├── .gitattributes
├── raw
├── .gitignore
├── gource
│ ├── logo.png
│ ├── avatar
│ │ ├── Alex Wolf.jpg
│ │ ├── Manu Setty.jpg
│ │ ├── Yvan Saeys.jpg
│ │ ├── Chuck Herring.jpg
│ │ ├── Helena Todorov.png
│ │ ├── Philipp Angerer.jpg
│ │ ├── Wouter Saelens.png
│ │ ├── Koen Van den Berge.jpg
│ │ ├── Robrecht Cannoodt.png
│ │ └── Daniel C Ellwanger.jpeg
│ ├── deprecated_repositories
│ │ ├── mlr_ti.txt
│ │ └── dynvaria.txt
│ └── gource_captions
├── 12-manuscript
│ ├── Software.pdf
│ ├── ReportingSummary.pdf
│ ├── linenumbers.sty
│ ├── manuscript.sty
│ ├── common.sty
│ └── nature-biotechnology.csl
├── README.md
├── overview_evaluation_v4.pdf
├── overview_evaluation_v5.pdf
├── 02-metrics
│ ├── trajectory_model_example.pdf
│ ├── 01-metric_characterisation
│ │ ├── metrics_geodesic.pdf
│ │ └── metrics_prediction.pdf
│ └── 03-aggregation
│ │ └── example.csv
└── 01-datasets
│ └── 02-synthetic
│ └── samplers
├── package
├── tests
│ ├── testthat
│ │ ├── test-format.R
│ │ ├── test-dataset_generation.R
│ │ ├── test-google_scholar.R
│ │ ├── test-plotting_trajectory_types.R
│ │ ├── test-labels.R
│ │ └── test-get_dynbenchmark_folder.R
│ └── testthat.R
├── LICENSE
├── data
│ ├── labels.rda
│ ├── id_mapper.rda
│ ├── prior_types.rda
│ ├── error_reasons.rda
│ ├── prior_usages.rda
│ ├── qc_categories.rda
│ ├── topinf_types.rda
│ ├── method_statuses.rda
│ ├── qc_applications.rda
│ ├── method_status_colours.rda
│ └── non_inclusion_reasons.rda
├── man
│ ├── figures
│ │ ├── logo.png
│ │ ├── logo2.png
│ │ ├── datasets.png
│ │ └── dependencies.png
│ ├── label_time.Rd
│ ├── label_long.Rd
│ ├── label_capitalise.Rd
│ ├── label_thousands.Rd
│ ├── label_method.Rd
│ ├── setup_singularity_methods.Rd
│ ├── limits_metric.Rd
│ ├── setup_refs_globally.Rd
│ ├── label_extrema.Rd
│ ├── label_vector.Rd
│ ├── tag_first.Rd
│ ├── label_memory.Rd
│ ├── label_short.Rd
│ ├── benchmark_qsub_fun.Rd
│ ├── print_url.Rd
│ ├── labels.Rd
│ ├── label_pvalue.Rd
│ ├── method_status_colours.Rd
│ ├── pdf_supplementary_note.Rd
│ ├── label_facet.Rd
│ ├── prior_types.Rd
│ ├── paramoptim_qsub_fun.Rd
│ ├── create_replacers.Rd
│ ├── error_reasons.Rd
│ ├── link_to_results.Rd
│ ├── qc_categories.Rd
│ ├── prior_usages.Rd
│ ├── method_statuses.Rd
│ ├── qc_applications.Rd
│ ├── google_scholar.Rd
│ ├── topinf_types.Rd
│ ├── metrics_evaluated.Rd
│ ├── setup_refs.Rd
│ ├── theme_empty.Rd
│ ├── id_mapper.Rd
│ ├── render_equations.Rd
│ ├── load_or_generate.Rd
│ ├── non_inclusion_reasons.Rd
│ ├── headtail.Rd
│ ├── metrics_characterised.Rd
│ ├── google_scholar_num_citations.Rd
│ ├── estimate_platform.Rd
│ ├── label_wrap.Rd
│ ├── replace_svg.Rd
│ ├── load_methods.Rd
│ ├── legend_at.Rd
│ ├── generate_prior_mini.Rd
│ ├── download_dataset_source_file.Rd
│ ├── reformat_tribbles.Rd
│ ├── word_manuscript.Rd
│ ├── get_dynbenchmark_folder.Rd
│ ├── github_markdown_nested.Rd
│ ├── ref.Rd
│ ├── load_dyneval_model.Rd
│ ├── load_dataset.Rd
│ ├── load_platforms.Rd
│ ├── label_metric.Rd
│ ├── knit_nest.Rd
│ ├── add_table.Rd
│ ├── dynbenchmark.Rd
│ ├── nacor.Rd
│ ├── paramoptim_bind_results.Rd
│ ├── benchmark_fetch_results.Rd
│ ├── common_dynbenchmark_format.Rd
│ ├── extract_scripts_documentation.Rd
│ ├── benchmark_bind_results.Rd
│ ├── paramoptim_fetch_results.Rd
│ ├── theme_pub.Rd
│ ├── fix_relative_paths.Rd
│ ├── experiment.Rd
│ ├── add_fig.Rd
│ ├── platform.Rd
│ ├── process_raw_dataset.Rd
│ ├── dataset_preprocessing.Rd
│ ├── benchmark_aggregate.Rd
│ ├── pdf_manuscript.Rd
│ ├── qsub_pmap.Rd
│ ├── make_obj_fun.Rd
│ ├── plot_trajectory_types.Rd
│ └── paramoptim_generate_design.Rd
├── R
│ ├── nacor.R
│ ├── metrics.R
│ ├── setup_pipeline.R
│ ├── headtail.R
│ ├── text_helpers.R
│ ├── qsub.R
│ ├── setup_methods.R
│ ├── topologies_with_same_n_milestones.R
│ ├── install_fontawesome.R
│ ├── data.R
│ ├── text_equations.R
│ ├── load_dyneval_model.R
│ ├── setup_get_dynbenchmark_folder.R
│ ├── package.R
│ ├── plotting_priors_mini.R
│ ├── reformat_tribbles.R
│ └── setup_experiment.R
├── inst
│ ├── tester.md
│ └── tester.Rmd
├── .Rbuildignore
├── data-raw
│ ├── priors.R
│ ├── qc_categorisation.R
│ ├── method_characterisation.R
│ ├── non_inclusion_reasons.R
│ ├── id_mapper.R
│ ├── method_status_colours.R
│ └── labels.R
└── LICENSE.md
├── scripts
├── varia
│ ├── manual_ti
│ │ ├── .gitignore
│ │ ├── arrow.png
│ │ ├── pen.png
│ │ └── select.png
│ ├── generate_hypotheses_mindmap
│ │ ├── mindmap.pdf
│ │ ├── hypotheses_script.R
│ │ └── mindmap.tex
│ ├── README.Rmd
│ ├── rename_experiment.sh
│ ├── README.md
│ └── gource
│ │ └── 2_gource_gits.R
├── 12-manuscript
│ ├── README.Rmd
│ ├── render_results.R
│ ├── render_readmes.R
│ ├── README.md
│ └── render_supplementary_notes.R
├── 03-methods
│ ├── 03-method_characterisation
│ │ ├── README.Rmd
│ │ ├── 02-tools_table.R
│ │ └── README.md
│ ├── 02-tool_qc
│ │ ├── README.Rmd
│ │ ├── 01-qc_aspects_table.R
│ │ └── README.md
│ ├── 01-gather_methods_information
│ │ ├── README.Rmd
│ │ ├── README.md
│ │ └── 03-add_quality_control.R
│ └── README.Rmd
├── 01-datasets
│ ├── 04-dataset_characterisation
│ │ ├── README.Rmd
│ │ └── README.md
│ ├── 01-real
│ │ ├── helpers-download_from_sources
│ │ │ ├── wip
│ │ │ │ ├── developing-thymus_kernfeld.R
│ │ │ │ ├── mammary-gland-epitheliam_pal.R
│ │ │ │ ├── tabula-muris.R
│ │ │ │ └── pbmc-citeseq_stoeckius.R
│ │ │ ├── dataset_macrophage-salmonella_saliba.R
│ │ │ ├── dataset_distal-lung-epithelium_treutlein.R
│ │ │ ├── dataset_mESC-differentation_hayashi.R
│ │ │ ├── dataset_fibroblast-reprogramming_treutlein.R
│ │ │ ├── dataset_cellbench_luyitian.R
│ │ │ ├── dataset_cortical-interneuron-differentiation_frazer.R
│ │ │ ├── dataset_developing-dendritic-cells_schlitzer.R
│ │ │ ├── dataset_stimulated-dendritic-cells_shalek.R
│ │ │ └── dataset_planaria-plass.R
│ │ ├── 04-datasets_table.R
│ │ ├── README.Rmd
│ │ ├── 01-download_from_sources.R
│ │ ├── 02-filter_and_normalise.R
│ │ └── README.md
│ ├── 02-synthetic
│ │ ├── 03-gather_metadata.R
│ │ ├── 04-dyngen_samplers_table.R
│ │ ├── README.Rmd
│ │ ├── 02d-simulate_dyntoy_datasets.R
│ │ ├── 02a-simulate_dyngen_datasets.R
│ │ ├── 02c-simulate_splatter_datasets.R
│ │ └── 02b-simulate_prosstt_datasets.R
│ ├── 03-download_from_prism.R
│ ├── README.Rmd
│ └── 00-download_from_zenodo.R
├── 08-summary
│ ├── 2a_columns_suppfig.R
│ ├── 2a_columns_summary.R
│ ├── README.Rmd
│ ├── 2a_columns_detailed.R
│ └── README.md
├── 06-benchmark
│ ├── README.Rmd
│ ├── 3d-compare_sources.R
│ ├── inspect_identity_timings.R
│ ├── README.md
│ ├── 3-diagnostic_figures.R
│ └── 3b-time_mem_predictions.R
├── 11-example_predictions
│ ├── README.Rmd
│ ├── README.md
│ └── 02-funky.R
├── 09-guidelines
│ ├── README.Rmd
│ ├── README.md
│ └── 01-aggregate.R
├── 10-benchmark_interpretation
│ ├── README.Rmd
│ ├── helper-04-complementarity.R
│ ├── 05-benchmark_interpretation_figure.R
│ └── 02-dataset_sources.R
├── 02-metrics
│ ├── 02-metric_conformity
│ │ ├── helper-create_perturbation_images.R
│ │ ├── 04-assess_similarity.R
│ │ ├── README.Rmd
│ │ └── 01-generate_reference_datasets.R
│ ├── 03-aggregation
│ │ ├── README.Rmd
│ │ └── README.md
│ ├── 01-metric_characterisation
│ │ ├── README.Rmd
│ │ ├── 03-clustering.R
│ │ └── README.md
│ └── README.Rmd
├── 04-method_testing
│ ├── README.Rmd
│ ├── 0-pull_singularity_images.R
│ └── README.md
├── 07-stability
│ ├── README.Rmd
│ └── README.md
├── README.Rmd
├── 05-scaling
│ └── README.Rmd
└── README.md
├── LICENSE
├── manuscript
├── .gitignore
├── README.md
├── README.Rmd
└── supplementary.Rmd
├── .gitmodules
├── derived
├── README.md
└── README.Rmd
├── .gitignore
├── dynbenchmark.Rproj
├── .travis.yml
└── LICENSE.md
/.gitattributes:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/raw/.gitignore:
--------------------------------------------------------------------------------
1 | *.pdf
2 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-format.R:
--------------------------------------------------------------------------------
1 | context("")
2 |
--------------------------------------------------------------------------------
/scripts/varia/manual_ti/.gitignore:
--------------------------------------------------------------------------------
1 | guide.html
2 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | YEAR: 2014-2020
2 | COPYRIGHT HOLDER: Robrecht Cannoodt, Wouter Saelens
3 |
--------------------------------------------------------------------------------
/package/LICENSE:
--------------------------------------------------------------------------------
1 | YEAR: 2014-2020
2 | COPYRIGHT HOLDER: Robrecht Cannoodt, Wouter Saelens
3 |
--------------------------------------------------------------------------------
/manuscript/.gitignore:
--------------------------------------------------------------------------------
1 | .scratch/
2 | *.bcf
3 | *.run.xml
4 | *.pdf
5 | *.tex
6 | *.md
7 | *.rds
8 |
--------------------------------------------------------------------------------
/raw/gource/logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/logo.png
--------------------------------------------------------------------------------
/package/data/labels.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/labels.rda
--------------------------------------------------------------------------------
/package/data/id_mapper.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/id_mapper.rda
--------------------------------------------------------------------------------
/package/data/prior_types.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/prior_types.rda
--------------------------------------------------------------------------------
/package/man/figures/logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/man/figures/logo.png
--------------------------------------------------------------------------------
/package/data/error_reasons.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/error_reasons.rda
--------------------------------------------------------------------------------
/package/data/prior_usages.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/prior_usages.rda
--------------------------------------------------------------------------------
/package/data/qc_categories.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/qc_categories.rda
--------------------------------------------------------------------------------
/package/data/topinf_types.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/topinf_types.rda
--------------------------------------------------------------------------------
/package/man/figures/logo2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/man/figures/logo2.png
--------------------------------------------------------------------------------
/raw/12-manuscript/Software.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/12-manuscript/Software.pdf
--------------------------------------------------------------------------------
/raw/README.md:
--------------------------------------------------------------------------------
1 | # Raw
2 |
3 | This folder contains all manually created files, such as spreadsheets or figures.
4 |
--------------------------------------------------------------------------------
/raw/overview_evaluation_v4.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/overview_evaluation_v4.pdf
--------------------------------------------------------------------------------
/raw/overview_evaluation_v5.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/overview_evaluation_v5.pdf
--------------------------------------------------------------------------------
/.gitmodules:
--------------------------------------------------------------------------------
1 | [submodule "results"]
2 | path = results
3 | url = https://github.com/dynverse/dynbenchmark_results.git
4 |
--------------------------------------------------------------------------------
/package/data/method_statuses.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/method_statuses.rda
--------------------------------------------------------------------------------
/package/data/qc_applications.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/qc_applications.rda
--------------------------------------------------------------------------------
/package/man/figures/datasets.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/man/figures/datasets.png
--------------------------------------------------------------------------------
/raw/gource/avatar/Alex Wolf.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Alex Wolf.jpg
--------------------------------------------------------------------------------
/raw/gource/avatar/Manu Setty.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Manu Setty.jpg
--------------------------------------------------------------------------------
/raw/gource/avatar/Yvan Saeys.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Yvan Saeys.jpg
--------------------------------------------------------------------------------
/scripts/varia/manual_ti/arrow.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/scripts/varia/manual_ti/arrow.png
--------------------------------------------------------------------------------
/scripts/varia/manual_ti/pen.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/scripts/varia/manual_ti/pen.png
--------------------------------------------------------------------------------
/raw/gource/avatar/Chuck Herring.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Chuck Herring.jpg
--------------------------------------------------------------------------------
/scripts/varia/manual_ti/select.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/scripts/varia/manual_ti/select.png
--------------------------------------------------------------------------------
/package/data/method_status_colours.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/method_status_colours.rda
--------------------------------------------------------------------------------
/package/data/non_inclusion_reasons.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/data/non_inclusion_reasons.rda
--------------------------------------------------------------------------------
/package/man/figures/dependencies.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/package/man/figures/dependencies.png
--------------------------------------------------------------------------------
/raw/12-manuscript/ReportingSummary.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/12-manuscript/ReportingSummary.pdf
--------------------------------------------------------------------------------
/raw/gource/avatar/Helena Todorov.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Helena Todorov.png
--------------------------------------------------------------------------------
/raw/gource/avatar/Philipp Angerer.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Philipp Angerer.jpg
--------------------------------------------------------------------------------
/raw/gource/avatar/Wouter Saelens.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Wouter Saelens.png
--------------------------------------------------------------------------------
/raw/gource/avatar/Koen Van den Berge.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Koen Van den Berge.jpg
--------------------------------------------------------------------------------
/raw/gource/avatar/Robrecht Cannoodt.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Robrecht Cannoodt.png
--------------------------------------------------------------------------------
/manuscript/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Manuscript
3 |
4 | The files to generate the manuscript: the main paper and the
5 | supplementary material.
6 |
--------------------------------------------------------------------------------
/raw/02-metrics/trajectory_model_example.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/02-metrics/trajectory_model_example.pdf
--------------------------------------------------------------------------------
/raw/gource/avatar/Daniel C Ellwanger.jpeg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/gource/avatar/Daniel C Ellwanger.jpeg
--------------------------------------------------------------------------------
/scripts/varia/generate_hypotheses_mindmap/mindmap.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/scripts/varia/generate_hypotheses_mindmap/mindmap.pdf
--------------------------------------------------------------------------------
/raw/02-metrics/01-metric_characterisation/metrics_geodesic.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/02-metrics/01-metric_characterisation/metrics_geodesic.pdf
--------------------------------------------------------------------------------
/raw/12-manuscript/linenumbers.sty:
--------------------------------------------------------------------------------
1 | \usepackage{xcolor}
2 | \definecolor{changes}{HTML}{85144b}
3 | \newcommand{\changes}[1]{{\leavevmode\color{changes}#1}}
4 | \usepackage{float}
5 |
--------------------------------------------------------------------------------
/raw/12-manuscript/manuscript.sty:
--------------------------------------------------------------------------------
1 | \usepackage{xcolor}
2 | \definecolor{changes}{HTML}{85144b}
3 | \newcommand{\changes}[1]{{\leavevmode\color{changes}#1}}
4 | \usepackage{float}
5 |
--------------------------------------------------------------------------------
/raw/02-metrics/01-metric_characterisation/metrics_prediction.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/dynverse/dynbenchmark/HEAD/raw/02-metrics/01-metric_characterisation/metrics_prediction.pdf
--------------------------------------------------------------------------------
/scripts/varia/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Varia scripts
6 |
7 | ```{r}
8 | dynbenchmark::render_scripts_documentation()
9 | ```
10 |
--------------------------------------------------------------------------------
/package/tests/testthat.R:
--------------------------------------------------------------------------------
1 | library(testthat)
2 | library(dynutils)
3 | library(dplyr)
4 | library(ggplot2)
5 | library(purrr)
6 |
7 | Sys.setenv("R_TESTS" = "")
8 |
9 | test_check("dynbenchmark")
10 |
11 |
--------------------------------------------------------------------------------
/scripts/12-manuscript/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Preparing the manuscript
6 |
7 | ```{r}
8 | dynbenchmark::render_scripts_documentation()
9 | ```
10 |
--------------------------------------------------------------------------------
/derived/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Derived data
3 |
4 | Here, we place all the data which is generated by a script. This
5 | includes:
6 |
7 | - The datasets
8 | - The method singularity images
9 | - The main benchmarking results
10 |
--------------------------------------------------------------------------------
/package/R/nacor.R:
--------------------------------------------------------------------------------
1 | #' Na removed cor
2 | #' @inheritParams stats::cor
3 | #' @importFrom stats cor
4 | #' @export
5 | nacor <- function(x, y, method = "spearman") {
6 | is_na <- !is.finite(x) | !is.finite(y)
7 | stats::cor(x[!is_na], y[!is_na], method = method)
8 | }
9 |
--------------------------------------------------------------------------------
/derived/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Derived data
6 |
7 | Here, we place all the data which is generated by a script. This includes:
8 |
9 | - The datasets
10 | - The method singularity images
11 | - The main benchmarking results
12 |
--------------------------------------------------------------------------------
/scripts/03-methods/03-method_characterisation/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Method characterisation
6 |
7 | Here we have a look at the diversity of TI methods
8 |
9 | ```{r}
10 | dynbenchmark::render_scripts_documentation()
11 | ```
12 |
--------------------------------------------------------------------------------
/package/inst/tester.md:
--------------------------------------------------------------------------------
1 |
2 | # Markdown tester
3 |
4 | 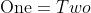
5 |
6 |
7 |
8 | | Three |
9 | |:------|
10 | | Four |
11 |
12 | **Table 1: Five**
13 |
14 | See [**Table 1**](#table_table) for Six
15 |
--------------------------------------------------------------------------------
/manuscript/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | ```{r, warning=FALSE, message=FALSE}
6 | library(tidyverse)
7 | library(dynbenchmark)
8 | ```
9 |
10 | # Manuscript
11 |
12 | The files to generate the manuscript: the main paper and the supplementary material.
13 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-dataset_generation.R:
--------------------------------------------------------------------------------
1 | # context("Testing dataset simulation")
2 | #
3 | # temp_dynbenchmark_folder <- tempdir()
4 | # write_file("", paste0(temp_dynbenchmark_folder, "/dynbenchmark.Rproj"))
5 | #
6 | # test_that("Simulation with dyngen", {
7 | # dataset <- simulate_dyngen("test")
8 | # })
9 |
--------------------------------------------------------------------------------
/package/.Rbuildignore:
--------------------------------------------------------------------------------
1 | ^.*\.Rproj$
2 | ^\.Rproj\.user$
3 | ^README\.Rmd$
4 | ^README-.*\.png$
5 | ^CONDUCT\.md$
6 | ^CONTRIBUTING\.md$
7 | ^analysis
8 | ^Dockerfile$
9 | ^\.travis
10 | ^\.httr-oauth$
11 | ^\.dockerignore$
12 | ^\.git$
13 | ^docker
14 | ^scripts
15 | ^data-raw$
16 | ^LICENSE\.md$
17 | ^LICENSE\.md$
18 |
--------------------------------------------------------------------------------
/scripts/03-methods/02-tool_qc/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Tool quality control
6 |
7 | Here we compare the user and developer friendliness of the different trajectory inference tools
8 |
9 | ```{r}
10 | dynbenchmark::render_scripts_documentation()
11 | ```
12 |
--------------------------------------------------------------------------------
/scripts/01-datasets/04-dataset_characterisation/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Dataset characterisation
6 |
7 | Characterisation of the datasets regarding the different topologies present.
8 |
9 | ```{r}
10 | dynbenchmark::render_scripts_documentation()
11 | ```
12 |
--------------------------------------------------------------------------------
/package/man/label_time.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_time}
4 | \alias{label_time}
5 | \title{Label time}
6 | \usage{
7 | label_time(time)
8 | }
9 | \arguments{
10 | \item{time}{Time}
11 | }
12 | \description{
13 | Label time
14 | }
15 |
--------------------------------------------------------------------------------
/scripts/08-summary/2a_columns_suppfig.R:
--------------------------------------------------------------------------------
1 | source(scripts_file("2a_columns_all.R", experiment = "08-summary"), local = TRUE)
2 |
3 | column_info <-
4 | column_info %>% filter(!id %in% c("method_most_complex_trajectory_type"))
5 |
6 | column_groups <-
7 | column_groups %>%
8 | filter(group %in% column_info$group)
9 |
10 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | .Rproj.user
2 | .Rhistory
3 | .RData
4 | derived/*
5 | *log
6 | analysis_output.tar
7 | .rds
8 | .httr-oauth
9 | manuscript/*.html
10 | manuscript/paper*.md
11 | manuscript/paper*.tex
12 | manuscript/.figures
13 | *.aux
14 | *.out
15 | *.bbl
16 | *.run.xml
17 | *tmp.pdf
18 | *tmp.png
19 | .DS_Store
20 | .Renviron
21 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Accuracy of TI methods on real and synthetic data
6 |
7 | ```{r}
8 | dynbenchmark::render_scripts_documentation()
9 | ```
10 |
11 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
12 |
--------------------------------------------------------------------------------
/package/inst/tester.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Markdown tester
6 |
7 | $\textrm{One} = Two$
8 |
9 | ```{r}
10 | dynbenchmark::add_table(
11 | tibble::tibble(Three = "Four"),
12 | "table",
13 | "Five"
14 | )
15 | ```
16 |
17 | See `r dynbenchmark::ref("table", "table")` for Six
18 |
--------------------------------------------------------------------------------
/package/man/label_long.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_long}
4 | \alias{label_long}
5 | \title{Long labelling function}
6 | \usage{
7 | label_long(x)
8 | }
9 | \arguments{
10 | \item{x}{The text}
11 | }
12 | \description{
13 | Long labelling function
14 | }
15 |
--------------------------------------------------------------------------------
/scripts/11-example_predictions/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Example predictions
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | Look at some predicted trajectories
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
--------------------------------------------------------------------------------
/package/man/label_capitalise.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_capitalise}
4 | \alias{label_capitalise}
5 | \title{Capitalise label}
6 | \usage{
7 | label_capitalise(x)
8 | }
9 | \arguments{
10 | \item{x}{The text}
11 | }
12 | \description{
13 | Capitalise label
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/label_thousands.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_thousands}
4 | \alias{label_thousands}
5 | \title{Label thousands}
6 | \usage{
7 | label_thousands(x)
8 | }
9 | \arguments{
10 | \item{x}{Numeric value}
11 | }
12 | \description{
13 | Label thousands
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/label_method.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_method}
4 | \alias{label_method}
5 | \title{Label a method}
6 | \usage{
7 | label_method(method_ids)
8 | }
9 | \arguments{
10 | \item{method_ids}{Vector of method ids}
11 | }
12 | \description{
13 | Label a method
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/setup_singularity_methods.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_methods.R
3 | \name{setup_singularity_methods}
4 | \alias{setup_singularity_methods}
5 | \title{Setup the singularity methods}
6 | \usage{
7 | setup_singularity_methods()
8 | }
9 | \description{
10 | Setup the singularity methods
11 | }
12 |
--------------------------------------------------------------------------------
/package/man/limits_metric.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{limits_metric}
4 | \alias{limits_metric}
5 | \title{Get the limits of a metric}
6 | \usage{
7 | limits_metric(metric_id)
8 | }
9 | \arguments{
10 | \item{metric_id}{metric id}
11 | }
12 | \description{
13 | Get the limits of a metric
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/setup_refs_globally.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{setup_refs_globally}
4 | \alias{setup_refs_globally}
5 | \title{Setup the refs for a markdown document, globally!}
6 | \usage{
7 | setup_refs_globally()
8 | }
9 | \description{
10 | Setup the refs for a markdown document, globally!
11 | }
12 |
--------------------------------------------------------------------------------
/scripts/09-guidelines/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Guidelines
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | Preparing the data for the dynguidelines app and creation of the guidelines figure.
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
--------------------------------------------------------------------------------
/package/man/label_extrema.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_extrema}
4 | \alias{label_extrema}
5 | \title{Labels only the extrema (and zero)}
6 | \usage{
7 | label_extrema(x)
8 | }
9 | \arguments{
10 | \item{x}{The values}
11 | }
12 | \description{
13 | Labels only the extrema (and zero)
14 | }
15 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Benchmark interpretation
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | Various attempts at interpreting the results of the benchmark.
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
--------------------------------------------------------------------------------
/package/man/label_vector.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_vector}
4 | \alias{label_vector}
5 | \title{Collapse a vector with commas and "ands"}
6 | \usage{
7 | label_vector(x)
8 | }
9 | \arguments{
10 | \item{x}{A vector}
11 | }
12 | \description{
13 | Collapse a vector with commas and "ands"
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/tag_first.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{tag_first}
4 | \alias{tag_first}
5 | \title{tag the first plot of an assemble}
6 | \usage{
7 | tag_first(x, tag)
8 | }
9 | \arguments{
10 | \item{x}{A ggassemble}
11 |
12 | \item{tag}{The tag}
13 | }
14 | \description{
15 | tag the first plot of an assemble
16 | }
17 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/wip/developing-thymus_kernfeld.R:
--------------------------------------------------------------------------------
1 | rm(list=ls())
2 | library(dynbenchmark)
3 | library(tidyverse)
4 | library(GEOquery)
5 | library(Biobase)
6 | options('download.file.method.GEOquery'='curl')
7 |
8 | dataset_preprocessing("real/developing-thymus_kernfeld")
9 |
10 | geo <- GEOquery::getGEO(GEO = "GSE107910", destdir = dataset_source_file(""))
11 |
--------------------------------------------------------------------------------
/scripts/02-metrics/02-metric_conformity/helper-create_perturbation_images.R:
--------------------------------------------------------------------------------
1 | #' Helper for creating small images for each rule
2 |
3 | library(dynbenchmark)
4 | library(esfiji)
5 |
6 | experiment("02-metrics/02-metric_conformity")
7 |
8 | svg_location <- raw_file("perturbations.svg")
9 |
10 | folder <- result_file("images")
11 | dir.create(folder)
12 | svg_groups_split(svg_location, folder = folder)
13 |
--------------------------------------------------------------------------------
/scripts/02-metrics/03-aggregation/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Aggregation
6 |
7 | Here, we create some examples here for why and how we aggregate the scores.
8 |
9 | ```{r, results = "asis"}
10 | dynbenchmark::render_scripts_documentation()
11 | ```
12 |
13 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
14 |
--------------------------------------------------------------------------------
/package/man/label_memory.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_memory}
4 | \alias{label_memory}
5 | \title{Label memory}
6 | \usage{
7 | label_memory(x, include_mb = FALSE)
8 | }
9 | \arguments{
10 | \item{x}{Memory in bytes}
11 |
12 | \item{include_mb}{Also include MB values.}
13 | }
14 | \description{
15 | Label memory
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/label_short.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_short}
4 | \alias{label_short}
5 | \title{Short labelling function}
6 | \usage{
7 | label_short(x, width = 10)
8 | }
9 | \arguments{
10 | \item{x}{The text}
11 |
12 | \item{width}{The width of the label}
13 | }
14 | \description{
15 | Short labelling function
16 | }
17 |
--------------------------------------------------------------------------------
/scripts/03-methods/02-tool_qc/01-qc_aspects_table.R:
--------------------------------------------------------------------------------
1 | #' Generate a table containing the qc scoresheet
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(xlsx)
6 |
7 | experiment("03-methods/02-tool_qc")
8 |
9 | qc_checks <- readRDS(result_file("qc_checks.rds", "03-methods"))
10 |
11 | qc_checks %>%
12 | as.data.frame() %>%
13 | write.xlsx(result_file("qc_scoresheet.xlsx"), row.names = FALSE)
14 |
--------------------------------------------------------------------------------
/package/man/benchmark_qsub_fun.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_benchmark_submit.R
3 | \name{benchmark_qsub_fun}
4 | \alias{benchmark_qsub_fun}
5 | \title{Helper function for benchmark suite}
6 | \usage{
7 | benchmark_qsub_fun(i)
8 | }
9 | \arguments{
10 | \item{i}{Row of a design dataframe}
11 | }
12 | \description{
13 | Helper function for benchmark suite
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/print_url.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_helpers.R
3 | \name{print_url}
4 | \alias{print_url}
5 | \title{Show a url}
6 | \usage{
7 | print_url(url, text = url, format = get_default_format())
8 | }
9 | \arguments{
10 | \item{url}{The url}
11 |
12 | \item{text}{The text}
13 |
14 | \item{format}{The format}
15 | }
16 | \description{
17 | Show a url
18 | }
19 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-google_scholar.R:
--------------------------------------------------------------------------------
1 | context("Testing google_scholar")
2 |
3 | test_that("Testing google_scholar", {
4 | skip_on_travis()
5 |
6 | sch_out <- google_scholar("6708368002535418936")
7 | expect_equal(colnames(sch_out), c("cluster_id", "title", "url", "num_citations", "web_of_science"))
8 |
9 | sch_out2 <- google_scholar_num_citations("14546114750226917320")
10 | expect_gte(sch_out2, 0)
11 | })
12 |
--------------------------------------------------------------------------------
/package/man/labels.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{labels}
5 | \alias{labels}
6 | \title{Common manual labelling}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 36 rows and 3 columns.}
8 | \usage{
9 | labels
10 | }
11 | \description{
12 | Common manual labelling
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/label_pvalue.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_pvalue}
4 | \alias{label_pvalue}
5 | \title{Label p_value}
6 | \usage{
7 | label_pvalue(p_values, cutoffs = c(0.1, 0.01, 1e-05))
8 | }
9 | \arguments{
10 | \item{p_values}{P values}
11 |
12 | \item{cutoffs}{Cutoffs for number of asterisks}
13 | }
14 | \description{
15 | Label p_value
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/method_status_colours.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{method_status_colours}
5 | \alias{method_status_colours}
6 | \title{Colours of method statuses}
7 | \format{An object of class \code{character} of length 7.}
8 | \usage{
9 | method_status_colours
10 | }
11 | \description{
12 | Colours of method statuses
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-plotting_trajectory_types.R:
--------------------------------------------------------------------------------
1 | context("Test plotting trajectory types")
2 |
3 |
4 | test_that("test plot_trajectory_type", {
5 | plot <- ggplot() +
6 | theme_void()
7 |
8 | new_plot <- plot %>%
9 | plot_trajectory_types(trajectory_types$id, ymin = seq_along(trajectory_types$id), ymax = seq_along(trajectory_types$id) + 1, size = 1.5)
10 |
11 | testthat::expect_true(is.ggplot(new_plot))
12 | })
13 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/3d-compare_sources.R:
--------------------------------------------------------------------------------
1 | #' Compare the different dataset sources
2 |
3 | dircom <-
4 | data_aggregations %>%
5 | filter(dataset_trajectory_type == "overall") %>%
6 | select(method_id:dataset_source, overall) %>%
7 | spread(dataset_source, overall)
8 |
9 | g <-
10 | GGally::ggpairs(dircom %>% select(-1:-4)) +
11 | theme_bw()
12 |
13 | ggsave(result_file("compare_sources.pdf"), g, width = 12, height = 12)
14 |
--------------------------------------------------------------------------------
/package/man/pdf_supplementary_note.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{pdf_supplementary_note}
4 | \alias{pdf_supplementary_note}
5 | \title{A supplementary note pdf document}
6 | \usage{
7 | pdf_supplementary_note(...)
8 | }
9 | \arguments{
10 | \item{...}{Parameters for rmarkdown::pdf_document}
11 | }
12 | \description{
13 | A supplementary note pdf document
14 | }
15 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/wip/mammary-gland-epitheliam_pal.R:
--------------------------------------------------------------------------------
1 | rm(list=ls())
2 | library(dynbenchmark)
3 | library(tidyverse)
4 | library(GEOquery)
5 | library(Biobase)
6 | options('download.file.method.GEOquery'='curl')
7 |
8 | id <- "real/mammary-gland-epitheliam_pal"
9 | dataset_preprocessing(id)
10 |
11 | geo <- GEOquery::getGEO(GEO = "GSE103272", destdir = dataset_source_file(""))
12 |
13 |
14 | geo
15 |
--------------------------------------------------------------------------------
/package/man/label_facet.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_facet}
4 | \alias{label_facet}
5 | \title{Labeller for facets}
6 | \usage{
7 | label_facet(label_func = label_long, ...)
8 | }
9 | \arguments{
10 | \item{label_func}{Which function to use for facet labelling}
11 |
12 | \item{...}{Passed to mutate_all}
13 | }
14 | \description{
15 | Labeller for facets
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/prior_types.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{prior_types}
5 | \alias{prior_types}
6 | \title{Metadata on prior types}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 8 rows and 2 columns.}
8 | \usage{
9 | prior_types
10 | }
11 | \description{
12 | Metadata on prior types
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/paramoptim_qsub_fun.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_submit.R
3 | \name{paramoptim_qsub_fun}
4 | \alias{paramoptim_qsub_fun}
5 | \title{Helper function for parameter optimisation suite}
6 | \usage{
7 | paramoptim_qsub_fun(i)
8 | }
9 | \arguments{
10 | \item{i}{Row of a design dataframe}
11 | }
12 | \description{
13 | Helper function for parameter optimisation suite
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/create_replacers.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_svg_replacement.R
3 | \name{create_replacers}
4 | \alias{create_replacers}
5 | \title{Create replacers
6 | Adds a replace_id column}
7 | \usage{
8 | create_replacers(to_replace)
9 | }
10 | \arguments{
11 | \item{to_replace}{Dataframe containing the data}
12 | }
13 | \description{
14 | Create replacers
15 | Adds a replace_id column
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/error_reasons.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{error_reasons}
5 | \alias{error_reasons}
6 | \title{Metadata on error reasons}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 4 rows and 3 columns.}
8 | \usage{
9 | error_reasons
10 | }
11 | \description{
12 | Metadata on error reasons
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/link_to_results.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_document_scripts.R
3 | \name{link_to_results}
4 | \alias{link_to_results}
5 | \title{Generates a link to the results}
6 | \usage{
7 | link_to_results(scripts_folder = getwd())
8 | }
9 | \arguments{
10 | \item{scripts_folder}{Folder from which to calculate the experiment id}
11 | }
12 | \description{
13 | Generates a link to the results
14 | }
15 |
--------------------------------------------------------------------------------
/package/man/qc_categories.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{qc_categories}
5 | \alias{qc_categories}
6 | \title{Metadata on QC categories}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 6 rows and 3 columns.}
8 | \usage{
9 | qc_categories
10 | }
11 | \description{
12 | Metadata on QC categories
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/prior_usages.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{prior_usages}
5 | \alias{prior_usages}
6 | \title{Metadata on prior usage types}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 3 rows and 2 columns.}
8 | \usage{
9 | prior_usages
10 | }
11 | \description{
12 | Metadata on prior usage types
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-labels.R:
--------------------------------------------------------------------------------
1 | context("Testing plotting_labels")
2 |
3 |
4 |
5 | test_that("Plotting labels works", {
6 | expect_equal(label_short("one_hundred_and_seven_thousand", width = 10), "One\nhundred\nand seven\nthousand")
7 | expect_equal(label_long("one_hundred_and_seven_thousand"), "One hundred and seven thousand")
8 | expect_equal(label_long("linear"), "Linear")
9 | expect_equal(label_extrema(c(0, 0.5, 1)), c("0", "", "1"))
10 | })
11 |
--------------------------------------------------------------------------------
/raw/01-datasets/02-synthetic/samplers:
--------------------------------------------------------------------------------
1 | parameter distribution
2 | r \mathcal{U}(10, 200)
3 | d \mathcal{U}(2, 8)
4 | p \mathcal{U}(2, 8)
5 | q \mathcal{U}(1, 5)
6 | a0 \begin{cases}1 & \text{if } e \in \{1\} \text{ or } e = \emptyset \\ 0 & \text{if } e \in \{-1\} \\ 0.5 & \text{otherwise}\end{cases}
7 | a_i \begin{cases}1 & \text{if } e_i \in \{-1\} \\ 0 & \text{otherwise}\end{cases}
8 | s \mathcal{U}(1, 20)
9 | k y_{max} / (2 * s)
10 | c \mathcal{U}(1, 4)
11 |
12 |
--------------------------------------------------------------------------------
/package/man/method_statuses.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{method_statuses}
5 | \alias{method_statuses}
6 | \title{Metadata on method statuses}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 7 rows and 4 columns.}
8 | \usage{
9 | method_statuses
10 | }
11 | \description{
12 | Metadata on method statuses
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/qc_applications.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{qc_applications}
5 | \alias{qc_applications}
6 | \title{Metadata on QC applications}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 3 rows and 1 columns.}
8 | \usage{
9 | qc_applications
10 | }
11 | \description{
12 | Metadata on QC applications
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/google_scholar.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/google_scholar.R
3 | \name{google_scholar}
4 | \alias{google_scholar}
5 | \title{Find the number of citations for a given google scholar id}
6 | \usage{
7 | google_scholar(cluster_id)
8 | }
9 | \arguments{
10 | \item{cluster_id}{The Google Scholar cluster id to query}
11 | }
12 | \description{
13 | Find the number of citations for a given google scholar id
14 | }
15 |
--------------------------------------------------------------------------------
/package/R/metrics.R:
--------------------------------------------------------------------------------
1 | #' Metrics used in the characterisation
2 | #' @export
3 | metrics_characterised <- dplyr::filter(
4 | dyneval::metrics,
5 | !metric_id %in% c("featureimp_ks", "featureimp_wilcox", "rf_rsq", "lm_rsq", "rf_mse", "lm_mse")
6 | )
7 |
8 |
9 | #' Metrics used in the evaluation
10 | #' @export
11 | metrics_evaluated <- dplyr::filter(
12 | dyneval::metrics,
13 | metric_id %in% c("correlation", "him", "featureimp_wcor", "F1_branches", "geom_mean")
14 | )
15 |
--------------------------------------------------------------------------------
/package/man/topinf_types.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{topinf_types}
5 | \alias{topinf_types}
6 | \title{Metadata on topology inference types}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 3 rows and 4 columns.}
8 | \usage{
9 | topinf_types
10 | }
11 | \description{
12 | Metadata on topology inference types
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/metrics_evaluated.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/metrics.R
3 | \docType{data}
4 | \name{metrics_evaluated}
5 | \alias{metrics_evaluated}
6 | \title{Metrics used in the evaluation}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 5 rows and 10 columns.}
8 | \usage{
9 | metrics_evaluated
10 | }
11 | \description{
12 | Metrics used in the evaluation
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/setup_refs.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{setup_refs}
4 | \alias{setup_refs}
5 | \alias{setup_figs}
6 | \alias{setup_sfigs}
7 | \alias{setup_tables}
8 | \alias{setup_stables}
9 | \title{Setup the text references}
10 | \usage{
11 | setup_refs()
12 |
13 | setup_figs()
14 |
15 | setup_sfigs()
16 |
17 | setup_tables()
18 |
19 | setup_stables()
20 | }
21 | \description{
22 | Setup the text references
23 | }
24 |
--------------------------------------------------------------------------------
/package/man/theme_empty.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_themes.R
3 | \docType{data}
4 | \name{theme_empty_x_axis}
5 | \alias{theme_empty_x_axis}
6 | \title{Themes which make certain parts of the plot empty}
7 | \format{An object of class \code{theme} (inherits from \code{gg}) of length 4.}
8 | \usage{
9 | theme_empty_x_axis
10 | }
11 | \description{
12 | Themes which make certain parts of the plot empty
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/scripts/01-datasets/04-dataset_characterisation/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Dataset characterisation
3 |
4 | Characterisation of the datasets regarding the different topologies
5 | present.
6 |
7 | | \# | script/folder | description |
8 | | :- | :----------------------------- | :-------------------------------------------------------- |
9 | | 1 | [📄`topology.R`](01-topology.R) | An overview of all the topologies present in the datasets |
10 |
--------------------------------------------------------------------------------
/package/man/id_mapper.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{id_mapper}
5 | \alias{id_mapper}
6 | \title{Mapping ensembl or entrez ids to symbols}
7 | \format{An object of class \code{grouped_df} (inherits from \code{tbl_df}, \code{tbl}, \code{data.frame}) with 129094 rows and 3 columns.}
8 | \usage{
9 | id_mapper
10 | }
11 | \description{
12 | Mapping ensembl or entrez ids to symbols
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/package/man/render_equations.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_equations.R
3 | \name{render_equations}
4 | \alias{render_equations}
5 | \title{Renders equations for github markdown}
6 | \usage{
7 | render_equations(x, format = get_default_format())
8 | }
9 | \arguments{
10 | \item{x}{A character vector}
11 |
12 | \item{format}{The format: html, markdown, latex, ...}
13 | }
14 | \description{
15 | Renders equations for github markdown
16 | }
17 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/04-datasets_table.R:
--------------------------------------------------------------------------------
1 | #' Creates a table of the datasets in, excuse me, excel (for supplementary material)
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(xlsx)
6 |
7 | experiment("01-datasets/01-real")
8 |
9 | metadata <- read_rds(result_file("metadata.rds"))
10 |
11 | metadata %>%
12 | mutate(notes = ifelse(is.na(notes), "", notes)) %>%
13 | as.data.frame() %>%
14 | write.xlsx(result_file("real_datasets.xlsx"), row.names = FALSE, showNA = FALSE)
15 |
--------------------------------------------------------------------------------
/package/man/load_or_generate.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_pipeline.R
3 | \name{load_or_generate}
4 | \alias{load_or_generate}
5 | \title{Load or generate an R object}
6 | \usage{
7 | load_or_generate(file_location, expression)
8 | }
9 | \arguments{
10 | \item{file_location}{Location of the RDS file}
11 |
12 | \item{expression}{Expression to run if the RDS file is not found}
13 | }
14 | \description{
15 | Load or generate an R object
16 | }
17 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Real datasets
6 |
7 |
8 | The generation of the real datasets is divided in two parts. We first download all the (annotated) expression files from sites such as GEO. Next, we filter and normalise all datasets, and wrap them into the common trajectory format of [dynwrap](https://www.github.com/dynverse/dynwrap).
9 |
10 | ```{r}
11 | dynbenchmark::render_scripts_documentation()
12 | ```
13 |
14 |
--------------------------------------------------------------------------------
/package/man/non_inclusion_reasons.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{non_inclusion_reasons}
5 | \alias{non_inclusion_reasons}
6 | \title{Metadata on non-inclusion reasons}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 9 rows and 3 columns.}
8 | \usage{
9 | non_inclusion_reasons
10 | }
11 | \description{
12 | Metadata on non-inclusion reasons
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/scripts/04-method_testing/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Method testing
6 |
7 | Each method is tested on a few small real and toy datasets. Ideally, each method should be able to execute successfully on each of these datasets.
8 | If a method is not able to infer a trajectory on any of the datasets, it will not be included in any of the next experiments.
9 |
10 | Overview experiment:
11 | ```{r}
12 | dynbenchmark::render_scripts_documentation()
13 | ```
14 |
--------------------------------------------------------------------------------
/scripts/07-stability/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Stability
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | Analysis of the stability of each method: how similar are the trajectories when rerunning a method on slightly perturbed data?
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
18 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
19 |
--------------------------------------------------------------------------------
/dynbenchmark.Rproj:
--------------------------------------------------------------------------------
1 | Version: 1.0
2 |
3 | RestoreWorkspace: No
4 | SaveWorkspace: No
5 | AlwaysSaveHistory: Default
6 |
7 | EnableCodeIndexing: Yes
8 | UseSpacesForTab: Yes
9 | NumSpacesForTab: 2
10 | Encoding: UTF-8
11 |
12 | RnwWeave: Sweave
13 | LaTeX: pdfLaTeX
14 |
15 | AutoAppendNewline: Yes
16 | StripTrailingWhitespace: Yes
17 |
18 | BuildType: Package
19 | PackageUseDevtools: Yes
20 | PackagePath: package
21 | PackageInstallArgs: --no-multiarch --with-keep.source
22 | PackageRoxygenize: rd,collate,namespace
23 |
--------------------------------------------------------------------------------
/package/man/headtail.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/headtail.R
3 | \name{headtail}
4 | \alias{headtail}
5 | \title{Retain the heads and tails of a character vector split by newlines}
6 | \usage{
7 | headtail(X, num)
8 | }
9 | \arguments{
10 | \item{X}{A character vector to process}
11 |
12 | \item{num}{The number of lines to retain at the head and at the tail}
13 | }
14 | \description{
15 | Retain the heads and tails of a character vector split by newlines
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/metrics_characterised.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/metrics.R
3 | \docType{data}
4 | \name{metrics_characterised}
5 | \alias{metrics_characterised}
6 | \title{Metrics used in the characterisation}
7 | \format{An object of class \code{tbl_df} (inherits from \code{tbl}, \code{data.frame}) with 13 rows and 10 columns.}
8 | \usage{
9 | metrics_characterised
10 | }
11 | \description{
12 | Metrics used in the characterisation
13 | }
14 | \keyword{datasets}
15 |
--------------------------------------------------------------------------------
/scripts/08-summary/2a_columns_summary.R:
--------------------------------------------------------------------------------
1 | source(scripts_file("2a_columns_all.R", experiment = "08-summary"), local = TRUE)
2 |
3 | selected_groups <- c("method_characteristic", "inferrable_trajtype", "score_overall")
4 | removed_columns <- c("method_most_complex_trajectory_type", "itt_convergence", "itt_acyclic_graph")
5 | column_info <-
6 | column_info %>%
7 | filter(group %in% selected_groups, !id %in% removed_columns)
8 |
9 | column_groups <-
10 | column_groups %>% filter(group %in% column_info$group)
11 |
12 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/03-gather_metadata.R:
--------------------------------------------------------------------------------
1 | #' Gathers some metadata about all the synthetic datasets
2 |
3 | library(dynbenchmark)
4 | library(googlesheets)
5 |
6 | experiment("01-datasets/02-synthetic")
7 |
8 | datasets_synthetic <- load_datasets(list_datasets() %>% filter(startsWith(source, "synthetic/")) %>% pull(id))
9 |
10 | datasets_synthetic_metadata <- datasets_synthetic$simulation_design %>%
11 | list_as_tibble()
12 |
13 | write_rds(datasets_synthetic_metadata, result_file("metadata.rds"))
14 |
15 |
--------------------------------------------------------------------------------
/scripts/03-methods/03-method_characterisation/02-tools_table.R:
--------------------------------------------------------------------------------
1 | #' Generate a table containing the methods
2 |
3 | library(googlesheets)
4 | library(tidyverse)
5 | library(dynbenchmark)
6 | library(xlsx)
7 |
8 | experiment("03-methods")
9 |
10 | tools <- read_rds(result_file("tools.rds"))
11 |
12 | tools <- tools %>%
13 | arrange(date)
14 |
15 | tools %>%
16 | select(tool_id, tool_name, platform, date, doi) %>%
17 | as.data.frame() %>%
18 | write.xlsx(result_file("tools.xlsx"), row.names = FALSE, showNA = FALSE)
19 |
--------------------------------------------------------------------------------
/scripts/08-summary/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Summarising the results into funky heatmaps
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | Here, we aggregate the results from the different experiments into a tidy format. This aggregation procedure is further discussed in our supplementary note.
13 |
14 | Next, we plot this into funky heatmaps.
15 |
16 | ```{r}
17 | dynbenchmark::render_scripts_documentation()
18 | ```
19 |
--------------------------------------------------------------------------------
/package/man/google_scholar_num_citations.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/google_scholar.R
3 | \name{google_scholar_num_citations}
4 | \alias{google_scholar_num_citations}
5 | \title{Find the number of citations for a given google scholar id}
6 | \usage{
7 | google_scholar_num_citations(cluster_id)
8 | }
9 | \arguments{
10 | \item{cluster_id}{The Google Scholar cluster id to query}
11 | }
12 | \description{
13 | Find the number of citations for a given google scholar id
14 | }
15 |
--------------------------------------------------------------------------------
/scripts/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | ```{r, warning=FALSE, message=FALSE}
6 | library(tidyverse)
7 | library(dynbenchmark)
8 | ```
9 |
10 | # Scripts
11 |
12 | The scripts folder contains all the scripts necessary to fully reproduce the benchmarking manuscript in chronologically ordered subfolders. Each subfolder contains a readme file with further explanations of the different scripts and what they do:
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
--------------------------------------------------------------------------------
/package/man/estimate_platform.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_platforms.R
3 | \name{estimate_platform}
4 | \alias{estimate_platform}
5 | \title{Estimate a platform}
6 | \usage{
7 | estimate_platform(dataset_id, subsample = 500)
8 | }
9 | \arguments{
10 | \item{dataset_id}{The dataset_id from which the platform will be estimated, using the files in `datasets_preproc/raw``}
11 |
12 | \item{subsample}{The number of cells to subsample.}
13 | }
14 | \description{
15 | Estimate a platform
16 | }
17 |
--------------------------------------------------------------------------------
/package/man/label_wrap.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_wrap}
4 | \alias{label_wrap}
5 | \title{Text wrapping}
6 | \usage{
7 | label_wrap(x, width = 10, collapse = "\\n")
8 | }
9 | \arguments{
10 | \item{x}{a character vector.}
11 |
12 | \item{width}{a positive integer giving the target column for wrapping lines in the output.}
13 |
14 | \item{collapse}{an optional character string to separate the different lines.}
15 | }
16 | \description{
17 | Text wrapping
18 | }
19 |
--------------------------------------------------------------------------------
/package/man/replace_svg.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_svg_replacement.R
3 | \name{replace_svg}
4 | \alias{replace_svg}
5 | \title{Replace
6 | Replaces the rectangles in a given svg with the svgs contained in the replacer}
7 | \usage{
8 | replace_svg(svg, replacer)
9 | }
10 | \arguments{
11 | \item{svg}{The svg to replace}
12 |
13 | \item{replacer}{Replacer dataframe}
14 | }
15 | \description{
16 | Replace
17 | Replaces the rectangles in a given svg with the svgs contained in the replacer
18 | }
19 |
--------------------------------------------------------------------------------
/scripts/02-metrics/01-metric_characterisation/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Metric characterisation
6 |
7 | A first characterisation of the metrics. For each metric we:
8 |
9 | - generate some examples to get some intuition on how the metric works
10 | - test the robustness to a metric to stochasticity or parameters when appropriate
11 |
12 | ```{r}
13 | dynbenchmark::render_scripts_documentation()
14 | ```
15 |
16 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`)
17 |
--------------------------------------------------------------------------------
/package/man/load_methods.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_methods.R
3 | \name{load_methods}
4 | \alias{load_methods}
5 | \title{Load the information on the methods}
6 | \usage{
7 | load_methods(default_filter = method_source == "tool" | method_id \%in\%
8 | c("angle", "comp1", "mst"), ...)
9 | }
10 | \arguments{
11 | \item{default_filter}{Default filtering for the benchmark.}
12 |
13 | \item{...}{Other filters for the method tibble.}
14 | }
15 | \description{
16 | Load the information on the methods
17 | }
18 |
--------------------------------------------------------------------------------
/scripts/08-summary/2a_columns_detailed.R:
--------------------------------------------------------------------------------
1 | source(scripts_file("2a_columns_all.R", experiment = "08-summary"), local = TRUE)
2 |
3 | selected_columns <- c("method_name")
4 | selected_groups <- c("benchmark_metric", "benchmark_source", "benchmark_trajtype", "scaling_predtime", "stability", "qc_category")
5 | removed_columns <- c()
6 |
7 | column_info <-
8 | column_info %>%
9 | filter(id %in% selected_columns | group %in% selected_groups, !id %in% removed_columns)
10 |
11 | column_groups <-
12 | column_groups %>%
13 | filter(group %in% column_info$group)
14 |
15 |
--------------------------------------------------------------------------------
/scripts/varia/rename_experiment.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | from=$1
4 | to=$2
5 |
6 | if [ -d raw/$from ]; then
7 | mv raw/$from raw/$to
8 | fi
9 |
10 | if [ -d figures/$from ]; then
11 | mv figures/$from figures/$to
12 | fi
13 |
14 | if [ -d results/$from ]; then
15 | mv results/$from results/$to
16 | fi
17 |
18 | if [ -d scripts/$from ]; then
19 | mv scripts/$from scripts/$to
20 | fi
21 |
22 | if [ -d derived/$from ]; then
23 | mv derived/$from derived/$to
24 | fi
25 |
26 | find . -type f -regex ".*.Rm?d?" -print0 | xargs -0 sed -i "s#$from#$to#g"
27 |
28 |
29 |
--------------------------------------------------------------------------------
/package/man/legend_at.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_themes.R
3 | \name{legend_at}
4 | \alias{legend_at}
5 | \title{Only retain the legend in the middle plot}
6 | \usage{
7 | legend_at(plots, at = floor(length(plots)/2), theme_legend = theme())
8 | }
9 | \arguments{
10 | \item{plots}{A list of plots}
11 |
12 | \item{at}{Integers at which a legend will be added}
13 |
14 | \item{theme_legend}{The theme to apply to the plot with a legend}
15 | }
16 | \description{
17 | Only retain the legend in the middle plot
18 | }
19 |
--------------------------------------------------------------------------------
/scripts/varia/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Varia scripts
3 |
4 | | \# | script/folder | description |
5 | | :- | :------------------------------------------------------------ | :---------- |
6 | | | [📁`generate_hypotheses_mindmap`](generate_hypotheses_mindmap) | |
7 | | | [📁`gource`](gource) | |
8 | | | [📁`manual_ti`](manual_ti) | |
9 | | | [📄`rename_experiment.sh`](rename_experiment.sh) | |
10 |
--------------------------------------------------------------------------------
/package/man/generate_prior_mini.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_priors_mini.R
3 | \name{generate_prior_mini}
4 | \alias{generate_prior_mini}
5 | \title{Creates a prior mini svg}
6 | \usage{
7 | generate_prior_mini(method_priors,
8 | svg = xml2::read_xml(result_file("priors.svg", experiment_id =
9 | "priors")))
10 | }
11 | \arguments{
12 | \item{method_priors}{Dataframe containing a prior_id and a prior_usage}
13 |
14 | \item{svg}{The base svg of the priors}
15 | }
16 | \description{
17 | Creates a prior mini svg
18 | }
19 |
--------------------------------------------------------------------------------
/package/man/download_dataset_source_file.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_datasets.R
3 | \name{download_dataset_source_file}
4 | \alias{download_dataset_source_file}
5 | \title{Download a file and return its location path}
6 | \usage{
7 | download_dataset_source_file(filename, url, id = NULL)
8 | }
9 | \arguments{
10 | \item{filename}{What name to give to the file}
11 |
12 | \item{url}{The url of the file to download}
13 |
14 | \item{id}{An optional id}
15 | }
16 | \description{
17 | Download a file and return its location path
18 | }
19 |
--------------------------------------------------------------------------------
/package/man/reformat_tribbles.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/reformat_tribbles.R
3 | \name{reformat_tribbles}
4 | \alias{reformat_tribbles}
5 | \title{Align tribbles in a file}
6 | \usage{
7 | reformat_tribbles(script_file, align = c("left", "center", "right"))
8 | }
9 | \arguments{
10 | \item{script_file}{The file in which to align the tribble}
11 |
12 | \item{align}{The alignment (left, center, right)}
13 | }
14 | \description{
15 | The start and end of a tribble need to be marked with a commend \code{# tribble_start} and \code{# tribble_end}.
16 | }
17 |
--------------------------------------------------------------------------------
/package/R/setup_pipeline.R:
--------------------------------------------------------------------------------
1 | #' Load or generate an R object
2 | #' @param file_location Location of the RDS file
3 | #' @param expression Expression to run if the RDS file is not found
4 | load_or_generate <- function(file_location, expression) {
5 | if (file.exists(file_location)) {
6 | message("Loaded ", basename(file_location))
7 | x <- read_rds(file_location)
8 | } else {
9 | message("Creating ", basename(file_location))
10 | x <- eval(expression)
11 | dir.create(dirname(file_location), showWarnings = FALSE, recursive = TRUE)
12 | write_rds(x, file_location)
13 | }
14 | x
15 | }
16 |
--------------------------------------------------------------------------------
/raw/12-manuscript/common.sty:
--------------------------------------------------------------------------------
1 | \usepackage{tabularx}
2 | \newcommand{\hideFromPandoc}[1]{#1}
3 | \hideFromPandoc{\newenvironment{myfigure}[1]{\begin{figure}[#1]}{\end{figure}}}
4 | \setmainfont{DejaVu Sans}
5 |
6 | \usepackage{geometry}
7 | \geometry{
8 | a4paper,
9 | total={170mm,257mm},
10 | left=20mm,
11 | top=20mm,
12 | }
13 |
14 | \usepackage[table]{xcolor}
15 | \definecolor{airforceblue}{rgb}{0.36, 0.54, 0.66}
16 | \definecolor{cerulean}{rgb}{0.0, 0.48, 0.65}
17 | \definecolor{burntorange}{rgb}{0.8, 0.33, 0.0}
18 | \hypersetup{colorlinks=true,linkcolor=airforceblue,citecolor=cerulean,urlcolor=burntorange}
19 |
--------------------------------------------------------------------------------
/scripts/11-example_predictions/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Example predictions
3 |
4 | Look at some predicted
5 | trajectories
6 |
7 | | \# | script/folder | description |
8 | | :- | :----------------------------------------------- | :---------------------------------------------------------------------------------------------------- |
9 | | 1 | [📄`example_consensus.R`](01-example_consensus.R) | Create a figure containing predicted trajectories from the top methods on a couple of simple datasets |
10 |
--------------------------------------------------------------------------------
/package/data-raw/priors.R:
--------------------------------------------------------------------------------
1 | prior_types <- tribble(
2 | ~prior_id, ~prior_name,
3 | "start_id", "Start cell",
4 | "end_id", "End cell(s)",
5 | "end_n", "# end states",
6 | "states_id", "Cell clustering",
7 | "states_n", "# states",
8 | "states_network", "State network",
9 | "time_id", "Time course",
10 | "genes_id", "Marker genes"
11 | )
12 |
13 | prior_usages <- tribble(
14 | ~prior_usage, ~color,
15 | "optional", "#0074D9",
16 | "no", "#EEEEEE",
17 | "required", "#FF4136"
18 | )
19 |
20 | devtools::use_data(prior_types, prior_usages, overwrite = TRUE)
21 |
22 | # also update data-raw/labels.R
23 |
--------------------------------------------------------------------------------
/package/R/headtail.R:
--------------------------------------------------------------------------------
1 | #' Retain the heads and tails of a character vector split by newlines
2 | #'
3 | #' @param X A character vector to process
4 | #' @param num The number of lines to retain at the head and at the tail
5 | #'
6 | #' @importFrom pbapply pbsapply
7 | #'
8 | #' @export
9 | headtail <- function(X, num) {
10 | pbapply::pbsapply(X, cl = 8, function(x) {
11 | xs <- strsplit(x, split = "\n")[[1]]
12 | if (length(xs) > 2 * num) {
13 | xs <- c(head(xs, num), paste0("... ", length(xs) - 2 * num, " lines omitted ..."), tail(xs, num))
14 | }
15 | paste0(c(xs, ""), collapse = "\n")
16 | })
17 | }
18 |
--------------------------------------------------------------------------------
/package/data-raw/qc_categorisation.R:
--------------------------------------------------------------------------------
1 | qc_applications <- tibble(
2 | application = c("developer_friendly", "user_friendly", "future_proof")
3 | )
4 |
5 | qc_categories <- tibble(
6 | category = c("availability", "code_quality", "code_assurance", "documentation", "behaviour", "paper"),
7 | color = c("#3498DB", "#E74C3C", "#A4CC2E", "#FEB308", "#B10DC9", "#85144b", "#EA8F10", "#2ECC49", "#CC2E63")[1:length(category)],
8 | label = c("Availability", "Code quality", "Code assurance", "Documentation", "Behaviour", "Study design")
9 | )
10 |
11 | devtools::use_data(qc_applications, qc_categories, overwrite = TRUE, pkg = "package")
12 |
--------------------------------------------------------------------------------
/scripts/05-scaling/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Scalability of methods
6 |
7 | Here we test how well a method scales with increasing number of features (genes) and/or cells.
8 |
9 | Each method is run on down- and upscaled datasets with increasing gene set and cell set sizes, and the execution times and memory usages are modelled using thin plate regressions splines within a generalised additivate model.
10 |
11 | ```{r}
12 | dynbenchmark::render_scripts_documentation()
13 | ```
14 |
15 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
16 |
--------------------------------------------------------------------------------
/package/man/word_manuscript.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{word_manuscript}
4 | \alias{word_manuscript}
5 | \title{A nested github markdown document}
6 | \usage{
7 | word_manuscript(bibliography = raw_file("references.bib",
8 | "12-manuscript"), csl = raw_file("nature-biotechnology.csl",
9 | "12-manuscript"), ...)
10 | }
11 | \arguments{
12 | \item{bibliography}{Bibliography location}
13 |
14 | \item{csl}{Csl file location}
15 |
16 | \item{...}{Parameters for rmarkdown::github_document}
17 | }
18 | \description{
19 | A nested github markdown document
20 | }
21 |
--------------------------------------------------------------------------------
/scripts/03-methods/02-tool_qc/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Tool quality control
3 |
4 | Here we compare the user and developer friendliness of the different
5 | trajectory inference
6 | tools
7 |
8 | | \# | script/folder | description |
9 | | :- | :------------------------------------------------- | :----------------------------------------------- |
10 | | 1 | [📄`qc_aspects_table.R`](01-qc_aspects_table.R) | Generate a table containing the qc scoresheet |
11 | | 2 | [📄`qc_scores_overview.R`](02-qc_scores_overview.R) | Create an overview figure of the quality control |
12 |
--------------------------------------------------------------------------------
/package/R/text_helpers.R:
--------------------------------------------------------------------------------
1 | #' Show a url
2 | #'
3 | #' @param url The url
4 | #' @param text The text
5 | #' @param format The format
6 | #'
7 | #' @export
8 | print_url <- function(url, text = url, format = get_default_format()) {
9 | # cut off www and http
10 | if (text == url) {
11 | text <- text %>%
12 | gsub("https:\\/\\/", "", .) %>%
13 | gsub("http:\\/\\/", "", .) %>%
14 | gsub("www\\.", "", .) %>%
15 | gsub("\\/$", "", .)
16 | }
17 |
18 | if(format == "latex") {
19 | paste0("\\textcolor{cyan}{\\href{", url, "}{", text, "}}")
20 | } else {
21 | stringr::str_glue("[{text}]({url})")
22 | }
23 | }
24 |
--------------------------------------------------------------------------------
/raw/02-metrics/03-aggregation/example.csv:
--------------------------------------------------------------------------------
1 | dataset_id,trajectory_type,dataset_source,method_id,metric_X,metric_Y
2 | A,linear,real/gold,a,0.15,0.1
3 | A,linear,real/gold,b,0.3,0.05
4 | A,linear,real/gold,c,0.4,0.2
5 | B,linear,real/gold,a,0.1,0
6 | B,linear,real/gold,b,0.25,0.05
7 | B,linear,real/gold,c,0.35,0.08
8 | C,linear,real/silver,a,0.25,0.1
9 | C,linear,real/silver,b,0.4,0.2
10 | C,linear,real/silver,c,0.85,0.4
11 | D,bifurcation,real/gold,a,0.2,0.15
12 | D,bifurcation,real/gold,b,0.5,0.6
13 | D,bifurcation,real/gold,c,0.7,0.8
14 | E,bifurcation,real/silver,a,0.8,0.9
15 | E,bifurcation,real/silver,b,0.9,0.95
16 | E,bifurcation,real/silver,c,0.8,1
17 |
--------------------------------------------------------------------------------
/scripts/02-metrics/03-aggregation/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Aggregation
3 |
4 | Here, we create some examples here for why and how we aggregate the
5 | scores.
6 |
7 | | \# | script/folder | description |
8 | | :- | :----------------------------------------------------------- | :---------- |
9 | | 1 | [📄`normalisation_reasoning.R`](01-normalisation_reasoning.R) | |
10 | | 2 | [📄`aggregation_example.R`](02-aggregation_example.R) | |
11 |
12 | The results of this experiment are available
13 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/02-metrics/03-aggregation).
14 |
--------------------------------------------------------------------------------
/scripts/12-manuscript/render_results.R:
--------------------------------------------------------------------------------
1 | #' Renders results READMEs
2 |
3 | library(dynbenchmark)
4 | library(fs)
5 | library(tidyverse)
6 |
7 | readme_paths <- c(
8 | fs::dir_ls("results", regexp = "README\\.Rmd", recursive = TRUE)
9 | # fs::dir_ls(result_file("", experiment = "03-methods"), regexp = "README\\.Rmd", recursive = TRUE)
10 | # "results/README.Rmd"
11 | )
12 |
13 | walk(readme_paths, function(readme_path) {
14 | print(paste0("Processing ", fs::path_rel(readme_path)))
15 | rmarkdown::render(
16 | readme_path,
17 | output_format = dynbenchmark::github_markdown_nested(html_preview = FALSE),
18 | quiet = TRUE
19 | )
20 | })
21 |
--------------------------------------------------------------------------------
/package/man/get_dynbenchmark_folder.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_get_dynbenchmark_folder.R
3 | \name{get_dynbenchmark_folder}
4 | \alias{get_dynbenchmark_folder}
5 | \title{Find the dynbenchmark path}
6 | \usage{
7 | get_dynbenchmark_folder(remote = FALSE)
8 | }
9 | \arguments{
10 | \item{remote}{The remote to access, "" if running locally, TRUE if using default qsub config remote}
11 | }
12 | \description{
13 | When executing some of the dynbenchmark functions, the git repository needs to be found.
14 | Either set the working directory to the repository, or define the DYNBENCHMARK_PATH variable.
15 | }
16 |
--------------------------------------------------------------------------------
/package/man/github_markdown_nested.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{github_markdown_nested}
4 | \alias{github_markdown_nested}
5 | \title{A nested github markdown document}
6 | \usage{
7 | github_markdown_nested(bibliography = raw_file("references.bib",
8 | "12-manuscript"), csl = raw_file("nature-biotechnology.csl",
9 | "12-manuscript"), ...)
10 | }
11 | \arguments{
12 | \item{bibliography}{Bibliography location}
13 |
14 | \item{csl}{Csl file location}
15 |
16 | \item{...}{Parameters for rmarkdown::github_document}
17 | }
18 | \description{
19 | A nested github markdown document
20 | }
21 |
--------------------------------------------------------------------------------
/scripts/02-metrics/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Metrics for comparing two trajectories
6 |
7 | Good metrics are crucial for an unbiased and comprehensive benchmarking. Here we test several possible metrics for comparing two trajectories.
8 |
9 | ```{r, results = "asis"}
10 | dynbenchmark::knit_nest("01-metric_characterisation")
11 | ```
12 |
13 | ```{r, results = "asis"}
14 | dynbenchmark::knit_nest("02-metric_conformity")
15 | ```
16 |
17 | ```{r, results = "asis"}
18 | dynbenchmark::knit_nest("03-aggregation")
19 | ```
20 |
21 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
22 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/inspect_identity_timings.R:
--------------------------------------------------------------------------------
1 | #' Check different benchmark component timings
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 |
6 | experiment("06-benchmark/inspect_identity_timings")
7 |
8 | raw_data <- read_rds(result_file("benchmark_results_unnormalised.rds", experiment_id = "06-benchmark"))
9 |
10 | timings <-
11 | raw_data %>%
12 | filter(method_id == "identity") %>%
13 | select(method_id, dataset_id, dataset_trajectory_type, starts_with("time_"))
14 |
15 | ggplot(timings %>% gather(step, time, starts_with("time_"))) +
16 | geom_density(aes(time, colour = step)) +
17 | facet_wrap(~ step, scales = "free") +
18 | labs(title = "Identity")
19 |
20 |
--------------------------------------------------------------------------------
/package/man/ref.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{ref}
4 | \alias{ref}
5 | \title{Refer to a figure or table}
6 | \usage{
7 | ref(ref_type, ref_id, suffix = "", prefix = "", anchor = FALSE,
8 | format = get_default_format())
9 | }
10 | \arguments{
11 | \item{ref_type}{fig, sfig, table, ...}
12 |
13 | \item{ref_id}{The identifier}
14 |
15 | \item{suffix}{Adding something to the index}
16 |
17 | \item{prefix}{Adding something before the index}
18 |
19 | \item{anchor}{Whether to anchor here}
20 |
21 | \item{format}{The output format (html, latex, markdown, ...)}
22 | }
23 | \description{
24 | Refer to a figure or table
25 | }
26 |
--------------------------------------------------------------------------------
/scripts/02-metrics/02-metric_conformity/04-assess_similarity.R:
--------------------------------------------------------------------------------
1 | #' Assess the similarity between metrics
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | experiment("02-metrics/02-metric_conformity")
7 |
8 | # load scores
9 | scores <-
10 | read_rds(derived_file("scores.rds"))
11 |
12 | metric_ids <- unique(scores$metric_id)
13 |
14 | scores_matrix <- scores %>%
15 | spread(metric_id, score) %>%
16 | select(metric_ids)
17 |
18 | metric_similarity <- cor(scores_matrix, method = "spearman") %>%
19 | reshape2::melt(value.name = "correlation", varnames = c("from", "to"))
20 |
21 | metric_similarity %>%
22 | ggplot(aes(from, to)) +
23 | geom_tile(aes(fill = correlation))
24 |
--------------------------------------------------------------------------------
/package/R/qsub.R:
--------------------------------------------------------------------------------
1 | #' pmap, but with qsub ^_^
2 | #'
3 | #' @inheritParams purrr::pmap
4 | #' @param ... Params given to [qsub::qsub_lapply()]
5 | #' @seealso qsub::qsub_lapply
6 | #' @export
7 | qsub_pmap <- function(.x, .f, ...) {
8 | number_of_elements <- unique(map_int(.x, length))
9 | testthat::expect_equal(length(number_of_elements), 1)
10 | .x2 <- map(seq_len(number_of_elements), function(ix) {
11 | .x2_element <- map(.x, ~.[[ix]]) %>% set_names(names(.x))
12 | .x2_element$.f <- .f
13 | .x2_element
14 | })
15 |
16 | .f2 <- function(.x, ...) {purrr::invoke(.x$.f, .x[names(.x) != ".f"], ...)}
17 |
18 | qsub::qsub_lapply(
19 | .x2,
20 | .f2,
21 | ...
22 | )
23 | }
24 |
--------------------------------------------------------------------------------
/scripts/03-methods/01-gather_methods_information/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Gathering all the information we have about the methods
6 |
7 | Most information of the methods are contained within their respective containers (see the [dynmethods](https://github.com/dynverse/dynmethods) repository, [https://github.com/dynverse/dynmethods](https://github.com/dynverse/dynmethods)). We gather additional information from our google sheets (https://docs.google.com/spreadsheets/d/1Mug0yz8BebzWt8cmEW306ie645SBh_tDHwjVw4OFhlE), which also contains the quality control for each methods.
8 |
9 | ```{r}
10 | dynbenchmark::render_scripts_documentation()
11 | ```
12 |
--------------------------------------------------------------------------------
/scripts/12-manuscript/render_readmes.R:
--------------------------------------------------------------------------------
1 | #' Renders all the READMEs in the repository
2 |
3 | library(dynbenchmark)
4 | library(fs)
5 | library(tidyverse)
6 |
7 | readme_paths <- c(
8 | fs::dir_ls("scripts", regexp = "README\\.Rmd", recursive = TRUE),
9 | fs::dir_ls("derived", regexp = "README\\.Rmd", recursive = TRUE),
10 | # fs::dir_ls("data", regexp = "README\\.Rmd", recursive = TRUE),
11 | c("./README.Rmd", "results/README.Rmd")
12 | )
13 |
14 | walk(readme_paths, function(readme_path) {
15 | print(paste0("Processing ", readme_path))
16 | rmarkdown::render(
17 | readme_path,
18 | output_format = dynbenchmark::github_markdown_nested(html_preview = FALSE),
19 | quiet = TRUE
20 | )
21 | })
22 |
--------------------------------------------------------------------------------
/package/man/load_dyneval_model.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/load_dyneval_model.R
3 | \name{load_dyneval_model}
4 | \alias{load_dyneval_model}
5 | \title{Load model from benchmark suite results}
6 | \usage{
7 | load_dyneval_model(method_id, df, auto_gc = TRUE, experiment_id = NULL)
8 | }
9 | \arguments{
10 | \item{method_id}{The short_name of a method}
11 |
12 | \item{df}{A data frame containing the crossing of which models to load}
13 |
14 | \item{auto_gc}{Whether or not to automatically garbage collect after loading models}
15 |
16 | \item{experiment_id}{The experiment id (default NULL}
17 | }
18 | \description{
19 | Load model from benchmark suite results
20 | }
21 |
--------------------------------------------------------------------------------
/package/man/load_dataset.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_datasets.R
3 | \name{list_datasets}
4 | \alias{list_datasets}
5 | \alias{load_dataset}
6 | \alias{load_datasets}
7 | \title{Load datasets}
8 | \usage{
9 | list_datasets(prefix = "")
10 |
11 | load_dataset(id, as_tibble = FALSE)
12 |
13 | load_datasets(ids = list_datasets()$id, as_tibble = TRUE)
14 | }
15 | \arguments{
16 | \item{prefix}{The prefix, eg. real or synthetic}
17 |
18 | \item{id}{The ID of the dataset to be used}
19 |
20 | \item{as_tibble}{Return the datasets as a tibble or as a list of datasets?}
21 |
22 | \item{ids}{Character vector of dataset identifiers}
23 | }
24 | \description{
25 | Load datasets
26 | }
27 |
--------------------------------------------------------------------------------
/package/data-raw/method_characterisation.R:
--------------------------------------------------------------------------------
1 | topinf_types <- tibble(
2 | name = c("free", "parameter", "fixed"),
3 | short_name = c("free", "param", "fixed"),
4 | colour = c("#00ab1b", "#edb600", "#cc2400"),
5 | explanation = c("inferred by algorithm", "determined by parameter", "fixed by algorithm")
6 | )
7 |
8 | error_reasons <- tibble(
9 | name = c("pct_memory_limit", "pct_time_limit", "pct_execution_error", "pct_method_error"),
10 | label = c("Memory limit exceeded", "Time limit exceeded", "Execution error", "Method error"),
11 | colour = RColorBrewer::brewer.pal(length(name), "Set3")
12 | )
13 | usethis::proj_set("package")
14 | usethis::use_data(error_reasons = error_reasons, topinf_types = topinf_types, overwrite = TRUE)
15 |
--------------------------------------------------------------------------------
/scripts/03-methods/03-method_characterisation/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Method characterisation
3 |
4 | Here we have a look at the diversity of TI
5 | methods
6 |
7 | | \# | script/folder | description |
8 | | :- | :------------------------------------------------------- | :--------------------------------------------------------------------------- |
9 | | 1 | [📄`tool_characterisation.R`](01-tool_characterisation.R) | Several figures for looking at the history and diversity of TI methods/tools |
10 | | 2 | [📄`tools_table.R`](02-tools_table.R) | Generate a table containing the methods |
11 |
--------------------------------------------------------------------------------
/package/data-raw/non_inclusion_reasons.R:
--------------------------------------------------------------------------------
1 | non_inclusion_reasons <- tribble(
2 | ~id, ~long,
3 | "not_free", "Not freely available",
4 | "unavailable", "No code available",
5 | "superseded", "Superseded by another method",
6 | "not_expression_based", "Requires data types other than expression",
7 | "gui_only", "No programming interface",
8 | "unwrappable", "Unresolved errors during wrapping",
9 | "speed", "Too slow (requires more than one hour on a 100x100 dataset)",
10 | "no_ordering", "Doesn't return an ordering",
11 | "user_input", "Requires additional user input during the algorithm (not prior information)"
12 | ) %>% mutate(footnote = letters[seq_along(id)])
13 |
14 | devtools::use_data(non_inclusion_reasons, overwrite = TRUE)
15 |
--------------------------------------------------------------------------------
/package/man/load_platforms.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_platforms.R
3 | \name{load_platforms}
4 | \alias{load_platforms}
5 | \alias{load_platform}
6 | \alias{get_platform_features}
7 | \alias{select_platforms}
8 | \title{List and load the platforms}
9 | \usage{
10 | load_platforms()
11 |
12 | load_platform(platform_id)
13 |
14 | get_platform_features(platforms)
15 |
16 | select_platforms(n_platforms)
17 | }
18 | \arguments{
19 | \item{platform_id}{Platform identifier}
20 |
21 | \item{platforms}{The platforms, loaded by load_platforms}
22 |
23 | \item{n_platforms}{Number of platforms to select}
24 | }
25 | \description{
26 | List and load the platforms
27 |
28 | Load a platform
29 | }
30 |
--------------------------------------------------------------------------------
/package/man/label_metric.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_labels.R
3 | \name{label_metric}
4 | \alias{label_metric}
5 | \alias{label_metrics}
6 | \title{Label the metrics}
7 | \usage{
8 | label_metric(metric_id, format = get_default_metric_format(),
9 | parse = FALSE)
10 |
11 | label_metrics(metric_ids, ...)
12 | }
13 | \arguments{
14 | \item{metric_id}{metric id}
15 |
16 | \item{format}{The format of the label to return, can be plotmath, latex, long_name or metric_id}
17 |
18 | \item{parse}{Whether to parse the label into an expression}
19 |
20 | \item{metric_ids}{The metric ids}
21 |
22 | \item{...}{Extra parameters for label_metric}
23 | }
24 | \description{
25 | Label the metrics
26 | }
27 |
--------------------------------------------------------------------------------
/scripts/01-datasets/03-download_from_prism.R:
--------------------------------------------------------------------------------
1 | #' Download the datasets from the cluster
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 |
6 | experiment("01-datasets")
7 |
8 | qsub::rsync_remote(
9 | remote_src = TRUE,
10 | path_src = derived_file(remote = TRUE, experiment = "01-datasets"),
11 | remote_dest = FALSE,
12 | path_dest = derived_file(remote = FALSE, experiment = "01-datasets"),
13 | verbose = TRUE,
14 | compress = FALSE
15 | )
16 |
17 | # Upload to prism
18 | # qsub::rsync_remote(
19 | # remote_src = FALSE,
20 | # path_src = derived_file(remote = FALSE, experiment = "01-datasets"),
21 | # remote_dest = TRUE,
22 | # path_dest = derived_file(remote = TRUE, experiment = "01-datasets"),
23 | # verbose = TRUE
24 | # )
25 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/wip/tabula-muris.R:
--------------------------------------------------------------------------------
1 | # https://figshare.com/articles/Single-cell_RNA-seq_data_from_Smart-seq2_sequencing_of_FACS_sorted_cells/5715040
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | dataset_preprocessing("real/tabula-muris")
7 |
8 | # cell info
9 | cell_assignments_file <- download_dataset_source_file("annotations_FACS.csv", "https://ndownloader.figshare.com/files/10039267")
10 | all_cell_info <- read_csv(cell_assignments_file) %>%
11 | mutate(group_id = cell_ontology_class)
12 |
13 | counts_zip <- download_dataset_source_file("FACS.zip", "https://ndownloader.figshare.com/files/10038307")
14 | all_cell_info <- read_csv(cell_assignments_file) %>%
15 | mutate(group_id = cell_ontology_class)
16 |
--------------------------------------------------------------------------------
/package/R/setup_methods.R:
--------------------------------------------------------------------------------
1 | #' Setup the singularity methods
2 | #'
3 | #' @export
4 | setup_singularity_methods <- function() {
5 | options(
6 | dynwrap_singularity_images_folder = derived_file("", experiment_id = "singularity_images"),
7 | dynwrap_run_environment = "singularity"
8 | )
9 | }
10 |
11 |
12 | #' Load the information on the methods
13 | #'
14 | #' @param default_filter Default filtering for the benchmark.
15 | #' @param ... Other filters for the method tibble.
16 | #'
17 | #' @export
18 | load_methods <- function(
19 | default_filter = method_source == "tool" | method_id %in% c("angle", "comp1", "mst"),
20 | ...
21 | ) {
22 | read_rds(result_file("methods.rds", "03-methods")) %>%
23 | filter(!!!rlang::enquos(default_filter, ...))
24 | }
25 |
--------------------------------------------------------------------------------
/package/man/knit_nest.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{knit_nest}
4 | \alias{knit_nest}
5 | \title{Knit a child, and add an extra level of headings + fix relative paths. Can be used both for latex and markdown output formats}
6 | \usage{
7 | knit_nest(file, format = get_default_format(), levels = 1)
8 | }
9 | \arguments{
10 | \item{file}{File to knit, can also be a directory in which case the README.Rmd will be knit}
11 |
12 | \item{format}{Which format to use.}
13 |
14 | \item{levels}{The number of levels to add onto section headings}
15 | }
16 | \description{
17 | Knit a child, and add an extra level of headings + fix relative paths. Can be used both for latex and markdown output formats
18 | }
19 |
--------------------------------------------------------------------------------
/package/man/add_table.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{add_table}
4 | \alias{add_table}
5 | \alias{add_stable}
6 | \title{Add a table}
7 | \usage{
8 | add_table(table, ref_id, caption_main, caption_text = "",
9 | format = get_default_format())
10 |
11 | add_stable(table, ref_id, caption_main, caption_text = "",
12 | format = get_default_format())
13 | }
14 | \arguments{
15 | \item{table}{Either a tibble, a path to a table or a named list with latex, html and markdown kables}
16 |
17 | \item{ref_id}{The identifier}
18 |
19 | \item{caption_main}{Caption title}
20 |
21 | \item{caption_text}{Caption text}
22 |
23 | \item{format}{The format, in html or latex}
24 | }
25 | \description{
26 | Add a table
27 | }
28 |
--------------------------------------------------------------------------------
/package/man/dynbenchmark.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/package.R
3 | \docType{package}
4 | \name{dynbenchmark}
5 | \alias{dynbenchmark}
6 | \alias{dynbenchmark-package}
7 | \title{Easy installing and loading of the dynverse}
8 | \description{
9 | The 'dynverse' is a set of packages with which to
10 | evaluate trajectory inference methods for dynamic processes.
11 | }
12 | \details{
13 | The dynbenchmark package contains the code to generate/preprocess datasets, evaluate TI methods on these datasets, and finally wrap all results together in a scientific paper.
14 | The code within this package can be run locally, but can also be run within a docker environment, for which the scripts are contained in the \code{scripts} folder.
15 | }
16 |
--------------------------------------------------------------------------------
/package/man/nacor.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/nacor.R
3 | \name{nacor}
4 | \alias{nacor}
5 | \title{Na removed cor}
6 | \usage{
7 | nacor(x, y, method = "spearman")
8 | }
9 | \arguments{
10 | \item{x}{a numeric vector, matrix or data frame.}
11 |
12 | \item{y}{\code{NULL} (default) or a vector, matrix or data frame with
13 | compatible dimensions to \code{x}. The default is equivalent to
14 | \code{y = x} (but more efficient).}
15 |
16 | \item{method}{a character string indicating which correlation
17 | coefficient (or covariance) is to be computed. One of
18 | \code{"pearson"} (default), \code{"kendall"}, or \code{"spearman"}:
19 | can be abbreviated.}
20 | }
21 | \description{
22 | Na removed cor
23 | }
24 |
--------------------------------------------------------------------------------
/package/man/paramoptim_bind_results.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_fetch_results.R
3 | \name{paramoptim_bind_results}
4 | \alias{paramoptim_bind_results}
5 | \title{Gather and bind the results of the benchmark jobs}
6 | \usage{
7 | paramoptim_bind_results(filter_fun = NULL,
8 | local_output_folder = derived_file("suite"))
9 | }
10 | \arguments{
11 | \item{filter_fun}{A function with which to filter the data as it is being read from files. Function must take a single data frame as input and return a filtered data frame as a result.}
12 |
13 | \item{local_output_folder}{A folder in which to output intermediate and final results.}
14 | }
15 | \description{
16 | Gather and bind the results of the benchmark jobs
17 | }
18 |
--------------------------------------------------------------------------------
/package/man/benchmark_fetch_results.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_benchmark_fetch_results.R
3 | \name{benchmark_fetch_results}
4 | \alias{benchmark_fetch_results}
5 | \title{Fetch the results of the benchmark jobs from the cluster.}
6 | \usage{
7 | benchmark_fetch_results(remote = NULL,
8 | local_output_folder = derived_file("suite"))
9 | }
10 | \arguments{
11 | \item{remote}{The host at which to check for running jobs.
12 | If \code{NULL}, each qsub handle will be checked individually.
13 | If \code{TRUE}, the default qsub handle will be used.}
14 |
15 | \item{local_output_folder}{A folder in which to output intermediate and final results.}
16 | }
17 | \description{
18 | Fetch the results of the benchmark jobs from the cluster.
19 | }
20 |
--------------------------------------------------------------------------------
/package/R/topologies_with_same_n_milestones.R:
--------------------------------------------------------------------------------
1 | # Different topologies with the same number of milestones
2 | topologies_with_same_n_milestones <- list(
3 | linear = tibble(from = c("A", "B", "C", "D", "E"), to = c("B", "C", "D", "E", "F")),
4 | bifurcation = tibble(from = c("A", "B", "B", "C", "D"), to = c("B", "C", "D", "E", "F")),
5 | multifurcating = tibble(from = c("A", "B", "B", "B", "C"), to = c("B", "C", "D", "E", "F")),
6 | tree = tibble(from = c("A", "B", "B", "C", "C"), to = c("B", "C", "D", "E", "F")),
7 | cycle = tibble(from = c("A", "B", "C", "D", "E"), to = c("B", "C", "D", "E", "A")),
8 | connected = tibble(from = c("A", "B", "B", "D", "E"), to = c("B", "C", "D", "E", "A")),
9 | disconnected = tibble(from = c("A", "B", "C", "D", "E"), to = c("B", "C", "A", "E", "F"))
10 | )
11 |
--------------------------------------------------------------------------------
/package/man/common_dynbenchmark_format.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{common_dynbenchmark_format}
4 | \alias{common_dynbenchmark_format}
5 | \title{Common dynbenchmark format}
6 | \usage{
7 | common_dynbenchmark_format(format,
8 | bibliography = raw_file("references.bib", "12-manuscript"),
9 | csl = raw_file("nature-biotechnology.csl", "12-manuscript"),
10 | reference_section_title = "References")
11 | }
12 | \arguments{
13 | \item{format}{The format on which to apply common changes}
14 |
15 | \item{bibliography}{Bibliography location}
16 |
17 | \item{csl}{Csl file location}
18 |
19 | \item{reference_section_title}{The title of the reference section}
20 | }
21 | \description{
22 | Common dynbenchmark format
23 | }
24 |
--------------------------------------------------------------------------------
/package/man/extract_scripts_documentation.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_document_scripts.R
3 | \name{extract_scripts_documentation}
4 | \alias{extract_scripts_documentation}
5 | \alias{render_scripts_documentation}
6 | \title{Will recursively extract documentations from the scripts and folders within a given folder}
7 | \usage{
8 | extract_scripts_documentation(folder = getwd(), recursive = TRUE)
9 |
10 | render_scripts_documentation(folder = ".", recursive = FALSE)
11 | }
12 | \arguments{
13 | \item{folder}{The folder from which to start, defaults to current working directory}
14 |
15 | \item{recursive}{Whether to extract scripts recursively}
16 | }
17 | \description{
18 | Will recursively extract documentations from the scripts and folders within a given folder
19 | }
20 |
--------------------------------------------------------------------------------
/package/data-raw/id_mapper.R:
--------------------------------------------------------------------------------
1 |
2 |
3 | ensembl_human <- org.Hs.eg.db::org.Hs.egENSEMBL2EG %>% as.list() %>% unlist() %>% tibble(ensembl = names(.), entrez = .)
4 | symbol_human <- org.Hs.eg.db::org.Hs.egSYMBOL %>% as.list() %>% unlist() %>% tibble(entrez = names(.), symbol = .)
5 | human_mapper <- full_join(ensembl_human, symbol_human, by = "entrez")
6 |
7 | ensembl_mouse <- org.Mm.eg.db::org.Mm.egENSEMBL2EG %>% as.list() %>% unlist() %>% tibble(ensembl = names(.), entrez = .)
8 | symbol_mouse <- org.Mm.eg.db::org.Mm.egSYMBOL %>% as.list() %>% unlist() %>% tibble(entrez = names(.), symbol = .)
9 | mouse_mapper <- full_join(ensembl_mouse, symbol_mouse, by = "entrez")
10 |
11 | id_mapper <- bind_rows(human_mapper, mouse_mapper) %>%
12 | group_by(entrez) %>%
13 | filter(row_number() == 1)
14 |
15 | devtools::use_data(id_mapper, overwrite = TRUE)
16 |
--------------------------------------------------------------------------------
/scripts/02-metrics/02-metric_conformity/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Metric conformity
6 |
7 | Differences between two datasets should be reflected in certain changes in the metrics. This can be formalised in a set of rules, for example:
8 |
9 | * If the position of some cells are different than in the reference, the score should decrease.
10 | * If the topology of the network is different than that in the reference, the score should not be perfect.
11 | * The more cells are filtered from the trajectory, the more the score should decrease.
12 |
13 | Here, we assess whether metrics conforms such rules empirically:
14 |
15 |
16 | ```{r, results = "asis"}
17 | dynbenchmark::render_scripts_documentation()
18 | ```
19 |
20 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
21 |
--------------------------------------------------------------------------------
/.travis.yml:
--------------------------------------------------------------------------------
1 | language: r
2 | sudo: required
3 | dist: trusty
4 | bioc_required: true
5 | services:
6 | - docker
7 | cache: packages
8 | warnings_are_errors: true
9 | addons:
10 | apt:
11 | packages:
12 | - libudunits2-dev # units > ggforce > ggraph > patchwork > dynplot
13 | - libssh-dev # ssh > qsub
14 | git:
15 | submodules: false
16 | before_install:
17 | - source <(curl -sSL https://raw.githubusercontent.com/dynverse/travis_scripts/master/helper.sh)
18 | - install_hdf5
19 | - install_dynverse
20 | - cd package
21 | install:
22 | - install_cran devtools covr
23 | - install_withdeps
24 | after_success:
25 | - R -e 'covr::codecov()'
26 |
27 | jobs:
28 | include:
29 | - stage: prepare cache
30 | script:
31 | - echo Skip
32 | after_success:
33 | - echo Skip
34 | - stage: test
35 |
--------------------------------------------------------------------------------
/package/man/benchmark_bind_results.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_benchmark_fetch_results.R
3 | \name{benchmark_bind_results}
4 | \alias{benchmark_bind_results}
5 | \title{Gather and bind the results of the benchmark jobs}
6 | \usage{
7 | benchmark_bind_results(load_models = FALSE, filter_fun = NULL,
8 | local_output_folder = derived_file("suite"))
9 | }
10 | \arguments{
11 | \item{load_models}{Whether or not to load the models as well.}
12 |
13 | \item{filter_fun}{A function with which to filter the data as it is being read from files. Function must take a single data frame as input and return a filtered data frame as a result.}
14 |
15 | \item{local_output_folder}{A folder in which to output intermediate and final results.}
16 | }
17 | \description{
18 | Gather and bind the results of the benchmark jobs
19 | }
20 |
--------------------------------------------------------------------------------
/scripts/12-manuscript/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Preparing the manuscript
3 |
4 | | \# | script/folder | description |
5 | | :- | :-------------------------------------------------------------- | :------------------------------------------------------------ |
6 | | | [📄`render_manuscript.R`](render_manuscript.R) | Renders the manuscript, and prepares the files for submission |
7 | | | [📄`render_readmes.R`](render_readmes.R) | Renders all the READMEs in the repository |
8 | | | [📄`render_results.R`](render_results.R) | Renders results READMEs |
9 | | | [📄`render_supplementary_notes.R`](render_supplementary_notes.R) | Renders the supplementary notes in pdf |
10 |
--------------------------------------------------------------------------------
/scripts/03-methods/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Trajectory inference methods
6 |
7 | Here we
8 |
9 | - Compile all the information we have about TI methods
10 | - Characterise the methods with regards to user and developer friendliness (method quality control)
11 | - Characterise the methods with regards to prior information, underlying algorithm, possible detectable trajectory types, ...
12 |
13 | ```{r}
14 | dynbenchmark::render_scripts_documentation()
15 | ```
16 |
17 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
18 |
19 |
20 | ```{r, results = "asis"}
21 | dynbenchmark::knit_nest("01-gather_methods_information")
22 | ```
23 |
24 | ```{r, results = "asis"}
25 | dynbenchmark::knit_nest("02-tool_qc")
26 | ```
27 |
28 | ```{r, results = "asis"}
29 | dynbenchmark::knit_nest("03-method_characterisation")
30 | ```
31 |
--------------------------------------------------------------------------------
/scripts/04-method_testing/0-pull_singularity_images.R:
--------------------------------------------------------------------------------
1 | #' Update singularity images on the cluster
2 |
3 | library(dynmethods)
4 | library(dynwrap)
5 | library(dynbenchmark)
6 | library(tidyverse)
7 |
8 | experiment("singularity_images")
9 |
10 | # If nothing is running on the cluster, run this on the login node to clear temporary folders:
11 | # rm -rf /tmp/* /data/*; mkdir /data/tmp
12 | # for i in $(seq 1 8); do ssh prismcls0$i 'rm -rf /tmp/* /data/*; mkdir /data/tmp'; done
13 |
14 | meth <- get_ti_methods()
15 | handle <- qsub::qsub_lapply(
16 | X = meth$fun,
17 | qsub_environment = c(),
18 | qsub_packages = c("babelwhale"),
19 | qsub_config = qsub::override_qsub_config(
20 | max_wall_time = "01:00:00",
21 | name = "singbuild",
22 | memory = "10G",
23 | num_cores = 1,
24 | wait = FALSE,
25 | stop_on_error = FALSE
26 | ),
27 | FUN = function(fun) {
28 | fun()
29 | }
30 | )
31 |
--------------------------------------------------------------------------------
/package/man/paramoptim_fetch_results.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_fetch_results.R
3 | \name{paramoptim_fetch_results}
4 | \alias{paramoptim_fetch_results}
5 | \title{Fetch the results of the benchmark jobs from the cluster.}
6 | \usage{
7 | paramoptim_fetch_results(remote = NULL,
8 | local_output_folder = derived_file("suite"), impatient = FALSE)
9 | }
10 | \arguments{
11 | \item{remote}{The host at which to check for running jobs.
12 | If \code{NULL}, each qsub handle will be checked individually.
13 | If \code{TRUE}, the default qsub handle will be used.}
14 |
15 | \item{local_output_folder}{A folder in which to output intermediate and final results.}
16 |
17 | \item{impatient}{If true, then the results will be fetched regardless of whether the optimisation has finished.}
18 | }
19 | \description{
20 | Fetch the results of the benchmark jobs from the cluster.
21 | }
22 |
--------------------------------------------------------------------------------
/package/R/install_fontawesome.R:
--------------------------------------------------------------------------------
1 | #' @importFrom extrafont fonts font_import
2 | #' @importFrom utils download.file unzip
3 | install_fontawesome <- function() {
4 | if (!"Font Awesome 5 Free" %in% extrafont::fonts()) {
5 | # install fontawesome
6 | release <- "fontawesome-free-5.3.1-web"
7 |
8 | # download and unzip
9 | url <- paste0("https://github.com/FortAwesome/Font-Awesome/releases/download/5.3.1/", release, ".zip")
10 | zip_file <- tempfile(fileext = '.zip')
11 | utils::download.file(url, destfile = zip_file)
12 |
13 | temp_dir <- tempfile()
14 | dir.create(temp_dir)
15 |
16 | utils::unzip(zipfile = zip_file, exdir = temp_dir)
17 |
18 | font_dir <- paste0(temp_dir, "/", release, "/webfonts/")
19 | extrafont::font_import(paths = font_dir, pattern = "fa-solid-900\\.ttf", prompt = FALSE)
20 | extrafont::font_import(paths = font_dir, pattern = "fa-brands-400\\.ttf", prompt = FALSE)
21 | }
22 | }
23 |
--------------------------------------------------------------------------------
/scripts/04-method_testing/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Method testing
3 |
4 | Each method is tested on a few small real and toy datasets. Ideally,
5 | each method should be able to execute successfully on each of these
6 | datasets. If a method is not able to infer a trajectory on any of the
7 | datasets, it will not be included in any of the next experiments.
8 |
9 | Overview
10 | experiment:
11 |
12 | | \# | script/folder | description |
13 | | :- | :---------------------------------------------------------- | :--------------------------------------- |
14 | | 0 | [📄`pull_singularity_images.R`](0-pull_singularity_images.R) | Update singularity images on the cluster |
15 | | 1 | [📄`submit_jobs.R`](1-submit_jobs.R) | Submit jobs for method testing |
16 | | 2 | [📄`retrieve_results.R`](2-retrieve_results.R) | Retrieve the results |
17 |
--------------------------------------------------------------------------------
/scripts/12-manuscript/render_supplementary_notes.R:
--------------------------------------------------------------------------------
1 | #' Renders the supplementary notes in pdf
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 |
6 | supp_notes <- tribble(
7 | ~rmd_path, ~final_path,
8 | result_file("README.Rmd", "02-metrics"), result_file("Supplementary Note 1.pdf", "12-manuscript"),
9 | result_file("README.Rmd", "01-datasets/02-synthetic"), result_file("Supplementary Note 2.pdf", "12-manuscript")
10 | ) %>%
11 | mutate(pdf_path = str_replace_all(rmd_path, "Rmd$", "pdf"))
12 |
13 |
14 | walk(supp_notes$rmd_path, function(supplementary_note_path) {
15 | print(paste0("Processing ", supplementary_note_path))
16 | rmarkdown::render(
17 | supplementary_note_path,
18 | output_format = dynbenchmark::pdf_supplementary_note(),
19 | quiet = TRUE
20 | )
21 | })
22 |
23 | pwalk(supp_notes, function(rmd_path, final_path, pdf_path) {
24 | if (file.exists(final_path)) file.remove(final_path)
25 | file.rename(pdf_path, final_path)
26 | })
27 |
--------------------------------------------------------------------------------
/scripts/07-stability/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Stability
3 |
4 | Analysis of the stability of each method: how similar are the
5 | trajectories when rerunning a method on slightly perturbed
6 | data?
7 |
8 | | \# | script/folder | description |
9 | | :- | :-------------------------------------------- | :-------------------------------------------------- |
10 | | 0 | [📄`generate_data.R`](0-generate_data.R) | Generate the subsampled datasets |
11 | | 1 | [📄`submit_jobs.R`](1-submit_jobs.R) | Submit the stability jobs |
12 | | 2 | [📄`retrieve_results.R`](2-retrieve_results.R) | Retrieve and process the stability results |
13 | | | [📄`generate_dataset.R`](generate_dataset.R) | Helper function for generating a subsampled dataset |
14 |
15 | The results of this experiment are available
16 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/07-stability).
17 |
--------------------------------------------------------------------------------
/scripts/09-guidelines/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Guidelines
3 |
4 | Preparing the data for the dynguidelines app and creation of the
5 | guidelines
6 | figure.
7 |
8 | | \# | script/folder | description |
9 | | :- | :----------------------------------------------- | :------------------------------------------------------------------------------------------------------------------------------------------------------- |
10 | | 1 | [📄`aggregate.R`](01-aggregate.R) | Aggregation of the different bnchmarking results for dynguidelines |
11 | | 2 | [📄`guidelines_figure.R`](02-guidelines_figure.R) | Generate the guidelines figure where we show the top 4 methods for every (set of) trajectory types, with some other information relevant to the end user |
12 |
--------------------------------------------------------------------------------
/package/man/theme_pub.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_themes.R
3 | \name{theme_pub}
4 | \alias{theme_pub}
5 | \title{Publication ready theme from the cowplot package}
6 | \usage{
7 | theme_pub(font_size = 14, font_family = "", line_size = 0.5,
8 | rel_small = 12/14, rel_tiny = 11/14, rel_large = 16/14)
9 | }
10 | \arguments{
11 | \item{font_size}{Overall font size.}
12 |
13 | \item{font_family}{Font family for plot title, axis titles and labels, legend texts, etc.}
14 |
15 | \item{line_size}{Line size for axis lines.}
16 |
17 | \item{rel_small}{Relative size of small text (e.g., axis tick labels)}
18 |
19 | \item{rel_tiny}{Relative size of tiny text (e.g., caption)}
20 |
21 | \item{rel_large}{Relative size of large text (e.g., title)}
22 | }
23 | \value{
24 | The theme.
25 | }
26 | \description{
27 | Publication ready theme from the cowplot package
28 | }
29 | \examples{
30 | library(ggplot2)
31 | qplot(1:10, (1:10)^2) + theme_pub(font_size = 12)
32 | }
33 |
--------------------------------------------------------------------------------
/package/R/data.R:
--------------------------------------------------------------------------------
1 | #' Mapping ensembl or entrez ids to symbols
2 | #'
3 | #' @docType data
4 | "id_mapper"
5 |
6 | #' Common manual labelling
7 | #'
8 | #' @docType data
9 | "labels"
10 |
11 | #' Metadata on error reasons
12 | #'
13 | #' @docType data
14 | "error_reasons"
15 |
16 | #' Metadata on topology inference types
17 | #'
18 | #' @docType data
19 | "topinf_types"
20 |
21 | #' Metadata on prior types
22 | #'
23 | #' @docType data
24 | "prior_types"
25 |
26 | #' Metadata on prior usage types
27 | #'
28 | #' @docType data
29 | "prior_usages"
30 |
31 | #' Metadata on non-inclusion reasons
32 | #'
33 | #' @docType data
34 | "non_inclusion_reasons"
35 |
36 | #' Metadata on QC applications
37 | #'
38 | #' @docType data
39 | "qc_applications"
40 |
41 | #' Metadata on QC categories
42 | #'
43 | #' @docType data
44 | "qc_categories"
45 |
46 | #' Metadata on method statuses
47 | #'
48 | #' @docType data
49 | "method_statuses"
50 |
51 | #' Colours of method statuses
52 | #'
53 | #' @docType data
54 | "method_status_colours"
55 |
--------------------------------------------------------------------------------
/package/man/fix_relative_paths.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_processors.R
3 | \name{fix_relative_paths}
4 | \alias{fix_relative_paths}
5 | \title{Process relative paths to links & figures
6 | First extract every link, determine whether it is a relative path and if yes, add folder to the front}
7 | \usage{
8 | fix_relative_paths(knit, folder)
9 | }
10 | \arguments{
11 | \item{knit}{Character vector}
12 |
13 | \item{folder}{The relative folder}
14 | }
15 | \description{
16 | Process relative paths to links & figures
17 | First extract every link, determine whether it is a relative path and if yes, add folder to the front
18 | }
19 | \examples{
20 | knit <- c(
21 | "hshlkjdsljkfdhg [i am a absolute path](/pompompom/dhkjhlkj/) kjfhlqkjsdhlkfjqsdf",
22 | "hshlkjdsljkfdhg [i am a relative path](pompompom/dhkjhlkj/) kjfhlqkjsdhlkfjqsdf",
23 | " ",
24 | "
",
24 | " "
25 | )
26 | dynbenchmark:::fix_relative_paths(knit, "IT WORKED :)")
27 | }
28 |
--------------------------------------------------------------------------------
/scripts/01-datasets/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Dataset processing and characterisation
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | The datasets are split in real datasets and synthetic datasets. The real datasets are downloaded and preprocessed first, and characteristics from these datasets (such as the number of cells and genes, library sizes, dropout probabilities, ...) are used to generate synthetic datasets. The datasets are then characterised, after which they are uploaded to Zenodo.
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
18 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
19 |
20 | ```{r results = 'asis'}
21 | dynbenchmark::knit_nest("01-real")
22 | ```
23 |
24 | ```{r results = 'asis'}
25 | dynbenchmark::knit_nest("02-synthetic")
26 | ```
27 | `
28 | ```{r results = 'asis'}
29 | dynbenchmark::knit_nest("04-dataset_characterisation")
30 | ```
31 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/04-dyngen_samplers_table.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("01-datasets/02-synthetic")
5 |
6 | # ____________________________________________________________________________
7 | # Samplers table ####
8 | samplers <- read_tsv(raw_file("samplers"))
9 | notes <- c("$y_{max} = r/d * p/q$")
10 |
11 | table <- map(c("latex", "html"), function(format) {
12 | table <- samplers %>% mutate(text = stringr::str_glue("${parameter} = {distribution}$")) %>%
13 | select(text) %>%
14 | mutate(text = kableExtra::cell_spec(text, format, escape = FALSE)) %>%
15 | knitr::kable(format, escape = FALSE, col.names = NULL) %>%
16 | kableExtra::kable_styling(bootstrap_options = "condensed") %>%
17 | kableExtra::footnote(notes, general_title = "where", escape = FALSE) %>%
18 | gsub("&", "&", .)
19 | table
20 | }) %>% set_names(c("latex", "html"))
21 |
22 | write_rds(table, result_file("samplers.rds"), compress = "xz")
23 |
--------------------------------------------------------------------------------
/scripts/08-summary/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Summarising the results into funky heatmaps
3 |
4 | Here, we aggregate the results from the different experiment into a
5 | “tidy” format. Next, we plot this into funky
6 | heatmaps.
7 |
8 | | \# | script/folder | description |
9 | | :- | :------------------------------------------------ | :----------------------------------------- |
10 | | 1 | [📄`aggregate_data.R`](1-aggregate_data.R) | Aggregation of the results |
11 | | 2 | [📄`main_figure.R`](2-main_figure.R) | Generation of the different funky heatmaps |
12 | | 2a | [📄`2a_columns_all.R`](2a_columns_all.R) | |
13 | | 2a | [📄`2a_columns_detailed.R`](2a_columns_detailed.R) | |
14 | | 2a | [📄`2a_columns_summary.R`](2a_columns_summary.R) | |
15 | | 2a | [📄`2a_columns_suppfig.R`](2a_columns_suppfig.R) | |
16 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Synthetic datasets
6 |
7 | Each synthetic dataset is based on some characteristics of some real datasets. These characteristics include:
8 |
9 | * The number of cells and features
10 | * The number of features which are differentially expressed in the trajectory
11 | * Estimates of the distribution of the library sizes, average expression, dropout probabilities, ... estimated by [Splatter](https://github.com/Oshlack/splatter).
12 |
13 | Here we estimate the parameters of these "platforms" and use them to simulate datasets using different simulators. Each simulation script first creates a design dataframe, which links particular platforms, different topologies, seeds and other parameters specific for a simulator.
14 |
15 | The data is then simulated using wrappers around the simulators (see [/package/R/simulators.R](/package/R/simulators.R)), so that they all return datasets in a format consistent with dynwrap.
16 |
17 | ```{r}
18 | dynbenchmark::render_scripts_documentation()
19 | ```
20 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/helper-04-complementarity.R:
--------------------------------------------------------------------------------
1 | #' Helper plotting functions the complementarity experiment
2 |
3 | quarter_circ <- function(xstart, ystart, from_angle, to_angle, size) {
4 | data_frame(
5 | angle = seq(from_angle, to_angle, length.out = 20) * 2 * pi,
6 | x = sin(angle),
7 | y = cos(angle)
8 | ) %>% mutate(
9 | x = (x - min(x)) / (max(x) - min(x)) * size + xstart,
10 | y = (y - min(y)) / (max(y) - min(y)) * size + ystart
11 | )
12 | }
13 |
14 | bracket <- function(xmin, xmax, ymin, ymax, flip = FALSE) {
15 | width <- xmax - xmin
16 | height <- ymax - ymin
17 | xmid <- xmin + width / 2
18 | ymid <- ymin + height / 2
19 | curly_size <- height / 2
20 |
21 | df <-
22 | bind_rows(
23 | quarter_circ(xmin, ymin, .75, 1, curly_size),
24 | quarter_circ(xmid - curly_size, ymid, .5, .25, curly_size),
25 | quarter_circ(xmid, ymid, .75, .5, curly_size),
26 | quarter_circ(xmax - curly_size, ymin, 0, .25, curly_size)
27 | )
28 |
29 | if (flip) {
30 | df <- df %>% mutate(y = ymin + ymax - y)
31 | }
32 |
33 | df
34 | }
35 |
--------------------------------------------------------------------------------
/scripts/02-metrics/01-metric_characterisation/03-clustering.R:
--------------------------------------------------------------------------------
1 | #' Characterisation of the `r dynbenchmark::label_metric("F1_branches")` and `r dynbenchmark::label_metric("F1_milestones")`
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(patchwork)
6 |
7 | experiment("02-metrics/01-metric_characterisation")
8 |
9 | set.seed(9)
10 | dataset <- dyntoy::generate_dataset(model = dyntoy::model_binary_tree(num_branchpoints = 2)) %>% simplify_trajectory()
11 |
12 | plot_clustering_scores_overview <- wrap_plots(
13 | plot_graph(dataset, label_milestones = TRUE) + ggtitle("Reference"),
14 | plot_graph(dataset, grouping = group_onto_nearest_milestones(dataset), plot_milestones = TRUE) + ggtitle("Cells mapped to milestones"),
15 | plot_graph(dataset, grouping = group_onto_trajectory_edges(dataset)) + ggtitle("Cells mapped to branches") + guides(color = guide_legend(ncol = 3))
16 | )
17 |
18 | plot_clustering_scores_overview
19 |
20 | write_rds(plot_clustering_scores_overview, result_file("clustering_scores_overview.rds"))
21 |
22 | ggsave(result_file("clustering_scores_overview.pdf"), width = 12, height = 5)
23 |
--------------------------------------------------------------------------------
/package/R/text_equations.R:
--------------------------------------------------------------------------------
1 | #' Renders equations for github markdown
2 | #'
3 | #' @param x A character vector
4 | #' @param format The format: html, markdown, latex, ...
5 | #' @export
6 | render_equations <- function(x, format = get_default_format()) {
7 | if (format == "markdown") {
8 | # inline equation
9 | x <- str_replace_all(
10 | x,
11 | "\\$[^\\$]+\\$",
12 | function(x) paste0("), ")")
13 | )
14 |
15 | # multiple line equations, e.g.
16 | # $$
17 | # 1 + 2 = 3
18 | # $$
19 |
20 | which_dd <- which(x == "$$")
21 | if (length(which_dd) %% 2 == 1) {
22 | stop("Uneven number of '$$' lines found!")
23 | }
24 |
25 | for (i in seq_len(length(which_dd) / 2)) {
26 | start <- which_dd[i * 2 - 1] + 1
27 | end <- which_dd[i * 2] - 1
28 |
29 | url <- paste0("), ")")
30 |
31 | x[start] <- url
32 | x[start - 1] <- ""
33 | x[(start):end+1] <- ""
34 | }
35 | }
36 |
37 | x
38 | }
39 |
--------------------------------------------------------------------------------
/package/man/experiment.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_experiment.R
3 | \name{experiment}
4 | \alias{experiment}
5 | \alias{derived_file}
6 | \alias{raw_file}
7 | \alias{result_file}
8 | \alias{scripts_file}
9 | \title{Helper function for controlling experiments}
10 | \usage{
11 | experiment(experiment_id)
12 |
13 | derived_file(filename = "", experiment_id = NULL, remote = FALSE)
14 |
15 | raw_file(filename = "", experiment_id = NULL, remote = FALSE)
16 |
17 | result_file(filename = "", experiment_id = NULL, remote = FALSE)
18 |
19 | scripts_file(filename = "", experiment_id = NULL, remote = FALSE)
20 | }
21 | \arguments{
22 | \item{experiment_id}{id for the experiment}
23 |
24 | \item{filename}{the filename}
25 |
26 | \item{remote}{The remote to access, "" if working locally, TRUE if using default qsub_config remote}
27 | }
28 | \description{
29 | Helper function for controlling experiments
30 | }
31 | \examples{
32 | \dontrun{
33 | experiment("test_plots")
34 |
35 | data <- matrix(runif(200), ncol = 2)
36 | pdf(result_file("testplot.pdf"), 5, 5)
37 | plot(data)
38 | dev.off()
39 | }
40 | }
41 |
--------------------------------------------------------------------------------
/raw/12-manuscript/nature-biotechnology.csl:
--------------------------------------------------------------------------------
1 |
2 |
18 |
19 |
--------------------------------------------------------------------------------
/package/man/add_fig.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{add_fig}
4 | \alias{add_fig}
5 | \alias{add_sfig}
6 | \alias{plot_fig}
7 | \title{Add a figure}
8 | \usage{
9 | add_fig(fig_path, ref_id, caption_main, caption_text = "", width = 5,
10 | height = 7, format = get_default_format())
11 |
12 | add_sfig(fig_path, ref_id, caption_main, caption_text = "", width = 5,
13 | height = 7, format = get_default_format(), integrate = TRUE)
14 |
15 | plot_fig(ref_type, ref_id, fig_path, caption_main, caption_text,
16 | width = 5, height = 7, format = "latex", integrate = TRUE)
17 | }
18 | \arguments{
19 | \item{fig_path}{Path to the figure}
20 |
21 | \item{ref_id}{The identifier}
22 |
23 | \item{caption_main}{Caption title}
24 |
25 | \item{caption_text}{Caption text}
26 |
27 | \item{width}{Width in inches}
28 |
29 | \item{height}{Height in inches}
30 |
31 | \item{format}{The format, in html or latex}
32 |
33 | \item{integrate}{Whether to integrate the figure, or direct the reader to a separate file}
34 |
35 | \item{ref_type}{fig, sfig, table, ...}
36 | }
37 | \description{
38 | Add a figure
39 | }
40 |
--------------------------------------------------------------------------------
/LICENSE.md:
--------------------------------------------------------------------------------
1 | # MIT License
2 |
3 | Copyright (c) 2014-2020 Robrecht Cannoodt, Wouter Saelens
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/package/man/platform.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_platforms.R
3 | \name{platform_from_counts}
4 | \alias{platform_from_counts}
5 | \alias{platform_simple}
6 | \title{Estimate platform parameters from a dataset}
7 | \usage{
8 | platform_from_counts(counts, grouping, subsample = 500)
9 |
10 | platform_simple(n_cells = 100L, n_features = 1000L,
11 | pct_main_features = 0.5, dropout_rate = 0.01, dropout_shape = 1)
12 | }
13 | \arguments{
14 | \item{counts}{The counts with cells in rows and genes in columns.}
15 |
16 | \item{grouping}{A named vector representing a grouping of the cells.}
17 |
18 | \item{subsample}{The number of cells to subsample.}
19 |
20 | \item{n_cells}{The number of cells}
21 |
22 | \item{n_features}{The number of features}
23 |
24 | \item{pct_main_features}{The percentage of features that are being driven by the trajectory (or vice versa)}
25 |
26 | \item{dropout_rate}{The mean rate of dropouts}
27 |
28 | \item{dropout_shape}{The shape of dropouts}
29 | }
30 | \description{
31 | Altenatively, \code{\link[=platform_simple]{platform_simple()}} returns a toy platform parameter configuration.
32 | }
33 |
--------------------------------------------------------------------------------
/package/LICENSE.md:
--------------------------------------------------------------------------------
1 | # MIT License
2 |
3 | Copyright (c) 2014-2020 Robrecht Cannoodt, Wouter Saelens
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/05-benchmark_interpretation_figure.R:
--------------------------------------------------------------------------------
1 | #' Combine the different figures of this experiment into one
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(patchwork)
6 |
7 | experiment("10-benchmark_interpretation")
8 |
9 | plot_variability_dataset_source <- read_rds(derived_file("variability_dataset_source.rds"))
10 | plot_variability_trajectory_type <- read_rds(derived_file("variability_trajectory_type.rds"))
11 | plot_dataset_source_correlation <- read_rds(derived_file("dataset_source_correlation.rds"))
12 | plot_topology_complexity_examples <- read_rds(derived_file("topology_complexity_examples.rds"))
13 |
14 | plot_benchmark_interpretation <- wrap_plots(
15 | plot_variability_dataset_source %>% patchwork::wrap_elements(),
16 | plot_dataset_source_correlation %>% patchwork::wrap_elements(),
17 | plot_variability_trajectory_type %>% patchwork::wrap_elements(),
18 | plot_topology_complexity_examples %>% patchwork::wrap_elements(),
19 | ncol = 1,
20 | heights = c(1, 0.7, 1, 1)
21 | ) + plot_annotation(tag_levels = "a")
22 |
23 | ggsave(result_file("benchmark_interpretation.pdf"), plot_benchmark_interpretation, width = 14, height = 18)
24 |
--------------------------------------------------------------------------------
/scripts/03-methods/01-gather_methods_information/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Gathering all the information we have about the methods
3 |
4 | Most information of the methods are contained within their respective
5 | containers (see the [dynmethods](https://github.com/dynverse/dynmethods)
6 | repository, ). We gather
7 | additional information from our google sheets
8 | (),
9 | which also contains the quality control for each
10 | methods.
11 |
12 | | \# | script/folder | description |
13 | | :- | :------------------------------------------------------------- | :------------------------------------------------------- |
14 | | 1 | [📄`group_methods_into_tools.R`](01-group_methods_into_tools.R) | Grouping methods into tools |
15 | | 2 | [📄`process_quality_control.R`](02-process_quality_control.R) | Downloading and processing the quality control worksheet |
16 | | 3 | [📄`add_quality_control.R`](03-add_quality_control.R) | Add QC scores to methods and tools tibble |
17 |
--------------------------------------------------------------------------------
/package/R/load_dyneval_model.R:
--------------------------------------------------------------------------------
1 | #' Load model from benchmark suite results
2 | #'
3 | #' @param method_id The short_name of a method
4 | #' @param df A data frame containing the crossing of which models to load
5 | #' @param auto_gc Whether or not to automatically garbage collect after loading models
6 | #' @param experiment_id The experiment id (default NULL
7 | #'
8 | #' @export
9 | load_dyneval_model <- function(method_id, df, auto_gc = TRUE, experiment_id = NULL) {
10 | models_filename <- derived_file(paste0("suite/", method_id, "/output_models.rds"), experiment_id = experiment_id)
11 | metrics_filename <- derived_file(paste0("suite/", method_id, "/output_metrics.rds"), experiment_id = experiment_id)
12 |
13 | if (!file.exists(models_filename)) {
14 | stop("Models of method ", sQuote(method_id), " not found.", sep = "")
15 | }
16 |
17 | models <- read_rds(models_filename)
18 | metrics <- read_rds(metrics_filename)
19 |
20 | ix <- metrics %>% mutate(i = row_number()) %>% inner_join(df, by = colnames(df)) %>% pull(i)
21 |
22 | if (length(ix) == 0) {
23 | stop("df did not match with any of the rows")
24 | }
25 |
26 | model <- models[ix]
27 |
28 | if (auto_gc) {
29 | gc()
30 | }
31 |
32 | model
33 | }
34 |
--------------------------------------------------------------------------------
/package/R/setup_get_dynbenchmark_folder.R:
--------------------------------------------------------------------------------
1 | #' Find the dynbenchmark path
2 | #'
3 | #' When executing some of the dynbenchmark functions, the git repository needs to be found.
4 | #' Either set the working directory to the repository, or define the DYNBENCHMARK_PATH variable.
5 | #'
6 | #' @param remote The remote to access, "" if running locally, TRUE if using default qsub config remote
7 | #'
8 | #' @export
9 | get_dynbenchmark_folder <- function(remote = FALSE) {
10 | if (is.logical(remote) && !remote) {
11 | pwd <- getOption("dynbenchmark_path")
12 |
13 | if (is.null(pwd)) {
14 | pwd <- Sys.getenv("DYNBENCHMARK_PATH")
15 | }
16 |
17 | if (is.null(pwd)) {
18 | pwd <- getwd()
19 | }
20 |
21 | if (pwd == "") {
22 | pwd <- paste0("./")
23 | }
24 |
25 | if (!file.exists(paste0(pwd, "/dynbenchmark.Rproj"))) {
26 | stop("dynbenchmark folder could not be found. Either set it as the working directory, or export a DYNBENCHMARK_PATH variable.")
27 | }
28 |
29 | pwd
30 | } else {
31 | pwd <- qsub::run_remote("echo $DYNBENCHMARK_PATH", remote)$stdout
32 |
33 | if (pwd == "") {
34 | stop("DYNBENCHMARK_PATH not found at remote")
35 | }
36 |
37 | pwd
38 | }
39 | }
40 |
--------------------------------------------------------------------------------
/package/man/process_raw_dataset.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/datasets_preprocessing.R
3 | \name{process_raw_dataset}
4 | \alias{process_raw_dataset}
5 | \title{Process a raw real or synthetic dataset}
6 | \usage{
7 | process_raw_dataset(id, counts, milestone_network, grouping,
8 | root_milestone_id = NULL, cell_info = tibble(cell_id =
9 | rownames(counts)), feature_info = tibble(feature_id =
10 | colnames(counts)))
11 | }
12 | \arguments{
13 | \item{id}{A unique identifier for the data. If \code{NULL}, a random string will be generated.}
14 |
15 | \item{counts}{The counts values of genes (columns) within cells (rows). This can be both a dense and sparse matrix.}
16 |
17 | \item{milestone_network}{A network of milestones.}
18 |
19 | \item{grouping}{A grouping of the cells, can be a named vector or a dataframe with \emph{group_id} and \emph{cell_id}}
20 |
21 | \item{root_milestone_id}{The root milestone, optional}
22 |
23 | \item{cell_info}{Optional meta-information pertaining the cells.}
24 |
25 | \item{feature_info}{Optional meta-information of the features, a dataframe with at least \emph{feature_id} as column}
26 | }
27 | \description{
28 | Process a raw real or synthetic dataset
29 | }
30 |
--------------------------------------------------------------------------------
/scripts/02-metrics/01-metric_characterisation/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Metric characterisation
3 |
4 | A first characterisation of the metrics. For each metric we:
5 |
6 | - generate some examples to get some intuition on how the metric works
7 | - test the robustness to a metric to stochasticity or parameters when
8 | appropriate
9 |
10 | | \# | script/folder | description |
11 | | :- | :----------------------------------- | :------------------------------------------------------------------------- |
12 | | 1 | [📄`correlation.R`](01-correlation.R) | Characterisation of the cordist |
13 | | 2 | [📄`topology.R`](02-topology.R) | Characterisation of the Isomorphic, edgeflip and HIM |
14 | | 3 | [📄`clustering.R`](03-clustering.R) | Characterisation of the F1branches and F1milestones |
15 | | 4 | [📄`featureimp.R`](04-featureimp.R) | Characterisation of the corfeatures and wcorfeatures |
16 |
17 | The results of this experiment are available
18 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/02-metrics/01-metric_characterisation)
19 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_macrophage-salmonella_saliba.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/gold/macrophage-salmonella_saliba")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "GSE79363_first_dataset_read_count.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE79363&format=file&file=GSE79363%5Ffirst%5Fdataset%5Fread%5Fcount%2Etxt%2Egz"
9 | )
10 |
11 | counts <- read_tsv(txt_location) %>% as.data.frame() %>% tibble::column_to_rownames("X1") %>% as.matrix() %>% t
12 | cell_info <- tibble(cell_id = rownames(counts), milestone_id = gsub(".*_([A-Za-z]*)$", "\\1", rownames(counts)))
13 |
14 | counts <- counts[cell_info$cell_id, ]
15 |
16 | milestone_network = tribble(
17 | ~from, ~to,
18 | "NNI", "MNGB",
19 | "NNI", "MGB",
20 | "NNI", "bystanders"
21 | ) %>% mutate(length = 1, directed = TRUE)
22 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
23 |
24 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
25 | counts <- counts[cell_info$cell_id, ]
26 |
27 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
28 |
29 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
30 |
--------------------------------------------------------------------------------
/package/man/dataset_preprocessing.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_datasets.R
3 | \name{dataset_preprocessing}
4 | \alias{dataset_preprocessing}
5 | \alias{get_dataset_preprocessing_id}
6 | \alias{dataset_source_file}
7 | \alias{dataset_raw_file}
8 | \alias{save_raw_dataset}
9 | \alias{dataset_file}
10 | \alias{save_dataset}
11 | \title{Helper functions for creating new datasets}
12 | \usage{
13 | dataset_preprocessing(id)
14 |
15 | get_dataset_preprocessing_id()
16 |
17 | dataset_source_file(filename = "", id = NULL, remote = FALSE)
18 |
19 | dataset_raw_file(id)
20 |
21 | save_raw_dataset(dataset, id = get_dataset_preprocessing_id())
22 |
23 | dataset_file(filename = "", id = NULL, remote = FALSE)
24 |
25 | save_dataset(dataset, id = NULL, lazy_load = TRUE)
26 | }
27 | \arguments{
28 | \item{id}{The ID of the dataset to be used}
29 |
30 | \item{filename}{Custom filename}
31 |
32 | \item{remote}{The remote to access, "" if working locally, TRUE if using default qsub_config remote}
33 |
34 | \item{dataset}{Dataset object to save}
35 |
36 | \item{lazy_load}{Whether or not to allow for lazy loading of large objects in the dataset}
37 | }
38 | \description{
39 | Helper functions for creating new datasets
40 | }
41 |
--------------------------------------------------------------------------------
/package/R/package.R:
--------------------------------------------------------------------------------
1 | #' Easy installing and loading of the dynverse
2 | #'
3 | #' The 'dynverse' is a set of packages with which to
4 | #' evaluate trajectory inference methods for dynamic processes.
5 | #'
6 | #' The dynbenchmark package contains the code to generate/preprocess datasets, evaluate TI methods on these datasets, and finally wrap all results together in a scientific paper.
7 | #' The code within this package can be run locally, but can also be run within a docker environment, for which the scripts are contained in the `scripts` folder.
8 | #'
9 | #' @import assertthat
10 | #' @importFrom tibble is_tibble as_tibble as_data_frame tibble data_frame enframe deframe lst tribble rownames_to_column column_to_rownames
11 | #' @import dplyr
12 | #' @import ggplot2
13 | #' @importFrom tidyr crossing gather spread gather
14 | #' @importFrom purrr %>% %||% walk set_names map map_dbl map_lgl map_chr map_df map2 map2_dbl map2_lgl map2_chr map2_df invoke
15 | #' @importFrom stringr str_replace_all str_replace str_detect str_sub str_subset str_glue
16 | #'
17 | #' @import dynutils
18 | #' @import dynplot
19 | #' @import dyneval
20 | #' @import dynmethods
21 | #' @import dynwrap
22 | #' @import dynparam
23 | #'
24 | #' @docType package
25 | #' @name dynbenchmark
26 | #'
27 | NULL
28 |
29 |
--------------------------------------------------------------------------------
/package/man/benchmark_aggregate.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_benchmark_aggregate.R
3 | \name{benchmark_aggregate}
4 | \alias{benchmark_aggregate}
5 | \alias{get_dataset_weighting}
6 | \title{Normalisation and aggregation function}
7 | \usage{
8 | benchmark_aggregate(data, metrics = c("correlation", "him",
9 | "featureimp_wcor", "F1_branches"), norm_fun = "normal",
10 | mean_fun = "geometric", mean_weights = set_names(rep(1,
11 | length(metrics)), metrics),
12 | dataset_source_weights = get_default_dataset_source_weights())
13 |
14 | get_dataset_weighting(datasets,
15 | dataset_source_weights = get_default_dataset_source_weights())
16 | }
17 | \arguments{
18 | \item{data}{Data as generated by the \code{06-benchmark/2-retrieve_results.R} script}
19 |
20 | \item{metrics}{Metrics to normalise}
21 |
22 | \item{norm_fun}{Which normalisation to use}
23 |
24 | \item{mean_fun}{Which mean function to use}
25 |
26 | \item{mean_weights}{Which metrics to use for the calculation of the mean, and which weights}
27 |
28 | \item{dataset_source_weights}{The weights of the dataset sources}
29 |
30 | \item{datasets}{Tibble with dataset_id, dataset_source and dataset_trajectory_type}
31 | }
32 | \description{
33 | Normalisation and aggregation function
34 | }
35 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02d-simulate_dyntoy_datasets.R:
--------------------------------------------------------------------------------
1 | #' [dyntoy](https://github.com/dynverse/dyntoy), simulations of toy data using random expression gradients in a reduced space
2 |
3 | library(dynbenchmark)
4 | library(dyntoy)
5 | library(tidyverse)
6 | library(qsub)
7 |
8 | experiment("01-datasets/02-synthetic")
9 |
10 | # remove all datasets
11 | # rm_remote(dataset_file(id = "synthetic/dyntoy", remote = TRUE), remote = TRUE, recursive = TRUE)
12 |
13 | # generate design
14 | set.seed(1)
15 | design <- crossing(
16 | topology_model = names(dyntoy::topology_models),
17 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
18 | ) %>%
19 | mutate(
20 | dataset_id = paste0("synthetic/dyntoy/", topology_model, "_", platform_ix),
21 | seed = sample(1:100000, n())
22 | ) %>%
23 | select(-platform_ix)
24 | write_rds(design, result_file("design_dyntoy.rds"))
25 |
26 | # simulate datasets
27 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "dyntoy", wait = F, stop_on_error = FALSE)
28 |
29 | handle <- qsub_pmap(
30 | design,
31 | simulate_dyntoy,
32 | qsub_config = qsub_config
33 | )
34 |
35 | write_rds(handle, "handle_dyntoy.rds")
36 |
37 | handle <- read_rds("handle_dyntoy.rds")
38 | qsub::qsub_retrieve(handle)
39 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02a-simulate_dyngen_datasets.R:
--------------------------------------------------------------------------------
1 | #' [dyngen](https://github.com/dynverse/dyngen), simulations of regulatory networks which will produce a particular trajectory
2 |
3 | library(qsub)
4 | library(dyngen)
5 | library(dynbenchmark)
6 | library(tidyverse)
7 |
8 | experiment("01-datasets/02-synthetic")
9 |
10 | # remove all datasets
11 | # rm_remote(dataset_file(id = "synthetic/dyngen", remote = TRUE), remote = TRUE, recursive = TRUE)
12 |
13 | # create design
14 | set.seed(1)
15 | design <- crossing(
16 | modulenet_name = dyngen::list_modulenets(),
17 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
18 | ) %>%
19 | mutate(
20 | dataset_id = paste0("synthetic/dyngen/", row_number()),
21 | seed = sample(1:100000, n())
22 | ) %>%
23 | select(-platform_ix)
24 | write_rds(design, result_file("design_dyngen.rds"))
25 |
26 | # simulate datasets
27 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "dyngen", wait = F, stop_on_error = FALSE)
28 |
29 | handle <- qsub_pmap(
30 | design,
31 | simulate_dyngen,
32 | use_cache = TRUE,
33 | qsub_config = qsub_config
34 | )
35 |
36 | write_rds(handle, "handle_dyngen.rds")
37 |
38 | ##
39 | handle <- read_rds("handle_dyngen.rds")
40 | qsub::qsub_retrieve(handle)
41 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-get_dynbenchmark_folder.R:
--------------------------------------------------------------------------------
1 | context("Testing get_dynbenchmark_folder")
2 |
3 | prev_path <- Sys.getenv("DYNBENCHMARK_PATH")
4 | prev_option <- getOption("dynbenchmark_path")
5 |
6 | # save old settings
7 | Sys.setenv(DYNBENCHMARK_PATH = "")
8 | options(dynbenchmark_path = NULL)
9 |
10 |
11 | test_that("get_dynbenchmark_folder works using variables", {
12 | tmp_path <- tempfile()
13 | dir.create(tmp_path)
14 |
15 | Sys.setenv(DYNBENCHMARK_PATH = tmp_path)
16 |
17 | file.create(paste0(tmp_path, "/dynbenchmark.Rproj"))
18 |
19 | expect_equal(get_dynbenchmark_folder(), tmp_path)
20 |
21 | Sys.setenv(DYNBENCHMARK_PATH = "")
22 |
23 | expect_error(get_dynbenchmark_folder(), regexp = "dynbenchmark folder could not be found")
24 | })
25 |
26 | test_that("get_dynbenchmark_folder works using options", {
27 | tmp_path <- tempfile()
28 | dir.create(tmp_path)
29 |
30 | options(dynbenchmark_path = tmp_path)
31 |
32 | file.create(paste0(tmp_path, "/dynbenchmark.Rproj"))
33 |
34 | expect_equal(get_dynbenchmark_folder(), tmp_path)
35 |
36 | options(dynbenchmark_path = NULL)
37 |
38 | expect_error(get_dynbenchmark_folder(), regexp = "dynbenchmark folder could not be found")
39 | })
40 |
41 | # revert to old settings
42 | Sys.setenv(DYNBENCHMARK_PATH = prev_path)
43 | options(dynbenchmark_path = prev_option)
44 |
--------------------------------------------------------------------------------
/package/man/pdf_manuscript.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{pdf_manuscript}
4 | \alias{pdf_manuscript}
5 | \title{The manuscript pdf document}
6 | \usage{
7 | pdf_manuscript(bibliography = raw_file("references.bib",
8 | "12-manuscript"), csl = raw_file("nature-biotechnology.csl",
9 | "12-manuscript"), render_changes = TRUE, toc = FALSE,
10 | linenumbers = FALSE, sty_header = NULL, sty_beforebody = NULL,
11 | sty_afterbody = NULL,
12 | reference_section_title = reference_section_title, ...)
13 | }
14 | \arguments{
15 | \item{bibliography}{Bibliography location}
16 |
17 | \item{csl}{Csl file location}
18 |
19 | \item{render_changes}{Whether to export a *_changes.pdf as well}
20 |
21 | \item{toc}{Whether to include a table of contents}
22 |
23 | \item{linenumbers}{Whether to add linenumbers to the left side}
24 |
25 | \item{sty_header}{Extra sty files to be included in the header of the document}
26 |
27 | \item{sty_beforebody}{Extra sty files to be included before the body of the document}
28 |
29 | \item{sty_afterbody}{Extra sty files to be included after the body of the document}
30 |
31 | \item{reference_section_title}{The title of the reference section}
32 |
33 | \item{...}{Parameters for rmarkdown::pdf_document}
34 | }
35 | \description{
36 | The manuscript pdf document
37 | }
38 |
--------------------------------------------------------------------------------
/manuscript/supplementary.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | title: "Supplementary material for: \nA comparison of single-cell trajectory inference methods"
3 | output:
4 | dynbenchmark::pdf_supplementary_note:
5 | render_change: FALSE
6 | ---
7 |
8 | ```{r setup, include=FALSE}
9 | library(dynbenchmark)
10 | library(tidyverse)
11 | ```
12 |
13 | ```{r}
14 | supplementary <- read_rds("supplementary.rds")
15 | sfigs <- supplementary$sfigs
16 | stables <- supplementary$stables
17 | refs <- supplementary$refs
18 | ```
19 |
20 | # Supplementary figures
21 | ```{r, results="asis"}
22 | sfigs %>%
23 | left_join(refs %>% select(ref_id, name), "ref_id") %>%
24 | arrange(name) %>%
25 | select(-name) %>%
26 | pmap(dynbenchmark:::plot_fig) %>% modify(paste0, "\n\n") %>% walk(cat)
27 | ```
28 |
29 | \clearpage
30 |
31 | # Supplementary tables
32 |
33 | ```{r, results="asis"}
34 | stables %>%
35 | left_join(refs %>% select(ref_id, name), "ref_id") %>%
36 | arrange(name) %>%
37 | select(-name) %>%
38 | pmap(dynbenchmark:::show_table) %>% modify(paste0, "\n\n") %>% walk(cat)
39 | ```
40 |
41 | \clearpage
42 |
43 | # Supplementary note 1
44 |
45 | ```{r}
46 | dynbenchmark::knit_nest("../results/02-metrics/")
47 | ```
48 | \clearpage
49 |
50 | # Supplementary note 2
51 |
52 | ```{r}
53 | dynbenchmark::knit_nest("../results/01-datasets/02-synthetic/")
54 | ```
55 |
56 | \clearpage
57 |
--------------------------------------------------------------------------------
/package/R/plotting_priors_mini.R:
--------------------------------------------------------------------------------
1 | #' Creates a prior mini svg
2 | #' @param method_priors Dataframe containing a prior_id and a prior_usage
3 | #' @param svg The base svg of the priors
4 | #' @export
5 | #'
6 | #' @importFrom xml2 read_xml xml_new_root xml_find_first xml_remove xml_add_child write_xml
7 | generate_prior_mini <- function(method_priors, svg = xml2::read_xml(result_file("priors.svg", experiment_id = "priors"))) {
8 | svg2 <- xml2::xml_new_root(svg, .copy = T)
9 |
10 | map2(method_priors$prior_id, method_priors$prior_usage, function(prior_id, usage) {
11 | color <- dynbenchmark::prior_usages$color[dynbenchmark::prior_usages$prior_usage == usage]
12 |
13 | layer <- svg2 %>% xml2::xml_find_first(stringr::str_glue(".//svg:g[@inkscape:label='{prior_id}']")) %>% xml2::xml_remove()
14 |
15 | if(!is.na(usage) && usage != "no") {
16 |
17 | print(color)
18 | # only add layer back if needed
19 | layer %>%
20 | as.character() %>%
21 | gsub("stroke:#fb1e00", stringr::str_glue("stroke:{color}"), .) %>%
22 | gsub("fill:#fb1e00", stringr::str_glue("fill:{color}"), .) %>%
23 | {suppressWarnings(xml2::read_xml(.))} %>%
24 | xml2::xml_add_child(svg2, .)
25 | }
26 | })
27 |
28 | svg2 %>% xml2::write_xml("~/test.svg")
29 |
30 | svg2 %>% as.character()# %>% charToRaw() %>% base64enc::base64encode()
31 | }
32 |
--------------------------------------------------------------------------------
/scripts/varia/generate_hypotheses_mindmap/hypotheses_script.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(googlesheets)
3 |
4 | # gs_auth()
5 |
6 | sheet <- gs_key("13jHFuMwie7oIxGcyLO95-onFri4M_GJjqgMEUnZliQI")
7 | hypotheses <- gs_read(sheet, ws = "Hypotheses") %>% mutate(
8 | parent_order = gsub("\\.[0-9]*$", "", order),
9 | parent_id = experiment_id[match(parent_order, order)],
10 | level = sapply(order, function(x) stringr::str_length(gsub("[^\\.]", "", x))),
11 | colstr = sapply(color, function(x) col2rgb(x)[,1] %>% paste(collapse = ","))
12 | )
13 |
14 | experiment_links_df <- hypotheses %>% select(parent_id, experiment_id) %>% filter(parent_id != experiment_id)
15 |
16 | textbox_tikz <- function(x) {
17 | ifelse(is.na(x), "", x %>%
18 | seqinr::stresc() %>%
19 | gsub("\n", "\\\\\\\\\n ", .) %>%
20 | gsub("@", "\\@", .))
21 | }
22 | hypotheses <- hypotheses %>% mutate(
23 | tikznode = paste0(
24 | " \\myhypothesis{", experiment_id, "}{",
25 | gsub("_", "\\\\_", experiment_id), "}{",
26 | colstr, "}{",
27 | textbox_tikz(statement), "}{",
28 | textbox_tikz(experiment_method), "}{",
29 | textbox_tikz(expected_results), "}{",
30 | textbox_tikz(motivation), "}"
31 | )
32 | )
33 |
34 | write_lines(
35 | paste(hypotheses$tikznode, collapse = "\n\n"),
36 | "analysis/analyses/generate_hypotheses_mindmap/hypotheses_tikz.tex"
37 | )
38 |
39 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/wip/pbmc-citeseq_stoeckius.R:
--------------------------------------------------------------------------------
1 | # extract the NK cells and ordering according to CD56??
2 | # cannot find the NK cells...
3 |
4 |
5 | rm(list=ls())
6 | library(dynbenchmark)
7 | library(tidyverse)
8 | library(GEOquery)
9 | library(Biobase)
10 | options('download.file.method.GEOquery'='curl')
11 |
12 | id <- "real/pbmc-citeseq_stoeckius"
13 | dataset_preprocessing(id)
14 |
15 | geo <- GEOquery::getGEO(GEO = "GSM2695382", destdir = dataset_source_file(""))
16 |
17 | rna_location <- download_dataset_source_file(
18 | "GSE100866_PBMC_vs_flow_10X-RNA_umi.csv.gz",
19 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE100866&format=file&file=GSE100866%5FPBMC%5Fvs%5Fflow%5F10X%2DRNA%5Fumi%2Ecsv%2Egz"
20 | )
21 |
22 | protein_location <- download_dataset_source_file(
23 | "GSE100866_PBMC_vs_flow_10X-ADT_umi.csv.gz",
24 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE100866&format=file&file=GSE100866%5FPBMC%5Fvs%5Fflow%5F10X%2DADT%5Fumi%2Ecsv%2Egz"
25 | )
26 |
27 | counts_rna <- read_csv(rna_location) %>% as.data.frame %>% column_to_rownames("X1") %>% as.matrix() %>% t
28 | colnames(counts_rna) <- gsub("HUMAN_(.*)", "\\1", colnames(counts_rna))
29 | counts_protein <- read_csv(protein_location) %>% as.data.frame %>% column_to_rownames("X1") %>% as.matrix() %>% t
30 |
31 |
32 |
33 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2695382
34 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_distal-lung-epithelium_treutlein.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/silver/distal-lung-epithelium_treutlein")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "nature13173-s4.txt",
8 | "http://www.nature.com/nature/journal/v509/n7500/extref/nature13173-s4.txt"
9 | )
10 |
11 | df <- read_tsv(txt_location, col_types = cols(cell_name = "c", time_point = "c", sample = "c", putative_cell_type = "c", .default = "d"))
12 | expression <- df[, -c(1:4)] %>% as.matrix() %>% magrittr::set_rownames(df$cell_name)
13 | cell_info <- df[, c(1:4)] %>% as.data.frame() %>% magrittr::set_rownames(df$cell_name) %>%
14 | rename(
15 | cell_id = cell_name,
16 | milestone_id = putative_cell_type
17 | ) %>% filter(sample == "singleCell")
18 | expression <- expression[cell_info$cell_id, ]
19 |
20 | milestone_network = tribble(
21 | ~from, ~to,
22 | "BP", "AT1",
23 | "BP", "AT2"
24 | ) %>% mutate(length = 1, directed = TRUE)
25 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
26 |
27 | counts <- 2^expression-1
28 |
29 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
30 | counts <- counts[cell_info$cell_id, ]
31 |
32 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
33 |
34 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
35 |
--------------------------------------------------------------------------------
/package/data-raw/method_status_colours.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(extrafont)
3 |
4 | dynbenchmark:::install_fontawesome()
5 |
6 | extrafont::loadfonts()
7 |
8 | method_statuses <- tribble(
9 | ~method_status, ~colour, ~fa, ~icon_colour,
10 | "execution_error", "#9d0549", "\uf00d", "white",
11 | "method_error", "#d51d14", "\uf057", "white",
12 | "memory_limit", "#f46b21", "\uf538", "black",
13 | "time_limit", "#f1be21", "\uf017", "black",
14 | "missing_prior", "#fff600", "\uf0cb", "black",
15 | "low_correlation", "#b4ef43", "\uf537", "black",
16 | "no_error", "#2ECC40", "\uf058", "black"
17 | ) %>%
18 | mutate(method_status = fct_inorder(method_status))
19 |
20 | method_status_colours <-
21 | method_statuses %>% deframe()
22 |
23 | devtools::use_data(method_statuses, method_status_colours, pkg = "package", overwrite = TRUE)
24 |
25 | ggplot(method_statuses, aes(y = fct_rev(method_status))) +
26 | geom_tile(aes(x = 1, fill = method_status), size = 1, height = .9, width = .9) +
27 | geom_text(aes(x = 1, label = fa, colour = method_status), family = "Font Awesome 5 Free", size = 8) +
28 | geom_text(aes(x = 0, label = method_status), hjust = 1, size = 8) +
29 | scale_fill_manual(values = method_status_colours) +
30 | scale_colour_manual(values = method_statuses %>% select(method_status, icon_colour) %>% deframe()) +
31 | expand_limits(x = -2) +
32 | coord_equal() +
33 | cowplot::theme_nothing()
34 |
--------------------------------------------------------------------------------
/package/man/qsub_pmap.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/qsub.R
3 | \name{qsub_pmap}
4 | \alias{qsub_pmap}
5 | \title{pmap, but with qsub ^_^}
6 | \usage{
7 | qsub_pmap(.x, .f, ...)
8 | }
9 | \arguments{
10 | \item{.x}{Vectors of the same length. A vector of length 1 will
11 | be recycled.}
12 |
13 | \item{.f}{A function, formula, or vector (not necessarily atomic).
14 |
15 | If a \strong{function}, it is used as is.
16 |
17 | If a \strong{formula}, e.g. \code{~ .x + 2}, it is converted to a function. There
18 | are three ways to refer to the arguments:
19 | \itemize{
20 | \item For a single argument function, use \code{.}
21 | \item For a two argument function, use \code{.x} and \code{.y}
22 | \item For more arguments, use \code{..1}, \code{..2}, \code{..3} etc
23 | }
24 |
25 | This syntax allows you to create very compact anonymous functions.
26 |
27 | If \strong{character vector}, \strong{numeric vector}, or \strong{list}, it is
28 | converted to an extractor function. Character vectors index by
29 | name and numeric vectors index by position; use a list to index
30 | by position and name at different levels. If a component is not
31 | present, the value of \code{.default} will be returned.}
32 |
33 | \item{...}{Params given to \code{\link[qsub:qsub_lapply]{qsub::qsub_lapply()}}}
34 | }
35 | \description{
36 | pmap, but with qsub ^_^
37 | }
38 | \seealso{
39 | qsub::qsub_lapply
40 | }
41 |
--------------------------------------------------------------------------------
/scripts/03-methods/01-gather_methods_information/03-add_quality_control.R:
--------------------------------------------------------------------------------
1 | #' Add QC scores to methods and tools tibble
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(googlesheets)
6 |
7 | experiment("03-methods")
8 |
9 | tools <- read_rds(result_file("tools.rds"))
10 | methods <- read_rds(result_file("methods.rds"))
11 |
12 | # combine with qc scores
13 | tool_qc_scores <- readRDS(result_file("tool_qc_scores.rds"))
14 |
15 | # merge qc scores with tools tibble
16 | tools <- tools %>%
17 | select(-tidyselect::matches("qc_score")) %>%
18 | left_join(tool_qc_scores, by = "tool_id")
19 |
20 | methods <- methods %>%
21 | select(-tidyselect::matches("qc_score")) %>%
22 | left_join(tool_qc_scores, by = c("method_tool_id" = "tool_id"))
23 |
24 | # filter evaluated
25 | methods_evaluated <- methods %>%
26 | filter(method_evaluated)
27 |
28 | tools_evaluated <- tools %>%
29 | filter(method_evaluated)
30 |
31 | # check that all non-control evaluated methods & tools have a QC!
32 | methods_evaluated_nonqc <- methods_evaluated %>% filter(method_source == "tool" & is.na(qc_score))
33 | if (nrow(methods_evaluated_nonqc) != 0) {
34 | stop("Methods ", methods_evaluated_nonqc$id %>% glue::glue_collapse(", "), " dont have a QC score")
35 | }
36 |
37 | # save
38 | write_rds(tools, result_file("tools.rds"))
39 | write_rds(tools_evaluated, result_file("tools_evaluated.rds"))
40 | write_rds(methods, result_file("methods.rds"))
41 | write_rds(methods_evaluated, result_file("methods_evaluated.rds"))
42 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02c-simulate_splatter_datasets.R:
--------------------------------------------------------------------------------
1 | #' [Splatter](https://github.com/Oshlack/splatter), simulations of non-linear paths between different states
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(qsub)
6 |
7 | experiment("01-datasets/02-synthetic")
8 |
9 | # remove all datasets
10 | # rm_remote(dataset_file(id = "synthetic/splatter", remote = TRUE), remote = TRUE, recursive = TRUE)
11 |
12 | # generate design
13 | set.seed(1)
14 | design <- crossing(
15 | topology_model = c("linear", "bifurcating", "multifurcating", "binary_tree", "tree"),
16 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
17 | ) %>%
18 | mutate(
19 | path.skew = runif(n(), 0, 1),
20 | path.nonlinearProb = runif(n(), 0, 1),
21 | path.sigmaFac = runif(n(), 0, 1),
22 | bcv.common.factor = runif(n(), 10, 200),
23 | dataset_id = paste0("synthetic/splatter/", topology_model, "_", platform_ix),
24 | seed = sample(1:100000, n())
25 | ) %>%
26 | select(-platform_ix)
27 | write_rds(design, result_file("design_splatter.rds"))
28 |
29 | # simulate datasets
30 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "splatter", wait = F, stop_on_error = FALSE)
31 |
32 | handle <- qsub_pmap(
33 | design,
34 | simulate_splatter,
35 | qsub_config = qsub_config
36 | )
37 |
38 | write_rds(handle, "handle_splatter.rds")
39 |
40 | ##
41 | handle <- read_rds("handle_splatter.rds")
42 | qsub::qsub_retrieve(handle)
43 |
--------------------------------------------------------------------------------
/package/man/make_obj_fun.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_submit.R
3 | \name{make_obj_fun}
4 | \alias{make_obj_fun}
5 | \title{Used for wrapping an evaluation function around a TI method}
6 | \usage{
7 | make_obj_fun(method, metrics, give_priors, seed, noisy = FALSE,
8 | verbose = FALSE)
9 | }
10 | \arguments{
11 | \item{method}{One or more methods. Must be one of:
12 | \itemize{
13 | \item{an object or list of ti_... objects (eg. \code{dynmethods::ti_comp1()}),}
14 | \item{a character vector containing the names of methods to execute (e.g. \code{"scorpius"}),}
15 | \item{a character vector containing dockerhub repositories (e.g. \code{dynverse/paga}), or}
16 | \item{a dynguidelines data frame.}
17 | }}
18 |
19 | \item{metrics}{Which metrics to evaluate. Check \code{dyneval::metrics} for a list of possible metrics.
20 | Passing a custom metric function with format \code{function(dataset, model) { 1 }} is also supported. The name of this function within the list will be used as the name of the metric.}
21 |
22 | \item{give_priors}{All the priors a method is allowed to receive.
23 | Must be a subset of all available priors (\link[dynwrap:priors]{dynwrap::priors}).}
24 |
25 | \item{seed}{A seed to be passed to the TI method.}
26 |
27 | \item{noisy}{Whether or not the metric is noisy or not}
28 |
29 | \item{verbose}{Whether or not to print extra information output}
30 | }
31 | \description{
32 | Used for wrapping an evaluation function around a TI method
33 | }
34 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_mESC-differentation_hayashi.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 | library(GEOquery)
4 | library(Biobase)
5 | options('download.file.method.GEOquery'='curl')
6 |
7 | id <- "real/gold/mESC-differentiation_hayashi"
8 | dataset_preprocessing(id)
9 |
10 | counts_location <- download_dataset_source_file(
11 | "GSE98664_tpm_sailfish_mergedGTF_RamDA_mESC_differentiation_time_course.txt.gz",
12 | "ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE98nnn/GSE98664/suppl/GSE98664_tpm_sailfish_mergedGTF_RamDA_mESC_differentiation_time_course.txt.gz"
13 | )
14 |
15 | counts_all <- read_tsv(counts_location) %>% as.data.frame %>% column_to_rownames("transcript_id") %>% as.matrix() %>% t %>% round()
16 |
17 | cell_info_all <- tibble(
18 | cell_id = rownames(counts_all),
19 | milestone_id = gsub("RamDA_mESC_(.*)_.*", "\\1", cell_id)
20 | )
21 |
22 | milestone_network <- tribble(
23 | ~from, ~to, ~length,
24 | "00h", "12h", 1,
25 | "12h", "24h", 1,
26 | "24h", "48h", 2,
27 | "48h", "72h", 2
28 | ) %>% mutate(directed = TRUE)
29 |
30 |
31 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
32 |
33 | cell_info <- slice(cell_info_all, match(rownames(counts_all), cell_id))
34 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
35 | counts <- counts_all[cell_info$cell_id, ]
36 |
37 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
38 |
39 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
40 |
--------------------------------------------------------------------------------
/scripts/varia/generate_hypotheses_mindmap/mindmap.tex:
--------------------------------------------------------------------------------
1 | \documentclass[a4paper,twocolumn]{minimal}
2 |
3 | \usepackage{bm}
4 | \usepackage[math]{iwona}
5 | \usepackage[T1]{fontenc}
6 | \usepackage[utf8]{inputenc}
7 |
8 | \usepackage{tikz}
9 | \usetikzlibrary{shapes,decorations}
10 | \usepackage{amsmath,amssymb}
11 |
12 |
13 | \usepackage[a4paper,left=1cm,right=1cm,top=1cm,bottom=1cm]{geometry}
14 |
15 | \usetikzlibrary{shapes.multipart}
16 |
17 | \usetikzlibrary{trees}
18 |
19 | \newcommand{\mysection}[3]{%
20 | \nodepart{#1}
21 | \begin{minipage}{.9\linewidth}
22 | \textbf{#2}\\
23 | #3
24 | \end{minipage}
25 | }
26 | \newcommand{\myhypothesis}[7]{%
27 | \definecolor{my_#1}{RGB}{#3}
28 | \begin{tikzpicture}
29 | \node [mybox,draw=my_#1,fill=my_#1!10!white] (#1) {
30 | \mysection{one}{Statement}{#4}
31 | \mysection{two}{Experiment}{#5}
32 | \mysection{three}{Expected results}{#6}
33 | \mysection{four}{Motivation}{#7}
34 | };
35 | \node [mytitle,fill=my_#1] at (#1.north){\textbf{\color{white}#2}};
36 | \end{tikzpicture}\\
37 | }
38 |
39 | \tikzstyle{mybox} = [
40 | fill=black!03, very thick,
41 | rectangle, rounded corners,
42 | inner sep=5pt, inner ysep=5pt,
43 | rectangle split, rectangle split parts=4,
44 | font=\fontsize{10}{11.4}\selectfont]
45 | \tikzstyle{mytitle} = [
46 | anchor=center, fill=black!05,
47 | very thick, rectangle,
48 | yshift=.1cm,
49 | font=\fontsize{14}{16.4}\selectfont]
50 |
51 | \begin{document}
52 |
53 | \input{hypotheses_tikz.tex}
54 |
55 | \end{document}
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02b-simulate_prosstt_datasets.R:
--------------------------------------------------------------------------------
1 | #' [PROSSTT](https://github.com/soedinglab/prosstt), expression is sampled from a linear model which depends on pseudotime
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(qsub)
6 |
7 | experiment("01-datasets/02-synthetic")
8 |
9 | # generate design of models, platforms and (randomised) splatter parameters
10 | design <- crossing(
11 | topology_model = c("linear", "bifurcating", "multifurcating", "binary_tree", "tree"),
12 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
13 | ) %>%
14 | mutate(
15 | a = runif(n(), 0.01, 0.1),
16 | intra_branch_tol = runif(n(), 0, 0.9),
17 | inter_branch_tol = runif(n(), 0, 0.9),
18 | alpha = exp(rnorm(n(), log(0.3), log(1.5))),
19 | beta = exp(rnorm(n(), log(2), log(1.5))) + 1
20 | ) %>%
21 | mutate(
22 | dataset_id = paste0("synthetic/prosstt/", topology_model, "_", platform_ix),
23 | seed = sample(1:100000, n())
24 | ) %>%
25 | select(-platform_ix)
26 | write_rds(design, result_file("design_prosstt.rds"))
27 |
28 | # simulate datasets
29 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "prosstt", wait = F, execute_before = "module load python/x86_64/3.6.5", stop_on_error = FALSE)
30 |
31 | handle <- qsub_pmap(
32 | design,
33 | simulate_prosstt,
34 | qsub_config = qsub_config
35 | )
36 |
37 | write_rds(handle, "handle_prosstt.rds")
38 |
39 | ##
40 | handle <- read_rds("handle_prosstt.rds")
41 | qsub::qsub_retrieve(handle)
42 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_fibroblast-reprogramming_treutlein.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/silver/fibroblast-reprogramming_treutlein")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "GSE67310_iN_data_log2FPKM_annotated.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE67310&format=file&file=GSE67310%5FiN%5Fdata%5Flog2FPKM%5Fannotated%2Etxt%2Egz"
9 | )
10 |
11 | df <- read_tsv(txt_location, col_types = cols(cell_name = "c", assignment = "c", experiment = "c", time_point = "c", .default = "d"))
12 | expression <- df[, -c(1:5)] %>% as.matrix() %>% magrittr::set_rownames(df$cell_name)
13 | cell_info <- df[, c(1:5)] %>% as.data.frame() %>% magrittr::set_rownames(df$cell_name) %>%
14 | rename(
15 | cell_id = cell_name,
16 | milestone_id = assignment
17 | )
18 |
19 | milestone_network = tribble(
20 | ~from, ~to,
21 | "MEF", "d2_intermediate",
22 | "d2_intermediate", "d2_induced",
23 | "d2_induced", "d5_intermediate",
24 | "d5_intermediate", "d5_earlyiN",
25 | "d5_earlyiN", "Neuron",
26 | "d5_earlyiN", "Myocyte"
27 | ) %>% mutate(length = 1, directed = TRUE)
28 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
29 |
30 | counts <- floor(2^expression-1)
31 |
32 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
33 | counts <- counts[cell_info$cell_id, ]
34 |
35 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
36 |
37 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
38 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/01-download_from_sources.R:
--------------------------------------------------------------------------------
1 | #' Downloading the real datasets from their sources (eg. GEO), and constructing the gold standard model, using the helpers in [helpers-download_from_sources](helpers-download_from_sources)
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | experiment("01-datasets/real/run_all_datasets")
7 |
8 | dataset_scripts <- list.files(path = "scripts/01-datasets/01-real/helpers-download_from_sources/", pattern = "^dataset_.*\\.R", full.names = TRUE)
9 | # dataset_scripts <- dataset_scripts[str_detect(dataset_scripts, "mouse-cell-atlas") | str_detect(dataset_scripts, "plass")]
10 |
11 | script_contents <- map_chr(dataset_scripts, read_file)
12 |
13 | qsub_config <- qsub::override_qsub_config(
14 | name = "dynreal",
15 | memory = "30G",
16 | max_wall_time = "02:00:00",
17 | wait = FALSE,
18 | stop_on_error = FALSE
19 | )
20 |
21 | # Make sure all packages are installed on the cluster; i.e. GEOquery, MultiAssayExperiment, tidyverse, and dynbenchmark.
22 | handle <- qsub::qsub_lapply(
23 | script_contents,
24 | function(script_content) {
25 | dataset_script <- tempfile()
26 | readr::write_file(script_content, dataset_script)
27 |
28 | source(dataset_script)
29 |
30 | TRUE
31 | },
32 | qsub_environment = new.env(),
33 | qsub_config = qsub_config
34 | )
35 |
36 | write_rds(handle, "handle.rds")
37 |
38 | ##
39 |
40 | handle <- read_rds("handle.rds")
41 | results <- qsub::qsub_retrieve(handle, wait = TRUE)
42 |
43 | names(results) <- dataset_scripts
44 |
45 | results <- results %>%
46 | keep(is.na) %>%
47 | enframe("dataset_script", "result")
48 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_cellbench_luyitian.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/gold/cellbench")
5 |
6 | rda_location <- download_dataset_source_file(
7 | "9cellmix_qc.RData",
8 | "https://raw.githubusercontent.com/LuyiTian/CellBench_data/master/data/9cellmix_qc.RData"
9 | )
10 |
11 | settings <- list(
12 | list(
13 | object = sce_SC1_qc,
14 | id = "real/gold/cellbench-SC1_luyitian"
15 | ),
16 | list(
17 | object = sce_SC2_qc,
18 | id = "real/gold/cellbench-SC2_luyitian"
19 | ),
20 | list(
21 | object = sce_SC3_qc,
22 | id = "real/gold/cellbench-SC3_luyitian"
23 | ),
24 | list(
25 | object = sce_SC4_qc,
26 | id = "real/gold/cellbench-SC4_luyitian"
27 | )
28 | )
29 |
30 | for (setting in settings) {
31 | cell_info <- colData(setting$object) %>%
32 | as.data.frame() %>%
33 | rownames_to_column("cell_id") %>%
34 | mutate_at(vars(H1975, H2228, HCC827), ~.>0) %>%
35 | filter(traj == "YES") # why isn't this a logical?
36 | cell_info$group_id <- cell_info[, c("H1975", "H2228", "HCC827")] %>% apply(1, function(x) glue::glue_collapse(names(x)[x], ","))
37 |
38 | grouping <- cell_info %>% select(cell_id, group_id) %>% deframe()
39 |
40 | milestone_network <- tribble(
41 | ~from, ~to,
42 | "H1975", "H1975,H2228,HCC827",
43 | "HCC827", "H1975,H2228,HCC827",
44 | "H2228", "H1975,H2228,HCC827"
45 | ) %>% mutate(length = 1, directed = FALSE)
46 |
47 | counts <- t(counts(setting$object))[cell_info$cell_id, ]
48 |
49 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts), setting$id)
50 | }
51 |
--------------------------------------------------------------------------------
/raw/gource/deprecated_repositories/mlr_ti.txt:
--------------------------------------------------------------------------------
1 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/R/ResampleDesc.R
2 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/R/utils_imbalancy.R
3 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/man/makeResampleDesc.Rd
4 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/NAMESPACE
5 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/Measure.R
6 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/Prediction.R
7 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/RLearner.R
8 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/TITask.R
9 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/helpers.R
10 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/measures.R
11 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/setPredictType.R
12 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/zzz.R
13 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/RLearner.Rd
14 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/Task.Rd
15 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/ti_geodesic_dist.Rd
16 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/mlr.Rproj
17 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/NAMESPACE
18 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/Prediction.R
19 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/TITask.R
20 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/measures.R
21 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/zzz.R
22 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/man/Task.Rd
23 | 1494492034|Robrecht Cannoodt|A|/libraries/mlr_ti/mlr_ti.Rproj
24 | 1494493345|Robrecht Cannoodt|M|/libraries/mlr_ti/R/measures.R
25 | 1494494000|Robrecht Cannoodt|D|/libraries/mlr_ti/
26 |
--------------------------------------------------------------------------------
/scripts/11-example_predictions/02-funky.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("11-example_predictions")
5 |
6 | source(scripts_file("helper-funky.R"))
7 |
8 | folder <- "../../dyndocs/funky_cover/data/embeddings"
9 | dir.create(folder, showWarnings = FALSE)
10 |
11 | dataset_ids <- c(
12 | "synthetic/dyntoy/bifurcating_3",
13 | "real/gold/developing-dendritic-cells_schlitzer",
14 | "real/silver/fibroblast-reprogramming_treutlein",
15 | "synthetic/dyntoy/disconnected_1",
16 | "synthetic/dyngen/72",
17 | "synthetic/dyntoy/linear_1",
18 | "synthetic/dyntoy/cyclic_7",
19 | "synthetic/dyntoy/diverging_with_loops_5",
20 | "synthetic/dyntoy/tree_5"
21 | )
22 |
23 | walk(seq_along(dataset_ids), function(dataset_ix) {
24 | dataset_id <- dataset_ids[dataset_ix]
25 |
26 | dataset <- load_dataset(dataset_id)
27 | plot_dimred(dataset)
28 |
29 | dimred <- dyndimred::dimred_landmark_mds(dataset$expression(), ndim = 3)
30 |
31 | cell_positions <- dimred %>%
32 | as.data.frame() %>%
33 | rownames_to_column("cell_id") %>%
34 | mutate_at(vars(comp_1, comp_2, comp_3), dynutils::scale_minmax)
35 | projection <- project_waypoints_multidim(dataset, cell_positions)
36 |
37 |
38 | cell_positions %>%
39 | sample_n(50) %>%
40 | write_csv(fs::path(folder, paste0("embedding_", dataset_ix, ".csv")))
41 |
42 | projection$edges$group = projection$edges %>% group_by(from_milestone_id, to_milestone_id) %>% group_indices()
43 | projection$edges %>%
44 | select(comp_1 = comp_1_from, comp_2 = comp_2_from, comp_3 = comp_3_from, group) %>%
45 | write_csv(fs::path(folder, paste0("trajectory_", dataset_ix, ".csv")))
46 | })
47 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Benchmarking of TI methods on real and synthetic data
3 |
4 | | \# | script/folder | description |
5 | | :- | :---------------------------------------------------------- | :----------------------------------------------------------------------- |
6 | | 1 | [📄`submit_jobs.R`](1-submit_jobs.R) | Submit the jobs to the gridengine cluster |
7 | | 2 | [📄`retrieve_results.R`](2-retrieve_results.R) | Retrieve the results from the gridengine cluster and aggregate the data |
8 | | 3 | [📄`diagnostic_figures.R`](3-diagnostic_figures.R) | Create some diagnostic figures to get an overview of the results |
9 | | 3a | [📄`overall_comparison.R`](3a-overall_comparison.R) | Overall comparison |
10 | | 3b | [📄`time_mem_predictions.R`](3b-time_mem_predictions.R) | Comparison between predicted versus actual running time and memory usage |
11 | | 3c | [📄`normalisation.R`](3c-normalisation.R) | Compare the effect of normalisation on the results |
12 | | 3d | [📄`compare_sources.R`](3d-compare_sources.R) | Compare the different dataset sources |
13 | | | [📄`inspect_identity_timings.R`](inspect_identity_timings.R) | Check different benchmark component timings |
14 |
15 | The results of this experiment are available
16 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/06-benchmark).
17 |
--------------------------------------------------------------------------------
/package/data-raw/labels.R:
--------------------------------------------------------------------------------
1 | common_labels <- tribble(
2 | ~id, ~long, ~short,
3 | "directed_acyclic_graph", "Directed acyclic graph", "DAG",
4 | "disconnected_directed_graph", "Disconnected directed graph", "DDG",
5 | "n_genes", "Number of genes", "# genes",
6 | "n_cells", "Number of cells", "# cells",
7 | "ngenes", "Number of genes", "# genes",
8 | "ncells", "Number of cells", "# cells",
9 | "n_methods", "Number of methods", "# methods",
10 | "n_tools", "Number of tools", "# tools",
11 | "silver", "Silver standard", "Silver",
12 | "gold", "Gold standard", "Gold",
13 | "gse", "Source(s)", "Source(s)",
14 | "gs", "Gold standard", "Gold standard",
15 | "rf_mse", "Random Forest MSE", "RF MSE",
16 | "maximal_trajectory_type", "Most complex trajectory type", "Trajectory type",
17 | "qc_score", "QC score", "QC score",
18 | "component_id", "Component", "Component",
19 | "p_value", "p-value", "p-value",
20 | "q_value", "Adjusted p-value", "p-value (adj.)",
21 | TRUE, "Yes", "Yes",
22 | FALSE, "No", "No",
23 | "trajectory_type_gold", "Gold standard trajectory type", "Gold trajectory type",
24 | "trajectory_type_predicted", "Predicted trajectory type", "Predicted trajectory type",
25 | "harm_mean", "Harmonic mean", "Harmonic mean",
26 | "dataset_source", "Dataset source", "Dataset source",
27 | "source", "Dataset source", "Dataset source",
28 | "qc", "Quality Control", "QC",
29 | "graph", "Connected graph", "Connected graph",
30 | "offtheshelf", "Off-the-shelf", "Off-the-shelf"
31 | )
32 |
33 |
34 | labels <- bind_rows(
35 | common_labels,
36 | prior_types %>% select(id = prior_id, long = prior_name, short = prior_name)
37 | )
38 |
39 | usethis::proj_set("package")
40 | usethis::use_data(labels, overwrite = TRUE)
41 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_cortical-interneuron-differentiation_frazer.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 | library(GEOquery)
4 | options('download.file.method.GEOquery'='curl')
5 |
6 | dataset_preprocessing("real/silver/cortical-interneuron-differentiation_frazer")
7 |
8 | txt_location <- download_dataset_source_file(
9 | "GSE90860_SupplementaryData1_revised_julien.tsv.gz",
10 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE90860&format=file&file=GSE90860%5FSupplementaryData1%5Frevised%5Fjulien%2Etsv%2Egz"
11 | )
12 |
13 | counts_df <- read_tsv(txt_location)
14 | counts <- counts_df %>% select(-X1) %>% as.matrix %>% t %>% magrittr::set_colnames(counts_df$X1)
15 |
16 | geo <- GEOquery::getGEO(GEO = "GSE90860", destdir = dataset_source_file(""))
17 | cell_info <- geo[[1]] %>%
18 | Biobase::phenoData() %>%
19 | as("data.frame") %>%
20 | mutate(
21 | cell_id = rownames(counts),
22 | milestone_id = paste0(gsub("age: (.*)", "\\1", characteristics_ch1), "#", gsub("assigned_subgroup: (.*)", "\\1", characteristics_ch1.1))
23 | ) %>%
24 | select(cell_id, milestone_id) %>%
25 | mutate_all(funs(as.character))
26 |
27 | milestone_network <- tribble(
28 | ~from, ~to, ~length,
29 | "E18#0", "P2#1", 4,
30 | "E18#0", "P2#2", 4,
31 | "E18#0", "P2#3", 4,
32 | "P2#1", "P5#1", 3,
33 | "P2#2", "P5#2", 3,
34 | "P2#3", "P5#3", 3
35 | ) %>% mutate(directed = TRUE)
36 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
37 |
38 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
39 | counts <- counts[cell_info$cell_id, ]
40 |
41 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
42 |
43 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
44 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/3-diagnostic_figures.R:
--------------------------------------------------------------------------------
1 | #' Create some diagnostic figures to get an overview of the results
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(dynplot)
6 |
7 | experiment("06-benchmark")
8 |
9 |
10 | ############################################################
11 | ############### PART THREE: GENERATE FIGURES ###############
12 | ############################################################
13 |
14 | num_replicates <- 1
15 |
16 | list2env(read_rds(result_file("benchmark_results_input.rds")), environment())
17 | list2env(read_rds(result_file("benchmark_results_normalised.rds")), environment())
18 |
19 | metrics_info <- dyneval::metrics %>%
20 | slice(match(metrics, metric_id)) %>%
21 | add_row(metric_id = "overall", plotmath = "overall", latex = "\\textrm{overall}", html = "overall", long_name = "Overall score", category = "average", type = "overall", perfect = 1, worst = 0)
22 |
23 | # get ordering of methods
24 | method_ord <-
25 | data_aggregations %>%
26 | filter(dataset_source == "mean", dataset_trajectory_type == "overall") %>%
27 | arrange(desc(overall)) %>%
28 | select(method_id, method_name) %>%
29 | mutate_all(factor)
30 |
31 | # create method_id_f factor in all data structures
32 | data_aggregations <- data_aggregations %>% mutate(method_id = factor(method_id, levels = method_ord$method_id), method_name = factor(method_name, levels = method_ord$method_name))
33 | data <- data %>% mutate(method_id = factor(method_id, levels = method_ord$method_id), method_name = factor(method_name, levels = method_ord$method_name))
34 |
35 | # execute plotting scripts
36 | source(scripts_file("3a-overall_comparison.R"))
37 | source(scripts_file("3b-time_mem_predictions.R"))
38 | source(scripts_file("3c-normalisation.R"))
39 | source(scripts_file("3d-compare_sources.R"))
40 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_developing-dendritic-cells_schlitzer.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 | library(GEOquery)
4 | options('download.file.method.GEOquery'='curl')
5 |
6 | dataset_preprocessing("real/gold/developing-dendritic-cells_schlitzer")
7 |
8 | # download and untar files
9 | file <- download_dataset_source_file("GSE60781_RAW.tar", "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE60781&format=file")
10 |
11 | untar(file, exdir = dataset_source_file(""))
12 |
13 | # read counts
14 | counts <- map_df(list.files(dataset_source_file(""), ".*read[cC]ount"), function(filename) {
15 | read_tsv(dataset_source_file(filename), col_names = c("gene", "count"), col_types = cols(gene = "c", count = "i")) %>%
16 | mutate(sample = gsub("_.*", "", filename))
17 | }) %>%
18 | spread(gene, count) %>%
19 | as.data.frame() %>%
20 | magrittr::set_rownames(NULL) %>%
21 | column_to_rownames("sample") %>%
22 | as.matrix
23 |
24 | counts <- counts[, !(colnames(counts) %in% c("alignment_not_unique", "no_feature", "ambiguous"))]
25 |
26 | # download cell info
27 | geo <- GEOquery::getGEO("GSE60781", destdir = dataset_source_file(""))
28 | cell_info <- geo[[1]] %>%
29 | Biobase::phenoData() %>%
30 | as("data.frame") %>%
31 | mutate(milestone_id = gsub("_[0-9]*", "", title)) %>%
32 | select(cell_id = geo_accession, milestone_id) %>%
33 | slice(match(rownames(counts), cell_id))
34 |
35 | milestone_network = tribble(
36 | ~from, ~to,
37 | "MDP", "CDP",
38 | "CDP", "PreDC"
39 | ) %>% mutate(length = 1, directed = TRUE)
40 |
41 | milestone_ids <- c("MDP", "CDP", "PreDC")
42 |
43 | cell_ids <- cell_info$cell_id
44 |
45 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
46 |
47 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
48 |
--------------------------------------------------------------------------------
/scripts/varia/gource/2_gource_gits.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("gource")
5 |
6 | gitlinks_folder <- derived_file("git_links/")
7 | output_folder <- derived_file("git_output/")
8 |
9 | dir.create(output_folder)
10 |
11 | git_df <-
12 | readr::read_lines("../.gitmodules") %>%
13 | keep(~ grepl("submodule", .)) %>%
14 | gsub(".*\"(.*)\".*", "\\1", .) %>%
15 | data_frame(path = ., prefix = paste0("/", .), filename = str_replace(., "/", "_"), gitmodule = TRUE) %>%
16 | add_row(path = "..", prefix = "", filename = "dynverse", gitmodule = FALSE) %>%
17 | add_row(path = "../../dyndocs", prefix = "/dyndocs", filename = "dyndocs", gitmodule = FALSE) %>%
18 | add_row(path = "../../projects/qsub/", prefix = "/libraries/qsub", filename = "libraries_qsub", gitmodule = FALSE)
19 |
20 | unlink(gitlinks_folder, recursive = T)
21 | for (i in seq_len(nrow(git_df))) {
22 | path <- git_df$path[[i]]
23 | prefix <- git_df$prefix[[i]]
24 | filename <- git_df$filename[[i]]
25 | gitmodule <- git_df$gitmodule[[i]]
26 |
27 | gource_output <- paste0(
28 | output_folder, "/",
29 | filename,
30 | ".txt"
31 | )
32 |
33 | if (gitmodule) {
34 | dir.create(paste0(gitlinks_folder, path), recursive = T, showWarnings = F)
35 | file.symlink(
36 | normalizePath(paste0("../.git/modules/", path)),
37 | paste0(gitlinks_folder, path, "/.git")
38 | )
39 | git_location <- paste0(gitlinks_folder, path)
40 | } else {
41 | git_location <- path
42 | }
43 | cmd <- paste0("gource --output-custom-log ", gource_output, " ", git_location)
44 | system(cmd)
45 |
46 | read_lines(gource_output) %>%
47 | str_replace("\\|/", paste0("|", prefix, "/")) %>%
48 | str_replace("\\.REMOVED\\.git-id$", "") %>%
49 | write_lines(gource_output)
50 | }
51 | unlink(gitlinks_folder, recursive = T)
52 |
--------------------------------------------------------------------------------
/scripts/02-metrics/02-metric_conformity/01-generate_reference_datasets.R:
--------------------------------------------------------------------------------
1 | #' Generation of toy datasets used as reference to assess metric conformity
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(dyntoy)
6 |
7 | experiment("02-metrics/02-metric_conformity")
8 |
9 | # construct all different combinations of topologies, # cells, ...
10 | dataset_design <-
11 | crossing(
12 | bind_rows(
13 | dynbenchmark:::topologies_with_same_n_milestones %>% enframe("topology_id", "topology_model") %>% mutate(allow_tented_progressions = FALSE),
14 | tribble(
15 | ~topology_id, ~topology_model, ~allow_tented_progressions,
16 | "bifurcation_simple", dyntoy::model_bifurcating(), FALSE,
17 | "bifurcation_tented", dyntoy::model_bifurcating(), TRUE
18 | )
19 | ),
20 | num_cells = c(10, 20, 50, 100, 200, 500),
21 | repeat_ix = 1,
22 | cell_positioning = c("edges", "milestones")
23 | ) %>%
24 | mutate(
25 | dataset_id = as.character(str_glue("{topology_id}_{num_cells}_{cell_positioning}_{repeat_ix}")),
26 | seed = repeat_ix
27 | )
28 |
29 | dataset_design$dataset <- pmap(dataset_design, function(dataset_id, topology_model, num_cells, seed, cell_positioning, allow_tented_progressions, ...) {
30 | function() {
31 | set.seed(seed)
32 |
33 | dataset <- dyntoy::generate_dataset(
34 | id = dataset_id,
35 | model = topology_model,
36 | num_cells = num_cells,
37 | num_features = 200,
38 | allow_tented_progressions = allow_tented_progressions,
39 | add_prior_information = FALSE,
40 | normalise = FALSE
41 | )
42 |
43 | if (cell_positioning == "milestones") {
44 | dataset <- dataset %>% dynwrap::gather_cells_at_milestones()
45 | }
46 |
47 | dataset
48 | }
49 | })
50 |
51 | write_rds(dataset_design, result_file("dataset_design.rds"))
52 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/3b-time_mem_predictions.R:
--------------------------------------------------------------------------------
1 | #' Comparison between predicted versus actual running time and memory usage
2 |
3 | join <-
4 | data %>%
5 | filter(error_status %in% c("no_error", "time_limit", "memory_limit")) %>%
6 | mutate(
7 | time = ifelse(error_status == "memory_limit", NA, time),
8 | mem = ifelse(error_status == "time_limit" | mem < log10(100e6), NA, mem),
9 | ltime = log10(time),
10 | lmem = log10(mem)
11 | )
12 |
13 | range_time <- quantile(abs(join$ltime - join$time_lpred), .9, na.rm = TRUE) %>% {2 * c(-., .)}
14 | range_mem <- quantile(abs(join$lmem - join$mem_lpred), .9, na.rm = TRUE) %>% {2 * c(-., .)}
15 |
16 | g1 <-
17 | ggplot(join) +
18 | geom_point(aes(time_lpred, ltime, colour = ltime - time_lpred)) +
19 | theme_bw() +
20 | facet_wrap(~method_id, scales = "free") +
21 | scale_color_distiller(palette = "RdYlBu", limits = range_time)
22 |
23 | g2 <-
24 | ggplot(join) +
25 | geom_point(aes(mem_lpred, lmem, colour = lmem - mem_lpred)) +
26 | theme_bw() +
27 | facet_wrap(~method_id, scales = "free") +
28 | scale_color_distiller(palette = "RdYlBu", limits = range_mem)
29 |
30 | pdf(result_file("compare_pred_ind.pdf"), width = 20, height = 15)
31 | print(g1 + labs(title = "Timings"))
32 | print(g2 + labs(title = "Memory"))
33 | dev.off()
34 |
35 |
36 | g1 <- ggplot(join) +
37 | geom_point(aes(time_lpred, ltime, colour = ltime - time_lpred)) +
38 | theme_bw() +
39 | scale_color_distiller(palette = "RdYlBu", limits = range_time) +
40 | theme(legend.position = "bottom")
41 | g2 <- ggplot(join %>% filter(lmem > 8)) +
42 | geom_point(aes(mem_lpred, lmem, colour = lmem - mem_lpred)) +
43 | theme_bw() +
44 | scale_color_distiller(palette = "RdYlBu", limits = range_mem) +
45 | theme(legend.position = "bottom")
46 | ggsave(result_file("compare_pred_all.pdf"), patchwork::wrap_plots(g1, g2, nrow = 1), width = 16, height = 8)
47 |
48 | rm(join, g1, g2)
49 |
--------------------------------------------------------------------------------
/package/man/plot_trajectory_types.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_trajectory_types.R
3 | \name{plot_trajectory_types}
4 | \alias{plot_trajectory_types}
5 | \title{Plot a trajectory type}
6 | \usage{
7 | plot_trajectory_types(plot = ggplot() + theme_void(), trajectory_types,
8 | xmins = 0, xmaxs = 1, ymins = 0, ymaxs = 1,
9 | node_colours = NULL, edge_colours = NULL, size = 4,
10 | geom = c("point", "circle"), circ_size = ifelse(geom == "circle",
11 | 0.05, 0))
12 | }
13 | \arguments{
14 | \item{plot}{Existing plot}
15 |
16 | \item{trajectory_types}{One or more trajectory types}
17 |
18 | \item{xmins}{Minimal x bound (length 1 or length of \code{trajectory_types})}
19 |
20 | \item{xmaxs}{Maximal y bound (likewise)}
21 |
22 | \item{ymins}{Minimal y bound (likewise)}
23 |
24 | \item{ymaxs}{Maximal y bound (likewise)}
25 |
26 | \item{node_colours}{The colours of the nodes.
27 | If \code{NULL}, the colours provided by \code{dynwrap::trajectory_types} will be used (recommended).
28 | Otherwise, \code{node_colours} is expected to be of length 1 or length of \code{trajectory_types}.}
29 |
30 | \item{edge_colours}{The colours of the edges.
31 | If \code{NULL}, the edges will be coloured black.
32 | Otherwise, \code{edge_colours} is expected to be of length 1 or length of \code{trajectory_types}.}
33 |
34 | \item{size}{Size of the arrows and nodes}
35 |
36 | \item{geom}{Which underlying geom to use. When using \code{"point"}, it is expected to also be
37 | using \code{scale_colour_identity()} with your plot. The nodes will go slightly outside the (x, y) boundaries,
38 | because the size of the points depend on the plot size. When using \code{"circle"}, it is expected to be using
39 | \code{scale_fill_identity()} and \code{coord_equal()}, otherwise the nodes might be elliptical.`}
40 |
41 | \item{circ_size}{Size of nodes, only used when \code{geom == "circle"}.}
42 | }
43 | \description{
44 | Plot a trajectory type
45 | }
46 |
--------------------------------------------------------------------------------
/scripts/09-guidelines/01-aggregate.R:
--------------------------------------------------------------------------------
1 | #' Aggregation of the different bnchmarking results for dynguidelines
2 | #' One big methods tibble is created, which contains the information for each method in each row
3 | #' This tibble is nested if a particular experiment generates multiple data points for a method
4 | #' Otherwise, the result is given by using {experiment_id}_{metric_id}
5 |
6 | library(dynbenchmark)
7 | library(tidyverse)
8 |
9 | experiment("09-guidelines")
10 |
11 | # gathers scaling, methods, benchmark results
12 | methods_aggr <- read_rds(result_file("results.rds", experiment_id = "08-summary"))
13 |
14 | # temporary fix for stringlytyped scaling columns
15 | methods_aggr <- methods_aggr %>%
16 | mutate_at(vars(matches("scaling_pred_(time|mem)_")), as.numeric)
17 |
18 | # benchmarking metrics
19 | benchmark_metrics <- dynbenchmark::metrics_evaluated %>% filter(type != "overall")
20 | benchmark_results <- read_rds(result_file("benchmark_results_unnormalised.rds", experiment_id = "06-benchmark"))
21 | benchmark <- benchmark_results %>%
22 | select(-prior_df) %>%
23 | select(method_id, dataset_id, dataset_trajectory_type, !!!benchmark_metrics$metric_id) %>%
24 | mutate_if(is.character, factor) %>%
25 | nest(-method_id, .key = "benchmark")
26 |
27 | methods_aggr <- methods_aggr %>%
28 | left_join(benchmark, "method_id")
29 |
30 | # benchmarking datasets
31 | benchmark_datasets_info <- load_datasets(unique(benchmark_results$dataset_id)) %>%
32 | select(id, source, trajectory_type)
33 |
34 | # metrics
35 | benchmark_metrics <- dyneval::metrics %>%
36 | filter(metric_id %in% c("correlation", "him", "F1_branches", "featureimp_wcor"))
37 | benchmark_metrics$description <- "todo"
38 |
39 | withr::with_dir(
40 | "../dynguidelines",
41 | {
42 | usethis::use_data(
43 | methods_aggr,
44 | benchmark_datasets_info,
45 | benchmark_metrics,
46 | internal = TRUE,
47 | compress = "xz",
48 | overwrite = TRUE
49 | )
50 | }
51 | )
52 |
53 |
--------------------------------------------------------------------------------
/package/man/paramoptim_generate_design.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_generate_design.R
3 | \name{paramoptim_generate_design}
4 | \alias{paramoptim_generate_design}
5 | \title{Generate a benchmarking design}
6 | \usage{
7 | paramoptim_generate_design(datasets, methods, priors = NULL,
8 | num_repeats = 1, crossing = NULL)
9 | }
10 | \arguments{
11 | \item{datasets}{The datasets to be used in the evaluation.
12 | Must be a named list consisting of dataset ids (character),
13 | dynwrap::data_wrapper's, or functions that generate dynwrap::data_wrapper's.}
14 |
15 | \item{methods}{The methods to be evaluated.
16 | Must be a named list consisting of method_ids (character) or dynwrap::ti_wrapper's.}
17 |
18 | \item{priors}{A list of lists. Each sublist contains a list of priors that each method is allowed to optionally use.
19 | Check \code{\link[dynwrap:priors]{dynwrap::priors}} for a list of possible priors. Default priors given is "none"}
20 |
21 | \item{num_repeats}{The number of times to repeat the evaluation.}
22 |
23 | \item{crossing}{A data frame containing the combinations of methods, datasets and priors to be evaluated.
24 | Must be a data frame containing the columns dataset_id, method_id, prior_id, repeat_ix and param_id}
25 | }
26 | \description{
27 | Generate a benchmarking design
28 | }
29 | \examples{
30 | \dontrun{
31 | library(tibble)
32 |
33 | datasets <- list(
34 | "synthetic/dyntoy/bifurcating_1",
35 | dyntoy::generate_dataset(id = "test1"),
36 | test2 = function() dyntoy::generate_dataset()
37 | )
38 |
39 | methods <- list(
40 | "comp1",
41 | dynmethods::ti_scorpius(),
42 | "tscan"
43 | )
44 |
45 | priors <- list(
46 | list(id = "none", method_id = c("comp1", "scorpius", "tscan")),
47 | list(id = "some", method_id = c("tscan"), set = c("start_id", "end_n"))
48 | )
49 |
50 | paramoptim_generate_design(
51 | datasets = datasets,
52 | methods = methods,
53 | priors = priors,
54 | num_repeats = 2
55 | )
56 | }
57 | }
58 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/02-dataset_sources.R:
--------------------------------------------------------------------------------
1 | #' Similarity of the results across dataset sources
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | experiment("10-benchmark_interpretation")
7 |
8 | methods <- load_methods()
9 | data <- read_rds(result_file("benchmark_results_normalised.rds", "06-benchmark"))$data %>% filter(method_id %in% methods$id)
10 |
11 | # aggregate without errors
12 | data_aggregations <- benchmark_aggregate(
13 | data = data %>% filter(error_status == "no_error")
14 | )$data_aggregations
15 |
16 | overall_dataset_source_scores <- data_aggregations %>%
17 | filter(dataset_trajectory_type == "overall", dataset_source != "mean") %>%
18 | select(method_id, dataset_source, overall) %>%
19 | spread(dataset_source, overall)
20 |
21 | gold_dataset_source_scores <- overall_dataset_source_scores %>%
22 | gather("dataset_source", "dataset_source_score", -method_id, -`real/gold`)
23 |
24 | correlation_dataset_source_scores <- gold_dataset_source_scores %>%
25 | group_by(dataset_source) %>%
26 | summarise(cor = nacor(`real/gold`, dataset_source_score)) %>%
27 | arrange(-cor) %>%
28 | mutate(dataset_source = fct_inorder(dataset_source))
29 |
30 | plot_dataset_source_correlation <- gold_dataset_source_scores %>%
31 | mutate(dataset_source = factor(dataset_source, levels = levels(correlation_dataset_source_scores$dataset_source))) %>%
32 | ggplot(aes(dataset_source_score, `real/gold`)) +
33 | geom_abline(intercept = 0, slope = 1) +
34 | geom_point() +
35 | geom_label(aes(label = sprintf("corr = %0.2f", cor)), x = 0.1, y = 0.9, data = correlation_dataset_source_scores, vjust = 0, hjust = 0) +
36 | facet_grid(.~dataset_source) +
37 | scale_x_continuous(limits = c(0, 1)) +
38 | scale_y_continuous(limits = c(0, 1)) +
39 | labs(x = "Overall score on datasets from source", y = "Overall score on real/gold datasets") +
40 | theme_pub()
41 |
42 | plot_dataset_source_correlation
43 |
44 | write_rds(plot_dataset_source_correlation, derived_file("dataset_source_correlation.rds"))
45 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/02-filter_and_normalise.R:
--------------------------------------------------------------------------------
1 | #' Filtering and normalising the real datasets using `dynbenchmark::process_raw_dataset` All datasets are then saved into the dynwrap format.
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(qsub)
6 |
7 | run_remote("find . -maxdepth 2 -type d -ls", args = c("-2", derived_file(experiment_id = '01-datasets_preproc/raw/real', remote = T)), remote = T)$stdout
8 |
9 | # determine datasets to normalise & filter
10 | list_raw_datasets_remote <- function() {
11 | run_remote(
12 | "find",
13 | args = c(
14 | derived_file(experiment_id = '01-datasets_preproc/raw/real', remote = T),
15 | "-maxdepth", "2",
16 | "-type", "f"
17 | ),
18 | remote = T
19 | )$stdout %>%
20 | gsub(".*(real/.*)\\.rds", "\\1", .)
21 | }
22 |
23 | list_datasets_remote <- function() {
24 | run_remote(
25 | "find",
26 | args = c(
27 | derived_file(experiment_id = '01-datasets/real', remote = T),
28 | "-maxdepth", "3",
29 | "-type", "f",
30 | "-regex", "'.*dataset\\.rds'"
31 | ),
32 | remote = T
33 | )$stdout %>%
34 | gsub(".*(real/.*)/dataset\\.rds", "\\1", .)
35 | }
36 |
37 | dataset_ids_to_process <- setdiff(list_raw_datasets_remote(), list_datasets_remote())
38 |
39 | process_raw_dataset_wrapper <- function(id) {
40 | raw_dataset <- readr::read_rds(dynbenchmark::dataset_raw_file(id))
41 | raw_dataset$id <- id
42 |
43 | do.call(dynbenchmark::process_raw_dataset, raw_dataset)
44 | TRUE
45 | }
46 |
47 | qsub_config <- qsub::override_qsub_config(
48 | name = "filter_norm",
49 | memory = "30G",
50 | wait = FALSE,
51 | stop_on_error = TRUE
52 | )
53 |
54 | handle <- qsub_lapply(dataset_ids_to_process, process_raw_dataset_wrapper, qsub_environment = character(), qsub_config = qsub_config)
55 |
56 |
57 | # sync locally
58 | local_folder_sync <- derived_file("", "01-datasets/real/")
59 | remote_folder_sync <- derived_file("", "01-datasets/real/", remote = TRUE)
60 |
61 | qsub::rsync_remote("prism", remote_folder_sync, "", local_folder_sync)
62 |
--------------------------------------------------------------------------------
/package/R/reformat_tribbles.R:
--------------------------------------------------------------------------------
1 | #' Align tribbles in a file
2 | #'
3 | #' The start and end of a tribble need to be marked with a commend `# tribble_start` and `# tribble_end`.
4 | #'
5 | #' @param script_file The file in which to align the tribble
6 | #' @param align The alignment (left, center, right)
7 | #'
8 | #' @importFrom readr read_lines write_lines
9 | #' @export
10 | reformat_tribbles <- function(script_file, align = c("left", "center", "right")) {
11 | align <- match.arg(align)
12 |
13 | pad_fun <- function(word, width) {
14 | padding <- width - nchar(word)
15 | nleft <- switch(
16 | align,
17 | left = 0,
18 | center = floor(padding / 2),
19 | right = padding
20 | )
21 | nright = switch(
22 | align,
23 | left = padding,
24 | center = ceiling(padding / 2),
25 | right = 0
26 | )
27 | paste(c(rep(" ", nleft), word, rep(" ", nright)), collapse = "")
28 | }
29 |
30 | lines <- readr::read_lines(script_file)
31 | tribble_start <- grep("tribble_start", lines)
32 | tribble_end <- grep("tribble_end", lines)
33 | for (i in seq_along(tribble_start)) {
34 | starti <- tribble_start[[i]] + 1
35 | endi <- tribble_end[[i]] - 1
36 | subset <-
37 | lines[starti:endi] %>%
38 | gsub(" *([\"`][^\"`]*[\"`]|[a-zA-Z_]*\\([^\\)]*\\)|[^ ,]*),? *", "\\1\U00B0", .) %>%
39 | map(~ strsplit(., "\U00B0")[[1]])
40 |
41 | numcols <- length(subset[[1]])
42 |
43 | maxwidths <- map_int(seq_len(numcols), function(j) {
44 | subset %>% map_chr(~ .[[j]]) %>% map_int(nchar) %>% max
45 | }) + 3
46 |
47 | new_subset <-
48 | map_chr(subset, function(line) {
49 | map2_chr(paste0(line, ","), maxwidths, pad_fun) %>%
50 | paste(collapse = "") %>%
51 | paste0(" ", .) %>%
52 | gsub(" *$", "", .)
53 | })
54 |
55 | new_subset[length(new_subset)] <-
56 | new_subset[length(new_subset)] %>%
57 | gsub(", *$", "", .)
58 |
59 | lines[starti:endi] <- new_subset
60 | }
61 | readr::write_lines(lines, script_file)
62 | }
63 |
--------------------------------------------------------------------------------
/raw/gource/deprecated_repositories/dynvaria.txt:
--------------------------------------------------------------------------------
1 | 1490793919|Robrecht Cannoodt|A|/dynvaria/.gitignore
2 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/experiments.Rproj
3 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/hypotheses_script.R
4 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/hypotheses_tikz.tex
5 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/mindmap.pdf
6 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/mindmap.tex
7 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_hypotheses-and-tasks_clean.xml
8 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_hypotheses-and-tasks_dirty.xml
9 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_methods_clean.xml
10 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_methods_dirty.xml
11 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/experiments.Rproj
12 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/gource.txt
13 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/process.R
14 | 1505671726|Robrecht Cannoodt|M|/dynvaria/drive/gource.txt
15 | 1505671742|Robrecht Cannoodt|M|/dynvaria/drive/process.R
16 | 1505672668|Robrecht Cannoodt|M|/dynvaria/drive/gource.txt
17 | 1505672668|Robrecht Cannoodt|M|/dynvaria/drive/process.R
18 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_hypotheses-and-tasks_clean.xml
19 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_hypotheses-and-tasks_dirty.xml
20 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_methods_clean.xml
21 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_methods_dirty.xml
22 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/experiments.Rproj
23 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/gource.txt
24 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/process.R
25 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/experiments.Rproj
26 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/hypotheses_script.R
27 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/hypotheses_tikz.tex
28 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/mindmap.pdf
29 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/mindmap.tex
30 | 1506950076|Robrecht Cannoodt|D|/dynvaria/
31 |
--------------------------------------------------------------------------------
/scripts/01-datasets/00-download_from_zenodo.R:
--------------------------------------------------------------------------------
1 | #' Downloading the processed datasets from Zenodo ([10.5281/zenodo.1443566](https://doi.org/10.5281/zenodo.1443566))
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(httr)
6 | library(Matrix)
7 |
8 | experiment("01-datasets")
9 |
10 | # download the rds files from zenodo
11 | deposit_id <- 1443566
12 |
13 | files <-
14 | GET(glue::glue("https://zenodo.org/api/records/{deposit_id}")) %>%
15 | httr::content() %>%
16 | .$files %>%
17 | map(unlist) %>%
18 | list_as_tibble() %>%
19 | mutate(
20 | dataset_id = str_replace(filename, "\\.rds$", ""),
21 | dest_folder = map_chr(dataset_id, derived_file)
22 | )
23 |
24 | pwalk(
25 | files,
26 | function(checksum, filename, filesize, links.download, dataset_id, dest_folder, ...) {
27 | download_info <- paste0(dest_folder, "/download_info.json")
28 | if (file.exists(download_info)) {
29 | didata <- jsonlite::read_json(download_info, simplifyVector = TRUE)
30 |
31 | if (didata$checksum == checksum) {
32 | return()
33 | }
34 | }
35 |
36 | tmp_download_file <- tempfile()
37 | on.exit(file.remove(tmp_download_file))
38 | download.file(links.download, tmp_download_file)
39 |
40 | if (tools::md5sum(tmp_download_file) != checksum) {
41 | stop("MD5 checksum after downloading file '", dataset_id, "' is wrong!\nAborting downloading procedure.")
42 | }
43 |
44 | dataset <- read_rds(tmp_download_file)
45 | dataset$expression <- as(dataset$expression, "dgCMatrix")
46 | dataset$counts <- as(dataset$counts, "dgCMatrix")
47 |
48 | save_dataset(dataset, id = dataset_id)
49 | jsonlite::write_json(
50 | lst(
51 | deposit_id,
52 | dataset_id,
53 | checksum,
54 | downloaded_on = as.character(Sys.time())
55 | ),
56 | download_info
57 | )
58 | }
59 | )
60 |
61 | # upload to remote, if relevant
62 | # this assumes you have a gridengine cluster at hand
63 | qsub::rsync_remote(
64 | remote_src = FALSE,
65 | path_src = derived_file(),
66 | remote_dest = TRUE,
67 | path_dest = derived_file(remote = TRUE)
68 | )
69 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_stimulated-dendritic-cells_shalek.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/gold/stimulated-dendritic-cells_shalek")
5 |
6 | file <- download_dataset_source_file(
7 | "GSM1406531_GSM1407094.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE48968&format=file&file=GSE48968%5FallgenesTPM%5FGSM1189042%5FGSM1190902%2Etxt%2Egz"
9 | )
10 | tab <- read.table(gzfile(file), header = TRUE, sep = "\t") %>% as.matrix %>% t
11 |
12 | allcell_info <- data_frame(
13 | cell_id = rownames(tab),
14 | time = ifelse(grepl("_[0-9]+h", cell_id), gsub(".*_([0-9]+h).*", "\\1", cell_id), "0h"),
15 | type = ifelse(grepl("_[0-9]+h", cell_id), gsub("(.*)_[0-9]+h.*", "\\1", cell_id), "Unstimulated"),
16 | sid = ifelse(grepl("_S[0-9]+$", cell_id), gsub("_(S[0-9]+)$", "\\1", cell_id), ""),
17 | milestone_id = paste0(type, "#", time)
18 | ) %>% filter(
19 | type %in% c("LPS", "PAM", "PIC", "Unstimulated")
20 | )
21 |
22 | settings <- lapply(c("LPS", "PAM", "PIC"), function(stim) {
23 | list(
24 | id = stringr::str_glue("real/gold/stimulated-dendritic-cells-{stim}_shalek"),
25 | milestone_network = tribble(
26 | ~from, ~to, ~length,
27 | "Unstimulated#0h", stringr::str_glue("{stim}#1h"), 1,
28 | stringr::str_glue("{stim}#1h"), stringr::str_glue("{stim}#2h"), 1,
29 | stringr::str_glue("{stim}#2h"), stringr::str_glue("{stim}#4h"), 2,
30 | stringr::str_glue("{stim}#4h"), stringr::str_glue("{stim}#6h"), 2
31 | ) %>% mutate(directed = TRUE)
32 | )
33 | })
34 |
35 | tab <- tab[, !grepl("^ERCC", colnames(tab))] # remove spike-ins, are not present in every cell
36 |
37 | for (setting in settings) {
38 | milestone_network <- setting$milestone_network
39 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
40 |
41 | cell_info <- allcell_info %>% filter(milestone_id %in% milestone_ids)
42 |
43 | counts <- tab[cell_info$cell_id, ]
44 |
45 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
46 |
47 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts), setting$id)
48 | }
49 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_planaria-plass.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/silver/whole-schmidtea-mediterranea_plass")
5 |
6 | # get settings
7 | source(paste0(dynbenchmark::get_dynbenchmark_folder(), "/scripts/01-datasets/01-real/helpers-download_from_sources/helper_planaria-plass.R"))
8 |
9 | # counts
10 | counts_file <- download_dataset_source_file(
11 | "dge.txt.gz",
12 | "http://bimsbstatic.mdc-berlin.de/rajewsky/PSCA/dge.txt.gz"
13 | )
14 |
15 | counts_file_unzipped <- gsub("\\.gz", "", counts_file)
16 | read_lines(counts_file) %>% {.[[1]] <- paste0("\"gene\"\t", .[[1]]); .} %>% write_lines(counts_file_unzipped)
17 |
18 | counts_all <- read_tsv(counts_file_unzipped) %>%
19 | as.data.frame() %>%
20 | column_to_rownames("gene") %>%
21 | as.matrix() %>%
22 | t
23 |
24 | # cell info
25 | cell_info_file <- download_dataset_source_file(
26 | "R_annotation.txt",
27 | "http://bimsbstatic.mdc-berlin.de/rajewsky/PSCA/R_annotation.txt"
28 | )
29 | cell_info_all <- tibble(group_id = read_lines(cell_info_file))
30 | cell_info_all$cell_id <- rownames(counts_all)
31 |
32 |
33 | for (setting in settings) {
34 | print(setting$id)
35 |
36 | milestone_network <- setting$milestone_network
37 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
38 |
39 | if (!all(milestone_ids %in% cell_info_all$group_id)) {stop(setdiff(milestone_ids, cell_info_all$group_id))}
40 |
41 | if (!"length" %in% names(milestone_network)) milestone_network$length <- 1
42 | milestone_network$length[is.na(milestone_network$length)] <- TRUE
43 | if (!"directed" %in% names(milestone_network)) milestone_network$directed <- TRUE
44 | milestone_network$directed[is.na(milestone_network$directed)] <- TRUE
45 |
46 | cell_info <- cell_info_all %>%
47 | filter(group_id %in% milestone_ids)
48 |
49 | grouping <- cell_info %>%
50 | select(cell_id, group_id) %>%
51 | deframe()
52 |
53 | counts <- counts_all[names(grouping), ]
54 |
55 | save_raw_dataset(
56 | lst(
57 | milestone_network,
58 | grouping,
59 | cell_info,
60 | counts
61 | ),
62 | setting$id)
63 | }
64 |
--------------------------------------------------------------------------------
/scripts/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Scripts
3 |
4 | The scripts folder contains all the scripts necessary to fully reproduce
5 | the benchmarking manuscript in chronologically ordered subfolders. Each
6 | subfolder contains a readme file with further explanations of the
7 | different scripts and what they
8 | do:
9 |
10 | | \# | script/folder | description |
11 | | :- | :--------------------------------------------------------- | :---------------------------------------------------- |
12 | | 1 | [📁`datasets`](01-datasets) | Dataset processing and characterisation |
13 | | 2 | [📁`metrics`](02-metrics) | Metrics for comparing two trajectories |
14 | | 3 | [📁`methods`](03-methods) | Trajectory inference methods |
15 | | 4 | [📁`method_testing`](04-method_testing) | Method testing |
16 | | 5 | [📁`scaling`](05-scaling) | Scalability of methods |
17 | | 6 | [📁`benchmark`](06-benchmark) | Benchmarking of TI methods on real and synthetic data |
18 | | 7 | [📁`stability`](07-stability) | Stability |
19 | | 8 | [📁`summary`](08-summary) | Summarising the results into funky heatmaps |
20 | | 9 | [📁`guidelines`](09-guidelines) | Guidelines |
21 | | 10 | [📁`benchmark_interpretation`](10-benchmark_interpretation) | Benchmark interpretation |
22 | | 11 | [📁`example_predictions`](11-example_predictions) | Example predictions |
23 | | 12 | [📁`manuscript`](12-manuscript) | Preparing the manuscript |
24 | | | [📁`varia`](varia) | Varia scripts |
25 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Real datasets
3 |
4 | The generation of the real datasets is divided in two parts. We first
5 | download all the (annotated) expression files from sites such as GEO.
6 | Next, we filter and normalise all datasets, and wrap them into the
7 | common trajectory format of
8 | [dynwrap](https://www.github.com/dynverse/dynwrap).
9 |
10 | | \# | script/folder | description |
11 | | :- | :---------------------------------------------------------------- | :------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
12 | | 1 | [📄`download_from_sources.R`](01-download_from_sources.R) | Downloading the real datasets from their sources (eg. GEO), and constructing the gold standard model, using the helpers in [helpers-download\_from\_sources](helpers-download_from_sources) |
13 | | 2 | [📄`filter_and_normalise.R`](02-filter_and_normalise.R) | Filtering and normalising the real datasets using `dynbenchmark::process_raw_dataset` All datasets are then saved into the dynwrap format. |
14 | | 3 | [📄`gather_metadata.R`](03-gather_metadata.R) | Gathers some metadata about all the real datasets |
15 | | 4 | [📄`datasets_table.R`](04-datasets_table.R) | Creates a table of the datasets in, excuse me, excel (for supplementary material) |
16 | | | [📁`helpers-download_from_sources`](helpers-download_from_sources) | |
17 |
--------------------------------------------------------------------------------
/raw/gource/gource_captions:
--------------------------------------------------------------------------------
1 | 2016/04/02|The birth of dynverse (dyngen and qsub)
2 | 2017/01/15|But first, what are our hypotheses regarding this project?
3 | 2017/02/25|Generating good synthetic data is fun
4 | 2017/03/12|Birth of dyneval, taking baby steps in evaluation
5 | 2017/03/20|Planning, planning, planning!
6 | 2017/03/27|Discussing, discussing, discussing!
7 | 2017/04/04|First TI methods are added to dyneval
8 | 2017/05/13|Initial experiments in performing parameter optimisation
9 | 2017/07/05|Collecting meta info on more TI methods
10 | 2017/07/28|Restructuring of dyngen
11 | 2017/07/31|Including code of other frameworks into our repository was not a good idea
12 | 2017/08/10|Cleaning up dyngen
13 | 2017/08/19|Adding wrappers and metrics
14 | 2017/08/23|Creation of dynplot
15 | 2017/08/27|Huge restructuring; bye bye monster matlab code
16 | 2017/09/06|Birth of dynbenchmark
17 | 2017/09/12|Doing many things! (can't keep up)
18 | 2017/09/16|Creation of dyntoy, cleaning up dyngen
19 | 2017/10/04|It's getting real; downloading lots of real datasets
20 | 2017/11/01|Developing code base for setting up experiments
21 | 2017/11/20|Moving TI methods from dyneval to dynmethods
22 | 2017/12/15|Making presentations
23 | 2018/02/01|Writing preprint manuscript
24 | 2018/02/24|Still writing...
25 | 2018/03/05|Submission to bioRxiv!
26 | 2018/03/25|Preparing for submission to NBT
27 | 2018/04/05|Submission to NBT
28 | 2018/04/10|Initial experiments with Docker containerisation
29 | 2018/04/11|First guest appearance: Koen Van den Berge
30 | 2018/04/21|Birth of dynguidelines
31 | 2018/04/28|Creating a decent dynplot
32 | 2018/05/11|Wouter decides logos are a thing
33 | 2018/06/10|Getting the reviews back, writing a rebuttal
34 | 2018/06/20|Creating TI method dockers
35 | 2018/06/21|with guests Philipp Angerer, Manu Setty and Alex Wolf
36 | 2018/07/01|Watch out for a large restructuring of dynbenchmark...
37 | 2018/08/14|Hello guests Daniel C Ellwanger and Chuck Herring!
38 | 2018/08/29|Heavily integrating containerisation into dynmethods
39 | 2018/09/11|Our avatars are getting hard to see
40 | 2018/10/25|Preparing for revision
41 | 2018/11/12|Revision submitted
42 | 2019/02/04|Final revision
43 | 2019/03/21|Preparing for release of dyno v1.0
44 | 2019/04/01|TODAY!
45 |
--------------------------------------------------------------------------------
/package/R/setup_experiment.R:
--------------------------------------------------------------------------------
1 | #' Helper function for controlling experiments
2 | #'
3 | #' @param experiment_id id for the experiment
4 | #' @param filename the filename
5 | #' @param remote The remote to access, "" if working locally, TRUE if using default qsub_config remote
6 | #'
7 | #' @export
8 | #'
9 | #' @rdname experiment
10 | #'
11 | #' @examples
12 | #' \dontrun{
13 | #' experiment("test_plots")
14 | #'
15 | #' data <- matrix(runif(200), ncol = 2)
16 | #' pdf(result_file("testplot.pdf"), 5, 5)
17 | #' plot(data)
18 | #' dev.off()
19 | #' }
20 | experiment <- function(experiment_id) {
21 | # check whether the working directory is indeed the dynbenchmark folder
22 | dyn_fold <- get_dynbenchmark_folder()
23 |
24 | # set option
25 | options(dynbenchmark_experiment_id = experiment_id)
26 | }
27 |
28 | # create a helper function
29 | experiment_subfolder <- function(path) {
30 | function(filename = "", experiment_id = NULL, remote = FALSE) {
31 | filename <- paste0(filename, collapse = "")
32 |
33 | dyn_folder <- get_dynbenchmark_folder(remote = remote)
34 |
35 | # check whether exp_id is given
36 | if (is.null(experiment_id)) {
37 | experiment_id <- getOption("dynbenchmark_experiment_id")
38 | }
39 |
40 | # check whether exp_fold could be found
41 | if (is.null(experiment_id)) {
42 | stop("No experiment folder found. Did you run experiment(...) yet?")
43 | }
44 |
45 | # determine the full path
46 | full_path <- paste0(dyn_folder, "/", path, "/", experiment_id, "/")
47 |
48 | # create if necessary
49 | if (is.logical(remote) && !remote) {
50 | dir.create(full_path, showWarnings = FALSE, recursive = TRUE)
51 | } else {
52 | qsub::mkdir_remote(full_path, remote = remote)
53 | }
54 |
55 | # get complete filename
56 | paste(full_path, filename, sep = "")
57 | }
58 | }
59 |
60 | #' @rdname experiment
61 | #' @export
62 | derived_file <- experiment_subfolder("derived")
63 |
64 | #' @rdname experiment
65 | #' @export
66 | raw_file <- experiment_subfolder("raw")
67 |
68 | #' @rdname experiment
69 | #' @export
70 | result_file <- experiment_subfolder("results")
71 |
72 | #' @rdname experiment
73 | #' @export
74 | scripts_file <- experiment_subfolder("scripts")
75 |
--------------------------------------------------------------------------------
"
25 | )
26 | dynbenchmark:::fix_relative_paths(knit, "IT WORKED :)")
27 | }
28 |
--------------------------------------------------------------------------------
/scripts/01-datasets/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Dataset processing and characterisation
6 |
7 | ```{r include=FALSE}
8 | library(dynbenchmark)
9 | library(tidyverse)
10 | ```
11 |
12 | The datasets are split in real datasets and synthetic datasets. The real datasets are downloaded and preprocessed first, and characteristics from these datasets (such as the number of cells and genes, library sizes, dropout probabilities, ...) are used to generate synthetic datasets. The datasets are then characterised, after which they are uploaded to Zenodo.
13 |
14 | ```{r}
15 | dynbenchmark::render_scripts_documentation()
16 | ```
17 |
18 | The results of this experiment are available [here](`r dynbenchmark::link_to_results()`).
19 |
20 | ```{r results = 'asis'}
21 | dynbenchmark::knit_nest("01-real")
22 | ```
23 |
24 | ```{r results = 'asis'}
25 | dynbenchmark::knit_nest("02-synthetic")
26 | ```
27 | `
28 | ```{r results = 'asis'}
29 | dynbenchmark::knit_nest("04-dataset_characterisation")
30 | ```
31 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/04-dyngen_samplers_table.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("01-datasets/02-synthetic")
5 |
6 | # ____________________________________________________________________________
7 | # Samplers table ####
8 | samplers <- read_tsv(raw_file("samplers"))
9 | notes <- c("$y_{max} = r/d * p/q$")
10 |
11 | table <- map(c("latex", "html"), function(format) {
12 | table <- samplers %>% mutate(text = stringr::str_glue("${parameter} = {distribution}$")) %>%
13 | select(text) %>%
14 | mutate(text = kableExtra::cell_spec(text, format, escape = FALSE)) %>%
15 | knitr::kable(format, escape = FALSE, col.names = NULL) %>%
16 | kableExtra::kable_styling(bootstrap_options = "condensed") %>%
17 | kableExtra::footnote(notes, general_title = "where", escape = FALSE) %>%
18 | gsub("&", "&", .)
19 | table
20 | }) %>% set_names(c("latex", "html"))
21 |
22 | write_rds(table, result_file("samplers.rds"), compress = "xz")
23 |
--------------------------------------------------------------------------------
/scripts/08-summary/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Summarising the results into funky heatmaps
3 |
4 | Here, we aggregate the results from the different experiment into a
5 | “tidy” format. Next, we plot this into funky
6 | heatmaps.
7 |
8 | | \# | script/folder | description |
9 | | :- | :------------------------------------------------ | :----------------------------------------- |
10 | | 1 | [📄`aggregate_data.R`](1-aggregate_data.R) | Aggregation of the results |
11 | | 2 | [📄`main_figure.R`](2-main_figure.R) | Generation of the different funky heatmaps |
12 | | 2a | [📄`2a_columns_all.R`](2a_columns_all.R) | |
13 | | 2a | [📄`2a_columns_detailed.R`](2a_columns_detailed.R) | |
14 | | 2a | [📄`2a_columns_summary.R`](2a_columns_summary.R) | |
15 | | 2a | [📄`2a_columns_suppfig.R`](2a_columns_suppfig.R) | |
16 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: dynbenchmark::github_markdown_nested
3 | ---
4 |
5 | # Synthetic datasets
6 |
7 | Each synthetic dataset is based on some characteristics of some real datasets. These characteristics include:
8 |
9 | * The number of cells and features
10 | * The number of features which are differentially expressed in the trajectory
11 | * Estimates of the distribution of the library sizes, average expression, dropout probabilities, ... estimated by [Splatter](https://github.com/Oshlack/splatter).
12 |
13 | Here we estimate the parameters of these "platforms" and use them to simulate datasets using different simulators. Each simulation script first creates a design dataframe, which links particular platforms, different topologies, seeds and other parameters specific for a simulator.
14 |
15 | The data is then simulated using wrappers around the simulators (see [/package/R/simulators.R](/package/R/simulators.R)), so that they all return datasets in a format consistent with dynwrap.
16 |
17 | ```{r}
18 | dynbenchmark::render_scripts_documentation()
19 | ```
20 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/helper-04-complementarity.R:
--------------------------------------------------------------------------------
1 | #' Helper plotting functions the complementarity experiment
2 |
3 | quarter_circ <- function(xstart, ystart, from_angle, to_angle, size) {
4 | data_frame(
5 | angle = seq(from_angle, to_angle, length.out = 20) * 2 * pi,
6 | x = sin(angle),
7 | y = cos(angle)
8 | ) %>% mutate(
9 | x = (x - min(x)) / (max(x) - min(x)) * size + xstart,
10 | y = (y - min(y)) / (max(y) - min(y)) * size + ystart
11 | )
12 | }
13 |
14 | bracket <- function(xmin, xmax, ymin, ymax, flip = FALSE) {
15 | width <- xmax - xmin
16 | height <- ymax - ymin
17 | xmid <- xmin + width / 2
18 | ymid <- ymin + height / 2
19 | curly_size <- height / 2
20 |
21 | df <-
22 | bind_rows(
23 | quarter_circ(xmin, ymin, .75, 1, curly_size),
24 | quarter_circ(xmid - curly_size, ymid, .5, .25, curly_size),
25 | quarter_circ(xmid, ymid, .75, .5, curly_size),
26 | quarter_circ(xmax - curly_size, ymin, 0, .25, curly_size)
27 | )
28 |
29 | if (flip) {
30 | df <- df %>% mutate(y = ymin + ymax - y)
31 | }
32 |
33 | df
34 | }
35 |
--------------------------------------------------------------------------------
/scripts/02-metrics/01-metric_characterisation/03-clustering.R:
--------------------------------------------------------------------------------
1 | #' Characterisation of the `r dynbenchmark::label_metric("F1_branches")` and `r dynbenchmark::label_metric("F1_milestones")`
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(patchwork)
6 |
7 | experiment("02-metrics/01-metric_characterisation")
8 |
9 | set.seed(9)
10 | dataset <- dyntoy::generate_dataset(model = dyntoy::model_binary_tree(num_branchpoints = 2)) %>% simplify_trajectory()
11 |
12 | plot_clustering_scores_overview <- wrap_plots(
13 | plot_graph(dataset, label_milestones = TRUE) + ggtitle("Reference"),
14 | plot_graph(dataset, grouping = group_onto_nearest_milestones(dataset), plot_milestones = TRUE) + ggtitle("Cells mapped to milestones"),
15 | plot_graph(dataset, grouping = group_onto_trajectory_edges(dataset)) + ggtitle("Cells mapped to branches") + guides(color = guide_legend(ncol = 3))
16 | )
17 |
18 | plot_clustering_scores_overview
19 |
20 | write_rds(plot_clustering_scores_overview, result_file("clustering_scores_overview.rds"))
21 |
22 | ggsave(result_file("clustering_scores_overview.pdf"), width = 12, height = 5)
23 |
--------------------------------------------------------------------------------
/package/R/text_equations.R:
--------------------------------------------------------------------------------
1 | #' Renders equations for github markdown
2 | #'
3 | #' @param x A character vector
4 | #' @param format The format: html, markdown, latex, ...
5 | #' @export
6 | render_equations <- function(x, format = get_default_format()) {
7 | if (format == "markdown") {
8 | # inline equation
9 | x <- str_replace_all(
10 | x,
11 | "\\$[^\\$]+\\$",
12 | function(x) paste0("), ")")
13 | )
14 |
15 | # multiple line equations, e.g.
16 | # $$
17 | # 1 + 2 = 3
18 | # $$
19 |
20 | which_dd <- which(x == "$$")
21 | if (length(which_dd) %% 2 == 1) {
22 | stop("Uneven number of '$$' lines found!")
23 | }
24 |
25 | for (i in seq_len(length(which_dd) / 2)) {
26 | start <- which_dd[i * 2 - 1] + 1
27 | end <- which_dd[i * 2] - 1
28 |
29 | url <- paste0("), ")")
30 |
31 | x[start] <- url
32 | x[start - 1] <- ""
33 | x[(start):end+1] <- ""
34 | }
35 | }
36 |
37 | x
38 | }
39 |
--------------------------------------------------------------------------------
/package/man/experiment.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_experiment.R
3 | \name{experiment}
4 | \alias{experiment}
5 | \alias{derived_file}
6 | \alias{raw_file}
7 | \alias{result_file}
8 | \alias{scripts_file}
9 | \title{Helper function for controlling experiments}
10 | \usage{
11 | experiment(experiment_id)
12 |
13 | derived_file(filename = "", experiment_id = NULL, remote = FALSE)
14 |
15 | raw_file(filename = "", experiment_id = NULL, remote = FALSE)
16 |
17 | result_file(filename = "", experiment_id = NULL, remote = FALSE)
18 |
19 | scripts_file(filename = "", experiment_id = NULL, remote = FALSE)
20 | }
21 | \arguments{
22 | \item{experiment_id}{id for the experiment}
23 |
24 | \item{filename}{the filename}
25 |
26 | \item{remote}{The remote to access, "" if working locally, TRUE if using default qsub_config remote}
27 | }
28 | \description{
29 | Helper function for controlling experiments
30 | }
31 | \examples{
32 | \dontrun{
33 | experiment("test_plots")
34 |
35 | data <- matrix(runif(200), ncol = 2)
36 | pdf(result_file("testplot.pdf"), 5, 5)
37 | plot(data)
38 | dev.off()
39 | }
40 | }
41 |
--------------------------------------------------------------------------------
/raw/12-manuscript/nature-biotechnology.csl:
--------------------------------------------------------------------------------
1 |
2 |
18 |
19 |
--------------------------------------------------------------------------------
/package/man/add_fig.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_ref.R
3 | \name{add_fig}
4 | \alias{add_fig}
5 | \alias{add_sfig}
6 | \alias{plot_fig}
7 | \title{Add a figure}
8 | \usage{
9 | add_fig(fig_path, ref_id, caption_main, caption_text = "", width = 5,
10 | height = 7, format = get_default_format())
11 |
12 | add_sfig(fig_path, ref_id, caption_main, caption_text = "", width = 5,
13 | height = 7, format = get_default_format(), integrate = TRUE)
14 |
15 | plot_fig(ref_type, ref_id, fig_path, caption_main, caption_text,
16 | width = 5, height = 7, format = "latex", integrate = TRUE)
17 | }
18 | \arguments{
19 | \item{fig_path}{Path to the figure}
20 |
21 | \item{ref_id}{The identifier}
22 |
23 | \item{caption_main}{Caption title}
24 |
25 | \item{caption_text}{Caption text}
26 |
27 | \item{width}{Width in inches}
28 |
29 | \item{height}{Height in inches}
30 |
31 | \item{format}{The format, in html or latex}
32 |
33 | \item{integrate}{Whether to integrate the figure, or direct the reader to a separate file}
34 |
35 | \item{ref_type}{fig, sfig, table, ...}
36 | }
37 | \description{
38 | Add a figure
39 | }
40 |
--------------------------------------------------------------------------------
/LICENSE.md:
--------------------------------------------------------------------------------
1 | # MIT License
2 |
3 | Copyright (c) 2014-2020 Robrecht Cannoodt, Wouter Saelens
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/package/man/platform.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_platforms.R
3 | \name{platform_from_counts}
4 | \alias{platform_from_counts}
5 | \alias{platform_simple}
6 | \title{Estimate platform parameters from a dataset}
7 | \usage{
8 | platform_from_counts(counts, grouping, subsample = 500)
9 |
10 | platform_simple(n_cells = 100L, n_features = 1000L,
11 | pct_main_features = 0.5, dropout_rate = 0.01, dropout_shape = 1)
12 | }
13 | \arguments{
14 | \item{counts}{The counts with cells in rows and genes in columns.}
15 |
16 | \item{grouping}{A named vector representing a grouping of the cells.}
17 |
18 | \item{subsample}{The number of cells to subsample.}
19 |
20 | \item{n_cells}{The number of cells}
21 |
22 | \item{n_features}{The number of features}
23 |
24 | \item{pct_main_features}{The percentage of features that are being driven by the trajectory (or vice versa)}
25 |
26 | \item{dropout_rate}{The mean rate of dropouts}
27 |
28 | \item{dropout_shape}{The shape of dropouts}
29 | }
30 | \description{
31 | Altenatively, \code{\link[=platform_simple]{platform_simple()}} returns a toy platform parameter configuration.
32 | }
33 |
--------------------------------------------------------------------------------
/package/LICENSE.md:
--------------------------------------------------------------------------------
1 | # MIT License
2 |
3 | Copyright (c) 2014-2020 Robrecht Cannoodt, Wouter Saelens
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/05-benchmark_interpretation_figure.R:
--------------------------------------------------------------------------------
1 | #' Combine the different figures of this experiment into one
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(patchwork)
6 |
7 | experiment("10-benchmark_interpretation")
8 |
9 | plot_variability_dataset_source <- read_rds(derived_file("variability_dataset_source.rds"))
10 | plot_variability_trajectory_type <- read_rds(derived_file("variability_trajectory_type.rds"))
11 | plot_dataset_source_correlation <- read_rds(derived_file("dataset_source_correlation.rds"))
12 | plot_topology_complexity_examples <- read_rds(derived_file("topology_complexity_examples.rds"))
13 |
14 | plot_benchmark_interpretation <- wrap_plots(
15 | plot_variability_dataset_source %>% patchwork::wrap_elements(),
16 | plot_dataset_source_correlation %>% patchwork::wrap_elements(),
17 | plot_variability_trajectory_type %>% patchwork::wrap_elements(),
18 | plot_topology_complexity_examples %>% patchwork::wrap_elements(),
19 | ncol = 1,
20 | heights = c(1, 0.7, 1, 1)
21 | ) + plot_annotation(tag_levels = "a")
22 |
23 | ggsave(result_file("benchmark_interpretation.pdf"), plot_benchmark_interpretation, width = 14, height = 18)
24 |
--------------------------------------------------------------------------------
/scripts/03-methods/01-gather_methods_information/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Gathering all the information we have about the methods
3 |
4 | Most information of the methods are contained within their respective
5 | containers (see the [dynmethods](https://github.com/dynverse/dynmethods)
6 | repository, ). We gather
7 | additional information from our google sheets
8 | (),
9 | which also contains the quality control for each
10 | methods.
11 |
12 | | \# | script/folder | description |
13 | | :- | :------------------------------------------------------------- | :------------------------------------------------------- |
14 | | 1 | [📄`group_methods_into_tools.R`](01-group_methods_into_tools.R) | Grouping methods into tools |
15 | | 2 | [📄`process_quality_control.R`](02-process_quality_control.R) | Downloading and processing the quality control worksheet |
16 | | 3 | [📄`add_quality_control.R`](03-add_quality_control.R) | Add QC scores to methods and tools tibble |
17 |
--------------------------------------------------------------------------------
/package/R/load_dyneval_model.R:
--------------------------------------------------------------------------------
1 | #' Load model from benchmark suite results
2 | #'
3 | #' @param method_id The short_name of a method
4 | #' @param df A data frame containing the crossing of which models to load
5 | #' @param auto_gc Whether or not to automatically garbage collect after loading models
6 | #' @param experiment_id The experiment id (default NULL
7 | #'
8 | #' @export
9 | load_dyneval_model <- function(method_id, df, auto_gc = TRUE, experiment_id = NULL) {
10 | models_filename <- derived_file(paste0("suite/", method_id, "/output_models.rds"), experiment_id = experiment_id)
11 | metrics_filename <- derived_file(paste0("suite/", method_id, "/output_metrics.rds"), experiment_id = experiment_id)
12 |
13 | if (!file.exists(models_filename)) {
14 | stop("Models of method ", sQuote(method_id), " not found.", sep = "")
15 | }
16 |
17 | models <- read_rds(models_filename)
18 | metrics <- read_rds(metrics_filename)
19 |
20 | ix <- metrics %>% mutate(i = row_number()) %>% inner_join(df, by = colnames(df)) %>% pull(i)
21 |
22 | if (length(ix) == 0) {
23 | stop("df did not match with any of the rows")
24 | }
25 |
26 | model <- models[ix]
27 |
28 | if (auto_gc) {
29 | gc()
30 | }
31 |
32 | model
33 | }
34 |
--------------------------------------------------------------------------------
/package/R/setup_get_dynbenchmark_folder.R:
--------------------------------------------------------------------------------
1 | #' Find the dynbenchmark path
2 | #'
3 | #' When executing some of the dynbenchmark functions, the git repository needs to be found.
4 | #' Either set the working directory to the repository, or define the DYNBENCHMARK_PATH variable.
5 | #'
6 | #' @param remote The remote to access, "" if running locally, TRUE if using default qsub config remote
7 | #'
8 | #' @export
9 | get_dynbenchmark_folder <- function(remote = FALSE) {
10 | if (is.logical(remote) && !remote) {
11 | pwd <- getOption("dynbenchmark_path")
12 |
13 | if (is.null(pwd)) {
14 | pwd <- Sys.getenv("DYNBENCHMARK_PATH")
15 | }
16 |
17 | if (is.null(pwd)) {
18 | pwd <- getwd()
19 | }
20 |
21 | if (pwd == "") {
22 | pwd <- paste0("./")
23 | }
24 |
25 | if (!file.exists(paste0(pwd, "/dynbenchmark.Rproj"))) {
26 | stop("dynbenchmark folder could not be found. Either set it as the working directory, or export a DYNBENCHMARK_PATH variable.")
27 | }
28 |
29 | pwd
30 | } else {
31 | pwd <- qsub::run_remote("echo $DYNBENCHMARK_PATH", remote)$stdout
32 |
33 | if (pwd == "") {
34 | stop("DYNBENCHMARK_PATH not found at remote")
35 | }
36 |
37 | pwd
38 | }
39 | }
40 |
--------------------------------------------------------------------------------
/package/man/process_raw_dataset.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/datasets_preprocessing.R
3 | \name{process_raw_dataset}
4 | \alias{process_raw_dataset}
5 | \title{Process a raw real or synthetic dataset}
6 | \usage{
7 | process_raw_dataset(id, counts, milestone_network, grouping,
8 | root_milestone_id = NULL, cell_info = tibble(cell_id =
9 | rownames(counts)), feature_info = tibble(feature_id =
10 | colnames(counts)))
11 | }
12 | \arguments{
13 | \item{id}{A unique identifier for the data. If \code{NULL}, a random string will be generated.}
14 |
15 | \item{counts}{The counts values of genes (columns) within cells (rows). This can be both a dense and sparse matrix.}
16 |
17 | \item{milestone_network}{A network of milestones.}
18 |
19 | \item{grouping}{A grouping of the cells, can be a named vector or a dataframe with \emph{group_id} and \emph{cell_id}}
20 |
21 | \item{root_milestone_id}{The root milestone, optional}
22 |
23 | \item{cell_info}{Optional meta-information pertaining the cells.}
24 |
25 | \item{feature_info}{Optional meta-information of the features, a dataframe with at least \emph{feature_id} as column}
26 | }
27 | \description{
28 | Process a raw real or synthetic dataset
29 | }
30 |
--------------------------------------------------------------------------------
/scripts/02-metrics/01-metric_characterisation/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Metric characterisation
3 |
4 | A first characterisation of the metrics. For each metric we:
5 |
6 | - generate some examples to get some intuition on how the metric works
7 | - test the robustness to a metric to stochasticity or parameters when
8 | appropriate
9 |
10 | | \# | script/folder | description |
11 | | :- | :----------------------------------- | :------------------------------------------------------------------------- |
12 | | 1 | [📄`correlation.R`](01-correlation.R) | Characterisation of the cordist |
13 | | 2 | [📄`topology.R`](02-topology.R) | Characterisation of the Isomorphic, edgeflip and HIM |
14 | | 3 | [📄`clustering.R`](03-clustering.R) | Characterisation of the F1branches and F1milestones |
15 | | 4 | [📄`featureimp.R`](04-featureimp.R) | Characterisation of the corfeatures and wcorfeatures |
16 |
17 | The results of this experiment are available
18 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/02-metrics/01-metric_characterisation)
19 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_macrophage-salmonella_saliba.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/gold/macrophage-salmonella_saliba")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "GSE79363_first_dataset_read_count.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE79363&format=file&file=GSE79363%5Ffirst%5Fdataset%5Fread%5Fcount%2Etxt%2Egz"
9 | )
10 |
11 | counts <- read_tsv(txt_location) %>% as.data.frame() %>% tibble::column_to_rownames("X1") %>% as.matrix() %>% t
12 | cell_info <- tibble(cell_id = rownames(counts), milestone_id = gsub(".*_([A-Za-z]*)$", "\\1", rownames(counts)))
13 |
14 | counts <- counts[cell_info$cell_id, ]
15 |
16 | milestone_network = tribble(
17 | ~from, ~to,
18 | "NNI", "MNGB",
19 | "NNI", "MGB",
20 | "NNI", "bystanders"
21 | ) %>% mutate(length = 1, directed = TRUE)
22 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
23 |
24 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
25 | counts <- counts[cell_info$cell_id, ]
26 |
27 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
28 |
29 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
30 |
--------------------------------------------------------------------------------
/package/man/dataset_preprocessing.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/setup_datasets.R
3 | \name{dataset_preprocessing}
4 | \alias{dataset_preprocessing}
5 | \alias{get_dataset_preprocessing_id}
6 | \alias{dataset_source_file}
7 | \alias{dataset_raw_file}
8 | \alias{save_raw_dataset}
9 | \alias{dataset_file}
10 | \alias{save_dataset}
11 | \title{Helper functions for creating new datasets}
12 | \usage{
13 | dataset_preprocessing(id)
14 |
15 | get_dataset_preprocessing_id()
16 |
17 | dataset_source_file(filename = "", id = NULL, remote = FALSE)
18 |
19 | dataset_raw_file(id)
20 |
21 | save_raw_dataset(dataset, id = get_dataset_preprocessing_id())
22 |
23 | dataset_file(filename = "", id = NULL, remote = FALSE)
24 |
25 | save_dataset(dataset, id = NULL, lazy_load = TRUE)
26 | }
27 | \arguments{
28 | \item{id}{The ID of the dataset to be used}
29 |
30 | \item{filename}{Custom filename}
31 |
32 | \item{remote}{The remote to access, "" if working locally, TRUE if using default qsub_config remote}
33 |
34 | \item{dataset}{Dataset object to save}
35 |
36 | \item{lazy_load}{Whether or not to allow for lazy loading of large objects in the dataset}
37 | }
38 | \description{
39 | Helper functions for creating new datasets
40 | }
41 |
--------------------------------------------------------------------------------
/package/R/package.R:
--------------------------------------------------------------------------------
1 | #' Easy installing and loading of the dynverse
2 | #'
3 | #' The 'dynverse' is a set of packages with which to
4 | #' evaluate trajectory inference methods for dynamic processes.
5 | #'
6 | #' The dynbenchmark package contains the code to generate/preprocess datasets, evaluate TI methods on these datasets, and finally wrap all results together in a scientific paper.
7 | #' The code within this package can be run locally, but can also be run within a docker environment, for which the scripts are contained in the `scripts` folder.
8 | #'
9 | #' @import assertthat
10 | #' @importFrom tibble is_tibble as_tibble as_data_frame tibble data_frame enframe deframe lst tribble rownames_to_column column_to_rownames
11 | #' @import dplyr
12 | #' @import ggplot2
13 | #' @importFrom tidyr crossing gather spread gather
14 | #' @importFrom purrr %>% %||% walk set_names map map_dbl map_lgl map_chr map_df map2 map2_dbl map2_lgl map2_chr map2_df invoke
15 | #' @importFrom stringr str_replace_all str_replace str_detect str_sub str_subset str_glue
16 | #'
17 | #' @import dynutils
18 | #' @import dynplot
19 | #' @import dyneval
20 | #' @import dynmethods
21 | #' @import dynwrap
22 | #' @import dynparam
23 | #'
24 | #' @docType package
25 | #' @name dynbenchmark
26 | #'
27 | NULL
28 |
29 |
--------------------------------------------------------------------------------
/package/man/benchmark_aggregate.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_benchmark_aggregate.R
3 | \name{benchmark_aggregate}
4 | \alias{benchmark_aggregate}
5 | \alias{get_dataset_weighting}
6 | \title{Normalisation and aggregation function}
7 | \usage{
8 | benchmark_aggregate(data, metrics = c("correlation", "him",
9 | "featureimp_wcor", "F1_branches"), norm_fun = "normal",
10 | mean_fun = "geometric", mean_weights = set_names(rep(1,
11 | length(metrics)), metrics),
12 | dataset_source_weights = get_default_dataset_source_weights())
13 |
14 | get_dataset_weighting(datasets,
15 | dataset_source_weights = get_default_dataset_source_weights())
16 | }
17 | \arguments{
18 | \item{data}{Data as generated by the \code{06-benchmark/2-retrieve_results.R} script}
19 |
20 | \item{metrics}{Metrics to normalise}
21 |
22 | \item{norm_fun}{Which normalisation to use}
23 |
24 | \item{mean_fun}{Which mean function to use}
25 |
26 | \item{mean_weights}{Which metrics to use for the calculation of the mean, and which weights}
27 |
28 | \item{dataset_source_weights}{The weights of the dataset sources}
29 |
30 | \item{datasets}{Tibble with dataset_id, dataset_source and dataset_trajectory_type}
31 | }
32 | \description{
33 | Normalisation and aggregation function
34 | }
35 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02d-simulate_dyntoy_datasets.R:
--------------------------------------------------------------------------------
1 | #' [dyntoy](https://github.com/dynverse/dyntoy), simulations of toy data using random expression gradients in a reduced space
2 |
3 | library(dynbenchmark)
4 | library(dyntoy)
5 | library(tidyverse)
6 | library(qsub)
7 |
8 | experiment("01-datasets/02-synthetic")
9 |
10 | # remove all datasets
11 | # rm_remote(dataset_file(id = "synthetic/dyntoy", remote = TRUE), remote = TRUE, recursive = TRUE)
12 |
13 | # generate design
14 | set.seed(1)
15 | design <- crossing(
16 | topology_model = names(dyntoy::topology_models),
17 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
18 | ) %>%
19 | mutate(
20 | dataset_id = paste0("synthetic/dyntoy/", topology_model, "_", platform_ix),
21 | seed = sample(1:100000, n())
22 | ) %>%
23 | select(-platform_ix)
24 | write_rds(design, result_file("design_dyntoy.rds"))
25 |
26 | # simulate datasets
27 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "dyntoy", wait = F, stop_on_error = FALSE)
28 |
29 | handle <- qsub_pmap(
30 | design,
31 | simulate_dyntoy,
32 | qsub_config = qsub_config
33 | )
34 |
35 | write_rds(handle, "handle_dyntoy.rds")
36 |
37 | handle <- read_rds("handle_dyntoy.rds")
38 | qsub::qsub_retrieve(handle)
39 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02a-simulate_dyngen_datasets.R:
--------------------------------------------------------------------------------
1 | #' [dyngen](https://github.com/dynverse/dyngen), simulations of regulatory networks which will produce a particular trajectory
2 |
3 | library(qsub)
4 | library(dyngen)
5 | library(dynbenchmark)
6 | library(tidyverse)
7 |
8 | experiment("01-datasets/02-synthetic")
9 |
10 | # remove all datasets
11 | # rm_remote(dataset_file(id = "synthetic/dyngen", remote = TRUE), remote = TRUE, recursive = TRUE)
12 |
13 | # create design
14 | set.seed(1)
15 | design <- crossing(
16 | modulenet_name = dyngen::list_modulenets(),
17 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
18 | ) %>%
19 | mutate(
20 | dataset_id = paste0("synthetic/dyngen/", row_number()),
21 | seed = sample(1:100000, n())
22 | ) %>%
23 | select(-platform_ix)
24 | write_rds(design, result_file("design_dyngen.rds"))
25 |
26 | # simulate datasets
27 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "dyngen", wait = F, stop_on_error = FALSE)
28 |
29 | handle <- qsub_pmap(
30 | design,
31 | simulate_dyngen,
32 | use_cache = TRUE,
33 | qsub_config = qsub_config
34 | )
35 |
36 | write_rds(handle, "handle_dyngen.rds")
37 |
38 | ##
39 | handle <- read_rds("handle_dyngen.rds")
40 | qsub::qsub_retrieve(handle)
41 |
--------------------------------------------------------------------------------
/package/tests/testthat/test-get_dynbenchmark_folder.R:
--------------------------------------------------------------------------------
1 | context("Testing get_dynbenchmark_folder")
2 |
3 | prev_path <- Sys.getenv("DYNBENCHMARK_PATH")
4 | prev_option <- getOption("dynbenchmark_path")
5 |
6 | # save old settings
7 | Sys.setenv(DYNBENCHMARK_PATH = "")
8 | options(dynbenchmark_path = NULL)
9 |
10 |
11 | test_that("get_dynbenchmark_folder works using variables", {
12 | tmp_path <- tempfile()
13 | dir.create(tmp_path)
14 |
15 | Sys.setenv(DYNBENCHMARK_PATH = tmp_path)
16 |
17 | file.create(paste0(tmp_path, "/dynbenchmark.Rproj"))
18 |
19 | expect_equal(get_dynbenchmark_folder(), tmp_path)
20 |
21 | Sys.setenv(DYNBENCHMARK_PATH = "")
22 |
23 | expect_error(get_dynbenchmark_folder(), regexp = "dynbenchmark folder could not be found")
24 | })
25 |
26 | test_that("get_dynbenchmark_folder works using options", {
27 | tmp_path <- tempfile()
28 | dir.create(tmp_path)
29 |
30 | options(dynbenchmark_path = tmp_path)
31 |
32 | file.create(paste0(tmp_path, "/dynbenchmark.Rproj"))
33 |
34 | expect_equal(get_dynbenchmark_folder(), tmp_path)
35 |
36 | options(dynbenchmark_path = NULL)
37 |
38 | expect_error(get_dynbenchmark_folder(), regexp = "dynbenchmark folder could not be found")
39 | })
40 |
41 | # revert to old settings
42 | Sys.setenv(DYNBENCHMARK_PATH = prev_path)
43 | options(dynbenchmark_path = prev_option)
44 |
--------------------------------------------------------------------------------
/package/man/pdf_manuscript.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/text_formats.R
3 | \name{pdf_manuscript}
4 | \alias{pdf_manuscript}
5 | \title{The manuscript pdf document}
6 | \usage{
7 | pdf_manuscript(bibliography = raw_file("references.bib",
8 | "12-manuscript"), csl = raw_file("nature-biotechnology.csl",
9 | "12-manuscript"), render_changes = TRUE, toc = FALSE,
10 | linenumbers = FALSE, sty_header = NULL, sty_beforebody = NULL,
11 | sty_afterbody = NULL,
12 | reference_section_title = reference_section_title, ...)
13 | }
14 | \arguments{
15 | \item{bibliography}{Bibliography location}
16 |
17 | \item{csl}{Csl file location}
18 |
19 | \item{render_changes}{Whether to export a *_changes.pdf as well}
20 |
21 | \item{toc}{Whether to include a table of contents}
22 |
23 | \item{linenumbers}{Whether to add linenumbers to the left side}
24 |
25 | \item{sty_header}{Extra sty files to be included in the header of the document}
26 |
27 | \item{sty_beforebody}{Extra sty files to be included before the body of the document}
28 |
29 | \item{sty_afterbody}{Extra sty files to be included after the body of the document}
30 |
31 | \item{reference_section_title}{The title of the reference section}
32 |
33 | \item{...}{Parameters for rmarkdown::pdf_document}
34 | }
35 | \description{
36 | The manuscript pdf document
37 | }
38 |
--------------------------------------------------------------------------------
/manuscript/supplementary.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | title: "Supplementary material for: \nA comparison of single-cell trajectory inference methods"
3 | output:
4 | dynbenchmark::pdf_supplementary_note:
5 | render_change: FALSE
6 | ---
7 |
8 | ```{r setup, include=FALSE}
9 | library(dynbenchmark)
10 | library(tidyverse)
11 | ```
12 |
13 | ```{r}
14 | supplementary <- read_rds("supplementary.rds")
15 | sfigs <- supplementary$sfigs
16 | stables <- supplementary$stables
17 | refs <- supplementary$refs
18 | ```
19 |
20 | # Supplementary figures
21 | ```{r, results="asis"}
22 | sfigs %>%
23 | left_join(refs %>% select(ref_id, name), "ref_id") %>%
24 | arrange(name) %>%
25 | select(-name) %>%
26 | pmap(dynbenchmark:::plot_fig) %>% modify(paste0, "\n\n") %>% walk(cat)
27 | ```
28 |
29 | \clearpage
30 |
31 | # Supplementary tables
32 |
33 | ```{r, results="asis"}
34 | stables %>%
35 | left_join(refs %>% select(ref_id, name), "ref_id") %>%
36 | arrange(name) %>%
37 | select(-name) %>%
38 | pmap(dynbenchmark:::show_table) %>% modify(paste0, "\n\n") %>% walk(cat)
39 | ```
40 |
41 | \clearpage
42 |
43 | # Supplementary note 1
44 |
45 | ```{r}
46 | dynbenchmark::knit_nest("../results/02-metrics/")
47 | ```
48 | \clearpage
49 |
50 | # Supplementary note 2
51 |
52 | ```{r}
53 | dynbenchmark::knit_nest("../results/01-datasets/02-synthetic/")
54 | ```
55 |
56 | \clearpage
57 |
--------------------------------------------------------------------------------
/package/R/plotting_priors_mini.R:
--------------------------------------------------------------------------------
1 | #' Creates a prior mini svg
2 | #' @param method_priors Dataframe containing a prior_id and a prior_usage
3 | #' @param svg The base svg of the priors
4 | #' @export
5 | #'
6 | #' @importFrom xml2 read_xml xml_new_root xml_find_first xml_remove xml_add_child write_xml
7 | generate_prior_mini <- function(method_priors, svg = xml2::read_xml(result_file("priors.svg", experiment_id = "priors"))) {
8 | svg2 <- xml2::xml_new_root(svg, .copy = T)
9 |
10 | map2(method_priors$prior_id, method_priors$prior_usage, function(prior_id, usage) {
11 | color <- dynbenchmark::prior_usages$color[dynbenchmark::prior_usages$prior_usage == usage]
12 |
13 | layer <- svg2 %>% xml2::xml_find_first(stringr::str_glue(".//svg:g[@inkscape:label='{prior_id}']")) %>% xml2::xml_remove()
14 |
15 | if(!is.na(usage) && usage != "no") {
16 |
17 | print(color)
18 | # only add layer back if needed
19 | layer %>%
20 | as.character() %>%
21 | gsub("stroke:#fb1e00", stringr::str_glue("stroke:{color}"), .) %>%
22 | gsub("fill:#fb1e00", stringr::str_glue("fill:{color}"), .) %>%
23 | {suppressWarnings(xml2::read_xml(.))} %>%
24 | xml2::xml_add_child(svg2, .)
25 | }
26 | })
27 |
28 | svg2 %>% xml2::write_xml("~/test.svg")
29 |
30 | svg2 %>% as.character()# %>% charToRaw() %>% base64enc::base64encode()
31 | }
32 |
--------------------------------------------------------------------------------
/scripts/varia/generate_hypotheses_mindmap/hypotheses_script.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(googlesheets)
3 |
4 | # gs_auth()
5 |
6 | sheet <- gs_key("13jHFuMwie7oIxGcyLO95-onFri4M_GJjqgMEUnZliQI")
7 | hypotheses <- gs_read(sheet, ws = "Hypotheses") %>% mutate(
8 | parent_order = gsub("\\.[0-9]*$", "", order),
9 | parent_id = experiment_id[match(parent_order, order)],
10 | level = sapply(order, function(x) stringr::str_length(gsub("[^\\.]", "", x))),
11 | colstr = sapply(color, function(x) col2rgb(x)[,1] %>% paste(collapse = ","))
12 | )
13 |
14 | experiment_links_df <- hypotheses %>% select(parent_id, experiment_id) %>% filter(parent_id != experiment_id)
15 |
16 | textbox_tikz <- function(x) {
17 | ifelse(is.na(x), "", x %>%
18 | seqinr::stresc() %>%
19 | gsub("\n", "\\\\\\\\\n ", .) %>%
20 | gsub("@", "\\@", .))
21 | }
22 | hypotheses <- hypotheses %>% mutate(
23 | tikznode = paste0(
24 | " \\myhypothesis{", experiment_id, "}{",
25 | gsub("_", "\\\\_", experiment_id), "}{",
26 | colstr, "}{",
27 | textbox_tikz(statement), "}{",
28 | textbox_tikz(experiment_method), "}{",
29 | textbox_tikz(expected_results), "}{",
30 | textbox_tikz(motivation), "}"
31 | )
32 | )
33 |
34 | write_lines(
35 | paste(hypotheses$tikznode, collapse = "\n\n"),
36 | "analysis/analyses/generate_hypotheses_mindmap/hypotheses_tikz.tex"
37 | )
38 |
39 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/wip/pbmc-citeseq_stoeckius.R:
--------------------------------------------------------------------------------
1 | # extract the NK cells and ordering according to CD56??
2 | # cannot find the NK cells...
3 |
4 |
5 | rm(list=ls())
6 | library(dynbenchmark)
7 | library(tidyverse)
8 | library(GEOquery)
9 | library(Biobase)
10 | options('download.file.method.GEOquery'='curl')
11 |
12 | id <- "real/pbmc-citeseq_stoeckius"
13 | dataset_preprocessing(id)
14 |
15 | geo <- GEOquery::getGEO(GEO = "GSM2695382", destdir = dataset_source_file(""))
16 |
17 | rna_location <- download_dataset_source_file(
18 | "GSE100866_PBMC_vs_flow_10X-RNA_umi.csv.gz",
19 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE100866&format=file&file=GSE100866%5FPBMC%5Fvs%5Fflow%5F10X%2DRNA%5Fumi%2Ecsv%2Egz"
20 | )
21 |
22 | protein_location <- download_dataset_source_file(
23 | "GSE100866_PBMC_vs_flow_10X-ADT_umi.csv.gz",
24 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE100866&format=file&file=GSE100866%5FPBMC%5Fvs%5Fflow%5F10X%2DADT%5Fumi%2Ecsv%2Egz"
25 | )
26 |
27 | counts_rna <- read_csv(rna_location) %>% as.data.frame %>% column_to_rownames("X1") %>% as.matrix() %>% t
28 | colnames(counts_rna) <- gsub("HUMAN_(.*)", "\\1", colnames(counts_rna))
29 | counts_protein <- read_csv(protein_location) %>% as.data.frame %>% column_to_rownames("X1") %>% as.matrix() %>% t
30 |
31 |
32 |
33 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2695382
34 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_distal-lung-epithelium_treutlein.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/silver/distal-lung-epithelium_treutlein")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "nature13173-s4.txt",
8 | "http://www.nature.com/nature/journal/v509/n7500/extref/nature13173-s4.txt"
9 | )
10 |
11 | df <- read_tsv(txt_location, col_types = cols(cell_name = "c", time_point = "c", sample = "c", putative_cell_type = "c", .default = "d"))
12 | expression <- df[, -c(1:4)] %>% as.matrix() %>% magrittr::set_rownames(df$cell_name)
13 | cell_info <- df[, c(1:4)] %>% as.data.frame() %>% magrittr::set_rownames(df$cell_name) %>%
14 | rename(
15 | cell_id = cell_name,
16 | milestone_id = putative_cell_type
17 | ) %>% filter(sample == "singleCell")
18 | expression <- expression[cell_info$cell_id, ]
19 |
20 | milestone_network = tribble(
21 | ~from, ~to,
22 | "BP", "AT1",
23 | "BP", "AT2"
24 | ) %>% mutate(length = 1, directed = TRUE)
25 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
26 |
27 | counts <- 2^expression-1
28 |
29 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
30 | counts <- counts[cell_info$cell_id, ]
31 |
32 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
33 |
34 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
35 |
--------------------------------------------------------------------------------
/package/data-raw/method_status_colours.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(extrafont)
3 |
4 | dynbenchmark:::install_fontawesome()
5 |
6 | extrafont::loadfonts()
7 |
8 | method_statuses <- tribble(
9 | ~method_status, ~colour, ~fa, ~icon_colour,
10 | "execution_error", "#9d0549", "\uf00d", "white",
11 | "method_error", "#d51d14", "\uf057", "white",
12 | "memory_limit", "#f46b21", "\uf538", "black",
13 | "time_limit", "#f1be21", "\uf017", "black",
14 | "missing_prior", "#fff600", "\uf0cb", "black",
15 | "low_correlation", "#b4ef43", "\uf537", "black",
16 | "no_error", "#2ECC40", "\uf058", "black"
17 | ) %>%
18 | mutate(method_status = fct_inorder(method_status))
19 |
20 | method_status_colours <-
21 | method_statuses %>% deframe()
22 |
23 | devtools::use_data(method_statuses, method_status_colours, pkg = "package", overwrite = TRUE)
24 |
25 | ggplot(method_statuses, aes(y = fct_rev(method_status))) +
26 | geom_tile(aes(x = 1, fill = method_status), size = 1, height = .9, width = .9) +
27 | geom_text(aes(x = 1, label = fa, colour = method_status), family = "Font Awesome 5 Free", size = 8) +
28 | geom_text(aes(x = 0, label = method_status), hjust = 1, size = 8) +
29 | scale_fill_manual(values = method_status_colours) +
30 | scale_colour_manual(values = method_statuses %>% select(method_status, icon_colour) %>% deframe()) +
31 | expand_limits(x = -2) +
32 | coord_equal() +
33 | cowplot::theme_nothing()
34 |
--------------------------------------------------------------------------------
/package/man/qsub_pmap.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/qsub.R
3 | \name{qsub_pmap}
4 | \alias{qsub_pmap}
5 | \title{pmap, but with qsub ^_^}
6 | \usage{
7 | qsub_pmap(.x, .f, ...)
8 | }
9 | \arguments{
10 | \item{.x}{Vectors of the same length. A vector of length 1 will
11 | be recycled.}
12 |
13 | \item{.f}{A function, formula, or vector (not necessarily atomic).
14 |
15 | If a \strong{function}, it is used as is.
16 |
17 | If a \strong{formula}, e.g. \code{~ .x + 2}, it is converted to a function. There
18 | are three ways to refer to the arguments:
19 | \itemize{
20 | \item For a single argument function, use \code{.}
21 | \item For a two argument function, use \code{.x} and \code{.y}
22 | \item For more arguments, use \code{..1}, \code{..2}, \code{..3} etc
23 | }
24 |
25 | This syntax allows you to create very compact anonymous functions.
26 |
27 | If \strong{character vector}, \strong{numeric vector}, or \strong{list}, it is
28 | converted to an extractor function. Character vectors index by
29 | name and numeric vectors index by position; use a list to index
30 | by position and name at different levels. If a component is not
31 | present, the value of \code{.default} will be returned.}
32 |
33 | \item{...}{Params given to \code{\link[qsub:qsub_lapply]{qsub::qsub_lapply()}}}
34 | }
35 | \description{
36 | pmap, but with qsub ^_^
37 | }
38 | \seealso{
39 | qsub::qsub_lapply
40 | }
41 |
--------------------------------------------------------------------------------
/scripts/03-methods/01-gather_methods_information/03-add_quality_control.R:
--------------------------------------------------------------------------------
1 | #' Add QC scores to methods and tools tibble
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(googlesheets)
6 |
7 | experiment("03-methods")
8 |
9 | tools <- read_rds(result_file("tools.rds"))
10 | methods <- read_rds(result_file("methods.rds"))
11 |
12 | # combine with qc scores
13 | tool_qc_scores <- readRDS(result_file("tool_qc_scores.rds"))
14 |
15 | # merge qc scores with tools tibble
16 | tools <- tools %>%
17 | select(-tidyselect::matches("qc_score")) %>%
18 | left_join(tool_qc_scores, by = "tool_id")
19 |
20 | methods <- methods %>%
21 | select(-tidyselect::matches("qc_score")) %>%
22 | left_join(tool_qc_scores, by = c("method_tool_id" = "tool_id"))
23 |
24 | # filter evaluated
25 | methods_evaluated <- methods %>%
26 | filter(method_evaluated)
27 |
28 | tools_evaluated <- tools %>%
29 | filter(method_evaluated)
30 |
31 | # check that all non-control evaluated methods & tools have a QC!
32 | methods_evaluated_nonqc <- methods_evaluated %>% filter(method_source == "tool" & is.na(qc_score))
33 | if (nrow(methods_evaluated_nonqc) != 0) {
34 | stop("Methods ", methods_evaluated_nonqc$id %>% glue::glue_collapse(", "), " dont have a QC score")
35 | }
36 |
37 | # save
38 | write_rds(tools, result_file("tools.rds"))
39 | write_rds(tools_evaluated, result_file("tools_evaluated.rds"))
40 | write_rds(methods, result_file("methods.rds"))
41 | write_rds(methods_evaluated, result_file("methods_evaluated.rds"))
42 |
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02c-simulate_splatter_datasets.R:
--------------------------------------------------------------------------------
1 | #' [Splatter](https://github.com/Oshlack/splatter), simulations of non-linear paths between different states
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(qsub)
6 |
7 | experiment("01-datasets/02-synthetic")
8 |
9 | # remove all datasets
10 | # rm_remote(dataset_file(id = "synthetic/splatter", remote = TRUE), remote = TRUE, recursive = TRUE)
11 |
12 | # generate design
13 | set.seed(1)
14 | design <- crossing(
15 | topology_model = c("linear", "bifurcating", "multifurcating", "binary_tree", "tree"),
16 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
17 | ) %>%
18 | mutate(
19 | path.skew = runif(n(), 0, 1),
20 | path.nonlinearProb = runif(n(), 0, 1),
21 | path.sigmaFac = runif(n(), 0, 1),
22 | bcv.common.factor = runif(n(), 10, 200),
23 | dataset_id = paste0("synthetic/splatter/", topology_model, "_", platform_ix),
24 | seed = sample(1:100000, n())
25 | ) %>%
26 | select(-platform_ix)
27 | write_rds(design, result_file("design_splatter.rds"))
28 |
29 | # simulate datasets
30 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "splatter", wait = F, stop_on_error = FALSE)
31 |
32 | handle <- qsub_pmap(
33 | design,
34 | simulate_splatter,
35 | qsub_config = qsub_config
36 | )
37 |
38 | write_rds(handle, "handle_splatter.rds")
39 |
40 | ##
41 | handle <- read_rds("handle_splatter.rds")
42 | qsub::qsub_retrieve(handle)
43 |
--------------------------------------------------------------------------------
/package/man/make_obj_fun.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_submit.R
3 | \name{make_obj_fun}
4 | \alias{make_obj_fun}
5 | \title{Used for wrapping an evaluation function around a TI method}
6 | \usage{
7 | make_obj_fun(method, metrics, give_priors, seed, noisy = FALSE,
8 | verbose = FALSE)
9 | }
10 | \arguments{
11 | \item{method}{One or more methods. Must be one of:
12 | \itemize{
13 | \item{an object or list of ti_... objects (eg. \code{dynmethods::ti_comp1()}),}
14 | \item{a character vector containing the names of methods to execute (e.g. \code{"scorpius"}),}
15 | \item{a character vector containing dockerhub repositories (e.g. \code{dynverse/paga}), or}
16 | \item{a dynguidelines data frame.}
17 | }}
18 |
19 | \item{metrics}{Which metrics to evaluate. Check \code{dyneval::metrics} for a list of possible metrics.
20 | Passing a custom metric function with format \code{function(dataset, model) { 1 }} is also supported. The name of this function within the list will be used as the name of the metric.}
21 |
22 | \item{give_priors}{All the priors a method is allowed to receive.
23 | Must be a subset of all available priors (\link[dynwrap:priors]{dynwrap::priors}).}
24 |
25 | \item{seed}{A seed to be passed to the TI method.}
26 |
27 | \item{noisy}{Whether or not the metric is noisy or not}
28 |
29 | \item{verbose}{Whether or not to print extra information output}
30 | }
31 | \description{
32 | Used for wrapping an evaluation function around a TI method
33 | }
34 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_mESC-differentation_hayashi.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 | library(GEOquery)
4 | library(Biobase)
5 | options('download.file.method.GEOquery'='curl')
6 |
7 | id <- "real/gold/mESC-differentiation_hayashi"
8 | dataset_preprocessing(id)
9 |
10 | counts_location <- download_dataset_source_file(
11 | "GSE98664_tpm_sailfish_mergedGTF_RamDA_mESC_differentiation_time_course.txt.gz",
12 | "ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE98nnn/GSE98664/suppl/GSE98664_tpm_sailfish_mergedGTF_RamDA_mESC_differentiation_time_course.txt.gz"
13 | )
14 |
15 | counts_all <- read_tsv(counts_location) %>% as.data.frame %>% column_to_rownames("transcript_id") %>% as.matrix() %>% t %>% round()
16 |
17 | cell_info_all <- tibble(
18 | cell_id = rownames(counts_all),
19 | milestone_id = gsub("RamDA_mESC_(.*)_.*", "\\1", cell_id)
20 | )
21 |
22 | milestone_network <- tribble(
23 | ~from, ~to, ~length,
24 | "00h", "12h", 1,
25 | "12h", "24h", 1,
26 | "24h", "48h", 2,
27 | "48h", "72h", 2
28 | ) %>% mutate(directed = TRUE)
29 |
30 |
31 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
32 |
33 | cell_info <- slice(cell_info_all, match(rownames(counts_all), cell_id))
34 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
35 | counts <- counts_all[cell_info$cell_id, ]
36 |
37 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
38 |
39 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
40 |
--------------------------------------------------------------------------------
/scripts/varia/generate_hypotheses_mindmap/mindmap.tex:
--------------------------------------------------------------------------------
1 | \documentclass[a4paper,twocolumn]{minimal}
2 |
3 | \usepackage{bm}
4 | \usepackage[math]{iwona}
5 | \usepackage[T1]{fontenc}
6 | \usepackage[utf8]{inputenc}
7 |
8 | \usepackage{tikz}
9 | \usetikzlibrary{shapes,decorations}
10 | \usepackage{amsmath,amssymb}
11 |
12 |
13 | \usepackage[a4paper,left=1cm,right=1cm,top=1cm,bottom=1cm]{geometry}
14 |
15 | \usetikzlibrary{shapes.multipart}
16 |
17 | \usetikzlibrary{trees}
18 |
19 | \newcommand{\mysection}[3]{%
20 | \nodepart{#1}
21 | \begin{minipage}{.9\linewidth}
22 | \textbf{#2}\\
23 | #3
24 | \end{minipage}
25 | }
26 | \newcommand{\myhypothesis}[7]{%
27 | \definecolor{my_#1}{RGB}{#3}
28 | \begin{tikzpicture}
29 | \node [mybox,draw=my_#1,fill=my_#1!10!white] (#1) {
30 | \mysection{one}{Statement}{#4}
31 | \mysection{two}{Experiment}{#5}
32 | \mysection{three}{Expected results}{#6}
33 | \mysection{four}{Motivation}{#7}
34 | };
35 | \node [mytitle,fill=my_#1] at (#1.north){\textbf{\color{white}#2}};
36 | \end{tikzpicture}\\
37 | }
38 |
39 | \tikzstyle{mybox} = [
40 | fill=black!03, very thick,
41 | rectangle, rounded corners,
42 | inner sep=5pt, inner ysep=5pt,
43 | rectangle split, rectangle split parts=4,
44 | font=\fontsize{10}{11.4}\selectfont]
45 | \tikzstyle{mytitle} = [
46 | anchor=center, fill=black!05,
47 | very thick, rectangle,
48 | yshift=.1cm,
49 | font=\fontsize{14}{16.4}\selectfont]
50 |
51 | \begin{document}
52 |
53 | \input{hypotheses_tikz.tex}
54 |
55 | \end{document}
--------------------------------------------------------------------------------
/scripts/01-datasets/02-synthetic/02b-simulate_prosstt_datasets.R:
--------------------------------------------------------------------------------
1 | #' [PROSSTT](https://github.com/soedinglab/prosstt), expression is sampled from a linear model which depends on pseudotime
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(qsub)
6 |
7 | experiment("01-datasets/02-synthetic")
8 |
9 | # generate design of models, platforms and (randomised) splatter parameters
10 | design <- crossing(
11 | topology_model = c("linear", "bifurcating", "multifurcating", "binary_tree", "tree"),
12 | tibble(platform = select_platforms(10)) %>% mutate(platform_ix = row_number())
13 | ) %>%
14 | mutate(
15 | a = runif(n(), 0.01, 0.1),
16 | intra_branch_tol = runif(n(), 0, 0.9),
17 | inter_branch_tol = runif(n(), 0, 0.9),
18 | alpha = exp(rnorm(n(), log(0.3), log(1.5))),
19 | beta = exp(rnorm(n(), log(2), log(1.5))) + 1
20 | ) %>%
21 | mutate(
22 | dataset_id = paste0("synthetic/prosstt/", topology_model, "_", platform_ix),
23 | seed = sample(1:100000, n())
24 | ) %>%
25 | select(-platform_ix)
26 | write_rds(design, result_file("design_prosstt.rds"))
27 |
28 | # simulate datasets
29 | qsub_config <- override_qsub_config(memory = "10G", max_wall_time = "24:00:00", num_cores = 1, name = "prosstt", wait = F, execute_before = "module load python/x86_64/3.6.5", stop_on_error = FALSE)
30 |
31 | handle <- qsub_pmap(
32 | design,
33 | simulate_prosstt,
34 | qsub_config = qsub_config
35 | )
36 |
37 | write_rds(handle, "handle_prosstt.rds")
38 |
39 | ##
40 | handle <- read_rds("handle_prosstt.rds")
41 | qsub::qsub_retrieve(handle)
42 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_fibroblast-reprogramming_treutlein.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/silver/fibroblast-reprogramming_treutlein")
5 |
6 | txt_location <- download_dataset_source_file(
7 | "GSE67310_iN_data_log2FPKM_annotated.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE67310&format=file&file=GSE67310%5FiN%5Fdata%5Flog2FPKM%5Fannotated%2Etxt%2Egz"
9 | )
10 |
11 | df <- read_tsv(txt_location, col_types = cols(cell_name = "c", assignment = "c", experiment = "c", time_point = "c", .default = "d"))
12 | expression <- df[, -c(1:5)] %>% as.matrix() %>% magrittr::set_rownames(df$cell_name)
13 | cell_info <- df[, c(1:5)] %>% as.data.frame() %>% magrittr::set_rownames(df$cell_name) %>%
14 | rename(
15 | cell_id = cell_name,
16 | milestone_id = assignment
17 | )
18 |
19 | milestone_network = tribble(
20 | ~from, ~to,
21 | "MEF", "d2_intermediate",
22 | "d2_intermediate", "d2_induced",
23 | "d2_induced", "d5_intermediate",
24 | "d5_intermediate", "d5_earlyiN",
25 | "d5_earlyiN", "Neuron",
26 | "d5_earlyiN", "Myocyte"
27 | ) %>% mutate(length = 1, directed = TRUE)
28 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
29 |
30 | counts <- floor(2^expression-1)
31 |
32 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
33 | counts <- counts[cell_info$cell_id, ]
34 |
35 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
36 |
37 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
38 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/01-download_from_sources.R:
--------------------------------------------------------------------------------
1 | #' Downloading the real datasets from their sources (eg. GEO), and constructing the gold standard model, using the helpers in [helpers-download_from_sources](helpers-download_from_sources)
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | experiment("01-datasets/real/run_all_datasets")
7 |
8 | dataset_scripts <- list.files(path = "scripts/01-datasets/01-real/helpers-download_from_sources/", pattern = "^dataset_.*\\.R", full.names = TRUE)
9 | # dataset_scripts <- dataset_scripts[str_detect(dataset_scripts, "mouse-cell-atlas") | str_detect(dataset_scripts, "plass")]
10 |
11 | script_contents <- map_chr(dataset_scripts, read_file)
12 |
13 | qsub_config <- qsub::override_qsub_config(
14 | name = "dynreal",
15 | memory = "30G",
16 | max_wall_time = "02:00:00",
17 | wait = FALSE,
18 | stop_on_error = FALSE
19 | )
20 |
21 | # Make sure all packages are installed on the cluster; i.e. GEOquery, MultiAssayExperiment, tidyverse, and dynbenchmark.
22 | handle <- qsub::qsub_lapply(
23 | script_contents,
24 | function(script_content) {
25 | dataset_script <- tempfile()
26 | readr::write_file(script_content, dataset_script)
27 |
28 | source(dataset_script)
29 |
30 | TRUE
31 | },
32 | qsub_environment = new.env(),
33 | qsub_config = qsub_config
34 | )
35 |
36 | write_rds(handle, "handle.rds")
37 |
38 | ##
39 |
40 | handle <- read_rds("handle.rds")
41 | results <- qsub::qsub_retrieve(handle, wait = TRUE)
42 |
43 | names(results) <- dataset_scripts
44 |
45 | results <- results %>%
46 | keep(is.na) %>%
47 | enframe("dataset_script", "result")
48 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_cellbench_luyitian.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/gold/cellbench")
5 |
6 | rda_location <- download_dataset_source_file(
7 | "9cellmix_qc.RData",
8 | "https://raw.githubusercontent.com/LuyiTian/CellBench_data/master/data/9cellmix_qc.RData"
9 | )
10 |
11 | settings <- list(
12 | list(
13 | object = sce_SC1_qc,
14 | id = "real/gold/cellbench-SC1_luyitian"
15 | ),
16 | list(
17 | object = sce_SC2_qc,
18 | id = "real/gold/cellbench-SC2_luyitian"
19 | ),
20 | list(
21 | object = sce_SC3_qc,
22 | id = "real/gold/cellbench-SC3_luyitian"
23 | ),
24 | list(
25 | object = sce_SC4_qc,
26 | id = "real/gold/cellbench-SC4_luyitian"
27 | )
28 | )
29 |
30 | for (setting in settings) {
31 | cell_info <- colData(setting$object) %>%
32 | as.data.frame() %>%
33 | rownames_to_column("cell_id") %>%
34 | mutate_at(vars(H1975, H2228, HCC827), ~.>0) %>%
35 | filter(traj == "YES") # why isn't this a logical?
36 | cell_info$group_id <- cell_info[, c("H1975", "H2228", "HCC827")] %>% apply(1, function(x) glue::glue_collapse(names(x)[x], ","))
37 |
38 | grouping <- cell_info %>% select(cell_id, group_id) %>% deframe()
39 |
40 | milestone_network <- tribble(
41 | ~from, ~to,
42 | "H1975", "H1975,H2228,HCC827",
43 | "HCC827", "H1975,H2228,HCC827",
44 | "H2228", "H1975,H2228,HCC827"
45 | ) %>% mutate(length = 1, directed = FALSE)
46 |
47 | counts <- t(counts(setting$object))[cell_info$cell_id, ]
48 |
49 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts), setting$id)
50 | }
51 |
--------------------------------------------------------------------------------
/raw/gource/deprecated_repositories/mlr_ti.txt:
--------------------------------------------------------------------------------
1 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/R/ResampleDesc.R
2 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/R/utils_imbalancy.R
3 | 1494411168|Robrecht Cannoodt|A|/libraries/mlr_ti/man/makeResampleDesc.Rd
4 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/NAMESPACE
5 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/Measure.R
6 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/Prediction.R
7 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/RLearner.R
8 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/TITask.R
9 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/helpers.R
10 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/measures.R
11 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/setPredictType.R
12 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/R/zzz.R
13 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/RLearner.Rd
14 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/Task.Rd
15 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/man/ti_geodesic_dist.Rd
16 | 1494420113|Robrecht Cannoodt|A|/libraries/mlr_ti/mlr.Rproj
17 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/NAMESPACE
18 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/Prediction.R
19 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/TITask.R
20 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/measures.R
21 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/R/zzz.R
22 | 1494492034|Robrecht Cannoodt|M|/libraries/mlr_ti/man/Task.Rd
23 | 1494492034|Robrecht Cannoodt|A|/libraries/mlr_ti/mlr_ti.Rproj
24 | 1494493345|Robrecht Cannoodt|M|/libraries/mlr_ti/R/measures.R
25 | 1494494000|Robrecht Cannoodt|D|/libraries/mlr_ti/
26 |
--------------------------------------------------------------------------------
/scripts/11-example_predictions/02-funky.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("11-example_predictions")
5 |
6 | source(scripts_file("helper-funky.R"))
7 |
8 | folder <- "../../dyndocs/funky_cover/data/embeddings"
9 | dir.create(folder, showWarnings = FALSE)
10 |
11 | dataset_ids <- c(
12 | "synthetic/dyntoy/bifurcating_3",
13 | "real/gold/developing-dendritic-cells_schlitzer",
14 | "real/silver/fibroblast-reprogramming_treutlein",
15 | "synthetic/dyntoy/disconnected_1",
16 | "synthetic/dyngen/72",
17 | "synthetic/dyntoy/linear_1",
18 | "synthetic/dyntoy/cyclic_7",
19 | "synthetic/dyntoy/diverging_with_loops_5",
20 | "synthetic/dyntoy/tree_5"
21 | )
22 |
23 | walk(seq_along(dataset_ids), function(dataset_ix) {
24 | dataset_id <- dataset_ids[dataset_ix]
25 |
26 | dataset <- load_dataset(dataset_id)
27 | plot_dimred(dataset)
28 |
29 | dimred <- dyndimred::dimred_landmark_mds(dataset$expression(), ndim = 3)
30 |
31 | cell_positions <- dimred %>%
32 | as.data.frame() %>%
33 | rownames_to_column("cell_id") %>%
34 | mutate_at(vars(comp_1, comp_2, comp_3), dynutils::scale_minmax)
35 | projection <- project_waypoints_multidim(dataset, cell_positions)
36 |
37 |
38 | cell_positions %>%
39 | sample_n(50) %>%
40 | write_csv(fs::path(folder, paste0("embedding_", dataset_ix, ".csv")))
41 |
42 | projection$edges$group = projection$edges %>% group_by(from_milestone_id, to_milestone_id) %>% group_indices()
43 | projection$edges %>%
44 | select(comp_1 = comp_1_from, comp_2 = comp_2_from, comp_3 = comp_3_from, group) %>%
45 | write_csv(fs::path(folder, paste0("trajectory_", dataset_ix, ".csv")))
46 | })
47 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Benchmarking of TI methods on real and synthetic data
3 |
4 | | \# | script/folder | description |
5 | | :- | :---------------------------------------------------------- | :----------------------------------------------------------------------- |
6 | | 1 | [📄`submit_jobs.R`](1-submit_jobs.R) | Submit the jobs to the gridengine cluster |
7 | | 2 | [📄`retrieve_results.R`](2-retrieve_results.R) | Retrieve the results from the gridengine cluster and aggregate the data |
8 | | 3 | [📄`diagnostic_figures.R`](3-diagnostic_figures.R) | Create some diagnostic figures to get an overview of the results |
9 | | 3a | [📄`overall_comparison.R`](3a-overall_comparison.R) | Overall comparison |
10 | | 3b | [📄`time_mem_predictions.R`](3b-time_mem_predictions.R) | Comparison between predicted versus actual running time and memory usage |
11 | | 3c | [📄`normalisation.R`](3c-normalisation.R) | Compare the effect of normalisation on the results |
12 | | 3d | [📄`compare_sources.R`](3d-compare_sources.R) | Compare the different dataset sources |
13 | | | [📄`inspect_identity_timings.R`](inspect_identity_timings.R) | Check different benchmark component timings |
14 |
15 | The results of this experiment are available
16 | [here](https://github.com/dynverse/dynbenchmark_results/tree/master/06-benchmark).
17 |
--------------------------------------------------------------------------------
/package/data-raw/labels.R:
--------------------------------------------------------------------------------
1 | common_labels <- tribble(
2 | ~id, ~long, ~short,
3 | "directed_acyclic_graph", "Directed acyclic graph", "DAG",
4 | "disconnected_directed_graph", "Disconnected directed graph", "DDG",
5 | "n_genes", "Number of genes", "# genes",
6 | "n_cells", "Number of cells", "# cells",
7 | "ngenes", "Number of genes", "# genes",
8 | "ncells", "Number of cells", "# cells",
9 | "n_methods", "Number of methods", "# methods",
10 | "n_tools", "Number of tools", "# tools",
11 | "silver", "Silver standard", "Silver",
12 | "gold", "Gold standard", "Gold",
13 | "gse", "Source(s)", "Source(s)",
14 | "gs", "Gold standard", "Gold standard",
15 | "rf_mse", "Random Forest MSE", "RF MSE",
16 | "maximal_trajectory_type", "Most complex trajectory type", "Trajectory type",
17 | "qc_score", "QC score", "QC score",
18 | "component_id", "Component", "Component",
19 | "p_value", "p-value", "p-value",
20 | "q_value", "Adjusted p-value", "p-value (adj.)",
21 | TRUE, "Yes", "Yes",
22 | FALSE, "No", "No",
23 | "trajectory_type_gold", "Gold standard trajectory type", "Gold trajectory type",
24 | "trajectory_type_predicted", "Predicted trajectory type", "Predicted trajectory type",
25 | "harm_mean", "Harmonic mean", "Harmonic mean",
26 | "dataset_source", "Dataset source", "Dataset source",
27 | "source", "Dataset source", "Dataset source",
28 | "qc", "Quality Control", "QC",
29 | "graph", "Connected graph", "Connected graph",
30 | "offtheshelf", "Off-the-shelf", "Off-the-shelf"
31 | )
32 |
33 |
34 | labels <- bind_rows(
35 | common_labels,
36 | prior_types %>% select(id = prior_id, long = prior_name, short = prior_name)
37 | )
38 |
39 | usethis::proj_set("package")
40 | usethis::use_data(labels, overwrite = TRUE)
41 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_cortical-interneuron-differentiation_frazer.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 | library(GEOquery)
4 | options('download.file.method.GEOquery'='curl')
5 |
6 | dataset_preprocessing("real/silver/cortical-interneuron-differentiation_frazer")
7 |
8 | txt_location <- download_dataset_source_file(
9 | "GSE90860_SupplementaryData1_revised_julien.tsv.gz",
10 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE90860&format=file&file=GSE90860%5FSupplementaryData1%5Frevised%5Fjulien%2Etsv%2Egz"
11 | )
12 |
13 | counts_df <- read_tsv(txt_location)
14 | counts <- counts_df %>% select(-X1) %>% as.matrix %>% t %>% magrittr::set_colnames(counts_df$X1)
15 |
16 | geo <- GEOquery::getGEO(GEO = "GSE90860", destdir = dataset_source_file(""))
17 | cell_info <- geo[[1]] %>%
18 | Biobase::phenoData() %>%
19 | as("data.frame") %>%
20 | mutate(
21 | cell_id = rownames(counts),
22 | milestone_id = paste0(gsub("age: (.*)", "\\1", characteristics_ch1), "#", gsub("assigned_subgroup: (.*)", "\\1", characteristics_ch1.1))
23 | ) %>%
24 | select(cell_id, milestone_id) %>%
25 | mutate_all(funs(as.character))
26 |
27 | milestone_network <- tribble(
28 | ~from, ~to, ~length,
29 | "E18#0", "P2#1", 4,
30 | "E18#0", "P2#2", 4,
31 | "E18#0", "P2#3", 4,
32 | "P2#1", "P5#1", 3,
33 | "P2#2", "P5#2", 3,
34 | "P2#3", "P5#3", 3
35 | ) %>% mutate(directed = TRUE)
36 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
37 |
38 | cell_info <- cell_info %>% filter(milestone_id %in% milestone_ids)
39 | counts <- counts[cell_info$cell_id, ]
40 |
41 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
42 |
43 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
44 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/3-diagnostic_figures.R:
--------------------------------------------------------------------------------
1 | #' Create some diagnostic figures to get an overview of the results
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(dynplot)
6 |
7 | experiment("06-benchmark")
8 |
9 |
10 | ############################################################
11 | ############### PART THREE: GENERATE FIGURES ###############
12 | ############################################################
13 |
14 | num_replicates <- 1
15 |
16 | list2env(read_rds(result_file("benchmark_results_input.rds")), environment())
17 | list2env(read_rds(result_file("benchmark_results_normalised.rds")), environment())
18 |
19 | metrics_info <- dyneval::metrics %>%
20 | slice(match(metrics, metric_id)) %>%
21 | add_row(metric_id = "overall", plotmath = "overall", latex = "\\textrm{overall}", html = "overall", long_name = "Overall score", category = "average", type = "overall", perfect = 1, worst = 0)
22 |
23 | # get ordering of methods
24 | method_ord <-
25 | data_aggregations %>%
26 | filter(dataset_source == "mean", dataset_trajectory_type == "overall") %>%
27 | arrange(desc(overall)) %>%
28 | select(method_id, method_name) %>%
29 | mutate_all(factor)
30 |
31 | # create method_id_f factor in all data structures
32 | data_aggregations <- data_aggregations %>% mutate(method_id = factor(method_id, levels = method_ord$method_id), method_name = factor(method_name, levels = method_ord$method_name))
33 | data <- data %>% mutate(method_id = factor(method_id, levels = method_ord$method_id), method_name = factor(method_name, levels = method_ord$method_name))
34 |
35 | # execute plotting scripts
36 | source(scripts_file("3a-overall_comparison.R"))
37 | source(scripts_file("3b-time_mem_predictions.R"))
38 | source(scripts_file("3c-normalisation.R"))
39 | source(scripts_file("3d-compare_sources.R"))
40 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_developing-dendritic-cells_schlitzer.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 | library(GEOquery)
4 | options('download.file.method.GEOquery'='curl')
5 |
6 | dataset_preprocessing("real/gold/developing-dendritic-cells_schlitzer")
7 |
8 | # download and untar files
9 | file <- download_dataset_source_file("GSE60781_RAW.tar", "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE60781&format=file")
10 |
11 | untar(file, exdir = dataset_source_file(""))
12 |
13 | # read counts
14 | counts <- map_df(list.files(dataset_source_file(""), ".*read[cC]ount"), function(filename) {
15 | read_tsv(dataset_source_file(filename), col_names = c("gene", "count"), col_types = cols(gene = "c", count = "i")) %>%
16 | mutate(sample = gsub("_.*", "", filename))
17 | }) %>%
18 | spread(gene, count) %>%
19 | as.data.frame() %>%
20 | magrittr::set_rownames(NULL) %>%
21 | column_to_rownames("sample") %>%
22 | as.matrix
23 |
24 | counts <- counts[, !(colnames(counts) %in% c("alignment_not_unique", "no_feature", "ambiguous"))]
25 |
26 | # download cell info
27 | geo <- GEOquery::getGEO("GSE60781", destdir = dataset_source_file(""))
28 | cell_info <- geo[[1]] %>%
29 | Biobase::phenoData() %>%
30 | as("data.frame") %>%
31 | mutate(milestone_id = gsub("_[0-9]*", "", title)) %>%
32 | select(cell_id = geo_accession, milestone_id) %>%
33 | slice(match(rownames(counts), cell_id))
34 |
35 | milestone_network = tribble(
36 | ~from, ~to,
37 | "MDP", "CDP",
38 | "CDP", "PreDC"
39 | ) %>% mutate(length = 1, directed = TRUE)
40 |
41 | milestone_ids <- c("MDP", "CDP", "PreDC")
42 |
43 | cell_ids <- cell_info$cell_id
44 |
45 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
46 |
47 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts))
48 |
--------------------------------------------------------------------------------
/scripts/varia/gource/2_gource_gits.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | experiment("gource")
5 |
6 | gitlinks_folder <- derived_file("git_links/")
7 | output_folder <- derived_file("git_output/")
8 |
9 | dir.create(output_folder)
10 |
11 | git_df <-
12 | readr::read_lines("../.gitmodules") %>%
13 | keep(~ grepl("submodule", .)) %>%
14 | gsub(".*\"(.*)\".*", "\\1", .) %>%
15 | data_frame(path = ., prefix = paste0("/", .), filename = str_replace(., "/", "_"), gitmodule = TRUE) %>%
16 | add_row(path = "..", prefix = "", filename = "dynverse", gitmodule = FALSE) %>%
17 | add_row(path = "../../dyndocs", prefix = "/dyndocs", filename = "dyndocs", gitmodule = FALSE) %>%
18 | add_row(path = "../../projects/qsub/", prefix = "/libraries/qsub", filename = "libraries_qsub", gitmodule = FALSE)
19 |
20 | unlink(gitlinks_folder, recursive = T)
21 | for (i in seq_len(nrow(git_df))) {
22 | path <- git_df$path[[i]]
23 | prefix <- git_df$prefix[[i]]
24 | filename <- git_df$filename[[i]]
25 | gitmodule <- git_df$gitmodule[[i]]
26 |
27 | gource_output <- paste0(
28 | output_folder, "/",
29 | filename,
30 | ".txt"
31 | )
32 |
33 | if (gitmodule) {
34 | dir.create(paste0(gitlinks_folder, path), recursive = T, showWarnings = F)
35 | file.symlink(
36 | normalizePath(paste0("../.git/modules/", path)),
37 | paste0(gitlinks_folder, path, "/.git")
38 | )
39 | git_location <- paste0(gitlinks_folder, path)
40 | } else {
41 | git_location <- path
42 | }
43 | cmd <- paste0("gource --output-custom-log ", gource_output, " ", git_location)
44 | system(cmd)
45 |
46 | read_lines(gource_output) %>%
47 | str_replace("\\|/", paste0("|", prefix, "/")) %>%
48 | str_replace("\\.REMOVED\\.git-id$", "") %>%
49 | write_lines(gource_output)
50 | }
51 | unlink(gitlinks_folder, recursive = T)
52 |
--------------------------------------------------------------------------------
/scripts/02-metrics/02-metric_conformity/01-generate_reference_datasets.R:
--------------------------------------------------------------------------------
1 | #' Generation of toy datasets used as reference to assess metric conformity
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(dyntoy)
6 |
7 | experiment("02-metrics/02-metric_conformity")
8 |
9 | # construct all different combinations of topologies, # cells, ...
10 | dataset_design <-
11 | crossing(
12 | bind_rows(
13 | dynbenchmark:::topologies_with_same_n_milestones %>% enframe("topology_id", "topology_model") %>% mutate(allow_tented_progressions = FALSE),
14 | tribble(
15 | ~topology_id, ~topology_model, ~allow_tented_progressions,
16 | "bifurcation_simple", dyntoy::model_bifurcating(), FALSE,
17 | "bifurcation_tented", dyntoy::model_bifurcating(), TRUE
18 | )
19 | ),
20 | num_cells = c(10, 20, 50, 100, 200, 500),
21 | repeat_ix = 1,
22 | cell_positioning = c("edges", "milestones")
23 | ) %>%
24 | mutate(
25 | dataset_id = as.character(str_glue("{topology_id}_{num_cells}_{cell_positioning}_{repeat_ix}")),
26 | seed = repeat_ix
27 | )
28 |
29 | dataset_design$dataset <- pmap(dataset_design, function(dataset_id, topology_model, num_cells, seed, cell_positioning, allow_tented_progressions, ...) {
30 | function() {
31 | set.seed(seed)
32 |
33 | dataset <- dyntoy::generate_dataset(
34 | id = dataset_id,
35 | model = topology_model,
36 | num_cells = num_cells,
37 | num_features = 200,
38 | allow_tented_progressions = allow_tented_progressions,
39 | add_prior_information = FALSE,
40 | normalise = FALSE
41 | )
42 |
43 | if (cell_positioning == "milestones") {
44 | dataset <- dataset %>% dynwrap::gather_cells_at_milestones()
45 | }
46 |
47 | dataset
48 | }
49 | })
50 |
51 | write_rds(dataset_design, result_file("dataset_design.rds"))
52 |
--------------------------------------------------------------------------------
/scripts/06-benchmark/3b-time_mem_predictions.R:
--------------------------------------------------------------------------------
1 | #' Comparison between predicted versus actual running time and memory usage
2 |
3 | join <-
4 | data %>%
5 | filter(error_status %in% c("no_error", "time_limit", "memory_limit")) %>%
6 | mutate(
7 | time = ifelse(error_status == "memory_limit", NA, time),
8 | mem = ifelse(error_status == "time_limit" | mem < log10(100e6), NA, mem),
9 | ltime = log10(time),
10 | lmem = log10(mem)
11 | )
12 |
13 | range_time <- quantile(abs(join$ltime - join$time_lpred), .9, na.rm = TRUE) %>% {2 * c(-., .)}
14 | range_mem <- quantile(abs(join$lmem - join$mem_lpred), .9, na.rm = TRUE) %>% {2 * c(-., .)}
15 |
16 | g1 <-
17 | ggplot(join) +
18 | geom_point(aes(time_lpred, ltime, colour = ltime - time_lpred)) +
19 | theme_bw() +
20 | facet_wrap(~method_id, scales = "free") +
21 | scale_color_distiller(palette = "RdYlBu", limits = range_time)
22 |
23 | g2 <-
24 | ggplot(join) +
25 | geom_point(aes(mem_lpred, lmem, colour = lmem - mem_lpred)) +
26 | theme_bw() +
27 | facet_wrap(~method_id, scales = "free") +
28 | scale_color_distiller(palette = "RdYlBu", limits = range_mem)
29 |
30 | pdf(result_file("compare_pred_ind.pdf"), width = 20, height = 15)
31 | print(g1 + labs(title = "Timings"))
32 | print(g2 + labs(title = "Memory"))
33 | dev.off()
34 |
35 |
36 | g1 <- ggplot(join) +
37 | geom_point(aes(time_lpred, ltime, colour = ltime - time_lpred)) +
38 | theme_bw() +
39 | scale_color_distiller(palette = "RdYlBu", limits = range_time) +
40 | theme(legend.position = "bottom")
41 | g2 <- ggplot(join %>% filter(lmem > 8)) +
42 | geom_point(aes(mem_lpred, lmem, colour = lmem - mem_lpred)) +
43 | theme_bw() +
44 | scale_color_distiller(palette = "RdYlBu", limits = range_mem) +
45 | theme(legend.position = "bottom")
46 | ggsave(result_file("compare_pred_all.pdf"), patchwork::wrap_plots(g1, g2, nrow = 1), width = 16, height = 8)
47 |
48 | rm(join, g1, g2)
49 |
--------------------------------------------------------------------------------
/package/man/plot_trajectory_types.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/plotting_trajectory_types.R
3 | \name{plot_trajectory_types}
4 | \alias{plot_trajectory_types}
5 | \title{Plot a trajectory type}
6 | \usage{
7 | plot_trajectory_types(plot = ggplot() + theme_void(), trajectory_types,
8 | xmins = 0, xmaxs = 1, ymins = 0, ymaxs = 1,
9 | node_colours = NULL, edge_colours = NULL, size = 4,
10 | geom = c("point", "circle"), circ_size = ifelse(geom == "circle",
11 | 0.05, 0))
12 | }
13 | \arguments{
14 | \item{plot}{Existing plot}
15 |
16 | \item{trajectory_types}{One or more trajectory types}
17 |
18 | \item{xmins}{Minimal x bound (length 1 or length of \code{trajectory_types})}
19 |
20 | \item{xmaxs}{Maximal y bound (likewise)}
21 |
22 | \item{ymins}{Minimal y bound (likewise)}
23 |
24 | \item{ymaxs}{Maximal y bound (likewise)}
25 |
26 | \item{node_colours}{The colours of the nodes.
27 | If \code{NULL}, the colours provided by \code{dynwrap::trajectory_types} will be used (recommended).
28 | Otherwise, \code{node_colours} is expected to be of length 1 or length of \code{trajectory_types}.}
29 |
30 | \item{edge_colours}{The colours of the edges.
31 | If \code{NULL}, the edges will be coloured black.
32 | Otherwise, \code{edge_colours} is expected to be of length 1 or length of \code{trajectory_types}.}
33 |
34 | \item{size}{Size of the arrows and nodes}
35 |
36 | \item{geom}{Which underlying geom to use. When using \code{"point"}, it is expected to also be
37 | using \code{scale_colour_identity()} with your plot. The nodes will go slightly outside the (x, y) boundaries,
38 | because the size of the points depend on the plot size. When using \code{"circle"}, it is expected to be using
39 | \code{scale_fill_identity()} and \code{coord_equal()}, otherwise the nodes might be elliptical.`}
40 |
41 | \item{circ_size}{Size of nodes, only used when \code{geom == "circle"}.}
42 | }
43 | \description{
44 | Plot a trajectory type
45 | }
46 |
--------------------------------------------------------------------------------
/scripts/09-guidelines/01-aggregate.R:
--------------------------------------------------------------------------------
1 | #' Aggregation of the different bnchmarking results for dynguidelines
2 | #' One big methods tibble is created, which contains the information for each method in each row
3 | #' This tibble is nested if a particular experiment generates multiple data points for a method
4 | #' Otherwise, the result is given by using {experiment_id}_{metric_id}
5 |
6 | library(dynbenchmark)
7 | library(tidyverse)
8 |
9 | experiment("09-guidelines")
10 |
11 | # gathers scaling, methods, benchmark results
12 | methods_aggr <- read_rds(result_file("results.rds", experiment_id = "08-summary"))
13 |
14 | # temporary fix for stringlytyped scaling columns
15 | methods_aggr <- methods_aggr %>%
16 | mutate_at(vars(matches("scaling_pred_(time|mem)_")), as.numeric)
17 |
18 | # benchmarking metrics
19 | benchmark_metrics <- dynbenchmark::metrics_evaluated %>% filter(type != "overall")
20 | benchmark_results <- read_rds(result_file("benchmark_results_unnormalised.rds", experiment_id = "06-benchmark"))
21 | benchmark <- benchmark_results %>%
22 | select(-prior_df) %>%
23 | select(method_id, dataset_id, dataset_trajectory_type, !!!benchmark_metrics$metric_id) %>%
24 | mutate_if(is.character, factor) %>%
25 | nest(-method_id, .key = "benchmark")
26 |
27 | methods_aggr <- methods_aggr %>%
28 | left_join(benchmark, "method_id")
29 |
30 | # benchmarking datasets
31 | benchmark_datasets_info <- load_datasets(unique(benchmark_results$dataset_id)) %>%
32 | select(id, source, trajectory_type)
33 |
34 | # metrics
35 | benchmark_metrics <- dyneval::metrics %>%
36 | filter(metric_id %in% c("correlation", "him", "F1_branches", "featureimp_wcor"))
37 | benchmark_metrics$description <- "todo"
38 |
39 | withr::with_dir(
40 | "../dynguidelines",
41 | {
42 | usethis::use_data(
43 | methods_aggr,
44 | benchmark_datasets_info,
45 | benchmark_metrics,
46 | internal = TRUE,
47 | compress = "xz",
48 | overwrite = TRUE
49 | )
50 | }
51 | )
52 |
53 |
--------------------------------------------------------------------------------
/package/man/paramoptim_generate_design.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/suite_paramoptim_generate_design.R
3 | \name{paramoptim_generate_design}
4 | \alias{paramoptim_generate_design}
5 | \title{Generate a benchmarking design}
6 | \usage{
7 | paramoptim_generate_design(datasets, methods, priors = NULL,
8 | num_repeats = 1, crossing = NULL)
9 | }
10 | \arguments{
11 | \item{datasets}{The datasets to be used in the evaluation.
12 | Must be a named list consisting of dataset ids (character),
13 | dynwrap::data_wrapper's, or functions that generate dynwrap::data_wrapper's.}
14 |
15 | \item{methods}{The methods to be evaluated.
16 | Must be a named list consisting of method_ids (character) or dynwrap::ti_wrapper's.}
17 |
18 | \item{priors}{A list of lists. Each sublist contains a list of priors that each method is allowed to optionally use.
19 | Check \code{\link[dynwrap:priors]{dynwrap::priors}} for a list of possible priors. Default priors given is "none"}
20 |
21 | \item{num_repeats}{The number of times to repeat the evaluation.}
22 |
23 | \item{crossing}{A data frame containing the combinations of methods, datasets and priors to be evaluated.
24 | Must be a data frame containing the columns dataset_id, method_id, prior_id, repeat_ix and param_id}
25 | }
26 | \description{
27 | Generate a benchmarking design
28 | }
29 | \examples{
30 | \dontrun{
31 | library(tibble)
32 |
33 | datasets <- list(
34 | "synthetic/dyntoy/bifurcating_1",
35 | dyntoy::generate_dataset(id = "test1"),
36 | test2 = function() dyntoy::generate_dataset()
37 | )
38 |
39 | methods <- list(
40 | "comp1",
41 | dynmethods::ti_scorpius(),
42 | "tscan"
43 | )
44 |
45 | priors <- list(
46 | list(id = "none", method_id = c("comp1", "scorpius", "tscan")),
47 | list(id = "some", method_id = c("tscan"), set = c("start_id", "end_n"))
48 | )
49 |
50 | paramoptim_generate_design(
51 | datasets = datasets,
52 | methods = methods,
53 | priors = priors,
54 | num_repeats = 2
55 | )
56 | }
57 | }
58 |
--------------------------------------------------------------------------------
/scripts/10-benchmark_interpretation/02-dataset_sources.R:
--------------------------------------------------------------------------------
1 | #' Similarity of the results across dataset sources
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 |
6 | experiment("10-benchmark_interpretation")
7 |
8 | methods <- load_methods()
9 | data <- read_rds(result_file("benchmark_results_normalised.rds", "06-benchmark"))$data %>% filter(method_id %in% methods$id)
10 |
11 | # aggregate without errors
12 | data_aggregations <- benchmark_aggregate(
13 | data = data %>% filter(error_status == "no_error")
14 | )$data_aggregations
15 |
16 | overall_dataset_source_scores <- data_aggregations %>%
17 | filter(dataset_trajectory_type == "overall", dataset_source != "mean") %>%
18 | select(method_id, dataset_source, overall) %>%
19 | spread(dataset_source, overall)
20 |
21 | gold_dataset_source_scores <- overall_dataset_source_scores %>%
22 | gather("dataset_source", "dataset_source_score", -method_id, -`real/gold`)
23 |
24 | correlation_dataset_source_scores <- gold_dataset_source_scores %>%
25 | group_by(dataset_source) %>%
26 | summarise(cor = nacor(`real/gold`, dataset_source_score)) %>%
27 | arrange(-cor) %>%
28 | mutate(dataset_source = fct_inorder(dataset_source))
29 |
30 | plot_dataset_source_correlation <- gold_dataset_source_scores %>%
31 | mutate(dataset_source = factor(dataset_source, levels = levels(correlation_dataset_source_scores$dataset_source))) %>%
32 | ggplot(aes(dataset_source_score, `real/gold`)) +
33 | geom_abline(intercept = 0, slope = 1) +
34 | geom_point() +
35 | geom_label(aes(label = sprintf("corr = %0.2f", cor)), x = 0.1, y = 0.9, data = correlation_dataset_source_scores, vjust = 0, hjust = 0) +
36 | facet_grid(.~dataset_source) +
37 | scale_x_continuous(limits = c(0, 1)) +
38 | scale_y_continuous(limits = c(0, 1)) +
39 | labs(x = "Overall score on datasets from source", y = "Overall score on real/gold datasets") +
40 | theme_pub()
41 |
42 | plot_dataset_source_correlation
43 |
44 | write_rds(plot_dataset_source_correlation, derived_file("dataset_source_correlation.rds"))
45 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/02-filter_and_normalise.R:
--------------------------------------------------------------------------------
1 | #' Filtering and normalising the real datasets using `dynbenchmark::process_raw_dataset` All datasets are then saved into the dynwrap format.
2 |
3 | library(dynbenchmark)
4 | library(tidyverse)
5 | library(qsub)
6 |
7 | run_remote("find . -maxdepth 2 -type d -ls", args = c("-2", derived_file(experiment_id = '01-datasets_preproc/raw/real', remote = T)), remote = T)$stdout
8 |
9 | # determine datasets to normalise & filter
10 | list_raw_datasets_remote <- function() {
11 | run_remote(
12 | "find",
13 | args = c(
14 | derived_file(experiment_id = '01-datasets_preproc/raw/real', remote = T),
15 | "-maxdepth", "2",
16 | "-type", "f"
17 | ),
18 | remote = T
19 | )$stdout %>%
20 | gsub(".*(real/.*)\\.rds", "\\1", .)
21 | }
22 |
23 | list_datasets_remote <- function() {
24 | run_remote(
25 | "find",
26 | args = c(
27 | derived_file(experiment_id = '01-datasets/real', remote = T),
28 | "-maxdepth", "3",
29 | "-type", "f",
30 | "-regex", "'.*dataset\\.rds'"
31 | ),
32 | remote = T
33 | )$stdout %>%
34 | gsub(".*(real/.*)/dataset\\.rds", "\\1", .)
35 | }
36 |
37 | dataset_ids_to_process <- setdiff(list_raw_datasets_remote(), list_datasets_remote())
38 |
39 | process_raw_dataset_wrapper <- function(id) {
40 | raw_dataset <- readr::read_rds(dynbenchmark::dataset_raw_file(id))
41 | raw_dataset$id <- id
42 |
43 | do.call(dynbenchmark::process_raw_dataset, raw_dataset)
44 | TRUE
45 | }
46 |
47 | qsub_config <- qsub::override_qsub_config(
48 | name = "filter_norm",
49 | memory = "30G",
50 | wait = FALSE,
51 | stop_on_error = TRUE
52 | )
53 |
54 | handle <- qsub_lapply(dataset_ids_to_process, process_raw_dataset_wrapper, qsub_environment = character(), qsub_config = qsub_config)
55 |
56 |
57 | # sync locally
58 | local_folder_sync <- derived_file("", "01-datasets/real/")
59 | remote_folder_sync <- derived_file("", "01-datasets/real/", remote = TRUE)
60 |
61 | qsub::rsync_remote("prism", remote_folder_sync, "", local_folder_sync)
62 |
--------------------------------------------------------------------------------
/package/R/reformat_tribbles.R:
--------------------------------------------------------------------------------
1 | #' Align tribbles in a file
2 | #'
3 | #' The start and end of a tribble need to be marked with a commend `# tribble_start` and `# tribble_end`.
4 | #'
5 | #' @param script_file The file in which to align the tribble
6 | #' @param align The alignment (left, center, right)
7 | #'
8 | #' @importFrom readr read_lines write_lines
9 | #' @export
10 | reformat_tribbles <- function(script_file, align = c("left", "center", "right")) {
11 | align <- match.arg(align)
12 |
13 | pad_fun <- function(word, width) {
14 | padding <- width - nchar(word)
15 | nleft <- switch(
16 | align,
17 | left = 0,
18 | center = floor(padding / 2),
19 | right = padding
20 | )
21 | nright = switch(
22 | align,
23 | left = padding,
24 | center = ceiling(padding / 2),
25 | right = 0
26 | )
27 | paste(c(rep(" ", nleft), word, rep(" ", nright)), collapse = "")
28 | }
29 |
30 | lines <- readr::read_lines(script_file)
31 | tribble_start <- grep("tribble_start", lines)
32 | tribble_end <- grep("tribble_end", lines)
33 | for (i in seq_along(tribble_start)) {
34 | starti <- tribble_start[[i]] + 1
35 | endi <- tribble_end[[i]] - 1
36 | subset <-
37 | lines[starti:endi] %>%
38 | gsub(" *([\"`][^\"`]*[\"`]|[a-zA-Z_]*\\([^\\)]*\\)|[^ ,]*),? *", "\\1\U00B0", .) %>%
39 | map(~ strsplit(., "\U00B0")[[1]])
40 |
41 | numcols <- length(subset[[1]])
42 |
43 | maxwidths <- map_int(seq_len(numcols), function(j) {
44 | subset %>% map_chr(~ .[[j]]) %>% map_int(nchar) %>% max
45 | }) + 3
46 |
47 | new_subset <-
48 | map_chr(subset, function(line) {
49 | map2_chr(paste0(line, ","), maxwidths, pad_fun) %>%
50 | paste(collapse = "") %>%
51 | paste0(" ", .) %>%
52 | gsub(" *$", "", .)
53 | })
54 |
55 | new_subset[length(new_subset)] <-
56 | new_subset[length(new_subset)] %>%
57 | gsub(", *$", "", .)
58 |
59 | lines[starti:endi] <- new_subset
60 | }
61 | readr::write_lines(lines, script_file)
62 | }
63 |
--------------------------------------------------------------------------------
/raw/gource/deprecated_repositories/dynvaria.txt:
--------------------------------------------------------------------------------
1 | 1490793919|Robrecht Cannoodt|A|/dynvaria/.gitignore
2 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/experiments.Rproj
3 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/hypotheses_script.R
4 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/hypotheses_tikz.tex
5 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/mindmap.pdf
6 | 1490793919|Robrecht Cannoodt|A|/dynvaria/mindmap/mindmap.tex
7 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_hypotheses-and-tasks_clean.xml
8 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_hypotheses-and-tasks_dirty.xml
9 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_methods_clean.xml
10 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/20170917_methods_dirty.xml
11 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/experiments.Rproj
12 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/gource.txt
13 | 1505671561|Robrecht Cannoodt|A|/dynvaria/drive/process.R
14 | 1505671726|Robrecht Cannoodt|M|/dynvaria/drive/gource.txt
15 | 1505671742|Robrecht Cannoodt|M|/dynvaria/drive/process.R
16 | 1505672668|Robrecht Cannoodt|M|/dynvaria/drive/gource.txt
17 | 1505672668|Robrecht Cannoodt|M|/dynvaria/drive/process.R
18 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_hypotheses-and-tasks_clean.xml
19 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_hypotheses-and-tasks_dirty.xml
20 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_methods_clean.xml
21 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/20170917_methods_dirty.xml
22 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/experiments.Rproj
23 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/gource.txt
24 | 1506950076|Robrecht Cannoodt|D|/dynvaria/drive/process.R
25 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/experiments.Rproj
26 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/hypotheses_script.R
27 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/hypotheses_tikz.tex
28 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/mindmap.pdf
29 | 1506950076|Robrecht Cannoodt|D|/dynvaria/mindmap/mindmap.tex
30 | 1506950076|Robrecht Cannoodt|D|/dynvaria/
31 |
--------------------------------------------------------------------------------
/scripts/01-datasets/00-download_from_zenodo.R:
--------------------------------------------------------------------------------
1 | #' Downloading the processed datasets from Zenodo ([10.5281/zenodo.1443566](https://doi.org/10.5281/zenodo.1443566))
2 |
3 | library(tidyverse)
4 | library(dynbenchmark)
5 | library(httr)
6 | library(Matrix)
7 |
8 | experiment("01-datasets")
9 |
10 | # download the rds files from zenodo
11 | deposit_id <- 1443566
12 |
13 | files <-
14 | GET(glue::glue("https://zenodo.org/api/records/{deposit_id}")) %>%
15 | httr::content() %>%
16 | .$files %>%
17 | map(unlist) %>%
18 | list_as_tibble() %>%
19 | mutate(
20 | dataset_id = str_replace(filename, "\\.rds$", ""),
21 | dest_folder = map_chr(dataset_id, derived_file)
22 | )
23 |
24 | pwalk(
25 | files,
26 | function(checksum, filename, filesize, links.download, dataset_id, dest_folder, ...) {
27 | download_info <- paste0(dest_folder, "/download_info.json")
28 | if (file.exists(download_info)) {
29 | didata <- jsonlite::read_json(download_info, simplifyVector = TRUE)
30 |
31 | if (didata$checksum == checksum) {
32 | return()
33 | }
34 | }
35 |
36 | tmp_download_file <- tempfile()
37 | on.exit(file.remove(tmp_download_file))
38 | download.file(links.download, tmp_download_file)
39 |
40 | if (tools::md5sum(tmp_download_file) != checksum) {
41 | stop("MD5 checksum after downloading file '", dataset_id, "' is wrong!\nAborting downloading procedure.")
42 | }
43 |
44 | dataset <- read_rds(tmp_download_file)
45 | dataset$expression <- as(dataset$expression, "dgCMatrix")
46 | dataset$counts <- as(dataset$counts, "dgCMatrix")
47 |
48 | save_dataset(dataset, id = dataset_id)
49 | jsonlite::write_json(
50 | lst(
51 | deposit_id,
52 | dataset_id,
53 | checksum,
54 | downloaded_on = as.character(Sys.time())
55 | ),
56 | download_info
57 | )
58 | }
59 | )
60 |
61 | # upload to remote, if relevant
62 | # this assumes you have a gridengine cluster at hand
63 | qsub::rsync_remote(
64 | remote_src = FALSE,
65 | path_src = derived_file(),
66 | remote_dest = TRUE,
67 | path_dest = derived_file(remote = TRUE)
68 | )
69 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_stimulated-dendritic-cells_shalek.R:
--------------------------------------------------------------------------------
1 | library(tidyverse)
2 | library(dynbenchmark)
3 |
4 | dataset_preprocessing("real/gold/stimulated-dendritic-cells_shalek")
5 |
6 | file <- download_dataset_source_file(
7 | "GSM1406531_GSM1407094.txt.gz",
8 | "https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE48968&format=file&file=GSE48968%5FallgenesTPM%5FGSM1189042%5FGSM1190902%2Etxt%2Egz"
9 | )
10 | tab <- read.table(gzfile(file), header = TRUE, sep = "\t") %>% as.matrix %>% t
11 |
12 | allcell_info <- data_frame(
13 | cell_id = rownames(tab),
14 | time = ifelse(grepl("_[0-9]+h", cell_id), gsub(".*_([0-9]+h).*", "\\1", cell_id), "0h"),
15 | type = ifelse(grepl("_[0-9]+h", cell_id), gsub("(.*)_[0-9]+h.*", "\\1", cell_id), "Unstimulated"),
16 | sid = ifelse(grepl("_S[0-9]+$", cell_id), gsub("_(S[0-9]+)$", "\\1", cell_id), ""),
17 | milestone_id = paste0(type, "#", time)
18 | ) %>% filter(
19 | type %in% c("LPS", "PAM", "PIC", "Unstimulated")
20 | )
21 |
22 | settings <- lapply(c("LPS", "PAM", "PIC"), function(stim) {
23 | list(
24 | id = stringr::str_glue("real/gold/stimulated-dendritic-cells-{stim}_shalek"),
25 | milestone_network = tribble(
26 | ~from, ~to, ~length,
27 | "Unstimulated#0h", stringr::str_glue("{stim}#1h"), 1,
28 | stringr::str_glue("{stim}#1h"), stringr::str_glue("{stim}#2h"), 1,
29 | stringr::str_glue("{stim}#2h"), stringr::str_glue("{stim}#4h"), 2,
30 | stringr::str_glue("{stim}#4h"), stringr::str_glue("{stim}#6h"), 2
31 | ) %>% mutate(directed = TRUE)
32 | )
33 | })
34 |
35 | tab <- tab[, !grepl("^ERCC", colnames(tab))] # remove spike-ins, are not present in every cell
36 |
37 | for (setting in settings) {
38 | milestone_network <- setting$milestone_network
39 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
40 |
41 | cell_info <- allcell_info %>% filter(milestone_id %in% milestone_ids)
42 |
43 | counts <- tab[cell_info$cell_id, ]
44 |
45 | grouping <- cell_info %>% select(cell_id, milestone_id) %>% deframe()
46 |
47 | save_raw_dataset(lst(milestone_network, cell_info, grouping, counts), setting$id)
48 | }
49 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/helpers-download_from_sources/dataset_planaria-plass.R:
--------------------------------------------------------------------------------
1 | library(dynbenchmark)
2 | library(tidyverse)
3 |
4 | dataset_preprocessing("real/silver/whole-schmidtea-mediterranea_plass")
5 |
6 | # get settings
7 | source(paste0(dynbenchmark::get_dynbenchmark_folder(), "/scripts/01-datasets/01-real/helpers-download_from_sources/helper_planaria-plass.R"))
8 |
9 | # counts
10 | counts_file <- download_dataset_source_file(
11 | "dge.txt.gz",
12 | "http://bimsbstatic.mdc-berlin.de/rajewsky/PSCA/dge.txt.gz"
13 | )
14 |
15 | counts_file_unzipped <- gsub("\\.gz", "", counts_file)
16 | read_lines(counts_file) %>% {.[[1]] <- paste0("\"gene\"\t", .[[1]]); .} %>% write_lines(counts_file_unzipped)
17 |
18 | counts_all <- read_tsv(counts_file_unzipped) %>%
19 | as.data.frame() %>%
20 | column_to_rownames("gene") %>%
21 | as.matrix() %>%
22 | t
23 |
24 | # cell info
25 | cell_info_file <- download_dataset_source_file(
26 | "R_annotation.txt",
27 | "http://bimsbstatic.mdc-berlin.de/rajewsky/PSCA/R_annotation.txt"
28 | )
29 | cell_info_all <- tibble(group_id = read_lines(cell_info_file))
30 | cell_info_all$cell_id <- rownames(counts_all)
31 |
32 |
33 | for (setting in settings) {
34 | print(setting$id)
35 |
36 | milestone_network <- setting$milestone_network
37 | milestone_ids <- unique(c(milestone_network$from, milestone_network$to))
38 |
39 | if (!all(milestone_ids %in% cell_info_all$group_id)) {stop(setdiff(milestone_ids, cell_info_all$group_id))}
40 |
41 | if (!"length" %in% names(milestone_network)) milestone_network$length <- 1
42 | milestone_network$length[is.na(milestone_network$length)] <- TRUE
43 | if (!"directed" %in% names(milestone_network)) milestone_network$directed <- TRUE
44 | milestone_network$directed[is.na(milestone_network$directed)] <- TRUE
45 |
46 | cell_info <- cell_info_all %>%
47 | filter(group_id %in% milestone_ids)
48 |
49 | grouping <- cell_info %>%
50 | select(cell_id, group_id) %>%
51 | deframe()
52 |
53 | counts <- counts_all[names(grouping), ]
54 |
55 | save_raw_dataset(
56 | lst(
57 | milestone_network,
58 | grouping,
59 | cell_info,
60 | counts
61 | ),
62 | setting$id)
63 | }
64 |
--------------------------------------------------------------------------------
/scripts/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Scripts
3 |
4 | The scripts folder contains all the scripts necessary to fully reproduce
5 | the benchmarking manuscript in chronologically ordered subfolders. Each
6 | subfolder contains a readme file with further explanations of the
7 | different scripts and what they
8 | do:
9 |
10 | | \# | script/folder | description |
11 | | :- | :--------------------------------------------------------- | :---------------------------------------------------- |
12 | | 1 | [📁`datasets`](01-datasets) | Dataset processing and characterisation |
13 | | 2 | [📁`metrics`](02-metrics) | Metrics for comparing two trajectories |
14 | | 3 | [📁`methods`](03-methods) | Trajectory inference methods |
15 | | 4 | [📁`method_testing`](04-method_testing) | Method testing |
16 | | 5 | [📁`scaling`](05-scaling) | Scalability of methods |
17 | | 6 | [📁`benchmark`](06-benchmark) | Benchmarking of TI methods on real and synthetic data |
18 | | 7 | [📁`stability`](07-stability) | Stability |
19 | | 8 | [📁`summary`](08-summary) | Summarising the results into funky heatmaps |
20 | | 9 | [📁`guidelines`](09-guidelines) | Guidelines |
21 | | 10 | [📁`benchmark_interpretation`](10-benchmark_interpretation) | Benchmark interpretation |
22 | | 11 | [📁`example_predictions`](11-example_predictions) | Example predictions |
23 | | 12 | [📁`manuscript`](12-manuscript) | Preparing the manuscript |
24 | | | [📁`varia`](varia) | Varia scripts |
25 |
--------------------------------------------------------------------------------
/scripts/01-datasets/01-real/README.md:
--------------------------------------------------------------------------------
1 |
2 | # Real datasets
3 |
4 | The generation of the real datasets is divided in two parts. We first
5 | download all the (annotated) expression files from sites such as GEO.
6 | Next, we filter and normalise all datasets, and wrap them into the
7 | common trajectory format of
8 | [dynwrap](https://www.github.com/dynverse/dynwrap).
9 |
10 | | \# | script/folder | description |
11 | | :- | :---------------------------------------------------------------- | :------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
12 | | 1 | [📄`download_from_sources.R`](01-download_from_sources.R) | Downloading the real datasets from their sources (eg. GEO), and constructing the gold standard model, using the helpers in [helpers-download\_from\_sources](helpers-download_from_sources) |
13 | | 2 | [📄`filter_and_normalise.R`](02-filter_and_normalise.R) | Filtering and normalising the real datasets using `dynbenchmark::process_raw_dataset` All datasets are then saved into the dynwrap format. |
14 | | 3 | [📄`gather_metadata.R`](03-gather_metadata.R) | Gathers some metadata about all the real datasets |
15 | | 4 | [📄`datasets_table.R`](04-datasets_table.R) | Creates a table of the datasets in, excuse me, excel (for supplementary material) |
16 | | | [📁`helpers-download_from_sources`](helpers-download_from_sources) | |
17 |
--------------------------------------------------------------------------------
/raw/gource/gource_captions:
--------------------------------------------------------------------------------
1 | 2016/04/02|The birth of dynverse (dyngen and qsub)
2 | 2017/01/15|But first, what are our hypotheses regarding this project?
3 | 2017/02/25|Generating good synthetic data is fun
4 | 2017/03/12|Birth of dyneval, taking baby steps in evaluation
5 | 2017/03/20|Planning, planning, planning!
6 | 2017/03/27|Discussing, discussing, discussing!
7 | 2017/04/04|First TI methods are added to dyneval
8 | 2017/05/13|Initial experiments in performing parameter optimisation
9 | 2017/07/05|Collecting meta info on more TI methods
10 | 2017/07/28|Restructuring of dyngen
11 | 2017/07/31|Including code of other frameworks into our repository was not a good idea
12 | 2017/08/10|Cleaning up dyngen
13 | 2017/08/19|Adding wrappers and metrics
14 | 2017/08/23|Creation of dynplot
15 | 2017/08/27|Huge restructuring; bye bye monster matlab code
16 | 2017/09/06|Birth of dynbenchmark
17 | 2017/09/12|Doing many things! (can't keep up)
18 | 2017/09/16|Creation of dyntoy, cleaning up dyngen
19 | 2017/10/04|It's getting real; downloading lots of real datasets
20 | 2017/11/01|Developing code base for setting up experiments
21 | 2017/11/20|Moving TI methods from dyneval to dynmethods
22 | 2017/12/15|Making presentations
23 | 2018/02/01|Writing preprint manuscript
24 | 2018/02/24|Still writing...
25 | 2018/03/05|Submission to bioRxiv!
26 | 2018/03/25|Preparing for submission to NBT
27 | 2018/04/05|Submission to NBT
28 | 2018/04/10|Initial experiments with Docker containerisation
29 | 2018/04/11|First guest appearance: Koen Van den Berge
30 | 2018/04/21|Birth of dynguidelines
31 | 2018/04/28|Creating a decent dynplot
32 | 2018/05/11|Wouter decides logos are a thing
33 | 2018/06/10|Getting the reviews back, writing a rebuttal
34 | 2018/06/20|Creating TI method dockers
35 | 2018/06/21|with guests Philipp Angerer, Manu Setty and Alex Wolf
36 | 2018/07/01|Watch out for a large restructuring of dynbenchmark...
37 | 2018/08/14|Hello guests Daniel C Ellwanger and Chuck Herring!
38 | 2018/08/29|Heavily integrating containerisation into dynmethods
39 | 2018/09/11|Our avatars are getting hard to see
40 | 2018/10/25|Preparing for revision
41 | 2018/11/12|Revision submitted
42 | 2019/02/04|Final revision
43 | 2019/03/21|Preparing for release of dyno v1.0
44 | 2019/04/01|TODAY!
45 |
--------------------------------------------------------------------------------
/package/R/setup_experiment.R:
--------------------------------------------------------------------------------
1 | #' Helper function for controlling experiments
2 | #'
3 | #' @param experiment_id id for the experiment
4 | #' @param filename the filename
5 | #' @param remote The remote to access, "" if working locally, TRUE if using default qsub_config remote
6 | #'
7 | #' @export
8 | #'
9 | #' @rdname experiment
10 | #'
11 | #' @examples
12 | #' \dontrun{
13 | #' experiment("test_plots")
14 | #'
15 | #' data <- matrix(runif(200), ncol = 2)
16 | #' pdf(result_file("testplot.pdf"), 5, 5)
17 | #' plot(data)
18 | #' dev.off()
19 | #' }
20 | experiment <- function(experiment_id) {
21 | # check whether the working directory is indeed the dynbenchmark folder
22 | dyn_fold <- get_dynbenchmark_folder()
23 |
24 | # set option
25 | options(dynbenchmark_experiment_id = experiment_id)
26 | }
27 |
28 | # create a helper function
29 | experiment_subfolder <- function(path) {
30 | function(filename = "", experiment_id = NULL, remote = FALSE) {
31 | filename <- paste0(filename, collapse = "")
32 |
33 | dyn_folder <- get_dynbenchmark_folder(remote = remote)
34 |
35 | # check whether exp_id is given
36 | if (is.null(experiment_id)) {
37 | experiment_id <- getOption("dynbenchmark_experiment_id")
38 | }
39 |
40 | # check whether exp_fold could be found
41 | if (is.null(experiment_id)) {
42 | stop("No experiment folder found. Did you run experiment(...) yet?")
43 | }
44 |
45 | # determine the full path
46 | full_path <- paste0(dyn_folder, "/", path, "/", experiment_id, "/")
47 |
48 | # create if necessary
49 | if (is.logical(remote) && !remote) {
50 | dir.create(full_path, showWarnings = FALSE, recursive = TRUE)
51 | } else {
52 | qsub::mkdir_remote(full_path, remote = remote)
53 | }
54 |
55 | # get complete filename
56 | paste(full_path, filename, sep = "")
57 | }
58 | }
59 |
60 | #' @rdname experiment
61 | #' @export
62 | derived_file <- experiment_subfolder("derived")
63 |
64 | #' @rdname experiment
65 | #' @export
66 | raw_file <- experiment_subfolder("raw")
67 |
68 | #' @rdname experiment
69 | #' @export
70 | result_file <- experiment_subfolder("results")
71 |
72 | #' @rdname experiment
73 | #' @export
74 | scripts_file <- experiment_subfolder("scripts")
75 |
--------------------------------------------------------------------------------