├── .github
└── workflows
│ └── deploy-docs.yml
├── .gitignore
├── COPYING.md
├── README.md
├── bun.lockb
├── dist
├── py_replay_bg-0.1.0-py3-none-any.whl
├── py_replay_bg-0.1.0.tar.gz
├── py_replay_bg-0.1.1-py3-none-any.whl
├── py_replay_bg-0.1.1.tar.gz
├── py_replay_bg-0.1.2-py3-none-any.whl
├── py_replay_bg-0.1.2.tar.gz
├── py_replay_bg-0.1.3-py3-none-any.whl
├── py_replay_bg-0.1.3.tar.gz
├── py_replay_bg-1.0.0-py3-none-any.whl
├── py_replay_bg-1.0.0.tar.gz
├── py_replay_bg-1.0.1-py3-none-any.whl
├── py_replay_bg-1.0.1.tar.gz
├── py_replay_bg-1.0.2-py3-none-any.whl
├── py_replay_bg-1.0.2.tar.gz
├── py_replay_bg-1.0.3-py3-none-any.whl
├── py_replay_bg-1.0.3.tar.gz
├── py_replay_bg-1.0.4-py3-none-any.whl
├── py_replay_bg-1.0.4.tar.gz
├── py_replay_bg-1.0.5-py3-none-any.whl
├── py_replay_bg-1.0.5.tar.gz
├── py_replay_bg-1.1.0-py3-none-any.whl
└── py_replay_bg-1.1.0.tar.gz
├── docs
├── .vuepress
│ ├── config.js
│ └── styles
│ │ ├── index.scss
│ │ └── palette.scss
├── README.md
└── documentation
│ ├── README.md
│ ├── analyzing_replay_results.md
│ ├── choosing_blueprint.md
│ ├── data_requirements.md
│ ├── get_started.md
│ ├── replaybg_object.md
│ ├── replaying.md
│ ├── results_folder.md
│ ├── twinning_procedure.md
│ └── visualizing_replay_results.md
├── package.json
├── py_replay_bg.egg-info
├── PKG-INFO
├── SOURCES.txt
├── dependency_links.txt
├── requires.txt
└── top_level.txt
├── py_replay_bg
├── __init__.py
├── analyzer
│ └── __init__.py
├── data
│ └── __init__.py
├── dss
│ ├── __init__.py
│ └── default_dss_handlers.py
├── environment
│ └── __init__.py
├── example
│ ├── code
│ │ ├── analysis_example.py
│ │ ├── analysis_intervals_example.py
│ │ ├── get_started.py
│ │ ├── replay_intervals_map.py
│ │ ├── replay_intervals_mcmc.py
│ │ ├── replay_map.py
│ │ ├── replay_map_dss.py
│ │ ├── replay_mcmc.py
│ │ ├── replay_mcmc_dss.py
│ │ ├── twin_intervals_map.py
│ │ ├── twin_intervals_mcmc.py
│ │ ├── twin_map.py
│ │ ├── twin_map_extended.py
│ │ ├── twin_mcmc.py
│ │ ├── twin_mcmc_extended.py
│ │ └── utils.py
│ └── data
│ │ ├── data_day_1.csv
│ │ ├── data_day_1_extended.csv
│ │ ├── data_day_2.csv
│ │ └── patient_info.csv
├── input_validation
│ ├── __init__.py
│ ├── input_validator_init.py
│ ├── input_validator_replay.py
│ └── input_validator_twin.py
├── model
│ ├── __init__.py
│ ├── logpriors_t1d.py
│ ├── model_parameters_t1d.py
│ ├── model_step_equations_t1d.py
│ ├── t1d_model_multi_meal.py
│ └── t1d_model_single_meal.py
├── py_replay_bg.py
├── replay
│ └── __init__.py
├── sensors
│ └── __init__.py
├── tests
│ ├── __init__.py
│ ├── test_analysis_example.py
│ ├── test_analysis_intervals_example.py
│ ├── test_replay_intervals_map.py
│ ├── test_replay_intervals_mcmc.py
│ ├── test_replay_map.py
│ ├── test_replay_map_dss.py
│ ├── test_replay_map_extended.py
│ ├── test_replay_mcmc.py
│ ├── test_replay_mcmc_dss.py
│ ├── test_replay_mcmc_extended.py
│ ├── test_twin_intervals_map.py
│ ├── test_twin_intervals_mcmc.py
│ ├── test_twin_map.py
│ ├── test_twin_map_extended.py
│ ├── test_twin_mcmc.py
│ └── test_twin_mcmc_extended.py
├── twinning
│ ├── __init__.py
│ ├── map.py
│ └── mcmc.py
├── utils
│ ├── __init__.py
│ └── stats.py
└── visualizer
│ └── __init__.py
├── pyproject.toml
├── requirements.txt
└── tsconfig.json
/.github/workflows/deploy-docs.yml:
--------------------------------------------------------------------------------
1 |
2 | name: Deploy Docs
3 |
4 | on:
5 | push:
6 | branches:
7 | - master
8 |
9 | permissions:
10 | contents: write
11 |
12 | jobs:
13 | deploy-gh-pages:
14 | runs-on: ubuntu-latest
15 | steps:
16 | - name: Checkout
17 | uses: actions/checkout@v4
18 | with:
19 | fetch-depth: 0
20 | # if your docs needs submodules, uncomment the following line
21 | # submodules: true
22 |
23 |
24 |
25 | - name: Setup bun
26 | uses: oven-sh/setup-bun@v2
27 |
28 | - name: Install Deps
29 | run: |
30 | bun install --frozen-lockfile
31 |
32 | - name: Build Docs
33 | env:
34 | NODE_OPTIONS: --max_old_space_size=8192
35 | run: |-
36 | bun run docs:build
37 | > docs/.vuepress/dist/.nojekyll
38 |
39 | - name: Deploy Docs
40 | uses: JamesIves/github-pages-deploy-action@v4
41 | with:
42 | # Deploy Docs
43 | branch: gh-pages
44 | folder: docs/.vuepress/dist
45 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 |
5 | # Virtual environment
6 | /venv
7 |
8 | #DS_stores
9 | .DS_Store
10 |
11 | # VS Code things
12 | .vscode/
13 |
14 | #Results folder
15 | /results
16 |
17 | node_modules/
18 | docs/.vuepress/.cache/
19 | docs/.vuepress/.temp/
20 | docs/.vuepress/dist/
21 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # ReplayBG
2 |
3 |  4 |
5 | [](https://github.com/gcappon/py_replay_bg/COPYING)
6 | [](https://github.com/gcappon/py_replay_bg/commits/master)
7 |
8 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
9 |
10 | # Reference
11 |
12 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
13 |
14 | # Get started
15 |
16 | ## Installation
17 |
18 | **ReplayBG** can be installed via pypi by simply
19 |
20 | ```python
21 | pip install py-replay-bg
22 | ```
23 |
24 | ### Requirements
25 |
26 | * Python >= 3.11
27 | * List of Python packages in `requirements.txt`
28 |
29 | ## Preparation: imports, setup, and data loading
30 |
31 | First of all import the core modules:
32 | ```python
33 | import os
34 | import numpy as np
35 | import pandas as pd
36 |
37 | from multiprocessing import freeze_support
38 | ```
39 |
40 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
41 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
42 | parallelized way.
43 |
44 | Then, we will import the necessary ReplayBG modules:
45 | ```python
46 | from py_replay_bg.py_replay_bg import ReplayBG
47 | from py_replay_bg.visualizer import Visualizer
48 | from py_replay_bg.analyzer import Analyzer
49 | ```
50 |
51 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
52 | while `Analyzer` and `Visualizer` are utility objects that will be used to
53 | respectively analyze and visualize the results that we will produce with ReplayBG
54 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
55 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
56 |
57 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
58 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
59 | ```python
60 | if __name__ == '__main__':
61 | freeze_support()
62 | ```
63 |
64 | Then, we will set the verbosity of ReplayBG:
65 | ```python
66 | verbose = True
67 | plot_mode = False
68 | ```
69 |
70 | Then, we need to decide what blueprint to use for twinning the data at hand.
71 | ```python
72 | blueprint = 'multi-meal'
73 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
74 | parallelize = True
75 | ```
76 |
77 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
78 |
79 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
80 | which contains a day of data of a patient with T1D:
81 |
82 | ```python
83 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
84 | data.t = pd.to_datetime(data['t'])
85 | ```
86 |
87 | ::: warning
88 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
89 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
90 | :::
91 |
92 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
93 |
94 | ```python
95 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
96 | p = np.where(patient_info['patient'] == 1)[0][0]
97 | # Set bw and u2ss
98 | bw = float(patient_info.bw.values[p])
99 | u2ss = float(patient_info.u2ss.values[p])
100 | ```
101 |
102 | Finally, instantiate a `ReplayBG` object:
103 |
104 | ```python
105 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
106 | yts=5, exercise=False,
107 | seed=1,
108 | verbose=verbose, plot_mode=plot_mode)
109 |
110 | ```
111 |
112 | ## Step 1: Creation of the digital twin
113 |
114 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
115 |
116 | ```python
117 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
118 | twinning_method='mcmc',
119 | parallelize=parallelize,
120 | n_steps=5000,
121 | u2ss=u2ss)
122 | ```
123 |
124 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
125 |
126 |
127 | ## Step 2: Run replay simulations
128 |
129 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
130 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
131 |
132 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
133 |
134 | ```python
135 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
136 | twinning_method='mcmc',
137 | save_workspace=True,
138 | save_suffix='_step_2a')
139 | ```
140 |
141 | It is possible to visualize the results of the simulation using:
142 |
143 | ```python
144 | Visualizer.plot_replay_results(replay_results, data=data)
145 | ```
146 |
147 | and analyzing the results using:
148 |
149 | ```python
150 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
151 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
152 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
153 | ```
154 |
155 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
156 | To do that run:
157 |
158 | ```python
159 | data.bolus = data.bolus * .7
160 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
161 | twinning_method='mcmc',
162 | save_workspace=True,
163 | save_suffix='_step_2b')
164 |
165 | # Visualize results
166 | Visualizer.plot_replay_results(replay_results)
167 | # Analyze results
168 | analysis = Analyzer.analyze_replay_results(replay_results)
169 |
170 | # Print, for example, the average glucose
171 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
172 | ```
173 |
174 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
175 |
176 | # Documentation
177 |
178 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
179 |
--------------------------------------------------------------------------------
/bun.lockb:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/bun.lockb
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.0.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.1-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.1-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.1.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.1.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.2-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.2-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.2.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.2.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.3-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.3-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.3.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.3.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.0.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.1-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.1-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.1.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.1.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.2-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.2-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.2.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.2.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.3-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.3-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.3.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.3.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.4-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.4-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.4.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.4.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.5-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.5-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.5.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.5.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.1.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.1.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.1.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.1.0.tar.gz
--------------------------------------------------------------------------------
/docs/.vuepress/config.js:
--------------------------------------------------------------------------------
1 | import { defineUserConfig } from 'vuepress/cli'
2 | import { viteBundler } from '@vuepress/bundler-vite'
3 | import { hopeTheme } from "vuepress-theme-hope";
4 |
5 | export default defineUserConfig({
6 | lang: 'en-US',
7 |
8 | title: 'ReplayBG',
9 | base: '/py_replay_bg/',
10 | description: 'A digital twin based framework for the development and assessment of new algorithms for type 1 ' +
11 | 'diabetes management',
12 |
13 | theme: hopeTheme({
14 | logo: 'https://i.postimg.cc/gJn8Sy0X/replay-bg-logo.png',
15 | navbar: ['/', '/documentation/get_started', '/documentation/'],
16 | repo: 'https://github.com/gcappon/py_replay_bg',
17 | docsDir: 'docs/',
18 | docsBranch: 'master',
19 | markdown: {
20 | highlighter: {
21 | type: "shiki",
22 | langs: ['python', 'json', 'md', 'bash', 'diff'],
23 | themes: {
24 | dark: 'catppuccin-mocha',
25 | light: 'catppuccin-latte'
26 | }
27 | },
28 | math: {
29 | type: "mathjax",
30 | },
31 | },
32 | sidebar: [
33 | {
34 | text: 'Get Started',
35 | link: 'documentation/get_started.md'
36 | },
37 | {
38 | text: 'Data Requirements',
39 | link: 'documentation/data_requirements.md'
40 | },
41 | {
42 | text: 'The ReplayBG Object',
43 | link: 'documentation/replaybg_object.md'
44 | },
45 | {
46 | text: 'Choosing Blueprint',
47 | link: 'documentation/choosing_blueprint.md'
48 | },
49 | {
50 | text: 'Twinning Procedure',
51 | link: 'documentation/twinning_procedure.md'
52 | },
53 | {
54 | text: 'Replaying',
55 | link: 'documentation/replaying.md'
56 | },
57 | {
58 | text: 'The _results/_ Folder',
59 | link: 'documentation/results_folder.md'
60 | },
61 | {
62 | text: 'Visualizing Replay Results',
63 | link: 'documentation/visualizing_replay_results.md'
64 | },
65 | {
66 | text: 'Analyzing Replay Results',
67 | link: 'documentation/analyzing_replay_results.md'
68 | }
69 | ],
70 | }),
71 |
72 | bundler: viteBundler(),
73 |
74 | plugins: [],
75 | })
76 |
--------------------------------------------------------------------------------
/docs/.vuepress/styles/index.scss:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/docs/.vuepress/styles/palette.scss:
--------------------------------------------------------------------------------
1 | $light-accent-color: #990000;
2 | $dark-accent-color: #fa0000;
3 | $vp-c-accent: (
4 | light: $light-accent-color,
5 | dark: $dark-accent-color,
6 | );
7 | $vp-c-accent-bg: (
8 | light: $light-accent-color,
9 | dark: $dark-accent-color,
10 | );
11 | $vp-c-accent-hover: (
12 | light: $light-accent-color,
13 | dark: $dark-accent-color,
14 | );
15 |
16 |
--------------------------------------------------------------------------------
/docs/README.md:
--------------------------------------------------------------------------------
1 | ---
2 | home: true
3 | title: Home
4 | heroImage: https://i.postimg.cc/gJn8Sy0X/replay-bg-logo.png
5 | actions:
6 | - text: Get Started
7 | link: /documentation/get_started
8 | type: primary
9 |
10 | features:
11 | - title: Digital Twin
12 | details: Leverage the power of digital twin to assess new therapies for type 1 diabetes
13 | - title: State of the Art
14 | details: ReplayBG is supported by solid scientific research from UNIPD
15 | - title: Easy to Use
16 | details: ReplayBG is easy to use and ready to be integrated in your research pipeline
17 |
18 | footer: Made by Giacomo Cappon with ❤️
19 | ---
20 |
21 | ## Supporting research
22 |
23 | ### Research on ReplayBG development
24 |
25 | #### Journal Papers
26 | - F. Prendin, A. Facchinetti, and G. Cappon “Data Augmentation via Digital Twins Enables the Development of Personalized
27 | Deep Learning Glucose Prediction Algorithms for Type 1 Diabetes in Poor Data Context” IEEE Journal of Biomedical
28 | Health Informatics, (under revision, submitted October 2024)
29 | - G. Cappon, and A. Facchinetti, “Digital twins in type 1 diabetes: A systematic review” Journal of Diabetes
30 | Science and Technology, online ahead of print, 2024, DOI: 10.1177/1932296824126211.
31 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “ReplayBG: a digital twin-based methodology
32 | to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative
33 | therapies”, IEEE Transactions on Biomedical Engineering, vol. 70, no. 11, pp. 3105-3115, Nov 2023,
34 | DOI: 10.1109/TBME.2023.3286856
35 |
36 |
37 | #### Conference papers and abstracts
38 | - G. Cappon, S. Del Favero, and A. Facchinetti “Extending the applicability of ReplayBG tool for digital
39 | twinning in type 1 diabetes from single-meal to multi-day scenarios” in the 24th Annual Diabetes Technology
40 | Meeting – DTM, Burlingame, CA, USA, October 15-17, 2024.
41 | - J. Leth, H. Ghazaleh, H. Peuscher, and G. Cappon “The potential of open-source software in diabetes research” in the
42 | 17th International Conference on Advanced Technology & Treatment for Diabetes – ATTD, Firenze, Italy, March 6-9,
43 | 2024 (invited oral presentation, speaker J. Leth).
44 | - H. Peuscher, G. Cappon, A. Cinar, J. Deichmann, H. Ghazaleh, H. Kaltenbach, L. Sandini, M. Siket, J. Xie, and
45 | X. Zhou “A survey on existing open-source projects in diabetes simulation” in the 17th International Conference on
46 | Advanced Technology & Treatment for Diabetes – ATTD, Firenze, Italy, March 6-9, 2024
47 | (invited oral presentation, speaker J. Leth).
48 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “Expanding ReplayBG simulation methodology
49 | domain of validity to single day multiple meal scenarios” in the 15th International Conference on Advanced
50 | Technology & Treatment for Diabetes – ATTD, Barcelona, Spain, April 27-30, 2022.
51 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “ReplayBG provides reliable indications when
52 | used to assess meal bolus alterations in type 1 diabetes” in the 14th International Conference on Advanced
53 | Technology & Treatment for Diabetes – ATTD, Paris, France, June 2-6, 2021.
54 | G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “ReplayBG: A novel in-silico framework to retrospectively
55 | assess new therapy guidelines for type 1 diabetes management” in the 20th Annual Diabetes Technology Meeting –
56 | DTM, Bethesda, Maryland, USA, November 12-14, 2020.
57 | - G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “A Bayesian framework to personalize glucose prediction
58 | in type 1 diabetes using a physiological model” in the 19th Annual Diabetes Technology Meeting –
59 | DTM, Bethesda, Maryland, USA, November 14-16, 2019.
60 | - G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “A Bayesian framework to identify type 1 diabetes
61 | physiological models using easily accessible patient data” in the 41th Annual International Conference of the IEEE
62 | Engineering in Medicine and Biology Society – EMBC, Berlin, Germany, July 23-27, 2019
63 | (accepted for oral presentation, speaker G.Cappon).
64 |
65 | ### Research using ReplayBG as component or validation tool
66 |
67 | #### Journal Papers
68 | - E. Pellizzari, G. Cappon, G. Nicolis, G. Sparacino, and A. Facchinetti “Developing effective machine learning models
69 | for insulin bolus calculation in type 1 diabetes exploiting real-world data and digital twins” IEEE Transactions on
70 | Biomedical Engineering, (under revision, submitted October 2024)
71 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti, “drCORRECT: An algorithm for the
72 | preventive administration of postprandial corrective insulin boluses in type 1 diabetes management” Journal of
73 | Diabetes Science and Technology, online ahead of print, 2023, DOI: 10.1177/19322968231221768
74 | - F. Prendin, J. Pavan, G. Cappon, S. Del Favero, G. Sparacino, and A. Facchinetti, “The importance of interpreting
75 | machine learning models for blood glucose prediction in diabetes: an analysis using SHAP” Scientific
76 | Reports, vol. 13, no. 1, pp. 16865, Oct 2023, DOI: 10.1038/s41598-023-44155-x.
77 | - G. Noaro, G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “Machine-learning based model
78 | to improve insulin bolus calculation in type 1 diabetes therapy” IEEE Transactions on Biomedical Engineering,
79 | vol. 68, no. 1, pp. 247-255, Jan 2021. DOI: 10.1109/TBME.2020.3004031
80 |
81 | #### Conference papers and abstracts
82 | - E. Pellizzari, G. Cappon, G. Nicolis, G. Sparacino, and A. Facchinetti “Exploiting real-world data and digital
83 | twins to develop effective formulas for dosing insulin boluses in type 1 diabetes therapy” in the 24th Annual
84 | Diabetes Technology Meeting – DTM, Burlingame, CA, USA, October 15-17, 2024.
85 | - F. Prendin, A. Facchinetti, and G. Cappon “Digital twin for data augmentation enables the development of accurate
86 | personalized deep glucose forecasting algorithms” in the 24th Annual Diabetes Technology Meeting –
87 | DTM, Burlingame, CA, USA, October 15-17, 2024.
88 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti “DR-CIB: An algorithm for the preventive
89 | administration of corrective insulin boluses in T1D based on dynamic risk concept and patient-specific timing” in
90 | the 23rd Annual Diabetes Technology Meeting – DTM, Online, November 1-4, 2023 (Gold Student Award, accepted for
91 | oral presentation, speaker E. Pellizzari).
92 | - G. Cappon, E. Pellizzari, L. Cossu, G. Sparacino, A. Deodati, R. Schiaffini, S. Cianfarani, and A. Facchinetti
93 | “System architecture of TWIN: A new digital twin-based clinical decision support system for type 1 diabetes
94 | management in children” in the 19th International Conference on Body Sensor Networks –
95 | BSN, Boston, MA, USA, October 9-11, 2023.
96 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti “A deep-learning based algorithm for the
97 | management of hyperglycemia in type 1 diabetes therapy” in the 19th International Conference on Body Sensor Networks
98 | – BSN, Boston, MA, USA, October 9-11, 2023.
99 | - G. Noaro, G. Cappon, G. Sparacino, and A. Facchinetti “An ensemble learning algorithm based on dynamic voting for
100 | targeting the optimal insulin dosage in type 1 diabetes management“ in the 43rd Annual International Conference of the
101 | IEEE Engineering in Medicine and Biology Society – EMBC, Guadalajara, Mexico, October 31-4, 2021 (accepted for
102 | oral presentation, speaker G. Noaro).
103 | - G. Cappon, E. Pighin, F. Prendin, G. Sparacino, and A. Facchinetti “A correction insulin bolus delivery strategy
104 | for decision support systems in type 1 diabetes“ in the 43rd Annual International Conference of the IEEE Engineering
105 | in Medicine and Biology Society – EMBC, Guadalajara, Mexico, October 31-4, 2021 (accepted for oral presentation,
106 | speaker G. Cappon).
107 | - G. Noaro, G. Cappon, M. Vettoretti, S. Del Favero, G. Sparacino, and A. Facchinetti “A new model for mealtime insulin
108 | dosing in type 1 diabetes: Retrospective validation on CTR3 dataset” in the 20th Annual Diabetes Technology Meeting –
109 | DTM, Bethesda, Maryland, USA, November 12-14, 2020 (accepted for oral presentation, speaker G. Noaro).
110 | - G. Noaro, G. Cappon, G. Sparacino, S. Del Favero, A. Facchinetti “Nonlinear machine learning models for insulin
111 | bolus estimation in type 1 diabetes therapy” in the 42nd Annual International Conference of the IEEE Engineering
112 | in Medicine and Biology Society – EMBC, Montreal, Canada, July 20-24, 2020 (accepted for oral presentation,
113 | speaker G. Noaro)
--------------------------------------------------------------------------------
/docs/documentation/README.md:
--------------------------------------------------------------------------------
1 | ---
2 | lang: en-US
3 | title: Docs
4 | description: Docs index
5 | ---
6 |
7 | # Docs
8 |
9 | ### [Get Started](./get_started.md)
10 |
11 | ---
12 |
13 | ### [Data Requirements](./data_requirements.md)
14 |
15 | ### [The ReplayBG Object](./replaybg_object.md)
16 |
17 | ### [Choosing Blueprint](./choosing_blueprint.md)
18 |
19 | ### [Twinning Procedure](./twinning_procedure.md)
20 |
21 | ### [Replaying](./replaying.md)
22 |
23 | ### [The _results/_ Folder](./results_folder.md)
24 |
25 | ---
26 |
27 | ### [Visualizing replay results](./visualizing_replay_results.md)
28 |
29 | ### [Analyzing replay results](./analyzing_replay_results.md)
30 |
31 | ---
--------------------------------------------------------------------------------
/docs/documentation/data_requirements.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: heading
3 | ---
4 | # Data Requirements
5 |
6 | Data provided to ReplayBG must comply to strict format requirements and should be selected following some best
7 | practices.
8 |
9 | ## Format requirements
10 |
11 | Format requirements depend on the selected `blueprint`, i.e., single-meal or multi-meal, as such,
12 | in the following, details about these aspects are presented for each possible blueprint separately.
13 |
14 | ### Single Meal blueprint
15 |
16 | By "single meal" one can refer to a specific period
17 | of time when a specific subject had only 1 main meal and a corresponding insulin
18 | basal-bolus administration. Usually, this period of time spans maximum 6/8 hours,
19 | starts near such main meal, and ends just before the subsequent main meal and/or after
20 | a reasonable amount of time.
21 |
22 | `data` must be saved in a `.csv` file and contain the following columns:
23 | * `t`: the timestamps when data of the corresponding row were recorded (format
24 | `DD-MMM-YYYY HH:mm:SS` for example `20-Dec-2013 10:35:00`). The sampling grid defined by the `t` column must be
25 | homogeneous, e.g., have a datapoint every 5 minutes.
26 | * `glucose`: the glucose concentration (mg/dl) at `t`. Can contain NaN values.
27 | * `cho`: the meal intake (g/min) at `t`. Can't contain NaN values. If no meals were recorded at `t` just put `0` there.
28 | * `bolus`: the insulin bolus (U/min) administered at `t`. Can't contain NaN values. If no insulin boluses were
29 | administered at `t` just put `0` there.
30 | * `basal`: the basal insulin (U/min) administered at `t`. Can't contain NaN values. If no basal insulin was
31 | administered at `t` just put `0` there.
32 | * `bolus_label`: the type of `bolus` at time `t`. This column Each `bolus` entry > 0 must have a label defined. Can be
33 | * `B` if it is the bolus of a breakfast.
34 | * `L` if it is the bolus of a lunch.
35 | * `D` if it is the bolus of a dinner.
36 | * `C` if it is a corrective bolus.
37 | * `S` if it is the bolus of a snack.
38 |
39 | If other columns are present in your data file, they will be ignored.
40 |
41 | ::: tip NOTE
42 | The total length of the simulation, `simulation_length`, is defined in minutes and determined by ReplayBG automatically
43 | using the `t` column of `data` and the `yts` input parameter provided to the `ReplayBG` object builder.
44 |
45 | For example, if `yts` is `5` minutes and `t` starts from `20-Dec-2013 10:36:00` and ends to `20-Dec-2013 10:46:00`,
46 | then `simulation_length` will be `10`.
47 | :::
48 |
49 | ::: tip
50 | If `bolus_label` is not important for you (e.g., you do not plan to use it during replay) or if you do not need that,
51 | just add an empty `bolus_label` column.
52 | :::
53 |
54 | ::: warning
55 | If more than 1 meal are present in the provided file, ReplayBG will consider the first meal as "main" meal. The others
56 | will be considered as "other" meals. The resulting `kabs` and $\beta$ parameters will be unique so their value will
57 | depend by ALL the meals you have on your data. SO, it is really not advised to have more than one meal when using the
58 | single-meal blueprint.
59 | :::
60 |
61 | #### Requirements during replay
62 | When replaying (using the `replay` method), the following requirements are no more valid under the following circumstances:
63 | - `glucose`: during replay this is simply ignored.
64 | - `cho`: if `cho_source` is `generated` since the CHO event will be generated by the provided handler during the replay simulation.
65 | - `bolus` and `bolus_label`: if `bolus_source` is `dss` since the insulin bolus events will be generated by the provided handler during the replay simulation.
66 | - `basal`: if `basal_source` is `dss` since the basal insulin will be generated by the provided handler during the replay simulation.
67 |
68 | ### Multi Meal blueprint
69 |
70 | By "multi meal" one can refer to a specific period
71 | of time when a specific subject had more than 1 main meal and a
72 | corresponding insulin basal-bolus administration regimen. One can think to such
73 | period of time by thinking to a day, when multiple meals occur, or even multiple days.
74 |
75 | `data` must be saved in a `.csv` file and contain (at least) the following columns:
76 | * `t`: the timestamps when data of the corresponding row were recorded (format
77 | `DD-MMM-YYYY HH:mm:SS` for example `20-Dec-2013 10:35:00`). The sampling grid defined by the `t` column must be
78 | homogeneous.
79 | * `glucose`: the glucose concentration (mg/dl) at `t`. Can contain NaN values.
80 | * `cho`: the meal intake (g/min) at `t`. Can't contain NaN values. If no meals were recorded at `t` just put `0` there.
81 | * `bolus`: the insulin bolus (U/min) administered at `t`. Can't contain NaN values. If no insulin boluses were
82 | administered at `t` just put `0` there.
83 | * `basal`: the basal insulin (U/min) administered at `t`. Can't contain NaN values. If no basal insulin was
84 | administered at `t` just put `0` there.
85 | * `cho_label`: the type of `cho` at time `t`. Each `cho` entry > 0 must have a label defined. Can be
86 | * `B` if it is a breakfast.
87 | * `L` if it is a lunch.
88 | * `D` if it is a dinner.
89 | * `H` if it is a hypotreatment.
90 | * `S` if it is a snack.
91 | * `bolus_label`: the type of `bolus` at time `t`. Each `bolus` entry > 0 must have a label defined. Can be
92 | * `B` if it is the bolus of a breakfast.
93 | * `L` if it is the bolus of a lunch.
94 | * `D` if it is the bolus of a dinner.
95 | * `C` if it is a corrective bolus.
96 | * `S` if it is the bolus of a snack.
97 |
98 | If other columns are present in your data file, they will be ignored.
99 |
100 | ::: warning
101 | The `cho` and `bolus` columns must contain at least one event when twinning.
102 | :::
103 |
104 | ::: tip
105 | If `bolus_label` is not important for you (e.g., you do not plan to use it during replay) or if you do not need that,
106 | just add an empty `bolus_label` column.
107 | :::
108 |
109 | ::: tip
110 | A representative data file of a single meal blueprint can be found in `example/data/multi-meal_example.csv`
111 | :::
112 |
113 | #### Requirements during replay
114 | When replaying (using the `replay` method), the following requirements are no more valid under the following circumstances:
115 | - `glucose`: during replay this is simply ignored.
116 | - `cho` and `cho_label`: if `cho_source` is `generated` since the CHO events will be generated by the provided handler during the replay simulation.
117 | - `bolus` and `bolus_label`: if `bolus_source` is `dss` since the insulin bolus events will be generated by the provided handler during the replay simulation.
118 | - `basal`: if `basal_source` is `dss` since the basal insulin will be generated by the provided handler during the replay simulation.
119 |
120 | ## Best practices
121 | The potential ReplayBG user should be aware of several practical aspects and be careful when selecting the portion
122 | of data to work with. Here's the details.
123 |
124 | ### Starting point
125 | The twinning procedure of ReplayBG does not estimate the states of the blueprint mathematical model and, more
126 | importantly, the corresponding initial conditions. Identifying such initial conditions is crucial to correctly
127 | estimating the unknown model parameter vector $\boldsymbol{\theta}_{phy}$ and, by product, reliably replaying the
128 | glucose profile with new, altered, inputs. To circumvent this issue, ReplayBG assumes all model state initial
129 | conditions at steady state. This assumption is granted to be valid when the actions of exogenous insulin and
130 | carbohydrate intake are “exhausted”, i.e., when the starting point of the portion of data is reasonably distant
131 | from the last meal and insulin boluses, e.g., 4 hours.
132 |
133 | ::: tip
134 | When twinning and replaying intervals (using the `twin` and the `replay` methods, respectively) instead of single
135 | portions of data, things change. Indeed, the problem of the starting point applies only to the first portion where we
136 | must assume initial steady state conditions. The other subsequent portions will start from the immediate next datapoint
137 | with initial conditions defined by `x0` and `previous_data_name`.
138 |
139 | For more information on the `x0` and
140 | `previous_data_name` parameters when twinning and replaying please refer to the [Twinning Procedure](./twinning_procedure.md)
141 | and the [Replaying](./replaying.md) pages.
142 | :::
143 |
144 | ### Minimum data length
145 | As a rule of thumb we suggest to use portions of data that span at least 6 hours.
146 | As demonstrated in the literature, this ensures to obtain better parameter estimates and simulation results.
147 |
148 | ### Data gaps
149 | To make the twinning procedure more reliable, data portions having significant data gaps (i.e., more that 10% of missing glucose
150 | readings) or without a single reported meal intake or insulin bolus, should be discarded to avoid the creation of
151 | digital twins not representing the actual underneath physiology.
--------------------------------------------------------------------------------
/docs/documentation/get_started.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 | # Get started
5 |
6 | ## Installation
7 |
8 | **ReplayBG** can be installed via pypi by simply
9 |
10 | ```python
11 | pip install py-replay-bg
12 | ```
13 |
14 | ### Requirements
15 |
16 | * Python >= 3.11
17 | * List of Python packages in `requirements.txt`
18 |
19 | ## Preparation: imports, setup, and data loading
20 |
21 | First of all import the core modules:
22 | ```python
23 | import os
24 | import numpy as np
25 | import pandas as pd
26 |
27 | from multiprocessing import freeze_support
28 | ```
29 |
30 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
31 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
32 | parallelized way.
33 |
34 | Then, we will import the necessary ReplayBG modules:
35 | ```python twoslash
36 | from py_replay_bg.py_replay_bg import ReplayBG
37 | from py_replay_bg.visualizer import Visualizer
38 | from py_replay_bg.analyzer import Analyzer
39 | ```
40 |
41 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](./replaybg_object.md) page),
42 | while `Analyzer` and `Visualizer` are utility objects that will be used to
43 | respectively analyze and visualize the results that we will produce with ReplayBG
44 | (more information in the ([Visualizing Replay Results](./visualizing_replay_results.md) and
45 | [Analyzing Replay Results](./analyzing_replay_results.md) pages).
46 |
47 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
48 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
49 | ```python
50 | if __name__ == '__main__':
51 | freeze_support()
52 | ```
53 |
54 | Then, we will set the verbosity of ReplayBG:
55 | ```python
56 | verbose = True
57 | plot_mode = False
58 | ```
59 |
60 | Then, we need to decide what blueprint to use for twinning the data at hand.
61 | ```python
62 | blueprint = 'multi-meal'
63 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
64 | parallelize = True
65 | ```
66 |
67 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](./choosing_blueprint.md) page.
68 |
69 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
70 | which contains a day of data of a patient with T1D:
71 |
72 | ```python
73 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
74 | data.t = pd.to_datetime(data['t'])
75 | ```
76 |
77 | ::: warning
78 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
79 | information see the [Data Requirements](./data_requirements) page.
80 | :::
81 |
82 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
83 |
84 | ```python
85 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
86 | p = np.where(patient_info['patient'] == 1)[0][0]
87 | # Set bw and u2ss
88 | bw = float(patient_info.bw.values[p])
89 | u2ss = float(patient_info.u2ss.values[p])
90 | ```
91 |
92 | Finally, instantiate a `ReplayBG` object:
93 |
94 | ```python
95 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
96 | yts=5, exercise=False,
97 | seed=1,
98 | verbose=verbose, plot_mode=plot_mode)
99 |
100 | ```
101 |
102 | ## Step 1: Creation of the digital twin
103 |
104 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
105 |
106 | ```python
107 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
108 | twinning_method='mcmc',
109 | parallelize=parallelize,
110 | n_steps=5000,
111 | u2ss=u2ss)
112 | ```
113 |
114 | For more information on the twinning procedure see the [Twinning Procedure](./twinning_procedure.md) page.
115 |
116 |
117 | ## Step 2: Run replay simulations
118 |
119 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
120 | see the [Replaying](./replaying.md) page.
121 |
122 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
123 |
124 | ```python
125 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
126 | twinning_method='mcmc',
127 | save_workspace=True,
128 | u2ss=u2ss,
129 | save_suffix='_step_2a')
130 | ```
131 |
132 | It is possible to visualize the results of the simulation using:
133 |
134 | ```python
135 | Visualizer.plot_replay_results(replay_results, data=data)
136 | ```
137 |
138 | and analyzing the results using:
139 |
140 | ```python
141 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
142 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
143 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
144 | ```
145 |
146 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
147 | To do that run:
148 |
149 | ```python
150 | data.bolus = data.bolus * .7
151 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
152 | twinning_method='mcmc',
153 | save_workspace=True,
154 | save_suffix='_step_2b')
155 |
156 | # Visualize results
157 | Visualizer.plot_replay_results(replay_results)
158 | # Analyze results
159 | analysis = Analyzer.analyze_replay_results(replay_results)
160 |
161 | # Print, for example, the average glucose
162 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
163 | ```
164 |
165 | ## Full example
166 |
167 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
168 |
169 | ```python
170 | import os
171 | import numpy as np
172 | import pandas as pd
173 |
174 | from multiprocessing import freeze_support
175 |
176 | from py_replay_bg.py_replay_bg import ReplayBG
177 | from py_replay_bg.visualizer import Visualizer
178 | from py_replay_bg.analyzer import Analyzer
179 |
180 |
181 | if __name__ == '__main__':
182 | freeze_support()
183 |
184 | # Set verbosity
185 | verbose = True
186 | plot_mode = False
187 |
188 | # Set other parameters for twinning
189 | blueprint = 'multi-meal'
190 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
191 | parallelize = True
192 |
193 | # Load data
194 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
195 | data.t = pd.to_datetime(data['t'])
196 |
197 | # Load patient_info
198 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
199 | p = np.where(patient_info['patient'] == 1)[0][0]
200 | # Set bw and u2ss
201 | bw = float(patient_info.bw.values[p])
202 | u2ss = float(patient_info.u2ss.values[p])
203 |
204 | # Instantiate ReplayBG
205 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
206 | yts=5, exercise=False,
207 | seed=1,
208 | verbose=verbose, plot_mode=plot_mode)
209 |
210 | # Set save name
211 | save_name = 'data_day_1'
212 |
213 | # Step 1. Run twinning procedure
214 | rbg.twin(data=data, bw=bw, save_name=save_name,
215 | twinning_method='mcmc',
216 | parallelize=parallelize,
217 | n_steps=5000,
218 | u2ss=u2ss)
219 |
220 | # Step 2a. Replay the twin with the same input data
221 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
222 | twinning_method='mcmc',
223 | save_workspace=True,
224 | save_suffix='_step_2a')
225 |
226 | # Visualize results and compare with the original glucose data
227 | Visualizer.plot_replay_results(replay_results, data=data)
228 | # Analyze results
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | # Print, for example, the fit MARD and the average glucose
231 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
232 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
233 |
234 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
235 | data.bolus = data.bolus * .7
236 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
237 | twinning_method='mcmc',
238 | save_workspace=True,
239 | save_suffix='_step_2b')
240 |
241 | # Visualize results
242 | Visualizer.plot_replay_results(replay_results)
243 | # Analyze results
244 | analysis = Analyzer.analyze_replay_results(replay_results)
245 |
246 | # Print, for example, the average glucose
247 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
248 | ```
249 |
--------------------------------------------------------------------------------
/docs/documentation/replaybg_object.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 |
5 | # The ReplayBG Object
6 |
7 | The `ReplayBG` object is the core, key, object to instatiate when starting to work with the ReplayBG framework.
8 |
9 | Its constructor is formally defined as:
10 |
11 | ```python
12 | from py_replay_bg.py_replay_bg import ReplayBG
13 |
14 | ReplayBG(save_folder: str, blueprint: str = 'single_meal',
15 | yts: int = 5, exercise: bool = False,
16 | seed: int = 1,
17 | plot_mode: bool = True, verbose: bool = True)
18 | ```
19 |
20 | ## Input parameters
21 |
22 | Let's inspect and describe each input parameter.
23 |
24 | - `save_folder`: a string defining the folder that will contain the results of the twinning procedure and the replay
25 | simulations. This parameter is mandatory. More information on how to set it can be found in
26 | [The _results/_ Folder](./results_folder.md) page.
27 | - `blueprint`, optional, `{'single-meal', 'multi-meal'}`, default: `'single-meal'`: a string that specifies the blueprint to be used to create
28 | the digital twin. More information on how to set it can be found in [Choosing Blueprint](./choosing_blueprint.md) page.
29 | - `yts`, optional, default: `5` : an integer that specifies the data sample time (in minutes).
30 | - `exercise`, optional, default: `False`: a boolean that specifies whether to simulate exercise or not.
31 | - `seed`, optional, default: `1`: an integer that specifies the random seed. For reproducibility.
32 | - `plot_mode`, optional, default: `True`: a boolean that specifies whether to show the plot of the results or not. More
33 | information on how to visualize the results of ReplayBG can be found in
34 | [Visualizing Replay Results](./visualizing_replay_results.md) page.
35 | - `verbose`, optional, default: `True`: a boolean that specifies the verbosity of ReplayBG.
36 |
37 |
38 |
39 |
--------------------------------------------------------------------------------
/docs/documentation/results_folder.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 |
5 | # The _results/_ Folder
6 |
7 | All results of ReplayBG, i.e., both model parameters obtained via twinning procedure and replayed
8 | glucose traces of a given scenario, will be saved in a folder named `results/`.
9 |
10 | ## Choosing the path of _results/_
11 |
12 | When creating the ReplayBG object, user must decide the location of `results/`.
13 | To do that, specify a custom location, relative to current ``
14 | using the `save_folder` parameter of the `ReplayBG` object builder. For example,
15 | to put `results/` in `/custom/path/location/`, simply:
16 |
17 | ```python
18 | rbg = ReplayBG(save_folder=os.path.join('custom', 'path', 'location'), ...)
19 | ```
20 |
21 | This means that if the user wants to put `results/` just in ``
22 | he/she must use:
23 |
24 | ```python
25 | rbg = ReplayBG(save_folder=os.path.join(''), ...)
26 | ```
27 |
28 | ## What's inside
29 |
30 | The `results/` folder is organized as follows:
31 |
32 | ```
33 | results/
34 | |--- mcmc/
35 | |--- map/
36 | |--- workspaces/
37 | ```
38 |
39 | where the `mcmc/` and `map/` subfolders contains the model parameters obtained via
40 | MCMC-based and MAP-based twinning procedures, respectively; and the `workspaces/` folder
41 | contains the results of the replayed scenarios simulated when the `rbg.replay()` method
42 | is called.
43 |
44 | ::: tip REMEMBER
45 | Results of the replayed scenarios are saved in the `workspace/` subfolder only if the
46 | `save_workspace` parameter of `rbg.replay()` is set to `True`:
47 | ```python
48 | rbg.replay(save_workspace=True, ...)
49 | ```
50 | If not set, being its default value `False`, nothing will be saved.
51 | :::
52 |
53 |
54 |
55 |
56 |
57 |
--------------------------------------------------------------------------------
/docs/documentation/visualizing_replay_results.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: heading
3 | ---
4 |
5 | # Visualizing replay results
6 |
7 | ReplayBG provides the possibility, to visualize in a painless way the results of `rbg.replay()`.
8 |
9 | This is done using the `Visualizer` class importable from `py_replay_bg.visualizer`.
10 |
11 | In the following, we show how to do that in the case of single portions of data and portions of data spanning
12 | more than one day (i.e., intervals).
13 |
14 | ## Visualizing replay results from single portions of data
15 |
16 | To visualize replay results from single portions of data use the `Visualizer.plot_replay_results()` static
17 | method, which is formerly defined as:
18 | ```python
19 | @staticmethod
20 | def plot_replay_results(
21 | replay_results: Dict,
22 | data: pd.DataFrame = None,
23 | title: str = '',
24 | ) -> None
25 | ```
26 |
27 | ### Input parameters
28 | - `replay_results`: the dictionary returned by or saved with the `rbg.replay()` method
29 | - `data`, optional, default: `None`: The `data` parameter passed to `rbg.replay()` . If present, the method will also
30 | compare the glucose fit vs the data.
31 | - `title`, optional, default: `None`: A string with an optional title to be added to the figure.
32 |
33 | ### Example
34 |
35 | ```python
36 | # Load previously saved results, e.g., ...
37 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'results.pkl'), 'rb') as file:
38 | replay_results = pickle.load(file)
39 |
40 | # Analyze them
41 | Visualizer.plot_replay_results(replay_results=replay_results)
42 | ```
43 |
44 | Will produce:
45 |
46 | 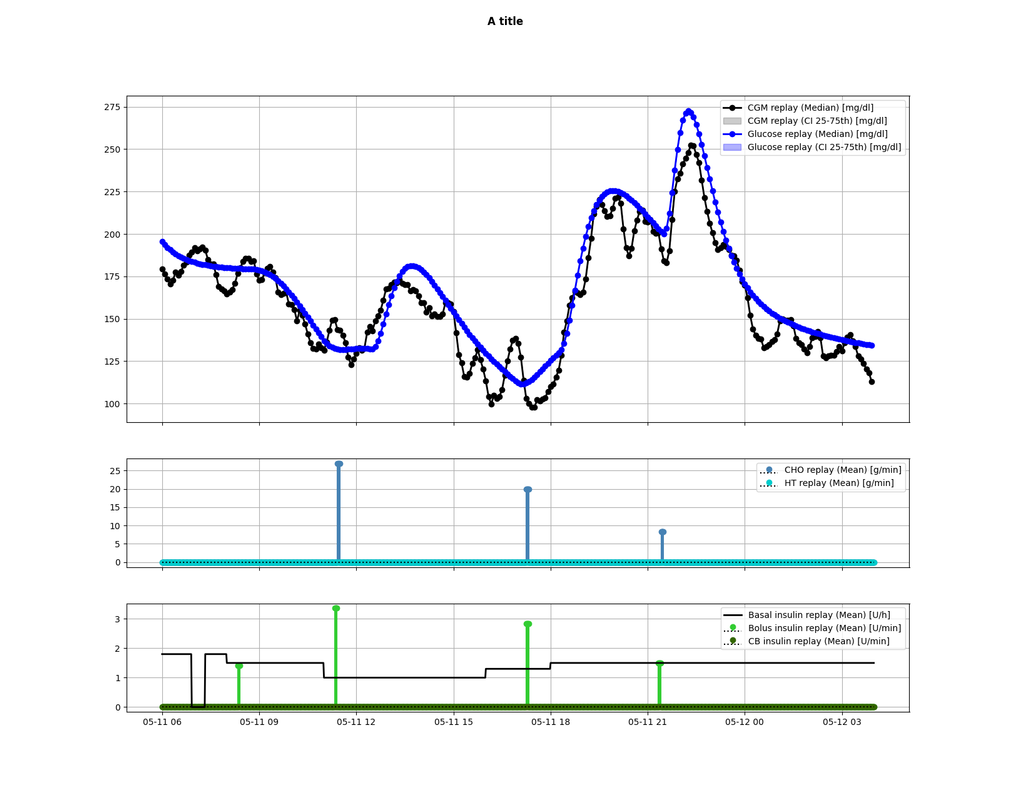
47 |
48 | The full code can be found in `/example/code/analysis_example.py`.
49 |
50 | ## Visualizing replay results from portions of data spanning more than one day (i.e., intervals)
51 |
52 | To visualize replay results from single portions of data use the `Visualizer.plot_replay_results_interval()` static
53 | method, which is formerly defined as:
54 | ```python
55 | @staticmethod
56 | def plot_replay_results_interval(
57 | replay_results_interval: list,
58 | data_interval: list = None,
59 | ) -> Dict
60 | ```
61 |
62 | ### Input parameters
63 | - `replay_results_interval`: a list of dictionaries returned by or saved with the `rbg.replay()` method
64 | - `data_interval`, optional, default: `None`: The list of `data` passed to `rbg.replay()` . If present, the method will also
65 | compare the glucose fit vs the data.
66 | - `title`, optional, default: `None`: A string with an optional title to be added to the figure.
67 |
68 | ### Example
69 |
70 | ```python
71 | # Initialize results list
72 | replay_results_interval = []
73 |
74 | # Load previously saved results, e.g., ...
75 | for day in range(start_day, end_day+1):
76 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'results_' + str(day) + '.pkl'), 'rb') as file:
77 | replay_results = pickle.load(file)
78 | replay_results_interval.append(replay_results)
79 |
80 | # Visualize them
81 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
82 | ```
83 |
84 | Will produce:
85 |
86 | 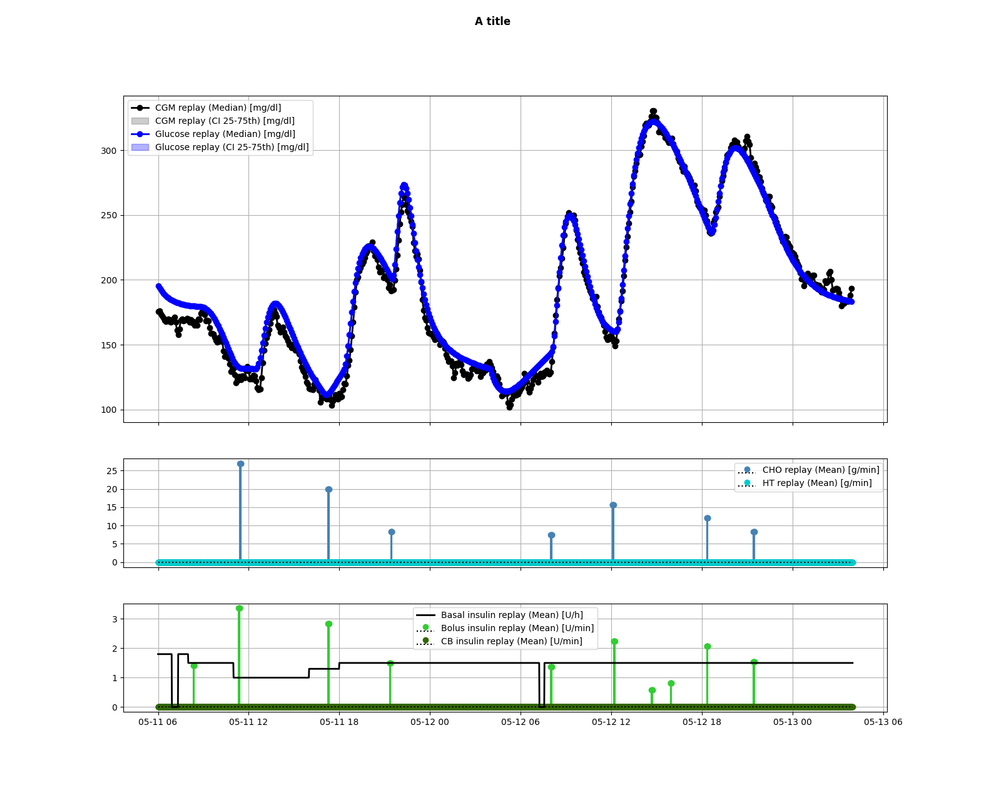
87 |
88 | The full code can be found in `/example/code/analysis_example_intervals.py`.
--------------------------------------------------------------------------------
/package.json:
--------------------------------------------------------------------------------
1 | {
2 | "name": "docs",
3 | "version": "1.0.0",
4 | "description": "A digital twin based framework for the development and assessment of new algorithms for type 1 diabetes management",
5 | "license": "MIT",
6 | "type": "module",
7 | "scripts": {
8 | "docs:build": "vuepress build docs",
9 | "docs:clean-dev": "vuepress dev docs --clean-cache",
10 | "docs:dev": "vuepress dev docs",
11 | "docs:update-package": "bunx vp-update"
12 | },
13 | "devDependencies": {
14 | "@vuepress/bundler-vite": "2.0.0-rc.18",
15 | "@vuepress/plugin-markdown-math": "2.0.0-rc.65",
16 | "@vuepress/plugin-shiki": "2.0.0-rc.65",
17 | "@vuepress/theme-default": "2.0.0-rc.65",
18 | "mathjax-full": "^3.2.2",
19 | "sass": "^1.82.0",
20 | "vue": "^3.5.13",
21 | "vuepress": "2.0.0-rc.18",
22 | "vuepress-theme-hope": "2.0.0-rc.63"
23 | }
24 | }
25 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/PKG-INFO:
--------------------------------------------------------------------------------
1 | Metadata-Version: 2.2
2 | Name: py_replay_bg
3 | Version: 1.1.0
4 | Summary: ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management.
5 | Author-email: Giacomo Cappon
6 | Project-URL: Homepage, https://github.com/gcappon/py_replay_bg
7 | Project-URL: Bug Tracker, https://github.com/gcappon/py_replay_bg/issues
8 | Classifier: Programming Language :: Python :: 3
9 | Classifier: License :: OSI Approved :: GNU General Public License v3 (GPLv3)
10 | Classifier: Operating System :: OS Independent
11 | Classifier: Intended Audience :: Science/Research
12 | Classifier: Topic :: Scientific/Engineering
13 | Requires-Python: >=3.11
14 | Description-Content-Type: text/markdown
15 | License-File: COPYING.md
16 | Requires-Dist: build==1.2.1
17 | Requires-Dist: celerite==0.4.2
18 | Requires-Dist: certifi==2024.2.2
19 | Requires-Dist: charset-normalizer==3.3.2
20 | Requires-Dist: contourpy==1.2.0
21 | Requires-Dist: corner==2.2.2
22 | Requires-Dist: coverage==7.4.4
23 | Requires-Dist: cycler==0.12.1
24 | Requires-Dist: docutils==0.20.1
25 | Requires-Dist: emcee==3.1.4
26 | Requires-Dist: et-xmlfile==1.1.0
27 | Requires-Dist: fonttools==4.50.0
28 | Requires-Dist: idna==3.6
29 | Requires-Dist: importlib_metadata==7.1.0
30 | Requires-Dist: iniconfig==2.0.0
31 | Requires-Dist: jaraco.classes==3.3.1
32 | Requires-Dist: jaraco.context==4.3.0
33 | Requires-Dist: jaraco.functools==4.0.0
34 | Requires-Dist: joblib==1.3.2
35 | Requires-Dist: keyring==25.0.0
36 | Requires-Dist: kiwisolver==1.4.5
37 | Requires-Dist: llvmlite==0.42.0
38 | Requires-Dist: markdown-it-py==3.0.0
39 | Requires-Dist: matplotlib==3.8.3
40 | Requires-Dist: mdurl==0.1.2
41 | Requires-Dist: more-itertools==10.2.0
42 | Requires-Dist: mpi4py==3.1.5
43 | Requires-Dist: nh3==0.2.17
44 | Requires-Dist: numba==0.59.1
45 | Requires-Dist: numpy==1.26.4
46 | Requires-Dist: openpyxl==3.1.2
47 | Requires-Dist: packaging==24.0
48 | Requires-Dist: pandas==2.2.1
49 | Requires-Dist: patsy==0.5.6
50 | Requires-Dist: pillow==10.2.0
51 | Requires-Dist: pkginfo==1.10.0
52 | Requires-Dist: plotly==5.20.0
53 | Requires-Dist: pluggy==1.4.0
54 | Requires-Dist: py-agata==0.0.8
55 | Requires-Dist: Pygments==2.17.2
56 | Requires-Dist: pyparsing==3.1.2
57 | Requires-Dist: pyproject_hooks==1.0.0
58 | Requires-Dist: pytest==8.1.1
59 | Requires-Dist: pytest-cov==5.0.0
60 | Requires-Dist: python-dateutil==2.9.0.post0
61 | Requires-Dist: pytz==2024.1
62 | Requires-Dist: readme_renderer==43.0
63 | Requires-Dist: requests==2.31.0
64 | Requires-Dist: requests-toolbelt==1.0.0
65 | Requires-Dist: rfc3986==2.0.0
66 | Requires-Dist: rich==13.7.1
67 | Requires-Dist: scikit-learn==1.4.1.post1
68 | Requires-Dist: scipy==1.12.0
69 | Requires-Dist: seaborn==0.13.2
70 | Requires-Dist: six==1.16.0
71 | Requires-Dist: statsmodels==0.14.1
72 | Requires-Dist: tenacity==8.2.3
73 | Requires-Dist: threadpoolctl==3.4.0
74 | Requires-Dist: tqdm==4.66.2
75 | Requires-Dist: twine==5.0.0
76 | Requires-Dist: tzdata==2024.1
77 | Requires-Dist: urllib3==2.2.1
78 | Requires-Dist: zipp==3.18.1
79 |
80 | # ReplayBG
81 |
82 |
4 |
5 | [](https://github.com/gcappon/py_replay_bg/COPYING)
6 | [](https://github.com/gcappon/py_replay_bg/commits/master)
7 |
8 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
9 |
10 | # Reference
11 |
12 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
13 |
14 | # Get started
15 |
16 | ## Installation
17 |
18 | **ReplayBG** can be installed via pypi by simply
19 |
20 | ```python
21 | pip install py-replay-bg
22 | ```
23 |
24 | ### Requirements
25 |
26 | * Python >= 3.11
27 | * List of Python packages in `requirements.txt`
28 |
29 | ## Preparation: imports, setup, and data loading
30 |
31 | First of all import the core modules:
32 | ```python
33 | import os
34 | import numpy as np
35 | import pandas as pd
36 |
37 | from multiprocessing import freeze_support
38 | ```
39 |
40 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
41 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
42 | parallelized way.
43 |
44 | Then, we will import the necessary ReplayBG modules:
45 | ```python
46 | from py_replay_bg.py_replay_bg import ReplayBG
47 | from py_replay_bg.visualizer import Visualizer
48 | from py_replay_bg.analyzer import Analyzer
49 | ```
50 |
51 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
52 | while `Analyzer` and `Visualizer` are utility objects that will be used to
53 | respectively analyze and visualize the results that we will produce with ReplayBG
54 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
55 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
56 |
57 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
58 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
59 | ```python
60 | if __name__ == '__main__':
61 | freeze_support()
62 | ```
63 |
64 | Then, we will set the verbosity of ReplayBG:
65 | ```python
66 | verbose = True
67 | plot_mode = False
68 | ```
69 |
70 | Then, we need to decide what blueprint to use for twinning the data at hand.
71 | ```python
72 | blueprint = 'multi-meal'
73 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
74 | parallelize = True
75 | ```
76 |
77 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
78 |
79 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
80 | which contains a day of data of a patient with T1D:
81 |
82 | ```python
83 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
84 | data.t = pd.to_datetime(data['t'])
85 | ```
86 |
87 | ::: warning
88 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
89 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
90 | :::
91 |
92 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
93 |
94 | ```python
95 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
96 | p = np.where(patient_info['patient'] == 1)[0][0]
97 | # Set bw and u2ss
98 | bw = float(patient_info.bw.values[p])
99 | u2ss = float(patient_info.u2ss.values[p])
100 | ```
101 |
102 | Finally, instantiate a `ReplayBG` object:
103 |
104 | ```python
105 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
106 | yts=5, exercise=False,
107 | seed=1,
108 | verbose=verbose, plot_mode=plot_mode)
109 |
110 | ```
111 |
112 | ## Step 1: Creation of the digital twin
113 |
114 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
115 |
116 | ```python
117 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
118 | twinning_method='mcmc',
119 | parallelize=parallelize,
120 | n_steps=5000,
121 | u2ss=u2ss)
122 | ```
123 |
124 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
125 |
126 |
127 | ## Step 2: Run replay simulations
128 |
129 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
130 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
131 |
132 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
133 |
134 | ```python
135 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
136 | twinning_method='mcmc',
137 | save_workspace=True,
138 | save_suffix='_step_2a')
139 | ```
140 |
141 | It is possible to visualize the results of the simulation using:
142 |
143 | ```python
144 | Visualizer.plot_replay_results(replay_results, data=data)
145 | ```
146 |
147 | and analyzing the results using:
148 |
149 | ```python
150 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
151 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
152 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
153 | ```
154 |
155 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
156 | To do that run:
157 |
158 | ```python
159 | data.bolus = data.bolus * .7
160 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
161 | twinning_method='mcmc',
162 | save_workspace=True,
163 | save_suffix='_step_2b')
164 |
165 | # Visualize results
166 | Visualizer.plot_replay_results(replay_results)
167 | # Analyze results
168 | analysis = Analyzer.analyze_replay_results(replay_results)
169 |
170 | # Print, for example, the average glucose
171 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
172 | ```
173 |
174 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
175 |
176 | # Documentation
177 |
178 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
179 |
--------------------------------------------------------------------------------
/bun.lockb:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/bun.lockb
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.0.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.1-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.1-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.1.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.1.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.2-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.2-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.2.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.2.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.3-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.3-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-0.1.3.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-0.1.3.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.0.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.1-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.1-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.1.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.1.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.2-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.2-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.2.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.2.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.3-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.3-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.3.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.3.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.4-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.4-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.4.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.4.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.5-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.5-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.0.5.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.0.5.tar.gz
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.1.0-py3-none-any.whl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.1.0-py3-none-any.whl
--------------------------------------------------------------------------------
/dist/py_replay_bg-1.1.0.tar.gz:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/dist/py_replay_bg-1.1.0.tar.gz
--------------------------------------------------------------------------------
/docs/.vuepress/config.js:
--------------------------------------------------------------------------------
1 | import { defineUserConfig } from 'vuepress/cli'
2 | import { viteBundler } from '@vuepress/bundler-vite'
3 | import { hopeTheme } from "vuepress-theme-hope";
4 |
5 | export default defineUserConfig({
6 | lang: 'en-US',
7 |
8 | title: 'ReplayBG',
9 | base: '/py_replay_bg/',
10 | description: 'A digital twin based framework for the development and assessment of new algorithms for type 1 ' +
11 | 'diabetes management',
12 |
13 | theme: hopeTheme({
14 | logo: 'https://i.postimg.cc/gJn8Sy0X/replay-bg-logo.png',
15 | navbar: ['/', '/documentation/get_started', '/documentation/'],
16 | repo: 'https://github.com/gcappon/py_replay_bg',
17 | docsDir: 'docs/',
18 | docsBranch: 'master',
19 | markdown: {
20 | highlighter: {
21 | type: "shiki",
22 | langs: ['python', 'json', 'md', 'bash', 'diff'],
23 | themes: {
24 | dark: 'catppuccin-mocha',
25 | light: 'catppuccin-latte'
26 | }
27 | },
28 | math: {
29 | type: "mathjax",
30 | },
31 | },
32 | sidebar: [
33 | {
34 | text: 'Get Started',
35 | link: 'documentation/get_started.md'
36 | },
37 | {
38 | text: 'Data Requirements',
39 | link: 'documentation/data_requirements.md'
40 | },
41 | {
42 | text: 'The ReplayBG Object',
43 | link: 'documentation/replaybg_object.md'
44 | },
45 | {
46 | text: 'Choosing Blueprint',
47 | link: 'documentation/choosing_blueprint.md'
48 | },
49 | {
50 | text: 'Twinning Procedure',
51 | link: 'documentation/twinning_procedure.md'
52 | },
53 | {
54 | text: 'Replaying',
55 | link: 'documentation/replaying.md'
56 | },
57 | {
58 | text: 'The _results/_ Folder',
59 | link: 'documentation/results_folder.md'
60 | },
61 | {
62 | text: 'Visualizing Replay Results',
63 | link: 'documentation/visualizing_replay_results.md'
64 | },
65 | {
66 | text: 'Analyzing Replay Results',
67 | link: 'documentation/analyzing_replay_results.md'
68 | }
69 | ],
70 | }),
71 |
72 | bundler: viteBundler(),
73 |
74 | plugins: [],
75 | })
76 |
--------------------------------------------------------------------------------
/docs/.vuepress/styles/index.scss:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/docs/.vuepress/styles/palette.scss:
--------------------------------------------------------------------------------
1 | $light-accent-color: #990000;
2 | $dark-accent-color: #fa0000;
3 | $vp-c-accent: (
4 | light: $light-accent-color,
5 | dark: $dark-accent-color,
6 | );
7 | $vp-c-accent-bg: (
8 | light: $light-accent-color,
9 | dark: $dark-accent-color,
10 | );
11 | $vp-c-accent-hover: (
12 | light: $light-accent-color,
13 | dark: $dark-accent-color,
14 | );
15 |
16 |
--------------------------------------------------------------------------------
/docs/README.md:
--------------------------------------------------------------------------------
1 | ---
2 | home: true
3 | title: Home
4 | heroImage: https://i.postimg.cc/gJn8Sy0X/replay-bg-logo.png
5 | actions:
6 | - text: Get Started
7 | link: /documentation/get_started
8 | type: primary
9 |
10 | features:
11 | - title: Digital Twin
12 | details: Leverage the power of digital twin to assess new therapies for type 1 diabetes
13 | - title: State of the Art
14 | details: ReplayBG is supported by solid scientific research from UNIPD
15 | - title: Easy to Use
16 | details: ReplayBG is easy to use and ready to be integrated in your research pipeline
17 |
18 | footer: Made by Giacomo Cappon with ❤️
19 | ---
20 |
21 | ## Supporting research
22 |
23 | ### Research on ReplayBG development
24 |
25 | #### Journal Papers
26 | - F. Prendin, A. Facchinetti, and G. Cappon “Data Augmentation via Digital Twins Enables the Development of Personalized
27 | Deep Learning Glucose Prediction Algorithms for Type 1 Diabetes in Poor Data Context” IEEE Journal of Biomedical
28 | Health Informatics, (under revision, submitted October 2024)
29 | - G. Cappon, and A. Facchinetti, “Digital twins in type 1 diabetes: A systematic review” Journal of Diabetes
30 | Science and Technology, online ahead of print, 2024, DOI: 10.1177/1932296824126211.
31 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “ReplayBG: a digital twin-based methodology
32 | to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative
33 | therapies”, IEEE Transactions on Biomedical Engineering, vol. 70, no. 11, pp. 3105-3115, Nov 2023,
34 | DOI: 10.1109/TBME.2023.3286856
35 |
36 |
37 | #### Conference papers and abstracts
38 | - G. Cappon, S. Del Favero, and A. Facchinetti “Extending the applicability of ReplayBG tool for digital
39 | twinning in type 1 diabetes from single-meal to multi-day scenarios” in the 24th Annual Diabetes Technology
40 | Meeting – DTM, Burlingame, CA, USA, October 15-17, 2024.
41 | - J. Leth, H. Ghazaleh, H. Peuscher, and G. Cappon “The potential of open-source software in diabetes research” in the
42 | 17th International Conference on Advanced Technology & Treatment for Diabetes – ATTD, Firenze, Italy, March 6-9,
43 | 2024 (invited oral presentation, speaker J. Leth).
44 | - H. Peuscher, G. Cappon, A. Cinar, J. Deichmann, H. Ghazaleh, H. Kaltenbach, L. Sandini, M. Siket, J. Xie, and
45 | X. Zhou “A survey on existing open-source projects in diabetes simulation” in the 17th International Conference on
46 | Advanced Technology & Treatment for Diabetes – ATTD, Firenze, Italy, March 6-9, 2024
47 | (invited oral presentation, speaker J. Leth).
48 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “Expanding ReplayBG simulation methodology
49 | domain of validity to single day multiple meal scenarios” in the 15th International Conference on Advanced
50 | Technology & Treatment for Diabetes – ATTD, Barcelona, Spain, April 27-30, 2022.
51 | - G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “ReplayBG provides reliable indications when
52 | used to assess meal bolus alterations in type 1 diabetes” in the 14th International Conference on Advanced
53 | Technology & Treatment for Diabetes – ATTD, Paris, France, June 2-6, 2021.
54 | G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “ReplayBG: A novel in-silico framework to retrospectively
55 | assess new therapy guidelines for type 1 diabetes management” in the 20th Annual Diabetes Technology Meeting –
56 | DTM, Bethesda, Maryland, USA, November 12-14, 2020.
57 | - G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “A Bayesian framework to personalize glucose prediction
58 | in type 1 diabetes using a physiological model” in the 19th Annual Diabetes Technology Meeting –
59 | DTM, Bethesda, Maryland, USA, November 14-16, 2019.
60 | - G. Cappon, A. Facchinetti, G. Sparacino, and S. Del Favero “A Bayesian framework to identify type 1 diabetes
61 | physiological models using easily accessible patient data” in the 41th Annual International Conference of the IEEE
62 | Engineering in Medicine and Biology Society – EMBC, Berlin, Germany, July 23-27, 2019
63 | (accepted for oral presentation, speaker G.Cappon).
64 |
65 | ### Research using ReplayBG as component or validation tool
66 |
67 | #### Journal Papers
68 | - E. Pellizzari, G. Cappon, G. Nicolis, G. Sparacino, and A. Facchinetti “Developing effective machine learning models
69 | for insulin bolus calculation in type 1 diabetes exploiting real-world data and digital twins” IEEE Transactions on
70 | Biomedical Engineering, (under revision, submitted October 2024)
71 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti, “drCORRECT: An algorithm for the
72 | preventive administration of postprandial corrective insulin boluses in type 1 diabetes management” Journal of
73 | Diabetes Science and Technology, online ahead of print, 2023, DOI: 10.1177/19322968231221768
74 | - F. Prendin, J. Pavan, G. Cappon, S. Del Favero, G. Sparacino, and A. Facchinetti, “The importance of interpreting
75 | machine learning models for blood glucose prediction in diabetes: an analysis using SHAP” Scientific
76 | Reports, vol. 13, no. 1, pp. 16865, Oct 2023, DOI: 10.1038/s41598-023-44155-x.
77 | - G. Noaro, G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, and A. Facchinetti “Machine-learning based model
78 | to improve insulin bolus calculation in type 1 diabetes therapy” IEEE Transactions on Biomedical Engineering,
79 | vol. 68, no. 1, pp. 247-255, Jan 2021. DOI: 10.1109/TBME.2020.3004031
80 |
81 | #### Conference papers and abstracts
82 | - E. Pellizzari, G. Cappon, G. Nicolis, G. Sparacino, and A. Facchinetti “Exploiting real-world data and digital
83 | twins to develop effective formulas for dosing insulin boluses in type 1 diabetes therapy” in the 24th Annual
84 | Diabetes Technology Meeting – DTM, Burlingame, CA, USA, October 15-17, 2024.
85 | - F. Prendin, A. Facchinetti, and G. Cappon “Digital twin for data augmentation enables the development of accurate
86 | personalized deep glucose forecasting algorithms” in the 24th Annual Diabetes Technology Meeting –
87 | DTM, Burlingame, CA, USA, October 15-17, 2024.
88 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti “DR-CIB: An algorithm for the preventive
89 | administration of corrective insulin boluses in T1D based on dynamic risk concept and patient-specific timing” in
90 | the 23rd Annual Diabetes Technology Meeting – DTM, Online, November 1-4, 2023 (Gold Student Award, accepted for
91 | oral presentation, speaker E. Pellizzari).
92 | - G. Cappon, E. Pellizzari, L. Cossu, G. Sparacino, A. Deodati, R. Schiaffini, S. Cianfarani, and A. Facchinetti
93 | “System architecture of TWIN: A new digital twin-based clinical decision support system for type 1 diabetes
94 | management in children” in the 19th International Conference on Body Sensor Networks –
95 | BSN, Boston, MA, USA, October 9-11, 2023.
96 | - E. Pellizzari, F. Prendin, G. Cappon, G. Sparacino, and A. Facchinetti “A deep-learning based algorithm for the
97 | management of hyperglycemia in type 1 diabetes therapy” in the 19th International Conference on Body Sensor Networks
98 | – BSN, Boston, MA, USA, October 9-11, 2023.
99 | - G. Noaro, G. Cappon, G. Sparacino, and A. Facchinetti “An ensemble learning algorithm based on dynamic voting for
100 | targeting the optimal insulin dosage in type 1 diabetes management“ in the 43rd Annual International Conference of the
101 | IEEE Engineering in Medicine and Biology Society – EMBC, Guadalajara, Mexico, October 31-4, 2021 (accepted for
102 | oral presentation, speaker G. Noaro).
103 | - G. Cappon, E. Pighin, F. Prendin, G. Sparacino, and A. Facchinetti “A correction insulin bolus delivery strategy
104 | for decision support systems in type 1 diabetes“ in the 43rd Annual International Conference of the IEEE Engineering
105 | in Medicine and Biology Society – EMBC, Guadalajara, Mexico, October 31-4, 2021 (accepted for oral presentation,
106 | speaker G. Cappon).
107 | - G. Noaro, G. Cappon, M. Vettoretti, S. Del Favero, G. Sparacino, and A. Facchinetti “A new model for mealtime insulin
108 | dosing in type 1 diabetes: Retrospective validation on CTR3 dataset” in the 20th Annual Diabetes Technology Meeting –
109 | DTM, Bethesda, Maryland, USA, November 12-14, 2020 (accepted for oral presentation, speaker G. Noaro).
110 | - G. Noaro, G. Cappon, G. Sparacino, S. Del Favero, A. Facchinetti “Nonlinear machine learning models for insulin
111 | bolus estimation in type 1 diabetes therapy” in the 42nd Annual International Conference of the IEEE Engineering
112 | in Medicine and Biology Society – EMBC, Montreal, Canada, July 20-24, 2020 (accepted for oral presentation,
113 | speaker G. Noaro)
--------------------------------------------------------------------------------
/docs/documentation/README.md:
--------------------------------------------------------------------------------
1 | ---
2 | lang: en-US
3 | title: Docs
4 | description: Docs index
5 | ---
6 |
7 | # Docs
8 |
9 | ### [Get Started](./get_started.md)
10 |
11 | ---
12 |
13 | ### [Data Requirements](./data_requirements.md)
14 |
15 | ### [The ReplayBG Object](./replaybg_object.md)
16 |
17 | ### [Choosing Blueprint](./choosing_blueprint.md)
18 |
19 | ### [Twinning Procedure](./twinning_procedure.md)
20 |
21 | ### [Replaying](./replaying.md)
22 |
23 | ### [The _results/_ Folder](./results_folder.md)
24 |
25 | ---
26 |
27 | ### [Visualizing replay results](./visualizing_replay_results.md)
28 |
29 | ### [Analyzing replay results](./analyzing_replay_results.md)
30 |
31 | ---
--------------------------------------------------------------------------------
/docs/documentation/data_requirements.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: heading
3 | ---
4 | # Data Requirements
5 |
6 | Data provided to ReplayBG must comply to strict format requirements and should be selected following some best
7 | practices.
8 |
9 | ## Format requirements
10 |
11 | Format requirements depend on the selected `blueprint`, i.e., single-meal or multi-meal, as such,
12 | in the following, details about these aspects are presented for each possible blueprint separately.
13 |
14 | ### Single Meal blueprint
15 |
16 | By "single meal" one can refer to a specific period
17 | of time when a specific subject had only 1 main meal and a corresponding insulin
18 | basal-bolus administration. Usually, this period of time spans maximum 6/8 hours,
19 | starts near such main meal, and ends just before the subsequent main meal and/or after
20 | a reasonable amount of time.
21 |
22 | `data` must be saved in a `.csv` file and contain the following columns:
23 | * `t`: the timestamps when data of the corresponding row were recorded (format
24 | `DD-MMM-YYYY HH:mm:SS` for example `20-Dec-2013 10:35:00`). The sampling grid defined by the `t` column must be
25 | homogeneous, e.g., have a datapoint every 5 minutes.
26 | * `glucose`: the glucose concentration (mg/dl) at `t`. Can contain NaN values.
27 | * `cho`: the meal intake (g/min) at `t`. Can't contain NaN values. If no meals were recorded at `t` just put `0` there.
28 | * `bolus`: the insulin bolus (U/min) administered at `t`. Can't contain NaN values. If no insulin boluses were
29 | administered at `t` just put `0` there.
30 | * `basal`: the basal insulin (U/min) administered at `t`. Can't contain NaN values. If no basal insulin was
31 | administered at `t` just put `0` there.
32 | * `bolus_label`: the type of `bolus` at time `t`. This column Each `bolus` entry > 0 must have a label defined. Can be
33 | * `B` if it is the bolus of a breakfast.
34 | * `L` if it is the bolus of a lunch.
35 | * `D` if it is the bolus of a dinner.
36 | * `C` if it is a corrective bolus.
37 | * `S` if it is the bolus of a snack.
38 |
39 | If other columns are present in your data file, they will be ignored.
40 |
41 | ::: tip NOTE
42 | The total length of the simulation, `simulation_length`, is defined in minutes and determined by ReplayBG automatically
43 | using the `t` column of `data` and the `yts` input parameter provided to the `ReplayBG` object builder.
44 |
45 | For example, if `yts` is `5` minutes and `t` starts from `20-Dec-2013 10:36:00` and ends to `20-Dec-2013 10:46:00`,
46 | then `simulation_length` will be `10`.
47 | :::
48 |
49 | ::: tip
50 | If `bolus_label` is not important for you (e.g., you do not plan to use it during replay) or if you do not need that,
51 | just add an empty `bolus_label` column.
52 | :::
53 |
54 | ::: warning
55 | If more than 1 meal are present in the provided file, ReplayBG will consider the first meal as "main" meal. The others
56 | will be considered as "other" meals. The resulting `kabs` and $\beta$ parameters will be unique so their value will
57 | depend by ALL the meals you have on your data. SO, it is really not advised to have more than one meal when using the
58 | single-meal blueprint.
59 | :::
60 |
61 | #### Requirements during replay
62 | When replaying (using the `replay` method), the following requirements are no more valid under the following circumstances:
63 | - `glucose`: during replay this is simply ignored.
64 | - `cho`: if `cho_source` is `generated` since the CHO event will be generated by the provided handler during the replay simulation.
65 | - `bolus` and `bolus_label`: if `bolus_source` is `dss` since the insulin bolus events will be generated by the provided handler during the replay simulation.
66 | - `basal`: if `basal_source` is `dss` since the basal insulin will be generated by the provided handler during the replay simulation.
67 |
68 | ### Multi Meal blueprint
69 |
70 | By "multi meal" one can refer to a specific period
71 | of time when a specific subject had more than 1 main meal and a
72 | corresponding insulin basal-bolus administration regimen. One can think to such
73 | period of time by thinking to a day, when multiple meals occur, or even multiple days.
74 |
75 | `data` must be saved in a `.csv` file and contain (at least) the following columns:
76 | * `t`: the timestamps when data of the corresponding row were recorded (format
77 | `DD-MMM-YYYY HH:mm:SS` for example `20-Dec-2013 10:35:00`). The sampling grid defined by the `t` column must be
78 | homogeneous.
79 | * `glucose`: the glucose concentration (mg/dl) at `t`. Can contain NaN values.
80 | * `cho`: the meal intake (g/min) at `t`. Can't contain NaN values. If no meals were recorded at `t` just put `0` there.
81 | * `bolus`: the insulin bolus (U/min) administered at `t`. Can't contain NaN values. If no insulin boluses were
82 | administered at `t` just put `0` there.
83 | * `basal`: the basal insulin (U/min) administered at `t`. Can't contain NaN values. If no basal insulin was
84 | administered at `t` just put `0` there.
85 | * `cho_label`: the type of `cho` at time `t`. Each `cho` entry > 0 must have a label defined. Can be
86 | * `B` if it is a breakfast.
87 | * `L` if it is a lunch.
88 | * `D` if it is a dinner.
89 | * `H` if it is a hypotreatment.
90 | * `S` if it is a snack.
91 | * `bolus_label`: the type of `bolus` at time `t`. Each `bolus` entry > 0 must have a label defined. Can be
92 | * `B` if it is the bolus of a breakfast.
93 | * `L` if it is the bolus of a lunch.
94 | * `D` if it is the bolus of a dinner.
95 | * `C` if it is a corrective bolus.
96 | * `S` if it is the bolus of a snack.
97 |
98 | If other columns are present in your data file, they will be ignored.
99 |
100 | ::: warning
101 | The `cho` and `bolus` columns must contain at least one event when twinning.
102 | :::
103 |
104 | ::: tip
105 | If `bolus_label` is not important for you (e.g., you do not plan to use it during replay) or if you do not need that,
106 | just add an empty `bolus_label` column.
107 | :::
108 |
109 | ::: tip
110 | A representative data file of a single meal blueprint can be found in `example/data/multi-meal_example.csv`
111 | :::
112 |
113 | #### Requirements during replay
114 | When replaying (using the `replay` method), the following requirements are no more valid under the following circumstances:
115 | - `glucose`: during replay this is simply ignored.
116 | - `cho` and `cho_label`: if `cho_source` is `generated` since the CHO events will be generated by the provided handler during the replay simulation.
117 | - `bolus` and `bolus_label`: if `bolus_source` is `dss` since the insulin bolus events will be generated by the provided handler during the replay simulation.
118 | - `basal`: if `basal_source` is `dss` since the basal insulin will be generated by the provided handler during the replay simulation.
119 |
120 | ## Best practices
121 | The potential ReplayBG user should be aware of several practical aspects and be careful when selecting the portion
122 | of data to work with. Here's the details.
123 |
124 | ### Starting point
125 | The twinning procedure of ReplayBG does not estimate the states of the blueprint mathematical model and, more
126 | importantly, the corresponding initial conditions. Identifying such initial conditions is crucial to correctly
127 | estimating the unknown model parameter vector $\boldsymbol{\theta}_{phy}$ and, by product, reliably replaying the
128 | glucose profile with new, altered, inputs. To circumvent this issue, ReplayBG assumes all model state initial
129 | conditions at steady state. This assumption is granted to be valid when the actions of exogenous insulin and
130 | carbohydrate intake are “exhausted”, i.e., when the starting point of the portion of data is reasonably distant
131 | from the last meal and insulin boluses, e.g., 4 hours.
132 |
133 | ::: tip
134 | When twinning and replaying intervals (using the `twin` and the `replay` methods, respectively) instead of single
135 | portions of data, things change. Indeed, the problem of the starting point applies only to the first portion where we
136 | must assume initial steady state conditions. The other subsequent portions will start from the immediate next datapoint
137 | with initial conditions defined by `x0` and `previous_data_name`.
138 |
139 | For more information on the `x0` and
140 | `previous_data_name` parameters when twinning and replaying please refer to the [Twinning Procedure](./twinning_procedure.md)
141 | and the [Replaying](./replaying.md) pages.
142 | :::
143 |
144 | ### Minimum data length
145 | As a rule of thumb we suggest to use portions of data that span at least 6 hours.
146 | As demonstrated in the literature, this ensures to obtain better parameter estimates and simulation results.
147 |
148 | ### Data gaps
149 | To make the twinning procedure more reliable, data portions having significant data gaps (i.e., more that 10% of missing glucose
150 | readings) or without a single reported meal intake or insulin bolus, should be discarded to avoid the creation of
151 | digital twins not representing the actual underneath physiology.
--------------------------------------------------------------------------------
/docs/documentation/get_started.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 | # Get started
5 |
6 | ## Installation
7 |
8 | **ReplayBG** can be installed via pypi by simply
9 |
10 | ```python
11 | pip install py-replay-bg
12 | ```
13 |
14 | ### Requirements
15 |
16 | * Python >= 3.11
17 | * List of Python packages in `requirements.txt`
18 |
19 | ## Preparation: imports, setup, and data loading
20 |
21 | First of all import the core modules:
22 | ```python
23 | import os
24 | import numpy as np
25 | import pandas as pd
26 |
27 | from multiprocessing import freeze_support
28 | ```
29 |
30 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
31 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
32 | parallelized way.
33 |
34 | Then, we will import the necessary ReplayBG modules:
35 | ```python twoslash
36 | from py_replay_bg.py_replay_bg import ReplayBG
37 | from py_replay_bg.visualizer import Visualizer
38 | from py_replay_bg.analyzer import Analyzer
39 | ```
40 |
41 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](./replaybg_object.md) page),
42 | while `Analyzer` and `Visualizer` are utility objects that will be used to
43 | respectively analyze and visualize the results that we will produce with ReplayBG
44 | (more information in the ([Visualizing Replay Results](./visualizing_replay_results.md) and
45 | [Analyzing Replay Results](./analyzing_replay_results.md) pages).
46 |
47 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
48 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
49 | ```python
50 | if __name__ == '__main__':
51 | freeze_support()
52 | ```
53 |
54 | Then, we will set the verbosity of ReplayBG:
55 | ```python
56 | verbose = True
57 | plot_mode = False
58 | ```
59 |
60 | Then, we need to decide what blueprint to use for twinning the data at hand.
61 | ```python
62 | blueprint = 'multi-meal'
63 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
64 | parallelize = True
65 | ```
66 |
67 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](./choosing_blueprint.md) page.
68 |
69 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
70 | which contains a day of data of a patient with T1D:
71 |
72 | ```python
73 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
74 | data.t = pd.to_datetime(data['t'])
75 | ```
76 |
77 | ::: warning
78 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
79 | information see the [Data Requirements](./data_requirements) page.
80 | :::
81 |
82 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
83 |
84 | ```python
85 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
86 | p = np.where(patient_info['patient'] == 1)[0][0]
87 | # Set bw and u2ss
88 | bw = float(patient_info.bw.values[p])
89 | u2ss = float(patient_info.u2ss.values[p])
90 | ```
91 |
92 | Finally, instantiate a `ReplayBG` object:
93 |
94 | ```python
95 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
96 | yts=5, exercise=False,
97 | seed=1,
98 | verbose=verbose, plot_mode=plot_mode)
99 |
100 | ```
101 |
102 | ## Step 1: Creation of the digital twin
103 |
104 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
105 |
106 | ```python
107 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
108 | twinning_method='mcmc',
109 | parallelize=parallelize,
110 | n_steps=5000,
111 | u2ss=u2ss)
112 | ```
113 |
114 | For more information on the twinning procedure see the [Twinning Procedure](./twinning_procedure.md) page.
115 |
116 |
117 | ## Step 2: Run replay simulations
118 |
119 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
120 | see the [Replaying](./replaying.md) page.
121 |
122 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
123 |
124 | ```python
125 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
126 | twinning_method='mcmc',
127 | save_workspace=True,
128 | u2ss=u2ss,
129 | save_suffix='_step_2a')
130 | ```
131 |
132 | It is possible to visualize the results of the simulation using:
133 |
134 | ```python
135 | Visualizer.plot_replay_results(replay_results, data=data)
136 | ```
137 |
138 | and analyzing the results using:
139 |
140 | ```python
141 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
142 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
143 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
144 | ```
145 |
146 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
147 | To do that run:
148 |
149 | ```python
150 | data.bolus = data.bolus * .7
151 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
152 | twinning_method='mcmc',
153 | save_workspace=True,
154 | save_suffix='_step_2b')
155 |
156 | # Visualize results
157 | Visualizer.plot_replay_results(replay_results)
158 | # Analyze results
159 | analysis = Analyzer.analyze_replay_results(replay_results)
160 |
161 | # Print, for example, the average glucose
162 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
163 | ```
164 |
165 | ## Full example
166 |
167 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
168 |
169 | ```python
170 | import os
171 | import numpy as np
172 | import pandas as pd
173 |
174 | from multiprocessing import freeze_support
175 |
176 | from py_replay_bg.py_replay_bg import ReplayBG
177 | from py_replay_bg.visualizer import Visualizer
178 | from py_replay_bg.analyzer import Analyzer
179 |
180 |
181 | if __name__ == '__main__':
182 | freeze_support()
183 |
184 | # Set verbosity
185 | verbose = True
186 | plot_mode = False
187 |
188 | # Set other parameters for twinning
189 | blueprint = 'multi-meal'
190 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
191 | parallelize = True
192 |
193 | # Load data
194 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
195 | data.t = pd.to_datetime(data['t'])
196 |
197 | # Load patient_info
198 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
199 | p = np.where(patient_info['patient'] == 1)[0][0]
200 | # Set bw and u2ss
201 | bw = float(patient_info.bw.values[p])
202 | u2ss = float(patient_info.u2ss.values[p])
203 |
204 | # Instantiate ReplayBG
205 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
206 | yts=5, exercise=False,
207 | seed=1,
208 | verbose=verbose, plot_mode=plot_mode)
209 |
210 | # Set save name
211 | save_name = 'data_day_1'
212 |
213 | # Step 1. Run twinning procedure
214 | rbg.twin(data=data, bw=bw, save_name=save_name,
215 | twinning_method='mcmc',
216 | parallelize=parallelize,
217 | n_steps=5000,
218 | u2ss=u2ss)
219 |
220 | # Step 2a. Replay the twin with the same input data
221 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
222 | twinning_method='mcmc',
223 | save_workspace=True,
224 | save_suffix='_step_2a')
225 |
226 | # Visualize results and compare with the original glucose data
227 | Visualizer.plot_replay_results(replay_results, data=data)
228 | # Analyze results
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | # Print, for example, the fit MARD and the average glucose
231 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
232 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
233 |
234 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
235 | data.bolus = data.bolus * .7
236 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
237 | twinning_method='mcmc',
238 | save_workspace=True,
239 | save_suffix='_step_2b')
240 |
241 | # Visualize results
242 | Visualizer.plot_replay_results(replay_results)
243 | # Analyze results
244 | analysis = Analyzer.analyze_replay_results(replay_results)
245 |
246 | # Print, for example, the average glucose
247 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
248 | ```
249 |
--------------------------------------------------------------------------------
/docs/documentation/replaybg_object.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 |
5 | # The ReplayBG Object
6 |
7 | The `ReplayBG` object is the core, key, object to instatiate when starting to work with the ReplayBG framework.
8 |
9 | Its constructor is formally defined as:
10 |
11 | ```python
12 | from py_replay_bg.py_replay_bg import ReplayBG
13 |
14 | ReplayBG(save_folder: str, blueprint: str = 'single_meal',
15 | yts: int = 5, exercise: bool = False,
16 | seed: int = 1,
17 | plot_mode: bool = True, verbose: bool = True)
18 | ```
19 |
20 | ## Input parameters
21 |
22 | Let's inspect and describe each input parameter.
23 |
24 | - `save_folder`: a string defining the folder that will contain the results of the twinning procedure and the replay
25 | simulations. This parameter is mandatory. More information on how to set it can be found in
26 | [The _results/_ Folder](./results_folder.md) page.
27 | - `blueprint`, optional, `{'single-meal', 'multi-meal'}`, default: `'single-meal'`: a string that specifies the blueprint to be used to create
28 | the digital twin. More information on how to set it can be found in [Choosing Blueprint](./choosing_blueprint.md) page.
29 | - `yts`, optional, default: `5` : an integer that specifies the data sample time (in minutes).
30 | - `exercise`, optional, default: `False`: a boolean that specifies whether to simulate exercise or not.
31 | - `seed`, optional, default: `1`: an integer that specifies the random seed. For reproducibility.
32 | - `plot_mode`, optional, default: `True`: a boolean that specifies whether to show the plot of the results or not. More
33 | information on how to visualize the results of ReplayBG can be found in
34 | [Visualizing Replay Results](./visualizing_replay_results.md) page.
35 | - `verbose`, optional, default: `True`: a boolean that specifies the verbosity of ReplayBG.
36 |
37 |
38 |
39 |
--------------------------------------------------------------------------------
/docs/documentation/results_folder.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: auto
3 | ---
4 |
5 | # The _results/_ Folder
6 |
7 | All results of ReplayBG, i.e., both model parameters obtained via twinning procedure and replayed
8 | glucose traces of a given scenario, will be saved in a folder named `results/`.
9 |
10 | ## Choosing the path of _results/_
11 |
12 | When creating the ReplayBG object, user must decide the location of `results/`.
13 | To do that, specify a custom location, relative to current ``
14 | using the `save_folder` parameter of the `ReplayBG` object builder. For example,
15 | to put `results/` in `/custom/path/location/`, simply:
16 |
17 | ```python
18 | rbg = ReplayBG(save_folder=os.path.join('custom', 'path', 'location'), ...)
19 | ```
20 |
21 | This means that if the user wants to put `results/` just in ``
22 | he/she must use:
23 |
24 | ```python
25 | rbg = ReplayBG(save_folder=os.path.join(''), ...)
26 | ```
27 |
28 | ## What's inside
29 |
30 | The `results/` folder is organized as follows:
31 |
32 | ```
33 | results/
34 | |--- mcmc/
35 | |--- map/
36 | |--- workspaces/
37 | ```
38 |
39 | where the `mcmc/` and `map/` subfolders contains the model parameters obtained via
40 | MCMC-based and MAP-based twinning procedures, respectively; and the `workspaces/` folder
41 | contains the results of the replayed scenarios simulated when the `rbg.replay()` method
42 | is called.
43 |
44 | ::: tip REMEMBER
45 | Results of the replayed scenarios are saved in the `workspace/` subfolder only if the
46 | `save_workspace` parameter of `rbg.replay()` is set to `True`:
47 | ```python
48 | rbg.replay(save_workspace=True, ...)
49 | ```
50 | If not set, being its default value `False`, nothing will be saved.
51 | :::
52 |
53 |
54 |
55 |
56 |
57 |
--------------------------------------------------------------------------------
/docs/documentation/visualizing_replay_results.md:
--------------------------------------------------------------------------------
1 | ---
2 | sidebar: heading
3 | ---
4 |
5 | # Visualizing replay results
6 |
7 | ReplayBG provides the possibility, to visualize in a painless way the results of `rbg.replay()`.
8 |
9 | This is done using the `Visualizer` class importable from `py_replay_bg.visualizer`.
10 |
11 | In the following, we show how to do that in the case of single portions of data and portions of data spanning
12 | more than one day (i.e., intervals).
13 |
14 | ## Visualizing replay results from single portions of data
15 |
16 | To visualize replay results from single portions of data use the `Visualizer.plot_replay_results()` static
17 | method, which is formerly defined as:
18 | ```python
19 | @staticmethod
20 | def plot_replay_results(
21 | replay_results: Dict,
22 | data: pd.DataFrame = None,
23 | title: str = '',
24 | ) -> None
25 | ```
26 |
27 | ### Input parameters
28 | - `replay_results`: the dictionary returned by or saved with the `rbg.replay()` method
29 | - `data`, optional, default: `None`: The `data` parameter passed to `rbg.replay()` . If present, the method will also
30 | compare the glucose fit vs the data.
31 | - `title`, optional, default: `None`: A string with an optional title to be added to the figure.
32 |
33 | ### Example
34 |
35 | ```python
36 | # Load previously saved results, e.g., ...
37 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'results.pkl'), 'rb') as file:
38 | replay_results = pickle.load(file)
39 |
40 | # Analyze them
41 | Visualizer.plot_replay_results(replay_results=replay_results)
42 | ```
43 |
44 | Will produce:
45 |
46 | 
47 |
48 | The full code can be found in `/example/code/analysis_example.py`.
49 |
50 | ## Visualizing replay results from portions of data spanning more than one day (i.e., intervals)
51 |
52 | To visualize replay results from single portions of data use the `Visualizer.plot_replay_results_interval()` static
53 | method, which is formerly defined as:
54 | ```python
55 | @staticmethod
56 | def plot_replay_results_interval(
57 | replay_results_interval: list,
58 | data_interval: list = None,
59 | ) -> Dict
60 | ```
61 |
62 | ### Input parameters
63 | - `replay_results_interval`: a list of dictionaries returned by or saved with the `rbg.replay()` method
64 | - `data_interval`, optional, default: `None`: The list of `data` passed to `rbg.replay()` . If present, the method will also
65 | compare the glucose fit vs the data.
66 | - `title`, optional, default: `None`: A string with an optional title to be added to the figure.
67 |
68 | ### Example
69 |
70 | ```python
71 | # Initialize results list
72 | replay_results_interval = []
73 |
74 | # Load previously saved results, e.g., ...
75 | for day in range(start_day, end_day+1):
76 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'results_' + str(day) + '.pkl'), 'rb') as file:
77 | replay_results = pickle.load(file)
78 | replay_results_interval.append(replay_results)
79 |
80 | # Visualize them
81 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
82 | ```
83 |
84 | Will produce:
85 |
86 | 
87 |
88 | The full code can be found in `/example/code/analysis_example_intervals.py`.
--------------------------------------------------------------------------------
/package.json:
--------------------------------------------------------------------------------
1 | {
2 | "name": "docs",
3 | "version": "1.0.0",
4 | "description": "A digital twin based framework for the development and assessment of new algorithms for type 1 diabetes management",
5 | "license": "MIT",
6 | "type": "module",
7 | "scripts": {
8 | "docs:build": "vuepress build docs",
9 | "docs:clean-dev": "vuepress dev docs --clean-cache",
10 | "docs:dev": "vuepress dev docs",
11 | "docs:update-package": "bunx vp-update"
12 | },
13 | "devDependencies": {
14 | "@vuepress/bundler-vite": "2.0.0-rc.18",
15 | "@vuepress/plugin-markdown-math": "2.0.0-rc.65",
16 | "@vuepress/plugin-shiki": "2.0.0-rc.65",
17 | "@vuepress/theme-default": "2.0.0-rc.65",
18 | "mathjax-full": "^3.2.2",
19 | "sass": "^1.82.0",
20 | "vue": "^3.5.13",
21 | "vuepress": "2.0.0-rc.18",
22 | "vuepress-theme-hope": "2.0.0-rc.63"
23 | }
24 | }
25 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/PKG-INFO:
--------------------------------------------------------------------------------
1 | Metadata-Version: 2.2
2 | Name: py_replay_bg
3 | Version: 1.1.0
4 | Summary: ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management.
5 | Author-email: Giacomo Cappon
6 | Project-URL: Homepage, https://github.com/gcappon/py_replay_bg
7 | Project-URL: Bug Tracker, https://github.com/gcappon/py_replay_bg/issues
8 | Classifier: Programming Language :: Python :: 3
9 | Classifier: License :: OSI Approved :: GNU General Public License v3 (GPLv3)
10 | Classifier: Operating System :: OS Independent
11 | Classifier: Intended Audience :: Science/Research
12 | Classifier: Topic :: Scientific/Engineering
13 | Requires-Python: >=3.11
14 | Description-Content-Type: text/markdown
15 | License-File: COPYING.md
16 | Requires-Dist: build==1.2.1
17 | Requires-Dist: celerite==0.4.2
18 | Requires-Dist: certifi==2024.2.2
19 | Requires-Dist: charset-normalizer==3.3.2
20 | Requires-Dist: contourpy==1.2.0
21 | Requires-Dist: corner==2.2.2
22 | Requires-Dist: coverage==7.4.4
23 | Requires-Dist: cycler==0.12.1
24 | Requires-Dist: docutils==0.20.1
25 | Requires-Dist: emcee==3.1.4
26 | Requires-Dist: et-xmlfile==1.1.0
27 | Requires-Dist: fonttools==4.50.0
28 | Requires-Dist: idna==3.6
29 | Requires-Dist: importlib_metadata==7.1.0
30 | Requires-Dist: iniconfig==2.0.0
31 | Requires-Dist: jaraco.classes==3.3.1
32 | Requires-Dist: jaraco.context==4.3.0
33 | Requires-Dist: jaraco.functools==4.0.0
34 | Requires-Dist: joblib==1.3.2
35 | Requires-Dist: keyring==25.0.0
36 | Requires-Dist: kiwisolver==1.4.5
37 | Requires-Dist: llvmlite==0.42.0
38 | Requires-Dist: markdown-it-py==3.0.0
39 | Requires-Dist: matplotlib==3.8.3
40 | Requires-Dist: mdurl==0.1.2
41 | Requires-Dist: more-itertools==10.2.0
42 | Requires-Dist: mpi4py==3.1.5
43 | Requires-Dist: nh3==0.2.17
44 | Requires-Dist: numba==0.59.1
45 | Requires-Dist: numpy==1.26.4
46 | Requires-Dist: openpyxl==3.1.2
47 | Requires-Dist: packaging==24.0
48 | Requires-Dist: pandas==2.2.1
49 | Requires-Dist: patsy==0.5.6
50 | Requires-Dist: pillow==10.2.0
51 | Requires-Dist: pkginfo==1.10.0
52 | Requires-Dist: plotly==5.20.0
53 | Requires-Dist: pluggy==1.4.0
54 | Requires-Dist: py-agata==0.0.8
55 | Requires-Dist: Pygments==2.17.2
56 | Requires-Dist: pyparsing==3.1.2
57 | Requires-Dist: pyproject_hooks==1.0.0
58 | Requires-Dist: pytest==8.1.1
59 | Requires-Dist: pytest-cov==5.0.0
60 | Requires-Dist: python-dateutil==2.9.0.post0
61 | Requires-Dist: pytz==2024.1
62 | Requires-Dist: readme_renderer==43.0
63 | Requires-Dist: requests==2.31.0
64 | Requires-Dist: requests-toolbelt==1.0.0
65 | Requires-Dist: rfc3986==2.0.0
66 | Requires-Dist: rich==13.7.1
67 | Requires-Dist: scikit-learn==1.4.1.post1
68 | Requires-Dist: scipy==1.12.0
69 | Requires-Dist: seaborn==0.13.2
70 | Requires-Dist: six==1.16.0
71 | Requires-Dist: statsmodels==0.14.1
72 | Requires-Dist: tenacity==8.2.3
73 | Requires-Dist: threadpoolctl==3.4.0
74 | Requires-Dist: tqdm==4.66.2
75 | Requires-Dist: twine==5.0.0
76 | Requires-Dist: tzdata==2024.1
77 | Requires-Dist: urllib3==2.2.1
78 | Requires-Dist: zipp==3.18.1
79 |
80 | # ReplayBG
81 |
82 |  83 |
84 | [](https://github.com/gcappon/py_replay_bg/COPYING)
85 | [](https://github.com/gcappon/py_replay_bg/commits/master)
86 |
87 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
88 |
89 | # Reference
90 |
91 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
92 |
93 | # Get started
94 |
95 | ## Installation
96 |
97 | **ReplayBG** can be installed via pypi by simply
98 |
99 | ```python
100 | pip install py-replay-bg
101 | ```
102 |
103 | ### Requirements
104 |
105 | * Python >= 3.11
106 | * List of Python packages in `requirements.txt`
107 |
108 | ## Preparation: imports, setup, and data loading
109 |
110 | First of all import the core modules:
111 | ```python
112 | import os
113 | import numpy as np
114 | import pandas as pd
115 |
116 | from multiprocessing import freeze_support
117 | ```
118 |
119 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
120 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
121 | parallelized way.
122 |
123 | Then, we will import the necessary ReplayBG modules:
124 | ```python
125 | from py_replay_bg.py_replay_bg import ReplayBG
126 | from py_replay_bg.visualizer import Visualizer
127 | from py_replay_bg.analyzer import Analyzer
128 | ```
129 |
130 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
131 | while `Analyzer` and `Visualizer` are utility objects that will be used to
132 | respectively analyze and visualize the results that we will produce with ReplayBG
133 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
134 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
135 |
136 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
137 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
138 | ```python
139 | if __name__ == '__main__':
140 | freeze_support()
141 | ```
142 |
143 | Then, we will set the verbosity of ReplayBG:
144 | ```python
145 | verbose = True
146 | plot_mode = False
147 | ```
148 |
149 | Then, we need to decide what blueprint to use for twinning the data at hand.
150 | ```python
151 | blueprint = 'multi-meal'
152 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
153 | parallelize = True
154 | ```
155 |
156 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
157 |
158 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
159 | which contains a day of data of a patient with T1D:
160 |
161 | ```python
162 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
163 | data.t = pd.to_datetime(data['t'])
164 | ```
165 |
166 | ::: warning
167 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
168 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
169 | :::
170 |
171 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
172 |
173 | ```python
174 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
175 | p = np.where(patient_info['patient'] == 1)[0][0]
176 | # Set bw and u2ss
177 | bw = float(patient_info.bw.values[p])
178 | u2ss = float(patient_info.u2ss.values[p])
179 | ```
180 |
181 | Finally, instantiate a `ReplayBG` object:
182 |
183 | ```python
184 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
185 | yts=5, exercise=False,
186 | seed=1,
187 | verbose=verbose, plot_mode=plot_mode)
188 |
189 | ```
190 |
191 | ## Step 1: Creation of the digital twin
192 |
193 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
194 |
195 | ```python
196 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
197 | twinning_method='mcmc',
198 | parallelize=parallelize,
199 | n_steps=5000,
200 | u2ss=u2ss)
201 | ```
202 |
203 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
204 |
205 |
206 | ## Step 2: Run replay simulations
207 |
208 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
209 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
210 |
211 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
212 |
213 | ```python
214 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
215 | twinning_method='mcmc',
216 | save_workspace=True,
217 | save_suffix='_step_2a')
218 | ```
219 |
220 | It is possible to visualize the results of the simulation using:
221 |
222 | ```python
223 | Visualizer.plot_replay_results(replay_results, data=data)
224 | ```
225 |
226 | and analyzing the results using:
227 |
228 | ```python
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
231 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
232 | ```
233 |
234 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
235 | To do that run:
236 |
237 | ```python
238 | data.bolus = data.bolus * .7
239 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
240 | twinning_method='mcmc',
241 | save_workspace=True,
242 | save_suffix='_step_2b')
243 |
244 | # Visualize results
245 | Visualizer.plot_replay_results(replay_results)
246 | # Analyze results
247 | analysis = Analyzer.analyze_replay_results(replay_results)
248 |
249 | # Print, for example, the average glucose
250 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
251 | ```
252 |
253 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
254 |
255 | # Documentation
256 |
257 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
258 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/SOURCES.txt:
--------------------------------------------------------------------------------
1 | COPYING.md
2 | README.md
3 | pyproject.toml
4 | requirements.txt
5 | py_replay_bg/__init__.py
6 | py_replay_bg/py_replay_bg.py
7 | py_replay_bg.egg-info/PKG-INFO
8 | py_replay_bg.egg-info/SOURCES.txt
9 | py_replay_bg.egg-info/dependency_links.txt
10 | py_replay_bg.egg-info/requires.txt
11 | py_replay_bg.egg-info/top_level.txt
12 | py_replay_bg/analyzer/__init__.py
13 | py_replay_bg/data/__init__.py
14 | py_replay_bg/dss/__init__.py
15 | py_replay_bg/dss/default_dss_handlers.py
16 | py_replay_bg/environment/__init__.py
17 | py_replay_bg/example/code/analysis_example.py
18 | py_replay_bg/example/code/analysis_intervals_example.py
19 | py_replay_bg/example/code/get_started.py
20 | py_replay_bg/example/code/replay_intervals_map.py
21 | py_replay_bg/example/code/replay_intervals_mcmc.py
22 | py_replay_bg/example/code/replay_map.py
23 | py_replay_bg/example/code/replay_map_dss.py
24 | py_replay_bg/example/code/replay_mcmc.py
25 | py_replay_bg/example/code/replay_mcmc_dss.py
26 | py_replay_bg/example/code/twin_intervals_map.py
27 | py_replay_bg/example/code/twin_intervals_mcmc.py
28 | py_replay_bg/example/code/twin_map.py
29 | py_replay_bg/example/code/twin_map_extended.py
30 | py_replay_bg/example/code/twin_mcmc.py
31 | py_replay_bg/example/code/twin_mcmc_extended.py
32 | py_replay_bg/example/code/utils.py
33 | py_replay_bg/input_validation/__init__.py
34 | py_replay_bg/input_validation/input_validator_init.py
35 | py_replay_bg/input_validation/input_validator_replay.py
36 | py_replay_bg/input_validation/input_validator_twin.py
37 | py_replay_bg/model/__init__.py
38 | py_replay_bg/model/logpriors_t1d.py
39 | py_replay_bg/model/model_parameters_t1d.py
40 | py_replay_bg/model/model_step_equations_t1d.py
41 | py_replay_bg/model/t1d_model_multi_meal.py
42 | py_replay_bg/model/t1d_model_single_meal.py
43 | py_replay_bg/replay/__init__.py
44 | py_replay_bg/sensors/__init__.py
45 | py_replay_bg/tests/__init__.py
46 | py_replay_bg/tests/test_analysis_example.py
47 | py_replay_bg/tests/test_analysis_intervals_example.py
48 | py_replay_bg/tests/test_replay_intervals_map.py
49 | py_replay_bg/tests/test_replay_intervals_mcmc.py
50 | py_replay_bg/tests/test_replay_map.py
51 | py_replay_bg/tests/test_replay_map_dss.py
52 | py_replay_bg/tests/test_replay_map_extended.py
53 | py_replay_bg/tests/test_replay_mcmc.py
54 | py_replay_bg/tests/test_replay_mcmc_dss.py
55 | py_replay_bg/tests/test_replay_mcmc_extended.py
56 | py_replay_bg/tests/test_twin_intervals_map.py
57 | py_replay_bg/tests/test_twin_intervals_mcmc.py
58 | py_replay_bg/tests/test_twin_map.py
59 | py_replay_bg/tests/test_twin_map_extended.py
60 | py_replay_bg/tests/test_twin_mcmc.py
61 | py_replay_bg/tests/test_twin_mcmc_extended.py
62 | py_replay_bg/twinning/__init__.py
63 | py_replay_bg/twinning/map.py
64 | py_replay_bg/twinning/mcmc.py
65 | py_replay_bg/utils/__init__.py
66 | py_replay_bg/utils/stats.py
67 | py_replay_bg/visualizer/__init__.py
68 | venv/bin/rst2html.py
69 | venv/bin/rst2html4.py
70 | venv/bin/rst2html5.py
71 | venv/bin/rst2latex.py
72 | venv/bin/rst2man.py
73 | venv/bin/rst2odt.py
74 | venv/bin/rst2odt_prepstyles.py
75 | venv/bin/rst2pseudoxml.py
76 | venv/bin/rst2s5.py
77 | venv/bin/rst2xetex.py
78 | venv/bin/rst2xml.py
79 | venv/bin/rstpep2html.py
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/dependency_links.txt:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/requires.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/top_level.txt:
--------------------------------------------------------------------------------
1 | py_replay_bg
2 |
--------------------------------------------------------------------------------
/py_replay_bg/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/dss/__init__.py:
--------------------------------------------------------------------------------
1 | from typing import Callable, Dict
2 |
3 | from py_replay_bg.dss.default_dss_handlers import default_meal_generator_handler, standard_bolus_calculator_handler, \
4 | default_basal_handler, ada_hypotreatments_handler, corrects_above_250_handler
5 |
6 |
7 | class DSS:
8 | """
9 | A class that represents the hyperparameters of the integrated decision support system.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | bw: double
15 | The patient's body weight.
16 | meal_generator_handler: Callable
17 | A callback function that implements a meal generator to be used during the replay of a given scenario.
18 | meal_generator_handler_params: dict
19 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
20 | bolus_calculator_handler: Callable
21 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
22 | bolus_calculator_handler_params: dict
23 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
24 | serves as memory area for the bolusCalculatorHandler function.
25 | basal_handler: Callable
26 | A callback function that implements a basal controller to be used during the replay of a given scenario.
27 | basal_handler_params: dict
28 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
29 | memory area for the basalHandler function.
30 | enable_hypotreatments: boolean
31 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
32 | hypotreatments_handler: Callable
33 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
34 | hypotreatments_handler_params: dict
35 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
36 | serves as memory area for the hypoTreatmentsHandler function.
37 | enable_correction_boluses: boolean
38 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
39 | correction_boluses_handler: Callable
40 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
41 | correction_boluses_handler_params: dict
42 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also
43 | serves as memory area for the correctionBolusesHandler function.
44 |
45 | Methods
46 | -------
47 | None
48 | """
49 |
50 | def __init__(self,
51 | bw: float,
52 | meal_generator_handler: Callable = default_meal_generator_handler,
53 | meal_generator_handler_params: Dict | None = None,
54 | bolus_calculator_handler: Callable = standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params: Dict | None = None,
56 | basal_handler: Callable = default_basal_handler,
57 | basal_handler_params: Dict | None = None,
58 | enable_hypotreatments: bool = False,
59 | hypotreatments_handler: Callable = ada_hypotreatments_handler,
60 | hypotreatments_handler_params: Dict | None = None,
61 | enable_correction_boluses: bool = False,
62 | correction_boluses_handler: Callable = corrects_above_250_handler,
63 | correction_boluses_handler_params: Dict | None = None,
64 | ):
65 | """
66 | Constructs all the necessary attributes for the DSS object.
67 |
68 | Parameters
69 | ----------
70 | bw: double
71 | The patient's body weight.
72 | meal_generator_handler: Callable, optional, default : default_meal_generator_handler
73 | A callback function that implements a meal generator to be used during the replay of a given scenario.
74 | meal_generator_handler_params: dict, optional, default : None
75 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
76 | bolus_calculator_handler: Callable, optional, default : standard_bolus_calculator_handler
77 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
78 | bolus_calculator_handler_params: dict, optional, default : None
79 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
80 | serves as memory area for the bolusCalculatorHandler function.
81 | basal_handler: function, Callable, default : default_basal_handler
82 | A callback function that implements a basal controller to be used during the replay of a given scenario.
83 | basal_handler_params: dict, optional, default : None
84 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
85 | memory area for the basalHandler function.
86 | enable_hypotreatments: boolean, optional, default : False
87 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
88 | hypotreatments_handler: Callable, optional, default : ada_hypotreatments_handler
89 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
90 | hypotreatments_handler_params: dict, optional, default : None
91 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
92 | serves as memory area for the hypoTreatmentsHandler function.
93 | enable_correction_boluses: boolean, optional, default : False
94 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
95 | correction_boluses_handler: Callable, optional, default : corrects_above_250_handler
96 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
97 | correction_boluses_handler_params: dict, optional, default : None
98 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function.
99 | It also serves as memory area for the correctionBolusesHandler function.
100 | """
101 |
102 | # Patient's body weight
103 | self.bw = bw
104 |
105 | # Meal Generator module parameters
106 | self.meal_generator_handler = meal_generator_handler
107 | self.meal_generator_handler_params = meal_generator_handler_params if meal_generator_handler_params is not None else {}
108 |
109 | # Bolus Calculator module parameters
110 | self.bolus_calculator_handler = bolus_calculator_handler
111 | self.bolus_calculator_handler_params = bolus_calculator_handler_params if bolus_calculator_handler_params is not None else {}
112 |
113 | # Basal module parameters
114 | self.basal_handler = basal_handler
115 | self.basal_handler_params = basal_handler_params if basal_handler_params is not None else {}
116 |
117 | # Hypotreatment module parameters
118 | self.enable_hypotreatments = enable_hypotreatments
119 | self.hypotreatments_handler = hypotreatments_handler
120 | self.hypotreatments_handler_params = hypotreatments_handler_params if hypotreatments_handler_params is not None else {}
121 |
122 | # Correction bolus module parameters
123 | self.enable_correction_boluses = enable_correction_boluses
124 | self.correction_boluses_handler = correction_boluses_handler
125 | self.correction_boluses_handler_params = correction_boluses_handler_params if correction_boluses_handler_params is not None else {}
126 |
--------------------------------------------------------------------------------
/py_replay_bg/environment/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 |
4 | class Environment:
5 | """
6 | A class that represents the hyperparameters to be used by ReplayBG.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | save_name : str
12 | A string used to label, thus identify, each output file and result.
13 | replay_bg_path: str
14 | The current absolute path of ReplayBG.
15 |
16 | blueprint: str, {'single-meal', 'multi-meal'}
17 | A string that specifies the blueprint to be used to create the digital twin.
18 | yts: int
19 | An integer that specifies the data sampling time (in minutes).
20 |
21 | seed : int
22 | An integer that specifies the random seed. For reproducibility.
23 |
24 | plot_mode : bool
25 | A boolean that specifies whether to show the plot of the results or not.
26 | verbose : bool
27 | A boolean that specifies the verbosity of ReplayBG.
28 |
29 | Methods
30 | -------
31 | None
32 | """
33 |
34 | def __init__(self,
35 | blueprint: str = 'single_meal',
36 | save_name: str = '',
37 | save_folder: str = '',
38 | yts: int = 5,
39 | exercise: bool = False,
40 | seed: int = 1,
41 | plot_mode: bool = True,
42 | verbose: bool = True
43 | ):
44 | """
45 | Constructs all the necessary attributes for the Environment object.
46 |
47 | Parameters
48 | ----------
49 | blueprint: str, {'single-meal', 'multi-meal'}, optional, default : 'single-meal'
50 | A string that specifies the blueprint to be used to create the digital twin.
51 |
52 | save_name : str, optional, default : ''
53 | A string used to label, thus identify, each output file and result.
54 | save_folder : str, optional, default : ''
55 | A string used to set the folder where the ReplayBG results will be saved.
56 |
57 | yts: int, optional, default : 5
58 | An integer that specifies the data sampling time (in minutes).
59 | exercise: boolean, optional, default : False
60 | A boolean that specifies whether to simulate exercise or not.
61 |
62 | seed : int, optional, default : 1
63 | An integer that specifies the random seed. For reproducibility.
64 |
65 | plot_mode : boolean, optional, default : True
66 | A boolean that specifies whether to show the plot of the results or not.
67 | verbose : boolean, optional, default : True
68 | A boolean that specifies the verbosity of ReplayBG.
69 | """
70 |
71 | # Set the save name and folder
72 | self.save_name = save_name
73 | self.replay_bg_path = save_folder
74 |
75 | # Create the results sub folders if they do not exist
76 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results'))):
77 | os.mkdir(os.path.join(self.replay_bg_path, 'results'))
78 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'mcmc'))):
79 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'mcmc'))
80 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'map'))):
81 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'map'))
82 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'workspaces'))):
83 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'workspaces'))
84 |
85 | # Single-meal or multi-meal blueprint?
86 | self.blueprint = blueprint
87 |
88 | # Set sample time
89 | self.yts = yts
90 |
91 | # Set whether to use the exercise model
92 | self.exercise = exercise
93 |
94 | # Set the seed
95 | self.seed = seed
96 |
97 | # Set plot mode and verbosity
98 | self.plot_mode = plot_mode
99 | self.verbose = verbose
100 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the location of the 'results' folder
9 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
10 |
11 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
12 | replay_results = pickle.load(file)
13 |
14 | # Visualize and analyze MCMC results
15 | Visualizer.plot_replay_results(replay_results)
16 | analysis = Analyzer.analyze_replay_results(replay_results)
17 | print(" ----- Analysis using MCMC-derived twins ----- ")
18 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
19 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
20 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
21 |
22 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
23 | replay_results = pickle.load(file)
24 |
25 | # Visualize and analyze MAP results
26 | Visualizer.plot_replay_results(replay_results)
27 | analysis = Analyzer.analyze_replay_results(replay_results)
28 | print(" ----- Analysis using MAP-derived twins ----- ")
29 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
30 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
31 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
32 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the interval to analyze
9 | start_day = 1
10 | end_day = 2
11 |
12 | # Set the location of the 'results' folder
13 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
14 |
15 | # Initialize results list
16 | replay_results_interval = []
17 |
18 | for day in range(start_day, end_day+1):
19 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
20 | replay_results = pickle.load(file)
21 | replay_results_interval.append(replay_results)
22 |
23 | # Visualize and analyze MCMC results
24 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
25 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
26 | print(" ----- Analysis using MAP-derived twins ----- ")
27 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
28 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
29 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
30 |
31 | # Initialize results list
32 | replay_results_interval = []
33 |
34 | for day in range(start_day, end_day + 1):
35 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
36 | replay_results = pickle.load(file)
37 | replay_results_interval.append(replay_results)
38 |
39 | # Visualize and analyze MAP results
40 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
41 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
42 | print(" ----- Analysis using MCMC-derived twins ----- ")
43 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
44 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
45 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
46 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/get_started.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | import pandas as pd
4 |
5 | from multiprocessing import freeze_support
6 |
7 | from py_replay_bg.py_replay_bg import ReplayBG
8 | from py_replay_bg.visualizer import Visualizer
9 | from py_replay_bg.analyzer import Analyzer
10 |
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # Load data
25 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
26 | data.t = pd.to_datetime(data['t'])
27 |
28 | # Load patient_info
29 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Set save name
42 | save_name = 'data_day_1'
43 |
44 | # Step 1. Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=5000,
49 | u2ss=u2ss)
50 |
51 | # Step 2a. Replay the twin with the same input data to get the initial conditions for the subsequent day
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_step_2a')
56 |
57 | # Visualize results and compare with the original glucose data
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | # Analyze results
60 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
61 | # Print, for example, the fit MARD and the average glucose
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
63 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
64 |
65 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
66 | data.bolus = data.bolus * .7
67 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
68 | twinning_method='mcmc',
69 | save_workspace=True,
70 | u2ss=u2ss,
71 | save_suffix='_step_2b')
72 |
73 | # Visualize results
74 | Visualizer.plot_replay_results(replay_results)
75 | # Analyze results
76 | analysis = Analyzer.analyze_replay_results(replay_results)
77 |
78 | # Print, for example, the average glucose
79 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='map',
53 | save_workspace=True,
54 | x0=x0, previous_data_name=previous_data_name,
55 | save_suffix='_replay_map',
56 | sensors=sensors)
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='mcmc',
53 | n_replay=100,
54 | save_workspace=True,
55 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
56 | save_suffix='_replay_mcmc')
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | twinning_method='map',
39 | save_workspace=True,
40 | save_suffix='_replay_map')
41 |
42 | # Visualize and analyze results
43 | Visualizer.plot_replay_results(replay_results, data=data)
44 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
45 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
46 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
47 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
48 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | n_replay=10,
39 | twinning_method='mcmc',
40 | save_workspace=True,
41 | save_suffix='_replay_mcmc')
42 |
43 | # Visualize and analyze results
44 | Visualizer.plot_replay_results(replay_results, data=data)
45 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
46 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
47 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
48 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
49 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='mcmc',
50 | save_workspace=True,
51 | save_suffix='_replay_mcmc_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 | x0 = None
32 | previous_data_name = None
33 |
34 | # Initialize the list of results
35 | replay_results_interval = []
36 | data_interval = []
37 |
38 | # Instantiate ReplayBG
39 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
40 | yts=5, exercise=False,
41 | seed=1,
42 | verbose=verbose, plot_mode=plot_mode)
43 |
44 | # Set interval to twin
45 | start_day = 1
46 | end_day = 2
47 |
48 | # Twin the interval
49 | for day in range(start_day, end_day+1):
50 |
51 | # Step 1: Load data and set save_name
52 | data = load_test_data(day=day)
53 | save_name = 'data_day_' + str(day) + '_interval'
54 |
55 | print("Twinning " + save_name)
56 |
57 | # Run twinning procedure
58 | rbg.twin(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | parallelize=parallelize,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='map',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_map')
69 | # Append results
70 | replay_results_interval.append(replay_results)
71 | data_interval.append(data)
72 |
73 | # Set initial conditions for next day equal to the "ending conditions" of the current day
74 | x0 = replay_results['x_end']['realizations'][0].tolist()
75 |
76 | # Set previous_data_name
77 | previous_data_name = save_name
78 |
79 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
80 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
82 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 | x0 = None
35 | previous_data_name = None
36 |
37 | # Initialize the list of results
38 | replay_results_interval = []
39 | data_interval = []
40 |
41 | # Instantiate ReplayBG
42 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
43 | yts=5, exercise=False,
44 | seed=1,
45 | verbose=verbose, plot_mode=plot_mode)
46 |
47 | # Set interval to twin
48 | start_day = 1
49 | end_day = 2
50 |
51 | # Twin the interval
52 | for day in range(start_day, end_day+1):
53 |
54 | # Step 1: Load data and set save_name
55 | data = load_test_data(day=day)
56 | save_name = 'data_day_' + str(day) + '_interval'
57 |
58 | print("Twinning " + save_name)
59 |
60 | # Run twinning procedure
61 | rbg.twin(data=data, bw=bw, save_name=save_name,
62 | twinning_method='mcmc',
63 | parallelize=parallelize,
64 | n_steps=n_steps,
65 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
66 |
67 | # Replay the twin with the same input data
68 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
69 | twinning_method='mcmc',
70 | save_workspace=True,
71 | x0=x0, previous_data_name=previous_data_name,
72 | save_suffix='_twin_mcmc')
73 | # Append results
74 | replay_results_interval.append(replay_results)
75 | data_interval.append(data)
76 |
77 | # Set initial conditions for next day equal to the "ending conditions" of the current day
78 | x0 = replay_results['x_end']['realizations'][0].tolist()
79 |
80 | # Set previous_data_name
81 | previous_data_name = save_name
82 |
83 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
84 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
85 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
86 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data(day=1)
39 | save_name = 'data_day_' + str(1)
40 |
41 | print("Twinning " + save_name)
42 |
43 | # Run twinning procedure
44 | rbg.twin(data=data, bw=bw, save_name=save_name,
45 | twinning_method='map',
46 | parallelize=parallelize,
47 | u2ss=u2ss)
48 |
49 | # Replay the twin with the same input data
50 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
51 | twinning_method='map',
52 | save_workspace=True,
53 | save_suffix='_twin_map')
54 |
55 | # Visualize and analyze results
56 | Visualizer.plot_replay_results(replay_results, data=data)
57 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
58 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
59 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data_extended(day=1)
39 | # Cut data up to 11:00
40 | data = data.iloc[0:348, :]
41 | # Modify label of second day
42 | data.loc[312, "cho_label"] = 'B2'
43 |
44 | save_name = 'data_day_' + str(1) + '_extended'
45 |
46 | # Run twinning procedure using also the burn-out period
47 | rbg.twin(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | parallelize=parallelize,
50 | u2ss=u2ss,
51 | extended=True,
52 | find_start_guess_first=True)
53 |
54 | # Cut data up to 4:00
55 | data = data.iloc[0:264, :]
56 |
57 | # Replay the twin with the data up to the start of the burn-out period
58 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | save_workspace=True,
61 | save_suffix='_twin_map_extended')
62 |
63 | # Visualize and analyze results
64 | Visualizer.plot_replay_results(replay_results, data=data)
65 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
66 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
67 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data(day=1)
43 | save_name = 'data_day_' + str(1)
44 |
45 | print("Twinning " + save_name)
46 |
47 | # Run twinning procedure
48 | rbg.twin(data=data, bw=bw, save_name=save_name,
49 | twinning_method='mcmc',
50 | parallelize=parallelize,
51 | n_steps=n_steps,
52 | u2ss=u2ss)
53 |
54 | # Replay the twin with the same input data
55 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
56 | twinning_method='mcmc',
57 | save_workspace=True,
58 | save_suffix='_twin_mcmc')
59 |
60 | Visualizer.plot_replay_results(replay_results, data=data)
61 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data_extended(day=1)
43 | # Cut data up to 11:00
44 | data = data.iloc[0:348, :]
45 | # Modify label of second day
46 | data.loc[312, "cho_label"] = 'B2'
47 |
48 | save_name = 'data_day_' + str(1) + '_extended'
49 |
50 | print("Twinning " + save_name)
51 |
52 | # Run twinning procedure using also the burn-out period
53 | rbg.twin(data=data, bw=bw, save_name=save_name,
54 | twinning_method='mcmc',
55 | parallelize=parallelize,
56 | n_steps=n_steps,
57 | u2ss=u2ss,
58 | extended=True,
59 | find_start_guess_first=True)
60 |
61 | # Cut data up to 4:00
62 | data = data.iloc[0:264, :]
63 |
64 | # Replay the twin with the data up to the start of the burn-out period
65 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
66 | twinning_method='mcmc',
67 | save_workspace=True,
68 | save_suffix='_twin_mcmc_extended')
69 |
70 | Visualizer.plot_replay_results(replay_results, data=data)
71 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
72 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/utils.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import os
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
20 | return df
21 |
--------------------------------------------------------------------------------
/py_replay_bg/example/data/patient_info.csv:
--------------------------------------------------------------------------------
1 | patient,u2ss,bw
2 | 1,0.2440484700275897,100
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_init.py:
--------------------------------------------------------------------------------
1 | from py_replay_bg.input_validation import *
2 |
3 |
4 | class InputValidatorInit:
5 | """
6 | Class for validating the input of ReplayBG __init__ method.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | blueprint: str, {'single-meal', 'multi-meal'}
12 | A string that specifies the blueprint to be used to create the digital twin.
13 | save_folder : str
14 | A string defining the folder that will contain the results.
15 |
16 | yts: int, optional, default : 5
17 | An integer that specifies the data sample time (in minutes).
18 | exercise: boolean, optional, default : False
19 | A boolean that specifies whether to simulate exercise or not.
20 | seed: int, optional, default: 1
21 | An integer that specifies the random seed. For reproducibility.
22 |
23 | plot_mode : boolean, optional, default : True
24 | A boolean that specifies whether to show the plot of the results or not.
25 | verbose : boolean, optional, default : True
26 | A boolean that specifies the verbosity of ReplayBG.
27 |
28 | Methods
29 | -------
30 | validate():
31 | Run the input validation process.
32 | """
33 |
34 | def __init__(self,
35 | save_folder: str,
36 | blueprint: str,
37 | yts: int,
38 | exercise: bool,
39 | seed: int,
40 | plot_mode: bool,
41 | verbose: bool,
42 | ):
43 | self.save_folder = save_folder
44 | self.blueprint = blueprint
45 | self.yts = yts
46 | self.exercise = exercise
47 | self.seed = seed
48 | self.plot_mode = plot_mode
49 | self.verbose = verbose
50 |
51 | def validate(self):
52 | """
53 | Run the input validation process.
54 | """
55 |
56 | # Validate the 'save_folder' input
57 | SaveFolderValidator(save_folder=self.save_folder).validate()
58 |
59 | # Validate the 'blueprint' input
60 | BlueprintValidator(blueprint=self.blueprint).validate()
61 |
62 | # Validate the 'yts' input
63 | YTSValidator(yts=self.yts).validate()
64 |
65 | # Validate the 'exercise' input
66 | ExerciseValidator(exercise=self.exercise).validate()
67 |
68 | # Validate the 'seed' input
69 | SeedValidator(seed=self.seed).validate()
70 |
71 | # Validate the 'plot_mode' input
72 | PlotModeValidator(plot_mode=self.plot_mode).validate()
73 |
74 | # Validate the 'verbose' input
75 | VerboseValidator(verbose=self.verbose).validate()
76 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_replay.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from typing import Callable, Dict
5 |
6 | from py_replay_bg.input_validation import *
7 |
8 |
9 | class InputValidatorReplay:
10 | """
11 | Class for validating the input of ReplayBG twin method.
12 |
13 | ...
14 | Attributes
15 | ----------
16 | data : pd.DataFrame
17 | Pandas dataframe which contains the data to be used by the tool.
18 | bw : float
19 | The patient's body weight.
20 | save_name : str
21 | A string used to label, thus identify, each output file and result.
22 |
23 | x0: list
24 | The initial model state.
25 | previous_data_name: str
26 | The name of the previous portion of data. To be used to initialize the initial conditions.
27 |
28 | twinning_method : str
29 | The method used to twin the model.
30 |
31 | bolus_source : str
32 | A string defining whether to use, during replay, the insulin bolus data contained in the 'data' timetable
33 | (if 'data'), or the boluses generated by the bolus calculator implemented via the provided
34 | 'bolusCalculatorHandler' function.
35 | basal_source : str
36 | A string defining whether to use, during replay, the insulin basal data contained in the 'data' timetable
37 | (if 'data'), or the basal generated by the controller implemented via the provided
38 | 'basalControllerHandler' function (if 'dss'), or fixed to the average basal rate used during
39 | twinning (if 'u2ss').
40 | cho_source : str
41 | A string defining whether to use, during replay, the CHO data contained in the 'data' timetable (if 'data'),
42 | or the CHO generated by the meal generator implemented via the provided 'mealGeneratorHandler' function.
43 |
44 | meal_generator_handler: Callable
45 | A callback function that implements a meal generator to be used during the replay of a given scenario.
46 | meal_generator_handler_params: dict
47 | A dictionary that contains the parameters to pass to the meal_generator_handler function.
48 | bolus_calculator_handler: Callable
49 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
50 | bolus_calculator_handler_params: dict
51 | A dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also serves
52 | as memory area for the bolusCalculatorHandler function.
53 | basal_handler: Callable
54 | A callback function that implements a basal controller to be used during the replay of a given scenario.
55 | basal_handler_params: dict
56 | A dictionary that contains the parameters to pass to the basalHandler function. It also serves as memory
57 | area for the basalHandler function.
58 | enable_hypotreatments: boolean
59 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
60 | hypotreatments_handler: Callable
61 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
62 | hypotreatments_handler_params: dict
63 | A dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also serves
64 | as memory area for the hypoTreatmentsHandler function.
65 | enable_correction_boluses: boolean
66 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
67 | correction_boluses_handler: Callable
68 | A callback function that implements a corrective bolus strategy during the replay of a given scenario.
69 | correction_boluses_handler_params: dict
70 | A dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also serves
71 | as memory area for the correctionBolusesHandler function.
72 |
73 | save_suffix : string
74 | A string to be attached as suffix to the resulting output files' name.
75 | save_workspace: bool
76 | A flag that specifies whether to save the resulting workspace.
77 |
78 | n_replay: int
79 | The number of Monte Carlo replays to be performed. Ignored if twinning_method is 'map'.
80 | sensors: list[Sensors]
81 | The sensors to be used in each of the replay simulations.
82 |
83 | blueprint: str
84 | A string that specifies the blueprint to be used to create the digital twin.
85 | exercise: bool
86 | A boolean that specifies whether to use exercise model or not.
87 |
88 | Methods
89 | -------
90 | validate():
91 | Run the input validation process.
92 | """
93 |
94 | def __init__(self,
95 | data: pd.DataFrame,
96 | bw: float,
97 | save_name: str,
98 | x0: np.ndarray,
99 | previous_data_name: str,
100 | twinning_method: str,
101 | bolus_source: str,
102 | basal_source: str,
103 | cho_source: str,
104 | meal_generator_handler: Callable,
105 | meal_generator_handler_params: Dict,
106 | bolus_calculator_handler: Callable,
107 | bolus_calculator_handler_params: Dict,
108 | basal_handler: Callable,
109 | basal_handler_params: Dict,
110 | enable_hypotreatments: bool,
111 | hypotreatments_handler: Callable,
112 | hypotreatments_handler_params: Dict,
113 | enable_correction_boluses: bool,
114 | correction_boluses_handler: Callable,
115 | correction_boluses_handler_params: Dict,
116 | save_suffix: str,
117 | save_workspace: bool,
118 | n_replay: int,
119 | sensors: list,
120 | blueprint: str,
121 | exercise: bool,
122 | ):
123 | self.data = data
124 | self.bw = bw
125 | self.save_name = save_name
126 | self.x0 = x0
127 | self.previous_data_name = previous_data_name
128 | self.twinning_method = twinning_method
129 | self.bolus_source = bolus_source
130 | self.basal_source = basal_source
131 | self.cho_source = cho_source
132 | self.meal_generator_handler = meal_generator_handler
133 | self.meal_generator_handler_params = meal_generator_handler_params
134 | self.bolus_calculator_handler = bolus_calculator_handler
135 | self.bolus_calculator_handler_params = bolus_calculator_handler_params
136 | self.basal_handler = basal_handler
137 | self.basal_handler_params = basal_handler_params
138 | self.enable_hypotreatments = enable_hypotreatments
139 | self.hypotreatments_handler = hypotreatments_handler
140 | self.hypotreatments_handler_params = hypotreatments_handler_params
141 | self.enable_correction_boluses = enable_correction_boluses
142 | self.correction_boluses_handler = correction_boluses_handler
143 | self.correction_boluses_handler_params = correction_boluses_handler_params
144 | self.save_suffix = save_suffix
145 | self.save_workspace = save_workspace
146 | self.n_replay = n_replay
147 | self.sensors = sensors
148 | self.blueprint = blueprint
149 | self.exercise = exercise
150 |
151 |
152 | def validate(self):
153 | """
154 | Run the input validation process.
155 | """
156 |
157 | # Validate the 'data' input
158 | DataValidator(modality='replay', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
159 | bolus_source=self.bolus_source, basal_source=self.basal_source, cho_source=self.cho_source).validate()
160 |
161 | # Validate the 'bw' input
162 | BWValidator(bw=self.bw).validate()
163 |
164 | # Validate the 'save_name' input
165 | SaveNameValidator(save_name=self.save_name).validate()
166 |
167 | # Validate the 'save_name' input
168 | X0Validator(x0=self.x0).validate()
169 |
170 | # Validate the 'previous_data_name' input
171 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
172 |
173 | # Validate the 'twinning_method' input
174 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
175 |
176 | # Validate the 'bolus_source' input
177 | BolusSourceValidator(bolus_source=self.bolus_source).validate()
178 |
179 | # Validate the 'basal_source' input
180 | BasalSourceValidator(basal_source=self.basal_source).validate()
181 |
182 | # Validate the 'cho_source' input
183 | CHOSourceValidator(cho_source=self.cho_source).validate()
184 |
185 | # Validate the 'meal_generator_handler' input
186 | MealGeneratorHandlerValidator(meal_generator_handler=self.meal_generator_handler).validate()
187 |
188 | # Validate the 'meal_generator_handler_params' input
189 | MealGeneratorHandlerParamsValidator(meal_generator_handler_params=self.meal_generator_handler_params).validate()
190 |
191 | # Validate the 'bolus_calculator_handler' input
192 | BolusCalculatorHandlerValidator(bolus_calculator_handler=self.bolus_calculator_handler).validate()
193 |
194 | # Validate the 'bolus_calculator_handler_params' input
195 | BolusCalculatorHandlerParamsValidator(bolus_calculator_handler_params=self.bolus_calculator_handler_params).validate()
196 |
197 | # Validate the 'basal_handler' input
198 | BasalHandlerValidator(basal_handler=self.basal_handler).validate()
199 |
200 | # Validate the 'basal_handler_params' input
201 | BasalHandlerParamsValidator(basal_handler_params=self.basal_handler_params).validate()
202 |
203 | # Validate the 'enable_hypotreatments' input
204 | EnableHypotreatmentsValidator(enable_hypotreatments=self.enable_hypotreatments).validate()
205 |
206 | # Validate the 'hypotreatments_handler' input
207 | HypotreatmentsHandlerValidator(hypotreatments_handler=self.hypotreatments_handler).validate()
208 |
209 | # Validate the 'hypotreatments_handler_params' input
210 | HypotreatmentsHandlerParamsValidator(hypotreatments_handler_params=self.hypotreatments_handler_params).validate()
211 |
212 | # Validate the 'enable_correction_boluses' input
213 | EnableCorrectionBolusesValidator(enable_correction_boluses=self.enable_correction_boluses).validate()
214 |
215 | # Validate the 'correction_boluses_handler' input
216 | CorrectionBolusesHandlerValidator(correction_boluses_handler=self.correction_boluses_handler).validate()
217 |
218 | # Validate the 'correction_boluses_handler_params' input
219 | CorrectionBolusesHandlerParamsValidator(correction_boluses_handler_params=self.correction_boluses_handler_params).validate()
220 |
221 | # Validate the 'save_suffix' input
222 | SaveSuffixValidator(save_suffix=self.save_suffix).validate()
223 |
224 | # Validate the 'save_workspace' input
225 | SaveWorkspaceValidator(save_workspace=self.save_workspace).validate()
226 |
227 | # Validate the 'n_replay' input
228 | NReplayValidator(n_replay=self.n_replay).validate()
229 |
230 | # Validate the 'sensors' input
231 | SensorsValidator(sensors=self.sensors).validate()
232 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_twin.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from py_replay_bg.input_validation import *
5 |
6 |
7 | class InputValidatorTwin:
8 | """
9 | Class for validating the input of ReplayBG twin method.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | data: pd.DataFrame
15 | Pandas dataframe which contains the data to be used by the tool.
16 | bw: float
17 | The patient's body weight.
18 | save_name : str
19 | A string used to label, thus identify, each output file and result.
20 |
21 |
22 | u2ss : float
23 | The steady state of the basal insulin infusion.
24 | x0 : np.ndarray
25 | The initial model conditions.
26 | previous_data_name : str
27 | The name of the previous data portion. This is used to correctly "transfer" the initial model conditions to
28 | the current portion of data.
29 |
30 | twinning_method : str
31 | The method to be used to twin the model.
32 |
33 | extended : bool
34 | A flag indicating whether to use the "extended" model for twinning
35 | find_start_guess_first : bool
36 | A flag indicating whether to find the start guess using MAP before twinning.
37 |
38 | n_steps: int
39 | Number of steps to use for the main chain. This is ignored if modality is 'replay'.
40 | save_chains: bool, optional, default : False
41 | A flag that specifies whether to save the resulting mcmc chains and copula samplers.
42 |
43 | parallelize : boolean
44 | A boolean that specifies whether to parallelize the twinning process.
45 | n_processes : int, optional, default : None
46 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
47 |
48 | blueprint: str
49 | A string that specifies the blueprint to be used to create the digital twin.
50 | exercise: bool
51 | A boolean that specifies whether to use exercise model or not.
52 |
53 | Methods
54 | -------
55 | validate():
56 | Run the input validation process.
57 | """
58 |
59 | def __init__(self,
60 | data: pd.DataFrame,
61 | bw: float,
62 | save_name: str,
63 | twinning_method: str,
64 | extended: bool,
65 | find_start_guess_first: bool,
66 | n_steps: int,
67 | save_chains: bool,
68 | u2ss: float | None,
69 | x0: np.ndarray | None,
70 | previous_data_name: str | None,
71 | parallelize: bool,
72 | n_processes: int | None,
73 | blueprint: str,
74 | exercise: bool
75 | ):
76 | self.data = data
77 | self.bw = bw
78 | self.save_name = save_name
79 | self.twinning_method = twinning_method
80 | self.extended = extended
81 | self.find_start_guess_first = find_start_guess_first
82 | self.n_steps = n_steps
83 | self.save_chains = save_chains
84 | self.u2ss = u2ss
85 | self.x0 = x0
86 | self.previous_data_name = previous_data_name
87 | self.parallelize = parallelize
88 | self.n_processes = n_processes
89 | self.blueprint = blueprint
90 | self.exercise = exercise
91 |
92 | def validate(self):
93 | """
94 | Run the input validation process.
95 | """
96 |
97 | # Validate the 'data' input
98 | DataValidator(modality='twin', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
99 | bolus_source='data', basal_source='data', cho_source='data').validate()
100 |
101 | # Validate the 'bw' input
102 | BWValidator(bw=self.bw).validate()
103 |
104 | # Validate the 'save_name' input
105 | SaveNameValidator(save_name=self.save_name).validate()
106 |
107 | # Validate the 'twinning_method' input
108 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
109 |
110 | # Validate the 'extended' input
111 | ExtendedValidator(extended=self.extended).validate()
112 |
113 | # Validate the 'find_start_guess_first' input
114 | FindStartGuessFirstValidator(find_start_guess_first=self.find_start_guess_first).validate()
115 |
116 | # Validate the 'n_steps' input
117 | NStepsValidator(n_steps=self.n_steps).validate()
118 |
119 | # Validate the 'u2ss' input
120 | U2SSValidator(u2ss=self.u2ss).validate()
121 |
122 | # Validate the 'save_name' input
123 | X0Validator(x0=self.x0).validate()
124 |
125 | # Validate the 'previous_data_name' input
126 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
127 |
128 | # Validate the 'parallelize' input
129 | ParallelizeValidator(parallelize=self.parallelize).validate()
130 |
131 | # Validate the 'n_processes' input
132 | NProcessesValidator(n_processes=self.n_processes).validate()
133 |
--------------------------------------------------------------------------------
/py_replay_bg/model/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/model/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/model/model_parameters_t1d.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 |
4 |
5 | class ModelParametersT1D:
6 | def __init__(self,
7 | data: pd.DataFrame,
8 | bw: float,
9 | u2ss: float,
10 | extended: bool
11 | ):
12 |
13 | """
14 | Function that returns the default parameters values of the t1d model.
15 |
16 | Parameters
17 | ----------
18 | data : pd.DataFrame
19 | Pandas dataframe which contains the data to be used by the tool.
20 | bw : float
21 | The patient's body weight.
22 | u2ss : float
23 | The steady state of the basal insulin infusion
24 | extended : bool
25 | A flag indicating whether to use the "extended" model for twinning
26 |
27 | Returns
28 | -------
29 | model_parameters: dict
30 | A dictionary containing the default model parameters.
31 |
32 | Raises
33 | ------
34 | None
35 |
36 | See Also
37 | --------
38 | None
39 |
40 | Examples
41 | --------
42 | None
43 | """
44 | # Initial conditions
45 | self.Xpb = 0.0 # Insulin action initial condition
46 | self.Qgutb = 0.0 # Intestinal content initial condition
47 |
48 | # Glucose-insulin submodel parameters
49 | self.VG = 1.45 # dl/kg
50 | self.SG = 2.5e-2 # 1/min
51 | self.Gb = 119.13 # mg/dL
52 | self.r1 = 1.4407 # unitless
53 | self.r2 = 0.8124 # unitless
54 | self.alpha = 7 # 1/min
55 | self.p2 = 0.012 # 1/min
56 |
57 | if u2ss is None:
58 | self.u2ss = np.mean(data.basal) * 1000 / bw # mU/(kg*min)
59 | else:
60 | self.u2ss = u2ss
61 |
62 | # Subcutaneous insulin absorption submodel parameters
63 | self.VI = 0.126 # L/kg
64 | self.ke = 0.127 # 1/min
65 | self.kd = 0.026 # 1/min
66 | # model_parameters['ka1'] = 0.0034 # 1/min (virtually 0 in 77% of the cases)
67 | # self.ka1 = 0.0
68 | self.ka2 = 0.014 # 1/min
69 | self.tau = 8 # min
70 | self.Ipb = self.u2ss / self.ke # from eq. 5 steady-state
71 |
72 |
73 | # Oral glucose absorption submodel parameters
74 | self.kgri = 0.18 # = kmax % 1/min
75 | self.kempt = 0.18 # 1/min
76 | self.f = 0.9 # dimensionless
77 |
78 | # Exercise submodel parameters
79 | self.vo2rest = 0.33 # dimensionless, VO2rest was derived from heart rate round(66/(220-30))
80 | self.vo2max = 1 # dimensionless, VO2max is normalized and estimated from heart rate (220-age) = 100%.
81 | self.e1 = 1.6 # dimensionless
82 | self.e2 = 0.78 # dimensionless
83 |
84 | # Patient specific parameters
85 | self.bw = bw # kg
86 | self.to_g = self.bw / 1000
87 | self.to_mgkg = 1000 / self.bw
88 |
89 | # Measurement noise specifics
90 | self.SDn = 5
91 |
92 | # Glucose starting point
93 | if 'glucose' in data:
94 | idx = np.where(data.glucose.isnull().values == False)[0][0]
95 | self.G0 = data.glucose[idx]
96 | else:
97 | self.G0 = self.Gb
98 |
99 |
100 | class ModelParametersT1DMultiMeal(ModelParametersT1D):
101 |
102 | def __init__(self,
103 | data: pd.DataFrame,
104 | bw: float,
105 | u2ss: float,
106 | extended: bool,
107 | ):
108 |
109 | """
110 | Function that returns the default parameters values of the t1d model in a multi-meal blueprint.
111 |
112 | Parameters
113 | ----------
114 | data : pd.DataFrame
115 | Pandas dataframe which contains the data to be used by the tool.
116 | bw : float
117 | The patient's body weight.
118 | u2ss : float
119 | The steady state of the basal insulin infusion
120 | extended : bool
121 | A flag indicating whether to use the "extended" model for twinning
122 |
123 | Returns
124 | -------
125 | model_parameters: dict
126 | A dictionary containing the default model parameters.
127 |
128 | Raises
129 | ------
130 | None
131 |
132 | See Also
133 | --------
134 | None
135 |
136 | Examples
137 | --------
138 | None
139 | """
140 | # Initialize common parameters
141 | super().__init__(data, bw, u2ss, extended)
142 |
143 | # Glucose-insulin submodel parameters
144 | self.SI_B = 10.35e-4 / self.VG # mL/(uU*min)
145 | self.SI_L = 10.35e-4 / self.VG # mL/(uU*min)
146 | self.SI_D = 10.35e-4 / self.VG # mL/(uU*min)
147 |
148 | if extended:
149 | self.SI_B2 = 10.35e-4 / self.VG # mL/(uU*min)
150 |
151 | # Oral glucose absorption submodel parameters
152 | self.kabs_B = 0.012 # 1/min
153 | self.kabs_L = 0.012 # 1/min
154 | self.kabs_D = 0.012 # 1/min
155 | self.kabs_S = 0.012 # 1/min
156 | self.kabs_H = 0.012 # 1/min
157 | self.beta_B = 0 # min
158 | self.beta_L = 0 # min
159 | self.beta_D = 0 # min
160 | self.beta_S = 0 # min
161 |
162 | if extended:
163 | self.kabs_B2 = 0.012 # 1/min
164 | self.kabs_L2 = 0.012 # 1/min
165 | self.kabs_S2 = 0.012 # 1/min
166 | self.beta_B2 = 0 # min
167 | self.beta_L2 = 0 # min
168 | self.beta_S2 = 0 # min
169 |
170 |
171 | class ModelParametersT1DSingleMeal(ModelParametersT1D):
172 |
173 | def __init__(self,
174 | data: pd.DataFrame,
175 | bw: float,
176 | u2ss: float,
177 | ):
178 |
179 | """
180 | Function that returns the default parameters values of the t1d model in a single-meal blueprint.
181 |
182 | Parameters
183 | ----------
184 | data : pd.DataFrame
185 | Pandas dataframe which contains the data to be used by the tool.
186 | bw : float
187 | The patient's body weight.
188 | u2ss : float
189 | The steady state of the basal insulin infusion
190 |
191 | Returns
192 | -------
193 | model_parameters: dict
194 | A dictionary containing the default model parameters.
195 |
196 | Raises
197 | ------
198 | None
199 |
200 | See Also
201 | --------
202 | None
203 |
204 | Examples
205 | --------
206 | None
207 | """
208 | # Initialize common parameters
209 | super().__init__(data, bw, u2ss, extended=False)
210 |
211 | # Glucose-insulin submodel parameters
212 | self.SI = 10.35e-4 / self.VG # mL/(uU*min)
213 |
214 | # Oral glucose absorption submodel parameters
215 | self.kabs = 0.012 # 1/min
216 | self.beta = 0 # 1/min
217 |
--------------------------------------------------------------------------------
/py_replay_bg/sensors/__init__.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 |
4 | class CGM:
5 | """
6 | A class that represents a CGM sensor.
7 |
8 | Attributes
9 | ----------
10 | ts: int
11 | The sample time of the cgm sensor (min).
12 | t_offset: float
13 | The time offset of the cgm sensor (when new this is 0). This is used if a cgm sensor is shared between more
14 | replay runs.
15 | cgm_error_parameters: np.ndarray
16 | An array containing the parameters of the CGM.
17 | output_noise_sd: float
18 | The output standard deviation of the CGM error model.
19 | ekm1: float
20 | Memory term for the CGM noise: e(k-1).
21 | ekm2: float
22 | Memory term for the CGM noise: e(k-2).
23 | max_lifetime: float
24 | The maximum lifetime of the CGM sensor (in minutes).
25 | connected_at: int
26 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
27 | ReplayBG calls).
28 |
29 | Methods
30 | -------
31 | connect_new_cgm(connected_at):
32 | Connects a new CGM sensor by sampling new error parameters.
33 | measure(ig, t):
34 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
35 | add_offset(to_add):
36 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
37 | multiple ReplayBG runs.
38 | """
39 |
40 | def __init__(self,
41 | ts: int = 5,
42 | ):
43 | """
44 | Constructs all the necessary attributes for the CGM object.
45 |
46 | Parameters
47 | ----------
48 | ts: int, optional, default = 5
49 | The sample time of the cgm sensor (min).
50 | """
51 | self.ts = ts
52 | self.t_offset = 0
53 |
54 | self.connect_new_cgm()
55 |
56 | # Set max lifetime (minutes)
57 | self.max_lifetime = 10 * 1440 # 10 days
58 | self.connected_at = 0
59 |
60 | # Set the parameters of the CGM
61 | self.cgm_error_parameters = []
62 | self.output_noise_sd = []
63 |
64 | # Set memory terms
65 | self.ekm1 = None
66 | self.ekm2 = None
67 |
68 | def connect_new_cgm(self, connected_at=0):
69 | """
70 | Connects a new CGM sensor by sampling new error parameters.
71 |
72 | Parameters
73 | ----------
74 | connected_at: int, optional, default = 0
75 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
76 | ReplayBG calls).
77 |
78 | Returns
79 | -------
80 | None
81 |
82 | Raises
83 | ------
84 | None
85 |
86 | See Also
87 | --------
88 | None
89 |
90 | Examples
91 | --------
92 | None
93 | """
94 | # Load Mean vector and covariance matrix of the parameter vector as
95 | # defined in Vettoretti et al., Sensors, 2019
96 | mu = np.array([0.94228767821000314341972625697963, 0.0049398821141803427384187052950892,
97 | -0.0005848748565491275084801681138913, 6.382602204050874306062723917421,
98 | 1.2604070417357611244568715846981, -0.4022228938823663169088717950217,
99 | 3.2516360856114072674927228945307])
100 | sigma = np.array([[0.013245827952891258902368143424155, -0.0039513025350735725416129184850433,
101 | 0.00031276743283791636970891936186945, 0.15717912467153988265167186000326,
102 | 0.0026876560011614997885986966252858, -0.0028904633825263671524641306831427,
103 | -0.0031801707001874032418320403792222],
104 | [-0.0039513025350735725416129184850433, 0.0018527975980744701509778105119608,
105 | -0.00015580332205794781403294935184789, -0.10288007693621757654423021222101,
106 | -0.0013902327543057948350258001823931, 0.0011591852212130876378232136048041,
107 | -0.0027284927011686846420879248853453],
108 | [0.00031276743283791636970891936186945, -0.00015580332205794781403294935184789,
109 | 0.000013745962164724157000697882247131, 0.0080685863688738888865881193623864,
110 | 0.00012074974710011031125631020266553, -0.00010042135441622312822841645019167,
111 | 0.00011130290033867137325027107941366],
112 | [0.15717912467153988265167186000326, -0.10288007693621757654423021222101,
113 | 0.0080685863688738888865881193623864, 29.005838188852990811028575990349,
114 | 0.12408344051778112671069465022811, -0.10193644943826736526393261783596,
115 | 0.60075381294204155402383094042307],
116 | [0.0026876560011614997885986966252858, -0.0013902327543057948350258001823931,
117 | 0.00012074974710011031125631020266553, 0.12408344051778112671069465022811,
118 | 0.02079352674233487727195601735275, -0.018431109170459980539646949182497,
119 | -0.015721846813032722134373386779771],
120 | [-0.0028904633825263671524641306831427, 0.0011591852212130876378232136048041,
121 | -0.00010042135441622312822841645019167, -0.10193644943826736526393261783596,
122 | -0.018431109170459980539646949182497, 0.018700867933453400870913441167431,
123 | 0.01552333576829629385729347745837],
124 | [-0.0031801707001874032418320403792222, -0.0027284927011686846420879248853453,
125 | 0.00011130290033867137325027107941366, 0.60075381294204155402383094042307,
126 | -0.015721846813032722134373386779771, 0.01552333576829629385729347745837,
127 | 0.72356838038463477946748980684788]])
128 |
129 | # Modulation factor of the covariance of the parameter vector not to generate too extreme realizations of
130 | # parameter vector
131 | f = 0.90
132 | # Modulate the covariance matrix
133 | sigma = sigma * f
134 | # Maximum allowed output noise SD (mg/dl)
135 | max_output_noise_sd = 10
136 | # Tolerance for model stability check
137 | toll = 0.02
138 |
139 | # Flag for stability of the AR(2) model
140 | stable = 0
141 | output_noise_sd = np.inf
142 | cgm_error_parameters = []
143 | while (~ stable) or (output_noise_sd > max_output_noise_sd):
144 | # Sample CGM error parameters
145 | cgm_error_parameters = np.random.multivariate_normal(mu, sigma)
146 |
147 | # Check the stability of the resulting AR(2) model
148 | stable = (cgm_error_parameters[5] >= -1) and (
149 | cgm_error_parameters[5] <= (1 - np.abs(cgm_error_parameters[4]) - toll))
150 |
151 | output_noise_var = cgm_error_parameters[6] ** 2 / (
152 | 1 - (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) - cgm_error_parameters[5] * (
153 | (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) + cgm_error_parameters[5]))
154 |
155 | if output_noise_var < 0:
156 | continue
157 |
158 | # Compute the output noise standard deviation
159 | output_noise_sd = np.sqrt(output_noise_var)
160 |
161 | # Set the parameters of the CGM
162 | self.cgm_error_parameters = cgm_error_parameters
163 | self.output_noise_sd = output_noise_sd
164 |
165 | # Set memory terms
166 | self.ekm1 = 0
167 | self.ekm2 = 0
168 |
169 | # Set the offset (minutes)
170 | self.t_offset = 0
171 |
172 | # Set the connection time
173 | self.connected_at = connected_at
174 |
175 | def measure(self, ig, t):
176 | """
177 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
178 |
179 | Parameters
180 | ----------
181 | ig: float
182 | The interstitial glucose concentration at current time (mg/dl).
183 | t: float
184 | Current time in days from the start of CGM sensor.
185 |
186 | Returns
187 | -------
188 | cgm: float
189 | The CGM measurement at current time (mg/dl).
190 |
191 | Raises
192 | ------
193 | None
194 |
195 | See Also
196 | --------
197 | None
198 |
199 | Examples
200 | --------
201 | None
202 | """
203 |
204 | # Apply calibration error
205 | ig_s = (self.cgm_error_parameters[0] + self.cgm_error_parameters[1] * (t + self.t_offset) +
206 | self.cgm_error_parameters[2] * ((t + self.t_offset) ** 2)) * ig + self.cgm_error_parameters[3]
207 |
208 | # Generate noise
209 | z = np.random.normal(0, 1)
210 | u = self.cgm_error_parameters[6] * z
211 | e = u + self.cgm_error_parameters[4] * self.ekm1 + self.cgm_error_parameters[5] * self.ekm2
212 |
213 | # Update memory terms
214 | self.ekm2 = self.ekm1
215 | self.ekm1 = e
216 |
217 | # Get final CGM
218 | return ig_s + e
219 |
220 | def add_offset(self,
221 | to_add: float) -> None:
222 | """
223 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
224 | multiple ReplayBG runs.
225 |
226 | Parameters
227 | ----------
228 | to_add: float
229 | The offset to add to the sensor life.
230 |
231 | Returns
232 | -------
233 | None
234 |
235 | Raises
236 | ------
237 | None
238 |
239 | See Also
240 | --------
241 | None
242 |
243 | Examples
244 | --------
245 | None
246 | """
247 | self.t_offset += to_add
248 |
249 |
250 | class Sensors:
251 | """
252 | A class that represents the sensors to be used by ReplayBG.
253 |
254 | ...
255 | Attributes
256 | ----------
257 | cgm: CGM
258 | The CGM sensor used to measure the interstitial glucose during simulations.
259 |
260 | Methods
261 | -------
262 | None
263 | """
264 |
265 | def __init__(self, cgm: CGM):
266 | """
267 | Constructs all the necessary attributes for the Sensors object.
268 |
269 | Parameters
270 | ----------
271 | cgm: CGM
272 | The CGM sensor used to measure the interstitial glucose during simulations.
273 | """
274 | self.cgm = cgm
275 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pandas as pd
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'patient_info.csv'))
20 | return df
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the location of the 'results' folder
10 | results_folder_location = os.path.join(os.path.abspath(''))
11 |
12 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
13 | replay_results = pickle.load(file)
14 |
15 | # Visualize and analyze MCMC results
16 | Visualizer.plot_replay_results(replay_results)
17 | analysis = Analyzer.analyze_replay_results(replay_results)
18 | print(" ----- Analysis using MCMC-derived twins ----- ")
19 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
20 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
21 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
22 |
23 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
24 | replay_results = pickle.load(file)
25 |
26 | # Visualize and analyze MAP results
27 | Visualizer.plot_replay_results(replay_results)
28 | analysis = Analyzer.analyze_replay_results(replay_results)
29 | print(" ----- Analysis using MAP-derived twins ----- ")
30 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
31 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
32 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
33 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the interval to analyze
10 | start_day = 1
11 | end_day = 2
12 |
13 | # Set the location of the 'results' folder
14 | results_folder_location = os.path.join(os.path.abspath(''))
15 |
16 | # Initialize results list
17 | replay_results_interval = []
18 |
19 | for day in range(start_day, end_day+1):
20 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
21 | replay_results = pickle.load(file)
22 | replay_results_interval.append(replay_results)
23 |
24 | # Visualize and analyze MCMC results
25 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
26 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
27 | print(" ----- Analysis using MAP-derived twins ----- ")
28 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
29 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
30 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
31 |
32 | # Initialize results list
33 | replay_results_interval = []
34 |
35 | for day in range(start_day, end_day + 1):
36 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
37 | replay_results = pickle.load(file)
38 | replay_results_interval.append(replay_results)
39 |
40 | # Visualize and analyze MAP results
41 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
42 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
43 | print(" ----- Analysis using MCMC-derived twins ----- ")
44 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
45 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
46 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | save_workspace=True,
57 | x0=x0, previous_data_name=previous_data_name,
58 | save_suffix='_replay_map',
59 | sensors=sensors)
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='mcmc',
56 | n_replay=10,
57 | save_workspace=True,
58 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
59 | save_suffix='_replay_mcmc')
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params)
55 |
56 | # Visualize and analyze results
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
60 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
61 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
62 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | n_replay=10,
50 | twinning_method='mcmc',
51 | save_workspace=True,
52 | save_suffix='_replay_mcmc_dss',
53 | enable_correction_boluses=True,
54 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params=bolus_calculator_handler_params)
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 | x0 = None
28 | previous_data_name = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Twinning " + save_name)
52 |
53 | # Run twinning procedure
54 | rbg.twin(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | parallelize=parallelize,
57 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
58 |
59 | # Replay the twin with the same input data
60 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
61 | twinning_method='map',
62 | save_workspace=True,
63 | x0=x0, previous_data_name=previous_data_name,
64 | save_suffix='_twin_map')
65 |
66 | # Append results
67 | replay_results_interval.append(replay_results)
68 | data_interval.append(data)
69 |
70 | # Set initial conditions for next day equal to the "ending conditions" of the current day
71 | x0 = replay_results['x_end']['realizations'][0].tolist()
72 |
73 | # Set previous_data_name
74 | previous_data_name = save_name
75 |
76 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
78 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set the number of steps for MCMC
17 | n_steps = 5000 # In production, this should be >= 50k
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''))
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 | x0 = None
31 | previous_data_name = None
32 |
33 | # Initialize the list of results
34 | replay_results_interval = []
35 | data_interval = []
36 |
37 | # Instantiate ReplayBG
38 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
39 | yts=5, exercise=False,
40 | seed=1,
41 | verbose=verbose, plot_mode=plot_mode)
42 |
43 | # Set interval to twin
44 | start_day = 1
45 | end_day = 2
46 |
47 | # Twin the interval
48 | for day in range(start_day, end_day+1):
49 |
50 | # Step 1: Load data and set save_name
51 | data = load_test_data(day=day)
52 | save_name = 'data_day_' + str(day) + '_interval'
53 |
54 | print("Twinning " + save_name)
55 |
56 | # Run twinning procedure
57 | rbg.twin(data=data, bw=bw, save_name=save_name,
58 | twinning_method='mcmc',
59 | parallelize=parallelize,
60 | n_steps=n_steps,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='mcmc',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_mcmc')
69 |
70 | # Append results
71 | replay_results_interval.append(replay_results)
72 | data_interval.append(data)
73 |
74 | # Set initial conditions for next day equal to the "ending conditions" of the current day
75 | x0 = replay_results['x_end']['realizations'][0].tolist()
76 |
77 | # Set previous_data_name
78 | previous_data_name = save_name
79 |
80 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
82 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
83 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | print("Twinning " + save_name)
39 |
40 | # Run twinning procedure
41 | rbg.twin(data=data, bw=bw, save_name=save_name,
42 | twinning_method='map',
43 | parallelize=parallelize,
44 | u2ss=u2ss)
45 |
46 | # Replay the twin with the same input data
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | save_workspace=True,
50 | save_suffix='_twin_map')
51 |
52 | # Visualize and analyze results
53 | Visualizer.plot_replay_results(replay_results, data=data)
54 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
55 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data_extended(day=1)
36 | # Cut data up to 11:00
37 | data = data.iloc[0:348, :]
38 | # Modify label of second day
39 | data.loc[312, "cho_label"] = 'B2'
40 |
41 | save_name = 'data_day_' + str(1) + '_extended'
42 |
43 | print("Twinning " + save_name)
44 |
45 | # Run twinning procedure using also the burn-out period
46 | rbg.twin(data=data, bw=bw, save_name=save_name,
47 | twinning_method='map',
48 | parallelize=parallelize,
49 | u2ss=u2ss,
50 | extended=True,
51 | find_start_guess_first=False)
52 |
53 | # Cut data up to 4:00
54 | data = data.iloc[0:264,:]
55 |
56 | # Replay the twin with the data up to the start of the burn-out period
57 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
58 | twinning_method='map',
59 | save_workspace=True,
60 | save_suffix='_twin_map_extended')
61 |
62 | # Visualize and analyze results
63 | Visualizer.plot_replay_results(replay_results, data=data)
64 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
65 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data(day=1)
40 | save_name = 'data_day_' + str(1)
41 |
42 | print("Twinning " + save_name)
43 |
44 | # Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=n_steps,
49 | u2ss=u2ss)
50 |
51 | # Replay the twin with the same input data
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_twin_mcmc')
56 |
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data_extended(day=1)
40 | # Cut data up to 11:00
41 | data = data.iloc[0:348, :]
42 | # Modify label of second day
43 | data.loc[312, "cho_label"] = 'B2'
44 |
45 | save_name = 'data_day_' + str(1) + '_extended'
46 |
47 | print("Twinning " + save_name)
48 |
49 | # Run twinning procedure using also the burn-out period
50 | rbg.twin(data=data, bw=bw, save_name=save_name,
51 | twinning_method='mcmc',
52 | parallelize=parallelize,
53 | n_steps=n_steps,
54 | u2ss=u2ss,
55 | extended=True,
56 | find_start_guess_first=True)
57 |
58 | # Cut data up to 4:00
59 | data = data.iloc[0:264, :]
60 |
61 | # Replay the twin with the data up to the start of the burn-out period
62 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
63 | twinning_method='mcmc',
64 | save_workspace=True,
65 | save_suffix='_twin_mcmc_extended')
66 |
67 | Visualizer.plot_replay_results(replay_results, data=data)
68 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
69 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/twinning/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/twinning/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/twinning/map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import warnings
3 | import pickle
4 | import numpy as np
5 |
6 | from typing import Dict, Callable
7 |
8 | from multiprocessing import Pool
9 | from tqdm import tqdm
10 | from scipy.optimize import minimize
11 |
12 | from py_replay_bg.data import ReplayBGData
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 |
16 | from py_replay_bg.environment import Environment
17 |
18 | # Suppress all RuntimeWarnings
19 | warnings.filterwarnings("ignore", category=RuntimeWarning)
20 |
21 |
22 | class MAP:
23 | """
24 | A class that orchestrates the twinning process via MAP.
25 |
26 | Attributes
27 | ----------
28 | max_iter: int
29 | Maximum number of iterations.
30 | parallelize : bool
31 | A boolean that specifies whether to parallelize the twinning process.
32 | n_processes : int
33 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
34 |
35 | Methods
36 | -------
37 | twin(rbg_data, model, save_name, environment)
38 | Runs the twinning procedure.
39 | """
40 |
41 | def __init__(self,
42 | max_iter: int = 100000,
43 | parallelize: bool = False,
44 | n_processes: int | None = None,
45 | ):
46 | """
47 | Constructs all the necessary attributes for the MCMC object.
48 |
49 | Parameters
50 | ----------
51 | max_iter: int, optional, default : 100000
52 | Maximum number of iterations.
53 | parallelize : bool, optional, default : False
54 | A boolean that specifies whether to parallelize the twinning process.
55 | n_processes : int, optional, default : None
56 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
57 |
58 | Returns
59 | -------
60 | None
61 |
62 | Raises
63 | ------
64 | None
65 |
66 | See Also
67 | --------
68 | None
69 |
70 | Examples
71 | --------
72 | None
73 | """
74 |
75 | # Number of times to re-run the procedure
76 | self.n_rerun = 64
77 |
78 | # Maximum number of iterations
79 | self.max_iter = max_iter
80 |
81 | # Maximum number of function evaluations
82 | self.max_fev = 1000000
83 |
84 | # Parallelization options
85 | self.parallelize = parallelize
86 | self.n_processes = n_processes
87 |
88 | def twin(self,
89 | rbg_data: ReplayBGData,
90 | model: T1DModelSingleMeal | T1DModelMultiMeal,
91 | save_name: str,
92 | environment: Environment,
93 | start_guess: Dict = None,
94 | for_start_guess: bool = False) -> Dict:
95 | """
96 | Runs the twinning procedure.
97 |
98 | Parameters
99 | ----------
100 | rbg_data: ReplayBGData
101 | An object containing the data to be used during the twinning procedure.
102 | model: T1DModelSingleMeal | T1DModelMultiMeal
103 | An object that represents the physiological model to be used by ReplayBG.
104 | environment: Environment
105 | An object that represents the hyperparameters to be used by ReplayBG.
106 | save_name : str
107 | A string used to label, thus identify, each output file and result.
108 | start_guess: Dict, optional, default : None
109 | The initial guess for the twinning process. If None, this is set to population values.
110 | for_start_guess: bool, optional, default : False
111 | Whether to use the twin method just to find the start guess to pass to the MCMC twinner.
112 |
113 | Returns
114 | -------
115 | draws: dict
116 | A dictionary containing the chain and the samples obtained from the MCMC procedure and the copula
117 | sampling, respectively.
118 |
119 | Raises
120 | ------
121 | None
122 |
123 | See Also
124 | --------
125 | None
126 |
127 | Examples
128 | --------
129 | None
130 | """
131 |
132 | # If this is being used to find the start_guess, do /4 less reruns
133 | if for_start_guess:
134 | self.n_rerun = int(self.n_rerun/16)
135 |
136 | # Number of unknown parameters to twin
137 | n_dim = len(model.unknown_parameters)
138 |
139 | if start_guess is None:
140 | sg = model.start_guess
141 | else:
142 | sg = np.array(list(start_guess.values()))
143 |
144 | # Set the initial positions of the walkers.
145 | start = sg + model.start_guess_sigma * np.random.randn(self.n_rerun, n_dim)
146 | start[start < 0] = 0
147 |
148 | # Set the pooler
149 | pool = None
150 | if self.parallelize:
151 | pool = Pool(processes=self.n_processes)
152 |
153 | # Set up the options
154 | options = dict()
155 | options['maxiter'] = self.max_iter
156 | options['maxfev'] = self.max_fev
157 | options['disp'] = False
158 |
159 | # Select the function to minimize
160 | neg_log_posterior_func = model.neg_log_posterior_extended if model.extended else model.neg_log_posterior
161 |
162 | # Initialize results
163 | results = []
164 |
165 | if pool is None:
166 |
167 | if environment.verbose:
168 | iterator = tqdm(range(self.n_rerun))
169 | iterator.set_description("Min loss: %f" % np.nan)
170 | else:
171 | iterator = range(self.n_rerun)
172 |
173 | # Initialize best
174 | best = -1
175 |
176 | for r in iterator:
177 | result = run_map(start[r], neg_log_posterior_func, rbg_data, options)
178 | results.append(result)
179 | if best == -1 or result['fun'] < results[best]['fun']:
180 | best = r
181 | if environment.verbose:
182 | iterator.set_description("Min loss %f" % results[best]['fun'])
183 |
184 | else:
185 | # Prepare input arguments as tuples for starmap
186 | args = [(start[r], neg_log_posterior_func, rbg_data, options) for r in range(self.n_rerun)]
187 |
188 | # Initialize best
189 | best = -1
190 |
191 | # Get results (verbosity not allowed for the moment)
192 | results = pool.starmap(run_map, args)
193 |
194 | # Get best
195 | for r, result in enumerate(results):
196 | if best == -1 or result['fun'] < results[best]['fun']:
197 | best = r
198 |
199 | draws = dict()
200 | for up in range(n_dim):
201 | draws[model.unknown_parameters[up]] = results[best]['x'][up]
202 |
203 | # If twin is being used just to find the start guess, just return draws without saving
204 | if for_start_guess:
205 | return draws
206 |
207 | # Clean-up draws from "extended" parameters
208 | if model.extended:
209 | if 'SI_B2' in draws:
210 | del draws['SI_B2']
211 | if 'kabs_B2' in draws:
212 | del draws['kabs_B2']
213 | if 'beta_B2' in draws:
214 | del draws['beta_B2']
215 | if 'kabs_L2' in draws:
216 | del draws['kabs_L2']
217 | if 'beta_L2' in draws:
218 | del draws['beta_L2']
219 | if 'kabs_S2' in draws:
220 | del draws['kabs_S2']
221 | if 'beta_S2' in draws:
222 | del draws['beta_S2']
223 |
224 | # Save results
225 | twinning_results = dict()
226 | twinning_results['draws'] = draws
227 | twinning_results['u2ss'] = model.model_parameters.u2ss
228 |
229 | saved_file = os.path.join(environment.replay_bg_path, 'results', 'map',
230 | 'map_' + save_name + '.pkl')
231 |
232 | with open(saved_file, 'wb') as file:
233 | pickle.dump(twinning_results, file)
234 |
235 | if environment.verbose:
236 | print('Parameters saved in ' + saved_file)
237 |
238 | return draws
239 |
240 |
241 | def run_map(start: np.ndarray,
242 | neg_log_posterior_func: Callable,
243 | rbg_data: ReplayBGData,
244 | options: Dict
245 | ) -> Dict:
246 | """
247 | Utility function used to run MAP twinning.
248 |
249 | Parameters
250 | ----------
251 | start: np.ndarray
252 | An object containing the data to be used during the twinning procedure.
253 | neg_log_posterior_func: Callable
254 | The function to minimize, i.e., the neg-loglikelihood.
255 | rbg_data: ReplayBGData
256 | An object containing the data to be used during the twinning procedure.
257 | options : Dict
258 | A dictionary with the options necessary to the minimization function.
259 |
260 | Returns
261 | -------
262 | ret: dict
263 | A dictionary containing the results of the MAP twinning and the final value of the objective function.
264 |
265 | Raises
266 | ------
267 | None
268 |
269 | See Also
270 | --------
271 | None
272 |
273 | Examples
274 | --------
275 | None
276 | """
277 | result = minimize(neg_log_posterior_func, start, method='Powell', args=(rbg_data,), options=options)
278 | ret = dict()
279 | ret['fun'] = result.fun
280 | ret['x'] = result.x
281 | return ret
282 |
--------------------------------------------------------------------------------
/py_replay_bg/utils/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/utils/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/utils/stats.py:
--------------------------------------------------------------------------------
1 | import math
2 | import numpy as np
3 | from numba import njit
4 |
5 |
6 | @njit(fastmath=True)
7 | def log_lognorm(x, mu, sigma):
8 | """
9 | Computes the logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
10 |
11 | Parameters
12 | ----------
13 | x: float
14 | The value where to evaluate the log-normal pdf.
15 | mu: float
16 | The mean of the log-normal distribution.
17 | sigma: float
18 | The standard deviation of the log-normal distribution.
19 |
20 | Returns
21 | -------
22 | ll_norm: float
23 | The logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
24 |
25 | Raises
26 | ------
27 | None
28 |
29 | See Also
30 | --------
31 | None
32 |
33 | Examples
34 | --------
35 | None
36 | """
37 | return np.log(1 / (x * sigma * np.sqrt(2 * np.pi)) * np.exp(- ((np.log(x) - mu) ** 2) / (2 * (sigma ** 2))))
38 |
39 |
40 | @njit(fastmath=True)
41 | def log_norm(x, mu, sigma):
42 | """
43 | Computes the logarithm of the normal pdf evaluated at given x with given mu and sigma.
44 |
45 | Parameters
46 | ----------
47 | x: float
48 | The value where to evaluate the normal pdf.
49 | mu: float
50 | The mean of the normal distribution.
51 | sigma: float
52 | The standard deviation of the normal distribution.
53 |
54 | Returns
55 | -------
56 | l_norm: float
57 | The logarithm of the normal pdf evaluated at given x with given mu and sigma.
58 |
59 | Raises
60 | ------
61 | None
62 |
63 | See Also
64 | --------
65 | None
66 |
67 | Examples
68 | --------
69 | None
70 | """
71 | return np.log(1 / (sigma * np.sqrt(2 * np.pi)) * np.exp(- 0.5 * ((x - mu) / sigma) ** 2))
72 |
73 |
74 | @njit(fastmath=True)
75 | def log_gamma(x, alpha, beta):
76 | """
77 | Computes the logarithm of the gamma pdf evaluated at given x with given alpha and beta.
78 |
79 | Parameters
80 | ----------
81 | x: float
82 | The value where to evaluate the normal pdf.
83 | alpha: float
84 | The alpha value of the gamma distribution at hand.
85 | beta: float
86 | The beta value of the gamma distribution at hand.
87 |
88 | Returns
89 | -------
90 | l_gam: float
91 | The logarithm of the gamma pdf evaluated at given x with given alpha and beta.
92 |
93 | Raises
94 | ------
95 | None
96 |
97 | See Also
98 | --------
99 | None
100 |
101 | Examples
102 | --------
103 | None
104 | """
105 | return -np.inf if x < 0 else np.log((beta ** alpha * x ** (alpha - 1) * math.exp(-beta * x)) / math.gamma(alpha))
106 |
--------------------------------------------------------------------------------
/py_replay_bg/visualizer/__init__.py:
--------------------------------------------------------------------------------
1 | from datetime import datetime, timedelta
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 |
6 | from typing import Dict
7 |
8 | import pandas as pd
9 | from tqdm import tqdm
10 |
11 | from py_replay_bg.data import ReplayBGData
12 | from py_replay_bg.environment import Environment
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 | from py_replay_bg.dss import DSS
16 |
17 |
18 | class Visualizer:
19 | """
20 | A class to be used by ReplayBG for plotting.
21 |
22 | ...
23 | Attributes
24 | ----------
25 | -
26 |
27 | Methods
28 | -------
29 | plot_replay_results(replay_results, data)
30 | Function that plots the results of ReplayBG replay simulation.
31 | """
32 |
33 | def __init__(self):
34 | """
35 | Constructs all the necessary attributes for the Visualizer object.
36 |
37 | Parameters
38 | ----------
39 | -
40 |
41 | Returns
42 | -------
43 | None
44 |
45 | Raises
46 | ------
47 | None
48 |
49 | See Also
50 | --------
51 | None
52 |
53 | Examples
54 | --------
55 | None
56 | """
57 | pass
58 |
59 | @staticmethod
60 | def plot_replay_results(
61 | replay_results: Dict,
62 | data: pd.DataFrame = None,
63 | title: str = '',
64 | ) -> None:
65 | """
66 | Function that plots the results of ReplayBG simulation.
67 |
68 | Parameters
69 | ----------
70 | replay_results: dict
71 | The replayed scenario results with fields:
72 | glucose: dict
73 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
74 | cgm: dict
75 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
76 | insulin_bolus: dict
77 | A dictionary which contains the insulin boluses simulated via ReplayBG.
78 | correction_bolus: dict
79 | A dictionary which contains the correction boluses simulated via ReplayBG.
80 | insulin_basal: dict
81 | A dictionary which contains the basal insulin simulated via ReplayBG.
82 | cho: dict
83 | A dictionary which contains the meals simulated via ReplayBG.
84 | hypotreatments: dict
85 | A dictionary which contains the hypotreatments simulated via ReplayBG.
86 | meal_announcement: dict
87 | A dictionary which contains the meal announcements simulated via ReplayBG.
88 | vo2: dict
89 | A dictionary which contains the vo2 simulated via ReplayBG.
90 | sensors: dict
91 | A dictionary which contains the sensors used during the replayed scenario.
92 | rbg_data: ReplayBGData
93 | The data to be used by ReplayBG during simulation.
94 | model: T1DModelSingleMeal | T1DModelMultiMeal
95 | An object that represents the physiological model to be used by ReplayBG.
96 | data: pd.DataFrame, optional, default: None
97 | Pandas dataframe which contains the data to be used by the tool. If present, adds glucose data to the
98 | glucose subplot.
99 |
100 | Returns
101 | -------
102 | None
103 |
104 | Raises
105 | ------
106 | None
107 |
108 | See Also
109 | --------
110 | None
111 |
112 | Examples
113 | --------
114 | None
115 | """
116 | # Subplot 1: Glucose
117 |
118 | fig, ax = None, None
119 | if replay_results['model'].exercise:
120 | fig, ax = plt.subplots(4, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1, 1]})
121 | else:
122 | fig, ax = plt.subplots(3, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1]})
123 |
124 | if data is not None:
125 | ax[0].plot(data.t, data.glucose, marker='*', color='red', linewidth=2, label='CGM data [mg/dl]')
126 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['cgm']['median'], marker='o', color='black', linewidth=2, label='CGM replay (Median) [mg/dl]')
127 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['cgm']['ci25th'], replay_results['cgm']['ci75th'], color='black', alpha=0.2,
128 | label='CGM replay (CI 25-75th) [mg/dl]')
129 |
130 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['glucose']['median'][::replay_results['model'].yts], marker='o', color='blue', linewidth=2,
131 | label='Glucose replay (Median) [mg/dl]')
132 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['glucose']['ci25th'][::replay_results['model'].yts], replay_results['glucose']['ci75th'][::replay_results['model'].yts], color='blue',
133 | alpha=0.3, label='Glucose replay (CI 25-75th) [mg/dl]')
134 |
135 | ax[0].grid()
136 | ax[0].legend()
137 |
138 | # Subplot 2: Meals
139 | t = np.arange(replay_results['rbg_data'].t_data[0]+ pd.Timedelta(minutes=0), replay_results['rbg_data'].t_data[-1] + pd.Timedelta(minutes=replay_results['model'].yts),
140 | timedelta(minutes=1)).astype(datetime)
141 |
142 | cho_events = np.sum(replay_results['cho']['realizations'], axis=0) / replay_results['cho']['realizations'].shape[0]
143 | ht_events = np.sum(replay_results['hypotreatments']['realizations'], axis=0) / replay_results['hypotreatments']['realizations'].shape[0]
144 |
145 | markerline, stemlines, baseline = ax[1].stem(t, cho_events, basefmt='k:', label='CHO replay (Mean) [g/min]')
146 | plt.setp(stemlines, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
147 | plt.setp(markerline, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
148 |
149 | markerline, stemlines, baseline = ax[1].stem(t, ht_events, basefmt='k:', label='HT replay (Mean) [g/min]')
150 | plt.setp(stemlines, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
151 | plt.setp(markerline, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
152 |
153 | ax[1].grid()
154 | ax[1].legend()
155 |
156 | # Subplot 3: Insulin
157 |
158 | bolus_events = np.sum(replay_results['insulin_bolus']['realizations'], axis=0) / replay_results['insulin_bolus']['realizations'].shape[0]
159 | cb_events = np.sum(replay_results['correction_bolus']['realizations'], axis=0) / replay_results['correction_bolus']['realizations'].shape[0]
160 | basal_rate = np.sum(replay_results['insulin_basal']['realizations'], axis=0) / replay_results['insulin_basal']['realizations'].shape[0]
161 |
162 | markerline, stemlines, baseline = ax[2].stem(t, bolus_events, basefmt='k:',
163 | label='Bolus insulin replay (Mean) [U/min]')
164 | plt.setp(stemlines, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
165 | plt.setp(markerline, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
166 |
167 | markerline, stemlines, baseline = ax[2].stem(t, cb_events, basefmt='k:',
168 | label='CB insulin replay (Mean) [U/min]')
169 | plt.setp(stemlines, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
170 | plt.setp(markerline, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
171 |
172 | ax[2].plot(t, basal_rate * 60, color='black', linewidth=2, label='Basal insulin replay (Mean) [U/h]')
173 |
174 | ax[2].grid()
175 | ax[2].legend()
176 |
177 | # Subplot 4: Exercise
178 |
179 | if replay_results['model'].exercise:
180 |
181 | vo2_events = np.sum(replay_results['vo2']['realizations'], axis=0) / replay_results['vo2']['realizations'].shape[0]
182 |
183 | markerline, stemlines, baseline = ax[3].stem(t, vo2_events, basefmt='k:', label='VO2 replay (Mean) [-]')
184 | plt.setp(stemlines, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
185 | plt.setp(markerline, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
186 |
187 | ax[3].grid()
188 | ax[3].legend()
189 |
190 | fig.suptitle(title, fontweight='bold')
191 | plt.show()
192 |
193 |
194 | @staticmethod
195 | def plot_replay_results_interval(
196 | replay_results_interval: list,
197 | data_interval: list = None,
198 | title: str = '',
199 | ) -> None:
200 | """
201 | Function that plots the results of ReplayBG simulation (intervals).
202 |
203 | Parameters
204 | ----------
205 | replay_results_interval: list
206 | A list dictionaries of replayed scenario results. Each element has fields:
207 | glucose: dict
208 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
209 | cgm: dict
210 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
211 | insulin_bolus: dict
212 | A dictionary which contains the insulin boluses simulated via ReplayBG.
213 | correction_bolus: dict
214 | A dictionary which contains the correction boluses simulated via ReplayBG.
215 | insulin_basal: dict
216 | A dictionary which contains the basal insulin simulated via ReplayBG.
217 | cho: dict
218 | A dictionary which contains the meals simulated via ReplayBG.
219 | hypotreatments: dict
220 | A dictionary which contains the hypotreatments simulated via ReplayBG.
221 | meal_announcement: dict
222 | A dictionary which contains the meal announcements simulated via ReplayBG.
223 | vo2: dict
224 | A dictionary which contains the vo2 simulated via ReplayBG.
225 | sensors: dict
226 | A dictionary which contains the sensors used during the replayed scenario.
227 | rbg_data: ReplayBGData
228 | The data to be used by ReplayBG during simulation.
229 | model: T1DModelSingleMeal | T1DModelMultiMeal
230 | An object that represents the physiological model to be used by ReplayBG.
231 | data_interval: list, optional, default: None
232 | A list of pandas dataframe which contains the data to be used by the tool. If present, adds glucose data
233 | to the glucose subplot.
234 |
235 | Returns
236 | -------
237 | None
238 |
239 | Raises
240 | ------
241 | None
242 |
243 | See Also
244 | --------
245 | None
246 |
247 | Examples
248 | --------
249 | None
250 | """
251 | data = None
252 | if data_interval is not None:
253 | data = pd.concat(data_interval, axis=0, ignore_index=True)
254 |
255 | # Pad each array to the maximum shape
256 | replay_results = dict()
257 |
258 |
259 | # Re-map glucose data
260 | fields = ['median', 'ci5th', 'ci25th', 'ci75th', 'ci95th']
261 | category = ['glucose', 'cgm']
262 |
263 | for c in category:
264 |
265 | replay_results[c] = dict()
266 |
267 | for f in fields:
268 | replay_results[c][f] = dict()
269 |
270 | replay_results[c][f] = [r[c][f] for r in replay_results_interval]
271 | replay_results[c][f] = np.concatenate(replay_results[c][f], axis=0)
272 |
273 | # Re-map cho, insulin, and vo2 data
274 | category = ['cho', 'hypotreatments', 'insulin_bolus', 'correction_bolus', 'insulin_basal', 'vo2']
275 |
276 | for c in category:
277 | replay_results[c] = dict()
278 | replay_results[c]['realizations'] = [r[c]['realizations'] for r in replay_results_interval]
279 | replay_results[c]['realizations'] = np.concatenate(replay_results[c]['realizations'], axis=1)
280 |
281 | replay_results['model'] = replay_results_interval[0]['model']
282 | t_data = [r['rbg_data'].t_data for r in replay_results_interval]
283 | replay_results['rbg_data'] = replay_results_interval[0]['rbg_data']
284 | replay_results['rbg_data'].t_data = np.concatenate(t_data, axis=0)
285 |
286 | Visualizer.plot_replay_results(replay_results=replay_results, data=data, title=title)
287 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.pytest.ini_options]
2 | pythonpath = [
3 | "."
4 | ]
5 |
6 | [build-system]
7 | requires = ["setuptools>=61.0.0", "wheel"]
8 | build-backend = "setuptools.build_meta"
9 |
10 | [project]
11 | name = "py_replay_bg"
12 | version = "1.1.0"
13 | authors = [
14 | { name="Giacomo Cappon", email="cappongiacomo@gmail.com" },
15 | ]
16 | description = "ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management."
17 | readme = "README.md"
18 | requires-python = ">=3.11"
19 | classifiers = [
20 | "Programming Language :: Python :: 3",
21 | "License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
22 | "Operating System :: OS Independent",
23 | "Intended Audience :: Science/Research",
24 | "Topic :: Scientific/Engineering",
25 | ]
26 | dynamic = ["dependencies"]
27 | [tool.setuptools.dynamic]
28 | dependencies = {file = ["requirements.txt"]}
29 | optional-dependencies = {dev = { file = ["requirements-dev.txt"] }}
30 |
31 | [project.urls]
32 | "Homepage" = "https://github.com/gcappon/py_replay_bg"
33 | "Bug Tracker" = "https://github.com/gcappon/py_replay_bg/issues"
34 |
35 | [tool.setuptools.packages.find]
36 | include = ["py_replay_bg*"]
37 | exclude = ["venv*"]
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/tsconfig.json:
--------------------------------------------------------------------------------
1 | {
2 | "compilerOptions": {
3 | "module": "NodeNext",
4 | "moduleResolution": "NodeNext",
5 | "target": "ES2022"
6 | },
7 | "include": [
8 | "docs/.vuepress/**/*.ts",
9 | "docs/.vuepress/**/*.vue"
10 | ],
11 | "exclude": [
12 | "node_modules"
13 | ]
14 | }
15 |
--------------------------------------------------------------------------------
83 |
84 | [](https://github.com/gcappon/py_replay_bg/COPYING)
85 | [](https://github.com/gcappon/py_replay_bg/commits/master)
86 |
87 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
88 |
89 | # Reference
90 |
91 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
92 |
93 | # Get started
94 |
95 | ## Installation
96 |
97 | **ReplayBG** can be installed via pypi by simply
98 |
99 | ```python
100 | pip install py-replay-bg
101 | ```
102 |
103 | ### Requirements
104 |
105 | * Python >= 3.11
106 | * List of Python packages in `requirements.txt`
107 |
108 | ## Preparation: imports, setup, and data loading
109 |
110 | First of all import the core modules:
111 | ```python
112 | import os
113 | import numpy as np
114 | import pandas as pd
115 |
116 | from multiprocessing import freeze_support
117 | ```
118 |
119 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
120 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
121 | parallelized way.
122 |
123 | Then, we will import the necessary ReplayBG modules:
124 | ```python
125 | from py_replay_bg.py_replay_bg import ReplayBG
126 | from py_replay_bg.visualizer import Visualizer
127 | from py_replay_bg.analyzer import Analyzer
128 | ```
129 |
130 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
131 | while `Analyzer` and `Visualizer` are utility objects that will be used to
132 | respectively analyze and visualize the results that we will produce with ReplayBG
133 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
134 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
135 |
136 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
137 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
138 | ```python
139 | if __name__ == '__main__':
140 | freeze_support()
141 | ```
142 |
143 | Then, we will set the verbosity of ReplayBG:
144 | ```python
145 | verbose = True
146 | plot_mode = False
147 | ```
148 |
149 | Then, we need to decide what blueprint to use for twinning the data at hand.
150 | ```python
151 | blueprint = 'multi-meal'
152 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
153 | parallelize = True
154 | ```
155 |
156 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
157 |
158 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
159 | which contains a day of data of a patient with T1D:
160 |
161 | ```python
162 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
163 | data.t = pd.to_datetime(data['t'])
164 | ```
165 |
166 | ::: warning
167 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
168 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
169 | :::
170 |
171 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
172 |
173 | ```python
174 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
175 | p = np.where(patient_info['patient'] == 1)[0][0]
176 | # Set bw and u2ss
177 | bw = float(patient_info.bw.values[p])
178 | u2ss = float(patient_info.u2ss.values[p])
179 | ```
180 |
181 | Finally, instantiate a `ReplayBG` object:
182 |
183 | ```python
184 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
185 | yts=5, exercise=False,
186 | seed=1,
187 | verbose=verbose, plot_mode=plot_mode)
188 |
189 | ```
190 |
191 | ## Step 1: Creation of the digital twin
192 |
193 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
194 |
195 | ```python
196 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
197 | twinning_method='mcmc',
198 | parallelize=parallelize,
199 | n_steps=5000,
200 | u2ss=u2ss)
201 | ```
202 |
203 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
204 |
205 |
206 | ## Step 2: Run replay simulations
207 |
208 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
209 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
210 |
211 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
212 |
213 | ```python
214 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
215 | twinning_method='mcmc',
216 | save_workspace=True,
217 | save_suffix='_step_2a')
218 | ```
219 |
220 | It is possible to visualize the results of the simulation using:
221 |
222 | ```python
223 | Visualizer.plot_replay_results(replay_results, data=data)
224 | ```
225 |
226 | and analyzing the results using:
227 |
228 | ```python
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
231 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
232 | ```
233 |
234 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
235 | To do that run:
236 |
237 | ```python
238 | data.bolus = data.bolus * .7
239 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
240 | twinning_method='mcmc',
241 | save_workspace=True,
242 | save_suffix='_step_2b')
243 |
244 | # Visualize results
245 | Visualizer.plot_replay_results(replay_results)
246 | # Analyze results
247 | analysis = Analyzer.analyze_replay_results(replay_results)
248 |
249 | # Print, for example, the average glucose
250 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
251 | ```
252 |
253 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
254 |
255 | # Documentation
256 |
257 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
258 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/SOURCES.txt:
--------------------------------------------------------------------------------
1 | COPYING.md
2 | README.md
3 | pyproject.toml
4 | requirements.txt
5 | py_replay_bg/__init__.py
6 | py_replay_bg/py_replay_bg.py
7 | py_replay_bg.egg-info/PKG-INFO
8 | py_replay_bg.egg-info/SOURCES.txt
9 | py_replay_bg.egg-info/dependency_links.txt
10 | py_replay_bg.egg-info/requires.txt
11 | py_replay_bg.egg-info/top_level.txt
12 | py_replay_bg/analyzer/__init__.py
13 | py_replay_bg/data/__init__.py
14 | py_replay_bg/dss/__init__.py
15 | py_replay_bg/dss/default_dss_handlers.py
16 | py_replay_bg/environment/__init__.py
17 | py_replay_bg/example/code/analysis_example.py
18 | py_replay_bg/example/code/analysis_intervals_example.py
19 | py_replay_bg/example/code/get_started.py
20 | py_replay_bg/example/code/replay_intervals_map.py
21 | py_replay_bg/example/code/replay_intervals_mcmc.py
22 | py_replay_bg/example/code/replay_map.py
23 | py_replay_bg/example/code/replay_map_dss.py
24 | py_replay_bg/example/code/replay_mcmc.py
25 | py_replay_bg/example/code/replay_mcmc_dss.py
26 | py_replay_bg/example/code/twin_intervals_map.py
27 | py_replay_bg/example/code/twin_intervals_mcmc.py
28 | py_replay_bg/example/code/twin_map.py
29 | py_replay_bg/example/code/twin_map_extended.py
30 | py_replay_bg/example/code/twin_mcmc.py
31 | py_replay_bg/example/code/twin_mcmc_extended.py
32 | py_replay_bg/example/code/utils.py
33 | py_replay_bg/input_validation/__init__.py
34 | py_replay_bg/input_validation/input_validator_init.py
35 | py_replay_bg/input_validation/input_validator_replay.py
36 | py_replay_bg/input_validation/input_validator_twin.py
37 | py_replay_bg/model/__init__.py
38 | py_replay_bg/model/logpriors_t1d.py
39 | py_replay_bg/model/model_parameters_t1d.py
40 | py_replay_bg/model/model_step_equations_t1d.py
41 | py_replay_bg/model/t1d_model_multi_meal.py
42 | py_replay_bg/model/t1d_model_single_meal.py
43 | py_replay_bg/replay/__init__.py
44 | py_replay_bg/sensors/__init__.py
45 | py_replay_bg/tests/__init__.py
46 | py_replay_bg/tests/test_analysis_example.py
47 | py_replay_bg/tests/test_analysis_intervals_example.py
48 | py_replay_bg/tests/test_replay_intervals_map.py
49 | py_replay_bg/tests/test_replay_intervals_mcmc.py
50 | py_replay_bg/tests/test_replay_map.py
51 | py_replay_bg/tests/test_replay_map_dss.py
52 | py_replay_bg/tests/test_replay_map_extended.py
53 | py_replay_bg/tests/test_replay_mcmc.py
54 | py_replay_bg/tests/test_replay_mcmc_dss.py
55 | py_replay_bg/tests/test_replay_mcmc_extended.py
56 | py_replay_bg/tests/test_twin_intervals_map.py
57 | py_replay_bg/tests/test_twin_intervals_mcmc.py
58 | py_replay_bg/tests/test_twin_map.py
59 | py_replay_bg/tests/test_twin_map_extended.py
60 | py_replay_bg/tests/test_twin_mcmc.py
61 | py_replay_bg/tests/test_twin_mcmc_extended.py
62 | py_replay_bg/twinning/__init__.py
63 | py_replay_bg/twinning/map.py
64 | py_replay_bg/twinning/mcmc.py
65 | py_replay_bg/utils/__init__.py
66 | py_replay_bg/utils/stats.py
67 | py_replay_bg/visualizer/__init__.py
68 | venv/bin/rst2html.py
69 | venv/bin/rst2html4.py
70 | venv/bin/rst2html5.py
71 | venv/bin/rst2latex.py
72 | venv/bin/rst2man.py
73 | venv/bin/rst2odt.py
74 | venv/bin/rst2odt_prepstyles.py
75 | venv/bin/rst2pseudoxml.py
76 | venv/bin/rst2s5.py
77 | venv/bin/rst2xetex.py
78 | venv/bin/rst2xml.py
79 | venv/bin/rstpep2html.py
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/dependency_links.txt:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/requires.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/top_level.txt:
--------------------------------------------------------------------------------
1 | py_replay_bg
2 |
--------------------------------------------------------------------------------
/py_replay_bg/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/dss/__init__.py:
--------------------------------------------------------------------------------
1 | from typing import Callable, Dict
2 |
3 | from py_replay_bg.dss.default_dss_handlers import default_meal_generator_handler, standard_bolus_calculator_handler, \
4 | default_basal_handler, ada_hypotreatments_handler, corrects_above_250_handler
5 |
6 |
7 | class DSS:
8 | """
9 | A class that represents the hyperparameters of the integrated decision support system.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | bw: double
15 | The patient's body weight.
16 | meal_generator_handler: Callable
17 | A callback function that implements a meal generator to be used during the replay of a given scenario.
18 | meal_generator_handler_params: dict
19 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
20 | bolus_calculator_handler: Callable
21 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
22 | bolus_calculator_handler_params: dict
23 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
24 | serves as memory area for the bolusCalculatorHandler function.
25 | basal_handler: Callable
26 | A callback function that implements a basal controller to be used during the replay of a given scenario.
27 | basal_handler_params: dict
28 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
29 | memory area for the basalHandler function.
30 | enable_hypotreatments: boolean
31 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
32 | hypotreatments_handler: Callable
33 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
34 | hypotreatments_handler_params: dict
35 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
36 | serves as memory area for the hypoTreatmentsHandler function.
37 | enable_correction_boluses: boolean
38 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
39 | correction_boluses_handler: Callable
40 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
41 | correction_boluses_handler_params: dict
42 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also
43 | serves as memory area for the correctionBolusesHandler function.
44 |
45 | Methods
46 | -------
47 | None
48 | """
49 |
50 | def __init__(self,
51 | bw: float,
52 | meal_generator_handler: Callable = default_meal_generator_handler,
53 | meal_generator_handler_params: Dict | None = None,
54 | bolus_calculator_handler: Callable = standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params: Dict | None = None,
56 | basal_handler: Callable = default_basal_handler,
57 | basal_handler_params: Dict | None = None,
58 | enable_hypotreatments: bool = False,
59 | hypotreatments_handler: Callable = ada_hypotreatments_handler,
60 | hypotreatments_handler_params: Dict | None = None,
61 | enable_correction_boluses: bool = False,
62 | correction_boluses_handler: Callable = corrects_above_250_handler,
63 | correction_boluses_handler_params: Dict | None = None,
64 | ):
65 | """
66 | Constructs all the necessary attributes for the DSS object.
67 |
68 | Parameters
69 | ----------
70 | bw: double
71 | The patient's body weight.
72 | meal_generator_handler: Callable, optional, default : default_meal_generator_handler
73 | A callback function that implements a meal generator to be used during the replay of a given scenario.
74 | meal_generator_handler_params: dict, optional, default : None
75 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
76 | bolus_calculator_handler: Callable, optional, default : standard_bolus_calculator_handler
77 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
78 | bolus_calculator_handler_params: dict, optional, default : None
79 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
80 | serves as memory area for the bolusCalculatorHandler function.
81 | basal_handler: function, Callable, default : default_basal_handler
82 | A callback function that implements a basal controller to be used during the replay of a given scenario.
83 | basal_handler_params: dict, optional, default : None
84 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
85 | memory area for the basalHandler function.
86 | enable_hypotreatments: boolean, optional, default : False
87 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
88 | hypotreatments_handler: Callable, optional, default : ada_hypotreatments_handler
89 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
90 | hypotreatments_handler_params: dict, optional, default : None
91 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
92 | serves as memory area for the hypoTreatmentsHandler function.
93 | enable_correction_boluses: boolean, optional, default : False
94 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
95 | correction_boluses_handler: Callable, optional, default : corrects_above_250_handler
96 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
97 | correction_boluses_handler_params: dict, optional, default : None
98 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function.
99 | It also serves as memory area for the correctionBolusesHandler function.
100 | """
101 |
102 | # Patient's body weight
103 | self.bw = bw
104 |
105 | # Meal Generator module parameters
106 | self.meal_generator_handler = meal_generator_handler
107 | self.meal_generator_handler_params = meal_generator_handler_params if meal_generator_handler_params is not None else {}
108 |
109 | # Bolus Calculator module parameters
110 | self.bolus_calculator_handler = bolus_calculator_handler
111 | self.bolus_calculator_handler_params = bolus_calculator_handler_params if bolus_calculator_handler_params is not None else {}
112 |
113 | # Basal module parameters
114 | self.basal_handler = basal_handler
115 | self.basal_handler_params = basal_handler_params if basal_handler_params is not None else {}
116 |
117 | # Hypotreatment module parameters
118 | self.enable_hypotreatments = enable_hypotreatments
119 | self.hypotreatments_handler = hypotreatments_handler
120 | self.hypotreatments_handler_params = hypotreatments_handler_params if hypotreatments_handler_params is not None else {}
121 |
122 | # Correction bolus module parameters
123 | self.enable_correction_boluses = enable_correction_boluses
124 | self.correction_boluses_handler = correction_boluses_handler
125 | self.correction_boluses_handler_params = correction_boluses_handler_params if correction_boluses_handler_params is not None else {}
126 |
--------------------------------------------------------------------------------
/py_replay_bg/environment/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 |
4 | class Environment:
5 | """
6 | A class that represents the hyperparameters to be used by ReplayBG.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | save_name : str
12 | A string used to label, thus identify, each output file and result.
13 | replay_bg_path: str
14 | The current absolute path of ReplayBG.
15 |
16 | blueprint: str, {'single-meal', 'multi-meal'}
17 | A string that specifies the blueprint to be used to create the digital twin.
18 | yts: int
19 | An integer that specifies the data sampling time (in minutes).
20 |
21 | seed : int
22 | An integer that specifies the random seed. For reproducibility.
23 |
24 | plot_mode : bool
25 | A boolean that specifies whether to show the plot of the results or not.
26 | verbose : bool
27 | A boolean that specifies the verbosity of ReplayBG.
28 |
29 | Methods
30 | -------
31 | None
32 | """
33 |
34 | def __init__(self,
35 | blueprint: str = 'single_meal',
36 | save_name: str = '',
37 | save_folder: str = '',
38 | yts: int = 5,
39 | exercise: bool = False,
40 | seed: int = 1,
41 | plot_mode: bool = True,
42 | verbose: bool = True
43 | ):
44 | """
45 | Constructs all the necessary attributes for the Environment object.
46 |
47 | Parameters
48 | ----------
49 | blueprint: str, {'single-meal', 'multi-meal'}, optional, default : 'single-meal'
50 | A string that specifies the blueprint to be used to create the digital twin.
51 |
52 | save_name : str, optional, default : ''
53 | A string used to label, thus identify, each output file and result.
54 | save_folder : str, optional, default : ''
55 | A string used to set the folder where the ReplayBG results will be saved.
56 |
57 | yts: int, optional, default : 5
58 | An integer that specifies the data sampling time (in minutes).
59 | exercise: boolean, optional, default : False
60 | A boolean that specifies whether to simulate exercise or not.
61 |
62 | seed : int, optional, default : 1
63 | An integer that specifies the random seed. For reproducibility.
64 |
65 | plot_mode : boolean, optional, default : True
66 | A boolean that specifies whether to show the plot of the results or not.
67 | verbose : boolean, optional, default : True
68 | A boolean that specifies the verbosity of ReplayBG.
69 | """
70 |
71 | # Set the save name and folder
72 | self.save_name = save_name
73 | self.replay_bg_path = save_folder
74 |
75 | # Create the results sub folders if they do not exist
76 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results'))):
77 | os.mkdir(os.path.join(self.replay_bg_path, 'results'))
78 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'mcmc'))):
79 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'mcmc'))
80 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'map'))):
81 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'map'))
82 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'workspaces'))):
83 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'workspaces'))
84 |
85 | # Single-meal or multi-meal blueprint?
86 | self.blueprint = blueprint
87 |
88 | # Set sample time
89 | self.yts = yts
90 |
91 | # Set whether to use the exercise model
92 | self.exercise = exercise
93 |
94 | # Set the seed
95 | self.seed = seed
96 |
97 | # Set plot mode and verbosity
98 | self.plot_mode = plot_mode
99 | self.verbose = verbose
100 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the location of the 'results' folder
9 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
10 |
11 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
12 | replay_results = pickle.load(file)
13 |
14 | # Visualize and analyze MCMC results
15 | Visualizer.plot_replay_results(replay_results)
16 | analysis = Analyzer.analyze_replay_results(replay_results)
17 | print(" ----- Analysis using MCMC-derived twins ----- ")
18 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
19 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
20 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
21 |
22 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
23 | replay_results = pickle.load(file)
24 |
25 | # Visualize and analyze MAP results
26 | Visualizer.plot_replay_results(replay_results)
27 | analysis = Analyzer.analyze_replay_results(replay_results)
28 | print(" ----- Analysis using MAP-derived twins ----- ")
29 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
30 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
31 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
32 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the interval to analyze
9 | start_day = 1
10 | end_day = 2
11 |
12 | # Set the location of the 'results' folder
13 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
14 |
15 | # Initialize results list
16 | replay_results_interval = []
17 |
18 | for day in range(start_day, end_day+1):
19 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
20 | replay_results = pickle.load(file)
21 | replay_results_interval.append(replay_results)
22 |
23 | # Visualize and analyze MCMC results
24 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
25 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
26 | print(" ----- Analysis using MAP-derived twins ----- ")
27 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
28 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
29 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
30 |
31 | # Initialize results list
32 | replay_results_interval = []
33 |
34 | for day in range(start_day, end_day + 1):
35 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
36 | replay_results = pickle.load(file)
37 | replay_results_interval.append(replay_results)
38 |
39 | # Visualize and analyze MAP results
40 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
41 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
42 | print(" ----- Analysis using MCMC-derived twins ----- ")
43 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
44 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
45 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
46 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/get_started.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | import pandas as pd
4 |
5 | from multiprocessing import freeze_support
6 |
7 | from py_replay_bg.py_replay_bg import ReplayBG
8 | from py_replay_bg.visualizer import Visualizer
9 | from py_replay_bg.analyzer import Analyzer
10 |
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # Load data
25 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
26 | data.t = pd.to_datetime(data['t'])
27 |
28 | # Load patient_info
29 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Set save name
42 | save_name = 'data_day_1'
43 |
44 | # Step 1. Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=5000,
49 | u2ss=u2ss)
50 |
51 | # Step 2a. Replay the twin with the same input data to get the initial conditions for the subsequent day
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_step_2a')
56 |
57 | # Visualize results and compare with the original glucose data
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | # Analyze results
60 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
61 | # Print, for example, the fit MARD and the average glucose
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
63 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
64 |
65 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
66 | data.bolus = data.bolus * .7
67 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
68 | twinning_method='mcmc',
69 | save_workspace=True,
70 | u2ss=u2ss,
71 | save_suffix='_step_2b')
72 |
73 | # Visualize results
74 | Visualizer.plot_replay_results(replay_results)
75 | # Analyze results
76 | analysis = Analyzer.analyze_replay_results(replay_results)
77 |
78 | # Print, for example, the average glucose
79 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='map',
53 | save_workspace=True,
54 | x0=x0, previous_data_name=previous_data_name,
55 | save_suffix='_replay_map',
56 | sensors=sensors)
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='mcmc',
53 | n_replay=100,
54 | save_workspace=True,
55 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
56 | save_suffix='_replay_mcmc')
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | twinning_method='map',
39 | save_workspace=True,
40 | save_suffix='_replay_map')
41 |
42 | # Visualize and analyze results
43 | Visualizer.plot_replay_results(replay_results, data=data)
44 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
45 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
46 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
47 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
48 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | n_replay=10,
39 | twinning_method='mcmc',
40 | save_workspace=True,
41 | save_suffix='_replay_mcmc')
42 |
43 | # Visualize and analyze results
44 | Visualizer.plot_replay_results(replay_results, data=data)
45 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
46 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
47 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
48 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
49 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='mcmc',
50 | save_workspace=True,
51 | save_suffix='_replay_mcmc_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 | x0 = None
32 | previous_data_name = None
33 |
34 | # Initialize the list of results
35 | replay_results_interval = []
36 | data_interval = []
37 |
38 | # Instantiate ReplayBG
39 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
40 | yts=5, exercise=False,
41 | seed=1,
42 | verbose=verbose, plot_mode=plot_mode)
43 |
44 | # Set interval to twin
45 | start_day = 1
46 | end_day = 2
47 |
48 | # Twin the interval
49 | for day in range(start_day, end_day+1):
50 |
51 | # Step 1: Load data and set save_name
52 | data = load_test_data(day=day)
53 | save_name = 'data_day_' + str(day) + '_interval'
54 |
55 | print("Twinning " + save_name)
56 |
57 | # Run twinning procedure
58 | rbg.twin(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | parallelize=parallelize,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='map',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_map')
69 | # Append results
70 | replay_results_interval.append(replay_results)
71 | data_interval.append(data)
72 |
73 | # Set initial conditions for next day equal to the "ending conditions" of the current day
74 | x0 = replay_results['x_end']['realizations'][0].tolist()
75 |
76 | # Set previous_data_name
77 | previous_data_name = save_name
78 |
79 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
80 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
82 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 | x0 = None
35 | previous_data_name = None
36 |
37 | # Initialize the list of results
38 | replay_results_interval = []
39 | data_interval = []
40 |
41 | # Instantiate ReplayBG
42 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
43 | yts=5, exercise=False,
44 | seed=1,
45 | verbose=verbose, plot_mode=plot_mode)
46 |
47 | # Set interval to twin
48 | start_day = 1
49 | end_day = 2
50 |
51 | # Twin the interval
52 | for day in range(start_day, end_day+1):
53 |
54 | # Step 1: Load data and set save_name
55 | data = load_test_data(day=day)
56 | save_name = 'data_day_' + str(day) + '_interval'
57 |
58 | print("Twinning " + save_name)
59 |
60 | # Run twinning procedure
61 | rbg.twin(data=data, bw=bw, save_name=save_name,
62 | twinning_method='mcmc',
63 | parallelize=parallelize,
64 | n_steps=n_steps,
65 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
66 |
67 | # Replay the twin with the same input data
68 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
69 | twinning_method='mcmc',
70 | save_workspace=True,
71 | x0=x0, previous_data_name=previous_data_name,
72 | save_suffix='_twin_mcmc')
73 | # Append results
74 | replay_results_interval.append(replay_results)
75 | data_interval.append(data)
76 |
77 | # Set initial conditions for next day equal to the "ending conditions" of the current day
78 | x0 = replay_results['x_end']['realizations'][0].tolist()
79 |
80 | # Set previous_data_name
81 | previous_data_name = save_name
82 |
83 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
84 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
85 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
86 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data(day=1)
39 | save_name = 'data_day_' + str(1)
40 |
41 | print("Twinning " + save_name)
42 |
43 | # Run twinning procedure
44 | rbg.twin(data=data, bw=bw, save_name=save_name,
45 | twinning_method='map',
46 | parallelize=parallelize,
47 | u2ss=u2ss)
48 |
49 | # Replay the twin with the same input data
50 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
51 | twinning_method='map',
52 | save_workspace=True,
53 | save_suffix='_twin_map')
54 |
55 | # Visualize and analyze results
56 | Visualizer.plot_replay_results(replay_results, data=data)
57 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
58 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
59 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data_extended(day=1)
39 | # Cut data up to 11:00
40 | data = data.iloc[0:348, :]
41 | # Modify label of second day
42 | data.loc[312, "cho_label"] = 'B2'
43 |
44 | save_name = 'data_day_' + str(1) + '_extended'
45 |
46 | # Run twinning procedure using also the burn-out period
47 | rbg.twin(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | parallelize=parallelize,
50 | u2ss=u2ss,
51 | extended=True,
52 | find_start_guess_first=True)
53 |
54 | # Cut data up to 4:00
55 | data = data.iloc[0:264, :]
56 |
57 | # Replay the twin with the data up to the start of the burn-out period
58 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | save_workspace=True,
61 | save_suffix='_twin_map_extended')
62 |
63 | # Visualize and analyze results
64 | Visualizer.plot_replay_results(replay_results, data=data)
65 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
66 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
67 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data(day=1)
43 | save_name = 'data_day_' + str(1)
44 |
45 | print("Twinning " + save_name)
46 |
47 | # Run twinning procedure
48 | rbg.twin(data=data, bw=bw, save_name=save_name,
49 | twinning_method='mcmc',
50 | parallelize=parallelize,
51 | n_steps=n_steps,
52 | u2ss=u2ss)
53 |
54 | # Replay the twin with the same input data
55 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
56 | twinning_method='mcmc',
57 | save_workspace=True,
58 | save_suffix='_twin_mcmc')
59 |
60 | Visualizer.plot_replay_results(replay_results, data=data)
61 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data_extended(day=1)
43 | # Cut data up to 11:00
44 | data = data.iloc[0:348, :]
45 | # Modify label of second day
46 | data.loc[312, "cho_label"] = 'B2'
47 |
48 | save_name = 'data_day_' + str(1) + '_extended'
49 |
50 | print("Twinning " + save_name)
51 |
52 | # Run twinning procedure using also the burn-out period
53 | rbg.twin(data=data, bw=bw, save_name=save_name,
54 | twinning_method='mcmc',
55 | parallelize=parallelize,
56 | n_steps=n_steps,
57 | u2ss=u2ss,
58 | extended=True,
59 | find_start_guess_first=True)
60 |
61 | # Cut data up to 4:00
62 | data = data.iloc[0:264, :]
63 |
64 | # Replay the twin with the data up to the start of the burn-out period
65 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
66 | twinning_method='mcmc',
67 | save_workspace=True,
68 | save_suffix='_twin_mcmc_extended')
69 |
70 | Visualizer.plot_replay_results(replay_results, data=data)
71 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
72 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/utils.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import os
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
20 | return df
21 |
--------------------------------------------------------------------------------
/py_replay_bg/example/data/patient_info.csv:
--------------------------------------------------------------------------------
1 | patient,u2ss,bw
2 | 1,0.2440484700275897,100
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_init.py:
--------------------------------------------------------------------------------
1 | from py_replay_bg.input_validation import *
2 |
3 |
4 | class InputValidatorInit:
5 | """
6 | Class for validating the input of ReplayBG __init__ method.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | blueprint: str, {'single-meal', 'multi-meal'}
12 | A string that specifies the blueprint to be used to create the digital twin.
13 | save_folder : str
14 | A string defining the folder that will contain the results.
15 |
16 | yts: int, optional, default : 5
17 | An integer that specifies the data sample time (in minutes).
18 | exercise: boolean, optional, default : False
19 | A boolean that specifies whether to simulate exercise or not.
20 | seed: int, optional, default: 1
21 | An integer that specifies the random seed. For reproducibility.
22 |
23 | plot_mode : boolean, optional, default : True
24 | A boolean that specifies whether to show the plot of the results or not.
25 | verbose : boolean, optional, default : True
26 | A boolean that specifies the verbosity of ReplayBG.
27 |
28 | Methods
29 | -------
30 | validate():
31 | Run the input validation process.
32 | """
33 |
34 | def __init__(self,
35 | save_folder: str,
36 | blueprint: str,
37 | yts: int,
38 | exercise: bool,
39 | seed: int,
40 | plot_mode: bool,
41 | verbose: bool,
42 | ):
43 | self.save_folder = save_folder
44 | self.blueprint = blueprint
45 | self.yts = yts
46 | self.exercise = exercise
47 | self.seed = seed
48 | self.plot_mode = plot_mode
49 | self.verbose = verbose
50 |
51 | def validate(self):
52 | """
53 | Run the input validation process.
54 | """
55 |
56 | # Validate the 'save_folder' input
57 | SaveFolderValidator(save_folder=self.save_folder).validate()
58 |
59 | # Validate the 'blueprint' input
60 | BlueprintValidator(blueprint=self.blueprint).validate()
61 |
62 | # Validate the 'yts' input
63 | YTSValidator(yts=self.yts).validate()
64 |
65 | # Validate the 'exercise' input
66 | ExerciseValidator(exercise=self.exercise).validate()
67 |
68 | # Validate the 'seed' input
69 | SeedValidator(seed=self.seed).validate()
70 |
71 | # Validate the 'plot_mode' input
72 | PlotModeValidator(plot_mode=self.plot_mode).validate()
73 |
74 | # Validate the 'verbose' input
75 | VerboseValidator(verbose=self.verbose).validate()
76 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_replay.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from typing import Callable, Dict
5 |
6 | from py_replay_bg.input_validation import *
7 |
8 |
9 | class InputValidatorReplay:
10 | """
11 | Class for validating the input of ReplayBG twin method.
12 |
13 | ...
14 | Attributes
15 | ----------
16 | data : pd.DataFrame
17 | Pandas dataframe which contains the data to be used by the tool.
18 | bw : float
19 | The patient's body weight.
20 | save_name : str
21 | A string used to label, thus identify, each output file and result.
22 |
23 | x0: list
24 | The initial model state.
25 | previous_data_name: str
26 | The name of the previous portion of data. To be used to initialize the initial conditions.
27 |
28 | twinning_method : str
29 | The method used to twin the model.
30 |
31 | bolus_source : str
32 | A string defining whether to use, during replay, the insulin bolus data contained in the 'data' timetable
33 | (if 'data'), or the boluses generated by the bolus calculator implemented via the provided
34 | 'bolusCalculatorHandler' function.
35 | basal_source : str
36 | A string defining whether to use, during replay, the insulin basal data contained in the 'data' timetable
37 | (if 'data'), or the basal generated by the controller implemented via the provided
38 | 'basalControllerHandler' function (if 'dss'), or fixed to the average basal rate used during
39 | twinning (if 'u2ss').
40 | cho_source : str
41 | A string defining whether to use, during replay, the CHO data contained in the 'data' timetable (if 'data'),
42 | or the CHO generated by the meal generator implemented via the provided 'mealGeneratorHandler' function.
43 |
44 | meal_generator_handler: Callable
45 | A callback function that implements a meal generator to be used during the replay of a given scenario.
46 | meal_generator_handler_params: dict
47 | A dictionary that contains the parameters to pass to the meal_generator_handler function.
48 | bolus_calculator_handler: Callable
49 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
50 | bolus_calculator_handler_params: dict
51 | A dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also serves
52 | as memory area for the bolusCalculatorHandler function.
53 | basal_handler: Callable
54 | A callback function that implements a basal controller to be used during the replay of a given scenario.
55 | basal_handler_params: dict
56 | A dictionary that contains the parameters to pass to the basalHandler function. It also serves as memory
57 | area for the basalHandler function.
58 | enable_hypotreatments: boolean
59 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
60 | hypotreatments_handler: Callable
61 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
62 | hypotreatments_handler_params: dict
63 | A dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also serves
64 | as memory area for the hypoTreatmentsHandler function.
65 | enable_correction_boluses: boolean
66 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
67 | correction_boluses_handler: Callable
68 | A callback function that implements a corrective bolus strategy during the replay of a given scenario.
69 | correction_boluses_handler_params: dict
70 | A dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also serves
71 | as memory area for the correctionBolusesHandler function.
72 |
73 | save_suffix : string
74 | A string to be attached as suffix to the resulting output files' name.
75 | save_workspace: bool
76 | A flag that specifies whether to save the resulting workspace.
77 |
78 | n_replay: int
79 | The number of Monte Carlo replays to be performed. Ignored if twinning_method is 'map'.
80 | sensors: list[Sensors]
81 | The sensors to be used in each of the replay simulations.
82 |
83 | blueprint: str
84 | A string that specifies the blueprint to be used to create the digital twin.
85 | exercise: bool
86 | A boolean that specifies whether to use exercise model or not.
87 |
88 | Methods
89 | -------
90 | validate():
91 | Run the input validation process.
92 | """
93 |
94 | def __init__(self,
95 | data: pd.DataFrame,
96 | bw: float,
97 | save_name: str,
98 | x0: np.ndarray,
99 | previous_data_name: str,
100 | twinning_method: str,
101 | bolus_source: str,
102 | basal_source: str,
103 | cho_source: str,
104 | meal_generator_handler: Callable,
105 | meal_generator_handler_params: Dict,
106 | bolus_calculator_handler: Callable,
107 | bolus_calculator_handler_params: Dict,
108 | basal_handler: Callable,
109 | basal_handler_params: Dict,
110 | enable_hypotreatments: bool,
111 | hypotreatments_handler: Callable,
112 | hypotreatments_handler_params: Dict,
113 | enable_correction_boluses: bool,
114 | correction_boluses_handler: Callable,
115 | correction_boluses_handler_params: Dict,
116 | save_suffix: str,
117 | save_workspace: bool,
118 | n_replay: int,
119 | sensors: list,
120 | blueprint: str,
121 | exercise: bool,
122 | ):
123 | self.data = data
124 | self.bw = bw
125 | self.save_name = save_name
126 | self.x0 = x0
127 | self.previous_data_name = previous_data_name
128 | self.twinning_method = twinning_method
129 | self.bolus_source = bolus_source
130 | self.basal_source = basal_source
131 | self.cho_source = cho_source
132 | self.meal_generator_handler = meal_generator_handler
133 | self.meal_generator_handler_params = meal_generator_handler_params
134 | self.bolus_calculator_handler = bolus_calculator_handler
135 | self.bolus_calculator_handler_params = bolus_calculator_handler_params
136 | self.basal_handler = basal_handler
137 | self.basal_handler_params = basal_handler_params
138 | self.enable_hypotreatments = enable_hypotreatments
139 | self.hypotreatments_handler = hypotreatments_handler
140 | self.hypotreatments_handler_params = hypotreatments_handler_params
141 | self.enable_correction_boluses = enable_correction_boluses
142 | self.correction_boluses_handler = correction_boluses_handler
143 | self.correction_boluses_handler_params = correction_boluses_handler_params
144 | self.save_suffix = save_suffix
145 | self.save_workspace = save_workspace
146 | self.n_replay = n_replay
147 | self.sensors = sensors
148 | self.blueprint = blueprint
149 | self.exercise = exercise
150 |
151 |
152 | def validate(self):
153 | """
154 | Run the input validation process.
155 | """
156 |
157 | # Validate the 'data' input
158 | DataValidator(modality='replay', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
159 | bolus_source=self.bolus_source, basal_source=self.basal_source, cho_source=self.cho_source).validate()
160 |
161 | # Validate the 'bw' input
162 | BWValidator(bw=self.bw).validate()
163 |
164 | # Validate the 'save_name' input
165 | SaveNameValidator(save_name=self.save_name).validate()
166 |
167 | # Validate the 'save_name' input
168 | X0Validator(x0=self.x0).validate()
169 |
170 | # Validate the 'previous_data_name' input
171 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
172 |
173 | # Validate the 'twinning_method' input
174 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
175 |
176 | # Validate the 'bolus_source' input
177 | BolusSourceValidator(bolus_source=self.bolus_source).validate()
178 |
179 | # Validate the 'basal_source' input
180 | BasalSourceValidator(basal_source=self.basal_source).validate()
181 |
182 | # Validate the 'cho_source' input
183 | CHOSourceValidator(cho_source=self.cho_source).validate()
184 |
185 | # Validate the 'meal_generator_handler' input
186 | MealGeneratorHandlerValidator(meal_generator_handler=self.meal_generator_handler).validate()
187 |
188 | # Validate the 'meal_generator_handler_params' input
189 | MealGeneratorHandlerParamsValidator(meal_generator_handler_params=self.meal_generator_handler_params).validate()
190 |
191 | # Validate the 'bolus_calculator_handler' input
192 | BolusCalculatorHandlerValidator(bolus_calculator_handler=self.bolus_calculator_handler).validate()
193 |
194 | # Validate the 'bolus_calculator_handler_params' input
195 | BolusCalculatorHandlerParamsValidator(bolus_calculator_handler_params=self.bolus_calculator_handler_params).validate()
196 |
197 | # Validate the 'basal_handler' input
198 | BasalHandlerValidator(basal_handler=self.basal_handler).validate()
199 |
200 | # Validate the 'basal_handler_params' input
201 | BasalHandlerParamsValidator(basal_handler_params=self.basal_handler_params).validate()
202 |
203 | # Validate the 'enable_hypotreatments' input
204 | EnableHypotreatmentsValidator(enable_hypotreatments=self.enable_hypotreatments).validate()
205 |
206 | # Validate the 'hypotreatments_handler' input
207 | HypotreatmentsHandlerValidator(hypotreatments_handler=self.hypotreatments_handler).validate()
208 |
209 | # Validate the 'hypotreatments_handler_params' input
210 | HypotreatmentsHandlerParamsValidator(hypotreatments_handler_params=self.hypotreatments_handler_params).validate()
211 |
212 | # Validate the 'enable_correction_boluses' input
213 | EnableCorrectionBolusesValidator(enable_correction_boluses=self.enable_correction_boluses).validate()
214 |
215 | # Validate the 'correction_boluses_handler' input
216 | CorrectionBolusesHandlerValidator(correction_boluses_handler=self.correction_boluses_handler).validate()
217 |
218 | # Validate the 'correction_boluses_handler_params' input
219 | CorrectionBolusesHandlerParamsValidator(correction_boluses_handler_params=self.correction_boluses_handler_params).validate()
220 |
221 | # Validate the 'save_suffix' input
222 | SaveSuffixValidator(save_suffix=self.save_suffix).validate()
223 |
224 | # Validate the 'save_workspace' input
225 | SaveWorkspaceValidator(save_workspace=self.save_workspace).validate()
226 |
227 | # Validate the 'n_replay' input
228 | NReplayValidator(n_replay=self.n_replay).validate()
229 |
230 | # Validate the 'sensors' input
231 | SensorsValidator(sensors=self.sensors).validate()
232 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_twin.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from py_replay_bg.input_validation import *
5 |
6 |
7 | class InputValidatorTwin:
8 | """
9 | Class for validating the input of ReplayBG twin method.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | data: pd.DataFrame
15 | Pandas dataframe which contains the data to be used by the tool.
16 | bw: float
17 | The patient's body weight.
18 | save_name : str
19 | A string used to label, thus identify, each output file and result.
20 |
21 |
22 | u2ss : float
23 | The steady state of the basal insulin infusion.
24 | x0 : np.ndarray
25 | The initial model conditions.
26 | previous_data_name : str
27 | The name of the previous data portion. This is used to correctly "transfer" the initial model conditions to
28 | the current portion of data.
29 |
30 | twinning_method : str
31 | The method to be used to twin the model.
32 |
33 | extended : bool
34 | A flag indicating whether to use the "extended" model for twinning
35 | find_start_guess_first : bool
36 | A flag indicating whether to find the start guess using MAP before twinning.
37 |
38 | n_steps: int
39 | Number of steps to use for the main chain. This is ignored if modality is 'replay'.
40 | save_chains: bool, optional, default : False
41 | A flag that specifies whether to save the resulting mcmc chains and copula samplers.
42 |
43 | parallelize : boolean
44 | A boolean that specifies whether to parallelize the twinning process.
45 | n_processes : int, optional, default : None
46 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
47 |
48 | blueprint: str
49 | A string that specifies the blueprint to be used to create the digital twin.
50 | exercise: bool
51 | A boolean that specifies whether to use exercise model or not.
52 |
53 | Methods
54 | -------
55 | validate():
56 | Run the input validation process.
57 | """
58 |
59 | def __init__(self,
60 | data: pd.DataFrame,
61 | bw: float,
62 | save_name: str,
63 | twinning_method: str,
64 | extended: bool,
65 | find_start_guess_first: bool,
66 | n_steps: int,
67 | save_chains: bool,
68 | u2ss: float | None,
69 | x0: np.ndarray | None,
70 | previous_data_name: str | None,
71 | parallelize: bool,
72 | n_processes: int | None,
73 | blueprint: str,
74 | exercise: bool
75 | ):
76 | self.data = data
77 | self.bw = bw
78 | self.save_name = save_name
79 | self.twinning_method = twinning_method
80 | self.extended = extended
81 | self.find_start_guess_first = find_start_guess_first
82 | self.n_steps = n_steps
83 | self.save_chains = save_chains
84 | self.u2ss = u2ss
85 | self.x0 = x0
86 | self.previous_data_name = previous_data_name
87 | self.parallelize = parallelize
88 | self.n_processes = n_processes
89 | self.blueprint = blueprint
90 | self.exercise = exercise
91 |
92 | def validate(self):
93 | """
94 | Run the input validation process.
95 | """
96 |
97 | # Validate the 'data' input
98 | DataValidator(modality='twin', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
99 | bolus_source='data', basal_source='data', cho_source='data').validate()
100 |
101 | # Validate the 'bw' input
102 | BWValidator(bw=self.bw).validate()
103 |
104 | # Validate the 'save_name' input
105 | SaveNameValidator(save_name=self.save_name).validate()
106 |
107 | # Validate the 'twinning_method' input
108 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
109 |
110 | # Validate the 'extended' input
111 | ExtendedValidator(extended=self.extended).validate()
112 |
113 | # Validate the 'find_start_guess_first' input
114 | FindStartGuessFirstValidator(find_start_guess_first=self.find_start_guess_first).validate()
115 |
116 | # Validate the 'n_steps' input
117 | NStepsValidator(n_steps=self.n_steps).validate()
118 |
119 | # Validate the 'u2ss' input
120 | U2SSValidator(u2ss=self.u2ss).validate()
121 |
122 | # Validate the 'save_name' input
123 | X0Validator(x0=self.x0).validate()
124 |
125 | # Validate the 'previous_data_name' input
126 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
127 |
128 | # Validate the 'parallelize' input
129 | ParallelizeValidator(parallelize=self.parallelize).validate()
130 |
131 | # Validate the 'n_processes' input
132 | NProcessesValidator(n_processes=self.n_processes).validate()
133 |
--------------------------------------------------------------------------------
/py_replay_bg/model/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/model/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/model/model_parameters_t1d.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 |
4 |
5 | class ModelParametersT1D:
6 | def __init__(self,
7 | data: pd.DataFrame,
8 | bw: float,
9 | u2ss: float,
10 | extended: bool
11 | ):
12 |
13 | """
14 | Function that returns the default parameters values of the t1d model.
15 |
16 | Parameters
17 | ----------
18 | data : pd.DataFrame
19 | Pandas dataframe which contains the data to be used by the tool.
20 | bw : float
21 | The patient's body weight.
22 | u2ss : float
23 | The steady state of the basal insulin infusion
24 | extended : bool
25 | A flag indicating whether to use the "extended" model for twinning
26 |
27 | Returns
28 | -------
29 | model_parameters: dict
30 | A dictionary containing the default model parameters.
31 |
32 | Raises
33 | ------
34 | None
35 |
36 | See Also
37 | --------
38 | None
39 |
40 | Examples
41 | --------
42 | None
43 | """
44 | # Initial conditions
45 | self.Xpb = 0.0 # Insulin action initial condition
46 | self.Qgutb = 0.0 # Intestinal content initial condition
47 |
48 | # Glucose-insulin submodel parameters
49 | self.VG = 1.45 # dl/kg
50 | self.SG = 2.5e-2 # 1/min
51 | self.Gb = 119.13 # mg/dL
52 | self.r1 = 1.4407 # unitless
53 | self.r2 = 0.8124 # unitless
54 | self.alpha = 7 # 1/min
55 | self.p2 = 0.012 # 1/min
56 |
57 | if u2ss is None:
58 | self.u2ss = np.mean(data.basal) * 1000 / bw # mU/(kg*min)
59 | else:
60 | self.u2ss = u2ss
61 |
62 | # Subcutaneous insulin absorption submodel parameters
63 | self.VI = 0.126 # L/kg
64 | self.ke = 0.127 # 1/min
65 | self.kd = 0.026 # 1/min
66 | # model_parameters['ka1'] = 0.0034 # 1/min (virtually 0 in 77% of the cases)
67 | # self.ka1 = 0.0
68 | self.ka2 = 0.014 # 1/min
69 | self.tau = 8 # min
70 | self.Ipb = self.u2ss / self.ke # from eq. 5 steady-state
71 |
72 |
73 | # Oral glucose absorption submodel parameters
74 | self.kgri = 0.18 # = kmax % 1/min
75 | self.kempt = 0.18 # 1/min
76 | self.f = 0.9 # dimensionless
77 |
78 | # Exercise submodel parameters
79 | self.vo2rest = 0.33 # dimensionless, VO2rest was derived from heart rate round(66/(220-30))
80 | self.vo2max = 1 # dimensionless, VO2max is normalized and estimated from heart rate (220-age) = 100%.
81 | self.e1 = 1.6 # dimensionless
82 | self.e2 = 0.78 # dimensionless
83 |
84 | # Patient specific parameters
85 | self.bw = bw # kg
86 | self.to_g = self.bw / 1000
87 | self.to_mgkg = 1000 / self.bw
88 |
89 | # Measurement noise specifics
90 | self.SDn = 5
91 |
92 | # Glucose starting point
93 | if 'glucose' in data:
94 | idx = np.where(data.glucose.isnull().values == False)[0][0]
95 | self.G0 = data.glucose[idx]
96 | else:
97 | self.G0 = self.Gb
98 |
99 |
100 | class ModelParametersT1DMultiMeal(ModelParametersT1D):
101 |
102 | def __init__(self,
103 | data: pd.DataFrame,
104 | bw: float,
105 | u2ss: float,
106 | extended: bool,
107 | ):
108 |
109 | """
110 | Function that returns the default parameters values of the t1d model in a multi-meal blueprint.
111 |
112 | Parameters
113 | ----------
114 | data : pd.DataFrame
115 | Pandas dataframe which contains the data to be used by the tool.
116 | bw : float
117 | The patient's body weight.
118 | u2ss : float
119 | The steady state of the basal insulin infusion
120 | extended : bool
121 | A flag indicating whether to use the "extended" model for twinning
122 |
123 | Returns
124 | -------
125 | model_parameters: dict
126 | A dictionary containing the default model parameters.
127 |
128 | Raises
129 | ------
130 | None
131 |
132 | See Also
133 | --------
134 | None
135 |
136 | Examples
137 | --------
138 | None
139 | """
140 | # Initialize common parameters
141 | super().__init__(data, bw, u2ss, extended)
142 |
143 | # Glucose-insulin submodel parameters
144 | self.SI_B = 10.35e-4 / self.VG # mL/(uU*min)
145 | self.SI_L = 10.35e-4 / self.VG # mL/(uU*min)
146 | self.SI_D = 10.35e-4 / self.VG # mL/(uU*min)
147 |
148 | if extended:
149 | self.SI_B2 = 10.35e-4 / self.VG # mL/(uU*min)
150 |
151 | # Oral glucose absorption submodel parameters
152 | self.kabs_B = 0.012 # 1/min
153 | self.kabs_L = 0.012 # 1/min
154 | self.kabs_D = 0.012 # 1/min
155 | self.kabs_S = 0.012 # 1/min
156 | self.kabs_H = 0.012 # 1/min
157 | self.beta_B = 0 # min
158 | self.beta_L = 0 # min
159 | self.beta_D = 0 # min
160 | self.beta_S = 0 # min
161 |
162 | if extended:
163 | self.kabs_B2 = 0.012 # 1/min
164 | self.kabs_L2 = 0.012 # 1/min
165 | self.kabs_S2 = 0.012 # 1/min
166 | self.beta_B2 = 0 # min
167 | self.beta_L2 = 0 # min
168 | self.beta_S2 = 0 # min
169 |
170 |
171 | class ModelParametersT1DSingleMeal(ModelParametersT1D):
172 |
173 | def __init__(self,
174 | data: pd.DataFrame,
175 | bw: float,
176 | u2ss: float,
177 | ):
178 |
179 | """
180 | Function that returns the default parameters values of the t1d model in a single-meal blueprint.
181 |
182 | Parameters
183 | ----------
184 | data : pd.DataFrame
185 | Pandas dataframe which contains the data to be used by the tool.
186 | bw : float
187 | The patient's body weight.
188 | u2ss : float
189 | The steady state of the basal insulin infusion
190 |
191 | Returns
192 | -------
193 | model_parameters: dict
194 | A dictionary containing the default model parameters.
195 |
196 | Raises
197 | ------
198 | None
199 |
200 | See Also
201 | --------
202 | None
203 |
204 | Examples
205 | --------
206 | None
207 | """
208 | # Initialize common parameters
209 | super().__init__(data, bw, u2ss, extended=False)
210 |
211 | # Glucose-insulin submodel parameters
212 | self.SI = 10.35e-4 / self.VG # mL/(uU*min)
213 |
214 | # Oral glucose absorption submodel parameters
215 | self.kabs = 0.012 # 1/min
216 | self.beta = 0 # 1/min
217 |
--------------------------------------------------------------------------------
/py_replay_bg/sensors/__init__.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 |
4 | class CGM:
5 | """
6 | A class that represents a CGM sensor.
7 |
8 | Attributes
9 | ----------
10 | ts: int
11 | The sample time of the cgm sensor (min).
12 | t_offset: float
13 | The time offset of the cgm sensor (when new this is 0). This is used if a cgm sensor is shared between more
14 | replay runs.
15 | cgm_error_parameters: np.ndarray
16 | An array containing the parameters of the CGM.
17 | output_noise_sd: float
18 | The output standard deviation of the CGM error model.
19 | ekm1: float
20 | Memory term for the CGM noise: e(k-1).
21 | ekm2: float
22 | Memory term for the CGM noise: e(k-2).
23 | max_lifetime: float
24 | The maximum lifetime of the CGM sensor (in minutes).
25 | connected_at: int
26 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
27 | ReplayBG calls).
28 |
29 | Methods
30 | -------
31 | connect_new_cgm(connected_at):
32 | Connects a new CGM sensor by sampling new error parameters.
33 | measure(ig, t):
34 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
35 | add_offset(to_add):
36 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
37 | multiple ReplayBG runs.
38 | """
39 |
40 | def __init__(self,
41 | ts: int = 5,
42 | ):
43 | """
44 | Constructs all the necessary attributes for the CGM object.
45 |
46 | Parameters
47 | ----------
48 | ts: int, optional, default = 5
49 | The sample time of the cgm sensor (min).
50 | """
51 | self.ts = ts
52 | self.t_offset = 0
53 |
54 | self.connect_new_cgm()
55 |
56 | # Set max lifetime (minutes)
57 | self.max_lifetime = 10 * 1440 # 10 days
58 | self.connected_at = 0
59 |
60 | # Set the parameters of the CGM
61 | self.cgm_error_parameters = []
62 | self.output_noise_sd = []
63 |
64 | # Set memory terms
65 | self.ekm1 = None
66 | self.ekm2 = None
67 |
68 | def connect_new_cgm(self, connected_at=0):
69 | """
70 | Connects a new CGM sensor by sampling new error parameters.
71 |
72 | Parameters
73 | ----------
74 | connected_at: int, optional, default = 0
75 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
76 | ReplayBG calls).
77 |
78 | Returns
79 | -------
80 | None
81 |
82 | Raises
83 | ------
84 | None
85 |
86 | See Also
87 | --------
88 | None
89 |
90 | Examples
91 | --------
92 | None
93 | """
94 | # Load Mean vector and covariance matrix of the parameter vector as
95 | # defined in Vettoretti et al., Sensors, 2019
96 | mu = np.array([0.94228767821000314341972625697963, 0.0049398821141803427384187052950892,
97 | -0.0005848748565491275084801681138913, 6.382602204050874306062723917421,
98 | 1.2604070417357611244568715846981, -0.4022228938823663169088717950217,
99 | 3.2516360856114072674927228945307])
100 | sigma = np.array([[0.013245827952891258902368143424155, -0.0039513025350735725416129184850433,
101 | 0.00031276743283791636970891936186945, 0.15717912467153988265167186000326,
102 | 0.0026876560011614997885986966252858, -0.0028904633825263671524641306831427,
103 | -0.0031801707001874032418320403792222],
104 | [-0.0039513025350735725416129184850433, 0.0018527975980744701509778105119608,
105 | -0.00015580332205794781403294935184789, -0.10288007693621757654423021222101,
106 | -0.0013902327543057948350258001823931, 0.0011591852212130876378232136048041,
107 | -0.0027284927011686846420879248853453],
108 | [0.00031276743283791636970891936186945, -0.00015580332205794781403294935184789,
109 | 0.000013745962164724157000697882247131, 0.0080685863688738888865881193623864,
110 | 0.00012074974710011031125631020266553, -0.00010042135441622312822841645019167,
111 | 0.00011130290033867137325027107941366],
112 | [0.15717912467153988265167186000326, -0.10288007693621757654423021222101,
113 | 0.0080685863688738888865881193623864, 29.005838188852990811028575990349,
114 | 0.12408344051778112671069465022811, -0.10193644943826736526393261783596,
115 | 0.60075381294204155402383094042307],
116 | [0.0026876560011614997885986966252858, -0.0013902327543057948350258001823931,
117 | 0.00012074974710011031125631020266553, 0.12408344051778112671069465022811,
118 | 0.02079352674233487727195601735275, -0.018431109170459980539646949182497,
119 | -0.015721846813032722134373386779771],
120 | [-0.0028904633825263671524641306831427, 0.0011591852212130876378232136048041,
121 | -0.00010042135441622312822841645019167, -0.10193644943826736526393261783596,
122 | -0.018431109170459980539646949182497, 0.018700867933453400870913441167431,
123 | 0.01552333576829629385729347745837],
124 | [-0.0031801707001874032418320403792222, -0.0027284927011686846420879248853453,
125 | 0.00011130290033867137325027107941366, 0.60075381294204155402383094042307,
126 | -0.015721846813032722134373386779771, 0.01552333576829629385729347745837,
127 | 0.72356838038463477946748980684788]])
128 |
129 | # Modulation factor of the covariance of the parameter vector not to generate too extreme realizations of
130 | # parameter vector
131 | f = 0.90
132 | # Modulate the covariance matrix
133 | sigma = sigma * f
134 | # Maximum allowed output noise SD (mg/dl)
135 | max_output_noise_sd = 10
136 | # Tolerance for model stability check
137 | toll = 0.02
138 |
139 | # Flag for stability of the AR(2) model
140 | stable = 0
141 | output_noise_sd = np.inf
142 | cgm_error_parameters = []
143 | while (~ stable) or (output_noise_sd > max_output_noise_sd):
144 | # Sample CGM error parameters
145 | cgm_error_parameters = np.random.multivariate_normal(mu, sigma)
146 |
147 | # Check the stability of the resulting AR(2) model
148 | stable = (cgm_error_parameters[5] >= -1) and (
149 | cgm_error_parameters[5] <= (1 - np.abs(cgm_error_parameters[4]) - toll))
150 |
151 | output_noise_var = cgm_error_parameters[6] ** 2 / (
152 | 1 - (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) - cgm_error_parameters[5] * (
153 | (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) + cgm_error_parameters[5]))
154 |
155 | if output_noise_var < 0:
156 | continue
157 |
158 | # Compute the output noise standard deviation
159 | output_noise_sd = np.sqrt(output_noise_var)
160 |
161 | # Set the parameters of the CGM
162 | self.cgm_error_parameters = cgm_error_parameters
163 | self.output_noise_sd = output_noise_sd
164 |
165 | # Set memory terms
166 | self.ekm1 = 0
167 | self.ekm2 = 0
168 |
169 | # Set the offset (minutes)
170 | self.t_offset = 0
171 |
172 | # Set the connection time
173 | self.connected_at = connected_at
174 |
175 | def measure(self, ig, t):
176 | """
177 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
178 |
179 | Parameters
180 | ----------
181 | ig: float
182 | The interstitial glucose concentration at current time (mg/dl).
183 | t: float
184 | Current time in days from the start of CGM sensor.
185 |
186 | Returns
187 | -------
188 | cgm: float
189 | The CGM measurement at current time (mg/dl).
190 |
191 | Raises
192 | ------
193 | None
194 |
195 | See Also
196 | --------
197 | None
198 |
199 | Examples
200 | --------
201 | None
202 | """
203 |
204 | # Apply calibration error
205 | ig_s = (self.cgm_error_parameters[0] + self.cgm_error_parameters[1] * (t + self.t_offset) +
206 | self.cgm_error_parameters[2] * ((t + self.t_offset) ** 2)) * ig + self.cgm_error_parameters[3]
207 |
208 | # Generate noise
209 | z = np.random.normal(0, 1)
210 | u = self.cgm_error_parameters[6] * z
211 | e = u + self.cgm_error_parameters[4] * self.ekm1 + self.cgm_error_parameters[5] * self.ekm2
212 |
213 | # Update memory terms
214 | self.ekm2 = self.ekm1
215 | self.ekm1 = e
216 |
217 | # Get final CGM
218 | return ig_s + e
219 |
220 | def add_offset(self,
221 | to_add: float) -> None:
222 | """
223 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
224 | multiple ReplayBG runs.
225 |
226 | Parameters
227 | ----------
228 | to_add: float
229 | The offset to add to the sensor life.
230 |
231 | Returns
232 | -------
233 | None
234 |
235 | Raises
236 | ------
237 | None
238 |
239 | See Also
240 | --------
241 | None
242 |
243 | Examples
244 | --------
245 | None
246 | """
247 | self.t_offset += to_add
248 |
249 |
250 | class Sensors:
251 | """
252 | A class that represents the sensors to be used by ReplayBG.
253 |
254 | ...
255 | Attributes
256 | ----------
257 | cgm: CGM
258 | The CGM sensor used to measure the interstitial glucose during simulations.
259 |
260 | Methods
261 | -------
262 | None
263 | """
264 |
265 | def __init__(self, cgm: CGM):
266 | """
267 | Constructs all the necessary attributes for the Sensors object.
268 |
269 | Parameters
270 | ----------
271 | cgm: CGM
272 | The CGM sensor used to measure the interstitial glucose during simulations.
273 | """
274 | self.cgm = cgm
275 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pandas as pd
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'patient_info.csv'))
20 | return df
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the location of the 'results' folder
10 | results_folder_location = os.path.join(os.path.abspath(''))
11 |
12 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
13 | replay_results = pickle.load(file)
14 |
15 | # Visualize and analyze MCMC results
16 | Visualizer.plot_replay_results(replay_results)
17 | analysis = Analyzer.analyze_replay_results(replay_results)
18 | print(" ----- Analysis using MCMC-derived twins ----- ")
19 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
20 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
21 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
22 |
23 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
24 | replay_results = pickle.load(file)
25 |
26 | # Visualize and analyze MAP results
27 | Visualizer.plot_replay_results(replay_results)
28 | analysis = Analyzer.analyze_replay_results(replay_results)
29 | print(" ----- Analysis using MAP-derived twins ----- ")
30 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
31 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
32 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
33 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the interval to analyze
10 | start_day = 1
11 | end_day = 2
12 |
13 | # Set the location of the 'results' folder
14 | results_folder_location = os.path.join(os.path.abspath(''))
15 |
16 | # Initialize results list
17 | replay_results_interval = []
18 |
19 | for day in range(start_day, end_day+1):
20 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
21 | replay_results = pickle.load(file)
22 | replay_results_interval.append(replay_results)
23 |
24 | # Visualize and analyze MCMC results
25 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
26 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
27 | print(" ----- Analysis using MAP-derived twins ----- ")
28 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
29 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
30 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
31 |
32 | # Initialize results list
33 | replay_results_interval = []
34 |
35 | for day in range(start_day, end_day + 1):
36 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
37 | replay_results = pickle.load(file)
38 | replay_results_interval.append(replay_results)
39 |
40 | # Visualize and analyze MAP results
41 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
42 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
43 | print(" ----- Analysis using MCMC-derived twins ----- ")
44 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
45 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
46 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | save_workspace=True,
57 | x0=x0, previous_data_name=previous_data_name,
58 | save_suffix='_replay_map',
59 | sensors=sensors)
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='mcmc',
56 | n_replay=10,
57 | save_workspace=True,
58 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
59 | save_suffix='_replay_mcmc')
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params)
55 |
56 | # Visualize and analyze results
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
60 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
61 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
62 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | n_replay=10,
50 | twinning_method='mcmc',
51 | save_workspace=True,
52 | save_suffix='_replay_mcmc_dss',
53 | enable_correction_boluses=True,
54 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params=bolus_calculator_handler_params)
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 | x0 = None
28 | previous_data_name = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Twinning " + save_name)
52 |
53 | # Run twinning procedure
54 | rbg.twin(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | parallelize=parallelize,
57 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
58 |
59 | # Replay the twin with the same input data
60 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
61 | twinning_method='map',
62 | save_workspace=True,
63 | x0=x0, previous_data_name=previous_data_name,
64 | save_suffix='_twin_map')
65 |
66 | # Append results
67 | replay_results_interval.append(replay_results)
68 | data_interval.append(data)
69 |
70 | # Set initial conditions for next day equal to the "ending conditions" of the current day
71 | x0 = replay_results['x_end']['realizations'][0].tolist()
72 |
73 | # Set previous_data_name
74 | previous_data_name = save_name
75 |
76 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
78 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set the number of steps for MCMC
17 | n_steps = 5000 # In production, this should be >= 50k
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''))
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 | x0 = None
31 | previous_data_name = None
32 |
33 | # Initialize the list of results
34 | replay_results_interval = []
35 | data_interval = []
36 |
37 | # Instantiate ReplayBG
38 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
39 | yts=5, exercise=False,
40 | seed=1,
41 | verbose=verbose, plot_mode=plot_mode)
42 |
43 | # Set interval to twin
44 | start_day = 1
45 | end_day = 2
46 |
47 | # Twin the interval
48 | for day in range(start_day, end_day+1):
49 |
50 | # Step 1: Load data and set save_name
51 | data = load_test_data(day=day)
52 | save_name = 'data_day_' + str(day) + '_interval'
53 |
54 | print("Twinning " + save_name)
55 |
56 | # Run twinning procedure
57 | rbg.twin(data=data, bw=bw, save_name=save_name,
58 | twinning_method='mcmc',
59 | parallelize=parallelize,
60 | n_steps=n_steps,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='mcmc',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_mcmc')
69 |
70 | # Append results
71 | replay_results_interval.append(replay_results)
72 | data_interval.append(data)
73 |
74 | # Set initial conditions for next day equal to the "ending conditions" of the current day
75 | x0 = replay_results['x_end']['realizations'][0].tolist()
76 |
77 | # Set previous_data_name
78 | previous_data_name = save_name
79 |
80 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
82 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
83 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | print("Twinning " + save_name)
39 |
40 | # Run twinning procedure
41 | rbg.twin(data=data, bw=bw, save_name=save_name,
42 | twinning_method='map',
43 | parallelize=parallelize,
44 | u2ss=u2ss)
45 |
46 | # Replay the twin with the same input data
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | save_workspace=True,
50 | save_suffix='_twin_map')
51 |
52 | # Visualize and analyze results
53 | Visualizer.plot_replay_results(replay_results, data=data)
54 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
55 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data_extended(day=1)
36 | # Cut data up to 11:00
37 | data = data.iloc[0:348, :]
38 | # Modify label of second day
39 | data.loc[312, "cho_label"] = 'B2'
40 |
41 | save_name = 'data_day_' + str(1) + '_extended'
42 |
43 | print("Twinning " + save_name)
44 |
45 | # Run twinning procedure using also the burn-out period
46 | rbg.twin(data=data, bw=bw, save_name=save_name,
47 | twinning_method='map',
48 | parallelize=parallelize,
49 | u2ss=u2ss,
50 | extended=True,
51 | find_start_guess_first=False)
52 |
53 | # Cut data up to 4:00
54 | data = data.iloc[0:264,:]
55 |
56 | # Replay the twin with the data up to the start of the burn-out period
57 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
58 | twinning_method='map',
59 | save_workspace=True,
60 | save_suffix='_twin_map_extended')
61 |
62 | # Visualize and analyze results
63 | Visualizer.plot_replay_results(replay_results, data=data)
64 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
65 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data(day=1)
40 | save_name = 'data_day_' + str(1)
41 |
42 | print("Twinning " + save_name)
43 |
44 | # Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=n_steps,
49 | u2ss=u2ss)
50 |
51 | # Replay the twin with the same input data
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_twin_mcmc')
56 |
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data_extended(day=1)
40 | # Cut data up to 11:00
41 | data = data.iloc[0:348, :]
42 | # Modify label of second day
43 | data.loc[312, "cho_label"] = 'B2'
44 |
45 | save_name = 'data_day_' + str(1) + '_extended'
46 |
47 | print("Twinning " + save_name)
48 |
49 | # Run twinning procedure using also the burn-out period
50 | rbg.twin(data=data, bw=bw, save_name=save_name,
51 | twinning_method='mcmc',
52 | parallelize=parallelize,
53 | n_steps=n_steps,
54 | u2ss=u2ss,
55 | extended=True,
56 | find_start_guess_first=True)
57 |
58 | # Cut data up to 4:00
59 | data = data.iloc[0:264, :]
60 |
61 | # Replay the twin with the data up to the start of the burn-out period
62 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
63 | twinning_method='mcmc',
64 | save_workspace=True,
65 | save_suffix='_twin_mcmc_extended')
66 |
67 | Visualizer.plot_replay_results(replay_results, data=data)
68 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
69 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/twinning/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/twinning/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/twinning/map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import warnings
3 | import pickle
4 | import numpy as np
5 |
6 | from typing import Dict, Callable
7 |
8 | from multiprocessing import Pool
9 | from tqdm import tqdm
10 | from scipy.optimize import minimize
11 |
12 | from py_replay_bg.data import ReplayBGData
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 |
16 | from py_replay_bg.environment import Environment
17 |
18 | # Suppress all RuntimeWarnings
19 | warnings.filterwarnings("ignore", category=RuntimeWarning)
20 |
21 |
22 | class MAP:
23 | """
24 | A class that orchestrates the twinning process via MAP.
25 |
26 | Attributes
27 | ----------
28 | max_iter: int
29 | Maximum number of iterations.
30 | parallelize : bool
31 | A boolean that specifies whether to parallelize the twinning process.
32 | n_processes : int
33 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
34 |
35 | Methods
36 | -------
37 | twin(rbg_data, model, save_name, environment)
38 | Runs the twinning procedure.
39 | """
40 |
41 | def __init__(self,
42 | max_iter: int = 100000,
43 | parallelize: bool = False,
44 | n_processes: int | None = None,
45 | ):
46 | """
47 | Constructs all the necessary attributes for the MCMC object.
48 |
49 | Parameters
50 | ----------
51 | max_iter: int, optional, default : 100000
52 | Maximum number of iterations.
53 | parallelize : bool, optional, default : False
54 | A boolean that specifies whether to parallelize the twinning process.
55 | n_processes : int, optional, default : None
56 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
57 |
58 | Returns
59 | -------
60 | None
61 |
62 | Raises
63 | ------
64 | None
65 |
66 | See Also
67 | --------
68 | None
69 |
70 | Examples
71 | --------
72 | None
73 | """
74 |
75 | # Number of times to re-run the procedure
76 | self.n_rerun = 64
77 |
78 | # Maximum number of iterations
79 | self.max_iter = max_iter
80 |
81 | # Maximum number of function evaluations
82 | self.max_fev = 1000000
83 |
84 | # Parallelization options
85 | self.parallelize = parallelize
86 | self.n_processes = n_processes
87 |
88 | def twin(self,
89 | rbg_data: ReplayBGData,
90 | model: T1DModelSingleMeal | T1DModelMultiMeal,
91 | save_name: str,
92 | environment: Environment,
93 | start_guess: Dict = None,
94 | for_start_guess: bool = False) -> Dict:
95 | """
96 | Runs the twinning procedure.
97 |
98 | Parameters
99 | ----------
100 | rbg_data: ReplayBGData
101 | An object containing the data to be used during the twinning procedure.
102 | model: T1DModelSingleMeal | T1DModelMultiMeal
103 | An object that represents the physiological model to be used by ReplayBG.
104 | environment: Environment
105 | An object that represents the hyperparameters to be used by ReplayBG.
106 | save_name : str
107 | A string used to label, thus identify, each output file and result.
108 | start_guess: Dict, optional, default : None
109 | The initial guess for the twinning process. If None, this is set to population values.
110 | for_start_guess: bool, optional, default : False
111 | Whether to use the twin method just to find the start guess to pass to the MCMC twinner.
112 |
113 | Returns
114 | -------
115 | draws: dict
116 | A dictionary containing the chain and the samples obtained from the MCMC procedure and the copula
117 | sampling, respectively.
118 |
119 | Raises
120 | ------
121 | None
122 |
123 | See Also
124 | --------
125 | None
126 |
127 | Examples
128 | --------
129 | None
130 | """
131 |
132 | # If this is being used to find the start_guess, do /4 less reruns
133 | if for_start_guess:
134 | self.n_rerun = int(self.n_rerun/16)
135 |
136 | # Number of unknown parameters to twin
137 | n_dim = len(model.unknown_parameters)
138 |
139 | if start_guess is None:
140 | sg = model.start_guess
141 | else:
142 | sg = np.array(list(start_guess.values()))
143 |
144 | # Set the initial positions of the walkers.
145 | start = sg + model.start_guess_sigma * np.random.randn(self.n_rerun, n_dim)
146 | start[start < 0] = 0
147 |
148 | # Set the pooler
149 | pool = None
150 | if self.parallelize:
151 | pool = Pool(processes=self.n_processes)
152 |
153 | # Set up the options
154 | options = dict()
155 | options['maxiter'] = self.max_iter
156 | options['maxfev'] = self.max_fev
157 | options['disp'] = False
158 |
159 | # Select the function to minimize
160 | neg_log_posterior_func = model.neg_log_posterior_extended if model.extended else model.neg_log_posterior
161 |
162 | # Initialize results
163 | results = []
164 |
165 | if pool is None:
166 |
167 | if environment.verbose:
168 | iterator = tqdm(range(self.n_rerun))
169 | iterator.set_description("Min loss: %f" % np.nan)
170 | else:
171 | iterator = range(self.n_rerun)
172 |
173 | # Initialize best
174 | best = -1
175 |
176 | for r in iterator:
177 | result = run_map(start[r], neg_log_posterior_func, rbg_data, options)
178 | results.append(result)
179 | if best == -1 or result['fun'] < results[best]['fun']:
180 | best = r
181 | if environment.verbose:
182 | iterator.set_description("Min loss %f" % results[best]['fun'])
183 |
184 | else:
185 | # Prepare input arguments as tuples for starmap
186 | args = [(start[r], neg_log_posterior_func, rbg_data, options) for r in range(self.n_rerun)]
187 |
188 | # Initialize best
189 | best = -1
190 |
191 | # Get results (verbosity not allowed for the moment)
192 | results = pool.starmap(run_map, args)
193 |
194 | # Get best
195 | for r, result in enumerate(results):
196 | if best == -1 or result['fun'] < results[best]['fun']:
197 | best = r
198 |
199 | draws = dict()
200 | for up in range(n_dim):
201 | draws[model.unknown_parameters[up]] = results[best]['x'][up]
202 |
203 | # If twin is being used just to find the start guess, just return draws without saving
204 | if for_start_guess:
205 | return draws
206 |
207 | # Clean-up draws from "extended" parameters
208 | if model.extended:
209 | if 'SI_B2' in draws:
210 | del draws['SI_B2']
211 | if 'kabs_B2' in draws:
212 | del draws['kabs_B2']
213 | if 'beta_B2' in draws:
214 | del draws['beta_B2']
215 | if 'kabs_L2' in draws:
216 | del draws['kabs_L2']
217 | if 'beta_L2' in draws:
218 | del draws['beta_L2']
219 | if 'kabs_S2' in draws:
220 | del draws['kabs_S2']
221 | if 'beta_S2' in draws:
222 | del draws['beta_S2']
223 |
224 | # Save results
225 | twinning_results = dict()
226 | twinning_results['draws'] = draws
227 | twinning_results['u2ss'] = model.model_parameters.u2ss
228 |
229 | saved_file = os.path.join(environment.replay_bg_path, 'results', 'map',
230 | 'map_' + save_name + '.pkl')
231 |
232 | with open(saved_file, 'wb') as file:
233 | pickle.dump(twinning_results, file)
234 |
235 | if environment.verbose:
236 | print('Parameters saved in ' + saved_file)
237 |
238 | return draws
239 |
240 |
241 | def run_map(start: np.ndarray,
242 | neg_log_posterior_func: Callable,
243 | rbg_data: ReplayBGData,
244 | options: Dict
245 | ) -> Dict:
246 | """
247 | Utility function used to run MAP twinning.
248 |
249 | Parameters
250 | ----------
251 | start: np.ndarray
252 | An object containing the data to be used during the twinning procedure.
253 | neg_log_posterior_func: Callable
254 | The function to minimize, i.e., the neg-loglikelihood.
255 | rbg_data: ReplayBGData
256 | An object containing the data to be used during the twinning procedure.
257 | options : Dict
258 | A dictionary with the options necessary to the minimization function.
259 |
260 | Returns
261 | -------
262 | ret: dict
263 | A dictionary containing the results of the MAP twinning and the final value of the objective function.
264 |
265 | Raises
266 | ------
267 | None
268 |
269 | See Also
270 | --------
271 | None
272 |
273 | Examples
274 | --------
275 | None
276 | """
277 | result = minimize(neg_log_posterior_func, start, method='Powell', args=(rbg_data,), options=options)
278 | ret = dict()
279 | ret['fun'] = result.fun
280 | ret['x'] = result.x
281 | return ret
282 |
--------------------------------------------------------------------------------
/py_replay_bg/utils/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/utils/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/utils/stats.py:
--------------------------------------------------------------------------------
1 | import math
2 | import numpy as np
3 | from numba import njit
4 |
5 |
6 | @njit(fastmath=True)
7 | def log_lognorm(x, mu, sigma):
8 | """
9 | Computes the logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
10 |
11 | Parameters
12 | ----------
13 | x: float
14 | The value where to evaluate the log-normal pdf.
15 | mu: float
16 | The mean of the log-normal distribution.
17 | sigma: float
18 | The standard deviation of the log-normal distribution.
19 |
20 | Returns
21 | -------
22 | ll_norm: float
23 | The logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
24 |
25 | Raises
26 | ------
27 | None
28 |
29 | See Also
30 | --------
31 | None
32 |
33 | Examples
34 | --------
35 | None
36 | """
37 | return np.log(1 / (x * sigma * np.sqrt(2 * np.pi)) * np.exp(- ((np.log(x) - mu) ** 2) / (2 * (sigma ** 2))))
38 |
39 |
40 | @njit(fastmath=True)
41 | def log_norm(x, mu, sigma):
42 | """
43 | Computes the logarithm of the normal pdf evaluated at given x with given mu and sigma.
44 |
45 | Parameters
46 | ----------
47 | x: float
48 | The value where to evaluate the normal pdf.
49 | mu: float
50 | The mean of the normal distribution.
51 | sigma: float
52 | The standard deviation of the normal distribution.
53 |
54 | Returns
55 | -------
56 | l_norm: float
57 | The logarithm of the normal pdf evaluated at given x with given mu and sigma.
58 |
59 | Raises
60 | ------
61 | None
62 |
63 | See Also
64 | --------
65 | None
66 |
67 | Examples
68 | --------
69 | None
70 | """
71 | return np.log(1 / (sigma * np.sqrt(2 * np.pi)) * np.exp(- 0.5 * ((x - mu) / sigma) ** 2))
72 |
73 |
74 | @njit(fastmath=True)
75 | def log_gamma(x, alpha, beta):
76 | """
77 | Computes the logarithm of the gamma pdf evaluated at given x with given alpha and beta.
78 |
79 | Parameters
80 | ----------
81 | x: float
82 | The value where to evaluate the normal pdf.
83 | alpha: float
84 | The alpha value of the gamma distribution at hand.
85 | beta: float
86 | The beta value of the gamma distribution at hand.
87 |
88 | Returns
89 | -------
90 | l_gam: float
91 | The logarithm of the gamma pdf evaluated at given x with given alpha and beta.
92 |
93 | Raises
94 | ------
95 | None
96 |
97 | See Also
98 | --------
99 | None
100 |
101 | Examples
102 | --------
103 | None
104 | """
105 | return -np.inf if x < 0 else np.log((beta ** alpha * x ** (alpha - 1) * math.exp(-beta * x)) / math.gamma(alpha))
106 |
--------------------------------------------------------------------------------
/py_replay_bg/visualizer/__init__.py:
--------------------------------------------------------------------------------
1 | from datetime import datetime, timedelta
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 |
6 | from typing import Dict
7 |
8 | import pandas as pd
9 | from tqdm import tqdm
10 |
11 | from py_replay_bg.data import ReplayBGData
12 | from py_replay_bg.environment import Environment
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 | from py_replay_bg.dss import DSS
16 |
17 |
18 | class Visualizer:
19 | """
20 | A class to be used by ReplayBG for plotting.
21 |
22 | ...
23 | Attributes
24 | ----------
25 | -
26 |
27 | Methods
28 | -------
29 | plot_replay_results(replay_results, data)
30 | Function that plots the results of ReplayBG replay simulation.
31 | """
32 |
33 | def __init__(self):
34 | """
35 | Constructs all the necessary attributes for the Visualizer object.
36 |
37 | Parameters
38 | ----------
39 | -
40 |
41 | Returns
42 | -------
43 | None
44 |
45 | Raises
46 | ------
47 | None
48 |
49 | See Also
50 | --------
51 | None
52 |
53 | Examples
54 | --------
55 | None
56 | """
57 | pass
58 |
59 | @staticmethod
60 | def plot_replay_results(
61 | replay_results: Dict,
62 | data: pd.DataFrame = None,
63 | title: str = '',
64 | ) -> None:
65 | """
66 | Function that plots the results of ReplayBG simulation.
67 |
68 | Parameters
69 | ----------
70 | replay_results: dict
71 | The replayed scenario results with fields:
72 | glucose: dict
73 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
74 | cgm: dict
75 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
76 | insulin_bolus: dict
77 | A dictionary which contains the insulin boluses simulated via ReplayBG.
78 | correction_bolus: dict
79 | A dictionary which contains the correction boluses simulated via ReplayBG.
80 | insulin_basal: dict
81 | A dictionary which contains the basal insulin simulated via ReplayBG.
82 | cho: dict
83 | A dictionary which contains the meals simulated via ReplayBG.
84 | hypotreatments: dict
85 | A dictionary which contains the hypotreatments simulated via ReplayBG.
86 | meal_announcement: dict
87 | A dictionary which contains the meal announcements simulated via ReplayBG.
88 | vo2: dict
89 | A dictionary which contains the vo2 simulated via ReplayBG.
90 | sensors: dict
91 | A dictionary which contains the sensors used during the replayed scenario.
92 | rbg_data: ReplayBGData
93 | The data to be used by ReplayBG during simulation.
94 | model: T1DModelSingleMeal | T1DModelMultiMeal
95 | An object that represents the physiological model to be used by ReplayBG.
96 | data: pd.DataFrame, optional, default: None
97 | Pandas dataframe which contains the data to be used by the tool. If present, adds glucose data to the
98 | glucose subplot.
99 |
100 | Returns
101 | -------
102 | None
103 |
104 | Raises
105 | ------
106 | None
107 |
108 | See Also
109 | --------
110 | None
111 |
112 | Examples
113 | --------
114 | None
115 | """
116 | # Subplot 1: Glucose
117 |
118 | fig, ax = None, None
119 | if replay_results['model'].exercise:
120 | fig, ax = plt.subplots(4, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1, 1]})
121 | else:
122 | fig, ax = plt.subplots(3, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1]})
123 |
124 | if data is not None:
125 | ax[0].plot(data.t, data.glucose, marker='*', color='red', linewidth=2, label='CGM data [mg/dl]')
126 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['cgm']['median'], marker='o', color='black', linewidth=2, label='CGM replay (Median) [mg/dl]')
127 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['cgm']['ci25th'], replay_results['cgm']['ci75th'], color='black', alpha=0.2,
128 | label='CGM replay (CI 25-75th) [mg/dl]')
129 |
130 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['glucose']['median'][::replay_results['model'].yts], marker='o', color='blue', linewidth=2,
131 | label='Glucose replay (Median) [mg/dl]')
132 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['glucose']['ci25th'][::replay_results['model'].yts], replay_results['glucose']['ci75th'][::replay_results['model'].yts], color='blue',
133 | alpha=0.3, label='Glucose replay (CI 25-75th) [mg/dl]')
134 |
135 | ax[0].grid()
136 | ax[0].legend()
137 |
138 | # Subplot 2: Meals
139 | t = np.arange(replay_results['rbg_data'].t_data[0]+ pd.Timedelta(minutes=0), replay_results['rbg_data'].t_data[-1] + pd.Timedelta(minutes=replay_results['model'].yts),
140 | timedelta(minutes=1)).astype(datetime)
141 |
142 | cho_events = np.sum(replay_results['cho']['realizations'], axis=0) / replay_results['cho']['realizations'].shape[0]
143 | ht_events = np.sum(replay_results['hypotreatments']['realizations'], axis=0) / replay_results['hypotreatments']['realizations'].shape[0]
144 |
145 | markerline, stemlines, baseline = ax[1].stem(t, cho_events, basefmt='k:', label='CHO replay (Mean) [g/min]')
146 | plt.setp(stemlines, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
147 | plt.setp(markerline, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
148 |
149 | markerline, stemlines, baseline = ax[1].stem(t, ht_events, basefmt='k:', label='HT replay (Mean) [g/min]')
150 | plt.setp(stemlines, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
151 | plt.setp(markerline, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
152 |
153 | ax[1].grid()
154 | ax[1].legend()
155 |
156 | # Subplot 3: Insulin
157 |
158 | bolus_events = np.sum(replay_results['insulin_bolus']['realizations'], axis=0) / replay_results['insulin_bolus']['realizations'].shape[0]
159 | cb_events = np.sum(replay_results['correction_bolus']['realizations'], axis=0) / replay_results['correction_bolus']['realizations'].shape[0]
160 | basal_rate = np.sum(replay_results['insulin_basal']['realizations'], axis=0) / replay_results['insulin_basal']['realizations'].shape[0]
161 |
162 | markerline, stemlines, baseline = ax[2].stem(t, bolus_events, basefmt='k:',
163 | label='Bolus insulin replay (Mean) [U/min]')
164 | plt.setp(stemlines, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
165 | plt.setp(markerline, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
166 |
167 | markerline, stemlines, baseline = ax[2].stem(t, cb_events, basefmt='k:',
168 | label='CB insulin replay (Mean) [U/min]')
169 | plt.setp(stemlines, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
170 | plt.setp(markerline, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
171 |
172 | ax[2].plot(t, basal_rate * 60, color='black', linewidth=2, label='Basal insulin replay (Mean) [U/h]')
173 |
174 | ax[2].grid()
175 | ax[2].legend()
176 |
177 | # Subplot 4: Exercise
178 |
179 | if replay_results['model'].exercise:
180 |
181 | vo2_events = np.sum(replay_results['vo2']['realizations'], axis=0) / replay_results['vo2']['realizations'].shape[0]
182 |
183 | markerline, stemlines, baseline = ax[3].stem(t, vo2_events, basefmt='k:', label='VO2 replay (Mean) [-]')
184 | plt.setp(stemlines, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
185 | plt.setp(markerline, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
186 |
187 | ax[3].grid()
188 | ax[3].legend()
189 |
190 | fig.suptitle(title, fontweight='bold')
191 | plt.show()
192 |
193 |
194 | @staticmethod
195 | def plot_replay_results_interval(
196 | replay_results_interval: list,
197 | data_interval: list = None,
198 | title: str = '',
199 | ) -> None:
200 | """
201 | Function that plots the results of ReplayBG simulation (intervals).
202 |
203 | Parameters
204 | ----------
205 | replay_results_interval: list
206 | A list dictionaries of replayed scenario results. Each element has fields:
207 | glucose: dict
208 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
209 | cgm: dict
210 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
211 | insulin_bolus: dict
212 | A dictionary which contains the insulin boluses simulated via ReplayBG.
213 | correction_bolus: dict
214 | A dictionary which contains the correction boluses simulated via ReplayBG.
215 | insulin_basal: dict
216 | A dictionary which contains the basal insulin simulated via ReplayBG.
217 | cho: dict
218 | A dictionary which contains the meals simulated via ReplayBG.
219 | hypotreatments: dict
220 | A dictionary which contains the hypotreatments simulated via ReplayBG.
221 | meal_announcement: dict
222 | A dictionary which contains the meal announcements simulated via ReplayBG.
223 | vo2: dict
224 | A dictionary which contains the vo2 simulated via ReplayBG.
225 | sensors: dict
226 | A dictionary which contains the sensors used during the replayed scenario.
227 | rbg_data: ReplayBGData
228 | The data to be used by ReplayBG during simulation.
229 | model: T1DModelSingleMeal | T1DModelMultiMeal
230 | An object that represents the physiological model to be used by ReplayBG.
231 | data_interval: list, optional, default: None
232 | A list of pandas dataframe which contains the data to be used by the tool. If present, adds glucose data
233 | to the glucose subplot.
234 |
235 | Returns
236 | -------
237 | None
238 |
239 | Raises
240 | ------
241 | None
242 |
243 | See Also
244 | --------
245 | None
246 |
247 | Examples
248 | --------
249 | None
250 | """
251 | data = None
252 | if data_interval is not None:
253 | data = pd.concat(data_interval, axis=0, ignore_index=True)
254 |
255 | # Pad each array to the maximum shape
256 | replay_results = dict()
257 |
258 |
259 | # Re-map glucose data
260 | fields = ['median', 'ci5th', 'ci25th', 'ci75th', 'ci95th']
261 | category = ['glucose', 'cgm']
262 |
263 | for c in category:
264 |
265 | replay_results[c] = dict()
266 |
267 | for f in fields:
268 | replay_results[c][f] = dict()
269 |
270 | replay_results[c][f] = [r[c][f] for r in replay_results_interval]
271 | replay_results[c][f] = np.concatenate(replay_results[c][f], axis=0)
272 |
273 | # Re-map cho, insulin, and vo2 data
274 | category = ['cho', 'hypotreatments', 'insulin_bolus', 'correction_bolus', 'insulin_basal', 'vo2']
275 |
276 | for c in category:
277 | replay_results[c] = dict()
278 | replay_results[c]['realizations'] = [r[c]['realizations'] for r in replay_results_interval]
279 | replay_results[c]['realizations'] = np.concatenate(replay_results[c]['realizations'], axis=1)
280 |
281 | replay_results['model'] = replay_results_interval[0]['model']
282 | t_data = [r['rbg_data'].t_data for r in replay_results_interval]
283 | replay_results['rbg_data'] = replay_results_interval[0]['rbg_data']
284 | replay_results['rbg_data'].t_data = np.concatenate(t_data, axis=0)
285 |
286 | Visualizer.plot_replay_results(replay_results=replay_results, data=data, title=title)
287 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.pytest.ini_options]
2 | pythonpath = [
3 | "."
4 | ]
5 |
6 | [build-system]
7 | requires = ["setuptools>=61.0.0", "wheel"]
8 | build-backend = "setuptools.build_meta"
9 |
10 | [project]
11 | name = "py_replay_bg"
12 | version = "1.1.0"
13 | authors = [
14 | { name="Giacomo Cappon", email="cappongiacomo@gmail.com" },
15 | ]
16 | description = "ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management."
17 | readme = "README.md"
18 | requires-python = ">=3.11"
19 | classifiers = [
20 | "Programming Language :: Python :: 3",
21 | "License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
22 | "Operating System :: OS Independent",
23 | "Intended Audience :: Science/Research",
24 | "Topic :: Scientific/Engineering",
25 | ]
26 | dynamic = ["dependencies"]
27 | [tool.setuptools.dynamic]
28 | dependencies = {file = ["requirements.txt"]}
29 | optional-dependencies = {dev = { file = ["requirements-dev.txt"] }}
30 |
31 | [project.urls]
32 | "Homepage" = "https://github.com/gcappon/py_replay_bg"
33 | "Bug Tracker" = "https://github.com/gcappon/py_replay_bg/issues"
34 |
35 | [tool.setuptools.packages.find]
36 | include = ["py_replay_bg*"]
37 | exclude = ["venv*"]
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/tsconfig.json:
--------------------------------------------------------------------------------
1 | {
2 | "compilerOptions": {
3 | "module": "NodeNext",
4 | "moduleResolution": "NodeNext",
5 | "target": "ES2022"
6 | },
7 | "include": [
8 | "docs/.vuepress/**/*.ts",
9 | "docs/.vuepress/**/*.vue"
10 | ],
11 | "exclude": [
12 | "node_modules"
13 | ]
14 | }
15 |
--------------------------------------------------------------------------------
 83 |
84 | [](https://github.com/gcappon/py_replay_bg/COPYING)
85 | [](https://github.com/gcappon/py_replay_bg/commits/master)
86 |
87 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
88 |
89 | # Reference
90 |
91 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
92 |
93 | # Get started
94 |
95 | ## Installation
96 |
97 | **ReplayBG** can be installed via pypi by simply
98 |
99 | ```python
100 | pip install py-replay-bg
101 | ```
102 |
103 | ### Requirements
104 |
105 | * Python >= 3.11
106 | * List of Python packages in `requirements.txt`
107 |
108 | ## Preparation: imports, setup, and data loading
109 |
110 | First of all import the core modules:
111 | ```python
112 | import os
113 | import numpy as np
114 | import pandas as pd
115 |
116 | from multiprocessing import freeze_support
117 | ```
118 |
119 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
120 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
121 | parallelized way.
122 |
123 | Then, we will import the necessary ReplayBG modules:
124 | ```python
125 | from py_replay_bg.py_replay_bg import ReplayBG
126 | from py_replay_bg.visualizer import Visualizer
127 | from py_replay_bg.analyzer import Analyzer
128 | ```
129 |
130 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
131 | while `Analyzer` and `Visualizer` are utility objects that will be used to
132 | respectively analyze and visualize the results that we will produce with ReplayBG
133 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
134 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
135 |
136 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
137 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
138 | ```python
139 | if __name__ == '__main__':
140 | freeze_support()
141 | ```
142 |
143 | Then, we will set the verbosity of ReplayBG:
144 | ```python
145 | verbose = True
146 | plot_mode = False
147 | ```
148 |
149 | Then, we need to decide what blueprint to use for twinning the data at hand.
150 | ```python
151 | blueprint = 'multi-meal'
152 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
153 | parallelize = True
154 | ```
155 |
156 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
157 |
158 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
159 | which contains a day of data of a patient with T1D:
160 |
161 | ```python
162 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
163 | data.t = pd.to_datetime(data['t'])
164 | ```
165 |
166 | ::: warning
167 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
168 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
169 | :::
170 |
171 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
172 |
173 | ```python
174 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
175 | p = np.where(patient_info['patient'] == 1)[0][0]
176 | # Set bw and u2ss
177 | bw = float(patient_info.bw.values[p])
178 | u2ss = float(patient_info.u2ss.values[p])
179 | ```
180 |
181 | Finally, instantiate a `ReplayBG` object:
182 |
183 | ```python
184 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
185 | yts=5, exercise=False,
186 | seed=1,
187 | verbose=verbose, plot_mode=plot_mode)
188 |
189 | ```
190 |
191 | ## Step 1: Creation of the digital twin
192 |
193 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
194 |
195 | ```python
196 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
197 | twinning_method='mcmc',
198 | parallelize=parallelize,
199 | n_steps=5000,
200 | u2ss=u2ss)
201 | ```
202 |
203 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
204 |
205 |
206 | ## Step 2: Run replay simulations
207 |
208 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
209 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
210 |
211 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
212 |
213 | ```python
214 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
215 | twinning_method='mcmc',
216 | save_workspace=True,
217 | save_suffix='_step_2a')
218 | ```
219 |
220 | It is possible to visualize the results of the simulation using:
221 |
222 | ```python
223 | Visualizer.plot_replay_results(replay_results, data=data)
224 | ```
225 |
226 | and analyzing the results using:
227 |
228 | ```python
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
231 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
232 | ```
233 |
234 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
235 | To do that run:
236 |
237 | ```python
238 | data.bolus = data.bolus * .7
239 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
240 | twinning_method='mcmc',
241 | save_workspace=True,
242 | save_suffix='_step_2b')
243 |
244 | # Visualize results
245 | Visualizer.plot_replay_results(replay_results)
246 | # Analyze results
247 | analysis = Analyzer.analyze_replay_results(replay_results)
248 |
249 | # Print, for example, the average glucose
250 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
251 | ```
252 |
253 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
254 |
255 | # Documentation
256 |
257 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
258 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/SOURCES.txt:
--------------------------------------------------------------------------------
1 | COPYING.md
2 | README.md
3 | pyproject.toml
4 | requirements.txt
5 | py_replay_bg/__init__.py
6 | py_replay_bg/py_replay_bg.py
7 | py_replay_bg.egg-info/PKG-INFO
8 | py_replay_bg.egg-info/SOURCES.txt
9 | py_replay_bg.egg-info/dependency_links.txt
10 | py_replay_bg.egg-info/requires.txt
11 | py_replay_bg.egg-info/top_level.txt
12 | py_replay_bg/analyzer/__init__.py
13 | py_replay_bg/data/__init__.py
14 | py_replay_bg/dss/__init__.py
15 | py_replay_bg/dss/default_dss_handlers.py
16 | py_replay_bg/environment/__init__.py
17 | py_replay_bg/example/code/analysis_example.py
18 | py_replay_bg/example/code/analysis_intervals_example.py
19 | py_replay_bg/example/code/get_started.py
20 | py_replay_bg/example/code/replay_intervals_map.py
21 | py_replay_bg/example/code/replay_intervals_mcmc.py
22 | py_replay_bg/example/code/replay_map.py
23 | py_replay_bg/example/code/replay_map_dss.py
24 | py_replay_bg/example/code/replay_mcmc.py
25 | py_replay_bg/example/code/replay_mcmc_dss.py
26 | py_replay_bg/example/code/twin_intervals_map.py
27 | py_replay_bg/example/code/twin_intervals_mcmc.py
28 | py_replay_bg/example/code/twin_map.py
29 | py_replay_bg/example/code/twin_map_extended.py
30 | py_replay_bg/example/code/twin_mcmc.py
31 | py_replay_bg/example/code/twin_mcmc_extended.py
32 | py_replay_bg/example/code/utils.py
33 | py_replay_bg/input_validation/__init__.py
34 | py_replay_bg/input_validation/input_validator_init.py
35 | py_replay_bg/input_validation/input_validator_replay.py
36 | py_replay_bg/input_validation/input_validator_twin.py
37 | py_replay_bg/model/__init__.py
38 | py_replay_bg/model/logpriors_t1d.py
39 | py_replay_bg/model/model_parameters_t1d.py
40 | py_replay_bg/model/model_step_equations_t1d.py
41 | py_replay_bg/model/t1d_model_multi_meal.py
42 | py_replay_bg/model/t1d_model_single_meal.py
43 | py_replay_bg/replay/__init__.py
44 | py_replay_bg/sensors/__init__.py
45 | py_replay_bg/tests/__init__.py
46 | py_replay_bg/tests/test_analysis_example.py
47 | py_replay_bg/tests/test_analysis_intervals_example.py
48 | py_replay_bg/tests/test_replay_intervals_map.py
49 | py_replay_bg/tests/test_replay_intervals_mcmc.py
50 | py_replay_bg/tests/test_replay_map.py
51 | py_replay_bg/tests/test_replay_map_dss.py
52 | py_replay_bg/tests/test_replay_map_extended.py
53 | py_replay_bg/tests/test_replay_mcmc.py
54 | py_replay_bg/tests/test_replay_mcmc_dss.py
55 | py_replay_bg/tests/test_replay_mcmc_extended.py
56 | py_replay_bg/tests/test_twin_intervals_map.py
57 | py_replay_bg/tests/test_twin_intervals_mcmc.py
58 | py_replay_bg/tests/test_twin_map.py
59 | py_replay_bg/tests/test_twin_map_extended.py
60 | py_replay_bg/tests/test_twin_mcmc.py
61 | py_replay_bg/tests/test_twin_mcmc_extended.py
62 | py_replay_bg/twinning/__init__.py
63 | py_replay_bg/twinning/map.py
64 | py_replay_bg/twinning/mcmc.py
65 | py_replay_bg/utils/__init__.py
66 | py_replay_bg/utils/stats.py
67 | py_replay_bg/visualizer/__init__.py
68 | venv/bin/rst2html.py
69 | venv/bin/rst2html4.py
70 | venv/bin/rst2html5.py
71 | venv/bin/rst2latex.py
72 | venv/bin/rst2man.py
73 | venv/bin/rst2odt.py
74 | venv/bin/rst2odt_prepstyles.py
75 | venv/bin/rst2pseudoxml.py
76 | venv/bin/rst2s5.py
77 | venv/bin/rst2xetex.py
78 | venv/bin/rst2xml.py
79 | venv/bin/rstpep2html.py
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/dependency_links.txt:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/requires.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/top_level.txt:
--------------------------------------------------------------------------------
1 | py_replay_bg
2 |
--------------------------------------------------------------------------------
/py_replay_bg/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/dss/__init__.py:
--------------------------------------------------------------------------------
1 | from typing import Callable, Dict
2 |
3 | from py_replay_bg.dss.default_dss_handlers import default_meal_generator_handler, standard_bolus_calculator_handler, \
4 | default_basal_handler, ada_hypotreatments_handler, corrects_above_250_handler
5 |
6 |
7 | class DSS:
8 | """
9 | A class that represents the hyperparameters of the integrated decision support system.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | bw: double
15 | The patient's body weight.
16 | meal_generator_handler: Callable
17 | A callback function that implements a meal generator to be used during the replay of a given scenario.
18 | meal_generator_handler_params: dict
19 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
20 | bolus_calculator_handler: Callable
21 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
22 | bolus_calculator_handler_params: dict
23 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
24 | serves as memory area for the bolusCalculatorHandler function.
25 | basal_handler: Callable
26 | A callback function that implements a basal controller to be used during the replay of a given scenario.
27 | basal_handler_params: dict
28 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
29 | memory area for the basalHandler function.
30 | enable_hypotreatments: boolean
31 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
32 | hypotreatments_handler: Callable
33 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
34 | hypotreatments_handler_params: dict
35 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
36 | serves as memory area for the hypoTreatmentsHandler function.
37 | enable_correction_boluses: boolean
38 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
39 | correction_boluses_handler: Callable
40 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
41 | correction_boluses_handler_params: dict
42 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also
43 | serves as memory area for the correctionBolusesHandler function.
44 |
45 | Methods
46 | -------
47 | None
48 | """
49 |
50 | def __init__(self,
51 | bw: float,
52 | meal_generator_handler: Callable = default_meal_generator_handler,
53 | meal_generator_handler_params: Dict | None = None,
54 | bolus_calculator_handler: Callable = standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params: Dict | None = None,
56 | basal_handler: Callable = default_basal_handler,
57 | basal_handler_params: Dict | None = None,
58 | enable_hypotreatments: bool = False,
59 | hypotreatments_handler: Callable = ada_hypotreatments_handler,
60 | hypotreatments_handler_params: Dict | None = None,
61 | enable_correction_boluses: bool = False,
62 | correction_boluses_handler: Callable = corrects_above_250_handler,
63 | correction_boluses_handler_params: Dict | None = None,
64 | ):
65 | """
66 | Constructs all the necessary attributes for the DSS object.
67 |
68 | Parameters
69 | ----------
70 | bw: double
71 | The patient's body weight.
72 | meal_generator_handler: Callable, optional, default : default_meal_generator_handler
73 | A callback function that implements a meal generator to be used during the replay of a given scenario.
74 | meal_generator_handler_params: dict, optional, default : None
75 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
76 | bolus_calculator_handler: Callable, optional, default : standard_bolus_calculator_handler
77 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
78 | bolus_calculator_handler_params: dict, optional, default : None
79 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
80 | serves as memory area for the bolusCalculatorHandler function.
81 | basal_handler: function, Callable, default : default_basal_handler
82 | A callback function that implements a basal controller to be used during the replay of a given scenario.
83 | basal_handler_params: dict, optional, default : None
84 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
85 | memory area for the basalHandler function.
86 | enable_hypotreatments: boolean, optional, default : False
87 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
88 | hypotreatments_handler: Callable, optional, default : ada_hypotreatments_handler
89 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
90 | hypotreatments_handler_params: dict, optional, default : None
91 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
92 | serves as memory area for the hypoTreatmentsHandler function.
93 | enable_correction_boluses: boolean, optional, default : False
94 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
95 | correction_boluses_handler: Callable, optional, default : corrects_above_250_handler
96 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
97 | correction_boluses_handler_params: dict, optional, default : None
98 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function.
99 | It also serves as memory area for the correctionBolusesHandler function.
100 | """
101 |
102 | # Patient's body weight
103 | self.bw = bw
104 |
105 | # Meal Generator module parameters
106 | self.meal_generator_handler = meal_generator_handler
107 | self.meal_generator_handler_params = meal_generator_handler_params if meal_generator_handler_params is not None else {}
108 |
109 | # Bolus Calculator module parameters
110 | self.bolus_calculator_handler = bolus_calculator_handler
111 | self.bolus_calculator_handler_params = bolus_calculator_handler_params if bolus_calculator_handler_params is not None else {}
112 |
113 | # Basal module parameters
114 | self.basal_handler = basal_handler
115 | self.basal_handler_params = basal_handler_params if basal_handler_params is not None else {}
116 |
117 | # Hypotreatment module parameters
118 | self.enable_hypotreatments = enable_hypotreatments
119 | self.hypotreatments_handler = hypotreatments_handler
120 | self.hypotreatments_handler_params = hypotreatments_handler_params if hypotreatments_handler_params is not None else {}
121 |
122 | # Correction bolus module parameters
123 | self.enable_correction_boluses = enable_correction_boluses
124 | self.correction_boluses_handler = correction_boluses_handler
125 | self.correction_boluses_handler_params = correction_boluses_handler_params if correction_boluses_handler_params is not None else {}
126 |
--------------------------------------------------------------------------------
/py_replay_bg/environment/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 |
4 | class Environment:
5 | """
6 | A class that represents the hyperparameters to be used by ReplayBG.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | save_name : str
12 | A string used to label, thus identify, each output file and result.
13 | replay_bg_path: str
14 | The current absolute path of ReplayBG.
15 |
16 | blueprint: str, {'single-meal', 'multi-meal'}
17 | A string that specifies the blueprint to be used to create the digital twin.
18 | yts: int
19 | An integer that specifies the data sampling time (in minutes).
20 |
21 | seed : int
22 | An integer that specifies the random seed. For reproducibility.
23 |
24 | plot_mode : bool
25 | A boolean that specifies whether to show the plot of the results or not.
26 | verbose : bool
27 | A boolean that specifies the verbosity of ReplayBG.
28 |
29 | Methods
30 | -------
31 | None
32 | """
33 |
34 | def __init__(self,
35 | blueprint: str = 'single_meal',
36 | save_name: str = '',
37 | save_folder: str = '',
38 | yts: int = 5,
39 | exercise: bool = False,
40 | seed: int = 1,
41 | plot_mode: bool = True,
42 | verbose: bool = True
43 | ):
44 | """
45 | Constructs all the necessary attributes for the Environment object.
46 |
47 | Parameters
48 | ----------
49 | blueprint: str, {'single-meal', 'multi-meal'}, optional, default : 'single-meal'
50 | A string that specifies the blueprint to be used to create the digital twin.
51 |
52 | save_name : str, optional, default : ''
53 | A string used to label, thus identify, each output file and result.
54 | save_folder : str, optional, default : ''
55 | A string used to set the folder where the ReplayBG results will be saved.
56 |
57 | yts: int, optional, default : 5
58 | An integer that specifies the data sampling time (in minutes).
59 | exercise: boolean, optional, default : False
60 | A boolean that specifies whether to simulate exercise or not.
61 |
62 | seed : int, optional, default : 1
63 | An integer that specifies the random seed. For reproducibility.
64 |
65 | plot_mode : boolean, optional, default : True
66 | A boolean that specifies whether to show the plot of the results or not.
67 | verbose : boolean, optional, default : True
68 | A boolean that specifies the verbosity of ReplayBG.
69 | """
70 |
71 | # Set the save name and folder
72 | self.save_name = save_name
73 | self.replay_bg_path = save_folder
74 |
75 | # Create the results sub folders if they do not exist
76 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results'))):
77 | os.mkdir(os.path.join(self.replay_bg_path, 'results'))
78 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'mcmc'))):
79 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'mcmc'))
80 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'map'))):
81 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'map'))
82 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'workspaces'))):
83 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'workspaces'))
84 |
85 | # Single-meal or multi-meal blueprint?
86 | self.blueprint = blueprint
87 |
88 | # Set sample time
89 | self.yts = yts
90 |
91 | # Set whether to use the exercise model
92 | self.exercise = exercise
93 |
94 | # Set the seed
95 | self.seed = seed
96 |
97 | # Set plot mode and verbosity
98 | self.plot_mode = plot_mode
99 | self.verbose = verbose
100 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the location of the 'results' folder
9 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
10 |
11 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
12 | replay_results = pickle.load(file)
13 |
14 | # Visualize and analyze MCMC results
15 | Visualizer.plot_replay_results(replay_results)
16 | analysis = Analyzer.analyze_replay_results(replay_results)
17 | print(" ----- Analysis using MCMC-derived twins ----- ")
18 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
19 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
20 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
21 |
22 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
23 | replay_results = pickle.load(file)
24 |
25 | # Visualize and analyze MAP results
26 | Visualizer.plot_replay_results(replay_results)
27 | analysis = Analyzer.analyze_replay_results(replay_results)
28 | print(" ----- Analysis using MAP-derived twins ----- ")
29 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
30 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
31 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
32 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the interval to analyze
9 | start_day = 1
10 | end_day = 2
11 |
12 | # Set the location of the 'results' folder
13 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
14 |
15 | # Initialize results list
16 | replay_results_interval = []
17 |
18 | for day in range(start_day, end_day+1):
19 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
20 | replay_results = pickle.load(file)
21 | replay_results_interval.append(replay_results)
22 |
23 | # Visualize and analyze MCMC results
24 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
25 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
26 | print(" ----- Analysis using MAP-derived twins ----- ")
27 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
28 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
29 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
30 |
31 | # Initialize results list
32 | replay_results_interval = []
33 |
34 | for day in range(start_day, end_day + 1):
35 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
36 | replay_results = pickle.load(file)
37 | replay_results_interval.append(replay_results)
38 |
39 | # Visualize and analyze MAP results
40 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
41 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
42 | print(" ----- Analysis using MCMC-derived twins ----- ")
43 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
44 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
45 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
46 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/get_started.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | import pandas as pd
4 |
5 | from multiprocessing import freeze_support
6 |
7 | from py_replay_bg.py_replay_bg import ReplayBG
8 | from py_replay_bg.visualizer import Visualizer
9 | from py_replay_bg.analyzer import Analyzer
10 |
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # Load data
25 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
26 | data.t = pd.to_datetime(data['t'])
27 |
28 | # Load patient_info
29 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Set save name
42 | save_name = 'data_day_1'
43 |
44 | # Step 1. Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=5000,
49 | u2ss=u2ss)
50 |
51 | # Step 2a. Replay the twin with the same input data to get the initial conditions for the subsequent day
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_step_2a')
56 |
57 | # Visualize results and compare with the original glucose data
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | # Analyze results
60 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
61 | # Print, for example, the fit MARD and the average glucose
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
63 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
64 |
65 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
66 | data.bolus = data.bolus * .7
67 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
68 | twinning_method='mcmc',
69 | save_workspace=True,
70 | u2ss=u2ss,
71 | save_suffix='_step_2b')
72 |
73 | # Visualize results
74 | Visualizer.plot_replay_results(replay_results)
75 | # Analyze results
76 | analysis = Analyzer.analyze_replay_results(replay_results)
77 |
78 | # Print, for example, the average glucose
79 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='map',
53 | save_workspace=True,
54 | x0=x0, previous_data_name=previous_data_name,
55 | save_suffix='_replay_map',
56 | sensors=sensors)
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='mcmc',
53 | n_replay=100,
54 | save_workspace=True,
55 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
56 | save_suffix='_replay_mcmc')
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | twinning_method='map',
39 | save_workspace=True,
40 | save_suffix='_replay_map')
41 |
42 | # Visualize and analyze results
43 | Visualizer.plot_replay_results(replay_results, data=data)
44 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
45 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
46 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
47 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
48 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | n_replay=10,
39 | twinning_method='mcmc',
40 | save_workspace=True,
41 | save_suffix='_replay_mcmc')
42 |
43 | # Visualize and analyze results
44 | Visualizer.plot_replay_results(replay_results, data=data)
45 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
46 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
47 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
48 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
49 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='mcmc',
50 | save_workspace=True,
51 | save_suffix='_replay_mcmc_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 | x0 = None
32 | previous_data_name = None
33 |
34 | # Initialize the list of results
35 | replay_results_interval = []
36 | data_interval = []
37 |
38 | # Instantiate ReplayBG
39 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
40 | yts=5, exercise=False,
41 | seed=1,
42 | verbose=verbose, plot_mode=plot_mode)
43 |
44 | # Set interval to twin
45 | start_day = 1
46 | end_day = 2
47 |
48 | # Twin the interval
49 | for day in range(start_day, end_day+1):
50 |
51 | # Step 1: Load data and set save_name
52 | data = load_test_data(day=day)
53 | save_name = 'data_day_' + str(day) + '_interval'
54 |
55 | print("Twinning " + save_name)
56 |
57 | # Run twinning procedure
58 | rbg.twin(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | parallelize=parallelize,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='map',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_map')
69 | # Append results
70 | replay_results_interval.append(replay_results)
71 | data_interval.append(data)
72 |
73 | # Set initial conditions for next day equal to the "ending conditions" of the current day
74 | x0 = replay_results['x_end']['realizations'][0].tolist()
75 |
76 | # Set previous_data_name
77 | previous_data_name = save_name
78 |
79 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
80 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
82 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 | x0 = None
35 | previous_data_name = None
36 |
37 | # Initialize the list of results
38 | replay_results_interval = []
39 | data_interval = []
40 |
41 | # Instantiate ReplayBG
42 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
43 | yts=5, exercise=False,
44 | seed=1,
45 | verbose=verbose, plot_mode=plot_mode)
46 |
47 | # Set interval to twin
48 | start_day = 1
49 | end_day = 2
50 |
51 | # Twin the interval
52 | for day in range(start_day, end_day+1):
53 |
54 | # Step 1: Load data and set save_name
55 | data = load_test_data(day=day)
56 | save_name = 'data_day_' + str(day) + '_interval'
57 |
58 | print("Twinning " + save_name)
59 |
60 | # Run twinning procedure
61 | rbg.twin(data=data, bw=bw, save_name=save_name,
62 | twinning_method='mcmc',
63 | parallelize=parallelize,
64 | n_steps=n_steps,
65 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
66 |
67 | # Replay the twin with the same input data
68 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
69 | twinning_method='mcmc',
70 | save_workspace=True,
71 | x0=x0, previous_data_name=previous_data_name,
72 | save_suffix='_twin_mcmc')
73 | # Append results
74 | replay_results_interval.append(replay_results)
75 | data_interval.append(data)
76 |
77 | # Set initial conditions for next day equal to the "ending conditions" of the current day
78 | x0 = replay_results['x_end']['realizations'][0].tolist()
79 |
80 | # Set previous_data_name
81 | previous_data_name = save_name
82 |
83 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
84 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
85 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
86 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data(day=1)
39 | save_name = 'data_day_' + str(1)
40 |
41 | print("Twinning " + save_name)
42 |
43 | # Run twinning procedure
44 | rbg.twin(data=data, bw=bw, save_name=save_name,
45 | twinning_method='map',
46 | parallelize=parallelize,
47 | u2ss=u2ss)
48 |
49 | # Replay the twin with the same input data
50 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
51 | twinning_method='map',
52 | save_workspace=True,
53 | save_suffix='_twin_map')
54 |
55 | # Visualize and analyze results
56 | Visualizer.plot_replay_results(replay_results, data=data)
57 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
58 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
59 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data_extended(day=1)
39 | # Cut data up to 11:00
40 | data = data.iloc[0:348, :]
41 | # Modify label of second day
42 | data.loc[312, "cho_label"] = 'B2'
43 |
44 | save_name = 'data_day_' + str(1) + '_extended'
45 |
46 | # Run twinning procedure using also the burn-out period
47 | rbg.twin(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | parallelize=parallelize,
50 | u2ss=u2ss,
51 | extended=True,
52 | find_start_guess_first=True)
53 |
54 | # Cut data up to 4:00
55 | data = data.iloc[0:264, :]
56 |
57 | # Replay the twin with the data up to the start of the burn-out period
58 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | save_workspace=True,
61 | save_suffix='_twin_map_extended')
62 |
63 | # Visualize and analyze results
64 | Visualizer.plot_replay_results(replay_results, data=data)
65 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
66 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
67 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data(day=1)
43 | save_name = 'data_day_' + str(1)
44 |
45 | print("Twinning " + save_name)
46 |
47 | # Run twinning procedure
48 | rbg.twin(data=data, bw=bw, save_name=save_name,
49 | twinning_method='mcmc',
50 | parallelize=parallelize,
51 | n_steps=n_steps,
52 | u2ss=u2ss)
53 |
54 | # Replay the twin with the same input data
55 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
56 | twinning_method='mcmc',
57 | save_workspace=True,
58 | save_suffix='_twin_mcmc')
59 |
60 | Visualizer.plot_replay_results(replay_results, data=data)
61 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data_extended(day=1)
43 | # Cut data up to 11:00
44 | data = data.iloc[0:348, :]
45 | # Modify label of second day
46 | data.loc[312, "cho_label"] = 'B2'
47 |
48 | save_name = 'data_day_' + str(1) + '_extended'
49 |
50 | print("Twinning " + save_name)
51 |
52 | # Run twinning procedure using also the burn-out period
53 | rbg.twin(data=data, bw=bw, save_name=save_name,
54 | twinning_method='mcmc',
55 | parallelize=parallelize,
56 | n_steps=n_steps,
57 | u2ss=u2ss,
58 | extended=True,
59 | find_start_guess_first=True)
60 |
61 | # Cut data up to 4:00
62 | data = data.iloc[0:264, :]
63 |
64 | # Replay the twin with the data up to the start of the burn-out period
65 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
66 | twinning_method='mcmc',
67 | save_workspace=True,
68 | save_suffix='_twin_mcmc_extended')
69 |
70 | Visualizer.plot_replay_results(replay_results, data=data)
71 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
72 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/utils.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import os
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
20 | return df
21 |
--------------------------------------------------------------------------------
/py_replay_bg/example/data/patient_info.csv:
--------------------------------------------------------------------------------
1 | patient,u2ss,bw
2 | 1,0.2440484700275897,100
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_init.py:
--------------------------------------------------------------------------------
1 | from py_replay_bg.input_validation import *
2 |
3 |
4 | class InputValidatorInit:
5 | """
6 | Class for validating the input of ReplayBG __init__ method.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | blueprint: str, {'single-meal', 'multi-meal'}
12 | A string that specifies the blueprint to be used to create the digital twin.
13 | save_folder : str
14 | A string defining the folder that will contain the results.
15 |
16 | yts: int, optional, default : 5
17 | An integer that specifies the data sample time (in minutes).
18 | exercise: boolean, optional, default : False
19 | A boolean that specifies whether to simulate exercise or not.
20 | seed: int, optional, default: 1
21 | An integer that specifies the random seed. For reproducibility.
22 |
23 | plot_mode : boolean, optional, default : True
24 | A boolean that specifies whether to show the plot of the results or not.
25 | verbose : boolean, optional, default : True
26 | A boolean that specifies the verbosity of ReplayBG.
27 |
28 | Methods
29 | -------
30 | validate():
31 | Run the input validation process.
32 | """
33 |
34 | def __init__(self,
35 | save_folder: str,
36 | blueprint: str,
37 | yts: int,
38 | exercise: bool,
39 | seed: int,
40 | plot_mode: bool,
41 | verbose: bool,
42 | ):
43 | self.save_folder = save_folder
44 | self.blueprint = blueprint
45 | self.yts = yts
46 | self.exercise = exercise
47 | self.seed = seed
48 | self.plot_mode = plot_mode
49 | self.verbose = verbose
50 |
51 | def validate(self):
52 | """
53 | Run the input validation process.
54 | """
55 |
56 | # Validate the 'save_folder' input
57 | SaveFolderValidator(save_folder=self.save_folder).validate()
58 |
59 | # Validate the 'blueprint' input
60 | BlueprintValidator(blueprint=self.blueprint).validate()
61 |
62 | # Validate the 'yts' input
63 | YTSValidator(yts=self.yts).validate()
64 |
65 | # Validate the 'exercise' input
66 | ExerciseValidator(exercise=self.exercise).validate()
67 |
68 | # Validate the 'seed' input
69 | SeedValidator(seed=self.seed).validate()
70 |
71 | # Validate the 'plot_mode' input
72 | PlotModeValidator(plot_mode=self.plot_mode).validate()
73 |
74 | # Validate the 'verbose' input
75 | VerboseValidator(verbose=self.verbose).validate()
76 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_replay.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from typing import Callable, Dict
5 |
6 | from py_replay_bg.input_validation import *
7 |
8 |
9 | class InputValidatorReplay:
10 | """
11 | Class for validating the input of ReplayBG twin method.
12 |
13 | ...
14 | Attributes
15 | ----------
16 | data : pd.DataFrame
17 | Pandas dataframe which contains the data to be used by the tool.
18 | bw : float
19 | The patient's body weight.
20 | save_name : str
21 | A string used to label, thus identify, each output file and result.
22 |
23 | x0: list
24 | The initial model state.
25 | previous_data_name: str
26 | The name of the previous portion of data. To be used to initialize the initial conditions.
27 |
28 | twinning_method : str
29 | The method used to twin the model.
30 |
31 | bolus_source : str
32 | A string defining whether to use, during replay, the insulin bolus data contained in the 'data' timetable
33 | (if 'data'), or the boluses generated by the bolus calculator implemented via the provided
34 | 'bolusCalculatorHandler' function.
35 | basal_source : str
36 | A string defining whether to use, during replay, the insulin basal data contained in the 'data' timetable
37 | (if 'data'), or the basal generated by the controller implemented via the provided
38 | 'basalControllerHandler' function (if 'dss'), or fixed to the average basal rate used during
39 | twinning (if 'u2ss').
40 | cho_source : str
41 | A string defining whether to use, during replay, the CHO data contained in the 'data' timetable (if 'data'),
42 | or the CHO generated by the meal generator implemented via the provided 'mealGeneratorHandler' function.
43 |
44 | meal_generator_handler: Callable
45 | A callback function that implements a meal generator to be used during the replay of a given scenario.
46 | meal_generator_handler_params: dict
47 | A dictionary that contains the parameters to pass to the meal_generator_handler function.
48 | bolus_calculator_handler: Callable
49 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
50 | bolus_calculator_handler_params: dict
51 | A dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also serves
52 | as memory area for the bolusCalculatorHandler function.
53 | basal_handler: Callable
54 | A callback function that implements a basal controller to be used during the replay of a given scenario.
55 | basal_handler_params: dict
56 | A dictionary that contains the parameters to pass to the basalHandler function. It also serves as memory
57 | area for the basalHandler function.
58 | enable_hypotreatments: boolean
59 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
60 | hypotreatments_handler: Callable
61 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
62 | hypotreatments_handler_params: dict
63 | A dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also serves
64 | as memory area for the hypoTreatmentsHandler function.
65 | enable_correction_boluses: boolean
66 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
67 | correction_boluses_handler: Callable
68 | A callback function that implements a corrective bolus strategy during the replay of a given scenario.
69 | correction_boluses_handler_params: dict
70 | A dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also serves
71 | as memory area for the correctionBolusesHandler function.
72 |
73 | save_suffix : string
74 | A string to be attached as suffix to the resulting output files' name.
75 | save_workspace: bool
76 | A flag that specifies whether to save the resulting workspace.
77 |
78 | n_replay: int
79 | The number of Monte Carlo replays to be performed. Ignored if twinning_method is 'map'.
80 | sensors: list[Sensors]
81 | The sensors to be used in each of the replay simulations.
82 |
83 | blueprint: str
84 | A string that specifies the blueprint to be used to create the digital twin.
85 | exercise: bool
86 | A boolean that specifies whether to use exercise model or not.
87 |
88 | Methods
89 | -------
90 | validate():
91 | Run the input validation process.
92 | """
93 |
94 | def __init__(self,
95 | data: pd.DataFrame,
96 | bw: float,
97 | save_name: str,
98 | x0: np.ndarray,
99 | previous_data_name: str,
100 | twinning_method: str,
101 | bolus_source: str,
102 | basal_source: str,
103 | cho_source: str,
104 | meal_generator_handler: Callable,
105 | meal_generator_handler_params: Dict,
106 | bolus_calculator_handler: Callable,
107 | bolus_calculator_handler_params: Dict,
108 | basal_handler: Callable,
109 | basal_handler_params: Dict,
110 | enable_hypotreatments: bool,
111 | hypotreatments_handler: Callable,
112 | hypotreatments_handler_params: Dict,
113 | enable_correction_boluses: bool,
114 | correction_boluses_handler: Callable,
115 | correction_boluses_handler_params: Dict,
116 | save_suffix: str,
117 | save_workspace: bool,
118 | n_replay: int,
119 | sensors: list,
120 | blueprint: str,
121 | exercise: bool,
122 | ):
123 | self.data = data
124 | self.bw = bw
125 | self.save_name = save_name
126 | self.x0 = x0
127 | self.previous_data_name = previous_data_name
128 | self.twinning_method = twinning_method
129 | self.bolus_source = bolus_source
130 | self.basal_source = basal_source
131 | self.cho_source = cho_source
132 | self.meal_generator_handler = meal_generator_handler
133 | self.meal_generator_handler_params = meal_generator_handler_params
134 | self.bolus_calculator_handler = bolus_calculator_handler
135 | self.bolus_calculator_handler_params = bolus_calculator_handler_params
136 | self.basal_handler = basal_handler
137 | self.basal_handler_params = basal_handler_params
138 | self.enable_hypotreatments = enable_hypotreatments
139 | self.hypotreatments_handler = hypotreatments_handler
140 | self.hypotreatments_handler_params = hypotreatments_handler_params
141 | self.enable_correction_boluses = enable_correction_boluses
142 | self.correction_boluses_handler = correction_boluses_handler
143 | self.correction_boluses_handler_params = correction_boluses_handler_params
144 | self.save_suffix = save_suffix
145 | self.save_workspace = save_workspace
146 | self.n_replay = n_replay
147 | self.sensors = sensors
148 | self.blueprint = blueprint
149 | self.exercise = exercise
150 |
151 |
152 | def validate(self):
153 | """
154 | Run the input validation process.
155 | """
156 |
157 | # Validate the 'data' input
158 | DataValidator(modality='replay', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
159 | bolus_source=self.bolus_source, basal_source=self.basal_source, cho_source=self.cho_source).validate()
160 |
161 | # Validate the 'bw' input
162 | BWValidator(bw=self.bw).validate()
163 |
164 | # Validate the 'save_name' input
165 | SaveNameValidator(save_name=self.save_name).validate()
166 |
167 | # Validate the 'save_name' input
168 | X0Validator(x0=self.x0).validate()
169 |
170 | # Validate the 'previous_data_name' input
171 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
172 |
173 | # Validate the 'twinning_method' input
174 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
175 |
176 | # Validate the 'bolus_source' input
177 | BolusSourceValidator(bolus_source=self.bolus_source).validate()
178 |
179 | # Validate the 'basal_source' input
180 | BasalSourceValidator(basal_source=self.basal_source).validate()
181 |
182 | # Validate the 'cho_source' input
183 | CHOSourceValidator(cho_source=self.cho_source).validate()
184 |
185 | # Validate the 'meal_generator_handler' input
186 | MealGeneratorHandlerValidator(meal_generator_handler=self.meal_generator_handler).validate()
187 |
188 | # Validate the 'meal_generator_handler_params' input
189 | MealGeneratorHandlerParamsValidator(meal_generator_handler_params=self.meal_generator_handler_params).validate()
190 |
191 | # Validate the 'bolus_calculator_handler' input
192 | BolusCalculatorHandlerValidator(bolus_calculator_handler=self.bolus_calculator_handler).validate()
193 |
194 | # Validate the 'bolus_calculator_handler_params' input
195 | BolusCalculatorHandlerParamsValidator(bolus_calculator_handler_params=self.bolus_calculator_handler_params).validate()
196 |
197 | # Validate the 'basal_handler' input
198 | BasalHandlerValidator(basal_handler=self.basal_handler).validate()
199 |
200 | # Validate the 'basal_handler_params' input
201 | BasalHandlerParamsValidator(basal_handler_params=self.basal_handler_params).validate()
202 |
203 | # Validate the 'enable_hypotreatments' input
204 | EnableHypotreatmentsValidator(enable_hypotreatments=self.enable_hypotreatments).validate()
205 |
206 | # Validate the 'hypotreatments_handler' input
207 | HypotreatmentsHandlerValidator(hypotreatments_handler=self.hypotreatments_handler).validate()
208 |
209 | # Validate the 'hypotreatments_handler_params' input
210 | HypotreatmentsHandlerParamsValidator(hypotreatments_handler_params=self.hypotreatments_handler_params).validate()
211 |
212 | # Validate the 'enable_correction_boluses' input
213 | EnableCorrectionBolusesValidator(enable_correction_boluses=self.enable_correction_boluses).validate()
214 |
215 | # Validate the 'correction_boluses_handler' input
216 | CorrectionBolusesHandlerValidator(correction_boluses_handler=self.correction_boluses_handler).validate()
217 |
218 | # Validate the 'correction_boluses_handler_params' input
219 | CorrectionBolusesHandlerParamsValidator(correction_boluses_handler_params=self.correction_boluses_handler_params).validate()
220 |
221 | # Validate the 'save_suffix' input
222 | SaveSuffixValidator(save_suffix=self.save_suffix).validate()
223 |
224 | # Validate the 'save_workspace' input
225 | SaveWorkspaceValidator(save_workspace=self.save_workspace).validate()
226 |
227 | # Validate the 'n_replay' input
228 | NReplayValidator(n_replay=self.n_replay).validate()
229 |
230 | # Validate the 'sensors' input
231 | SensorsValidator(sensors=self.sensors).validate()
232 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_twin.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from py_replay_bg.input_validation import *
5 |
6 |
7 | class InputValidatorTwin:
8 | """
9 | Class for validating the input of ReplayBG twin method.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | data: pd.DataFrame
15 | Pandas dataframe which contains the data to be used by the tool.
16 | bw: float
17 | The patient's body weight.
18 | save_name : str
19 | A string used to label, thus identify, each output file and result.
20 |
21 |
22 | u2ss : float
23 | The steady state of the basal insulin infusion.
24 | x0 : np.ndarray
25 | The initial model conditions.
26 | previous_data_name : str
27 | The name of the previous data portion. This is used to correctly "transfer" the initial model conditions to
28 | the current portion of data.
29 |
30 | twinning_method : str
31 | The method to be used to twin the model.
32 |
33 | extended : bool
34 | A flag indicating whether to use the "extended" model for twinning
35 | find_start_guess_first : bool
36 | A flag indicating whether to find the start guess using MAP before twinning.
37 |
38 | n_steps: int
39 | Number of steps to use for the main chain. This is ignored if modality is 'replay'.
40 | save_chains: bool, optional, default : False
41 | A flag that specifies whether to save the resulting mcmc chains and copula samplers.
42 |
43 | parallelize : boolean
44 | A boolean that specifies whether to parallelize the twinning process.
45 | n_processes : int, optional, default : None
46 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
47 |
48 | blueprint: str
49 | A string that specifies the blueprint to be used to create the digital twin.
50 | exercise: bool
51 | A boolean that specifies whether to use exercise model or not.
52 |
53 | Methods
54 | -------
55 | validate():
56 | Run the input validation process.
57 | """
58 |
59 | def __init__(self,
60 | data: pd.DataFrame,
61 | bw: float,
62 | save_name: str,
63 | twinning_method: str,
64 | extended: bool,
65 | find_start_guess_first: bool,
66 | n_steps: int,
67 | save_chains: bool,
68 | u2ss: float | None,
69 | x0: np.ndarray | None,
70 | previous_data_name: str | None,
71 | parallelize: bool,
72 | n_processes: int | None,
73 | blueprint: str,
74 | exercise: bool
75 | ):
76 | self.data = data
77 | self.bw = bw
78 | self.save_name = save_name
79 | self.twinning_method = twinning_method
80 | self.extended = extended
81 | self.find_start_guess_first = find_start_guess_first
82 | self.n_steps = n_steps
83 | self.save_chains = save_chains
84 | self.u2ss = u2ss
85 | self.x0 = x0
86 | self.previous_data_name = previous_data_name
87 | self.parallelize = parallelize
88 | self.n_processes = n_processes
89 | self.blueprint = blueprint
90 | self.exercise = exercise
91 |
92 | def validate(self):
93 | """
94 | Run the input validation process.
95 | """
96 |
97 | # Validate the 'data' input
98 | DataValidator(modality='twin', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
99 | bolus_source='data', basal_source='data', cho_source='data').validate()
100 |
101 | # Validate the 'bw' input
102 | BWValidator(bw=self.bw).validate()
103 |
104 | # Validate the 'save_name' input
105 | SaveNameValidator(save_name=self.save_name).validate()
106 |
107 | # Validate the 'twinning_method' input
108 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
109 |
110 | # Validate the 'extended' input
111 | ExtendedValidator(extended=self.extended).validate()
112 |
113 | # Validate the 'find_start_guess_first' input
114 | FindStartGuessFirstValidator(find_start_guess_first=self.find_start_guess_first).validate()
115 |
116 | # Validate the 'n_steps' input
117 | NStepsValidator(n_steps=self.n_steps).validate()
118 |
119 | # Validate the 'u2ss' input
120 | U2SSValidator(u2ss=self.u2ss).validate()
121 |
122 | # Validate the 'save_name' input
123 | X0Validator(x0=self.x0).validate()
124 |
125 | # Validate the 'previous_data_name' input
126 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
127 |
128 | # Validate the 'parallelize' input
129 | ParallelizeValidator(parallelize=self.parallelize).validate()
130 |
131 | # Validate the 'n_processes' input
132 | NProcessesValidator(n_processes=self.n_processes).validate()
133 |
--------------------------------------------------------------------------------
/py_replay_bg/model/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/model/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/model/model_parameters_t1d.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 |
4 |
5 | class ModelParametersT1D:
6 | def __init__(self,
7 | data: pd.DataFrame,
8 | bw: float,
9 | u2ss: float,
10 | extended: bool
11 | ):
12 |
13 | """
14 | Function that returns the default parameters values of the t1d model.
15 |
16 | Parameters
17 | ----------
18 | data : pd.DataFrame
19 | Pandas dataframe which contains the data to be used by the tool.
20 | bw : float
21 | The patient's body weight.
22 | u2ss : float
23 | The steady state of the basal insulin infusion
24 | extended : bool
25 | A flag indicating whether to use the "extended" model for twinning
26 |
27 | Returns
28 | -------
29 | model_parameters: dict
30 | A dictionary containing the default model parameters.
31 |
32 | Raises
33 | ------
34 | None
35 |
36 | See Also
37 | --------
38 | None
39 |
40 | Examples
41 | --------
42 | None
43 | """
44 | # Initial conditions
45 | self.Xpb = 0.0 # Insulin action initial condition
46 | self.Qgutb = 0.0 # Intestinal content initial condition
47 |
48 | # Glucose-insulin submodel parameters
49 | self.VG = 1.45 # dl/kg
50 | self.SG = 2.5e-2 # 1/min
51 | self.Gb = 119.13 # mg/dL
52 | self.r1 = 1.4407 # unitless
53 | self.r2 = 0.8124 # unitless
54 | self.alpha = 7 # 1/min
55 | self.p2 = 0.012 # 1/min
56 |
57 | if u2ss is None:
58 | self.u2ss = np.mean(data.basal) * 1000 / bw # mU/(kg*min)
59 | else:
60 | self.u2ss = u2ss
61 |
62 | # Subcutaneous insulin absorption submodel parameters
63 | self.VI = 0.126 # L/kg
64 | self.ke = 0.127 # 1/min
65 | self.kd = 0.026 # 1/min
66 | # model_parameters['ka1'] = 0.0034 # 1/min (virtually 0 in 77% of the cases)
67 | # self.ka1 = 0.0
68 | self.ka2 = 0.014 # 1/min
69 | self.tau = 8 # min
70 | self.Ipb = self.u2ss / self.ke # from eq. 5 steady-state
71 |
72 |
73 | # Oral glucose absorption submodel parameters
74 | self.kgri = 0.18 # = kmax % 1/min
75 | self.kempt = 0.18 # 1/min
76 | self.f = 0.9 # dimensionless
77 |
78 | # Exercise submodel parameters
79 | self.vo2rest = 0.33 # dimensionless, VO2rest was derived from heart rate round(66/(220-30))
80 | self.vo2max = 1 # dimensionless, VO2max is normalized and estimated from heart rate (220-age) = 100%.
81 | self.e1 = 1.6 # dimensionless
82 | self.e2 = 0.78 # dimensionless
83 |
84 | # Patient specific parameters
85 | self.bw = bw # kg
86 | self.to_g = self.bw / 1000
87 | self.to_mgkg = 1000 / self.bw
88 |
89 | # Measurement noise specifics
90 | self.SDn = 5
91 |
92 | # Glucose starting point
93 | if 'glucose' in data:
94 | idx = np.where(data.glucose.isnull().values == False)[0][0]
95 | self.G0 = data.glucose[idx]
96 | else:
97 | self.G0 = self.Gb
98 |
99 |
100 | class ModelParametersT1DMultiMeal(ModelParametersT1D):
101 |
102 | def __init__(self,
103 | data: pd.DataFrame,
104 | bw: float,
105 | u2ss: float,
106 | extended: bool,
107 | ):
108 |
109 | """
110 | Function that returns the default parameters values of the t1d model in a multi-meal blueprint.
111 |
112 | Parameters
113 | ----------
114 | data : pd.DataFrame
115 | Pandas dataframe which contains the data to be used by the tool.
116 | bw : float
117 | The patient's body weight.
118 | u2ss : float
119 | The steady state of the basal insulin infusion
120 | extended : bool
121 | A flag indicating whether to use the "extended" model for twinning
122 |
123 | Returns
124 | -------
125 | model_parameters: dict
126 | A dictionary containing the default model parameters.
127 |
128 | Raises
129 | ------
130 | None
131 |
132 | See Also
133 | --------
134 | None
135 |
136 | Examples
137 | --------
138 | None
139 | """
140 | # Initialize common parameters
141 | super().__init__(data, bw, u2ss, extended)
142 |
143 | # Glucose-insulin submodel parameters
144 | self.SI_B = 10.35e-4 / self.VG # mL/(uU*min)
145 | self.SI_L = 10.35e-4 / self.VG # mL/(uU*min)
146 | self.SI_D = 10.35e-4 / self.VG # mL/(uU*min)
147 |
148 | if extended:
149 | self.SI_B2 = 10.35e-4 / self.VG # mL/(uU*min)
150 |
151 | # Oral glucose absorption submodel parameters
152 | self.kabs_B = 0.012 # 1/min
153 | self.kabs_L = 0.012 # 1/min
154 | self.kabs_D = 0.012 # 1/min
155 | self.kabs_S = 0.012 # 1/min
156 | self.kabs_H = 0.012 # 1/min
157 | self.beta_B = 0 # min
158 | self.beta_L = 0 # min
159 | self.beta_D = 0 # min
160 | self.beta_S = 0 # min
161 |
162 | if extended:
163 | self.kabs_B2 = 0.012 # 1/min
164 | self.kabs_L2 = 0.012 # 1/min
165 | self.kabs_S2 = 0.012 # 1/min
166 | self.beta_B2 = 0 # min
167 | self.beta_L2 = 0 # min
168 | self.beta_S2 = 0 # min
169 |
170 |
171 | class ModelParametersT1DSingleMeal(ModelParametersT1D):
172 |
173 | def __init__(self,
174 | data: pd.DataFrame,
175 | bw: float,
176 | u2ss: float,
177 | ):
178 |
179 | """
180 | Function that returns the default parameters values of the t1d model in a single-meal blueprint.
181 |
182 | Parameters
183 | ----------
184 | data : pd.DataFrame
185 | Pandas dataframe which contains the data to be used by the tool.
186 | bw : float
187 | The patient's body weight.
188 | u2ss : float
189 | The steady state of the basal insulin infusion
190 |
191 | Returns
192 | -------
193 | model_parameters: dict
194 | A dictionary containing the default model parameters.
195 |
196 | Raises
197 | ------
198 | None
199 |
200 | See Also
201 | --------
202 | None
203 |
204 | Examples
205 | --------
206 | None
207 | """
208 | # Initialize common parameters
209 | super().__init__(data, bw, u2ss, extended=False)
210 |
211 | # Glucose-insulin submodel parameters
212 | self.SI = 10.35e-4 / self.VG # mL/(uU*min)
213 |
214 | # Oral glucose absorption submodel parameters
215 | self.kabs = 0.012 # 1/min
216 | self.beta = 0 # 1/min
217 |
--------------------------------------------------------------------------------
/py_replay_bg/sensors/__init__.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 |
4 | class CGM:
5 | """
6 | A class that represents a CGM sensor.
7 |
8 | Attributes
9 | ----------
10 | ts: int
11 | The sample time of the cgm sensor (min).
12 | t_offset: float
13 | The time offset of the cgm sensor (when new this is 0). This is used if a cgm sensor is shared between more
14 | replay runs.
15 | cgm_error_parameters: np.ndarray
16 | An array containing the parameters of the CGM.
17 | output_noise_sd: float
18 | The output standard deviation of the CGM error model.
19 | ekm1: float
20 | Memory term for the CGM noise: e(k-1).
21 | ekm2: float
22 | Memory term for the CGM noise: e(k-2).
23 | max_lifetime: float
24 | The maximum lifetime of the CGM sensor (in minutes).
25 | connected_at: int
26 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
27 | ReplayBG calls).
28 |
29 | Methods
30 | -------
31 | connect_new_cgm(connected_at):
32 | Connects a new CGM sensor by sampling new error parameters.
33 | measure(ig, t):
34 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
35 | add_offset(to_add):
36 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
37 | multiple ReplayBG runs.
38 | """
39 |
40 | def __init__(self,
41 | ts: int = 5,
42 | ):
43 | """
44 | Constructs all the necessary attributes for the CGM object.
45 |
46 | Parameters
47 | ----------
48 | ts: int, optional, default = 5
49 | The sample time of the cgm sensor (min).
50 | """
51 | self.ts = ts
52 | self.t_offset = 0
53 |
54 | self.connect_new_cgm()
55 |
56 | # Set max lifetime (minutes)
57 | self.max_lifetime = 10 * 1440 # 10 days
58 | self.connected_at = 0
59 |
60 | # Set the parameters of the CGM
61 | self.cgm_error_parameters = []
62 | self.output_noise_sd = []
63 |
64 | # Set memory terms
65 | self.ekm1 = None
66 | self.ekm2 = None
67 |
68 | def connect_new_cgm(self, connected_at=0):
69 | """
70 | Connects a new CGM sensor by sampling new error parameters.
71 |
72 | Parameters
73 | ----------
74 | connected_at: int, optional, default = 0
75 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
76 | ReplayBG calls).
77 |
78 | Returns
79 | -------
80 | None
81 |
82 | Raises
83 | ------
84 | None
85 |
86 | See Also
87 | --------
88 | None
89 |
90 | Examples
91 | --------
92 | None
93 | """
94 | # Load Mean vector and covariance matrix of the parameter vector as
95 | # defined in Vettoretti et al., Sensors, 2019
96 | mu = np.array([0.94228767821000314341972625697963, 0.0049398821141803427384187052950892,
97 | -0.0005848748565491275084801681138913, 6.382602204050874306062723917421,
98 | 1.2604070417357611244568715846981, -0.4022228938823663169088717950217,
99 | 3.2516360856114072674927228945307])
100 | sigma = np.array([[0.013245827952891258902368143424155, -0.0039513025350735725416129184850433,
101 | 0.00031276743283791636970891936186945, 0.15717912467153988265167186000326,
102 | 0.0026876560011614997885986966252858, -0.0028904633825263671524641306831427,
103 | -0.0031801707001874032418320403792222],
104 | [-0.0039513025350735725416129184850433, 0.0018527975980744701509778105119608,
105 | -0.00015580332205794781403294935184789, -0.10288007693621757654423021222101,
106 | -0.0013902327543057948350258001823931, 0.0011591852212130876378232136048041,
107 | -0.0027284927011686846420879248853453],
108 | [0.00031276743283791636970891936186945, -0.00015580332205794781403294935184789,
109 | 0.000013745962164724157000697882247131, 0.0080685863688738888865881193623864,
110 | 0.00012074974710011031125631020266553, -0.00010042135441622312822841645019167,
111 | 0.00011130290033867137325027107941366],
112 | [0.15717912467153988265167186000326, -0.10288007693621757654423021222101,
113 | 0.0080685863688738888865881193623864, 29.005838188852990811028575990349,
114 | 0.12408344051778112671069465022811, -0.10193644943826736526393261783596,
115 | 0.60075381294204155402383094042307],
116 | [0.0026876560011614997885986966252858, -0.0013902327543057948350258001823931,
117 | 0.00012074974710011031125631020266553, 0.12408344051778112671069465022811,
118 | 0.02079352674233487727195601735275, -0.018431109170459980539646949182497,
119 | -0.015721846813032722134373386779771],
120 | [-0.0028904633825263671524641306831427, 0.0011591852212130876378232136048041,
121 | -0.00010042135441622312822841645019167, -0.10193644943826736526393261783596,
122 | -0.018431109170459980539646949182497, 0.018700867933453400870913441167431,
123 | 0.01552333576829629385729347745837],
124 | [-0.0031801707001874032418320403792222, -0.0027284927011686846420879248853453,
125 | 0.00011130290033867137325027107941366, 0.60075381294204155402383094042307,
126 | -0.015721846813032722134373386779771, 0.01552333576829629385729347745837,
127 | 0.72356838038463477946748980684788]])
128 |
129 | # Modulation factor of the covariance of the parameter vector not to generate too extreme realizations of
130 | # parameter vector
131 | f = 0.90
132 | # Modulate the covariance matrix
133 | sigma = sigma * f
134 | # Maximum allowed output noise SD (mg/dl)
135 | max_output_noise_sd = 10
136 | # Tolerance for model stability check
137 | toll = 0.02
138 |
139 | # Flag for stability of the AR(2) model
140 | stable = 0
141 | output_noise_sd = np.inf
142 | cgm_error_parameters = []
143 | while (~ stable) or (output_noise_sd > max_output_noise_sd):
144 | # Sample CGM error parameters
145 | cgm_error_parameters = np.random.multivariate_normal(mu, sigma)
146 |
147 | # Check the stability of the resulting AR(2) model
148 | stable = (cgm_error_parameters[5] >= -1) and (
149 | cgm_error_parameters[5] <= (1 - np.abs(cgm_error_parameters[4]) - toll))
150 |
151 | output_noise_var = cgm_error_parameters[6] ** 2 / (
152 | 1 - (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) - cgm_error_parameters[5] * (
153 | (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) + cgm_error_parameters[5]))
154 |
155 | if output_noise_var < 0:
156 | continue
157 |
158 | # Compute the output noise standard deviation
159 | output_noise_sd = np.sqrt(output_noise_var)
160 |
161 | # Set the parameters of the CGM
162 | self.cgm_error_parameters = cgm_error_parameters
163 | self.output_noise_sd = output_noise_sd
164 |
165 | # Set memory terms
166 | self.ekm1 = 0
167 | self.ekm2 = 0
168 |
169 | # Set the offset (minutes)
170 | self.t_offset = 0
171 |
172 | # Set the connection time
173 | self.connected_at = connected_at
174 |
175 | def measure(self, ig, t):
176 | """
177 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
178 |
179 | Parameters
180 | ----------
181 | ig: float
182 | The interstitial glucose concentration at current time (mg/dl).
183 | t: float
184 | Current time in days from the start of CGM sensor.
185 |
186 | Returns
187 | -------
188 | cgm: float
189 | The CGM measurement at current time (mg/dl).
190 |
191 | Raises
192 | ------
193 | None
194 |
195 | See Also
196 | --------
197 | None
198 |
199 | Examples
200 | --------
201 | None
202 | """
203 |
204 | # Apply calibration error
205 | ig_s = (self.cgm_error_parameters[0] + self.cgm_error_parameters[1] * (t + self.t_offset) +
206 | self.cgm_error_parameters[2] * ((t + self.t_offset) ** 2)) * ig + self.cgm_error_parameters[3]
207 |
208 | # Generate noise
209 | z = np.random.normal(0, 1)
210 | u = self.cgm_error_parameters[6] * z
211 | e = u + self.cgm_error_parameters[4] * self.ekm1 + self.cgm_error_parameters[5] * self.ekm2
212 |
213 | # Update memory terms
214 | self.ekm2 = self.ekm1
215 | self.ekm1 = e
216 |
217 | # Get final CGM
218 | return ig_s + e
219 |
220 | def add_offset(self,
221 | to_add: float) -> None:
222 | """
223 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
224 | multiple ReplayBG runs.
225 |
226 | Parameters
227 | ----------
228 | to_add: float
229 | The offset to add to the sensor life.
230 |
231 | Returns
232 | -------
233 | None
234 |
235 | Raises
236 | ------
237 | None
238 |
239 | See Also
240 | --------
241 | None
242 |
243 | Examples
244 | --------
245 | None
246 | """
247 | self.t_offset += to_add
248 |
249 |
250 | class Sensors:
251 | """
252 | A class that represents the sensors to be used by ReplayBG.
253 |
254 | ...
255 | Attributes
256 | ----------
257 | cgm: CGM
258 | The CGM sensor used to measure the interstitial glucose during simulations.
259 |
260 | Methods
261 | -------
262 | None
263 | """
264 |
265 | def __init__(self, cgm: CGM):
266 | """
267 | Constructs all the necessary attributes for the Sensors object.
268 |
269 | Parameters
270 | ----------
271 | cgm: CGM
272 | The CGM sensor used to measure the interstitial glucose during simulations.
273 | """
274 | self.cgm = cgm
275 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pandas as pd
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'patient_info.csv'))
20 | return df
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the location of the 'results' folder
10 | results_folder_location = os.path.join(os.path.abspath(''))
11 |
12 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
13 | replay_results = pickle.load(file)
14 |
15 | # Visualize and analyze MCMC results
16 | Visualizer.plot_replay_results(replay_results)
17 | analysis = Analyzer.analyze_replay_results(replay_results)
18 | print(" ----- Analysis using MCMC-derived twins ----- ")
19 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
20 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
21 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
22 |
23 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
24 | replay_results = pickle.load(file)
25 |
26 | # Visualize and analyze MAP results
27 | Visualizer.plot_replay_results(replay_results)
28 | analysis = Analyzer.analyze_replay_results(replay_results)
29 | print(" ----- Analysis using MAP-derived twins ----- ")
30 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
31 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
32 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
33 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the interval to analyze
10 | start_day = 1
11 | end_day = 2
12 |
13 | # Set the location of the 'results' folder
14 | results_folder_location = os.path.join(os.path.abspath(''))
15 |
16 | # Initialize results list
17 | replay_results_interval = []
18 |
19 | for day in range(start_day, end_day+1):
20 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
21 | replay_results = pickle.load(file)
22 | replay_results_interval.append(replay_results)
23 |
24 | # Visualize and analyze MCMC results
25 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
26 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
27 | print(" ----- Analysis using MAP-derived twins ----- ")
28 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
29 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
30 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
31 |
32 | # Initialize results list
33 | replay_results_interval = []
34 |
35 | for day in range(start_day, end_day + 1):
36 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
37 | replay_results = pickle.load(file)
38 | replay_results_interval.append(replay_results)
39 |
40 | # Visualize and analyze MAP results
41 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
42 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
43 | print(" ----- Analysis using MCMC-derived twins ----- ")
44 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
45 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
46 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | save_workspace=True,
57 | x0=x0, previous_data_name=previous_data_name,
58 | save_suffix='_replay_map',
59 | sensors=sensors)
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='mcmc',
56 | n_replay=10,
57 | save_workspace=True,
58 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
59 | save_suffix='_replay_mcmc')
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params)
55 |
56 | # Visualize and analyze results
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
60 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
61 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
62 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | n_replay=10,
50 | twinning_method='mcmc',
51 | save_workspace=True,
52 | save_suffix='_replay_mcmc_dss',
53 | enable_correction_boluses=True,
54 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params=bolus_calculator_handler_params)
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 | x0 = None
28 | previous_data_name = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Twinning " + save_name)
52 |
53 | # Run twinning procedure
54 | rbg.twin(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | parallelize=parallelize,
57 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
58 |
59 | # Replay the twin with the same input data
60 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
61 | twinning_method='map',
62 | save_workspace=True,
63 | x0=x0, previous_data_name=previous_data_name,
64 | save_suffix='_twin_map')
65 |
66 | # Append results
67 | replay_results_interval.append(replay_results)
68 | data_interval.append(data)
69 |
70 | # Set initial conditions for next day equal to the "ending conditions" of the current day
71 | x0 = replay_results['x_end']['realizations'][0].tolist()
72 |
73 | # Set previous_data_name
74 | previous_data_name = save_name
75 |
76 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
78 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set the number of steps for MCMC
17 | n_steps = 5000 # In production, this should be >= 50k
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''))
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 | x0 = None
31 | previous_data_name = None
32 |
33 | # Initialize the list of results
34 | replay_results_interval = []
35 | data_interval = []
36 |
37 | # Instantiate ReplayBG
38 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
39 | yts=5, exercise=False,
40 | seed=1,
41 | verbose=verbose, plot_mode=plot_mode)
42 |
43 | # Set interval to twin
44 | start_day = 1
45 | end_day = 2
46 |
47 | # Twin the interval
48 | for day in range(start_day, end_day+1):
49 |
50 | # Step 1: Load data and set save_name
51 | data = load_test_data(day=day)
52 | save_name = 'data_day_' + str(day) + '_interval'
53 |
54 | print("Twinning " + save_name)
55 |
56 | # Run twinning procedure
57 | rbg.twin(data=data, bw=bw, save_name=save_name,
58 | twinning_method='mcmc',
59 | parallelize=parallelize,
60 | n_steps=n_steps,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='mcmc',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_mcmc')
69 |
70 | # Append results
71 | replay_results_interval.append(replay_results)
72 | data_interval.append(data)
73 |
74 | # Set initial conditions for next day equal to the "ending conditions" of the current day
75 | x0 = replay_results['x_end']['realizations'][0].tolist()
76 |
77 | # Set previous_data_name
78 | previous_data_name = save_name
79 |
80 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
82 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
83 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | print("Twinning " + save_name)
39 |
40 | # Run twinning procedure
41 | rbg.twin(data=data, bw=bw, save_name=save_name,
42 | twinning_method='map',
43 | parallelize=parallelize,
44 | u2ss=u2ss)
45 |
46 | # Replay the twin with the same input data
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | save_workspace=True,
50 | save_suffix='_twin_map')
51 |
52 | # Visualize and analyze results
53 | Visualizer.plot_replay_results(replay_results, data=data)
54 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
55 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data_extended(day=1)
36 | # Cut data up to 11:00
37 | data = data.iloc[0:348, :]
38 | # Modify label of second day
39 | data.loc[312, "cho_label"] = 'B2'
40 |
41 | save_name = 'data_day_' + str(1) + '_extended'
42 |
43 | print("Twinning " + save_name)
44 |
45 | # Run twinning procedure using also the burn-out period
46 | rbg.twin(data=data, bw=bw, save_name=save_name,
47 | twinning_method='map',
48 | parallelize=parallelize,
49 | u2ss=u2ss,
50 | extended=True,
51 | find_start_guess_first=False)
52 |
53 | # Cut data up to 4:00
54 | data = data.iloc[0:264,:]
55 |
56 | # Replay the twin with the data up to the start of the burn-out period
57 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
58 | twinning_method='map',
59 | save_workspace=True,
60 | save_suffix='_twin_map_extended')
61 |
62 | # Visualize and analyze results
63 | Visualizer.plot_replay_results(replay_results, data=data)
64 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
65 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data(day=1)
40 | save_name = 'data_day_' + str(1)
41 |
42 | print("Twinning " + save_name)
43 |
44 | # Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=n_steps,
49 | u2ss=u2ss)
50 |
51 | # Replay the twin with the same input data
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_twin_mcmc')
56 |
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data_extended(day=1)
40 | # Cut data up to 11:00
41 | data = data.iloc[0:348, :]
42 | # Modify label of second day
43 | data.loc[312, "cho_label"] = 'B2'
44 |
45 | save_name = 'data_day_' + str(1) + '_extended'
46 |
47 | print("Twinning " + save_name)
48 |
49 | # Run twinning procedure using also the burn-out period
50 | rbg.twin(data=data, bw=bw, save_name=save_name,
51 | twinning_method='mcmc',
52 | parallelize=parallelize,
53 | n_steps=n_steps,
54 | u2ss=u2ss,
55 | extended=True,
56 | find_start_guess_first=True)
57 |
58 | # Cut data up to 4:00
59 | data = data.iloc[0:264, :]
60 |
61 | # Replay the twin with the data up to the start of the burn-out period
62 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
63 | twinning_method='mcmc',
64 | save_workspace=True,
65 | save_suffix='_twin_mcmc_extended')
66 |
67 | Visualizer.plot_replay_results(replay_results, data=data)
68 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
69 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/twinning/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/twinning/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/twinning/map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import warnings
3 | import pickle
4 | import numpy as np
5 |
6 | from typing import Dict, Callable
7 |
8 | from multiprocessing import Pool
9 | from tqdm import tqdm
10 | from scipy.optimize import minimize
11 |
12 | from py_replay_bg.data import ReplayBGData
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 |
16 | from py_replay_bg.environment import Environment
17 |
18 | # Suppress all RuntimeWarnings
19 | warnings.filterwarnings("ignore", category=RuntimeWarning)
20 |
21 |
22 | class MAP:
23 | """
24 | A class that orchestrates the twinning process via MAP.
25 |
26 | Attributes
27 | ----------
28 | max_iter: int
29 | Maximum number of iterations.
30 | parallelize : bool
31 | A boolean that specifies whether to parallelize the twinning process.
32 | n_processes : int
33 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
34 |
35 | Methods
36 | -------
37 | twin(rbg_data, model, save_name, environment)
38 | Runs the twinning procedure.
39 | """
40 |
41 | def __init__(self,
42 | max_iter: int = 100000,
43 | parallelize: bool = False,
44 | n_processes: int | None = None,
45 | ):
46 | """
47 | Constructs all the necessary attributes for the MCMC object.
48 |
49 | Parameters
50 | ----------
51 | max_iter: int, optional, default : 100000
52 | Maximum number of iterations.
53 | parallelize : bool, optional, default : False
54 | A boolean that specifies whether to parallelize the twinning process.
55 | n_processes : int, optional, default : None
56 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
57 |
58 | Returns
59 | -------
60 | None
61 |
62 | Raises
63 | ------
64 | None
65 |
66 | See Also
67 | --------
68 | None
69 |
70 | Examples
71 | --------
72 | None
73 | """
74 |
75 | # Number of times to re-run the procedure
76 | self.n_rerun = 64
77 |
78 | # Maximum number of iterations
79 | self.max_iter = max_iter
80 |
81 | # Maximum number of function evaluations
82 | self.max_fev = 1000000
83 |
84 | # Parallelization options
85 | self.parallelize = parallelize
86 | self.n_processes = n_processes
87 |
88 | def twin(self,
89 | rbg_data: ReplayBGData,
90 | model: T1DModelSingleMeal | T1DModelMultiMeal,
91 | save_name: str,
92 | environment: Environment,
93 | start_guess: Dict = None,
94 | for_start_guess: bool = False) -> Dict:
95 | """
96 | Runs the twinning procedure.
97 |
98 | Parameters
99 | ----------
100 | rbg_data: ReplayBGData
101 | An object containing the data to be used during the twinning procedure.
102 | model: T1DModelSingleMeal | T1DModelMultiMeal
103 | An object that represents the physiological model to be used by ReplayBG.
104 | environment: Environment
105 | An object that represents the hyperparameters to be used by ReplayBG.
106 | save_name : str
107 | A string used to label, thus identify, each output file and result.
108 | start_guess: Dict, optional, default : None
109 | The initial guess for the twinning process. If None, this is set to population values.
110 | for_start_guess: bool, optional, default : False
111 | Whether to use the twin method just to find the start guess to pass to the MCMC twinner.
112 |
113 | Returns
114 | -------
115 | draws: dict
116 | A dictionary containing the chain and the samples obtained from the MCMC procedure and the copula
117 | sampling, respectively.
118 |
119 | Raises
120 | ------
121 | None
122 |
123 | See Also
124 | --------
125 | None
126 |
127 | Examples
128 | --------
129 | None
130 | """
131 |
132 | # If this is being used to find the start_guess, do /4 less reruns
133 | if for_start_guess:
134 | self.n_rerun = int(self.n_rerun/16)
135 |
136 | # Number of unknown parameters to twin
137 | n_dim = len(model.unknown_parameters)
138 |
139 | if start_guess is None:
140 | sg = model.start_guess
141 | else:
142 | sg = np.array(list(start_guess.values()))
143 |
144 | # Set the initial positions of the walkers.
145 | start = sg + model.start_guess_sigma * np.random.randn(self.n_rerun, n_dim)
146 | start[start < 0] = 0
147 |
148 | # Set the pooler
149 | pool = None
150 | if self.parallelize:
151 | pool = Pool(processes=self.n_processes)
152 |
153 | # Set up the options
154 | options = dict()
155 | options['maxiter'] = self.max_iter
156 | options['maxfev'] = self.max_fev
157 | options['disp'] = False
158 |
159 | # Select the function to minimize
160 | neg_log_posterior_func = model.neg_log_posterior_extended if model.extended else model.neg_log_posterior
161 |
162 | # Initialize results
163 | results = []
164 |
165 | if pool is None:
166 |
167 | if environment.verbose:
168 | iterator = tqdm(range(self.n_rerun))
169 | iterator.set_description("Min loss: %f" % np.nan)
170 | else:
171 | iterator = range(self.n_rerun)
172 |
173 | # Initialize best
174 | best = -1
175 |
176 | for r in iterator:
177 | result = run_map(start[r], neg_log_posterior_func, rbg_data, options)
178 | results.append(result)
179 | if best == -1 or result['fun'] < results[best]['fun']:
180 | best = r
181 | if environment.verbose:
182 | iterator.set_description("Min loss %f" % results[best]['fun'])
183 |
184 | else:
185 | # Prepare input arguments as tuples for starmap
186 | args = [(start[r], neg_log_posterior_func, rbg_data, options) for r in range(self.n_rerun)]
187 |
188 | # Initialize best
189 | best = -1
190 |
191 | # Get results (verbosity not allowed for the moment)
192 | results = pool.starmap(run_map, args)
193 |
194 | # Get best
195 | for r, result in enumerate(results):
196 | if best == -1 or result['fun'] < results[best]['fun']:
197 | best = r
198 |
199 | draws = dict()
200 | for up in range(n_dim):
201 | draws[model.unknown_parameters[up]] = results[best]['x'][up]
202 |
203 | # If twin is being used just to find the start guess, just return draws without saving
204 | if for_start_guess:
205 | return draws
206 |
207 | # Clean-up draws from "extended" parameters
208 | if model.extended:
209 | if 'SI_B2' in draws:
210 | del draws['SI_B2']
211 | if 'kabs_B2' in draws:
212 | del draws['kabs_B2']
213 | if 'beta_B2' in draws:
214 | del draws['beta_B2']
215 | if 'kabs_L2' in draws:
216 | del draws['kabs_L2']
217 | if 'beta_L2' in draws:
218 | del draws['beta_L2']
219 | if 'kabs_S2' in draws:
220 | del draws['kabs_S2']
221 | if 'beta_S2' in draws:
222 | del draws['beta_S2']
223 |
224 | # Save results
225 | twinning_results = dict()
226 | twinning_results['draws'] = draws
227 | twinning_results['u2ss'] = model.model_parameters.u2ss
228 |
229 | saved_file = os.path.join(environment.replay_bg_path, 'results', 'map',
230 | 'map_' + save_name + '.pkl')
231 |
232 | with open(saved_file, 'wb') as file:
233 | pickle.dump(twinning_results, file)
234 |
235 | if environment.verbose:
236 | print('Parameters saved in ' + saved_file)
237 |
238 | return draws
239 |
240 |
241 | def run_map(start: np.ndarray,
242 | neg_log_posterior_func: Callable,
243 | rbg_data: ReplayBGData,
244 | options: Dict
245 | ) -> Dict:
246 | """
247 | Utility function used to run MAP twinning.
248 |
249 | Parameters
250 | ----------
251 | start: np.ndarray
252 | An object containing the data to be used during the twinning procedure.
253 | neg_log_posterior_func: Callable
254 | The function to minimize, i.e., the neg-loglikelihood.
255 | rbg_data: ReplayBGData
256 | An object containing the data to be used during the twinning procedure.
257 | options : Dict
258 | A dictionary with the options necessary to the minimization function.
259 |
260 | Returns
261 | -------
262 | ret: dict
263 | A dictionary containing the results of the MAP twinning and the final value of the objective function.
264 |
265 | Raises
266 | ------
267 | None
268 |
269 | See Also
270 | --------
271 | None
272 |
273 | Examples
274 | --------
275 | None
276 | """
277 | result = minimize(neg_log_posterior_func, start, method='Powell', args=(rbg_data,), options=options)
278 | ret = dict()
279 | ret['fun'] = result.fun
280 | ret['x'] = result.x
281 | return ret
282 |
--------------------------------------------------------------------------------
/py_replay_bg/utils/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/utils/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/utils/stats.py:
--------------------------------------------------------------------------------
1 | import math
2 | import numpy as np
3 | from numba import njit
4 |
5 |
6 | @njit(fastmath=True)
7 | def log_lognorm(x, mu, sigma):
8 | """
9 | Computes the logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
10 |
11 | Parameters
12 | ----------
13 | x: float
14 | The value where to evaluate the log-normal pdf.
15 | mu: float
16 | The mean of the log-normal distribution.
17 | sigma: float
18 | The standard deviation of the log-normal distribution.
19 |
20 | Returns
21 | -------
22 | ll_norm: float
23 | The logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
24 |
25 | Raises
26 | ------
27 | None
28 |
29 | See Also
30 | --------
31 | None
32 |
33 | Examples
34 | --------
35 | None
36 | """
37 | return np.log(1 / (x * sigma * np.sqrt(2 * np.pi)) * np.exp(- ((np.log(x) - mu) ** 2) / (2 * (sigma ** 2))))
38 |
39 |
40 | @njit(fastmath=True)
41 | def log_norm(x, mu, sigma):
42 | """
43 | Computes the logarithm of the normal pdf evaluated at given x with given mu and sigma.
44 |
45 | Parameters
46 | ----------
47 | x: float
48 | The value where to evaluate the normal pdf.
49 | mu: float
50 | The mean of the normal distribution.
51 | sigma: float
52 | The standard deviation of the normal distribution.
53 |
54 | Returns
55 | -------
56 | l_norm: float
57 | The logarithm of the normal pdf evaluated at given x with given mu and sigma.
58 |
59 | Raises
60 | ------
61 | None
62 |
63 | See Also
64 | --------
65 | None
66 |
67 | Examples
68 | --------
69 | None
70 | """
71 | return np.log(1 / (sigma * np.sqrt(2 * np.pi)) * np.exp(- 0.5 * ((x - mu) / sigma) ** 2))
72 |
73 |
74 | @njit(fastmath=True)
75 | def log_gamma(x, alpha, beta):
76 | """
77 | Computes the logarithm of the gamma pdf evaluated at given x with given alpha and beta.
78 |
79 | Parameters
80 | ----------
81 | x: float
82 | The value where to evaluate the normal pdf.
83 | alpha: float
84 | The alpha value of the gamma distribution at hand.
85 | beta: float
86 | The beta value of the gamma distribution at hand.
87 |
88 | Returns
89 | -------
90 | l_gam: float
91 | The logarithm of the gamma pdf evaluated at given x with given alpha and beta.
92 |
93 | Raises
94 | ------
95 | None
96 |
97 | See Also
98 | --------
99 | None
100 |
101 | Examples
102 | --------
103 | None
104 | """
105 | return -np.inf if x < 0 else np.log((beta ** alpha * x ** (alpha - 1) * math.exp(-beta * x)) / math.gamma(alpha))
106 |
--------------------------------------------------------------------------------
/py_replay_bg/visualizer/__init__.py:
--------------------------------------------------------------------------------
1 | from datetime import datetime, timedelta
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 |
6 | from typing import Dict
7 |
8 | import pandas as pd
9 | from tqdm import tqdm
10 |
11 | from py_replay_bg.data import ReplayBGData
12 | from py_replay_bg.environment import Environment
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 | from py_replay_bg.dss import DSS
16 |
17 |
18 | class Visualizer:
19 | """
20 | A class to be used by ReplayBG for plotting.
21 |
22 | ...
23 | Attributes
24 | ----------
25 | -
26 |
27 | Methods
28 | -------
29 | plot_replay_results(replay_results, data)
30 | Function that plots the results of ReplayBG replay simulation.
31 | """
32 |
33 | def __init__(self):
34 | """
35 | Constructs all the necessary attributes for the Visualizer object.
36 |
37 | Parameters
38 | ----------
39 | -
40 |
41 | Returns
42 | -------
43 | None
44 |
45 | Raises
46 | ------
47 | None
48 |
49 | See Also
50 | --------
51 | None
52 |
53 | Examples
54 | --------
55 | None
56 | """
57 | pass
58 |
59 | @staticmethod
60 | def plot_replay_results(
61 | replay_results: Dict,
62 | data: pd.DataFrame = None,
63 | title: str = '',
64 | ) -> None:
65 | """
66 | Function that plots the results of ReplayBG simulation.
67 |
68 | Parameters
69 | ----------
70 | replay_results: dict
71 | The replayed scenario results with fields:
72 | glucose: dict
73 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
74 | cgm: dict
75 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
76 | insulin_bolus: dict
77 | A dictionary which contains the insulin boluses simulated via ReplayBG.
78 | correction_bolus: dict
79 | A dictionary which contains the correction boluses simulated via ReplayBG.
80 | insulin_basal: dict
81 | A dictionary which contains the basal insulin simulated via ReplayBG.
82 | cho: dict
83 | A dictionary which contains the meals simulated via ReplayBG.
84 | hypotreatments: dict
85 | A dictionary which contains the hypotreatments simulated via ReplayBG.
86 | meal_announcement: dict
87 | A dictionary which contains the meal announcements simulated via ReplayBG.
88 | vo2: dict
89 | A dictionary which contains the vo2 simulated via ReplayBG.
90 | sensors: dict
91 | A dictionary which contains the sensors used during the replayed scenario.
92 | rbg_data: ReplayBGData
93 | The data to be used by ReplayBG during simulation.
94 | model: T1DModelSingleMeal | T1DModelMultiMeal
95 | An object that represents the physiological model to be used by ReplayBG.
96 | data: pd.DataFrame, optional, default: None
97 | Pandas dataframe which contains the data to be used by the tool. If present, adds glucose data to the
98 | glucose subplot.
99 |
100 | Returns
101 | -------
102 | None
103 |
104 | Raises
105 | ------
106 | None
107 |
108 | See Also
109 | --------
110 | None
111 |
112 | Examples
113 | --------
114 | None
115 | """
116 | # Subplot 1: Glucose
117 |
118 | fig, ax = None, None
119 | if replay_results['model'].exercise:
120 | fig, ax = plt.subplots(4, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1, 1]})
121 | else:
122 | fig, ax = plt.subplots(3, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1]})
123 |
124 | if data is not None:
125 | ax[0].plot(data.t, data.glucose, marker='*', color='red', linewidth=2, label='CGM data [mg/dl]')
126 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['cgm']['median'], marker='o', color='black', linewidth=2, label='CGM replay (Median) [mg/dl]')
127 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['cgm']['ci25th'], replay_results['cgm']['ci75th'], color='black', alpha=0.2,
128 | label='CGM replay (CI 25-75th) [mg/dl]')
129 |
130 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['glucose']['median'][::replay_results['model'].yts], marker='o', color='blue', linewidth=2,
131 | label='Glucose replay (Median) [mg/dl]')
132 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['glucose']['ci25th'][::replay_results['model'].yts], replay_results['glucose']['ci75th'][::replay_results['model'].yts], color='blue',
133 | alpha=0.3, label='Glucose replay (CI 25-75th) [mg/dl]')
134 |
135 | ax[0].grid()
136 | ax[0].legend()
137 |
138 | # Subplot 2: Meals
139 | t = np.arange(replay_results['rbg_data'].t_data[0]+ pd.Timedelta(minutes=0), replay_results['rbg_data'].t_data[-1] + pd.Timedelta(minutes=replay_results['model'].yts),
140 | timedelta(minutes=1)).astype(datetime)
141 |
142 | cho_events = np.sum(replay_results['cho']['realizations'], axis=0) / replay_results['cho']['realizations'].shape[0]
143 | ht_events = np.sum(replay_results['hypotreatments']['realizations'], axis=0) / replay_results['hypotreatments']['realizations'].shape[0]
144 |
145 | markerline, stemlines, baseline = ax[1].stem(t, cho_events, basefmt='k:', label='CHO replay (Mean) [g/min]')
146 | plt.setp(stemlines, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
147 | plt.setp(markerline, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
148 |
149 | markerline, stemlines, baseline = ax[1].stem(t, ht_events, basefmt='k:', label='HT replay (Mean) [g/min]')
150 | plt.setp(stemlines, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
151 | plt.setp(markerline, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
152 |
153 | ax[1].grid()
154 | ax[1].legend()
155 |
156 | # Subplot 3: Insulin
157 |
158 | bolus_events = np.sum(replay_results['insulin_bolus']['realizations'], axis=0) / replay_results['insulin_bolus']['realizations'].shape[0]
159 | cb_events = np.sum(replay_results['correction_bolus']['realizations'], axis=0) / replay_results['correction_bolus']['realizations'].shape[0]
160 | basal_rate = np.sum(replay_results['insulin_basal']['realizations'], axis=0) / replay_results['insulin_basal']['realizations'].shape[0]
161 |
162 | markerline, stemlines, baseline = ax[2].stem(t, bolus_events, basefmt='k:',
163 | label='Bolus insulin replay (Mean) [U/min]')
164 | plt.setp(stemlines, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
165 | plt.setp(markerline, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
166 |
167 | markerline, stemlines, baseline = ax[2].stem(t, cb_events, basefmt='k:',
168 | label='CB insulin replay (Mean) [U/min]')
169 | plt.setp(stemlines, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
170 | plt.setp(markerline, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
171 |
172 | ax[2].plot(t, basal_rate * 60, color='black', linewidth=2, label='Basal insulin replay (Mean) [U/h]')
173 |
174 | ax[2].grid()
175 | ax[2].legend()
176 |
177 | # Subplot 4: Exercise
178 |
179 | if replay_results['model'].exercise:
180 |
181 | vo2_events = np.sum(replay_results['vo2']['realizations'], axis=0) / replay_results['vo2']['realizations'].shape[0]
182 |
183 | markerline, stemlines, baseline = ax[3].stem(t, vo2_events, basefmt='k:', label='VO2 replay (Mean) [-]')
184 | plt.setp(stemlines, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
185 | plt.setp(markerline, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
186 |
187 | ax[3].grid()
188 | ax[3].legend()
189 |
190 | fig.suptitle(title, fontweight='bold')
191 | plt.show()
192 |
193 |
194 | @staticmethod
195 | def plot_replay_results_interval(
196 | replay_results_interval: list,
197 | data_interval: list = None,
198 | title: str = '',
199 | ) -> None:
200 | """
201 | Function that plots the results of ReplayBG simulation (intervals).
202 |
203 | Parameters
204 | ----------
205 | replay_results_interval: list
206 | A list dictionaries of replayed scenario results. Each element has fields:
207 | glucose: dict
208 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
209 | cgm: dict
210 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
211 | insulin_bolus: dict
212 | A dictionary which contains the insulin boluses simulated via ReplayBG.
213 | correction_bolus: dict
214 | A dictionary which contains the correction boluses simulated via ReplayBG.
215 | insulin_basal: dict
216 | A dictionary which contains the basal insulin simulated via ReplayBG.
217 | cho: dict
218 | A dictionary which contains the meals simulated via ReplayBG.
219 | hypotreatments: dict
220 | A dictionary which contains the hypotreatments simulated via ReplayBG.
221 | meal_announcement: dict
222 | A dictionary which contains the meal announcements simulated via ReplayBG.
223 | vo2: dict
224 | A dictionary which contains the vo2 simulated via ReplayBG.
225 | sensors: dict
226 | A dictionary which contains the sensors used during the replayed scenario.
227 | rbg_data: ReplayBGData
228 | The data to be used by ReplayBG during simulation.
229 | model: T1DModelSingleMeal | T1DModelMultiMeal
230 | An object that represents the physiological model to be used by ReplayBG.
231 | data_interval: list, optional, default: None
232 | A list of pandas dataframe which contains the data to be used by the tool. If present, adds glucose data
233 | to the glucose subplot.
234 |
235 | Returns
236 | -------
237 | None
238 |
239 | Raises
240 | ------
241 | None
242 |
243 | See Also
244 | --------
245 | None
246 |
247 | Examples
248 | --------
249 | None
250 | """
251 | data = None
252 | if data_interval is not None:
253 | data = pd.concat(data_interval, axis=0, ignore_index=True)
254 |
255 | # Pad each array to the maximum shape
256 | replay_results = dict()
257 |
258 |
259 | # Re-map glucose data
260 | fields = ['median', 'ci5th', 'ci25th', 'ci75th', 'ci95th']
261 | category = ['glucose', 'cgm']
262 |
263 | for c in category:
264 |
265 | replay_results[c] = dict()
266 |
267 | for f in fields:
268 | replay_results[c][f] = dict()
269 |
270 | replay_results[c][f] = [r[c][f] for r in replay_results_interval]
271 | replay_results[c][f] = np.concatenate(replay_results[c][f], axis=0)
272 |
273 | # Re-map cho, insulin, and vo2 data
274 | category = ['cho', 'hypotreatments', 'insulin_bolus', 'correction_bolus', 'insulin_basal', 'vo2']
275 |
276 | for c in category:
277 | replay_results[c] = dict()
278 | replay_results[c]['realizations'] = [r[c]['realizations'] for r in replay_results_interval]
279 | replay_results[c]['realizations'] = np.concatenate(replay_results[c]['realizations'], axis=1)
280 |
281 | replay_results['model'] = replay_results_interval[0]['model']
282 | t_data = [r['rbg_data'].t_data for r in replay_results_interval]
283 | replay_results['rbg_data'] = replay_results_interval[0]['rbg_data']
284 | replay_results['rbg_data'].t_data = np.concatenate(t_data, axis=0)
285 |
286 | Visualizer.plot_replay_results(replay_results=replay_results, data=data, title=title)
287 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.pytest.ini_options]
2 | pythonpath = [
3 | "."
4 | ]
5 |
6 | [build-system]
7 | requires = ["setuptools>=61.0.0", "wheel"]
8 | build-backend = "setuptools.build_meta"
9 |
10 | [project]
11 | name = "py_replay_bg"
12 | version = "1.1.0"
13 | authors = [
14 | { name="Giacomo Cappon", email="cappongiacomo@gmail.com" },
15 | ]
16 | description = "ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management."
17 | readme = "README.md"
18 | requires-python = ">=3.11"
19 | classifiers = [
20 | "Programming Language :: Python :: 3",
21 | "License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
22 | "Operating System :: OS Independent",
23 | "Intended Audience :: Science/Research",
24 | "Topic :: Scientific/Engineering",
25 | ]
26 | dynamic = ["dependencies"]
27 | [tool.setuptools.dynamic]
28 | dependencies = {file = ["requirements.txt"]}
29 | optional-dependencies = {dev = { file = ["requirements-dev.txt"] }}
30 |
31 | [project.urls]
32 | "Homepage" = "https://github.com/gcappon/py_replay_bg"
33 | "Bug Tracker" = "https://github.com/gcappon/py_replay_bg/issues"
34 |
35 | [tool.setuptools.packages.find]
36 | include = ["py_replay_bg*"]
37 | exclude = ["venv*"]
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/tsconfig.json:
--------------------------------------------------------------------------------
1 | {
2 | "compilerOptions": {
3 | "module": "NodeNext",
4 | "moduleResolution": "NodeNext",
5 | "target": "ES2022"
6 | },
7 | "include": [
8 | "docs/.vuepress/**/*.ts",
9 | "docs/.vuepress/**/*.vue"
10 | ],
11 | "exclude": [
12 | "node_modules"
13 | ]
14 | }
15 |
--------------------------------------------------------------------------------
83 |
84 | [](https://github.com/gcappon/py_replay_bg/COPYING)
85 | [](https://github.com/gcappon/py_replay_bg/commits/master)
86 |
87 | ReplayBG is a digital twin-based methodology to develop and assess new strategies for type 1 diabetes management.
88 |
89 | # Reference
90 |
91 | [G. Cappon, M. Vettoretti, G. Sparacino, S. Del Favero, A. Facchinetti, "ReplayBG: a digital twin-based methodology to identify a personalized model from type 1 diabetes data and simulate glucose concentrations to assess alternative therapies", IEEE Transactions on Biomedical Engineering, 2023, DOI: 10.1109/TBME.2023.3286856.](https://ieeexplore.ieee.org/document/10164140)
92 |
93 | # Get started
94 |
95 | ## Installation
96 |
97 | **ReplayBG** can be installed via pypi by simply
98 |
99 | ```python
100 | pip install py-replay-bg
101 | ```
102 |
103 | ### Requirements
104 |
105 | * Python >= 3.11
106 | * List of Python packages in `requirements.txt`
107 |
108 | ## Preparation: imports, setup, and data loading
109 |
110 | First of all import the core modules:
111 | ```python
112 | import os
113 | import numpy as np
114 | import pandas as pd
115 |
116 | from multiprocessing import freeze_support
117 | ```
118 |
119 | Here, `os` will be used to manage the filesystem, `numpy` and `pandas` to manipulate and manage the data to be used, and
120 | `multiprocessing.freeze_support` to enable multiprocessing functionalities and run the twinning procedure in a faster,
121 | parallelized way.
122 |
123 | Then, we will import the necessary ReplayBG modules:
124 | ```python
125 | from py_replay_bg.py_replay_bg import ReplayBG
126 | from py_replay_bg.visualizer import Visualizer
127 | from py_replay_bg.analyzer import Analyzer
128 | ```
129 |
130 | Here, `ReplayBG` is the core ReplayBG object (more information in the [The ReplayBG Object](https://gcappon.github.io/py_replay_bg/documentation/replaybg_object.html) page),
131 | while `Analyzer` and `Visualizer` are utility objects that will be used to
132 | respectively analyze and visualize the results that we will produce with ReplayBG
133 | (more information in the ([Visualizing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/visualizing_replay_results.html) and
134 | [Analyzing Replay Results](https://gcappon.github.io/py_replay_bg/documentation/analyzing_replay_results.html) pages).
135 |
136 | Next steps consist of setting up some variables that will be used by ReplayBG environment.
137 | First of all, we will run the twinning procedure in a parallelized way so let's start with:
138 | ```python
139 | if __name__ == '__main__':
140 | freeze_support()
141 | ```
142 |
143 | Then, we will set the verbosity of ReplayBG:
144 | ```python
145 | verbose = True
146 | plot_mode = False
147 | ```
148 |
149 | Then, we need to decide what blueprint to use for twinning the data at hand.
150 | ```python
151 | blueprint = 'multi-meal'
152 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
153 | parallelize = True
154 | ```
155 |
156 | For more information on how to choose a blueprint, please refer to the [Choosing Blueprint](https://gcappon.github.io/py_replay_bg/documentation/choosing_blueprint.html) page.
157 |
158 | Now, let's load some data to play with. In this example, we will use the data stored in `example/data/data_day_1.csv`
159 | which contains a day of data of a patient with T1D:
160 |
161 | ```python
162 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
163 | data.t = pd.to_datetime(data['t'])
164 | ```
165 |
166 | ::: warning
167 | Be careful, data in PyReplayBG must be provided in a `.csv.` file that must follow some strict requirements. For more
168 | information see the [Data Requirements](https://gcappon.github.io/py_replay_bg/documentation/data_requirements.html) page.
169 | :::
170 |
171 | Let's also load the patient information (i.e., body weight and basal insulin `u2ss`) stored in the `example/data/patient_info.csv` file.
172 |
173 | ```python
174 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
175 | p = np.where(patient_info['patient'] == 1)[0][0]
176 | # Set bw and u2ss
177 | bw = float(patient_info.bw.values[p])
178 | u2ss = float(patient_info.u2ss.values[p])
179 | ```
180 |
181 | Finally, instantiate a `ReplayBG` object:
182 |
183 | ```python
184 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
185 | yts=5, exercise=False,
186 | seed=1,
187 | verbose=verbose, plot_mode=plot_mode)
188 |
189 | ```
190 |
191 | ## Step 1: Creation of the digital twin
192 |
193 | To create the digital twin, i.e., run the twinning procedure, using the MCMC method, use the `rbg.twin()` method:
194 |
195 | ```python
196 | rbg.twin(data=data, bw=bw, save_name='data_day_1',
197 | twinning_method='mcmc',
198 | parallelize=parallelize,
199 | n_steps=5000,
200 | u2ss=u2ss)
201 | ```
202 |
203 | For more information on the twinning procedure see the [Twinning Procedure](https://gcappon.github.io/py_replay_bg/documentation/twinning_procedure.html) page.
204 |
205 |
206 | ## Step 2: Run replay simulations
207 |
208 | Now that we have the digital twin created, it's time to replay using the `rbg.replay()` method. For more details
209 | see the [Replaying](https://gcappon.github.io/py_replay_bg/documentation/replaying.html) page.
210 |
211 | The possibilities are several, but for now let's just see what happens if we run a replay using the same input data used for twinning:
212 |
213 | ```python
214 | replay_results = rbg.replay(data=data, bw=bw, save_name='data_day_1',
215 | twinning_method='mcmc',
216 | save_workspace=True,
217 | save_suffix='_step_2a')
218 | ```
219 |
220 | It is possible to visualize the results of the simulation using:
221 |
222 | ```python
223 | Visualizer.plot_replay_results(replay_results, data=data)
224 | ```
225 |
226 | and analyzing the results using:
227 |
228 | ```python
229 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
230 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
231 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
232 | ```
233 |
234 | As a second example, we can simulate what happens with different inputs, for example when we reduce insulin by 30%.
235 | To do that run:
236 |
237 | ```python
238 | data.bolus = data.bolus * .7
239 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
240 | twinning_method='mcmc',
241 | save_workspace=True,
242 | save_suffix='_step_2b')
243 |
244 | # Visualize results
245 | Visualizer.plot_replay_results(replay_results)
246 | # Analyze results
247 | analysis = Analyzer.analyze_replay_results(replay_results)
248 |
249 | # Print, for example, the average glucose
250 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
251 | ```
252 |
253 | A `.py` file with the full code of the get started example can be found in `example/code/get_started.py`.
254 |
255 | # Documentation
256 |
257 | Full documentation at [https://gcappon.github.io/py_replay_bg/](https://gcappon.github.io/py_replay_bg/).
258 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/SOURCES.txt:
--------------------------------------------------------------------------------
1 | COPYING.md
2 | README.md
3 | pyproject.toml
4 | requirements.txt
5 | py_replay_bg/__init__.py
6 | py_replay_bg/py_replay_bg.py
7 | py_replay_bg.egg-info/PKG-INFO
8 | py_replay_bg.egg-info/SOURCES.txt
9 | py_replay_bg.egg-info/dependency_links.txt
10 | py_replay_bg.egg-info/requires.txt
11 | py_replay_bg.egg-info/top_level.txt
12 | py_replay_bg/analyzer/__init__.py
13 | py_replay_bg/data/__init__.py
14 | py_replay_bg/dss/__init__.py
15 | py_replay_bg/dss/default_dss_handlers.py
16 | py_replay_bg/environment/__init__.py
17 | py_replay_bg/example/code/analysis_example.py
18 | py_replay_bg/example/code/analysis_intervals_example.py
19 | py_replay_bg/example/code/get_started.py
20 | py_replay_bg/example/code/replay_intervals_map.py
21 | py_replay_bg/example/code/replay_intervals_mcmc.py
22 | py_replay_bg/example/code/replay_map.py
23 | py_replay_bg/example/code/replay_map_dss.py
24 | py_replay_bg/example/code/replay_mcmc.py
25 | py_replay_bg/example/code/replay_mcmc_dss.py
26 | py_replay_bg/example/code/twin_intervals_map.py
27 | py_replay_bg/example/code/twin_intervals_mcmc.py
28 | py_replay_bg/example/code/twin_map.py
29 | py_replay_bg/example/code/twin_map_extended.py
30 | py_replay_bg/example/code/twin_mcmc.py
31 | py_replay_bg/example/code/twin_mcmc_extended.py
32 | py_replay_bg/example/code/utils.py
33 | py_replay_bg/input_validation/__init__.py
34 | py_replay_bg/input_validation/input_validator_init.py
35 | py_replay_bg/input_validation/input_validator_replay.py
36 | py_replay_bg/input_validation/input_validator_twin.py
37 | py_replay_bg/model/__init__.py
38 | py_replay_bg/model/logpriors_t1d.py
39 | py_replay_bg/model/model_parameters_t1d.py
40 | py_replay_bg/model/model_step_equations_t1d.py
41 | py_replay_bg/model/t1d_model_multi_meal.py
42 | py_replay_bg/model/t1d_model_single_meal.py
43 | py_replay_bg/replay/__init__.py
44 | py_replay_bg/sensors/__init__.py
45 | py_replay_bg/tests/__init__.py
46 | py_replay_bg/tests/test_analysis_example.py
47 | py_replay_bg/tests/test_analysis_intervals_example.py
48 | py_replay_bg/tests/test_replay_intervals_map.py
49 | py_replay_bg/tests/test_replay_intervals_mcmc.py
50 | py_replay_bg/tests/test_replay_map.py
51 | py_replay_bg/tests/test_replay_map_dss.py
52 | py_replay_bg/tests/test_replay_map_extended.py
53 | py_replay_bg/tests/test_replay_mcmc.py
54 | py_replay_bg/tests/test_replay_mcmc_dss.py
55 | py_replay_bg/tests/test_replay_mcmc_extended.py
56 | py_replay_bg/tests/test_twin_intervals_map.py
57 | py_replay_bg/tests/test_twin_intervals_mcmc.py
58 | py_replay_bg/tests/test_twin_map.py
59 | py_replay_bg/tests/test_twin_map_extended.py
60 | py_replay_bg/tests/test_twin_mcmc.py
61 | py_replay_bg/tests/test_twin_mcmc_extended.py
62 | py_replay_bg/twinning/__init__.py
63 | py_replay_bg/twinning/map.py
64 | py_replay_bg/twinning/mcmc.py
65 | py_replay_bg/utils/__init__.py
66 | py_replay_bg/utils/stats.py
67 | py_replay_bg/visualizer/__init__.py
68 | venv/bin/rst2html.py
69 | venv/bin/rst2html4.py
70 | venv/bin/rst2html5.py
71 | venv/bin/rst2latex.py
72 | venv/bin/rst2man.py
73 | venv/bin/rst2odt.py
74 | venv/bin/rst2odt_prepstyles.py
75 | venv/bin/rst2pseudoxml.py
76 | venv/bin/rst2s5.py
77 | venv/bin/rst2xetex.py
78 | venv/bin/rst2xml.py
79 | venv/bin/rstpep2html.py
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/dependency_links.txt:
--------------------------------------------------------------------------------
1 |
2 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/requires.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/py_replay_bg.egg-info/top_level.txt:
--------------------------------------------------------------------------------
1 | py_replay_bg
2 |
--------------------------------------------------------------------------------
/py_replay_bg/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/dss/__init__.py:
--------------------------------------------------------------------------------
1 | from typing import Callable, Dict
2 |
3 | from py_replay_bg.dss.default_dss_handlers import default_meal_generator_handler, standard_bolus_calculator_handler, \
4 | default_basal_handler, ada_hypotreatments_handler, corrects_above_250_handler
5 |
6 |
7 | class DSS:
8 | """
9 | A class that represents the hyperparameters of the integrated decision support system.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | bw: double
15 | The patient's body weight.
16 | meal_generator_handler: Callable
17 | A callback function that implements a meal generator to be used during the replay of a given scenario.
18 | meal_generator_handler_params: dict
19 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
20 | bolus_calculator_handler: Callable
21 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
22 | bolus_calculator_handler_params: dict
23 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
24 | serves as memory area for the bolusCalculatorHandler function.
25 | basal_handler: Callable
26 | A callback function that implements a basal controller to be used during the replay of a given scenario.
27 | basal_handler_params: dict
28 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
29 | memory area for the basalHandler function.
30 | enable_hypotreatments: boolean
31 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
32 | hypotreatments_handler: Callable
33 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
34 | hypotreatments_handler_params: dict
35 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
36 | serves as memory area for the hypoTreatmentsHandler function.
37 | enable_correction_boluses: boolean
38 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
39 | correction_boluses_handler: Callable
40 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
41 | correction_boluses_handler_params: dict
42 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also
43 | serves as memory area for the correctionBolusesHandler function.
44 |
45 | Methods
46 | -------
47 | None
48 | """
49 |
50 | def __init__(self,
51 | bw: float,
52 | meal_generator_handler: Callable = default_meal_generator_handler,
53 | meal_generator_handler_params: Dict | None = None,
54 | bolus_calculator_handler: Callable = standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params: Dict | None = None,
56 | basal_handler: Callable = default_basal_handler,
57 | basal_handler_params: Dict | None = None,
58 | enable_hypotreatments: bool = False,
59 | hypotreatments_handler: Callable = ada_hypotreatments_handler,
60 | hypotreatments_handler_params: Dict | None = None,
61 | enable_correction_boluses: bool = False,
62 | correction_boluses_handler: Callable = corrects_above_250_handler,
63 | correction_boluses_handler_params: Dict | None = None,
64 | ):
65 | """
66 | Constructs all the necessary attributes for the DSS object.
67 |
68 | Parameters
69 | ----------
70 | bw: double
71 | The patient's body weight.
72 | meal_generator_handler: Callable, optional, default : default_meal_generator_handler
73 | A callback function that implements a meal generator to be used during the replay of a given scenario.
74 | meal_generator_handler_params: dict, optional, default : None
75 | A mutable dictionary that contains the parameters to pass to the meal_generator_handler function.
76 | bolus_calculator_handler: Callable, optional, default : standard_bolus_calculator_handler
77 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
78 | bolus_calculator_handler_params: dict, optional, default : None
79 | A mutable dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also
80 | serves as memory area for the bolusCalculatorHandler function.
81 | basal_handler: function, Callable, default : default_basal_handler
82 | A callback function that implements a basal controller to be used during the replay of a given scenario.
83 | basal_handler_params: dict, optional, default : None
84 | A mutable dictionary that contains the parameters to pass to the basalHandler function. It also serves as
85 | memory area for the basalHandler function.
86 | enable_hypotreatments: boolean, optional, default : False
87 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
88 | hypotreatments_handler: Callable, optional, default : ada_hypotreatments_handler
89 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
90 | hypotreatments_handler_params: dict, optional, default : None
91 | A mutable dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also
92 | serves as memory area for the hypoTreatmentsHandler function.
93 | enable_correction_boluses: boolean, optional, default : False
94 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
95 | correction_boluses_handler: Callable, optional, default : corrects_above_250_handler
96 | A callback function that implements a corrective bolusing strategy during the replay of a given scenario.
97 | correction_boluses_handler_params: dict, optional, default : None
98 | A mutable dictionary that contains the parameters to pass to the correctionBolusesHandler function.
99 | It also serves as memory area for the correctionBolusesHandler function.
100 | """
101 |
102 | # Patient's body weight
103 | self.bw = bw
104 |
105 | # Meal Generator module parameters
106 | self.meal_generator_handler = meal_generator_handler
107 | self.meal_generator_handler_params = meal_generator_handler_params if meal_generator_handler_params is not None else {}
108 |
109 | # Bolus Calculator module parameters
110 | self.bolus_calculator_handler = bolus_calculator_handler
111 | self.bolus_calculator_handler_params = bolus_calculator_handler_params if bolus_calculator_handler_params is not None else {}
112 |
113 | # Basal module parameters
114 | self.basal_handler = basal_handler
115 | self.basal_handler_params = basal_handler_params if basal_handler_params is not None else {}
116 |
117 | # Hypotreatment module parameters
118 | self.enable_hypotreatments = enable_hypotreatments
119 | self.hypotreatments_handler = hypotreatments_handler
120 | self.hypotreatments_handler_params = hypotreatments_handler_params if hypotreatments_handler_params is not None else {}
121 |
122 | # Correction bolus module parameters
123 | self.enable_correction_boluses = enable_correction_boluses
124 | self.correction_boluses_handler = correction_boluses_handler
125 | self.correction_boluses_handler_params = correction_boluses_handler_params if correction_boluses_handler_params is not None else {}
126 |
--------------------------------------------------------------------------------
/py_replay_bg/environment/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 |
4 | class Environment:
5 | """
6 | A class that represents the hyperparameters to be used by ReplayBG.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | save_name : str
12 | A string used to label, thus identify, each output file and result.
13 | replay_bg_path: str
14 | The current absolute path of ReplayBG.
15 |
16 | blueprint: str, {'single-meal', 'multi-meal'}
17 | A string that specifies the blueprint to be used to create the digital twin.
18 | yts: int
19 | An integer that specifies the data sampling time (in minutes).
20 |
21 | seed : int
22 | An integer that specifies the random seed. For reproducibility.
23 |
24 | plot_mode : bool
25 | A boolean that specifies whether to show the plot of the results or not.
26 | verbose : bool
27 | A boolean that specifies the verbosity of ReplayBG.
28 |
29 | Methods
30 | -------
31 | None
32 | """
33 |
34 | def __init__(self,
35 | blueprint: str = 'single_meal',
36 | save_name: str = '',
37 | save_folder: str = '',
38 | yts: int = 5,
39 | exercise: bool = False,
40 | seed: int = 1,
41 | plot_mode: bool = True,
42 | verbose: bool = True
43 | ):
44 | """
45 | Constructs all the necessary attributes for the Environment object.
46 |
47 | Parameters
48 | ----------
49 | blueprint: str, {'single-meal', 'multi-meal'}, optional, default : 'single-meal'
50 | A string that specifies the blueprint to be used to create the digital twin.
51 |
52 | save_name : str, optional, default : ''
53 | A string used to label, thus identify, each output file and result.
54 | save_folder : str, optional, default : ''
55 | A string used to set the folder where the ReplayBG results will be saved.
56 |
57 | yts: int, optional, default : 5
58 | An integer that specifies the data sampling time (in minutes).
59 | exercise: boolean, optional, default : False
60 | A boolean that specifies whether to simulate exercise or not.
61 |
62 | seed : int, optional, default : 1
63 | An integer that specifies the random seed. For reproducibility.
64 |
65 | plot_mode : boolean, optional, default : True
66 | A boolean that specifies whether to show the plot of the results or not.
67 | verbose : boolean, optional, default : True
68 | A boolean that specifies the verbosity of ReplayBG.
69 | """
70 |
71 | # Set the save name and folder
72 | self.save_name = save_name
73 | self.replay_bg_path = save_folder
74 |
75 | # Create the results sub folders if they do not exist
76 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results'))):
77 | os.mkdir(os.path.join(self.replay_bg_path, 'results'))
78 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'mcmc'))):
79 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'mcmc'))
80 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'map'))):
81 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'map'))
82 | if not (os.path.exists(os.path.join(self.replay_bg_path, 'results', 'workspaces'))):
83 | os.mkdir(os.path.join(self.replay_bg_path, 'results', 'workspaces'))
84 |
85 | # Single-meal or multi-meal blueprint?
86 | self.blueprint = blueprint
87 |
88 | # Set sample time
89 | self.yts = yts
90 |
91 | # Set whether to use the exercise model
92 | self.exercise = exercise
93 |
94 | # Set the seed
95 | self.seed = seed
96 |
97 | # Set plot mode and verbosity
98 | self.plot_mode = plot_mode
99 | self.verbose = verbose
100 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the location of the 'results' folder
9 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
10 |
11 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
12 | replay_results = pickle.load(file)
13 |
14 | # Visualize and analyze MCMC results
15 | Visualizer.plot_replay_results(replay_results)
16 | analysis = Analyzer.analyze_replay_results(replay_results)
17 | print(" ----- Analysis using MCMC-derived twins ----- ")
18 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
19 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
20 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
21 |
22 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
23 | replay_results = pickle.load(file)
24 |
25 | # Visualize and analyze MAP results
26 | Visualizer.plot_replay_results(replay_results)
27 | analysis = Analyzer.analyze_replay_results(replay_results)
28 | print(" ----- Analysis using MAP-derived twins ----- ")
29 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
30 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
31 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
32 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 |
8 | # Set the interval to analyze
9 | start_day = 1
10 | end_day = 2
11 |
12 | # Set the location of the 'results' folder
13 | results_folder_location = os.path.join(os.path.abspath(''),'..','..','..')
14 |
15 | # Initialize results list
16 | replay_results_interval = []
17 |
18 | for day in range(start_day, end_day+1):
19 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
20 | replay_results = pickle.load(file)
21 | replay_results_interval.append(replay_results)
22 |
23 | # Visualize and analyze MCMC results
24 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
25 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
26 | print(" ----- Analysis using MAP-derived twins ----- ")
27 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
28 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
29 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
30 |
31 | # Initialize results list
32 | replay_results_interval = []
33 |
34 | for day in range(start_day, end_day + 1):
35 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
36 | replay_results = pickle.load(file)
37 | replay_results_interval.append(replay_results)
38 |
39 | # Visualize and analyze MAP results
40 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
41 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
42 | print(" ----- Analysis using MCMC-derived twins ----- ")
43 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
44 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
45 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
46 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/get_started.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 | import pandas as pd
4 |
5 | from multiprocessing import freeze_support
6 |
7 | from py_replay_bg.py_replay_bg import ReplayBG
8 | from py_replay_bg.visualizer import Visualizer
9 | from py_replay_bg.analyzer import Analyzer
10 |
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # Load data
25 | data = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'data_day_1.csv'))
26 | data.t = pd.to_datetime(data['t'])
27 |
28 | # Load patient_info
29 | patient_info = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Set save name
42 | save_name = 'data_day_1'
43 |
44 | # Step 1. Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=5000,
49 | u2ss=u2ss)
50 |
51 | # Step 2a. Replay the twin with the same input data to get the initial conditions for the subsequent day
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_step_2a')
56 |
57 | # Visualize results and compare with the original glucose data
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | # Analyze results
60 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
61 | # Print, for example, the fit MARD and the average glucose
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
63 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
64 |
65 | # Step 2b. Replay the twin with different input data (-30% bolus insulin) to experiment how glucose changes
66 | data.bolus = data.bolus * .7
67 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
68 | twinning_method='mcmc',
69 | save_workspace=True,
70 | u2ss=u2ss,
71 | save_suffix='_step_2b')
72 |
73 | # Visualize results
74 | Visualizer.plot_replay_results(replay_results)
75 | # Analyze results
76 | analysis = Analyzer.analyze_replay_results(replay_results)
77 |
78 | # Print, for example, the average glucose
79 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='map',
53 | save_workspace=True,
54 | x0=x0, previous_data_name=previous_data_name,
55 | save_suffix='_replay_map',
56 | sensors=sensors)
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 | x0 = None
24 | previous_data_name = None
25 | sensors = None
26 |
27 | # Initialize the list of results
28 | replay_results_interval = []
29 | data_interval = []
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Set interval to twin
38 | start_day = 1
39 | end_day = 2
40 |
41 | # Twin the interval
42 | for day in range(start_day, end_day+1):
43 |
44 | # Load data and set save_name
45 | data = load_test_data(day=day)
46 | save_name = 'data_day_' + str(day) + '_interval'
47 |
48 | print("Replaying " + save_name)
49 |
50 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
51 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
52 | twinning_method='mcmc',
53 | n_replay=100,
54 | save_workspace=True,
55 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
56 | save_suffix='_replay_mcmc')
57 |
58 | # Append results
59 | replay_results_interval.append(replay_results)
60 | data_interval.append(data)
61 |
62 | # Set initial conditions for next day equal to the "ending conditions" of the current day
63 | x0 = replay_results['x_end']['realizations'][0].tolist()
64 |
65 | # Set sensors to use the same sensors during the next portion of data
66 | sensors = replay_results['sensors']
67 |
68 | # Set previous_data_name
69 | previous_data_name = save_name
70 |
71 | # Visualize and analyze results
72 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
73 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
74 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
75 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
76 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
77 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | twinning_method='map',
39 | save_workspace=True,
40 | save_suffix='_replay_map')
41 |
42 | # Visualize and analyze results
43 | Visualizer.plot_replay_results(replay_results, data=data)
44 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
45 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
46 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
47 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
48 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | # Set verbosity
11 | verbose = True
12 | plot_mode = False
13 |
14 | # Set other parameters for twinning
15 | blueprint = 'multi-meal'
16 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
17 |
18 | # load patient_info
19 | patient_info = load_patient_info()
20 | p = np.where(patient_info['patient'] == 1)[0][0]
21 | # Set bw and u2ss
22 | bw = float(patient_info.bw.values[p])
23 |
24 | # Instantiate ReplayBG
25 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
26 | yts=5, exercise=False,
27 | seed=1,
28 | verbose=verbose, plot_mode=plot_mode)
29 |
30 | # Load data and set save_name
31 | data = load_test_data(day=1)
32 | save_name = 'data_day_' + str(1)
33 |
34 | print("Replaying " + save_name)
35 |
36 | # Replay the twin with the same input data used for twinning
37 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
38 | n_replay=10,
39 | twinning_method='mcmc',
40 | save_workspace=True,
41 | save_suffix='_replay_mcmc')
42 |
43 | # Visualize and analyze results
44 | Visualizer.plot_replay_results(replay_results, data=data)
45 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
46 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
47 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
48 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
49 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from utils import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | # Set params of the bolus calculator
37 | bolus_calculator_handler_params = dict()
38 | bolus_calculator_handler_params['cr'] = 7
39 | bolus_calculator_handler_params['cf'] = 25
40 | bolus_calculator_handler_params['gt'] = 110
41 |
42 | print("Replaying " + save_name)
43 |
44 |
45 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
46 | # meal insulin boluses
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | n_replay=10,
49 | twinning_method='mcmc',
50 | save_workspace=True,
51 | save_suffix='_replay_mcmc_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params
55 | )
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 | x0 = None
32 | previous_data_name = None
33 |
34 | # Initialize the list of results
35 | replay_results_interval = []
36 | data_interval = []
37 |
38 | # Instantiate ReplayBG
39 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
40 | yts=5, exercise=False,
41 | seed=1,
42 | verbose=verbose, plot_mode=plot_mode)
43 |
44 | # Set interval to twin
45 | start_day = 1
46 | end_day = 2
47 |
48 | # Twin the interval
49 | for day in range(start_day, end_day+1):
50 |
51 | # Step 1: Load data and set save_name
52 | data = load_test_data(day=day)
53 | save_name = 'data_day_' + str(day) + '_interval'
54 |
55 | print("Twinning " + save_name)
56 |
57 | # Run twinning procedure
58 | rbg.twin(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | parallelize=parallelize,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='map',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_map')
69 | # Append results
70 | replay_results_interval.append(replay_results)
71 | data_interval.append(data)
72 |
73 | # Set initial conditions for next day equal to the "ending conditions" of the current day
74 | x0 = replay_results['x_end']['realizations'][0].tolist()
75 |
76 | # Set previous_data_name
77 | previous_data_name = save_name
78 |
79 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
80 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
82 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 | x0 = None
35 | previous_data_name = None
36 |
37 | # Initialize the list of results
38 | replay_results_interval = []
39 | data_interval = []
40 |
41 | # Instantiate ReplayBG
42 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
43 | yts=5, exercise=False,
44 | seed=1,
45 | verbose=verbose, plot_mode=plot_mode)
46 |
47 | # Set interval to twin
48 | start_day = 1
49 | end_day = 2
50 |
51 | # Twin the interval
52 | for day in range(start_day, end_day+1):
53 |
54 | # Step 1: Load data and set save_name
55 | data = load_test_data(day=day)
56 | save_name = 'data_day_' + str(day) + '_interval'
57 |
58 | print("Twinning " + save_name)
59 |
60 | # Run twinning procedure
61 | rbg.twin(data=data, bw=bw, save_name=save_name,
62 | twinning_method='mcmc',
63 | parallelize=parallelize,
64 | n_steps=n_steps,
65 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
66 |
67 | # Replay the twin with the same input data
68 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
69 | twinning_method='mcmc',
70 | save_workspace=True,
71 | x0=x0, previous_data_name=previous_data_name,
72 | save_suffix='_twin_mcmc')
73 | # Append results
74 | replay_results_interval.append(replay_results)
75 | data_interval.append(data)
76 |
77 | # Set initial conditions for next day equal to the "ending conditions" of the current day
78 | x0 = replay_results['x_end']['realizations'][0].tolist()
79 |
80 | # Set previous_data_name
81 | previous_data_name = save_name
82 |
83 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
84 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
85 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
86 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data(day=1)
39 | save_name = 'data_day_' + str(1)
40 |
41 | print("Twinning " + save_name)
42 |
43 | # Run twinning procedure
44 | rbg.twin(data=data, bw=bw, save_name=save_name,
45 | twinning_method='map',
46 | parallelize=parallelize,
47 | u2ss=u2ss)
48 |
49 | # Replay the twin with the same input data
50 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
51 | twinning_method='map',
52 | save_workspace=True,
53 | save_suffix='_twin_map')
54 |
55 | # Visualize and analyze results
56 | Visualizer.plot_replay_results(replay_results, data=data)
57 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
58 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
59 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | from multiprocessing import freeze_support
3 |
4 | import numpy as np
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 | if __name__ == '__main__':
13 | freeze_support()
14 |
15 | # Set verbosity
16 | verbose = True
17 | plot_mode = False
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 |
31 | # Instantiate ReplayBG
32 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
33 | yts=5, exercise=False,
34 | seed=1,
35 | verbose=verbose, plot_mode=plot_mode)
36 |
37 | # Load data and set save_name
38 | data = load_test_data_extended(day=1)
39 | # Cut data up to 11:00
40 | data = data.iloc[0:348, :]
41 | # Modify label of second day
42 | data.loc[312, "cho_label"] = 'B2'
43 |
44 | save_name = 'data_day_' + str(1) + '_extended'
45 |
46 | # Run twinning procedure using also the burn-out period
47 | rbg.twin(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | parallelize=parallelize,
50 | u2ss=u2ss,
51 | extended=True,
52 | find_start_guess_first=True)
53 |
54 | # Cut data up to 4:00
55 | data = data.iloc[0:264, :]
56 |
57 | # Replay the twin with the data up to the start of the burn-out period
58 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
59 | twinning_method='map',
60 | save_workspace=True,
61 | save_suffix='_twin_map_extended')
62 |
63 | # Visualize and analyze results
64 | Visualizer.plot_replay_results(replay_results, data=data)
65 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
66 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
67 |
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data(day=1)
43 | save_name = 'data_day_' + str(1)
44 |
45 | print("Twinning " + save_name)
46 |
47 | # Run twinning procedure
48 | rbg.twin(data=data, bw=bw, save_name=save_name,
49 | twinning_method='mcmc',
50 | parallelize=parallelize,
51 | n_steps=n_steps,
52 | u2ss=u2ss)
53 |
54 | # Replay the twin with the same input data
55 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
56 | twinning_method='mcmc',
57 | save_workspace=True,
58 | save_suffix='_twin_mcmc')
59 |
60 | Visualizer.plot_replay_results(replay_results, data=data)
61 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
62 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from multiprocessing import freeze_support
5 |
6 | from utils import load_test_data_extended, load_patient_info
7 |
8 | from py_replay_bg.py_replay_bg import ReplayBG
9 | from py_replay_bg.visualizer import Visualizer
10 | from py_replay_bg.analyzer import Analyzer
11 |
12 |
13 | if __name__ == '__main__':
14 | freeze_support()
15 |
16 | # Set verbosity
17 | verbose = True
18 | plot_mode = False
19 |
20 | # Set the number of steps for MCMC
21 | n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
22 |
23 | # Set other parameters for twinning
24 | blueprint = 'multi-meal'
25 | save_folder = os.path.join(os.path.abspath(''),'..','..','..')
26 | parallelize = True
27 |
28 | # load patient_info
29 | patient_info = load_patient_info()
30 | p = np.where(patient_info['patient'] == 1)[0][0]
31 | # Set bw and u2ss
32 | bw = float(patient_info.bw.values[p])
33 | u2ss = float(patient_info.u2ss.values[p])
34 |
35 | # Instantiate ReplayBG
36 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
37 | yts=5, exercise=False,
38 | seed=1,
39 | verbose=verbose, plot_mode=plot_mode)
40 |
41 | # Load data and set save_name
42 | data = load_test_data_extended(day=1)
43 | # Cut data up to 11:00
44 | data = data.iloc[0:348, :]
45 | # Modify label of second day
46 | data.loc[312, "cho_label"] = 'B2'
47 |
48 | save_name = 'data_day_' + str(1) + '_extended'
49 |
50 | print("Twinning " + save_name)
51 |
52 | # Run twinning procedure using also the burn-out period
53 | rbg.twin(data=data, bw=bw, save_name=save_name,
54 | twinning_method='mcmc',
55 | parallelize=parallelize,
56 | n_steps=n_steps,
57 | u2ss=u2ss,
58 | extended=True,
59 | find_start_guess_first=True)
60 |
61 | # Cut data up to 4:00
62 | data = data.iloc[0:264, :]
63 |
64 | # Replay the twin with the data up to the start of the burn-out period
65 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
66 | twinning_method='mcmc',
67 | save_workspace=True,
68 | save_suffix='_twin_mcmc_extended')
69 |
70 | Visualizer.plot_replay_results(replay_results, data=data)
71 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
72 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/example/code/utils.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import os
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..' , 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), '..', 'data', 'patient_info.csv'))
20 | return df
21 |
--------------------------------------------------------------------------------
/py_replay_bg/example/data/patient_info.csv:
--------------------------------------------------------------------------------
1 | patient,u2ss,bw
2 | 1,0.2440484700275897,100
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_init.py:
--------------------------------------------------------------------------------
1 | from py_replay_bg.input_validation import *
2 |
3 |
4 | class InputValidatorInit:
5 | """
6 | Class for validating the input of ReplayBG __init__ method.
7 |
8 | ...
9 | Attributes
10 | ----------
11 | blueprint: str, {'single-meal', 'multi-meal'}
12 | A string that specifies the blueprint to be used to create the digital twin.
13 | save_folder : str
14 | A string defining the folder that will contain the results.
15 |
16 | yts: int, optional, default : 5
17 | An integer that specifies the data sample time (in minutes).
18 | exercise: boolean, optional, default : False
19 | A boolean that specifies whether to simulate exercise or not.
20 | seed: int, optional, default: 1
21 | An integer that specifies the random seed. For reproducibility.
22 |
23 | plot_mode : boolean, optional, default : True
24 | A boolean that specifies whether to show the plot of the results or not.
25 | verbose : boolean, optional, default : True
26 | A boolean that specifies the verbosity of ReplayBG.
27 |
28 | Methods
29 | -------
30 | validate():
31 | Run the input validation process.
32 | """
33 |
34 | def __init__(self,
35 | save_folder: str,
36 | blueprint: str,
37 | yts: int,
38 | exercise: bool,
39 | seed: int,
40 | plot_mode: bool,
41 | verbose: bool,
42 | ):
43 | self.save_folder = save_folder
44 | self.blueprint = blueprint
45 | self.yts = yts
46 | self.exercise = exercise
47 | self.seed = seed
48 | self.plot_mode = plot_mode
49 | self.verbose = verbose
50 |
51 | def validate(self):
52 | """
53 | Run the input validation process.
54 | """
55 |
56 | # Validate the 'save_folder' input
57 | SaveFolderValidator(save_folder=self.save_folder).validate()
58 |
59 | # Validate the 'blueprint' input
60 | BlueprintValidator(blueprint=self.blueprint).validate()
61 |
62 | # Validate the 'yts' input
63 | YTSValidator(yts=self.yts).validate()
64 |
65 | # Validate the 'exercise' input
66 | ExerciseValidator(exercise=self.exercise).validate()
67 |
68 | # Validate the 'seed' input
69 | SeedValidator(seed=self.seed).validate()
70 |
71 | # Validate the 'plot_mode' input
72 | PlotModeValidator(plot_mode=self.plot_mode).validate()
73 |
74 | # Validate the 'verbose' input
75 | VerboseValidator(verbose=self.verbose).validate()
76 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_replay.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from typing import Callable, Dict
5 |
6 | from py_replay_bg.input_validation import *
7 |
8 |
9 | class InputValidatorReplay:
10 | """
11 | Class for validating the input of ReplayBG twin method.
12 |
13 | ...
14 | Attributes
15 | ----------
16 | data : pd.DataFrame
17 | Pandas dataframe which contains the data to be used by the tool.
18 | bw : float
19 | The patient's body weight.
20 | save_name : str
21 | A string used to label, thus identify, each output file and result.
22 |
23 | x0: list
24 | The initial model state.
25 | previous_data_name: str
26 | The name of the previous portion of data. To be used to initialize the initial conditions.
27 |
28 | twinning_method : str
29 | The method used to twin the model.
30 |
31 | bolus_source : str
32 | A string defining whether to use, during replay, the insulin bolus data contained in the 'data' timetable
33 | (if 'data'), or the boluses generated by the bolus calculator implemented via the provided
34 | 'bolusCalculatorHandler' function.
35 | basal_source : str
36 | A string defining whether to use, during replay, the insulin basal data contained in the 'data' timetable
37 | (if 'data'), or the basal generated by the controller implemented via the provided
38 | 'basalControllerHandler' function (if 'dss'), or fixed to the average basal rate used during
39 | twinning (if 'u2ss').
40 | cho_source : str
41 | A string defining whether to use, during replay, the CHO data contained in the 'data' timetable (if 'data'),
42 | or the CHO generated by the meal generator implemented via the provided 'mealGeneratorHandler' function.
43 |
44 | meal_generator_handler: Callable
45 | A callback function that implements a meal generator to be used during the replay of a given scenario.
46 | meal_generator_handler_params: dict
47 | A dictionary that contains the parameters to pass to the meal_generator_handler function.
48 | bolus_calculator_handler: Callable
49 | A callback function that implements a bolus calculator to be used during the replay of a given scenario.
50 | bolus_calculator_handler_params: dict
51 | A dictionary that contains the parameters to pass to the bolusCalculatorHandler function. It also serves
52 | as memory area for the bolusCalculatorHandler function.
53 | basal_handler: Callable
54 | A callback function that implements a basal controller to be used during the replay of a given scenario.
55 | basal_handler_params: dict
56 | A dictionary that contains the parameters to pass to the basalHandler function. It also serves as memory
57 | area for the basalHandler function.
58 | enable_hypotreatments: boolean
59 | A flag that specifies whether to enable hypotreatments during the replay of a given scenario.
60 | hypotreatments_handler: Callable
61 | A callback function that implements a hypotreatment strategy during the replay of a given scenario.
62 | hypotreatments_handler_params: dict
63 | A dictionary that contains the parameters to pass to the hypoTreatmentsHandler function. It also serves
64 | as memory area for the hypoTreatmentsHandler function.
65 | enable_correction_boluses: boolean
66 | A flag that specifies whether to enable correction boluses during the replay of a given scenario.
67 | correction_boluses_handler: Callable
68 | A callback function that implements a corrective bolus strategy during the replay of a given scenario.
69 | correction_boluses_handler_params: dict
70 | A dictionary that contains the parameters to pass to the correctionBolusesHandler function. It also serves
71 | as memory area for the correctionBolusesHandler function.
72 |
73 | save_suffix : string
74 | A string to be attached as suffix to the resulting output files' name.
75 | save_workspace: bool
76 | A flag that specifies whether to save the resulting workspace.
77 |
78 | n_replay: int
79 | The number of Monte Carlo replays to be performed. Ignored if twinning_method is 'map'.
80 | sensors: list[Sensors]
81 | The sensors to be used in each of the replay simulations.
82 |
83 | blueprint: str
84 | A string that specifies the blueprint to be used to create the digital twin.
85 | exercise: bool
86 | A boolean that specifies whether to use exercise model or not.
87 |
88 | Methods
89 | -------
90 | validate():
91 | Run the input validation process.
92 | """
93 |
94 | def __init__(self,
95 | data: pd.DataFrame,
96 | bw: float,
97 | save_name: str,
98 | x0: np.ndarray,
99 | previous_data_name: str,
100 | twinning_method: str,
101 | bolus_source: str,
102 | basal_source: str,
103 | cho_source: str,
104 | meal_generator_handler: Callable,
105 | meal_generator_handler_params: Dict,
106 | bolus_calculator_handler: Callable,
107 | bolus_calculator_handler_params: Dict,
108 | basal_handler: Callable,
109 | basal_handler_params: Dict,
110 | enable_hypotreatments: bool,
111 | hypotreatments_handler: Callable,
112 | hypotreatments_handler_params: Dict,
113 | enable_correction_boluses: bool,
114 | correction_boluses_handler: Callable,
115 | correction_boluses_handler_params: Dict,
116 | save_suffix: str,
117 | save_workspace: bool,
118 | n_replay: int,
119 | sensors: list,
120 | blueprint: str,
121 | exercise: bool,
122 | ):
123 | self.data = data
124 | self.bw = bw
125 | self.save_name = save_name
126 | self.x0 = x0
127 | self.previous_data_name = previous_data_name
128 | self.twinning_method = twinning_method
129 | self.bolus_source = bolus_source
130 | self.basal_source = basal_source
131 | self.cho_source = cho_source
132 | self.meal_generator_handler = meal_generator_handler
133 | self.meal_generator_handler_params = meal_generator_handler_params
134 | self.bolus_calculator_handler = bolus_calculator_handler
135 | self.bolus_calculator_handler_params = bolus_calculator_handler_params
136 | self.basal_handler = basal_handler
137 | self.basal_handler_params = basal_handler_params
138 | self.enable_hypotreatments = enable_hypotreatments
139 | self.hypotreatments_handler = hypotreatments_handler
140 | self.hypotreatments_handler_params = hypotreatments_handler_params
141 | self.enable_correction_boluses = enable_correction_boluses
142 | self.correction_boluses_handler = correction_boluses_handler
143 | self.correction_boluses_handler_params = correction_boluses_handler_params
144 | self.save_suffix = save_suffix
145 | self.save_workspace = save_workspace
146 | self.n_replay = n_replay
147 | self.sensors = sensors
148 | self.blueprint = blueprint
149 | self.exercise = exercise
150 |
151 |
152 | def validate(self):
153 | """
154 | Run the input validation process.
155 | """
156 |
157 | # Validate the 'data' input
158 | DataValidator(modality='replay', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
159 | bolus_source=self.bolus_source, basal_source=self.basal_source, cho_source=self.cho_source).validate()
160 |
161 | # Validate the 'bw' input
162 | BWValidator(bw=self.bw).validate()
163 |
164 | # Validate the 'save_name' input
165 | SaveNameValidator(save_name=self.save_name).validate()
166 |
167 | # Validate the 'save_name' input
168 | X0Validator(x0=self.x0).validate()
169 |
170 | # Validate the 'previous_data_name' input
171 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
172 |
173 | # Validate the 'twinning_method' input
174 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
175 |
176 | # Validate the 'bolus_source' input
177 | BolusSourceValidator(bolus_source=self.bolus_source).validate()
178 |
179 | # Validate the 'basal_source' input
180 | BasalSourceValidator(basal_source=self.basal_source).validate()
181 |
182 | # Validate the 'cho_source' input
183 | CHOSourceValidator(cho_source=self.cho_source).validate()
184 |
185 | # Validate the 'meal_generator_handler' input
186 | MealGeneratorHandlerValidator(meal_generator_handler=self.meal_generator_handler).validate()
187 |
188 | # Validate the 'meal_generator_handler_params' input
189 | MealGeneratorHandlerParamsValidator(meal_generator_handler_params=self.meal_generator_handler_params).validate()
190 |
191 | # Validate the 'bolus_calculator_handler' input
192 | BolusCalculatorHandlerValidator(bolus_calculator_handler=self.bolus_calculator_handler).validate()
193 |
194 | # Validate the 'bolus_calculator_handler_params' input
195 | BolusCalculatorHandlerParamsValidator(bolus_calculator_handler_params=self.bolus_calculator_handler_params).validate()
196 |
197 | # Validate the 'basal_handler' input
198 | BasalHandlerValidator(basal_handler=self.basal_handler).validate()
199 |
200 | # Validate the 'basal_handler_params' input
201 | BasalHandlerParamsValidator(basal_handler_params=self.basal_handler_params).validate()
202 |
203 | # Validate the 'enable_hypotreatments' input
204 | EnableHypotreatmentsValidator(enable_hypotreatments=self.enable_hypotreatments).validate()
205 |
206 | # Validate the 'hypotreatments_handler' input
207 | HypotreatmentsHandlerValidator(hypotreatments_handler=self.hypotreatments_handler).validate()
208 |
209 | # Validate the 'hypotreatments_handler_params' input
210 | HypotreatmentsHandlerParamsValidator(hypotreatments_handler_params=self.hypotreatments_handler_params).validate()
211 |
212 | # Validate the 'enable_correction_boluses' input
213 | EnableCorrectionBolusesValidator(enable_correction_boluses=self.enable_correction_boluses).validate()
214 |
215 | # Validate the 'correction_boluses_handler' input
216 | CorrectionBolusesHandlerValidator(correction_boluses_handler=self.correction_boluses_handler).validate()
217 |
218 | # Validate the 'correction_boluses_handler_params' input
219 | CorrectionBolusesHandlerParamsValidator(correction_boluses_handler_params=self.correction_boluses_handler_params).validate()
220 |
221 | # Validate the 'save_suffix' input
222 | SaveSuffixValidator(save_suffix=self.save_suffix).validate()
223 |
224 | # Validate the 'save_workspace' input
225 | SaveWorkspaceValidator(save_workspace=self.save_workspace).validate()
226 |
227 | # Validate the 'n_replay' input
228 | NReplayValidator(n_replay=self.n_replay).validate()
229 |
230 | # Validate the 'sensors' input
231 | SensorsValidator(sensors=self.sensors).validate()
232 |
--------------------------------------------------------------------------------
/py_replay_bg/input_validation/input_validator_twin.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 |
4 | from py_replay_bg.input_validation import *
5 |
6 |
7 | class InputValidatorTwin:
8 | """
9 | Class for validating the input of ReplayBG twin method.
10 |
11 | ...
12 | Attributes
13 | ----------
14 | data: pd.DataFrame
15 | Pandas dataframe which contains the data to be used by the tool.
16 | bw: float
17 | The patient's body weight.
18 | save_name : str
19 | A string used to label, thus identify, each output file and result.
20 |
21 |
22 | u2ss : float
23 | The steady state of the basal insulin infusion.
24 | x0 : np.ndarray
25 | The initial model conditions.
26 | previous_data_name : str
27 | The name of the previous data portion. This is used to correctly "transfer" the initial model conditions to
28 | the current portion of data.
29 |
30 | twinning_method : str
31 | The method to be used to twin the model.
32 |
33 | extended : bool
34 | A flag indicating whether to use the "extended" model for twinning
35 | find_start_guess_first : bool
36 | A flag indicating whether to find the start guess using MAP before twinning.
37 |
38 | n_steps: int
39 | Number of steps to use for the main chain. This is ignored if modality is 'replay'.
40 | save_chains: bool, optional, default : False
41 | A flag that specifies whether to save the resulting mcmc chains and copula samplers.
42 |
43 | parallelize : boolean
44 | A boolean that specifies whether to parallelize the twinning process.
45 | n_processes : int, optional, default : None
46 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
47 |
48 | blueprint: str
49 | A string that specifies the blueprint to be used to create the digital twin.
50 | exercise: bool
51 | A boolean that specifies whether to use exercise model or not.
52 |
53 | Methods
54 | -------
55 | validate():
56 | Run the input validation process.
57 | """
58 |
59 | def __init__(self,
60 | data: pd.DataFrame,
61 | bw: float,
62 | save_name: str,
63 | twinning_method: str,
64 | extended: bool,
65 | find_start_guess_first: bool,
66 | n_steps: int,
67 | save_chains: bool,
68 | u2ss: float | None,
69 | x0: np.ndarray | None,
70 | previous_data_name: str | None,
71 | parallelize: bool,
72 | n_processes: int | None,
73 | blueprint: str,
74 | exercise: bool
75 | ):
76 | self.data = data
77 | self.bw = bw
78 | self.save_name = save_name
79 | self.twinning_method = twinning_method
80 | self.extended = extended
81 | self.find_start_guess_first = find_start_guess_first
82 | self.n_steps = n_steps
83 | self.save_chains = save_chains
84 | self.u2ss = u2ss
85 | self.x0 = x0
86 | self.previous_data_name = previous_data_name
87 | self.parallelize = parallelize
88 | self.n_processes = n_processes
89 | self.blueprint = blueprint
90 | self.exercise = exercise
91 |
92 | def validate(self):
93 | """
94 | Run the input validation process.
95 | """
96 |
97 | # Validate the 'data' input
98 | DataValidator(modality='twin', data=self.data, blueprint=self.blueprint, exercise=self.exercise,
99 | bolus_source='data', basal_source='data', cho_source='data').validate()
100 |
101 | # Validate the 'bw' input
102 | BWValidator(bw=self.bw).validate()
103 |
104 | # Validate the 'save_name' input
105 | SaveNameValidator(save_name=self.save_name).validate()
106 |
107 | # Validate the 'twinning_method' input
108 | TwinningMethodValidator(twinning_method=self.twinning_method).validate()
109 |
110 | # Validate the 'extended' input
111 | ExtendedValidator(extended=self.extended).validate()
112 |
113 | # Validate the 'find_start_guess_first' input
114 | FindStartGuessFirstValidator(find_start_guess_first=self.find_start_guess_first).validate()
115 |
116 | # Validate the 'n_steps' input
117 | NStepsValidator(n_steps=self.n_steps).validate()
118 |
119 | # Validate the 'u2ss' input
120 | U2SSValidator(u2ss=self.u2ss).validate()
121 |
122 | # Validate the 'save_name' input
123 | X0Validator(x0=self.x0).validate()
124 |
125 | # Validate the 'previous_data_name' input
126 | PreviousDataNameValidator(previous_data_name=self.previous_data_name).validate()
127 |
128 | # Validate the 'parallelize' input
129 | ParallelizeValidator(parallelize=self.parallelize).validate()
130 |
131 | # Validate the 'n_processes' input
132 | NProcessesValidator(n_processes=self.n_processes).validate()
133 |
--------------------------------------------------------------------------------
/py_replay_bg/model/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/model/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/model/model_parameters_t1d.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pandas as pd
3 |
4 |
5 | class ModelParametersT1D:
6 | def __init__(self,
7 | data: pd.DataFrame,
8 | bw: float,
9 | u2ss: float,
10 | extended: bool
11 | ):
12 |
13 | """
14 | Function that returns the default parameters values of the t1d model.
15 |
16 | Parameters
17 | ----------
18 | data : pd.DataFrame
19 | Pandas dataframe which contains the data to be used by the tool.
20 | bw : float
21 | The patient's body weight.
22 | u2ss : float
23 | The steady state of the basal insulin infusion
24 | extended : bool
25 | A flag indicating whether to use the "extended" model for twinning
26 |
27 | Returns
28 | -------
29 | model_parameters: dict
30 | A dictionary containing the default model parameters.
31 |
32 | Raises
33 | ------
34 | None
35 |
36 | See Also
37 | --------
38 | None
39 |
40 | Examples
41 | --------
42 | None
43 | """
44 | # Initial conditions
45 | self.Xpb = 0.0 # Insulin action initial condition
46 | self.Qgutb = 0.0 # Intestinal content initial condition
47 |
48 | # Glucose-insulin submodel parameters
49 | self.VG = 1.45 # dl/kg
50 | self.SG = 2.5e-2 # 1/min
51 | self.Gb = 119.13 # mg/dL
52 | self.r1 = 1.4407 # unitless
53 | self.r2 = 0.8124 # unitless
54 | self.alpha = 7 # 1/min
55 | self.p2 = 0.012 # 1/min
56 |
57 | if u2ss is None:
58 | self.u2ss = np.mean(data.basal) * 1000 / bw # mU/(kg*min)
59 | else:
60 | self.u2ss = u2ss
61 |
62 | # Subcutaneous insulin absorption submodel parameters
63 | self.VI = 0.126 # L/kg
64 | self.ke = 0.127 # 1/min
65 | self.kd = 0.026 # 1/min
66 | # model_parameters['ka1'] = 0.0034 # 1/min (virtually 0 in 77% of the cases)
67 | # self.ka1 = 0.0
68 | self.ka2 = 0.014 # 1/min
69 | self.tau = 8 # min
70 | self.Ipb = self.u2ss / self.ke # from eq. 5 steady-state
71 |
72 |
73 | # Oral glucose absorption submodel parameters
74 | self.kgri = 0.18 # = kmax % 1/min
75 | self.kempt = 0.18 # 1/min
76 | self.f = 0.9 # dimensionless
77 |
78 | # Exercise submodel parameters
79 | self.vo2rest = 0.33 # dimensionless, VO2rest was derived from heart rate round(66/(220-30))
80 | self.vo2max = 1 # dimensionless, VO2max is normalized and estimated from heart rate (220-age) = 100%.
81 | self.e1 = 1.6 # dimensionless
82 | self.e2 = 0.78 # dimensionless
83 |
84 | # Patient specific parameters
85 | self.bw = bw # kg
86 | self.to_g = self.bw / 1000
87 | self.to_mgkg = 1000 / self.bw
88 |
89 | # Measurement noise specifics
90 | self.SDn = 5
91 |
92 | # Glucose starting point
93 | if 'glucose' in data:
94 | idx = np.where(data.glucose.isnull().values == False)[0][0]
95 | self.G0 = data.glucose[idx]
96 | else:
97 | self.G0 = self.Gb
98 |
99 |
100 | class ModelParametersT1DMultiMeal(ModelParametersT1D):
101 |
102 | def __init__(self,
103 | data: pd.DataFrame,
104 | bw: float,
105 | u2ss: float,
106 | extended: bool,
107 | ):
108 |
109 | """
110 | Function that returns the default parameters values of the t1d model in a multi-meal blueprint.
111 |
112 | Parameters
113 | ----------
114 | data : pd.DataFrame
115 | Pandas dataframe which contains the data to be used by the tool.
116 | bw : float
117 | The patient's body weight.
118 | u2ss : float
119 | The steady state of the basal insulin infusion
120 | extended : bool
121 | A flag indicating whether to use the "extended" model for twinning
122 |
123 | Returns
124 | -------
125 | model_parameters: dict
126 | A dictionary containing the default model parameters.
127 |
128 | Raises
129 | ------
130 | None
131 |
132 | See Also
133 | --------
134 | None
135 |
136 | Examples
137 | --------
138 | None
139 | """
140 | # Initialize common parameters
141 | super().__init__(data, bw, u2ss, extended)
142 |
143 | # Glucose-insulin submodel parameters
144 | self.SI_B = 10.35e-4 / self.VG # mL/(uU*min)
145 | self.SI_L = 10.35e-4 / self.VG # mL/(uU*min)
146 | self.SI_D = 10.35e-4 / self.VG # mL/(uU*min)
147 |
148 | if extended:
149 | self.SI_B2 = 10.35e-4 / self.VG # mL/(uU*min)
150 |
151 | # Oral glucose absorption submodel parameters
152 | self.kabs_B = 0.012 # 1/min
153 | self.kabs_L = 0.012 # 1/min
154 | self.kabs_D = 0.012 # 1/min
155 | self.kabs_S = 0.012 # 1/min
156 | self.kabs_H = 0.012 # 1/min
157 | self.beta_B = 0 # min
158 | self.beta_L = 0 # min
159 | self.beta_D = 0 # min
160 | self.beta_S = 0 # min
161 |
162 | if extended:
163 | self.kabs_B2 = 0.012 # 1/min
164 | self.kabs_L2 = 0.012 # 1/min
165 | self.kabs_S2 = 0.012 # 1/min
166 | self.beta_B2 = 0 # min
167 | self.beta_L2 = 0 # min
168 | self.beta_S2 = 0 # min
169 |
170 |
171 | class ModelParametersT1DSingleMeal(ModelParametersT1D):
172 |
173 | def __init__(self,
174 | data: pd.DataFrame,
175 | bw: float,
176 | u2ss: float,
177 | ):
178 |
179 | """
180 | Function that returns the default parameters values of the t1d model in a single-meal blueprint.
181 |
182 | Parameters
183 | ----------
184 | data : pd.DataFrame
185 | Pandas dataframe which contains the data to be used by the tool.
186 | bw : float
187 | The patient's body weight.
188 | u2ss : float
189 | The steady state of the basal insulin infusion
190 |
191 | Returns
192 | -------
193 | model_parameters: dict
194 | A dictionary containing the default model parameters.
195 |
196 | Raises
197 | ------
198 | None
199 |
200 | See Also
201 | --------
202 | None
203 |
204 | Examples
205 | --------
206 | None
207 | """
208 | # Initialize common parameters
209 | super().__init__(data, bw, u2ss, extended=False)
210 |
211 | # Glucose-insulin submodel parameters
212 | self.SI = 10.35e-4 / self.VG # mL/(uU*min)
213 |
214 | # Oral glucose absorption submodel parameters
215 | self.kabs = 0.012 # 1/min
216 | self.beta = 0 # 1/min
217 |
--------------------------------------------------------------------------------
/py_replay_bg/sensors/__init__.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 |
4 | class CGM:
5 | """
6 | A class that represents a CGM sensor.
7 |
8 | Attributes
9 | ----------
10 | ts: int
11 | The sample time of the cgm sensor (min).
12 | t_offset: float
13 | The time offset of the cgm sensor (when new this is 0). This is used if a cgm sensor is shared between more
14 | replay runs.
15 | cgm_error_parameters: np.ndarray
16 | An array containing the parameters of the CGM.
17 | output_noise_sd: float
18 | The output standard deviation of the CGM error model.
19 | ekm1: float
20 | Memory term for the CGM noise: e(k-1).
21 | ekm2: float
22 | Memory term for the CGM noise: e(k-2).
23 | max_lifetime: float
24 | The maximum lifetime of the CGM sensor (in minutes).
25 | connected_at: int
26 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
27 | ReplayBG calls).
28 |
29 | Methods
30 | -------
31 | connect_new_cgm(connected_at):
32 | Connects a new CGM sensor by sampling new error parameters.
33 | measure(ig, t):
34 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
35 | add_offset(to_add):
36 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
37 | multiple ReplayBG runs.
38 | """
39 |
40 | def __init__(self,
41 | ts: int = 5,
42 | ):
43 | """
44 | Constructs all the necessary attributes for the CGM object.
45 |
46 | Parameters
47 | ----------
48 | ts: int, optional, default = 5
49 | The sample time of the cgm sensor (min).
50 | """
51 | self.ts = ts
52 | self.t_offset = 0
53 |
54 | self.connect_new_cgm()
55 |
56 | # Set max lifetime (minutes)
57 | self.max_lifetime = 10 * 1440 # 10 days
58 | self.connected_at = 0
59 |
60 | # Set the parameters of the CGM
61 | self.cgm_error_parameters = []
62 | self.output_noise_sd = []
63 |
64 | # Set memory terms
65 | self.ekm1 = None
66 | self.ekm2 = None
67 |
68 | def connect_new_cgm(self, connected_at=0):
69 | """
70 | Connects a new CGM sensor by sampling new error parameters.
71 |
72 | Parameters
73 | ----------
74 | connected_at: int, optional, default = 0
75 | The time at which the CGM sensor is connected (utility variable to manage sensor lifetime through multiple
76 | ReplayBG calls).
77 |
78 | Returns
79 | -------
80 | None
81 |
82 | Raises
83 | ------
84 | None
85 |
86 | See Also
87 | --------
88 | None
89 |
90 | Examples
91 | --------
92 | None
93 | """
94 | # Load Mean vector and covariance matrix of the parameter vector as
95 | # defined in Vettoretti et al., Sensors, 2019
96 | mu = np.array([0.94228767821000314341972625697963, 0.0049398821141803427384187052950892,
97 | -0.0005848748565491275084801681138913, 6.382602204050874306062723917421,
98 | 1.2604070417357611244568715846981, -0.4022228938823663169088717950217,
99 | 3.2516360856114072674927228945307])
100 | sigma = np.array([[0.013245827952891258902368143424155, -0.0039513025350735725416129184850433,
101 | 0.00031276743283791636970891936186945, 0.15717912467153988265167186000326,
102 | 0.0026876560011614997885986966252858, -0.0028904633825263671524641306831427,
103 | -0.0031801707001874032418320403792222],
104 | [-0.0039513025350735725416129184850433, 0.0018527975980744701509778105119608,
105 | -0.00015580332205794781403294935184789, -0.10288007693621757654423021222101,
106 | -0.0013902327543057948350258001823931, 0.0011591852212130876378232136048041,
107 | -0.0027284927011686846420879248853453],
108 | [0.00031276743283791636970891936186945, -0.00015580332205794781403294935184789,
109 | 0.000013745962164724157000697882247131, 0.0080685863688738888865881193623864,
110 | 0.00012074974710011031125631020266553, -0.00010042135441622312822841645019167,
111 | 0.00011130290033867137325027107941366],
112 | [0.15717912467153988265167186000326, -0.10288007693621757654423021222101,
113 | 0.0080685863688738888865881193623864, 29.005838188852990811028575990349,
114 | 0.12408344051778112671069465022811, -0.10193644943826736526393261783596,
115 | 0.60075381294204155402383094042307],
116 | [0.0026876560011614997885986966252858, -0.0013902327543057948350258001823931,
117 | 0.00012074974710011031125631020266553, 0.12408344051778112671069465022811,
118 | 0.02079352674233487727195601735275, -0.018431109170459980539646949182497,
119 | -0.015721846813032722134373386779771],
120 | [-0.0028904633825263671524641306831427, 0.0011591852212130876378232136048041,
121 | -0.00010042135441622312822841645019167, -0.10193644943826736526393261783596,
122 | -0.018431109170459980539646949182497, 0.018700867933453400870913441167431,
123 | 0.01552333576829629385729347745837],
124 | [-0.0031801707001874032418320403792222, -0.0027284927011686846420879248853453,
125 | 0.00011130290033867137325027107941366, 0.60075381294204155402383094042307,
126 | -0.015721846813032722134373386779771, 0.01552333576829629385729347745837,
127 | 0.72356838038463477946748980684788]])
128 |
129 | # Modulation factor of the covariance of the parameter vector not to generate too extreme realizations of
130 | # parameter vector
131 | f = 0.90
132 | # Modulate the covariance matrix
133 | sigma = sigma * f
134 | # Maximum allowed output noise SD (mg/dl)
135 | max_output_noise_sd = 10
136 | # Tolerance for model stability check
137 | toll = 0.02
138 |
139 | # Flag for stability of the AR(2) model
140 | stable = 0
141 | output_noise_sd = np.inf
142 | cgm_error_parameters = []
143 | while (~ stable) or (output_noise_sd > max_output_noise_sd):
144 | # Sample CGM error parameters
145 | cgm_error_parameters = np.random.multivariate_normal(mu, sigma)
146 |
147 | # Check the stability of the resulting AR(2) model
148 | stable = (cgm_error_parameters[5] >= -1) and (
149 | cgm_error_parameters[5] <= (1 - np.abs(cgm_error_parameters[4]) - toll))
150 |
151 | output_noise_var = cgm_error_parameters[6] ** 2 / (
152 | 1 - (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) - cgm_error_parameters[5] * (
153 | (cgm_error_parameters[4] ** 2) / (1 - cgm_error_parameters[5]) + cgm_error_parameters[5]))
154 |
155 | if output_noise_var < 0:
156 | continue
157 |
158 | # Compute the output noise standard deviation
159 | output_noise_sd = np.sqrt(output_noise_var)
160 |
161 | # Set the parameters of the CGM
162 | self.cgm_error_parameters = cgm_error_parameters
163 | self.output_noise_sd = output_noise_sd
164 |
165 | # Set memory terms
166 | self.ekm1 = 0
167 | self.ekm2 = 0
168 |
169 | # Set the offset (minutes)
170 | self.t_offset = 0
171 |
172 | # Set the connection time
173 | self.connected_at = connected_at
174 |
175 | def measure(self, ig, t):
176 | """
177 | Function that provides a CGM measure using the model of Vettoretti et al., Sensors, 2019.
178 |
179 | Parameters
180 | ----------
181 | ig: float
182 | The interstitial glucose concentration at current time (mg/dl).
183 | t: float
184 | Current time in days from the start of CGM sensor.
185 |
186 | Returns
187 | -------
188 | cgm: float
189 | The CGM measurement at current time (mg/dl).
190 |
191 | Raises
192 | ------
193 | None
194 |
195 | See Also
196 | --------
197 | None
198 |
199 | Examples
200 | --------
201 | None
202 | """
203 |
204 | # Apply calibration error
205 | ig_s = (self.cgm_error_parameters[0] + self.cgm_error_parameters[1] * (t + self.t_offset) +
206 | self.cgm_error_parameters[2] * ((t + self.t_offset) ** 2)) * ig + self.cgm_error_parameters[3]
207 |
208 | # Generate noise
209 | z = np.random.normal(0, 1)
210 | u = self.cgm_error_parameters[6] * z
211 | e = u + self.cgm_error_parameters[4] * self.ekm1 + self.cgm_error_parameters[5] * self.ekm2
212 |
213 | # Update memory terms
214 | self.ekm2 = self.ekm1
215 | self.ekm1 = e
216 |
217 | # Get final CGM
218 | return ig_s + e
219 |
220 | def add_offset(self,
221 | to_add: float) -> None:
222 | """
223 | Utility function that adds an offset to the sensor life. Used when the sensor object must be shared through
224 | multiple ReplayBG runs.
225 |
226 | Parameters
227 | ----------
228 | to_add: float
229 | The offset to add to the sensor life.
230 |
231 | Returns
232 | -------
233 | None
234 |
235 | Raises
236 | ------
237 | None
238 |
239 | See Also
240 | --------
241 | None
242 |
243 | Examples
244 | --------
245 | None
246 | """
247 | self.t_offset += to_add
248 |
249 |
250 | class Sensors:
251 | """
252 | A class that represents the sensors to be used by ReplayBG.
253 |
254 | ...
255 | Attributes
256 | ----------
257 | cgm: CGM
258 | The CGM sensor used to measure the interstitial glucose during simulations.
259 |
260 | Methods
261 | -------
262 | None
263 | """
264 |
265 | def __init__(self, cgm: CGM):
266 | """
267 | Constructs all the necessary attributes for the Sensors object.
268 |
269 | Parameters
270 | ----------
271 | cgm: CGM
272 | The CGM sensor used to measure the interstitial glucose during simulations.
273 | """
274 | self.cgm = cgm
275 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/__init__.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pandas as pd
3 |
4 |
5 | def load_test_data(day):
6 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) + '.csv'))
7 | df.t = pd.to_datetime(df['t'])
8 | return df
9 |
10 |
11 | def load_test_data_extended(day):
12 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'data_day_' + str(day) +
13 | '_extended.csv'))
14 | df.t = pd.to_datetime(df['t'])
15 | return df
16 |
17 |
18 | def load_patient_info():
19 | df = pd.read_csv(os.path.join(os.path.abspath(''), 'py_replay_bg', 'example', 'data', 'patient_info.csv'))
20 | return df
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the location of the 'results' folder
10 | results_folder_location = os.path.join(os.path.abspath(''))
11 |
12 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_mcmc.pkl'), 'rb') as file:
13 | replay_results = pickle.load(file)
14 |
15 | # Visualize and analyze MCMC results
16 | Visualizer.plot_replay_results(replay_results)
17 | analysis = Analyzer.analyze_replay_results(replay_results)
18 | print(" ----- Analysis using MCMC-derived twins ----- ")
19 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
20 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
21 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
22 |
23 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_1_replay_map.pkl'), 'rb') as file:
24 | replay_results = pickle.load(file)
25 |
26 | # Visualize and analyze MAP results
27 | Visualizer.plot_replay_results(replay_results)
28 | analysis = Analyzer.analyze_replay_results(replay_results)
29 | print(" ----- Analysis using MAP-derived twins ----- ")
30 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
31 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
32 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
33 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_analysis_intervals_example.py:
--------------------------------------------------------------------------------
1 | import os
2 | import pickle
3 |
4 | from py_replay_bg.visualizer import Visualizer
5 | from py_replay_bg.analyzer import Analyzer
6 |
7 | def test_replay_bg():
8 |
9 | # Set the interval to analyze
10 | start_day = 1
11 | end_day = 2
12 |
13 | # Set the location of the 'results' folder
14 | results_folder_location = os.path.join(os.path.abspath(''))
15 |
16 | # Initialize results list
17 | replay_results_interval = []
18 |
19 | for day in range(start_day, end_day+1):
20 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_map.pkl'), 'rb') as file:
21 | replay_results = pickle.load(file)
22 | replay_results_interval.append(replay_results)
23 |
24 | # Visualize and analyze MCMC results
25 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
26 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
27 | print(" ----- Analysis using MAP-derived twins ----- ")
28 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
29 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
30 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
31 |
32 | # Initialize results list
33 | replay_results_interval = []
34 |
35 | for day in range(start_day, end_day + 1):
36 | with open(os.path.join(results_folder_location, 'results', 'workspaces', 'data_day_' + str(day) + '_replay_intervals_mcmc.pkl'), 'rb') as file:
37 | replay_results = pickle.load(file)
38 | replay_results_interval.append(replay_results)
39 |
40 | # Visualize and analyze MAP results
41 | Visualizer.plot_replay_results_interval(replay_results_interval=replay_results_interval)
42 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval=replay_results_interval)
43 | print(" ----- Analysis using MCMC-derived twins ----- ")
44 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
45 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
46 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | save_workspace=True,
57 | x0=x0, previous_data_name=previous_data_name,
58 | save_suffix='_replay_map',
59 | sensors=sensors)
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 | u2ss = float(patient_info.u2ss.values[p])
26 | x0 = None
27 | previous_data_name = None
28 | sensors = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Replaying " + save_name)
52 |
53 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
54 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
55 | twinning_method='mcmc',
56 | n_replay=10,
57 | save_workspace=True,
58 | x0=x0, previous_data_name=previous_data_name, sensors=sensors,
59 | save_suffix='_replay_mcmc')
60 |
61 | # Append results
62 | replay_results_interval.append(replay_results)
63 | data_interval.append(data)
64 |
65 | # Set initial conditions for next day equal to the "ending conditions" of the current day
66 | x0 = replay_results['x_end']['realizations'][0].tolist()
67 |
68 | # Set sensors to use the same sensors during the next portion of data
69 | sensors = replay_results['sensors']
70 |
71 | # Set previous_data_name
72 | previous_data_name = save_name
73 |
74 | # Visualize and analyze results
75 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
76 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
78 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
79 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
80 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | twinning_method='map',
50 | save_workspace=True,
51 | save_suffix='_replay_map_dss',
52 | enable_correction_boluses=True,
53 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
54 | bolus_calculator_handler_params=bolus_calculator_handler_params)
55 |
56 | # Visualize and analyze results
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
60 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
61 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
62 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | twinning_method='map',
41 | save_workspace=True,
42 | save_suffix='_replay_map')
43 |
44 | # Visualize and analyze results
45 | Visualizer.plot_replay_results(replay_results, data=data)
46 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
47 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
48 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
49 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
50 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1)
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_dss.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | from py_replay_bg.dss.default_dss_handlers import standard_bolus_calculator_handler
11 |
12 | def test_replay_bg():
13 |
14 | # Set verbosity
15 | verbose = True
16 | plot_mode = False
17 |
18 | # Set other parameters for twinning
19 | blueprint = 'multi-meal'
20 | save_folder = os.path.join(os.path.abspath(''))
21 |
22 | # load patient_info
23 | patient_info = load_patient_info()
24 | p = np.where(patient_info['patient'] == 1)[0][0]
25 | # Set bw and u2ss
26 | bw = float(patient_info.bw.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | # Set params of the bolus calculator
39 | bolus_calculator_handler_params = dict()
40 | bolus_calculator_handler_params['cr'] = 7
41 | bolus_calculator_handler_params['cf'] = 25
42 | bolus_calculator_handler_params['gt'] = 110
43 |
44 | print("Replaying " + save_name)
45 |
46 | # Replay the twin using a correction insulin bolus injection strategy and the standard formula for
47 | # meal insulin boluses
48 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
49 | n_replay=10,
50 | twinning_method='mcmc',
51 | save_workspace=True,
52 | save_suffix='_replay_mcmc_dss',
53 | enable_correction_boluses=True,
54 | bolus_source='dss', bolus_calculator_handler=standard_bolus_calculator_handler,
55 | bolus_calculator_handler_params=bolus_calculator_handler_params)
56 |
57 | # Visualize and analyze results
58 | Visualizer.plot_replay_results(replay_results, data=data)
59 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
60 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
61 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
62 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
63 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_replay_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 |
20 | # load patient_info
21 | patient_info = load_patient_info()
22 | p = np.where(patient_info['patient'] == 1)[0][0]
23 | # Set bw and u2ss
24 | bw = float(patient_info.bw.values[p])
25 |
26 | # Instantiate ReplayBG
27 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
28 | yts=5, exercise=False,
29 | seed=1,
30 | verbose=verbose, plot_mode=plot_mode)
31 |
32 | # Load data and set save_name
33 | data = load_test_data(day=1)
34 | save_name = 'data_day_' + str(1) + '_extended'
35 |
36 | print("Replaying " + save_name)
37 |
38 | # Replay the twin with the same input data to get the initial conditions for the subsequent day
39 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
40 | n_replay=10,
41 | twinning_method='mcmc',
42 | save_workspace=True,
43 | save_suffix='_replay_mcmc')
44 |
45 | # Visualize and analyze results
46 | Visualizer.plot_replay_results(replay_results, data=data)
47 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
48 | print('Mean glucose: %.2f mg/dl' % analysis['median']['glucose']['variability']['mean_glucose'])
49 | print('TIR: %.2f %%' % analysis['median']['glucose']['time_in_ranges']['time_in_target'])
50 | print('N Days: %.2f days' % analysis['median']['glucose']['data_quality']['number_days_of_observation'])
51 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 | x0 = None
28 | previous_data_name = None
29 |
30 | # Initialize the list of results
31 | replay_results_interval = []
32 | data_interval = []
33 |
34 | # Instantiate ReplayBG
35 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
36 | yts=5, exercise=False,
37 | seed=1,
38 | verbose=verbose, plot_mode=plot_mode)
39 |
40 | # Set interval to twin
41 | start_day = 1
42 | end_day = 2
43 |
44 | # Twin the interval
45 | for day in range(start_day, end_day+1):
46 |
47 | # Step 1: Load data and set save_name
48 | data = load_test_data(day=day)
49 | save_name = 'data_day_' + str(day) + '_interval'
50 |
51 | print("Twinning " + save_name)
52 |
53 | # Run twinning procedure
54 | rbg.twin(data=data, bw=bw, save_name=save_name,
55 | twinning_method='map',
56 | parallelize=parallelize,
57 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
58 |
59 | # Replay the twin with the same input data
60 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
61 | twinning_method='map',
62 | save_workspace=True,
63 | x0=x0, previous_data_name=previous_data_name,
64 | save_suffix='_twin_map')
65 |
66 | # Append results
67 | replay_results_interval.append(replay_results)
68 | data_interval.append(data)
69 |
70 | # Set initial conditions for next day equal to the "ending conditions" of the current day
71 | x0 = replay_results['x_end']['realizations'][0].tolist()
72 |
73 | # Set previous_data_name
74 | previous_data_name = save_name
75 |
76 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
77 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
78 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_intervals_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set the number of steps for MCMC
17 | n_steps = 5000 # In production, this should be >= 50k
18 |
19 | # Set other parameters for twinning
20 | blueprint = 'multi-meal'
21 | save_folder = os.path.join(os.path.abspath(''))
22 | parallelize = True
23 |
24 | # load patient_info
25 | patient_info = load_patient_info()
26 | p = np.where(patient_info['patient'] == 1)[0][0]
27 | # Set bw and u2ss
28 | bw = float(patient_info.bw.values[p])
29 | u2ss = float(patient_info.u2ss.values[p])
30 | x0 = None
31 | previous_data_name = None
32 |
33 | # Initialize the list of results
34 | replay_results_interval = []
35 | data_interval = []
36 |
37 | # Instantiate ReplayBG
38 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
39 | yts=5, exercise=False,
40 | seed=1,
41 | verbose=verbose, plot_mode=plot_mode)
42 |
43 | # Set interval to twin
44 | start_day = 1
45 | end_day = 2
46 |
47 | # Twin the interval
48 | for day in range(start_day, end_day+1):
49 |
50 | # Step 1: Load data and set save_name
51 | data = load_test_data(day=day)
52 | save_name = 'data_day_' + str(day) + '_interval'
53 |
54 | print("Twinning " + save_name)
55 |
56 | # Run twinning procedure
57 | rbg.twin(data=data, bw=bw, save_name=save_name,
58 | twinning_method='mcmc',
59 | parallelize=parallelize,
60 | n_steps=n_steps,
61 | x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
62 |
63 | # Replay the twin with the same input data
64 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
65 | twinning_method='mcmc',
66 | save_workspace=True,
67 | x0=x0, previous_data_name=previous_data_name,
68 | save_suffix='_twin_mcmc')
69 |
70 | # Append results
71 | replay_results_interval.append(replay_results)
72 | data_interval.append(data)
73 |
74 | # Set initial conditions for next day equal to the "ending conditions" of the current day
75 | x0 = replay_results['x_end']['realizations'][0].tolist()
76 |
77 | # Set previous_data_name
78 | previous_data_name = save_name
79 |
80 | Visualizer.plot_replay_results_interval(replay_results_interval, data_interval=data_interval)
81 | analysis = Analyzer.analyze_replay_results_interval(replay_results_interval, data_interval=data_interval)
82 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
83 |
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data(day=1)
36 | save_name = 'data_day_' + str(1)
37 |
38 | print("Twinning " + save_name)
39 |
40 | # Run twinning procedure
41 | rbg.twin(data=data, bw=bw, save_name=save_name,
42 | twinning_method='map',
43 | parallelize=parallelize,
44 | u2ss=u2ss)
45 |
46 | # Replay the twin with the same input data
47 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
48 | twinning_method='map',
49 | save_workspace=True,
50 | save_suffix='_twin_map')
51 |
52 | # Visualize and analyze results
53 | Visualizer.plot_replay_results(replay_results, data=data)
54 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
55 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_map_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 | def test_replay_bg():
11 |
12 | # Set verbosity
13 | verbose = True
14 | plot_mode = False
15 |
16 | # Set other parameters for twinning
17 | blueprint = 'multi-meal'
18 | save_folder = os.path.join(os.path.abspath(''))
19 | parallelize = True
20 |
21 | # load patient_info
22 | patient_info = load_patient_info()
23 | p = np.where(patient_info['patient'] == 1)[0][0]
24 | # Set bw and u2ss
25 | bw = float(patient_info.bw.values[p])
26 | u2ss = float(patient_info.u2ss.values[p])
27 |
28 | # Instantiate ReplayBG
29 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
30 | yts=5, exercise=False,
31 | seed=1,
32 | verbose=verbose, plot_mode=plot_mode)
33 |
34 | # Load data and set save_name
35 | data = load_test_data_extended(day=1)
36 | # Cut data up to 11:00
37 | data = data.iloc[0:348, :]
38 | # Modify label of second day
39 | data.loc[312, "cho_label"] = 'B2'
40 |
41 | save_name = 'data_day_' + str(1) + '_extended'
42 |
43 | print("Twinning " + save_name)
44 |
45 | # Run twinning procedure using also the burn-out period
46 | rbg.twin(data=data, bw=bw, save_name=save_name,
47 | twinning_method='map',
48 | parallelize=parallelize,
49 | u2ss=u2ss,
50 | extended=True,
51 | find_start_guess_first=False)
52 |
53 | # Cut data up to 4:00
54 | data = data.iloc[0:264,:]
55 |
56 | # Replay the twin with the data up to the start of the burn-out period
57 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
58 | twinning_method='map',
59 | save_workspace=True,
60 | save_suffix='_twin_map_extended')
61 |
62 | # Visualize and analyze results
63 | Visualizer.plot_replay_results(replay_results, data=data)
64 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
65 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data(day=1)
40 | save_name = 'data_day_' + str(1)
41 |
42 | print("Twinning " + save_name)
43 |
44 | # Run twinning procedure
45 | rbg.twin(data=data, bw=bw, save_name=save_name,
46 | twinning_method='mcmc',
47 | parallelize=parallelize,
48 | n_steps=n_steps,
49 | u2ss=u2ss)
50 |
51 | # Replay the twin with the same input data
52 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
53 | twinning_method='mcmc',
54 | save_workspace=True,
55 | save_suffix='_twin_mcmc')
56 |
57 | Visualizer.plot_replay_results(replay_results, data=data)
58 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
59 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/tests/test_twin_mcmc_extended.py:
--------------------------------------------------------------------------------
1 | import os
2 | import numpy as np
3 |
4 | from py_replay_bg.tests import load_test_data_extended, load_patient_info
5 |
6 | from py_replay_bg.py_replay_bg import ReplayBG
7 | from py_replay_bg.visualizer import Visualizer
8 | from py_replay_bg.analyzer import Analyzer
9 |
10 |
11 | def test_replay_bg():
12 |
13 | # Set verbosity
14 | verbose = True
15 | plot_mode = False
16 |
17 | # Set the number of steps for MCMC
18 | n_steps = 5000 # In production, this should be >= 50k
19 |
20 | # Set other parameters for twinning
21 | blueprint = 'multi-meal'
22 | save_folder = os.path.join(os.path.abspath(''))
23 | parallelize = True
24 |
25 | # load patient_info
26 | patient_info = load_patient_info()
27 | p = np.where(patient_info['patient'] == 1)[0][0]
28 | # Set bw and u2ss
29 | bw = float(patient_info.bw.values[p])
30 | u2ss = float(patient_info.u2ss.values[p])
31 |
32 | # Instantiate ReplayBG
33 | rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
34 | yts=5, exercise=False,
35 | seed=1,
36 | verbose=verbose, plot_mode=plot_mode)
37 |
38 | # Load data and set save_name
39 | data = load_test_data_extended(day=1)
40 | # Cut data up to 11:00
41 | data = data.iloc[0:348, :]
42 | # Modify label of second day
43 | data.loc[312, "cho_label"] = 'B2'
44 |
45 | save_name = 'data_day_' + str(1) + '_extended'
46 |
47 | print("Twinning " + save_name)
48 |
49 | # Run twinning procedure using also the burn-out period
50 | rbg.twin(data=data, bw=bw, save_name=save_name,
51 | twinning_method='mcmc',
52 | parallelize=parallelize,
53 | n_steps=n_steps,
54 | u2ss=u2ss,
55 | extended=True,
56 | find_start_guess_first=True)
57 |
58 | # Cut data up to 4:00
59 | data = data.iloc[0:264, :]
60 |
61 | # Replay the twin with the data up to the start of the burn-out period
62 | replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
63 | twinning_method='mcmc',
64 | save_workspace=True,
65 | save_suffix='_twin_mcmc_extended')
66 |
67 | Visualizer.plot_replay_results(replay_results, data=data)
68 | analysis = Analyzer.analyze_replay_results(replay_results, data=data)
69 | print('Fit MARD: %.2f %%' % analysis['median']['twin']['mard'])
--------------------------------------------------------------------------------
/py_replay_bg/twinning/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/twinning/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/twinning/map.py:
--------------------------------------------------------------------------------
1 | import os
2 | import warnings
3 | import pickle
4 | import numpy as np
5 |
6 | from typing import Dict, Callable
7 |
8 | from multiprocessing import Pool
9 | from tqdm import tqdm
10 | from scipy.optimize import minimize
11 |
12 | from py_replay_bg.data import ReplayBGData
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 |
16 | from py_replay_bg.environment import Environment
17 |
18 | # Suppress all RuntimeWarnings
19 | warnings.filterwarnings("ignore", category=RuntimeWarning)
20 |
21 |
22 | class MAP:
23 | """
24 | A class that orchestrates the twinning process via MAP.
25 |
26 | Attributes
27 | ----------
28 | max_iter: int
29 | Maximum number of iterations.
30 | parallelize : bool
31 | A boolean that specifies whether to parallelize the twinning process.
32 | n_processes : int
33 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
34 |
35 | Methods
36 | -------
37 | twin(rbg_data, model, save_name, environment)
38 | Runs the twinning procedure.
39 | """
40 |
41 | def __init__(self,
42 | max_iter: int = 100000,
43 | parallelize: bool = False,
44 | n_processes: int | None = None,
45 | ):
46 | """
47 | Constructs all the necessary attributes for the MCMC object.
48 |
49 | Parameters
50 | ----------
51 | max_iter: int, optional, default : 100000
52 | Maximum number of iterations.
53 | parallelize : bool, optional, default : False
54 | A boolean that specifies whether to parallelize the twinning process.
55 | n_processes : int, optional, default : None
56 | The number of processes to be spawn if `parallelize` is `True`. If None, the number of CPU cores is used.
57 |
58 | Returns
59 | -------
60 | None
61 |
62 | Raises
63 | ------
64 | None
65 |
66 | See Also
67 | --------
68 | None
69 |
70 | Examples
71 | --------
72 | None
73 | """
74 |
75 | # Number of times to re-run the procedure
76 | self.n_rerun = 64
77 |
78 | # Maximum number of iterations
79 | self.max_iter = max_iter
80 |
81 | # Maximum number of function evaluations
82 | self.max_fev = 1000000
83 |
84 | # Parallelization options
85 | self.parallelize = parallelize
86 | self.n_processes = n_processes
87 |
88 | def twin(self,
89 | rbg_data: ReplayBGData,
90 | model: T1DModelSingleMeal | T1DModelMultiMeal,
91 | save_name: str,
92 | environment: Environment,
93 | start_guess: Dict = None,
94 | for_start_guess: bool = False) -> Dict:
95 | """
96 | Runs the twinning procedure.
97 |
98 | Parameters
99 | ----------
100 | rbg_data: ReplayBGData
101 | An object containing the data to be used during the twinning procedure.
102 | model: T1DModelSingleMeal | T1DModelMultiMeal
103 | An object that represents the physiological model to be used by ReplayBG.
104 | environment: Environment
105 | An object that represents the hyperparameters to be used by ReplayBG.
106 | save_name : str
107 | A string used to label, thus identify, each output file and result.
108 | start_guess: Dict, optional, default : None
109 | The initial guess for the twinning process. If None, this is set to population values.
110 | for_start_guess: bool, optional, default : False
111 | Whether to use the twin method just to find the start guess to pass to the MCMC twinner.
112 |
113 | Returns
114 | -------
115 | draws: dict
116 | A dictionary containing the chain and the samples obtained from the MCMC procedure and the copula
117 | sampling, respectively.
118 |
119 | Raises
120 | ------
121 | None
122 |
123 | See Also
124 | --------
125 | None
126 |
127 | Examples
128 | --------
129 | None
130 | """
131 |
132 | # If this is being used to find the start_guess, do /4 less reruns
133 | if for_start_guess:
134 | self.n_rerun = int(self.n_rerun/16)
135 |
136 | # Number of unknown parameters to twin
137 | n_dim = len(model.unknown_parameters)
138 |
139 | if start_guess is None:
140 | sg = model.start_guess
141 | else:
142 | sg = np.array(list(start_guess.values()))
143 |
144 | # Set the initial positions of the walkers.
145 | start = sg + model.start_guess_sigma * np.random.randn(self.n_rerun, n_dim)
146 | start[start < 0] = 0
147 |
148 | # Set the pooler
149 | pool = None
150 | if self.parallelize:
151 | pool = Pool(processes=self.n_processes)
152 |
153 | # Set up the options
154 | options = dict()
155 | options['maxiter'] = self.max_iter
156 | options['maxfev'] = self.max_fev
157 | options['disp'] = False
158 |
159 | # Select the function to minimize
160 | neg_log_posterior_func = model.neg_log_posterior_extended if model.extended else model.neg_log_posterior
161 |
162 | # Initialize results
163 | results = []
164 |
165 | if pool is None:
166 |
167 | if environment.verbose:
168 | iterator = tqdm(range(self.n_rerun))
169 | iterator.set_description("Min loss: %f" % np.nan)
170 | else:
171 | iterator = range(self.n_rerun)
172 |
173 | # Initialize best
174 | best = -1
175 |
176 | for r in iterator:
177 | result = run_map(start[r], neg_log_posterior_func, rbg_data, options)
178 | results.append(result)
179 | if best == -1 or result['fun'] < results[best]['fun']:
180 | best = r
181 | if environment.verbose:
182 | iterator.set_description("Min loss %f" % results[best]['fun'])
183 |
184 | else:
185 | # Prepare input arguments as tuples for starmap
186 | args = [(start[r], neg_log_posterior_func, rbg_data, options) for r in range(self.n_rerun)]
187 |
188 | # Initialize best
189 | best = -1
190 |
191 | # Get results (verbosity not allowed for the moment)
192 | results = pool.starmap(run_map, args)
193 |
194 | # Get best
195 | for r, result in enumerate(results):
196 | if best == -1 or result['fun'] < results[best]['fun']:
197 | best = r
198 |
199 | draws = dict()
200 | for up in range(n_dim):
201 | draws[model.unknown_parameters[up]] = results[best]['x'][up]
202 |
203 | # If twin is being used just to find the start guess, just return draws without saving
204 | if for_start_guess:
205 | return draws
206 |
207 | # Clean-up draws from "extended" parameters
208 | if model.extended:
209 | if 'SI_B2' in draws:
210 | del draws['SI_B2']
211 | if 'kabs_B2' in draws:
212 | del draws['kabs_B2']
213 | if 'beta_B2' in draws:
214 | del draws['beta_B2']
215 | if 'kabs_L2' in draws:
216 | del draws['kabs_L2']
217 | if 'beta_L2' in draws:
218 | del draws['beta_L2']
219 | if 'kabs_S2' in draws:
220 | del draws['kabs_S2']
221 | if 'beta_S2' in draws:
222 | del draws['beta_S2']
223 |
224 | # Save results
225 | twinning_results = dict()
226 | twinning_results['draws'] = draws
227 | twinning_results['u2ss'] = model.model_parameters.u2ss
228 |
229 | saved_file = os.path.join(environment.replay_bg_path, 'results', 'map',
230 | 'map_' + save_name + '.pkl')
231 |
232 | with open(saved_file, 'wb') as file:
233 | pickle.dump(twinning_results, file)
234 |
235 | if environment.verbose:
236 | print('Parameters saved in ' + saved_file)
237 |
238 | return draws
239 |
240 |
241 | def run_map(start: np.ndarray,
242 | neg_log_posterior_func: Callable,
243 | rbg_data: ReplayBGData,
244 | options: Dict
245 | ) -> Dict:
246 | """
247 | Utility function used to run MAP twinning.
248 |
249 | Parameters
250 | ----------
251 | start: np.ndarray
252 | An object containing the data to be used during the twinning procedure.
253 | neg_log_posterior_func: Callable
254 | The function to minimize, i.e., the neg-loglikelihood.
255 | rbg_data: ReplayBGData
256 | An object containing the data to be used during the twinning procedure.
257 | options : Dict
258 | A dictionary with the options necessary to the minimization function.
259 |
260 | Returns
261 | -------
262 | ret: dict
263 | A dictionary containing the results of the MAP twinning and the final value of the objective function.
264 |
265 | Raises
266 | ------
267 | None
268 |
269 | See Also
270 | --------
271 | None
272 |
273 | Examples
274 | --------
275 | None
276 | """
277 | result = minimize(neg_log_posterior_func, start, method='Powell', args=(rbg_data,), options=options)
278 | ret = dict()
279 | ret['fun'] = result.fun
280 | ret['x'] = result.x
281 | return ret
282 |
--------------------------------------------------------------------------------
/py_replay_bg/utils/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/gcappon/py_replay_bg/f13e057811b591f6d3ce1dfd46bcc074c9dabfc9/py_replay_bg/utils/__init__.py
--------------------------------------------------------------------------------
/py_replay_bg/utils/stats.py:
--------------------------------------------------------------------------------
1 | import math
2 | import numpy as np
3 | from numba import njit
4 |
5 |
6 | @njit(fastmath=True)
7 | def log_lognorm(x, mu, sigma):
8 | """
9 | Computes the logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
10 |
11 | Parameters
12 | ----------
13 | x: float
14 | The value where to evaluate the log-normal pdf.
15 | mu: float
16 | The mean of the log-normal distribution.
17 | sigma: float
18 | The standard deviation of the log-normal distribution.
19 |
20 | Returns
21 | -------
22 | ll_norm: float
23 | The logarithm of the log-normal pdf evaluated at given x with given mu and sigma.
24 |
25 | Raises
26 | ------
27 | None
28 |
29 | See Also
30 | --------
31 | None
32 |
33 | Examples
34 | --------
35 | None
36 | """
37 | return np.log(1 / (x * sigma * np.sqrt(2 * np.pi)) * np.exp(- ((np.log(x) - mu) ** 2) / (2 * (sigma ** 2))))
38 |
39 |
40 | @njit(fastmath=True)
41 | def log_norm(x, mu, sigma):
42 | """
43 | Computes the logarithm of the normal pdf evaluated at given x with given mu and sigma.
44 |
45 | Parameters
46 | ----------
47 | x: float
48 | The value where to evaluate the normal pdf.
49 | mu: float
50 | The mean of the normal distribution.
51 | sigma: float
52 | The standard deviation of the normal distribution.
53 |
54 | Returns
55 | -------
56 | l_norm: float
57 | The logarithm of the normal pdf evaluated at given x with given mu and sigma.
58 |
59 | Raises
60 | ------
61 | None
62 |
63 | See Also
64 | --------
65 | None
66 |
67 | Examples
68 | --------
69 | None
70 | """
71 | return np.log(1 / (sigma * np.sqrt(2 * np.pi)) * np.exp(- 0.5 * ((x - mu) / sigma) ** 2))
72 |
73 |
74 | @njit(fastmath=True)
75 | def log_gamma(x, alpha, beta):
76 | """
77 | Computes the logarithm of the gamma pdf evaluated at given x with given alpha and beta.
78 |
79 | Parameters
80 | ----------
81 | x: float
82 | The value where to evaluate the normal pdf.
83 | alpha: float
84 | The alpha value of the gamma distribution at hand.
85 | beta: float
86 | The beta value of the gamma distribution at hand.
87 |
88 | Returns
89 | -------
90 | l_gam: float
91 | The logarithm of the gamma pdf evaluated at given x with given alpha and beta.
92 |
93 | Raises
94 | ------
95 | None
96 |
97 | See Also
98 | --------
99 | None
100 |
101 | Examples
102 | --------
103 | None
104 | """
105 | return -np.inf if x < 0 else np.log((beta ** alpha * x ** (alpha - 1) * math.exp(-beta * x)) / math.gamma(alpha))
106 |
--------------------------------------------------------------------------------
/py_replay_bg/visualizer/__init__.py:
--------------------------------------------------------------------------------
1 | from datetime import datetime, timedelta
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 |
6 | from typing import Dict
7 |
8 | import pandas as pd
9 | from tqdm import tqdm
10 |
11 | from py_replay_bg.data import ReplayBGData
12 | from py_replay_bg.environment import Environment
13 | from py_replay_bg.model.t1d_model_single_meal import T1DModelSingleMeal
14 | from py_replay_bg.model.t1d_model_multi_meal import T1DModelMultiMeal
15 | from py_replay_bg.dss import DSS
16 |
17 |
18 | class Visualizer:
19 | """
20 | A class to be used by ReplayBG for plotting.
21 |
22 | ...
23 | Attributes
24 | ----------
25 | -
26 |
27 | Methods
28 | -------
29 | plot_replay_results(replay_results, data)
30 | Function that plots the results of ReplayBG replay simulation.
31 | """
32 |
33 | def __init__(self):
34 | """
35 | Constructs all the necessary attributes for the Visualizer object.
36 |
37 | Parameters
38 | ----------
39 | -
40 |
41 | Returns
42 | -------
43 | None
44 |
45 | Raises
46 | ------
47 | None
48 |
49 | See Also
50 | --------
51 | None
52 |
53 | Examples
54 | --------
55 | None
56 | """
57 | pass
58 |
59 | @staticmethod
60 | def plot_replay_results(
61 | replay_results: Dict,
62 | data: pd.DataFrame = None,
63 | title: str = '',
64 | ) -> None:
65 | """
66 | Function that plots the results of ReplayBG simulation.
67 |
68 | Parameters
69 | ----------
70 | replay_results: dict
71 | The replayed scenario results with fields:
72 | glucose: dict
73 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
74 | cgm: dict
75 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
76 | insulin_bolus: dict
77 | A dictionary which contains the insulin boluses simulated via ReplayBG.
78 | correction_bolus: dict
79 | A dictionary which contains the correction boluses simulated via ReplayBG.
80 | insulin_basal: dict
81 | A dictionary which contains the basal insulin simulated via ReplayBG.
82 | cho: dict
83 | A dictionary which contains the meals simulated via ReplayBG.
84 | hypotreatments: dict
85 | A dictionary which contains the hypotreatments simulated via ReplayBG.
86 | meal_announcement: dict
87 | A dictionary which contains the meal announcements simulated via ReplayBG.
88 | vo2: dict
89 | A dictionary which contains the vo2 simulated via ReplayBG.
90 | sensors: dict
91 | A dictionary which contains the sensors used during the replayed scenario.
92 | rbg_data: ReplayBGData
93 | The data to be used by ReplayBG during simulation.
94 | model: T1DModelSingleMeal | T1DModelMultiMeal
95 | An object that represents the physiological model to be used by ReplayBG.
96 | data: pd.DataFrame, optional, default: None
97 | Pandas dataframe which contains the data to be used by the tool. If present, adds glucose data to the
98 | glucose subplot.
99 |
100 | Returns
101 | -------
102 | None
103 |
104 | Raises
105 | ------
106 | None
107 |
108 | See Also
109 | --------
110 | None
111 |
112 | Examples
113 | --------
114 | None
115 | """
116 | # Subplot 1: Glucose
117 |
118 | fig, ax = None, None
119 | if replay_results['model'].exercise:
120 | fig, ax = plt.subplots(4, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1, 1]})
121 | else:
122 | fig, ax = plt.subplots(3, 1, sharex=True, gridspec_kw={'height_ratios': [3, 1, 1]})
123 |
124 | if data is not None:
125 | ax[0].plot(data.t, data.glucose, marker='*', color='red', linewidth=2, label='CGM data [mg/dl]')
126 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['cgm']['median'], marker='o', color='black', linewidth=2, label='CGM replay (Median) [mg/dl]')
127 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['cgm']['ci25th'], replay_results['cgm']['ci75th'], color='black', alpha=0.2,
128 | label='CGM replay (CI 25-75th) [mg/dl]')
129 |
130 | ax[0].plot(replay_results['rbg_data'].t_data, replay_results['glucose']['median'][::replay_results['model'].yts], marker='o', color='blue', linewidth=2,
131 | label='Glucose replay (Median) [mg/dl]')
132 | ax[0].fill_between(replay_results['rbg_data'].t_data, replay_results['glucose']['ci25th'][::replay_results['model'].yts], replay_results['glucose']['ci75th'][::replay_results['model'].yts], color='blue',
133 | alpha=0.3, label='Glucose replay (CI 25-75th) [mg/dl]')
134 |
135 | ax[0].grid()
136 | ax[0].legend()
137 |
138 | # Subplot 2: Meals
139 | t = np.arange(replay_results['rbg_data'].t_data[0]+ pd.Timedelta(minutes=0), replay_results['rbg_data'].t_data[-1] + pd.Timedelta(minutes=replay_results['model'].yts),
140 | timedelta(minutes=1)).astype(datetime)
141 |
142 | cho_events = np.sum(replay_results['cho']['realizations'], axis=0) / replay_results['cho']['realizations'].shape[0]
143 | ht_events = np.sum(replay_results['hypotreatments']['realizations'], axis=0) / replay_results['hypotreatments']['realizations'].shape[0]
144 |
145 | markerline, stemlines, baseline = ax[1].stem(t, cho_events, basefmt='k:', label='CHO replay (Mean) [g/min]')
146 | plt.setp(stemlines, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
147 | plt.setp(markerline, 'color', (70.0 / 255, 130.0 / 255, 180.0 / 255))
148 |
149 | markerline, stemlines, baseline = ax[1].stem(t, ht_events, basefmt='k:', label='HT replay (Mean) [g/min]')
150 | plt.setp(stemlines, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
151 | plt.setp(markerline, 'color', (0.0 / 255, 204.0 / 255, 204.0 / 255))
152 |
153 | ax[1].grid()
154 | ax[1].legend()
155 |
156 | # Subplot 3: Insulin
157 |
158 | bolus_events = np.sum(replay_results['insulin_bolus']['realizations'], axis=0) / replay_results['insulin_bolus']['realizations'].shape[0]
159 | cb_events = np.sum(replay_results['correction_bolus']['realizations'], axis=0) / replay_results['correction_bolus']['realizations'].shape[0]
160 | basal_rate = np.sum(replay_results['insulin_basal']['realizations'], axis=0) / replay_results['insulin_basal']['realizations'].shape[0]
161 |
162 | markerline, stemlines, baseline = ax[2].stem(t, bolus_events, basefmt='k:',
163 | label='Bolus insulin replay (Mean) [U/min]')
164 | plt.setp(stemlines, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
165 | plt.setp(markerline, 'color', (50.0 / 255, 205.0 / 255, 50.0 / 255))
166 |
167 | markerline, stemlines, baseline = ax[2].stem(t, cb_events, basefmt='k:',
168 | label='CB insulin replay (Mean) [U/min]')
169 | plt.setp(stemlines, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
170 | plt.setp(markerline, 'color', (51.0 / 255, 102.0 / 255, 0.0 / 255))
171 |
172 | ax[2].plot(t, basal_rate * 60, color='black', linewidth=2, label='Basal insulin replay (Mean) [U/h]')
173 |
174 | ax[2].grid()
175 | ax[2].legend()
176 |
177 | # Subplot 4: Exercise
178 |
179 | if replay_results['model'].exercise:
180 |
181 | vo2_events = np.sum(replay_results['vo2']['realizations'], axis=0) / replay_results['vo2']['realizations'].shape[0]
182 |
183 | markerline, stemlines, baseline = ax[3].stem(t, vo2_events, basefmt='k:', label='VO2 replay (Mean) [-]')
184 | plt.setp(stemlines, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
185 | plt.setp(markerline, 'color', (249.0 / 255, 115.0 / 255, 6.0 / 255))
186 |
187 | ax[3].grid()
188 | ax[3].legend()
189 |
190 | fig.suptitle(title, fontweight='bold')
191 | plt.show()
192 |
193 |
194 | @staticmethod
195 | def plot_replay_results_interval(
196 | replay_results_interval: list,
197 | data_interval: list = None,
198 | title: str = '',
199 | ) -> None:
200 | """
201 | Function that plots the results of ReplayBG simulation (intervals).
202 |
203 | Parameters
204 | ----------
205 | replay_results_interval: list
206 | A list dictionaries of replayed scenario results. Each element has fields:
207 | glucose: dict
208 | A dictionary which contains the obtained glucose traces simulated via ReplayBG.
209 | cgm: dict
210 | A dictionary which contains the obtained cgm traces simulated via ReplayBG.
211 | insulin_bolus: dict
212 | A dictionary which contains the insulin boluses simulated via ReplayBG.
213 | correction_bolus: dict
214 | A dictionary which contains the correction boluses simulated via ReplayBG.
215 | insulin_basal: dict
216 | A dictionary which contains the basal insulin simulated via ReplayBG.
217 | cho: dict
218 | A dictionary which contains the meals simulated via ReplayBG.
219 | hypotreatments: dict
220 | A dictionary which contains the hypotreatments simulated via ReplayBG.
221 | meal_announcement: dict
222 | A dictionary which contains the meal announcements simulated via ReplayBG.
223 | vo2: dict
224 | A dictionary which contains the vo2 simulated via ReplayBG.
225 | sensors: dict
226 | A dictionary which contains the sensors used during the replayed scenario.
227 | rbg_data: ReplayBGData
228 | The data to be used by ReplayBG during simulation.
229 | model: T1DModelSingleMeal | T1DModelMultiMeal
230 | An object that represents the physiological model to be used by ReplayBG.
231 | data_interval: list, optional, default: None
232 | A list of pandas dataframe which contains the data to be used by the tool. If present, adds glucose data
233 | to the glucose subplot.
234 |
235 | Returns
236 | -------
237 | None
238 |
239 | Raises
240 | ------
241 | None
242 |
243 | See Also
244 | --------
245 | None
246 |
247 | Examples
248 | --------
249 | None
250 | """
251 | data = None
252 | if data_interval is not None:
253 | data = pd.concat(data_interval, axis=0, ignore_index=True)
254 |
255 | # Pad each array to the maximum shape
256 | replay_results = dict()
257 |
258 |
259 | # Re-map glucose data
260 | fields = ['median', 'ci5th', 'ci25th', 'ci75th', 'ci95th']
261 | category = ['glucose', 'cgm']
262 |
263 | for c in category:
264 |
265 | replay_results[c] = dict()
266 |
267 | for f in fields:
268 | replay_results[c][f] = dict()
269 |
270 | replay_results[c][f] = [r[c][f] for r in replay_results_interval]
271 | replay_results[c][f] = np.concatenate(replay_results[c][f], axis=0)
272 |
273 | # Re-map cho, insulin, and vo2 data
274 | category = ['cho', 'hypotreatments', 'insulin_bolus', 'correction_bolus', 'insulin_basal', 'vo2']
275 |
276 | for c in category:
277 | replay_results[c] = dict()
278 | replay_results[c]['realizations'] = [r[c]['realizations'] for r in replay_results_interval]
279 | replay_results[c]['realizations'] = np.concatenate(replay_results[c]['realizations'], axis=1)
280 |
281 | replay_results['model'] = replay_results_interval[0]['model']
282 | t_data = [r['rbg_data'].t_data for r in replay_results_interval]
283 | replay_results['rbg_data'] = replay_results_interval[0]['rbg_data']
284 | replay_results['rbg_data'].t_data = np.concatenate(t_data, axis=0)
285 |
286 | Visualizer.plot_replay_results(replay_results=replay_results, data=data, title=title)
287 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [tool.pytest.ini_options]
2 | pythonpath = [
3 | "."
4 | ]
5 |
6 | [build-system]
7 | requires = ["setuptools>=61.0.0", "wheel"]
8 | build-backend = "setuptools.build_meta"
9 |
10 | [project]
11 | name = "py_replay_bg"
12 | version = "1.1.0"
13 | authors = [
14 | { name="Giacomo Cappon", email="cappongiacomo@gmail.com" },
15 | ]
16 | description = "ReplayBG is a digital twin-based methodology to assess new strategies for type 1 diabetes management."
17 | readme = "README.md"
18 | requires-python = ">=3.11"
19 | classifiers = [
20 | "Programming Language :: Python :: 3",
21 | "License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
22 | "Operating System :: OS Independent",
23 | "Intended Audience :: Science/Research",
24 | "Topic :: Scientific/Engineering",
25 | ]
26 | dynamic = ["dependencies"]
27 | [tool.setuptools.dynamic]
28 | dependencies = {file = ["requirements.txt"]}
29 | optional-dependencies = {dev = { file = ["requirements-dev.txt"] }}
30 |
31 | [project.urls]
32 | "Homepage" = "https://github.com/gcappon/py_replay_bg"
33 | "Bug Tracker" = "https://github.com/gcappon/py_replay_bg/issues"
34 |
35 | [tool.setuptools.packages.find]
36 | include = ["py_replay_bg*"]
37 | exclude = ["venv*"]
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | build==1.2.1
2 | celerite==0.4.2
3 | certifi==2024.2.2
4 | charset-normalizer==3.3.2
5 | contourpy==1.2.0
6 | corner==2.2.2
7 | coverage==7.4.4
8 | cycler==0.12.1
9 | docutils==0.20.1
10 | emcee==3.1.4
11 | et-xmlfile==1.1.0
12 | fonttools==4.50.0
13 | idna==3.6
14 | importlib_metadata==7.1.0
15 | iniconfig==2.0.0
16 | jaraco.classes==3.3.1
17 | jaraco.context==4.3.0
18 | jaraco.functools==4.0.0
19 | joblib==1.3.2
20 | keyring==25.0.0
21 | kiwisolver==1.4.5
22 | llvmlite==0.42.0
23 | markdown-it-py==3.0.0
24 | matplotlib==3.8.3
25 | mdurl==0.1.2
26 | more-itertools==10.2.0
27 | mpi4py==3.1.5
28 | nh3==0.2.17
29 | numba==0.59.1
30 | numpy==1.26.4
31 | openpyxl==3.1.2
32 | packaging==24.0
33 | pandas==2.2.1
34 | patsy==0.5.6
35 | pillow==10.2.0
36 | pkginfo==1.10.0
37 | plotly==5.20.0
38 | pluggy==1.4.0
39 | py-agata==0.0.8
40 | Pygments==2.17.2
41 | pyparsing==3.1.2
42 | pyproject_hooks==1.0.0
43 | pytest==8.1.1
44 | pytest-cov==5.0.0
45 | python-dateutil==2.9.0.post0
46 | pytz==2024.1
47 | readme_renderer==43.0
48 | requests==2.31.0
49 | requests-toolbelt==1.0.0
50 | rfc3986==2.0.0
51 | rich==13.7.1
52 | scikit-learn==1.4.1.post1
53 | scipy==1.12.0
54 | seaborn==0.13.2
55 | six==1.16.0
56 | statsmodels==0.14.1
57 | tenacity==8.2.3
58 | threadpoolctl==3.4.0
59 | tqdm==4.66.2
60 | twine==5.0.0
61 | tzdata==2024.1
62 | urllib3==2.2.1
63 | zipp==3.18.1
64 |
--------------------------------------------------------------------------------
/tsconfig.json:
--------------------------------------------------------------------------------
1 | {
2 | "compilerOptions": {
3 | "module": "NodeNext",
4 | "moduleResolution": "NodeNext",
5 | "target": "ES2022"
6 | },
7 | "include": [
8 | "docs/.vuepress/**/*.ts",
9 | "docs/.vuepress/**/*.vue"
10 | ],
11 | "exclude": [
12 | "node_modules"
13 | ]
14 | }
15 |
--------------------------------------------------------------------------------