hist(rnorm(1000))
29 |
31 | 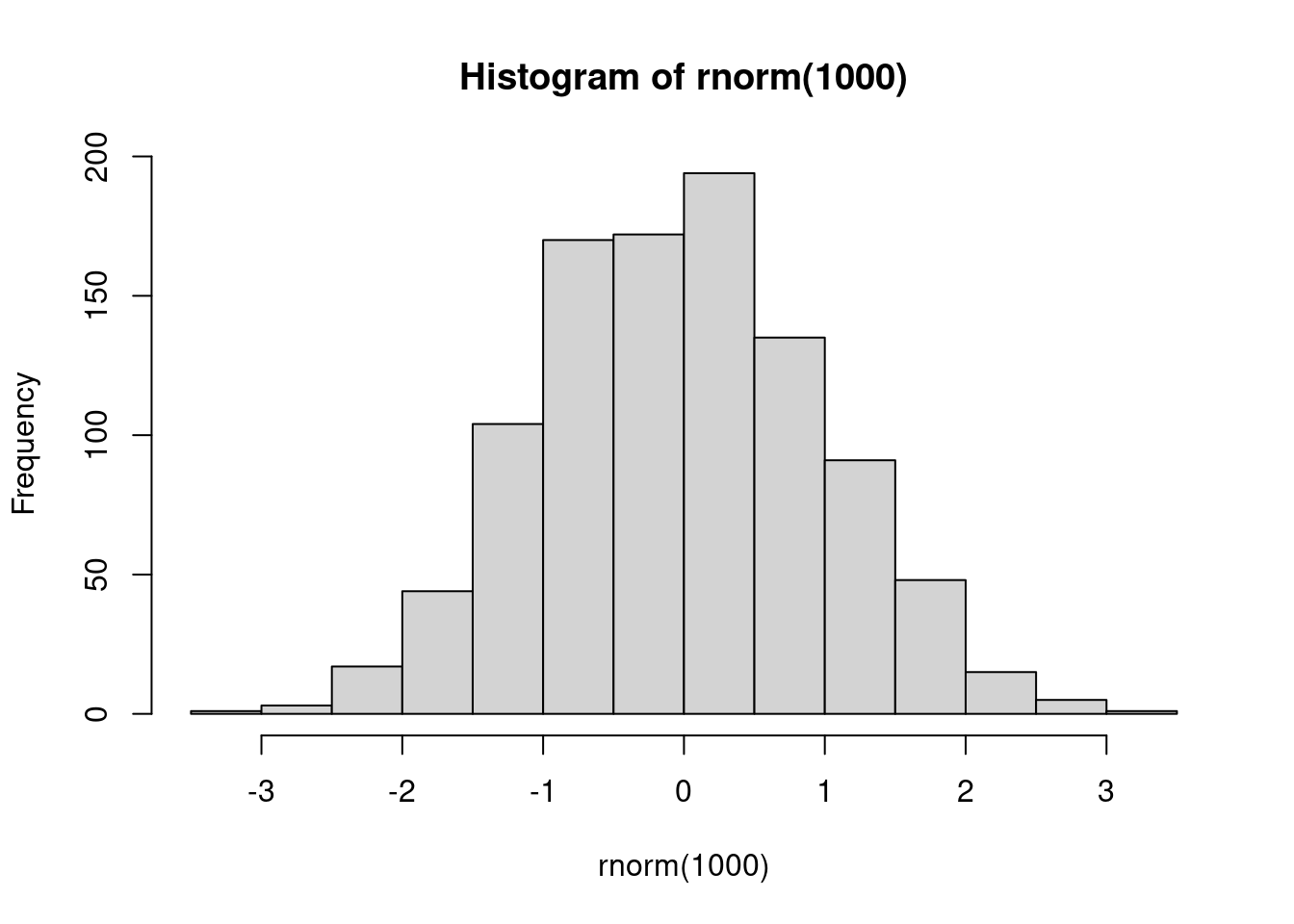
A test post including some R code.
26 |hist(rnorm(1000))
Blah.
33 | 34 | 35 | 36 | 37 | ]]>A test post including some python code
55 |x = [1,2,3]
57 | print(x[1])
58 |
59 | import matplotlib.pyplot as plt
60 | fig, axis = plt.subplots()
61 | axis.plot([0,1,2,3,4,5], [3,4,6,5,2,4])2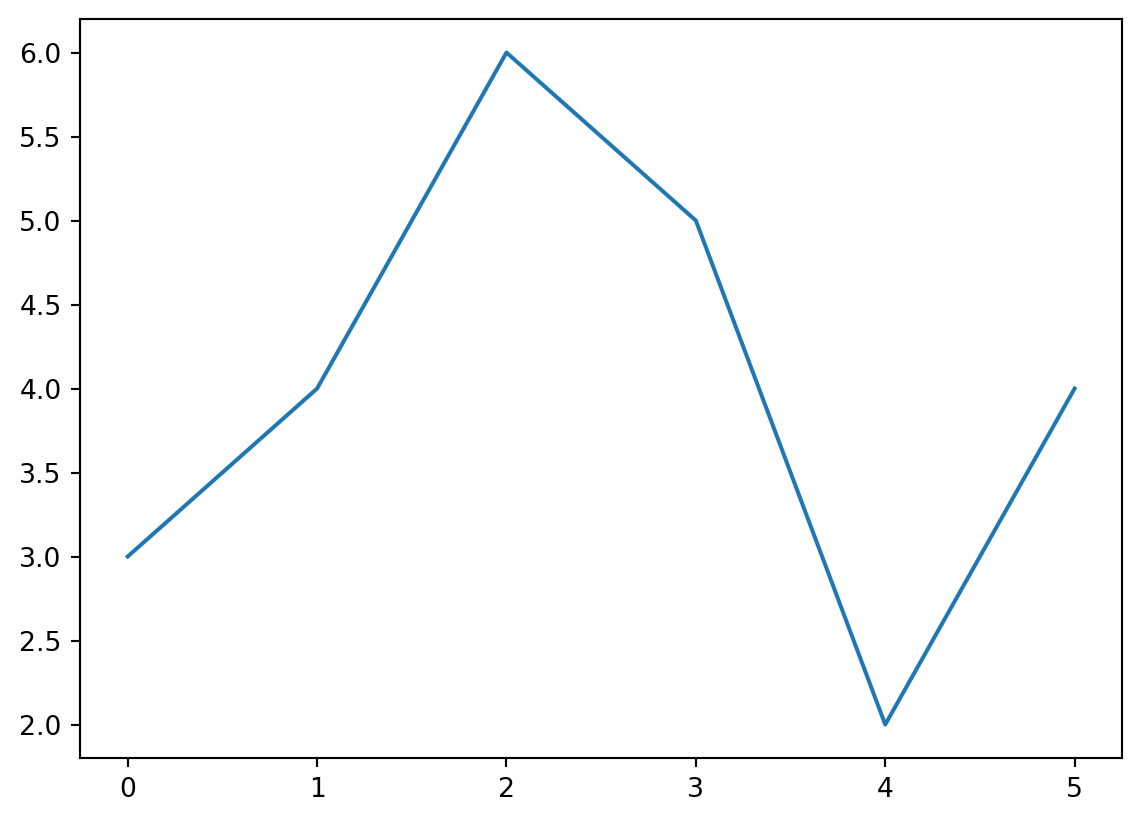
Blah.
70 | 71 | 72 | 73 | 74 | ]]>/[sourcecode language="scala" light="true"]\n/g' | \
11 | sed 's|/[sourcecode language="scala" light="true"]\n/g' | \
10 | sed 's||\n[/sourcecode]|g' | \
11 | sed 's/"/"/g' | \
12 | sed 's/>/>/g' | \
13 | sed 's/</ meanVar}
10 | import breeze.linalg.DenseVector
11 |
12 | // Normal random sample
13 | def example1 = {
14 | val mod = for {
15 | mu <- Normal(0,100)
16 | v <- Gamma(1,0.1)
17 | _ <- Normal(mu,v).fitQ(List(8.0,9,7,7,8,10))
18 | } yield (mu,v)
19 | val modEmp = mod.empirical
20 | print("mu : ")
21 | println(meanVar(modEmp map (_._1)))
22 | print("v : ")
23 | println(meanVar(modEmp map (_._2)))
24 | }
25 |
26 | // Normal random sample - IG on v
27 | def example2 = {
28 | val mod = for {
29 | mu <- Normal(0, 100)
30 | tau <- Gamma(1, 0.1)
31 | _ <- Normal(mu, 1.0/tau).fitQ(List(8.0,9,7,7,8,10))
32 | } yield (mu,tau)
33 | val modEmp = mod.empirical
34 | print("mu : ")
35 | println(meanVar(modEmp map (_._1)))
36 | print("tau : ")
37 | println(meanVar(modEmp map (_._2)))
38 | }
39 |
40 | // Poisson DGLM
41 | def example3 = {
42 |
43 | val data = List(2,1,0,2,3,4,5,4,3,2,1)
44 |
45 | val prior = for {
46 | w <- Gamma(1, 1)
47 | state0 <- Normal(0.0, 2.0)
48 | } yield (w, List(state0))
49 |
50 | def addTimePoint(current: Prob[(Double, List[Double])],
51 | obs: Int): Prob[(Double, List[Double])] = {
52 | println(s"Conditioning on observation: $obs")
53 | for {
54 | tup <- current
55 | (w, states) = tup
56 | os = states.head
57 | ns <- Normal(os, w)
58 | _ <- Poisson(math.exp(ns)).fitQ(obs)
59 | } yield (w, ns :: states)
60 | }
61 |
62 | val mod = data.foldLeft(prior)(addTimePoint(_,_)).empirical
63 | print("w : ")
64 | println(meanVar(mod map (_._1)))
65 | print("s0 : ")
66 | println(meanVar(mod map (_._2.reverse.head)))
67 | print("sN : ")
68 | println(meanVar(mod map (_._2.head)))
69 |
70 | }
71 |
72 | // Linear model
73 | def example4 = {
74 | val x = List(1.0,2,3,4,5,6)
75 | val y = List(3.0,2,4,5,5,6)
76 | val xy = x zip y
77 | case class Param(alpha: Double, beta: Double, v: Double)
78 | println("Forming prior distribution")
79 | val prior = for {
80 | alpha <- Normal(0,10)
81 | beta <- Normal(0,4)
82 | v <- Gamma(1,0.1)
83 | } yield Param(alpha, beta, v)
84 | def addPoint(current: Prob[Param], obs: (Double, Double)): Prob[Param] = {
85 | println(s"Conditioning on $obs")
86 | for {
87 | p <- current
88 | (x, y) = obs
89 | _ <- Normal(p.alpha + p.beta * x, p.v).fitQ(y)
90 | } yield p
91 | }
92 | val mod = xy.foldLeft(prior)(addPoint(_,_)).empirical
93 | print("a : ")
94 | println(meanVar(mod map (_.alpha)))
95 | print("b : ")
96 | println(meanVar(mod map (_.beta)))
97 | print("v : ")
98 | println(meanVar(mod map (_.v)))
99 | }
100 |
101 | // Noisy observations of a count
102 | def example5 = {
103 | val mod = for {
104 | count <- Poisson(10)
105 | tau <- Gamma(1,0.1)
106 | _ <- Normal(count,1.0/tau).fitQ(List(4.2,5.1,4.6,3.3,4.7,5.3))
107 | } yield (count,tau)
108 | val modEmp = mod.empirical
109 | print("count : ")
110 | println(meanVar(modEmp map (_._1.toDouble)))
111 | print("tau : ")
112 | println(meanVar(modEmp map (_._2)))
113 | }
114 |
115 |
116 | // Main entry point
117 |
118 | def main(args: Array[String]): Unit = {
119 | println("Hi")
120 | example1
121 | example2
122 | example3
123 | example4
124 | example5
125 | println("Bye")
126 | }
127 |
128 | }
129 |
130 | // eof
131 |
132 |
--------------------------------------------------------------------------------
/min-ppl/src/main/scala/min-ppl.scala:
--------------------------------------------------------------------------------
1 | object MinPpl {
2 |
3 | import breeze.stats.{distributions => bdist}
4 | import breeze.linalg.DenseVector
5 |
6 | implicit val numParticles = 300
7 |
8 | case class Particle[T](v: T, lw: Double) { // value and log-weight
9 | def map[S](f: T => S): Particle[S] = Particle(f(v), lw)
10 | }
11 |
12 | trait Prob[T] {
13 | val particles: Vector[Particle[T]]
14 | def map[S](f: T => S): Prob[S] = Empirical(particles map (_ map f))
15 | def flatMap[S](f: T => Prob[S]): Prob[S] = {
16 | Empirical((particles map (p => {
17 | f(p.v).particles.map(psi => Particle(psi.v, p.lw + psi.lw))
18 | })).flatten).resample
19 | }
20 | def resample(implicit N: Int): Prob[T] = {
21 | val lw = particles map (_.lw)
22 | val mx = lw reduce (math.max(_,_))
23 | val rw = lw map (lwi => math.exp(lwi - mx))
24 | val law = mx + math.log(rw.sum/(rw.length))
25 | val ind = bdist.Multinomial(DenseVector(rw.toArray)).sample(N)
26 | val newParticles = ind map (i => particles(i))
27 | Empirical(newParticles.toVector map (pi => Particle(pi.v, law)))
28 | }
29 | def cond(ll: T => Double): Prob[T] =
30 | Empirical(particles map (p => Particle(p.v, p.lw + ll(p.v))))
31 | def empirical: Vector[T] = resample.particles.map(_.v)
32 | }
33 |
34 | case class Empirical[T](particles: Vector[Particle[T]]) extends Prob[T]
35 |
36 | def unweighted[T](ts: Vector[T], lw: Double = 0.0): Prob[T] =
37 | Empirical(ts map (Particle(_, lw)))
38 |
39 | trait Dist[T] extends Prob[T] {
40 | def ll(obs: T): Double
41 | def ll(obs: Seq[T]): Double = obs map (ll) reduce (_+_)
42 | def fit(obs: Seq[T]): Prob[T] =
43 | Empirical(particles map (p => Particle(p.v, p.lw + ll(obs))))
44 | def fitQ(obs: Seq[T]): Prob[T] = Empirical(Vector(Particle(obs.head, ll(obs))))
45 | def fit(obs: T): Prob[T] = fit(List(obs))
46 | def fitQ(obs: T): Prob[T] = fitQ(List(obs))
47 | }
48 |
49 | case class Normal(mu: Double, v: Double)(implicit N: Int) extends Dist[Double] {
50 | lazy val particles = unweighted(bdist.Gaussian(mu, math.sqrt(v)).sample(N).toVector).particles
51 | def ll(obs: Double) = bdist.Gaussian(mu, math.sqrt(v)).logPdf(obs)
52 | }
53 |
54 | case class Gamma(a: Double, b: Double)(implicit N: Int) extends Dist[Double] {
55 | lazy val particles = unweighted(bdist.Gamma(a, 1.0/b).sample(N).toVector).particles
56 | def ll(obs: Double) = bdist.Gamma(a, 1.0/b).logPdf(obs)
57 | }
58 |
59 | case class Poisson(mu: Double)(implicit N: Int) extends Dist[Int] {
60 | lazy val particles = unweighted(bdist.Poisson(mu).sample(N).toVector).particles
61 | def ll(obs: Int) = bdist.Poisson(mu).logProbabilityOf(obs)
62 | }
63 |
64 | }
65 |
--------------------------------------------------------------------------------
/min-ppl/src/test/scala/min-ppl-test.scala:
--------------------------------------------------------------------------------
1 | /*
2 | min-ppl-test.scala
3 |
4 | Some basic sanity checks on the language
5 |
6 | */
7 |
8 | import org.scalatest.flatspec.AnyFlatSpec
9 | import org.scalactic._
10 | import MinPpl._
11 | import breeze.stats.{meanAndVariance => meanVar}
12 |
13 | class PplSpec extends AnyFlatSpec with Tolerance {
14 |
15 | "A linear Gaussian" should "flatMap correctly" in {
16 | System.err.println("**These tests take a LONG time, and it is normal for a couple to fail**")
17 | val xy = for {
18 | x <- Normal(5,4)
19 | y <- Normal(x,1)
20 | } yield (x,y)
21 | val y = xy.map(_._2).empirical
22 | val mv = meanVar(y)
23 | assert(mv.mean === 5.0 +- 0.5)
24 | assert(mv.variance === 5.0 +- 1.0)
25 | }
26 |
27 | it should "cond correctly" in {
28 | val xy = for {
29 | x <- Normal(5,4)
30 | y <- Normal(x,1)
31 | } yield (x,y)
32 | val y = xy.map(_._2)
33 | val yGz = y.cond(yi => Normal(yi, 9).ll(8.0)).empirical
34 | val mv = meanVar(yGz)
35 | assert(mv.mean === 5.857 +- 0.5)
36 | assert(mv.variance === 2.867 +- 1.0)
37 | val xyGz = xy.cond{case (x,y) => Normal(y,9).ll(8.0)}.empirical
38 | val mvx = meanVar(xyGz.map(_._1))
39 | assert(mvx.mean === 5.857 +- 0.5)
40 | assert(mvx.variance === 2.867 +- 1.0)
41 | val mvy = meanVar(xyGz.map(_._2))
42 | assert(mvy.mean === 6.071 +- 0.5)
43 | assert(mvy.variance === 3.214 +- 1.0)
44 | }
45 |
46 | it should "cond correctly in a for" in {
47 | val wxyz = for {
48 | w <- Normal(5,2)
49 | x <- Normal(w,2)
50 | y <- Normal(x,1).cond(y => Normal(y,9).ll(8.0))
51 | } yield (w,x,y)
52 | val wxyze = wxyz.empirical
53 | val mvw = meanVar(wxyze.map(_._1))
54 | assert(mvw.mean === 5.429 +- 0.5)
55 | assert(mvw.variance === 1.714 +- 1.0)
56 | val mvx = meanVar(wxyze.map(_._2))
57 | assert(mvx.mean === 5.857 +- 0.5)
58 | assert(mvx.variance === 2.867 +- 1.0)
59 | val mvy = meanVar(wxyze.map(_._3))
60 | assert(mvy.mean === 6.071 +- 0.5)
61 | assert(mvy.variance === 3.214 +- 1.0)
62 | }
63 |
64 | it should "fit correctly" in {
65 | val xyzf = for {
66 | x <- Normal(5,4)
67 | y <- Normal(x,1)
68 | z <- Normal(y,9).fit(8.0)
69 | } yield (x,y,z)
70 | val xyzfe = xyzf.empirical
71 | val mvx = meanVar(xyzfe.map(_._1))
72 | assert(mvx.mean === 5.857 +- 0.5)
73 | assert(mvx.variance === 2.867 +- 1.0)
74 | val mvy = meanVar(xyzfe.map(_._2))
75 | assert(mvy.mean === 6.071 +- 0.5)
76 | assert(mvy.variance === 3.214 +- 1.0)
77 | val mvz = meanVar(xyzfe.map(_._3))
78 | assert(mvz.mean === 6.071 +- 0.5)
79 | assert(mvz.variance === 12.214 +- 2.0)
80 | }
81 |
82 | it should "fitQ correctly" in {
83 | val xyzfq = for {
84 | x <- Normal(5,4)
85 | y <- Normal(x,1)
86 | z <- Normal(y,9).fitQ(8.0)
87 | } yield (x,y,z)
88 | val xyzfqe = xyzfq.empirical
89 | val mvx = meanVar(xyzfqe.map(_._1))
90 | assert(mvx.mean === 5.857 +- 0.5)
91 | assert(mvx.variance === 2.867 +- 1.0)
92 | val mvy = meanVar(xyzfqe.map(_._2))
93 | assert(mvy.mean === 6.071 +- 0.5)
94 | assert(mvy.variance === 3.214 +- 1.0)
95 | val mvz = meanVar(xyzfqe.map(_._3))
96 | assert(mvz.mean === 8.000 +- 0.001)
97 | assert(mvz.variance === 0.000 +- 0.001)
98 | }
99 |
100 | it should "fit marginalised correctly" in {
101 | val yzf = for {
102 | y <- Normal(5,5)
103 | z <- Normal(y,9).fit(8.0)
104 | } yield (y,z)
105 | val yzfe = yzf.empirical
106 | val mvy = meanVar(yzfe.map(_._1))
107 | assert(mvy.mean === 6.071 +- 0.5)
108 | assert(mvy.variance === 3.213 +- 1.0)
109 | val mvz = meanVar(yzfe.map(_._2))
110 | assert(mvz.mean === 6.071 +- 0.5)
111 | assert(mvz.variance === 12.214 +- 2.0)
112 | }
113 |
114 | it should "fit multiple iid observations correctly" in {
115 | val yzf2 = for {
116 | y <- Normal(5,5)
117 | z <- Normal(y,18).fit(List(6.0,10.0))
118 | } yield (y,z)
119 | val yzfe2 = yzf2.empirical
120 | val mvy = meanVar(yzfe2.map(_._1))
121 | assert(mvy.mean === 6.071 +- 0.5)
122 | assert(mvy.variance === 3.214 +- 1.0)
123 | val mvz = meanVar(yzfe2.map(_._2))
124 | assert(mvz.mean === 6.071 +- 0.5)
125 | assert(mvz.variance === 21.214 +- 2.5)
126 | }
127 |
128 | it should "deep chain correctly" in {
129 | val deep = for {
130 | w <- Normal(2.0,1.0)
131 | x <- Normal(w,1)
132 | y <- Normal(x,2)
133 | z <- Normal(y,1)
134 | } yield z
135 | val mvz = meanVar(deep.empirical)
136 | assert(mvz.mean === 2.0 +- 0.5)
137 | assert(mvz.variance === 5.0 +- 1.0)
138 | }
139 |
140 | }
141 |
142 |
--------------------------------------------------------------------------------
/min-ppl2/.gitignore:
--------------------------------------------------------------------------------
1 | # .gitignore for scala projects
2 |
3 | # Classes and logs
4 | *.class
5 | *.log
6 | *~
7 |
8 | # SBT-specific
9 | .cache
10 | .history
11 | .classpath
12 | .project
13 | .settings
14 |
15 | .lib/
16 | dist/*
17 | target/
18 | lib_managed/
19 | src_managed/
20 | project/boot/
21 | project/plugins/project/
22 |
23 | # Ensime specific
24 | .ensime
25 |
26 | # Scala-IDE specific
27 | .scala_dependencies
28 | .worksheet
29 |
30 |
31 |
--------------------------------------------------------------------------------
/min-ppl2/Makefile:
--------------------------------------------------------------------------------
1 | # Makefile
2 |
3 |
4 | DraftPost.html: src/main/tut/DraftPost.md
5 | make tut
6 | pandoc -t html5 target/scala-2.13/tut/DraftPost.md -o DraftPost.html
7 |
8 | tut:
9 | sbt tut
10 |
11 | edit:
12 | emacs Makefile build.sbt *.md src/test/scala/*.scala src/main/scala/*.scala src/main/tut/*.md &
13 |
14 |
15 | # eof
16 |
--------------------------------------------------------------------------------
/min-ppl2/Readme.md:
--------------------------------------------------------------------------------
1 | # min-ppl2
2 |
3 | ## [A probability monad for the bootstrap particle filter](https://darrenjw.wordpress.com/2019/08/10/a-probability-monad-for-the-bootstrap-particle-filter/)
4 |
5 | If you have (a recent JDK, and) [sbt](https://www.scala-sbt.org/) installed, you can compile and run the examples with `sbt run`, or run some tests with `sbt test` (slow), or compile the [tut](http://tpolecat.github.io/tut/) document that formed the draft of the post with `sbt tut`.
6 |
7 | If you are a statistician or data scientist interested to learn more about Scala, note that I have a free on-line course available: [Scala for statistical computing and data science](https://github.com/darrenjw/scala-course/blob/master/StartHere.md)
8 |
9 |
10 | Copyright (C) 2019 [Darren J Wilkinson](https://darrenjw.github.io/)
11 |
12 |
--------------------------------------------------------------------------------
/min-ppl2/build.sbt:
--------------------------------------------------------------------------------
1 | name := "min-ppl2"
2 |
3 | version := "0.1-SNAPSHOT"
4 |
5 | scalacOptions ++= Seq(
6 | "-unchecked", "-deprecation", "-feature"
7 | )
8 |
9 | libraryDependencies ++= Seq(
10 | "org.scalatest" %% "scalatest" % "3.1.0-SNAP13" % "test",
11 | "org.scalactic" %% "scalactic" % "3.0.8" % "test",
12 | "org.typelevel" %% "cats-core" % "2.0.0-RC1",
13 | "org.scalanlp" %% "breeze" % "1.0-RC4",
14 | //"org.scalanlp" %% "breeze-viz" % "1.0-RC4",
15 | "org.scalanlp" %% "breeze-natives" % "1.0-RC4"
16 | )
17 |
18 | resolvers ++= Seq(
19 | "Sonatype Snapshots" at
20 | "https://oss.sonatype.org/content/repositories/snapshots/",
21 | "Sonatype Releases" at
22 | "https://oss.sonatype.org/content/repositories/releases/"
23 | )
24 |
25 | enablePlugins(TutPlugin)
26 |
27 | scalaVersion := "2.13.0"
28 |
29 |
--------------------------------------------------------------------------------
/min-ppl2/md2wp:
--------------------------------------------------------------------------------
1 | #!/bin/sh
2 | # md2wp
3 | # convert github flavoured markdown to wordpress html
4 |
5 | cat $1 | \
6 | sed 's/```scala/```/g' | \
7 | sed 's/```bash/```/g' | \
8 | pandoc -f markdown_github -t html5 | \
9 | sed 's//[sourcecode language="scala" light="true"]\n/g' | \
10 | sed 's|
11 |  12 |
12 |
15 | Latest movie 16 |
17 | 18 | 19 | 20 | 21 | 22 | -------------------------------------------------------------------------------- /pi-cam/web-setup.sh: -------------------------------------------------------------------------------- 1 | # web-setup.sh 2 | # run-once web setup script 3 | 4 | sudo apt-get -y update 5 | sudo apt-get -y install lighttpd 6 | 7 | sudo chown www-data:www-data /var/www/html 8 | sudo chmod 775 /var/www/html 9 | sudo usermod -a -G www-data pi 10 | 11 | sudo cp index.html /var/www/html/ 12 | sudo chown www-data:www-data /var/www/html/index.html 13 | sudo chmod g+w /var/www/html/index.html 14 | 15 | # eof 16 | 17 | 18 | -------------------------------------------------------------------------------- /pi-cluster/README.md: -------------------------------------------------------------------------------- 1 | # Raspberry Pi 2 cluster with NAT routing head node 2 | 3 | Scripts and config files associated with my blog post "Raspberry Pi 2 cluster with NAT routing": 4 | 5 | https://darrenjw2.wordpress.com/2015/09/07/raspberry-pi-2-cluster-with-nat-routing/ 6 | 7 | The brief summary is as follows: 8 | 9 | Create a cluster by connecting a bunch of Pis to a switch via the Pis ethernet port. Pick one of the Pis to be a head node and NAT router. Connect a USB ethernet dongle to this Pi, and use the dongle port as the internet uplink. 10 | 11 | Stick Raspbian on each node, with SSH server enabled. 12 | 13 | Boot up the head node. 14 | 15 | ```bash 16 | wget https://github.com/darrenjw/blog/archive/master.zip 17 | unzip master.zip 18 | cd blog-master/pi-cluster 19 | sudo sh install-packages 20 | ``` 21 | 22 | will reboot when done. On reboot, re-enter same directory, and then do: 23 | 24 | ```sudo sh setup-network``` 25 | 26 | when done, will reboot. 27 | 28 | On reboot, re-enter same directory. Boot up the other nodes and then run 29 | 30 | ```sh setup-cluster``` 31 | 32 | on the head node. 33 | 34 | 35 | -------------------------------------------------------------------------------- /pi-cluster/copy-keys: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | awk '{print "ssh-copy-id " $1}' < workers.txt > /tmp/copy-keys 4 | sh /tmp/copy-keys 5 | 6 | # eof 7 | 8 | -------------------------------------------------------------------------------- /pi-cluster/dhcpd.conf: -------------------------------------------------------------------------------- 1 | # /etc/dhcp/dhcpd.conf 2 | 3 | authoritative; 4 | 5 | subnet 192.168.0.0 netmask 255.255.255.0 { 6 | range 192.168.0.10 192.168.0.250; 7 | option broadcast-address 192.168.0.255; 8 | option routers 192.168.0.1; 9 | default-lease-time 600; 10 | max-lease-time 7200; 11 | option domain-name "local"; 12 | option domain-name-servers 8.8.8.8, 8.8.4.4; 13 | } 14 | 15 | 16 | # eof 17 | 18 | 19 | -------------------------------------------------------------------------------- /pi-cluster/install-packages: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | apt-get update && sudo apt-get -y upgrade 4 | apt-get -y install nmap isc-dhcp-server pssh 5 | reboot 6 | 7 | # eof 8 | 9 | -------------------------------------------------------------------------------- /pi-cluster/interfaces: -------------------------------------------------------------------------------- 1 | # /etc/network/interfaces 2 | 3 | auto lo 4 | iface lo inet loopback 5 | 6 | # dongle uplink 7 | auto eth1 8 | iface eth1 inet dhcp 9 | 10 | # internal gateway 11 | auto eth0 12 | iface eth0 inet static 13 | address 192.168.0.1 14 | netmask 255.255.255.0 15 | network 192.168.0.0 16 | broadcast 192.168.0.255 17 | 18 | # eof 19 | 20 | -------------------------------------------------------------------------------- /pi-cluster/iptables: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | iptables-restore < /etc/iptables.up.rules 4 | 5 | exit 0 6 | 7 | # eof 8 | 9 | -------------------------------------------------------------------------------- /pi-cluster/map-network: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | nmap -sn 192.168.0.0/24 -oG - | grep -v "^#" | cut -d " " -f 2 > all-hosts.txt 4 | grep -v "^192.168.0.1$" < all-hosts.txt > workers.txt 5 | cp workers.txt ~/ 6 | 7 | # eof 8 | 9 | -------------------------------------------------------------------------------- /pi-cluster/setup-cluster: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | ssh-keygen 4 | 5 | sh map-network 6 | sh copy-keys 7 | sh upgrade-workers 8 | 9 | # eof 10 | 11 | 12 | -------------------------------------------------------------------------------- /pi-cluster/setup-network: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | # basic interfaces 4 | cp interfaces /etc/network/ 5 | /etc/init.d/networking restart 6 | 7 | # DHCP server 8 | cp /etc/dhcp/dhcpd.conf /etc/dhcp/dhcpd.conf.old 9 | cp dhcpd.conf /etc/dhcp/ 10 | echo 'INTERFACES="eth0"' >> /etc/default/isc-dhcp-server 11 | /etc/init.d/isc-dhcp-server restart 12 | 13 | # NAT routing 14 | echo 1 > /proc/sys/net/ipv4/ip_forward 15 | echo "net.ipv4.ip_forward=1" >> /etc/sysctl.conf 16 | iptables -t nat -A POSTROUTING -o eth1 -j MASQUERADE 17 | # TODO: add more rules here for extra security... 18 | 19 | ifdown eth1 && ifup eth1 20 | iptables-save > /etc/iptables.up.rules 21 | 22 | cp iptables /etc/network/if-pre-up.d/ 23 | chown root:root /etc/network/if-pre-up.d/iptables 24 | chmod 755 /etc/network/if-pre-up.d/iptables 25 | 26 | reboot 27 | 28 | # eof 29 | 30 | -------------------------------------------------------------------------------- /pi-cluster/shutdown-workers: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | parallel-ssh -h workers.txt -t 0 -p 100 -P sudo shutdown -h now 4 | 5 | # eof 6 | 7 | 8 | -------------------------------------------------------------------------------- /pi-cluster/upgrade-workers: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | 3 | parallel-ssh -h workers.txt -t 0 -p 100 -P sudo apt-get update 4 | parallel-ssh -h workers.txt -t 0 -p 100 -P sudo apt-get -y upgrade 5 | parallel-ssh -h workers.txt -t 0 -p 100 -P sudo apt-get clean 6 | parallel-ssh -h workers.txt -t 0 -p 100 -P sudo reboot 7 | 8 | # eof 9 | 10 | 11 | -------------------------------------------------------------------------------- /qblog/.gitignore: -------------------------------------------------------------------------------- 1 | /.quarto/ 2 | -------------------------------------------------------------------------------- /qblog/Makefile: -------------------------------------------------------------------------------- 1 | # Makefile 2 | 3 | CURRENT=draft 4 | 5 | FORCE: 6 | make render 7 | 8 | preview: 9 | quarto preview 10 | 11 | render: 12 | quarto render 13 | 14 | view: 15 | make render 16 | xdg-open _site/index.html 17 | 18 | publish: 19 | make render 20 | cp -r _site/* ../docs/ 21 | git add ../docs/* 22 | git commit -a && git push 23 | 24 | edit: 25 | emacs Makefile *.yml *.qmd posts/$(CURRENT)/*.qmd & 26 | 27 | 28 | # eof 29 | 30 | -------------------------------------------------------------------------------- /qblog/_quarto.yml: -------------------------------------------------------------------------------- 1 | project: 2 | type: website 3 | 4 | website: 5 | title: "DJW's blog" 6 | site-url: https://darrenjw.github.io/blog/ 7 | description: "Darren Wilkinson's blog" 8 | navbar: 9 | right: 10 | - about.qmd 11 | - icon: rss 12 | href: index.xml 13 | - icon: github 14 | href: https://github.com/darrenjw/blog/ 15 | - icon: twitter 16 | href: https://twitter.com/darrenjw 17 | format: 18 | html: 19 | theme: united 20 | css: styles.css 21 | 22 | 23 | 24 | -------------------------------------------------------------------------------- /qblog/about.qmd: -------------------------------------------------------------------------------- 1 | --- 2 | title: "About" 3 | image: profile.jpg 4 | about: 5 | template: jolla 6 | links: 7 | - icon: twitter 8 | text: Twitter 9 | href: https://twitter.com/darrenjw 10 | - icon: linkedin 11 | text: LinkedIn 12 | href: https://linkedin.com/in/darrenjwilkinson/ 13 | - icon: github 14 | text: Github 15 | href: https://github.com/darrenjw 16 | 17 | --- 18 | 19 | This is the blog of [Darren Wilkinson](https://darrenjw.github.io/). 20 | 21 | This blog is intended to cover my reflections on mathematics, statistics, machine learning, AI, computing and biology, and especially their interactions, and relationship with “big data” and data science. This blog replaces my two wordpress blogs: my [main blog](https://darrenjw.wordpress.com/) and my [personal blog](https://darrenjw2.wordpress.com). [Wordpress](https://wordpress.com/) served me well for more than a decade, but these days I dislike having to write anything other than [Quarto](http://quarto.org/), so I've taken the plunge and moved to a Quarto based blog. 22 | 23 | -------------------------------------------------------------------------------- /qblog/index.qmd: -------------------------------------------------------------------------------- 1 | --- 2 | title: "Darren Wilkinson's blog" 3 | listing: 4 | contents: posts 5 | sort: "date desc" 6 | type: default 7 | categories: true 8 | sort-ui: false 9 | filter-ui: false 10 | feed: true 11 | page-layout: full 12 | title-block-banner: true 13 | --- 14 | 15 | 16 | -------------------------------------------------------------------------------- /qblog/posts/_metadata.yml: -------------------------------------------------------------------------------- 1 | # options specified here will apply to all posts in this folder 2 | 3 | # freeze computational output 4 | # (see https://quarto.org/docs/projects/code-execution.html#freeze) 5 | freeze: true 6 | 7 | # Enable banner style title blocks 8 | title-block-banner: true 9 | -------------------------------------------------------------------------------- /qblog/posts/draft/index.qmd: -------------------------------------------------------------------------------- 1 | --- 2 | title: "Draft post" 3 | author: "Darren Wilkinson" 4 | date: "2024-03-05" 5 | categories: [stats] 6 | draft: true 7 | --- 8 | 9 | # Draft post 10 | 11 | This shouldn't appear in the published site. Blah. A draft post. 12 | 13 | -------------------------------------------------------------------------------- /qblog/posts/py-test/index.qmd: -------------------------------------------------------------------------------- 1 | --- 2 | title: "Python test" 3 | author: "Darren Wilkinson" 4 | date: "2024-03-04" 5 | categories: [python, code] 6 | --- 7 | 8 | # Python test 9 | 10 | A test post including some python code 11 | ```{python} 12 | x = [1,2,3] 13 | print(x[1]) 14 | 15 | import matplotlib.pyplot as plt 16 | fig, axis = plt.subplots() 17 | axis.plot([0,1,2,3,4,5], [3,4,6,5,2,4]) 18 | ``` 19 | Blah. 20 | -------------------------------------------------------------------------------- /qblog/posts/r-test/index.qmd: -------------------------------------------------------------------------------- 1 | --- 2 | title: "R test" 3 | author: "Darren Wilkinson" 4 | date: "2024-03-04" 5 | categories: [R, code] 6 | --- 7 | 8 | # R test 9 | 10 | A test post including some R code. 11 | ```{r} 12 | hist(rnorm(1000)) 13 | ``` 14 | Blah. 15 | 16 | -------------------------------------------------------------------------------- /qblog/profile.jpg: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/qblog/profile.jpg -------------------------------------------------------------------------------- /qblog/styles.css: -------------------------------------------------------------------------------- 1 | /* css styles */ 2 | -------------------------------------------------------------------------------- /rainier/.gitignore: -------------------------------------------------------------------------------- 1 | # .gitignore for scala projects 2 | 3 | # Classes and logs 4 | *.class 5 | *.log 6 | *~ 7 | 8 | # SBT-specific 9 | .cache 10 | .history 11 | .classpath 12 | .project 13 | .settings 14 | 15 | .lib/ 16 | dist/* 17 | target/ 18 | lib_managed/ 19 | src_managed/ 20 | project/boot/ 21 | project/plugins/project/ 22 | 23 | # Ensime specific 24 | .ensime 25 | 26 | # Scala-IDE specific 27 | .scala_dependencies 28 | .worksheet 29 | 30 | 31 | -------------------------------------------------------------------------------- /rainier/Makefile: -------------------------------------------------------------------------------- 1 | # Makefile 2 | 3 | 4 | FORCE: 5 | make doc 6 | 7 | doc: docs/DraftPost.md docs/Tutorial.md 8 | sbt mdoc 9 | cd target/mdoc ; pandoc DraftPost.md -o DraftPost.html 10 | cd target/mdoc ; pandoc Tutorial.md -o Tutorial.html 11 | 12 | edit: 13 | emacs *.md Makefile build.sbt src/test/scala/*.scala docs/*.md src/main/scala/*.scala & 14 | 15 | commit: 16 | git commit -a && git push 17 | 18 | update: 19 | git pull 20 | git log|less 21 | 22 | 23 | # eof 24 | 25 | 26 | -------------------------------------------------------------------------------- /rainier/Readme.md: -------------------------------------------------------------------------------- 1 | # Rainier 0.3.0 2 | 3 | Materials supporting a blog post about Rainier 0.3.0, which is a major update of the functional probabilistic programming language, [Rainier](https://rainier.fit/). 4 | 5 | A draft post, using **mdoc**, will gradually appear in [docs](docs/). 6 | 7 | 8 | 9 | #### eof 10 | 11 | 12 | -------------------------------------------------------------------------------- /rainier/build.sbt: -------------------------------------------------------------------------------- 1 | // build.sbt 2 | 3 | name := "rainier" 4 | 5 | version := "0.1-SNAPSHOT" 6 | 7 | scalacOptions ++= Seq( 8 | "-unchecked", "-deprecation", "-feature", "-language:higherKinds", 9 | "-language:implicitConversions", "-Ypartial-unification" 10 | ) 11 | 12 | addCompilerPlugin("org.typelevel" %% "kind-projector" % "0.11.0" cross CrossVersion.full) 13 | addCompilerPlugin("org.scalamacros" %% "paradise" % "2.1.1" cross CrossVersion.full) 14 | 15 | enablePlugins(MdocPlugin) 16 | 17 | libraryDependencies ++= Seq( 18 | "org.scalatest" %% "scalatest" % "3.0.8" % "test", 19 | "org.scalactic" %% "scalactic" % "3.0.8" % "test", 20 | "org.typelevel" %% "cats-core" % "2.0.0", 21 | "org.typelevel" %% "discipline-core" % "1.0.0", 22 | "org.typelevel" %% "discipline-scalatest" % "1.0.0", 23 | "org.typelevel" %% "simulacrum" % "1.0.0", 24 | "com.cibo" %% "evilplot" % "0.6.3", // 0.7.0 25 | "com.cibo" %% "evilplot-repl" % "0.6.3", // 0.7.0 26 | // "com.stripe" %% "rainier-core" % "0.3.0", 27 | // "com.stripe" %% "rainier-notebook" % "0.3.0" 28 | "com.stripe" %% "rainier-core" % "0.3.2+2-8a01736f", 29 | "com.stripe" %% "rainier-notebook" % "0.3.2+2-8a01736f" 30 | ) 31 | 32 | 33 | resolvers += Resolver.bintrayRepo("cibotech", "public") // for EvilPlot 34 | 35 | resolvers ++= Seq( 36 | "Sonatype Snapshots" at 37 | "https://oss.sonatype.org/content/repositories/snapshots/", 38 | "Sonatype Releases" at 39 | "https://oss.sonatype.org/content/repositories/releases/", 40 | "jitpack" at "https://jitpack.io" // for Jupiter/notebook 41 | ) 42 | 43 | scalaVersion := "2.12.10" 44 | 45 | 46 | // eof 47 | 48 | -------------------------------------------------------------------------------- /rainier/docs/DraftPost.md: -------------------------------------------------------------------------------- 1 | # Probabilistic programming with Rainier 0.3.0 2 | 3 | 4 | ## Setup 5 | 6 | 7 | **Start with setting up an SBT console from scratch? Bit messy due to non-standard resolvers...** 8 | 9 | 10 | ```scala mdoc 11 | import com.stripe.rainier.core._ 12 | import com.stripe.rainier.compute._ 13 | import com.stripe.rainier.notebook._ 14 | import com.stripe.rainier.sampler._ 15 | 16 | implicit val rng = ScalaRNG(3) 17 | val sampler = EHMC(warmupIterations = 5000, iterations = 5000) 18 | ``` 19 | 20 | # Normal random sample 21 | 22 | Let's start by looking at inferring the mean and standard deviation of a normal random sample. 23 | 24 | 25 | ```scala mdoc 26 | // first simulate some data 27 | val n = 1000 28 | val mu = 3.0 29 | val sig = 5.0 30 | val x = Vector.fill(n)(mu + sig*rng.standardNormal) 31 | // now build Rainier model 32 | val m = Normal(0,100).latent 33 | val s = Gamma(1,10).latent 34 | val nrs = Model.observe(x, Normal(m,s)) 35 | // now sample from the model 36 | val out = nrs.sample(sampler) 37 | ``` 38 | 39 | ```scala mdoc:image:nrs-mu.png 40 | val mut = out.predict(m) 41 | show("mu", density(mut)) 42 | ``` 43 | 44 | ```scala mdoc:image:nrs-sig.png 45 | val sigt = out.predict(s) 46 | show("sig", density(sigt)) 47 | ``` 48 | 49 | # Logistic regression 50 | 51 | Now let's fit a basic logistic regression model. 52 | 53 | ```scala mdoc:silent:reset 54 | import com.stripe.rainier.core._ 55 | import com.stripe.rainier.compute._ 56 | import com.stripe.rainier.notebook._ 57 | import com.stripe.rainier.sampler._ 58 | 59 | implicit val rng = ScalaRNG(3) 60 | val sampler = EHMC(warmupIterations = 5000, iterations = 5000) 61 | ``` 62 | 63 | ```scala mdoc 64 | val N = 1000 65 | val beta0 = 0.1 66 | val beta1 = 0.3 67 | val x = (1 to N) map { _ => 2.0 * rng.standardNormal } 68 | val theta = x map { xi => beta0 + beta1 * xi } 69 | def expit(x: Double): Double = 1.0 / (1.0 + math.exp(-x)) 70 | val p = theta map expit 71 | val yb = p map (pi => (rng.standardUniform < pi)) 72 | val y = yb map (b => if (b) 1L else 0L) 73 | println("Proportion of successes: " + (y.filter(_ > 0L).length.toDouble/N)) 74 | // now build Rainier model 75 | val b0 = Normal(0, 2).latent 76 | val b1 = Normal(0, 2).latent 77 | val model = Model.observe(y, Vec.from(x).map(xi => { 78 | val theta = b0 + b1*xi 79 | val p = 1.0 / (1.0 + (-theta).exp) 80 | Bernoulli(p) 81 | })) 82 | // now sample from the model 83 | val bt = model.sample(sampler) 84 | ``` 85 | 86 | ```scala mdoc:image:lr-b0.png 87 | val b0t = bt.predict(b0) 88 | show("b0", density(b0t)) 89 | ``` 90 | 91 | ```scala mdoc:image:lr-b1.png 92 | val b1t = bt.predict(b1) 93 | show("b1", density(b1t)) 94 | ``` 95 | 96 | 97 | # ANOVA model 98 | 99 | Let's now turn attention to a very basic normal random effects model. 100 | 101 | ```scala mdoc:silent:reset 102 | import com.stripe.rainier.core._ 103 | import com.stripe.rainier.compute._ 104 | import com.stripe.rainier.notebook._ 105 | import com.stripe.rainier.sampler._ 106 | 107 | implicit val rng = ScalaRNG(3) 108 | val sampler = EHMC(warmupIterations = 5000, iterations = 5000) 109 | ``` 110 | 111 | ```scala mdoc 112 | // simulate synthetic data 113 | //val n = 50 // groups 114 | //val N = 150 // obs per group 115 | val n = 15 // groups 116 | val N = 50 // obs per group 117 | val mu = 5.0 // overall mean 118 | val sigE = 2.0 // random effect SD 119 | val sigD = 3.0 // obs SD 120 | val effects = Vector.fill(n)(sigE * rng.standardNormal) 121 | val data = effects map (e => 122 | Vector.fill(N)(mu + e + sigD * rng.standardNormal)) 123 | // build model 124 | val m = Normal(0, 100).latent 125 | val sD = LogNormal(0, 10).latent 126 | val sE = LogNormal(1, 5).latent 127 | val eff = Vector.fill(n)(Normal(m, sE).latent) 128 | val models = (0 until n).map(i => 129 | Model.observe(data(i), Normal(eff(i), sD))) 130 | val anova = models.reduce{(m1, m2) => m1.merge(m2)} 131 | // now sample the model 132 | val trace = anova.sample(sampler) 133 | val mt = trace.predict(m) 134 | ``` 135 | 136 | ```scala mdoc:image:anova-mu.png 137 | show("mu", density(mt)) 138 | ``` 139 | 140 | ```scala mdoc:image:anova-sd.png 141 | val sDt = trace.predict(sD) 142 | show("sigD", density(sDt)) 143 | ``` 144 | 145 | ```scala mdoc:image:anova-se.png 146 | val sEt = trace.predict(sE) 147 | show("sigE", density(sEt)) 148 | ``` 149 | 150 | 151 | 152 | #### eof 153 | 154 | -------------------------------------------------------------------------------- /rainier/docs/Tutorial.md: -------------------------------------------------------------------------------- 1 | # Tutorial for Rainier 0.3 2 | 3 | 4 | ```scala mdoc 5 | import com.stripe.rainier.core._ 6 | import com.stripe.rainier.compute._ 7 | 8 | val a = Uniform(0,1).latent 9 | val b = a + 1 10 | 11 | val c = Normal(b, a).latent 12 | Model.sample((a,c)).take(10) 13 | ``` 14 | 15 | ```scala mdoc:image:scatter.png 16 | import com.stripe.rainier.notebook._ 17 | val ac = Model.sample((a,c)) 18 | show("a", "c", scatter(ac)) 19 | ``` 20 | 21 | ```scala mdoc 22 | val eggs = List[Long](45, 52, 45, 47, 41, 42, 44, 42, 46, 38, 36, 35, 41, 48, 42, 29, 45, 43, 45, 40, 42, 53, 31, 48, 40, 45, 39, 29, 45, 42) 23 | val lambda = Gamma(0.5, 100).latent 24 | ``` 25 | 26 | ```scala mdoc:image:lambda.png 27 | show("lambda", density(Model.sample(lambda))) 28 | ``` 29 | 30 | ```scala mdoc 31 | val eggModel = Model.observe(eggs, Poisson(lambda)) 32 | eggModel.optimize(lambda) 33 | val dozens = eggModel.optimize(lambda / 12) 34 | import com.stripe.rainier.sampler._ 35 | 36 | val sampler = EHMC(warmupIterations = 5000, iterations = 500) 37 | val eggTrace = eggModel.sample(sampler) 38 | eggTrace.diagnostics 39 | val thinTrace = eggTrace.thin(2) 40 | thinTrace.diagnostics 41 | val posterior = eggTrace.predict(lambda) 42 | ``` 43 | 44 | ```scala mdoc:image:lambdap.png 45 | show("lambda", density(posterior)) 46 | ``` 47 | 48 | 49 | #### eof 50 | 51 | -------------------------------------------------------------------------------- /rainier/project/build.properties: -------------------------------------------------------------------------------- 1 | sbt.version=1.3.2 2 | -------------------------------------------------------------------------------- /rainier/project/plugins.sbt: -------------------------------------------------------------------------------- 1 | addSbtPlugin("org.scalameta" % "sbt-mdoc" % "1.3.6") 2 | 3 | 4 | -------------------------------------------------------------------------------- /rainier/src/main/scala/rainier.scala: -------------------------------------------------------------------------------- 1 | /* 2 | rainier.scala 3 | Simple example rainier app 4 | */ 5 | 6 | object RainierApp { 7 | 8 | import cats._ 9 | import cats.implicits._ 10 | import com.stripe.rainier.core._ 11 | import com.stripe.rainier.compute._ 12 | import com.stripe.rainier.sampler._ 13 | import com.stripe.rainier.notebook._ 14 | import com.cibo.evilplot._ 15 | import com.cibo.evilplot.plot._ 16 | 17 | // rainier tutorial 18 | def tutorial: Unit = { 19 | println("Tutorial") 20 | val a = Uniform(0,1).latent 21 | val b = a + 1 22 | 23 | val c = Normal(b, a).latent 24 | Model.sample((a,c)).take(10) 25 | 26 | val ac = Model.sample((a,c)) 27 | show("a", "c", scatter(ac)) // produces an almond Image, but then what? 28 | displayPlot(scatter(ac).render()) // use Evilplot to display on console 29 | 30 | val eggs = List[Long](45, 52, 45, 47, 41, 42, 44, 42, 46, 38, 36, 35, 41, 48, 42, 29, 45, 43, 31 | 45, 40, 42, 53, 31, 48, 40, 45, 39, 29, 45, 42) 32 | val lambda = Gamma(0.5, 100).latent 33 | 34 | show("lambda", density(Model.sample(lambda))) // show 35 | mcmcSummary("lambda", Model.sample(lambda)) 36 | 37 | val eggModel = Model.observe(eggs, Poisson(lambda)) 38 | eggModel.optimize(lambda) 39 | val dozens = eggModel.optimize(lambda / 12) 40 | 41 | val sampler = EHMC(5000, 500) 42 | val eggTrace = eggModel.sample(sampler) 43 | eggTrace.diagnostics 44 | val thinTrace = eggTrace.thin(2) 45 | thinTrace.diagnostics 46 | val posterior = eggTrace.predict(lambda) 47 | 48 | show("lambda", density(posterior)) // show 49 | mcmcSummary("lambda", posterior) 50 | } 51 | 52 | def mcmcSummary(name: String, chain: Seq[Double]): Unit = { 53 | println(name) 54 | val dens = density(chain). 55 | standard(). 56 | xLabel(name). 57 | yLabel("Density") 58 | val trace = line(chain.zipWithIndex map (_.swap)). 59 | standard(). 60 | xLabel("Iteration"). 61 | yLabel(name) 62 | //displayPlot(dens.render()) 63 | //displayPlot(trace.render()) 64 | displayPlot(Facets(Vector(Vector(trace, dens))).render()) 65 | } 66 | 67 | def mcmcSummary(chain: Seq[Double]): Unit = mcmcSummary("Variable", chain) 68 | 69 | 70 | // normal random sample 71 | def nrs: Unit = { 72 | // first simulate some synthetic data 73 | val n = 1000 74 | val mu = 3.0 75 | val sig = 5.0 76 | implicit val rng = ScalaRNG(3) 77 | val x = Vector.fill(n)(mu + sig*rng.standardNormal) 78 | // now build Rainier model 79 | val m = Normal(0,100).latent 80 | val s = Gamma(1,10).latent 81 | val model = Model.observe(x, Normal(m,s)) 82 | // now sample from the model 83 | val sampler = EHMC(5000, 5000) 84 | println("sampling...") 85 | val out = model.sample(sampler) 86 | println("finished sampling.") 87 | println(out.diagnostics) 88 | val mut = out.predict(m) 89 | show("mu", density(mut)) 90 | val sigt = out.predict(s) 91 | show("sig", density(sigt)) 92 | // try some diagnostic plots 93 | mcmcSummary("mu", mut) 94 | mcmcSummary("sig", sigt) 95 | } 96 | 97 | // logistic regression 98 | def logReg: Unit = { 99 | println("logReg") 100 | // first simulate some data from a logistic regression model 101 | implicit val rng = ScalaRNG(3) 102 | val N = 1000 103 | val beta0 = 0.1 104 | val beta1 = 0.3 105 | val x = (1 to N) map { _ => 106 | 3.0 * rng.standardNormal 107 | } 108 | val theta = x map { xi => 109 | beta0 + beta1 * xi 110 | } 111 | def expit(x: Double): Double = 1.0 / (1.0 + math.exp(-x)) 112 | val p = theta map expit 113 | val yb = p map (pi => (rng.standardUniform < pi)) 114 | val y = yb map (b => if (b) 1L else 0L) 115 | println(y.take(10)) 116 | println(x.take(10)) 117 | // now build Rainier model 118 | val b0 = Normal(0, 5).latent 119 | val b1 = Normal(0, 5).latent 120 | val model = Model.observe(y, Vec.from(x).map{xi => 121 | val theta = b0 + b1*xi 122 | val p = 1.0 / (1.0 + (-theta).exp) 123 | Bernoulli(p) 124 | }) 125 | // now sample from the model 126 | //val sampler = EHMC(10000, 1000) 127 | val sampler = HMC(5000, 1000, 50) 128 | println("sampling...") 129 | val bt = model.sample(sampler) 130 | println("finished sampling.") 131 | println(bt.diagnostics) 132 | val b0t = bt.predict(b0) 133 | show("b0", density(b0t)) 134 | val b1t = bt.predict(b1) 135 | show("b1", density(b1t)) 136 | mcmcSummary("b0", b0t) 137 | mcmcSummary("b1", b1t) 138 | } 139 | 140 | // one-way anova model 141 | def anova: Unit = { 142 | println("anova") 143 | // simulate synthetic data 144 | implicit val rng = ScalaRNG(3) 145 | //val n = 50 // groups 146 | //val N = 150 // obs per group 147 | val n = 10 // groups 148 | val N = 20 // obs per group 149 | val mu = 5.0 // overall mean 150 | val sigE = 2.0 // random effect SD 151 | val sigD = 3.0 // obs SD 152 | val effects = Vector.fill(n)(sigE * rng.standardNormal) 153 | val data = effects map (e => 154 | Vector.fill(N)(mu + e + sigD * rng.standardNormal)) 155 | // build model 156 | val m = Normal(0, 100).latent 157 | val sD = LogNormal(0, 10).latent 158 | val sE = LogNormal(1, 5).latent 159 | val eff = Vector.fill(n)(Normal(m, sE).latent) 160 | val models = (0 until n).map(i => 161 | Model.observe(data(i), Normal(eff(i), sD))) 162 | val model = models.reduce{(m1, m2) => m1.merge(m2)} 163 | // now sample the model 164 | val sampler = EHMC(5000, 5000) 165 | println("sampling...") 166 | val trace = model.sample(sampler) 167 | println("finished sampling.") 168 | println(trace.diagnostics) 169 | val mt = trace.predict(m) 170 | show("mu", density(mt)) 171 | mcmcSummary("mu", mt) 172 | mcmcSummary("sigE", trace.predict(sE)) 173 | mcmcSummary("sigD", trace.predict(sD)) 174 | } 175 | 176 | // repeat one-way anova, but without merging 177 | def anova2: Unit = { 178 | 179 | } 180 | 181 | 182 | def main(args: Array[String]): Unit = { 183 | println("main starting") 184 | 185 | //tutorial 186 | //nrs 187 | logReg 188 | //anova 189 | 190 | 191 | println("main finishing") 192 | } 193 | 194 | } 195 | -------------------------------------------------------------------------------- /rainier/src/test/scala/rainier-test.scala: -------------------------------------------------------------------------------- 1 | import org.scalatest.flatspec.AnyFlatSpec 2 | import org.scalatest.matchers.should.Matchers 3 | 4 | 5 | // Example unit tests 6 | class CatsSpec extends AnyFlatSpec with Matchers { 7 | 8 | import cats._ 9 | import cats.implicits._ 10 | 11 | "A List" should "combine" in { 12 | val l = List(1,2) |+| List(3,4) 13 | l should be (List(1,2,3,4)) 14 | } 15 | 16 | } 17 | 18 | 19 | // Example property-based tests 20 | import org.scalatestplus.scalacheck._ 21 | class MyPropertyTests extends AnyFlatSpec with Matchers with ScalaCheckPropertyChecks { 22 | 23 | import cats._ 24 | import cats.implicits._ 25 | 26 | "An Int" should "combine commutatively" in { 27 | forAll { (a: Int, b: Int) => 28 | (a |+| b) should be (b |+| a) 29 | } 30 | } 31 | 32 | it should "invert" in { 33 | forAll { (a: Int) => 34 | (a |+| a.inverse) shouldBe Monoid[Int].empty 35 | } 36 | } 37 | 38 | } 39 | 40 | // eof 41 | 42 | 43 | -------------------------------------------------------------------------------- /reaction-diffusion/.gitignore: -------------------------------------------------------------------------------- 1 | # .gitignore for scala projects 2 | 3 | # Classes and logs 4 | *.class 5 | *.log 6 | *~ 7 | 8 | # SBT-specific 9 | .cache 10 | .history 11 | .classpath 12 | .project 13 | .settings 14 | 15 | .lib/ 16 | dist/* 17 | target/ 18 | lib_managed/ 19 | src_managed/ 20 | project/boot/ 21 | project/plugins/project/ 22 | 23 | # Ensime specific 24 | .ensime 25 | 26 | # Scala-IDE specific 27 | .scala_dependencies 28 | .worksheet 29 | 30 | 31 | -------------------------------------------------------------------------------- /reaction-diffusion/Makefile: -------------------------------------------------------------------------------- 1 | # Makefile 2 | 3 | 4 | 5 | 6 | view: DraftPost.html 7 | xdg-open DraftPost.html 8 | 9 | DraftPost.html: DraftPost.md 10 | pandoc DraftPost.md -o DraftPost.html 11 | 12 | clean: 13 | sbt clean 14 | rm -f siv-*.png 15 | 16 | 17 | 18 | # eof 19 | 20 | -------------------------------------------------------------------------------- /reaction-diffusion/Readme.md: -------------------------------------------------------------------------------- 1 | # Stochastic reaction-diffusion modelling (in Scala) 2 | 3 | This directory contains the source code associated with the blog post: 4 | 5 | [Stochastic reaction-diffusion modelling](https://darrenjw.wordpress.com/2019/01/22/stochastic-reaction-diffusion-modelling/) 6 | 7 | This code should run on any system with a recent [Java JDK](https://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html) installation, and [sbt](https://www.scala-sbt.org/). Note that this code relies on JavaFx libraries, so if you are running OpenJdk, you will also need to install OpenJfx. On most Linux (and similar) systems, this should be as easy as installing the `openjfx` package in addition to (say) `openjdk-8-jdk` using your OS package manager. 8 | 9 | Once you have Java and sbt installed, you should be able to compile and run the examples by typing: 10 | ```bash 11 | sbt run 12 | ``` 13 | at your OS prompt from *this* directory (that is, the directory containing the file `build.sbt`). Then just select the number of the example you want to run. The animation should auto-start. 14 | 15 | 16 | 17 | #### eof 18 | 19 | -------------------------------------------------------------------------------- /reaction-diffusion/build.sbt: -------------------------------------------------------------------------------- 1 | name := "reaction-diffusion" 2 | 3 | version := "0.1-SNAPSHOT" 4 | 5 | scalacOptions ++= Seq( 6 | "-unchecked", "-deprecation", "-feature" 7 | ) 8 | 9 | libraryDependencies ++= Seq( 10 | "org.scalatest" %% "scalatest" % "3.0.1" % "test", 11 | "org.scalanlp" %% "breeze" % "0.13.2", 12 | "org.scalanlp" %% "breeze-viz" % "0.13.2", 13 | "org.scalanlp" %% "breeze-natives" % "0.13.2", 14 | "com.github.darrenjw" %% "scala-view" % "0.5", 15 | "com.github.darrenjw" %% "scala-smfsb" % "0.6" 16 | ) 17 | 18 | resolvers ++= Seq( 19 | "Sonatype Snapshots" at 20 | "https://oss.sonatype.org/content/repositories/snapshots/", 21 | "Sonatype Releases" at 22 | "https://oss.sonatype.org/content/repositories/releases/" 23 | ) 24 | 25 | scalaVersion := "2.12.8" 26 | 27 | scalaVersion in ThisBuild := "2.12.8" // for ensime 28 | 29 | 30 | -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle2.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle2.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle2.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle2.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle3.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle3.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-cle3.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-cle3.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-exact.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-exact.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-exact.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-exact.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre2.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre2.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre2.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre2.png -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre3.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre3.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/lv-rre3.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/lv-rre3.png -------------------------------------------------------------------------------- /reaction-diffusion/make-movie.sh: -------------------------------------------------------------------------------- 1 | #!/bin/sh 2 | # make-movie.sh 3 | 4 | rm -f siv-??????-s.png 5 | 6 | for name in siv-??????.png 7 | do 8 | short="${name%.*}" 9 | echo $short 10 | #pngtopnm "$name" | pnmscale 20 | pnmtopng > "${short}-s.png" 11 | convert "$name" -scale 1200x600 -define png:color-type=2 "${short}-s.png" 12 | done 13 | 14 | rm -f movie.mp4 15 | 16 | #avconv -r 20 -i siv-%06d-s.png movie.mp4 17 | ffmpeg -f image2 -r 10 -pattern_type glob -i 'siv-*-s.png' movie.mp4 18 | 19 | # make a version that should play on Android devices... 20 | ffmpeg -i movie.mp4 -codec:v libx264 -profile:v main -preset slow -b:v 400k -maxrate 400k -bufsize 800k -vf scale=-1:480 -threads 0 -codec:a libfdk_aac -b:a 128k -pix_fmt yuv420p movie-a.mp4 21 | 22 | # Animated GIF... 23 | # ffmpeg -i movie.mp4 -s 200x100 movie.gif 24 | 25 | 26 | # eof 27 | 28 | 29 | 30 | -------------------------------------------------------------------------------- /reaction-diffusion/project/build.properties: -------------------------------------------------------------------------------- 1 | sbt.version=1.2.8 2 | 3 | -------------------------------------------------------------------------------- /reaction-diffusion/sir-cle.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/sir-cle.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/sir-cle.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/sir-cle.png -------------------------------------------------------------------------------- /reaction-diffusion/sir-rre.mp4: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/sir-rre.mp4 -------------------------------------------------------------------------------- /reaction-diffusion/sir-rre.png: -------------------------------------------------------------------------------- https://raw.githubusercontent.com/darrenjw/blog/be123278cc20048ab8458b895ccb5ccca798ea59/reaction-diffusion/sir-rre.png -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvCle.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvCle.scala 3 | 4 | Chemical Langevin approximation for a Lotka-Volterra system 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object LvCle { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | 18 | def main(args: Array[String]): Unit = { 19 | val r = 250; val c = 300 20 | val model = SpnModels.lv[DoubleState]() 21 | val step = Spatial.cle2d(model, DenseVector(0.6, 0.6), 0.05) 22 | val x00 = DenseVector(0.0, 0.0) 23 | val x0 = DenseVector(50.0, 100.0) 24 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 25 | val xx0 = xx00.updated(c/2, r/2, x0) 26 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 27 | val si = s map (toSfxI(_)) 28 | scalaview.SfxImageViewer(si, 1, autoStart=true) 29 | } 30 | 31 | } 32 | 33 | // eof 34 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvCle2.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvCle2.scala 3 | 4 | Chemical Langevin approximation for a Lotka-Volterra system 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object LvCle2 { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | 18 | def main(args: Array[String]): Unit = { 19 | val r = 300; val c = 600 20 | val model = SpnModels.lv[DoubleState]() 21 | val step = Spatial.cle2d(model, DenseVector(0.6, 0.6), 0.05) 22 | val x00 = DenseVector(0.0, 0.0) 23 | val x0 = DenseVector(50.0, 100.0) 24 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 25 | val xx0 = xx00. 26 | updated(c/3, r/2, x0). 27 | updated(2*c/3,r/2,x0). 28 | updated(c/2,2*r/3,x0) 29 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 30 | val si = s map (toSfxI(_)) 31 | scalaview.SfxImageViewer(si, 1, autoStart=true) 32 | } 33 | 34 | } 35 | 36 | // eof 37 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvCle3.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvCle3.scala 3 | 4 | Chemical Langevin approximation for a Lotka-Volterra system 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object LvCle3 { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | import breeze.stats.distributions.Uniform 18 | 19 | def main(args: Array[String]): Unit = { 20 | val r = 300; val c = 400 21 | val model = SpnModels.lv[DoubleState]() 22 | val step = Spatial.cle2d(model, DenseVector(0.6, 0.6), 0.05) 23 | val xx0 = PMatrix(r, c, Vector.fill(r*c)(DenseVector( 24 | Uniform(100,300).draw, 25 | Uniform(100,300).draw))) 26 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 27 | val si = s map (toSfxI(_)) 28 | scalaview.SfxImageViewer(si, 1, autoStart=true) 29 | } 30 | 31 | } 32 | 33 | // eof 34 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvExact.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvExact.scala 3 | 4 | Exact Gillespie simulation of the RDME for a Lotka-Volterra system 5 | 6 | */ 7 | 8 | package rd 9 | 10 | object LvExact { 11 | 12 | import smfsb._ 13 | import breeze.linalg.{Vector => BVec, _} 14 | import breeze.numerics._ 15 | 16 | def main(args: Array[String]): Unit = { 17 | val r = 100; val c = 120 18 | val model = SpnModels.lv[IntState]() 19 | val step = Spatial.gillespie2d(model, DenseVector(0.6, 0.6), maxH=1e12) 20 | val x00 = DenseVector(0, 0) 21 | val x0 = DenseVector(50, 100) 22 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 23 | val xx0 = xx00.updated(c/2, r/2, x0) 24 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 25 | val si = s map (toSfxIi(_)) 26 | scalaview.SfxImageViewer(si, 1, autoStart=true) 27 | } 28 | 29 | } 30 | 31 | // eof 32 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvRre.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvRre.scala 3 | 4 | Reaction rate equations for the Lotka-Volterra model 5 | Numerical solution of a PDE 6 | 7 | */ 8 | 9 | package rd 10 | 11 | object LvRre { 12 | 13 | import smfsb._ 14 | import breeze.linalg.{Vector => BVec, _} 15 | import breeze.numerics._ 16 | 17 | def main(args: Array[String]): Unit = { 18 | val r = 250; val c = 300 19 | val model = SpnModels.lv[DoubleState]() 20 | val step = Spatial.euler2d(model, DenseVector(0.6, 0.6), 0.02) 21 | val x00 = DenseVector(0.0, 0.0) 22 | val x0 = DenseVector(50.0, 100.0) 23 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 24 | val xx0 = xx00.updated(c/2, r/2, x0) 25 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 26 | val si = s map (toSfxI(_)) 27 | scalaview.SfxImageViewer(si, 1, autoStart=true) 28 | } 29 | 30 | } 31 | 32 | // eof 33 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvRre2.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvRre2.scala 3 | 4 | PDE approximation for a Lotka-Volterra system 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object LvRre2 { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | 18 | def main(args: Array[String]): Unit = { 19 | val r = 300; val c = 600 20 | val model = SpnModels.lv[DoubleState]() 21 | val step = Spatial.euler2d(model, DenseVector(0.6, 0.6), 0.05) 22 | val x00 = DenseVector(0.0, 0.0) 23 | val x0 = DenseVector(50.0, 100.0) 24 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 25 | val xx0 = xx00. 26 | updated(c/3, r/2, x0). 27 | updated(2*c/3,r/2,x0). 28 | updated(c/2,2*r/3,x0) 29 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 30 | val si = s map (toSfxI(_)) 31 | scalaview.SfxImageViewer(si, 1, autoStart=true) 32 | } 33 | 34 | } 35 | 36 | // eof 37 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/LvRre3.scala: -------------------------------------------------------------------------------- 1 | /* 2 | LvRre3.scala 3 | 4 | PDE approximation for a Lotka-Volterra system 5 | 6 | Numerical solution of a PDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object LvRre3 { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | import breeze.stats.distributions.Uniform 18 | 19 | def main(args: Array[String]): Unit = { 20 | val r = 300; val c = 400 21 | val model = SpnModels.lv[DoubleState]() 22 | val step = Spatial.euler2d(model, DenseVector(0.6, 0.6), 0.05) 23 | val xx0 = PMatrix(r, c, Vector.fill(r*c)(DenseVector( 24 | Uniform(100,300).draw, 25 | Uniform(100,300).draw))) 26 | val s = Stream.iterate(xx0)(step(_,0.0,0.1)) 27 | val si = s map (toSfxI(_)) 28 | scalaview.SfxImageViewer(si, 1, autoStart=true) 29 | } 30 | 31 | } 32 | 33 | // eof 34 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/SirCle.scala: -------------------------------------------------------------------------------- 1 | /* 2 | SirCle.scala 3 | 4 | Chemical Langevin approximation for a SIR epidemic model 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object SirCle { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | 18 | 19 | def main(args: Array[String]): Unit = { 20 | val r = 250; val c = 300 21 | val model = sir[DoubleState]() 22 | val step = Spatial.cle2d(model, DenseVector(3.0, 2.0, 0.0), 0.005) 23 | val x00 = DenseVector(100.0, 0.0, 0.0) 24 | val x0 = DenseVector(50.0, 50.0, 0.0) 25 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 26 | val xx0 = xx00.updated(c/2, r/2, x0) 27 | val s = Stream.iterate(xx0)(step(_,0.0,0.05)) 28 | val si = s map (toSfxI3(_)) 29 | scalaview.SfxImageViewer(si, 1, autoStart=true) 30 | } 31 | 32 | } 33 | 34 | // eof 35 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/SirRre.scala: -------------------------------------------------------------------------------- 1 | /* 2 | SirRre.scala 3 | 4 | PDE approximation for a SIR epidemic model 5 | 6 | Numerical solution of an SPDE 7 | 8 | */ 9 | 10 | package rd 11 | 12 | object SirRre { 13 | 14 | import smfsb._ 15 | import breeze.linalg.{Vector => BVec, _} 16 | import breeze.numerics._ 17 | 18 | 19 | def main(args: Array[String]): Unit = { 20 | val r = 250; val c = 300 21 | val model = sir[DoubleState]() 22 | val step = Spatial.euler2d(model, DenseVector(3.0, 2.0, 0.0), 0.005) 23 | val x00 = DenseVector(100.0, 0.0, 0.0) 24 | val x0 = DenseVector(50.0, 50.0, 0.0) 25 | val xx00 = PMatrix(r, c, Vector.fill(r*c)(x00)) 26 | val xx0 = xx00.updated(c/2, r/2, x0) 27 | val s = Stream.iterate(xx0)(step(_,0.0,0.05)) 28 | val si = s map (toSfxI3(_)) 29 | scalaview.SfxImageViewer(si, 1, autoStart=true) 30 | } 31 | 32 | } 33 | 34 | // eof 35 | -------------------------------------------------------------------------------- /reaction-diffusion/src/main/scala/rd/package.scala: -------------------------------------------------------------------------------- 1 | /* 2 | package.scala 3 | 4 | Shared code 5 | 6 | */ 7 | 8 | package object rd { 9 | 10 | import smfsb._ 11 | import breeze.linalg.{Vector => BVec, _} 12 | import breeze.numerics._ 13 | import scalafx.scene.image.WritableImage 14 | import scalafx.scene.paint._ 15 | 16 | def toSfxI(im: PMatrix[DenseVector[Double]]): WritableImage = { 17 | val wi = new WritableImage(im.c, im.r) 18 | val pw = wi.pixelWriter 19 | val m = im.data.aggregate(0.0)((acc,v) => math.max(acc,max(v)), math.max(_,_)) 20 | val rsi = im map (_ / m) 21 | (0 until im.c).par foreach (i => 22 | (0 until im.r).par foreach (j => 23 | pw.setColor(i, j, Color.rgb((rsi(i,j)(1)*255).toInt, 0, (rsi(i,j)(0)*255).toInt)) 24 | )) 25 | wi 26 | } 27 | 28 | def toSfxIi(im: PMatrix[DenseVector[Int]]): WritableImage = 29 | toSfxI(im map (v => v map (_.toDouble))) 30 | 31 | def sir[S: State](p: DenseVector[Double] = DenseVector(0.1, 0.5)): Spn[S] = 32 | UnmarkedSpn[S]( 33 | List("S", "I", "R"), 34 | DenseMatrix((1, 1, 0), (0, 1, 0)), 35 | DenseMatrix((0, 2, 0), (0, 0, 1)), 36 | (x, t) => { 37 | val xd = x.toDvd 38 | DenseVector( 39 | xd(0) * xd(1) * p(0), xd(1) * p(1) 40 | )} 41 | ) 42 | 43 | def toSfxI3(im: PMatrix[DenseVector[Double]]): WritableImage = { 44 | val wi = new WritableImage(im.c, im.r) 45 | val pw = wi.pixelWriter 46 | val m = im.data.aggregate(0.0)((acc,v) => math.max(acc,max(v)), math.max(_,_)) 47 | val rsi = im map (_ / m) 48 | (0 until im.c).par foreach (i => 49 | (0 until im.r).par foreach (j => 50 | pw.setColor(i, j, Color.rgb((rsi(i,j)(1)*255).toInt, (rsi(i,j)(0)*255).toInt, (rsi(i,j)(2)*255).toInt)) 51 | )) 52 | wi 53 | } 54 | 55 | 56 | 57 | } 58 | 59 | // eof 60 | 61 | -------------------------------------------------------------------------------- /reaction-diffusion/src/test/scala/reaction-diffusion-test.scala: -------------------------------------------------------------------------------- 1 | import org.scalatest.FlatSpec 2 | 3 | import smfsb._ 4 | import breeze.linalg._ 5 | 6 | class SmfsbSpec extends FlatSpec { 7 | 8 | "Step.gillespie" should "create and step LV model" in { 9 | val model = SpnModels.lv[IntState]() 10 | val step = Step.gillespie(model) 11 | val output = step(DenseVector(50, 100), 0.0, 1.0) 12 | assert(output.length === 2) 13 | } 14 | 15 | 16 | } 17 | 18 | -------------------------------------------------------------------------------- /sbml-scala/README.md: -------------------------------------------------------------------------------- 1 | # Working with SBML using Scala 2 | 3 | Code example for the blog post: 4 | 5 | https://darrenjw.wordpress.com/2016/12/17/working-with-sbml-using-scala/ 6 | 7 | Note that this repo contains everything that is needed to build and run the Scala code examples on any system that has Java installed. Any recent version of Java is fine. You do not need to "install" Scala or any Scala "packages" in order to run the code. If you have Java and a decent internet connection, you are good to go. This is one of the benefits of Scala - you can run it anywhere, on any system with a Java installation. 8 | 9 | To check if you have Java installed, just run: 10 | 11 | ```bash 12 | java -version 13 | ``` 14 | 15 | at your system command prompt. If you get an error, Java is absent or incorrectly installed. Installing Java is very easy on any platform, but the best way to install it depends on exactly what OS you are running, so search the internet for advice on the best way to install Java on your OS. 16 | 17 | The code uses `sbt` (the simple build tool) as the build tool. The sbt launcher has been included in the repo for the benefit of those new to Scala. It should be possible to run sbt from this directory by typing: 18 | 19 | ```bash 20 | ..\sbt 21 | ``` 22 | 23 | on Windows (which should run `..\sbt.bat`), or 24 | 25 | ```bash 26 | ../sbt 27 | ``` 28 | 29 | on Linux and similar systems (including Macs). If you want to be able to experiment with Scala yourself, you should copy the script and the file `sbt-launch.jar` to the same directory somewhere in your path, but this isn't necessary to run these examples. 30 | 31 | The sbt launcher script will download and run sbt, which will then download scala, the scala compiler, scala standard libraries and all dependencies needed to compile and run the code. All the downloaded files will be cached on your system for future use. Therefore, make sure you have a good internet connection and a bit of free disk space before running sbt for the first time. 32 | 33 | Assuming you can run sbt, just typing `run` at the sbt prompt will compile and run the example code. Typing `console` will give a Scala REPL with a properly configured classpath including all dependencies. You can type scala expressions directly into the REPL just as you would in your favourite dynamic math/stat language. Type `help` at the sbt prompt for help on sbt. Type `:help` at the Scala REPL for help on the REPL. 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | -------------------------------------------------------------------------------- /sbml-scala/build.sbt: -------------------------------------------------------------------------------- 1 | name := "jsbml" 2 | 3 | version := "0.1" 4 | 5 | scalacOptions ++= Seq("-unchecked", "-deprecation", "-feature") 6 | 7 | libraryDependencies ++= Seq( 8 | "org.sbml.jsbml" % "jsbml" % "1.2", 9 | "org.apache.logging.log4j" % "log4j-1.2-api" % "2.3", 10 | "org.apache.logging.log4j" % "log4j-api" % "2.3", 11 | "org.apache.logging.log4j" % "log4j-core" % "2.3" 12 | ) 13 | 14 | scalaVersion := "2.11.7" 15 | 16 | 17 | -------------------------------------------------------------------------------- /sbml-scala/ch07-mm-stoch.xml: -------------------------------------------------------------------------------- 1 | 2 |/[sourcecode language="r" light="true"]\n/g' | \
11 | sed 's|