.jpg
364 | fps (float): Output frame rate.
365 |
366 | Returns:
367 | Output video is stored in the path: folder_p/video.mp4
368 | '''
369 | vid_p = osp.join(folder_p, 'video.mp4')
370 | cmd = ['ffmpeg', '-r', str(int(fps)), '-i',
371 | osp.join(folder_p, 'frames', '%d.jpg'), '-y', vid_p]
372 | FNULL = open(os.devnull, 'w')
373 | retcode = subprocess.call(cmd, stdout=FNULL, stderr=subprocess.STDOUT)
374 | if not 0 == retcode:
375 | print('*******ValueError(Error {0} executing command: {1}*********'.format(retcode, ' '.join(cmd)))
376 | shutil.rmtree(osp.join(folder_p, 'frames'))
377 |
378 | def viz_seq(seq, folder_p, sk_type, orig_fps=30.0, debug=False):
379 | '''1. Dumps sequence of skeleton images for the given sequence of joints.
380 | 2. Collates the sequence of images into an mp4 video.
381 |
382 | Args:
383 | seq (np.array): Array of joint positions.
384 | folder_p (str): Path to root folder that will contain frames folder.
385 | sk_type (str): {'smpl', 'nturgbd'}
386 |
387 | Return:
388 | None. Path of mp4 video: folder_p/video.mp4
389 | '''
390 | # Delete folder if exists
391 | if osp.exists(folder_p):
392 | print('Deleting existing folder ', folder_p)
393 | shutil.rmtree(folder_p)
394 |
395 | # Create folder for frames
396 | os.makedirs(osp.join(folder_p, 'frames'))
397 |
398 | # Dump frames into folder. Args: (data, radius, frames path)
399 | viz_skeleton(seq, folder_p=folder_p, sk_type=sk_type, radius=1.2, debug=debug)
400 | write_vid_from_imgs(folder_p, orig_fps)
401 |
402 | return None
403 |
404 | def viz_rand_seq(X, Y, dtype, epoch, wb, urls=None,
405 | k=3, pred_labels=None):
406 | '''
407 | Args:
408 | X (np.array): Array (frames) of SMPL joint positions.
409 | Y (np.array): Multiple labels for each frame in x \in X.
410 | dtype (str): {'input', 'pred'}
411 | k (int): # samples to viz.
412 | urls (tuple): Tuple of URLs of the rendered videos from original mocap.
413 | wb (dict): Wandb log dict.
414 | Returns:
415 | viz_ds (dict): Data structure containing all viz. info so far.
416 | '''
417 | import wandb
418 | # `idx2al`: idx --> action label string
419 | al2idx = dutils.read_json('data/action_label_to_idx.json')
420 | idx2al = {al2idx[k]: k for k in al2idx}
421 |
422 | # Sample k random seqs. to viz.
423 | for s_idx in random.sample(list(range(X.shape[0])), k):
424 | # Visualize a single seq. in path `folder_p`
425 | folder_p = osp.join('viz', str(uuid.uuid4()))
426 | viz_seq(seq=X[s_idx], folder_p=folder_p)
427 | title='{0} seq. {1}: '.format(dtype, s_idx)
428 | acts_str = ', '.join([idx2al[l] for l in torch.unique(Y[s_idx])])
429 | wb[title+urls[s_idx]] = wandb.Video(osp.join(folder_p, 'video.mp4'),
430 | caption='Actions: '+acts_str)
431 |
432 | if 'pred' == dtype or 'preds'==dtype:
433 | raise NotImplementedError
434 |
435 | print('Done viz. {0} seqs.'.format(k))
436 | return wb
437 |
--------------------------------------------------------------------------------
/notebooks/BABEL_visualization.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "## Visualizing BABEL labels\n",
8 | "[BABEL](https://babel.is.tue.mpg.de/) labels mocap sequences from [AMASS](https://amass.is.tue.mpg.de) with action labels. \n",
9 | "A single sequence in BABEL can have multiple action labels associated with it, from multiple annotators. \n",
10 | "Here, we present code to load data from BABEL, visualize the mocap sequence rendered as a 2D video, and view the action labels corresponding to the sequence. "
11 | ]
12 | },
13 | {
14 | "cell_type": "code",
15 | "execution_count": 1,
16 | "metadata": {},
17 | "outputs": [],
18 | "source": [
19 | "# Preparing the environment\n",
20 | "%load_ext autoreload\n",
21 | "%autoreload 2\n",
22 | "%matplotlib notebook\n",
23 | "%matplotlib inline"
24 | ]
25 | },
26 | {
27 | "cell_type": "code",
28 | "execution_count": 2,
29 | "metadata": {},
30 | "outputs": [],
31 | "source": [
32 | "import json\n",
33 | "from os.path import join as ospj\n",
34 | "\n",
35 | "import numpy as np\n",
36 | "\n",
37 | "import pprint\n",
38 | "pp = pprint.PrettyPrinter()\n",
39 | "\n",
40 | "from IPython.display import HTML"
41 | ]
42 | },
43 | {
44 | "cell_type": "markdown",
45 | "metadata": {},
46 | "source": [
47 | "### Load BABEL\n",
48 | "We assume that you have downloaded BABEL annotations from the [website](https://babel.is.tue.mpg.de/data.html) and placed the downloaded `babel_v1.0_release` folder in `data/`. The BABEL data is provided as two sets -- BABEL dense and BABEL extra. "
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "execution_count": 3,
54 | "metadata": {},
55 | "outputs": [],
56 | "source": [
57 | "d_folder = '../data/babel_v1.0_release' # Data folder\n",
58 | "l_babel_dense_files = ['train', 'val', 'test']\n",
59 | "l_babel_extra_files = ['extra_train', 'extra_val']\n",
60 | "\n",
61 | "# BABEL Dataset \n",
62 | "babel = {}\n",
63 | "for file in l_babel_dense_files:\n",
64 | " babel[file] = json.load(open(ospj(d_folder, file+'.json')))\n",
65 | " \n",
66 | "for file in l_babel_extra_files:\n",
67 | " babel[file] = json.load(open(ospj(d_folder, file+'.json'))) "
68 | ]
69 | },
70 | {
71 | "cell_type": "markdown",
72 | "metadata": {},
73 | "source": [
74 | "### View random annotation"
75 | ]
76 | },
77 | {
78 | "cell_type": "markdown",
79 | "metadata": {},
80 | "source": [
81 | "Now, let us view an annotation data structure from the BABEL. \n",
82 | "The overall data structure is a dictionary, with a unique sequence ID as key and the annotation as value. "
83 | ]
84 | },
85 | {
86 | "cell_type": "code",
87 | "execution_count": 4,
88 | "metadata": {},
89 | "outputs": [],
90 | "source": [
91 | "def get_random_babel_ann():\n",
92 | " '''Get annotation from random sequence from a random file'''\n",
93 | " file = np.random.choice(l_babel_dense_files + l_babel_extra_files)\n",
94 | " seq_id = np.random.choice(list(babel[file].keys()))\n",
95 | " print('We are visualizing annotations for seq ID: {0} in \"{1}.json\"'.format(seq_id, file))\n",
96 | " ann = babel[file][seq_id]\n",
97 | " return ann, file"
98 | ]
99 | },
100 | {
101 | "cell_type": "code",
102 | "execution_count": 5,
103 | "metadata": {},
104 | "outputs": [
105 | {
106 | "name": "stdout",
107 | "output_type": "stream",
108 | "text": [
109 | "We are visualizing annotations for seq ID: 3312 in \"test.json\"\n",
110 | "{'babel_sid': 3312,\n",
111 | " 'dur': 76.73,\n",
112 | " 'feat_p': 'CMU/CMU/86/86_08_poses.npz',\n",
113 | " 'frame_ann': {'anntr_id': 'c6065e9c-1652-46df-a45f-fe8b8158428f',\n",
114 | " 'babel_lid': 'a642048f-7fa9-402f-a4c1-d7e9e7f696d1',\n",
115 | " 'labels': [{'act_cat': None,\n",
116 | " 'end_t': 68.093,\n",
117 | " 'proc_label': None,\n",
118 | " 'raw_label': None,\n",
119 | " 'seg_id': 'ad703788-bd17-42d4-854b-2b64cb58ee16',\n",
120 | " 'start_t': 59.51},\n",
121 | " {'act_cat': None,\n",
122 | " 'end_t': 32.82,\n",
123 | " 'proc_label': None,\n",
124 | " 'raw_label': None,\n",
125 | " 'seg_id': '1785aeca-53ce-4a33-a249-8a5d3466ea95',\n",
126 | " 'start_t': 27.445},\n",
127 | " {'act_cat': None,\n",
128 | " 'end_t': 52.426,\n",
129 | " 'proc_label': None,\n",
130 | " 'raw_label': None,\n",
131 | " 'seg_id': '12768b82-b342-46ee-ae60-e158f8b1dd47',\n",
132 | " 'start_t': 47.843},\n",
133 | " {'act_cat': None,\n",
134 | " 'end_t': 59.51,\n",
135 | " 'proc_label': None,\n",
136 | " 'raw_label': None,\n",
137 | " 'seg_id': '435bd5a6-01e9-4fc4-abee-642954466832',\n",
138 | " 'start_t': 53.26},\n",
139 | " {'act_cat': None,\n",
140 | " 'end_t': 40.007,\n",
141 | " 'proc_label': None,\n",
142 | " 'raw_label': None,\n",
143 | " 'seg_id': 'd3911406-ad83-4438-941c-919bf296d5e1',\n",
144 | " 'start_t': 33.382},\n",
145 | " {'act_cat': None,\n",
146 | " 'end_t': 76.733,\n",
147 | " 'proc_label': None,\n",
148 | " 'raw_label': None,\n",
149 | " 'seg_id': 'f222a4d9-a8d5-4002-893b-4df102e1e0fa',\n",
150 | " 'start_t': 70.593},\n",
151 | " {'act_cat': None,\n",
152 | " 'end_t': 2.252,\n",
153 | " 'proc_label': None,\n",
154 | " 'raw_label': None,\n",
155 | " 'seg_id': '35e605ec-c9f8-4c9d-8320-680de71837ce',\n",
156 | " 'start_t': 0.294},\n",
157 | " {'act_cat': None,\n",
158 | " 'end_t': 6.961,\n",
159 | " 'proc_label': None,\n",
160 | " 'raw_label': None,\n",
161 | " 'seg_id': 'fdaead4c-0a37-4579-a42a-4a94145570b9',\n",
162 | " 'start_t': 4.232},\n",
163 | " {'act_cat': None,\n",
164 | " 'end_t': 70.593,\n",
165 | " 'proc_label': None,\n",
166 | " 'raw_label': None,\n",
167 | " 'seg_id': '52d3c3e9-102b-4cf0-b082-cd416a7b5f64',\n",
168 | " 'start_t': 68.093},\n",
169 | " {'act_cat': None,\n",
170 | " 'end_t': 4.232,\n",
171 | " 'proc_label': None,\n",
172 | " 'raw_label': None,\n",
173 | " 'seg_id': 'f524e2df-36e2-45ce-a54e-892fdb7353d0',\n",
174 | " 'start_t': 2.252},\n",
175 | " {'act_cat': None,\n",
176 | " 'end_t': 9.336,\n",
177 | " 'proc_label': None,\n",
178 | " 'raw_label': None,\n",
179 | " 'seg_id': '7f265bed-f445-4b6b-a41f-c62106d7be3b',\n",
180 | " 'start_t': 6.961},\n",
181 | " {'act_cat': None,\n",
182 | " 'end_t': 47.843,\n",
183 | " 'proc_label': None,\n",
184 | " 'raw_label': None,\n",
185 | " 'seg_id': '1aa33355-a669-45a6-86a9-19ae862a47e9',\n",
186 | " 'start_t': 40.007},\n",
187 | " {'act_cat': None,\n",
188 | " 'end_t': 15.523,\n",
189 | " 'proc_label': None,\n",
190 | " 'raw_label': None,\n",
191 | " 'seg_id': 'd9c310f5-fc1e-47d8-b2f7-075c31a2eb6d',\n",
192 | " 'start_t': 9.523},\n",
193 | " {'act_cat': None,\n",
194 | " 'end_t': 22.507,\n",
195 | " 'proc_label': None,\n",
196 | " 'raw_label': None,\n",
197 | " 'seg_id': 'f7a71a16-2807-49f7-8a66-7df3e678e161',\n",
198 | " 'start_t': 15.523},\n",
199 | " {'act_cat': None,\n",
200 | " 'end_t': 0.294,\n",
201 | " 'proc_label': None,\n",
202 | " 'raw_label': None,\n",
203 | " 'seg_id': '3f57a657-2c8f-4995-87a4-965bcf8ea2a6',\n",
204 | " 'start_t': 0},\n",
205 | " {'act_cat': None,\n",
206 | " 'end_t': 9.523,\n",
207 | " 'proc_label': None,\n",
208 | " 'raw_label': None,\n",
209 | " 'seg_id': 'c9f97199-97eb-463c-a04e-a511413ad5ba',\n",
210 | " 'start_t': 9.336},\n",
211 | " {'act_cat': None,\n",
212 | " 'end_t': 33.382,\n",
213 | " 'proc_label': None,\n",
214 | " 'raw_label': None,\n",
215 | " 'seg_id': 'dac4fabe-e96c-411c-ad2e-29211e8c212a',\n",
216 | " 'start_t': 32.82},\n",
217 | " {'act_cat': None,\n",
218 | " 'end_t': 53.26,\n",
219 | " 'proc_label': None,\n",
220 | " 'raw_label': None,\n",
221 | " 'seg_id': 'ed99bf22-3ea5-45a6-9df3-17e67e49f119',\n",
222 | " 'start_t': 52.426},\n",
223 | " {'act_cat': None,\n",
224 | " 'end_t': 27.445,\n",
225 | " 'proc_label': None,\n",
226 | " 'raw_label': None,\n",
227 | " 'seg_id': '5c459b13-35e6-4c36-8ec4-9eb1536bfe95',\n",
228 | " 'start_t': 22.507}],\n",
229 | " 'mul_act': True},\n",
230 | " 'seq_ann': {'anntr_id': 'a217bb6b-93ae-4611-8e53-d4318ed5be00',\n",
231 | " 'babel_lid': '037dc092-28d5-4537-9632-9a91fc9f7fb9',\n",
232 | " 'labels': [{'act_cat': None,\n",
233 | " 'proc_label': None,\n",
234 | " 'raw_label': None,\n",
235 | " 'seg_id': 'f7d4b8fa-de77-487f-a08c-84bbc05c3148'}],\n",
236 | " 'mul_act': True},\n",
237 | " 'url': 'https://babel-renders.s3.eu-central-1.amazonaws.com/003312.mp4'}\n"
238 | ]
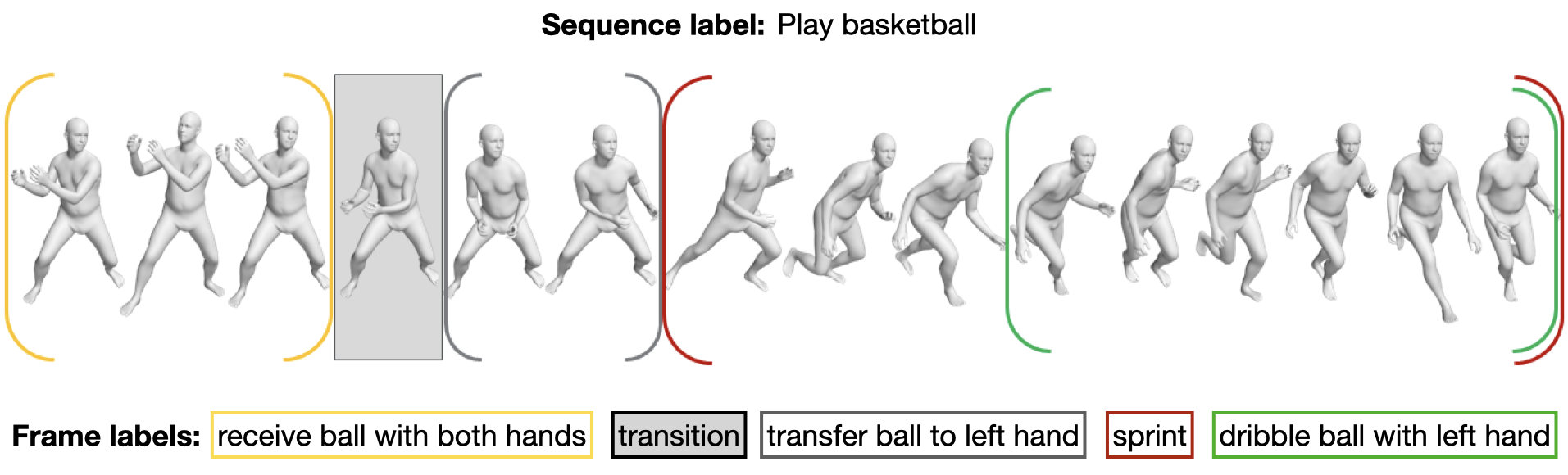
239 | }
240 | ],
241 | "source": [
242 | "ann, _ = get_random_babel_ann()\n",
243 | "pp.pprint(ann)"
244 | ]
245 | },
246 | {
247 | "cell_type": "markdown",
248 | "metadata": {},
249 | "source": [
250 | "Note that the action labels from `test.json` are not available publicly. \n",
251 | "Also note that the internal data structures of BABEL dense and BABEL extra differ slightly. \n",
252 | "For a detailed description of the annotation, see [BABEL's data page](https://babel.is.tue.mpg.de/data.html)."
253 | ]
254 | },
255 | {
256 | "cell_type": "markdown",
257 | "metadata": {},
258 | "source": [
259 | "### Visualize a mocap seq. and its action labels "
260 | ]
261 | },
262 | {
263 | "cell_type": "code",
264 | "execution_count": 6,
265 | "metadata": {},
266 | "outputs": [],
267 | "source": [
268 | "def get_vid_html(url):\n",
269 | " '''Helper code to embed a URL in a notebook'''\n",
270 | " html_code = ''\n",
273 | " return html_code"
274 | ]
275 | },
276 | {
277 | "cell_type": "code",
278 | "execution_count": 7,
279 | "metadata": {},
280 | "outputs": [],
281 | "source": [
282 | "def get_labels(ann, file):\n",
283 | " # Get sequence labels and frame labels if they exist\n",
284 | " seq_l, frame_l = None, None\n",
285 | " if 'extra' not in file:\n",
286 | " if ann['seq_ann'] is not None:\n",
287 | " seq_l = [seg['raw_label'] for seg in ann['seq_ann']['labels']]\n",
288 | " if ann['frame_ann'] is not None:\n",
289 | " frame_l = [(seg['raw_label'], seg['start_t'], seg['end_t']) for seg in ann['frame_ann']['labels']]\n",
290 | " else:\n",
291 | " # Load labels from 1st annotator (random) if there are multiple annotators\n",
292 | " if ann['seq_anns'] is not None:\n",
293 | " seq_l = [seg['raw_label'] for seg in ann['seq_anns'][0]['labels']]\n",
294 | " if ann['frame_anns'] is not None:\n",
295 | " frame_l = [(seg['raw_label'], seg['start_t'], seg['end_t']) for seg in ann['frame_anns'][0]['labels']]\n",
296 | " return seq_l, frame_l"
297 | ]
298 | },
299 | {
300 | "cell_type": "markdown",
301 | "metadata": {},
302 | "source": [
303 | "#### Visualize a random mocap and its annotation from BABEL, by running the cell below. "
304 | ]
305 | },
306 | {
307 | "cell_type": "code",
308 | "execution_count": 8,
309 | "metadata": {},
310 | "outputs": [
311 | {
312 | "name": "stdout",
313 | "output_type": "stream",
314 | "text": [
315 | "We are visualizing annotations for seq ID: 7536 in \"train.json\"\n",
316 | "Sequence labels: ['pace and shake hand']\n",
317 | "Frame labels: (action label, start time, end time)\n",
318 | "[('walk', 0, 2.106),\n",
319 | " ('transition', 2.106, 2.845),\n",
320 | " ('make a knocking gesture', 2.845, 3.507),\n",
321 | " ('transition', 3.466, 4.6),\n",
322 | " ('turn around', 4.519, 5.519),\n",
323 | " ('walk back', 5.424, 7.734)]\n"
324 | ]
325 | },
326 | {
327 | "data": {
328 | "text/html": [
329 | ""
330 | ],
331 | "text/plain": [
332 | ""
333 | ]
334 | },
335 | "execution_count": 8,
336 | "metadata": {},
337 | "output_type": "execute_result"
338 | }
339 | ],
340 | "source": [
341 | "ann, file = get_random_babel_ann()\n",
342 | "seq_l, frame_l = get_labels(ann, file)\n",
343 | "print('Sequence labels: ', seq_l)\n",
344 | "print('Frame labels: (action label, start time, end time)')\n",
345 | "pp.pprint(frame_l) \n",
346 | "HTML(get_vid_html(ann['url']))"
347 | ]
348 | },
349 | {
350 | "cell_type": "markdown",
351 | "metadata": {},
352 | "source": [
353 | "- If you are interested in loading the mocap sequence in 3D, please refer to the tutorials in [AMASS](https://github.com/nghorbani/amass/tree/master/notebooks)"
354 | ]
355 | }
356 | ],
357 | "metadata": {
358 | "kernelspec": {
359 | "display_name": "Python 3",
360 | "language": "python",
361 | "name": "python3"
362 | },
363 | "language_info": {
364 | "codemirror_mode": {
365 | "name": "ipython",
366 | "version": 3
367 | },
368 | "file_extension": ".py",
369 | "mimetype": "text/x-python",
370 | "name": "python",
371 | "nbconvert_exporter": "python",
372 | "pygments_lexer": "ipython3",
373 | "version": "3.8.3"
374 | }
375 | },
376 | "nbformat": 4,
377 | "nbformat_minor": 4
378 | }
379 |
--------------------------------------------------------------------------------
/action_recognition/data_gen/create_dataset.py:
--------------------------------------------------------------------------------
1 | #! /usr/bin/env python
2 | # -*- coding: utf-8 -*-
3 | # vim:fenc=utf-8
4 | #
5 | # Copyright © 2021 achandrasekaran
6 | #
7 | # Distributed under terms of the MIT license.
8 |
9 |
10 | import sys, os, pdb
11 | from os.path import join as ospj
12 | from os.path import basename as ospb
13 | from os.path import dirname as ospd
14 | import numpy as np
15 | import torch
16 | from collections import *
17 | from itertools import *
18 | import pandas as pd
19 | import pickle, json, csv
20 | from tqdm import tqdm
21 | from pandas.core.common import flatten
22 | import ipdb
23 | import pickle
24 |
25 | # Custom
26 | import preprocess
27 | import dutils
28 | import viz
29 |
30 | """
31 | Script to load BABEL segments with NTU skeleton format and pre-process.
32 | """
33 |
34 |
35 | def ntu_style_preprocessing(b_dset_path):
36 | '''
37 | '''
38 | pdb.set_trace()
39 | print('Load BABEL v1.0 dataset subset', b_dset_path)
40 | b_dset = dutils.read_pkl(b_dset_path)
41 | # Get unnormalized 5-sec. samples
42 | X = np.array(b_dset['X'])
43 | print('X (old) = ', np.shape(X)) # N, T, V, C
44 |
45 | # Prep. data for normalization
46 | X = X.transpose(0, 3, 1, 2) # N, C, T, V
47 | X = X[:, :, :, :, np.newaxis] # N, C, T, V, M
48 | print('Shape of prepped X: ', X.shape)
49 |
50 | # Normalize (pre-process) in NTU RGBD-style
51 | ntu_sk_spine_bone = np.array([0, 1])
52 | ntu_sk_shoulder_bone = np.array([8, 4])
53 | X, l_m_sk = preprocess.pre_normalization(X, zaxis=ntu_sk_spine_bone,
54 | xaxis=ntu_sk_shoulder_bone)
55 | print('Shape of normalized X: ', X.shape)
56 | print('Skipping {0} samples because "skeleton is missing"'.format(len(l_m_sk)))
57 | print('Skipped idxs = ', l_m_sk)
58 |
59 | # Dataset w/ processed seg. chunks. (Skip samples w/ missing skeletons)
60 | b_AR_dset = {k: np.delete(b_dset[k], l_m_sk) for k in b_dset if k!='X'}
61 | b_AR_dset['X'] = np.delete(X, l_m_sk, axis=0)
62 | print('Shape of dataset = ', b_AR_dset['X'].shape)
63 |
64 | fp = b_dset_path.replace('samples', 'ntu_sk_ntu-style_preprocessed' )
65 | # fp = '../data/babel_v1.0/babel_v1.0_ntu_sk_ntu-style_preprocessed.pkl'
66 | # dutils.write_pkl(b_AR_dset, fp)
67 | with open(fp, 'wb') as of:
68 | pickle.dump(b_AR_dset, of, protocol=4)
69 |
70 | def get_act_idx(y, act2idx, n_classes):
71 | '''
72 | '''
73 | if y in act2idx:
74 | return act2idx[y]
75 | else:
76 | return n_classes
77 |
78 | def store_splits_subsets(n_classes, spl, plus_extra = True, w_folder = '../data/babel_v1.0/'):
79 | '''
80 | '''
81 | # Get splits
82 | splits = dutils.read_json('../data/amass_splits.json')
83 | sid2split = {int(ospb(u).replace('.mp4', '')): spl for spl in splits \
84 | for u in splits[spl] }
85 |
86 | # In labels, act. cat. --> idx
87 | act2idx_150 = dutils.read_json('../data/action_label_2_idx.json')
88 | act2idx = {k: act2idx_150[k] for k in act2idx_150 if act2idx_150[k] < n_classes}
89 | print('{0} actions in label set: {1}'.format(len(act2idx), act2idx))
90 |
91 | if plus_extra :
92 | fp = w_folder + 'babel_v1.0_'+spl+'_extra_ntu_sk_ntu-style_preprocessed.pkl'
93 | else:
94 | fp = w_folder + 'babel_v1.0_'+spl+'_ntu_sk_ntu-style_preprocessed.pkl'
95 |
96 | # Get full dataset
97 | b_AR_dset = dutils.read_pkl(fp)
98 |
99 | # Store idxs of samples to include in learning
100 | split_idxs = defaultdict(list)
101 | for i, y1 in enumerate(b_AR_dset['Y1']):
102 |
103 | # Check if action category in list of classes
104 | if y1 not in act2idx:
105 | continue
106 |
107 | sid = b_AR_dset['sid'][i]
108 | split_idxs[sid2split[sid]].append(i) # Include idx in dataset
109 |
110 | # Save features that'll be loaded by dataloader
111 | ar_idxs = np.array(split_idxs[spl])
112 | X = b_AR_dset['X'][ar_idxs]
113 | if plus_extra:
114 | fn = w_folder + f'{spl}_extra_ntu_sk_{n_classes}.npy'
115 | else:
116 | fn = w_folder + f'{spl}_ntu_sk_{n_classes}.npy'
117 | np.save(fn, X)
118 |
119 | # labels
120 | labels = {k: np.array(b_AR_dset[k])[ar_idxs] for k in b_AR_dset if k!='X'}
121 |
122 | # Create, save label data structure that'll be loaded by dataloader
123 | label_idxs = defaultdict(list)
124 | for i, y1 in enumerate(labels['Y1']):
125 | # y1
126 | label_idxs['Y1'].append(act2idx[y1])
127 | # yk
128 | yk = [get_act_idx(y, act2idx, n_classes) for y in labels['Yk'][i]]
129 | label_idxs['Yk'].append(yk)
130 | # yov
131 | yov_o = labels['Yov'][i]
132 | yov = {get_act_idx(y, act2idx, n_classes): yov_o[y] for y in yov_o}
133 | label_idxs['Yov'].append(yov)
134 | #
135 | label_idxs['seg_id'].append(labels['seg_id'][i])
136 | label_idxs['sid'].append(labels['sid'][i])
137 | label_idxs['chunk_n'].append(labels['chunk_n'][i])
138 | label_idxs['anntr_id'].append(labels['anntr_id'][i])
139 |
140 | if plus_extra:
141 | wr_f = w_folder + f'{spl}_extra_label_{n_classes}.pkl'
142 | else:
143 | wr_f = w_folder + f'{spl}_label_{n_classes}.pkl'
144 | dutils.write_pkl(\
145 | (label_idxs['seg_id'], (label_idxs['Y1'], label_idxs['sid'],
146 | label_idxs['chunk_n'], label_idxs['anntr_id'])), \
147 | wr_f)
148 |
149 | class Babel_AR:
150 | '''Object containing data, methods for Action Recognition.
151 |
152 | Task
153 | -----
154 | Given: x (Segment from Babel)
155 | Predict: \hat{p}(x) (Distribution over action categories)
156 |
157 | GT
158 | ---
159 | How to compute GT for a given segment?
160 | - yk: All action categories that are labeled for the entirety of segment
161 | - y1: One of yk
162 | - yov: Any y that belongs to part of a segment is considered to be GT.
163 | Fraction of segment covered by an action: {'walk': 1.0, 'wave': 0.5}
164 |
165 | '''
166 | def __init__(self, dataset, dense=True, seq_dense_ann_type={}):
167 | '''Dataset with (samples, different GTs)
168 | '''
169 | # Load dataset
170 | self.babel = dataset
171 | self.dense = dense
172 | self.seq_dense_ann_type = seq_dense_ann_type
173 | self.jpos_p = '../../../../../amass/'
174 |
175 | # Get frame-rate for each seq. in AMASS
176 | f_p = '../data/featp_2_fps.json'
177 | self.ft_p_2_fps = dutils.read_json(f_p)
178 |
179 | # Dataset w/ keys = {'X', 'Y1', 'Yk', 'Yov', 'seg_id', 'sid',
180 | # 'seg_dur'}
181 | self.d = defaultdict(list)
182 | for ann in tqdm(self.babel):
183 | self._update_dataset(ann)
184 |
185 | def _subsample_to_30fps(self, orig_ft, orig_fps):
186 | '''Get features at 30fps frame-rate
187 | Args:
188 | orig_ft (T, 25*3): Feats. @ `orig_fps` frame-rate
189 | orig_fps : Frame-rate in original (ft) seq.
190 | Return:
191 | ft (T', 25*3): Feats. @ 30fps
192 | '''
193 | T, n_j, _ = orig_ft.shape
194 | out_fps = 30.0
195 | # Matching the sub-sampling used for rendering
196 | if int(orig_fps)%int(out_fps):
197 | sel_fr = np.floor(orig_fps / out_fps * np.arange(int(out_fps))).astype(int)
198 | n_duration = int(T/int(orig_fps))
199 | t_idxs = []
200 | for i in range(n_duration):
201 | t_idxs += list(i * int(orig_fps) + sel_fr)
202 | if int(T % int(orig_fps)):
203 | last_sec_frame_idx = n_duration*int(orig_fps)
204 | t_idxs += [x+ last_sec_frame_idx for x in sel_fr if x + last_sec_frame_idx < T ]
205 | else:
206 | t_idxs = np.arange(0, T, orig_fps/out_fps, dtype=int)
207 |
208 | ft = orig_ft[t_idxs, :, :]
209 | return ft
210 |
211 | def _viz_x(self, ft, fn='test_sample'):
212 | '''Wraper to Viz. the given sample (w/ NTU RGBD skeleton)'''
213 | viz.viz_seq(seq=ft, folder_p=f'test_viz/{fn}', sk_type='nturgbd',
214 | debug=True)
215 | return None
216 |
217 | def _load_seq_feats(self, ft_p, sk_type):
218 | '''Given path to joint position features, return them in 30fps'''
219 | # Identify appropriate feature directory path on disk
220 | if 'smpl_wo_hands' == sk_type: # SMPL w/o hands (T, 22*3)
221 | jpos_p = ospj(self.jpos_p, 'joint_pos')
222 | if 'nturgbd' == sk_type: # NTU (T, 219)
223 | jpos_p = ospj(self.jpos_p, 'babel_joint_pos')
224 |
225 | # Get the correct dataset folder name
226 | ddir_n = ospb(ospd(ospd(ft_p)))
227 | ddir_map = {'BioMotionLab_NTroje': 'BMLrub', 'DFaust_67': 'DFaust'}
228 | ddir_n = ddir_map[ddir_n] if ddir_n in ddir_map else ddir_n
229 | # Get the subject folder name
230 | sub_fol_n = ospb(ospd(ft_p))
231 |
232 | # Sanity check
233 | fft_p = ospj(jpos_p, ddir_n, sub_fol_n, ospb(ft_p))
234 | assert os.path.exists(fft_p)
235 |
236 | # Load seq. fts.
237 | ft = np.load(fft_p)['joint_pos']
238 | T, ft_sz = ft.shape
239 |
240 | # Get NTU skeleton joints
241 | ntu_js = dutils.smpl_to_nturgbd(model_type='smplh', out_format='nturgbd')
242 | ft = ft.reshape(T, -1, 3)
243 | ft = ft[:, ntu_js, :]
244 |
245 | # Sub-sample to 30fps
246 | orig_fps = self.ft_p_2_fps[ft_p]
247 | ft = self._subsample_to_30fps(ft, orig_fps)
248 | # print(f'Feat. shape = {ft.shape}, fps = {orig_fps}')
249 | # if orig_fps != 30.0:
250 | # self._viz_x(ft)
251 | return ft
252 |
253 | def _get_per_f_labels(self, ann, ann_type, seq_dur):
254 | ''' '''
255 | # Per-frame labels: {0: ['walk'], 1: ['walk', 'wave'], ... T: ['stand']}
256 | yf = defaultdict(list)
257 | T = int(30.0*seq_dur)
258 | for n_f in range(T):
259 | cur_t = float(n_f/30.0)
260 | for seg in ann['labels']:
261 |
262 | if seg['act_cat'] is None:
263 | continue
264 |
265 | if 'seq_ann' == ann_type:
266 | seg['start_t'] = 0.0
267 | seg['end_t'] = seq_dur
268 |
269 | if cur_t >= float(seg['start_t']) and cur_t < float(seg['end_t']):
270 | yf[n_f] += seg['act_cat']
271 | return yf
272 |
273 | def _compute_dur_samples(self, ann, ann_type, seq_ft, seq_dur, dur=5.0):
274 | '''Return each GT action, corresponding to the fraction of the
275 | segment that it overlaps with.
276 | There are 2 conditions that we need to handle:
277 | 1. Multiple action categories in 'act_cat'
278 | 2. Simultaneous (overlapping action segments).
279 |
280 | Example Input:
281 | Seq. => frames [0, 1, 2, 3, 4, 5]
282 | GT acts. => [[2,3], [2,3], [2], [0], [0,1], [0,1]]
283 |
284 | Segs, GT:
285 | 1. seg_x = seq[0: 3], y1 = 2, yall = {2: 1.0, 3: 0.66}
286 | 2. seg_x = seq[0: 2], y1 = 3, yall = {2: 1.0, 3: 1.0}
287 | 3. seg_x = seq[3: ], y1 = 0, yall = {0: 1.0, 1: 0.66}
288 | 4. seg_x = seq[4: ], y1 = 1, yall = {0: 1.0, 1: 1.0}
289 |
290 | - Note that we should do the above for each chunk in a segment,
291 | each of duration = seconds.
292 |
293 | Return:
294 | [ { 'x': [st_t, end_t],
295 | 'y1': ,
296 | 'yall': { : , ...}},
297 | { ... }, ...
298 | ]
299 | '''
300 | #
301 | yf = self._get_per_f_labels(ann, ann_type, seq_dur)
302 |
303 | # Compute, store all samples for each segment
304 | seq_samples = []
305 | for seg in ann['labels']:
306 |
307 | # If no labeled act. cats. for current seg., skip it
308 | if seg['act_cat'] is None or 0 == len(seg['act_cat']):
309 | continue
310 |
311 | # Handle stage 1 missing durs.
312 | if 'seq_ann' == ann_type:
313 | seg['start_t'] = 0.0
314 | seg['end_t'] = seq_dur
315 |
316 | # Get segment feats.
317 | seg_st_f, seg_end_f = int(30.0*seg['start_t']), int(30.0*seg['end_t'])
318 | seg_x = seq_ft[seg_st_f: seg_end_f, :, :]
319 |
320 | # Split segment into -second chunks
321 | n_f_pc = 30.0 * dur
322 | n_chunks = int(np.ceil(seg_x.shape[0]/n_f_pc))
323 | for n_ch in range(n_chunks):
324 |
325 | # Single -sec. chunk in segment
326 | ch_st_f = int(n_f_pc * n_ch)
327 | ch_end_f = int(min(ch_st_f + n_f_pc, seg_x.shape[0]))
328 | x = seg_x[ch_st_f: ch_end_f, :, :]
329 |
330 | # Handle case where chunk_T < n_f_pc
331 | x_T, nj, xyz = x.shape
332 | x_ch = np.concatenate((x, np.zeros((int(n_f_pc)- x_T, nj, xyz))), axis=0)
333 |
334 | # Labels for this chunk
335 | yov = Counter(flatten([yf[seg_st_f + n_f] for n_f in range(ch_st_f, ch_end_f)]))
336 |
337 | # Sanity check -- is segment smaller than 1 frame?
338 | if seg['act_cat'][0] not in yov:
339 | # print('Skipping seg:', seg)
340 | # print(f'Chunk # {n_ch}, Yov: ', yov)
341 | continue
342 |
343 | yov = {k: round(yov[k]/x_T, 3) for k in yov}
344 |
345 | # For each act_cat in segment, create a separate sample
346 | for cat in seg['act_cat']:
347 | # Add to samples GTs
348 | seq_samples.append({'seg_id': seg['seg_id'],
349 | 'chunk_n': n_ch,
350 | 'chunk_dur': round(x_T/n_f_pc, 3),
351 | 'x': x_ch,
352 | 'y1': cat,
353 | 'yk': seg['act_cat'],
354 | 'yov': yov,
355 | 'anntr_id': ann['anntr_id']
356 | })
357 | return seq_samples
358 |
359 | def _sample_at_seg_chunk_level(self, ann, seq_samples):
360 | # Samples at segment-chunk-level
361 | for i, sample in enumerate(seq_samples):
362 |
363 | self.d['sid'].append(ann['babel_sid']) # Seq. info
364 | self.d['seg_id'].append(sample['seg_id']) # Seg. info
365 | self.d['chunk_n'].append(sample['chunk_n']) # Seg. chunk info
366 | self.d['anntr_id'].append(sample['anntr_id']) # Annotator id (useful in rebuttal exp.)
367 | self.d['chunk_dur'].append(sample['chunk_dur']) # Seg. chunk info
368 | self.d['X'].append(sample['x']) # Seg. chunk feats.
369 | self.d['Y1'].append(sample['y1']) # 1 out of k GT act. cats.
370 | self.d['Yk'].append(sample['yk']) # List of k GT act. cats.
371 | # : fractions of overlapping act. cats.
372 | self.d['Yov'].append(sample['yov'])
373 | return
374 |
375 | def _update_dataset(self, ann):
376 | '''Return one sample (one segment) = (X, Y1, Yall)'''
377 |
378 | # Get feats. for seq.
379 | seq_ft = self._load_seq_feats(ann['feat_p'], 'nturgbd')
380 |
381 | # To keep track of type of annotation for loading 'extra'
382 | # Compute all GT labels for this seq.
383 | seq_samples = None
384 | if self.dense:
385 | if ann['frame_ann'] is not None:
386 | ann_ar = ann['frame_ann']
387 | self.seq_dense_ann_type[ann['babel_sid']] = 'frame_ann'
388 | seq_samples = self._compute_dur_samples(ann_ar, 'frame_ann', seq_ft, ann['dur'])
389 | else:
390 | ann_ar = ann['seq_ann']
391 | self.seq_dense_ann_type[ann['babel_sid']] = 'seq_ann'
392 | seq_samples = self._compute_dur_samples(ann_ar, 'seq_ann', seq_ft, ann['dur'])
393 | self._sample_at_seg_chunk_level(ann, seq_samples)
394 | else:
395 | # check if extra exists
396 | if 'frame_anns' in ann.keys() or 'seq_anns' in ann.keys():
397 | ann_type = None
398 | if ann['babel_sid'] in self.seq_dense_ann_type:
399 | ann_type = self.seq_dense_ann_type[ann['babel_sid']]
400 | else:
401 | if ann['frame_anns'] is not None:
402 | ann_type = 'frame_ann'
403 | elif ann['seq_anns'] is not None:
404 | ann_type = 'seq_ann'
405 | else:
406 | ipdb.set_trace()
407 | self.seq_dense_ann_type['babel_sid'] = ann_type
408 | ann_ar = None
409 | if ann_type == 'frame_ann':
410 | if ann['frame_anns'] is not None:
411 | ann_ar = ann['frame_anns']
412 | elif ann_type == 'seq_ann':
413 | if ann['seq_anns'] is not None:
414 | ann_ar = ann['seq_anns']
415 | else:
416 | ipdb.set_trace()
417 | if ann_ar:
418 | for an in ann_ar:
419 | seq_samples = self._compute_dur_samples(an, ann_type, \
420 | seq_ft, ann['dur'])
421 | self._sample_at_seg_chunk_level(ann, seq_samples)
422 | else:
423 | print('Unexpected format for extra!')
424 | return
425 |
426 |
427 | # Create dataset
428 | # --------------------------
429 | d_folder = '../../data/babel_v1.0_release/'
430 | w_folder = '../data/babel_v1.0/'
431 | for spl in ['train', 'val']:
432 |
433 | # Load Dense BABEL
434 | data = dutils.read_json(ospj(d_folder, f'{spl}.json'))
435 | dataset = [data[sid] for sid in data]
436 | dense_babel = Babel_AR(dataset, dense=True)
437 | # Store Dense BABEL
438 | d_filename = w_folder + 'babel_v1.0_'+ spl + '_samples.pkl'
439 | dutils.write_pkl(dense_babel.d, d_filename)
440 |
441 | # Load Extra BABEL
442 | data = dutils.read_json(ospj(d_folder, f'extra_{spl}.json'))
443 | dataset = [data[sid] for sid in data]
444 | extra_babel = Babel_AR(dataset, dense=False,
445 | seq_dense_ann_type=dense_babel.seq_dense_ann_type)
446 | # Store Dense + Extra
447 | de = {}
448 | for k in dense_babel.d.keys():
449 | de[k] = dense_babel.d[k] + extra_babel.d[k]
450 | ex_filename = w_folder + 'babel_v1.0_' + spl + '_extra_samples.pkl'
451 | dutils.write_pkl(de, ex_filename)
452 |
453 | # Pre-process, Store data in dataset
454 | print('NTU-style preprocessing')
455 | babel_dataset_AR = ntu_style_preprocessing(d_filename)
456 | babel_dataset_AR = ntu_style_preprocessing(ex_filename)
457 |
458 | for ex, C in product(('', '_extra'), (120, 60)):
459 |
460 | # Split, store data in npy file, labels in pkl
461 | store_splits_subsets(n_classes=C, spl=spl, plus_extra=True)
462 | store_splits_subsets(n_classes=C, spl=spl, plus_extra=False)
463 |
464 | # Store counts of samples for training with class-balanced focal loss
465 | label_fp = ospj(w_folder, f'{spl}{ex}_label_{C}.pkl')
466 | dutils.store_counts(label_fp)
467 |
468 |
--------------------------------------------------------------------------------
/action_recognition/train_test.py:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env python
2 |
3 | # -*- coding: utf-8 -*-
4 | #
5 | # Adapted from https://github.com/lshiwjx/2s-AGCN for BABEL (https://babel.is.tue.mpg.de/)
6 |
7 | from __future__ import print_function
8 |
9 | import argparse
10 | import inspect
11 | import os
12 | import pickle
13 | import random
14 | import shutil
15 | import time
16 | from collections import *
17 | import numpy as np
18 |
19 | # torch
20 | import torch
21 | import torch.backends.cudnn as cudnn
22 | import torch.nn as nn
23 | import torch.optim as optim
24 | import torch.nn.functional as F
25 |
26 | import yaml
27 | from tensorboardX import SummaryWriter
28 | from torch.autograd import Variable
29 | from torch.optim.lr_scheduler import _LRScheduler
30 | from tqdm import tqdm
31 |
32 | import pdb
33 | import ipdb
34 |
35 | # Custom
36 | from class_balanced_loss import CB_loss
37 |
38 |
39 | # class GradualWarmupScheduler(_LRScheduler):
40 | # def __init__(self, optimizer, total_epoch, after_scheduler=None):
41 | # self.total_epoch = total_epoch
42 | # self.after_scheduler = after_scheduler
43 | # self.finished = False
44 | # self.last_epoch = -1
45 | # super().__init__(optimizer)
46 |
47 | # def get_lr(self):
48 | # return [base_lr * (self.last_epoch + 1) / self.total_epoch for base_lr in self.base_lrs]
49 |
50 | # def step(self, epoch=None, metric=None):
51 | # if self.last_epoch >= self.total_epoch - 1:
52 | # if metric is None:
53 | # return self.after_scheduler.step(epoch)
54 | # else:
55 | # return self.after_scheduler.step(metric, epoch)
56 | # else:
57 | # return super(GradualWarmupScheduler, self).step(epoch)
58 |
59 |
60 | def init_seed(_):
61 | torch.cuda.manual_seed_all(1)
62 | torch.manual_seed(1)
63 | np.random.seed(1)
64 | random.seed(1)
65 | # torch.backends.cudnn.enabled = False
66 | torch.backends.cudnn.deterministic = True
67 | torch.backends.cudnn.benchmark = False
68 |
69 |
70 | def get_parser():
71 | # parameter priority: command line > config > default
72 | parser = argparse.ArgumentParser(
73 | description='Spatial Temporal Graph Convolution Network')
74 | parser.add_argument(
75 | '--work-dir',
76 | default='./work_dir/temp',
77 | help='the work folder for storing results')
78 |
79 | parser.add_argument('-model_saved_name', default='')
80 | parser.add_argument(
81 | '--config',
82 | default='./config/nturgbd-cross-view/test_bone.yaml',
83 | help='path to the configuration file')
84 |
85 | # processor
86 | parser.add_argument(
87 | '--phase', default='train', help='must be train or test')
88 | parser.add_argument(
89 | '--save-score',
90 | type=str2bool,
91 | default=True,
92 | help='if ture, the classification score will be stored')
93 |
94 | # visulize and debug
95 | parser.add_argument(
96 | '--seed', type=int, default=1, help='random seed for pytorch')
97 | parser.add_argument(
98 | '--log-interval',
99 | type=int,
100 | default=100,
101 | help='the interval for printing messages (#iteration)')
102 | parser.add_argument(
103 | '--save-interval',
104 | type=int,
105 | default=2,
106 | help='the interval for storing models (#iteration)')
107 | parser.add_argument(
108 | '--eval-interval',
109 | type=int,
110 | default=5,
111 | help='the interval for evaluating models (#iteration)')
112 | parser.add_argument(
113 | '--print-log',
114 | type=str2bool,
115 | default=True,
116 | help='print logging or not')
117 | parser.add_argument(
118 | '--show-topk',
119 | type=int,
120 | default=[1, 5],
121 | nargs='+',
122 | help='which Top K accuracy will be shown')
123 |
124 | # feeder
125 | parser.add_argument(
126 | '--feeder', default='feeder.feeder', help='data loader will be used')
127 | parser.add_argument(
128 | '--num-worker',
129 | type=int,
130 | default=32,
131 | help='the number of worker for data loader')
132 | parser.add_argument(
133 | '--train-feeder-args',
134 | default=dict(),
135 | help='the arguments of data loader for training')
136 | parser.add_argument(
137 | '--test-feeder-args',
138 | default=dict(),
139 | help='the arguments of data loader for test')
140 |
141 | # model
142 | parser.add_argument('--model', default=None, help='the model will be used')
143 | parser.add_argument(
144 | '--model-args',
145 | type=dict,
146 | default=dict(),
147 | help='the arguments of model')
148 | parser.add_argument(
149 | '--weights',
150 | default=None,
151 | help='the weights for network initialization')
152 | parser.add_argument(
153 | '--ignore-weights',
154 | type=str,
155 | default=[],
156 | nargs='+',
157 | help='the name of weights which will be ignored in the initialization')
158 |
159 | # optim
160 | parser.add_argument(
161 | '--base-lr', type=float, default=0.01, help='initial learning rate')
162 | parser.add_argument(
163 | '--step',
164 | type=int,
165 | default=[20, 40, 60],
166 | nargs='+',
167 | help='the epoch where optimizer reduce the learning rate')
168 |
169 | #training
170 | parser.add_argument(
171 | '--device',
172 | type=int,
173 | default=0,
174 | nargs='+',
175 | help='the indexes of GPUs for training or testing')
176 | parser.add_argument('--optimizer', default='SGD', help='type of optimizer')
177 | parser.add_argument(
178 | '--nesterov', type=str2bool, default=False, help='use nesterov or not')
179 | parser.add_argument(
180 | '--batch-size', type=int, default=256, help='training batch size')
181 | parser.add_argument(
182 | '--test-batch-size', type=int, default=256, help='test batch size')
183 | parser.add_argument(
184 | '--start-epoch',
185 | type=int,
186 | default=0,
187 | help='start training from which epoch')
188 | parser.add_argument(
189 | '--num-epoch',
190 | type=int,
191 | default=80,
192 | help='stop training in which epoch')
193 | parser.add_argument(
194 | '--weight-decay',
195 | type=float,
196 | default=0.0005,
197 | help='weight decay for optimizer')
198 | # loss
199 | parser.add_argument(
200 | '--loss',
201 | type=str,
202 | default='CE',
203 | help='loss type(CE or focal)')
204 | parser.add_argument(
205 | '--label_count_path',
206 | default=None,
207 | type=str,
208 | help='Path to label counts (used in loss weighting)')
209 | parser.add_argument(
210 | '---beta',
211 | type=float,
212 | default=0.9999,
213 | help='Hyperparameter for Class balanced loss')
214 | parser.add_argument(
215 | '--gamma',
216 | type=float,
217 | default=2.0,
218 | help='Hyperparameter for Focal loss')
219 |
220 | parser.add_argument('--only_train_part', default=False)
221 | parser.add_argument('--only_train_epoch', default=0)
222 | parser.add_argument('--warm_up_epoch', default=0)
223 | return parser
224 |
225 |
226 | class Processor():
227 | """
228 | Processor for Skeleton-based Action Recgnition
229 | """
230 | def __init__(self, arg):
231 | self.arg = arg
232 | self.save_arg()
233 | if arg.phase == 'train':
234 | if not arg.train_feeder_args['debug']:
235 | if os.path.isdir(arg.model_saved_name):
236 | print('log_dir: ', arg.model_saved_name, 'already exist')
237 | # answer = input('delete it? y/n:')
238 | answer = 'y'
239 | if answer == 'y':
240 | print('Deleting dir...')
241 | shutil.rmtree(arg.model_saved_name)
242 | print('Dir removed: ', arg.model_saved_name)
243 | # input('Refresh the website of tensorboard by pressing any keys')
244 | else:
245 | print('Dir not removed: ', arg.model_saved_name)
246 | self.train_writer = SummaryWriter(os.path.join(arg.model_saved_name, 'train'), 'train')
247 | self.val_writer = SummaryWriter(os.path.join(arg.model_saved_name, 'val'), 'val')
248 | else:
249 | self.train_writer = self.val_writer = SummaryWriter(os.path.join(arg.model_saved_name, 'test'), 'test')
250 | self.global_step = 0
251 | self.load_model()
252 | self.load_optimizer()
253 | self.load_data()
254 | self.lr = self.arg.base_lr
255 | self.best_acc = 0
256 | self.best_per_class_acc = 0
257 |
258 | def load_data(self):
259 | Feeder = import_class(self.arg.feeder)
260 | self.data_loader = dict()
261 | if self.arg.phase == 'train':
262 | self.data_loader['train'] = torch.utils.data.DataLoader(
263 | dataset=Feeder(**self.arg.train_feeder_args),
264 | batch_size=self.arg.batch_size,
265 | shuffle=True,
266 | num_workers=self.arg.num_worker,
267 | drop_last=True,
268 | worker_init_fn=init_seed)
269 | self.data_loader['test'] = torch.utils.data.DataLoader(
270 | dataset=Feeder(**self.arg.test_feeder_args),
271 | batch_size=self.arg.test_batch_size,
272 | shuffle=False,
273 | num_workers=self.arg.num_worker,
274 | drop_last=False,
275 | worker_init_fn=init_seed)
276 |
277 | def load_class_weights(self):
278 | if arg.label_count_path == None:
279 | raise Exception('No label count path..!!!')
280 | with open(arg.label_count_path, 'rb') as f:
281 | label_count = pickle.load(f)
282 | img_num_per_cls = []
283 | # ipdb.set_trace()
284 | for cls_idx in range(len(label_count)):
285 | img_num_per_cls.append(int(label_count[cls_idx]))
286 | self.samples_per_class = img_num_per_cls
287 |
288 | def load_model(self):
289 | output_device = self.arg.device[0] if type(self.arg.device) is list else self.arg.device
290 | self.output_device = output_device
291 | Model = import_class(self.arg.model)
292 | shutil.copy2(inspect.getfile(Model), self.arg.work_dir)
293 | print(Model)
294 | self.model = Model(**self.arg.model_args).cuda(output_device)
295 | print(self.model)
296 | self.loss_type = arg.loss
297 | if self.loss_type != 'CE':
298 | self.load_class_weights()
299 |
300 | if self.arg.weights:
301 | self.global_step = int(arg.weights[:-3].split('-')[-1])
302 | self.print_log('Load weights from {}.'.format(self.arg.weights))
303 | if '.pkl' in self.arg.weights:
304 | with open(self.arg.weights, 'r') as f:
305 | weights = pickle.load(f)
306 | else:
307 | weights = torch.load(self.arg.weights)
308 |
309 | weights = OrderedDict(
310 | [[k.split('module.')[-1],
311 | v.cuda(output_device)] for k, v in weights.items()])
312 |
313 | keys = list(weights.keys())
314 | for w in self.arg.ignore_weights:
315 | for key in keys:
316 | if w in key:
317 | if weights.pop(key, None) is not None:
318 | self.print_log('Sucessfully Remove Weights: {}.'.format(key))
319 | else:
320 | self.print_log('Can Not Remove Weights: {}.'.format(key))
321 |
322 | try:
323 | self.model.load_state_dict(weights)
324 | except:
325 | state = self.model.state_dict()
326 | diff = list(set(state.keys()).difference(set(weights.keys())))
327 | print('Can not find these weights:')
328 | for d in diff:
329 | print(' ' + d)
330 | state.update(weights)
331 | self.model.load_state_dict(state)
332 |
333 | if type(self.arg.device) is list:
334 | if len(self.arg.device) > 1:
335 | self.model = nn.DataParallel(
336 | self.model,
337 | device_ids=self.arg.device,
338 | output_device=output_device)
339 |
340 | def load_optimizer(self):
341 | if self.arg.optimizer == 'SGD':
342 | self.optimizer = optim.SGD(
343 | self.model.parameters(),
344 | lr=self.arg.base_lr,
345 | momentum=0.9,
346 | nesterov=self.arg.nesterov,
347 | weight_decay=self.arg.weight_decay)
348 | elif self.arg.optimizer == 'Adam':
349 | self.optimizer = optim.Adam(
350 | self.model.parameters(),
351 | lr=self.arg.base_lr,
352 | weight_decay=self.arg.weight_decay)
353 | else:
354 | raise ValueError()
355 |
356 | def save_arg(self):
357 | # save arg
358 | arg_dict = vars(self.arg)
359 | if not os.path.exists(self.arg.work_dir):
360 | os.makedirs(self.arg.work_dir)
361 | with open('{}/config.yaml'.format(self.arg.work_dir), 'w') as f:
362 | yaml.dump(arg_dict, f)
363 |
364 | def adjust_learning_rate(self, epoch):
365 | if self.arg.optimizer == 'SGD' or self.arg.optimizer == 'Adam':
366 | if epoch < self.arg.warm_up_epoch:
367 | lr = self.arg.base_lr * (epoch + 1) / self.arg.warm_up_epoch
368 | else:
369 | lr = self.arg.base_lr * (
370 | 0.1 ** np.sum(epoch >= np.array(self.arg.step)))
371 | for param_group in self.optimizer.param_groups:
372 | param_group['lr'] = lr
373 |
374 | return lr
375 | else:
376 | raise ValueError()
377 |
378 | def print_time(self):
379 | localtime = time.asctime(time.localtime(time.time()))
380 | self.print_log("Local current time : " + localtime)
381 |
382 | def print_log(self, str, print_time=True):
383 | if print_time:
384 | localtime = time.asctime(time.localtime(time.time()))
385 | str = "[ " + localtime + ' ] ' + str
386 | print(str)
387 | if self.arg.print_log:
388 | with open('{}/log.txt'.format(self.arg.work_dir), 'a') as f:
389 | print(str, file=f)
390 |
391 | def record_time(self):
392 | self.cur_time = time.time()
393 | return self.cur_time
394 |
395 | def split_time(self):
396 | split_time = time.time() - self.cur_time
397 | self.record_time()
398 | return split_time
399 |
400 | def train(self, epoch, wb_dict, save_model=False):
401 | self.model.train()
402 | self.print_log('Training epoch: {}'.format(epoch + 1))
403 | loader = self.data_loader['train']
404 | self.adjust_learning_rate(epoch)
405 |
406 | loss_value, batch_acc, batch_per_class_acc = [], [], []
407 | self.train_writer.add_scalar('epoch', epoch, self.global_step)
408 | self.record_time()
409 | timer = dict(dataloader=0.001, model=0.001, statistics=0.001)
410 | process = tqdm(loader)
411 | if self.arg.only_train_part:
412 | if epoch > self.arg.only_train_epoch:
413 | print('only train part, require grad')
414 | for key, value in self.model.named_parameters():
415 | if 'PA' in key:
416 | value.requires_grad = True
417 | else:
418 | print('only train part, do not require grad')
419 | for key, value in self.model.named_parameters():
420 | if 'PA' in key:
421 | value.requires_grad = False
422 |
423 | nb_classes = self.arg.model_args['num_class']
424 | confusion_matrix = torch.zeros(nb_classes, nb_classes)
425 | for batch_idx, (data, label, sid, seg_id, chunk_n, anntr_id, index) in enumerate(process):
426 |
427 | self.global_step += 1
428 | # get data
429 | data = Variable(data.float().cuda(self.output_device), requires_grad=False)
430 | label = Variable(label.long().cuda(self.output_device), requires_grad=False)
431 | timer['dataloader'] += self.split_time()
432 |
433 | # forward
434 | output = self.model(data)
435 |
436 | if self.loss_type == "CE":
437 | l_type = nn.CrossEntropyLoss()
438 | loss = l_type(output, label)
439 | else:
440 | loss = CB_loss(label, output,
441 | self.samples_per_class,
442 | nb_classes, self.loss_type,
443 | self.arg.beta,
444 | self.arg.gamma,
445 | self.arg.device[0]
446 | )
447 |
448 | # backward
449 | self.optimizer.zero_grad()
450 | loss.backward()
451 | self.optimizer.step()
452 | loss_value.append(loss.data.item())
453 | timer['model'] += self.split_time()
454 |
455 | # Compute per-class acc.
456 | value, predict_label = torch.max(output.data, 1)
457 | for t, p in zip(label.view(-1), predict_label.view(-1)):
458 | confusion_matrix[t.long(), p.long()] += 1
459 |
460 | # Acc.
461 | acc = torch.mean((predict_label == label.data).float())
462 | batch_acc.append(acc.item())
463 | self.train_writer.add_scalar('acc', acc, self.global_step)
464 | self.train_writer.add_scalar('loss', loss.data.item(), self.global_step)
465 |
466 | # statistics
467 | self.lr = self.optimizer.param_groups[0]['lr']
468 | self.train_writer.add_scalar('lr', self.lr, self.global_step)
469 | # if self.global_step % self.arg.log_interval == 0:
470 | # self.print_log(
471 | # '\tBatch({}/{}) done. Loss: {:.4f} lr:{:.6f}'.format(
472 | # batch_idx, len(loader), loss.data[0], lr))
473 | timer['statistics'] += self.split_time()
474 |
475 | per_class_acc_vals = confusion_matrix.diag()/confusion_matrix.sum(1)
476 | per_class_acc = torch.mean(per_class_acc_vals).float()
477 |
478 | # statistics of time consumption and loss
479 | proportion = {
480 | k: '{:02d}%'.format(int(round(v * 100 / sum(timer.values()))))
481 | for k, v in timer.items()
482 | }
483 | self.print_log(
484 | '\tMean training loss: {:.4f}.'.format(np.mean(loss_value)))
485 | self.print_log('\tTop-1-norm: {:.3f}%'.format(100*per_class_acc))
486 |
487 | # Log
488 | wb_dict['train loss'] = np.mean(loss_value)
489 | wb_dict['train acc'] = np.mean(batch_acc)
490 |
491 | if save_model:
492 | state_dict = self.model.state_dict()
493 | weights = OrderedDict([[k.split('module.')[-1],
494 | v.cpu()] for k, v in state_dict.items()])
495 |
496 | torch.save(weights, self.arg.model_saved_name + '-' + str(epoch) + '-' + str(int(self.global_step)) + '.pt')
497 |
498 | return wb_dict
499 |
500 | @torch.no_grad()
501 | def eval(self, epoch,
502 | wb_dict,
503 | save_score=True,
504 | loader_name=['test'],
505 | wrong_file=None,
506 | result_file=None
507 | ):
508 | if wrong_file is not None:
509 | f_w = open(wrong_file, 'w')
510 | if result_file is not None:
511 | f_r = open(result_file, 'w')
512 | self.model.eval()
513 | self.print_log('Eval epoch: {}'.format(epoch + 1))
514 | for ln in loader_name:
515 | loss_value = []
516 | score_frag = []
517 | pred_label_list = []
518 | step = 0
519 | nb_classes = self.arg.model_args['num_class']
520 | confusion_matrix = torch.zeros(nb_classes, nb_classes)
521 | process = tqdm(self.data_loader[ln])
522 | for batch_idx, (data, label, sid, seg_id, chunk_n, anntr_id, index) in enumerate(process):
523 | data = Variable(
524 | data.float().cuda(self.output_device),
525 | requires_grad=False)
526 | # volatile=True)
527 | label = Variable(
528 | label.long().cuda(self.output_device),
529 | requires_grad=False)
530 | # volatile=True)
531 | output = self.model(data)
532 |
533 | if self.loss_type == "CE":
534 | l_type = nn.CrossEntropyLoss()
535 | loss = l_type(output, label)
536 | else:
537 | loss = CB_loss(label, output,
538 | self.samples_per_class,
539 | nb_classes, self.loss_type,
540 | self.arg.beta,
541 | self.arg.gamma,
542 | self.arg.device[0]
543 | )
544 | # Store outputs
545 | logits = output.data.cpu().numpy()
546 | score_frag.append(logits)
547 | loss_value.append(loss.data.item())
548 |
549 | _, predict_label = torch.max(output.data, 1)
550 | pred_label_list.append(predict_label)
551 |
552 | step += 1
553 |
554 | # Compute per-class acc.
555 | for t, p in zip(label.view(-1), predict_label.view(-1)):
556 | confusion_matrix[t.long(), p.long()] += 1
557 | if wrong_file is not None or result_file is not None:
558 | predict = list(predict_label.cpu().numpy())
559 | true = list(label.data.cpu().numpy())
560 | for i, x in enumerate(predict):
561 | if result_file is not None:
562 | f_r.write(str(x) + ',' + str(true[i]) + '\n')
563 | if x != true[i] and wrong_file is not None:
564 | f_w.write(str(index[i]) + ',' + str(x) + ',' + str(true[i]) + '\n')
565 | per_class_acc_vals = confusion_matrix.diag()/confusion_matrix.sum(1)
566 | per_class_acc = torch.mean(per_class_acc_vals).float()
567 | score = np.concatenate(score_frag)
568 | loss = np.mean(loss_value)
569 |
570 | accuracy = self.data_loader[ln].dataset.top_k(score, 1)
571 | topk_scores = { k: self.data_loader[ln].dataset.top_k(score, k) \
572 | for k in self.arg.show_topk }
573 |
574 | wb_dict['val loss'] = loss
575 | wb_dict['val acc'] = accuracy
576 | wb_dict['val per class acc'] = per_class_acc
577 | for k in topk_scores:
578 | wb_dict['val top{0} score'.format(k)] = topk_scores[k]
579 |
580 | if accuracy > self.best_acc:
581 | self.best_acc = accuracy
582 | if per_class_acc > self.best_per_class_acc:
583 | self.best_per_class_acc = per_class_acc
584 |
585 | print('Accuracy: ', accuracy, ' model: ', self.arg.model_saved_name)
586 | if self.arg.phase == 'train':
587 | self.val_writer.add_scalar('loss', loss, self.global_step)
588 | self.val_writer.add_scalar('acc', accuracy, self.global_step)
589 | self.val_writer.add_scalar('per_class_acc', per_class_acc , self.global_step)
590 |
591 | pred_scores = list(zip(
592 | self.data_loader[ln].dataset.label[1], # sid
593 | self.data_loader[ln].dataset.sample_name, # seg_id
594 | self.data_loader[ln].dataset.label[2], # chunk_id
595 | score))
596 |
597 | self.print_log('\tMean {} loss of {} batches: {}.'.format(
598 | ln, len(self.data_loader[ln]), np.mean(loss_value)))
599 | self.print_log('\tTop-1-norm: {:.3f}%'.format(100*per_class_acc))
600 | for k in topk_scores:
601 | self.print_log('\tTop{}: {:.3f}%'.format(k, 100*topk_scores[k]))
602 |
603 | if save_score:
604 | with open('{}/epoch{}_{}_score.pkl'.format(
605 | self.arg.work_dir, epoch + 1, ln), 'wb') as f:
606 | pickle.dump(pred_scores, f)
607 | return wb_dict
608 |
609 | def start(self):
610 | wb_dict = {}
611 | if self.arg.phase == 'train':
612 | self.print_log('Parameters:\n{}\n'.format(str(vars(self.arg))))
613 | self.global_step = self.arg.start_epoch * len(self.data_loader['train']) / self.arg.batch_size
614 |
615 | for epoch in range(self.arg.start_epoch, self.arg.num_epoch):
616 |
617 | save_model = ((epoch + 1) % self.arg.save_interval == 0) or (
618 | epoch + 1 == self.arg.num_epoch)
619 |
620 | # Wandb logging
621 | wb_dict = {'lr': self.lr}
622 |
623 | # Train
624 | wb_dict = self.train(epoch, wb_dict, save_model=save_model)
625 |
626 | # Eval. on val set
627 | wb_dict = self.eval(
628 | epoch,
629 | wb_dict,
630 | save_score=self.arg.save_score,
631 | loader_name=['test'])
632 | # Log stats. for this epoch
633 | print('Epoch: {0}\nMetrics: {1}'.format(epoch, wb_dict))
634 |
635 | print('best accuracy: ', self.best_acc, ' model_name: ', self.arg.model_saved_name)
636 |
637 | elif self.arg.phase == 'test':
638 | if not self.arg.test_feeder_args['debug']:
639 | wf = self.arg.model_saved_name + '_wrong.txt'
640 | rf = self.arg.model_saved_name + '_right.txt'
641 | else:
642 | wf = rf = None
643 | if self.arg.weights is None:
644 | raise ValueError('Please appoint --weights.')
645 | self.arg.print_log = False
646 | self.print_log('Model: {}.'.format(self.arg.model))
647 | self.print_log('Weights: {}.'.format(self.arg.weights))
648 |
649 | wb_dict = self.eval(epoch=0, wb_dict=wb_dict,
650 | save_score=self.arg.save_score,
651 | loader_name=['test'],
652 | wrong_file=wf,
653 | result_file=rf
654 | )
655 | print('Inference metrics: ', wb_dict)
656 | self.print_log('Done.\n')
657 |
658 |
659 | def str2bool(v):
660 | if v.lower() in ('yes', 'true', 't', 'y', '1'):
661 | return True
662 | elif v.lower() in ('no', 'false', 'f', 'n', '0'):
663 | return False
664 | else:
665 | raise argparse.ArgumentTypeError('Boolean value expected.')
666 |
667 |
668 | def import_class(name):
669 | components = name.split('.')
670 | mod = __import__(components[0])
671 | for comp in components[1:]:
672 | mod = getattr(mod, comp)
673 | return mod
674 |
675 |
676 | if __name__ == '__main__':
677 | parser = get_parser()
678 |
679 | # load arg form config file
680 | p = parser.parse_args()
681 | if p.config is not None:
682 | with open(p.config, 'r') as f:
683 | default_arg = yaml.load(f)
684 | key = vars(p).keys()
685 | for k in default_arg.keys():
686 | if k not in key:
687 | print('WRONG ARG: {}'.format(k))
688 | assert (k in key)
689 | parser.set_defaults(**default_arg)
690 |
691 | arg = parser.parse_args()
692 | print('BABEL Action Recognition')
693 | print('Config: ', arg)
694 | init_seed(0)
695 | processor = Processor(arg)
696 | processor.start()
697 |
--------------------------------------------------------------------------------
 6 |
6 |  6 |
6 |