├── README.md
├── deeplearning4j
└── logistic
│ ├── .classpath
│ ├── .project
│ ├── .settings
│ ├── org.eclipse.jdt.core.prefs
│ └── org.eclipse.m2e.core.prefs
│ ├── compile.sh
│ ├── pom.xml
│ ├── run.sh
│ ├── src
│ └── LogisticExample.java
│ └── target
│ └── .gitignore
├── general
├── Modified_KTx_75_patients_April_17_2019.csv
├── Survival-Decision-Tree-Example1.ipynb
└── simplex-softmax.ipynb
├── mxnet
├── logistic
│ └── mxnet-logistic-example.ipynb
├── mxnet-vary-inputs-slideexamples.ipynb
├── mxnet-vary-inputs.ipynb
└── randomout
│ ├── data
│ └── crater
│ │ ├── All-Fold1-test.rec
│ │ └── All-Fold1-train.rec
│ ├── get_data.py
│ ├── randomout-cifar-inception.ipynb
│ ├── symbol_cratercnn.py
│ ├── symbol_inception-28-small.py
│ └── symbol_inception-bn-28-small.py
├── pytorch
├── pytorch-logistic-regression.ipynb
├── pytorch-mnist.ipynb
└── segmentation
│ ├── 10279_500_f00182_original.jpg
│ ├── 8917.png
│ ├── PatchWiseSegmentation.ipynb
│ └── reconstruct-slides.ipynb
└── theano
├── cnn
├── Interactive-Spatial-Transformer-Network-Layer-Lasagne.ipynb
├── lasagne-cifar10-autoencoder.ipynb
├── lasagne-cifar10-gan.ipynb
├── lasagne-cifar10-inception-example.ipynb
├── lasagne-mnist-autoencoder.ipynb
├── lasagne-mnist-gan.ipynb
└── lasagne-mnist-small-example.ipynb
├── counting
├── README.md
├── count-ception.ipynb
├── count-ception.py
└── models.py
├── logistic
└── theano-logistic-example.ipynb
└── visualization
├── inception-example-basic.ipynb
└── unet-example-basic.ipynb
/README.md:
--------------------------------------------------------------------------------
1 | # NeuralNetwork-Examples
2 | The same small networks implemented in different frameworks
3 |
4 | ### Citation:
5 |
6 | If you use these examples in your research please cite this repository as follows:
7 |
8 | Joseph Paul Cohen. Neural Network Examples. GitHub. 2016. [Online] Available:https://github.com/ieee8023/NeuralNetwork-Examples/
9 |
10 |
11 | ```
12 | @misc{CohenGitHub,
13 | author = {Cohen, Joseph Paul},
14 | booktitle = {GitHub},
15 | title = {Neural Network Examples},
16 | url = {https://github.com/ieee8023/NeuralNetwork-Examples/},
17 | year = {2016}
18 | }
19 | ```
20 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/.classpath:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
21 |
22 |
23 |
24 |
25 |
26 |
27 |
28 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/.project:
--------------------------------------------------------------------------------
1 |
2 |
3 | deeplearning4j-logistic-example
4 |
5 |
6 |
7 |
8 |
9 | org.eclipse.jdt.core.javabuilder
10 |
11 |
12 |
13 |
14 | org.eclipse.m2e.core.maven2Builder
15 |
16 |
17 |
18 |
19 |
20 | org.eclipse.jdt.core.javanature
21 | org.eclipse.m2e.core.maven2Nature
22 |
23 |
24 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/.settings/org.eclipse.jdt.core.prefs:
--------------------------------------------------------------------------------
1 | eclipse.preferences.version=1
2 | org.eclipse.jdt.core.compiler.codegen.targetPlatform=1.7
3 | org.eclipse.jdt.core.compiler.compliance=1.7
4 | org.eclipse.jdt.core.compiler.problem.forbiddenReference=warning
5 | org.eclipse.jdt.core.compiler.source=1.7
6 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/.settings/org.eclipse.m2e.core.prefs:
--------------------------------------------------------------------------------
1 | activeProfiles=
2 | eclipse.preferences.version=1
3 | resolveWorkspaceProjects=true
4 | version=1
5 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/compile.sh:
--------------------------------------------------------------------------------
1 | mkdir -p classes
2 |
3 | javac -J-Xms512m -J-Xmx512m -cp target/deeplearning4j-0.4-rc3.8.jar -d classes `find src -type f -name "*.java"`
4 |
5 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/pom.xml:
--------------------------------------------------------------------------------
1 |
2 | 4.0.0
3 |
4 | org.deeplearning4j
5 | deeplearning4j
6 | 0.4-rc3.8
7 |
8 | DeepLearning4j
9 |
10 |
11 | 0.4-rc3.8
12 | 0.4-rc3.8
13 | 0.0.0.14
14 | 2.5.1
15 |
16 |
17 |
18 |
19 |
20 |
21 | sonatype-nexus-snapshots
22 | Sonatype Nexus snapshot repository
23 | https://oss.sonatype.org/content/repositories/snapshots
24 |
25 |
26 | nexus-releases
27 | Nexus Release Repository
28 | http://oss.sonatype.org/service/local/staging/deploy/maven2/
29 |
30 |
31 |
32 |
33 |
34 | org.nd4j
35 | nd4j-x86
36 | ${nd4j.version}
37 |
38 |
39 |
40 |
41 |
42 | org.deeplearning4j
43 | deeplearning4j-nlp
44 | ${dl4j.version}
45 |
46 |
47 |
48 | org.deeplearning4j
49 | deeplearning4j-core
50 | ${dl4j.version}
51 |
52 |
53 | org.deeplearning4j
54 | deeplearning4j-ui
55 | ${dl4j.version}
56 |
57 |
58 | com.google.guava
59 | guava
60 | 19.0
61 |
62 |
63 | org.nd4j
64 | nd4j-x86
65 | ${nd4j.version}
66 |
67 |
68 | canova-nd4j-image
69 | org.nd4j
70 | ${canova.version}
71 |
72 |
73 | canova-nd4j-codec
74 | org.nd4j
75 | ${canova.version}
76 |
77 |
78 | com.fasterxml.jackson.dataformat
79 | jackson-dataformat-yaml
80 | ${jackson.version}
81 |
82 |
83 |
84 |
85 |
86 |
87 |
88 | org.codehaus.mojo
89 | exec-maven-plugin
90 | 1.4.0
91 |
92 |
93 |
94 | exec
95 |
96 |
97 |
98 |
99 | java
100 |
101 |
102 |
103 | org.apache.maven.plugins
104 | maven-shade-plugin
105 | 1.6
106 |
107 | true
108 |
109 |
110 | *:*
111 |
112 | org/datanucleus/**
113 | META-INF/*.SF
114 | META-INF/*.DSA
115 | META-INF/*.RSA
116 |
117 |
118 |
119 |
120 |
121 |
122 | package
123 |
124 | shade
125 |
126 |
127 |
128 |
129 | reference.conf
130 |
131 |
132 |
133 |
134 |
135 |
136 |
137 |
138 |
139 |
140 |
141 | org.apache.maven.plugins
142 | maven-compiler-plugin
143 |
144 | 1.7
145 | 1.7
146 |
147 |
148 |
149 |
150 |
151 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/run.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 |
3 | java -cp classes:target/deeplearning4j-0.4-rc3.8.jar LogisticExample $@
4 |
5 | echo java finished
6 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/src/LogisticExample.java:
--------------------------------------------------------------------------------
1 |
2 | import java.io.IOException;
3 | import java.util.List;
4 |
5 | import org.deeplearning4j.nn.api.OptimizationAlgorithm;
6 | import org.deeplearning4j.nn.conf.MultiLayerConfiguration;
7 | import org.deeplearning4j.nn.conf.NeuralNetConfiguration;
8 | import org.deeplearning4j.nn.conf.layers.OutputLayer;
9 | import org.deeplearning4j.nn.multilayer.MultiLayerNetwork;
10 | import org.nd4j.linalg.api.ndarray.INDArray;
11 | import org.nd4j.linalg.factory.Nd4j;
12 | import org.nd4j.linalg.lossfunctions.LossFunctions;
13 | import org.slf4j.LoggerFactory;
14 |
15 | import ch.qos.logback.classic.Level;
16 | import ch.qos.logback.classic.Logger;
17 |
18 | public class LogisticExample {
19 |
20 |
21 | public static void main(String[] args) throws IOException, ClassNotFoundException {
22 |
23 | // disable logging
24 | Logger root = (Logger) LoggerFactory.getLogger(ch.qos.logback.classic.Logger.ROOT_LOGGER_NAME);
25 | root.setLevel(Level.ERROR);
26 |
27 | /* build the graph. Only one layer is created because this is
28 | * the lowest dl4j can go. 2 inputs are specified and 1 output.
29 | * The sigmoid activation will perform the dot product of the
30 | * weights and bias.
31 | */
32 | MultiLayerConfiguration.Builder builder = new NeuralNetConfiguration.Builder()
33 | .optimizationAlgo(OptimizationAlgorithm.STOCHASTIC_GRADIENT_DESCENT)
34 | .list(1)
35 | .layer(0, new OutputLayer.Builder()
36 | .nIn(2)
37 | .nOut(1)
38 | .activation("sigmoid")
39 | .build())
40 | .backprop(true);
41 | MultiLayerNetwork model = new MultiLayerNetwork(builder.build());
42 | model.init();
43 |
44 | // specify the x0 and x2 values
45 | INDArray x = Nd4j.create(new double[][] { { -1.0, -2.0}, });
46 |
47 | // specify the weights. w0 and w1 are contained in W
48 | model.getLayers()[0].setParam("W", Nd4j.create(new double[][] {{2},{-3}}));
49 | model.getLayers()[0].setParam("b", Nd4j.create(new double[] {-3}));
50 |
51 | // process input data
52 | List results = model.feedForward(x.getRow(0));
53 |
54 | // get weights of the sigmoid
55 | INDArray weights = model.getLayers()[0].getParam("W");
56 | INDArray bias = model.getLayers()[0].getParam("b");
57 |
58 | System.out.println("x0, x1: " + x);
59 | System.out.println("w0, w1: " + weights);
60 | System.out.println("w3 (bias): " + bias);
61 |
62 | System.out.println("Result: " + results.get(1));
63 |
64 | }
65 | }
66 |
--------------------------------------------------------------------------------
/deeplearning4j/logistic/target/.gitignore:
--------------------------------------------------------------------------------
1 | /classes/
2 |
--------------------------------------------------------------------------------
/general/simplex-softmax.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 1,
6 | "metadata": {
7 | "collapsed": true
8 | },
9 | "outputs": [],
10 | "source": [
11 | "\"\"\"\n",
12 | "Softmax simplex visulaizer\n",
13 | "To show the relationshp between presoftmax \n",
14 | "values and the simplex projection\n",
15 | "\n",
16 | "Joseph Paul Cohen 2018\n",
17 | "Using code by David Andrzejewski\n",
18 | "\n",
19 | "\"\"\"\n",
20 | "\n",
21 | "import matplotlib\n",
22 | "import matplotlib.pyplot as plt\n",
23 | "%matplotlib inline\n",
24 | "import numpy as np"
25 | ]
26 | },
27 | {
28 | "cell_type": "code",
29 | "execution_count": 2,
30 | "metadata": {
31 | "collapsed": true
32 | },
33 | "outputs": [],
34 | "source": [
35 | "\"\"\"\n",
36 | "Visualize points on the 3-simplex (eg, the parameters of a\n",
37 | "3-dimensional multinomial distributions) as a scatter plot \n",
38 | "contained within a 2D triangle.\n",
39 | "\n",
40 | "David Andrzejewski (david.andrzej@gmail.com)\n",
41 | "\"\"\"\n",
42 | "import matplotlib.ticker as MT\n",
43 | "import matplotlib.lines as L\n",
44 | "\n",
45 | "def plotSimplex(points, fig=None, vertexlabels=['Class 1','Class 2','Class 3'], **kwargs):\n",
46 | " \"\"\"\n",
47 | " Plot Nx3 points array on the 3-simplex \n",
48 | " (with optionally labeled vertices) \n",
49 | " \n",
50 | " kwargs will be passed along directly to matplotlib.pyplot.scatter \n",
51 | "\n",
52 | " Returns Figure, caller must .show()\n",
53 | " \"\"\"\n",
54 | " if(fig == None): \n",
55 | " fig = plt.figure()\n",
56 | " # Draw the triangle\n",
57 | " l1 = L.Line2D([0, 0.5, 1.0, 0], # xcoords\n",
58 | " [0, np.sqrt(3) / 2, 0, 0], # ycoords\n",
59 | " color='k')\n",
60 | " fig.gca().add_line(l1)\n",
61 | " fig.gca().xaxis.set_major_locator(MT.NullLocator())\n",
62 | " fig.gca().yaxis.set_major_locator(MT.NullLocator())\n",
63 | " # Draw vertex labels\n",
64 | " fig.gca().text(-0.05, -0.05, vertexlabels[0])\n",
65 | " fig.gca().text(1.05, -0.05, vertexlabels[1])\n",
66 | " fig.gca().text(0.5, np.sqrt(3) / 2 + 0.05, vertexlabels[2])\n",
67 | " # Project and draw the actual points\n",
68 | " projected = projectSimplex(points)\n",
69 | " plt.scatter(projected[:,0], projected[:,1], **kwargs) ; \n",
70 | " # Leave some buffer around the triangle for vertex labels\n",
71 | " fig.gca().set_xlim(-0.2, 1.2)\n",
72 | " fig.gca().set_ylim(-0.2, 1.2)\n",
73 | "\n",
74 | " return fig \n",
75 | "\n",
76 | "def projectSimplex(points):\n",
77 | " \"\"\" \n",
78 | " Project probabilities on the 3-simplex to a 2D triangle\n",
79 | " \n",
80 | " N points are given as N x 3 array\n",
81 | " \"\"\"\n",
82 | " # Convert points one at a time\n",
83 | " tripts = np.zeros((points.shape[0],2))\n",
84 | " for idx in range(points.shape[0]):\n",
85 | " # Init to triangle centroid\n",
86 | " x = 1.0 / 2\n",
87 | " y = 1.0 / (2 * np.sqrt(3))\n",
88 | " # Vector 1 - bisect out of lower left vertex \n",
89 | " p1 = points[idx, 0]\n",
90 | " x = x - (1.0 / np.sqrt(3)) * p1 * np.cos(np.pi / 6)\n",
91 | " y = y - (1.0 / np.sqrt(3)) * p1 * np.sin(np.pi / 6)\n",
92 | " # Vector 2 - bisect out of lower right vertex \n",
93 | " p2 = points[idx, 1] \n",
94 | " x = x + (1.0 / np.sqrt(3)) * p2 * np.cos(np.pi / 6)\n",
95 | " y = y - (1.0 / np.sqrt(3)) * p2 * np.sin(np.pi / 6) \n",
96 | " # Vector 3 - bisect out of top vertex\n",
97 | " p3 = points[idx, 2]\n",
98 | " y = y + (1.0 / np.sqrt(3) * p3)\n",
99 | " \n",
100 | " tripts[idx,:] = (x,y)\n",
101 | "\n",
102 | " return tripts\n",
103 | "\n",
104 | "def softmax(x):\n",
105 | " e_x = np.exp(x - np.max(x))\n",
106 | " return e_x / e_x.sum(axis=0)"
107 | ]
108 | },
109 | {
110 | "cell_type": "code",
111 | "execution_count": null,
112 | "metadata": {
113 | "collapsed": true
114 | },
115 | "outputs": [],
116 | "source": []
117 | },
118 | {
119 | "cell_type": "code",
120 | "execution_count": null,
121 | "metadata": {
122 | "collapsed": true
123 | },
124 | "outputs": [],
125 | "source": []
126 | },
127 | {
128 | "cell_type": "code",
129 | "execution_count": null,
130 | "metadata": {
131 | "collapsed": true
132 | },
133 | "outputs": [],
134 | "source": []
135 | },
136 | {
137 | "cell_type": "code",
138 | "execution_count": 3,
139 | "metadata": {
140 | "collapsed": true
141 | },
142 | "outputs": [],
143 | "source": [
144 | "def plot(x, y, z):\n",
145 | " step=0\n",
146 | " dot = np.asarray([x, y, z])\n",
147 | " testpoints = [softmax([x, y, z])]\n",
148 | " \n",
149 | " steps = np.arange(0.1,1,0.1)\n",
150 | " for i1 in steps:\n",
151 | " i1 = -np.log(i1)\n",
152 | " testpoints.append(softmax([x+i1, y, z]))\n",
153 | " for i1 in steps:\n",
154 | " i1 = -np.log(i1)\n",
155 | " testpoints.append(softmax([x, y+i1, z]))\n",
156 | " for i1 in steps:\n",
157 | " i1 = -np.log(i1)\n",
158 | " testpoints.append(softmax([x, y, z+i1]))\n",
159 | " \n",
160 | "# steps = np.arange(0.0,step,0.1)\n",
161 | "# for i1 in steps:\n",
162 | "# for i2 in steps:\n",
163 | "# for i3 in steps:\n",
164 | "# if np.linalg.norm(np.subtract(dot, [x+i1, y+i2, z+i3]), 1).sum()"
212 | ]
213 | },
214 | "metadata": {},
215 | "output_type": "display_data"
216 | }
217 | ],
218 | "source": [
219 | "plt.rcParams['figure.figsize'] = (7, 7)\n",
220 | "x_widget = ipywidgets.FloatSlider(min=-4.0, max=4.0, step=0.05);\n",
221 | "y_widget = ipywidgets.FloatSlider(min=-4.0, max=4.0, step=0.05);\n",
222 | "z_widget = ipywidgets.FloatSlider(min=-4.0, max=4.0, step=0.05);\n",
223 | "#step = ipywidgets.FloatSlider(min=0, max=5, step=0.1);\n",
224 | "interact(plot,x=x_widget, y=y_widget, z=z_widget)#, step=step);"
225 | ]
226 | },
227 | {
228 | "cell_type": "code",
229 | "execution_count": null,
230 | "metadata": {
231 | "collapsed": true
232 | },
233 | "outputs": [],
234 | "source": []
235 | },
236 | {
237 | "cell_type": "code",
238 | "execution_count": null,
239 | "metadata": {
240 | "collapsed": true
241 | },
242 | "outputs": [],
243 | "source": []
244 | },
245 | {
246 | "cell_type": "code",
247 | "execution_count": null,
248 | "metadata": {
249 | "collapsed": true
250 | },
251 | "outputs": [],
252 | "source": []
253 | },
254 | {
255 | "cell_type": "code",
256 | "execution_count": null,
257 | "metadata": {
258 | "collapsed": true
259 | },
260 | "outputs": [],
261 | "source": []
262 | },
263 | {
264 | "cell_type": "code",
265 | "execution_count": null,
266 | "metadata": {
267 | "collapsed": true
268 | },
269 | "outputs": [],
270 | "source": []
271 | }
272 | ],
273 | "metadata": {
274 | "kernelspec": {
275 | "display_name": "Python 2",
276 | "language": "python",
277 | "name": "python2"
278 | },
279 | "language_info": {
280 | "codemirror_mode": {
281 | "name": "ipython",
282 | "version": 2
283 | },
284 | "file_extension": ".py",
285 | "mimetype": "text/x-python",
286 | "name": "python",
287 | "nbconvert_exporter": "python",
288 | "pygments_lexer": "ipython2",
289 | "version": "2.7.13"
290 | },
291 | "widgets": {
292 | "state": {},
293 | "version": "2.0.10"
294 | }
295 | },
296 | "nbformat": 4,
297 | "nbformat_minor": 2
298 | }
299 |
--------------------------------------------------------------------------------
/mxnet/logistic/mxnet-logistic-example.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 2,
6 | "metadata": {
7 | "collapsed": true
8 | },
9 | "outputs": [],
10 | "source": [
11 | "import mxnet as mx\n",
12 | "import cmath\n",
13 | "import numpy as np"
14 | ]
15 | },
16 | {
17 | "cell_type": "code",
18 | "execution_count": 100,
19 | "metadata": {
20 | "collapsed": false
21 | },
22 | "outputs": [
23 | {
24 | "name": "stdout",
25 | "output_type": "stream",
26 | "text": [
27 | " w0=2.000\n",
28 | " x0=-1.000\n",
29 | " w1=-3.000\n",
30 | " x1=-2.000\n",
31 | " w2=-3.000\n",
32 | " _divscalar34_output=0.731\n",
33 | " dout/dw0=-0.197\n",
34 | " dout/dx0=0.393\n",
35 | " dout/dw1=-0.393\n",
36 | " dout/dx1=-0.590\n",
37 | " dout/dw2=0.197\n"
38 | ]
39 | }
40 | ],
41 | "source": [
42 | "# Declare input values in mxnet type\n",
43 | "w0 = mx.symbol.Variable('w0')\n",
44 | "x0 = mx.symbol.Variable('x0')\n",
45 | "w1 = mx.symbol.Variable('w1')\n",
46 | "x1 = mx.symbol.Variable('x1')\n",
47 | "w2 = mx.symbol.Variable('w2')\n",
48 | "\n",
49 | "# Form expression using overloaded +,-,*, and / operators\n",
50 | "# Use special mx methods to achieve other operations\n",
51 | "s = 1/(1 + (mx.symbol.pow(cmath.e, -1*(w0*x0 + w1*x1 + w2))))\n",
52 | "\n",
53 | "# Specify inputs we declared \n",
54 | "args={'x0': mx.nd.array([-1.0]),\n",
55 | " 'x1': mx.nd.array([-2]),\n",
56 | " 'w0': mx.nd.array([2.0]),\n",
57 | " 'w1': mx.nd.array([-3.0]),\n",
58 | " 'w2': mx.nd.array([-3.0])\n",
59 | " }\n",
60 | "\n",
61 | "# Bind to symbol and create executor\n",
62 | "c_exec = s.simple_bind(\n",
63 | " ctx=mx.cpu(),\n",
64 | " x0 = args['x0'].shape,\n",
65 | " x1 = args['x1'].shape,\n",
66 | " w0 = args['w0'].shape, \n",
67 | " w1 = args['w1'].shape, \n",
68 | " w2 = args['w2'].shape)\n",
69 | "\n",
70 | "# Copy input values into executor memory\n",
71 | "c_exec.copy_params_from(arg_params = args)\n",
72 | "\n",
73 | "# Perform computation forward to populate outputs\n",
74 | "c_exec.forward()\n",
75 | "\n",
76 | "# Backpropagate to calculate gradients \n",
77 | "c_exec.backward(out_grads=mx.nd.array([1.0]))\n",
78 | "\n",
79 | "for k,v in zip(s.list_arguments(),c_exec.arg_arrays): print \"%20s=%.03f\"% (k,v.asnumpy())\n",
80 | "for k,v in zip(s.list_outputs(),c_exec.outputs): print \"%20s=%.03f\"% (k,v.asnumpy())\n",
81 | "for k,v in zip(s.list_arguments(),c_exec.grad_arrays): print \"%20s=%.03f\"% (\"dout/d%s\"%k,v.asnumpy())\n",
82 | " "
83 | ]
84 | },
85 | {
86 | "cell_type": "code",
87 | "execution_count": null,
88 | "metadata": {
89 | "collapsed": false
90 | },
91 | "outputs": [],
92 | "source": []
93 | },
94 | {
95 | "cell_type": "code",
96 | "execution_count": 101,
97 | "metadata": {
98 | "collapsed": false
99 | },
100 | "outputs": [
101 | {
102 | "data": {
103 | "image/svg+xml": [
104 | "\n",
105 | "\n",
107 | "\n",
109 | "\n",
110 | "\n"
242 | ],
243 | "text/plain": [
244 | ""
245 | ]
246 | },
247 | "execution_count": 101,
248 | "metadata": {},
249 | "output_type": "execute_result"
250 | }
251 | ],
252 | "source": [
253 | "# Print computation graph of s.get_internals() because is shows more nodes\n",
254 | "a = mx.viz.plot_network(s.get_internals(), shape={\"w0\":data_shape, \n",
255 | " \"x0\":data_shape,\n",
256 | " \"w1\":data_shape,\n",
257 | " \"x1\":data_shape,\n",
258 | " \"w2\":data_shape},\n",
259 | " node_attrs={\"shape\":'rect',\"fixedsize\":'false'})\n",
260 | "#Rotate the graphviz object that is returned\n",
261 | "a.body.extend(['rankdir=RL', 'size=\"10,5\"'])\n",
262 | "\n",
263 | "#Show it. Use a.render() to write it to disk\n",
264 | "a"
265 | ]
266 | },
267 | {
268 | "cell_type": "code",
269 | "execution_count": null,
270 | "metadata": {
271 | "collapsed": true
272 | },
273 | "outputs": [],
274 | "source": []
275 | },
276 | {
277 | "cell_type": "code",
278 | "execution_count": null,
279 | "metadata": {
280 | "collapsed": true
281 | },
282 | "outputs": [],
283 | "source": []
284 | }
285 | ],
286 | "metadata": {
287 | "kernelspec": {
288 | "display_name": "Python 2",
289 | "language": "python",
290 | "name": "python2"
291 | },
292 | "language_info": {
293 | "codemirror_mode": {
294 | "name": "ipython",
295 | "version": 2
296 | },
297 | "file_extension": ".py",
298 | "mimetype": "text/x-python",
299 | "name": "python",
300 | "nbconvert_exporter": "python",
301 | "pygments_lexer": "ipython2",
302 | "version": "2.7.10"
303 | }
304 | },
305 | "nbformat": 4,
306 | "nbformat_minor": 0
307 | }
308 |
--------------------------------------------------------------------------------
/mxnet/mxnet-vary-inputs.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "At the bottom of this notebook there is an interactive MXNet example. It Lets you vary the inputs with sliders and will compute outputs and gradients. You can even edit the network to make it more complex!\n",
8 | "\n",
9 | "A video of this working is here: https://www.youtube.com/watch?v=-KmImwP5eGk\n",
10 | "Joseph Paul Cohen 2016 (Code free for non-commercial use)\n",
11 | "\n",
12 | "# This version is old!\n",
13 | "The next version makes it easy to increase and decrease the number of parameters. Check out the next version here: [mxnet-vary-inputs-slideexamples.ipynb](https://github.com/ieee8023/NeuralNetwork-Examples/blob/master/mxnet/mxnet-vary-inputs-slideexamples.ipynb) "
14 | ]
15 | },
16 | {
17 | "cell_type": "code",
18 | "execution_count": 5,
19 | "metadata": {
20 | "collapsed": true
21 | },
22 | "outputs": [],
23 | "source": [
24 | "import mxnet as mx\n",
25 | "import cmath\n",
26 | "import numpy as np\n",
27 | "\n",
28 | "from __future__ import print_function\n",
29 | "from ipywidgets import interact, interactive, fixed\n",
30 | "import ipywidgets as widgets"
31 | ]
32 | },
33 | {
34 | "cell_type": "code",
35 | "execution_count": 6,
36 | "metadata": {
37 | "collapsed": false
38 | },
39 | "outputs": [],
40 | "source": [
41 | "def compute(s=None, x0=1, x1=1, x2=1, w0=1, w1=1, w2=1):\n",
42 | "\n",
43 | " # Specify inputs we declared \n",
44 | " args={'x0': mx.nd.array([x0]),\n",
45 | " 'x1': mx.nd.array([x1]),\n",
46 | " 'x2': mx.nd.array([x2]),\n",
47 | " 'w0': mx.nd.array([w0]),\n",
48 | " 'w1': mx.nd.array([w1]),\n",
49 | " 'w2': mx.nd.array([w2])\n",
50 | " }\n",
51 | "\n",
52 | "\n",
53 | " sym = s.get_internals()\n",
54 | " blob_names = sym.list_outputs()\n",
55 | " sym_group = []\n",
56 | " for i in range(len(blob_names)):\n",
57 | " if blob_names[i] not in args:\n",
58 | " x = sym[i]\n",
59 | " if blob_names[i] not in sym.list_outputs():\n",
60 | " x = mx.symbol.BlockGrad(x, name=blob_names[i])\n",
61 | " sym_group.append(x)\n",
62 | " sym = mx.symbol.Group(sym_group)\n",
63 | "\n",
64 | "\n",
65 | " # Bind to symbol and create executor\n",
66 | " c_exec = sym.simple_bind(\n",
67 | " ctx=mx.cpu(),\n",
68 | " x0 = args['x0'].shape,\n",
69 | " x1 = args['x1'].shape,\n",
70 | " w0 = args['w0'].shape, \n",
71 | " w1 = args['w1'].shape, \n",
72 | " x2 = args['x2'].shape,\n",
73 | " w2 = args['w2'].shape)\n",
74 | "\n",
75 | " # Copy input values into executor memory\n",
76 | " c_exec.copy_params_from(arg_params = args)\n",
77 | "\n",
78 | " # Perform computation forward to populate outputs\n",
79 | " c_exec.forward()\n",
80 | "\n",
81 | " values = []\n",
82 | " values = values + [(k,v.asnumpy()[0]) for k,v in zip(sym.list_arguments(),c_exec.arg_arrays)]\n",
83 | " values = values + [(k,v.asnumpy()[0]) for k,v in zip(sym.list_outputs(),c_exec.outputs)]\n",
84 | "\n",
85 | " # Bind to symbol and create executor\n",
86 | " c_exec = s.simple_bind(\n",
87 | " ctx=mx.cpu(),\n",
88 | " x0 = args['x0'].shape,\n",
89 | " x1 = args['x1'].shape,\n",
90 | " w0 = args['w0'].shape, \n",
91 | " w1 = args['w1'].shape, \n",
92 | " x2 = args['x2'].shape, \n",
93 | " w2 = args['w2'].shape)\n",
94 | "\n",
95 | " # Copy input values into executor memory\n",
96 | " c_exec.copy_params_from(arg_params = args)\n",
97 | "\n",
98 | " # Perform computation forward to populate outputs\n",
99 | " c_exec.forward()\n",
100 | "\n",
101 | " # Backpropagate to calculate gradients \n",
102 | " c_exec.backward(out_grads=mx.nd.array([1]))\n",
103 | "\n",
104 | " grads = []\n",
105 | " grads = grads + [(k,v.asnumpy()[0]) for k,v in zip(s.list_arguments(),c_exec.grad_arrays)]\n",
106 | "\n",
107 | " # Use these for debugging\n",
108 | " #for k,v in values: print(\"%20s=%.03f\"% (k,v))\n",
109 | " #for k,v in grads: print(\"%20s=%.03f\"% (\"dout/d%s\"%k,v))\n",
110 | " \n",
111 | " values_dict = dict(values)\n",
112 | " grads_dict = dict(grads)\n",
113 | "\n",
114 | " # Print computation graph of s.get_internals() because is shows more nodes\n",
115 | " a = plot_network2(sym, shape={\n",
116 | " \"w0\":(1,), \n",
117 | " \"x0\":(1,),\n",
118 | " \"w1\":(1,),\n",
119 | " \"x1\":(1,),\n",
120 | " \"x2\":(1,),\n",
121 | " \"w2\":(1,)},\n",
122 | " node_attrs={\"shape\":'rect',\"fixedsize\":'false'},\n",
123 | " values_dict=values_dict, grads_dict=grads_dict)\n",
124 | " #Rotate the graphviz object that is returned\n",
125 | " a.body.extend(['rankdir=RL', 'size=\"10,5\"'])\n",
126 | "\n",
127 | " del c_exec\n",
128 | " del sym\n",
129 | " \n",
130 | " #Show it. Use a.render() to write it to disk\n",
131 | " return a"
132 | ]
133 | },
134 | {
135 | "cell_type": "code",
136 | "execution_count": 7,
137 | "metadata": {
138 | "collapsed": false
139 | },
140 | "outputs": [],
141 | "source": [
142 | "## Here we define a new print network function\n",
143 | "\n",
144 | "from __future__ import absolute_import\n",
145 | "from mxnet.symbol import Symbol\n",
146 | "import json\n",
147 | "import re\n",
148 | "import copy\n",
149 | "\n",
150 | "def plot_network2(symbol, title=\"plot\", shape=None, node_attrs={}, values_dict=None, grads_dict=None):\n",
151 | " try:\n",
152 | " from graphviz import Digraph\n",
153 | " except:\n",
154 | " raise ImportError(\"Draw network requires graphviz library\")\n",
155 | " if not isinstance(symbol, Symbol):\n",
156 | " raise TypeError(\"symbol must be Symbol\")\n",
157 | " draw_shape = False\n",
158 | " if shape != None:\n",
159 | " draw_shape = True\n",
160 | " interals = symbol.get_internals()\n",
161 | " _, out_shapes, _ = interals.infer_shape(**shape)\n",
162 | " if out_shapes == None:\n",
163 | " raise ValueError(\"Input shape is incompete\")\n",
164 | " shape_dict = dict(zip(interals.list_outputs(), out_shapes))\n",
165 | " conf = json.loads(symbol.tojson())\n",
166 | " nodes = conf[\"nodes\"]\n",
167 | " #print(conf)\n",
168 | " heads = set([x[0] for x in conf[\"heads\"]]) # TODO(xxx): check careful\n",
169 | " #print(heads)\n",
170 | " # default attributes of node\n",
171 | " node_attr = {\"shape\": \"box\", \"fixedsize\": \"true\",\n",
172 | " \"width\": \"1.3\", \"height\": \"0.8034\", \"style\": \"filled\"}\n",
173 | " # merge the dcit provided by user and the default one\n",
174 | " node_attr.update(node_attrs)\n",
175 | " dot = Digraph(name=title)\n",
176 | " dot.body.extend(['rankdir=RL', 'size=\"10,5\"'])\n",
177 | " # color map\n",
178 | " cm = (\"#8dd3c7\", \"#fb8072\", \"#ffffb3\", \"#bebada\", \"#80b1d3\",\n",
179 | " \"#fdb462\", \"#b3de69\", \"#fccde5\")\n",
180 | "\n",
181 | " # make nodes\n",
182 | " for i in range(len(nodes)):\n",
183 | " node = nodes[i]\n",
184 | " op = node[\"op\"]\n",
185 | " name = node[\"name\"]\n",
186 | " # input data\n",
187 | " attr = copy.deepcopy(node_attr)\n",
188 | " label = op\n",
189 | "\n",
190 | " if op == \"null\":\n",
191 | " label = node[\"name\"]\n",
192 | " if grads_dict != None and label in grads_dict:\n",
193 | " label = label + (\"\\n d%s: %.2f\" % (label, grads_dict[label]))\n",
194 | " \n",
195 | " attr[\"fillcolor\"] = cm[1]\n",
196 | " \n",
197 | " if op == \"Convolution\":\n",
198 | " label = \"Convolution\\n%sx%s/%s, %s\" % (_str2tuple(node[\"param\"][\"kernel\"])[0],\n",
199 | " _str2tuple(node[\"param\"][\"kernel\"])[1],\n",
200 | " _str2tuple(node[\"param\"][\"stride\"])[0],\n",
201 | " node[\"param\"][\"num_filter\"])\n",
202 | " attr[\"fillcolor\"] = cm[1]\n",
203 | " elif op == \"FullyConnected\":\n",
204 | " label = \"FullyConnected\\n%s\" % node[\"param\"][\"num_hidden\"]\n",
205 | " attr[\"fillcolor\"] = cm[1]\n",
206 | " elif op == \"BatchNorm\":\n",
207 | " attr[\"fillcolor\"] = cm[3]\n",
208 | " elif op == \"Activation\" or op == \"LeakyReLU\":\n",
209 | " label = \"%s\\n%s\" % (op, node[\"param\"][\"act_type\"])\n",
210 | " attr[\"fillcolor\"] = cm[2]\n",
211 | " elif op == \"Pooling\":\n",
212 | " label = \"Pooling\\n%s, %sx%s/%s\" % (node[\"param\"][\"pool_type\"],\n",
213 | " _str2tuple(node[\"param\"][\"kernel\"])[0],\n",
214 | " _str2tuple(node[\"param\"][\"kernel\"])[1],\n",
215 | " _str2tuple(node[\"param\"][\"stride\"])[0])\n",
216 | " attr[\"fillcolor\"] = cm[4]\n",
217 | " elif op == \"Concat\" or op == \"Flatten\" or op == \"Reshape\":\n",
218 | " attr[\"fillcolor\"] = cm[5]\n",
219 | " elif op == \"Softmax\":\n",
220 | " attr[\"fillcolor\"] = cm[6]\n",
221 | " else:\n",
222 | " attr[\"fillcolor\"] = cm[0]\n",
223 | "\n",
224 | " dot.node(name=name, label=label, **attr)\n",
225 | " \n",
226 | " # add edges\n",
227 | " for i in range(len(nodes)):\n",
228 | " node = nodes[i]\n",
229 | " op = node[\"op\"]\n",
230 | " name = node[\"name\"]\n",
231 | " inputs = node[\"inputs\"]\n",
232 | " for item in inputs:\n",
233 | " input_node = nodes[item[0]]\n",
234 | " input_name = input_node[\"name\"]\n",
235 | " attr = {\"dir\": \"back\", 'arrowtail':'open'}\n",
236 | " \n",
237 | " label = \"\"\n",
238 | " if values_dict != None and input_name in values_dict:\n",
239 | " label = \"%.2f\" % values_dict[input_name] \n",
240 | " \n",
241 | " if values_dict != None and input_name + \"_output\" in values_dict:\n",
242 | " label = \"%.2f\" % values_dict[input_name + \"_output\"]\n",
243 | " \n",
244 | " #if grads_dict != None and input_name in grads_dict:\n",
245 | " # label = label + (\"/%.2f\" %grads_dict[input_name])\n",
246 | " \n",
247 | " attr[\"label\"] = label.replace(\"_\",\"\")\n",
248 | " dot.edge(tail_name=name, head_name=input_name, **attr)\n",
249 | " return dot"
250 | ]
251 | },
252 | {
253 | "cell_type": "code",
254 | "execution_count": 8,
255 | "metadata": {
256 | "collapsed": false
257 | },
258 | "outputs": [
259 | {
260 | "data": {
261 | "image/svg+xml": [
262 | "\n",
263 | "\n",
265 | "\n",
267 | "\n",
268 | "\n"
419 | ],
420 | "text/plain": [
421 | ""
422 | ]
423 | },
424 | "metadata": {},
425 | "output_type": "display_data"
426 | },
427 | {
428 | "data": {
429 | "text/plain": [
430 | ""
431 | ]
432 | },
433 | "execution_count": 8,
434 | "metadata": {},
435 | "output_type": "execute_result"
436 | }
437 | ],
438 | "source": [
439 | "## Outputs from a node are shown on the edges and the gradients are shown in the box\n",
440 | "## Modify the s object in compute different functions\n",
441 | "\n",
442 | "# Declare input values in mxnet type\n",
443 | "w0 = mx.symbol.Variable('w0')\n",
444 | "x0 = mx.symbol.Variable('x0')\n",
445 | "w1 = mx.symbol.Variable('w1')\n",
446 | "x1 = mx.symbol.Variable('x1')\n",
447 | "w2 = mx.symbol.Variable('w2')\n",
448 | "x2 = mx.symbol.Variable('x2')\n",
449 | "\n",
450 | "# Form expression using overloaded +,-,*, and / operators\n",
451 | "# Use special mx methods to achieve other operations\n",
452 | "n = ((w0*x0 + w1*x1) * x2 + w2)\n",
453 | "s = mx.symbol.Activation(data=n, name='relu1', act_type=\"relu\")+0\n",
454 | "\n",
455 | "interact(compute, s=fixed(s), x0=1.0, w0=1.0, x1=1.0, w1=1.0, x2=1.0, w2=1.0)"
456 | ]
457 | },
458 | {
459 | "cell_type": "code",
460 | "execution_count": null,
461 | "metadata": {
462 | "collapsed": false
463 | },
464 | "outputs": [],
465 | "source": []
466 | },
467 | {
468 | "cell_type": "code",
469 | "execution_count": 9,
470 | "metadata": {
471 | "collapsed": false
472 | },
473 | "outputs": [
474 | {
475 | "data": {
476 | "image/svg+xml": [
477 | "\n",
478 | "\n",
480 | "\n",
482 | "\n",
483 | "\n"
685 | ],
686 | "text/plain": [
687 | ""
688 | ]
689 | },
690 | "metadata": {},
691 | "output_type": "display_data"
692 | },
693 | {
694 | "data": {
695 | "text/plain": [
696 | ""
697 | ]
698 | },
699 | "execution_count": 9,
700 | "metadata": {},
701 | "output_type": "execute_result"
702 | }
703 | ],
704 | "source": [
705 | "## Outputs from a node are shown on the edges and the gradients are shown in the box\n",
706 | "## Modify the s object in compute different functions\n",
707 | "\n",
708 | "# Declare input values in mxnet type\n",
709 | "w0 = mx.symbol.Variable('w0')\n",
710 | "x0 = mx.symbol.Variable('x0')\n",
711 | "w1 = mx.symbol.Variable('w1')\n",
712 | "x1 = mx.symbol.Variable('x1')\n",
713 | "w2 = mx.symbol.Variable('w2')\n",
714 | "x2 = mx.symbol.Variable('x2')\n",
715 | "\n",
716 | "# Form expression using overloaded +,-,*, and / operators\n",
717 | "# Use special mx methods to achieve other operations\n",
718 | "n = ((w0*x0*x2 + w1*x1*x2*x0) * x2*w2)\n",
719 | "s = mx.symbol.Activation(data=n, name='relu1', act_type=\"relu\")+0\n",
720 | "\n",
721 | "interact(compute, s=fixed(s), x0=1.0, w0=1.0, x1=1.0, w1=1.0, x2=1.0, w2=1.0)"
722 | ]
723 | },
724 | {
725 | "cell_type": "code",
726 | "execution_count": null,
727 | "metadata": {
728 | "collapsed": true
729 | },
730 | "outputs": [],
731 | "source": []
732 | },
733 | {
734 | "cell_type": "code",
735 | "execution_count": null,
736 | "metadata": {
737 | "collapsed": true
738 | },
739 | "outputs": [],
740 | "source": []
741 | },
742 | {
743 | "cell_type": "code",
744 | "execution_count": null,
745 | "metadata": {
746 | "collapsed": true
747 | },
748 | "outputs": [],
749 | "source": []
750 | }
751 | ],
752 | "metadata": {
753 | "kernelspec": {
754 | "display_name": "Python 2",
755 | "language": "python",
756 | "name": "python2"
757 | },
758 | "language_info": {
759 | "codemirror_mode": {
760 | "name": "ipython",
761 | "version": 2
762 | },
763 | "file_extension": ".py",
764 | "mimetype": "text/x-python",
765 | "name": "python",
766 | "nbconvert_exporter": "python",
767 | "pygments_lexer": "ipython2",
768 | "version": "2.7.10"
769 | }
770 | },
771 | "nbformat": 4,
772 | "nbformat_minor": 0
773 | }
774 |
--------------------------------------------------------------------------------
/mxnet/randomout/data/crater/All-Fold1-test.rec:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ieee8023/NeuralNetwork-Examples/98677d0eb06eba8b511251c99826319fd0b17bc1/mxnet/randomout/data/crater/All-Fold1-test.rec
--------------------------------------------------------------------------------
/mxnet/randomout/data/crater/All-Fold1-train.rec:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ieee8023/NeuralNetwork-Examples/98677d0eb06eba8b511251c99826319fd0b17bc1/mxnet/randomout/data/crater/All-Fold1-train.rec
--------------------------------------------------------------------------------
/mxnet/randomout/get_data.py:
--------------------------------------------------------------------------------
1 | # pylint: skip-file
2 | import os, gzip
3 | import pickle as pickle
4 | import sys
5 |
6 | # download mnist.pkl.gz
7 | def GetMNIST_pkl():
8 | if not os.path.isdir("data/"):
9 | os.system("mkdir data/")
10 | if not os.path.exists('data/mnist.pkl.gz'):
11 | os.system("wget http://deeplearning.net/data/mnist/mnist.pkl.gz -P data/")
12 |
13 | # download ubyte version of mnist and untar
14 | def GetMNIST_ubyte():
15 | if not os.path.isdir("data/"):
16 | os.system("mkdir data/")
17 | if (not os.path.exists('data/train-images-idx3-ubyte')) or \
18 | (not os.path.exists('data/train-labels-idx1-ubyte')) or \
19 | (not os.path.exists('data/t10k-images-idx3-ubyte')) or \
20 | (not os.path.exists('data/t10k-labels-idx1-ubyte')):
21 | os.system("wget http://webdocs.cs.ualberta.ca/~bx3/data/mnist.zip -P data/")
22 | os.chdir("./data")
23 | os.system("unzip -u mnist.zip")
24 | os.chdir("..")
25 |
26 | # download cifar
27 | def GetCifar10():
28 | if not os.path.isdir("data/"):
29 | os.system("mkdir data/")
30 | if not os.path.exists('data/cifar10.zip'):
31 | os.system("wget http://webdocs.cs.ualberta.ca/~bx3/data/cifar10.zip -P data/")
32 | os.chdir("./data")
33 | os.system("unzip -u cifar10.zip")
34 | os.chdir("..")
35 |
--------------------------------------------------------------------------------
/mxnet/randomout/randomout-cifar-inception.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "metadata": {},
6 | "source": [
7 | "## RandomOut implementation for MXNet\n",
8 | "This notebook is a demo of the RandomOut algorithm. It is implemented as a Monitor that can be passed to the fit method of FeedForward model object. Every epoch the monitor will be invoked and test that every convolutional filter has a CGN value greater than the tau value passed in. If a filter fails the check then it is reinitialized using the initializer from the model.\n",
9 | "\n",
10 | "The code is set up to train the 28x28 inception arch on the CIFAR-10 dataset. It can be run on multiple GPUs by setting the num_devs variable.\n",
11 | "\n",
12 | "Using the default script parameters (on 8 GPUs) after 20 epochs we achieve the following testing accuracy:\n",
13 | "+ wo/RandomOut = 0.7075\n",
14 | "+ w/RandomOut = 0.7929\n",
15 | "\n",
16 | "Paper: https://arxiv.org/abs/1602.05931\n",
17 | "\n",
18 | "ShortScience.org: http://www.shortscience.org/paper?bibtexKey=journals/corr/CohenL016\n",
19 | "\n",
20 | "This nodebook can be run from the command line using: \n",
21 | "\n",
22 | " jupyter nbconvert randomout-cifar-inception.ipynb --to script\n",
23 | " python randomout-cifar-inception.py\n"
24 | ]
25 | },
26 | {
27 | "cell_type": "code",
28 | "execution_count": null,
29 | "metadata": {
30 | "collapsed": false
31 | },
32 | "outputs": [],
33 | "source": [
34 | "import mxnet as mx"
35 | ]
36 | },
37 | {
38 | "cell_type": "code",
39 | "execution_count": null,
40 | "metadata": {
41 | "collapsed": false
42 | },
43 | "outputs": [],
44 | "source": [
45 | "import numpy as np\n",
46 | "import cmath\n",
47 | "import graphviz\n",
48 | "import argparse\n",
49 | "import os, sys"
50 | ]
51 | },
52 | {
53 | "cell_type": "code",
54 | "execution_count": null,
55 | "metadata": {
56 | "collapsed": false
57 | },
58 | "outputs": [],
59 | "source": [
60 | "parser = argparse.ArgumentParser()\n",
61 | "parser.add_argument('--seed', type=int, default=1)\n",
62 | "parser.add_argument('--epochs', type=int, default=4)\n",
63 | "parser.add_argument('--batch-size', type=int, default=128)\n",
64 | "parser.add_argument('--tau', type=float, default=1e-30)\n",
65 | "parser.add_argument('--randomout', type=str, default=\"True\")\n",
66 | "parser.add_argument('--network', type=str, default=\"inception-28-small\")\n",
67 | "parser.add_argument('-f', type=str, default='')\n",
68 | "args = parser.parse_args()\n",
69 | "args.f = ''\n",
70 | "\n",
71 | "# setup logging\n",
72 | "import logging\n",
73 | "logging.getLogger().handlers = []\n",
74 | "logger = logging.getLogger()\n",
75 | "logger.setLevel(logging.DEBUG)\n",
76 | "#logging.root = logging.getLogger(str(args))\n",
77 | "logging.root = logging.getLogger()\n",
78 | "logging.debug(\"test\")"
79 | ]
80 | },
81 | {
82 | "cell_type": "code",
83 | "execution_count": null,
84 | "metadata": {
85 | "collapsed": false
86 | },
87 | "outputs": [],
88 | "source": [
89 | "import importlib\n",
90 | "softmax = importlib.import_module('symbol_' + args.network).get_symbol(10)"
91 | ]
92 | },
93 | {
94 | "cell_type": "code",
95 | "execution_count": null,
96 | "metadata": {
97 | "collapsed": false
98 | },
99 | "outputs": [],
100 | "source": [
101 | "# If you'd like to see the network structure, run the plot_network function\n",
102 | "a = mx.viz.plot_network(symbol=softmax.get_internals(),node_attrs={'shape':'rect','fixedsize':'false'},\n",
103 | " shape={\"data\":(1,3, 28, 28)}) \n",
104 | "\n",
105 | "a.body.extend(['rankdir=RL', 'size=\"40,5\"'])\n",
106 | "#a"
107 | ]
108 | },
109 | {
110 | "cell_type": "code",
111 | "execution_count": null,
112 | "metadata": {
113 | "collapsed": false
114 | },
115 | "outputs": [],
116 | "source": [
117 | "mx.random.seed(args.seed)\n",
118 | "num_epoch = args.epochs\n",
119 | "batch_size = args.batch_size\n",
120 | "num_devs = 1\n",
121 | "model = mx.model.FeedForward(ctx=[mx.gpu(i) for i in range(num_devs)], symbol=softmax, num_epoch = num_epoch,\n",
122 | " learning_rate=0.1, momentum=0.9, wd=0.00001\n",
123 | " ,optimizer=mx.optimizer.Adam()\n",
124 | " )"
125 | ]
126 | },
127 | {
128 | "cell_type": "code",
129 | "execution_count": null,
130 | "metadata": {
131 | "collapsed": false
132 | },
133 | "outputs": [],
134 | "source": [
135 | "import get_data\n",
136 | "get_data.GetCifar10()\n",
137 | "\n",
138 | "train_dataiter = mx.io.ImageRecordIter(\n",
139 | " shuffle=True,\n",
140 | " path_imgrec=\"data/cifar/train.rec\",\n",
141 | " mean_img=\"data/cifar/cifar_mean.bin\",\n",
142 | " rand_crop=False,\n",
143 | " rand_mirror=False,\n",
144 | " data_shape=(3,28,28),\n",
145 | " batch_size=batch_size,\n",
146 | " preprocess_threads=4)\n",
147 | "# test iterator make batch of 128 image, and center crop each image into 3x28x28 from original 3x32x32\n",
148 | "# Note: We don't need round batch in test because we only test once at one time\n",
149 | "test_dataiter = mx.io.ImageRecordIter(\n",
150 | " path_imgrec=\"data/cifar/test.rec\",\n",
151 | " mean_img=\"data/cifar/cifar_mean.bin\",\n",
152 | " rand_crop=False,\n",
153 | " rand_mirror=False,\n",
154 | " data_shape=(3,28,28),\n",
155 | " batch_size=batch_size,\n",
156 | " round_batch=False,\n",
157 | " preprocess_threads=4)\n"
158 | ]
159 | },
160 | {
161 | "cell_type": "code",
162 | "execution_count": null,
163 | "metadata": {
164 | "collapsed": false
165 | },
166 | "outputs": [],
167 | "source": []
168 | },
169 | {
170 | "cell_type": "code",
171 | "execution_count": null,
172 | "metadata": {
173 | "collapsed": false
174 | },
175 | "outputs": [],
176 | "source": [
177 | "from mxnet.ndarray import NDArray\n",
178 | "from mxnet.base import NDArrayHandle\n",
179 | "from mxnet import ndarray\n",
180 | "\n",
181 | "class RandomOutMonitor(mx.monitor.Monitor):\n",
182 | " \n",
183 | " def __init__(self, initializer, network, tau=0.000001, *args,**kwargs):\n",
184 | " mx.monitor.Monitor.__init__(self, 1, *args, **kwargs) \n",
185 | " self.tau = tau\n",
186 | " self.initializer = initializer\n",
187 | " \n",
188 | " # here the layers we want to subject to the threshold are specified\n",
189 | " targetlayers = [x for x in network.list_arguments() if x.startswith(\"conv\") and x.endswith(\"weight\")]\n",
190 | " self.targetlayers = targetlayers\n",
191 | " \n",
192 | " logging.info(\"RandomOut active on layers: %s\" % self.targetlayers)\n",
193 | " \n",
194 | " def toc(self):\n",
195 | " for exe in self.exes:\n",
196 | " for array in exe.arg_arrays:\n",
197 | " array.wait_to_read()\n",
198 | " for exe in self.exes:\n",
199 | " for name, array in zip(exe._symbol.list_arguments(), exe.arg_arrays):\n",
200 | " self.queue.append((self.step, name, self.stat_func(array)))\n",
201 | " \n",
202 | " for exe in self.exes:\n",
203 | " weights = dict(zip(softmax.list_arguments(), exe.arg_arrays))\n",
204 | " grads = dict(zip(softmax.list_arguments(), exe.grad_arrays))\n",
205 | " numFilters = 0\n",
206 | " for name in self.targetlayers:\n",
207 | " \n",
208 | " filtersg = grads[name].asnumpy()\n",
209 | " filtersw = weights[name].asnumpy()\n",
210 | "\n",
211 | " #get random array to copy over\n",
212 | " filtersw_rand = mx.nd.array(filtersw.copy())\n",
213 | " self.initializer(name, filtersw_rand)\n",
214 | " filtersw_rand = filtersw_rand.asnumpy()\n",
215 | " \n",
216 | " agrads = [0.0] * len(filtersg)\n",
217 | " for i in range(len(filtersg)):\n",
218 | " agrads[i] = np.absolute(filtersg[i]).sum()\n",
219 | " if agrads[i] < self.tau:\n",
220 | " numFilters = numFilters+1\n",
221 | " #logging.info(\"RandomOut: filter %i of %s has been randomized because CGN=%f\" % (i,name,agrads[i]))\n",
222 | " filtersw[i] = filtersw_rand[i]\n",
223 | "\n",
224 | " #logging.info(\"%s, %s, %s\" % (name, min(agrads),np.mean(agrads)))\n",
225 | " \n",
226 | " weights[name] = mx.nd.array(filtersw)\n",
227 | " #print filtersw\n",
228 | " if numFilters >0:\n",
229 | " #logging.info(\"numFilters replaced: %i\"%numFilters) \n",
230 | " exe.copy_params_from(arg_params=weights)\n",
231 | " \n",
232 | " self.activated = False\n",
233 | " return []\n",
234 | " "
235 | ]
236 | },
237 | {
238 | "cell_type": "code",
239 | "execution_count": null,
240 | "metadata": {
241 | "collapsed": false
242 | },
243 | "outputs": [],
244 | "source": []
245 | },
246 | {
247 | "cell_type": "code",
248 | "execution_count": null,
249 | "metadata": {
250 | "collapsed": false
251 | },
252 | "outputs": [],
253 | "source": [
254 | "train_dataiter.reset()\n",
255 | "if args.randomout == \"True\":\n",
256 | " model.fit(X=train_dataiter,\n",
257 | " eval_data=test_dataiter,\n",
258 | " eval_metric=\"accuracy\",\n",
259 | " batch_end_callback=mx.callback.Speedometer(batch_size)\n",
260 | " ,monitor=RandomOutMonitor(initializer = model.initializer, network=softmax, tau=args.tau)\n",
261 | " )\n",
262 | "else:\n",
263 | " model.fit(X=train_dataiter,\n",
264 | " eval_data=test_dataiter,\n",
265 | " eval_metric=\"accuracy\",\n",
266 | " batch_end_callback=mx.callback.Speedometer(batch_size)\n",
267 | " )\n"
268 | ]
269 | },
270 | {
271 | "cell_type": "code",
272 | "execution_count": null,
273 | "metadata": {
274 | "collapsed": false
275 | },
276 | "outputs": [],
277 | "source": []
278 | },
279 | {
280 | "cell_type": "code",
281 | "execution_count": null,
282 | "metadata": {
283 | "collapsed": true
284 | },
285 | "outputs": [],
286 | "source": []
287 | }
288 | ],
289 | "metadata": {
290 | "kernelspec": {
291 | "display_name": "Python 2",
292 | "language": "python",
293 | "name": "python2"
294 | },
295 | "language_info": {
296 | "codemirror_mode": {
297 | "name": "ipython",
298 | "version": 2
299 | },
300 | "file_extension": ".py",
301 | "mimetype": "text/x-python",
302 | "name": "python",
303 | "nbconvert_exporter": "python",
304 | "pygments_lexer": "ipython2",
305 | "version": "2.7.6"
306 | }
307 | },
308 | "nbformat": 4,
309 | "nbformat_minor": 0
310 | }
311 |
--------------------------------------------------------------------------------
/mxnet/randomout/symbol_cratercnn.py:
--------------------------------------------------------------------------------
1 | """
2 | simplified inception-bn.py for images has size around 15 x 15
3 | """
4 |
5 | import mxnet as mx
6 |
7 | def ConvFactory(data, num_filter, kernel, stride=(1,1), pad=(0, 0), act_type="relu"):
8 | conv = mx.symbol.Convolution(data=data, num_filter=num_filter, kernel=kernel, stride=stride, pad=pad)
9 | #bn = mx.symbol.BatchNorm(data=conv)
10 | act = mx.symbol.Activation(data = conv, act_type=act_type)
11 | return act
12 |
13 | def get_symbol(num_classes = 2, num_filter=20):
14 | data = mx.symbol.Variable(name="data")
15 | #data = data/255
16 | conv1 = ConvFactory(data=data, kernel=(4,4), pad=(1,1), num_filter=num_filter, act_type="relu")
17 | conv2 = ConvFactory(data=conv1, kernel=(4,4), pad=(1,1), num_filter=num_filter, act_type="relu")
18 | flatten = mx.symbol.Flatten(data=conv2, name="flatten1")
19 | fc1 = mx.symbol.FullyConnected(data=flatten, num_hidden=500, name="fc1")
20 | fc2 = mx.symbol.FullyConnected(data=fc1, num_hidden=num_classes, name="fc2")
21 | softmax = mx.symbol.SoftmaxOutput(data=fc2, name="softmax")
22 | return softmax
23 |
--------------------------------------------------------------------------------
/mxnet/randomout/symbol_inception-28-small.py:
--------------------------------------------------------------------------------
1 | """
2 | simplified inception-.py for images has size around 28 x 28
3 | """
4 |
5 |
6 | import mxnet as mx
7 |
8 | # Basic Conv + BN + ReLU factory
9 | def ConvFactory(data, num_filter, kernel, stride=(1,1), pad=(0, 0), act_type="relu"):
10 | conv = mx.symbol.Convolution(data=data, num_filter=num_filter, kernel=kernel, stride=stride, pad=pad)
11 | #bn = mx.symbol.BatchNorm(data=conv)
12 | act = mx.symbol.Activation(data = conv, act_type=act_type)
13 | return act
14 |
15 | # A Simple Downsampling Factory
16 | def DownsampleFactory(data, ch_3x3):
17 | # conv 3x3
18 | conv = ConvFactory(data=data, kernel=(3, 3), stride=(2, 2), num_filter=ch_3x3, pad=(1, 1))
19 | # pool
20 | pool = mx.symbol.Pooling(data=data, kernel=(3, 3), stride=(2, 2), pool_type='max')
21 | # concat
22 | concat = mx.symbol.Concat(*[conv, pool])
23 | return concat
24 |
25 | # A Simple module
26 | def SimpleFactory(data, ch_1x1, ch_3x3):
27 | # 1x1
28 | conv1x1 = ConvFactory(data=data, kernel=(1, 1), pad=(0, 0), num_filter=ch_1x1)
29 | # 3x3
30 | conv3x3 = ConvFactory(data=data, kernel=(3, 3), pad=(1, 1), num_filter=ch_3x3)
31 | #concat
32 | concat = mx.symbol.Concat(*[conv1x1, conv3x3])
33 | return concat
34 |
35 | def get_symbol(num_classes = 10):
36 | data = mx.symbol.Variable(name="data")
37 | conv1 = ConvFactory(data=data, kernel=(3,3), pad=(1,1), num_filter=96, act_type="relu")
38 | in3a = SimpleFactory(conv1, 32, 32)
39 | in3b = SimpleFactory(in3a, 32, 48)
40 | in3c = DownsampleFactory(in3b, 80)
41 | in4a = SimpleFactory(in3c, 112, 48)
42 | in4b = SimpleFactory(in4a, 96, 64)
43 | in4c = SimpleFactory(in4b, 80, 80)
44 | in4d = SimpleFactory(in4c, 48, 96)
45 | in4e = DownsampleFactory(in4d, 96)
46 | in5a = SimpleFactory(in4e, 176, 160)
47 | in5b = SimpleFactory(in5a, 176, 160)
48 | pool = mx.symbol.Pooling(data=in5b, pool_type="avg", kernel=(7,7), name="global_pool")
49 | flatten = mx.symbol.Flatten(data=pool, name="flatten1")
50 | fc = mx.symbol.FullyConnected(data=flatten, num_hidden=num_classes, name="fc1")
51 | softmax = mx.symbol.SoftmaxOutput(data=fc, name="softmax")
52 | return softmax
53 |
54 |
--------------------------------------------------------------------------------
/mxnet/randomout/symbol_inception-bn-28-small.py:
--------------------------------------------------------------------------------
1 | """
2 | simplified inception-bn.py for images has size around 28 x 28
3 | """
4 |
5 |
6 | import mxnet as mx
7 |
8 | # Basic Conv + BN + ReLU factory
9 | def ConvFactory(data, num_filter, kernel, stride=(1,1), pad=(0, 0), act_type="relu"):
10 | conv = mx.symbol.Convolution(data=data, num_filter=num_filter, kernel=kernel, stride=stride, pad=pad)
11 | bn = mx.symbol.BatchNorm(data=conv)
12 | act = mx.symbol.Activation(data = bn, act_type=act_type)
13 | return act
14 |
15 | # A Simple Downsampling Factory
16 | def DownsampleFactory(data, ch_3x3):
17 | # conv 3x3
18 | conv = ConvFactory(data=data, kernel=(3, 3), stride=(2, 2), num_filter=ch_3x3, pad=(1, 1))
19 | # pool
20 | pool = mx.symbol.Pooling(data=data, kernel=(3, 3), stride=(2, 2), pool_type='max')
21 | # concat
22 | concat = mx.symbol.Concat(*[conv, pool])
23 | return concat
24 |
25 | # A Simple module
26 | def SimpleFactory(data, ch_1x1, ch_3x3):
27 | # 1x1
28 | conv1x1 = ConvFactory(data=data, kernel=(1, 1), pad=(0, 0), num_filter=ch_1x1)
29 | # 3x3
30 | conv3x3 = ConvFactory(data=data, kernel=(3, 3), pad=(1, 1), num_filter=ch_3x3)
31 | #concat
32 | concat = mx.symbol.Concat(*[conv1x1, conv3x3])
33 | return concat
34 |
35 | def get_symbol(num_classes = 10):
36 | data = mx.symbol.Variable(name="data")
37 | conv1 = ConvFactory(data=data, kernel=(3,3), pad=(1,1), num_filter=96, act_type="relu")
38 | in3a = SimpleFactory(conv1, 32, 32)
39 | in3b = SimpleFactory(in3a, 32, 48)
40 | in3c = DownsampleFactory(in3b, 80)
41 | in4a = SimpleFactory(in3c, 112, 48)
42 | in4b = SimpleFactory(in4a, 96, 64)
43 | in4c = SimpleFactory(in4b, 80, 80)
44 | in4d = SimpleFactory(in4c, 48, 96)
45 | in4e = DownsampleFactory(in4d, 96)
46 | in5a = SimpleFactory(in4e, 176, 160)
47 | in5b = SimpleFactory(in5a, 176, 160)
48 | pool = mx.symbol.Pooling(data=in5b, pool_type="avg", kernel=(7,7), name="global_pool")
49 | flatten = mx.symbol.Flatten(data=pool, name="flatten1")
50 | fc = mx.symbol.FullyConnected(data=flatten, num_hidden=num_classes, name="fc1")
51 | softmax = mx.symbol.SoftmaxOutput(data=fc, name="softmax")
52 | return softmax
53 |
54 |
--------------------------------------------------------------------------------
/pytorch/pytorch-logistic-regression.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 1,
6 | "metadata": {
7 | "collapsed": true
8 | },
9 | "outputs": [],
10 | "source": [
11 | "import torch\n",
12 | "from torch.autograd import Variable\n",
13 | "import numpy as np"
14 | ]
15 | },
16 | {
17 | "cell_type": "code",
18 | "execution_count": null,
19 | "metadata": {
20 | "collapsed": false
21 | },
22 | "outputs": [],
23 | "source": []
24 | },
25 | {
26 | "cell_type": "code",
27 | "execution_count": 2,
28 | "metadata": {
29 | "collapsed": false
30 | },
31 | "outputs": [
32 | {
33 | "name": "stdout",
34 | "output_type": "stream",
35 | "text": [
36 | "x1=[-1.], df/dx1=0.393\n",
37 | "x2=[-2.], df/dx2=-0.590\n",
38 | "w1=[ 2.], df/dw1=-0.197\n",
39 | "w2=[-3.], df/dw2=-0.393\n",
40 | "w3=[-3.], df/dw3=0.197\n"
41 | ]
42 | }
43 | ],
44 | "source": [
45 | "# Declare input values\n",
46 | "x1 = Variable(torch.Tensor([-1]), requires_grad=True)\n",
47 | "x1.name = \"x1\"\n",
48 | "x2 = Variable(torch.Tensor([-2]), requires_grad=True)\n",
49 | "x2.name = \"x2\"\n",
50 | "w1 = Variable(torch.Tensor([2]), requires_grad=True)\n",
51 | "w1.name = \"w1\"\n",
52 | "w2 = Variable(torch.Tensor([-3]), requires_grad=True)\n",
53 | "w2.name = \"w2\"\n",
54 | "w3 = Variable(torch.Tensor([-3]), requires_grad=True)\n",
55 | "w3.name = \"w3\"\n",
56 | "\n",
57 | "# Form expression using overloaded +,-,*, and / operators\n",
58 | "# Use special T methods to achieve other operations\n",
59 | "s = 1 / (1 + torch.exp(-1*(x1*w1+x2*w2+w3)))\n",
60 | "s.name= \"out\" # just here so the printouts looks better\n",
61 | "\n",
62 | "#compute gradients\n",
63 | "s.backward()\n",
64 | "\n",
65 | "# Specify inputs we declared \n",
66 | "inputs = [x1,x2,w1,w2,w3]\n",
67 | "\n",
68 | "# Call T.grad(s,_) on every input to represent gradient\n",
69 | "gradients = [i.grad for i in inputs]\n",
70 | "\n",
71 | "for (k,v) in zip(inputs, gradients): print \"%s=%5s, df/d%s=%.03f\"% (k.name, k.data.numpy(),k.name, v.data.numpy())"
72 | ]
73 | },
74 | {
75 | "cell_type": "code",
76 | "execution_count": null,
77 | "metadata": {
78 | "collapsed": false
79 | },
80 | "outputs": [],
81 | "source": []
82 | },
83 | {
84 | "cell_type": "code",
85 | "execution_count": null,
86 | "metadata": {
87 | "collapsed": false
88 | },
89 | "outputs": [],
90 | "source": []
91 | },
92 | {
93 | "cell_type": "code",
94 | "execution_count": null,
95 | "metadata": {
96 | "collapsed": true
97 | },
98 | "outputs": [],
99 | "source": []
100 | },
101 | {
102 | "cell_type": "code",
103 | "execution_count": null,
104 | "metadata": {
105 | "collapsed": true
106 | },
107 | "outputs": [],
108 | "source": []
109 | },
110 | {
111 | "cell_type": "code",
112 | "execution_count": null,
113 | "metadata": {
114 | "collapsed": true
115 | },
116 | "outputs": [],
117 | "source": []
118 | },
119 | {
120 | "cell_type": "code",
121 | "execution_count": null,
122 | "metadata": {
123 | "collapsed": true
124 | },
125 | "outputs": [],
126 | "source": []
127 | },
128 | {
129 | "cell_type": "code",
130 | "execution_count": 31,
131 | "metadata": {
132 | "collapsed": false
133 | },
134 | "outputs": [],
135 | "source": [
136 | "from graphviz import Digraph\n",
137 | "\n",
138 | "def make_dot(var, params):\n",
139 | " \"\"\" Produces Graphviz representation of PyTorch autograd graph\n",
140 | " \n",
141 | " Blue nodes are the Variables that require grad, orange are Tensors\n",
142 | " saved for backward in torch.autograd.Function\n",
143 | " \n",
144 | " Args:\n",
145 | " var: output Variable\n",
146 | " params: dict of (name, Variable) to add names to node that\n",
147 | " require grad (TODO: make optional)\n",
148 | " \n",
149 | " From here:\n",
150 | " https://github.com/szagoruyko/functional-zoo/blob/master/visualize.py\n",
151 | " \"\"\"\n",
152 | " param_map = {id(v): k for k, v in params.items()}\n",
153 | " print(param_map)\n",
154 | " \n",
155 | " node_attr = dict(style='filled',\n",
156 | " shape='box',\n",
157 | " align='left',\n",
158 | " fontsize='12',\n",
159 | " ranksep='0.1',\n",
160 | " height='0.2'\n",
161 | " )\n",
162 | " dot = Digraph(node_attr=node_attr, graph_attr=dict(size=\"12,12\",rankdir=\"LR\"))\n",
163 | " seen = set()\n",
164 | " \n",
165 | " def size_to_str(size):\n",
166 | " return '('+(', ').join(['%d'% v for v in size])+')'\n",
167 | "\n",
168 | " def add_nodes(var):\n",
169 | " if var not in seen:\n",
170 | " if torch.is_tensor(var):\n",
171 | " dot.node(str(id(var)), size_to_str(var.size()), fillcolor='orange')\n",
172 | " elif hasattr(var, 'variable'):\n",
173 | " u = var.variable\n",
174 | " node_name = '%s\\n %s' % (param_map.get(id(u)), size_to_str(u.size()))\n",
175 | " dot.node(str(id(var)), node_name, fillcolor='lightblue')\n",
176 | " else:\n",
177 | " dot.node(str(id(var)), str(type(var).__name__))\n",
178 | " seen.add(var)\n",
179 | " if hasattr(var, 'next_functions'):\n",
180 | " for u in var.next_functions:\n",
181 | " if u[0] is not None:\n",
182 | " dot.edge(str(id(u[0])), str(id(var)))\n",
183 | " add_nodes(u[0])\n",
184 | " if hasattr(var, 'saved_tensors'):\n",
185 | " for t in var.saved_tensors:\n",
186 | " dot.edge(str(id(t)), str(id(var)))\n",
187 | " add_nodes(t)\n",
188 | " add_nodes(var.grad_fn)\n",
189 | " return dot"
190 | ]
191 | },
192 | {
193 | "cell_type": "code",
194 | "execution_count": null,
195 | "metadata": {
196 | "collapsed": false
197 | },
198 | "outputs": [],
199 | "source": []
200 | },
201 | {
202 | "cell_type": "code",
203 | "execution_count": null,
204 | "metadata": {
205 | "collapsed": false
206 | },
207 | "outputs": [],
208 | "source": []
209 | },
210 | {
211 | "cell_type": "code",
212 | "execution_count": 32,
213 | "metadata": {
214 | "collapsed": false
215 | },
216 | "outputs": [
217 | {
218 | "name": "stdout",
219 | "output_type": "stream",
220 | "text": [
221 | "{140426904914352: 'x2', 140426904912624: 'x1', 140426904915888: 'w2', 140426904957008: 'w3', 140426904912720: 'w1'}\n"
222 | ]
223 | },
224 | {
225 | "data": {
226 | "image/svg+xml": [
227 | "\n",
228 | "\n",
230 | "\n",
232 | "\n",
233 | "\n"
370 | ],
371 | "text/plain": [
372 | ""
373 | ]
374 | },
375 | "execution_count": 32,
376 | "metadata": {},
377 | "output_type": "execute_result"
378 | }
379 | ],
380 | "source": [
381 | "mapping = dict([(k.name,k) for k in inputs])\n",
382 | "g = make_dot(s, mapping)\n",
383 | "g"
384 | ]
385 | },
386 | {
387 | "cell_type": "code",
388 | "execution_count": null,
389 | "metadata": {
390 | "collapsed": false
391 | },
392 | "outputs": [],
393 | "source": []
394 | },
395 | {
396 | "cell_type": "code",
397 | "execution_count": null,
398 | "metadata": {
399 | "collapsed": false
400 | },
401 | "outputs": [],
402 | "source": []
403 | },
404 | {
405 | "cell_type": "code",
406 | "execution_count": null,
407 | "metadata": {
408 | "collapsed": false
409 | },
410 | "outputs": [],
411 | "source": []
412 | },
413 | {
414 | "cell_type": "code",
415 | "execution_count": null,
416 | "metadata": {
417 | "collapsed": true
418 | },
419 | "outputs": [],
420 | "source": []
421 | }
422 | ],
423 | "metadata": {
424 | "kernelspec": {
425 | "display_name": "Python 2",
426 | "language": "python",
427 | "name": "python2"
428 | },
429 | "language_info": {
430 | "codemirror_mode": {
431 | "name": "ipython",
432 | "version": 2
433 | },
434 | "file_extension": ".py",
435 | "mimetype": "text/x-python",
436 | "name": "python",
437 | "nbconvert_exporter": "python",
438 | "pygments_lexer": "ipython2",
439 | "version": "2.7.13"
440 | }
441 | },
442 | "nbformat": 4,
443 | "nbformat_minor": 2
444 | }
445 |
--------------------------------------------------------------------------------
/pytorch/pytorch-mnist.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "code",
5 | "execution_count": 1,
6 | "metadata": {
7 | "collapsed": true
8 | },
9 | "outputs": [],
10 | "source": [
11 | "#modified from https://github.com/floydhub/quick-start-pytorch/blob/master/mnist.ipynb\n",
12 | "\n",
13 | "import numpy as np\n",
14 | "import torch\n",
15 | "import torch.nn as nn\n",
16 | "import torch.nn.functional as F\n",
17 | "import torch.utils.data.dataloader as dataloader\n",
18 | "import torch.optim as optim\n",
19 | "import torchvision\n",
20 | "\n",
21 | "from torch.autograd import Variable\n",
22 | "cuda = torch.cuda.is_available()\n",
23 | "\n",
24 | "torch.manual_seed(0)\n",
25 | "if cuda:\n",
26 | " torch.cuda.manual_seed(0)"
27 | ]
28 | },
29 | {
30 | "cell_type": "code",
31 | "execution_count": 2,
32 | "metadata": {
33 | "collapsed": false
34 | },
35 | "outputs": [],
36 | "source": [
37 | "train = torchvision.datasets.MNIST('./data', train=True, download=True, transform=torchvision.transforms.Compose([\n",
38 | " torchvision.transforms.ToTensor(), # ToTensor does min-max normalization. \n",
39 | "]), )\n",
40 | "\n",
41 | "test = torchvision.datasets.MNIST('./data', train=False, download=True, transform=torchvision.transforms.Compose([\n",
42 | " torchvision.transforms.ToTensor(), # ToTensor does min-max normalization. \n",
43 | "]), )\n",
44 | "\n",
45 | "# Create DataLoader\n",
46 | "dataloader_args = dict(shuffle=True, batch_size=10000,num_workers=4, pin_memory=True) if cuda else dict(shuffle=True, batch_size=64)\n",
47 | "train_loader = dataloader.DataLoader(train, **dataloader_args)\n",
48 | "test_loader = dataloader.DataLoader(test, **dataloader_args)\n",
49 | "\n",
50 | "train_data = train.train_data\n",
51 | "train_data = train.transform(train_data.numpy())"
52 | ]
53 | },
54 | {
55 | "cell_type": "code",
56 | "execution_count": 3,
57 | "metadata": {
58 | "collapsed": true
59 | },
60 | "outputs": [],
61 | "source": [
62 | "class Model(nn.Module):\n",
63 | " def __init__(self):\n",
64 | " super(Model, self).__init__()\n",
65 | " self.fc = nn.Linear(784, 1000)\n",
66 | " self.fc2 = nn.Linear(1000, 10)\n",
67 | "\n",
68 | " def forward(self, x):\n",
69 | " x = x.view((-1, 784))\n",
70 | " h = F.relu(self.fc(x))\n",
71 | " h = self.fc2(h)\n",
72 | " return F.log_softmax(h, dim=1) \n",
73 | " \n",
74 | "model = Model()\n",
75 | "if cuda:\n",
76 | " model.cuda()\n",
77 | " \n",
78 | "optimizer = optim.Adam(model.parameters(), lr=1e-4)"
79 | ]
80 | },
81 | {
82 | "cell_type": "code",
83 | "execution_count": null,
84 | "metadata": {
85 | "collapsed": false
86 | },
87 | "outputs": [],
88 | "source": []
89 | },
90 | {
91 | "cell_type": "code",
92 | "execution_count": null,
93 | "metadata": {
94 | "collapsed": true
95 | },
96 | "outputs": [],
97 | "source": []
98 | },
99 | {
100 | "cell_type": "code",
101 | "execution_count": 4,
102 | "metadata": {
103 | "collapsed": false,
104 | "scrolled": false
105 | },
106 | "outputs": [
107 | {
108 | "name": "stdout",
109 | "output_type": "stream",
110 | "text": [
111 | " Train Epoch: 1/20 [0/60000 (0%)]\tLoss: 2.301791\n",
112 | " Train Epoch: 1/20 [10000/60000 (17%)]\tLoss: 2.281543\n",
113 | " Train Epoch: 1/20 [20000/60000 (33%)]\tLoss: 2.262036\n",
114 | " Train Epoch: 1/20 [30000/60000 (50%)]\tLoss: 2.243205\n",
115 | " Train Epoch: 1/20 [40000/60000 (67%)]\tLoss: 2.222202\n",
116 | " Train Epoch: 1/20 [50000/60000 (83%)]\tLoss: 2.202651\n",
117 | " Train Epoch: 1/20 [60000/60000 (83%)]\tLoss: 2.202651\t Test Accuracy: 51.54%\n",
118 | " Train Epoch: 2/20 [0/60000 (0%)]\tLoss: 2.180925\n",
119 | " Train Epoch: 2/20 [10000/60000 (17%)]\tLoss: 2.163620\n",
120 | " Train Epoch: 2/20 [20000/60000 (33%)]\tLoss: 2.141855\n",
121 | " Train Epoch: 2/20 [30000/60000 (50%)]\tLoss: 2.122058\n",
122 | " Train Epoch: 2/20 [40000/60000 (67%)]\tLoss: 2.102757\n",
123 | " Train Epoch: 2/20 [50000/60000 (83%)]\tLoss: 2.082337\n",
124 | " Train Epoch: 2/20 [60000/60000 (83%)]\tLoss: 2.082337\t Test Accuracy: 68.20%\n",
125 | " Train Epoch: 3/20 [0/60000 (0%)]\tLoss: 2.060444\n",
126 | " Train Epoch: 3/20 [10000/60000 (17%)]\tLoss: 2.045146\n",
127 | " Train Epoch: 3/20 [20000/60000 (33%)]\tLoss: 2.022069\n",
128 | " Train Epoch: 3/20 [30000/60000 (50%)]\tLoss: 2.000584\n",
129 | " Train Epoch: 3/20 [40000/60000 (67%)]\tLoss: 1.981953\n",
130 | " Train Epoch: 3/20 [50000/60000 (83%)]\tLoss: 1.961174\n",
131 | " Train Epoch: 3/20 [60000/60000 (83%)]\tLoss: 1.961174\t Test Accuracy: 73.91%\n",
132 | " Train Epoch: 4/20 [0/60000 (0%)]\tLoss: 1.943693\n",
133 | " Train Epoch: 4/20 [10000/60000 (17%)]\tLoss: 1.920084\n",
134 | " Train Epoch: 4/20 [20000/60000 (33%)]\tLoss: 1.891357\n",
135 | " Train Epoch: 4/20 [30000/60000 (50%)]\tLoss: 1.878343\n",
136 | " Train Epoch: 4/20 [40000/60000 (67%)]\tLoss: 1.854145\n",
137 | " Train Epoch: 4/20 [50000/60000 (83%)]\tLoss: 1.831880\n",
138 | " Train Epoch: 4/20 [60000/60000 (83%)]\tLoss: 1.831880\t Test Accuracy: 77.37%\n",
139 | " Train Epoch: 5/20 [0/60000 (0%)]\tLoss: 1.810800\n",
140 | " Train Epoch: 5/20 [10000/60000 (17%)]\tLoss: 1.790708\n",
141 | " Train Epoch: 5/20 [20000/60000 (33%)]\tLoss: 1.765454\n",
142 | " Train Epoch: 5/20 [30000/60000 (50%)]\tLoss: 1.746305\n",
143 | " Train Epoch: 5/20 [40000/60000 (67%)]\tLoss: 1.720231\n",
144 | " Train Epoch: 5/20 [50000/60000 (83%)]\tLoss: 1.700785\n",
145 | " Train Epoch: 5/20 [60000/60000 (83%)]\tLoss: 1.700785\t Test Accuracy: 79.34%\n",
146 | " Train Epoch: 6/20 [0/60000 (0%)]\tLoss: 1.676791\n",
147 | " Train Epoch: 6/20 [10000/60000 (17%)]\tLoss: 1.659807\n",
148 | " Train Epoch: 6/20 [20000/60000 (33%)]\tLoss: 1.637383\n",
149 | " Train Epoch: 6/20 [30000/60000 (50%)]\tLoss: 1.603419\n",
150 | " Train Epoch: 6/20 [40000/60000 (67%)]\tLoss: 1.584017\n",
151 | " Train Epoch: 6/20 [50000/60000 (83%)]\tLoss: 1.568018\n",
152 | " Train Epoch: 6/20 [60000/60000 (83%)]\tLoss: 1.568018\t Test Accuracy: 81.17%\n",
153 | " Train Epoch: 7/20 [0/60000 (0%)]\tLoss: 1.542853\n",
154 | " Train Epoch: 7/20 [10000/60000 (17%)]\tLoss: 1.528000\n",
155 | " Train Epoch: 7/20 [20000/60000 (33%)]\tLoss: 1.499194\n",
156 | " Train Epoch: 7/20 [30000/60000 (50%)]\tLoss: 1.476696\n",
157 | " Train Epoch: 7/20 [40000/60000 (67%)]\tLoss: 1.452573\n",
158 | " Train Epoch: 7/20 [50000/60000 (83%)]\tLoss: 1.431361\n",
159 | " Train Epoch: 7/20 [60000/60000 (83%)]\tLoss: 1.431361\t Test Accuracy: 82.18%\n",
160 | " Train Epoch: 8/20 [0/60000 (0%)]\tLoss: 1.407419\n",
161 | " Train Epoch: 8/20 [10000/60000 (17%)]\tLoss: 1.395671\n",
162 | " Train Epoch: 8/20 [20000/60000 (33%)]\tLoss: 1.371859\n",
163 | " Train Epoch: 8/20 [30000/60000 (50%)]\tLoss: 1.355923\n",
164 | " Train Epoch: 8/20 [40000/60000 (67%)]\tLoss: 1.326650\n",
165 | " Train Epoch: 8/20 [50000/60000 (83%)]\tLoss: 1.306829\n",
166 | " Train Epoch: 8/20 [60000/60000 (83%)]\tLoss: 1.306829\t Test Accuracy: 83.19%\n",
167 | " Train Epoch: 9/20 [0/60000 (0%)]\tLoss: 1.295578\n",
168 | " Train Epoch: 9/20 [10000/60000 (17%)]\tLoss: 1.261904\n",
169 | " Train Epoch: 9/20 [20000/60000 (33%)]\tLoss: 1.247882\n",
170 | " Train Epoch: 9/20 [30000/60000 (50%)]\tLoss: 1.238853\n",
171 | " Train Epoch: 9/20 [40000/60000 (67%)]\tLoss: 1.215009\n",
172 | " Train Epoch: 9/20 [50000/60000 (83%)]\tLoss: 1.190309\n",
173 | " Train Epoch: 9/20 [60000/60000 (83%)]\tLoss: 1.190309\t Test Accuracy: 83.64%\n",
174 | " Train Epoch: 10/20 [0/60000 (0%)]\tLoss: 1.171327\n",
175 | " Train Epoch: 10/20 [10000/60000 (17%)]\tLoss: 1.165096\n",
176 | " Train Epoch: 10/20 [20000/60000 (33%)]\tLoss: 1.142629\n",
177 | " Train Epoch: 10/20 [30000/60000 (50%)]\tLoss: 1.123629\n",
178 | " Train Epoch: 10/20 [40000/60000 (67%)]\tLoss: 1.104114\n",
179 | " Train Epoch: 10/20 [50000/60000 (83%)]\tLoss: 1.094527\n",
180 | " Train Epoch: 10/20 [60000/60000 (83%)]\tLoss: 1.094527\t Test Accuracy: 84.20%\n",
181 | " Train Epoch: 11/20 [0/60000 (0%)]\tLoss: 1.078334\n",
182 | " Train Epoch: 11/20 [10000/60000 (17%)]\tLoss: 1.056460\n",
183 | " Train Epoch: 11/20 [20000/60000 (33%)]\tLoss: 1.045727\n",
184 | " Train Epoch: 11/20 [30000/60000 (50%)]\tLoss: 1.029339\n",
185 | " Train Epoch: 11/20 [40000/60000 (67%)]\tLoss: 1.007853\n",
186 | " Train Epoch: 11/20 [50000/60000 (83%)]\tLoss: 1.005731\n",
187 | " Train Epoch: 11/20 [60000/60000 (83%)]\tLoss: 1.005731\t Test Accuracy: 84.61%\n",
188 | " Train Epoch: 12/20 [0/60000 (0%)]\tLoss: 0.981822\n",
189 | " Train Epoch: 12/20 [10000/60000 (17%)]\tLoss: 0.965761\n",
190 | " Train Epoch: 12/20 [20000/60000 (33%)]\tLoss: 0.962455\n",
191 | " Train Epoch: 12/20 [30000/60000 (50%)]\tLoss: 0.951225\n",
192 | " Train Epoch: 12/20 [40000/60000 (67%)]\tLoss: 0.934389\n",
193 | " Train Epoch: 12/20 [50000/60000 (83%)]\tLoss: 0.922147\n",
194 | " Train Epoch: 12/20 [60000/60000 (83%)]\tLoss: 0.922147\t Test Accuracy: 85.09%\n",
195 | " Train Epoch: 13/20 [0/60000 (0%)]\tLoss: 0.897114\n",
196 | " Train Epoch: 13/20 [10000/60000 (17%)]\tLoss: 0.896791\n",
197 | " Train Epoch: 13/20 [20000/60000 (33%)]\tLoss: 0.893328\n",
198 | " Train Epoch: 13/20 [30000/60000 (50%)]\tLoss: 0.877841\n",
199 | " Train Epoch: 13/20 [40000/60000 (67%)]\tLoss: 0.868895\n",
200 | " Train Epoch: 13/20 [50000/60000 (83%)]\tLoss: 0.844944\n",
201 | " Train Epoch: 13/20 [60000/60000 (83%)]\tLoss: 0.844944\t Test Accuracy: 85.65%\n",
202 | " Train Epoch: 14/20 [0/60000 (0%)]\tLoss: 0.835898\n",
203 | " Train Epoch: 14/20 [10000/60000 (17%)]\tLoss: 0.835512\n",
204 | " Train Epoch: 14/20 [20000/60000 (33%)]\tLoss: 0.823491\n",
205 | " Train Epoch: 14/20 [30000/60000 (50%)]\tLoss: 0.816006\n",
206 | " Train Epoch: 14/20 [40000/60000 (67%)]\tLoss: 0.792492\n",
207 | " Train Epoch: 14/20 [50000/60000 (83%)]\tLoss: 0.796947\n",
208 | " Train Epoch: 14/20 [60000/60000 (83%)]\tLoss: 0.796947\t Test Accuracy: 86.31%\n",
209 | " Train Epoch: 15/20 [0/60000 (0%)]\tLoss: 0.779239\n",
210 | " Train Epoch: 15/20 [10000/60000 (17%)]\tLoss: 0.778198\n",
211 | " Train Epoch: 15/20 [20000/60000 (33%)]\tLoss: 0.767019\n",
212 | " Train Epoch: 15/20 [30000/60000 (50%)]\tLoss: 0.750927\n",
213 | " Train Epoch: 15/20 [40000/60000 (67%)]\tLoss: 0.754814\n",
214 | " Train Epoch: 15/20 [50000/60000 (83%)]\tLoss: 0.743768\n",
215 | " Train Epoch: 15/20 [60000/60000 (83%)]\tLoss: 0.743768\t Test Accuracy: 86.62%\n",
216 | " Train Epoch: 16/20 [0/60000 (0%)]\tLoss: 0.737757\n",
217 | " Train Epoch: 16/20 [10000/60000 (17%)]\tLoss: 0.730100\n",
218 | " Train Epoch: 16/20 [20000/60000 (33%)]\tLoss: 0.721721\n",
219 | " Train Epoch: 16/20 [30000/60000 (50%)]\tLoss: 0.710267\n",
220 | " Train Epoch: 16/20 [40000/60000 (67%)]\tLoss: 0.699788\n",
221 | " Train Epoch: 16/20 [50000/60000 (83%)]\tLoss: 0.692801\n",
222 | " Train Epoch: 16/20 [60000/60000 (83%)]\tLoss: 0.692801\t Test Accuracy: 86.87%\n",
223 | " Train Epoch: 17/20 [0/60000 (0%)]\tLoss: 0.708184\n",
224 | " Train Epoch: 17/20 [10000/60000 (17%)]\tLoss: 0.684190\n",
225 | " Train Epoch: 17/20 [20000/60000 (33%)]\tLoss: 0.667487\n",
226 | " Train Epoch: 17/20 [30000/60000 (50%)]\tLoss: 0.670384\n",
227 | " Train Epoch: 17/20 [40000/60000 (67%)]\tLoss: 0.664699\n",
228 | " Train Epoch: 17/20 [50000/60000 (83%)]\tLoss: 0.653951\n",
229 | " Train Epoch: 17/20 [60000/60000 (83%)]\tLoss: 0.653951\t Test Accuracy: 87.40%\n",
230 | " Train Epoch: 18/20 [0/60000 (0%)]\tLoss: 0.660717\n",
231 | " Train Epoch: 18/20 [10000/60000 (17%)]\tLoss: 0.639914\n",
232 | " Train Epoch: 18/20 [20000/60000 (33%)]\tLoss: 0.638611\n",
233 | " Train Epoch: 18/20 [30000/60000 (50%)]\tLoss: 0.644234\n",
234 | " Train Epoch: 18/20 [40000/60000 (67%)]\tLoss: 0.640062\n",
235 | " Train Epoch: 18/20 [50000/60000 (83%)]\tLoss: 0.613140\n",
236 | " Train Epoch: 18/20 [60000/60000 (83%)]\tLoss: 0.613140\t Test Accuracy: 87.69%\n",
237 | " Train Epoch: 19/20 [0/60000 (0%)]\tLoss: 0.625232\n",
238 | " Train Epoch: 19/20 [10000/60000 (17%)]\tLoss: 0.607092\n",
239 | " Train Epoch: 19/20 [20000/60000 (33%)]\tLoss: 0.617470\n",
240 | " Train Epoch: 19/20 [30000/60000 (50%)]\tLoss: 0.605471\n",
241 | " Train Epoch: 19/20 [40000/60000 (67%)]\tLoss: 0.596517\n",
242 | " Train Epoch: 19/20 [50000/60000 (83%)]\tLoss: 0.599279\n",
243 | " Train Epoch: 19/20 [60000/60000 (83%)]\tLoss: 0.599279\t Test Accuracy: 88.07%\n",
244 | " Train Epoch: 20/20 [0/60000 (0%)]\tLoss: 0.589844\n",
245 | " Train Epoch: 20/20 [10000/60000 (17%)]\tLoss: 0.584870\n",
246 | " Train Epoch: 20/20 [20000/60000 (33%)]\tLoss: 0.593216\n",
247 | " Train Epoch: 20/20 [30000/60000 (50%)]\tLoss: 0.570595\n",
248 | " Train Epoch: 20/20 [40000/60000 (67%)]\tLoss: 0.583682\n",
249 | " Train Epoch: 20/20 [50000/60000 (83%)]\tLoss: 0.565770\n",
250 | " Train Epoch: 20/20 [60000/60000 (83%)]\tLoss: 0.565770\t Test Accuracy: 88.36%\n"
251 | ]
252 | }
253 | ],
254 | "source": [
255 | "EPOCHS = 20\n",
256 | "losses = []\n",
257 | "\n",
258 | "model.train()\n",
259 | "for epoch in range(EPOCHS):\n",
260 | " for batch_idx, (data, target) in enumerate(train_loader):\n",
261 | "\n",
262 | " data, target = Variable(data), Variable(target)\n",
263 | " \n",
264 | " if cuda:\n",
265 | " data, target = data.cuda(), target.cuda()\n",
266 | " \n",
267 | " optimizer.zero_grad()\n",
268 | " y_pred = model(data) \n",
269 | "\n",
270 | " loss = F.cross_entropy(y_pred, target)\n",
271 | " losses.append(loss.cpu().data.item())\n",
272 | " loss.backward()\n",
273 | " optimizer.step()\n",
274 | "\n",
275 | " #if batch_idx % 100 == 1:\n",
276 | " print('\\r Train Epoch: {}/{} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n",
277 | " epoch+1,\n",
278 | " EPOCHS,\n",
279 | " batch_idx * len(data), \n",
280 | " len(train_loader.dataset),\n",
281 | " 100. * batch_idx / len(train_loader), \n",
282 | " loss.cpu().data.item()))\n",
283 | "\n",
284 | " evaluate_x = Variable(test_loader.dataset.test_data.type_as(torch.FloatTensor()))\n",
285 | " evaluate_y = Variable(test_loader.dataset.test_labels)\n",
286 | " if cuda:\n",
287 | " evaluate_x, evaluate_y = evaluate_x.cuda(), evaluate_y.cuda()\n",
288 | "\n",
289 | " model.eval()\n",
290 | " output = model(evaluate_x)\n",
291 | " pred = output.data.max(1)[1]\n",
292 | " d = pred.eq(evaluate_y.data).cpu()\n",
293 | " accuracy = d.sum().item()*1./d.size()[0]\n",
294 | " \n",
295 | " print('\\r Train Epoch: {}/{} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}\\t Test Accuracy: {:.2f}%'.format(\n",
296 | " epoch+1,\n",
297 | " EPOCHS,\n",
298 | " len(train_loader.dataset), \n",
299 | " len(train_loader.dataset),\n",
300 | " 100. * batch_idx / len(train_loader), \n",
301 | " loss.cpu().data.item(),\n",
302 | " accuracy*100.0))"
303 | ]
304 | },
305 | {
306 | "cell_type": "code",
307 | "execution_count": null,
308 | "metadata": {
309 | "collapsed": true
310 | },
311 | "outputs": [],
312 | "source": []
313 | },
314 | {
315 | "cell_type": "code",
316 | "execution_count": null,
317 | "metadata": {
318 | "collapsed": false
319 | },
320 | "outputs": [],
321 | "source": []
322 | },
323 | {
324 | "cell_type": "code",
325 | "execution_count": null,
326 | "metadata": {
327 | "collapsed": true
328 | },
329 | "outputs": [],
330 | "source": []
331 | }
332 | ],
333 | "metadata": {
334 | "kernelspec": {
335 | "display_name": "Python 2",
336 | "language": "python",
337 | "name": "python2"
338 | },
339 | "language_info": {
340 | "codemirror_mode": {
341 | "name": "ipython",
342 | "version": 2
343 | },
344 | "file_extension": ".py",
345 | "mimetype": "text/x-python",
346 | "name": "python",
347 | "nbconvert_exporter": "python",
348 | "pygments_lexer": "ipython2",
349 | "version": "2.7.13"
350 | }
351 | },
352 | "nbformat": 4,
353 | "nbformat_minor": 2
354 | }
355 |
--------------------------------------------------------------------------------
/pytorch/segmentation/10279_500_f00182_original.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ieee8023/NeuralNetwork-Examples/98677d0eb06eba8b511251c99826319fd0b17bc1/pytorch/segmentation/10279_500_f00182_original.jpg
--------------------------------------------------------------------------------
/pytorch/segmentation/8917.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/ieee8023/NeuralNetwork-Examples/98677d0eb06eba8b511251c99826319fd0b17bc1/pytorch/segmentation/8917.png
--------------------------------------------------------------------------------
/theano/counting/README.md:
--------------------------------------------------------------------------------
1 | ## [The primary location of code for this paper is now here https://github.com/ieee8023/countception](https://github.com/ieee8023/countception)
2 |
3 |
4 |
5 |
6 | J. P. Cohen, H. Z. Lo, and Y. Bengio, “Count-ception: Counting by Fully Convolutional Redundant Counting,” 2017.
7 | https://arxiv.org/abs/1703.08710
8 |
--------------------------------------------------------------------------------
/theano/counting/count-ception.py:
--------------------------------------------------------------------------------
1 |
2 | # coding: utf-8
3 |
4 | # This code is for:
5 | #
6 | # J. P. Cohen, H. Z. Lo, and Y. Bengio, “Count-ception: Counting by Fully Convolutional Redundant Counting,” 2017.
7 | # https://arxiv.org/abs/1703.08710
8 | #
9 | #
10 | # Here is a video of the learning in progress:
11 | # [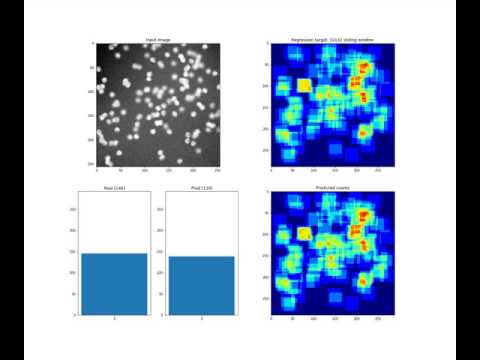](https://www.youtube.com/watch?v=ej5bj0mlQq8)
12 | #
13 | # The cell dataset used in this work is available from [VGG](http://www.robots.ox.ac.uk/~vgg/research/counting/cells.zip) and [Academic Torrents](http://academictorrents.com/details/b32305598175bb8e03c5f350e962d772a910641c)
14 | #
15 |
16 | # In[ ]:
17 |
18 |
19 |
20 |
21 | # In[1]:
22 |
23 | import sys,os,time,random
24 | import numpy as np
25 | import matplotlib
26 | matplotlib.use('Agg');
27 | import matplotlib.pyplot as plt
28 | plt.set_cmap('jet');
29 |
30 | import theano
31 | import theano.tensor as T
32 | import lasagne
33 |
34 | import skimage
35 | from skimage.io import imread, imsave
36 | import pickle
37 | import scipy
38 |
39 | from os import walk
40 | print "theano",theano.version.full_version
41 | print "lasagne",lasagne.__version__
42 |
43 |
44 | # In[ ]:
45 |
46 |
47 |
48 |
49 | # In[2]:
50 |
51 |
52 |
53 | # In[3]:
54 |
55 | import sys
56 | import argparse
57 |
58 | if len(sys.argv) == 3 and sys.argv[1] == "-f": #on jupyter
59 | sys.argv = ['']
60 |
61 | parser = argparse.ArgumentParser(description='Count-ception')
62 |
63 | parser.add_argument('-seed', type=int, nargs='?',default=0, help='random seed for split and init')
64 | parser.add_argument('-nsamples', type=int, nargs='?',default=32, help='Number of samples (N) in train and valid')
65 | parser.add_argument('-stride', type=int, nargs='?',default=1, help='The args.stride at the initial layer')
66 | parser.add_argument('-lr', type=float, nargs='?',default=0.00005, help='This will set the learning rate ')
67 | parser.add_argument('-kern', type=str, nargs='?',default="sq", help='This can be gaus or sq')
68 | parser.add_argument('-cov', type=float, nargs='?',default=1, help='This is the covariance when kern=gaus')
69 | parser.add_argument('-scale', type=int, nargs='?',default=1, help='Scale the input image and labels')
70 | parser.add_argument('-data', type=str, nargs='?',default="cells", help='Dataset folder')
71 | parser.add_argument('-framesize', type=int, nargs='?',default=256, help='Size of the images processed at once')
72 |
73 |
74 | args = parser.parse_args()
75 |
76 |
77 | # In[4]:
78 |
79 | print args
80 |
81 |
82 | # In[5]:
83 |
84 | job_id = os.environ.get('SLURM_JOB_ID')
85 |
86 | if job_id == None:
87 | job_id = os.environ.get('PBS_JOBID')
88 |
89 | print "job_id",job_id
90 |
91 |
92 | # In[ ]:
93 |
94 |
95 |
96 |
97 | # In[6]:
98 |
99 | patch_size = 32
100 | framesize = int(args.framesize/args.scale)
101 | framesize_h = framesize_w = framesize
102 | noutputs = 1
103 | channels = 3
104 |
105 |
106 | # In[ ]:
107 |
108 |
109 |
110 |
111 | # In[7]:
112 |
113 | paramfilename = str(args.scale) + "-" + str(patch_size) + "-" + args.data + "-" + args.kern + str(args.cov) + "_params.p"
114 | datasetfilename = str(args.scale) + "-" + str(patch_size) + "-" + str(framesize) + "-" + args.kern + str(args.stride) + "-" + args.data + "-" + str(args.cov) + "-dataset.p"
115 | print paramfilename
116 | print datasetfilename
117 |
118 |
119 | # In[8]:
120 |
121 | random.seed(args.seed)
122 | np.random.seed(args.seed)
123 | lasagne.random.set_rng(np.random.RandomState(args.seed))
124 |
125 |
126 | # In[9]:
127 |
128 | from lasagne.layers.normalization import BatchNormLayer
129 | from lasagne.layers import InputLayer, ConcatLayer, Conv2DLayer
130 |
131 | input_var = T.tensor4('inputs')
132 | input_var_ex = T.ivector('input_var_ex')
133 |
134 | def ConvFactory(data, num_filter, filter_size, stride=1, pad=0, nonlinearity=lasagne.nonlinearities.leaky_rectify):
135 | data = lasagne.layers.batch_norm(Conv2DLayer(
136 | data, num_filters=num_filter,
137 | filter_size=filter_size,
138 | stride=stride, pad=pad,
139 | nonlinearity=nonlinearity,
140 | W=lasagne.init.GlorotUniform(gain='relu')))
141 | return data
142 |
143 | def SimpleFactory(data, ch_1x1, ch_3x3):
144 | conv1x1 = ConvFactory(data=data, filter_size=1, pad=0, num_filter=ch_1x1)
145 | conv3x3 = ConvFactory(data=data, filter_size=3, pad=1, num_filter=ch_3x3)
146 | concat = ConcatLayer([conv1x1, conv3x3])
147 | return concat
148 |
149 |
150 | input_shape = (None, channels, framesize, framesize)
151 | img = InputLayer(shape=input_shape, input_var=input_var[input_var_ex])
152 | net = img
153 |
154 |
155 | net = ConvFactory(net, filter_size=3, num_filter=64, pad=patch_size)
156 | print net.output_shape
157 | net = SimpleFactory(net, 16, 16)
158 | print net.output_shape
159 | net = SimpleFactory(net, 16, 32)
160 | print net.output_shape
161 | net = ConvFactory(net, filter_size=14, num_filter=16)
162 | print net.output_shape
163 | net = SimpleFactory(net, 112, 48)
164 | print net.output_shape
165 | net = SimpleFactory(net, 64, 32)
166 | print net.output_shape
167 | net = SimpleFactory(net, 40, 40)
168 | print net.output_shape
169 | net = SimpleFactory(net, 32, 96)
170 | print net.output_shape
171 | net = ConvFactory(net, filter_size=18, num_filter=32)
172 | print net.output_shape
173 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
174 | print net.output_shape
175 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
176 | print net.output_shape
177 | net = ConvFactory(net, filter_size=1, num_filter=1, stride=args.stride)
178 | print net.output_shape
179 |
180 |
181 | # In[10]:
182 |
183 | output_shape = lasagne.layers.get_output_shape(net)
184 | real_input_shape = (None, input_shape[1], input_shape[2]+2*patch_size, input_shape[3]+2*patch_size)
185 | print "real_input_shape:",real_input_shape,"-> output_shape:",output_shape
186 |
187 |
188 | # In[ ]:
189 |
190 |
191 |
192 |
193 | # In[11]:
194 |
195 | print "network output size should be",(input_shape[2]+2*patch_size)-(patch_size)
196 |
197 |
198 | # In[ ]:
199 |
200 |
201 |
202 |
203 | # In[12]:
204 |
205 | if (args.kern == "sq"):
206 | ef = ((patch_size/args.stride)**2.0)
207 | elif (args.kern == "gaus"):
208 | ef = 1.0
209 | print "ef", ef
210 |
211 | prediction = lasagne.layers.get_output(net, deterministic=True)
212 | prediction_count = (prediction/ef).sum(axis=(2,3))
213 |
214 | classify = theano.function([input_var, input_var_ex], prediction)
215 |
216 |
217 | # In[13]:
218 |
219 | train_start_time = time.time()
220 | print classify(np.zeros((1,channels,framesize,framesize), dtype=theano.config.floatX), [0]).shape
221 | print time.time() - train_start_time, "sec"
222 |
223 | train_start_time = time.time()
224 | print classify(np.zeros((1,channels,framesize,framesize), dtype=theano.config.floatX), [0]).shape
225 | print time.time() - train_start_time, "sec"
226 |
227 |
228 | # In[ ]:
229 |
230 |
231 |
232 |
233 | # In[ ]:
234 |
235 |
236 |
237 |
238 | # In[14]:
239 |
240 | def genGausImage(framesize, mx, my, cov=1):
241 | x, y = np.mgrid[0:framesize, 0:framesize]
242 | pos = np.dstack((x, y))
243 | mean = [mx, my]
244 | cov = [[cov, 0], [0, cov]]
245 | rv = scipy.stats.multivariate_normal(mean, cov).pdf(pos)
246 | return rv/rv.sum()
247 |
248 | def getDensity(width, markers):
249 | gaus_img = np.zeros((width,width))
250 | for k in range(width):
251 | for l in range(width):
252 | if (markers[k,l] > 0.5):
253 | gaus_img += genGausImage(len(markers),k-patch_size/2,l-patch_size/2,cov)
254 | return gaus_img
255 |
256 | def getMarkersCells(labelPath, scale, size):
257 | labs = imread(labelPath)
258 | if len(labs.shape) == 2:
259 | lab = labs[:,:]/255
260 | elif len(labs.shape) == 3:
261 | lab = labs[:,:,0]/255
262 | else:
263 | print "unknown label format"
264 |
265 | binsize = [scale,scale]
266 | out = np.zeros(size)
267 | for i in xrange(binsize[0]):
268 | for j in xrange(binsize[1]):
269 | out = np.maximum(lab[i::binsize[0], j::binsize[1]], out)
270 |
271 | print lab.sum(),out.sum()
272 | assert np.allclose(lab.sum(),out.sum(), 1)
273 |
274 | return out
275 |
276 | def getCellCountCells(markers,(x,y,h,w)):
277 | types = [0] * noutputs
278 | for i in range(noutputs):
279 | types[i] = (markers[y:y+h,x:x+w] == 1).sum()
280 | #types[i] = (markers[y:y+h,x:x+w] != -1).sum()
281 | return types
282 |
283 | def getLabelsCells(markers, img_pad, base_x, base_y, stride, scale):
284 |
285 | height = ((img_pad.shape[0])/args.stride)
286 | width = ((img_pad.shape[1])/args.stride)
287 | print "label size: ", height, width
288 | labels = np.zeros((noutputs, height, width))
289 | if (args.kern == "sq"):
290 | for y in range(0,height):
291 | for x in range(0,width):
292 | count = getCellCountCells(markers,(x*args.stride,y*args.stride,patch_size,patch_size))
293 | for i in range(0,noutputs):
294 | labels[i][y][x] = count[i]
295 |
296 |
297 | elif (args.kern == "gaus"):
298 | for i in range(0,noutputs):
299 | labels[i] = getDensity(width, markers[base_y:base_y+width,base_x:base_x+width])
300 |
301 |
302 | count_total = getCellCountCells(markers,(0,0,framesize_h+patch_size,framesize_w+patch_size))
303 | return labels, count_total
304 |
305 | def getTrainingExampleCells(img_raw, framesize_w, framesize_h, labelPath, base_x, base_y, stride, scale):
306 |
307 | img = img_raw[base_y:base_y+framesize_h, base_x:base_x+framesize_w]
308 | img_pad = np.pad(img[:,:,0],(patch_size)/2, "constant")
309 |
310 | markers = getMarkersCells(labelPath, scale, img_raw.shape[0:2])
311 | markers = markers[base_y:base_y+framesize_h, base_x:base_x+framesize_w]
312 | markers = np.pad(markers, patch_size, "constant", constant_values=-1)
313 |
314 | labels, count = getLabelsCells(markers, img_pad, base_x, base_y, args.stride, scale)
315 | return img, labels, count
316 |
317 |
318 | # In[ ]:
319 |
320 |
321 |
322 |
323 | # In[15]:
324 |
325 | import glob
326 | imgs = []
327 | for filename in glob.iglob(args.data + "/*dots.png"):
328 | imgg = filename.replace("dots","cell")
329 | imgs.append([imgg,filename])
330 |
331 | if len(imgs) == 0:
332 | print "Issue with dataset"
333 | sys.exit()
334 |
335 |
336 | # In[ ]:
337 |
338 |
339 |
340 |
341 | # In[16]:
342 |
343 | #print imgs
344 |
345 |
346 | # In[ ]:
347 |
348 |
349 |
350 |
351 | # In[ ]:
352 |
353 |
354 |
355 |
356 | # In[17]:
357 |
358 | ## code to debug data generation
359 | plt.rcParams['figure.figsize'] = (18, 9)
360 | imgPath,labelPath,x,y = imgs[9][0],imgs[9][1], 0, 0
361 | #imgPath,labelPath,x,y = imgs[0][0], imgs[0][1], 100,200
362 |
363 | print imgPath, labelPath
364 |
365 | im = imread(imgPath)
366 | img_raw_raw = im #grayscale
367 |
368 | img_raw = scipy.misc.imresize(img_raw_raw, (int(img_raw_raw.shape[0]/args.scale), int(img_raw_raw.shape[1]/args.scale)))
369 | print img_raw_raw.shape," ->>>>", img_raw.shape
370 |
371 | print "img_raw",img_raw.shape
372 | img, lab, count = getTrainingExampleCells(img_raw, framesize_w, framesize_h, labelPath, x, y, args.stride, args.scale)
373 | print "count", count
374 |
375 | markers = getMarkersCells(labelPath, args.scale, img_raw.shape[0:2])
376 | markers = markers[y:y+framesize_h, x:x+framesize_w]
377 | count = getCellCountCells(markers, (0, 0, framesize_w,framesize_h))
378 | print "count", count, 'markers max', markers.max()
379 |
380 | pcount = classify([img.transpose((2,0,1))], [0])[0]
381 |
382 | lab_est = [(l.sum()/ef).astype(np.int) for l in lab]
383 | pred_est = [(l.sum()/ef).astype(np.int) for l in pcount]
384 |
385 | print "img shape", img.shape
386 | print "label shape", lab.shape
387 | print "label est ",lab_est," --> predicted est ",pred_est
388 |
389 |
390 | # In[18]:
391 |
392 | fig = plt.Figure(figsize=(18, 9), dpi=160)
393 | gcf = plt.gcf()
394 | gcf.set_size_inches(18, 15)
395 | fig.set_canvas(gcf.canvas)
396 |
397 | ax2 = plt.subplot2grid((2,4), (0, 0), colspan=2)
398 | ax3 = plt.subplot2grid((2,4), (0, 2), colspan=3)
399 | ax4 = plt.subplot2grid((2,4), (1, 2), colspan=3)
400 | ax5 = plt.subplot2grid((2,4), (1, 0), rowspan=1)
401 | ax6 = plt.subplot2grid((2,4), (1, 1), rowspan=1)
402 |
403 | ax2.set_title("Input Image")
404 | ax2.imshow(img, interpolation='none',cmap='Greys_r');
405 | ax3.set_title("Regression target, {}x{} sliding window.".format(patch_size, patch_size))
406 | ax3.imshow(np.concatenate((lab),axis=1), interpolation='none');
407 | #ax3.imshow(lab[0], interpolation='none')
408 |
409 | ax4.set_title("Predicted counts")
410 | ax4.imshow(np.concatenate((pcount),axis=1), interpolation='none');
411 |
412 | ax5.set_title("Real " + str(lab_est))
413 | ax5.set_ylim((0, np.max(lab_est)*2))
414 | ax5.set_xticks(np.arange(0, noutputs, 1.0))
415 | ax5.bar(range(noutputs),lab_est, align='center')
416 | ax6.set_title("Pred " + str(pred_est))
417 | ax6.set_ylim((0, np.max(lab_est)*2))
418 | ax6.set_xticks(np.arange(0, noutputs, 1.0))
419 | ax6.bar(range(noutputs),pred_est, align='center')
420 |
421 |
422 | # In[ ]:
423 |
424 |
425 |
426 |
427 | # In[19]:
428 |
429 | img_pad = np.asarray([np.pad(img[:,:,i],(patch_size-1)/2, "constant", constant_values=255) for i in range(img[0,0].shape[0])])
430 | img_pad = img_pad.transpose((1,2,0))
431 | plt.imshow(img_pad);
432 | plt.imshow(lab[0], alpha=0.5);
433 |
434 |
435 | # In[ ]:
436 |
437 |
438 |
439 |
440 | # In[ ]:
441 |
442 |
443 |
444 |
445 | # In[ ]:
446 |
447 |
448 |
449 |
450 | # In[20]:
451 |
452 | for path in imgs:
453 | if (not os.path.isfile(path[0])):
454 | print path, "bad", path[0]
455 | if (not os.path.isfile(path[1])):
456 | print path, "bad", path[1]
457 |
458 |
459 | # In[21]:
460 |
461 | dataset = []
462 | if (os.path.isfile(datasetfilename)):
463 | print "reading", datasetfilename
464 | dataset = pickle.load(open(datasetfilename, "rb" ))
465 | else:
466 | dataset_x = []
467 | dataset_y = []
468 | dataset_c = []
469 | print len(imgs)
470 | for path in imgs:
471 |
472 | imgPath = path[0]
473 | print imgPath
474 |
475 | im = imread(imgPath)
476 | img_raw_raw = im
477 |
478 | img_raw = scipy.misc.imresize(img_raw_raw, (int(img_raw_raw.shape[0]/args.scale),int(img_raw_raw.shape[1]/args.scale)))
479 | print img_raw_raw.shape," ->>>>", img_raw.shape
480 |
481 | labelPath = path[1]
482 | for base_x in range(0,img_raw.shape[0],framesize_h):
483 | for base_y in range(0,img_raw.shape[1],framesize_w):
484 |
485 | if (img_raw.shape[1] - base_y < framesize_w) or (img_raw.shape[0] - base_x < framesize_h):
486 | print "!!!! Not adding image because size is" , img_raw.shape[1] - base_y, img_raw.shape[0] - base_x
487 | continue
488 |
489 | img, lab, count = getTrainingExampleCells(img_raw, framesize_w, framesize_h, labelPath, base_y, base_x, args.stride, args.scale)
490 | print "count ", count
491 |
492 | if img.shape[0:2] != (framesize_w,framesize_h):
493 | print "!!!! Not adding image because size is" , img.shape[0:2]
494 |
495 | else :
496 | lab_est = [(l.sum()/ef).astype(np.int) for l in lab]
497 |
498 | assert np.allclose(count,lab_est, 0)
499 |
500 | dataset.append((img,lab,count))
501 |
502 | print "lab_est", lab_est, "img shape", img.shape, "label shape", lab.shape
503 | sys.stdout.flush()
504 |
505 | print "dataset size", len(dataset)
506 |
507 | print "writing", datasetfilename
508 | out = open(datasetfilename, "wb",0)
509 | pickle.dump(dataset, out)
510 | out.close()
511 | print "DONE"
512 |
513 |
514 | # In[ ]:
515 |
516 |
517 |
518 |
519 | # In[22]:
520 |
521 | # %matplotlib inline
522 | # plt.rcParams['figure.figsize'] = (18, 9)
523 | # plt.imshow(lab[0])
524 |
525 |
526 | # In[23]:
527 |
528 | #np_dataset = np.asarray(dataset)
529 |
530 | np.random.shuffle(dataset)
531 |
532 | np_dataset_x = np.asarray([d[0] for d in dataset], dtype=theano.config.floatX)
533 | np_dataset_y = np.asarray([d[1] for d in dataset], dtype=theano.config.floatX)
534 | np_dataset_c = np.asarray([d[2] for d in dataset], dtype=theano.config.floatX)
535 |
536 |
537 | np_dataset_x = np_dataset_x.transpose((0,3,1,2))
538 |
539 | print "np_dataset_x", np_dataset_x.shape
540 | print "np_dataset_y", np_dataset_y.shape
541 | print "np_dataset_c", np_dataset_c.shape
542 |
543 |
544 | # In[ ]:
545 |
546 |
547 |
548 |
549 | # In[24]:
550 |
551 | length = len(np_dataset_x)
552 |
553 | n = args.nsamples
554 |
555 | np_dataset_x_train = np_dataset_x[0:n]
556 | np_dataset_y_train = np_dataset_y[0:n]
557 | np_dataset_c_train = np_dataset_c[0:n]
558 | print "np_dataset_x_train", len(np_dataset_x_train)
559 |
560 | np_dataset_x_valid = np_dataset_x[n:2*n]
561 | np_dataset_y_valid = np_dataset_y[n:2*n]
562 | np_dataset_c_valid = np_dataset_c[n:2*n]
563 | print "np_dataset_x_valid", len(np_dataset_x_valid)
564 |
565 | np_dataset_x_test = np_dataset_x[-100:]
566 | np_dataset_y_test = np_dataset_y[-100:]
567 | np_dataset_c_test = np_dataset_c[-100:]
568 | print "np_dataset_x_test", len(np_dataset_x_test)
569 |
570 |
571 | # In[25]:
572 |
573 | print "number of counts total ", np_dataset_c.sum()
574 | print "number of counts on average ", np_dataset_c.mean(), "+-", np_dataset_c.std()
575 | print "counts min:", np_dataset_c.min(), "max:", np_dataset_c.max()
576 |
577 |
578 | # In[ ]:
579 |
580 |
581 |
582 |
583 | # In[ ]:
584 |
585 |
586 |
587 |
588 | # In[ ]:
589 |
590 |
591 |
592 |
593 | # In[26]:
594 |
595 | plt.rcParams['figure.figsize'] = (15, 5)
596 | plt.title("Example images")
597 | plt.imshow(np.concatenate(np_dataset_x_train[:5].astype(np.uint8).transpose((0,2,3,1)),axis=1), interpolation='none');
598 |
599 |
600 | # In[27]:
601 |
602 | plt.title("Example images")
603 | plt.imshow(np.concatenate(np_dataset_y_train[:5,0],axis=1), interpolation='none');
604 |
605 |
606 | # In[28]:
607 |
608 | plt.rcParams['figure.figsize'] = (15, 5)
609 | plt.title("Counts in each image")
610 | plt.bar(range(len(np_dataset_c_train)),np_dataset_c_train);
611 |
612 |
613 | # In[29]:
614 |
615 | print "Total cells in training", np.sum(np_dataset_c_train[0:], axis=0)
616 | print "Total cells in validation", np.sum(np_dataset_c_valid[0:], axis=0)
617 | print "Total cells in testing", np.sum(np_dataset_c_test[0:], axis=0)
618 |
619 |
620 | # In[ ]:
621 |
622 |
623 |
624 |
625 | # In[30]:
626 |
627 | #to make video: ffmpeg -i images-cell/image-0-%d-cell.png -vcodec libx264 aout.mp4
628 | def processImages(name, i):
629 | fig = plt.Figure(figsize=(18, 9), dpi=160)
630 | gcf = plt.gcf()
631 | gcf.set_size_inches(18, 15)
632 | fig.set_canvas(gcf.canvas)
633 |
634 | (img, lab, count) = dataset[i]
635 |
636 | #print str(i),count
637 | pcount = classify([img.transpose((2,0,1))], [0])[0]
638 |
639 | lab_est = [(l.sum()/(ef)).astype(np.int) for l in lab]
640 |
641 | #print lab_est
642 |
643 | pred_est = [(l.sum()/(ef)).astype(np.int) for l in pcount]
644 |
645 | print str(i),"label est ",lab_est," --> predicted est ",pred_est
646 |
647 | ax2 = plt.subplot2grid((2,6), (0, 0), colspan=2)
648 | ax3 = plt.subplot2grid((2,6), (0, 2), colspan=5)
649 | ax4 = plt.subplot2grid((2,6), (1, 2), colspan=5)
650 | ax5 = plt.subplot2grid((2,6), (1, 0), rowspan=1)
651 | ax6 = plt.subplot2grid((2,6), (1, 1), rowspan=1)
652 |
653 | ax2.set_title("Input Image")
654 | ax2.imshow(img, interpolation='none', cmap='Greys_r')
655 | ax3.set_title("Regression target, {}x{} sliding window.".format(patch_size, patch_size))
656 | ax3.imshow(np.concatenate((lab),axis=1), interpolation='none')
657 | ax4.set_title("Predicted counts")
658 | ax4.imshow(np.concatenate((pcount),axis=1), interpolation='none')
659 |
660 | ax5.set_title("Real " + str(lab_est))
661 | ax5.set_ylim((0, np.max(lab_est)*2))
662 | ax5.set_xticks(np.arange(0, noutputs, 1.0))
663 | ax5.bar(range(noutputs),lab_est, align='center')
664 | ax6.set_title("Pred " + str(pred_est))
665 | ax6.set_ylim((0, np.max(lab_est)*2))
666 | ax6.set_xticks(np.arange(0, noutputs, 1.0))
667 | ax6.bar(range(noutputs),pred_est, align='center')
668 | if not os.path.exists('images-cell'):
669 | os.mkdir('images-cell')
670 | fig.savefig('images-cell/image-' + str(i) + "-" + name + '.png', bbox_inches='tight', pad_inches=0)
671 |
672 |
673 | # In[ ]:
674 |
675 |
676 |
677 |
678 | # In[ ]:
679 |
680 |
681 |
682 |
683 | # In[31]:
684 |
685 | import pickle, os
686 |
687 | directory = "network-temp/"
688 | ext = "countception.p"
689 |
690 | if not os.path.exists(directory):
691 | os.makedirs(directory)
692 |
693 | def save_network(net,name):
694 | pkl_params = lasagne.layers.get_all_param_values(net, trainable=True)
695 | out = open(directory + str(name) + ext, "w", 0) #bufsize=0
696 | pickle.dump(pkl_params, out)
697 | out.close()
698 |
699 | def load_network(net,name):
700 | all_param_values = pickle.load(open(directory + str(name) + ext, "r" ))
701 | lasagne.layers.set_all_param_values(net, all_param_values, trainable=True)
702 |
703 |
704 | # In[ ]:
705 |
706 |
707 |
708 |
709 | # In[ ]:
710 |
711 |
712 |
713 |
714 | # In[32]:
715 |
716 | #test accuracy
717 | def test_perf(dataset_x, dataset_y, dataset_c):
718 |
719 | testpixelerrors = []
720 | testerrors = []
721 | bs = 1
722 | for i in range(0,len(dataset_x), bs):
723 |
724 | pcount = classify(dataset_x,range(i,i+bs))
725 | pixelerr = np.abs(pcount - dataset_y[i:i+bs]).mean(axis=(2,3))
726 | testpixelerrors.append(pixelerr)
727 |
728 | pred_est = (pcount/(ef)).sum(axis=(1,2,3))
729 | err = np.abs(dataset_c[i:i+bs].flatten()-pred_est)
730 |
731 | testerrors.append(err)
732 |
733 | return np.abs(testpixelerrors).mean(), np.abs(testerrors).mean()
734 |
735 |
736 | # In[33]:
737 |
738 | print "Random performance"
739 | print test_perf(np_dataset_x_train, np_dataset_y_train, np_dataset_c_train)
740 | print test_perf(np_dataset_x_valid, np_dataset_y_valid, np_dataset_c_valid)
741 | print test_perf(np_dataset_x_test, np_dataset_y_test, np_dataset_c_test)
742 |
743 |
744 | # In[ ]:
745 |
746 |
747 |
748 |
749 | # In[ ]:
750 |
751 |
752 |
753 |
754 | # In[ ]:
755 |
756 |
757 |
758 |
759 | # In[34]:
760 |
761 | target_var = T.tensor4('target')
762 | lr = theano.shared(np.array(args.lr, dtype=theano.config.floatX))
763 |
764 | #Mean Absolute Error is computed between each count of the count map
765 | l1_loss = T.abs_(prediction - target_var[input_var_ex])
766 |
767 | #Mean Absolute Error is computed for the overall image prediction
768 | prediction_count2 =(prediction/ef).sum(axis=(2,3))
769 | mae_loss = T.abs_(prediction_count2 - (target_var[input_var_ex]/ef).sum(axis=(2,3)))
770 |
771 | loss = l1_loss.mean()
772 |
773 | params = lasagne.layers.get_all_params(net, trainable=True)
774 | updates = lasagne.updates.adam(loss, params, learning_rate=lr)
775 |
776 | train_fn = theano.function([input_var_ex], [loss,mae_loss], updates=updates,
777 | givens={input_var:np_dataset_x_train, target_var:np_dataset_y_train})
778 |
779 | print "DONE compiling theano functons"
780 |
781 |
782 | # In[35]:
783 |
784 | #lr.set_value(0.00005)
785 | best_valid_err = 99999999
786 | best_test_err = 99999999
787 | epoch = 0
788 |
789 |
790 | # In[36]:
791 |
792 | batch_size = 2
793 |
794 | print "batch_size", batch_size
795 | print "lr", lr.eval()
796 |
797 | datasetlength = len(np_dataset_x_train)
798 | print "datasetlength",datasetlength
799 |
800 | for epoch in range(epoch, 1000):
801 | start_time = time.time()
802 |
803 | epoch_err_pix = []
804 | epoch_err_pred = []
805 | todo = range(datasetlength)
806 |
807 | for i in range(0,datasetlength, batch_size):
808 | ex = todo[i:i+batch_size]
809 |
810 | train_start_time = time.time()
811 | err_pix,err_pred = train_fn(ex)
812 | train_elapsed_time = time.time() - train_start_time
813 |
814 | epoch_err_pix.append(err_pix)
815 | epoch_err_pred.append(err_pred)
816 |
817 | valid_pix_err, valid_err = test_perf(np_dataset_x_valid, np_dataset_y_valid, np_dataset_c_valid)
818 |
819 | # a threshold is used to reduce processing when we are far from the goal
820 | if (valid_err < 10 and valid_err < best_valid_err):
821 | best_valid_err = valid_err
822 | best_test_err = test_perf(np_dataset_x_test, np_dataset_y_test,np_dataset_c_test)
823 | print "OOO best test (err_pix, err_pred)", best_test_err, ",epoch",epoch
824 | save_network(net,"best_valid_err" + job_id)
825 |
826 |
827 | elapsed_time = time.time() - start_time
828 | err = np.mean(epoch_err_pix)
829 | acc = np.mean(np.concatenate(epoch_err_pred))
830 |
831 | if epoch % 5 == 0:
832 | print "#" + str(epoch) + "#(err_pix:" + str(np.around(err,3)) + ",err_pred:" + str(np.around(acc,3)) + "),valid(err_pix:" + str(np.around(valid_pix_err,3)) + ",err_pred:" + str(np.around(valid_err,3)) +"),(time:" + str(np.around(elapsed_time,3)) + "sec)"
833 |
834 | #visualize training
835 | #processImages(str(epoch) + '-cell',0)
836 |
837 | print "#####", "best_test_acc", best_test_err, args
838 |
839 |
840 | # In[37]:
841 |
842 | print "Done"
843 |
844 |
845 | # In[ ]:
846 |
847 |
848 |
849 |
850 | # In[38]:
851 |
852 | #load best network
853 | load_network(net,"best_valid_err" + job_id)
854 |
855 |
856 | # In[ ]:
857 |
858 |
859 |
860 |
861 | # In[ ]:
862 |
863 |
864 |
865 |
866 | # In[39]:
867 |
868 | def compute_counts(dataset_x):
869 |
870 | bs = 1
871 | ests = []
872 | for i in range(0,len(dataset_x), bs):
873 | pcount = classify(dataset_x,range(i,i+bs))
874 | pred_est = (pcount/(ef)).sum(axis=(1,2,3))
875 | ests.append(pred_est)
876 | return ests
877 |
878 |
879 | # In[ ]:
880 |
881 |
882 |
883 |
884 | # In[40]:
885 |
886 | plt.rcParams['figure.figsize'] = (15, 5)
887 | plt.title("Training Data")
888 |
889 | pcounts = compute_counts(np_dataset_x_train)
890 | plt.bar(np.arange(len(np_dataset_c_train))-0.1,np_dataset_c_train, width=0.5, label="Real Count");
891 | plt.bar(np.arange(len(np_dataset_c_train))+0.1,pcounts, width=0.5,label="Predicted Count");
892 | plt.tight_layout()
893 | plt.legend()
894 |
895 |
896 | # In[41]:
897 |
898 | plt.rcParams['figure.figsize'] = (15, 5)
899 | plt.title("Valid Data")
900 |
901 | pcounts = compute_counts(np_dataset_x_valid)
902 | plt.bar(np.arange(len(np_dataset_c_valid))-0.1,np_dataset_c_valid, width=0.5, label="Real Count");
903 | plt.bar(np.arange(len(np_dataset_c_valid))+0.1,pcounts, width=0.5,label="Predicted Count");
904 | plt.tight_layout()
905 | plt.legend()
906 |
907 |
908 | # In[42]:
909 |
910 | plt.rcParams['figure.figsize'] = (15, 5)
911 | plt.title("Test Data")
912 |
913 | pcounts = compute_counts(np_dataset_x_test)
914 | plt.bar(np.arange(len(np_dataset_c_test))-0.1,np_dataset_c_test, width=0.5, label="Real Count");
915 | plt.bar(np.arange(len(np_dataset_c_test))+0.1,pcounts, width=0.5,label="Predicted Count");
916 | plt.tight_layout()
917 | plt.legend()
918 |
919 |
920 | # In[ ]:
921 |
922 |
923 |
924 |
925 | # In[43]:
926 |
927 | processImages('test',0)
928 |
929 |
930 | # In[44]:
931 |
932 | processImages('test',1)
933 |
934 |
935 | # In[45]:
936 |
937 | processImages('test',2)
938 |
939 |
940 | # In[46]:
941 |
942 | processImages('test',3)
943 |
944 |
945 | # In[47]:
946 |
947 | processImages('test',4)
948 |
949 |
950 | # In[48]:
951 |
952 | processImages('test',5)
953 |
954 |
955 | # In[49]:
956 |
957 | processImages('test',10)
958 |
959 |
960 | # In[ ]:
961 |
962 |
963 |
964 |
965 | # In[ ]:
966 |
967 |
968 |
969 |
970 | # In[ ]:
971 |
972 |
973 |
974 |
975 | # In[ ]:
976 |
977 |
978 |
979 |
980 | # In[ ]:
981 |
982 |
983 |
984 |
--------------------------------------------------------------------------------
/theano/counting/models.py:
--------------------------------------------------------------------------------
1 | #
2 | # This code can be added to switch between multiple size models using this code:
3 | #
4 | # import models
5 | # input_var, input_var_ex, net = models.model(patch_size,framesize, noutputs, stride)
6 | #
7 |
8 |
9 | import sys,os,time,random
10 | import numpy as np
11 |
12 | import theano
13 | import theano.tensor as T
14 | import lasagne
15 | from lasagne.layers.normalization import BatchNormLayer
16 | from lasagne.layers import InputLayer, ConcatLayer, Conv2DLayer
17 |
18 | import pickle
19 | import scipy
20 |
21 | def ConvFactory(data, num_filter, filter_size, stride=1, pad=0, nonlinearity=lasagne.nonlinearities.leaky_rectify):
22 | data = lasagne.layers.batch_norm(Conv2DLayer(
23 | data, num_filters=num_filter,
24 | filter_size=filter_size,
25 | stride=stride, pad=pad,
26 | nonlinearity=nonlinearity,
27 | W=lasagne.init.GlorotUniform(gain='relu')))
28 | return data
29 |
30 | def SimpleFactory(data, ch_1x1, ch_3x3):
31 | conv1x1 = ConvFactory(data=data, filter_size=1, pad=0, num_filter=ch_1x1)
32 | conv3x3 = ConvFactory(data=data, filter_size=3, pad=1, num_filter=ch_3x3)
33 | concat = ConcatLayer([conv1x1, conv3x3])
34 | return concat
35 |
36 |
37 | def model(patch_size,framesize, noutputs, stride):
38 | if patch_size == 32:
39 | return model_32x32(framesize, noutputs, stride)
40 | elif patch_size == 64:
41 | return model_64x64(framesize, noutputs, stride)
42 | elif patch_size == 128:
43 | return model_128x128(framesize, noutputs, stride)
44 | else:
45 | raise Exception('No network for that size')
46 |
47 |
48 | def model_32x32(framesize, noutputs, stride):
49 |
50 | patch_size = 32
51 |
52 | input_var = T.tensor4('inputs')
53 | input_var_ex = T.ivector('input_var_ex')
54 |
55 | input_shape = (None, 1, framesize, framesize)
56 | img = InputLayer(shape=input_shape, input_var=input_var[input_var_ex])
57 | net = img
58 |
59 | net = ConvFactory(net, filter_size=3, num_filter=64, pad=patch_size)
60 | print net.output_shape
61 | net = SimpleFactory(net, 16, 16)
62 | print net.output_shape
63 | net = SimpleFactory(net, 16, 32)
64 | print net.output_shape
65 | net = ConvFactory(net, filter_size=14, num_filter=16)
66 | print net.output_shape
67 | net = SimpleFactory(net, 112, 48)
68 | print net.output_shape
69 | net = SimpleFactory(net, 64, 32)
70 | print net.output_shape
71 | net = SimpleFactory(net, 40, 40)
72 | print net.output_shape
73 | net = SimpleFactory(net, 32, 96)
74 | print net.output_shape
75 | net = ConvFactory(net, filter_size=20, num_filter=32)
76 | print net.output_shape
77 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
78 | print net.output_shape
79 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
80 | print net.output_shape
81 | net = ConvFactory(net, filter_size=1, num_filter=1, stride=stride)
82 | print net.output_shape
83 |
84 | return input_var, input_var_ex, net
85 |
86 |
87 | def model_64x64(framesize, noutputs, stride):
88 |
89 | patch_size = 64
90 | input_var = T.tensor4('inputs')
91 | input_var_ex = T.ivector('input_var_ex')
92 |
93 | input_shape = (None, 1, framesize, framesize)
94 | img = InputLayer(shape=input_shape, input_var=input_var[input_var_ex])
95 | net = img
96 |
97 | net = ConvFactory(net, filter_size=3, num_filter=64, pad=patch_size)
98 | print net.output_shape
99 | net = SimpleFactory(net, 16, 16)
100 | print net.output_shape
101 | net = SimpleFactory(net, 16, 32)
102 | print net.output_shape
103 | net = ConvFactory(net, filter_size=24, num_filter=16)
104 | print net.output_shape
105 | net = SimpleFactory(net, 112, 48)
106 | print net.output_shape
107 | net = SimpleFactory(net, 64, 32)
108 | print net.output_shape
109 | net = SimpleFactory(net, 40, 40)
110 | print net.output_shape
111 | net = SimpleFactory(net, 32, 96)
112 | print net.output_shape
113 | net = ConvFactory(net, filter_size=42, num_filter=32)
114 | print net.output_shape
115 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
116 | print net.output_shape
117 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
118 | print net.output_shape
119 | net = ConvFactory(net, filter_size=1, num_filter=1, stride=stride)
120 | print net.output_shape
121 |
122 | return input_var, input_var_ex, net
123 |
124 |
125 |
126 | def model_128x128(framesize, noutputs, stride):
127 |
128 | patch_size =128
129 | input_var = T.tensor4('inputs')
130 | input_var_ex = T.ivector('input_var_ex')
131 |
132 | input_shape = (None, 1, framesize, framesize)
133 | img = InputLayer(shape=input_shape, input_var=input_var[input_var_ex])
134 | net = img
135 |
136 | net = ConvFactory(net, filter_size=3, num_filter=64, pad=patch_size)
137 | print net.output_shape
138 | net = SimpleFactory(net, 16, 16)
139 | print net.output_shape
140 | net = SimpleFactory(net, 16, 32)
141 | print net.output_shape
142 | net = ConvFactory(net, filter_size=64, num_filter=16)
143 | print net.output_shape
144 | net = SimpleFactory(net, 112, 48)
145 | print net.output_shape
146 | net = SimpleFactory(net, 64, 32)
147 | print net.output_shape
148 | net = SimpleFactory(net, 40, 40)
149 | print net.output_shape
150 | net = SimpleFactory(net, 32, 96)
151 | print net.output_shape
152 | net = ConvFactory(net, filter_size=66, num_filter=32)
153 | print net.output_shape
154 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
155 | print net.output_shape
156 | net = ConvFactory(net, filter_size=1, pad=0, num_filter=64)
157 | print net.output_shape
158 | net = ConvFactory(net, filter_size=1, num_filter=1, stride=stride)
159 | print net.output_shape
160 |
161 | return input_var, input_var_ex, net
162 |
163 |
164 |
--------------------------------------------------------------------------------