├── .gitignore
├── LICENSE
├── contributors.md
├── future.md
├── imgs
├── alphafold_preds.png
├── angle_preds.png
├── elu_resnet_2d.png
├── our_preds.png
└── ramachandran_plot.png
├── implementation_details.md
├── models
├── angles
│ ├── predicting_angles.ipynb
│ ├── resnet_1d_angles.h5
│ └── resnet_1d_angles.py
├── distance_pipeline
│ ├── Tutorials
│ │ ├── README.pdf
│ │ ├── RR_format.py
│ │ └── modify_pssm.py
│ ├── distance_generator_data.py
│ ├── elu_resnet_2d_distances.py
│ ├── evaluation_pipeline.py
│ ├── func_utils.py
│ ├── images
│ │ ├── FM_T0869_CASP12.jpg
│ │ ├── golden_img_v91_17.png
│ │ ├── golden_img_v91_20.png
│ │ ├── golden_img_v91_32.png
│ │ ├── golden_img_v91_45.png
│ │ ├── golden_img_v91_50.png
│ │ ├── golden_img_v91_54.png
│ │ ├── golden_img_v91_55.png
│ │ └── golden_img_v91_56.png
│ ├── models
│ │ └── tester_28_lxl_golden.h5
│ ├── pipeline_caller.py
│ ├── pretrain_model_pssm_l_x_l.ipynb
│ ├── record.txt
│ └── training_pipeline.py
├── new_distances
│ ├── distance_generator_changes.py
│ ├── elu_resnet_2d_distances.py
│ ├── pretrain_model.ipynb
│ ├── resume_training.ipynb
│ └── trained_models_h5
│ │ ├── 17_test.h5
│ │ └── tester_28.h5
└── old_distances
│ ├── elu_resnet_2d_distances.h5
│ ├── elu_resnet_2d_distances.py
│ └── predicting_distances.ipynb
├── preprocessing
├── angle_data_preparation_py.ipynb
├── get_angles_from_coords_py.ipynb
├── get_proteins_under_200aa.jl
└── julia_get_proteins_under_200aa.ipynb
├── readme.md
└── requirements.txt
/.gitignore:
--------------------------------------------------------------------------------
1 | # exclude everything
2 | data/*
3 | minifold_journey.md
4 | models/__pycache__/*
5 | models/.ipynb_checkpoints/*
6 | preprocessing/.ipynb_checkpoints/*
7 | # exception to the rule
8 | # !somefolder/.gitkeep
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2019 Eric Alcaide
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/contributors.md:
--------------------------------------------------------------------------------
1 | # Contributors
2 | Here's a list of people who have made code contributions to this project:
3 | * [@EricAlcaide](https://github.com/EricAlcaide)
4 | * [@McMenemy](https://github.com/McMenemy)
5 | * [@roberCO](https://github.com/roberCO)
6 | * [@pabloAMC](https://github.com/PabloAMC)
7 |
--------------------------------------------------------------------------------
/future.md:
--------------------------------------------------------------------------------
1 | # Project Future - MiniFold
2 |
3 | In a brief way, some promising ideas:
4 |
5 | * Train with crops of 64x64, not windows of 200x200 (and average at prediction time).
6 | * Use data from Multiple Sequence Alignments (MSA) such as paired changes bewteen AAs.
7 | * Use distance map as potential input for angle prediction (or vice versa?).
8 | * Use Physicochemical features of AAs as input.
9 | * Train with more data (in the cloud?)
10 | * Use predictions as constraints to the Rosetta Method for Protein Structure Prediction
11 | * Set up a prediction script/function from raw text/FASTA file
12 | * ...
13 |
14 | *"Science is a Work In Progress."*
15 |
16 | ## Contribute
17 | Hey there! New ideas are welcome: open/close issues, fork the repo and share your code with a Pull Request.
18 | Clone this project to your computer:
19 |
20 | `git clone https://github.com/EricAlcaide/MiniFold`
21 |
22 | By participating in this project, you agree to abide by the thoughtbot [code of conduct](https://thoughtbot.com/open-source-code-of-conduct)
23 |
24 | ## Meta
25 |
26 | * **Author's GitHub Profile**: [Eric Alcaide](https://github.com/EricAlcaide/)
27 | * **Twitter**: [@eric_alcaide](https://twitter.com/eric_alcaide)
28 | * **Email**: ericalcaide1@gmail.com
--------------------------------------------------------------------------------
/imgs/alphafold_preds.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/hypnopump/MiniFold/6eb47c5600c22c7dabbb1294adbd8c6704a185cb/imgs/alphafold_preds.png
--------------------------------------------------------------------------------
/imgs/angle_preds.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/hypnopump/MiniFold/6eb47c5600c22c7dabbb1294adbd8c6704a185cb/imgs/angle_preds.png
--------------------------------------------------------------------------------
/imgs/elu_resnet_2d.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/hypnopump/MiniFold/6eb47c5600c22c7dabbb1294adbd8c6704a185cb/imgs/elu_resnet_2d.png
--------------------------------------------------------------------------------
/imgs/our_preds.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/hypnopump/MiniFold/6eb47c5600c22c7dabbb1294adbd8c6704a185cb/imgs/our_preds.png
--------------------------------------------------------------------------------
/imgs/ramachandran_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/hypnopump/MiniFold/6eb47c5600c22c7dabbb1294adbd8c6704a185cb/imgs/ramachandran_plot.png
--------------------------------------------------------------------------------
/implementation_details.md:
--------------------------------------------------------------------------------
1 | # Implementation Details
2 |
3 | ## Proposed Architecture
4 |
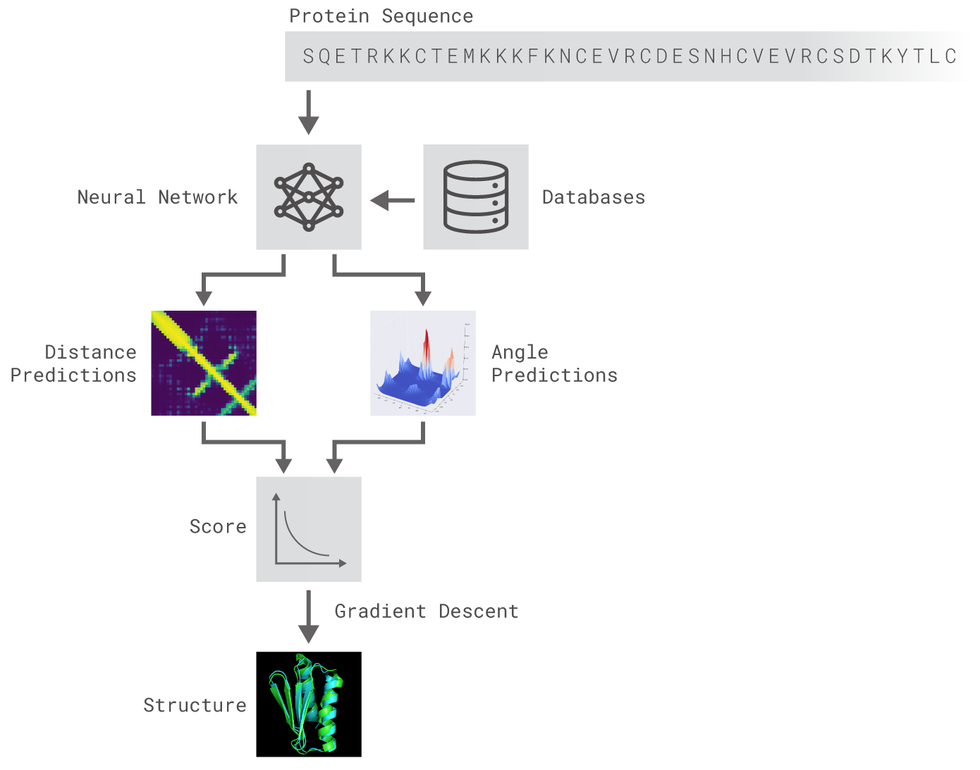
5 | The methods implemented are inspired by DeepMind's original post. Two different residual neural networks (ResNets) are used to predict **angles** between adjacent aminoacids (AAs) and **distance** between every pair of AAs of a protein. For distance prediction a 2D Resnet was used while for angles prediction a 1D Resnet was used.
6 |
7 |
8 |
9 |
22 |

23 |
32 |

33 |
37 |

38 |
55 |

56 |
61 |

62 |
44 |

45 |
58 |

59 |
68 |

69 |
73 |

74 |
91 |

92 |
97 |

98 |
 9 |
9 |  9 |
9 |  23 |
23 |  33 |
33 |  38 |
38 |  56 |
56 |  62 |
62 |  45 |
45 |  59 |
59 |  69 |
69 |  74 |
74 |  92 |
92 |  98 |
98 |