├── magiccluster
├── __init__.py
├── main.py
├── base.py
├── cli.py
├── magic_clustering.py
├── clustering.py
└── utils.py
├── docs
├── _config.yml
├── images
│ └── magic.png
└── index.md
├── data
├── magic.png
├── participant.tsv
└── test_covariate.tsv
├── requirements.txt
├── .gitignore
├── install_requirements.sh
├── CITATION.cff
├── setup.py
├── LICENSE
└── README.md
/magiccluster/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/docs/_config.yml:
--------------------------------------------------------------------------------
1 | theme: jekyll-theme-leap-day
--------------------------------------------------------------------------------
/data/magic.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/anbai106/MAGIC/HEAD/data/magic.png
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | numpy
2 | scikit-learn==0.21.3
3 | pandas
4 | nibabel
5 |
--------------------------------------------------------------------------------
/docs/images/magic.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/anbai106/MAGIC/HEAD/docs/images/magic.png
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | /.idea/
2 | # Compiled python modules.
3 | *.pyc
4 |
5 | # Setuptools distribution folder.
6 | /dist/
7 | /build/
8 | /venv/
9 |
10 | test.py
11 |
12 | # Python egg metadata, regenerated from source files by setuptools.
13 | /*.egg-info
--------------------------------------------------------------------------------
/install_requirements.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | while read requirement;
4 | do
5 | if conda install --yes $requirement; then
6 | echo "Successfully install: ${requirement}"
7 | else
8 | conda install --yes -c conda-forge $requirement
9 | fi
10 | done < requirements.txt
11 |
--------------------------------------------------------------------------------
/CITATION.cff:
--------------------------------------------------------------------------------
1 | abstract: "This is my MAGIC software for research purposes only..."
2 | authors:
3 | - family-names: Wen
4 | given-names: Junhao
5 | orcid: "https://orcid.org/0000-0003-2077-3070"
6 | cff-version: 1.2.0
7 | version: 0.0.3
8 | date-released: "2023-09-24"
9 | keywords:
10 | - "multi-scale clustering"
11 | - research
12 | license: MIT
13 | message: "If you use this software, please cite it using these metadata."
14 | repository-code: "https://github.com/anbai106/MAGIC"
15 | title: "MAGIC"
--------------------------------------------------------------------------------
/magiccluster/main.py:
--------------------------------------------------------------------------------
1 | from magiccluster import cli
2 |
3 | __author__ = "Junhao Wen"
4 | __copyright__ = "Copyright 2023"
5 | __credits__ = ["Junhao Wen"]
6 | __license__ = "See LICENSE file"
7 | __version__ = "0.0.3"
8 | __maintainer__ = "Junhao Wen"
9 | __email__ = "junhao.wen89@gmail.com"
10 | __status__ = "Development"
11 |
12 | def main():
13 |
14 | parser = cli.parse_command_line()
15 | args = parser.parse_args()
16 | args.func(args)
17 |
18 |
19 | if __name__ == '__main__':
20 | main()
21 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | import setuptools
2 |
3 | with open("README.md", "r") as fh:
4 | long_description = fh.read()
5 |
6 | setuptools.setup(
7 | name="magiccluster",
8 | version="0.0.3",
9 | author="junhao.wen",
10 | author_email="junhao.wen89@email.com",
11 | description="Multi-scale semi-supervised clustering",
12 | long_description=long_description,
13 | long_description_content_type="text/markdown",
14 | url="https://github.com/anbai106/MAGIC",
15 | packages=setuptools.find_packages(),
16 | entry_points={
17 | 'console_scripts': [
18 | 'magiccluster = magiccluster.main:main',
19 | ],
20 | },
21 | classifiers=(
22 | "Programming Language :: Python :: 3",
23 | "License :: OSI Approved :: MIT License",
24 | "Operating System :: OS Independent",
25 | ),
26 | )
27 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2022 Junhao WEN
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |
3 | 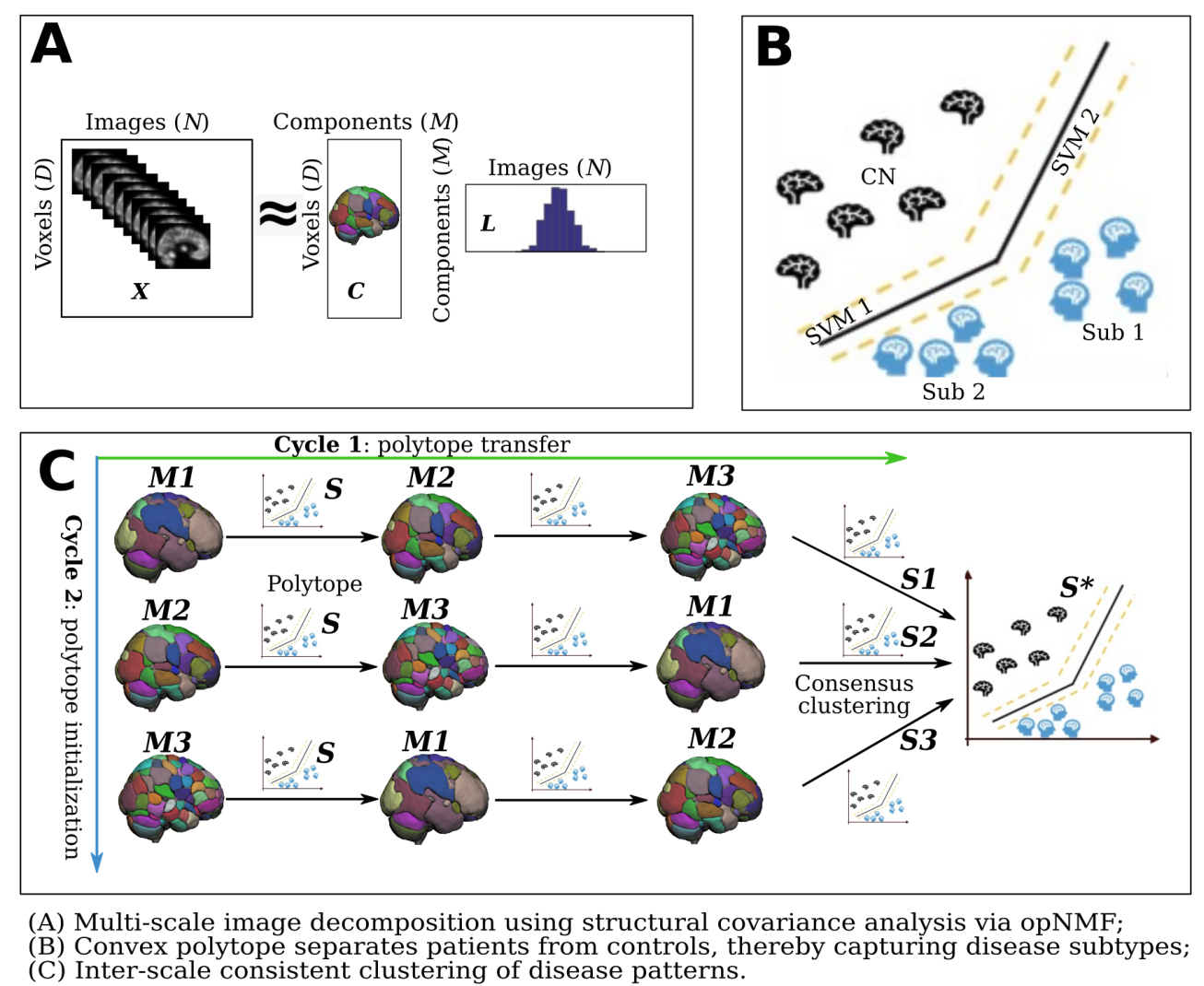 4 |
5 |
4 |
5 |
6 | MAGIC
7 |
8 |
9 | Multi-scAle heteroGeneity analysIs and Clustering
10 |
11 |
12 | Documentation
13 |
14 |
15 | ## `MAGIC`
16 | **MAGIC**, Multi-scAle heteroGeneity analysIs and Clustering, is a multi-scale semi-supervised clustering method that aims to derive robust clustering solutions across different scales for brain diseases.
17 |
18 | > :warning: **The documentation of this software is currently under development**
19 |
20 | ## Citing this work
21 | > :warning: Please let me know if you use this package for your publication; I will update your papers in the section of **Publication using MAGIC**...
22 |
23 | > :warning: Please cite the software using the **Cite this repository** button on the right sidebar menu, as well as the original papers below ...
24 |
25 | ### Original papers
26 | > Wen J., Varol E., Chand G., Sotiras A., Davatzikos C. (2020) **MAGIC: Multi-scale Heterogeneity Analysis and Clustering for Brain Diseases**. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol 12267. Springer, Cham. https://doi.org/10.1007/978-3-030-59728-3_66
27 |
28 | > Wen J., Varol E., Chand G., Sotiras A., Davatzikos C. (2022) **Multi-scale semi-supervised clustering of brain images: Deriving disease subtypes**. Medical Image Analysis, 2022. https://doi.org/10.1016/j.media.2021.102304 - [Link](https://www.sciencedirect.com/science/article/pii/S1361841521003492)
29 |
--------------------------------------------------------------------------------
/data/participant.tsv:
--------------------------------------------------------------------------------
1 | participant_id session_id diagnosis
2 | sub-80010 ses-M0 -1
3 | sub-80179 ses-M0 -1
4 | sub-80199 ses-M0 -1
5 | sub-80208 ses-M0 1
6 | sub-80249 ses-M0 -1
7 | sub-80265 ses-M0 1
8 | sub-80289 ses-M0 1
9 | sub-80396 ses-M0 -1
10 | sub-80425 ses-M0 1
11 | sub-80498 ses-M0 1
12 | sub-80537 ses-M0 -1
13 | sub-80557 ses-M0 -1
14 | sub-80575 ses-M0 -1
15 | sub-80607 ses-M0 1
16 | sub-80680 ses-M0 1
17 | sub-80688 ses-M0 -1
18 | sub-80765 ses-M0 -1
19 | sub-80812 ses-M0 1
20 | sub-80854 ses-M0 -1
21 | sub-80889 ses-M0 -1
22 | sub-81043 ses-M0 -1
23 | sub-81222 ses-M0 -1
24 | sub-81231 ses-M0 1
25 | sub-81287 ses-M0 -1
26 | sub-81323 ses-M0 -1
27 | sub-81353 ses-M0 -1
28 | sub-81456 ses-M0 -1
29 | sub-81528 ses-M0 -1
30 | sub-81533 ses-M0 1
31 | sub-81544 ses-M0 -1
32 | sub-81644 ses-M0 -1
33 | sub-81659 ses-M0 -1
34 | sub-81662 ses-M0 1
35 | sub-81754 ses-M0 1
36 | sub-81826 ses-M0 -1
37 | sub-81865 ses-M0 -1
38 | sub-81876 ses-M0 -1
39 | sub-81903 ses-M0 -1
40 | sub-81906 ses-M0 1
41 | sub-81989 ses-M0 -1
42 | sub-81992 ses-M0 -1
43 | sub-82003 ses-M0 1
44 | sub-82021 ses-M0 -1
45 | sub-82063 ses-M0 1
46 | sub-82066 ses-M0 1
47 | sub-82096 ses-M0 -1
48 | sub-82124 ses-M0 1
49 | sub-82155 ses-M0 1
50 | sub-82202 ses-M0 1
51 | sub-82208 ses-M0 -1
52 | sub-82217 ses-M0 -1
53 | sub-82229 ses-M0 1
54 | sub-82232 ses-M0 -1
55 | sub-82281 ses-M0 1
56 | sub-82293 ses-M0 1
57 | sub-82311 ses-M0 -1
58 | sub-82359 ses-M0 -1
59 | sub-82373 ses-M0 -1
60 | sub-82423 ses-M0 -1
61 | sub-82453 ses-M0 1
62 | sub-82458 ses-M0 -1
63 | sub-82467 ses-M0 -1
64 | sub-82492 ses-M0 1
65 | sub-82511 ses-M0 1
66 | sub-82587 ses-M0 -1
67 | sub-82674 ses-M0 1
68 | sub-82709 ses-M0 1
69 | sub-82754 ses-M0 -1
70 | sub-82784 ses-M0 1
71 | sub-82877 ses-M0 -1
72 | sub-82962 ses-M0 1
73 | sub-82982 ses-M0 -1
74 | sub-82985 ses-M0 1
75 | sub-82989 ses-M0 1
76 | sub-83010 ses-M0 1
77 | sub-83013 ses-M0 1
78 | sub-83044 ses-M0 1
79 | sub-83080 ses-M0 -1
80 | sub-83103 ses-M0 -1
81 | sub-83113 ses-M0 1
82 | sub-83207 ses-M0 -1
83 | sub-83260 ses-M0 1
84 | sub-83358 ses-M0 1
85 | sub-83372 ses-M0 -1
86 | sub-83423 ses-M0 -1
87 | sub-83429 ses-M0 1
88 | sub-83454 ses-M0 -1
89 | sub-83525 ses-M0 -1
90 | sub-83531 ses-M0 1
91 | sub-83580 ses-M0 -1
92 | sub-83612 ses-M0 1
93 | sub-83616 ses-M0 1
94 | sub-83632 ses-M0 1
95 | sub-83648 ses-M0 1
96 | sub-83835 ses-M0 -1
97 | sub-83972 ses-M0 -1
98 | sub-83987 ses-M0 -1
99 | sub-83999 ses-M0 1
100 | sub-84002 ses-M0 -1
101 |
--------------------------------------------------------------------------------
/data/test_covariate.tsv:
--------------------------------------------------------------------------------
1 | participant_id session_id diagnosis age sex
2 | sub-80010 ses-M0 -1 21.75 0
3 | sub-80179 ses-M0 -1 21.1666666666667 1
4 | sub-80199 ses-M0 -1 20.3333333333333 0
5 | sub-80208 ses-M0 1 20.5 0
6 | sub-80249 ses-M0 -1 20.8333333333333 1
7 | sub-80265 ses-M0 1 20.5 1
8 | sub-80289 ses-M0 1 20.0833333333333 0
9 | sub-80396 ses-M0 -1 20.8333333333333 0
10 | sub-80425 ses-M0 1 20 1
11 | sub-80498 ses-M0 1 20.9166666666667 0
12 | sub-80537 ses-M0 -1 20.9166666666667 1
13 | sub-80557 ses-M0 -1 21.5 1
14 | sub-80575 ses-M0 -1 21.75 0
15 | sub-80607 ses-M0 1 21 0
16 | sub-80680 ses-M0 1 21.0833333333333 0
17 | sub-80688 ses-M0 -1 21.9166666666667 1
18 | sub-80765 ses-M0 -1 20.5833333333333 1
19 | sub-80812 ses-M0 1 20.5833333333333 1
20 | sub-80854 ses-M0 -1 20.1666666666667 0
21 | sub-80889 ses-M0 -1 21.75 0
22 | sub-81043 ses-M0 -1 20.75 1
23 | sub-81222 ses-M0 -1 20.25 1
24 | sub-81231 ses-M0 1 21.75 1
25 | sub-81287 ses-M0 -1 20 1
26 | sub-81323 ses-M0 -1 20.5833333333333 1
27 | sub-81353 ses-M0 -1 20.0833333333333 0
28 | sub-81456 ses-M0 -1 21.6666666666667 1
29 | sub-81528 ses-M0 -1 19.5833333333333 1
30 | sub-81533 ses-M0 1 21.5833333333333 1
31 | sub-81544 ses-M0 -1 19.3333333333333 1
32 | sub-81644 ses-M0 -1 19.25 1
33 | sub-81659 ses-M0 -1 19.25 0
34 | sub-81662 ses-M0 1 19.3333333333333 1
35 | sub-81754 ses-M0 1 19.3333333333333 1
36 | sub-81826 ses-M0 -1 19.0833333333333 1
37 | sub-81865 ses-M0 -1 21.75 0
38 | sub-81876 ses-M0 -1 21.25 0
39 | sub-81903 ses-M0 -1 19.25 1
40 | sub-81906 ses-M0 1 21.3333333333333 1
41 | sub-81989 ses-M0 -1 19.0833333333333 1
42 | sub-81992 ses-M0 -1 21.1666666666667 0
43 | sub-82003 ses-M0 1 21.1666666666667 1
44 | sub-82021 ses-M0 -1 19.3333333333333 1
45 | sub-82063 ses-M0 1 20.1666666666667 0
46 | sub-82066 ses-M0 1 21.4166666666667 1
47 | sub-82096 ses-M0 -1 20.0833333333333 1
48 | sub-82124 ses-M0 1 20.6666666666667 1
49 | sub-82155 ses-M0 1 19.5 1
50 | sub-82202 ses-M0 1 19.5833333333333 1
51 | sub-82208 ses-M0 -1 21.1666666666667 1
52 | sub-82217 ses-M0 -1 19.5 1
53 | sub-82229 ses-M0 1 21.3333333333333 1
54 | sub-82232 ses-M0 -1 19 1
55 | sub-82281 ses-M0 1 20 1
56 | sub-82293 ses-M0 1 19.8333333333333 1
57 | sub-82311 ses-M0 -1 20.6666666666667 1
58 | sub-82359 ses-M0 -1 21.6666666666667 0
59 | sub-82373 ses-M0 -1 19.6666666666667 0
60 | sub-82423 ses-M0 -1 21.25 0
61 | sub-82453 ses-M0 1 21.4166666666667 0
62 | sub-82458 ses-M0 -1 19.0833333333333 0
63 | sub-82467 ses-M0 -1 20.0833333333333 0
64 | sub-82492 ses-M0 1 19.5 0
65 | sub-82511 ses-M0 1 19.6666666666667 1
66 | sub-82587 ses-M0 -1 19.5833333333333 1
67 | sub-82674 ses-M0 1 19.6666666666667 1
68 | sub-82709 ses-M0 1 22.5 1
69 | sub-82754 ses-M0 -1 20.8333333333333 0
70 | sub-82784 ses-M0 1 19.5833333333333 1
71 | sub-82877 ses-M0 -1 20.25 0
72 | sub-82962 ses-M0 1 19.1666666666667 0
73 | sub-82982 ses-M0 -1 18.8333333333333 1
74 | sub-82985 ses-M0 1 20.8333333333333 1
75 | sub-82989 ses-M0 1 18.8333333333333 0
76 | sub-83010 ses-M0 1 19.5833333333333 0
77 | sub-83013 ses-M0 1 20.25 1
78 | sub-83044 ses-M0 1 20.8333333333333 1
79 | sub-83080 ses-M0 -1 18.5 1
80 | sub-83103 ses-M0 -1 18.1666666666667 0
81 | sub-83113 ses-M0 1 18.6666666666667 1
82 | sub-83207 ses-M0 -1 19.25 0
83 | sub-83260 ses-M0 1 22.6666666666667 0
84 | sub-83358 ses-M0 1 19.8333333333333 1
85 | sub-83372 ses-M0 -1 18.25 0
86 | sub-83423 ses-M0 -1 20.75 1
87 | sub-83429 ses-M0 1 18.8333333333333 1
88 | sub-83454 ses-M0 -1 20.1666666666667 1
89 | sub-83525 ses-M0 -1 18.9166666666667 1
90 | sub-83531 ses-M0 1 19.9166666666667 0
91 | sub-83580 ses-M0 -1 19 0
92 | sub-83612 ses-M0 1 20 1
93 | sub-83616 ses-M0 1 18.5833333333333 1
94 | sub-83632 ses-M0 1 20.9166666666667 1
95 | sub-83648 ses-M0 1 19.5833333333333 1
96 | sub-83835 ses-M0 -1 20.6666666666667 0
97 | sub-83972 ses-M0 -1 19.0833333333333 0
98 | sub-83987 ses-M0 -1 18.4166666666667 1
99 | sub-83999 ses-M0 1 18.6666666666667 0
100 | sub-84002 ses-M0 -1 22.25 0
101 |
--------------------------------------------------------------------------------
/docs/index.md:
--------------------------------------------------------------------------------
1 |
2 |  3 |
3 |
4 |
5 | # MAGIC documentation
6 | **MAGIC**, Multi-scAle heteroGeneity analysIs and Clustering, is a multi-scale semi-supervised clustering method that aims to derive robust clustering solutions across different scales for brain diseases.
7 | Compared to original HYDRA method, MAGIC has the following advantages:
8 | - Multi-scale feature extractions via opNMF;
9 | - Inter-scale consistent clustering solution.
10 |
11 | ## Installation
12 | ### Prerequisites
13 | In order to run MAGIC, one must have already installed and ran [SOPNMF (https://github.com/anbai106/SOPNMF) with the voxel-wise image data. After this, please follow the following steps for installation.
14 |
15 | There are three choices to install MAGIC.
16 | ### Use MAGIC as a python package
17 | We recommend the users to use Conda virtual environment:
18 | ```
19 | 1) conda create --name MAGIC python=3.6
20 | ```
21 | Activate the virtual environment:
22 | ```
23 | 2) source activate MAGIC
24 | ```
25 | Install other python package dependencies (go to the root folder of MAGIC):
26 | ```
27 | 3) ./install_requirements.sh
28 | ```
29 | Finally, we need install MAGIC from PyPi:
30 | ```
31 | 3) pip install magiccluster==0.0.3
32 | ```
33 |
34 | ### Use MAGIC from commandline:
35 | After installing all dependencies in the **requirements.txt** file, go to the root folder of MAGIC where the **setup.py** locates:
36 | ```
37 | pip install -e .
38 | ```
39 |
40 | ### Use MAGIC as a developer version:
41 | ```
42 | python -m pip install git+https://github.com/anbai106/MAGIC.git
43 | ```
44 |
45 | ## Input structure
46 | MAGIC requires a specific input structure inspired by [BIDS](https://bids.neuroimaging.io/).
47 | Some conventions for the group label/diagnosis: -1 represents healthy control (**CN**) and 1 represents patient (**PT**); categorical variables, such as sex, should be encoded to numbers: Female for 0 and Male for 1, for instance.
48 |
49 | ### participant and covariate tsv
50 | The first 3 columns are **participant_id**, **session_id** and **diagnosis**.
51 |
52 | Example for feature tsv:
53 | ```
54 | participant_id session_id diagnosis
55 | sub-CLNC0001 ses-M00 -1 432.1
56 | sub-CLNC0002 ses-M00 1 398.2
57 | sub-CLNC0003 ses-M00 -1 412.0
58 | sub-CLNC0004 ses-M00 -1 487.4
59 | sub-CLNC0005 ses-M00 1 346.5
60 | sub-CLNC0006 ses-M00 1 443.2
61 | sub-CLNC0007 ses-M00 -1 450.2
62 | sub-CLNC0008 ses-M00 1 443.2
63 | ```
64 | Example for covariate tsv:
65 | ```
66 | participant_id session_id diagnosis age sex ...
67 | sub-CLNC0001 ses-M00 -1 56.1 0
68 | sub-CLNC0002 ses-M00 1 57.2 0
69 | sub-CLNC0003 ses-M00 -1 43.0 1
70 | sub-CLNC0004 ses-M00 -1 25.4 1
71 | sub-CLNC0005 ses-M00 1 74.5 1
72 | sub-CLNC0006 ses-M00 1 44.2 0
73 | sub-CLNC0007 ses-M00 -1 40.2 0
74 | sub-CLNC0008 ses-M00 1 43.2 1
75 | ```
76 |
77 | ## Example
78 | We offer a fake dataset in the folder of **MAGIC/data**. Users should follow the same data structure.
79 |
80 | ### Running MAGIC for clustering CN vs Subtype1 vs Subtype2 vs ...:
81 | ```
82 | from from magic.magic_clustering import clustering
83 | participant_tsv="MAGIC/data/participant.tsv"
84 | opnmf_dir = "PATH_OPNMF_DIR"
85 | output_dir = "PATH_OUTPUT_DIR"
86 | k_min=2
87 | k_max=8
88 | cv_repetition=100
89 | clustering(participant_tsv, opnmf_dir, output_dir, k_min, k_max, 25, 60, 5, cv_repetition)
90 | ```
91 |
92 | ## Citing this work
93 | > :warning: Please let me know if you use this package for your publication; I will update your papers in the section of **Publication using MAGIC**...
94 |
95 | > :warning: Please cite the software using the **Cite this repository** button on the right sidebar menu, as well as the original papers below ...
96 |
97 | ### Original papers
98 | > Wen J., Varol E., Chand G., Sotiras A., Davatzikos C. (2020) **MAGIC: Multi-scale Heterogeneity Analysis and Clustering for Brain Diseases**. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol 12267. Springer, Cham. https://doi.org/10.1007/978-3-030-59728-3_66
99 |
100 | > Wen J., Varol E., Chand G., Sotiras A., Davatzikos C. (2022) **Multi-scale semi-supervised clustering of brain images: Deriving disease subtypes**. Medical Image Analysis, 2022. https://doi.org/10.1016/j.media.2021.102304 - [Link](https://www.sciencedirect.com/science/article/pii/S1361841521003492)
--------------------------------------------------------------------------------
/magiccluster/base.py:
--------------------------------------------------------------------------------
1 | import abc
2 | import pandas as pd
3 | from utils import GLMcorrection
4 | import numpy as np
5 | import os
6 | from sklearn.preprocessing import StandardScaler
7 |
8 | __author__ = "Junhao Wen"
9 | __copyright__ = "Copyright 2023"
10 | __credits__ = ["Junhao Wen, Erdem Varol"]
11 | __license__ = "See LICENSE file"
12 | __version__ = "0.0.3"
13 | __maintainer__ = "Junhao Wen"
14 | __email__ = "junhao.wen89@gmail.com"
15 | __status__ = "Development"
16 |
17 |
18 | class WorkFlow:
19 | __metaclass__ = abc.ABCMeta
20 |
21 | @abc.abstractmethod
22 | def run(self):
23 | pass
24 |
25 |
26 | class Input:
27 | __metaclass__ = abc.ABCMeta
28 |
29 | @abc.abstractmethod
30 | def get_x(self):
31 | pass
32 |
33 | @abc.abstractmethod
34 | def get_y(self):
35 | pass
36 |
37 | class OPNMF_Input(Input):
38 |
39 | def __init__(self, opnmf_dir, participant_tsv, covariate_tsv=None):
40 | self._opnmf_dir = opnmf_dir
41 | self._participant_tsv = participant_tsv
42 | self._covariate_tsv = covariate_tsv
43 | self._x = None
44 | self._y = None
45 |
46 | ## check the participant_tsv & covariate_tsv, the header, the order of the columns, etc
47 | self._df_feature = pd.read_csv(participant_tsv, sep='\t')
48 | if ('participant_id' != list(self._df_feature.columns.values)[0]) or (
49 | 'session_id' != list(self._df_feature.columns.values)[1]) or \
50 | ('diagnosis' != list(self._df_feature.columns.values)[2]):
51 | raise Exception("the data file is not in the correct format."
52 | "Columns should include ['participant_id', 'session_id', 'diagnosis']")
53 | self._subjects = list(self._df_feature['participant_id'])
54 | self._sessions = list(self._df_feature['session_id'])
55 | self._diagnosis = list(self._df_feature['diagnosis'])
56 |
57 | def get_x(self, num_component, opnmf_dir):
58 |
59 | ## alternatively, we use here the output of pyOPNMF loading coefficient
60 | loading_coefficient_csv = os.path.join(opnmf_dir, 'NMF', 'component_' + str(num_component),
61 | 'loading_coefficient.tsv')
62 | ## read the tsv
63 | df_opnmf = pd.read_csv(loading_coefficient_csv, sep='\t')
64 | df_opnmf = df_opnmf.loc[df_opnmf['participant_id'].isin(self._df_feature['participant_id'])]
65 | ### adjust the order of the rows to match the original tsv files
66 | df_opnmf = df_opnmf.set_index('participant_id')
67 | df_opnmf = df_opnmf.reindex(index=self._df_feature['participant_id'])
68 | df_opnmf = df_opnmf.reset_index()
69 |
70 | self._x = df_opnmf[['component_' + str(i + 1) for i in range(num_component)]].to_numpy()
71 |
72 | ### normalize the data, note the normalization should be done for each component, not across component
73 | scaler = StandardScaler()
74 | self._x = scaler.fit_transform(self._x)
75 |
76 | if self._covariate_tsv is not None:
77 | df_covariate = pd.read_csv(self._covariate_tsv, sep='\t')
78 | if ('participant_id' != list(self._df_feature.columns.values)[0]) or (
79 | 'session_id' != list(self._df_feature.columns.values)[1]) or \
80 | ('diagnosis' != list(self._df_feature.columns.values)[2]):
81 | raise Exception("the data file is not in the correct format."
82 | "Columns should include ['participant_id', 'session_id', 'diagnosis']")
83 | participant_covariate = list(df_covariate['participant_id'])
84 | session_covariate = list(df_covariate['session_id'])
85 | label_covariate = list(df_covariate['diagnosis'])
86 |
87 | # check that the participant_tsv and covariate_tsv have the same orders for the first three column
88 | if (not self._subjects == participant_covariate) or (not self._sessions == session_covariate) or (
89 | not self._diagnosis == label_covariate):
90 | raise Exception(
91 | "the first three columns in the feature csv and covariate csv should be exactly the same.")
92 |
93 | ## normalize the covariate z-scoring
94 | data_covariate = df_covariate.iloc[:, 3:]
95 | data_covariate = ((data_covariate - data_covariate.mean()) / data_covariate.std()).values
96 |
97 | ## correction for the covariate, only retain the pathodological correspondance
98 | self._x, _ = GLMcorrection(self._x, np.asarray(self._diagnosis), data_covariate, self._x, data_covariate)
99 |
100 | return self._x
101 |
102 | def get_y(self):
103 | """
104 | Do not change the label's representation
105 | :return:
106 | """
107 |
108 | if self._y is not None:
109 | return self._y
110 |
111 | self._y = np.array(self._diagnosis)
112 | return self._y
113 |
114 |
--------------------------------------------------------------------------------
/magiccluster/cli.py:
--------------------------------------------------------------------------------
1 | import argparse
2 |
3 | __author__ = "Junhao Wen"
4 | __copyright__ = "Copyright 2023"
5 | __credits__ = ["Junhao Wen"]
6 | __license__ = "See LICENSE file"
7 | __version__ = "0.0.3"
8 | __maintainer__ = "Junhao Wen"
9 | __email__ = "junhao.wen89@gmail.com"
10 | __status__ = "Development"
11 |
12 | def magic_func(args):
13 | """
14 | The default function to run classification.
15 | Args:
16 | args: args from parser
17 |
18 | Returns:
19 |
20 | """
21 | from magiccluster.magic_clustering import clustering
22 | clustering(
23 | args.participant_tsv,
24 | args.opnmf_dir,
25 | args.output_dir,
26 | args.k_min,
27 | args.k_max,
28 | args.num_components_min,
29 | args.num_components_max,
30 | args.num_components_step,
31 | args.cv_repetition,
32 | args.covariate_tsv,
33 | args.cv_strategy,
34 | args.save_models,
35 | args.cluster_predefined_c,
36 | args.class_weight_balanced,
37 | args.weight_initialization_type,
38 | args.num_iteration,
39 | args.num_consensus,
40 | args.tol,
41 | args.multiscale_tol,
42 | args.n_threads,
43 | args.verbose
44 | )
45 |
46 | def parse_command_line():
47 | """
48 | Definition for the commandline parser
49 | Returns:
50 |
51 | """
52 |
53 | parser = argparse.ArgumentParser(
54 | prog='magiccluster-cluster',
55 | description='Perform multi-scale semi-supervised clustering using MAGIC...')
56 |
57 | subparser = parser.add_subparsers(
58 | title='''Task to perform...''',
59 | description='''We now only allow to use MAGIC for clustering''',
60 | dest='task',
61 | help='''****** Tasks proposed by MAGIC ******''')
62 |
63 | subparser.required = True
64 |
65 | ########################################################################################################################
66 |
67 | ## Add arguments for ADML ROI classification
68 | clustering_parser = subparser.add_parser(
69 | 'cluster',
70 | help='Perform clustering with MAGIC.')
71 |
72 | clustering_parser.add_argument(
73 | 'participant_tsv',
74 | help="Path to the tsv containing the following first columns:"
75 | "i) the first column is the participant_id. "
76 | "ii) the second column should be the session_id. "
77 | "iii) the third column should be the diagnosis. ",
78 | default=None
79 | )
80 |
81 | clustering_parser.add_argument(
82 | 'opnmf_dir',
83 | help='Path to the directory of where SOPNMF was run (the voxel-wise images should be run first with SOPNMF).',
84 | default=None
85 | )
86 |
87 | clustering_parser.add_argument(
88 | 'output_dir',
89 | help='Path to the directory of where to store the final output.',
90 | default=None
91 | )
92 |
93 | clustering_parser.add_argument(
94 | 'k_min',
95 | help='Number of cluster (k) minimum value.',
96 | default=None, type=int
97 | )
98 |
99 | clustering_parser.add_argument(
100 | 'k_max',
101 | help='Number of cluster (k) maximum value.',
102 | default=None, type=int
103 | )
104 |
105 | clustering_parser.add_argument(
106 | 'num_components_min',
107 | help='Number of the min PSC for the SOPNMF',
108 | default=None, type=int
109 | )
110 |
111 | clustering_parser.add_argument(
112 | 'num_components_max',
113 | help='Number of the max PSC for the SOPNMF',
114 | default=None, type=int
115 | )
116 |
117 | clustering_parser.add_argument(
118 | 'num_components_step',
119 | help='The step size between the min and the max PSC for the SOPNMF',
120 | default=None, type=int
121 | )
122 |
123 | clustering_parser.add_argument(
124 | 'cv_repetition',
125 | help='Number of repetitions for the chosen cross-validation (CV).',
126 | default=None, type=int
127 | )
128 |
129 | clustering_parser.add_argument(
130 | '--covariate_tsv',
131 | help="Path to the tsv containing covariates, following the BIDS convention. The first 3 columns is the same as feature_tsv",
132 | default=None,
133 | type=str
134 | )
135 |
136 | clustering_parser.add_argument(
137 | '-cs', '--cv_strategy',
138 | help='Chosen CV strategy, default is hold_out. ',
139 | type=str, default='hold_out',
140 | choices=['k_fold', 'hold_out'],
141 | )
142 |
143 | clustering_parser.add_argument(
144 | '-sm', '--save_models',
145 | help='If save modles during all repetitions of CV. ',
146 | default=False, action="store_true"
147 | )
148 |
149 | clustering_parser.add_argument(
150 | '--cluster_predefined_c',
151 | type=float,
152 | default=0.25,
153 | help="Predefined hyperparameter C of SVM. Default is 0.25. "
154 | "Better choice may be guided by HYDRA global classification with nested CV for optimal C searching. "
155 | )
156 |
157 | clustering_parser.add_argument(
158 | '-cwb', '--class_weight_balanced',
159 | help='If group samples are balanced, default is True. ',
160 | default=False, action="store_true"
161 | )
162 |
163 | clustering_parser.add_argument(
164 | '-wit', '--weight_initialization_type',
165 | help='Strategy for initializing the weighted sample matrix of the polytope. ',
166 | type=str, default='DPP',
167 | choices=['DPP', 'random_assign'],
168 | )
169 |
170 | clustering_parser.add_argument(
171 | '--num_iteration',
172 | help='Number of iteration to converge each SVM.',
173 | default=50, type=int
174 | )
175 |
176 | clustering_parser.add_argument(
177 | '--num_consensus',
178 | help='Number of iteration for inner consensus clusetering.',
179 | default=20, type=int
180 | )
181 |
182 | clustering_parser.add_argument(
183 | '--tol',
184 | help='Clustering stopping criterion, until the polytope becomes stable',
185 | default=1e-8, type=float
186 | )
187 |
188 | clustering_parser.add_argument(
189 | '--multiscale_tol',
190 | help='Clustering stopping criterion, until the multi-scale clustering solution stable',
191 | default=0.85, type=float
192 | )
193 |
194 | clustering_parser.add_argument(
195 | '-nt', '--n_threads',
196 | help='Number of cores used, default is 4',
197 | type=int, default=4
198 | )
199 |
200 | clustering_parser.add_argument(
201 | '-v', '--verbose',
202 | help='Increase output verbosity',
203 | default=False, action="store_true"
204 | )
205 |
206 | clustering_parser.set_defaults(func=magic_func)
207 |
208 |
209 |
210 |
--------------------------------------------------------------------------------
/magiccluster/magic_clustering.py:
--------------------------------------------------------------------------------
1 | from .clustering import DualSVM_Subtype, DualSVM_Subtype_transfer_learning
2 | from .base import OPNMF_Input
3 | import os, pickle
4 | from .utils import cluster_stability_across_resolution, summary_clustering_result_multiscale, shift_list, consensus_clustering_across_c, make_cv_partition
5 | import numpy as np
6 |
7 | __author__ = "Junhao Wen"
8 | __copyright__ = "Copyright 2023"
9 | __credits__ = ["Junhao Wen, Erdem Varol"]
10 | __license__ = "See LICENSE file"
11 | __version__ = "0.0.3"
12 | __maintainer__ = "Junhao Wen"
13 | __email__ = "junhao.wen89@gmail.com"
14 | __status__ = "Development"
15 |
16 | def clustering(participant_tsv, opnmf_dir, output_dir, k_min, k_max, num_components_min, num_components_max, num_components_step, cv_repetition, covariate_tsv=None, cv_strategy='hold_out', save_models=False,
17 | cluster_predefined_c=0.25, class_weight_balanced=True, weight_initialization_type='DPP', num_iteration=50,
18 | num_consensus=20, tol=1e-8, multiscale_tol=0.85, n_threads=8, verbose=False):
19 | """
20 | pyhydra core function for clustering
21 | Args:
22 | participant_tsv:str, path to the participant_tsv tsv, following the BIDS convention. The tsv contains
23 | the following headers: "
24 | "i) the first column is the participant_id;"
25 | "ii) the second column should be the session_id;"
26 | "iii) the third column should be the diagnosis;"
27 | opnmf_dir: str, path to store the OPNMF results
28 | output_dir: str, path to store the clustering results
29 | k_min: int, minimum k (number of clusters)
30 | k_max: int, maximum k (number of clusters)
31 | cv_repetition: int, number of repetitions for cross-validation (CV)
32 | covariate_tsv: str, path to the tsv containing the covaria`tes, eg., age or sex. The header (first 3 columns) of
33 | the tsv file is the same as the feature_tsv, following the BIDS convention.
34 | cv_strategy: str, cross validation strategy used. Default is hold_out. choices=['k_fold', 'hold_out']
35 | save_models: Bool, if save all models during CV. Default is False to save space.
36 | Set true only if you are going to apply the trained model to unseen data.
37 | cluster_predefined_c: Float, default is 0.25. The predefined best c if you do not want to perform a nested CV to
38 | find it. If used, it should be a float number
39 | class_weight_balanced: Bool, default is True. If the two groups are balanced.
40 | weight_initialization_type: str, default is DPP. The strategy for initializing the weight to control the
41 | hyperplances and the subpopulation of patients. choices=["random_hyperplane", "random_assign", "k_means", "DPP"]
42 | num_iteration: int, default is 50. The number of iterations to iteratively optimize the polytope.
43 | num_consensus: int, default is 20. The number of repeats for consensus clustering to eliminate the unstable clustering.
44 | tol: float, default is 1e-8. Clustering stopping criterion.
45 | multiscale_tol: float, default is 0.85. Double cyclic optimization stopping criterion.
46 | n_threads: int, default is 8. The number of threads to run model in parallel.
47 | verbose: Bool, default is False. If the output message is verbose.
48 |
49 | Returns: clustering outputs.
50 |
51 | """
52 | ### For voxel approach
53 | print('MAGIC for semi-supervised clustering...')
54 | if covariate_tsv == None:
55 | input_data = OPNMF_Input(opnmf_dir, participant_tsv, covariate_tsv=None)
56 | else:
57 | input_data = OPNMF_Input(opnmf_dir, participant_tsv, covariate_tsv=covariate_tsv)
58 |
59 | ## data split
60 | print('Data split was performed based on validation strategy: %s...\n' % cv_strategy)
61 | if cv_strategy == "hold_out":
62 | ## check if data split has been done, if yes, the pickle file is there
63 | if os.path.isfile(os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-holdout.pkl')):
64 | split_index = pickle.load(open(os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-holdout.pkl'), 'rb'))
65 | else:
66 | split_index, _ = make_cv_partition(input_data.get_y(), cv_strategy, output_dir, cv_repetition)
67 | elif cv_strategy == "k_fold":
68 | ## check if data split has been done, if yes, the pickle file is there
69 | if os.path.isfile(os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-fold.pkl')):
70 | split_index = pickle.load(open(os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-fold.pkl'), 'rb'))
71 | else:
72 | split_index, _ = make_cv_partition(input_data.get_y(), cv_strategy, output_dir, cv_repetition)
73 |
74 | print('Data split has been done!\n')
75 |
76 | print('Starts semi-supervised clustering...')
77 | ### Here, semi-supervised clustering with multi-scale feature reduction learning
78 | if (num_components_max - num_components_min) % num_components_step != 0:
79 | raise Exception('Number of componnets step should be divisible!')
80 |
81 | ## C lists
82 | C_list = list(range(num_components_min, num_components_max+num_components_step, num_components_step))
83 | ## first loop on different initial C.
84 | for i in range(len(C_list)):
85 | c_list = shift_list(C_list, i)
86 | num_run = 0
87 | loop = True
88 | print('Initialize C == %d\n' % C_list[i])
89 | while loop:
90 | for j in range(len(c_list)):
91 | if num_run == 0:
92 | num_run += 1
93 | k_continuing = np.arange(k_min, k_max+1).tolist()
94 | print('First C == %d\n' % c_list[j])
95 | output_dir_loop = os.path.join(output_dir, 'initialization_c_' + str(C_list[i]), 'clustering_run' + str(num_run))
96 | wf_clustering = DualSVM_Subtype(input_data,
97 | participant_tsv,
98 | split_index,
99 | cv_repetition,
100 | k_min,
101 | k_max,
102 | output_dir_loop,
103 | opnmf_dir,
104 | balanced=class_weight_balanced,
105 | num_consensus=num_consensus,
106 | num_iteration=num_iteration,
107 | tol=tol,
108 | predefined_c=cluster_predefined_c,
109 | weight_initialization_type=weight_initialization_type,
110 | n_threads=n_threads,
111 | num_components_min=c_list[j],
112 | num_components_max=c_list[j],
113 | num_components_step=num_components_step,
114 | save_models=save_models,
115 | verbose=verbose)
116 |
117 | wf_clustering.run()
118 | else: ## initialize the model from the former resolution
119 | num_run += 1
120 | print('Transfer learning on resolution C == %d for run == %d\n' % (c_list[j], num_run))
121 | output_dir_tl = os.path.join(output_dir, 'initialization_c_' + str(C_list[i]))
122 | wf_clustering = DualSVM_Subtype_transfer_learning(input_data,
123 | participant_tsv,
124 | split_index,
125 | cv_repetition,
126 | k_continuing,
127 | output_dir_tl,
128 | opnmf_dir,
129 | balanced=class_weight_balanced,
130 | num_iteration=num_iteration,
131 | tol=tol,
132 | predefined_c=cluster_predefined_c,
133 | weight_initialization_type=weight_initialization_type,
134 | n_threads=n_threads,
135 | num_component=c_list[j],
136 | num_component_former=c_list[j-1],
137 | num_run=num_run)
138 |
139 | wf_clustering.run()
140 |
141 | ### check the clustering stability between the current C and former C
142 | k_continuing, k_converged = cluster_stability_across_resolution(c_list[j], c_list[j-1], os.path.join(output_dir, 'initialization_c_' + str(C_list[i])), k_continuing, num_run, stop_tol=multiscale_tol)
143 |
144 | if not k_continuing:
145 | loop = False
146 | break
147 |
148 | ## After cross validate the hyperparameter k & num_components, summarize the results into a single tsv file.
149 | if not k_continuing:
150 | summary_clustering_result_multiscale(os.path.join(output_dir, 'initialization_c_' + str(C_list[i])), k_min, k_max)
151 |
152 | ## consensus learning based on different initialization Cs
153 | print('Computing the final consensus group membership!\n')
154 | consensus_clustering_across_c(output_dir, C_list, k_min, k_max)
155 | print('Finish...')
--------------------------------------------------------------------------------
/magiccluster/clustering.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | import pandas as pd
4 | from .utils import consensus_clustering, cv_cluster_stability, hydra_solver_svm_tl
5 | from .base import WorkFlow
6 | from utils import hydra_solver_svm
7 |
8 | __author__ = "Junhao Wen"
9 | __copyright__ = "Copyright 2023"
10 | __credits__ = ["Junhao Wen, Erdem Varol"]
11 | __license__ = "See LICENSE file"
12 | __version__ = "0.0.3"
13 | __maintainer__ = "Junhao Wen"
14 | __email__ = "junhao.wen89@gmail.com"
15 | __status__ = "Development"
16 | class DualSVM_Subtype(WorkFlow):
17 |
18 | def __init__(self, input, participant_tsv, split_index, cv_repetition, k_min, k_max, output_dir, opnmf_dir, balanced=True,
19 | test_size=0.2, num_consensus=20, num_iteration=50, tol=1e-6, predefined_c=None, weight_initialization_type='DPP',
20 | n_threads=8, num_components_min=10, num_components_max=100, num_components_step=10, save_models=False,
21 | verbose=True):
22 |
23 | self._input = input
24 | self._participant_tsv = participant_tsv
25 | self._split_index = split_index
26 | self._cv_repetition = cv_repetition
27 | self._output_dir = output_dir
28 | self._opnmf_dir = opnmf_dir
29 | self._k_min = k_min
30 | self._k_max = k_max
31 | self._balanced = balanced
32 | self._test_size = test_size
33 | self._num_consensus = num_consensus
34 | self._num_iteration = num_iteration
35 | self._tol = tol
36 | self._predefined_c = predefined_c
37 | self._weight_initialization_type = weight_initialization_type
38 | self._k_range_list = list(range(k_min, k_max + 1))
39 | self._n_threads = n_threads
40 | self._num_components_min = num_components_min
41 | self._num_components_max = num_components_max
42 | self._num_components_step = num_components_step
43 | self._save_models = save_models
44 | self._verbose = verbose
45 |
46 |
47 | def run(self):
48 |
49 | ## by default, we solve the problem using dual solver with a linear kernel.
50 | for num_component in range(self._num_components_min, self._num_components_max + self._num_components_step, self._num_components_step):
51 |
52 | if os.path.exists(os.path.join(self._output_dir, 'component_' + str(num_component), "adjusted_rand_index.tsv")):
53 | print("This number of component have been trained and converged: %d" % num_component)
54 | else:
55 | x = self._input.get_x(num_component, self._opnmf_dir)
56 | y = self._input.get_y_raw()
57 | data_label_folds_ks = np.zeros((y.shape[0], self._cv_repetition, self._k_max - self._k_min + 1)).astype(int)
58 |
59 | for i in range(self._cv_repetition):
60 | for j in self._k_range_list:
61 | print('Applying pyHRDRA for finding %d clusters. Repetition: %d / %d...\n' % (j, i+1, self._cv_repetition))
62 | training_final_prediction = hydra_solver_svm(i, x[self._split_index[i][0]], y[self._split_index[i][0]], j, self._output_dir,
63 | self._num_consensus, self._num_iteration, self._tol, self._balanced, self._predefined_c,
64 | self._weight_initialization_type, self._n_threads, self._save_models, self._verbose)
65 |
66 |
67 | # change the final prediction's label: test data to be 0, the rest training data will b e updated by the model's prediction
68 | data_label_fold = y.copy()
69 | data_label_fold[self._split_index[i][1]] = 0 # all test data to be 0
70 | data_label_fold[self._split_index[i][0]] = training_final_prediction ## assign the training prediction
71 | data_label_folds_ks[:, i, j - self._k_min] = data_label_fold
72 |
73 | print('Estimating clustering stability...\n')
74 | ## for the adjusted rand index, only consider the PT results
75 | adjusted_rand_index_results = np.zeros(self._k_max - self._k_min + 1)
76 | index_pt = np.where(y == 1)[0] # index for PTs
77 | for m in range(self._k_max - self._k_min + 1):
78 | result = data_label_folds_ks[:, :, m][index_pt]
79 | adjusted_rand_index_result = cv_cluster_stability(result, self._k_range_list[m])
80 | # saving each k result into the final adjusted_rand_index_results
81 | adjusted_rand_index_results[m] = adjusted_rand_index_result
82 |
83 | print('Computing the final consensus group membership...\n')
84 | final_assignment_ks = -np.ones((self._input.get_y_raw().shape[0], self._k_max - self._k_min + 1)).astype(int)
85 | for n in range(self._k_max - self._k_min + 1):
86 | result = data_label_folds_ks[:, :, n][index_pt]
87 | final_assignment_ks_pt = consensus_clustering(result, n + self._k_min)
88 | final_assignment_ks[index_pt, n] = final_assignment_ks_pt + 1
89 |
90 | print('Saving the final results...\n')
91 | # save_cluster_results(adjusted_rand_index_results, final_assignment_ks)
92 | columns = ['ari_' + str(i) + '_subtypes' for i in self._k_range_list]
93 | ari_df = pd.DataFrame(adjusted_rand_index_results[:, np.newaxis].transpose(), columns=columns)
94 | ari_df.to_csv(os.path.join(self._output_dir, 'adjusted_rand_index.tsv'), index=False, sep='\t',
95 | encoding='utf-8')
96 |
97 | # save the final assignment for consensus clustering across different folds
98 | participant_df = pd.read_csv(self._participant_tsv, sep='\t')
99 | columns = ['assignment_' + str(i) for i in self._k_range_list]
100 | cluster_df = pd.DataFrame(final_assignment_ks, columns=columns)
101 | all_df = pd.concat([participant_df, cluster_df], axis=1)
102 | all_df.to_csv(os.path.join(self._output_dir, 'clustering_assignment.tsv'), index=False,
103 | sep='\t', encoding='utf-8')
104 |
105 | class DualSVM_Subtype_transfer_learning(WorkFlow):
106 | """
107 | Instead of training from scratch, we initialize the polytope from the former C
108 | """
109 | def __init__(self, input, participant_tsv, split_index, cv_repetition, k_list, output_dir, opnmf_output, balanced=True,
110 | test_size=0.2, num_iteration=50, tol=1e-6, predefined_c=None,
111 | weight_initialization_type='DPP', n_threads=8, num_component=10, num_component_former=10, num_run=None):
112 |
113 | self._input = input
114 | self._participant_tsv = participant_tsv
115 | self._split_index = split_index
116 | self._cv_repetition = cv_repetition
117 | self._output_dir = output_dir
118 | self._opnmf_output = opnmf_output
119 | self._k_list = k_list
120 | self._balanced = balanced
121 | self._test_size = test_size
122 | self._num_iteration = num_iteration
123 | self._tol = tol
124 | self._predefined_c = predefined_c
125 | self._weight_initialization_type = weight_initialization_type
126 | self._n_threads = n_threads
127 | self._num_component = num_component
128 | self._num_component_former = num_component_former
129 | self._num_run = num_run
130 |
131 | def run(self):
132 |
133 | if os.path.exists(os.path.join(self._output_dir, 'clustering_run' + str(self._num_run), 'component_' + str(self._num_component), "adjusted_rand_index.tsv")):
134 | print("This number of component have been trained and converged: %d" % self._num_component)
135 | else:
136 | print("cross validate for num_component, running for %d components for feature selection" % self._num_component)
137 | x = self._input.get_x(self._num_component, self._opnmf_output)
138 |
139 | y = self._input.get_y_raw()
140 | data_label_folds_ks = np.zeros((y.shape[0], self._cv_repetition, len(self._k_list))).astype(int)
141 |
142 | for i in range(self._cv_repetition):
143 | for j in range(len(self._k_list)):
144 | print('Applying HRDRA for finding %d clusters. Repetition: %d / %d...\n' % (self._k_list[j], i+1, self._cv_repetition))
145 | training_final_prediction = hydra_solver_svm_tl(self._num_component, self._num_component_former, i, x[self._split_index[i][0]], y[self._split_index[i][0]], self._k_list[j], self._output_dir,
146 | self._num_iteration, self._tol, self._balanced, self._predefined_c,
147 | self._n_threads, self._num_run)
148 |
149 |
150 | # change the final prediction's label: test data to be 0, the rest training data will be updated by the model's prediction
151 | data_label_fold = y.copy()
152 | data_label_fold[self._split_index[i][1]] = 0 # all test data to be 0
153 | data_label_fold[self._split_index[i][0]] = training_final_prediction ## assign the training prediction
154 | data_label_folds_ks[:, i, j] = data_label_fold
155 |
156 | print('Finish the clustering procedure!\n')
157 |
158 | print('Estimating clustering stability!\n')
159 | ## for the adjusted rand index, only consider the PT results
160 | adjusted_rand_index_results = np.zeros(len(self._k_list))
161 | index_pt = np.where(y == 1)[0] # index for PTs
162 | for m in range(len(self._k_list)):
163 | result = data_label_folds_ks[:, :, m][index_pt] ## the result of each K during all runs of CV
164 | adjusted_rand_index_result = cv_cluster_stability(result, self._k_list[m])
165 |
166 | # saving each k result into the final adjusted_rand_index_results
167 | adjusted_rand_index_results[m] = adjusted_rand_index_result
168 | print('Done!\n')
169 |
170 | print('Computing the final consensus group membership!\n')
171 | final_assignment_ks = -np.ones((self._input.get_y_raw().shape[0], len(self._k_list))).astype(int)
172 | for n in range(len(self._k_list)):
173 | result = data_label_folds_ks[:, :, n][index_pt]
174 | final_assignment_ks_pt = consensus_clustering(result, n + self._k_list[0]) ## the final subtype assignment is performed with consensus clustering with KMeans
175 | final_assignment_ks[index_pt, n] = final_assignment_ks_pt + 1
176 | print('Done!\n')
177 |
178 | print('Saving the final results!\n')
179 | # save_cluster_results(adjusted_rand_index_results, final_assignment_ks)
180 | columns = ['ari_' + str(i) + '_subtypes' for i in self._k_list]
181 | ari_df = pd.DataFrame(adjusted_rand_index_results[:, np.newaxis].transpose(), columns=columns)
182 | ari_df.to_csv(os.path.join(self._output_dir, 'clustering_run' + str(self._num_run), 'component_' + str(self._num_component), 'adjusted_rand_index.tsv'), index=False, sep='\t',
183 | encoding='utf-8')
184 |
185 | # save the final assignment for consensus clustering across different folds
186 | df_feature = pd.read_csv(self._participant_tsv, sep='\t')
187 | columns = ['assignment_' + str(i) for i in self._k_list]
188 | participant_df = df_feature.iloc[:, :3]
189 | cluster_df = pd.DataFrame(final_assignment_ks, columns=columns)
190 | all_df = pd.concat([participant_df, cluster_df], axis=1)
191 | all_df.to_csv(os.path.join(self._output_dir, 'clustering_run' + str(self._num_run), 'component_' + str(self._num_component), 'clustering_assignment.tsv'), index=False,
192 | sep='\t', encoding='utf-8')
193 |
194 | print('Done!\n')
--------------------------------------------------------------------------------
/magiccluster/utils.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import numpy as np
3 | import scipy
4 | import os, pickle
5 | from sklearn.cluster import KMeans
6 | from sklearn.metrics import adjusted_rand_score
7 | from sklearn.model_selection import StratifiedKFold, StratifiedShuffleSplit, KFold, ShuffleSplit
8 | from joblib import dump
9 | import pandas as pd
10 | from multiprocessing.pool import ThreadPool
11 | from sklearn.svm import SVC
12 |
13 | __author__ = "Junhao Wen"
14 | __copyright__ = "Copyright 2023"
15 | __credits__ = ["Junhao Wen, Erdem Varol"]
16 | __license__ = "See LICENSE file"
17 | __version__ = "0.0.3"
18 | __maintainer__ = "Junhao Wen"
19 | __email__ = "junhao.wen89@gmail.com"
20 | __status__ = "Development"
21 |
22 | def elem_sym_poly(lambda_value, k):
23 | """
24 | given a vector of lambdas and a maximum size k, determine the value of
25 | the elementary symmetric polynomials:
26 | E(l+1,n+1) = sum_{J \subseteq 1..n,|J| = l} prod_{i \in J} lambda(i)
27 | :param lambda_value: the corresponding eigenvalues
28 | :param k: number of clusters
29 | :return:
30 | """
31 | N = lambda_value.shape[0]
32 | E = np.zeros((k + 1, N + 1))
33 | E[0, :] = 1
34 |
35 | for i in range(1, k+1):
36 | for j in range(1, N+1):
37 | E[i, j] = E[i, j - 1] + lambda_value[j-1] * E[i - 1, j - 1]
38 |
39 | return E
40 |
41 |
42 | def sample_k(lambda_value, k):
43 | """
44 | Pick k lambdas according to p(S) \propto prod(lambda \in S)
45 | :param lambda_value: the corresponding eigenvalues
46 | :param k: the number of clusters
47 | :return:
48 | """
49 |
50 | ## compute elementary symmetric polynomials
51 | E = elem_sym_poly(lambda_value, k)
52 |
53 | ## ietrate over the lambda value
54 | num = lambda_value.shape[0]
55 | remaining = k
56 | S = np.zeros(k)
57 | while remaining > 0:
58 | #compute marginal of num given that we choose remaining values from 0:num-1
59 | if num == remaining:
60 | marg = 1
61 | else:
62 | marg = lambda_value[num-1] * E[remaining-1, num-1] / E[remaining, num]
63 |
64 | # sample marginal
65 | if np.random.rand(1) < marg:
66 | S[remaining-1] = num
67 | remaining = remaining - 1
68 | num = num - 1
69 | return S
70 |
71 | def sample_dpp(evalue, evector, k=None):
72 | """
73 | sample a set Y from a dpp. evalue, evector are a decomposed kernel, and k is (optionally) the size of the set to return
74 | :param evalue: eigenvalue
75 | :param evector: normalized eigenvector
76 | :param k: number of cluster
77 | :return:
78 | """
79 | if k == None:
80 | # choose eigenvectors randomly
81 | evalue = np.divide(evalue, (1 + evalue))
82 | evector = np.where(np.random.random(evalue.shape[0]) <= evalue)[0]
83 | else:
84 | v = sample_k(evalue, k) ## v here is a 1d array with size: k
85 |
86 | k = v.shape[0]
87 | v = v.astype(int)
88 | v = [i - 1 for i in v.tolist()] ## due to the index difference between matlab & python, here, the element of v is for matlab
89 | V = evector[:, v]
90 |
91 | ## iterate
92 | y = np.zeros(k)
93 | for i in range(k, 0, -1):
94 | ## compute probabilities for each item
95 | P = np.sum(np.square(V), axis=1)
96 | P = P / np.sum(P)
97 |
98 | # choose a new item to include
99 | y[i-1] = np.where(np.random.rand(1) < np.cumsum(P))[0][0]
100 | y = y.astype(int)
101 |
102 | # choose a vector to eliminate
103 | j = np.where(V[y[i-1], :])[0][0]
104 | Vj = V[:, j]
105 | V = np.delete(V, j, 1)
106 |
107 | ## Update V
108 | if V.size == 0:
109 | pass
110 | else:
111 | V = np.subtract(V, np.multiply(Vj, (V[y[i-1], :] / Vj[y[i-1]])[:, np.newaxis]).transpose()) ## watch out the dimension here
112 |
113 | ## orthogonalize
114 | for m in range(i - 1):
115 | for n in range(m):
116 | V[:, m] = np.subtract(V[:, m], np.matmul(V[:, m].transpose(), V[:, n]) * V[:, n])
117 |
118 | V[:, m] = V[:, m] / np.linalg.norm(V[:, m])
119 |
120 | y = np.sort(y)
121 |
122 | return y
123 |
124 | def proportional_assign(l, d):
125 | """
126 | Proportional assignment based on margin
127 | :param l: int
128 | :param d: int
129 | :return:
130 | """
131 | np.seterr(divide='ignore', invalid='ignore')
132 | invL = np.divide(1, l)
133 | idx = np.isinf(invL)
134 | invL[idx] = d[idx]
135 |
136 | for i in range(l.shape[0]):
137 | pos = np.where(invL[i, :] > 0)[0]
138 | neg = np.where(invL[i, :] < 0)[0]
139 | if pos.size != 0:
140 | invL[i, neg] = 0
141 | else:
142 | invL[i, :] = np.divide(invL[i, :], np.amin(invL[i, :]))
143 | invL[i, invL[i, :] < 1] = 0
144 |
145 | S = np.multiply(invL, np.divide(1, np.sum(invL, axis=1))[:, np.newaxis])
146 |
147 | return S
148 |
149 | def random_init_dirichlet(k, num_pt):
150 | """
151 | a sample from a dirichlet distribution
152 | :param k: number of clusters
153 | :param num_pt: number of PT
154 | :return:
155 | """
156 | a = np.ones(k)
157 | s = np.random.dirichlet(a, num_pt)
158 |
159 | return s
160 |
161 | def hydra_init_weight(X, y, k, index_pt, index_cn, weight_initialization_type):
162 | """
163 | Function performs initialization for the polytope of mlni
164 | Args:
165 | X: the input features
166 | y: the label
167 | k: number of predefined clusters
168 | index_pt: list, the index for patient subjects
169 | index_cn: list, the index for control subjects
170 | weight_initialization_type: the type of chosen initialization method
171 | Returns:
172 |
173 | """
174 | if weight_initialization_type == "DPP": ##
175 | num_subject = y.shape[0]
176 | W = np.zeros((num_subject, X.shape[1]))
177 | for j in range(num_subject):
178 | ipt = np.random.randint(index_pt.shape[0])
179 | icn = np.random.randint(index_cn.shape[0])
180 | W[j, :] = X[index_pt[ipt], :] - X[index_cn[icn], :]
181 |
182 | KW = np.matmul(W, W.transpose())
183 | KW = np.divide(KW, np.sqrt(np.multiply(np.diag(KW)[:, np.newaxis], np.diag(KW)[:, np.newaxis].transpose())))
184 | evalue, evector = np.linalg.eig(KW)
185 | Widx = sample_dpp(np.real(evalue), np.real(evector), k)
186 | prob = np.zeros((len(index_pt), k)) # only consider the PTs
187 |
188 | for i in range(k):
189 | prob[:, i] = np.matmul(np.multiply(X[index_pt, :], np.divide(1, np.linalg.norm(X[index_pt, :], axis=1))[:, np.newaxis]), W[Widx[i], :].transpose())
190 |
191 | l = np.minimum(prob - 1, 0)
192 | d = prob - 1
193 | S = proportional_assign(l, d)
194 |
195 | elif weight_initialization_type == "random_hyperplane":
196 | print("TODO")
197 |
198 | elif weight_initialization_type == "random_assign":
199 | S = random_init_dirichlet(k, len(index_pt))
200 |
201 | elif weight_initialization_type == "k_means":
202 | print("TODO")
203 | else:
204 | raise Exception("Not implemented yet!")
205 |
206 | return S
207 |
208 | def hydra_solver_svm(num_repetition, X, y, k, output_dir, num_consensus, num_iteration, tol, balanced, predefined_c,
209 | weight_initialization_type, n_threads, save_models, verbose):
210 | """
211 | This is the main function of HYDRA, which find the convex polytope using a supervised classification fashion.
212 | Args:
213 | num_repetition: int, number of repetitions for CV
214 | X: input matrix for features
215 | y: input for group label
216 | k: number of clusters
217 | output_dir: the path for output
218 | num_consensus: int, number of runs for consensus clustering
219 | num_iteration: int, number of maximum iterations for running HYDRA
220 | tol: float, tolerance value for model convergence
221 | balanced: if sample imbalance should be considered during model optimization
222 | predefined_c: predefined c for SVM for clustering
223 | weight_initialization_type: the type of initialization of the weighted sample matrix
224 | n_threads: number of threads used
225 | save_models: if save all models during CV
226 | verbose: if output is verbose
227 |
228 | Returns:
229 |
230 | """
231 | censensus_assignment = np.zeros((y[y == 1].shape[0], num_consensus)) ## only consider the PTs
232 |
233 | index_pt = np.where(y == 1)[0] # index for PTs
234 | index_cn = np.where(y == -1)[0] # index for CNs
235 |
236 | for i in range(num_consensus):
237 | weight_sample = np.ones((y.shape[0], k)) / k
238 | ## depending on the weight initialization strategy, random hyperplanes were initialized with maximum diversity to constitute the convex polytope
239 | weight_sample_pt = hydra_init_weight(X, y, k, index_pt, index_cn, weight_initialization_type)
240 | weight_sample[index_pt] = weight_sample_pt ## only replace the sample weight of the PT group
241 | ## cluster assignment is based on this svm scores across different SVM/hyperplanes
242 | svm_scores = np.zeros((weight_sample.shape[0], weight_sample.shape[1]))
243 | update_weights_pool = ThreadPool(processes=n_threads)
244 |
245 | for j in range(num_iteration):
246 | for m in range(k):
247 | sample_weight = np.ascontiguousarray(weight_sample[:, m])
248 | if np.count_nonzero(sample_weight[index_pt]) == 0:
249 | if verbose == True:
250 | print(
251 | "Cluster dropped, meaning that all PT has been assigned to one single hyperplane in iteration: %d" % (
252 | j - 1))
253 | print(
254 | "Be careful, this could cause problem because of the ill-posed solution. Especially when k==2")
255 | else:
256 | results = update_weights_pool.apply_async(launch_svc,
257 | args=(X, y, predefined_c, sample_weight, balanced))
258 | weight_coef = results.get()[0]
259 | intesept = results.get()[1]
260 | ## Apply the data again the trained model to get the final SVM scores
261 | svm_scores[:, m] = (np.matmul(weight_coef, X.transpose()) + intesept).transpose().squeeze()
262 |
263 | cluster_index = np.argmax(svm_scores[index_pt], axis=1)
264 |
265 | ## decide the converge of the polytope based on the toleration
266 | weight_sample_hold = weight_sample.copy()
267 | # after each iteration, first set the weight of patient rows to be 0
268 | weight_sample[index_pt, :] = 0
269 | # then set the pt's weight to be 1 for the assigned hyperplane
270 | for n in range(len(index_pt)):
271 | weight_sample[index_pt[n], cluster_index[n]] = 1
272 |
273 | ## check the loss comparted to the tolorence for stopping criteria
274 | loss = np.linalg.norm(np.subtract(weight_sample, weight_sample_hold), ord='fro')
275 | if verbose == True:
276 | print("The loss is: %f" % loss)
277 | if loss < tol:
278 | if verbose == True:
279 | print(
280 | "The polytope has been converged for iteration %d in finding %d clusters in consensus running: %d" % (
281 | j, k, i))
282 | break
283 | update_weights_pool.close()
284 | update_weights_pool.join()
285 |
286 | ## update the cluster index for the consensus clustering
287 | censensus_assignment[:, i] = cluster_index + 1
288 |
289 | ## do censensus clustering
290 | final_predict = consensus_clustering(censensus_assignment.astype(int), k)
291 |
292 | ## after deciding the final convex polytope, we refit the training data once to save the best model
293 | weight_sample_final = np.zeros((y.shape[0], k))
294 | ## change the weight of PTs to be 1, CNs to be 1/k
295 |
296 | # then set the pt's weight to be 1 for the assigned hyperplane
297 | for n in range(len(index_pt)):
298 | weight_sample_final[index_pt[n], final_predict[n]] = 1
299 |
300 | weight_sample_final[index_cn] = 1 / k
301 | update_weights_pool_final = ThreadPool(processes=n_threads)
302 | ## create the final polytope by applying all weighted subjects
303 | for o in range(k):
304 | sample_weight = np.ascontiguousarray(weight_sample_final[:, o])
305 | results = update_weights_pool_final.apply_async(launch_svc, args=(X, y, predefined_c, sample_weight, balanced))
306 |
307 | if not os.path.exists(os.path.join(output_dir, str(k) + '_clusters', 'models')):
308 | os.makedirs(os.path.join(output_dir, str(k) + '_clusters', 'models'))
309 |
310 | ## save the final model for the k SVMs/hyperplanes
311 | if save_models == True:
312 | if not os.path.exists(os.path.join(output_dir, str(k) + '_clusters', 'models')):
313 | os.makedirs(os.path.join(output_dir, str(k) + '_clusters', 'models'))
314 |
315 | dump(results.get()[2], os.path.join(output_dir, str(k) + '_clusters', 'models',
316 | 'svm-' + str(o) + '_cv_' + str(num_repetition) + '.joblib'))
317 | else:
318 | ## only save the last repetition
319 | if not os.path.isfile(os.path.join(output_dir, str(k) + '_clusters', 'models',
320 | 'svm-' + str(o) + '_last_repetition.joblib')):
321 | dump(results.get()[2], os.path.join(output_dir, str(k) + '_clusters', 'models',
322 | 'svm-' + str(o) + '_last_repetition.joblib'))
323 | update_weights_pool_final.close()
324 | update_weights_pool_final.join()
325 |
326 | y[index_pt] = final_predict + 1
327 |
328 | if not os.path.exists(os.path.join(output_dir, str(k) + '_clusters', 'tsv')):
329 | os.makedirs(os.path.join(output_dir, str(k) + '_clusters', 'tsv'))
330 |
331 | ### save results also in tsv file for each repetition
332 | ## save the assigned weight for each subject across k-fold
333 | columns = ['hyperplane' + str(i) for i in range(k)]
334 | weight_sample_df = pd.DataFrame(weight_sample_final, columns=columns)
335 | weight_sample_df.to_csv(
336 | os.path.join(output_dir, str(k) + '_clusters', 'tsv', 'weight_sample_cv_' + str(num_repetition) + '.tsv'),
337 | index=False, sep='\t', encoding='utf-8')
338 |
339 | ## save the final_predict_all
340 | columns = ['y_hat']
341 | y_hat_df = pd.DataFrame(y, columns=columns)
342 | y_hat_df.to_csv(os.path.join(output_dir, str(k) + '_clusters', 'tsv', 'y_hat_cv_' + str(num_repetition) + '.tsv'),
343 | index=False, sep='\t', encoding='utf-8')
344 |
345 | ## save the pt index
346 | columns = ['pt_index']
347 | pt_df = pd.DataFrame(index_pt, columns=columns)
348 | pt_df.to_csv(os.path.join(output_dir, str(k) + '_clusters', 'tsv', 'pt_index_cv_' + str(num_repetition) + '.tsv'),
349 | index=False, sep='\t', encoding='utf-8')
350 |
351 | return y
352 |
353 | def GLMcorrection(X_train, Y_train, covar_train, X_test, covar_test):
354 | """
355 | Eliminate the confound of covariate, such as age and sex, from the disease-based changes.
356 | Ref: "Age Correction in Dementia Matching to a Healthy Brain"
357 | :param X_train: array, training features
358 | :param Y_train: array, training labels
359 | :param covar_train: array, ttraining covariate data
360 | :param X_test: array, test labels
361 | :param covar_test: array, ttest covariate data
362 | :return: corrected training & test feature data
363 | """
364 | Yc = X_train[Y_train == -1]
365 | Xc = covar_train[Y_train == -1]
366 | Xc = np.concatenate((Xc, np.ones((Xc.shape[0], 1))), axis=1)
367 | beta = np.matmul(np.matmul(Yc.transpose(), Xc), np.linalg.inv(np.matmul(Xc.transpose(), Xc)))
368 | num_col = beta.shape[1]
369 | X_train_cor = (X_train.transpose() - np.matmul(beta[:, : num_col - 1], covar_train.transpose())).transpose()
370 | X_test_cor = (X_test.transpose() - np.matmul(beta[:, : num_col - 1], covar_test.transpose())).transpose()
371 |
372 | return X_train_cor, X_test_cor
373 |
374 | def launch_svc(X, y, predefined_c, sample_weight, balanced):
375 | """
376 | Lauch svc classifier of sklearn
377 | Args:
378 | X: input matrix for features
379 | y: input matrix for label
380 | predefined_c: predefined C
381 | sample_weight: the weighted sample matrix
382 | balanced:
383 |
384 | Returns:
385 |
386 | """
387 | if not balanced:
388 | model = SVC(kernel='linear', C=predefined_c)
389 | else:
390 | model = SVC(kernel='linear', C=predefined_c, class_weight='balanced')
391 |
392 | ## fit the different SVM/hyperplanes
393 | model.fit(X, y, sample_weight=sample_weight)
394 |
395 | weight_coef = model.coef_
396 | intesept = model.intercept_
397 |
398 | return weight_coef, intesept, model
399 |
400 | def check_symmetric(a, rtol=1e-05, atol=1e-08):
401 | """
402 | Check if the numpy array is symmetric or not
403 | Args:
404 | a:
405 | rtol:
406 | atol:
407 |
408 | Returns:
409 |
410 | """
411 | result = np.allclose(a, a.T, rtol=rtol, atol=atol)
412 | return result
413 |
414 | def make_cv_partition(diagnosis, cv_strategy, output_dir, cv_repetition, seed=None):
415 | """
416 | Randomly generate the data split index for different CV strategy.
417 |
418 | :param diagnosis: the list for labels
419 | :param cv_repetition: the number of repetitions or folds
420 | :param output_dir: the output folder path
421 | :param cv_repetition: the number of repetitions for CV
422 | :param seed: random seed for sklearn split generator. Default is None
423 | :return:
424 | """
425 | unique = list(set(diagnosis))
426 | y = np.array(diagnosis)
427 | if len(unique) == 2: ### CV for classification and clustering
428 | if cv_strategy == 'k_fold':
429 | splits_indices_pickle = os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-fold.pkl')
430 | ## try to see if the shuffle has been done
431 | if os.path.isfile(splits_indices_pickle):
432 | splits_indices = pickle.load(open(splits_indices_pickle, 'rb'))
433 | else:
434 | splits = StratifiedKFold(n_splits=cv_repetition, random_state=seed)
435 | splits_indices = list(splits.split(np.zeros(len(y)), y))
436 | elif cv_strategy == 'hold_out':
437 | splits_indices_pickle = os.path.join(output_dir, 'data_split_stratified_' + str(cv_repetition) + '-holdout.pkl')
438 | ## try to see if the shuffle has been done
439 | if os.path.isfile(splits_indices_pickle):
440 | splits_indices = pickle.load(open(splits_indices_pickle, 'rb'))
441 | else:
442 | splits = StratifiedShuffleSplit(n_splits=cv_repetition, test_size=0.2, random_state=seed)
443 | splits_indices = list(splits.split(np.zeros(len(y)), y))
444 | else:
445 | raise Exception("this cross validation strategy has not been implemented!")
446 | elif len(unique) == 1:
447 | raise Exception("Diagnosis cannot be the same for all participants...")
448 | else: ### CV for regression, no need to be stratified
449 | if cv_strategy == 'k_fold':
450 | splits_indices_pickle = os.path.join(output_dir, 'data_split_' + str(cv_repetition) + '-fold.pkl')
451 |
452 | ## try to see if the shuffle has been done
453 | if os.path.isfile(splits_indices_pickle):
454 | splits_indices = pickle.load(open(splits_indices_pickle, 'rb'))
455 | else:
456 | splits = KFold(n_splits=cv_repetition, random_state=seed)
457 | splits_indices = list(splits.split(np.zeros(len(y)), y))
458 | elif cv_strategy == 'hold_out':
459 | splits_indices_pickle = os.path.join(output_dir, 'data_split_' + str(cv_repetition) + '-holdout.pkl')
460 | ## try to see if the shuffle has been done
461 | if os.path.isfile(splits_indices_pickle):
462 | splits_indices = pickle.load(open(splits_indices_pickle, 'rb'))
463 | else:
464 | splits = ShuffleSplit(n_splits=cv_repetition, test_size=0.2, random_state=seed)
465 | splits_indices = list(splits.split(np.zeros(len(y)), y))
466 | else:
467 | raise Exception("this cross validation strategy has not been implemented!")
468 |
469 | with open(splits_indices_pickle, 'wb') as s:
470 | pickle.dump(splits_indices, s)
471 |
472 | return splits_indices, splits_indices_pickle
473 |
474 | def consensus_clustering(clustering_results, k):
475 | """
476 | This function performs consensus clustering on a co-occurence matrix
477 | :param clustering_results: an array containing all the clustering results across different iterations, in order to
478 | perform
479 | :param k:
480 | :return:
481 | """
482 |
483 | num_pt = clustering_results.shape[0]
484 | cooccurence_matrix = np.zeros((num_pt, num_pt))
485 |
486 | for i in range(num_pt - 1):
487 | for j in range(i + 1, num_pt):
488 | cooccurence_matrix[i, j] = sum(clustering_results[i, :] == clustering_results[j, :])
489 |
490 | cooccurence_matrix = np.add(cooccurence_matrix, cooccurence_matrix.transpose())
491 | ## here is to compute the Laplacian matrix
492 | Laplacian = np.subtract(np.diag(np.sum(cooccurence_matrix, axis=1)), cooccurence_matrix)
493 |
494 | Laplacian_norm = np.subtract(np.eye(num_pt), np.matmul(np.matmul(np.diag(1 / np.sqrt(np.sum(cooccurence_matrix, axis=1))), cooccurence_matrix), np.diag(1 / np.sqrt(np.sum(cooccurence_matrix, axis=1)))))
495 | ## replace the nan with 0
496 | Laplacian_norm = np.nan_to_num(Laplacian_norm)
497 |
498 | ## check if the Laplacian norm is symmetric or not, because matlab eig function will automatically check this, but not in numpy or scipy
499 | if check_symmetric(Laplacian_norm):
500 | ## extract the eigen value and vector
501 | ## matlab eig equivalence is eigh, not eig from numpy or scipy, see this post: https://stackoverflow.com/questions/8765310/scipy-linalg-eig-return-complex-eigenvalues-for-covariance-matrix
502 | ## Note, the eigenvector is not unique, thus the matlab and python eigenvector may be different, but this will not affect the results.
503 | evalue, evector = scipy.linalg.eigh(Laplacian_norm)
504 | else:
505 | # evalue, evector = np.linalg.eig(Laplacian_norm)
506 | raise Exception("The Laplacian matrix should be symmetric here...")
507 |
508 | ## check if the eigen vector is complex
509 | if np.any(np.iscomplex(evector)):
510 | evalue, evector = scipy.linalg.eigh(Laplacian)
511 |
512 | ## create the kmean algorithm with sklearn
513 | kmeans = KMeans(n_clusters=k, n_init=20).fit(evector.real[:, 0: k])
514 | final_predict = kmeans.labels_
515 |
516 | return final_predict
517 |
518 | def cv_cluster_stability(result, k):
519 | """
520 | To compute the adjusted rand index across different pair of 2 folds cross CV
521 | :param result:
522 | :return:
523 | """
524 |
525 | num_pair = 0
526 | aris = []
527 | if k == 1:
528 | adjusted_rand_index = 0 ## note, here, we manually set it to be 0, because it does not make sense when k==1. TODO, need to clarify if there is really heterogeneity in the data, i.e., k == 1 or k>1
529 | else:
530 | for i in range(result.shape[1] - 1):
531 | for j in range(i+1, result.shape[1]):
532 | num_pair += 1
533 | non_zero_index = np.all(result[:, [i, j]], axis=1)
534 | pair_result = result[:, [i, j]][non_zero_index]
535 | ari = adjusted_rand_score(pair_result[:, 0], pair_result[:, 1])
536 | aris.append(ari)
537 |
538 | adjusted_rand_index = np.mean(np.asarray(aris))
539 |
540 | return adjusted_rand_index
541 |
542 | def hydra_solver_svm_tl(num_component, num_component_former, num_repetition, X, y, k, output_dir, num_iteration, tol, balanced, predefined_c, n_threads, num_run):
543 | """
544 | This is the main function of HYDRA, which find the convex polytope using a supervised classification fashion.
545 | :param num_repetition: the number of iteration of CV currently. This is helpful to reconstruct the model and also moniter the processing
546 | :param X: corrected training data feature
547 | :param y: traing data label
548 | :param k: hyperparameter for desired number of clusters in patients
549 | :param options: commandline parameters
550 | :return: the optimal model
551 | """
552 | index_pt = np.where(y == 1)[0] # index for PTs
553 | index_cn = np.where(y == -1)[0] # index for CNs
554 |
555 | ### initialize the final weight for the polytope from the former C
556 | weight_file = os.path.join(output_dir, 'clustering_run' + str(num_run-1), 'component_' + str(num_component_former), str(k) + '_clusters', 'tsv', 'weight_sample_cv_' + str(num_repetition) + '.tsv')
557 | weight_sample = pd.read_csv(weight_file, sep='\t').to_numpy()
558 |
559 | ## cluster assignment is based on this svm scores across different SVM/hyperplanes
560 | svm_scores = np.zeros((weight_sample.shape[0], weight_sample.shape[1]))
561 | update_weights_pool = ThreadPool(n_threads)
562 | for j in range(num_iteration):
563 | for m in range(k):
564 | sample_weight = np.ascontiguousarray(weight_sample[:, m])
565 |
566 | if np.count_nonzero(sample_weight[index_pt]) == 0:
567 | print("Cluster dropped, meaning that all PT has been assigned to one single hyperplane in iteration: %d" % (j-1))
568 | svm_scores[:, m] = np.asarray([np.NINF] * (y.shape[0]))
569 | else:
570 |
571 | results = update_weights_pool.apply_async(launch_svc, args=(X, y, predefined_c, sample_weight, balanced))
572 | weight_coef = results.get()[0]
573 | intesept = results.get()[1]
574 | ## Apply the data again the trained model to get the final SVM scores
575 | svm_scores[:, m] = (np.matmul(weight_coef, X.transpose()) + intesept).transpose().squeeze()
576 |
577 |
578 | final_predict = np.argmax(svm_scores[index_pt], axis=1)
579 |
580 | ## decide the converge of the polytope based on the toleration
581 | weight_sample_hold = weight_sample.copy()
582 | # after each iteration, first set the weight of patient rows to be 0
583 | weight_sample[index_pt, :] = 0
584 | # then set the pt's weight to be 1 for the assigned hyperplane
585 | for n in range(len(index_pt)):
586 | weight_sample[index_pt[n], final_predict[n]] = 1
587 |
588 | ## check the loss comparted to the tolorence for stopping criteria
589 | loss = np.linalg.norm(np.subtract(weight_sample, weight_sample_hold), ord='fro')
590 | print("The loss is: %f" % loss)
591 | if loss < tol:
592 | print("The polytope has been converged for iteration %d in finding %d clusters" % (j, k))
593 | break
594 | update_weights_pool.close()
595 | update_weights_pool.join()
596 |
597 | ## after deciding the final convex polytope, we refit the training data once to save the best model
598 | weight_sample_final = np.zeros((y.shape[0], k))
599 | ## change the weight of PTs to be 1, CNs to be 1/k
600 |
601 | # then set the pt's weight to be 1 for the assigned hyperplane
602 | for n in range(len(index_pt)):
603 | weight_sample_final[index_pt[n], final_predict[n]] = 1
604 |
605 | weight_sample_final[index_cn] = 1 / k
606 | update_weights_pool_final = ThreadPool(n_threads)
607 |
608 | for o in range(k):
609 | sample_weight = np.ascontiguousarray(weight_sample_final[:, o])
610 | if np.count_nonzero(sample_weight[index_pt]) == 0:
611 | print("Cluster dropped, meaning that the %d th hyperplane is useless!" % (o))
612 | else:
613 | results = update_weights_pool_final.apply_async(launch_svc, args=(X, y, predefined_c, sample_weight, balanced))
614 |
615 | ## save the final model for the k SVMs/hyperplanes
616 | if not os.path.exists(
617 | os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component),
618 | str(k) + '_clusters', 'models')):
619 | os.makedirs(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component),
620 | str(k) + '_clusters', 'models'))
621 |
622 | dump(results.get()[2],
623 | os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component),
624 | str(k) + '_clusters', 'models',
625 | 'svm-' + str(o) + '_last_repetition.joblib'))
626 |
627 | update_weights_pool_final.close()
628 | update_weights_pool_final.join()
629 |
630 | y[index_pt] = final_predict + 1
631 |
632 | if not os.path.exists(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component), str(k) + '_clusters', 'tsv')):
633 | os.makedirs(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component), str(k) + '_clusters', 'tsv'))
634 |
635 | ## save the assigned weight for each subject across k-fold
636 | columns = ['hyperplane' + str(i) for i in range(k)]
637 | weight_sample_df = pd.DataFrame(weight_sample_final, columns=columns)
638 | weight_sample_df.to_csv(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component), str(k) + '_clusters', 'tsv', 'weight_sample_cv_' + str(num_repetition) + '.tsv'), index=False, sep='\t', encoding='utf-8')
639 |
640 | ## save the final_predict_all
641 | columns = ['y_hat']
642 | y_hat_df = pd.DataFrame(y, columns=columns)
643 | y_hat_df.to_csv(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component), str(k) + '_clusters', 'tsv', 'y_hat_cv_' + str(num_repetition) + '.tsv'), index=False, sep='\t', encoding='utf-8')
644 |
645 | ## save the pt index
646 | columns = ['pt_index']
647 | pt_df = pd.DataFrame(index_pt, columns=columns)
648 | pt_df.to_csv(os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(num_component), str(k) + '_clusters', 'tsv', 'pt_index_cv_' + str(num_repetition) + '.tsv'), index=False, sep='\t', encoding='utf-8')
649 |
650 | return y
651 |
652 | def cluster_stability_across_resolution(c, c_former, output_dir, k_continuing, num_run, stop_tol=0.98):
653 | """
654 | To evaluate the stability of clustering across two different C for stopping criterion.
655 | Args:
656 | c:
657 | c_former:
658 | output_dir:
659 | k_continuing:
660 | num_run:
661 | stop_tol:
662 | max_num_iter:
663 |
664 | Returns:
665 |
666 | """
667 | ## read the output of current C and former Cs
668 | cluster_ass1 = os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(c), 'clustering_assignment.tsv')
669 | ass1_df = pd.read_csv(cluster_ass1, sep='\t')
670 | ass1_df = ass1_df.loc[ass1_df['diagnosis'] == 1]
671 |
672 | cluster_ass2 = os.path.join(output_dir, 'clustering_run' + str(num_run-1), 'component_' + str(c_former), 'clustering_assignment.tsv')
673 | ass2_df = pd.read_csv(cluster_ass2, sep='\t')
674 | ass2_df = ass2_df.loc[ass2_df['diagnosis'] == 1]
675 |
676 | df_final = pd.DataFrame(columns=['C', 'K', 'num_run'])
677 |
678 | k_continuing_update = []
679 | k_converged = []
680 | for i in k_continuing:
681 | ari = adjusted_rand_score(ass1_df['assignment_' + str(i)], ass2_df['assignment_' + str(i)])
682 | print("For k == %d, run %d got ARI == %f compared to former run" % (i, num_run, ari))
683 | if ari < stop_tol and num_run:
684 | k_continuing_update.append(i)
685 | else:
686 | print("Model has been converged or stop at the max iteration: C == %d, K == %d and run == %d" % (c, i, num_run))
687 | k_converged.append(i)
688 | df_row = pd.DataFrame(columns=['C', 'K', 'num_run'])
689 | df_row.loc[len(['C', 'K', 'num_run'])] = [c, i, num_run]
690 | df_final = df_final.append(df_row)
691 |
692 | if len(k_converged) != 0:
693 | df_final.to_csv(os.path.join(output_dir, 'results_convergence_run' + str(num_run) + '.tsv'), index=False, sep='\t', encoding='utf-8')
694 |
695 | return k_continuing_update, k_converged
696 |
697 | def summary_clustering_result_multiscale(output_dir, k_min, k_max):
698 | """
699 | This is a function to summarize the clustering results
700 | :param num_components_min:
701 | :param num_components_max:
702 | :param num_components_step:
703 | :param output_dir:
704 | :return:
705 | """
706 | clu_col_list = ['assignment_' + str(e) for e in range(k_min, k_max)]
707 | df_clusters = pd.DataFrame(columns=clu_col_list)
708 |
709 | ## read the convergence tsv
710 | convergence_tsvs = [f for f in glob.glob(output_dir + "/results_convergence_*.tsv", recursive=True)]
711 |
712 | for tsv in convergence_tsvs:
713 | df_convergence = pd.read_csv(tsv, sep='\t')
714 |
715 | ## sorf by K
716 | df_convergence = df_convergence.sort_values(by=['K'])
717 |
718 | for i in range(df_convergence.shape[0]):
719 | k = df_convergence['K'].tolist()[i]
720 | num_run = df_convergence['num_run'].tolist()[i]
721 | C = df_convergence['C'].tolist()[i]
722 | cluster_file = os.path.join(output_dir, 'clustering_run' + str(num_run), 'component_' + str(C), 'clustering_assignment.tsv')
723 |
724 | df_cluster = pd.read_csv(cluster_file, sep='\t')
725 | if i == 0:
726 | df_header = df_cluster.iloc[:, 0:3]
727 | assign = df_cluster['assignment_' + str(k)]
728 | df_clusters['assignment_' + str(k)] = assign
729 |
730 | ## concatenqte the header
731 | df_assignment = pd.concat((df_header, df_clusters), axis=1)
732 |
733 | ## save the result
734 | df_assignment.to_csv(os.path.join(output_dir, 'results_cluster_assignment_final.tsv'), index=False, sep='\t', encoding='utf-8')
735 |
736 | def shift_list(c_list, index):
737 | """
738 | This is a function to reorder a list to have all posibility by putting each element in the first place
739 | Args:
740 | c_list: list to shift
741 | index: the index of which element to shift
742 |
743 | Returns:
744 |
745 | """
746 | new_list = c_list[index:] + c_list[:index]
747 |
748 | return new_list
749 |
750 | def consensus_clustering_across_c(output_dir, c_list, k_min, k_max):
751 | """
752 | This is for consensus learning at the end across different Cs
753 | Args:
754 | output_dir:
755 | c_list:
756 |
757 | Returns:
758 |
759 | """
760 | k_list = list(range(k_min, k_max+1))
761 | for k in k_list:
762 | for i in c_list:
763 | clu_col_list = ['c_' + str(i) + '_assignment_' + str(e) for e in k_list]
764 | df_clusters = pd.DataFrame(columns=clu_col_list)
765 |
766 | tsv = os.path.join(output_dir, 'initialization_c_' + str(i), 'results_cluster_assignment_final.tsv')
767 | df = pd.read_csv(tsv, sep='\t')
768 |
769 | if i == c_list[0]:
770 | df_header = df.iloc[:, 0:3]
771 | df_clusters['c_' + str(i) + '_assignment_' + str(k)] = df['assignment_' + str(k)]
772 | if i == c_list[0]:
773 | df_final = df_clusters
774 | else:
775 | df_final = pd.concat([df_final, df_clusters], axis=1)
776 |
777 | ## concatenate the header and the results
778 | df_final = pd.concat([df_header, df_final], axis=1)
779 | df_final_pt = df_final.loc[df_final['diagnosis'] == 1]
780 | df_final_cn = df_final.loc[df_final['diagnosis'] == -1]
781 | num_cn = df_final_cn.shape[0]
782 |
783 | ## create the final dataframe to store the final assignment
784 | col_list = ['assignment_' + str(e) for e in k_list]
785 | df_final_assign = pd.DataFrame(columns=col_list)
786 |
787 | ## read the final clustering assignment for each C
788 | for m in k_list:
789 | columns_names = ['c_' + str(e) + '_assignment_' + str(m) for e in c_list]
790 | assignment_pt = df_final_pt[columns_names]
791 | final_predict_pt = consensus_clustering(assignment_pt.to_numpy(), m)
792 | final_predict_cn = -2 * np.ones(num_cn)
793 | final_predict = np.concatenate((final_predict_cn, final_predict_pt)).astype(int)
794 | df_final_assign['assignment_' + str(m)] = final_predict + 1
795 |
796 | df_final_assign = pd.concat([df_header, df_final_assign], axis=1)
797 | ## save the final results into tsv file.
798 | df_final_assign.to_csv(os.path.join(output_dir, 'results_cluster_assignment_final.tsv'), index=False, sep='\t',
799 | encoding='utf-8')
--------------------------------------------------------------------------------
 4 |
5 |
4 |
5 |  4 |
5 |
4 |
5 |  3 |
3 |