├── .gitignore
├── README.md
├── docs
├── README.md
├── _config.yml
├── _layouts
│ └── default.html
└── images
│ └── Figure1.png
├── ref
└── protVec_100d_3grams.csv
└── src
├── DeepFam
├── dataset.py
├── draw_logo.py
├── experiment.py
├── model.py
├── run.py
├── test.py
├── train.py
└── utils.py
├── Kmer
├── dataset.py
├── experiment.py
├── model.py
├── run.py
├── test.py
├── train.py
└── utils.py
├── ProtVec
├── dataset.py
├── experiment.py
├── model.py
├── preprocess.py
├── run.py
├── test.py
├── train.py
└── utils.py
└── util
├── draw_logo.py
├── draw_logo.sh
├── preprocess.py
├── prosite_purity.py
└── run_weblogo.sh
/.gitignore:
--------------------------------------------------------------------------------
1 | /data/*
2 | /result/*
3 | /tmp/
4 | /Kmer/
5 | /Protvec/
6 | /result_adam/
7 | /testsrc/
8 | /gpcrsrc/
9 |
10 | *.py[cod]
11 | __pycache__/
12 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | docs/README.md
--------------------------------------------------------------------------------
/docs/README.md:
--------------------------------------------------------------------------------
1 | # About DeepFam
2 |
3 | DeepFam is a deep learning based alignment-free protein function prediction method. DeepFam first extracts features of conserved regions from a raw sequence by convolution layer and makes a prediction based on the features.
4 |
5 | 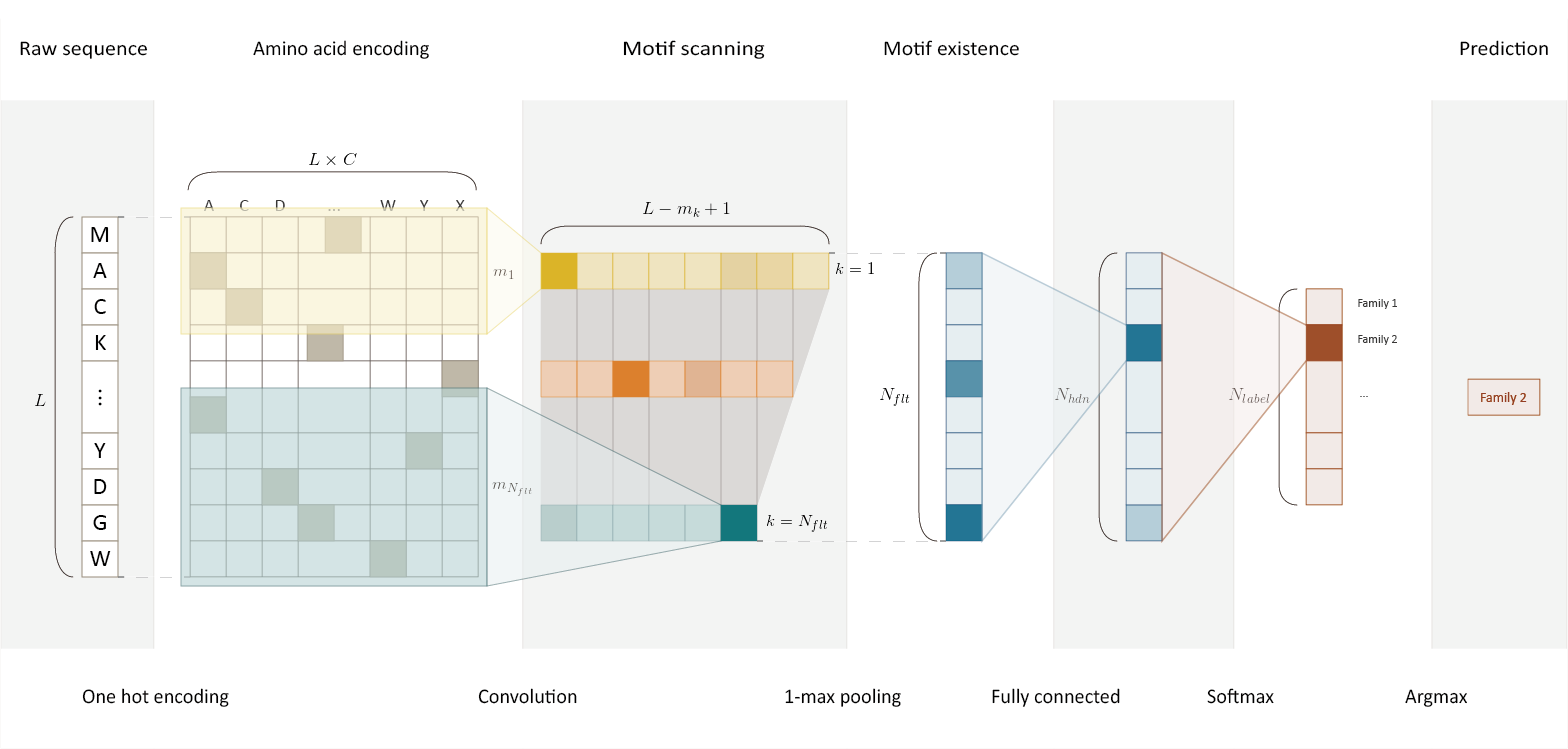
6 |
7 |
8 | # Features
9 |
10 | * Alignment-free: Do not need multiple or pairwise sequence alignment to train family model.
11 | * Instead, locally conserved regions within a family are trained by convolution units and 1-max pooling. Convolution unit works similar as PSSM.
12 | * Utilizing variable-size convolution unit (multiscale convolution unit) to train family specific conserved regions whose lengths are usually various.
13 |
14 |
15 | # Installation
16 |
17 | DeepFam is implemented in with [Tensorflow](https://www.tensorflow.org/) library. Both CPU and GPU machines are supported. For detail instruction of installing Tensorflow, see the [guide on official website](https://www.tensorflow.org/install).
18 |
19 | ## Requirements
20 |
21 | * Python: 2.7
22 | * Tensorflow: over 1.0
23 |
24 | # Usage
25 |
26 | First, clone the repository or download compressed source code files.
27 | ```
28 | $ git clone https://github.com/bhi-kimlab/DeepFam.git
29 | $ cd DeepFam
30 | ```
31 | You can see the valid paramenters for DeepFam by help option:
32 | ```
33 | $ python src/DeepFam/run.py --help
34 | ```
35 | One example of parameter setting is like:
36 | ```
37 | $ python src/DeepFam/run.py \
38 | --num_windows [256, 256, 256, 256, 256, 256, 256, 256] \
39 | --window_lengths [8, 12, 16, 20, 24, 28, 32, 36] \
40 | --num_hidden 2000 \
41 | --batch_size 100 \
42 | --keep_prob 0.7 \
43 | --learning_rate 0.001 \
44 | --regularizer 0.001 \
45 | --max_epoch 25 \
46 | --seq_len 1000 \
47 | --num_classes 1074 \
48 | --log_interval 100 \
49 | --save_interval 100 \
50 | --log_dir '/tmp/logs' \
51 | --test_file '/data/test.txt' \
52 | --train_file '/data/train.txt'
53 | ```
54 |
55 |
56 | # Data
57 | All data used by experiments described in manuscript is available at [here](http://epigenomics.snu.ac.kr/DeepFam/data.zip).
58 |
59 |
60 | # Contact
61 | If you have any question or problem, please send a email to [dane2522@gmail.com](mailto:dane2522@gmail.com)
62 |
--------------------------------------------------------------------------------
/docs/_config.yml:
--------------------------------------------------------------------------------
1 | theme: jekyll-theme-cayman
2 | title: DeepFam
3 | description: Deep learning based alignment-free method for protein family modeling and prediction.
4 | show_downloads: true
5 |
--------------------------------------------------------------------------------
/docs/_layouts/default.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 | {{ page.title | default: site.title }}
6 |
7 |
8 |
9 |
10 |
11 |
12 |
23 |
24 |
25 | {{ content }}
26 |
27 |
33 |
34 |
35 | {% if site.google_analytics %}
36 |
45 | {% endif %}
46 |
47 |
48 |
--------------------------------------------------------------------------------
/docs/images/Figure1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/bhi-kimlab/DeepFam/c13dad20c0d411e5e851f9d62a8524c8b52ca54a/docs/images/Figure1.png
--------------------------------------------------------------------------------
/src/DeepFam/dataset.py:
--------------------------------------------------------------------------------
1 | """
2 | " Dataset module
3 | " Handling protein sequence data.
4 | " Batch control, encoding, etc.
5 | """
6 | import sys
7 | import os
8 |
9 | import tensorflow as tf
10 | import numpy as np
11 |
12 |
13 | ## ######################## ##
14 | #
15 | # Define CHARSET, CHARLEN
16 | #
17 | ## ######################## ##
18 | CHARSET = { 'A': 0, 'C': 1, 'D': 2, 'E': 3, 'F': 4, 'G': 5, 'H': 6, \
19 | 'I': 7, 'K': 8, 'L': 9, 'M': 10, 'N': 11, 'P': 12, 'Q': 13, \
20 | 'R': 14, 'S': 15, 'T': 16, 'V': 17, 'W': 18, 'Y': 19, 'X': 20, \
21 | 'O': 20, 'U': 20,
22 | 'B': (2, 11),
23 | 'Z': (3, 13),
24 | 'J': (7, 9) }
25 | CHARLEN = 21
26 |
27 |
28 |
29 | ## ######################## ##
30 | #
31 | # Encoding Helpers
32 | #
33 | ## ######################## ##
34 | def encoding_seq_np( seq, arr):
35 | for i, c in enumerate(seq):
36 | if c == "_":
37 | # let them zero
38 | continue
39 | elif isinstance(CHARSET[ c ], int):
40 | idx = CHARLEN * i + CHARSET[ c ]
41 | arr[ idx ] = 1
42 | else:
43 | idx1 = CHARLEN * i + CHARSET[ c ][0]
44 | idx2 = CHARLEN * i + CHARSET[ c ][1]
45 | arr[ idx1 ] = 0.5
46 | arr[ idx2 ] = 0.5
47 | # raise Exception("notreachhere")
48 |
49 |
50 | def encoding_label_np( l, arr):
51 | arr[ int(l) ] = 1
52 |
53 |
54 |
55 |
56 | ## ######################## ##
57 | #
58 | # DATASET Class
59 | #
60 | ## ######################## ##
61 | # works for large dataset
62 | class DataSet(object):
63 | def __init__(self, fpath, seqlen, n_classes, need_shuffle = True):
64 | self.SEQLEN = seqlen
65 | self.NCLASSES = n_classes
66 | self.charset = CHARSET
67 | self.charset_size = CHARLEN
68 |
69 | # read raw file
70 | self._raw = self.read_raw( fpath )
71 |
72 | # iteration flags
73 | self._num_data = len(self._raw)
74 | self._epochs_completed = 0
75 | self._index_in_epoch = 0
76 |
77 | self._perm = np.arange(self._num_data)
78 | if need_shuffle:
79 | # shuffle data
80 | print("Needs shuffle")
81 | np.random.shuffle(self._perm)
82 | print("Reading data done")
83 |
84 |
85 | def next_batch(self, batch_size):
86 | start = self._index_in_epoch
87 | self._index_in_epoch += batch_size
88 |
89 | if self._index_in_epoch > self._num_data:
90 | # print("%d epoch finish!" % self._epochs_completed)
91 | # finished epoch
92 | self._epochs_completed += 1
93 | # shuffle the data
94 | np.random.shuffle(self._perm)
95 |
96 | # start next epoch
97 | start = 0
98 | self._index_in_epoch = batch_size
99 | assert batch_size <= self._num_data
100 |
101 | end = self._index_in_epoch
102 | idxs = self._perm[start:end]
103 | return self.parse_data( idxs )
104 |
105 |
106 | def iter_batch(self, batch_size, max_iter):
107 | while True:
108 | batch = self.next_batch( batch_size )
109 |
110 | if self._epochs_completed >= max_iter:
111 | break
112 | elif len(batch) == 0:
113 | continue
114 | else:
115 | yield batch
116 |
117 |
118 | def iter_once(self, batch_size, with_raw=False):
119 | while True:
120 | start = self._index_in_epoch
121 | self._index_in_epoch += batch_size
122 |
123 | if self._index_in_epoch > self._num_data:
124 | end = self._num_data
125 | idxs = self._perm[start:end]
126 | if len(idxs) > 0:
127 | yield self.parse_data( idxs, with_raw )
128 | break
129 |
130 | end = self._index_in_epoch
131 | idxs = self._perm[start:end]
132 | yield self.parse_data( idxs, with_raw )
133 |
134 |
135 | def full_batch(self):
136 | return self.parse_data( self._perm )
137 |
138 |
139 | def read_raw(self, fpath):
140 | print("Read %s start" % fpath)
141 | res = []
142 |
143 | with open( fpath, 'r') as tr:
144 | for row in tr.readlines():
145 | (label, seq) = row.strip().split("\t")
146 | seqlen = len(seq)
147 |
148 | if (seqlen != self.SEQLEN):
149 | raise Exception("SEQLEN is different from input data (%d / %d)"
150 | % (seqlen, self.SEQLEN))
151 |
152 | res.append( (label, seq) )
153 | return res
154 |
155 |
156 | def parse_data(self, idxs, with_raw=False):
157 | isize = len(idxs)
158 |
159 | data = np.zeros( (isize, CHARLEN * self.SEQLEN), dtype=np.float32 )
160 | labels = np.zeros( (isize, self.NCLASSES), dtype=np.uint8 )
161 | raw = []
162 |
163 | for i, idx in enumerate(idxs):
164 | label, seq = self._raw[ idx ]
165 |
166 | encoding_label_np(label, labels[i] )
167 | encoding_seq_np(seq, data[i] )
168 | raw.append( (label, seq) )
169 |
170 | if with_raw:
171 | return ( data, labels, raw )
172 | else:
173 | return ( data, labels )
174 |
175 |
--------------------------------------------------------------------------------
/src/DeepFam/draw_logo.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 | import math
10 | from heapq import *
11 | from collections import defaultdict
12 |
13 | import tensorflow as tf
14 | import numpy as np
15 | import tensorflow.contrib.slim as slim
16 |
17 |
18 | from utils import argparser
19 | from dataset import DataSet
20 | from model import get_placeholders, inference
21 |
22 |
23 |
24 |
25 | def test( FLAGS ):
26 | # read data
27 | dataset = DataSet( fpath = FLAGS.test_file,
28 | seqlen = FLAGS.seq_len,
29 | n_classes = FLAGS.num_classes,
30 | need_shuffle = False )
31 |
32 | FLAGS.charset_size = dataset.charset_size
33 |
34 | with tf.Graph().as_default():
35 | # placeholder
36 | placeholders = get_placeholders(FLAGS)
37 |
38 | # get inference
39 | pred, layers = inference( placeholders['data'], FLAGS,
40 | for_training=False )
41 |
42 | # calculate prediction
43 | _hit_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
44 | hit_op = tf.reduce_sum( tf.cast( _hit_op ,tf.float32 ) )
45 |
46 | # create saver
47 | saver = tf.train.Saver()
48 |
49 | # argmax of hidden1
50 | h1_argmax_ops = []
51 | for op in layers['conv']:
52 | h1_argmax_ops.append(tf.argmax(op, axis=2))
53 |
54 |
55 | with tf.Session() as sess:
56 | # load model
57 | ckpt = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
58 | if tf.train.checkpoint_exists( ckpt ):
59 | saver.restore( sess, ckpt )
60 | global_step = ckpt.split('/')[-1].split('-')[-1]
61 | print('Succesfully loaded model from %s at step=%s.' %
62 | (ckpt, global_step))
63 | else:
64 | print("[ERROR] Checkpoint not exist")
65 | return
66 |
67 |
68 | # iter batch
69 | hit_count = 0.0
70 | total_count = 0
71 | # top_matches = [ ([], []) ] * FLAGS.hidden1 # top 100 matching proteins
72 | wlens = [4, 8, 12, 16, 20]
73 | hsize = int(FLAGS.hidden1 / 5)
74 | motif_matches = (defaultdict(list), defaultdict(list))
75 |
76 | print("%s: starting test." % (datetime.now()))
77 | start_time = time.time()
78 | total_batch_size = math.ceil( dataset._num_data / FLAGS.batch_size )
79 |
80 | for step, (data, labels, raws) in enumerate(dataset.iter_once( FLAGS.batch_size, with_raw=True )):
81 | res_run = sess.run( [hit_op, h1_argmax_ops] + layers['conv'], feed_dict={
82 | placeholders['data']: data,
83 | placeholders['labels']: labels
84 | })
85 |
86 | hits = res_run[0]
87 | max_idxs = res_run[1] # shape = (wlens, N, 1, # of filters)
88 | motif_filters = res_run[2:]
89 |
90 |
91 | # mf.shape = (N, 1, l-w+1, # of filters)
92 | for i in range(len(motif_filters)):
93 | s = motif_filters[i].shape
94 | motif_filters[i] = np.transpose( motif_filters[i], (0, 1, 3, 2) ).reshape( (s[0], s[3], s[2]) )
95 |
96 | # mf.shape = (N, # of filters, l-w+1)
97 | for gidx, mf in enumerate(motif_filters):
98 | wlen = wlens[gidx]
99 | for ridx, row in enumerate(mf):
100 | for fidx, vals in enumerate(row):
101 | # for each filter, get max value and it's index
102 | max_idx = max_idxs[gidx][ridx][0][fidx]
103 | # max_idx = np.argmax(vals)
104 | max_val = vals[ max_idx ]
105 |
106 | hidx = gidx * hsize + fidx
107 |
108 | if max_val > 0:
109 | # get sequence
110 | rawseq = raws[ridx][1]
111 | subseq = rawseq[ max_idx : max_idx+wlen ]
112 | # heappush( top_matches[hidx], (max_val, subseq) )
113 | motif_matches[0][hidx].append( max_val )
114 | motif_matches[1][hidx].append( subseq )
115 | # motif_matches[gidx][fidx][0].append( max_val )
116 | # motif_matches[gidx][fidx][1].append( subseq )
117 |
118 |
119 | hit_count += np.sum( hits )
120 | total_count += len( data )
121 | # print("total:%d" % total_count)
122 |
123 | if step % FLAGS.log_interval == 0:
124 | duration = time.time() - start_time
125 | sec_per_batch = duration / FLAGS.log_interval
126 | examples_per_sec = FLAGS.batch_size / sec_per_batch
127 | print('%s: [%d batches out of %d] (%.1f examples/sec; %.3f'
128 | 'sec/batch)' % (datetime.now(), step, total_batch_size,
129 | examples_per_sec, sec_per_batch))
130 | start_time = time.time()
131 |
132 | # if step > 10:
133 | # break
134 |
135 |
136 | # # micro precision
137 | # print("%s: micro-precision = %.5f" %
138 | # (datetime.now(), (hit_count/total_count)))
139 |
140 |
141 | ### sort top lists
142 | print('%s: write result to file' % (datetime.now()) )
143 | for fidx in motif_matches[0]:
144 | val_lst = motif_matches[0][fidx]
145 | seq_lst = motif_matches[1][fidx]
146 | # top k
147 | k = wlens[ int(fidx / hsize) ] * 25
148 | l = min(k, len(val_lst)) * -1
149 | tidxs = np.argpartition(val_lst, l)[l:]
150 | with open("/home/kimlab/project/CCC/tmp/logos/test/p%d.txt"%fidx, 'w') as fw:
151 | for idx in tidxs:
152 | fw.write("%f\t%s\n" % (val_lst[idx], seq_lst[idx]) )
153 |
154 | if fidx % 50 == 0:
155 | print('%s: [%d filters out of %d]' % (datetime.now(), fidx, FLAGS.hidden1))
156 | # print(len(val_lst))
157 |
158 |
159 |
160 |
161 |
162 | if __name__ == '__main__':
163 | FLAGS = argparser()
164 | FLAGS.is_training = False
165 |
166 | test( FLAGS )
167 |
--------------------------------------------------------------------------------
/src/DeepFam/experiment.py:
--------------------------------------------------------------------------------
1 | from Queue import Queue
2 | from threading import Thread
3 | from subprocess import call
4 |
5 |
6 | import sys
7 | import os
8 |
9 | ## get argv
10 | num_classes = 2892
11 | seqlen = 1000
12 |
13 | # num_classes = 1796
14 | # seqlen = 1000
15 |
16 | # num_classes = 5
17 | # num_classes = 85
18 | # seqlen = 999
19 | # num_classes = 2
20 | # seqlen = 1000
21 |
22 |
23 |
24 | ## globals
25 | num_threads = 3
26 | enclosure_queue = Queue()
27 |
28 |
29 | ## helpers
30 | def mkdir(d):
31 | if not os.path.exists(d):
32 | os.makedirs(d)
33 |
34 | def run(i, queue):
35 | while True:
36 | # print("%d: Looking for next command" % i)
37 | cmd = queue.get()
38 | cmd = "export TF_CPP_MIN_LOG_LEVEL=2; export CUDA_VISIBLE_DEVICES=%d; %s" % (i, cmd)
39 | # cmd = "export CUDA_VISIBLE_DEVICES=%d; %s" % (7, cmd)
40 | call(cmd, shell=True)
41 | queue.task_done()
42 |
43 |
44 |
45 | # ####### hyperpara search #########
46 |
47 | # # hidden1 = [ '100', '150', '200', '250', '300' ]
48 | # hidden1 = [ '150', '200', '250', '300' ]
49 | # hidden2 = [ 1000, 1500, 2000 ]
50 | # indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "cog_cv", "dataset0" )
51 | # outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "hyper_search" )
52 |
53 | # for h1 in hidden1:
54 | # for h2 in hidden2:
55 | # expname = os.path.join( outdir, "%s-%d" % (h1, h2) )
56 | # logdir = os.path.join( expname, "logs" )
57 | # mkdir( os.path.join( logdir, "train" ) )
58 | # mkdir( os.path.join( logdir, "test" ) )
59 |
60 | # ckptdir = os.path.join( expname, "save" )
61 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
62 | # mkdir( ckptdir )
63 |
64 |
65 | # # cmd
66 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
67 | # s+= " --num_classes %s --seq_len %s" % ( num_classes, seqlen )
68 | # s+= " --window_lengths %s" % ( " ".join(['8', '12', '16', '20', '24', '28', '32', '36']) )
69 | # s+= " --num_windows %s" % ( " ".join([h1, h1, h1, h1, h1, h1, h1, h1]) )
70 | # s+= " --num_hidden %d --max_epoch %d" % ( h2, 25 )
71 | # s+= " --train_file %s" % os.path.join( indir, "train.txt" )
72 | # s+= " --test_file %s" % os.path.join( indir, "test.txt" )
73 | # s+= " --checkpoint_path %s --log_dir %s" % (ckptfile, logdir)
74 | # s+= " --save_interval %d" % (10000)
75 | # s+= " --batch_size %d" % (100)
76 |
77 | # enclosure_queue.put( s )
78 |
79 |
80 |
81 |
82 |
83 |
84 |
85 |
86 |
87 |
88 |
89 | ###### general task #########
90 | tests = ["dataset1", "dataset2", "dataset0"]
91 | # tests = ["dataset0"]
92 |
93 | for exp in tests:
94 | indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "l_1000_s_100", exp )
95 | outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "l_1000_s_100_try2" )
96 | expname = os.path.join( outdir, exp )
97 |
98 | logdir = os.path.join( expname, "logs" )
99 | mkdir( os.path.join( logdir, "train" ) )
100 | mkdir( os.path.join( logdir, "test" ) )
101 |
102 | ckptdir = os.path.join( expname, "save" )
103 | ckptfile = os.path.join( ckptdir, "model.ckpt" )
104 | mkdir( ckptdir )
105 |
106 |
107 | # cmd
108 | s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
109 | s+= " --num_classes %s --seq_len %s" % ( num_classes, seqlen )
110 | s+= " --window_lengths %s" % ( " ".join(['8', '12', '16', '20', '24', '28', '32', '36']) )
111 | s+= " --num_windows %s" % ( " ".join(['250', '250', '250', '250', '250', '250', '250', '250']) )
112 | s+= " --num_hidden %d --max_epoch %d" % ( 2000, 20 )
113 | s+= " --train_file %s" % os.path.join( indir, "train.txt" )
114 | s+= " --test_file %s" % os.path.join( indir, "test.txt" )
115 | s+= " --checkpoint_path %s --log_dir %s" % (ckptfile, logdir)
116 | s+= " --batch_size %d" % (100)
117 | s+= " --log_interval %d --save_interval %d" % (1000, 10000)
118 | # s+= " --prev_checkpoint_path=/home/kimlab/project/CCC/RES/otest-1665-1536-0.001000/save-158019"
119 | # s+= " --fine_tuning=True --fine_tuning_layers fc2"
120 | enclosure_queue.put( s )
121 |
122 |
123 |
124 |
125 | # ###### re- task #########
126 | # tests = ["dataset0", "dataset1", "dataset2"]
127 | # paths = [ "/home/kimlab/project/DeepFam/result/l_1000_s_250/dataset0/save/model.ckpt-138869",
128 | # "/home/kimlab/project/DeepFam/result/l_1000_s_250/dataset1/save/model.ckpt-138959",
129 | # "/home/kimlab/project/DeepFam/result/l_1000_s_250/dataset2/save/model.ckpt-139064" ]
130 |
131 | # for idx, exp in enumerate(tests):
132 | # indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "l_1000_s_250", exp )
133 | # outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "l_1000_s_250_re" )
134 | # expname = os.path.join( outdir, exp )
135 |
136 | # logdir = os.path.join( expname, "logs" )
137 | # mkdir( os.path.join( logdir, "train" ) )
138 | # mkdir( os.path.join( logdir, "test" ) )
139 |
140 | # ckptdir = os.path.join( expname, "save" )
141 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
142 | # mkdir( ckptdir )
143 |
144 |
145 | # # cmd
146 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
147 | # s+= " --num_classes %s --seq_len %s" % ( num_classes, seqlen )
148 | # s+= " --window_lengths %s" % ( " ".join(['8', '12', '16', '20', '24', '28', '32', '36']) )
149 | # s+= " --num_windows %s" % ( " ".join(['250', '250', '250', '250', '250', '250', '250', '250']) )
150 | # s+= " --num_hidden %d --max_epoch %d" % ( 2000, 7 )
151 | # s+= " --train_file %s" % os.path.join( indir, "train.txt" )
152 | # s+= " --test_file %s" % os.path.join( indir, "test.txt" )
153 | # s+= " --checkpoint_path %s --log_dir %s" % (ckptfile, logdir)
154 | # s+= " --batch_size %d" % (100)
155 | # s+= " --log_interval %d --save_interval %d" % (100, 10000)
156 | # s+= " --prev_checkpoint_path %s" % (paths[idx])
157 | # # s+= " --fine_tuning=True --fine_tuning_layers fc2"
158 | # enclosure_queue.put( s )
159 |
160 |
161 |
162 |
163 |
164 |
165 |
166 |
167 |
168 | # ####### fine-tuning task #########
169 | # # tests = ["class_level"]
170 | # tests = ["family_level"]
171 | # indirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/data", p ) for p in tests ]
172 | # outdirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/result", p ) for p in tests ]
173 |
174 | # for idx, expname in enumerate(outdirs):
175 | # indir = indirs[idx]
176 |
177 | # logdir = os.path.join( expname, "logs" )
178 | # mkdir( os.path.join( logdir, "train" ) )
179 | # mkdir( os.path.join( logdir, "test" ) )
180 |
181 | # ckptdir = os.path.join( expname, "save" )
182 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
183 | # mkdir( ckptdir )
184 |
185 |
186 | # # for cv in [1, 2]:
187 | # # cmd
188 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
189 | # s+= " --num_classes %s --seq_len %s" % ( num_classes, seqlen )
190 | # s+= " --window_lengths %s" % ( " ".join(['8', '12', '16', '20', '24', '28', '32', '36']) )
191 | # s+= " --num_windows %s" % ( " ".join(['250', '250', '250', '250', '250', '250', '250', '250']) )
192 | # s+= " --num_hidden %s --max_epoch %d" % ( 2000, 10 )
193 | # s+= " --train_file %s" % os.path.join( indir, "train.txt" )
194 | # s+= " --test_file %s" % os.path.join( indir, "test.txt" )
195 | # s+= " --checkpoint_path %s --log_dir %s" % (ckptfile, logdir)
196 | # s+= " --batch_size %d" % (100)
197 | # s+= " --prev_checkpoint_path /home/kimlab/project/DeepFam/result/cog_cv/90percent/save/model.ckpt-25402"
198 | # s+= " --fine_tuning True --fine_tuning_layers %s" % ( " ".join(['fc2']) )
199 | # s+= " --log_interval %d" % (10)
200 | # s+= " --save_interval %d" % (10000)
201 | # s+= " --learning_rate %f" % (0.001)
202 |
203 | # enclosure_queue.put( s )
204 |
205 |
206 |
207 | # ####### csss task #########
208 | # # tests = ['1.27.1.1', '1.27.1.2', '1.36.1.2', '1.36.1.5', '1.4.1.1', '1.41.1.2', '1.41.1.5', '1.4.1.2', '1.4.1.3', '1.45.1.2', '2.1.1.1', '2.1.1.2', '2.1.1.3', '2.1.1.4', '2.1.1.5', '2.28.1.1', '2.28.1.3', '2.38.4.1', '2.38.4.3', '2.38.4.5', '2.44.1.2', '2.5.1.1', '2.5.1.3', '2.52.1.2', '2.56.1.2', '2.9.1.2', '2.9.1.3', '2.9.1.4', '3.1.8.1', '3.1.8.3', '3.2.1.2', '3.2.1.3', '3.2.1.4', '3.2.1.5', '3.2.1.6', '3.2.1.7', '3.3.1.2', '3.3.1.5', '3.32.1.1', '3.32.1.11', '3.32.1.13', '3.32.1.8', '3.42.1.1', '3.42.1.5', '3.42.1.8', '7.3.10.1', '7.3.5.2', '7.3.6.1', '7.3.6.2', '7.3.6.4', '7.39.1.2', '7.39.1.3', '7.41.5.1', '7.41.5.2']
209 | # tests = ['2.44.1.2','2.28.1.1','3.3.1.5','3.42.1.5','2.28.1.3','2.38.4.5','2.38.4.3','3.2.1.3','3.42.1.8','2.1.1.5','2.52.1.2','2.38.4.1','3.32.1.8','3.32.1.1','3.2.1.4','3.3.1.2','3.2.1.5','2.5.1.3','1.41.1.5','2.5.1.1','3.2.1.2','7.39.1.3','3.2.1.6','1.36.1.5','2.1.1.3','7.39.1.2','2.9.1.2','3.42.1.1','2.1.1.1','3.32.1.13','2.1.1.4','1.36.1.2','3.32.1.11','7.41.5.1','1.27.1.2','1.4.1.2','3.2.1.7','1.27.1.1','7.3.10.1','3.1.8.3','1.45.1.2','2.9.1.4','7.41.5.2','2.56.1.2','2.1.1.2','7.3.6.2','1.4.1.3','3.1.8.1','1.41.1.2','1.4.1.1','7.3.5.2','2.9.1.3','7.3.6.1','7.3.6.4']
210 | # # tests = ['2.44.1.2','2.28.1.1','3.3.1.5']
211 | # indirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/csss/data2", p ) for p in tests ]
212 | # outdirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/csss/result", p ) for p in tests ]
213 |
214 | # for idx, expname in enumerate(outdirs):
215 | # indir = indirs[idx]
216 |
217 | # logdir = os.path.join( expname, "logs" )
218 | # mkdir( os.path.join( logdir, "train" ) )
219 | # mkdir( os.path.join( logdir, "test" ) )
220 |
221 | # ckptdir = os.path.join( expname, "save" )
222 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
223 | # mkdir( ckptdir )
224 |
225 |
226 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
227 | # s+= " --num_classes %s --seq_len %s" % ( num_classes, seqlen )
228 | # s+= " --window_lengths %s" % ( " ".join(['8', '12', '16', '20', '24', '28', '32', '36']) )
229 | # s+= " --num_windows %s" % ( " ".join(['250', '250', '250', '250', '250', '250', '250', '250']) )
230 | # s+= " --num_hidden %s --max_epoch %d" % ( 2000, 10 )
231 | # s+= " --train_file %s" % os.path.join( indir, "train.txt" )
232 | # s+= " --test_file %s" % os.path.join( indir, "test.txt" )
233 | # s+= " --checkpoint_path %s --log_dir %s" % (ckptfile, logdir)
234 | # s+= " --batch_size %d" % (100)
235 | # # s+= " --prev_checkpoint_path /home/kimlab/project/DeepFam/result/cog_cv/90percent/save/model.ckpt-254024"
236 | # # s+= " --fine_tuning True --fine_tuning_layers %s" % ( " ".join(['fc1', 'fc2']) )
237 | # s+= " --log_interval %d" % (10)
238 | # s+= " --save_interval %d" % (10000)
239 | # s+= " --learning_rate %f" % (0.001)
240 |
241 | # enclosure_queue.put( s )
242 |
243 |
244 |
245 |
246 |
247 |
248 |
249 |
250 | ## main
251 | # for i in range( num_threads ):
252 | for i in range( 1, 1+num_threads ):
253 | worker = Thread( target=run, args=(i, enclosure_queue,) )
254 | worker.setDaemon(True)
255 | worker.start()
256 |
257 |
258 | ## run
259 | enclosure_queue.join()
--------------------------------------------------------------------------------
/src/DeepFam/model.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 | import numpy as np
3 | import tensorflow.contrib.slim as slim
4 |
5 | import re
6 |
7 |
8 |
9 | def inference(data, FLAGS, for_training=False, scope=None):
10 | pred, layers = network( data, FLAGS,
11 | is_training=for_training,
12 | scope=scope)
13 |
14 | return pred, layers
15 |
16 |
17 | def get_placeholders(FLAGS):
18 | placeholders = {}
19 | placeholders['data'] = tf.placeholder(tf.float32, [None, FLAGS.seq_len * FLAGS.charset_size])
20 | placeholders['labels'] = tf.placeholder(tf.float32, [None, FLAGS.num_classes])
21 | return placeholders
22 |
23 |
24 |
25 |
26 | def network(data, FLAGS, is_training=True, scope=''):
27 | batch_norm_params = {
28 | 'decay': 0.9, # might problem if too small updates
29 | 'is_training': FLAGS.is_training,
30 | 'updates_collections': None
31 | }
32 |
33 | layers = {}
34 |
35 | x_data = tf.reshape( data, [-1, 1, FLAGS.seq_len, FLAGS.charset_size] )
36 |
37 |
38 | ###
39 | # define layers
40 | ###
41 |
42 | with tf.name_scope(scope, 'v1', [x_data]):
43 | with slim.arg_scope([slim.conv2d, slim.fully_connected],
44 | activation_fn=tf.nn.relu,
45 | biases_initializer=tf.constant_initializer(0.1),
46 | normalizer_fn=slim.batch_norm,
47 | normalizer_params=batch_norm_params,
48 | weights_initializer=tf.contrib.layers.xavier_initializer() ):
49 |
50 | layers['conv'] = []
51 | layers['hidden1'] = []
52 | for i, wlen in enumerate(FLAGS.window_lengths):
53 | layers['conv'].append ( slim.conv2d( x_data,
54 | FLAGS.num_windows[i],
55 | [1, wlen],
56 | # padding='SAME',
57 | padding='VALID',
58 | weights_initializer=tf.contrib.layers.xavier_initializer_conv2d(),
59 | scope="conv%d" % i ) )
60 | # max pooling
61 | max_pooled = slim.max_pool2d( layers['conv'][i],
62 | # [1, FLAGS.seq_len],
63 | [1, FLAGS.seq_len - wlen + 1],
64 | stride=[1, FLAGS.seq_len],
65 | padding='VALID',
66 | scope="pool%d" % i )
67 | # reshape
68 | layers['hidden1'].append( slim.flatten( max_pooled, scope="flatten%d" % i) )
69 |
70 | # concat
71 | layers['concat'] = tf.concat( layers['hidden1'], 1 )
72 |
73 | # dropout
74 | dropped = slim.dropout( layers['concat'],
75 | keep_prob=FLAGS.keep_prob,
76 | is_training=FLAGS.is_training,
77 | scope="dropout" )
78 |
79 | # fc layers
80 | layers['hidden2'] = slim.fully_connected( dropped,
81 | FLAGS.num_hidden,
82 | weights_regularizer=None,
83 | scope="fc1" )
84 |
85 | layers['pred'] = slim.fully_connected( layers['hidden2'],
86 | FLAGS.num_classes,
87 | activation_fn=None,
88 | normalizer_fn=None,
89 | normalizer_params=None,
90 | weights_regularizer=slim.l2_regularizer(FLAGS.regularizer) if FLAGS.regularizer > 0 else None,
91 | scope="fc2" )
92 |
93 | return layers['pred'], layers
94 |
95 |
96 |
97 |
--------------------------------------------------------------------------------
/src/DeepFam/run.py:
--------------------------------------------------------------------------------
1 | import re
2 | import time
3 | from datetime import datetime
4 | import os
5 |
6 | import tensorflow as tf
7 |
8 | from utils import argparser, logging
9 | from train import train
10 | from test import test
11 |
12 |
13 | if __name__ == '__main__':
14 | FLAGS = argparser()
15 | logging(str(FLAGS), FLAGS)
16 |
17 | logdir = FLAGS.log_dir
18 |
19 | ### iterate over max_epoch
20 | for i in range( FLAGS.max_epoch ):
21 | logging("%s: Run epoch %d" % (datetime.now(), i), FLAGS)
22 | # for training
23 | FLAGS.is_training = True
24 | FLAGS.keep_prob = 0.7
25 | FLAGS.log_dir = os.path.join( logdir, "train" )
26 | train( FLAGS )
27 |
28 | # for test
29 | FLAGS.is_training = False
30 | FLAGS.keep_prob = 1.0
31 | FLAGS.log_dir = os.path.join( logdir, "test" )
32 | test( FLAGS )
33 |
34 | FLAGS.prev_checkpoint_path = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
35 | FLAGS.fine_tuning = False
--------------------------------------------------------------------------------
/src/DeepFam/test.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 | import math
10 | from sklearn.metrics import roc_auc_score
11 |
12 | import tensorflow as tf
13 | import numpy as np
14 | import tensorflow.contrib.slim as slim
15 |
16 |
17 | from utils import argparser, logging
18 | from dataset import DataSet
19 | from model import get_placeholders, inference
20 |
21 |
22 |
23 | def test( FLAGS ):
24 | # read data
25 | dataset = DataSet( fpath = FLAGS.test_file,
26 | seqlen = FLAGS.seq_len,
27 | n_classes = FLAGS.num_classes,
28 | need_shuffle = False )
29 | # set character set size
30 | FLAGS.charset_size = dataset.charset_size
31 |
32 | with tf.Graph().as_default():
33 | # placeholder
34 | placeholders = get_placeholders(FLAGS)
35 |
36 | # get inference
37 | pred, layers = inference( placeholders['data'], FLAGS,
38 | for_training=False )
39 |
40 | prob = tf.nn.softmax(pred)
41 | # calculate prediction
42 | _hit_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
43 | hit_op = tf.reduce_sum( tf.cast( _hit_op ,tf.float32 ) )
44 |
45 | # create saver
46 | saver = tf.train.Saver()
47 |
48 | # summary
49 | summary_op = tf.summary.merge_all()
50 |
51 | with tf.Session() as sess:
52 | # load model
53 | ckpt = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
54 | if tf.train.checkpoint_exists( ckpt ):
55 | saver.restore( sess, ckpt )
56 | global_step = ckpt.split('/')[-1].split('-')[-1]
57 | logging('Succesfully loaded model from %s at step=%s.' %
58 | (ckpt, global_step), FLAGS)
59 | else:
60 | logging("[ERROR] Checkpoint not exist", FLAGS)
61 | return
62 |
63 | # iter batch
64 | hit_count = 0.0
65 | total_count = 0
66 | pred_list = []
67 | label_list = []
68 |
69 | logging("%s: starting test." % (datetime.now()), FLAGS)
70 | start_time = time.time()
71 | total_batch_size = math.ceil( dataset._num_data / FLAGS.batch_size )
72 |
73 | for step, (data, labels) in enumerate(dataset.iter_once( FLAGS.batch_size )):
74 | hits, pred_val = sess.run( [hit_op, prob], feed_dict={
75 | placeholders['data']: data,
76 | placeholders['labels']: labels
77 | })
78 |
79 | hit_count += np.sum( hits )
80 | total_count += len( data )
81 |
82 | for i, p in enumerate(pred_val):
83 | pred_list.append( p[0] )
84 | label_list.append( labels[i][0] )
85 |
86 | if step % FLAGS.log_interval == 0:
87 | duration = time.time() - start_time
88 | sec_per_batch = duration / FLAGS.log_interval
89 | examples_per_sec = FLAGS.batch_size / sec_per_batch
90 | logging('%s: [%d batches out of %d] (%.1f examples/sec; %.3f'
91 | 'sec/batch)' % (datetime.now(), step, total_batch_size,
92 | examples_per_sec, sec_per_batch), FLAGS)
93 | start_time = time.time()
94 |
95 |
96 | # micro precision
97 | # logging("%s: micro-precision = %.5f" %
98 | # (datetime.now(), (hit_count/total_count)), FLAGS)
99 | auc_val = roc_auc_score(label_list, pred_list)
100 | logging("%s: micro-precision = %.5f, auc = %.5f" %
101 | (datetime.now(), (hit_count/total_count), auc_val), FLAGS)
102 |
103 |
104 |
105 |
106 |
107 | if __name__ == '__main__':
108 | FLAGS = argparser()
109 | FLAGS.is_training = False
110 | logging(str(FLAGS), FLAGS)
111 | test( FLAGS )
112 |
--------------------------------------------------------------------------------
/src/DeepFam/train.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 |
10 |
11 | import tensorflow as tf
12 | import numpy as np
13 | import tensorflow.contrib.slim as slim
14 |
15 | from utils import argparser, logging
16 | from dataset import DataSet
17 | from model import get_placeholders, inference
18 |
19 |
20 |
21 | # ########## #
22 | #
23 | # Single-gpu version training.
24 | #
25 | # ########## #
26 | def train( FLAGS ):
27 | # read data
28 | dataset = DataSet( fpath = FLAGS.train_file,
29 | seqlen = FLAGS.seq_len,
30 | n_classes = FLAGS.num_classes,
31 | need_shuffle = True )
32 | # set character set size
33 | FLAGS.charset_size = dataset.charset_size
34 |
35 | with tf.Graph().as_default():
36 | # get placeholders
37 | global_step = tf.placeholder( tf.int32 )
38 | placeholders = get_placeholders(FLAGS)
39 |
40 | # prediction
41 | pred, layers = inference( placeholders['data'], FLAGS,
42 | for_training=True )
43 | # loss
44 | # slim.losses.softmax_cross_entropy(pred, placeholders['labels'])
45 | # class_weight = tf.constant([[1.0, 5.0]])
46 | # weight_per_label = tf.transpose( tf.matmul(placeholders['labels']
47 | # , tf.transpose(class_weight)) )
48 | # loss = tf.multiply(weight_per_label,
49 | # tf.nn.softmax_cross_entropy_with_logits(labels=placeholders['labels'], logits=pred))
50 | # loss = tf.losses.compute_weighted_loss(loss)
51 |
52 | tf.losses.softmax_cross_entropy(placeholders['labels'], pred)
53 | loss = tf.losses.get_total_loss()
54 |
55 | # accuracy
56 | _acc_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
57 | acc_op = tf.reduce_mean( tf.cast( _acc_op ,tf.float32 ) )
58 |
59 | # optimization
60 | train_op = tf.train.AdamOptimizer( FLAGS.learning_rate ).minimize( loss )
61 | # train_op = tf.train.RMSPropOptimizer( FLAGS.learning_rate ).minimize( loss )
62 |
63 | # Create a saver.
64 | saver = tf.train.Saver(max_to_keep=None)
65 |
66 | with tf.Session() as sess:
67 | sess.run( tf.global_variables_initializer() )
68 |
69 | if tf.train.checkpoint_exists( FLAGS.prev_checkpoint_path ):
70 | if FLAGS.fine_tuning:
71 | logging('%s: Fine Tuning Experiment!' %
72 | (datetime.now()), FLAGS)
73 | restore_variables = slim.get_variables_to_restore(exclude=FLAGS.fine_tuning_layers)
74 | restorer = tf.train.Saver( restore_variables )
75 | else:
76 | restorer = tf.train.Saver()
77 | restorer.restore(sess, FLAGS.prev_checkpoint_path)
78 | logging('%s: Pre-trained model restored from %s' %
79 | (datetime.now(), FLAGS.prev_checkpoint_path), FLAGS)
80 | step = int(FLAGS.prev_checkpoint_path.split('/')[-1].split('-')[-1]) + 1
81 | else:
82 | step = 0
83 |
84 | # iter epoch
85 | # for data, labels in dataset.iter_batch( FLAGS.batch_size, 5 ):
86 | for data, labels in dataset.iter_once( FLAGS.batch_size ):
87 | start_time = time.time()
88 | _, loss_val, acc_val = sess.run([train_op, loss, acc_op], feed_dict={
89 | placeholders['data']: data,
90 | placeholders['labels']: labels,
91 | global_step: step
92 | })
93 | duration = time.time() - start_time
94 |
95 | assert not np.isnan(loss_val), 'Model diverge'
96 |
97 | # logging
98 | if step > 0 and step % FLAGS.log_interval == 0:
99 | examples_per_sec = FLAGS.batch_size / float(duration)
100 | format_str = ('%s: step %d, loss = %.2f, acc = %.2f (%.1f examples/sec; %.3f '
101 | 'sec/batch)')
102 | logging(format_str % (datetime.now(), step, loss_val, acc_val,
103 | examples_per_sec, duration), FLAGS)
104 |
105 | # save model
106 | if step > 0 and step % FLAGS.save_interval == 0:
107 | saver.save(sess, FLAGS.checkpoint_path, global_step=step)
108 |
109 | # counter
110 | step += 1
111 |
112 | # save for last
113 | saver.save(sess, FLAGS.checkpoint_path, global_step=step-1)
114 |
115 |
116 |
117 |
118 | if __name__ == '__main__':
119 | FLAGS = argparser()
120 | FLAGS.is_training = True
121 | logging(str(FLAGS), FLAGS)
122 |
123 | train( FLAGS )
124 |
125 |
126 |
127 |
128 |
--------------------------------------------------------------------------------
/src/DeepFam/utils.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import os
3 |
4 |
5 |
6 | def argparser():
7 | parser = argparse.ArgumentParser()
8 | # for model
9 | parser.add_argument(
10 | '--window_lengths',
11 | type=int,
12 | nargs='+',
13 | help='Space seperated list of motif filter lengths. (ex, --window_lengths 4 8 12)'
14 | )
15 | parser.add_argument(
16 | '--num_windows',
17 | type=int,
18 | nargs='+',
19 | help='Space seperated list of the number of motif filters corresponding to length list. (ex, --num_windows 100 200 100)'
20 | )
21 | parser.add_argument(
22 | '--num_hidden',
23 | type=int,
24 | default=0,
25 | help='Number of neurons in hidden layer.'
26 | )
27 | parser.add_argument(
28 | '--regularizer',
29 | type=float,
30 | default=0.001,
31 | help='(Lambda value / 2) of L2 regularizer on weights connected to last layer (0 to exclude).'
32 | )
33 | parser.add_argument(
34 | '--keep_prob',
35 | type=float,
36 | default=0.7,
37 | help='Rate to be kept for dropout.'
38 | )
39 | parser.add_argument(
40 | '--num_classes',
41 | type=int,
42 | default=0,
43 | help='Number of classes (families).'
44 | )
45 | parser.add_argument(
46 | '--seq_len',
47 | type=int,
48 | default=0,
49 | help='Length of input sequences.'
50 | )
51 | # for learning
52 | parser.add_argument(
53 | '--learning_rate',
54 | type=float,
55 | default=0.001,
56 | help='Initial learning rate.'
57 | )
58 | parser.add_argument(
59 | '--max_epoch',
60 | type=int,
61 | default=1,
62 | help='Number of epochs to train.'
63 | )
64 | parser.add_argument(
65 | '--batch_size',
66 | type=int,
67 | default=64,
68 | help='Batch size. Must divide evenly into the dataset sizes.'
69 | )
70 | parser.add_argument(
71 | '--train_file',
72 | type=str,
73 | default='/tmp/tensorflow/mnist/input_data',
74 | help='Directory for input data.'
75 | )
76 | parser.add_argument(
77 | '--test_file',
78 | type=str,
79 | default='/tmp/tensorflow/mnist/input_data',
80 | help='Directory for input data.'
81 | )
82 | parser.add_argument(
83 | '--prev_checkpoint_path',
84 | type=str,
85 | default='',
86 | help='Restore from pre-trained model if specified.'
87 | )
88 | parser.add_argument(
89 | '--checkpoint_path',
90 | type=str,
91 | default='',
92 | help='Path to write checkpoint file.'
93 | )
94 | parser.add_argument(
95 | '--log_dir',
96 | type=str,
97 | default='/tmp',
98 | help='Directory for log data.'
99 | )
100 | parser.add_argument(
101 | '--log_interval',
102 | type=int,
103 | default=100,
104 | help='Interval of steps for logging.'
105 | )
106 | parser.add_argument(
107 | '--save_interval',
108 | type=int,
109 | default=100,
110 | help='Interval of steps for save model.'
111 | )
112 | # test
113 | parser.add_argument(
114 | '--fine_tuning',
115 | type=bool,
116 | default=False,
117 | help='If true, weight on last layer will not be restored.'
118 | )
119 | parser.add_argument(
120 | '--fine_tuning_layers',
121 | type=str,
122 | nargs='+',
123 | default=["fc2"],
124 | help='Which layers should be restored. Default is ["fc2"].'
125 | )
126 |
127 | FLAGS, unparsed = parser.parse_known_args()
128 |
129 | # check validity
130 | assert( len(FLAGS.window_lengths) == len(FLAGS.num_windows) )
131 |
132 | return FLAGS
133 |
134 |
135 |
136 |
137 | def logging(msg, FLAGS):
138 | fpath = os.path.join( FLAGS.log_dir, "log.txt" )
139 | with open( fpath, "a" ) as fw:
140 | fw.write("%s\n" % msg)
141 | print(msg)
--------------------------------------------------------------------------------
/src/Kmer/dataset.py:
--------------------------------------------------------------------------------
1 | """
2 | " Dataset module
3 | " Handling protein sequence data.
4 | " Batch control, encoding, etc.
5 | """
6 | import sys
7 | import os
8 | import itertools
9 |
10 | import tensorflow as tf
11 | import numpy as np
12 |
13 |
14 |
15 | # # truncated version
16 | # class WordDict(object):
17 | # def __init__(self, files, k, logpath):
18 | # self._kmers = set()
19 | # self.k = k
20 | # self.w2i = dict()
21 | # self.size = 0
22 |
23 | # # check if file exist
24 | # w2i_path = os.path.join(logpath, "w2i.txt")
25 |
26 | # if os.path.exists(w2i_path):
27 | # # load
28 | # print("Load W2I")
29 | # with open(w2i_path, 'r') as fr:
30 | # for line in fr.readlines():
31 | # self.size += 1
32 | # word, i = line.strip().split("\t")
33 | # i = int(i)
34 | # self.w2i[word] = i
35 | # else:
36 | # print("Create new W2I")
37 | # # parse files
38 | # for f in files:
39 | # self.parse_file(f)
40 |
41 | # # set to sorted list -> dict
42 | # for i, word in enumerate(sorted(self._kmers)):
43 | # self.w2i[ word ] = i
44 | # self.size += 1
45 |
46 | # del self._kmers
47 | # # save w2i
48 | # with open(w2i_path, 'w') as fw:
49 | # for word, i in self.w2i.iteritems():
50 | # fw.write("%s\t%d\n" % (word, i))
51 |
52 |
53 | # def parse_file(self, fpath):
54 | # with open( fpath, 'r') as tr:
55 | # for row in tr.readlines():
56 | # (label, seq) = row.strip().split("\t")
57 | # seq = seq.strip("_")
58 | # seqlen = len(seq)
59 |
60 | # for i in range( seqlen - self.k + 1 ):
61 | # self._kmers.add( seq[i:i+self.k] )
62 |
63 |
64 | # use all version
65 | class WordDict(object):
66 | def __init__(self, files, k, logpath):
67 | self._kmers = set()
68 | self.k = k
69 | self.w2i = dict()
70 | self.size = 0
71 |
72 | # check if file exist
73 | w2i_path = os.path.join(logpath, "w2i.txt")
74 |
75 | if os.path.exists(w2i_path):
76 | # load
77 | print("Load W2I")
78 | with open(w2i_path, 'r') as fr:
79 | for line in fr.readlines():
80 | self.size += 1
81 | word, i = line.strip().split("\t")
82 | i = int(i)
83 | self.w2i[word] = i
84 | else:
85 | print("Create new W2I")
86 | # generate kmer dict
87 | self.gen_kdict()
88 |
89 | # set to sorted list -> dict
90 | for i, word in enumerate(sorted(self._kmers)):
91 | self.w2i[ word ] = i
92 | self.size += 1
93 |
94 | del self._kmers
95 | # save w2i
96 | with open(w2i_path, 'w') as fw:
97 | for word, i in self.w2i.iteritems():
98 | fw.write("%s\t%d\n" % (word, i))
99 |
100 |
101 | def parse_file(self, fpath):
102 | with open( fpath, 'r') as tr:
103 | for row in tr.readlines():
104 | (label, seq) = row.strip().split("\t")
105 | seq = seq.strip("_")
106 | seqlen = len(seq)
107 |
108 | for i in range( seqlen - self.k + 1 ):
109 | self._kmers.add( seq[i:i+self.k] )
110 |
111 |
112 | def gen_kdict(self):
113 | CHAR = 'ABCDEFGHIJKLMNOPQRSTUVWXYZ'
114 | lst = [ CHAR for i in range(self.k) ]
115 |
116 | for elem in itertools.product(*lst):
117 | kmer = ''.join(elem)
118 | self._kmers.add( kmer )
119 |
120 |
121 |
122 |
123 |
124 |
125 |
126 |
127 |
128 | ## ######################## ##
129 | #
130 | # DATASET Class
131 | #
132 | ## ######################## ##
133 | # works for large dataset
134 | class DataSet(object):
135 | def __init__(self, fpath, n_classes, wd, need_shuffle = True):
136 | self.NCLASSES = n_classes
137 | self.worddict = wd
138 |
139 | # read raw file
140 | self._raw = self.read_raw( fpath, wd.k )
141 |
142 | # iteration flags
143 | self._num_data = len(self._raw)
144 | self._epochs_completed = 0
145 | self._index_in_epoch = 0
146 |
147 | self._perm = np.arange(self._num_data)
148 | if need_shuffle:
149 | # shuffle data
150 | print("Needs shuffle")
151 | np.random.shuffle(self._perm)
152 | print("Reading data done")
153 |
154 |
155 | def next_batch(self, batch_size):
156 | start = self._index_in_epoch
157 | self._index_in_epoch += batch_size
158 |
159 | if self._index_in_epoch > self._num_data:
160 | print("%d epoch finish!" % self._epochs_completed)
161 | # finished epoch
162 | self._epochs_completed += 1
163 | # shuffle the data
164 | np.random.shuffle(self._perm)
165 |
166 | # start next epoch
167 | start = 0
168 | self._index_in_epoch = batch_size
169 | assert batch_size <= self._num_data
170 |
171 | end = self._index_in_epoch
172 | idxs = self._perm[start:end]
173 | return self.parse_data( idxs )
174 |
175 |
176 | def iter_batch(self, batch_size, max_iter):
177 | while True:
178 | batch = self.next_batch( batch_size )
179 |
180 | if self._epochs_completed >= max_iter:
181 | break
182 | elif len(batch) == 0:
183 | continue
184 | else:
185 | yield batch
186 |
187 |
188 | def iter_once(self, batch_size):
189 | while True:
190 | start = self._index_in_epoch
191 | self._index_in_epoch += batch_size
192 |
193 | if self._index_in_epoch > self._num_data:
194 | end = self._num_data
195 | idxs = self._perm[start:end]
196 | if len(idxs) > 0:
197 | yield self.parse_data( idxs )
198 | break
199 |
200 | end = self._index_in_epoch
201 | idxs = self._perm[start:end]
202 | yield self.parse_data( idxs )
203 |
204 |
205 | def full_batch(self):
206 | return self.parse_data( self._perm )
207 |
208 |
209 | def read_raw(self, fpath, k):
210 | # read raw files
211 | print("Read %s start" % fpath)
212 | res = []
213 |
214 | with open( fpath, 'r') as tr:
215 | for row in tr.readlines():
216 | (label, seq) = row.strip().split("\t")
217 | seq = seq.strip("_")
218 |
219 | res.append( (label, seq) )
220 | return res
221 |
222 |
223 | def parse_data(self, idxs):
224 | isize = len(idxs)
225 |
226 | data = np.zeros( (isize, self.worddict.size), dtype=np.float32 )
227 | labels = np.zeros( (isize, self.NCLASSES), dtype=np.uint8 )
228 |
229 | for i, idx in enumerate(idxs):
230 | label, seq = self._raw[ idx ]
231 |
232 | ### encoding label
233 | labels[i][ int(label) ] = 1
234 | ### encoding seq
235 | seqlen = len(seq)
236 | # count frequency
237 | for j in range( seqlen - self.worddict.k + 1 ):
238 | word = seq[j:j+self.worddict.k]
239 | widx = self.worddict.w2i[ word ]
240 | data[i][ widx ] += (1.0 / seqlen - self.worddict.k + 1)
241 |
242 |
243 | return ( data, labels )
--------------------------------------------------------------------------------
/src/Kmer/experiment.py:
--------------------------------------------------------------------------------
1 | from Queue import Queue
2 | from threading import Thread
3 | from subprocess import call
4 |
5 |
6 | import sys
7 | import os
8 |
9 | ## get argv
10 | # outdir = sys.argv[1]
11 | # indir = sys.argv[2]
12 | num_classes = 1074
13 | # num_classes = 107
14 |
15 |
16 |
17 | ## globals
18 | num_threads = 3
19 | enclosure_queue = Queue()
20 |
21 |
22 | ## helpers
23 | def mkdir(d):
24 | if not os.path.exists(d):
25 | os.makedirs(d)
26 |
27 | def run(i, queue):
28 | while True:
29 | # print("%d: Looking for next command" % i)
30 | cmd = queue.get()
31 | cmd = "export TF_CPP_MIN_LOG_LEVEL=2; export CUDA_VISIBLE_DEVICES=%d; %s" % (i, cmd)
32 | # cmd = "export CUDA_VISIBLE_DEVICES=%d; %s" % (0, cmd)
33 | call(cmd, shell=True)
34 | queue.task_done()
35 |
36 |
37 |
38 |
39 | # ## general
40 | # k = 3
41 | # tests = ["90percent", "dataset1", "dataset2", "dataset0"]
42 | # tests = ["dataset0"]
43 |
44 | # for exp in tests:
45 | # indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "cog_cv", exp )
46 | # outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "Kmer" )
47 | # expname = os.path.join( outdir, exp )
48 |
49 | # logdir = os.path.join( expname, "logs" )
50 | # mkdir( os.path.join( logdir, "train" ) )
51 | # mkdir( os.path.join( logdir, "test" ) )
52 |
53 | # ckptdir = os.path.join( expname, "save" )
54 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
55 | # mkdir( ckptdir )
56 |
57 |
58 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
59 | # s+= " --num_classes=%s " % ( num_classes )
60 | # s+= " --k=%d " % ( k )
61 | # s+= " --max_epoch %d" % (25)
62 | # s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
63 | # s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
64 | # s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
65 | # s+= " --batch_size=100"
66 | # s+= " --log_interval %d --save_interval %d" % (100, 10000)
67 |
68 | # enclosure_queue.put( s )
69 |
70 |
71 |
72 |
73 | # num_classes = 1796
74 | # seqlen = 1000
75 |
76 | num_classes = 2892
77 | seqlen = 1000
78 |
79 | k = 3
80 | tests = ["dataset0", "dataset1", "dataset2"]
81 |
82 | for exp in tests:
83 | indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "l_1000_s_100", exp )
84 | outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "l_1000_s_100_kmer" )
85 | expname = os.path.join( outdir, exp )
86 |
87 | logdir = os.path.join( expname, "logs" )
88 | mkdir( os.path.join( logdir, "train" ) )
89 | mkdir( os.path.join( logdir, "test" ) )
90 |
91 | ckptdir = os.path.join( expname, "save" )
92 | ckptfile = os.path.join( ckptdir, "model.ckpt" )
93 | mkdir( ckptdir )
94 |
95 |
96 | s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
97 | s+= " --num_classes=%s " % ( num_classes )
98 | s+= " --k=%d " % ( k )
99 | s+= " --max_epoch %d" % (20)
100 | s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
101 | s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
102 | s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
103 | s+= " --batch_size=100"
104 | s+= " --log_interval %d --save_interval %d" % (100, 10000)
105 |
106 | enclosure_queue.put( s )
107 |
108 |
109 |
110 |
111 |
112 |
113 |
114 |
115 |
116 | # ## GPCR
117 | # k = 3
118 | # tests = [ "cv_%d" % i for i in range(10) ]
119 | # indirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/data/subfamily_level", p ) for p in tests ]
120 | # outdirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/result_kmer", p ) for p in tests ]
121 |
122 | # for idx, expname in enumerate(outdirs):
123 | # indir = indirs[idx]
124 |
125 | # logdir = os.path.join( expname, "logs" )
126 | # mkdir( os.path.join( logdir, "train" ) )
127 | # mkdir( os.path.join( logdir, "test" ) )
128 |
129 | # ckptdir = os.path.join( expname, "save" )
130 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
131 | # mkdir( ckptdir )
132 |
133 |
134 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
135 | # s+= " --num_classes=%s " % ( num_classes )
136 | # s+= " --k=%d " % ( k )
137 | # s+= " --max_epoch %d" % (20)
138 | # s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
139 | # s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
140 | # s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
141 | # s+= " --batch_size=100"
142 |
143 | # enclosure_queue.put( s )
144 |
145 |
146 |
147 |
148 |
149 |
150 |
151 |
152 | ## main
153 | # for i in range( num_threads ):
154 | for i in range( 4, 4+num_threads ):
155 | worker = Thread( target=run, args=(i, enclosure_queue,) )
156 | worker.setDaemon(True)
157 | worker.start()
158 |
159 |
160 | ## run
161 | enclosure_queue.join()
--------------------------------------------------------------------------------
/src/Kmer/model.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 | import numpy as np
3 | import tensorflow.contrib.slim as slim
4 |
5 | import re
6 |
7 |
8 |
9 | def inference(data, FLAGS, for_training=False, scope=None):
10 | pred = network( data, FLAGS,
11 | is_training=for_training,
12 | scope=scope)
13 |
14 | return pred
15 |
16 |
17 | def get_placeholders(FLAGS):
18 | placeholders = {}
19 | placeholders['data'] = tf.placeholder(tf.float32, [None, FLAGS.word_size])
20 | placeholders['labels'] = tf.placeholder(tf.float32, [None, FLAGS.num_classes])
21 | return placeholders
22 |
23 |
24 |
25 |
26 |
27 | def network(data, FLAGS, is_training=True, scope=''):
28 | ###
29 | # define layers
30 | ###
31 | net = slim.fully_connected( data,
32 | FLAGS.num_classes,
33 | activation_fn=None,
34 | biases_initializer=tf.constant_initializer(0.1),
35 | normalizer_fn=None,
36 | normalizer_params=None,
37 | weights_initializer=tf.contrib.layers.xavier_initializer(),
38 | weights_regularizer=slim.l2_regularizer(FLAGS.regularizer) if FLAGS.regularizer > 0 else None,
39 | scope="fc" )
40 |
41 | return net
42 |
43 |
--------------------------------------------------------------------------------
/src/Kmer/run.py:
--------------------------------------------------------------------------------
1 | import re
2 | import time
3 | from datetime import datetime
4 | import os
5 |
6 | import tensorflow as tf
7 |
8 | from utils import argparser, logging
9 | from train import train
10 | from test import test
11 |
12 |
13 | if __name__ == '__main__':
14 | FLAGS = argparser()
15 | logging(str(FLAGS), FLAGS)
16 |
17 | logdir = FLAGS.log_dir
18 |
19 |
20 | ### iterate over max_epoch
21 | for i in range( FLAGS.max_epoch ):
22 | logging("%s: Run epoch %d" % (datetime.now(), i), FLAGS)
23 | # for training
24 | FLAGS.is_training = True
25 | FLAGS.log_dir = os.path.join( logdir, "train" )
26 | train( FLAGS )
27 |
28 | # for test
29 | FLAGS.is_training = False
30 | FLAGS.log_dir = os.path.join( logdir, "test" )
31 | test( FLAGS )
32 |
33 | FLAGS.prev_checkpoint_path = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
--------------------------------------------------------------------------------
/src/Kmer/test.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 | import math
10 |
11 | import tensorflow as tf
12 | import numpy as np
13 | import tensorflow.contrib.slim as slim
14 |
15 |
16 | from utils import argparser, logging
17 | from dataset import DataSet, WordDict
18 | from model import get_placeholders, inference
19 |
20 |
21 |
22 | def test( FLAGS ):
23 | # first make bag of words
24 | worddict = WordDict( files = [FLAGS.train_file, FLAGS.test_file],
25 | k = FLAGS.k,
26 | logpath = FLAGS.log_dir )
27 |
28 | FLAGS.word_size = worddict.size
29 |
30 | # read data
31 | dataset = DataSet( fpath = FLAGS.test_file,
32 | n_classes = FLAGS.num_classes,
33 | wd = worddict,
34 | need_shuffle = False )
35 |
36 |
37 |
38 | with tf.Graph().as_default():
39 | # placeholder
40 | placeholders = get_placeholders(FLAGS)
41 |
42 | # get inference
43 | pred = inference( placeholders['data'], FLAGS,
44 | for_training=False )
45 |
46 | # calculate prediction
47 | pred_label_op = tf.argmax(pred, 1)
48 | label_op = tf.argmax(placeholders['labels'], 1)
49 | _hit_op = tf.equal( pred_label_op, label_op)
50 | hit_op = tf.reduce_sum( tf.cast( _hit_op ,tf.float32 ) )
51 |
52 | # create saver
53 | saver = tf.train.Saver()
54 |
55 | # summary
56 | summary_op = tf.summary.merge_all()
57 |
58 | with tf.Session() as sess:
59 | # load model
60 | ckpt = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
61 | if tf.train.checkpoint_exists( ckpt ):
62 | saver.restore( sess, ckpt )
63 | global_step = ckpt.split('/')[-1].split('-')[-1]
64 | logging('Succesfully loaded model from %s at step=%s.' %
65 | (ckpt, global_step), FLAGS)
66 | else:
67 | logging("[ERROR] Checkpoint not exist", FLAGS)
68 | return
69 |
70 | # summary writer
71 | summary_writer = tf.summary.FileWriter( FLAGS.log_dir,
72 | graph=sess.graph )
73 |
74 | # iter batch
75 | hit_count = 0.0
76 | total_count = 0
77 | results = []
78 |
79 | logging("%s: starting test." % (datetime.now()), FLAGS)
80 | start_time = time.time()
81 | total_batch_size = math.ceil( dataset._num_data / FLAGS.batch_size )
82 |
83 | for step, (data, labels) in enumerate(dataset.iter_once( FLAGS.batch_size )):
84 | hits, pred, lb = sess.run( [hit_op, pred_label_op, label_op], feed_dict={
85 | placeholders['data']: data,
86 | placeholders['labels']: labels
87 | })
88 |
89 | hit_count += np.sum( hits )
90 | total_count += len( data )
91 |
92 | for i, p in enumerate(pred):
93 | results.append( (p, lb[i]) )
94 |
95 | if step % FLAGS.log_interval == 0:
96 | duration = time.time() - start_time
97 | sec_per_batch = duration / FLAGS.log_interval
98 | examples_per_sec = FLAGS.batch_size / sec_per_batch
99 | logging('%s: [%d batches out of %d] (%.1f examples/sec; %.3f'
100 | 'sec/batch)' % (datetime.now(), step, total_batch_size,
101 | examples_per_sec, sec_per_batch), FLAGS)
102 | start_time = time.time()
103 |
104 | # micro precision
105 | logging("%s: micro-precision = %.5f" %
106 | (datetime.now(), (hit_count/total_count)), FLAGS)
107 |
108 |
109 | # write result
110 | outpath = os.path.join( FLAGS.log_dir, "out.txt" )
111 | with open(outpath, 'w') as fw:
112 | for p, l in results:
113 | fw.write("%d\t%d\n" % (int(l), int(p)))
114 |
115 |
116 |
117 |
118 | if __name__ == '__main__':
119 | FLAGS = argparser()
120 | FLAGS.is_training = False
121 | logging(str(FLAGS), FLAGS)
122 | test( FLAGS )
123 |
--------------------------------------------------------------------------------
/src/Kmer/train.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 |
10 |
11 | import tensorflow as tf
12 | import numpy as np
13 | import tensorflow.contrib.slim as slim
14 |
15 | from utils import argparser, logging
16 | from dataset import DataSet, WordDict
17 | from model import get_placeholders, inference
18 |
19 |
20 |
21 | # ########## #
22 | #
23 | # Single-gpu version training.
24 | #
25 | # ########## #
26 | def train( FLAGS ):
27 | # first make bag of words
28 | worddict = WordDict( files = [FLAGS.train_file, FLAGS.test_file],
29 | k = FLAGS.k,
30 | logpath = FLAGS.log_dir )
31 |
32 | FLAGS.word_size = worddict.size
33 |
34 | # read data
35 | dataset = DataSet( fpath = FLAGS.train_file,
36 | n_classes = FLAGS.num_classes,
37 | wd = worddict,
38 | need_shuffle = True )
39 |
40 |
41 |
42 | with tf.Graph().as_default():
43 | global_step = tf.placeholder( tf.int32 )
44 | placeholders = get_placeholders(FLAGS)
45 |
46 | pred = inference( placeholders['data'], FLAGS,
47 | for_training=True )
48 |
49 |
50 | tf.losses.softmax_cross_entropy(placeholders['labels'], pred)
51 | loss = tf.losses.get_total_loss()
52 |
53 |
54 | _acc_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
55 | acc_op = tf.reduce_mean( tf.cast( _acc_op ,tf.float32 ) )
56 |
57 |
58 | train_op = tf.train.AdamOptimizer( FLAGS.learning_rate ).minimize( loss )
59 |
60 |
61 |
62 | # Create a saver.
63 | saver = tf.train.Saver(max_to_keep=None)
64 |
65 | with tf.Session() as sess:
66 | sess.run( tf.global_variables_initializer() )
67 |
68 | if tf.train.checkpoint_exists( FLAGS.prev_checkpoint_path ):
69 | restorer = tf.train.Saver()
70 | restorer.restore(sess, FLAGS.prev_checkpoint_path)
71 | logging('%s: Pre-trained model restored from %s' %
72 | (datetime.now(), FLAGS.prev_checkpoint_path), FLAGS)
73 | step = int(FLAGS.prev_checkpoint_path.split('/')[-1].split('-')[-1]) + 1
74 | else:
75 | step = 0
76 |
77 | for data, labels in dataset.iter_once( FLAGS.batch_size ):
78 | start_time = time.time()
79 | _, loss_val, acc_val = sess.run([train_op, loss, acc_op], feed_dict={
80 | placeholders['data']: data,
81 | placeholders['labels']: labels,
82 | global_step: step
83 | })
84 | duration = time.time() - start_time
85 |
86 | assert not np.isnan(loss_val), 'Model diverge'
87 |
88 | if step > 0 and step % FLAGS.log_interval == 0:
89 | examples_per_sec = FLAGS.batch_size / float(duration)
90 | format_str = ('%s: step %d, loss = %.2f, acc = %.2f (%.1f examples/sec; %.3f '
91 | 'sec/batch)')
92 | logging(format_str % (datetime.now(), step, loss_val, acc_val,
93 | examples_per_sec, duration), FLAGS)
94 |
95 | if step > 0 and step % FLAGS.save_interval == 0:
96 | saver.save(sess, FLAGS.checkpoint_path, global_step=step)
97 |
98 | # counter
99 | step += 1
100 |
101 | # save for last
102 | saver.save(sess, FLAGS.checkpoint_path, global_step=step-1)
103 |
104 |
105 |
106 | if __name__ == '__main__':
107 | FLAGS = argparser()
108 | FLAGS.is_training = True
109 | logging(str(FLAGS), FLAGS)
110 |
111 | train( FLAGS )
112 |
113 |
114 |
115 |
116 |
--------------------------------------------------------------------------------
/src/Kmer/utils.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import os
3 |
4 |
5 |
6 | def argparser():
7 | parser = argparse.ArgumentParser()
8 | # for model
9 | parser.add_argument(
10 | '--k',
11 | type=int,
12 | default=3,
13 | help='Which k.'
14 | )
15 | parser.add_argument(
16 | '--regularizer',
17 | type=float,
18 | default=0.001,
19 | help='(Lambda value / 2) of L2 regularizer on weights connected to last layer (0 to exclude).'
20 | )
21 | parser.add_argument(
22 | '--num_classes',
23 | type=int,
24 | default=0,
25 | help='Number of classes.'
26 | )
27 | # for learning
28 | parser.add_argument(
29 | '--learning_rate',
30 | type=float,
31 | default=0.001,
32 | help='Initial learning rate.'
33 | )
34 | parser.add_argument(
35 | '--max_epoch',
36 | type=int,
37 | default=1,
38 | help='Number of epochs to train.'
39 | )
40 | parser.add_argument(
41 | '--batch_size',
42 | type=int,
43 | default=64,
44 | help='Batch size.'
45 | )
46 | parser.add_argument(
47 | '--train_file',
48 | type=str,
49 | default='/tmp/tensorflow/mnist/input_data',
50 | help='Directory for input data.'
51 | )
52 | parser.add_argument(
53 | '--test_file',
54 | type=str,
55 | default='/tmp/tensorflow/mnist/input_data',
56 | help='Directory for input data.'
57 | )
58 | parser.add_argument(
59 | '--prev_checkpoint_path',
60 | type=str,
61 | default='',
62 | help='Restore from pre-trained model if specified.'
63 | )
64 | parser.add_argument(
65 | '--checkpoint_path',
66 | type=str,
67 | default='',
68 | help='Path to write checkpoint file.'
69 | )
70 | parser.add_argument(
71 | '--log_dir',
72 | type=str,
73 | default='/tmp/tensorflow/mnist/logs/fully_connected_feed',
74 | help='Directory for log data.'
75 | )
76 | parser.add_argument(
77 | '--log_interval',
78 | type=int,
79 | default=100,
80 | help='Number of gpus to use'
81 | )
82 | parser.add_argument(
83 | '--save_interval',
84 | type=int,
85 | default=4000,
86 | help='Number of gpus to use'
87 | )
88 |
89 | FLAGS, unparsed = parser.parse_known_args()
90 |
91 | return FLAGS
92 |
93 |
94 |
95 |
96 |
97 |
98 | def logging(msg, FLAGS):
99 | fpath = os.path.join( FLAGS.log_dir, "log.txt" )
100 | with open( fpath, "a" ) as fw:
101 | fw.write("%s\n" % msg)
102 | print(msg)
--------------------------------------------------------------------------------
/src/ProtVec/dataset.py:
--------------------------------------------------------------------------------
1 | """
2 | " Dataset module
3 | " Handling protein sequence data.
4 | " Batch control, encoding, etc.
5 | """
6 | import sys
7 | import os
8 |
9 | import tensorflow as tf
10 | import numpy as np
11 |
12 | from preprocess import getProtVec
13 |
14 |
15 | class WordDict(object):
16 | def __init__(self, embedding_file):
17 | self.k = 3
18 | self.protVec = getProtVec(embedding_file) # 3mer -> vector
19 | self.size = 100
20 |
21 |
22 |
23 |
24 |
25 |
26 |
27 | ## ######################## ##
28 | #
29 | # DATASET Class
30 | #
31 | ## ######################## ##
32 | # works for large dataset
33 | class DataSet(object):
34 | def __init__(self, fpath, n_classes, wd, need_shuffle = True):
35 | self.NCLASSES = n_classes
36 | self.worddict = wd
37 |
38 | # read raw file
39 | self._raw = self.read_raw( fpath, wd.k )
40 |
41 | # iteration flags
42 | self._num_data = len(self._raw)
43 | self._epochs_completed = 0
44 | self._index_in_epoch = 0
45 |
46 | self._perm = np.arange(self._num_data)
47 | if need_shuffle:
48 | # shuffle data
49 | print("Needs shuffle")
50 | np.random.shuffle(self._perm)
51 | print("Reading data done")
52 |
53 |
54 | def next_batch(self, batch_size):
55 | start = self._index_in_epoch
56 | self._index_in_epoch += batch_size
57 |
58 | if self._index_in_epoch > self._num_data:
59 | print("%d epoch finish!" % self._epochs_completed)
60 | # finished epoch
61 | self._epochs_completed += 1
62 | # shuffle the data
63 | np.random.shuffle(self._perm)
64 |
65 | # start next epoch
66 | start = 0
67 | self._index_in_epoch = batch_size
68 | assert batch_size <= self._num_data
69 |
70 | end = self._index_in_epoch
71 | idxs = self._perm[start:end]
72 | return self.parse_data( idxs )
73 |
74 |
75 | def iter_batch(self, batch_size, max_iter):
76 | while True:
77 | batch = self.next_batch( batch_size )
78 |
79 | if self._epochs_completed >= max_iter:
80 | break
81 | elif len(batch) == 0:
82 | continue
83 | else:
84 | yield batch
85 |
86 |
87 | def iter_once(self, batch_size):
88 | while True:
89 | start = self._index_in_epoch
90 | self._index_in_epoch += batch_size
91 |
92 | if self._index_in_epoch > self._num_data:
93 | end = self._num_data
94 | idxs = self._perm[start:end]
95 | if len(idxs) > 0:
96 | yield self.parse_data( idxs )
97 | break
98 |
99 | end = self._index_in_epoch
100 | idxs = self._perm[start:end]
101 | yield self.parse_data( idxs )
102 |
103 |
104 | def full_batch(self):
105 | return self.parse_data( self._perm )
106 |
107 |
108 | def read_raw(self, fpath, k):
109 | # read raw files
110 | print("Read %s start" % fpath)
111 | res = []
112 |
113 | with open( fpath, 'r') as tr:
114 | for row in tr.readlines():
115 | (label, seq) = row.strip().split("\t")
116 | seq = seq.strip("_")
117 |
118 | res.append( (label, seq) )
119 | return res
120 |
121 |
122 | def parse_data(self, idxs):
123 | isize = len(idxs)
124 |
125 | data = np.zeros( (isize, self.worddict.size), dtype=np.float32 )
126 | labels = np.zeros( (isize, self.NCLASSES), dtype=np.uint8 )
127 |
128 | for i, idx in enumerate(idxs):
129 | label, seq = self._raw[ idx ]
130 |
131 | ### encoding label
132 | labels[i][ int(label) ] = 1
133 | ### encoding seq
134 | seqlen = len(seq)
135 | # count frequency
136 | for j in range( seqlen - self.worddict.k + 1 ):
137 | word = seq[j:j+self.worddict.k] # 3mer
138 | v = self.worddict.protVec[ word ]
139 | data[i] += v # sum of all 3mer
140 | # normalize by length
141 | # data[i] /= (1.0 / seqlen - self.worddict.k + 1)
142 |
143 |
144 | return ( data, labels )
--------------------------------------------------------------------------------
/src/ProtVec/experiment.py:
--------------------------------------------------------------------------------
1 | from Queue import Queue
2 | from threading import Thread
3 | from subprocess import call
4 |
5 |
6 | import sys
7 | import os
8 |
9 | ## get argv
10 | # outdir = sys.argv[1]
11 | # indir = sys.argv[2]
12 | num_classes = 1074
13 | # num_classes = 107
14 |
15 |
16 | ## globals
17 | num_threads = 3
18 | enclosure_queue = Queue()
19 |
20 |
21 | ## helpers
22 | def mkdir(d):
23 | if not os.path.exists(d):
24 | os.makedirs(d)
25 |
26 | def run(i, queue):
27 | while True:
28 | # print("%d: Looking for next command" % i)
29 | cmd = queue.get()
30 | cmd = "export TF_CPP_MIN_LOG_LEVEL=2; export CUDA_VISIBLE_DEVICES=%d; %s" % (i, cmd)
31 | # cmd = "export CUDA_VISIBLE_DEVICES=%d; %s" % (7, cmd)
32 | call(cmd, shell=True)
33 | queue.task_done()
34 |
35 |
36 |
37 | # ## general
38 | # k = 3
39 | # # tests = ["90percent", "dataset1", "dataset2", "dataset0"]
40 | # embedding_file = "/home/kimlab/project/DeepFam/ref/protVec_100d_3grams.csv"
41 |
42 | # for exp in tests:
43 | # indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "cog_cv", exp )
44 | # outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "Protvec" )
45 | # expname = os.path.join( outdir, exp )
46 |
47 | # logdir = os.path.join( expname, "logs" )
48 | # mkdir( os.path.join( logdir, "train" ) )
49 | # mkdir( os.path.join( logdir, "test" ) )
50 |
51 | # ckptdir = os.path.join( expname, "save" )
52 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
53 | # mkdir( ckptdir )
54 |
55 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
56 | # s+= " --num_classes=%s " % ( num_classes )
57 | # s+= " --embedding_file=%s " % ( embedding_file )
58 | # s+= " --max_epoch %d" % 25
59 | # s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
60 | # s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
61 | # s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
62 | # s+= " --batch_size=100"
63 |
64 | # enclosure_queue.put( s )
65 |
66 |
67 |
68 |
69 |
70 |
71 | num_classes = 1796
72 | seqlen = 1000
73 |
74 | # num_classes = 2892
75 | # seqlen = 1000
76 |
77 | k = 3
78 | tests = ["dataset0", "dataset1", "dataset2"]
79 | embedding_file = "/home/kimlab/project/DeepFam/ref/protVec_100d_3grams.csv"
80 |
81 | for exp in tests:
82 | indir = os.path.join( "/home/kimlab/project/DeepFam", "data", "l_1000_s_250", exp )
83 | outdir = os.path.join( "/home/kimlab/project/DeepFam", "result", "l_1000_s_250_protvec" )
84 | expname = os.path.join( outdir, exp )
85 |
86 | logdir = os.path.join( expname, "logs" )
87 | mkdir( os.path.join( logdir, "train" ) )
88 | mkdir( os.path.join( logdir, "test" ) )
89 |
90 | ckptdir = os.path.join( expname, "save" )
91 | ckptfile = os.path.join( ckptdir, "model.ckpt" )
92 | mkdir( ckptdir )
93 |
94 | s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
95 | s+= " --num_classes=%s " % ( num_classes )
96 | s+= " --embedding_file=%s " % ( embedding_file )
97 | s+= " --max_epoch %d" % 25
98 | s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
99 | s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
100 | s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
101 | s+= " --batch_size=100"
102 | s+= " --log_interval %d --save_interval %d" % (100, 10000)
103 |
104 | enclosure_queue.put( s )
105 |
106 |
107 |
108 |
109 |
110 |
111 |
112 |
113 | # ## GPCR
114 | # k = 3
115 | # tests = [ "cv_%d" % i for i in range(10) ]
116 | # indirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/data/subfamily_level", p ) for p in tests ]
117 | # outdirs = [ os.path.join( "/home/kimlab/project/DeepFam/tmp/gpcr/result_protvec", p ) for p in tests ]
118 | # embedding_file = "/home/kimlab/project/DeepFam/ref/protVec_100d_3grams.csv"
119 |
120 | # for idx, expname in enumerate(outdirs):
121 | # indir = indirs[idx]
122 | # logdir = os.path.join( expname, "logs" )
123 | # mkdir( os.path.join( logdir, "train" ) )
124 | # mkdir( os.path.join( logdir, "test" ) )
125 |
126 | # ckptdir = os.path.join( expname, "save" )
127 | # ckptfile = os.path.join( ckptdir, "model.ckpt" )
128 | # mkdir( ckptdir )
129 |
130 | # # cmd
131 | # s="python %s" % os.path.join( os.path.dirname(os.path.realpath(__file__)), "run.py" )
132 | # s+= " --num_classes=%s " % ( num_classes )
133 | # s+= " --embedding_file=%s " % ( embedding_file )
134 | # s+= " --max_epoch %d" % 20
135 | # s+= " --train_file=%s" % os.path.join( indir, "train.txt" )
136 | # s+= " --test_file=%s" % os.path.join( indir, "test.txt" )
137 | # s+= " --checkpoint_path=%s --log_dir=%s" % (ckptfile, logdir)
138 | # s+= " --batch_size=100"
139 |
140 | # enclosure_queue.put( s )
141 |
142 |
143 |
144 |
145 |
146 |
147 | ## main
148 | # for i in range( num_threads ):
149 | for i in range( 1, 1+num_threads ):
150 | worker = Thread( target=run, args=(i, enclosure_queue,) )
151 | worker.setDaemon(True)
152 | worker.start()
153 |
154 |
155 | ## run
156 | enclosure_queue.join()
--------------------------------------------------------------------------------
/src/ProtVec/model.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 | import numpy as np
3 | import tensorflow.contrib.slim as slim
4 |
5 | import re
6 |
7 |
8 |
9 | def inference(data, FLAGS, for_training=False, scope=None):
10 | pred = network( data, FLAGS,
11 | is_training=for_training,
12 | scope=scope)
13 |
14 | return pred
15 |
16 |
17 | def get_placeholders(FLAGS):
18 | placeholders = {}
19 | placeholders['data'] = tf.placeholder(tf.float32, [None, FLAGS.word_size])
20 | placeholders['labels'] = tf.placeholder(tf.float32, [None, FLAGS.num_classes])
21 | return placeholders
22 |
23 |
24 |
25 |
26 |
27 | def network(data, FLAGS, is_training=True, scope=''):
28 | ###
29 | # define layers
30 | ###
31 | net = slim.fully_connected( data,
32 | FLAGS.num_classes,
33 | activation_fn=None,
34 | biases_initializer=tf.constant_initializer(0.1),
35 | normalizer_fn=None,
36 | normalizer_params=None,
37 | weights_initializer=tf.contrib.layers.xavier_initializer(),
38 | weights_regularizer=slim.l2_regularizer(FLAGS.regularizer) if FLAGS.regularizer > 0 else None,
39 | scope="fc" )
40 |
41 | return net
42 |
43 |
--------------------------------------------------------------------------------
/src/ProtVec/preprocess.py:
--------------------------------------------------------------------------------
1 | from collections import defaultdict
2 | import numpy as np
3 |
4 |
5 | def getProtVec(embedding_file):
6 | zeroVec = np.zeros(100, dtype=np.float16)
7 | protVec = defaultdict(lambda: zeroVec)
8 |
9 | with open(embedding_file, 'r') as fr:
10 | for row in fr.readlines():
11 | row = row.replace("\"", "").rstrip()
12 | elems = row.split("\t")
13 | assert len(elems) == 101
14 |
15 | protVec[ elems[0] ] = np.asarray( [ float(x) for x in elems[1:] ], dtype=np.float32 )
16 |
17 | return protVec
18 |
19 |
20 |
21 |
22 |
23 | # ### #
24 | # parse raw seq into protVec
25 | # ### #
26 | def parse(seq, protVec=None):
27 | # read 3gram
28 | if protVec == None:
29 | protVec = getProtVec()
30 |
31 | # parse seqs
32 | seq = seq.rstrip("_")
33 | vec = np.zeros( 100, dtype=np.float16 )
34 |
35 | for sidx in range(1):
36 | tmpseq = seq[sidx:]
37 | l = len(tmpseq) - 3 + 1
38 |
39 | for i in range(l):
40 | vec = vec + protVec[ tmpseq[i:i+3] ]
41 |
42 | return vec
43 |
44 |
45 | def save_to_file( seqfile, outfile ):
46 | protVec = getProtVec()
47 |
48 | data = []
49 | labels = []
50 | familyset = set()
51 | with open(seqfile, 'r') as fr:
52 | for row in fr.readlines():
53 | (family, seq) = row.rstrip().split("\t")
54 | embedded = parse( seq, protVec )
55 | labels.append( family )
56 | data.append( embedded )
57 | familyset.add( family )
58 |
59 | with open(outfile, 'w') as fw:
60 | ### write header
61 | # # of data, # of features, classes
62 | header = [ str(len(data)), str(len(data[0])) ] + [ str(x) for x in range(len(familyset)) ]
63 | fw.write("%s\n" % ",".join( header ))
64 |
65 | ### write row
66 | for i, d in enumerate(data):
67 | l = labels[i]
68 | row = [ "%.3f" % x for x in list(d) ]
69 | row.append( l )
70 | fw.write("%s\n" % ",".join( row ))
71 |
72 |
73 |
74 |
75 |
76 |
77 |
78 | ### main
79 | if __name__ == '__main__':
80 | train_file = "/home/kimlab/project/CCC/tmp/swissprot/data/example/train.txt"
81 | test_file = "/home/kimlab/project/CCC/tmp/swissprot/data/example/test.txt"
82 | save_to_file(train_file, "train.txt")
83 | save_to_file(test_file, "test.txt")
--------------------------------------------------------------------------------
/src/ProtVec/run.py:
--------------------------------------------------------------------------------
1 | import re
2 | import time
3 | from datetime import datetime
4 | import os
5 |
6 | import tensorflow as tf
7 |
8 | from utils import argparser, logging
9 | from train import train
10 | from test import test
11 |
12 |
13 | if __name__ == '__main__':
14 | FLAGS = argparser()
15 | logging(str(FLAGS), FLAGS)
16 |
17 | logdir = FLAGS.log_dir
18 |
19 | ### iterate over max_epoch
20 | for i in range( FLAGS.max_epoch ):
21 | logging("%s: Run epoch %d" % (datetime.now(), i), FLAGS)
22 | # for training
23 | FLAGS.is_training = True
24 | FLAGS.log_dir = os.path.join( logdir, "train" )
25 | train( FLAGS )
26 |
27 | # for test
28 | FLAGS.is_training = False
29 | FLAGS.log_dir = os.path.join( logdir, "test" )
30 | test( FLAGS )
31 |
32 | FLAGS.prev_checkpoint_path = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
--------------------------------------------------------------------------------
/src/ProtVec/test.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 | import math
10 |
11 | import tensorflow as tf
12 | import numpy as np
13 | import tensorflow.contrib.slim as slim
14 |
15 |
16 | from utils import argparser, logging
17 | from dataset import DataSet, WordDict
18 | from model import get_placeholders, inference
19 |
20 |
21 |
22 | def test( FLAGS ):
23 | # first make bag of words
24 | worddict = WordDict(FLAGS.embedding_file)
25 |
26 | FLAGS.word_size = worddict.size
27 |
28 | # read data
29 | dataset = DataSet( fpath = FLAGS.test_file,
30 | n_classes = FLAGS.num_classes,
31 | wd = worddict,
32 | need_shuffle = False )
33 |
34 |
35 |
36 | with tf.Graph().as_default():
37 | # placeholder
38 | placeholders = get_placeholders(FLAGS)
39 |
40 | # get inference
41 | pred = inference( placeholders['data'], FLAGS,
42 | for_training=False )
43 |
44 | # calculate prediction
45 | pred_label_op = tf.argmax(pred, 1)
46 | label_op = tf.argmax(placeholders['labels'], 1)
47 | _hit_op = tf.equal( pred_label_op, label_op)
48 | hit_op = tf.reduce_sum( tf.cast( _hit_op ,tf.float32 ) )
49 |
50 | # create saver
51 | saver = tf.train.Saver()
52 |
53 | # summary
54 | summary_op = tf.summary.merge_all()
55 |
56 | with tf.Session() as sess:
57 | # load model
58 | ckpt = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
59 | if tf.train.checkpoint_exists( ckpt ):
60 | saver.restore( sess, ckpt )
61 | global_step = ckpt.split('/')[-1].split('-')[-1]
62 | logging('Succesfully loaded model from %s at step=%s.' %
63 | (ckpt, global_step), FLAGS)
64 | else:
65 | logging("[ERROR] Checkpoint not exist", FLAGS)
66 | return
67 |
68 | # summary writer
69 | summary_writer = tf.summary.FileWriter( FLAGS.log_dir,
70 | graph=sess.graph )
71 |
72 | # iter batch
73 | hit_count = 0.0
74 | total_count = 0
75 | results = []
76 |
77 | logging("%s: starting test." % (datetime.now()), FLAGS)
78 | start_time = time.time()
79 | total_batch_size = math.ceil( dataset._num_data / FLAGS.batch_size )
80 |
81 | for step, (data, labels) in enumerate(dataset.iter_once( FLAGS.batch_size )):
82 | hits, pred, lb = sess.run( [hit_op, pred_label_op, label_op], feed_dict={
83 | placeholders['data']: data,
84 | placeholders['labels']: labels
85 | })
86 |
87 | hit_count += np.sum( hits )
88 | total_count += len( data )
89 |
90 | for i, p in enumerate(pred):
91 | results.append( (p, lb[i]) )
92 |
93 | if step % FLAGS.log_interval == 0:
94 | duration = time.time() - start_time
95 | sec_per_batch = duration / FLAGS.log_interval
96 | examples_per_sec = FLAGS.batch_size / sec_per_batch
97 | logging('%s: [%d batches out of %d] (%.1f examples/sec; %.3f'
98 | 'sec/batch)' % (datetime.now(), step, total_batch_size,
99 | examples_per_sec, sec_per_batch), FLAGS)
100 | start_time = time.time()
101 |
102 |
103 | # micro precision
104 | logging("%s: micro-precision = %.5f" %
105 | (datetime.now(), (hit_count/total_count)), FLAGS)
106 |
107 |
108 | # write result
109 | outpath = os.path.join( FLAGS.log_dir, "out.txt" )
110 | with open(outpath, 'w') as fw:
111 | for p, l in results:
112 | fw.write("%d\t%d\n" % (int(l), int(p)))
113 |
114 |
115 |
116 |
117 | if __name__ == '__main__':
118 | FLAGS = argparser()
119 | FLAGS.is_training = False
120 | logging(str(FLAGS), FLAGS)
121 | test( FLAGS )
122 |
--------------------------------------------------------------------------------
/src/ProtVec/train.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 |
10 |
11 | import tensorflow as tf
12 | import numpy as np
13 | import tensorflow.contrib.slim as slim
14 |
15 | from utils import argparser, logging
16 | from dataset import DataSet, WordDict
17 | from model import get_placeholders, inference
18 |
19 |
20 |
21 | # ########## #
22 | #
23 | # Single-gpu version training.
24 | #
25 | # ########## #
26 | def train( FLAGS ):
27 | # first make bag of words
28 | worddict = WordDict(FLAGS.embedding_file)
29 |
30 | FLAGS.word_size = worddict.size
31 |
32 | # read data
33 | dataset = DataSet( fpath = FLAGS.train_file,
34 | n_classes = FLAGS.num_classes,
35 | wd = worddict,
36 | need_shuffle = True )
37 |
38 |
39 |
40 | with tf.Graph().as_default():
41 | global_step = tf.placeholder( tf.int32 )
42 | placeholders = get_placeholders(FLAGS)
43 |
44 | pred = inference( placeholders['data'], FLAGS,
45 | for_training=True )
46 |
47 | tf.losses.softmax_cross_entropy(placeholders['labels'], pred)
48 | loss = tf.losses.get_total_loss()
49 |
50 |
51 | _acc_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
52 | acc_op = tf.reduce_mean( tf.cast( _acc_op ,tf.float32 ) )
53 |
54 |
55 | train_op = tf.train.AdamOptimizer( FLAGS.learning_rate ).minimize( loss )
56 |
57 | # Create a saver.
58 | saver = tf.train.Saver(max_to_keep=None)
59 |
60 |
61 | with tf.Session() as sess:
62 | sess.run( tf.global_variables_initializer() )
63 |
64 | if tf.train.checkpoint_exists( FLAGS.prev_checkpoint_path ):
65 | restorer = tf.train.Saver()
66 | restorer.restore(sess, FLAGS.prev_checkpoint_path)
67 | logging('%s: Pre-trained model restored from %s' %
68 | (datetime.now(), FLAGS.prev_checkpoint_path), FLAGS)
69 | step = int(FLAGS.prev_checkpoint_path.split('/')[-1].split('-')[-1]) + 1
70 | else:
71 | step = 0
72 |
73 | for data, labels in dataset.iter_once( FLAGS.batch_size ):
74 | start_time = time.time()
75 | _, loss_val, acc_val = sess.run([train_op, loss, acc_op], feed_dict={
76 | placeholders['data']: data,
77 | placeholders['labels']: labels,
78 | global_step: step
79 | })
80 | duration = time.time() - start_time
81 |
82 | assert not np.isnan(loss_val), 'Model diverge'
83 |
84 | if step > 0 and step % FLAGS.log_interval == 0:
85 | examples_per_sec = FLAGS.batch_size / float(duration)
86 | format_str = ('%s: step %d, loss = %.2f, acc = %.2f (%.1f examples/sec; %.3f '
87 | 'sec/batch)')
88 | logging(format_str % (datetime.now(), step, loss_val, acc_val,
89 | examples_per_sec, duration), FLAGS)
90 |
91 | if step > 0 and step % FLAGS.save_interval == 0:
92 | saver.save(sess, FLAGS.checkpoint_path, global_step=step)
93 |
94 | # counter
95 | step += 1
96 |
97 | # save for last
98 | saver.save(sess, FLAGS.checkpoint_path, global_step=step-1)
99 |
100 |
101 |
102 | if __name__ == '__main__':
103 | FLAGS = argparser()
104 | FLAGS.is_training = True
105 | logging(str(FLAGS), FLAGS)
106 |
107 | train( FLAGS )
108 |
109 |
110 |
111 |
112 |
--------------------------------------------------------------------------------
/src/ProtVec/utils.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import os
3 |
4 |
5 |
6 | def argparser():
7 | parser = argparse.ArgumentParser()
8 | # for model
9 | parser.add_argument(
10 | '--embedding_file',
11 | type=str,
12 | help='ProtVec embedding file.'
13 | )
14 | parser.add_argument(
15 | '--regularizer',
16 | type=float,
17 | default=0.001,
18 | help='(Lambda value / 2) of L2 regularizer on weights connected to last layer (0 to exclude).'
19 | )
20 | parser.add_argument(
21 | '--num_classes',
22 | type=int,
23 | default=0,
24 | help='Number of classes.'
25 | )
26 | # for learning
27 | parser.add_argument(
28 | '--learning_rate',
29 | type=float,
30 | default=0.001,
31 | help='Initial learning rate.'
32 | )
33 | parser.add_argument(
34 | '--max_epoch',
35 | type=int,
36 | default=1,
37 | help='Number of epochs to train.'
38 | )
39 | parser.add_argument(
40 | '--batch_size',
41 | type=int,
42 | default=64,
43 | help='Batch size.'
44 | )
45 | parser.add_argument(
46 | '--train_file',
47 | type=str,
48 | default='/tmp/tensorflow/mnist/input_data',
49 | help='Directory for input data.'
50 | )
51 | parser.add_argument(
52 | '--test_file',
53 | type=str,
54 | default='/tmp/tensorflow/mnist/input_data',
55 | help='Directory for input data.'
56 | )
57 | parser.add_argument(
58 | '--prev_checkpoint_path',
59 | type=str,
60 | default='',
61 | help='Restore from pre-trained model if specified.'
62 | )
63 | parser.add_argument(
64 | '--checkpoint_path',
65 | type=str,
66 | default='',
67 | help='Path to write checkpoint file.'
68 | )
69 | parser.add_argument(
70 | '--log_dir',
71 | type=str,
72 | default='/tmp/tensorflow/mnist/logs/fully_connected_feed',
73 | help='Directory for log data.'
74 | )
75 | parser.add_argument(
76 | '--log_interval',
77 | type=int,
78 | default=100,
79 | help='Number of gpus to use'
80 | )
81 | parser.add_argument(
82 | '--save_interval',
83 | type=int,
84 | default=4000,
85 | help='Number of gpus to use'
86 | )
87 |
88 | FLAGS, unparsed = parser.parse_known_args()
89 |
90 | return FLAGS
91 |
92 |
93 |
94 |
95 |
96 |
97 | def logging(msg, FLAGS):
98 | fpath = os.path.join( FLAGS.log_dir, "log.txt" )
99 | with open( fpath, "a" ) as fw:
100 | fw.write("%s\n" % msg)
101 | print(msg)
--------------------------------------------------------------------------------
/src/util/draw_logo.py:
--------------------------------------------------------------------------------
1 | from __future__ import absolute_import
2 | from __future__ import division
3 | from __future__ import print_function
4 |
5 | import re
6 | import time
7 | from datetime import datetime
8 | import os
9 | import math
10 | import sys
11 | from collections import defaultdict
12 |
13 | import tensorflow as tf
14 | import numpy as np
15 | import tensorflow.contrib.slim as slim
16 |
17 |
18 | sys.path.append( os.path.join( os.path.dirname(__file__), "..", "DeepFam" ) )
19 | from utils import argparser
20 | from dataset import DataSet
21 | from model import get_placeholders, inference
22 |
23 |
24 |
25 |
26 | def test( FLAGS ):
27 | # read data
28 | dataset = DataSet( fpath = FLAGS.test_file,
29 | seqlen = FLAGS.seq_len,
30 | n_classes = FLAGS.num_classes,
31 | need_shuffle = False )
32 |
33 | FLAGS.charset_size = dataset.charset_size
34 |
35 | with tf.Graph().as_default():
36 | # placeholder
37 | placeholders = get_placeholders(FLAGS)
38 |
39 | # get inference
40 | pred, layers = inference( placeholders['data'], FLAGS,
41 | for_training=False )
42 |
43 | # calculate prediction
44 | label_op = tf.argmax(pred, 1)
45 | prob_op = tf.nn.softmax(pred)
46 | # _hit_op = tf.equal( tf.argmax(pred, 1), tf.argmax(placeholders['labels'], 1))
47 | # hit_op = tf.reduce_sum( tf.cast( _hit_op ,tf.float32 ) )
48 |
49 | # create saver
50 | saver = tf.train.Saver()
51 |
52 | # argmax of hidden1
53 | h1_argmax_ops = []
54 | for op in layers['conv']:
55 | h1_argmax_ops.append(tf.argmax(op, axis=2))
56 |
57 |
58 | with tf.Session() as sess:
59 | # load model
60 | # ckpt = tf.train.latest_checkpoint( os.path.dirname( FLAGS.checkpoint_path ) )
61 | ckpt = FLAGS.checkpoint_path
62 | if tf.train.checkpoint_exists( ckpt ):
63 | saver.restore( sess, ckpt )
64 | global_step = ckpt.split('/')[-1].split('-')[-1]
65 | print('Succesfully loaded model from %s at step=%s.' %
66 | (ckpt, global_step))
67 | else:
68 | print("[ERROR] Checkpoint not exist")
69 | return
70 |
71 |
72 | # iter batch
73 | hit_count = 0.0
74 | total_count = 0
75 |
76 | wlens = FLAGS.window_lengths
77 | hsizes = FLAGS.num_windows
78 | motif_matches = (defaultdict(list), defaultdict(list))
79 | pred_labels = []
80 | pred_prob = []
81 |
82 | print("%s: starting test." % (datetime.now()))
83 | start_time = time.time()
84 | total_batch_size = math.ceil( dataset._num_data / FLAGS.batch_size )
85 |
86 | for step, (data, labels, raws) in enumerate(dataset.iter_once( FLAGS.batch_size, with_raw=True )):
87 | res_run = sess.run( [label_op, prob_op, h1_argmax_ops] + layers['conv'], feed_dict={
88 | placeholders['data']: data,
89 | placeholders['labels']: labels
90 | })
91 |
92 | pred_label = res_run[0]
93 | pred_arr = res_run[1]
94 | max_idxs = res_run[2] # shape = (wlens, N, 1, # of filters)
95 | motif_filters = res_run[3:]
96 |
97 | for i, l in enumerate(pred_label):
98 | pred_labels.append(l)
99 | pred_prob.append( pred_arr[i][ 388 ] )
100 |
101 | # mf.shape = (N, 1, l-w+1, # of filters)
102 | for i in range(len(motif_filters)):
103 | s = motif_filters[i].shape
104 | motif_filters[i] = np.transpose( motif_filters[i], (0, 1, 3, 2) ).reshape( (s[0], s[3], s[2]) )
105 |
106 | # mf.shape = (N, # of filters, l-w+1)
107 | for gidx, mf in enumerate(motif_filters):
108 | wlen = wlens[gidx]
109 | hsize = hsizes[gidx]
110 | for ridx, row in enumerate(mf):
111 | for fidx, vals in enumerate(row):
112 | # for each filter, get max value and it's index
113 | max_idx = max_idxs[gidx][ridx][0][fidx]
114 | # max_idx = np.argmax(vals)
115 | max_val = vals[ max_idx ]
116 |
117 | hidx = gidx * hsize + fidx
118 |
119 | if max_val > 0:
120 | # get sequence
121 | rawseq = raws[ridx][1]
122 | subseq = rawseq[ max_idx : max_idx+wlen ]
123 | # heappush( top_matches[hidx], (max_val, subseq) )
124 | motif_matches[0][hidx].append( max_val )
125 | motif_matches[1][hidx].append( subseq )
126 | # motif_matches[gidx][fidx][0].append( max_val )
127 | # motif_matches[gidx][fidx][1].append( subseq )
128 |
129 |
130 | # hit_count += np.sum( hits )
131 | total_count += len( data )
132 | # print("total:%d" % total_count)
133 |
134 | if step % FLAGS.log_interval == 0:
135 | duration = time.time() - start_time
136 | sec_per_batch = duration / FLAGS.log_interval
137 | examples_per_sec = FLAGS.batch_size / sec_per_batch
138 | print('%s: [%d batches out of %d] (%.1f examples/sec; %.3f'
139 | 'sec/batch)' % (datetime.now(), step, total_batch_size,
140 | examples_per_sec, sec_per_batch))
141 | start_time = time.time()
142 |
143 | # if step > 10:
144 | # break
145 |
146 |
147 | # # micro precision
148 | # print("%s: micro-precision = %.5f" %
149 | # (datetime.now(), (hit_count/total_count)))
150 |
151 | print(pred_labels)
152 |
153 | ### sort top lists
154 | # report whose activation was higher
155 | mean_acts = {}
156 | on_acts = {}
157 | print('%s: write result to file' % (datetime.now()) )
158 | for fidx in motif_matches[0]:
159 | val_lst = motif_matches[0][fidx]
160 | seq_lst = motif_matches[1][fidx]
161 | # top k
162 | # k = wlens[ int(fidx / hsize) ] * 25
163 | k = 30
164 | # l = min(k, len(val_lst)) * -1
165 | l = len(val_lst) * -1
166 | tidxs = np.argpartition(val_lst, l)[l:]
167 | # tracking acts
168 | acts = 0.0
169 |
170 | opath = os.path.join(FLAGS.motif_outpath, "p%d.txt"%fidx)
171 | with open(opath, 'w') as fw:
172 | for idx in tidxs:
173 | fw.write("%f\t%s\n" % (val_lst[idx], seq_lst[idx]) )
174 | acts += val_lst[idx]
175 |
176 | mean_acts[fidx] = acts / l * -1
177 | on_acts[fidx] = len(val_lst)
178 |
179 | if fidx % 50 == 0:
180 | print('%s: [%d filters out of %d]' % (datetime.now(), fidx, sum(FLAGS.num_windows)))
181 | # print(len(val_lst))
182 |
183 | # report mean acts
184 | with open(os.path.join(FLAGS.motif_outpath, "report.txt"), 'w') as fw:
185 | for i in sorted(on_acts, key=on_acts.get, reverse=True):
186 | fw.write("%f\t%f\t%d\n" % (on_acts[i] / total_count, mean_acts[i], i))
187 |
188 | with open(os.path.join(FLAGS.motif_outpath, "predictions.txt"), 'w') as fw:
189 | for i, p in enumerate(pred_labels):
190 | fw.write("%s\t%f\n" % (str(p), pred_prob[i]))
191 |
192 |
193 |
194 |
195 |
196 |
197 |
198 | if __name__ == '__main__':
199 | FLAGS = argparser()
200 | FLAGS.is_training = False
201 | FLAGS.motif_outpath = FLAGS.log_dir
202 | if not os.path.exists(FLAGS.motif_outpath):
203 | os.mkdir(FLAGS.motif_outpath)
204 |
205 | test( FLAGS )
206 |
--------------------------------------------------------------------------------
/src/util/draw_logo.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | # $1: ps name
4 |
5 | basepath="/home/kimlab/project/DeepFam/data/prosite"
6 | utilpath=$(dirname "$0")
7 |
8 | # iFile="${basepath}/$1/data.txt"
9 | iFile="${basepath}/$1/pure_data.txt"
10 | oPath="${basepath}/$1/res"
11 | lPath="${basepath}/$1/logo"
12 |
13 | mkdir -p $lPath
14 |
15 | seqlen=$(head -n 1 "${basepath}/$1/trans.txt" | cut -f 2)
16 |
17 |
18 | export CUDA_VISIBLE_DEVICES=0; \
19 | python "${utilpath}/draw_logo.py" \
20 | --num_classes 1074 \
21 | --seq_len $seqlen \
22 | --window_lengths 8 12 16 20 24 28 32 36 \
23 | --num_windows 250 250 250 250 250 250 250 250 \
24 | --num_hidden 2000 \
25 | --max_epoch=25 \
26 | --train_file=$iFile \
27 | --test_file=$iFile \
28 | --batch_size=10 \
29 | --checkpoint_pat /home/kimlab/project/DeepFam/result/cog_cv/90percent/save/model.ckpt-203219 \
30 | --log_dir=$oPath
31 |
32 |
33 |
34 | # draw logo
35 | cat "$oPath/report.txt" | \
36 | awk -v OFS="\t" '{if($1>=0.7) print $2, $3}' | \
37 | sort -rn | cut -f 2 | head -n 30 | \
38 | xargs -P 10 -I {} sh -c "cut -f 2 $oPath/p{}.txt | grep -v _ | weblogo -F pdf -A protein -s large -t '[DeepFam] {}' > $lPath/l{}.pdf"
39 | #head -n 20 "$oPath/report.txt" | cut -f 2 | xargs -P 10 -I {} sh -c "echo {}.txt"
40 |
41 |
--------------------------------------------------------------------------------
/src/util/preprocess.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import os
3 | import sys
4 | import numpy as np
5 | import csv
6 | from collections import defaultdict
7 | import re
8 |
9 |
10 | def read_fasta(fasta_file):
11 | input = open(fasta_file, 'r')
12 |
13 | chrom_seq = ''
14 | chrom_id = None
15 |
16 | for line in input:
17 | if line[0] == '>':
18 | if chrom_id is not None:
19 | yield chrom_id, chrom_seq
20 |
21 | chrom_seq = ''
22 | chrom_id = line.split()[0][1:].replace("|", "_")
23 | else:
24 | chrom_seq += line.strip().upper()
25 |
26 | yield chrom_id, chrom_seq
27 |
28 | input.close()
29 |
30 |
31 |
32 |
33 | def read_data(sample_file):
34 | maxlen = 0
35 | data = []
36 | for sid, seq in read_fasta(sample_file):
37 | data.append( (sid, seq) )
38 | maxlen = max(maxlen, len(seq))
39 |
40 | return data, maxlen
41 |
42 |
43 |
44 |
45 | if __name__ == '__main__':
46 | target_path = sys.argv[1]
47 | sample_file = os.path.join(target_path, "prot.fasta")
48 |
49 |
50 | data, maxlen = read_data( sample_file )
51 |
52 | # parsing data
53 | trans_path = os.path.join(target_path, "trans.txt")
54 | data_path = os.path.join(target_path, "data.txt")
55 |
56 | maxlen = 1000
57 | with open(trans_path, 'w') as transw, open(data_path, 'w') as dataw:
58 | # first write maxlen
59 | transw.write("MAXLEN\t%d\n" % (maxlen))
60 | for idx, (pid, seq) in enumerate(data):
61 | if len(seq) <= maxlen:
62 | transw.write("%s\t%d\n" % (pid, idx))
63 |
64 | padded_seq = seq + "_"*(maxlen - len(seq))
65 | dataw.write("0\t%s\n" % (padded_seq) )
66 |
67 |
--------------------------------------------------------------------------------
/src/util/prosite_purity.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import os
3 | import sys
4 | import numpy as np
5 | import csv
6 | from collections import defaultdict
7 | import re
8 |
9 |
10 | def read_data(sample_file):
11 | data = []
12 |
13 | with open(sample_file, 'r') as fr:
14 | for line in fr.readlines():
15 | fam, seq = line.strip().split("\t")
16 | data.append( (fam, seq.replace("_", "")) )
17 |
18 | return data
19 |
20 |
21 |
22 |
23 | if __name__ == '__main__':
24 | f1 = sys.argv[1] # f1 is reference
25 | f2 = sys.argv[2]
26 | f3 = sys.argv[3] # f3 is output
27 | f4 = sys.argv[4] # f4 is filtered seqs
28 |
29 | d1 = read_data(f1)
30 | d2 = read_data(f2)
31 |
32 | intersect = []
33 |
34 | for _, s1 in d1:
35 | for _, s2 in d2:
36 | if s1 == s2:
37 | intersect.append(s1)
38 |
39 |
40 | print("Intersection : %d" % len(intersect))
41 |
42 | with open(f3, 'w') as fw, open(f4, 'w') as flfw:
43 | with open(f2, 'r') as fr:
44 | for line in fr.readlines():
45 | fam, seq = line.strip().split("\t")
46 | rawseq = seq.replace("_", "")
47 |
48 | if rawseq not in intersect:
49 | fw.write("%s\t%s\n" % (fam, seq))
50 | else:
51 | flfw.write("%s\t%s\n" % (fam, seq))
52 |
--------------------------------------------------------------------------------
/src/util/run_weblogo.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | weblogo -F png_print -D array -A protein -s large -t logo-737 <(cut -f 2 ps51657/res/p737.txt | grep -v _ )> tlogo.png
4 |
--------------------------------------------------------------------------------