├── .github
└── workflows
│ ├── buildsdist.yml
│ ├── ciwheels.yml
│ ├── draft-pdf.yml
│ ├── scripts
│ └── windows-setup.sh
│ └── test.yml
├── .gitignore
├── .readthedocs.yml
├── CMakeLists.txt
├── LICENSE
├── README.md
├── build.sh
├── cmake
└── Modules
│ ├── FindGLPK.cmake
│ └── FindSphinx.cmake
├── docs
├── CMakeLists.txt
├── Doxyfile
├── code_design1.png
├── conf.py
├── cpp_dense_walk.rst
├── cpp_init.rst
├── cpp_sparse_walk.rst
├── cpp_utils.rst
├── index.rst
├── installation.rst
├── logo1.png
├── make.bat
├── py_dense_walk.rst
├── py_init.rst
├── py_modules.rst
├── py_sparse_walk.rst
├── py_utils.rst
├── requirements.txt
└── support.rst
├── examples
├── data
│ ├── ADLITTLE.pkl
│ ├── ISRAEL.pkl
│ └── SCTAP1.pkl
├── dense_walks.ipynb
└── sparse_walks.ipynb
├── paper
├── images
│ └── Code_Design.pdf
├── paper.bib
└── paper.md
├── pybind11
└── PybindExt.cpp
├── pyproject.toml
├── requirements.txt
├── src
├── CMakeLists.txt
├── dense
│ ├── BallWalk.cpp
│ ├── BallWalk.hpp
│ ├── BarrierWalk.cpp
│ ├── BarrierWalk.hpp
│ ├── Common.hpp
│ ├── DikinLSWalk.cpp
│ ├── DikinLSWalk.hpp
│ ├── DikinWalk.cpp
│ ├── DikinWalk.hpp
│ ├── HitRun.cpp
│ ├── HitRun.hpp
│ ├── JohnWalk.cpp
│ ├── JohnWalk.hpp
│ ├── RandomWalk.cpp
│ ├── RandomWalk.hpp
│ ├── VaidyaWalk.cpp
│ └── VaidyaWalk.hpp
├── sparse
│ ├── Common.hpp
│ ├── LeverageScore.cpp
│ ├── LeverageScore.hpp
│ ├── SparseBallWalk.cpp
│ ├── SparseBallWalk.hpp
│ ├── SparseBarrierWalk.cpp
│ ├── SparseBarrierWalk.hpp
│ ├── SparseDikinLSWalk.cpp
│ ├── SparseDikinLSWalk.hpp
│ ├── SparseDikinWalk.cpp
│ ├── SparseDikinWalk.hpp
│ ├── SparseHitRun.cpp
│ ├── SparseHitRun.hpp
│ ├── SparseJohnWalk.cpp
│ ├── SparseJohnWalk.hpp
│ ├── SparseRandomWalk.cpp
│ ├── SparseRandomWalk.hpp
│ ├── SparseVaidyaWalk.cpp
│ └── SparseVaidyaWalk.hpp
└── utils
│ ├── Common.hpp
│ ├── DenseCenter.cpp
│ ├── DenseCenter.hpp
│ ├── FacialReduction.cpp
│ ├── FacialReduction.hpp
│ ├── FullWalkRun.hpp
│ ├── SparseCenter.cpp
│ ├── SparseCenter.hpp
│ ├── SparseLP.cpp
│ └── SparseLP.hpp
└── tests
├── CMakeLists.txt
├── cpp
├── test_dense_walk.cpp
├── test_fr.cpp
├── test_init.cpp
├── test_sparse_walk.cpp
└── test_weights.cpp
└── python
├── test_dense_walk.py
├── test_fr.py
├── test_init.py
├── test_sparse_walk.py
└── test_weights.py
/.github/workflows/buildsdist.yml:
--------------------------------------------------------------------------------
1 | name: Build source distribution
2 | # Build the source distribution under Linux
3 |
4 | on:
5 | push:
6 | branches:
7 | - main
8 | tags:
9 | - 'v*' # Triggers on tag pushes that match the pattern (e.g., v1.0.0, v2.0.1, etc.)
10 | pull_request:
11 | branches:
12 | - main
13 |
14 | jobs:
15 | build_sdist:
16 | name: Source distribution
17 | runs-on: ubuntu-latest
18 |
19 | steps:
20 | - name: Checkout repository
21 | uses: actions/checkout@v4
22 |
23 | - name: Set up Python
24 | uses: actions/setup-python@v5
25 | with:
26 | python-version: 3.9
27 |
28 | - name: Install dependencies
29 | shell: bash
30 | run: |
31 | sudo apt-get install -y libeigen3-dev libglpk-dev
32 | python -m pip install numpy scipy
33 | python -m pip install twine build
34 |
35 | - name: Build source distribution
36 | run: |
37 | python -m build --sdist
38 | # Check whether the source distribution will render correctly
39 | twine check dist/*.tar.gz
40 |
41 | - name: Store artifacts
42 | uses: actions/upload-artifact@v4
43 | with:
44 | name: cibw-sdist
45 | path: dist/*.tar.gz
46 |
47 | publish-to-pypi:
48 | name: >-
49 | Publish Python 🐍 distribution 📦 to PyPI
50 | if: startsWith(github.ref, 'refs/tags/') # only publish on tag pushes
51 | needs:
52 | - build_sdist
53 | runs-on: ubuntu-latest
54 |
55 | environment:
56 | name: pypi
57 | url: https://pypi.org/p/polytopewalk
58 |
59 | permissions:
60 | id-token: write # IMPORTANT: mandatory for trusted publishing
61 |
62 | steps:
63 | - name: Download the sdist
64 | uses: actions/download-artifact@v4
65 | with:
66 | pattern: cibw-*
67 | path: dist
68 | merge-multiple: true
69 |
70 | - name: Publish distribution 📦 to PyPI
71 | uses: pypa/gh-action-pypi-publish@release/v1
72 |
73 | publish-to-testpypi:
74 | name: >-
75 | Publish Python 🐍 distribution 📦 to TestPyPI
76 | if: startsWith(github.ref, 'refs/tags/') # only publish on tag pushes
77 | needs:
78 | - build_sdist
79 | runs-on: ubuntu-latest
80 |
81 | environment:
82 | name: testpypi
83 | url: https://test.pypi.org/p/polytopewalk
84 |
85 | permissions:
86 | id-token: write # IMPORTANT: mandatory for trusted publishing
87 |
88 | steps:
89 | - name: Download the sdist
90 | uses: actions/download-artifact@v4
91 | with:
92 | pattern: cibw-*
93 | path: dist
94 | merge-multiple: true
95 |
96 | - name: Publish distribution 📦 to TestPyPI
97 | uses: pypa/gh-action-pypi-publish@release/v1
98 | with:
99 | repository-url: https://test.pypi.org/legacy/
100 | verbose: true
101 |
102 |
103 |
104 |
105 |
--------------------------------------------------------------------------------
/.github/workflows/ciwheels.yml:
--------------------------------------------------------------------------------
1 | name: Build CI wheels
2 |
3 | on:
4 | push:
5 | branches:
6 | - main
7 | tags:
8 | - 'v*' # Triggers on tag pushes that match the pattern (e.g., v1.0.0, v2.0.1, etc.)
9 | pull_request:
10 | branches:
11 | - main
12 |
13 | jobs:
14 | build_wheels:
15 | name: Build wheel for ${{ matrix.os }} on cp${{ matrix.python }}-${{ matrix.platform_id }}-${{ matrix.manylinux_image }}
16 | runs-on: ${{ matrix.os }}
17 | strategy:
18 | matrix:
19 | include:
20 | # Window 64 bit
21 | - os: windows-latest
22 | python: 39
23 | platform_id: win_amd64
24 | - os: windows-latest
25 | python: 310
26 | platform_id: win_amd64
27 | - os: windows-latest
28 | python: 311
29 | platform_id: win_amd64
30 | - os: windows-latest
31 | python: 312
32 | platform_id: win_amd64
33 | - os: windows-latest
34 | python: 313
35 | platform_id: win_amd64
36 |
37 | # Linux 64 bit manylinux2014
38 | - os: ubuntu-latest
39 | python: 39
40 | platform_id: manylinux_x86_64
41 | manylinux_image: manylinux2014
42 | - os: ubuntu-latest
43 | python: 310
44 | platform_id: manylinux_x86_64
45 | manylinux_image: manylinux2014
46 | - os: ubuntu-latest
47 | python: 311
48 | platform_id: manylinux_x86_64
49 | manylinux_image: manylinux2014
50 | - os: ubuntu-latest
51 | python: 312
52 | platform_id: manylinux_x86_64
53 | manylinux_image: manylinux2014
54 | - os: ubuntu-latest
55 | python: 313
56 | platform_id: manylinux_x86_64
57 | manylinux_image: manylinux2014
58 |
59 | # MacOS macos-12 x86_64 is deprecated
60 | # Macos macos-13 x86_64
61 | - os: macos-13
62 | python: 39
63 | platform_id: macosx_x86_64
64 | deployment_target: "13"
65 | - os: macos-13
66 | python: 310
67 | platform_id: macosx_x86_64
68 | deployment_target: "13"
69 | - os: macos-13
70 | python: 311
71 | platform_id: macosx_x86_64
72 | deployment_target: "13"
73 | - os: macos-13
74 | python: 312
75 | platform_id: macosx_x86_64
76 | deployment_target: "13"
77 | - os: macos-13
78 | python: 313
79 | platform_id: macosx_x86_64
80 | deployment_target: "13"
81 | # MacOS macos-14 arm64
82 | - os: macos-latest
83 | python: 39
84 | platform_id: macosx_arm64
85 | deployment_target: "14"
86 | - os: macos-latest
87 | python: 310
88 | platform_id: macosx_arm64
89 | deployment_target: "14"
90 | - os: macos-latest
91 | python: 311
92 | platform_id: macosx_arm64
93 | deployment_target: "14"
94 | - os: macos-latest
95 | python: 312
96 | platform_id: macosx_arm64
97 | deployment_target: "14"
98 | - os: macos-latest
99 | python: 313
100 | platform_id: macosx_arm64
101 | deployment_target: "14"
102 |
103 | steps:
104 | - name: Checkout code
105 | uses: actions/checkout@v4
106 |
107 | - name: Install packages (Windows)
108 | if: runner.os == 'Windows'
109 | shell: bash
110 | run: |
111 | curl -owinglpk-4.65.zip -L --insecure https://jaist.dl.sourceforge.net/project/winglpk/winglpk/GLPK-4.65/winglpk-4.65.zip
112 | 7z x winglpk-4.65.zip
113 | cp glpk-4.65/w64/glpk_4_65.lib glpk-4.65/w64/glpk.lib
114 | mkdir lib
115 | cp glpk-4.65/w64/glpk_4_65.dll lib/glpk_4_65.dll
116 | echo GLPK_LIB_DIR=${GITHUB_WORKSPACE}\\glpk-4.65\\w64 >> $GITHUB_ENV
117 | echo GLPK_INCLUDE_DIR=${GITHUB_WORKSPACE}\\glpk-4.65\\src >> $GITHUB_ENV
118 |
119 | - name: Build wheels
120 | uses: pypa/cibuildwheel@v2.20.0
121 | env:
122 | # Skip 32-bit builds and musllinux
123 | CIBW_SKIP: "*-win32 *-manylinux_i686 *musllinux*"
124 | CIBW_BUILD: cp${{ matrix.python }}-${{ matrix.platform_id }}
125 | BUILD_DOCS: "OFF"
126 | MACOSX_DEPLOYMENT_TARGET: ${{ matrix.deployment_target }}
127 | CIBW_BEFORE_ALL_MACOS: |
128 | brew install eigen glpk
129 | CIBW_BEFORE_ALL_LINUX: |
130 | yum install -y epel-release eigen3-devel glpk-devel
131 | CIBW_BEFORE_ALL_WINDOWS: >
132 | echo "OK starts Windows build" &&
133 | choco install eigen -y
134 | CIBW_BEFORE_BUILD_WINDOWS: "pip install delvewheel"
135 | CIBW_REPAIR_WHEEL_COMMAND_WINDOWS: "delvewheel repair --add-path \"glpk-4.65/w64/\" -w {dest_dir} {wheel}"
136 | with:
137 | package-dir: .
138 | output-dir: wheelhouse
139 | config-file: '{package}/pyproject.toml'
140 |
141 | - uses: actions/upload-artifact@v4
142 | with:
143 | name: cibw-wheels-${{ matrix.os }}-cp${{ matrix.python }}-${{ matrix.platform_id }}
144 | path: ./wheelhouse/*.whl
145 |
146 | publish-to-pypi:
147 | name: >-
148 | Publish Python 🐍 distribution 📦 to PyPI
149 | if: startsWith(github.ref, 'refs/tags/') # only publish on tag pushes
150 | needs:
151 | - build_wheels
152 | runs-on: ubuntu-latest
153 |
154 | environment:
155 | name: pypi
156 | url: https://pypi.org/p/polytopewalk
157 |
158 | permissions:
159 | id-token: write # IMPORTANT: mandatory for trusted publishing
160 |
161 | steps:
162 | - name: Download all the wheels
163 | uses: actions/download-artifact@v4
164 | with:
165 | pattern: cibw-*
166 | path: dist
167 | merge-multiple: true
168 |
169 | - name: Publish distribution 📦 to PyPI
170 | uses: pypa/gh-action-pypi-publish@release/v1
171 |
172 | publish-to-testpypi:
173 | name: >-
174 | Publish Python 🐍 distribution 📦 to TestPyPI
175 | if: startsWith(github.ref, 'refs/tags/') # only publish on tag pushes
176 | needs:
177 | - build_wheels
178 | runs-on: ubuntu-latest
179 |
180 | environment:
181 | name: testpypi
182 | url: https://test.pypi.org/p/polytopewalk
183 |
184 | permissions:
185 | id-token: write # IMPORTANT: mandatory for trusted publishing
186 |
187 | steps:
188 | - name: Download all the wheels
189 | uses: actions/download-artifact@v4
190 | with:
191 | pattern: cibw-*
192 | path: dist
193 | merge-multiple: true
194 |
195 | - name: Publish distribution 📦 to TestPyPI
196 | uses: pypa/gh-action-pypi-publish@release/v1
197 | with:
198 | repository-url: https://test.pypi.org/legacy/
199 |
--------------------------------------------------------------------------------
/.github/workflows/draft-pdf.yml:
--------------------------------------------------------------------------------
1 | name: Draft PDF
2 | on:
3 | push:
4 | branches:
5 | - main
6 | paths:
7 | - 'paper/**'
8 |
9 | jobs:
10 | paper:
11 | runs-on: ubuntu-latest
12 | name: Paper Draft
13 | steps:
14 | - name: Checkout
15 | uses: actions/checkout@v4
16 | - name: Build draft PDF
17 | uses: openjournals/openjournals-draft-action@master
18 | with:
19 | journal: joss
20 | # This should be the path to the paper within your repo.
21 | paper-path: paper/paper.md
22 | - name: Upload
23 | uses: actions/upload-artifact@v4
24 | with:
25 | name: paper
26 | # This is the output path where Pandoc will write the compiled
27 | # PDF. Note, this should be the same directory as the input
28 | # paper.md
29 | path: paper/paper.pdf
30 |
--------------------------------------------------------------------------------
/.github/workflows/scripts/windows-setup.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | echo "Let's start windows-setup"
4 | # export PATH="/c/msys64/mingw64/bin:/c/Program Files/Git/bin:$PATH"
5 | export PATH="/mingw64/bin:$PATH"
6 |
7 | # # install GLPK for Windows from binary
8 | # wget https://downloads.sourceforge.net/project/winglpk/winglpk/GLPK-4.65/winglpk-4.65.zip
9 | # upzip winglpk-4.65.zip
10 | # mkdir /mingw64/local
11 | # cp -r winglpk-4.65/* /mingw64/
12 |
13 | # # install ipopt from binary
14 | # wget https://github.com/coin-or/Ipopt/releases/download/releases%2F3.14.16/Ipopt-3.14.16-win64-msvs2019-md.zip
15 | # unzip Ipopt-3.14.16-win64-msvs2019-md.zip
16 | # mkdir /mingw64/local
17 | # cp -r Ipopt-3.14.16-win64-msvs2019-md/* /mingw64/
18 |
19 | # # # install ipopt via https://coin-or.github.io/Ipopt/INSTALL.html
20 | # # # install Mumps
21 | # # git clone https://github.com/coin-or-tools/ThirdParty-Mumps.git
22 | # # cd ThirdParty-Mumps
23 | # # ./get.Mumps
24 | # # ./configure --prefix=/mingw64/
25 | # # make
26 | # # make install
27 | # # cd ..
28 |
29 | # # # install ipopt from source
30 | # # git clone https://github.com/coin-or/Ipopt.git
31 | # # cd Ipopt
32 | # # mkdir build
33 | # # cd build
34 | # # ../configure --prefix=/mingw64/
35 | # # make
36 | # # make install
37 | # # cd ..
38 | # # cd ..
39 |
40 | # # get FindIPOPT_DIR from casadi, which is better written
41 | # git clone --depth 1 --branch 3.6.5 https://github.com/casadi/casadi.git
42 |
43 | # # install ifopt from source
44 | # git clone https://github.com/ethz-adrl/ifopt.git
45 | # cd ifopt
46 | # # move FindIPOPT.cmake around
47 | # mv ifopt_ipopt/cmake/FindIPOPT.cmake ifopt_ipopt/cmake/FindIPOPT.cmakeold
48 | # cp ../casadi/cmake/FindIPOPT.cmake ifopt_ipopt/cmake/
49 | # cp ../casadi/cmake/canonicalize_paths.cmake ifopt_ipopt/cmake/
50 | # cmake -A x64 -B build \
51 | # -DCMAKE_VERBOSE_MAKEFILE=ON \
52 | # -DCMAKE_INSTALL_PREFIX="/mingw64/local" \
53 | # -DCMAKE_PREFIX_PATH="/mingw64" \
54 | # -G "Visual Studio 17 2022"
55 |
56 | # cmake --build build --config Release
57 | # cmake --install build --config Release
58 | # cd ..
59 |
60 | # # mkdir build
61 | # # cd build
62 | # # cmake .. -DCMAKE_VERBOSE_MAKEFILE=ON \
63 | # # -DCMAKE_INSTALL_PREFIX="/mingw64/local" \
64 | # # -DCMAKE_PREFIX_PATH="/mingw64" \
65 | # # -DIPOPT_LIBRARIES="/mingw64/lib/libipopt.dll.a" \
66 | # # -DIPOPT_INCLUDE_DIRS="/mingw64/include/coin-or" \
67 | # # -G "Unix Makefiles"
68 |
69 | # # make VERBOSE=1
70 | # # make install
71 | # # cd ..
72 | # # cd ..
73 |
74 | # eigen_dir=$(cygpath -w /mingw64/share/eigen3/cmake)
75 | # echo $eigen_dir
76 | # echo "Eigen3_DIR=$eigen_dir" >> $GITHUB_ENV
77 | # ifopt_dir=$(cygpath -w /mingw64/local/share/ifopt/cmake)
78 | # echo `ls /mingw64/local/share/ifopt/cmake`
79 | # echo $ifopt_dir
80 | # echo "ifopt_DIR=$ifopt_dir" >> $GITHUB_ENV
81 | eigen_dir=$(cygpath -w /mingw64/share/eigen3/cmake)
82 | echo $eigen_dir
83 | echo "Eigen3_DIR=$eigen_dir" >> $GITHUB_ENV
84 |
85 | # glpk_include_dir=$(cygpath -w /mingw64/include)
86 | # glpk_library=$(cygpath -w /mingw64/lib)
87 | # echo "GLPK_INCLUDE_DIR=$glpk_include_dir" >> $GITHUB_ENV
88 | # echo "GLPK_LIBRARY=$glpk_library" >> $GITHUB_ENV
--------------------------------------------------------------------------------
/.github/workflows/test.yml:
--------------------------------------------------------------------------------
1 | name: Run automated test suite
2 |
3 | on:
4 | push:
5 | branches:
6 | - main
7 |
8 | jobs:
9 | test:
10 | runs-on: windows-latest
11 | strategy:
12 | matrix:
13 | python-version: ["3.9", "3.10", "3.11", "3.12", "3.13"]
14 |
15 | steps:
16 | - name: Checkout repository
17 | uses: actions/checkout@v4
18 |
19 | - name: Set up Python ${{ matrix.python-version }}
20 | uses: actions/setup-python@v4
21 | with:
22 | python-version: ${{ matrix.python-version }}
23 |
24 | - name: Install dependencies
25 | shell: bash
26 | run: |
27 | python -m pip install --upgrade pip
28 | pip install -r requirements.txt
29 | pip install polytopewalk
30 |
31 | - name: Run tests
32 | run: python -m unittest discover -s tests/python -p "*.py"
33 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | CMakeLists.txt.user
2 | CMakeCache.txt
3 | CMakeFiles
4 | CMakeScripts
5 | Testing
6 | Makefile
7 | cmake_install.cmake
8 | install_manifest.txt
9 | compile_commands.json
10 | CTestTestfile.cmake
11 | _deps
12 | .vscode
13 |
14 | .DS_Store
15 |
16 | test.ipynb
17 | # Byte-compiled / optimized / DLL files
18 | __pycache__/
19 | *.py[cod]
20 | *$py.class
21 |
22 | # C extensions
23 | *.so
24 |
25 | # Distribution / packaging
26 | .Python
27 | build/
28 | develop-eggs/
29 | dist/
30 | downloads/
31 | eggs/
32 | .eggs/
33 | lib/
34 | lib64/
35 | parts/
36 | sdist/
37 | var/
38 | wheels/
39 | share/python-wheels/
40 | *.egg-info/
41 | .installed.cfg
42 | *.egg
43 | MANIFEST
44 |
45 | # PyInstaller
46 | # Usually these files are written by a python script from a template
47 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
48 | *.manifest
49 | *.spec
50 |
51 | # Installer logs
52 | pip-log.txt
53 | pip-delete-this-directory.txt
54 |
55 | # Unit test / coverage reports
56 | htmlcov/
57 | .tox/
58 | .nox/
59 | .coverage
60 | .coverage.*
61 | .cache
62 | nosetests.xml
63 | coverage.xml
64 | *.cover
65 | *.py,cover
66 | .hypothesis/
67 | .pytest_cache/
68 | cover/
69 |
70 | # Translations
71 | *.mo
72 | *.pot
73 |
74 | # Django stuff:
75 | *.log
76 | local_settings.py
77 | db.sqlite3
78 | db.sqlite3-journal
79 |
80 | # Flask stuff:
81 | instance/
82 | .webassets-cache
83 |
84 | # Scrapy stuff:
85 | .scrapy
86 |
87 | # Sphinx documentation
88 | docs/_build/
89 |
90 | # PyBuilder
91 | .pybuilder/
92 | target/
93 |
94 | # Jupyter Notebook
95 | .ipynb_checkpoints
96 |
97 | # IPython
98 | profile_default/
99 | ipython_config.py
100 |
101 | # pyenv
102 | # For a library or package, you might want to ignore these files since the code is
103 | # intended to run in multiple environments; otherwise, check them in:
104 | # .python-version
105 |
106 | # pipenv
107 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
108 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
109 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
110 | # install all needed dependencies.
111 | #Pipfile.lock
112 |
113 | # poetry
114 | # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
115 | # This is especially recommended for binary packages to ensure reproducibility, and is more
116 | # commonly ignored for libraries.
117 | # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
118 | #poetry.lock
119 |
120 | # pdm
121 | # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
122 | #pdm.lock
123 | # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
124 | # in version control.
125 | # https://pdm.fming.dev/#use-with-ide
126 | .pdm.toml
127 |
128 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
129 | __pypackages__/
130 |
131 | # Celery stuff
132 | celerybeat-schedule
133 | celerybeat.pid
134 |
135 | # SageMath parsed files
136 | *.sage.py
137 |
138 | # Environments
139 | .env
140 | .venv
141 | env/
142 | venv/
143 | ENV/
144 | env.bak/
145 | venv.bak/
146 |
147 | # Spyder project settings
148 | .spyderproject
149 | .spyproject
150 |
151 | # Rope project settings
152 | .ropeproject
153 |
154 | # mkdocs documentation
155 | /site
156 |
157 | # mypy
158 | .mypy_cache/
159 | .dmypy.json
160 | dmypy.json
161 |
162 | # Pyre type checker
163 | .pyre/
164 |
165 | # pytype static type analyzer
166 | .pytype/
167 |
168 | # Cython debug symbols
169 | cython_debug/
170 |
171 | # PyCharm
172 | # JetBrains specific template is maintained in a separate JetBrains.gitignore that can
173 | # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
174 | # and can be added to the global gitignore or merged into this file. For a more nuclear
175 | # option (not recommended) you can uncomment the following to ignore the entire idea folder.
176 | #.idea/
177 |
178 | .pypirc
--------------------------------------------------------------------------------
/.readthedocs.yml:
--------------------------------------------------------------------------------
1 | version: 2
2 |
3 | build:
4 | os: ubuntu-22.04
5 | apt_packages:
6 | - cmake
7 | tools:
8 | python: "3.10"
9 | # You can also specify other tool versions:
10 | # nodejs: "16"
11 | # commands:
12 | # - cmake -B build -S . -DBUILD_DOCS=ON # Configure CMake
13 | # - cmake --build build --target Doxygen Sphinx # Build project
14 |
15 | # Build documentation in the docs/ directory with Sphinx
16 | sphinx:
17 | configuration: docs/conf.py
18 |
19 | # Dependencies required to build your docs
20 | python:
21 | install:
22 | - requirements: docs/requirements.txt
--------------------------------------------------------------------------------
/CMakeLists.txt:
--------------------------------------------------------------------------------
1 | cmake_minimum_required(VERSION 3.15)
2 | project(polytopewalk)
3 |
4 | set(CMAKE_CXX_STANDARD 11)
5 | if(UNIX)
6 | # For Unix-like systems, add the -fPIC option
7 | set(CMAKE_CXX_FLAGS "-O2 -fPIC")
8 | elseif(WIN32)
9 | set(CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -D_USE_MATH_DEFINES")
10 | # For Windows, specific Windows options can be set here
11 | get_property(dirs DIRECTORY ${CMAKE_CURRENT_SOURCE_DIR} PROPERTY INCLUDE_DIRECTORIES)
12 | foreach(dir ${dirs})
13 | message(STATUS "dir='${dir}'")
14 | endforeach()
15 | endif()
16 |
17 | set(CMAKE_MODULE_PATH ${CMAKE_MODULE_PATH} "${CMAKE_SOURCE_DIR}/cmake/Modules/")
18 | message(STATUS "CMAKE_MODULE_PATH: ${CMAKE_MODULE_PATH}")
19 |
20 | # Set project directories
21 | set(POLYTOPEWALK_DIR ${CMAKE_CURRENT_SOURCE_DIR})
22 | set(SOURCES_DIR ${POLYTOPEWALK_DIR}/src)
23 | set(TESTS_DIR ${POLYTOPEWALK_DIR}/tests/cpp)
24 |
25 | # Documentation configuration
26 | if (BUILD_DOCS)
27 | add_subdirectory("docs")
28 | return() # Stop processing further targets for BUILD_DOCS
29 | endif()
30 |
31 | # Main project dependencies (only when not building docs)
32 | find_package(Python COMPONENTS Interpreter Development.Module REQUIRED)
33 | find_package(pybind11 CONFIG REQUIRED)
34 | find_package(Eigen3 CONFIG REQUIRED)
35 | find_package(GLPK REQUIRED)
36 |
37 | add_subdirectory("src")
38 | # set(CMAKE_MODULE_PATH "${PROJECT_SOURCE_DIR}/cmake" ${CMAKE_MODULE_PATH})
39 |
40 | pybind11_add_module(${PROJECT_NAME} MODULE pybind11/PybindExt.cpp)
41 | target_link_libraries(${PROJECT_NAME} PUBLIC utils dense sparse pybind11::module Eigen3::Eigen ${GLPK_LIBRARY})
42 | target_include_directories(${PROJECT_NAME} PRIVATE ${Eigen3_INCLUDE_DIRS} ${GLPK_INCLUDE_DIR})
43 | install(TARGETS ${PROJECT_NAME} LIBRARY DESTINATION .)
44 |
45 | # # Windows-specific DLL installation
46 | # if(WIN32)
47 | # file(GLOB DLL_FILES "${CMAKE_CURRENT_SOURCE_DIR}/lib/*.dll")
48 | # install(FILES ${DLL_FILES} DESTINATION ${PROJECT_NAME})
49 | # endif()
50 |
51 | # if (BUILD_TESTS)
52 | add_subdirectory("tests")
53 | # endif()
54 |
55 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2023 Yuansi Chen, Benny Sun
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |  3 |
3 |
4 |
5 | 
6 | 
7 | 
8 |
9 | # PolytopeWalk
10 | **PolytopeWalk** is a `C++` library for running MCMC sampling algorithms to generate samples from a uniform distribution over a polytope with a `Python` interface. It handles preprocessing of the polytope (Facial Reduction algorithm) and initialization as well. Current implementations include the Dikin Walk, John Walk, Vaidya Walk, Ball Walk, Lee Sidford Walk, and Hit-and-Run in both the full-dimensional formulation and the sparse constrained formulation. For documentation on all functions/methods, please visit our webpage: https://polytopewalk.readthedocs.io/en/latest/ and read our paper on arXiv here: https://arxiv.org/abs/2412.06629. Finally, for example inputs and outputs, please visit the examples folder, which includes code to uniformly sample from both real-world polytopes from the `Netlib` dataset and structured polytopes.
11 |
12 | ## Code Structure
13 |
14 |
15 | 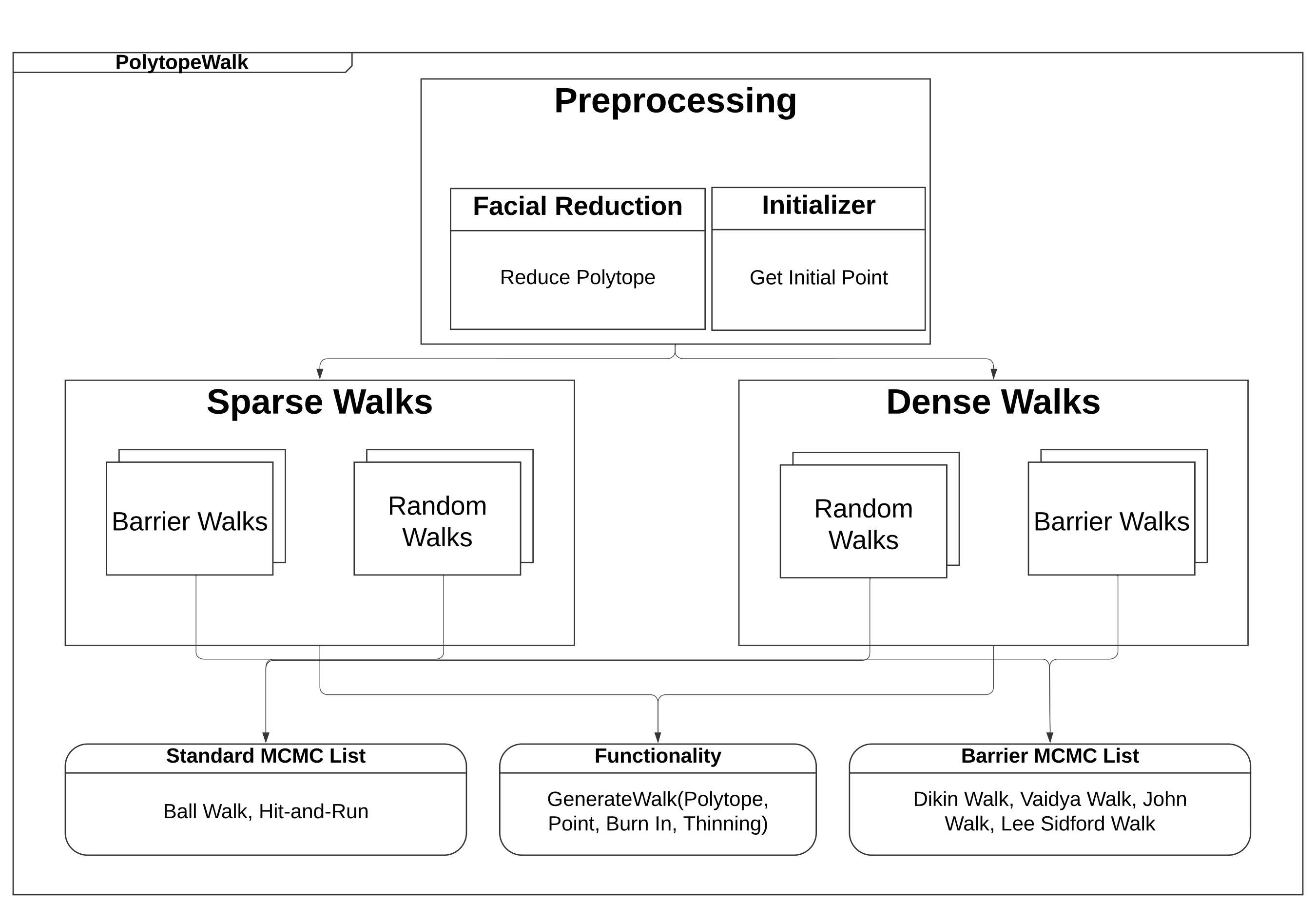 16 |
16 |
17 |
18 | ## Implemented Algorithms
19 | Let `d` be the dimension of the polytope, `n` be the number of boundaries, and `R/r` be where the convex body contains a ball of radius `r` and is mostly contained in a ball of radius `R`. We implement the following 6 MCMC sampling algorithms for uniform sampling over polytopes.
20 |
21 | | Name | Mixing Time | Author |
22 | | ------------ | ----------------- | ------------------- |
23 | | `Ball Walk` | $O(d^2R^2/r^2)$ | [Vempala (2005)](https://faculty.cc.gatech.edu/~vempala/papers/survey.pdf) |
24 | | `Hit and Run` | $O(d^2R^2/r^2)$ | [Lovasz (1999)](https://link.springer.com/content/pdf/10.1007/s101070050099.pdf) |
25 | | `Dikin Walk` | $O(nd)$ | [Sachdeva and Vishnoi (2015)](https://arxiv.org/pdf/1508.01977) |
26 | | `Vaidya Walk` | $O(n^{1/2}d^{3/2})$ | [Chen et al. (2018)](https://jmlr.org/papers/v19/18-158.html) |
27 | | `John Walk` | $O(d^{2.5})$ | [Chen et al. (2018)](https://jmlr.org/papers/v19/18-158.html) |
28 | | `Lee Sidford Walk` | $\tau(d^{2})$ | [Laddha et al. (2019)](https://arxiv.org/abs/1911.05656) (conjectured, proof incomplete) |
29 |
30 | For each implemented algorithm, we provide the full-dimensional formulation and the sparse constrained formulation. Each polytope can be expressed from 1 formulation to the other. The main benefit of utilizing the constrained formulation is that it maintains sparse operations in A, ensuring scalability in higher dimensions. Many of the `netlib` dataset sparse polytopes are represented in this formulation. The formulations are specified below.
31 |
32 | In the full-dimensional formulation with dense matrix A ($n$ x $d$ matrix) and vector b ($n$ dimensional vector), we specify the following:
33 |
34 | ```math
35 | \mathcal{K}_1 = \{x \in \mathbb{R}^{d} | Ax \le b\}
36 | ```
37 |
38 | where the polytope is specified with $n$ constraints.
39 |
40 | In the constrained formulation with sparse matrix A ($n$ x $d$ matrix) and vector b ($n$ dimensional vector), we specify the following:
41 |
42 | ```math
43 | \mathcal{K}_2 = \{x \in \mathbb{R}^{d} | Ax = b, x \succeq_k 0\}
44 | ```
45 |
46 | where the polytope is specified with $n$ equality constraints and $k$ coordinate-wise inequality constraints.
47 |
48 | In **PolytopeWalk**, we implement the MCMC algorithms in both the dense, full-dimensional and the sparse, constrained polytope formulation.
49 |
50 |
51 | ## Installation
52 |
53 | ### Dependencies
54 | **PolytopeWalk** requires:
55 | - Python (>= 3.9)
56 | - NumPy (>= 1.20)
57 | - SciPy (>= 1.6.0)
58 |
59 | ### User installation
60 | If you already have a working installation of NumPy and SciPy, the easiest way to install **PolytopeWalk** is using `pip`:
61 | ```bash
62 | pip install -U polytopewalk
63 | ```
64 |
65 |
66 | ## Developer Installation Instructions
67 |
68 | ### Important links
69 | - Official source code repo: https://github.com/ethz-randomwalk/polytopewalk
70 | - Download releases: https://pypi.org/project/polytopewalk/
71 |
72 | ### Install prerequisites
73 | (listed in each of the operating systems)
74 | - macOS: ``brew install eigen glpk``
75 | - Linux:
76 | - Ubuntu ``sudo apt-get install -y libeigen3-dev libglpk-dev``
77 | - CentOS ``yum install -y epel-release eigen3-devel glpk-devel``
78 | - Windows: ``choco install eigen -y``
79 | - Then, install winglpk from sourceforge
80 |
81 | ### Local install from source via pip
82 | ```bash
83 | git clone https://github.com/ethz-randomwalk/polytopewalk.git
84 | cd polytopewalk
85 | pip install .
86 | ```
87 |
88 |
89 | ### Compile C++ from source (not necessary)
90 | Only do this, if there is need to run and test C++ code directly. For normal users, we recommend only using the Python interface.
91 |
92 | Build with cmake
93 | ```bash
94 | git clone https://github.com/ethz-randomwalk/polytopewalk.git && cd polytopewalk
95 | cmake -B build -S . & cd build
96 | make
97 | sudo make install
98 | ```
99 |
100 | ## Examples
101 | The `examples` folder provides examples of sampling from both sparse (constrained) and dense (full-dimensional) formulations of the MCMC sampling algorithms. We test our random walk algorithms on family of 3 structured polytopes and 3 polytopes from `netlib` for real-world analysis. The lines below show a quick demonstration of sampling from a polytope using a sparse MCMC algorithm.
102 | ```python
103 | import numpy as np
104 | from scipy.sparse import csr_matrix, lil_matrix, csr_array
105 | from polytopewalk.sparse import SparseDikinWalk
106 |
107 | def generate_simplex(d):
108 | return np.array([1/d] * d), np.array([[1] * d]), np.array([1]), d, 'simplex'
109 |
110 | x, A, b, k, name = generate_simplex(5)
111 | sparse_dikin = SparseDikinWalk(r = 0.9, thin = 1)
112 | dikin_res = sparse_dikin.generateCompleteWalk(10_000, x, A, b, k, burn = 100)
113 | ```
114 | We also demonstrate how to sample from a polytope in a dense, full-dimensional formulation. We additionally introduce the Facial Reduction algorithm, used to simplify the constrained polytope into the full-dimensional form.
115 | ```python
116 | import numpy as np
117 | from scipy.sparse import csr_matrix, lil_matrix, csr_array
118 | from polytopewalk.dense import DikinWalk
119 | from polytopewalk import FacialReduction
120 |
121 | def generate_simplex(d):
122 | return np.array([1/d] * d), np.array([[1] * d]), np.array([1]), d, 'simplex'
123 |
124 | fr = FacialReduction()
125 | _, A, b, k, name = generate_simplex(5)
126 |
127 | polytope = fr.reduce(A, b, k, sparse = False)
128 | dense_A = polytope.dense_A
129 | dense_b = polytope.dense_b
130 |
131 | dc = DenseCenter()
132 | init = dc.getInitialPoint(dense_A, dense_b)
133 |
134 | dikin_res = dikin.generateCompleteWalk(1_000, init, dense_A, dense_b, burn = 100)
135 | ```
136 |
137 | ## Testing
138 | The `tests` folder includes comprehensives tests of the Facial Reduction algorithm, Initialization, Weights from MCMC algorithms, and Sparse/Dense Random Walk algorithms in both Python and C++. Our Github package page comes with an automated test suite hooked up to continuous integration after push requests to the main branch.
139 |
140 | We provide instructions for locally testing **PolytopeWalk** in both Python and C++. For both, we must locally clone the repository (assuming we have installed the package already):
141 |
142 | ```bash
143 | git clone https://github.com/ethz-randomwalk/polytopewalk.git
144 | cd polytopewalk
145 | ```
146 |
147 | ### Python Testing
148 | We can simply run the command:
149 | ```bash
150 | python -m unittest discover -s tests/python -p "*.py"
151 | ```
152 |
153 | ### C++ Testing
154 | First, we must compile the C++ code :
155 | ```bash
156 | cmake -B build -S . && cd build
157 | make
158 | ```
159 | Then, we can individually run the test files:
160 | ```bash
161 | ./tests/test_weights
162 | ./tests/test_fr
163 | ./tests/test_dense_walk
164 | ./tests/test_sparse_walk
165 | ./tests/test_init
166 | ```
167 |
168 | ## Community Guidelines
169 |
170 | For those wishing to contribute to the software, please feel free to use the pull-request feature on our Github page, alongside a brief description of the improvements to the code. For those who have any issues with our software, please let us know in the issues section of our Github page. Finally, if you have any questions, feel free to contact the authors of this page at this email address: bys7@duke.edu.
171 |
--------------------------------------------------------------------------------
/build.sh:
--------------------------------------------------------------------------------
1 | cmake -B build -S .
2 | # rm -r build; mkdir build ; cd build; cmake ..
3 | # get source distribution
4 | # python3 -m build --sdist
5 | # upload to pypi
6 | # python3 -m twine upload dist/*
7 |
--------------------------------------------------------------------------------
/cmake/Modules/FindGLPK.cmake:
--------------------------------------------------------------------------------
1 | find_path(GLPK_INCLUDE_DIR glpk.h
2 | PATHS
3 | D:/a/polytopewalk/polytopewalk/glpk-4.65/src
4 | glpk-4.65/src
5 | )
6 | find_library(GLPK_LIBRARY NAMES glpk
7 | PATHS
8 | D:/a/polytopewalk/polytopewalk/glpk-4.65/w64
9 | glpk-4.65/w64
10 | )
11 |
12 | # Handle finding status with CMake standard arguments
13 | include(FindPackageHandleStandardArgs)
14 | find_package_handle_standard_args(GLPK DEFAULT_MSG GLPK_LIBRARY GLPK_INCLUDE_DIR)
15 |
16 | mark_as_advanced(GLPK_INCLUDE_DIR GLPK_LIBRARY)
--------------------------------------------------------------------------------
/cmake/Modules/FindSphinx.cmake:
--------------------------------------------------------------------------------
1 | #Look for an executable called sphinx-build
2 | find_program(SPHINX_EXECUTABLE
3 | NAMES sphinx-build
4 | DOC "Path to sphinx-build executable")
5 |
6 | include(FindPackageHandleStandardArgs)
7 |
8 | #Handle standard arguments to find_package like REQUIRED and QUIET
9 | find_package_handle_standard_args(Sphinx
10 | "Failed to find sphinx-build executable"
11 | SPHINX_EXECUTABLE)
--------------------------------------------------------------------------------
/docs/CMakeLists.txt:
--------------------------------------------------------------------------------
1 | find_package(Doxygen REQUIRED)
2 |
3 | # Find all the public headers
4 | # get_target_property(MY_PUBLIC_HEADER_DIR polytopewalk INTERFACE_INCLUDE_DIRECTORIES)
5 | set(MY_PUBLIC_HEADER_DIR "${CMAKE_SOURCE_DIR}/src")
6 | file(GLOB_RECURSE MY_PUBLIC_HEADERS ${MY_PUBLIC_HEADER_DIR}/*.hpp)
7 |
8 | set(DOXYGEN_INPUT_DIR ${PROJECT_SOURCE_DIR}/src)
9 | set(DOXYGEN_RECURSIVE YES)
10 | set(DOXYGEN_USE_MDFILE_AS_MAINPAGE ${PROJECT_SOURCE_DIR}/README.md)
11 | set(DOXYGEN_EXCLUDE_PATTERNS */tests/*)
12 | set(DOXYGEN_OUTPUT_DIR ${CMAKE_CURRENT_BINARY_DIR})
13 | set(DOXYGEN_INDEX_FILE ${DOXYGEN_OUTPUT_DIR}/html/index.html)
14 | set(DOXYFILE_IN ${CMAKE_CURRENT_SOURCE_DIR}/Doxyfile)
15 | set(DOXYFILE_OUT ${CMAKE_CURRENT_BINARY_DIR}/Doxyfile.out)
16 |
17 | #Replace variables inside @@ with the current values
18 | configure_file(${DOXYFILE_IN} ${DOXYFILE_OUT} @ONLY)
19 |
20 | file(MAKE_DIRECTORY ${DOXYGEN_OUTPUT_DIR}) #Doxygen won't create this for us
21 | add_custom_command(OUTPUT ${DOXYGEN_INDEX_FILE}
22 | DEPENDS ${MY_PUBLIC_HEADERS}
23 | COMMAND ${DOXYGEN_EXECUTABLE} ${DOXYFILE_OUT}
24 | MAIN_DEPENDENCY ${DOXYFILE_OUT} ${DOXYFILE_IN}
25 | COMMENT "Generating docs")
26 |
27 | add_custom_target(Doxygen ALL DEPENDS ${DOXYGEN_INDEX_FILE})
28 |
29 | find_package(Sphinx REQUIRED)
30 |
31 | set(SPHINX_SOURCE ${CMAKE_CURRENT_SOURCE_DIR})
32 | set(SPHINX_BUILD ${CMAKE_CURRENT_BINARY_DIR}/sphinx)
33 |
34 | add_custom_target(Sphinx ALL
35 | COMMAND ${SPHINX_EXECUTABLE} -b html
36 | # Tell Breathe where to find the Doxygen output
37 | -Dbreathe_projects.polytopewalk=${DOXYGEN_OUTPUT_DIR}/xml
38 | ${SPHINX_SOURCE} ${SPHINX_BUILD}
39 | WORKING_DIRECTORY ${CMAKE_CURRENT_BINARY_DIR}
40 | COMMENT "Generating documentation with Sphinx")
41 |
--------------------------------------------------------------------------------
/docs/code_design1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/docs/code_design1.png
--------------------------------------------------------------------------------
/docs/conf.py:
--------------------------------------------------------------------------------
1 | # Configuration file for the Sphinx documentation builder.
2 | #
3 | # This file only contains a selection of the most common options. For a full
4 | # list see the documentation:
5 | # https://www.sphinx-doc.org/en/master/usage/configuration.html
6 |
7 | # -- Path setup --------------------------------------------------------------
8 |
9 | # If extensions (or modules to document with autodoc) are in another directory,
10 | # add these directories to sys.path here. If the directory is relative to the
11 | # documentation root, use os.path.abspath to make it absolute, like shown here.
12 | #
13 | import os
14 | import subprocess
15 | import toml
16 | # import sys
17 | # sys.path.insert(0, os.path.abspath('.'))
18 |
19 |
20 |
21 | # -- Project information -----------------------------------------------------
22 | # obtain project information from pyproject.toml
23 | # Path to your pyproject.toml file
24 | pyproject_path = os.path.join(os.path.dirname(__file__), '..', 'pyproject.toml')
25 |
26 | # Load the pyproject.toml file
27 | with open(pyproject_path, 'r') as f:
28 | pyproject_data = toml.load(f)
29 |
30 | # Extract the relevant fields from the pyproject.toml
31 | project = pyproject_data['project']['name']
32 | authors = pyproject_data['project']['authors']
33 | author = ', '.join([author['name'] for author in authors]) # Comma-separated list of author names
34 | release = pyproject_data['project']['version']
35 | copyright = f"2024, {author}" # Use author and release for copyright
36 |
37 | # -- General configuration ---------------------------------------------------
38 |
39 | # Settings to determine if we are building on readthedocs
40 | read_the_docs_build = os.environ.get('READTHEDOCS', None) == 'True'
41 | if read_the_docs_build:
42 | subprocess.call('pwd', shell=True)
43 | subprocess.call('ls', shell=True)
44 | subprocess.call('cmake -B ../build -S .. -DBUILD_DOCS=ON', shell=True)
45 | subprocess.call('cmake --build ../build --target Doxygen', shell=True)
46 | xml_dir = os.path.abspath("../build/docs/xml")
47 |
48 |
49 |
50 | # Add any Sphinx extension module names here, as strings. They can be
51 | # extensions coming with Sphinx (named 'sphinx.ext.*') or your custom
52 | # ones.
53 | extensions = [
54 | 'sphinx.ext.autodoc',
55 | 'sphinx.ext.napoleon', # For Google/NumPy-style docstrings
56 | 'sphinx.ext.viewcode', # Links to source code
57 | 'breathe', # For C++ API
58 | ]
59 |
60 | # Breathe Configuration
61 | breathe_default_project = "polytopewalk"
62 | if read_the_docs_build:
63 | breathe_projects = {"polytopewalk": xml_dir}
64 |
65 |
66 | # Add any paths that contain templates here, relative to this directory.
67 | templates_path = ['_templates']
68 |
69 | # List of patterns, relative to source directory, that match files and
70 | # directories to ignore when looking for source files.
71 | # This pattern also affects html_static_path and html_extra_path.
72 | exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store']
73 |
74 | autoclass_content = 'both'
75 |
76 |
77 | # -- Options for HTML output -------------------------------------------------
78 |
79 | # The theme to use for HTML and HTML Help pages. See the documentation for
80 | # a list of builtin themes.
81 | #
82 | html_theme = 'sphinx_rtd_theme'
83 |
84 | # Add any paths that contain custom static files (such as style sheets) here,
85 | # relative to this directory. They are copied after the builtin static files,
86 | # so a file named "default.css" will overwrite the builtin "default.css".
87 | html_static_path = ['_static']
--------------------------------------------------------------------------------
/docs/cpp_dense_walk.rst:
--------------------------------------------------------------------------------
1 | C++ Dense Random Walks
2 | ==========
3 |
4 | .. doxygenclass:: RandomWalk
5 | :members:
6 |
7 | .. doxygenclass:: BallWalk
8 | :members:
9 |
10 | .. doxygenclass:: HitAndRun
11 | :members:
12 |
13 | .. doxygenclass:: BarrierWalk
14 | :members:
15 |
16 | .. doxygenclass:: DikinWalk
17 | :members:
18 |
19 | .. doxygenclass:: VaidyaWalk
20 | :members:
21 |
22 | .. doxygenclass:: JohnWalk
23 | :members:
24 |
25 | .. doxygenclass:: DikinLSWalk
26 | :members:
--------------------------------------------------------------------------------
/docs/cpp_init.rst:
--------------------------------------------------------------------------------
1 | C++ Initialization Classes
2 | ==========
3 |
4 | .. doxygenstruct:: FROutput
5 | :members:
6 |
7 | .. doxygenclass:: FacialReduction
8 | :members:
9 |
10 | .. doxygenclass:: SparseCenter
11 | :members:
12 |
13 | .. doxygenclass:: DenseCenter
14 | :members:
--------------------------------------------------------------------------------
/docs/cpp_sparse_walk.rst:
--------------------------------------------------------------------------------
1 | C++ Sparse Random Walks
2 | ==========
3 |
4 | .. doxygenclass:: SparseRandomWalk
5 | :members:
6 |
7 | .. doxygenclass:: SparseBallWalk
8 | :members:
9 |
10 | .. doxygenclass:: SparseHitAndRun
11 | :members:
12 |
13 | .. doxygenclass:: SparseBarrierWalk
14 | :members:
15 |
16 | .. doxygenclass:: SparseDikinWalk
17 | :members:
18 |
19 | .. doxygenclass:: SparseVaidyaWalk
20 | :members:
21 |
22 | .. doxygenclass:: SparseJohnWalk
23 | :members:
24 |
25 | .. doxygenclass:: SparseDikinLSWalk
26 | :members:
--------------------------------------------------------------------------------
/docs/cpp_utils.rst:
--------------------------------------------------------------------------------
1 | C++ PolytopeWalk Utils
2 | ==========
3 |
4 | .. doxygenclass:: LeverageScore
5 | :members:
6 |
7 | .. doxygenfunction:: sparseFullWalkRun
8 |
9 | .. doxygenfunction:: denseFullWalkRun
--------------------------------------------------------------------------------
/docs/index.rst:
--------------------------------------------------------------------------------
1 | .. polytopewalk documentation master file, created by
2 | sphinx-quickstart on Tue May 9 22:50:40 2023.
3 | You can adapt this file completely to your liking, but it should at least
4 | contain the root `toctree` directive.
5 |
6 | Welcome to polytopewalk's documentation!
7 | =======================================
8 |
9 | .. toctree::
10 | :maxdepth: 2
11 | :caption: User Guide
12 |
13 | installation
14 | python_api
15 | cpp_api
16 | support
17 |
18 |
19 | Indices and tables
20 | ==================
21 |
22 | * :ref:`genindex`
23 | * :ref:`modindex`
24 | * :ref:`search`
25 |
26 | Python API
27 | ===========
28 |
29 | The Python bindings are created using `pybind11` and are documented here. This section contains the Python API documentation for the PolytopeWalk library.
30 |
31 | .. toctree::
32 | :maxdepth: 4
33 | :caption: Python Modules
34 |
35 | py_init
36 | py_dense_walk
37 | py_sparse_walk
38 | py_utils
39 |
40 |
41 | C++ API
42 | ========
43 |
44 | The C++ API is documented using Doxygen.
45 | This section provides an overview of the C++ API.
46 |
47 | Here is a list of important classes in the C++ API:
48 |
49 | * `RandomWalk`
50 | * `BallWalk`
51 | * `HitAndRun`
52 | * `BarrierWalk`
53 | * `DikinWalk`
54 | * `VaidyaWalk`
55 | * `JohnWalk`
56 | * `DikinLSWalk`
57 | * `SparseRandomWalk`
58 | * `SparseBallWalk`
59 | * `SparseHitAndRun`
60 | * `SparseBarrierWalk`
61 | * `SparseDikinWalk`
62 | * `SparseVaidyaWalk`
63 | * `SparseJohnWalk`
64 | * `SparseDikinLSWalk`
65 |
66 |
67 | For more detailed documentation on these classes, see the following pages.
68 |
69 | .. toctree::
70 | :maxdepth: 4
71 | :caption: C++ Modules
72 |
73 | cpp_init
74 | cpp_dense_walk
75 | cpp_sparse_walk
76 | cpp_utils
--------------------------------------------------------------------------------
/docs/installation.rst:
--------------------------------------------------------------------------------
1 | ============
2 | Installation
3 | ============
4 |
5 | Developer instructions listed at https://github.com/ethz-randomwalk/polytopewalk
6 |
7 | Or, if you have pip installed:
8 |
9 | .. code-block:: bash
10 |
11 | pip install polytopewalk
--------------------------------------------------------------------------------
/docs/logo1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/docs/logo1.png
--------------------------------------------------------------------------------
/docs/make.bat:
--------------------------------------------------------------------------------
1 | @ECHO OFF
2 |
3 | pushd %~dp0
4 |
5 | REM Command file for Sphinx documentation
6 |
7 | if "%SPHINXBUILD%" == "" (
8 | set SPHINXBUILD=sphinx-build
9 | )
10 | set SOURCEDIR=source
11 | set BUILDDIR=build
12 |
13 | if "%1" == "" goto help
14 |
15 | %SPHINXBUILD% >NUL 2>NUL
16 | if errorlevel 9009 (
17 | echo.
18 | echo.The 'sphinx-build' command was not found. Make sure you have Sphinx
19 | echo.installed, then set the SPHINXBUILD environment variable to point
20 | echo.to the full path of the 'sphinx-build' executable. Alternatively you
21 | echo.may add the Sphinx directory to PATH.
22 | echo.
23 | echo.If you don't have Sphinx installed, grab it from
24 | echo.https://www.sphinx-doc.org/
25 | exit /b 1
26 | )
27 |
28 | %SPHINXBUILD% -M %1 %SOURCEDIR% %BUILDDIR% %SPHINXOPTS% %O%
29 | goto end
30 |
31 | :help
32 | %SPHINXBUILD% -M help %SOURCEDIR% %BUILDDIR% %SPHINXOPTS% %O%
33 |
34 | :end
35 | popd
36 |
--------------------------------------------------------------------------------
/docs/py_dense_walk.rst:
--------------------------------------------------------------------------------
1 | Python Dense Random Walks
2 | ==========
3 |
4 | .. autoclass:: polytopewalk.dense.RandomWalk
5 | :members:
6 | :undoc-members:
7 | :show-inheritance:
8 |
9 | .. autoclass:: polytopewalk.dense.BarrierWalk
10 | :members:
11 | :undoc-members:
12 | :inherited-members:
13 | :show-inheritance:
14 |
15 | .. autoclass:: polytopewalk.dense.DikinWalk
16 | :members:
17 | :undoc-members:
18 |
19 | .. autoclass:: polytopewalk.dense.VaidyaWalk
20 | :members:
21 | :undoc-members:
22 |
23 | .. autoclass:: polytopewalk.dense.JohnWalk
24 | :members:
25 | :undoc-members:
26 |
27 | .. autoclass:: polytopewalk.dense.DikinLSWalk
28 | :members:
29 | :undoc-members:
30 |
31 | .. autoclass:: polytopewalk.dense.BallWalk
32 | :members:
33 | :undoc-members:
34 |
35 | .. autoclass:: polytopewalk.dense.HitAndRun
36 | :members:
37 | :undoc-members:
--------------------------------------------------------------------------------
/docs/py_init.rst:
--------------------------------------------------------------------------------
1 | Python Initialization Classes
2 | ==========
3 |
4 | .. autoclass:: polytopewalk.FROutput
5 | :members:
6 | :undoc-members:
7 | :show-inheritance:
8 |
9 | .. autoclass:: polytopewalk.FacialReduction
10 | :members:
11 | :undoc-members:
12 | :show-inheritance:
13 |
14 | .. autoclass:: polytopewalk.dense.DenseCenter
15 | :members:
16 | :undoc-members:
17 | :show-inheritance:
18 |
19 | .. autoclass:: polytopewalk.sparse.SparseCenter
20 | :members:
21 | :undoc-members:
22 | :show-inheritance:
--------------------------------------------------------------------------------
/docs/py_modules.rst:
--------------------------------------------------------------------------------
1 | Python API Documentation
2 | ========================
3 |
4 | This section contains the Python API documentation for the PolytopeWalk library.
5 |
6 | Module: polytopewalk
7 | ---------------------
8 |
9 | .. automodule:: polytopewalk
10 | :members:
11 | :undoc-members:
12 | :show-inheritance:
13 |
14 | Initialization Classes
15 | --------
16 |
17 | .. autoclass:: polytopewalk.FacialReduction
18 | :members:
19 | :undoc-members:
20 | :show-inheritance:
21 |
22 | .. autoclass:: polytopewalk.dense.DenseCenter
23 | :members:
24 | :undoc-members:
25 | :show-inheritance:
26 |
27 | .. autoclass:: polytopewalk.sparse.SparseCenter
28 | :members:
29 | :undoc-members:
30 | :show-inheritance:
31 |
32 | RandomWalk Classes
33 | -------------------
34 |
35 | .. autoclass:: polytopewalk.dense.RandomWalk
36 | :members:
37 | :undoc-members:
38 | :show-inheritance:
39 |
40 | .. autoclass:: polytopewalk.sparse.SparseRandomWalk
41 | :members:
42 | :undoc-members:
43 | :show-inheritance:
44 |
45 | Subclasses of RandomWalk
46 | ------------------------
47 |
48 | .. autoclass:: polytopewalk.dense.BarrierWalk
49 | :members:
50 | :undoc-members:
51 | :inherited-members:
52 | :show-inheritance:
53 |
54 | .. autoclass:: polytopewalk.dense.DikinWalk
55 | :members:
56 | :undoc-members:
57 |
58 | .. autoclass:: polytopewalk.dense.VaidyaWalk
59 | :members:
60 | :undoc-members:
61 |
62 | .. autoclass:: polytopewalk.dense.JohnWalk
63 | :members:
64 | :undoc-members:
65 |
66 | .. autoclass:: polytopewalk.dense.DikinLSWalk
67 | :members:
68 | :undoc-members:
69 |
70 | .. autoclass:: polytopewalk.dense.BallWalk
71 | :members:
72 | :undoc-members:
73 |
74 | .. autoclass:: polytopewalk.dense.HitAndRun
75 | :members:
76 | :undoc-members:
77 |
78 | Subclasses of SparseRandomWalk
79 | ------------------------
80 |
81 | .. autoclass:: polytopewalk.sparse.SparseBarrierWalk
82 | :members:
83 | :undoc-members:
84 | :inherited-members:
85 | :show-inheritance:
86 |
87 | .. autoclass:: polytopewalk.sparse.SparseDikinWalk
88 | :members:
89 | :undoc-members:
90 |
91 | .. autoclass:: polytopewalk.sparse.SparseVaidyaWalk
92 | :members:
93 | :undoc-members:
94 |
95 | .. autoclass:: polytopewalk.sparse.SparseJohnWalk
96 | :members:
97 | :undoc-members:
98 |
99 | .. autoclass:: polytopewalk.sparse.SparseDikinLSWalk
100 | :members:
101 | :undoc-members:
102 |
103 | .. autoclass:: polytopewalk.sparse.SparseBallWalk
104 | :members:
105 | :undoc-members:
106 |
107 | .. autoclass:: polytopewalk.sparse.SparseHitAndRun
108 | :members:
109 | :undoc-members:
--------------------------------------------------------------------------------
/docs/py_sparse_walk.rst:
--------------------------------------------------------------------------------
1 | Python Sparse Random Walks
2 | ==========
3 |
4 | .. autoclass:: polytopewalk.sparse.SparseRandomWalk
5 | :members:
6 | :undoc-members:
7 | :show-inheritance:
8 |
9 | .. autoclass:: polytopewalk.sparse.SparseBarrierWalk
10 | :members:
11 | :undoc-members:
12 | :inherited-members:
13 | :show-inheritance:
14 |
15 | .. autoclass:: polytopewalk.sparse.SparseDikinWalk
16 | :members:
17 | :undoc-members:
18 |
19 | .. autoclass:: polytopewalk.sparse.SparseVaidyaWalk
20 | :members:
21 | :undoc-members:
22 |
23 | .. autoclass:: polytopewalk.sparse.SparseJohnWalk
24 | :members:
25 | :undoc-members:
26 |
27 | .. autoclass:: polytopewalk.sparse.SparseDikinLSWalk
28 | :members:
29 | :undoc-members:
30 |
31 | .. autoclass:: polytopewalk.sparse.SparseBallWalk
32 | :members:
33 | :undoc-members:
34 |
35 | .. autoclass:: polytopewalk.sparse.SparseHitAndRun
36 | :members:
37 | :undoc-members:
--------------------------------------------------------------------------------
/docs/py_utils.rst:
--------------------------------------------------------------------------------
1 | Python PolytopeWalk Utils
2 | ==========
3 |
4 | .. autofunction:: polytopewalk.sparseFullWalkRun
5 |
6 | .. autofunction:: polytopewalk.denseFullWalkRun
7 |
--------------------------------------------------------------------------------
/docs/requirements.txt:
--------------------------------------------------------------------------------
1 | toml
2 | sphinx
3 | breathe
4 | sphinx-rtd-theme
5 | polytopewalk # to have prebuilt python polytopewalk package, so that docs from pybind11 can be generated
--------------------------------------------------------------------------------
/docs/support.rst:
--------------------------------------------------------------------------------
1 | =======
2 | Support
3 | =======
4 |
5 | Contact us via benny.sun@duke.edu.
6 |
7 | Or, via Github Issues
--------------------------------------------------------------------------------
/examples/data/ADLITTLE.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/examples/data/ADLITTLE.pkl
--------------------------------------------------------------------------------
/examples/data/ISRAEL.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/examples/data/ISRAEL.pkl
--------------------------------------------------------------------------------
/examples/data/SCTAP1.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/examples/data/SCTAP1.pkl
--------------------------------------------------------------------------------
/paper/images/Code_Design.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ethz-randomwalk/polytopewalk/c6ff026acff73c09c678d8394fa327778b0a8ed5/paper/images/Code_Design.pdf
--------------------------------------------------------------------------------
/paper/paper.bib:
--------------------------------------------------------------------------------
1 |
2 | @Article{chow:68,
3 | author = {C. K. Chow and C. N. Liu},
4 | title = {Approximating discrete probability distributions with dependence trees},
5 | journal = {IEEE Transactions on Information Theory},
6 | year = {1968},
7 | volume = {IT-14},
8 | number = {3},
9 | pages = {462--467}}
10 |
11 |
12 | @article{10.1093/biomet/57.1.97,
13 | author = {Hastings, W. K.},
14 | title = {{Monte Carlo sampling methods using Markov chains and their applications}},
15 | journal = {Biometrika},
16 | volume = {57},
17 | number = {1},
18 | pages = {97-109},
19 | year = {1970},
20 | month = {04},
21 | abstract = {A generalization of the sampling method introduced by Metropolis et al. (1953) is presented along with an exposition of the relevant theory, techniques of application and methods and difficulties of assessing the error in Monte Carlo estimates. Examples of the methods, including the generation of random orthogonal matrices and potential applications of the methods to numerical problems arising in statistics, are discussed.},
22 | issn = {0006-3444},
23 | doi = {10.1093/biomet/57.1.97},
24 | url = {https://doi.org/10.1093/biomet/57.1.97},
25 | eprint = {https://academic.oup.com/biomet/article-pdf/57/1/97/23940249/57-1-97.pdf},
26 | }
27 |
28 |
29 | @misc{pybind11,
30 | author = {Wenzel Jakob and Jason Rhinelander and Dean Moldovan},
31 | year = {2017},
32 | note = {https://github.com/pybind/pybind11},
33 | title = {pybind11 -- Seamless operability between C++11 and Python}
34 | }
35 | @MISC{eigenweb,
36 | author = {Ga\"{e}l Guennebaud and Beno\^{i}t Jacob and others},
37 | title = {Eigen v3},
38 | howpublished = {http://eigen.tuxfamily.org},
39 | year = {2010}
40 | }
41 |
42 | @misc{glpk,
43 | author = {Andrew Makhorin},
44 | year = {2012},
45 | note = {https://www.gnu.org/software/glpk/glpk.html},
46 | title = {(GNU Linear Programming Kit) package}
47 | }
48 |
49 | @Article{ harris2020array,
50 | title = {Array programming with {NumPy}},
51 | author = {Charles R. Harris and K. Jarrod Millman and St{\'{e}}fan J.
52 | van der Walt and Ralf Gommers and Pauli Virtanen and David
53 | Cournapeau and Eric Wieser and Julian Taylor and Sebastian
54 | Berg and Nathaniel J. Smith and Robert Kern and Matti Picus
55 | and Stephan Hoyer and Marten H. van Kerkwijk and Matthew
56 | Brett and Allan Haldane and Jaime Fern{\'{a}}ndez del
57 | R{\'{i}}o and Mark Wiebe and Pearu Peterson and Pierre
58 | G{\'{e}}rard-Marchant and Kevin Sheppard and Tyler Reddy and
59 | Warren Weckesser and Hameer Abbasi and Christoph Gohlke and
60 | Travis E. Oliphant},
61 | year = {2020},
62 | month = sep,
63 | journal = {Nature},
64 | volume = {585},
65 | number = {7825},
66 | pages = {357--362},
67 | doi = {10.1038/s41586-020-2649-2},
68 | publisher = {Springer Science and Business Media {LLC}},
69 | url = {https://doi.org/10.1038/s41586-020-2649-2}
70 | }
71 |
72 | @ARTICLE{2020SciPy-NMeth,
73 | author = {Virtanen, Pauli and Gommers, Ralf and Oliphant, Travis E. and
74 | Haberland, Matt and Reddy, Tyler and Cournapeau, David and
75 | Burovski, Evgeni and Peterson, Pearu and Weckesser, Warren and
76 | Bright, Jonathan and {van der Walt}, St{\'e}fan J. and
77 | Brett, Matthew and Wilson, Joshua and Millman, K. Jarrod and
78 | Mayorov, Nikolay and Nelson, Andrew R. J. and Jones, Eric and
79 | Kern, Robert and Larson, Eric and Carey, C J and
80 | Polat, {\.I}lhan and Feng, Yu and Moore, Eric W. and
81 | {VanderPlas}, Jake and Laxalde, Denis and Perktold, Josef and
82 | Cimrman, Robert and Henriksen, Ian and Quintero, E. A. and
83 | Harris, Charles R. and Archibald, Anne M. and
84 | Ribeiro, Ant{\^o}nio H. and Pedregosa, Fabian and
85 | {van Mulbregt}, Paul and {SciPy 1.0 Contributors}},
86 | title = {{{SciPy} 1.0: Fundamental Algorithms for Scientific
87 | Computing in Python}},
88 | journal = {Nature Methods},
89 | year = {2020},
90 | volume = {17},
91 | pages = {261--272},
92 | adsurl = {https://rdcu.be/b08Wh},
93 | doi = {10.1038/s41592-019-0686-2},
94 | }
95 |
96 | @article{Metropolis1953,
97 | added-at = {2010-08-02T15:41:00.000+0200},
98 | author = {Metropolis, Nicholas and Rosenbluth, Arianna W. and Rosenbluth, Marshall N. and Teller, Augusta H. and Teller, Edward},
99 | biburl = {https://www.bibsonomy.org/bibtex/25bdc169acdc743b5f9946748d3ce587b/lopusz},
100 | doi = {10.1063/1.1699114},
101 | interhash = {b67019ed11f34441c67cc69ee5683945},
102 | intrahash = {5bdc169acdc743b5f9946748d3ce587b},
103 | journal = {The Journal of Chemical Physics},
104 | keywords = {MonteCarlo},
105 | number = 6,
106 | pages = {1087-1092},
107 | publisher = {AIP},
108 | timestamp = {2010-08-02T15:41:00.000+0200},
109 | title = {Equation of State Calculations by Fast Computing Machines},
110 | url = {http://link.aip.org/link/?JCP/21/1087/1},
111 | volume = 21,
112 | year = 1953
113 | }
114 |
115 |
116 | @Book{pearl:88,
117 | author = {Judea Pearl},
118 | title = {Probabilistic {R}easoning in {I}ntelligent {S}ystems:
119 | {N}etworks of {P}lausible {I}nference},
120 | publisher = {Morgan Kaufman Publishers},
121 | year = {1988},
122 | address = {San Mateo, CA}
123 | }
124 |
125 | @article{DBLP:journals/corr/abs-1911-05656,
126 | author = {Aditi Laddha and
127 | Yin Tat Lee and
128 | Santosh S. Vempala},
129 | title = {Strong Self-Concordance and Sampling},
130 | journal = {CoRR},
131 | volume = {abs/1911.05656},
132 | year = {2019},
133 | url = {http://arxiv.org/abs/1911.05656},
134 | eprinttype = {arXiv},
135 | eprint = {1911.05656},
136 | timestamp = {Mon, 02 Dec 2019 13:44:01 +0100},
137 | biburl = {https://dblp.org/rec/journals/corr/abs-1911-05656.bib},
138 | bibsource = {dblp computer science bibliography, https://dblp.org}
139 | }
140 |
141 | @article{DBLP:journals/corr/abs-2202-01908,
142 | author = {Yunbum Kook and
143 | Yin Tat Lee and
144 | Ruoqi Shen and
145 | Santosh S. Vempala},
146 | title = {Sampling with Riemannian Hamiltonian Monte Carlo in a Constrained
147 | Space},
148 | journal = {CoRR},
149 | volume = {abs/2202.01908},
150 | year = {2022},
151 | url = {https://arxiv.org/abs/2202.01908},
152 | eprinttype = {arXiv},
153 | eprint = {2202.01908},
154 | timestamp = {Wed, 09 Feb 2022 15:43:34 +0100},
155 | biburl = {https://dblp.org/rec/journals/corr/abs-2202-01908.bib},
156 | bibsource = {dblp computer science bibliography, https://dblp.org}
157 | }
158 |
159 | @article{JMLR:v19:18-158,
160 | author = {Yuansi Chen and Raaz Dwivedi and Martin J. Wainwright and Bin Yu},
161 | title = {Fast MCMC Sampling Algorithms on Polytopes},
162 | journal = {Journal of Machine Learning Research},
163 | year = {2018},
164 | volume = {19},
165 | number = {55},
166 | pages = {1--86},
167 | url = {http://jmlr.org/papers/v19/18-158.html}
168 | }
169 |
170 | @article{drusvyatskiy2017many,
171 | author = {Drusvyatskiy, Dmitriy and Wolkowicz, Henry and others},
172 | date-added = {2022-11-20 21:00:56 -0500},
173 | date-modified = {2022-11-20 21:00:56 -0500},
174 | journal = {Foundations and Trends{\textregistered} in Optimization},
175 | number = {2},
176 | pages = {77--170},
177 | publisher = {Now Publishers, Inc.},
178 | title = {The many faces of degeneracy in conic optimization},
179 | volume = {3},
180 | year = {2017}}
181 |
182 | @article{DBLP:journals/corr/SachdevaV15,
183 | author = {Sushant Sachdeva and
184 | Nisheeth K. Vishnoi},
185 | title = {A Simple Analysis of the Dikin Walk},
186 | journal = {CoRR},
187 | volume = {abs/1508.01977},
188 | year = {2015},
189 | url = {http://arxiv.org/abs/1508.01977},
190 | eprinttype = {arXiv},
191 | eprint = {1508.01977},
192 | timestamp = {Mon, 13 Aug 2018 16:46:57 +0200},
193 | biburl = {https://dblp.org/rec/journals/corr/SachdevaV15.bib},
194 | bibsource = {dblp computer science bibliography, https://dblp.org}
195 | }
196 |
197 | @article{vempala2005,
198 | author = {Santosh Vempala},
199 | title = {Geometric Random Walks: A Survey},
200 | journal = {Combinatorial and Computational Geometry},
201 | volume = {52},
202 | year = {2005},
203 | url = {https://faculty.cc.gatech.edu/~vempala/papers/survey.pdf}
204 | }
205 |
206 | @article{lovasz1999,
207 | author = {László Lovász},
208 | title = {Hit-and-run mixes fast},
209 | journal = {Mathematical Programming},
210 | volume = {86},
211 | year = {1999},
212 | url = {https://link.springer.com/content/pdf/10.1007/s101070050099.pdf}
213 | }
214 |
215 | @article{Simonovits2003,
216 | author = {Mikl´os Simonovits},
217 | title = {How to compute the volume in high dimension?},
218 | journal = {Mathematical Programming},
219 | volume = {97},
220 | year = {2003},
221 | url = {https://link.springer.com/article/10.1007/s10107-003-0447-x}
222 | }
223 |
224 | @article{DBLP:journals/corr/abs-1803-05861,
225 | author = {Ludovic Cal{\`{e}}s and
226 | Apostolos Chalkis and
227 | Ioannis Z. Emiris and
228 | Vissarion Fisikopoulos},
229 | title = {Practical volume computation of structured convex bodies, and an application
230 | to modeling portfolio dependencies and financial crises},
231 | journal = {CoRR},
232 | volume = {abs/1803.05861},
233 | year = {2018},
234 | url = {http://arxiv.org/abs/1803.05861},
235 | eprinttype = {arXiv},
236 | eprint = {1803.05861},
237 | timestamp = {Mon, 13 Aug 2018 16:45:57 +0200},
238 | biburl = {https://dblp.org/rec/journals/corr/abs-1803-05861.bib},
239 | bibsource = {dblp computer science bibliography, https://dblp.org}
240 | }
241 |
242 |
243 | @misc{COBRA,
244 | title={Creation and analysis of biochemical constraint-based models: the COBRA Toolbox v3.0},
245 | author={Laurent Heirendt and Sylvain Arreckx and Thomas Pfau and Sebastián N. Mendoza and Anne Richelle and Almut Heinken and Hulda S. Haraldsdóttir and Jacek Wachowiak and Sarah M. Keating and Vanja Vlasov and Stefania Magnusdóttir and Chiam Yu Ng and German Preciat and Alise Žagare and Siu H. J. Chan and Maike K. Aurich and Catherine M. Clancy and Jennifer Modamio and John T. Sauls and Alberto Noronha and Aarash Bordbar and Benjamin Cousins and Diana C. El Assal and Luis V. Valcarcel and Iñigo Apaolaza and Susan Ghaderi and Masoud Ahookhosh and Marouen Ben Guebila and Andrejs Kostromins and Nicolas Sompairac and Hoai M. Le and Ding Ma and Yuekai Sun and Lin Wang and James T. Yurkovich and Miguel A. P. Oliveira and Phan T. Vuong and Lemmer P. El Assal and Inna Kuperstein and Andrei Zinovyev and H. Scott Hinton and William A. Bryant and Francisco J. Aragón Artacho and Francisco J. Planes and Egils Stalidzans and Alejandro Maass and Santosh Vempala and Michael Hucka and Michael A. Saunders and Costas D. Maranas and Nathan E. Lewis and Thomas Sauter and Bernhard Ø. Palsson and Ines Thiele and Ronan M. T. Fleming},
246 | year={2018},
247 | eprint={1710.04038},
248 | archivePrefix={arXiv},
249 | primaryClass={q-bio.QM}
250 | }
251 |
252 | @article{10.1093/nar/gkv1049,
253 | author = {King, Zachary A. and Lu, Justin and Dräger, Andreas and Miller, Philip and Federowicz, Stephen and Lerman, Joshua A. and Ebrahim, Ali and Palsson, Bernhard O. and Lewis, Nathan E.},
254 | title = "{BiGG Models: A platform for integrating, standardizing and sharing genome-scale models}",
255 | journal = {Nucleic Acids Research},
256 | volume = {44},
257 | number = {D1},
258 | pages = {D515-D522},
259 | year = {2015},
260 | month = {10},
261 | abstract = "{Genome-scale metabolic models are mathematically-structured knowledge bases that can be used to predict metabolic pathway usage and growth phenotypes. Furthermore, they can generate and test hypotheses when integrated with experimental data. To maximize the value of these models, centralized repositories of high-quality models must be established, models must adhere to established standards and model components must be linked to relevant databases. Tools for model visualization further enhance their utility. To meet these needs, we present BiGG Models (http://bigg.ucsd.edu), a completely redesigned Biochemical, Genetic and Genomic knowledge base. BiGG Models contains more than 75 high-quality, manually-curated genome-scale metabolic models. On the website, users can browse, search and visualize models. BiGG Models connects genome-scale models to genome annotations and external databases. Reaction and metabolite identifiers have been standardized across models to conform to community standards and enable rapid comparison across models. Furthermore, BiGG Models provides a comprehensive application programming interface for accessing BiGG Models with modeling and analysis tools. As a resource for highly curated, standardized and accessible models of metabolism, BiGG Models will facilitate diverse systems biology studies and support knowledge-based analysis of diverse experimental data.}",
262 | issn = {0305-1048},
263 | doi = {10.1093/nar/gkv1049},
264 | url = {https://doi.org/10.1093/nar/gkv1049},
265 | eprint = {https://academic.oup.com/nar/article-pdf/44/D1/D515/16661243/gkv1049.pdf},
266 | }
267 |
268 |
269 | @article{im2023revisiting,
270 | author = {Im, Haesol and Wolkowicz, Henry},
271 | date-added = {2023-11-15 15:48:48 -0500},
272 | date-modified = {2023-11-15 15:48:48 -0500},
273 | journal = {European Journal of Operational Research},
274 | number = {2},
275 | pages = {495--510},
276 | publisher = {Elsevier},
277 | title = {Revisiting degeneracy, strict feasibility, stability, in linear programming},
278 | volume = {310},
279 | year = {2023}}
280 |
281 |
282 | @article{Chalkis_2021,
283 | title={volesti: Volume Approximation and Sampling for Convex Polytopes in R},
284 | volume={13},
285 | ISSN={2073-4859},
286 | url={http://dx.doi.org/10.32614/RJ-2021-077},
287 | DOI={10.32614/rj-2021-077},
288 | number={2},
289 | journal={The R Journal},
290 | publisher={The R Foundation},
291 | author={Chalkis, Apostolos and Fisikopoulos, Vissarion},
292 | year={2021},
293 | pages={561} }
294 |
295 | @article{Yao2017, doi = {10.21105/joss.00061}, url = {https://doi.org/10.21105/joss.00061}, year = {2017}, publisher = {The Open Journal}, volume = {2}, number = {11}, pages = {61}, author = {Andy Yu Zhu Yao and David Kane}, title = {walkr: MCMC Sampling from Non-Negative Convex Polytopes}, journal = {Journal of Open Source Software} }
296 |
297 |
298 | @article{CIOMEK2021100659,
299 | author = {Krzysztof Ciomek and Mi{\l}osz Kadzi{\'n}ski},
300 | journal = {SoftwareX},
301 | pages = {100659},
302 | title = {{Polyrun: A Java library for sampling from the bounded convex polytopes}},
303 | volume = {13},
304 | year = {2021}
305 | }
306 |
307 | @article {Emanuel:2007,
308 | author = "Kerry Emanuel",
309 | title = "Environmental Factors Affecting Tropical Cyclone Power Dissipation",
310 | journal = "Journal of Climate",
311 | year = "2007",
312 | publisher = "American Meteorological Society",
313 | address = "Boston MA, USA",
314 | volume = "20",
315 | number = "22",
316 | doi = "10.1175/2007JCLI1571.1",
317 | pages= "5497 - 5509",
318 | url = "https://journals.ametsoc.org/view/journals/clim/20/22/2007jcli1571.1.xml"
319 | }
--------------------------------------------------------------------------------
/paper/paper.md:
--------------------------------------------------------------------------------
1 | ---
2 | title: 'PolytopeWalk: Sparse MCMC Sampling over Polytopes'
3 | tags:
4 | - MCMC methods
5 | - sparsity
6 | - interior-point methods
7 | - polytopes
8 | - facial reduction
9 | authors:
10 | - name: Benny Sun
11 | affiliation: 1
12 | - name: Yuansi Chen
13 | affiliation: 2
14 |
15 |
16 | affiliations:
17 | - name: Department of Statistics, Duke University
18 | index: 1
19 | - name: Department of Mathematics, ETH Zurich
20 | index: 2
21 | date: 4 March 2025
22 | bibliography: paper.bib
23 | ---
24 |
25 | # Summary
26 |
27 | High dimensional sampling is an important computational tool in statistics, with applications in stochastic simulation, volume computation, and fast randomized algorithms. We present ``PolytopeWalk``, a scalable library designed for sampling from a uniform distribution over polytopes, which are bounded geometric objects formed by linear inequalities. For sampling, we use Markov chain Monte Carlo (MCMC) methods, defined as a family of algorithms for generating approximate samples from a target probability distribution. Six state-of-the-art MCMC algorithms are implemented, including the Dikin, Vaidya, and John Walk. Additionally, we introduce novel sparse constrained formulations of these algorithms, enabling efficient sampling from sparse polytopes of the form $\mathcal{K}_2 = \{x \in \mathbb{R}^d \ | \ Ax = b, x \succeq_k 0\}$. This implementation maintains sparsity in $A$, ensuring scalability to high dimensional settings $(d > 10^4)$. Finally, ``PolytopeWalk`` includes novel implementations of preprocessing algorithms such as facial reduction and initialization, thus providing an end-to-end solution.
28 |
29 | # Statement of Need
30 |
31 | High dimensional sampling is a fundamental problem in many computational disciplines such as statistics, probability, and operation research. For example, sampling is applied in portfolio optimization [@DBLP:journals/corr/abs-1803-05861], metabolic networks in biology [@COBRA] and volume approximation over convex shapes [@Simonovits2003]. Markov chain Monte Carlo (MCMC) sampling algorithms offer a natural and scalable solution to this problem. These algorithms construct a Markov chain whose stationary distribution matches the target distribution. By running the chain for a large number of steps to ensure mixing, MCMC algorithms can efficiently generate approximately independent samples close to the target distribution, while not suffering from the curse of dimension issues.
32 |
33 | This package focuses on sampling from a uniform distribution over a user-specified polytope. We define the polytope as the following. Let $A \in \mathbb{R}^{n \times d}$, $b \in \mathbb{R}^n$ and let $x \succeq_k y$ mean that the last $k$-coordinates of $x$ are greater than or equal to the corresponding coordinates of $y$, i.e., $\{x_{d-k+1} - y_{d-k+1} \ge 0, ... , x_{d} - y_{d} \ge 0\}$. Depending on whether we allow equality constraints, the sampling problem can be formalized in two forms:
34 |
35 | \begin{enumerate}
36 | \item The full-dimensional form:
37 | \begin{align}
38 | \mathcal{K}_1 = \{x \in \mathbb{R}^d \ | Ax \le b\},
39 | \label{eq:full_dim}
40 | \end{align}
41 | where $\mathcal{K}_1$ is specified via $n$ inequality constraints.
42 | \item The constrained form:
43 | \begin{align}
44 | \mathcal{K}_2 = \{x \in \mathbb{R}^d \ | \ Ax = b, x \succeq_k 0\},
45 | \label{eq:constrained}
46 | \end{align}
47 | where $\mathcal{K}_2$ is specified via $n$ equality constraints and $k$ coordinate inequality constraints.
48 | \end{enumerate}
49 |
50 | Large polytopes with sparse constraints are common in many applications. The largest human metabolic network RECON3D is modeled as a $13543$-dimensional sparse polytope [@10.1093/nar/gkv1049]. Moreover, linear programming datasets from `NetLib` are naturally in the constrained form, where $A$ matrix is sparse. These applications motivate the need for MCMC algorithms that leverage $\mathcal{K}_2$ form. We implement novel interior-point-method-based MCMC algorithms optimized for large and sparse constrained polytopes. By exploiting sparsity, our algorithms scale well in per-iteration cost and sampling efficiency as a function of increasing dimension, enabling effective sampling from polytopes with dimensions exceeding $10^4$.
51 |
52 | Interior-point-method-based MCMC sampling algorithms on a polytope are modifications of the Ball Walk [@vempala2005], incorporating key concepts from interior-point methods in optimization. These algorithms operate in two primary steps. First, the algorithm generates a proposal distribution whose covariance matrix is state-dependent and equal to the inverse of the Hessian matrix of a specified barrier function, capturing the local geometry of the polytope. Second, the algorithm employs the Metropolis-Hastings accept-reject step to ensure that its stationary distribution is uniform on the polytope [@Metropolis1953; @10.1093/biomet/57.1.97]. Using a state-dependent proposal distribution that adapts to the polytope's local geometry, these MCMC algorithms achieve an improved mixing rate.
53 |
54 | In ``PolytopeWalk``, we implement 4 interior-point-method-based MCMC sampling algorithms in both the sparse-constrained and full-dimensional formulation. ``PolytopeWalk`` makes meaningful strides in the open-source development of MCMC, speeding up calculations for sparse high-dimensional sampling. Finally, we provide an an open-source implementation of the Facial Reduction algorithm, described in detail in the Preprocessing Algorithms section.
55 |
56 | # Package Overview
57 |
58 | ``PolytopeWalk`` is an open-source library written in C++ with Python wrapper code, providing accelerated MCMC sampling algorithms in both $\mathcal{K}_1$ and $\mathcal{K}_2$ formulation. The source code is written in C++ with `Eigen` for linear algebra [@eigenweb], `glpk` for linear programming [@glpk], and `pybind` for Python binding [@pybind11]. In Python, ``PolytopeWalk`` relies on both NumPy [@harris2020array] and SciPy [@2020SciPy-NMeth].
59 |
60 | { width=80% }
61 |
62 | ## Random Walk Algorithms
63 |
64 | Mixing times refer to the required number of steps to converge to stationary distribution. In each, $d$ refers to the dimension of the polytope and $n$ refers to the number of boundaries ($\mathcal{K}_1$ dimensions). In the first 2 walks, $R^2/r^2$ means where the convex body contains a ball of radius $r$ and is mostly contained in a ball of radius $R$.
65 |
66 | | Name | Mixing Time | Author |
67 | |:----------------:|:---------------------:|:-------------------:|
68 | | `Ball Walk` | $O(d^2R^2/r^2)$ | [Vempala (2005)](https://faculty.cc.gatech.edu/~vempala/papers/survey.pdf) |
69 | | `Hit and Run` | $O(d^2R^2/r^2)$ | [Lovasz (1999)](https://link.springer.com/content/pdf/10.1007/s101070050099.pdf) |
70 | | `Dikin Walk` | $O(nd)$ | [Sachdeva et al. (2015)](https://arxiv.org/pdf/1508.01977) |
71 | | `Vaidya Walk` | $O(n^{1/2}d^{3/2})$ | [Chen et al. (2018)](https://jmlr.org/papers/v19/18-158.html) |
72 | | `John Walk` | $O(d^{2.5})$ | [Chen et al. (2018)](https://jmlr.org/papers/v19/18-158.html) |
73 | | `Lee Sidford Walk`| $O(d^{2})$ | [Laddha et al. (2019)](https://arxiv.org/abs/1911.05656) |
74 |
75 | ## Preprocessing Algorithms
76 |
77 | ``PolytopeWalk`` comes with 2 preprocessing algorithms: initialization and facial reduction.
78 |
79 | **Initialization:** If the user cannot specify a point inside of the polytope to start, ``PolytopeWalk`` provides a class to compute an initial point well within the polytope for both the full-dimensional formulation and constrained formulation.
80 |
81 | **Facial Reduction:** We adopt the facial reduction algorithm implementation from Drusvyatskiy's research [@drusvyatskiy2017many; @im2023revisiting]. In the constrained formulation $\mathcal{K}_2 = \{x \in \mathbb{R}^d \ | \ Ax = b, x \succeq_k 0\}$, degeneracy occurs when there is a lack of strict feasibility in the polytope: there does not exist an $x \in \mathbb{R}^d$ such that $Ax = b$ and $x \succ_k 0$. Thus, degeneracy exists in polytopes when the lower-dimensional polytope is embedded in a higher dimension. The facial reduction algorithm eliminates variables in the last k dimensions fixed at $0$, thus ensuring numerical stability for sampling.
82 |
83 | ## Package Comparison
84 |
85 | | Feature | ``PolytopeWalk`` | `Volesti` | `WalkR` | `Polyrun` |
86 | |--------------------------|:-------------:|:--------:|:------:|:--------:|
87 | | Constrained Formulation | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{green}Y}$$ | $${\color{green}Y}$$ |
88 | | Sparse Friendly | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
89 | | C++ Implementation | $${\color{green}Y}$$ | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
90 | | Facial Reduction | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
91 | | Dikin Walk | $${\color{green}Y}$$ | $${\color{green}Y}$$ | $${\color{green}Y}$$ | $${\color{red}N}$$ |
92 | | Vaidya Walk | $${\color{green}Y}$$ | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
93 | | John Walk | $${\color{green}Y}$$ | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
94 | | Lee-Sidford Walk | $${\color{green}Y}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ | $${\color{red}N}$$ |
95 |
96 |
97 | Table II contrasts the features of ``PolytopeWalk`` with `Volesti` [@Chalkis_2021], `WalkR` [@Yao2017], and `Polyrun` [@CIOMEK2021100659]. `Volesti` is implemented in C++ with some of its code represented in the Python library `Dingo`. `Polyrun` only works on Java and `WalkR` on R. Notably, `WalkR` was removed from the CRAN repository, motivating further open source MCMC sampling development.
98 |
99 | # Acknowledgements
100 |
101 | Much work was done while Yuansi Chen was an assistant professor in the Department of Statistical Science at Duke University. Both authors are partially supported by NSF CAREER Award DMS-2237322, Sloan Research Fellowship and Ralph E. Powe Junior Faculty Enhancement Awards.
102 |
103 | # References
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = [
3 | 'scikit-build-core',
4 | 'pybind11',
5 | ]
6 | build-backend = 'scikit_build_core.build'
7 |
8 | [project]
9 | name = 'polytopewalk'
10 | version = '1.0.24'
11 | description='Open Source Implementation of MCMC Polytope Walks'
12 | readme = "README.md"
13 | keywords = ["MCMC", "Dikin Walk", "Vaidya Walk", "John Walk", "Lee Sidford Walk", "Sparse", "Sampling"]
14 | authors = [
15 | { name = 'Benny Sun', email = 'benny.sun@duke.edu' },
16 | { name = 'Yuansi Chen', email = 'yuansi.chen@stat.math.ethz.ch' },
17 | ]
18 |
19 | dependencies = [
20 | "numpy>=1.20",
21 | "scipy>=1.6.0",
22 | ]
23 |
24 | requires-python = '>=3.9'
25 | classifiers = [
26 | 'Intended Audience :: Science/Research',
27 | 'Intended Audience :: Developers',
28 | 'License :: OSI Approved :: MIT License',
29 | 'Programming Language :: C',
30 | 'Programming Language :: Python',
31 | 'Programming Language :: Python :: 3',
32 | 'Programming Language :: Python :: 3.9',
33 | 'Programming Language :: Python :: 3.10',
34 | 'Programming Language :: Python :: 3.11',
35 | 'Programming Language :: Python :: 3.12',
36 | 'Programming Language :: Python :: 3.13',
37 | 'Programming Language :: Python :: 3 :: Only',
38 | 'Programming Language :: Python :: Implementation :: CPython',
39 | 'Topic :: Software Development',
40 | 'Topic :: Scientific/Engineering',
41 | 'Operating System :: Microsoft :: Windows',
42 | 'Operating System :: Unix',
43 | 'Operating System :: MacOS',
44 | ]
45 |
46 | [tool.scikit-build]
47 | # A list of args to pass to CMake when configuring the project. Setting this in
48 | # config or envvar will override toml. See also ``cmake.define``.
49 | cmake.args = []
50 |
51 | # A table of defines to pass to CMake when configuring the project. Additive.
52 | cmake.define = {BUILD_DOCS = "OFF"}
53 |
54 | # Verbose printout when building.
55 | build.verbose = true
56 |
57 | # The build type to use when building the project. Valid options are: "Debug",
58 | # "Release", "RelWithDebInfo", "MinSizeRel", "", etc.
59 | cmake.build-type = "Release"
60 |
61 |
62 | [tool.scikit-build-core.env]
63 | # Allow overriding CMake options via environment variables
64 | BUILD_DOCS = { default = "OFF" }
65 |
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | numpy
2 | pandas
3 | scipy
--------------------------------------------------------------------------------
/src/CMakeLists.txt:
--------------------------------------------------------------------------------
1 |
2 | file(GLOB DENSE_SOURCES "${SOURCES_DIR}/dense/*.cpp")
3 | file(GLOB DENSE_HEADERS "${SOURCES_DIR}/dense/*.hpp")
4 |
5 | file(GLOB SPARSE_SOURCES "${SOURCES_DIR}/sparse/*.cpp")
6 | file(GLOB SPARSE_HEADERS "${SOURCES_DIR}/sparse/*.hpp")
7 |
8 | file(GLOB UTILS_SOURCES "${SOURCES_DIR}/utils/*.cpp")
9 | file(GLOB UTILS_HEADERS "${SOURCES_DIR}/utils/*.hpp")
10 |
11 | set(DENSE_FILES ${DENSE_SOURCES} ${DENSE_HEADERS})
12 | set(SPARSE_FILES ${SPARSE_SOURCES} ${SPARSE_HEADERS})
13 | set(UTILS_FILES ${UTILS_SOURCES} ${UTILS_HEADERS})
14 |
15 | set(ALL_FILES
16 | ${DENSE_FILES}
17 | ${SPARSE_FILES}
18 | ${UTILS_FILES}
19 | )
20 |
21 | add_library(dense ${DENSE_FILES})
22 | target_link_libraries(dense PUBLIC Eigen3::Eigen)
23 | target_include_directories(dense PUBLIC . Eigen3::Eigen)
24 |
25 | add_library(sparse ${SPARSE_FILES})
26 | target_link_libraries(sparse PUBLIC Eigen3::Eigen)
27 | target_include_directories(sparse PUBLIC . Eigen3::Eigen)
28 |
29 | add_library(utils ${UTILS_FILES})
30 | target_link_libraries(utils PUBLIC Eigen3::Eigen dense sparse ${GLPK_LIBRARY})
31 | target_include_directories(utils PUBLIC . Eigen3::Eigen PRIVATE ${GLPK_INCLUDE_DIR})
32 |
--------------------------------------------------------------------------------
/src/dense/BallWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "BallWalk.hpp"
2 |
3 |
4 | MatrixXd BallWalk::generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn = 0){
5 | int n = x.rows();
6 | int d = A.cols();
7 | MatrixXd results = MatrixXd::Zero(num_steps, n);
8 | int total = (burn + num_steps) * THIN;
9 | for (int i = 1; i <= total; i++){
10 | // proposal x_new = x + R /sqrt(d) * Gaussian
11 | VectorXd new_x = generateGaussianRVNorm(n) * R/sqrt(d) + x;
12 | // accept if the proposal is in the polytope
13 | if (inPolytope(new_x, A, b)){
14 | x = new_x;
15 | }
16 | // if THIN != 1, then record one for every THIN samples
17 | if (i % THIN == 0 && i/THIN > burn){
18 | results.row((int)i/THIN - burn - 1) = x;
19 | }
20 | }
21 | return results;
22 | }
23 |
24 | void BallWalk::printType(){
25 | cout << "Ball Walk" << endl;
26 | }
--------------------------------------------------------------------------------
/src/dense/BallWalk.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef BALLWALK_HPP
3 | #define BALLWALK_HPP
4 |
5 | #include "RandomWalk.hpp"
6 |
7 | class BallWalk: public RandomWalk{
8 |
9 |
10 | public:
11 |
12 | /**
13 | * @brief initialization of Ball Walk class
14 | * @param r spread parameter
15 | * @param thin thin constant
16 | */
17 | BallWalk(double r, int thin = 1) : R(r), RandomWalk(thin) {
18 |
19 | }
20 |
21 | /**
22 | * @brief generate values from Ball Walk
23 | * @param num_steps number of steps wanted to take
24 | * @param x initial starting point
25 | * @param A polytope matrixd (Ax <= b)
26 | * @param b polytope vector (Ax <= b)
27 | * @param burn number of initial steps to cut
28 | * @return num_steps by d (dimension of x) matrix
29 | */
30 | MatrixXd generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn) override;
31 |
32 | /**
33 | * @brief print general type
34 | */
35 | void printType() override;
36 |
37 | protected:
38 | /**
39 | * @brief spread parameter
40 | */
41 | const double R;
42 |
43 |
44 | };
45 |
46 | #endif
--------------------------------------------------------------------------------
/src/dense/BarrierWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "BarrierWalk.hpp"
2 |
3 | void BarrierWalk::setDistTerm(int d, int n){
4 | DIST_TERM = R*R/n;
5 | }
6 |
7 | VectorXd BarrierWalk::generateGaussianRV(int d){
8 | VectorXd v(d);
9 | random_device rd;
10 | mt19937 gen(rd());
11 | normal_distribution dis(0.0, 1.0);
12 | for(int i = 0; i < d; i++){
13 | v(i) = dis(gen);
14 | }
15 | return v;
16 | }
17 |
18 | void BarrierWalk::generateSlack(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
19 | slack = (b - (A * x));
20 | }
21 |
22 | double BarrierWalk::localNorm(VectorXd v, const MatrixXd& m){
23 | return ((v.transpose() * m) * v)(0);
24 | }
25 |
26 | void BarrierWalk::generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
27 | // always overwrite
28 | }
29 |
30 | void BarrierWalk::generateHessian(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
31 | generateWeight(x, A, b);
32 | generateSlack(x, A, b);
33 | VectorXd slack_inv = slack.cwiseInverse();
34 | DiagonalMatrix middle = slack_inv.cwiseProduct(weights.diagonal()).cwiseProduct(slack_inv).asDiagonal();
35 | hess = A.transpose() * middle * A;
36 | }
37 |
38 | void BarrierWalk::generateSample(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
39 | random_device rd;
40 | mt19937 gen(rd());
41 | uniform_real_distribution<> dis(0.0, 1.0);
42 |
43 | generateHessian(x, A, b); // sets global hess
44 | // cholesky decomposition to compute inverse of hess
45 | LLT cholesky1(hess);
46 | MatrixXd L = cholesky1.matrixL();
47 | FullPivLU lu(L);
48 | VectorXd direction = generateGaussianRV(x.rows());
49 | prop = x + sqrt(DIST_TERM) * (lu.solve(direction));
50 |
51 | if(!inPolytope(prop, A, b)){
52 | prop = x;
53 | return;
54 | }
55 | double det = L.diagonal().array().log().sum();
56 | double dist = -(0.5/DIST_TERM) * localNorm(x - prop, hess);
57 | double g_x_z = det + dist;
58 |
59 | generateHessian(prop, A, b);
60 | LLT cholesky2(hess);
61 | L = cholesky2.matrixL();
62 | det = L.diagonal().array().log().sum();
63 | dist = -(0.5/DIST_TERM) * localNorm(x - prop, hess);
64 | double g_z_x = det + dist;
65 |
66 | // accept reject step
67 | double alpha = min(1.0, exp(g_z_x-g_x_z));
68 | double val = dis(gen);
69 | prop = val < alpha ? prop : x;
70 | }
71 |
72 | MatrixXd BarrierWalk::generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn = 0){
73 | MatrixXd results = MatrixXd::Zero(num_steps, A.cols());
74 |
75 | setDistTerm(A.cols(), A.rows());
76 | int total = (burn + num_steps) * THIN;
77 | for(int i = 1; i <= total; i++){
78 | generateSample(x, A, b);
79 | x = prop;
80 |
81 | if (i % THIN == 0 && i/THIN > burn){
82 | results.row((int)i/THIN - burn - 1) = x.transpose();
83 | }
84 | }
85 | return results;
86 | }
--------------------------------------------------------------------------------
/src/dense/BarrierWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef BARRIER_HPP
2 | #define BARRIER_HPP
3 |
4 | #include "RandomWalk.hpp"
5 |
6 | class BarrierWalk : public RandomWalk{
7 | public:
8 |
9 | /**
10 | * @brief initialization of BarrierWalk class
11 | * @param r spread parameter
12 | * @param thin thin constant
13 | */
14 | BarrierWalk(double r, int thin = 1) : R(r), RandomWalk(thin){
15 |
16 | }
17 |
18 | /**
19 | * @brief weights generated from generateWeights function
20 | */
21 | DiagonalMatrix weights{};
22 |
23 | /**
24 | * @brief generate weights when calculating Hessian matrix
25 | * @param x point in polytope to generate weight
26 | * @param A polytope matrix (Ax <= b)
27 | * @param b polytope vector (Ax <= b)
28 | */
29 | virtual void generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b);
30 |
31 | /**
32 | * @brief generate values from the walk
33 | * @param num_steps number of steps wanted to take
34 | * @param x initial starting point
35 | * @param A polytope matrix
36 | * @param b polytope vector
37 | * @param burn number of initial steps to cut
38 | * @return num_steps by d (dimension of x) matrix
39 | */
40 | MatrixXd generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn) override;
41 |
42 | /**
43 | * @brief set distribution constant
44 | * @param d (dimension)
45 | * @param n (number of constraints)
46 | */
47 | virtual void setDistTerm(int d, int n);
48 |
49 | protected:
50 |
51 | /**
52 | * @brief spread parameter
53 | */
54 | const double R;
55 |
56 |
57 | /**
58 | * @brief distribution constant
59 | */
60 | double DIST_TERM;
61 |
62 | /**
63 | * @brief represents global variable b - Ax
64 | */
65 | VectorXd slack{};
66 |
67 | /**
68 | * @brief hessian matrix from global variable from generateHessian
69 | */
70 | MatrixXd hess{};
71 |
72 | /**
73 | * @brief new proposal point generated from generateSample function
74 | */
75 | VectorXd prop{};
76 |
77 | /**
78 | * @brief generates a gaussian random vector with d dimension
79 | * @param d dimension
80 | * @return Vector
81 | */
82 | VectorXd generateGaussianRV(int d);
83 |
84 | /**
85 | * @brief generates b - Ax (called slack) and
86 | * makes global variable slack equal to it
87 | * @param x point
88 | * @param A polytope matrix (Ax <= b)
89 | * @param b polytope vector (Ax <= b)
90 | */
91 | void generateSlack(const VectorXd& x, const MatrixXd& A, const VectorXd& b);
92 |
93 | /**
94 | * @brief calculates Mahalanobis distance weighted by Hessian matrix m
95 | * @param m Weighted Hessian Matrix
96 | * @param v vector to be measured
97 | * @return norm distance (double)
98 | */
99 | double localNorm(VectorXd v, const MatrixXd& m);
100 |
101 | /**
102 | * @brief generates Hessian of Log Barrier
103 | * @param x centered at x
104 | * @param A polytope matrix (Ax <= b)
105 | * @param b polytope vector (Ax <= b)
106 | */
107 | void generateHessian(const VectorXd& x, const MatrixXd& A, const VectorXd& b);
108 |
109 | /**
110 | * @brief generates a point drawn from a Multivariate Gaussian N(x, f(Hessian(x)))
111 | * @param x centered point in the polytope
112 | * @param A polytope matrix (Ax <= b)
113 | * @param b polytope vector (Ax <= b)
114 | */
115 | void generateSample(const VectorXd& x, const MatrixXd& A, const VectorXd& b);
116 | };
117 |
118 | #endif
--------------------------------------------------------------------------------
/src/dense/Common.hpp:
--------------------------------------------------------------------------------
1 | #ifndef COMMON_HPP
2 | #define COMMON_HPP
3 |
4 | #include
5 | #include
6 | #include
7 |
8 | #include
9 | #include
10 | #include
11 | #include
12 | #include
13 | #include
14 | #include
15 |
16 | using namespace Eigen;
17 | using namespace std;
18 |
19 | #endif
--------------------------------------------------------------------------------
/src/dense/DikinLSWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "DikinLSWalk.hpp"
2 |
3 | void DikinLSWalk::setDistTerm(int d, int n){
4 | w_i = VectorXd::Ones(n);
5 | double q = 2.0 * (1.0 + log(n));
6 | double term = (1.0 + q) * (1.0 + q * q);
7 | DIST_TERM = R*R/term;
8 | }
9 |
10 | void DikinLSWalk::generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
11 |
12 | double q = 2.0 * (1.0 + log(A.rows()));
13 | double alpha = 1.0 - (2.0/q);
14 |
15 | if (w_i.coeffRef(0) == -1 || w_i.rows() != A.rows()){
16 | w_i = VectorXd::Ones(A.rows());
17 | }
18 |
19 | generateSlack(x, A, b);
20 | DiagonalMatrix slack_inv = slack.cwiseInverse().asDiagonal();
21 | MatrixXd A_x = slack_inv * A;
22 |
23 | DiagonalMatrix W;
24 | MatrixXd WAX (A.rows(), A.cols());
25 | VectorXd term2a (A.rows());
26 | VectorXd term2b (A.rows());
27 | VectorXd term2(A.rows());
28 | VectorXd gradient (A.rows());
29 | VectorXd proposal (A.rows());
30 | VectorXd term3 (A.rows());
31 | VectorXd error = 0.00001 * VectorXd::Ones(A.rows());
32 |
33 | // gradient descent to compute LS weights
34 | for(int i = 0; i < MAXITER; i++){
35 | W = VectorXd(w_i.array().pow(alpha * 0.5)).asDiagonal();
36 | term2a = alpha * w_i.cwiseInverse();
37 |
38 | WAX = W * A_x;
39 | // leverage score based on previous W
40 | term2b = (WAX * (WAX.transpose() * WAX).inverse()).cwiseProduct(WAX).rowwise().sum();
41 |
42 | term2 = term2a.cwiseProduct(term2b);
43 |
44 | gradient = (alpha) * VectorXd::Ones(A.rows()) - term2;
45 | if(gradient.norm() < GRADLIM){
46 | break;
47 | }
48 | w_i = (w_i - STEPSIZE * gradient).cwiseMax(error);
49 | }
50 | weights = w_i.asDiagonal();
51 |
52 | }
53 |
54 | void DikinLSWalk::printType(){
55 | cout << "DikinLSWalk" << endl;
56 | }
--------------------------------------------------------------------------------
/src/dense/DikinLSWalk.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef DIKINLSWALK_HPP
3 | #define DIKINLSWALK_HPP
4 |
5 | #include "BarrierWalk.hpp"
6 |
7 | class DikinLSWalk: public BarrierWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of Lee Sidford Walk class
12 | * @param r spread parameter
13 | * @param thin thin constant
14 | * @param g_lim gradient descent norm limit
15 | * @param step_size size of gradient descent step
16 | * @param max_iter maximum number of iterations in gradient descent
17 | */
18 | DikinLSWalk(double r, int thin = 1, double g_lim = 0.01, double step_size = 0.1, int max_iter = 1000) : STEPSIZE(step_size), MAXITER(max_iter), GRADLIM(g_lim), BarrierWalk(r, thin){
19 |
20 | }
21 |
22 | /**
23 | * @brief print dikinls
24 | */
25 | void printType() override;
26 |
27 | /**
28 | * @brief generate weights when calculating Hessian matrix
29 | * @param x point in polytope to generate DikinLS weight
30 | * @param A polytope matrix (Ax <= b)
31 | * @param b polytope vector (Ax <= b)
32 | */
33 | void generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b) override;
34 |
35 | protected:
36 | /**
37 | * @brief step size for gradient descent
38 | */
39 | const double STEPSIZE;
40 |
41 | /**
42 | * @brief max number of iterations in gradient descent
43 | */
44 | const int MAXITER;
45 |
46 | /**
47 | * @brief stops gradient descent if it reaches under this number
48 | */
49 | const double GRADLIM;
50 |
51 | /**
52 | * @brief saves current weight for iteration
53 | */
54 | VectorXd w_i = VectorXd::Zero(1) - VectorXd::Ones(1);
55 |
56 | /**
57 | * @brief set distribution constant
58 | * @param d (dimension)
59 | * @param n (number of constraints)
60 | */
61 | void setDistTerm(int d, int n) override;
62 | };
63 |
64 | #endif
--------------------------------------------------------------------------------
/src/dense/DikinWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "DikinWalk.hpp"
2 |
3 | void DikinWalk::setDistTerm(int d, int n){
4 | DIST_TERM = R*R/d;
5 | }
6 |
7 | void DikinWalk::generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
8 | int d = b.rows();
9 | weights = VectorXd::Ones(d).asDiagonal();
10 | }
11 |
12 | void DikinWalk::printType(){
13 | cout << "Dikin Walk" << endl;
14 | }
--------------------------------------------------------------------------------
/src/dense/DikinWalk.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef DIKINWALK_HPP
3 | #define DIKINWALK_HPP
4 |
5 | #include "BarrierWalk.hpp"
6 |
7 | class DikinWalk: public BarrierWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of Dikin Walk class
12 | * @param r spread parameter
13 | * @param thin thin parameter
14 | */
15 | DikinWalk(double r, int thin = 1) : BarrierWalk(r, thin){}
16 |
17 | /**
18 | * @brief print dikin
19 | */
20 | void printType() override;
21 |
22 | /**
23 | * @brief returns weight for DikinWalk (Identity Matrix)
24 | * @param x point
25 | * @param A polytope matrix (Ax <= b)
26 | * @param b polytope vector (Ax <= b)
27 | */
28 | void generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd&b) override;
29 |
30 | protected:
31 |
32 | /**
33 | * @brief set distribution constant
34 | * @param d (dimension)
35 | * @param n (number of constraints)
36 | */
37 | void setDistTerm(int d, int n) override;
38 |
39 | };
40 |
41 |
42 | #endif
--------------------------------------------------------------------------------
/src/dense/HitRun.cpp:
--------------------------------------------------------------------------------
1 | #include "HitRun.hpp"
2 |
3 | double HitAndRun::distance(VectorXd& x, VectorXd&y){
4 | return (x - y).norm();
5 | }
6 |
7 | double HitAndRun::binarySearch(VectorXd direction, VectorXd& x, const MatrixXd& A, const VectorXd& b){
8 |
9 | VectorXd farth = x + R * direction;
10 | double dist = 0;
11 |
12 | while(true){
13 | dist = distance(x, farth);
14 | farth = x + 2 * dist * direction;
15 | if (!inPolytope(farth, A, b)){
16 | break;
17 | }
18 | }
19 | VectorXd left = x;
20 | VectorXd right = farth;
21 | VectorXd mid = (x + farth)/2;
22 |
23 | while (distance(left, right) > ERR || ! inPolytope(mid, A, b)){
24 | mid = (left + right)/2;
25 | if (inPolytope(mid, A, b)){
26 | left = mid;
27 | } else {
28 | right = mid;
29 | }
30 | }
31 | // return the distance bewteen the intersection of direction and polytope
32 | // and x
33 | return distance(mid, x);
34 | }
35 |
36 | MatrixXd HitAndRun::generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn = 0){
37 | int n = x.rows();
38 | MatrixXd results = MatrixXd::Zero(num_steps, n);
39 | random_device rd;
40 | mt19937 gen(rd());
41 | uniform_real_distribution<> dis(0.0, 1.0);
42 | int total = (burn + num_steps) * THIN;
43 | for (int i = 1; i <= total; i++){
44 | VectorXd new_direct = generateGaussianRVNorm(n);
45 | double pos_side = binarySearch(new_direct, x, A, b);
46 | double neg_side = binarySearch(new_direct * -1, x, A, b) * -1;
47 | double val = dis(gen);
48 | double random_point = val * (pos_side - neg_side) + neg_side;
49 | // the next iterate is uniform on the segment passing x
50 | x = random_point * new_direct + x;
51 |
52 | if (i % THIN == 0 && i/THIN > burn){
53 | results.row((int)i/THIN - burn - 1) = x;
54 | }
55 | }
56 | return results;
57 | }
58 |
59 | void HitAndRun::printType(){
60 | cout << "HitAndRunWalk" << endl;
61 | }
--------------------------------------------------------------------------------
/src/dense/HitRun.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef HITRUN_HPP
3 | #define HITRUN_HPP
4 |
5 | #include "RandomWalk.hpp"
6 |
7 | class HitAndRun: public RandomWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of Hit and Run class
12 | * @param r spread hyperparamter
13 | * @param err error hyperparameter
14 | * @param thin thin parameter (record every ith value)
15 | */
16 | HitAndRun(double r, double err = 1e-6, int thin = 1) : ERR(err), R(r), RandomWalk(thin) {
17 |
18 | }
19 |
20 | /**
21 | * @brief Generate values from the walk
22 | * @param num_steps number of steps wanted to take
23 | * @param x initial starting point
24 | * @param A polytope matrix
25 | * @param b polytope matrix
26 | * @param burn number of steps to burn
27 | * @return num_steps by d (dimension of x) matrix
28 | */
29 | MatrixXd generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn) override;
30 |
31 | /**
32 | * @brief print general type
33 | */
34 | void printType() override;
35 |
36 | protected:
37 | /**
38 | * @brief relative error of the binary search operation
39 | */
40 | const double ERR;
41 |
42 | /**
43 | * @brief initial starting value
44 | */
45 | const double R;
46 |
47 | /**
48 | * @brief get distance between vectors x and y
49 | * @param x
50 | * @param y
51 | * @return double
52 | */
53 | double distance(VectorXd& x, VectorXd&y);
54 |
55 | /**
56 | * @brief runs binary search to find a suitable chord intersection with the polytope
57 | * @param direction (random direction variable)
58 | * @param x (starting point)
59 | * @param A polytope matrix
60 | * @param b polytope vector
61 | * @return double
62 | */
63 | double binarySearch(VectorXd direction, VectorXd& x, const MatrixXd& A, const VectorXd& b);
64 |
65 | };
66 |
67 | #endif
--------------------------------------------------------------------------------
/src/dense/JohnWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "JohnWalk.hpp"
2 |
3 | void JohnWalk::setDistTerm(int d, int n){
4 | w_i = VectorXd::Ones(n);
5 | DIST_TERM = R*R/(pow(d, 1.5));
6 | }
7 |
8 | void JohnWalk::generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b ){
9 | double alpha = 1 - 1/(log2(2 * A.rows() / A.cols()));
10 | double beta = (double)A.cols() / (2 * A.rows());
11 |
12 | generateSlack(x, A, b);
13 | DiagonalMatrix slack_inv = slack.cwiseInverse().asDiagonal();
14 |
15 | if (w_i.coeffRef(0) == -1 || w_i.rows() != A.rows()){
16 | w_i = VectorXd::Ones(A.rows());
17 | }
18 |
19 | MatrixXd A_x = slack_inv * A;
20 |
21 | DiagonalMatrix W;
22 | MatrixXd WAX (A.rows(), A.cols());
23 | VectorXd gradient (A.rows());
24 | VectorXd score;
25 |
26 | VectorXd beta_ones = beta * VectorXd::Ones(A.rows());
27 | VectorXd next_weight = w_i;
28 |
29 | for(int i = 0; i < MAXITER; i++){
30 | w_i = next_weight;
31 |
32 | W = VectorXd(w_i.array().pow(alpha * 0.5)).asDiagonal();
33 | WAX = W * A_x;
34 | score = (WAX * (WAX.transpose() * WAX).inverse()).cwiseProduct(WAX).rowwise().sum();
35 |
36 | next_weight = 0.5 * (w_i + score + beta_ones).cwiseMax(beta_ones);
37 | if((next_weight - w_i).cwiseAbs().maxCoeff() < LIM){
38 | break;
39 | }
40 | }
41 |
42 | weights = w_i.asDiagonal();
43 |
44 | }
45 |
46 |
47 | void JohnWalk::printType(){
48 | cout << "John Walk" << endl;
49 | }
--------------------------------------------------------------------------------
/src/dense/JohnWalk.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef JOHNWALK_HPP
3 | #define JOHNWALK_HPP
4 |
5 | #include "BarrierWalk.hpp"
6 |
7 | class JohnWalk: public BarrierWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of John Walk class
12 | * @param r spread parameter
13 | * @param thin thin constant
14 | * @param lim norm limit for fixed point iteration
15 | * @param max_iter maximum number of iterations in gradient descent
16 | */
17 | JohnWalk(double r, int thin = 1, double lim = 1e-5, int max_iter = 1000) : MAXITER(max_iter), LIM(lim), BarrierWalk(r, thin){
18 |
19 | }
20 |
21 | /**
22 | * @brief print john walk
23 | */
24 | void printType() override;
25 |
26 | /**
27 | * @brief generates John weight by solving convex optimization problem
28 | * @param x point in polytope to generate DikinLS weight
29 | * @param A polytope matrix
30 | * @param b polytope matrix
31 | */
32 | void generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b) override;
33 |
34 |
35 | protected:
36 |
37 | /**
38 | * @brief max number of iterations in fixed point iteration
39 | */
40 | const double MAXITER;
41 |
42 | /**
43 | * @brief stops if it reaches under this number in fixed iteration
44 | */
45 | const double LIM;
46 |
47 | /**
48 | * @brief saves current weight for iteration
49 | */
50 | VectorXd w_i = VectorXd::Zero(1) - VectorXd::Ones(1);
51 |
52 | /**
53 | * @brief set distribution constant
54 | * @param d (dimension)
55 | * @param n (number of constraints)
56 | */
57 | void setDistTerm(int d, int n) override;
58 |
59 | };
60 |
61 | #endif
--------------------------------------------------------------------------------
/src/dense/RandomWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "RandomWalk.hpp"
2 |
3 | bool RandomWalk::inPolytope(const VectorXd& vec, const MatrixXd& A, const VectorXd&b){
4 | return ((A * vec) - b).maxCoeff() <= 0;
5 | }
6 |
7 | VectorXd RandomWalk::generateGaussianRVNorm(const int d){
8 | VectorXd v(d);
9 | random_device rd;
10 | mt19937 gen(rd());
11 | normal_distribution dis(0.0, 1.0);
12 | for(int i = 0; i < d; i++){
13 | v(i) = dis(gen);
14 | }
15 | return v/v.norm();
16 | }
17 |
18 | MatrixXd RandomWalk::generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn = 0){
19 | cout << "oops" << endl;
20 | return MatrixXd::Zero(1,1);
21 | }
22 |
23 | void RandomWalk::printType(){
24 | cout << "oops" << endl;
25 | }
--------------------------------------------------------------------------------
/src/dense/RandomWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef RANDOMWALK_HPP
2 | #define RANDOMWALK_HPP
3 | #include "Common.hpp"
4 |
5 | class RandomWalk{
6 |
7 | public:
8 |
9 | /**
10 | * @brief initialization of Random Walk super class
11 | * @param thin thin constant
12 | */
13 | RandomWalk(int thin = 1) : THIN(thin){}
14 |
15 | /**
16 | * @brief generate values from the walk
17 | * @param num_steps number of steps wanted to take
18 | * @param x initial starting point
19 | * @param A polytope matrix (Ax <= b)
20 | * @param b polytope vector (Ax <= b)
21 | * @param burn number of initial steps to cut
22 | * @return num_steps by d (dimension of x) matrix
23 | */
24 | virtual MatrixXd generateCompleteWalk(const int num_steps, VectorXd& x, const MatrixXd& A, const VectorXd& b, int burn);
25 |
26 | protected:
27 |
28 | /**
29 | * @brief checks Az <= b
30 | * @param z vector
31 | * @param A polytope matrix (Ax <= b)
32 | * @param b polytope vector (Ax <= b)
33 | * @return bool (inside polytope or not)
34 | */
35 | bool inPolytope(const VectorXd& z, const MatrixXd& A, const VectorXd& b);
36 |
37 | /**
38 | * @brief returns normalized Gaussian vector of dimension d
39 | * @param d
40 | * @return vector (normalized vector)
41 | */
42 | VectorXd generateGaussianRVNorm(const int d);
43 |
44 | /**
45 | * @brief prints unique identifier of the walk
46 | */
47 | virtual void printType();
48 |
49 | /**
50 | * @brief only include every __ sample
51 | */
52 | const int THIN;
53 | };
54 |
55 | #endif
--------------------------------------------------------------------------------
/src/dense/VaidyaWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "VaidyaWalk.hpp"
2 |
3 | void VaidyaWalk::setDistTerm(int d, int n){
4 | DIST_TERM = R*R/(sqrt(d * n));
5 | }
6 |
7 | void VaidyaWalk::generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b){
8 | generateSlack(x, A, b); // sets global slack
9 | DiagonalMatrix slack_inv = slack.cwiseInverse().asDiagonal();
10 | MatrixXd half_hess = slack_inv * A;
11 |
12 | // leverage score computation
13 | VectorXd wi = (half_hess * (half_hess.transpose() * half_hess).inverse()).cwiseProduct(half_hess).rowwise().sum();
14 |
15 | // leverage score + constants
16 | wi = wi.array() + (double)A.cols()/A.rows();
17 | weights = wi.asDiagonal();
18 | }
19 |
20 | void VaidyaWalk::printType(){
21 | cout << "Vaidya Walk" << endl;
22 | }
--------------------------------------------------------------------------------
/src/dense/VaidyaWalk.hpp:
--------------------------------------------------------------------------------
1 |
2 | #ifndef VAIDYAWALK_HPP
3 | #define VAIDYAWALK_HPP
4 |
5 | #include "BarrierWalk.hpp"
6 |
7 | class VaidyaWalk: public BarrierWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of Sparse Vaidya Walk class
12 | * @param r spread parameter
13 | * @param thin thin constant
14 | */
15 | VaidyaWalk(double r, int thin = 1) : BarrierWalk(r, thin){}
16 |
17 | /**
18 | * @brief print general type
19 | */
20 | void printType() override;
21 |
22 | /**
23 | * @brief returns weight for Vaidya Walk (leverage score calculation)
24 | * @param x center vector
25 | * @param A polytope matrix
26 | * @param b polytope vector
27 | */
28 | void generateWeight(const VectorXd& x, const MatrixXd& A, const VectorXd& b) override;
29 |
30 | protected:
31 |
32 | /**
33 | * @brief set distribution constant
34 | * @param d (dimension)
35 | * @param n (number of constraints)
36 | */
37 | void setDistTerm(int d, int n) override;
38 |
39 | };
40 |
41 | #endif
--------------------------------------------------------------------------------
/src/sparse/Common.hpp:
--------------------------------------------------------------------------------
1 | #ifndef COMMON_HPP
2 | #define COMMON_HPP
3 |
4 | #include
5 | #include
6 | #include
7 | #include
8 | #include
9 | typedef Eigen::SparseMatrix SparseMatrixXd;
10 | typedef Eigen::Triplet T;
11 |
12 | #include
13 | #include
14 | #include
15 | #include
16 | #include
17 | #include
18 | #include
19 |
20 | using namespace Eigen;
21 | using namespace std;
22 |
23 | #endif
--------------------------------------------------------------------------------
/src/sparse/LeverageScore.cpp:
--------------------------------------------------------------------------------
1 | #include "LeverageScore.hpp"
2 |
3 |
4 | VectorXd LeverageScore::generate(const SparseMatrixXd& A, const SparseMatrixXd& W, const VectorXd& x, const double ERR, const int k){
5 | // Efficient Computation of Leverage Score in B.2.3 of Kook et al. 2022,
6 | // Sampling with Riemannian Hamiltonian Monte Carlo in a Constrained Space
7 | // Takahashi, Fagan, and Chin method for sparse matrix inversion
8 | // via sparse Cholesky decomposition
9 | // W (A\tp W^2 A)^{-1} W
10 | VectorXd S_inv(x.rows());
11 | for(int i = x.rows() - k; i < x.rows(); i++){
12 | S_inv.coeffRef(i) = 1/x(i);
13 | }
14 | VectorXd G_sqrt = W * S_inv;
15 | for(int i = 0; i < x.rows() - k; i++){
16 | G_sqrt.coeffRef(i) = ERR;
17 | }
18 |
19 | SparseMatrixXd G_inv_sqrt = SparseMatrixXd(G_sqrt.cwiseInverse().asDiagonal());
20 | SparseMatrixXd AG_inv_sqrt = A * G_inv_sqrt;
21 |
22 | SparseMatrixXd hess = AG_inv_sqrt * AG_inv_sqrt.transpose();
23 |
24 | SimplicialLDLT, Eigen::Lower, Eigen::NaturalOrdering> cholesky;
25 | cholesky.analyzePattern(hess);
26 | cholesky.factorize(hess);
27 | SparseMatrixXd L0 = cholesky.matrixL();
28 | VectorXd D = cholesky.vectorD();
29 |
30 | // permutation matrix rearranges the row and columns
31 | // the reason is that sparse matrix does not allow backward access
32 | // of row entries
33 | Eigen::PermutationMatrix perm (L0.rows());
34 | for(int i = 0; i < L0.rows(); i++){
35 | perm.indices()(i) = perm.rows() - 1 - i;
36 | }
37 |
38 | SparseMatrixXd L_col = perm * L0 * perm;
39 |
40 | SparseMatrixXd inv(L0.rows(), L0.rows());
41 |
42 | // get the sparsity pattern of L
43 | // computer the inverse of (A g^{-1} A^T) restricted to L's sparsity pattern
44 | VectorXd nnz (L0.rows());
45 | for(int i = 0; i < L_col.rows(); i++){
46 | nnz(i) = L_col.col(i).nonZeros();
47 | }
48 | inv.reserve(nnz);
49 |
50 | // the inverse can be computed top row -> bottom row
51 | // left -> right, one by one
52 | // Takahashi, Fagan, and Chin method
53 | for(int i = 0; i < L_col.outerSize(); i++){
54 | for(SparseMatrixXd::InnerIterator it(L_col, i); it; ++it){
55 | int j = it.row();
56 | double z = (i == j) ? (double)1/D(L_col.outerSize() - 1 - i) : 0;
57 |
58 | for(SparseMatrixXd::InnerIterator it2(L_col, i); it2; ++it2){
59 | if (it2.row() >= i) break;
60 | double val = it2.row() <= j ? inv.coeff(it2.row(), j) : inv.coeff(j, it2.row());
61 | z -= it2.value() * val;
62 | }
63 | if (i <= j) inv.insert(i, j) = z;
64 | else inv.insert(j, i) = z;
65 | }
66 | }
67 | // the permutation matrix rearranges the row and column
68 | inv = perm * inv * perm;
69 |
70 | // P = (A g^{-1} A^T)^{-1} * A g^{-1/2}
71 | SparseMatrixXd P = inv.selfadjointView() * AG_inv_sqrt;
72 | VectorXd result (AG_inv_sqrt.cols());
73 | for(int i = 0; i < AG_inv_sqrt.cols(); i++){
74 | double val = AG_inv_sqrt.col(i).dot(P.col(i));
75 | // i-th leverage score
76 | result.coeffRef(i) = val;
77 | }
78 |
79 | for(int i = 0; i < x.rows() - k; i++){
80 | result(i) = 0;
81 | }
82 |
83 | // leverage score of I - g^{-1/2} A^T (A g^{-1} A^T)^{-1} A g^{-1/2}
84 | for(int i = x.rows() - k; i < x.rows(); i++){
85 | result(i) = 1 - result(i);
86 | }
87 |
88 | return result;
89 | }
--------------------------------------------------------------------------------
/src/sparse/LeverageScore.hpp:
--------------------------------------------------------------------------------
1 | #ifndef LEVSCORE_HPP
2 | #define LEVSCORE_HPP
3 | #include "Common.hpp"
4 |
5 | class LeverageScore{
6 | public:
7 |
8 | LeverageScore (){};
9 | /**
10 | * @brief get the Leverage Score approximate calculation
11 | * @param A polytope matrix (Ax = b)
12 | * @param W Weight Matrix for slack
13 | * @param x polytope vector (Ax = b)
14 | * @param ERR error term
15 | * @param k last k values have inequality constraint
16 | * @return Vector
17 | */
18 | VectorXd generate(const SparseMatrixXd& A, const SparseMatrixXd& W, const VectorXd& x, const double ERR, const int k);
19 | };
20 |
21 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseBallWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseBallWalk.hpp"
2 |
3 | MatrixXd SparseBallWalk::generateCompleteWalk(
4 | const int num_steps,
5 | const VectorXd& init,
6 | const SparseMatrixXd& A,
7 | const VectorXd& b,
8 | int k,
9 | int burn = 0
10 | ){
11 | MatrixXd results = MatrixXd::Zero(num_steps, A.cols());
12 |
13 | SparseLU A_solver (A * A.transpose());
14 | SparseMatrixXd I = SparseMatrixXd(VectorXd::Ones(A.cols()).asDiagonal());
15 |
16 | VectorXd x = init;
17 | int d = A.cols() - A.rows();
18 | int total = (burn + num_steps) * THIN;
19 | for (int i = 1; i <= total; i++){
20 | VectorXd rand = generateGaussianRV(A.cols());
21 | VectorXd z;
22 | z = A * rand;
23 | z = rand - A.transpose() * A_solver.solve(z);
24 | z /= z.norm();
25 | z = R/sqrt(d) * z + x;
26 |
27 | if (inPolytope(z, k)){
28 | x = z;
29 | }
30 | if (i % THIN == 0 && i/THIN > burn){
31 | results.row((int)i/THIN - burn - 1) = x;
32 | }
33 | }
34 | return results;
35 | }
--------------------------------------------------------------------------------
/src/sparse/SparseBallWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSBALLWALK_HPP
2 | #define CONSBALLWALK_HPP
3 |
4 | #include "SparseRandomWalk.hpp"
5 |
6 | class SparseBallWalk : public SparseRandomWalk{
7 | public:
8 | /**
9 | * @brief initialization of Sparse Ball Walk class
10 | * @param r spread parameter
11 | * @param thin thin parameter
12 | */
13 | SparseBallWalk(double r, int thin = 1) : R(r), SparseRandomWalk(thin, 0.0){}
14 |
15 | /**
16 | * @brief generate values from the Ball walk (constrained)
17 | * @param num_steps number of steps wanted to take
18 | * @param init initial starting point
19 | * @param A polytope matrix
20 | * @param b polytope vector
21 | * @param k k values >= 0 constraint
22 | * @param burn number of initial steps to cut
23 | * @return num_steps by d (dimension of x) matrix
24 | */
25 | MatrixXd generateCompleteWalk(
26 | const int num_steps,
27 | const VectorXd& init,
28 | const SparseMatrixXd& A,
29 | const VectorXd& b,
30 | int k,
31 | int burn
32 | ) override;
33 |
34 | protected:
35 | /**
36 | * @brief spread parameter
37 | */
38 | const double R;
39 | };
40 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseBarrierWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseBarrierWalk.hpp"
2 |
3 | SparseMatrixXd SparseBarrierWalk::generateWeight(
4 | const VectorXd& x,
5 | const SparseMatrixXd& A,
6 | int k
7 | ){
8 | return SparseMatrixXd(VectorXd::Ones(A.cols()).asDiagonal());
9 | }
10 |
11 | void SparseBarrierWalk::setDistTerm(int d, int n){
12 | DIST_TERM = 0;
13 | }
14 |
15 | SparseMatrixXd SparseBarrierWalk::generateSlackInverse(const VectorXd& x, int k){
16 | VectorXd slack_inv (x.rows());
17 | for(int i = x.rows() - k; i < x.rows(); i++) slack_inv(i) = 1/x(i);
18 |
19 | return SparseMatrixXd(slack_inv.asDiagonal());
20 | }
21 |
22 | VectorXd SparseBarrierWalk::generateSample(
23 | const VectorXd& x,
24 | const SparseMatrixXd& A,
25 | int k
26 | ){
27 | SparseMatrixXd slack_inv = generateSlackInverse(x, k);
28 | SparseMatrixXd W = generateWeight(x, A, k);
29 | SparseMatrixXd G = slack_inv * W * slack_inv;
30 | for(int i = 0; i < x.rows() - k; i++) G.coeffRef(i, i) = ERR;
31 |
32 | SparseMatrixXd G_inv_sqrt = SparseMatrixXd(VectorXd(G.diagonal()).cwiseInverse().cwiseSqrt().asDiagonal());
33 |
34 | SparseMatrixXd AG_inv_sqrt = A * G_inv_sqrt;
35 |
36 | VectorXd rand = generateGaussianRV(A.cols());
37 | SparseMatrixXd res = AG_inv_sqrt * AG_inv_sqrt.transpose();
38 | SimplicialLLT chol;
39 | chol.analyzePattern(res);
40 | chol.factorize(res);
41 |
42 | VectorXd z = AG_inv_sqrt * rand;
43 |
44 | z = AG_inv_sqrt.transpose() * chol.solve(z);
45 | z = G_inv_sqrt * (rand - z);
46 | z = x + sqrt(DIST_TERM) * z;
47 |
48 | return z;
49 | }
50 |
51 | double SparseBarrierWalk::generateProposalDensity(
52 | const VectorXd& x,
53 | const VectorXd& z,
54 | const SparseMatrixXd& A,
55 | int k
56 | ){
57 | SparseMatrixXd slack_inv = generateSlackInverse(x, k);
58 | SparseMatrixXd W = generateWeight(x, A, k);
59 | SparseMatrixXd G = slack_inv * W * slack_inv;
60 | for(int i = 0; i < x.rows() - k; i++) G.coeffRef(i, i) = ERR;
61 |
62 | SparseMatrixXd G_inv_sqrt = SparseMatrixXd(VectorXd(G.diagonal()).cwiseInverse().cwiseSqrt().asDiagonal());
63 | SparseMatrixXd AG_inv_sqrt = A * G_inv_sqrt;
64 |
65 | // determinant of S^{-1} W S^{-1}
66 | double det1 = G.diagonal().array().log().sum();
67 |
68 | SimplicialLLT d2;
69 | SparseMatrixXd mat = AG_inv_sqrt * AG_inv_sqrt.transpose();
70 | d2.analyzePattern(mat);
71 | d2.factorize(mat);
72 |
73 | double det2 = 2 * SparseMatrixXd(d2.matrixL()).diagonal().array().log().sum();
74 | // -logdet of the matrix g^{-1/2} A^T (A g A^T )^{-1} A g^{-1/2}
75 | // equals to logdet(g) + logdet(A g A^T) - \logdet(AA^T)
76 | // but - \logdet(AA^T) is shared at x or z, so ignored
77 | double det = det1 + det2;
78 |
79 | VectorXd diff = z - x;
80 | VectorXd Qx = A * diff;
81 | Qx = A_solver.solve(Qx);
82 | Qx = A.transpose() * Qx;
83 | Qx = diff - Qx;
84 |
85 | double dist = Qx.transpose() * (G * Qx);
86 | // return the log proposal density
87 | return 0.5 * det - 0.5/DIST_TERM * dist;
88 | }
89 |
90 | MatrixXd SparseBarrierWalk::generateCompleteWalk(
91 | const int num_steps,
92 | const VectorXd& init,
93 | const SparseMatrixXd& A,
94 | const VectorXd& b,
95 | int k,
96 | int burn = 0
97 | ){
98 | MatrixXd results = MatrixXd::Zero(num_steps, A.cols());
99 | random_device rd;
100 | mt19937 gen(rd());
101 | uniform_real_distribution<> dis(0.0, 1.0);
102 | setDistTerm(A.cols() - A.rows(), k);
103 | VectorXd x = init;
104 | A_solver.compute(A * A.transpose());
105 | int total = (burn + num_steps) * THIN;
106 | for(int i = 1; i <= total; i++){
107 | VectorXd z = generateSample(x, A, k);
108 | if (inPolytope(z, k)){
109 | double g_x_z = generateProposalDensity(x, z, A, k);
110 | double g_z_x = generateProposalDensity(z, x, A, k);
111 | double alpha = min(1.0, exp(g_z_x - g_x_z));
112 | double val = dis(gen);
113 | x = val < alpha ? z : x;
114 | }
115 | if (i % THIN == 0 && i/THIN > burn){
116 | results.row((int)i/THIN - burn - 1) = x.transpose();
117 | }
118 | }
119 |
120 | return results;
121 |
122 | }
--------------------------------------------------------------------------------
/src/sparse/SparseBarrierWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSBARRIERWALK_HPP
2 | #define CONSBARRIERWALK_HPP
3 | #include "SparseRandomWalk.hpp"
4 |
5 | class SparseBarrierWalk : public SparseRandomWalk{
6 |
7 | public:
8 | /**
9 | * @brief initialization of Sparse Barrier Walk class
10 | * @param r spread parameter
11 | * @param thin thin parameter
12 | * @param err error term parameter
13 | */
14 | SparseBarrierWalk(double r, int thin = 1, double err = 1e-6) : R(r), SparseRandomWalk(thin, err) {}
15 |
16 | /**
17 | * @brief generate weight for slack inverse
18 | * @param x slack variable
19 | * @param A polytope constraint
20 | * @param k k values >= 0 constraint
21 | * @return SparseMatrixXd
22 | */
23 | virtual SparseMatrixXd generateWeight(
24 | const VectorXd& x,
25 | const SparseMatrixXd& A,
26 | int k
27 | );
28 |
29 | /**
30 | * @brief generate values from the SparseBarrierWalk
31 | * @param num_steps number of steps wanted to take
32 | * @param init initial starting point
33 | * @param A polytope matrix
34 | * @param b polytope vector
35 | * @param k k values >= 0 constraint
36 | * @param burn number of initial steps to cut
37 | * @return num_steps by d (dimension of x) matrix
38 | */
39 | MatrixXd generateCompleteWalk(
40 | const int num_steps,
41 | const VectorXd& init,
42 | const SparseMatrixXd& A,
43 | const VectorXd& b,
44 | int k,
45 | int burn
46 | ) override;
47 |
48 | /**
49 | * @brief set distribution constant
50 | * @param d polytope matrix
51 | * @param n polytope vector
52 | */
53 | virtual void setDistTerm(int d, int n);
54 |
55 | protected:
56 | /**
57 | * @brief distribution constant
58 | */
59 | double DIST_TERM;
60 |
61 | /**
62 | * @brief spread parameter
63 | */
64 | double R;
65 |
66 | /**
67 | * @brief inverse solver
68 | */
69 | SparseLU A_solver;
70 |
71 | /**
72 | * @brief generate slack inverse (1/x)
73 | * @param x vector value
74 | * @param k k values >= 0 constraint
75 | * @return SparseMatrixXd
76 | */
77 | SparseMatrixXd generateSlackInverse(
78 | const VectorXd& x,
79 | int k
80 | );
81 |
82 | /**
83 | * @brief generate sample from distribution
84 | * @param x vector value
85 | * @param A polytope matrix (Ax = b)
86 | * @param k values >= 0 constraint
87 | * @return VectorXd
88 | */
89 | VectorXd generateSample(
90 | const VectorXd& x,
91 | const SparseMatrixXd& A,
92 | int k
93 | );
94 |
95 | /**
96 | * @brief generate density term
97 | * @param x center of distribution
98 | * @param z value from distribution
99 | * @param A polytope matrix (Ax = b)
100 | * @param k values >= 0 constraint
101 | * @return double
102 | */
103 | double generateProposalDensity(
104 | const VectorXd& x,
105 | const VectorXd& z,
106 | const SparseMatrixXd& A,
107 | int k
108 | );
109 | };
110 |
111 | #endif
112 |
113 |
114 |
115 |
116 |
--------------------------------------------------------------------------------
/src/sparse/SparseDikinLSWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseDikinLSWalk.hpp"
2 | SparseMatrixXd SparseDikinLSWalk::generateWeight(
3 | const VectorXd& x,
4 | const SparseMatrixXd& A,
5 | int k
6 | ){
7 | LeverageScore L;
8 |
9 | double d = A.cols() - A.rows();
10 | double n = k;
11 | double q = 2.0 * (1.0 + log(n));
12 | double alpha = 1.0 - 2.0/q;
13 |
14 | if (w_i.coeffRef(0) == -1 || w_i.rows() != x.rows()){
15 | w_i = VectorXd::Ones(x.rows());
16 | }
17 |
18 | // term1 is all 1 vect on the first k coordinates
19 | VectorXd term1 = (alpha) * VectorXd::Ones(x.rows());
20 | VectorXd errors = ERR * VectorXd::Ones(x.rows());
21 |
22 | for(int i = 0; i < w_i.rows() - k; i++){
23 | w_i(i) = 0;
24 | term1(i) = 0;
25 | }
26 | // gradient descent to optimize the LS barrier
27 | for(int i = 0; i < MAX_ITER; i++){
28 | SparseMatrixXd W (x.rows(), x.rows());
29 | VectorXd term2a = VectorXd::Zero(x.rows());
30 | for(int j = x.rows() - k; j < x.rows(); j++){
31 | // term2a = alpha / w
32 | term2a(j) = (double)alpha/w_i(j);
33 | W.coeffRef(j, j) = pow(w_i(j), alpha * 0.5);
34 | }
35 |
36 | // term2b is leverage score
37 | VectorXd term2b = L.generate(A, W, x, ERR, k);
38 | // term2 is gradient log det
39 | // which is the ratio between leverage score and w
40 | VectorXd term2 = term2a.cwiseProduct(term2b);
41 | VectorXd grad = term1 - term2;
42 | if (grad.norm() < G_LIM){
43 | break;
44 | }
45 | w_i = (w_i - STEP_SIZE * grad);
46 | for(int j = x.rows() - k; j < x.rows(); j++){
47 | w_i(j) = max(w_i(j), ERR);
48 | }
49 | }
50 | return SparseMatrixXd(w_i.asDiagonal());
51 |
52 | }
53 |
54 | void SparseDikinLSWalk::setDistTerm(int d, int n){
55 | w_i = VectorXd::Ones(d);
56 | double q = 2.0 * (1.0 + log(n));
57 | double term = (1.0 + q) * (1.0 + q * q);
58 | DIST_TERM = (R * R)/term;
59 | }
--------------------------------------------------------------------------------
/src/sparse/SparseDikinLSWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSDIKINLS_HPP
2 | #define CONSDIKINLS_HPP
3 |
4 | #include "SparseBarrierWalk.hpp"
5 | #include "LeverageScore.hpp"
6 |
7 | class SparseDikinLSWalk : public SparseBarrierWalk{
8 |
9 | public:
10 | /**
11 | * @brief initialization of Sparse Lee Sidford Walk class
12 | * @param r spread parameter
13 | * @param thin thin parameter
14 | * @param g_lim gradient descent norm limit
15 | * @param step_size size of gradient descent step
16 | * @param max_iter maximum number of iterations in gradient descent
17 | * @param err error constant
18 | */
19 | SparseDikinLSWalk(double r, int thin = 1, double g_lim = 0.01, double step_size = 0.1, int max_iter = 1000, double err = 1e-6) : G_LIM(g_lim), STEP_SIZE(step_size), MAX_ITER(max_iter), SparseBarrierWalk(r, thin, err) {}
20 |
21 | /**
22 | * @brief generate weight by solving convex optimization task
23 | * @param x slack variable
24 | * @param A polytope constraint
25 | * @param k k values >= 0 constraint
26 | * @return SparseMatrixXd
27 | */
28 | SparseMatrixXd generateWeight(
29 | const VectorXd& x,
30 | const SparseMatrixXd& A,
31 | int k
32 | ) override;
33 |
34 | protected:
35 |
36 | /**
37 | * @brief set distribution constant
38 | * @param d polytope matrix
39 | * @param n polytope vector
40 | */
41 | void setDistTerm(int d, int n) override;
42 |
43 | /**
44 | * @brief stops gradient descent if it reaches under this number
45 | */
46 | const double G_LIM;
47 |
48 | /**
49 | * @brief step size for gradient descent
50 | */
51 | const double STEP_SIZE;
52 |
53 | /**
54 | * @brief max number of iterations in gradient descent
55 | */
56 | const int MAX_ITER;
57 |
58 | /**
59 | * @brief saves current weight for iteration
60 | */
61 | VectorXd w_i = VectorXd::Zero(1) - VectorXd::Ones(1);
62 |
63 | };
64 |
65 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseDikinWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseDikinWalk.hpp"
2 |
3 | SparseMatrixXd SparseDikinWalk::generateWeight(
4 | const VectorXd& x,
5 | const SparseMatrixXd& A,
6 | int k
7 | ){
8 |
9 | return SparseMatrixXd(VectorXd::Ones(A.cols()).asDiagonal());
10 | }
11 |
12 | void SparseDikinWalk::setDistTerm(int d, int n){
13 | DIST_TERM = (R * R)/d;
14 | }
15 |
--------------------------------------------------------------------------------
/src/sparse/SparseDikinWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSDIKIN_HPP
2 | #define CONSDIKIN_HPP
3 |
4 | #include "SparseBarrierWalk.hpp"
5 |
6 | class SparseDikinWalk : public SparseBarrierWalk{
7 |
8 | public:
9 | /**
10 | * @brief initialization of Sparse Dikin Walk class
11 | * @param r spread parameter
12 | * @param thin thin parameter
13 | * @param err error constant
14 | */
15 | SparseDikinWalk(double r, int thin = 1, double err = 1e-6) : SparseBarrierWalk(r, thin, err) {}
16 |
17 | /**
18 | * @brief generate weight (identity matrix)
19 | * @param x slack variable
20 | * @param A polytope constraint
21 | * @param k k values >= 0 constraint
22 | * @return SparseMatrixXd
23 | */
24 | SparseMatrixXd generateWeight(
25 | const VectorXd& x,
26 | const SparseMatrixXd& A,
27 | int k
28 | ) override;
29 |
30 | protected:
31 |
32 | /**
33 | * @brief set distribution constant
34 | * @param d polytope matrix
35 | * @param n polytope vector
36 | */
37 | void setDistTerm(int d, int n) override;
38 |
39 | };
40 |
41 | #endif
42 |
43 |
--------------------------------------------------------------------------------
/src/sparse/SparseHitRun.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseHitRun.hpp"
2 |
3 | double SparseHitAndRun::binarySearch(
4 | VectorXd direction,
5 | VectorXd& x,
6 | int k
7 | ){
8 | VectorXd farth = x + R * direction;
9 | double dist = 0;
10 |
11 | const int MAXITER = 10000;
12 | int iter = 0;
13 |
14 | while(iter < MAXITER){
15 | dist = (x - farth).norm();
16 | farth = x + 2 * dist * direction;
17 | if (!inPolytope(farth, k)){
18 | break;
19 | }
20 | iter++;
21 | }
22 |
23 | if (iter == MAXITER){
24 | return 0.0;
25 | }
26 | VectorXd left = x;
27 | VectorXd right = farth;
28 | VectorXd mid = (x + farth)/2;
29 | while ((left - right).norm() > ERR || !inPolytope(mid, k)){
30 | mid = (left + right)/2;
31 | if (inPolytope(mid, k)){
32 | left = mid;
33 | } else {
34 | right = mid;
35 | }
36 |
37 | }
38 | return (mid - x).norm();
39 | }
40 |
41 | MatrixXd SparseHitAndRun::generateCompleteWalk(
42 | const int num_steps,
43 | const VectorXd& init,
44 | const SparseMatrixXd& A,
45 | const VectorXd& b,
46 | int k,
47 | int burn = 0
48 | ){
49 |
50 | MatrixXd results = MatrixXd::Zero(num_steps, A.cols());
51 | random_device rd;
52 | mt19937 gen(rd());
53 | uniform_real_distribution<> dis(0.0, 1.0);
54 |
55 | SparseLU A_solver (A * A.transpose());
56 | VectorXd x = init;
57 | int total = (burn + num_steps) * THIN;
58 | for (int i = 1; i <= total; i++){
59 | VectorXd rand = generateGaussianRV(A.cols());
60 | VectorXd z = A * rand;
61 | z = rand - A.transpose() * A_solver.solve(z);
62 | z /= z.norm();
63 | double pos_side = binarySearch(z, x, k);
64 | double neg_side = -binarySearch(-z, x, k);
65 | double val = dis(gen);
66 | double random_point = val * (pos_side - neg_side) + neg_side;
67 | x = random_point * z + x;
68 |
69 | if (i % THIN == 0 && i/THIN > burn){
70 | results.row((int)i/THIN - burn - 1) = x;
71 | }
72 | }
73 | return results;
74 |
75 | }
--------------------------------------------------------------------------------
/src/sparse/SparseHitRun.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSHITRUN_HPP
2 | #define CONSHITRUN_HPP
3 |
4 | #include "SparseRandomWalk.hpp"
5 |
6 | class SparseHitAndRun : public SparseRandomWalk{
7 | public:
8 | /**
9 | * @brief initialization of Sparse Hit and Run class
10 | * @param r spread parameter
11 | * @param thin thin parameter
12 | * @param err error constant
13 | */
14 | SparseHitAndRun(double r, int thin = 1, double err = 1e-6) : R(r), SparseRandomWalk(thin, err) {}
15 |
16 | /**
17 | * @brief generate values from the Hit and Run
18 | * @param num_steps number of steps wanted to take
19 | * @param init initial starting point
20 | * @param A polytope matrix
21 | * @param b polytope vector
22 | * @param k k values >= 0 constraint
23 | * @param burn number of initial steps to cut
24 | * @return Matrix
25 | */
26 | MatrixXd generateCompleteWalk(
27 | const int num_steps,
28 | const VectorXd& init,
29 | const SparseMatrixXd& A,
30 | const VectorXd& b,
31 | int k,
32 | int burn
33 | ) override;
34 |
35 |
36 | protected:
37 | /**
38 | * @brief spread parameter
39 | */
40 | const double R;
41 |
42 | /**
43 | * @brief runs binary search to find a suitable chord intersection with the polytope
44 | * @param direction (random direction variable)
45 | * @param x (starting point)
46 | * @param k k values >= 0 constraint
47 | * @return double
48 | */
49 | double binarySearch(
50 | VectorXd direction,
51 | VectorXd& x,
52 | int k);
53 | };
54 |
55 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseJohnWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseJohnWalk.hpp"
2 | #include "LeverageScore.hpp"
3 |
4 | SparseMatrixXd SparseJohnWalk::generateWeight(
5 | const VectorXd& x,
6 | const SparseMatrixXd& A,
7 | int k
8 | ){
9 | double d = A.cols() - A.rows();
10 | double n = k;
11 | double alpha = 1 - 1/(log2(2.0 * n / d));
12 | double beta = (double)d / (2.0 * n);
13 |
14 | if (w_i.coeffRef(0) == -1 || w_i.rows() != x.rows()){
15 | w_i = VectorXd::Ones(x.rows());
16 | }
17 |
18 | LeverageScore L;
19 | VectorXd beta_ones = beta * VectorXd::Ones(x.rows());
20 |
21 | for(int i = 0; i < w_i.rows() - k; i++){
22 | w_i(i) = 0;
23 | beta_ones.coeffRef(i) = 0;
24 | }
25 | VectorXd next_weight = w_i;
26 |
27 | // fixed point iteration
28 | for(int i = 0; i < MAX_ITER; i++){
29 | w_i = next_weight;
30 | SparseMatrixXd W (w_i.rows(), w_i.rows());
31 | for(int j = x.rows() - k; j < x.rows(); j++){
32 | W.coeffRef(j, j) = pow(w_i(j), alpha * 0.5);
33 | }
34 | VectorXd score = L.generate(A, W, x, ERR, k);
35 | next_weight = 0.5 * (w_i + score + beta_ones).cwiseMax(beta_ones);
36 |
37 | if ((w_i - next_weight).cwiseAbs().maxCoeff() < LIM){
38 | break;
39 | }
40 | }
41 |
42 | return SparseMatrixXd(w_i.asDiagonal());
43 |
44 | }
45 |
46 | void SparseJohnWalk::setDistTerm(int d, int n){
47 | w_i = VectorXd::Ones(d);
48 | DIST_TERM = (R * R)/pow(d, 1.5);
49 | }
--------------------------------------------------------------------------------
/src/sparse/SparseJohnWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSJOHN_HPP
2 | #define CONSJOHN_HPP
3 |
4 | #include "SparseBarrierWalk.hpp"
5 |
6 | class SparseJohnWalk : public SparseBarrierWalk{
7 |
8 | public:
9 | /**
10 | * @brief initialization of Sparse John Walk class
11 | * @param r spread parameter
12 | * @param thin thin parameter
13 | * @param lim limit in l-infinity norm
14 | * @param max_iter maximum number of iterations in fixed iteration
15 | * @param err error constant
16 | */
17 | SparseJohnWalk(double r, int thin = 1, double lim = 1e-5, int max_iter = 1000, double err = 1e-5) : LIM(lim), MAX_ITER(max_iter), SparseBarrierWalk(r, thin, err) {}
18 |
19 | /**
20 | * @brief generate weight by solving fixed point iteration
21 | * @param x slack variable
22 | * @param A polytope constraint
23 | * @param k k values >= 0 constraint
24 | * @return SparseMatrixXd
25 | */
26 | SparseMatrixXd generateWeight(
27 | const VectorXd& x,
28 | const SparseMatrixXd& A,
29 | int k
30 | ) override;
31 |
32 | protected:
33 |
34 | /**
35 | * @brief set distribution constant
36 | * @param d polytope matrix
37 | * @param n polytope vector
38 | */
39 | void setDistTerm(int d, int n) override;
40 |
41 | /**
42 | * @brief stops if it reaches under this number during fixed iteration
43 | */
44 | const double LIM;
45 |
46 | /**
47 | * @brief max number of iterations in fixed iteration
48 | */
49 | const int MAX_ITER;
50 |
51 | /**
52 | * @brief saves current weight for iteration
53 | */
54 | VectorXd w_i = VectorXd::Zero(1) - VectorXd::Ones(1);
55 |
56 |
57 |
58 | };
59 |
60 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseRandomWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseRandomWalk.hpp"
2 |
3 | VectorXd SparseRandomWalk::generateGaussianRV(int d){
4 | VectorXd v(d);
5 | random_device rd;
6 | mt19937 gen(rd());
7 | normal_distribution dis(0.0, 1.0);
8 | for(int i = 0; i < d; i++){
9 | v(i) = dis(gen);
10 | }
11 | return v;
12 | }
13 |
14 | MatrixXd SparseRandomWalk::generateCompleteWalk(
15 | const int num_steps,
16 | const VectorXd& init,
17 | const SparseMatrixXd& A,
18 | const VectorXd& b,
19 | int k,
20 | int burn = 0
21 | ){
22 | cout << "Oops" << endl;
23 | return MatrixXd::Zero(1,1);
24 |
25 | }
26 |
27 | bool SparseRandomWalk::inPolytope(
28 | const VectorXd&z,
29 | int k
30 | ){
31 | return z.tail(k).minCoeff() >= 0;
32 | }
--------------------------------------------------------------------------------
/src/sparse/SparseRandomWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSTRAINTWALK_HPP
2 | #define CONSTRAINTWALK_HPP
3 | #include "Common.hpp"
4 |
5 | class SparseRandomWalk{
6 |
7 | public:
8 | /**
9 | * @brief initialization of Sparse Random Walk class
10 | * @param thin thin constant
11 | * @param err error constant
12 | */
13 | SparseRandomWalk(int thin = 1, double err = 1e-6) : ERR(err), THIN(thin) {}
14 |
15 | /**
16 | * @brief Generate values from the RandomWalk
17 | * @param num_steps number of steps wanted to take
18 | * @param init initial starting point
19 | * @param A polytope matrix
20 | * @param b polytope vector
21 | * @param k k values >= 0 constraint
22 | * @param burn number of steps to burn
23 | * @return Matrix
24 | */
25 | virtual MatrixXd generateCompleteWalk(
26 | const int num_steps,
27 | const VectorXd& init,
28 | const SparseMatrixXd& A,
29 | const VectorXd& b,
30 | int k,
31 | int burn
32 | );
33 |
34 | protected:
35 | /**
36 | * @brief check if value is in polytope
37 | * @param z proposal vector (assuming sampled from Ax = 0)
38 | * @param k k values >= 0 constraint
39 | * @return Matrix
40 | */
41 | bool inPolytope(const VectorXd& z, int k);
42 |
43 | /**
44 | * @brief returns Gaussian vector of dimension d
45 | * @param d
46 | * @return vector
47 | */
48 | VectorXd generateGaussianRV(const int d);
49 |
50 | /**
51 | * @brief error constant
52 | */
53 | const double ERR;
54 |
55 | /**
56 | * @brief include every ___ sample
57 | */
58 | const int THIN;
59 | };
60 |
61 | #endif
--------------------------------------------------------------------------------
/src/sparse/SparseVaidyaWalk.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseVaidyaWalk.hpp"
2 | #include "LeverageScore.hpp"
3 |
4 | SparseMatrixXd SparseVaidyaWalk::generateWeight(
5 | const VectorXd& x,
6 | const SparseMatrixXd& A,
7 | int k
8 | ){
9 | LeverageScore L;
10 | SparseMatrixXd W (x.rows(), x.rows());
11 | for(int i = x.rows() - k; i < x.rows(); i++){
12 | W.coeffRef(i, i) = 1;
13 | }
14 | VectorXd weights = L.generate(A, W, x, ERR, k);
15 | for (int i = weights.rows() - k; i < weights.rows(); i++){
16 | // in the full-rank form of the polytope
17 | // # constraints = A.cols() - A.rows()
18 | // # variables = k
19 | weights(i) += ((double)(A.cols() - A.rows())/k);
20 | }
21 | return SparseMatrixXd(weights.asDiagonal());
22 | }
23 |
24 | void SparseVaidyaWalk::setDistTerm(int d, int n){
25 | DIST_TERM = (R * R)/sqrt(n * d);
26 | }
--------------------------------------------------------------------------------
/src/sparse/SparseVaidyaWalk.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CONSVAIDYA_HPP
2 | #define CONSVAIDYA_HPP
3 |
4 | #include "SparseBarrierWalk.hpp"
5 |
6 | class SparseVaidyaWalk : public SparseBarrierWalk{
7 |
8 | public:
9 | /**
10 | * @brief constructor for Vaidya Walk class
11 | * @param r spread parameter
12 | * @param thin thin parameter
13 | * @param err error constant
14 | */
15 | SparseVaidyaWalk(double r, int thin = 1, double err = 1e-6) : SparseBarrierWalk(r, thin, err) {}
16 |
17 | /**
18 | * @brief generate weight (leverage score calculation)
19 | * @param x slack variable
20 | * @param A polytope constraint
21 | * @param k k values >= 0 constraint
22 | * @return SparseMatrixXd
23 | */
24 | SparseMatrixXd generateWeight(
25 | const VectorXd& x,
26 | const SparseMatrixXd& A,
27 | int k
28 | ) override;
29 |
30 | protected:
31 |
32 | /**
33 | * @brief Distribution constant
34 | * @param d polytope matrix
35 | * @param n polytope vector
36 | */
37 | void setDistTerm(int d, int n) override;
38 |
39 | };
40 |
41 | #endif
--------------------------------------------------------------------------------
/src/utils/Common.hpp:
--------------------------------------------------------------------------------
1 | #ifndef COMMON1_HPP
2 | #define COMMON1_HPP
3 |
4 | #include
5 | #include
6 | #include
7 | #include
8 | #include
9 | typedef Eigen::SparseMatrix SparseMatrixXd;
10 | typedef Eigen::Triplet T;
11 |
12 | #include
13 | #include
14 | #include
15 | #include
16 | #include
17 | #include
18 | #include
19 | #include
20 |
21 | using namespace Eigen;
22 | using namespace std;
23 |
24 | #endif
--------------------------------------------------------------------------------
/src/utils/DenseCenter.cpp:
--------------------------------------------------------------------------------
1 | #include "DenseCenter.hpp"
2 |
3 | VectorXd DenseCenter::getInitialPoint(MatrixXd& A, VectorXd& b){
4 | // Solve the linear program
5 | // max delta
6 | // s.t. A x + delta * 1 <= b
7 | glp_prob *lp;
8 | lp = glp_create_prob();
9 | glp_term_out(GLP_OFF);
10 | int amount = 1 + (A.rows() * (A.cols() + 1));
11 | vector ia (amount);
12 | vector ja (amount);
13 | vector ar (amount);
14 |
15 | int row_length = A.rows();
16 | // delta is stored at the last column
17 | int col_length = A.cols() + 1;
18 |
19 | glp_add_rows(lp, row_length);
20 | glp_add_cols(lp, col_length);
21 | // maximize delta * 1
22 | glp_set_obj_coef(lp, col_length , 1);
23 | glp_set_obj_dir(lp, GLP_MAX);
24 |
25 | for(int i = 0; i < b.rows(); i++){
26 | glp_set_row_bnds(lp, i + 1, GLP_UP, b(i), b(i));
27 | }
28 | for(int i = 0; i < col_length - 1; i++){
29 | glp_set_col_bnds(lp, i + 1, GLP_FR, 0, 0);
30 | }
31 | glp_set_col_bnds(lp, col_length, GLP_LO, 0, 0);
32 |
33 | int ind = 1;
34 | for(int i = 0; i < A.rows(); i++){
35 | // for A * x
36 | for(int j = 0; j < A.cols(); j++){
37 | ia[ind] = i + 1;
38 | ja[ind] = j + 1;
39 | ar[ind] = A.coeff(i, j);
40 | ind ++;
41 | }
42 | // for + delta * 1
43 | ia[ind] = i + 1;
44 | ja[ind] = A.cols() + 1;
45 | ar[ind] = 1.0;
46 | ind ++;
47 | }
48 |
49 | glp_load_matrix(lp, ind-1, ia.data(), ja.data(), ar.data());
50 | glp_simplex(lp, NULL);
51 | double val = glp_get_obj_val(lp);
52 |
53 | // retrieve x
54 | VectorXd ans(A.cols());
55 | for(int i = 0; i < A.cols(); i++){
56 | ans.coeffRef(i) = glp_get_col_prim(lp, i + 1);
57 | }
58 | glp_delete_prob(lp);
59 | return ans;
60 |
61 | }
--------------------------------------------------------------------------------
/src/utils/DenseCenter.hpp:
--------------------------------------------------------------------------------
1 | #ifndef CPF_HPP
2 | #define CPF_HPP
3 |
4 | #include "Common.hpp"
5 |
6 |
7 | class DenseCenter {
8 | public:
9 |
10 | /**
11 | * @brief initialization for Center Algorithm
12 | */
13 | DenseCenter(){};
14 |
15 | /**
16 | * @brief finds analytical center Ax <= b
17 | * @param A polytope matrix (Ax <= b)
18 | * @param b polytope vector (Ax <= b)
19 | * @return VectorXd
20 | */
21 | VectorXd getInitialPoint(MatrixXd& A, VectorXd& b);
22 |
23 | };
24 |
25 | #endif
--------------------------------------------------------------------------------
/src/utils/FacialReduction.cpp:
--------------------------------------------------------------------------------
1 | #include "FacialReduction.hpp"
2 |
3 |
4 | ZResult FacialReduction::findZ(const SparseMatrixXd& A, const VectorXd& b, int x_dim){
5 | // A size n * d
6 | // b size n
7 | // x_dim = d-k
8 |
9 | // finds a vector y satisfying
10 | // A^Ty = [0 z]
11 | // s.t. = 0
12 | // z in R^k, z >= 0, z != 0
13 | // first n-k terms is 0, last k terms is z
14 | ZResult ans;
15 | ans.found_sol = false;
16 | SparseLP sparse_lp;
17 |
18 | int row_length = A.cols() + 1;
19 | int col_length = A.rows();
20 | SparseMatrixXd obj_mat(row_length, col_length);
21 | VectorXd obj_vec = VectorXd::Zero(col_length);
22 | obj_vec(obj_vec.rows() - 1) = 1;
23 |
24 | VectorXd row_bnds = VectorXd::Zero(row_length);
25 | VectorXd row_rel = VectorXd::Zero(row_length);
26 | VectorXd col_bnds = VectorXd::Zero(col_length);
27 | VectorXd col_rel = VectorXd::Zero(col_length);
28 |
29 | // construct [0 z]
30 | // z in R^k, z >= 0, z != 0
31 | // first d-k terms is 0, last k terms is z
32 | for(int i = 0; i < A.cols(); i++){
33 | if (i < x_dim) {
34 | row_rel(i) = GLP_FX;
35 | } else {
36 | row_rel(i) = GLP_LO;
37 | }
38 | }
39 |
40 | // = 0
41 | row_rel(A.cols()) = GLP_FX;
42 |
43 | // y is free
44 | for(int i = 0; i < col_length; i++){
45 | col_rel(i) = GLP_FR;
46 | }
47 |
48 | // copy A into obj_mat
49 | for(int i = 0; i < A.outerSize(); i++){
50 | for(SparseMatrixXd::InnerIterator it(A, i); it; ++it){
51 | int row = it.row();
52 | int col = it.col();
53 | double val = it.value();
54 |
55 | obj_mat.insert(col, row) = val;
56 | }
57 | }
58 | // copy b into obj_mat
59 | for(int i = 0; i < b.rows(); i++){
60 | obj_mat.insert(A.cols(), i) = b.coeff(i);
61 | }
62 |
63 | // loop over index i where z_i is nonzero
64 | // global_index is the previous known index that works
65 | // always start with global_index to save computation
66 | for(int i = global_index; i < A.cols(); i++){
67 | // at least one coordinate of z is nonzero
68 | // the problem is scale-invariant, can make it 1
69 | row_rel(i) = GLP_FX;
70 | row_bnds(i) = 1;
71 |
72 | // solve the LP via one LP solver
73 | // A^Ty = [0 z]
74 | // s.t. = 0
75 | // z_i = 1
76 | // z in R^k, z >= 0,
77 | VectorXd sol = sparse_lp.findOptimalVector(obj_mat, row_bnds, obj_vec, row_rel, col_bnds, col_rel);
78 | if (sol.cwiseAbs().sum() != 0){
79 | ans.found_sol = true;
80 | ans.z = (A.transpose() * sol);
81 | return ans;
82 | }
83 | // increment global_index if we didn't find a solution
84 | global_index++;
85 | row_rel(i) = GLP_LO;
86 | row_bnds(i) = 0;
87 | }
88 | return ans;
89 | }
90 |
91 | SparseMatrixXd FacialReduction::pickV(const VectorXd& z, int x_dim){
92 | // z size d
93 | // first d-k coordinate are always 0
94 | int d = z.rows();
95 | vector indices;
96 | for(int i = 0; i < x_dim; i++){
97 | indices.push_back(T(indices.size(), i, 1));
98 | }
99 | // find indices where z == 0
100 | for(int i = x_dim; i < d; i++){
101 | if(z(i) < ERR_DC) indices.push_back(T(indices.size(), i, 1));
102 | }
103 | // outputs a matrix selecting the coordinates corresponds to zero of z
104 | SparseMatrixXd mat(indices.size(), d);
105 | mat.setFromTriplets(indices.begin(), indices.end());
106 | return mat.transpose();
107 | }
108 |
109 | SparseMatrixXd FacialReduction::pickP(const SparseMatrixXd& AV){
110 | // sparse QR decomposition to find redundant constraints
111 | SparseQR::StorageIndex>> solver;
112 | solver.compute(AV.transpose());
113 | SparseMatrixXd R = solver.matrixR();
114 |
115 | vector indices;
116 | for (int i = 0; i < min(R.cols(), R.rows()); i++){
117 | if (abs(R.coeffRef(i, i)) > ERR_DC){
118 | // nonzero R(i,i) means linearly independent row
119 | indices.push_back(T(indices.size(), solver.colsPermutation().indices()(i), 1));
120 | }
121 | }
122 | // proj is a projection that projects into linearly independent rows
123 | SparseMatrixXd proj (indices.size(), AV.rows());
124 | proj.setFromTriplets(indices.begin(), indices.end());
125 | return proj;
126 | }
127 |
128 | FRResult FacialReduction::entireFacialReductionStep(SparseMatrixXd& A, VectorXd& b, int x_dim, SparseMatrixXd& savedV){
129 | // findZ->pickV->pickP
130 | ZResult z_ans = findZ(A, b, x_dim);
131 |
132 | // if findZ is not successful, then the original form is strictly feasible
133 | if(!z_ans.found_sol){
134 | FRResult ans;
135 | ans.A = A;
136 | ans.b = b;
137 | ans.savedV = savedV;
138 | return ans;
139 | }
140 | SparseMatrixXd V = pickV(z_ans.z, x_dim);
141 | // savedV stores the multiplication of all Vs in all pickV steps
142 | savedV = savedV * V;
143 | SparseMatrixXd AV = A * V;
144 | SparseMatrixXd P = pickP(AV);
145 | A = P * AV;
146 | b = P * b;
147 | return entireFacialReductionStep(A, b, x_dim, savedV);
148 | }
149 |
150 | FROutput FacialReduction::reduce(SparseMatrixXd A, VectorXd b, int k, bool sparse){
151 | int x_dim = A.cols() - k;
152 | SparseMatrixXd savedV = SparseMatrixXd(VectorXd::Ones(A.cols()).asDiagonal());
153 | global_index = x_dim;
154 | //remove dependent rows
155 | SparseMatrixXd P = pickP(A);
156 | A = P * A;
157 | b = P * b;
158 | FRResult result = entireFacialReductionStep(A, b, x_dim, savedV);
159 | FROutput final_res;
160 |
161 | final_res.sparse_A = result.A;
162 | final_res.sparse_b = result.b;
163 | final_res.saved_V = result.savedV;
164 |
165 | if(!sparse){
166 | // get full-dim formulation after facial reduction via QR decomp
167 | // Ax <= b
168 | // but A and b can be dense
169 | HouseholderQR qr(result.A.cols(), result.A.rows());
170 | qr.compute(MatrixXd(result.A.transpose()));
171 | MatrixXd Q = qr.householderQ();
172 | MatrixXd R = qr.matrixQR().triangularView();
173 | int d = R.rows();
174 | int n = R.cols();
175 |
176 | MatrixXd newR = R.block(0, 0, R.cols(), R.cols());
177 | VectorXd z1 = newR.transpose().inverse() * result.b;
178 |
179 | MatrixXd Q1 = Q.block(0, 0, Q.rows(), n);
180 | MatrixXd Q2 = Q.block(0, n, Q.rows(), d - n);
181 | MatrixXd reduced_A = -1 * Q2.block(x_dim, 0, d - x_dim, d - n);
182 | VectorXd reduced_b = (Q1 * z1).tail(d - x_dim);
183 |
184 | final_res.dense_A = reduced_A;
185 | final_res.dense_b = reduced_b;
186 |
187 | // z1 and Q are saved so that we can convert back to original form
188 | final_res.z1 = z1;
189 | final_res.Q = Q;
190 | }
191 | return final_res;
192 | }
193 |
194 |
--------------------------------------------------------------------------------
/src/utils/FacialReduction.hpp:
--------------------------------------------------------------------------------
1 | #ifndef SPARSE_FR_HPP
2 | #define SPARSE_FR_HPP
3 |
4 | #include "Common.hpp"
5 | #include "SparseLP.hpp"
6 |
7 | /**
8 | * @brief result of Find Z algorithm
9 | * @param found_sol if the algorithm found a z
10 | * @param z the vector z
11 | */
12 | struct ZResult{
13 | bool found_sol;
14 | VectorXd z;
15 | };
16 |
17 | /**
18 | * @brief result of Facial Reduction step
19 | * @param A Ax = b
20 | * @param b Ax = b
21 | * @param savedV PAVv = Pb decomposition
22 | */
23 | struct FRResult{
24 | SparseMatrixXd A;
25 | VectorXd b;
26 | SparseMatrixXd savedV;
27 | };
28 |
29 | /**
30 | * @brief final output of Facial Reduction algorithm
31 | * @param sparse_A constrained form Ax = b, x >=_k 0
32 | * @param sparse_b constrained form Ax = b, x >=_k 0
33 | * @param sparse_V PAVv = Pb decomposition
34 | * @param dense_A full-dim form Ax <= b
35 | * @param dense_b full-dim form Ax <= b
36 | * @param Q matrix used to go between forms
37 | * @param z1 vector used to go between forms
38 | */
39 | struct FROutput{
40 | SparseMatrixXd sparse_A;
41 | VectorXd sparse_b;
42 | SparseMatrixXd saved_V;
43 | MatrixXd dense_A;
44 | VectorXd dense_b;
45 | MatrixXd Q;
46 | VectorXd z1;
47 | };
48 |
49 | class FacialReduction {
50 | public:
51 | /**
52 | * @brief initialization for Facial Reduction class
53 | * @param err_dc error sensitivity for decomposition calculation
54 | */
55 | FacialReduction(double err_dc = 1e-5) : ERR_DC(err_dc){}
56 | /**
57 | * @brief completes facial reduction on Ax = b, x >=_k 0
58 | * @param A polytope matrix (Ax = b)
59 | * @param b polytope vector (Ax = b)
60 | * @param k k values >= 0 constraint
61 | * @param sparse decision to choose full-dimensional or constraint formulation
62 | * @return FROutput
63 | */
64 | FROutput reduce(SparseMatrixXd A, VectorXd b, int k, bool sparse);
65 |
66 | protected:
67 | /**
68 | * @brief finds a vector z satisfying A^Ty = [0 z], z in R^n, z >= 0, z != 0, = 0
69 | * @param A polytope matrix (Ax = b)
70 | * @param b polytope vector (Ax = b)
71 | * @param k values >= 0 constraint
72 | * @return ZResult
73 | */
74 | ZResult findZ(const SparseMatrixXd& A, const VectorXd& b, int k);
75 |
76 | /**
77 | * @brief finds supports with z vector
78 | * @param z vector
79 | * @param k values >= 0 constraint
80 | * @return SparseMatrixXd
81 | */
82 | SparseMatrixXd pickV(const VectorXd& z, int k);
83 |
84 | /**
85 | * @brief removes redundant constraints in AV
86 | * @param AV matrix to remove redundant constraints
87 | * @return SparseMatrixXd
88 | */
89 | SparseMatrixXd pickP(const SparseMatrixXd& AV);
90 |
91 | /**
92 | * @brief iteratively reduces dimension of the problem using recursion
93 | * @param A polytope matrix (Ax = b)
94 | * @param b polytope vector (Ax = b)
95 | * @param k values >= 0 constraint
96 | * @param savedV V in AVv = b
97 | * @return FRResult
98 | */
99 | FRResult entireFacialReductionStep(SparseMatrixXd& A, VectorXd& b, int k, SparseMatrixXd& savedV);
100 |
101 | /**
102 | * @brief DC error parameter
103 | */
104 | const double ERR_DC;
105 |
106 | /**
107 | * @brief save last index
108 | */
109 | int global_index;
110 | };
111 |
112 | #endif
--------------------------------------------------------------------------------
/src/utils/FullWalkRun.hpp:
--------------------------------------------------------------------------------
1 | #include "FacialReduction.hpp"
2 | #include "DenseCenter.hpp"
3 | #include "SparseCenter.hpp"
4 | #include "dense/DikinWalk.hpp"
5 | #include "dense/DikinLSWalk.hpp"
6 | #include "dense/JohnWalk.hpp"
7 | #include "dense/VaidyaWalk.hpp"
8 | #include "dense/HitRun.hpp"
9 | #include "dense/BallWalk.hpp"
10 | #include "sparse/SparseDikinWalk.hpp"
11 | #include "sparse/SparseDikinLSWalk.hpp"
12 | #include "sparse/SparseJohnWalk.hpp"
13 | #include "sparse/SparseVaidyaWalk.hpp"
14 | #include "sparse/SparseBallWalk.hpp"
15 | #include "sparse/SparseHitRun.hpp"
16 |
17 |
18 | /**
19 | * @brief runs full preprocessing, walk, and post-processing steps in dense formulation
20 | * @param A polytope matrix (Ax = b)
21 | * @param b polytope vector (Ax = b)
22 | * @param k values >= 0 constraint
23 | * @param num_sim number of steps
24 | * @param walk dense random walk implementation
25 | * @param fr facial reduction algorithm
26 | * @param init initialization algorithm
27 | * @param burn how many to exclude
28 | * @return Matrix
29 | */
30 | MatrixXd denseFullWalkRun(SparseMatrixXd A, VectorXd b, int k, int num_sim, RandomWalk* walk, FacialReduction* fr, DenseCenter* init, int burn = 0){
31 | FROutput fr_result = fr->reduce(A, b, k, false);
32 | VectorXd x = init->getInitialPoint(fr_result.dense_A, fr_result.dense_b);
33 | MatrixXd steps = walk->generateCompleteWalk(num_sim, x, fr_result.dense_A, fr_result.dense_b, burn);
34 | MatrixXd res(num_sim, A.cols());
35 | for(int i = 0; i < num_sim; i++){
36 | VectorXd val (steps.cols() + fr_result.z1.rows());
37 | VectorXd row = steps.row(i);
38 | val << fr_result.z1, row;
39 | res.row(i) = (fr_result.Q * val).head(A.cols());
40 | }
41 | return res;
42 | }
43 |
44 | /**
45 | * @brief runs full preprocessing, walk, and post-processing steps in sparse formulation
46 | * @param A polytope matrix (Ax <= b)
47 | * @param b polytope vector (Ax <= b)
48 | * @param k last k coordinates >= 0
49 | * @param num_sim number of steps
50 | * @param walk sparse random walk implementation
51 | * @param fr facial reduction algorithm
52 | * @param init initialization algorithm
53 | * @param burn how many to exclude
54 | * @return Matrix
55 | */
56 | MatrixXd sparseFullWalkRun(SparseMatrixXd A, VectorXd b, int k, int num_sim, SparseRandomWalk* walk, FacialReduction* fr, SparseCenter* init, int burn = 0){
57 | FROutput fr_result = fr->reduce(A, b, k, true);
58 | int new_k = fr_result.sparse_A.rows() - (A.rows() - k);
59 | VectorXd x = init->getInitialPoint(fr_result.sparse_A, fr_result.sparse_b, new_k);
60 | MatrixXd steps = walk->generateCompleteWalk(num_sim, x, fr_result.sparse_A, fr_result.sparse_b, new_k, burn);
61 | MatrixXd res(num_sim, A.cols());
62 | for(int i = 0; i < num_sim; i++){
63 | res.row(i) = fr_result.saved_V * steps.row(i).transpose();
64 | }
65 | return res;
66 | }
--------------------------------------------------------------------------------
/src/utils/SparseCenter.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseCenter.hpp"
2 |
3 | VectorXd SparseCenter::getInitialPoint(SparseMatrixXd& A, VectorXd& b, int k){
4 |
5 | // Solve the linear program
6 | // max delta
7 | // s.t. A x = b
8 | // and x>= delta, on the last k coordinates
9 | SparseLP sparse_lp;
10 | int row_length = A.rows() + k;
11 | int col_length = A.cols() + 1;
12 |
13 | SparseMatrixXd obj_mat(row_length, col_length);
14 | VectorXd obj_vec = VectorXd::Zero(col_length);
15 | obj_vec(obj_vec.rows() - 1) = 1;
16 |
17 | VectorXd row_bnds = VectorXd::Zero(row_length);
18 | VectorXd row_rel = VectorXd::Zero(row_length);
19 | VectorXd col_bnds = VectorXd::Zero(col_length);
20 | VectorXd col_rel = VectorXd::Zero(col_length);
21 | vector coefficients;
22 |
23 | for(int i = 0; i < b.rows(); i++){
24 | row_bnds(i) = b(i);
25 | row_rel(i) = GLP_FX;
26 | }
27 | for(int i = b.rows(); i < row_length; i++){
28 | row_rel(i) = GLP_LO;
29 | }
30 | for(int i = 0; i < col_length - k - 1; i++){
31 | col_rel(i) = GLP_FR;
32 | }
33 | for(int i = col_length - k - 1; i < col_length; i++){
34 | col_rel(i) = GLP_LO;
35 | }
36 |
37 | for(int i = 0; i < A.outerSize(); i++){
38 | for(SparseMatrixXd::InnerIterator it(A, i); it; ++it){
39 | int row = it.row();
40 | int col = it.col();
41 | double val = it.value();
42 | coefficients.push_back(T(row, col, val));
43 | }
44 | }
45 | for(int i = 0; i < k; i++){
46 | int row_val = A.rows() + i;
47 | int col_val = A.cols() - k + i;
48 | coefficients.push_back(T(row_val, col_val, 1));
49 | coefficients.push_back(T(row_val, A.cols(), -1));
50 | }
51 | obj_mat.setFromTriplets(coefficients.begin(), coefficients.end());
52 |
53 | // call the lp solver
54 | VectorXd sol = sparse_lp.findOptimalVector(obj_mat, row_bnds, obj_vec, row_rel, col_bnds, col_rel);
55 |
56 | // retrieve x
57 | VectorXd ans = VectorXd::Zero(sol.rows() - 1);
58 | for(int i = 0; i < ans.rows(); i++){
59 | ans(i) = sol(i);
60 | }
61 | return ans;
62 |
63 | }
--------------------------------------------------------------------------------
/src/utils/SparseCenter.hpp:
--------------------------------------------------------------------------------
1 | #ifndef SPARSE_CENTER_HPP
2 | #define SPARSE_CENTER_HPP
3 |
4 | #include "Common.hpp"
5 | #include "SparseLP.hpp"
6 |
7 | class SparseCenter {
8 | public:
9 | /**
10 | * @brief initialization for Sparse Center Algorithm
11 | */
12 | SparseCenter(){};
13 |
14 | /**
15 | * @brief finds analytical center Ax = b, x >=_k 0
16 | * @param A polytope matrix (Ax = b)
17 | * @param b polytope vector (Ax = b)
18 | * @param k k values >= 0 constraint
19 | * @return VectorXd
20 | */
21 | VectorXd getInitialPoint(SparseMatrixXd& A, VectorXd& b, int k);
22 |
23 | };
24 |
25 | #endif
--------------------------------------------------------------------------------
/src/utils/SparseLP.cpp:
--------------------------------------------------------------------------------
1 | #include "SparseLP.hpp"
2 |
3 | VectorXd SparseLP::findOptimalVector(SparseMatrixXd& A, VectorXd& b, VectorXd& c, VectorXd& row_rel, VectorXd& col_cons, VectorXd& col_rel){
4 |
5 | glp_prob *lp;
6 | glp_term_out(GLP_OFF);
7 | lp = glp_create_prob();
8 | int amount = 1 + A.nonZeros();
9 |
10 | vector ia (amount);
11 | vector ja (amount);
12 | vector ar (amount);
13 |
14 | int row_length = A.rows();
15 | int col_length = A.cols();
16 | glp_add_rows(lp, row_length);
17 | glp_add_cols(lp, col_length);
18 | for(int i = 0; i < col_length; i++){
19 | glp_set_obj_coef(lp, i + 1, c(i));
20 | }
21 | glp_set_obj_dir(lp, GLP_MAX);
22 | for(int i = 0; i < row_length; i++){
23 | glp_set_row_bnds(lp, i + 1, row_rel(i), b(i), b(i));
24 | }
25 | for(int i = 0; i < col_length; i++){
26 | glp_set_col_bnds(lp, i + 1, col_rel(i), col_cons(i), col_cons(i));
27 | }
28 | int ind = 1;
29 | for(int i = 0; i < A.outerSize(); i++){
30 | for(SparseMatrixXd::InnerIterator it(A, i); it; ++it){
31 | int row = it.row();
32 | int col = it.col();
33 | double val = it.value();
34 |
35 | ia[ind] = row + 1;
36 | ja[ind] = col + 1;
37 | ar[ind] = val;
38 |
39 | ind ++;
40 | }
41 | }
42 |
43 | glp_load_matrix(lp, amount-1, ia.data(), ja.data(), ar.data());
44 | glp_simplex(lp, NULL);
45 | VectorXd ans(A.cols());
46 | for(int i = 0; i < A.cols(); i++){
47 | ans.coeffRef(i) = glp_get_col_prim(lp, i + 1);
48 | }
49 | glp_delete_prob(lp);
50 | return ans;
51 |
52 |
53 | }
--------------------------------------------------------------------------------
/src/utils/SparseLP.hpp:
--------------------------------------------------------------------------------
1 | #ifndef SPARSE_LP_HPP
2 | #define SPARSE_LP_HPP
3 |
4 | #include "Common.hpp"
5 |
6 | class SparseLP{
7 | public:
8 | /**
9 | * @brief initialization for Sparse Linear Programming Solver
10 | */
11 | SparseLP(){};
12 |
13 | /**
14 | * @brief finds analytical center Ax = b, x >=_k 0
15 | * @param A constraint matrix
16 | * @param b constriant vector
17 | * @param c objective vector
18 | * @param row_rel relationship for A and b
19 | * @param col_cons constraint for columns
20 | * @param col_rel relation for columns
21 | * @return VectorXd
22 | */
23 | VectorXd findOptimalVector(SparseMatrixXd& A, VectorXd& b, VectorXd& c, VectorXd& row_rel,
24 | VectorXd& col_cons, VectorXd& col_rel);
25 |
26 | };
27 |
28 | #endif
--------------------------------------------------------------------------------
/tests/CMakeLists.txt:
--------------------------------------------------------------------------------
1 | #find_package(Catch2 REQUIRED)
2 | Include(FetchContent)
3 |
4 | FetchContent_Declare(
5 | Catch2
6 | GIT_REPOSITORY https://github.com/catchorg/Catch2.git
7 | GIT_TAG v3.8.1 # or a later release
8 | )
9 |
10 | FetchContent_MakeAvailable(Catch2)
11 |
12 | add_executable(test_dense_walk ${TESTS_DIR}/test_dense_walk.cpp)
13 | target_link_libraries(test_dense_walk PRIVATE utils dense sparse Catch2::Catch2WithMain ${GLPK_LIBRARY})
14 | target_include_directories(test_dense_walk PRIVATE ${GLPK_INCLUDE_DIR})
15 |
16 | add_executable(test_sparse_walk ${TESTS_DIR}/test_sparse_walk.cpp)
17 | target_link_libraries(test_sparse_walk PRIVATE utils dense sparse Catch2::Catch2WithMain ${GLPK_LIBRARY})
18 | target_include_directories(test_sparse_walk PRIVATE ${GLPK_INCLUDE_DIR})
19 |
20 | add_executable(test_fr ${TESTS_DIR}/test_fr.cpp)
21 | target_link_libraries(test_fr PRIVATE utils dense sparse Catch2::Catch2WithMain ${GLPK_LIBRARY})
22 | target_include_directories(test_fr PRIVATE ${GLPK_INCLUDE_DIR})
23 |
24 | add_executable(test_weights ${TESTS_DIR}/test_weights.cpp)
25 | target_link_libraries(test_weights PRIVATE utils dense sparse Catch2::Catch2WithMain ${GLPK_LIBRARY})
26 | target_include_directories(test_weights PRIVATE ${GLPK_INCLUDE_DIR})
27 |
28 | add_executable(test_init ${TESTS_DIR}/test_init.cpp)
29 | target_link_libraries(test_init PRIVATE utils dense sparse Catch2::Catch2WithMain ${GLPK_LIBRARY})
30 | target_include_directories(test_init PRIVATE ${GLPK_INCLUDE_DIR})
31 |
--------------------------------------------------------------------------------
/tests/cpp/test_dense_walk.cpp:
--------------------------------------------------------------------------------
1 | #define CATCH_CONFIG_MAIN
2 | #include
3 | #include
4 | #include
5 | #include "utils/FullWalkRun.hpp"
6 | #include
7 |
8 | struct sparse_polytope{
9 | SparseMatrixXd A;
10 | VectorXd b;
11 | int k;
12 | };
13 |
14 | sparse_polytope generate_simplex(){
15 | SparseMatrixXd simplex_A (1, 3);
16 | simplex_A.coeffRef(0, 0) = 1;
17 | simplex_A.coeffRef(0, 1) = 1;
18 | simplex_A.coeffRef(0, 2) = 1;
19 | VectorXd simplex_b (1);
20 | simplex_b << 1;
21 | sparse_polytope result;
22 | result.A = simplex_A;
23 | result.b = simplex_b;
24 | result.k = 3;
25 | return result;
26 | }
27 |
28 | sparse_polytope generate_hc(){
29 | SparseMatrixXd hc_A (4, 6);
30 | hc_A.coeffRef(0, 0) = 1;
31 | hc_A.coeffRef(0, 2) = 1;
32 | hc_A.coeffRef(1, 1) = 1;
33 | hc_A.coeffRef(1, 3) = 1;
34 | hc_A.coeffRef(2, 0) = -1;
35 | hc_A.coeffRef(2, 4) = 1;
36 | hc_A.coeffRef(3, 1) = -1;
37 | hc_A.coeffRef(3, 5) = 1;
38 |
39 | VectorXd hc_b (4);
40 | hc_b << 1, 1, 1, 1;
41 | sparse_polytope result;
42 | result.A = hc_A;
43 | result.b = hc_b;
44 | result.k = 4;
45 | return result;
46 | }
47 |
48 | sparse_polytope generate_birkhoff(){
49 | SparseMatrixXd birk_A (3, 4);
50 | birk_A.coeffRef(0, 0) = 1;
51 | birk_A.coeffRef(0, 1) = 1;
52 | birk_A.coeffRef(1, 2) = 1;
53 | birk_A.coeffRef(1, 3) = 1;
54 | birk_A.coeffRef(2, 0) = 1;
55 | birk_A.coeffRef(2, 2) = 1;
56 |
57 | VectorXd birk_b (3);
58 | birk_b << 1, 1, 1;
59 | sparse_polytope result;
60 | result.A = birk_A;
61 | result.b = birk_b;
62 | result.k = 4;
63 | return result;
64 | }
65 |
66 | sparse_polytope simplex = generate_simplex();
67 | sparse_polytope hc = generate_hc();
68 | sparse_polytope birk = generate_birkhoff();
69 |
70 | TEST_CASE( "Test All Dense Combinations", "[require]" ){
71 | JohnWalk john(0.5, 1, 1e-5, 1000);
72 | DikinLSWalk dikinls(3.0, 1, 0.001, 0.01, 100);
73 | VaidyaWalk vaidya(0.5);
74 | DikinWalk dikin(0.5);
75 | BallWalk ball(0.5);
76 | HitAndRun hitrun(0.5, 0.001);
77 | DenseCenter dc;
78 | FacialReduction fr;
79 |
80 | MatrixXd walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &john, &fr, &dc);
81 | walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &dikinls, &fr, &dc);
82 | REQUIRE(walk_res.rows() == 100);
83 | walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &vaidya, &fr, &dc);
84 | REQUIRE(walk_res.rows() == 100);
85 | walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &dikin, &fr, &dc);
86 | REQUIRE(walk_res.rows() == 100);
87 | walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &ball, &fr, &dc);
88 | REQUIRE(walk_res.rows() == 100);
89 | walk_res = denseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &hitrun, &fr, &dc);
90 | REQUIRE(walk_res.rows() == 100);
91 |
92 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &john, &fr, &dc);
93 | REQUIRE(walk_res.rows() == 100);
94 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &dikinls, &fr, &dc);
95 | REQUIRE(walk_res.rows() == 100);
96 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &vaidya, &fr, &dc);
97 | REQUIRE(walk_res.rows() == 100);
98 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &dikin, &fr, &dc);
99 | REQUIRE(walk_res.rows() == 100);
100 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &ball, &fr, &dc);
101 | REQUIRE(walk_res.rows() == 100);
102 | walk_res = denseFullWalkRun(hc.A, hc.b, hc.k, 100, &hitrun, &fr, &dc);
103 | REQUIRE(walk_res.rows() == 100);
104 |
105 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &john, &fr, &dc);
106 | REQUIRE(walk_res.rows() == 100);
107 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &dikinls, &fr, &dc);
108 | REQUIRE(walk_res.rows() == 100);
109 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &vaidya, &fr, &dc);
110 | REQUIRE(walk_res.rows() == 100);
111 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &dikin, &fr, &dc);
112 | REQUIRE(walk_res.rows() == 100);
113 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &ball, &fr, &dc);
114 | REQUIRE(walk_res.rows() == 100);
115 | walk_res = denseFullWalkRun(birk.A, birk.b, birk.k, 100, &hitrun, &fr, &dc);
116 | REQUIRE(walk_res.rows() == 100);
117 | }
--------------------------------------------------------------------------------
/tests/cpp/test_fr.cpp:
--------------------------------------------------------------------------------
1 | #define CATCH_CONFIG_MAIN
2 | #include
3 | #include
4 | #include
5 | #include "utils/FullWalkRun.hpp"
6 | #include
7 |
8 | struct sparse_polytope{
9 | SparseMatrixXd A;
10 | VectorXd b;
11 | int k;
12 | };
13 |
14 | sparse_polytope generate_simplex(){
15 | SparseMatrixXd simplex_A (1, 3);
16 | simplex_A.coeffRef(0, 0) = 1;
17 | simplex_A.coeffRef(0, 1) = 1;
18 | simplex_A.coeffRef(0, 2) = 1;
19 | VectorXd simplex_b (1);
20 | simplex_b << 1;
21 | sparse_polytope result;
22 | result.A = simplex_A;
23 | result.b = simplex_b;
24 | result.k = 3;
25 | return result;
26 | }
27 |
28 | sparse_polytope generate_hc(){
29 | SparseMatrixXd hc_A (4, 6);
30 | hc_A.coeffRef(0, 0) = 1;
31 | hc_A.coeffRef(0, 2) = 1;
32 | hc_A.coeffRef(1, 1) = 1;
33 | hc_A.coeffRef(1, 3) = 1;
34 | hc_A.coeffRef(2, 0) = -1;
35 | hc_A.coeffRef(2, 4) = 1;
36 | hc_A.coeffRef(3, 1) = -1;
37 | hc_A.coeffRef(3, 5) = 1;
38 |
39 | VectorXd hc_b (4);
40 | hc_b << 1, 1, 1, 1;
41 | sparse_polytope result;
42 | result.A = hc_A;
43 | result.b = hc_b;
44 | result.k = 4;
45 | return result;
46 | }
47 |
48 | sparse_polytope generate_birkhoff(){
49 | SparseMatrixXd birk_A (3, 4);
50 | birk_A.coeffRef(0, 0) = 1;
51 | birk_A.coeffRef(0, 1) = 1;
52 | birk_A.coeffRef(1, 2) = 1;
53 | birk_A.coeffRef(1, 3) = 1;
54 | birk_A.coeffRef(2, 0) = 1;
55 | birk_A.coeffRef(2, 2) = 1;
56 |
57 | VectorXd birk_b (3);
58 | birk_b << 1, 1, 1;
59 | sparse_polytope result;
60 | result.A = birk_A;
61 | result.b = birk_b;
62 | result.k = 4;
63 | return result;
64 | }
65 |
66 | sparse_polytope simplex = generate_simplex();
67 | sparse_polytope hc = generate_hc();
68 | sparse_polytope birk = generate_birkhoff();
69 |
70 | TEST_CASE( "Check Facial Reduction Algorithm", "[require]" ) {
71 |
72 | FacialReduction fr;
73 |
74 | FROutput simplex_dense = fr.reduce(simplex.A, simplex.b, simplex.k, false);
75 | FROutput hc_dense = fr.reduce(hc.A, hc.b, hc.k, false);
76 | FROutput birk_dense = fr.reduce(birk.A, birk.b, birk.k, false);
77 |
78 | int dense_A_row, dense_A_col, dense_b_row;
79 | dense_A_row = simplex_dense.dense_A.rows();
80 | dense_A_col = simplex_dense.dense_A.cols();
81 | dense_b_row = simplex_dense.dense_b.rows();
82 |
83 | REQUIRE(((dense_A_row == 3) && (dense_A_col == 2)));
84 | REQUIRE(dense_b_row == 3);
85 |
86 | dense_A_row = hc_dense.dense_A.rows();
87 | dense_A_col = hc_dense.dense_A.cols();
88 | dense_b_row = hc_dense.dense_b.rows();
89 |
90 | REQUIRE(((dense_A_row == 4) && (dense_A_col == 2)));
91 | REQUIRE(dense_b_row == 4);
92 |
93 | dense_A_row = birk_dense.dense_A.rows();
94 | dense_A_col = birk_dense.dense_A.cols();
95 | dense_b_row = birk_dense.dense_b.rows();
96 |
97 | REQUIRE(((dense_A_row == 4) && (dense_A_col == 1)));
98 | REQUIRE(dense_b_row == 4);
99 |
100 | MatrixXd A1 (6, 3);
101 | A1 << 1, 1, 0, -1, -1, 0, 0, 1, 0, 0, -1, 0, 0, 0, 1, 0, 0, -1;
102 | MatrixXd temp (6, 9);
103 | temp << A1, VectorXd::Ones(6).asDiagonal().toDenseMatrix();
104 | SparseMatrixXd SA1 = temp.sparseView();
105 | VectorXd b1(6);
106 | b1 << 1, -1, 1, 1, 1, 1;
107 |
108 |
109 | FROutput test1a = fr.reduce(SA1, b1, 6, true);
110 | REQUIRE((test1a.sparse_A.rows() == 5 && test1a.sparse_A.cols() == 7));
111 | FROutput test1b = fr.reduce(SA1, b1, 6, false);
112 | dense_A_row = test1b.dense_A.rows();
113 | dense_A_col = test1b.dense_A.cols();
114 | REQUIRE((dense_A_row == 4 && dense_A_col == 2));
115 | MatrixXd A2(6,3);
116 | A2 << 1, 0, 0, -1, 0, 0, 0, 1, 0, 0, -1, 0, 0, 0, 1, 0, 0, -1;
117 | MatrixXd temp2 (6, 9);
118 | temp2 << A2, VectorXd::Ones(6).asDiagonal().toDenseMatrix();
119 | SparseMatrixXd SA2 = temp2.sparseView();
120 | VectorXd b2(6);
121 | b2 << 1, 1, 0, 0, 0, 0;
122 |
123 | FROutput test2a = fr.reduce(SA2, b2, 6, true);
124 | int sparse_rows = test2a.sparse_A.rows();
125 | int sparse_cols = test2a.sparse_A.cols();
126 |
127 | REQUIRE_THAT(sparse_rows, Catch::Matchers::WithinAbs(4, 0.01));
128 | REQUIRE_THAT(sparse_cols, Catch::Matchers::WithinAbs(5, 0.01));
129 | FROutput test2b = fr.reduce(SA2, b2, 6, false);
130 | dense_A_row = test2b.dense_A.rows();
131 | dense_A_col = test2b.dense_A.cols();
132 | REQUIRE((dense_A_row == 2 && dense_A_col == 1));
133 |
134 |
135 | MatrixXd A3(4,2);
136 | A3 << 1, 0, -1, 0, 0, 1, 0, -1;
137 | MatrixXd temp3 (4, 6);
138 | temp3 << A3, VectorXd::Ones(4).asDiagonal().toDenseMatrix();
139 | SparseMatrixXd SA3 = temp3.sparseView();
140 | VectorXd b3(4);
141 | b3 << 1, 0, 1, 0;
142 |
143 | FROutput test3a = fr.reduce(SA3, b3, 4, true);
144 | int sparse_A_rows = test3a.sparse_A.rows();
145 | int sparse_A_cols = test3a.sparse_A.cols();
146 |
147 | REQUIRE_THAT(sparse_A_rows, Catch::Matchers::WithinAbs(4, 0.0001));
148 | REQUIRE_THAT(sparse_A_cols, Catch::Matchers::WithinAbs(6, 0.0001));
149 | FROutput test3b = fr.reduce(SA3, b3, 4, false);
150 | dense_A_row = test3b.dense_A.rows();
151 | dense_A_col = test3b.dense_A.cols();
152 | REQUIRE((dense_A_row == 4 && dense_A_col == 2));
153 |
154 |
155 | }
156 |
--------------------------------------------------------------------------------
/tests/cpp/test_init.cpp:
--------------------------------------------------------------------------------
1 | #define CATCH_CONFIG_MAIN
2 | #include
3 | #include
4 | #include
5 | #include "utils/FullWalkRun.hpp"
6 | #include
7 |
8 | struct sparse_polytope{
9 | SparseMatrixXd A;
10 | VectorXd b;
11 | int k;
12 | };
13 |
14 | sparse_polytope generate_simplex(){
15 | SparseMatrixXd simplex_A (1, 3);
16 | simplex_A.coeffRef(0, 0) = 1;
17 | simplex_A.coeffRef(0, 1) = 1;
18 | simplex_A.coeffRef(0, 2) = 1;
19 | VectorXd simplex_b (1);
20 | simplex_b << 1;
21 | sparse_polytope result;
22 | result.A = simplex_A;
23 | result.b = simplex_b;
24 | result.k = 3;
25 | return result;
26 | }
27 |
28 | sparse_polytope generate_hc(){
29 | SparseMatrixXd hc_A (4, 6);
30 | hc_A.coeffRef(0, 0) = 1;
31 | hc_A.coeffRef(0, 2) = 1;
32 | hc_A.coeffRef(1, 1) = 1;
33 | hc_A.coeffRef(1, 3) = 1;
34 | hc_A.coeffRef(2, 0) = -1;
35 | hc_A.coeffRef(2, 4) = 1;
36 | hc_A.coeffRef(3, 1) = -1;
37 | hc_A.coeffRef(3, 5) = 1;
38 |
39 | VectorXd hc_b (4);
40 | hc_b << 1, 1, 1, 1;
41 | sparse_polytope result;
42 | result.A = hc_A;
43 | result.b = hc_b;
44 | result.k = 4;
45 | return result;
46 | }
47 |
48 | sparse_polytope generate_birkhoff(){
49 | SparseMatrixXd birk_A (3, 4);
50 | birk_A.coeffRef(0, 0) = 1;
51 | birk_A.coeffRef(0, 1) = 1;
52 | birk_A.coeffRef(1, 2) = 1;
53 | birk_A.coeffRef(1, 3) = 1;
54 | birk_A.coeffRef(2, 0) = 1;
55 | birk_A.coeffRef(2, 2) = 1;
56 |
57 | VectorXd birk_b (3);
58 | birk_b << 1, 1, 1;
59 | sparse_polytope result;
60 | result.A = birk_A;
61 | result.b = birk_b;
62 | result.k = 4;
63 | return result;
64 | }
65 |
66 | sparse_polytope simplex = generate_simplex();
67 | sparse_polytope hc = generate_hc();
68 | sparse_polytope birk = generate_birkhoff();
69 |
70 | TEST_CASE( "Check Initialization Algorithm", "[require]" ){
71 | SparseCenter sc;
72 | VectorXd simplex_x = sc.getInitialPoint(simplex.A, simplex.b, simplex.k);
73 | REQUIRE(simplex_x.rows() == 3);
74 | REQUIRE(simplex_x(0) == Catch::Approx(0.3333333).epsilon(0.01));
75 | REQUIRE(simplex_x(1) == Catch::Approx(0.3333333).epsilon(0.01));
76 | REQUIRE(simplex_x(2) == Catch::Approx(0.3333333).epsilon(0.01));
77 |
78 | VectorXd hc_x = sc.getInitialPoint(hc.A, hc.b, hc.k);
79 | REQUIRE(hc_x.rows() == 6);
80 | REQUIRE_THAT(hc_x(0), Catch::Matchers::WithinAbs(0, 0.0001));
81 | REQUIRE_THAT(hc_x(1), Catch::Matchers::WithinAbs(0, 0.0001));
82 | REQUIRE_THAT(hc_x(2), Catch::Matchers::WithinAbs(1, 0.0001));
83 | REQUIRE_THAT(hc_x(3), Catch::Matchers::WithinAbs(1, 0.0001));
84 | REQUIRE_THAT(hc_x(4), Catch::Matchers::WithinAbs(1, 0.0001));
85 | REQUIRE_THAT(hc_x(5), Catch::Matchers::WithinAbs(1, 0.0001));
86 |
87 | VectorXd birk_x = sc.getInitialPoint(birk.A, birk.b, birk.k);
88 | REQUIRE(birk_x.rows() == 4);
89 | REQUIRE_THAT(birk_x(0), Catch::Matchers::WithinAbs(0.5, 0.0001));
90 | REQUIRE_THAT(birk_x(1), Catch::Matchers::WithinAbs(0.5, 0.0001));
91 | REQUIRE_THAT(birk_x(2), Catch::Matchers::WithinAbs(0.5, 0.0001));
92 | REQUIRE_THAT(birk_x(3), Catch::Matchers::WithinAbs(0.5, 0.0001));
93 |
94 | DenseCenter dc;
95 |
96 | MatrixXd A1 (4, 2);
97 | A1 << 1, 0, 0, 1, -1, 0, 0, -1;
98 | VectorXd b1 (4);
99 | b1 << 1, 1, 0, 0;
100 |
101 | VectorXd center1 = dc.getInitialPoint(A1, b1);
102 | REQUIRE_THAT(center1(0), Catch::Matchers::WithinAbs(0.5, 0.0001));
103 | REQUIRE_THAT(center1(1), Catch::Matchers::WithinAbs(0.5, 0.0001));
104 |
105 | MatrixXd A2 (3, 2);
106 | A2 << -1, 0, 0, -1, 1, 1;
107 | VectorXd b2 (3);
108 | b2 << 0, 0, 1;
109 |
110 | VectorXd center2 = dc.getInitialPoint(A2, b2);
111 | REQUIRE_THAT(center2(0), Catch::Matchers::WithinAbs(0.33, 0.01));
112 | REQUIRE_THAT(center2(1), Catch::Matchers::WithinAbs(0.33, 0.01));
113 |
114 | }
--------------------------------------------------------------------------------
/tests/cpp/test_sparse_walk.cpp:
--------------------------------------------------------------------------------
1 | #define CATCH_CONFIG_MAIN
2 | #include
3 | #include
4 | #include
5 | #include "utils/FullWalkRun.hpp"
6 | #include
7 |
8 | struct sparse_polytope{
9 | SparseMatrixXd A;
10 | VectorXd b;
11 | int k;
12 | };
13 |
14 | sparse_polytope generate_simplex(){
15 | SparseMatrixXd simplex_A (1, 3);
16 | simplex_A.coeffRef(0, 0) = 1;
17 | simplex_A.coeffRef(0, 1) = 1;
18 | simplex_A.coeffRef(0, 2) = 1;
19 | VectorXd simplex_b (1);
20 | simplex_b << 1;
21 | sparse_polytope result;
22 | result.A = simplex_A;
23 | result.b = simplex_b;
24 | result.k = 3;
25 | return result;
26 | }
27 |
28 | sparse_polytope generate_hc(){
29 | SparseMatrixXd hc_A (4, 6);
30 | hc_A.coeffRef(0, 0) = 1;
31 | hc_A.coeffRef(0, 2) = 1;
32 | hc_A.coeffRef(1, 1) = 1;
33 | hc_A.coeffRef(1, 3) = 1;
34 | hc_A.coeffRef(2, 0) = -1;
35 | hc_A.coeffRef(2, 4) = 1;
36 | hc_A.coeffRef(3, 1) = -1;
37 | hc_A.coeffRef(3, 5) = 1;
38 |
39 | VectorXd hc_b (4);
40 | hc_b << 1, 1, 1, 1;
41 | sparse_polytope result;
42 | result.A = hc_A;
43 | result.b = hc_b;
44 | result.k = 4;
45 | return result;
46 | }
47 |
48 | sparse_polytope generate_birkhoff(){
49 | SparseMatrixXd birk_A (3, 4);
50 | birk_A.coeffRef(0, 0) = 1;
51 | birk_A.coeffRef(0, 1) = 1;
52 | birk_A.coeffRef(1, 2) = 1;
53 | birk_A.coeffRef(1, 3) = 1;
54 | birk_A.coeffRef(2, 0) = 1;
55 | birk_A.coeffRef(2, 2) = 1;
56 |
57 | VectorXd birk_b (3);
58 | birk_b << 1, 1, 1;
59 | sparse_polytope result;
60 | result.A = birk_A;
61 | result.b = birk_b;
62 | result.k = 4;
63 | return result;
64 | }
65 |
66 | sparse_polytope simplex = generate_simplex();
67 | sparse_polytope hc = generate_hc();
68 | sparse_polytope birk = generate_birkhoff();
69 |
70 | TEST_CASE( "Test All Sparse Combinations", "[require]" ){
71 | SparseJohnWalk john(0.5, 2);
72 | SparseDikinLSWalk dikinls(3.0, 2);
73 | SparseVaidyaWalk vaidya(0.5, 2);
74 | SparseDikinWalk dikin(0.5, 2);
75 | SparseBallWalk ball(0.5, 2);
76 | SparseHitAndRun hitrun(0.5, 2);
77 | SparseCenter sc;
78 | FacialReduction fr;
79 |
80 | MatrixXd walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &john, &fr, &sc, 1);
81 | REQUIRE(walk_res.rows() == 100);
82 | walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &dikinls, &fr, &sc, 1);
83 | REQUIRE(walk_res.rows() == 100);
84 | walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &vaidya, &fr, &sc, 1);
85 | REQUIRE(walk_res.rows() == 100);
86 | walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &dikin, &fr, &sc, 1);
87 | REQUIRE(walk_res.rows() == 100);
88 | walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &ball, &fr, &sc, 1);
89 | REQUIRE(walk_res.rows() == 100);
90 | walk_res = sparseFullWalkRun(simplex.A, simplex.b, simplex.k, 100, &hitrun, &fr, &sc, 1);
91 | REQUIRE(walk_res.rows() == 100);
92 |
93 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &john, &fr, &sc, 1);
94 | REQUIRE(walk_res.rows() == 100);
95 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &dikinls, &fr, &sc, 1);
96 | REQUIRE(walk_res.rows() == 100);
97 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &vaidya, &fr, &sc, 1);
98 | REQUIRE(walk_res.rows() == 100);
99 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &dikin, &fr, &sc, 1);
100 | REQUIRE(walk_res.rows() == 100);
101 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &ball, &fr, &sc, 1);;
102 | REQUIRE(walk_res.rows() == 100);
103 | walk_res = sparseFullWalkRun(hc.A, hc.b, hc.k, 100, &hitrun, &fr, &sc, 1);
104 | REQUIRE(walk_res.rows() == 100);
105 |
106 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &john, &fr, &sc, 1);
107 | REQUIRE(walk_res.rows() == 100);
108 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &dikinls, &fr, &sc, 1);
109 | REQUIRE(walk_res.rows() == 100);
110 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &vaidya, &fr, &sc, 1);
111 | REQUIRE(walk_res.rows() == 100);
112 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &dikin, &fr, &sc, 1);
113 | REQUIRE(walk_res.rows() == 100);
114 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &ball, &fr, &sc, 1);
115 | REQUIRE(walk_res.rows() == 100);
116 | walk_res = sparseFullWalkRun(birk.A, birk.b, birk.k, 100, &hitrun, &fr, &sc, 1);
117 | REQUIRE(walk_res.rows() == 100);
118 | }
--------------------------------------------------------------------------------
/tests/cpp/test_weights.cpp:
--------------------------------------------------------------------------------
1 | #define CATCH_CONFIG_MAIN
2 | #include
3 | #include
4 | #include
5 | #include "utils/FullWalkRun.hpp"
6 | #include
7 |
8 | struct sparse_polytope{
9 | SparseMatrixXd A;
10 | VectorXd b;
11 | int k;
12 | };
13 |
14 | sparse_polytope generate_simplex(){
15 | SparseMatrixXd simplex_A (1, 3);
16 | simplex_A.coeffRef(0, 0) = 1;
17 | simplex_A.coeffRef(0, 1) = 1;
18 | simplex_A.coeffRef(0, 2) = 1;
19 | VectorXd simplex_b (1);
20 | simplex_b << 1;
21 | sparse_polytope result;
22 | result.A = simplex_A;
23 | result.b = simplex_b;
24 | result.k = 3;
25 | return result;
26 | }
27 |
28 | sparse_polytope generate_hc(){
29 | SparseMatrixXd hc_A (4, 6);
30 | hc_A.coeffRef(0, 0) = 1;
31 | hc_A.coeffRef(0, 2) = 1;
32 | hc_A.coeffRef(1, 1) = 1;
33 | hc_A.coeffRef(1, 3) = 1;
34 | hc_A.coeffRef(2, 0) = -1;
35 | hc_A.coeffRef(2, 4) = 1;
36 | hc_A.coeffRef(3, 1) = -1;
37 | hc_A.coeffRef(3, 5) = 1;
38 |
39 | VectorXd hc_b (4);
40 | hc_b << 1, 1, 1, 1;
41 | sparse_polytope result;
42 | result.A = hc_A;
43 | result.b = hc_b;
44 | result.k = 4;
45 | return result;
46 | }
47 |
48 | sparse_polytope generate_birkhoff(){
49 | SparseMatrixXd birk_A (3, 4);
50 | birk_A.coeffRef(0, 0) = 1;
51 | birk_A.coeffRef(0, 1) = 1;
52 | birk_A.coeffRef(1, 2) = 1;
53 | birk_A.coeffRef(1, 3) = 1;
54 | birk_A.coeffRef(2, 0) = 1;
55 | birk_A.coeffRef(2, 2) = 1;
56 |
57 | VectorXd birk_b (3);
58 | birk_b << 1, 1, 1;
59 | sparse_polytope result;
60 | result.A = birk_A;
61 | result.b = birk_b;
62 | result.k = 4;
63 | return result;
64 | }
65 |
66 | sparse_polytope simplex = generate_simplex();
67 | sparse_polytope hc = generate_hc();
68 | sparse_polytope birk = generate_birkhoff();
69 |
70 |
71 | TEST_CASE( "Check Weight Properties", "[require]" ){
72 | //Vaidya, John, DikinLS
73 | SparseVaidyaWalk vaidya_sparse(0.5);
74 | SparseDikinLSWalk dikinls_sparse(1.0, 1, 0.001, 0.01, 20000);
75 | SparseJohnWalk john_sparse(0.5, 1, 1e-5, 10000);
76 |
77 | VectorXd simplex_start (3);
78 | simplex_start << 0.33, 0.34, 0.33;
79 | SparseMatrixXd w = dikinls_sparse.generateWeight(simplex_start, simplex.A, simplex.k);
80 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(2, 0.01));
81 | w = john_sparse.generateWeight(simplex_start, simplex.A, simplex.k);
82 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(3, 0.01));
83 | w = vaidya_sparse.generateWeight(simplex_start, simplex.A, simplex.k);
84 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(4, 0.01));
85 |
86 | VectorXd hc_start (6);
87 | hc_start << 0, 0, 1, 1, 1, 1;
88 | w = dikinls_sparse.generateWeight(hc_start, hc.A, hc.k);
89 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(2, 0.01));
90 | w = john_sparse.generateWeight(hc_start, hc.A, hc.k);
91 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(3, 0.01));
92 | w = vaidya_sparse.generateWeight(hc_start, hc.A, hc.k);
93 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(4, 0.01));
94 |
95 | VectorXd birk_start (4);
96 | birk_start << 0.5, 0.5, 0.5, 0.5;
97 | w = dikinls_sparse.generateWeight(birk_start, birk.A, birk.k);
98 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(1, 0.01));
99 | w = john_sparse.generateWeight(birk_start, birk.A, birk.k);
100 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(1.5, 0.01));
101 | w = vaidya_sparse.generateWeight(birk_start, birk.A, birk.k);
102 | REQUIRE_THAT(w.diagonal().sum(), Catch::Matchers::WithinAbs(2, 0.01));
103 |
104 | FacialReduction fr;
105 | DenseCenter dc;
106 | FROutput simplex_dense = fr.reduce(simplex.A, simplex.b, simplex.k, false);
107 | FROutput hc_dense = fr.reduce(hc.A, hc.b, hc.k, false);
108 | FROutput birk_dense = fr.reduce(birk.A, birk.b, birk.k, false);
109 | VectorXd sd_x = dc.getInitialPoint(simplex_dense.dense_A, simplex_dense.dense_b);
110 | VectorXd hc_x = dc.getInitialPoint(hc_dense.dense_A, hc_dense.dense_b);
111 | VectorXd birk_x = dc.getInitialPoint(birk_dense.dense_A, birk_dense.dense_b);
112 |
113 | JohnWalk john(0.5, 1, 0.001, 10000);
114 | DikinLSWalk dikinls(0.5, 1, 0.001, 0.01, 10000);
115 | VaidyaWalk vaidya(0.5);
116 |
117 | john.generateWeight(sd_x, simplex_dense.dense_A, simplex_dense.dense_b);
118 | dikinls.generateWeight(sd_x, simplex_dense.dense_A, simplex_dense.dense_b);
119 | vaidya.generateWeight(sd_x, simplex_dense.dense_A, simplex_dense.dense_b);
120 | double dw, jw, vw;
121 | dw = dikinls.weights.diagonal().sum();
122 | jw = john.weights.diagonal().sum();
123 | vw = vaidya.weights.diagonal().sum();
124 |
125 | REQUIRE_THAT(dw, Catch::Matchers::WithinAbs(2, 0.01));
126 | REQUIRE_THAT(jw, Catch::Matchers::WithinAbs(3, 0.01));
127 | REQUIRE_THAT(vw, Catch::Matchers::WithinAbs(4, 0.01));
128 |
129 | john.generateWeight(hc_x, hc_dense.dense_A, hc_dense.dense_b);
130 | dikinls.generateWeight(hc_x, hc_dense.dense_A, hc_dense.dense_b);
131 | vaidya.generateWeight(hc_x, hc_dense.dense_A, hc_dense.dense_b);
132 | dw = dikinls.weights.diagonal().sum();
133 | jw = john.weights.diagonal().sum();
134 | vw = vaidya.weights.diagonal().sum();
135 |
136 | REQUIRE_THAT(dw, Catch::Matchers::WithinAbs(2, 0.01));
137 | REQUIRE_THAT(jw, Catch::Matchers::WithinAbs(3, 0.01));
138 | REQUIRE_THAT(vw, Catch::Matchers::WithinAbs(4, 0.01));
139 |
140 | john.generateWeight(birk_x, birk_dense.dense_A, birk_dense.dense_b);
141 | dikinls.generateWeight(birk_x, birk_dense.dense_A, birk_dense.dense_b);
142 | vaidya.generateWeight(birk_x, birk_dense.dense_A, birk_dense.dense_b);
143 | dw = dikinls.weights.diagonal().sum();
144 | jw = john.weights.diagonal().sum();
145 | vw = vaidya.weights.diagonal().sum();
146 |
147 | REQUIRE_THAT(dw, Catch::Matchers::WithinAbs(1, 0.01));
148 | REQUIRE_THAT(jw, Catch::Matchers::WithinAbs(1.5, 0.01));
149 | REQUIRE_THAT(vw, Catch::Matchers::WithinAbs(2, 0.01));
150 |
151 | }
--------------------------------------------------------------------------------
/tests/python/test_dense_walk.py:
--------------------------------------------------------------------------------
1 | import unittest
2 | from polytopewalk import FacialReduction
3 | import numpy as np
4 | import pandas as pd
5 | from scipy.sparse import lil_matrix
6 | from polytopewalk.dense import *
7 |
8 | simplex_A = lil_matrix((1, 3))
9 | simplex_A[(0, 0)] = 1
10 | simplex_A[(0, 1)] = 1
11 | simplex_A[(0, 2)] = 1
12 | simplex_b = np.array([1])
13 |
14 | hc_A = lil_matrix((4,6))
15 | hc_A[(0, 0)] = 1
16 | hc_A[(0, 2)] = 1
17 | hc_A[(1, 1)] = 1
18 | hc_A[(1, 3)] = 1
19 | hc_A[(2, 0)] = -1
20 | hc_A[(2, 4)] = 1
21 | hc_A[(3, 1)] = -1
22 | hc_A[(3, 5)] = 1
23 |
24 | hc_b = np.array([1,1,1,1])
25 |
26 | birk_A = lil_matrix((3, 4))
27 | birk_A[(0, 0)] = 1
28 | birk_A[(0, 1)] = 1
29 | birk_A[(1, 2)] = 1
30 | birk_A[(1, 3)] = 1
31 | birk_A[(2, 0)] = 1
32 | birk_A[(2, 2)] = 1
33 |
34 | birk_b = np.array([1,1,1])
35 |
36 |
37 |
38 | class TestDenseWalk(unittest.TestCase):
39 | def test_run(self):
40 | dikin = DikinWalk(r = 0.8, thin = 2)
41 | vaidya = VaidyaWalk(r = 0.8, thin = 2)
42 | john = JohnWalk(r = 0.8, thin = 2)
43 |
44 | fr = FacialReduction()
45 | dc = DenseCenter()
46 | fr_res_simplex = fr.reduce(simplex_A, simplex_b, simplex_A.shape[1], False)
47 | fr_res_hc = fr.reduce(hc_A, hc_b, 4, False)
48 | fr_res_birk = fr.reduce(birk_A, birk_b, birk_A.shape[1], False)
49 |
50 | init_simplex = dc.getInitialPoint(fr_res_simplex.dense_A, fr_res_simplex.dense_b)
51 | init_hc = dc.getInitialPoint(fr_res_hc.dense_A, fr_res_hc.dense_b)
52 | init_birk = dc.getInitialPoint(fr_res_birk.dense_A, fr_res_birk.dense_b)
53 |
54 | res1 = dikin.generateCompleteWalk(100, init_simplex, fr_res_simplex.dense_A, fr_res_simplex.dense_b)
55 | self.assertTrue(res1.shape == (100, 2))
56 | res2 = vaidya.generateCompleteWalk(100, init_simplex, fr_res_simplex.dense_A, fr_res_simplex.dense_b)
57 | self.assertTrue(res2.shape == (100, 2))
58 | res3 = john.generateCompleteWalk(100, init_simplex, fr_res_simplex.dense_A, fr_res_simplex.dense_b)
59 | self.assertTrue(res3.shape == (100, 2))
60 |
61 |
62 | res1 = dikin.generateCompleteWalk(100, init_hc, fr_res_hc.dense_A, fr_res_hc.dense_b)
63 | self.assertTrue(res1.shape[0] == 100)
64 | res2 = vaidya.generateCompleteWalk(100, init_hc, fr_res_hc.dense_A, fr_res_hc.dense_b)
65 | self.assertTrue(res2.shape[0] == 100)
66 | res3 = john.generateCompleteWalk(100, init_hc, fr_res_hc.dense_A, fr_res_hc.dense_b)
67 | self.assertTrue(res3.shape[0] == 100)
68 |
69 | res1 = dikin.generateCompleteWalk(100, init_birk, fr_res_birk.dense_A, fr_res_birk.dense_b)
70 | self.assertTrue(res1.shape[0] == 100)
71 | res2 = vaidya.generateCompleteWalk(100, init_birk, fr_res_birk.dense_A, fr_res_birk.dense_b)
72 | self.assertTrue(res2.shape[0] == 100)
73 | res3 = john.generateCompleteWalk(100, init_birk, fr_res_birk.dense_A, fr_res_birk.dense_b)
74 | self.assertTrue(res3.shape[0] == 100)
75 |
76 | if __name__ == '__main__':
77 | unittest.main()
78 |
79 |
--------------------------------------------------------------------------------
/tests/python/test_fr.py:
--------------------------------------------------------------------------------
1 | import unittest
2 | from polytopewalk import FacialReduction
3 | import numpy as np
4 | import pandas as pd
5 | from scipy.sparse import lil_matrix
6 | from polytopewalk.sparse import *
7 | from polytopewalk.dense import *
8 |
9 | simplex_A = lil_matrix((1, 3))
10 | simplex_A[(0, 0)] = 1
11 | simplex_A[(0, 1)] = 1
12 | simplex_A[(0, 2)] = 1
13 | simplex_b = np.array([1])
14 |
15 | hc_A = lil_matrix((4,6))
16 | hc_A[(0, 0)] = 1
17 | hc_A[(0, 2)] = 1
18 | hc_A[(1, 1)] = 1
19 | hc_A[(1, 3)] = 1
20 | hc_A[(2, 0)] = -1
21 | hc_A[(2, 4)] = 1
22 | hc_A[(3, 1)] = -1
23 | hc_A[(3, 5)] = 1
24 |
25 | hc_b = np.array([1,1,1,1])
26 |
27 | birk_A = lil_matrix((3, 4))
28 | birk_A[(0, 0)] = 1
29 | birk_A[(0, 1)] = 1
30 | birk_A[(1, 2)] = 1
31 | birk_A[(1, 3)] = 1
32 | birk_A[(2, 0)] = 1
33 | birk_A[(2, 2)] = 1
34 |
35 | birk_b = np.array([1,1,1])
36 |
37 | class TestFacialReduction(unittest.TestCase):
38 |
39 | def test_fr(self):
40 | fr = FacialReduction()
41 | simplex_dense = fr.reduce(simplex_A, simplex_b, 3, False)
42 | hc_dense = fr.reduce(hc_A, hc_b, 4, False)
43 | birk_dense = fr.reduce(birk_A, birk_b, 4, False)
44 |
45 | self.assertTrue(simplex_dense.dense_A.shape == (3,2))
46 | self.assertTrue(len(simplex_dense.dense_b) == 3)
47 |
48 | self.assertTrue(hc_dense.dense_A.shape == (4,2))
49 | self.assertTrue(len(hc_dense.dense_b) == 4)
50 |
51 | self.assertTrue(birk_dense.dense_A.shape == (4,1))
52 | self.assertTrue(len(birk_dense.dense_b) == 4)
53 |
54 | A = np.array([[1, 1, 0], [-1, -1, 0], [0, 1, 0], [0, -1, 0], [0, 0, 1], [0, 0, -1]])
55 | A = np.hstack((A, np.eye(6)))
56 | b = np.array([1, -1, 1, 1, 1, 1])
57 |
58 | fr_res = fr.reduce(A, b, 6, False)
59 | self.assertTrue(fr_res.sparse_A.shape == (5, 7))
60 | self.assertTrue(fr_res.dense_A.shape == (4, 2))
61 |
62 |
63 | A = np.array([[1, 0, 0], [-1, 0, 0], [0, 1, 0], [0, -1, 0], [0, 0, 1], [0, 0, -1]])
64 | A = np.hstack((A, np.eye(6)))
65 | b = np.array([1, 1, 0, 0, 0, 0])
66 |
67 | fr_res = fr.reduce(A, b, 6, False)
68 | self.assertTrue(fr_res.sparse_A.shape == (4, 5))
69 | self.assertTrue(fr_res.dense_A.shape == (2, 1))
70 |
71 | A = np.array([[1, 0], [-1, 0], [0, 1], [0, -1]])
72 | A = np.hstack((A, np.eye(4)))
73 | b = np.array([1, 0, 1, 0])
74 |
75 | fr_res = fr.reduce(A, b, 4, False)
76 | self.assertTrue(fr_res.sparse_A.shape == (4, 6))
77 | self.assertTrue(fr_res.dense_A.shape == (4, 2))
78 |
79 |
80 |
81 | if __name__ == '__main__':
82 | unittest.main()
83 |
84 |
--------------------------------------------------------------------------------
/tests/python/test_init.py:
--------------------------------------------------------------------------------
1 | import unittest
2 | from polytopewalk import FacialReduction
3 | import numpy as np
4 | import pandas as pd
5 | from scipy.sparse import lil_matrix
6 | from polytopewalk.sparse import *
7 | from polytopewalk.dense import *
8 |
9 | simplex_A = lil_matrix((1, 3))
10 | simplex_A[(0, 0)] = 1
11 | simplex_A[(0, 1)] = 1
12 | simplex_A[(0, 2)] = 1
13 | simplex_b = np.array([1])
14 |
15 | hc_A = lil_matrix((4,6))
16 | hc_A[(0, 0)] = 1
17 | hc_A[(0, 2)] = 1
18 | hc_A[(1, 1)] = 1
19 | hc_A[(1, 3)] = 1
20 | hc_A[(2, 0)] = -1
21 | hc_A[(2, 4)] = 1
22 | hc_A[(3, 1)] = -1
23 | hc_A[(3, 5)] = 1
24 |
25 | hc_b = np.array([1,1,1,1])
26 |
27 | birk_A = lil_matrix((3, 4))
28 | birk_A[(0, 0)] = 1
29 | birk_A[(0, 1)] = 1
30 | birk_A[(1, 2)] = 1
31 | birk_A[(1, 3)] = 1
32 | birk_A[(2, 0)] = 1
33 | birk_A[(2, 2)] = 1
34 |
35 | birk_b = np.array([1,1,1])
36 |
37 |
38 | class TestInitialization(unittest.TestCase):
39 | def test_init(self):
40 | sc = SparseCenter()
41 | dc = DenseCenter()
42 |
43 | x = sc.getInitialPoint(simplex_A, simplex_b, 3)
44 | self.assertAlmostEqual(max(abs(x - np.array([1/3, 1/3, 1/3]))), 0)
45 |
46 | x = sc.getInitialPoint(hc_A, hc_b, 4)
47 | self.assertAlmostEqual(max(abs(x - np.array([0, 0, 1, 1, 1, 1]))), 0)
48 |
49 | x = sc.getInitialPoint(birk_A, birk_b, 4)
50 | self.assertAlmostEqual(max(abs(x - np.array([0.5, 0.5, 0.5, 0.5]))), 0)
51 |
52 | A = np.array([[1, 0], [0, 1], [-1, 0], [0, -1]])
53 | b = np.array([1,1,0,0])
54 | x = dc.getInitialPoint(A, b)
55 | self.assertAlmostEqual(max(abs(x - np.array([0.5, 0.5]))), 0)
56 |
57 | A = np.array([[-1, 0], [0, -1], [1, 1]])
58 | b = np.array([0, 0, 1])
59 | x = dc.getInitialPoint(A, b)
60 | self.assertAlmostEqual(max(abs(x - np.array([1/3, 1/3]))), 0)
61 |
62 |
63 |
64 | if __name__ == '__main__':
65 | unittest.main()
--------------------------------------------------------------------------------
/tests/python/test_sparse_walk.py:
--------------------------------------------------------------------------------
1 | import unittest
2 | from polytopewalk import FacialReduction
3 | import numpy as np
4 | import pandas as pd
5 | from scipy.sparse import lil_matrix
6 | from polytopewalk.sparse import *
7 |
8 | simplex_A = lil_matrix((1, 3))
9 | simplex_A[(0, 0)] = 1
10 | simplex_A[(0, 1)] = 1
11 | simplex_A[(0, 2)] = 1
12 | simplex_b = np.array([1])
13 |
14 | hc_A = lil_matrix((4,6))
15 | hc_A[(0, 0)] = 1
16 | hc_A[(0, 2)] = 1
17 | hc_A[(1, 1)] = 1
18 | hc_A[(1, 3)] = 1
19 | hc_A[(2, 0)] = -1
20 | hc_A[(2, 4)] = 1
21 | hc_A[(3, 1)] = -1
22 | hc_A[(3, 5)] = 1
23 |
24 | hc_b = np.array([1,1,1,1])
25 |
26 | birk_A = lil_matrix((3, 4))
27 | birk_A[(0, 0)] = 1
28 | birk_A[(0, 1)] = 1
29 | birk_A[(1, 2)] = 1
30 | birk_A[(1, 3)] = 1
31 | birk_A[(2, 0)] = 1
32 | birk_A[(2, 2)] = 1
33 |
34 | birk_b = np.array([1,1,1])
35 |
36 |
37 |
38 | class TestSparseWalk(unittest.TestCase):
39 | def test_run(self):
40 | dikin = SparseDikinWalk(r = 0.8, thin = 2)
41 | vaidya = SparseVaidyaWalk(r = 0.8, thin = 2)
42 | john = SparseJohnWalk(r = 0.8, thin = 2)
43 |
44 | fr = FacialReduction()
45 | sc = SparseCenter()
46 | fr_res_simplex = fr.reduce(simplex_A, simplex_b, simplex_A.shape[1], True)
47 | fr_res_hc = fr.reduce(hc_A, hc_b, 4, True)
48 | fr_res_birk = fr.reduce(birk_A, birk_b, birk_A.shape[1], True)
49 |
50 | init_simplex = sc.getInitialPoint(fr_res_simplex.sparse_A, fr_res_simplex.sparse_b, 3)
51 | init_hc = sc.getInitialPoint(fr_res_hc.sparse_A, fr_res_hc.sparse_b, 4)
52 | init_birk = sc.getInitialPoint(fr_res_birk.sparse_A, fr_res_birk.sparse_b, 4)
53 |
54 | res1 = dikin.generateCompleteWalk(100, init_simplex, fr_res_simplex.sparse_A, fr_res_simplex.sparse_b, 3)
55 | self.assertTrue(res1.shape == (100, 3))
56 | res2 = vaidya.generateCompleteWalk(100, init_simplex, fr_res_simplex.sparse_A, fr_res_simplex.sparse_b, 3)
57 | self.assertTrue(res2.shape == (100, 3))
58 | res3 = john.generateCompleteWalk(100, init_simplex, fr_res_simplex.sparse_A, fr_res_simplex.sparse_b, 3)
59 | self.assertTrue(res3.shape == (100, 3))
60 |
61 |
62 | res1 = dikin.generateCompleteWalk(100, init_hc, fr_res_hc.sparse_A, fr_res_hc.sparse_b, 4)
63 | self.assertTrue(res1.shape[0] == 100)
64 | res2 = vaidya.generateCompleteWalk(100, init_hc, fr_res_hc.sparse_A, fr_res_hc.sparse_b, 4)
65 | self.assertTrue(res2.shape[0] == 100)
66 | res3 = john.generateCompleteWalk(100, init_hc, fr_res_hc.sparse_A, fr_res_hc.sparse_b, 4)
67 | self.assertTrue(res3.shape[0] == 100)
68 |
69 | res1 = dikin.generateCompleteWalk(100, init_birk, fr_res_birk.sparse_A, fr_res_birk.sparse_b, 4)
70 | self.assertTrue(res1.shape[0] == 100)
71 | res2 = vaidya.generateCompleteWalk(100, init_birk, fr_res_birk.sparse_A, fr_res_birk.sparse_b, 4)
72 | self.assertTrue(res2.shape[0] == 100)
73 | res3 = john.generateCompleteWalk(100, init_birk, fr_res_birk.sparse_A, fr_res_birk.sparse_b, 4)
74 | self.assertTrue(res3.shape[0] == 100)
75 |
76 | if __name__ == '__main__':
77 | unittest.main()
78 |
--------------------------------------------------------------------------------
/tests/python/test_weights.py:
--------------------------------------------------------------------------------
1 | import unittest
2 | from polytopewalk import FacialReduction
3 | import numpy as np
4 | import pandas as pd
5 | from scipy.sparse import lil_matrix
6 | from polytopewalk.sparse import *
7 | from polytopewalk.dense import *
8 |
9 | simplex_A = lil_matrix((1, 3))
10 | simplex_A[(0, 0)] = 1
11 | simplex_A[(0, 1)] = 1
12 | simplex_A[(0, 2)] = 1
13 | simplex_b = np.array([1])
14 |
15 | hc_A = lil_matrix((4,6))
16 | hc_A[(0, 0)] = 1
17 | hc_A[(0, 2)] = 1
18 | hc_A[(1, 1)] = 1
19 | hc_A[(1, 3)] = 1
20 | hc_A[(2, 0)] = -1
21 | hc_A[(2, 4)] = 1
22 | hc_A[(3, 1)] = -1
23 | hc_A[(3, 5)] = 1
24 |
25 | hc_b = np.array([1,1,1,1])
26 |
27 | birk_A = lil_matrix((3, 4))
28 | birk_A[(0, 0)] = 1
29 | birk_A[(0, 1)] = 1
30 | birk_A[(1, 2)] = 1
31 | birk_A[(1, 3)] = 1
32 | birk_A[(2, 0)] = 1
33 | birk_A[(2, 2)] = 1
34 |
35 | birk_b = np.array([1,1,1])
36 |
37 | class TestWeights(unittest.TestCase):
38 | def test_weights(self):
39 | sparse_vaidya = SparseVaidyaWalk(r = 0.9, thin = 1)
40 | sparse_john = SparseJohnWalk(r = 0.9, thin = 1)
41 | sparse_dikinls = SparseDikinLSWalk(r = 0.9, thin = 1)
42 |
43 | simplex_start = np.array([0.33, 0.34, 0.33])
44 | w = sparse_dikinls.generateWeight(simplex_start, simplex_A, 3)
45 | self.assertAlmostEqual(w.sum(), 2, places = 1)
46 | w = sparse_john.generateWeight(simplex_start, simplex_A, 3)
47 | self.assertAlmostEqual(w.sum(), 3, places = 2)
48 | w = sparse_vaidya.generateWeight(simplex_start, simplex_A, 3)
49 | self.assertAlmostEqual(w.sum(), 4, places = 2)
50 |
51 | hc_start = np.array([0, 0, 1, 1, 1, 1])
52 | w = sparse_dikinls.generateWeight(hc_start, hc_A, 4)
53 | self.assertAlmostEqual(w.sum(), 2, places = 1)
54 | w = sparse_john.generateWeight(hc_start, hc_A, 4)
55 | self.assertAlmostEqual(w.sum(), 3, places = 2)
56 | w = sparse_vaidya.generateWeight(hc_start, hc_A, 4)
57 | self.assertAlmostEqual(w.sum(), 4, places = 2)
58 |
59 | birk_start = np.array([0.5, 0.5, 0.5, 0.5])
60 | w = sparse_dikinls.generateWeight(birk_start, birk_A, 4)
61 | self.assertAlmostEqual(w.sum(), 1, places = 1)
62 | w = sparse_john.generateWeight(birk_start, birk_A, 4)
63 | self.assertAlmostEqual(w.sum(), 1.5, places = 2)
64 | w = sparse_vaidya.generateWeight(birk_start, birk_A, 4)
65 | self.assertAlmostEqual(w.sum(), 2, places = 2)
66 |
67 |
68 | if __name__ == '__main__':
69 | unittest.main()
--------------------------------------------------------------------------------
 3 |
3 |  3 |
3 |  16 |
16 |