, 0.72730137,\n",
126 | " 0.37124926, 0.90060496, 0.29506503, 0.14570761, 0.05080957,\n",
127 | " 0.84666837, 0.89391903, 0.24643966, 0.44699371, 0.64934757,\n",
128 | " 0.67063548, 0.75535129, 0.75834603, 0.36194954, 0.35376051,\n",
129 | " 0.86358269, 0.67616135, 0.0427738 , 0.70108469, 0.27322976,\n",
130 | " 0.30507254, 0.39729602, 0.43346906, 0.20055928, 0.0809176 ,\n",
131 | " 0.05108797, 0.16594553, 0.60731689, 0.25100448, 0.60585635,\n",
132 | " 0.18616766, 0.35069704, 0.88805346, 0.07440514, 0.91451748,\n",
133 | " 0.14379966, 0.30309109, 0.6960067 , 0.5342824 , 0.90332074,\n",
134 | " 0.95252648, 0.74792365, 0.09408956, 0.44708244, 0.75941983,\n",
135 | " 0.49066048, 0.6441386 , 0.63012995, 0.98668287, 0.60327176,\n",
136 | " 0.62091147, 0.72095104, 0.59418854, 0.84847164, 0.18768005,\n",
137 | " 0.50059785, 0.83818945, 0.58380742, 0.79375409, 0.38927237,\n",
138 | " 0.30760971, 0.90913216, 0.75463438, 0.32010021, 0.71195468]]), array([[0.77078444, 0.08654989, 0.53784898, 0.27799896, 0.8002419 ,\n",
139 | " 0.3718589 , 0.79333253, 0.93137511, 0.62069102, 0.24775348,\n",
140 | " 0.4272542 , 0.98292296, 0.8470418 , 0.52637634, 0.80102169,\n",
141 | " 0.9440776 , 0.68466439, 0.86571266, 0.27882657, 0.71765609,\n",
142 | " 0.75582515, 0.94039075, 0.20125464, 0.64221553, 0.83386962,\n",
143 | " 0.73863021, 0.93711781, 0.4794255 , 0.48793874, 0.86123148,\n",
144 | " 0.92146684, 0.81077912, 0.50759451, 0.18685755, 0.98628992,\n",
145 | " 0.97781323, 0.38057898, 0.87204991, 0.30697755, 0.49756831,\n",
146 | " 0.21495361, 0.38075046, 0.30418495, 0.02362606, 0.96563469,\n",
147 | " 0.80356302, 0.60805212, 0.30632618, 0.59294981, 0.08821319,\n",
148 | " 0.19220448, 0.17337163, 0.93316608, 0.46086827, 0.17464549,\n",
149 | " 0.16804619, 0.05238805, 0.95753146, 0.87498728, 0.5543621 ,\n",
150 | " 0.727997 , 0.54389248, 0.38181964, 0.47099822, 0.59929861,\n",
151 | " 0.35783896, 0.27050514, 0.98113893, 0.49137662, 0.05494532]])]\n"
152 | ]
153 | }
154 | ],
155 | "source": [
156 | "a=np.random.random((1, 70))\n",
157 | "b=np.random.random((1, 70))\n",
158 | "c=np.random.random((1, 70))\n",
159 | "# Repeat the array to generate a 1x70 array\n",
160 | "print(a)\n",
161 | "print(b)\n",
162 | "dataset=[a,b,c]\n",
163 | "print(dataset)\n"
164 | ]
165 | },
166 | {
167 | "cell_type": "code",
168 | "execution_count": 22,
169 | "metadata": {},
170 | "outputs": [
171 | {
172 | "data": {
173 | "text/plain": [
174 | "array([[-0.98219519, -1.00134517, 0.23751343]])"
175 | ]
176 | },
177 | "execution_count": 22,
178 | "metadata": {},
179 | "output_type": "execute_result"
180 | }
181 | ],
182 | "source": [
183 | "# calculate the dot product for each of these\n",
184 | "a_dot = np.dot(a, plane_norms.T)\n",
185 | "b_dot = np.dot(b, plane_norms.T)\n",
186 | "c_dot = np.dot(c, plane_norms.T)\n",
187 | "a_dot"
188 | ]
189 | },
190 | {
191 | "cell_type": "code",
192 | "execution_count": 23,
193 | "metadata": {},
194 | "outputs": [
195 | {
196 | "data": {
197 | "text/plain": [
198 | "array([[False, False, True]])"
199 | ]
200 | },
201 | "execution_count": 23,
202 | "metadata": {},
203 | "output_type": "execute_result"
204 | }
205 | ],
206 | "source": [
207 | " #! Dataset\n",
208 | "# we know that a positive dot product == +ve side of hyperplane\n",
209 | "# and negative dot product == -ve side of hyperplane\n",
210 | "a_dot = a_dot > 0\n",
211 | "b_dot = b_dot > 0\n",
212 | "c_dot = c_dot > 0\n",
213 | "a_dot"
214 | ]
215 | },
216 | {
217 | "cell_type": "code",
218 | "execution_count": 24,
219 | "metadata": {},
220 | "outputs": [
221 | {
222 | "data": {
223 | "text/plain": [
224 | "array([0, 0, 1])"
225 | ]
226 | },
227 | "execution_count": 24,
228 | "metadata": {},
229 | "output_type": "execute_result"
230 | }
231 | ],
232 | "source": [
233 | "# convert our boolean arrays to int arrays to make bucketing\n",
234 | "# easier (although is okay to use boolean for Hamming distance)\n",
235 | "a_dot = a_dot.astype(int)[0]\n",
236 | "b_dot = b_dot.astype(int)[0]\n",
237 | "c_dot = c_dot.astype(int)[0]\n",
238 | "a_dot"
239 | ]
240 | },
241 | {
242 | "cell_type": "code",
243 | "execution_count": 25,
244 | "metadata": {},
245 | "outputs": [
246 | {
247 | "name": "stdout",
248 | "output_type": "stream",
249 | "text": [
250 | "001\n",
251 | "000\n",
252 | "000\n",
253 | "{'001': [0], '000': [1, 2]}\n"
254 | ]
255 | }
256 | ],
257 | "source": [
258 | "vectors = [a_dot, b_dot, c_dot]\n",
259 | "buckets = {}\n",
260 | "i = 0\n",
261 | "\n",
262 | "for i in range(len(vectors)):\n",
263 | " # convert from array to string\n",
264 | " hash_str = ''.join(vectors[i].astype(str))\n",
265 | " print(hash_str)\n",
266 | " # create bucket if it doesn't exist\n",
267 | " if hash_str not in buckets.keys():\n",
268 | " buckets[hash_str] = []\n",
269 | " # add vector position to bucket\n",
270 | " buckets[hash_str].append(i)\n",
271 | "\n",
272 | "print(buckets)"
273 | ]
274 | },
275 | {
276 | "cell_type": "code",
277 | "execution_count": 26,
278 | "metadata": {},
279 | "outputs": [
280 | {
281 | "name": "stdout",
282 | "output_type": "stream",
283 | "text": [
284 | "[[0.98217599 0.0740474 0.60518903 0.61501807 0.37022865 0.59778156\n",
285 | " 0.37753566 0.44307262 0.00959564 0.43188369 0.77591898 0.40505507\n",
286 | " 0.40793131 0.30983161 0.20962565 0.47749091 0.64407774 0.07951662\n",
287 | " 0.21950209 0.5617165 0.27452938 0.73727983 0.10188798 0.15470781\n",
288 | " 0.9668675 0.63906846 0.76027852 0.59622264 0.90857345 0.49078738\n",
289 | " 0.17966669 0.29728534 0.76605938 0.89432476 0.17330878 0.91669595\n",
290 | " 0.327371 0.89819097 0.98084002 0.44954373 0.04567631 0.18179054\n",
291 | " 0.97601994 0.11805953 0.42048999 0.19162843 0.69235512 0.48898102\n",
292 | " 0.89062367 0.27993955 0.90024359 0.15118375 0.81479348 0.42161774\n",
293 | " 0.78538813 0.84025532 0.42421928 0.63838074 0.74416435 0.76683117\n",
294 | " 0.63133131 0.36330796 0.24528589 0.42388808 0.53415259 0.58918953\n",
295 | " 0.50997522 0.88050857 0.5938881 0.76094727]]\n",
296 | "Query belongs to bucket: [1, 2]\n"
297 | ]
298 | }
299 | ],
300 | "source": [
301 | " #! for tesing input query\n",
302 | "\n",
303 | "query=np.random.random((1, 70))\n",
304 | "print(query)\n",
305 | "\n",
306 | "query_dot = np.dot(query, plane_norms.T)\n",
307 | "query_dot = query_dot > 0\n",
308 | "query_dot = query_dot.astype(int)[0]\n",
309 | "\n",
310 | "# Convert the query array to a string\n",
311 | "query_hash_str = ''.join(query_dot.astype(str))\n",
312 | "\n",
313 | "# Check which bucket the query belongs to\n",
314 | "if query_hash_str in buckets.keys():\n",
315 | " bucket_containing_query = buckets[query_hash_str]\n",
316 | " print(\"Query belongs to bucket:\", bucket_containing_query)\n",
317 | "else:\n",
318 | " print(\"Query doesn't match any existing buckets\")\n",
319 | " print(0)\n",
320 | "\n"
321 | ]
322 | },
323 | {
324 | "cell_type": "code",
325 | "execution_count": 27,
326 | "metadata": {},
327 | "outputs": [
328 | {
329 | "name": "stdout",
330 | "output_type": "stream",
331 | "text": [
332 | "[[0.24346879 0.20847865 0.09572784 0.55319155 0.07226476 0.87621505\n",
333 | " 0.58913737 0.46246208 0.3333929 0.96904595 0.89600564 0.59480015\n",
334 | " 0.77746555 0.54283576 0.67569602 0.23497124 0.07950847 0.31373656\n",
335 | " 0.14113477 0.408343 0.81078156 0.57261091 0.35708278 0.41438797\n",
336 | " 0.44477819 0.65966904 0.12290202 0.22656549 0.07834719 0.83130173\n",
337 | " 0.4011682 0.72348172 0.58621532 0.97192664 0.12448728 0.19814639\n",
338 | " 0.84652828 0.19786014 0.70758209 0.91906712 0.93421752 0.70310207\n",
339 | " 0.27726954 0.96761661 0.06944151 0.32237123 0.95214921 0.55453475\n",
340 | " 0.22046089 0.75767774 0.76303426 0.11226826 0.96778584 0.18364252\n",
341 | " 0.86959947 0.35425893 0.05710316 0.67524358 0.84838244 0.84003201\n",
342 | " 0.71003473 0.74628258 0.48265117 0.48526062 0.41682251 0.24458015\n",
343 | " 0.56040784 0.47847743 0.11747069 0.2841326 ]]\n",
344 | "[[0.14634457 0.20457996 0.24741999 0.92396483 0.02756294 0.55294575\n",
345 | " 0.53786271 0.25648871 0.87692497 0.0504492 0.78309208 0.78032385\n",
346 | " 0.60510748 0.50301495 0.33663068 0.48482831 0.31022081 0.22420917\n",
347 | " 0.03656832 0.0576642 0.02176892 0.41431776 0.97839866 0.36627294\n",
348 | " 0.90726783 0.942912 0.10056186 0.27157551 0.35269652 0.65476284\n",
349 | " 0.3802201 0.10174655 0.59332797 0.41950798 0.51371538 0.66586081\n",
350 | " 0.86951989 0.30015139 0.39627816 0.90903098 0.71667395 0.27805884\n",
351 | " 0.43209455 0.17558953 0.55876165 0.44371013 0.54468845 0.95573234\n",
352 | " 0.8846391 0.69004526 0.78832344 0.97466743 0.01227129 0.26130364\n",
353 | " 0.46660631 0.64762417 0.39275623 0.7934695 0.29527433 0.66989054\n",
354 | " 0.73093381 0.26445753 0.17682595 0.24664441 0.41129241 0.83235075\n",
355 | " 0.72809304 0.64075518 0.98233083 0.85216486]]\n",
356 | "[[18.0160595]]\n",
357 | "0\n",
358 | "Query belongs to bucket: [0]\n"
359 | ]
360 | }
361 | ],
362 | "source": [
363 | "query=np.random.random((1, 70))\n",
364 | "# print(query)\n",
365 | "\n",
366 | "# Assuming 'query' is your query vector\n",
367 | "query_dot = np.dot(query, plane_norms.T)\n",
368 | "query_dot = query_dot > 0\n",
369 | "query_dot = query_dot.astype(int)[0]\n",
370 | "\n",
371 | "# Convert the query array to a string\n",
372 | "query_hash_str = ''.join(query_dot.astype(str))\n",
373 | "\n",
374 | "# Check which bucket the query belongs to\n",
375 | "if query_hash_str in buckets.keys():\n",
376 | " bucket_containing_query = buckets[query_hash_str]\n",
377 | " min_dist=100\n",
378 | " index=-1\n",
379 | " for vec in bucket_containing_query:\n",
380 | " print(dataset[vec])\n",
381 | " print(query)\n",
382 | " dot_res=np.dot(query,dataset[vec].T)\n",
383 | " print(dot_res)\n",
384 | " res = dot_res / (np.linalg.norm(query) * np.linalg.norm(dataset[vec].T))\n",
385 | " if res list2[j]:
28 | j += 1
29 | else:
30 | intersection.append(list1[i])
31 | i += 1
32 | j += 1
33 |
34 | return intersection
35 |
36 |
37 | def split_on_sign(data:[[float]],split_on)->int:
38 | '''
39 | @param data: Data to categorize
40 | @split_on: The max count to split on
41 |
42 | @return dictionary of the data splitted by sign +ve and -ve
43 | '''
44 | if(split_on is None or split_on>np.shape(data)[1]):
45 | #split on the whole size
46 | # split_on=np.shape(data)[1]
47 | split_on=10
48 |
49 | regions = []
50 | for col in data[:,:split_on].T: # Transpose the matrix to iterate over columns
51 | positive_region = (col >= 0)
52 | negative_region = (col < 0)
53 | regions.append(np.where(positive_region)[0])
54 | regions.append(np.where(negative_region)[0])
55 | return regions
56 |
57 | def search_on_sign(q:[float],regions:[[int]]):
58 | # O(m * n), where m is the average length of the input lists, and n is the number of input lists.
59 | # Check on sign of the feature
60 | intersect=None
61 | split_on=np.shape(regions)[0]//2
62 | for ind,feature in enumerate(q[:split_on]):
63 | if(ind==0):
64 | intersect=regions[0] if feature>=0 else regions[1]
65 | continue
66 | if(feature>=0):
67 | # positive
68 | intersect=sorted_list_intersection(intersect, regions[2*ind])
69 | else:
70 | #negative
71 | intersect=sorted_list_intersection(intersect, regions[2*ind+1])
72 | return intersect
--------------------------------------------------------------------------------
 Get Started
24 | ### Infernce Mode
25 | ***Check Final Notebook***
26 | ```
27 | https://github.com/ZiadSheriif/IntelliQuery/blob/main/Evaluate_ADB_Project.ipynb
28 | ```
29 | ### Run Locally
30 |
31 | ***Clone Repo***
32 | ```
33 | git clone https://github.com/ZiadSheriif/IntelliQuery.git
34 | ```
35 | ***Install dependencies***
36 | ```
37 | pip install -r requirements.txt
38 | ```
39 | ***Run Indexer***
40 | ```
41 | $ python ./src/evaluation.py
42 | ```
43 |
44 |
45 | ##
Get Started
24 | ### Infernce Mode
25 | ***Check Final Notebook***
26 | ```
27 | https://github.com/ZiadSheriif/IntelliQuery/blob/main/Evaluate_ADB_Project.ipynb
28 | ```
29 | ### Run Locally
30 |
31 | ***Clone Repo***
32 | ```
33 | git clone https://github.com/ZiadSheriif/IntelliQuery.git
34 | ```
35 | ***Install dependencies***
36 | ```
37 | pip install -r requirements.txt
38 | ```
39 | ***Run Indexer***
40 | ```
41 | $ python ./src/evaluation.py
42 | ```
43 |
44 |
45 | ##  Methods
46 | ### Inverted File Inedex (IVF)
47 | This is out final Approach with Some Enhancements
48 | 1. Changed MiniBatchKMeans to regular KMeans
49 | 2. We calculate initial centroids with just the first chunk of data
50 | 3. Introduced parallel processing for different regions
51 |
Methods
46 | ### Inverted File Inedex (IVF)
47 | This is out final Approach with Some Enhancements
48 | 1. Changed MiniBatchKMeans to regular KMeans
49 | 2. We calculate initial centroids with just the first chunk of data
50 | 3. Introduced parallel processing for different regions
51 |  52 |
53 | ### Local Sensitive Hashing (LSH)
54 |
52 |
53 | ### Local Sensitive Hashing (LSH)
54 |  55 |
56 | ### Product Qunatization (PQ)
57 |
55 |
56 | ### Product Qunatization (PQ)
57 |  58 |
59 | ### PQ-LSH
60 | It Combines both LSH & PQ
61 |
62 | ## 🕴 Contributors
63 |
64 |
65 |
58 |
59 | ### PQ-LSH
60 | It Combines both LSH & PQ
61 |
62 | ## 🕴 Contributors
63 |
64 |
65 |