 3 |
3 |
2 |  3 |
3 |
 39 |
40 | * `!cov ghelp` Help for graphical commands
41 |
42 | *
39 |
40 | * `!cov ghelp` Help for graphical commands
41 |
42 | * 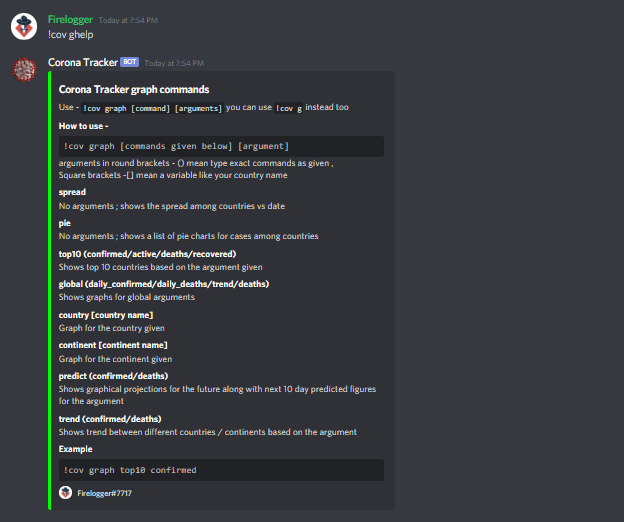 43 |
44 | * you can find dozens of other commands and features using `!cov help`
45 |
46 |
47 | # Installation
48 |
49 | ### Discord Bot
50 | 1. Fork and clone the git repository
51 | 2. In the new folder you will find a `example.env` file, duplicate it and rename it to `.env` only.
52 | 3. in the new `.env` file, change `DISCORD_TOKEN` to to your bot token
53 | 4. Cd to the directory and `pip install -r requirements.txt`
54 | 5. Run `bot.py`
55 | 6. You can now see the bot connected to the server
56 |
57 | ### Visualisation [Machine Learning] part
58 | 1. Fork and clone the git repository
59 | 2. Cd to the directory and `pip install -r requirements.txt`
60 | 3. Run `python nb_og.py` and all the files should be created in the same directory
61 | #### Warning: It is resource intensive and can take 15-20 minutes to complete, it generates over 100 visuals and html files
62 |
63 | ### Support or Contact
64 |
65 | Discord: Firelogger#7717
66 |
--------------------------------------------------------------------------------
/Bot/bot.py:
--------------------------------------------------------------------------------
1 | '''
2 | This project was made by https://github.com/himanshu2406 , incase of cloning / modifying or release of any bot based on this source code,
3 | You are obligated to give credits to the original author.
4 |
5 | Original repo: https://github.com/himanshu2406/Corona-Tracker-Bot
6 |
7 | Original Bot Support Server: https://discord.gg/kdj6DMr
8 | '''
9 |
10 | import uuid
11 | from dotenv import load_dotenv
12 | import discord
13 | from discord.ext import commands
14 | import os
15 | import diseaseapi
16 | from helper_functions import *
17 |
18 |
19 | class CovidCommands(commands.Cog):
20 | def __init__(self, client):
21 | self.client = client
22 | self.api_client = diseaseapi.Client().covid19
23 |
24 | @commands.command(help="Restarts the bot, requires admin permission")
25 | @commands.has_role('admin')
26 | async def restart(self, ctx):
27 | if ctx.message.author.id == 550965528936710154:
28 | await ctx.send('Restarting...')
29 | print('Restarting...')
30 | try:
31 | await self.client.logout()
32 | finally:
33 | os.system("py -3 bot.py")
34 |
35 |
36 | @commands.command(help="Shuts Down The bot, Requires admin permission")
37 | @commands.has_role('admin')
38 | async def shutdown(self, ctx):
39 | if ctx.message.author.id == 550965528936710154:
40 | await ctx.send('Bot is Shut Down')
41 | print('Shut Down Command Received')
42 | await self.client.logout()
43 | return
44 |
45 |
46 | @commands.command()
47 | @commands.has_role('admin')
48 | async def botservers(self, ctx):
49 | if ctx.message.author.id == 550965528936710154:
50 | list_guilds = []
51 | total_mem = 0
52 | await self.client.change_presence(activity=discord.Activity(type=discord.ActivityType.listening, name=" a !cov help in " + str(len(self.client.guilds)) + " server(s)"))
53 | await ctx.send("I'm in " + str(len(self.client.guilds)) + " servers")
54 | for guild in self.client.guilds:
55 | total_mem += len(guild.members)
56 | list_guilds.append(guild.name + ' : ' + str(guild.id))
57 | list_guilds.append('Total members: ' + str(total_mem))
58 |
59 | # await ctx.send(list_guilds)
60 | print(list_guilds)
61 | await ctx.send('Total members: ' + str(total_mem))
62 | return
63 |
64 |
65 | @commands.command()
66 | @commands.has_role('admin')
67 | async def leave(self, ctx, id):
68 | if ctx.message.author.id == 550965528936710154:
69 | to_leave = self.client.get_guild(int(id))
70 | await ctx.send('Leaving ' + str(to_leave))
71 | try:
72 | await to_leave.leave()
73 | await ctx.send('Left ' + str(to_leave))
74 | except:
75 | await ctx.send('Failed leaving ' + str(to_leave))
76 | return
77 |
78 |
79 | @commands.group(name='graph', aliases=['g'])
80 | async def graph(self, ctx):
81 | if ctx.invoked_subcommand is None:
82 | await ctx.invoke(self.ghelp)
83 |
84 |
85 | @graph.command(name='top10')
86 | async def graphtop10(self, ctx, arg2):
87 | if arg2 not in ['confirmed', 'active', 'deaths', 'recovered']:
88 | return
89 |
90 | newcountry = arg2.lower()
91 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/top_10_countries_' + \
92 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
93 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
94 | description="Graph for Top 10 " + newcountry + 'Cases', color=0x00ff00)
95 | embedVar.set_footer(
96 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
97 | embedVar.set_image(url=urltobe)
98 | await ctx.send(embed=embedVar)
99 |
100 |
101 | @graph.command(name='country')
102 | async def graphcountry(self, ctx, arg2='all'):
103 | if arg2 == 'all':
104 | urltobe = 'https://github.com/resoucesforcorona/Resources/blob/master/All_countries.png?raw=true'
105 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
106 | description="Graph for all countries", color=0x00ff00)
107 |
108 | else:
109 | newcountry = (arg2.lower()).capitalize()
110 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/' + \
111 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
112 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
113 | description="Graph for: " + newcountry, color=0x00ff00)
114 |
115 | embedVar.set_footer(
116 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
117 | embedVar.set_image(url=urltobe)
118 | await ctx.send(embed=embedVar)
119 |
120 |
121 | @graph.command(name='continent')
122 | async def graphcontinent(self, ctx, arg2='all'):
123 | if arg2 == 'all':
124 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/All_continents.png'
125 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
126 | description="Graph for all continents", color=0x00ff00)
127 |
128 | else:
129 | newcountry = (arg2.lower()).capitalize()
130 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/' + \
131 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
132 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
133 | description="Graph for: " + newcountry, color=0x00ff00)
134 |
135 | embedVar.set_footer(
136 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
137 | embedVar.set_image(url=urltobe)
138 | await ctx.send(embed=embedVar)
139 |
140 |
141 | @graph.command(name='pie')
142 | async def graphpie(self, ctx):
143 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/all_countrywise_pie.png'
144 | embedVar = discord.Embed(
145 | title="Corona Tracker", url='https://anondoser.xyz', description="Graph for Pie", color=0x00ff00)
146 | embedVar.set_footer(
147 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
148 | embedVar.set_image(url=urltobe)
149 | await ctx.send(embed=embedVar)
150 |
151 |
152 | @graph.command(name='predict')
153 | async def predict(self, ctx, arg2):
154 | newcountry = (arg2.lower())
155 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/world_prediction_curve_' + \
156 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
157 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
158 | description="Graph for global prediction curve for " + newcountry, color=0x00ff00)
159 | embedVar.set_footer(
160 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
161 | embedVar.set_image(url=urltobe)
162 | await ctx.send(embed=embedVar)
163 |
164 |
165 | @graph.command(name='global')
166 | async def graphglobal(self, ctx, arg2):
167 | if arg2 not in ['daily_confirmed', 'daily_deaths', 'trend', 'deaths']:
168 | return
169 |
170 | if arg2 == 'daily_confirmed':
171 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/daily_confirmed_cases_global.png'
172 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
173 | description="Graph for global daily confirmed", color=0x00ff00)
174 |
175 | elif arg2 == 'daily_deaths':
176 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/daily_deaths_cases_global.png'
177 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
178 | description="Graph for global daily deaths", color=0x00ff00)
179 |
180 | elif arg2 == 'trend':
181 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/world_trend_confirmed_cases.png'
182 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
183 | description="Graph for global trend", color=0x00ff00)
184 |
185 | else:
186 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/worldwide_cases_deaths.png'
187 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
188 | description="Graph for global total deaths", color=0x00ff00)
189 |
190 | embedVar.set_footer(
191 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
192 | embedVar.set_image(url=urltobe)
193 | await ctx.send(embed=embedVar)
194 |
195 |
196 | @graph.command(name='trend')
197 | async def graphtrend(self, ctx, arg2):
198 | if arg2 not in ['confirmed', 'deaths']:
199 | return
200 |
201 | if arg2 == 'confirmed':
202 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/trend_comparison_continents_confirmed.png'
203 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
204 | description="Graph for trend comparison b/w continents [confirmed]", color=0x00ff00)
205 |
206 | else:
207 | newcountry = (arg2.lower()).capitalize()
208 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/trend_comparison_countries_deaths.png'
209 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
210 | description="Graph for trend comparison b/w countries [deaths]" + newcountry, color=0x00ff00)
211 |
212 | embedVar.set_footer(
213 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
214 | embedVar.set_image(url=urltobe)
215 | await ctx.send(embed=embedVar)
216 |
217 |
218 | @graph.command(name='spread')
219 | async def graphspread(self, ctx):
220 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/countries_vs_date_spread.png'
221 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
222 | description="Graph for number of countries infected vs date", color=0x00ff00)
223 | embedVar.set_footer(
224 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
225 | embedVar.set_image(url=urltobe)
226 | await ctx.send(embed=embedVar)
227 |
228 |
229 | @commands.command()
230 | async def help(self, ctx):
231 | embedVar = discord.Embed(title="Corona Tracker Help Panel",
232 | description="Here's some help for you :heart:", color=0x00ff00)
233 | embedVar.add_field(
234 | name="!cov graph [commands]", value="Consists of graphical predictions, analysis , statistics and more see below for command", inline=False)
235 | embedVar.add_field(
236 | name="!cov g [commands]", value="Same as above, use !cov ghelp for list of commands", inline=False)
237 | embedVar.add_field(
238 | name="!cov ghelp", value="Help and commands list for `!cov graph`", inline=False)
239 | embedVar.add_field(
240 | name="!cov all", value="Shows global Covid-19 statistics", inline=False)
241 | embedVar.add_field(name="!cov interactive", value="Sends the best live interactive maps \n See how the Covid spread on the world map from the very start \n See live status of Covid on the world map \n See Mortality Progression from the very beginning", inline=False)
242 | embedVar.add_field(name="!cov country {your country} {full/micro}",
243 | value="Shows you a particular country's stats (Optional- use 'micro' for a simplified report)", inline=False)
244 | embedVar.add_field(
245 | name="!cov help", value="Shows you this message", inline=False)
246 | embedVar.add_field(
247 | name="!cov invite", value="Sends you the links to invite the bot to your own server & the official bot server", inline=False)
248 | embedVar.add_field(
249 | name="github", value="https://github.com/himanshu2406/Corona-Tracker-Bot", inline=False)
250 | embedVar.add_field(
251 | name="tip :heart: ", value="Buy me a Coffee [sends addresses for tipping]", inline=False)
252 | embedVar.add_field(name="Dev Contact",
253 | value="Firelogger#7717", inline=False)
254 | await ctx.send(embed=embedVar)
255 |
256 |
257 | @commands.command()
258 | async def invite(self, ctx):
259 | embedVar = discord.Embed(title="Invite me", url='https://discord.com/api/oauth2/authorize?client_id=731855425145798668&permissions=121856&scope=bot',
260 | description="Click the title to invite to your own server", color=0x00ff00)
261 | embedVar.set_footer(text="Firelogger#7717",
262 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
263 | await ctx.send(embed=embedVar)
264 |
265 |
266 | @commands.command()
267 | async def tip(self, ctx):
268 | embedVar = discord.Embed(title="Tip :heart:", url='https://www.buymeacoffee.com/anondoser/shop',
269 | description="Donate for improving the services and help running the bot", color=0x00ff00)
270 | embedVar.add_field(
271 | name="Btc address", value="```37btgSzgWdywmSPeBN5rUH8W5G9EYJoRoA```", inline=False)
272 | embedVar.add_field(
273 | name="Paypal", value="```https://www.paypal.me/firelogger```", inline=False)
274 | embedVar.add_field(
275 | name="For more", value="For more methods please dm me", inline=False)
276 | embedVar.set_footer(text="Firelogger#7717",
277 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
278 | await ctx.send(embed=embedVar)
279 |
280 |
281 | @commands.command()
282 | async def interactive(self, ctx):
283 | embedVar = discord.Embed(title="Interactive and playable Maps and statistics",
284 | description="Play with these live maps , statistics and visuals", color=0x00ff00)
285 | embedVar.add_field(name="World Map Live Progression",
286 | value="See how COVID spread from the very beginning on the world map, seekable and playable", inline=False)
287 | embedVar.add_field(
288 | name="Link", value="https://corona.anondoser.xyz/worldmap_progression.html", inline=False)
289 | embedVar.add_field(name="World Map static interactive ",
290 | value="see the current spread of Covid-19 on the world map", inline=False)
291 | embedVar.add_field(
292 | name="Link", value="https://corona.anondoser.xyz/worldmap_cases_interactive.html", inline=False)
293 | embedVar.add_field(name="Mortality rate Live progression",
294 | value="A beautiful playable and seekable representation of the mortality rate progression from the beginning , see those balls bounce", inline=False)
295 | embedVar.add_field(
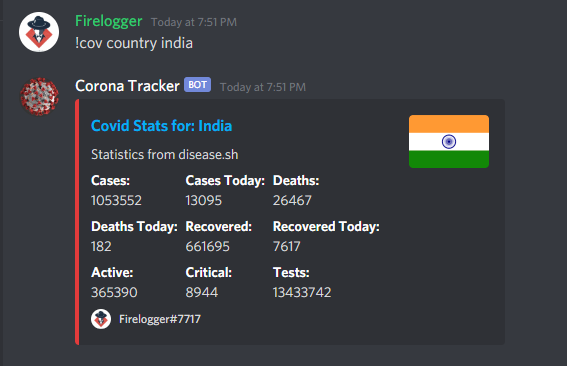
296 | name="Link", value="https://corona.anondoser.xyz/mortalityrate_progression.html", inline=False)
297 | embedVar.set_footer(text="Firelogger#7717",
298 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
299 | await ctx.send(embed=embedVar)
300 |
301 |
302 | @commands.command()
303 | async def ghelp(self, ctx):
304 | embedVar = discord.Embed(title="Corona Tracker graph commands ",
305 | description="Use - `!cov graph [command] [arguments]` you can use `!cov g` instead too", color=0x00ff00)
306 | embedVar.add_field(name="How to use - ",
307 | value="```!cov graph [commands given below] [argument]``` arguments in round brackets - () mean type exact commands as given , \n Square brackets -[] mean a variable like your country name", inline=False)
308 | embedVar.add_field(
309 | name="spread", value="No arguments ; shows the spread among countries vs date", inline=False)
310 | embedVar.add_field(
311 | name="pie", value="No arguments ; shows a list of pie charts for cases among countries", inline=False)
312 | embedVar.add_field(name="top10 (confirmed/active/deaths/recovered)",
313 | value="Shows top 10 countries based on the argument given", inline=False)
314 | embedVar.add_field(name="global (daily_confirmed/daily_deaths/trend/deaths)",
315 | value="Shows graphs for global arguments", inline=False)
316 | embedVar.add_field(
317 | name="country [country name]", value="Graph for the country given", inline=False)
318 | embedVar.add_field(name="continent [continent name]",
319 | value="Graph for the continent given", inline=False)
320 | embedVar.add_field(name="predict (confirmed/deaths)",
321 | value="Shows graphical projections for the future along with next 10 day predicted figures for the argument", inline=False)
322 | embedVar.add_field(name="trend (confirmed/deaths)",
323 | value="Shows trend between different countries / continents based on the argument", inline=False)
324 | #embedVar.add_field(name="github", value="https://github.com/himanshu2406/Corona-Tracker", inline=False)
325 | #embedVar.add_field(name="tip :heart: ", value="Buy me a Coffee [sends addresses for tipping]", inline=False)
326 | embedVar.add_field(
327 | name="Example", value="```!cov graph top10 confirmed```", inline=False)
328 | embedVar.set_footer(text="Firelogger#7717",
329 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
330 | await ctx.send(embed=embedVar)
331 |
332 |
333 | @commands.command(aliases=['c'])
334 | async def country(self, ctx, args, complete='full'):
335 | if complete not in ['full', 'micro']:
336 | return
337 |
338 | try:
339 | i = await self.api_client.country(args)
340 | yday_c = await self.api_client.country(i.name, yesterday=True)
341 | except diseaseapi.NotFound:
342 | embedVar = discord.Embed(title="Invalid Country: " + args.capitalize(), description="Error, the country doesn't exist in the database", color=0xe33b3b, url='https://anondoser.xyz')
343 | return await ctx.send(embed=embedVar)
344 |
345 | embedVar = discord.Embed(description="Statistics from disease.py",
346 | color=0xe33b3b, url='https://anondoser.xyz')
347 |
348 | yday_c = await self.api_client.country(i.name, yesterday=True)
349 |
350 | cases_diff = cleaned_diff(i.cases, yday_c.cases)
351 | death_diff = cleaned_diff(i.deaths, yday_c.deaths)
352 | re_diff = cleaned_diff(i.recoveries, yday_c.recoveries)
353 | active_diff = cleaned_diff(i.active, yday_c.active)
354 | crit_diff = cleaned_diff(i.critical, yday_c.critical)
355 | test_diff = cleaned_diff(i.tests, yday_c.tests)
356 |
357 | embedVar.add_field(name='Cases:', value=check_na(i.cases)+cases_diff, inline=True)
358 | embedVar.add_field(name='Cases Today:', value=check_na(i.today.cases), inline=True)
359 | embedVar.add_field(name='Deaths:', value=check_na(i.deaths)+death_diff, inline=True)
360 | embedVar.add_field(name='Deaths Today:', value=check_na(i.today.deaths), inline=True)
361 | embedVar.add_field(name='Recovered:', value=check_na(i.recoveries)+re_diff, inline=True)
362 | embedVar.add_field(name='Recovered Today:', value=check_na(i.today.recoveries), inline=True)
363 | embedVar.add_field(name='Active:', value=check_na(i.active)+active_diff, inline=True)
364 | embedVar.add_field(name='Critical:', value=check_na(i.critical)+crit_diff, inline=True)
365 | embedVar.add_field(name='Tests:', value=check_na(i.tests)+test_diff, inline=True)
366 | urltobe = str(i.info.flag)
367 |
368 | if complete == 'full':
369 | embedVar.add_field(name ='Cases per Million', value= i.per_million.cases, inline=True)
370 | embedVar.add_field(name ='Active per Million', value= i.per_million.active, inline=True)
371 | embedVar.add_field(name ='Recoveries per Million', value= i.per_million.recoveries, inline=True)
372 | embedVar.add_field(name ='Tests per Million', value= i.per_million.tests, inline=True)
373 | embedVar.add_field(name ='Deaths per Million', value= i.per_million.deaths, inline=True)
374 | embedVar.add_field(name ='Critical per Million', value= i.per_million.critical, inline=True)
375 | embedVar.add_field(name ='One Case per Person', value= i.per_people.case, inline=True)
376 | embedVar.add_field(name ='One Death per Person', value= i.per_people.death, inline=True)
377 | embedVar.add_field(name ='One Test per Person', value= i.per_people.test, inline=True)
378 | embedVar.add_field(name ='Updated', value= i.updated, inline=True)
379 |
380 | embedVar.title = "Covid Stats for: " + i.name
381 | embedVar.set_thumbnail(url=urltobe)
382 | embedVar.set_footer(
383 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
384 | await ctx.send(embed=embedVar)
385 |
386 |
387 | @commands.command()
388 | async def all(self, ctx):
389 | response = await self.api_client.all()
390 | embedVar = discord.Embed(title="Covid Worldwide Stats",
391 | description="Statistics from disease.py", color=0xe33b3b, url='https://anondoser.xyz')
392 |
393 |
394 | yday_c = await self.api_client.all(yesterday=True)
395 | cases_diff = cleaned_diff(response.cases, yday_c.cases)
396 | death_diff = cleaned_diff(response.deaths, yday_c.deaths)
397 | re_diff = cleaned_diff(response.recoveries, yday_c.recoveries)
398 | active_diff = cleaned_diff(response.active, yday_c.active)
399 | crit_diff = cleaned_diff(response.critical, yday_c.critical)
400 | test_diff = cleaned_diff(response.tests, yday_c.tests)
401 | embedVar.add_field(name='Cases:', value=check_na(response.cases)+cases_diff, inline=True)
402 | embedVar.add_field(name='Cases Today:', value=check_na(response.today.cases), inline=True)
403 | embedVar.add_field(name='Deaths:', value=check_na(response.deaths)+death_diff, inline=True)
404 | embedVar.add_field(name='Deaths Today:', value=check_na(response.today.deaths), inline=True)
405 | embedVar.add_field(name='Recovered:', value=check_na(response.recoveries)+re_diff, inline=True)
406 | embedVar.add_field(name='Recovered Today:', value=check_na(response.today.recoveries), inline=True)
407 | embedVar.add_field(name='Active:', value=check_na(response.active)+active_diff, inline=True)
408 | embedVar.add_field(name='Critical:', value=check_na(response.critical)+crit_diff, inline=True)
409 | embedVar.add_field(name='Tests:', value=check_na(response.tests)+test_diff, inline=True)
410 | embedVar.add_field(name ='Cases per Million', value= response.per_million.cases, inline=True)
411 | embedVar.add_field(name ='Active per Million', value= response.per_million.active, inline=True)
412 | embedVar.add_field(name ='Recoveries per Million', value= response.per_million.recoveries, inline=True)
413 | embedVar.add_field(name ='Tests per Million', value= response.per_million.tests, inline=True)
414 | embedVar.add_field(name ='Deaths per Million', value= response.per_million.deaths, inline=True)
415 | embedVar.add_field(name ='Critical per Million', value= response.per_million.critical, inline=True)
416 | embedVar.add_field(name ='One Case per Person', value= response.per_people.case, inline=True)
417 | embedVar.add_field(name ='One Death per Person', value= response.per_people.death, inline=True)
418 | embedVar.add_field(name ='One Test per Person', value= response.per_people.test, inline=True)
419 | embedVar.add_field(name ='Updated', value= response.updated, inline=True)
420 |
421 | embedVar.set_footer(text="Firelogger#7717",icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
422 | await ctx.send(embed=embedVar)
423 |
424 |
425 | def setup(client):
426 | client.add_cog(CovidCommands(client))
--------------------------------------------------------------------------------
/Visualisations and Ai predictions/nb_og.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 | import matplotlib.pyplot as plt
4 | from matplotlib import ticker
5 | import pycountry_convert as pc
6 | import folium
7 | import branca
8 | from datetime import datetime, timedelta,date
9 | from scipy.interpolate import make_interp_spline, BSpline
10 | import plotly.express as px
11 | import json, requests
12 | import calmap
13 | import urllib
14 | import base64
15 | from base64 import b64encode
16 | from keras.layers import Input, Dense, Activation, LeakyReLU, Dropout
17 | from keras import models
18 | from keras.optimizers import RMSprop, Adam
19 |
20 | import warnings
21 | warnings.filterwarnings('ignore')
22 |
23 |
24 |
25 | # Retriving Dataset
26 |
27 | while True:
28 | try:
29 | df_table = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/web-data/data/cases_time.csv",parse_dates=['Last_Update'])
30 | df_confirmed = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv')
31 | df_deaths = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv')
32 |
33 | # Depricated
34 | # df_recovered = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Recovered.csv')
35 | df_covid19 = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/web-data/data/cases_country.csv")
36 | except:
37 | continue
38 | else:
39 | break
40 |
41 |
42 | # new dataset
43 | df_covid19 = df_covid19.drop(["People_Tested","People_Hospitalized","UID","ISO3","Mortality_Rate"],axis =1)
44 | df_covid19.head(2)
45 | df_confirmed.head(2)
46 |
47 | df_confirmed = df_confirmed.rename(columns={"Province/State":"state","Country/Region": "country"})
48 | df_deaths = df_deaths.rename(columns={"Province/State":"state","Country/Region": "country"})
49 | df_covid19 = df_covid19.rename(columns={"Country_Region": "country"})
50 | df_covid19["Active"] = df_covid19["Confirmed"]-df_covid19["Recovered"]-df_covid19["Deaths"]
51 | # df_recovered = df_recovered.rename(columns={"Province/State":"state","Country/Region": "country"})
52 | df_confirmed.loc[df_confirmed['country'] == "US", "country"] = "USA"
53 | df_deaths.loc[df_deaths['country'] == "US", "country"] = "USA"
54 | df_covid19.loc[df_covid19['country'] == "US", "country"] = "USA"
55 | df_table.loc[df_table['Country_Region'] == "US", "Country_Region"] = "USA"
56 | # df_recovered.loc[df_recovered['country'] == "US", "country"] = "USA"
57 |
58 |
59 | df_confirmed.loc[df_confirmed['country'] == 'Korea, South', "country"] = 'South Korea'
60 | df_deaths.loc[df_deaths['country'] == 'Korea, South', "country"] = 'South Korea'
61 | df_covid19.loc[df_covid19['country'] == "Korea, South", "country"] = "South Korea"
62 | df_table.loc[df_table['Country_Region'] == "Korea, South", "Country_Region"] = "South Korea"
63 | # df_recovered.loc[df_recovered['country'] == 'Korea, South', "country"] = 'South Korea'
64 |
65 | df_confirmed.loc[df_confirmed['country'] == 'Taiwan*', "country"] = 'Taiwan'
66 | df_deaths.loc[df_deaths['country'] == 'Taiwan*', "country"] = 'Taiwan'
67 | df_covid19.loc[df_covid19['country'] == "Taiwan*", "country"] = "Taiwan"
68 | df_table.loc[df_table['Country_Region'] == "Taiwan*", "Country_Region"] = "Taiwan"
69 | # df_recovered.loc[df_recovered['country'] == 'Taiwan*', "country"] = 'Taiwan'
70 |
71 | df_confirmed.loc[df_confirmed['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
72 | df_deaths.loc[df_deaths['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
73 | df_covid19.loc[df_covid19['country'] == "Congo (Kinshasa)", "country"] = "Democratic Republic of the Congo"
74 | df_table.loc[df_table['Country_Region'] == "Congo (Kinshasa)", "Country_Region"] = "Democratic Republic of the Congo"
75 | # df_recovered.loc[df_recovered['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
76 |
77 | df_confirmed.loc[df_confirmed['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
78 | df_deaths.loc[df_deaths['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
79 | df_covid19.loc[df_covid19['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
80 | df_table.loc[df_table['Country_Region'] == "Cote d'Ivoire", "Country_Region"] = "Côte d'Ivoire"
81 | # df_recovered.loc[df_recovered['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
82 |
83 | df_confirmed.loc[df_confirmed['country'] == "Reunion", "country"] = "Réunion"
84 | df_deaths.loc[df_deaths['country'] == "Reunion", "country"] = "Réunion"
85 | df_covid19.loc[df_covid19['country'] == "Reunion", "country"] = "Réunion"
86 | df_table.loc[df_table['Country_Region'] == "Reunion", "Country_Region"] = "Réunion"
87 | # df_recovered.loc[df_recovered['country'] == "Reunion", "country"] = "Réunion"
88 |
89 | df_confirmed.loc[df_confirmed['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
90 | df_deaths.loc[df_deaths['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
91 | df_covid19.loc[df_covid19['country'] == "Congo (Brazzaville)", "country"] = "Republic of the Congo"
92 | df_table.loc[df_table['Country_Region'] == "Congo (Brazzaville)", "Country_Region"] = "Republic of the Congo"
93 | # df_recovered.loc[df_recovered['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
94 |
95 | df_confirmed.loc[df_confirmed['country'] == 'Bahamas, The', "country"] = 'Bahamas'
96 | df_deaths.loc[df_deaths['country'] == 'Bahamas, The', "country"] = 'Bahamas'
97 | df_covid19.loc[df_covid19['country'] == "Bahamas, The", "country"] = "Bahamas"
98 | df_table.loc[df_table['Country_Region'] == "Bahamas, The", "Country_Region"] = "Bahamas"
99 | # df_recovered.loc[df_recovered['country'] == 'Bahamas, The', "country"] = 'Bahamas'
100 |
101 | df_confirmed.loc[df_confirmed['country'] == 'Gambia, The', "country"] = 'Gambia'
102 | df_deaths.loc[df_deaths['country'] == 'Gambia, The', "country"] = 'Gambia'
103 | df_covid19.loc[df_covid19['country'] == "Gambia, The", "country"] = "Gambia"
104 | df_table.loc[df_table['Country_Region'] == "Gambia", "Country_Region"] = "Gambia"
105 | # df_recovered.loc[df_recovered['country'] == 'Gambia, The', "country"] = 'Gambia'
106 |
107 | # getting all countries
108 | countries = np.asarray(df_confirmed["country"])
109 | countries1 = np.asarray(df_covid19["country"])
110 | # Continent_code to Continent_names
111 | continents = {
112 | 'NA': 'North America',
113 | 'SA': 'South America',
114 | 'AS': 'Asia',
115 | 'OC': 'Australia',
116 | 'AF': 'Africa',
117 | 'EU' : 'Europe',
118 | 'na' : 'Others'
119 | }
120 |
121 | # Defininng Function for getting continent code for country.
122 | def country_to_continent_code(country):
123 | try:
124 | return pc.country_alpha2_to_continent_code(pc.country_name_to_country_alpha2(country))
125 | except :
126 | return 'na'
127 |
128 | #Collecting Continent Information
129 | df_confirmed.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]])
130 | df_deaths.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]])
131 | df_covid19.insert(1,"continent", [continents[country_to_continent_code(country)] for country in countries1[:]])
132 | df_table.insert(1,"continent", [continents[country_to_continent_code(country)] for country in df_table["Country_Region"].values])
133 | # df_recovered.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]] )
134 |
135 | df_table = df_table[df_table["continent"] != "Others"]
136 |
137 | df_deaths[df_deaths["continent" ]== 'Others']
138 |
139 | df_confirmed = df_confirmed.replace(np.nan, '', regex=True)
140 | df_deaths = df_deaths.replace(np.nan, '', regex=True)
141 | # df_recovered = df_recovered.replace(np.nan, '', regex=True)
142 | # df_active = df_active.replace(np.nan, '', regex=True)
143 |
144 | """# FUNCTIONS BELOW HERE"""
145 |
146 | def plot_params(ax,axis_label= None, plt_title = None,label_size=15, axis_fsize = 15, title_fsize = 20, scale = 'linear' ):
147 | # Tick-Parameters
148 | ax.xaxis.set_minor_locator(ticker.AutoMinorLocator())
149 | ax.yaxis.set_minor_locator(ticker.AutoMinorLocator())
150 | ax.tick_params(which='both', width=1,labelsize=label_size)
151 | ax.tick_params(which='major', length=6)
152 | ax.tick_params(which='minor', length=3, color='0.8')
153 |
154 | # Grid
155 | plt.grid(lw = 1, ls = '-', c = "0.7", which = 'major')
156 | plt.grid(lw = 1, ls = '-', c = "0.9", which = 'minor')
157 |
158 | # Plot Title
159 | plt.title( plt_title,{'fontsize':title_fsize})
160 |
161 | # Yaxis sacle
162 | plt.yscale(scale)
163 | plt.minorticks_on()
164 | # Plot Axes Labels
165 | xl = plt.xlabel(axis_label[0],fontsize = axis_fsize)
166 | yl = plt.ylabel(axis_label[1],fontsize = axis_fsize)
167 |
168 | def visualize_covid_cases(confirmed, deaths, continent=None , country = None , state = None, period = None, figure = None, scale = "linear"):
169 | x = 0
170 | if figure == None:
171 | f = plt.figure(figsize=(10,10))
172 | # Sub plot
173 | ax = f.add_subplot(111)
174 | else :
175 | f = figure[0]
176 | # Sub plot
177 | ax = f.add_subplot(figure[1],figure[2],figure[3])
178 | ax.set_axisbelow(True)
179 | plt.tight_layout(pad=10, w_pad=5, h_pad=5)
180 |
181 | stats = [confirmed, deaths]
182 | label = ["Confirmed", "Deaths"]
183 |
184 | if continent != None:
185 | params = ["continent",continent]
186 | elif country != None:
187 | params = ["country",country]
188 | else:
189 | params = ["All", "All"]
190 | color = ["darkcyan","crimson"]
191 | marker_style = dict(linewidth=3, linestyle='-', marker='o',markersize=4, markerfacecolor='#ffffff')
192 | for i,stat in enumerate(stats):
193 | if params[1] == "All" :
194 | cases = np.sum(np.asarray(stat.iloc[:,5:]),axis = 0)[x:]
195 | else :
196 | cases = np.sum(np.asarray(stat[stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
197 | date = np.arange(1,cases.shape[0]+1)[x:]
198 | plt.plot(date,cases,label = label[i]+" (Total : "+str(cases[-1])+")",color=color[i],**marker_style)
199 | plt.fill_between(date,cases,color=color[i],alpha=0.3)
200 |
201 | if params[1] == "All" :
202 | Total_confirmed = np.sum(np.asarray(stats[0].iloc[:,5:]),axis = 0)[x:]
203 | Total_deaths = np.sum(np.asarray(stats[1].iloc[:,5:]),axis = 0)[x:]
204 | else :
205 | Total_confirmed = np.sum(np.asarray(stats[0][stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
206 | Total_deaths = np.sum(np.asarray(stats[1][stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
207 |
208 | text = "From "+stats[0].columns[5]+" to "+stats[0].columns[-1]+"\n"
209 | text += "Mortality rate : "+ str(int(Total_deaths[-1]/(Total_confirmed[-1])*10000)/100)+"\n"
210 | text += "Last 5 Days:\n"
211 | text += "Confirmed : " + str(Total_confirmed[-1] - Total_confirmed[-6])+"\n"
212 | text += "Deaths : " + str(Total_deaths[-1] - Total_deaths[-6])+"\n"
213 | text += "Last 24 Hours:\n"
214 | text += "Confirmed : " + str(Total_confirmed[-1] - Total_confirmed[-2])+"\n"
215 | text += "Deaths : " + str(Total_deaths[-1] - Total_deaths[-2])+"\n"
216 |
217 | plt.text(0.02, 0.78, text, fontsize=15, horizontalalignment='left', verticalalignment='top', transform=ax.transAxes,bbox=dict(facecolor='white', alpha=0.4))
218 |
219 | # Plot Axes Labels
220 | axis_label = ["Days ("+df_confirmed.columns[5]+" - "+df_confirmed.columns[-1]+")","No of Cases"]
221 |
222 | # Plot Parameters
223 | plot_params(ax,axis_label,scale = scale)
224 |
225 | # Plot Title
226 | if params[1] == "All" :
227 | plt.title("COVID-19 Cases World",{'fontsize':25})
228 | else:
229 | plt.title("COVID-19: "+params[1] ,{'fontsize':25})
230 |
231 | # Legend Location
232 | l = plt.legend(loc= "best",fontsize = 15)
233 |
234 | if figure == None:
235 | #plt.show()
236 | pass
237 |
238 | def get_total_cases(cases, country = "All"):
239 | if(country == "All") :
240 | return np.sum(np.asarray(cases.iloc[:,5:]),axis = 0)[-1]
241 | else :
242 | return np.sum(np.asarray(cases[cases["country"] == country].iloc[:,5:]),axis = 0)[-1]
243 |
244 | def get_mortality_rate(confirmed,deaths, continent = None, country = None):
245 | if continent != None:
246 | params = ["continent",continent]
247 | elif country != None:
248 | params = ["country",country]
249 | else :
250 | params = ["All", "All"]
251 |
252 | if params[1] == "All" :
253 | Total_confirmed = np.sum(np.asarray(confirmed.iloc[:,5:]),axis = 0)
254 | Total_deaths = np.sum(np.asarray(deaths.iloc[:,5:]),axis = 0)
255 | mortality_rate = np.round((Total_deaths/(Total_confirmed+1.01))*100,2)
256 | else :
257 | Total_confirmed = np.sum(np.asarray(confirmed[confirmed[params[0]] == params[1]].iloc[:,5:]),axis = 0)
258 | Total_deaths = np.sum(np.asarray(deaths[deaths[params[0]] == params[1]].iloc[:,5:]),axis = 0)
259 | mortality_rate = np.round((Total_deaths/(Total_confirmed+1.01))*100,2)
260 |
261 | return np.nan_to_num(mortality_rate)
262 | def dd(date1,date2):
263 | return (datetime.strptime(date1,'%m/%d/%y') - datetime.strptime(date2,'%m/%d/%y')).days
264 |
265 |
266 | out = ""#+"output/"
267 |
268 | df_countries_cases = df_covid19.copy().drop(['Lat','Long_','continent','Last_Update'],axis =1)
269 | df_countries_cases.index = df_countries_cases["country"]
270 | df_countries_cases = df_countries_cases.drop(['country'],axis=1)
271 |
272 | df_continents_cases = df_covid19.copy().drop(['Lat','Long_','country','Last_Update'],axis =1)
273 | df_continents_cases = df_continents_cases.groupby(["continent"]).sum()
274 |
275 | df_countries_cases.fillna(0,inplace=True)
276 | df_continents_cases.fillna(0,inplace=True)
277 |
278 |
279 |
280 | f = plt.figure(figsize=(10,5))
281 | f.add_subplot(111)
282 |
283 | plt.axes(axisbelow=True)
284 | plt.barh(df_countries_cases.sort_values('Confirmed')["Confirmed"].index[-10:],df_countries_cases.sort_values('Confirmed')["Confirmed"].values[-10:],color="darkcyan")
285 | plt.tick_params(size=5,labelsize = 13)
286 | plt.xlabel("Confirmed Cases",fontsize=18)
287 | plt.title("Top 10 Countries (Confirmed Cases)",fontsize=20)
288 | plt.grid(alpha=0.3)
289 | plt.savefig(out+'top_10_countries_confirmed.png')
290 |
291 |
292 | f = plt.figure(figsize=(10,5))
293 | f.add_subplot(111)
294 |
295 | plt.axes(axisbelow=True)
296 | plt.barh(df_countries_cases.sort_values('Deaths')["Deaths"].index[-10:],df_countries_cases.sort_values('Deaths')["Deaths"].values[-10:],color="crimson")
297 | plt.tick_params(size=5,labelsize = 13)
298 | plt.xlabel("Deaths Cases",fontsize=18)
299 | plt.title("Top 10 Countries (Deaths Cases)",fontsize=20)
300 | plt.grid(alpha=0.3,which='both')
301 | plt.savefig(out+'top_10_countries_deaths.png')
302 |
303 |
304 |
305 | f = plt.figure(figsize=(10,5))
306 | f.add_subplot(111)
307 |
308 | plt.axes(axisbelow=True)
309 | plt.barh(df_countries_cases.sort_values('Active')["Active"].index[-10:],df_countries_cases.sort_values('Active')["Active"].values[-10:],color="darkorange")
310 | plt.tick_params(size=5,labelsize = 13)
311 | plt.xlabel("Active Cases",fontsize=18)
312 | plt.title("Top 10 Countries (Active Cases)",fontsize=20)
313 | plt.grid(alpha=0.3,which='both')
314 | plt.savefig(out+'top_10_countries_active.png')
315 |
316 |
317 |
318 | f = plt.figure(figsize=(10,5))
319 | f.add_subplot(111)
320 |
321 | plt.axes(axisbelow=True)
322 | plt.barh(df_countries_cases.sort_values('Recovered')["Recovered"].index[-10:],df_countries_cases.sort_values('Recovered')["Recovered"].values[-10:],color="limegreen")
323 | plt.tick_params(size=5,labelsize = 13)
324 | plt.xlabel("Recovered Cases",fontsize=18)
325 | plt.title("Top 10 Countries (Recovered Cases)",fontsize=20)
326 | plt.grid(alpha=0.3,which='both')
327 | plt.savefig(out+'top_10_countries_recovered.png')
328 |
329 |
330 | world_map = folium.Map(location=[10,0], tiles="cartodbpositron", zoom_start=2,max_zoom=6,min_zoom=2)
331 | for i in range(0,len(df_confirmed)):
332 | folium.Circle(
333 | location=[df_confirmed.iloc[i]['Lat'], df_confirmed.iloc[i]['Long']],
334 | tooltip = "
43 |
44 | * you can find dozens of other commands and features using `!cov help`
45 |
46 |
47 | # Installation
48 |
49 | ### Discord Bot
50 | 1. Fork and clone the git repository
51 | 2. In the new folder you will find a `example.env` file, duplicate it and rename it to `.env` only.
52 | 3. in the new `.env` file, change `DISCORD_TOKEN` to to your bot token
53 | 4. Cd to the directory and `pip install -r requirements.txt`
54 | 5. Run `bot.py`
55 | 6. You can now see the bot connected to the server
56 |
57 | ### Visualisation [Machine Learning] part
58 | 1. Fork and clone the git repository
59 | 2. Cd to the directory and `pip install -r requirements.txt`
60 | 3. Run `python nb_og.py` and all the files should be created in the same directory
61 | #### Warning: It is resource intensive and can take 15-20 minutes to complete, it generates over 100 visuals and html files
62 |
63 | ### Support or Contact
64 |
65 | Discord: Firelogger#7717
66 |
--------------------------------------------------------------------------------
/Bot/bot.py:
--------------------------------------------------------------------------------
1 | '''
2 | This project was made by https://github.com/himanshu2406 , incase of cloning / modifying or release of any bot based on this source code,
3 | You are obligated to give credits to the original author.
4 |
5 | Original repo: https://github.com/himanshu2406/Corona-Tracker-Bot
6 |
7 | Original Bot Support Server: https://discord.gg/kdj6DMr
8 | '''
9 |
10 | import uuid
11 | from dotenv import load_dotenv
12 | import discord
13 | from discord.ext import commands
14 | import os
15 | import diseaseapi
16 | from helper_functions import *
17 |
18 |
19 | class CovidCommands(commands.Cog):
20 | def __init__(self, client):
21 | self.client = client
22 | self.api_client = diseaseapi.Client().covid19
23 |
24 | @commands.command(help="Restarts the bot, requires admin permission")
25 | @commands.has_role('admin')
26 | async def restart(self, ctx):
27 | if ctx.message.author.id == 550965528936710154:
28 | await ctx.send('Restarting...')
29 | print('Restarting...')
30 | try:
31 | await self.client.logout()
32 | finally:
33 | os.system("py -3 bot.py")
34 |
35 |
36 | @commands.command(help="Shuts Down The bot, Requires admin permission")
37 | @commands.has_role('admin')
38 | async def shutdown(self, ctx):
39 | if ctx.message.author.id == 550965528936710154:
40 | await ctx.send('Bot is Shut Down')
41 | print('Shut Down Command Received')
42 | await self.client.logout()
43 | return
44 |
45 |
46 | @commands.command()
47 | @commands.has_role('admin')
48 | async def botservers(self, ctx):
49 | if ctx.message.author.id == 550965528936710154:
50 | list_guilds = []
51 | total_mem = 0
52 | await self.client.change_presence(activity=discord.Activity(type=discord.ActivityType.listening, name=" a !cov help in " + str(len(self.client.guilds)) + " server(s)"))
53 | await ctx.send("I'm in " + str(len(self.client.guilds)) + " servers")
54 | for guild in self.client.guilds:
55 | total_mem += len(guild.members)
56 | list_guilds.append(guild.name + ' : ' + str(guild.id))
57 | list_guilds.append('Total members: ' + str(total_mem))
58 |
59 | # await ctx.send(list_guilds)
60 | print(list_guilds)
61 | await ctx.send('Total members: ' + str(total_mem))
62 | return
63 |
64 |
65 | @commands.command()
66 | @commands.has_role('admin')
67 | async def leave(self, ctx, id):
68 | if ctx.message.author.id == 550965528936710154:
69 | to_leave = self.client.get_guild(int(id))
70 | await ctx.send('Leaving ' + str(to_leave))
71 | try:
72 | await to_leave.leave()
73 | await ctx.send('Left ' + str(to_leave))
74 | except:
75 | await ctx.send('Failed leaving ' + str(to_leave))
76 | return
77 |
78 |
79 | @commands.group(name='graph', aliases=['g'])
80 | async def graph(self, ctx):
81 | if ctx.invoked_subcommand is None:
82 | await ctx.invoke(self.ghelp)
83 |
84 |
85 | @graph.command(name='top10')
86 | async def graphtop10(self, ctx, arg2):
87 | if arg2 not in ['confirmed', 'active', 'deaths', 'recovered']:
88 | return
89 |
90 | newcountry = arg2.lower()
91 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/top_10_countries_' + \
92 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
93 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
94 | description="Graph for Top 10 " + newcountry + 'Cases', color=0x00ff00)
95 | embedVar.set_footer(
96 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
97 | embedVar.set_image(url=urltobe)
98 | await ctx.send(embed=embedVar)
99 |
100 |
101 | @graph.command(name='country')
102 | async def graphcountry(self, ctx, arg2='all'):
103 | if arg2 == 'all':
104 | urltobe = 'https://github.com/resoucesforcorona/Resources/blob/master/All_countries.png?raw=true'
105 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
106 | description="Graph for all countries", color=0x00ff00)
107 |
108 | else:
109 | newcountry = (arg2.lower()).capitalize()
110 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/' + \
111 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
112 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
113 | description="Graph for: " + newcountry, color=0x00ff00)
114 |
115 | embedVar.set_footer(
116 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
117 | embedVar.set_image(url=urltobe)
118 | await ctx.send(embed=embedVar)
119 |
120 |
121 | @graph.command(name='continent')
122 | async def graphcontinent(self, ctx, arg2='all'):
123 | if arg2 == 'all':
124 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/All_continents.png'
125 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
126 | description="Graph for all continents", color=0x00ff00)
127 |
128 | else:
129 | newcountry = (arg2.lower()).capitalize()
130 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/' + \
131 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
132 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
133 | description="Graph for: " + newcountry, color=0x00ff00)
134 |
135 | embedVar.set_footer(
136 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
137 | embedVar.set_image(url=urltobe)
138 | await ctx.send(embed=embedVar)
139 |
140 |
141 | @graph.command(name='pie')
142 | async def graphpie(self, ctx):
143 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/all_countrywise_pie.png'
144 | embedVar = discord.Embed(
145 | title="Corona Tracker", url='https://anondoser.xyz', description="Graph for Pie", color=0x00ff00)
146 | embedVar.set_footer(
147 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
148 | embedVar.set_image(url=urltobe)
149 | await ctx.send(embed=embedVar)
150 |
151 |
152 | @graph.command(name='predict')
153 | async def predict(self, ctx, arg2):
154 | newcountry = (arg2.lower())
155 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/world_prediction_curve_' + \
156 | newcountry + '.png' + '?' + str(uuid.uuid4().hex[:15])
157 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
158 | description="Graph for global prediction curve for " + newcountry, color=0x00ff00)
159 | embedVar.set_footer(
160 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
161 | embedVar.set_image(url=urltobe)
162 | await ctx.send(embed=embedVar)
163 |
164 |
165 | @graph.command(name='global')
166 | async def graphglobal(self, ctx, arg2):
167 | if arg2 not in ['daily_confirmed', 'daily_deaths', 'trend', 'deaths']:
168 | return
169 |
170 | if arg2 == 'daily_confirmed':
171 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/daily_confirmed_cases_global.png'
172 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
173 | description="Graph for global daily confirmed", color=0x00ff00)
174 |
175 | elif arg2 == 'daily_deaths':
176 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/daily_deaths_cases_global.png'
177 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
178 | description="Graph for global daily deaths", color=0x00ff00)
179 |
180 | elif arg2 == 'trend':
181 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/world_trend_confirmed_cases.png'
182 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
183 | description="Graph for global trend", color=0x00ff00)
184 |
185 | else:
186 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/worldwide_cases_deaths.png'
187 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
188 | description="Graph for global total deaths", color=0x00ff00)
189 |
190 | embedVar.set_footer(
191 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
192 | embedVar.set_image(url=urltobe)
193 | await ctx.send(embed=embedVar)
194 |
195 |
196 | @graph.command(name='trend')
197 | async def graphtrend(self, ctx, arg2):
198 | if arg2 not in ['confirmed', 'deaths']:
199 | return
200 |
201 | if arg2 == 'confirmed':
202 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/trend_comparison_continents_confirmed.png'
203 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
204 | description="Graph for trend comparison b/w continents [confirmed]", color=0x00ff00)
205 |
206 | else:
207 | newcountry = (arg2.lower()).capitalize()
208 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/trend_comparison_countries_deaths.png'
209 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
210 | description="Graph for trend comparison b/w countries [deaths]" + newcountry, color=0x00ff00)
211 |
212 | embedVar.set_footer(
213 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
214 | embedVar.set_image(url=urltobe)
215 | await ctx.send(embed=embedVar)
216 |
217 |
218 | @graph.command(name='spread')
219 | async def graphspread(self, ctx):
220 | urltobe = 'https://raw.githubusercontent.com/resoucesforcorona/Resources/master/countries_vs_date_spread.png'
221 | embedVar = discord.Embed(title="Corona Tracker", url='https://anondoser.xyz',
222 | description="Graph for number of countries infected vs date", color=0x00ff00)
223 | embedVar.set_footer(
224 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
225 | embedVar.set_image(url=urltobe)
226 | await ctx.send(embed=embedVar)
227 |
228 |
229 | @commands.command()
230 | async def help(self, ctx):
231 | embedVar = discord.Embed(title="Corona Tracker Help Panel",
232 | description="Here's some help for you :heart:", color=0x00ff00)
233 | embedVar.add_field(
234 | name="!cov graph [commands]", value="Consists of graphical predictions, analysis , statistics and more see below for command", inline=False)
235 | embedVar.add_field(
236 | name="!cov g [commands]", value="Same as above, use !cov ghelp for list of commands", inline=False)
237 | embedVar.add_field(
238 | name="!cov ghelp", value="Help and commands list for `!cov graph`", inline=False)
239 | embedVar.add_field(
240 | name="!cov all", value="Shows global Covid-19 statistics", inline=False)
241 | embedVar.add_field(name="!cov interactive", value="Sends the best live interactive maps \n See how the Covid spread on the world map from the very start \n See live status of Covid on the world map \n See Mortality Progression from the very beginning", inline=False)
242 | embedVar.add_field(name="!cov country {your country} {full/micro}",
243 | value="Shows you a particular country's stats (Optional- use 'micro' for a simplified report)", inline=False)
244 | embedVar.add_field(
245 | name="!cov help", value="Shows you this message", inline=False)
246 | embedVar.add_field(
247 | name="!cov invite", value="Sends you the links to invite the bot to your own server & the official bot server", inline=False)
248 | embedVar.add_field(
249 | name="github", value="https://github.com/himanshu2406/Corona-Tracker-Bot", inline=False)
250 | embedVar.add_field(
251 | name="tip :heart: ", value="Buy me a Coffee [sends addresses for tipping]", inline=False)
252 | embedVar.add_field(name="Dev Contact",
253 | value="Firelogger#7717", inline=False)
254 | await ctx.send(embed=embedVar)
255 |
256 |
257 | @commands.command()
258 | async def invite(self, ctx):
259 | embedVar = discord.Embed(title="Invite me", url='https://discord.com/api/oauth2/authorize?client_id=731855425145798668&permissions=121856&scope=bot',
260 | description="Click the title to invite to your own server", color=0x00ff00)
261 | embedVar.set_footer(text="Firelogger#7717",
262 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
263 | await ctx.send(embed=embedVar)
264 |
265 |
266 | @commands.command()
267 | async def tip(self, ctx):
268 | embedVar = discord.Embed(title="Tip :heart:", url='https://www.buymeacoffee.com/anondoser/shop',
269 | description="Donate for improving the services and help running the bot", color=0x00ff00)
270 | embedVar.add_field(
271 | name="Btc address", value="```37btgSzgWdywmSPeBN5rUH8W5G9EYJoRoA```", inline=False)
272 | embedVar.add_field(
273 | name="Paypal", value="```https://www.paypal.me/firelogger```", inline=False)
274 | embedVar.add_field(
275 | name="For more", value="For more methods please dm me", inline=False)
276 | embedVar.set_footer(text="Firelogger#7717",
277 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
278 | await ctx.send(embed=embedVar)
279 |
280 |
281 | @commands.command()
282 | async def interactive(self, ctx):
283 | embedVar = discord.Embed(title="Interactive and playable Maps and statistics",
284 | description="Play with these live maps , statistics and visuals", color=0x00ff00)
285 | embedVar.add_field(name="World Map Live Progression",
286 | value="See how COVID spread from the very beginning on the world map, seekable and playable", inline=False)
287 | embedVar.add_field(
288 | name="Link", value="https://corona.anondoser.xyz/worldmap_progression.html", inline=False)
289 | embedVar.add_field(name="World Map static interactive ",
290 | value="see the current spread of Covid-19 on the world map", inline=False)
291 | embedVar.add_field(
292 | name="Link", value="https://corona.anondoser.xyz/worldmap_cases_interactive.html", inline=False)
293 | embedVar.add_field(name="Mortality rate Live progression",
294 | value="A beautiful playable and seekable representation of the mortality rate progression from the beginning , see those balls bounce", inline=False)
295 | embedVar.add_field(
296 | name="Link", value="https://corona.anondoser.xyz/mortalityrate_progression.html", inline=False)
297 | embedVar.set_footer(text="Firelogger#7717",
298 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
299 | await ctx.send(embed=embedVar)
300 |
301 |
302 | @commands.command()
303 | async def ghelp(self, ctx):
304 | embedVar = discord.Embed(title="Corona Tracker graph commands ",
305 | description="Use - `!cov graph [command] [arguments]` you can use `!cov g` instead too", color=0x00ff00)
306 | embedVar.add_field(name="How to use - ",
307 | value="```!cov graph [commands given below] [argument]``` arguments in round brackets - () mean type exact commands as given , \n Square brackets -[] mean a variable like your country name", inline=False)
308 | embedVar.add_field(
309 | name="spread", value="No arguments ; shows the spread among countries vs date", inline=False)
310 | embedVar.add_field(
311 | name="pie", value="No arguments ; shows a list of pie charts for cases among countries", inline=False)
312 | embedVar.add_field(name="top10 (confirmed/active/deaths/recovered)",
313 | value="Shows top 10 countries based on the argument given", inline=False)
314 | embedVar.add_field(name="global (daily_confirmed/daily_deaths/trend/deaths)",
315 | value="Shows graphs for global arguments", inline=False)
316 | embedVar.add_field(
317 | name="country [country name]", value="Graph for the country given", inline=False)
318 | embedVar.add_field(name="continent [continent name]",
319 | value="Graph for the continent given", inline=False)
320 | embedVar.add_field(name="predict (confirmed/deaths)",
321 | value="Shows graphical projections for the future along with next 10 day predicted figures for the argument", inline=False)
322 | embedVar.add_field(name="trend (confirmed/deaths)",
323 | value="Shows trend between different countries / continents based on the argument", inline=False)
324 | #embedVar.add_field(name="github", value="https://github.com/himanshu2406/Corona-Tracker", inline=False)
325 | #embedVar.add_field(name="tip :heart: ", value="Buy me a Coffee [sends addresses for tipping]", inline=False)
326 | embedVar.add_field(
327 | name="Example", value="```!cov graph top10 confirmed```", inline=False)
328 | embedVar.set_footer(text="Firelogger#7717",
329 | icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
330 | await ctx.send(embed=embedVar)
331 |
332 |
333 | @commands.command(aliases=['c'])
334 | async def country(self, ctx, args, complete='full'):
335 | if complete not in ['full', 'micro']:
336 | return
337 |
338 | try:
339 | i = await self.api_client.country(args)
340 | yday_c = await self.api_client.country(i.name, yesterday=True)
341 | except diseaseapi.NotFound:
342 | embedVar = discord.Embed(title="Invalid Country: " + args.capitalize(), description="Error, the country doesn't exist in the database", color=0xe33b3b, url='https://anondoser.xyz')
343 | return await ctx.send(embed=embedVar)
344 |
345 | embedVar = discord.Embed(description="Statistics from disease.py",
346 | color=0xe33b3b, url='https://anondoser.xyz')
347 |
348 | yday_c = await self.api_client.country(i.name, yesterday=True)
349 |
350 | cases_diff = cleaned_diff(i.cases, yday_c.cases)
351 | death_diff = cleaned_diff(i.deaths, yday_c.deaths)
352 | re_diff = cleaned_diff(i.recoveries, yday_c.recoveries)
353 | active_diff = cleaned_diff(i.active, yday_c.active)
354 | crit_diff = cleaned_diff(i.critical, yday_c.critical)
355 | test_diff = cleaned_diff(i.tests, yday_c.tests)
356 |
357 | embedVar.add_field(name='Cases:', value=check_na(i.cases)+cases_diff, inline=True)
358 | embedVar.add_field(name='Cases Today:', value=check_na(i.today.cases), inline=True)
359 | embedVar.add_field(name='Deaths:', value=check_na(i.deaths)+death_diff, inline=True)
360 | embedVar.add_field(name='Deaths Today:', value=check_na(i.today.deaths), inline=True)
361 | embedVar.add_field(name='Recovered:', value=check_na(i.recoveries)+re_diff, inline=True)
362 | embedVar.add_field(name='Recovered Today:', value=check_na(i.today.recoveries), inline=True)
363 | embedVar.add_field(name='Active:', value=check_na(i.active)+active_diff, inline=True)
364 | embedVar.add_field(name='Critical:', value=check_na(i.critical)+crit_diff, inline=True)
365 | embedVar.add_field(name='Tests:', value=check_na(i.tests)+test_diff, inline=True)
366 | urltobe = str(i.info.flag)
367 |
368 | if complete == 'full':
369 | embedVar.add_field(name ='Cases per Million', value= i.per_million.cases, inline=True)
370 | embedVar.add_field(name ='Active per Million', value= i.per_million.active, inline=True)
371 | embedVar.add_field(name ='Recoveries per Million', value= i.per_million.recoveries, inline=True)
372 | embedVar.add_field(name ='Tests per Million', value= i.per_million.tests, inline=True)
373 | embedVar.add_field(name ='Deaths per Million', value= i.per_million.deaths, inline=True)
374 | embedVar.add_field(name ='Critical per Million', value= i.per_million.critical, inline=True)
375 | embedVar.add_field(name ='One Case per Person', value= i.per_people.case, inline=True)
376 | embedVar.add_field(name ='One Death per Person', value= i.per_people.death, inline=True)
377 | embedVar.add_field(name ='One Test per Person', value= i.per_people.test, inline=True)
378 | embedVar.add_field(name ='Updated', value= i.updated, inline=True)
379 |
380 | embedVar.title = "Covid Stats for: " + i.name
381 | embedVar.set_thumbnail(url=urltobe)
382 | embedVar.set_footer(
383 | text="Firelogger#7717", icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
384 | await ctx.send(embed=embedVar)
385 |
386 |
387 | @commands.command()
388 | async def all(self, ctx):
389 | response = await self.api_client.all()
390 | embedVar = discord.Embed(title="Covid Worldwide Stats",
391 | description="Statistics from disease.py", color=0xe33b3b, url='https://anondoser.xyz')
392 |
393 |
394 | yday_c = await self.api_client.all(yesterday=True)
395 | cases_diff = cleaned_diff(response.cases, yday_c.cases)
396 | death_diff = cleaned_diff(response.deaths, yday_c.deaths)
397 | re_diff = cleaned_diff(response.recoveries, yday_c.recoveries)
398 | active_diff = cleaned_diff(response.active, yday_c.active)
399 | crit_diff = cleaned_diff(response.critical, yday_c.critical)
400 | test_diff = cleaned_diff(response.tests, yday_c.tests)
401 | embedVar.add_field(name='Cases:', value=check_na(response.cases)+cases_diff, inline=True)
402 | embedVar.add_field(name='Cases Today:', value=check_na(response.today.cases), inline=True)
403 | embedVar.add_field(name='Deaths:', value=check_na(response.deaths)+death_diff, inline=True)
404 | embedVar.add_field(name='Deaths Today:', value=check_na(response.today.deaths), inline=True)
405 | embedVar.add_field(name='Recovered:', value=check_na(response.recoveries)+re_diff, inline=True)
406 | embedVar.add_field(name='Recovered Today:', value=check_na(response.today.recoveries), inline=True)
407 | embedVar.add_field(name='Active:', value=check_na(response.active)+active_diff, inline=True)
408 | embedVar.add_field(name='Critical:', value=check_na(response.critical)+crit_diff, inline=True)
409 | embedVar.add_field(name='Tests:', value=check_na(response.tests)+test_diff, inline=True)
410 | embedVar.add_field(name ='Cases per Million', value= response.per_million.cases, inline=True)
411 | embedVar.add_field(name ='Active per Million', value= response.per_million.active, inline=True)
412 | embedVar.add_field(name ='Recoveries per Million', value= response.per_million.recoveries, inline=True)
413 | embedVar.add_field(name ='Tests per Million', value= response.per_million.tests, inline=True)
414 | embedVar.add_field(name ='Deaths per Million', value= response.per_million.deaths, inline=True)
415 | embedVar.add_field(name ='Critical per Million', value= response.per_million.critical, inline=True)
416 | embedVar.add_field(name ='One Case per Person', value= response.per_people.case, inline=True)
417 | embedVar.add_field(name ='One Death per Person', value= response.per_people.death, inline=True)
418 | embedVar.add_field(name ='One Test per Person', value= response.per_people.test, inline=True)
419 | embedVar.add_field(name ='Updated', value= response.updated, inline=True)
420 |
421 | embedVar.set_footer(text="Firelogger#7717",icon_url='https://avatars2.githubusercontent.com/u/37951606?s=460&u=f45b1c7a7f0eddbe0036a7cf79b47d7dfa889321&v=4')
422 | await ctx.send(embed=embedVar)
423 |
424 |
425 | def setup(client):
426 | client.add_cog(CovidCommands(client))
--------------------------------------------------------------------------------
/Visualisations and Ai predictions/nb_og.py:
--------------------------------------------------------------------------------
1 | import pandas as pd
2 | import numpy as np
3 | import matplotlib.pyplot as plt
4 | from matplotlib import ticker
5 | import pycountry_convert as pc
6 | import folium
7 | import branca
8 | from datetime import datetime, timedelta,date
9 | from scipy.interpolate import make_interp_spline, BSpline
10 | import plotly.express as px
11 | import json, requests
12 | import calmap
13 | import urllib
14 | import base64
15 | from base64 import b64encode
16 | from keras.layers import Input, Dense, Activation, LeakyReLU, Dropout
17 | from keras import models
18 | from keras.optimizers import RMSprop, Adam
19 |
20 | import warnings
21 | warnings.filterwarnings('ignore')
22 |
23 |
24 |
25 | # Retriving Dataset
26 |
27 | while True:
28 | try:
29 | df_table = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/web-data/data/cases_time.csv",parse_dates=['Last_Update'])
30 | df_confirmed = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv')
31 | df_deaths = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv')
32 |
33 | # Depricated
34 | # df_recovered = pd.read_csv('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Recovered.csv')
35 | df_covid19 = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/web-data/data/cases_country.csv")
36 | except:
37 | continue
38 | else:
39 | break
40 |
41 |
42 | # new dataset
43 | df_covid19 = df_covid19.drop(["People_Tested","People_Hospitalized","UID","ISO3","Mortality_Rate"],axis =1)
44 | df_covid19.head(2)
45 | df_confirmed.head(2)
46 |
47 | df_confirmed = df_confirmed.rename(columns={"Province/State":"state","Country/Region": "country"})
48 | df_deaths = df_deaths.rename(columns={"Province/State":"state","Country/Region": "country"})
49 | df_covid19 = df_covid19.rename(columns={"Country_Region": "country"})
50 | df_covid19["Active"] = df_covid19["Confirmed"]-df_covid19["Recovered"]-df_covid19["Deaths"]
51 | # df_recovered = df_recovered.rename(columns={"Province/State":"state","Country/Region": "country"})
52 | df_confirmed.loc[df_confirmed['country'] == "US", "country"] = "USA"
53 | df_deaths.loc[df_deaths['country'] == "US", "country"] = "USA"
54 | df_covid19.loc[df_covid19['country'] == "US", "country"] = "USA"
55 | df_table.loc[df_table['Country_Region'] == "US", "Country_Region"] = "USA"
56 | # df_recovered.loc[df_recovered['country'] == "US", "country"] = "USA"
57 |
58 |
59 | df_confirmed.loc[df_confirmed['country'] == 'Korea, South', "country"] = 'South Korea'
60 | df_deaths.loc[df_deaths['country'] == 'Korea, South', "country"] = 'South Korea'
61 | df_covid19.loc[df_covid19['country'] == "Korea, South", "country"] = "South Korea"
62 | df_table.loc[df_table['Country_Region'] == "Korea, South", "Country_Region"] = "South Korea"
63 | # df_recovered.loc[df_recovered['country'] == 'Korea, South', "country"] = 'South Korea'
64 |
65 | df_confirmed.loc[df_confirmed['country'] == 'Taiwan*', "country"] = 'Taiwan'
66 | df_deaths.loc[df_deaths['country'] == 'Taiwan*', "country"] = 'Taiwan'
67 | df_covid19.loc[df_covid19['country'] == "Taiwan*", "country"] = "Taiwan"
68 | df_table.loc[df_table['Country_Region'] == "Taiwan*", "Country_Region"] = "Taiwan"
69 | # df_recovered.loc[df_recovered['country'] == 'Taiwan*', "country"] = 'Taiwan'
70 |
71 | df_confirmed.loc[df_confirmed['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
72 | df_deaths.loc[df_deaths['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
73 | df_covid19.loc[df_covid19['country'] == "Congo (Kinshasa)", "country"] = "Democratic Republic of the Congo"
74 | df_table.loc[df_table['Country_Region'] == "Congo (Kinshasa)", "Country_Region"] = "Democratic Republic of the Congo"
75 | # df_recovered.loc[df_recovered['country'] == 'Congo (Kinshasa)', "country"] = 'Democratic Republic of the Congo'
76 |
77 | df_confirmed.loc[df_confirmed['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
78 | df_deaths.loc[df_deaths['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
79 | df_covid19.loc[df_covid19['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
80 | df_table.loc[df_table['Country_Region'] == "Cote d'Ivoire", "Country_Region"] = "Côte d'Ivoire"
81 | # df_recovered.loc[df_recovered['country'] == "Cote d'Ivoire", "country"] = "Côte d'Ivoire"
82 |
83 | df_confirmed.loc[df_confirmed['country'] == "Reunion", "country"] = "Réunion"
84 | df_deaths.loc[df_deaths['country'] == "Reunion", "country"] = "Réunion"
85 | df_covid19.loc[df_covid19['country'] == "Reunion", "country"] = "Réunion"
86 | df_table.loc[df_table['Country_Region'] == "Reunion", "Country_Region"] = "Réunion"
87 | # df_recovered.loc[df_recovered['country'] == "Reunion", "country"] = "Réunion"
88 |
89 | df_confirmed.loc[df_confirmed['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
90 | df_deaths.loc[df_deaths['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
91 | df_covid19.loc[df_covid19['country'] == "Congo (Brazzaville)", "country"] = "Republic of the Congo"
92 | df_table.loc[df_table['Country_Region'] == "Congo (Brazzaville)", "Country_Region"] = "Republic of the Congo"
93 | # df_recovered.loc[df_recovered['country'] == 'Congo (Brazzaville)', "country"] = 'Republic of the Congo'
94 |
95 | df_confirmed.loc[df_confirmed['country'] == 'Bahamas, The', "country"] = 'Bahamas'
96 | df_deaths.loc[df_deaths['country'] == 'Bahamas, The', "country"] = 'Bahamas'
97 | df_covid19.loc[df_covid19['country'] == "Bahamas, The", "country"] = "Bahamas"
98 | df_table.loc[df_table['Country_Region'] == "Bahamas, The", "Country_Region"] = "Bahamas"
99 | # df_recovered.loc[df_recovered['country'] == 'Bahamas, The', "country"] = 'Bahamas'
100 |
101 | df_confirmed.loc[df_confirmed['country'] == 'Gambia, The', "country"] = 'Gambia'
102 | df_deaths.loc[df_deaths['country'] == 'Gambia, The', "country"] = 'Gambia'
103 | df_covid19.loc[df_covid19['country'] == "Gambia, The", "country"] = "Gambia"
104 | df_table.loc[df_table['Country_Region'] == "Gambia", "Country_Region"] = "Gambia"
105 | # df_recovered.loc[df_recovered['country'] == 'Gambia, The', "country"] = 'Gambia'
106 |
107 | # getting all countries
108 | countries = np.asarray(df_confirmed["country"])
109 | countries1 = np.asarray(df_covid19["country"])
110 | # Continent_code to Continent_names
111 | continents = {
112 | 'NA': 'North America',
113 | 'SA': 'South America',
114 | 'AS': 'Asia',
115 | 'OC': 'Australia',
116 | 'AF': 'Africa',
117 | 'EU' : 'Europe',
118 | 'na' : 'Others'
119 | }
120 |
121 | # Defininng Function for getting continent code for country.
122 | def country_to_continent_code(country):
123 | try:
124 | return pc.country_alpha2_to_continent_code(pc.country_name_to_country_alpha2(country))
125 | except :
126 | return 'na'
127 |
128 | #Collecting Continent Information
129 | df_confirmed.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]])
130 | df_deaths.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]])
131 | df_covid19.insert(1,"continent", [continents[country_to_continent_code(country)] for country in countries1[:]])
132 | df_table.insert(1,"continent", [continents[country_to_continent_code(country)] for country in df_table["Country_Region"].values])
133 | # df_recovered.insert(2,"continent", [continents[country_to_continent_code(country)] for country in countries[:]] )
134 |
135 | df_table = df_table[df_table["continent"] != "Others"]
136 |
137 | df_deaths[df_deaths["continent" ]== 'Others']
138 |
139 | df_confirmed = df_confirmed.replace(np.nan, '', regex=True)
140 | df_deaths = df_deaths.replace(np.nan, '', regex=True)
141 | # df_recovered = df_recovered.replace(np.nan, '', regex=True)
142 | # df_active = df_active.replace(np.nan, '', regex=True)
143 |
144 | """# FUNCTIONS BELOW HERE"""
145 |
146 | def plot_params(ax,axis_label= None, plt_title = None,label_size=15, axis_fsize = 15, title_fsize = 20, scale = 'linear' ):
147 | # Tick-Parameters
148 | ax.xaxis.set_minor_locator(ticker.AutoMinorLocator())
149 | ax.yaxis.set_minor_locator(ticker.AutoMinorLocator())
150 | ax.tick_params(which='both', width=1,labelsize=label_size)
151 | ax.tick_params(which='major', length=6)
152 | ax.tick_params(which='minor', length=3, color='0.8')
153 |
154 | # Grid
155 | plt.grid(lw = 1, ls = '-', c = "0.7", which = 'major')
156 | plt.grid(lw = 1, ls = '-', c = "0.9", which = 'minor')
157 |
158 | # Plot Title
159 | plt.title( plt_title,{'fontsize':title_fsize})
160 |
161 | # Yaxis sacle
162 | plt.yscale(scale)
163 | plt.minorticks_on()
164 | # Plot Axes Labels
165 | xl = plt.xlabel(axis_label[0],fontsize = axis_fsize)
166 | yl = plt.ylabel(axis_label[1],fontsize = axis_fsize)
167 |
168 | def visualize_covid_cases(confirmed, deaths, continent=None , country = None , state = None, period = None, figure = None, scale = "linear"):
169 | x = 0
170 | if figure == None:
171 | f = plt.figure(figsize=(10,10))
172 | # Sub plot
173 | ax = f.add_subplot(111)
174 | else :
175 | f = figure[0]
176 | # Sub plot
177 | ax = f.add_subplot(figure[1],figure[2],figure[3])
178 | ax.set_axisbelow(True)
179 | plt.tight_layout(pad=10, w_pad=5, h_pad=5)
180 |
181 | stats = [confirmed, deaths]
182 | label = ["Confirmed", "Deaths"]
183 |
184 | if continent != None:
185 | params = ["continent",continent]
186 | elif country != None:
187 | params = ["country",country]
188 | else:
189 | params = ["All", "All"]
190 | color = ["darkcyan","crimson"]
191 | marker_style = dict(linewidth=3, linestyle='-', marker='o',markersize=4, markerfacecolor='#ffffff')
192 | for i,stat in enumerate(stats):
193 | if params[1] == "All" :
194 | cases = np.sum(np.asarray(stat.iloc[:,5:]),axis = 0)[x:]
195 | else :
196 | cases = np.sum(np.asarray(stat[stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
197 | date = np.arange(1,cases.shape[0]+1)[x:]
198 | plt.plot(date,cases,label = label[i]+" (Total : "+str(cases[-1])+")",color=color[i],**marker_style)
199 | plt.fill_between(date,cases,color=color[i],alpha=0.3)
200 |
201 | if params[1] == "All" :
202 | Total_confirmed = np.sum(np.asarray(stats[0].iloc[:,5:]),axis = 0)[x:]
203 | Total_deaths = np.sum(np.asarray(stats[1].iloc[:,5:]),axis = 0)[x:]
204 | else :
205 | Total_confirmed = np.sum(np.asarray(stats[0][stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
206 | Total_deaths = np.sum(np.asarray(stats[1][stat[params[0]] == params[1]].iloc[:,5:]),axis = 0)[x:]
207 |
208 | text = "From "+stats[0].columns[5]+" to "+stats[0].columns[-1]+"\n"
209 | text += "Mortality rate : "+ str(int(Total_deaths[-1]/(Total_confirmed[-1])*10000)/100)+"\n"
210 | text += "Last 5 Days:\n"
211 | text += "Confirmed : " + str(Total_confirmed[-1] - Total_confirmed[-6])+"\n"
212 | text += "Deaths : " + str(Total_deaths[-1] - Total_deaths[-6])+"\n"
213 | text += "Last 24 Hours:\n"
214 | text += "Confirmed : " + str(Total_confirmed[-1] - Total_confirmed[-2])+"\n"
215 | text += "Deaths : " + str(Total_deaths[-1] - Total_deaths[-2])+"\n"
216 |
217 | plt.text(0.02, 0.78, text, fontsize=15, horizontalalignment='left', verticalalignment='top', transform=ax.transAxes,bbox=dict(facecolor='white', alpha=0.4))
218 |
219 | # Plot Axes Labels
220 | axis_label = ["Days ("+df_confirmed.columns[5]+" - "+df_confirmed.columns[-1]+")","No of Cases"]
221 |
222 | # Plot Parameters
223 | plot_params(ax,axis_label,scale = scale)
224 |
225 | # Plot Title
226 | if params[1] == "All" :
227 | plt.title("COVID-19 Cases World",{'fontsize':25})
228 | else:
229 | plt.title("COVID-19: "+params[1] ,{'fontsize':25})
230 |
231 | # Legend Location
232 | l = plt.legend(loc= "best",fontsize = 15)
233 |
234 | if figure == None:
235 | #plt.show()
236 | pass
237 |
238 | def get_total_cases(cases, country = "All"):
239 | if(country == "All") :
240 | return np.sum(np.asarray(cases.iloc[:,5:]),axis = 0)[-1]
241 | else :
242 | return np.sum(np.asarray(cases[cases["country"] == country].iloc[:,5:]),axis = 0)[-1]
243 |
244 | def get_mortality_rate(confirmed,deaths, continent = None, country = None):
245 | if continent != None:
246 | params = ["continent",continent]
247 | elif country != None:
248 | params = ["country",country]
249 | else :
250 | params = ["All", "All"]
251 |
252 | if params[1] == "All" :
253 | Total_confirmed = np.sum(np.asarray(confirmed.iloc[:,5:]),axis = 0)
254 | Total_deaths = np.sum(np.asarray(deaths.iloc[:,5:]),axis = 0)
255 | mortality_rate = np.round((Total_deaths/(Total_confirmed+1.01))*100,2)
256 | else :
257 | Total_confirmed = np.sum(np.asarray(confirmed[confirmed[params[0]] == params[1]].iloc[:,5:]),axis = 0)
258 | Total_deaths = np.sum(np.asarray(deaths[deaths[params[0]] == params[1]].iloc[:,5:]),axis = 0)
259 | mortality_rate = np.round((Total_deaths/(Total_confirmed+1.01))*100,2)
260 |
261 | return np.nan_to_num(mortality_rate)
262 | def dd(date1,date2):
263 | return (datetime.strptime(date1,'%m/%d/%y') - datetime.strptime(date2,'%m/%d/%y')).days
264 |
265 |
266 | out = ""#+"output/"
267 |
268 | df_countries_cases = df_covid19.copy().drop(['Lat','Long_','continent','Last_Update'],axis =1)
269 | df_countries_cases.index = df_countries_cases["country"]
270 | df_countries_cases = df_countries_cases.drop(['country'],axis=1)
271 |
272 | df_continents_cases = df_covid19.copy().drop(['Lat','Long_','country','Last_Update'],axis =1)
273 | df_continents_cases = df_continents_cases.groupby(["continent"]).sum()
274 |
275 | df_countries_cases.fillna(0,inplace=True)
276 | df_continents_cases.fillna(0,inplace=True)
277 |
278 |
279 |
280 | f = plt.figure(figsize=(10,5))
281 | f.add_subplot(111)
282 |
283 | plt.axes(axisbelow=True)
284 | plt.barh(df_countries_cases.sort_values('Confirmed')["Confirmed"].index[-10:],df_countries_cases.sort_values('Confirmed')["Confirmed"].values[-10:],color="darkcyan")
285 | plt.tick_params(size=5,labelsize = 13)
286 | plt.xlabel("Confirmed Cases",fontsize=18)
287 | plt.title("Top 10 Countries (Confirmed Cases)",fontsize=20)
288 | plt.grid(alpha=0.3)
289 | plt.savefig(out+'top_10_countries_confirmed.png')
290 |
291 |
292 | f = plt.figure(figsize=(10,5))
293 | f.add_subplot(111)
294 |
295 | plt.axes(axisbelow=True)
296 | plt.barh(df_countries_cases.sort_values('Deaths')["Deaths"].index[-10:],df_countries_cases.sort_values('Deaths')["Deaths"].values[-10:],color="crimson")
297 | plt.tick_params(size=5,labelsize = 13)
298 | plt.xlabel("Deaths Cases",fontsize=18)
299 | plt.title("Top 10 Countries (Deaths Cases)",fontsize=20)
300 | plt.grid(alpha=0.3,which='both')
301 | plt.savefig(out+'top_10_countries_deaths.png')
302 |
303 |
304 |
305 | f = plt.figure(figsize=(10,5))
306 | f.add_subplot(111)
307 |
308 | plt.axes(axisbelow=True)
309 | plt.barh(df_countries_cases.sort_values('Active')["Active"].index[-10:],df_countries_cases.sort_values('Active')["Active"].values[-10:],color="darkorange")
310 | plt.tick_params(size=5,labelsize = 13)
311 | plt.xlabel("Active Cases",fontsize=18)
312 | plt.title("Top 10 Countries (Active Cases)",fontsize=20)
313 | plt.grid(alpha=0.3,which='both')
314 | plt.savefig(out+'top_10_countries_active.png')
315 |
316 |
317 |
318 | f = plt.figure(figsize=(10,5))
319 | f.add_subplot(111)
320 |
321 | plt.axes(axisbelow=True)
322 | plt.barh(df_countries_cases.sort_values('Recovered')["Recovered"].index[-10:],df_countries_cases.sort_values('Recovered')["Recovered"].values[-10:],color="limegreen")
323 | plt.tick_params(size=5,labelsize = 13)
324 | plt.xlabel("Recovered Cases",fontsize=18)
325 | plt.title("Top 10 Countries (Recovered Cases)",fontsize=20)
326 | plt.grid(alpha=0.3,which='both')
327 | plt.savefig(out+'top_10_countries_recovered.png')
328 |
329 |
330 | world_map = folium.Map(location=[10,0], tiles="cartodbpositron", zoom_start=2,max_zoom=6,min_zoom=2)
331 | for i in range(0,len(df_confirmed)):
332 | folium.Circle(
333 | location=[df_confirmed.iloc[i]['Lat'], df_confirmed.iloc[i]['Long']],
334 | tooltip = "