├── scripts
├── att_test.sh
├── test.sh
├── train.sh
└── att_train.sh
├── imgs
├── gcn_web.png
└── pytorch.png
├── LICENSE
├── .gitignore
├── test.py
├── opts.py
├── models.py
├── layers.py
├── train.py
├── utils.py
└── README.md
/scripts/att_test.sh:
--------------------------------------------------------------------------------
1 | python test.py \
2 | --dataset cora \
3 | --model attention \
4 |
--------------------------------------------------------------------------------
/scripts/test.sh:
--------------------------------------------------------------------------------
1 | python test.py \
2 | --dataset pubmed \
3 | --model residual \
4 |
--------------------------------------------------------------------------------
/imgs/gcn_web.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/bmsookim/graph-cnn.pytorch/HEAD/imgs/gcn_web.png
--------------------------------------------------------------------------------
/imgs/pytorch.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/bmsookim/graph-cnn.pytorch/HEAD/imgs/pytorch.png
--------------------------------------------------------------------------------
/scripts/train.sh:
--------------------------------------------------------------------------------
1 | python train.py \
2 | --dataset pubmed \
3 | --num_hidden 32 \
4 | --dropout 0.5 \
5 | --weight_decay 0 \
6 | --model basic \
7 | --lr 1e-2 \
8 | --optimizer sgd \
9 | --epoch 10000 \
10 | --lr_decay_epoch 2500
11 |
--------------------------------------------------------------------------------
/scripts/att_train.sh:
--------------------------------------------------------------------------------
1 | python train.py \
2 | --dataset cora \
3 | --num_hidden 8 \
4 | --nb_heads 8 \
5 | --dropout 0.6 \
6 | --weight_decay 5e-4 \
7 | --model res_attention \
8 | --lr 5e-3 \
9 | --optimizer adam \
10 | --epoch 800
11 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2018 Bumsoo Kim
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | *.egg-info/
24 | .installed.cfg
25 | *.egg
26 | MANIFEST
27 |
28 | # PyInstaller
29 | # Usually these files are written by a python script from a template
30 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
31 | *.manifest
32 | *.spec
33 |
34 | # Installer logs
35 | pip-log.txt
36 | pip-delete-this-directory.txt
37 |
38 | # Unit test / coverage reports
39 | htmlcov/
40 | .tox/
41 | .coverage
42 | .coverage.*
43 | .cache
44 | nosetests.xml
45 | coverage.xml
46 | *.cover
47 | .hypothesis/
48 | .pytest_cache/
49 |
50 | # Translations
51 | *.mo
52 | *.pot
53 |
54 | # Django stuff:
55 | *.log
56 | local_settings.py
57 | db.sqlite3
58 |
59 | # Flask stuff:
60 | instance/
61 | .webassets-cache
62 |
63 | # Scrapy stuff:
64 | .scrapy
65 |
66 | # Sphinx documentation
67 | docs/_build/
68 |
69 | # PyBuilder
70 | target/
71 |
72 | # Jupyter Notebook
73 | .ipynb_checkpoints
74 |
75 | # pyenv
76 | .python-version
77 |
78 | # celery beat schedule file
79 | celerybeat-schedule
80 |
81 | # SageMath parsed files
82 | *.sage.py
83 |
84 | # Environments
85 | .env
86 | .venv
87 | env/
88 | venv/
89 | ENV/
90 | env.bak/
91 | venv.bak/
92 |
93 | # Spyder project settings
94 | .spyderproject
95 | .spyproject
96 |
97 | # Rope project settings
98 | .ropeproject
99 |
100 | # mkdocs documentation
101 | /site
102 |
103 | # mypy
104 | .mypy_cache/
105 |
106 | # checkpoint
107 | ./checkpoint/*
108 | *.t7
109 |
--------------------------------------------------------------------------------
/test.py:
--------------------------------------------------------------------------------
1 | # ************************************************************
2 | # Author : Bumsoo Kim, 2018
3 | # Github : https://github.com/meliketoy/graph-cnn.pytorch
4 | #
5 | # Korea University, Data-Mining Lab
6 | # Graph Convolutional Neural Network
7 | #
8 | # Description : test.py
9 | # The main code for testing graph classification networks.
10 | # ***********************************************************
11 |
12 | import time
13 | import os
14 | import numpy as np
15 | import torch
16 | import torch.nn.functional as F
17 | import torch.optim as optim
18 |
19 | from utils import *
20 | from models import GCN
21 | from opts import TestOptions
22 |
23 | """
24 | N : number of nodes
25 | D : number of features per node
26 | E : number of classes
27 |
28 | @ input :
29 | - adjacency matrix (N x N)
30 | - feature matrix (N x D)
31 | - label matrix (N x E)
32 |
33 | @ dataset :
34 | - citeseer
35 | - cora
36 | - pubmed
37 | """
38 | opt = TestOptions().parse()
39 |
40 | adj, features, labels, idx_train, idx_val, idx_test = load_data(path=opt.dataroot, dataset=opt.dataset)
41 | use_gpu = torch.cuda.is_available()
42 |
43 | print("\n[STEP 2] : Obtain (adjacency, feature, label) matrix")
44 | print("| Adjacency matrix : {}".format(adj.shape))
45 | print("| Feature matrix : {}".format(features.shape))
46 | print("| Label matrix : {}".format(labels.shape))
47 |
48 | load_model = torch.load(os.path.join('checkpoint', opt.dataset, '%s.t7' %(opt.model)))
49 | model = load_model['model'].cpu()
50 | acc_val = load_model['acc']
51 |
52 | if use_gpu:
53 | _, features, adj, labels, idx_test = \
54 | list(map(lambda x: x.cuda(), [model, features, adj, labels, idx_test]))
55 |
56 | def test():
57 | print("\n[STEP 4] : Testing")
58 |
59 | model.eval()
60 | output = model(features, adj)
61 |
62 | print(output[idx_test].shape)
63 | print(labels[idx_test].shape)
64 |
65 | acc_test = accuracy(output[idx_test], labels[idx_test])
66 | print("| Validation acc : {}%".format(acc_val.data.cpu().numpy() * 100))
67 | print("| Test acc : {}%\n".format(acc_test.data.cpu().numpy() * 100))
68 |
69 | if __name__ == "__main__":
70 | test()
71 |
--------------------------------------------------------------------------------
/opts.py:
--------------------------------------------------------------------------------
1 | import argparse
2 | import os
3 | import torch
4 |
5 | class BaseOptions():
6 | def __init__(self):
7 | self.parser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
8 | self.initialized = False
9 |
10 | def initialize(self):
11 | self.parser.add_argument('--dataroot', type=str, default='/home/bumsoo/Data/Planetoid', help='path')
12 | self.parser.add_argument('--dataset', type=str, default='pubmed', help='[cora | citeseer | pubmed]')
13 | self.parser.add_argument('--num_hidden', type=int, default=8, help='number of features')
14 | self.parser.add_argument('--dropout', type=float, default=0.6, help='dropout')
15 | self.parser.add_argument('--weight_decay', type=float, default=5e-4, help='weight decay')
16 | self.parser.add_argument('--init_type', type=str, default='uniform', help='[uniform | xavier]')
17 | self.parser.add_argument('--model', type=str, default='basic', help='[basic | drop_in | attention | res_attention]')
18 |
19 | def parse(self):
20 | if not self.initialized:
21 | self.initialize()
22 |
23 | self.opt = self.parser.parse_args()
24 | self.opt.isTrain = self.isTrain
25 | args = vars(self.opt)
26 |
27 | return self.opt
28 |
29 | class TrainOptions(BaseOptions):
30 | # Override
31 | def initialize(self):
32 | BaseOptions.initialize(self)
33 | self.parser.add_argument('--lr', type=float, default=5e-3, help='initial learning rate')
34 | self.parser.add_argument('--optimizer', type=str, default='adam', help='[sgd | adam]')
35 | self.parser.add_argument('--epoch', type=int, default=800, help='number of training epochs')
36 | self.parser.add_argument('--lr_decay_epoch', type=int, default=5000, help='multiply by a gamma every set iter')
37 | self.parser.add_argument('--nb_heads', type=int, default=8, help='number of head attentions')
38 | self.parser.add_argument('--alpha', type=float, default=0.2, help='Alpha value for the leaky_relu')
39 | self.isTrain = True
40 |
41 | class TestOptions(BaseOptions):
42 | def initialize(self):
43 | BaseOptions.initialize(self)
44 | self.isTrain = False
45 |
--------------------------------------------------------------------------------
/models.py:
--------------------------------------------------------------------------------
1 | import torch
2 | import torch.nn as nn
3 | import torch.nn.functional as F
4 | from layers import GraphConvolution, GraphAttention
5 |
6 | class GCN(nn.Module):
7 | def __init__(self, nfeat, nhid, nclass, dropout, init):
8 | super(GCN, self).__init__()
9 |
10 | self.gc1 = GraphConvolution(nfeat, nhid, init=init)
11 | self.gc2 = GraphConvolution(nhid, nclass, init=init)
12 | self.dropout = dropout
13 |
14 | def bottleneck(self, path1, path2, path3, adj, in_x):
15 | return F.relu(path3(F.relu(path2(F.relu(path1(in_x, adj)), adj)), adj))
16 |
17 | def forward(self, x, adj):
18 | x = F.dropout(F.relu(self.gc1(x, adj)), self.dropout, training=self.training)
19 | x = self.gc2(x, adj)
20 |
21 | return F.log_softmax(x, dim=1)
22 |
23 | class GCN_drop_in(nn.Module):
24 | def __init__(self, nfeat, nhid, nclass, dropout, init):
25 | super(GCN_drop_in, self).__init__()
26 |
27 | self.gc1 = GraphConvolution(nfeat, nhid, init=init)
28 | self.gc2 = GraphConvolution(nhid, nclass, init=init)
29 | self.dropout = dropout
30 |
31 | def bottleneck(self, path1, path2, path3, adj, in_x):
32 | return F.relu(path3(F.relu(path2(F.relu(path1(in_x, adj)), adj)), adj))
33 |
34 | def forward(self, x, adj):
35 | x = F.dropout(x, self.dropout, training=self.training)
36 | x = F.dropout(F.relu(self.gc1(x, adj)), self.dropout, training=self.training)
37 | x = self.gc2(x, adj)
38 |
39 | return F.log_softmax(x, dim=1)
40 |
41 | class GAT(nn.Module):
42 | def __init__(self, nfeat, nhid, nclass, dropout, alpha, nheads):
43 | super(GAT, self).__init__()
44 | self.dropout = dropout
45 |
46 | self.attentions = [GraphAttention(nfeat, nhid, dropout=dropout, alpha=alpha, concat=True) for _ in range(nheads)]
47 | for i, attention in enumerate(self.attentions):
48 | self.add_module('attention_{}'.format(i), attention)
49 |
50 | self.out_att = GraphAttention(nhid * nheads, nclass, dropout=dropout, alpha=alpha, concat=False)
51 |
52 | def forward(self, x, adj):
53 | x = F.dropout(x, self.dropout, training=self.training)
54 | x = torch.cat([att(x, adj) for att in self.attentions], dim=1)
55 | x = F.dropout(x, self.dropout, training=self.training)
56 | x = F.elu(self.out_att(x, adj))
57 | return F.log_softmax(x, dim=1)
58 |
--------------------------------------------------------------------------------
/layers.py:
--------------------------------------------------------------------------------
1 | import math
2 | import torch
3 | import numpy as np
4 | import torch.nn as nn
5 | import torch.nn.functional as F
6 | from torch.nn.parameter import Parameter
7 | from torch.nn.modules.module import Module
8 |
9 | class GraphConvolution(Module):
10 | """
11 | Simple GCN layer, similar to https://arxiv.org/abs/1609.02907
12 | """
13 |
14 | def __init__(self, in_features, out_features, bias=True, init='xavier'):
15 | super(GraphConvolution, self).__init__()
16 | self.in_features = in_features
17 | self.out_features = out_features
18 | self.weight = Parameter(torch.FloatTensor(in_features, out_features))
19 | if bias:
20 | self.bias = Parameter(torch.FloatTensor(out_features))
21 | else:
22 | self.register_parameter('bias', None)
23 | if init == 'uniform':
24 | print("| Uniform Initialization")
25 | self.reset_parameters_uniform()
26 | elif init == 'xavier':
27 | print("| Xavier Initialization")
28 | self.reset_parameters_xavier()
29 | elif init == 'kaiming':
30 | print("| Kaiming Initialization")
31 | self.reset_parameters_kaiming()
32 | else:
33 | raise NotImplementedError
34 |
35 | def reset_parameters_uniform(self):
36 | stdv = 1. / math.sqrt(self.weight.size(1))
37 | self.weight.data.uniform_(-stdv, stdv)
38 | if self.bias is not None:
39 | self.bias.data.uniform_(-stdv, stdv)
40 |
41 | def reset_parameters_xavier(self):
42 | nn.init.xavier_normal_(self.weight.data, gain=0.02) # Implement Xavier Uniform
43 | if self.bias is not None:
44 | nn.init.constant_(self.bias.data, 0.0)
45 |
46 | def reset_parameters_kaiming(self):
47 | nn.init.kaiming_normal_(self.weight.data, a=0, mode='fan_in')
48 | if self.bias is not None:
49 | nn.init.constant_(self.bias.data, 0.0)
50 |
51 | def forward(self, input, adj):
52 | support = torch.mm(input, self.weight)
53 | output = torch.spmm(adj, support)
54 | if self.bias is not None:
55 | return output + self.bias
56 | else:
57 | return output
58 |
59 | def __repr__(self):
60 | return self.__class__.__name__ + ' (' \
61 | + str(self.in_features) + ' -> ' \
62 | + str(self.out_features) + ')'
63 |
64 |
65 | class GraphAttention(nn.Module):
66 | """
67 | Simple GAT layer, similar to https://arxiv.org/abs/1710.10903
68 | """
69 |
70 | def __init__(self, in_features, out_features, dropout, alpha, concat=True):
71 | super(GraphAttention, self).__init__()
72 | self.dropout = dropout
73 | self.in_features = in_features
74 | self.out_features = out_features
75 | self.alpha = alpha
76 | self.concat = concat
77 |
78 | self.W = nn.Parameter(nn.init.xavier_normal_(torch.Tensor(in_features, out_features).type(torch.cuda.FloatTensor if torch.cuda.is_available() else torch.FloatTensor), gain=np.sqrt(2.0)), requires_grad=True)
79 | self.a1 = nn.Parameter(nn.init.xavier_normal_(torch.Tensor(out_features, 1).type(torch.cuda.FloatTensor if torch.cuda.is_available() else torch.FloatTensor), gain=np.sqrt(2.0)), requires_grad=True)
80 | self.a2 = nn.Parameter(nn.init.xavier_normal_(torch.Tensor(out_features, 1).type(torch.cuda.FloatTensor if torch.cuda.is_available() else torch.FloatTensor), gain=np.sqrt(2.0)), requires_grad=True)
81 |
82 | self.leakyrelu = nn.LeakyReLU(self.alpha)
83 |
84 | def forward(self, input, adj):
85 | h = torch.mm(input, self.W)
86 | N = h.size()[0]
87 |
88 | f_1 = torch.matmul(h, self.a1)
89 | f_2 = torch.matmul(h, self.a2)
90 | e = self.leakyrelu(f_1 + f_2.transpose(0,1))
91 |
92 | zero_vec = -9e15*torch.ones_like(e)
93 | attention = torch.where(adj > 0, e, zero_vec)
94 | attention = F.softmax(attention, dim=1)
95 | attention = F.dropout(attention, self.dropout, training=self.training)
96 | h_prime = torch.matmul(attention, h)
97 |
98 | if self.concat:
99 | return F.elu(h_prime)
100 | else:
101 | return h_prime
102 |
103 | def __repr__(self):
104 | return self.__class__.__name__ + ' (' + str(self.in_features) + ' -> ' + str(self.out_features) + ')'
105 |
--------------------------------------------------------------------------------
/train.py:
--------------------------------------------------------------------------------
1 | # ************************************************************
2 | # Author : Bumsoo Kim, 2018
3 | # Github : https://github.com/meliketoy/graph-cnn.pytorch
4 | #
5 | # Korea University, Data-Mining Lab
6 | # Graph Convolutional Neural Network
7 | #
8 | # Description : train.py
9 | # The main code for training classification networks.
10 | # ***********************************************************
11 |
12 | import time
13 | import random
14 | import os
15 | import sys
16 | import numpy as np
17 | import torch
18 | import torch.nn.functional as F
19 | import torch.optim as optim
20 |
21 | from torch.autograd import Variable
22 | from utils import *
23 | from models import GCN, GAT
24 | from opts import TrainOptions
25 |
26 | """
27 | N : number of nodes
28 | D : number of features per node
29 | E : number of classes
30 |
31 | @ input :
32 | - adjacency matrix (N x N)
33 | - feature matrix (N x D)
34 | - label matrix (N x E)
35 |
36 | @ dataset :
37 | - citeseer
38 | - cora
39 | - pubmed
40 | """
41 | opt = TrainOptions().parse()
42 |
43 | # Data upload
44 | adj, features, labels, idx_train, idx_val, idx_test = load_data(path=opt.dataroot, dataset=opt.dataset)
45 | use_gpu = torch.cuda.is_available()
46 |
47 | random.seed(42)
48 | np.random.seed(42)
49 | torch.manual_seed(42)
50 | if use_gpu:
51 | torch.cuda.manual_seed(42)
52 |
53 | model, optimizer = None, None
54 | best_acc = 0
55 |
56 | # Define the model and optimizer
57 | if (opt.model == 'basic'):
58 | print("| Constructing basic GCN model...")
59 | model = GCN(
60 | nfeat = features.shape[1],
61 | nhid = opt.num_hidden,
62 | nclass = labels.max().item() + 1,
63 | dropout = opt.dropout,
64 | init = opt.init_type
65 | )
66 | elif (opt.model == 'attention'):
67 | print("| Constructing Attention GCN model...")
68 | model = GAT(
69 | nfeat = features.shape[1],
70 | nhid = opt.num_hidden,
71 | nclass = int(labels.max().item()) + 1,
72 | dropout = opt.dropout,
73 | nheads = opt.nb_heads,
74 | alpha = opt.alpha
75 | )
76 | else:
77 | raise NotImplementedError
78 |
79 | if (opt.optimizer == 'sgd'):
80 | optimizer = optim.SGD(
81 | model.parameters(),
82 | lr = opt.lr,
83 | weight_decay = opt.weight_decay,
84 | momentum = 0.9
85 | )
86 | elif (opt.optimizer == 'adam'):
87 | optimizer = optim.Adam(
88 | model.parameters(),
89 | lr = opt.lr,
90 | weight_decay = opt.weight_decay
91 | )
92 | else:
93 | raise NotImplementedError
94 |
95 | if use_gpu:
96 | model.cuda()
97 | features, adj, labels, idx_train, idx_val, idx_test = \
98 | list(map(lambda x: x.cuda(), [features, adj, labels, idx_train, idx_val, idx_test]))
99 |
100 | features, adj, labels = list(map(lambda x : Variable(x), [features, adj, labels]))
101 |
102 | if not os.path.isdir('checkpoint'):
103 | os.mkdir('checkpoint')
104 |
105 | save_point = os.path.join('./checkpoint', opt.dataset)
106 |
107 | if not os.path.isdir(save_point):
108 | os.mkdir(save_point)
109 |
110 | def lr_scheduler(epoch, opt):
111 | return opt.lr * (0.5 ** (epoch / opt.lr_decay_epoch))
112 |
113 | # Train
114 | def train(epoch):
115 | global best_acc
116 |
117 | t = time.time()
118 | model.train()
119 | optimizer.lr = lr_scheduler(epoch, opt)
120 | optimizer.zero_grad()
121 |
122 | output = model(features, adj)
123 | loss_train = F.nll_loss(output[idx_train], labels[idx_train])
124 | acc_train = accuracy(output[idx_train], labels[idx_train])
125 |
126 | loss_train.backward()
127 | optimizer.step()
128 |
129 | # Validation for each epoch
130 | model.eval()
131 | output = model(features, adj)
132 | loss_val = F.nll_loss(output[idx_val], labels[idx_val])
133 | acc_val = accuracy(output[idx_val], labels[idx_val])
134 |
135 | if acc_val > best_acc:
136 | best_acc = acc_val
137 | state = {
138 | 'model': model,
139 | 'acc': best_acc,
140 | 'epoch': epoch,
141 | }
142 |
143 | torch.save(state, os.path.join(save_point, '%s.t7' %(opt.model)))
144 |
145 | sys.stdout.flush()
146 | sys.stdout.write('\r')
147 | sys.stdout.write("=> Training Epoch #{} : lr = {:.4f}".format(epoch, optimizer.lr))
148 | sys.stdout.write(" | Training acc : {:6.2f}%".format(acc_train.data.cpu().numpy() * 100))
149 | sys.stdout.write(" | Best acc : {:.2f}%". format(best_acc.data.cpu().numpy() * 100))
150 |
151 |
152 | # Main code for training
153 | if __name__ == "__main__":
154 | print("\n[STEP 2] : Obtain (adjacency, feature, label) matrix")
155 | print("| Adjacency matrix : {}".format(adj.shape))
156 | print("| Feature matrix : {}".format(features.shape))
157 | print("| Label matrix : {}".format(labels.shape))
158 |
159 | # Training
160 | print("\n[STEP 3] : Training")
161 | for epoch in range(1, opt.epoch+1):
162 | train(epoch)
163 | print("\n=> Training finished!")
164 |
--------------------------------------------------------------------------------
/utils.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pickle as pkl
3 | import networkx as nx
4 | import scipy.sparse as sp
5 | import torch
6 | from scipy.sparse import csgraph
7 |
8 | def parse_index_file(filename):
9 | index = []
10 |
11 | for line in open(filename):
12 | index.append(int(line.strip()))
13 |
14 | return index
15 |
16 | def normalize(mx):
17 | """Row-normalize sparse matrix"""
18 | rowsum = np.array(mx.sum(1))
19 | r_inv = np.power(rowsum, -1).flatten()
20 | r_inv[np.isinf(r_inv)] = 0.
21 | r_mat_inv = sp.diags(r_inv)
22 | mx = r_mat_inv.dot(mx)
23 |
24 | return mx
25 |
26 | def normalize_adj(mx):
27 | """Row-normalize sparse matrix"""
28 | rowsum = np.array(mx.sum(1))
29 | r_inv_sqrt = np.power(rowsum, -0.5).flatten()
30 | r_inv_sqrt[np.isinf(r_inv_sqrt)] = 0.
31 | r_mat_inv_sqrt = sp.diags(r_inv_sqrt)
32 |
33 | return mx.dot(r_mat_inv_sqrt).transpose().dot(r_mat_inv_sqrt).tocoo()
34 |
35 | def laplacian(mx, norm):
36 | """Laplacian-normalize sparse matrix"""
37 | assert (all (len(row) == len(mx) for row in mx)), "Input should be a square matrix"
38 |
39 | return csgraph.laplacian(adj, normed = norm)
40 |
41 | def accuracy(output, labels):
42 | preds = output.max(1)[1].type_as(labels)
43 | correct = preds.eq(labels).double()
44 | correct = correct.sum()
45 | return correct / len(labels)
46 |
47 | def load_data(path="/home/bumsoo/Data/Planetoid", dataset="cora"):
48 | """
49 | ind.[:dataset].x => the feature vectors of the training instances (scipy.sparse.csr.csr_matrix)
50 | ind.[:dataset].y => the one-hot labels of the labeled training instances (numpy.ndarray)
51 | ind.[:dataset].allx => the feature vectors of both labeled and unlabeled training instances (csr_matrix)
52 | ind.[:dataset].ally => the labels for instances in ind.dataset_str.allx (numpy.ndarray)
53 | ind.[:dataset].graph => the dict in the format {index: [index of neighbor nodes]} (collections.defaultdict)
54 |
55 | ind.[:dataset].tx => the feature vectors of the test instances (scipy.sparse.csr.csr_matrix)
56 | ind.[:dataset].ty => the one-hot labels of the test instances (numpy.ndarray)
57 |
58 | ind.[:dataset].test.index => indices of test instances in graph, for the inductive setting
59 | """
60 | print("\n[STEP 1]: Upload {} dataset.".format(dataset))
61 |
62 | names = ['x', 'y', 'tx', 'ty', 'allx', 'ally', 'graph']
63 | objects = []
64 |
65 | for i in range(len(names)):
66 | with open("{}/ind.{}.{}".format(path, dataset, names[i]), 'rb') as f:
67 | objects.append(pkl.load(f))
68 |
69 | x, y, tx, ty, allx, ally, graph = tuple(objects)
70 |
71 | test_idx_reorder = parse_index_file("{}/ind.{}.test.index".format(path, dataset))
72 | test_idx_range = np.sort(test_idx_reorder)
73 |
74 | if dataset == 'citeseer':
75 | #Citeseer dataset contains some isolated nodes in the graph
76 | test_idx_range_full = range(min(test_idx_reorder), max(test_idx_reorder)+1)
77 | tx_extended = sp.lil_matrix((len(test_idx_range_full), x.shape[1]))
78 | tx_extended[test_idx_range-min(test_idx_range), :] = tx
79 | tx = tx_extended
80 |
81 | ty_extended = np.zeros((len(test_idx_range_full), y.shape[1]))
82 | ty_extended[test_idx_range-min(test_idx_range), :] = ty
83 | ty = ty_extended
84 |
85 | features = sp.vstack((allx, tx)).tolil()
86 | features[test_idx_reorder, :] = features[test_idx_range, :]
87 |
88 | adj = nx.adjacency_matrix(nx.from_dict_of_lists(graph))
89 | print("| # of nodes : {}".format(adj.shape[0]))
90 | print("| # of edges : {}".format(adj.sum().sum()/2))
91 |
92 | features = normalize(features)
93 | adj = normalize_adj(adj + sp.eye(adj.shape[0]))
94 | print("| # of features : {}".format(features.shape[1]))
95 | print("| # of clases : {}".format(ally.shape[1]))
96 |
97 | features = torch.FloatTensor(np.array(features.todense()))
98 | sparse_mx = adj.tocoo().astype(np.float32)
99 | adj = torch.FloatTensor(np.array(adj.todense()))
100 |
101 | labels = np.vstack((ally, ty))
102 | labels[test_idx_reorder, :] = labels[test_idx_range, :]
103 |
104 | if dataset == 'citeseer':
105 | save_label = np.where(labels)[1]
106 | labels = torch.LongTensor(np.where(labels)[1])
107 |
108 | idx_train = range(len(y))
109 | idx_val = range(len(y), len(y)+500)
110 | idx_test = test_idx_range.tolist()
111 |
112 | print("| # of train set : {}".format(len(idx_train)))
113 | print("| # of val set : {}".format(len(idx_val)))
114 | print("| # of test set : {}".format(len(idx_test)))
115 |
116 | idx_train, idx_val, idx_test = list(map(lambda x: torch.LongTensor(x), [idx_train, idx_val, idx_test]))
117 |
118 | def missing_elements(L):
119 | start, end = L[0], L[-1]
120 | return sorted(set(range(start, end+1)).difference(L))
121 |
122 | if dataset == 'citeseer':
123 | L = np.sort(idx_test)
124 | missing = missing_elements(L)
125 |
126 | for element in missing:
127 | save_label = np.insert(save_label, element, 0)
128 |
129 | labels = torch.LongTensor(save_label)
130 |
131 | return adj, features, labels, idx_train, idx_val, idx_test
132 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | 
2 |
3 | Pytorch implementation of Graph Convolution Networks & Graph Attention Convolutional Networks.
4 |
5 | This project is made by Bumsoo Kim, Ph.D Candidate in Korea University.
6 | This repo has been forked from [https://github.com/tkipf/pygcn](https://github.com/tkipf/pygcn).
7 |
8 | ## Graph Convolutional Networks
9 | Many important real-world datasets come in the form of graphs or networks: social networks, knowledge graphs, protein-interaction networks, the World Wide Web, etc. In this repository, we introduce a basic tutorial for generalizing neural netowrks to work on arbitrarily structured graphs, along with Graph Attention Convolutional Networks([Attention GCN](https://arxiv.org/abs/1710.10903)).
10 |
11 | Currently, most graph neural network models have a somewhat universal architecture in common. They are referred as Graph Convoutional Networks(GCNs) since filter parameters are typically shared over all locations in the graph.
12 |
13 | 
14 |
15 | For these models, the goal is to learn a function of signals/features on a graph G=(V, E), which takes as
16 |
17 | **Input**
18 | - N x D feature matrix (N : Number of nodes, D : number of input features)
19 | - representative description of the graph structure in matrix form; typically in the form of adjacency matrix A
20 |
21 | **Output**
22 | - N x F feature matrix (N : Number of nodes, F : number of output features)
23 |
24 | Graph-level outputs can be modeled by introducing some form of pooling operation.
25 |
26 | Every neural network layer can then be written as a non-linear function
27 |
28 | 
29 |
30 | with  and 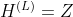, where ***L*** is the number of layers. The specific models then differ only in how function ***f*** is chosen and parameterized.
31 |
32 | In this repo, the layer-wise propagation is consisted as
33 |
34 | 
35 |
36 | As the activation function is a non-linear ReLU (Rectified Linear Unit), this becomes
37 |
38 | 
39 |
40 | **Implementation detail #1 :**
41 |
42 | Multiplication with ***A*** means that, for every node, we sum up all the feature vectors of all neighboring nodes but not the node itself. To address this, we add the identity matrix to ***A***.
43 |
44 | **Implementation detail #2 :**
45 |
46 | ***A*** is typically not normalized and therfore the multiplication and therefore the multiplication with ***A*** will completely change the scale of the feature vectors. Normalizing A such that all rows sum to one, i.e. 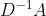.
47 |
48 |
49 | **Final Implementation :**
50 |
51 | Combining the two implementation details above gives us a final propagation rule introduced in [Kipf & Welling](http://arxiv.org/abs/1609.02907) (ICLR 2017).
52 |
53 | 
54 |
55 | For more details, see [here](https://tkipf.github.io/graph-convolutional-networks/).
56 |
57 | ## Requirements
58 | See the [installation instruction](INSTALL.md) for a step-by-step installation guide.
59 | See the [server instruction](SERVER.md) for server settup.
60 | - Install [cuda-8.0](https://developer.nvidia.com/cuda-downloads)
61 | - Install [cudnn v5.1](https://developer.nvidia.com/cudnn)
62 | - Download [Pytorch for python-2.7](https://pytorch.org) and clone the repository.
63 | - Install python package 'networkx'
64 |
65 | ```bash
66 | pip install http://download.pytorch.org/whl/cu80/torch-0.1.12.post2-cp27-none-linux_x86_64.whl
67 | pip install torchvision
68 | git clone https://github.com/meliketoy/graph-cnn.pytorch
69 | pip install networkx
70 | ```
71 |
72 | ## Planetoid Dataset
73 | In this repo, we use an implementation of Planetoid, a graph-based sem-supervised learning method proposed in the following paper: [Revisiting Semi-Supervised Learning with Graph Embeddings](https://arxiv.org/abs/1603.08861).
74 |
75 | This dataset is consisted of 3 sub-datasets ('pubmed', 'cora', 'citeseer')
76 |
77 | Each node in the dataset represents a document, and the edge represents the 'reference' relationship between the documents.
78 |
79 | The data
80 |
81 | ### Transductive learning
82 | - x : the feature vectors of the training instances
83 | - y : the one-hot labels of the training instances
84 | - graph : {index: [index of neighber nodes]}, where the neighbor nodes are given as a list.
85 |
86 | ### Inductive learning
87 | - x : the feature vectors of the labeled training instances
88 | - y : the one-hot labels of the training instances
89 | - allx : the feature vectors of both labeled and unlabeled training instances.
90 | - graph : {index: [index of neighber nodes]}, where the neighbor nodes are given as a list.
91 |
92 | For more details, see [here](https://github.com/kimiyoung/planetoid)
93 |
94 | ## Train network
95 | After you have cloned the repository, you can train the dataset by running the script below.
96 |
97 | Download the planetoid datset above and give the [:dir to dataset] the directory to the downloaded datset.
98 |
99 | ```bash
100 | python train.py --dataroot [:dir to dataset] --datset [:cora | citeseer | pubmed] --model [:basic|drop_in]
101 | ```
102 |
103 | ## Test (Inference) various networks
104 | After you have finished training, you can test out your network by
105 |
106 | ```bash
107 | python test.py --dataroot [:dir to dataset] --dataset [:cora | citeseer | pubmed] --model [:basic|drop_in]
108 | ```
109 |
110 | Enjoy :-)
111 |
--------------------------------------------------------------------------------


