├── object-locator
├── checkpoints
│ └── .gitignore
├── __init__.py
├── models
│ ├── __init__.py
│ ├── utils.py
│ ├── unet_parts.py
│ └── unet_model.py
├── __main__.py
├── make_metric_plots.py
├── paint.py
├── metrics_from_results.py
├── bmm.py
├── find_lr.py
├── logger.py
├── losses.py
├── data_plant_stuff.py
├── utils.py
├── get_image_size.py
├── locate.py
├── metrics.py
├── data.py
└── train.py
├── environment.yml
├── .gitignore
├── setup.py
├── scripts_dataset_and_results
├── parseResults.py
├── generate_csv.py
└── spacing_stats_to_csv.py
├── README.md
└── COPYRIGHT.txt
/object-locator/checkpoints/.gitignore:
--------------------------------------------------------------------------------
1 | # https://stackoverflow.com/questions/115983/how-can-i-add-an-empty-directory-to-a-git-repository#932982
2 | # Ignore everything in this directory
3 | *
4 | # Except this file
5 | !.gitignore
6 |
--------------------------------------------------------------------------------
/object-locator/__init__.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
--------------------------------------------------------------------------------
/object-locator/models/__init__.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
--------------------------------------------------------------------------------
/environment.yml:
--------------------------------------------------------------------------------
1 | name: object-locator
2 | channels:
3 | - pytorch
4 | - conda-forge
5 | - defaults
6 | dependencies:
7 | - imageio=2.3.0
8 | - ipdb=0.11
9 | - ipython=6.3.1
10 | - ipython_genutils=0.2.0
11 | - matplotlib=2.2.2
12 | - numpy=1.14.3
13 | - opencv=3.4.1
14 | - pandas=0.22.0
15 | - parse=1.8.2
16 | - pip=9.0.3
17 | - python=3.6.5
18 | - python-dateutil=2.7.2
19 | - scikit-image=0.13.1
20 | - scikit-learn=0.19.1
21 | - scipy=1.0.1
22 | - setuptools=39.1.0
23 | - tqdm=4.23.1
24 | - xmltodict=0.11.0
25 | - pytorch=1.0.0
26 | - pip:
27 | - ballpark==1.4.0
28 | - visdom==0.1.8.5
29 | - peterpy
30 | - torchvision==0.2.1
31 |
32 |
--------------------------------------------------------------------------------
/object-locator/__main__.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 | # Allow printing Unicode characters
18 | import os
19 | os.environ["PYTHONIOENCODING"] = 'UTF-8'
20 |
21 | # Execute locate.py script
22 | from . import locate as object_locator
23 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | *.egg-info/
24 | .installed.cfg
25 | *.egg
26 | MANIFEST
27 |
28 | # PyInstaller
29 | # Usually these files are written by a python script from a template
30 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
31 | *.manifest
32 | *.spec
33 |
34 | # Installer logs
35 | pip-log.txt
36 | pip-delete-this-directory.txt
37 |
38 | # Unit test / coverage reports
39 | htmlcov/
40 | .tox/
41 | .coverage
42 | .coverage.*
43 | .cache
44 | nosetests.xml
45 | coverage.xml
46 | *.cover
47 | .hypothesis/
48 |
49 | # Translations
50 | *.mo
51 | *.pot
52 |
53 | # Django stuff:
54 | *.log
55 | .static_storage/
56 | .media/
57 | local_settings.py
58 |
59 | # Flask stuff:
60 | instance/
61 | .webassets-cache
62 |
63 | # Scrapy stuff:
64 | .scrapy
65 |
66 | # Sphinx documentation
67 | docs/_build/
68 |
69 | # PyBuilder
70 | target/
71 |
72 | # Jupyter Notebook
73 | .ipynb_checkpoints
74 |
75 | # pyenv
76 | .python-version
77 |
78 | # celery beat schedule file
79 | celerybeat-schedule
80 |
81 | # SageMath parsed files
82 | *.sage.py
83 |
84 | # Environments
85 | .env
86 | .venv
87 | env/
88 | venv/
89 | ENV/
90 | env.bak/
91 | venv.bak/
92 |
93 | # Spyder project settings
94 | .spyderproject
95 | .spyproject
96 |
97 | # Rope project settings
98 | .ropeproject
99 |
100 | # mkdocs documentation
101 | /site
102 |
103 | # mypy
104 | .mypy_cache/
105 |
106 | # vim
107 | *.swp
108 |
--------------------------------------------------------------------------------
/object-locator/models/utils.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import h5py
19 | import torch
20 | import shutil
21 |
22 | def save_net(fname, net):
23 | with h5py.File(fname, 'w') as h5f:
24 | for k, v in net.state_dict().items():
25 | h5f.create_dataset(k, data=v.cpu().numpy())
26 | def load_net(fname, net):
27 | with h5py.File(fname, 'r') as h5f:
28 | for k, v in net.state_dict().items():
29 | param = torch.from_numpy(np.asarray(h5f[k]))
30 | v.copy_(param)
31 |
32 | def save_checkpoint(state, is_best,task_id, filename='checkpoint.pth.tar'):
33 | torch.save(state, task_id+filename)
34 | if is_best:
35 | shutil.copyfile(task_id+filename, task_id+'model_best.pth.tar')
36 |
37 |
38 | """
39 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
40 | All rights reserved.
41 |
42 | This software is covered by US patents and copyright.
43 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
44 |
45 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
46 |
47 | Last Modified: 10/02/2019
48 | """
49 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | from setuptools import setup, find_packages
2 |
3 | setup(
4 | name='object-locator',
5 | version='1.6.0',
6 | description='Object Location using PyTorch.',
7 |
8 | # The project's main homepage.
9 | url='https://engineering.purdue.edu/~sorghum',
10 |
11 | # Author details
12 | author='Javier Ribera, David Guera, Yuhao Chen, and Edward J. Delp',
13 | author_email='ace@ecn.purdue.edu',
14 | # See https://pypi.python.org/pypi?%3Aaction=list_classifiers
15 | classifiers=[
16 | # How mature is this project? Common values are
17 | # 3 - Alpha
18 | # 4 - Beta
19 | # 5 - Production/Stable
20 | 'Development Status :: 4 - Beta',
21 |
22 | # Specify the Python versions you support here. In particular, ensure
23 | # that you indicate whether you support Python 2, Python 3 or both.
24 | 'Programming Language :: Python :: 3.6',
25 | ],
26 | python_requires='~=3.6',
27 | # What does your project relate to?
28 | keywords='object localization location purdue',
29 |
30 | # You can just specify the packages manually here if your project is
31 | # simple. Or you can use find_packages().
32 | packages=['object-locator', 'object-locator.models'],
33 | package_dir={'object-locator': 'object-locator'},
34 |
35 | package_data={'object-locator': ['checkpoints/*.ckpt',
36 | '../COPYRIGHT.txt',
37 | '../README.md']},
38 | include_package_data=True,
39 |
40 | # List run-time dependencies here. These will be installed by pip when
41 | # your project is installed. For an analysis of "install_requires" vs pip's
42 | # requirements files see:
43 | # https://packaging.python.org/en/latest/requirements.html
44 | # (We actually use conda for dependency management)
45 | # install_requires=['matplotlib', 'numpy',
46 | # 'scikit-image', 'tqdm', 'argparse', 'parse',
47 | # 'scikit-learn', 'pandas'],
48 | )
49 |
--------------------------------------------------------------------------------
/object-locator/make_metric_plots.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import os
19 | import numpy as np
20 | import pandas as pd
21 | import argparse
22 |
23 | from . import metrics
24 |
25 | # Parse command-line arguments
26 | parser = argparse.ArgumentParser(
27 | description='Create a bunch of plot from the metrics in a CSV.',
28 | formatter_class=argparse.ArgumentDefaultsHelpFormatter)
29 | parser.add_argument('csv',
30 | help='CSV file with the precision and recall results.')
31 | parser.add_argument('out',
32 | help='Output directory.')
33 | parser.add_argument('--title',
34 | default='',
35 | help='Title of the plot in the figure.')
36 | parser.add_argument('--taus',
37 | type=str,
38 | required=True,
39 | help='Detection threshold taus. '
40 | 'For each of these taus, a precision(r) and recall(r) will be created.'
41 | 'The closest to these values will be used.')
42 | parser.add_argument('--radii',
43 | type=str,
44 | required=True,
45 | help='List of values, each with different colors in the scatter plot. '
46 | 'Maximum distance to consider a True Positive. '
47 | 'The closest to this value will be used.')
48 | args = parser.parse_args()
49 |
50 |
51 | os.makedirs(args.out, exist_ok=True)
52 |
53 | taus = [float(tau) for tau in args.taus.replace('[', '').replace(']', '').split(',')]
54 | radii = [int(r) for r in args.radii.replace('[', '').replace(']', '').split(',')]
55 |
56 | figs = metrics.make_metric_plots(csv_path=args.csv,
57 | taus=taus,
58 | radii=radii,
59 | title=args.title)
60 |
61 | for label, fig in figs.items():
62 | # Save to disk
63 | fig.savefig(os.path.join(args.out, f'{label}.png'))
64 |
65 |
66 | """
67 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
68 | All rights reserved.
69 |
70 | This software is covered by US patents and copyright.
71 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
72 |
73 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
74 |
75 | Last Modified: 10/02/2019
76 | """
77 |
--------------------------------------------------------------------------------
/object-locator/paint.py:
--------------------------------------------------------------------------------
1 | from __future__ import print_function
2 |
3 | __copyright__ = \
4 | """
5 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
6 | All rights reserved.
7 |
8 | This software is covered by US patents and copyright.
9 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
10 |
11 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
12 |
13 | Last Modified: 10/02/2019
14 | """
15 | __license__ = "CC BY-NC-SA 4.0"
16 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
17 | __version__ = "1.6.0"
18 |
19 |
20 | import os
21 | import sys
22 |
23 | import cv2
24 | from tqdm import tqdm
25 | import numpy as np
26 | import torch
27 | from torchvision import transforms
28 | from torch.utils import data
29 |
30 | from .data import CSVDataset
31 | from .data import csv_collator
32 | from . import argparser
33 | from . import utils
34 |
35 |
36 | # Parse command line arguments
37 | args = argparser.parse_command_args('testing')
38 |
39 | # Tensor type to use, select CUDA or not
40 | torch.set_default_dtype(torch.float32)
41 | device_cpu = torch.device('cpu')
42 |
43 | # Set seeds
44 | np.random.seed(args.seed)
45 | torch.manual_seed(args.seed)
46 | if args.cuda:
47 | torch.cuda.manual_seed_all(args.seed)
48 |
49 | # Data loading code
50 | try:
51 | testset = CSVDataset(args.dataset,
52 | transforms=transforms.Compose([
53 | transforms.ToTensor(),
54 | ]),
55 | max_dataset_size=args.max_testset_size)
56 | except ValueError as e:
57 | print(f'E: {e}')
58 | exit(-1)
59 | dataset_loader = data.DataLoader(testset,
60 | batch_size=1,
61 | num_workers=args.nThreads,
62 | collate_fn=csv_collator)
63 |
64 | os.makedirs(os.path.join(args.out), exist_ok=True)
65 |
66 | for img, dictionary in tqdm(dataset_loader):
67 |
68 | # Move to device
69 | img = img.to(device_cpu)
70 |
71 | # One image at a time (BS=1)
72 | img = img[0]
73 | dictionary = dictionary[0]
74 |
75 | # Tensor -> float & numpy
76 | target_locs = dictionary['locations'].to(device_cpu).numpy().reshape(-1, 2)
77 | img = img.to(device_cpu).numpy()
78 |
79 | img *= 255
80 |
81 | # Paint circles on top of image
82 | img_with_x = utils.paint_circles(img=img,
83 | points=target_locs,
84 | color='white')
85 | img_with_x = np.moveaxis(img_with_x, 0, 2)
86 | img_with_x = img_with_x[:, :, ::-1]
87 |
88 | cv2.imwrite(os.path.join(args.out, dictionary['filename']),
89 | img_with_x)
90 |
91 |
92 | """
93 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
94 | All rights reserved.
95 |
96 | This software is covered by US patents and copyright.
97 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
98 |
99 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
100 |
101 | Last Modified: 10/02/2019
102 | """
103 |
--------------------------------------------------------------------------------
/scripts_dataset_and_results/parseResults.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import pandas as pd
19 | import numpy as np
20 | import sys
21 | import ast
22 | import cv2
23 | from sklearn.cluster import KMeans
24 | from sklearn.metrics.pairwise import pairwise_distances
25 | from sklearn import mixture
26 |
27 | CSV_FILE = "estimations.csv"
28 |
29 | def eval_plant_locations(estimated, gt):
30 | """
31 | Distance function between the estimated plant locations and the ground
32 | truth.
33 | This function is a symmetric function which parameter is the estimated
34 | plant locations and which is the ground truth should not matter.
35 | The returned value is guaranteed to be always positive,
36 | and is only zero if both lists are exactly equal.
37 |
38 | :param estimated: List of (x, y) or (y,x) plant locations.
39 | :param gt: List of (x, y) or (y, x) plant locations.
40 | :return: Distance between two sets.
41 | """
42 |
43 | estimated = np.array(estimated)
44 | gt = np.array(gt)
45 |
46 | # Check dimension
47 | assert estimated.ndim == gt.ndim == 2, \
48 | 'Both estimated and GT plant locations must be 2D, i.e, (x, y) or (y, x)'
49 |
50 | d2_matrix = pairwise_distances(estimated, gt, metric='euclidean')
51 |
52 | res = np.average(np.min(d2_matrix, axis=0)) + \
53 | np.average(np.min(d2_matrix, axis=1))
54 |
55 | return res
56 |
57 | def processImg(image, n, GMM=False):
58 | #extract mask from the image
59 | mask = cv2.inRange(image, (5,5,5), (255,255,255))
60 | coord = np.where(mask > 0)
61 | y = coord[0].reshape((-1, 1))
62 | x = coord[1].reshape((-1, 1))
63 |
64 | c = np.concatenate((y, x), axis=1)
65 |
66 | if GMM:
67 | gmm = mixture.GaussianMixture(n_components=n, n_init=1, covariance_type='full').fit(c)
68 | return gmm.means_.astype(np.int)
69 |
70 | else:
71 |
72 | #find kmean cluster
73 | kmeans = KMeans(n_clusters=n, random_state=0).fit(c)
74 | return kmeans.cluster_centers_
75 |
76 | def processCSV(csvfile):
77 |
78 | df = pd.read_csv(csvfile)
79 | res_array = []
80 | for i in range(len(df.iloc[:])):
81 | filename = df.iloc[:, 1][i]
82 |
83 | plant_count = df.iloc[:, 2][i]
84 | plant_count = float(plant_count.split('\n')[1].strip())
85 |
86 | gt = df.iloc[:, 3][i]
87 | gt = ast.literal_eval(gt)
88 |
89 | image = cv2.imread(filename)
90 | detected = processImg(image, int(plant_count), GMM=True)
91 |
92 | res = eval_plant_locations(detected, gt)
93 | res_array.append(res)
94 | print(res)
95 | break
96 | return res_array
97 |
98 |
99 | #Note the script needs to be put into the data directory with the CSV file
100 | res = processCSV(CSV_FILE)
101 |
102 |
103 | """
104 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
105 | All rights reserved.
106 |
107 | This software is covered by US patents and copyright.
108 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
109 |
110 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
111 |
112 | Last Modified: 10/02/2019
113 | """
114 |
--------------------------------------------------------------------------------
/object-locator/models/unet_parts.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | # sub-parts of the U-Net model
19 |
20 | import math
21 | import warnings
22 |
23 | import torch
24 | import torch.nn as nn
25 | import torch.nn.functional as F
26 |
27 |
28 | class double_conv(nn.Module):
29 | def __init__(self, in_ch, out_ch, normaliz=True, activ=True):
30 | super(double_conv, self).__init__()
31 |
32 | ops = []
33 | ops += [nn.Conv2d(in_ch, out_ch, 3, padding=1)]
34 | # ops += [nn.Dropout(p=0.1)]
35 | if normaliz:
36 | ops += [nn.BatchNorm2d(out_ch)]
37 | if activ:
38 | ops += [nn.ReLU(inplace=True)]
39 | ops += [nn.Conv2d(out_ch, out_ch, 3, padding=1)]
40 | # ops += [nn.Dropout(p=0.1)]
41 | if normaliz:
42 | ops += [nn.BatchNorm2d(out_ch)]
43 | if activ:
44 | ops += [nn.ReLU(inplace=True)]
45 |
46 | self.conv = nn.Sequential(*ops)
47 |
48 | def forward(self, x):
49 | x = self.conv(x)
50 | return x
51 |
52 |
53 | class inconv(nn.Module):
54 | def __init__(self, in_ch, out_ch):
55 | super(inconv, self).__init__()
56 | self.conv = double_conv(in_ch, out_ch)
57 |
58 | def forward(self, x):
59 | x = self.conv(x)
60 | return x
61 |
62 |
63 | class down(nn.Module):
64 | def __init__(self, in_ch, out_ch, normaliz=True):
65 | super(down, self).__init__()
66 | self.mpconv = nn.Sequential(
67 | nn.MaxPool2d(2),
68 | double_conv(in_ch, out_ch, normaliz=normaliz)
69 | )
70 |
71 | def forward(self, x):
72 | x = self.mpconv(x)
73 | return x
74 |
75 |
76 | class up(nn.Module):
77 | def __init__(self, in_ch, out_ch, normaliz=True, activ=True):
78 | super(up, self).__init__()
79 | self.up = nn.Upsample(scale_factor=2,
80 | mode='bilinear',
81 | align_corners=True)
82 | # self.up = nn.ConvTranspose2d(in_ch, out_ch, 2, stride=2)
83 | self.conv = double_conv(in_ch, out_ch,
84 | normaliz=normaliz, activ=activ)
85 |
86 | def forward(self, x1, x2):
87 | with warnings.catch_warnings():
88 | warnings.simplefilter("ignore") # Upsample is deprecated
89 | x1 = self.up(x1)
90 | diffY = x2.size()[2] - x1.size()[2]
91 | diffX = x2.size()[3] - x1.size()[3]
92 | x1 = F.pad(x1, (diffX // 2, int(math.ceil(diffX / 2)),
93 | diffY // 2, int(math.ceil(diffY / 2))))
94 | x = torch.cat([x2, x1], dim=1)
95 | x = self.conv(x)
96 | return x
97 |
98 |
99 | class outconv(nn.Module):
100 | def __init__(self, in_ch, out_ch):
101 | super(outconv, self).__init__()
102 | self.conv = nn.Conv2d(in_ch, out_ch, 1)
103 | # self.conv = nn.Sequential(
104 | # nn.Conv2d(in_ch, out_ch, 1),
105 | # )
106 |
107 | def forward(self, x):
108 | x = self.conv(x)
109 | return x
110 |
111 |
112 | """
113 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
114 | All rights reserved.
115 |
116 | This software is covered by US patents and copyright.
117 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
118 |
119 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
120 |
121 | Last Modified: 10/02/2019
122 | """
123 |
--------------------------------------------------------------------------------
/scripts_dataset_and_results/generate_csv.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import pandas as pd
19 | import cv2
20 | import numpy as np
21 | import sys
22 | import os
23 | import ast
24 | import random
25 | import shutil
26 | from tqdm import tqdm
27 |
28 | np.random.seed(0)
29 |
30 | train_df = pd.DataFrame(columns=['plant_count'])
31 | test_df = pd.DataFrame(columns=['plant_count'])
32 | validate_df = pd.DataFrame(columns=['plant_count'])
33 |
34 | if not os.path.exists('train'):
35 | os.makedirs('train')
36 | if not os.path.exists('test'):
37 | os.makedirs('test')
38 | if not os.path.exists('validate'):
39 | os.makedirs('validate')
40 |
41 | dirs = [i for i in range(1, 18)]
42 | dirs.pop(11)
43 |
44 | filecounter = 0
45 | for dirnum in dirs:
46 | dirname = 'dataset' + str(dirnum).zfill(2)
47 |
48 | fd = open(os.path.join(dirname,'gt.txt'))
49 |
50 | data = []
51 | for line in fd:
52 | line = line.strip()
53 | imgnum = line.split(' ')[1]
54 | x = line.split(' ')[2]
55 | if (x == 'X'):
56 | continue
57 | y = line.split(' ')[3]
58 |
59 | imagename = imgnum.zfill(10)+'.png'
60 | if not os.path.exists(os.path.join(dirname,imagename)):
61 | continue
62 | image = cv2.imread(os.path.join(dirname,imagename))
63 |
64 | h = image.shape[0]
65 | x = int(x)/2
66 | y = h - int(y)/2
67 | data.append([imagename, y, x])
68 |
69 | #print(imagename)
70 | #print(x, y)
71 |
72 | random.shuffle(data)
73 | for i in range(len(data)):
74 | item = data[i]
75 | imagename = item[0]
76 | y = item[1]
77 | x = item[2]
78 |
79 | # newname = str(filecounter).zfill(10) + '.png'
80 | newname = dirname + '_' + imagename
81 | df = pd.DataFrame(data=[[1, [[y, x]]]],
82 | index=[newname],
83 | columns=['plant_count', 'plant_locations'])
84 | if (i < len(data)*0.8):
85 | if os.path.isfile('train/'+newname):
86 | print('%s exists' % 'train/'+newname)
87 | exit(-1)
88 | shutil.move(os.path.join(dirname,imagename), 'train/'+newname)
89 | train_df = train_df.append(df)
90 | elif (i < len(data)*0.9):
91 | if os.path.isfile('train/'+newname):
92 | print('%s exists' % 'test/'+newname)
93 | exit(-1)
94 | shutil.move(os.path.join(dirname,imagename), 'test/'+newname)
95 | test_df = test_df.append(df)
96 | else:
97 | if os.path.isfile('train/'+newname):
98 | print('%s exists' % 'test/'+newname)
99 | exit(-1)
100 | shutil.move(os.path.join(dirname,imagename), 'validate/'+newname)
101 | validate_df = validate_df.append(df)
102 |
103 | train_df.to_csv('train.csv')
104 | shutil.move('train.csv', 'train')
105 | test_df.to_csv('test.csv')
106 | shutil.move('test.csv', 'test')
107 | validate_df.to_csv('validate.csv')
108 | shutil.move('validate.csv', 'validate')
109 |
110 |
111 | """
112 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
113 | All rights reserved.
114 |
115 | This software is covered by US patents and copyright.
116 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
117 |
118 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
119 |
120 | Last Modified: 10/02/2019
121 | """
122 |
--------------------------------------------------------------------------------
/object-locator/metrics_from_results.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import os
19 | import argparse
20 | import ast
21 | import math
22 |
23 | from tqdm import tqdm
24 | import numpy as np
25 | import pandas as pd
26 |
27 | from . import metrics
28 | from . import get_image_size
29 |
30 | # Parse command-line arguments
31 | parser = argparse.ArgumentParser(
32 | description='Compute metrics from results and GT.',
33 | formatter_class=argparse.ArgumentDefaultsHelpFormatter)
34 | required_args = parser.add_argument_group('MANDATORY arguments')
35 | optional_args = parser._action_groups.pop()
36 | required_args.add_argument('results',
37 | help='Input CSV file with the estimated locations.')
38 | required_args.add_argument('gt',
39 | help='Input CSV file with the groundtruthed locations.')
40 | required_args.add_argument('metrics',
41 | help='Output CSV file with the metrics '
42 | '(MAE, AHD, Precision, Recall...)')
43 | required_args.add_argument('--dataset',
44 | type=str,
45 | required=True,
46 | help='Dataset directory with the images. '
47 | 'This is used only to get the image diagonal, '
48 | 'as the worst estimate for the AHD.')
49 | optional_args.add_argument('--radii',
50 | type=str,
51 | default=range(0, 15 + 1),
52 | metavar='Rs',
53 | help='Detections at dist <= R to a GT pt are True Positives.')
54 | args = parser.parse_args()
55 |
56 |
57 | # Prepare Judges that will compute P/R as fct of r and th
58 | judges = [metrics.Judge(r=r) for r in args.radii]
59 |
60 | df_results = pd.read_csv(args.results)
61 | df_gt = pd.read_csv(args.gt)
62 |
63 | df_metrics = pd.DataFrame(columns=['r',
64 | 'precision', 'recall', 'fscore', 'MAHD',
65 | 'MAPE', 'ME', 'MPE', 'MAE',
66 | 'MSE', 'RMSE', 'r', 'R2'])
67 |
68 | for j, judge in enumerate(tqdm(judges)):
69 |
70 | for idx, row_result in df_results.iterrows():
71 | filename = row_result['filename']
72 | row_gt = df_gt[df_gt['filename'] == filename].iloc()[0]
73 |
74 | w, h = get_image_size.get_image_size(os.path.join(args.dataset, filename))
75 | diagonal = math.sqrt(w**2 + h**2)
76 |
77 | judge.feed_count(row_result['count'],
78 | row_gt['count'])
79 | judge.feed_points(ast.literal_eval(row_result['locations']),
80 | ast.literal_eval(row_gt['locations']),
81 | max_ahd=diagonal)
82 |
83 | df = pd.DataFrame(data=[[judge.r,
84 | judge.precision,
85 | judge.recall,
86 | judge.fscore,
87 | judge.mahd,
88 | judge.mape,
89 | judge.me,
90 | judge.mpe,
91 | judge.mae,
92 | judge.mse,
93 | judge.rmse,
94 | judge.pearson_corr \
95 | if not np.isnan(judge.pearson_corr) else 1,
96 | judge.coeff_of_determination]],
97 | columns=['r',
98 | 'precision', 'recall', 'fscore', 'MAHD',
99 | 'MAPE', 'ME', 'MPE', 'MAE',
100 | 'MSE', 'RMSE', 'r', 'R2'],
101 | index=[j])
102 | df.index.name = 'idx'
103 | df_metrics = df_metrics.append(df)
104 |
105 | # Write CSV of metrics to disk
106 | df_metrics.to_csv(args.metrics)
107 |
108 |
109 | """
110 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
111 | All rights reserved.
112 |
113 | This software is covered by US patents and copyright.
114 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
115 |
116 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
117 |
118 | Last Modified: 10/02/2019
119 | """
120 |
--------------------------------------------------------------------------------
/object-locator/bmm.py:
--------------------------------------------------------------------------------
1 | """
2 | Code from paper
3 | "A hybrid parameter estimation algorithm for beta mixtures
4 | and applications to methylation state classification"
5 | https://doi.org/10.1186/s13015-017-0112-1
6 | https://bitbucket.org/genomeinformatics/betamix
7 | """
8 |
9 | import numpy as np
10 |
11 | from itertools import count
12 | from argparse import ArgumentParser
13 |
14 | import numpy as np
15 | from scipy.stats import beta
16 |
17 |
18 | def _get_values(x, left, right):
19 | y = x[np.logical_and(x>=left, x<=right)]
20 | n = len(y)
21 | if n == 0:
22 | m = (left+right) / 2.0
23 | v = (right-left) / 12.0
24 | else:
25 | m = np.mean(y)
26 | v = np.var(y)
27 | if v == 0.0:
28 | v = (right-left) / (12.0*(n+1))
29 | return m, v, n
30 |
31 |
32 | def get_initialization(x, ncomponents, limit=0.8):
33 | # TODO: work with specific components instead of just their number
34 | points = np.linspace(0.0, 1.0, ncomponents+2)
35 | means = np.zeros(ncomponents)
36 | variances = np.zeros(ncomponents)
37 | pi = np.zeros(ncomponents)
38 | # init first component
39 | means[0], variances[0], pi[0] = _get_values(x, points[0], points[1])
40 | # init intermediate components
41 | N = ncomponents - 1
42 | for j in range(1, N):
43 | means[j], variances[j], pi[j] = _get_values(x, points[j], points[j+2])
44 | # init last component
45 | means[N], variances[N], pi[N] = _get_values(x, points[N+1], points[N+2])
46 |
47 | # compute parameters ab, pi
48 | ab = [ab_from_mv(m,v) for (m,v) in zip(means,variances)]
49 | pi = pi / pi.sum()
50 |
51 | # adjust first and last
52 | if ab[0][0] >= limit: ab[0] = (limit, ab[0][1])
53 | if ab[-1][1] >= limit: ab[-1] = (ab[-1][0], limit)
54 | return ab, pi
55 |
56 |
57 | def ab_from_mv(m, v):
58 | """
59 | estimate beta parameters (a,b) from given mean and variance;
60 | return (a,b).
61 |
62 | Note, for uniform distribution on [0,1], (m,v)=(0.5,1/12)
63 | """
64 | phi = m*(1-m)/v - 1 # z = 2 for uniform distribution
65 | return (phi*m, phi*(1-m)) # a = b = 1 for uniform distribution
66 |
67 |
68 | def get_weights(x, ab, pi):
69 | """return nsamples X ncomponents matrix with association weights"""
70 | bpdf = beta.pdf
71 | n, c = len(x), len(ab)
72 | y = np.zeros((n,c), dtype=float)
73 | s = np.zeros((n,1), dtype=float)
74 | for (j, p,(a,b)) in zip(count(), pi, ab):
75 | y[:,j] = p * bpdf(x, a, b)
76 | s = np.sum(y,1).reshape((n,1))
77 | with np.warnings.catch_warnings():
78 | np.warnings.filterwarnings('ignore')
79 | w = y / s # this may produce inf or nan; this is o.k.!
80 | # clean up weights w, remove infs, nans, etc.
81 | wfirst = np.array([1] + [0]*(c-1), dtype=float)
82 | wlast = np.array([0]*(c-1) + [1], dtype=float)
83 | bad = (~np.isfinite(w)).any(axis=1)
84 | badfirst = np.logical_and(bad, x<0.5)
85 | badlast = np.logical_and(bad, x>=0.5)
86 | w[badfirst,:] = wfirst
87 | w[badlast,:] = wlast
88 | # now all weights are valid finite values and sum to 1 for each row
89 | assert np.all(np.isfinite(w)), (w, np.isfinite(w))
90 | assert np.allclose(np.sum(w,1), 1.0), np.max(np.abs(np.sum(w,1)-1.0))
91 | return w
92 |
93 |
94 | def relerror(x,y):
95 | if x==y: return 0.0
96 | return abs(x-y)/max(abs(x),abs(y))
97 |
98 | def get_delta(ab, abold, pi, piold):
99 | epi = max(relerror(p,po) for (p,po) in zip(pi,piold))
100 | ea = max(relerror(a,ao) for (a,_), (ao,_) in zip(ab,abold))
101 | eb = max(relerror(b,bo) for (_,b), (_,bo) in zip(ab,abold))
102 | return max(epi,ea,eb)
103 |
104 |
105 | def estimate_mixture(x, init, steps=1000, tolerance=1E-5):
106 | """
107 | estimate a beta mixture model from the given data x
108 | with the given number of components and component types
109 | """
110 | (ab, pi) = init
111 | n, ncomponents = len(x), len(ab)
112 |
113 | for step in count():
114 | if step >= steps:
115 | break
116 | abold = list(ab)
117 | piold = pi[:]
118 | # E-step: compute component memberships for each x
119 | w = get_weights(x, ab, pi)
120 | # compute component means and variances and parameters
121 | for j in range(ncomponents):
122 | wj = w[:,j]
123 | pij = np.sum(wj)
124 | m = np.dot(wj,x) / pij
125 | v = np.dot(wj,(x-m)**2) / pij
126 | if np.isnan(m) or np.isnan(v):
127 | m = 0.5; v = 1/12 # uniform
128 | ab[j]=(1,1) # uniform
129 | assert pij == 0.0
130 | else:
131 | assert np.isfinite(m) and np.isfinite(v), (j,m,v,pij)
132 | ab[j] = ab_from_mv(m,v)
133 | pi[j] = pij / n
134 | delta = get_delta(ab, abold, pi, piold)
135 | if delta < tolerance:
136 | break
137 | usedsteps = step + 1
138 | return (ab, pi, usedsteps)
139 |
140 |
141 | def estimate(x, components, steps=1000, tolerance=1E-4):
142 | init = get_initialization(x, len(components))
143 | (ab, pi, usedsteps) = estimate_mixture(x, init, steps=steps, tolerance=tolerance)
144 | return (ab, pi, usedsteps)
145 |
146 |
147 | class AccumHistogram1D():
148 | """https://raw.githubusercontent.com/NichtJens/numpy-accumulative-histograms/master/accuhist.py"""
149 |
150 | def __init__(self, nbins, xlow, xhigh):
151 | self.nbins = nbins
152 | self.xlow = xlow
153 | self.xhigh = xhigh
154 |
155 | self.range = (xlow, xhigh)
156 |
157 | self.hist, edges = np.histogram([], bins=nbins, range=self.range)
158 | self.bins = (edges[:-1] + edges[1:]) / 2.
159 |
160 | def fill(self, arr):
161 | hist, _ = np.histogram(arr, bins=self.nbins, range=self.range)
162 | self.hist += hist
163 |
164 | @property
165 | def data(self):
166 | return self.bins, self.hist
167 |
168 |
169 |
--------------------------------------------------------------------------------
/scripts_dataset_and_results/spacing_stats_to_csv.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import argparse
19 | import os
20 | import pandas as pd
21 | from tqdm import tqdm
22 | from scipy.spatial.distance import euclidean as distance
23 | import statistics
24 | import matplotlib.mlab as mlab
25 | import matplotlib.pyplot as plt
26 | import numpy as np
27 |
28 | if __name__ == '__main__':
29 | # Parse command-line arguments
30 | parser = argparse.ArgumentParser(
31 | description='Compute intra-row spacing stats of a CSV. '
32 | 'Add mean, median, and stdev of each row. '
33 | 'Optional: plot histograms',
34 | formatter_class=argparse.ArgumentDefaultsHelpFormatter)
35 | parser.add_argument('in_csv',
36 | help='Input CSV with plant location info.')
37 | parser.add_argument('out_csv',
38 | help='Output CSV with the added stats.')
39 | parser.add_argument('--hist',
40 | metavar='DIR',
41 | help='Directory with histograms.')
42 | parser.add_argument('--res',
43 | metavar='DIR',
44 | type=float,
45 | default=1,
46 | help='Resolution in centimeters.')
47 | args = parser.parse_args()
48 |
49 | # Import GT from CSV
50 | df = pd.read_csv(args.in_csv)
51 |
52 | # Store stats of each single-row plot
53 | means, medians, stds = [], [], []
54 |
55 | for idx, row in tqdm(df.iterrows(), total=len(df.index)):
56 | if row['locations_wrt_orthophoto'] is np.nan:
57 | continue
58 | locs = eval(row['locations_wrt_orthophoto'])
59 |
60 | # 1. Sort by row coordinate
61 | locs = sorted(locs, key=lambda x: x[0])
62 |
63 | # 2. Compute distances (chain-like) between plants
64 | dists = list(map(distance, locs[:-1], locs[1:]))
65 |
66 | # 3. pixels -> centimeters
67 | dists = [d * args.res for d in dists]
68 |
69 | # 4. Statistics!
70 | mean = statistics.mean(dists)
71 | median = statistics.median(dists)
72 | std = statistics.stdev(dists)
73 | means.append(mean)
74 | medians.append(median)
75 | stds.append(std)
76 |
77 | # 5. Put in CSV
78 | df.loc[idx, 'mean_intrarow_spacing_in_cm'] = mean
79 | df.loc[idx, 'median_intrarow_spacing_in_cm'] = median
80 | df.loc[idx, 'stdev_intrarow_spacing_in_cm'] = std
81 |
82 | # Save to disk as CSV
83 | df.to_csv(args.out_csv)
84 |
85 | if args.hist is not None:
86 | os.makedirs(args.hist, exist_ok=True)
87 |
88 | # 6. Generate nice graphs for presentation
89 | # Means
90 | fig = plt.figure()
91 | n, bins, patches = plt.hist(
92 | means, 30, normed=1, facecolor='green', alpha=0.75, label='Histogram')
93 | # add a 'best fit' norm line
94 | y = mlab.normpdf(bins, statistics.mean(means), statistics.stdev(means))
95 | l = plt.plot(bins, y, 'r--', linewidth=1, label='Fitted Gaussian')
96 | plt.xlabel('Average intra-row spacing [cm]')
97 | plt.ylabel('Probability')
98 | plt.title('Histogram of average intra-row spacing')
99 | plt.axis([5, 30, 0, 0.3])
100 | plt.grid(True)
101 | plt.legend()
102 | fig.savefig(os.path.join(
103 | args.hist, 'histogram_averages.png'), dpi=fig.dpi)
104 |
105 | # Medians
106 | fig = plt.figure()
107 | n, bins, patches = plt.hist(
108 | medians, 30, normed=1, facecolor='green', alpha=0.75, label='Histogram')
109 | # add a 'best fit' norm line
110 | y = mlab.normpdf(bins, statistics.mean(
111 | medians), statistics.stdev(medians))
112 | l = plt.plot(bins, y, 'r--', linewidth=1, label='Fitted Gaussian')
113 | plt.xlabel('Median of intra-row spacing [cm]')
114 | plt.ylabel('Probability')
115 | plt.title('Histogram of medians intra-row spacing')
116 | plt.axis([5, 30, 0, 0.3])
117 | plt.grid(True)
118 | plt.legend()
119 | fig.savefig(os.path.join(
120 | args.hist, 'histogram_medians.png'), dpi=fig.dpi)
121 |
122 | # Standard deviations
123 | fig = plt.figure()
124 | n, bins, patches = plt.hist(
125 | stds, 30, normed=1, facecolor='green', alpha=0.75, label='Histogram')
126 | # add a 'best fit' norm line
127 | y = mlab.normpdf(bins, statistics.mean(stds), statistics.stdev(stds))
128 | l = plt.plot(bins, y, 'r--', linewidth=1, label='Fitted Gaussian')
129 | plt.xlabel('Standard deviation of intra-row spacing [cm]')
130 | plt.ylabel('Probability')
131 | plt.title('Histogram of standard deviations of intra-row spacing')
132 | plt.axis([0, 25, 0, 0.3])
133 | plt.grid(True)
134 | plt.legend()
135 | fig.savefig(os.path.join(
136 | args.hist, 'histogram_stdevs.png'), dpi=fig.dpi)
137 |
138 |
139 | """
140 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
141 | All rights reserved.
142 |
143 | This software is covered by US patents and copyright.
144 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
145 |
146 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
147 |
148 | Last Modified: 10/02/2019
149 | """
150 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Locating Objects Without Bounding Boxes
2 | PyTorch code for "Locating Objects Without Bounding Boxes" , CVPR 2019 - Oral, Best Paper Finalist (Top 1 %) [[Paper]](http://openaccess.thecvf.com/content_CVPR_2019/html/Ribera_Locating_Objects_Without_Bounding_Boxes_CVPR_2019_paper.html) [[Youtube]](https://youtu.be/8qkrPSjONhA?t=2620)
3 |
4 |  5 |
5 | 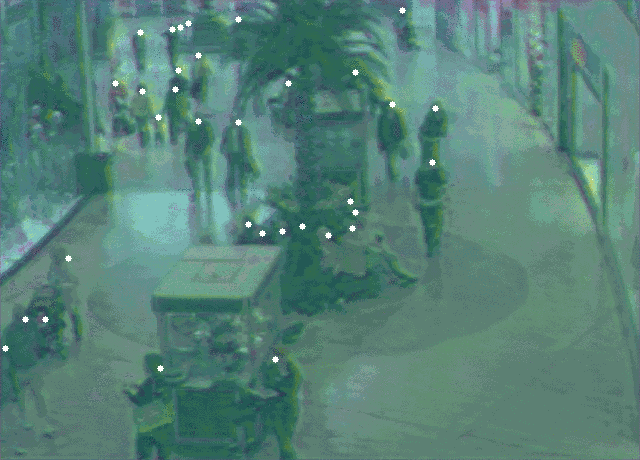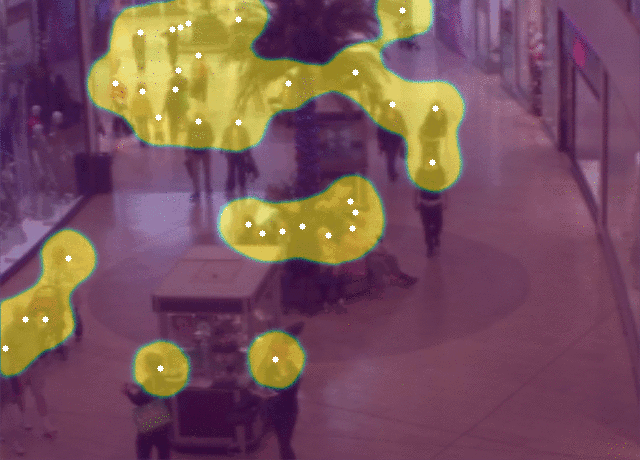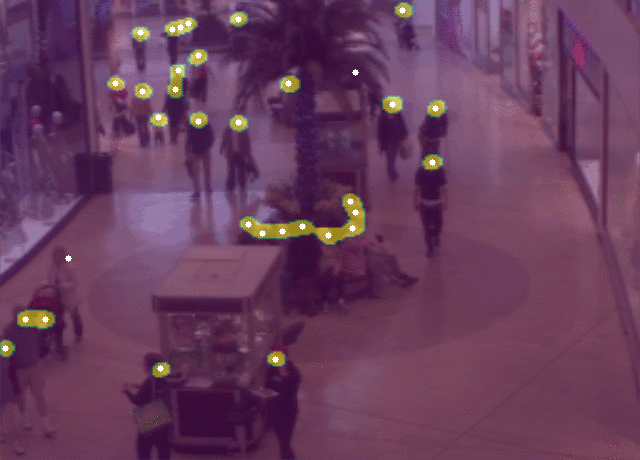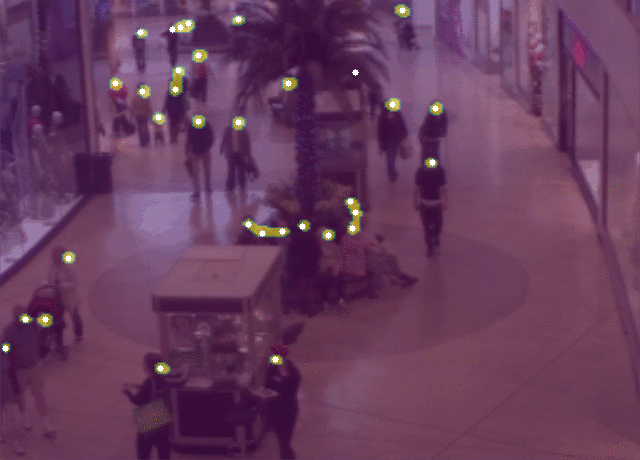 6 |
7 |
8 | ## Citing this work
9 | ```
10 | @article{ribera2019,
11 | title={Locating Objects Without Bounding Boxes},
12 | author={Javier Ribera and David G\"{u}era and Yuhao Chen and Edward J. Delp},
13 | journal={Proceedings of the Computer Vision and Pattern Recognition (CVPR)},
14 | month={June},
15 | year={2019},
16 | note={{Long Beach, CA}}
17 | }
18 | ```
19 |
20 | ## Datasets
21 | The datasets used in the paper can be downloaded from:
22 | - [Mall dataset](http://personal.ie.cuhk.edu.hk/~ccloy/downloads_mall_dataset.html)
23 | - [Pupil dataset](http://www.ti.uni-tuebingen.de/Pupil-detection.1827.0.html)
24 | - [Plant dataset](https://engineering.purdue.edu/~sorghum/dataset-plant-centers-2016)
25 |
26 | ## Installation
27 | Use conda to recreate the environment provided with the code:
28 |
6 |
7 |
8 | ## Citing this work
9 | ```
10 | @article{ribera2019,
11 | title={Locating Objects Without Bounding Boxes},
12 | author={Javier Ribera and David G\"{u}era and Yuhao Chen and Edward J. Delp},
13 | journal={Proceedings of the Computer Vision and Pattern Recognition (CVPR)},
14 | month={June},
15 | year={2019},
16 | note={{Long Beach, CA}}
17 | }
18 | ```
19 |
20 | ## Datasets
21 | The datasets used in the paper can be downloaded from:
22 | - [Mall dataset](http://personal.ie.cuhk.edu.hk/~ccloy/downloads_mall_dataset.html)
23 | - [Pupil dataset](http://www.ti.uni-tuebingen.de/Pupil-detection.1827.0.html)
24 | - [Plant dataset](https://engineering.purdue.edu/~sorghum/dataset-plant-centers-2016)
25 |
26 | ## Installation
27 | Use conda to recreate the environment provided with the code:
28 |
29 | conda env create -f environment.yml
30 |
31 |
32 | Activate the environment:
33 |
34 | conda activate object-locator
35 |
36 |
37 | Install the tool:
38 |
39 | pip install .
40 |
41 | (do not forget the period)
42 |
43 | ## Usage
44 | If you are only interested in the code of the Weighted Hausdorff Distance (which is the loss used in the paper and the main contribution), you can just get the [losses.py](object-locator/losses.py) file. If you want to use the entire object location tool:
45 |
46 | Activate the environment:
47 |
48 | conda activate object-locator
49 |
50 |
51 | Run this to get help (usage instructions):
52 |
53 | python -m object-locator.locate -h
54 | python -m object-locator.train -h
55 |
56 |
57 | Example:
58 |
59 |
60 | python -m object-locator.locate \
61 | --dataset DIRECTORY \
62 | --out DIRECTORY \
63 | --model CHECKPOINTS \
64 | --evaluate \
65 | --no-gpu \
66 | --radius 5
67 |
68 |
69 |
70 | python -m object-locator.train \
71 | --train-dir TRAINING_DIRECTORY \
72 | --batch-size 32 \
73 | --visdom-env mytrainingsession \
74 | --visdom-server localhost \
75 | --lr 1e-3 \
76 | --val-dir TRAINING_DIRECTORY \
77 | --optim Adam \
78 | --save saved_model.ckpt
79 |
80 |
81 | ## Dataset format
82 | The options `--dataset` and `--train-dir` should point to a directory.
83 | This directory must contain your dataset, meaning:
84 | 1. One file per image to analyze (png, jpg, jpeg, tiff or tif).
85 | 2. One ground truth file called `gt.csv` with the following format:
86 | ```
87 | filename,count,locations
88 | img1.png,3,"[(28, 52), (58, 53), (135, 50)]"
89 | img2.png,2,"[(92, 47), (33, 82)]"
90 | ```
91 | Each row of the CSV must describe the ground truth of an image: the count (number) and location of all objects in that image.
92 | The locations are in (y, x) format, being the origin the most top left pixel, y being the pixel row number, and x being the pixel column number.
93 |
94 | Optionally, if you are working on precision agriculture or plant phenotyping you can use an XML file `gt.xml` instead of a CSV.
95 | The required XML specifications can be found in
96 | [https://communityhub.purdue.edu/groups/phenosorg/wiki/APIspecs](https://communityhub.purdue.edu/groups/phenosorg/wiki/APIspecs)
97 | (accessible only to Purdue users) and in [this](https://hammer.figshare.com/articles/Image-based_Plant_Phenotyping_Using_Machine_Learning/7774313) thesis, but this is only useful in agronomy/phenotyping applications.
98 | The XML file is parsed by the file `data_plant_stuff.py`.
99 |
100 | ## Pre-trained models
101 | Models are trained separately for each of the four datasets, as described in the paper:
102 | 1. [Mall dataset](https://lorenz.ecn.purdue.edu/~cvpr2019/pretrained_models/mall,lambdaa=1,BS=32,Adam,LR1e-4.ckpt)
103 | 2. [Pupil dataset](https://lorenz.ecn.purdue.edu/~cvpr2019/pretrained_models/pupil,lambdaa=1,BS=64,SGD,LR1e-3,p=-1,ultrasmallNet.ckpt)
104 | 3. [Plant dataset](https://lorenz.ecn.purdue.edu/~cvpr2019/pretrained_models/plants_20160613_F54,BS=32,Adam,LR1e-5,p=-1.ckpt)
105 | 4. [ShanghaiTechB dataset](https://lorenz.ecn.purdue.edu/~cvpr2019/pretrained_models/shanghai,lambdaa=1,p=-1,BS=32,Adam,LR=1e-4.ckpt)
106 |
107 | The [COPYRIGHT](COPYRIGHT.txt) of the pre-trained models is the same as in this repository.
108 |
109 | As described in the paper, the pre-trained model for the pupil dataset excludes the five central layers. Thus if you want to use this model you will have to use the option `--ultrasmallnet`.
110 |
111 | ## Uninstall
112 |
113 | conda deactivate object-locator
114 | conda env remove --name object-locator
115 |
116 |
117 |
118 | ## Code Versioning
119 | The code used in the paper corresponds to the tag `used-for-cvpr2019-submission`.
120 | If you want to reproduce the results, checkout that tag with `git checkout used-for-cvpr2019-submission`.
121 | The master branch is the latest version available, with convenient bug fixes and better documentation.
122 | If you want to develop or retrain your models, we recommend the master branch.
123 | Versions numbers follow [semantic versioning](https://semver.org) and the changelog is in [CHANGELOG.md](CHANGELOG.md).
124 |
125 |
126 | ## Creating an issue
127 | If you're experiencing a problem or a bug, creating a GitHub issue is encouraged, but please include the following:
128 | 1. The commit version of this repository that you ran (`git show | head -n 1`)
129 | 2. The dataset you used (including images and the CSV with groundtruth with the [appropriate format](#datasetformat))
130 | 3. CPU and GPU model(s) you are using
131 | 4. The full standard output of the training log if you are training, and the testing log if you are evaluating (you can upload it to https://pastebin.com)
132 | 5. The operating system you are using
133 | 6. The command you run to train and evaluate
134 |
--------------------------------------------------------------------------------
/object-locator/models/unet_model.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 11/11/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import torch
19 | import torch.nn as nn
20 | import torch.nn.functional as F
21 | from torch.autograd import Variable
22 |
23 | from .unet_parts import *
24 |
25 |
26 | class UNet(nn.Module):
27 | def __init__(self, n_channels, n_classes,
28 | height, width,
29 | known_n_points=None,
30 | ultrasmall=False,
31 | device=torch.device('cuda')):
32 | """
33 | Instantiate a UNet network.
34 | :param n_channels: Number of input channels (e.g, 3 for RGB)
35 | :param n_classes: Number of output classes
36 | :param height: Height of the input images
37 | :param known_n_points: If you know the number of points,
38 | (e.g, one pupil), then set it.
39 | Otherwise it will be estimated by a lateral NN.
40 | If provided, no lateral network will be build
41 | and the resulting UNet will be a FCN.
42 | :param ultrasmall: If True, the 5 central layers are removed,
43 | resulting in a much smaller UNet.
44 | :param device: Which torch device to use. Default: CUDA (GPU).
45 | """
46 | super(UNet, self).__init__()
47 |
48 | self.ultrasmall = ultrasmall
49 | self.device = device
50 |
51 | # With this network depth, there is a minimum image size
52 | if height < 256 or width < 256:
53 | raise ValueError('Minimum input image size is 256x256, got {}x{}'.\
54 | format(height, width))
55 |
56 | self.inc = inconv(n_channels, 64)
57 | self.down1 = down(64, 128)
58 | self.down2 = down(128, 256)
59 | if self.ultrasmall:
60 | self.down3 = down(256, 512, normaliz=False)
61 | self.up1 = up(768, 128)
62 | self.up2 = up(256, 64)
63 | self.up3 = up(128, 64, activ=False)
64 | else:
65 | self.down3 = down(256, 512)
66 | self.down4 = down(512, 512)
67 | self.down5 = down(512, 512)
68 | self.down6 = down(512, 512)
69 | self.down7 = down(512, 512)
70 | self.down8 = down(512, 512, normaliz=False)
71 | self.up1 = up(1024, 512)
72 | self.up2 = up(1024, 512)
73 | self.up3 = up(1024, 512)
74 | self.up4 = up(1024, 512)
75 | self.up5 = up(1024, 256)
76 | self.up6 = up(512, 128)
77 | self.up7 = up(256, 64)
78 | self.up8 = up(128, 64, activ=False)
79 | self.outc = outconv(64, n_classes)
80 | self.out_nonlin = nn.Sigmoid()
81 |
82 | self.known_n_points = known_n_points
83 | if known_n_points is None:
84 | steps = 3 if self.ultrasmall else 8
85 | height_mid_features = height//(2**steps)
86 | width_mid_features = width//(2**steps)
87 | self.branch_1 = nn.Sequential(nn.Linear(height_mid_features*\
88 | width_mid_features*\
89 | 512,
90 | 64),
91 | nn.ReLU(inplace=True),
92 | nn.Dropout(p=0.5))
93 | self.branch_2 = nn.Sequential(nn.Linear(height*width, 64),
94 | nn.ReLU(inplace=True),
95 | nn.Dropout(p=0.5))
96 | self.regressor = nn.Sequential(nn.Linear(64 + 64, 1),
97 | nn.ReLU())
98 |
99 | # This layer is not connected anywhere
100 | # It is only here for backward compatibility
101 | self.lin = nn.Linear(1, 1, bias=False)
102 |
103 | def forward(self, x):
104 |
105 | batch_size = x.shape[0]
106 |
107 | x1 = self.inc(x)
108 | x2 = self.down1(x1)
109 | x3 = self.down2(x2)
110 | x4 = self.down3(x3)
111 | if self.ultrasmall:
112 | x = self.up1(x4, x3)
113 | x = self.up2(x, x2)
114 | x = self.up3(x, x1)
115 | else:
116 | x5 = self.down4(x4)
117 | x6 = self.down5(x5)
118 | x7 = self.down6(x6)
119 | x8 = self.down7(x7)
120 | x9 = self.down8(x8)
121 | x = self.up1(x9, x8)
122 | x = self.up2(x, x7)
123 | x = self.up3(x, x6)
124 | x = self.up4(x, x5)
125 | x = self.up5(x, x4)
126 | x = self.up6(x, x3)
127 | x = self.up7(x, x2)
128 | x = self.up8(x, x1)
129 | x = self.outc(x)
130 | x = self.out_nonlin(x)
131 |
132 | # Reshape Bx1xHxW -> BxHxW
133 | # because probability map is real-valued by definition
134 | x = x.squeeze(1)
135 |
136 | if self.known_n_points is None:
137 | middle_layer = x4 if self.ultrasmall else x9
138 | middle_layer_flat = middle_layer.view(batch_size, -1)
139 | x_flat = x.view(batch_size, -1)
140 |
141 | lateral_flat = self.branch_1(middle_layer_flat)
142 | x_flat = self.branch_2(x_flat)
143 |
144 | regression_features = torch.cat((x_flat, lateral_flat), dim=1)
145 | regression = self.regressor(regression_features)
146 |
147 | return x, regression
148 | else:
149 | n_pts = torch.tensor([self.known_n_points]*batch_size,
150 | dtype=torch.get_default_dtype())
151 | n_pts = n_pts.to(self.device)
152 | return x, n_pts
153 | # summ = torch.sum(x)
154 | # count = self.lin(summ)
155 |
156 | # count = torch.abs(count)
157 |
158 | # if self.known_n_points is not None:
159 | # count = Variable(torch.cuda.FloatTensor([self.known_n_points]))
160 |
161 | # return x, count
162 |
163 |
164 | """
165 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
166 | All rights reserved.
167 |
168 | This software is covered by US patents and copyright.
169 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
170 |
171 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
172 |
173 | Last Modified: 11/11/2019
174 | """
175 |
--------------------------------------------------------------------------------
/object-locator/find_lr.py:
--------------------------------------------------------------------------------
1 | from __future__ import print_function
2 |
3 | __copyright__ = \
4 | """
5 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
6 | All rights reserved.
7 |
8 | This software is covered by US patents and copyright.
9 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
10 |
11 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
12 |
13 | Last Modified: 10/02/2019
14 | """
15 | __license__ = "CC BY-NC-SA 4.0"
16 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
17 | __version__ = "1.6.0"
18 |
19 |
20 | import math
21 | import os
22 | from itertools import chain
23 | from tqdm import tqdm
24 |
25 | import numpy as np
26 | import torch
27 | import torch.optim as optim
28 | from torch import nn
29 | from torch.autograd import Variable
30 | from torchvision import transforms
31 | from torch.utils.data import DataLoader
32 | import torch.optim.lr_scheduler
33 | import matplotlib
34 | matplotlib.use('Agg')
35 | import skimage.transform
36 | from peterpy import peter
37 | from ballpark import ballpark
38 | from matplotlib import pyplot as plt
39 |
40 | from . import losses
41 | from .models import unet_model
42 | from .data import CSVDataset
43 | from .data import csv_collator
44 | from .data import RandomHorizontalFlipImageAndLabel
45 | from .data import RandomVerticalFlipImageAndLabel
46 | from .data import ScaleImageAndLabel
47 | from . import argparser
48 |
49 |
50 | # Parse command line arguments
51 | args = argparser.parse_command_args('training')
52 |
53 | # Tensor type to use, select CUDA or not
54 | torch.set_default_dtype(torch.float32)
55 | device_cpu = torch.device('cpu')
56 | device = torch.device('cuda') if args.cuda else device_cpu
57 |
58 | # Set seeds
59 | np.random.seed(args.seed)
60 | torch.manual_seed(args.seed)
61 | if args.cuda:
62 | torch.cuda.manual_seed_all(args.seed)
63 |
64 | # Data loading code
65 | training_transforms = []
66 | if not args.no_data_augm:
67 | training_transforms += [RandomHorizontalFlipImageAndLabel(p=0.5)]
68 | training_transforms += [RandomVerticalFlipImageAndLabel(p=0.5)]
69 | training_transforms += [ScaleImageAndLabel(size=(args.height, args.width))]
70 | training_transforms += [transforms.ToTensor()]
71 | training_transforms += [transforms.Normalize((0.5, 0.5, 0.5),
72 | (0.5, 0.5, 0.5))]
73 | trainset = CSVDataset(args.train_dir,
74 | transforms=transforms.Compose(training_transforms),
75 | max_dataset_size=args.max_trainset_size)
76 | trainset_loader = DataLoader(trainset,
77 | batch_size=args.batch_size,
78 | drop_last=args.drop_last_batch,
79 | shuffle=True,
80 | num_workers=args.nThreads,
81 | collate_fn=csv_collator)
82 |
83 | # Model
84 | with peter('Building network'):
85 | model = unet_model.UNet(3, 1,
86 | height=args.height,
87 | width=args.width,
88 | known_n_points=args.n_points)
89 | num_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
90 | print(f" with {ballpark(num_params)} trainable parameters. ", end='')
91 | model = nn.DataParallel(model)
92 | model.to(device)
93 |
94 |
95 | # Loss function
96 | loss_regress = nn.SmoothL1Loss()
97 | loss_loc = losses.WeightedHausdorffDistance(resized_height=args.height,
98 | resized_width=args.width,
99 | p=args.p,
100 | return_2_terms=True,
101 | device=device)
102 | l1_loss = nn.L1Loss(size_average=False)
103 | mse_loss = nn.MSELoss(reduce=False)
104 |
105 | optimizer = optim.SGD(model.parameters(),
106 | lr=999) # will be set later
107 |
108 |
109 | def find_lr(init_value = 1e-6, final_value=1e-3, beta = 0.7):
110 | num = len(trainset_loader)-1
111 | mult = (final_value / init_value) ** (1/num)
112 | lr = init_value
113 | optimizer.param_groups[0]['lr'] = lr
114 | avg_loss = 0.
115 | best_loss = 0.
116 | batch_num = 0
117 | losses = []
118 | log_lrs = []
119 | for imgs, dicts in tqdm(trainset_loader):
120 | batch_num += 1
121 |
122 | # Pull info from this batch and move to device
123 | imgs = imgs.to(device)
124 | imgs = Variable(imgs)
125 | target_locations = [dictt['locations'].to(device)
126 | for dictt in dicts]
127 | target_counts = [dictt['count'].to(device)
128 | for dictt in dicts]
129 | target_orig_heights = [dictt['orig_height'].to(device)
130 | for dictt in dicts]
131 | target_orig_widths = [dictt['orig_width'].to(device)
132 | for dictt in dicts]
133 |
134 | # Lists -> Tensor batches
135 | target_counts = torch.stack(target_counts)
136 | target_orig_heights = torch.stack(target_orig_heights)
137 | target_orig_widths = torch.stack(target_orig_widths)
138 | target_orig_sizes = torch.stack((target_orig_heights,
139 | target_orig_widths)).transpose(0, 1)
140 | # As before, get the loss for this mini-batch of inputs/outputs
141 | optimizer.zero_grad()
142 | est_maps, est_counts = model.forward(imgs)
143 | term1, term2 = loss_loc.forward(est_maps,
144 | target_locations,
145 | target_orig_sizes)
146 | target_counts = target_counts.view(-1)
147 | est_counts = est_counts.view(-1)
148 | target_counts = target_counts.view(-1)
149 | term3 = loss_regress.forward(est_counts, target_counts)
150 | term3 *= args.lambdaa
151 | loss = term1 + term2 + term3
152 |
153 | # Compute the smoothed loss
154 | avg_loss = beta * avg_loss + (1-beta) *loss.item()
155 | smoothed_loss = avg_loss / (1 - beta**batch_num)
156 |
157 | # Stop if the loss is exploding
158 | if (batch_num > 1 and smoothed_loss > 4 * best_loss):
159 | return log_lrs, losses
160 |
161 | # Record the best loss
162 | if smoothed_loss < best_loss or batch_num==1:
163 | best_loss = smoothed_loss
164 |
165 | # Store the values
166 | losses.append(smoothed_loss)
167 | log_lrs.append(math.log10(lr))
168 |
169 | # Do the SGD step
170 | loss.backward()
171 | optimizer.step()
172 |
173 | # Update the lr for the next step
174 | lr *= mult
175 | optimizer.param_groups[0]['lr'] = lr
176 | return log_lrs, losses
177 |
178 | logs, losses = find_lr()
179 | plt.plot(logs, losses)
180 | plt.savefig('/data/jprat/plot_beta0.7.png')
181 |
182 |
183 | """
184 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
185 | All rights reserved.
186 |
187 | This software is covered by US patents and copyright.
188 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
189 |
190 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
191 |
192 | Last Modified: 10/02/2019
193 | """
194 |
--------------------------------------------------------------------------------
/object-locator/logger.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import visdom
19 | import torch
20 | import numbers
21 | from . import utils
22 |
23 | from torch.autograd import Variable
24 |
25 | class Logger():
26 | def __init__(self,
27 | server=None,
28 | port=8989,
29 | env_name='main'):
30 | """
31 | Logger that connects to a Visdom server
32 | and sends training losses/metrics and images of any kind.
33 |
34 | :param server: Host name of the server (e.g, http://localhost),
35 | without the port number. If None,
36 | this Logger will do nothing at all
37 | (it will not connect to any server,
38 | and the functions here will do nothing).
39 | :param port: Port number of the Visdom server.
40 | :param env_name: Name of the environment within the Visdom
41 | server where everything you sent to it will go.
42 | :param terms_legends: Legend of each term.
43 | """
44 |
45 | if server is None:
46 | self.train_losses = utils.nothing
47 | self.val_losses = utils.nothing
48 | self.image = utils.nothing
49 | print('W: Not connected to any Visdom server. '
50 | 'You will not visualize any training/validation plot '
51 | 'or intermediate image')
52 | else:
53 | # Connect to Visdom
54 | self.client = visdom.Visdom(server=server,
55 | env=env_name,
56 | port=port)
57 | if self.client.check_connection():
58 | print(f'Connected to Visdom server '
59 | f'{server}:{port}')
60 | else:
61 | print(f'E: cannot connect to Visdom server '
62 | f'{server}:{port}')
63 | exit(-1)
64 |
65 | # Each of the 'windows' in visdom web panel
66 | self.viz_train_input_win = None
67 | self.viz_train_loss_win = None
68 | self.viz_train_gt_win = None
69 | self.viz_train_est_win = None
70 | self.viz_val_input_win = None
71 | self.viz_val_loss_win = None
72 | self.viz_val_gt_win = None
73 | self.viz_val_est_win = None
74 |
75 | # Visdom only supports CPU Tensors
76 | self.device = torch.device("cpu")
77 |
78 |
79 | def train_losses(self, terms, iteration_number, terms_legends=None):
80 | """
81 | Plot a new point of the training losses (scalars) to Visdom.
82 | All losses will be plotted in the same figure/window.
83 |
84 | :param terms: List of scalar losses.
85 | Each element will be a different plot in the y axis.
86 | :param iteration_number: Value of the x axis in the plot.
87 | :param terms_legends: Legend of each term.
88 | """
89 |
90 | # Watch dog
91 | if terms_legends is not None and \
92 | len(terms) != len(terms_legends):
93 | raise ValueError('The number of "terms" and "terms_legends" must be equal, got %s and %s, respectively'
94 | % (len(terms), len(terms_legends)))
95 | if not isinstance(iteration_number, numbers.Number):
96 | raise ValueError('iteration_number must be a number, got %s'
97 | % iteration_number)
98 |
99 | # Make terms CPU Tensors
100 | curated_terms = []
101 | for term in terms:
102 | if isinstance(term, numbers.Number):
103 | curated_term = torch.tensor([term])
104 | elif isinstance(term, torch.Tensor):
105 | curated_term = term
106 | else:
107 | raise ValueError('there is a term with an unsupported type'
108 | f'({type(term)}')
109 | curated_term = curated_term.to(self.device)
110 | curated_term = curated_term.view(1)
111 | curated_terms.append(curated_term)
112 |
113 | y = torch.cat(curated_terms).view(1, -1).data
114 | x = torch.Tensor([iteration_number]).repeat(1, len(terms))
115 | if terms_legends is None:
116 | terms_legends = ['Term %s' % t
117 | for t in range(1, len(terms) + 1)]
118 |
119 | # Send training loss to Visdom
120 | self.win_train_loss = \
121 | self.client.line(Y=y,
122 | X=x,
123 | opts=dict(title='Training',
124 | legend=terms_legends,
125 | ylabel='Loss',
126 | xlabel='Epoch'),
127 | update='append',
128 | win='train_losses')
129 | if self.win_train_loss == 'win does not exist':
130 | self.win_train_loss = \
131 | self.client.line(Y=y,
132 | X=x,
133 | opts=dict(title='Training',
134 | legend=terms_legends,
135 | ylabel='Loss',
136 | xlabel='Epoch'),
137 | win='train_losses')
138 |
139 | def image(self, imgs, titles, window_ids):
140 | """Send images to Visdom.
141 | Each image will be shown in a different window/plot.
142 |
143 | :param imgs: List of numpy images.
144 | :param titles: List of titles of each image.
145 | :param window_ids: List of window IDs.
146 | """
147 |
148 | # Watchdog
149 | if not(len(imgs) == len(titles) == len(window_ids)):
150 | raise ValueError('The number of "imgs", "titles" and '
151 | '"window_ids" must be equal, got '
152 | '%s, %s and %s, respectively'
153 | % (len(imgs), len(titles), len(window_ids)))
154 |
155 | for img, title, win in zip(imgs, titles, window_ids):
156 | self.client.image(img,
157 | opts=dict(title=title),
158 | win=str(win))
159 |
160 | def val_losses(self, terms, iteration_number, terms_legends=None):
161 | """
162 | Plot a new point of the training losses (scalars) to Visdom. All losses will be plotted in the same figure/window.
163 |
164 | :param terms: List of scalar losses.
165 | Each element will be a different plot in the y axis.
166 | :param iteration_number: Value of the x axis in the plot.
167 | :param terms_legends: Legend of each term.
168 | """
169 |
170 | # Watchdog
171 | if terms_legends is not None and \
172 | len(terms) != len(terms_legends):
173 | raise ValueError('The number of "terms" and "terms_legends" must be equal, got %s and %s, respectively'
174 | % (len(terms), len(terms_legends)))
175 | if not isinstance(iteration_number, numbers.Number):
176 | raise ValueError('iteration_number must be a number, got %s'
177 | % iteration_number)
178 |
179 | # Make terms CPU Tensors
180 | curated_terms = []
181 | for term in terms:
182 | if isinstance(term, numbers.Number):
183 | curated_term = torch.tensor([term],
184 | dtype=torch.get_default_dtype())
185 | elif isinstance(term, torch.Tensor):
186 | curated_term = term
187 | else:
188 | raise ValueError('there is a term with an unsupported type'
189 | f'({type(term)}')
190 | curated_term = curated_term.to(self.device)
191 | curated_term = curated_term.view(1)
192 | curated_terms.append(curated_term)
193 |
194 | y = torch.stack(curated_terms).view(1, -1)
195 | x = torch.Tensor([iteration_number]).repeat(1, len(terms))
196 | if terms_legends is None:
197 | terms_legends = ['Term %s' % t for t in range(1, len(terms) + 1)]
198 |
199 | # Send validation loss to Visdom
200 | self.win_val_loss = \
201 | self.client.line(Y=y,
202 | X=x,
203 | opts=dict(title='Validation',
204 | legend=terms_legends,
205 | ylabel='Loss',
206 | xlabel='Epoch'),
207 | update='append',
208 | win='val_metrics')
209 | if self.win_val_loss == 'win does not exist':
210 | self.win_val_loss = \
211 | self.client.line(Y=y,
212 | X=x,
213 | opts=dict(title='Validation',
214 | legend=terms_legends,
215 | ylabel='Loss',
216 | xlabel='Epoch'),
217 | win='val_metrics')
218 |
219 |
220 | """
221 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
222 | All rights reserved.
223 |
224 | This software is covered by US patents and copyright.
225 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
226 |
227 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
228 |
229 | Last Modified: 10/02/2019
230 | """
231 |
--------------------------------------------------------------------------------
/object-locator/losses.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import math
19 | import torch
20 | from sklearn.utils.extmath import cartesian
21 | import numpy as np
22 | from torch.nn import functional as F
23 | import os
24 | import time
25 | from sklearn.metrics.pairwise import pairwise_distances
26 | from sklearn.neighbors.kde import KernelDensity
27 | import skimage.io
28 | from matplotlib import pyplot as plt
29 | from torch import nn
30 |
31 |
32 | torch.set_default_dtype(torch.float32)
33 |
34 |
35 | def _assert_no_grad(variables):
36 | for var in variables:
37 | assert not var.requires_grad, \
38 | "nn criterions don't compute the gradient w.r.t. targets - please " \
39 | "mark these variables as volatile or not requiring gradients"
40 |
41 |
42 | def cdist(x, y):
43 | """
44 | Compute distance between each pair of the two collections of inputs.
45 | :param x: Nxd Tensor
46 | :param y: Mxd Tensor
47 | :res: NxM matrix where dist[i,j] is the norm between x[i,:] and y[j,:],
48 | i.e. dist[i,j] = ||x[i,:]-y[j,:]||
49 |
50 | """
51 | differences = x.unsqueeze(1) - y.unsqueeze(0)
52 | distances = torch.sum(differences**2, -1).sqrt()
53 | return distances
54 |
55 |
56 | def averaged_hausdorff_distance(set1, set2, max_ahd=np.inf):
57 | """
58 | Compute the Averaged Hausdorff Distance function
59 | between two unordered sets of points (the function is symmetric).
60 | Batches are not supported, so squeeze your inputs first!
61 | :param set1: Array/list where each row/element is an N-dimensional point.
62 | :param set2: Array/list where each row/element is an N-dimensional point.
63 | :param max_ahd: Maximum AHD possible to return if any set is empty. Default: inf.
64 | :return: The Averaged Hausdorff Distance between set1 and set2.
65 | """
66 |
67 | if len(set1) == 0 or len(set2) == 0:
68 | return max_ahd
69 |
70 | set1 = np.array(set1)
71 | set2 = np.array(set2)
72 |
73 | assert set1.ndim == 2, 'got %s' % set1.ndim
74 | assert set2.ndim == 2, 'got %s' % set2.ndim
75 |
76 | assert set1.shape[1] == set2.shape[1], \
77 | 'The points in both sets must have the same number of dimensions, got %s and %s.'\
78 | % (set2.shape[1], set2.shape[1])
79 |
80 | d2_matrix = pairwise_distances(set1, set2, metric='euclidean')
81 |
82 | res = np.average(np.min(d2_matrix, axis=0)) + \
83 | np.average(np.min(d2_matrix, axis=1))
84 |
85 | return res
86 |
87 |
88 | class AveragedHausdorffLoss(nn.Module):
89 | def __init__(self):
90 | super(nn.Module, self).__init__()
91 |
92 | def forward(self, set1, set2):
93 | """
94 | Compute the Averaged Hausdorff Distance function
95 | between two unordered sets of points (the function is symmetric).
96 | Batches are not supported, so squeeze your inputs first!
97 | :param set1: Tensor where each row is an N-dimensional point.
98 | :param set2: Tensor where each row is an N-dimensional point.
99 | :return: The Averaged Hausdorff Distance between set1 and set2.
100 | """
101 |
102 | assert set1.ndimension() == 2, 'got %s' % set1.ndimension()
103 | assert set2.ndimension() == 2, 'got %s' % set2.ndimension()
104 |
105 | assert set1.size()[1] == set2.size()[1], \
106 | 'The points in both sets must have the same number of dimensions, got %s and %s.'\
107 | % (set2.size()[1], set2.size()[1])
108 |
109 | d2_matrix = cdist(set1, set2)
110 |
111 | # Modified Chamfer Loss
112 | term_1 = torch.mean(torch.min(d2_matrix, 1)[0])
113 | term_2 = torch.mean(torch.min(d2_matrix, 0)[0])
114 |
115 | res = term_1 + term_2
116 |
117 | return res
118 |
119 |
120 | class WeightedHausdorffDistance(nn.Module):
121 | def __init__(self,
122 | resized_height, resized_width,
123 | p=-9,
124 | return_2_terms=False,
125 | device=torch.device('cpu')):
126 | """
127 | :param resized_height: Number of rows in the image.
128 | :param resized_width: Number of columns in the image.
129 | :param p: Exponent in the generalized mean. -inf makes it the minimum.
130 | :param return_2_terms: Whether to return the 2 terms
131 | of the WHD instead of their sum.
132 | Default: False.
133 | :param device: Device where all Tensors will reside.
134 | """
135 | super(nn.Module, self).__init__()
136 |
137 | # Prepare all possible (row, col) locations in the image
138 | self.height, self.width = resized_height, resized_width
139 | self.resized_size = torch.tensor([resized_height,
140 | resized_width],
141 | dtype=torch.get_default_dtype(),
142 | device=device)
143 | self.max_dist = math.sqrt(resized_height**2 + resized_width**2)
144 | self.n_pixels = resized_height * resized_width
145 | self.all_img_locations = torch.from_numpy(cartesian([np.arange(resized_height),
146 | np.arange(resized_width)]))
147 | # Convert to appropiate type
148 | self.all_img_locations = self.all_img_locations.to(device=device,
149 | dtype=torch.get_default_dtype())
150 |
151 | self.return_2_terms = return_2_terms
152 | self.p = p
153 |
154 | def forward(self, prob_map, gt, orig_sizes):
155 | """
156 | Compute the Weighted Hausdorff Distance function
157 | between the estimated probability map and ground truth points.
158 | The output is the WHD averaged through all the batch.
159 |
160 | :param prob_map: (B x H x W) Tensor of the probability map of the estimation.
161 | B is batch size, H is height and W is width.
162 | Values must be between 0 and 1.
163 | :param gt: List of Tensors of the Ground Truth points.
164 | Must be of size B as in prob_map.
165 | Each element in the list must be a 2D Tensor,
166 | where each row is the (y, x), i.e, (row, col) of a GT point.
167 | :param orig_sizes: Bx2 Tensor containing the size
168 | of the original images.
169 | B is batch size.
170 | The size must be in (height, width) format.
171 | :param orig_widths: List of the original widths for each image
172 | in the batch.
173 | :return: Single-scalar Tensor with the Weighted Hausdorff Distance.
174 | If self.return_2_terms=True, then return a tuple containing
175 | the two terms of the Weighted Hausdorff Distance.
176 | """
177 |
178 | _assert_no_grad(gt)
179 |
180 | assert prob_map.dim() == 3, 'The probability map must be (B x H x W)'
181 | assert prob_map.size()[1:3] == (self.height, self.width), \
182 | 'You must configure the WeightedHausdorffDistance with the height and width of the ' \
183 | 'probability map that you are using, got a probability map of size %s'\
184 | % str(prob_map.size())
185 |
186 | batch_size = prob_map.shape[0]
187 | assert batch_size == len(gt)
188 |
189 | terms_1 = []
190 | terms_2 = []
191 | for b in range(batch_size):
192 |
193 | # One by one
194 | prob_map_b = prob_map[b, :, :]
195 | gt_b = gt[b]
196 | orig_size_b = orig_sizes[b, :]
197 | norm_factor = (orig_size_b/self.resized_size).unsqueeze(0)
198 | n_gt_pts = gt_b.size()[0]
199 |
200 | # Corner case: no GT points
201 | if gt_b.ndimension() == 1 and (gt_b < 0).all().item() == 0:
202 | terms_1.append(torch.tensor([0],
203 | dtype=torch.get_default_dtype()))
204 | terms_2.append(torch.tensor([self.max_dist],
205 | dtype=torch.get_default_dtype()))
206 | continue
207 |
208 | # Pairwise distances between all possible locations and the GTed locations
209 | n_gt_pts = gt_b.size()[0]
210 | normalized_x = norm_factor.repeat(self.n_pixels, 1) *\
211 | self.all_img_locations
212 | normalized_y = norm_factor.repeat(len(gt_b), 1)*gt_b
213 | d_matrix = cdist(normalized_x, normalized_y)
214 |
215 | # Reshape probability map as a long column vector,
216 | # and prepare it for multiplication
217 | p = prob_map_b.view(prob_map_b.nelement())
218 | n_est_pts = p.sum()
219 | p_replicated = p.view(-1, 1).repeat(1, n_gt_pts)
220 |
221 | # Weighted Hausdorff Distance
222 | term_1 = (1 / (n_est_pts + 1e-6)) * \
223 | torch.sum(p * torch.min(d_matrix, 1)[0])
224 | weighted_d_matrix = (1 - p_replicated)*self.max_dist + p_replicated*d_matrix
225 | minn = generaliz_mean(weighted_d_matrix,
226 | p=self.p,
227 | dim=0, keepdim=False)
228 | term_2 = torch.mean(minn)
229 |
230 | # terms_1[b] = term_1
231 | # terms_2[b] = term_2
232 | terms_1.append(term_1)
233 | terms_2.append(term_2)
234 |
235 | terms_1 = torch.stack(terms_1)
236 | terms_2 = torch.stack(terms_2)
237 |
238 | if self.return_2_terms:
239 | res = terms_1.mean(), terms_2.mean()
240 | else:
241 | res = terms_1.mean() + terms_2.mean()
242 |
243 | return res
244 |
245 |
246 | def generaliz_mean(tensor, dim, p=-9, keepdim=False):
247 | # """
248 | # Computes the softmin along some axes.
249 | # Softmin is the same as -softmax(-x), i.e,
250 | # softmin(x) = -log(sum_i(exp(-x_i)))
251 |
252 | # The smoothness of the operator is controlled with k:
253 | # softmin(x) = -log(sum_i(exp(-k*x_i)))/k
254 |
255 | # :param input: Tensor of any dimension.
256 | # :param dim: (int or tuple of ints) The dimension or dimensions to reduce.
257 | # :param keepdim: (bool) Whether the output tensor has dim retained or not.
258 | # :param k: (float>0) How similar softmin is to min (the lower the more smooth).
259 | # """
260 | # return -torch.log(torch.sum(torch.exp(-k*input), dim, keepdim))/k
261 | """
262 | The generalized mean. It corresponds to the minimum when p = -inf.
263 | https://en.wikipedia.org/wiki/Generalized_mean
264 | :param tensor: Tensor of any dimension.
265 | :param dim: (int or tuple of ints) The dimension or dimensions to reduce.
266 | :param keepdim: (bool) Whether the output tensor has dim retained or not.
267 | :param p: (float<0).
268 | """
269 | assert p < 0
270 | res= torch.mean((tensor + 1e-6)**p, dim, keepdim=keepdim)**(1./p)
271 | return res
272 |
273 |
274 | """
275 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
276 | All rights reserved.
277 |
278 | This software is covered by US patents and copyright.
279 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
280 |
281 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
282 |
283 | Last Modified: 10/02/2019
284 | """
285 |
--------------------------------------------------------------------------------
/object-locator/data_plant_stuff.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import os
19 | import random
20 | from collections import OrderedDict
21 |
22 | from PIL import Image
23 | import numpy as np
24 | import torch
25 | from torchvision import datasets
26 | from torchvision import transforms
27 | import xmltodict
28 | from parse import parse
29 |
30 | from . import get_image_size
31 |
32 | IMG_EXTENSIONS = ['.png', '.jpeg', '.jpg', '.tiff']
33 |
34 | torch.set_default_dtype(torch.float32)
35 |
36 |
37 | class XMLDataset(torch.utils.data.Dataset):
38 | def __init__(self,
39 | directory,
40 | transforms=None,
41 | max_dataset_size=float('inf'),
42 | ignore_gt=False,
43 | seed=0):
44 | """XMLDataset.
45 | The sample images of this dataset must be all inside one directory.
46 | Inside the same directory, there must be one XML file as described by
47 | https://communityhub.purdue.edu/groups/phenosorg/wiki/APIspecs
48 | (minimum XML API version is v.0.4.0).
49 | If there is no XML file, metrics will not be computed,
50 | and only estimations will be provided.
51 | :param directory: Directory with all the images and the XML file.
52 | :param transform: Transform to be applied to each image.
53 | :param max_dataset_size: Only use the first N images in the directory.
54 | :param ignore_gt: Ignore the GT in the XML file,
55 | i.e, provide samples without plant locations or counts.
56 | :param seed: Random seed.
57 | """
58 |
59 | self.root_dir = directory
60 | self.transforms = transforms
61 |
62 | # Get list of files in the dataset directory,
63 | # and the filename of the XML
64 | listfiles = os.listdir(directory)
65 | xml_filenames = [f for f in listfiles if f.endswith('.xml')]
66 | if len(xml_filenames) == 1:
67 | xml_filename = xml_filenames[0]
68 | elif len(xml_filenames) == 0:
69 | xml_filename = None
70 | else:
71 | print(f"E: there is more than one XML file in '{directory}'")
72 | exit(-1)
73 |
74 | # Ignore files that are not images
75 | listfiles = [f for f in listfiles
76 | if any(f.lower().endswith(ext) for ext in IMG_EXTENSIONS)]
77 |
78 | # Shuffle list of files
79 | np.random.seed(seed)
80 | random.shuffle(listfiles)
81 |
82 | if len(listfiles) == 0:
83 | raise ValueError(f"There are no images in '{directory}'")
84 |
85 | if xml_filename is None:
86 | print('W: The dataset directory %s does not contain '
87 | 'a XML file with groundtruth. Metrics will not be evaluated.'
88 | 'Only estimations will be returned.' % directory)
89 |
90 | self.there_is_gt = (xml_filename is not None) and (not ignore_gt)
91 |
92 | # Read all XML as a string
93 | with open(os.path.join(directory, xml_filename), 'r') as fd:
94 | xml_str = fd.read()
95 |
96 | # Convert to dictionary
97 | # (some elements we expect to have multiple repetitions,
98 | # so put them in a list)
99 | xml_dict = xmltodict.parse(xml_str,
100 | force_list=['field',
101 | 'panel',
102 | 'plot',
103 | 'plant'])
104 |
105 | # Check API version number
106 | try:
107 | api_version = xml_dict['fields']['@apiversion']
108 | except:
109 | # An unknown version number means it's the very first one
110 | # when we did not have api version numbers
111 | api_version = '0.1.0'
112 | major_version, minor_version, _ = parse('{}.{}.{}', api_version)
113 | major_version = int(major_version)
114 | minor_version = int(minor_version)
115 | if not(major_version == 0 and minor_version == 4):
116 | raise ValueError('An XML with API v0.4 is required.')
117 |
118 | # Create the dictionary with the entire dataset
119 | dictt = {}
120 | for field in xml_dict['fields']['field']:
121 | for panel in field['panels']['panel']:
122 | for plot in panel['plots']['plot']:
123 |
124 | if self.there_is_gt and \
125 | not('plant_count' in plot and \
126 | 'plants' in plot):
127 | # There is GT for some plots but not this one
128 | continue
129 |
130 | filename = plot['orthophoto_chop_filename']
131 | if 'plot_number' in plot:
132 | plot_number = plot['plot_number']
133 | else:
134 | plot_number = 'unknown'
135 | if 'subrow_grid_location' in plot:
136 | subrow_grid_x = \
137 | int(plot['subrow_grid_location']['x']['#text'])
138 | subrow_grid_y = \
139 | int(plot['subrow_grid_location']['y']['#text'])
140 | else:
141 | subrow_grid_x = 'unknown'
142 | subrow_grid_y = 'unknown'
143 | if 'row_number' in plot:
144 | row_number = plot['row_number']
145 | else:
146 | row_number = 'unknown'
147 | if 'range_number' in plot:
148 | range_number = plot['range_number']
149 | else:
150 | range_number = 'unknown'
151 | img_abspath = os.path.join(self.root_dir, filename)
152 | orig_width, orig_height = \
153 | get_image_size.get_image_size(img_abspath)
154 | with torch.no_grad():
155 | orig_height = torch.tensor(
156 | orig_height, dtype=torch.get_default_dtype())
157 | orig_width = torch.tensor(
158 | orig_width, dtype=torch.get_default_dtype())
159 | dictt[filename] = {'filename': filename,
160 | 'plot_number': plot_number,
161 | 'subrow_grid_location_x': subrow_grid_x,
162 | 'subrow_grid_location_y': subrow_grid_y,
163 | 'row_number': row_number,
164 | 'range_number': range_number,
165 | 'orig_width': orig_width,
166 | 'orig_height': orig_height}
167 | if self.there_is_gt:
168 | count = int(plot['plant_count'])
169 | locations = []
170 | for plant in plot['plants']['plant']:
171 | for y in plant['location']['y']:
172 | if y['@units'] == 'pixels' and \
173 | y['@wrt'] == 'plot':
174 | y = float(y['#text'])
175 | break
176 | for x in plant['location']['x']:
177 | if x['@units'] == 'pixels' and \
178 | x['@wrt'] == 'plot':
179 | x = float(x['#text'])
180 | break
181 | locations.append([y, x])
182 | dictt[filename]['count'] = count
183 | dictt[filename]['locations'] = locations
184 |
185 | # Use an Ordered Dictionary to allow random access
186 | dictt = OrderedDict(dictt.items())
187 | self.dict_list = list(dictt.items())
188 |
189 | # Make dataset smaller
190 | new_dataset_length = min(len(dictt), max_dataset_size)
191 | dictt = {key: elem_dict
192 | for key, elem_dict in
193 | self.dict_list[:new_dataset_length]}
194 | self.dict_list = list(dictt.items())
195 |
196 | def __len__(self):
197 | return len(self.dict_list)
198 |
199 | def __getitem__(self, idx):
200 | """Get one element of the dataset.
201 | Returns a tuple. The first element is the image.
202 | The second element is a dictionary containing the labels of that image.
203 | The dictionary may not contain the location and count if the original
204 | XML did not include it.
205 |
206 | :param idx: Index of the image in the dataset to get.
207 | """
208 |
209 | filename, dictionary = self.dict_list[idx]
210 | img_abspath = os.path.join(self.root_dir, filename)
211 |
212 | if self.there_is_gt:
213 | # list --> Tensors
214 | with torch.no_grad():
215 | dictionary['locations'] = torch.tensor(
216 | dictionary['locations'],
217 | dtype=torch.get_default_dtype())
218 | dictionary['count'] = torch.tensor(
219 | dictionary['count'],
220 | dtype=torch.get_default_dtype())
221 | # else:
222 | # filename = self.listfiles[idx]
223 | # img_abspath = os.path.join(self.root_dir, filename)
224 | # orig_width, orig_height = \
225 | # get_image_size.get_image_size(img_abspath)
226 | # with torch.no_grad():
227 | # orig_height = torch.tensor(
228 | # orig_height, dtype=torch.get_default_dtype())
229 | # orig_width = torch.tensor(
230 | # orig_width, dtype=torch.get_default_dtype())
231 | # dictionary = {'filename': self.listfiles[idx],
232 | # 'orig_width': orig_width,
233 | # 'orig_height': orig_height}
234 |
235 | img = Image.open(img_abspath)
236 |
237 | img_transformed = img
238 | transformed_dictionary = dictionary
239 |
240 | # Apply all transformations provided
241 | if self.transforms is not None:
242 | for transform in self.transforms.transforms:
243 | if hasattr(transform, 'modifies_label'):
244 | img_transformed, transformed_dictionary = \
245 | transform(img_transformed, transformed_dictionary)
246 | else:
247 | img_transformed = transform(img_transformed)
248 |
249 | # Prevents crash when making a batch out of an empty tensor
250 | if self.there_is_gt and dictionary['count'].item() == 0:
251 | with torch.no_grad():
252 | dictionary['locations'] = torch.tensor([-1, -1],
253 | dtype=torch.get_default_dtype())
254 |
255 | return (img_transformed, transformed_dictionary)
256 |
257 |
258 | """
259 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
260 | All rights reserved.
261 |
262 | This software is covered by US patents and copyright.
263 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
264 |
265 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
266 |
267 | Last Modified: 10/02/2019
268 | """
269 |
--------------------------------------------------------------------------------
/object-locator/utils.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import torch
19 | import numpy as np
20 | import sklearn.mixture
21 | import scipy.stats
22 | import cv2
23 | from . import bmm
24 | from matplotlib import pyplot as plt
25 | import matplotlib.cm
26 | import scipy.stats
27 |
28 | class Normalizer():
29 | def __init__(self, new_size_height, new_size_width):
30 | """
31 | Normalizer.
32 | Converts coordinates in an original image size
33 | to a new image size (resized/normalized).
34 |
35 | :param new_size_height: (int) Height of the new (resized) image size.
36 | :param new_size_width: (int) Width of the new (resized) image size.
37 | """
38 | new_size_height = int(new_size_height)
39 | new_size_width = int(new_size_width)
40 |
41 | self.new_size = np.array([new_size_height, new_size_width])
42 |
43 | def unnormalize(self, coordinates_yx_normalized, orig_img_size):
44 | """
45 | Unnormalize coordinates,
46 | i.e, make them with respect to the original image.
47 |
48 | :param coordinates_yx_normalized:
49 | :param orig_size: Original image size ([height, width]).
50 | :return: Unnormalized coordinates
51 | """
52 |
53 | orig_img_size = np.array(orig_img_size)
54 | assert orig_img_size.ndim == 1
55 | assert len(orig_img_size) == 2
56 |
57 | norm_factor = orig_img_size / self.new_size
58 | norm_factor = np.tile(norm_factor, (len(coordinates_yx_normalized),1))
59 | coordinates_yx_unnormalized = norm_factor*coordinates_yx_normalized
60 |
61 | return coordinates_yx_unnormalized
62 |
63 | def threshold(array, tau):
64 | """
65 | Threshold an array using either hard thresholding, Otsu thresholding or beta-fitting.
66 |
67 | If the threshold value is fixed, this function returns
68 | the mask and the threshold used to obtain the mask.
69 | When using tau=-1, the threshold is obtained as described in the Otsu method.
70 | When using tau=-2, it also returns the fitted 2-beta Mixture Model.
71 |
72 |
73 | :param array: Array to threshold.
74 | :param tau: (float) Threshold to use.
75 | Values above tau become 1, and values below tau become 0.
76 | If -1, use Otsu thresholding.
77 | If -2, fit a mixture of 2 beta distributions, and use
78 | the average of the two means.
79 | :return: The tuple (mask, threshold).
80 | If tau==-2, returns the tuple (mask, otsu_tau, ((rv1, rv2), (pi1, pi2))).
81 |
82 | """
83 | if tau == -1:
84 | # Otsu thresholding
85 | minn, maxx = array.min(), array.max()
86 | array_scaled = ((array - minn)/(maxx - minn)*255) \
87 | .round().astype(np.uint8).squeeze()
88 | tau, mask = cv2.threshold(array_scaled,
89 | 0, 255, cv2.THRESH_BINARY + cv2.THRESH_OTSU)
90 | tau = minn + (tau/255)*(maxx - minn)
91 | # print(f'Otsu selected tau={tau_otsu}')
92 | elif tau == -2:
93 | array_flat = array.flatten()
94 | ((a1, b1), (a2, b2)), (pi1, pi2), niter = bmm.estimate(array_flat, list(range(2)))
95 | rv1 = scipy.stats.beta(a1, b1)
96 | rv2 = scipy.stats.beta(a2, b2)
97 |
98 | tau = rv2.mean()
99 | mask = cv2.inRange(array, tau, 1)
100 |
101 | return mask, tau, ((rv1, pi1), (rv2, pi2))
102 | else:
103 | # Thresholding with a fixed threshold tau
104 | mask = cv2.inRange(array, tau, 1)
105 |
106 | return mask, tau
107 |
108 |
109 | class AccBetaMixtureModel():
110 |
111 | def __init__(self, n_components=2, n_pts=1000):
112 | """
113 | Accumulator that tracks multiple Mixture Models based on Beta distributions.
114 | Each mixture is a tuple (scipy.RV, weight).
115 |
116 | :param n_components: (int) Number of components in the mixtures.
117 | :param n_pts: Number of points in the x axis (values the RV can take in [0, 1])
118 | """
119 | self.n_components = n_components
120 | self.mixtures = []
121 | self.x = np.linspace(0, 1, n_pts)
122 |
123 | def feed(self, mixture):
124 | """
125 | Accumulate another mixture so that this AccBetaMixtureModel can track it.
126 |
127 | :param mixture: List/Tuple of mixtures, i.e, ((RV, weight), (RV, weight), ...)
128 | """
129 | assert len(mixture) == self.n_components
130 |
131 | self.mixtures.append(mixture)
132 |
133 | def plot(self):
134 | """
135 | Create and return plots showing a variety of stats
136 | of the mixtures feeded into this object.
137 | """
138 | assert len(self.mixtures) > 0
139 |
140 | figs = {}
141 |
142 | # Compute the mean of the pdf of each component
143 | pdf_means = [(1/len(self.mixtures))*np.clip(rv.pdf(self.x), a_min=0, a_max=8)\
144 | for rv, w in self.mixtures[0]]
145 | for mix in self.mixtures[1:]:
146 | for c, (rv, w) in enumerate(mix):
147 | pdf_means[c] += (1/len(self.mixtures))*np.clip(rv.pdf(self.x), a_min=0, a_max=8)
148 |
149 | # Compute the stdev of the pdf of each component
150 | if len(self.mixtures) > 1:
151 | pdfs_sq_err_sum = [(np.clip(rv.pdf(self.x), a_min=0, a_max=8) - pdf_means[c])**2 \
152 | for c, (rv, w) in enumerate(self.mixtures[0])]
153 | for mix in self.mixtures[1:]:
154 | for c, (rv, w) in enumerate(mix):
155 | pdfs_sq_err_sum[c] += (np.clip(rv.pdf(self.x), a_min=0, a_max=8) - pdf_means[c])**2
156 | pdf_stdevs = [np.sqrt(pdf_sq_err_sum)/(len(self.mixtures) - 1) \
157 | for pdf_sq_err_sum in pdfs_sq_err_sum]
158 |
159 | # Plot the means of the pdfs
160 | fig, ax = plt.subplots()
161 | colors = ['r', 'g', 'b', 'c', 'm', 'y', 'k']
162 | for c, (pdf_mean, color) in enumerate(zip(pdf_means, colors)):
163 | ax.plot(self.x, pdf_mean, c=color, label=f'BMM Component #{c}')
164 | ax.set_xlabel('Pixel value / $\\tau$')
165 | ax.set_ylabel('Probability Density')
166 | plt.legend()
167 |
168 | if len(self.mixtures) > 1:
169 | # # Plot the standard deviations of the pdfs
170 | # fig, ax = plt.subplots()
171 | # colors = ['r', 'g', 'b', 'c', 'm', 'y', 'k']
172 | # max_stdev = 0
173 | # for c, (pdf_stdev, color) in enumerate(zip(pdf_stdevs, colors)):
174 | # ax.plot(self.x, pdf_stdev, c=color, label=f'Component #{c}')
175 | # max_stdev = max(max_stdev, max(pdf_stdev))
176 | # ax.set_title('Standard Deviation of the\nProbability Density Functions\n'
177 | # 'of the fitted bimodal Beta Mixture Model')
178 | # ax.set_xlabel('Pixel value')
179 | # ax.set_ylabel('Standard Deviation')
180 | # ax.set_ylim([0, max_stdev])
181 | # figs['std_bmm'] = fig
182 | # plt.close(fig)
183 |
184 | # Plot the KDE of the histogram of the threshold (the mean of last RV)
185 | thresholds = [mix[-1][0].mean() for mix in self.mixtures]
186 | thresholds = np.array(thresholds)[np.bitwise_not(np.isnan(thresholds))]
187 | kde = scipy.stats.gaussian_kde(thresholds.reshape(1, -1))
188 | ax.plot(self.x, kde.pdf(self.x),
189 | '--',
190 | label='KDE of $\\tau$ selected by BMM method')
191 | ax.set_xlabel('Pixel value / $\\tau$')
192 | ax.set_ylabel('Probability Density')
193 | plt.legend()
194 | figs['bmm_stats'] = fig
195 | plt.close(fig)
196 |
197 | return figs
198 |
199 | def cluster(array, n_clusters, max_mask_pts=np.infty):
200 | """
201 | Cluster a 2-D binary array.
202 | Applies a Gaussian Mixture Model on the positive elements of the array,
203 | and returns the number of clusters.
204 |
205 | :param array: Binary array.
206 | :param n_clusters: Number of clusters (Gaussians) to fit,
207 | :param max_mask_pts: Randomly subsample "max_pts" points
208 | from the array before fitting.
209 | :return: Centroids in the input array.
210 | """
211 |
212 | array = np.array(array)
213 |
214 | assert array.ndim == 2
215 |
216 | coord = np.where(array > 0)
217 | y = coord[0].reshape((-1, 1))
218 | x = coord[1].reshape((-1, 1))
219 | c = np.concatenate((y, x), axis=1)
220 | if len(c) == 0:
221 | centroids = np.array([])
222 | else:
223 | # Subsample our points randomly so it is faster
224 | if max_mask_pts != np.infty:
225 | n_pts = min(len(c), max_mask_pts)
226 | np.random.shuffle(c)
227 | c = c[:n_pts]
228 |
229 | # If the estimation is horrible, we cannot fit a GMM if n_components > n_samples
230 | n_components = max(min(n_clusters, x.size), 1)
231 | centroids = sklearn.mixture.GaussianMixture(n_components=n_components,
232 | n_init=1,
233 | covariance_type='full').\
234 | fit(c).means_.astype(np.int)
235 |
236 | return centroids
237 |
238 |
239 | class RunningAverage():
240 |
241 | def __init__(self, size):
242 | self.list = []
243 | self.size = size
244 |
245 | def put(self, elem):
246 | if len(self.list) >= self.size:
247 | self.list.pop(0)
248 | self.list.append(elem)
249 |
250 | def pop(self):
251 | self.list.pop(0)
252 |
253 | @property
254 | def avg(self):
255 | return np.average(self.list)
256 |
257 |
258 | def overlay_heatmap(img, map, colormap=matplotlib.cm.viridis):
259 | """
260 | Overlay a scalar map onto an image by using a heatmap
261 |
262 | :param img: RGB image (numpy array).
263 | Must be between 0 and 255.
264 | First dimension must be color.
265 | :param map: Scalar image (numpy array)
266 | Must be a 2D array between 0 and 1.
267 | :param colormap: Colormap to use to convert grayscale values
268 | to pseudo-color.
269 | :return: Heatmap on top of the original image in [0, 255]
270 | """
271 | assert img.ndim == 3
272 | assert map.ndim == 2
273 | assert img.shape[0] == 3
274 |
275 | # Convert image to CHW->HWC
276 | img = img.transpose(1, 2, 0)
277 |
278 | # Generate pseudocolor
279 | heatmap = colormap(map)[:, :, :3]
280 |
281 | # Scale heatmap [0, 1] -> [0, 255]
282 | heatmap *= 255
283 |
284 | # Fusion!
285 | img_w_heatmap = (img + heatmap)/2
286 |
287 | # Convert output to HWC->CHW
288 | img_w_heatmap = img_w_heatmap.transpose(2, 0, 1)
289 |
290 | return img_w_heatmap
291 |
292 |
293 | def paint_circles(img, points, color='red', crosshair=False):
294 | """
295 | Paint points as circles on top of an image.
296 |
297 | :param img: RGB image (numpy array).
298 | Must be between 0 and 255.
299 | First dimension must be color.
300 | :param centroids: List of centroids in (y, x) format.
301 | :param color: String of the color used to paint centroids.
302 | Default: 'red'.
303 | :param crosshair: Paint crosshair instead of circle.
304 | Default: False.
305 | :return: Image with painted circles centered on the points.
306 | First dimension is be color.
307 | """
308 |
309 | if color == 'red':
310 | color = [255, 0, 0]
311 | elif color == 'white':
312 | color = [255, 255, 255]
313 | else:
314 | raise NotImplementedError(f'color {color} not implemented')
315 |
316 | points = points.round().astype(np.uint16)
317 |
318 | img = np.moveaxis(img, 0, 2).copy()
319 | if not crosshair:
320 | for y, x in points:
321 | img = cv2.circle(img, (x, y), 3, color, -1)
322 | else:
323 | for y, x in points:
324 | img = cv2.drawMarker(img,
325 | (x, y),
326 | color, cv2.MARKER_TILTED_CROSS, 7, 1, cv2.LINE_AA)
327 | img = np.moveaxis(img, 2, 0)
328 |
329 | return img
330 |
331 |
332 | def nothing(*args, **kwargs):
333 | """ A useless function that does nothing at all. """
334 | pass
335 |
336 |
337 | """
338 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
339 | All rights reserved.
340 |
341 | This software is covered by US patents and copyright.
342 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
343 |
344 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
345 |
346 | Last Modified: 10/02/2019
347 | """
348 |
--------------------------------------------------------------------------------
/object-locator/get_image_size.py:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env python
2 | # -*- coding: utf-8 -*-
3 | from __future__ import print_function
4 | """
5 |
6 | get_image_size.py
7 | ====================
8 |
9 | :Name: get_image_size
10 | :Purpose: extract image dimensions given a file path
11 |
12 | :Author: Paulo Scardine (based on code from Emmanuel VAÏSSE)
13 |
14 | :Created: 26/09/2013
15 | :Copyright: (c) Paulo Scardine 2013
16 | :Licence: MIT
17 |
18 | """
19 | import collections
20 | import json
21 | import os
22 | import struct
23 |
24 | FILE_UNKNOWN = "Sorry, don't know how to get size for this file."
25 |
26 |
27 | class UnknownImageFormat(Exception):
28 | pass
29 |
30 |
31 | types = collections.OrderedDict()
32 | BMP = types['BMP'] = 'BMP'
33 | GIF = types['GIF'] = 'GIF'
34 | ICO = types['ICO'] = 'ICO'
35 | JPEG = types['JPEG'] = 'JPEG'
36 | PNG = types['PNG'] = 'PNG'
37 | TIFF = types['TIFF'] = 'TIFF'
38 |
39 | image_fields = ['path', 'type', 'file_size', 'width', 'height']
40 |
41 |
42 | class Image(collections.namedtuple('Image', image_fields)):
43 |
44 | def to_str_row(self):
45 | return ("%d\t%d\t%d\t%s\t%s" % (
46 | self.width,
47 | self.height,
48 | self.file_size,
49 | self.type,
50 | self.path.replace('\t', '\\t'),
51 | ))
52 |
53 | def to_str_row_verbose(self):
54 | return ("%d\t%d\t%d\t%s\t%s\t##%s" % (
55 | self.width,
56 | self.height,
57 | self.file_size,
58 | self.type,
59 | self.path.replace('\t', '\\t'),
60 | self))
61 |
62 | def to_str_json(self, indent=None):
63 | return json.dumps(self._asdict(), indent=indent)
64 |
65 |
66 | def get_image_size(file_path):
67 | """
68 | Return (width, height) for a given img file content - no external
69 | dependencies except the os and struct builtin modules
70 | """
71 | img = get_image_metadata(file_path)
72 | return (img.width, img.height)
73 |
74 |
75 | def get_image_metadata(file_path):
76 | """
77 | Return an `Image` object for a given img file content - no external
78 | dependencies except the os and struct builtin modules
79 |
80 | Args:
81 | file_path (str): path to an image file
82 |
83 | Returns:
84 | Image: (path, type, file_size, width, height)
85 | """
86 | size = os.path.getsize(file_path)
87 |
88 | # be explicit with open arguments - we need binary mode
89 | with open(file_path, "rb") as input:
90 | height = -1
91 | width = -1
92 | data = input.read(26)
93 | msg = " raised while trying to decode as JPEG."
94 |
95 | if (size >= 10) and data[:6] in (b'GIF87a', b'GIF89a'):
96 | # GIFs
97 | imgtype = GIF
98 | w, h = struct.unpack("= 24) and data.startswith(b'\211PNG\r\n\032\n')
102 | and (data[12:16] == b'IHDR')):
103 | # PNGs
104 | imgtype = PNG
105 | w, h = struct.unpack(">LL", data[16:24])
106 | width = int(w)
107 | height = int(h)
108 | elif (size >= 16) and data.startswith(b'\211PNG\r\n\032\n'):
109 | # older PNGs
110 | imgtype = PNG
111 | w, h = struct.unpack(">LL", data[8:16])
112 | width = int(w)
113 | height = int(h)
114 | elif (size >= 2) and data.startswith(b'\377\330'):
115 | # JPEG
116 | imgtype = JPEG
117 | input.seek(0)
118 | input.read(2)
119 | b = input.read(1)
120 | try:
121 | while (b and ord(b) != 0xDA):
122 | while (ord(b) != 0xFF):
123 | b = input.read(1)
124 | while (ord(b) == 0xFF):
125 | b = input.read(1)
126 | if (ord(b) >= 0xC0 and ord(b) <= 0xC3):
127 | input.read(3)
128 | h, w = struct.unpack(">HH", input.read(4))
129 | break
130 | else:
131 | input.read(
132 | int(struct.unpack(">H", input.read(2))[0]) - 2)
133 | b = input.read(1)
134 | width = int(w)
135 | height = int(h)
136 | except struct.error:
137 | raise UnknownImageFormat("StructError" + msg)
138 | except ValueError:
139 | raise UnknownImageFormat("ValueError" + msg)
140 | except Exception as e:
141 | raise UnknownImageFormat(e.__class__.__name__ + msg)
142 | elif (size >= 26) and data.startswith(b'BM'):
143 | # BMP
144 | imgtype = 'BMP'
145 | headersize = struct.unpack("= 40:
151 | w, h = struct.unpack("= 8) and data[:4] in (b"II\052\000", b"MM\000\052"):
160 | # Standard TIFF, big- or little-endian
161 | # BigTIFF and other different but TIFF-like formats are not
162 | # supported currently
163 | imgtype = TIFF
164 | byteOrder = data[:2]
165 | boChar = ">" if byteOrder == "MM" else "<"
166 | # maps TIFF type id to size (in bytes)
167 | # and python format char for struct
168 | tiffTypes = {
169 | 1: (1, boChar + "B"), # BYTE
170 | 2: (1, boChar + "c"), # ASCII
171 | 3: (2, boChar + "H"), # SHORT
172 | 4: (4, boChar + "L"), # LONG
173 | 5: (8, boChar + "LL"), # RATIONAL
174 | 6: (1, boChar + "b"), # SBYTE
175 | 7: (1, boChar + "c"), # UNDEFINED
176 | 8: (2, boChar + "h"), # SSHORT
177 | 9: (4, boChar + "l"), # SLONG
178 | 10: (8, boChar + "ll"), # SRATIONAL
179 | 11: (4, boChar + "f"), # FLOAT
180 | 12: (8, boChar + "d") # DOUBLE
181 | }
182 | ifdOffset = struct.unpack(boChar + "L", data[4:8])[0]
183 | try:
184 | countSize = 2
185 | input.seek(ifdOffset)
186 | ec = input.read(countSize)
187 | ifdEntryCount = struct.unpack(boChar + "H", ec)[0]
188 | # 2 bytes: TagId + 2 bytes: type + 4 bytes: count of values + 4
189 | # bytes: value offset
190 | ifdEntrySize = 12
191 | for i in range(ifdEntryCount):
192 | entryOffset = ifdOffset + countSize + i * ifdEntrySize
193 | input.seek(entryOffset)
194 | tag = input.read(2)
195 | tag = struct.unpack(boChar + "H", tag)[0]
196 | if(tag == 256 or tag == 257):
197 | # if type indicates that value fits into 4 bytes, value
198 | # offset is not an offset but value itself
199 | type = input.read(2)
200 | type = struct.unpack(boChar + "H", type)[0]
201 | if type not in tiffTypes:
202 | raise UnknownImageFormat(
203 | "Unkown TIFF field type:" +
204 | str(type))
205 | typeSize = tiffTypes[type][0]
206 | typeChar = tiffTypes[type][1]
207 | input.seek(entryOffset + 8)
208 | value = input.read(typeSize)
209 | value = int(struct.unpack(typeChar, value)[0])
210 | if tag == 256:
211 | width = value
212 | else:
213 | height = value
214 | if width > -1 and height > -1:
215 | break
216 | except Exception as e:
217 | raise UnknownImageFormat(str(e))
218 | elif size >= 2:
219 | # see http://en.wikipedia.org/wiki/ICO_(file_format)
220 | imgtype = 'ICO'
221 | input.seek(0)
222 | reserved = input.read(2)
223 | if 0 != struct.unpack(" 1:
230 | import warnings
231 | warnings.warn("ICO File contains more than one image")
232 | # http://msdn.microsoft.com/en-us/library/ms997538.aspx
233 | w = input.read(1)
234 | h = input.read(1)
235 | width = ord(w)
236 | height = ord(h)
237 | else:
238 | raise UnknownImageFormat(FILE_UNKNOWN)
239 |
240 | return Image(path=file_path,
241 | type=imgtype,
242 | file_size=size,
243 | width=width,
244 | height=height)
245 |
246 |
247 | import unittest
248 |
249 |
250 | class Test_get_image_size(unittest.TestCase):
251 | data = [{
252 | 'path': 'lookmanodeps.png',
253 | 'width': 251,
254 | 'height': 208,
255 | 'file_size': 22228,
256 | 'type': 'PNG'}]
257 |
258 | def setUp(self):
259 | pass
260 |
261 | def test_get_image_metadata(self):
262 | img = self.data[0]
263 | output = get_image_metadata(img['path'])
264 | self.assertTrue(output)
265 | self.assertEqual(output.path, img['path'])
266 | self.assertEqual(output.width, img['width'])
267 | self.assertEqual(output.height, img['height'])
268 | self.assertEqual(output.type, img['type'])
269 | self.assertEqual(output.file_size, img['file_size'])
270 | for field in image_fields:
271 | self.assertEqual(getattr(output, field), img[field])
272 |
273 | def test_get_image_metadata__ENOENT_OSError(self):
274 | with self.assertRaises(OSError):
275 | get_image_metadata('THIS_DOES_NOT_EXIST')
276 |
277 | def test_get_image_metadata__not_an_image_UnknownImageFormat(self):
278 | with self.assertRaises(UnknownImageFormat):
279 | get_image_metadata('README.rst')
280 |
281 | def test_get_image_size(self):
282 | img = self.data[0]
283 | output = get_image_size(img['path'])
284 | self.assertTrue(output)
285 | self.assertEqual(output,
286 | (img['width'],
287 | img['height']))
288 |
289 | def tearDown(self):
290 | pass
291 |
292 |
293 | def main(argv=None):
294 | """
295 | Print image metadata fields for the given file path.
296 |
297 | Keyword Arguments:
298 | argv (list): commandline arguments (e.g. sys.argv[1:])
299 | Returns:
300 | int: zero for OK
301 | """
302 | import logging
303 | import optparse
304 | import sys
305 |
306 | prs = optparse.OptionParser(
307 | usage="%prog [-v|--verbose] [--json|--json-indent] []",
308 | description="Print metadata for the given image paths "
309 | "(without image library bindings).")
310 |
311 | prs.add_option('--json',
312 | dest='json',
313 | action='store_true')
314 | prs.add_option('--json-indent',
315 | dest='json_indent',
316 | action='store_true')
317 |
318 | prs.add_option('-v', '--verbose',
319 | dest='verbose',
320 | action='store_true',)
321 | prs.add_option('-q', '--quiet',
322 | dest='quiet',
323 | action='store_true',)
324 | prs.add_option('-t', '--test',
325 | dest='run_tests',
326 | action='store_true',)
327 |
328 | argv = list(argv) if argv is not None else sys.argv[1:]
329 | (opts, args) = prs.parse_args(args=argv)
330 | loglevel = logging.INFO
331 | if opts.verbose:
332 | loglevel = logging.DEBUG
333 | elif opts.quiet:
334 | loglevel = logging.ERROR
335 | logging.basicConfig(level=loglevel)
336 | log = logging.getLogger()
337 | log.debug('argv: %r', argv)
338 | log.debug('opts: %r', opts)
339 | log.debug('args: %r', args)
340 |
341 | if opts.run_tests:
342 | import sys
343 | sys.argv = [sys.argv[0]] + args
344 | import unittest
345 | return unittest.main()
346 |
347 | output_func = Image.to_str_row
348 | if opts.json_indent:
349 | import functools
350 | output_func = functools.partial(Image.to_str_json, indent=2)
351 | elif opts.json:
352 | output_func = Image.to_str_json
353 | elif opts.verbose:
354 | output_func = Image.to_str_row_verbose
355 |
356 | EX_OK = 0
357 | EX_NOT_OK = 2
358 |
359 | if len(args) < 1:
360 | prs.print_help()
361 | print('')
362 | prs.error("You must specify one or more paths to image files")
363 |
364 | errors = []
365 | for path_arg in args:
366 | try:
367 | img = get_image_metadata(path_arg)
368 | print(output_func(img))
369 | except KeyboardInterrupt:
370 | raise
371 | except OSError as e:
372 | log.error((path_arg, e))
373 | errors.append((path_arg, e))
374 | except Exception as e:
375 | log.exception(e)
376 | errors.append((path_arg, e))
377 | pass
378 | if len(errors):
379 | import pprint
380 | print("ERRORS", file=sys.stderr)
381 | print("======", file=sys.stderr)
382 | print(pprint.pformat(errors, indent=2), file=sys.stderr)
383 | return EX_NOT_OK
384 | return EX_OK
385 |
386 |
387 | if __name__ == "__main__":
388 | import sys
389 | sys.exit(main(argv=sys.argv[1:]))
390 |
--------------------------------------------------------------------------------
/object-locator/locate.py:
--------------------------------------------------------------------------------
1 | from __future__ import print_function
2 |
3 | __copyright__ = \
4 | """
5 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
6 | All rights reserved.
7 |
8 | This software is covered by US patents and copyright.
9 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
10 |
11 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
12 |
13 | Last Modified: 10/02/2019
14 | """
15 | __license__ = "CC BY-NC-SA 4.0"
16 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
17 | __version__ = "1.6.0"
18 |
19 |
20 | import argparse
21 | import os
22 | import sys
23 | import time
24 | import shutil
25 | from parse import parse
26 | import math
27 | from collections import OrderedDict
28 | import itertools
29 |
30 | import matplotlib
31 | matplotlib.use('Agg')
32 | import cv2
33 | from tqdm import tqdm
34 | import numpy as np
35 | import pandas as pd
36 | import skimage.io
37 | import torch
38 | from torch import nn
39 | from torch.autograd import Variable
40 | from torch.utils import data
41 | from torchvision import datasets
42 | from torchvision import transforms
43 | import torchvision as tv
44 | from torchvision.models import inception_v3
45 | import skimage.transform
46 | from peterpy import peter
47 | from ballpark import ballpark
48 |
49 | from .data import csv_collator
50 | from .data import ScaleImageAndLabel
51 | from .data import build_dataset
52 | from . import losses
53 | from . import argparser
54 | from .models import unet_model
55 | from .metrics import Judge
56 | from .metrics import make_metric_plots
57 | from . import utils
58 |
59 |
60 | # Parse command line arguments
61 | args = argparser.parse_command_args('testing')
62 |
63 | # Tensor type to use, select CUDA or not
64 | torch.set_default_dtype(torch.float32)
65 | device_cpu = torch.device('cpu')
66 | device = torch.device('cuda') if args.cuda else device_cpu
67 |
68 | # Set seeds
69 | np.random.seed(args.seed)
70 | torch.manual_seed(args.seed)
71 | if args.cuda:
72 | torch.cuda.manual_seed_all(args.seed)
73 |
74 | # Data loading code
75 | try:
76 | testset = build_dataset(args.dataset,
77 | transforms=transforms.Compose([
78 | ScaleImageAndLabel(size=(args.height,
79 | args.width)),
80 | transforms.ToTensor(),
81 | transforms.Normalize((0.5, 0.5, 0.5),
82 | (0.5, 0.5, 0.5)),
83 | ]),
84 | ignore_gt=not args.evaluate,
85 | max_dataset_size=args.max_testset_size)
86 | except ValueError as e:
87 | print(f'E: {e}')
88 | exit(-1)
89 | testset_loader = data.DataLoader(testset,
90 | batch_size=1,
91 | num_workers=args.nThreads,
92 | collate_fn=csv_collator)
93 |
94 | # Array with [height, width] of the new size
95 | resized_size = np.array([args.height, args.width])
96 |
97 | # Loss function
98 | criterion_training = losses.WeightedHausdorffDistance(resized_height=args.height,
99 | resized_width=args.width,
100 | return_2_terms=True,
101 | device=device)
102 |
103 | # Restore saved checkpoint (model weights)
104 | with peter("Loading checkpoint"):
105 |
106 | if os.path.isfile(args.model):
107 | if args.cuda:
108 | checkpoint = torch.load(args.model)
109 | else:
110 | checkpoint = torch.load(
111 | args.model, map_location=lambda storage, loc: storage)
112 | # Model
113 | if args.n_points is None:
114 | if 'n_points' not in checkpoint:
115 | # Model will also estimate # of points
116 | model = unet_model.UNet(3, 1,
117 | known_n_points=None,
118 | height=args.height,
119 | width=args.width,
120 | ultrasmall=args.ultrasmallnet)
121 |
122 | else:
123 | # The checkpoint tells us the # of points to estimate
124 | model = unet_model.UNet(3, 1,
125 | known_n_points=checkpoint['n_points'],
126 | height=args.height,
127 | width=args.width,
128 | ultrasmall=args.ultrasmallnet)
129 | else:
130 | # The user tells us the # of points to estimate
131 | model = unet_model.UNet(3, 1,
132 | known_n_points=args.n_points,
133 | height=args.height,
134 | width=args.width,
135 | ultrasmall=args.ultrasmallnet)
136 |
137 | # Parallelize
138 | if args.cuda:
139 | model = nn.DataParallel(model)
140 | model = model.to(device)
141 |
142 | # Load model in checkpoint
143 | if args.cuda:
144 | state_dict = checkpoint['model']
145 | else:
146 | # remove 'module.' of DataParallel
147 | state_dict = OrderedDict()
148 | for k, v in checkpoint['model'].items():
149 | name = k[7:]
150 | state_dict[name] = v
151 | model.load_state_dict(state_dict)
152 | num_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
153 | print(f"\n\__ loaded checkpoint '{args.model}' "
154 | f"with {ballpark(num_params)} trainable parameters")
155 | # print(model)
156 | else:
157 | print(f"\n\__ E: no checkpoint found at '{args.model}'")
158 | exit(-1)
159 |
160 | tic = time.time()
161 |
162 |
163 | # Set the module in evaluation mode
164 | model.eval()
165 |
166 | # Accumulative histogram of estimated maps
167 | bmm_tracker = utils.AccBetaMixtureModel()
168 |
169 |
170 | if testset.there_is_gt:
171 | # Prepare Judges that will compute P/R as fct of r and th
172 | judges = []
173 | for r, th in itertools.product(args.radii, args.taus):
174 | judge = Judge(r=r)
175 | judge.th = th
176 | judges.append(judge)
177 |
178 | # Empty output CSV (one per threshold)
179 | df_outs = [pd.DataFrame() for _ in args.taus]
180 |
181 | # --force will overwrite output directory
182 | if args.force:
183 | shutil.rmtree(args.out)
184 |
185 | for batch_idx, (imgs, dictionaries) in tqdm(enumerate(testset_loader),
186 | total=len(testset_loader)):
187 |
188 | # Move to device
189 | imgs = imgs.to(device)
190 |
191 | # Pull info from this batch and move to device
192 | if testset.there_is_gt:
193 | target_locations = [dictt['locations'].to(device)
194 | for dictt in dictionaries]
195 | target_count = [dictt['count'].to(device)
196 | for dictt in dictionaries]
197 |
198 | target_orig_heights = [dictt['orig_height'].to(device)
199 | for dictt in dictionaries]
200 | target_orig_widths = [dictt['orig_width'].to(device)
201 | for dictt in dictionaries]
202 |
203 | # Lists -> Tensor batches
204 | if testset.there_is_gt:
205 | target_count = torch.stack(target_count)
206 | target_orig_heights = torch.stack(target_orig_heights)
207 | target_orig_widths = torch.stack(target_orig_widths)
208 | target_orig_sizes = torch.stack((target_orig_heights,
209 | target_orig_widths)).transpose(0, 1)
210 | origsize = (dictionaries[0]['orig_height'].item(),
211 | dictionaries[0]['orig_width'].item())
212 |
213 | # Tensor -> float & numpy
214 | if testset.there_is_gt:
215 | target_count = target_count.item()
216 | target_locations = \
217 | target_locations[0].to(device_cpu).numpy().reshape(-1, 2)
218 | target_orig_size = \
219 | target_orig_sizes[0].to(device_cpu).numpy().reshape(2)
220 |
221 | normalzr = utils.Normalizer(args.height, args.width)
222 |
223 | # Feed forward
224 | with torch.no_grad():
225 | est_maps, est_count = model.forward(imgs)
226 |
227 | # Convert to original size
228 | est_map_np = est_maps[0, :, :].to(device_cpu).numpy()
229 | est_map_np_origsize = \
230 | skimage.transform.resize(est_map_np,
231 | output_shape=origsize,
232 | mode='constant')
233 | orig_img_np = imgs[0].to(device_cpu).squeeze().numpy()
234 | orig_img_np_origsize = ((skimage.transform.resize(orig_img_np.transpose((1, 2, 0)),
235 | output_shape=origsize,
236 | mode='constant') + 1) / 2.0 * 255.0).\
237 | astype(np.float32).transpose((2, 0, 1))
238 |
239 | # Overlay output on original image as a heatmap
240 | orig_img_w_heatmap_origsize = utils.overlay_heatmap(img=orig_img_np_origsize,
241 | map=est_map_np_origsize).\
242 | astype(np.float32)
243 |
244 | # Save estimated map to disk
245 | os.makedirs(os.path.join(args.out, 'intermediate', 'estimated_map'),
246 | exist_ok=True)
247 | cv2.imwrite(os.path.join(args.out,
248 | 'intermediate',
249 | 'estimated_map',
250 | dictionaries[0]['filename']),
251 | orig_img_w_heatmap_origsize.transpose((1, 2, 0))[:, :, ::-1])
252 |
253 | # Tensor -> int
254 | est_count_int = int(round(est_count.item()))
255 |
256 | # The estimated map must be thresholded to obtain estimated points
257 | for t, tau in enumerate(args.taus):
258 | if tau != -2:
259 | mask, _ = utils.threshold(est_map_np_origsize, tau)
260 | else:
261 | mask, _, mix = utils.threshold(est_map_np_origsize, tau)
262 | bmm_tracker.feed(mix)
263 | centroids_wrt_orig = utils.cluster(mask, est_count_int,
264 | max_mask_pts=args.max_mask_pts)
265 |
266 | # Save thresholded map to disk
267 | os.makedirs(os.path.join(args.out,
268 | 'intermediate',
269 | 'estimated_map_thresholded',
270 | f'tau={round(tau, 4)}'),
271 | exist_ok=True)
272 | cv2.imwrite(os.path.join(args.out,
273 | 'intermediate',
274 | 'estimated_map_thresholded',
275 | f'tau={round(tau, 4)}',
276 | dictionaries[0]['filename']),
277 | mask)
278 |
279 | # Paint red dots if user asked for it
280 | if args.paint:
281 | # Paint a cross at the estimated centroids
282 | img_with_x_n_map = utils.paint_circles(img=orig_img_w_heatmap_origsize,
283 | points=centroids_wrt_orig,
284 | color='red',
285 | crosshair=True)
286 | # Save to disk
287 | os.makedirs(os.path.join(args.out,
288 | 'intermediate',
289 | 'painted_on_estimated_map',
290 | f'tau={round(tau, 4)}'), exist_ok=True)
291 | cv2.imwrite(os.path.join(args.out,
292 | 'intermediate',
293 | 'painted_on_estimated_map',
294 | f'tau={round(tau, 4)}',
295 | dictionaries[0]['filename']),
296 | img_with_x_n_map.transpose((1, 2, 0))[:, :, ::-1])
297 | # Paint a cross at the estimated centroids
298 | img_with_x = utils.paint_circles(img=orig_img_np_origsize,
299 | points=centroids_wrt_orig,
300 | color='red',
301 | crosshair=True)
302 | # Save to disk
303 | os.makedirs(os.path.join(args.out,
304 | 'intermediate',
305 | 'painted_on_original',
306 | f'tau={round(tau, 4)}'), exist_ok=True)
307 | cv2.imwrite(os.path.join(args.out,
308 | 'intermediate',
309 | 'painted_on_original',
310 | f'tau={round(tau, 4)}',
311 | dictionaries[0]['filename']),
312 | img_with_x.transpose((1, 2, 0))[:, :, ::-1])
313 |
314 |
315 | if args.evaluate:
316 | target_locations_wrt_orig = normalzr.unnormalize(target_locations,
317 | orig_img_size=target_orig_size)
318 |
319 | # Compute metrics for each value of r (for each Judge)
320 | for judge in judges:
321 | if judge.th != tau:
322 | continue
323 | judge.feed_points(centroids_wrt_orig, target_locations_wrt_orig,
324 | max_ahd=criterion_training.max_dist)
325 | judge.feed_count(est_count_int, target_count)
326 |
327 | # Save a new line in the CSV corresonding to the resuls of this img
328 | res_dict = dictionaries[0]
329 | res_dict['count'] = est_count_int
330 | res_dict['locations'] = str(centroids_wrt_orig.tolist())
331 | for key, val in res_dict.copy().items():
332 | if 'height' in key or 'width' in key:
333 | del res_dict[key]
334 | df = pd.DataFrame(data={idx: [val] for idx, val in res_dict.items()})
335 | df = df.set_index('filename')
336 | df_outs[t] = df_outs[t].append(df)
337 |
338 | # Write CSVs to disk
339 | os.makedirs(os.path.join(args.out, 'estimations'), exist_ok=True)
340 | for df_out, tau in zip(df_outs, args.taus):
341 | df_out.to_csv(os.path.join(args.out,
342 | 'estimations',
343 | f'estimations_tau={round(tau, 4)}.csv'))
344 |
345 | os.makedirs(os.path.join(args.out, 'intermediate', 'metrics_plots'),
346 | exist_ok=True)
347 |
348 | if args.evaluate:
349 |
350 | with peter("Evauating metrics"):
351 |
352 | # Output CSV where we will put
353 | # all our metrics as a function of r and the threshold
354 | df_metrics = pd.DataFrame(columns=['r', 'th',
355 | 'precision', 'recall', 'fscore', 'MAHD',
356 | 'MAPE', 'ME', 'MPE', 'MAE',
357 | 'MSE', 'RMSE', 'r', 'R2'])
358 | df_metrics.index.name = 'idx'
359 |
360 | for j, judge in enumerate(tqdm(judges)):
361 | # Accumulate precision and recall in the CSV dataframe
362 | df = pd.DataFrame(data=[[judge.r,
363 | judge.th,
364 | judge.precision,
365 | judge.recall,
366 | judge.fscore,
367 | judge.mahd,
368 | judge.mape,
369 | judge.me,
370 | judge.mpe,
371 | judge.mae,
372 | judge.mse,
373 | judge.rmse,
374 | judge.pearson_corr,
375 | judge.coeff_of_determination]],
376 | columns=['r', 'th',
377 | 'precision', 'recall', 'fscore', 'MAHD',
378 | 'MAPE', 'ME', 'MPE', 'MAE',
379 | 'MSE', 'RMSE', 'r', 'R2'],
380 | index=[j])
381 | df.index.name = 'idx'
382 | df_metrics = df_metrics.append(df)
383 |
384 | # Write CSV of metrics to disk
385 | df_metrics.to_csv(os.path.join(args.out, 'metrics.csv'))
386 |
387 | # Generate plots
388 | figs = make_metric_plots(csv_path=os.path.join(args.out, 'metrics.csv'),
389 | taus=args.taus,
390 | radii=args.radii)
391 | for label, fig in figs.items():
392 | # Save to disk
393 | fig.savefig(os.path.join(args.out,
394 | 'intermediate',
395 | 'metrics_plots',
396 | f'{label}.png'))
397 |
398 |
399 | # Save plot figures of the statistics of the BMM-based threshold
400 | if -2 in args.taus:
401 | for label, fig in bmm_tracker.plot().items():
402 | fig.savefig(os.path.join(args.out,
403 | 'intermediate',
404 | 'metrics_plots',
405 | f'{label}.png'))
406 |
407 |
408 | elapsed_time = int(time.time() - tic)
409 | print(f'It took {elapsed_time} seconds to evaluate all this dataset.')
410 |
411 |
412 | """
413 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
414 | All rights reserved.
415 |
416 | This software is covered by US patents and copyright.
417 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
418 |
419 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
420 |
421 | Last Modified: 10/02/2019
422 | """
423 |
--------------------------------------------------------------------------------
/object-locator/metrics.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import math
19 |
20 | import pandas as pd
21 | import numpy as np
22 | import matplotlib
23 | matplotlib.use('Agg')
24 | from matplotlib import pyplot as plt
25 | import sklearn.metrics
26 | import sklearn.neighbors
27 | import scipy.stats
28 | from . import losses
29 |
30 | class Judge():
31 | """
32 | A Judge computes the following metrics:
33 | (Location metrics)
34 | - Precision
35 | - Recall
36 | - Fscore
37 | - Mean Average Hausdorff Distance (MAHD)
38 | (Count metrics)
39 | - Mean Error (ME)
40 | - Mean Absolute Error (MAE)
41 | - Mean Percent Error (MPE)
42 | - Mean Absolute Percent Error (MAPE)
43 | - Mean Squared Error (MSE)
44 | - Root Mean Squared Error (RMSE)
45 | - Pearson correlation (r)
46 | - Coefficient of determination (R^2)
47 | """
48 |
49 | def __init__(self, r):
50 | """
51 | Create a Judge that will compute metrics with a particular r
52 | (r is only used to compute Precision, Recall, and Fscore).
53 |
54 | :param r: If an estimated point and a ground truth point
55 | are at a distance <= r, then a True Positive is counted.
56 | """
57 | # Location metrics
58 | self.r = r
59 | self.tp = 0
60 | self.fp = 0
61 | self.fn = 0
62 |
63 | # Count data points
64 | self._predicted_counts = []
65 | self._true_counts = []
66 |
67 | # Internal variables
68 | self._sum_ahd = 0
69 | self._sum_e = 0
70 | self._sum_pe = 0
71 | self._sum_ae = 0
72 | self._sum_se = 0
73 | self._sum_ape = 0
74 | self._n_calls_to_feed_points = 0
75 | self._n_calls_to_feed_count = 0
76 |
77 | def feed_points(self, pts, gt, max_ahd=np.inf):
78 | """
79 | Evaluate the location metrics of one set of estimations.
80 | This set can correspond to the estimated points and
81 | the groundtruthed points of one image.
82 | The TP, FP, FN, Precision, Recall, Fscore, and AHD will be
83 | accumulated into this Judge.
84 |
85 | :param pts: List of estmated points.
86 | :param gt: List of ground truth points.
87 | :param max_ahd: Maximum AHD possible to return if any set is empty. Default: inf.
88 | """
89 |
90 | if len(pts) == 0:
91 | tp = 0
92 | fp = 0
93 | fn = len(gt)
94 | else:
95 | nbr = sklearn.neighbors.NearestNeighbors(n_neighbors=1, metric='euclidean').fit(gt)
96 | dis, idx = nbr.kneighbors(pts)
97 | detected_pts = (dis[:, 0] <= self.r).astype(np.uint8)
98 |
99 | nbr = sklearn.neighbors.NearestNeighbors(n_neighbors=1, metric='euclidean').fit(pts)
100 | dis, idx = nbr.kneighbors(gt)
101 | detected_gt = (dis[:, 0] <= self.r).astype(np.uint8)
102 |
103 | tp = np.sum(detected_pts)
104 | fp = len(pts) - tp
105 | fn = len(gt) - np.sum(detected_gt)
106 |
107 | self.tp += tp
108 | self.fp += fp
109 | self.fn += fn
110 |
111 | # Evaluation using the Averaged Hausdorff Distance
112 | ahd = losses.averaged_hausdorff_distance(pts, gt,
113 | max_ahd=max_ahd)
114 | self._sum_ahd += ahd
115 | self._n_calls_to_feed_points += 1
116 |
117 | def feed_count(self, estim_count, gt_count):
118 | """
119 | Evaluate count metrics for a count estimation.
120 | This count can correspond to the estimated and groundtruthed count
121 | of one image. The ME, MAE, MPE, MAPE, MSE, and RMSE will be updated
122 | accordignly.
123 |
124 | :param estim_count: (positive number) Estimated count.
125 | :param gt_count: (positive number) Groundtruthed count.
126 | """
127 |
128 | if estim_count < 0:
129 | raise ValueError(f'estim_count < 0, got {estim_count}')
130 | if gt_count < 0:
131 | raise ValueError(f'gt_count < 0, got {gt_count}')
132 |
133 | self._predicted_counts.append(estim_count)
134 | self._true_counts.append(gt_count)
135 |
136 | e = estim_count - gt_count
137 | ae = abs(e)
138 | if gt_count == 0:
139 | ape = 100*ae
140 | pe = 100*e
141 | else:
142 | ape = 100 * ae / gt_count
143 | pe = 100 * e / gt_count
144 | se = e**2
145 |
146 | self._sum_e += e
147 | self._sum_pe += pe
148 | self._sum_ae += ae
149 | self._sum_se += se
150 | self._sum_ape += ape
151 |
152 | self._n_calls_to_feed_count += 1
153 |
154 | @property
155 | def me(self):
156 | """ Mean Error (float) """
157 | return float(self._sum_e / self._n_calls_to_feed_count)
158 |
159 | @property
160 | def mae(self):
161 | """ Mean Absolute Error (positive float) """
162 | return float(self._sum_ae / self._n_calls_to_feed_count)
163 |
164 | @property
165 | def mpe(self):
166 | """ Mean Percent Error (float) """
167 | return float(self._sum_pe / self._n_calls_to_feed_count)
168 |
169 | @property
170 | def mape(self):
171 | """ Mean Absolute Percent Error (positive float) """
172 | return float(self._sum_ape / self._n_calls_to_feed_count)
173 |
174 | @property
175 | def mse(self):
176 | """ Mean Squared Error (positive float)"""
177 | return float(self._sum_se / self._n_calls_to_feed_count)
178 |
179 | @property
180 | def rmse(self):
181 | """ Root Mean Squared Error (positive float)"""
182 | return float(math.sqrt(self.mse))
183 |
184 | @property
185 | def coeff_of_determination(self):
186 | """ Coefficient of Determination (-inf, 1]"""
187 | return sklearn.metrics.r2_score(self._true_counts,
188 | self._predicted_counts)
189 |
190 | @property
191 | def pearson_corr(self):
192 | """ Pearson coefficient of Correlation [-1, 1]"""
193 | return scipy.stats.pearsonr(self._true_counts,
194 | self._predicted_counts)[0]
195 |
196 | @property
197 | def mahd(self):
198 | """ Mean Average Hausdorff Distance (positive float)"""
199 | return float(self._sum_ahd / self._n_calls_to_feed_points)
200 |
201 | @property
202 | def precision(self):
203 | """ Precision (positive float) """
204 | return float(100*self.tp / (self.tp + self.fp)) \
205 | if self.tp > 0 else 0
206 |
207 | @property
208 | def recall(self):
209 | """ Recall (positive float) """
210 | return float(100*self.tp / (self.tp + self.fn)) \
211 | if self.tp > 0 else 0
212 |
213 | @property
214 | def fscore(self):
215 | """ F-score (positive float) """
216 | return float(2 * (self.precision*self.recall /
217 | (self.precision+self.recall))) \
218 | if self.tp > 0 else 0
219 |
220 |
221 | def make_metric_plots(csv_path, taus, radii, title=''):
222 | """
223 | Create a bunch of plots from the metrics contained in a CSV file.
224 |
225 | :param csv_path: Path to a CSV file containing metrics.
226 | :param taus: Detection thresholds tau.
227 | For each of these taus, a precision(r) and recall(r) will be created.
228 | The closest to each of these values will be used.
229 | :param radii: List of values, each with different colors in the scatter plot.
230 | Maximum distance to consider a True Positive.
231 | The closest to each of these values will be used.
232 | :param title: (optional) Title of the plot in the figure.
233 | :return: Dictionary with matplotlib figures.
234 | """
235 |
236 | dic = {}
237 |
238 | # Data extraction
239 | df = pd.read_csv(csv_path)
240 |
241 | plt.ioff()
242 |
243 | # ==== Precision and Recall as a function of R, fixing t ====
244 | for tau in taus:
245 | # Find closest threshold
246 | tau_selected = df.th.values[np.argmin(np.abs(df.th.values - tau))]
247 | print(f'Making Precision(r) and Recall(r) using tau={tau_selected}')
248 |
249 | # Use only a particular r
250 | precision = df.precision.values[df.th.values == tau_selected]
251 | recall = df.recall.values[df.th.values == tau_selected]
252 | r = df.r.values[df.th.values == tau_selected]
253 |
254 | # Create the figure for "Crowd" Dataset
255 | fig, ax = plt.subplots()
256 | precision = ax.plot(r, precision, 'r--',label='Precision')
257 | recall = ax.plot(r, recall, 'b:',label='Recall')
258 | ax.legend()
259 | ax.set_ylabel('%')
260 | ax.set_xlabel(r'$r$ (in pixels)')
261 | ax.grid(True)
262 | plt.title(title + f' tau={round(tau_selected, 4)}')
263 |
264 | # Hide grid lines below the plot
265 | ax.set_axisbelow(True)
266 |
267 | # Add figure to dictionary
268 | dic[f'precision_and_recall_vs_r,_tau={round(tau_selected, 4)}'] = fig
269 | plt.close(fig)
270 |
271 | # ==== Precision vs Recall ====
272 | colors = ['b', 'g', 'r', 'c', 'm', 'y', 'k']
273 | if len(radii) > len(colors):
274 | print(f'W: {len(radii)} are too many radii to plot, '
275 | f'taking {len(colors)} randomly.')
276 | radii = list(radii)
277 | np.random.shuffle(radii)
278 | radii = radii[:len(colors)]
279 | radii = sorted(radii)
280 |
281 | # Create figure
282 | fig, ax = plt.subplots()
283 | plt.ioff()
284 | ax.set_ylabel('Precision')
285 | ax.set_xlabel('Recall')
286 | ax.grid(True)
287 | plt.title(title)
288 |
289 | for r, c in zip(radii, colors):
290 | # Find closest R
291 | r_selected = df.r.values[np.argmin(np.abs(df.r.values - r))]
292 |
293 | # Use only a particular r for all fixed thresholds
294 | selection = (df.r.values == r_selected) & (df.th.values >= 0)
295 | if selection.any():
296 | precision = df.precision.values[selection]
297 | recall = df.recall.values[selection]
298 |
299 | # Sort by ascending recall
300 | idxs = np.argsort(recall)
301 | recall = recall[idxs]
302 | precision = precision[idxs]
303 |
304 | # Plot precision vs. recall for this r
305 | ax.scatter(recall, precision,
306 | c=c, s=2, label=f'$r={r}$')
307 |
308 | # Otsu threshold (tau = -1)
309 | selection = (df.r.values == r_selected) & (df.th.values == -1)
310 | if selection.any():
311 | precision = df.precision.values[selection]
312 | recall = df.recall.values[selection]
313 | ax.scatter(recall, precision,
314 | c=c, s=8, marker='+', label=f'$r={r}$, Otsu')
315 |
316 | # BMM threshold (tau = -2)
317 | selection = (df.r.values == r_selected) & (df.th.values == -2)
318 | if selection.any():
319 | precision = df.precision.values[selection]
320 | recall = df.recall.values[selection]
321 | ax.scatter(recall, precision,
322 | c=c, s=8, marker='s', label=f'$r={r}$, BMM')
323 |
324 | # Invert legend order
325 | handles, labels = ax.get_legend_handles_labels()
326 | handles, labels = handles[::-1], labels[::-1]
327 |
328 | # Put legend outside the plot
329 | box = ax.get_position()
330 | ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
331 | ax.legend(handles, labels, loc='upper left', bbox_to_anchor=(1, 1.03))
332 |
333 | # Hide grid lines below the plot
334 | ax.set_axisbelow(True)
335 |
336 | # Add figure to dictionary
337 | dic['precision_vs_recall'] = fig
338 | plt.close(fig)
339 |
340 |
341 | # ==== Precision as a function of tau for all provided R ====
342 | # Create figure
343 | fig, ax = plt.subplots()
344 | plt.ioff()
345 | ax.set_ylabel('Precision')
346 | ax.set_xlabel(r'$\tau$')
347 | ax.grid(True)
348 | plt.title(title)
349 |
350 | list_of_precisions = []
351 |
352 | for r, c in zip(radii, colors):
353 | # Find closest R
354 | r_selected = df.r.values[np.argmin(np.abs(df.r.values - r))]
355 |
356 | # Use only a particular r for all fixed thresholds
357 | selection = (df.r.values == r_selected) & (df.th.values >= 0)
358 | if selection.any():
359 | precision = df.precision.values[selection]
360 | list_of_precisions.append(precision)
361 | taus = df.th.values[selection]
362 |
363 | # Plot precision vs tau for this r
364 | ax.scatter(taus, precision, c=c, s=2, label=f'$r={r}$')
365 |
366 | # Otsu threshold (tau = -1)
367 | selection = (df.r.values == r_selected) & (df.th.values == -1)
368 | if selection.any():
369 | precision = df.precision.values[selection]
370 | ax.axhline(y=precision,
371 | linestyle='-',
372 | c=c, label=f'$r={r}$, Otsu')
373 |
374 | # BMM threshold (tau = -1)
375 | selection = (df.r.values == r_selected) & (df.th.values == -2)
376 | if selection.any():
377 | precision = df.precision.values[selection]
378 | ax.axhline(y=precision,
379 | linestyle='--',
380 | c=c, label=f'$r={r}$, BMM')
381 |
382 | if len(list_of_precisions) > 0:
383 | # Plot average precision for all r's
384 | ax.scatter(taus, np.average(np.stack(list_of_precisions), axis=0),
385 | c='k', marker='x', s=7, label='avg along r')
386 |
387 |
388 |
389 | # Invert legend order
390 | handles, labels = ax.get_legend_handles_labels()
391 | handles, labels = handles[::-1], labels[::-1]
392 |
393 | # Put legend outside the plot
394 | box = ax.get_position()
395 | ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
396 | ax.legend(handles, labels, loc='upper left', bbox_to_anchor=(1, 1.03))
397 |
398 | # Hide grid lines below the plot
399 | ax.set_axisbelow(True)
400 |
401 | # Add figure to dictionary
402 | dic['precision_vs_th'] = fig
403 | plt.close(fig)
404 |
405 | # ==== Recall as a function of tau for all provided R ====
406 | # Create figure
407 | fig, ax = plt.subplots()
408 | plt.ioff()
409 | ax.set_ylabel('Recall')
410 | ax.set_xlabel(r'$\tau$')
411 | ax.grid(True)
412 | plt.title(title)
413 |
414 | list_of_recalls = []
415 |
416 | for r, c in zip(radii, colors):
417 | # Find closest R
418 | r_selected = df.r.values[np.argmin(np.abs(df.r.values - r))]
419 |
420 | # Use only a particular r
421 | selection = (df.r.values == r_selected) & (df.th.values >= 0)
422 | if selection.any():
423 | recall = df.recall.values[selection]
424 | list_of_recalls.append(recall)
425 | taus = df.th.values[selection]
426 |
427 | # Plot precision vs tau for this r
428 | ax.scatter(taus, recall, c=c, s=2, label=f'$r={r}$')
429 |
430 | # Otsu threshold (tau = -1)
431 | selection = (df.r.values == r_selected) & (df.th.values == -1)
432 | if selection.any():
433 | recall = df.recall.values[selection]
434 | ax.axhline(y=recall,
435 | linestyle='-',
436 | c=c, label=f'$r={r}$, Otsu')
437 |
438 | # BMM threshold (tau = -2)
439 | selection = (df.r.values == r_selected) & (df.th.values == -2)
440 | if selection.any():
441 | recall = df.recall.values[selection]
442 | ax.axhline(y=recall,
443 | linestyle='--',
444 | c=c, label=f'$r={r}$, BMM')
445 |
446 |
447 | if len(list_of_recalls) > 0:
448 | ax.scatter(taus, np.average(np.stack(list_of_recalls), axis=0),
449 | c='k', marker='x', s=7, label='avg along $r$')
450 |
451 | # Invert legend order
452 | handles, labels = ax.get_legend_handles_labels()
453 | handles, labels = handles[::-1], labels[::-1]
454 |
455 | # Put legend outside the plot
456 | box = ax.get_position()
457 | ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
458 | ax.legend(handles, labels, loc='upper left', bbox_to_anchor=(1, 1.03))
459 |
460 | # Hide grid lines below the plot
461 | ax.set_axisbelow(True)
462 |
463 | # Add figure to dictionary
464 | dic['recall_vs_tau'] = fig
465 | plt.close(fig)
466 |
467 |
468 | # ==== F-score as a function of tau for all provided R ====
469 | # Create figure
470 | fig, ax = plt.subplots()
471 | plt.ioff()
472 | ax.set_ylabel('F-score')
473 | ax.set_xlabel(r'$\tau$')
474 | ax.grid(True)
475 | plt.title(title)
476 |
477 | list_of_fscores = []
478 |
479 | for r, c in zip(radii, colors):
480 | # Find closest R
481 | r_selected = df.r.values[np.argmin(np.abs(df.r.values - r))]
482 |
483 | # Use only a particular r
484 | selection = (df.r.values == r_selected) & (df.th.values >= 0)
485 | if selection.any():
486 | fscore = df.fscore.values[selection]
487 | list_of_fscores.append(fscore)
488 | taus = df.th.values[selection]
489 |
490 | # Plot precision vs tau for this r
491 | ax.scatter(taus, fscore, c=c, s=2, label=f'$r={r}$')
492 |
493 | # Otsu threshold (tau = -1)
494 | selection = (df.r.values == r_selected) & (df.th.values == -1)
495 | if selection.any():
496 | fscore = df.fscore.values[selection]
497 | ax.axhline(y=fscore,
498 | linestyle='-',
499 | c=c, label=f'$r={r}$, Otsu')
500 |
501 | # BMM threshold (tau = -2)
502 | selection = (df.r.values == r_selected) & (df.th.values == -2)
503 | if selection.any():
504 | fscore = df.fscore.values[selection]
505 | ax.axhline(y=fscore,
506 | linestyle='--',
507 | c=c, label=f'$r={r}$, BMM')
508 |
509 | if len(list_of_fscores) > 0:
510 | ax.scatter(taus, np.average(np.stack(list_of_fscores), axis=0),
511 | c='k', marker='x', s=7, label='avg along r')
512 |
513 | # Invert legend order
514 | handles, labels = ax.get_legend_handles_labels()
515 | handles, labels = handles[::-1], labels[::-1]
516 |
517 | # Put legend outside the plot
518 | box = ax.get_position()
519 | ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
520 | ax.legend(handles, labels, loc='upper left', bbox_to_anchor=(1, 1.03))
521 |
522 | # Hide grid lines below the plot
523 | ax.set_axisbelow(True)
524 |
525 | # Add figure to dictionary
526 | dic['fscore_vs_tau'] = fig
527 | plt.close(fig)
528 |
529 | return dic
530 |
531 |
532 | """
533 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
534 | All rights reserved.
535 |
536 | This software is covered by US patents and copyright.
537 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
538 |
539 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
540 |
541 | Last Modified: 10/02/2019
542 | """
543 |
--------------------------------------------------------------------------------
/object-locator/data.py:
--------------------------------------------------------------------------------
1 | __copyright__ = \
2 | """
3 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
4 | All rights reserved.
5 |
6 | This software is covered by US patents and copyright.
7 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
8 |
9 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
10 |
11 | Last Modified: 10/02/2019
12 | """
13 | __license__ = "CC BY-NC-SA 4.0"
14 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
15 | __version__ = "1.6.0"
16 |
17 |
18 | import os
19 | import random
20 |
21 | from PIL import Image
22 | import numpy as np
23 | import pandas as pd
24 | import torch
25 | import torchvision
26 | from ballpark import ballpark
27 |
28 | from . import get_image_size
29 |
30 | IMG_EXTENSIONS = ['.png', '.jpeg', '.jpg', '.tiff', '.tif']
31 |
32 | torch.set_default_dtype(torch.float32)
33 |
34 |
35 | def build_dataset(directory,
36 | transforms=None,
37 | max_dataset_size=float('inf'),
38 | ignore_gt=False,
39 | seed=0):
40 | """
41 | Build a dataset from a directory.
42 | Depending if the directory contains a CSV or an XML dataset,
43 | it builds an XMLDataset or a CSVDataset, which are subclasses
44 | of torch.utils.data.Dataset.
45 | :param directory: Directory with all the images and the CSV file.
46 | :param transform: Transform to be applied to each image.
47 | :param max_dataset_size: Only use the first N images in the directory.
48 | :param ignore_gt: Ignore the GT of the dataset,
49 | i.e, provide samples without locations or counts.
50 | :param seed: Random seed.
51 | :return: An XMLDataset or CSVDataset instance.
52 | """
53 | if any(fn.endswith('.csv') for fn in os.listdir(directory)) \
54 | or ignore_gt:
55 | dset = CSVDataset(directory=directory,
56 | transforms=transforms,
57 | max_dataset_size=max_dataset_size,
58 | ignore_gt=ignore_gt,
59 | seed=seed)
60 | else:
61 | from . import data_plant_stuff
62 | dset = data_plant_stuff.\
63 | XMLDataset(directory=directory,
64 | transforms=transforms,
65 | max_dataset_size=max_dataset_size,
66 | ignore_gt=ignore_gt,
67 | seed=seed)
68 |
69 | return dset
70 |
71 |
72 | def get_train_val_loaders(train_dir,
73 | collate_fn,
74 | height,
75 | width,
76 | no_data_augmentation=False,
77 | max_trainset_size=np.infty,
78 | seed=0,
79 | batch_size=1,
80 | drop_last_batch=False,
81 | shuffle=True,
82 | num_workers=0,
83 | val_dir=None,
84 | max_valset_size=np.infty):
85 | """
86 | Create a training loader and a validation set.
87 | If the validation directory is 'auto',
88 | 20% of the dataset is used for validation.
89 |
90 | :param train_dir: Directory with all the training images and the CSV file.
91 | :param train_transforms: Transform to be applied to each training image.
92 | :param max_trainset_size: Only use first N images for training.
93 | :param collate_fn: Function to assemble samples into batches.
94 | :param height: Resize the images to this height.
95 | :param width: Resize the images to this width.
96 | :param no_data_augmentation: Do not perform data augmentation.
97 | :param seed: Random seed.
98 | :param batch_size: Number of samples in a batch, for training.
99 | :param drop_last_batch: Drop the last incomplete batch during training
100 | :param shuffle: Randomly shuffle the dataset before each epoch.
101 | :param num_workers: Number of subprocesses dedicated for data loading.
102 | :param val_dir: Directory with all the training images and the CSV file.
103 | :param max_valset_size: Only use first N images for validation.
104 | """
105 |
106 | # Data augmentation for training
107 | training_transforms = []
108 | if not no_data_augmentation:
109 | training_transforms += [RandomHorizontalFlipImageAndLabel(p=0.5,
110 | seed=seed)]
111 | training_transforms += [RandomVerticalFlipImageAndLabel(p=0.5,
112 | seed=seed)]
113 | training_transforms += [ScaleImageAndLabel(size=(height, width))]
114 | training_transforms += [torchvision.transforms.ToTensor()]
115 | training_transforms += [torchvision.transforms.Normalize((0.5, 0.5, 0.5),
116 | (0.5, 0.5, 0.5))]
117 | training_transforms = torchvision.transforms.Compose(training_transforms)
118 |
119 | # Data augmentation for validation
120 | validation_transforms = torchvision.transforms.Compose([
121 | ScaleImageAndLabel(size=(height, width)),
122 | torchvision.transforms.ToTensor(),
123 | torchvision.transforms.\
124 | Normalize((0.5, 0.5, 0.5),
125 | (0.5, 0.5, 0.5)),
126 | ])
127 |
128 | # Training dataset
129 | trainset = build_dataset(directory=train_dir,
130 | transforms=training_transforms,
131 | max_dataset_size=max_trainset_size,
132 | seed=seed)
133 |
134 | # Validation dataset
135 | if val_dir is not None:
136 | if val_dir == 'auto':
137 | # Create a dataset just as in training
138 | valset = build_dataset(directory=train_dir,
139 | transforms=validation_transforms,
140 | max_dataset_size=max_trainset_size,
141 | seed=seed)
142 |
143 | # Split 80% for training, 20% for validation
144 | n_imgs_for_training = int(round(0.8*len(trainset)))
145 | if isinstance(trainset, CSVDataset):
146 | if trainset.there_is_gt:
147 | trainset.csv_df = \
148 | trainset.csv_df[:n_imgs_for_training]
149 | valset.csv_df = \
150 | valset.csv_df[n_imgs_for_training:].reset_index()
151 | else:
152 | trainset.listfiles = \
153 | trainset.listfiles[:n_imgs_for_training]
154 | valset.listfiles = \
155 | valset.listfiles[n_imgs_for_training:]
156 | else: # isinstance(trainset, XMLDataset):
157 | trainset.dict_list = trainset.dict_list[:n_imgs_for_training]
158 | valset.dict_list = valset.dict_list[n_imgs_for_training:]
159 |
160 | else:
161 | valset = build_dataset(val_dir,
162 | transforms=validation_transforms,
163 | max_dataset_size=max_valset_size,
164 | seed=seed)
165 | valset_loader = torch.utils.data.DataLoader(valset,
166 | batch_size=1,
167 | shuffle=True,
168 | num_workers=num_workers,
169 | collate_fn=csv_collator)
170 | else:
171 | valset, valset_loader = None, None
172 |
173 | print(f'# images for training: '
174 | f'{ballpark(len(trainset))}')
175 | if valset is not None:
176 | print(f'# images for validation: '
177 | f'{ballpark(len(valset))}')
178 | else:
179 | print('W: no validation set was selected!')
180 |
181 | # Build data loaders from the datasets
182 | trainset_loader = torch.utils.data.DataLoader(trainset,
183 | batch_size=batch_size,
184 | drop_last=drop_last_batch,
185 | shuffle=True,
186 | num_workers=num_workers,
187 | collate_fn=csv_collator)
188 | if valset is not None:
189 | valset_loader = torch.utils.data.DataLoader(valset,
190 | batch_size=1,
191 | shuffle=True,
192 | num_workers=num_workers,
193 | collate_fn=csv_collator)
194 |
195 | return trainset_loader, valset_loader
196 |
197 |
198 | class CSVDataset(torch.utils.data.Dataset):
199 | def __init__(self,
200 | directory,
201 | transforms=None,
202 | max_dataset_size=float('inf'),
203 | ignore_gt=False,
204 | seed=0):
205 | """CSVDataset.
206 | The sample images of this dataset must be all inside one directory.
207 | Inside the same directory, there must be one CSV file.
208 | This file must contain one row per image.
209 | It can contain as many columns as wanted, i.e, filename, count...
210 |
211 | :param directory: Directory with all the images and the CSV file.
212 | :param transform: Transform to be applied to each image.
213 | :param max_dataset_size: Only use the first N images in the directory.
214 | :param ignore_gt: Ignore the GT of the dataset,
215 | i.e, provide samples without locations or counts.
216 | :param seed: Random seed.
217 | """
218 |
219 | self.root_dir = directory
220 | self.transforms = transforms
221 |
222 | # Get groundtruth from CSV file
223 | listfiles = os.listdir(directory)
224 | csv_filename = None
225 | for filename in listfiles:
226 | if filename.endswith('.csv'):
227 | csv_filename = filename
228 | break
229 |
230 | # Ignore files that are not images
231 | listfiles = [f for f in listfiles
232 | if any(f.lower().endswith(ext) for ext in IMG_EXTENSIONS)]
233 |
234 | # Shuffle list of files
235 | np.random.seed(seed)
236 | random.shuffle(listfiles)
237 |
238 | if len(listfiles) == 0:
239 | raise ValueError(f"There are no images in '{directory}'")
240 |
241 | self.there_is_gt = (csv_filename is not None) and (not ignore_gt)
242 |
243 | # CSV does not exist (no GT available)
244 | if not self.there_is_gt:
245 | print('W: The dataset directory %s does not contain a CSV file with groundtruth. \n'
246 | ' Metrics will not be evaluated. Only estimations will be returned.' % directory)
247 | self.csv_df = None
248 | self.listfiles = listfiles
249 |
250 | # Make dataset smaller
251 | self.listfiles = self.listfiles[0:min(len(self.listfiles),
252 | max_dataset_size)]
253 |
254 | # CSV does exist (GT is available)
255 | else:
256 | self.csv_df = pd.read_csv(os.path.join(directory, csv_filename))
257 |
258 | # Shuffle CSV dataframe
259 | self.csv_df = self.csv_df.sample(frac=1).reset_index(drop=True)
260 |
261 | # Make dataset smaller
262 | self.csv_df = self.csv_df[0:min(
263 | len(self.csv_df), max_dataset_size)]
264 |
265 | def __len__(self):
266 | if self.there_is_gt:
267 | return len(self.csv_df)
268 | else:
269 | return len(self.listfiles)
270 |
271 | def __getitem__(self, idx):
272 | """Get one element of the dataset.
273 | Returns a tuple. The first element is the image.
274 | The second element is a dictionary where the keys are the columns of the CSV.
275 | If the CSV did not exist in the dataset directory,
276 | the dictionary will only contain the filename of the image.
277 | :param idx: Index of the image in the dataset to get.
278 | """
279 |
280 | if self.there_is_gt:
281 | img_abspath = os.path.join(self.root_dir, self.csv_df.ix[idx].filename)
282 | dictionary = dict(self.csv_df.ix[idx])
283 | else:
284 | img_abspath = os.path.join(self.root_dir, self.listfiles[idx])
285 | dictionary = {'filename': self.listfiles[idx]}
286 |
287 | img = Image.open(img_abspath)
288 |
289 | if self.there_is_gt:
290 | # str -> lists
291 | dictionary['locations'] = eval(dictionary['locations'])
292 | dictionary['locations'] = [
293 | list(loc) for loc in dictionary['locations']]
294 |
295 | # list --> Tensors
296 | with torch.no_grad():
297 | dictionary['locations'] = torch.tensor(
298 | dictionary['locations'], dtype=torch.get_default_dtype())
299 | dictionary['count'] = torch.tensor(
300 | [dictionary['count']], dtype=torch.get_default_dtype())
301 |
302 | # Record original size
303 | orig_width, orig_height = get_image_size.get_image_size(img_abspath)
304 | with torch.no_grad():
305 | orig_height = torch.tensor(orig_height,
306 | dtype=torch.get_default_dtype())
307 | orig_width = torch.tensor(orig_width,
308 | dtype=torch.get_default_dtype())
309 | dictionary['orig_width'] = orig_width
310 | dictionary['orig_height'] = orig_height
311 |
312 | img_transformed = img
313 | transformed_dictionary = dictionary
314 |
315 | # Apply all transformations provided
316 | if self.transforms is not None:
317 | for transform in self.transforms.transforms:
318 | if hasattr(transform, 'modifies_label'):
319 | img_transformed, transformed_dictionary = \
320 | transform(img_transformed, transformed_dictionary)
321 | else:
322 | img_transformed = transform(img_transformed)
323 |
324 | # Prevents crash when making a batch out of an empty tensor
325 | if self.there_is_gt:
326 | if dictionary['count'][0] == 0:
327 | with torch.no_grad():
328 | dictionary['locations'] = torch.tensor([-1, -1],
329 | dtype=torch.get_default_dtype())
330 |
331 | return (img_transformed, transformed_dictionary)
332 |
333 |
334 | def csv_collator(samples):
335 | """Merge a list of samples to form a batch.
336 | The batch is a 2-element tuple, being the first element
337 | the BxHxW tensor and the second element a list of dictionaries.
338 |
339 | :param samples: List of samples returned by CSVDataset as (img, dict) tuples.
340 | """
341 |
342 | imgs = []
343 | dicts = []
344 |
345 | for sample in samples:
346 | img = sample[0]
347 | dictt = sample[1]
348 |
349 | # # We cannot deal with images with 0 objects (WHD is not defined)
350 | # if dictt['count'][0] == 0:
351 | # continue
352 |
353 | imgs.append(img)
354 | dicts.append(dictt)
355 |
356 | data = torch.stack(imgs)
357 |
358 | return data, dicts
359 |
360 |
361 | class RandomHorizontalFlipImageAndLabel(object):
362 | """ Horizontally flip a numpy array image and the GT with probability p """
363 |
364 | def __init__(self, p, seed=0):
365 | self.modifies_label = True
366 | self.p = p
367 | np.random.seed(seed)
368 |
369 | def __call__(self, img, dictionary):
370 | transformed_img = img
371 | transformed_dictionary = dictionary
372 |
373 | if random.random() < self.p:
374 | transformed_img = hflip(img)
375 | width = img.size[0]
376 | for l, loc in enumerate(dictionary['locations']):
377 | dictionary['locations'][l][1] = (width - 1) - loc[1]
378 |
379 | return transformed_img, transformed_dictionary
380 |
381 |
382 | class RandomVerticalFlipImageAndLabel(object):
383 | """ Vertically flip a numpy array image and the GT with probability p """
384 |
385 | def __init__(self, p, seed=0):
386 | self.modifies_label = True
387 | self.p = p
388 | np.random.seed(seed)
389 |
390 | def __call__(self, img, dictionary):
391 | transformed_img = img
392 | transformed_dictionary = dictionary
393 |
394 | if random.random() < self.p:
395 | transformed_img = vflip(img)
396 | height = img.size[1]
397 | for l, loc in enumerate(dictionary['locations']):
398 | dictionary['locations'][l][0] = (height - 1) - loc[0]
399 |
400 | return transformed_img, transformed_dictionary
401 |
402 |
403 | class ScaleImageAndLabel(torchvision.transforms.Resize):
404 | """
405 | Scale a PIL Image and the GT to a given size.
406 | If there is no GT, then only scale the PIL Image.
407 |
408 | Args:
409 | size: Desired output size (h, w).
410 | interpolation (int, optional): Desired interpolation.
411 | Default is ``PIL.Image.BILINEAR``.
412 | """
413 |
414 | def __init__(self, size, interpolation=Image.BILINEAR):
415 | self.modifies_label = True
416 | self.size = size
417 | super(ScaleImageAndLabel, self).__init__(size, interpolation)
418 |
419 | def __call__(self, img, dictionary):
420 |
421 | old_width, old_height = img.size
422 | scale_h = self.size[0]/old_height
423 | scale_w = self.size[1]/old_width
424 |
425 | # Scale image to new size
426 | img = super(ScaleImageAndLabel, self).__call__(img)
427 |
428 | # Scale GT
429 | if 'locations' in dictionary and len(dictionary['locations']) > 0:
430 | # print(dictionary['locations'].type())
431 | # print(torch.tensor([scale_h, scale_w]).type())
432 | with torch.no_grad():
433 | dictionary['locations'] *= torch.tensor([scale_h, scale_w])
434 | dictionary['locations'] = torch.round(dictionary['locations'])
435 | ys = torch.clamp(
436 | dictionary['locations'][:, 0], 0, self.size[0])
437 | xs = torch.clamp(
438 | dictionary['locations'][:, 1], 0, self.size[1])
439 | dictionary['locations'] = torch.cat((ys.view(-1, 1),
440 | xs.view(-1, 1)),
441 | 1)
442 |
443 | # Indicate new size in dictionary
444 | with torch.no_grad():
445 | dictionary['resized_height'] = self.size[0]
446 | dictionary['resized_width'] = self.size[1]
447 |
448 | return img, dictionary
449 |
450 |
451 | def hflip(img):
452 | """Horizontally flip the given PIL Image.
453 | Args:
454 | img (PIL Image): Image to be flipped.
455 | Returns:
456 | PIL Image: Horizontall flipped image.
457 | """
458 | if not _is_pil_image(img):
459 | raise TypeError('img should be PIL Image. Got {}'.format(type(img)))
460 |
461 | return img.transpose(Image.FLIP_LEFT_RIGHT)
462 |
463 |
464 | def vflip(img):
465 | """Vertically flip the given PIL Image.
466 | Args:
467 | img (PIL Image): Image to be flipped.
468 | Returns:
469 | PIL Image: Vertically flipped image.
470 | """
471 | if not _is_pil_image(img):
472 | raise TypeError('img should be PIL Image. Got {}'.format(type(img)))
473 |
474 | return img.transpose(Image.FLIP_TOP_BOTTOM)
475 |
476 |
477 | def _is_pil_image(img):
478 | return isinstance(img, Image.Image)
479 |
480 |
481 | """
482 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
483 | All rights reserved.
484 |
485 | This software is covered by US patents and copyright.
486 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
487 |
488 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
489 |
490 | Last Modified: 10/02/2019
491 | """
492 |
--------------------------------------------------------------------------------
/object-locator/train.py:
--------------------------------------------------------------------------------
1 | from __future__ import print_function
2 |
3 | __copyright__ = \
4 | """
5 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
6 | All rights reserved.
7 |
8 | This software is covered by US patents and copyright.
9 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
10 |
11 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
12 |
13 | Last Modified: 10/02/2019
14 | """
15 | __license__ = "CC BY-NC-SA 4.0"
16 | __authors__ = "Javier Ribera, David Guera, Yuhao Chen, Edward J. Delp"
17 | __version__ = "1.6.0"
18 |
19 |
20 | import math
21 | import cv2
22 | import os
23 | import sys
24 | import time
25 | import shutil
26 | from itertools import chain
27 | from tqdm import tqdm
28 |
29 | import numpy as np
30 | import torch
31 | import torch.optim as optim
32 | from torch import nn
33 | from torch.autograd import Variable
34 | import torchvision as tv
35 | from torchvision.models import inception_v3
36 | from torchvision import transforms
37 | from torch.utils.data import DataLoader
38 | import matplotlib
39 | matplotlib.use('Agg')
40 | import skimage.transform
41 | from peterpy import peter
42 | from ballpark import ballpark
43 |
44 | from . import losses
45 | from .models import unet_model
46 | from .metrics import Judge
47 | from . import logger
48 | from . import argparser
49 | from . import utils
50 | from . import data
51 | from .data import csv_collator
52 | from .data import RandomHorizontalFlipImageAndLabel
53 | from .data import RandomVerticalFlipImageAndLabel
54 | from .data import ScaleImageAndLabel
55 |
56 |
57 | # Parse command line arguments
58 | args = argparser.parse_command_args('training')
59 |
60 | # Tensor type to use, select CUDA or not
61 | torch.set_default_dtype(torch.float32)
62 | device_cpu = torch.device('cpu')
63 | device = torch.device('cuda') if args.cuda else device_cpu
64 |
65 | # Create directory for checkpoint to be saved
66 | if args.save:
67 | os.makedirs(os.path.split(args.save)[0], exist_ok=True)
68 |
69 | # Set seeds
70 | np.random.seed(args.seed)
71 | torch.manual_seed(args.seed)
72 | if args.cuda:
73 | torch.cuda.manual_seed_all(args.seed)
74 |
75 | # Visdom setup
76 | log = logger.Logger(server=args.visdom_server,
77 | port=args.visdom_port,
78 | env_name=args.visdom_env)
79 |
80 |
81 | # Create data loaders (return data in batches)
82 | trainset_loader, valset_loader = \
83 | data.get_train_val_loaders(train_dir=args.train_dir,
84 | max_trainset_size=args.max_trainset_size,
85 | collate_fn=csv_collator,
86 | height=args.height,
87 | width=args.width,
88 | seed=args.seed,
89 | batch_size=args.batch_size,
90 | drop_last_batch=args.drop_last_batch,
91 | num_workers=args.nThreads,
92 | val_dir=args.val_dir,
93 | max_valset_size=args.max_valset_size)
94 |
95 | # Model
96 | with peter('Building network'):
97 | model = unet_model.UNet(3, 1,
98 | height=args.height,
99 | width=args.width,
100 | known_n_points=args.n_points,
101 | device=device,
102 | ultrasmall=args.ultrasmallnet)
103 | num_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
104 | print(f" with {ballpark(num_params)} trainable parameters. ", end='')
105 | model = nn.DataParallel(model)
106 | model.to(device)
107 |
108 | # Loss functions
109 | loss_regress = nn.SmoothL1Loss()
110 | loss_loc = losses.WeightedHausdorffDistance(resized_height=args.height,
111 | resized_width=args.width,
112 | p=args.p,
113 | return_2_terms=True,
114 | device=device)
115 |
116 | # Optimization strategy
117 | if args.optimizer == 'sgd':
118 | optimizer = optim.SGD(model.parameters(),
119 | lr=args.lr,
120 | momentum=0.9)
121 | elif args.optimizer == 'adam':
122 | optimizer = optim.Adam(model.parameters(),
123 | lr=args.lr,
124 | amsgrad=True)
125 |
126 | start_epoch = 0
127 | lowest_mahd = np.infty
128 |
129 | # Restore saved checkpoint (model weights + epoch + optimizer state)
130 | if args.resume:
131 | with peter('Loading checkpoint'):
132 | if os.path.isfile(args.resume):
133 | checkpoint = torch.load(args.resume)
134 | start_epoch = checkpoint['epoch']
135 | try:
136 | lowest_mahd = checkpoint['mahd']
137 | except KeyError:
138 | lowest_mahd = np.infty
139 | print('W: Loaded checkpoint has not been validated. ', end='')
140 | model.load_state_dict(checkpoint['model'])
141 | if not args.replace_optimizer:
142 | optimizer.load_state_dict(checkpoint['optimizer'])
143 | print(f"\n\__ loaded checkpoint '{args.resume}'"
144 | f"(now on epoch {checkpoint['epoch']})")
145 | else:
146 | print(f"\n\__ E: no checkpoint found at '{args.resume}'")
147 | exit(-1)
148 |
149 | running_avg = utils.RunningAverage(len(trainset_loader))
150 |
151 | normalzr = utils.Normalizer(args.height, args.width)
152 |
153 | # Time at the last evaluation
154 | tic_train = -np.infty
155 | tic_val = -np.infty
156 |
157 | epoch = start_epoch
158 | it_num = 0
159 | while epoch < args.epochs:
160 |
161 | loss_avg_this_epoch = 0
162 | iter_train = tqdm(trainset_loader,
163 | desc=f'Epoch {epoch} ({len(trainset_loader.dataset)} images)')
164 |
165 | # === TRAIN ===
166 |
167 | # Set the module in training mode
168 | model.train()
169 |
170 | for batch_idx, (imgs, dictionaries) in enumerate(iter_train):
171 |
172 | # Pull info from this batch and move to device
173 | imgs = imgs.to(device)
174 | target_locations = [dictt['locations'].to(device)
175 | for dictt in dictionaries]
176 | target_counts = [dictt['count'].to(device)
177 | for dictt in dictionaries]
178 | target_orig_heights = [dictt['orig_height'].to(device)
179 | for dictt in dictionaries]
180 | target_orig_widths = [dictt['orig_width'].to(device)
181 | for dictt in dictionaries]
182 |
183 | # Lists -> Tensor batches
184 | target_counts = torch.stack(target_counts)
185 | target_orig_heights = torch.stack(target_orig_heights)
186 | target_orig_widths = torch.stack(target_orig_widths)
187 | target_orig_sizes = torch.stack((target_orig_heights,
188 | target_orig_widths)).transpose(0, 1)
189 |
190 | # One training step
191 | optimizer.zero_grad()
192 | est_maps, est_counts = model.forward(imgs)
193 | term1, term2 = loss_loc.forward(est_maps,
194 | target_locations,
195 | target_orig_sizes)
196 | est_counts = est_counts.view(-1)
197 | target_counts = target_counts.view(-1)
198 | term3 = loss_regress.forward(est_counts, target_counts)
199 | term3 *= args.lambdaa
200 | loss = term1 + term2 + term3
201 | loss.backward()
202 | optimizer.step()
203 |
204 | # Update progress bar
205 | running_avg.put(loss.item())
206 | iter_train.set_postfix(running_avg=f'{round(running_avg.avg/3, 1)}')
207 |
208 | # Log training error
209 | if time.time() > tic_train + args.log_interval:
210 | tic_train = time.time()
211 |
212 | # Log training losses
213 | log.train_losses(terms=[term1, term2, term3, loss / 3, running_avg.avg / 3],
214 | iteration_number=epoch +
215 | batch_idx/len(trainset_loader),
216 | terms_legends=['Term1',
217 | 'Term2',
218 | 'Term3*%s' % args.lambdaa,
219 | 'Sum/3',
220 | 'Sum/3 runn avg'])
221 |
222 | # Resize images to original size
223 | orig_shape = target_orig_sizes[0].data.to(device_cpu).numpy().tolist()
224 | orig_img_origsize = ((skimage.transform.resize(imgs[0].data.squeeze().to(device_cpu).numpy().transpose((1, 2, 0)),
225 | output_shape=orig_shape,
226 | mode='constant') + 1) / 2.0 * 255.0).\
227 | astype(np.float32).transpose((2, 0, 1))
228 | est_map_origsize = skimage.transform.resize(est_maps[0].data.unsqueeze(0).to(device_cpu).numpy().transpose((1, 2, 0)),
229 | output_shape=orig_shape,
230 | mode='constant').\
231 | astype(np.float32).transpose((2, 0, 1)).squeeze(0)
232 |
233 | # Overlay output on heatmap
234 | orig_img_w_heatmap_origsize = utils.overlay_heatmap(img=orig_img_origsize,
235 | map=est_map_origsize).\
236 | astype(np.float32)
237 |
238 | # Send heatmap with circles at the labeled points to Visdom
239 | target_locs_np = target_locations[0].\
240 | to(device_cpu).numpy().reshape(-1, 2)
241 | target_orig_size_np = target_orig_sizes[0].\
242 | to(device_cpu).numpy().reshape(2)
243 | target_locs_wrt_orig = normalzr.unnormalize(target_locs_np,
244 | orig_img_size=target_orig_size_np)
245 | img_with_x = utils.paint_circles(img=orig_img_w_heatmap_origsize,
246 | points=target_locs_wrt_orig,
247 | color='white')
248 | log.image(imgs=[img_with_x],
249 | titles=['(Training) Image w/ output heatmap and labeled points'],
250 | window_ids=[1])
251 |
252 | # # Read image with GT dots from disk
253 | # gt_img_numpy = skimage.io.imread(
254 | # os.path.join('/home/jprat/projects/phenosorg/data/plant_counts_dots/20160613_F54_training_256x256_white_bigdots',
255 | # dictionary['filename'][0]))
256 | # # dots_img_tensor = torch.from_numpy(gt_img_numpy).permute(
257 | # # 2, 0, 1)[0, :, :].type(torch.FloatTensor) / 255
258 | # # Send GT image to Visdom
259 | # viz.image(np.moveaxis(gt_img_numpy, 2, 0),
260 | # opts=dict(title='(Training) Ground Truth'),
261 | # win=3)
262 |
263 | it_num += 1
264 |
265 | # Never do validation?
266 | if not args.val_dir or \
267 | not valset_loader or \
268 | len(valset_loader) == 0 or \
269 | args.val_freq == 0:
270 |
271 | # Time to save checkpoint?
272 | if args.save and (epoch + 1) % args.val_freq == 0:
273 | torch.save({'epoch': epoch,
274 | 'model': model.state_dict(),
275 | 'optimizer': optimizer.state_dict(),
276 | 'n_points': args.n_points,
277 | }, args.save)
278 | epoch += 1
279 | continue
280 |
281 | # Time to do validation?
282 | if (epoch + 1) % args.val_freq != 0:
283 | epoch += 1
284 | continue
285 |
286 | # === VALIDATION ===
287 |
288 | # Set the module in evaluation mode
289 | model.eval()
290 |
291 | judge = Judge(r=args.radius)
292 | sum_term1 = 0
293 | sum_term2 = 0
294 | sum_term3 = 0
295 | sum_loss = 0
296 | iter_val = tqdm(valset_loader,
297 | desc=f'Validating Epoch {epoch} ({len(valset_loader.dataset)} images)')
298 | for batch_idx, (imgs, dictionaries) in enumerate(iter_val):
299 |
300 | # Pull info from this batch and move to device
301 | imgs = imgs.to(device)
302 | target_locations = [dictt['locations'].to(device)
303 | for dictt in dictionaries]
304 | target_counts = [dictt['count'].to(device)
305 | for dictt in dictionaries]
306 | target_orig_heights = [dictt['orig_height'].to(device)
307 | for dictt in dictionaries]
308 | target_orig_widths = [dictt['orig_width'].to(device)
309 | for dictt in dictionaries]
310 |
311 | with torch.no_grad():
312 | target_counts = torch.stack(target_counts)
313 | target_orig_heights = torch.stack(target_orig_heights)
314 | target_orig_widths = torch.stack(target_orig_widths)
315 | target_orig_sizes = torch.stack((target_orig_heights,
316 | target_orig_widths)).transpose(0, 1)
317 | orig_shape = (dictionaries[0]['orig_height'].item(),
318 | dictionaries[0]['orig_width'].item())
319 |
320 | # Tensor -> float & numpy
321 | target_count_int = int(round(target_counts.item()))
322 | target_locations_np = \
323 | target_locations[0].to(device_cpu).numpy().reshape(-1, 2)
324 | target_orig_size_np = \
325 | target_orig_sizes[0].to(device_cpu).numpy().reshape(2)
326 |
327 | normalzr = utils.Normalizer(args.height, args.width)
328 |
329 | if target_count_int == 0:
330 | continue
331 |
332 | # Feed-forward
333 | with torch.no_grad():
334 | est_maps, est_counts = model.forward(imgs)
335 |
336 | # Tensor -> int
337 | est_count_int = int(round(est_counts.item()))
338 |
339 | # The 3 terms
340 | with torch.no_grad():
341 | est_counts = est_counts.view(-1)
342 | target_counts = target_counts.view(-1)
343 | term1, term2 = loss_loc.forward(est_maps,
344 | target_locations,
345 | target_orig_sizes)
346 | term3 = loss_regress.forward(est_counts, target_counts)
347 | term3 *= args.lambdaa
348 | sum_term1 += term1.item()
349 | sum_term2 += term2.item()
350 | sum_term3 += term3.item()
351 | sum_loss += term1 + term2 + term3
352 |
353 | # Update progress bar
354 | loss_avg_this_epoch = sum_loss.item() / (batch_idx + 1)
355 | iter_val.set_postfix(
356 | avg_val_loss_this_epoch=f'{loss_avg_this_epoch:.1f}-----')
357 |
358 | # The estimated map must be thresholed to obtain estimated points
359 | # BMM thresholding
360 | est_map_numpy = est_maps[0, :, :].to(device_cpu).numpy()
361 | est_map_numpy_origsize = skimage.transform.resize(est_map_numpy,
362 | output_shape=orig_shape,
363 | mode='constant')
364 | mask, _ = utils.threshold(est_map_numpy_origsize, tau=-1)
365 | # Obtain centroids of the mask
366 | centroids_wrt_orig = utils.cluster(mask, est_count_int,
367 | max_mask_pts=args.max_mask_pts)
368 |
369 | # Validation metrics
370 | target_locations_wrt_orig = normalzr.unnormalize(target_locations_np,
371 | orig_img_size=target_orig_size_np)
372 | judge.feed_points(centroids_wrt_orig, target_locations_wrt_orig,
373 | max_ahd=loss_loc.max_dist)
374 | judge.feed_count(est_count_int, target_count_int)
375 |
376 | if time.time() > tic_val + args.log_interval:
377 | tic_val = time.time()
378 |
379 | # Resize to original size
380 | orig_img_origsize = ((skimage.transform.resize(imgs[0].to(device_cpu).squeeze().numpy().transpose((1, 2, 0)),
381 | output_shape=target_orig_size_np.tolist(),
382 | mode='constant') + 1) / 2.0 * 255.0).\
383 | astype(np.float32).transpose((2, 0, 1))
384 | est_map_origsize = skimage.transform.resize(est_maps[0].to(device_cpu).unsqueeze(0).numpy().transpose((1, 2, 0)),
385 | output_shape=orig_shape,
386 | mode='constant').\
387 | astype(np.float32).transpose((2, 0, 1)).squeeze(0)
388 |
389 | # Overlay output on heatmap
390 | orig_img_w_heatmap_origsize = utils.overlay_heatmap(img=orig_img_origsize,
391 | map=est_map_origsize).\
392 | astype(np.float32)
393 |
394 | # # Read image with GT dots from disk
395 | # gt_img_numpy = skimage.io.imread(
396 | # os.path.join('/home/jprat/projects/phenosorg/data/plant_counts_dots/20160613_F54_validation_256x256_white_bigdots',
397 | # dictionary['filename'][0]))
398 | # # dots_img_tensor = torch.from_numpy(gt_img_numpy).permute(
399 | # # 2, 0, 1)[0, :, :].type(torch.FloatTensor) / 255
400 | # # Send GT image to Visdom
401 | # viz.image(np.moveaxis(gt_img_numpy, 2, 0),
402 | # opts=dict(title='(Validation) Ground Truth'),
403 | # win=7)
404 | if not args.paint:
405 | # Send input and output heatmap (first one in the batch)
406 | log.image(imgs=[orig_img_w_heatmap_origsize],
407 | titles=['(Validation) Image w/ output heatmap'],
408 | window_ids=[5])
409 | else:
410 | # Send heatmap with a cross at the estimated centroids to Visdom
411 | img_with_x = utils.paint_circles(img=orig_img_w_heatmap_origsize,
412 | points=centroids_wrt_orig,
413 | color='red',
414 | crosshair=True )
415 | log.image(imgs=[img_with_x],
416 | titles=['(Validation) Image w/ output heatmap '
417 | 'and point estimations'],
418 | window_ids=[8])
419 |
420 | avg_term1_val = sum_term1 / len(valset_loader)
421 | avg_term2_val = sum_term2 / len(valset_loader)
422 | avg_term3_val = sum_term3 / len(valset_loader)
423 | avg_loss_val = sum_loss / len(valset_loader)
424 |
425 | # Log validation metrics
426 | log.val_losses(terms=(avg_term1_val,
427 | avg_term2_val,
428 | avg_term3_val,
429 | avg_loss_val / 3,
430 | judge.mahd,
431 | judge.mae,
432 | judge.rmse,
433 | judge.mape,
434 | judge.coeff_of_determination,
435 | judge.pearson_corr \
436 | if not np.isnan(judge.pearson_corr) else 1,
437 | judge.precision,
438 | judge.recall),
439 | iteration_number=epoch,
440 | terms_legends=['Term 1',
441 | 'Term 2',
442 | 'Term3*%s' % args.lambdaa,
443 | 'Sum/3',
444 | 'AHD',
445 | 'MAE',
446 | 'RMSE',
447 | 'MAPE (%)',
448 | 'R^2',
449 | 'r',
450 | f'r{args.radius}-Precision (%)',
451 | f'r{args.radius}-Recall (%)'])
452 |
453 | # If this is the best epoch (in terms of validation error)
454 | if judge.mahd < lowest_mahd:
455 | # Keep the best model
456 | lowest_mahd = judge.mahd
457 | if args.save:
458 | torch.save({'epoch': epoch + 1, # when resuming, we will start at the next epoch
459 | 'model': model.state_dict(),
460 | 'mahd': lowest_mahd,
461 | 'optimizer': optimizer.state_dict(),
462 | 'n_points': args.n_points,
463 | }, args.save)
464 | print("Saved best checkpoint so far in %s " % args.save)
465 |
466 | epoch += 1
467 |
468 |
469 | """
470 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
471 | All rights reserved.
472 |
473 | This software is covered by US patents and copyright.
474 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
475 |
476 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
477 |
478 | Last Modified: 10/02/2019
479 | """
480 |
--------------------------------------------------------------------------------
/COPYRIGHT.txt:
--------------------------------------------------------------------------------
1 | Copyright ©right © (c) 2019 The Board of Trustees of Purdue University and the Purdue Research Foundation.
2 | All rights reserved.
3 |
4 | This software is covered by US patents and copyright.
5 | This source code is to be used for academic research purposes only, and no commercial use is allowed.
6 |
7 | For any questions, please contact Edward J. Delp (ace@ecn.purdue.edu) at Purdue University.
8 |
9 | Last Modified: 10/02/2019
10 |
11 |
12 | Attribution-NonCommercial-ShareAlike 4.0 International
13 |
14 | =======================================================================
15 |
16 | Creative Commons Corporation ("Creative Commons") is not a law firm and
17 | does not provide legal services or legal advice. Distribution of
18 | Creative Commons public licenses does not create a lawyer-client or
19 | other relationship. Creative Commons makes its licenses and related
20 | information available on an "as-is" basis. Creative Commons gives no
21 | warranties regarding its licenses, any material licensed under their
22 | terms and conditions, or any related information. Creative Commons
23 | disclaims all liability for damages resulting from their use to the
24 | fullest extent possible.
25 |
26 | Using Creative Commons Public Licenses
27 |
28 | Creative Commons public licenses provide a standard set of terms and
29 | conditions that creators and other rights holders may use to share
30 | original works of authorship and other material subject to copyright
31 | and certain other rights specified in the public license below. The
32 | following considerations are for informational purposes only, are not
33 | exhaustive, and do not form part of our licenses.
34 |
35 | Considerations for licensors: Our public licenses are
36 | intended for use by those authorized to give the public
37 | permission to use material in ways otherwise restricted by
38 | copyright and certain other rights. Our licenses are
39 | irrevocable. Licensors should read and understand the terms
40 | and conditions of the license they choose before applying it.
41 | Licensors should also secure all rights necessary before
42 | applying our licenses so that the public can reuse the
43 | material as expected. Licensors should clearly mark any
44 | material not subject to the license. This includes other CC-
45 | licensed material, or material used under an exception or
46 | limitation to copyright. More considerations for licensors:
47 | wiki.creativecommons.org/Considerations_for_licensors
48 |
49 | Considerations for the public: By using one of our public
50 | licenses, a licensor grants the public permission to use the
51 | licensed material under specified terms and conditions. If
52 | the licensor's permission is not necessary for any reason--for
53 | example, because of any applicable exception or limitation to
54 | copyright--then that use is not regulated by the license. Our
55 | licenses grant only permissions under copyright and certain
56 | other rights that a licensor has authority to grant. Use of
57 | the licensed material may still be restricted for other
58 | reasons, including because others have copyright or other
59 | rights in the material. A licensor may make special requests,
60 | such as asking that all changes be marked or described.
61 | Although not required by our licenses, you are encouraged to
62 | respect those requests where reasonable. More considerations
63 | for the public:
64 | wiki.creativecommons.org/Considerations_for_licensees
65 |
66 | =======================================================================
67 |
68 | Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International
69 | Public License
70 |
71 | By exercising the Licensed Rights (defined below), You accept and agree
72 | to be bound by the terms and conditions of this Creative Commons
73 | Attribution-NonCommercial-ShareAlike 4.0 International Public License
74 | ("Public License"). To the extent this Public License may be
75 | interpreted as a contract, You are granted the Licensed Rights in
76 | consideration of Your acceptance of these terms and conditions, and the
77 | Licensor grants You such rights in consideration of benefits the
78 | Licensor receives from making the Licensed Material available under
79 | these terms and conditions.
80 |
81 |
82 | Section 1 -- Definitions.
83 |
84 | a. Adapted Material means material subject to Copyright and Similar
85 | Rights that is derived from or based upon the Licensed Material
86 | and in which the Licensed Material is translated, altered,
87 | arranged, transformed, or otherwise modified in a manner requiring
88 | permission under the Copyright and Similar Rights held by the
89 | Licensor. For purposes of this Public License, where the Licensed
90 | Material is a musical work, performance, or sound recording,
91 | Adapted Material is always produced where the Licensed Material is
92 | synched in timed relation with a moving image.
93 |
94 | b. Adapter's License means the license You apply to Your Copyright
95 | and Similar Rights in Your contributions to Adapted Material in
96 | accordance with the terms and conditions of this Public License.
97 |
98 | c. BY-NC-SA Compatible License means a license listed at
99 | creativecommons.org/compatiblelicenses, approved by Creative
100 | Commons as essentially the equivalent of this Public License.
101 |
102 | d. Copyright and Similar Rights means copyright and/or similar rights
103 | closely related to copyright including, without limitation,
104 | performance, broadcast, sound recording, and Sui Generis Database
105 | Rights, without regard to how the rights are labeled or
106 | categorized. For purposes of this Public License, the rights
107 | specified in Section 2(b)(1)-(2) are not Copyright and Similar
108 | Rights.
109 |
110 | e. Effective Technological Measures means those measures that, in the
111 | absence of proper authority, may not be circumvented under laws
112 | fulfilling obligations under Article 11 of the WIPO Copyright
113 | Treaty adopted on December 20, 1996, and/or similar international
114 | agreements.
115 |
116 | f. Exceptions and Limitations means fair use, fair dealing, and/or
117 | any other exception or limitation to Copyright and Similar Rights
118 | that applies to Your use of the Licensed Material.
119 |
120 | g. License Elements means the license attributes listed in the name
121 | of a Creative Commons Public License. The License Elements of this
122 | Public License are Attribution, NonCommercial, and ShareAlike.
123 |
124 | h. Licensed Material means the artistic or literary work, database,
125 | or other material to which the Licensor applied this Public
126 | License.
127 |
128 | i. Licensed Rights means the rights granted to You subject to the
129 | terms and conditions of this Public License, which are limited to
130 | all Copyright and Similar Rights that apply to Your use of the
131 | Licensed Material and that the Licensor has authority to license.
132 |
133 | j. Licensor means the individual(s) or entity(ies) granting rights
134 | under this Public License.
135 |
136 | k. NonCommercial means not primarily intended for or directed towards
137 | commercial advantage or monetary compensation. For purposes of
138 | this Public License, the exchange of the Licensed Material for
139 | other material subject to Copyright and Similar Rights by digital
140 | file-sharing or similar means is NonCommercial provided there is
141 | no payment of monetary compensation in connection with the
142 | exchange.
143 |
144 | l. Share means to provide material to the public by any means or
145 | process that requires permission under the Licensed Rights, such
146 | as reproduction, public display, public performance, distribution,
147 | dissemination, communication, or importation, and to make material
148 | available to the public including in ways that members of the
149 | public may access the material from a place and at a time
150 | individually chosen by them.
151 |
152 | m. Sui Generis Database Rights means rights other than copyright
153 | resulting from Directive 96/9/EC of the European Parliament and of
154 | the Council of 11 March 1996 on the legal protection of databases,
155 | as amended and/or succeeded, as well as other essentially
156 | equivalent rights anywhere in the world.
157 |
158 | n. You means the individual or entity exercising the Licensed Rights
159 | under this Public License. Your has a corresponding meaning.
160 |
161 |
162 | Section 2 -- Scope.
163 |
164 | a. License grant.
165 |
166 | 1. Subject to the terms and conditions of this Public License,
167 | the Licensor hereby grants You a worldwide, royalty-free,
168 | non-sublicensable, non-exclusive, irrevocable license to
169 | exercise the Licensed Rights in the Licensed Material to:
170 |
171 | a. reproduce and Share the Licensed Material, in whole or
172 | in part, for NonCommercial purposes only; and
173 |
174 | b. produce, reproduce, and Share Adapted Material for
175 | NonCommercial purposes only.
176 |
177 | 2. Exceptions and Limitations. For the avoidance of doubt, where
178 | Exceptions and Limitations apply to Your use, this Public
179 | License does not apply, and You do not need to comply with
180 | its terms and conditions.
181 |
182 | 3. Term. The term of this Public License is specified in Section
183 | 6(a).
184 |
185 | 4. Media and formats; technical modifications allowed. The
186 | Licensor authorizes You to exercise the Licensed Rights in
187 | all media and formats whether now known or hereafter created,
188 | and to make technical modifications necessary to do so. The
189 | Licensor waives and/or agrees not to assert any right or
190 | authority to forbid You from making technical modifications
191 | necessary to exercise the Licensed Rights, including
192 | technical modifications necessary to circumvent Effective
193 | Technological Measures. For purposes of this Public License,
194 | simply making modifications authorized by this Section 2(a)
195 | (4) never produces Adapted Material.
196 |
197 | 5. Downstream recipients.
198 |
199 | a. Offer from the Licensor -- Licensed Material. Every
200 | recipient of the Licensed Material automatically
201 | receives an offer from the Licensor to exercise the
202 | Licensed Rights under the terms and conditions of this
203 | Public License.
204 |
205 | b. Additional offer from the Licensor -- Adapted Material.
206 | Every recipient of Adapted Material from You
207 | automatically receives an offer from the Licensor to
208 | exercise the Licensed Rights in the Adapted Material
209 | under the conditions of the Adapter's License You apply.
210 |
211 | c. No downstream restrictions. You may not offer or impose
212 | any additional or different terms or conditions on, or
213 | apply any Effective Technological Measures to, the
214 | Licensed Material if doing so restricts exercise of the
215 | Licensed Rights by any recipient of the Licensed
216 | Material.
217 |
218 | 6. No endorsement. Nothing in this Public License constitutes or
219 | may be construed as permission to assert or imply that You
220 | are, or that Your use of the Licensed Material is, connected
221 | with, or sponsored, endorsed, or granted official status by,
222 | the Licensor or others designated to receive attribution as
223 | provided in Section 3(a)(1)(A)(i).
224 |
225 | b. Other rights.
226 |
227 | 1. Moral rights, such as the right of integrity, are not
228 | licensed under this Public License, nor are publicity,
229 | privacy, and/or other similar personality rights; however, to
230 | the extent possible, the Licensor waives and/or agrees not to
231 | assert any such rights held by the Licensor to the limited
232 | extent necessary to allow You to exercise the Licensed
233 | Rights, but not otherwise.
234 |
235 | 2. Patent and trademark rights are not licensed under this
236 | Public License.
237 |
238 | 3. To the extent possible, the Licensor waives any right to
239 | collect royalties from You for the exercise of the Licensed
240 | Rights, whether directly or through a collecting society
241 | under any voluntary or waivable statutory or compulsory
242 | licensing scheme. In all other cases the Licensor expressly
243 | reserves any right to collect such royalties, including when
244 | the Licensed Material is used other than for NonCommercial
245 | purposes.
246 |
247 |
248 | Section 3 -- License Conditions.
249 |
250 | Your exercise of the Licensed Rights is expressly made subject to the
251 | following conditions.
252 |
253 | a. Attribution.
254 |
255 | 1. If You Share the Licensed Material (including in modified
256 | form), You must:
257 |
258 | a. retain the following if it is supplied by the Licensor
259 | with the Licensed Material:
260 |
261 | i. identification of the creator(s) of the Licensed
262 | Material and any others designated to receive
263 | attribution, in any reasonable manner requested by
264 | the Licensor (including by pseudonym if
265 | designated);
266 |
267 | ii. a copyright notice;
268 |
269 | iii. a notice that refers to this Public License;
270 |
271 | iv. a notice that refers to the disclaimer of
272 | warranties;
273 |
274 | v. a URI or hyperlink to the Licensed Material to the
275 | extent reasonably practicable;
276 |
277 | b. indicate if You modified the Licensed Material and
278 | retain an indication of any previous modifications; and
279 |
280 | c. indicate the Licensed Material is licensed under this
281 | Public License, and include the text of, or the URI or
282 | hyperlink to, this Public License.
283 |
284 | 2. You may satisfy the conditions in Section 3(a)(1) in any
285 | reasonable manner based on the medium, means, and context in
286 | which You Share the Licensed Material. For example, it may be
287 | reasonable to satisfy the conditions by providing a URI or
288 | hyperlink to a resource that includes the required
289 | information.
290 | 3. If requested by the Licensor, You must remove any of the
291 | information required by Section 3(a)(1)(A) to the extent
292 | reasonably practicable.
293 |
294 | b. ShareAlike.
295 |
296 | In addition to the conditions in Section 3(a), if You Share
297 | Adapted Material You produce, the following conditions also apply.
298 |
299 | 1. The Adapter's License You apply must be a Creative Commons
300 | license with the same License Elements, this version or
301 | later, or a BY-NC-SA Compatible License.
302 |
303 | 2. You must include the text of, or the URI or hyperlink to, the
304 | Adapter's License You apply. You may satisfy this condition
305 | in any reasonable manner based on the medium, means, and
306 | context in which You Share Adapted Material.
307 |
308 | 3. You may not offer or impose any additional or different terms
309 | or conditions on, or apply any Effective Technological
310 | Measures to, Adapted Material that restrict exercise of the
311 | rights granted under the Adapter's License You apply.
312 |
313 |
314 | Section 4 -- Sui Generis Database Rights.
315 |
316 | Where the Licensed Rights include Sui Generis Database Rights that
317 | apply to Your use of the Licensed Material:
318 |
319 | a. for the avoidance of doubt, Section 2(a)(1) grants You the right
320 | to extract, reuse, reproduce, and Share all or a substantial
321 | portion of the contents of the database for NonCommercial purposes
322 | only;
323 |
324 | b. if You include all or a substantial portion of the database
325 | contents in a database in which You have Sui Generis Database
326 | Rights, then the database in which You have Sui Generis Database
327 | Rights (but not its individual contents) is Adapted Material,
328 | including for purposes of Section 3(b); and
329 |
330 | c. You must comply with the conditions in Section 3(a) if You Share
331 | all or a substantial portion of the contents of the database.
332 |
333 | For the avoidance of doubt, this Section 4 supplements and does not
334 | replace Your obligations under this Public License where the Licensed
335 | Rights include other Copyright and Similar Rights.
336 |
337 |
338 | Section 5 -- Disclaimer of Warranties and Limitation of Liability.
339 |
340 | a. UNLESS OTHERWISE SEPARATELY UNDERTAKEN BY THE LICENSOR, TO THE
341 | EXTENT POSSIBLE, THE LICENSOR OFFERS THE LICENSED MATERIAL AS-IS
342 | AND AS-AVAILABLE, AND MAKES NO REPRESENTATIONS OR WARRANTIES OF
343 | ANY KIND CONCERNING THE LICENSED MATERIAL, WHETHER EXPRESS,
344 | IMPLIED, STATUTORY, OR OTHER. THIS INCLUDES, WITHOUT LIMITATION,
345 | WARRANTIES OF TITLE, MERCHANTABILITY, FITNESS FOR A PARTICULAR
346 | PURPOSE, NON-INFRINGEMENT, ABSENCE OF LATENT OR OTHER DEFECTS,
347 | ACCURACY, OR THE PRESENCE OR ABSENCE OF ERRORS, WHETHER OR NOT
348 | KNOWN OR DISCOVERABLE. WHERE DISCLAIMERS OF WARRANTIES ARE NOT
349 | ALLOWED IN FULL OR IN PART, THIS DISCLAIMER MAY NOT APPLY TO YOU.
350 |
351 | b. TO THE EXTENT POSSIBLE, IN NO EVENT WILL THE LICENSOR BE LIABLE
352 | TO YOU ON ANY LEGAL THEORY (INCLUDING, WITHOUT LIMITATION,
353 | NEGLIGENCE) OR OTHERWISE FOR ANY DIRECT, SPECIAL, INDIRECT,
354 | INCIDENTAL, CONSEQUENTIAL, PUNITIVE, EXEMPLARY, OR OTHER LOSSES,
355 | COSTS, EXPENSES, OR DAMAGES ARISING OUT OF THIS PUBLIC LICENSE OR
356 | USE OF THE LICENSED MATERIAL, EVEN IF THE LICENSOR HAS BEEN
357 | ADVISED OF THE POSSIBILITY OF SUCH LOSSES, COSTS, EXPENSES, OR
358 | DAMAGES. WHERE A LIMITATION OF LIABILITY IS NOT ALLOWED IN FULL OR
359 | IN PART, THIS LIMITATION MAY NOT APPLY TO YOU.
360 |
361 | c. The disclaimer of warranties and limitation of liability provided
362 | above shall be interpreted in a manner that, to the extent
363 | possible, most closely approximates an absolute disclaimer and
364 | waiver of all liability.
365 |
366 |
367 | Section 6 -- Term and Termination.
368 |
369 | a. This Public License applies for the term of the Copyright and
370 | Similar Rights licensed here. However, if You fail to comply with
371 | this Public License, then Your rights under this Public License
372 | terminate automatically.
373 |
374 | b. Where Your right to use the Licensed Material has terminated under
375 | Section 6(a), it reinstates:
376 |
377 | 1. automatically as of the date the violation is cured, provided
378 | it is cured within 30 days of Your discovery of the
379 | violation; or
380 |
381 | 2. upon express reinstatement by the Licensor.
382 |
383 | For the avoidance of doubt, this Section 6(b) does not affect any
384 | right the Licensor may have to seek remedies for Your violations
385 | of this Public License.
386 |
387 | c. For the avoidance of doubt, the Licensor may also offer the
388 | Licensed Material under separate terms or conditions or stop
389 | distributing the Licensed Material at any time; however, doing so
390 | will not terminate this Public License.
391 |
392 | d. Sections 1, 5, 6, 7, and 8 survive termination of this Public
393 | License.
394 |
395 |
396 | Section 7 -- Other Terms and Conditions.
397 |
398 | a. The Licensor shall not be bound by any additional or different
399 | terms or conditions communicated by You unless expressly agreed.
400 |
401 | b. Any arrangements, understandings, or agreements regarding the
402 | Licensed Material not stated herein are separate from and
403 | independent of the terms and conditions of this Public License.
404 |
405 |
406 | Section 8 -- Interpretation.
407 |
408 | a. For the avoidance of doubt, this Public License does not, and
409 | shall not be interpreted to, reduce, limit, restrict, or impose
410 | conditions on any use of the Licensed Material that could lawfully
411 | be made without permission under this Public License.
412 |
413 | b. To the extent possible, if any provision of this Public License is
414 | deemed unenforceable, it shall be automatically reformed to the
415 | minimum extent necessary to make it enforceable. If the provision
416 | cannot be reformed, it shall be severed from this Public License
417 | without affecting the enforceability of the remaining terms and
418 | conditions.
419 |
420 | c. No term or condition of this Public License will be waived and no
421 | failure to comply consented to unless expressly agreed to by the
422 | Licensor.
423 |
424 | d. Nothing in this Public License constitutes or may be interpreted
425 | as a limitation upon, or waiver of, any privileges and immunities
426 | that apply to the Licensor or You, including from the legal
427 | processes of any jurisdiction or authority.
428 |
429 | =======================================================================
430 |
431 | Creative Commons is not a party to its public
432 | licenses. Notwithstanding, Creative Commons may elect to apply one of
433 | its public licenses to material it publishes and in those instances
434 | will be considered the “Licensor.” The text of the Creative Commons
435 | public licenses is dedicated to the public domain under the CC0 Public
436 | Domain Dedication. Except for the limited purpose of indicating that
437 | material is shared under a Creative Commons public license or as
438 | otherwise permitted by the Creative Commons policies published at
439 | creativecommons.org/policies, Creative Commons does not authorize the
440 | use of the trademark "Creative Commons" or any other trademark or logo
441 | of Creative Commons without its prior written consent including,
442 | without limitation, in connection with any unauthorized modifications
443 | to any of its public licenses or any other arrangements,
444 | understandings, or agreements concerning use of licensed material. For
445 | the avoidance of doubt, this paragraph does not form part of the
446 | public licenses.
447 |
448 | Creative Commons may be contacted at creativecommons.org.
449 |
450 |
--------------------------------------------------------------------------------
 5 |
5 | 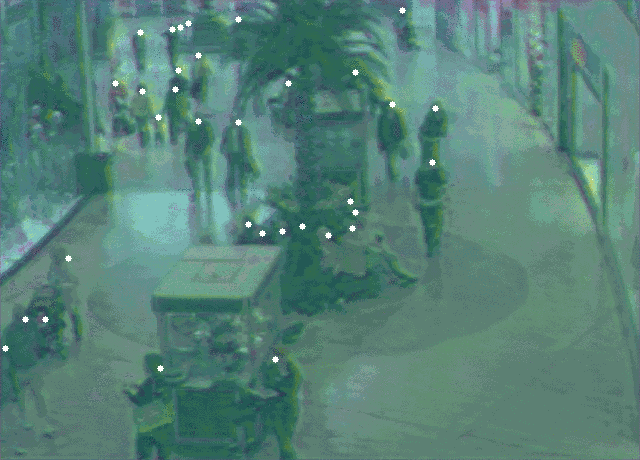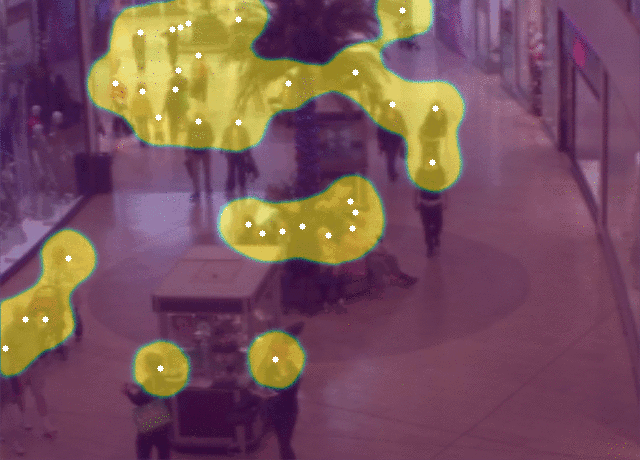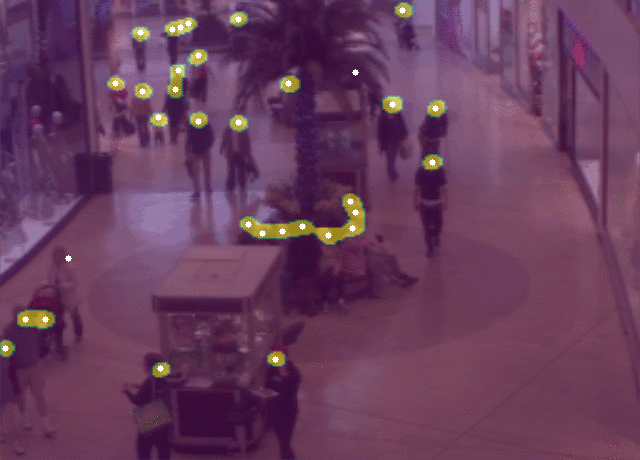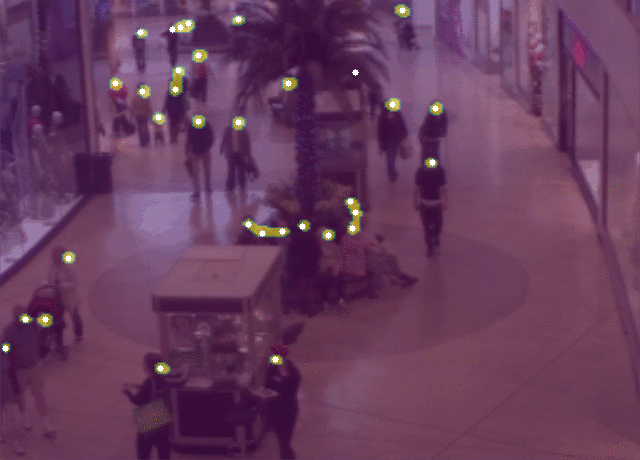 6 |
7 |
8 | ## Citing this work
9 | ```
10 | @article{ribera2019,
11 | title={Locating Objects Without Bounding Boxes},
12 | author={Javier Ribera and David G\"{u}era and Yuhao Chen and Edward J. Delp},
13 | journal={Proceedings of the Computer Vision and Pattern Recognition (CVPR)},
14 | month={June},
15 | year={2019},
16 | note={{Long Beach, CA}}
17 | }
18 | ```
19 |
20 | ## Datasets
21 | The datasets used in the paper can be downloaded from:
22 | - [Mall dataset](http://personal.ie.cuhk.edu.hk/~ccloy/downloads_mall_dataset.html)
23 | - [Pupil dataset](http://www.ti.uni-tuebingen.de/Pupil-detection.1827.0.html)
24 | - [Plant dataset](https://engineering.purdue.edu/~sorghum/dataset-plant-centers-2016)
25 |
26 | ## Installation
27 | Use conda to recreate the environment provided with the code:
28 |
6 |
7 |
8 | ## Citing this work
9 | ```
10 | @article{ribera2019,
11 | title={Locating Objects Without Bounding Boxes},
12 | author={Javier Ribera and David G\"{u}era and Yuhao Chen and Edward J. Delp},
13 | journal={Proceedings of the Computer Vision and Pattern Recognition (CVPR)},
14 | month={June},
15 | year={2019},
16 | note={{Long Beach, CA}}
17 | }
18 | ```
19 |
20 | ## Datasets
21 | The datasets used in the paper can be downloaded from:
22 | - [Mall dataset](http://personal.ie.cuhk.edu.hk/~ccloy/downloads_mall_dataset.html)
23 | - [Pupil dataset](http://www.ti.uni-tuebingen.de/Pupil-detection.1827.0.html)
24 | - [Plant dataset](https://engineering.purdue.edu/~sorghum/dataset-plant-centers-2016)
25 |
26 | ## Installation
27 | Use conda to recreate the environment provided with the code:
28 |