├── .gitattributes
├── .github
└── workflows
│ └── pytest.yaml
├── .gitignore
├── LICENCE
├── README.md
├── assets
└── screencast_resized.gif
├── environment.yml
├── notebooks
├── 1_pciSeq.ipynb
├── 2_viewer.ipynb
├── 3_pciSeq_without_singleCell_data.ipynb
├── 4_diagnostics.ipynb
└── img
│ └── diagnostics.gif
├── pciSeq
├── __init__.py
├── _version.py
├── app.py
├── config.py
├── data
│ └── mouse
│ │ └── ca1
│ │ ├── iss
│ │ └── spots.csv
│ │ ├── scRNA
│ │ └── scRNAseq.csv.gz
│ │ └── segmentation
│ │ └── label_image.coo.npz
├── src
│ ├── __init__.py
│ ├── core
│ │ ├── README.md
│ │ ├── __init__.py
│ │ ├── analysis.py
│ │ ├── datatypes.py
│ │ ├── logger.py
│ │ ├── main.py
│ │ ├── summary.py
│ │ └── utils.py
│ ├── diagnostics
│ │ ├── README.md
│ │ ├── __init__.py
│ │ ├── config.py
│ │ ├── constants.py
│ │ ├── controller
│ │ │ ├── __init__.py
│ │ │ └── diagnostic_controller.py
│ │ ├── model
│ │ │ ├── __init__.py
│ │ │ └── diagnostic_model.py
│ │ ├── utils.py
│ │ └── view
│ │ │ ├── __init__.py
│ │ │ └── dashboard.py
│ ├── preprocess
│ │ ├── __init__.py
│ │ ├── cell_borders.py
│ │ ├── spot_labels.py
│ │ └── utils.py
│ ├── validation
│ │ ├── README.md
│ │ ├── __init__.py
│ │ ├── config_manager.py
│ │ └── input_validation.py
│ └── viewer
│ │ ├── __init__.py
│ │ ├── run_flask.py
│ │ ├── stage_image.py
│ │ └── utils.py
└── static
│ ├── 2D
│ ├── index.html
│ └── viewer
│ │ ├── css
│ │ ├── index.css
│ │ └── progress.css
│ │ ├── genes_datatable.html
│ │ └── js
│ │ ├── classConfig.js
│ │ ├── config.js
│ │ ├── customControl.js
│ │ ├── dapi.js
│ │ ├── dataLoader.js
│ │ ├── donut.js
│ │ ├── dt.js
│ │ ├── glyphConfig.js
│ │ ├── glyphPaths.js
│ │ ├── glyphs.js
│ │ ├── index.js
│ │ ├── lib
│ │ ├── css
│ │ │ ├── L.Control.Layers.Tree.css
│ │ │ ├── Leaflet.Coordinates-0.1.3.css
│ │ │ ├── bootstrap.min.css
│ │ │ ├── index.css
│ │ │ ├── keen-dashboards.css
│ │ │ └── screen.css
│ │ └── js
│ │ │ ├── L.Control.Layers.Tree.js
│ │ │ ├── Leaflet.Coordinates-0.1.3.src.js
│ │ │ ├── bootstrap.min.js
│ │ │ ├── keen.min.js
│ │ │ ├── leaflet-pip.js
│ │ │ ├── leaflet.textpath.js
│ │ │ ├── pixiOverlay
│ │ │ ├── MarkerContainer.js
│ │ │ ├── bezier-easing.js
│ │ │ ├── example.min.js
│ │ │ └── tools.min.js
│ │ │ └── preloader.js
│ │ ├── progress.js
│ │ ├── stage_cells.js
│ │ ├── stage_glyphs.js
│ │ ├── stage_markers_patched.js
│ │ ├── stage_polygons.js
│ │ ├── streaming-tsv-parser.js
│ │ └── viewerUtils.js
│ ├── cell_analysis
│ ├── __init__ .py
│ └── dashboard
│ │ ├── cell_index.html
│ │ └── static
│ │ ├── css
│ │ └── styles.css
│ │ └── js
│ │ ├── distanceProbabilityPlot.js
│ │ ├── interpretationGuide.js
│ │ ├── plotConfig.js
│ │ ├── scatterPlot.js
│ │ └── tooltip.js
│ └── memurai
│ └── Memurai-Developer-v3.1.4.msi
├── pdf
├── Qian_et_al_2020.pdf
└── appendix_mu.pdf
├── requirements.txt
├── setup.py
└── tests
├── conftest.py
├── constants.py
├── data
└── test_scRNAseq.csv
├── test_app.py
└── utils.py
/.gitattributes:
--------------------------------------------------------------------------------
1 | *.ipynb linguist-language=Python
--------------------------------------------------------------------------------
/.github/workflows/pytest.yaml:
--------------------------------------------------------------------------------
1 | name: Run tests
2 |
3 | on:
4 | push:
5 | branches: [ master ]
6 | pull_request:
7 | branches: [ master ]
8 |
9 | jobs:
10 | build:
11 | name: ${{ matrix.platform }} py${{ matrix.python-version }}
12 | runs-on: ${{ matrix.platform }}
13 | strategy:
14 | fail-fast: true
15 | matrix:
16 | platform: [ubuntu-latest]
17 | python-version: ["3.8", "3.9", "3.10", "3.11"]
18 |
19 | steps:
20 | - uses: actions/checkout@v3
21 | - name: Set up Python ${{ matrix.python-version }}
22 | uses: actions/setup-python@v4
23 | with:
24 | python-version: ${{ matrix.python-version }}
25 | - name: Install dependencies
26 | run: |
27 | python -m pip install pytest
28 | pip install -r requirements.txt
29 | python -m pip install -e .
30 | - name: Test with pytest
31 | run: |
32 | pytest
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | .vscode/
2 | .idea/
3 | __pycache__/
4 | .pytest_cache
5 | /tests/.pytest_cache/
6 | .ipynb_checkpoints
7 |
8 | /dist/
9 | /.eggs/
10 | /build/
11 | /pciSeq.egg-info/
12 | /pciSeq/static/2D/data/
13 |
--------------------------------------------------------------------------------
/LICENCE:
--------------------------------------------------------------------------------
1 | Copyright (c) 2018 The Python Packaging Authority
2 |
3 | Permission is hereby granted, free of charge, to any person obtaining a copy
4 | of this software and associated documentation files (the "Software"), to deal
5 | in the Software without restriction, including without limitation the rights
6 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
7 | copies of the Software, and to permit persons to whom the Software is
8 | furnished to do so, subject to the following conditions:
9 |
10 | The above copyright notice and this permission notice shall be included in all
11 | copies or substantial portions of the Software.
12 |
13 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
14 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
15 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
16 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
17 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
18 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
19 | SOFTWARE.
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # pciSeq: Probabilistic Cell typing by In situ Sequencing
2 |
3 | [](https://github.com/acycliq/pciSeq/actions)
4 | [](https://github.com/acycliq/pciSeq/)
5 | [](https://pepy.tech/project/pciSeq)
6 | [](https://pypistats.org/packages/pciSeq)
7 | [](https://github.com/acycliq/pciSeq/blob/master/LICENSE)
8 | [](https://github.com/acycliq/pciSeq/graphs/contributors)
9 |
10 | [](https://github.com/acycliq/pciSeq/)
11 | [](https://github.com/acycliq/pciSeq/)
12 |
13 |
14 | A Python package that implements the cell calling algorithm as described in [Qian, X., et al. Nature Methods (2020)](https://www.nature.com/articles/s41592-019-0631-4)
15 |
16 | 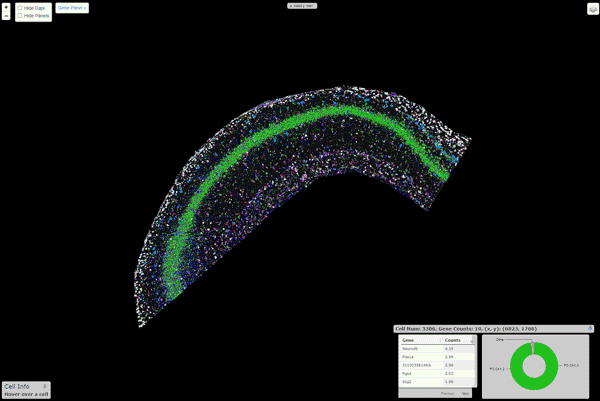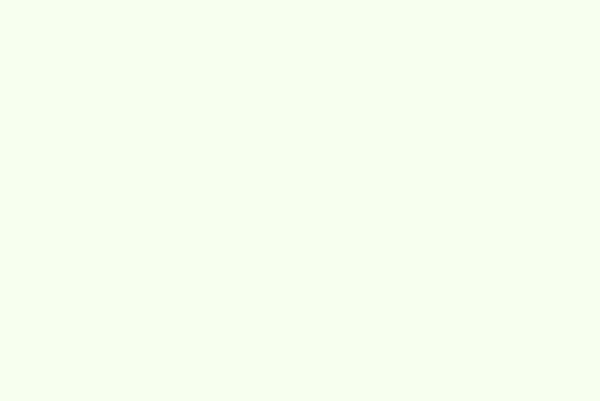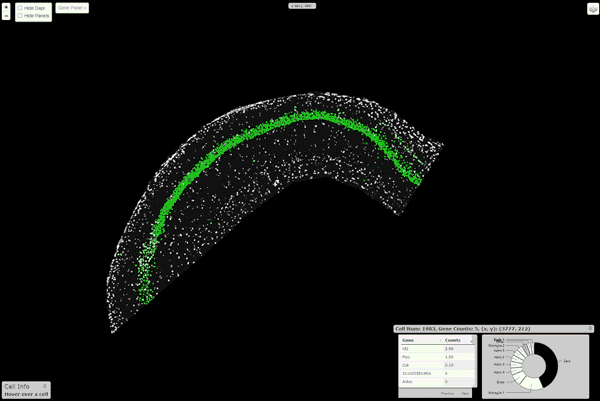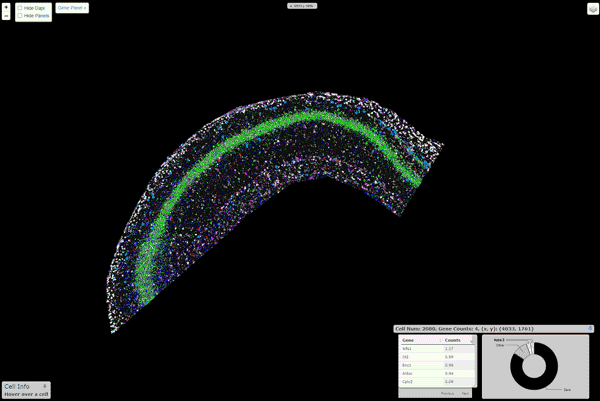 17 |
17 |
18 |
19 | ## Installation
20 | ```
21 | python -m pip install pciSeq
22 | ```
23 | Requirement: Python >= 3.8
24 |
25 | If you want to work with the source code you can download the repo and then replicate the python environment by
26 | ```
27 | conda env create -n pciSeq -f /path/to/environment.yml
28 | ```
29 |
30 | That will create a conda environment with the name `pciSeq` containing all the necessary packages to run the algorithm. To activate it run
31 | ```
32 | conda activate pciSeq
33 | ```
34 | or, if you open the project in your IDE, then in your project settings, switch your interpreter to the interpreter of the `pciSeq` env.
35 | ## Usage
36 | You need to create two `pandas dataframes` for the spots and the single cell data and a `coo_matrix` for the label image (which in
37 | most cases will be the output of some image segmentation application). Then you pass them into the `pciSeq.fit()` method as follows:
38 | ```
39 | import pciSeq
40 |
41 | res = pciSeq.fit(spots=spots_df, coo=label_image, scRNAseq=scRNA_df)
42 | ```
43 | See the demo below for a more detailed explanation about the arguments of `pciSeq.fit()` and its return values.
44 |
45 | There is also a fourth argument (optional) to override the default hyperparameter values which are initialised
46 | by the [config.py](https://github.com/acycliq/pciSeq/blob/master/pciSeq/config.py) module. To pass user-defined hyperparameter values, create a `dictionary` with `keys` the
47 | hyperparameter names and `values` their new values. For example, to exclude all Npy and Vip spots you can do:
48 |
49 | ```
50 | import pciSeq
51 |
52 | opts = { 'exclude_genes': ['Npy', 'Vip'] }
53 | res = pciSeq.fit(spots=spots_df, coo=label_image, scRNAseq=scRNA_df, opts=opts)
54 | ```
55 |
56 | ## Demo
57 | You can run a pciSeq demo in google colab: [](https://colab.research.google.com/github/acycliq/pciSeq/blob/master/notebooks/1_pciSeq.ipynb)
58 |
59 | ## Viewer
60 | An interactive viewer to explore the data runs on this [url](https://acycliq.github.io/visage/). Instructions about
61 | building this viewer with your own data are [here](https://github.com/acycliq/visage). \
62 | If you have `v 0.0.49` or greater you can also launch the viewer automatically by
63 | setting `opts = {'launch_viewer': True}` and passing it to `pciSeq.fit()`, see [](https://colab.research.google.com/github/acycliq/pciSeq/blob/master/notebooks/2_viewer.ipynb)
64 |
65 | ## Diagnostics
66 | Diagnostics will help you understand whether pciSeq has been misconfigured, the algorithm has taken the
67 | wrong path and will produce meaningless results when it finishes. You will need however to install redis (or Memurai if you are using Windows).
68 |
69 | For linux do:
70 | `sudo apt-get install redis-server redis-tools`
71 | and then start the service:
72 | `sudo service redis-server start`
73 |
74 | You can get the free version of memurai from [here](https://www.memurai.com/get-memurai). Once installed, the service should start automatically but you can manually do that by:
75 | `memurai.exe –service-start`
76 |
77 | If redis (or memurai) is missing from your system, the call to launch the diagnostics dashboard will be
78 | ignored. If you are interested in this feature you may find this notebook [](https://colab.research.google.com/github/acycliq/pciSeq/blob/master/notebooks/4_diagnostics.ipynb)
79 | useful
80 |
81 | ## Change Log
82 | ### [0.0.59] - 2023-09-22
83 | - Fixed a SIGSERV error under pandas '2.1.1'
84 |
85 | ### [0.0.56] - 2023-07-03
86 | - Diagnostics dashboard
87 |
88 | - Baselayers on the viewer. You can have multiple background images and switch between them.
89 |
90 | ### [0.0.50] - 2023-05-27
91 | - Single cell data are optional, more info can be found here [](https://colab.research.google.com/github/acycliq/pciSeq/blob/master/notebooks/3_pciSeq_without_singleCell_data.ipynb)
92 |
93 | - `pciSeq.fit()` takes keyword arguments
94 |
95 |
96 | ## References
97 | Qian, X., et al. (2020). Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat
98 | Methods 17, 101 - 106.
99 |
100 |
101 |
--------------------------------------------------------------------------------
/assets/screencast_resized.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/assets/screencast_resized.gif
--------------------------------------------------------------------------------
/environment.yml:
--------------------------------------------------------------------------------
1 | name: pciSeq
2 | channels:
3 | - conda-forge
4 | dependencies:
5 | - pip
6 | - python=3.8
7 | - pip:
8 | - -r requirements.txt
--------------------------------------------------------------------------------
/notebooks/img/diagnostics.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/notebooks/img/diagnostics.gif
--------------------------------------------------------------------------------
/pciSeq/__init__.py:
--------------------------------------------------------------------------------
1 | import subprocess
2 | import sys
3 | import os
4 | from pciSeq._version import __version__
5 | from pciSeq.app import fit
6 | from pciSeq.app import cell_type
7 | from pciSeq.src.preprocess.spot_labels import stage_data
8 | import pciSeq.src.core.utils as utils
9 | from pciSeq.src.core.logger import attach_to_log, setup_logger
10 | from pciSeq.src.core.analysis import CellExplorer
11 | import logging
12 |
13 | init_logger = logging.getLogger(__name__)
14 |
15 |
16 | def confirm_prompt(question):
17 | reply = None
18 | while reply not in ("", "y", "n"):
19 | reply = input(f"{question} (y/n): ").lower()
20 | return (reply in ("", "y"))
21 |
22 |
23 | def install(package):
24 | subprocess.check_call([sys.executable, "-m", "pip", "install", package])
25 |
26 |
27 | def install_libvips():
28 | try:
29 | subprocess.check_call("apt-get update", shell=True)
30 | subprocess.check_call("apt-get install -y libvips", shell=True) # Combined command with -y flag
31 | subprocess.check_call([sys.executable, "-m", "pip", "install", "pyvips"])
32 | return True
33 | except subprocess.CalledProcessError as e:
34 | init_logger.error(f"Failed to install libvips: {str(e)}")
35 | return False
36 |
37 | def check_libvips():
38 | try:
39 | import pyvips
40 | return True
41 | except ImportError: # More specific exception
42 | init_logger.warning('libvips not found. Please install it manually or run with sudo privileges.')
43 | return False
44 | except Exception as err:
45 | init_logger.error(f"Unexpected error checking libvips: {str(err)}")
46 | return False
47 |
48 |
49 | if check_libvips():
50 | from pciSeq.src.viewer.stage_image import tile_maker
51 | else:
52 | def tile_maker():
53 | init_logger.warning('>>>> tile_maker() because libvips is not installed. Please see https://www.libvips.org/install.html <<<<')
54 | init_logger.warning('>>>> If you are on Linux you can install it by calling: sudo apt install libvips <<<<')
55 |
56 |
57 |
58 |
--------------------------------------------------------------------------------
/pciSeq/_version.py:
--------------------------------------------------------------------------------
1 | __version__ = '0.0.61'
--------------------------------------------------------------------------------
/pciSeq/app.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | from typing import Tuple, Optional, Dict, Any
3 | from .src.validation.config_manager import ConfigManager
4 | from .src.validation.input_validation import InputValidator
5 | from .src.core.main import VarBayes
6 | from .src.core.utils import write_data, pre_launch
7 | from .src.viewer.run_flask import flask_app_start
8 | from .src.preprocess.spot_labels import stage_data
9 | import logging
10 |
11 | app_logger = logging.getLogger(__name__)
12 |
13 |
14 | def fit(*args, **kwargs) -> Tuple[pd.DataFrame, pd.DataFrame]:
15 | """
16 | Main entry point for pciSeq cell typing analysis.
17 |
18 | Parameters
19 | ----------
20 | *args : tuple
21 | Positional arguments:
22 | - args[0]: pd.DataFrame containing spot data
23 | - args[1]: scipy.sparse.coo_matrix containing gene expression data
24 |
25 | **kwargs : dict
26 | Keyword arguments:
27 | - spots: pd.DataFrame
28 | Spot data with gene expressions and coordinates
29 | - coo: scipy.sparse.coo_matrix
30 | Sparse matrix of gene expression data
31 | - scRNAseq: pd.DataFrame, optional
32 | Single-cell RNA sequencing reference data
33 | - opts: dict, optional
34 | Configuration options for the analysis
35 |
36 | Returns
37 | -------
38 | Tuple[pd.DataFrame, pd.DataFrame]
39 | - cellData: DataFrame containing cell typing results and metadata
40 | - geneData: DataFrame containing gene assignment results
41 |
42 | Raises
43 | ------
44 | ValueError
45 | If required arguments (spots and coo) are missing or invalid
46 | RuntimeError
47 | If cell typing algorithm fails to converge
48 |
49 | Notes
50 | -----
51 | The function can be called either with positional arguments (spots, coo)
52 | or with keyword arguments. If both are provided, keyword arguments take precedence.
53 | """
54 | try:
55 | # 1. parse/check the arguments
56 | spots, coo, scRNAseq, opts = parse_args(*args, **kwargs)
57 |
58 | # 2. Create and validate config
59 | cfg_man = ConfigManager.from_opts(opts)
60 | cfg_man.set_runtime_attributes()
61 | spots, coo, scdata, cfg = InputValidator.validate(spots, coo, scRNAseq, cfg_man)
62 |
63 | # 3. Use validated inputs and prepare the data
64 | app_logger.info('Preprocessing data')
65 | _cells, cellBoundaries, _spots = stage_data(spots, coo)
66 |
67 | # 4. cell typing (diagnostics are now handled inside VarBayes)
68 | cellData, geneData, varBayes = cell_type(_cells, _spots, scdata, cfg)
69 |
70 | # 5. Save data and launch viewer if needed
71 | if (cfg['save_data'] and varBayes.has_converged) or cfg['launch_viewer']:
72 | write_data(cellData, geneData, cellBoundaries, varBayes, cfg)
73 |
74 | if cfg['launch_viewer']:
75 | dst = pre_launch(cellData, coo, scRNAseq, cfg)
76 | flask_app_start(dst)
77 |
78 | app_logger.info('Done')
79 | return cellData, geneData
80 |
81 | except Exception as e:
82 | app_logger.error(f"Error in fit function: {str(e)}")
83 | raise
84 |
85 |
86 | def cell_type(
87 | cells: pd.DataFrame,
88 | spots: pd.DataFrame,
89 | scRNAseq: Optional[pd.DataFrame],
90 | config: Dict[str, Any]
91 | ) -> Tuple[pd.DataFrame, pd.DataFrame, VarBayes]:
92 | """
93 | Perform cell typing using Variational Bayes algorithm.
94 |
95 | Parameters
96 | ----------
97 | cells : pd.DataFrame
98 | Preprocessed cell data containing cell locations and boundaries
99 | spots : pd.DataFrame
100 | Preprocessed spot data containing gene expressions and coordinates
101 | scRNAseq : Optional[pd.DataFrame]
102 | Single-cell RNA sequencing reference data. Can be None if not using reference data
103 | config : Dict[str, Any]
104 | Configuration dictionary containing algorithm parameters
105 |
106 | Returns
107 | -------
108 | Tuple[pd.DataFrame, pd.DataFrame, VarBayes]
109 | - cellData: DataFrame containing cell typing results
110 | - geneData: DataFrame containing gene assignment results
111 | - varBayes: The fitted VarBayes model instance
112 |
113 | Raises
114 | ------
115 | ValueError
116 | If input data is invalid or incompatible
117 | RuntimeError
118 | If cell typing algorithm fails to converge
119 | """
120 | try:
121 | app_logger.info('Initializing VarBayes model')
122 | varBayes = VarBayes(cells, spots, scRNAseq, config)
123 |
124 | app_logger.info('Starting cell typing algorithm')
125 | cellData, geneData = varBayes.run()
126 |
127 | if not varBayes.has_converged:
128 | app_logger.warning('Cell typing algorithm did not fully converge')
129 |
130 | return cellData, geneData, varBayes
131 |
132 | except Exception as e:
133 | app_logger.error(f"Error during cell typing: {str(e)}")
134 | raise RuntimeError(f"Cell typing failed: {str(e)}") from e
135 |
136 |
137 | def parse_args(*args, **kwargs) -> Tuple[pd.DataFrame, Any, Optional[pd.DataFrame], Optional[Dict]]:
138 | """Parse and validate input arguments.
139 |
140 | Returns:
141 | Tuple containing (spots, coo, scRNAseq, opts)
142 |
143 | Raises:
144 | ValueError: If required arguments are missing or invalid
145 | """
146 | # Check if we have the minimum required arguments
147 | if not {'spots', 'coo'}.issubset(set(kwargs)) and len(args) < 2:
148 | raise ValueError('Need to provide the spots and the coo matrix either as keyword '
149 | 'arguments or as the first and second positional arguments.')
150 |
151 | # Get spots from kwargs if present, otherwise from args
152 | spots = kwargs['spots'] if 'spots' in kwargs else args[0]
153 |
154 | # Get coo from kwargs if present, otherwise from args
155 | coo = kwargs['coo'] if 'coo' in kwargs else args[1]
156 |
157 | # Optional arguments
158 | scRNAseq = kwargs.get('scRNAseq', None)
159 | opts = kwargs.get('opts', None)
160 |
161 | if spots is None or coo is None:
162 | raise ValueError("Both 'spots' and 'coo' must be provided")
163 |
164 | return spots, coo, scRNAseq, opts
165 |
166 |
167 |
168 |
169 |
170 |
171 |
--------------------------------------------------------------------------------
/pciSeq/config.py:
--------------------------------------------------------------------------------
1 | """
2 | hyperparameters for the pciSeq method
3 | """
4 | import numpy as np
5 |
6 | DEFAULT = {
7 |
8 | # list of genes to be excluded during cell-typing, e.g ['Aldoc', 'Id2'] to exclude all spots from Aldoc and Id2
9 | 'exclude_genes': [],
10 |
11 | # Maximum number of loops allowed for the Variational Bayes to run
12 | 'max_iter': 1000,
13 |
14 | # Convergence achieved if assignment probabilities between two successive loops is less than the tolerance
15 | 'CellCallTolerance': 0.02,
16 |
17 | # A gamma distribution expresses the efficiency of the in-situ sequencing for each gene. It tries to capture
18 | # the ratio of the observed over the theoretical counts for a given gene. rGene controls the variance and
19 | # Inefficiency is the average of this assumed Gamma distribution
20 | 'rGene': 20,
21 | 'Inefficiency': 0.2,
22 |
23 | # If a spot is inside the cell boundaries this bonus will give the likelihood an extra boost
24 | # in order to make the spot more probable to get assigned to the cell than another spot positioned
25 | # outside the cell boundaries
26 | 'InsideCellBonus': 2,
27 |
28 | # To account for spots far from the some a uniform distribution is introduced to describe those misreads.
29 | # By default this uniform distribution has a density of 1e-5 misreads per pixel.
30 | 'MisreadDensity': 0.00001,
31 |
32 | # Gene detection might come with irregularities due to technical errors. A small value is introduced
33 | # here to account for these errors. It is an additive factor, applied to the single cell expression
34 | # counts when the mean counts per class and per gene are calculated.
35 | 'SpotReg': 0.1,
36 |
37 | # By default only the 3 nearest cells will be considered as possible parent cells for any given spot.
38 | # There is also one extra 'super-neighbor', which is always a neighbor to the spots so we can assign
39 | # the misreads to. Could be seen as the background. Hence, by default the algorithm tries examines
40 | # whether any of the 3 nearest cells is a possible parent cell to a given cell or whether the spot is
41 | # a misread
42 | 'nNeighbors': 3,
43 |
44 | # A gamma distributed variate from Gamma(rSpot, 1) is applied to the mean expression, hence the counts
45 | # are distributed according to a Negative Binomial distribution.
46 | # The value for rSpot will control the variance/dispersion of the counts
47 | 'rSpot': 2,
48 |
49 | # Boolean, if True the output will be saved as tsv files in a folder named 'pciSeq' in your system's temp dir.
50 | 'save_data': True,
51 |
52 | # Set here where the results will be saved. If default then they will be saved at your system's temp folder
53 | 'output_path': 'default',
54 |
55 | # if true the viewer will be launched once convergence has been achieved
56 | 'launch_viewer': False,
57 |
58 | 'launch_diagnostics': False,
59 |
60 | # Runtime attribute (automatically set during execution)
61 | 'is_redis_running': False,
62 |
63 | # cell radius. If None then pciSeq will calc that as the mean radius across all cells.
64 | # Otherwise it will use the value provided below
65 | 'cell_radius': None,
66 |
67 | # cell type prior: The prior distribution on the classes. It expresses the view on
68 | # how likely each class is to occur a-priori. It can be either 'uniform' or 'weighted'
69 | # 'uniform' means that the Zero class gets 50% and the remaining 50% is equally split
70 | # on the cell classes.
71 | # 'weighted' means that the cell type which is more likely to occur will be given more
72 | # weight. These weights are calculated dynamically within the algorithm based on
73 | # a Dirichlet distribution assumption.
74 | 'cell_type_prior': 'uniform',
75 |
76 |
77 | # *******************************************************************************
78 | # Hyperparameters below come into action **ONLY** if single cell data are missing
79 | # *******************************************************************************
80 | 'mean_gene_counts_per_class': 60,
81 | 'mean_gene_counts_per_cell': 15,
82 |
83 | }
84 |
85 |
--------------------------------------------------------------------------------
/pciSeq/data/mouse/ca1/scRNA/scRNAseq.csv.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/data/mouse/ca1/scRNA/scRNAseq.csv.gz
--------------------------------------------------------------------------------
/pciSeq/data/mouse/ca1/segmentation/label_image.coo.npz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/data/mouse/ca1/segmentation/label_image.coo.npz
--------------------------------------------------------------------------------
/pciSeq/src/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/core/README.md:
--------------------------------------------------------------------------------
1 | # pciSeq Core Module
2 |
3 | This module contains the core functionality of pciSeq, implementing the Variational Bayes algorithm for spatial transcriptomics analysis.
4 |
5 | ## Key Components
6 |
7 | ### 1. Main Algorithm (`main.py`)
8 | - Implements the VarBayes class for iterative optimization
9 | - Handles convergence and parameter updates
10 | - Manages the overall analysis pipeline
11 |
12 | ### 2. Data Types (`datatypes.py`)
13 | - Defines fundamental data structures:
14 | * Cells: Segmented cells data and features
15 | * Genes: Gene-specific features, like inefficiency.
16 | * Spots: Gene-read features, like spot to cell assignment probabilities
17 | * SingleCell: scRNA-seq reference
18 | * CellType: Cell type prior probabilities
19 |
20 | ### 3. Summary Functions (`summary.py`)
21 | - Processes analysis results
22 | - Formats data for visualization
23 | - Generates cell and spot summaries
24 |
25 | ### 4. Utilities (`utils.py`)
26 | - Helper functions for computation
27 | - File handling utilities
28 | - Convergence checking
29 |
30 | ### 5. Analysis (`analysis.py`)
31 | - Data analysis dashboard
32 |
33 | ### 6. Logging (`logger.py`)
34 | - Standardized logging setup
35 | - Color-coded output levels
36 | - Consistent formatting
37 |
38 | ## Dependencies
39 | - numpy: Numerical computations
40 | - pandas: Data management
41 | - scipy: Statistical operations
42 | - dask: Delayed computations
43 | - sklearn: Nearest neighbor calculations
44 | - numpy_groupies: Group operations
45 |
46 |
--------------------------------------------------------------------------------
/pciSeq/src/core/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/core/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/core/logger.py:
--------------------------------------------------------------------------------
1 | """
2 | Logging Configuration Module for pciSeq
3 |
4 | This module provides a standardized logging setup for the pciSeq package,
5 | with colored output and consistent formatting across all modules.
6 |
7 | Key Functions:
8 | ------------
9 | setup_logger:
10 | Main configuration function that:
11 | - Sets up colored console output
12 | - Configures log levels and formats
13 | - Returns a configured logger instance
14 |
15 | attach_to_log:
16 | Legacy function maintained for backwards compatibility
17 | (Redirects to setup_logger)
18 |
19 | Features:
20 | --------
21 | - Color-coded output by log level:
22 | * DEBUG: Cyan
23 | * INFO: Green
24 | * WARNING: Yellow
25 | * ERROR: Red
26 | * CRITICAL: Red with white background
27 |
28 | - Standardized timestamp format
29 | - Console output to stdout
30 | - Configurable log levels
31 | - Thread-safe logging
32 |
33 | Usage:
34 | -----
35 | >>> logger = setup_logger()
36 | >>> logger.info("Analysis started")
37 | >>> logger.error("Error occurred")
38 |
39 | Notes:
40 | -----
41 | - Default log level is INFO
42 | - All handlers are cleared before setup to avoid duplication
43 | - Uses colorlog for ANSI color support
44 | - Format: "timestamp - level - message"
45 | """
46 |
47 | import sys
48 | import colorlog
49 | import logging
50 |
51 |
52 | def attach_to_log():
53 | """
54 | exists only for backwards compatibility.
55 | Replaced by logger_setup
56 | """
57 | setup_logger()
58 |
59 |
60 | def setup_logger(level=None):
61 |
62 | if level is None:
63 | level = logging.INFO
64 |
65 | # Remove all handlers associated with the root logger object.
66 | for handler in logging.root.handlers[:]:
67 | logging.root.removeHandler(handler)
68 |
69 | # Create color formatter
70 | color_formatter = colorlog.ColoredFormatter(
71 | "%(log_color)s%(asctime)s - %(levelname)s - %(message)s",
72 | log_colors={
73 | 'DEBUG': 'cyan',
74 | 'INFO': 'green',
75 | 'WARNING': 'yellow',
76 | 'ERROR': 'red',
77 | 'CRITICAL': 'red,bg_white',
78 | },
79 | reset=True,
80 | style='%'
81 | )
82 |
83 | # Console handler
84 | console_handler = colorlog.StreamHandler(sys.stdout)
85 | console_handler.setFormatter(color_formatter)
86 |
87 | logging.basicConfig(
88 | level=logging.INFO,
89 | format="%(asctime)s [%(levelname)s] %(message)s",
90 | handlers=[
91 | console_handler
92 | ]
93 | )
94 | logger = logging.getLogger('pciSeq')
95 |
96 | return logger
97 |
--------------------------------------------------------------------------------
/pciSeq/src/core/summary.py:
--------------------------------------------------------------------------------
1 | """
2 | Data Summary Module for pciSeq
3 |
4 | This module provides functionality to summarize and format the results of the
5 | pciSeq analysis for visualization and reporting. It processes both cell and spot
6 | data into structured DataFrames.
7 |
8 | Key Functions:
9 | ------------
10 | cells_summary:
11 | Summarizes cell-level information including:
12 | - Spatial coordinates
13 | - Gene counts
14 | - Cell type probabilities
15 | - Gaussian contours for visualization
16 | Returns a DataFrame with one row per cell.
17 |
18 | spots_summary:
19 | Summarizes spot-level information including:
20 | - Gene identities
21 | - Spatial coordinates
22 | - Cell assignments and probabilities
23 | Returns a DataFrame with one row per spot.
24 |
25 | collect_data:
26 | Main interface function that combines cell and spot summaries
27 | for downstream visualization and analysis.
28 |
29 | Data Structure:
30 | -------------
31 | Cell Summary DataFrame:
32 | - Cell_Num: Unique cell identifier
33 | - X, Y: Spatial coordinates
34 | - Genenames: List of detected genes
35 | - CellGeneCount: Corresponding gene counts
36 | - ClassName: Assigned cell types
37 | - Prob: Cell type probabilities
38 | - gaussian_contour: Cell boundary coordinates
39 |
40 | Spot Summary DataFrame:
41 | - Gene: Gene name
42 | - Gene_id: Numeric gene identifier
43 | - x, y: Spatial coordinates
44 | - neighbour: Most likely parent cell
45 | - neighbour_array: Array of nearby cells
46 | - neighbour_prob: Assignment probabilities
47 |
48 | Notes:
49 | -----
50 | - First cell (index 0) is treated as background and excluded from summaries
51 | - Gene counts and probabilities below tolerance (0.001) are filtered
52 | - Gaussian contours are computed at 3 standard deviations
53 | """
54 |
55 | import numpy as np
56 | import pandas as pd
57 | from pciSeq.src.core.utils import gaussian_contour
58 | import logging
59 |
60 | summary_logger = logging.getLogger(__name__)
61 |
62 |
63 | def cells_summary(cells, genes, single_cell):
64 | '''
65 | returns a dataframe summarising the main features of each cell, ie gene counts and cell types
66 | :param spots:
67 | :return:
68 | '''
69 | iCounts = np.argsort(-1 * cells.geneCount, axis=1)
70 | gene_names = genes.gene_panel[iCounts]
71 | gene_count = np.take_along_axis(cells.geneCount, iCounts, axis=1)
72 |
73 | iProb = np.argsort(-1 * cells.classProb, axis=1)
74 | class_names = single_cell.classes[iProb]
75 | class_prob = np.take_along_axis(cells.classProb, iProb, axis=1)
76 |
77 | tol = 0.001
78 |
79 | summary_logger.info('Start collecting data ...')
80 |
81 | isCount_nonZero = [d > tol for d in gene_count]
82 | name_list = [list(gene_names[i][d]) for (i, d) in enumerate(isCount_nonZero)]
83 | count_list = [gene_count[i][d].tolist() for (i, d) in enumerate(isCount_nonZero)]
84 |
85 | isProb_nonZero = [d > tol for d in class_prob]
86 | class_name_list = [list(class_names[i][d]) for (i, d) in enumerate(isProb_nonZero)]
87 | prob_list = [class_prob[i][d].tolist() for (i, d) in enumerate(isProb_nonZero)]
88 |
89 | contour = []
90 | for i in range(cells.nC):
91 | # ea = cells.ellipsoid_attributes[i]
92 | mu = cells.centroid.iloc[i].values
93 | cov = cells.cov[i]
94 | ellipsis = gaussian_contour(mu[:2], cov[:2, :2], 3).astype(np.int64)
95 | contour.append(ellipsis.tolist())
96 |
97 | iss_df = pd.DataFrame({'Cell_Num': cells.centroid.index.tolist(),
98 | 'X': cells.centroid['x'].round(3).tolist(),
99 | 'Y': cells.centroid['y'].round(3).tolist(),
100 | 'Genenames': name_list,
101 | 'CellGeneCount': count_list,
102 | 'ClassName': class_name_list,
103 | 'Prob': prob_list,
104 | 'gaussian_contour': contour
105 | })
106 | iss_df.set_index(['Cell_Num'])
107 |
108 | # Ignore the first row. It is the pseudocell to keep the misreads (ie the background)
109 | iss_df = iss_df[1:]
110 | summary_logger.info('Data collected!')
111 | return iss_df
112 |
113 |
114 | def spots_summary(spots):
115 | # check for duplicates (ie spots with the same coordinates with or without the same gene name).
116 | # is_duplicate = spots.data.duplicated(subset=['x', 'y'])
117 |
118 | idx = np.argsort(-1 * spots.parent_cell_prob, axis=1)
119 | p = np.take_along_axis(spots.parent_cell_prob, idx, axis=1).round(3)
120 | nbrs = np.take_along_axis(spots.parent_cell_id, idx, axis=1)
121 | max_nbrs = nbrs[:, 0]
122 |
123 | out = pd.DataFrame({'Gene': spots.data.gene_name.tolist(),
124 | 'Gene_id': spots.gene_id.tolist(),
125 | 'x': spots.data.x.tolist(),
126 | 'y': spots.data.y.tolist(),
127 | 'neighbour': max_nbrs.tolist(),
128 | 'neighbour_array': nbrs.tolist(),
129 | 'neighbour_prob': p.tolist()
130 | })
131 |
132 | return out
133 |
134 |
135 | def collect_data(cells, spots, genes, single_cell):
136 | '''
137 | Collects data for the viewer
138 | :param cells:
139 | :param spots:
140 | :return:
141 | '''
142 | cell_df = cells_summary(cells, genes, single_cell)
143 | gene_df = spots_summary(spots)
144 | return cell_df, gene_df
145 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/README.md:
--------------------------------------------------------------------------------
1 | # pciSeq Diagnostics System
2 |
3 | The pciSeq Diagnostics System implements a Model-View-Controller (MVC) architecture with Redis pub/sub for real-time communication between components. This design enables efficient monitoring and visualization of the pciSeq algorithm's diagnostic data.
4 |
5 | ## Architecture Overview
6 |
7 | ┌─────────────────────┐
8 | │ Redis Pub/Sub │
9 | │ Message Broker │
10 | └─────────┬───────────┘
11 | │
12 | ┌──────────┼──────────┐
13 | │ │
14 | Publish Subscribe
15 | ▲ ▲
16 | │ │
17 | │ │
18 | ┌──────────┐ ┌─────────┐ ┌──────────────┐
19 | │ │ │ │ │ │
20 | │Algorithm ├───►│ Model │ │ View │
21 | │ │ │ │ │ Dashboard │
22 | └──────────┘ └────┬────┘ └──────┬───────┘
23 | ▲ ▲
24 | │ ┌──────────┐ │
25 | └─── │Controller│───┘
26 | │ │
27 | └──────────┘
28 |
29 |
30 |
31 | ## Components
32 |
33 | ### Model (`model/diagnostic_model.py`)
34 | - Takes algorithm data (gene efficiency and cell type data)
35 | - Converts it to JSON strings with metadata
36 | - Stores in Redis and notifies subscribers
37 | - Initializes basic Redis connection
38 |
39 | ### View (`view/dashboard.py`)
40 | - Implements Streamlit dashboard
41 | - Subscribes to Redis channels
42 | - Updates visualizations when new data arrives
43 |
44 | ### Controller (`controller/diagnostic_controller.py`)
45 | Simple coordinator that:
46 | - Launches the dashboard
47 | - Passes algorithm updates to Model
48 |
49 | ### Redis
50 | Separate database service that:
51 | - Stores data as strings
52 | - Enables pub/sub messaging between Model and View
53 |
54 | ## How It Works
55 |
56 | 1. **Startup**
57 | - Controller launches Streamlit dashboard
58 | - View subscribes to Redis channels
59 | - Redis server must be running
60 |
61 | 2. **During Algorithm Run**
62 | - pciSeq produces new data
63 | - Controller tells Model to publish it
64 | - Model converts to JSON and stores in Redis
65 | - Model notifies subscribers
66 | - View updates dashboard
67 |
68 | ## Redis Channels
69 |
70 | Two main channels:
71 | - `gene_efficiency`: Gene efficiency metrics
72 | - `cell_type_posterior`: Cell type distribution data
73 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/diagnostics/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/config.py:
--------------------------------------------------------------------------------
1 | import redis
2 |
3 | SETTINGS = {
4 | # 'DB_URL': os.path.join(get_out_dir(pciSeq_cfg._BASE['output_path'], ''), 'pciSeq.db'),
5 | 'POOL': redis.ConnectionPool(host='localhost', port=6379, db=0)
6 | }
7 |
8 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/constants.py:
--------------------------------------------------------------------------------
1 | from enum import Enum, auto
2 |
3 |
4 | class DiagnosticKeys(Enum):
5 | """
6 | This enumeration serves as the centralised source for all keys used in
7 | Redis publish/subscribe operations throughout the diagnostics package:
8 | - RedisPublisher uses these keys to publish diagnostic data
9 | - DiagnosticsModel uses these keys to subscribe to and retrieve data
10 |
11 | Current keys:
12 | - GENE_EFFICIENCY: Used for gene efficiency metrics
13 | - CELL_TYPE_POSTERIOR: Used for cell type distribution data
14 | """
15 | GENE_EFFICIENCY = "gene_efficiency"
16 | CELL_TYPE_POSTERIOR = "cell_type_posterior"
17 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/controller/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/diagnostics/controller/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/controller/diagnostic_controller.py:
--------------------------------------------------------------------------------
1 | """
2 | Controller component of the MVC pattern for pciSeq diagnostics.
3 |
4 | This module implements the Controller layer, responsible for:
5 | 1. Coordinating between Model and View components
6 | 2. Managing dashboard lifecycle (start/stop)
7 | 3. Handling data flow from algorithm to diagnostics
8 | 4. System initialization and shutdown
9 |
10 | The Controller acts as the central coordinator, managing the flow of data
11 | and control between the Model and View components.
12 | """
13 |
14 | import os
15 | import subprocess
16 | import logging
17 | import signal
18 | from pathlib import Path
19 | from typing import Optional
20 | import tomlkit
21 |
22 | from pciSeq.src.diagnostics.model.diagnostic_model import DiagnosticModel
23 |
24 | controller_logger = logging.getLogger(__name__)
25 |
26 |
27 | class DiagnosticController:
28 | """
29 | Main controller class coordinating the diagnostics system.
30 |
31 | This class manages the interaction between the Model (diagnostic_model.py)
32 | and View (dashboard.py) components, handling system lifecycle and data flow.
33 | """
34 |
35 | def __init__(self):
36 | """Initialize controller with required components."""
37 | self.model = DiagnosticModel()
38 | self.dashboard_process: Optional[subprocess.Popen] = None
39 | self._setup_signal_handlers()
40 |
41 | def _setup_signal_handlers(self) -> None:
42 | """Setup handlers for clean shutdown on system signals."""
43 | signal.signal(signal.SIGINT, self._signal_handler)
44 | signal.signal(signal.SIGTERM, self._signal_handler)
45 |
46 | def _signal_handler(self, signum: int, frame) -> None:
47 | """
48 | Handle system signals for clean shutdown.
49 |
50 | Args:
51 | signum: Signal number
52 | frame: Current stack frame
53 | """
54 | controller_logger.info(f"Received signal {signum}, initiating shutdown...")
55 | self.shutdown()
56 |
57 | def initialize(self) -> bool:
58 | """
59 | Initialize the diagnostics system.
60 | Returns:
61 | bool: True if critical initialization successful
62 | """
63 | try:
64 | self._setup_streamlit_credentials()
65 | controller_logger.info("Diagnostics system initialized successfully")
66 | return True
67 | except Exception as e:
68 | controller_logger.error(f"Failed to initialize diagnostics: {e}")
69 | return False
70 |
71 | def launch_dashboard(self) -> bool:
72 | """
73 | Launch the Streamlit dashboard (View component).
74 |
75 | Returns:
76 | bool: True if dashboard launched successfully, False otherwise
77 | """
78 | if not self.initialize():
79 | return False
80 |
81 | try:
82 | # Get path to dashboard script
83 | dirname = os.path.dirname(os.path.dirname(__file__))
84 | dashboard_path = os.path.join(dirname, 'view', 'dashboard.py')
85 |
86 | # Launch dashboard process
87 | self.dashboard_process = subprocess.Popen([
88 | "streamlit", "run", dashboard_path, " --server.headless true"
89 | ])
90 |
91 | controller_logger.info(f'Started dashboard with PID: {self.dashboard_process.pid}')
92 | return True
93 | except subprocess.SubprocessError as e:
94 | controller_logger.error(f"Failed to start dashboard: {e}")
95 | return False
96 |
97 | def update_diagnostics(self, algorithm_model, iteration: int, has_converged: bool) -> None:
98 | """
99 | Update diagnostic data through the Model.
100 |
101 | Args:
102 | algorithm_model: Current state of the algorithm
103 | iteration: Current iteration number
104 | has_converged: Whether the algorithm has converged
105 | """
106 | try:
107 | self.model.publish_diagnostics(
108 | algorithm_model=algorithm_model,
109 | iteration=iteration,
110 | has_converged=has_converged
111 | )
112 | controller_logger.debug(f"Updated diagnostics for iteration {iteration}")
113 | except Exception as e:
114 | controller_logger.error(f"Failed to update diagnostics: {e}")
115 |
116 | def shutdown(self) -> None:
117 | """Clean shutdown of all components."""
118 | if self.dashboard_process:
119 | try:
120 | self.dashboard_process.terminate()
121 | self.dashboard_process.wait(timeout=5)
122 | controller_logger.info(f'Terminated dashboard with PID: {self.dashboard_process.pid}')
123 | except subprocess.TimeoutExpired:
124 | self.dashboard_process.kill()
125 | controller_logger.warning(f'Forced dashboard termination with PID: {self.dashboard_process.pid}')
126 | finally:
127 | self.dashboard_process = None
128 |
129 | @staticmethod
130 | def _setup_streamlit_credentials() -> bool:
131 | """
132 | Setup Streamlit credentials to bypass welcome message.
133 | Returns:
134 | bool: True if setup successful, False otherwise
135 | """
136 | credentials_path = os.path.join(Path.home(), '.streamlit', 'credentials.toml')
137 | Path(credentials_path).parent.mkdir(parents=True, exist_ok=True)
138 |
139 | try:
140 | mode = 'rt' if os.path.exists(credentials_path) else 'w+'
141 | with open(credentials_path, mode) as fp:
142 | doc = tomlkit.load(fp) if mode == 'rt' else tomlkit.document()
143 |
144 | if 'general' not in doc:
145 | doc['general'] = tomlkit.table()
146 | if 'email' not in doc['general']:
147 | doc['general']['email'] = ""
148 |
149 | with open(credentials_path, 'w') as outfile:
150 | tomlkit.dump(doc, outfile)
151 |
152 | controller_logger.debug("Streamlit credentials configured successfully")
153 | return True
154 | except Exception as e:
155 | controller_logger.warning(f"Failed to setup Streamlit credentials: {e}")
156 | controller_logger.warning("Diagnostics dashboard may show welcome screen")
157 | return False # Continue without proper credentials
158 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/model/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/diagnostics/model/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/model/diagnostic_model.py:
--------------------------------------------------------------------------------
1 | """
2 | Model component of the MVC pattern for pciSeq diagnostics.
3 | """
4 |
5 | import redis
6 | import pandas as pd
7 | import numpy as np
8 | import logging
9 | from typing import Optional, Dict, Any
10 | from pciSeq.src.diagnostics.constants import DiagnosticKeys
11 |
12 | model_logger = logging.getLogger(__name__)
13 |
14 |
15 | class DiagnosticModel:
16 | def __init__(self, redis_host: str = 'localhost', redis_port: int = 6379):
17 | """Initialize Redis connection."""
18 | self.redis_client = redis.Redis(
19 | host=redis_host,
20 | port=redis_port,
21 | decode_responses=True
22 | )
23 | self._verify_connection()
24 |
25 | def publish_diagnostics(self, algorithm_model: Any, iteration: int, has_converged: bool) -> None:
26 | """Publish diagnostic data to Redis."""
27 | try:
28 | # Publish both gene efficiency and cell type data using the same pattern
29 | self._publish_data_to_redis(
30 | key=DiagnosticKeys.GENE_EFFICIENCY,
31 | data=self._prepare_gene_data(algorithm_model),
32 | metadata=self._create_metadata(iteration, has_converged)
33 | )
34 |
35 | self._publish_data_to_redis(
36 | key=DiagnosticKeys.CELL_TYPE_POSTERIOR,
37 | data=self._prepare_cell_type_data(algorithm_model),
38 | metadata=self._create_metadata(iteration, has_converged)
39 | )
40 | except Exception as e:
41 | model_logger.error(f"Failed to publish diagnostics: {e}")
42 |
43 | def _publish_data_to_redis(self, key: DiagnosticKeys, data: pd.DataFrame, metadata: dict) -> None:
44 | """Helper method to publish data to Redis with a consistent pattern."""
45 | formatted_data = {
46 | 'data': data.to_json(),
47 | 'metadata': metadata
48 | }
49 | # Store the data in Redis
50 | self.redis_client.set(key.value, str(formatted_data))
51 | # Notify subscribers that new data are available
52 | self.redis_client.publish(key.value, 'update')
53 |
54 | @staticmethod
55 | def _prepare_gene_data(algorithm_model: Any) -> pd.DataFrame:
56 | """Prepare gene efficiency data."""
57 | return pd.DataFrame({
58 | 'gene_efficiency': algorithm_model.genes.eta_bar,
59 | 'gene': algorithm_model.genes.gene_panel
60 | })
61 |
62 | @staticmethod
63 | def _prepare_cell_type_data(algorithm_model: Any) -> pd.DataFrame:
64 | """Prepare cell type distribution data."""
65 | cell_type_indices = np.argmax(algorithm_model.cells.classProb[1:, :], axis=1)
66 | counts = np.bincount(
67 | cell_type_indices,
68 | minlength=len(algorithm_model.cellTypes.names)
69 | )
70 | return pd.DataFrame({
71 | 'class_name': algorithm_model.cellTypes.names,
72 | 'counts': counts
73 | })
74 |
75 | @staticmethod
76 | def _create_metadata(iteration: int, has_converged: bool) -> dict:
77 | """Create consistent metadata structure."""
78 | return {
79 | 'iteration': iteration,
80 | 'has_converged': has_converged,
81 | 'timestamp': pd.Timestamp.now().isoformat()
82 | }
83 |
84 | def get_diagnostic_data(self, key: DiagnosticKeys) -> Optional[Dict]:
85 | """Retrieve data from Redis."""
86 | try:
87 | data = self.redis_client.get(key.value)
88 | return eval(data) if data else None
89 | except Exception as e:
90 | model_logger.error(f"Failed to retrieve data: {e}")
91 | return None
92 |
93 | def _verify_connection(self) -> None:
94 | """Verify Redis connection is working."""
95 | try:
96 | self.redis_client.ping()
97 | except redis.ConnectionError as e:
98 | model_logger.error(f"Redis connection failed: {e}")
99 | raise
100 |
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/view/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/diagnostics/view/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/diagnostics/view/dashboard.py:
--------------------------------------------------------------------------------
1 | """
2 | View component of the MVC pattern for pciSeq diagnostics.
3 |
4 | This module is responsible for presenting data to the user through a Streamlit dashboard.
5 | It maintains the original visualization style while working with the new MVC structure.
6 | """
7 |
8 | import streamlit as st
9 | import pandas as pd
10 | from io import StringIO
11 | import altair as alt
12 | import logging
13 | from pciSeq.src.diagnostics.constants import DiagnosticKeys
14 | from pciSeq.src.diagnostics.model.diagnostic_model import DiagnosticModel
15 |
16 | dashboard_logger = logging.getLogger(__name__)
17 |
18 |
19 | class DiagnosticDashboard:
20 | def __init__(self, model: DiagnosticModel):
21 | """
22 | Initialize dashboard with model connection.
23 |
24 | The dashboard subscribes to Redis channels that the Model publishes to:
25 | - gene_efficiency: Model publishes gene efficiency metrics
26 | - cell_type_posterior: Model publishes cell type distribution data
27 |
28 | This pub/sub mechanism enables real-time updates as the Model
29 | publishes new diagnostic data to Redis.
30 | """
31 | self.model = model
32 | self._setup_page()
33 |
34 | # Subscribe to the Redis channels that the Model publishes to
35 | self.pubsub = self.model.redis_client.pubsub()
36 |
37 | # These channels match the Model's publishing channels defined in DiagnosticKeys
38 | # Model -> Redis -> View data flow
39 | self.pubsub.subscribe(
40 | DiagnosticKeys.GENE_EFFICIENCY.value, # Model publishes gene metrics here
41 | DiagnosticKeys.CELL_TYPE_POSTERIOR.value # Model publishes cell data here
42 | )
43 |
44 | def _setup_page(self) -> None:
45 | """Configure Streamlit page settings."""
46 | st.set_page_config(
47 | page_title="Diagnostics: pciSeq",
48 | page_icon="✅",
49 | layout="wide",
50 | )
51 |
52 | @staticmethod
53 | def _create_barchart(df: pd.DataFrame, nominal_col: str, val_col: str) -> alt.Chart:
54 | """Create a bar chart using Altair."""
55 | chart = alt.Chart(df).mark_bar().encode(
56 | y=alt.Y(f'{nominal_col}:N', title=nominal_col),
57 | x=f'{val_col}:Q',
58 | color=alt.Color(f'{nominal_col}:N', legend=None),
59 | tooltip=[
60 | alt.Tooltip(f'{nominal_col}:N'),
61 | alt.Tooltip(f'{val_col}:Q')
62 | ]
63 | ).properties(height=1200)
64 | return chart
65 |
66 | def render(self) -> None:
67 | """
68 | Main rendering loop for the dashboard.
69 | Listens for Model's publications to Redis and updates visualizations.
70 | """
71 | title = st.title("Convergence screen. (Waiting for data....)")
72 | placeholder = st.empty()
73 |
74 | while True:
75 | with placeholder.container():
76 | fig_col1, fig_col2 = st.columns(2)
77 |
78 | with fig_col1:
79 | self._render_gene_efficiency(title) # Visualize Model's gene efficiency data
80 |
81 | with fig_col2:
82 | self._render_cell_distribution(title) # Visualize Model's cell type data
83 |
84 | # Check for new messages from Model's publications. This is the heart of the LIVE UPDATE MECHANISM
85 | message = self.pubsub.get_message() # Check if Model published new data
86 | if message and message['type'] == 'message': # If it's a real message (not control message)
87 | st.rerun() # Tell Streamlit to refresh the dashboard
88 |
89 | def _render_gene_efficiency(self, title) -> None:
90 | """Render gene efficiency visualization."""
91 | data = self.model.get_diagnostic_data(DiagnosticKeys.GENE_EFFICIENCY)
92 | if data:
93 | df = pd.read_json(StringIO(data['data']))
94 | metadata = data['metadata']
95 |
96 | title.title("Convergence screen")
97 | st.markdown(f"#### Gene efficiency after iteration {metadata['iteration']}")
98 |
99 | bar_chart = self._create_barchart(df, nominal_col='gene', val_col='gene_efficiency')
100 | st.altair_chart(bar_chart, use_container_width=True)
101 |
102 | def _render_cell_distribution(self, title) -> None:
103 | """Render cell type distribution visualization."""
104 | data = self.model.get_diagnostic_data(DiagnosticKeys.CELL_TYPE_POSTERIOR)
105 | if data:
106 | df = pd.read_json(StringIO(data['data']))
107 | metadata = data['metadata']
108 |
109 | title.title("Convergence screen")
110 | st.markdown(f"#### Cell counts per cell class after iteration {metadata['iteration']}")
111 |
112 | bar_chart = self._create_barchart(df, nominal_col='class_name', val_col='counts')
113 | bar_chart = bar_chart.properties(
114 | title=alt.TitleParams(
115 | [f'#cells: {df.counts.sum()}'],
116 | baseline='bottom',
117 | orient='bottom',
118 | anchor='end',
119 | fontWeight='normal',
120 | fontSize=12,
121 | ),
122 | height=1200
123 | )
124 | st.altair_chart(bar_chart, use_container_width=True)
125 |
126 |
127 | def main():
128 | """Entry point for the Streamlit dashboard."""
129 | try:
130 | model = DiagnosticModel()
131 | dashboard = DiagnosticDashboard(model)
132 | dashboard.render()
133 | except Exception as e:
134 | st.error(f"Dashboard initialization failed: {e}")
135 | dashboard_logger.error(f"Dashboard error: {e}", exc_info=True)
136 |
137 |
138 | if __name__ == "__main__":
139 | main()
140 |
--------------------------------------------------------------------------------
/pciSeq/src/preprocess/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/preprocess/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/preprocess/cell_borders.py:
--------------------------------------------------------------------------------
1 | """ Functions to extract the cell boundaries """
2 | import pandas as pd
3 | import numpy as np
4 | from multiprocessing import Pool, cpu_count
5 | from multiprocessing.dummy import Pool as ThreadPool
6 |
7 | # All that below is to avoid diplib to show a welcome msg on import. If hasattr(sys,'ps1') return True, then
8 | # import diplib will print out the diplib name, version, description which I find annoying. I am deleting ps1
9 | # and then reinstate it after the import.
10 | try:
11 | import sys
12 | ps1 = sys.__dict__['ps1']
13 | del sys.ps1
14 | import diplib as dip
15 | sys.__dict__['ps1'] = ps1
16 | except KeyError:
17 | import diplib as dip
18 |
19 |
20 | def extract_borders_dip(label_image, offset_x=0, offset_y=0, exclude_labels=(0,)):
21 | """
22 | Extracts the cell boundaries from the label image array. The background is
23 | assumed to have label=0 and it will be ignored by default.
24 | Parameters
25 | ----------
26 | label_image: The label image array, typically obtained from some image segmentation
27 | application and maps every pixel on the image to a cell label.
28 | offset_x: Amount to shift the boundaries along the x-axis
29 | offset_y: Amount to shift the boundaries along the y-axis
30 | exclude_labels: Array-like, contains the labels to be ignored.
31 |
32 | Returns
33 | -------
34 | Returns a dataframe with columns ['labels', 'coords'] where column 'coords' keeps a
35 | list like [[x0, y0], [x1, y2],...,[x0, y0]] of the (closed-loop) boundaries coordinates
36 | for corresponding cell label
37 | """
38 |
39 | if exclude_labels is None:
40 | exclude_labels = [0]
41 | labels = sorted(set(label_image.flatten()) - set(exclude_labels))
42 | cc = dip.GetImageChainCodes(label_image) # input must be an unsigned integer type
43 | d = {}
44 | for c in cc:
45 | if c.objectID in labels:
46 | # p = np.array(c.Polygon())
47 | p = c.Polygon().Simplify()
48 | p = p + np.array([offset_x, offset_y])
49 | p = np.uint64(p).tolist()
50 | p.append(p[0]) # append the first pair at the end to close the polygon
51 | d[np.uint64(c.objectID)] = p
52 | else:
53 | pass
54 | df = pd.DataFrame([d]).T
55 | df = df.reset_index()
56 | df.columns = ['label', 'coords']
57 | return df
58 |
59 |
60 | def extract_borders(cell_labels):
61 | '''
62 | Extracts the cell boundaries from the label image array. Same as 'extract_borders_dip()' but a lot faster.
63 | Parameters
64 | ----------
65 | label_image: The label image array, typically obtained from some image segmentation
66 | application and maps every pixel on the image to a cell label.
67 |
68 | Returns
69 | -------
70 | Returns a dataframe with columns 'labels' and 'coords'
71 | """
72 | '''
73 | cell_boundaries = pd.DataFrame()
74 | borders_list = _extract_borders(cell_labels)
75 | d = dict(borders_list)
76 | cell_boundaries['label'] = d.keys()
77 | cell_boundaries['coords'] = d.values()
78 | return cell_boundaries
79 |
80 |
81 | def _extract_borders(label_image):
82 | """
83 | Extracts the cell boundaries from the label image array.
84 | Returns a dict where keys are the cell label and values the corresponding cell boundaries
85 | """
86 |
87 | # labels = sorted(set(label_image.flatten()) - set(exclude_labels))
88 | cc = dip.GetImageChainCodes(label_image) # input must be an unsigned integer type
89 |
90 | pool = ThreadPool(cpu_count())
91 | # it would be nice to process only those cc whose cc.objectID is in labels
92 | results = pool.map(parse_chaincode, cc)
93 | # close the pool and wait for the work to finish
94 | pool.close()
95 | pool.join()
96 |
97 | return dict(results)
98 |
99 |
100 | def parse_chaincode(c):
101 | p = c.Polygon().Simplify()
102 | p = p + np.array([0, 0])
103 | p = np.uint64(p).tolist()
104 | p.append(p[0]) # append the first pair at the end to close the polygon
105 | return np.uint64(c.objectID), p
106 |
107 |
108 |
109 |
--------------------------------------------------------------------------------
/pciSeq/src/preprocess/spot_labels.py:
--------------------------------------------------------------------------------
1 | """
2 | Cell and Spot Label Processing Module for pciSeq
3 |
4 | This module provides core functionality for processing and analyzing spatial transcriptomics data,
5 | specifically handling the relationship between cell segmentation and RNA spot detection.
6 |
7 | Key Functions:
8 | -------------
9 | - inside_cell: Maps RNA spots to their containing cells
10 | - reorder_labels: Normalizes cell labels to sequential integers
11 | - stage_data: Main processing pipeline that integrates all functionality
12 |
13 | Data Processing Steps:
14 | --------------------
15 | 1. Cell Label Processing:
16 | - Validates and normalizes cell segmentation labels
17 | - Ensures sequential integer labeling
18 | - Computes cell properties (centroids, areas)
19 |
20 | 2. Spot Assignment:
21 | - Maps each RNA spot to its containing cell
22 | - Validates spot coordinates against image boundaries
23 | - Links spots with cell metadata
24 |
25 | 3. Boundary Processing:
26 | - Extracts and validates cell boundaries
27 | - Ensures consistency between properties and boundaries
28 |
29 | """
30 |
31 | from dataclasses import dataclass
32 |
33 |
34 | import numpy as np
35 | import pandas as pd
36 | from typing import Tuple, Union
37 | import skimage.measure as skmeas
38 | from scipy.sparse import coo_matrix, csr_matrix, spmatrix
39 | from pciSeq.src.preprocess.cell_borders import extract_borders
40 | import logging
41 |
42 |
43 | spot_labels_logger = logging.getLogger(__name__)
44 |
45 |
46 | def inside_cell(label_image: Union[spmatrix, np.ndarray],
47 | spots: pd.DataFrame) -> np.ndarray:
48 | """
49 | Determine which cell contains each spot.
50 |
51 | Args:
52 | label_image: Cell segmentation mask (sparse matrix or array)
53 | spots: DataFrame with spot coordinates ('x' and 'y' columns)
54 |
55 | Returns:
56 | Array of cell labels for each spot
57 |
58 | Raises:
59 | TypeError: If label_image is not of supported type
60 | """
61 | if isinstance(label_image, np.ndarray):
62 | label_image = csr_matrix(label_image)
63 | elif isinstance(label_image, coo_matrix):
64 | label_image = label_image.tocsr()
65 | elif not isinstance(label_image, csr_matrix):
66 | raise TypeError('label_image must be ndarray, coo_matrix, or csr_matrix')
67 |
68 | return np.asarray(label_image[spots.y, spots.x], dtype=np.uint32)[0]
69 |
70 |
71 | def remap_labels(coo: coo_matrix) -> coo_matrix:
72 | """
73 | Randomly reshuffle label assignments (for testing/debugging).
74 |
75 | Args:
76 | coo: Input label matrix
77 |
78 | Returns:
79 | Matrix with randomly remapped labels
80 | """
81 | coo_max = coo.data.max()
82 | original_labels = 1 + np.arange(coo_max)
83 | new_labels = original_labels.copy()
84 | np.random.shuffle(new_labels)
85 |
86 | label_map = dict(zip(original_labels, new_labels))
87 | new_data = np.array([label_map[x] for x in coo.data]).astype(np.uint64)
88 |
89 | return coo_matrix((new_data, (coo.row, coo.col)), shape=coo.shape)
90 |
91 |
92 | def reorder_labels(coo: coo_matrix) -> Tuple[coo_matrix, pd.DataFrame]:
93 | """
94 | Normalize labels to be sequential integers starting from 1.
95 |
96 | Args:
97 | coo: Sparse matrix containing cell labels
98 |
99 | Returns:
100 | Tuple containing:
101 | - Relabeled sparse matrix
102 | - DataFrame mapping old labels to new labels
103 | """
104 | label_image = coo.toarray()
105 | unique_labels, idx = np.unique(label_image.flatten(), return_inverse=True)
106 |
107 | label_map = pd.DataFrame({
108 | 'old_label': unique_labels,
109 | 'new_label': np.arange(len(unique_labels))
110 | }, dtype=np.uint32).sort_values(by='old_label', ignore_index=True)
111 |
112 | return coo_matrix(idx.reshape(label_image.shape)), label_map
113 |
114 |
115 | def stage_data(spots: pd.DataFrame, coo: coo_matrix) -> Tuple[pd.DataFrame, pd.DataFrame, pd.DataFrame]:
116 | """
117 | Process spot and cell segmentation data to establish spot-cell relationships.
118 |
119 | Args:
120 | spots: DataFrame with columns ['x', 'y', 'Gene']
121 | coo: Sparse matrix containing cell segmentation labels
122 |
123 | Returns:
124 | Tuple containing:
125 | - cells: DataFrame with cell properties
126 | - boundaries: DataFrame with cell boundary coordinates
127 | - spots: DataFrame with spot locations and cell assignments
128 |
129 | Raises:
130 | ValueError: If required columns are missing or data validation fails
131 | """
132 | # Validate inputs
133 | required_columns = {'x', 'y', 'Gene'}
134 | missing_columns = required_columns - set(spots.columns)
135 | if missing_columns:
136 | raise ValueError(f"Missing required columns in spots DataFrame: {missing_columns}")
137 |
138 | spot_labels_logger.info(f'Number of spots passed-in: {spots.shape[0]}')
139 | spot_labels_logger.info(f'Number of segmented cells: {len(set(coo.data))}')
140 | spot_labels_logger.info(
141 | f'Segmentation array implies that image has width: {coo.shape[1]}px and height: {coo.shape[0]}px')
142 |
143 | # Normalize labels if needed
144 | label_map = None
145 | unique_labels = set(coo.data)
146 | max_label = coo.data.max()
147 |
148 | if coo.data.max() != len(set(coo.data)):
149 | spot_labels_logger.info(
150 | f'Detected non-sequential cell labels: found {len(unique_labels)} unique labels '
151 | f'with maximum value of {max_label}. Normalizing labels to range [1, {len(unique_labels)}]'
152 | )
153 | coo, label_map = reorder_labels(coo)
154 |
155 | # Filter spots to image bounds
156 | mask_x = (spots.x >= 0) & (spots.x <= coo.shape[1])
157 | mask_y = (spots.y >= 0) & (spots.y <= coo.shape[0])
158 | spots = spots[mask_x & mask_y].copy()
159 |
160 | # Assign spots to cells
161 | spots = spots.assign(label=inside_cell(coo, spots))
162 |
163 | # Calculate cell properties
164 | props = skmeas.regionprops_table(
165 | coo.toarray().astype(np.int32),
166 | properties=['label', 'area', 'centroid']
167 | )
168 | props_df = pd.DataFrame(props).rename(
169 | columns={'centroid-0': 'y_cell', 'centroid-1': 'x_cell'}
170 | )
171 |
172 | # Apply label mapping if exists
173 | if label_map is not None:
174 | props_df = pd.merge(
175 | props_df,
176 | label_map,
177 | left_on='label',
178 | right_on='new_label',
179 | how='left'
180 | ).drop(['new_label'], axis=1)
181 |
182 | # Set datatypes
183 | props_df = props_df.astype({
184 | "label": np.uint32,
185 | "area": np.uint32,
186 | 'y_cell': np.float32,
187 | 'x_cell': np.float32
188 | })
189 |
190 | # Extract cell boundaries
191 | cell_boundaries = extract_borders(coo.toarray().astype(np.uint32))
192 |
193 | # Validate cell data consistency
194 | if not (props_df.shape[0] == cell_boundaries.shape[0] == np.unique(coo.data).shape[0]):
195 | raise ValueError("Inconsistency detected between cell properties and boundaries")
196 |
197 | # Ensure spots are assigned to valid cells
198 | if not set(spots.label[spots.label > 0]).issubset(set(props_df.label)):
199 | raise ValueError("Spots assigned to non-existent cell labels")
200 |
201 | # Prepare final data structures
202 | cells = props_df.merge(cell_boundaries)
203 | cells.sort_values(by=['label', 'x_cell', 'y_cell'], inplace=True)
204 | spots = spots.merge(cells, how='left', on=['label'])
205 |
206 | return (
207 | cells.drop(columns=['coords']).rename(columns={
208 | 'x_cell': 'x0',
209 | 'y_cell': 'y0'
210 | }),
211 | cells[['label', 'coords']].rename(columns={
212 | 'label': 'cell_id'
213 | }),
214 | spots[['x', 'y', 'label', 'Gene', 'x_cell', 'y_cell']].rename(columns={
215 | 'Gene': 'target',
216 | 'x': 'x_global',
217 | 'y': 'y_global'
218 | })
219 | )
220 |
--------------------------------------------------------------------------------
/pciSeq/src/preprocess/utils.py:
--------------------------------------------------------------------------------
1 |
2 | def get_img_shape(coo):
3 | w = coo.shape[1]
4 | h = coo.shape[0]
5 | return [h, w]
--------------------------------------------------------------------------------
/pciSeq/src/validation/README.md:
--------------------------------------------------------------------------------
1 | # pciSeq Validation Module
2 |
3 | This module handles validation and configuration management for the pciSeq pipeline.
4 |
5 | ## Files and Their Functions
6 |
7 | ### 1. `input_validation.py`
8 | - Primary input validation module using `ValidatedInputs` dataclass
9 | - Validates and processes core data structures:
10 | * Spots DataFrame (Gene, x, y coordinates)
11 | * COO matrix for segmentation masks
12 | * Single cell data (optional)
13 |
14 | ### 2. `config_manager.py`
15 | - Manages configuration settings through `ConfigManager` dataclass
16 | - Provides type hints for configuration parameters
17 |
--------------------------------------------------------------------------------
/pciSeq/src/validation/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/validation/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/validation/config_manager.py:
--------------------------------------------------------------------------------
1 | from dataclasses import dataclass
2 | from typing import List, Dict, Optional, Union
3 | from numbers import Number
4 | import logging
5 | from pciSeq import config
6 | from pciSeq.src.core.utils import log_file, check_redis_server
7 | config_manager_logger = logging.getLogger(__name__)
8 |
9 |
10 | @dataclass
11 | class ConfigManager:
12 | exclude_genes: List[str]
13 | max_iter: int
14 | CellCallTolerance: float
15 | rGene: int
16 | Inefficiency: float
17 | InsideCellBonus: Union[bool, int, float]
18 | MisreadDensity: Union[float, Dict[str, float]]
19 | SpotReg: float
20 | nNeighbors: int
21 | rSpot: int
22 | save_data: bool
23 | output_path: str
24 | launch_viewer: bool
25 | launch_diagnostics: bool
26 | is_redis_running: bool
27 | cell_radius: Optional[float]
28 | cell_type_prior: str
29 | mean_gene_counts_per_class: int
30 | mean_gene_counts_per_cell: int
31 |
32 | @classmethod
33 | def from_opts(cls, opts: Optional[Dict] = None) -> 'ConfigManager':
34 | """Create configuration from default values and optional overrides"""
35 | if opts is None:
36 | opts = config.DEFAULT.copy()
37 |
38 | # Start with default configuration
39 | cfg_dict = config.DEFAULT.copy()
40 |
41 | # # Override with user options if provided
42 | for key in opts:
43 | if key in cfg_dict:
44 | cfg_dict[key] = opts[key]
45 | config_manager_logger.info(f'{key} is set to {opts[key]}')
46 |
47 | log_file(cfg_dict)
48 |
49 | # Create instance
50 | instance = cls(**cfg_dict)
51 | # instance._validate()
52 | return instance
53 |
54 | def to_dict(self) -> Dict:
55 | """Convert configuration back to dictionary format"""
56 | return {k: v for k, v in self.__dict__.items() if not k.startswith('_')}
57 |
58 | def set_runtime_attributes(self):
59 | """Set configuration attributes that can only be determined at runtime.
60 |
61 | Returns:
62 | config: Updated configuration with runtime attributes
63 | """
64 | self.is_redis_running = check_redis_server()
--------------------------------------------------------------------------------
/pciSeq/src/validation/input_validation.py:
--------------------------------------------------------------------------------
1 | from __future__ import annotations
2 |
3 | from dataclasses import dataclass
4 | from typing import Dict, Optional, Union, List
5 | from typing import get_origin, get_args, get_type_hints
6 | from .config_manager import ConfigManager
7 | import pandas as pd
8 | import numpy as np
9 | from pandas import DataFrame
10 | from scipy.sparse import coo_matrix
11 | import logging
12 |
13 | validation_logger = logging.getLogger(__name__)
14 |
15 |
16 | @dataclass
17 | class InputValidator:
18 | """Container for validated inputs"""
19 | spots: pd.DataFrame
20 | coo: coo_matrix
21 | scdata: Optional[pd.DataFrame]
22 | config: ConfigManager
23 |
24 | @classmethod
25 | def validate(
26 | cls,
27 | spots: pd.DataFrame,
28 | coo: coo_matrix,
29 | scdata: Optional[pd.DataFrame],
30 | config: ConfigManager
31 | ) -> tuple[DataFrame, coo_matrix, Union[DataFrame, None], Dict]:
32 | """Validate all inputs and return validated versions"""
33 |
34 | # Validate spots
35 | cls._validate_spots(spots)
36 | spots = cls._process_spots(spots.copy(), scdata, config)
37 |
38 | # Validate coo matrix
39 | cls._validate_coo(coo)
40 |
41 | # Validate single cell data if present
42 | if scdata is not None:
43 | cls._validate_scdata(scdata)
44 |
45 | # Validate config
46 | config = cls._validate_config(config)
47 |
48 | out = cls(spots=spots, coo=coo, scdata=scdata, config=config)
49 |
50 | return out.spots, out.coo, out.scdata, out.config.to_dict()
51 |
52 | @staticmethod
53 | def _validate_spots(spots: pd.DataFrame) -> None:
54 | """Validate spots dataframe structure"""
55 | if not isinstance(spots, pd.DataFrame):
56 | raise TypeError("Spots should be passed-in as a dataframe")
57 |
58 | if set(spots.columns) != {'Gene', 'x', 'y'}:
59 | raise ValueError("Spots dataframe must have columns ['Gene', 'x', 'y']")
60 |
61 | @staticmethod
62 | def _validate_coo(coo: coo_matrix) -> None:
63 | """Validate coo matrix"""
64 | if not isinstance(coo, coo_matrix):
65 | raise TypeError('The segmentation masks should be passed-in as a coo_matrix')
66 |
67 | @staticmethod
68 | def _validate_scdata(scdata: pd.DataFrame) -> None:
69 | """Validate single cell data"""
70 | if not isinstance(scdata, pd.DataFrame):
71 | raise TypeError("Single cell data should be passed-in as a dataframe")
72 |
73 | @staticmethod
74 | def _process_spots(spots: pd.DataFrame, scdata: Optional[pd.DataFrame], config: 'ConfigManager') -> pd.DataFrame:
75 | """Process and validate spots data"""
76 | # Cast datatypes
77 | spots = spots.astype({
78 | 'Gene': str,
79 | 'x': np.float32,
80 | 'y': np.float32
81 | })
82 |
83 | # Remove genes not in single cell data if present
84 | if scdata is not None:
85 | if not set(spots.Gene).issubset(scdata.index):
86 | spots = InputValidator._purge_spots(spots, scdata)
87 |
88 | return spots
89 |
90 | @staticmethod
91 | def _purge_spots(spots: pd.DataFrame, scdata: pd.DataFrame) -> pd.DataFrame:
92 | """Remove spots with genes not found in single cell data"""
93 | drop_spots = list(set(spots.Gene) - set(scdata.index))
94 | validation_logger.warning(f'Found {len(drop_spots)} genes that are not included in the single cell data')
95 | idx = ~np.in1d(spots.Gene, drop_spots)
96 | spots = spots.iloc[idx]
97 | validation_logger.warning(f'Removed from spots: {drop_spots}')
98 | return spots
99 |
100 | @staticmethod
101 | def _validate_config(config: 'ConfigManager') -> 'ConfigManager':
102 | """Process and validate configuration"""
103 |
104 | # Fetch type hints directly from ConfigManager dataclass to ensure type consistency
105 | type_validations = get_type_hints(ConfigManager)
106 |
107 | # Validate each configuration parameter matches its expected type from the dataclass definition
108 | for attr_name, expected_type in type_validations.items():
109 | value = getattr(config, attr_name)
110 | origin_type = get_origin(expected_type)
111 |
112 | # Handle Union types (e.g., Union[bool, int, float]) separately since they can't be used
113 | # directly with isinstance
114 | if origin_type is Union:
115 | allowed_types = get_args(expected_type)
116 | allowed_types = tuple([get_origin(d) if get_origin(d) else d for d in allowed_types])
117 | if not isinstance(value, allowed_types):
118 | raise TypeError(f"'{attr_name}' must be one of {allowed_types}, got {type(value)}")
119 | else:
120 | # For non-Union types, handle both simple types and generics (e.g., List[str])
121 | origin_type = origin_type or expected_type
122 | if not isinstance(value, origin_type):
123 | raise TypeError(f"'{attr_name}' must be of type {expected_type}, got {type(value)}")
124 |
125 | # Validate specific value constraints
126 | # Validate cell_type_prior
127 | if config.cell_type_prior.lower() not in ['uniform', 'weighted']:
128 | raise ValueError("'cell_type_prior' should be either 'uniform' or 'weighted'")
129 |
130 | # Convert to lowercase
131 | config.cell_type_prior = config.cell_type_prior.lower()
132 |
133 | # Convert boolean InsideCellBonus to its numeric equivalent
134 | if config.InsideCellBonus is True:
135 | config.InsideCellBonus = 2
136 | validation_logger.warning('InsideCellBonus was passed-in as True. Overriding with the default value of 2')
137 |
138 | # Process MisreadDensity to ensure it has the required structure
139 | if isinstance(config.MisreadDensity, dict):
140 | if 'default' not in config.MisreadDensity:
141 | raise ValueError("When MisreadDensity is a dictionary, it must contain a 'default' key")
142 | elif isinstance(config.MisreadDensity, (int, float)):
143 | config.MisreadDensity = {'default': config.MisreadDensity}
144 | else:
145 | raise ValueError("MisreadDensity must be either a number or a dictionary with a 'default' key")
146 |
147 | return config
148 |
--------------------------------------------------------------------------------
/pciSeq/src/viewer/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/src/viewer/__init__.py
--------------------------------------------------------------------------------
/pciSeq/src/viewer/run_flask.py:
--------------------------------------------------------------------------------
1 | from flask import Flask, render_template
2 | import os
3 | import webbrowser
4 | import platform
5 | import random
6 | from threading import Timer
7 | import logging
8 |
9 | run_flask_logger = logging.getLogger(__name__)
10 |
11 |
12 | def get_browser(port_num):
13 | url = 'http://127.0.0.1:%d' % port_num
14 | my_os = platform.system()
15 |
16 | if my_os == 'Windows':
17 | chrome_path = 'C:/Program Files (x86)/Google/Chrome/Application/chrome.exe'
18 | elif my_os == 'Darwin': # Is this always the case for MacOS?
19 | chrome_path = 'open -a /Applications/Google\ Chrome.app'
20 | elif my_os == 'Linux':
21 | chrome_path = '/usr/bin/google-chrome'
22 | else:
23 | chrome_path = None
24 |
25 | run_flask_logger.info('Platform is %s' % my_os)
26 | if chrome_path:
27 | run_flask_logger.info('Chrome path: %s' % chrome_path)
28 |
29 | if chrome_path and os.path.isfile(chrome_path):
30 | webbrowser.register('chrome', None, webbrowser.BackgroundBrowser(chrome_path), preferred=True)
31 | wb = webbrowser.get('chrome').open_new_tab(url)
32 | else:
33 | wb = webbrowser.open_new(url)
34 |
35 | if not wb:
36 | run_flask_logger.info('Could not open browser')
37 |
38 |
39 | def open_browser():
40 | webbrowser.open_new('http://127.0.0.1:5000/')
41 |
42 |
43 | def flask_app_start(dir):
44 | run_flask_logger.info('Launching viewer. Serving directory %s ' % dir)
45 | port = 5000 + random.randint(0, 999)
46 | flask_app = Flask(__name__,
47 | static_url_path='', # remove the static folder path
48 | static_folder=dir,
49 | # set here the path of the folder to be served. The js files referenced in your html file are with respect to this folder. Adjust the paths in your html file (look for the tag, so that the js libraries will be parsed
50 | template_folder=dir) # set here the path to the folder where your html page lives. Absolute and relative paths both work fine
51 | flask_app.config['EXPLAIN_TEMPLATE_LOADING'] = True
52 |
53 | @flask_app.route("/")
54 | def index():
55 | return render_template("index.html", data=None)
56 |
57 | Timer(1, get_browser, [port]).start()
58 | flask_app.run(port=port, debug=False)
59 |
60 |
61 |
--------------------------------------------------------------------------------
/pciSeq/src/viewer/stage_image.py:
--------------------------------------------------------------------------------
1 | import shutil
2 | import os
3 | import pyvips
4 | import logging
5 |
6 | stage_image_logger = logging.getLogger(__name__)
7 |
8 | def split_image(im):
9 | # DEPRECATED to be removed
10 | '''
11 | you can just do:
12 | im.dzsave('./out', suffix='.tif', skip_blanks=-1, background=0, depth='one', overlap=0, tile_size=2000, layout='google')
13 | to split the image to smaller squares. However you need to write a couple of line to rename and move the file to the correct
14 | folders
15 | :param im:
16 | :return:
17 | '''
18 | im = pyvips.Image.new_from_file(im, access='random')
19 | tile_size = 2000;
20 |
21 | if im.width % tile_size == 0:
22 | tiles_across = int(im.width / tile_size)
23 | else:
24 | tiles_across = im.width // tile_size + 1
25 |
26 |
27 | if im.width % tile_size == 0:
28 | tiles_down = int(im.height/tile_size)
29 | else:
30 | tiles_down = im.height // tile_size + 1
31 |
32 | image = im.gravity('north-west', tiles_across * tile_size, tiles_down * tile_size)
33 |
34 | for j in range(tiles_down):
35 | stage_image_logger.info('Moving to the next row: %d/%d '% (j, tiles_down-1) )
36 | y_top_left = j * tile_size
37 | for i in range(tiles_across):
38 | x_top_left = i * tile_size
39 | tile = image.crop(x_top_left, y_top_left, tile_size, tile_size)
40 | tile_num = j * tiles_across + i
41 | fov_id = 'fov_' + str(tile_num)
42 |
43 | out_dir = os.path.join(stage_image_logger.ROOT_DIR, 'fov', fov_id, 'img')
44 | full_path = os.path.join(out_dir, fov_id +'.tif')
45 | if not os.path.exists(os.path.dirname(full_path)):
46 | os.makedirs(os.path.dirname(full_path))
47 | tile.write_to_file(full_path)
48 | stage_image_logger.info('tile: %s saved at %s' % (fov_id, full_path) )
49 |
50 |
51 | def map_image_size(z):
52 | '''
53 | returns the image size for each zoom level. Assumes that each map tile is 256x256 pixels
54 | :param z:
55 | :return:
56 | '''
57 |
58 | return 256 * 2 ** z
59 |
60 |
61 | def tile_maker(img_path, zoom_levels=8, out_dir=r"./tiles"):
62 | """
63 | Makes a pyramid of tiles.
64 | img_path:(str) The path to the image
65 | zoom_levels: (int) Specifies how many zoom levels will be produced. Default value is 8.
66 | out_dir: (str) The path to the folder where the output (the pyramid of map tiles) will be saved to. If the folder
67 | does not exist, it will be created automatically. If it exists, it will be deleted before being populated
68 | with the new tiles. Dy default the tiles will be saved inside the current
69 | directory in a folder named "tiles".
70 | """
71 | # img_path = os.path.join(dir_path, 'demo_data', 'background_boundaries.tif')
72 |
73 | dim = map_image_size(zoom_levels)
74 | # remove the dir if it exists
75 | if os.path.exists(out_dir):
76 | shutil.rmtree(out_dir)
77 |
78 | # now make a fresh one
79 | if not os.path.exists(out_dir):
80 | os.makedirs(out_dir)
81 |
82 | im = pyvips.Image.new_from_file(img_path, access='sequential')
83 |
84 | # The following two lines add an alpha component to rgb which allows for transparency.
85 | # Is this worth it? It adds quite a bit on the execution time, about x2 increase
86 | # im = im.colourspace('srgb')
87 | # im = im.addalpha()
88 |
89 | stage_image_logger.info('Resizing image: %s' % img_path)
90 | factor = dim / max(im.width, im.height)
91 | im = im.resize(factor)
92 | stage_image_logger.info('Done! Image is now %d by %d' % (im.width, im.height))
93 | pixel_dims = [im.width, im.height]
94 |
95 | # sanity check
96 | assert max(im.width, im.height) == dim, 'Something went wrong. Image isnt scaled up properly. ' \
97 | 'It should be %d pixels in its longest side' % dim
98 |
99 | # im = im.gravity('south-west', dim, dim) # <---- Uncomment this if the origin is the bottomleft corner
100 |
101 | # now you can create a fresh one and populate it with tiles
102 | stage_image_logger.info('Started doing the image tiles ')
103 | im.dzsave(out_dir, layout='google', suffix='.jpg', background=0)
104 | stage_image_logger.info('Done. Pyramid of tiles saved at: %s' % out_dir)
105 |
106 | return pixel_dims
107 |
108 |
109 |
110 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/css/progress.css:
--------------------------------------------------------------------------------
1 | @import url(https://fonts.googleapis.com/css?family=Lato:700);
2 | *,
3 | *:before,
4 | *:after {
5 | box-sizing: border-box;
6 | }
7 |
8 | html,
9 | body {
10 | background: #ECF0F1;
11 | }
12 |
13 | #preloader {
14 | /*background: #ECF0F1;*/
15 | padding: 0px;
16 | color: #444;
17 | font-family: "Lato", Tahoma, Geneva, sans-serif;
18 | font-size: 16px;
19 | width: 100%;
20 | height: 100%;
21 | }
22 |
23 | /*from https://stackoverflow.com/questions/5482677/how-to-apply-two-css-classes-to-a-single-element*/
24 | .midplaced {
25 | position: fixed;
26 | top: 50%;
27 | left: 50%;
28 | -webkit-transform: translate(-50%, -50%);
29 | transform: translate(-50%, -50%);
30 | }
31 |
32 | #percent, #percent2, #percent3 {
33 | /*display: block;*/
34 | /*width: 160px;*/
35 | /*border: none;//1px solid #CCC;*/
36 | /*border-radius: 5px;*/
37 | /*margin: 50px;*/
38 | /*!*padding: 10px;*!*/
39 | color: #1ABC9C;
40 | font-family: "Lato", Tahoma, Geneva, sans-serif;
41 | font-size: 15px;
42 | text-align: center;
43 | }
44 |
45 | .meter {
46 | color: #1ABC9C;
47 | font-family: "Lato", Tahoma, Geneva, sans-serif;
48 | font-size: 14px; /* controls the format of the numbers under the donut */
49 | text-align: center;
50 | }
51 |
52 | #donut {
53 | display: block;
54 | margin: 0px auto;
55 | color: #1ABC9C;
56 | font-size: 20px;
57 | text-align: center;
58 | }
59 |
60 | p.pieLabel {
61 | max-width: 600px;
62 | margin: 12px auto;
63 | font-weight: normal;
64 | /*font-family: sans-serif;*/
65 | font-family: "Lato", Tahoma, Geneva, sans-serif;
66 | /*font-size: 16px;*/
67 | font-size: 16px; /* controls the font-size of the text under the donut */
68 | }
69 |
70 | code {
71 | background: #FAFAFA;
72 | border: 1px solid #DDD;
73 | border-radius: 3px;
74 | padding: 0px 4px;
75 | }
76 |
77 | .donut-size {
78 | font-size: 6em; /*controls the progress donut size*/
79 | }
80 |
81 | .pie-wrapper {
82 | position: relative;
83 | width: 1em;
84 | height: 1em;
85 | margin: 0px auto;
86 | }
87 | .pie-wrapper .pie {
88 | position: absolute;
89 | top: 0px;
90 | left: 0px;
91 | width: 100%;
92 | height: 100%;
93 | clip: rect(0, 1em, 1em, 0.5em);
94 | }
95 | .pie-wrapper .half-circle {
96 | position: absolute;
97 | top: 0px;
98 | left: 0px;
99 | width: 100%;
100 | height: 100%;
101 | border: 0.1em solid #1abc9c;
102 | border-radius: 50%;
103 | clip: rect(0em, 0.5em, 1em, 0em);
104 | }
105 | .pie-wrapper .right-side {
106 | -webkit-transform: rotate(0deg);
107 | transform: rotate(0deg);
108 | }
109 | .pie-wrapper .label {
110 | position: absolute;
111 | top: 0.4em;
112 | right: 0.4em;
113 | bottom: 0.4em;
114 | left: 0.4em;
115 | display: block;
116 | background: none;
117 | border-radius: 50%;
118 | color: #7F8C8D;
119 | font-size: 0.25em;
120 | line-height: 2.6em;
121 | text-align: center;
122 | cursor: default;
123 | z-index: 2;
124 | }
125 | .pie-wrapper .smaller {
126 | padding-bottom: 20px;
127 | color: #BDC3C7;
128 | font-size: 0.45em;
129 | vertical-align: super;
130 | }
131 | .pie-wrapper .shadow {
132 | width: 100%;
133 | height: 100%;
134 | border: 0.1em solid #BDC3C7;
135 | border-radius: 50%;
136 | }
137 |
138 | #specificChart, specificChart2{
139 | /*float:left;*/
140 | }
141 |
142 | .aligned-middle {
143 | margin-top: 25% !important;
144 | margin-bottom: 25% !important;
145 | }
146 |
147 | .stack-top {
148 | z-index: 9;
149 | margin: 20px; /* for demo purpose */
150 | }
151 |
152 | #wait_chart {
153 | height: 90px;
154 | line-height: 90px;
155 | text-align: center;
156 | font-size: 25px;
157 | color: #1ABC9C;
158 | /*border: 2px dashed #f69c55;*/
159 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/genes_datatable.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 | Gene Panel
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
16 |
18 |
19 |
20 |
21 |
22 |
23 |
30 |
31 |
32 |
33 |
34 |
35 |
36 |
37 |
38 | |
39 | Gene Name |
40 |
41 | Glyph Name |
42 | Glyph Color |
43 | Glyph |
44 |
45 |
46 |
47 |
48 |
49 |
50 |
188 |
189 |
190 |
191 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/classConfig.js:
--------------------------------------------------------------------------------
1 | // Color scheme for the cell classes.
2 | // IdentifiedType is some wider categorization. On the viewer, the donut chart at the bottom right is aggregated
3 | // by IdentifiedType and these aggregated probabilities are shown on the table at the bottom left.
4 | // Also, on the viewer, at the top right, there is a collapsible control to switch on/off the cells by the
5 | // cell class. The control has the classes in a nested class/subClass manner. For the nesting operation to
6 | // work, the separator in the className string should be a dot.
7 |
8 | function classColorsCodes()
9 | {
10 | var out = [
11 |
12 | {className: 'Pvalb.C1ql1.Cpne5', IdentifiedType: 'Axo-axonic', color: '#3A19BD'},
13 | {className: 'Pvalb.C1ql1.Npy', IdentifiedType: 'Axo-axonic', color: '#3A19BD'},
14 | {className: 'Pvalb.C1ql1.Pvalb', IdentifiedType: 'Axo-axonic', color: '#3A19BD'},
15 | {className: 'Sst.Nos1', IdentifiedType: 'Backprojection', color: '#3DCCFF'},
16 | {className: 'Pvalb.Tac1.Nr4a2', IdentifiedType: 'Basket', color: '#7757FA'},
17 | {className: 'Pvalb.Tac1.Syt2', IdentifiedType: 'Basket', color: '#7757FA'},
18 | {className: 'Pvalb.Tac1.Akr1c18', IdentifiedType: 'Bistratified', color: '#5938DB'},
19 | {className: 'Pvalb.Tac1.Sst', IdentifiedType: 'Bistratified', color: '#5938DB'},
20 | {className: 'Cck.Lypd1', IdentifiedType: 'Cck Calb1/Slc17a8*', color: '#FF0000'},
21 | {className: 'Cck.Calca', IdentifiedType: 'Cck Cxcl14-', color: '#FF0000'},
22 | {className: 'Cck.Lmo1.Npy', IdentifiedType: 'Cck Cxcl14-', color: '#FF0000'},
23 | {className: 'Cck.Sema5a', IdentifiedType: 'Cck Cxcl14-', color: '#FF0000'},
24 | {className: 'Cck.Cxcl14.Calb1.Igfbp5', IdentifiedType: 'Cck Cxcl14+', color: '#996E00'},
25 | {className: 'Cck.Cxcl14.Calb1.Kctd12', IdentifiedType: 'Cck Cxcl14+', color: '#996E00'},
26 | {className: 'Cck.Cxcl14.Calb1.Tac2', IdentifiedType: 'Cck Cxcl14+', color: '#996E00'},
27 | {className: 'Cck.Cxcl14.Calb1.Tnfaip8l3', IdentifiedType: 'Cck Cxcl14+', color: '#996E00'},
28 | {className: 'Cck.Cxcl14.Slc17a8', IdentifiedType: 'Cck Cxcl14+', color: '#996E00'},
29 | {className: 'Cck.Lmo1.Vip.Crh', IdentifiedType: 'Cck Vip Cxcl14-', color: '#FF0000'},
30 | {className: 'Cck.Lmo1.Vip.Fam19a2', IdentifiedType: 'Cck Vip Cxcl14-', color: '#FF0000'},
31 | {className: 'Cck.Lmo1.Vip.Tac2', IdentifiedType: 'Cck Vip Cxcl14-', color: '#FF0000'},
32 | {className: 'Cck.Cxcl14.Vip', IdentifiedType: 'Cck Vip Cxcl14+', color: '#996E00'},
33 | {className: 'Cacna2d1.Ndnf.Cxcl14', IdentifiedType: 'CGE NGF', color: '#FF00E6'},
34 | {className: 'Cacna2d1.Ndnf.Npy', IdentifiedType: 'CGE NGF', color: '#FF00E6'},
35 | {className: 'Cacna2d1.Ndnf.Rgs10', IdentifiedType: 'CGE NGF', color: '#FF00E6'},
36 | {className: 'Sst.Npy.Cort', IdentifiedType: 'Hippocamposeptal', color: '#1FADEB'},
37 | {className: 'Sst.Npy.Zbtb20', IdentifiedType: 'Hippocamposeptal', color: '#1FADEB'},
38 | {className: 'Sst.Npy.Mgat4c', IdentifiedType: 'Hippocamposeptal', color: '#1FADEB'},
39 | {className: 'Sst.Npy.Serpine2', IdentifiedType: 'Hippocamposeptal', color: '#1FADEB'},
40 | {className: 'Calb2.Cntnap5a.Igfbp6', IdentifiedType: 'IS1', color: '#BDA800'},
41 | {className: 'Calb2.Cntnap5a.Rspo3', IdentifiedType: 'IS1', color: '#BDA800'},
42 | {className: 'Calb2.Cntnap5a.Vip', IdentifiedType: 'IS1', color: '#BDA800'},
43 | {className: 'Vip.Crh.C1ql1', IdentifiedType: 'IS2', color: '#FAE52E'},
44 | {className: 'Vip.Crh.Pcp4', IdentifiedType: 'IS2', color: '#FAE52E'},
45 | {className: 'Calb2.Vip.Gpd1', IdentifiedType: 'IS3', color: '#DBC70F'},
46 | {className: 'Calb2.Vip.Igfbp4', IdentifiedType: 'IS3', color: '#DBC70F'},
47 | {className: 'Calb2.Vip.Nos1', IdentifiedType: 'IS3', color: '#DBC70F'},
48 | {className: 'Cacna2d1.Lhx6.Vwa5a', IdentifiedType: 'Ivy', color: '#994D91'},
49 | {className: 'Cacna2d1.Lhx6.Reln', IdentifiedType: 'MGE NGF', color: '#994D91'},
50 | {className: 'Calb2.Cryab', IdentifiedType: 'NGF/I-S transition', color: '#FF00E6'},
51 | {className: 'Astro.1', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
52 | {className: 'Astro.2', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
53 | {className: 'Astro.3', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
54 | {className: 'Astro.4', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
55 | {className: 'Astro.5', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
56 | {className: 'Choroid', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
57 | {className: 'Endo', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
58 | {className: 'Eryth.1', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
59 | {className: 'Eryth.2', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
60 | {className: 'Microglia.1', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
61 | {className: 'Microglia.2', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
62 | {className: 'Oligo.1', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
63 | {className: 'Oligo.2', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
64 | {className: 'Oligo.3', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
65 | {className: 'Oligo.4', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
66 | {className: 'Oligo.5', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
67 | {className: 'Vsmc', IdentifiedType: 'Non Neuron', color: '#FFFFFF'},
68 | {className: 'Sst.Pnoc.Calb1.Igfbp5', IdentifiedType: 'O/LM', color: '#008FCC'},
69 | {className: 'Sst.Pnoc.Calb1.Pvalb', IdentifiedType: 'O/LM', color: '#008FCC'},
70 | {className: 'Sst.Pnoc.Pvalb', IdentifiedType: 'O/LM', color: '#008FCC'},
71 | {className: 'Sst.Erbb4.Crh', IdentifiedType: 'O-Bi', color: '#0070AD'},
72 | {className: 'Sst.Erbb4.Rgs10', IdentifiedType: 'O-Bi', color: '#0070AD'},
73 | {className: 'Sst.Erbb4.Th', IdentifiedType: 'O-Bi', color: '#0070AD'},
74 | {className: 'PC.CA1.1', IdentifiedType: 'PC', color: '#00FF00'},
75 | {className: 'PC.CA1.2', IdentifiedType: 'PC', color: '#00FF00'},
76 | {className: 'PC.CA1.3', IdentifiedType: 'PC', color: '#00FF00'},
77 | {className: 'PC.Other1', IdentifiedType: 'PC Other1', color: '#73E500'},
78 | {className: 'PC.Other2', IdentifiedType: 'PC Other2', color: '#73E500'},
79 | {className: 'Ntng1.Rgs10', IdentifiedType: 'Radiatum retrohip', color: '#3D855A'},
80 | {className: 'Ntng1.Synpr', IdentifiedType: 'Radiatum retrohip', color: '#3D855A'},
81 | {className: 'Ntng1.Chrm2', IdentifiedType: 'Trilaminar', color: '#1F663B'},
82 | {className: 'Sst.Cryab', IdentifiedType: 'Unidentified', color: '#A6A6A6'},
83 |
84 | // ****************************************************************************************
85 | // ********************** Do not remove the lines below. **********************************
86 | // ****************************************************************************************
87 |
88 | // Zero class is the "none from the above" class. If the algorithm cannot find a good
89 | // transcriptomic class from the single cell data then the cell will be labelled as Zero class
90 | {className: 'Zero', IdentifiedType: 'Zero', color: '#000000'},
91 |
92 | // If a class is missing from the settings above, use these default settings
93 | {className: 'Generic', IdentifiedType: 'Generic', color: '#C0C0C0'},
94 |
95 | // The donut chart at the bottom right will aggregate all classes with prob < 2% under the Other
96 | // label (see line 284, donut.js)
97 | {className: 'Other', IdentifiedType: 'Other', color: '#C0C0C0'},
98 |
99 |
100 |
101 | ];
102 |
103 | return out
104 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/config.js:
--------------------------------------------------------------------------------
1 | // NOTES:
2 | // 1. paths in 'cellData', 'geneData' and 'cellBoundaries' are with respect to the location of
3 | // 'streaming-tsv-parser.js'
4 | // 2. size is the tsv size in bytes. I use os.path.getsize() to get it. Not crucial if you
5 | // don't get it right, ie the full tsv will still be parsed despite this being wrong. It
6 | // is used by the loading page piecharts to calc how far we are.
7 | // 3. roi is the image size in pixels. Leave x0 and y0 at zero and set x1 to the width and y1 to the height.
8 | // 4. layers is a dict. Each key/value pair contains the string (the name) of the background image and the
9 | // location of the folder that the corresponding pyramid of tiles. If the tiles are stored locally, they
10 | // should be kept in a folder which is served, for example next to the tsv flatfiles. The path should be
11 | // in relation to the location of the index.html If you do not have a pyramid of tiles just
12 | // change the link to a blind one (change the jpg extension for example or just use an empty string).
13 | // The viewer should work without the dapi background though.
14 | // If the dict has more than one entries then a small control with radio button will appear at the top
15 | // right of the viewer to switch between different background images.
16 | // 5. maxZoom: maximum zoom levels. In most cases a value of 8 if good enough. If you have a big image, like
17 | // full coronal section for example then a value of 10 would make sense. Note that this should be typically
18 | // inline with the zoom level you used when you did
19 | // the pyramid of tiles. No harm is it less. If it is greater, then for these extra zoom levels there will
20 | // be no background image.
21 | // 6. spotSize: Scalar. Use this to adjust the screen-size of your spots before they morph into glyphs.
22 | function config() {
23 | return {
24 | "cellData": { "mediaLink": "../../data/cellData.tsv", "size": "2180603"},
25 | "geneData": { "mediaLink": "../../data/geneData.tsv", "size": "9630820"},
26 | "cellBoundaries": {"mediaLink": "../../data/cellBoundaries.tsv", "size": "1306209"},
27 | "roi": {"x0": 0, "x1": 7602, "y0": 0, "y1": 5471},
28 | "maxZoom": 8,
29 | "layers": {
30 | // "empty": "",
31 | "dapi": "https://storage.googleapis.com/ca1-data/img/262144px/{z}/{y}/{x}.jpg"
32 | },
33 | "spotSize": 1/16
34 | }
35 | }
36 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/customControl.js:
--------------------------------------------------------------------------------
1 | L.Control.Custom = L.Control.Layers.extend({
2 | onAdd: function () {
3 | this._controlInputs = [];
4 | this._initLayout();
5 | this._base();
6 | this._addButton();
7 | this._update();
8 | this._refresh();
9 | this._isEnabled = true;
10 | this._allSelected = false;
11 | this._noneSelected = false;
12 | this._checkBoxesCounts = 0;
13 | return this._container;
14 | },
15 |
16 | _radioController: function () {
17 | var selected = this._getSelected();
18 | if (selected.length === this._checkBoxesCounts) {
19 | $("#1").prop("checked", true)
20 | console.log('All checked')
21 | }
22 |
23 | if (selected.length === 0) {
24 | $("#0").prop("checked", true)
25 | console.log('All empty')
26 | }
27 |
28 | if ([1, this._checkBoxesCounts - 1].includes(selected.length)) {
29 | $('.control_radio').prop('checked', false);
30 | }
31 |
32 | },
33 |
34 | _radioMaker: function (id, isChecked) {
35 | var rb = document.createElement('input');
36 | rb.type = 'radio';
37 | rb.className = 'leaflet-control-layers-selector control_radio';
38 | rb.name = 'control_radio';
39 | rb.id = id;
40 | if (isChecked) {
41 | rb.setAttribute('checked', 'checked')
42 | }
43 |
44 | return rb
45 | },
46 |
47 | _base: function () {
48 | var elements = this._container.getElementsByClassName('leaflet-control-layers-list');
49 | var baseDiv = L.DomUtil.create('div', 'leaflet-control-layers-base', elements[0]);
50 |
51 | baseDiv.innerHTML = ' ' +
54 | '' +
57 | ' ';
58 |
59 | // this._controlInputs.push(cb);
60 | L.DomEvent.on(baseDiv, 'click', this._onRadioClick, this);
61 | },
62 |
63 | _onRadioClick: function (e) {
64 | var id = +$("input[name='control_radio']:checked").attr('id');
65 | id === 1 ? this._checkAll() : this._unCheckAll();
66 |
67 | this._radioController();
68 | this._refresh();
69 | },
70 |
71 | _checkAll: function () {
72 | var inputs = this._getAllInputs();
73 | for (var i = 0; i < inputs.length; i++) {
74 | var input = inputs[i];
75 | if (!input.checked) {
76 | input.checked = true
77 | }
78 | }
79 | },
80 |
81 | _unCheckAll: function () {
82 | var inputs = this._getAllInputs();
83 | for (var i = 0; i < inputs.length; i++) {
84 | var input = inputs[i];
85 | if (input.checked) {
86 | input.checked = false
87 | }
88 | }
89 |
90 | },
91 |
92 | _getAllInputs: function () {
93 | return document.querySelectorAll('input.leaflet-customcontrol-layers-selector' +
94 | ':not([name="control_radio"])');
95 | },
96 |
97 | _addButton: function (containerName) {
98 | if (this._isEnabled) {
99 |
100 | var elements = this._container.getElementsByClassName('leaflet-control-layers-list');
101 | var div = L.DomUtil.create('div', 'leaflet-control-layers-overlays', elements[0]);
102 | var cb = document.createElement('input');
103 | cb.type = 'checkbox';
104 | cb.className = 'leaflet-customcontrol-layers-selector';
105 | cb.defaultChecked = 'checked';
106 | cb.value = containerName;
107 | cb.name = 'control_checkbox';
108 | cb.checked = true;
109 |
110 | div.innerHTML = ' ';
113 |
114 | // this._controlInputs.push(cb);
115 | L.DomEvent.on(div, 'click', this._onInputClick, this);
116 |
117 | this._checkBoxesCounts = this._checkBoxesCounts + 1
118 |
119 | }
120 | },
121 |
122 | _contentToggle: function (target) {
123 | var tempContainer;
124 | var containerName = target.value;
125 | if (target.checked) {
126 | tempContainer = cellContainer_array.filter(d => d.name === containerName)[0];
127 | // pixiContainer.addChild(tempContainer);
128 | tempContainer.visible = true
129 | masterCellRenderer.render(masterCellContainer)
130 | } else {
131 | tempContainer = masterCellContainer.getChildByName(containerName);
132 | tempContainer.visible = false
133 | // pixiContainer.removeChild(tempContainer);
134 | masterCellRenderer.render(masterCellContainer)
135 | }
136 | },
137 |
138 | _onInputClick: function (e) {
139 | // var containerName = e.target.value;
140 | masterCellRenderer.render(masterCellContainer); // Not sure if I need that here. I cant remember if its on purpose or not...it is called a few lines below anyway
141 | this._contentToggle(e.target);
142 |
143 | this._radioController()
144 |
145 | },
146 |
147 | _refresh: function () {
148 | //This deserves its own space, it is not a simple function to sit inside the code for this layer control
149 | var inputs = this._getAllInputs();
150 |
151 | for (var i = 0; i < inputs.length; i++) {
152 | var input = inputs[i];
153 | this._contentToggle(input);
154 | }
155 | },
156 |
157 | _getSelected: function () {
158 | var inputs = this._getAllInputs();
159 |
160 | var selected = Array.from(inputs).filter(d => d.checked === true).map(d => d.value);
161 | return selected
162 | },
163 |
164 | });
165 |
166 | L.control.custom = function() {
167 | return new L.Control.Custom();
168 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/dataLoader.js:
--------------------------------------------------------------------------------
1 | function data_loader(workPackage) {
2 | var data = [],
3 | agg_data,
4 | previous_avg = 0;
5 | var average = list => list.reduce((prev, curr) => prev + curr) / list.length;
6 | workPackage = workPackage.sort((a,b) => a.size-b.size); //order by size (hemmm...doest really matter, does it?? Everything happens in parallel)
7 |
8 | function aggregate_stats(workPackage){
9 | out = [];
10 | var names = [... new Set(workPackage.map(d => d.root_name))].sort();
11 | names.forEach(name => {
12 | var temp = workPackage.filter(d => d.root_name === name);
13 | // console.log(temp)
14 | var size = temp.map(d => d.size).reduce((a, b) => a + b, 0);
15 | var bytes_streamed = temp.map(d => d.bytes_streamed).reduce((a, b) => a + b, 0);
16 | // var data = temp.map(d => d.data).reduce((a,b)=>a.concat(b));
17 | var data_length = temp.map(d => d.data_length).reduce((a, b) => a + b, 0);
18 | var progress = bytes_streamed / size;
19 | out.push({name, size, bytes_streamed, progress, data_length})
20 | // console.log(out)
21 | });
22 | return out
23 | }
24 |
25 | function aggregate_data(workPackage) {
26 | out = {};
27 | var names = [...new Set(workPackage.map(d => d.root_name))].sort();
28 | names.forEach(name => {
29 | var temp = workPackage.filter(d => d.root_name === name);
30 | var data = temp.map(d => d.data).reduce((a,b)=>a.concat(b));
31 | out[name] = data
32 | });
33 | return out
34 | }
35 |
36 | function makeInnerHtml(data) {
37 | var innerHtml = "";
38 | for (var i = 0; i < data.length; i++) {
39 | innerHtml = innerHtml + data[i] + "/"
40 | }
41 | innerHtml = innerHtml.slice(0, -1); //remove the last slash from the end of the string
42 | innerHtml = " Loaded: " + innerHtml + "";
43 |
44 | return innerHtml
45 | }
46 |
47 | function setupWorker() {
48 | // create a web worker that streams the chart data
49 | worker = new Worker("viewer/js/streaming-tsv-parser.js");
50 | worker.onmessage = function (event) {
51 | if (event.data.finished) {
52 | console.log(agg_data);
53 | data = aggregate_data(workPackage);
54 | onDataLoaded(data);
55 | // redraw(stats);
56 | return
57 | }
58 | var i = event.data.i;
59 | workPackage[i].bytes_streamed += event.data.bytes_streamed;
60 | workPackage[i].data = workPackage[i].data.concat(event.data.items);
61 | workPackage[i].data_length += event.data.items.length;
62 | // if (i === 0) {
63 | // console.log('i: ' + i)
64 | // console.log('bytes_streamed: ' + workPackage[i].bytes_streamed)
65 | // console.log('size: ' + workPackage[i].size)
66 | // console.log('data length: ' + workPackage[i].data.length)
67 | // console.log('')
68 | // }
69 | agg_data = aggregate_stats(workPackage);
70 |
71 | redraw(agg_data);
72 | };
73 | }
74 |
75 | function redraw(data) {
76 | // console.log(data[0])
77 | // console.log(data[1])
78 | innerHtml = makeInnerHtml(data.map(d => d3.format(",")(d.data_length)));
79 | document.getElementById("loading").innerHTML = innerHtml;
80 |
81 | innerHtml = makeInnerHtml(data.map(d => d3.format(".0%")(d.progress) ));
82 | innerHtml = innerHtml.replace(/% \//g, '% ');
83 | document.getElementById("loading_perc").innerHTML = innerHtml;
84 |
85 | innerHtml = makeInnerHtml(data.map(d => (d.bytes_streamed/(1024*1024)).toFixed() + 'MB' ));
86 | document.getElementById("loading_mb").innerHTML = innerHtml;
87 |
88 |
89 | var avg = average(data.map(d => d.progress));
90 | var avg_mb = average(data.map(d => (d.bytes_streamed/(1024*1024)).toFixed() ));
91 | var progress_1 = data[0].progress,
92 | progress_2 = data[1].progress;
93 | progress_3 = data[2].progress;
94 |
95 | var inc = 0.0; // controls how often it will be update. Every 2% set inc = 0.02
96 | if (avg >= Math.min(1, previous_avg + inc)) {
97 | if (avg > 0.99){
98 | $('#wait_chart').show();
99 | }
100 | // refresh the progress animation
101 | updateDonutChart('#specificChart', progress_1*100, true);
102 | var mb_1 = (data[0].bytes_streamed/(1024*1024)).toFixed();
103 | $('#mb').html(mb_1 + 'MB');
104 | $('#datapoints').html(d3.format(",")(data[0].data_length));
105 |
106 | updateDonutChart('#specificChart2', progress_2*100, true);
107 | $('#mb2').html((data[1].bytes_streamed/(1024*1024)).toFixed() + 'MB');
108 | $('#datapoints2').html(d3.format(",")(data[1].data_length));
109 |
110 | updateDonutChart('#specificChart3', progress_3*100, true);
111 | $('#mb3').html((data[2].bytes_streamed/(1024*1024)).toFixed() + 'MB');
112 | $('#datapoints3').html(d3.format(",")(data[2].data_length));
113 |
114 | previous_avg = avg;
115 | }
116 | }
117 |
118 | function onDataLoaded(data) {
119 | [cellBoundaries, cellData, genepanel] = postLoad([data.cellBoundaries, data.cellData, data.geneData]);
120 |
121 | // sort cellBoundaries and cellData. These two should be aligned
122 | cellBoundaries.sort((a, b) => a.cell_id - b.cell_id);
123 | cellData.sort((a, b) => a.cell_id - b.cell_id);
124 |
125 | all_geneData = data.geneData;
126 | console.log('loading data finished');
127 | console.log('num of genes loaded: ' + all_geneData.length);
128 | console.log('num of cells loaded: ' + cellData.length);
129 |
130 |
131 | //finaly make a spatial index on the spots. We will need that to filter them if/when needed
132 | console.log('Creating the index');
133 | spotsIndex = new KDBush(all_geneData, p => p.x, p => p.y, 64, Int32Array);
134 |
135 | // do now the chart
136 | dapiChart(configSettings);
137 |
138 | }
139 |
140 | setupWorker();
141 | worker.postMessage(workPackage);
142 | }
143 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/dt.js:
--------------------------------------------------------------------------------
1 | function renderDataTable(d) {
2 |
3 |
4 | var mydata = [];
5 | var mydata2 = [];
6 |
7 | // var str = "Cell Num: " + d.Cell_Num +
8 | // ", (x, y): (" + d.x.toFixed(2) + ", " + d.y.toFixed(2) + ")";
9 | // document.getElementById('dtTitle').innerHTML = str;
10 | var n = d3.max([d.CellGeneCount.length, d.Genenames.length]);
11 | for (i = 0; i < n; i++) {
12 | mydata.push({
13 | "Genenames": (d.Genenames[i] === undefined) ? "" : d.Genenames[i],
14 | "CellGeneCount": (d.CellGeneCount[i] === undefined) ? "" : +d.CellGeneCount[i].toFixed(2),
15 | })
16 | }
17 |
18 | var n = d3.max([d.ClassName.length, d.Prob.length]);
19 | for (i = 0; i < n; i++) {
20 | mydata2.push({
21 | "ClassName": (d.ClassName[i] === undefined) ? "" : d.ClassName[i],
22 | "Prob": (d.ClassName[i] === undefined) ? "" : (!Array.isArray(d.Prob)) ? [d.Prob] : d.Prob[i], // d.Prob can be just a float, make sure it is an array
23 | })
24 | }
25 |
26 |
27 | // check if a there is a reference to a datatable.
28 | // If yes, refresh with the new data
29 | // Otherwise create and populate a datatable
30 | if ($.fn.dataTable.isDataTable('#dtTable')) {
31 | table = $('#dtTable').DataTable();
32 | table.clear().rows.add(mydata).draw();
33 | } else {
34 | table = $('#dtTable').DataTable({
35 | //bFilter: false,
36 | "lengthChange": false,
37 | searching: false,
38 | //"scrollY": "200px",
39 | //"scrollCollapse": true,
40 | "paging": true,
41 | "pagingType": "simple",
42 | // "dom": 't',
43 | "bInfo": false, //Dont display info e.g. "Showing 1 to 4 of 4 entries"
44 | // "paging": false,//Dont want paging
45 | "bPaginate": false,//Dont want paging
46 |
47 | "data": mydata,
48 | "columns": [
49 | {
50 | title: "Gene",
51 | data: "Genenames"
52 | },
53 | {
54 | title: "Counts",
55 | data: "CellGeneCount"
56 | },
57 | ],
58 | });
59 |
60 | }
61 |
62 | function getTotal(table){
63 | var total = table.column(1).data().reduce(function (a,b) {return a+b}, 0)
64 | $(table.column(1).footer()).html('Total: ' +total )
65 | console.log('Total number of gene counts: ' + total )
66 |
67 | return total
68 | }
69 |
70 |
71 | if ($.fn.dataTable.isDataTable('#dtTable2')) {
72 | table2 = $('#dtTable2').DataTable();
73 | table2.clear().rows.add(mydata2).draw();
74 | } else {
75 | table2 = $('#dtTable2').DataTable({
76 | //bFilter: false,
77 | "lengthChange": false,
78 | searching: false,
79 | //"scrollY": "200px",
80 | //"scrollCollapse": true,
81 | "paging": true,
82 | //dom: 't',
83 | "data": mydata2,
84 | "columns": [
85 | {
86 | title: "Class Name",
87 | data: "ClassName"
88 | },
89 | {
90 | title: "Prob",
91 | data: "Prob"
92 | },
93 | ]
94 | });
95 | }
96 |
97 | // Sort by column 1 and then re-draw
98 | table
99 | .order([1, 'desc'])
100 | .draw();
101 |
102 | table2
103 | .order([1, 'desc'])
104 | .draw();
105 |
106 | var total = getTotal(table);
107 | // centroid = dapiConfig.t.untransform(d.centroid);
108 |
109 | var str = " Cell Num: " + d.cell_id
110 | + ", Gene Counts: " + total.toFixed(0)
111 | + ", (x, y): (" + d.centroid[0].toFixed(0) + ", " + d.centroid[1].toFixed(0) + ") ";
112 |
113 | if (pinnedControls){
114 | str = str + " ";
115 | }
116 | else
117 | str = str + "
";
115 | }
116 | else
117 | str = str + " ";
118 | document.getElementById('dtTitle').innerHTML = str
119 |
120 | }
121 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/glyphPaths.js:
--------------------------------------------------------------------------------
1 |
2 | function ctxPath(glyphName, ctx, p, r) {
3 |
4 | if (glyphName === 'star6') {
5 | ctx.beginPath();
6 | ctx.moveTo(p.x + r, p.y);
7 | ctx.lineTo(p.x + 0.43 * r, p.y + 0.25 * r);
8 | ctx.lineTo(p.x + 0.50 * r, p.y + 0.87 * r);
9 | ctx.lineTo(p.x, p.y + 0.50 * r);
10 | ctx.lineTo(p.x - 0.50 * r, p.y + 0.87 * r);
11 | ctx.lineTo(p.x - 0.43 * r, p.y + 0.25 * r);
12 | ctx.lineTo(p.x - r, p.y);
13 | ctx.lineTo(p.x - 0.43 * r, p.y - 0.25 * r);
14 | ctx.lineTo(p.x - 0.50 * r, p.y - 0.87 * r);
15 | ctx.lineTo(p.x, p.y - 0.50 * r);
16 | ctx.lineTo(p.x + 0.50 * r, p.y - 0.87 * r);
17 | ctx.lineTo(p.x + 0.43 * r, p.y - 0.25 * r);
18 | ctx.closePath();
19 | }
20 |
21 |
22 | else if (glyphName === 'star5') {
23 | ctx.beginPath();
24 | ctx.moveTo(p.x, p.y + 0.658351875 * r);
25 | ctx.lineTo(p.x + 0.618027443 * r, p.y + 1 * r);
26 | ctx.lineTo(p.x + 0.5 * r, p.y + 0.27637816 * r);
27 | ctx.lineTo(p.x + 1 * r, p.y - 0.236080209 * r);
28 | ctx.lineTo(p.x + 0.309026865 * r, p.y - 0.341634306 * r);
29 | ctx.lineTo(p.x + 0 * r, p.y - 1 * r);
30 | ctx.lineTo(p.x - 0.309026865 * r, p.y - 0.341634306 * r);
31 | ctx.lineTo(p.x - 1 * r, p.y - 0.236080209 * r);
32 | ctx.lineTo(p.x - 0.5 * r, p.y + 0.27637816 * r);
33 | ctx.lineTo(p.x - 0.618027443 * r, p.y + 1 * r);
34 | ctx.lineTo(p.x, p.y + 0.658351875 * r);
35 | ctx.closePath();
36 | }
37 |
38 |
39 | else if (glyphName === 'diamond') {
40 | ctx.beginPath();
41 | ctx.moveTo(p.x - r, p.y);
42 | ctx.lineTo(p.x, p.y - r);
43 | ctx.lineTo(p.x + r, p.y);
44 | ctx.lineTo(p.x, p.y + r);
45 | ctx.lineTo(p.x - r, p.y);
46 | ctx.closePath();
47 | }
48 |
49 |
50 | else if (glyphName === 'square') {
51 | ctx.beginPath();
52 | ctx.moveTo(p.x - r, p.y - r);
53 | ctx.lineTo(p.x + r, p.y - r);
54 | ctx.lineTo(p.x + r, p.y + r);
55 | ctx.lineTo(p.x - r, p.y + r);
56 | ctx.lineTo(p.x - r, p.y - r);
57 | ctx.closePath();
58 | }
59 |
60 |
61 | else if (glyphName === 'triangleUp') {
62 | ctx.beginPath();
63 | ctx.moveTo(p.x - r, p.y + r);
64 | ctx.lineTo(p.x, p.y - r);
65 | ctx.lineTo(p.x + r, p.y + r);
66 | ctx.closePath();
67 | }
68 |
69 |
70 | else if (glyphName === 'triangleDown') {
71 | ctx.beginPath();
72 | ctx.moveTo(p.x - r, p.y - r);
73 | ctx.lineTo(p.x, p.y + r);
74 | ctx.lineTo(p.x + r, p.y - r);
75 | ctx.closePath();
76 | }
77 |
78 |
79 | else if (glyphName === 'triangleRight') {
80 | ctx.beginPath();
81 | ctx.moveTo(p.x - r, p.y - r);
82 | ctx.lineTo(p.x + r, p.y);
83 | ctx.lineTo(p.x - r, p.y + r);
84 | ctx.closePath();
85 | }
86 |
87 |
88 | else if (glyphName === 'triangleLeft') {
89 | ctx.beginPath();
90 | ctx.moveTo(p.x + r, p.y - r);
91 | ctx.lineTo(p.x - r, p.y);
92 | ctx.lineTo(p.x + r, p.y + r);
93 | ctx.closePath();
94 | }
95 |
96 |
97 | else if (glyphName === 'cross') {
98 | ctx.beginPath();
99 | ctx.moveTo(p.x + r, p.y + r);
100 | ctx.lineTo(p.x - r, p.y - r);
101 | ctx.moveTo(p.x - r, p.y + r);
102 | ctx.lineTo(p.x + r, p.y - r);
103 | ctx.closePath();
104 | }
105 |
106 |
107 | else if (glyphName === 'plus') {
108 | ctx.beginPath();
109 | ctx.moveTo(p.x, p.y + r);
110 | ctx.lineTo(p.x, p.y - r);
111 | ctx.moveTo(p.x - r, p.y);
112 | ctx.lineTo(p.x + r, p.y);
113 | ctx.closePath();
114 | }
115 |
116 |
117 | else if (glyphName === 'asterisk') {
118 | ctx.beginPath();
119 | ctx.moveTo(p.x, p.y + r);
120 | ctx.lineTo(p.x, p.y - r);
121 | ctx.moveTo(p.x - r, p.y);
122 | ctx.lineTo(p.x + r, p.y);
123 | ctx.moveTo(p.x + 0.5 * r, p.y + 0.5 * r);
124 | ctx.lineTo(p.x - 0.5 * r, p.y - 0.5 * r);
125 | ctx.moveTo(p.x - 0.5 * r, p.y + 0.5 * r);
126 | ctx.lineTo(p.x + 0.5 * r, p.y - 0.5 * r);
127 | ctx.arc(p.x, p.y, 2, 0, Math.PI * 2, true);
128 | ctx.closePath();
129 | }
130 |
131 |
132 | else if (glyphName === 'circle') {
133 | ctx.beginPath();
134 | ctx.arc(p.x, p.y, r, 0, Math.PI * 2, true);
135 | ctx.closePath();
136 | }
137 |
138 |
139 | else if (glyphName === 'point') {
140 | ctx.beginPath();
141 | ctx.arc(p.x, p.y, 3, 0, Math.PI * 2, true);
142 | ctx.arc(p.x, p.y, 2, 0, Math.PI * 2, true);
143 | ctx.closePath();
144 | }
145 |
146 | else {
147 | console.log('glyph: "' + glyphName + '" not implemented.')
148 | }
149 |
150 | return ctx
151 | }
152 |
153 |
154 |
155 |
156 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/glyphs.js:
--------------------------------------------------------------------------------
1 |
2 | L.Canvas.include({
3 | _updateMarkerStar6: function (layer) {
4 | if (!this._drawing || layer._empty()) { return; }
5 |
6 | var p = layer._point,
7 | ctx = this._ctx,
8 | r = Math.max(Math.round(layer._radius), 8);
9 |
10 | this._layers[layer._leaflet_id] = layer;
11 | ctx = ctxPath('star6', ctx, p, r);
12 | this._fillStroke(ctx, layer);
13 | }
14 | });
15 |
16 |
17 | L.Canvas.include({
18 | _updateMarkerStar: function (layer) {
19 | if (!this._drawing || layer._empty()) { return; }
20 |
21 | var p = layer._point,
22 | ctx = this._ctx,
23 | r = Math.max(Math.round(layer._radius), 8);
24 |
25 | this._layers[layer._leaflet_id] = layer;
26 | ctx = ctxPath('star5', ctx, p, r);
27 | this._fillStroke(ctx, layer);
28 |
29 | }
30 | });
31 |
32 | L.Canvas.include({
33 | _updateMarkerDiamond: function (layer) {
34 | if (!this._drawing || layer._empty()) { return; }
35 |
36 | var p = layer._point,
37 | ctx = this._ctx,
38 | r = Math.max(Math.round(layer._radius), 6);
39 |
40 | this._layers[layer._leaflet_id] = layer;
41 | ctx = ctxPath('diamond', ctx, p, r);
42 | this._fillStroke(ctx, layer);
43 | }
44 | });
45 |
46 |
47 | L.Canvas.include({
48 | _updateMarkerSquare: function (layer) {
49 | if (!this._drawing || layer._empty()) { return; }
50 |
51 | var p = layer._point,
52 | ctx = this._ctx,
53 | r = Math.max(Math.round(layer._radius), 5);
54 |
55 | this._layers[layer._leaflet_id] = layer;
56 | ctx = ctxPath('square', ctx, p, r);
57 | this._fillStroke(ctx, layer);
58 | }
59 | });
60 |
61 | L.Canvas.include({
62 | _updateMarkerTriangleUp: function (layer) {
63 | if (!this._drawing || layer._empty()) { return; }
64 |
65 | var p = layer._point,
66 | ctx = this._ctx,
67 | r = Math.max(Math.round(layer._radius), 5);
68 |
69 | this._layers[layer._leaflet_id] = layer;
70 | ctx = ctxPath('triangleUp', ctx, p, r);
71 | this._fillStroke(ctx, layer);
72 | }
73 | });
74 |
75 |
76 | L.Canvas.include({
77 | _updateMarkerTriangleDown: function (layer) {
78 | if (!this._drawing || layer._empty()) { return; }
79 |
80 | var p = layer._point,
81 | ctx = this._ctx,
82 | r = Math.max(Math.round(layer._radius), 5);
83 |
84 | this._layers[layer._leaflet_id] = layer;
85 | ctx = ctxPath('triangleDown', ctx, p, r);
86 | this._fillStroke(ctx, layer);
87 | }
88 | });
89 |
90 |
91 |
92 | L.Canvas.include({
93 | _updateMarkerTriangleRight: function (layer) {
94 | if (!this._drawing || layer._empty()) { return; }
95 |
96 | var p = layer._point,
97 | ctx = this._ctx,
98 | r = Math.max(Math.round(layer._radius), 5);
99 |
100 | this._layers[layer._leaflet_id] = layer;
101 | ctx = ctxPath('triangleRight', ctx, p, r);
102 | this._fillStroke(ctx, layer);
103 | }
104 | });
105 |
106 |
107 | L.Canvas.include({
108 | _updateMarkerTriangleLeft: function (layer) {
109 | if (!this._drawing || layer._empty()) { return; }
110 |
111 | var p = layer._point,
112 | ctx = this._ctx,
113 | r = Math.max(Math.round(layer._radius), 5);
114 |
115 | this._layers[layer._leaflet_id] = layer;
116 | ctx = ctxPath('triangleLeft', ctx, p, r);
117 | this._fillStroke(ctx, layer);
118 | }
119 | });
120 |
121 |
122 | L.Canvas.include({
123 | _updateMarkerCross: function (layer) {
124 | if (!this._drawing || layer._empty()) { return; }
125 |
126 | var p = layer._point,
127 | ctx = this._ctx,
128 | r = Math.max(Math.round(layer._radius), 5);
129 |
130 | this._layers[layer._leaflet_id] = layer;
131 | ctx = ctxPath('cross', ctx, p, r);
132 | this._fillStroke(ctx, layer);
133 | }
134 | });
135 |
136 |
137 |
138 | L.Canvas.include({
139 | _updateMarkerPlus: function (layer) {
140 | if (!this._drawing || layer._empty()) { return; }
141 |
142 | var p = layer._point,
143 | ctx = this._ctx,
144 | r = Math.max(Math.round(layer._radius), 7);
145 |
146 | this._layers[layer._leaflet_id] = layer;
147 | ctx = ctxPath('plus', ctx, p, r);
148 | this._fillStroke(ctx, layer);
149 | }
150 | });
151 |
152 |
153 | L.Canvas.include({
154 | _updateMarkerAsterisk: function (layer) {
155 | if (!this._drawing || layer._empty()) { return; }
156 |
157 | var p = layer._point,
158 | ctx = this._ctx,
159 | r = Math.max(Math.round(layer._radius), 7);
160 |
161 | this._layers[layer._leaflet_id] = layer;
162 | ctx = ctxPath('asterisk', ctx, p, r);
163 | this._fillStroke(ctx, layer);
164 | }
165 | });
166 |
167 |
168 | L.Canvas.include({
169 | _updateMarkerCircle: function (layer) {
170 | if (!this._drawing || layer._empty()) { return; }
171 |
172 | var p = layer._point,
173 | ctx = this._ctx,
174 | r = Math.max(Math.round(layer._radius), 6);
175 |
176 | this._layers[layer._leaflet_id] = layer;
177 | ctx = ctxPath('circle', ctx, p, r);
178 | this._fillStroke(ctx, layer);
179 | }
180 | });
181 |
182 |
183 | L.Canvas.include({
184 | _updateMarkerDot: function (layer) {
185 | if (!this._drawing || layer._empty()) { return; }
186 |
187 | var p = layer._point,
188 | ctx = this._ctx;
189 | //r = Math.max(Math.round(layer._radius), 6);
190 |
191 | this._layers[layer._leaflet_id] = layer;
192 | ctx = ctxPath('point', ctx, p);
193 | this._fillStroke(ctx, layer);
194 | }
195 | });
196 |
197 | var svgGlyph = L.CircleMarker.extend({
198 | _updatePath: function() {
199 | if ((this.options.shape === "star5") && (this._renderer._updateMarkerStar))
200 | this._renderer._updateMarkerStar(this);
201 | if ((this.options.shape === "star6") && (this._renderer._updateMarkerStar6))
202 | this._renderer._updateMarkerStar6(this);
203 | if ((this.options.shape === "diamond") && (this._renderer._updateMarkerDiamond))
204 | this._renderer._updateMarkerDiamond(this);
205 | if ((this.options.shape === "square") && (this._renderer._updateMarkerSquare))
206 | this._renderer._updateMarkerSquare(this);
207 | if ((this.options.shape === "triangleUp") && (this._renderer._updateMarkerTriangleUp))
208 | this._renderer._updateMarkerTriangleUp(this);
209 | if ((this.options.shape === "triangleDown") && (this._renderer._updateMarkerTriangleDown))
210 | this._renderer._updateMarkerTriangleDown(this);
211 | if ((this.options.shape === "triangleLeft") && (this._renderer._updateMarkerTriangleLeft))
212 | this._renderer._updateMarkerTriangleLeft(this);
213 | if ((this.options.shape === "triangleRight") && (this._renderer._updateMarkerTriangleRight))
214 | this._renderer._updateMarkerTriangleRight(this);
215 | if ((this.options.shape === "cross") && (this._renderer._updateMarkerCross))
216 | this._renderer._updateMarkerCross(this);
217 | if ((this.options.shape === "plus") && (this._renderer._updateMarkerPlus))
218 | this._renderer._updateMarkerPlus(this);
219 | if ((this.options.shape === "asterisk") && (this._renderer._updateMarkerAsterisk))
220 | this._renderer._updateMarkerAsterisk(this);
221 | if ((this.options.shape === "circle") && (this._renderer._updateMarkerCircle))
222 | this._renderer._updateMarkerCircle(this);
223 | if ((this.options.shape === "point") && (this._renderer._updateMarkerDot))
224 | this._renderer._updateMarkerDot(this);
225 | }
226 | });
227 |
228 |
229 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/L.Control.Layers.Tree.css:
--------------------------------------------------------------------------------
1 | .leaflet-control-layers-toggle.leaflet-layerstree-named-toggle {
2 | margin: 2px 5px;
3 | width: auto;
4 | height: auto;
5 | background-image: none;
6 | }
7 |
8 | .leaflet-layerstree-node {
9 | }
10 |
11 | .leaflet-layerstree-header input{
12 | margin-left: 0px;
13 | margin-right: 5px;
14 | }
15 |
16 |
17 | .leaflet-layerstree-header {
18 | }
19 |
20 | .leaflet-layerstree-header-pointer {
21 | cursor: pointer;
22 | }
23 |
24 | .leaflet-layerstree-header label {
25 | display: inline-block;
26 | cursor: pointer;
27 | }
28 |
29 | .leaflet-layerstree-header-label {
30 | }
31 |
32 | .leaflet-layerstree-header-name {
33 | }
34 |
35 | .leaflet-layerstree-header-space {
36 | }
37 |
38 | .leaflet-layerstree-children {
39 | /*padding-left: 10px;*/
40 | padding-left: 15px;
41 | }
42 |
43 | .leaflet-layerstree-children-nopad {
44 | padding-left: 0px;
45 | }
46 |
47 | .leaflet-layerstree-closed {
48 | }
49 |
50 | .leaflet-layerstree-opened {
51 | }
52 |
53 | .leaflet-layerstree-hide {
54 | display: none;
55 | }
56 |
57 | .leaflet-layerstree-nevershow {
58 | display: none;
59 | }
60 |
61 | .leaflet-layerstree-expand-collapse {
62 | cursor: pointer;
63 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/Leaflet.Coordinates-0.1.3.css:
--------------------------------------------------------------------------------
1 | .leaflet-control-coordinates{background-color:#D8D8D8;background-color:rgba(255,255,255,.8);cursor:pointer}.leaflet-control-coordinates,.leaflet-control-coordinates .uiElement input{-webkit-border-radius:5px;-moz-border-radius:5px;border-radius:5px}.leaflet-control-coordinates .uiElement{margin:4px}.leaflet-control-coordinates .uiElement .labelFirst{margin-right:4px}.leaflet-control-coordinates .uiHidden{display:none}
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/keen-dashboards.css:
--------------------------------------------------------------------------------
1 |
2 | /*
3 | * Full-page application style
4 | */
5 |
6 | body.application {
7 | background: #f2f2f2;
8 | padding: 60px 20px 0;
9 | }
10 | body.application > .container-fluid {
11 | padding-left: 0px;
12 | padding-right: 0px;
13 | }
14 | body.application div[class^="col-"] {
15 | padding-left: 5px;
16 | padding-right: 5px;
17 | }
18 | body.application div[class^="col-"] div[class^="col-"] {
19 | padding-left: 15px;
20 | padding-right: 15px;
21 | }
22 |
23 | body.application hr {
24 | border-color: #d7d7d7;
25 | margin: 10px 0;
26 | }
27 |
28 |
29 |
30 | .navbar-inverse {
31 | background-color: #3d4a57;
32 | border-color: #333;
33 | }
34 | .navbar-inverse .navbar-nav > li > a,
35 | .navbar a.navbar-brand {
36 | color: #fbfbfb;
37 | text-decoration: none;
38 | }
39 |
40 | .chart-wrapper {
41 | background: #fff;
42 | border: 1px solid #e2e2e2;
43 | border-radius: 3px;
44 | margin-bottom: 10px;
45 | }
46 | .chart-wrapper .chart-title {
47 | border-bottom: 1px solid #d7d7d7;
48 | color: #666;
49 | font-size: 14px;
50 | font-weight: 200;
51 | padding: 7px 10px 4px;
52 | }
53 |
54 | .chart-wrapper .chart-stage {
55 | /*min-height: 240px;*/
56 | overflow: hidden;
57 | padding: 5px 10px;
58 | position: relative;
59 | }
60 |
61 | .chart-wrapper .chart-notes {
62 | background: #fbfbfb;
63 | border-top: 1px solid #e2e2e2;
64 | color: #808080;
65 | font-size: 12px;
66 | padding: 8px 10px 5px;
67 | }
68 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/screen.css:
--------------------------------------------------------------------------------
1 | * {
2 | font-size: 100.01%;
3 | margin: 0;
4 | padding: 0;
5 | }
6 | html {
7 | font: 62.5%/1.5 Verdana, Tahoma, sans-serif;
8 | }
9 | html, body {
10 | width: 100%;
11 | height: 100%;
12 | overflow: hidden;
13 | }
14 | #map {
15 | position: absolute;
16 | top: 100px;
17 | bottom: 22px;
18 | left: 0;
19 | right: 0;
20 | overflow: hidden;
21 | border-top: 1px solid #ccc;
22 | border-bottom: 1px solid #ccc;
23 | }
24 | #footer {
25 | position: absolute;
26 | bottom: 0;
27 | left: 0;
28 | height: 25px;
29 | line-height: 25px;
30 | padding-left: 20px;
31 | font-size: 1.1em;
32 | color: #777;
33 | }
34 | #footer a, .ui-widget-content a, a {
35 | color: #0083CB;
36 | }
37 | #welcome {
38 | }
39 | .ui-widget-content {
40 | background: white;
41 | }
42 | .ui-widget-header {
43 | color: black;
44 | }
45 | .ui-dialog-content p {
46 | font-size: 1.2em;
47 | margin: 1em 0;
48 | }
49 | #header {
50 | height: 70px;
51 | width: 100%;
52 | position: absolute;
53 | }
54 | #header h1 {
55 | font: bold 3.2em Georgia, serif;
56 | float: left;
57 | height: 60px;
58 | margin: 9px 0 0 10px;
59 | display: inline;
60 | }
61 | #header h1 a {
62 | text-decoration: none;
63 | color: black;
64 | padding-top: 47px;
65 | height: 0;
66 | width: 143px;
67 | overflow: hidden;
68 | display: block;
69 | background: url(../images/logo.png) no-repeat;
70 | }
71 | #settings {
72 | font-size: 1.4em;
73 | margin: 6px 0 0 0;
74 | white-space: nowrap;
75 | }
76 | #date-container, #time-container, #location-container {
77 | float: left;
78 | display: inline;
79 | }
80 | #date-container {
81 | margin-right: 7px;
82 | }
83 | input[type=text], input.text {
84 | border: 1px solid #bbb;
85 | margin: 10px 0;
86 | padding: 4px;
87 | font-size: 1.4em;
88 | }
89 | input[type=text]:focus, input.text:focus, input[type=text]:hover, input.text:hover {
90 | border-color: #666;
91 | }
92 | #date {
93 | width: 120px;
94 | }
95 | #time {
96 | width: 55px;
97 | }
98 | #location {
99 | width: 250px;
100 | }
101 | #location-container {
102 | float: left;
103 | display: inline;
104 | margin-right: 7px;
105 | }
106 | #tagline {
107 | margin-right: 7px;
108 | margin-top: 20px;
109 | float: left;
110 | display: inline;
111 | }
112 | #location-container button, #time-container button {
113 | display: none;
114 | }
115 | #location-container.extended, #time-container.extended {
116 | background: white;
117 | z-index: 2000;
118 | position: relative;
119 | text-align: center;
120 | white-space: normal;
121 | padding: 0 10px 0;
122 | border: 1px solid #bbb;
123 | margin-top: -1px;
124 | }
125 | #location-container.extended button, #time-container.extended button {
126 | display: inline;
127 | }
128 |
129 | /* button styles taken from the lovely Blueprint CSS framework */
130 |
131 | a.button, button {
132 | margin: 0;
133 | margin-bottom: 10px;
134 | padding:4px 10px 5px 7px; /* Links */
135 |
136 | border:1px solid #dedede;
137 | border-top:1px solid #eee;
138 | border-left:1px solid #eee;
139 |
140 | background-color:#f0f0f0;
141 | line-height:130%;
142 | text-decoration:none;
143 | color:#565656;
144 | cursor:pointer;
145 | }
146 | button {
147 | width:auto;
148 | overflow:visible;
149 | padding:4px 7px 3px 7px; /* IE6 */
150 | }
151 | button[type] {
152 | padding:4px 7px 4px 7px; /* Firefox */
153 | line-height:17px; /* Safari */
154 | }
155 | *:first-child+html button[type] {
156 | padding:4px 7px 3px 7px; /* IE7 */
157 | }
158 | button:hover, a.button:hover{
159 | background-color:#dff4ff;
160 | border:1px solid #c2e1ef;
161 | color:#336699;
162 | }
163 | a.button:active{
164 | background-color:#6299c5;
165 | border:1px solid #6299c5;
166 | color:#fff;
167 | }
168 |
169 | .time-span, #welcome .sunrise, #welcome .sun, #welcome .sunset {
170 | padding: 0px 4px 1px;
171 | border-radius: 5px;
172 | -moz-border-radius: 5px;
173 | -webkit-border-radius: 5px;
174 | }
175 | #before-sunrise, #after-sunset, #daylight {
176 | display: none;
177 | }
178 | .sunrise {
179 | background: #FFED9E;
180 | }
181 | .sunset {
182 | background: #FFC3AD;
183 | }
184 | .sun {
185 | background: #ffdc9c;
186 | }
187 | .transit {
188 | background: #FFFC9C;
189 | }
190 | .dark {
191 | background: #bfd5dd;
192 | }
193 | .twilight {
194 | background: #d0e5ff;
195 | }
196 | acronym {
197 | cursor: help;
198 | }
199 |
200 | #now-link {
201 | position: relative;
202 | top: 17px;
203 | }
204 |
205 | #legend {
206 | top: 160px;
207 | right: 5px;
208 | }
209 | #forecast {
210 | bottom: 45px;
211 | right: 60px;
212 | }
213 | #links {
214 | top: 110px;
215 | left: 80px;
216 | }
217 | .map-content {
218 | position: absolute;
219 | z-index: 1000;
220 | padding: 4px 7px 5px;
221 | border-top: 1px solid #777;
222 | border-left: 1px solid #777;
223 | border-right: 2px solid #444;
224 | border-bottom: 2px solid #444;
225 | line-height: 1.6;
226 | font-size: 1.2em;
227 | background: white;
228 | -moz-border-radius: 5px;
229 | -webkit-border-radius: 5px;
230 | border-radius: 5px;
231 | }
232 |
233 | #time-slider-container {
234 | position: absolute;
235 | top: 60px;
236 | width: 100%;
237 | }
238 | #time-slider-2 {
239 | margin: 0px 15px 0;
240 | background: #91a1be;
241 | }
242 | #time-slider-2 .ui-slider-handle {
243 | width: 16px;
244 | height: 16px;
245 | margin-left: -10px;
246 | margin-top: -2px;
247 | border-radius: 10px;
248 | -moz-border-radius: 9px;
249 | -webkit-border-radius: 9px;
250 | background: #ffaa52;
251 | border-color: #fdcf94;
252 | z-index: 1;
253 | }
254 | #time-slider-2 .ui-slider-handle:hover, #time-slider-2 .ui-slider-handle:focus {
255 | background: #ffCC33;
256 | border-color: white;
257 | }
258 | #time-scale-container {
259 | margin: 0 15px 0 15px;
260 | font-size: 0.9em;
261 | }
262 | #time-scale {
263 | width: 100%;
264 | border-collapse: collapse;
265 | }
266 | #time-scale td {
267 | width: 4.1666666666%;
268 | padding-left: 5px;
269 | /*height: 30px;
270 | vertical-align: top;
271 | padding-top: 3px;*/
272 | height: 20px;
273 | border-left: 1px solid #ccc;
274 | }
275 | #time-scale td span {
276 | color: #999;
277 | }
278 | #time-scale-sunlight, #time-scale-twilight, #time-scale-sunlight-2, #time-scale-twilight-2 {
279 | height: 100%;
280 | position: absolute;
281 | background: #FFED9E;
282 | }
283 | #time-scale-twilight , #time-scale-twilight-2 {
284 | background: #c0ddF0;
285 | }
286 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/leaflet.textpath.js:
--------------------------------------------------------------------------------
1 | /*
2 | * Leaflet.TextPath - Shows text along a polyline
3 | * Inspired by Tom Mac Wright article :
4 | * http://mapbox.com/osmdev/2012/11/20/getting-serious-about-svg/
5 | */
6 |
7 | (function () {
8 |
9 | var __onAdd = L.Polyline.prototype.onAdd,
10 | __onRemove = L.Polyline.prototype.onRemove,
11 | __updatePath = L.Polyline.prototype._updatePath,
12 | __bringToFront = L.Polyline.prototype.bringToFront;
13 |
14 |
15 | var PolylineTextPath = {
16 |
17 | onAdd: function (map) {
18 | __onAdd.call(this, map);
19 | this._textRedraw();
20 | },
21 |
22 | onRemove: function (map) {
23 | map = map || this._map;
24 | if (map && this._textNode && map._renderer._container)
25 | map._renderer._container.removeChild(this._textNode);
26 | __onRemove.call(this, map);
27 | },
28 |
29 | bringToFront: function () {
30 | __bringToFront.call(this);
31 | this._textRedraw();
32 | },
33 |
34 | _updatePath: function () {
35 | __updatePath.call(this);
36 | this._textRedraw();
37 | },
38 |
39 | _textRedraw: function () {
40 | var text = this._text,
41 | options = this._textOptions;
42 | if (text) {
43 | this.setText(null).setText(text, options);
44 | }
45 | },
46 |
47 | setText: function (text, options) {
48 | this._text = text;
49 | this._textOptions = options;
50 |
51 | /* If not in SVG mode or Polyline not added to map yet return */
52 | /* setText will be called by onAdd, using value stored in this._text */
53 | if (!L.Browser.svg || typeof this._map === 'undefined') {
54 | return this;
55 | }
56 |
57 | var defaults = {
58 | repeat: false,
59 | fillColor: 'black',
60 | attributes: {},
61 | below: false,
62 | };
63 | options = L.Util.extend(defaults, options);
64 |
65 | /* If empty text, hide */
66 | if (!text) {
67 | if (this._textNode && this._textNode.parentNode) {
68 | this._map._renderer._container.removeChild(this._textNode);
69 |
70 | /* delete the node, so it will not be removed a 2nd time if the layer is later removed from the map */
71 | delete this._textNode;
72 | }
73 | return this;
74 | }
75 |
76 | text = text.replace(/ /g, '\u00A0'); // Non breakable spaces
77 | var id = 'pathdef-' + L.Util.stamp(this);
78 | var svg = this._map._renderer._container;
79 | this._path.setAttribute('id', id);
80 |
81 | if (options.repeat) {
82 | /* Compute single pattern length */
83 | var pattern = L.SVG.create('text');
84 | for (var attr in options.attributes)
85 | pattern.setAttribute(attr, options.attributes[attr]);
86 | pattern.appendChild(document.createTextNode(text));
87 | svg.appendChild(pattern);
88 | var alength = pattern.getComputedTextLength();
89 | svg.removeChild(pattern);
90 |
91 | /* Create string as long as path */
92 | text = new Array(Math.ceil(isNaN(this._path.getTotalLength() / alength) ? 0 : this._path.getTotalLength() / alength)).join(text);
93 | }
94 |
95 | /* Put it along the path using textPath */
96 | var textNode = L.SVG.create('text'),
97 | textPath = L.SVG.create('textPath');
98 |
99 | var dy = options.offset || this._path.getAttribute('stroke-width');
100 |
101 | textPath.setAttributeNS("http://www.w3.org/1999/xlink", "xlink:href", '#'+id);
102 | textNode.setAttribute('dy', dy);
103 | for (var attr in options.attributes)
104 | textNode.setAttribute(attr, options.attributes[attr]);
105 | textPath.appendChild(document.createTextNode(text));
106 | textNode.appendChild(textPath);
107 | this._textNode = textNode;
108 |
109 | if (options.below) {
110 | svg.insertBefore(textNode, svg.firstChild);
111 | }
112 | else {
113 | svg.appendChild(textNode);
114 | }
115 |
116 | /* Center text according to the path's bounding box */
117 | if (options.center) {
118 | var textLength = textNode.getComputedTextLength();
119 | var pathLength = this._path.getTotalLength();
120 | /* Set the position for the left side of the textNode */
121 | textNode.setAttribute('dx', ((pathLength / 2) - (textLength / 2)));
122 | }
123 |
124 | /* Change label rotation (if required) */
125 | if (options.orientation) {
126 | var rotateAngle = 0;
127 | switch (options.orientation) {
128 | case 'flip':
129 | rotateAngle = 180;
130 | break;
131 | case 'perpendicular':
132 | rotateAngle = 90;
133 | break;
134 | default:
135 | rotateAngle = options.orientation;
136 | }
137 |
138 | var rotatecenterX = (textNode.getBBox().x + textNode.getBBox().width / 2);
139 | var rotatecenterY = (textNode.getBBox().y + textNode.getBBox().height / 2);
140 | textNode.setAttribute('transform','rotate(' + rotateAngle + ' ' + rotatecenterX + ' ' + rotatecenterY + ')');
141 | }
142 |
143 | /* Initialize mouse events for the additional nodes */
144 | if (this.options.interactive) {
145 | if (L.Browser.svg || !L.Browser.vml) {
146 | textPath.setAttribute('class', 'leaflet-interactive');
147 | }
148 |
149 | var events = ['click', 'dblclick', 'mousedown', 'mouseover',
150 | 'mouseout', 'mousemove', 'contextmenu'];
151 | for (var i = 0; i < events.length; i++) {
152 | L.DomEvent.on(textNode, events[i], this.fire, this);
153 | }
154 | }
155 |
156 | return this;
157 | }
158 | };
159 |
160 | L.Polyline.include(PolylineTextPath);
161 |
162 | L.LayerGroup.include({
163 | setText: function(text, options) {
164 | for (var layer in this._layers) {
165 | if (typeof this._layers[layer].setText === 'function') {
166 | this._layers[layer].setText(text, options);
167 | }
168 | }
169 | return this;
170 | }
171 | });
172 |
173 |
174 |
175 | })();
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/pixiOverlay/bezier-easing.js:
--------------------------------------------------------------------------------
1 | (function(f){if(typeof exports==="object"&&typeof module!=="undefined"){module.exports=f()}else if(typeof define==="function"&&define.amd){define([],f)}else{var g;if(typeof window!=="undefined"){g=window}else if(typeof global!=="undefined"){g=global}else if(typeof self!=="undefined"){g=self}else{g=this}g.BezierEasing = f()}})(function(){var define,module,exports;return (function(){function r(e,n,t){function o(i,f){if(!n[i]){if(!e[i]){var c="function"==typeof require&&require;if(!f&&c)return c(i,!0);if(u)return u(i,!0);var a=new Error("Cannot find module '"+i+"'");throw a.code="MODULE_NOT_FOUND",a}var p=n[i]={exports:{}};e[i][0].call(p.exports,function(r){var n=e[i][1][r];return o(n||r)},p,p.exports,r,e,n,t)}return n[i].exports}for(var u="function"==typeof require&&require,i=0;i 0.0) {
35 | aB = currentT;
36 | } else {
37 | aA = currentT;
38 | }
39 | } while (Math.abs(currentX) > SUBDIVISION_PRECISION && ++i < SUBDIVISION_MAX_ITERATIONS);
40 | return currentT;
41 | }
42 |
43 | function newtonRaphsonIterate (aX, aGuessT, mX1, mX2) {
44 | for (var i = 0; i < NEWTON_ITERATIONS; ++i) {
45 | var currentSlope = getSlope(aGuessT, mX1, mX2);
46 | if (currentSlope === 0.0) {
47 | return aGuessT;
48 | }
49 | var currentX = calcBezier(aGuessT, mX1, mX2) - aX;
50 | aGuessT -= currentX / currentSlope;
51 | }
52 | return aGuessT;
53 | }
54 |
55 | function LinearEasing (x) {
56 | return x;
57 | }
58 |
59 | module.exports = function bezier (mX1, mY1, mX2, mY2) {
60 | if (!(0 <= mX1 && mX1 <= 1 && 0 <= mX2 && mX2 <= 1)) {
61 | throw new Error('bezier x values must be in [0, 1] range');
62 | }
63 |
64 | if (mX1 === mY1 && mX2 === mY2) {
65 | return LinearEasing;
66 | }
67 |
68 | // Precompute samples table
69 | var sampleValues = float32ArraySupported ? new Float32Array(kSplineTableSize) : new Array(kSplineTableSize);
70 | for (var i = 0; i < kSplineTableSize; ++i) {
71 | sampleValues[i] = calcBezier(i * kSampleStepSize, mX1, mX2);
72 | }
73 |
74 | function getTForX (aX) {
75 | var intervalStart = 0.0;
76 | var currentSample = 1;
77 | var lastSample = kSplineTableSize - 1;
78 |
79 | for (; currentSample !== lastSample && sampleValues[currentSample] <= aX; ++currentSample) {
80 | intervalStart += kSampleStepSize;
81 | }
82 | --currentSample;
83 |
84 | // Interpolate to provide an initial guess for t
85 | var dist = (aX - sampleValues[currentSample]) / (sampleValues[currentSample + 1] - sampleValues[currentSample]);
86 | var guessForT = intervalStart + dist * kSampleStepSize;
87 |
88 | var initialSlope = getSlope(guessForT, mX1, mX2);
89 | if (initialSlope >= NEWTON_MIN_SLOPE) {
90 | return newtonRaphsonIterate(aX, guessForT, mX1, mX2);
91 | } else if (initialSlope === 0.0) {
92 | return guessForT;
93 | } else {
94 | return binarySubdivide(aX, intervalStart, intervalStart + kSampleStepSize, mX1, mX2);
95 | }
96 | }
97 |
98 | return function BezierEasing (x) {
99 | // Because JavaScript number are imprecise, we should guarantee the extremes are right.
100 | if (x === 0) {
101 | return 0;
102 | }
103 | if (x === 1) {
104 | return 1;
105 | }
106 | return calcBezier(getTForX(x), mY1, mY2);
107 | };
108 | };
109 |
110 | },{}]},{},[1])(1)
111 | });
112 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/preloader.js:
--------------------------------------------------------------------------------
1 | $(window).on('load', function () {
2 | $(".loader").delay(2000).fadeOut("slow");
3 | $("#overlayer").delay(2000).fadeOut("slow");
4 | })
5 |
6 | // window.addEventListener ("load", function() {
7 | // loader.style.display = 'none';
8 | // console.log('Done...')
9 | // });
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/progress.js:
--------------------------------------------------------------------------------
1 | function updateDonutChart (el, percent, donut) {
2 | percent = Math.round(percent);
3 | if (percent > 100) {
4 | percent = 100;
5 | } else if (percent < 0) {
6 | percent = 0;
7 | }
8 | var deg = Math.round(360 * (percent / 100));
9 |
10 | if (percent > 50) {
11 | $(el + ' .pie').css('clip', 'rect(auto, auto, auto, auto)');
12 | $(el + ' .right-side').css('transform', 'rotate(180deg)');
13 | } else {
14 | $(el + ' .pie').css('clip', 'rect(0, 1em, 1em, 0.5em)');
15 | $(el + ' .right-side').css('transform', 'rotate(0deg)');
16 | }
17 | if (donut) {
18 | $(el + ' .right-side').css('border-width', '0.1em');
19 | $(el + ' .left-side').css('border-width', '0.1em');
20 | $(el + ' .shadow').css('border-width', '0.1em');
21 | } else {
22 | $(el + ' .right-side').css('border-width', '0.5em');
23 | $(el + ' .left-side').css('border-width', '0.5em');
24 | $(el + ' .shadow').css('border-width', '0.5em');
25 | }
26 | $(el + ' .num').text(percent);
27 | $(el + ' .left-side').css('transform', 'rotate(' + deg + 'deg)');
28 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/stage_cells.js:
--------------------------------------------------------------------------------
1 | // *******************************************************
2 | // DEPRECATED CODE. NOT USED ANYMORE
3 | // *******************************************************
4 |
5 | function drawCellSprites(resources, markers, map) {
6 | var textures = [resources.plane.texture, resources.circle.texture, resources.bicycle.texture];
7 | var focusTextures = [resources.focusPlane.texture, resources.focusCircle.texture, resources.focusBicycle.texture];
8 |
9 | var pixiLayer = cellOverlay(markers, textures, focusTextures);
10 | // pixiLayer().addTo(map);
11 | return pixiLayer()
12 | }
13 |
14 | function cellOverlay(markers, textures, focusTextures) {
15 | return function () {
16 | var pixiContainer = new PIXI.Container();
17 | var doubleBuffering = /iPad|iPhone|iPod/.test(navigator.userAgent) && !window.MSStream;
18 |
19 | var _drawCallback = drawCallback(markers, textures, focusTextures);
20 | return L.pixiOverlay(_drawCallback, pixiContainer, {
21 | doubleBuffering: doubleBuffering,
22 | destroyInteractionManager: true
23 | });
24 | };
25 |
26 |
27 | function drawCallback(markers, textures, focusTextures) {
28 | var firstDraw = true;
29 | var prevZoom;
30 | var markerSprites = [];
31 | var colorScale = d3.scaleLinear()
32 | .domain([0, 50, 100])
33 | .range(["#c6233c", "#ffd300", "#008000"]);
34 |
35 | var frame = null;
36 | var focus = null;
37 | return function (utils, event) {
38 | var zoom = utils.getMap().getZoom();
39 | if (frame) {
40 | cancelAnimationFrame(frame);
41 | frame = null;
42 | }
43 | var container = utils.getContainer();
44 | var renderer = utils.getRenderer();
45 | var project = utils.latLngToLayerPoint;
46 | var scale = utils.getScale();
47 | var invScale = 1 / scale;
48 | var initialScale = invScale / 4;
49 | if (firstDraw) {
50 | prevZoom = zoom;
51 | markers.forEach(marker => {
52 | var tempSpot = [marker.Y, marker.X],
53 | lp = dapiConfig.t.transform(L.point(tempSpot)),
54 | coords = project([lp.x, lp.y]);
55 | markerSprite = createMarker(coords, textures, scale);
56 | var tint = d3.color(colorScale(marker.avancement || Math.random() * 100)).rgb();
57 | markerSprite.tint = 256 * (tint.r * 256 + tint.g) + tint.b;
58 | container.addChild(markerSprite);
59 | markerSprites.push(markerSprite);
60 | markerSprite.legend = marker.city || marker.label;
61 | });
62 |
63 | // create zoom quad tree
64 | let zoomQuadTree = {};
65 | for (var z = map.getMinZoom(); z <= map.getMaxZoom(); z++) {
66 | const initialRadius = 8 / utils.getScale(z);
67 | zoomQuadTree[z] = solveCollision(markerSprites, {r0: initialRadius, zoom: z});
68 | }
69 |
70 | var self = this;
71 | map.on('mousemove', L.Util.throttle(function (e) {
72 | onMouseMove(e);
73 | // console.log('mouse move fired')
74 | }, 32));
75 |
76 | function onMouseMove(e) {
77 | // console.log('clicked');
78 | var redraw = false;
79 | var my_target = findMarker(project(e.latlng), zoomQuadTree, zoom);
80 | if (my_target) {
81 | console.log('Event fired, marker found');
82 | my_target.texture = textures[1]
83 | redraw = true
84 | }
85 | if (my_target) {
86 | L.DomUtil.addClass(self._container, 'leaflet-interactive');
87 | } else {
88 | L.DomUtil.removeClass(self._container, 'leaflet-interactive');
89 | }
90 | // console.log(my_target);
91 | if (redraw) utils.getRenderer().render(container)
92 | }
93 |
94 | } // first draw ends here
95 |
96 |
97 | if (firstDraw || prevZoom !== zoom) {
98 | markerSprites.forEach(markerSprite => {
99 | rescaleMarker(markerSprite, invScale, zoom)
100 | });
101 | }
102 |
103 | firstDraw = false;
104 | prevZoom = zoom;
105 | renderer.render(container);
106 | }
107 | }
108 |
109 | function createMarker(coords, textures, scale) {
110 | var index = Math.floor(Math.random() * textures.length);
111 | var sprite = new PIXI.Sprite(textures[index]);
112 | sprite.textureIndex = index;
113 | sprite.x0 = coords.x;
114 | sprite.y0 = coords.y;
115 | sprite.anchor.set(0.5, 0.5);
116 | sprite.scale.set(scale);
117 | sprite.currentScale = scale;
118 | return sprite;
119 | }
120 |
121 | function rescaleMarker(marker, scale, zoom, redraw) {
122 | const position = marker.cache[zoom];
123 | if (!redraw) { // 1st draw
124 | marker.x = position.x;
125 | marker.y = position.y;
126 | marker.scale.set((position.r * scale < 8) ? position.r / 8 : scale/16); // 16
127 | } else {
128 | marker.currentX = marker.x;
129 | marker.currentY = marker.y;
130 | marker.targetX = position.x;
131 | marker.targetY = position.y;
132 | marker.currentScale = marker.scale.x;
133 | marker.targetScale = (position.r * scale < 8) ? position.r / 16 : scale; // 16
134 | }
135 | }
136 |
137 | function findMarker(layerPoint, quad, zoom) {
138 | const quadTree = quad[zoom];
139 | const maxR = quadTree.rMax;
140 | var marker;
141 | var found = false;
142 | quadTree.visit((quad, x1, y1, x2, y2) => {
143 | if (!quad.length) {
144 | const dx = quad.data.x - layerPoint.x;
145 | const dy = quad.data.y - layerPoint.y;
146 | const r = quad.data.scale.x * 8; // 16;
147 | if (dx * dx + dy * dy <= r * r) {
148 | marker = quad.data;
149 | found = true;
150 | }
151 | }
152 | return found ||
153 | x1 > layerPoint.x + maxR ||
154 | x2 + maxR < layerPoint.x ||
155 | y1 > layerPoint.y + maxR ||
156 | y2 + maxR < layerPoint.y;
157 | });
158 | return marker;
159 | }
160 |
161 | }
162 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/stage_markers_patched.js:
--------------------------------------------------------------------------------
1 | function add_spots_patched(all_geneData, map) {
2 | // this is used to simulate leaflet zoom animation timing:
3 | var easing = BezierEasing(0, 0, 0.25, 1);
4 |
5 | function generateCircleTexture(color, radius, renderer) {
6 | const gfx = new PIXI.Graphics();
7 | const tileSize = radius * 2;
8 | const texture = PIXI.RenderTexture.create(tileSize, tileSize);
9 | gfx.beginFill(color); // color base
10 | gfx.alpha = 0.8;
11 | gfx.drawCircle(tileSize / 2, tileSize / 2, radius);
12 | gfx.endFill();
13 | renderer.render(gfx, texture);
14 | return texture;
15 | }
16 |
17 | var glyph_map = d3.map(glyphSettings(), function (d) {return d.gene; });
18 |
19 | function markerColor(geneName) {
20 | var t = glyph_map.get(geneName)
21 | if (!t){
22 | console.log('Cannot get color for ' + geneName + '. Defaulting to generic')
23 | t = glyph_map.get('Generic')
24 | }
25 | var hexCode = t.color
26 | // var colorCode = glyphSettings().filter(d => d.gene === geneName)[0].color;
27 | var out = myUtils().string2hex(hexCode);
28 | return out
29 | }
30 |
31 | const groupBy = (array, key) => {
32 | // from https://learnwithparam.com/blog/how-to-group-by-array-of-objects-using-a-key/
33 | // Return the end result
34 | return array.reduce((result, currentValue) => {
35 | // If an array already present for key, push it to the array. Else create an array and push the object
36 | (result[currentValue[key]] = result[currentValue[key]] || []).push(
37 | currentValue
38 | );
39 | // Return the current iteration `result` value, this will be taken as next iteration `result` value and accumulate
40 | return result;
41 | }, {}); // empty object is the initial value for result object
42 | };
43 |
44 | function scaleRamp(z) {
45 | // var scale = 1 / 64;
46 | var scale = config().spotSize;
47 | return z === 0 ? scaleRampHelper(z, scale) :
48 | z === 1 ? scaleRampHelper(z, 2 * scale) :
49 | z === 2 ? scaleRampHelper(z, 2 * scale) : scaleRampHelper(z, 4 * scale)
50 |
51 | // return z === 0 ? 0.03 * 2**3 :
52 | // z === 1 ? 0.03 * 2**3 :
53 | // z === 2 ? 0.03 * 2**3 :
54 | // z === 3 ? 0.03 * 2**3 :
55 | // z === 4 ? 0.03 * 2**3 :
56 | // z === 5 ? (2**z)/7602 :
57 | // z === 6 ? 0.03 * 2**1 : // every time you zoom in, leaflet scales up by 2. Divide here by 2 to keep the marker the same as in zoom level 5
58 | // z === 7 ? 0.03 * 2**0 :
59 | // z === 8 ? 0.03 : (2**z)/7602
60 | }
61 |
62 | function scaleRampHelper(z, scale){
63 | // makes a tiny dot and then its scales it up based on the map and the dapi dimensions
64 | // As a general remark also, keep in mind that every time you zoom in, leaflet (I think) scales up by 2.
65 | // Divide by 2 to keep the marker the same as size. Hence if for zoom level = 3 the return value from
66 | // this function is lets say zo 10, then when to keep the same size on the screen for the dot, at zoom = 4
67 | // the return value should be 5
68 | var map_side = mapSide(configSettings.maxZoom),
69 | dapi_size = [configSettings.roi.x1 - configSettings.roi.x0, configSettings.roi.y1 - configSettings.roi.y0],
70 | max_dapi = Math.max(...dapi_size),
71 | c = map_side / max_dapi,
72 | tiny_dot = 1 / (2**z),
73 | dot = c * tiny_dot;
74 | return dot * scale
75 | }
76 |
77 |
78 | var pixiLayer = (function () {
79 | masterMarkerContainer = new PIXI.Graphics();
80 |
81 | // group by gene name
82 | var data = groupBy(all_geneData, 'Gene');
83 |
84 | // get all the gene names
85 | var geneNames = Object.keys(data).sort();
86 |
87 | // populate an array with empty particle containers. One for each gene
88 | geneNames.forEach(gene => {
89 | var n = data[gene].length;
90 | var pc = new PIXI.particles.ParticleContainer(n, {vertices: true});
91 | pc.anchor = {x: 0.5, y: 0.5};
92 | pc.x = 0;
93 | pc.y = 0;
94 | pc.name = gene;
95 | masterMarkerContainer.addChild(pc);
96 | geneContainer_array.push(pc) // I think I can get rid of this. I can access the pc via masterMarkerContainer.getChildByName
97 | });
98 |
99 | // pixiContainer.addChild(innerContainer);
100 | var doubleBuffering = /iPad|iPhone|iPod/.test(navigator.userAgent) && !window.MSStream;
101 | var initialScale;
102 | var firstDraw = true;
103 | var prevZoom;
104 | return L.pixiOverlay(function (utils, event) {
105 | var zoom = utils.getMap().getZoom();
106 | var container = utils.getContainer();
107 | masterMarkerRenderer = utils.getRenderer();
108 | var project = utils.latLngToLayerPoint;
109 | var getScale = utils.getScale;
110 | var invScale = 1 / getScale();
111 |
112 |
113 | geneNames.forEach(gene => {
114 | var my_color = markerColor(gene);
115 | var radius = 16;
116 | var texture = generateCircleTexture(my_color, radius, masterMarkerRenderer);
117 | var pc = geneContainer_array.filter(d => d.name === gene)[0];
118 | pc.texture = texture;
119 | pc.baseTexture = texture.baseTexture;
120 | })
121 |
122 | // var innerContainer = geneContainer_array.filter(d => d.name === dummy_gene)[0];
123 | if (firstDraw) {
124 | if (event.type === 'add') {
125 |
126 | // initialScale = invScale / 8;
127 | // initialScale = 0.125;
128 | var targetScale = zoom <= zoomSwitch ? scaleRamp(zoom) : 1;
129 | for (var i = 0; i < geneNames.length; i++) {
130 | var gene = geneNames[i];
131 | var innerContainer = geneContainer_array.filter(d => d.name === gene)[0];
132 | innerContainer.localScale = targetScale;
133 | var _data = data[gene];
134 | for (var j = 0; j < _data.length; j++) {
135 | // our patched particleContainer accepts simple {x: ..., y: ...} objects as children:
136 | var x = _data[j].x;
137 | var y = _data[j].y;
138 | var point = dapiConfig.t.transform(L.point([x, y]));
139 | var coords = project([point.y, point.x]);
140 | innerContainer.addChild({
141 | x: coords.x,
142 | y: coords.y,
143 | });
144 | }
145 | }
146 | }
147 | }
148 |
149 | if (prevZoom !== zoom) {
150 | // console.log('zoom: From ' + prevZoom + ' to ' + zoom);
151 | geneNames.forEach(gene => {
152 | var innerContainer = masterMarkerContainer.getChildByName(gene);
153 | innerContainer.localScale = scaleRamp(zoom)
154 | })
155 | }
156 | firstDraw = false;
157 | prevZoom = zoom;
158 |
159 |
160 | masterMarkerRenderer.render(masterMarkerContainer);
161 | }, masterMarkerContainer, {
162 | doubleBuffering: true,
163 | destroyInteractionManager: true
164 | }); // L.pixiOverlay closes
165 | })();
166 |
167 | pixiLayer.addTo(map);
168 |
169 | // All done, hide the preloader now.
170 | removePreloader();
171 |
172 | };
173 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/streaming-tsv-parser.js:
--------------------------------------------------------------------------------
1 | function json_parse(d) {
2 | if (d === '\r') {
3 | // Add the reasoning for this ASAP cause I will forget it in a month's time
4 | return null
5 | }
6 | try {
7 | return JSON.parse(d.replace(/'/g, '"'))
8 | } catch {
9 | return d
10 | }
11 | }
12 |
13 | const getRootName = (d) => d.split('/').pop().split('_')[0];
14 | const count = (d) => {
15 | var out = {};
16 | var key;
17 | d.forEach(function (i) {
18 | key = getRootName(i);
19 | out[key] = (out[key] || 0) + 1;
20 | });
21 | return out
22 | };
23 |
24 | // a TSV parser that parses the data incrementally in chunks
25 | const tsvChunkedParser = () => {
26 | const textDecoder = new TextDecoder("utf-8");
27 | let columnHeadings;
28 | let previousChunk = "";
29 |
30 | return {
31 | parseChunk(chunk) {
32 | // decode and split into lines
33 | const textData = previousChunk + textDecoder.decode(chunk);
34 | const lines = textData.split("\n");
35 |
36 | // the first line is our column headings
37 | if (!columnHeadings) {
38 | columnHeadings = lines[0].split("\t");
39 | lines.shift();
40 | }
41 | // the last line is probably partial - so append to the next chunk
42 | previousChunk = lines.pop();
43 |
44 | // convert each row to an object
45 | const items = lines
46 | .map(row => {
47 | const cells = row.split("\t");
48 | if (cells.length !== columnHeadings.length) {
49 | return null;
50 | }
51 | let rowValue = {};
52 | columnHeadings.forEach((h, i) => {
53 | rowValue[h.trim()] = json_parse(cells[i]); //I am using trim() because I have no idea why I got whitespace at the end!!
54 | });
55 | return rowValue;
56 | })
57 | .filter(i => i);
58 |
59 | return items;
60 | }
61 | };
62 | };
63 |
64 |
65 | const fetchExternalData = (data) => {
66 | // the input data is actually the workpackage
67 | console.log(encodeURIComponent(data[0].mediaLink))
68 | var filenames = data.map(d => d.mediaLink);
69 | return Promise.all(
70 | // filenames.forEach(d => d.map(el => fetch(el)))
71 | filenames.map(d => fetch(d))
72 | )
73 | };
74 |
75 |
76 | onmessage = async function (event) {
77 | //onmessage: receives messages from the UI thread
78 |
79 | var totalBytes = Array(event.data.length).fill(0);
80 | var perc = Array(event.data.length).fill(0);
81 |
82 | const tsvParser = [];
83 | for (var i=0; i< event.data.length; i++){
84 | // make an array where each element is a **NEW** instance of the parser
85 | tsvParser.push(tsvChunkedParser())
86 | }
87 |
88 | var results = await fetchExternalData(event.data);
89 |
90 | // if (!response.body) {
91 | // throw Error("ReadableStream not yet supported in this browser.");
92 | // }
93 |
94 | const streamResponses = (responses) => {
95 | return Promise.all(
96 | responses.map((d,i) => streamedResponse(d, i).text())
97 | )
98 | };
99 |
100 |
101 | function streamedResponse(my_response, i) {
102 | return new Response(
103 | new ReadableStream({
104 | start(controller) {
105 | const reader = my_response.body.getReader();
106 |
107 | const read = async () => {
108 | const {done, value} = await reader.read();
109 | if (done) {
110 | console.log('ok, ' + i + ' is done')
111 | controller.close();
112 | return;
113 | }
114 |
115 | const items = tsvParser[i].parseChunk(value);
116 |
117 |
118 | totalBytes[i] += value.byteLength;
119 | // console.log('File num: ' + i)
120 | // var len = my_response.headers.get('content-length')
121 | // perc[i] = totalBytes[i]/my_response.headers.get('content-length');
122 | postMessage({i, items, url: my_response.url, bytes_streamed: +value.byteLength});
123 |
124 | controller.enqueue(value);
125 | read();
126 | };
127 |
128 | read();
129 | }
130 | })
131 | );
132 | };
133 |
134 |
135 | myData = await streamResponses(results);
136 |
137 |
138 | // call postMessage to send a message back to the UI thread
139 | postMessage({ items: [], totalBytes: [myData.length, null], finished: true });
140 |
141 | };
142 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/viewerUtils.js:
--------------------------------------------------------------------------------
1 | function myUtils() {
2 | function poly_collection(data, t) {
3 | console.log('Doing geojson object for polygons');
4 | var dots = {
5 | type: "FeatureCollection",
6 | features: []
7 | };
8 | for (var i = 0; i < data.length; ++i) {
9 | var c = data[i].coords,
10 | temp = [];
11 | // if (data[i].cell_id === 737) {
12 | // console.log('stop')
13 | // }
14 | if (c) {
15 | // That needs some attention. It leaves an open bug
16 | // if c is null (ie a cell doesnt have boundary coords for some reason) then
17 | // the geometry g which will be attached to data[i] will be the previous point's (ie data[i-1]) geometry,
18 | // I am turning a blind eye cause I dont have too many such cells (in fact all cells should have boundaries)
19 | // but it is a bug!
20 | for (var j = 0; j < c.length; ++j) {
21 | var x = c[j][0],
22 | y = c[j][1];
23 | var lp = t.transform(L.point([x, y]));
24 | temp.push([lp.x, lp.y])
25 | }
26 | var g = {
27 | "type": "Polygon",
28 | "coordinates": [temp]
29 | };
30 | }
31 |
32 | var target_cell = get_cell(+data[i].cell_id, i);
33 | //create feature properties
34 | var p = {
35 | // "fov": getFov(+target_cell.X, +target_cell.Y), // <-- Do not use that if you can. Browser will load a bit faster if removed
36 | "id": i,
37 | "cell_id": +data[i].cell_id,
38 | "centroid": get_centroid(target_cell),
39 | "X": +target_cell.X,
40 | "Y": +target_cell.Y,
41 | "Genenames": target_cell.Genenames,
42 | "CellGeneCount": target_cell.CellGeneCount,
43 | "ClassName": target_cell.ClassName,
44 | "topClass": target_cell.topClass,
45 | "Prob": target_cell.Prob,
46 | "agg": target_cell.agg,
47 | };
48 |
49 | //create features with proper geojson structure
50 | dots.features.push({
51 | "geometry": g,
52 | "type": "Feature",
53 | "properties": p
54 | });
55 | }
56 | console.log('geojson done');
57 | return dots;
58 | }
59 |
60 | function get_cell(cell_num, i) {
61 | // that can be done better i think!
62 | if (cellData[i].Cell_Num === cell_num){
63 | return cellData[i]
64 | }
65 | else{
66 | console.log('"cellData" and "data" arrays arent aligned');
67 | return null
68 | }
69 | //
70 | // That is a safer way to do the same but takes longer
71 | // return cellData.filter(d => d.Cell_Num === cell_num)[0];
72 | }
73 |
74 | function get_centroid(target_cell) {
75 | // var target_cell = cellData.filter(d => d.Cell_Num === cell_num)[0];
76 | return [target_cell.X, target_cell.Y]
77 | }
78 |
79 | function getFov(x, y) {
80 | var turfPoint = turf.point([x, y]);
81 |
82 | var p = block_boundaries.features.filter(d => turf.booleanPointInPolygon(turfPoint, turf.polygon(d.geometry.coordinates)));
83 | if (p) {
84 | return p[0].properties.id
85 | } else {
86 | return null
87 | }
88 | }
89 |
90 | /**
91 | * Converts a hexadecimal string to a hexadecimal color number.
92 | *
93 | * @example
94 | * PIXI.utils.string2hex("#ffffff"); // returns 0xffffff
95 | * @memberof PIXI.utils
96 | * @function string2hex
97 | * @param {string} The string color (e.g., `"#ffffff"`)
98 | * @return {number} Number in hexadecimal.
99 | */
100 | function string2hex(string) {
101 | if (typeof string === 'string' && string[0] === '#') {
102 | string = string.substr(1);
103 | }
104 | return parseInt(string, 16);
105 | }
106 |
107 | function stripper(d, k) {
108 | for (i = 0; i < k; ++i) {
109 | if (d.lastIndexOf('.') > 0) {
110 | d = d.substring(0, d.lastIndexOf('.'))
111 | }
112 | }
113 | return d
114 | }
115 |
116 | // find the position of the i-th occurrence of substring m in string str
117 | function getPosition(str, m, i) { return str.split(m, i).join(m).length; }
118 |
119 | // fw_stripper('this.is.a.test', 2) = 'this.is'
120 | // fw_stripper('this.is.a.test', 3) = 'this.is.a'
121 | function fw_stripper(d, k) {
122 | var out = d.substring(0, getPosition(d, '.', k));
123 | return out
124 | }
125 |
126 |
127 | var res = {};
128 | res.poly_collection = poly_collection;
129 | res.string2hex = string2hex;
130 | res.stripper = stripper;
131 | res.fw_stripper = fw_stripper
132 |
133 | return res
134 | }
135 |
136 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/__init__ .py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/static/cell_analysis/__init__ .py
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/cell_index.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | Cell Analysis Plot
5 |
6 |
7 |
8 |
9 |
";
118 | document.getElementById('dtTitle').innerHTML = str
119 |
120 | }
121 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/glyphPaths.js:
--------------------------------------------------------------------------------
1 |
2 | function ctxPath(glyphName, ctx, p, r) {
3 |
4 | if (glyphName === 'star6') {
5 | ctx.beginPath();
6 | ctx.moveTo(p.x + r, p.y);
7 | ctx.lineTo(p.x + 0.43 * r, p.y + 0.25 * r);
8 | ctx.lineTo(p.x + 0.50 * r, p.y + 0.87 * r);
9 | ctx.lineTo(p.x, p.y + 0.50 * r);
10 | ctx.lineTo(p.x - 0.50 * r, p.y + 0.87 * r);
11 | ctx.lineTo(p.x - 0.43 * r, p.y + 0.25 * r);
12 | ctx.lineTo(p.x - r, p.y);
13 | ctx.lineTo(p.x - 0.43 * r, p.y - 0.25 * r);
14 | ctx.lineTo(p.x - 0.50 * r, p.y - 0.87 * r);
15 | ctx.lineTo(p.x, p.y - 0.50 * r);
16 | ctx.lineTo(p.x + 0.50 * r, p.y - 0.87 * r);
17 | ctx.lineTo(p.x + 0.43 * r, p.y - 0.25 * r);
18 | ctx.closePath();
19 | }
20 |
21 |
22 | else if (glyphName === 'star5') {
23 | ctx.beginPath();
24 | ctx.moveTo(p.x, p.y + 0.658351875 * r);
25 | ctx.lineTo(p.x + 0.618027443 * r, p.y + 1 * r);
26 | ctx.lineTo(p.x + 0.5 * r, p.y + 0.27637816 * r);
27 | ctx.lineTo(p.x + 1 * r, p.y - 0.236080209 * r);
28 | ctx.lineTo(p.x + 0.309026865 * r, p.y - 0.341634306 * r);
29 | ctx.lineTo(p.x + 0 * r, p.y - 1 * r);
30 | ctx.lineTo(p.x - 0.309026865 * r, p.y - 0.341634306 * r);
31 | ctx.lineTo(p.x - 1 * r, p.y - 0.236080209 * r);
32 | ctx.lineTo(p.x - 0.5 * r, p.y + 0.27637816 * r);
33 | ctx.lineTo(p.x - 0.618027443 * r, p.y + 1 * r);
34 | ctx.lineTo(p.x, p.y + 0.658351875 * r);
35 | ctx.closePath();
36 | }
37 |
38 |
39 | else if (glyphName === 'diamond') {
40 | ctx.beginPath();
41 | ctx.moveTo(p.x - r, p.y);
42 | ctx.lineTo(p.x, p.y - r);
43 | ctx.lineTo(p.x + r, p.y);
44 | ctx.lineTo(p.x, p.y + r);
45 | ctx.lineTo(p.x - r, p.y);
46 | ctx.closePath();
47 | }
48 |
49 |
50 | else if (glyphName === 'square') {
51 | ctx.beginPath();
52 | ctx.moveTo(p.x - r, p.y - r);
53 | ctx.lineTo(p.x + r, p.y - r);
54 | ctx.lineTo(p.x + r, p.y + r);
55 | ctx.lineTo(p.x - r, p.y + r);
56 | ctx.lineTo(p.x - r, p.y - r);
57 | ctx.closePath();
58 | }
59 |
60 |
61 | else if (glyphName === 'triangleUp') {
62 | ctx.beginPath();
63 | ctx.moveTo(p.x - r, p.y + r);
64 | ctx.lineTo(p.x, p.y - r);
65 | ctx.lineTo(p.x + r, p.y + r);
66 | ctx.closePath();
67 | }
68 |
69 |
70 | else if (glyphName === 'triangleDown') {
71 | ctx.beginPath();
72 | ctx.moveTo(p.x - r, p.y - r);
73 | ctx.lineTo(p.x, p.y + r);
74 | ctx.lineTo(p.x + r, p.y - r);
75 | ctx.closePath();
76 | }
77 |
78 |
79 | else if (glyphName === 'triangleRight') {
80 | ctx.beginPath();
81 | ctx.moveTo(p.x - r, p.y - r);
82 | ctx.lineTo(p.x + r, p.y);
83 | ctx.lineTo(p.x - r, p.y + r);
84 | ctx.closePath();
85 | }
86 |
87 |
88 | else if (glyphName === 'triangleLeft') {
89 | ctx.beginPath();
90 | ctx.moveTo(p.x + r, p.y - r);
91 | ctx.lineTo(p.x - r, p.y);
92 | ctx.lineTo(p.x + r, p.y + r);
93 | ctx.closePath();
94 | }
95 |
96 |
97 | else if (glyphName === 'cross') {
98 | ctx.beginPath();
99 | ctx.moveTo(p.x + r, p.y + r);
100 | ctx.lineTo(p.x - r, p.y - r);
101 | ctx.moveTo(p.x - r, p.y + r);
102 | ctx.lineTo(p.x + r, p.y - r);
103 | ctx.closePath();
104 | }
105 |
106 |
107 | else if (glyphName === 'plus') {
108 | ctx.beginPath();
109 | ctx.moveTo(p.x, p.y + r);
110 | ctx.lineTo(p.x, p.y - r);
111 | ctx.moveTo(p.x - r, p.y);
112 | ctx.lineTo(p.x + r, p.y);
113 | ctx.closePath();
114 | }
115 |
116 |
117 | else if (glyphName === 'asterisk') {
118 | ctx.beginPath();
119 | ctx.moveTo(p.x, p.y + r);
120 | ctx.lineTo(p.x, p.y - r);
121 | ctx.moveTo(p.x - r, p.y);
122 | ctx.lineTo(p.x + r, p.y);
123 | ctx.moveTo(p.x + 0.5 * r, p.y + 0.5 * r);
124 | ctx.lineTo(p.x - 0.5 * r, p.y - 0.5 * r);
125 | ctx.moveTo(p.x - 0.5 * r, p.y + 0.5 * r);
126 | ctx.lineTo(p.x + 0.5 * r, p.y - 0.5 * r);
127 | ctx.arc(p.x, p.y, 2, 0, Math.PI * 2, true);
128 | ctx.closePath();
129 | }
130 |
131 |
132 | else if (glyphName === 'circle') {
133 | ctx.beginPath();
134 | ctx.arc(p.x, p.y, r, 0, Math.PI * 2, true);
135 | ctx.closePath();
136 | }
137 |
138 |
139 | else if (glyphName === 'point') {
140 | ctx.beginPath();
141 | ctx.arc(p.x, p.y, 3, 0, Math.PI * 2, true);
142 | ctx.arc(p.x, p.y, 2, 0, Math.PI * 2, true);
143 | ctx.closePath();
144 | }
145 |
146 | else {
147 | console.log('glyph: "' + glyphName + '" not implemented.')
148 | }
149 |
150 | return ctx
151 | }
152 |
153 |
154 |
155 |
156 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/glyphs.js:
--------------------------------------------------------------------------------
1 |
2 | L.Canvas.include({
3 | _updateMarkerStar6: function (layer) {
4 | if (!this._drawing || layer._empty()) { return; }
5 |
6 | var p = layer._point,
7 | ctx = this._ctx,
8 | r = Math.max(Math.round(layer._radius), 8);
9 |
10 | this._layers[layer._leaflet_id] = layer;
11 | ctx = ctxPath('star6', ctx, p, r);
12 | this._fillStroke(ctx, layer);
13 | }
14 | });
15 |
16 |
17 | L.Canvas.include({
18 | _updateMarkerStar: function (layer) {
19 | if (!this._drawing || layer._empty()) { return; }
20 |
21 | var p = layer._point,
22 | ctx = this._ctx,
23 | r = Math.max(Math.round(layer._radius), 8);
24 |
25 | this._layers[layer._leaflet_id] = layer;
26 | ctx = ctxPath('star5', ctx, p, r);
27 | this._fillStroke(ctx, layer);
28 |
29 | }
30 | });
31 |
32 | L.Canvas.include({
33 | _updateMarkerDiamond: function (layer) {
34 | if (!this._drawing || layer._empty()) { return; }
35 |
36 | var p = layer._point,
37 | ctx = this._ctx,
38 | r = Math.max(Math.round(layer._radius), 6);
39 |
40 | this._layers[layer._leaflet_id] = layer;
41 | ctx = ctxPath('diamond', ctx, p, r);
42 | this._fillStroke(ctx, layer);
43 | }
44 | });
45 |
46 |
47 | L.Canvas.include({
48 | _updateMarkerSquare: function (layer) {
49 | if (!this._drawing || layer._empty()) { return; }
50 |
51 | var p = layer._point,
52 | ctx = this._ctx,
53 | r = Math.max(Math.round(layer._radius), 5);
54 |
55 | this._layers[layer._leaflet_id] = layer;
56 | ctx = ctxPath('square', ctx, p, r);
57 | this._fillStroke(ctx, layer);
58 | }
59 | });
60 |
61 | L.Canvas.include({
62 | _updateMarkerTriangleUp: function (layer) {
63 | if (!this._drawing || layer._empty()) { return; }
64 |
65 | var p = layer._point,
66 | ctx = this._ctx,
67 | r = Math.max(Math.round(layer._radius), 5);
68 |
69 | this._layers[layer._leaflet_id] = layer;
70 | ctx = ctxPath('triangleUp', ctx, p, r);
71 | this._fillStroke(ctx, layer);
72 | }
73 | });
74 |
75 |
76 | L.Canvas.include({
77 | _updateMarkerTriangleDown: function (layer) {
78 | if (!this._drawing || layer._empty()) { return; }
79 |
80 | var p = layer._point,
81 | ctx = this._ctx,
82 | r = Math.max(Math.round(layer._radius), 5);
83 |
84 | this._layers[layer._leaflet_id] = layer;
85 | ctx = ctxPath('triangleDown', ctx, p, r);
86 | this._fillStroke(ctx, layer);
87 | }
88 | });
89 |
90 |
91 |
92 | L.Canvas.include({
93 | _updateMarkerTriangleRight: function (layer) {
94 | if (!this._drawing || layer._empty()) { return; }
95 |
96 | var p = layer._point,
97 | ctx = this._ctx,
98 | r = Math.max(Math.round(layer._radius), 5);
99 |
100 | this._layers[layer._leaflet_id] = layer;
101 | ctx = ctxPath('triangleRight', ctx, p, r);
102 | this._fillStroke(ctx, layer);
103 | }
104 | });
105 |
106 |
107 | L.Canvas.include({
108 | _updateMarkerTriangleLeft: function (layer) {
109 | if (!this._drawing || layer._empty()) { return; }
110 |
111 | var p = layer._point,
112 | ctx = this._ctx,
113 | r = Math.max(Math.round(layer._radius), 5);
114 |
115 | this._layers[layer._leaflet_id] = layer;
116 | ctx = ctxPath('triangleLeft', ctx, p, r);
117 | this._fillStroke(ctx, layer);
118 | }
119 | });
120 |
121 |
122 | L.Canvas.include({
123 | _updateMarkerCross: function (layer) {
124 | if (!this._drawing || layer._empty()) { return; }
125 |
126 | var p = layer._point,
127 | ctx = this._ctx,
128 | r = Math.max(Math.round(layer._radius), 5);
129 |
130 | this._layers[layer._leaflet_id] = layer;
131 | ctx = ctxPath('cross', ctx, p, r);
132 | this._fillStroke(ctx, layer);
133 | }
134 | });
135 |
136 |
137 |
138 | L.Canvas.include({
139 | _updateMarkerPlus: function (layer) {
140 | if (!this._drawing || layer._empty()) { return; }
141 |
142 | var p = layer._point,
143 | ctx = this._ctx,
144 | r = Math.max(Math.round(layer._radius), 7);
145 |
146 | this._layers[layer._leaflet_id] = layer;
147 | ctx = ctxPath('plus', ctx, p, r);
148 | this._fillStroke(ctx, layer);
149 | }
150 | });
151 |
152 |
153 | L.Canvas.include({
154 | _updateMarkerAsterisk: function (layer) {
155 | if (!this._drawing || layer._empty()) { return; }
156 |
157 | var p = layer._point,
158 | ctx = this._ctx,
159 | r = Math.max(Math.round(layer._radius), 7);
160 |
161 | this._layers[layer._leaflet_id] = layer;
162 | ctx = ctxPath('asterisk', ctx, p, r);
163 | this._fillStroke(ctx, layer);
164 | }
165 | });
166 |
167 |
168 | L.Canvas.include({
169 | _updateMarkerCircle: function (layer) {
170 | if (!this._drawing || layer._empty()) { return; }
171 |
172 | var p = layer._point,
173 | ctx = this._ctx,
174 | r = Math.max(Math.round(layer._radius), 6);
175 |
176 | this._layers[layer._leaflet_id] = layer;
177 | ctx = ctxPath('circle', ctx, p, r);
178 | this._fillStroke(ctx, layer);
179 | }
180 | });
181 |
182 |
183 | L.Canvas.include({
184 | _updateMarkerDot: function (layer) {
185 | if (!this._drawing || layer._empty()) { return; }
186 |
187 | var p = layer._point,
188 | ctx = this._ctx;
189 | //r = Math.max(Math.round(layer._radius), 6);
190 |
191 | this._layers[layer._leaflet_id] = layer;
192 | ctx = ctxPath('point', ctx, p);
193 | this._fillStroke(ctx, layer);
194 | }
195 | });
196 |
197 | var svgGlyph = L.CircleMarker.extend({
198 | _updatePath: function() {
199 | if ((this.options.shape === "star5") && (this._renderer._updateMarkerStar))
200 | this._renderer._updateMarkerStar(this);
201 | if ((this.options.shape === "star6") && (this._renderer._updateMarkerStar6))
202 | this._renderer._updateMarkerStar6(this);
203 | if ((this.options.shape === "diamond") && (this._renderer._updateMarkerDiamond))
204 | this._renderer._updateMarkerDiamond(this);
205 | if ((this.options.shape === "square") && (this._renderer._updateMarkerSquare))
206 | this._renderer._updateMarkerSquare(this);
207 | if ((this.options.shape === "triangleUp") && (this._renderer._updateMarkerTriangleUp))
208 | this._renderer._updateMarkerTriangleUp(this);
209 | if ((this.options.shape === "triangleDown") && (this._renderer._updateMarkerTriangleDown))
210 | this._renderer._updateMarkerTriangleDown(this);
211 | if ((this.options.shape === "triangleLeft") && (this._renderer._updateMarkerTriangleLeft))
212 | this._renderer._updateMarkerTriangleLeft(this);
213 | if ((this.options.shape === "triangleRight") && (this._renderer._updateMarkerTriangleRight))
214 | this._renderer._updateMarkerTriangleRight(this);
215 | if ((this.options.shape === "cross") && (this._renderer._updateMarkerCross))
216 | this._renderer._updateMarkerCross(this);
217 | if ((this.options.shape === "plus") && (this._renderer._updateMarkerPlus))
218 | this._renderer._updateMarkerPlus(this);
219 | if ((this.options.shape === "asterisk") && (this._renderer._updateMarkerAsterisk))
220 | this._renderer._updateMarkerAsterisk(this);
221 | if ((this.options.shape === "circle") && (this._renderer._updateMarkerCircle))
222 | this._renderer._updateMarkerCircle(this);
223 | if ((this.options.shape === "point") && (this._renderer._updateMarkerDot))
224 | this._renderer._updateMarkerDot(this);
225 | }
226 | });
227 |
228 |
229 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/L.Control.Layers.Tree.css:
--------------------------------------------------------------------------------
1 | .leaflet-control-layers-toggle.leaflet-layerstree-named-toggle {
2 | margin: 2px 5px;
3 | width: auto;
4 | height: auto;
5 | background-image: none;
6 | }
7 |
8 | .leaflet-layerstree-node {
9 | }
10 |
11 | .leaflet-layerstree-header input{
12 | margin-left: 0px;
13 | margin-right: 5px;
14 | }
15 |
16 |
17 | .leaflet-layerstree-header {
18 | }
19 |
20 | .leaflet-layerstree-header-pointer {
21 | cursor: pointer;
22 | }
23 |
24 | .leaflet-layerstree-header label {
25 | display: inline-block;
26 | cursor: pointer;
27 | }
28 |
29 | .leaflet-layerstree-header-label {
30 | }
31 |
32 | .leaflet-layerstree-header-name {
33 | }
34 |
35 | .leaflet-layerstree-header-space {
36 | }
37 |
38 | .leaflet-layerstree-children {
39 | /*padding-left: 10px;*/
40 | padding-left: 15px;
41 | }
42 |
43 | .leaflet-layerstree-children-nopad {
44 | padding-left: 0px;
45 | }
46 |
47 | .leaflet-layerstree-closed {
48 | }
49 |
50 | .leaflet-layerstree-opened {
51 | }
52 |
53 | .leaflet-layerstree-hide {
54 | display: none;
55 | }
56 |
57 | .leaflet-layerstree-nevershow {
58 | display: none;
59 | }
60 |
61 | .leaflet-layerstree-expand-collapse {
62 | cursor: pointer;
63 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/Leaflet.Coordinates-0.1.3.css:
--------------------------------------------------------------------------------
1 | .leaflet-control-coordinates{background-color:#D8D8D8;background-color:rgba(255,255,255,.8);cursor:pointer}.leaflet-control-coordinates,.leaflet-control-coordinates .uiElement input{-webkit-border-radius:5px;-moz-border-radius:5px;border-radius:5px}.leaflet-control-coordinates .uiElement{margin:4px}.leaflet-control-coordinates .uiElement .labelFirst{margin-right:4px}.leaflet-control-coordinates .uiHidden{display:none}
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/keen-dashboards.css:
--------------------------------------------------------------------------------
1 |
2 | /*
3 | * Full-page application style
4 | */
5 |
6 | body.application {
7 | background: #f2f2f2;
8 | padding: 60px 20px 0;
9 | }
10 | body.application > .container-fluid {
11 | padding-left: 0px;
12 | padding-right: 0px;
13 | }
14 | body.application div[class^="col-"] {
15 | padding-left: 5px;
16 | padding-right: 5px;
17 | }
18 | body.application div[class^="col-"] div[class^="col-"] {
19 | padding-left: 15px;
20 | padding-right: 15px;
21 | }
22 |
23 | body.application hr {
24 | border-color: #d7d7d7;
25 | margin: 10px 0;
26 | }
27 |
28 |
29 |
30 | .navbar-inverse {
31 | background-color: #3d4a57;
32 | border-color: #333;
33 | }
34 | .navbar-inverse .navbar-nav > li > a,
35 | .navbar a.navbar-brand {
36 | color: #fbfbfb;
37 | text-decoration: none;
38 | }
39 |
40 | .chart-wrapper {
41 | background: #fff;
42 | border: 1px solid #e2e2e2;
43 | border-radius: 3px;
44 | margin-bottom: 10px;
45 | }
46 | .chart-wrapper .chart-title {
47 | border-bottom: 1px solid #d7d7d7;
48 | color: #666;
49 | font-size: 14px;
50 | font-weight: 200;
51 | padding: 7px 10px 4px;
52 | }
53 |
54 | .chart-wrapper .chart-stage {
55 | /*min-height: 240px;*/
56 | overflow: hidden;
57 | padding: 5px 10px;
58 | position: relative;
59 | }
60 |
61 | .chart-wrapper .chart-notes {
62 | background: #fbfbfb;
63 | border-top: 1px solid #e2e2e2;
64 | color: #808080;
65 | font-size: 12px;
66 | padding: 8px 10px 5px;
67 | }
68 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/css/screen.css:
--------------------------------------------------------------------------------
1 | * {
2 | font-size: 100.01%;
3 | margin: 0;
4 | padding: 0;
5 | }
6 | html {
7 | font: 62.5%/1.5 Verdana, Tahoma, sans-serif;
8 | }
9 | html, body {
10 | width: 100%;
11 | height: 100%;
12 | overflow: hidden;
13 | }
14 | #map {
15 | position: absolute;
16 | top: 100px;
17 | bottom: 22px;
18 | left: 0;
19 | right: 0;
20 | overflow: hidden;
21 | border-top: 1px solid #ccc;
22 | border-bottom: 1px solid #ccc;
23 | }
24 | #footer {
25 | position: absolute;
26 | bottom: 0;
27 | left: 0;
28 | height: 25px;
29 | line-height: 25px;
30 | padding-left: 20px;
31 | font-size: 1.1em;
32 | color: #777;
33 | }
34 | #footer a, .ui-widget-content a, a {
35 | color: #0083CB;
36 | }
37 | #welcome {
38 | }
39 | .ui-widget-content {
40 | background: white;
41 | }
42 | .ui-widget-header {
43 | color: black;
44 | }
45 | .ui-dialog-content p {
46 | font-size: 1.2em;
47 | margin: 1em 0;
48 | }
49 | #header {
50 | height: 70px;
51 | width: 100%;
52 | position: absolute;
53 | }
54 | #header h1 {
55 | font: bold 3.2em Georgia, serif;
56 | float: left;
57 | height: 60px;
58 | margin: 9px 0 0 10px;
59 | display: inline;
60 | }
61 | #header h1 a {
62 | text-decoration: none;
63 | color: black;
64 | padding-top: 47px;
65 | height: 0;
66 | width: 143px;
67 | overflow: hidden;
68 | display: block;
69 | background: url(../images/logo.png) no-repeat;
70 | }
71 | #settings {
72 | font-size: 1.4em;
73 | margin: 6px 0 0 0;
74 | white-space: nowrap;
75 | }
76 | #date-container, #time-container, #location-container {
77 | float: left;
78 | display: inline;
79 | }
80 | #date-container {
81 | margin-right: 7px;
82 | }
83 | input[type=text], input.text {
84 | border: 1px solid #bbb;
85 | margin: 10px 0;
86 | padding: 4px;
87 | font-size: 1.4em;
88 | }
89 | input[type=text]:focus, input.text:focus, input[type=text]:hover, input.text:hover {
90 | border-color: #666;
91 | }
92 | #date {
93 | width: 120px;
94 | }
95 | #time {
96 | width: 55px;
97 | }
98 | #location {
99 | width: 250px;
100 | }
101 | #location-container {
102 | float: left;
103 | display: inline;
104 | margin-right: 7px;
105 | }
106 | #tagline {
107 | margin-right: 7px;
108 | margin-top: 20px;
109 | float: left;
110 | display: inline;
111 | }
112 | #location-container button, #time-container button {
113 | display: none;
114 | }
115 | #location-container.extended, #time-container.extended {
116 | background: white;
117 | z-index: 2000;
118 | position: relative;
119 | text-align: center;
120 | white-space: normal;
121 | padding: 0 10px 0;
122 | border: 1px solid #bbb;
123 | margin-top: -1px;
124 | }
125 | #location-container.extended button, #time-container.extended button {
126 | display: inline;
127 | }
128 |
129 | /* button styles taken from the lovely Blueprint CSS framework */
130 |
131 | a.button, button {
132 | margin: 0;
133 | margin-bottom: 10px;
134 | padding:4px 10px 5px 7px; /* Links */
135 |
136 | border:1px solid #dedede;
137 | border-top:1px solid #eee;
138 | border-left:1px solid #eee;
139 |
140 | background-color:#f0f0f0;
141 | line-height:130%;
142 | text-decoration:none;
143 | color:#565656;
144 | cursor:pointer;
145 | }
146 | button {
147 | width:auto;
148 | overflow:visible;
149 | padding:4px 7px 3px 7px; /* IE6 */
150 | }
151 | button[type] {
152 | padding:4px 7px 4px 7px; /* Firefox */
153 | line-height:17px; /* Safari */
154 | }
155 | *:first-child+html button[type] {
156 | padding:4px 7px 3px 7px; /* IE7 */
157 | }
158 | button:hover, a.button:hover{
159 | background-color:#dff4ff;
160 | border:1px solid #c2e1ef;
161 | color:#336699;
162 | }
163 | a.button:active{
164 | background-color:#6299c5;
165 | border:1px solid #6299c5;
166 | color:#fff;
167 | }
168 |
169 | .time-span, #welcome .sunrise, #welcome .sun, #welcome .sunset {
170 | padding: 0px 4px 1px;
171 | border-radius: 5px;
172 | -moz-border-radius: 5px;
173 | -webkit-border-radius: 5px;
174 | }
175 | #before-sunrise, #after-sunset, #daylight {
176 | display: none;
177 | }
178 | .sunrise {
179 | background: #FFED9E;
180 | }
181 | .sunset {
182 | background: #FFC3AD;
183 | }
184 | .sun {
185 | background: #ffdc9c;
186 | }
187 | .transit {
188 | background: #FFFC9C;
189 | }
190 | .dark {
191 | background: #bfd5dd;
192 | }
193 | .twilight {
194 | background: #d0e5ff;
195 | }
196 | acronym {
197 | cursor: help;
198 | }
199 |
200 | #now-link {
201 | position: relative;
202 | top: 17px;
203 | }
204 |
205 | #legend {
206 | top: 160px;
207 | right: 5px;
208 | }
209 | #forecast {
210 | bottom: 45px;
211 | right: 60px;
212 | }
213 | #links {
214 | top: 110px;
215 | left: 80px;
216 | }
217 | .map-content {
218 | position: absolute;
219 | z-index: 1000;
220 | padding: 4px 7px 5px;
221 | border-top: 1px solid #777;
222 | border-left: 1px solid #777;
223 | border-right: 2px solid #444;
224 | border-bottom: 2px solid #444;
225 | line-height: 1.6;
226 | font-size: 1.2em;
227 | background: white;
228 | -moz-border-radius: 5px;
229 | -webkit-border-radius: 5px;
230 | border-radius: 5px;
231 | }
232 |
233 | #time-slider-container {
234 | position: absolute;
235 | top: 60px;
236 | width: 100%;
237 | }
238 | #time-slider-2 {
239 | margin: 0px 15px 0;
240 | background: #91a1be;
241 | }
242 | #time-slider-2 .ui-slider-handle {
243 | width: 16px;
244 | height: 16px;
245 | margin-left: -10px;
246 | margin-top: -2px;
247 | border-radius: 10px;
248 | -moz-border-radius: 9px;
249 | -webkit-border-radius: 9px;
250 | background: #ffaa52;
251 | border-color: #fdcf94;
252 | z-index: 1;
253 | }
254 | #time-slider-2 .ui-slider-handle:hover, #time-slider-2 .ui-slider-handle:focus {
255 | background: #ffCC33;
256 | border-color: white;
257 | }
258 | #time-scale-container {
259 | margin: 0 15px 0 15px;
260 | font-size: 0.9em;
261 | }
262 | #time-scale {
263 | width: 100%;
264 | border-collapse: collapse;
265 | }
266 | #time-scale td {
267 | width: 4.1666666666%;
268 | padding-left: 5px;
269 | /*height: 30px;
270 | vertical-align: top;
271 | padding-top: 3px;*/
272 | height: 20px;
273 | border-left: 1px solid #ccc;
274 | }
275 | #time-scale td span {
276 | color: #999;
277 | }
278 | #time-scale-sunlight, #time-scale-twilight, #time-scale-sunlight-2, #time-scale-twilight-2 {
279 | height: 100%;
280 | position: absolute;
281 | background: #FFED9E;
282 | }
283 | #time-scale-twilight , #time-scale-twilight-2 {
284 | background: #c0ddF0;
285 | }
286 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/leaflet.textpath.js:
--------------------------------------------------------------------------------
1 | /*
2 | * Leaflet.TextPath - Shows text along a polyline
3 | * Inspired by Tom Mac Wright article :
4 | * http://mapbox.com/osmdev/2012/11/20/getting-serious-about-svg/
5 | */
6 |
7 | (function () {
8 |
9 | var __onAdd = L.Polyline.prototype.onAdd,
10 | __onRemove = L.Polyline.prototype.onRemove,
11 | __updatePath = L.Polyline.prototype._updatePath,
12 | __bringToFront = L.Polyline.prototype.bringToFront;
13 |
14 |
15 | var PolylineTextPath = {
16 |
17 | onAdd: function (map) {
18 | __onAdd.call(this, map);
19 | this._textRedraw();
20 | },
21 |
22 | onRemove: function (map) {
23 | map = map || this._map;
24 | if (map && this._textNode && map._renderer._container)
25 | map._renderer._container.removeChild(this._textNode);
26 | __onRemove.call(this, map);
27 | },
28 |
29 | bringToFront: function () {
30 | __bringToFront.call(this);
31 | this._textRedraw();
32 | },
33 |
34 | _updatePath: function () {
35 | __updatePath.call(this);
36 | this._textRedraw();
37 | },
38 |
39 | _textRedraw: function () {
40 | var text = this._text,
41 | options = this._textOptions;
42 | if (text) {
43 | this.setText(null).setText(text, options);
44 | }
45 | },
46 |
47 | setText: function (text, options) {
48 | this._text = text;
49 | this._textOptions = options;
50 |
51 | /* If not in SVG mode or Polyline not added to map yet return */
52 | /* setText will be called by onAdd, using value stored in this._text */
53 | if (!L.Browser.svg || typeof this._map === 'undefined') {
54 | return this;
55 | }
56 |
57 | var defaults = {
58 | repeat: false,
59 | fillColor: 'black',
60 | attributes: {},
61 | below: false,
62 | };
63 | options = L.Util.extend(defaults, options);
64 |
65 | /* If empty text, hide */
66 | if (!text) {
67 | if (this._textNode && this._textNode.parentNode) {
68 | this._map._renderer._container.removeChild(this._textNode);
69 |
70 | /* delete the node, so it will not be removed a 2nd time if the layer is later removed from the map */
71 | delete this._textNode;
72 | }
73 | return this;
74 | }
75 |
76 | text = text.replace(/ /g, '\u00A0'); // Non breakable spaces
77 | var id = 'pathdef-' + L.Util.stamp(this);
78 | var svg = this._map._renderer._container;
79 | this._path.setAttribute('id', id);
80 |
81 | if (options.repeat) {
82 | /* Compute single pattern length */
83 | var pattern = L.SVG.create('text');
84 | for (var attr in options.attributes)
85 | pattern.setAttribute(attr, options.attributes[attr]);
86 | pattern.appendChild(document.createTextNode(text));
87 | svg.appendChild(pattern);
88 | var alength = pattern.getComputedTextLength();
89 | svg.removeChild(pattern);
90 |
91 | /* Create string as long as path */
92 | text = new Array(Math.ceil(isNaN(this._path.getTotalLength() / alength) ? 0 : this._path.getTotalLength() / alength)).join(text);
93 | }
94 |
95 | /* Put it along the path using textPath */
96 | var textNode = L.SVG.create('text'),
97 | textPath = L.SVG.create('textPath');
98 |
99 | var dy = options.offset || this._path.getAttribute('stroke-width');
100 |
101 | textPath.setAttributeNS("http://www.w3.org/1999/xlink", "xlink:href", '#'+id);
102 | textNode.setAttribute('dy', dy);
103 | for (var attr in options.attributes)
104 | textNode.setAttribute(attr, options.attributes[attr]);
105 | textPath.appendChild(document.createTextNode(text));
106 | textNode.appendChild(textPath);
107 | this._textNode = textNode;
108 |
109 | if (options.below) {
110 | svg.insertBefore(textNode, svg.firstChild);
111 | }
112 | else {
113 | svg.appendChild(textNode);
114 | }
115 |
116 | /* Center text according to the path's bounding box */
117 | if (options.center) {
118 | var textLength = textNode.getComputedTextLength();
119 | var pathLength = this._path.getTotalLength();
120 | /* Set the position for the left side of the textNode */
121 | textNode.setAttribute('dx', ((pathLength / 2) - (textLength / 2)));
122 | }
123 |
124 | /* Change label rotation (if required) */
125 | if (options.orientation) {
126 | var rotateAngle = 0;
127 | switch (options.orientation) {
128 | case 'flip':
129 | rotateAngle = 180;
130 | break;
131 | case 'perpendicular':
132 | rotateAngle = 90;
133 | break;
134 | default:
135 | rotateAngle = options.orientation;
136 | }
137 |
138 | var rotatecenterX = (textNode.getBBox().x + textNode.getBBox().width / 2);
139 | var rotatecenterY = (textNode.getBBox().y + textNode.getBBox().height / 2);
140 | textNode.setAttribute('transform','rotate(' + rotateAngle + ' ' + rotatecenterX + ' ' + rotatecenterY + ')');
141 | }
142 |
143 | /* Initialize mouse events for the additional nodes */
144 | if (this.options.interactive) {
145 | if (L.Browser.svg || !L.Browser.vml) {
146 | textPath.setAttribute('class', 'leaflet-interactive');
147 | }
148 |
149 | var events = ['click', 'dblclick', 'mousedown', 'mouseover',
150 | 'mouseout', 'mousemove', 'contextmenu'];
151 | for (var i = 0; i < events.length; i++) {
152 | L.DomEvent.on(textNode, events[i], this.fire, this);
153 | }
154 | }
155 |
156 | return this;
157 | }
158 | };
159 |
160 | L.Polyline.include(PolylineTextPath);
161 |
162 | L.LayerGroup.include({
163 | setText: function(text, options) {
164 | for (var layer in this._layers) {
165 | if (typeof this._layers[layer].setText === 'function') {
166 | this._layers[layer].setText(text, options);
167 | }
168 | }
169 | return this;
170 | }
171 | });
172 |
173 |
174 |
175 | })();
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/pixiOverlay/bezier-easing.js:
--------------------------------------------------------------------------------
1 | (function(f){if(typeof exports==="object"&&typeof module!=="undefined"){module.exports=f()}else if(typeof define==="function"&&define.amd){define([],f)}else{var g;if(typeof window!=="undefined"){g=window}else if(typeof global!=="undefined"){g=global}else if(typeof self!=="undefined"){g=self}else{g=this}g.BezierEasing = f()}})(function(){var define,module,exports;return (function(){function r(e,n,t){function o(i,f){if(!n[i]){if(!e[i]){var c="function"==typeof require&&require;if(!f&&c)return c(i,!0);if(u)return u(i,!0);var a=new Error("Cannot find module '"+i+"'");throw a.code="MODULE_NOT_FOUND",a}var p=n[i]={exports:{}};e[i][0].call(p.exports,function(r){var n=e[i][1][r];return o(n||r)},p,p.exports,r,e,n,t)}return n[i].exports}for(var u="function"==typeof require&&require,i=0;i 0.0) {
35 | aB = currentT;
36 | } else {
37 | aA = currentT;
38 | }
39 | } while (Math.abs(currentX) > SUBDIVISION_PRECISION && ++i < SUBDIVISION_MAX_ITERATIONS);
40 | return currentT;
41 | }
42 |
43 | function newtonRaphsonIterate (aX, aGuessT, mX1, mX2) {
44 | for (var i = 0; i < NEWTON_ITERATIONS; ++i) {
45 | var currentSlope = getSlope(aGuessT, mX1, mX2);
46 | if (currentSlope === 0.0) {
47 | return aGuessT;
48 | }
49 | var currentX = calcBezier(aGuessT, mX1, mX2) - aX;
50 | aGuessT -= currentX / currentSlope;
51 | }
52 | return aGuessT;
53 | }
54 |
55 | function LinearEasing (x) {
56 | return x;
57 | }
58 |
59 | module.exports = function bezier (mX1, mY1, mX2, mY2) {
60 | if (!(0 <= mX1 && mX1 <= 1 && 0 <= mX2 && mX2 <= 1)) {
61 | throw new Error('bezier x values must be in [0, 1] range');
62 | }
63 |
64 | if (mX1 === mY1 && mX2 === mY2) {
65 | return LinearEasing;
66 | }
67 |
68 | // Precompute samples table
69 | var sampleValues = float32ArraySupported ? new Float32Array(kSplineTableSize) : new Array(kSplineTableSize);
70 | for (var i = 0; i < kSplineTableSize; ++i) {
71 | sampleValues[i] = calcBezier(i * kSampleStepSize, mX1, mX2);
72 | }
73 |
74 | function getTForX (aX) {
75 | var intervalStart = 0.0;
76 | var currentSample = 1;
77 | var lastSample = kSplineTableSize - 1;
78 |
79 | for (; currentSample !== lastSample && sampleValues[currentSample] <= aX; ++currentSample) {
80 | intervalStart += kSampleStepSize;
81 | }
82 | --currentSample;
83 |
84 | // Interpolate to provide an initial guess for t
85 | var dist = (aX - sampleValues[currentSample]) / (sampleValues[currentSample + 1] - sampleValues[currentSample]);
86 | var guessForT = intervalStart + dist * kSampleStepSize;
87 |
88 | var initialSlope = getSlope(guessForT, mX1, mX2);
89 | if (initialSlope >= NEWTON_MIN_SLOPE) {
90 | return newtonRaphsonIterate(aX, guessForT, mX1, mX2);
91 | } else if (initialSlope === 0.0) {
92 | return guessForT;
93 | } else {
94 | return binarySubdivide(aX, intervalStart, intervalStart + kSampleStepSize, mX1, mX2);
95 | }
96 | }
97 |
98 | return function BezierEasing (x) {
99 | // Because JavaScript number are imprecise, we should guarantee the extremes are right.
100 | if (x === 0) {
101 | return 0;
102 | }
103 | if (x === 1) {
104 | return 1;
105 | }
106 | return calcBezier(getTForX(x), mY1, mY2);
107 | };
108 | };
109 |
110 | },{}]},{},[1])(1)
111 | });
112 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/lib/js/preloader.js:
--------------------------------------------------------------------------------
1 | $(window).on('load', function () {
2 | $(".loader").delay(2000).fadeOut("slow");
3 | $("#overlayer").delay(2000).fadeOut("slow");
4 | })
5 |
6 | // window.addEventListener ("load", function() {
7 | // loader.style.display = 'none';
8 | // console.log('Done...')
9 | // });
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/progress.js:
--------------------------------------------------------------------------------
1 | function updateDonutChart (el, percent, donut) {
2 | percent = Math.round(percent);
3 | if (percent > 100) {
4 | percent = 100;
5 | } else if (percent < 0) {
6 | percent = 0;
7 | }
8 | var deg = Math.round(360 * (percent / 100));
9 |
10 | if (percent > 50) {
11 | $(el + ' .pie').css('clip', 'rect(auto, auto, auto, auto)');
12 | $(el + ' .right-side').css('transform', 'rotate(180deg)');
13 | } else {
14 | $(el + ' .pie').css('clip', 'rect(0, 1em, 1em, 0.5em)');
15 | $(el + ' .right-side').css('transform', 'rotate(0deg)');
16 | }
17 | if (donut) {
18 | $(el + ' .right-side').css('border-width', '0.1em');
19 | $(el + ' .left-side').css('border-width', '0.1em');
20 | $(el + ' .shadow').css('border-width', '0.1em');
21 | } else {
22 | $(el + ' .right-side').css('border-width', '0.5em');
23 | $(el + ' .left-side').css('border-width', '0.5em');
24 | $(el + ' .shadow').css('border-width', '0.5em');
25 | }
26 | $(el + ' .num').text(percent);
27 | $(el + ' .left-side').css('transform', 'rotate(' + deg + 'deg)');
28 | }
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/stage_cells.js:
--------------------------------------------------------------------------------
1 | // *******************************************************
2 | // DEPRECATED CODE. NOT USED ANYMORE
3 | // *******************************************************
4 |
5 | function drawCellSprites(resources, markers, map) {
6 | var textures = [resources.plane.texture, resources.circle.texture, resources.bicycle.texture];
7 | var focusTextures = [resources.focusPlane.texture, resources.focusCircle.texture, resources.focusBicycle.texture];
8 |
9 | var pixiLayer = cellOverlay(markers, textures, focusTextures);
10 | // pixiLayer().addTo(map);
11 | return pixiLayer()
12 | }
13 |
14 | function cellOverlay(markers, textures, focusTextures) {
15 | return function () {
16 | var pixiContainer = new PIXI.Container();
17 | var doubleBuffering = /iPad|iPhone|iPod/.test(navigator.userAgent) && !window.MSStream;
18 |
19 | var _drawCallback = drawCallback(markers, textures, focusTextures);
20 | return L.pixiOverlay(_drawCallback, pixiContainer, {
21 | doubleBuffering: doubleBuffering,
22 | destroyInteractionManager: true
23 | });
24 | };
25 |
26 |
27 | function drawCallback(markers, textures, focusTextures) {
28 | var firstDraw = true;
29 | var prevZoom;
30 | var markerSprites = [];
31 | var colorScale = d3.scaleLinear()
32 | .domain([0, 50, 100])
33 | .range(["#c6233c", "#ffd300", "#008000"]);
34 |
35 | var frame = null;
36 | var focus = null;
37 | return function (utils, event) {
38 | var zoom = utils.getMap().getZoom();
39 | if (frame) {
40 | cancelAnimationFrame(frame);
41 | frame = null;
42 | }
43 | var container = utils.getContainer();
44 | var renderer = utils.getRenderer();
45 | var project = utils.latLngToLayerPoint;
46 | var scale = utils.getScale();
47 | var invScale = 1 / scale;
48 | var initialScale = invScale / 4;
49 | if (firstDraw) {
50 | prevZoom = zoom;
51 | markers.forEach(marker => {
52 | var tempSpot = [marker.Y, marker.X],
53 | lp = dapiConfig.t.transform(L.point(tempSpot)),
54 | coords = project([lp.x, lp.y]);
55 | markerSprite = createMarker(coords, textures, scale);
56 | var tint = d3.color(colorScale(marker.avancement || Math.random() * 100)).rgb();
57 | markerSprite.tint = 256 * (tint.r * 256 + tint.g) + tint.b;
58 | container.addChild(markerSprite);
59 | markerSprites.push(markerSprite);
60 | markerSprite.legend = marker.city || marker.label;
61 | });
62 |
63 | // create zoom quad tree
64 | let zoomQuadTree = {};
65 | for (var z = map.getMinZoom(); z <= map.getMaxZoom(); z++) {
66 | const initialRadius = 8 / utils.getScale(z);
67 | zoomQuadTree[z] = solveCollision(markerSprites, {r0: initialRadius, zoom: z});
68 | }
69 |
70 | var self = this;
71 | map.on('mousemove', L.Util.throttle(function (e) {
72 | onMouseMove(e);
73 | // console.log('mouse move fired')
74 | }, 32));
75 |
76 | function onMouseMove(e) {
77 | // console.log('clicked');
78 | var redraw = false;
79 | var my_target = findMarker(project(e.latlng), zoomQuadTree, zoom);
80 | if (my_target) {
81 | console.log('Event fired, marker found');
82 | my_target.texture = textures[1]
83 | redraw = true
84 | }
85 | if (my_target) {
86 | L.DomUtil.addClass(self._container, 'leaflet-interactive');
87 | } else {
88 | L.DomUtil.removeClass(self._container, 'leaflet-interactive');
89 | }
90 | // console.log(my_target);
91 | if (redraw) utils.getRenderer().render(container)
92 | }
93 |
94 | } // first draw ends here
95 |
96 |
97 | if (firstDraw || prevZoom !== zoom) {
98 | markerSprites.forEach(markerSprite => {
99 | rescaleMarker(markerSprite, invScale, zoom)
100 | });
101 | }
102 |
103 | firstDraw = false;
104 | prevZoom = zoom;
105 | renderer.render(container);
106 | }
107 | }
108 |
109 | function createMarker(coords, textures, scale) {
110 | var index = Math.floor(Math.random() * textures.length);
111 | var sprite = new PIXI.Sprite(textures[index]);
112 | sprite.textureIndex = index;
113 | sprite.x0 = coords.x;
114 | sprite.y0 = coords.y;
115 | sprite.anchor.set(0.5, 0.5);
116 | sprite.scale.set(scale);
117 | sprite.currentScale = scale;
118 | return sprite;
119 | }
120 |
121 | function rescaleMarker(marker, scale, zoom, redraw) {
122 | const position = marker.cache[zoom];
123 | if (!redraw) { // 1st draw
124 | marker.x = position.x;
125 | marker.y = position.y;
126 | marker.scale.set((position.r * scale < 8) ? position.r / 8 : scale/16); // 16
127 | } else {
128 | marker.currentX = marker.x;
129 | marker.currentY = marker.y;
130 | marker.targetX = position.x;
131 | marker.targetY = position.y;
132 | marker.currentScale = marker.scale.x;
133 | marker.targetScale = (position.r * scale < 8) ? position.r / 16 : scale; // 16
134 | }
135 | }
136 |
137 | function findMarker(layerPoint, quad, zoom) {
138 | const quadTree = quad[zoom];
139 | const maxR = quadTree.rMax;
140 | var marker;
141 | var found = false;
142 | quadTree.visit((quad, x1, y1, x2, y2) => {
143 | if (!quad.length) {
144 | const dx = quad.data.x - layerPoint.x;
145 | const dy = quad.data.y - layerPoint.y;
146 | const r = quad.data.scale.x * 8; // 16;
147 | if (dx * dx + dy * dy <= r * r) {
148 | marker = quad.data;
149 | found = true;
150 | }
151 | }
152 | return found ||
153 | x1 > layerPoint.x + maxR ||
154 | x2 + maxR < layerPoint.x ||
155 | y1 > layerPoint.y + maxR ||
156 | y2 + maxR < layerPoint.y;
157 | });
158 | return marker;
159 | }
160 |
161 | }
162 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/stage_markers_patched.js:
--------------------------------------------------------------------------------
1 | function add_spots_patched(all_geneData, map) {
2 | // this is used to simulate leaflet zoom animation timing:
3 | var easing = BezierEasing(0, 0, 0.25, 1);
4 |
5 | function generateCircleTexture(color, radius, renderer) {
6 | const gfx = new PIXI.Graphics();
7 | const tileSize = radius * 2;
8 | const texture = PIXI.RenderTexture.create(tileSize, tileSize);
9 | gfx.beginFill(color); // color base
10 | gfx.alpha = 0.8;
11 | gfx.drawCircle(tileSize / 2, tileSize / 2, radius);
12 | gfx.endFill();
13 | renderer.render(gfx, texture);
14 | return texture;
15 | }
16 |
17 | var glyph_map = d3.map(glyphSettings(), function (d) {return d.gene; });
18 |
19 | function markerColor(geneName) {
20 | var t = glyph_map.get(geneName)
21 | if (!t){
22 | console.log('Cannot get color for ' + geneName + '. Defaulting to generic')
23 | t = glyph_map.get('Generic')
24 | }
25 | var hexCode = t.color
26 | // var colorCode = glyphSettings().filter(d => d.gene === geneName)[0].color;
27 | var out = myUtils().string2hex(hexCode);
28 | return out
29 | }
30 |
31 | const groupBy = (array, key) => {
32 | // from https://learnwithparam.com/blog/how-to-group-by-array-of-objects-using-a-key/
33 | // Return the end result
34 | return array.reduce((result, currentValue) => {
35 | // If an array already present for key, push it to the array. Else create an array and push the object
36 | (result[currentValue[key]] = result[currentValue[key]] || []).push(
37 | currentValue
38 | );
39 | // Return the current iteration `result` value, this will be taken as next iteration `result` value and accumulate
40 | return result;
41 | }, {}); // empty object is the initial value for result object
42 | };
43 |
44 | function scaleRamp(z) {
45 | // var scale = 1 / 64;
46 | var scale = config().spotSize;
47 | return z === 0 ? scaleRampHelper(z, scale) :
48 | z === 1 ? scaleRampHelper(z, 2 * scale) :
49 | z === 2 ? scaleRampHelper(z, 2 * scale) : scaleRampHelper(z, 4 * scale)
50 |
51 | // return z === 0 ? 0.03 * 2**3 :
52 | // z === 1 ? 0.03 * 2**3 :
53 | // z === 2 ? 0.03 * 2**3 :
54 | // z === 3 ? 0.03 * 2**3 :
55 | // z === 4 ? 0.03 * 2**3 :
56 | // z === 5 ? (2**z)/7602 :
57 | // z === 6 ? 0.03 * 2**1 : // every time you zoom in, leaflet scales up by 2. Divide here by 2 to keep the marker the same as in zoom level 5
58 | // z === 7 ? 0.03 * 2**0 :
59 | // z === 8 ? 0.03 : (2**z)/7602
60 | }
61 |
62 | function scaleRampHelper(z, scale){
63 | // makes a tiny dot and then its scales it up based on the map and the dapi dimensions
64 | // As a general remark also, keep in mind that every time you zoom in, leaflet (I think) scales up by 2.
65 | // Divide by 2 to keep the marker the same as size. Hence if for zoom level = 3 the return value from
66 | // this function is lets say zo 10, then when to keep the same size on the screen for the dot, at zoom = 4
67 | // the return value should be 5
68 | var map_side = mapSide(configSettings.maxZoom),
69 | dapi_size = [configSettings.roi.x1 - configSettings.roi.x0, configSettings.roi.y1 - configSettings.roi.y0],
70 | max_dapi = Math.max(...dapi_size),
71 | c = map_side / max_dapi,
72 | tiny_dot = 1 / (2**z),
73 | dot = c * tiny_dot;
74 | return dot * scale
75 | }
76 |
77 |
78 | var pixiLayer = (function () {
79 | masterMarkerContainer = new PIXI.Graphics();
80 |
81 | // group by gene name
82 | var data = groupBy(all_geneData, 'Gene');
83 |
84 | // get all the gene names
85 | var geneNames = Object.keys(data).sort();
86 |
87 | // populate an array with empty particle containers. One for each gene
88 | geneNames.forEach(gene => {
89 | var n = data[gene].length;
90 | var pc = new PIXI.particles.ParticleContainer(n, {vertices: true});
91 | pc.anchor = {x: 0.5, y: 0.5};
92 | pc.x = 0;
93 | pc.y = 0;
94 | pc.name = gene;
95 | masterMarkerContainer.addChild(pc);
96 | geneContainer_array.push(pc) // I think I can get rid of this. I can access the pc via masterMarkerContainer.getChildByName
97 | });
98 |
99 | // pixiContainer.addChild(innerContainer);
100 | var doubleBuffering = /iPad|iPhone|iPod/.test(navigator.userAgent) && !window.MSStream;
101 | var initialScale;
102 | var firstDraw = true;
103 | var prevZoom;
104 | return L.pixiOverlay(function (utils, event) {
105 | var zoom = utils.getMap().getZoom();
106 | var container = utils.getContainer();
107 | masterMarkerRenderer = utils.getRenderer();
108 | var project = utils.latLngToLayerPoint;
109 | var getScale = utils.getScale;
110 | var invScale = 1 / getScale();
111 |
112 |
113 | geneNames.forEach(gene => {
114 | var my_color = markerColor(gene);
115 | var radius = 16;
116 | var texture = generateCircleTexture(my_color, radius, masterMarkerRenderer);
117 | var pc = geneContainer_array.filter(d => d.name === gene)[0];
118 | pc.texture = texture;
119 | pc.baseTexture = texture.baseTexture;
120 | })
121 |
122 | // var innerContainer = geneContainer_array.filter(d => d.name === dummy_gene)[0];
123 | if (firstDraw) {
124 | if (event.type === 'add') {
125 |
126 | // initialScale = invScale / 8;
127 | // initialScale = 0.125;
128 | var targetScale = zoom <= zoomSwitch ? scaleRamp(zoom) : 1;
129 | for (var i = 0; i < geneNames.length; i++) {
130 | var gene = geneNames[i];
131 | var innerContainer = geneContainer_array.filter(d => d.name === gene)[0];
132 | innerContainer.localScale = targetScale;
133 | var _data = data[gene];
134 | for (var j = 0; j < _data.length; j++) {
135 | // our patched particleContainer accepts simple {x: ..., y: ...} objects as children:
136 | var x = _data[j].x;
137 | var y = _data[j].y;
138 | var point = dapiConfig.t.transform(L.point([x, y]));
139 | var coords = project([point.y, point.x]);
140 | innerContainer.addChild({
141 | x: coords.x,
142 | y: coords.y,
143 | });
144 | }
145 | }
146 | }
147 | }
148 |
149 | if (prevZoom !== zoom) {
150 | // console.log('zoom: From ' + prevZoom + ' to ' + zoom);
151 | geneNames.forEach(gene => {
152 | var innerContainer = masterMarkerContainer.getChildByName(gene);
153 | innerContainer.localScale = scaleRamp(zoom)
154 | })
155 | }
156 | firstDraw = false;
157 | prevZoom = zoom;
158 |
159 |
160 | masterMarkerRenderer.render(masterMarkerContainer);
161 | }, masterMarkerContainer, {
162 | doubleBuffering: true,
163 | destroyInteractionManager: true
164 | }); // L.pixiOverlay closes
165 | })();
166 |
167 | pixiLayer.addTo(map);
168 |
169 | // All done, hide the preloader now.
170 | removePreloader();
171 |
172 | };
173 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/streaming-tsv-parser.js:
--------------------------------------------------------------------------------
1 | function json_parse(d) {
2 | if (d === '\r') {
3 | // Add the reasoning for this ASAP cause I will forget it in a month's time
4 | return null
5 | }
6 | try {
7 | return JSON.parse(d.replace(/'/g, '"'))
8 | } catch {
9 | return d
10 | }
11 | }
12 |
13 | const getRootName = (d) => d.split('/').pop().split('_')[0];
14 | const count = (d) => {
15 | var out = {};
16 | var key;
17 | d.forEach(function (i) {
18 | key = getRootName(i);
19 | out[key] = (out[key] || 0) + 1;
20 | });
21 | return out
22 | };
23 |
24 | // a TSV parser that parses the data incrementally in chunks
25 | const tsvChunkedParser = () => {
26 | const textDecoder = new TextDecoder("utf-8");
27 | let columnHeadings;
28 | let previousChunk = "";
29 |
30 | return {
31 | parseChunk(chunk) {
32 | // decode and split into lines
33 | const textData = previousChunk + textDecoder.decode(chunk);
34 | const lines = textData.split("\n");
35 |
36 | // the first line is our column headings
37 | if (!columnHeadings) {
38 | columnHeadings = lines[0].split("\t");
39 | lines.shift();
40 | }
41 | // the last line is probably partial - so append to the next chunk
42 | previousChunk = lines.pop();
43 |
44 | // convert each row to an object
45 | const items = lines
46 | .map(row => {
47 | const cells = row.split("\t");
48 | if (cells.length !== columnHeadings.length) {
49 | return null;
50 | }
51 | let rowValue = {};
52 | columnHeadings.forEach((h, i) => {
53 | rowValue[h.trim()] = json_parse(cells[i]); //I am using trim() because I have no idea why I got whitespace at the end!!
54 | });
55 | return rowValue;
56 | })
57 | .filter(i => i);
58 |
59 | return items;
60 | }
61 | };
62 | };
63 |
64 |
65 | const fetchExternalData = (data) => {
66 | // the input data is actually the workpackage
67 | console.log(encodeURIComponent(data[0].mediaLink))
68 | var filenames = data.map(d => d.mediaLink);
69 | return Promise.all(
70 | // filenames.forEach(d => d.map(el => fetch(el)))
71 | filenames.map(d => fetch(d))
72 | )
73 | };
74 |
75 |
76 | onmessage = async function (event) {
77 | //onmessage: receives messages from the UI thread
78 |
79 | var totalBytes = Array(event.data.length).fill(0);
80 | var perc = Array(event.data.length).fill(0);
81 |
82 | const tsvParser = [];
83 | for (var i=0; i< event.data.length; i++){
84 | // make an array where each element is a **NEW** instance of the parser
85 | tsvParser.push(tsvChunkedParser())
86 | }
87 |
88 | var results = await fetchExternalData(event.data);
89 |
90 | // if (!response.body) {
91 | // throw Error("ReadableStream not yet supported in this browser.");
92 | // }
93 |
94 | const streamResponses = (responses) => {
95 | return Promise.all(
96 | responses.map((d,i) => streamedResponse(d, i).text())
97 | )
98 | };
99 |
100 |
101 | function streamedResponse(my_response, i) {
102 | return new Response(
103 | new ReadableStream({
104 | start(controller) {
105 | const reader = my_response.body.getReader();
106 |
107 | const read = async () => {
108 | const {done, value} = await reader.read();
109 | if (done) {
110 | console.log('ok, ' + i + ' is done')
111 | controller.close();
112 | return;
113 | }
114 |
115 | const items = tsvParser[i].parseChunk(value);
116 |
117 |
118 | totalBytes[i] += value.byteLength;
119 | // console.log('File num: ' + i)
120 | // var len = my_response.headers.get('content-length')
121 | // perc[i] = totalBytes[i]/my_response.headers.get('content-length');
122 | postMessage({i, items, url: my_response.url, bytes_streamed: +value.byteLength});
123 |
124 | controller.enqueue(value);
125 | read();
126 | };
127 |
128 | read();
129 | }
130 | })
131 | );
132 | };
133 |
134 |
135 | myData = await streamResponses(results);
136 |
137 |
138 | // call postMessage to send a message back to the UI thread
139 | postMessage({ items: [], totalBytes: [myData.length, null], finished: true });
140 |
141 | };
142 |
--------------------------------------------------------------------------------
/pciSeq/static/2D/viewer/js/viewerUtils.js:
--------------------------------------------------------------------------------
1 | function myUtils() {
2 | function poly_collection(data, t) {
3 | console.log('Doing geojson object for polygons');
4 | var dots = {
5 | type: "FeatureCollection",
6 | features: []
7 | };
8 | for (var i = 0; i < data.length; ++i) {
9 | var c = data[i].coords,
10 | temp = [];
11 | // if (data[i].cell_id === 737) {
12 | // console.log('stop')
13 | // }
14 | if (c) {
15 | // That needs some attention. It leaves an open bug
16 | // if c is null (ie a cell doesnt have boundary coords for some reason) then
17 | // the geometry g which will be attached to data[i] will be the previous point's (ie data[i-1]) geometry,
18 | // I am turning a blind eye cause I dont have too many such cells (in fact all cells should have boundaries)
19 | // but it is a bug!
20 | for (var j = 0; j < c.length; ++j) {
21 | var x = c[j][0],
22 | y = c[j][1];
23 | var lp = t.transform(L.point([x, y]));
24 | temp.push([lp.x, lp.y])
25 | }
26 | var g = {
27 | "type": "Polygon",
28 | "coordinates": [temp]
29 | };
30 | }
31 |
32 | var target_cell = get_cell(+data[i].cell_id, i);
33 | //create feature properties
34 | var p = {
35 | // "fov": getFov(+target_cell.X, +target_cell.Y), // <-- Do not use that if you can. Browser will load a bit faster if removed
36 | "id": i,
37 | "cell_id": +data[i].cell_id,
38 | "centroid": get_centroid(target_cell),
39 | "X": +target_cell.X,
40 | "Y": +target_cell.Y,
41 | "Genenames": target_cell.Genenames,
42 | "CellGeneCount": target_cell.CellGeneCount,
43 | "ClassName": target_cell.ClassName,
44 | "topClass": target_cell.topClass,
45 | "Prob": target_cell.Prob,
46 | "agg": target_cell.agg,
47 | };
48 |
49 | //create features with proper geojson structure
50 | dots.features.push({
51 | "geometry": g,
52 | "type": "Feature",
53 | "properties": p
54 | });
55 | }
56 | console.log('geojson done');
57 | return dots;
58 | }
59 |
60 | function get_cell(cell_num, i) {
61 | // that can be done better i think!
62 | if (cellData[i].Cell_Num === cell_num){
63 | return cellData[i]
64 | }
65 | else{
66 | console.log('"cellData" and "data" arrays arent aligned');
67 | return null
68 | }
69 | //
70 | // That is a safer way to do the same but takes longer
71 | // return cellData.filter(d => d.Cell_Num === cell_num)[0];
72 | }
73 |
74 | function get_centroid(target_cell) {
75 | // var target_cell = cellData.filter(d => d.Cell_Num === cell_num)[0];
76 | return [target_cell.X, target_cell.Y]
77 | }
78 |
79 | function getFov(x, y) {
80 | var turfPoint = turf.point([x, y]);
81 |
82 | var p = block_boundaries.features.filter(d => turf.booleanPointInPolygon(turfPoint, turf.polygon(d.geometry.coordinates)));
83 | if (p) {
84 | return p[0].properties.id
85 | } else {
86 | return null
87 | }
88 | }
89 |
90 | /**
91 | * Converts a hexadecimal string to a hexadecimal color number.
92 | *
93 | * @example
94 | * PIXI.utils.string2hex("#ffffff"); // returns 0xffffff
95 | * @memberof PIXI.utils
96 | * @function string2hex
97 | * @param {string} The string color (e.g., `"#ffffff"`)
98 | * @return {number} Number in hexadecimal.
99 | */
100 | function string2hex(string) {
101 | if (typeof string === 'string' && string[0] === '#') {
102 | string = string.substr(1);
103 | }
104 | return parseInt(string, 16);
105 | }
106 |
107 | function stripper(d, k) {
108 | for (i = 0; i < k; ++i) {
109 | if (d.lastIndexOf('.') > 0) {
110 | d = d.substring(0, d.lastIndexOf('.'))
111 | }
112 | }
113 | return d
114 | }
115 |
116 | // find the position of the i-th occurrence of substring m in string str
117 | function getPosition(str, m, i) { return str.split(m, i).join(m).length; }
118 |
119 | // fw_stripper('this.is.a.test', 2) = 'this.is'
120 | // fw_stripper('this.is.a.test', 3) = 'this.is.a'
121 | function fw_stripper(d, k) {
122 | var out = d.substring(0, getPosition(d, '.', k));
123 | return out
124 | }
125 |
126 |
127 | var res = {};
128 | res.poly_collection = poly_collection;
129 | res.string2hex = string2hex;
130 | res.stripper = stripper;
131 | res.fw_stripper = fw_stripper
132 |
133 | return res
134 | }
135 |
136 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/__init__ .py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/static/cell_analysis/__init__ .py
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/cell_index.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | Cell Analysis Plot
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
94 |
95 |
96 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/static/css/styles.css:
--------------------------------------------------------------------------------
1 | :root {
2 | --primary-color: #2563eb;
3 | --secondary-color: #4a90e2;
4 | --background-color: #f8fafc;
5 | --card-background: #ffffff;
6 | --text-color: #1e293b;
7 | --border-color: #e2e8f0;
8 | --border-radius: 12px;
9 | --shadow: 0 4px 6px -1px rgb(0 0 0 / 0.1), 0 2px 4px -2px rgb(0 0 0 / 0.1);
10 | --shadow-hover: 0 10px 15px -3px rgb(0 0 0 / 0.1), 0 4px 6px -4px rgb(0 0 0 / 0.1);
11 | }
12 |
13 | body {
14 | margin: 0;
15 | padding: 24px;
16 | font-family: -apple-system, BlinkMacSystemFont, 'Inter', 'Segoe UI', Roboto, Oxygen, Ubuntu, sans-serif;
17 | background-color: var(--background-color);
18 | color: var(--text-color);
19 | line-height: 1.5;
20 | }
21 |
22 | /* Plot Containers */
23 | #plot-area, #bottom-plot {
24 | position: relative;
25 | background-color: var(--card-background);
26 | border-radius: var(--border-radius);
27 | padding: 24px;
28 | margin: 24px 0;
29 | box-shadow: var(--shadow);
30 | border: 1px solid var(--border-color);
31 | transition: all 0.2s ease-in-out;
32 | }
33 |
34 | #plot-area:hover, #bottom-plot:hover {
35 | box-shadow: var(--shadow-hover);
36 | }
37 |
38 | /* Select Container Styles */
39 | .select-container {
40 | margin: 24px 0;
41 | display: flex;
42 | align-items: center;
43 | gap: 16px;
44 | }
45 |
46 | .select-label {
47 | font-weight: 600;
48 | color: var(--text-color);
49 | text-transform: uppercase;
50 | letter-spacing: 0.5px;
51 | font-size: 0.875rem;
52 | }
53 |
54 | .select-input {
55 | padding: 8px 16px;
56 | border: 2px solid var(--border-color);
57 | border-radius: 8px;
58 | font-size: 0.95rem;
59 | background-color: var(--card-background);
60 | cursor: pointer;
61 | transition: all 0.2s ease;
62 | min-width: 200px;
63 | }
64 |
65 | .select-input:hover {
66 | border-color: var(--secondary-color);
67 | }
68 |
69 | .select-input:focus {
70 | outline: none;
71 | border-color: var(--primary-color);
72 | box-shadow: 0 0 0 3px rgba(37, 99, 235, 0.1);
73 | }
74 |
75 | /* Tooltip */
76 | .tooltip {
77 | position: absolute;
78 | padding: 8px 10px;
79 | background: linear-gradient(145deg, #f8fafc, #e2e8f0);
80 | color: #334155;
81 | border: 1px solid #e2e8f0;
82 | border-radius: 6px;
83 | pointer-events: none;
84 | font-size: 0.75rem;
85 | box-shadow: 0 4px 6px rgba(0, 0, 0, 0.1);
86 | z-index: 1000;
87 | max-width: 200px;
88 | line-height: 1.3;
89 | transform: translate(-50%, -100%);
90 | margin-top: 0px; /* Changed from -8px to 0px */
91 | }
92 |
93 | .tooltip::after {
94 | content: '';
95 | position: absolute;
96 | bottom: -5px;
97 | left: 50%;
98 | transform: translateX(-50%);
99 | width: 0;
100 | height: 0;
101 | border-left: 5px solid transparent;
102 | border-right: 5px solid transparent;
103 | border-top: 5px solid #e2e8f0;
104 | }
105 |
106 | /* Plot Elements */
107 | .plot-title {
108 | font-size: 1.25rem;
109 | font-weight: 600;
110 | margin-bottom: 16px;
111 | color: var(--text-color);
112 | }
113 |
114 | .plot-subtitle {
115 | font-size: 1rem;
116 | color: #64748b;
117 | margin-bottom: 24px;
118 | }
119 |
120 | .axis-label {
121 | font-size: 0.875rem;
122 | font-weight: 500;
123 | fill: #64748b;
124 | }
125 |
126 | .diagonal-line {
127 | pointer-events: none;
128 | stroke-opacity: 0.6;
129 | }
130 |
131 | /* Interpretation Guide */
132 | .interpretation-guide rect {
133 | fill: rgba(241, 245, 249, 0.95);
134 | rx: 8px;
135 | stroke: var(--border-color);
136 | stroke-width: 1;
137 | }
138 |
139 | .interpretation-guide text {
140 | font-size: 0.75rem;
141 | fill: #475569;
142 | font-weight: 500;
143 | }
144 |
145 | /* Error and Loading States */
146 | .error {
147 | color: #dc2626;
148 | background-color: #fee2e2;
149 | border: 1px solid #fecaca;
150 | padding: 16px;
151 | margin: 16px;
152 | border-radius: 8px;
153 | text-align: center;
154 | font-weight: 500;
155 | }
156 |
157 | .loading {
158 | text-align: center;
159 | padding: 32px;
160 | color: #6b7280;
161 | font-weight: 500;
162 | }
163 |
164 | /* Responsive Adjustments */
165 | @media (max-width: 768px) {
166 | body {
167 | padding: 16px;
168 | }
169 |
170 | .select-container {
171 | flex-direction: column;
172 | align-items: flex-start;
173 | }
174 |
175 | .select-input {
176 | width: 100%;
177 | }
178 | }
179 |
180 |
181 | .gene-selector-container {
182 | margin: 20px;
183 | padding: 15px;
184 | border: 1px solid #ddd;
185 | border-radius: 8px;
186 | background-color: white;
187 | }
188 |
189 | .gene-selector-header {
190 | display: flex;
191 | justify-content: space-between;
192 | align-items: center;
193 | margin-bottom: 15px;
194 | }
195 |
196 | .gene-selector-header h3 {
197 | margin: 0;
198 | color: #333;
199 | }
200 |
201 | .gene-selector-controls {
202 | display: flex;
203 | gap: 10px;
204 | }
205 |
206 | .gene-selector-controls button {
207 | padding: 5px 10px;
208 | border: 1px solid #ccc;
209 | border-radius: 4px;
210 | background: #f5f5f5;
211 | cursor: pointer;
212 | }
213 |
214 | .gene-selector-controls button:hover {
215 | background: #e5e5e5;
216 | }
217 |
218 | .gene-checkbox-grid {
219 | display: grid;
220 | grid-template-columns: repeat(auto-fill, minmax(150px, 1fr));
221 | gap: 10px;
222 | max-height: 200px;
223 | overflow-y: auto;
224 | padding: 10px;
225 | border: 1px solid #eee;
226 | border-radius: 4px;
227 | }
228 |
229 | .gene-checkbox-item {
230 | display: flex;
231 | align-items: center;
232 | gap: 5px;
233 | }
234 |
235 | .gene-checkbox-item label {
236 | font-size: 14px;
237 | color: #444;
238 | cursor: pointer;
239 | }
240 |
241 | .gene-checkbox-item input[type="checkbox"] {
242 | cursor: pointer;
243 | }
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/static/js/distanceProbabilityPlot.js:
--------------------------------------------------------------------------------
1 | import {calculateDimensions, PLOT_CONFIG} from "./plotConfig.js";
2 | import { InterpretationGuide } from './interpretationGuide.js';
3 |
4 | export class DistanceProbabilityPlot {
5 | constructor(containerId, data, tooltip) {
6 | this.containerId = containerId;
7 | this.data = data;
8 | this.tooltip = tooltip;
9 | this.margin = {top: 60, right: 80, bottom: 50, left: 100};
10 | this.guide = null;
11 | this.sizeByCount = false;
12 | this.defaultRadius = 5.5;
13 | this.visibleGenes = new Set(data.labels);
14 |
15 | this.radiusScale = d3.scaleSqrt()
16 | .domain([0, d3.max(Object.values(data.gene_counts)) || 1])
17 | .range([2, 8]);
18 |
19 | this.initializePlot();
20 | }
21 |
22 | initializePlot() {
23 | const { width, height } = calculateDimensions();
24 | this.width = width;
25 | this.height = height;
26 |
27 | this.svg = d3.select(`#${this.containerId}`)
28 | .append('svg')
29 | .attr('width', width + PLOT_CONFIG.margin.left + PLOT_CONFIG.margin.right)
30 | .attr('height', height + PLOT_CONFIG.margin.top + PLOT_CONFIG.margin.bottom)
31 | .append('g')
32 | .attr('transform', `translate(${PLOT_CONFIG.margin.left},${PLOT_CONFIG.margin.top})`);
33 |
34 | this.x = d3.scaleLinear()
35 | .domain(d3.extent(this.data.x))
36 | .range([0, width]);
37 |
38 | this.y = d3.scaleLinear()
39 | .domain(d3.extent(this.data.y))
40 | .range([height, 0]);
41 |
42 | this.setupAxes();
43 | this.setupLabels();
44 | this.setupGuide();
45 | this.setupSizeControl();
46 | this.updatePlot();
47 | }
48 |
49 | setupAxes() {
50 | this.svg.append('g')
51 | .attr('class', 'x-axis')
52 | .attr('transform', `translate(0,${this.height})`)
53 | .call(d3.axisBottom(this.x)
54 | .ticks(5)
55 | .tickSize(-5)
56 | .tickPadding(5));
57 |
58 | this.svg.append('g')
59 | .attr('class', 'y-axis')
60 | .call(d3.axisLeft(this.y)
61 | .ticks(5)
62 | .tickSize(-5)
63 | .tickPadding(5));
64 | }
65 |
66 | setupLabels() {
67 | this.svg.append("text")
68 | .attr("class", "plot-title")
69 | .attr("x", this.width / 2)
70 | .attr("y", -this.margin.top / 2)
71 | .style("text-anchor", "middle")
72 | .style("font-size", "16px")
73 | .text(this.data.title);
74 |
75 | this.svg.append("text")
76 | .attr("class", "axis-label x-axis-label")
77 | .attr("x", this.width / 2)
78 | .attr("y", this.height + this.margin.bottom - 10)
79 | .style("text-anchor", "middle")
80 | .text(this.data.xlabel);
81 |
82 | this.svg.append("text")
83 | .attr("class", "axis-label y-axis-label")
84 | .attr("transform", "rotate(-90)")
85 | .attr("x", -this.height / 2)
86 | .attr("y", -this.margin.left + 40)
87 | .style("text-anchor", "middle")
88 | .text(this.data.ylabel);
89 | }
90 |
91 | setupGuide() {
92 | this.guide = new InterpretationGuide(this.svg, this.width, this.height);
93 | this.guide.update('', '');
94 |
95 | const guideText = [
96 | `• Shows all spots in neighborhood of cell ${this.data.cell_num}`,
97 | `• Points close to x=0: Spots near cell ${this.data.cell_num} centroid`,
98 | `• Points with high y-values: Spots likely assigned to cell ${this.data.cell_num}`,
99 | `• Points with low y-values: Spots likely assigned to neighboring cells`
100 | ];
101 |
102 | let guide = this.svg.select('.interpretation-guide');
103 | guide.selectAll("text").remove();
104 | guide.selectAll("text")
105 | .data(guideText)
106 | .enter()
107 | .append("text")
108 | .attr("x", 0)
109 | .attr("y", (d, i) => i * 15)
110 | .style("text-anchor", "start")
111 | .style("font-size", "12px")
112 | .text(d => d);
113 |
114 | const guideBBox = guide.node().getBBox();
115 | guide.attr("transform", `translate(${this.width - guideBBox.width - 20}, 40)`);
116 | }
117 |
118 | setupSizeControl() {
119 | const checkboxGroup = this.svg.append("g")
120 | .attr("class", "checkbox-group")
121 | .attr("transform", `translate(${this.width - 200}, -5)`);
122 |
123 | checkboxGroup.append("rect")
124 | .attr("width", 180)
125 | .attr("height", 20)
126 | .attr("fill", "white")
127 | .attr("opacity", 0.8)
128 | .attr("rx", 4);
129 |
130 | checkboxGroup.append("foreignObject")
131 | .attr("width", 180)
132 | .attr("height", 20)
133 | .append("xhtml:div")
134 | .style("font-size", "12px")
135 | .html(`
136 |
137 |
138 | `);
139 |
140 | d3.select(`#size-toggle-${this.containerId}`)
141 | .on("change", (event) => {
142 | this.sizeByCount = event.target.checked;
143 | this.updatePlot();
144 | });
145 | }
146 |
147 | updateVisibleGenes(visibleGenes) {
148 | this.visibleGenes = new Set(visibleGenes);
149 | this.updatePlot();
150 | }
151 |
152 | updatePlot() {
153 | const points = this.data.x.map((x, i) => ({
154 | x: x,
155 | y: this.data.y[i],
156 | label: this.data.labels[i],
157 | geneCount: this.data.gene_counts[this.data.labels[i]] || 0
158 | })).filter(d => this.visibleGenes.has(d.label));
159 |
160 | const dots = this.svg.selectAll("circle")
161 | .data(points, d => d.label);
162 |
163 | dots.exit()
164 | .transition()
165 | .duration(PLOT_CONFIG.animation.duration)
166 | .attr("r", 0)
167 | .remove();
168 |
169 | const dotsEnter = dots.enter()
170 | .append("circle")
171 | .attr("fill", PLOT_CONFIG.point.color)
172 | .attr("stroke", "white")
173 | .attr("stroke-width", "0.5")
174 | .attr("cx", d => this.x(d.x))
175 | .attr("cy", d => this.y(d.y))
176 | .attr("r", 0);
177 |
178 | dots.merge(dotsEnter)
179 | .transition()
180 | .duration(PLOT_CONFIG.animation.duration)
181 | .attr("cx", d => this.x(d.x))
182 | .attr("cy", d => this.y(d.y))
183 | .attr("r", d => this.sizeByCount ?
184 | this.radiusScale(d.geneCount) :
185 | this.defaultRadius);
186 |
187 | this.svg.selectAll("circle")
188 | .on("mouseenter", (event, d) => {
189 | d3.select(event.target)
190 | .transition()
191 | .duration(PLOT_CONFIG.animation.tooltip.fadeIn)
192 | .attr("r", d => this.sizeByCount ?
193 | this.radiusScale(d.geneCount) * 1.6 :
194 | this.defaultRadius * 1.6)
195 | .attr("fill", PLOT_CONFIG.point.hoverColor);
196 | })
197 | .on("mouseleave", (event, d) => {
198 | d3.select(event.target)
199 | .transition()
200 | .duration(PLOT_CONFIG.animation.tooltip.fadeOut)
201 | .attr("r", d => this.sizeByCount ?
202 | this.radiusScale(d.geneCount) :
203 | this.defaultRadius)
204 | .attr("fill", PLOT_CONFIG.point.color);
205 | })
206 | .on("mouseover", (event, d) => {
207 | this.tooltip.transition()
208 | .duration(200)
209 | .style("opacity", .9);
210 |
211 | this.tooltip.html(
212 | `${d.label}
` +
213 | `Distance: ${d.x.toFixed(2)}
` +
214 | `Probability: ${d.y.toFixed(2)}
` +
215 | `Gene Count: ${d.geneCount.toFixed(2)}`
216 | )
217 | .style("left", (event.pageX + 10) + "px")
218 | .style("top", (event.pageY - 28) + "px");
219 | })
220 | .on("mouseout", () => {
221 | this.tooltip.transition()
222 | .duration(500)
223 | .style("opacity", 0);
224 | });
225 | }
226 |
227 | resize() {
228 | const { width, height } = calculateDimensions();
229 | this.width = width;
230 | this.height = height;
231 |
232 | const svg = d3.select(`#${this.containerId}`).select('svg')
233 | .attr('width', width + PLOT_CONFIG.margin.left + PLOT_CONFIG.margin.right)
234 | .attr('height', height + PLOT_CONFIG.margin.top + PLOT_CONFIG.margin.bottom);
235 |
236 | this.x.range([0, width]);
237 | this.y.range([height, 0]);
238 |
239 | this.svg.select('.x-axis')
240 | .attr('transform', `translate(0,${height})`)
241 | .call(d3.axisBottom(this.x));
242 |
243 | this.svg.select('.y-axis')
244 | .call(d3.axisLeft(this.y));
245 |
246 | this.updatePlot();
247 | }
248 | }
249 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/static/js/interpretationGuide.js:
--------------------------------------------------------------------------------
1 | // Save this as: static/js/components/InterpretationGuide.js
2 |
3 | /**
4 | * Component for rendering and managing the interpretation guide overlay
5 | */
6 |
7 | export class InterpretationGuide {
8 | constructor(svg, width, height) {
9 | this.svg = svg;
10 | this.width = width;
11 | this.height = height;
12 | this.padding = 5;
13 | this.lineHeight = 15;
14 | }
15 |
16 | getText(currentUserClass, currentAssignedClass) {
17 | // Defines the guide text with dynamic class names
18 | return [
19 | `• Genes on diagonal: Contribute equally to both cell types`,
20 | `• Genes above diagonal: Support classification as ${currentUserClass}`,
21 | `• Genes below diagonal: Support classification as ${currentAssignedClass}`,
22 | `• Distance from diagonal: Strength of support for one type over the other`
23 | ];
24 | }
25 |
26 | update(currentUserClass, currentAssignedClass) {
27 | const guideText = this.getText(currentUserClass, currentAssignedClass);
28 | let guide = this.svg.select('.interpretation-guide');
29 |
30 | if (guide.empty()) {
31 | guide = this.createGuide(guideText);
32 | } else {
33 | this.updateGuideText(guide, guideText);
34 | }
35 | }
36 |
37 | createGuide(guideText) {
38 | // Creates the initial guide group
39 | const guide = this.svg.append("g")
40 | .attr("class", "interpretation-guide")
41 | .attr("transform", `translate(${this.width - 10}, ${this.height - 10})`);
42 |
43 | // Add text elements
44 | guide.selectAll("text")
45 | .data(guideText)
46 | .enter()
47 | .append("text")
48 | .attr("x", 0)
49 | .attr("y", (d, i) => i * this.lineHeight)
50 | .style("text-anchor", "start")
51 | .style("font-size", "12px")
52 | .text(d => d);
53 |
54 | this.addBackgroundRect(guide);
55 | return guide;
56 | }
57 |
58 | addBackgroundRect(guide) {
59 | // Add semi-transparent background
60 | const guideBBox = guide.node().getBBox();
61 | guide.insert("rect", ":first-child")
62 | .attr("x", guideBBox.x - this.padding)
63 | .attr("y", guideBBox.y - this.padding)
64 | .attr("width", guideBBox.width + (this.padding * 2))
65 | .attr("height", guideBBox.height + (this.padding * 2))
66 | .attr("fill", "rgba(255, 223, 186, 0.7)");
67 |
68 | // Position the guide in the bottom-right corner
69 | guide.attr("transform",
70 | `translate(${this.width - guideBBox.width - 15}, ${this.height - guideBBox.height - 15})`);
71 | }
72 |
73 | updateGuideText(guide, guideText) {
74 | // Updates existing guide text
75 | guide.selectAll("text")
76 | .data(guideText)
77 | .text(d => d);
78 | }
79 | }
80 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/static/js/plotConfig.js:
--------------------------------------------------------------------------------
1 | export const PLOT_CONFIG = {
2 | margin: {
3 | top: 60,
4 | right: 80,
5 | bottom: 50,
6 | left: 100
7 | },
8 | point: {
9 | radius: 5,
10 | // color: '#0ea5e9',
11 | // color: '#69b3a2',
12 | color: '#4a90e2',
13 | hoverColor: '#4a90e2',
14 | // color: '#64748b',
15 |
16 | },
17 | diagonalLine: {
18 | color: 'red',
19 | width: 2,
20 | dashArray: '5,5'
21 | },
22 | animation: {
23 | duration: 1000,
24 | tooltip: {
25 | fadeIn: 200,
26 | fadeOut: 500
27 | }
28 | },
29 | guide: {
30 | backgroundColor: 'rgba(255, 223, 186, 0.7)',
31 | padding: 5,
32 | lineHeight: 15,
33 | fontSize: '12px'
34 | },
35 | axis: {
36 | fontSize: '12px',
37 | tickSize: 5,
38 | tickPadding: 5
39 | }
40 | };
41 |
42 | export function calculateDimensions() {
43 | return {
44 | width: window.innerWidth - PLOT_CONFIG.margin.left - PLOT_CONFIG.margin.right,
45 | height: window.innerHeight * 0.34 - PLOT_CONFIG.margin.top - PLOT_CONFIG.margin.bottom
46 | };
47 | }
48 |
--------------------------------------------------------------------------------
/pciSeq/static/cell_analysis/dashboard/static/js/tooltip.js:
--------------------------------------------------------------------------------
1 | export function createTooltip() {
2 | return d3.select("body")
3 | .append("div")
4 | .attr("class", "tooltip")
5 | .style("opacity", 0);
6 | }
7 |
8 | export function formatProbability(prob) {
9 | return `${(prob * 100).toFixed(2)}%`;
10 | }
11 |
12 | export function createScales(width, height, xData, yData) {
13 | const xExtent = d3.extent(xData);
14 | const yExtent = d3.extent(yData);
15 |
16 | // Add a small padding to the domain
17 | const xPadding = (xExtent[1] - xExtent[0]) * 0.05;
18 | const yPadding = (yExtent[1] - yExtent[0]) * 0.05;
19 |
20 | return {
21 | x: d3.scaleLinear()
22 | .domain([xExtent[0] - xPadding, xExtent[1] + xPadding])
23 | .range([0, width]),
24 | y: d3.scaleLinear()
25 | .domain([yExtent[0] - yPadding, yExtent[1] + yPadding])
26 | .range([height, 0])
27 | };
28 | }
29 |
30 | export function handleTooltip(tooltip, config) {
31 | return {
32 | mouseOver: (event, d) => {
33 | tooltip.transition()
34 | .duration(config.animation.tooltip.fadeIn)
35 | .style("opacity", 0.9);
36 |
37 | tooltip.html(
38 | `${d.name}
` +
39 | `X: ${d.x.toFixed(3)}
` +
40 | `Y: ${d.y.toFixed(3)}`
41 | )
42 | .style("left", `${event.pageX + 10}px`)
43 | .style("top", `${event.pageY - 28}px`);
44 | },
45 | mouseOut: () => {
46 | tooltip.transition()
47 | .duration(config.animation.tooltip.fadeOut)
48 | .style("opacity", 0);
49 | }
50 | };
51 | }
52 |
53 | export function createAxis(scale, orientation) {
54 | const axis = orientation === 'bottom' ? d3.axisBottom(scale) : d3.axisLeft(scale);
55 | return axis
56 | .tickSize(PLOT_CONFIG.axis.tickSize)
57 | .tickPadding(PLOT_CONFIG.axis.tickPadding);
58 | }
59 |
--------------------------------------------------------------------------------
/pciSeq/static/memurai/Memurai-Developer-v3.1.4.msi:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pciSeq/static/memurai/Memurai-Developer-v3.1.4.msi
--------------------------------------------------------------------------------
/pdf/Qian_et_al_2020.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pdf/Qian_et_al_2020.pdf
--------------------------------------------------------------------------------
/pdf/appendix_mu.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/acycliq/pciSeq/c3c47553ff5b380c5746716d4984302e4ea92814/pdf/appendix_mu.pdf
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | pytest
2 | jupyterlab
3 | -e .
4 |
5 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | """
2 | pciSeq: Probabilistic Cell Typing for Spatial Transcriptomics
3 |
4 | A Python package for analyzing spatial transcriptomics data with probabilistic cell typing.
5 | This setup script handles package configuration and dependencies.
6 |
7 | For development installation:
8 | pip install -e .
9 |
10 | For building distribution:
11 | python -m build
12 | """
13 |
14 | import os
15 | from pathlib import Path
16 | from setuptools import setup, find_packages
17 |
18 |
19 | def get_static_files(root: str) -> list[str]:
20 | """
21 | Collect all static files needed for web interface.
22 |
23 | Args:
24 | root: Root directory to search for static files
25 |
26 | Returns:
27 | List of file paths relative to pciSeq package
28 | """
29 | static_extensions = {'.html', '.js', '.css', '.msi'}
30 | root_path = Path(root)
31 |
32 | static_files = []
33 | for path in root_path.rglob('*'):
34 | if path.suffix in static_extensions:
35 | relative_path = str(path.relative_to('pciSeq'))
36 | static_files.append(relative_path)
37 |
38 | return static_files
39 |
40 |
41 | # Core dependencies required for basic functionality
42 | INSTALL_REQUIRES = [
43 | 'altair', # Data visualization
44 | 'dask', # Parallel computing
45 | 'diplib', # Image processing
46 | 'fastremap', # Fast array operations
47 | 'flask', # Web framework
48 | 'natsort', # Natural sorting
49 | 'numexpr', # Fast numerical expressions
50 | 'numpy_groupies', # Group operations
51 | 'pandas', # Data manipulation
52 | 'pyvips', # Image processing
53 | 'redis', # Caching
54 | 'scikit-image', # Image analysis
55 | 'scikit-learn', # Machine learning
56 | 'scipy', # Scientific computing
57 | 'streamlit', # Web apps
58 | 'tomlkit', # TOML parsing
59 | 'tqdm', # Progress bars
60 | 'colorlog', # Colored logging
61 | ]
62 |
63 | # Optional dependencies for interactive use
64 | EXTRAS_REQUIRE = {
65 | 'interactive': [
66 | 'matplotlib>=2.2.0',
67 | 'jupyter'
68 | ],
69 | }
70 |
71 |
72 | # Get version from _version.py
73 | def get_version() -> str:
74 | """Extract version from _version.py."""
75 | version_file = Path('pciSeq/_version.py')
76 | if not version_file.exists():
77 | raise RuntimeError('Version file not found')
78 |
79 | version = None
80 | with open(version_file) as f:
81 | for line in f:
82 | if line.startswith('__version__'):
83 | version = line.split('=')[1].strip().strip("'")
84 | break
85 |
86 | if version is None:
87 | raise RuntimeError('Version not found in _version.py')
88 |
89 | return version
90 |
91 |
92 | # Read long description from README
93 | def get_long_description() -> str:
94 | """Read long description from README.md."""
95 | with open("README.md", encoding="utf-8") as f:
96 | return f.read()
97 |
98 |
99 | setup(
100 | name="pciSeq",
101 | version=get_version(),
102 | license="BSD",
103 | author="Dimitris Nicoloutsopoulos",
104 | author_email="dimitris.nicoloutsopoulos@gmail.com",
105 | description="Probabilistic cell typing for spatial transcriptomics",
106 | long_description=get_long_description(),
107 | long_description_content_type="text/markdown",
108 | url="https://github.com/acycliq/pciSeq",
109 | packages=find_packages(),
110 | python_requires=">=3.8", # Specify minimum Python version
111 | install_requires=INSTALL_REQUIRES,
112 | extras_require=EXTRAS_REQUIRE,
113 | include_package_data=True,
114 | package_data={
115 | 'pciSeq': get_static_files(os.path.join('pciSeq', 'static'))
116 | },
117 | classifiers=[
118 | "Development Status :: 4 - Beta", # Add development status
119 | "Intended Audience :: Science/Research",
120 | "Programming Language :: Python :: 3",
121 | "Programming Language :: Python :: 3.8",
122 | "Programming Language :: Python :: 3.9",
123 | "Programming Language :: Python :: 3.10",
124 | "License :: OSI Approved :: BSD License",
125 | "Operating System :: OS Independent",
126 | "Topic :: Scientific/Engineering :: Bio-Informatics",
127 | "Topic :: Scientific/Engineering :: Image Processing",
128 | ],
129 | keywords=[
130 | "spatial-transcriptomics",
131 | "cell-typing",
132 | "bioinformatics",
133 | "image-analysis",
134 | "single-cell"
135 | ],
136 | project_urls={
137 | "Bug Reports": "https://github.com/acycliq/pciSeq/issues",
138 | "Source": "https://github.com/acycliq/pciSeq",
139 | "Documentation": "https://github.com/acycliq/pciSeq#readme"
140 | }
141 | )
142 |
--------------------------------------------------------------------------------
/tests/conftest.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pytest
3 | import tempfile
4 | import pandas as pd
5 | from scipy.sparse import load_npz
6 | from pciSeq.src.core.utils import load_from_url
7 | from utils import setup_logger
8 |
9 | conftest_logger = setup_logger(__name__)
10 |
11 | bbox = [
12 | (4238, 364), # bottomleft, [x0, y0]
13 | (5160, 933) # topright, [x1, y1]
14 | ]
15 |
16 |
17 | def pytest_configure():
18 | """
19 | https://docs.pytest.org/en/7.1.x/deprecations.html#pytest-namespace
20 | """
21 | pytest.fspots = None
22 | pytest.fcells = None
23 | pytest.fscData = None
24 |
25 |
26 | def get_out_dir():
27 | out_dir = os.path.join(tempfile.gettempdir(), 'pciSeq', 'tests')
28 | if not os.path.exists(out_dir):
29 | os.makedirs(out_dir)
30 | return out_dir
31 |
32 |
33 | @pytest.fixture(scope='module')
34 | def read_demo_data(bbox=None):
35 | ROOT = r'https://github.com/acycliq/pciSeq/raw/master'
36 | path_str = "{}".format("/".join([ROOT, 'pciSeq', 'data', 'mouse', 'ca1', 'iss', 'spots.csv']))
37 | spots = pd.read_csv(os.path.join(path_str))
38 |
39 | coo_file = load_from_url(
40 | 'https://github.com/acycliq/pciSeq/blob/dev/pciSeq/data/mouse/ca1/segmentation/label_image.coo.npz?raw=true')
41 | label_image = load_npz(coo_file)
42 |
43 | path_str = "{}".format("/".join([ROOT, 'tests', 'data', 'test_scRNAseq.csv']))
44 | scData = pd.read_csv(path_str).set_index('gene_name')
45 | if bbox is not None:
46 | spots, label_image = clip_data(spots.copy(), label_image.copy, bbox)
47 | return spots, label_image, scData
48 |
49 |
50 | def clip_data(spots, img, bbox):
51 | spots_out = clip_spots(spots, bbox)
52 | img_out = clip_label_image(img, bbox)
53 | return spots_out, img_out
54 |
55 |
56 | def clip_spots(spots, bbox):
57 | spots = clip_dataframe(spots.copy(), bbox)
58 |
59 | # Save the test spots
60 | x0, y0 = bbox[0]
61 | spots.x = spots.x - x0
62 | spots.y = spots.y - y0
63 | return spots
64 |
65 |
66 | def clip_dataframe(df, bbox):
67 | x0, y0 = bbox[0]
68 | x1, y1 = bbox[1]
69 | idx_x = (df.x >= x0) & (df.x <= x1)
70 | idx_y = (df.y >= y0) & (df.y <= y1)
71 |
72 | idx = idx_x & idx_y
73 | return df[idx]
74 |
75 |
76 | def clip_label_image(im, bbox):
77 | x0, y0 = bbox[0]
78 | x1, y1 = bbox[1]
79 | return im[y0:y1, x0:x1]
80 |
81 |
82 |
83 |
--------------------------------------------------------------------------------
/tests/constants.py:
--------------------------------------------------------------------------------
1 | # Test data constants
2 | EXPECTED_AREA_METRICS = {
3 | 'area_sum': 3721912.0,
4 | 'max_label': 3481,
5 | 'column_sums': [231498829.0, 150952363.0, 75868924.0, 151411232.0, 82555688.0]
6 | }
7 |
8 | # VarBayes iteration deltas
9 | EXPECTED_ITER_DELTAS = [
10 | 1.0, 0.8998741, 0.6707933, 0.4703735, 0.62746835, 0.58092755,
11 | 0.604991, 0.28188822, 0.26233155, 0.19196033, 0.5576391, 0.19038722,
12 | 0.24835205, 0.28241628, 0.06873486, 0.16072749, 0.5829934, 0.16633023,
13 | 0.14863184, 0.12343879, 0.099603266, 0.05200758, 0.06741488, 0.1034664,
14 | 0.18765935, 0.16917473, 0.06632929, 0.046282567, 0.033078548, 0.020875283,
15 | 0.012404523
16 | ]
17 |
--------------------------------------------------------------------------------
/tests/test_app.py:
--------------------------------------------------------------------------------
1 | import pytest
2 | import pathlib
3 | import hashlib
4 | import numpy as np
5 | from pciSeq.src.core.main import VarBayes
6 | from pciSeq.app import parse_args, stage_data
7 | from pciSeq.src.validation.config_manager import ConfigManager
8 | from pciSeq.src.validation.input_validation import InputValidator
9 | from constants import EXPECTED_AREA_METRICS, EXPECTED_ITER_DELTAS
10 | from utils import setup_logger
11 |
12 | test_app_logger = setup_logger(__name__)
13 |
14 |
15 | def calculate_checksum(str_path):
16 | """
17 | Calculate SHA256 hash/checksum of a file
18 | """
19 | hasher = hashlib.sha256()
20 | path = pathlib.Path(str_path)
21 | with open(path, mode="rb") as kgo_file:
22 | while True:
23 | # read 1 megabyte binary chunks from file and feed them to hasher
24 | kgo_chunk = kgo_file.read(2 ** 20)
25 | if not kgo_chunk:
26 | break
27 | hasher.update(kgo_chunk)
28 | checksum = hasher.hexdigest()
29 | return checksum
30 |
31 |
32 | class TestPciSeq:
33 | """Test suite for data processing functionality"""
34 |
35 | def test_parse_args(self, read_demo_data):
36 | """Test argument parsing with various input combinations"""
37 | test_app_logger.info('Testing argument parsing')
38 | spots = read_demo_data[0]
39 | coo = read_demo_data[1]
40 |
41 | with pytest.raises(ValueError) as excinfo:
42 | parse_args(spots)
43 | assert str(excinfo.value) == ('Need to provide the spots and the coo matrix either as keyword arguments or as the first and second positional arguments.')
44 |
45 | _, _, scData, opts = parse_args(spots, coo)
46 | assert scData is None
47 | assert opts is None
48 |
49 | def test_validate(self, read_demo_data):
50 | """Test data validation functionality"""
51 | test_app_logger.info('Testing data validation')
52 | spots = read_demo_data[0]
53 | coo = read_demo_data[1]
54 | scData = read_demo_data[2]
55 |
56 | with pytest.raises(TypeError) as excinfo:
57 | cfg_man = ConfigManager.from_opts(None)
58 | InputValidator.validate(coo, coo, scData, cfg_man)
59 | assert str(excinfo.value) == "Spots should be passed-in as a dataframe"
60 |
61 | @pytest.mark.parametrize('filename, expected', [
62 | ('read_demo_data', EXPECTED_AREA_METRICS)
63 | ])
64 | def test_stage_data(self, filename, expected, request):
65 | """Test data staging and processing"""
66 | test_app_logger.info('Testing data staging and preprocessing')
67 | read_demo_data = request.getfixturevalue(filename)
68 | spots = read_demo_data[0]
69 | coo = read_demo_data[1]
70 |
71 | cells, cell_boundaries, spots = stage_data(spots, coo)
72 | pytest.fspots = spots
73 | pytest.fcells = cells
74 |
75 | assert len(cells.label) == len(np.unique(cells.label))
76 | assert cells.area.sum() == expected['area_sum']
77 | assert cells.label.max() == expected['max_label']
78 | assert np.all(
79 | spots[['x_global', 'y_global', 'label', 'x_cell', 'y_cell']]
80 | .sum().round(5).values == expected['column_sums'])
81 |
82 | @pytest.mark.parametrize('filename, expected', [
83 | ('read_demo_data', EXPECTED_ITER_DELTAS)
84 | ])
85 | def test_varBayes(self, filename, expected, request):
86 | """Test VarBayes algorithm convergence"""
87 | test_app_logger.info('Testing algorithm convergence')
88 | read_demo_data = request.getfixturevalue(filename)
89 | spots, coo, scData = read_demo_data
90 |
91 | # Create config
92 | opts = {
93 | 'launch_viewer': True,
94 | 'launch_diagnostics': True,
95 | 'max_iter': 31,
96 | }
97 | cfg_man = ConfigManager.from_opts(opts)
98 |
99 | # validate inputs
100 | spots, coo, scdata, cfg = InputValidator.validate(spots, coo, scData, cfg_man)
101 |
102 | _cells, cellBoundaries, _spots = stage_data(spots, coo)
103 | varBayes = VarBayes(_cells, _spots, scData, cfg)
104 | cellData, geneData = varBayes.run()
105 |
106 | arr_1 = np.array(varBayes.iter_delta, dtype=np.float32).round(11)
107 | arr_2 = np.array(expected[: cfg['max_iter']], dtype=np.float32).round(11)
108 | assert np.allclose(arr_1, arr_2, rtol=0, atol=1e-05)
109 |
--------------------------------------------------------------------------------
/tests/utils.py:
--------------------------------------------------------------------------------
1 | import logging
2 | import sys
3 |
4 |
5 | def setup_logger(name):
6 | """
7 | Set up a logger that writes to standard output
8 | """
9 | logger = logging.getLogger(name)
10 | logger.setLevel(logging.INFO)
11 |
12 | # Check if logger already has handlers to avoid duplicates
13 | if not logger.handlers:
14 | handler = logging.StreamHandler(sys.stdout)
15 | formatter = logging.Formatter('%(message)s')
16 | handler.setFormatter(formatter)
17 | logger.addHandler(handler)
18 |
19 | return logger
20 |
--------------------------------------------------------------------------------
 17 |
17 |  17 |
17 |