├── LICENSE
├── .gitignore
├── README.md
├── group_norm.py
├── model.py
└── Example_on_BRATS2018.ipynb
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2019 Suyog Jadhav
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | *.egg-info/
24 | .installed.cfg
25 | *.egg
26 | MANIFEST
27 |
28 | # PyInstaller
29 | # Usually these files are written by a python script from a template
30 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
31 | *.manifest
32 | *.spec
33 |
34 | # Installer logs
35 | pip-log.txt
36 | pip-delete-this-directory.txt
37 |
38 | # Unit test / coverage reports
39 | htmlcov/
40 | .tox/
41 | .coverage
42 | .coverage.*
43 | .cache
44 | nosetests.xml
45 | coverage.xml
46 | *.cover
47 | .hypothesis/
48 | .pytest_cache/
49 |
50 | # Translations
51 | *.mo
52 | *.pot
53 |
54 | # Django stuff:
55 | *.log
56 | local_settings.py
57 | db.sqlite3
58 |
59 | # Flask stuff:
60 | instance/
61 | .webassets-cache
62 |
63 | # Scrapy stuff:
64 | .scrapy
65 |
66 | # Sphinx documentation
67 | docs/_build/
68 |
69 | # PyBuilder
70 | target/
71 |
72 | # Jupyter Notebook
73 | .ipynb_checkpoints
74 |
75 | # pyenv
76 | .python-version
77 |
78 | # celery beat schedule file
79 | celerybeat-schedule
80 |
81 | # SageMath parsed files
82 | *.sage.py
83 |
84 | # Environments
85 | .env
86 | .venv
87 | env/
88 | venv/
89 | ENV/
90 | env.bak/
91 | venv.bak/
92 |
93 | # Spyder project settings
94 | .spyderproject
95 | .spyproject
96 |
97 | # Rope project settings
98 | .ropeproject
99 |
100 | # mkdocs documentation
101 | /site
102 |
103 | # mypy
104 | .mypy_cache/
105 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization
2 |
3 | [](https://paperswithcode.com/sota/brain-tumor-segmentation-on-brats-2018?p=3d-mri-brain-tumor-segmentation-using)
4 | 
5 |
6 | 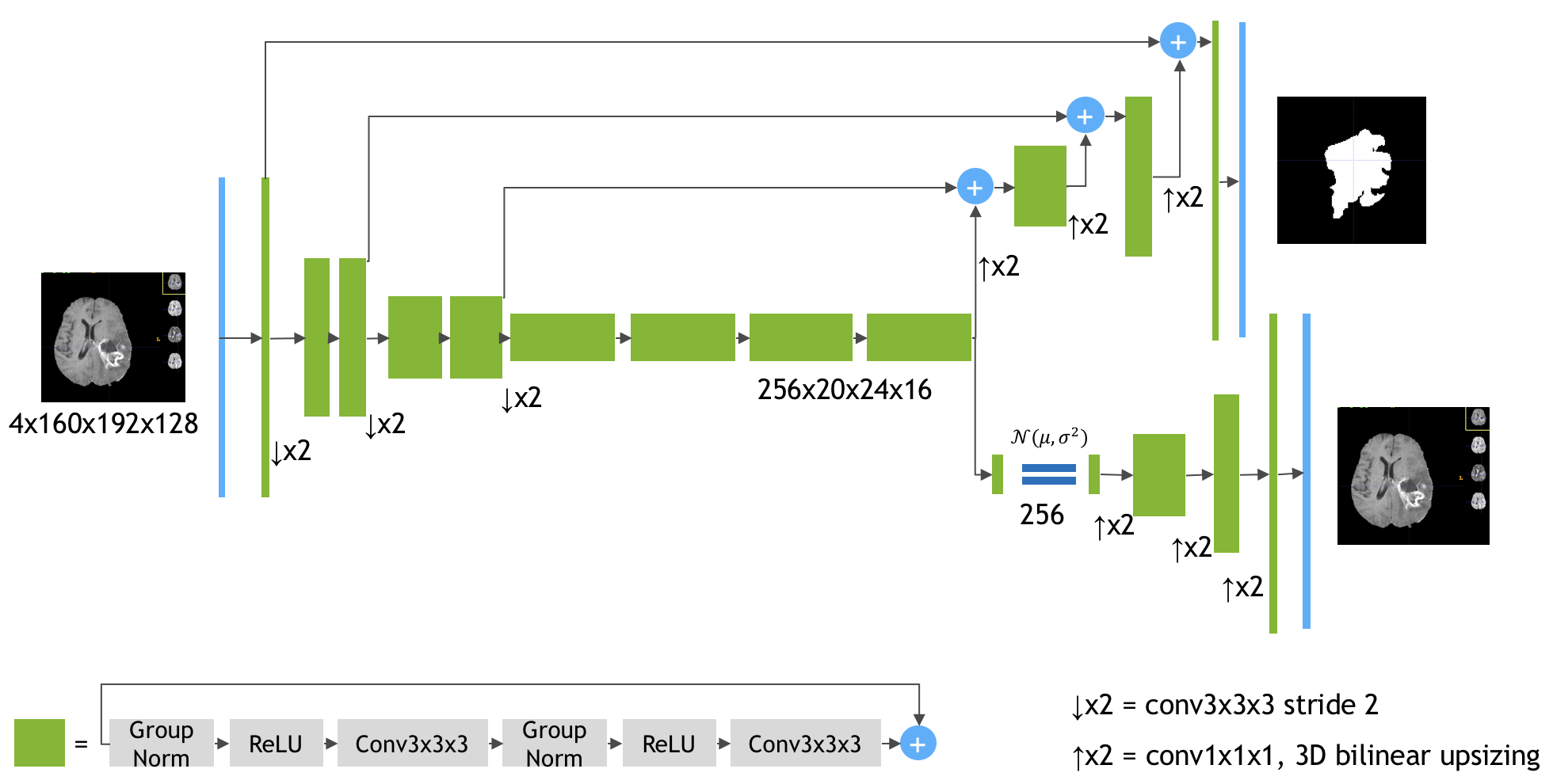
7 | The Model Architecture
Source: https://arxiv.org/pdf/1810.11654.pdf
8 |
9 |
10 | Keras implementation of the paper 3D MRI brain tumor segmentation using autoencoder regularization by Myronenko A. (https://arxiv.org/abs/1810.11654). The author (team name: NVDLMED) ranked #1 on the BraTS 2018 leaderboard using the model described in the paper.
11 |
12 | This repository contains the model complete with the loss function, all implemented end-to-end in Keras. The usage is described in the next section.
13 |
14 | # Usage
15 | 1. Download the file [`model.py`](model.py) and keep in the same folder as your project notebook/script.
16 |
17 | 2. In your python script, import `build_model` function from `model.py`.
18 |
19 | ```python
20 | from model import build_model
21 | ```
22 |
23 | It will automatically download an additional script needed for the implementation, namely [`group_norm.py`](https://github.com/titu1994/Keras-Group-Normalization/blob/master/group_norm.py), which contains keras implementation for the group normalization layer.
24 |
25 | 3. Note that the input MRI scans you are going to feed need to have 4 dimensions, with channels-first format. i.e., the shape should look like (c, H, W, D), where:
26 | - `c`, the no.of channels are divisible by 4.
27 | - `H`, `W`, `D`, which are height, width and depth, respectively, are _all_ divisible by 24, i.e., 16.
28 | This is to get correct output shape according to the model.
29 |
30 | 4. Now to create the model, simply run:
31 |
32 | ```python
33 | model = build_model(input_shape, output_channels)
34 | ```
35 |
36 | where, `input_shape` is a 4-tuple (channels, Height, Width, Depth) and `output_channels` is the no. of channels in the output of the model.
37 | The output of the model will be the segmentation map generated by the model with the shape (output_channels, Height, Width, Depth), where Height, Width and Depth will be same as that of the input.
38 |
39 | # Example on BraTS2018 dataset
40 |
41 | Go through the [Example_on_BRATS2018](Example_on_BRATS2018.ipynb) notebook to see an example where this model is used on the BraTS2018 dataset.
42 |
43 | You can also test-run the example on Google Colaboratory by clicking the following button.
44 |
45 | [](https://colab.research.google.com/github/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/blob/master/Example_on_BRATS2018.ipynb)
46 |
47 | However, note that you will need to have access to the BraTS2018 dataset before running the example on Google Colaboratory. If you already have access to the dataset, You can simply upload the dataset to Google Drive and input the dataset path in the example notebook.
48 |
49 | # Issues
50 |
51 | If you encounter any issue or have a feedback, please don't hesitate to [raise an issue](https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/issues/new).
52 |
53 | # Updates
54 | - Thanks to [@Crispy13](https://github.com/Crispy13), issues #29 and #24 are now fixed. VAE branch output was earlier not being included in the model's output. The current format model gives out two outputs: the segmentation map and the VAE output. The VAE branch weights were not being trained for some reason. The issue should be fixed now. Dice score calculation is slightly modified to work for any batch size. SpatialDropout3D is now used instead of Dropout, as specified in the paper.
55 | - Added an [example notebook](Example_on_BRATS2018.ipynb) showing how to run the model on the BraTS2018 dataset.
56 | - Added a minus term before `loss_dice` in the loss function. From discussion in #7 with [@woodywff](https://github.com/woodywff) and [@doc78](https://github.com/doc78).
57 | - Thanks to [@doc78](https://github.com/doc78) , the NaN loss problem has been permanently fixed.
58 | - The NaN loss problem has now been fixed (clipping the activations for now).
59 | - Added an argument in the `build_model` function to allow for different no. of channels in the output.
60 |
--------------------------------------------------------------------------------
/group_norm.py:
--------------------------------------------------------------------------------
1 | from keras.engine import Layer, InputSpec
2 | from keras import initializers
3 | from keras import regularizers
4 | from keras import constraints
5 | from keras import backend as K
6 |
7 | from keras.utils.generic_utils import get_custom_objects
8 |
9 |
10 | class GroupNormalization(Layer):
11 | """Group normalization layer
12 |
13 | Group Normalization divides the channels into groups and computes within each group
14 | the mean and variance for normalization. GN's computation is independent of batch sizes,
15 | and its accuracy is stable in a wide range of batch sizes
16 |
17 | # Arguments
18 | groups: Integer, the number of groups for Group Normalization.
19 | axis: Integer, the axis that should be normalized
20 | (typically the features axis).
21 | For instance, after a `Conv2D` layer with
22 | `data_format="channels_first"`,
23 | set `axis=1` in `BatchNormalization`.
24 | epsilon: Small float added to variance to avoid dividing by zero.
25 | center: If True, add offset of `beta` to normalized tensor.
26 | If False, `beta` is ignored.
27 | scale: If True, multiply by `gamma`.
28 | If False, `gamma` is not used.
29 | When the next layer is linear (also e.g. `nn.relu`),
30 | this can be disabled since the scaling
31 | will be done by the next layer.
32 | beta_initializer: Initializer for the beta weight.
33 | gamma_initializer: Initializer for the gamma weight.
34 | beta_regularizer: Optional regularizer for the beta weight.
35 | gamma_regularizer: Optional regularizer for the gamma weight.
36 | beta_constraint: Optional constraint for the beta weight.
37 | gamma_constraint: Optional constraint for the gamma weight.
38 |
39 | # Input shape
40 | Arbitrary. Use the keyword argument `input_shape`
41 | (tuple of integers, does not include the samples axis)
42 | when using this layer as the first layer in a model.
43 |
44 | # Output shape
45 | Same shape as input.
46 |
47 | # References
48 | - [Group Normalization](https://arxiv.org/abs/1803.08494)
49 | """
50 |
51 | def __init__(self,
52 | groups=32,
53 | axis=-1,
54 | epsilon=1e-5,
55 | center=True,
56 | scale=True,
57 | beta_initializer='zeros',
58 | gamma_initializer='ones',
59 | beta_regularizer=None,
60 | gamma_regularizer=None,
61 | beta_constraint=None,
62 | gamma_constraint=None,
63 | **kwargs):
64 | super(GroupNormalization, self).__init__(**kwargs)
65 | self.supports_masking = True
66 | self.groups = groups
67 | self.axis = axis
68 | self.epsilon = epsilon
69 | self.center = center
70 | self.scale = scale
71 | self.beta_initializer = initializers.get(beta_initializer)
72 | self.gamma_initializer = initializers.get(gamma_initializer)
73 | self.beta_regularizer = regularizers.get(beta_regularizer)
74 | self.gamma_regularizer = regularizers.get(gamma_regularizer)
75 | self.beta_constraint = constraints.get(beta_constraint)
76 | self.gamma_constraint = constraints.get(gamma_constraint)
77 |
78 | def build(self, input_shape):
79 | dim = input_shape[self.axis]
80 |
81 | if dim is None:
82 | raise ValueError('Axis ' + str(self.axis) + ' of '
83 | 'input tensor should have a defined dimension '
84 | 'but the layer received an input with shape ' +
85 | str(input_shape) + '.')
86 |
87 | if dim < self.groups:
88 | raise ValueError('Number of groups (' + str(self.groups) + ') cannot be '
89 | 'more than the number of channels (' +
90 | str(dim) + ').')
91 |
92 | if dim % self.groups != 0:
93 | raise ValueError('Number of groups (' + str(self.groups) + ') must be a '
94 | 'multiple of the number of channels (' +
95 | str(dim) + ').')
96 |
97 | self.input_spec = InputSpec(ndim=len(input_shape),
98 | axes={self.axis: dim})
99 | shape = (dim,)
100 |
101 | if self.scale:
102 | self.gamma = self.add_weight(shape=shape,
103 | name='gamma',
104 | initializer=self.gamma_initializer,
105 | regularizer=self.gamma_regularizer,
106 | constraint=self.gamma_constraint)
107 | else:
108 | self.gamma = None
109 | if self.center:
110 | self.beta = self.add_weight(shape=shape,
111 | name='beta',
112 | initializer=self.beta_initializer,
113 | regularizer=self.beta_regularizer,
114 | constraint=self.beta_constraint)
115 | else:
116 | self.beta = None
117 | self.built = True
118 |
119 | def call(self, inputs, **kwargs):
120 | input_shape = K.int_shape(inputs)
121 | tensor_input_shape = K.shape(inputs)
122 |
123 | # Prepare broadcasting shape.
124 | reduction_axes = list(range(len(input_shape)))
125 | del reduction_axes[self.axis]
126 | broadcast_shape = [1] * len(input_shape)
127 | broadcast_shape[self.axis] = input_shape[self.axis] // self.groups
128 | broadcast_shape.insert(1, self.groups)
129 |
130 | reshape_group_shape = K.shape(inputs)
131 | group_axes = [reshape_group_shape[i] for i in range(len(input_shape))]
132 | group_axes[self.axis] = input_shape[self.axis] // self.groups
133 | group_axes.insert(1, self.groups)
134 |
135 | # reshape inputs to new group shape
136 | group_shape = [group_axes[0], self.groups] + group_axes[2:]
137 | group_shape = K.stack(group_shape)
138 | inputs = K.reshape(inputs, group_shape)
139 |
140 | group_reduction_axes = list(range(len(group_axes)))
141 | group_reduction_axes = group_reduction_axes[2:]
142 |
143 | mean = K.mean(inputs, axis=group_reduction_axes, keepdims=True)
144 | variance = K.var(inputs, axis=group_reduction_axes, keepdims=True)
145 |
146 | inputs = (inputs - mean) / (K.sqrt(variance + self.epsilon))
147 |
148 | # prepare broadcast shape
149 | inputs = K.reshape(inputs, group_shape)
150 | outputs = inputs
151 |
152 | # In this case we must explicitly broadcast all parameters.
153 | if self.scale:
154 | broadcast_gamma = K.reshape(self.gamma, broadcast_shape)
155 | outputs = outputs * broadcast_gamma
156 |
157 | if self.center:
158 | broadcast_beta = K.reshape(self.beta, broadcast_shape)
159 | outputs = outputs + broadcast_beta
160 |
161 | outputs = K.reshape(outputs, tensor_input_shape)
162 |
163 | return outputs
164 |
165 | def get_config(self):
166 | config = {
167 | 'groups': self.groups,
168 | 'axis': self.axis,
169 | 'epsilon': self.epsilon,
170 | 'center': self.center,
171 | 'scale': self.scale,
172 | 'beta_initializer': initializers.serialize(self.beta_initializer),
173 | 'gamma_initializer': initializers.serialize(self.gamma_initializer),
174 | 'beta_regularizer': regularizers.serialize(self.beta_regularizer),
175 | 'gamma_regularizer': regularizers.serialize(self.gamma_regularizer),
176 | 'beta_constraint': constraints.serialize(self.beta_constraint),
177 | 'gamma_constraint': constraints.serialize(self.gamma_constraint)
178 | }
179 | base_config = super(GroupNormalization, self).get_config()
180 | return dict(list(base_config.items()) + list(config.items()))
181 |
182 | def compute_output_shape(self, input_shape):
183 | return input_shape
184 |
185 |
186 | get_custom_objects().update({'GroupNormalization': GroupNormalization})
187 |
188 |
189 | if __name__ == '__main__':
190 | from keras.layers import Input
191 | from keras.models import Model
192 | ip = Input(shape=(None, None, 4))
193 | #ip = Input(batch_shape=(100, None, None, 2))
194 | x = GroupNormalization(groups=2, axis=-1, epsilon=0.1)(ip)
195 | model = Model(ip, x)
196 | model.summary()
197 |
198 |
--------------------------------------------------------------------------------
/model.py:
--------------------------------------------------------------------------------
1 | # Keras implementation of the paper:
2 | # 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization
3 | # by Myronenko A. (https://arxiv.org/pdf/1810.11654.pdf)
4 | # Author of this code: Suyog Jadhav (https://github.com/IAmSUyogJadhav)
5 |
6 | import keras.backend as K
7 | from keras.losses import mse
8 | from keras.layers import Conv3D, Activation, Add, UpSampling3D, Lambda, Dense
9 | from keras.layers import Input, Reshape, Flatten, Dropout, SpatialDropout3D
10 | from keras.optimizers import adam
11 | from keras.models import Model
12 | try:
13 | from group_norm import GroupNormalization
14 | except ImportError:
15 | import urllib.request
16 | print('Downloading group_norm.py in the current directory...')
17 | url = 'https://raw.githubusercontent.com/titu1994/Keras-Group-Normalization/master/group_norm.py'
18 | urllib.request.urlretrieve(url, "group_norm.py")
19 | from group_norm import GroupNormalization
20 |
21 |
22 | def green_block(inp, filters, data_format='channels_first', name=None):
23 | """
24 | green_block(inp, filters, name=None)
25 | ------------------------------------
26 | Implementation of the special residual block used in the paper. The block
27 | consists of two (GroupNorm --> ReLu --> 3x3x3 non-strided Convolution)
28 | units, with a residual connection from the input `inp` to the output. Used
29 | internally in the model. Can be used independently as well.
30 |
31 | Parameters
32 | ----------
33 | `inp`: An keras.layers.layer instance, required
34 | The keras layer just preceding the green block.
35 | `filters`: integer, required
36 | No. of filters to use in the 3D convolutional block. The output
37 | layer of this green block will have this many no. of channels.
38 | `data_format`: string, optional
39 | The format of the input data. Must be either 'chanels_first' or
40 | 'channels_last'. Defaults to `channels_first`, as used in the paper.
41 | `name`: string, optional
42 | The name to be given to this green block. Defaults to None, in which
43 | case, keras uses generated names for the involved layers. If a string
44 | is provided, the names of individual layers are generated by attaching
45 | a relevant prefix from [GroupNorm_, Res_, Conv3D_, Relu_, ], followed
46 | by _1 or _2.
47 |
48 | Returns
49 | -------
50 | `out`: A keras.layers.Layer instance
51 | The output of the green block. Has no. of channels equal to `filters`.
52 | The size of the rest of the dimensions remains same as in `inp`.
53 | """
54 | inp_res = Conv3D(
55 | filters=filters,
56 | kernel_size=(1, 1, 1),
57 | strides=1,
58 | data_format=data_format,
59 | name=f'Res_{name}' if name else None)(inp)
60 |
61 | # axis=1 for channels_first data format
62 | # No. of groups = 8, as given in the paper
63 | x = GroupNormalization(
64 | groups=8,
65 | axis=1 if data_format == 'channels_first' else 0,
66 | name=f'GroupNorm_1_{name}' if name else None)(inp)
67 | x = Activation('relu', name=f'Relu_1_{name}' if name else None)(x)
68 | x = Conv3D(
69 | filters=filters,
70 | kernel_size=(3, 3, 3),
71 | strides=1,
72 | padding='same',
73 | data_format=data_format,
74 | name=f'Conv3D_1_{name}' if name else None)(x)
75 |
76 | x = GroupNormalization(
77 | groups=8,
78 | axis=1 if data_format == 'channels_first' else 0,
79 | name=f'GroupNorm_2_{name}' if name else None)(x)
80 | x = Activation('relu', name=f'Relu_2_{name}' if name else None)(x)
81 | x = Conv3D(
82 | filters=filters,
83 | kernel_size=(3, 3, 3),
84 | strides=1,
85 | padding='same',
86 | data_format=data_format,
87 | name=f'Conv3D_2_{name}' if name else None)(x)
88 |
89 | out = Add(name=f'Out_{name}' if name else None)([x, inp_res])
90 | return out
91 |
92 |
93 | # From keras-team/keras/blob/master/examples/variational_autoencoder.py
94 | def sampling(args):
95 | """Reparameterization trick by sampling from an isotropic unit Gaussian.
96 | # Arguments
97 | args (tensor): mean and log of variance of Q(z|X)
98 | # Returns
99 | z (tensor): sampled latent vector
100 | """
101 | z_mean, z_var = args
102 | batch = K.shape(z_mean)[0]
103 | dim = K.int_shape(z_mean)[1]
104 | # by default, random_normal has mean = 0 and std = 1.0
105 | epsilon = K.random_normal(shape=(batch, dim))
106 | return z_mean + K.exp(0.5 * z_var) * epsilon
107 |
108 |
109 | def dice_coefficient(y_true, y_pred):
110 | intersection = K.sum(K.abs(y_true * y_pred), axis=[-3,-2,-1])
111 | dn = K.sum(K.square(y_true) + K.square(y_pred), axis=[-3,-2,-1]) + 1e-8
112 | return K.mean(2 * intersection / dn, axis=[0,1])
113 |
114 |
115 | def loss_gt(e=1e-8):
116 | """
117 | loss_gt(e=1e-8)
118 | ------------------------------------------------------
119 | Since keras does not allow custom loss functions to have arguments
120 | other than the true and predicted labels, this function acts as a wrapper
121 | that allows us to implement the custom loss used in the paper. This function
122 | only calculates - L term of the following equation. (i.e. GT Decoder part loss)
123 |
124 | L = - L + weight_L2 ∗ L + weight_KL ∗ L
125 |
126 | Parameters
127 | ----------
128 | `e`: Float, optional
129 | A small epsilon term to add in the denominator to avoid dividing by
130 | zero and possible gradient explosion.
131 |
132 | Returns
133 | -------
134 | loss_gt_(y_true, y_pred): A custom keras loss function
135 | This function takes as input the predicted and ground labels, uses them
136 | to calculate the dice loss.
137 |
138 | """
139 | def loss_gt_(y_true, y_pred):
140 | intersection = K.sum(K.abs(y_true * y_pred), axis=[-3,-2,-1])
141 | dn = K.sum(K.square(y_true) + K.square(y_pred), axis=[-3,-2,-1]) + e
142 |
143 | return - K.mean(2 * intersection / dn, axis=[0,1])
144 |
145 | return loss_gt_
146 |
147 | def loss_VAE(input_shape, z_mean, z_var, weight_L2=0.1, weight_KL=0.1):
148 | """
149 | loss_VAE(input_shape, z_mean, z_var, weight_L2=0.1, weight_KL=0.1)
150 | ------------------------------------------------------
151 | Since keras does not allow custom loss functions to have arguments
152 | other than the true and predicted labels, this function acts as a wrapper
153 | that allows us to implement the custom loss used in the paper. This function
154 | calculates the following equation, except for -L term. (i.e. VAE decoder part loss)
155 |
156 | L = - L + weight_L2 ∗ L + weight_KL ∗ L
157 |
158 | Parameters
159 | ----------
160 | `input_shape`: A 4-tuple, required
161 | The shape of an image as the tuple (c, H, W, D), where c is
162 | the no. of channels; H, W and D is the height, width and depth of the
163 | input image, respectively.
164 | `z_mean`: An keras.layers.Layer instance, required
165 | The vector representing values of mean for the learned distribution
166 | in the VAE part. Used internally.
167 | `z_var`: An keras.layers.Layer instance, required
168 | The vector representing values of variance for the learned distribution
169 | in the VAE part. Used internally.
170 | `weight_L2`: A real number, optional
171 | The weight to be given to the L2 loss term in the loss function. Adjust to get best
172 | results for your task. Defaults to 0.1.
173 | `weight_KL`: A real number, optional
174 | The weight to be given to the KL loss term in the loss function. Adjust to get best
175 | results for your task. Defaults to 0.1.
176 |
177 | Returns
178 | -------
179 | loss_VAE_(y_true, y_pred): A custom keras loss function
180 | This function takes as input the predicted and ground labels, uses them
181 | to calculate the L2 and KL loss.
182 |
183 | """
184 | def loss_VAE_(y_true, y_pred):
185 | c, H, W, D = input_shape
186 | n = c * H * W * D
187 |
188 | loss_L2 = K.mean(K.square(y_true - y_pred), axis=(1, 2, 3, 4)) # original axis value is (1,2,3,4).
189 |
190 | loss_KL = (1 / n) * K.sum(
191 | K.exp(z_var) + K.square(z_mean) - 1. - z_var,
192 | axis=-1
193 | )
194 |
195 | return weight_L2 * loss_L2 + weight_KL * loss_KL
196 |
197 | return loss_VAE_
198 |
199 | def build_model(input_shape=(4, 160, 192, 128), output_channels=3, weight_L2=0.1, weight_KL=0.1, dice_e=1e-8):
200 | """
201 | build_model(input_shape=(4, 160, 192, 128), output_channels=3, weight_L2=0.1, weight_KL=0.1)
202 | -------------------------------------------
203 | Creates the model used in the BRATS2018 winning solution
204 | by Myronenko A. (https://arxiv.org/pdf/1810.11654.pdf)
205 |
206 | Parameters
207 | ----------
208 | `input_shape`: A 4-tuple, optional.
209 | Shape of the input image. Must be a 4D image of shape (c, H, W, D),
210 | where, each of H, W and D are divisible by 2^4, and c is divisible by 4.
211 | Defaults to the crop size used in the paper, i.e., (4, 160, 192, 128).
212 | `output_channels`: An integer, optional.
213 | The no. of channels in the output. Defaults to 3 (BraTS 2018 format).
214 | `weight_L2`: A real number, optional
215 | The weight to be given to the L2 loss term in the loss function. Adjust to get best
216 | results for your task. Defaults to 0.1.
217 | `weight_KL`: A real number, optional
218 | The weight to be given to the KL loss term in the loss function. Adjust to get best
219 | results for your task. Defaults to 0.1.
220 | `dice_e`: Float, optional
221 | A small epsilon term to add in the denominator of dice loss to avoid dividing by
222 | zero and possible gradient explosion. This argument will be passed to loss_gt function.
223 |
224 |
225 | Returns
226 | -------
227 | `model`: A keras.models.Model instance
228 | The created model.
229 | """
230 | c, H, W, D = input_shape
231 | assert len(input_shape) == 4, "Input shape must be a 4-tuple"
232 | assert (c % 4) == 0, "The no. of channels must be divisible by 4"
233 | assert (H % 16) == 0 and (W % 16) == 0 and (D % 16) == 0, \
234 | "All the input dimensions must be divisible by 16"
235 |
236 |
237 | # -------------------------------------------------------------------------
238 | # Encoder
239 | # -------------------------------------------------------------------------
240 |
241 | ## Input Layer
242 | inp = Input(input_shape)
243 |
244 | ## The Initial Block
245 | x = Conv3D(
246 | filters=32,
247 | kernel_size=(3, 3, 3),
248 | strides=1,
249 | padding='same',
250 | data_format='channels_first',
251 | name='Input_x1')(inp)
252 |
253 | ## Dropout (0.2)
254 | x = SpatialDropout3D(0.2, data_format='channels_first')(x)
255 |
256 | ## Green Block x1 (output filters = 32)
257 | x1 = green_block(x, 32, name='x1')
258 | x = Conv3D(
259 | filters=32,
260 | kernel_size=(3, 3, 3),
261 | strides=2,
262 | padding='same',
263 | data_format='channels_first',
264 | name='Enc_DownSample_32')(x1)
265 |
266 | ## Green Block x2 (output filters = 64)
267 | x = green_block(x, 64, name='Enc_64_1')
268 | x2 = green_block(x, 64, name='x2')
269 | x = Conv3D(
270 | filters=64,
271 | kernel_size=(3, 3, 3),
272 | strides=2,
273 | padding='same',
274 | data_format='channels_first',
275 | name='Enc_DownSample_64')(x2)
276 |
277 | ## Green Blocks x2 (output filters = 128)
278 | x = green_block(x, 128, name='Enc_128_1')
279 | x3 = green_block(x, 128, name='x3')
280 | x = Conv3D(
281 | filters=128,
282 | kernel_size=(3, 3, 3),
283 | strides=2,
284 | padding='same',
285 | data_format='channels_first',
286 | name='Enc_DownSample_128')(x3)
287 |
288 | ## Green Blocks x4 (output filters = 256)
289 | x = green_block(x, 256, name='Enc_256_1')
290 | x = green_block(x, 256, name='Enc_256_2')
291 | x = green_block(x, 256, name='Enc_256_3')
292 | x4 = green_block(x, 256, name='x4')
293 |
294 | # -------------------------------------------------------------------------
295 | # Decoder
296 | # -------------------------------------------------------------------------
297 |
298 | ## GT (Groud Truth) Part
299 | # -------------------------------------------------------------------------
300 |
301 | ### Green Block x1 (output filters=128)
302 | x = Conv3D(
303 | filters=128,
304 | kernel_size=(1, 1, 1),
305 | strides=1,
306 | data_format='channels_first',
307 | name='Dec_GT_ReduceDepth_128')(x4)
308 | x = UpSampling3D(
309 | size=2,
310 | data_format='channels_first',
311 | name='Dec_GT_UpSample_128')(x)
312 | x = Add(name='Input_Dec_GT_128')([x, x3])
313 | x = green_block(x, 128, name='Dec_GT_128')
314 |
315 | ### Green Block x1 (output filters=64)
316 | x = Conv3D(
317 | filters=64,
318 | kernel_size=(1, 1, 1),
319 | strides=1,
320 | data_format='channels_first',
321 | name='Dec_GT_ReduceDepth_64')(x)
322 | x = UpSampling3D(
323 | size=2,
324 | data_format='channels_first',
325 | name='Dec_GT_UpSample_64')(x)
326 | x = Add(name='Input_Dec_GT_64')([x, x2])
327 | x = green_block(x, 64, name='Dec_GT_64')
328 |
329 | ### Green Block x1 (output filters=32)
330 | x = Conv3D(
331 | filters=32,
332 | kernel_size=(1, 1, 1),
333 | strides=1,
334 | data_format='channels_first',
335 | name='Dec_GT_ReduceDepth_32')(x)
336 | x = UpSampling3D(
337 | size=2,
338 | data_format='channels_first',
339 | name='Dec_GT_UpSample_32')(x)
340 | x = Add(name='Input_Dec_GT_32')([x, x1])
341 | x = green_block(x, 32, name='Dec_GT_32')
342 |
343 | ### Blue Block x1 (output filters=32)
344 | x = Conv3D(
345 | filters=32,
346 | kernel_size=(3, 3, 3),
347 | strides=1,
348 | padding='same',
349 | data_format='channels_first',
350 | name='Input_Dec_GT_Output')(x)
351 |
352 | ### Output Block
353 | out_GT = Conv3D(
354 | filters=output_channels, # No. of tumor classes is 3
355 | kernel_size=(1, 1, 1),
356 | strides=1,
357 | data_format='channels_first',
358 | activation='sigmoid',
359 | name='Dec_GT_Output')(x)
360 |
361 | ## VAE (Variational Auto Encoder) Part
362 | # -------------------------------------------------------------------------
363 |
364 | ### VD Block (Reducing dimensionality of the data)
365 | x = GroupNormalization(groups=8, axis=1, name='Dec_VAE_VD_GN')(x4)
366 | x = Activation('relu', name='Dec_VAE_VD_relu')(x)
367 | x = Conv3D(
368 | filters=16,
369 | kernel_size=(3, 3, 3),
370 | strides=2,
371 | padding='same',
372 | data_format='channels_first',
373 | name='Dec_VAE_VD_Conv3D')(x)
374 |
375 | # Not mentioned in the paper, but the author used a Flattening layer here.
376 | x = Flatten(name='Dec_VAE_VD_Flatten')(x)
377 | x = Dense(256, name='Dec_VAE_VD_Dense')(x)
378 |
379 | ### VDraw Block (Sampling)
380 | z_mean = Dense(128, name='Dec_VAE_VDraw_Mean')(x)
381 | z_var = Dense(128, name='Dec_VAE_VDraw_Var')(x)
382 | x = Lambda(sampling, name='Dec_VAE_VDraw_Sampling')([z_mean, z_var])
383 |
384 | ### VU Block (Upsizing back to a depth of 256)
385 | x = Dense((c//4) * (H//16) * (W//16) * (D//16))(x)
386 | x = Activation('relu')(x)
387 | x = Reshape(((c//4), (H//16), (W//16), (D//16)))(x)
388 | x = Conv3D(
389 | filters=256,
390 | kernel_size=(1, 1, 1),
391 | strides=1,
392 | data_format='channels_first',
393 | name='Dec_VAE_ReduceDepth_256')(x)
394 | x = UpSampling3D(
395 | size=2,
396 | data_format='channels_first',
397 | name='Dec_VAE_UpSample_256')(x)
398 |
399 | ### Green Block x1 (output filters=128)
400 | x = Conv3D(
401 | filters=128,
402 | kernel_size=(1, 1, 1),
403 | strides=1,
404 | data_format='channels_first',
405 | name='Dec_VAE_ReduceDepth_128')(x)
406 | x = UpSampling3D(
407 | size=2,
408 | data_format='channels_first',
409 | name='Dec_VAE_UpSample_128')(x)
410 | x = green_block(x, 128, name='Dec_VAE_128')
411 |
412 | ### Green Block x1 (output filters=64)
413 | x = Conv3D(
414 | filters=64,
415 | kernel_size=(1, 1, 1),
416 | strides=1,
417 | data_format='channels_first',
418 | name='Dec_VAE_ReduceDepth_64')(x)
419 | x = UpSampling3D(
420 | size=2,
421 | data_format='channels_first',
422 | name='Dec_VAE_UpSample_64')(x)
423 | x = green_block(x, 64, name='Dec_VAE_64')
424 |
425 | ### Green Block x1 (output filters=32)
426 | x = Conv3D(
427 | filters=32,
428 | kernel_size=(1, 1, 1),
429 | strides=1,
430 | data_format='channels_first',
431 | name='Dec_VAE_ReduceDepth_32')(x)
432 | x = UpSampling3D(
433 | size=2,
434 | data_format='channels_first',
435 | name='Dec_VAE_UpSample_32')(x)
436 | x = green_block(x, 32, name='Dec_VAE_32')
437 |
438 | ### Blue Block x1 (output filters=32)
439 | x = Conv3D(
440 | filters=32,
441 | kernel_size=(3, 3, 3),
442 | strides=1,
443 | padding='same',
444 | data_format='channels_first',
445 | name='Input_Dec_VAE_Output')(x)
446 |
447 | ### Output Block
448 | out_VAE = Conv3D(
449 | filters=4,
450 | kernel_size=(1, 1, 1),
451 | strides=1,
452 | data_format='channels_first',
453 | name='Dec_VAE_Output')(x)
454 |
455 | # Build and Compile the model
456 | out = out_GT
457 | model = Model(inp, outputs=[out, out_VAE]) # Create the model
458 | model.compile(
459 | adam(lr=1e-4),
460 | [loss_gt(dice_e), loss_VAE(input_shape, z_mean, z_var, weight_L2=weight_L2, weight_KL=weight_KL)],
461 | metrics=[dice_coefficient]
462 | )
463 |
464 | return model
465 |
--------------------------------------------------------------------------------
/Example_on_BRATS2018.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "nbformat": 4,
3 | "nbformat_minor": 0,
4 | "metadata": {
5 | "colab": {
6 | "name": "Example on BRATS2018",
7 | "version": "0.3.2",
8 | "provenance": [],
9 | "collapsed_sections": [],
10 | "include_colab_link": true
11 | },
12 | "kernelspec": {
13 | "name": "python3",
14 | "display_name": "Python 3"
15 | },
16 | "accelerator": "GPU"
17 | },
18 | "cells": [
19 | {
20 | "cell_type": "markdown",
21 | "metadata": {
22 | "id": "view-in-github",
23 | "colab_type": "text"
24 | },
25 | "source": [
26 | " "
27 | ]
28 | },
29 | {
30 | "cell_type": "markdown",
31 | "metadata": {
32 | "id": "0Uld1IQgwKIL",
33 | "colab_type": "text"
34 | },
35 | "source": [
36 | "Note\n",
37 | "- [Colab Only] Means the associated cell is required only if you are running the model on Google Colaboratory"
38 | ]
39 | },
40 | {
41 | "cell_type": "markdown",
42 | "metadata": {
43 | "id": "m-oHUemNtm59",
44 | "colab_type": "text"
45 | },
46 | "source": [
47 | "## Mount Drive [Colab Only]\n",
48 | "Mount the google drive to access the dataset stored on drive."
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "metadata": {
54 | "id": "zYkOcDYNtf-e",
55 | "colab_type": "code",
56 | "outputId": "607d7128-cc53-4b85-fa43-bb2ca4727b03",
57 | "colab": {
58 | "base_uri": "https://localhost:8080/",
59 | "height": 121
60 | }
61 | },
62 | "source": [
63 | "from google.colab import drive\n",
64 | "drive.mount('/gdrive')"
65 | ],
66 | "execution_count": 2,
67 | "outputs": [
68 | {
69 | "output_type": "stream",
70 | "text": [
71 | "Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n",
72 | "\n",
73 | "Enter your authorization code:\n",
74 | "··········\n",
75 | "Mounted at /gdrive\n"
76 | ],
77 | "name": "stdout"
78 | }
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {
84 | "id": "Psq2s15_wlfJ",
85 | "colab_type": "text"
86 | },
87 | "source": [
88 | "## Extract the Dataset"
89 | ]
90 | },
91 | {
92 | "cell_type": "code",
93 | "metadata": {
94 | "id": "DuUurNXQtmXT",
95 | "colab_type": "code",
96 | "colab": {}
97 | },
98 | "source": [
99 | "import zipfile # For faster extraction\n",
100 | "dataset_path = \"/gdrive/My Drive/MICCAI_BraTS_2018_Data_Training.zip\" # Replace with your dataset path\n",
101 | "zfile = zipfile.ZipFile(dataset_path)\n",
102 | "zfile.extractall()"
103 | ],
104 | "execution_count": 0,
105 | "outputs": []
106 | },
107 | {
108 | "cell_type": "markdown",
109 | "metadata": {
110 | "id": "4NA9UG5zw938",
111 | "colab_type": "text"
112 | },
113 | "source": [
114 | "## Get required packages\n",
115 | "- **SimpleITK**: For loading the dataset\n",
116 | "- **[model.py](https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/)**: The model from BRATS2018 winning paper"
117 | ]
118 | },
119 | {
120 | "cell_type": "code",
121 | "metadata": {
122 | "id": "SqMFpmc6uCjQ",
123 | "colab_type": "code",
124 | "outputId": "3cc182ee-0bd0-49f5-c381-f2dfa8cebd6a",
125 | "colab": {
126 | "base_uri": "https://localhost:8080/",
127 | "height": 390
128 | }
129 | },
130 | "source": [
131 | "!pip install simpleitk\n",
132 | "!wget https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/raw/master/model.py"
133 | ],
134 | "execution_count": 4,
135 | "outputs": [
136 | {
137 | "output_type": "stream",
138 | "text": [
139 | "Collecting simpleitk\n",
140 | "\u001b[?25l Downloading https://files.pythonhosted.org/packages/45/ee/8ddd251cf447e1fc131622e925d538c2542780195ae75e26c122587630d1/SimpleITK-1.2.0-cp36-cp36m-manylinux1_x86_64.whl (42.5MB)\n",
141 | "\u001b[K |████████████████████████████████| 42.5MB 33.9MB/s \n",
142 | "\u001b[?25hInstalling collected packages: simpleitk\n",
143 | "Successfully installed simpleitk-1.2.0\n",
144 | "--2019-07-08 09:15:00-- https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/raw/master/model.py\n",
145 | "Resolving github.com (github.com)... 140.82.113.3\n",
146 | "Connecting to github.com (github.com)|140.82.113.3|:443... connected.\n",
147 | "HTTP request sent, awaiting response... 302 Found\n",
148 | "Location: https://raw.githubusercontent.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/master/model.py [following]\n",
149 | "--2019-07-08 09:15:01-- https://raw.githubusercontent.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/master/model.py\n",
150 | "Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 151.101.0.133, 151.101.64.133, 151.101.128.133, ...\n",
151 | "Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.\n",
152 | "HTTP request sent, awaiting response... 200 OK\n",
153 | "Length: 15603 (15K) [text/plain]\n",
154 | "Saving to: ‘model.py’\n",
155 | "\n",
156 | "model.py 100%[===================>] 15.24K --.-KB/s in 0.01s \n",
157 | "\n",
158 | "2019-07-08 09:15:01 (1.24 MB/s) - ‘model.py’ saved [15603/15603]\n",
159 | "\n"
160 | ],
161 | "name": "stdout"
162 | }
163 | ]
164 | },
165 | {
166 | "cell_type": "markdown",
167 | "metadata": {
168 | "id": "09ARH9U4D2Wy",
169 | "colab_type": "text"
170 | },
171 | "source": [
172 | "## Imports and helper functions"
173 | ]
174 | },
175 | {
176 | "cell_type": "code",
177 | "metadata": {
178 | "id": "4uAsBShgB3v_",
179 | "colab_type": "code",
180 | "outputId": "f1b7fe72-b0d7-4f63-ba15-44c16775e725",
181 | "colab": {
182 | "base_uri": "https://localhost:8080/",
183 | "height": 34
184 | }
185 | },
186 | "source": [
187 | "import SimpleITK as sitk # For loading the dataset\n",
188 | "import numpy as np # For data manipulation\n",
189 | "from model import build_model # For creating the model\n",
190 | "import glob # For populating the list of files\n",
191 | "from scipy.ndimage import zoom # For resizing\n",
192 | "import re # For parsing the filenames (to know their modality)"
193 | ],
194 | "execution_count": 1,
195 | "outputs": [
196 | {
197 | "output_type": "stream",
198 | "text": [
199 | "Using TensorFlow backend.\n"
200 | ],
201 | "name": "stderr"
202 | }
203 | ]
204 | },
205 | {
206 | "cell_type": "code",
207 | "metadata": {
208 | "id": "nNNtloQ9B3sA",
209 | "colab_type": "code",
210 | "colab": {}
211 | },
212 | "source": [
213 | "def read_img(img_path):\n",
214 | " \"\"\"\n",
215 | " Reads a .nii.gz image and returns as a numpy array.\n",
216 | " \"\"\"\n",
217 | " return sitk.GetArrayFromImage(sitk.ReadImage(img_path))"
218 | ],
219 | "execution_count": 0,
220 | "outputs": []
221 | },
222 | {
223 | "cell_type": "code",
224 | "metadata": {
225 | "id": "fEIRGXpdfssT",
226 | "colab_type": "code",
227 | "colab": {}
228 | },
229 | "source": [
230 | "def resize(img, shape, mode='constant', orig_shape=(155, 240, 240)):\n",
231 | " \"\"\"\n",
232 | " Wrapper for scipy.ndimage.zoom suited for MRI images.\n",
233 | " \"\"\"\n",
234 | " assert len(shape) == 3, \"Can not have more than 3 dimensions\"\n",
235 | " factors = (\n",
236 | " shape[0]/orig_shape[0],\n",
237 | " shape[1]/orig_shape[1], \n",
238 | " shape[2]/orig_shape[2]\n",
239 | " )\n",
240 | " \n",
241 | " # Resize to the given shape\n",
242 | " return zoom(img, factors, mode=mode)\n",
243 | "\n",
244 | "\n",
245 | "def preprocess(img, out_shape=None):\n",
246 | " \"\"\"\n",
247 | " Preprocess the image.\n",
248 | " Just an example, you can add more preprocessing steps if you wish to.\n",
249 | " \"\"\"\n",
250 | " if out_shape is not None:\n",
251 | " img = resize(img, out_shape, mode='constant')\n",
252 | " \n",
253 | " # Normalize the image\n",
254 | " mean = img.mean()\n",
255 | " std = img.std()\n",
256 | " return (img - mean) / std\n",
257 | "\n",
258 | "\n",
259 | "def preprocess_label(img, out_shape=None, mode='nearest'):\n",
260 | " \"\"\"\n",
261 | " Separates out the 3 labels from the segmentation provided, namely:\n",
262 | " GD-enhancing tumor (ET — label 4), the peritumoral edema (ED — label 2))\n",
263 | " and the necrotic and non-enhancing tumor core (NCR/NET — label 1)\n",
264 | " \"\"\"\n",
265 | " ncr = img == 1 # Necrotic and Non-Enhancing Tumor (NCR/NET)\n",
266 | " ed = img == 2 # Peritumoral Edema (ED)\n",
267 | " et = img == 4 # GD-enhancing Tumor (ET)\n",
268 | " \n",
269 | " if out_shape is not None:\n",
270 | " ncr = resize(ncr, out_shape, mode=mode)\n",
271 | " ed = resize(ed, out_shape, mode=mode)\n",
272 | " et = resize(et, out_shape, mode=mode)\n",
273 | "\n",
274 | " return np.array([ncr, ed, et], dtype=np.uint8)\n",
275 | " "
276 | ],
277 | "execution_count": 0,
278 | "outputs": []
279 | },
280 | {
281 | "cell_type": "markdown",
282 | "metadata": {
283 | "id": "piRsc9rYYRzl",
284 | "colab_type": "text"
285 | },
286 | "source": [
287 | "## Loading Data\n"
288 | ]
289 | },
290 | {
291 | "cell_type": "code",
292 | "metadata": {
293 | "id": "7adjWzdCuECK",
294 | "colab_type": "code",
295 | "colab": {}
296 | },
297 | "source": [
298 | "# Get a list of files for all modalities individually\n",
299 | "t1 = glob.glob('*GG/*/*t1.nii.gz')\n",
300 | "t2 = glob.glob('*GG/*/*t2.nii.gz')\n",
301 | "flair = glob.glob('*GG/*/*flair.nii.gz')\n",
302 | "t1ce = glob.glob('*GG/*/*t1ce.nii.gz')\n",
303 | "seg = glob.glob('*GG/*/*seg.nii.gz') # Ground Truth"
304 | ],
305 | "execution_count": 0,
306 | "outputs": []
307 | },
308 | {
309 | "cell_type": "markdown",
310 | "metadata": {
311 | "id": "x_WhT40dzlHO",
312 | "colab_type": "text"
313 | },
314 | "source": [
315 | "Parse all the filenames and create a dictionary for each patient with structure:\n",
316 | "\n",
317 | "{
"
27 | ]
28 | },
29 | {
30 | "cell_type": "markdown",
31 | "metadata": {
32 | "id": "0Uld1IQgwKIL",
33 | "colab_type": "text"
34 | },
35 | "source": [
36 | "Note\n",
37 | "- [Colab Only] Means the associated cell is required only if you are running the model on Google Colaboratory"
38 | ]
39 | },
40 | {
41 | "cell_type": "markdown",
42 | "metadata": {
43 | "id": "m-oHUemNtm59",
44 | "colab_type": "text"
45 | },
46 | "source": [
47 | "## Mount Drive [Colab Only]\n",
48 | "Mount the google drive to access the dataset stored on drive."
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "metadata": {
54 | "id": "zYkOcDYNtf-e",
55 | "colab_type": "code",
56 | "outputId": "607d7128-cc53-4b85-fa43-bb2ca4727b03",
57 | "colab": {
58 | "base_uri": "https://localhost:8080/",
59 | "height": 121
60 | }
61 | },
62 | "source": [
63 | "from google.colab import drive\n",
64 | "drive.mount('/gdrive')"
65 | ],
66 | "execution_count": 2,
67 | "outputs": [
68 | {
69 | "output_type": "stream",
70 | "text": [
71 | "Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n",
72 | "\n",
73 | "Enter your authorization code:\n",
74 | "··········\n",
75 | "Mounted at /gdrive\n"
76 | ],
77 | "name": "stdout"
78 | }
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {
84 | "id": "Psq2s15_wlfJ",
85 | "colab_type": "text"
86 | },
87 | "source": [
88 | "## Extract the Dataset"
89 | ]
90 | },
91 | {
92 | "cell_type": "code",
93 | "metadata": {
94 | "id": "DuUurNXQtmXT",
95 | "colab_type": "code",
96 | "colab": {}
97 | },
98 | "source": [
99 | "import zipfile # For faster extraction\n",
100 | "dataset_path = \"/gdrive/My Drive/MICCAI_BraTS_2018_Data_Training.zip\" # Replace with your dataset path\n",
101 | "zfile = zipfile.ZipFile(dataset_path)\n",
102 | "zfile.extractall()"
103 | ],
104 | "execution_count": 0,
105 | "outputs": []
106 | },
107 | {
108 | "cell_type": "markdown",
109 | "metadata": {
110 | "id": "4NA9UG5zw938",
111 | "colab_type": "text"
112 | },
113 | "source": [
114 | "## Get required packages\n",
115 | "- **SimpleITK**: For loading the dataset\n",
116 | "- **[model.py](https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/)**: The model from BRATS2018 winning paper"
117 | ]
118 | },
119 | {
120 | "cell_type": "code",
121 | "metadata": {
122 | "id": "SqMFpmc6uCjQ",
123 | "colab_type": "code",
124 | "outputId": "3cc182ee-0bd0-49f5-c381-f2dfa8cebd6a",
125 | "colab": {
126 | "base_uri": "https://localhost:8080/",
127 | "height": 390
128 | }
129 | },
130 | "source": [
131 | "!pip install simpleitk\n",
132 | "!wget https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/raw/master/model.py"
133 | ],
134 | "execution_count": 4,
135 | "outputs": [
136 | {
137 | "output_type": "stream",
138 | "text": [
139 | "Collecting simpleitk\n",
140 | "\u001b[?25l Downloading https://files.pythonhosted.org/packages/45/ee/8ddd251cf447e1fc131622e925d538c2542780195ae75e26c122587630d1/SimpleITK-1.2.0-cp36-cp36m-manylinux1_x86_64.whl (42.5MB)\n",
141 | "\u001b[K |████████████████████████████████| 42.5MB 33.9MB/s \n",
142 | "\u001b[?25hInstalling collected packages: simpleitk\n",
143 | "Successfully installed simpleitk-1.2.0\n",
144 | "--2019-07-08 09:15:00-- https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/raw/master/model.py\n",
145 | "Resolving github.com (github.com)... 140.82.113.3\n",
146 | "Connecting to github.com (github.com)|140.82.113.3|:443... connected.\n",
147 | "HTTP request sent, awaiting response... 302 Found\n",
148 | "Location: https://raw.githubusercontent.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/master/model.py [following]\n",
149 | "--2019-07-08 09:15:01-- https://raw.githubusercontent.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/master/model.py\n",
150 | "Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 151.101.0.133, 151.101.64.133, 151.101.128.133, ...\n",
151 | "Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.\n",
152 | "HTTP request sent, awaiting response... 200 OK\n",
153 | "Length: 15603 (15K) [text/plain]\n",
154 | "Saving to: ‘model.py’\n",
155 | "\n",
156 | "model.py 100%[===================>] 15.24K --.-KB/s in 0.01s \n",
157 | "\n",
158 | "2019-07-08 09:15:01 (1.24 MB/s) - ‘model.py’ saved [15603/15603]\n",
159 | "\n"
160 | ],
161 | "name": "stdout"
162 | }
163 | ]
164 | },
165 | {
166 | "cell_type": "markdown",
167 | "metadata": {
168 | "id": "09ARH9U4D2Wy",
169 | "colab_type": "text"
170 | },
171 | "source": [
172 | "## Imports and helper functions"
173 | ]
174 | },
175 | {
176 | "cell_type": "code",
177 | "metadata": {
178 | "id": "4uAsBShgB3v_",
179 | "colab_type": "code",
180 | "outputId": "f1b7fe72-b0d7-4f63-ba15-44c16775e725",
181 | "colab": {
182 | "base_uri": "https://localhost:8080/",
183 | "height": 34
184 | }
185 | },
186 | "source": [
187 | "import SimpleITK as sitk # For loading the dataset\n",
188 | "import numpy as np # For data manipulation\n",
189 | "from model import build_model # For creating the model\n",
190 | "import glob # For populating the list of files\n",
191 | "from scipy.ndimage import zoom # For resizing\n",
192 | "import re # For parsing the filenames (to know their modality)"
193 | ],
194 | "execution_count": 1,
195 | "outputs": [
196 | {
197 | "output_type": "stream",
198 | "text": [
199 | "Using TensorFlow backend.\n"
200 | ],
201 | "name": "stderr"
202 | }
203 | ]
204 | },
205 | {
206 | "cell_type": "code",
207 | "metadata": {
208 | "id": "nNNtloQ9B3sA",
209 | "colab_type": "code",
210 | "colab": {}
211 | },
212 | "source": [
213 | "def read_img(img_path):\n",
214 | " \"\"\"\n",
215 | " Reads a .nii.gz image and returns as a numpy array.\n",
216 | " \"\"\"\n",
217 | " return sitk.GetArrayFromImage(sitk.ReadImage(img_path))"
218 | ],
219 | "execution_count": 0,
220 | "outputs": []
221 | },
222 | {
223 | "cell_type": "code",
224 | "metadata": {
225 | "id": "fEIRGXpdfssT",
226 | "colab_type": "code",
227 | "colab": {}
228 | },
229 | "source": [
230 | "def resize(img, shape, mode='constant', orig_shape=(155, 240, 240)):\n",
231 | " \"\"\"\n",
232 | " Wrapper for scipy.ndimage.zoom suited for MRI images.\n",
233 | " \"\"\"\n",
234 | " assert len(shape) == 3, \"Can not have more than 3 dimensions\"\n",
235 | " factors = (\n",
236 | " shape[0]/orig_shape[0],\n",
237 | " shape[1]/orig_shape[1], \n",
238 | " shape[2]/orig_shape[2]\n",
239 | " )\n",
240 | " \n",
241 | " # Resize to the given shape\n",
242 | " return zoom(img, factors, mode=mode)\n",
243 | "\n",
244 | "\n",
245 | "def preprocess(img, out_shape=None):\n",
246 | " \"\"\"\n",
247 | " Preprocess the image.\n",
248 | " Just an example, you can add more preprocessing steps if you wish to.\n",
249 | " \"\"\"\n",
250 | " if out_shape is not None:\n",
251 | " img = resize(img, out_shape, mode='constant')\n",
252 | " \n",
253 | " # Normalize the image\n",
254 | " mean = img.mean()\n",
255 | " std = img.std()\n",
256 | " return (img - mean) / std\n",
257 | "\n",
258 | "\n",
259 | "def preprocess_label(img, out_shape=None, mode='nearest'):\n",
260 | " \"\"\"\n",
261 | " Separates out the 3 labels from the segmentation provided, namely:\n",
262 | " GD-enhancing tumor (ET — label 4), the peritumoral edema (ED — label 2))\n",
263 | " and the necrotic and non-enhancing tumor core (NCR/NET — label 1)\n",
264 | " \"\"\"\n",
265 | " ncr = img == 1 # Necrotic and Non-Enhancing Tumor (NCR/NET)\n",
266 | " ed = img == 2 # Peritumoral Edema (ED)\n",
267 | " et = img == 4 # GD-enhancing Tumor (ET)\n",
268 | " \n",
269 | " if out_shape is not None:\n",
270 | " ncr = resize(ncr, out_shape, mode=mode)\n",
271 | " ed = resize(ed, out_shape, mode=mode)\n",
272 | " et = resize(et, out_shape, mode=mode)\n",
273 | "\n",
274 | " return np.array([ncr, ed, et], dtype=np.uint8)\n",
275 | " "
276 | ],
277 | "execution_count": 0,
278 | "outputs": []
279 | },
280 | {
281 | "cell_type": "markdown",
282 | "metadata": {
283 | "id": "piRsc9rYYRzl",
284 | "colab_type": "text"
285 | },
286 | "source": [
287 | "## Loading Data\n"
288 | ]
289 | },
290 | {
291 | "cell_type": "code",
292 | "metadata": {
293 | "id": "7adjWzdCuECK",
294 | "colab_type": "code",
295 | "colab": {}
296 | },
297 | "source": [
298 | "# Get a list of files for all modalities individually\n",
299 | "t1 = glob.glob('*GG/*/*t1.nii.gz')\n",
300 | "t2 = glob.glob('*GG/*/*t2.nii.gz')\n",
301 | "flair = glob.glob('*GG/*/*flair.nii.gz')\n",
302 | "t1ce = glob.glob('*GG/*/*t1ce.nii.gz')\n",
303 | "seg = glob.glob('*GG/*/*seg.nii.gz') # Ground Truth"
304 | ],
305 | "execution_count": 0,
306 | "outputs": []
307 | },
308 | {
309 | "cell_type": "markdown",
310 | "metadata": {
311 | "id": "x_WhT40dzlHO",
312 | "colab_type": "text"
313 | },
314 | "source": [
315 | "Parse all the filenames and create a dictionary for each patient with structure:\n",
316 | "\n",
317 | "{
\n",
318 | " 't1': _\n",
319 | " 't2': _\n",
320 | " 'flair': _\n",
321 | " 't1ce': _\n",
322 | " 'seg': _\n",
323 | "}
"
324 | ]
325 | },
326 | {
327 | "cell_type": "code",
328 | "metadata": {
329 | "id": "4dssK9Nmwojp",
330 | "colab_type": "code",
331 | "colab": {}
332 | },
333 | "source": [
334 | "pat = re.compile('.*_(\\w*)\\.nii\\.gz')\n",
335 | "\n",
336 | "data_paths = [{\n",
337 | " pat.findall(item)[0]:item\n",
338 | " for item in items\n",
339 | "}\n",
340 | "for items in list(zip(t1, t2, t1ce, flair, seg))]"
341 | ],
342 | "execution_count": 0,
343 | "outputs": []
344 | },
345 | {
346 | "cell_type": "markdown",
347 | "metadata": {
348 | "id": "wHqqrbnH0n2E",
349 | "colab_type": "text"
350 | },
351 | "source": [
352 | "## Load the data in a Numpy array\n",
353 | "Creating an empty Numpy array beforehand and then filling up the data helps you gauge beforehand if the data fits in your memory.\n",
354 | "\n"
355 | ]
356 | },
357 | {
358 | "cell_type": "markdown",
359 | "metadata": {
360 | "id": "tBgbr-R-7Zat",
361 | "colab_type": "text"
362 | },
363 | "source": [
364 | "_Loading only the first 4 images here, to save time._"
365 | ]
366 | },
367 | {
368 | "cell_type": "code",
369 | "metadata": {
370 | "id": "nvbbddauz8ij",
371 | "colab_type": "code",
372 | "colab": {}
373 | },
374 | "source": [
375 | "input_shape = (4, 80, 96, 64)\n",
376 | "output_channels = 3\n",
377 | "data = np.empty((len(data_paths[:4]),) + input_shape, dtype=np.float32)\n",
378 | "labels = np.empty((len(data_paths[:4]), output_channels) + input_shape[1:], dtype=np.uint8)"
379 | ],
380 | "execution_count": 0,
381 | "outputs": []
382 | },
383 | {
384 | "cell_type": "code",
385 | "metadata": {
386 | "id": "G4u_yrMf30k4",
387 | "colab_type": "code",
388 | "outputId": "81c1cb4d-8339-41bb-971c-a386bf3c7f69",
389 | "colab": {
390 | "base_uri": "https://localhost:8080/",
391 | "height": 34
392 | }
393 | },
394 | "source": [
395 | "import math\n",
396 | "\n",
397 | "# Parameters for the progress bar\n",
398 | "total = len(data_paths[:4])\n",
399 | "step = 25 / total\n",
400 | "\n",
401 | "for i, imgs in enumerate(data_paths[:4]):\n",
402 | " try:\n",
403 | " data[i] = np.array([preprocess(read_img(imgs[m]), input_shape[1:]) for m in ['t1', 't2', 't1ce', 'flair']], dtype=np.float32)\n",
404 | " labels[i] = preprocess_label(read_img(imgs['seg']), input_shape[1:])[None, ...]\n",
405 | " \n",
406 | " # Print the progress bar\n",
407 | " print('\\r' + f'Progress: '\n",
408 | " f\"[{'=' * int((i+1) * step) + ' ' * (24 - int((i+1) * step))}]\"\n",
409 | " f\"({math.ceil((i+1) * 100 / (total))} %)\",\n",
410 | " end='')\n",
411 | " except Exception as e:\n",
412 | " print(f'Something went wrong with {imgs[\"t1\"]}, skipping...\\n Exception:\\n{str(e)}')\n",
413 | " continue"
414 | ],
415 | "execution_count": 67,
416 | "outputs": [
417 | {
418 | "output_type": "stream",
419 | "text": [
420 | "Progress: [=========================](100 %)"
421 | ],
422 | "name": "stdout"
423 | }
424 | ]
425 | },
426 | {
427 | "cell_type": "markdown",
428 | "metadata": {
429 | "id": "Ojc7YgwC305t",
430 | "colab_type": "text"
431 | },
432 | "source": [
433 | "## Model"
434 | ]
435 | },
436 | {
437 | "cell_type": "markdown",
438 | "metadata": {
439 | "id": "uR_SDmgn4Lrd",
440 | "colab_type": "text"
441 | },
442 | "source": [
443 | "build the model"
444 | ]
445 | },
446 | {
447 | "cell_type": "code",
448 | "metadata": {
449 | "id": "vI53S6yZJN2V",
450 | "colab_type": "code",
451 | "colab": {}
452 | },
453 | "source": [
454 | "model = build_model(input_shape=input_shape, output_channels=3)"
455 | ],
456 | "execution_count": 0,
457 | "outputs": []
458 | },
459 | {
460 | "cell_type": "markdown",
461 | "metadata": {
462 | "id": "s1u107O74NOP",
463 | "colab_type": "text"
464 | },
465 | "source": [
466 | "Train the model"
467 | ]
468 | },
469 | {
470 | "cell_type": "code",
471 | "metadata": {
472 | "id": "b0y7NfU4JBgi",
473 | "colab_type": "code",
474 | "outputId": "9f506516-2c7b-460e-aa5a-7abdab4eceb1",
475 | "colab": {
476 | "base_uri": "https://localhost:8080/",
477 | "height": 87
478 | }
479 | },
480 | "source": [
481 | "model.fit(data, [labels, data], batch_size=1, epochs=1)"
482 | ],
483 | "execution_count": 100,
484 | "outputs": [
485 | {
486 | "output_type": "stream",
487 | "text": [
488 | "Epoch 1/1\n",
489 | "4/4 [==============================] - 29s 7s/step - loss: 0.3766 - acc: 0.1188\n"
490 | ],
491 | "name": "stdout"
492 | },
493 | {
494 | "output_type": "execute_result",
495 | "data": {
496 | "text/plain": [
497 | ""
498 | ]
499 | },
500 | "metadata": {
501 | "tags": []
502 | },
503 | "execution_count": 100
504 | }
505 | ]
506 | },
507 | {
508 | "cell_type": "markdown",
509 | "metadata": {
510 | "id": "VlrqEF7m7ist",
511 | "colab_type": "text"
512 | },
513 | "source": [
514 | "That's it!"
515 | ]
516 | },
517 | {
518 | "cell_type": "markdown",
519 | "metadata": {
520 | "id": "ZKerVpLQTRck",
521 | "colab_type": "text"
522 | },
523 | "source": [
524 | "## Closing Regards"
525 | ]
526 | },
527 | {
528 | "cell_type": "markdown",
529 | "metadata": {
530 | "id": "bkSxJbRx4SsO",
531 | "colab_type": "text"
532 | },
533 | "source": [
534 | "If you are resizing the segmentation mask, the resized segmentation mask retains the overall shape, but loses a lot of pixels and becomes somewhat 'grainy'. See the illustration below."
535 | ]
536 | },
537 | {
538 | "cell_type": "markdown",
539 | "metadata": {
540 | "id": "eAau6Z7hznFC",
541 | "colab_type": "text"
542 | },
543 | "source": [
544 | "1. Original segmentation mask:"
545 | ]
546 | },
547 | {
548 | "cell_type": "code",
549 | "metadata": {
550 | "id": "CXRW65_hkL9n",
551 | "colab_type": "code",
552 | "colab": {
553 | "base_uri": "https://localhost:8080/",
554 | "height": 336
555 | },
556 | "outputId": "4c42e648-b9fe-4176-a324-b15ca9be7db5"
557 | },
558 | "source": [
559 | "import matplotlib.pyplot as plt\n",
560 | "img = (read_img(seg[0])[100] == 2).astype(np.uint8)\n",
561 | "print(img.shape)\n",
562 | "print(np.unique(img))\n",
563 | "print(img.sum())\n",
564 | "plt.imshow(img, cmap='Greys_r')"
565 | ],
566 | "execution_count": 109,
567 | "outputs": [
568 | {
569 | "output_type": "stream",
570 | "text": [
571 | "(240, 240)\n",
572 | "[0 1]\n",
573 | "2111\n"
574 | ],
575 | "name": "stdout"
576 | },
577 | {
578 | "output_type": "execute_result",
579 | "data": {
580 | "text/plain": [
581 | ""
582 | ]
583 | },
584 | "metadata": {
585 | "tags": []
586 | },
587 | "execution_count": 109
588 | },
589 | {
590 | "output_type": "display_data",
591 | "data": {
592 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAQUAAAD8CAYAAAB+fLH0AAAABHNCSVQICAgIfAhkiAAAAAlwSFlz\nAAALEgAACxIB0t1+/AAAADl0RVh0U29mdHdhcmUAbWF0cGxvdGxpYiB2ZXJzaW9uIDMuMC4zLCBo\ndHRwOi8vbWF0cGxvdGxpYi5vcmcvnQurowAADPZJREFUeJzt3U+sXOV9xvHvUwgsEiSgtJZl3EIi\nb+iGWFeUBYroogmwMdkgusGKkNwFSInULJxkEZZtpaQSaovkKCimSqFICcKLtA21ItENBDsixkAB\nNwFhy9iKqAhqpKTAr4s5F+Y193rm/pk5Z+79fqSjOfPOmZnfnHvPc9/3Pefem6pCkpb9Xt8FSBoW\nQ0FSw1CQ1DAUJDUMBUkNQ0FSY2ahkOS2JK8kOZXk4KzeR9LmyiyuU0hyCfAq8OfAaeA54C+q6qVN\nfzNJm2pWPYWbgFNV9Yuq+h3wGLBvRu8laRNdOqPX3QW8OXb/NPCnq22cxMsqpdn7VVX9waSNZhUK\nEyU5ABzo6/2lbeiNaTaaVSicAXaP3b+2a/tQVR0CDoE9BWlIZjWn8BywJ8n1SS4D7gaOzOi9JG2i\nmfQUquq9JPcD/w5cAjxcVS/O4r0kba6ZnJJccxEOH6R5OF5VS5M28opGSQ1DQVLDUJDUMBQkNQwF\nSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQk\nNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDU\nMBQkNS7dyJOTvA68C7wPvFdVS0muBv4FuA54Hbirqv5nY2VKmpfN6Cn8WVXdWFVL3f2DwNGq2gMc\n7e5LWhCzGD7sAw5364eBO2fwHpJmZKOhUMCPkxxPcqBr21FVZ7v1t4AdG3wPSXO0oTkF4JaqOpPk\nD4GnkvzX+INVVUlqpSd2IXJgpcck9WdDPYWqOtPdngeeAG4CziXZCdDdnl/luYeqamlsLkLSAKw7\nFJJ8MskVy+vA54GTwBFgf7fZfuDJjRYpaX42MnzYATyRZPl1/rmq/i3Jc8DjSe4F3gDu2niZkuYl\nVSsO+edbxCrzDpI21fFphute0SipYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgK\nmpshXFKvyQwFzU0Sg2EBGAqaq+63ajVghoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoqFdetzA8hoJ6\n5XULw2MoqHcX6y1U1YeL5sNQkNQwFNS71YYQF/YO7C3Mh6Gg3q12sK8UFgbD7BkK6p2TjcNiKGiw\n7BX0YyP/YFbadAZB/+wpSGoYCpIahoIGw6HDMBgKGgQDYTgMBUkNQ0ELx17FbBkKkhqGgqSGoaCF\n42XRszUxFJI8nOR8kpNjbVcneSrJa93tVV17kjyY5FSSE0n2zrJ4bQ3OEQzLND2F7wG3XdB2EDha\nVXuAo919gNuBPd1yAHhoc8qUNC8TQ6GqngbevqB5H3C4Wz8M3DnW/kiNPANcmWTnZhWrrcnhwLCs\nd05hR1Wd7dbfAnZ067uAN8e2O921fUySA0mOJTm2zhq0RTh8GJYN/5ZkVVWSNX9Vq+oQcAhgPc/X\n4ltPGNirmL319hTOLQ8LutvzXfsZYPfYdtd2bZqCf6BUQ7DeUDgC7O/W9wNPjrXf052FuBl4Z2yY\noYsYD4Pxv2A8aVlUi1z7Vjdx+JDkUeBW4Jokp4FvAn8NPJ7kXuAN4K5u8x8BdwCngN8AX5pBzRpz\n4cG1CN3r9QbCIny2rSBDSOztPqcwq6/BUA8iQ6E3x6tqadJGXtHYk3l0/xd9iKF+GAo9uHD+QBoS\nQ0Fz5dBh+AwFSQ1DQSuaxXyEQ6XFYCjM2aIdGJtV76J97u3MUNBEG+01bDQQnE+YL/9D1JwlmetP\nzfUcUKvVt1L7tP8xWovDUOjBPIOhqtYUDGuty4N/63H40JOhdYmHeqHT0PbTdmAo9Ghe3/BDPNg1\nXA4ferbSUGIWw4vl11sOIoNCqzEUBmClHsOs5h2GGgarhaPmz+GDeufBPyyGgqSGoaBeOccxPIaC\nBskhRX+caByw8QNjKD9JN6umi72OgdAvewoLYogHynprutjzhvg5txtDYYEk2ZIHzfJn2oqfbREZ\nCpraLA9aA2E4DIUFNKQDaC21bNWezlZjKCyoeR9ck+YBJtVjGCwOzz4ssHn/bYZJPPC3BnsKkhqG\nwoLz16+12QwFTcWhwfZhKGwB4wesM/zaKCcat4gLg2CIl0hrMdhT2AbsOWgtDIVtwmDQtAwFTWSg\nbC+Gwjbiwa1pGArbjGcnNImhsE0ZDFqNpyS3MU9baiUTewpJHk5yPsnJsbYHkpxJ8ny33DH22NeS\nnErySpIvzKpwbS6HFVo2zfDhe8BtK7T/XVXd2C0/AkhyA3A38Cfdc/4xySWbVaxmbzkcxhdtLxND\noaqeBt6e8vX2AY9V1W+r6pfAKeCmDdQnac42MtF4f5IT3fDiqq5tF/Dm2DanuzZJC2K9ofAQ8Bng\nRuAs8K21vkCSA0mOJTm2zhokzcC6QqGqzlXV+1X1AfAdPhoinAF2j216bde20mscqqqlqlpaTw2S\nZmNdoZBk59jdLwLLZyaOAHcnuTzJ9cAe4KcbK1HSPE28TiHJo8CtwDVJTgPfBG5NciNQwOvAXwJU\n1YtJHgdeAt4D7quq92dTuqRZyBAuWknSfxHS1nd8muG6lzlLahgKkhqGgqSGoSCpYShIahgKkhqG\ngqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgK\nkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpMTEU\nkuxO8pMkLyV5McmXu/arkzyV5LXu9qquPUkeTHIqyYkke2f9ISRtnml6Cu8Bf1VVNwA3A/cluQE4\nCBytqj3A0e4+wO3Anm45ADy06VVLmpmJoVBVZ6vqZ936u8DLwC5gH3C42+wwcGe3vg94pEaeAa5M\nsnPTK5c0E2uaU0hyHfBZ4FlgR1Wd7R56C9jRre8C3hx72umuTdICuHTaDZN8CvgB8JWq+nWSDx+r\nqkpSa3njJAcYDS8kDchUPYUkn2AUCN+vqh92zeeWhwXd7fmu/Qywe+zp13Ztjao6VFVLVbW03uIl\nbb5pzj4E+C7wclV9e+yhI8D+bn0/8ORY+z3dWYibgXfGhhmSBi5VF+/1J7kF+E/gBeCDrvnrjOYV\nHgf+CHgDuKuq3u5C5O+B24DfAF+qqmMT3mNNQw9J63J8mp75xFCYB0NBmoupQsErGiU1DAVJDUNB\nUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJ\nDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUuPSvgvo/Ar4\n3+52kVyDNc/LItY9tJr/eJqNUlWzLmQqSY5V1VLfdayFNc/PIta9iDWDwwdJFzAUJDWGFAqH+i5g\nHax5fhax7kWseThzCpKGYUg9BUkD0HsoJLktyStJTiU52Hc9q0nyepIXkjyf5FjXdnWSp5K81t1e\nNYA6H05yPsnJsbYV68zIg92+P5Fk74BqfiDJmW5/P5/kjrHHvtbV/EqSL/RU8+4kP0nyUpIXk3y5\nax/0vp5KVfW2AJcA/w18GrgM+DlwQ581XaTW14FrLmj7W+Bgt34Q+JsB1Pk5YC9wclKdwB3AvwIB\nbgaeHVDNDwBfXWHbG7rvk8uB67vvn0t6qHknsLdbvwJ4tatt0Pt6mqXvnsJNwKmq+kVV/Q54DNjX\nc01rsQ843K0fBu7ssRYAqupp4O0Lmlercx/wSI08A1yZZOd8Kv3IKjWvZh/wWFX9tqp+CZxi9H00\nV1V1tqp+1q2/C7wM7GLg+3oafYfCLuDNsfunu7YhKuDHSY4nOdC17aiqs936W8COfkqbaLU6h77/\n7++62g+PDc0GV3OS64DPAs+yuPv6Q32HwiK5par2ArcD9yX53PiDNeojDv5UzqLUCTwEfAa4ETgL\nfKvfclaW5FPAD4CvVNWvxx9boH3d6DsUzgC7x+5f27UNTlWd6W7PA08w6rKeW+4Cdrfn+6vwolar\nc7D7v6rOVdX7VfUB8B0+GiIMpuYkn2AUCN+vqh92zQu3ry/Udyg8B+xJcn2Sy4C7gSM91/QxST6Z\n5IrldeDzwElGte7vNtsPPNlPhROtVucR4J5uZvxm4J2xrm+vLhhvf5HR/oZRzXcnuTzJ9cAe4Kc9\n1Bfgu8DLVfXtsYcWbl9/TN8znYxmZV9lNIv8jb7rWaXGTzOa8f458OJyncDvA0eB14D/AK4eQK2P\nMupu/x+jceu9q9XJaCb8H7p9/wKwNKCa/6mr6QSjA2rn2Pbf6Gp+Bbi9p5pvYTQ0OAE83y13DH1f\nT7N4RaOkRt/DB0kDYyhIahgKkhqGgqSGoSCpYShIahgKkhqGgqTG/wPODMdY3yiYIwAAAABJRU5E\nrkJggg==\n",
593 | "text/plain": [
594 | ""
595 | ]
596 | },
597 | "metadata": {
598 | "tags": []
599 | }
600 | }
601 | ]
602 | },
603 | {
604 | "cell_type": "markdown",
605 | "metadata": {
606 | "id": "soDTIJFn5CE9",
607 | "colab_type": "text"
608 | },
609 | "source": [
610 | "After resizing to (80, 96, 64)"
611 | ]
612 | },
613 | {
614 | "cell_type": "code",
615 | "metadata": {
616 | "id": "-kmgpXjsAMzj",
617 | "colab_type": "code",
618 | "colab": {
619 | "base_uri": "https://localhost:8080/",
620 | "height": 336
621 | },
622 | "outputId": "7eac8776-abc2-4334-a5b2-ffba151be588"
623 | },
624 | "source": [
625 | "img = preprocess_label(read_img(seg[0]), out_shape=(80, 96, 64), mode='nearest')[1][50]\n",
626 | "print(img.shape)\n",
627 | "print(np.unique(img))\n",
628 | "print(img.sum())\n",
629 | "plt.imshow(img, cmap='Greys_r')"
630 | ],
631 | "execution_count": 105,
632 | "outputs": [
633 | {
634 | "output_type": "stream",
635 | "text": [
636 | "(96, 64)\n",
637 | "[0 1]\n",
638 | "116\n"
639 | ],
640 | "name": "stdout"
641 | },
642 | {
643 | "output_type": "execute_result",
644 | "data": {
645 | "text/plain": [
646 | ""
647 | ]
648 | },
649 | "metadata": {
650 | "tags": []
651 | },
652 | "execution_count": 105
653 | },
654 | {
655 | "output_type": "display_data",
656 | "data": {
657 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAALYAAAD8CAYAAADaM14OAAAABHNCSVQICAgIfAhkiAAAAAlwSFlz\nAAALEgAACxIB0t1+/AAAADl0RVh0U29mdHdhcmUAbWF0cGxvdGxpYiB2ZXJzaW9uIDMuMC4zLCBo\ndHRwOi8vbWF0cGxvdGxpYi5vcmcvnQurowAACzBJREFUeJzt3d+LXPUZx/H3p1lTq1KT2BK22bSm\nGJRQsJFQFL0oWsFaUS+kRLxYSiA3to1V0KT9C4Si5qIIi6nkQqo2ShNyodiYi15t3WipTWLMVqvZ\nkBilWot3wacX800dN7OZMzNnfuyznxcMO+fMmTkPJx8fv+ec2f0qIjDL5ivDLsCsHxxsS8nBtpQc\nbEvJwbaUHGxLycG2lHoKtqTbJB2TNCtpe11FmfVK3d6gkbQMeBu4FZgDXgPujYgj9ZVn1p2xHt77\nA2A2It4BkPQscBewYLAl+Tan9eqjiPhmu416GYqsAU40Lc+VdV8iaaukGUkzPezL7Jz3qmzUS8eu\nJCKmgClwx7bB6aVjnwTWNi1PlHVmQ9dLsF8D1ktaJ2k5sBnYV09ZZr3peigSEWcl/Rx4GVgG/D4i\nDtdWmVkPur7c19XOPMa23h2KiE3tNvKdR0vJwbaUHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2\nlBxsS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpQcbEvJwbaU\nHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpTaBlvSWkkHJR2RdFjStrJ+laRXJB0v\nP1f2v1yzaqp07LPAQxGxAbgeuF/SBmA7cCAi1gMHyrLZSGgb7Ig4FRGvl+f/BY7SmDP9LmB32Ww3\ncHe/ijTrVEdjbElXAhuBaWB1RJwqL50GVtdamVkPKs/MK+ky4AXggYj4VNL/X4uIWGhyUklbga29\nFmrWiUodW9JFNEL9TES8WFZ/IGm8vD4OnGn13oiYiohNVWZTNatLlasiAnYBRyPisaaX9gGT5fkk\nsLf+8sy603YudUk3AX8B3gQ+L6t/TWOc/TzwbeA94KcR8e82n+W51K1XleZSbxvsOjnYVoNKwfad\nR0vJwbaUHGxLycG2lBxsS8nBtpQcbEup8ndFbDha3Wdo/p6OteaObSm5Y4+oQd4Rzsgd21JysC0l\nD0VGXJUTxXPDFp9UfsEd21Jyxx5R7r69cce2lNyxE3B3P587tqXkYFtKDral5GBbSg52UhGxpL9v\n4mBbSr7cl9RSvwTojm0puWMncKGx9FLt3O7YlpI7diJLtTu34o5tKTnYlpKHIgmcG4I0n0Qu9WGJ\nO7al5GAn5VvqZgl5jJ3IUh9XN3PHtpQqB1vSMklvSNpfltdJmpY0K+k5Scv7V6ZZZzrp2NtozKN+\nzqPA4xFxFfAxsKXOwsx6UXVm3gngJ8BTZVnAzcCesslu4O5+FGjWjaod+wngYb6YwPQK4JOIOFuW\n54A1NddmHTp3ia/5sVRVmXL6DuBMRBzqZgeStkqakTTTzfvNulHlct+NwJ2SbgcuBr4O7ARWSBor\nXXsCONnqzRExBUzB4pyZ17epF6e2HTsidkTERERcCWwGXo2I+4CDwD1ls0lgb9+qNOtQL9exHwEe\nlDRLY8y9q56SFo9RG9NKOu+xVGmQ/xjZhiKe+GgoDkXEpnYb+c6jpeTvinRg2EON+TyTwcLcsS0l\nd+x5qnTBVr+x0sn7ezV/Hx7rn88d21Jyx17AhTrv/A5ZV3esegVmoU691Lt0M3dsS8kdu41WnfJC\nY9teNHfcbj7bt/+/4I5tKTnYlpKHIl2o+5Jep5frqlzuW+rcsS0ld+x55nfBVid0vXbq+Z9T5YZP\nFUv9hLGZO7al5I69gFbdr5cbIxe6wTK/c7fah3XGHdtScrAtJQ9FulDXZbYqQ5gqf/vaJ43nc8e2\nlNyxe9BJ565y2bDKCaZV445tKblj16yT32S/0OU+6407tqXkjl2DC91Y6fX3Et3Fu+OObSk52JaS\nhyJD4Et5/eeObSm5Y9dsfjf2t/SGwx3bUnLH7hOPo4fLHdtScsfusypfO7X6uWNbSg62peShyBD4\nhLL/3LEtpapzqa+QtEfSW5KOSrpB0ipJr0g6Xn6u7Hexi5mnqhusqh17J/BSRFwDXAscBbYDByJi\nPXCgLJuNhlaTcM6bkPNy4F3KnJBN648B4+X5OHCswmeFH370+Jhpl7OIqNSx1wEfAk9LekPSU5Iu\nBVZHxKmyzWlgdas3S9oqaUbSTIV9mdWiSrDHgOuAJyNiI/AZ84Yd0WjH0erNETEVEZuqzKZqVpcq\nwZ4D5iJiuizvoRH0DySNA5SfZ/pTolnn2gY7Ik4DJyRdXVbdAhwB9gGTZd0ksLcvFZp1oeoNml8A\nz0haDrwD/IzGfxTPS9oCvAf8tD8lmnVOg/wivKTB7cyyOlTlfM13Hi0lB9tScrAtJQfbUnKwLSUH\n21JysC0lB9tScrAtJQfbUnKwLSUH21JysC0lB9tScrAtJQfbUnKwLSUH21JysC0lB9tScrAtJQfb\nUnKwLSUH21JysC0lB9tScrAtJQfbUnKwLSUH21JysC0lB9tScrAtJQfbUnKwLSUH21KqFGxJv5J0\nWNI/JP1B0sWS1kmaljQr6bkyo5jZSGgbbElrgF8CmyLie8AyYDPwKPB4RFwFfAxs6WehZp2oOhQZ\nA74maQy4BDgF3Exjll6A3cDd9Zdn1p0qM/OeBH4LvE8j0P8BDgGfRMTZstkcsKZfRZp1qspQZCVw\nF7AO+BZwKXBb1R1I2ippRtJM11WadajKlNM/At6NiA8BJL0I3AiskDRWuvYEcLLVmyNiCpgq7/XM\nvDYQVcbY7wPXS7pEkoBbgCPAQeCess0ksLc/JZp1rsoYe5rGSeLrwJvlPVPAI8CDkmaBK4BdfazT\nrCOKGNzowEMRq8GhiNjUbiPfebSUHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpQc\nbEvJwbaUHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpQcbEvJwbaUHGxLycG2lBxs\nS8nBtpQcbEupyowGdfoI+Kz8XEy+weKrGRZn3e1q/k6VDxno38cGkDRT5e8bj5LFWDMszrrrqtlD\nEUvJwbaUhhHsqSHss1eLsWZYnHXXUvPAx9hmg+ChiKU0sGBLuk3SMUmzkrYPar+dkrRW0kFJRyQd\nlrStrF8l6RVJx8vPlcOudT5JyyS9IWl/WV4naboc8+ckLR92jc0krZC0R9Jbko5KuqGu4zyQYEta\nBvwO+DGwAbhX0oZB7LsLZ4GHImIDcD1wf6l1O3AgItYDB8ryqNkGHG1afhR4PCKuAj4GtgylqoXt\nBF6KiGuAa2nUXs9xjoi+P4AbgJeblncAOwax7xpq3wvcChwDxsu6ceDYsGubV+dECcLNwH5ANG50\njLX6Nxj2A7gceJdynte0vpbjPKihyBrgRNPyXFk30iRdCWwEpoHVEXGqvHQaWD2kshbyBPAw8HlZ\nvgL4JBpz3cPoHfN1wIfA02X49JSkS6npOPvkcQGSLgNeAB6IiE+bX4tGOxmZy0mS7gDORMShYdfS\ngTHgOuDJiNhI46sWXxp29HKcBxXsk8DapuWJsm4kSbqIRqifiYgXy+oPJI2X18eBM8Oqr4UbgTsl\n/Qt4lsZwZCewQtK57wON2jGfA+YiYros76ER9FqO86CC/RqwvpylLwc2A/sGtO+OSBKwCzgaEY81\nvbQPmCzPJ2mMvUdCROyIiImIuJLGsX01Iu4DDgL3lM1GrebTwAlJV5dVtwBHqOs4D/Bk4XbgbeCf\nwG+GffJygTpvovG/v78DfyuP22mMWQ8Ax4E/A6uGXesC9f8Q2F+efxf4KzAL/BH46rDrm1fr94GZ\ncqz/BKys6zj7zqOl5JNHS8nBtpQcbEvJwbaUHGxLycG2lBxsS8nBtpT+B2PtlHm34J5qAAAAAElF\nTkSuQmCC\n",
658 | "text/plain": [
659 | ""
660 | ]
661 | },
662 | "metadata": {
663 | "tags": []
664 | }
665 | }
666 | ]
667 | },
668 | {
669 | "cell_type": "markdown",
670 | "metadata": {
671 | "id": "FiGSgdqA5LGd",
672 | "colab_type": "text"
673 | },
674 | "source": [
675 | "One can clearly notice that there are now a lot of black pixels in the region where there should have been only white pixels. This can potentially hurt our model. So, it is best to not resize the image too much. But, due to computational constraints and the model requirements, it is unavoidable. \n",
676 | "\n",
677 | "However, given below are a few things one could try to reduce the downsampling noise as much as possible."
678 | ]
679 | },
680 | {
681 | "cell_type": "code",
682 | "metadata": {
683 | "id": "01FtsxQi6dts",
684 | "colab_type": "code",
685 | "colab": {}
686 | },
687 | "source": [
688 | "import cv2"
689 | ],
690 | "execution_count": 0,
691 | "outputs": []
692 | },
693 | {
694 | "cell_type": "markdown",
695 | "metadata": {
696 | "id": "vbflvVYqz2yu",
697 | "colab_type": "text"
698 | },
699 | "source": [
700 | "- Original Image > preprocess_label > Morphological Closing"
701 | ]
702 | },
703 | {
704 | "cell_type": "code",
705 | "metadata": {
706 | "id": "vFn6tiXPsUdl",
707 | "colab_type": "code",
708 | "colab": {
709 | "base_uri": "https://localhost:8080/",
710 | "height": 319
711 | },
712 | "outputId": "b7474d73-2f00-4c25-8968-4f740e5a2510"
713 | },
714 | "source": [
715 | "kernel = np.ones((3, 3))\n",
716 | "img_closed = cv2.morphologyEx(img, cv2.MORPH_CLOSE, kernel, iterations=3)\n",
717 | "print(np.unique(img_closed))\n",
718 | "print(img_closed.sum())\n",
719 | "plt.imshow(img_closed, cmap='Greys_r')"
720 | ],
721 | "execution_count": 97,
722 | "outputs": [
723 | {
724 | "output_type": "stream",
725 | "text": [
726 | "[0 1]\n",
727 | "491\n"
728 | ],
729 | "name": "stdout"
730 | },
731 | {
732 | "output_type": "execute_result",
733 | "data": {
734 | "text/plain": [
735 | ""
736 | ]
737 | },
738 | "metadata": {
739 | "tags": []
740 | },
741 | "execution_count": 97
742 | },
743 | {
744 | "output_type": "display_data",
745 | "data": {
746 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAQUAAAD8CAYAAAB+fLH0AAAABHNCSVQICAgIfAhkiAAAAAlwSFlz\nAAALEgAACxIB0t1+/AAAADl0RVh0U29mdHdhcmUAbWF0cGxvdGxpYiB2ZXJzaW9uIDMuMC4zLCBo\ndHRwOi8vbWF0cGxvdGxpYi5vcmcvnQurowAADTBJREFUeJzt3V+MXOV9xvHvUzuEQtTYppXl2LQ4\nwkqEIlHQqjIiFxEkKtAocIEiUKRYlSXfpA35IyWmveolUhRCJIRqBRK3QoTUQbXFRSLiULU3dVkn\nERgbYicUbMvGRAFS5QqLXy/mbDSvWbPL/Lf3+5GOZs6ZM3N+++7ss+/7nrOzqSokacEfTbsASbPF\nUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNcYSCkluSfJikmNJdo7jGJLGI6O+eCnJKuCXwKeAE8AzwN1V\ndXikB5I0FqvH8Jp/BRyrql8DJPk+cDtw3lBI4mWV0vj9pqr+bKmdxjF82Agc71s/0W1rJNmRZD7J\n/BhqkPROLy9np3H0FJalqnYBu8CegjRLxtFTOAlc2be+qdsm6QIwjlB4BtiSZHOSS4C7gH1jOI6k\nMRj58KGqzib5O+DHwCrgkap6ftTHkTQeIz8lOVARzilIk3CwquaW2skrGiU1DAVJDUNBUsNQkNQw\nFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUsNQ\nkNQwFCQ1DAVJDUNBUsNQkNQwFCQ1DAVJDUNBUmPgUEhyZZKnkxxO8nySe7rt65I8leRod7t2dOVK\nGrdhegpnga9W1TXAVuALSa4BdgL7q2oLsL9bl3SBGDgUqupUVf2su/9/wBFgI3A7sLvbbTdwx7BF\nSpqckcwpJLkKuA44AKyvqlPdQ6eB9aM4hqTJWD3sCyT5APBD4EtV9bskf3isqipJned5O4Adwx5f\n0mgN1VNI8j56gfBoVT3RbX41yYbu8Q3AmcWeW1W7qmququaGqUHSaA1z9iHAw8CRqvpm30P7gG3d\n/W3A3sHLkzRpqVq0d7/0E5OPA/8FPAe83W3+B3rzCj8A/hx4GfhsVf12idcarAhJ78XB5fTMBw6F\nUTIUpIlYVih4RaOkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoYCpIahoKk\nhqEgqWEoSGoYCpIahoKkxtCf5iwtWO6nePV/4rdmjz0FSQ1DQVLDUJDUcE5BIzfMnEH/vIRzD9Nh\nKGhkRv1DvBAQhsNkOXyQ1DAUJDUMBUkNQ0FSY+hQSLIqyc+TPNmtb05yIMmxJI8nuWT4MiVNyih6\nCvcAR/rW7wPur6qrgdeB7SM4hqQJGSoUkmwC/gb4Trce4CZgT7fLbuCOYY4habKG7Sl8C/ga8Ha3\nfgXwRlWd7dZPABuHPIakCRo4FJJ8GjhTVQcHfP6OJPNJ5getQStDVTWLxmuYKxpvBD6T5DbgUuBP\ngAeANUlWd72FTcDJxZ5cVbuAXQBJ/E5LM2LgnkJV3VtVm6rqKuAu4KdV9TngaeDObrdtwN6hq5Q0\nMeO4TuHrwFeSHKM3x/DwGI4haUwyC2M0hw9asJz3o38gNbCDVTW31E5e0SipYShIahgKkhqGgqSG\noSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIavi/JGfESv+/if7J9OwwFGbMIJ9v\n4Q+LRsnhg6SGPYWLwGK9iwul9zALn/yllj0FSQ17CpoKewizy56CpIY9hSmYxG/JWT7F+V6//ln8\nGi5m9hQkNewpTMHCb74LeVx9Ideud2dPQVLDUJDUcPig92SSwwYnGKdjqJ5CkjVJ9iR5IcmRJDck\nWZfkqSRHu9u1oypW0vgNO3x4APhRVX0UuBY4AuwE9lfVFmB/ty7pAjHwf51O8kHgF8CHq+9FkrwI\nfKKqTiXZAPxHVX1kiddakVPZk+iKj7oL7vDhgras/zo9zJzCZuA14LtJrgUOAvcA66vqVLfPaWD9\nEMe4qE3i1KSnDvVeDTN8WA1cDzxUVdcBv+ecoULXg1j0XZlkR5L5JPND1CBpxIYJhRPAiao60K3v\noRcSr3bDBrrbM4s9uap2VdXccrozF7sk71hWOttgegYOhao6DRxPsjBfcDNwGNgHbOu2bQP2DlWh\npIka9jqFvwceTXIJ8Gvgb+kFzQ+SbAdeBj475DFWpHN/Uzo3oEkZ+OzDSItYoWcfhjUL37t34xBg\n5izr7IOXOUtqGAqSGoaCpIahIKlhKEhq+KfTF7BJfoKTZxJWDnsKkhr2FC4C477QyV7CymJPQVLD\nnsJFaLHf7O+l92DPYGWzpyCpYShIajh8WCH8q0stlz0FSQ1DYYVyMlHnYyhIahgKkhqGgqSGZx9W\nMM9IaDGGgv7AyUeBwwdJ5zAUJDUMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSY6hQSPLlJM8nOZTksSSX\nJtmc5ECSY0ke7/4jtaQLxMChkGQj8EVgrqo+BqwC7gLuA+6vqquB14HtoyhU0mQMO3xYDfxxktXA\nZcAp4CZgT/f4buCOIY8haYIGDoWqOgl8A3iFXhi8CRwE3qiqs91uJ4CNiz0/yY4k80nmB61B0ugN\nM3xYC9wObAY+BFwO3LLc51fVrqqaq6q5QWuQNHrDDB8+CbxUVa9V1VvAE8CNwJpuOAGwCTg5ZI2S\nJmiYUHgF2JrksvT+5vZm4DDwNHBnt882YO9wJUqapGHmFA7Qm1D8GfBc91q7gK8DX0lyDLgCeHgE\ndUqakMzCp+0kmX4R0sXv4HLm8LyiUVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDU\nMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLD\nUJDUMBQkNZYMhSSPJDmT5FDftnVJnkpytLtd221Pkm8nOZbk2STXj7N4SaO3nJ7C94Bbztm2E9hf\nVVuA/d06wK3Alm7ZATw0mjIlTcqSoVBV/wn89pzNtwO7u/u7gTv6tv9L9fw3sCbJhlEVK2n8Bp1T\nWF9Vp7r7p4H13f2NwPG+/U50294hyY4k80nmB6xB0hisHvYFqqqS1ADP2wXsAhjk+ZLGY9CewqsL\nw4Lu9ky3/SRwZd9+m7ptki4Qg4bCPmBbd38bsLdv++e7sxBbgTf7hhmSLgRV9a4L8BhwCniL3hzB\nduAKemcdjgI/AdZ1+wZ4EPgV8Bwwt9Trd88rFxeXsS/zy/l5TPdDOVXOKUgTcbCq5pbaySsaJTUM\nBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1DAU\nJDUMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1DAUJDUMBUmN1dMuoPMb\n4Pfd7az4U6xnKbNWk/W8u79Yzk6pqnEXsixJ5qtqbtp1LLCepc1aTdYzGg4fJDUMBUmNWQqFXdMu\n4BzWs7RZq8l6RmBm5hQkzYZZ6ilImgFTD4UktyR5McmxJDunVMOVSZ5OcjjJ80nu6bavS/JUkqPd\n7doJ17Uqyc+TPNmtb05yoGurx5NcMsFa1iTZk+SFJEeS3DDN9kny5e57dSjJY0kunXT7JHkkyZkk\nh/q2Ldom6fl2V9uzSa4fZ23DmGooJFkFPAjcClwD3J3kmimUchb4alVdA2wFvtDVsRPYX1VbgP3d\n+iTdAxzpW78PuL+qrgZeB7ZPsJYHgB9V1UeBa7u6ptI+STYCXwTmqupjwCrgLibfPt8Dbjln2/na\n5FZgS7fsAB4ac22Dq6qpLcANwI/71u8F7p1mTV0de4FPAS8CG7ptG4AXJ1jDJnpvqpuAJ4HQuxBm\n9WJtN+ZaPgi8RDcH1bd9Ku0DbASOA+voXYD3JPDX02gf4Crg0FJtAvwzcPdi+83aMu3hw8I3d8GJ\nbtvUJLkKuA44AKyvqlPdQ6eB9RMs5VvA14C3u/UrgDeq6my3Psm22gy8Bny3G858J8nlTKl9quok\n8A3gFeAU8CZwkOm1T7/ztcnMvdfPZ9qhMFOSfAD4IfClqvpd/2PVi/eJnKpJ8mngTFUdnMTxlmE1\ncD3wUFVdR++S9GaoMOH2WQvcTi+sPgRczju78VM3yTYZpWmHwkngyr71Td22iUvyPnqB8GhVPdFt\nfjXJhu7xDcCZCZVzI/CZJP8LfJ/eEOIBYE2Shb9XmWRbnQBOVNWBbn0PvZCYVvt8Enipql6rqreA\nJ+i12bTap9/52mRm3utLmXYoPANs6WaNL6E3WbRv0kUkCfAwcKSqvtn30D5gW3d/G725hrGrqnur\nalNVXUWvTX5aVZ8DngbunEI9p4HjST7SbboZOMyU2ofesGFrksu6791CPVNpn3Ocr032AZ/vzkJs\nBd7sG2bMlmlPagC3Ab8EfgX845Rq+Di9bt6zwC+65TZ64/j9wFHgJ8C6KdT2CeDJ7v6Hgf8BjgH/\nBrx/gnX8JTDftdG/A2un2T7APwEvAIeAfwXeP+n2AR6jN6fxFr3e1PbztQm9ieIHu/f5c/TOnEz8\nvb6cxSsaJTWmPXyQNGMMBUkNQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1Ph/ynia1WfnoHgAAAAASUVO\nRK5CYII=\n",
747 | "text/plain": [
748 | ""
749 | ]
750 | },
751 | "metadata": {
752 | "tags": []
753 | }
754 | }
755 | ]
756 | },
757 | {
758 | "cell_type": "markdown",
759 | "metadata": {
760 | "id": "17CKK5zQz7hs",
761 | "colab_type": "text"
762 | },
763 | "source": [
764 | "- Original Image > preprocess_label > Morphological Dilation"
765 | ]
766 | },
767 | {
768 | "cell_type": "code",
769 | "metadata": {
770 | "id": "bxhKa3kKtCLZ",
771 | "colab_type": "code",
772 | "colab": {
773 | "base_uri": "https://localhost:8080/",
774 | "height": 319
775 | },
776 | "outputId": "7ee710b6-c5ed-4e9c-e2f0-935357e1bf0c"
777 | },
778 | "source": [
779 | "kernel = np.ones((3, 3))\n",
780 | "img_dilated = cv2.dilate(img, kernel, iterations=1)\n",
781 | "print(np.unique(img_dilated))\n",
782 | "print(img_dilated.sum())\n",
783 | "plt.imshow(img_dilated, cmap='Greys_r')"
784 | ],
785 | "execution_count": 98,
786 | "outputs": [
787 | {
788 | "output_type": "stream",
789 | "text": [
790 | "[0 1]\n",
791 | "660\n"
792 | ],
793 | "name": "stdout"
794 | },
795 | {
796 | "output_type": "execute_result",
797 | "data": {
798 | "text/plain": [
799 | ""
800 | ]
801 | },
802 | "metadata": {
803 | "tags": []
804 | },
805 | "execution_count": 98
806 | },
807 | {
808 | "output_type": "display_data",
809 | "data": {
810 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAAQUAAAD8CAYAAAB+fLH0AAAABHNCSVQICAgIfAhkiAAAAAlwSFlz\nAAALEgAACxIB0t1+/AAAADl0RVh0U29mdHdhcmUAbWF0cGxvdGxpYiB2ZXJzaW9uIDMuMC4zLCBo\ndHRwOi8vbWF0cGxvdGxpYi5vcmcvnQurowAADZlJREFUeJzt3V+MXOV9xvHvUzuEQtTYppXl2LQ4\nwkqEIqUgqzIiFxEkKtAocBFFoEixKku+SRvyR0pMe9VLpCiESAjVAhK3QoTUQcXiIhFxiNqbuqxD\nBMaG2AkFbNmYKECqXGHx68UcV/uatXc9f4/X3480mjlnzsz57buzz7zve87MpqqQpNP+aNYFSOoX\nQ0FSw1CQ1DAUJDUMBUkNQ0FSw1CQ1JhIKCS5OclLSY4k2TGJfUiajIz75KUkK4BfAZ8GjgLPAHdW\n1cGx7kjSRKycwHP+FXCkqn4DkOQHwG3AWUMhiadVSpP326r6s8U2msTwYT3w2rzlo926RpLtSeaS\nzE2gBknv9cpSNppET2FJqmonsBPsKUh9MomewjHgynnLG7p1ki4AkwiFZ4BNSTYmuQS4A9gzgf1I\nmoCxDx+q6lSSvwN+AqwAHq6qF8a9H0mTMfZDkkMV4ZyCNA37q2rzYht5RqOkhqEgqWEoSGoYCpIa\nhoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoY\nCpIahoKkhqEgqWEoSGoYCpIahoKkhqEgqWEoSGoMHQpJrkzydJKDSV5Icle3fk2Sp5Ic7q5Xj69c\nSZM2Sk/hFPD1qroG2AJ8Kck1wA5gb1VtAvZ2y5IuEEOHQlUdr6pfdLf/FzgErAduA3Z1m+0Cbh+1\nSEnTM5Y5hSRXAdcC+4C1VXW8u+sEsHYc+5A0HStHfYIkHwB+BHylqn6f5P/vq6pKUmd53HZg+6j7\nlzReI/UUkryPQSA8UlWPd6tfT7Kuu38dcHKhx1bVzqraXFWbR6lB0niNcvQhwEPAoar69ry79gBb\nu9tbgSeGL0/StKVqwd794g9MPgH8J/A88G63+h8YzCv8EPhz4BXg81X1u0Wea7giJJ2P/UvpmQ8d\nCuNkKEhTsaRQ8IxGSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwFSQ1DQVLDUJDUMBQkNQwF\nSQ1DQVLDUJDUGPk7GiWA8/1ejvnf5al+sacgqWEoSGoYCpIazimoF841J+H8w3TZU5DUMBTUe1V1\n3kc3NDyHD5oYu/0XJnsKkhr2FDQW9gqWD3sKkhqGgqTGyKGQZEWSZ5M82S1vTLIvyZEkjyW5ZPQy\nJU3LOHoKdwGH5i3fA9xbVVcDbwLbxrAPLXNJnJfoiZFCIckG4G+AB7vlADcCu7tNdgG3j7IPSdM1\nak/hO8A3gHe75SuAt6rqVLd8FFg/4j50ETlXj+H0SUyezDRZQ4dCks8AJ6tq/5CP355kLsncsDVo\n+fEPfvZGOU/hBuCzSW4FLgX+BLgPWJVkZddb2AAcW+jBVbUT2AmQxFeB1BND9xSq6u6q2lBVVwF3\nAD+rqi8ATwOf6zbbCjwxcpVa9uwh9MckzlP4JvC1JEcYzDE8NIF9SJqQ9CGdHT5omNehhzDP2/6q\n2rzYRp7RKKlhKEhqGAqSGn50Wr1w5vxAH+a6Llb2FCQ17CmoV+whzJ6hoF4wDPrD4YOkhqEgqWEo\nSGoYCpIahoKkhkcfemChmfeL5cM+HnXoH3sKkhr2FHrKjxJrVgyFZeRCGoaMMmzo68+0XDh8kNQw\nFHrgYnvnc3Kx3wwFSQ3nFKZoFu+Qp/fZh96IPYQLgz0FSQ17Cj1xoXzz0NnqOldPpK8/ixZmT0FS\nw56CxmIavYE+zItcDOwpSGrYU5ii0+90C72rnrnuXNsOs89ROS9w8TAUeso/Qs3KSMOHJKuS7E7y\nYpJDSa5PsibJU0kOd9erx1WspMkbdU7hPuDHVfVR4OPAIWAHsLeqNgF7u2VNWZJFDxMu9TJri/0s\nGq+h/+t0kg8CvwQ+XPOeJMlLwCer6niSdcDPq+ojizzX7F95U9SHmfrzqSHJTMPBQBibif/X6Y3A\nG8D3kjyb5MEklwNrq+p4t80JYO0I+9CY9eXdX/01SiisBK4DHqiqa4E/cMZQoetBLPgKTLI9yVyS\nuRFqkDRmo4TCUeBoVe3rlnczCInXu2ED3fXJhR5cVTuravNSujPLzaTHyAs9tz0ELdXQoVBVJ4DX\nkpyeL7gJOAjsAbZ267YCT4xUoaSpGvU8hb8HHklyCfAb4G8ZBM0Pk2wDXgE+P+I+lq2zvaOPatw9\ngln1MJxgnI2hjz6MtYiL7OjD+ejD72dUw36C0lAYu4kffZC0DHmac89NaojRF/YG+seegqSGoSCp\nYShIahgKOi9+OGn5MxQkNTz6cAE6n29lmtS3RNtbWL7sKUhq2FO4gC2lF2DPQOfLUFhGxn2ik0Fw\ncXL4IKlhKEhqGAqSGs4pLHMXyj+uVX/YU5DUMBQuMp6mrMUYCpIahoKkhhONF6n5QwgnHzWfPQVJ\nDXsKcuJRDXsKkhqGgqSGoSCpYShIahgKkhqGgqTGSKGQ5KtJXkhyIMmjSS5NsjHJviRHkjzW/Udq\nSReIoUMhyXrgy8DmqvoYsAK4A7gHuLeqrgbeBLaNo1BJ0zHq8GEl8MdJVgKXAceBG4Hd3f27gNtH\n3IekKRo6FKrqGPAt4FUGYfA2sB94q6pOdZsdBdYv9Pgk25PMJZkbtgZJ4zfK8GE1cBuwEfgQcDlw\n81IfX1U7q2pzVW0etgZJ4zfK8OFTwMtV9UZVvQM8DtwArOqGEwAbgGMj1ihpikYJhVeBLUkuy+AT\nNTcBB4Gngc9122wFnhitREnTNMqcwj4GE4q/AJ7vnmsn8E3ga0mOAFcAD42hTklTkj58wUaS2Rch\nLX/7lzKH5xmNkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqG\ngqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpsWgoJHk4\nyckkB+atW5PkqSSHu+vV3fok+W6SI0meS3LdJIuXNH5L6Sl8H7j5jHU7gL1VtQnY2y0D3AJs6i7b\ngQfGU6akaVk0FKrqP4DfnbH6NmBXd3sXcPu89f9SA/8FrEqyblzFSpq8YecU1lbV8e72CWBtd3s9\n8Nq87Y52694jyfYkc0nmhqxB0gSsHPUJqqqS1BCP2wnsBBjm8ZImY9iewuunhwXd9clu/THgynnb\nbejWSbpADBsKe4Ct3e2twBPz1n+xOwqxBXh73jBD0oWgqs55AR4FjgPvMJgj2AZcweCow2Hgp8Ca\nbtsA9wO/Bp4HNi/2/N3jyosXLxO/zC3l7zHdH+VMOacgTcX+qtq82Eae0SipYShIahgKkhqGgqSG\noSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqG\ngqSGoSCpYShIahgKkhqGgqSGoSCpYShIahgKkhqGgqSGoSCpYShIaqycdQGd3wJ/6K774k+xnsX0\nrSbrObe/WMpGqapJF7IkSeaqavOs6zjNehbXt5qsZzwcPkhqGAqSGn0KhZ2zLuAM1rO4vtVkPWPQ\nmzkFSf3Qp56CpB6YeSgkuTnJS0mOJNkxoxquTPJ0koNJXkhyV7d+TZKnkhzurldPua4VSZ5N8mS3\nvDHJvq6tHktyyRRrWZVkd5IXkxxKcv0s2yfJV7vf1YEkjya5dNrtk+ThJCeTHJi3bsE2ycB3u9qe\nS3LdJGsbxUxDIckK4H7gFuAa4M4k18yglFPA16vqGmAL8KWujh3A3qraBOztlqfpLuDQvOV7gHur\n6mrgTWDbFGu5D/hxVX0U+HhX10zaJ8l64MvA5qr6GLACuIPpt8/3gZvPWHe2NrkF2NRdtgMPTLi2\n4VXVzC7A9cBP5i3fDdw9y5q6Op4APg28BKzr1q0DXppiDRsYvKhuBJ4EwuBEmJULtd2Ea/kg8DLd\nHNS89TNpH2A98BqwhsEJeE8Cfz2L9gGuAg4s1ibAPwN3LrRd3y6zHj6c/uWedrRbNzNJrgKuBfYB\na6vqeHfXCWDtFEv5DvAN4N1u+Qrgrao61S1Ps602Am8A3+uGMw8muZwZtU9VHQO+BbwKHAfeBvYz\nu/aZ72xt0rvX+tnMOhR6JckHgB8BX6mq38+/rwbxPpVDNUk+A5ysqv3T2N8SrASuAx6oqmsZnJLe\nDBWm3D6rgdsYhNWHgMt5bzd+5qbZJuM061A4Blw5b3lDt27qkryPQSA8UlWPd6tfT7Kuu38dcHJK\n5dwAfDbJ/wA/YDCEuA9YleT051Wm2VZHgaNVta9b3s0gJGbVPp8CXq6qN6rqHeBxBm02q/aZ72xt\n0pvX+mJmHQrPAJu6WeNLGEwW7Zl2EUkCPAQcqqpvz7trD7C1u72VwVzDxFXV3VW1oaquYtAmP6uq\nLwBPA5+bQT0ngNeSfKRbdRNwkBm1D4Nhw5Ykl3W/u9P1zKR9znC2NtkDfLE7CrEFeHveMKNfZj2p\nAdwK/Ar4NfCPM6rhEwy6ec8Bv+wutzIYx+8FDgM/BdbMoLZPAk92tz8M/DdwBPg34P1TrOMvgbmu\njf4dWD3L9gH+CXgROAD8K/D+abcP8CiDOY13GPSmtp2tTRhMFN/fvc6fZ3DkZOqv9aVcPKNRUmPW\nwwdJPWMoSGoYCpIahoKkhqEgqWEoSGoYCpIahoKkxv8BvsUAzCLmrd4AAAAASUVORK5CYII=\n",
811 | "text/plain": [
812 | ""
813 | ]
814 | },
815 | "metadata": {
816 | "tags": []
817 | }
818 | }
819 | ]
820 | },
821 | {
822 | "cell_type": "markdown",
823 | "metadata": {
824 | "id": "gG3vfTyS0BVe",
825 | "colab_type": "text"
826 | },
827 | "source": [
828 | "You could try these things to get even better results."
829 | ]
830 | },
831 | {
832 | "cell_type": "markdown",
833 | "metadata": {
834 | "id": "CX6Cj1VO6vrZ",
835 | "colab_type": "text"
836 | },
837 | "source": [
838 | "## Feedback"
839 | ]
840 | },
841 | {
842 | "cell_type": "markdown",
843 | "metadata": {
844 | "id": "ZPt_15Ps6vO1",
845 | "colab_type": "text"
846 | },
847 | "source": [
848 | "If you have any feedback, queries, bug reports to send, please feel free to [raise an issue](https://github.com/IAmSuyogJadhav/3d-mri-brain-tumor-segmentation-using-autoencoder-regularization/issues/new) on github. It would be really helpful!"
849 | ]
850 | }
851 | ]
852 | }
853 |
--------------------------------------------------------------------------------