├── environment.yml
├── example_input_output_files
├── example_input_file.csv
└── example_output_file.csv
├── license.txt
├── test_spacial_score.py
├── README.md
└── spacial_score.py
/environment.yml:
--------------------------------------------------------------------------------
1 | name: my_sps_env
2 | channels:
3 | - conda-forge
4 | - defaults
5 | dependencies:
6 | - python>=3.8
7 | - rdkit=2021.09.3
8 | - numpy=1.21.5
9 | - pytest
--------------------------------------------------------------------------------
/example_input_output_files/example_input_file.csv:
--------------------------------------------------------------------------------
1 | Smiles,ID
2 | C1COCC(=O)N1c1ccc(cc1)N1C[C@@H](OC1=O)CNC(=O)c1ccc(s1)Cl,1
3 | C[C@@H]1CC[C@@]23CCC(=O)[C@H]2[C@@]1([C@@H](C[C@@]([C@H]([C@@H]3C)O)(C)C=C)OC(=O)CO)C,2
4 | C[C@@H]1CCC23[C@H]1CC[C@]2([C@H]1C[C@]2(CC[C@H]([C@@H]2CC1=C3C(=O)O)C(C)C)C)C,3
5 | CC(C)CCCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C,4
6 | O=C(OCC1=CC=CC=C1)C=P(C2=CC=CC=C2)(C3=CC=CC=C3)C4=CC=CC=C4,5
7 | BrC(C(OCC1=CC=CC=C1)=O)/C(C(OC)OC)=C/C(OCC2=CC=CC=C2)=O,6
8 | O[C@H]([C@H](C(OCC1=CC=CC=C1)=O)/C(C(OC)OC)=C/C(OCC2=CC=CC=C2)=O)/C=C(C(C)(C)C)\Br,7
9 | O[C@H]1C=C(C(C)(C)C)[C@](CC(OCC2=CC=CC=C2)=O)(C(OC)OC)[C@H]1C(OCC3=CC=CC=C3)=O,8
10 | O[C@H]1C[C@@](O)(C(C)(C)C)[C@](CC(OCC2=CC=CC=C2)=O)(C(OC)OC)[C@H]1C(OCC3=CC=CC=C3)=O,9
11 | O[C@H]1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@H]1C(OCC4=CC=CC=C4)=O,10
12 | O=C1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@H]1C(OCC4=CC=CC=C4)=O,11
13 | O=C1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@]1(C#C)C(OCC4=CC=CC=C4)=O,12
14 | O[C@H]1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@]1(C#C)C(OCC4=CC=CC=C4)=O,13
15 | O=C1C[C@]2(C(OCC3=CC=CC=C3)=O)[C@]4(CC(OCC5=CC=CC=C5)=O)[C@](O[C@@H]4OC)(C(C)(C)C)C[C@]2([H])O1,14
16 | O=C1C[C@]23C([C@]4([H])OC3=O)(CC(O4)=O)[C@](O)(C(C)(C)C)C[C@]2([H])O1,15
17 | O=C1C[C@]23C([C@]4([H])OC3=O)([C@](O)([H])C(O4)=O)[C@](O)(C(C)(C)C)C[C@]2([H])O1,16
18 |
--------------------------------------------------------------------------------
/license.txt:
--------------------------------------------------------------------------------
1 | BSD 3-Clause License
2 |
3 | Copyright (c) 2023, Waldmann Lab.
4 |
5 | Redistribution and use in source and binary forms, with or without
6 | modification, are permitted provided that the following conditions are met:
7 |
8 | 1. Redistributions of source code must retain the above copyright notice, this
9 | list of conditions and the following disclaimer.
10 |
11 | 2. Redistributions in binary form must reproduce the above copyright notice,
12 | this list of conditions and the following disclaimer in the documentation
13 | and/or other materials provided with the distribution.
14 |
15 | 3. Neither the name of the copyright holder nor the names of its
16 | contributors may be used to endorse or promote products derived from
17 | this software without specific prior written permission.
18 |
19 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
20 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
21 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
22 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
23 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
24 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
25 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
26 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
27 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
28 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
29 |
--------------------------------------------------------------------------------
/example_input_output_files/example_output_file.csv:
--------------------------------------------------------------------------------
1 | Smiles,ID,nSPS
2 | C1COCC(=O)N1c1ccc(cc1)N1C[C@@H](OC1=O)CNC(=O)c1ccc(s1)Cl,1,19.413793103448278
3 | C[C@@H]1CC[C@@]23CCC(=O)[C@H]2[C@@]1([C@@H](C[C@@]([C@H]([C@@H]3C)O)(C)C=C)OC(=O)CO)C,2,49.7037037037037
4 | C[C@@H]1CCC23[C@H]1CC[C@]2([C@H]1C[C@]2(CC[C@H]([C@@H]2CC1=C3C(=O)O)C(C)C)C)C,3,53.48148148148148
5 | CC(C)CCCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C,4,46.535714285714285
6 | O=C(OCC1=CC=CC=C1)C=P(C2=CC=CC=C2)(C3=CC=CC=C3)C4=CC=CC=C4,5,10.933333333333334
7 | BrC(C(OCC1=CC=CC=C1)=O)/C(C(OC)OC)=C/C(OCC2=CC=CC=C2)=O,6,12.482758620689655
8 | O[C@H]([C@H](C(OCC1=CC=CC=C1)=O)/C(C(OC)OC)=C/C(OCC2=CC=CC=C2)=O)/C=C(C(C)(C)C)\Br,7,14.27027027027027
9 | O[C@H]1C=C(C(C)(C)C)[C@](CC(OCC2=CC=CC=C2)=O)(C(OC)OC)[C@H]1C(OCC3=CC=CC=C3)=O,8,21.805555555555557
10 | O[C@H]1C[C@@](O)(C(C)(C)C)[C@](CC(OCC2=CC=CC=C2)=O)(C(OC)OC)[C@H]1C(OCC3=CC=CC=C3)=O,9,25.72972972972973
11 | O[C@H]1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@H]1C(OCC4=CC=CC=C4)=O,10,29.685714285714287
12 | O=C1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@H]1C(OCC4=CC=CC=C4)=O,11,27.6
13 | O=C1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@]1(C#C)C(OCC4=CC=CC=C4)=O,12,28.513513513513512
14 | O[C@H]1C[C@@](O[C@@H]2OC)(C(C)(C)C)[C@@]2(CC(OCC3=CC=CC=C3)=O)[C@]1(C#C)C(OCC4=CC=CC=C4)=O,13,30.486486486486488
15 | O=C1C[C@]2(C(OCC3=CC=CC=C3)=O)[C@]4(CC(OCC5=CC=CC=C5)=O)[C@](O[C@@H]4OC)(C(C)(C)C)C[C@]2([H])O1,14,31.526315789473685
16 | O=C1C[C@]23C([C@]4([H])OC3=O)(CC(O4)=O)[C@](O)(C(C)(C)C)C[C@]2([H])O1,15,49.36363636363637
17 | O=C1C[C@]23C([C@]4([H])OC3=O)([C@](O)([H])C(O4)=O)[C@](O)(C(C)(C)C)C[C@]2([H])O1,16,51.0
18 |
--------------------------------------------------------------------------------
/test_spacial_score.py:
--------------------------------------------------------------------------------
1 | # Script for testing of spacial_score.py
2 |
3 | import numpy as np
4 | import rdkit
5 | import spacial_score as sps
6 |
7 |
8 | valid_smiles = r"C/C=C\C1C=CCC(Br)C1C2=CC(C#C)=CC=C2"
9 | invalid_smiles = r"abcx--a"
10 |
11 |
12 | def test_smiles_to_mol():
13 | """ Test conversion of SMILES to RDKit mol """
14 | valid_mol = sps.smiles_to_mol(valid_smiles)
15 | invalid_mol = sps.smiles_to_mol(invalid_smiles)
16 |
17 | assert isinstance(valid_mol, rdkit.Chem.rdchem.Mol)
18 | assert invalid_mol is np.nan
19 |
20 |
21 | def create_sps_object():
22 | """ Create an instance of sps object based on a valid SMILES representation of a molecule """
23 | mol = sps.smiles_to_mol(valid_smiles)
24 | sps_object = sps.SpacialScore(valid_smiles, mol)
25 | return sps_object
26 |
27 |
28 | def calc_sum_score(score_type):
29 | """ Calculate a summed score for values in a dictionary """
30 | sum_score = 0
31 | for atom_score in score_type.values():
32 | sum_score += atom_score
33 | return sum_score
34 |
35 |
36 | def test_hybridisation_score():
37 | """ Test if the summed hybridisation score for a molecule is calculated properly """
38 | hyb_score = calc_sum_score(create_sps_object().hyb_score)
39 | assert hyb_score == 40
40 |
41 |
42 | def test_stereo_score():
43 | """ Test if the summed stereo score for a molecule is calculated properly """
44 |
45 | stereo_score = calc_sum_score(create_sps_object().stereo_score)
46 | assert stereo_score == 23
47 |
48 |
49 | def test_ring_score():
50 | """ Test if the summed ring score for a molecule is calculated properly """
51 | ring_score = calc_sum_score(create_sps_object().ring_score)
52 | assert ring_score == 24
53 |
54 |
55 | def test_bond_score():
56 | """ Test if the summed bond score for a molecule is calculated properly """
57 | bond_score = calc_sum_score(create_sps_object().bond_score)
58 | assert bond_score == 88
59 |
60 |
61 | def test_SPS_from_smiles():
62 | """ Test if the un-normilised SPS is calculated properly for a test molecule """
63 | sps_score = sps.calculate_score_from_smiles(valid_smiles)
64 | assert sps_score == 491
65 |
66 |
67 | def test_nSPS_from_smiles():
68 | """ Test if the normilised SPS (nSPS) is calculated properly for a test molecule """
69 | sps_score = sps.calculate_score_from_smiles(valid_smiles, per_atom=True).__round__(2)
70 | assert sps_score == 27.28
71 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Spacial-Score
2 | A Comprehensive Topological Indicator for Small Molecule Complexity
3 |
(Created by the Waldmann Lab at the Max Planck Institute of Molecular Physiology, Dortmund)
4 |
5 | The score is intended for assessing molecular topology of organic molecules and to improve upon the idea of the fraction of stereo and sp3 carbons. The score is described in our J. Med. Chem. paper: [Spacial Score – A Comprehensive Topological Indicator for Small Molecule Complexity](https://doi.org/10.1021/acs.jmedchem.3c00689).
6 |
7 | > [!IMPORTANT]
8 | > **The SPS indicator is now also a part of the RDKit package and available through the [rdkit.Chem.SpacialScore](https://www.rdkit.org/docs/source/rdkit.Chem.SpacialScore.html#module-rdkit.Chem.SpacialScore) module. SPS script is actively mantained as part of the RDKit package, and thus, it is recommended to calculate the SPS through RDKit.**
9 |
10 | 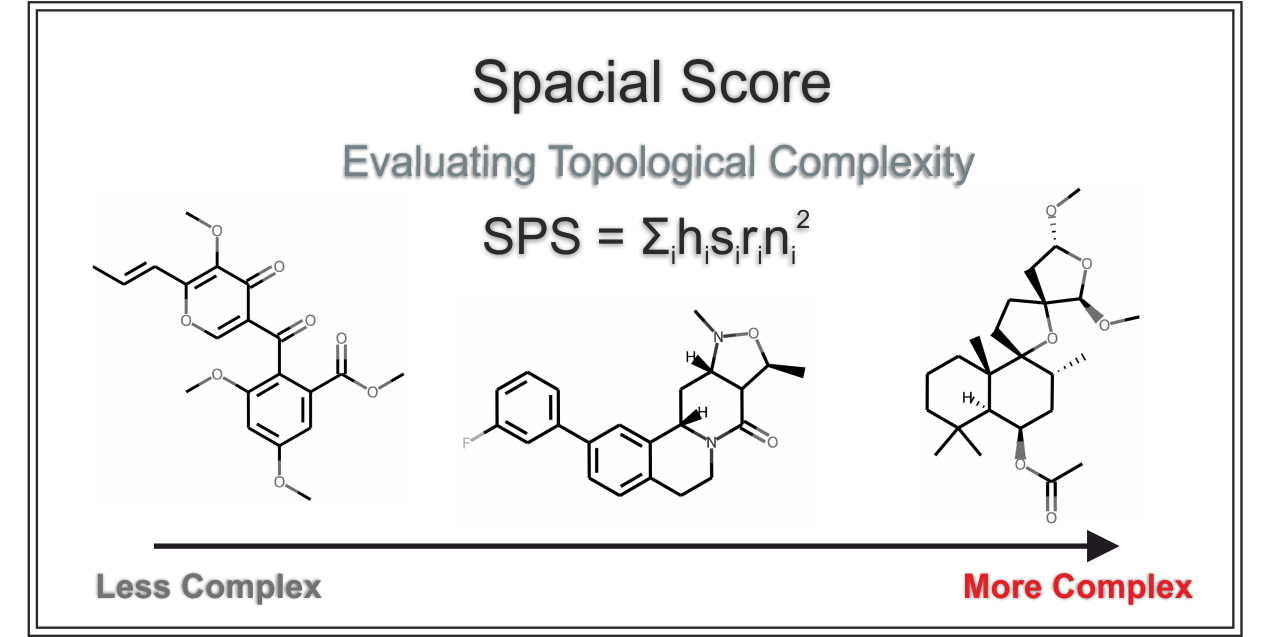 11 |
12 | ***
13 | ### Required Python Packages
14 | The script requires [RDKit package](https://www.rdkit.org/) and [NumPy](https://numpy.org/).
15 | To install the required packages through [Conda](https://docs.conda.io/en/latest/miniconda.html), simply use the `environment.yml` file:
16 | ```
17 | conda env create -f environment.yml
18 | ```
19 | The script can be used after activation of the just created conda environment:
20 | ```
21 | conda activate my_sps_env
22 | ```
23 | To display the options of `spacial_score.py`, type:
24 | ```
25 | python spacial_score.py -h
26 | ```
27 | (Please remember that `spacial_score.py` needs to be in your current directory)
28 | ***
29 | ### Using the Script Through Command Line
30 | The script can be used directly from a command line, reading either a directly provided SMILES string `(-s)` or a .csv/.tsv file `(-i)`:
31 | ```
32 | usage: spacial_score.py [-h] [-s SMILES string] [-i filename.ext] [-o filename.ext] [-t] [-v] [-p]
33 |
34 | Script for calculating Spacial Score (SPS) or normalised SPS (nSPS) for small molecules.
35 | The script can calculate the scores for a direct SMILES input or for a .csv or .tsv file containing a list of SMILES.
36 | nSPS is calculated by deafult.
37 |
38 | optional arguments:
39 | -h, --help show this help message and exit
40 | -s SMILES string Your input SMILES string for which to calculate the score
41 | -i filename.ext Your .csv or .tsv file containing column called "Smiles" which contains SMILES strings. Resutls will be
42 | saved in a new .csv file
43 | -o filename.csv You can specify name of the output .csv file. Not required.
44 | -t Option to calculate total SPS (no normalisation).
45 | -v Option to print verbose results, with information for each atom index.
46 | -p Option to print confirmation after processing of each SMILES string in a file.
47 | ```
48 |
49 | To calculate nSPS directly from a SMILES string you can just type:
50 | ```
51 | python spacial_score.py -s CC(C)CBr
52 | ```
53 | Where CC(C)CBr is just an example of a SMILES string (on Linux you may need to use quotation marks "CC(C)CBr").
54 | This returns:
55 | ```
56 | Normalisation Applied: True
57 | SMILES: CC(C)CBr
58 | Calculated nSPS: 9.6
59 | ```
60 |
61 | To calculate the un-normalised (total) SPS you need to add option `-t`:
62 | ```
63 | python spacial_score.py -s CC(C)CBr -t
64 | ```
65 | This returns:
66 | ```
67 | Normalisation Applied: False
68 | SMILES: CC(C)CBr
69 | Calculated SPS: 48
70 | ```
71 |
72 | A more verbose output can be achieved with the option `-v`:
73 | ```
74 | python spacial_score.py -s CC(C)CBr -v
75 | ```
76 | The output:
77 | ```
78 | SMILES: CC(C)CBr
79 | Atom Idx Element Hybrid Stereo Ring Neighbs
80 | ------------------------------------------------------------
81 | 0 C 3 1 1 1
82 | 1 C 3 1 1 9
83 | 2 C 3 1 1 1
84 | 3 C 3 1 1 4
85 | 4 Br 3 1 1 1
86 | ------------------------------------------------------------
87 | Total Spacial Score: 48
88 | Per-Atom Score: 9.6
89 | ```
90 |
91 | To read in a .csv or .tsv file, please type:
92 | ```
93 | python spacial_score.py -i your_input_file_name.csv -o your_output_file_name.csv
94 | ```
95 | nSPS is calculated by default, and option `-t` can be used to calculate un-normalised SPS.
96 | Please, remember that your input file needs to contain a column named `Smiles` containing SMILES which will be used for the calculation of the scores.
97 | Examples of input and output files can be found in the folder named `example_input_output_files`.
98 |
99 | ***
100 | ### Calculate the Score with a Python Function
101 | The scores can also be calculated by using function:
102 | ```
103 | def calculate_score_from_smiles(smiles: str, per_atom=False, verbose=False) -> float:
104 | """ Calculates the spacial score as a total SPS or size-normalised, per-atom nSPS for a molecule.
105 |
106 | Parameters:
107 | ===========
108 | smiles: valid SMILES string
109 | per_atom: flag to denote if the normalised per-atom result (nSPS) should be returned
110 | verbose: flag to denote if the detailed scores for each atom should be printed
111 |
112 | Returns:
113 | ========
114 | Total or per-atom numeric spacial score for the provided molecule.
115 | """
116 | ```
117 | ***
118 | ### Testing the Script
119 | The script in `spacial_score.py` can be tested by running [pytest](https://docs.pytest.org/en/7.2.x/contents.html). In the active `my_sps_env` conda environment type:
120 | ```
121 | pytest
122 | ```
123 | Correct output will look like this:
124 | ```
125 | collected 7 items
126 |
127 | test_spacial_score.py ....... [100%]
128 |
129 | ================================================== 7 passed in 0.48s ==================================================
130 |
131 | ```
132 |
--------------------------------------------------------------------------------
/spacial_score.py:
--------------------------------------------------------------------------------
1 | # Version 1.0
2 |
3 | from rdkit import Chem
4 | import rdkit.Chem.Descriptors as Desc
5 | import numpy as np
6 | import argparse
7 | import sys
8 | import csv
9 |
10 |
11 | class SpacialScore:
12 | """Class intended for calculating spacial score (SPS) and size-normalised SPS (nSPS) for small organic molecules"""
13 | def __init__(self, smiles, mol, verbose=False):
14 | self.smiles = smiles
15 | self.mol = mol
16 | self.verbose = verbose
17 |
18 | self.hyb_score = {}
19 | self.stereo_score = {}
20 | self.ring_score = {}
21 | self.bond_score = {}
22 | self.chiral_idxs = self.find_stereo_atom_idxs()
23 | self.doublebonds_stereo = self.find_doublebonds_stereo()
24 | self.score = self.calculate_spacial_score()
25 | self.per_atom_score = self.score/Desc.HeavyAtomCount(self.mol)
26 |
27 | if self.verbose:
28 | self.display_scores()
29 |
30 |
31 | def display_scores(self):
32 | """Displays the individual scores for each molecule atom"""
33 |
34 | print("SMILES:", self.smiles)
35 | print("Atom Idx".ljust(10, " "), end="")
36 | print("Element".ljust(10, " "), end="")
37 | print("Hybrid".ljust(10, " "), end="")
38 | print("Stereo".ljust(10, " "), end="")
39 | print("Ring".ljust(10, " "), end="")
40 | print("Neighbs".ljust(10, " "))
41 | print("".ljust(60, "-"))
42 |
43 | for atom in self.mol.GetAtoms():
44 | atom_idx = atom.GetIdx()
45 | print(str(atom_idx).ljust(10, " "), end="")
46 | print(str(Chem.rdchem.Atom.GetSymbol(atom)).ljust(10, " "), end="")
47 | print(str(self.hyb_score[atom_idx]).ljust(10, " "), end="")
48 | print(str(self.stereo_score[atom_idx]).ljust(10, " "), end="")
49 | print(str(self.ring_score[atom_idx]).ljust(10, " "), end="")
50 | print(str(self.bond_score[atom_idx]).ljust(10, " "))
51 |
52 | print("".ljust(60, "-"))
53 | print("Total Spacial Score:", self.score)
54 | print("Per-Atom Score:", self.per_atom_score.__round__(2), "\n")
55 |
56 |
57 | def find_stereo_atom_idxs(self, includeUnassigned=True):
58 | """Finds indeces of atoms that are (pseudo)stereo/chiralcentres, in repsect to the attached groups (does not account for double bond isomers)"""
59 | stereo_centers = Chem.FindMolChiralCenters(self.mol, includeUnassigned=includeUnassigned, includeCIP=False, useLegacyImplementation=False)
60 | stereo_idxs = [atom_idx for atom_idx, _ in stereo_centers]
61 | return stereo_idxs
62 |
63 |
64 | def find_doublebonds_stereo(self):
65 | """Finds indeces of stereo double bond atoms (E/Z)"""
66 | db_stereo = {}
67 | for bond in self.mol.GetBonds():
68 | if str(bond.GetBondType()) == "DOUBLE":
69 | db_stereo[(bond.GetBeginAtomIdx(), bond.GetEndAtomIdx())] = bond.GetStereo()
70 | return db_stereo
71 |
72 |
73 | def calculate_spacial_score(self):
74 | """Calculates the total spacial score for a molecule"""
75 | score = 0
76 | for atom in self.mol.GetAtoms():
77 | atom_idx = atom.GetIdx()
78 | self.hyb_score[atom_idx] = self._account_for_hybridisation(atom)
79 | self.stereo_score[atom_idx] = self._account_for_stereo(atom_idx)
80 | self.ring_score[atom_idx] = self._account_for_ring(atom)
81 | self.bond_score[atom_idx] = self._account_for_neighbours(atom)
82 | score += self._calculate_score_for_atom(atom_idx)
83 | return score
84 |
85 |
86 | def _calculate_score_for_atom(self, atom_idx):
87 | """Calculates the total score for a single atom in a molecule"""
88 | atom_score = self.hyb_score[atom_idx] * self.stereo_score[atom_idx] * self.ring_score[atom_idx] * self.bond_score[atom_idx]
89 | return atom_score
90 |
91 |
92 | def _account_for_hybridisation(self, atom):
93 | """Calculates the hybridisation score for a single atom in a molecule"""
94 | hybridisations = {"SP": 1, "SP2": 2, "SP3": 3}
95 | hyb_type = str(atom.GetHybridization())
96 |
97 | if hyb_type in hybridisations.keys():
98 | return hybridisations[hyb_type]
99 | return 4 # h score for any other hybridisation than sp, sp2 or sp3
100 |
101 |
102 | def _account_for_stereo(self, atom_idx):
103 | """Calculates the stereo score for a single atom in a molecule"""
104 | if atom_idx in self.chiral_idxs:
105 | return 2
106 | for bond_atom_idxs, stereo in self.doublebonds_stereo.items():
107 | if atom_idx in bond_atom_idxs and not(str(stereo).endswith("NONE")):
108 | return 2
109 | return 1
110 |
111 |
112 | def _account_for_ring(self, atom):
113 | """Calculates the ring score for a single atom in a molecule"""
114 | if atom.GetIsAromatic(): # aromatic rings are not promoted
115 | return 1
116 | if atom.IsInRing():
117 | return 2
118 | return 1
119 |
120 |
121 | def _account_for_neighbours(self, atom):
122 | """Calculates the neighbour score for a single atom in a molecule
123 | The second power allows to account for branching in the molecular structure"""
124 | return (len(atom.GetNeighbors()))**2
125 |
126 |
127 | def smiles_to_mol(smiles: str):
128 | """ Generate a RDKit Molecule from SMILES.
129 |
130 | Parameters:
131 | ===========
132 | smiles: the input string
133 |
134 | Returns:
135 | ========
136 | The RDKit Molecule. If the Smiles parsing failed, NAN is returned instead.
137 | """
138 | try:
139 | mol = Chem.MolFromSmiles(smiles)
140 | if mol is not None:
141 | return mol
142 | return np.nan
143 | except:
144 | return np.nan
145 |
146 |
147 | def close_files(open_files:tuple):
148 | """Closes open files"""
149 | for file in open_files:
150 | file.close()
151 |
152 |
153 | def process_input(smiles:str, filename:str, output_name:str, total_score:bool, verbose:bool, confirmation:False):
154 | """Processes the command line input to print out the resulting score or create a file with added results"""
155 |
156 | if smiles: # process a directly provided SMILES string
157 | result = calculate_score_from_smiles(smiles, per_atom=(not total_score), verbose=verbose)
158 | if not verbose:
159 | score_type = "SPS" if total_score else "nSPS"
160 | print(f"\nNormalisation Applied: {not total_score}\nSMILES: {smiles}\nCalculated {score_type}: {result}")
161 | if result is np.nan:
162 | print("\nPlease double-check your input SMILES string...")
163 |
164 | elif filename: # process provided file
165 | provided_filename_base = filename.split(".")[0]
166 | output_filename = output_name if output_name else provided_filename_base + "_SPS.csv"
167 | outfile = open(output_filename, "w")
168 |
169 | # read the input .csv or .tsv file
170 | if filename.endswith("csv"):
171 | infile = open(filename, "r")
172 | reader = csv.DictReader(infile, dialect="excel")
173 | elif filename.endswith("tsv"):
174 | infile = open(filename, "r")
175 | reader = csv.DictReader(infile, dialect="excel-tab")
176 | else:
177 | raise ValueError(f"Unknown input file format: {filename}")
178 |
179 | print("\nProcessing, please wait...")
180 | for idx, row in enumerate(reader):
181 | if idx == 0:
182 | header = [column_name for column_name in row] # read existing headers
183 | # add SPS or nSPS column to the file
184 | if total_score:
185 | header.append("SPS")
186 | header.append("nSPS")
187 | outfile.write(",".join(header) + "\n")
188 |
189 | try:
190 | if total_score:
191 | row["SPS"] = calculate_score_from_smiles(row["Smiles"], per_atom=False, verbose=verbose)
192 | row["nSPS"] = calculate_score_from_smiles(row["Smiles"], per_atom=True, verbose=verbose)
193 | except KeyError:
194 | close_files((outfile, infile))
195 | raise KeyError("Please make sure that your file contains column called 'Smiles' with SMILES strings")
196 |
197 | line = [str(row[x]) for x in row] # reconstruct the row
198 | outfile.write(",".join(line) + "\n")
199 | if confirmation:
200 | print("Finished calculations for:", row["Smiles"])
201 |
202 | close_files((outfile, infile))
203 | print(f"Finished. {output_filename} was saved.")
204 | else:
205 | raise ValueError(f"No input was provided")
206 |
207 |
208 | def calculate_score_from_smiles(smiles: str, per_atom=False, verbose=False) -> float:
209 | """ Calculates the spacial score as a total SPS or size-normalised, per-atom nSPS for a molecule.
210 |
211 | Parameters:
212 | ===========
213 | smiles: valid SMILES string
214 | per_atom: flag to denote if the normalised per-atom result (nSPS) should be returned
215 | verbose: flag to denote if the detailed scores for each atom should be printed
216 |
217 | Returns:
218 | ========
219 | Total or per-atom numeric spacial score for the provided molecule.
220 | """
221 | mol = smiles_to_mol(smiles)
222 | if mol is np.nan:
223 | return np.nan
224 | sps = SpacialScore(smiles, mol, verbose)
225 | if per_atom:
226 | return sps.per_atom_score
227 | return sps.score
228 |
229 |
230 | if __name__ == "__main__":
231 |
232 | parser = argparse.ArgumentParser(description=
233 | 'Script for calculating Spacial Score (SPS) or normalised SPS (nSPS) for small molecules.\
234 | \nThe script can calculate the scores for a direct SMILES input or for a .csv or .tsv file containing a list of SMILES.\
235 | \nnSPS is calculated by deafult.',
236 | usage=None, formatter_class=argparse.RawDescriptionHelpFormatter)
237 | parser.add_argument('-s', action="store",
238 | metavar="SMILES string",
239 | help='Your input SMILES string for which to calculate the score', default=None)
240 | parser.add_argument('-i', action="store",
241 | help='Your .csv or .tsv file containing column called "Smiles" which contains SMILES strings. Resutls will be saved in a new .csv file',
242 | metavar='filename.ext',
243 | default=None)
244 | parser.add_argument('-o', action="store",
245 | help='You can specify name of the output .csv file. Not required.',
246 | metavar='filename.csv',
247 | default=None)
248 | parser.add_argument('-t', action="store_true",
249 | help='Option to calculate total SPS (no normalisation).',
250 | default=False)
251 | parser.add_argument('-v', action="store_true",
252 | help='Option to print verbose results, with information for each atom index.',
253 | default=False)
254 | parser.add_argument('-p', action="store_true",
255 | help='Option to print confirmation after processing of each SMILES string in a file.',
256 | default=False)
257 |
258 | if len(sys.argv) < 2:
259 | parser.print_help()
260 | sys.exit(1)
261 |
262 | ARGS = parser.parse_args()
263 | process_input(smiles=ARGS.s, filename=ARGS.i, output_name=ARGS.o, total_score=ARGS.t, verbose=ARGS.v, confirmation=ARGS.p)
264 |
--------------------------------------------------------------------------------
11 |
12 | ***
13 | ### Required Python Packages
14 | The script requires [RDKit package](https://www.rdkit.org/) and [NumPy](https://numpy.org/).
15 | To install the required packages through [Conda](https://docs.conda.io/en/latest/miniconda.html), simply use the `environment.yml` file:
16 | ```
17 | conda env create -f environment.yml
18 | ```
19 | The script can be used after activation of the just created conda environment:
20 | ```
21 | conda activate my_sps_env
22 | ```
23 | To display the options of `spacial_score.py`, type:
24 | ```
25 | python spacial_score.py -h
26 | ```
27 | (Please remember that `spacial_score.py` needs to be in your current directory)
28 | ***
29 | ### Using the Script Through Command Line
30 | The script can be used directly from a command line, reading either a directly provided SMILES string `(-s)` or a .csv/.tsv file `(-i)`:
31 | ```
32 | usage: spacial_score.py [-h] [-s SMILES string] [-i filename.ext] [-o filename.ext] [-t] [-v] [-p]
33 |
34 | Script for calculating Spacial Score (SPS) or normalised SPS (nSPS) for small molecules.
35 | The script can calculate the scores for a direct SMILES input or for a .csv or .tsv file containing a list of SMILES.
36 | nSPS is calculated by deafult.
37 |
38 | optional arguments:
39 | -h, --help show this help message and exit
40 | -s SMILES string Your input SMILES string for which to calculate the score
41 | -i filename.ext Your .csv or .tsv file containing column called "Smiles" which contains SMILES strings. Resutls will be
42 | saved in a new .csv file
43 | -o filename.csv You can specify name of the output .csv file. Not required.
44 | -t Option to calculate total SPS (no normalisation).
45 | -v Option to print verbose results, with information for each atom index.
46 | -p Option to print confirmation after processing of each SMILES string in a file.
47 | ```
48 |
49 | To calculate nSPS directly from a SMILES string you can just type:
50 | ```
51 | python spacial_score.py -s CC(C)CBr
52 | ```
53 | Where CC(C)CBr is just an example of a SMILES string (on Linux you may need to use quotation marks "CC(C)CBr").
54 | This returns:
55 | ```
56 | Normalisation Applied: True
57 | SMILES: CC(C)CBr
58 | Calculated nSPS: 9.6
59 | ```
60 |
61 | To calculate the un-normalised (total) SPS you need to add option `-t`:
62 | ```
63 | python spacial_score.py -s CC(C)CBr -t
64 | ```
65 | This returns:
66 | ```
67 | Normalisation Applied: False
68 | SMILES: CC(C)CBr
69 | Calculated SPS: 48
70 | ```
71 |
72 | A more verbose output can be achieved with the option `-v`:
73 | ```
74 | python spacial_score.py -s CC(C)CBr -v
75 | ```
76 | The output:
77 | ```
78 | SMILES: CC(C)CBr
79 | Atom Idx Element Hybrid Stereo Ring Neighbs
80 | ------------------------------------------------------------
81 | 0 C 3 1 1 1
82 | 1 C 3 1 1 9
83 | 2 C 3 1 1 1
84 | 3 C 3 1 1 4
85 | 4 Br 3 1 1 1
86 | ------------------------------------------------------------
87 | Total Spacial Score: 48
88 | Per-Atom Score: 9.6
89 | ```
90 |
91 | To read in a .csv or .tsv file, please type:
92 | ```
93 | python spacial_score.py -i your_input_file_name.csv -o your_output_file_name.csv
94 | ```
95 | nSPS is calculated by default, and option `-t` can be used to calculate un-normalised SPS.
96 | Please, remember that your input file needs to contain a column named `Smiles` containing SMILES which will be used for the calculation of the scores.
97 | Examples of input and output files can be found in the folder named `example_input_output_files`.
98 |
99 | ***
100 | ### Calculate the Score with a Python Function
101 | The scores can also be calculated by using function:
102 | ```
103 | def calculate_score_from_smiles(smiles: str, per_atom=False, verbose=False) -> float:
104 | """ Calculates the spacial score as a total SPS or size-normalised, per-atom nSPS for a molecule.
105 |
106 | Parameters:
107 | ===========
108 | smiles: valid SMILES string
109 | per_atom: flag to denote if the normalised per-atom result (nSPS) should be returned
110 | verbose: flag to denote if the detailed scores for each atom should be printed
111 |
112 | Returns:
113 | ========
114 | Total or per-atom numeric spacial score for the provided molecule.
115 | """
116 | ```
117 | ***
118 | ### Testing the Script
119 | The script in `spacial_score.py` can be tested by running [pytest](https://docs.pytest.org/en/7.2.x/contents.html). In the active `my_sps_env` conda environment type:
120 | ```
121 | pytest
122 | ```
123 | Correct output will look like this:
124 | ```
125 | collected 7 items
126 |
127 | test_spacial_score.py ....... [100%]
128 |
129 | ================================================== 7 passed in 0.48s ==================================================
130 |
131 | ```
132 |
--------------------------------------------------------------------------------
/spacial_score.py:
--------------------------------------------------------------------------------
1 | # Version 1.0
2 |
3 | from rdkit import Chem
4 | import rdkit.Chem.Descriptors as Desc
5 | import numpy as np
6 | import argparse
7 | import sys
8 | import csv
9 |
10 |
11 | class SpacialScore:
12 | """Class intended for calculating spacial score (SPS) and size-normalised SPS (nSPS) for small organic molecules"""
13 | def __init__(self, smiles, mol, verbose=False):
14 | self.smiles = smiles
15 | self.mol = mol
16 | self.verbose = verbose
17 |
18 | self.hyb_score = {}
19 | self.stereo_score = {}
20 | self.ring_score = {}
21 | self.bond_score = {}
22 | self.chiral_idxs = self.find_stereo_atom_idxs()
23 | self.doublebonds_stereo = self.find_doublebonds_stereo()
24 | self.score = self.calculate_spacial_score()
25 | self.per_atom_score = self.score/Desc.HeavyAtomCount(self.mol)
26 |
27 | if self.verbose:
28 | self.display_scores()
29 |
30 |
31 | def display_scores(self):
32 | """Displays the individual scores for each molecule atom"""
33 |
34 | print("SMILES:", self.smiles)
35 | print("Atom Idx".ljust(10, " "), end="")
36 | print("Element".ljust(10, " "), end="")
37 | print("Hybrid".ljust(10, " "), end="")
38 | print("Stereo".ljust(10, " "), end="")
39 | print("Ring".ljust(10, " "), end="")
40 | print("Neighbs".ljust(10, " "))
41 | print("".ljust(60, "-"))
42 |
43 | for atom in self.mol.GetAtoms():
44 | atom_idx = atom.GetIdx()
45 | print(str(atom_idx).ljust(10, " "), end="")
46 | print(str(Chem.rdchem.Atom.GetSymbol(atom)).ljust(10, " "), end="")
47 | print(str(self.hyb_score[atom_idx]).ljust(10, " "), end="")
48 | print(str(self.stereo_score[atom_idx]).ljust(10, " "), end="")
49 | print(str(self.ring_score[atom_idx]).ljust(10, " "), end="")
50 | print(str(self.bond_score[atom_idx]).ljust(10, " "))
51 |
52 | print("".ljust(60, "-"))
53 | print("Total Spacial Score:", self.score)
54 | print("Per-Atom Score:", self.per_atom_score.__round__(2), "\n")
55 |
56 |
57 | def find_stereo_atom_idxs(self, includeUnassigned=True):
58 | """Finds indeces of atoms that are (pseudo)stereo/chiralcentres, in repsect to the attached groups (does not account for double bond isomers)"""
59 | stereo_centers = Chem.FindMolChiralCenters(self.mol, includeUnassigned=includeUnassigned, includeCIP=False, useLegacyImplementation=False)
60 | stereo_idxs = [atom_idx for atom_idx, _ in stereo_centers]
61 | return stereo_idxs
62 |
63 |
64 | def find_doublebonds_stereo(self):
65 | """Finds indeces of stereo double bond atoms (E/Z)"""
66 | db_stereo = {}
67 | for bond in self.mol.GetBonds():
68 | if str(bond.GetBondType()) == "DOUBLE":
69 | db_stereo[(bond.GetBeginAtomIdx(), bond.GetEndAtomIdx())] = bond.GetStereo()
70 | return db_stereo
71 |
72 |
73 | def calculate_spacial_score(self):
74 | """Calculates the total spacial score for a molecule"""
75 | score = 0
76 | for atom in self.mol.GetAtoms():
77 | atom_idx = atom.GetIdx()
78 | self.hyb_score[atom_idx] = self._account_for_hybridisation(atom)
79 | self.stereo_score[atom_idx] = self._account_for_stereo(atom_idx)
80 | self.ring_score[atom_idx] = self._account_for_ring(atom)
81 | self.bond_score[atom_idx] = self._account_for_neighbours(atom)
82 | score += self._calculate_score_for_atom(atom_idx)
83 | return score
84 |
85 |
86 | def _calculate_score_for_atom(self, atom_idx):
87 | """Calculates the total score for a single atom in a molecule"""
88 | atom_score = self.hyb_score[atom_idx] * self.stereo_score[atom_idx] * self.ring_score[atom_idx] * self.bond_score[atom_idx]
89 | return atom_score
90 |
91 |
92 | def _account_for_hybridisation(self, atom):
93 | """Calculates the hybridisation score for a single atom in a molecule"""
94 | hybridisations = {"SP": 1, "SP2": 2, "SP3": 3}
95 | hyb_type = str(atom.GetHybridization())
96 |
97 | if hyb_type in hybridisations.keys():
98 | return hybridisations[hyb_type]
99 | return 4 # h score for any other hybridisation than sp, sp2 or sp3
100 |
101 |
102 | def _account_for_stereo(self, atom_idx):
103 | """Calculates the stereo score for a single atom in a molecule"""
104 | if atom_idx in self.chiral_idxs:
105 | return 2
106 | for bond_atom_idxs, stereo in self.doublebonds_stereo.items():
107 | if atom_idx in bond_atom_idxs and not(str(stereo).endswith("NONE")):
108 | return 2
109 | return 1
110 |
111 |
112 | def _account_for_ring(self, atom):
113 | """Calculates the ring score for a single atom in a molecule"""
114 | if atom.GetIsAromatic(): # aromatic rings are not promoted
115 | return 1
116 | if atom.IsInRing():
117 | return 2
118 | return 1
119 |
120 |
121 | def _account_for_neighbours(self, atom):
122 | """Calculates the neighbour score for a single atom in a molecule
123 | The second power allows to account for branching in the molecular structure"""
124 | return (len(atom.GetNeighbors()))**2
125 |
126 |
127 | def smiles_to_mol(smiles: str):
128 | """ Generate a RDKit Molecule from SMILES.
129 |
130 | Parameters:
131 | ===========
132 | smiles: the input string
133 |
134 | Returns:
135 | ========
136 | The RDKit Molecule. If the Smiles parsing failed, NAN is returned instead.
137 | """
138 | try:
139 | mol = Chem.MolFromSmiles(smiles)
140 | if mol is not None:
141 | return mol
142 | return np.nan
143 | except:
144 | return np.nan
145 |
146 |
147 | def close_files(open_files:tuple):
148 | """Closes open files"""
149 | for file in open_files:
150 | file.close()
151 |
152 |
153 | def process_input(smiles:str, filename:str, output_name:str, total_score:bool, verbose:bool, confirmation:False):
154 | """Processes the command line input to print out the resulting score or create a file with added results"""
155 |
156 | if smiles: # process a directly provided SMILES string
157 | result = calculate_score_from_smiles(smiles, per_atom=(not total_score), verbose=verbose)
158 | if not verbose:
159 | score_type = "SPS" if total_score else "nSPS"
160 | print(f"\nNormalisation Applied: {not total_score}\nSMILES: {smiles}\nCalculated {score_type}: {result}")
161 | if result is np.nan:
162 | print("\nPlease double-check your input SMILES string...")
163 |
164 | elif filename: # process provided file
165 | provided_filename_base = filename.split(".")[0]
166 | output_filename = output_name if output_name else provided_filename_base + "_SPS.csv"
167 | outfile = open(output_filename, "w")
168 |
169 | # read the input .csv or .tsv file
170 | if filename.endswith("csv"):
171 | infile = open(filename, "r")
172 | reader = csv.DictReader(infile, dialect="excel")
173 | elif filename.endswith("tsv"):
174 | infile = open(filename, "r")
175 | reader = csv.DictReader(infile, dialect="excel-tab")
176 | else:
177 | raise ValueError(f"Unknown input file format: {filename}")
178 |
179 | print("\nProcessing, please wait...")
180 | for idx, row in enumerate(reader):
181 | if idx == 0:
182 | header = [column_name for column_name in row] # read existing headers
183 | # add SPS or nSPS column to the file
184 | if total_score:
185 | header.append("SPS")
186 | header.append("nSPS")
187 | outfile.write(",".join(header) + "\n")
188 |
189 | try:
190 | if total_score:
191 | row["SPS"] = calculate_score_from_smiles(row["Smiles"], per_atom=False, verbose=verbose)
192 | row["nSPS"] = calculate_score_from_smiles(row["Smiles"], per_atom=True, verbose=verbose)
193 | except KeyError:
194 | close_files((outfile, infile))
195 | raise KeyError("Please make sure that your file contains column called 'Smiles' with SMILES strings")
196 |
197 | line = [str(row[x]) for x in row] # reconstruct the row
198 | outfile.write(",".join(line) + "\n")
199 | if confirmation:
200 | print("Finished calculations for:", row["Smiles"])
201 |
202 | close_files((outfile, infile))
203 | print(f"Finished. {output_filename} was saved.")
204 | else:
205 | raise ValueError(f"No input was provided")
206 |
207 |
208 | def calculate_score_from_smiles(smiles: str, per_atom=False, verbose=False) -> float:
209 | """ Calculates the spacial score as a total SPS or size-normalised, per-atom nSPS for a molecule.
210 |
211 | Parameters:
212 | ===========
213 | smiles: valid SMILES string
214 | per_atom: flag to denote if the normalised per-atom result (nSPS) should be returned
215 | verbose: flag to denote if the detailed scores for each atom should be printed
216 |
217 | Returns:
218 | ========
219 | Total or per-atom numeric spacial score for the provided molecule.
220 | """
221 | mol = smiles_to_mol(smiles)
222 | if mol is np.nan:
223 | return np.nan
224 | sps = SpacialScore(smiles, mol, verbose)
225 | if per_atom:
226 | return sps.per_atom_score
227 | return sps.score
228 |
229 |
230 | if __name__ == "__main__":
231 |
232 | parser = argparse.ArgumentParser(description=
233 | 'Script for calculating Spacial Score (SPS) or normalised SPS (nSPS) for small molecules.\
234 | \nThe script can calculate the scores for a direct SMILES input or for a .csv or .tsv file containing a list of SMILES.\
235 | \nnSPS is calculated by deafult.',
236 | usage=None, formatter_class=argparse.RawDescriptionHelpFormatter)
237 | parser.add_argument('-s', action="store",
238 | metavar="SMILES string",
239 | help='Your input SMILES string for which to calculate the score', default=None)
240 | parser.add_argument('-i', action="store",
241 | help='Your .csv or .tsv file containing column called "Smiles" which contains SMILES strings. Resutls will be saved in a new .csv file',
242 | metavar='filename.ext',
243 | default=None)
244 | parser.add_argument('-o', action="store",
245 | help='You can specify name of the output .csv file. Not required.',
246 | metavar='filename.csv',
247 | default=None)
248 | parser.add_argument('-t', action="store_true",
249 | help='Option to calculate total SPS (no normalisation).',
250 | default=False)
251 | parser.add_argument('-v', action="store_true",
252 | help='Option to print verbose results, with information for each atom index.',
253 | default=False)
254 | parser.add_argument('-p', action="store_true",
255 | help='Option to print confirmation after processing of each SMILES string in a file.',
256 | default=False)
257 |
258 | if len(sys.argv) < 2:
259 | parser.print_help()
260 | sys.exit(1)
261 |
262 | ARGS = parser.parse_args()
263 | process_input(smiles=ARGS.s, filename=ARGS.i, output_name=ARGS.o, total_score=ARGS.t, verbose=ARGS.v, confirmation=ARGS.p)
264 |
--------------------------------------------------------------------------------