├── FullRun.png
├── LICENSE
├── NodeLibrary
├── SimulationLoop(WuhanCoronaVirus).koi
└── episim.koi
├── README.md
├── demo.gif
├── episim.koi
├── episim.py
├── graphgenerator.koi
├── graphgenerator.py
├── legend.png
├── legend.svg
├── nodegraph.pkl
├── nodegraph40000.pkl
├── nodegraph6000.pkl
├── nodegraph8000.pkl
├── summarydata.xlsx
├── summarydataChina.xlsx
├── summarydataLosAngelesBestCase.xlsx
├── summarydataLosAngelesFullRun.xlsx
├── summarydataLosAngelesPossibleCase.xlsx
└── summarydataLosAngelesUndetectedCase.xlsx
/FullRun.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/FullRun.png
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2019 Dave Pena
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/NodeLibrary/SimulationLoop(WuhanCoronaVirus).koi:
--------------------------------------------------------------------------------
1 | {"nodes":[{"nid":25,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus)","x":464,"y":183,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.nodes[0]['Status'] = 'Infected Symptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n asymptomatic=1\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Undetected Infections\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n if deaths >= 60:\n running = False\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],100),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(totaldetected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Undetected Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(totaldetected)\n datacontainer['Current Undetected Infections'].append(asymptomatic)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n \n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus","rnaught":"0","infprob":"0.045","inflength":"20","mortality":"0.13","infincubation":"5","infdetected":"10"},"description":"\n\n","showExecution":false,"showFunction":true,"executionClass":"","descriptionClass":"","importsClass":"","functionClass":"uk-active","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":false,"headerStyle":{"background":"#ad1562"}}],"connections":[],"title":"Simulation Loop (Wuhan Corona Virus)","showPopup":false,"current":-1,"count":1,"servers":{},"serverselection":null}
--------------------------------------------------------------------------------
/NodeLibrary/episim.koi:
--------------------------------------------------------------------------------
1 | {"nodes":[{"nid":4,"version":"0.0.1","uid":5,"category":"logical","type":"Data2Terminal","x":71,"y":458,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef data2Terminal|||(*args,**kwargs):\n global {userinput}\n skwargs=kwargs.copy()\n print('kwargs from this node available as global variable {userinput}')\n return kwargs","python_import":"","python_exec":"data2Terminal|||","settings":[{"key":"userinput","label":"Global Variable Name"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"skwargs"},"kwargs":{},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Global Variable Name\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#15ad60"}},{"nid":0,"version":"1.0.0","uid":1,"category":"function","type":"Start","x":10,"y":10,"headerStyle":{"background":""},"fields":{"in":[],"out":[{"name":"start"}]},"python":"def start():\n return {'Start':True,'Settings':{'Verbose':True},'Status':{},'Threads':[]}\n","python_import":"","python_exec":"start","settings":[],"icon":"./static/media/start.c59e7ea6.svg","data":{},"kwargs":{},"description":"\n\nThe Start Node \n\n_This node is required to run a standard nodegraph that use the onstart decorator_\n\n\n## This Node Changes\n\nThis node initializes all standard kwargs\n\n## Getting This Node to Work\n\nThis node takes no inputs and only one can be connected in any given nodegraph. It sets Verbose to True by default, this setting outputs nodes run in the terminal when they are running and other debugging information. \n\n\n### Input State Requirements\n\nNone\n\n### Output State\n\n```python\nkwargs={\n Start: {},\n Settings: {Verbose:True},\n Data:{},\n Status: {}\n }\n```\n\n## Example Usage\n\n\n\n## Todo\n\n## Known Issues\n\nNo known Issues\n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n\nThis node signifies the beginning of your node graph by the streme server. The start node passes true from the start output pin initiating the nodes it is connected to.","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[]","stringPins":"{\"in\":[],\"out\":[{\"name\":\"start\"}]}","togglePins":false},{"nid":1,"version":"1.0.0","uid":10,"category":"function","type":"Stop","x":1037,"y":118,"headerStyle":{"background":"#ad1562"},"fields":{"in":[{"name":"start"}],"out":[]},"python":"@on_start\ndef stop(*args,**kwargs):\n print('exiting')\n sys.exit()\n ","python_import":"import sys","python_exec":"stop","settings":[],"icon":"./static/media/stop.a2171794.svg","data":{},"kwargs":{},"description":"This node ends the existing node experiment. It will not stop processes that have started in parallel using split nodes.","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[]}","togglePins":false},{"nid":3,"version":"0.0.1","uid":5,"category":"logical","type":"Generate nodeGraph","x":91,"y":113,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef nodeGraph(*args,**kwargs):\n nodes = {nodes}\n k = {k}\n prob = {prob}\n G = nx.newman_watts_strogatz_graph(nodes, k, prob)\n kwargs['Settings']['NetworkX']={{\"nodes\":nodes, \"k\":k,\"prob\":prob}}\n pos = nx.spring_layout(G)\n kwargs['Settings']['NetworkX']['Pos']= pos\n #nx.draw(G, with_labels=True, font_weight='bold',pos=pos)\n #plt.show()\n \n kwargs['Data'] = G\n return kwargs","python_import":"import networkx as nx\nimport matplotlib.pyplot as plt","python_exec":"nodeGraph","settings":[{"key":"nodes","label":"Number of Nodes (Integer)"},{"key":"k","label":"Mean number of edges per node (Integer)"},{"key":"prob","label":"Probability nodes will have an extra edge (0-1)"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"Filename6kl5hte":"","nodes":"6000","k":"20","prob":"0.1","anodes":"10"},"kwargs":{},"description":"\n\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"nodes\",\"label\":\"Number of Nodes (Integer)\"},{\"key\":\"k\",\"label\":\"Mean number of edges per node (Integer)\"},{\"key\":\"prob\",\"label\":\"Probability nodes will have an extra edge (0-1)\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#15ad60"},"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":7,"uid":"custom","category":"function","type":"Simulation Loop (Ebola)","x":912,"y":584,"fields":{"in":[{"name":"start"},{"name":"action1"},{"name":"action2"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[15,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (1000,900)\n inflength = 16\n mortality = 0.7\n rnaught = 2.0\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n G.graph['colors'] = {\"Naive\":(52, 101, 164),\"Infected\":(78, 154, 6),\"Immune\":(237, 212, 0),\"Dead\":(204, 0, 0)}\n G.graph['disease'] = {\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}\n # Add Parameters to Nodegraph\n for nid in G:\n G.node[nid]['Status'] = 'Naive'\n G.node[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.node[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pygame.image.load(\"logo32x32.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"Pygame Simulation\")\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((250, 250, 250))\n green = (0, 255, 0) \n blue = (0, 0, 128) \n deaths = 0\n infected = 1\n font = pg.font.Font('freesansbold.ttf', 32)\n text = font.render('R0:{}'.format(rnaught), True, green, blue)\n textinfected = font.render('Infected:{}'.format(infected), True, green, blue)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n screen.blit(background, (0, 0))\n screen.blit(text, (0,0))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(85, 87, 83))\n for nid in pos:\n color = G.graph['colors'][G.node[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.node[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.node[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.node[nid]['Day'] = G.node[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.node[cnode]['Status'] != \"Dead\": \n if G.node[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.node[cnode]['Status'] = 'Infected'\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n textdeath = font.render('Deaths:{}'.format(deaths), True, green, blue)\n screen.blit(textdeath, (0,30))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random","python_exec":"simLoop|||","settings":[],"icon":"./static/media/for.e1841ce0.svg","data":{},"description":"\n\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[]","stringPins":"{\"in\":[{\"name\":\"start\"},{\"name\":\"action1\"},{\"name\":\"action2\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false},{"nid":11,"uid":"custom","category":"function","type":"Simulation Loop (Meningococcal Disease)","x":731,"y":79,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (1000,900)\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.node[nid]['Status'] = 'Naive'\n G.node[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.node[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.node[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.node[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.node[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.node[nid]['Day'] = G.node[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.node[cnode]['Status'] != \"Dead\": \n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.node[cnode]['Status'] = 'Infected'\n G.node[cnode]['Day'] = 0\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(825,20))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Bacterial Meningitis","rnaught":"2.5","infprob":"0","inflength":"21","mortality":"0.1"},"description":"\n\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"},"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":12,"uid":"custom","category":"logical","type":"data2Pickle","x":56,"y":527,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef data2Pickle|||(*args,**kwargs):\n pickle.dump(kwargs,open(\"{userinput}\",\"wb\"))\n return kwargs","python_import":"import pickle","python_exec":"data2Pickle|||","settings":[{"key":"userinput","label":"Save Path including Filename"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"nodegraph6000.pkl"},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Save Path including Filename\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#15ad60"},"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":14,"uid":"custom","category":"logical","type":"loadPickle","x":252,"y":572,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef loadPickle|||(*args,**kwargs):\n kwargs = pickle.load(open(\"{userinput}\",\"rb\"))\n return kwargs","python_import":"import pickle","python_exec":"loadPickle|||","settings":[{"key":"userinput","label":"Load Path including Filename"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"nodegraph.pkl"},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Load Path including Filename\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#15ad60"}},{"nid":15,"uid":"custom","category":"logical","type":"Load NodeGraph 1200 People","x":93,"y":174,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef loadPickle|||(*args,**kwargs):\n kwargs = pickle.load(open(\"{userinput}\",\"rb\"))\n kwargs['Threads'] = []\n\n return kwargs","python_import":"import pickle","python_exec":"loadPickle|||","settings":[{"key":"userinput","label":"Load Path including Filename"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"nodegraph.pkl"},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Load Path including Filename\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":17,"uid":"custom","category":"function","type":"Simulation Loop (Polio)","x":736,"y":228,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (1000,900)\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.node[nid]['Status'] = 'Naive'\n G.node[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.node[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.node[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.node[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.node[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.node[nid]['Day'] = G.node[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.node[cnode]['Status'] != \"Dead\":\n if G.node[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.node[cnode]['Status'] = 'Infected'\n G.node[cnode]['Day'] = 0\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(825,20))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Polio","rnaught":"2.5","infprob":"0","inflength":"42","mortality":"0.001"},"description":"\n\n","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"}},{"nid":18,"uid":"custom","category":"function","type":"Simulation Loop (Measles UnVaccinated)","x":745,"y":300,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n#############################################################################\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.nodes[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n if infday == 366:\n running = False\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.nodes[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected'\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(625,-15))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random\n","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Measles","rnaught":"15","infprob":"0","inflength":"8","mortality":"0.002"},"description":"\n\n","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"}},{"nid":19,"uid":"custom","category":"function","type":"Simulation Loop (Ebola)","x":865,"y":438,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (1000,900)\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.node[nid]['Status'] = 'Naive'\n G.node[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.node[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.node[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.node[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.node[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.node[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.node[nid]['Day'] = G.node[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.node[cnode]['Status'] != \"Dead\":\n if G.node[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.node[cnode]['Status'] = 'Infected'\n G.node[cnode]['Day'] = 0\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(825,20))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Ebola","rnaught":"2","infprob":"0","inflength":"16","mortality":"0.8"},"description":"\n\n","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"}},{"nid":23,"uid":"custom","category":"function","type":"Simulation Loop (Ebola PreVaccine)","x":954,"y":198,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n#############################################################################\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.nodes[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n if infday == 366:\n running = False\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.nodes[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected'\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(625,-15))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport numpy as np\nimport random\n","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Ebola","rnaught":"2","infprob":"0","inflength":"16","mortality":"0.8"},"description":"\n\n","showExecution":false,"showDescription":true,"showImports":false,"showFunction":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"},"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":24,"uid":"custom","category":"function","type":"Simulation Loop (Normal Flu)","x":943,"y":320,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected\":(145, 238, 7),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob, \"Infection Length\" : inflength, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n G.nodes[0]['Status'] = 'Infected'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = 1\n infday = 0 \n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n infected = 0\n if infday == 366:\n running = False\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],80),0)\n screen.blit(text, (10,10))\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,40))\n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n if G.nodes[nid]['Status'] == \"Infected\":\n infected = infected+1\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n roll = random.random()\n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected'\n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n screen.blit(legend,(625,-15))\n pg.display.flip()\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\n","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Normal Flu","rnaught":"2","infprob":"0","inflength":"8","mortality":"0.0015"},"description":"\n\n","showExecution":false,"showDescription":false,"showImports":false,"showFunction":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"headerStyle":{"background":"#adac15"}},{"nid":26,"uid":"custom","category":"logical","type":"Load NodeGraph 6000 People","x":87,"y":256,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef loadPickle|||(*args,**kwargs):\n kwargs = pickle.load(open(\"{userinput}\",\"rb\"))\n kwargs['Threads'] = []\n\n return kwargs","python_import":"import pickle","python_exec":"loadPickle|||","settings":[{"key":"userinput","label":"Load Path including Filename"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"nodegraph6000.pkl"},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showFunction":false,"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","showDescription":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Load Path including Filename\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":27,"uid":"custom","category":"logical","type":"Load NodeGraph Los Angeles","x":98,"y":324,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"@on_start\ndef loadPickle|||(*args,**kwargs):\n kwargs = pickle.load(open(\"{userinput}\",\"rb\"))\n kwargs['Threads'] = []\n\n return kwargs","python_import":"import pickle","python_exec":"loadPickle|||","settings":[{"key":"userinput","label":"Load Path including Filename"}],"icon":"./static/media/blankNode.38260d5b.svg","data":{"undefined":"","userinput":"nodegraph40000.pkl"},"description":"\n\nDumps kwargs in a node to a user specified global variable\n\n_Allows you to introspect within nodes after your algorithm completes_\n\n[YOUR GITHUB PAGE FOR THE NODE SOURCE](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet#links)\n\n## This Node Changes\n\n\n\nThis node takes a Pandas Dataframe in outputs and excel file and returns kwargs unchanged to the next node.\n\n## Getting This Node to Work\n\nThis node will work if it provided with a Dataframe in kwargs{Data} and Pandas is installed\n\n### Installing Dependencies\n\nThis node requires Pandas. Pandas can be installed using pip in your terminal \n\n```bash\npip install pandas\n```\n\nif you have python2 and python3 installed\n\n```bash\npip3 install pandas\n```\n\nIf you are using Anaconda (Pandas is already installed in the default setup):\n\n```bash\nconda install pandas\n```\n\n### Input State Requirements\n\nThis node requires that kwargs{Data} be a [Pandas Dataframe](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html)\n\n```python\nkwargs={\n Start: True,\n Settings: Any,\n Data: Any Pandas Dataframe,\n Status: Any,\n }\n```\n\n### Output State\n\n```python\nkwargs={\n Start: No Change,\n Settings: No Change,\n Data: No Change,\n Status: No Change\n }\n```\n\n## Example Usage\n\nCheck out this simple demo of this node working : [Download Koi](https://mykoilocation.com)\n\n\n\n## Todo\n\n- Something\n\n- Another Thing\n\n- Something Else\n\n## Known Issues\n\n- We coded it poorly\n\n- Works only on the first of every month that is also within 5 days of the summer solstice \n\n## License\n\nUsage of this node is provided under the [MIT License](http://http//opensource.org/licenses/mit-license.php). See LICENSE for the full details.\n","showExecution":false,"showFunction":false,"executionClass":"","descriptionClass":"uk-active","importsClass":"","functionClass":"","showDescription":true,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"userinput\",\"label\":\"Load Path including Filename\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false},{"nid":28,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus) China","x":417,"y":499,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n #rng = default_rng()\n #initialinf = rng.choice({startinf}*2, size={startinf}, replace=False)\n initialinf = random.sample(range({startinf}*2),{startinf})\n\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n for nid in initialinf:\n G.nodes[nid]['Status'] = 'Infected Asymptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = {startinf}\n asymptomatic={startinf}\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Asymptomatic Infections\":[],\"Current Infectious Carriers\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n############################### STOP CRITERIA ########################################################\n# if infday >= 6:\n# running = False\n# totaldetected = infected - (asymptomatic+immune+deaths)\n# if detected >= 2:\n# running = False\n if deaths >=56:\n running = False\n \n#######################################################################################################\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n #totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(detected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Asymptomatic Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n carriers = infected-asymptomatic-detected\n textcarriers = font.render('Current Infectious Carriers: {{}}'.format(carriers), True, green)\n screen.blit(textcarriers, (10,100))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(detected)\n datacontainer['Current Asymptomatic Infections'].append(asymptomatic)\n datacontainer['Current Infectious Carriers'].append(carriers)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n df.to_excel('summarydataChina.xlsx')\n kwargs['Data'] = df\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"},{"key":"startinf","label":"Number of Initial Seed infections"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus China","rnaught":"0","infprob":"0.08","inflength":"20","mortality":"0.16","infincubation":"5","infdetected":"10","startinf":"21"},"description":"\n\n","showExecution":false,"showFunction":false,"executionClass":"","descriptionClass":"","importsClass":"uk-active ","functionClass":"","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"},{\"key\":\"startinf\",\"label\":\"Number of Initial Seed infections\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":true},{"nid":32,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus) Los Angeles Possible Case","x":409,"y":153,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n #initialinf = rng.choice({startinf}*2, size={startinf}, replace=False)\n initialinf = random.sample(range({startinf}*2),{startinf})\n\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n for nid in initialinf:\n G.nodes[nid]['Status'] = 'Infected Asymptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = {startinf}\n asymptomatic={startinf}\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Asymptomatic Infections\":[],\"Current Infectious Carriers\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n############################### STOP CRITERIA ########################################################\n# if infday >= 6:\n# running = False\n# totaldetected = infected - (asymptomatic+immune+deaths)\n if detected >= 2:\n running = False\n# if deaths >=56:\n# running = False\n \n#######################################################################################################\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n #totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(detected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Asymptomatic Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n carriers = infected-asymptomatic-detected\n textcarriers = font.render('Current Infectious Carriers: {{}}'.format(carriers), True, green)\n screen.blit(textcarriers, (10,100))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(detected)\n datacontainer['Current Asymptomatic Infections'].append(asymptomatic)\n datacontainer['Current Infectious Carriers'].append(carriers)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n df.to_excel('summarydataLosAngelesPossibleCase.xlsx')\n kwargs['Data'] = df\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"},{"key":"startinf","label":"Number of Initial Seed infections"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus Los Angeles","rnaught":"0","infprob":"0.08","inflength":"20","mortality":"0.16","infincubation":"5","infdetected":"10","startinf":"18"},"description":"\n\n","showExecution":false,"showFunction":true,"executionClass":"","descriptionClass":"","importsClass":"","functionClass":"uk-active","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"},{\"key\":\"startinf\",\"label\":\"Number of Initial Seed infections\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":false},{"nid":33,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus) Los Angeles Best Case","x":411,"y":46,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n #initialinf = rng.choice({startinf}*2, size={startinf}, replace=False)\n initialinf = random.sample(range({startinf}*2),{startinf})\n\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n for nid in initialinf:\n G.nodes[nid]['Status'] = 'Infected Asymptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = {startinf}\n asymptomatic={startinf}\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Asymptomatic Infections\":[],\"Current Infectious Carriers\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n############################### STOP CRITERIA ########################################################\n# if infday >= 6:\n# running = False\n# totaldetected = infected - (asymptomatic+immune+deaths)\n if detected >= 2:\n running = False\n# if deaths >=56:\n# running = False\n \n#######################################################################################################\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n #totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(detected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Asymptomatic Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n carriers = infected-asymptomatic-detected\n textcarriers = font.render('Current Infectious Carriers: {{}}'.format(carriers), True, green)\n screen.blit(textcarriers, (10,100))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(detected)\n datacontainer['Current Asymptomatic Infections'].append(asymptomatic)\n datacontainer['Current Infectious Carriers'].append(carriers)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n df.to_excel('summarydataLosAngelesBestCase.xlsx')\n kwargs['Data'] = df\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"},{"key":"startinf","label":"Number of Initial Seed infections"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus Los Angeles","rnaught":"0","infprob":"0.08","inflength":"20","mortality":"0.16","infincubation":"5","infdetected":"10","startinf":"2"},"description":"\n\n","showExecution":false,"showFunction":true,"executionClass":"","descriptionClass":"","importsClass":"","functionClass":"uk-active","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"},{\"key\":\"startinf\",\"label\":\"Number of Initial Seed infections\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":false},{"nid":34,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus) Los Angeles Worst Case","x":411,"y":283,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n #rng = default_rng()\n #initialinf = rng.choice({startinf}*2, size={startinf}, replace=False)\n initialinf = random.sample(range({startinf}*2),{startinf})\n\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n for nid in initialinf:\n G.nodes[nid]['Status'] = 'Infected Asymptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = {startinf}\n asymptomatic={startinf}\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Asymptomatic Infections\":[],\"Current Infectious Carriers\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n############################### STOP CRITERIA ########################################################\n if infday >= 30:\n running = False\n# totaldetected = infected - (asymptomatic+immune+deaths)\n# if detected >= 2:\n# running = False\n# if deaths >=56:\n# running = False\n \n#######################################################################################################\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n #totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(detected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Asymptomatic Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n carriers = infected-asymptomatic-detected\n textcarriers = font.render('Current Infectious Carriers: {{}}'.format(carriers), True, green)\n screen.blit(textcarriers, (10,100))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(detected)\n datacontainer['Current Asymptomatic Infections'].append(asymptomatic)\n datacontainer['Current Infectious Carriers'].append(carriers)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n df.to_excel('summarydataLosAngelesUndetectedCase.xlsx')\n kwargs['Data'] = df\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"},{"key":"startinf","label":"Number of Initial Seed infections"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus Los Angeles","rnaught":"0","infprob":"0.08","inflength":"20","mortality":"0.16","infincubation":"5","infdetected":"10","startinf":"1"},"description":"\n\n","showExecution":false,"showFunction":true,"executionClass":"","descriptionClass":"","importsClass":"","functionClass":"uk-active","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"},{\"key\":\"startinf\",\"label\":\"Number of Initial Seed infections\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":false},{"nid":35,"uid":"custom","category":"function","type":"Simulation Loop (Wuhan Corona Virus) Los Angeles Complete Run","x":401,"y":390,"fields":{"in":[{"name":"start"}],"out":[{"name":"pass"}]},"python":"def remap(narray,resolution):\n x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])\n y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])\n return np.array([int(x),int(y)])\n\n\n@on_start\ndef simLoop|||(*args, **kwargs):\n############################## EDIT THESE PARAMETERS ########################\n inflength = {inflength}\n mortality = {mortality}\n rnaught = {rnaught}\n infprob = {infprob}\n infincubation={infincubation}\n infdetected = {infdetected}\n#############################################################################\n\n if infprob == 0:\n infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)\n if rnaught == 0:\n rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength\n \n pos = kwargs['Settings']['NetworkX']['Pos']\n initialinf = random.sample(range({startinf}*2),{startinf})\n G = kwargs['Data']\n screensize = (800,800)\n G.graph['colors'] = {{\"Naive\":(99, 7, 238),\"Infected Symptomatic\":(145, 238, 7),\"Infected Asymptomatic\":(159, 190, 126),\"Immune\":(255, 235, 59),\"Dead\":(239, 99, 7)}}\n G.graph['disease'] = {{\"Infection Probability\" : infprob,\"Infection Detection\":infdetected, \"Infection Length\" : inflength, \"Incubation Length\":infincubation, \"Mortality\": mortality}}\n # Add Parameters to Nodegraph\n for nid in G:\n G.nodes[nid]['Status'] = 'Naive'\n G.nodes[nid]['Day'] = 0\n # Remap -1 1 square to pygame resolution\n for nid in pos:\n pos[nid] = remap(pos[nid],screensize) \n for nid in initialinf:\n G.nodes[nid]['Status'] = 'Infected Asymptomatic'\n # initialize the pygame module\n pg.init()\n # load and set the logo\n #logo = pg.image.load(\"logo.png\")\n #pg.display.set_icon(logo)\n pg.display.set_caption(\"{diseasename}\")\n legend = pg.image.load('legend.png')\n \n # create a surface on screen that has the size of 240 x 180\n screen = pg.display.set_mode(screensize)\n background = pg.Surface(screen.get_size())\n background = background.convert()\n background.fill((236, 239, 241))\n \n green = (255, 255, 255) \n deaths = 0\n infected = {startinf}\n asymptomatic={startinf}\n symptomatic=0 \n willdie=0\n infday = 0 \n detected = 0\n immune = 0 \n datacontainer={{\"Day\":[],\"Deaths\":[],\"Cumulative Infections\":[],\"Detected Infections\":[],\"Current Asymptomatic Infections\":[],\"Current Infectious Carriers\":[]}}\n font = pg.font.Font(pg.font.match_font('lato'), 20)\n text = font.render('R0: {{0}}'.format(rnaught), True, green)\n textinfected = font.render('Infected: {{0}}'.format(infected), True, green)\n # define a variable to control the main loop\n running = True\n \n # main loop\n while running:\n #infected = 0\n############################### STOP CRITERIA ########################################################\n if infday >= 365:\n running = False\n# totaldetected = infected - (asymptomatic+immune+deaths)\n# if detected >= 2:\n# running = False\n# if deaths >=56:\n# running = False\n \n#######################################################################################################\n\n screen.blit(background, (0, 0))\n pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)\n\n textdeath = font.render('Deaths: {{}}'.format(deaths), True, green)\n screen.blit(textdeath, (10,10))\n textinfected = font.render('Cumulative Infections: {{}}'.format(infected), True, green)\n screen.blit(textinfected, (10,25))\n \n textinfected = font.render('Recoveries: {{}}'.format(immune), True, green)\n screen.blit(textinfected, (10,55))\n textinfected = font.render('Naive: {{}}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)\n screen.blit(textinfected, (10,40))\n #totaldetected = infected - (asymptomatic+immune+deaths)\n textsymptomatic = font.render('Cumulative Detected Infections: {{}}'.format(detected), True, green)\n screen.blit(textsymptomatic, (10,70))\n \n textasymptomatic = font.render('Current Asymptomatic Infections: {{}}'.format(asymptomatic), True, green)\n screen.blit(textasymptomatic, (10,85))\n carriers = infected-asymptomatic-detected\n textcarriers = font.render('Current Infectious Carriers: {{}}'.format(carriers), True, green)\n screen.blit(textcarriers, (10,100))\n \n textdays = font.render('Epidemic Day: {{}}'.format(infday), True, green)\n screen.blit(textdays, ((screensize[0]/2)-40,10))\n textrnaught = font.render('R0: {{}}'.format(rnaught), True, green)\n screen.blit(textrnaught, ((screensize[0]/2)-40,25))\n \n textmortality = font.render('Mortality Rate: {{}} %'.format(mortality*100), True, green)\n screen.blit(textmortality, ((screensize[0]/2)+40,25))\n datacontainer['Day'].append(infday)\n datacontainer['Deaths'].append(deaths)\n datacontainer['Cumulative Infections'].append(infected)\n datacontainer['Detected Infections'].append(detected)\n datacontainer['Current Asymptomatic Infections'].append(asymptomatic)\n datacontainer['Current Infectious Carriers'].append(carriers)\n \n for nid in pos:\n for e in G.edges(nid):\n cnode= e[1]\n gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))\n for nid in pos:\n color = G.graph['colors'][G.nodes[nid]['Status']]\n gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)\n gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)\n \n if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:\n detected = detected + 1 \n if G.nodes[nid]['Status'] == \"Infected Symptomatic\":\n if G.nodes[nid]['Day'] == G.graph['disease'][\"Infection Length\"]:\n roll = random.random()\n if roll >= G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Immune\"\n immune = immune+1\n symptomatic = symptomatic-1\n if roll < G.graph['disease']['Mortality']:\n G.nodes[nid]['Status'] = \"Dead\"\n symptomatic = symptomatic-1\n deaths=deaths+1\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1\n for e in G.edges(nid):\n cnode = e[1]\n if G.nodes[cnode]['Status'] != \"Dead\": \n if G.nodes[cnode]['Status'] != \"Immune\":\n if G.nodes[cnode]['Status'] != \"Infected Asymptomatic\":\n if G.nodes[cnode]['Status'] != \"Infected Symptomatic\":\n roll = random.random()\n \n if roll < G.graph['disease']['Infection Probability']:\n G.nodes[cnode]['Status'] = 'Infected Asymptomatic'\n infected = infected+1\n asymptomatic = asymptomatic+1\n \n \n if G.nodes[nid]['Status'] == \"Infected Asymptomatic\":\n if G.nodes[nid]['Day'] >= G.graph['disease'][\"Incubation Length\"]:\n asymptomatic = asymptomatic-1\n symptomatic = symptomatic+1\n G.nodes[nid]['Status'] = \"Infected Symptomatic\"\n G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1 \n \n # event handling, gets all event from the event queue\n for event in pg.event.get():\n # only do something if the event is of type QUIT\n if event.type == pg.QUIT:\n # change the value to False, to exit the main loop\n running = False\n infday = infday + 1\n \n screen.blit(legend,(625,-15))\n pg.display.flip()\n time.sleep(10)\n df=pd.DataFrame(datacontainer)\n print(df)\n df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()\n #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)\n #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)\n sns.lineplot(data=df, linewidth=2)\n\n\n plt.show()\n df.to_excel('summarydataLosAngelesFullRun.xlsx')\n kwargs['Data'] = df\n return kwargs\n ","python_import":"import pygame as pg\nimport pygame.gfxdraw as gfx \nimport random\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nimport time","python_exec":"simLoop|||","settings":[{"key":"diseasename","label":"Disease Name For Pygame Caption"},{"key":"rnaught","label":"R0 for disease (Set to zero to calculate from infection probablility)"},{"key":"infprob","label":"Contact Infection Probability (Set this value to 0 to have it calculated from R0)"},{"key":"infincubation","label":"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day "},{"key":"infdetected","label":"Number of Simulation Cycles before illness Detection"},{"key":"inflength","label":"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day "},{"key":"mortality","label":"What is the Mortality Rate of this disease 1 is equivalent to 100%"},{"key":"startinf","label":"Number of Initial Seed infections"}],"icon":"./static/media/for.e1841ce0.svg","data":{"diseasename":"Novel coronavirus (2019-nCoV) Wuhan Virus Los Angeles","rnaught":"0","infprob":"0.08","inflength":"20","mortality":"0.16","infincubation":"5","infdetected":"10","startinf":"2"},"description":"\n\n","showExecution":false,"showFunction":true,"executionClass":"","descriptionClass":"","importsClass":"","functionClass":"uk-active","showDescription":false,"toggleMarkdown":false,"toggleUserDialog":false,"stringDialog":"[{\"key\":\"diseasename\",\"label\":\"Disease Name For Pygame Caption\"},{\"key\":\"rnaught\",\"label\":\"R0 for disease (Set to zero to calculate from infection probablility)\"},{\"key\":\"infprob\",\"label\":\"Contact Infection Probability (Set this value to 0 to have it calculated from R0)\"},{\"key\":\"infincubation\",\"label\":\"Number of Simulation Cycles a Person Remains Incubated. One cycle for example could represent a day \"},{\"key\":\"infdetected\",\"label\":\"Number of Simulation Cycles before illness Detection\"},{\"key\":\"inflength\",\"label\":\"Number of Simulation Cycles a Person Remains infectious after cycle zero, incubation is excluded. One cycle for example could represent a day \"},{\"key\":\"mortality\",\"label\":\"What is the Mortality Rate of this disease 1 is equivalent to 100%\"},{\"key\":\"startinf\",\"label\":\"Number of Initial Seed infections\"}]","stringPins":"{\"in\":[{\"name\":\"start\"}],\"out\":[{\"name\":\"pass\"}]}","togglePins":false,"pintracker":false,"dialogtracker":false,"descriptiontracker":false,"showImports":false}],"connections":[{"from_node":0,"from":"start","to_node":27,"to":"start"},{"from_node":27,"from":"pass","to_node":34,"to":"start"}],"title":"episim","showPopup":false,"current":-1,"count":35,"servers":{},"serverselection":null}
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | 
2 |
3 | # Pygame Epidemic Simulator
4 |
5 | ALTHOUGH I TAKE GREAT PAIN TO TRY TO REPRESENT WHAT IS BEING REPORTED TAKE THE OUTPUT OF THIS SIMULATOR WITH PLENTY OF SALT.
6 |
7 | Simulation of infection progression given a reasonably natural network of connected individuals. [StremeCoder Desktop](https://gumroad.com/pluri) is the IDE used to maintain this code and generate different infection parameters, but is not required and the python source can be modified directly in the simloop function to simulate different diseases.
8 |
9 |
10 |
11 | You can also use the webversion for free [StremeCoder Live](https://pluricorp.com/stremecoder) it runs on your browser so it lacks some system functions that can only work on desktop apps, but it will still get the job done.
12 |
13 |
14 |
15 |
16 |
17 | This simulation uses a social network generated in networkx
18 |
19 | nx.newman_watts_strogatz_graph(nodes, k, prob)
20 |
21 | where
22 |
23 | nodes = 1200
24 |
25 | k = 20
26 |
27 | prob = 0.1
28 |
29 | To reduce runtime and reproduce the same population for each disease node a pickle was generated of that random starter population and run through a pygame simulation node.
30 |
31 | The simulation takes in
32 |
33 | R0, infection probability, mortality rate, infection length, or infection probability if R0 is set to 0. Otherwise, infection probability is estimated from R0 algebraically. Each cycle of the simulation for each infected node connection infection and mortality are simulated by random number selection against provided probabilities. The simulation is set to end at 365 days. To use this simulation in a montecarlo remove the pygame dependencies to eliminate the visualizations and perform your desired statistical calculation on the resulting network then repeat the process n times.
34 |
35 | ## Demo
36 |
37 | 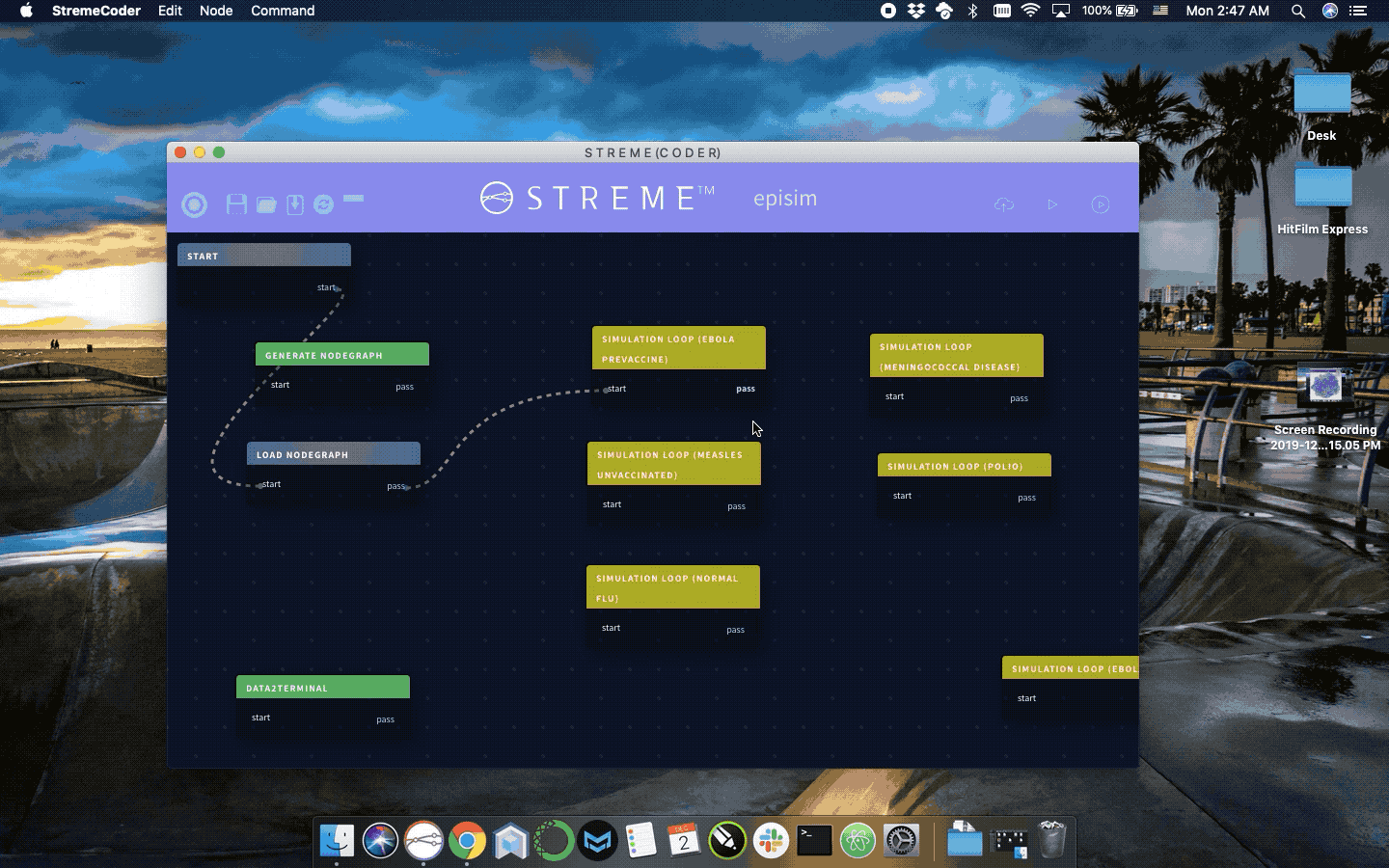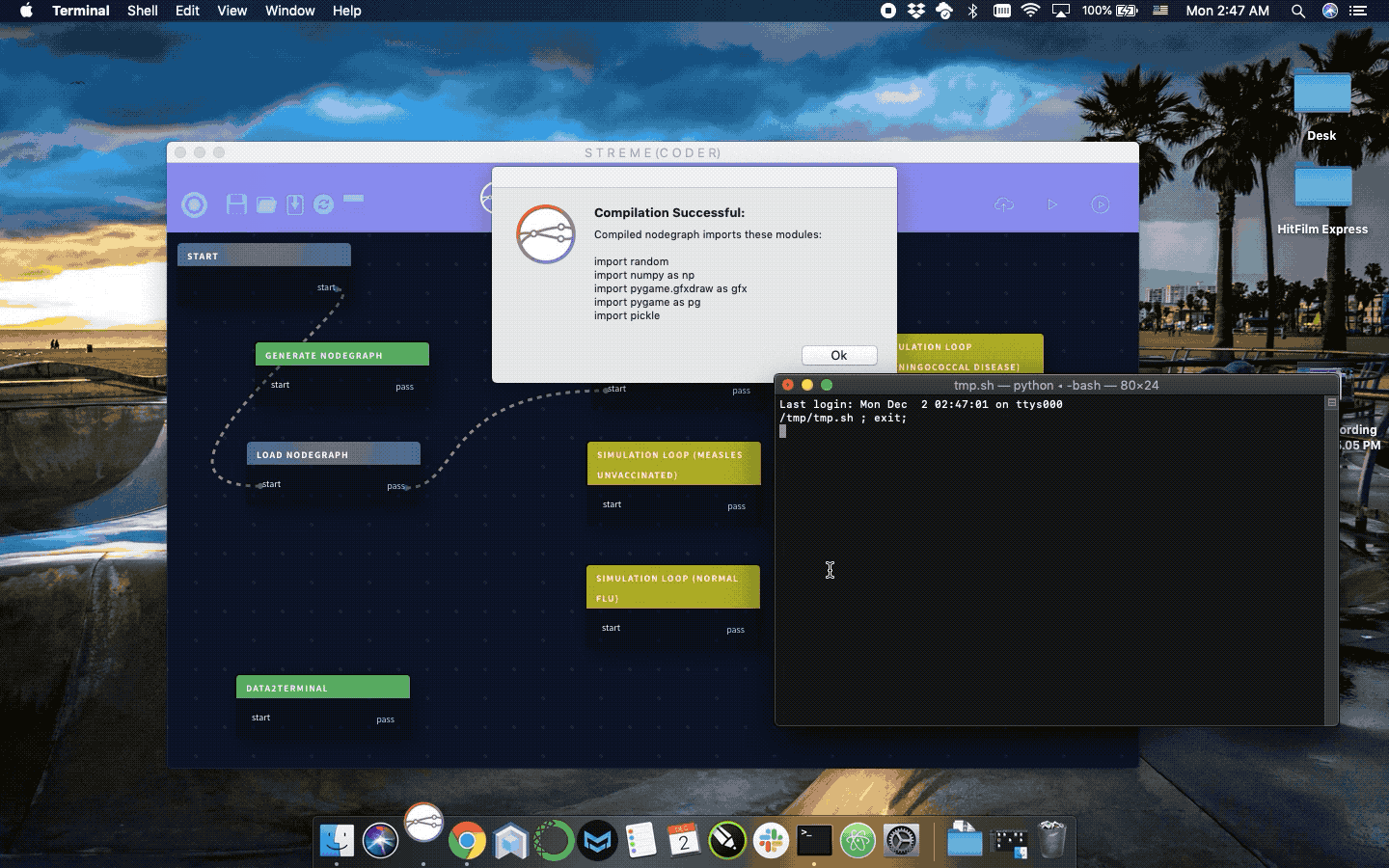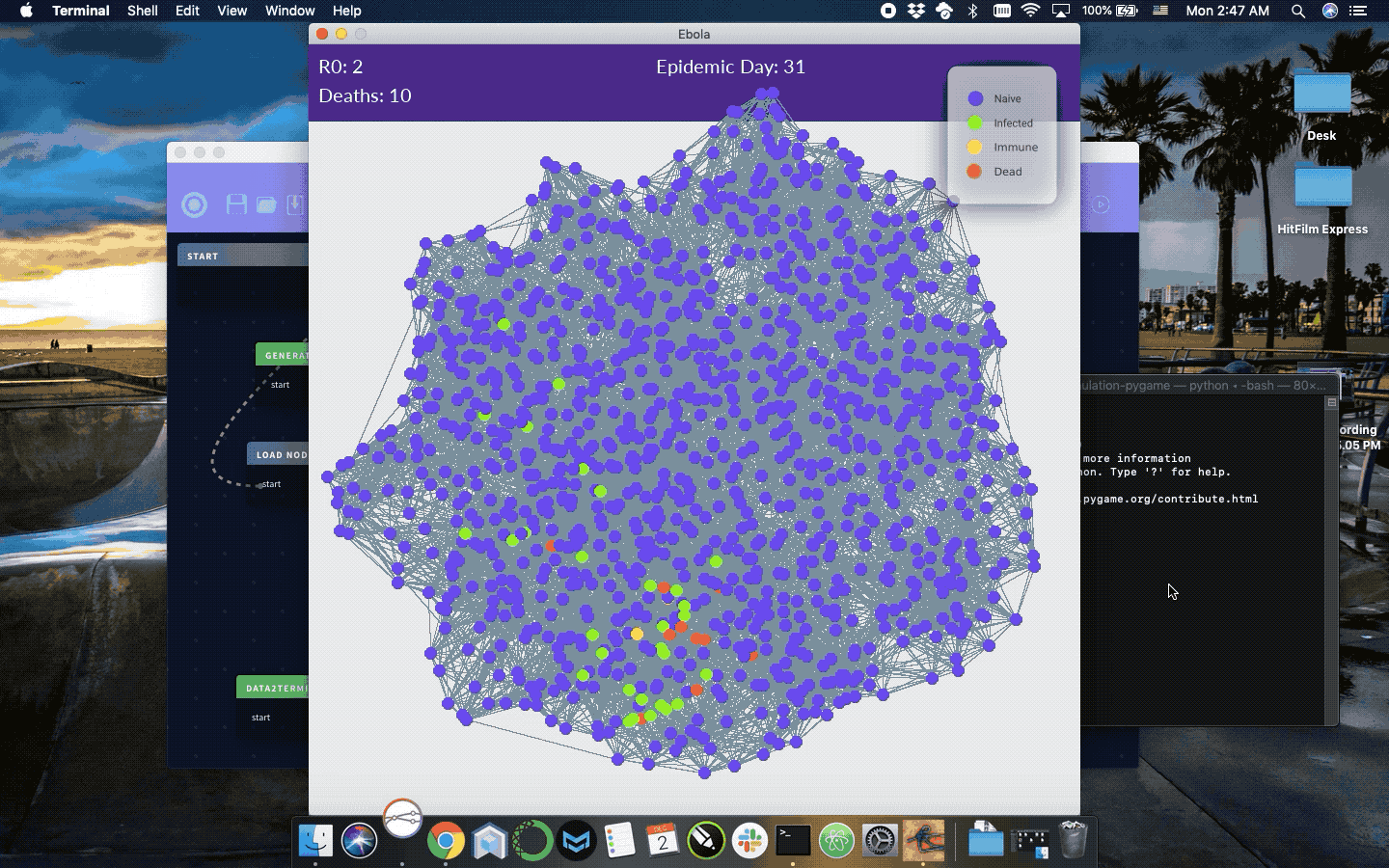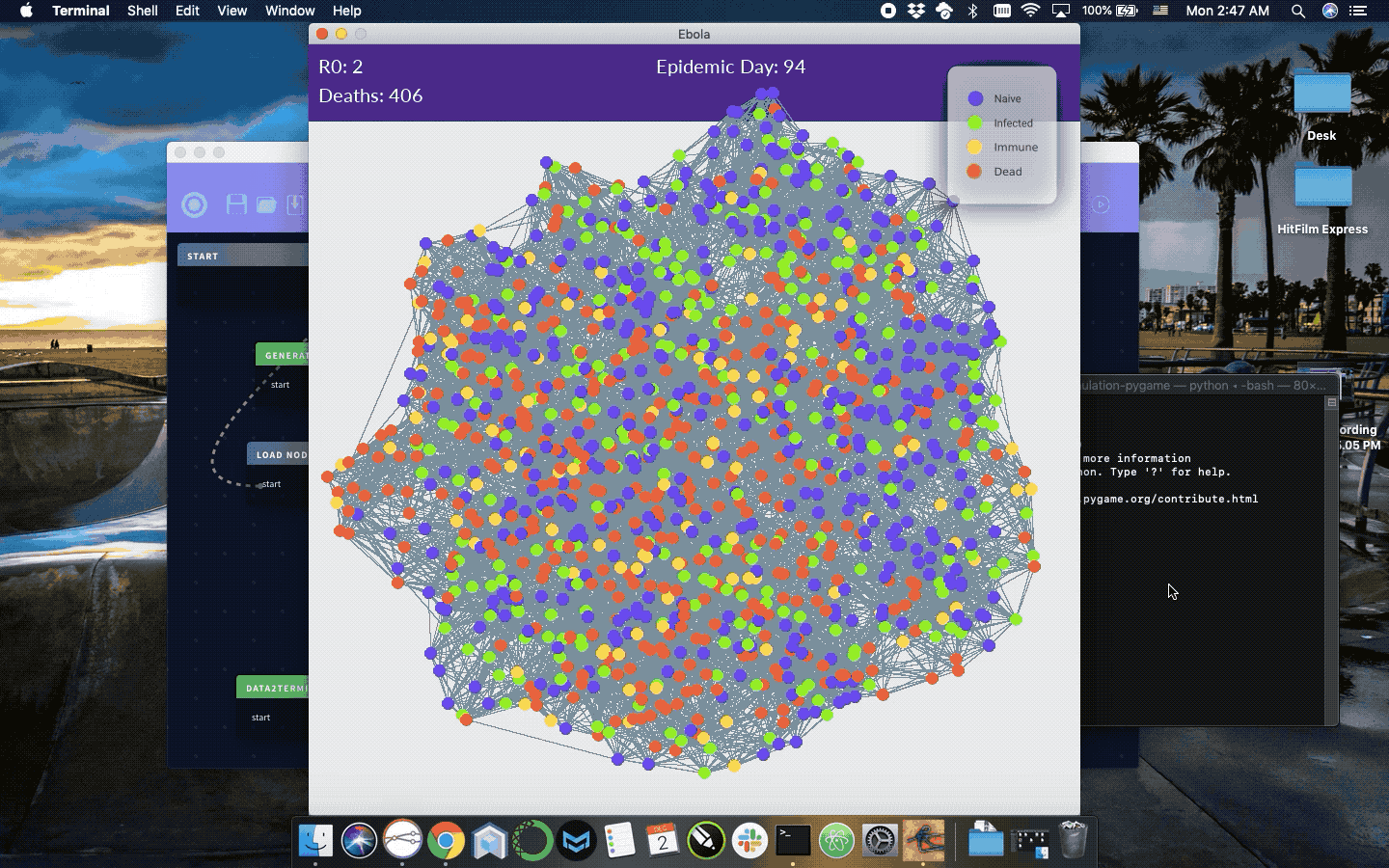
38 |
39 | ## Install
40 |
41 | ## Requirements
42 |
43 | This simulation requires python3, pygame, networkx, and numpy
44 |
45 | ### Pygame
46 |
47 | Runs the visualization of the node state changes
48 |
49 | ```
50 | pip install pygame
51 | or
52 | pip3 install pygame
53 | ```
54 |
55 | ### numpy
56 |
57 | To map the node network into the screen resolution space. Refer to [SCIPY](https://www.scipy.org/install.html)
58 |
59 | ```
60 | conda install numpy
61 | ```
62 |
63 | ### NetworkX
64 |
65 | Generates the social network of nodes to run in the simulation You need the latest version or you will get an error
66 |
67 | ```
68 | pip install networkx
69 | or
70 | pip3 install networkx
71 | ```
72 |
73 | ### Seaborn
74 |
75 | ```
76 | pip install seaborn
77 | ```
78 |
79 | ### Pandas
80 |
81 | ```
82 | pip install pandas
83 | ```
84 |
85 | In a terminal run
86 |
87 | ```
88 | python episim.py
89 | ```
90 |
91 | ## License
92 |
93 | [MIT](https://opensource.org/licenses/MIT)
94 |
--------------------------------------------------------------------------------
/demo.gif:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/demo.gif
--------------------------------------------------------------------------------
/episim.py:
--------------------------------------------------------------------------------
1 |
2 | #!/usr/bin/env python
3 | from decorator import decorator
4 |
5 | import time
6 | import random
7 | import matplotlib.pyplot as plt
8 | import numpy as np
9 | import pygame as pg
10 | import seaborn as sns
11 | import pygame.gfxdraw as gfx
12 | import pandas as pd
13 | import pickle
14 |
15 |
16 | @decorator
17 | def on_start(func,*args, **kwargs):
18 | if kwargs !={}:
19 | try:
20 | if kwargs['Start']:
21 | if 'Verbose' in kwargs['Settings']:
22 | if kwargs['Settings']['Verbose']:
23 | print(func)
24 | pass
25 | response= func(*args,**kwargs)
26 | return response
27 | else:
28 | kwargs['Start'] = False
29 | print(func,"DID NOT START")
30 | return(kwargs)
31 | except Exception as e:
32 | print('NODE ERROR OCCURED TRYING TO START NODE FUNCTION:')
33 | print('===========================================')
34 | print(func,e)
35 | print('===========================================')
36 | print('LAST STATE SET TO:')
37 | print('===========================================')
38 | print('ekwargs')
39 | print('===========================================')
40 | print('LAST NODE FUNCTION SET TO:')
41 | print('===========================================')
42 | print('efunc')
43 | print('===========================================')
44 | global ekwargs
45 | global efunc
46 | ekwargs = kwargs
47 | efunc = func
48 | print('HALTING')
49 | raise
50 | else:
51 | print('Empty kwargs')
52 | return ()
53 |
54 |
55 |
56 | def start():
57 | return {'Start':True,'Settings':{'Verbose':True},'Status':{},'Threads':[]}
58 |
59 |
60 | @on_start
61 | def loadPickle_node_27(*args,**kwargs):
62 | kwargs = pickle.load(open("nodegraph40000.pkl","rb"))
63 | kwargs['Threads'] = []
64 |
65 | return kwargs
66 |
67 | def remap(narray,resolution):
68 | x = np.interp(narray[0],[-1,1],[15,resolution[0]-15])
69 | y = np.interp(narray[1],[-1,1],[50,resolution[1]- 15])
70 | return np.array([int(x),int(y)])
71 |
72 |
73 | @on_start
74 | def simLoop_node_35(*args, **kwargs):
75 | ############################## EDIT THESE PARAMETERS ########################
76 | inflength = 20
77 | mortality = 0.0815
78 | rnaught = 0
79 | infprob = 0.049
80 | infincubation=5
81 | infdetected = 10
82 | #############################################################################
83 |
84 | if infprob == 0:
85 | infprob = rnaught/(kwargs['Settings']['NetworkX']['k']*inflength)
86 | if rnaught == 0:
87 | rnaught = infprob*kwargs['Settings']['NetworkX']['k']*inflength
88 |
89 | pos = kwargs['Settings']['NetworkX']['Pos']
90 | initialinf = random.sample(range(2*2),2)
91 | G = kwargs['Data']
92 | screensize = (800,800)
93 | G.graph['colors'] = {"Naive":(99, 7, 238),"Infected Symptomatic":(145, 238, 7),"Infected Asymptomatic":(159, 190, 126),"Immune":(255, 235, 59),"Dead":(239, 99, 7)}
94 | G.graph['disease'] = {"Infection Probability" : infprob,"Infection Detection":infdetected, "Infection Length" : inflength, "Incubation Length":infincubation, "Mortality": mortality}
95 | # Add Parameters to Nodegraph
96 | for nid in G:
97 | G.nodes[nid]['Status'] = 'Naive'
98 | G.nodes[nid]['Day'] = 0
99 | # Remap -1 1 square to pygame resolution
100 | for nid in pos:
101 | pos[nid] = remap(pos[nid],screensize)
102 | for nid in initialinf:
103 | G.nodes[nid]['Status'] = 'Infected Asymptomatic'
104 | # initialize the pygame module
105 | pg.init()
106 | # load and set the logo
107 | #logo = pg.image.load("logo.png")
108 | #pg.display.set_icon(logo)
109 | pg.display.set_caption("Novel coronavirus (2019-nCoV) Wuhan Virus Los Angeles")
110 | legend = pg.image.load('legend.png')
111 |
112 | # create a surface on screen that has the size of 240 x 180
113 | screen = pg.display.set_mode(screensize)
114 | background = pg.Surface(screen.get_size())
115 | background = background.convert()
116 | background.fill((236, 239, 241))
117 |
118 | green = (255, 255, 255)
119 | deaths = 0
120 | infected = 2
121 | asymptomatic=2
122 | symptomatic=0
123 | willdie=0
124 | infday = 0
125 | detected = 0
126 | immune = 0
127 | datacontainer={"Day":[],"Deaths":[],"Cumulative Infections":[],"Detected Infections":[],"Current Asymptomatic Infections":[],"Current Infectious Carriers":[]}
128 | font = pg.font.Font(pg.font.match_font('lato'), 20)
129 | text = font.render('R0: {0}'.format(rnaught), True, green)
130 | textinfected = font.render('Infected: {0}'.format(infected), True, green)
131 | # define a variable to control the main loop
132 | running = True
133 |
134 | # main loop
135 | while running:
136 | #infected = 0
137 | ############################### STOP CRITERIA ########################################################
138 | if infday >= 200:
139 | running = False
140 | # totaldetected = infected - (asymptomatic+immune+deaths)
141 | # if detected >= 2:
142 | # running = False
143 | # if deaths >=56:
144 | # running = False
145 |
146 | #######################################################################################################
147 |
148 | screen.blit(background, (0, 0))
149 | pg.draw.rect(background,(74,20,140),(0,0,screensize[0],120),0)
150 |
151 | textdeath = font.render('Deaths: {}'.format(deaths), True, green)
152 | screen.blit(textdeath, (10,10))
153 | textinfected = font.render('Cumulative Infections: {}'.format(infected), True, green)
154 | screen.blit(textinfected, (10,25))
155 |
156 | textinfected = font.render('Recoveries: {}'.format(immune), True, green)
157 | screen.blit(textinfected, (10,55))
158 | textinfected = font.render('Naive: {}'.format(kwargs['Settings']['NetworkX']['nodes']-infected), True, green)
159 | screen.blit(textinfected, (10,40))
160 | #totaldetected = infected - (asymptomatic+immune+deaths)
161 | textsymptomatic = font.render('Cumulative Detected Infections: {}'.format(detected), True, green)
162 | screen.blit(textsymptomatic, (10,70))
163 |
164 | textasymptomatic = font.render('Current Asymptomatic Infections: {}'.format(asymptomatic), True, green)
165 | screen.blit(textasymptomatic, (10,85))
166 | carriers = infected-asymptomatic-detected
167 | textcarriers = font.render('Current Infectious Carriers: {}'.format(carriers), True, green)
168 | screen.blit(textcarriers, (10,100))
169 |
170 | textdays = font.render('Epidemic Day: {}'.format(infday), True, green)
171 | screen.blit(textdays, ((screensize[0]/2)-40,10))
172 | textrnaught = font.render('R0: {}'.format(rnaught), True, green)
173 | screen.blit(textrnaught, ((screensize[0]/2)-40,25))
174 |
175 | textmortality = font.render('Mortality Rate: {} %'.format(mortality*100), True, green)
176 | screen.blit(textmortality, ((screensize[0]/2)+40,25))
177 | datacontainer['Day'].append(infday)
178 | datacontainer['Deaths'].append(deaths)
179 | datacontainer['Cumulative Infections'].append(infected)
180 | datacontainer['Detected Infections'].append(detected)
181 | datacontainer['Current Asymptomatic Infections'].append(asymptomatic)
182 | datacontainer['Current Infectious Carriers'].append(carriers)
183 |
184 | for nid in pos:
185 | for e in G.edges(nid):
186 | cnode= e[1]
187 | gfx.line(screen,pos[nid][0],pos[nid][1],pos[cnode][0],pos[cnode][1],(120,144,156))
188 | for nid in pos:
189 | color = G.graph['colors'][G.nodes[nid]['Status']]
190 | gfx.aacircle(screen, pos[nid][0],pos[nid][1], 6, color)
191 | gfx.filled_circle(screen, pos[nid][0],pos[nid][1], 6, color)
192 |
193 | if G.nodes[nid]['Day'] == G.graph['disease']['Infection Detection']:
194 | detected = detected + 1
195 | if G.nodes[nid]['Status'] == "Infected Symptomatic":
196 | if G.nodes[nid]['Day'] == G.graph['disease']["Infection Length"]:
197 | roll = random.random()
198 | if roll >= G.graph['disease']['Mortality']:
199 | G.nodes[nid]['Status'] = "Immune"
200 | immune = immune+1
201 | symptomatic = symptomatic-1
202 | if roll < G.graph['disease']['Mortality']:
203 | G.nodes[nid]['Status'] = "Dead"
204 | symptomatic = symptomatic-1
205 | deaths=deaths+1
206 | G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1
207 | for e in G.edges(nid):
208 | cnode = e[1]
209 | if G.nodes[cnode]['Status'] != "Dead":
210 | if G.nodes[cnode]['Status'] != "Immune":
211 | if G.nodes[cnode]['Status'] != "Infected Asymptomatic":
212 | if G.nodes[cnode]['Status'] != "Infected Symptomatic":
213 | roll = random.random()
214 |
215 | if roll < G.graph['disease']['Infection Probability']:
216 | G.nodes[cnode]['Status'] = 'Infected Asymptomatic'
217 | infected = infected+1
218 | asymptomatic = asymptomatic+1
219 |
220 |
221 | if G.nodes[nid]['Status'] == "Infected Asymptomatic":
222 | if G.nodes[nid]['Day'] >= G.graph['disease']["Incubation Length"]:
223 | asymptomatic = asymptomatic-1
224 | symptomatic = symptomatic+1
225 | G.nodes[nid]['Status'] = "Infected Symptomatic"
226 | G.nodes[nid]['Day'] = G.nodes[nid]['Day'] + 1
227 |
228 | # event handling, gets all event from the event queue
229 | for event in pg.event.get():
230 | # only do something if the event is of type QUIT
231 | if event.type == pg.QUIT:
232 | # change the value to False, to exit the main loop
233 | running = False
234 | infday = infday + 1
235 |
236 | screen.blit(legend,(625,-15))
237 | pg.display.flip()
238 | time.sleep(10)
239 | df=pd.DataFrame(datacontainer)
240 | print(df)
241 | df = df[['Deaths','Detected Infections','Cumulative Infections','Current Asymptomatic Infections','Current Infectious Carriers']].rolling(1).sum()
242 | #sns.barplot(x = 'Day', y = 'Cumulative Infections', data = df)
243 | #sns.barplot(x = 'Day', y = 'Detected Infections', palette = 'magma', data = df)
244 | sns.lineplot(data=df, linewidth=2)
245 |
246 |
247 | plt.show()
248 | df.to_excel('summarydataLosAngelesFullRun.xlsx')
249 | kwargs['Data'] = df
250 | return kwargs
251 |
252 |
253 |
254 |
255 | class StremeNode:
256 | def __init__(self):
257 | pass
258 |
259 | def run(self,*args,**kwargs):
260 | self.kwargs=simLoop_node_35(**loadPickle_node_27(**kwargs))
261 | return (self.kwargs)
262 |
263 | class liveprocess:
264 | def __init__(self):
265 | pass
266 |
267 | def run(self,expname="Local"):
268 | self.response=simLoop_node_35(**loadPickle_node_27(**start()))
269 | return(self.response)
270 |
271 | if __name__ == '__main__':
272 | process = liveprocess()
273 | process.run()
274 |
--------------------------------------------------------------------------------
/graphgenerator.py:
--------------------------------------------------------------------------------
1 |
2 | #!/usr/bin/env python
3 | from decorator import decorator
4 |
5 | import matplotlib.pyplot as plt
6 | import pickle
7 | import networkx as nx
8 |
9 |
10 | @decorator
11 | def on_start(func,*args, **kwargs):
12 | if kwargs !={}:
13 | try:
14 | if kwargs['Start']:
15 | if 'Verbose' in kwargs['Settings']:
16 | if kwargs['Settings']['Verbose']:
17 | print(func)
18 | pass
19 | response= func(*args,**kwargs)
20 | return response
21 | else:
22 | kwargs['Start'] = False
23 | print(func,"DID NOT START")
24 | return(kwargs)
25 | except Exception as e:
26 | print('NODE ERROR OCCURED TRYING TO START NODE FUNCTION:')
27 | print('===========================================')
28 | print(func,e)
29 | print('===========================================')
30 | print('LAST STATE SET TO:')
31 | print('===========================================')
32 | print('ekwargs')
33 | print('===========================================')
34 | print('LAST NODE FUNCTION SET TO:')
35 | print('===========================================')
36 | print('efunc')
37 | print('===========================================')
38 | global ekwargs
39 | global efunc
40 | ekwargs = kwargs
41 | efunc = func
42 | print('HALTING')
43 | raise
44 | else:
45 | print('Empty kwargs')
46 | return ()
47 |
48 |
49 |
50 | def start():
51 | return {'Start':True,'Settings':{'Verbose':True},'Status':{},'Threads':[]}
52 |
53 |
54 | @on_start
55 | def nodeGraph(*args,**kwargs):
56 | nodes = 8000
57 | k = 20
58 | prob = 0.1
59 | G = nx.newman_watts_strogatz_graph(nodes, k, prob)
60 | kwargs['Settings']['NetworkX']={"nodes":nodes, "k":k,"prob":prob}
61 | pos = nx.spring_layout(G)
62 | kwargs['Settings']['NetworkX']['Pos']= pos
63 | #nx.draw(G, with_labels=True, font_weight='bold',pos=pos)
64 | #plt.show()
65 |
66 | kwargs['Data'] = G
67 | return kwargs
68 |
69 | @on_start
70 | def data2Pickle_node_12(*args,**kwargs):
71 | pickle.dump(kwargs,open("nodegraph8000.pkl","wb"))
72 | return kwargs
73 |
74 |
75 |
76 | class StremeNode:
77 | def __init__(self):
78 | pass
79 |
80 | def run(self,*args,**kwargs):
81 | self.kwargs=data2Pickle_node_12(**nodeGraph(**kwargs))
82 | return (self.kwargs)
83 |
84 | class liveprocess:
85 | def __init__(self):
86 | pass
87 |
88 | def run(self,expname="Local"):
89 | self.response=data2Pickle_node_12(**nodeGraph(**start()))
90 | return(self.response)
91 |
92 | if __name__ == '__main__':
93 | process = liveprocess()
94 | process.run()
95 |
--------------------------------------------------------------------------------
/legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/legend.png
--------------------------------------------------------------------------------
/legend.svg:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
190 |
--------------------------------------------------------------------------------
/nodegraph.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/nodegraph.pkl
--------------------------------------------------------------------------------
/nodegraph40000.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/nodegraph40000.pkl
--------------------------------------------------------------------------------
/nodegraph6000.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/nodegraph6000.pkl
--------------------------------------------------------------------------------
/nodegraph8000.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/nodegraph8000.pkl
--------------------------------------------------------------------------------
/summarydata.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydata.xlsx
--------------------------------------------------------------------------------
/summarydataChina.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydataChina.xlsx
--------------------------------------------------------------------------------
/summarydataLosAngelesBestCase.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydataLosAngelesBestCase.xlsx
--------------------------------------------------------------------------------
/summarydataLosAngelesFullRun.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydataLosAngelesFullRun.xlsx
--------------------------------------------------------------------------------
/summarydataLosAngelesPossibleCase.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydataLosAngelesPossibleCase.xlsx
--------------------------------------------------------------------------------
/summarydataLosAngelesUndetectedCase.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dfpena/epidemic-simulation-pygame/7e952ccddeacaa5cbcb7c3abb02ee46617a567ff/summarydataLosAngelesUndetectedCase.xlsx
--------------------------------------------------------------------------------