├── bio_plot.png
├── bio_inactive-active_plot.png
├── bio-machine-learning-emirhan-project.jpg
├── LICENSE
├── .gitignore
├── README.md
└── bio-chemist-lab-machine-learning-project.py
/bio_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/emirhanai/AID362-Bioassay-Classification-and-Regression-Neuronal-Network-and-Extra-Tree-with-Machine-Learnin/HEAD/bio_plot.png
--------------------------------------------------------------------------------
/bio_inactive-active_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/emirhanai/AID362-Bioassay-Classification-and-Regression-Neuronal-Network-and-Extra-Tree-with-Machine-Learnin/HEAD/bio_inactive-active_plot.png
--------------------------------------------------------------------------------
/bio-machine-learning-emirhan-project.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/emirhanai/AID362-Bioassay-Classification-and-Regression-Neuronal-Network-and-Extra-Tree-with-Machine-Learnin/HEAD/bio-machine-learning-emirhan-project.jpg
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2021 Emirhan BULUT
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | pip-wheel-metadata/
24 | share/python-wheels/
25 | *.egg-info/
26 | .installed.cfg
27 | *.egg
28 | MANIFEST
29 |
30 | # PyInstaller
31 | # Usually these files are written by a python script from a template
32 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
33 | *.manifest
34 | *.spec
35 |
36 | # Installer logs
37 | pip-log.txt
38 | pip-delete-this-directory.txt
39 |
40 | # Unit test / coverage reports
41 | htmlcov/
42 | .tox/
43 | .nox/
44 | .coverage
45 | .coverage.*
46 | .cache
47 | nosetests.xml
48 | coverage.xml
49 | *.cover
50 | *.py,cover
51 | .hypothesis/
52 | .pytest_cache/
53 |
54 | # Translations
55 | *.mo
56 | *.pot
57 |
58 | # Django stuff:
59 | *.log
60 | local_settings.py
61 | db.sqlite3
62 | db.sqlite3-journal
63 |
64 | # Flask stuff:
65 | instance/
66 | .webassets-cache
67 |
68 | # Scrapy stuff:
69 | .scrapy
70 |
71 | # Sphinx documentation
72 | docs/_build/
73 |
74 | # PyBuilder
75 | target/
76 |
77 | # Jupyter Notebook
78 | .ipynb_checkpoints
79 |
80 | # IPython
81 | profile_default/
82 | ipython_config.py
83 |
84 | # pyenv
85 | .python-version
86 |
87 | # pipenv
88 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
89 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
90 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
91 | # install all needed dependencies.
92 | #Pipfile.lock
93 |
94 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
95 | __pypackages__/
96 |
97 | # Celery stuff
98 | celerybeat-schedule
99 | celerybeat.pid
100 |
101 | # SageMath parsed files
102 | *.sage.py
103 |
104 | # Environments
105 | .env
106 | .venv
107 | env/
108 | venv/
109 | ENV/
110 | env.bak/
111 | venv.bak/
112 |
113 | # Spyder project settings
114 | .spyderproject
115 | .spyproject
116 |
117 | # Rope project settings
118 | .ropeproject
119 |
120 | # mkdocs documentation
121 | /site
122 |
123 | # mypy
124 | .mypy_cache/
125 | .dmypy.json
126 | dmypy.json
127 |
128 | # Pyre type checker
129 | .pyre/
130 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # **AID362 Bioassay Classification and Regression (Neuronal Network and Extra Tree) with Machine Learning**
2 | I developed Machine Learning Software with multiple models that predict and classify AID362 biology lab data. Accuracy values are 99% and above, and F1, Recall and Precision scores are average (average of 3) 78.33%. The purpose of this study is to prove that we can establish an artificial intelligence (machine learning) system in health. With my regression model, you can predict whether it is Inactive or Inactive (Neural Network or Extra Trees). In classification (Neural Network or Extra Trees), you can easily classify the provided data whether it is Inactive or Active.
3 |
4 | _Example:_
5 |
6 | `###Regressor Model
7 |
8 | model_emir_regress_predict = ExtraTreesRegressor(criterion="mse",max_features="auto",
9 | n_jobs=-1,n_estimators=1)
10 |
11 | model_emir_regress_predict = MLPRegressor(hidden_layer_sizes=(200,),activation="relu",
12 | #solver="adam",batch_size="auto")`
13 |
14 | ###Classifier Model
15 |
16 | `model_ml_emir = ExtraTreesClassifier(n_estimators=23,criterion="gini",max_features="auto",random_state=131)
17 |
18 | model_ml_emir = MLPClassifier(activation="relu",
19 | #solver="adam",
20 | #batch_size=200,
21 | #hidden_layer_sizes=(100,),random_state=17,
22 | #learning_rate='constant',
23 | #alpha=0.0006,
24 | #beta_1 = 0.9,
25 | #beta_2=0.4)`
26 |
27 | **I am happy to present this software to you!**
28 |
29 | ###**The coding language used:**
30 |
31 | `Python 3.9.6`
32 |
33 | ###**Libraries Used:**
34 |
35 | `Sklearn`
36 |
37 | `Pandas`
38 |
39 | `Numpy`
40 |
41 | `Matplotlib`
42 |
43 | `Pylab`
44 |
45 | `Plotly`
46 |
47 | ### **Tags**
48 |
49 | _business, earth and nature, health, biology, chemistry, biotechnology, Machine Learning, Python, Artificial Intelligence, Neural Networks, Extra Tree Classifier, Extra Tree Regressor, Software_
50 |
51 |
52 | ### **Developer Information:**
53 |
54 | Name-Surname: **Emirhan BULUT**
55 |
56 | Contact (Email) : **emirhan.bulut@turkiyeyapayzeka.com**
57 |
58 | LinkedIn : **[https://www.linkedin.com/in/artificialintelligencebulut/][LinkedinAccount]**
59 |
60 | Data Source: [DataSource]
61 |
62 | [LinkedinAccount]: https://www.linkedin.com/in/artificialintelligencebulut/
63 |
64 | Official Website: **[https://www.emirhanbulut.com.tr][OfficialWebSite]**
65 |
66 | [OfficialWebSite]: https://www.emirhanbulut.com.tr
67 |
68 | [DataSource]: https://kaggle.com
69 |
70 |
71 |
72 | 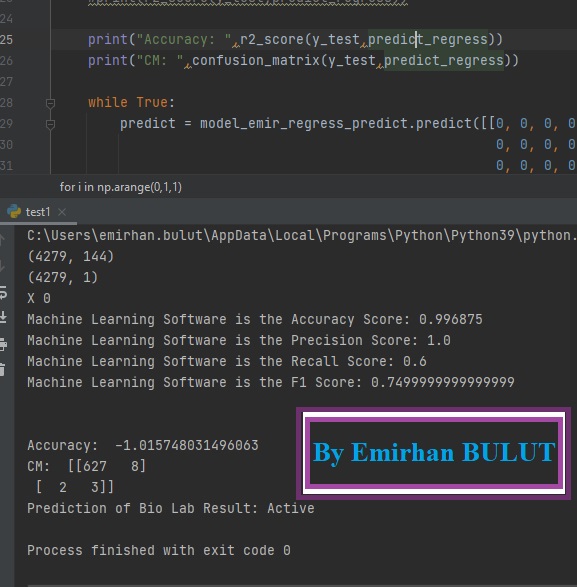 73 |
74 |
73 |
74 | 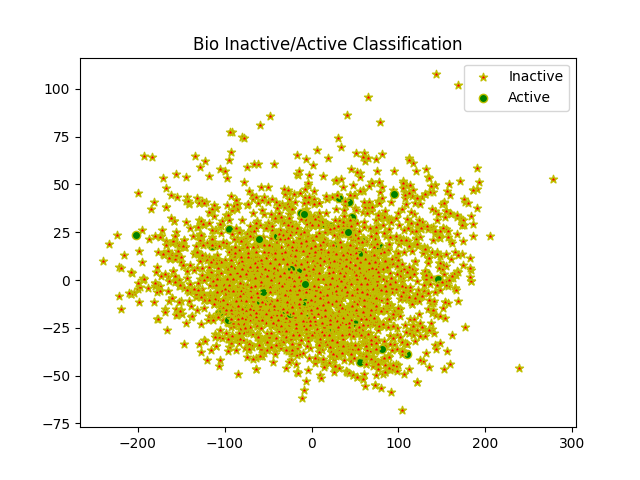 75 |
76 |
75 |
76 |  77 |
--------------------------------------------------------------------------------
/bio-chemist-lab-machine-learning-project.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from sklearn.ensemble import *
3 | from sklearn.metrics import *
4 | from sklearn.model_selection import *
5 | import pandas as pd
6 |
7 | df = pd.read_csv('bio_chemist_data.csv')
8 |
9 |
10 | def numerical_class(i):
11 | if i == 'Inactive':
12 | return 0
13 | else:
14 | return 1
15 |
16 |
17 | df['label'] = df['Outcome'].apply(numerical_class)
18 |
19 | X = df.drop(['Outcome','label'],axis='columns')
20 | XX = X.iloc[:4000,:].values
21 | y = df[['label']]
22 | yy = y.iloc[:4000,:].values
23 |
24 | print(X.shape)
25 | print(y.shape)
26 |
27 | #0.146
28 | #0.083
29 | for i in np.arange(0,1,1):
30 | X_train, X_test, y_train, y_test = train_test_split(XX, yy, test_size=0.16,random_state=7,shuffle=True,stratify=None)
31 | #18
32 |
33 | from sklearn.neural_network import *
34 |
35 | model_ml_emir = ExtraTreesClassifier(n_estimators=23,criterion="gini",max_features="auto",random_state=131)
36 |
37 | #model_ml_emir = MLPClassifier(activation="relu",
38 | #solver="adam",
39 | #batch_size=200,
40 | #hidden_layer_sizes=(100,),random_state=17,
41 | #learning_rate='constant',
42 | #alpha=0.0006,
43 | #beta_1 = 0.9,
44 | #beta_2=0.4)
45 |
46 | model_ml_emir.fit(X_train, y_train.ravel())
47 |
48 | prediction = model_ml_emir.predict(X_test)
49 |
50 | accuracy_score(y_pred=prediction, y_true=y_test)
51 |
52 | print("X",i)
53 |
54 | print("Machine Learning Software is the Accuracy Score: {0} "
55 | .format(accuracy_score(y_pred=prediction, y_true=y_test)))
56 | print("Machine Learning Software is the Precision Score: {0} "
57 | .format(precision_score(y_pred=prediction, y_true=y_test)))
58 | print("Machine Learning Software is the Recall Score: {0} "
59 | .format(recall_score(y_pred=prediction, y_true=y_test)))
60 | print("Machine Learning Software is the F1 Score: {0} "
61 | .format(f1_score(y_pred=prediction, y_true=y_test)))
62 |
63 | import matplotlib.pyplot as plt
64 |
65 | feature_import = model_ml_emir.feature_importances_
66 |
67 | a = np.std([h.feature_importances_ for h in
68 | model_ml_emir.estimators_],
69 | axis=0)
70 |
71 | df_x = pd.DataFrame(X_test,columns=X.columns)
72 | df_y = pd.DataFrame(y_test,columns=y.columns)
73 | #print(df_y)
74 |
75 | #print(len(prediction))
76 | #print(len(X_test))
77 |
78 |
79 | #plt.scatter(X_test,y_test)
80 | #plt.plot(prediction,df_x,color = "red")
81 | #plt.xlabel('Feature Labels')
82 | #plt.ylabel('Feature Importances')
83 | #plt.title('Comparison of different Feature Importances')
84 | #plt.show()
85 |
86 | import pylab as pl
87 |
88 | from sklearn.decomposition import PCA
89 |
90 | model_ozone = PCA(n_components=2).fit(X_train)
91 | model_ozone_2d = model_ozone.transform(X_train)
92 |
93 | for i in range(0, model_ozone_2d.shape[0]):
94 | if y_train[i] == 0:
95 | c1 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='r', edgecolors='y', marker='*',
96 | linewidths=1)
97 |
98 | elif y_train[i] == 1:
99 | c2 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='g', edgecolors='y', marker='o',
100 | linewidths=1)
101 | import matplotlib.pyplot as plt
102 |
103 | pl.legend([c1, c2], ['Inactive', 'Active'])
104 | plt.title('Bio Inactive/Active Classification')
105 | #pl.show()
106 |
107 | import plotly.express as px
108 |
109 | #print(X.shape)
110 |
111 | #model creating of regression in Extra Tree Regressor of prediction
112 |
113 | model_emir_regress_predict = ExtraTreesRegressor(criterion="mse",max_features="auto",
114 | n_jobs=-1,n_estimators=1)
115 | #model_emir_regress_predict = MLPRegressor(hidden_layer_sizes=(200,),activation="relu",
116 | #solver="adam",batch_size="auto")
117 |
118 | model_emir_regress_predict.fit(X_train,y_train)
119 |
120 | predict_regress = model_emir_regress_predict.predict(X_test)
121 |
122 | #print(r2_score(y_test,predict_regress))
123 |
124 | print("Accuracy: ",r2_score(y_test,predict_regress))
125 | print("CM: ",confusion_matrix(y_test,predict_regress))
126 |
127 | while True:
128 | predict = model_emir_regress_predict.predict([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
129 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
130 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
131 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
132 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
133 | 0, 0, -1.1569, 1.1837, -1.9082, 2.0213, -2.7385, 2.9148, -3.5948,
134 | 3.8259, -0.7602, 1.5808, -1.5435, 2.4587, -2.3527, 3.3599,
135 | -3.1856, 4.2714, -1.0911, 1.1333, -1.9381, 1.9813, -2.8029,
136 | 2.8675, -3.6753, 3.773, 2.704, 119.85, 4, 6, 0, 424.569, 0, 0]])
137 | predict_to_np = np.array(predict)
138 | np_to_list = predict_to_np.tolist()
139 |
140 | if np_to_list == [0]:
141 | print("Prediction of Bio Lab Result: Inactive")
142 | break
143 | elif np_to_list == [1]:
144 | print("Prediction of Bio Lab Result: Active")
145 | break
146 |
147 |
148 | fig = px.sunburst(df, path=['MW', 'BBB'],
149 | values='label',
150 | color_discrete_map={'(?)':'black', 0:'gold', 1:'darkblue'})
151 | fig.show()
--------------------------------------------------------------------------------
77 |
--------------------------------------------------------------------------------
/bio-chemist-lab-machine-learning-project.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from sklearn.ensemble import *
3 | from sklearn.metrics import *
4 | from sklearn.model_selection import *
5 | import pandas as pd
6 |
7 | df = pd.read_csv('bio_chemist_data.csv')
8 |
9 |
10 | def numerical_class(i):
11 | if i == 'Inactive':
12 | return 0
13 | else:
14 | return 1
15 |
16 |
17 | df['label'] = df['Outcome'].apply(numerical_class)
18 |
19 | X = df.drop(['Outcome','label'],axis='columns')
20 | XX = X.iloc[:4000,:].values
21 | y = df[['label']]
22 | yy = y.iloc[:4000,:].values
23 |
24 | print(X.shape)
25 | print(y.shape)
26 |
27 | #0.146
28 | #0.083
29 | for i in np.arange(0,1,1):
30 | X_train, X_test, y_train, y_test = train_test_split(XX, yy, test_size=0.16,random_state=7,shuffle=True,stratify=None)
31 | #18
32 |
33 | from sklearn.neural_network import *
34 |
35 | model_ml_emir = ExtraTreesClassifier(n_estimators=23,criterion="gini",max_features="auto",random_state=131)
36 |
37 | #model_ml_emir = MLPClassifier(activation="relu",
38 | #solver="adam",
39 | #batch_size=200,
40 | #hidden_layer_sizes=(100,),random_state=17,
41 | #learning_rate='constant',
42 | #alpha=0.0006,
43 | #beta_1 = 0.9,
44 | #beta_2=0.4)
45 |
46 | model_ml_emir.fit(X_train, y_train.ravel())
47 |
48 | prediction = model_ml_emir.predict(X_test)
49 |
50 | accuracy_score(y_pred=prediction, y_true=y_test)
51 |
52 | print("X",i)
53 |
54 | print("Machine Learning Software is the Accuracy Score: {0} "
55 | .format(accuracy_score(y_pred=prediction, y_true=y_test)))
56 | print("Machine Learning Software is the Precision Score: {0} "
57 | .format(precision_score(y_pred=prediction, y_true=y_test)))
58 | print("Machine Learning Software is the Recall Score: {0} "
59 | .format(recall_score(y_pred=prediction, y_true=y_test)))
60 | print("Machine Learning Software is the F1 Score: {0} "
61 | .format(f1_score(y_pred=prediction, y_true=y_test)))
62 |
63 | import matplotlib.pyplot as plt
64 |
65 | feature_import = model_ml_emir.feature_importances_
66 |
67 | a = np.std([h.feature_importances_ for h in
68 | model_ml_emir.estimators_],
69 | axis=0)
70 |
71 | df_x = pd.DataFrame(X_test,columns=X.columns)
72 | df_y = pd.DataFrame(y_test,columns=y.columns)
73 | #print(df_y)
74 |
75 | #print(len(prediction))
76 | #print(len(X_test))
77 |
78 |
79 | #plt.scatter(X_test,y_test)
80 | #plt.plot(prediction,df_x,color = "red")
81 | #plt.xlabel('Feature Labels')
82 | #plt.ylabel('Feature Importances')
83 | #plt.title('Comparison of different Feature Importances')
84 | #plt.show()
85 |
86 | import pylab as pl
87 |
88 | from sklearn.decomposition import PCA
89 |
90 | model_ozone = PCA(n_components=2).fit(X_train)
91 | model_ozone_2d = model_ozone.transform(X_train)
92 |
93 | for i in range(0, model_ozone_2d.shape[0]):
94 | if y_train[i] == 0:
95 | c1 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='r', edgecolors='y', marker='*',
96 | linewidths=1)
97 |
98 | elif y_train[i] == 1:
99 | c2 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='g', edgecolors='y', marker='o',
100 | linewidths=1)
101 | import matplotlib.pyplot as plt
102 |
103 | pl.legend([c1, c2], ['Inactive', 'Active'])
104 | plt.title('Bio Inactive/Active Classification')
105 | #pl.show()
106 |
107 | import plotly.express as px
108 |
109 | #print(X.shape)
110 |
111 | #model creating of regression in Extra Tree Regressor of prediction
112 |
113 | model_emir_regress_predict = ExtraTreesRegressor(criterion="mse",max_features="auto",
114 | n_jobs=-1,n_estimators=1)
115 | #model_emir_regress_predict = MLPRegressor(hidden_layer_sizes=(200,),activation="relu",
116 | #solver="adam",batch_size="auto")
117 |
118 | model_emir_regress_predict.fit(X_train,y_train)
119 |
120 | predict_regress = model_emir_regress_predict.predict(X_test)
121 |
122 | #print(r2_score(y_test,predict_regress))
123 |
124 | print("Accuracy: ",r2_score(y_test,predict_regress))
125 | print("CM: ",confusion_matrix(y_test,predict_regress))
126 |
127 | while True:
128 | predict = model_emir_regress_predict.predict([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
129 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
130 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
131 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
132 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
133 | 0, 0, -1.1569, 1.1837, -1.9082, 2.0213, -2.7385, 2.9148, -3.5948,
134 | 3.8259, -0.7602, 1.5808, -1.5435, 2.4587, -2.3527, 3.3599,
135 | -3.1856, 4.2714, -1.0911, 1.1333, -1.9381, 1.9813, -2.8029,
136 | 2.8675, -3.6753, 3.773, 2.704, 119.85, 4, 6, 0, 424.569, 0, 0]])
137 | predict_to_np = np.array(predict)
138 | np_to_list = predict_to_np.tolist()
139 |
140 | if np_to_list == [0]:
141 | print("Prediction of Bio Lab Result: Inactive")
142 | break
143 | elif np_to_list == [1]:
144 | print("Prediction of Bio Lab Result: Active")
145 | break
146 |
147 |
148 | fig = px.sunburst(df, path=['MW', 'BBB'],
149 | values='label',
150 | color_discrete_map={'(?)':'black', 0:'gold', 1:'darkblue'})
151 | fig.show()
--------------------------------------------------------------------------------
 73 |
74 |
73 |
74 |  75 |
76 |
75 |
76 |  77 |
--------------------------------------------------------------------------------
/bio-chemist-lab-machine-learning-project.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from sklearn.ensemble import *
3 | from sklearn.metrics import *
4 | from sklearn.model_selection import *
5 | import pandas as pd
6 |
7 | df = pd.read_csv('bio_chemist_data.csv')
8 |
9 |
10 | def numerical_class(i):
11 | if i == 'Inactive':
12 | return 0
13 | else:
14 | return 1
15 |
16 |
17 | df['label'] = df['Outcome'].apply(numerical_class)
18 |
19 | X = df.drop(['Outcome','label'],axis='columns')
20 | XX = X.iloc[:4000,:].values
21 | y = df[['label']]
22 | yy = y.iloc[:4000,:].values
23 |
24 | print(X.shape)
25 | print(y.shape)
26 |
27 | #0.146
28 | #0.083
29 | for i in np.arange(0,1,1):
30 | X_train, X_test, y_train, y_test = train_test_split(XX, yy, test_size=0.16,random_state=7,shuffle=True,stratify=None)
31 | #18
32 |
33 | from sklearn.neural_network import *
34 |
35 | model_ml_emir = ExtraTreesClassifier(n_estimators=23,criterion="gini",max_features="auto",random_state=131)
36 |
37 | #model_ml_emir = MLPClassifier(activation="relu",
38 | #solver="adam",
39 | #batch_size=200,
40 | #hidden_layer_sizes=(100,),random_state=17,
41 | #learning_rate='constant',
42 | #alpha=0.0006,
43 | #beta_1 = 0.9,
44 | #beta_2=0.4)
45 |
46 | model_ml_emir.fit(X_train, y_train.ravel())
47 |
48 | prediction = model_ml_emir.predict(X_test)
49 |
50 | accuracy_score(y_pred=prediction, y_true=y_test)
51 |
52 | print("X",i)
53 |
54 | print("Machine Learning Software is the Accuracy Score: {0} "
55 | .format(accuracy_score(y_pred=prediction, y_true=y_test)))
56 | print("Machine Learning Software is the Precision Score: {0} "
57 | .format(precision_score(y_pred=prediction, y_true=y_test)))
58 | print("Machine Learning Software is the Recall Score: {0} "
59 | .format(recall_score(y_pred=prediction, y_true=y_test)))
60 | print("Machine Learning Software is the F1 Score: {0} "
61 | .format(f1_score(y_pred=prediction, y_true=y_test)))
62 |
63 | import matplotlib.pyplot as plt
64 |
65 | feature_import = model_ml_emir.feature_importances_
66 |
67 | a = np.std([h.feature_importances_ for h in
68 | model_ml_emir.estimators_],
69 | axis=0)
70 |
71 | df_x = pd.DataFrame(X_test,columns=X.columns)
72 | df_y = pd.DataFrame(y_test,columns=y.columns)
73 | #print(df_y)
74 |
75 | #print(len(prediction))
76 | #print(len(X_test))
77 |
78 |
79 | #plt.scatter(X_test,y_test)
80 | #plt.plot(prediction,df_x,color = "red")
81 | #plt.xlabel('Feature Labels')
82 | #plt.ylabel('Feature Importances')
83 | #plt.title('Comparison of different Feature Importances')
84 | #plt.show()
85 |
86 | import pylab as pl
87 |
88 | from sklearn.decomposition import PCA
89 |
90 | model_ozone = PCA(n_components=2).fit(X_train)
91 | model_ozone_2d = model_ozone.transform(X_train)
92 |
93 | for i in range(0, model_ozone_2d.shape[0]):
94 | if y_train[i] == 0:
95 | c1 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='r', edgecolors='y', marker='*',
96 | linewidths=1)
97 |
98 | elif y_train[i] == 1:
99 | c2 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='g', edgecolors='y', marker='o',
100 | linewidths=1)
101 | import matplotlib.pyplot as plt
102 |
103 | pl.legend([c1, c2], ['Inactive', 'Active'])
104 | plt.title('Bio Inactive/Active Classification')
105 | #pl.show()
106 |
107 | import plotly.express as px
108 |
109 | #print(X.shape)
110 |
111 | #model creating of regression in Extra Tree Regressor of prediction
112 |
113 | model_emir_regress_predict = ExtraTreesRegressor(criterion="mse",max_features="auto",
114 | n_jobs=-1,n_estimators=1)
115 | #model_emir_regress_predict = MLPRegressor(hidden_layer_sizes=(200,),activation="relu",
116 | #solver="adam",batch_size="auto")
117 |
118 | model_emir_regress_predict.fit(X_train,y_train)
119 |
120 | predict_regress = model_emir_regress_predict.predict(X_test)
121 |
122 | #print(r2_score(y_test,predict_regress))
123 |
124 | print("Accuracy: ",r2_score(y_test,predict_regress))
125 | print("CM: ",confusion_matrix(y_test,predict_regress))
126 |
127 | while True:
128 | predict = model_emir_regress_predict.predict([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
129 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
130 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
131 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
132 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
133 | 0, 0, -1.1569, 1.1837, -1.9082, 2.0213, -2.7385, 2.9148, -3.5948,
134 | 3.8259, -0.7602, 1.5808, -1.5435, 2.4587, -2.3527, 3.3599,
135 | -3.1856, 4.2714, -1.0911, 1.1333, -1.9381, 1.9813, -2.8029,
136 | 2.8675, -3.6753, 3.773, 2.704, 119.85, 4, 6, 0, 424.569, 0, 0]])
137 | predict_to_np = np.array(predict)
138 | np_to_list = predict_to_np.tolist()
139 |
140 | if np_to_list == [0]:
141 | print("Prediction of Bio Lab Result: Inactive")
142 | break
143 | elif np_to_list == [1]:
144 | print("Prediction of Bio Lab Result: Active")
145 | break
146 |
147 |
148 | fig = px.sunburst(df, path=['MW', 'BBB'],
149 | values='label',
150 | color_discrete_map={'(?)':'black', 0:'gold', 1:'darkblue'})
151 | fig.show()
--------------------------------------------------------------------------------
77 |
--------------------------------------------------------------------------------
/bio-chemist-lab-machine-learning-project.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | from sklearn.ensemble import *
3 | from sklearn.metrics import *
4 | from sklearn.model_selection import *
5 | import pandas as pd
6 |
7 | df = pd.read_csv('bio_chemist_data.csv')
8 |
9 |
10 | def numerical_class(i):
11 | if i == 'Inactive':
12 | return 0
13 | else:
14 | return 1
15 |
16 |
17 | df['label'] = df['Outcome'].apply(numerical_class)
18 |
19 | X = df.drop(['Outcome','label'],axis='columns')
20 | XX = X.iloc[:4000,:].values
21 | y = df[['label']]
22 | yy = y.iloc[:4000,:].values
23 |
24 | print(X.shape)
25 | print(y.shape)
26 |
27 | #0.146
28 | #0.083
29 | for i in np.arange(0,1,1):
30 | X_train, X_test, y_train, y_test = train_test_split(XX, yy, test_size=0.16,random_state=7,shuffle=True,stratify=None)
31 | #18
32 |
33 | from sklearn.neural_network import *
34 |
35 | model_ml_emir = ExtraTreesClassifier(n_estimators=23,criterion="gini",max_features="auto",random_state=131)
36 |
37 | #model_ml_emir = MLPClassifier(activation="relu",
38 | #solver="adam",
39 | #batch_size=200,
40 | #hidden_layer_sizes=(100,),random_state=17,
41 | #learning_rate='constant',
42 | #alpha=0.0006,
43 | #beta_1 = 0.9,
44 | #beta_2=0.4)
45 |
46 | model_ml_emir.fit(X_train, y_train.ravel())
47 |
48 | prediction = model_ml_emir.predict(X_test)
49 |
50 | accuracy_score(y_pred=prediction, y_true=y_test)
51 |
52 | print("X",i)
53 |
54 | print("Machine Learning Software is the Accuracy Score: {0} "
55 | .format(accuracy_score(y_pred=prediction, y_true=y_test)))
56 | print("Machine Learning Software is the Precision Score: {0} "
57 | .format(precision_score(y_pred=prediction, y_true=y_test)))
58 | print("Machine Learning Software is the Recall Score: {0} "
59 | .format(recall_score(y_pred=prediction, y_true=y_test)))
60 | print("Machine Learning Software is the F1 Score: {0} "
61 | .format(f1_score(y_pred=prediction, y_true=y_test)))
62 |
63 | import matplotlib.pyplot as plt
64 |
65 | feature_import = model_ml_emir.feature_importances_
66 |
67 | a = np.std([h.feature_importances_ for h in
68 | model_ml_emir.estimators_],
69 | axis=0)
70 |
71 | df_x = pd.DataFrame(X_test,columns=X.columns)
72 | df_y = pd.DataFrame(y_test,columns=y.columns)
73 | #print(df_y)
74 |
75 | #print(len(prediction))
76 | #print(len(X_test))
77 |
78 |
79 | #plt.scatter(X_test,y_test)
80 | #plt.plot(prediction,df_x,color = "red")
81 | #plt.xlabel('Feature Labels')
82 | #plt.ylabel('Feature Importances')
83 | #plt.title('Comparison of different Feature Importances')
84 | #plt.show()
85 |
86 | import pylab as pl
87 |
88 | from sklearn.decomposition import PCA
89 |
90 | model_ozone = PCA(n_components=2).fit(X_train)
91 | model_ozone_2d = model_ozone.transform(X_train)
92 |
93 | for i in range(0, model_ozone_2d.shape[0]):
94 | if y_train[i] == 0:
95 | c1 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='r', edgecolors='y', marker='*',
96 | linewidths=1)
97 |
98 | elif y_train[i] == 1:
99 | c2 = pl.scatter(model_ozone_2d[i, 0], model_ozone_2d[i, 1], color='g', edgecolors='y', marker='o',
100 | linewidths=1)
101 | import matplotlib.pyplot as plt
102 |
103 | pl.legend([c1, c2], ['Inactive', 'Active'])
104 | plt.title('Bio Inactive/Active Classification')
105 | #pl.show()
106 |
107 | import plotly.express as px
108 |
109 | #print(X.shape)
110 |
111 | #model creating of regression in Extra Tree Regressor of prediction
112 |
113 | model_emir_regress_predict = ExtraTreesRegressor(criterion="mse",max_features="auto",
114 | n_jobs=-1,n_estimators=1)
115 | #model_emir_regress_predict = MLPRegressor(hidden_layer_sizes=(200,),activation="relu",
116 | #solver="adam",batch_size="auto")
117 |
118 | model_emir_regress_predict.fit(X_train,y_train)
119 |
120 | predict_regress = model_emir_regress_predict.predict(X_test)
121 |
122 | #print(r2_score(y_test,predict_regress))
123 |
124 | print("Accuracy: ",r2_score(y_test,predict_regress))
125 | print("CM: ",confusion_matrix(y_test,predict_regress))
126 |
127 | while True:
128 | predict = model_emir_regress_predict.predict([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
129 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
130 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
131 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
132 | 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
133 | 0, 0, -1.1569, 1.1837, -1.9082, 2.0213, -2.7385, 2.9148, -3.5948,
134 | 3.8259, -0.7602, 1.5808, -1.5435, 2.4587, -2.3527, 3.3599,
135 | -3.1856, 4.2714, -1.0911, 1.1333, -1.9381, 1.9813, -2.8029,
136 | 2.8675, -3.6753, 3.773, 2.704, 119.85, 4, 6, 0, 424.569, 0, 0]])
137 | predict_to_np = np.array(predict)
138 | np_to_list = predict_to_np.tolist()
139 |
140 | if np_to_list == [0]:
141 | print("Prediction of Bio Lab Result: Inactive")
142 | break
143 | elif np_to_list == [1]:
144 | print("Prediction of Bio Lab Result: Active")
145 | break
146 |
147 |
148 | fig = px.sunburst(df, path=['MW', 'BBB'],
149 | values='label',
150 | color_discrete_map={'(?)':'black', 0:'gold', 1:'darkblue'})
151 | fig.show()
--------------------------------------------------------------------------------