├── MANIFEST.in
├── manifest.json
├── src
├── sagemaker_defect_detection
│ ├── dataset
│ │ ├── __init__.py
│ │ └── neu.py
│ ├── models
│ │ ├── __init__.py
│ │ └── ddn.py
│ ├── __init__.py
│ ├── utils
│ │ ├── __init__.py
│ │ ├── coco_utils.py
│ │ ├── visualize.py
│ │ └── coco_eval.py
│ ├── transforms.py
│ └── classifier.py
├── prepare_data
│ ├── test_neu.py
│ └── neu.py
├── utils.py
├── prepare_RecordIO.py
└── xml2json.py
├── .gitattributes
├── cloudformation
├── solution-assistant
│ ├── requirements.in
│ ├── requirements.txt
│ ├── solution-assistant.yaml
│ └── src
│ │ └── lambda_fn.py
├── defect-detection-sagemaker-notebook-instance.yaml
├── defect-detection-permissions.yaml
└── defect-detection.yaml
├── .viperlightrc
├── NOTICE
├── docs
├── arch.png
├── data.png
├── sample1.png
├── sample2.png
├── sample3.png
├── data_flow.png
├── numerical.png
├── sagemaker.png
├── train_arch.png
└── launch.svg
├── requirements.in
├── .github
├── ISSUE_TEMPLATE
│ ├── questions-or-general-feedbacks.md
│ ├── bug_report.md
│ └── feature_request.md
└── workflows
│ ├── codeql.yaml
│ └── ci.yml
├── .viperlightignore
├── THIRD_PARTY
├── .gitignore
├── setup.cfg
├── CODE_OF_CONDUCT.md
├── scripts
├── train_classifier.sh
├── set_kernelspec.py
├── find_best_ckpt.py
├── build.sh
└── train_detector.sh
├── pyproject.toml
├── .pre-commit-config.yaml
├── setup.py
├── CONTRIBUTING.md
├── requirements.txt
├── LICENSE
├── notebooks
├── 3_classification_from_scratch.ipynb
├── 0_demo.ipynb
├── 1_retrain_from_checkpoint.ipynb
└── 2_detection_from_scratch.ipynb
└── README.md
/MANIFEST.in:
--------------------------------------------------------------------------------
1 | include requirements.txt
2 |
--------------------------------------------------------------------------------
/manifest.json:
--------------------------------------------------------------------------------
1 | {
2 | "version": "1.1.1"
3 | }
4 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/dataset/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/models/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/.gitattributes:
--------------------------------------------------------------------------------
1 | *.py text diff=python
2 | *.ipynb text
3 |
--------------------------------------------------------------------------------
/cloudformation/solution-assistant/requirements.in:
--------------------------------------------------------------------------------
1 | crhelper
2 |
--------------------------------------------------------------------------------
/.viperlightrc:
--------------------------------------------------------------------------------
1 | {
2 | "failOn": "medium",
3 | "all": true
4 | }
5 |
--------------------------------------------------------------------------------
/NOTICE:
--------------------------------------------------------------------------------
1 | Copyright Amazon.com, Inc. or its affiliates. All Rights Reserved.
2 |
--------------------------------------------------------------------------------
/docs/arch.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/arch.png
--------------------------------------------------------------------------------
/docs/data.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/data.png
--------------------------------------------------------------------------------
/docs/sample1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/sample1.png
--------------------------------------------------------------------------------
/docs/sample2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/sample2.png
--------------------------------------------------------------------------------
/docs/sample3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/sample3.png
--------------------------------------------------------------------------------
/docs/data_flow.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/data_flow.png
--------------------------------------------------------------------------------
/docs/numerical.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/numerical.png
--------------------------------------------------------------------------------
/docs/sagemaker.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/sagemaker.png
--------------------------------------------------------------------------------
/docs/train_arch.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/awslabs/sagemaker-defect-detection/HEAD/docs/train_arch.png
--------------------------------------------------------------------------------
/requirements.in:
--------------------------------------------------------------------------------
1 | patool
2 | pyunpack
3 | torch
4 | torchvision
5 | albumentations
6 | pytorch_lightning
7 | pycocotools
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/questions-or-general-feedbacks.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Questions or general feedbacks
3 | about: Question and feedbacks
4 | title: "[General]"
5 | labels: question
6 | assignees: ehsanmok
7 |
8 | ---
9 |
--------------------------------------------------------------------------------
/cloudformation/solution-assistant/requirements.txt:
--------------------------------------------------------------------------------
1 | #
2 | # This file is autogenerated by pip-compile
3 | # To update, run:
4 | #
5 | # pip-compile requirements.in
6 | #
7 | crhelper==2.0.6 # via -r requirements.in
8 |
--------------------------------------------------------------------------------
/.viperlightignore:
--------------------------------------------------------------------------------
1 | ^dist/
2 | docs/launch.svg:4
3 | CODE_OF_CONDUCT.md:4
4 | CONTRIBUTING.md:50
5 | build/solution-assistant.zip
6 | cloudformation/defect-detection-endpoint.yaml:42
7 | cloudformation/defect-detection-permissions.yaml:114
8 | docs/NEU_surface_defect_database.pdf
9 |
--------------------------------------------------------------------------------
/THIRD_PARTY:
--------------------------------------------------------------------------------
1 | Modifications Copyright Amazon.com, Inc. or its affiliates. All Rights Reserved.
2 | SPDX-License-Identifier: Apache-2.0
3 |
4 | Original Copyright 2016 Soumith Chintala. All Rights Reserved.
5 | SPDX-License-Identifier: BSD-3
6 | https://github.com/pytorch/vision/blob/master/LICENSE
7 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | *.egg-info
2 | __pycache__
3 | .vscode
4 | .mypy*
5 | .ipynb*
6 | .pytest*
7 | *output
8 | *logs*
9 | *checkpoints*
10 | *.ckpt*
11 | *.tar.gz
12 | dist
13 | build
14 | site-packages
15 | stack_outputs.json
16 | docs/*.py
17 | raw_neu_det
18 | neu_det
19 | raw_neu_cls
20 | neu_cls
21 |
--------------------------------------------------------------------------------
/setup.cfg:
--------------------------------------------------------------------------------
1 | [tool:pytest]
2 | norecursedirs =
3 | .git
4 | dist
5 | build
6 | python_files =
7 | test_*.py
8 |
9 | [metadata]
10 | # license_files = LICENSE
11 |
12 | [check-manifest]
13 | ignore =
14 | *.yaml
15 | .github
16 | .github/*
17 | build
18 | deploy
19 | notebook
20 |
--------------------------------------------------------------------------------
/CODE_OF_CONDUCT.md:
--------------------------------------------------------------------------------
1 | ## Code of Conduct
2 | This project has adopted the [Amazon Open Source Code of Conduct](https://aws.github.io/code-of-conduct).
3 | For more information see the [Code of Conduct FAQ](https://aws.github.io/code-of-conduct-faq) or contact
4 | opensource-codeofconduct@amazon.com with any additional questions or comments.
5 |
--------------------------------------------------------------------------------
/scripts/train_classifier.sh:
--------------------------------------------------------------------------------
1 | CLASSIFICATION_DATA_PATH=$1
2 | BACKBONE=$2
3 | BASE_DIR="$(dirname $(dirname $(readlink -f $0)))"
4 | CLASSIFIER_SCRIPT="${BASE_DIR}/src/sagemaker_defect_detection/classifier.py"
5 | MFN_LOGS="${BASE_DIR}/logs/"
6 |
7 | python ${CLASSIFIER_SCRIPT} \
8 | --data-path=${CLASSIFICATION_DATA_PATH} \
9 | --save-path=${MFN_LOGS} \
10 | --backbone=${BACKBONE} \
11 | --gpus=1 \
12 | --learning-rate=1e-3 \
13 | --epochs=50
14 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/bug_report.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Bug report
3 | about: Create a report to help us improve
4 | title: "[Bug]"
5 | labels: bug
6 | assignees: ehsanmok
7 |
8 | ---
9 |
10 | **Describe the bug**
11 | A clear and concise description of what the bug is.
12 |
13 | **To Reproduce**
14 | Steps to reproduce the behavior and error details
15 |
16 | **Expected behavior**
17 | A clear and concise description of what you expected to happen.
18 |
19 | **Additional context**
20 | Add any other context about the problem here.
21 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = [
3 | "setuptools",
4 | "wheel",
5 | ]
6 |
7 | [tool.black]
8 | line-length = 119
9 | exclude = '''
10 | (
11 | /(
12 | \.eggs
13 | | \.git
14 | | \.mypy_cache
15 | | build
16 | | dist
17 | )
18 | )
19 | '''
20 |
21 | [tool.portray]
22 | modules = ["src/sagemaker_defect_detection", "src/prepare_data/neu.py"]
23 | docs_dir = "docs"
24 |
25 | [tool.portray.mkdocs.theme]
26 | favicon = "docs/sagemaker.png"
27 | logo = "docs/sagemaker.png"
28 | name = "material"
29 | palette = {primary = "dark blue"}
30 |
--------------------------------------------------------------------------------
/docs/launch.svg:
--------------------------------------------------------------------------------
1 |
7 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/feature_request.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Feature request
3 | about: Suggest an idea for this project
4 | title: "[Feature]"
5 | labels: enhancement
6 | assignees: ehsanmok
7 |
8 | ---
9 |
10 | **Is your feature request related to a problem? Please describe.**
11 | A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
12 |
13 | **Describe the solution you'd like**
14 | A clear and concise description of what you want to happen.
15 |
16 | **Describe alternatives you've considered**
17 | A clear and concise description of any alternative solutions or features you've considered.
18 |
19 | **Additional context**
20 | Add any other context or screenshots about the feature request here.
21 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/__init__.py:
--------------------------------------------------------------------------------
1 | try:
2 | import pytorch_lightning
3 | except ModuleNotFoundError:
4 | print("installing the dependencies for sagemaker_defect_detection package ...")
5 | import subprocess
6 |
7 | subprocess.run(
8 | "python -m pip install -q albumentations==0.4.6 pytorch_lightning==0.8.5 pycocotools==2.0.1", shell=True
9 | )
10 |
11 | from sagemaker_defect_detection.models.ddn import Classification, Detection, RoI, RPN
12 | from sagemaker_defect_detection.dataset.neu import NEUCLS, NEUDET
13 | from sagemaker_defect_detection.transforms import get_transform, get_augmentation, get_preprocess
14 |
15 | __all__ = [
16 | "Classification",
17 | "Detection",
18 | "RoI",
19 | "RPN",
20 | "NEUCLS",
21 | "NEUDET",
22 | "get_transform",
23 | "get_augmentation",
24 | "get_preprocess",
25 | ]
26 |
--------------------------------------------------------------------------------
/scripts/set_kernelspec.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import json
3 |
4 |
5 | def set_kernel_spec(notebook_filepath, display_name, kernel_name):
6 | with open(notebook_filepath, "r") as openfile:
7 | notebook = json.load(openfile)
8 | kernel_spec = {"display_name": display_name, "language": "python", "name": kernel_name}
9 | if "metadata" not in notebook:
10 | notebook["metadata"] = {}
11 | notebook["metadata"]["kernelspec"] = kernel_spec
12 | with open(notebook_filepath, "w") as openfile:
13 | json.dump(notebook, openfile)
14 |
15 |
16 | if __name__ == "__main__":
17 | parser = argparse.ArgumentParser()
18 | parser.add_argument("--notebook")
19 | parser.add_argument("--display-name")
20 | parser.add_argument("--kernel")
21 | args = parser.parse_args()
22 | set_kernel_spec(args.notebook, args.display_name, args.kernel)
23 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | default_language_version:
2 | python: python3.6
3 |
4 | repos:
5 | - repo: https://github.com/pre-commit/pre-commit-hooks
6 | rev: v2.3.0
7 | hooks:
8 | - id: end-of-file-fixer
9 | - id: trailing-whitespace
10 | - repo: https://github.com/psf/black
11 | rev: 20.8b1
12 | hooks:
13 | - id: black
14 | - repo: https://github.com/kynan/nbstripout
15 | rev: 0.3.7
16 | hooks:
17 | - id: nbstripout
18 | name: nbstripout
19 | description: "nbstripout: strip output from Jupyter and IPython notebooks"
20 | entry: nbstripout notebooks
21 | language: python
22 | types: [jupyter]
23 | - repo: https://github.com/tomcatling/black-nb
24 | rev: 0.3.0
25 | hooks:
26 | - id: black-nb
27 | name: black-nb
28 | entry: black-nb
29 | language: python
30 | args: ["--include", '\.ipynb$']
31 |
--------------------------------------------------------------------------------
/.github/workflows/codeql.yaml:
--------------------------------------------------------------------------------
1 | name: "CodeQL"
2 |
3 | on:
4 | push:
5 | pull_request:
6 | schedule:
7 | - cron: '0 21 * * 1'
8 |
9 | jobs:
10 | CodeQL-Build:
11 |
12 | runs-on: ubuntu-latest
13 | strategy:
14 | fail-fast: false
15 | matrix:
16 | language: ['python']
17 |
18 | steps:
19 | - name: Checkout repository
20 | uses: actions/checkout@v2
21 | with:
22 | # We must fetch at least the immediate parents so that if this is
23 | # a pull request then we can checkout the head.
24 | fetch-depth: 2
25 |
26 | # If this run was triggered by a pull request event, then checkout
27 | # the head of the pull request instead of the merge commit.
28 | - run: git checkout HEAD^2
29 | if: ${{ github.event_name == 'pull_request' }}
30 |
31 | - name: Initialize CodeQL
32 | uses: github/codeql-action/init@v1
33 | with:
34 | languages: ${{ matrix.language }}
35 |
36 | - name: Perform CodeQL Analysis
37 | uses: github/codeql-action/analyze@v1
38 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | from setuptools import setup, find_packages
4 |
5 | ROOT = Path(__file__).parent.resolve()
6 |
7 | long_description = (ROOT / "README.md").read_text(encoding="utf-8")
8 |
9 | dev_dependencies = ["pre-commit", "mypy==0.781", "black==20.8b1", "nbstripout==0.3.7", "black-nb==0.3.0"]
10 | test_dependencies = ["pytest>=6.0"]
11 | doc_dependencies = ["portray>=1.4.0"]
12 |
13 | setup(
14 | name="sagemaker_defect_detection",
15 | version="0.1",
16 | description="Detect Defects in Products from their Images using Amazon SageMaker ",
17 | long_description=long_description,
18 | author="Ehsan M. Kermani",
19 | python_requires=">=3.6",
20 | package_dir={"": "src"},
21 | packages=find_packages("src", exclude=["tests", "tests/*"]),
22 | install_requires=open(str(ROOT / "requirements.txt"), "r").read(),
23 | extras_require={"dev": dev_dependencies, "test": test_dependencies, "doc": doc_dependencies},
24 | license="Apache License 2.0",
25 | classifiers=[
26 | "Development Status :: 3 - Alpha",
27 | "Programming Language :: Python :: 3.6",
28 | "Programming Language :: Python :: 3.7",
29 | "Programming Language :: Python :: 3.8",
30 | "Programming Language :: Python :: 3 :: Only",
31 | ],
32 | )
33 |
--------------------------------------------------------------------------------
/scripts/find_best_ckpt.py:
--------------------------------------------------------------------------------
1 | import re
2 | from pathlib import Path
3 |
4 |
5 | def get_score(s: str) -> float:
6 | """Gets the criterion score from .ckpt formated with ModelCheckpoint
7 |
8 | Parameters
9 | ----------
10 | s : str
11 | Assumption is the last number is the desired number carved in .ckpt
12 |
13 | Returns
14 | -------
15 | float
16 | The criterion float
17 | """
18 | return float(re.findall(r"(\d+.\d+).ckpt", s)[0])
19 |
20 |
21 | def main(path: str, op: str) -> str:
22 | """Finds the best ckpt path
23 |
24 | Parameters
25 | ----------
26 | path : str
27 | ckpt path
28 | op : str
29 | "max" (for mAP for example) or "min" (for loss)
30 |
31 | Returns
32 | -------
33 | str
34 | A ckpt path
35 | """
36 | ckpts = list(map(str, Path(path).glob("*.ckpt")))
37 | if not len(ckpts):
38 | return
39 |
40 | ckpt_score_dict = {ckpt: get_score(ckpt) for ckpt in ckpts}
41 | op = max if op == "max" else min
42 | out = op(ckpt_score_dict, key=ckpt_score_dict.get)
43 | print(out) # need to flush for bash
44 | return out
45 |
46 |
47 | if __name__ == "__main__":
48 | import sys
49 |
50 | if len(sys.argv) < 3:
51 | print("provide checkpoint path and op either max or min")
52 | sys.exit(1)
53 |
54 | main(sys.argv[1], sys.argv[2])
55 |
--------------------------------------------------------------------------------
/src/prepare_data/test_neu.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 | import pytest
4 |
5 | import neu
6 |
7 |
8 | def tmp_fill(tmpdir_, classes, ext):
9 | for cls_ in classes.values():
10 | tmpdir_.join(cls_ + "_0" + ext)
11 | # add one more image
12 | tmpdir_.join(classes["crazing"] + "_1" + ext)
13 | return
14 |
15 |
16 | @pytest.fixture()
17 | def tmp_neu():
18 | def _create(tmpdir, filename):
19 | tmpneu = tmpdir.mkdir(filename)
20 | if filename == "NEU-CLS":
21 | tmp_fill(tmpneu, neu.CLASSES, ".png")
22 | elif filename == "NEU-DET":
23 | imgs = tmpneu.mkdir("IMAGES")
24 | tmp_fill(imgs, neu.CLASSES, ".png")
25 | anns = tmpneu.mkdir("ANNOTATIONS")
26 | tmp_fill(anns, neu.CLASSES, ".xml")
27 | else:
28 | raise ValueError("Not supported")
29 | return tmpneu
30 |
31 | return _create
32 |
33 |
34 | @pytest.mark.parametrize("filename", ["NEU-CLS", "NEU-DET"])
35 | def test_main(tmpdir, tmp_neu, filename) -> None:
36 | data_path = tmp_neu(tmpdir, filename)
37 | output_path = tmpdir.mkdir("output_path")
38 | neu.main(data_path, output_path, archived=False)
39 | assert len(os.listdir(output_path)) == len(neu.CLASSES), "failed to match number of classes in output"
40 | for p in output_path.visit():
41 | assert p.check(), "correct path was not created"
42 | return
43 |
--------------------------------------------------------------------------------
/cloudformation/solution-assistant/solution-assistant.yaml:
--------------------------------------------------------------------------------
1 | AWSTemplateFormatVersion: "2010-09-09"
2 | Description: "(SA0015) - sagemaker-defect-detection solution assistant stack"
3 |
4 | Parameters:
5 | SolutionPrefix:
6 | Type: String
7 | SolutionName:
8 | Type: String
9 | StackName:
10 | Type: String
11 | S3Bucket:
12 | Type: String
13 | SolutionS3Bucket:
14 | Type: String
15 | RoleArn:
16 | Type: String
17 |
18 | Mappings:
19 | Function:
20 | SolutionAssistant:
21 | S3Key: "build/solution-assistant.zip"
22 |
23 | Resources:
24 | SolutionAssistant:
25 | Type: "Custom::SolutionAssistant"
26 | Properties:
27 | SolutionPrefix: !Ref SolutionPrefix
28 | ServiceToken: !GetAtt SolutionAssistantLambda.Arn
29 | S3Bucket: !Ref S3Bucket
30 | StackName: !Ref StackName
31 |
32 | SolutionAssistantLambda:
33 | Type: AWS::Lambda::Function
34 | Properties:
35 | Handler: "lambda_fn.handler"
36 | FunctionName: !Sub "${SolutionPrefix}-solution-assistant"
37 | Role: !Ref RoleArn
38 | Runtime: "python3.8"
39 | Code:
40 | S3Bucket: !Ref SolutionS3Bucket
41 | S3Key: !Sub
42 | - "${SolutionName}/${LambdaS3Key}"
43 | - LambdaS3Key: !FindInMap [Function, SolutionAssistant, S3Key]
44 | Timeout: 60
45 | Metadata:
46 | cfn_nag:
47 | rules_to_suppress:
48 | - id: W58
49 | reason: >-
50 | The required permissions are provided in the permissions stack.
51 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/utils/__init__.py:
--------------------------------------------------------------------------------
1 | from typing import Optional, Union
2 | from pathlib import Path
3 | import tarfile
4 | import logging
5 | from logging.config import fileConfig
6 |
7 | import torch
8 | import torch.nn as nn
9 |
10 |
11 | logger = logging.getLogger(__name__)
12 | logger.setLevel(logging.INFO)
13 |
14 |
15 | def get_logger(config_path: str) -> logging.Logger:
16 | fileConfig(config_path, disable_existing_loggers=False)
17 | logger = logging.getLogger()
18 | return logger
19 |
20 |
21 | def str2bool(flag: Union[str, bool]) -> bool:
22 | if not isinstance(flag, bool):
23 | if flag.lower() == "false":
24 | flag = False
25 | elif flag.lower() == "true":
26 | flag = True
27 | else:
28 | raise ValueError("Wrong boolean argument!")
29 | return flag
30 |
31 |
32 | def freeze(m: nn.Module) -> None:
33 | assert isinstance(m, nn.Module), "freeze only is applied to modules"
34 | for param in m.parameters():
35 | param.requires_grad = False
36 |
37 | return
38 |
39 |
40 | def load_checkpoint(model: nn.Module, path: str, prefix: Optional[str]) -> nn.Module:
41 | path = Path(path)
42 | logger.info(f"path: {path}")
43 | if path.is_dir():
44 | path_str = str(list(path.rglob("*.ckpt"))[0])
45 | else:
46 | path_str = str(path)
47 |
48 | device = "cuda" if torch.cuda.is_available() else "cpu"
49 | state_dict = torch.load(path_str, map_location=torch.device(device))["state_dict"]
50 | if prefix is not None:
51 | if prefix[-1] != ".":
52 | prefix += "."
53 |
54 | state_dict = {k[len(prefix) :]: v for k, v in state_dict.items() if k.startswith(prefix)}
55 |
56 | model.load_state_dict(state_dict, strict=True)
57 | return model
58 |
--------------------------------------------------------------------------------
/.github/workflows/ci.yml:
--------------------------------------------------------------------------------
1 | name: CI
2 |
3 | on:
4 | pull_request:
5 | push:
6 | branches: [ mainline ]
7 |
8 | jobs:
9 | build:
10 |
11 | strategy:
12 | max-parallel: 4
13 | fail-fast: false
14 | matrix:
15 | python-version: [3.7, 3.8]
16 | platform: [ubuntu-latest]
17 |
18 | runs-on: ${{ matrix.platform }}
19 |
20 | steps:
21 | - uses: actions/checkout@v2
22 | - name: Set up Python ${{ matrix.python-version }}
23 | uses: actions/setup-python@v2
24 | with:
25 | python-version: ${{ matrix.python-version }}
26 | - name: CloudFormation lint
27 | run: |
28 | python -m pip install --upgrade pip
29 | python -m pip install cfn-lint
30 | for y in `find deploy/* -name "*.yaml" -o -name "*.template" -o -name "*.json"`; do
31 | echo "============= $y ================"
32 | cfn-lint --fail-on-warnings $y || ec1=$?
33 | done
34 | if [ "$ec1" -ne "0" ]; then echo 'ERROR-1'; else echo 'SUCCESS-1'; ec1=0; fi
35 | echo "Exit Code 1 `echo $ec1`"
36 | if [ "$ec1" -ne "0" ]; then echo 'ERROR'; ec=1; else echo 'SUCCESS'; ec=0; fi;
37 | echo "Exit Code Final `echo $ec`"
38 | exit $ec
39 | - name: Build the package
40 | run: |
41 | python -m pip install -e '.[dev,test,doc]'
42 | - name: Code style check
43 | run: |
44 | black --check src
45 | - name: Notebook style check
46 | run: |
47 | black-nb notebooks/*.ipynb --check
48 | - name: Type check
49 | run: |
50 | mypy --ignore-missing-imports --allow-redefinition --pretty --show-error-context src/
51 | - name: Run tests
52 | run: |
53 | pytest --pyargs src/ -s
54 | - name: Update docs
55 | run: |
56 | portray on_github_pages -m "Update gh-pages" -f
57 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/utils/coco_utils.py:
--------------------------------------------------------------------------------
1 | """
2 | BSD 3-Clause License
3 |
4 | Copyright (c) Soumith Chintala 2016,
5 | All rights reserved.
6 |
7 | Redistribution and use in source and binary forms, with or without
8 | modification, are permitted provided that the following conditions are met:
9 |
10 | * Redistributions of source code must retain the above copyright notice, this
11 | list of conditions and the following disclaimer.
12 |
13 | * Redistributions in binary form must reproduce the above copyright notice,
14 | this list of conditions and the following disclaimer in the documentation
15 | and/or other materials provided with the distribution.
16 |
17 | * Neither the name of the copyright holder nor the names of its
18 | contributors may be used to endorse or promote products derived from
19 | this software without specific prior written permission.
20 |
21 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
22 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
23 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
24 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
25 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
26 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
27 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
28 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
29 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
30 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
31 |
32 |
33 | This module is a modified version of https://github.com/pytorch/vision/tree/03b1d38ba3c67703e648fb067570eeb1a1e61265/references/detection
34 | """
35 |
36 | from pycocotools.coco import COCO
37 |
38 |
39 | def convert_to_coco_api(ds):
40 | coco_ds = COCO()
41 | # annotation IDs need to start at 1, not 0, see torchvision issue #1530
42 | ann_id = 1
43 | dataset = {"images": [], "categories": [], "annotations": []}
44 | categories = set()
45 | for img_idx in range(len(ds)):
46 | # find better way to get target
47 | # targets = ds.get_annotations(img_idx)
48 | img, targets, _ = ds[img_idx]
49 | image_id = targets["image_id"].item()
50 | img_dict = {}
51 | img_dict["id"] = image_id
52 | img_dict["height"] = img.shape[-2]
53 | img_dict["width"] = img.shape[-1]
54 | dataset["images"].append(img_dict)
55 | bboxes = targets["boxes"]
56 | bboxes[:, 2:] -= bboxes[:, :2]
57 | bboxes = bboxes.tolist()
58 | labels = targets["labels"].tolist()
59 | areas = targets["area"].tolist()
60 | iscrowd = targets["iscrowd"].tolist()
61 | num_objs = len(bboxes)
62 | for i in range(num_objs):
63 | ann = {}
64 | ann["image_id"] = image_id
65 | ann["bbox"] = bboxes[i]
66 | ann["category_id"] = labels[i]
67 | categories.add(labels[i])
68 | ann["area"] = areas[i]
69 | ann["iscrowd"] = iscrowd[i]
70 | ann["id"] = ann_id
71 | dataset["annotations"].append(ann)
72 | ann_id += 1
73 | dataset["categories"] = [{"id": i} for i in sorted(categories)]
74 | coco_ds.dataset = dataset
75 | coco_ds.createIndex()
76 | return coco_ds
77 |
--------------------------------------------------------------------------------
/CONTRIBUTING.md:
--------------------------------------------------------------------------------
1 | # Contributing Guidelines
2 |
3 | Thank you for your interest in contributing to our project. Whether it's a bug report, new feature, correction, or additional
4 | documentation, we greatly value feedback and contributions from our community.
5 |
6 | Please read through this document before submitting any issues or pull requests to ensure we have all the necessary
7 | information to effectively respond to your bug report or contribution.
8 |
9 |
10 | ## Reporting Bugs/Feature Requests
11 |
12 | We welcome you to use the GitHub issue tracker to report bugs or suggest features.
13 |
14 | When filing an issue, please check existing open, or recently closed, issues to make sure somebody else hasn't already
15 | reported the issue. Please try to include as much information as you can. Details like these are incredibly useful:
16 |
17 | * A reproducible test case or series of steps
18 | * The version of our code being used
19 | * Any modifications you've made relevant to the bug
20 | * Anything unusual about your environment or deployment
21 |
22 |
23 | ## Contributing via Pull Requests
24 | Contributions via pull requests are much appreciated. Before sending us a pull request, please ensure that:
25 |
26 | 1. You are working against the latest source on the *mainline* branch.
27 | 2. You check existing open, and recently merged, pull requests to make sure someone else hasn't addressed the problem already.
28 | 3. You open an issue to discuss any significant work - we would hate for your time to be wasted.
29 |
30 | To send us a pull request, please:
31 |
32 | 1. Fork the repository.
33 | 2. Modify the source; please focus on the specific change you are contributing. If you also reformat all the code, it will be hard for us to focus on your change.
34 | 3. Ensure local tests pass.
35 | 4. Commit to your fork using clear commit messages.
36 | 5. Send us a pull request, answering any default questions in the pull request interface.
37 | 6. Pay attention to any automated CI failures reported in the pull request, and stay involved in the conversation.
38 |
39 | GitHub provides additional document on [forking a repository](https://help.github.com/articles/fork-a-repo/) and
40 | [creating a pull request](https://help.github.com/articles/creating-a-pull-request/).
41 |
42 |
43 | ## Finding contributions to work on
44 | Looking at the existing issues is a great way to find something to contribute on. As our projects, by default, use the default GitHub issue labels (enhancement/bug/duplicate/help wanted/invalid/question/wontfix), looking at any 'help wanted' issues is a great place to start.
45 |
46 |
47 | ## Code of Conduct
48 | This project has adopted the [Amazon Open Source Code of Conduct](https://aws.github.io/code-of-conduct).
49 | For more information see the [Code of Conduct FAQ](https://aws.github.io/code-of-conduct-faq) or contact
50 | opensource-codeofconduct@amazon.com with any additional questions or comments.
51 |
52 |
53 | ## Security issue notifications
54 | If you discover a potential security issue in this project we ask that you notify AWS/Amazon Security via our [vulnerability reporting page](http://aws.amazon.com/security/vulnerability-reporting/). Please do **not** create a public github issue.
55 |
56 |
57 | ## Licensing
58 |
59 | See the [LICENSE](LICENSE) file for our project's licensing. We will ask you to confirm the licensing of your contribution.
60 |
61 | We may ask you to sign a [Contributor License Agreement (CLA)](http://en.wikipedia.org/wiki/Contributor_License_Agreement) for larger changes.
62 |
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | #
2 | # This file is autogenerated by pip-compile with python 3.8
3 | # To update, run:
4 | #
5 | # pip-compile requirements.in
6 | #
7 | absl-py
8 | # via tensorboard
9 | albumentations
10 | # via -r requirements.in
11 | cachetools
12 | # via google-auth
13 | certifi
14 | # via requests
15 | charset-normalizer
16 | # via requests
17 | cycler
18 | # via matplotlib
19 | cython
20 | # via pycocotools
21 | easyprocess
22 | # via pyunpack
23 | entrypoint2
24 | # via pyunpack
25 | fonttools
26 | # via matplotlib
27 | future
28 | # via
29 | # pytorch-lightning
30 | # torch
31 | google-auth
32 | # via
33 | # google-auth-oauthlib
34 | # tensorboard
35 | google-auth-oauthlib
36 | # via tensorboard
37 | grpcio
38 | # via tensorboard
39 | idna
40 | # via requests
41 | imageio
42 | # via
43 | # imgaug
44 | # scikit-image
45 | imgaug
46 | # via albumentations
47 | importlib-metadata
48 | # via markdown
49 | kiwisolver
50 | # via matplotlib
51 | markdown
52 | # via tensorboard
53 | matplotlib
54 | # via
55 | # imgaug
56 | # pycocotools
57 | networkx
58 | # via scikit-image
59 | numpy
60 | # via
61 | # albumentations
62 | # imageio
63 | # imgaug
64 | # matplotlib
65 | # opencv-python
66 | # opencv-python-headless
67 | # pytorch-lightning
68 | # pywavelets
69 | # scikit-image
70 | # scipy
71 | # tensorboard
72 | # tifffile
73 | # torch
74 | # torchvision
75 | oauthlib
76 | # via requests-oauthlib
77 | opencv-python
78 | # via imgaug
79 | opencv-python-headless
80 | # via albumentations
81 | packaging
82 | # via
83 | # matplotlib
84 | # scikit-image
85 | patool
86 | # via -r requirements.in
87 | pillow
88 | # via
89 | # imageio
90 | # imgaug
91 | # matplotlib
92 | # scikit-image
93 | # torchvision
94 | protobuf
95 | # via tensorboard
96 | pyasn1
97 | # via
98 | # pyasn1-modules
99 | # rsa
100 | pyasn1-modules
101 | # via google-auth
102 | pycocotools
103 | # via -r requirements.in
104 | pyparsing
105 | # via
106 | # matplotlib
107 | # packaging
108 | python-dateutil
109 | # via matplotlib
110 | pytorch-lightning
111 | # via -r requirements.in
112 | pyunpack
113 | # via -r requirements.in
114 | pywavelets
115 | # via scikit-image
116 | pyyaml
117 | # via
118 | # albumentations
119 | # pytorch-lightning
120 | requests
121 | # via
122 | # requests-oauthlib
123 | # tensorboard
124 | requests-oauthlib
125 | # via google-auth-oauthlib
126 | rsa
127 | # via google-auth
128 | scikit-image
129 | # via imgaug
130 | scipy
131 | # via
132 | # albumentations
133 | # imgaug
134 | # scikit-image
135 | shapely
136 | # via imgaug
137 | six
138 | # via
139 | # google-auth
140 | # grpcio
141 | # imgaug
142 | # python-dateutil
143 | tensorboard
144 | # via pytorch-lightning
145 | tensorboard-data-server
146 | # via tensorboard
147 | tensorboard-plugin-wit
148 | # via tensorboard
149 | tifffile
150 | # via scikit-image
151 | torch
152 | # via
153 | # -r requirements.in
154 | # pytorch-lightning
155 | # torchvision
156 | torchvision
157 | # via -r requirements.in
158 | tqdm
159 | # via pytorch-lightning
160 | urllib3
161 | # via requests
162 | werkzeug
163 | # via tensorboard
164 | wheel
165 | # via tensorboard
166 | zipp
167 | # via importlib-metadata
168 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/transforms.py:
--------------------------------------------------------------------------------
1 | from typing import Callable
2 | import torchvision.transforms as transforms
3 |
4 | import albumentations as albu
5 | import albumentations.pytorch.transforms as albu_transforms
6 |
7 |
8 | PROBABILITY = 0.5
9 | ROTATION_ANGLE = 90
10 | NUM_CHANNELS = 3
11 | # required for resnet
12 | IMAGE_RESIZE_HEIGHT = 256
13 | IMAGE_RESIZE_WIDTH = 256
14 | IMAGE_HEIGHT = 224
15 | IMAGE_WIDTH = 224
16 | # standard imagenet1k mean and standard deviation of RGB channels
17 | MEAN_RED = 0.485
18 | MEAN_GREEN = 0.456
19 | MEAN_BLUE = 0.406
20 | STD_RED = 0.229
21 | STD_GREEN = 0.224
22 | STD_BLUE = 0.225
23 |
24 |
25 | def get_transform(split: str) -> Callable:
26 | """
27 | Image data transformations such as normalization for train split for classification task
28 |
29 | Parameters
30 | ----------
31 | split : str
32 | train or else
33 |
34 | Returns
35 | -------
36 | Callable
37 | Image transformation function
38 | """
39 | normalize = transforms.Normalize(mean=[MEAN_RED, MEAN_GREEN, MEAN_BLUE], std=[STD_RED, STD_GREEN, STD_BLUE])
40 | if split == "train":
41 | return transforms.Compose(
42 | [
43 | transforms.RandomResizedCrop(IMAGE_HEIGHT),
44 | transforms.RandomRotation(ROTATION_ANGLE),
45 | transforms.RandomHorizontalFlip(),
46 | transforms.ToTensor(),
47 | normalize,
48 | ]
49 | )
50 |
51 | else:
52 | return transforms.Compose(
53 | [
54 | transforms.Resize(IMAGE_RESIZE_HEIGHT),
55 | transforms.CenterCrop(IMAGE_HEIGHT),
56 | transforms.ToTensor(),
57 | normalize,
58 | ]
59 | )
60 |
61 |
62 | def get_augmentation(split: str) -> Callable:
63 | """
64 | Obtains proper image augmentation in train split for detection task.
65 | We have splitted transformations done for detection task into augmentation and preprocessing
66 | for clarity
67 |
68 | Parameters
69 | ----------
70 | split : str
71 | train or else
72 |
73 | Returns
74 | -------
75 | Callable
76 | Image augmentation function
77 | """
78 | if split == "train":
79 | return albu.Compose(
80 | [
81 | albu.Resize(IMAGE_RESIZE_HEIGHT, IMAGE_RESIZE_WIDTH, always_apply=True),
82 | albu.RandomCrop(IMAGE_HEIGHT, IMAGE_WIDTH, always_apply=True),
83 | albu.RandomRotate90(p=PROBABILITY),
84 | albu.HorizontalFlip(p=PROBABILITY),
85 | albu.RandomBrightness(p=PROBABILITY),
86 | ],
87 | bbox_params=albu.BboxParams(
88 | format="pascal_voc",
89 | label_fields=["labels"],
90 | min_visibility=0.2,
91 | ),
92 | )
93 | else:
94 | return albu.Compose(
95 | [albu.Resize(IMAGE_HEIGHT, IMAGE_WIDTH)],
96 | bbox_params=albu.BboxParams(format="pascal_voc", label_fields=["labels"]),
97 | )
98 |
99 |
100 | def get_preprocess() -> Callable:
101 | """
102 | Image normalization using albumentation for detection task that aligns well with image augmentation
103 |
104 | Returns
105 | -------
106 | Callable
107 | Image normalization function
108 | """
109 | return albu.Compose(
110 | [
111 | albu.Normalize(mean=[MEAN_RED, MEAN_GREEN, MEAN_BLUE], std=[STD_RED, STD_GREEN, STD_BLUE]),
112 | albu_transforms.ToTensorV2(),
113 | ]
114 | )
115 |
--------------------------------------------------------------------------------
/cloudformation/defect-detection-sagemaker-notebook-instance.yaml:
--------------------------------------------------------------------------------
1 | AWSTemplateFormatVersion: "2010-09-09"
2 | Description: "(SA0015) - sagemaker-defect-detection notebook stack"
3 | Parameters:
4 | SolutionPrefix:
5 | Type: String
6 | SolutionName:
7 | Type: String
8 | S3Bucket:
9 | Type: String
10 | SageMakerIAMRoleArn:
11 | Type: String
12 | SageMakerNotebookInstanceType:
13 | Type: String
14 | StackVersion:
15 | Type: String
16 |
17 | Mappings:
18 | S3:
19 | release:
20 | BucketPrefix: "sagemaker-solutions-prod"
21 | development:
22 | BucketPrefix: "sagemaker-solutions-devo"
23 |
24 | Resources:
25 | NotebookInstance:
26 | Type: AWS::SageMaker::NotebookInstance
27 | Properties:
28 | DirectInternetAccess: Enabled
29 | InstanceType: !Ref SageMakerNotebookInstanceType
30 | LifecycleConfigName: !GetAtt LifeCycleConfig.NotebookInstanceLifecycleConfigName
31 | NotebookInstanceName: !Sub "${SolutionPrefix}"
32 | RoleArn: !Sub "${SageMakerIAMRoleArn}"
33 | VolumeSizeInGB: 100
34 | Metadata:
35 | cfn_nag:
36 | rules_to_suppress:
37 | - id: W1201
38 | reason: Solution does not have KMS encryption enabled by default
39 | LifeCycleConfig:
40 | Type: AWS::SageMaker::NotebookInstanceLifecycleConfig
41 | Properties:
42 | NotebookInstanceLifecycleConfigName: !Sub "${SolutionPrefix}-nb-lifecycle-config"
43 | OnStart:
44 | - Content:
45 | Fn::Base64: |
46 | set -e
47 | sudo -u ec2-user -i <> stack_outputs.json
69 | echo ' "AccountID": "${AWS::AccountId}",' >> stack_outputs.json
70 | echo ' "AWSRegion": "${AWS::Region}",' >> stack_outputs.json
71 | echo ' "IamRole": "${SageMakerIAMRoleArn}",' >> stack_outputs.json
72 | echo ' "SolutionPrefix": "${SolutionPrefix}",' >> stack_outputs.json
73 | echo ' "SolutionName": "${SolutionName}",' >> stack_outputs.json
74 | echo ' "SolutionS3Bucket": "${SolutionsRefBucketBase}",' >> stack_outputs.json
75 | echo ' "S3Bucket": "${S3Bucket}"' >> stack_outputs.json
76 | echo '}' >> stack_outputs.json
77 | cat stack_outputs.json
78 | sudo chown -R ec2-user:ec2-user *
79 | EOF
80 | - SolutionsRefBucketBase:
81 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
82 |
83 | Outputs:
84 | SourceCode:
85 | Description: "Open Jupyter IDE. This authenticate you against Jupyter."
86 | Value: !Sub "https://${NotebookInstance.NotebookInstanceName}.notebook.${AWS::Region}.sagemaker.aws/"
87 |
88 | SageMakerNotebookInstanceSignOn:
89 | Description: "Link to the SageMaker notebook instance"
90 | Value: !Sub "https://${NotebookInstance.NotebookInstanceName}.notebook.${AWS::Region}.sagemaker.aws/notebooks/notebooks"
91 |
--------------------------------------------------------------------------------
/cloudformation/solution-assistant/src/lambda_fn.py:
--------------------------------------------------------------------------------
1 | import boto3
2 | import sys
3 |
4 | sys.path.append("./site-packages")
5 | from crhelper import CfnResource
6 |
7 | helper = CfnResource()
8 |
9 |
10 | @helper.create
11 | def on_create(_, __):
12 | pass
13 |
14 |
15 | @helper.update
16 | def on_update(_, __):
17 | pass
18 |
19 |
20 | def delete_sagemaker_endpoint(endpoint_name):
21 | sagemaker_client = boto3.client("sagemaker")
22 | try:
23 | sagemaker_client.delete_endpoint(EndpointName=endpoint_name)
24 | print("Successfully deleted endpoint " "called '{}'.".format(endpoint_name))
25 | except sagemaker_client.exceptions.ClientError as e:
26 | if "Could not find endpoint" in str(e):
27 | print("Could not find endpoint called '{}'. " "Skipping delete.".format(endpoint_name))
28 | else:
29 | raise e
30 |
31 |
32 | def delete_sagemaker_endpoint_config(endpoint_config_name):
33 | sagemaker_client = boto3.client("sagemaker")
34 | try:
35 | sagemaker_client.delete_endpoint_config(EndpointConfigName=endpoint_config_name)
36 | print("Successfully deleted endpoint configuration " "called '{}'.".format(endpoint_config_name))
37 | except sagemaker_client.exceptions.ClientError as e:
38 | if "Could not find endpoint configuration" in str(e):
39 | print(
40 | "Could not find endpoint configuration called '{}'. " "Skipping delete.".format(endpoint_config_name)
41 | )
42 | else:
43 | raise e
44 |
45 |
46 | def delete_sagemaker_model(model_name):
47 | sagemaker_client = boto3.client("sagemaker")
48 | try:
49 | sagemaker_client.delete_model(ModelName=model_name)
50 | print("Successfully deleted model called '{}'.".format(model_name))
51 | except sagemaker_client.exceptions.ClientError as e:
52 | if "Could not find model" in str(e):
53 | print("Could not find model called '{}'. " "Skipping delete.".format(model_name))
54 | else:
55 | raise e
56 |
57 |
58 | def delete_s3_objects(bucket_name):
59 | s3_resource = boto3.resource("s3")
60 | try:
61 | s3_resource.Bucket(bucket_name).objects.all().delete()
62 | print("Successfully deleted objects in bucket " "called '{}'.".format(bucket_name))
63 | except s3_resource.meta.client.exceptions.NoSuchBucket:
64 | print("Could not find bucket called '{}'. " "Skipping delete.".format(bucket_name))
65 |
66 |
67 | def delete_s3_bucket(bucket_name):
68 | s3_resource = boto3.resource("s3")
69 | try:
70 | s3_resource.Bucket(bucket_name).delete()
71 | print("Successfully deleted bucket " "called '{}'.".format(bucket_name))
72 | except s3_resource.meta.client.exceptions.NoSuchBucket:
73 | print("Could not find bucket called '{}'. " "Skipping delete.".format(bucket_name))
74 |
75 |
76 | @helper.delete
77 | def on_delete(event, __):
78 | # remove sagemaker endpoints
79 | solution_prefix = event["ResourceProperties"]["SolutionPrefix"]
80 | endpoint_names = [
81 | "{}-demo-endpoint".format(solution_prefix), # make sure it is the same as your endpoint name
82 | "{}-demo-model".format(solution_prefix),
83 | "{}-finetuned-endpoint".format(solution_prefix),
84 | "{}-detector-from-scratch-endpoint".format(solution_prefix),
85 | "{}-classification-endpoint".format(solution_prefix),
86 | ]
87 | for endpoint_name in endpoint_names:
88 | delete_sagemaker_model(endpoint_name)
89 | delete_sagemaker_endpoint_config(endpoint_name)
90 | delete_sagemaker_endpoint(endpoint_name)
91 |
92 | # remove files in s3
93 | output_bucket = event["ResourceProperties"]["S3Bucket"]
94 | delete_s3_objects(output_bucket)
95 |
96 | # delete buckets
97 | delete_s3_bucket(output_bucket)
98 |
99 |
100 | def handler(event, context):
101 | helper(event, context)
102 |
--------------------------------------------------------------------------------
/src/prepare_data/neu.py:
--------------------------------------------------------------------------------
1 | """
2 | Dependencies: unzip unrar
3 | python -m pip install patool pyunpack
4 | """
5 |
6 | from pathlib import Path
7 | import shutil
8 | import re
9 | import os
10 |
11 | try:
12 | from pyunpack import Archive
13 | except ModuleNotFoundError:
14 | print("installing the dependencies `patool` and `pyunpack` for unzipping the data")
15 | import subprocess
16 |
17 | subprocess.run("python -m pip install patool==1.12 pyunpack==0.2.1 -q", shell=True)
18 | from pyunpack import Archive
19 |
20 | CLASSES = {
21 | "crazing": "Cr",
22 | "inclusion": "In",
23 | "pitted_surface": "PS",
24 | "patches": "Pa",
25 | "rolled-in_scale": "RS",
26 | "scratches": "Sc",
27 | }
28 |
29 |
30 | def unpack(path: str) -> None:
31 | path = Path(path)
32 | Archive(str(path)).extractall(str(path.parent))
33 | return
34 |
35 |
36 | def cp_class_images(data_path: Path, class_name: str, class_path_dest: Path) -> None:
37 | lst = list(data_path.rglob(f"{class_name}_*"))
38 | for img_file in lst:
39 | shutil.copy2(str(img_file), str(class_path_dest / img_file.name))
40 |
41 | assert len(lst) == len(list(class_path_dest.glob("*")))
42 | return
43 |
44 |
45 | def cp_image_annotation(data_path: Path, class_name: str, image_path_dest: Path, annotation_path_dest: Path) -> None:
46 | img_lst = sorted(list((data_path / "IMAGES").rglob(f"{class_name}_*")))
47 | ann_lst = sorted(list((data_path / "ANNOTATIONS").rglob(f"{class_name}_*")))
48 | assert len(img_lst) == len(
49 | ann_lst
50 | ), f"images count {len(img_lst)} does not match with annotations count {len(ann_lst)} for class {class_name}"
51 | for (img_file, ann_file) in zip(img_lst, ann_lst):
52 | shutil.copy2(str(img_file), str(image_path_dest / img_file.name))
53 | shutil.copy2(str(ann_file), str(annotation_path_dest / ann_file.name))
54 |
55 | assert len(list(image_path_dest.glob("*"))) == len(list(annotation_path_dest.glob("*")))
56 | return
57 |
58 |
59 | def main(data_path: str, output_path: str, archived: bool = True) -> None:
60 | """
61 | Data preparation
62 |
63 | Parameters
64 | ----------
65 | data_path : str
66 | Raw data path
67 | output_path : str
68 | Output data path
69 | archived: bool
70 | Whether the file is archived or not (for testing)
71 |
72 | Raises
73 | ------
74 | ValueError
75 | If the packed data file is different from NEU-CLS or NEU-DET
76 | """
77 | data_path = Path(data_path)
78 | if archived:
79 | unpack(data_path)
80 |
81 | data_path = data_path.parent / re.search(r"^[^.]*", str(data_path.name)).group(0)
82 | try:
83 | os.remove(str(data_path / "Thumbs.db"))
84 | except FileNotFoundError:
85 | print(f"Thumbs.db is not found. Continuing ...")
86 | pass
87 | except Exception as e:
88 | print(f"{e}: Unknown error!")
89 | raise e
90 |
91 | output_path = Path(output_path)
92 | if data_path.name == "NEU-CLS":
93 | for cls_ in CLASSES.values():
94 | cls_path = output_path / cls_
95 | cls_path.mkdir(exist_ok=True)
96 | cp_class_images(data_path, cls_, cls_path)
97 | elif data_path.name == "NEU-DET":
98 | for cls_ in CLASSES:
99 | cls_path = output_path / CLASSES[cls_]
100 | image_path = cls_path / "images"
101 | image_path.mkdir(parents=True, exist_ok=True)
102 | annotation_path = cls_path / "annotations"
103 | annotation_path.mkdir(exist_ok=True)
104 | cp_image_annotation(data_path, cls_, image_path, annotation_path)
105 | else:
106 | raise ValueError(f"Unknown data. Choose between `NEU-CLS` and `NEU-DET`. Given {data_path.name}")
107 |
108 | return
109 |
110 |
111 | if __name__ == "__main__":
112 | import sys
113 |
114 | if len(sys.argv) < 3:
115 | print("Provide `data_path` and `output_path`")
116 | sys.exit(1)
117 |
118 | main(sys.argv[1], sys.argv[2])
119 | print("Done")
120 |
--------------------------------------------------------------------------------
/scripts/build.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | set -euxo pipefail
3 |
4 | NOW=$(date +"%x %r %Z")
5 | echo "Time: $NOW"
6 |
7 | if [ $# -lt 3 ]; then
8 | echo "Please provide the solution name as well as the base S3 bucket name and the region to run build script."
9 | echo "For example: bash ./scripts/build.sh trademarked-solution-name sagemaker-solutions-build us-west-2"
10 | exit 1
11 | fi
12 |
13 | REGION=$3
14 | SOURCE_REGION="us-west-2"
15 | echo "Region: $REGION, source region: $SOURCE_REGION"
16 | BASE_DIR="$(dirname "$(dirname "$(readlink -f "$0")")")"
17 | echo "Base dir: $BASE_DIR"
18 |
19 | rm -rf build
20 |
21 | # build python package
22 | echo "Python build and package"
23 | python -m pip install --upgrade pip
24 | python -m pip install --upgrade wheel setuptools
25 | python setup.py build sdist bdist_wheel
26 |
27 | find . | grep -E "(__pycache__|\.pyc|\.pyo$|\.egg*|\lightning_logs)" | xargs rm -rf
28 |
29 | echo "Add requirements for SageMaker"
30 | cp requirements.txt build/lib/
31 | cd build/lib || exit
32 | mv sagemaker_defect_detection/{classifier.py,detector.py} .
33 | touch source_dir.tar.gz
34 | tar --exclude=source_dir.tar.gz -czvf source_dir.tar.gz .
35 | echo "Only keep source_dir.tar.gz for SageMaker"
36 | find . ! -name "source_dir.tar.gz" -type f -exec rm -r {} +
37 | rm -rf sagemaker_defect_detection
38 |
39 | cd - || exit
40 |
41 | mv dist build
42 |
43 | # add notebooks to build
44 | echo "Prepare notebooks and add to build"
45 | cp -r notebooks build
46 | rm -rf build/notebooks/*neu* # remove local datasets for build
47 | for nb in build/notebooks/*.ipynb; do
48 | python "$BASE_DIR"/scripts/set_kernelspec.py --notebook "$nb" --display-name "Python 3 (PyTorch JumpStart)" --kernel "HUB_1P_IMAGE"
49 | done
50 |
51 | echo "Copy src to build"
52 | cp -r src build
53 |
54 | # add solution assistant
55 | echo "Solution assistant lambda function"
56 | cd cloudformation/solution-assistant/ || exit
57 | python -m pip install -r requirements.txt -t ./src/site-packages
58 |
59 | cd - || exit
60 |
61 | echo "Clean up pyc files, needed to avoid security issues. See: https://blog.jse.li/posts/pyc/"
62 | find cloudformation | grep -E "(__pycache__|\.pyc|\.pyo$)" | xargs rm -rf
63 | cp -r cloudformation/solution-assistant build/
64 | cd build/solution-assistant/src || exit
65 | zip -q -r9 "$BASE_DIR"/build/solution-assistant.zip -- *
66 |
67 | cd - || exit
68 | rm -rf build/solution-assistant
69 |
70 | if [ -z "$4" ] || [ "$4" == 'mainline' ]; then
71 | s3_prefix="s3://$2-$3/$1"
72 | else
73 | s3_prefix="s3://$2-$3/$1-$4"
74 | fi
75 |
76 | # cleanup and copy the build artifacts
77 | echo "Removing the existing objects under $s3_prefix"
78 | aws s3 rm --recursive "$s3_prefix" --region "$REGION"
79 | echo "Copying new objects to $s3_prefix"
80 | aws s3 sync . "$s3_prefix" --delete --region "$REGION" \
81 | --exclude ".git/*" \
82 | --exclude ".vscode/*" \
83 | --exclude ".mypy_cache/*" \
84 | --exclude "logs/*" \
85 | --exclude "stack_outputs.json" \
86 | --exclude "src/sagemaker_defect_detection/lightning_logs/*" \
87 | --exclude "notebooks/*neu*/*"
88 |
89 | echo "Copying solution artifacts"
90 | aws s3 cp "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/demo/model.tar.gz" "$s3_prefix"/demo/model.tar.gz --source-region "$SOURCE_REGION"
91 |
92 | mkdir -p build/pretrained/
93 | aws s3 cp "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/pretrained/model.tar.gz" build/pretrained --source-region "$SOURCE_REGION" &&
94 | cd build/pretrained/ && tar -xf model.tar.gz && cd .. &&
95 | aws s3 sync pretrained "$s3_prefix"/pretrained/ --delete --region "$REGION"
96 |
97 | aws s3 cp "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/data/NEU-CLS.zip" "$s3_prefix"/data/ --source-region "$SOURCE_REGION"
98 | aws s3 cp "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/data/NEU-DET.zip" "$s3_prefix"/data/ --source-region "$SOURCE_REGION"
99 |
100 | echo "Add docs to build"
101 | aws s3 sync "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/docs" "$s3_prefix"/docs --delete --region "$REGION"
102 | aws s3 sync "s3://sagemaker-solutions-artifacts/sagemaker-defect-detection/docs" "$s3_prefix"/build/docs --delete --region "$REGION"
103 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/utils/visualize.py:
--------------------------------------------------------------------------------
1 | from typing import Iterable, List, Union, Tuple
2 |
3 | import numpy as np
4 |
5 | import torch
6 |
7 | from matplotlib import pyplot as plt
8 |
9 | import cv2
10 |
11 | import torch
12 |
13 |
14 | TEXT_COLOR = (255, 255, 255) # White
15 | CLASSES = {
16 | "crazing": "Cr",
17 | "inclusion": "In",
18 | "pitted_surface": "PS",
19 | "patches": "Pa",
20 | "rolled-in_scale": "RS",
21 | "scratches": "Sc",
22 | }
23 | CATEGORY_ID_TO_NAME = {i: name for i, name in enumerate(CLASSES.keys(), start=1)}
24 |
25 |
26 | def unnormalize_to_hwc(
27 | image: torch.Tensor, mean: List[float] = [0.485, 0.456, 0.406], std: List[float] = [0.229, 0.224, 0.225]
28 | ) -> np.ndarray:
29 | """
30 | Unnormalizes and a normlized image tensor [0, 1] CHW -> HWC [0, 255]
31 |
32 | Parameters
33 | ----------
34 | image : torch.Tensor
35 | Normalized image
36 | mean : List[float], optional
37 | RGB averages used in normalization, by default [0.485, 0.456, 0.406] from imagenet1k
38 | std : List[float], optional
39 | RGB standard deviations used in normalization, by default [0.229, 0.224, 0.225] from imagenet1k
40 |
41 | Returns
42 | -------
43 | np.ndarray
44 | Unnormalized image as numpy array
45 | """
46 | image = image.numpy().transpose(1, 2, 0) # HWC
47 | image = (image * std + mean).clip(0, 1)
48 | image = (image * 255).astype(np.uint8)
49 | return image
50 |
51 |
52 | def visualize_bbox(img: np.ndarray, bbox: np.ndarray, class_name: str, color, thickness: int = 2) -> np.ndarray:
53 | """
54 | Uses cv2 to draw colored bounding boxes and class names in an image
55 |
56 | Parameters
57 | ----------
58 | img : np.ndarray
59 | [description]
60 | bbox : np.ndarray
61 | [description]
62 | class_name : str
63 | Class name
64 | color : tuple
65 | BGR tuple
66 | thickness : int, optional
67 | Bouding box thickness, by default 2
68 | """
69 | x_min, y_min, x_max, y_max = tuple(map(int, bbox))
70 |
71 | cv2.rectangle(img, (x_min, y_min), (x_max, y_max), color=color, thickness=thickness)
72 |
73 | ((text_width, text_height), _) = cv2.getTextSize(class_name, cv2.FONT_HERSHEY_SIMPLEX, 0.35, 1)
74 | cv2.rectangle(img, (x_min, y_min - int(1.3 * text_height)), (x_min + text_width, y_min), color, -1)
75 | cv2.putText(
76 | img,
77 | text=class_name,
78 | org=(x_min, y_min - int(0.3 * text_height)),
79 | fontFace=cv2.FONT_HERSHEY_SIMPLEX,
80 | fontScale=0.35,
81 | color=TEXT_COLOR,
82 | lineType=cv2.LINE_AA,
83 | )
84 | return img
85 |
86 |
87 | def visualize(
88 | image: np.ndarray,
89 | bboxes: Iterable[Union[torch.Tensor, np.ndarray]] = [],
90 | category_ids: Iterable[Union[torch.Tensor, np.ndarray]] = [],
91 | colors: Iterable[Tuple[int, int, int]] = [],
92 | titles: Iterable[str] = [],
93 | category_id_to_name=CATEGORY_ID_TO_NAME,
94 | dpi=150,
95 | ) -> None:
96 | """
97 | Applies the bounding boxes and category ids to an image
98 |
99 | Parameters
100 | ----------
101 | image : np.ndarray
102 | Image as numpy array
103 | bboxes : Iterable[Union[torch.Tensor, np.ndarray]], optional
104 | Bouding boxes, by default []
105 | category_ids : Iterable[Union[torch.Tensor, np.ndarray]], optional
106 | Category ids, by default []
107 | colors : Iterable[Tuple[int, int, int]], optional
108 | Colors for each bounding box, by default [()]
109 | titles : Iterable[str], optional
110 | Titles for each image, by default []
111 | category_id_to_name : Dict[str, str], optional
112 | Dictionary of category ids to names, by default CATEGORY_ID_TO_NAME
113 | dpi : int, optional
114 | DPI for clarity, by default 150
115 | """
116 | bboxes, category_ids, colors, titles = list(map(list, [bboxes, category_ids, colors, titles])) # type: ignore
117 | n = len(bboxes)
118 | assert (
119 | n == len(category_ids) == len(colors) == len(titles) - 1
120 | ), f"number of bboxes, category ids, colors and titles (minus one) do not match"
121 |
122 | plt.figure(dpi=dpi)
123 | ncols = n + 1
124 | plt.subplot(1, ncols, 1)
125 | img = image.copy()
126 | plt.axis("off")
127 | plt.title(titles[0])
128 | plt.imshow(image)
129 | if not len(bboxes):
130 | return
131 |
132 | titles = titles[1:]
133 | for i in range(2, ncols + 1):
134 | img = image.copy()
135 | plt.subplot(1, ncols, i)
136 | plt.axis("off")
137 | j = i - 2
138 | plt.title(titles[j])
139 | for bbox, category_id in zip(bboxes[j], category_ids[j]): # type: ignore
140 | if isinstance(bbox, torch.Tensor):
141 | bbox = bbox.numpy()
142 |
143 | if isinstance(category_id, torch.Tensor):
144 | category_id = category_id.numpy()
145 |
146 | if isinstance(category_id, np.ndarray):
147 | category_id = category_id.item()
148 |
149 | class_name = category_id_to_name[category_id]
150 | img = visualize_bbox(img, bbox, class_name, color=colors[j])
151 |
152 | plt.imshow(img)
153 | return
154 |
--------------------------------------------------------------------------------
/scripts/train_detector.sh:
--------------------------------------------------------------------------------
1 | NOW=$(date +"%x %r %Z")
2 | echo "Time: ${NOW}"
3 | DETECTION_DATA_PATH=$1
4 | BACKBONE=$2
5 | BASE_DIR="$(dirname $(dirname $(readlink -f $0)))"
6 | DETECTOR_SCRIPT="${BASE_DIR}/src/sagemaker_defect_detection/detector.py"
7 | LOG_DIR="${BASE_DIR}/logs"
8 | MFN_LOGS="${LOG_DIR}/classification_logs"
9 | RPN_LOGS="${LOG_DIR}/rpn_logs"
10 | ROI_LOGS="${LOG_DIR}/roi_logs"
11 | FINETUNED_RPN_LOGS="${LOG_DIR}/finetune_rpn_logs"
12 | FINETUNED_ROI_LOGS="${LOG_DIR}/finetune_roi_logs"
13 | FINETUNED_FINAL_LOGS="${LOG_DIR}/finetune_final_logs"
14 | EXTRA_FINETUNED_RPN_LOGS="${LOG_DIR}/extra_finetune_rpn_logs"
15 | EXTRA_FINETUNED_ROI_LOGS="${LOG_DIR}/extra_finetune_roi_logs"
16 | EXTRA_FINETUNING_STEPS=3
17 |
18 | function find_best_ckpt() {
19 | python ${BASE_DIR}/scripts/find_best_ckpt.py $1 $2
20 | }
21 |
22 | function train_step() {
23 | echo "training step $1"
24 | case $1 in
25 | "1")
26 | echo "skipping step 1 and use 'train_classifier.sh'"
27 | ;;
28 | "2") # train rpn
29 | python ${DETECTOR_SCRIPT} \
30 | --data-path=${DETECTION_DATA_PATH} \
31 | --backbone=${BACKBONE} \
32 | --train-rpn \
33 | --pretrained-mfn-ckpt=$(find_best_ckpt "${MFN_LOGS}" "max") \
34 | --save-path=${RPN_LOGS} \
35 | --gpus=-1 --distributed-backend=ddp \
36 | --epochs=100
37 | ;;
38 | "3") # train roi
39 | python ${DETECTOR_SCRIPT} \
40 | --data-path=${DETECTION_DATA_PATH} \
41 | --backbone=${BACKBONE} \
42 | --train-roi \
43 | --pretrained-rpn-ckpt=$(find_best_ckpt "${RPN_LOGS}" "min") \
44 | --save-path=${ROI_LOGS} \
45 | --gpus=-1 --distributed-backend=ddp \
46 | --epochs=100
47 | ;;
48 | "4") # finetune rpn
49 | python ${DETECTOR_SCRIPT} \
50 | --data-path=${DETECTION_DATA_PATH} \
51 | --backbone=${BACKBONE} \
52 | --finetune-rpn \
53 | --pretrained-rpn-ckpt=$(find_best_ckpt "${RPN_LOGS}" "min") \
54 | --pretrained-roi-ckpt=$(find_best_ckpt "${ROI_LOGS}" "min") \
55 | --save-path=${FINETUNED_RPN_LOGS} \
56 | --gpus=-1 --distributed-backend=ddp \
57 | --learning-rate=1e-4 \
58 | --epochs=100
59 | # --resume-from-checkpoint=$(find_best_ckpt "${FINETUNED_RPN_LOGS}" "max")
60 | ;;
61 | "5") # finetune roi
62 | python ${DETECTOR_SCRIPT} \
63 | --data-path=${DETECTION_DATA_PATH} \
64 | --backbone=${BACKBONE} \
65 | --finetune-roi \
66 | --finetuned-rpn-ckpt=$(find_best_ckpt "${FINETUNED_RPN_LOGS}" "max") \
67 | --pretrained-roi-ckpt=$(find_best_ckpt "${ROI_LOGS}" "min") \
68 | --save-path=${FINETUNED_ROI_LOGS} \
69 | --gpus=-1 --distributed-backend=ddp \
70 | --learning-rate=1e-4 \
71 | --epochs=100
72 | # --resume-from-checkpoint=$(find_best_ckpt "${FINETUNED_ROI_LOGS}" "max")
73 | ;;
74 | "extra_rpn") # initially EXTRA_FINETUNED_*_LOGS is a copy of FINETUNED_*_LOGS
75 | python ${DETECTOR_SCRIPT} \
76 | --data-path=${DETECTION_DATA_PATH} \
77 | --backbone=${BACKBONE} \
78 | --finetune-rpn \
79 | --finetuned-rpn-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_RPN_LOGS}" "max") \
80 | --finetuned-roi-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_ROI_LOGS}" "max") \
81 | --save-path=${EXTRA_FINETUNED_RPN_LOGS} \
82 | --gpus=-1 --distributed-backend=ddp \
83 | --learning-rate=1e-4 \
84 | --epochs=100
85 | ;;
86 | "extra_roi") # initially EXTRA_FINETUNED_*_LOGS is a copy of FINETUNED_*_LOGS
87 | python ${DETECTOR_SCRIPT} \
88 | --data-path=${DETECTION_DATA_PATH} \
89 | --backbone=${BACKBONE} \
90 | --finetune-roi \
91 | --finetuned-rpn-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_RPN_LOGS}" "max") \
92 | --finetuned-roi-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_ROI_LOGS}" "max") \
93 | --save-path=${EXTRA_FINETUNED_ROI_LOGS} \
94 | --gpus=-1 --distributed-backend=ddp \
95 | --learning-rate=1e-4 \
96 | --epochs=100
97 | ;;
98 | "joint") # final
99 | python ${DETECTOR_SCRIPT} \
100 | --data-path=${DETECTION_DATA_PATH} \
101 | --backbone=${BACKBONE} \
102 | --finetuned-rpn-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_RPN_LOGS}" "max") \
103 | --finetuned-roi-ckpt=$(find_best_ckpt "${EXTRA_FINETUNED_ROI_LOGS}" "max") \
104 | --save-path="${FINETUNED_FINAL_LOGS}" \
105 | --gpus=-1 --distributed-backend=ddp \
106 | --learning-rate=1e-3 \
107 | --epochs=300
108 | # --resume-from-checkpoint=$(find_best_ckpt "${FINETUNED_FINAL_LOGS}" "max")
109 | ;;
110 |
111 | *) ;;
112 | esac
113 | }
114 |
115 | function train_wait_to_finish() {
116 | train_step $1 &
117 | BPID=$!
118 | wait $BPID
119 | }
120 |
121 | function run() {
122 | if [ "$1" != "" ]; then
123 | train_step $1
124 | else
125 | nvidia-smi | grep python | awk '{ print $3 }' | xargs -n1 kill -9 >/dev/null 2>&1
126 | read -p "Training all steps from scratch? (Y/N): " confirm && [[ $confirm == [yY] || $confirm == [yY][eE][sS] ]]
127 | if [ "$confirm" == "Y" ]; then
128 | for i in {1..5}; do
129 | train_wait_to_finish $i
130 | done
131 | echo "finished all the training steps"
132 | mkdir -p "${EXTRA_FINETUNED_RPN_LOGS}" && cp -r "${FINETUNED_RPN_LOGS}/"* "${EXTRA_FINETUNED_RPN_LOGS}"
133 | mkdir -p "${EXTRA_FINETUNED_ROI_LOGS}" && cp -r "${FINETUNED_ROI_LOGS}/"* "${EXTRA_FINETUNED_ROI_LOGS}"
134 | fi
135 | echo "repeating extra finetuning steps ${EXTRA_FINETUNING_STEPS} more times"
136 | for i in {1..${EXTRA_FINETUNING_STEPS}}; do
137 | train_wait_to_finish "extra_rpn"
138 | train_wait_to_finish "extra_roi"
139 | done
140 | echo "final joint training"
141 | train_step "joint"
142 | fi
143 | exit 0
144 | }
145 |
146 | run $1
147 |
--------------------------------------------------------------------------------
/cloudformation/defect-detection-permissions.yaml:
--------------------------------------------------------------------------------
1 | AWSTemplateFormatVersion: "2010-09-09"
2 | Description: "(SA0015) - sagemaker-defect-detection permission stack"
3 | Parameters:
4 | SolutionPrefix:
5 | Type: String
6 | SolutionName:
7 | Type: String
8 | S3Bucket:
9 | Type: String
10 | StackVersion:
11 | Type: String
12 |
13 | Mappings:
14 | S3:
15 | release:
16 | BucketPrefix: "sagemaker-solutions-prod"
17 | development:
18 | BucketPrefix: "sagemaker-solutions-devo"
19 |
20 | Resources:
21 | SageMakerIAMRole:
22 | Type: AWS::IAM::Role
23 | Properties:
24 | RoleName: !Sub "${SolutionPrefix}-${AWS::Region}-nb-role"
25 | AssumeRolePolicyDocument:
26 | Version: "2012-10-17"
27 | Statement:
28 | - Effect: Allow
29 | Principal:
30 | AWS:
31 | - !Sub "arn:aws:iam::${AWS::AccountId}:root"
32 | Service:
33 | - sagemaker.amazonaws.com
34 | - lambda.amazonaws.com # for solution assistant resource cleanup
35 | Action:
36 | - "sts:AssumeRole"
37 | Metadata:
38 | cfn_nag:
39 | rules_to_suppress:
40 | - id: W28
41 | reason: Needs to be explicitly named to tighten launch permissions policy
42 |
43 | SageMakerIAMPolicy:
44 | Type: AWS::IAM::Policy
45 | Properties:
46 | PolicyName: !Sub "${SolutionPrefix}-nb-instance-policy"
47 | Roles:
48 | - !Ref SageMakerIAMRole
49 | PolicyDocument:
50 | Version: "2012-10-17"
51 | Statement:
52 | - Effect: Allow
53 | Action:
54 | - sagemaker:CreateTrainingJob
55 | - sagemaker:DescribeTrainingJob
56 | - sagemaker:CreateProcessingJob
57 | - sagemaker:DescribeProcessingJob
58 | - sagemaker:CreateModel
59 | - sagemaker:DescribeEndpointConfig
60 | - sagemaker:DescribeEndpoint
61 | - sagemaker:CreateEndpointConfig
62 | - sagemaker:CreateEndpoint
63 | - sagemaker:DeleteEndpointConfig
64 | - sagemaker:DeleteEndpoint
65 | - sagemaker:DeleteModel
66 | - sagemaker:InvokeEndpoint
67 | Resource:
68 | - !Sub "arn:aws:sagemaker:${AWS::Region}:${AWS::AccountId}:*"
69 | - Effect: Allow
70 | Action:
71 | - cloudwatch:GetMetricData
72 | - cloudwatch:GetMetricStatistics
73 | - cloudwatch:ListMetrics
74 | - cloudwatch:PutMetricData
75 | Resource:
76 | - !Sub "arn:aws:cloudwatch:${AWS::Region}:${AWS::AccountId}:*"
77 | - Effect: Allow
78 | Action:
79 | - logs:CreateLogGroup
80 | - logs:CreateLogStream
81 | - logs:DescribeLogStreams
82 | - logs:GetLogEvents
83 | - logs:PutLogEvents
84 | Resource:
85 | - !Sub "arn:aws:logs:${AWS::Region}:${AWS::AccountId}:log-group:/aws/sagemaker/*"
86 | - Effect: Allow
87 | Action:
88 | - iam:PassRole
89 | Resource:
90 | - !GetAtt SageMakerIAMRole.Arn
91 | Condition:
92 | StringEquals:
93 | iam:PassedToService: sagemaker.amazonaws.com
94 | - Effect: Allow
95 | Action:
96 | - iam:GetRole
97 | Resource:
98 | - !GetAtt SageMakerIAMRole.Arn
99 | - Effect: Allow
100 | Action:
101 | - ecr:GetAuthorizationToken
102 | - ecr:GetDownloadUrlForLayer
103 | - ecr:BatchGetImage
104 | - ecr:BatchCheckLayerAvailability
105 | - ecr:CreateRepository
106 | - ecr:DescribeRepositories
107 | - ecr:InitiateLayerUpload
108 | - ecr:CompleteLayerUpload
109 | - ecr:UploadLayerPart
110 | - ecr:TagResource
111 | - ecr:PutImage
112 | - ecr:DescribeImages
113 | - ecr:BatchDeleteImage
114 | Resource:
115 | - "*"
116 | - !Sub "arn:aws:ecr:${AWS::Region}:${AWS::AccountId}:repository/*"
117 | - Effect: Allow
118 | Action:
119 | - s3:ListBucket
120 | Resource: !Sub
121 | - "arn:aws:s3:::${SolutionS3BucketName}-${AWS::Region}"
122 | - SolutionS3BucketName:
123 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
124 | - Effect: Allow
125 | Action:
126 | - s3:GetObject
127 | Resource: !Sub
128 | - "arn:aws:s3:::${SolutionS3BucketName}-${AWS::Region}/${SolutionName}/*"
129 | - SolutionS3BucketName:
130 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
131 | SolutionName: !Ref SolutionName
132 | - Effect: Allow
133 | Action:
134 | - s3:ListBucket
135 | - s3:DeleteBucket
136 | - s3:GetBucketLocation
137 | - s3:ListBucketMultipartUploads
138 | Resource:
139 | - !Sub "arn:aws:s3:::${S3Bucket}"
140 | - Effect: Allow
141 | Action:
142 | - s3:AbortMultipartUpload
143 | - s3:ListObject
144 | - s3:GetObject
145 | - s3:PutObject
146 | - s3:DeleteObject
147 | Resource:
148 | - !Sub "arn:aws:s3:::${S3Bucket}"
149 | - !Sub "arn:aws:s3:::${S3Bucket}/*"
150 | - Effect: Allow
151 | Action:
152 | - s3:CreateBucket
153 | - s3:ListBucket
154 | - s3:GetObject
155 | - s3:GetObjectVersion
156 | - s3:PutObject
157 | - s3:DeleteObject
158 | Resource:
159 | - !Sub "arn:aws:s3:::sagemaker-${AWS::Region}-${AWS::AccountId}"
160 | - !Sub "arn:aws:s3:::sagemaker-${AWS::Region}-${AWS::AccountId}/*"
161 |
162 | Metadata:
163 | cfn_nag:
164 | rules_to_suppress:
165 | - id: W12
166 | reason: ECR GetAuthorizationToken is non resource-specific action
167 |
168 | Outputs:
169 | SageMakerRoleArn:
170 | Description: "SageMaker Execution Role for the solution"
171 | Value: !GetAtt SageMakerIAMRole.Arn
172 |
--------------------------------------------------------------------------------
/src/utils.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import json
3 | import boto3
4 | import copy
5 | import matplotlib.patches as patches
6 | from matplotlib import pyplot as plt
7 | from PIL import Image, ImageColor
8 |
9 |
10 | def query_Type2(image_file_name, endpoint_name, num_predictions=4):
11 |
12 | with open(image_file_name, "rb") as file:
13 | input_img_rb = file.read()
14 |
15 | client = boto3.client("runtime.sagemaker")
16 | query_response = client.invoke_endpoint(
17 | EndpointName=endpoint_name,
18 | ContentType="application/x-image",
19 | Body=input_img_rb,
20 | Accept=f'application/json;verbose;n_predictions={num_predictions}'
21 | )
22 | # If we remove ';n_predictions={}' from Accept, we get all the predicted boxes.
23 | query_response = query_response['Body'].read()
24 |
25 | model_predictions = json.loads(query_response)
26 | normalized_boxes, classes, scores, labels = (
27 | model_predictions["normalized_boxes"],
28 | model_predictions["classes"],
29 | model_predictions["scores"],
30 | model_predictions["labels"],
31 | )
32 | # Substitute the classes index with the classes name
33 | class_names = [labels[int(idx)] for idx in classes]
34 | return normalized_boxes, class_names, scores
35 |
36 |
37 | # Copied from albumentations/augmentations/functional.py
38 | # Follow albumentations.Normalize, which is used in sagemaker_defect_detection/detector.py
39 | def normalize(img, mean, std, max_pixel_value=255.0):
40 | mean = np.array(mean, dtype=np.float32)
41 | mean *= max_pixel_value
42 |
43 | std = np.array(std, dtype=np.float32)
44 | std *= max_pixel_value
45 |
46 | denominator = np.reciprocal(std, dtype=np.float32)
47 |
48 | img = img.astype(np.float32)
49 | img -= mean
50 | img *= denominator
51 | return img
52 |
53 |
54 | def query_DDN(image_file_name, endpoint_name, num_predictions=4):
55 |
56 | with Image.open(image_file_name) as im:
57 | image_np = np.array(im)
58 |
59 | # Follow albumentations.Normalize, which is used in sagemaker_defect_detection/detector.py
60 | mean = (0.485, 0.456, 0.406)
61 | std = (0.229, 0.224, 0.225)
62 | max_pixel_value = 255.0
63 |
64 | image_np = normalize(image_np, mean, std, max_pixel_value)
65 | image_np = image_np.transpose(2, 0, 1)
66 | image_np = np.expand_dims(image_np, 0) # CHW
67 |
68 | client = boto3.client("runtime.sagemaker")
69 | query_response = client.invoke_endpoint(
70 | EndpointName=endpoint_name,
71 | ContentType="application/json",
72 | Body=json.dumps(image_np.tolist())
73 | )
74 | query_response = query_response["Body"].read()
75 |
76 | model_predictions = json.loads(query_response.decode())[0]
77 | unnormalized_boxes = model_predictions['boxes'][:num_predictions]
78 | class_names = model_predictions['labels'][:num_predictions]
79 | scores = model_predictions['scores'][:num_predictions]
80 | return unnormalized_boxes, class_names, scores
81 |
82 |
83 |
84 | def query_Type1(image_file_name, endpoint_name, num_predictions=4):
85 |
86 | with open(image_file_name, "rb") as file:
87 | input_img_rb = file.read()
88 |
89 | client = boto3.client(service_name="runtime.sagemaker")
90 | query_response = client.invoke_endpoint(
91 | EndpointName=endpoint_name,
92 | ContentType="application/x-image",
93 | Body=input_img_rb
94 | )
95 | query_response = query_response["Body"].read()
96 |
97 | model_predictions = json.loads(query_response)['prediction'][:num_predictions]
98 | class_names = [int(pred[0])+1 for pred in model_predictions] # +1 for index starts from 1
99 | scores = [pred[1] for pred in model_predictions]
100 | normalized_boxes = [pred[2:] for pred in model_predictions]
101 | return normalized_boxes, class_names, scores
102 |
103 |
104 | def plot_results(image, bboxes, categories, d):
105 | # d - dictionary of endpoint responses
106 |
107 | colors = list(ImageColor.colormap.values())
108 | with Image.open(image) as im:
109 | image_np = np.array(im)
110 | fig = plt.figure(figsize=(20, 14))
111 |
112 | n = len(d)
113 |
114 | # Ground truth
115 | ax1 = fig.add_subplot(2, 3, 1)
116 | plt.axis('off')
117 | plt.title('Ground Truth')
118 |
119 | for bbox in bboxes:
120 | left, bot, right, top = bbox['bbox']
121 | x, y, w, h = left, bot, right - left, top - bot
122 |
123 | color = colors[hash(bbox['category_id']) % len(colors)]
124 | rect = patches.Rectangle((x, y), w, h, linewidth=3, edgecolor=color, facecolor="none")
125 | ax1.add_patch(rect)
126 | ax1.text(x, y, "{}".format(categories[bbox['category_id']]),
127 | bbox=dict(facecolor="white", alpha=0.5))
128 |
129 | ax1.imshow(image_np)
130 |
131 | # Predictions
132 | counter = 2
133 | for k, v in d.items():
134 | axi = fig.add_subplot(2, 3, counter)
135 | counter += 1
136 |
137 | if "Type2-HPO" in k:
138 | k = "Type2-HPO"
139 | elif "Type2" in k:
140 | k = "Type2"
141 | elif "Type1-HPO" in k:
142 | k = "Type1-HPO"
143 | elif "Type1" in k:
144 | k = "Type1"
145 | elif k.endswith("finetuned-endpoint"):
146 | k = "DDN"
147 |
148 | plt.title(f'Prediction: {k}')
149 | plt.axis('off')
150 |
151 | for idx in range(len(v['normalized_boxes'])):
152 | left, bot, right, top = v['normalized_boxes'][idx]
153 | if k == 'DDN':
154 | x, w = left, right - left
155 | y, h = bot, top - bot
156 | else:
157 | x, w = [val * image_np.shape[1] for val in [left, right - left]]
158 | y, h = [val * image_np.shape[0] for val in [bot, top - bot]]
159 | color = colors[hash(v['classes_names'][idx]) % len(colors)]
160 | rect = patches.Rectangle((x, y), w, h, linewidth=3, edgecolor=color, facecolor="none")
161 | axi.add_patch(rect)
162 | axi.text(x, y,
163 | "{} {:.0f}%".format(categories[v['classes_names'][idx]], v['confidences'][idx] * 100),
164 | bbox=dict(facecolor="white", alpha=0.5),
165 | )
166 |
167 | axi.imshow(image_np)
168 |
169 | plt.tight_layout()
170 | plt.savefig("results/"+ image.split('/')[-1])
171 |
172 | plt.show()
173 |
--------------------------------------------------------------------------------
/src/prepare_RecordIO.py:
--------------------------------------------------------------------------------
1 | import json
2 | import os
3 | import random
4 | import argparse
5 | import numpy as np
6 | from collections import defaultdict
7 | from pathlib import Path
8 |
9 |
10 | def write_line(img_path, width, height, boxes, ids, idx):
11 | """Create a line for each image with annotations, width, height and image name."""
12 | # for header, we use minimal length 2, plus width and height
13 | # with A: 4, B: 5, C: width, D: height
14 | A = 4
15 | B = 5
16 | C = width
17 | D = height
18 | # concat id and bboxes
19 | labels = np.hstack((ids.reshape(-1, 1), boxes)).astype("float")
20 | # normalized bboxes (recommanded)
21 | labels[:, (1, 3)] /= float(width)
22 | labels[:, (2, 4)] /= float(height)
23 | # flatten
24 | labels = labels.flatten().tolist()
25 | str_idx = [str(idx)]
26 | str_header = [str(x) for x in [A, B, C, D]]
27 | str_labels = [str(x) for x in labels]
28 | str_path = [img_path]
29 | line = "\t".join(str_idx + str_header + str_labels + str_path) + "\n"
30 | return line
31 |
32 |
33 | # adapt from __main__ from im2rec.py
34 | def write_lst(output_file, ids, images_annotations):
35 |
36 | all_labels = set()
37 | image_info = {}
38 | for entry in images_annotations['images']:
39 | if entry["id"] in ids:
40 | image_info[entry["id"]] = entry
41 | annotations_info = {} # one annotation for each id (ie., image)
42 | for entry in images_annotations['annotations']:
43 | image_id = entry['image_id']
44 | if image_id in ids:
45 | if image_id not in annotations_info:
46 | annotations_info[image_id] = {'boxes': [], 'labels': []}
47 | annotations_info[image_id]['boxes'].append(entry['bbox'])
48 | annotations_info[image_id]['labels'].append(entry['category_id'])
49 | all_labels.add(entry['category_id'])
50 | labels_list = [label for label in all_labels]
51 | class_to_idx_mapping = {label: idx for idx, label in enumerate(labels_list)}
52 | with open(output_file, "w") as fw:
53 | for i, image_id in enumerate(annotations_info):
54 | im_info = image_info[image_id]
55 | image_file = im_info['file_name']
56 | height = im_info['height']
57 | width = im_info['width']
58 | an_info = annotations_info[image_id]
59 | boxes = np.array(an_info['boxes'])
60 | labels = np.array([class_to_idx_mapping[label] for label in an_info['labels']])
61 | line = write_line(image_file, width, height, boxes, labels, i)
62 | fw.write(line)
63 |
64 |
65 | def create_lst(data_dir, args, rnd_seed=100):
66 | """Generate an lst file based on annotations file which is used to convert the input data to .rec format."""
67 | with open(os.path.join(data_dir, 'annotations.json')) as f:
68 | images_annotations = json.loads(f.read())
69 |
70 | # Size of each class
71 | class_ids = defaultdict(list)
72 | for entry in images_annotations['images']:
73 | cls_ = entry['file_name'].split('_')[0]
74 | class_ids[cls_].append(entry['id'])
75 | print('\ncategory\tnum of images')
76 | print('---------------')
77 | for cls_ in class_ids.keys():
78 | print(f"{cls_}\t{len(class_ids[cls_])}")
79 |
80 | random.seed(rnd_seed)
81 |
82 | # Split train/val/test image ids
83 | if args.test_ratio:
84 | test_ids = []
85 | if args.train_ratio + args.test_ratio < 1.0:
86 | val_ids = []
87 | train_ids = []
88 | for cls_ in class_ids.keys():
89 | random.shuffle(class_ids[cls_])
90 | N = len(class_ids[cls_])
91 | ids = class_ids[cls_]
92 |
93 | sep = int(N * args.train_ratio)
94 | sep_test = int(N * args.test_ratio)
95 | if args.train_ratio == 1.0:
96 | train_ids.extend(ids)
97 | else:
98 | if args.test_ratio:
99 | test_ids.extend(ids[:sep_test])
100 | if args.train_ratio + args.test_ratio < 1.0:
101 | val_ids.extend(ids[sep_test + sep:])
102 | train_ids.extend(ids[sep_test: sep_test + sep])
103 |
104 | write_lst(args.prefix + "_train.lst", train_ids, images_annotations)
105 | lsts = [args.prefix + "_train.lst"]
106 | if args.test_ratio:
107 | write_lst(args.prefix + "_test.lst", test_ids, images_annotations)
108 | lsts.append(args.prefix + "_test.lst")
109 | if args.train_ratio + args.test_ratio < 1.0:
110 | write_lst(args.prefix + "_val.lst", val_ids, images_annotations)
111 | lsts.append(args.prefix + "_val.lst")
112 |
113 | return lsts
114 |
115 |
116 | def parse_args():
117 | """Defines all arguments.
118 |
119 | Returns:
120 | args object that contains all the params
121 | """
122 | parser = argparse.ArgumentParser(
123 | formatter_class=argparse.ArgumentDefaultsHelpFormatter,
124 | description="Create an image list or \
125 | make a record database by reading from an image list",
126 | )
127 | parser.add_argument("prefix", help="prefix of input/output lst and rec files.")
128 | parser.add_argument("root", help="path to folder containing images.")
129 |
130 | cgroup = parser.add_argument_group("Options for creating image lists")
131 | cgroup.add_argument(
132 | "--exts", nargs="+", default=[".jpeg", ".jpg", ".png"], help="list of acceptable image extensions."
133 | )
134 | cgroup.add_argument("--train-ratio", type=float, default=0.8, help="Ratio of images to use for training.")
135 | cgroup.add_argument("--test-ratio", type=float, default=0, help="Ratio of images to use for testing.")
136 | cgroup.add_argument(
137 | "--recursive",
138 | action="store_true",
139 | help="If true recursively walk through subdirs and assign an unique label\
140 | to images in each folder. Otherwise only include images in the root folder\
141 | and give them label 0.",
142 | )
143 | args = parser.parse_args()
144 |
145 | args.prefix = os.path.abspath(args.prefix)
146 | args.root = os.path.abspath(args.root)
147 | return args
148 |
149 |
150 | if __name__ == '__main__':
151 |
152 | args = parse_args()
153 | data_dir = Path(args.root).parent
154 |
155 | lsts = create_lst(data_dir, args)
156 | print()
157 |
158 | for lst in lsts:
159 | os.system(f"python3 ../src/im2rec.py {lst} {os.path.join(data_dir, 'images')} --pass-through --pack-label")
160 | print()
161 |
--------------------------------------------------------------------------------
/src/xml2json.py:
--------------------------------------------------------------------------------
1 | # Use this script to convert annotation xmls to a single annotations.json file that will be taken by Jumpstart OD model
2 | # Reference: XML2JSON.py https://linuxtut.com/en/e391e5e6924945b8a852/
3 |
4 | import random
5 | import xmltodict
6 | import copy
7 | import json
8 | import glob
9 | import os
10 | from collections import defaultdict
11 |
12 |
13 | categories = [
14 | {"id": 1, "name": "crazing"},
15 | {"id": 2, "name": "inclusion"},
16 | {"id": 3, "name": "pitted_surface"},
17 | {"id": 4, "name": "patches"},
18 | {"id": 5, "name": "rolled-in_scale"},
19 | {"id": 6, "name": "scratches"},
20 | ]
21 |
22 |
23 | def XML2JSON(xmlFiles, test_ratio=None, rnd_seed=100):
24 | """ Convert all xmls to annotations.json
25 |
26 | If the test_ratio is not None, convert to two annotations.json files,
27 | one for train+val, another one for test.
28 | """
29 |
30 | images = list()
31 | annotations = list()
32 | image_id = 1
33 | annotation_id = 1
34 | for file in xmlFiles:
35 | annotation_path = file
36 | image = dict()
37 | with open(annotation_path) as fd:
38 | doc = xmltodict.parse(fd.read(), force_list=('object'))
39 | filename = str(doc['annotation']['filename'])
40 | image['file_name'] = filename if filename.endswith('.jpg') else filename + '.jpg'
41 | image['height'] = int(doc['annotation']['size']['height'])
42 | image['width'] = int(doc['annotation']['size']['width'])
43 | image['id'] = image_id
44 | # print("File Name: {} and image_id {}".format(file, image_id))
45 | images.append(image)
46 | if 'object' in doc['annotation']:
47 | for obj in doc['annotation']['object']:
48 | for value in categories:
49 | annotation = dict()
50 | if str(obj['name']) == value["name"]:
51 | annotation["image_id"] = image_id
52 | xmin = int(obj["bndbox"]["xmin"])
53 | ymin = int(obj["bndbox"]["ymin"])
54 | xmax = int(obj["bndbox"]["xmax"])
55 | ymax = int(obj["bndbox"]["ymax"])
56 | annotation["bbox"] = [xmin, ymin, xmax, ymax]
57 | annotation["category_id"] = value["id"]
58 | annotation["id"] = annotation_id
59 | annotation_id += 1
60 | annotations.append(annotation)
61 |
62 | else:
63 | print("File: {} doesn't have any object".format(file))
64 |
65 | image_id += 1
66 |
67 | if test_ratio is None:
68 | attrDict = dict()
69 | attrDict["images"] = images
70 | attrDict["annotations"] = annotations
71 |
72 | jsonString = json.dumps(attrDict)
73 | with open("annotations.json", "w") as f:
74 | f.write(jsonString)
75 | else:
76 | assert test_ratio < 1.0
77 |
78 | # Size of each class
79 | category_ids = defaultdict(list)
80 | for img in images:
81 | category = img['file_name'].split('_')[0]
82 | category_ids[category].append(img['id'])
83 | print('\ncategory\tnum of images')
84 | print('-' * 20)
85 |
86 | random.seed(rnd_seed)
87 |
88 | train_val_images = []
89 | test_images = []
90 | train_val_annotations = []
91 | test_annotations = []
92 |
93 | for category in category_ids.keys():
94 | print(f"{category}:\t{len(category_ids[category])}")
95 |
96 | random.shuffle(category_ids[category])
97 | N = len(category_ids[category])
98 | ids = category_ids[category]
99 |
100 | sep = int(N * test_ratio)
101 |
102 | category_images = [img for img in images if img['id'] in ids[:sep]]
103 | test_images.extend(category_images)
104 | category_images = [img for img in images if img['id'] in ids[sep:]]
105 | train_val_images.extend(category_images)
106 |
107 | category_annotations = [ann for ann in annotations if ann['image_id'] in ids[:sep]]
108 | test_annotations.extend(category_annotations)
109 | category_annotations = [ann for ann in annotations if ann['image_id'] in ids[sep:]]
110 | train_val_annotations.extend(category_annotations)
111 |
112 | print('-' * 20)
113 |
114 | train_val_attrDict = dict()
115 | train_val_attrDict["images"] = train_val_images
116 | train_val_attrDict["annotations"] = train_val_annotations
117 | print(f"\ntrain_val:\t{len(train_val_images)}")
118 |
119 | train_val_jsonString = json.dumps(train_val_attrDict)
120 | with open("annotations.json", "w") as f:
121 | f.write(train_val_jsonString)
122 |
123 | test_attDict = dict()
124 | test_attDict["images"] = test_images
125 | test_attDict["annotations"] = test_annotations
126 | print(f"test:\t{len(test_images)}")
127 |
128 | test_jsonString = json.dumps(test_attDict)
129 | with open("test_annotations.json", "w") as f:
130 | f.write(test_jsonString)
131 |
132 |
133 |
134 | def convert_to_pycocotools_ground_truth(annotations_file):
135 | """
136 | Given the annotation json file for the test data generated during
137 | initial data preparatoin, convert it to the input format pycocotools
138 | can consume.
139 | """
140 |
141 | with open(annotations_file) as f:
142 | images_annotations = json.loads(f.read())
143 |
144 | attrDict = dict()
145 | attrDict["images"] = images_annotations["images"]
146 | attrDict["categories"] = categories
147 |
148 | annotations = []
149 | for entry in images_annotations['annotations']:
150 | ann = copy.deepcopy(entry)
151 | xmin, ymin, xmax, ymax = ann["bbox"]
152 | ann["bbox"] = [xmin, ymin, xmax-xmin, ymax-ymin] # convert to [x, y, W, H]
153 | ann["area"] = (xmax - xmin) * (ymax - ymin)
154 | ann["iscrowd"] = 0
155 | annotations.append(ann)
156 |
157 | attrDict["annotations"] = annotations
158 |
159 | jsonString = json.dumps(attrDict)
160 | ground_truth_annotations = "results/ground_truth_annotations.json"
161 |
162 | with open(ground_truth_annotations, "w") as f:
163 | f.write(jsonString)
164 |
165 | return ground_truth_annotations
166 |
167 |
168 | if __name__ == "__main__":
169 | data_path = '../../NEU-DET/ANNOTATIONS'
170 | xmlfiles = glob.glob(os.path.join(data_path, '*.xml'))
171 | xmlfiles.sort()
172 |
173 | XML2JSON(xmlfiles, test_ratio=0.2)
174 |
175 |
176 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/dataset/neu.py:
--------------------------------------------------------------------------------
1 | from typing import List, Tuple, Optional, Callable
2 | import os
3 | from pathlib import Path
4 | from collections import namedtuple
5 |
6 | from xml.etree.ElementTree import ElementTree
7 |
8 | import numpy as np
9 |
10 | import cv2
11 |
12 | import torch
13 | from torch.utils.data.dataset import Dataset
14 |
15 | from torchvision.datasets import ImageFolder
16 |
17 |
18 | class NEUCLS(ImageFolder):
19 | """
20 | NEU-CLS dataset processing and loading
21 | """
22 |
23 | def __init__(
24 | self,
25 | root: str,
26 | split: str,
27 | augmentation: Optional[Callable] = None,
28 | preprocessing: Optional[Callable] = None,
29 | seed: int = 123,
30 | **kwargs,
31 | ) -> None:
32 | """
33 | NEU-CLS dataset
34 |
35 | Parameters
36 | ----------
37 | root : str
38 | Dataset root path
39 | split : str
40 | Data split from train, val and test
41 | augmentation : Optional[Callable], optional
42 | Image augmentation function, by default None
43 | preprocess : Optional[Callable], optional
44 | Image preprocessing function, by default None
45 | seed : int, optional
46 | Random number generator seed, by default 123
47 |
48 | Raises
49 | ------
50 | ValueError
51 | If unsupported split is used
52 | """
53 | super().__init__(root, **kwargs)
54 | self.samples: List[Tuple[str, int]]
55 | self.split = split
56 | self.augmentation = augmentation
57 | self.preprocessing = preprocessing
58 | n_items = len(self.samples)
59 | np.random.seed(seed)
60 | perm = np.random.permutation(list(range(n_items)))
61 | # TODO: add split ratios as parameters
62 | train_end = int(0.6 * n_items)
63 | val_end = int(0.2 * n_items) + train_end
64 | if split == "train":
65 | self.samples = [self.samples[i] for i in perm[:train_end]]
66 | elif split == "val":
67 | self.samples = [self.samples[i] for i in perm[train_end:val_end]]
68 | elif split == "test":
69 | self.samples = [self.samples[i] for i in perm[val_end:]]
70 | else:
71 | raise ValueError(f"Unknown split mode. Choose from `train`, `val` or `test`. Given {split}")
72 |
73 |
74 | DetectionSample = namedtuple("DetectionSample", ["image_path", "class_idx", "annotations"])
75 |
76 |
77 | class NEUDET(Dataset):
78 | """
79 | NEU-DET dataset processing and loading
80 | """
81 |
82 | def __init__(
83 | self,
84 | root: str,
85 | split: str,
86 | augmentation: Optional[Callable] = None,
87 | preprocess: Optional[Callable] = None,
88 | seed: int = 123,
89 | ):
90 | """
91 | NEU-DET dataset
92 |
93 | Parameters
94 | ----------
95 | root : str
96 | Dataset root path
97 | split : str
98 | Data split from train, val and test
99 | augmentation : Optional[Callable], optional
100 | Image augmentation function, by default None
101 | preprocess : Optional[Callable], optional
102 | Image preprocessing function, by default None
103 | seed : int, optional
104 | Random number generator seed, by default 123
105 |
106 | Raises
107 | ------
108 | ValueError
109 | If unsupported split is used
110 | """
111 | super().__init__()
112 | self.root = Path(root)
113 | self.split = split
114 | self.classes, self.class_to_idx = self._find_classes()
115 | self.samples: List[DetectionSample] = self._make_dataset()
116 | self.augmentation = augmentation
117 | self.preprocess = preprocess
118 | n_items = len(self.samples)
119 | np.random.seed(seed)
120 | perm = np.random.permutation(list(range(n_items)))
121 | train_end = int(0.6 * n_items)
122 | val_end = int(0.2 * n_items) + train_end
123 | if split == "train":
124 | self.samples = [self.samples[i] for i in perm[:train_end]]
125 | elif split == "val":
126 | self.samples = [self.samples[i] for i in perm[train_end:val_end]]
127 | elif split == "test":
128 | self.samples = [self.samples[i] for i in perm[val_end:]]
129 | else:
130 | raise ValueError(f"Unknown split mode. Choose from `train`, `val` or `test`. Given {split}")
131 |
132 | def _make_dataset(self) -> List[DetectionSample]:
133 | instances = []
134 | base_dir = self.root.expanduser()
135 | for target_cls in sorted(self.class_to_idx.keys()):
136 | cls_idx = self.class_to_idx[target_cls]
137 | target_dir = base_dir / target_cls
138 | if not target_dir.is_dir():
139 | continue
140 |
141 | images = sorted(list((target_dir / "images").glob("*.jpg")))
142 | annotations = sorted(list((target_dir / "annotations").glob("*.xml")))
143 | assert len(images) == len(annotations), f"something is wrong. Mismatched number of images and annotations"
144 | for path, ann in zip(images, annotations):
145 | instances.append(DetectionSample(str(path), int(cls_idx), str(ann)))
146 |
147 | return instances
148 |
149 | def _find_classes(self):
150 | classes = sorted([d.name for d in os.scandir(str(self.root)) if d.is_dir()])
151 | class_to_idx = {cls_name: i for i, cls_name in enumerate(classes, 1)} # no bg label in NEU
152 | return classes, class_to_idx
153 |
154 | @staticmethod
155 | def _get_bboxes(ann: str) -> List[List[int]]:
156 | tree = ElementTree().parse(ann)
157 | bboxes = []
158 | for bndbox in tree.iterfind("object/bndbox"):
159 | # should subtract 1 like coco?
160 | bbox = [int(bndbox.findtext(t)) - 1 for t in ("xmin", "ymin", "xmax", "ymax")] # type: ignore
161 | assert bbox[2] > bbox[0] and bbox[3] > bbox[1], f"box size error, given {bbox}"
162 | bboxes.append(bbox)
163 |
164 | return bboxes
165 |
166 | def __len__(self):
167 | return len(self.samples)
168 |
169 | def __getitem__(self, idx: int):
170 | # Note: images are grayscaled BUT resnet needs 3 channels
171 | image = cv2.imread(self.samples[idx].image_path) # BGR channel last
172 | image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
173 | boxes = self._get_bboxes(self.samples[idx].annotations)
174 | num_objs = len(boxes)
175 | boxes = torch.as_tensor(boxes, dtype=torch.float32)
176 | labels = torch.tensor([self.samples[idx].class_idx] * num_objs, dtype=torch.int64)
177 | image_id = torch.tensor([idx], dtype=torch.int64)
178 | iscrowd = torch.zeros((len(boxes),), dtype=torch.int64)
179 |
180 | target = {}
181 | target["boxes"] = boxes
182 | target["labels"] = labels
183 | target["image_id"] = image_id
184 | target["iscrowd"] = iscrowd
185 |
186 | if self.augmentation is not None:
187 | sample = self.augmentation(**{"image": image, "bboxes": boxes, "labels": labels})
188 | image = sample["image"]
189 | target["boxes"] = torch.as_tensor(sample["bboxes"], dtype=torch.float32)
190 | # guards against crops that don't pass the min_visibility augmentation threshold
191 | if not target["boxes"].numel():
192 | return None
193 |
194 | target["labels"] = torch.as_tensor(sample["labels"], dtype=torch.int64)
195 |
196 | if self.preprocess is not None:
197 | image = self.preprocess(image=image)["image"]
198 |
199 | boxes = target["boxes"]
200 | target["area"] = (boxes[:, 3] - boxes[:, 1]) * (boxes[:, 2] - boxes[:, 0])
201 | return image, target, image_id
202 |
203 | def collate_fn(self, batch):
204 | batch = filter(lambda x: x is not None, batch)
205 | return tuple(zip(*batch))
206 |
--------------------------------------------------------------------------------
/cloudformation/defect-detection.yaml:
--------------------------------------------------------------------------------
1 | AWSTemplateFormatVersion: "2010-09-09"
2 | Description: "(SA0015) - sagemaker-defect-detection:
3 | Solution for training and deploying deep learning models for defect detection in images using Amazon SageMaker.
4 | Version 1"
5 |
6 | Parameters:
7 | SolutionPrefix:
8 | Type: String
9 | Default: "sagemaker-soln-dfd-"
10 | Description: |
11 | Used to name resources created as part of this stack (and inside nested stacks too).
12 | Can be the same as the stack name used by AWS CloudFormation, but this field has extra
13 | constraints because it's used to name resources with restrictions (e.g. Amazon S3 bucket
14 | names cannot contain capital letters).
15 | AllowedPattern: '^sagemaker-soln-dfd[a-z0-9\-]{1,20}$'
16 | ConstraintDescription: |
17 | Only allowed to use lowercase letters, hyphens and/or numbers.
18 | Should also start with 'sagemaker-soln-dfd-' for permission management.
19 |
20 | SolutionName:
21 | Description: |
22 | Prefix for the solution name. Needs to be 'sagemaker-defect-detection'
23 | or begin with 'sagemaker-defect-detection-' followed by a set of letters and hyphens.
24 | Used to specify a particular directory on S3, that can correspond to a development branch.
25 | Type: String
26 | Default: "sagemaker-defect-detection/1.1.0"
27 | AllowedPattern: '^sagemaker-defect-detection(/[0-9]+\.[0-9]+\.[0-9]+-?[a-zA-Z-0-9\.+]*)?$'
28 |
29 | IamRole:
30 | Type: String
31 | Default: ""
32 | Description: |
33 | IAM Role that will be attached to the resources created by this CloudFormation to grant them permissions to
34 | perform their required functions. This role should allow SageMaker and Lambda perform the required actions like
35 | creating training jobs and processing jobs. If left blank, the template will attempt to create a role for you.
36 | This can cause a stack creation error if you don't have privileges to create new roles.
37 |
38 | SageMakerNotebookInstanceType:
39 | Description: SageMaker notebook instance type.
40 | Type: String
41 | Default: "ml.t3.medium"
42 |
43 | CreateSageMakerNotebookInstance:
44 | Description: Whether to launch sagemaker notebook instance
45 | Type: String
46 | AllowedValues:
47 | - "true"
48 | - "false"
49 | Default: "true"
50 |
51 | StackVersion:
52 | Description: |
53 | CloudFormation Stack version.
54 | Use "release" version unless you are customizing the
55 | CloudFormation templates and the solution artifacts in S3 bucket
56 | Type: String
57 | Default: release
58 | AllowedValues:
59 | - release
60 | - development
61 |
62 | Metadata:
63 | AWS::CloudFormation::Interface:
64 | ParameterGroups:
65 | - Label:
66 | default: "Solution Configuration"
67 | Parameters:
68 | - SolutionPrefix
69 | - SolutionName
70 | - IamRole
71 | - Label:
72 | default: "Advanced Configuration"
73 | Parameters:
74 | - SageMakerNotebookInstanceType
75 | - CreateSageMakerNotebookInstance
76 |
77 | ParameterLabels:
78 | SolutionPrefix:
79 | default: "Solution Resources Name Prefix"
80 | SolutionName:
81 | default: "Name of the solution"
82 | IamRole:
83 | default: "Solution IAM Role Arn"
84 | CreateSageMakerNotebookInstance:
85 | default: "Launch SageMaker Notebook Instance"
86 | SageMakerNotebookInstanceType:
87 | default: "SageMaker Notebook Instance Type"
88 | StackVersion:
89 | default: "Solution Stack Version"
90 |
91 | Conditions:
92 | CreateClassicSageMakerResources:
93 | !Equals [!Ref CreateSageMakerNotebookInstance, "true"]
94 | CreateCustomSolutionRole: !Equals [!Ref IamRole, ""]
95 |
96 | Mappings:
97 | S3:
98 | release:
99 | BucketPrefix: "sagemaker-solutions-prod"
100 | development:
101 | BucketPrefix: "sagemaker-solutions-devo"
102 |
103 | Resources:
104 | S3Bucket:
105 | Type: AWS::S3::Bucket
106 | DeletionPolicy: Retain
107 | UpdateReplacePolicy: "Retain"
108 | Properties:
109 | BucketName: !Sub "${SolutionPrefix}-${AWS::AccountId}-${AWS::Region}"
110 | PublicAccessBlockConfiguration:
111 | BlockPublicAcls: true
112 | BlockPublicPolicy: true
113 | IgnorePublicAcls: true
114 | RestrictPublicBuckets: true
115 | BucketEncryption:
116 | ServerSideEncryptionConfiguration:
117 | - ServerSideEncryptionByDefault:
118 | SSEAlgorithm: AES256
119 |
120 | Metadata:
121 | cfn_nag:
122 | rules_to_suppress:
123 | - id: W35
124 | reason: Configuring logging requires supplying an existing customer S3 bucket to store logs.
125 | - id: W51
126 | reason: Default access policy is sufficient.
127 |
128 | SageMakerPermissionsStack:
129 | Type: "AWS::CloudFormation::Stack"
130 | Condition: CreateCustomSolutionRole
131 | Properties:
132 | TemplateURL: !Sub
133 | - "https://${SolutionRefBucketBase}-${AWS::Region}.s3.${AWS::Region}.amazonaws.com/${SolutionName}/cloudformation/defect-detection-permissions.yaml"
134 | - SolutionRefBucketBase:
135 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
136 | Parameters:
137 | SolutionPrefix: !Ref SolutionPrefix
138 | SolutionName: !Ref SolutionName
139 | S3Bucket: !Ref S3Bucket
140 | StackVersion: !Ref StackVersion
141 |
142 | SageMakerStack:
143 | Type: "AWS::CloudFormation::Stack"
144 | Condition: CreateClassicSageMakerResources
145 | Properties:
146 | TemplateURL: !Sub
147 | - "https://${SolutionRefBucketBase}-${AWS::Region}.s3.${AWS::Region}.amazonaws.com/${SolutionName}/cloudformation/defect-detection-sagemaker-notebook-instance.yaml"
148 | - SolutionRefBucketBase:
149 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
150 | Parameters:
151 | SolutionPrefix: !Ref SolutionPrefix
152 | SolutionName: !Ref SolutionName
153 | S3Bucket: !Ref S3Bucket
154 | SageMakerIAMRoleArn:
155 | !If [
156 | CreateCustomSolutionRole,
157 | !GetAtt SageMakerPermissionsStack.Outputs.SageMakerRoleArn,
158 | !Ref IamRole,
159 | ]
160 | SageMakerNotebookInstanceType: !Ref SageMakerNotebookInstanceType
161 | StackVersion: !Ref StackVersion
162 |
163 | SolutionAssistantStack:
164 | Type: "AWS::CloudFormation::Stack"
165 | Properties:
166 | TemplateURL: !Sub
167 | - "https://${SolutionRefBucketBase}-${AWS::Region}.s3.${AWS::Region}.amazonaws.com/${SolutionName}/cloudformation/solution-assistant/solution-assistant.yaml"
168 | - SolutionRefBucketBase:
169 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
170 | Parameters:
171 | SolutionPrefix: !Ref SolutionPrefix
172 | SolutionName: !Ref SolutionName
173 | StackName: !Ref AWS::StackName
174 | S3Bucket: !Ref S3Bucket
175 | SolutionS3Bucket: !Sub
176 | - "${SolutionRefBucketBase}-${AWS::Region}"
177 | - SolutionRefBucketBase:
178 | !FindInMap [S3, !Ref StackVersion, BucketPrefix]
179 | RoleArn:
180 | !If [
181 | CreateCustomSolutionRole,
182 | !GetAtt SageMakerPermissionsStack.Outputs.SageMakerRoleArn,
183 | !Ref IamRole,
184 | ]
185 |
186 | Outputs:
187 | SolutionName:
188 | Description: "The name for the solution, can be used to deploy different versions of the solution"
189 | Value: !Ref SolutionName
190 |
191 | SourceCode:
192 | Condition: CreateClassicSageMakerResources
193 | Description: "Open Jupyter IDE. This authenticate you against Jupyter"
194 | Value: !GetAtt SageMakerStack.Outputs.SourceCode
195 |
196 | NotebookInstance:
197 | Description: "SageMaker Notebook instance to manually orchestrate data preprocessing, model training and deploying an endpoint"
198 | Value:
199 | !If [
200 | CreateClassicSageMakerResources,
201 | !GetAtt SageMakerStack.Outputs.SageMakerNotebookInstanceSignOn,
202 | "",
203 | ]
204 |
205 | AccountID:
206 | Description: "AWS Account ID to be passed downstream to the notebook instance"
207 | Value: !Ref AWS::AccountId
208 |
209 | AWSRegion:
210 | Description: "AWS Region to be passed downstream to the notebook instance"
211 | Value: !Ref AWS::Region
212 |
213 | IamRole:
214 | Description: "Arn of SageMaker Execution Role"
215 | Value:
216 | !If [
217 | CreateCustomSolutionRole,
218 | !GetAtt SageMakerPermissionsStack.Outputs.SageMakerRoleArn,
219 | !Ref IamRole,
220 | ]
221 |
222 | SolutionPrefix:
223 | Description: "Solution Prefix for naming SageMaker transient resources"
224 | Value: !Ref SolutionPrefix
225 |
226 | S3Bucket:
227 | Description: "S3 bucket name used in the solution to store artifacts"
228 | Value: !Ref S3Bucket
229 |
230 | SolutionS3Bucket:
231 | Description: "Solution S3 bucket"
232 | Value: !FindInMap [S3, !Ref StackVersion, BucketPrefix]
233 |
234 | SageMakerMode:
235 | Value: !If [CreateClassicSageMakerResources, "NotebookInstance", "Studio"]
236 |
237 | StackName:
238 | Value: !Ref AWS::StackName
239 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/classifier.py:
--------------------------------------------------------------------------------
1 | # mypy: ignore-errors
2 | from typing import Dict
3 | import os
4 | from collections import OrderedDict
5 | from argparse import ArgumentParser, Namespace
6 | from multiprocessing import cpu_count
7 |
8 | import torch
9 | import torch.optim as optim
10 | import torch.nn.functional as F
11 | from torch.utils.data import DataLoader
12 |
13 | import pytorch_lightning as pl
14 | from pytorch_lightning.callbacks import EarlyStopping, ModelCheckpoint
15 | import pytorch_lightning.metrics.functional as plm
16 |
17 | from sagemaker_defect_detection import Classification, NEUCLS, get_transform
18 | from sagemaker_defect_detection.utils import load_checkpoint, freeze
19 |
20 |
21 | def metrics(name: str, out: torch.Tensor, target: torch.Tensor) -> Dict[str, torch.Tensor]:
22 | pred = torch.argmax(out, 1).detach()
23 | target = target.detach()
24 | metrics = {}
25 | metrics[name + "_acc"] = plm.accuracy(pred, target)
26 | metrics[name + "_prec"] = plm.precision(pred, target)
27 | metrics[name + "_recall"] = plm.recall(pred, target)
28 | metrics[name + "_f1_score"] = plm.recall(pred, target)
29 | return metrics
30 |
31 |
32 | class DDNClassification(pl.LightningModule):
33 | def __init__(
34 | self,
35 | data_path: str,
36 | backbone: str,
37 | freeze_backbone: bool,
38 | num_classes: int,
39 | learning_rate: float,
40 | batch_size: int,
41 | momentum: float,
42 | weight_decay: float,
43 | seed: int,
44 | **kwargs
45 | ) -> None:
46 | super().__init__()
47 | self.data_path = data_path
48 | self.backbone = backbone
49 | self.freeze_backbone = freeze_backbone
50 | self.num_classes = num_classes
51 | self.learning_rate = learning_rate
52 | self.batch_size = batch_size
53 | self.momentum = momentum
54 | self.weight_decay = weight_decay
55 | self.seed = seed
56 |
57 | self.train_dataset = NEUCLS(self.data_path, split="train", transform=get_transform("train"), seed=self.seed)
58 | self.val_dataset = NEUCLS(self.data_path, split="val", transform=get_transform("val"), seed=self.seed)
59 | self.test_dataset = NEUCLS(self.data_path, split="test", transform=get_transform("test"), seed=self.seed)
60 |
61 | self.model = Classification(self.backbone, self.num_classes)
62 | if self.freeze_backbone:
63 | for param in self.model.mfn.backbone.parameters():
64 | param.requires_grad = False
65 |
66 | def forward(self, x): # ignore
67 | return self.model(x)
68 |
69 | def training_step(self, batch, batch_idx):
70 | images, target = batch
71 | output = self(images)
72 | loss_val = F.cross_entropy(output, target)
73 | metrics_dict = metrics("train", output, target)
74 | tqdm_dict = {"train_loss": loss_val, **metrics_dict}
75 | output = OrderedDict({"loss": loss_val, "progress_bar": tqdm_dict, "log": tqdm_dict})
76 | return output
77 |
78 | def validation_step(self, batch, batch_idx):
79 | images, target = batch
80 | output = self(images)
81 | loss_val = F.cross_entropy(output, target)
82 | metrics_dict = metrics("val", output, target)
83 | output = OrderedDict({"val_loss": loss_val, **metrics_dict})
84 | return output
85 |
86 | def validation_epoch_end(self, outputs):
87 | log_dict = {}
88 | for metric_name in outputs[0]:
89 | log_dict[metric_name] = torch.stack([x[metric_name] for x in outputs]).mean()
90 |
91 | return {"log": log_dict, "progress_bar": log_dict, **log_dict}
92 |
93 | def test_step(self, batch, batch_idx):
94 | images, target = batch
95 | output = self(images)
96 | loss_val = F.cross_entropy(output, target)
97 | metrics_dict = metrics("test", output, target)
98 | output = OrderedDict({"test_loss": loss_val, **metrics_dict})
99 | return output
100 |
101 | def test_epoch_end(self, outputs):
102 | log_dict = {}
103 | for metric_name in outputs[0]:
104 | log_dict[metric_name] = torch.stack([x[metric_name] for x in outputs]).mean()
105 |
106 | return {"log": log_dict, "progress_bar": log_dict, **log_dict}
107 |
108 | def configure_optimizers(self):
109 | optimizer = optim.SGD(
110 | self.parameters(), lr=self.learning_rate, momentum=self.momentum, weight_decay=self.weight_decay
111 | )
112 | return optimizer
113 |
114 | def train_dataloader(self):
115 | train_loader = DataLoader(
116 | dataset=self.train_dataset,
117 | batch_size=self.batch_size,
118 | shuffle=True,
119 | num_workers=cpu_count(),
120 | )
121 | return train_loader
122 |
123 | def val_dataloader(self):
124 | val_loader = DataLoader(
125 | self.val_dataset,
126 | batch_size=self.batch_size,
127 | shuffle=False,
128 | num_workers=cpu_count() // 2,
129 | )
130 | return val_loader
131 |
132 | def test_dataloader(self):
133 | test_loader = DataLoader(
134 | self.test_dataset,
135 | batch_size=self.batch_size,
136 | shuffle=False,
137 | num_workers=cpu_count(),
138 | )
139 | return test_loader
140 |
141 | @staticmethod
142 | def add_model_specific_args(parent_parser): # pragma: no-cover
143 | parser = ArgumentParser(parents=[parent_parser], add_help=False)
144 | aa = parser.add_argument
145 | aa(
146 | "--data-path",
147 | metavar="DIR",
148 | type=str,
149 | default=os.getenv("SM_CHANNEL_TRAINING", ""),
150 | )
151 | aa(
152 | "--backbone",
153 | default="resnet34",
154 | )
155 | aa(
156 | "--freeze-backbone",
157 | action="store_true",
158 | )
159 | aa(
160 | "--num-classes",
161 | default=6,
162 | type=int,
163 | metavar="N",
164 | )
165 | aa(
166 | "-b",
167 | "--batch-size",
168 | default=64,
169 | type=int,
170 | metavar="N",
171 | )
172 | aa(
173 | "--lr",
174 | "--learning-rate",
175 | default=1e-3,
176 | type=float,
177 | metavar="LR",
178 | dest="learning_rate",
179 | )
180 | aa("--momentum", default=0.9, type=float, metavar="M", help="momentum")
181 | aa(
182 | "--wd",
183 | "--weight-decay",
184 | default=1e-4,
185 | type=float,
186 | metavar="W",
187 | dest="weight_decay",
188 | )
189 | aa(
190 | "--seed",
191 | type=int,
192 | default=42,
193 | )
194 | return parser
195 |
196 |
197 | def get_args() -> Namespace:
198 | parent_parser = ArgumentParser(add_help=False)
199 | aa = parent_parser.add_argument
200 | aa("--epochs", type=int, default=100, help="number of training epochs")
201 | aa("--save-path", metavar="DIR", default=os.getenv("SM_MODEL_DIR", ""), type=str, help="path to save output")
202 | aa("--gpus", type=int, default=os.getenv("SM_NUM_GPUS", 1), help="how many gpus")
203 | aa(
204 | "--distributed-backend",
205 | type=str,
206 | default="",
207 | choices=("dp", "ddp", "ddp2"),
208 | help="supports three options dp, ddp, ddp2",
209 | )
210 | aa("--use-16bit", dest="use_16bit", action="store_true", help="if true uses 16 bit precision")
211 |
212 | parser = DDNClassification.add_model_specific_args(parent_parser)
213 | return parser.parse_args()
214 |
215 |
216 | def model_fn(model_dir):
217 | # TODO: `model_fn` doesn't get more args
218 | # see: https://github.com/aws/sagemaker-inference-toolkit/issues/65
219 | backbone = "resnet34"
220 | num_classes = 6
221 |
222 | model = load_checkpoint(Classification(backbone, num_classes), model_dir, prefix="model")
223 | model = model.eval()
224 | freeze(model)
225 | return model
226 |
227 |
228 | def main(args: Namespace) -> None:
229 | model = DDNClassification(**vars(args))
230 |
231 | if args.seed is not None:
232 | pl.seed_everything(args.seed)
233 | if torch.cuda.device_count() > 1:
234 | torch.cuda.manual_seed_all(args.seed)
235 |
236 | # TODO: add deterministic training
237 | # torch.backends.cudnn.deterministic = True
238 |
239 | checkpoint_callback = ModelCheckpoint(
240 | filepath=os.path.join(args.save_path, "{epoch}-{val_loss:.3f}-{val_acc:.3f}"),
241 | save_top_k=1,
242 | verbose=True,
243 | monitor="val_acc",
244 | mode="max",
245 | )
246 | early_stop_callback = EarlyStopping("val_loss", patience=10)
247 | trainer = pl.Trainer(
248 | default_root_dir=args.save_path,
249 | gpus=args.gpus,
250 | max_epochs=args.epochs,
251 | early_stop_callback=early_stop_callback,
252 | checkpoint_callback=checkpoint_callback,
253 | gradient_clip_val=10,
254 | num_sanity_val_steps=0,
255 | distributed_backend=args.distributed_backend or None,
256 | # precision=16 if args.use_16bit else 32, # TODO: amp apex support
257 | )
258 |
259 | trainer.fit(model)
260 | trainer.test()
261 | return
262 |
263 |
264 | if __name__ == "__main__":
265 | main(get_args())
266 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 |
2 | Apache License
3 | Version 2.0, January 2004
4 | http://www.apache.org/licenses/
5 |
6 | TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
7 |
8 | 1. Definitions.
9 |
10 | "License" shall mean the terms and conditions for use, reproduction,
11 | and distribution as defined by Sections 1 through 9 of this document.
12 |
13 | "Licensor" shall mean the copyright owner or entity authorized by
14 | the copyright owner that is granting the License.
15 |
16 | "Legal Entity" shall mean the union of the acting entity and all
17 | other entities that control, are controlled by, or are under common
18 | control with that entity. For the purposes of this definition,
19 | "control" means (i) the power, direct or indirect, to cause the
20 | direction or management of such entity, whether by contract or
21 | otherwise, or (ii) ownership of fifty percent (50%) or more of the
22 | outstanding shares, or (iii) beneficial ownership of such entity.
23 |
24 | "You" (or "Your") shall mean an individual or Legal Entity
25 | exercising permissions granted by this License.
26 |
27 | "Source" form shall mean the preferred form for making modifications,
28 | including but not limited to software source code, documentation
29 | source, and configuration files.
30 |
31 | "Object" form shall mean any form resulting from mechanical
32 | transformation or translation of a Source form, including but
33 | not limited to compiled object code, generated documentation,
34 | and conversions to other media types.
35 |
36 | "Work" shall mean the work of authorship, whether in Source or
37 | Object form, made available under the License, as indicated by a
38 | copyright notice that is included in or attached to the work
39 | (an example is provided in the Appendix below).
40 |
41 | "Derivative Works" shall mean any work, whether in Source or Object
42 | form, that is based on (or derived from) the Work and for which the
43 | editorial revisions, annotations, elaborations, or other modifications
44 | represent, as a whole, an original work of authorship. For the purposes
45 | of this License, Derivative Works shall not include works that remain
46 | separable from, or merely link (or bind by name) to the interfaces of,
47 | the Work and Derivative Works thereof.
48 |
49 | "Contribution" shall mean any work of authorship, including
50 | the original version of the Work and any modifications or additions
51 | to that Work or Derivative Works thereof, that is intentionally
52 | submitted to Licensor for inclusion in the Work by the copyright owner
53 | or by an individual or Legal Entity authorized to submit on behalf of
54 | the copyright owner. For the purposes of this definition, "submitted"
55 | means any form of electronic, verbal, or written communication sent
56 | to the Licensor or its representatives, including but not limited to
57 | communication on electronic mailing lists, source code control systems,
58 | and issue tracking systems that are managed by, or on behalf of, the
59 | Licensor for the purpose of discussing and improving the Work, but
60 | excluding communication that is conspicuously marked or otherwise
61 | designated in writing by the copyright owner as "Not a Contribution."
62 |
63 | "Contributor" shall mean Licensor and any individual or Legal Entity
64 | on behalf of whom a Contribution has been received by Licensor and
65 | subsequently incorporated within the Work.
66 |
67 | 2. Grant of Copyright License. Subject to the terms and conditions of
68 | this License, each Contributor hereby grants to You a perpetual,
69 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
70 | copyright license to reproduce, prepare Derivative Works of,
71 | publicly display, publicly perform, sublicense, and distribute the
72 | Work and such Derivative Works in Source or Object form.
73 |
74 | 3. Grant of Patent License. Subject to the terms and conditions of
75 | this License, each Contributor hereby grants to You a perpetual,
76 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
77 | (except as stated in this section) patent license to make, have made,
78 | use, offer to sell, sell, import, and otherwise transfer the Work,
79 | where such license applies only to those patent claims licensable
80 | by such Contributor that are necessarily infringed by their
81 | Contribution(s) alone or by combination of their Contribution(s)
82 | with the Work to which such Contribution(s) was submitted. If You
83 | institute patent litigation against any entity (including a
84 | cross-claim or counterclaim in a lawsuit) alleging that the Work
85 | or a Contribution incorporated within the Work constitutes direct
86 | or contributory patent infringement, then any patent licenses
87 | granted to You under this License for that Work shall terminate
88 | as of the date such litigation is filed.
89 |
90 | 4. Redistribution. You may reproduce and distribute copies of the
91 | Work or Derivative Works thereof in any medium, with or without
92 | modifications, and in Source or Object form, provided that You
93 | meet the following conditions:
94 |
95 | (a) You must give any other recipients of the Work or
96 | Derivative Works a copy of this License; and
97 |
98 | (b) You must cause any modified files to carry prominent notices
99 | stating that You changed the files; and
100 |
101 | (c) You must retain, in the Source form of any Derivative Works
102 | that You distribute, all copyright, patent, trademark, and
103 | attribution notices from the Source form of the Work,
104 | excluding those notices that do not pertain to any part of
105 | the Derivative Works; and
106 |
107 | (d) If the Work includes a "NOTICE" text file as part of its
108 | distribution, then any Derivative Works that You distribute must
109 | include a readable copy of the attribution notices contained
110 | within such NOTICE file, excluding those notices that do not
111 | pertain to any part of the Derivative Works, in at least one
112 | of the following places: within a NOTICE text file distributed
113 | as part of the Derivative Works; within the Source form or
114 | documentation, if provided along with the Derivative Works; or,
115 | within a display generated by the Derivative Works, if and
116 | wherever such third-party notices normally appear. The contents
117 | of the NOTICE file are for informational purposes only and
118 | do not modify the License. You may add Your own attribution
119 | notices within Derivative Works that You distribute, alongside
120 | or as an addendum to the NOTICE text from the Work, provided
121 | that such additional attribution notices cannot be construed
122 | as modifying the License.
123 |
124 | You may add Your own copyright statement to Your modifications and
125 | may provide additional or different license terms and conditions
126 | for use, reproduction, or distribution of Your modifications, or
127 | for any such Derivative Works as a whole, provided Your use,
128 | reproduction, and distribution of the Work otherwise complies with
129 | the conditions stated in this License.
130 |
131 | 5. Submission of Contributions. Unless You explicitly state otherwise,
132 | any Contribution intentionally submitted for inclusion in the Work
133 | by You to the Licensor shall be under the terms and conditions of
134 | this License, without any additional terms or conditions.
135 | Notwithstanding the above, nothing herein shall supersede or modify
136 | the terms of any separate license agreement you may have executed
137 | with Licensor regarding such Contributions.
138 |
139 | 6. Trademarks. This License does not grant permission to use the trade
140 | names, trademarks, service marks, or product names of the Licensor,
141 | except as required for reasonable and customary use in describing the
142 | origin of the Work and reproducing the content of the NOTICE file.
143 |
144 | 7. Disclaimer of Warranty. Unless required by applicable law or
145 | agreed to in writing, Licensor provides the Work (and each
146 | Contributor provides its Contributions) on an "AS IS" BASIS,
147 | WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
148 | implied, including, without limitation, any warranties or conditions
149 | of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
150 | PARTICULAR PURPOSE. You are solely responsible for determining the
151 | appropriateness of using or redistributing the Work and assume any
152 | risks associated with Your exercise of permissions under this License.
153 |
154 | 8. Limitation of Liability. In no event and under no legal theory,
155 | whether in tort (including negligence), contract, or otherwise,
156 | unless required by applicable law (such as deliberate and grossly
157 | negligent acts) or agreed to in writing, shall any Contributor be
158 | liable to You for damages, including any direct, indirect, special,
159 | incidental, or consequential damages of any character arising as a
160 | result of this License or out of the use or inability to use the
161 | Work (including but not limited to damages for loss of goodwill,
162 | work stoppage, computer failure or malfunction, or any and all
163 | other commercial damages or losses), even if such Contributor
164 | has been advised of the possibility of such damages.
165 |
166 | 9. Accepting Warranty or Additional Liability. While redistributing
167 | the Work or Derivative Works thereof, You may choose to offer,
168 | and charge a fee for, acceptance of support, warranty, indemnity,
169 | or other liability obligations and/or rights consistent with this
170 | License. However, in accepting such obligations, You may act only
171 | on Your own behalf and on Your sole responsibility, not on behalf
172 | of any other Contributor, and only if You agree to indemnify,
173 | defend, and hold each Contributor harmless for any liability
174 | incurred by, or claims asserted against, such Contributor by reason
175 | of your accepting any such warranty or additional liability.
176 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/models/ddn.py:
--------------------------------------------------------------------------------
1 | import torch
2 | import torch.nn as nn
3 | import torch.nn.functional as F
4 |
5 | import torchvision
6 | from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
7 | from torchvision.models.detection.transform import GeneralizedRCNNTransform
8 | from torchvision.models.detection.generalized_rcnn import GeneralizedRCNN
9 | from torchvision.models.detection.roi_heads import RoIHeads
10 | from torchvision.models.detection.rpn import AnchorGenerator, RPNHead, RegionProposalNetwork
11 | from torchvision.ops import MultiScaleRoIAlign
12 |
13 |
14 | def get_backbone(name: str) -> nn.Module:
15 | """
16 | Get official pretrained ResNet34 and ResNet50 as backbones
17 |
18 | Parameters
19 | ----------
20 | name : str
21 | Either `resnet34` or `resnet50`
22 |
23 | Returns
24 | -------
25 | nn.Module
26 | resnet34 or resnet50 pytorch modules
27 |
28 | Raises
29 | ------
30 | ValueError
31 | If unsupported name is used
32 | """
33 | if name == "resnet34":
34 | return torchvision.models.resnet34(pretrained=True)
35 | elif name == "resnet50":
36 | return torchvision.models.resnet50(pretrained=True)
37 | else:
38 | raise ValueError("Unsupported backbone")
39 |
40 |
41 | def init_weights(m) -> None:
42 | """
43 | Weight initialization
44 |
45 | Parameters
46 | ----------
47 | m : [type]
48 | Module used in recursive call
49 | """
50 | if isinstance(m, nn.Conv2d):
51 | nn.init.xavier_normal_(m.weight)
52 |
53 | elif isinstance(m, nn.Linear):
54 | m.weight.data.normal_(0.0, 0.02)
55 | m.bias.data.fill_(0.0)
56 |
57 | return
58 |
59 |
60 | class MFN(nn.Module):
61 | def __init__(self, backbone: str):
62 | """
63 | Implementation of MFN model as described in
64 |
65 | Yu He, Kechen Song, Qinggang Meng, Yunhui Yan,
66 | “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,”
67 | IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
68 |
69 | Parameters
70 | ----------
71 | backbone : str
72 | Either `resnet34` or `resnet50`
73 | """
74 | super().__init__()
75 | self.backbone = get_backbone(backbone)
76 | # input 224x224 -> conv1 output size 112x112
77 | self.start_layer = nn.Sequential(
78 | self.backbone.conv1, # type: ignore
79 | self.backbone.bn1, # type: ignore
80 | self.backbone.relu, # type: ignore
81 | self.backbone.maxpool, # type: ignore
82 | )
83 | self.r2 = self.backbone.layer1 # 64/256x56x56 <- (resnet34/resnet50)
84 | self.r3 = self.backbone.layer2 # 128/512x28x28
85 | self.r4 = self.backbone.layer3 # 256/1024x14x14
86 | self.r5 = self.backbone.layer4 # 512/2048x7x7
87 | in_channel = 64 if backbone == "resnet34" else 256
88 | self.b2 = nn.Sequential(
89 | nn.Conv2d(
90 | in_channel, in_channel, kernel_size=3, padding=1, stride=2
91 | ), # 56 -> 28 without Relu or batchnorm not in the paper ???
92 | nn.BatchNorm2d(in_channel),
93 | nn.ReLU(inplace=True),
94 | nn.Conv2d(in_channel, in_channel, kernel_size=3, padding=1, stride=2), # 28 -> 14
95 | nn.BatchNorm2d(in_channel),
96 | nn.ReLU(inplace=True),
97 | nn.Conv2d(in_channel, in_channel * 2, kernel_size=1, padding=0),
98 | nn.BatchNorm2d(in_channel * 2),
99 | nn.ReLU(inplace=True),
100 | ).apply(
101 | init_weights
102 | ) # after r2: 128/512x14x14 <-
103 | self.b3 = nn.MaxPool2d(2) # after r3: 128/512x14x14 <-
104 | in_channel *= 2 # 128/512
105 | self.b4 = nn.Sequential(
106 | nn.Conv2d(in_channel * 2, in_channel, kernel_size=1, padding=0),
107 | nn.BatchNorm2d(in_channel),
108 | nn.ReLU(inplace=True),
109 | ).apply(
110 | init_weights
111 | ) # after r4: 128/512x14x14
112 | in_channel *= 4 # 512 / 2048
113 | self.b5 = nn.Sequential(
114 | nn.ConvTranspose2d(
115 | in_channel, in_channel, kernel_size=3, stride=2, padding=1, output_padding=1
116 | ), # <- after r5 which is 512x7x7 -> 512x14x14
117 | nn.BatchNorm2d(in_channel),
118 | nn.ReLU(inplace=True),
119 | nn.Conv2d(in_channel, in_channel // 4, kernel_size=1, padding=0),
120 | nn.BatchNorm2d(in_channel // 4),
121 | nn.ReLU(inplace=True),
122 | ).apply(init_weights)
123 |
124 | self.out_channels = 512 if backbone == "resnet34" else 2048 # required for FasterRCNN
125 |
126 | def forward(self, x):
127 | x = self.start_layer(x)
128 | x = self.r2(x)
129 | b2_out = self.b2(x)
130 | x = self.r3(x)
131 | b3_out = self.b3(x)
132 | x = self.r4(x)
133 | b4_out = self.b4(x)
134 | x = self.r5(x)
135 | b5_out = self.b5(x)
136 | # BatchNorm works better than L2 normalize
137 | # out = torch.cat([F.normalize(o, p=2, dim=1) for o in (b2_out, b3_out, b4_out, b5_out)], dim=1)
138 | out = torch.cat((b2_out, b3_out, b4_out, b5_out), dim=1)
139 | return out
140 |
141 |
142 | class Classification(nn.Module):
143 | """
144 | Classification network
145 |
146 | Parameters
147 | ----------
148 | backbone : str
149 | Either `resnet34` or `resnet50`
150 |
151 | num_classes : int
152 | Number of classes
153 | """
154 |
155 | def __init__(self, backbone: str, num_classes: int) -> None:
156 | super().__init__()
157 | self.mfn = MFN(backbone)
158 | self.flatten = nn.Flatten()
159 | self.fc = nn.Linear(self.mfn.out_channels * 14 ** 2, num_classes)
160 |
161 | def forward(self, x):
162 | return self.fc(self.flatten(self.mfn(x)))
163 |
164 |
165 | class RPN(nn.Module):
166 | """
167 | RPN Module as described in
168 |
169 | Yu He, Kechen Song, Qinggang Meng, Yunhui Yan,
170 | “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,”
171 | IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
172 | """
173 |
174 | def __init__(
175 | self,
176 | out_channels: int = 512,
177 | rpn_pre_nms_top_n_train: int = 1000, # torchvision default 2000,
178 | rpn_pre_nms_top_n_test: int = 500, # torchvision default 1000,
179 | rpn_post_nms_top_n_train: int = 1000, # torchvision default 2000,
180 | rpn_post_nms_top_n_test: int = 500, # torchvision default 1000,
181 | rpn_nms_thresh: float = 0.7,
182 | rpn_fg_iou_thresh: float = 0.7,
183 | rpn_bg_iou_thresh: float = 0.3,
184 | rpn_batch_size_per_image: int = 256,
185 | rpn_positive_fraction: float = 0.5,
186 | ) -> None:
187 | super().__init__()
188 | rpn_anchor_generator = AnchorGenerator(sizes=((64, 128, 256, 512),), aspect_ratios=((0.5, 1.0, 2.0),))
189 | rpn_head = RPNHead(out_channels, rpn_anchor_generator.num_anchors_per_location()[0])
190 | rpn_pre_nms_top_n = dict(training=rpn_pre_nms_top_n_train, testing=rpn_pre_nms_top_n_test)
191 | rpn_post_nms_top_n = dict(training=rpn_post_nms_top_n_train, testing=rpn_post_nms_top_n_test)
192 | self.rpn = RegionProposalNetwork(
193 | rpn_anchor_generator,
194 | rpn_head,
195 | rpn_fg_iou_thresh,
196 | rpn_bg_iou_thresh,
197 | rpn_batch_size_per_image,
198 | rpn_positive_fraction,
199 | rpn_pre_nms_top_n,
200 | rpn_post_nms_top_n,
201 | rpn_nms_thresh,
202 | )
203 |
204 | def forward(self, *args, **kwargs):
205 | return self.rpn(*args, **kwargs)
206 |
207 |
208 | class CustomTwoMLPHead(nn.Module):
209 | def __init__(self, in_channels: int, representation_size: int):
210 | super().__init__()
211 | self.avgpool = nn.AdaptiveAvgPool2d(7)
212 | self.mlp = nn.Sequential(
213 | nn.Linear(in_channels, representation_size),
214 | nn.ReLU(inplace=True),
215 | nn.Linear(representation_size, representation_size),
216 | nn.ReLU(inplace=True),
217 | )
218 |
219 | def forward(self, x):
220 | x = self.avgpool(x)
221 | x = x.flatten(start_dim=1)
222 | x = self.mlp(x)
223 | return x
224 |
225 |
226 | class RoI(nn.Module):
227 | """
228 | ROI Module as described in
229 |
230 | Yu He, Kechen Song, Qinggang Meng, Yunhui Yan,
231 | “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,”
232 | IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
233 | """
234 |
235 | def __init__(
236 | self,
237 | num_classes: int,
238 | box_fg_iou_thresh=0.5,
239 | box_bg_iou_thresh=0.5,
240 | box_batch_size_per_image=512,
241 | box_positive_fraction=0.25,
242 | bbox_reg_weights=None,
243 | box_score_thresh=0.05,
244 | box_nms_thresh=0.5,
245 | box_detections_per_img=100,
246 | ) -> None:
247 | super().__init__()
248 | roi_pooler = MultiScaleRoIAlign(featmap_names=["0"], output_size=7, sampling_ratio=2)
249 | box_head = CustomTwoMLPHead(512 * 7 ** 2, 1024)
250 | box_predictor = FastRCNNPredictor(1024, num_classes=num_classes)
251 | self.roi_head = RoIHeads(
252 | roi_pooler,
253 | box_head,
254 | box_predictor,
255 | box_fg_iou_thresh,
256 | box_bg_iou_thresh,
257 | box_batch_size_per_image,
258 | box_positive_fraction,
259 | bbox_reg_weights,
260 | box_score_thresh,
261 | box_nms_thresh,
262 | box_detections_per_img,
263 | )
264 |
265 | def forward(self, *args, **kwargs):
266 | return self.roi_head(*args, **kwargs)
267 |
268 |
269 | class Detection(GeneralizedRCNN):
270 | """
271 | Detection network as described in
272 |
273 | Yu He, Kechen Song, Qinggang Meng, Yunhui Yan,
274 | “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,”
275 | IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
276 | """
277 |
278 | def __init__(self, mfn, rpn, roi):
279 | dummy_transform = GeneralizedRCNNTransform(800, 1333, [00.0, 0.0, 0.0], [1.0, 1.0, 1.0])
280 | super().__init__(mfn, rpn, roi, dummy_transform)
281 |
--------------------------------------------------------------------------------
/notebooks/3_classification_from_scratch.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "## Training our Classifier from scratch\n",
18 | "\n",
19 | "Depending on an application, sometimes image classification is enough. In this notebook, we see how to train and deploy an accurate classifier from scratch on **NEU-CLS** dataset"
20 | ]
21 | },
22 | {
23 | "cell_type": "code",
24 | "execution_count": null,
25 | "metadata": {
26 | "tags": []
27 | },
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import numpy as np\n",
33 | "\n",
34 | "import sagemaker\n",
35 | "from sagemaker.s3 import S3Downloader\n",
36 | "\n",
37 | "sagemaker_session = sagemaker.Session()\n",
38 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
39 | "role = sagemaker_config[\"IamRole\"]\n",
40 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
41 | "region = sagemaker_config[\"AWSRegion\"]\n",
42 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
43 | "bucket = sagemaker_config[\"S3Bucket\"]"
44 | ]
45 | },
46 | {
47 | "cell_type": "markdown",
48 | "metadata": {},
49 | "source": [
50 | "First, we download our **NEU-CLS** dataset from our public S3 bucket"
51 | ]
52 | },
53 | {
54 | "cell_type": "code",
55 | "execution_count": null,
56 | "metadata": {},
57 | "outputs": [],
58 | "source": [
59 | "from sagemaker.s3 import S3Downloader\n",
60 | "\n",
61 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
62 | "original_data = f\"{original_bucket}/data/NEU-CLS.zip\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\"\n",
64 | "print(\"original data: \")\n",
65 | "S3Downloader.list(original_data)"
66 | ]
67 | },
68 | {
69 | "cell_type": "markdown",
70 | "metadata": {},
71 | "source": [
72 | "For easiler data processing, depending on the dataset, we unify the class and label names using the scripts from `prepare_data`"
73 | ]
74 | },
75 | {
76 | "cell_type": "code",
77 | "execution_count": null,
78 | "metadata": {
79 | "tags": []
80 | },
81 | "outputs": [],
82 | "source": [
83 | "%%time\n",
84 | "RAW_DATA_PATH= !echo $PWD/raw_neu_cls\n",
85 | "RAW_DATA_PATH = RAW_DATA_PATH.n\n",
86 | "DATA_PATH = !echo $PWD/neu_cls\n",
87 | "DATA_PATH = DATA_PATH.n\n",
88 | "\n",
89 | "!mkdir -p $RAW_DATA_PATH\n",
90 | "!aws s3 cp $original_data $RAW_DATA_PATH\n",
91 | "\n",
92 | "!mkdir -p $DATA_PATH\n",
93 | "!python ../src/prepare_data/neu.py $RAW_DATA_PATH/NEU-CLS.zip $DATA_PATH"
94 | ]
95 | },
96 | {
97 | "cell_type": "markdown",
98 | "metadata": {},
99 | "source": [
100 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
101 | ]
102 | },
103 | {
104 | "cell_type": "code",
105 | "execution_count": null,
106 | "metadata": {},
107 | "outputs": [],
108 | "source": [
109 | "prefix = \"neu-cls\"\n",
110 | "neu_cls_s3 = f\"s3://{bucket}/{prefix}\"\n",
111 | "sources = f\"{neu_cls_s3}/code/\"\n",
112 | "train_output = f\"{neu_cls_s3}/output/\"\n",
113 | "neu_cls_prepared_s3 = f\"{neu_cls_s3}/data/\"\n",
114 | "!aws s3 sync $DATA_PATH $neu_cls_prepared_s3 --quiet # remove the --quiet flag to view sync outputs\n",
115 | "s3_checkpoint = f\"{neu_cls_s3}/checkpoint/\"\n",
116 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
117 | "!aws s3 cp $original_sources $sources"
118 | ]
119 | },
120 | {
121 | "cell_type": "markdown",
122 | "metadata": {},
123 | "source": [
124 | "## Visualization\n",
125 | "\n",
126 | "Let examine some datasets that we will use later by providing an `ID`"
127 | ]
128 | },
129 | {
130 | "cell_type": "code",
131 | "execution_count": null,
132 | "metadata": {
133 | "tags": []
134 | },
135 | "outputs": [],
136 | "source": [
137 | "import matplotlib.pyplot as plt\n",
138 | "\n",
139 | "%matplotlib inline\n",
140 | "import numpy as np\n",
141 | "import torch\n",
142 | "from PIL import Image\n",
143 | "from torch.utils.data import DataLoader\n",
144 | "\n",
145 | "try:\n",
146 | " import sagemaker_defect_detection\n",
147 | "except ImportError:\n",
148 | " import sys\n",
149 | " from pathlib import Path\n",
150 | "\n",
151 | " ROOT = Path(\"../src\").resolve()\n",
152 | " sys.path.insert(0, str(ROOT))\n",
153 | "\n",
154 | "from sagemaker_defect_detection import NEUCLS\n",
155 | "\n",
156 | "\n",
157 | "def visualize(image, label, predicted=None):\n",
158 | " if not isinstance(image, Image.Image):\n",
159 | " image = Image.fromarray(image)\n",
160 | "\n",
161 | " plt.figure(dpi=120)\n",
162 | " if predicted is not None:\n",
163 | " plt.title(f\"label: {label}, prediction: {predicted}\")\n",
164 | " else:\n",
165 | " plt.title(f\"label: {label}\")\n",
166 | "\n",
167 | " plt.axis(\"off\")\n",
168 | " plt.imshow(image)\n",
169 | " return\n",
170 | "\n",
171 | "\n",
172 | "dataset = NEUCLS(DATA_PATH, split=\"train\")\n",
173 | "ID = 0\n",
174 | "assert 0 <= ID <= 300\n",
175 | "image, label = dataset[ID]\n",
176 | "visualize(image, label)"
177 | ]
178 | },
179 | {
180 | "cell_type": "markdown",
181 | "metadata": {},
182 | "source": [
183 | "We train our model with `resnet34` backbone for **50 epochs** and obtains about **99%** test accuracy, f1-score, precision and recall as follows"
184 | ]
185 | },
186 | {
187 | "cell_type": "code",
188 | "execution_count": null,
189 | "metadata": {
190 | "tags": [
191 | "outputPrepend"
192 | ]
193 | },
194 | "outputs": [],
195 | "source": [
196 | "%%time\n",
197 | "import logging\n",
198 | "from os import path as osp\n",
199 | "\n",
200 | "from sagemaker.pytorch import PyTorch\n",
201 | "\n",
202 | "NUM_CLASSES = 6\n",
203 | "BACKBONE = \"resnet34\"\n",
204 | "assert BACKBONE in [\n",
205 | " \"resnet34\",\n",
206 | " \"resnet50\",\n",
207 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in classifier.py\"\n",
208 | "EPOCHS = 50\n",
209 | "SEED = 123\n",
210 | "\n",
211 | "hyperparameters = {\n",
212 | " \"backbone\": BACKBONE,\n",
213 | " \"num-classes\": NUM_CLASSES,\n",
214 | " \"epochs\": EPOCHS,\n",
215 | " \"seed\": SEED,\n",
216 | "}\n",
217 | "\n",
218 | "assert not isinstance(sagemaker_session, sagemaker.LocalSession), \"local session as share memory cannot be altered\"\n",
219 | "\n",
220 | "model = PyTorch(\n",
221 | " entry_point=\"classifier.py\",\n",
222 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
223 | " role=role,\n",
224 | " train_instance_count=1,\n",
225 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
226 | " hyperparameters=hyperparameters,\n",
227 | " py_version=\"py3\",\n",
228 | " framework_version=\"1.5\",\n",
229 | " sagemaker_session=sagemaker_session, # Note: Do not use local session as share memory cannot be altered\n",
230 | " output_path=train_output,\n",
231 | " checkpoint_s3_uri=s3_checkpoint,\n",
232 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
233 | " # container_log_level=logging.DEBUG,\n",
234 | ")\n",
235 | "\n",
236 | "model.fit(neu_cls_prepared_s3)"
237 | ]
238 | },
239 | {
240 | "cell_type": "markdown",
241 | "metadata": {},
242 | "source": [
243 | "Then, we deploy our model which takes about **8 minutes** to complete"
244 | ]
245 | },
246 | {
247 | "cell_type": "code",
248 | "execution_count": null,
249 | "metadata": {
250 | "tags": []
251 | },
252 | "outputs": [],
253 | "source": [
254 | "%%time\n",
255 | "predictor = model.deploy(\n",
256 | " initial_instance_count=1,\n",
257 | " instance_type=\"ml.m5.xlarge\",\n",
258 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-classification-endpoint\",\n",
259 | ")"
260 | ]
261 | },
262 | {
263 | "cell_type": "markdown",
264 | "metadata": {},
265 | "source": [
266 | "## Inference\n",
267 | "\n",
268 | "We are ready to test our model by providing some test data and compare the actual labels with the predicted one"
269 | ]
270 | },
271 | {
272 | "cell_type": "code",
273 | "execution_count": null,
274 | "metadata": {
275 | "tags": []
276 | },
277 | "outputs": [],
278 | "source": [
279 | "from sagemaker_defect_detection import get_transform\n",
280 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc\n",
281 | "\n",
282 | "ID = 100\n",
283 | "assert 0 <= ID <= 300\n",
284 | "test_dataset = NEUCLS(DATA_PATH, split=\"test\", transform=get_transform(\"test\"), seed=SEED)\n",
285 | "image, label = test_dataset[ID]\n",
286 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
287 | "np_load_old = np.load\n",
288 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
289 | "outputs = predictor.predict(image.unsqueeze(0).numpy())\n",
290 | "np.load = np_load_old\n",
291 | "\n",
292 | "_, predicted = torch.max(torch.from_numpy(np.array(outputs)), 1)\n",
293 | "\n",
294 | "image_unnorm = unnormalize_to_hwc(image)\n",
295 | "visualize(image_unnorm, label, predicted.item())"
296 | ]
297 | },
298 | {
299 | "cell_type": "markdown",
300 | "metadata": {},
301 | "source": [
302 | "## Optional: Delete the endpoint and model\n",
303 | "\n",
304 | "When you are done with the endpoint, you should clean it up.\n",
305 | "\n",
306 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
307 | ]
308 | },
309 | {
310 | "cell_type": "code",
311 | "execution_count": null,
312 | "metadata": {},
313 | "outputs": [],
314 | "source": [
315 | "predictor.delete_model()\n",
316 | "predictor.delete_endpoint()"
317 | ]
318 | }
319 | ],
320 | "metadata": {
321 | "kernelspec": {
322 | "display_name": "Python 3.6.10 64-bit",
323 | "metadata": {
324 | "interpreter": {
325 | "hash": "4c1e195df8d07db5ee7a78f454b46c3f2e14214bf8c9489d2db5cf8f372ff2ed"
326 | }
327 | },
328 | "name": "python3"
329 | },
330 | "language_info": {
331 | "codemirror_mode": {
332 | "name": "ipython",
333 | "version": 3
334 | },
335 | "file_extension": ".py",
336 | "mimetype": "text/x-python",
337 | "name": "python",
338 | "nbconvert_exporter": "python",
339 | "pygments_lexer": "ipython3",
340 | "version": "3.6.10-final"
341 | }
342 | },
343 | "nbformat": 4,
344 | "nbformat_minor": 2
345 | }
346 |
--------------------------------------------------------------------------------
/notebooks/0_demo.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## SageMaker Defect Detection Demo\n",
20 | "\n",
21 | "In this notebook, we deploy an endpoint from a provided pretrained detection model that was already trained on **NEU-DET** dataset. Then, we send some image samples with defects for detection and visual the results"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": null,
27 | "metadata": {},
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import numpy as np\n",
33 | "\n",
34 | "import sagemaker\n",
35 | "from sagemaker.s3 import S3Downloader\n",
36 | "\n",
37 | "sagemaker_session = sagemaker.Session()\n",
38 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
39 | "role = sagemaker_config[\"IamRole\"]\n",
40 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
41 | "region = sagemaker_config[\"AWSRegion\"]\n",
42 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
43 | "bucket = sagemaker_config[\"S3Bucket\"]"
44 | ]
45 | },
46 | {
47 | "cell_type": "markdown",
48 | "metadata": {},
49 | "source": [
50 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
51 | ]
52 | },
53 | {
54 | "cell_type": "code",
55 | "execution_count": null,
56 | "metadata": {},
57 | "outputs": [],
58 | "source": [
59 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
60 | "original_data_prefix = \"data/NEU-DET.zip\"\n",
61 | "original_data = f\"{original_bucket}/{original_data_prefix}\"\n",
62 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\"\n",
64 | "print(\"original data: \")\n",
65 | "S3Downloader.list(original_data)"
66 | ]
67 | },
68 | {
69 | "cell_type": "markdown",
70 | "metadata": {},
71 | "source": [
72 | "For easiler data processing, depending on the dataset, we unify the class and label names using the scripts from `prepare_data` which should take less than **5 minutes** to complete. This is done once throughout all our notebooks"
73 | ]
74 | },
75 | {
76 | "cell_type": "code",
77 | "execution_count": null,
78 | "metadata": {},
79 | "outputs": [],
80 | "source": [
81 | "%%time\n",
82 | "\n",
83 | "RAW_DATA_PATH = !echo $PWD/raw_neu_det\n",
84 | "RAW_DATA_PATH = RAW_DATA_PATH.n\n",
85 | "DATA_PATH = !echo $PWD/neu_det\n",
86 | "DATA_PATH = DATA_PATH.n\n",
87 | "\n",
88 | "!mkdir -p $RAW_DATA_PATH\n",
89 | "!aws s3 cp $original_data $RAW_DATA_PATH\n",
90 | "\n",
91 | "!mkdir -p $DATA_PATH\n",
92 | "!python ../src/prepare_data/neu.py $RAW_DATA_PATH/NEU-DET.zip $DATA_PATH"
93 | ]
94 | },
95 | {
96 | "cell_type": "markdown",
97 | "metadata": {},
98 | "source": [
99 | "After data preparation, we need upload the prepare data to S3 and setup some paths that will be used throughtout the notebooks"
100 | ]
101 | },
102 | {
103 | "cell_type": "code",
104 | "execution_count": null,
105 | "metadata": {},
106 | "outputs": [],
107 | "source": [
108 | "%%time\n",
109 | "prefix = \"neu-det\"\n",
110 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
111 | "sources = f\"{neu_det_s3}/code/\"\n",
112 | "train_output = f\"{neu_det_s3}/output/\"\n",
113 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
114 | "!aws s3 sync $DATA_PATH $neu_det_prepared_s3 --quiet # remove the --quiet flag to view the sync logs\n",
115 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
116 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
117 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\"\n",
118 | "!aws s3 sync $original_pretained_checkpoint $s3_pretrained\n",
119 | "!aws s3 ls $s3_pretrained\n",
120 | "!aws s3 cp $original_sources $sources"
121 | ]
122 | },
123 | {
124 | "cell_type": "markdown",
125 | "metadata": {},
126 | "source": [
127 | "## Visualization\n",
128 | "\n",
129 | "Let examine some datasets that we will use later by providing an `ID`"
130 | ]
131 | },
132 | {
133 | "cell_type": "code",
134 | "execution_count": null,
135 | "metadata": {},
136 | "outputs": [],
137 | "source": [
138 | "import copy\n",
139 | "\n",
140 | "import numpy as np\n",
141 | "import torch\n",
142 | "from PIL import Image\n",
143 | "from torch.utils.data import DataLoader\n",
144 | "\n",
145 | "try:\n",
146 | " import sagemaker_defect_detection\n",
147 | "except ImportError:\n",
148 | " import sys\n",
149 | " from pathlib import Path\n",
150 | "\n",
151 | " ROOT = Path(\"../src\").resolve()\n",
152 | " sys.path.insert(0, str(ROOT))\n",
153 | "\n",
154 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
155 | "\n",
156 | "SPLIT = \"test\"\n",
157 | "ID = 10\n",
158 | "assert 0 <= ID <= 300\n",
159 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
160 | "images, targets, _ = dataset[ID]\n",
161 | "original_image = copy.deepcopy(images)\n",
162 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
163 | "original_labels = targets[\"labels\"].numpy().copy()\n",
164 | "print(f\"first images size: {original_image.shape}\")\n",
165 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
166 | "print(f\"target labels: {original_labels}\")"
167 | ]
168 | },
169 | {
170 | "cell_type": "markdown",
171 | "metadata": {},
172 | "source": [
173 | "And we can now visualize it using the provided utilities as follows"
174 | ]
175 | },
176 | {
177 | "cell_type": "code",
178 | "execution_count": null,
179 | "metadata": {},
180 | "outputs": [],
181 | "source": [
182 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
183 | "\n",
184 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
185 | "\n",
186 | "visualize(\n",
187 | " original_image_unnorm,\n",
188 | " [original_boxes],\n",
189 | " [original_labels],\n",
190 | " colors=[(255, 0, 0)],\n",
191 | " titles=[\"original\", \"ground truth\"],\n",
192 | ")"
193 | ]
194 | },
195 | {
196 | "cell_type": "markdown",
197 | "metadata": {},
198 | "source": [
199 | "For our demo, we deploy an endpoint using a provided pretrained checkpoint. It takes about **10 minutes** to finish"
200 | ]
201 | },
202 | {
203 | "cell_type": "code",
204 | "execution_count": null,
205 | "metadata": {},
206 | "outputs": [],
207 | "source": [
208 | "%%time\n",
209 | "from os import path as osp\n",
210 | "\n",
211 | "from sagemaker.pytorch import PyTorchModel\n",
212 | "\n",
213 | "demo_model = PyTorchModel(\n",
214 | " osp.join(s3_pretrained, \"model.tar.gz\"),\n",
215 | " role,\n",
216 | " entry_point=\"detector.py\",\n",
217 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
218 | " framework_version=\"1.5\",\n",
219 | " py_version=\"py3\",\n",
220 | " sagemaker_session=sagemaker_session,\n",
221 | " name=sagemaker_config[\"SolutionPrefix\"] + \"-demo-model\",\n",
222 | ")\n",
223 | "\n",
224 | "demo_detector = demo_model.deploy(\n",
225 | " initial_instance_count=1,\n",
226 | " instance_type=\"ml.m5.xlarge\",\n",
227 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\",\n",
228 | ")"
229 | ]
230 | },
231 | {
232 | "cell_type": "markdown",
233 | "metadata": {},
234 | "source": [
235 | "## Inference\n",
236 | "\n",
237 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
238 | ]
239 | },
240 | {
241 | "cell_type": "code",
242 | "execution_count": null,
243 | "metadata": {},
244 | "outputs": [],
245 | "source": [
246 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
247 | ]
248 | },
249 | {
250 | "cell_type": "markdown",
251 | "metadata": {},
252 | "source": [
253 | "Now the input is ready and we can get some results"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "%%time\n",
263 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
264 | "np_load_old = np.load\n",
265 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
266 | "demo_predictions = demo_detector.predict(input)\n",
267 | "np.load = np_load_old"
268 | ]
269 | },
270 | {
271 | "cell_type": "markdown",
272 | "metadata": {},
273 | "source": [
274 | "And finally, we visualize them as follows "
275 | ]
276 | },
277 | {
278 | "cell_type": "code",
279 | "execution_count": null,
280 | "metadata": {
281 | "tags": []
282 | },
283 | "outputs": [],
284 | "source": [
285 | "visualize(\n",
286 | " original_image_unnorm,\n",
287 | " [original_boxes, demo_predictions[0][\"boxes\"]],\n",
288 | " [original_labels, demo_predictions[0][\"labels\"]],\n",
289 | " colors=[(255, 0, 0), (0, 0, 255)],\n",
290 | " titles=[\"original\", \"ground truth\", \"pretrained demo\"],\n",
291 | " dpi=200,\n",
292 | ")"
293 | ]
294 | },
295 | {
296 | "cell_type": "markdown",
297 | "metadata": {},
298 | "source": [
299 | "## Optional: Delete the endpoint and model\n",
300 | "\n",
301 | "**Note:** to follow all the notebooks, it is required to keep demo model and the demo endpoint. It will be automatically deleted when you delete the entire resources/stack. However, if you need to, please uncomment and run the next cell\n",
302 | "\n",
303 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
304 | ]
305 | },
306 | {
307 | "cell_type": "code",
308 | "execution_count": null,
309 | "metadata": {},
310 | "outputs": [],
311 | "source": [
312 | "# demo_detector.delete_model()\n",
313 | "# demo_detector.delete_endpoint()"
314 | ]
315 | },
316 | {
317 | "cell_type": "markdown",
318 | "metadata": {},
319 | "source": [
320 | "### [Click here to continue](./1_retrain_from_checkpoint.ipynb)"
321 | ]
322 | }
323 | ],
324 | "metadata": {
325 | "kernelspec": {
326 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
327 | "name": "python361064bitpytorchlatestp36conda2dfac45b320c45f3a4d1e89ca46b60d1"
328 | },
329 | "language_info": {
330 | "codemirror_mode": {
331 | "name": "ipython",
332 | "version": 3
333 | },
334 | "file_extension": ".py",
335 | "mimetype": "text/x-python",
336 | "name": "python",
337 | "nbconvert_exporter": "python",
338 | "pygments_lexer": "ipython3",
339 | "version": "3.6.10-final"
340 | }
341 | },
342 | "nbformat": 4,
343 | "nbformat_minor": 2
344 | }
345 |
--------------------------------------------------------------------------------
/src/sagemaker_defect_detection/utils/coco_eval.py:
--------------------------------------------------------------------------------
1 | """
2 | BSD 3-Clause License
3 |
4 | Copyright (c) Soumith Chintala 2016,
5 | All rights reserved.
6 |
7 | Redistribution and use in source and binary forms, with or without
8 | modification, are permitted provided that the following conditions are met:
9 |
10 | * Redistributions of source code must retain the above copyright notice, this
11 | list of conditions and the following disclaimer.
12 |
13 | * Redistributions in binary form must reproduce the above copyright notice,
14 | this list of conditions and the following disclaimer in the documentation
15 | and/or other materials provided with the distribution.
16 |
17 | * Neither the name of the copyright holder nor the names of its
18 | contributors may be used to endorse or promote products derived from
19 | this software without specific prior written permission.
20 |
21 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
22 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
23 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
24 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
25 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
26 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
27 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
28 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
29 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
30 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
31 |
32 |
33 | This module is a modified version of https://github.com/pytorch/vision/tree/03b1d38ba3c67703e648fb067570eeb1a1e61265/references/detection
34 | """
35 |
36 | import json
37 |
38 | import numpy as np
39 | import copy
40 | import torch
41 | import pickle
42 |
43 | import torch.distributed as dist
44 |
45 | from pycocotools.cocoeval import COCOeval
46 | from pycocotools.coco import COCO
47 | import pycocotools.mask as mask_util
48 |
49 | from collections import defaultdict
50 |
51 |
52 | def is_dist_avail_and_initialized():
53 | if not dist.is_available():
54 | return False
55 | if not dist.is_initialized():
56 | return False
57 | return True
58 |
59 |
60 | def get_world_size():
61 | if not is_dist_avail_and_initialized():
62 | return 1
63 | return dist.get_world_size()
64 |
65 |
66 | def all_gather(data):
67 | """
68 | Run all_gather on arbitrary picklable data (not necessarily tensors)
69 | Args:
70 | data: any picklable object
71 | Returns:
72 | list[data]: list of data gathered from each rank
73 | """
74 | world_size = get_world_size()
75 | if world_size == 1:
76 | return [data]
77 |
78 | # serialized to a Tensor
79 | buffer = pickle.dumps(data)
80 | storage = torch.ByteStorage.from_buffer(buffer)
81 | tensor = torch.ByteTensor(storage).to("cuda")
82 |
83 | # obtain Tensor size of each rank
84 | local_size = torch.tensor([tensor.numel()], device="cuda")
85 | size_list = [torch.tensor([0], device="cuda") for _ in range(world_size)]
86 | dist.all_gather(size_list, local_size)
87 | size_list = [int(size.item()) for size in size_list]
88 | max_size = max(size_list)
89 |
90 | # receiving Tensor from all ranks
91 | # we pad the tensor because torch all_gather does not support
92 | # gathering tensors of different shapes
93 | tensor_list = []
94 | for _ in size_list:
95 | tensor_list.append(torch.empty((max_size,), dtype=torch.uint8, device="cuda"))
96 | if local_size != max_size:
97 | padding = torch.empty(size=(max_size - local_size,), dtype=torch.uint8, device="cuda")
98 | tensor = torch.cat((tensor, padding), dim=0)
99 | dist.all_gather(tensor_list, tensor)
100 |
101 | data_list = []
102 | for size, tensor in zip(size_list, tensor_list):
103 | buffer = tensor.cpu().numpy().tobytes()[:size]
104 | data_list.append(pickle.loads(buffer))
105 |
106 | return data_list
107 |
108 |
109 | class CocoEvaluator(object):
110 | def __init__(self, coco_gt, iou_types):
111 | assert isinstance(iou_types, (list, tuple))
112 | coco_gt = copy.deepcopy(coco_gt)
113 | self.coco_gt = coco_gt
114 |
115 | self.iou_types = iou_types
116 | self.coco_eval = {}
117 | for iou_type in iou_types:
118 | self.coco_eval[iou_type] = COCOeval(coco_gt, iouType=iou_type)
119 |
120 | self.img_ids = []
121 | self.eval_imgs = {k: [] for k in iou_types}
122 |
123 | def update(self, predictions):
124 | img_ids = list(np.unique(list(predictions.keys())))
125 | self.img_ids.extend(img_ids)
126 |
127 | for iou_type in self.iou_types:
128 | results = self.prepare(predictions, iou_type)
129 | coco_dt = loadRes(self.coco_gt, results) if results else COCO()
130 | coco_eval = self.coco_eval[iou_type]
131 |
132 | coco_eval.cocoDt = coco_dt
133 | coco_eval.params.imgIds = list(img_ids)
134 | img_ids, eval_imgs = evaluate(coco_eval)
135 | if isinstance(self.eval_imgs[iou_type], np.ndarray):
136 | self.eval_imgs[iou_type] = self.eval_imgs[iou_type].tolist()
137 |
138 | self.eval_imgs[iou_type].append(eval_imgs)
139 |
140 | def synchronize_between_processes(self):
141 | for iou_type in self.iou_types:
142 | self.eval_imgs[iou_type] = np.concatenate(self.eval_imgs[iou_type], 2)
143 | create_common_coco_eval(self.coco_eval[iou_type], self.img_ids, self.eval_imgs[iou_type])
144 |
145 | def accumulate(self):
146 | for coco_eval in self.coco_eval.values():
147 | coco_eval.accumulate()
148 |
149 | def summarize(self):
150 | for iou_type, coco_eval in self.coco_eval.items():
151 | print("IoU metric: {}".format(iou_type))
152 | coco_eval.summarize()
153 |
154 | def prepare(self, predictions, iou_type):
155 | return self.prepare_for_coco_detection(predictions)
156 |
157 | def prepare_for_coco_detection(self, predictions):
158 | coco_results = []
159 | for original_id, prediction in predictions.items():
160 | if len(prediction) == 0:
161 | continue
162 |
163 | boxes = prediction["boxes"]
164 | boxes = convert_to_xywh(boxes).tolist()
165 | scores = prediction["scores"].tolist()
166 | labels = prediction["labels"].tolist()
167 |
168 | coco_results.extend(
169 | [
170 | {
171 | "image_id": original_id,
172 | "category_id": labels[k],
173 | "bbox": box,
174 | "score": scores[k],
175 | }
176 | for k, box in enumerate(boxes)
177 | ]
178 | )
179 | return coco_results
180 |
181 |
182 | def convert_to_xywh(boxes):
183 | xmin, ymin, xmax, ymax = boxes.unbind(1)
184 | return torch.stack((xmin, ymin, xmax - xmin, ymax - ymin), dim=1)
185 |
186 |
187 | def merge(img_ids, eval_imgs):
188 | all_img_ids = all_gather(img_ids)
189 | all_eval_imgs = all_gather(eval_imgs)
190 |
191 | merged_img_ids = []
192 | for p in all_img_ids:
193 | merged_img_ids.extend(p)
194 |
195 | merged_eval_imgs = []
196 | for p in all_eval_imgs:
197 | merged_eval_imgs.append(p)
198 |
199 | merged_img_ids = np.array(merged_img_ids)
200 | merged_eval_imgs = np.concatenate(merged_eval_imgs, 2)
201 |
202 | # keep only unique (and in sorted order) images
203 | merged_img_ids, idx = np.unique(merged_img_ids, return_index=True)
204 | merged_eval_imgs = merged_eval_imgs[..., idx]
205 |
206 | return merged_img_ids, merged_eval_imgs
207 |
208 |
209 | def create_common_coco_eval(coco_eval, img_ids, eval_imgs):
210 | img_ids, eval_imgs = merge(img_ids, eval_imgs)
211 | img_ids = list(img_ids)
212 | eval_imgs = list(eval_imgs.flatten())
213 |
214 | coco_eval.evalImgs = eval_imgs
215 | coco_eval.params.imgIds = img_ids
216 | coco_eval._paramsEval = copy.deepcopy(coco_eval.params)
217 |
218 |
219 | #################################################################
220 | # From pycocotools, just removed the prints and fixed
221 | # a Python3 bug about unicode not defined
222 | #################################################################
223 |
224 | # Ideally, pycocotools wouldn't have hard-coded prints
225 | # so that we could avoid copy-pasting those two functions
226 |
227 |
228 | def createIndex(self):
229 | # create index
230 | # print('creating index...')
231 | anns, cats, imgs = {}, {}, {}
232 | imgToAnns, catToImgs = defaultdict(list), defaultdict(list)
233 | if "annotations" in self.dataset:
234 | for ann in self.dataset["annotations"]:
235 | imgToAnns[ann["image_id"]].append(ann)
236 | anns[ann["id"]] = ann
237 |
238 | if "images" in self.dataset:
239 | for img in self.dataset["images"]:
240 | imgs[img["id"]] = img
241 |
242 | if "categories" in self.dataset:
243 | for cat in self.dataset["categories"]:
244 | cats[cat["id"]] = cat
245 |
246 | if "annotations" in self.dataset and "categories" in self.dataset:
247 | for ann in self.dataset["annotations"]:
248 | catToImgs[ann["category_id"]].append(ann["image_id"])
249 |
250 | # print('index created!')
251 |
252 | # create class members
253 | self.anns = anns
254 | self.imgToAnns = imgToAnns
255 | self.catToImgs = catToImgs
256 | self.imgs = imgs
257 | self.cats = cats
258 |
259 |
260 | maskUtils = mask_util
261 |
262 |
263 | def loadRes(self, resFile):
264 | """
265 | Load result file and return a result api object.
266 | :param resFile (str) : file name of result file
267 | :return: res (obj) : result api object
268 | """
269 | res = COCO()
270 | res.dataset["images"] = [img for img in self.dataset["images"]]
271 |
272 | # print('Loading and preparing results...')
273 | # tic = time.time()
274 | if isinstance(resFile, torch._six.string_classes):
275 | anns = json.load(open(resFile))
276 | elif type(resFile) == np.ndarray:
277 | anns = self.loadNumpyAnnotations(resFile)
278 | else:
279 | anns = resFile
280 | assert type(anns) == list, "results in not an array of objects"
281 | annsImgIds = [ann["image_id"] for ann in anns]
282 | assert set(annsImgIds) == (
283 | set(annsImgIds) & set(self.getImgIds())
284 | ), "Results do not correspond to current coco set"
285 | if "caption" in anns[0]:
286 | imgIds = set([img["id"] for img in res.dataset["images"]]) & set([ann["image_id"] for ann in anns])
287 | res.dataset["images"] = [img for img in res.dataset["images"] if img["id"] in imgIds]
288 | for id, ann in enumerate(anns):
289 | ann["id"] = id + 1
290 | elif "bbox" in anns[0] and not anns[0]["bbox"] == []:
291 | res.dataset["categories"] = copy.deepcopy(self.dataset["categories"])
292 | for id, ann in enumerate(anns):
293 | bb = ann["bbox"]
294 | x1, x2, y1, y2 = [bb[0], bb[0] + bb[2], bb[1], bb[1] + bb[3]]
295 | if "segmentation" not in ann:
296 | ann["segmentation"] = [[x1, y1, x1, y2, x2, y2, x2, y1]]
297 | ann["area"] = bb[2] * bb[3]
298 | ann["id"] = id + 1
299 | ann["iscrowd"] = 0
300 |
301 | res.dataset["annotations"] = anns
302 | createIndex(res)
303 | return res
304 |
305 |
306 | def evaluate(self):
307 | """
308 | Run per image evaluation on given images and store results (a list of dict) in self.evalImgs
309 | :return: None
310 | """
311 | # tic = time.time()
312 | # print('Running per image evaluation...')
313 | p = self.params
314 | # add backward compatibility if useSegm is specified in params
315 | if p.useSegm is not None:
316 | p.iouType = "segm" if p.useSegm == 1 else "bbox"
317 | print("useSegm (deprecated) is not None. Running {} evaluation".format(p.iouType))
318 | # print('Evaluate annotation type *{}*'.format(p.iouType))
319 | p.imgIds = list(np.unique(p.imgIds))
320 | if p.useCats:
321 | p.catIds = list(np.unique(p.catIds))
322 | p.maxDets = sorted(p.maxDets)
323 | self.params = p
324 |
325 | self._prepare()
326 | # loop through images, area range, max detection number
327 | catIds = p.catIds if p.useCats else [-1]
328 |
329 | if p.iouType == "segm" or p.iouType == "bbox":
330 | computeIoU = self.computeIoU
331 | elif p.iouType == "keypoints":
332 | computeIoU = self.computeOks
333 | self.ious = {(imgId, catId): computeIoU(imgId, catId) for imgId in p.imgIds for catId in catIds}
334 |
335 | evaluateImg = self.evaluateImg
336 | maxDet = p.maxDets[-1]
337 | evalImgs = [
338 | evaluateImg(imgId, catId, areaRng, maxDet) for catId in catIds for areaRng in p.areaRng for imgId in p.imgIds
339 | ]
340 | # this is NOT in the pycocotools code, but could be done outside
341 | evalImgs = np.asarray(evalImgs).reshape(len(catIds), len(p.areaRng), len(p.imgIds))
342 | self._paramsEval = copy.deepcopy(self.params)
343 | # toc = time.time()
344 | # print('DONE (t={:0.2f}s).'.format(toc-tic))
345 | return p.imgIds, evalImgs
346 |
347 |
348 | #################################################################
349 | # end of straight copy from pycocotools, just removing the prints
350 | #################################################################
351 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Visual Inspection Automation with Amazon SageMaker
2 |
3 |
4 |  5 |
5 |  6 |
6 |  7 |
7 |
8 |  9 |
9 |  10 |
10 |  11 |
11 |
12 |
13 | This solution detects product defects with an end-to-end Deep Learning workflow for quality control in manufacturing process. The solution takes input of product images and identifies defect regions with bounding boxes. In particular, this solution takes two distinct approaches:
14 | 1. Use an implementation of the *Defect Detection Network (DDN)* algorithm following [An End-to-End Steel Surface Defect Detection](https://ieeexplore.ieee.org/document/8709818) on [NEU surface defect database](http://faculty.neu.edu.cn/yunhyan/NEU_surface_defect_database.html) (see [resources](#resources)) in PyTorch using [PyTorch-lightning](https://github.com/PyTorchLightning/pytorch-lightning).
15 | 2. Use a pre-trained Sagemaker object detection model and fine-tune on the target dataset.
16 |
17 | This solution will demonstrate the immense advantage of fine-tuning a high-quality pre-trained model on the target dataset, both visually and numerically.
18 |
19 | ### Contents
20 | 1. [Overview](#overview)
21 | 1. [What Does the Input Data Look Like?](#input)
22 | 2. [How to Prepare Your Data to Feed into the Model?](#preparedata)
23 | 3. [What are the Outputs?](#output)
24 | 4. [What is the Estimated Cost?](#cost)
25 | 5. [What Algorithms & Models are Used?](#algorithms)
26 | 6. [What Does the Data Flow Look Like?](#dataflow)
27 | 2. [Solution Details](#solution)
28 | 1. [Background](#background)
29 | 2. [What is Visual Inspection?](#inspection)
30 | 3. [What are the Problems?](#problems)
31 | 4. [What Does this Solution Offer?](#offer)
32 | 3. [Architecture Overview](#architecture)
33 | 4. [Cleaning up](#cleaning-up)
34 | 5. [Customization](#customization)
35 |
36 |
37 | ## 1. Overview
38 |
39 | ### 1.1. What Does the Input Data Look Like?
40 |
41 | Input is an image of a defective / non-defective product. The training data should have relatively balanced classes, with annotations for ground truth defects (locations and defect types) per image. Here are examples of annotations used in the demo, they show some "inclusion" defects on the surface:
42 |
43 | 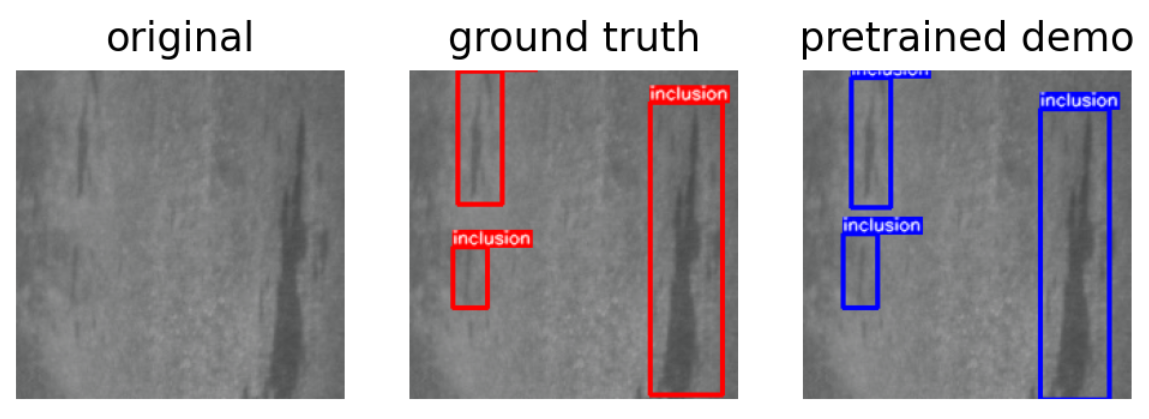
44 |
45 | The NEU surface defect database (see [references](#references)) is a *balanced* dataset which contains
46 |
47 | > Six kinds of typical surface defects of the hot-rolled steel strip are collected, i.e., rolled-in scale (RS), patches (Pa), crazing (Cr), pitted surface (PS), inclusion (In) and scratches (Sc). The database includes 1,800 grayscale images: 300 samples each of six different kinds of typical surface defects
48 |
49 | Here is a sample image of the six classes
50 |
51 | 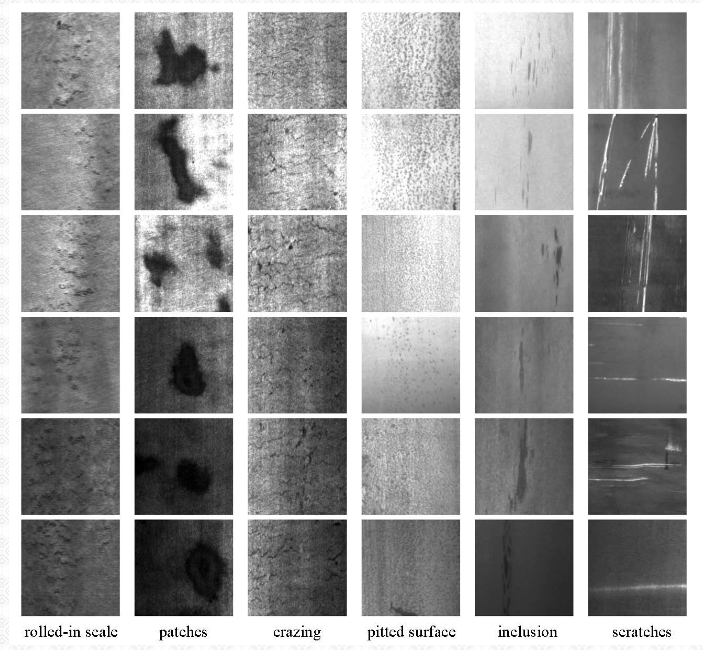
52 |
53 | ### 1.2. How to Prepare Your Data to Feed into the Model?
54 |
55 | There are data preparation and preprocessing steps and should be followed in the notebooks. It's critical to prepare your image annotations beforehand.
56 | * For training the DDN model, please prepare one xml file for each image with defect annotations. Check notebook 0,1,2,3 for details.
57 | * For finetuning pretrained Sagemaker models, you need to prepare either a single `annotation.json` for all data, or a `RecordIO` file for both all images and all annotations. Check notebook 4 for details.
58 |
59 | ### 1.3. What are the Outputs?
60 |
61 | * For each image, the trained model will produce bounding boxes of detected visual defects (if any), the predicted defect type, and prediction confidence score (0~1).
62 | * If you have a labeled test dataset, you could obtain the mean Average Precision (mAP) score for each model and compare among all the models.
63 | * For example, the mAP scores on a test set of the NEU dataset
64 |
65 | | | DDN | Type1 | Type1+HPO | Type2 | Type2+HPO|
66 | | --- | --- | --- | --- | --- | ---|
67 | | mAP | 0.08 | 0.067 | 0.226 | 0.371 | 0.375|
68 |
69 |
70 | ### 1.4. What is the Estimated Cost?
71 |
72 | * Running solution notebook 0~3 end-to-end costs around $8 USD and less than an hour, assuming using p3.2xlarge EC2 instance, and $3.06 on-demand hourly rate in US East. These notebooks only train DDN models for a few iterations for demonstration purpose, which is **far from convergence**. It would take around 8 hours to train from scratch till convergence and cost $25+.
73 | * Running solution notebook 4 costs around $130~140 USD. This notebook provides advanced materials, including finetuning two types of pretrained Sagemaker models **till convergence**, with and without hyperparameter optimization (HPO), and result in four models for inference. You could choose to train either one model, or all four models according to your budget and requirements. The cost and runtime for training each model are:
74 |
75 | | Model | Cost (USD) | Runtime (Hours) | Billable time (Hours)|
76 | |:----------:|:---------------:|:----:|:-----:|
77 | |Type 1| 1.5 | 0.5 | 0.5|
78 | |Type 1 with HPO (20 jobs)| 30.6 | 1* | 10|
79 | |Type 2| 4.6 | 1.5 | 1.5|
80 | |Type 2 with HPO (20 jobs)| 92 | 3* | 30|
81 | (*) HPO tasks in this solution consider 20 jobs in total and 10 jobs in parallel. So 1 actual runtime hour amounts to 10 billable cost hours.
82 | * Please make sure you have read the cleaning up part in [Section 4](#cleaning-up) after training to avoid incurred cost from deployed models.
83 |
84 |
85 |
86 | ### 1.5. What Algorithms & Models are Used?
87 |
88 | * The DDN model is based on [Faster RCNN](https://arxiv.org/abs/1506.01497). For more details, please read the paper [An End-to-End Steel Surface Defect Detection](https://ieeexplore.ieee.org/document/8709818).
89 | * The pretrained Sagemaker models include SSD models and FasterRCNN model, using either VGG, ResNet, or MobileNet as backbone, pretrained on either ImageNet, COCO, VOC, or FPN dataset.
90 |
91 | ### 1.6. What Does the Data Flow Look Like?
92 |
93 | 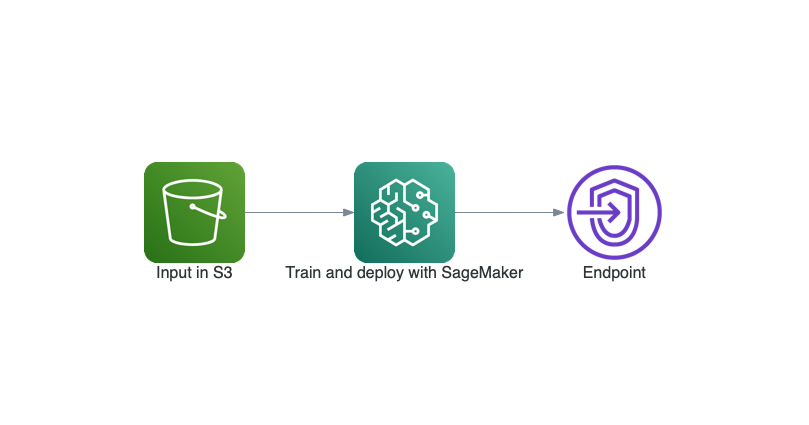
94 |
95 | ## 2. Solution Details
96 |
97 | ### 2.1. Background
98 |
99 | According to the [Gartner study on the top 10 strategic tech trends for 2020](https://www.gartner.com/smarterwithgartner/gartner-top-10-strategic-technology-trends-for-2020/), hyper-automation is the number one trend in 2020 and will continue advancing in future. When it comes to manufacturing, one of the main barriers to hyper-automation is in areas where Human involvements is still struggling to be reduced and intelligent systems have hard times to become on-par with Human visual recognition abilities and become mainstream, despite great advancement of Deep Learning in Computer Vision. This is mainly due to lack of enough annotated data (or when data is sparse) in areas such as _Quality Control_ sections where trained Human eyes still dominates.
100 |
101 |
102 | ### 2.2. What is Visual Inspection?
103 |
104 | The **analysis of products on the production line for the purpose of Quality Control**. According to [Everything you need to know about Visual Inspection with AI](https://nanonets.com/blog/ai-visual-inspection/), visual inspection can also be used for internal and external assessment of the various equipment in a production facility such as storage tanks, pressure vessels, piping, and other equipment which expands to many industries from Electronics, Medical, Food and Raw Materials.
105 |
106 | ### 2.3. What are the Problems?
107 |
108 | * *Human visual inspection error* is a major factor in this area. According to the report [The Role of Visual Inspection in the 21st Century](https://www.osti.gov/servlets/purl/1476816)
109 |
110 | > Most inspection tasks are much more complex and typically exhibit error rates of 20% to 30% (Drury & Fox, 1975)
111 |
112 | which directly translates to *cost*.
113 | * Cost: according to [glassdoor estimate](https://www.glassdoor.co.in/Salaries/us-quality-control-inspector-salary-SRCH_IL.0,2_IN1_KO3,28.htm), a trained quality inspector salary varies between 29K (US) - 64K per year.
114 |
115 | ### 2.4. What Does this Solution Offer?
116 |
117 | This solution offers
118 | 1. an implementation of the state-of-the-art Deep Learning approach for automatic *Steel Surface Defect Detection* using **Amazon SageMaker**. The [model](https://ieeexplore.ieee.org/document/8709818) enhances [Faster RCNN](https://arxiv.org/abs/1506.01497) and outputs possible defects in an steel surface image.
119 | This solution trains a classifier on **NEU-CLS** dataset as well as a detector on **NEU-DET** dataset.
120 | 2. a complete solution using high-quality pretrained Sagemaker models to finetune on the target dataset with and without hyperparameter optimization (HPO).
121 |
122 | The **most important** information this solution delivers, is that training a deep learning model from scratch on a small dataset can be both time-consuming and less effective, whereas finetuning a high-quality pretrained model, which was trained on large-scale dataset, could be both cost- and runtime-efficient and highly performant. Here are the sample detection results
123 |
124 |  125 |
126 | ## 3. Architecture Overview
127 |
128 | The following illustration is the architecture for the end-to-end training and deployment process
129 |
130 | 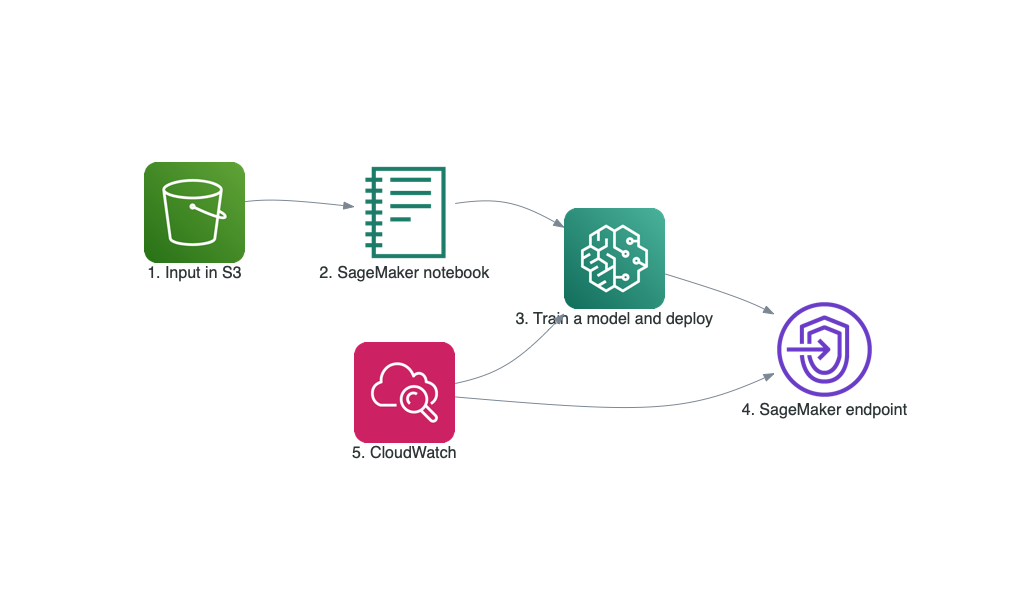
131 |
132 | 1. The input data located in an [Amazon S3](https://aws.amazon.com/s3/) bucket
133 | 2. The provided [SageMaker notebook](source/deep_demand_forecast.ipynb) that gets the input data and launches the later stages below
134 | 3. **Training Classifier and Detector models** and evaluating its results using Amazon SageMaker. If desired, one can deploy the trained models and create SageMaker endpoints
135 | 4. **SageMaker endpoint** created from the previous step is an [HTTPS endpoint](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html) and is capable of producing predictions
136 | 5. Monitoring the training and deployed model via [Amazon CloudWatch](https://aws.amazon.com/cloudwatch/)
137 |
138 | ## 4. Cleaning up
139 |
140 | When you've finished with this solution, make sure that you delete all unwanted AWS resources. AWS CloudFormation can be used to automatically delete all standard resources that have been created by the solution and notebook. Go to the AWS CloudFormation Console, and delete the parent stack. Choosing to delete the parent stack will automatically delete the nested stacks.
141 |
142 | **Caution:** You need to manually delete any extra resources that you may have created in this notebook. Some examples include, extra Amazon S3 buckets (to the solution's default bucket), extra Amazon SageMaker endpoints (using a custom name).
143 |
144 | ## 5. Customization
145 |
146 | For using your own data, make sure it is labeled and is a *relatively* balanced dataset. Also make sure the image annotations follow the required format.
147 |
148 |
149 |
150 | ### Useful Links
151 |
152 | * [Amazon SageMaker Getting Started](https://aws.amazon.com/sagemaker/getting-started/)
153 | * [Amazon SageMaker Developer Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/whatis.html)
154 | * [Amazon SageMaker Python SDK Documentation](https://sagemaker.readthedocs.io/en/stable/)
155 | * [AWS CloudFormation User Guide](https://docs.aws.amazon.com/AWSCloudFormation/latest/UserGuide/Welcome.html)
156 |
157 | ### References
158 |
159 | * K. Song and Y. Yan, “A noise robust method based on completed local binary patterns for hot-rolled steel strip surface defects,” Applied Surface Science, vol. 285, pp. 858-864, Nov. 2013.
160 |
161 | * Yu He, Kechen Song, Qinggang Meng, Yunhui Yan, “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,” IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
162 |
163 | * Hongwen Dong, Kechen Song, Yu He, Jing Xu, Yunhui Yan, Qinggang Meng, “PGA-Net: Pyramid Feature Fusion and Global Context Attention Network for Automated Surface Defect Detection,” IEEE Transactions on Industrial Informatics, 2020.
164 |
165 | ### Security
166 |
167 | See [CONTRIBUTING](CONTRIBUTING.md#security-issue-notifications) for more information.
168 |
169 | ### License
170 |
171 | This project is licensed under the Apache-2.0 License.
172 |
--------------------------------------------------------------------------------
/notebooks/1_retrain_from_checkpoint.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Resume Training\n",
20 | "\n",
21 | "In this notebook, we retrain our pretrained detector for a few more epochs and compare its results. The same process can be applied when finetuning on another dataset. For the purpose of this notebook, we use the same **NEU-DET** dataset.\n",
22 | "\n",
23 | "## Finetuning\n",
24 | "\n",
25 | "Finetuning is one way to do Transfer Learning. Finetuning a Deep Learning model on one particular task, involves using the learned weights from a particular dataset to enhance the performace of the model on usually another dataset. In a sense, finetuning can be done over the same dataset used in the intial training but perhaps with different hyperparameters.\n"
26 | ]
27 | },
28 | {
29 | "cell_type": "code",
30 | "execution_count": null,
31 | "metadata": {},
32 | "outputs": [],
33 | "source": [
34 | "import json\n",
35 | "\n",
36 | "import sagemaker\n",
37 | "from sagemaker.s3 import S3Downloader\n",
38 | "\n",
39 | "sagemaker_session = sagemaker.Session()\n",
40 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
41 | "role = sagemaker_config[\"IamRole\"]\n",
42 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
43 | "region = sagemaker_config[\"AWSRegion\"]\n",
44 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
45 | "bucket = sagemaker_config[\"S3Bucket\"]"
46 | ]
47 | },
48 | {
49 | "cell_type": "markdown",
50 | "metadata": {},
51 | "source": [
52 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
53 | ]
54 | },
55 | {
56 | "cell_type": "code",
57 | "execution_count": null,
58 | "metadata": {},
59 | "outputs": [],
60 | "source": [
61 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
62 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
64 | ]
65 | },
66 | {
67 | "cell_type": "markdown",
68 | "metadata": {},
69 | "source": [
70 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
71 | ]
72 | },
73 | {
74 | "cell_type": "code",
75 | "execution_count": null,
76 | "metadata": {},
77 | "outputs": [],
78 | "source": [
79 | "DATA_PATH = !echo $PWD/neu_det\n",
80 | "DATA_PATH = DATA_PATH.n"
81 | ]
82 | },
83 | {
84 | "cell_type": "markdown",
85 | "metadata": {},
86 | "source": [
87 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
88 | ]
89 | },
90 | {
91 | "cell_type": "code",
92 | "execution_count": null,
93 | "metadata": {
94 | "tags": []
95 | },
96 | "outputs": [],
97 | "source": [
98 | "prefix = \"neu-det\"\n",
99 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
100 | "sources = f\"{neu_det_s3}/code/\"\n",
101 | "train_output = f\"{neu_det_s3}/output/\"\n",
102 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
103 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
104 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
105 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
106 | ]
107 | },
108 | {
109 | "cell_type": "markdown",
110 | "metadata": {},
111 | "source": [
112 | "## Visualization\n",
113 | "\n",
114 | "Let examine some datasets that we will use later by providing an `ID`"
115 | ]
116 | },
117 | {
118 | "cell_type": "code",
119 | "execution_count": null,
120 | "metadata": {},
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "\n",
125 | "import numpy as np\n",
126 | "import torch\n",
127 | "from PIL import Image\n",
128 | "from torch.utils.data import DataLoader\n",
129 | "\n",
130 | "try:\n",
131 | " import sagemaker_defect_detection\n",
132 | "except ImportError:\n",
133 | " import sys\n",
134 | " from pathlib import Path\n",
135 | "\n",
136 | " ROOT = Path(\"../src\").resolve()\n",
137 | " sys.path.insert(0, str(ROOT))\n",
138 | "\n",
139 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
140 | "\n",
141 | "SPLIT = \"test\"\n",
142 | "ID = 30\n",
143 | "assert 0 <= ID <= 300\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "Here we resume from a provided pretrained checkpoint `epoch=294-loss=0.654-main_score=0.349.ckpt` that we have copied into our `s3_pretrained`. This takes about **10 minutes** to complete"
185 | ]
186 | },
187 | {
188 | "cell_type": "code",
189 | "execution_count": null,
190 | "metadata": {
191 | "tags": [
192 | "outputPrepend"
193 | ]
194 | },
195 | "outputs": [],
196 | "source": [
197 | "%%time\n",
198 | "import logging\n",
199 | "from os import path as osp\n",
200 | "\n",
201 | "from sagemaker.pytorch import PyTorch\n",
202 | "\n",
203 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
204 | "# Note: resnet34 was used in the pretrained model and it has to match the pretrained model backbone\n",
205 | "# if need resnet50, need to train from scratch\n",
206 | "BACKBONE = \"resnet34\"\n",
207 | "assert BACKBONE in [\n",
208 | " \"resnet34\",\n",
209 | " \"resnet50\",\n",
210 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
211 | "EPOCHS = 5\n",
212 | "LEARNING_RATE = 1e-4\n",
213 | "SEED = 123\n",
214 | "\n",
215 | "hyperparameters = {\n",
216 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
217 | " \"num-classes\": NUM_CLASSES, # number of classes + background\n",
218 | " \"epochs\": EPOCHS, # number of epochs to finetune\n",
219 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
220 | " \"seed\": SEED, # random number generator seed\n",
221 | "}\n",
222 | "\n",
223 | "assert not isinstance(sagemaker_session, sagemaker.LocalSession), \"local session as share memory cannot be altered\"\n",
224 | "\n",
225 | "finetuned_model = PyTorch(\n",
226 | " entry_point=\"detector.py\",\n",
227 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
228 | " role=role,\n",
229 | " train_instance_count=1,\n",
230 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
231 | " hyperparameters=hyperparameters,\n",
232 | " py_version=\"py3\",\n",
233 | " framework_version=\"1.5\",\n",
234 | " sagemaker_session=sagemaker_session,\n",
235 | " output_path=train_output,\n",
236 | " checkpoint_s3_uri=s3_checkpoint,\n",
237 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
238 | " # container_log_level=logging.DEBUG,\n",
239 | ")\n",
240 | "\n",
241 | "finetuned_model.fit(\n",
242 | " {\n",
243 | " \"training\": neu_det_prepared_s3,\n",
244 | " \"pretrained_checkpoint\": osp.join(s3_pretrained, \"epoch=294-loss=0.654-main_score=0.349.ckpt\"),\n",
245 | " }\n",
246 | ")"
247 | ]
248 | },
249 | {
250 | "cell_type": "markdown",
251 | "metadata": {},
252 | "source": [
253 | "Then, we deploy our new model which takes about **10 minutes** to complete"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "%%time\n",
263 | "finetuned_detector = finetuned_model.deploy(\n",
264 | " initial_instance_count=1,\n",
265 | " instance_type=\"ml.m5.xlarge\",\n",
266 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-finetuned-endpoint\",\n",
267 | ")"
268 | ]
269 | },
270 | {
271 | "cell_type": "markdown",
272 | "metadata": {},
273 | "source": [
274 | "## Inference\n",
275 | "\n",
276 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
277 | ]
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": null,
282 | "metadata": {},
283 | "outputs": [],
284 | "source": [
285 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
286 | ]
287 | },
288 | {
289 | "cell_type": "markdown",
290 | "metadata": {},
291 | "source": [
292 | "Now the input is ready and we can get some results"
293 | ]
294 | },
295 | {
296 | "cell_type": "code",
297 | "execution_count": null,
298 | "metadata": {},
299 | "outputs": [],
300 | "source": [
301 | "%%time\n",
302 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
303 | "np_load_old = np.load\n",
304 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
305 | "finetuned_predictions = finetuned_detector.predict(input)\n",
306 | "np.load = np_load_old"
307 | ]
308 | },
309 | {
310 | "cell_type": "markdown",
311 | "metadata": {},
312 | "source": [
313 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
314 | ]
315 | },
316 | {
317 | "cell_type": "code",
318 | "execution_count": null,
319 | "metadata": {},
320 | "outputs": [],
321 | "source": [
322 | "import boto3\n",
323 | "import botocore\n",
324 | "\n",
325 | "config = botocore.config.Config(read_timeout=200)\n",
326 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
327 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
328 | "response = runtime.invoke_endpoint(\n",
329 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
330 | ")\n",
331 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
332 | ]
333 | },
334 | {
335 | "cell_type": "markdown",
336 | "metadata": {},
337 | "source": [
338 | "Here comes the slight changes in inference"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": null,
344 | "metadata": {},
345 | "outputs": [],
346 | "source": [
347 | "visualize(\n",
348 | " original_image_unnorm,\n",
349 | " [original_boxes, demo_predictions[0][\"boxes\"], finetuned_predictions[0][\"boxes\"]],\n",
350 | " [original_labels, demo_predictions[0][\"labels\"], finetuned_predictions[0][\"labels\"]],\n",
351 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
352 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"finetuned\"],\n",
353 | " dpi=250,\n",
354 | ")"
355 | ]
356 | },
357 | {
358 | "cell_type": "markdown",
359 | "metadata": {},
360 | "source": [
361 | "## Optional: Delete the endpoint and model\n",
362 | "\n",
363 | "When you are done with the endpoint, you should clean it up.\n",
364 | "\n",
365 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account.\n"
366 | ]
367 | },
368 | {
369 | "cell_type": "code",
370 | "execution_count": null,
371 | "metadata": {},
372 | "outputs": [],
373 | "source": [
374 | "finetuned_detector.delete_model()\n",
375 | "finetuned_detector.delete_endpoint()"
376 | ]
377 | },
378 | {
379 | "cell_type": "markdown",
380 | "metadata": {},
381 | "source": [
382 | "### [Click here to continue](./2_detection_from_scratch.ipynb)"
383 | ]
384 | }
385 | ],
386 | "metadata": {
387 | "kernelspec": {
388 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
389 | "name": "python361064bitpytorchlatestp36conda2dfac45b320c45f3a4d1e89ca46b60d1"
390 | },
391 | "language_info": {
392 | "codemirror_mode": {
393 | "name": "ipython",
394 | "version": 3
395 | },
396 | "file_extension": ".py",
397 | "mimetype": "text/x-python",
398 | "name": "python",
399 | "nbconvert_exporter": "python",
400 | "pygments_lexer": "ipython3",
401 | "version": "3.6.10-final"
402 | }
403 | },
404 | "nbformat": 4,
405 | "nbformat_minor": 2
406 | }
407 |
--------------------------------------------------------------------------------
/notebooks/2_detection_from_scratch.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Training our Detector from Scratch\n",
20 | "\n",
21 | "In this notebook, we will see how to train our detector from scratch"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": null,
27 | "metadata": {},
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import sagemaker\n",
33 | "from sagemaker.s3 import S3Downloader\n",
34 | "\n",
35 | "sagemaker_session = sagemaker.Session()\n",
36 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
37 | "role = sagemaker_config[\"IamRole\"]\n",
38 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
39 | "region = sagemaker_config[\"AWSRegion\"]\n",
40 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
41 | "bucket = sagemaker_config[\"S3Bucket\"]"
42 | ]
43 | },
44 | {
45 | "cell_type": "markdown",
46 | "metadata": {},
47 | "source": [
48 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "execution_count": null,
54 | "metadata": {},
55 | "outputs": [],
56 | "source": [
57 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
58 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
59 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
60 | ]
61 | },
62 | {
63 | "cell_type": "markdown",
64 | "metadata": {},
65 | "source": [
66 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
67 | ]
68 | },
69 | {
70 | "cell_type": "code",
71 | "execution_count": null,
72 | "metadata": {
73 | "tags": []
74 | },
75 | "outputs": [],
76 | "source": [
77 | "DATA_PATH = !echo $PWD/neu_det\n",
78 | "DATA_PATH = DATA_PATH.n"
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {},
84 | "source": [
85 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": null,
91 | "metadata": {
92 | "tags": []
93 | },
94 | "outputs": [],
95 | "source": [
96 | "prefix = \"neu-det\"\n",
97 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
98 | "sources = f\"{neu_det_s3}/code/\"\n",
99 | "train_output = f\"{neu_det_s3}/output/\"\n",
100 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
101 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
102 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
103 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
104 | ]
105 | },
106 | {
107 | "cell_type": "markdown",
108 | "metadata": {},
109 | "source": [
110 | "## Visualization\n",
111 | "\n",
112 | "Let examine some datasets that we will use later by providing an `ID`"
113 | ]
114 | },
115 | {
116 | "cell_type": "code",
117 | "execution_count": null,
118 | "metadata": {
119 | "tags": []
120 | },
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "from typing import List\n",
125 | "\n",
126 | "import numpy as np\n",
127 | "import torch\n",
128 | "from PIL import Image\n",
129 | "from torch.utils.data import DataLoader\n",
130 | "\n",
131 | "try:\n",
132 | " import sagemaker_defect_detection\n",
133 | "except ImportError:\n",
134 | " import sys\n",
135 | " from pathlib import Path\n",
136 | "\n",
137 | " ROOT = Path(\"../src\").resolve()\n",
138 | " sys.path.insert(0, str(ROOT))\n",
139 | "\n",
140 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
141 | "\n",
142 | "SPLIT = \"test\"\n",
143 | "ID = 50\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "In order to get high **Mean Average Percision (mAP)** and **Mean Average Recall (mAR)** for **Intersection Over Union (IOU)** thresholds of 0.5 when training from scratch, it requires more than **300 epochs**. That is why we have provided a pretrained model and recommend finetuning whenever is possible. For demostration, we train the model from scratch for 10 epochs which takes about **16 minutes** and it results in the following mAP, mAR and the accumulated `main_score` of\n",
185 | "\n",
186 | "* `Average Precision (AP) @[ IoU=0.50:0.95 ] ~ 0.048`\n",
187 | "* `Average Recall (AR) @[ IoU=0.50:0.95] ~ 0.153`\n",
188 | "* `main_score=0.0509`\n",
189 | "\n",
190 | "To get higher mAP, mAR and overall `main_score`, you can train for more epochs"
191 | ]
192 | },
193 | {
194 | "cell_type": "code",
195 | "execution_count": null,
196 | "metadata": {
197 | "tags": [
198 | "outputPrepend"
199 | ]
200 | },
201 | "outputs": [],
202 | "source": [
203 | "%%time\n",
204 | "import logging\n",
205 | "from os import path as osp\n",
206 | "\n",
207 | "from sagemaker.pytorch import PyTorch\n",
208 | "\n",
209 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
210 | "BACKBONE = \"resnet34\" # has to match the pretrained model backbone\n",
211 | "assert BACKBONE in [\n",
212 | " \"resnet34\",\n",
213 | " \"resnet50\",\n",
214 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
215 | "EPOCHS = 10\n",
216 | "LEARNING_RATE = 1e-3\n",
217 | "SEED = 123\n",
218 | "\n",
219 | "hyperparameters = {\n",
220 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
221 | " \"num-classes\": NUM_CLASSES, # number of classes 6 + 1 background\n",
222 | " \"epochs\": EPOCHS, # number of epochs to train\n",
223 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
224 | " \"seed\": SEED, # random number generator seed\n",
225 | "}\n",
226 | "\n",
227 | "model = PyTorch(\n",
228 | " entry_point=\"detector.py\",\n",
229 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
230 | " role=role,\n",
231 | " train_instance_count=1,\n",
232 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
233 | " hyperparameters=hyperparameters,\n",
234 | " py_version=\"py3\",\n",
235 | " framework_version=\"1.5\",\n",
236 | " sagemaker_session=sagemaker_session,\n",
237 | " output_path=train_output,\n",
238 | " checkpoint_s3_uri=s3_checkpoint,\n",
239 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
240 | " # container_log_level=logging.DEBUG,\n",
241 | ")\n",
242 | "\n",
243 | "model.fit(neu_det_prepared_s3)"
244 | ]
245 | },
246 | {
247 | "cell_type": "markdown",
248 | "metadata": {},
249 | "source": [
250 | "Then, we deploy our model which takes about **10 minutes** to complete"
251 | ]
252 | },
253 | {
254 | "cell_type": "code",
255 | "execution_count": null,
256 | "metadata": {
257 | "tags": []
258 | },
259 | "outputs": [],
260 | "source": [
261 | "%%time\n",
262 | "detector = model.deploy(\n",
263 | " initial_instance_count=1,\n",
264 | " instance_type=\"ml.m5.xlarge\",\n",
265 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-detector-from-scratch-endpoint\",\n",
266 | ")"
267 | ]
268 | },
269 | {
270 | "cell_type": "markdown",
271 | "metadata": {},
272 | "source": [
273 | "## Inference\n",
274 | "\n",
275 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
276 | ]
277 | },
278 | {
279 | "cell_type": "code",
280 | "execution_count": null,
281 | "metadata": {},
282 | "outputs": [],
283 | "source": [
284 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
285 | ]
286 | },
287 | {
288 | "cell_type": "markdown",
289 | "metadata": {},
290 | "source": [
291 | "Now the input is ready and we can get some results"
292 | ]
293 | },
294 | {
295 | "cell_type": "code",
296 | "execution_count": null,
297 | "metadata": {
298 | "tags": []
299 | },
300 | "outputs": [],
301 | "source": [
302 | "%%time\n",
303 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
304 | "np_load_old = np.load\n",
305 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
306 | "predictions = detector.predict(input)\n",
307 | "np.load = np_load_old"
308 | ]
309 | },
310 | {
311 | "cell_type": "markdown",
312 | "metadata": {},
313 | "source": [
314 | "We use our `visualize` utility to check the detection results"
315 | ]
316 | },
317 | {
318 | "cell_type": "code",
319 | "execution_count": null,
320 | "metadata": {},
321 | "outputs": [],
322 | "source": [
323 | "visualize(\n",
324 | " original_image_unnorm,\n",
325 | " [original_boxes, predictions[0][\"boxes\"]],\n",
326 | " [original_labels, predictions[0][\"labels\"]],\n",
327 | " colors=[(255, 0, 0), (0, 0, 255)],\n",
328 | " titles=[\"original\", \"ground truth\", \"trained from scratch\"],\n",
329 | " dpi=200,\n",
330 | ")"
331 | ]
332 | },
333 | {
334 | "cell_type": "markdown",
335 | "metadata": {},
336 | "source": [
337 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
338 | ]
339 | },
340 | {
341 | "cell_type": "code",
342 | "execution_count": null,
343 | "metadata": {},
344 | "outputs": [],
345 | "source": [
346 | "import boto3\n",
347 | "import botocore\n",
348 | "\n",
349 | "config = botocore.config.Config(read_timeout=200)\n",
350 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
351 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
352 | "response = runtime.invoke_endpoint(\n",
353 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
354 | ")\n",
355 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
356 | ]
357 | },
358 | {
359 | "cell_type": "markdown",
360 | "metadata": {},
361 | "source": [
362 | "Finally, we compare the results of the provided pretrained model and our trained from scratch"
363 | ]
364 | },
365 | {
366 | "cell_type": "code",
367 | "execution_count": null,
368 | "metadata": {},
369 | "outputs": [],
370 | "source": [
371 | "visualize(\n",
372 | " original_image_unnorm,\n",
373 | " [original_boxes, demo_predictions[0][\"boxes\"], predictions[0][\"boxes\"]],\n",
374 | " [original_labels, demo_predictions[0][\"labels\"], predictions[0][\"labels\"]],\n",
375 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
376 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"from scratch\"],\n",
377 | " dpi=250,\n",
378 | ")"
379 | ]
380 | },
381 | {
382 | "cell_type": "markdown",
383 | "metadata": {},
384 | "source": [
385 | "## Optional: Delete the endpoint and model\n",
386 | "\n",
387 | "When you are done with the endpoint, you should clean it up.\n",
388 | "\n",
389 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
390 | ]
391 | },
392 | {
393 | "cell_type": "code",
394 | "execution_count": null,
395 | "metadata": {},
396 | "outputs": [],
397 | "source": [
398 | "detector.delete_model()\n",
399 | "detector.delete_endpoint()"
400 | ]
401 | },
402 | {
403 | "cell_type": "markdown",
404 | "metadata": {},
405 | "source": [
406 | "### [Click here to continue](./3_classification_from_scratch.ipynb)"
407 | ]
408 | }
409 | ],
410 | "metadata": {
411 | "kernelspec": {
412 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
413 | "metadata": {
414 | "interpreter": {
415 | "hash": "4c1e195df8d07db5ee7a78f454b46c3f2e14214bf8c9489d2db5cf8f372ff2ed"
416 | }
417 | },
418 | "name": "python3"
419 | },
420 | "language_info": {
421 | "codemirror_mode": {
422 | "name": "ipython",
423 | "version": 3
424 | },
425 | "file_extension": ".py",
426 | "mimetype": "text/x-python",
427 | "name": "python",
428 | "nbconvert_exporter": "python",
429 | "pygments_lexer": "ipython3",
430 | "version": "3.6.10-final"
431 | }
432 | },
433 | "nbformat": 4,
434 | "nbformat_minor": 2
435 | }
436 |
--------------------------------------------------------------------------------
125 |
126 | ## 3. Architecture Overview
127 |
128 | The following illustration is the architecture for the end-to-end training and deployment process
129 |
130 | 
131 |
132 | 1. The input data located in an [Amazon S3](https://aws.amazon.com/s3/) bucket
133 | 2. The provided [SageMaker notebook](source/deep_demand_forecast.ipynb) that gets the input data and launches the later stages below
134 | 3. **Training Classifier and Detector models** and evaluating its results using Amazon SageMaker. If desired, one can deploy the trained models and create SageMaker endpoints
135 | 4. **SageMaker endpoint** created from the previous step is an [HTTPS endpoint](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html) and is capable of producing predictions
136 | 5. Monitoring the training and deployed model via [Amazon CloudWatch](https://aws.amazon.com/cloudwatch/)
137 |
138 | ## 4. Cleaning up
139 |
140 | When you've finished with this solution, make sure that you delete all unwanted AWS resources. AWS CloudFormation can be used to automatically delete all standard resources that have been created by the solution and notebook. Go to the AWS CloudFormation Console, and delete the parent stack. Choosing to delete the parent stack will automatically delete the nested stacks.
141 |
142 | **Caution:** You need to manually delete any extra resources that you may have created in this notebook. Some examples include, extra Amazon S3 buckets (to the solution's default bucket), extra Amazon SageMaker endpoints (using a custom name).
143 |
144 | ## 5. Customization
145 |
146 | For using your own data, make sure it is labeled and is a *relatively* balanced dataset. Also make sure the image annotations follow the required format.
147 |
148 |
149 |
150 | ### Useful Links
151 |
152 | * [Amazon SageMaker Getting Started](https://aws.amazon.com/sagemaker/getting-started/)
153 | * [Amazon SageMaker Developer Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/whatis.html)
154 | * [Amazon SageMaker Python SDK Documentation](https://sagemaker.readthedocs.io/en/stable/)
155 | * [AWS CloudFormation User Guide](https://docs.aws.amazon.com/AWSCloudFormation/latest/UserGuide/Welcome.html)
156 |
157 | ### References
158 |
159 | * K. Song and Y. Yan, “A noise robust method based on completed local binary patterns for hot-rolled steel strip surface defects,” Applied Surface Science, vol. 285, pp. 858-864, Nov. 2013.
160 |
161 | * Yu He, Kechen Song, Qinggang Meng, Yunhui Yan, “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,” IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
162 |
163 | * Hongwen Dong, Kechen Song, Yu He, Jing Xu, Yunhui Yan, Qinggang Meng, “PGA-Net: Pyramid Feature Fusion and Global Context Attention Network for Automated Surface Defect Detection,” IEEE Transactions on Industrial Informatics, 2020.
164 |
165 | ### Security
166 |
167 | See [CONTRIBUTING](CONTRIBUTING.md#security-issue-notifications) for more information.
168 |
169 | ### License
170 |
171 | This project is licensed under the Apache-2.0 License.
172 |
--------------------------------------------------------------------------------
/notebooks/1_retrain_from_checkpoint.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Resume Training\n",
20 | "\n",
21 | "In this notebook, we retrain our pretrained detector for a few more epochs and compare its results. The same process can be applied when finetuning on another dataset. For the purpose of this notebook, we use the same **NEU-DET** dataset.\n",
22 | "\n",
23 | "## Finetuning\n",
24 | "\n",
25 | "Finetuning is one way to do Transfer Learning. Finetuning a Deep Learning model on one particular task, involves using the learned weights from a particular dataset to enhance the performace of the model on usually another dataset. In a sense, finetuning can be done over the same dataset used in the intial training but perhaps with different hyperparameters.\n"
26 | ]
27 | },
28 | {
29 | "cell_type": "code",
30 | "execution_count": null,
31 | "metadata": {},
32 | "outputs": [],
33 | "source": [
34 | "import json\n",
35 | "\n",
36 | "import sagemaker\n",
37 | "from sagemaker.s3 import S3Downloader\n",
38 | "\n",
39 | "sagemaker_session = sagemaker.Session()\n",
40 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
41 | "role = sagemaker_config[\"IamRole\"]\n",
42 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
43 | "region = sagemaker_config[\"AWSRegion\"]\n",
44 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
45 | "bucket = sagemaker_config[\"S3Bucket\"]"
46 | ]
47 | },
48 | {
49 | "cell_type": "markdown",
50 | "metadata": {},
51 | "source": [
52 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
53 | ]
54 | },
55 | {
56 | "cell_type": "code",
57 | "execution_count": null,
58 | "metadata": {},
59 | "outputs": [],
60 | "source": [
61 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
62 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
64 | ]
65 | },
66 | {
67 | "cell_type": "markdown",
68 | "metadata": {},
69 | "source": [
70 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
71 | ]
72 | },
73 | {
74 | "cell_type": "code",
75 | "execution_count": null,
76 | "metadata": {},
77 | "outputs": [],
78 | "source": [
79 | "DATA_PATH = !echo $PWD/neu_det\n",
80 | "DATA_PATH = DATA_PATH.n"
81 | ]
82 | },
83 | {
84 | "cell_type": "markdown",
85 | "metadata": {},
86 | "source": [
87 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
88 | ]
89 | },
90 | {
91 | "cell_type": "code",
92 | "execution_count": null,
93 | "metadata": {
94 | "tags": []
95 | },
96 | "outputs": [],
97 | "source": [
98 | "prefix = \"neu-det\"\n",
99 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
100 | "sources = f\"{neu_det_s3}/code/\"\n",
101 | "train_output = f\"{neu_det_s3}/output/\"\n",
102 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
103 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
104 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
105 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
106 | ]
107 | },
108 | {
109 | "cell_type": "markdown",
110 | "metadata": {},
111 | "source": [
112 | "## Visualization\n",
113 | "\n",
114 | "Let examine some datasets that we will use later by providing an `ID`"
115 | ]
116 | },
117 | {
118 | "cell_type": "code",
119 | "execution_count": null,
120 | "metadata": {},
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "\n",
125 | "import numpy as np\n",
126 | "import torch\n",
127 | "from PIL import Image\n",
128 | "from torch.utils.data import DataLoader\n",
129 | "\n",
130 | "try:\n",
131 | " import sagemaker_defect_detection\n",
132 | "except ImportError:\n",
133 | " import sys\n",
134 | " from pathlib import Path\n",
135 | "\n",
136 | " ROOT = Path(\"../src\").resolve()\n",
137 | " sys.path.insert(0, str(ROOT))\n",
138 | "\n",
139 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
140 | "\n",
141 | "SPLIT = \"test\"\n",
142 | "ID = 30\n",
143 | "assert 0 <= ID <= 300\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "Here we resume from a provided pretrained checkpoint `epoch=294-loss=0.654-main_score=0.349.ckpt` that we have copied into our `s3_pretrained`. This takes about **10 minutes** to complete"
185 | ]
186 | },
187 | {
188 | "cell_type": "code",
189 | "execution_count": null,
190 | "metadata": {
191 | "tags": [
192 | "outputPrepend"
193 | ]
194 | },
195 | "outputs": [],
196 | "source": [
197 | "%%time\n",
198 | "import logging\n",
199 | "from os import path as osp\n",
200 | "\n",
201 | "from sagemaker.pytorch import PyTorch\n",
202 | "\n",
203 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
204 | "# Note: resnet34 was used in the pretrained model and it has to match the pretrained model backbone\n",
205 | "# if need resnet50, need to train from scratch\n",
206 | "BACKBONE = \"resnet34\"\n",
207 | "assert BACKBONE in [\n",
208 | " \"resnet34\",\n",
209 | " \"resnet50\",\n",
210 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
211 | "EPOCHS = 5\n",
212 | "LEARNING_RATE = 1e-4\n",
213 | "SEED = 123\n",
214 | "\n",
215 | "hyperparameters = {\n",
216 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
217 | " \"num-classes\": NUM_CLASSES, # number of classes + background\n",
218 | " \"epochs\": EPOCHS, # number of epochs to finetune\n",
219 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
220 | " \"seed\": SEED, # random number generator seed\n",
221 | "}\n",
222 | "\n",
223 | "assert not isinstance(sagemaker_session, sagemaker.LocalSession), \"local session as share memory cannot be altered\"\n",
224 | "\n",
225 | "finetuned_model = PyTorch(\n",
226 | " entry_point=\"detector.py\",\n",
227 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
228 | " role=role,\n",
229 | " train_instance_count=1,\n",
230 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
231 | " hyperparameters=hyperparameters,\n",
232 | " py_version=\"py3\",\n",
233 | " framework_version=\"1.5\",\n",
234 | " sagemaker_session=sagemaker_session,\n",
235 | " output_path=train_output,\n",
236 | " checkpoint_s3_uri=s3_checkpoint,\n",
237 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
238 | " # container_log_level=logging.DEBUG,\n",
239 | ")\n",
240 | "\n",
241 | "finetuned_model.fit(\n",
242 | " {\n",
243 | " \"training\": neu_det_prepared_s3,\n",
244 | " \"pretrained_checkpoint\": osp.join(s3_pretrained, \"epoch=294-loss=0.654-main_score=0.349.ckpt\"),\n",
245 | " }\n",
246 | ")"
247 | ]
248 | },
249 | {
250 | "cell_type": "markdown",
251 | "metadata": {},
252 | "source": [
253 | "Then, we deploy our new model which takes about **10 minutes** to complete"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "%%time\n",
263 | "finetuned_detector = finetuned_model.deploy(\n",
264 | " initial_instance_count=1,\n",
265 | " instance_type=\"ml.m5.xlarge\",\n",
266 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-finetuned-endpoint\",\n",
267 | ")"
268 | ]
269 | },
270 | {
271 | "cell_type": "markdown",
272 | "metadata": {},
273 | "source": [
274 | "## Inference\n",
275 | "\n",
276 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
277 | ]
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": null,
282 | "metadata": {},
283 | "outputs": [],
284 | "source": [
285 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
286 | ]
287 | },
288 | {
289 | "cell_type": "markdown",
290 | "metadata": {},
291 | "source": [
292 | "Now the input is ready and we can get some results"
293 | ]
294 | },
295 | {
296 | "cell_type": "code",
297 | "execution_count": null,
298 | "metadata": {},
299 | "outputs": [],
300 | "source": [
301 | "%%time\n",
302 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
303 | "np_load_old = np.load\n",
304 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
305 | "finetuned_predictions = finetuned_detector.predict(input)\n",
306 | "np.load = np_load_old"
307 | ]
308 | },
309 | {
310 | "cell_type": "markdown",
311 | "metadata": {},
312 | "source": [
313 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
314 | ]
315 | },
316 | {
317 | "cell_type": "code",
318 | "execution_count": null,
319 | "metadata": {},
320 | "outputs": [],
321 | "source": [
322 | "import boto3\n",
323 | "import botocore\n",
324 | "\n",
325 | "config = botocore.config.Config(read_timeout=200)\n",
326 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
327 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
328 | "response = runtime.invoke_endpoint(\n",
329 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
330 | ")\n",
331 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
332 | ]
333 | },
334 | {
335 | "cell_type": "markdown",
336 | "metadata": {},
337 | "source": [
338 | "Here comes the slight changes in inference"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": null,
344 | "metadata": {},
345 | "outputs": [],
346 | "source": [
347 | "visualize(\n",
348 | " original_image_unnorm,\n",
349 | " [original_boxes, demo_predictions[0][\"boxes\"], finetuned_predictions[0][\"boxes\"]],\n",
350 | " [original_labels, demo_predictions[0][\"labels\"], finetuned_predictions[0][\"labels\"]],\n",
351 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
352 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"finetuned\"],\n",
353 | " dpi=250,\n",
354 | ")"
355 | ]
356 | },
357 | {
358 | "cell_type": "markdown",
359 | "metadata": {},
360 | "source": [
361 | "## Optional: Delete the endpoint and model\n",
362 | "\n",
363 | "When you are done with the endpoint, you should clean it up.\n",
364 | "\n",
365 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account.\n"
366 | ]
367 | },
368 | {
369 | "cell_type": "code",
370 | "execution_count": null,
371 | "metadata": {},
372 | "outputs": [],
373 | "source": [
374 | "finetuned_detector.delete_model()\n",
375 | "finetuned_detector.delete_endpoint()"
376 | ]
377 | },
378 | {
379 | "cell_type": "markdown",
380 | "metadata": {},
381 | "source": [
382 | "### [Click here to continue](./2_detection_from_scratch.ipynb)"
383 | ]
384 | }
385 | ],
386 | "metadata": {
387 | "kernelspec": {
388 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
389 | "name": "python361064bitpytorchlatestp36conda2dfac45b320c45f3a4d1e89ca46b60d1"
390 | },
391 | "language_info": {
392 | "codemirror_mode": {
393 | "name": "ipython",
394 | "version": 3
395 | },
396 | "file_extension": ".py",
397 | "mimetype": "text/x-python",
398 | "name": "python",
399 | "nbconvert_exporter": "python",
400 | "pygments_lexer": "ipython3",
401 | "version": "3.6.10-final"
402 | }
403 | },
404 | "nbformat": 4,
405 | "nbformat_minor": 2
406 | }
407 |
--------------------------------------------------------------------------------
/notebooks/2_detection_from_scratch.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Training our Detector from Scratch\n",
20 | "\n",
21 | "In this notebook, we will see how to train our detector from scratch"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": null,
27 | "metadata": {},
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import sagemaker\n",
33 | "from sagemaker.s3 import S3Downloader\n",
34 | "\n",
35 | "sagemaker_session = sagemaker.Session()\n",
36 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
37 | "role = sagemaker_config[\"IamRole\"]\n",
38 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
39 | "region = sagemaker_config[\"AWSRegion\"]\n",
40 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
41 | "bucket = sagemaker_config[\"S3Bucket\"]"
42 | ]
43 | },
44 | {
45 | "cell_type": "markdown",
46 | "metadata": {},
47 | "source": [
48 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "execution_count": null,
54 | "metadata": {},
55 | "outputs": [],
56 | "source": [
57 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
58 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
59 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
60 | ]
61 | },
62 | {
63 | "cell_type": "markdown",
64 | "metadata": {},
65 | "source": [
66 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
67 | ]
68 | },
69 | {
70 | "cell_type": "code",
71 | "execution_count": null,
72 | "metadata": {
73 | "tags": []
74 | },
75 | "outputs": [],
76 | "source": [
77 | "DATA_PATH = !echo $PWD/neu_det\n",
78 | "DATA_PATH = DATA_PATH.n"
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {},
84 | "source": [
85 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": null,
91 | "metadata": {
92 | "tags": []
93 | },
94 | "outputs": [],
95 | "source": [
96 | "prefix = \"neu-det\"\n",
97 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
98 | "sources = f\"{neu_det_s3}/code/\"\n",
99 | "train_output = f\"{neu_det_s3}/output/\"\n",
100 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
101 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
102 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
103 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
104 | ]
105 | },
106 | {
107 | "cell_type": "markdown",
108 | "metadata": {},
109 | "source": [
110 | "## Visualization\n",
111 | "\n",
112 | "Let examine some datasets that we will use later by providing an `ID`"
113 | ]
114 | },
115 | {
116 | "cell_type": "code",
117 | "execution_count": null,
118 | "metadata": {
119 | "tags": []
120 | },
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "from typing import List\n",
125 | "\n",
126 | "import numpy as np\n",
127 | "import torch\n",
128 | "from PIL import Image\n",
129 | "from torch.utils.data import DataLoader\n",
130 | "\n",
131 | "try:\n",
132 | " import sagemaker_defect_detection\n",
133 | "except ImportError:\n",
134 | " import sys\n",
135 | " from pathlib import Path\n",
136 | "\n",
137 | " ROOT = Path(\"../src\").resolve()\n",
138 | " sys.path.insert(0, str(ROOT))\n",
139 | "\n",
140 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
141 | "\n",
142 | "SPLIT = \"test\"\n",
143 | "ID = 50\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "In order to get high **Mean Average Percision (mAP)** and **Mean Average Recall (mAR)** for **Intersection Over Union (IOU)** thresholds of 0.5 when training from scratch, it requires more than **300 epochs**. That is why we have provided a pretrained model and recommend finetuning whenever is possible. For demostration, we train the model from scratch for 10 epochs which takes about **16 minutes** and it results in the following mAP, mAR and the accumulated `main_score` of\n",
185 | "\n",
186 | "* `Average Precision (AP) @[ IoU=0.50:0.95 ] ~ 0.048`\n",
187 | "* `Average Recall (AR) @[ IoU=0.50:0.95] ~ 0.153`\n",
188 | "* `main_score=0.0509`\n",
189 | "\n",
190 | "To get higher mAP, mAR and overall `main_score`, you can train for more epochs"
191 | ]
192 | },
193 | {
194 | "cell_type": "code",
195 | "execution_count": null,
196 | "metadata": {
197 | "tags": [
198 | "outputPrepend"
199 | ]
200 | },
201 | "outputs": [],
202 | "source": [
203 | "%%time\n",
204 | "import logging\n",
205 | "from os import path as osp\n",
206 | "\n",
207 | "from sagemaker.pytorch import PyTorch\n",
208 | "\n",
209 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
210 | "BACKBONE = \"resnet34\" # has to match the pretrained model backbone\n",
211 | "assert BACKBONE in [\n",
212 | " \"resnet34\",\n",
213 | " \"resnet50\",\n",
214 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
215 | "EPOCHS = 10\n",
216 | "LEARNING_RATE = 1e-3\n",
217 | "SEED = 123\n",
218 | "\n",
219 | "hyperparameters = {\n",
220 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
221 | " \"num-classes\": NUM_CLASSES, # number of classes 6 + 1 background\n",
222 | " \"epochs\": EPOCHS, # number of epochs to train\n",
223 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
224 | " \"seed\": SEED, # random number generator seed\n",
225 | "}\n",
226 | "\n",
227 | "model = PyTorch(\n",
228 | " entry_point=\"detector.py\",\n",
229 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
230 | " role=role,\n",
231 | " train_instance_count=1,\n",
232 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
233 | " hyperparameters=hyperparameters,\n",
234 | " py_version=\"py3\",\n",
235 | " framework_version=\"1.5\",\n",
236 | " sagemaker_session=sagemaker_session,\n",
237 | " output_path=train_output,\n",
238 | " checkpoint_s3_uri=s3_checkpoint,\n",
239 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
240 | " # container_log_level=logging.DEBUG,\n",
241 | ")\n",
242 | "\n",
243 | "model.fit(neu_det_prepared_s3)"
244 | ]
245 | },
246 | {
247 | "cell_type": "markdown",
248 | "metadata": {},
249 | "source": [
250 | "Then, we deploy our model which takes about **10 minutes** to complete"
251 | ]
252 | },
253 | {
254 | "cell_type": "code",
255 | "execution_count": null,
256 | "metadata": {
257 | "tags": []
258 | },
259 | "outputs": [],
260 | "source": [
261 | "%%time\n",
262 | "detector = model.deploy(\n",
263 | " initial_instance_count=1,\n",
264 | " instance_type=\"ml.m5.xlarge\",\n",
265 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-detector-from-scratch-endpoint\",\n",
266 | ")"
267 | ]
268 | },
269 | {
270 | "cell_type": "markdown",
271 | "metadata": {},
272 | "source": [
273 | "## Inference\n",
274 | "\n",
275 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
276 | ]
277 | },
278 | {
279 | "cell_type": "code",
280 | "execution_count": null,
281 | "metadata": {},
282 | "outputs": [],
283 | "source": [
284 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
285 | ]
286 | },
287 | {
288 | "cell_type": "markdown",
289 | "metadata": {},
290 | "source": [
291 | "Now the input is ready and we can get some results"
292 | ]
293 | },
294 | {
295 | "cell_type": "code",
296 | "execution_count": null,
297 | "metadata": {
298 | "tags": []
299 | },
300 | "outputs": [],
301 | "source": [
302 | "%%time\n",
303 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
304 | "np_load_old = np.load\n",
305 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
306 | "predictions = detector.predict(input)\n",
307 | "np.load = np_load_old"
308 | ]
309 | },
310 | {
311 | "cell_type": "markdown",
312 | "metadata": {},
313 | "source": [
314 | "We use our `visualize` utility to check the detection results"
315 | ]
316 | },
317 | {
318 | "cell_type": "code",
319 | "execution_count": null,
320 | "metadata": {},
321 | "outputs": [],
322 | "source": [
323 | "visualize(\n",
324 | " original_image_unnorm,\n",
325 | " [original_boxes, predictions[0][\"boxes\"]],\n",
326 | " [original_labels, predictions[0][\"labels\"]],\n",
327 | " colors=[(255, 0, 0), (0, 0, 255)],\n",
328 | " titles=[\"original\", \"ground truth\", \"trained from scratch\"],\n",
329 | " dpi=200,\n",
330 | ")"
331 | ]
332 | },
333 | {
334 | "cell_type": "markdown",
335 | "metadata": {},
336 | "source": [
337 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
338 | ]
339 | },
340 | {
341 | "cell_type": "code",
342 | "execution_count": null,
343 | "metadata": {},
344 | "outputs": [],
345 | "source": [
346 | "import boto3\n",
347 | "import botocore\n",
348 | "\n",
349 | "config = botocore.config.Config(read_timeout=200)\n",
350 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
351 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
352 | "response = runtime.invoke_endpoint(\n",
353 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
354 | ")\n",
355 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
356 | ]
357 | },
358 | {
359 | "cell_type": "markdown",
360 | "metadata": {},
361 | "source": [
362 | "Finally, we compare the results of the provided pretrained model and our trained from scratch"
363 | ]
364 | },
365 | {
366 | "cell_type": "code",
367 | "execution_count": null,
368 | "metadata": {},
369 | "outputs": [],
370 | "source": [
371 | "visualize(\n",
372 | " original_image_unnorm,\n",
373 | " [original_boxes, demo_predictions[0][\"boxes\"], predictions[0][\"boxes\"]],\n",
374 | " [original_labels, demo_predictions[0][\"labels\"], predictions[0][\"labels\"]],\n",
375 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
376 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"from scratch\"],\n",
377 | " dpi=250,\n",
378 | ")"
379 | ]
380 | },
381 | {
382 | "cell_type": "markdown",
383 | "metadata": {},
384 | "source": [
385 | "## Optional: Delete the endpoint and model\n",
386 | "\n",
387 | "When you are done with the endpoint, you should clean it up.\n",
388 | "\n",
389 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
390 | ]
391 | },
392 | {
393 | "cell_type": "code",
394 | "execution_count": null,
395 | "metadata": {},
396 | "outputs": [],
397 | "source": [
398 | "detector.delete_model()\n",
399 | "detector.delete_endpoint()"
400 | ]
401 | },
402 | {
403 | "cell_type": "markdown",
404 | "metadata": {},
405 | "source": [
406 | "### [Click here to continue](./3_classification_from_scratch.ipynb)"
407 | ]
408 | }
409 | ],
410 | "metadata": {
411 | "kernelspec": {
412 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
413 | "metadata": {
414 | "interpreter": {
415 | "hash": "4c1e195df8d07db5ee7a78f454b46c3f2e14214bf8c9489d2db5cf8f372ff2ed"
416 | }
417 | },
418 | "name": "python3"
419 | },
420 | "language_info": {
421 | "codemirror_mode": {
422 | "name": "ipython",
423 | "version": 3
424 | },
425 | "file_extension": ".py",
426 | "mimetype": "text/x-python",
427 | "name": "python",
428 | "nbconvert_exporter": "python",
429 | "pygments_lexer": "ipython3",
430 | "version": "3.6.10-final"
431 | }
432 | },
433 | "nbformat": 4,
434 | "nbformat_minor": 2
435 | }
436 |
--------------------------------------------------------------------------------
5 |
6 |




 125 |
126 | ## 3. Architecture Overview
127 |
128 | The following illustration is the architecture for the end-to-end training and deployment process
129 |
130 | 
131 |
132 | 1. The input data located in an [Amazon S3](https://aws.amazon.com/s3/) bucket
133 | 2. The provided [SageMaker notebook](source/deep_demand_forecast.ipynb) that gets the input data and launches the later stages below
134 | 3. **Training Classifier and Detector models** and evaluating its results using Amazon SageMaker. If desired, one can deploy the trained models and create SageMaker endpoints
135 | 4. **SageMaker endpoint** created from the previous step is an [HTTPS endpoint](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html) and is capable of producing predictions
136 | 5. Monitoring the training and deployed model via [Amazon CloudWatch](https://aws.amazon.com/cloudwatch/)
137 |
138 | ## 4. Cleaning up
139 |
140 | When you've finished with this solution, make sure that you delete all unwanted AWS resources. AWS CloudFormation can be used to automatically delete all standard resources that have been created by the solution and notebook. Go to the AWS CloudFormation Console, and delete the parent stack. Choosing to delete the parent stack will automatically delete the nested stacks.
141 |
142 | **Caution:** You need to manually delete any extra resources that you may have created in this notebook. Some examples include, extra Amazon S3 buckets (to the solution's default bucket), extra Amazon SageMaker endpoints (using a custom name).
143 |
144 | ## 5. Customization
145 |
146 | For using your own data, make sure it is labeled and is a *relatively* balanced dataset. Also make sure the image annotations follow the required format.
147 |
148 |
149 |
150 | ### Useful Links
151 |
152 | * [Amazon SageMaker Getting Started](https://aws.amazon.com/sagemaker/getting-started/)
153 | * [Amazon SageMaker Developer Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/whatis.html)
154 | * [Amazon SageMaker Python SDK Documentation](https://sagemaker.readthedocs.io/en/stable/)
155 | * [AWS CloudFormation User Guide](https://docs.aws.amazon.com/AWSCloudFormation/latest/UserGuide/Welcome.html)
156 |
157 | ### References
158 |
159 | * K. Song and Y. Yan, “A noise robust method based on completed local binary patterns for hot-rolled steel strip surface defects,” Applied Surface Science, vol. 285, pp. 858-864, Nov. 2013.
160 |
161 | * Yu He, Kechen Song, Qinggang Meng, Yunhui Yan, “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,” IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
162 |
163 | * Hongwen Dong, Kechen Song, Yu He, Jing Xu, Yunhui Yan, Qinggang Meng, “PGA-Net: Pyramid Feature Fusion and Global Context Attention Network for Automated Surface Defect Detection,” IEEE Transactions on Industrial Informatics, 2020.
164 |
165 | ### Security
166 |
167 | See [CONTRIBUTING](CONTRIBUTING.md#security-issue-notifications) for more information.
168 |
169 | ### License
170 |
171 | This project is licensed under the Apache-2.0 License.
172 |
--------------------------------------------------------------------------------
/notebooks/1_retrain_from_checkpoint.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Resume Training\n",
20 | "\n",
21 | "In this notebook, we retrain our pretrained detector for a few more epochs and compare its results. The same process can be applied when finetuning on another dataset. For the purpose of this notebook, we use the same **NEU-DET** dataset.\n",
22 | "\n",
23 | "## Finetuning\n",
24 | "\n",
25 | "Finetuning is one way to do Transfer Learning. Finetuning a Deep Learning model on one particular task, involves using the learned weights from a particular dataset to enhance the performace of the model on usually another dataset. In a sense, finetuning can be done over the same dataset used in the intial training but perhaps with different hyperparameters.\n"
26 | ]
27 | },
28 | {
29 | "cell_type": "code",
30 | "execution_count": null,
31 | "metadata": {},
32 | "outputs": [],
33 | "source": [
34 | "import json\n",
35 | "\n",
36 | "import sagemaker\n",
37 | "from sagemaker.s3 import S3Downloader\n",
38 | "\n",
39 | "sagemaker_session = sagemaker.Session()\n",
40 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
41 | "role = sagemaker_config[\"IamRole\"]\n",
42 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
43 | "region = sagemaker_config[\"AWSRegion\"]\n",
44 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
45 | "bucket = sagemaker_config[\"S3Bucket\"]"
46 | ]
47 | },
48 | {
49 | "cell_type": "markdown",
50 | "metadata": {},
51 | "source": [
52 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
53 | ]
54 | },
55 | {
56 | "cell_type": "code",
57 | "execution_count": null,
58 | "metadata": {},
59 | "outputs": [],
60 | "source": [
61 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
62 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
64 | ]
65 | },
66 | {
67 | "cell_type": "markdown",
68 | "metadata": {},
69 | "source": [
70 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
71 | ]
72 | },
73 | {
74 | "cell_type": "code",
75 | "execution_count": null,
76 | "metadata": {},
77 | "outputs": [],
78 | "source": [
79 | "DATA_PATH = !echo $PWD/neu_det\n",
80 | "DATA_PATH = DATA_PATH.n"
81 | ]
82 | },
83 | {
84 | "cell_type": "markdown",
85 | "metadata": {},
86 | "source": [
87 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
88 | ]
89 | },
90 | {
91 | "cell_type": "code",
92 | "execution_count": null,
93 | "metadata": {
94 | "tags": []
95 | },
96 | "outputs": [],
97 | "source": [
98 | "prefix = \"neu-det\"\n",
99 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
100 | "sources = f\"{neu_det_s3}/code/\"\n",
101 | "train_output = f\"{neu_det_s3}/output/\"\n",
102 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
103 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
104 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
105 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
106 | ]
107 | },
108 | {
109 | "cell_type": "markdown",
110 | "metadata": {},
111 | "source": [
112 | "## Visualization\n",
113 | "\n",
114 | "Let examine some datasets that we will use later by providing an `ID`"
115 | ]
116 | },
117 | {
118 | "cell_type": "code",
119 | "execution_count": null,
120 | "metadata": {},
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "\n",
125 | "import numpy as np\n",
126 | "import torch\n",
127 | "from PIL import Image\n",
128 | "from torch.utils.data import DataLoader\n",
129 | "\n",
130 | "try:\n",
131 | " import sagemaker_defect_detection\n",
132 | "except ImportError:\n",
133 | " import sys\n",
134 | " from pathlib import Path\n",
135 | "\n",
136 | " ROOT = Path(\"../src\").resolve()\n",
137 | " sys.path.insert(0, str(ROOT))\n",
138 | "\n",
139 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
140 | "\n",
141 | "SPLIT = \"test\"\n",
142 | "ID = 30\n",
143 | "assert 0 <= ID <= 300\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "Here we resume from a provided pretrained checkpoint `epoch=294-loss=0.654-main_score=0.349.ckpt` that we have copied into our `s3_pretrained`. This takes about **10 minutes** to complete"
185 | ]
186 | },
187 | {
188 | "cell_type": "code",
189 | "execution_count": null,
190 | "metadata": {
191 | "tags": [
192 | "outputPrepend"
193 | ]
194 | },
195 | "outputs": [],
196 | "source": [
197 | "%%time\n",
198 | "import logging\n",
199 | "from os import path as osp\n",
200 | "\n",
201 | "from sagemaker.pytorch import PyTorch\n",
202 | "\n",
203 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
204 | "# Note: resnet34 was used in the pretrained model and it has to match the pretrained model backbone\n",
205 | "# if need resnet50, need to train from scratch\n",
206 | "BACKBONE = \"resnet34\"\n",
207 | "assert BACKBONE in [\n",
208 | " \"resnet34\",\n",
209 | " \"resnet50\",\n",
210 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
211 | "EPOCHS = 5\n",
212 | "LEARNING_RATE = 1e-4\n",
213 | "SEED = 123\n",
214 | "\n",
215 | "hyperparameters = {\n",
216 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
217 | " \"num-classes\": NUM_CLASSES, # number of classes + background\n",
218 | " \"epochs\": EPOCHS, # number of epochs to finetune\n",
219 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
220 | " \"seed\": SEED, # random number generator seed\n",
221 | "}\n",
222 | "\n",
223 | "assert not isinstance(sagemaker_session, sagemaker.LocalSession), \"local session as share memory cannot be altered\"\n",
224 | "\n",
225 | "finetuned_model = PyTorch(\n",
226 | " entry_point=\"detector.py\",\n",
227 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
228 | " role=role,\n",
229 | " train_instance_count=1,\n",
230 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
231 | " hyperparameters=hyperparameters,\n",
232 | " py_version=\"py3\",\n",
233 | " framework_version=\"1.5\",\n",
234 | " sagemaker_session=sagemaker_session,\n",
235 | " output_path=train_output,\n",
236 | " checkpoint_s3_uri=s3_checkpoint,\n",
237 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
238 | " # container_log_level=logging.DEBUG,\n",
239 | ")\n",
240 | "\n",
241 | "finetuned_model.fit(\n",
242 | " {\n",
243 | " \"training\": neu_det_prepared_s3,\n",
244 | " \"pretrained_checkpoint\": osp.join(s3_pretrained, \"epoch=294-loss=0.654-main_score=0.349.ckpt\"),\n",
245 | " }\n",
246 | ")"
247 | ]
248 | },
249 | {
250 | "cell_type": "markdown",
251 | "metadata": {},
252 | "source": [
253 | "Then, we deploy our new model which takes about **10 minutes** to complete"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "%%time\n",
263 | "finetuned_detector = finetuned_model.deploy(\n",
264 | " initial_instance_count=1,\n",
265 | " instance_type=\"ml.m5.xlarge\",\n",
266 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-finetuned-endpoint\",\n",
267 | ")"
268 | ]
269 | },
270 | {
271 | "cell_type": "markdown",
272 | "metadata": {},
273 | "source": [
274 | "## Inference\n",
275 | "\n",
276 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
277 | ]
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": null,
282 | "metadata": {},
283 | "outputs": [],
284 | "source": [
285 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
286 | ]
287 | },
288 | {
289 | "cell_type": "markdown",
290 | "metadata": {},
291 | "source": [
292 | "Now the input is ready and we can get some results"
293 | ]
294 | },
295 | {
296 | "cell_type": "code",
297 | "execution_count": null,
298 | "metadata": {},
299 | "outputs": [],
300 | "source": [
301 | "%%time\n",
302 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
303 | "np_load_old = np.load\n",
304 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
305 | "finetuned_predictions = finetuned_detector.predict(input)\n",
306 | "np.load = np_load_old"
307 | ]
308 | },
309 | {
310 | "cell_type": "markdown",
311 | "metadata": {},
312 | "source": [
313 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
314 | ]
315 | },
316 | {
317 | "cell_type": "code",
318 | "execution_count": null,
319 | "metadata": {},
320 | "outputs": [],
321 | "source": [
322 | "import boto3\n",
323 | "import botocore\n",
324 | "\n",
325 | "config = botocore.config.Config(read_timeout=200)\n",
326 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
327 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
328 | "response = runtime.invoke_endpoint(\n",
329 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
330 | ")\n",
331 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
332 | ]
333 | },
334 | {
335 | "cell_type": "markdown",
336 | "metadata": {},
337 | "source": [
338 | "Here comes the slight changes in inference"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": null,
344 | "metadata": {},
345 | "outputs": [],
346 | "source": [
347 | "visualize(\n",
348 | " original_image_unnorm,\n",
349 | " [original_boxes, demo_predictions[0][\"boxes\"], finetuned_predictions[0][\"boxes\"]],\n",
350 | " [original_labels, demo_predictions[0][\"labels\"], finetuned_predictions[0][\"labels\"]],\n",
351 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
352 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"finetuned\"],\n",
353 | " dpi=250,\n",
354 | ")"
355 | ]
356 | },
357 | {
358 | "cell_type": "markdown",
359 | "metadata": {},
360 | "source": [
361 | "## Optional: Delete the endpoint and model\n",
362 | "\n",
363 | "When you are done with the endpoint, you should clean it up.\n",
364 | "\n",
365 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account.\n"
366 | ]
367 | },
368 | {
369 | "cell_type": "code",
370 | "execution_count": null,
371 | "metadata": {},
372 | "outputs": [],
373 | "source": [
374 | "finetuned_detector.delete_model()\n",
375 | "finetuned_detector.delete_endpoint()"
376 | ]
377 | },
378 | {
379 | "cell_type": "markdown",
380 | "metadata": {},
381 | "source": [
382 | "### [Click here to continue](./2_detection_from_scratch.ipynb)"
383 | ]
384 | }
385 | ],
386 | "metadata": {
387 | "kernelspec": {
388 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
389 | "name": "python361064bitpytorchlatestp36conda2dfac45b320c45f3a4d1e89ca46b60d1"
390 | },
391 | "language_info": {
392 | "codemirror_mode": {
393 | "name": "ipython",
394 | "version": 3
395 | },
396 | "file_extension": ".py",
397 | "mimetype": "text/x-python",
398 | "name": "python",
399 | "nbconvert_exporter": "python",
400 | "pygments_lexer": "ipython3",
401 | "version": "3.6.10-final"
402 | }
403 | },
404 | "nbformat": 4,
405 | "nbformat_minor": 2
406 | }
407 |
--------------------------------------------------------------------------------
/notebooks/2_detection_from_scratch.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Training our Detector from Scratch\n",
20 | "\n",
21 | "In this notebook, we will see how to train our detector from scratch"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": null,
27 | "metadata": {},
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import sagemaker\n",
33 | "from sagemaker.s3 import S3Downloader\n",
34 | "\n",
35 | "sagemaker_session = sagemaker.Session()\n",
36 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
37 | "role = sagemaker_config[\"IamRole\"]\n",
38 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
39 | "region = sagemaker_config[\"AWSRegion\"]\n",
40 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
41 | "bucket = sagemaker_config[\"S3Bucket\"]"
42 | ]
43 | },
44 | {
45 | "cell_type": "markdown",
46 | "metadata": {},
47 | "source": [
48 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "execution_count": null,
54 | "metadata": {},
55 | "outputs": [],
56 | "source": [
57 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
58 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
59 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
60 | ]
61 | },
62 | {
63 | "cell_type": "markdown",
64 | "metadata": {},
65 | "source": [
66 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
67 | ]
68 | },
69 | {
70 | "cell_type": "code",
71 | "execution_count": null,
72 | "metadata": {
73 | "tags": []
74 | },
75 | "outputs": [],
76 | "source": [
77 | "DATA_PATH = !echo $PWD/neu_det\n",
78 | "DATA_PATH = DATA_PATH.n"
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {},
84 | "source": [
85 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": null,
91 | "metadata": {
92 | "tags": []
93 | },
94 | "outputs": [],
95 | "source": [
96 | "prefix = \"neu-det\"\n",
97 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
98 | "sources = f\"{neu_det_s3}/code/\"\n",
99 | "train_output = f\"{neu_det_s3}/output/\"\n",
100 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
101 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
102 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
103 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
104 | ]
105 | },
106 | {
107 | "cell_type": "markdown",
108 | "metadata": {},
109 | "source": [
110 | "## Visualization\n",
111 | "\n",
112 | "Let examine some datasets that we will use later by providing an `ID`"
113 | ]
114 | },
115 | {
116 | "cell_type": "code",
117 | "execution_count": null,
118 | "metadata": {
119 | "tags": []
120 | },
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "from typing import List\n",
125 | "\n",
126 | "import numpy as np\n",
127 | "import torch\n",
128 | "from PIL import Image\n",
129 | "from torch.utils.data import DataLoader\n",
130 | "\n",
131 | "try:\n",
132 | " import sagemaker_defect_detection\n",
133 | "except ImportError:\n",
134 | " import sys\n",
135 | " from pathlib import Path\n",
136 | "\n",
137 | " ROOT = Path(\"../src\").resolve()\n",
138 | " sys.path.insert(0, str(ROOT))\n",
139 | "\n",
140 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
141 | "\n",
142 | "SPLIT = \"test\"\n",
143 | "ID = 50\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "In order to get high **Mean Average Percision (mAP)** and **Mean Average Recall (mAR)** for **Intersection Over Union (IOU)** thresholds of 0.5 when training from scratch, it requires more than **300 epochs**. That is why we have provided a pretrained model and recommend finetuning whenever is possible. For demostration, we train the model from scratch for 10 epochs which takes about **16 minutes** and it results in the following mAP, mAR and the accumulated `main_score` of\n",
185 | "\n",
186 | "* `Average Precision (AP) @[ IoU=0.50:0.95 ] ~ 0.048`\n",
187 | "* `Average Recall (AR) @[ IoU=0.50:0.95] ~ 0.153`\n",
188 | "* `main_score=0.0509`\n",
189 | "\n",
190 | "To get higher mAP, mAR and overall `main_score`, you can train for more epochs"
191 | ]
192 | },
193 | {
194 | "cell_type": "code",
195 | "execution_count": null,
196 | "metadata": {
197 | "tags": [
198 | "outputPrepend"
199 | ]
200 | },
201 | "outputs": [],
202 | "source": [
203 | "%%time\n",
204 | "import logging\n",
205 | "from os import path as osp\n",
206 | "\n",
207 | "from sagemaker.pytorch import PyTorch\n",
208 | "\n",
209 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
210 | "BACKBONE = \"resnet34\" # has to match the pretrained model backbone\n",
211 | "assert BACKBONE in [\n",
212 | " \"resnet34\",\n",
213 | " \"resnet50\",\n",
214 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
215 | "EPOCHS = 10\n",
216 | "LEARNING_RATE = 1e-3\n",
217 | "SEED = 123\n",
218 | "\n",
219 | "hyperparameters = {\n",
220 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
221 | " \"num-classes\": NUM_CLASSES, # number of classes 6 + 1 background\n",
222 | " \"epochs\": EPOCHS, # number of epochs to train\n",
223 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
224 | " \"seed\": SEED, # random number generator seed\n",
225 | "}\n",
226 | "\n",
227 | "model = PyTorch(\n",
228 | " entry_point=\"detector.py\",\n",
229 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
230 | " role=role,\n",
231 | " train_instance_count=1,\n",
232 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
233 | " hyperparameters=hyperparameters,\n",
234 | " py_version=\"py3\",\n",
235 | " framework_version=\"1.5\",\n",
236 | " sagemaker_session=sagemaker_session,\n",
237 | " output_path=train_output,\n",
238 | " checkpoint_s3_uri=s3_checkpoint,\n",
239 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
240 | " # container_log_level=logging.DEBUG,\n",
241 | ")\n",
242 | "\n",
243 | "model.fit(neu_det_prepared_s3)"
244 | ]
245 | },
246 | {
247 | "cell_type": "markdown",
248 | "metadata": {},
249 | "source": [
250 | "Then, we deploy our model which takes about **10 minutes** to complete"
251 | ]
252 | },
253 | {
254 | "cell_type": "code",
255 | "execution_count": null,
256 | "metadata": {
257 | "tags": []
258 | },
259 | "outputs": [],
260 | "source": [
261 | "%%time\n",
262 | "detector = model.deploy(\n",
263 | " initial_instance_count=1,\n",
264 | " instance_type=\"ml.m5.xlarge\",\n",
265 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-detector-from-scratch-endpoint\",\n",
266 | ")"
267 | ]
268 | },
269 | {
270 | "cell_type": "markdown",
271 | "metadata": {},
272 | "source": [
273 | "## Inference\n",
274 | "\n",
275 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
276 | ]
277 | },
278 | {
279 | "cell_type": "code",
280 | "execution_count": null,
281 | "metadata": {},
282 | "outputs": [],
283 | "source": [
284 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
285 | ]
286 | },
287 | {
288 | "cell_type": "markdown",
289 | "metadata": {},
290 | "source": [
291 | "Now the input is ready and we can get some results"
292 | ]
293 | },
294 | {
295 | "cell_type": "code",
296 | "execution_count": null,
297 | "metadata": {
298 | "tags": []
299 | },
300 | "outputs": [],
301 | "source": [
302 | "%%time\n",
303 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
304 | "np_load_old = np.load\n",
305 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
306 | "predictions = detector.predict(input)\n",
307 | "np.load = np_load_old"
308 | ]
309 | },
310 | {
311 | "cell_type": "markdown",
312 | "metadata": {},
313 | "source": [
314 | "We use our `visualize` utility to check the detection results"
315 | ]
316 | },
317 | {
318 | "cell_type": "code",
319 | "execution_count": null,
320 | "metadata": {},
321 | "outputs": [],
322 | "source": [
323 | "visualize(\n",
324 | " original_image_unnorm,\n",
325 | " [original_boxes, predictions[0][\"boxes\"]],\n",
326 | " [original_labels, predictions[0][\"labels\"]],\n",
327 | " colors=[(255, 0, 0), (0, 0, 255)],\n",
328 | " titles=[\"original\", \"ground truth\", \"trained from scratch\"],\n",
329 | " dpi=200,\n",
330 | ")"
331 | ]
332 | },
333 | {
334 | "cell_type": "markdown",
335 | "metadata": {},
336 | "source": [
337 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
338 | ]
339 | },
340 | {
341 | "cell_type": "code",
342 | "execution_count": null,
343 | "metadata": {},
344 | "outputs": [],
345 | "source": [
346 | "import boto3\n",
347 | "import botocore\n",
348 | "\n",
349 | "config = botocore.config.Config(read_timeout=200)\n",
350 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
351 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
352 | "response = runtime.invoke_endpoint(\n",
353 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
354 | ")\n",
355 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
356 | ]
357 | },
358 | {
359 | "cell_type": "markdown",
360 | "metadata": {},
361 | "source": [
362 | "Finally, we compare the results of the provided pretrained model and our trained from scratch"
363 | ]
364 | },
365 | {
366 | "cell_type": "code",
367 | "execution_count": null,
368 | "metadata": {},
369 | "outputs": [],
370 | "source": [
371 | "visualize(\n",
372 | " original_image_unnorm,\n",
373 | " [original_boxes, demo_predictions[0][\"boxes\"], predictions[0][\"boxes\"]],\n",
374 | " [original_labels, demo_predictions[0][\"labels\"], predictions[0][\"labels\"]],\n",
375 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
376 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"from scratch\"],\n",
377 | " dpi=250,\n",
378 | ")"
379 | ]
380 | },
381 | {
382 | "cell_type": "markdown",
383 | "metadata": {},
384 | "source": [
385 | "## Optional: Delete the endpoint and model\n",
386 | "\n",
387 | "When you are done with the endpoint, you should clean it up.\n",
388 | "\n",
389 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
390 | ]
391 | },
392 | {
393 | "cell_type": "code",
394 | "execution_count": null,
395 | "metadata": {},
396 | "outputs": [],
397 | "source": [
398 | "detector.delete_model()\n",
399 | "detector.delete_endpoint()"
400 | ]
401 | },
402 | {
403 | "cell_type": "markdown",
404 | "metadata": {},
405 | "source": [
406 | "### [Click here to continue](./3_classification_from_scratch.ipynb)"
407 | ]
408 | }
409 | ],
410 | "metadata": {
411 | "kernelspec": {
412 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
413 | "metadata": {
414 | "interpreter": {
415 | "hash": "4c1e195df8d07db5ee7a78f454b46c3f2e14214bf8c9489d2db5cf8f372ff2ed"
416 | }
417 | },
418 | "name": "python3"
419 | },
420 | "language_info": {
421 | "codemirror_mode": {
422 | "name": "ipython",
423 | "version": 3
424 | },
425 | "file_extension": ".py",
426 | "mimetype": "text/x-python",
427 | "name": "python",
428 | "nbconvert_exporter": "python",
429 | "pygments_lexer": "ipython3",
430 | "version": "3.6.10-final"
431 | }
432 | },
433 | "nbformat": 4,
434 | "nbformat_minor": 2
435 | }
436 |
--------------------------------------------------------------------------------
125 |
126 | ## 3. Architecture Overview
127 |
128 | The following illustration is the architecture for the end-to-end training and deployment process
129 |
130 | 
131 |
132 | 1. The input data located in an [Amazon S3](https://aws.amazon.com/s3/) bucket
133 | 2. The provided [SageMaker notebook](source/deep_demand_forecast.ipynb) that gets the input data and launches the later stages below
134 | 3. **Training Classifier and Detector models** and evaluating its results using Amazon SageMaker. If desired, one can deploy the trained models and create SageMaker endpoints
135 | 4. **SageMaker endpoint** created from the previous step is an [HTTPS endpoint](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html) and is capable of producing predictions
136 | 5. Monitoring the training and deployed model via [Amazon CloudWatch](https://aws.amazon.com/cloudwatch/)
137 |
138 | ## 4. Cleaning up
139 |
140 | When you've finished with this solution, make sure that you delete all unwanted AWS resources. AWS CloudFormation can be used to automatically delete all standard resources that have been created by the solution and notebook. Go to the AWS CloudFormation Console, and delete the parent stack. Choosing to delete the parent stack will automatically delete the nested stacks.
141 |
142 | **Caution:** You need to manually delete any extra resources that you may have created in this notebook. Some examples include, extra Amazon S3 buckets (to the solution's default bucket), extra Amazon SageMaker endpoints (using a custom name).
143 |
144 | ## 5. Customization
145 |
146 | For using your own data, make sure it is labeled and is a *relatively* balanced dataset. Also make sure the image annotations follow the required format.
147 |
148 |
149 |
150 | ### Useful Links
151 |
152 | * [Amazon SageMaker Getting Started](https://aws.amazon.com/sagemaker/getting-started/)
153 | * [Amazon SageMaker Developer Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/whatis.html)
154 | * [Amazon SageMaker Python SDK Documentation](https://sagemaker.readthedocs.io/en/stable/)
155 | * [AWS CloudFormation User Guide](https://docs.aws.amazon.com/AWSCloudFormation/latest/UserGuide/Welcome.html)
156 |
157 | ### References
158 |
159 | * K. Song and Y. Yan, “A noise robust method based on completed local binary patterns for hot-rolled steel strip surface defects,” Applied Surface Science, vol. 285, pp. 858-864, Nov. 2013.
160 |
161 | * Yu He, Kechen Song, Qinggang Meng, Yunhui Yan, “An End-to-end Steel Surface Defect Detection Approach via Fusing Multiple Hierarchical Features,” IEEE Transactions on Instrumentation and Measuremente, 2020,69(4),1493-1504.
162 |
163 | * Hongwen Dong, Kechen Song, Yu He, Jing Xu, Yunhui Yan, Qinggang Meng, “PGA-Net: Pyramid Feature Fusion and Global Context Attention Network for Automated Surface Defect Detection,” IEEE Transactions on Industrial Informatics, 2020.
164 |
165 | ### Security
166 |
167 | See [CONTRIBUTING](CONTRIBUTING.md#security-issue-notifications) for more information.
168 |
169 | ### License
170 |
171 | This project is licensed under the Apache-2.0 License.
172 |
--------------------------------------------------------------------------------
/notebooks/1_retrain_from_checkpoint.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Resume Training\n",
20 | "\n",
21 | "In this notebook, we retrain our pretrained detector for a few more epochs and compare its results. The same process can be applied when finetuning on another dataset. For the purpose of this notebook, we use the same **NEU-DET** dataset.\n",
22 | "\n",
23 | "## Finetuning\n",
24 | "\n",
25 | "Finetuning is one way to do Transfer Learning. Finetuning a Deep Learning model on one particular task, involves using the learned weights from a particular dataset to enhance the performace of the model on usually another dataset. In a sense, finetuning can be done over the same dataset used in the intial training but perhaps with different hyperparameters.\n"
26 | ]
27 | },
28 | {
29 | "cell_type": "code",
30 | "execution_count": null,
31 | "metadata": {},
32 | "outputs": [],
33 | "source": [
34 | "import json\n",
35 | "\n",
36 | "import sagemaker\n",
37 | "from sagemaker.s3 import S3Downloader\n",
38 | "\n",
39 | "sagemaker_session = sagemaker.Session()\n",
40 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
41 | "role = sagemaker_config[\"IamRole\"]\n",
42 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
43 | "region = sagemaker_config[\"AWSRegion\"]\n",
44 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
45 | "bucket = sagemaker_config[\"S3Bucket\"]"
46 | ]
47 | },
48 | {
49 | "cell_type": "markdown",
50 | "metadata": {},
51 | "source": [
52 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
53 | ]
54 | },
55 | {
56 | "cell_type": "code",
57 | "execution_count": null,
58 | "metadata": {},
59 | "outputs": [],
60 | "source": [
61 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
62 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
63 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
64 | ]
65 | },
66 | {
67 | "cell_type": "markdown",
68 | "metadata": {},
69 | "source": [
70 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
71 | ]
72 | },
73 | {
74 | "cell_type": "code",
75 | "execution_count": null,
76 | "metadata": {},
77 | "outputs": [],
78 | "source": [
79 | "DATA_PATH = !echo $PWD/neu_det\n",
80 | "DATA_PATH = DATA_PATH.n"
81 | ]
82 | },
83 | {
84 | "cell_type": "markdown",
85 | "metadata": {},
86 | "source": [
87 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
88 | ]
89 | },
90 | {
91 | "cell_type": "code",
92 | "execution_count": null,
93 | "metadata": {
94 | "tags": []
95 | },
96 | "outputs": [],
97 | "source": [
98 | "prefix = \"neu-det\"\n",
99 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
100 | "sources = f\"{neu_det_s3}/code/\"\n",
101 | "train_output = f\"{neu_det_s3}/output/\"\n",
102 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
103 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
104 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
105 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
106 | ]
107 | },
108 | {
109 | "cell_type": "markdown",
110 | "metadata": {},
111 | "source": [
112 | "## Visualization\n",
113 | "\n",
114 | "Let examine some datasets that we will use later by providing an `ID`"
115 | ]
116 | },
117 | {
118 | "cell_type": "code",
119 | "execution_count": null,
120 | "metadata": {},
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "\n",
125 | "import numpy as np\n",
126 | "import torch\n",
127 | "from PIL import Image\n",
128 | "from torch.utils.data import DataLoader\n",
129 | "\n",
130 | "try:\n",
131 | " import sagemaker_defect_detection\n",
132 | "except ImportError:\n",
133 | " import sys\n",
134 | " from pathlib import Path\n",
135 | "\n",
136 | " ROOT = Path(\"../src\").resolve()\n",
137 | " sys.path.insert(0, str(ROOT))\n",
138 | "\n",
139 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
140 | "\n",
141 | "SPLIT = \"test\"\n",
142 | "ID = 30\n",
143 | "assert 0 <= ID <= 300\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "Here we resume from a provided pretrained checkpoint `epoch=294-loss=0.654-main_score=0.349.ckpt` that we have copied into our `s3_pretrained`. This takes about **10 minutes** to complete"
185 | ]
186 | },
187 | {
188 | "cell_type": "code",
189 | "execution_count": null,
190 | "metadata": {
191 | "tags": [
192 | "outputPrepend"
193 | ]
194 | },
195 | "outputs": [],
196 | "source": [
197 | "%%time\n",
198 | "import logging\n",
199 | "from os import path as osp\n",
200 | "\n",
201 | "from sagemaker.pytorch import PyTorch\n",
202 | "\n",
203 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
204 | "# Note: resnet34 was used in the pretrained model and it has to match the pretrained model backbone\n",
205 | "# if need resnet50, need to train from scratch\n",
206 | "BACKBONE = \"resnet34\"\n",
207 | "assert BACKBONE in [\n",
208 | " \"resnet34\",\n",
209 | " \"resnet50\",\n",
210 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
211 | "EPOCHS = 5\n",
212 | "LEARNING_RATE = 1e-4\n",
213 | "SEED = 123\n",
214 | "\n",
215 | "hyperparameters = {\n",
216 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
217 | " \"num-classes\": NUM_CLASSES, # number of classes + background\n",
218 | " \"epochs\": EPOCHS, # number of epochs to finetune\n",
219 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
220 | " \"seed\": SEED, # random number generator seed\n",
221 | "}\n",
222 | "\n",
223 | "assert not isinstance(sagemaker_session, sagemaker.LocalSession), \"local session as share memory cannot be altered\"\n",
224 | "\n",
225 | "finetuned_model = PyTorch(\n",
226 | " entry_point=\"detector.py\",\n",
227 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
228 | " role=role,\n",
229 | " train_instance_count=1,\n",
230 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
231 | " hyperparameters=hyperparameters,\n",
232 | " py_version=\"py3\",\n",
233 | " framework_version=\"1.5\",\n",
234 | " sagemaker_session=sagemaker_session,\n",
235 | " output_path=train_output,\n",
236 | " checkpoint_s3_uri=s3_checkpoint,\n",
237 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
238 | " # container_log_level=logging.DEBUG,\n",
239 | ")\n",
240 | "\n",
241 | "finetuned_model.fit(\n",
242 | " {\n",
243 | " \"training\": neu_det_prepared_s3,\n",
244 | " \"pretrained_checkpoint\": osp.join(s3_pretrained, \"epoch=294-loss=0.654-main_score=0.349.ckpt\"),\n",
245 | " }\n",
246 | ")"
247 | ]
248 | },
249 | {
250 | "cell_type": "markdown",
251 | "metadata": {},
252 | "source": [
253 | "Then, we deploy our new model which takes about **10 minutes** to complete"
254 | ]
255 | },
256 | {
257 | "cell_type": "code",
258 | "execution_count": null,
259 | "metadata": {},
260 | "outputs": [],
261 | "source": [
262 | "%%time\n",
263 | "finetuned_detector = finetuned_model.deploy(\n",
264 | " initial_instance_count=1,\n",
265 | " instance_type=\"ml.m5.xlarge\",\n",
266 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-finetuned-endpoint\",\n",
267 | ")"
268 | ]
269 | },
270 | {
271 | "cell_type": "markdown",
272 | "metadata": {},
273 | "source": [
274 | "## Inference\n",
275 | "\n",
276 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
277 | ]
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": null,
282 | "metadata": {},
283 | "outputs": [],
284 | "source": [
285 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
286 | ]
287 | },
288 | {
289 | "cell_type": "markdown",
290 | "metadata": {},
291 | "source": [
292 | "Now the input is ready and we can get some results"
293 | ]
294 | },
295 | {
296 | "cell_type": "code",
297 | "execution_count": null,
298 | "metadata": {},
299 | "outputs": [],
300 | "source": [
301 | "%%time\n",
302 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
303 | "np_load_old = np.load\n",
304 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
305 | "finetuned_predictions = finetuned_detector.predict(input)\n",
306 | "np.load = np_load_old"
307 | ]
308 | },
309 | {
310 | "cell_type": "markdown",
311 | "metadata": {},
312 | "source": [
313 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
314 | ]
315 | },
316 | {
317 | "cell_type": "code",
318 | "execution_count": null,
319 | "metadata": {},
320 | "outputs": [],
321 | "source": [
322 | "import boto3\n",
323 | "import botocore\n",
324 | "\n",
325 | "config = botocore.config.Config(read_timeout=200)\n",
326 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
327 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
328 | "response = runtime.invoke_endpoint(\n",
329 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
330 | ")\n",
331 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
332 | ]
333 | },
334 | {
335 | "cell_type": "markdown",
336 | "metadata": {},
337 | "source": [
338 | "Here comes the slight changes in inference"
339 | ]
340 | },
341 | {
342 | "cell_type": "code",
343 | "execution_count": null,
344 | "metadata": {},
345 | "outputs": [],
346 | "source": [
347 | "visualize(\n",
348 | " original_image_unnorm,\n",
349 | " [original_boxes, demo_predictions[0][\"boxes\"], finetuned_predictions[0][\"boxes\"]],\n",
350 | " [original_labels, demo_predictions[0][\"labels\"], finetuned_predictions[0][\"labels\"]],\n",
351 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
352 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"finetuned\"],\n",
353 | " dpi=250,\n",
354 | ")"
355 | ]
356 | },
357 | {
358 | "cell_type": "markdown",
359 | "metadata": {},
360 | "source": [
361 | "## Optional: Delete the endpoint and model\n",
362 | "\n",
363 | "When you are done with the endpoint, you should clean it up.\n",
364 | "\n",
365 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account.\n"
366 | ]
367 | },
368 | {
369 | "cell_type": "code",
370 | "execution_count": null,
371 | "metadata": {},
372 | "outputs": [],
373 | "source": [
374 | "finetuned_detector.delete_model()\n",
375 | "finetuned_detector.delete_endpoint()"
376 | ]
377 | },
378 | {
379 | "cell_type": "markdown",
380 | "metadata": {},
381 | "source": [
382 | "### [Click here to continue](./2_detection_from_scratch.ipynb)"
383 | ]
384 | }
385 | ],
386 | "metadata": {
387 | "kernelspec": {
388 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
389 | "name": "python361064bitpytorchlatestp36conda2dfac45b320c45f3a4d1e89ca46b60d1"
390 | },
391 | "language_info": {
392 | "codemirror_mode": {
393 | "name": "ipython",
394 | "version": 3
395 | },
396 | "file_extension": ".py",
397 | "mimetype": "text/x-python",
398 | "name": "python",
399 | "nbconvert_exporter": "python",
400 | "pygments_lexer": "ipython3",
401 | "version": "3.6.10-final"
402 | }
403 | },
404 | "nbformat": 4,
405 | "nbformat_minor": 2
406 | }
407 |
--------------------------------------------------------------------------------
/notebooks/2_detection_from_scratch.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "**Jupyter Kernel**:\n",
8 | "\n",
9 | "* If you are in SageMaker Notebook instance, please make sure you are using **conda_pytorch_latest_p36** kernel\n",
10 | "* If you are on SageMaker Studio, please make sure you are using **SageMaker JumpStart PyTorch 1.0** kernel\n",
11 | "\n",
12 | "**Run All**:\n",
13 | "\n",
14 | "* If you are in SageMaker notebook instance, you can go to *Cell tab -> Run All*\n",
15 | "* If you are in SageMaker Studio, you can go to *Run tab -> Run All Cells*\n",
16 | "\n",
17 | "**Note**: To *Run All* successfully, make sure you have executed the entire demo notebook `0_demo.ipynb` first.\n",
18 | "\n",
19 | "## Training our Detector from Scratch\n",
20 | "\n",
21 | "In this notebook, we will see how to train our detector from scratch"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": null,
27 | "metadata": {},
28 | "outputs": [],
29 | "source": [
30 | "import json\n",
31 | "\n",
32 | "import sagemaker\n",
33 | "from sagemaker.s3 import S3Downloader\n",
34 | "\n",
35 | "sagemaker_session = sagemaker.Session()\n",
36 | "sagemaker_config = json.load(open(\"../stack_outputs.json\"))\n",
37 | "role = sagemaker_config[\"IamRole\"]\n",
38 | "solution_bucket = sagemaker_config[\"SolutionS3Bucket\"]\n",
39 | "region = sagemaker_config[\"AWSRegion\"]\n",
40 | "solution_name = sagemaker_config[\"SolutionName\"]\n",
41 | "bucket = sagemaker_config[\"S3Bucket\"]"
42 | ]
43 | },
44 | {
45 | "cell_type": "markdown",
46 | "metadata": {},
47 | "source": [
48 | "First, we download our **NEU-DET** dataset from our public S3 bucket"
49 | ]
50 | },
51 | {
52 | "cell_type": "code",
53 | "execution_count": null,
54 | "metadata": {},
55 | "outputs": [],
56 | "source": [
57 | "original_bucket = f\"s3://{solution_bucket}-{region}/{solution_name}\"\n",
58 | "original_pretained_checkpoint = f\"{original_bucket}/pretrained\"\n",
59 | "original_sources = f\"{original_bucket}/build/lib/source_dir.tar.gz\""
60 | ]
61 | },
62 | {
63 | "cell_type": "markdown",
64 | "metadata": {},
65 | "source": [
66 | "Note that for easiler data processing, we have already executed `prepare_data` once in our `0_demo.ipynb` and have already uploaded the prepared data to our S3 bucket"
67 | ]
68 | },
69 | {
70 | "cell_type": "code",
71 | "execution_count": null,
72 | "metadata": {
73 | "tags": []
74 | },
75 | "outputs": [],
76 | "source": [
77 | "DATA_PATH = !echo $PWD/neu_det\n",
78 | "DATA_PATH = DATA_PATH.n"
79 | ]
80 | },
81 | {
82 | "cell_type": "markdown",
83 | "metadata": {},
84 | "source": [
85 | "After data preparation, we need to setup some paths that will be used throughtout the notebook"
86 | ]
87 | },
88 | {
89 | "cell_type": "code",
90 | "execution_count": null,
91 | "metadata": {
92 | "tags": []
93 | },
94 | "outputs": [],
95 | "source": [
96 | "prefix = \"neu-det\"\n",
97 | "neu_det_s3 = f\"s3://{bucket}/{prefix}\"\n",
98 | "sources = f\"{neu_det_s3}/code/\"\n",
99 | "train_output = f\"{neu_det_s3}/output/\"\n",
100 | "neu_det_prepared_s3 = f\"{neu_det_s3}/data/\"\n",
101 | "s3_checkpoint = f\"{neu_det_s3}/checkpoint/\"\n",
102 | "sm_local_checkpoint_dir = \"/opt/ml/checkpoints/\"\n",
103 | "s3_pretrained = f\"{neu_det_s3}/pretrained/\""
104 | ]
105 | },
106 | {
107 | "cell_type": "markdown",
108 | "metadata": {},
109 | "source": [
110 | "## Visualization\n",
111 | "\n",
112 | "Let examine some datasets that we will use later by providing an `ID`"
113 | ]
114 | },
115 | {
116 | "cell_type": "code",
117 | "execution_count": null,
118 | "metadata": {
119 | "tags": []
120 | },
121 | "outputs": [],
122 | "source": [
123 | "import copy\n",
124 | "from typing import List\n",
125 | "\n",
126 | "import numpy as np\n",
127 | "import torch\n",
128 | "from PIL import Image\n",
129 | "from torch.utils.data import DataLoader\n",
130 | "\n",
131 | "try:\n",
132 | " import sagemaker_defect_detection\n",
133 | "except ImportError:\n",
134 | " import sys\n",
135 | " from pathlib import Path\n",
136 | "\n",
137 | " ROOT = Path(\"../src\").resolve()\n",
138 | " sys.path.insert(0, str(ROOT))\n",
139 | "\n",
140 | "from sagemaker_defect_detection import NEUDET, get_preprocess\n",
141 | "\n",
142 | "SPLIT = \"test\"\n",
143 | "ID = 50\n",
144 | "dataset = NEUDET(DATA_PATH, split=SPLIT, preprocess=get_preprocess())\n",
145 | "images, targets, _ = dataset[ID]\n",
146 | "original_image = copy.deepcopy(images)\n",
147 | "original_boxes = targets[\"boxes\"].numpy().copy()\n",
148 | "original_labels = targets[\"labels\"].numpy().copy()\n",
149 | "print(f\"first images size: {original_image.shape}\")\n",
150 | "print(f\"target bounding boxes: \\n {original_boxes}\")\n",
151 | "print(f\"target labels: {original_labels}\")"
152 | ]
153 | },
154 | {
155 | "cell_type": "markdown",
156 | "metadata": {},
157 | "source": [
158 | "And we can now visualize it using the provided utilities as follows"
159 | ]
160 | },
161 | {
162 | "cell_type": "code",
163 | "execution_count": null,
164 | "metadata": {},
165 | "outputs": [],
166 | "source": [
167 | "from sagemaker_defect_detection.utils.visualize import unnormalize_to_hwc, visualize\n",
168 | "\n",
169 | "original_image_unnorm = unnormalize_to_hwc(original_image)\n",
170 | "\n",
171 | "visualize(\n",
172 | " original_image_unnorm,\n",
173 | " [original_boxes],\n",
174 | " [original_labels],\n",
175 | " colors=[(255, 0, 0)],\n",
176 | " titles=[\"original\", \"ground truth\"],\n",
177 | ")"
178 | ]
179 | },
180 | {
181 | "cell_type": "markdown",
182 | "metadata": {},
183 | "source": [
184 | "In order to get high **Mean Average Percision (mAP)** and **Mean Average Recall (mAR)** for **Intersection Over Union (IOU)** thresholds of 0.5 when training from scratch, it requires more than **300 epochs**. That is why we have provided a pretrained model and recommend finetuning whenever is possible. For demostration, we train the model from scratch for 10 epochs which takes about **16 minutes** and it results in the following mAP, mAR and the accumulated `main_score` of\n",
185 | "\n",
186 | "* `Average Precision (AP) @[ IoU=0.50:0.95 ] ~ 0.048`\n",
187 | "* `Average Recall (AR) @[ IoU=0.50:0.95] ~ 0.153`\n",
188 | "* `main_score=0.0509`\n",
189 | "\n",
190 | "To get higher mAP, mAR and overall `main_score`, you can train for more epochs"
191 | ]
192 | },
193 | {
194 | "cell_type": "code",
195 | "execution_count": null,
196 | "metadata": {
197 | "tags": [
198 | "outputPrepend"
199 | ]
200 | },
201 | "outputs": [],
202 | "source": [
203 | "%%time\n",
204 | "import logging\n",
205 | "from os import path as osp\n",
206 | "\n",
207 | "from sagemaker.pytorch import PyTorch\n",
208 | "\n",
209 | "NUM_CLASSES = 7 # 6 classes + 1 for background\n",
210 | "BACKBONE = \"resnet34\" # has to match the pretrained model backbone\n",
211 | "assert BACKBONE in [\n",
212 | " \"resnet34\",\n",
213 | " \"resnet50\",\n",
214 | "], \"either resnet34 or resnet50. Make sure to be consistent with model_fn in detector.py\"\n",
215 | "EPOCHS = 10\n",
216 | "LEARNING_RATE = 1e-3\n",
217 | "SEED = 123\n",
218 | "\n",
219 | "hyperparameters = {\n",
220 | " \"backbone\": BACKBONE, # the backbone resnet model for feature extraction\n",
221 | " \"num-classes\": NUM_CLASSES, # number of classes 6 + 1 background\n",
222 | " \"epochs\": EPOCHS, # number of epochs to train\n",
223 | " \"learning-rate\": LEARNING_RATE, # learning rate for optimizer\n",
224 | " \"seed\": SEED, # random number generator seed\n",
225 | "}\n",
226 | "\n",
227 | "model = PyTorch(\n",
228 | " entry_point=\"detector.py\",\n",
229 | " source_dir=osp.join(sources, \"source_dir.tar.gz\"),\n",
230 | " role=role,\n",
231 | " train_instance_count=1,\n",
232 | " train_instance_type=\"ml.g4dn.2xlarge\",\n",
233 | " hyperparameters=hyperparameters,\n",
234 | " py_version=\"py3\",\n",
235 | " framework_version=\"1.5\",\n",
236 | " sagemaker_session=sagemaker_session,\n",
237 | " output_path=train_output,\n",
238 | " checkpoint_s3_uri=s3_checkpoint,\n",
239 | " checkpoint_local_path=sm_local_checkpoint_dir,\n",
240 | " # container_log_level=logging.DEBUG,\n",
241 | ")\n",
242 | "\n",
243 | "model.fit(neu_det_prepared_s3)"
244 | ]
245 | },
246 | {
247 | "cell_type": "markdown",
248 | "metadata": {},
249 | "source": [
250 | "Then, we deploy our model which takes about **10 minutes** to complete"
251 | ]
252 | },
253 | {
254 | "cell_type": "code",
255 | "execution_count": null,
256 | "metadata": {
257 | "tags": []
258 | },
259 | "outputs": [],
260 | "source": [
261 | "%%time\n",
262 | "detector = model.deploy(\n",
263 | " initial_instance_count=1,\n",
264 | " instance_type=\"ml.m5.xlarge\",\n",
265 | " endpoint_name=sagemaker_config[\"SolutionPrefix\"] + \"-detector-from-scratch-endpoint\",\n",
266 | ")"
267 | ]
268 | },
269 | {
270 | "cell_type": "markdown",
271 | "metadata": {},
272 | "source": [
273 | "## Inference\n",
274 | "\n",
275 | "We change the input depending on whether we are providing a list of images or a single image. Also the model requires a four dimensional array / tensor (with the first dimension as batch)"
276 | ]
277 | },
278 | {
279 | "cell_type": "code",
280 | "execution_count": null,
281 | "metadata": {},
282 | "outputs": [],
283 | "source": [
284 | "input = list(img.numpy() for img in images) if isinstance(images, list) else images.unsqueeze(0).numpy()"
285 | ]
286 | },
287 | {
288 | "cell_type": "markdown",
289 | "metadata": {},
290 | "source": [
291 | "Now the input is ready and we can get some results"
292 | ]
293 | },
294 | {
295 | "cell_type": "code",
296 | "execution_count": null,
297 | "metadata": {
298 | "tags": []
299 | },
300 | "outputs": [],
301 | "source": [
302 | "%%time\n",
303 | "# SageMaker 1.x doesn't allow_pickle=True by default\n",
304 | "np_load_old = np.load\n",
305 | "np.load = lambda *args, **kwargs: np_load_old(*args, allow_pickle=True, **kwargs)\n",
306 | "predictions = detector.predict(input)\n",
307 | "np.load = np_load_old"
308 | ]
309 | },
310 | {
311 | "cell_type": "markdown",
312 | "metadata": {},
313 | "source": [
314 | "We use our `visualize` utility to check the detection results"
315 | ]
316 | },
317 | {
318 | "cell_type": "code",
319 | "execution_count": null,
320 | "metadata": {},
321 | "outputs": [],
322 | "source": [
323 | "visualize(\n",
324 | " original_image_unnorm,\n",
325 | " [original_boxes, predictions[0][\"boxes\"]],\n",
326 | " [original_labels, predictions[0][\"labels\"]],\n",
327 | " colors=[(255, 0, 0), (0, 0, 255)],\n",
328 | " titles=[\"original\", \"ground truth\", \"trained from scratch\"],\n",
329 | " dpi=200,\n",
330 | ")"
331 | ]
332 | },
333 | {
334 | "cell_type": "markdown",
335 | "metadata": {},
336 | "source": [
337 | "Here we want to compare the results of the new model and the pretrained model that we already deployed in `0_demo.ipynb` visually by calling our endpoint from SageMaker runtime using `boto3`"
338 | ]
339 | },
340 | {
341 | "cell_type": "code",
342 | "execution_count": null,
343 | "metadata": {},
344 | "outputs": [],
345 | "source": [
346 | "import boto3\n",
347 | "import botocore\n",
348 | "\n",
349 | "config = botocore.config.Config(read_timeout=200)\n",
350 | "runtime = boto3.client(\"runtime.sagemaker\", config=config)\n",
351 | "payload = json.dumps(input.tolist() if isinstance(input, np.ndarray) else input)\n",
352 | "response = runtime.invoke_endpoint(\n",
353 | " EndpointName=sagemaker_config[\"SolutionPrefix\"] + \"-demo-endpoint\", ContentType=\"application/json\", Body=payload\n",
354 | ")\n",
355 | "demo_predictions = json.loads(response[\"Body\"].read().decode())"
356 | ]
357 | },
358 | {
359 | "cell_type": "markdown",
360 | "metadata": {},
361 | "source": [
362 | "Finally, we compare the results of the provided pretrained model and our trained from scratch"
363 | ]
364 | },
365 | {
366 | "cell_type": "code",
367 | "execution_count": null,
368 | "metadata": {},
369 | "outputs": [],
370 | "source": [
371 | "visualize(\n",
372 | " original_image_unnorm,\n",
373 | " [original_boxes, demo_predictions[0][\"boxes\"], predictions[0][\"boxes\"]],\n",
374 | " [original_labels, demo_predictions[0][\"labels\"], predictions[0][\"labels\"]],\n",
375 | " colors=[(255, 0, 0), (0, 0, 255), (127, 0, 127)],\n",
376 | " titles=[\"original\", \"ground truth\", \"pretrained\", \"from scratch\"],\n",
377 | " dpi=250,\n",
378 | ")"
379 | ]
380 | },
381 | {
382 | "cell_type": "markdown",
383 | "metadata": {},
384 | "source": [
385 | "## Optional: Delete the endpoint and model\n",
386 | "\n",
387 | "When you are done with the endpoint, you should clean it up.\n",
388 | "\n",
389 | "All of the training jobs, models and endpoints we created can be viewed through the SageMaker console of your AWS account."
390 | ]
391 | },
392 | {
393 | "cell_type": "code",
394 | "execution_count": null,
395 | "metadata": {},
396 | "outputs": [],
397 | "source": [
398 | "detector.delete_model()\n",
399 | "detector.delete_endpoint()"
400 | ]
401 | },
402 | {
403 | "cell_type": "markdown",
404 | "metadata": {},
405 | "source": [
406 | "### [Click here to continue](./3_classification_from_scratch.ipynb)"
407 | ]
408 | }
409 | ],
410 | "metadata": {
411 | "kernelspec": {
412 | "display_name": "Python 3.6.10 64-bit ('pytorch_latest_p36': conda)",
413 | "metadata": {
414 | "interpreter": {
415 | "hash": "4c1e195df8d07db5ee7a78f454b46c3f2e14214bf8c9489d2db5cf8f372ff2ed"
416 | }
417 | },
418 | "name": "python3"
419 | },
420 | "language_info": {
421 | "codemirror_mode": {
422 | "name": "ipython",
423 | "version": 3
424 | },
425 | "file_extension": ".py",
426 | "mimetype": "text/x-python",
427 | "name": "python",
428 | "nbconvert_exporter": "python",
429 | "pygments_lexer": "ipython3",
430 | "version": "3.6.10-final"
431 | }
432 | },
433 | "nbformat": 4,
434 | "nbformat_minor": 2
435 | }
436 |
--------------------------------------------------------------------------------