 12 |
13 |
12 |
13 |
10 |
11 |  12 |
13 |
12 |
13 |
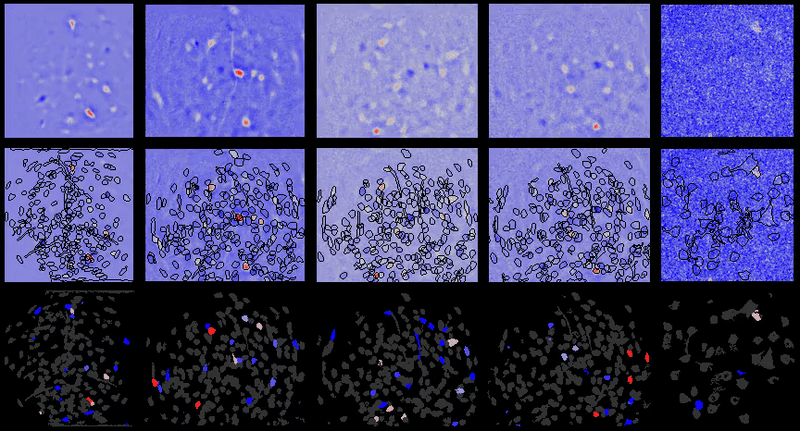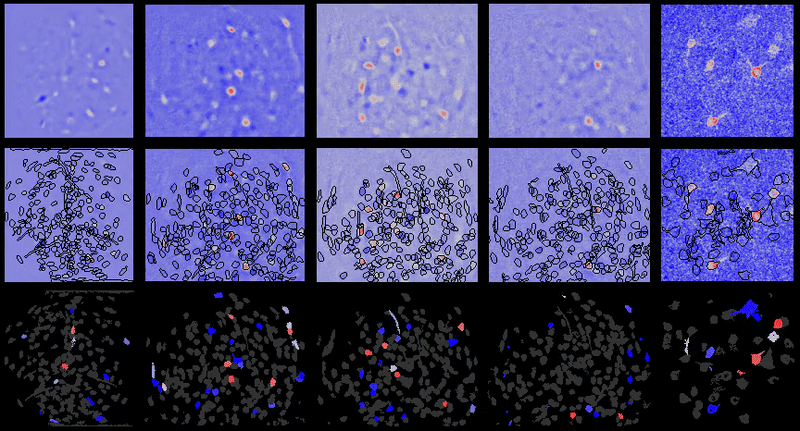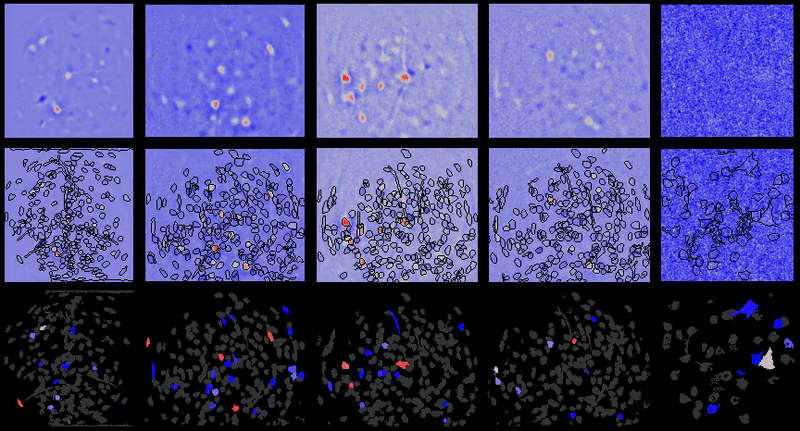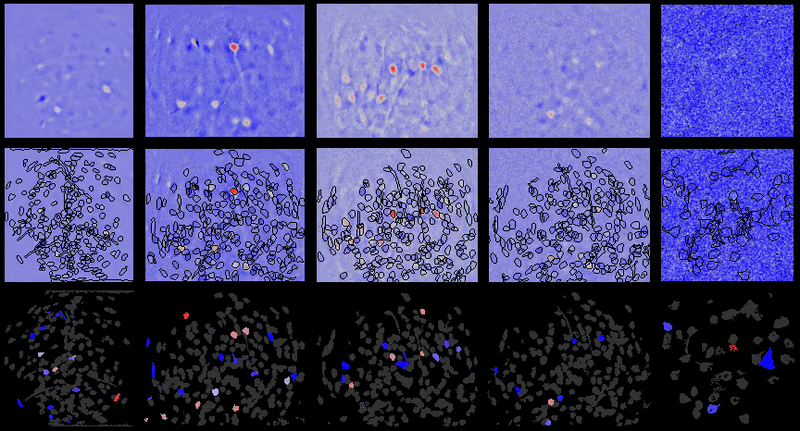
Calcium imaging analysis with CIAtah (https://github.com/bahanonu/ciatah). 14 |
15 | 16 | 17 | The table can also be found at: 18 | - https://bahanonu.com/brain/imaging_tools/ 19 | - https://bahanonu.com/syscarut/articles/233/ 20 | - If you would like to cite this table, see `Cite this repository` in the right `About` section or https://zenodo.org/record/8349533. 21 | 22 | _Notes_: 23 | - I use _cell extraction_ to refer to algorithms that perform cell segmentation and extract neural activity traces. 24 | - In cases where the publication did not explicitly give the algorithm a name, made one based on the underlying method used. 25 | - This table includes algorithms that simultaneously extract cell images/contours and reconstruct cell activity traces along with ones mainly focused on determining one or the other. 26 | - Several calcium imaging related packages have also been included along with algorithms dealing with post-hoc handling of data or cell activity traces. 27 | - Future versions of the repository will include table file (e.g. CSV) and basic LaTeX code so others can import or modify the table more easily going forward. 28 | - Depending on monitor size and browser, scroll horizontally to see right-most table columns (e.g. websites/URLs). 29 | - Any additional papers or algorithms that should be added or suggested updates to the table, leave a comment on the associated blog post or open an issue on the GitHub page, I want to make sure everyone’s brilliant work is acknowledged! 30 | 31 | 32 || # | Method | Year | Analysis pipeline | Notes/Code | Citation | 53 |
| 1 | PhaseCorrelation | 1996 | Motion correction. | • Phase correlation for motion correction, to include translation, rotation, and scale-invariance. | Reddy and Chatterji 1996 | 64 |
| 2 | Turboreg | 1998 | Motion correction. | • Motion correction. • http://bigwww.epfl.ch/thevenaz/turboreg/ | Thevenaz et al. 1998 | 76 |
| 3 | subPixelPhase | 2002 | Motion correction. | • Closed-form solution to subpixel translation estimation using phase correlation. | Foroosh et al. 2002 | 86 |
| 4 | ROI | 2005 | Cell extraction | • Matrix multiplication; in some methods neuropil/background subtraction implemented. | Kerr et al. 2005; Kuchibhotla et al. 2014; Peron et al. 2015 | 100 |
| 5 | CellProfiler | 2006 | Cell segmentation | • Multi-algorithm pipeline for cell segmentation. • https://cellprofiler.org | Carpenter et al. 2006; McQuin et al. 2018; Lamprecht et al. 2007 | 116 |
| 6 | PCA-ICA | 2009 | Cell extraction | • Cell extraction using principal component analysis (PCA) followed by independent component analysis (ICA). | Mukamel et al. 2009 | 126 |
| 7 | ANTs | 2009 | Image analysis | • Suite of tools for registering and analyzing imaging data. • http://stnava.github.io/ANTs/ | Avants et al. 2009 | 138 |
| 8 | elastix | 2009 | Motion correction | • A general toolbox for rigid and non-rigid image registration. • https://elastix.lumc.nl | Klein et al. 2009 | 150 |
| 9 | Lucas–Kanade framework | 2009 | Motion correction | • Lucas-Kanade framework for non-uniform motion image registration. | Greenberg and Kerr 2009 | 160 |
| 10 | CIRF (calcium-behavior) | 2011 | Cell extraction | • Regressive model to obtain Ca2+ signal based on behavior. | Miri et al. 2011 | 171 |
| 11 | openBIS | 2011 | Data handling | • FAIR data management. • https://openbis.ch | Bauch et al. 2011 | 183 |
| 12 | Automated ROI analysis | 2012 | Cell extraction | • Automatic ellipses based ROI detection. | Francis et al. 2012 | 193 |
| 13 | OMERO | 2012 | Data handling | • Microscopy data handling. • https://www.openmicroscopy.org | Allan et al. 2012 | 205 |
| 14 | ADINA | 2013 | Cell extraction | • Sparse dictionary learning. | Diego et al. 2013 | 215 |
| 15 | TPP | 2013 | Analysis pipeline | • Tool for processing two-photon calcium imaging data, e.g. finding cells with SeNeCA. • http://uemweb.biomed.cas.cz/tpp/ | Tomek et al. 2013 | 227 |
| 16 | NMF | 2014 | Cell extraction | • Cell extraction using nonnegative matrix factorization (NMF). Followed by CNMF. | Pnevmatikakis et al. 2014; Maruyama et al. 2014 | 239 |
| 17 | SIMA | 2014 | Analysis pipeline | • Normalized cut segmentation, motion correction, etc. • https://github.com/losonczylab/sima | Kaifosh et al. 2014 | 251 |
| 18 | DataJoint | 2015 | Data handling | • Schema for data handling. • https://github.com/datajoint/datajoint-matlab | Yatsenko et al. 2015 | 263 |
| 19 | NWB | 2015 | Data handling | • Neurodata Without Borders (NWB) initiative to produce a common data format for electrophysiology and imaging studies. • https://github.com/NeurodataWithoutBorders | Teeters et al. 2015 | 275 |
| 20 | Suite2p | 2016 | Cell extraction | • Generative model along with GUIs. | Pachitariu et al. 2016 | 285 |
| 21 | CNMF (CaImAn) | 2016 | Cell extraction | • Constrained NMF (CNMF). • https://github.com/flatironinstitute/CaImAn-MATLAB | Pnevmatikakis et al. 2016 | 297 |
| 22 | CNMF-E | 2016 | Cell extraction | • CNMF + background model to handle one-photon data. • https://github.com/zhoupc/CNMF_E | Zhou et al. 2016, 2018 | 310 |
| 23 | Apthorpe CNN | 2016 | Cell segmentation | • Convolutional neural network (CNN). | Apthorpe et al. 2016 | 320 |
| 24 | moco | 2016 | Motion correction | • Fourier-transform based motion correction. • https://github.com/NTCColumbia/moco | Dubbs et al. 2016 |
| 25 | Cytomine | 2016 | Analysis GUI | • Analysis of large-scale imaging data. • https://cytomine.be | Marée et al. 2016 | 343 |
| 26 | ROI clustering | 2016 | Cell extraction | • Select high-intensity pixels then perform clustering to segment. • https://www.bu.edu/hanlab/files/2016/02/pfgc.zip | Mohammed et al. 2016 | 355 |
| 27 | CELLMax (conference) | 2017 | Cell extraction | • Cell segmentation and activity trace extraction using a maximum likelihood approach. | Ahanonu et al. 2018, 2017; Ahanonu 2018 | 369 |
| 28 | sc-CNMF | 2017 | Cell extraction | • CNMF + GMM/RNN seed cleansing. | Lu et al. 2017 | 379 |
| 29 | OASIS | 2017 | Trace analysis | • Generalized pool adjacent violators algorithm. • https://github.com/zhoupc/OASIS_matlab | Friedrich et al. 2017 | 391 |
| 30 | ABLE | 2017 | Cell segmentation | • Multiple active contours and a cost function to identify cells in 2P data. • https://github.com/StephanieRey/ABLE | Reynolds et al. 2017 | 403 |
| 31 | SCALPEL | 2017 | Cell extraction | • Dictionary learning, dissimilarity, and clustering. • https://cran.r-project.org/web/packages/scalpel/index.html | Petersen et al. 2017 | 415 |
| 32 | HNCcorr | 2017 | Cell segmentation | • Combinatorial optimization (correlation space analysis). • https://github.com/hochbaumGroup/HNCcorr | Spaen et al. 2017 | 427 |
| 33 | OnACID | 2017 | Cell extraction (online) | • NMF variant for online Ca2+ imaging processing. | Giovannucci et al. 2017 | 438 |
| 34 | EXTRACT | 2017 | Cell extraction | • Robust statistical estimation. | Inan et al. 2017 |
| 35 | NETCAL | 2017 | Analysis pipeline | • Calcium imaging analysis GUI. • https://github.com/orlandi/netcal | Orlandi et al. | 458 |
| 36 | NoRMCorre | 2017 | Motion correction. | • Piecewise rigid motion correction. • https://github.com/simonsfoundation/NoRMCorre | Pnevmatikakis and Giovannucci 2017 | 470 |
| 37 | CellReg | 2017 | Cross-session alignment | • Alignment of cells across days using a probabilistic approach. • https://github.com/zivlab/CellReg | Sheintuch et al. 2017 | 482 |
| 38 | NeuroSeg | 2017 | Cell segmentation | • Filtering and seed/clustering based cell segmentation. • https://github.com/baidatong/NeuroSeg | Guan et al. 2018 | 494 |
| 39 | CNMF-E+ | 2017 | Cell extraction | • Shrinkage estimation to improve CNMF-E initialization. | Takekawa et al. 2017 | 504 |
| 40 | Toolbox-Romano | 2017 | Analysis pipeline | • Full analysis pipeline with ROI-based segmentation • https://github.com/zebrain-lab/Toolbox-Romano-et-al | Romano et al. 2017 | 516 |
| 41 | SamuROI | 2017 | Analysis GUI | • GUI for data visualization • https://github.com/samuroi/SamuROI | Rueckl et al. 2017 | 528 |
| 42 | KNIME | 2017 | Analysis pipeline | • Workflow manager for data analysis. • https://www.knime.com | Fillbrunn et al. 2017 | 540 |
| 43 | U-Net2DS | 2017 | Cell segmentation | • Evaluated several deep learning models on Neurofinder, U-Net2DS best. • https://github.com/alexklibisz/deep-calcium | Klibisz et al. 2017 | 552 |
| 44 | CLEAN (conference) | 2018 | Cell sorting | • Machine learning based cell sorting of cell extraction outputs based on image and activity trace features. | Ahanonu et al. 2018; Ahanonu 2018 | 565 |
| 45 | FISSA | 2018 | Trace analysis | • Neuropil decontamination using local region around cell. • https://github.com/rochefort-lab/fissa | Keemink et al. 2018 | 577 |
| 46 | LSSC | 2018 | Cell segmentation | • Spectral clustering; variant to find local subset of eigenvectors. | Mishne et al. 2018 | 587 |
| 47 | PMD - PCA | 2018 | Denoising | • Spatially-localized penalized matrix decomposition for denoising; compression; and improved demixing. • https://github.com/paninski-lab/funimag | Buchanan et al. 2018 | 599 |
| 48 | MIN1PIPE | 2018 | Analysis pipeline | • Pre-processing to enhance neural signals then sc-CNMF for cell extraction. | Lu et al. 2018 | 609 |
| 49 | CaImAn (preprint) | 2018 | Analysis pipeline | • CNMF + several other processing tools. | Giovannucci et al. 2018 | 619 |
| 50 | SEUDO (preprint) | 2018 | Trace analysis | • Mixture of Gaussians + maximum likelihood; post-hoc activity trace correction. | Gauthier et al. 2018 | 629 |
| 51 | ACSAT | 2018 | Cell segmentation | • Global and local adaptive thresholding to identify neurons. • https://github.com/sshen8/acsat | Shen et al. 2018 | 641 |
| 52 | onlineMotionCorrection | 2018 | Motion correction | • Tested multiple algorithms and developed an online motion correction pipeline. • https://github.com/amitani/onlineMotionCorrection | Mitani and Komiyama 2018 | 653 |
| 53 | CIAtah | 2019 | Analysis pipeline | • 1P and 2P Imaging analysis pipeline supporting PCA-ICA, CNMF, CELLMax, EXTRACT, etc. • https://github.com/bahanonu/ciatah | Corder et al. 2019; Ahanonu 2018; Ahanonu and Corder 2022 | 670 |
| 54 | NAOMi (bioRxiv) | 2019 | Simulator | • Generative model for creating simulated calcium imaging movies. | Charles et al. 2019 | 680 |
| 55 | CALIMA | 2019 | Analysis pipeline | • Calcium imaging analysis GUI. | Radstake et al. 2019 | 690 |
| 56 | STNeuroNet | 2019 | Cell segmentation | • Convolutional neural network to detect and segment cells. | Soltanian-Zadeh et al. 2019 | 700 |
| 57 | AQuA | 2019 | Cell extraction | • Astrocyte imaging focused. Non-ROI cluster and propagation based detection of events. | Wang et al. 2019 | 710 |
| 58 | CaImAn | 2019 | Analysis pipeline | • Popular calcium imaging pipeline that includes CNMF + several other processing tools. • https://github.com/flatironinstitute/CaImAn | Giovannucci et al. 2019 | 722 |
| 59 | DL+RWL1-SF | 2019 | Cell extraction | • Dictionary learning and spatial correlation based cell extraction. | Mishne and Charles 2019 | 732 |
| 60 | Segment2P | 2019 | Cell segmentation | • Pre-process images and run through DeepLabV3. • https://github.com/NoahDolev/Segment2P | Dolev et al. 2019 | 744 |
| 61 | LANMC | 2019 | Motion correction | • Long short-term memory non-rigid motion correction, reduce computational cost by predicting non-rigid motion. | Chen et al. 2019 | 754 |
| 62 | marked point processes | 2020 | Cell extraction | • Probabilistic generative model, specifically a marked point process, to extract activity traces. | Shibue and Komaki 2020 | 764 |
| 63 | LocaNMF | 2020 | Region extraction | • Localized semi-nonnegative matrix factorization for extracting active regions. • https://github.com/ikinsella/locaNMF | Saxena et al. 2020 | 776 |
| 64 | EZcalcium | 2020 | Analysis pipeline | • Calcium imaging analysis toolbox. • https://github.com/porteralab/EZcalcium | Cantu et al. 2020 | 788 |
| 65 | OnACID-E + ring CNN | 2020 | Cell extraction (online) | • OnACID for miniscope and new ring CNN background model to improve accuracy. • https://github.com/flatironinstitute/CaImAn | Friedrich et al. 2020 | 800 |
| 66 | Auto CNMF-E sorting | 2020 | Cell sorting | • Machine learning (AutoML) based curation of CNMF-E outputs. • https://github.com/jf-lab/cnmfe-reviewer | Tran et al. 2020a,b | 813 |
| 67 | DeepInterpolation | 2020 | Denoising | • Encoder-decoder architecture with 2D conv. to denoise imaging data. • https://github.com/AllenInstitute/deepinterpolation | Lecoq et al. 2020 | 825 |
| 68 | BIAFLOWS | 2020 | Benchmarking | • Framework for benchmarking imaging analysis workflows. • https://biaflows.neubias.org | Rubens et al. 2020 | 837 |
| 69 | FIBSI | 2020 | Trace analysis | • Extension of Ramer-Douglas-Peucker algorithm to identify baseline that is used for signal detection. • https://github.com/rmcassidy/FIBSI_program | Cassidy et al. 2020; Alles et al. 2021 | 851 |
| 70 | DISCo | 2020 | Cell segmentation | • Pixel correlation and deep learning (CNN) + graph based segmentation. • https://github.com/EKirschbaum/DISCo | Kirschbaum et al. 2020 | 863 |
| 71 | DeepCINAC | 2020 | Trace analysis | • Trace analysis after human labeling followed by CNNs + bidirectional long-short term memory (LSTM) network. • https://gitlab.com/cossartlab/deepcinac | Denis et al. 2020 | 875 |
| 72 | NDSEP | 2020 | Cell extraction | • Dataflow framework for real-time calcium imaging processing. • http://dspcad-www.iacs.umd.edu/bcnm/index.html | Lee et al. 2020 | 887 |
| 73 | DeepBrainSeg | 2020 | Segmentation | • Dual-pathway CNN to learn local and contextual features. | Tan et al. 2020 | 897 |
| 74 | RT-3DMC | 2020 | Motion correction | • Bead or soma tracking for real-time motion correction during 2P imaging. • https://github.com/SilverLabUCL/SilverLab-Microscope | Griffiths et al. 2020 | 909 |
| 75 | Cellpose | 2021 | Cell segmentation | • Neural network and gradient-based cell segmentation. • https://github.com/mouseland/cellpose | Stringer et al. 2021 | 921 |
| 76 | NAOMi | 2021 | Simulator | • Detailed model simulation for benchmarking calcium imaging algorithms. • https://bitbucket.org/adamshch/naomi_sim/src/master/ | Song et al. 2021 | 933 |
| 77 | OnACID-E + ring CNN | 2021 | Cell extraction (online) | • OnACID for 1P data and ring CNN background model. • https://github.com/flatironinstitute/CaImAn | Friedrich et al. 2021 | 945 |
| 78 | EXTRACT | 2021 | Cell extraction | • Robust statistics based cell extraction. • https://github.com/schnitzer-lab/EXTRACT-public | Inan et al. 2021 | 957 |
| 79 | Minian | 2021 | Analysis pipeline | • Imaging analysis pipeline with CNMF for cell extraction, in part using Jupyter notebooks with GUI elements. • https://github.com/DeniseCaiLab/minian | Dong et al. 2021 | 969 |
| 80 | Mesmerize | 2021 | Analysis pipeline | • Imaging analysis platform with CaImAn for cell extraction, import support for other cell extraction algorithms. • https://github.com/kushalkolar/MESmerize | Kolar et al. 2021 | 981 |
| 81 | DeepInterpolation | 2021 | Denoising | • Encoder-decoder architecture with 2D conv. to denoise imaging data. • https://github.com/AllenInstitute/deepinterpolation | Lecoq et al. 2021 | 993 |
| 82 | BEAR | 2021 | Cell extraction | • Neural network approximation of PCA for cell extraction. • https://github.com/NICALab/BEAR | Han et al. 2021 | 1005 |
| 83 | CaPTure | 2021 | Cell extraction | • ROI segmentation and activity extraction. • https://github.com/LieberInstitute/CaPTure | Tippani et al. 2021 | 1017 |
| 84 | CASCADE | 2021 | Trace analysis | • Spike inference based on dual ephys/calcium imaging recordings. • https://github.com/HelmchenLabSoftware/Cascade | Rupprecht et al. 2021 | 1029 |
| 85 | VolPy | 2021 | Analysis pipeline | • Voltage imaging analysis pipeline integrated into CaImAn. • https://github.com/flatironinstitute/CaImAn | Cai et al. 2021 | 1041 |
| 86 | DeepCAD | 2021 | Denoising | • Deep neural network based denoising. • https://github.com/cabooster/DeepCAD-RT | Li et al. 2021 | 1053 |
| 87 | SpecSeg | 2021 | Cell extraction | • Spectral density of pixels to identify ROIs. Also incorporates motion correction and cross-session matching. • https://github.com/Leveltlab/SpectralSegmentation | de Kraker et al. 2021 | 1065 |
| 88 | FIOLA | 2021 | Cell extraction (online) | • GPU- and computational graph-based speed-ups along with non-negative least squares for post-initialization signal extraction. • https://github.com/nel-lab/FIOLA | Giovannucci et al. 2021 | 1077 |
| 89 | PatchWarp | 2021 | Motion correction | • Affine transformation of subfields followed by stitching subfields together. • https://github.com/ryhattori/PatchWarp | Hattori and Komiyama 2021 | 1089 |
| 90 | MVG-CNN | 2021 | Region extraction | • Automated sleep states classification using multiplex visibility graphs and deep learning. Data URL. • https://github.com/comp-imaging-sci/MVG-CNN | Zhang et al. 2021 | 1102 |
| 91 | Flow-Registration | 2021 | Motion correction | • Variational optical flow for non-uniform motion correction • https://github.com/phflot/flow_registration | Flotho et al. 2022 | 1114 |
| 92 | SUNS | 2021 | Cell segmentation | • Cell segmentation using shallow U-Nets. • https://github.com/YijunBao/Shallow-UNet-Neuron-Segmentation_SUNS | Bao et al. 2021 | 1126 |
| 93 | Carignan | 2021 | Cell extraction | • Online cell extraction and triggering based on OnACID and CaImAn. • https://github.com/tzklab/carignan | Taniguchi et al. 2021 | 1138 |
| 94 | MullenClassifier | 2021 | Cell sorting | • Feature extraction from cell images and tracs followed by supervised learning classifier. | Mullen et al. 2021 | 1148 |
| 95 | timeUnet | 2021 | Denoising | • Deep learning for denoising with temporal information added in. • https://github.com/BoHuangLab/Transfer-Learning-Denoising/ | Wang et al. 2021 | 1160 |
| 96 | EMC2 | 2021 | Motion correction | • Wavelet decomposition to detect bright spots followed by motion correction with multiple hypothesis tracking and computing elastic deformation. • https://icy.bioimageanalysis.org/plugin/elastic-motion-correction-concatenation-emc2-of-tracks/ | Lagache et al. 2021 | 1173 |
| 97 | GraFT | 2022 | Cell extraction | • Dictionary-based learning of activity traces followed by graph-based segmentation. • https://github.com/adamshch/GraFT-analysis | Charles et al. 2022 | 1185 |
| 98 | CaPTure | 2022 | Analysis pipeline | • Binary/watershed segmentation followed by ROI-based mean traces. • https://github.com/LieberInstitute/CaPTure | Tippani et al. 2022 | 1197 |
| 99 | SpecSeg | 2022 | Cell segmentation | • Cross spectral power-based segmentation of neurons and neurites. • https://github.com/Leveltlab/SpectralSegmentation | de Kraker et al. 2022 | 1209 |
| 100 | CITE-On | 2022 | Cell extraction | • Online cell detection and trace extraction using CNNs. • https://gitlab.iit.it/fellin-public/cite-on | Sità et al. 2022 | 1221 |
| 101 | DL-assisted 2P fiberscope | 2022 | Denoising | • Denoising 2P fiberscope data using deep neural network (conditional generative adversarial network). • https://figshare.com/articles/dataset/Data/19193792 | Guan et al. 2022 | 1233 |
| 102 | 4SM | 2022 | Cell extraction | • Generative adversarial network for image segmentation. • https://github.com/SharifAmit/4SM | Kamran et al. 2022 | 1245 |
| 103 | DeepCAD-RT | 2022 | Denoising | • Improved version of DeepCAD for real time performance. • https://github.com/cabooster/DeepCAD-RT/ | Li et al. 2023a | 1257 |
| 104 | SEUDO | 2022 | Trace analysis | • Mixture of Gaussians + maximum likelihood; post-hoc activity trace correction. • https://github.com/adamshch/SEUDO | Gauthier et al. 2022 | 1269 |
| 105 | AxialMotionCorrect | 2022 | Motion correction | • Axial motion correction via multi-plane scanning plus maximum likelihood optimization. • https://gitlab.com/anflores/axial_motion_correction | Flores-Valle and Seelig 2022 | 1281 |
| 106 | FIFER | 2022 | Motion correction | • Feature-based motion correction, finding features using a density-based estimating and clustering algorithm and matching features with a similarity metric for registration. • https://github.com/Weiyi-Liu-Unique/FIFER | Liu et al. 2022 | 1293 |
| 107 | NWB | 2022 | Data handling | • Neurodata Without Borders (NWB) to standardize ephys and imaging data across tools. • https://github.com/NeurodataWithoutBorders | Rübel et al. 2022 | 1305 |
| 108 | DeCalciOn | 2023 | Online analysis pipeline | • Integrate hardware and software to online decode calcium signals. • https://github.com/zhe-ch/ACTEV | Chen et al. 2023 | 1317 |
| 109 | jGCaMP8 | 2023 | Calcium indicator | • Improved calcium indicators with increased sensitivity and reduced background. | Zhang et al. 2023a | 1327 |
| 110 | NeuroSeg-II | 2023 | Cell segmentation | • 2P cell segmentation using region-based convolutional neural network with modifications. • https://github.com/XZH-James/NeuroSeg2 | Xu et al. 2023 | 1339 |
| 111 | CaliAli | 2023 | Cross-session alignment | • Cross-session alignment using vasculature information. • https://github.com/CaliAli-PV/CaliAli | Vergara et al. 2023 | 1351 |
| 112 | DeepWonder | 2023 | Cell extraction | • Deep-learning-based cell finding for widefield datasets. • https://github.com/yuanlong-o/Deep_widefield_cal_inferece | Zhang et al. 2023b | 1363 |
| 113 | ASTRA | 2023 | Cell segmentation | • Deep neural network for astrocyte segmentation. • https://gitlab.iit.it/fellin-public/astra | Bonato et al. 2023 | 1375 |
| 114 | SRDTrans | 2023 | Denoise | • Spatial redundancy for training followed by spatiotemporal transformer architecture to reduce CNN bias/issues. • https://github.com/cabooster/SRDTrans | Li et al. 2023b | 1387 |
| 115 | REALS | 2023 | Motion correction | • Motion correction via simultaneous transformation and low rank and sparse decomposition with gradient-based updates. • https://openaccess.thecvf.com/content/WACV2023/supplemental/Cho_Robust_and_Efficient_WACV_2023_supplemental.zip | Cho et al. 2023 | 1399 |
| 116 | LD-MCM | 2023 | Motion correction | • Motion correction using deep learning feature identification and control point registration. • https://github.com/bahanonu/ciatah | Ahanonu et al. 2023 | 1412 |
1439 | B Srinivasa Reddy and Biswanath N Chatterji. An fft-based technique 1441 | for translation, rotation, and scale-invariant image registration. IEEE 1443 | transactions on image processing, 5(8):1266–1271, 1996. 1445 |
1446 |1447 | Philippe Thevenaz, Urs E Ruttimann, and Michael Unser. A pyramid 1449 | approach to subpixel registration based on intensity. Image Processing, 1451 | IEEE Transactions on, 7(1):27–41, 1998. 1453 |
1454 |1455 | Hassan Foroosh, Josiane B Zerubia, and Marc Berthod. Extension of 1457 | phase correlation to subpixel registration. IEEE transactions on image 1459 | processing, 11(3):188–200, 2002. 1461 |
1462 |1463 | J N Kerr, D Greenberg, and F Helmchen. Imaging input and output 1465 | of neocortical networks in vivo. Proc Natl Acad Sci U S A, 102(39): 1467 | 14063–14068, 2005. ISSN 0027-8424 (Print) 0027-8424. doi: 10.1073/pnas. 1468 | 0506029102. 1469 |
1470 |1471 | K V Kuchibhotla, S Wegmann, K J 1473 | Kopeikina, J Hawkes, N Rudinskiy, M L Andermann, T L Spires-Jones, 1474 | B J Bacskai, and B T Hyman. Neurofibrillary tangle-bearing neurons are 1475 | functionally integrated in cortical circuits in vivo. Proc Natl Acad Sci U S 1477 | A, 111(1):510–514, 2014. ISSN 0027-8424. doi: 10.1073/pnas.1318807111. 1479 | 1480 | 1481 | 1482 |
1483 |1484 | Simon P. Peron, Jeremy Freeman, Vijay Iyer, Caiying Guo, and Karel 1486 | Svoboda. A Cellular Resolution 1487 | Map of Barrel Cortex Activity during Tactile Behavior. Neuron, 86(3): 1489 | 783–799, 2015. ISSN 10974199. doi: 10.1016/j.neuron.2015.03.027. URL 1490 | http://dx.doi.org/10.1016/j.neuron.2015.03.027. 1493 |
1494 |1495 | Anne E Carpenter, Thouis R Jones, Michael R Lamprecht, Colin 1497 | Clarke, In Han Kang, Ola Friman, David A Guertin, Joo Han Chang, 1498 | Robert A Lindquist, Jason Moffat, et al. Cellprofiler: image analysis 1499 | software for identifying and quantifying cell phenotypes. Genome biology, 1501 | 7(10):1–11, 2006. 1502 |
1503 |1504 | Claire McQuin, Allen Goodman, Vasiliy Chernyshev, Lee Kamentsky, 1506 | Beth A Cimini, Kyle W Karhohs, Minh Doan, Liya Ding, Susanne M 1507 | Rafelski, Derek Thirstrup, et al. Cellprofiler 3.0: Next-generation image 1508 | processing for biology. PLoS biology, 16(7):e2005970, 2018. 1510 |
1511 |1512 | Michael R Lamprecht, David M Sabatini, and Anne E Carpenter. 1514 | Cellprofiler™: free, versatile software for automated biological image 1515 | analysis. Biotechniques, 42(1):71–75, 2007. 1517 |
1518 |1519 | Eran A Mukamel, Axel Nimmerjahn, and Mark J Schnitzer. Automated 1521 | analysis of cellular signals from large-scale calcium imaging data. Neuron, 1523 | 63(6):747–760, 2009. 1524 |
1525 |1526 | Brian B Avants, Nick Tustison, Gang Song, et al. Advanced 1528 | normalization tools (ants). Insight j, 2(365):1–35, 2009. 1530 |
1531 |1532 | Stefan Klein, Marius Staring, Keelin Murphy, Max A Viergever, and 1534 | 1535 | 1536 | 1537 | Josien PW Pluim. Elastix: a toolbox for intensity-based medical image 1538 | registration. IEEE transactions on medical imaging, 29(1):196–205, 2009. 1540 |
1541 |1542 | David S Greenberg and Jason ND Kerr. Automated correction of fast 1544 | motion artifacts for two-photon imaging of awake animals. Journal of 1546 | neuroscience methods, 176(1):1–15, 2009. 1548 |
1549 |1550 | A Miri, K Daie, 1552 | R D Burdine, E Aksay, and D W Tank. Regression-based identification 1553 | of behavior-encoding neurons during large-scale optical imaging of neural 1554 | activity at cellular resolution. J Neurophysiol, 105(2):964–980, 2011. ISSN 1556 | 1522-1598 (Electronic) 0022-3077 (Linking). doi: 10.1152/jn.00702.2010. 1557 | URL http://www.ncbi.nlm.nih.gov/pubmed/21084686. 1560 |
1561 |1562 | Angela Bauch, Izabela Adamczyk, Piotr Buczek, Franz-Josef Elmer, 1564 | Kaloyan Enimanev, Pawel Glyzewski, Manuel Kohler, Tomasz Pylak, 1565 | Andreas Quandt, Chandrasekhar Ramakrishnan, et al. openbis: a flexible 1566 | framework for managing and analyzing complex data in biology research. 1567 | BMC bioinformatics, 12(1):1–19, 2011. 1569 |
1570 |1571 | M Francis, X Qian, C Charbel, J Ledoux, J C Parker, and M S Taylor. 1573 | Automated region of interest analysis of dynamic Ca(2)+ signals in image 1574 | sequences. Am J Physiol Cell Physiol, 303(3):C236–43, 2012. ISSN 1576 | 0363-6143. doi: 10.1152/ajpcell.00016.2012. 1577 |
1578 |1579 | Chris Allan, Jean-Marie Burel, Josh Moore, Colin Blackburn, Melissa 1581 | Linkert, Scott Loynton, Donald MacDonald, William J Moore, Carlos 1582 | Neves, Andrew Patterson, et al. Omero: flexible, model-driven data 1583 | management for experimental biology. Nature methods, 9(3):245–253, 2012. 1585 |
1586 |1587 | Ferran Diego, Susanne Reichinnek, Martin Both, and Fred A Hamprecht. 1589 | 1590 | 1591 | 1592 | Automated identification of neuronal activity from calcium imaging by 1593 | sparse dictionary learning. In Biomedical Imaging (ISBI), 2013 IEEE 10th 1595 | International Symposium on, pages 1058–1061. IEEE, 2013. 1597 |
1598 |1599 | Jakub Tomek, Ondrej Novak, and Josef Syka. Two-photon processor and 1601 | seneca: a freely available software package to process data from two-photon 1602 | calcium imaging at speeds down to several milliseconds per frame. Journal 1604 | of neurophysiology, 110(1):243–256, 2013. 1606 |
1607 |1608 | Eftychios A Pnevmatikakis, Yuanjun Gao, Daniel Soudry, David Pfau, 1610 | Clay Lacefield, Kira Poskanzer, Randy Bruno, Rafael Yuste, and Liam 1611 | Paninski. A structured matrix factorization framework for large scale 1612 | calcium imaging data analysis. arXiv preprint arXiv:1409.2903, 2014. 1614 |
1615 |1616 | Ryuichi Maruyama, Kazuma Maeda, Hajime Moroda, Ichiro Kato, 1618 | Masashi Inoue, Hiroyoshi Miyakawa, and Toru Aonishi. Detecting cells 1619 | using non-negative matrix factorization on calcium imaging data. Neural 1621 | Netw, 55:11–19, mar 2014. ISSN 0893-6080. doi: 10.1016/j.neunet.2014. 1623 | 03.007. URL http://www.ncbi.nlm.nih.gov/pubmed/24705544. 1626 |
1627 |1628 | Patrick Kaifosh, Jeffrey D Zaremba, Nathan B Danielson, and Attila 1630 | Losonczy. SIMA: Python software for analysis of dynamic fluorescence 1631 | imaging data. Frontiers in neuroinformatics, 8:80, 2014. 1633 |
1634 |1635 | Dimitri Yatsenko, Jacob Reimer, Alexander S Ecker, Edgar Y Walker, 1637 | Fabian Sinz, Philipp Berens, Andreas Hoenselaar, R James Cotton, 1638 | Athanassios S Siapas, and Andreas S Tolias. Datajoint: managing big 1639 | scientific data using matlab or python. BioRxiv, page 031658, 2015. 1641 |
1642 |1643 | Jeffery L Teeters, Keith Godfrey, Rob Young, Chinh Dang, Claudia 1645 | Friedsam, Barry Wark, Hiroki Asari, Simon Peron, Nuo Li, and Adrien 1646 | 1647 | 1648 | 1649 | Peyrache. Neurodata without borders: creating a common data format for 1650 | neurophysiology. Neuron, 88(4):629–634, 2015. 1652 |
1653 |1654 | Marius Pachitariu, Carsen Stringer, Sylvia Schröder, Mario Dipoppa, 1656 | L Federico Rossi, Matteo Carandini, and Kenneth D Harris. Suite2p: 1657 | beyond 10,000 neurons with standard two-photon microscopy. Biorxiv, 1659 | page 061507, 2016. 1660 |
1661 |1662 | Eftychios A Pnevmatikakis, Daniel Soudry, Yuanjun Gao, Timothy A 1664 | Machado, Josh Merel, David Pfau, Thomas Reardon, Yu Mu, Clay 1665 | Lacefield, Weijian Yang, et al. Simultaneous denoising, deconvolution, and 1666 | demixing of calcium imaging data. Neuron, 89(2):285–299, 2016. 1668 |
1669 |1670 | P Zhou, SL Resendez, GD Stuber, RE Kass, and L Paninski. Efficient 1672 | and accurate extraction of in vivo calcium signals from microendoscope 1673 | video data. arXiv preprint arXiv:1605.07266, 2016. 1675 |
1676 |1677 | Pengcheng Zhou, Shanna L Resendez, Jose Rodriguez-Romaguera, 1679 | Jessica C Jimenez, Shay Q Neufeld, Andrea Giovannucci, Johannes 1680 | Friedrich, Eftychios A Pnevmatikakis, Garret D Stuber, Rene Hen, 1681 | et al. Efficient and accurate extraction of in vivo calcium signals from 1682 | microendoscopic video data. ELife, 7:e28728, 2018. 1684 |
1685 |1686 | Noah Apthorpe, Alexander Riordan, Robert Aguilar, Jan Homann, 1688 | Yi Gu, David Tank, and H Sebastian Seung. Automatic neuron detection 1689 | in calcium imaging data using convolutional networks. In Advances in 1691 | Neural Information Processing Systems, pages 3270–3278, 2016. 1693 |
1694 |1695 | Alexander Dubbs, James Guevara, and Rafael Yuste. moco: Fast motion 1697 | correction for calcium imaging. Frontiers in neuroinformatics, 10:6, 2016. 1699 | 1700 | 1701 | 1702 |
1703 |1704 | Raphaël Marée, Loïc Rollus, Benjamin Stévens, Renaud Hoyoux, 1706 | Gilles Louppe, Rémy Vandaele, Jean-Michel Begon, Philipp Kainz, Pierre 1707 | Geurts, and Louis Wehenkel. Collaborative analysis of multi-gigapixel 1708 | imaging data using cytomine. Bioinformatics, 32(9):1395–1401, 2016. 1710 |
1711 |1712 | Ali I Mohammed, Howard J Gritton, Hua-an Tseng, Mark E Bucklin, 1714 | Zhaojie Yao, and Xue Han. An integrative approach for analyzing hundreds 1715 | of neurons in task performing mice using wide-field calcium imaging. 1716 | Scientific reports, 6(1):20986, 2016. 1718 |
1719 |1720 | B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin, E. O. Hamel, 1722 | J. Lecoq, D. E. Aldarondo, and M. J. Schnitzer. Maximum likelihood and 1723 | machine learning based methods for automated cell sorting of large-scale 1724 | neural calcium imaging data. Society for Neuroscience, 2018. URL 1725 | https://abstractsonline.com/pp8/#!/4649/presentation/41917. 1728 |
1729 |1730 | B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin, 1732 | E. O. Hamel, J. Lecoq, and M. J. Schnitzer. Maximum 1733 | likelihood based cell sorting of large-scale neural calcium 1734 | imaging data. Society for Neuroscience, 2017. URL 1735 | http://www.abstractsonline.com/pp8/index.html#!/4376/presentation/18520. 1738 |
1739 |1740 | Biafra Owowonta Ahanonu. Neural Ensemble Dynamics in Behaving 1743 | Animals: Computational Approaches and Applications in Amygdala and 1745 | Striatum. Stanford University, 2018. 1747 |
1748 |1749 | Jinghao Lu, Chunyuan Li, and Fan Wang. Seeds cleansing cnmf for 1751 | spatiotemporal neural signals extraction of miniscope imaging data. arXiv 1753 | preprint arXiv:1704.00793, 2017. 1755 |
1756 | 1757 | 1758 | 1759 |1760 | Johannes Friedrich, Pengcheng Zhou, and Liam Paninski. Fast online 1762 | deconvolution of calcium imaging data. PLoS computational biology, 13 1764 | (3):e1005423, 2017. 1765 |
1766 |1767 | Stephanie Reynolds, Therese Abrahamsson, Renaud Schuck, P Jesper 1769 | Sjöström, Simon R Schultz, and Pier Luigi Dragotti. Able: An 1770 | activity-based level set segmentation algorithm for two-photon calcium 1771 | imaging data. eNeuro, pages ENEURO–0012, 2017. 1773 |
1774 |1775 | Ashley Petersen, Noah Simon, and Daniela Witten. SCALPEL: 1777 | Extracting Neurons from Calcium Imaging Data. ArXiv e-prints, art. 1779 | arXiv:1703.06946, March 2017. 1780 |
1781 |1782 | Quico Spaen, Dorit S Hochbaum, and Roberto Asín-Achá. Hnccorr: 1784 | A novel combinatorial approach for cell identification in calcium-imaging 1785 | movies. arXiv preprint arXiv:1703.01999, 2017. 1787 |
1788 |1789 | Andrea Giovannucci, Johannes Friedrich, Matt Kaufman, Anne 1791 | Churchland, Dmitri Chklovskii, Liam Paninski, and Eftychios A 1792 | Pnevmatikakis. Onacid: Online analysis of calcium imaging data in real 1793 | time. In Advances in Neural Information Processing Systems, pages 1795 | 2381–2391, 2017. 1796 |
1797 |1798 | Hakan Inan, Murat A Erdogdu, and Mark Schnitzer. Robust estimation 1800 | of neural signals in calcium imaging. In Advances in Neural Information 1802 | Processing Systems, pages 2901–2910, 2017. 1804 |
1805 |1806 | JG Orlandi, 1808 | S Fernández-García, A Comella-Bolla, M Masana, G García-Díaz 1809 | Barriga, M Yaghoubi, A Kipp, JM Canals, MA Colicos, J Davidsen, 1810 | 1811 | 1812 | 1813 | et al. Netcal: An interactive platform for large-scale, network and 1814 | population dynamics analysis of calcium imaging recordings, zenodo 1815 | (2017). 1816 |
1817 |1818 | Eftychios A Pnevmatikakis and Andrea Giovannucci. Normcorre: An 1820 | online algorithm for piecewise rigid motion correction of calcium imaging 1821 | data. Journal of neuroscience methods, 291:83–94, 2017. 1823 |
1824 |1825 | Liron Sheintuch, Alon Rubin, Noa Brande-Eilat, Nitzan Geva, Noa Sadeh, 1827 | Or Pinchasof, and Yaniv Ziv. Tracking the same neurons across multiple 1828 | days in ca2+ imaging data. Cell reports, 21(4):1102–1115, 2017. 1830 |
1831 |1832 | Jiangheng Guan, Jingcheng Li, Shanshan Liang, Ruijie Li, Xingyi Li, 1834 | Xiaozhe Shi, Ciyu Huang, Jianxiong Zhang, Junxia Pan, Hongbo Jia, 1835 | et al. Neuroseg: automated cell detection and segmentation for in vivo 1836 | two-photon ca 2+ imaging data. Brain Structure and Function, 223(1): 1838 | 519–533, 2018. 1839 |
1840 |1841 | Takashi Takekawa, Hirotaka Asai, Noriaki Ohkawa, Masanori Nomoto, 1843 | Reiko Okubo-Suzuki, Khaled Ghandour, Masaaki Sato, Yasunori Hayashi, 1844 | Kaoru Inokuchi, and Tomoki Fukai. Automatic sorting system for large 1845 | calcium imaging data. bioRxiv, page 215145, 2017. 1847 |
1848 |1849 | Sebastián A Romano, Verónica Pérez-Schuster, Adrien Jouary, 1851 | Jonathan Boulanger-Weill, Alessia Candeo, Thomas Pietri, and Germán 1852 | Sumbre. An integrated calcium imaging processing toolbox for the analysis 1853 | of neuronal population dynamics. PLoS computational biology, 13(6): 1855 | e1005526, 2017. 1856 |
1857 |1858 | Martin Rueckl, Stephen C Lenzi, Laura Moreno-Velasquez, Daniel 1860 | Parthier, Dietmar Schmitz, Sten Ruediger, and Friedrich W Johenning. 1861 | 1862 | 1863 | 1864 | Samuroi, a python-based software tool for visualization and analysis of 1865 | dynamic time series imaging at multiple spatial scales. Frontiers in 1867 | neuroinformatics, 11:44, 2017. 1869 |
1870 |1871 | Alexander Fillbrunn, Christian Dietz, Julianus Pfeuffer, René Rahn, 1873 | Gregory A Landrum, and Michael R Berthold. Knime for reproducible 1874 | cross-domain analysis of life science data. Journal of biotechnology, 261: 1876 | 149–156, 2017. 1877 |
1878 |1879 | Aleksander Klibisz, Derek Rose, Matthew Eicholtz, Jay Blundon, and 1881 | Stanislav Zakharenko. Fast, simple calcium imaging segmentation with 1882 | fully convolutional networks. In International Workshop on Deep Learning 1884 | in Medical Image Analysis, pages 285–293. Springer, 2017. 1886 |
1887 |1888 | Sander W Keemink, Scott C Lowe, Janelle MP Pakan, Evelyn Dylda, 1890 | Mark CW Van Rossum, and Nathalie L Rochefort. Fissa: A neuropil 1891 | decontamination toolbox for calcium imaging signals. Scientific reports, 8 1893 | (1):1–12, 2018. 1894 |
1895 |1896 | Gal Mishne, Ronald R Coifman, Maria Lavzin, and Jackie Schiller. 1898 | Automated cellular structure extraction in biological images with 1899 | applications to calcium imaging data. bioRxiv, page 313981, 2018. 1901 |
1902 |1903 | E Kelly Buchanan, Ian Kinsella, Ding Zhou, Rong Zhu, Pengcheng Zhou, 1905 | Felipe Gerhard, John Ferrante, Ying Ma, Sharon Kim, Mohammed Shaik, 1906 | et al. Penalized matrix decomposition for denoising, compression, and 1907 | improved demixing of functional imaging data. bioRxiv, page 334706, 2018. 1909 |
1910 |1911 | Jinghao Lu, Chunyuan Li, Jonnathan Singh-Alvarado, Zhe Charles 1913 | Zhou, Flavio Fröhlich, Richard Mooney, and Fan Wang. MIN1PIPE: A 1914 | 1915 | 1916 | 1917 | Miniscope 1-Photon-Based Calcium Imaging Signal Extraction Pipeline. 1918 | Cell Reports, 23(12):3673–3684, 2018. ISSN 2211-1247. 1920 |
1921 |1922 | Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon, 1924 | Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi, Jeffrey L Gauthier, 1925 | Pengcheng Zhou, and David W Tank. CaImAn: An open source tool for 1926 | scalable Calcium Imaging data Analysis. bioRxiv, page 339564, 2018. 1928 |
1929 |1930 | Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank, 1932 | Jonathan W Pillow, and Adam S Charles. Detecting and correcting false 1933 | transients in calcium imaging. bioRxiv, page 473470, 2018. 1935 |
1936 |1937 | Simon P Shen, Hua-an Tseng, Kyle R Hansen, Ruofan Wu, Howard J 1939 | Gritton, Jennie Si, and Xue Han. Automatic cell segmentation by adaptive 1940 | thresholding (acsat) for large-scale calcium imaging datasets. eneuro, 5(5), 1942 | 2018. 1943 |
1944 |1945 | Akinori Mitani and Takaki Komiyama. Real-time processing of 1947 | two-photon calcium imaging data including lateral motion artifact 1948 | correction. Frontiers in neuroinformatics, 12:98, 2018. 1950 |
1951 |1952 | Gregory Corder, Biafra Ahanonu, Benjamin F Grewe, Dong Wang, 1954 | Mark J Schnitzer, and Grégory Scherrer. An amygdalar neural ensemble 1955 | that encodes the unpleasantness of pain. Science, 363(6424):276–281, 2019. 1957 |
1958 |1959 | Biafra Ahanonu and Gregory Corder. Recording pain-related brain 1961 | activity in behaving animals using calcium imaging calcium imaging and 1962 | miniature microscopes. In Contemporary Approaches to the Study of Pain: 1964 | From Molecules to Neural Networks, pages 217–276. Springer, 2022. 1966 |
1967 | 1968 | 1969 | 1970 |1971 | Adam S Charles, Alex Song, Jeffrey L Gauthier, Jonathan W Pillow, 1973 | and David W Tank. Neural anatomy and optical microscopy (naomi) 1974 | simulation for evaluating calcium imaging methods. bioRxiv, page 726174, 1976 | 2019. 1977 |
1978 |1979 | FDW Radstake, EAL Raaijmakers, R Luttge, Svitlana Zinger, and 1981 | Jean-Philippe Frimat. Calima: The semi-automated open-source calcium 1982 | imaging analyzer. Computer methods and programs in biomedicine, 179: 1984 | 104991, 2019. 1985 |
1986 |1987 | Somayyeh Soltanian-Zadeh, Kaan Sahingur, Sarah Blau, Yiyang Gong, 1989 | and Sina Farsiu. Fast and robust active neuron segmentation in two-photon 1990 | calcium imaging using spatiotemporal deep learning. Proceedings of the 1992 | National Academy of Sciences, 116(17):8554–8563, 2019. 1994 |
1995 |1996 | Yizhi Wang, Nicole V DelRosso, Trisha V Vaidyanathan, Michelle K 1998 | Cahill, Michael E Reitman, Silvia Pittolo, Xuelong Mi, Guoqiang Yu, 1999 | and Kira E Poskanzer. Accurate quantification of astrocyte and 2000 | neurotransmitter fluorescence dynamics for single-cell and population-level 2001 | physiology. Nature Neuroscience, 22(11):1936–1944, 2019. 2003 |
2004 |2005 | Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon, 2007 | Brandon L Brown, Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi, 2008 | Jeffrey L Gauthier, Pengcheng Zhou, et al. Caiman an open source tool 2009 | for scalable calcium imaging data analysis. Elife, 8:e38173, 2019. 2011 |
2012 |2013 | Gal Mishne and Adam S Charles. Learning spatially-correlated 2015 | temporal dictionaries for calcium imaging. In ICASSP 2019-2019 IEEE 2017 | International Conference on Acoustics, Speech and Signal Processing 2019 | (ICASSP), pages 1065–1069. IEEE, 2019. 2021 |
2022 | 2023 | 2024 | 2025 |2026 | Noah Dolev, Lior Pinkus, and Michal Rivlin-Etzion. Segment2p: 2028 | Parameter-free automated segmentation of cellular fluorescent signals. 2029 | BioRxiv, page 832188, 2019. 2031 |
2032 |2033 | Zhe Chen, Hugh T Blair, and Jason Cong. Lanmc: Lstm-assisted 2035 | non-rigid motion correction on fpga for calcium image stabilization. 2036 | In Proceedings of the 2019 ACM/SIGDA International Symposium on 2038 | Field-Programmable Gate Arrays, pages 104–109, 2019. 2040 |
2041 |2042 | Ryohei Shibue and Fumiyasu Komaki. Deconvolution of calcium imaging 2044 | data using marked point processes. PLoS computational biology, 16(3): 2046 | e1007650, 2020. 2047 |
2048 |2049 | Shreya Saxena, Ian Kinsella, Simon 2051 | Musall, Sharon H Kim, Jozsef Meszaros, David N Thibodeaux, Carla 2052 | Kim, John Cunningham, Elizabeth MC Hillman, Anne Churchland, et al. 2053 | Localized semi-nonnegative matrix factorization (locanmf) of widefield 2054 | calcium imaging data. PLOS Computational Biology, 16(4):e1007791, 2056 | 2020. 2057 |
2058 |2059 | Daniel A Cantu, Bo Wang, Michael W Gongwer, Cynthia X He, 2061 | Anubhuti Goel, Anand Suresh, Nazim Kourdougli, Erica D Arroyo, 2062 | William Zeiger, and Carlos Portera-Cailliau. Ezcalcium: Open source 2063 | toolbox for analysis of calcium imaging data. bioRxiv, 2020. 2065 |
2066 |2067 | Johannes Friedrich, Andrea Giovannucci, and 2069 | Eftychios A Pnevmatikakis. Online analysis of microendoscopic 1-photon 2070 | calcium imaging data streams. bioRxiv, 2020. 2072 |
2073 |2074 | Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alex D Jacob, 2076 | 2077 | 2078 | 2079 | Paul W Frankland, and Sheena A Josselyn. Automated curation 2080 | of cnmf-e-extracted roi spatial footprints and calcium traces using 2081 | open-source automl tools. bioRxiv, 2020a. 2083 |
2084 |2085 | Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alexander D 2087 | Jacob, Paul W Frankland, and Sheena A Josselyn. Automated curation 2088 | of cnmf-e-extracted roi spatial footprints and calcium traces using 2089 | open-source automl tools. Frontiers in Neural Circuits, 14:42, 2020b. 2091 |
2092 |2093 | Jerome Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, and 2095 | Christof Koch. Removing independent noise in systems neuroscience data 2096 | using deepinterpolation. bioRxiv, 2020. 2098 |
2099 |2100 | Ulysse Rubens, Romain Mormont, Lassi Paavolainen, Volker Bäcker, 2102 | Benjamin Pavie, Leandro A Scholz, Gino Michiels, Martin Maška, 2103 | Devrim Ünay, Graeme Ball, et al. Biaflows: A collaborative framework 2104 | to reproducibly deploy and benchmark bioimage analysis workflows. 2105 | Patterns, 1(3):100040, 2020. 2107 |
2108 |2109 | Ryan M Cassidy, Alexis G Bavencoffe, Elia R Lopez, Sai S Cheruvu, 2111 | Edgar T Walters, Rosa A Uribe, Anne Marie Krachler, and Max A 2112 | Odem. Frequency-independent biological signal identification (fibsi): A 2113 | free program that simplifies intensive analysis of non-stationary time series 2114 | data. bioRxiv, 2020. 2116 |
2117 |2118 | Sascha RA Alles, Max A Odem, Van B Lu, Ryan M Cassidy, and 2120 | Peter A Smith. Chronic bdnf simultaneously inhibits and unmasks 2121 | superficial dorsal horn neuronal activity. Scientific reports, 11(1):1–14, 2123 | 2021. 2124 |
2125 |2126 | Elke Kirschbaum, Alberto Bailoni, and Fred A Hamprecht. Disco: deep 2128 | 2129 | 2130 | 2131 | learning, instance segmentation, and correlations for cell segmentation in 2132 | calcium imaging. In Medical Image Computing and Computer Assisted 2134 | Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 2136 | October 4–8, 2020, Proceedings, Part V 23, pages 151–162. Springer, 2020. 2138 |
2139 |2140 | Julien Denis, Robin F Dard, Eleonora Quiroli, Rosa Cossart, and 2142 | Michel A Picardo. Deepcinac: a deep-learning-based python toolbox for 2143 | inferring calcium imaging neuronal activity based on movie visualization. 2144 | eneuro, 7(4), 2020. 2146 |
2147 |2148 | Yaesop Lee, Jing Xie, Eungjoo Lee, Srijesh Sudarsanan, Da-Ting Lin, 2150 | Rong Chen, and Shuvra S Bhattacharyya. Real-time neuron detection and 2151 | neural signal extraction platform for miniature calcium imaging. Frontiers 2153 | in Computational Neuroscience, 14:43, 2020. 2155 |
2156 |2157 | Chaozhen Tan, Yue Guan, Zhao Feng, Hong Ni, Zoutao Zhang, Zhiguang 2159 | Wang, Xiangning Li, Jing Yuan, Hui Gong, Qingming Luo, et al. 2160 | Deepbrainseg: Automated brain region segmentation for micro-optical 2161 | images with a convolutional neural network. Frontiers in neuroscience, 14: 2163 | 179, 2020. 2164 |
2165 |2166 | Victoria A Griffiths, Antoine M Valera, Joanna YN Lau, Hana 2168 | Roš, Thomas J Younts, Bóris Marin, Chiara Baragli, Diccon Coyle, 2169 | Geoffrey J Evans, George Konstantinou, et al. Real-time 3d movement 2170 | correction for two-photon imaging in behaving animals. Nature methods, 2172 | 17(7):741–748, 2020. 2173 |
2174 |2175 | Carsen Stringer, Tim Wang, Michalis Michaelos, and Marius Pachitariu. 2177 | Cellpose: a generalist algorithm for cellular segmentation. Nature Methods, 2179 | 18(1):100–106, 2021. 2180 |
2181 | 2182 | 2183 | 2184 |2185 | Alexander Song, Jeff L Gauthier, Jonathan W Pillow, David W Tank, 2187 | and Adam S Charles. Neural anatomy and optical microscopy (naomi) 2188 | simulation for evaluating calcium imaging methods. Journal of 2190 | Neuroscience Methods, 358:109173, 2021. 2192 |
2193 |2194 | Johannes 2196 | Friedrich, Andrea Giovannucci, and Eftychios A Pnevmatikakis. Online 2197 | analysis of microendoscopic 1-photon calcium imaging data streams. PLoS 2199 | computational biology, 17(1):e1008565, 2021. 2201 |
2202 |2203 | Hakan Inan, Claudia Schmuckermair, Tugce Tasci, Biafra Ahanonu, 2205 | Oscar Hernandez, Jérôme Lecoq, Fatih Dinç, Mark J Wagner, Murat 2206 | Erdogdu, and Mark J Schnitzer. Fast and statistically robust cell 2207 | extraction from large-scale neural calcium imaging datasets. bioRxiv, 2021. 2209 |
2210 |2211 | Zhe Dong, William Mau, Yu Susie Feng, Zachary T Pennington, 2213 | Lingxuan Chen, Yosif Zaki, Kanaka Rajan, Tristan Shuman, Daniel 2214 | Aharoni, and Denise J Cai. Minian: An open-source miniscope analysis 2215 | pipeline. bioRxiv, 2021. 2217 |
2218 |2219 | Kushal Kolar, Daniel Dondorp, Jordi Cornelis Zwiggelaar, Jørgen 2221 | Høyer, and Marios Chatzigeorgiou. Mesmerize: a dynamically adaptable 2222 | user-friendly analysis platform for 2d & 3d calcium imaging data. bioRxiv, 2224 | page 840488, 2021. 2225 |
2226 |2227 | Jérôme Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, Peter 2229 | Ledochowitsch, and Christof Koch. Removing independent noise in 2230 | systems neuroscience data using deepinterpolation. Nature Methods, pages 2232 | 1–8, 2021. 2233 |
2234 | 2235 | 2236 | 2237 |2238 | Seungjae Han, Eun-Seo Cho, Inkyu Park, Kijung Shin, and Young-Gyu 2240 | Yoon. Efficient neural network approximation of robust pca for automated 2241 | analysis of calcium imaging data. In International Conference on Medical 2243 | Image Computing and Computer-Assisted Intervention, pages 595–604. 2245 | Springer, 2021. 2246 |
2247 |2248 | Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V 2250 | Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri 2251 | Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture: 2252 | Calcium peak toolbox for analysis of in vitro calcium imaging data. 2253 | bioRxiv, 2021. 2255 |
2256 |2257 | Peter Rupprecht, Stefano Carta, Adrian Hoffmann, Mayumi Echizen, 2259 | Antonin Blot, Alex C Kwan, Yang Dan, Sonja B Hofer, Kazuo Kitamura, 2260 | Fritjof Helmchen, et al. A database and deep learning toolbox for 2261 | noise-optimized, generalized spike inference from calcium imaging. Nature 2263 | Neuroscience, 24(9):1324–1337, 2021. 2265 |
2266 |2267 | Changjia Cai, Johannes Friedrich, Amrita Singh, M Hossein Eybposh, 2269 | Eftychios A Pnevmatikakis, Kaspar Podgorski, and Andrea Giovannucci. 2270 | Volpy: automated and scalable analysis pipelines for voltage imaging 2271 | datasets. PLoS computational biology, 17(4):e1008806, 2021. 2273 |
2274 |2275 | Xinyang Li, Guoxun Zhang, Jiamin Wu, Yuanlong Zhang, Zhifeng Zhao, 2277 | Xing Lin, Hui Qiao, Hao Xie, Haoqian Wang, Lu Fang, et al. Reinforcing 2278 | neuron extraction and spike inference in calcium imaging using deep 2279 | self-supervised denoising. Nature Methods, pages 1–6, 2021. 2281 |
2282 |2283 | Leander 2285 | de Kraker, Koen Seignette, Premnath Thamizharasu, Bastijn JG van den 2286 | Boom, Ildefonso Ferreira Pica, Ingo Willuhn, Christiaan N Levelt, and 2287 | 2288 | 2289 | 2290 | Chris van der Togt. Specseg: cross spectral power-based segmentation of 2291 | neurons and neurites in chronic calcium imaging datasets. bioRxiv, pages 2293 | 2020–10, 2021. 2294 |
2295 |2296 | Andrea Giovannucci, Changjia Cai, Cynthia Dong, Marton Rozsa, and 2298 | Eftychios Pnevmatikakis. Fiola: An accelerated pipeline for fluorescence 2299 | imaging online analysis. 2021. 2300 |
2301 |2302 | Ryoma Hattori and Takaki Komiyama. Patchwarp: 2304 | Corrections of non-uniform image distortions in two-photon 2305 | calcium imaging data by patchwork affine transformations. 2306 | bioRxiv, 2021. doi: 10.1101/2021.11.10.468164. URL 2308 | https://www.biorxiv.org/content/early/2021/11/13/2021.11.10.468164. 2311 |
2312 |2313 | Xiaohui Zhang, Eric C Landsness, Wei Chen, Hanyang Miao, Michelle 2315 | Tang, Lindsey M Brier, Joseph P Culver, Jin-Moo Lee, and Mark A 2316 | Anastasio. Automated sleep state classification of wide-field calcium 2317 | imaging data via multiplex visibility graphs and deep learning. Journal of 2319 | Neuroscience Methods, page 109421, 2021. 2321 |
2322 |2323 | Philipp Flotho, Shinobu Nomura, Bernd Kuhn, and Daniel J Strauss. 2325 | Software for non-parametric image registration of 2-photon imaging data. 2326 | Journal of Biophotonics, 15(8):e202100330, 2022. 2328 |
2329 |2330 | Yijun Bao, Somayyeh Soltanian-Zadeh, Sina Farsiu, and Yiyang Gong. 2332 | Segmentation of neurons from fluorescence calcium recordings beyond real 2333 | time. Nature machine intelligence, 3(7):590–600, 2021. 2335 |
2336 |2337 | Masaki Taniguchi, Taro Tezuka, Pablo Vergara, Sakthivel Srinivasan, 2339 | Takuma Hosokawa, Yoan Chérasse, Toshie Naoi, Takeshi Sakurai, 2340 | and Masanori Sakaguchi. Open-source software for real-time calcium 2341 | 2342 | 2343 | 2344 | imaging and synchronized neuron firing detection. In 2021 43rd Annual 2346 | International Conference of the IEEE Engineering in Medicine & Biology 2348 | Society (EMBC), pages 2997–3003. IEEE, 2021. 2350 |
2351 |2352 | Brian R Mullen, Sydney C Weiser, Desiderio Ascencio, and James B 2354 | Ackman. Automated classification of signal sources in mesoscale calcium 2355 | imaging. bioRxiv, pages 2021–02, 2021. 2357 |
2358 |2359 | Yina Wang, Henry Pinkard, Emaad Khwaja, Shuqin Zhou, Laura 2361 | Waller, and Bo Huang. Image denoising for fluorescence microscopy by 2362 | self-supervised transfer learning. bioRxiv, pages 2021–02, 2021. 2364 |
2365 |2366 | Thibault Lagache, Alison Hanson, Jesús E Pérez-Ortega, Adrienne 2368 | Fairhall, and Rafael Yuste. Tracking calcium dynamics from individual 2369 | neurons in behaving animals. PLOS Computational Biology, 17(10): 2371 | e1009432, 2021. 2372 |
2373 |2374 | Adam S Charles, Nathan Cermak, Rifqi O Affan, Benjamin B Scott, 2376 | Jackie Schiller, and Gal Mishne. Graft: graph filtered temporal dictionary 2377 | learning for functional neural imaging. IEEE Transactions on Image 2379 | Processing, 31:3509–3524, 2022. 2381 |
2382 |2383 | Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V 2385 | Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri 2386 | Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture: 2387 | Calcium peaktoolbox for analysis of in vitro calcium imaging data. BMC 2389 | neuroscience, 23(1):71, 2022. 2391 |
2392 |2393 | Leander de Kraker, Koen Seignette, 2395 | Premnath Thamizharasu, Bastijn JG van den Boom, Ildefonso Ferreira 2396 | Pica, Ingo Willuhn, Christiaan N Levelt, and Chris van der Togt. Specseg 2397 | 2398 | 2399 | 2400 | is a versatile toolbox that segments neurons and neurites in chronic calcium 2401 | imaging datasets based on low-frequency cross-spectral power. Cell reports 2403 | methods, 2(10), 2022. 2405 |
2406 |2407 | Luca Sità, Marco Brondi, Pedro Lagomarsino de Leon Roig, Sebastiano 2409 | Curreli, Mariangela Panniello, Dania Vecchia, and Tommaso Fellin. A 2410 | deep-learning approach for online cell identification and trace extraction in 2411 | functional two-photon calcium imaging. Nature Communications, 13(1): 2413 | 1529, 2022. 2414 |
2415 |2416 | Honghua Guan, Dawei Li, Hyeon-cheol Park, Ang Li, Yuanlei Yue, 2418 | Yung-Tian A Gau, Ming-Jun Li, Dwight E Bergles, Hui Lu, and Xingde 2419 | Li. Deep-learning two-photon fiberscopy for video-rate brain imaging in 2420 | freely-behaving mice. Nature communications, 13(1):1534, 2022. 2422 |
2423 |2424 | Sharif Amit Kamran, Khondker Fariha Hossain, Hussein Moghnieh, 2426 | Sarah Riar, Allison Bartlett, Alireza Tavakkoli, Kenton M Sanders, and 2427 | Salah A Baker. New open-source software for subcellular segmentation 2428 | and analysis of spatiotemporal fluorescence signals using deep learning. 2429 | Iscience, 25(5), 2022. 2431 |
2432 |2433 | Xinyang Li, Yixin Li, Yiliang Zhou, Jiamin Wu, Zhifeng Zhao, Jiaqi 2435 | Fan, Fei Deng, Zhaofa Wu, Guihua Xiao, Jing He, et al. Real-time 2436 | denoising enables high-sensitivity fluorescence time-lapse imaging beyond 2437 | the shot-noise limit. Nature Biotechnology, 41(2):282–292, 2023a. 2439 |
2440 |2441 | Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank, 2443 | Jonathan W Pillow, and Adam S Charles. Detecting and correcting false 2444 | transients in calcium imaging. Nature Methods, 19(4):470–478, 2022. 2446 |
2447 |2448 | Andres Flores-Valle and Johannes D Seelig. Axial motion estimation 2450 | 2451 | 2452 | 2453 | and correction for simultaneous multi-plane two-photon calcium imaging. 2454 | Biomedical Optics Express, 13(4):2035–2049, 2022. 2456 |
2457 |2458 | Weiyi Liu, Junxia Pan, Yuanxu Xu, Meng Wang, Hongbo Jia, Kuan 2460 | Zhang, Xiaowei Chen, Xingyi Li, and Xiang Liao. Fast and accurate motion 2461 | correction for two-photon ca2+ imaging in behaving mice. Frontiers in 2463 | Neuroinformatics, 16:851188, 2022. 2465 |
2466 |2467 | Oliver Rübel, Andrew Tritt, Ryan Ly, Benjamin K Dichter, Satrajit 2469 | Ghosh, Lawrence Niu, Pamela Baker, Ivan Soltesz, Lydia Ng, Karel 2470 | Svoboda, et al. The neurodata without borders ecosystem for 2471 | neurophysiological data science. Elife, 11:e78362, 2022. 2473 |
2474 |2475 | Zhe Chen, Garrett J Blair, Changliang Guo, Jim Zhou, Juan-Luis 2477 | Romero-Sosa, Alicia Izquierdo, Peyman Golshani, Jason Cong, Daniel 2478 | Aharoni, and Hugh T Blair. A hardware system for real-time decoding of 2479 | in vivo calcium imaging data. Elife, 12:e78344, 2023. 2481 |
2482 |2483 | Yan Zhang, Márton Rózsa, Yajie Liang, Daniel Bushey, Ziqiang 2485 | Wei, Jihong Zheng, Daniel Reep, Gerard Joey Broussard, Arthur Tsang, 2486 | Getahun Tsegaye, et al. Fast and sensitive gcamp calcium indicators for 2487 | imaging neural populations. Nature, 615(7954):884–891, 2023a. 2489 |
2490 |2491 | Zhehao Xu, Yukun Wu, Jiangheng Guan, Shanshan Liang, Junxia Pan, 2493 | Meng Wang, Qianshuo Hu, Hongbo Jia, Xiaowei Chen, and Xiang Liao. 2494 | Neuroseg-ii: A deep learning approach for generalized neuron segmentation 2495 | in two-photon ca2+ imaging. Frontiers in Cellular Neuroscience, 17: 2497 | 1127847, 2023. 2498 |
2499 |2500 | Pablo Vergara, Yuteng Wang, Sakthivel Srinivasan, Yoan Cherasse, 2502 | Toshie Naoi, Yuki Sugaya, Takeshi Sakurai, Masanobu Kano, and Masanori 2503 | 2504 | 2505 | 2506 | Sakaguchi. The caliali tool for long-term tracking of neuronal population 2507 | dynamics in calcium imaging. bioRxiv, pages 2023–05, 2023. 2509 |
2510 |2511 | Yuanlong Zhang, Guoxun Zhang, Xiaofei Han, Jiamin Wu, Ziwei Li, 2513 | Xinyang Li, Guihua Xiao, Hao Xie, Lu Fang, and Qionghai Dai. Rapid 2514 | detection of neurons in widefield calcium imaging datasets after training 2515 | with synthetic data. Nature Methods, 20(5):747–754, 2023b. 2517 |
2518 |2519 | Jacopo Bonato, Sebastiano Curreli, Sara Romanzi, Stefano Panzeri, and 2521 | Tommaso Fellin. Astra: a deep learning algorithm for fast semantic 2522 | segmentation of large-scale astrocytic networks. bioRxiv, pages 2023–05, 2524 | 2023. 2525 |
2526 |2527 | Xinyang Li, Xiaowan Hu, Xingye Chen, Jiaqi Fan, Zhifeng Zhao, Jiamin 2529 | Wu, Haoqian Wang, and Qionghai Dai. Spatial redundancy transformer 2530 | for self-supervised fluorescence image denoising. bioRxiv, pages 2023–06, 2532 | 2023b. 2533 |
2534 |2535 | Junmo Cho, Seungjae Han, Eun-Seo Cho, Kijung Shin, and Young-Gyu 2537 | Yoon. Robust and efficient alignment of calcium imaging data through 2538 | simultaneous low rank and sparse decomposition. In Proceedings of the 2540 | IEEE/CVF Winter Conference on Applications of Computer Vision, pages 2542 | 1939–1948, 2023. 2543 |
2544 |2545 | Biafra Ahanonu, Andrew Crowther, Artur Kania, Mariela Rosa Casillas, 2547 | and Allan Basbaum. Long-term optical imaging of the spinal cord in awake, 2548 | behaving animals. bioRxiv, pages 2023–05, 2023. 2550 |
2551 |