├── .github
├── .gitignore
├── CODEOWNERS
├── move.yml
├── ISSUE_TEMPLATE
│ └── issue_template.md
├── CODE_OF_CONDUCT.md
├── SUPPORT.md
└── CONTRIBUTING.md
├── vignettes
├── .gitignore
└── forcats.Rmd
├── .covrignore
├── R
├── forcats-package.R
├── shuffle.R
├── rev.R
├── unique.R
├── expand.R
├── data.R
├── anon.R
├── utils.R
├── shift.R
├── explicit_na.R
├── count.R
├── drop.R
├── other.R
├── match.R
├── collapse.R
├── relabel.R
├── c.R
├── fct_cross.R
├── as_factor.R
├── recode.R
├── relevel.R
├── lvls.R
├── reorder.R
└── lump.R
├── .gitignore
├── data-raw
├── GSS.xls
└── gss-cat.R
├── data
└── gss_cat.rda
├── tests
├── testthat.R
└── testthat
│ ├── helper-lump.R
│ ├── test-rev.R
│ ├── test-shuffle.R
│ ├── test-utils.R
│ ├── test-anon.R
│ ├── test-match.R
│ ├── test-as_factor.R
│ ├── test-fct_c.R
│ ├── test-shift.R
│ ├── test-explicit_na.R
│ ├── test-fct_other.R
│ ├── test-fct_drop.R
│ ├── test-count.R
│ ├── test-relevel.R
│ ├── test-lvls_reorder.R
│ ├── test-fct_cross.R
│ ├── test-fct_recode.R
│ ├── test-fct_relabel.R
│ ├── test-lvls.R
│ ├── test-collapse.R
│ ├── test-reorder.R
│ └── test-fct_lump.R
├── man
├── figures
│ ├── logo.png
│ ├── README-ordered-plot-1.png
│ └── README-unordered-plot-1.png
├── pipe.Rd
├── fct_rev.Rd
├── fct_shuffle.Rd
├── lvls_union.Rd
├── fct_unique.Rd
├── fct_unify.Rd
├── fct_anon.Rd
├── fct_explicit_na.Rd
├── fct_expand.Rd
├── fct_c.Rd
├── fct_shift.Rd
├── fct_count.Rd
├── fct_drop.Rd
├── fct_inorder.Rd
├── gss_cat.Rd
├── fct_collapse.Rd
├── fct_cross.Rd
├── fct_match.Rd
├── fct_other.Rd
├── forcats-package.Rd
├── fct_recode.Rd
├── as_factor.Rd
├── fct_relabel.Rd
├── lvls.Rd
├── fct_relevel.Rd
├── fct_reorder.Rd
└── fct_lump.Rd
├── revdep
├── .gitignore
├── email.yml
├── README.md
└── problems.md
├── pkgdown
└── favicon
│ ├── favicon.ico
│ ├── favicon-16x16.png
│ ├── favicon-32x32.png
│ ├── apple-touch-icon.png
│ ├── apple-touch-icon-60x60.png
│ ├── apple-touch-icon-76x76.png
│ ├── apple-touch-icon-120x120.png
│ ├── apple-touch-icon-152x152.png
│ └── apple-touch-icon-180x180.png
├── codecov.yml
├── .Rbuildignore
├── forcats.Rproj
├── .travis.yml
├── cran-comments.md
├── NAMESPACE
├── DESCRIPTION
├── _pkgdown.yml
├── README.Rmd
├── NEWS.md
├── README.md
└── LICENSE.md
/.github/.gitignore:
--------------------------------------------------------------------------------
1 | *.html
2 |
--------------------------------------------------------------------------------
/vignettes/.gitignore:
--------------------------------------------------------------------------------
1 | *.html
2 | *.R
3 |
--------------------------------------------------------------------------------
/.covrignore:
--------------------------------------------------------------------------------
1 | R/deprec-*.R
2 | R/compat-*.R
3 |
--------------------------------------------------------------------------------
/R/forcats-package.R:
--------------------------------------------------------------------------------
1 | #' @keywords internal
2 | "_PACKAGE"
3 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | Meta
2 | doc
3 | inst/doc
4 | docs
5 | .Rproj.user
6 | .Rhistory
7 | .RData
8 |
--------------------------------------------------------------------------------
/data-raw/GSS.xls:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/data-raw/GSS.xls
--------------------------------------------------------------------------------
/data/gss_cat.rda:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/data/gss_cat.rda
--------------------------------------------------------------------------------
/tests/testthat.R:
--------------------------------------------------------------------------------
1 | library(testthat)
2 | library(forcats)
3 |
4 | test_check("forcats")
5 |
--------------------------------------------------------------------------------
/man/figures/logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/man/figures/logo.png
--------------------------------------------------------------------------------
/revdep/.gitignore:
--------------------------------------------------------------------------------
1 | checks
2 | library
3 | checks.noindex
4 | library.noindex
5 | data.sqlite
6 | *.html
7 |
--------------------------------------------------------------------------------
/pkgdown/favicon/favicon.ico:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/favicon.ico
--------------------------------------------------------------------------------
/revdep/email.yml:
--------------------------------------------------------------------------------
1 | release_date: ???
2 | rel_release_date: ???
3 | my_news_url: ???
4 | release_version: ???
5 |
--------------------------------------------------------------------------------
/pkgdown/favicon/favicon-16x16.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/favicon-16x16.png
--------------------------------------------------------------------------------
/pkgdown/favicon/favicon-32x32.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/favicon-32x32.png
--------------------------------------------------------------------------------
/man/figures/README-ordered-plot-1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/man/figures/README-ordered-plot-1.png
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon.png
--------------------------------------------------------------------------------
/man/figures/README-unordered-plot-1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/man/figures/README-unordered-plot-1.png
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon-60x60.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon-60x60.png
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon-76x76.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon-76x76.png
--------------------------------------------------------------------------------
/.github/CODEOWNERS:
--------------------------------------------------------------------------------
1 | # CODEOWNERS for forcats
2 | # https://www.tidyverse.org/development/understudies
3 | .github/CODEOWNERS @hadley @jennybc
4 |
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon-120x120.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon-120x120.png
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon-152x152.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon-152x152.png
--------------------------------------------------------------------------------
/pkgdown/favicon/apple-touch-icon-180x180.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/jonocarroll/forcats/master/pkgdown/favicon/apple-touch-icon-180x180.png
--------------------------------------------------------------------------------
/tests/testthat/helper-lump.R:
--------------------------------------------------------------------------------
1 | lump_test <- function(x) {
2 | paste(ifelse(in_smallest(x), "X", letters[seq_along(x)]), collapse = "")
3 | }
4 |
--------------------------------------------------------------------------------
/tests/testthat/test-rev.R:
--------------------------------------------------------------------------------
1 | context("test-rev.R")
2 |
3 | test_that("reverses levels", {
4 | f1 <- factor(c("a", "b", "a"))
5 | f2 <- fct_rev(f1)
6 |
7 | expect_equal(levels(f2), c("b", "a"))
8 | })
9 |
--------------------------------------------------------------------------------
/codecov.yml:
--------------------------------------------------------------------------------
1 | comment: false
2 |
3 | coverage:
4 | status:
5 | project:
6 | default:
7 | target: auto
8 | threshold: 1%
9 | patch:
10 | default:
11 | target: auto
12 | threshold: 1%
13 |
--------------------------------------------------------------------------------
/tests/testthat/test-shuffle.R:
--------------------------------------------------------------------------------
1 | context("test-shuffle.R")
2 |
3 | test_that("reproducibility shuffles", {
4 | set.seed(1014)
5 |
6 | f1 <- factor(c("a", "b"))
7 | f2 <- fct_shuffle(f1)
8 |
9 | expect_equal(levels(f2), c("a", "b"))
10 | })
11 |
--------------------------------------------------------------------------------
/man/pipe.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/utils.R
3 | \name{\%>\%}
4 | \alias{\%>\%}
5 | \title{Pipe operator}
6 | \usage{
7 | lhs \%>\% rhs
8 | }
9 | \description{
10 | See \code{\link[magrittr]{\%>\%}} for more details.
11 | }
12 | \keyword{internal}
13 |
--------------------------------------------------------------------------------
/.Rbuildignore:
--------------------------------------------------------------------------------
1 | ^CRAN-RELEASE$
2 | ^Meta$
3 | ^doc$
4 | ^pkgdown$
5 | ^\.covrignore$
6 | ^.*\.Rproj$
7 | ^\.Rproj\.user$
8 | ^\.travis\.yml$
9 | ^codecov\.yml$
10 | ^data-raw$
11 | ^cran-comments\.md$
12 | ^docs$
13 | ^README\.Rmd$
14 | ^README-.*\.png$
15 | ^_pkgdown\.yml$
16 | ^revdep$
17 | ^\.github$
18 | ^LICENSE\.md$
19 |
--------------------------------------------------------------------------------
/R/shuffle.R:
--------------------------------------------------------------------------------
1 | #' Randomly permute factor levels
2 | #'
3 | #' @param f A factor (or character vector).
4 | #' @export
5 | #' @examples
6 | #' f <- factor(c("a", "b", "c"))
7 | #' fct_shuffle(f)
8 | #' fct_shuffle(f)
9 | fct_shuffle <- function(f) {
10 | f <- check_factor(f)
11 |
12 | lvls_reorder(f, sample(lvls_seq(f)))
13 | }
14 |
--------------------------------------------------------------------------------
/tests/testthat/test-utils.R:
--------------------------------------------------------------------------------
1 | context("test-utils.R")
2 |
3 | test_that("check_factor() fails when needed", {
4 | expect_error(check_factor(NA), "factor")
5 | })
6 |
7 | test_that("check_factor_list() fails when needed", {
8 | expect_error(check_factor_list(1), "list")
9 | expect_error(check_factor_list(list(1)), "factor")
10 | })
11 |
--------------------------------------------------------------------------------
/R/rev.R:
--------------------------------------------------------------------------------
1 | #' Reverse order of factor levels
2 | #'
3 | #' This is sometimes useful when plotting a factor.
4 | #'
5 | #' @param f A factor (or character vector).
6 | #' @export
7 | #' @examples
8 | #' f <- factor(c("a", "b", "c"))
9 | #' fct_rev(f)
10 | fct_rev <- function(f) {
11 | f <- check_factor(f)
12 |
13 | lvls_reorder(f, rev(lvls_seq(f)))
14 | }
15 |
--------------------------------------------------------------------------------
/R/unique.R:
--------------------------------------------------------------------------------
1 | #' Unique values of a factor
2 | #'
3 | #' @param f A factor.
4 | #' @export
5 | #' @examples
6 | #' f <- factor(letters[rpois(100, 10)])
7 | #'
8 | #' unique(f) # in order of appearance
9 | #' fct_unique(f) # in order of levels
10 | fct_unique <- function(f) {
11 | factor(levels(f), levels(f), exclude = NULL, ordered = is.ordered(f))
12 | }
13 |
14 |
15 |
--------------------------------------------------------------------------------
/tests/testthat/test-anon.R:
--------------------------------------------------------------------------------
1 | context("test-anon.R")
2 |
3 | test_that("new levels are padded numerics", {
4 | f1 <- factor(letters[1:10])
5 | f2 <- fct_anon(f1)
6 |
7 | expect_equal(levels(f2), sprintf("%02d", 1:10))
8 | })
9 |
10 | test_that("prefix added to start of level", {
11 | f1 <- factor("x")
12 | f2 <- fct_anon(f1, prefix = "X")
13 |
14 | expect_equal(levels(f2), "X1")
15 | })
16 |
--------------------------------------------------------------------------------
/tests/testthat/test-match.R:
--------------------------------------------------------------------------------
1 | context("test-fct_match.R")
2 |
3 | test_that("equivalent to %in% when levels present", {
4 | f <- factor(c("a", "b", "c", NA))
5 | expect_equal(fct_match(f, "a"), f %in% "a")
6 | expect_equal(fct_match(f, NA), f %in% NA)
7 | })
8 |
9 | test_that("error when levels are missing", {
10 | f <- factor(c("a", "b", "c"))
11 | expect_error(fct_match(f, "d"), "not present")
12 | })
13 |
--------------------------------------------------------------------------------
/man/fct_rev.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/rev.R
3 | \name{fct_rev}
4 | \alias{fct_rev}
5 | \title{Reverse order of factor levels}
6 | \usage{
7 | fct_rev(f)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 | }
12 | \description{
13 | This is sometimes useful when plotting a factor.
14 | }
15 | \examples{
16 | f <- factor(c("a", "b", "c"))
17 | fct_rev(f)
18 | }
19 |
--------------------------------------------------------------------------------
/man/fct_shuffle.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/shuffle.R
3 | \name{fct_shuffle}
4 | \alias{fct_shuffle}
5 | \title{Randomly permute factor levels}
6 | \usage{
7 | fct_shuffle(f)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 | }
12 | \description{
13 | Randomly permute factor levels
14 | }
15 | \examples{
16 | f <- factor(c("a", "b", "c"))
17 | fct_shuffle(f)
18 | fct_shuffle(f)
19 | }
20 |
--------------------------------------------------------------------------------
/man/lvls_union.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/lvls.R
3 | \name{lvls_union}
4 | \alias{lvls_union}
5 | \title{Find all levels in a list of factors}
6 | \usage{
7 | lvls_union(fs)
8 | }
9 | \arguments{
10 | \item{fs}{A list of factors.}
11 | }
12 | \description{
13 | Find all levels in a list of factors
14 | }
15 | \examples{
16 | fs <- list(factor("a"), factor("b"), factor(c("a", "b")))
17 | lvls_union(fs)
18 | }
19 |
--------------------------------------------------------------------------------
/tests/testthat/test-as_factor.R:
--------------------------------------------------------------------------------
1 | context("as_factor")
2 |
3 | test_that("equivalent to fct_inorder", {
4 | x <- c("a", "z", "g")
5 | expect_equal(as_factor(x), fct_inorder(x))
6 | })
7 |
8 | test_that("leaves factors as is", {
9 | f1 <- factor(letters)
10 | f2 <- as_factor(f1)
11 |
12 | expect_identical(f1, f2)
13 | })
14 |
15 | test_that("supports NA (#89)", {
16 | x <- c("a", "z", "g", NA)
17 | expect_equal(as_factor(x), fct_inorder(x))
18 | })
19 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_c.R:
--------------------------------------------------------------------------------
1 | context("fct_c")
2 |
3 | test_that("uses tidy_dots", {
4 | fs <- list(factor("a"), factor("b"))
5 | fab <- factor(c("a", "b"))
6 |
7 | expect_equal(fct_c(!!!fs), fab)
8 | expect_equal(fct_c(fs[[1]], fs[[2]]), fab)
9 | })
10 |
11 | test_that("all inputs must be factors", {
12 | expect_error(fct_c("a"), "must be factors")
13 | })
14 |
15 | test_that("empty input yields empty factor", {
16 | expect_equal(fct_c(), factor())
17 | })
18 |
--------------------------------------------------------------------------------
/forcats.Rproj:
--------------------------------------------------------------------------------
1 | Version: 1.0
2 |
3 | RestoreWorkspace: No
4 | SaveWorkspace: No
5 | AlwaysSaveHistory: Default
6 |
7 | EnableCodeIndexing: Yes
8 | UseSpacesForTab: Yes
9 | NumSpacesForTab: 2
10 | Encoding: UTF-8
11 |
12 | RnwWeave: knitr
13 | LaTeX: XeLaTeX
14 |
15 | AutoAppendNewline: Yes
16 | StripTrailingWhitespace: Yes
17 |
18 | BuildType: Package
19 | PackageUseDevtools: Yes
20 | PackageInstallArgs: --no-multiarch --with-keep.source
21 | PackageRoxygenize: rd,collate,namespace
22 |
--------------------------------------------------------------------------------
/man/fct_unique.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/unique.R
3 | \name{fct_unique}
4 | \alias{fct_unique}
5 | \title{Unique values of a factor}
6 | \usage{
7 | fct_unique(f)
8 | }
9 | \arguments{
10 | \item{f}{A factor.}
11 | }
12 | \description{
13 | Unique values of a factor
14 | }
15 | \examples{
16 | f <- factor(letters[rpois(100, 10)])
17 |
18 | unique(f) # in order of appearance
19 | fct_unique(f) # in order of levels
20 | }
21 |
--------------------------------------------------------------------------------
/tests/testthat/test-shift.R:
--------------------------------------------------------------------------------
1 | context("test-shift.R")
2 |
3 | test_that("can shift in either direction", {
4 | f1 <- factor(c("a", "b", "c"))
5 |
6 | f2_l <- fct_shift(f1, 1)
7 | expect_equal(levels(f2_l), c("b", "c", "a"))
8 |
9 | f2_r <- fct_shift(f1, -1)
10 | expect_equal(levels(f2_r), c("c", "a", "b"))
11 | })
12 |
13 | test_that("0 shift leaves unchanged", {
14 | f1 <- factor(c("a", "b", "c"))
15 | f2 <- fct_shift(f1, 0)
16 |
17 | expect_identical(f1, f2)
18 | })
19 |
--------------------------------------------------------------------------------
/.github/move.yml:
--------------------------------------------------------------------------------
1 | # Configuration for move-issues bot - https://github.com/dessant/move-issues
2 |
3 | # Delete the command comment when it contains no other content
4 | deleteCommand: true
5 |
6 | # Close the source issue after moving
7 | closeSourceIssue: true
8 |
9 | # Lock the source issue after moving
10 | lockSourceIssue: false
11 |
12 | # Mention issue and comment authors
13 | mentionAuthors: true
14 |
15 | # Preserve mentions in the issue content
16 | keepContentMentions: true
17 |
18 | # Set custom aliases for targets
19 | # aliases:
20 | # r: repo
21 | # or: owner/repo
22 |
--------------------------------------------------------------------------------
/man/fct_unify.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/c.R
3 | \name{fct_unify}
4 | \alias{fct_unify}
5 | \title{Unify the levels in a list of factors}
6 | \usage{
7 | fct_unify(fs, levels = lvls_union(fs))
8 | }
9 | \arguments{
10 | \item{fs}{A list of factors}
11 |
12 | \item{levels}{Set of levels to apply to every factor. Default to
13 | union of all factor levels}
14 | }
15 | \description{
16 | Unify the levels in a list of factors
17 | }
18 | \examples{
19 | fs <- list(factor("a"), factor("b"), factor(c("a", "b")))
20 | fct_unify(fs)
21 | }
22 |
--------------------------------------------------------------------------------
/tests/testthat/test-explicit_na.R:
--------------------------------------------------------------------------------
1 | context("test-explicit_na.R")
2 |
3 | test_that("factor unchanged if no missing levels", {
4 | f1 <- factor(letters[1:3])
5 | f2 <- fct_explicit_na(f1)
6 |
7 | expect_identical(f1, f2)
8 | })
9 |
10 | test_that("converts implicit NA", {
11 | f1 <- factor(c("a", NA))
12 | f2 <- fct_explicit_na(f1)
13 |
14 | expect_equal(f2, fct_inorder(c("a", "(Missing)")))
15 | })
16 |
17 | test_that("converts explicit NA", {
18 | f1 <- factor(c("a", NA), exclude = NULL)
19 | f2 <- fct_explicit_na(f1)
20 |
21 | expect_equal(f2, fct_inorder(c("a", "(Missing)")))
22 | })
23 |
--------------------------------------------------------------------------------
/R/expand.R:
--------------------------------------------------------------------------------
1 | #' Add additional levels to a factor
2 | #'
3 | #' @param f A factor (or character vector).
4 | #' @param ... Additional levels to add to the factor. Levels that already
5 | #' exist will be silently ignored.
6 | #' @export
7 | #' @seealso [fct_drop()] to drop unused factor levels.
8 | #' @examples
9 | #' f <- factor(sample(letters[1:3], 20, replace = TRUE))
10 | #' f
11 | #' fct_expand(f, "d", "e", "f")
12 | #' fct_expand(f, letters[1:6])
13 | fct_expand <- function(f, ...) {
14 | f <- check_factor(f)
15 |
16 | new_levels <- rlang::chr(...)
17 | lvls_expand(f, union(levels(f), new_levels))
18 | }
19 |
--------------------------------------------------------------------------------
/.travis.yml:
--------------------------------------------------------------------------------
1 | # R for travis: see documentation at https://docs.travis-ci.com/user/languages/r
2 |
3 | language: R
4 | cache: packages
5 |
6 | matrix:
7 | include:
8 | - r: devel
9 | - r: release
10 | after_success:
11 | - Rscript -e 'covr::codecov()'
12 | before_cache:
13 | - Rscript -e 'remotes::install_github("r-lib/pkgdown")'
14 | - Rscript -e 'remotes::install_github("tidyverse/tidytemplate")'
15 | deploy:
16 | provider: script
17 | script: Rscript -e 'pkgdown::deploy_site_github(verbose = TRUE)'
18 | skip_cleanup: true
19 | - r: oldrel
20 | - r: 3.4
21 | - r: 3.3
22 | - r: 3.2
23 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/issue_template.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Bug report or feature request
3 | about: Describe a bug you've seen or make a case for a new feature
4 | ---
5 |
6 | Please briefly describe your problem and what output you expect. If you have a question, please don't use this form. Instead, ask on or .
7 |
8 | Please include a minimal reproducible example (AKA a reprex). If you've never heard of a [reprex](http://reprex.tidyverse.org/) before, start by reading .
9 |
10 | Brief description of the problem
11 |
12 | ```r
13 | # insert reprex here
14 | ```
15 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_other.R:
--------------------------------------------------------------------------------
1 | context("fct_other")
2 |
3 | test_that("keeps levels in keep", {
4 | x1 <- factor(c("a", "b"))
5 | x2 <- fct_other(x1, keep = "a")
6 |
7 | expect_equal(levels(x2), c("a", "Other"))
8 | })
9 |
10 | test_that("drops levels in drop", {
11 | x1 <- factor(c("a", "b"))
12 | x2 <- fct_other(x1, drop = "a")
13 |
14 | # other always placed at end
15 | expect_equal(levels(x2), c("b", "Other"))
16 | })
17 |
18 | test_that("must supply exactly one of drop and keep", {
19 | f <- factor(c("a", "b"))
20 |
21 | expect_error(fct_other(f), "exactly one")
22 | expect_error(fct_other(f, keep = "a", drop = "a"), "exactly one")
23 | })
24 |
--------------------------------------------------------------------------------
/man/fct_anon.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/anon.R
3 | \name{fct_anon}
4 | \alias{fct_anon}
5 | \title{Anonymise factor levels}
6 | \usage{
7 | fct_anon(f, prefix = "")
8 | }
9 | \arguments{
10 | \item{f}{A factor.}
11 |
12 | \item{prefix}{A character prefix to insert in front of the random labels.}
13 | }
14 | \description{
15 | Replaces factor levels with arbitary numeric identifiers. Neither
16 | the values nor the order of the levels are preserved.
17 | }
18 | \examples{
19 | gss_cat$relig \%>\% fct_count()
20 | gss_cat$relig \%>\% fct_anon() \%>\% fct_count()
21 | gss_cat$relig \%>\% fct_anon("X") \%>\% fct_count()
22 | }
23 |
--------------------------------------------------------------------------------
/R/data.R:
--------------------------------------------------------------------------------
1 | #' A sample of categorical variables from the General Social survey
2 | #'
3 | #' @source Downloaded from \url{https://gssdataexplorer.norc.org/}.
4 | #' @format
5 | #' \describe{
6 | #' \item{year}{year of survey, 2000--2014}
7 | #' \item{age}{age. Maximum age truncated to 89.}
8 | #' \item{marital}{marital status}
9 | #' \item{race}{race}
10 | #' \item{rincome}{reported income}
11 | #' \item{partyid}{party affiliation}
12 | #' \item{relig}{religion}
13 | #' \item{denom}{denomination}
14 | #' \item{tvhours}{hours per day watching tv}
15 | #' }
16 | #' @examples
17 | #' gss_cat
18 | #'
19 | #' fct_count(gss_cat$relig)
20 | #' fct_count(fct_lump(gss_cat$relig))
21 | "gss_cat"
22 |
--------------------------------------------------------------------------------
/man/fct_explicit_na.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/explicit_na.R
3 | \name{fct_explicit_na}
4 | \alias{fct_explicit_na}
5 | \title{Make missing values explicit}
6 | \usage{
7 | fct_explicit_na(f, na_level = "(Missing)")
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{na_level}{Level to use for missing values.}
13 | }
14 | \description{
15 | This gives missing value an explicit factor level, ensuring that they
16 | appear in summaries and on plots.
17 | }
18 | \examples{
19 | f1 <- factor(c("a", "a", NA, NA, "a", "b", NA, "c", "a", "c", "b"))
20 | table(f1)
21 |
22 | f2 <- fct_explicit_na(f1)
23 | table(f2)
24 | }
25 |
--------------------------------------------------------------------------------
/cran-comments.md:
--------------------------------------------------------------------------------
1 | ## Test environments
2 |
3 | * local: darwin15.6.0-3.5.1
4 | * travis: 3.1, 3.2, 3.3, oldrel, release, devel
5 | * r-hub: windows-x86_64-devel, ubuntu-gcc-release, fedora-clang-devel
6 | * win-builder: windows-x86_64-devel
7 |
8 | ## R CMD check results
9 |
10 | 0 errors | 0 warnings | 0 notes
11 |
12 | ## revdepcheck results
13 |
14 | We checked 52 reverse dependencies (49 from CRAN + 3 from BioConductor), comparing R CMD check results across CRAN and dev versions of this package.
15 |
16 | * We saw 0 new problems
17 | * We failed to check 1 packages
18 |
19 | Issues with CRAN packages are summarised below.
20 |
21 | ### Failed to check
22 |
23 | * circumplex (failed to install)
24 |
--------------------------------------------------------------------------------
/man/fct_expand.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/expand.R
3 | \name{fct_expand}
4 | \alias{fct_expand}
5 | \title{Add additional levels to a factor}
6 | \usage{
7 | fct_expand(f, ...)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{...}{Additional levels to add to the factor. Levels that already
13 | exist will be silently ignored.}
14 | }
15 | \description{
16 | Add additional levels to a factor
17 | }
18 | \examples{
19 | f <- factor(sample(letters[1:3], 20, replace = TRUE))
20 | f
21 | fct_expand(f, "d", "e", "f")
22 | fct_expand(f, letters[1:6])
23 | }
24 | \seealso{
25 | \code{\link[=fct_drop]{fct_drop()}} to drop unused factor levels.
26 | }
27 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_drop.R:
--------------------------------------------------------------------------------
1 | context("fct_drop")
2 |
3 | test_that("doesn't add NA level", {
4 | f1 <- factor(c("a", NA), levels = c("a", "b"))
5 | f2 <- fct_drop(f1)
6 |
7 | expect_equal(levels(f2), "a")
8 | })
9 |
10 | test_that("can optionally restrict levels to drop", {
11 | f1 <- factor("a", levels = c("a", "b", "c"))
12 |
13 | expect_equal(levels(fct_drop(f1, only = "b")), c("a", "c"))
14 | expect_equal(levels(fct_drop(f1, only = "a")), c("a", "b", "c"))
15 | })
16 |
17 | test_that("preserves ordered class and attributes", {
18 | f1 <- ordered(letters[1:2], letters[1:3])
19 | attr(f1, "x") <- "test"
20 |
21 | f2 <- fct_drop(f1)
22 | expect_s3_class(f2, "ordered")
23 | expect_equal(attr(f2, "x"), attr(f1, "x"))
24 | })
25 |
--------------------------------------------------------------------------------
/man/fct_c.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/c.R
3 | \name{fct_c}
4 | \alias{fct_c}
5 | \title{Concatenate factors, combining levels}
6 | \usage{
7 | fct_c(...)
8 | }
9 | \arguments{
10 | \item{...}{Individual factors. Uses tidy dots, so you can splice in a list
11 | of factors with \verb{!!!}.}
12 | }
13 | \description{
14 | This is a useful way of patching together factors from multiple sources
15 | that really should have the same levels but don't.
16 | }
17 | \examples{
18 | fa <- factor("a")
19 | fb <- factor("b")
20 | fab <- factor(c("a", "b"))
21 |
22 | c(fa, fb, fab)
23 | fct_c(fa, fb, fab)
24 |
25 | # You can also pass a list of factors with !!!
26 | fs <- list(fa, fb, fab)
27 | fct_c(!!!fs)
28 | }
29 |
--------------------------------------------------------------------------------
/man/fct_shift.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/shift.R
3 | \name{fct_shift}
4 | \alias{fct_shift}
5 | \title{Shift factor levels to left or right, wrapping around at end}
6 | \usage{
7 | fct_shift(f, n = 1L)
8 | }
9 | \arguments{
10 | \item{f}{A factor.}
11 |
12 | \item{n}{Positive values shift to the left; negative values shift to
13 | the right.}
14 | }
15 | \description{
16 | This is useful when the levels of an ordered factor are actually cyclical,
17 | with different conventions on the starting point.
18 | }
19 | \examples{
20 | x <- factor(
21 | c("Mon", "Tue", "Wed"),
22 | levels = c("Sun", "Mon", "Tue", "Wed", "Thu", "Fri", "Sat"),
23 | ordered = TRUE
24 | )
25 | x
26 | fct_shift(x)
27 | fct_shift(x, 2)
28 | fct_shift(x, -1)
29 | }
30 |
--------------------------------------------------------------------------------
/R/anon.R:
--------------------------------------------------------------------------------
1 | #' Anonymise factor levels
2 | #'
3 | #' Replaces factor levels with arbitary numeric identifiers. Neither
4 | #' the values nor the order of the levels are preserved.
5 | #'
6 | #' @param f A factor.
7 | #' @param prefix A character prefix to insert in front of the random labels.
8 | #' @export
9 | #' @examples
10 | #' gss_cat$relig %>% fct_count()
11 | #' gss_cat$relig %>% fct_anon() %>% fct_count()
12 | #' gss_cat$relig %>% fct_anon("X") %>% fct_count()
13 | fct_anon <- function(f, prefix = "") {
14 | levels <- paste0(prefix, zero_pad(seq_len(nlevels(f))))
15 |
16 | f <- lvls_revalue(f, sample(levels))
17 | lvls_reorder(f, match(levels, levels(f)))
18 | }
19 |
20 | digits <- function(x) nchar(max(x, na.rm = TRUE))
21 |
22 | zero_pad <- function(x) {
23 | sprintf(paste0("%0", digits(x), "d"), x)
24 | }
25 |
--------------------------------------------------------------------------------
/tests/testthat/test-count.R:
--------------------------------------------------------------------------------
1 | context("test-count.R")
2 |
3 | test_that("0 count for empty levels", {
4 | f <- factor(levels = c("a", "b"))
5 | out <- fct_count(f)
6 |
7 | expect_equal(out$n, c(0, 0))
8 | })
9 |
10 |
11 | test_that("counts NA even when not in levels", {
12 | f <- factor(c("a", "a", NA))
13 | out <- fct_count(f)
14 |
15 | expect_equal(out$n, c(2, 1))
16 | })
17 |
18 | test_that("returns marginal table", {
19 | f <- factor(c("a", "a", "b"))
20 | out <- fct_count(f, prop = TRUE)
21 |
22 | expect_equal(out$n, c(2, 1))
23 | expect_equal(out$p, c(2, 1) / 3)
24 | })
25 |
26 | test_that("sort = TRUE brings most frequent values to top", {
27 | f <- factor(c("a", "b", "b"))
28 | out <- fct_count(f, sort = TRUE)
29 |
30 | expect_equal(out$f, factor(c("b", "a"), levels = c("a", "b")))
31 | })
32 |
--------------------------------------------------------------------------------
/R/utils.R:
--------------------------------------------------------------------------------
1 | #' Pipe operator
2 | #'
3 | #' See \code{\link[magrittr]{\%>\%}} for more details.
4 | #'
5 | #' @name %>%
6 | #' @rdname pipe
7 | #' @keywords internal
8 | #' @export
9 | #' @importFrom magrittr %>%
10 | #' @usage lhs \%>\% rhs
11 | NULL
12 |
13 |
14 | check_factor <- function(f) {
15 | if (is.character(f)) {
16 | factor(f)
17 | } else if (is.factor(f)) {
18 | f
19 | } else {
20 | stop("`f` must be a factor (or character vector).", call. = FALSE)
21 | }
22 | }
23 |
24 | check_factor_list <- function(fs, arg_name = "fs") {
25 | if (!is.list(fs)) {
26 | stop("`fs` must be a list", call. = FALSE)
27 | }
28 |
29 | is_factor <- vapply(fs, is.factor, logical(1))
30 | if (any(!is_factor)) {
31 | stop("All elements of `", arg_name, "` must be factors", call. = FALSE)
32 | }
33 |
34 | fs
35 | }
36 |
--------------------------------------------------------------------------------
/man/fct_count.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/count.R
3 | \name{fct_count}
4 | \alias{fct_count}
5 | \title{Count entries in a factor}
6 | \usage{
7 | fct_count(f, sort = FALSE, prop = FALSE)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{sort}{If \code{TRUE}, sort the result so that the most common values
13 | float to the top.}
14 |
15 | \item{prop}{If \code{TRUE}, compute the fraction of marginal table.}
16 | }
17 | \value{
18 | A tibble with columns \code{f}, \code{n} and \code{p}, if prop is \code{TRUE}.
19 | }
20 | \description{
21 | Count entries in a factor
22 | }
23 | \examples{
24 | f <- factor(sample(letters)[rpois(1000, 10)])
25 | table(f)
26 | fct_count(f)
27 | fct_count(f, sort = TRUE)
28 | fct_count(f, sort = TRUE, prop = TRUE)
29 | }
30 |
--------------------------------------------------------------------------------
/R/shift.R:
--------------------------------------------------------------------------------
1 | #' Shift factor levels to left or right, wrapping around at end
2 | #'
3 | #' This is useful when the levels of an ordered factor are actually cyclical,
4 | #' with different conventions on the starting point.
5 | #'
6 | #' @param f A factor.
7 | #' @param n Positive values shift to the left; negative values shift to
8 | #' the right.
9 | #' @export
10 | #' @examples
11 | #' x <- factor(
12 | #' c("Mon", "Tue", "Wed"),
13 | #' levels = c("Sun", "Mon", "Tue", "Wed", "Thu", "Fri", "Sat"),
14 | #' ordered = TRUE
15 | #' )
16 | #' x
17 | #' fct_shift(x)
18 | #' fct_shift(x, 2)
19 | #' fct_shift(x, -1)
20 | fct_shift <- function(f, n = 1L) {
21 | lvls_reorder(f, shift(nlevels(f), n))
22 | }
23 |
24 | shift <- function(m, n) {
25 | stopifnot(is.numeric(m), length(m) == 1L)
26 | stopifnot(is.numeric(n), length(n) == 1L)
27 |
28 | ((seq_len(m) - 1) + n) %% m + 1
29 | }
30 |
--------------------------------------------------------------------------------
/man/fct_drop.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/drop.R
3 | \name{fct_drop}
4 | \alias{fct_drop}
5 | \title{Drop unused levels}
6 | \usage{
7 | fct_drop(f, only)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{only}{A character vector restricting the set of levels to be dropped.
13 | If supplied, only levels that have no entries and appear in this vector
14 | will be removed.}
15 | }
16 | \description{
17 | Compared to \code{base::droplevels()}, does not drop \code{NA} levels that have values.
18 | }
19 | \examples{
20 | f <- factor(c("a", "b"), levels = c("a", "b", "c"))
21 | f
22 | fct_drop(f)
23 |
24 | # Set only to restrict which levels to drop
25 | fct_drop(f, only = "a")
26 | fct_drop(f, only = "c")
27 | }
28 | \seealso{
29 | \code{\link[=fct_expand]{fct_expand()}} to add additional levels to a factor.
30 | }
31 |
--------------------------------------------------------------------------------
/man/fct_inorder.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/reorder.R
3 | \name{fct_inorder}
4 | \alias{fct_inorder}
5 | \alias{fct_infreq}
6 | \alias{fct_inseq}
7 | \title{Reorder factors levels by first appearance, frequency, or numeric order.}
8 | \usage{
9 | fct_inorder(f, ordered = NA)
10 |
11 | fct_infreq(f, ordered = NA)
12 |
13 | fct_inseq(f, ordered = NA)
14 | }
15 | \arguments{
16 | \item{f}{A factor}
17 |
18 | \item{ordered}{A logical which determines the "ordered" status of the
19 | output factor. \code{NA} preserves the existing status of the factor.}
20 | }
21 | \description{
22 | Reorder factors levels by first appearance, frequency, or numeric order.
23 | }
24 | \examples{
25 | f <- factor(c("b", "b", "a", "c", "c", "c"))
26 | f
27 | fct_inorder(f)
28 | fct_infreq(f)
29 |
30 | fct_inorder(f, ordered = TRUE)
31 |
32 | f <- factor(sample(1:10))
33 | fct_inseq(f)
34 | }
35 |
--------------------------------------------------------------------------------
/R/explicit_na.R:
--------------------------------------------------------------------------------
1 | #' Make missing values explicit
2 | #'
3 | #' This gives missing value an explicit factor level, ensuring that they
4 | #' appear in summaries and on plots.
5 | #'
6 | #' @param f A factor (or character vector).
7 | #' @param na_level Level to use for missing values.
8 | #' @export
9 | #' @examples
10 | #' f1 <- factor(c("a", "a", NA, NA, "a", "b", NA, "c", "a", "c", "b"))
11 | #' table(f1)
12 | #'
13 | #' f2 <- fct_explicit_na(f1)
14 | #' table(f2)
15 | fct_explicit_na <- function(f, na_level = "(Missing)") {
16 | f <- check_factor(f)
17 |
18 | is_missing <- is.na(f)
19 | is_missing_level <- is.na(levels(f))
20 |
21 | if (any(is_missing)) {
22 | f <- fct_expand(f, na_level)
23 | f[is_missing] <- na_level
24 |
25 | f
26 | } else if (any(is_missing_level)) {
27 | levs <- levels(f)
28 | levs[is.na(levs)] <- na_level
29 |
30 | lvls_revalue(f, levs)
31 | } else {
32 | f
33 | }
34 | }
35 |
--------------------------------------------------------------------------------
/man/gss_cat.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/data.R
3 | \docType{data}
4 | \name{gss_cat}
5 | \alias{gss_cat}
6 | \title{A sample of categorical variables from the General Social survey}
7 | \format{\describe{
8 | \item{year}{year of survey, 2000--2014}
9 | \item{age}{age. Maximum age truncated to 89.}
10 | \item{marital}{marital status}
11 | \item{race}{race}
12 | \item{rincome}{reported income}
13 | \item{partyid}{party affiliation}
14 | \item{relig}{religion}
15 | \item{denom}{denomination}
16 | \item{tvhours}{hours per day watching tv}
17 | }}
18 | \source{
19 | Downloaded from \url{https://gssdataexplorer.norc.org/}.
20 | }
21 | \usage{
22 | gss_cat
23 | }
24 | \description{
25 | A sample of categorical variables from the General Social survey
26 | }
27 | \examples{
28 | gss_cat
29 |
30 | fct_count(gss_cat$relig)
31 | fct_count(fct_lump(gss_cat$relig))
32 | }
33 | \keyword{datasets}
34 |
--------------------------------------------------------------------------------
/R/count.R:
--------------------------------------------------------------------------------
1 | #' Count entries in a factor
2 | #'
3 | #' @param f A factor (or character vector).
4 | #' @param sort If `TRUE`, sort the result so that the most common values
5 | #' float to the top.
6 | #' @param prop If `TRUE`, compute the fraction of marginal table.
7 | #' @return A tibble with columns `f`, `n` and `p`, if prop is `TRUE`.
8 | #' @export
9 | #' @examples
10 | #' f <- factor(sample(letters)[rpois(1000, 10)])
11 | #' table(f)
12 | #' fct_count(f)
13 | #' fct_count(f, sort = TRUE)
14 | #' fct_count(f, sort = TRUE, prop = TRUE)
15 | fct_count <- function(f, sort = FALSE, prop = FALSE) {
16 | f <- check_factor(f)
17 | f2 <- addNA(f, ifany = TRUE)
18 |
19 | df <- tibble::tibble(
20 | f = fct_unique(f2),
21 | n = as.integer(table(f2))
22 | )
23 |
24 | if (sort) {
25 | df <- df[order(df$n, decreasing = TRUE), ]
26 | }
27 |
28 | if (prop) {
29 | df$p <- prop.table(df$n)
30 | }
31 |
32 | df
33 | }
34 |
--------------------------------------------------------------------------------
/NAMESPACE:
--------------------------------------------------------------------------------
1 | # Generated by roxygen2: do not edit by hand
2 |
3 | S3method(as_factor,character)
4 | S3method(as_factor,factor)
5 | S3method(as_factor,numeric)
6 | export("%>%")
7 | export(as_factor)
8 | export(fct_anon)
9 | export(fct_c)
10 | export(fct_collapse)
11 | export(fct_count)
12 | export(fct_cross)
13 | export(fct_drop)
14 | export(fct_expand)
15 | export(fct_explicit_na)
16 | export(fct_infreq)
17 | export(fct_inorder)
18 | export(fct_inseq)
19 | export(fct_lump)
20 | export(fct_lump_min)
21 | export(fct_match)
22 | export(fct_other)
23 | export(fct_recode)
24 | export(fct_relabel)
25 | export(fct_relevel)
26 | export(fct_reorder)
27 | export(fct_reorder2)
28 | export(fct_rev)
29 | export(fct_shift)
30 | export(fct_shuffle)
31 | export(fct_unify)
32 | export(fct_unique)
33 | export(first2)
34 | export(last2)
35 | export(lvls_expand)
36 | export(lvls_reorder)
37 | export(lvls_revalue)
38 | export(lvls_union)
39 | importFrom(magrittr,"%>%")
40 | importFrom(stats,median)
41 |
--------------------------------------------------------------------------------
/R/drop.R:
--------------------------------------------------------------------------------
1 | #' Drop unused levels
2 | #'
3 | #' Compared to `base::droplevels()`, does not drop `NA` levels that have values.
4 | #'
5 | #' @param f A factor (or character vector).
6 | #' @param only A character vector restricting the set of levels to be dropped.
7 | #' If supplied, only levels that have no entries and appear in this vector
8 | #' will be removed.
9 | #' @export
10 | #' @seealso [fct_expand()] to add additional levels to a factor.
11 | #' @examples
12 | #' f <- factor(c("a", "b"), levels = c("a", "b", "c"))
13 | #' f
14 | #' fct_drop(f)
15 | #'
16 | #' # Set only to restrict which levels to drop
17 | #' fct_drop(f, only = "a")
18 | #' fct_drop(f, only = "c")
19 | fct_drop <- function(f, only) {
20 | f <- check_factor(f)
21 |
22 | levels <- levels(f)
23 | count <- table(f)
24 |
25 | to_drop <- levels[count == 0]
26 | if (!missing(only)) {
27 | to_drop <- intersect(to_drop, only)
28 | }
29 |
30 | refactor(f, new_levels = setdiff(levels, to_drop))
31 | }
32 |
--------------------------------------------------------------------------------
/man/fct_collapse.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/collapse.R
3 | \name{fct_collapse}
4 | \alias{fct_collapse}
5 | \title{Collapse factor levels into manually defined groups}
6 | \usage{
7 | fct_collapse(.f, ..., group_other = FALSE)
8 | }
9 | \arguments{
10 | \item{.f}{A factor (or character vector).}
11 |
12 | \item{...}{A series of named character vectors. The levels in
13 | each vector will be replaced with the name.}
14 |
15 | \item{group_other}{Replace all levels not named in \code{...} with "Other"?}
16 | }
17 | \description{
18 | Collapse factor levels into manually defined groups
19 | }
20 | \examples{
21 | fct_count(gss_cat$partyid)
22 |
23 | partyid2 <- fct_collapse(gss_cat$partyid,
24 | missing = c("No answer", "Don't know"),
25 | other = "Other party",
26 | rep = c("Strong republican", "Not str republican"),
27 | ind = c("Ind,near rep", "Independent", "Ind,near dem"),
28 | dem = c("Not str democrat", "Strong democrat")
29 | )
30 | fct_count(partyid2)
31 | }
32 |
--------------------------------------------------------------------------------
/man/fct_cross.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/fct_cross.R
3 | \name{fct_cross}
4 | \alias{fct_cross}
5 | \title{Combine levels from two or more factors to create a new factor}
6 | \usage{
7 | fct_cross(.f, ..., sep = ":", keep_empty = FALSE)
8 | }
9 | \arguments{
10 | \item{.f}{A factor or character vector}
11 |
12 | \item{...}{Additional factors or character vectors}
13 |
14 | \item{sep}{A character string to separate the levels}
15 |
16 | \item{keep_empty}{If TRUE, keep combinations with no observations as levels}
17 | }
18 | \value{
19 | The new factor
20 | }
21 | \description{
22 | Computes a factor whose levels are all the combinations of the levels of the input factors.
23 | }
24 | \examples{
25 | fruit <- factor(c("apple", "kiwi", "apple", "apple"))
26 | colour <- factor(c("green", "green", "red", "green"))
27 | eaten <- c("yes", "no", "yes", "no")
28 | fct_cross(fruit, colour)
29 | fct_cross(fruit, colour, eaten)
30 | fct_cross(fruit, colour, keep_empty = TRUE)
31 | }
32 |
--------------------------------------------------------------------------------
/man/fct_match.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/match.R
3 | \name{fct_match}
4 | \alias{fct_match}
5 | \title{Test for presence of levels in a factor}
6 | \usage{
7 | fct_match(f, lvls)
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{lvls}{A character vector specifying levels to look for.}
13 | }
14 | \value{
15 | A logical vector
16 | }
17 | \description{
18 | Do any of \code{lvls} occur in \code{f}? Compared to \link{\%in\%}, this function validates
19 | \code{lvls} to ensure that they're actually present in \code{f}. In other words,
20 | \code{x \%in\% "not present"} will return \code{FALSE}, but \code{fct_match(x, "not present")}
21 | will throw an error.
22 | }
23 | \examples{
24 | table(fct_match(gss_cat$marital, c("Married", "Divorced")))
25 |
26 | # Compare to \%in\%, misspelled levels throw an error
27 | table(gss_cat$marital \%in\% c("Maried", "Davorced"))
28 | \dontrun{
29 | table(fct_match(gss_cat$marital, c("Maried", "Davorced")))
30 | }
31 |
32 | }
33 |
--------------------------------------------------------------------------------
/man/fct_other.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/other.R
3 | \name{fct_other}
4 | \alias{fct_other}
5 | \title{Replace levels with "other"}
6 | \usage{
7 | fct_other(f, keep, drop, other_level = "Other")
8 | }
9 | \arguments{
10 | \item{f}{A factor (or character vector).}
11 |
12 | \item{keep, drop}{Pick one of \code{keep} and \code{drop}:
13 | \itemize{
14 | \item \code{keep} will preserve listed levels, replacing all others with
15 | \code{other_level}.
16 | \item \code{drop} will replace listed levels with \code{other_level}, keeping all
17 | as is.
18 | }}
19 |
20 | \item{other_level}{Value of level used for "other" values. Always
21 | placed at end of levels.}
22 | }
23 | \description{
24 | Replace levels with "other"
25 | }
26 | \examples{
27 | x <- factor(rep(LETTERS[1:9], times = c(40, 10, 5, 27, 1, 1, 1, 1, 1)))
28 |
29 | fct_other(x, keep = c("A", "B"))
30 | fct_other(x, drop = c("A", "B"))
31 | }
32 | \seealso{
33 | \code{\link[=fct_lump]{fct_lump()}} to automatically convert the rarest (or most
34 | common) levels to "other".
35 | }
36 |
--------------------------------------------------------------------------------
/R/other.R:
--------------------------------------------------------------------------------
1 | #' Replace levels with "other"
2 | #'
3 | #' @inheritParams fct_lump
4 | #' @param keep,drop Pick one of `keep` and `drop`:
5 | #' * `keep` will preserve listed levels, replacing all others with
6 | #' `other_level`.
7 | #' * `drop` will replace listed levels with `other_level`, keeping all
8 | #' as is.
9 | #' @seealso [fct_lump()] to automatically convert the rarest (or most
10 | #' common) levels to "other".
11 | #' @export
12 | #' @examples
13 | #' x <- factor(rep(LETTERS[1:9], times = c(40, 10, 5, 27, 1, 1, 1, 1, 1)))
14 | #'
15 | #' fct_other(x, keep = c("A", "B"))
16 | #' fct_other(x, drop = c("A", "B"))
17 | fct_other <- function(f, keep, drop, other_level = "Other") {
18 | f <- check_factor(f)
19 |
20 | if (!xor(missing(keep), missing(drop))) {
21 | stop("Must supply exactly one of `keep` and `drop`", call. = FALSE)
22 | }
23 |
24 | levels <- levels(f)
25 | if (!missing(keep)) {
26 | levels[!levels %in% keep] <- other_level
27 | } else {
28 | levels[levels %in% drop] <- other_level
29 | }
30 |
31 | f <- lvls_revalue(f, levels)
32 | fct_relevel(f, other_level, after = Inf)
33 | }
34 |
--------------------------------------------------------------------------------
/man/forcats-package.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/forcats-package.R
3 | \docType{package}
4 | \name{forcats-package}
5 | \alias{forcats}
6 | \alias{forcats-package}
7 | \title{forcats: Tools for Working with Categorical Variables (Factors)}
8 | \description{
9 | \if{html}{\figure{logo.png}{options: align='right' alt='logo' width='120'}}
10 |
11 | Helpers for reordering factor levels (including
12 | moving specified levels to front, ordering by first appearance,
13 | reversing, and randomly shuffling), and tools for modifying factor
14 | levels (including collapsing rare levels into other, 'anonymising',
15 | and manually 'recoding').
16 | }
17 | \seealso{
18 | Useful links:
19 | \itemize{
20 | \item \url{http://forcats.tidyverse.org}
21 | \item \url{https://github.com/tidyverse/forcats}
22 | \item Report bugs at \url{https://github.com/tidyverse/forcats/issues}
23 | }
24 |
25 | }
26 | \author{
27 | \strong{Maintainer}: Hadley Wickham \email{hadley@rstudio.com}
28 |
29 | Other contributors:
30 | \itemize{
31 | \item RStudio [copyright holder, funder]
32 | }
33 |
34 | }
35 | \keyword{internal}
36 |
--------------------------------------------------------------------------------
/man/fct_recode.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/recode.R
3 | \name{fct_recode}
4 | \alias{fct_recode}
5 | \title{Change factor levels by hand}
6 | \usage{
7 | fct_recode(.f, ...)
8 | }
9 | \arguments{
10 | \item{.f}{A factor (or character vector).}
11 |
12 | \item{...}{A sequence of named character vectors where the
13 | name gives the new level, and the value gives the old level.
14 | Levels not otherwise mentioned will be left as is. Levels can
15 | be removed by naming them \code{NULL}. Uses tidy dots.}
16 | }
17 | \description{
18 | Change factor levels by hand
19 | }
20 | \examples{
21 | x <- factor(c("apple", "bear", "banana", "dear"))
22 | fct_recode(x, fruit = "apple", fruit = "banana")
23 |
24 | # If you make a mistake you'll get a warning
25 | fct_recode(x, fruit = "apple", fruit = "bananana")
26 |
27 | # If you name the level NULL it will be removed
28 | fct_recode(x, NULL = "apple", fruit = "banana")
29 |
30 | # When passing a named vector to rename levels use !!! to splice
31 | x <- factor(c("apple", "bear", "banana", "dear"))
32 | levels <- c(fruit = "apple", fruit = "banana")
33 | fct_recode(x, !!!levels)
34 | }
35 |
--------------------------------------------------------------------------------
/tests/testthat/test-relevel.R:
--------------------------------------------------------------------------------
1 | context("fct_relevel")
2 |
3 | test_that("warns about unknown levels", {
4 | f1 <- factor(c("a", "b"))
5 |
6 | expect_warning(f2 <- fct_relevel(f1, "d"), "Unknown levels")
7 | expect_equal(levels(f2), levels(f1))
8 | })
9 |
10 | test_that("moves supplied levels to front", {
11 | f1 <- factor(c("a", "b", "c", "d"))
12 |
13 | f2 <- fct_relevel(f1, "c", "b")
14 | expect_equal(levels(f2), c("c", "b", "a", "d"))
15 | })
16 |
17 | test_that("can moves supplied levels to end", {
18 | f1 <- factor(c("a", "b", "c", "d"))
19 |
20 | f2 <- fct_relevel(f1, "a", "b", after = 2)
21 | f3 <- fct_relevel(f1, "a", "b", after = Inf)
22 | expect_equal(levels(f2), c("c", "d", "a", "b"))
23 | expect_equal(levels(f3), c("c", "d", "a", "b"))
24 | })
25 |
26 | test_that("can relevel with function ", {
27 | f1 <- fct_rev(factor(c("a", "b")))
28 | f2a <- fct_relevel(f1, rev)
29 | f2b <- fct_relevel(f1, ~ rev(.))
30 |
31 | expect_equal(levels(f2a), c("a", "b"))
32 | expect_equal(levels(f2b), c("a", "b"))
33 | })
34 |
35 | test_that("function must return character vector", {
36 | f <- factor(c("a", "b"))
37 | expect_error(fct_relevel(f, ~ 1), "character vector")
38 | })
39 |
--------------------------------------------------------------------------------
/DESCRIPTION:
--------------------------------------------------------------------------------
1 | Package: forcats
2 | Title: Tools for Working with Categorical Variables (Factors)
3 | Version: 0.4.0.9000
4 | Authors@R:
5 | c(person(given = "Hadley",
6 | family = "Wickham",
7 | role = c("aut", "cre"),
8 | email = "hadley@rstudio.com"),
9 | person(given = "RStudio",
10 | role = c("cph", "fnd")))
11 | Description: Helpers for reordering factor levels (including
12 | moving specified levels to front, ordering by first appearance,
13 | reversing, and randomly shuffling), and tools for modifying factor
14 | levels (including collapsing rare levels into other, 'anonymising',
15 | and manually 'recoding').
16 | License: GPL-3
17 | URL: http://forcats.tidyverse.org,
18 | https://github.com/tidyverse/forcats
19 | BugReports: https://github.com/tidyverse/forcats/issues
20 | Depends:
21 | R (>= 3.2)
22 | Imports:
23 | ellipsis,
24 | magrittr,
25 | rlang,
26 | tibble
27 | Suggests:
28 | covr,
29 | ggplot2,
30 | testthat,
31 | readr,
32 | knitr,
33 | rmarkdown,
34 | dplyr
35 | Encoding: UTF-8
36 | LazyData: true
37 | Roxygen: list(markdown = TRUE)
38 | RoxygenNote: 7.0.2
39 | VignetteBuilder: knitr
40 |
--------------------------------------------------------------------------------
/R/match.R:
--------------------------------------------------------------------------------
1 | #' Test for presence of levels in a factor
2 | #'
3 | #' Do any of `lvls` occur in `f`? Compared to [%in%], this function validates
4 | #' `lvls` to ensure that they're actually present in `f`. In other words,
5 | #' `x %in% "not present"` will return `FALSE`, but `fct_match(x, "not present")`
6 | #' will throw an error.
7 | #'
8 | #' @rdname fct_match
9 | #' @param f A factor (or character vector).
10 | #' @param lvls A character vector specifying levels to look for.
11 | #' @return A logical vector
12 | #' @export
13 | #' @examples
14 | #' table(fct_match(gss_cat$marital, c("Married", "Divorced")))
15 | #'
16 | #' # Compare to %in%, misspelled levels throw an error
17 | #' table(gss_cat$marital %in% c("Maried", "Davorced"))
18 | #' \dontrun{
19 | #' table(fct_match(gss_cat$marital, c("Maried", "Davorced")))
20 | #' }
21 | #'
22 | fct_match <- function(f, lvls) {
23 | f <- check_factor(f)
24 |
25 | bad_lvls <- setdiff(lvls, levels(f))
26 | bad_lvls <- bad_lvls[!is.na(bad_lvls)]
27 | if (length(bad_lvls) > 0) {
28 | stop(
29 | "Levels not present in factor: ",
30 | paste0(encodeString(bad_lvls, quote = '"'), collapse = ", "),
31 | call. = FALSE
32 | )
33 | }
34 |
35 | f %in% lvls
36 | }
37 |
--------------------------------------------------------------------------------
/tests/testthat/test-lvls_reorder.R:
--------------------------------------------------------------------------------
1 | context("lvls_reorder")
2 |
3 | test_that("changes levels, not values", {
4 | f1 <- factor(c("a", "b"))
5 | f2 <- factor(c("a", "b"), levels = c("b", "a"))
6 |
7 | expect_equal(lvls_reorder(f1, 2:1), f2)
8 | })
9 |
10 | test_that("idx must be numeric", {
11 | f <- factor(c("a", "b"))
12 | expect_error(lvls_reorder(f, "a"), "must be numeric")
13 | })
14 |
15 | test_that("must have one integer per level", {
16 | f <- factor(c("a", "b", "c"))
17 |
18 | expect_error(lvls_reorder(f, c(1, 2)), "one integer for each level")
19 | expect_error(lvls_reorder(f, c(1, 2, 2)), "one integer for each level")
20 | expect_error(lvls_reorder(f, c(1, 2.5)), "one integer for each level")
21 | })
22 |
23 | test_that("can change ordered status of output", {
24 | f1 <- factor(letters[1:3])
25 | f2 <- ordered(f1)
26 |
27 | expect_equal(is.ordered(lvls_reorder(f1, 1:3)), FALSE)
28 | expect_equal(is.ordered(lvls_reorder(f1, 1:3, ordered = FALSE)), FALSE)
29 | expect_equal(is.ordered(lvls_reorder(f1, 1:3, ordered = TRUE)), TRUE)
30 |
31 | expect_equal(is.ordered(lvls_reorder(f2, 1:3)), TRUE)
32 | expect_equal(is.ordered(lvls_reorder(f2, 1:3, ordered = FALSE)), FALSE)
33 | expect_equal(is.ordered(lvls_reorder(f2, 1:3, ordered = TRUE)), TRUE)
34 | })
35 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_cross.R:
--------------------------------------------------------------------------------
1 | context("fct_cross")
2 |
3 | test_that("gives correct levels", {
4 | fruit <- as_factor(c("apple", "kiwi", "apple", "apple"))
5 | colour <- as_factor(c("green", "green", "red", "green"))
6 | f2 <- fct_cross(fruit, colour)
7 |
8 | expect_setequal(levels(f2), c("apple:green", "kiwi:green", "apple:red"))
9 | })
10 |
11 | test_that("keeps empty levels when requested", {
12 | fruit <- as_factor(c("apple", "kiwi", "apple", "apple"))
13 | colour <- as_factor(c("green", "green", "red", "green"))
14 | f2 <- fct_cross(fruit, colour, keep_empty = TRUE)
15 |
16 | expect_setequal(levels(f2), c("apple:green", "kiwi:green", "apple:red", "kiwi:red"))
17 | })
18 |
19 | test_that("gives NA output on NA input", {
20 | fruit <- as_factor(c("apple", "kiwi", "apple", "apple"))
21 | colour <- as_factor(c("green", "green", "red", "green"))
22 | fruit[1] <- NA
23 | f2 <- fct_cross(fruit, colour)
24 |
25 | expect_true(is.na(f2[1]))
26 | })
27 |
28 |
29 | test_that("gives NA output on NA input, when keeping empty levels", {
30 | fruit <- as_factor(c("apple", "kiwi", "apple", "apple"))
31 | colour <- as_factor(c("green", "green", "red", "green"))
32 | fruit[1] <- NA
33 | f2 <- fct_cross(fruit, colour, keep_empty = TRUE)
34 |
35 | expect_true(is.na(f2[1]))
36 | })
37 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_recode.R:
--------------------------------------------------------------------------------
1 | context("fct_recode")
2 |
3 | test_that("warns about unknown levels", {
4 | f1 <- factor(c("a", "b"))
5 |
6 | expect_warning(f2 <- fct_recode(f1, d = "e"), "Unknown levels")
7 | expect_equal(levels(f2), levels(f1))
8 | })
9 |
10 | test_that("can collapse levels", {
11 | f1 <- factor(c("a1", "a2", "b1", "b2"))
12 | f2 <- factor(c("a", "a", "b", "b"))
13 |
14 | expect_equal(fct_recode(f1, a = "a1", a = "a2", b = "b1", b = "b2"), f2)
15 | })
16 |
17 | test_that("can recode multiple levels to NA", {
18 | f1 <- factor(c("a1", "empty", "a2", "b", "missing"))
19 | f2 <- factor(c("a", NA, "a", "b", NA))
20 |
21 | expect_equal(fct_recode(f1, NULL = "missing", NULL = "empty", a = "a1", a = "a2"), f2)
22 | })
23 |
24 | test_that("can just remove levels", {
25 | f1 <- factor(c("a", "missing"))
26 | f2 <- factor(c("a", NA))
27 |
28 | expect_equal(fct_recode(f1, NULL = "missing"), f2)
29 | })
30 |
31 |
32 | # check_recode_levels -----------------------------------------------------
33 |
34 | test_that("new levels must be character", {
35 | expect_error(check_recode_levels(a = 1), "Problems at positions: 1")
36 | })
37 |
38 | test_that("new levels must be length 1", {
39 | expect_error(check_recode_levels(a = c("a", "b")), "Problems at positions: 1")
40 | })
41 |
--------------------------------------------------------------------------------
/man/as_factor.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/as_factor.R
3 | \name{as_factor}
4 | \alias{as_factor}
5 | \alias{as_factor.factor}
6 | \alias{as_factor.character}
7 | \alias{as_factor.numeric}
8 | \title{Convert input to a factor.}
9 | \usage{
10 | as_factor(x, ...)
11 |
12 | \method{as_factor}{factor}(x, ...)
13 |
14 | \method{as_factor}{character}(x, ...)
15 |
16 | \method{as_factor}{numeric}(x, ...)
17 | }
18 | \arguments{
19 | \item{x}{Object to coerce to a factor.}
20 |
21 | \item{...}{Other arguments passed down to method.}
22 | }
23 | \description{
24 | Compared to base R, when \code{x} is a character, this function creates
25 | levels in the order in which they appear, which will be the same on every
26 | platform. (Base R sorts in the current locale which can vary from place
27 | to place.) When \code{x} is numeric, the ordering is based on the numeric
28 | value and consistent with base R.
29 | }
30 | \details{
31 | This is a generic function.
32 | }
33 | \examples{
34 | # Character object
35 | x <- c("a", "z", "g")
36 | as_factor(x)
37 | as.factor(x)
38 |
39 | # Character object containing numbers
40 | y <- c("1.1", "11", "2.2", "22")
41 | as_factor(y)
42 | as.factor(y)
43 |
44 | # Numeric object
45 | z <- as.numeric(y)
46 | as_factor(z)
47 | as.factor(z)
48 | }
49 |
--------------------------------------------------------------------------------
/R/collapse.R:
--------------------------------------------------------------------------------
1 | #' Collapse factor levels into manually defined groups
2 | #'
3 | #' @param .f A factor (or character vector).
4 | #' @param ... A series of named character vectors. The levels in
5 | #' each vector will be replaced with the name.
6 | #' @param group_other Replace all levels not named in `...` with "Other"?
7 | #' @export
8 | #' @examples
9 | #' fct_count(gss_cat$partyid)
10 | #'
11 | #' partyid2 <- fct_collapse(gss_cat$partyid,
12 | #' missing = c("No answer", "Don't know"),
13 | #' other = "Other party",
14 | #' rep = c("Strong republican", "Not str republican"),
15 | #' ind = c("Ind,near rep", "Independent", "Ind,near dem"),
16 | #' dem = c("Not str democrat", "Strong democrat")
17 | #' )
18 | #' fct_count(partyid2)
19 | fct_collapse <- function(.f, ..., group_other = FALSE) {
20 | new <- rlang::dots_list(...)
21 | levs <- as.list(unlist(new, use.names = FALSE))
22 |
23 | if (group_other) {
24 | f <- check_factor(.f)
25 | levels <- levels(f)
26 | new[["Other"]] <- levels[!levels %in% levs]
27 | levs <- c(levs, new[["Other"]])
28 | }
29 |
30 | names(levs) <- names(new)[rep(seq_along(new), vapply(new, length, integer(1)))]
31 | out <- fct_recode(.f, !!!levs)
32 |
33 | if (group_other) {
34 | fct_relevel(out, "Other", after = Inf)
35 | } else {
36 | out
37 | }
38 | }
39 |
--------------------------------------------------------------------------------
/R/relabel.R:
--------------------------------------------------------------------------------
1 | #' Automatically relabel factor levels, collapse as necessary
2 | #'
3 | #' @param .f A factor (or character vector).
4 | #' @param .fun A function to be applied to each level. Must accept one
5 | #' character argument and return a character vector of the same length as
6 | #' its input.
7 | #'
8 | #' You can also use `~` to create as shorthand (in the style of purrr).
9 | #' `~ paste(., "x")` is equivalent to `function(.) paste(., "x")`
10 | #' @param ... Additional arguments to `fun`.
11 | #' @export
12 | #' @examples
13 | #' gss_cat$partyid %>% fct_count()
14 | #' gss_cat$partyid %>% fct_relabel(~ gsub(",", ", ", .x)) %>% fct_count()
15 | #'
16 | #' convert_income <- function(x) {
17 | #' regex <- "^(?:Lt |)[$]([0-9]+).*$"
18 | #' is_range <- grepl(regex, x)
19 | #' num_income <- as.numeric(gsub(regex, "\\1", x[is_range]))
20 | #' num_income <- trunc(num_income / 5000) * 5000
21 | #' x[is_range] <- paste0("Gt $", num_income)
22 | #' x
23 | #' }
24 | #' fct_count(gss_cat$rincome)
25 | #' convert_income(levels(gss_cat$rincome))
26 | #' rincome2 <- fct_relabel(gss_cat$rincome, convert_income)
27 | #' fct_count(rincome2)
28 | fct_relabel <- function(.f, .fun, ...) {
29 | f <- check_factor(.f)
30 | .fun <- rlang::as_function(.fun)
31 |
32 | old_levels <- levels(f)
33 | new_levels <- .fun(old_levels, ...)
34 |
35 | lvls_revalue(f, new_levels)
36 | }
37 |
--------------------------------------------------------------------------------
/R/c.R:
--------------------------------------------------------------------------------
1 | #' Concatenate factors, combining levels
2 | #'
3 | #' This is a useful way of patching together factors from multiple sources
4 | #' that really should have the same levels but don't.
5 | #'
6 | #' @param ... Individual factors. Uses tidy dots, so you can splice in a list
7 | #' of factors with `!!!`.
8 | #' @export
9 | #' @examples
10 | #' fa <- factor("a")
11 | #' fb <- factor("b")
12 | #' fab <- factor(c("a", "b"))
13 | #'
14 | #' c(fa, fb, fab)
15 | #' fct_c(fa, fb, fab)
16 | #'
17 | #' # You can also pass a list of factors with !!!
18 | #' fs <- list(fa, fb, fab)
19 | #' fct_c(!!!fs)
20 | fct_c <- function(...) {
21 | fs <- rlang::dots_list(...)
22 | fs <- check_factor_list(fs, "...")
23 |
24 | if (length(fs) == 0) {
25 | return(factor())
26 | }
27 |
28 | levels <- lvls_union(fs)
29 | all <- unlist(fct_unify(fs, levels), use.names = FALSE)
30 | factor(all, levels = levels, exclude = NULL)
31 | }
32 |
33 | #' Unify the levels in a list of factors
34 | #'
35 | #' @param fs A list of factors
36 | #' @param levels Set of levels to apply to every factor. Default to
37 | #' union of all factor levels
38 | #' @export
39 | #' @examples
40 | #' fs <- list(factor("a"), factor("b"), factor(c("a", "b")))
41 | #' fct_unify(fs)
42 | fct_unify <- function(fs, levels = lvls_union(fs)) {
43 | fs <- check_factor_list(fs)
44 |
45 | lapply(fs, lvls_expand, new_levels = levels)
46 | }
47 |
--------------------------------------------------------------------------------
/man/fct_relabel.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/relabel.R
3 | \name{fct_relabel}
4 | \alias{fct_relabel}
5 | \title{Automatically relabel factor levels, collapse as necessary}
6 | \usage{
7 | fct_relabel(.f, .fun, ...)
8 | }
9 | \arguments{
10 | \item{.f}{A factor (or character vector).}
11 |
12 | \item{.fun}{A function to be applied to each level. Must accept one
13 | character argument and return a character vector of the same length as
14 | its input.
15 |
16 | You can also use \code{~} to create as shorthand (in the style of purrr).
17 | \code{~ paste(., "x")} is equivalent to \code{function(.) paste(., "x")}}
18 |
19 | \item{...}{Additional arguments to \code{fun}.}
20 | }
21 | \description{
22 | Automatically relabel factor levels, collapse as necessary
23 | }
24 | \examples{
25 | gss_cat$partyid \%>\% fct_count()
26 | gss_cat$partyid \%>\% fct_relabel(~ gsub(",", ", ", .x)) \%>\% fct_count()
27 |
28 | convert_income <- function(x) {
29 | regex <- "^(?:Lt |)[$]([0-9]+).*$"
30 | is_range <- grepl(regex, x)
31 | num_income <- as.numeric(gsub(regex, "\\\\1", x[is_range]))
32 | num_income <- trunc(num_income / 5000) * 5000
33 | x[is_range] <- paste0("Gt $", num_income)
34 | x
35 | }
36 | fct_count(gss_cat$rincome)
37 | convert_income(levels(gss_cat$rincome))
38 | rincome2 <- fct_relabel(gss_cat$rincome, convert_income)
39 | fct_count(rincome2)
40 | }
41 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_relabel.R:
--------------------------------------------------------------------------------
1 | context("fct_relabel")
2 |
3 | test_that("identity", {
4 | f1 <- factor(c("a", "b"))
5 |
6 | expect_identical(fct_relabel(f1, identity), f1)
7 | })
8 |
9 | test_that("error if not function", {
10 | f1 <- factor("a")
11 |
12 | expect_error(fct_relabel(f1, 1), "function")
13 | })
14 |
15 | test_that("error if level not character", {
16 | f1 <- factor("a")
17 |
18 | expect_error(fct_relabel(f1, function(x) 1), "character")
19 | })
20 |
21 | test_that("error if level has different length", {
22 | f1 <- factor(letters)
23 |
24 | expect_error(fct_relabel(f1, function(x) x[-1]), "expected 26.*got 25")
25 | })
26 |
27 | test_that("total collapse", {
28 | f1 <- factor(letters)
29 | new_levels <- function(x) rep("1", length(x))
30 |
31 | expect_identical(fct_relabel(f1, new_levels), factor(new_levels(letters)))
32 | })
33 |
34 | test_that("additional arguments", {
35 | f1 <- factor(letters)
36 |
37 | expect_identical(fct_relabel(f1, paste0, "."), factor(paste0(letters, ".")))
38 | })
39 |
40 | test_that("formulas are coerced to functions", {
41 | f1 <- factor(letters)
42 |
43 | expect_identical(
44 | fct_relabel(f1, ~paste0(.x, ".")),
45 | factor(paste0(letters, "."))
46 | )
47 | })
48 |
49 | test_that("string input is coerced to a factor", {
50 | expect_identical(
51 | fct_relabel(LETTERS[1:2], .fun=function(x) x),
52 | factor(LETTERS[1:2])
53 | )
54 | })
--------------------------------------------------------------------------------
/R/fct_cross.R:
--------------------------------------------------------------------------------
1 | #' Combine levels from two or more factors to create a new factor
2 | #'
3 | #' Computes a factor whose levels are all the combinations of the levels of the input factors.
4 | #'
5 | #' @param .f A factor or character vector
6 | #' @param ... Additional factors or character vectors
7 | #' @param sep A character string to separate the levels
8 | #' @param keep_empty If TRUE, keep combinations with no observations as levels
9 | #' @return The new factor
10 | #'
11 | #' @export
12 | #' @examples
13 | #' fruit <- factor(c("apple", "kiwi", "apple", "apple"))
14 | #' colour <- factor(c("green", "green", "red", "green"))
15 | #' eaten <- c("yes", "no", "yes", "no")
16 | #' fct_cross(fruit, colour)
17 | #' fct_cross(fruit, colour, eaten)
18 | #' fct_cross(fruit, colour, keep_empty = TRUE)
19 | fct_cross <- function(.f, ..., sep = ":", keep_empty = FALSE) {
20 | .f <- check_factor(.f)
21 |
22 | flist <- rlang::list2(...)
23 | if (length(flist) == 0) {
24 | return(.f)
25 | }
26 |
27 | .data <- lapply(tibble::tibble(.f, !!!flist), check_factor)
28 | newf <- rlang::invoke(paste, .data, sep = sep)
29 |
30 | if (keep_empty) {
31 | all_levels <- lapply(.data, levels)
32 | factor(newf, levels = rlang::invoke(paste,
33 | rlang::invoke(expand.grid, all_levels),
34 | sep = sep
35 | ))
36 | } else {
37 | anyNA <- Reduce("|", lapply(.data, is.na), FALSE)
38 | newf[anyNA] <- NA
39 | as.factor(newf)
40 | }
41 | }
42 |
--------------------------------------------------------------------------------
/R/as_factor.R:
--------------------------------------------------------------------------------
1 | #' Convert input to a factor.
2 | #'
3 | #' Compared to base R, when `x` is a character, this function creates
4 | #' levels in the order in which they appear, which will be the same on every
5 | #' platform. (Base R sorts in the current locale which can vary from place
6 | #' to place.) When `x` is numeric, the ordering is based on the numeric
7 | #' value and consistent with base R.
8 | #'
9 | #' This is a generic function.
10 | #'
11 | #' @param x Object to coerce to a factor.
12 | #' @param ... Other arguments passed down to method.
13 | #' @export
14 | #' @examples
15 | #' # Character object
16 | #' x <- c("a", "z", "g")
17 | #' as_factor(x)
18 | #' as.factor(x)
19 | #'
20 | #' # Character object containing numbers
21 | #' y <- c("1.1", "11", "2.2", "22")
22 | #' as_factor(y)
23 | #' as.factor(y)
24 | #'
25 | #' # Numeric object
26 | #' z <- as.numeric(y)
27 | #' as_factor(z)
28 | #' as.factor(z)
29 | as_factor <- function(x, ...) {
30 | ellipsis::check_dots_used()
31 | UseMethod("as_factor")
32 | }
33 |
34 | #' @rdname as_factor
35 | #' @export
36 | as_factor.factor <- function(x, ...) {
37 | x
38 | }
39 |
40 | #' @rdname as_factor
41 | #' @export
42 | as_factor.character <- function(x, ...) {
43 | # Preserve label for future haven compatibility
44 | structure(
45 | fct_inorder(x),
46 | label = attr(x, "label", exact = TRUE)
47 | )
48 | }
49 |
50 | #' @rdname as_factor
51 | #' @export
52 | as_factor.numeric <- function(x, ...) {
53 | factor(x)
54 | }
55 |
--------------------------------------------------------------------------------
/.github/CODE_OF_CONDUCT.md:
--------------------------------------------------------------------------------
1 | # Contributor Code of Conduct
2 |
3 | As contributors and maintainers of this project, we pledge to respect all people who
4 | contribute through reporting issues, posting feature requests, updating documentation,
5 | submitting pull requests or patches, and other activities.
6 |
7 | We are committed to making participation in this project a harassment-free experience for
8 | everyone, regardless of level of experience, gender, gender identity and expression,
9 | sexual orientation, disability, personal appearance, body size, race, ethnicity, age, or religion.
10 |
11 | Examples of unacceptable behavior by participants include the use of sexual language or
12 | imagery, derogatory comments or personal attacks, trolling, public or private harassment,
13 | insults, or other unprofessional conduct.
14 |
15 | Project maintainers have the right and responsibility to remove, edit, or reject comments,
16 | commits, code, wiki edits, issues, and other contributions that are not aligned to this
17 | Code of Conduct. Project maintainers who do not follow the Code of Conduct may be removed
18 | from the project team.
19 |
20 | Instances of abusive, harassing, or otherwise unacceptable behavior may be reported by
21 | opening an issue or contacting one or more of the project maintainers.
22 |
23 | This Code of Conduct is adapted from the Contributor Covenant
24 | (http://contributor-covenant.org), version 1.0.0, available at
25 | http://contributor-covenant.org/version/1/0/0/
26 |
--------------------------------------------------------------------------------
/man/lvls.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/lvls.R
3 | \name{lvls}
4 | \alias{lvls}
5 | \alias{lvls_reorder}
6 | \alias{lvls_revalue}
7 | \alias{lvls_expand}
8 | \title{Low-level functions for manipulating levels}
9 | \usage{
10 | lvls_reorder(f, idx, ordered = NA)
11 |
12 | lvls_revalue(f, new_levels)
13 |
14 | lvls_expand(f, new_levels)
15 | }
16 | \arguments{
17 | \item{f}{A factor (or character vector).}
18 |
19 | \item{idx}{A integer index, with one integer for each existing level.}
20 |
21 | \item{ordered}{A logical which determines the "ordered" status of the
22 | output factor. \code{NA} preserves the existing status of the factor.}
23 |
24 | \item{new_levels}{A character vector of new levels.}
25 | }
26 | \description{
27 | \code{lvls_reorder} leaves values as they are, but changes the order.
28 | \code{lvls_revalue} changes the values of existing levels; there must
29 | be one new level for each old level.
30 | \code{lvls_expand} expands the set of levels; the new levels must
31 | include the old levels.
32 | }
33 | \details{
34 | These functions are less helpful than the higher-level \code{fct_} functions,
35 | but are safer than the very low-level manipulation of levels directly,
36 | because they are more specific, and hence can more carefully check their
37 | arguments.
38 | }
39 | \examples{
40 | f <- factor(c("a", "b", "c"))
41 | lvls_reorder(f, 3:1)
42 | lvls_revalue(f, c("apple", "banana", "carrot"))
43 | lvls_expand(f, c("a", "b", "c", "d"))
44 | }
45 |
--------------------------------------------------------------------------------
/tests/testthat/test-lvls.R:
--------------------------------------------------------------------------------
1 | context("lvls")
2 |
3 | # lvls_expand -------------------------------------------------------------
4 |
5 | test_that("changes levels, not values", {
6 | f1 <- factor(c("a"))
7 | f2 <- factor(c("a"), levels = c("a", "b"))
8 |
9 | expect_equal(lvls_expand(f1, c("a", "b")), f2)
10 | })
11 |
12 | test_that("must include all existing levels", {
13 | f1 <- factor(c("a", "b"))
14 |
15 | expect_error(lvls_expand(f1, c("a", "c")), "include all existing levels")
16 | })
17 |
18 | # lvls_revalue ------------------------------------------------------------
19 |
20 | test_that("changes values and levels", {
21 | f1 <- factor(c("a", "b"))
22 | f2 <- factor(c("b", "a"), levels = c("b", "a"))
23 |
24 | expect_equal(lvls_revalue(f1, c("b", "a")), f2)
25 | })
26 |
27 | test_that("can collapse values", {
28 | f1 <- factor(c("a", "b"))
29 | f2 <- factor(c("a", "a"))
30 |
31 | expect_equal(lvls_revalue(f1, c("a", "a")), f2)
32 | })
33 |
34 | test_that("preserves missing values", {
35 | f1 <- factor(c("a", NA), exclude = NULL)
36 | f2 <- lvls_revalue(f1, levels(f1))
37 | expect_equal(levels(f2), levels(f1))
38 | })
39 |
40 | test_that("`new_levels` must be a character", {
41 | f1 <- factor(c("a", "b"))
42 | expect_error(lvls_revalue(f1, 1:5), "character vector")
43 | })
44 |
45 | test_that("`new_levels` must be same length as existing levels", {
46 | f1 <- factor(c("a", "b"))
47 |
48 | expect_error(lvls_revalue(f1, c("a")), "same length")
49 | expect_error(lvls_revalue(f1, c("a", "b", "c")), "same length")
50 | })
51 |
--------------------------------------------------------------------------------
/man/fct_relevel.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/relevel.R
3 | \name{fct_relevel}
4 | \alias{fct_relevel}
5 | \title{Reorder factor levels by hand}
6 | \usage{
7 | fct_relevel(.f, ..., after = 0L)
8 | }
9 | \arguments{
10 | \item{.f}{A factor (or character vector).}
11 |
12 | \item{...}{Either a function (or formula), or character levels.
13 |
14 | A function will be called with the current levels, and the return
15 | value (which must be a character vector) will be used to relevel the
16 | function.
17 |
18 | Any levels not mentioned will be left in existing order, after the
19 | explicitly mentioned levels. Supports tidy dots.}
20 |
21 | \item{after}{Where should the new values be placed?}
22 | }
23 | \description{
24 | This is a generalisaton of \code{\link[stats:relevel]{stats::relevel()}} that allows you to move any
25 | number of levels to any location.
26 | }
27 | \examples{
28 | f <- factor(c("a", "b", "c", "d"), levels = c("b", "c", "d", "a"))
29 | fct_relevel(f)
30 | fct_relevel(f, "a")
31 | fct_relevel(f, "b", "a")
32 |
33 | # Move to the third position
34 | fct_relevel(f, "a", after = 2)
35 |
36 | # Relevel to the end
37 | fct_relevel(f, "a", after = Inf)
38 | fct_relevel(f, "a", after = 3)
39 |
40 | # Relevel with a function
41 | fct_relevel(f, sort)
42 | fct_relevel(f, sample)
43 | fct_relevel(f, rev)
44 |
45 | # Using 'Inf' allows you to relevel to the end when the number
46 | # of levels is unknown or variable (e.g. vectorised operations)
47 | df <- forcats::gss_cat[, c("rincome", "denom")]
48 | lapply(df, levels)
49 |
50 | df2 <- lapply(df, fct_relevel, "Don't know", after = Inf)
51 | lapply(df2, levels)
52 |
53 | # You'll get a warning if the levels don't exist
54 | fct_relevel(f, "e")
55 | }
56 |
--------------------------------------------------------------------------------
/tests/testthat/test-collapse.R:
--------------------------------------------------------------------------------
1 | context("test-collapse.R")
2 |
3 | test_that("can collapse multiple values", {
4 | f1 <- factor(letters[1:3])
5 | f2 <- fct_collapse(f1, x = c("a", "b"), y = "c")
6 |
7 | expect_equal(f2, factor(c("x", "x", "y")))
8 | })
9 |
10 | test_that("empty dots yields unchanged factor", {
11 | f1 <- factor(letters[1:3])
12 | f2 <- fct_collapse(f1)
13 |
14 | expect_identical(f1, f2)
15 | })
16 |
17 | test_that("can collapse missing levels", {
18 | f1 <- factor(c("x", NA), exclude = NULL)
19 | f2 <- fct_collapse(f1, y = NA_character_)
20 |
21 | expect_equal(f2, factor(c("x", "y")))
22 | })
23 |
24 | test_that("can collapse un-named levels to Other", {
25 | f1 <- factor(letters[1:3])
26 | f2 <- fct_collapse(f1, xy = c("a", "b"), group_other = TRUE)
27 |
28 | expect_equal(f2, factor(c("xy", "xy", "Other"), levels = c("xy", "Other")))
29 | })
30 |
31 | test_that("collapses levels correctly when group_other is TRUE but no other variables to group", {
32 | f1 <- factor(letters[1:4])
33 | f2 <- expect_warning(fct_collapse(f1, x1 = c("a", "b", "d"), x2 = "c", group_other = TRUE), "Unknown levels")
34 |
35 | expect_equal(f2, factor(c("x1", "x1", "x2", "x1"), levels = c("x1", "x2")))
36 | })
37 |
38 | test_that("collapses levels correctly when group_other is TRUE and some Other variables to group", {
39 | f1 <- factor(letters[1:4])
40 | f2 <- fct_collapse(f1, x1 = c("a", "d"), x2 = "c", group_other = TRUE)
41 |
42 | expect_equal(f2, factor(c("x1", "Other", "x2", "x1"), levels = c("x1", "x2", "Other")))
43 | })
44 |
45 | test_that("does not automatically collapse unnamed levels to Other", {
46 | f1 <- factor(letters[1:3])
47 | f2 <- fct_collapse(f1, xy = c("a", "b"))
48 |
49 | expect_equal(f2, factor(c("xy", "xy", "c"), levels = c("xy", "c")))
50 | })
51 |
--------------------------------------------------------------------------------
/tests/testthat/test-reorder.R:
--------------------------------------------------------------------------------
1 | context("test-reorder.R")
2 |
3 | test_that("can reorder by 2d summary", {
4 | df <- tibble::tribble(
5 | ~g, ~x,
6 | "a", 3,
7 | "a", 3,
8 | "b", 2,
9 | "b", 2,
10 | "b", 1
11 | )
12 |
13 | f1 <- fct_reorder(df$g, df$x)
14 | expect_equal(levels(f1), c("b", "a"))
15 |
16 | f2 <- fct_reorder(df$g, df$x, .desc = TRUE)
17 | expect_equal(levels(f2), c("a", "b"))
18 | })
19 |
20 | test_that("can reorder by 2d summary", {
21 | df <- tibble::tribble(
22 | ~g, ~x, ~y,
23 | "a", 1, 10,
24 | "a", 2, 5,
25 | "b", 1, 5,
26 | "b", 2, 10
27 | )
28 |
29 | f1 <- fct_reorder2(df$g, df$x, df$y)
30 | expect_equal(levels(f1), c("b", "a"))
31 |
32 | f2 <- fct_reorder(df$g, df$x, .desc = TRUE)
33 | expect_equal(levels(f2), c("a", "b"))
34 | })
35 |
36 | test_that("complains if summary doesn't return single value", {
37 | fun1 <- function(x, y) c(1, 2)
38 | fun2 <- function(x, y) integer()
39 |
40 | expect_error(fct_reorder("a", 1, fun1), "single value per group")
41 | expect_error(fct_reorder("a", 1, fun2), "single value per group")
42 | expect_error(fct_reorder2("a", 1, 2, fun1), "single value per group")
43 | expect_error(fct_reorder2("a", 1, 2, fun2), "single value per group")
44 | })
45 |
46 | test_that("fct_infreq respects missing values", {
47 | f <- factor(c("a", "b", "b", NA, NA, NA), exclude = FALSE)
48 | expect_equal(levels(fct_infreq(f)), c(NA, "b", "a"))
49 | })
50 |
51 | test_that("fct_inseq sorts in numeric order", {
52 | f <- factor(c("3", "1", "1", "2"))
53 | expect_equal(levels(fct_inseq(f)), c("1", "2", "3"))
54 | })
55 |
56 | test_that("fct_inseq gives error for non-numeric levels", {
57 | f <- factor(c("c", "a", "a", "b"))
58 | expect_error(levels(fct_inseq(f)), "level must be coercible to numeric")
59 | })
60 |
--------------------------------------------------------------------------------
/_pkgdown.yml:
--------------------------------------------------------------------------------
1 | template:
2 | package: tidytemplate
3 |
4 | url: https://forcats.tidyverse.org
5 |
6 | development:
7 | mode: auto
8 |
9 | home:
10 | strip_header: true
11 | links:
12 | - text: Learn more
13 | href: http://r4ds.had.co.nz/factors.html
14 |
15 | reference:
16 | - title: Change order of levels
17 | desc: >
18 | Keep the values of the levels the same, but change their order.
19 | These are particularly useful for models, tables, and visualisations.
20 | contents:
21 | - fct_relevel
22 | - fct_inorder
23 | - fct_reorder
24 | - fct_infreq
25 | - fct_shuffle
26 | - fct_rev
27 | - fct_shift
28 |
29 | - title: Change value of levels
30 | desc: >
31 | Change factor levels, while preserving order (as much as possible).
32 | contents:
33 | - fct_anon
34 | - fct_collapse
35 | - fct_lump
36 | - fct_other
37 | - fct_recode
38 | - fct_relabel
39 |
40 | - title: Add/remove levels
41 | desc: >
42 | Leave existing data as is, but add or remove levels.
43 | contents:
44 | - fct_expand
45 | - fct_explicit_na
46 | - fct_drop
47 | - fct_unify
48 |
49 | - title: Combine multiple factors
50 | contents:
51 | - fct_c
52 | - fct_cross
53 |
54 | - title: Other helpers
55 | contents:
56 | - as_factor
57 | - fct_count
58 | - fct_match
59 | - fct_unique
60 | - lvls
61 | - lvls_union
62 |
63 | - title: Data
64 | contents:
65 | - gss_cat
66 |
67 | navbar:
68 | components:
69 | home: ~
70 | news:
71 | text: News
72 | menu:
73 | - text: "Release notes"
74 | - text: "Version 0.4.0"
75 | href: https://www.tidyverse.org/articles/2019/05/forcats-0-4-0/
76 | - text: "Version 0.3.0"
77 | href: https://www.tidyverse.org/articles/2018/02/forcats-0-3-0/
78 | - text: "Version 0.2.0"
79 | href: https://blog.rstudio.com/2017/04/12/tidyverse-updates/
80 | - text: "Version 0.1.0"
81 | href: https://blog.rstudio.com/2016/08/31/forcats-0-1-0/
82 | - text: "------------------"
83 | - text: "Change log"
84 | href: news/index.html
85 |
--------------------------------------------------------------------------------
/R/recode.R:
--------------------------------------------------------------------------------
1 | #' Change factor levels by hand
2 | #'
3 | #' @param .f A factor (or character vector).
4 | #' @param ... A sequence of named character vectors where the
5 | #' name gives the new level, and the value gives the old level.
6 | #' Levels not otherwise mentioned will be left as is. Levels can
7 | #' be removed by naming them `NULL`. Uses tidy dots.
8 | #' @export
9 | #' @examples
10 | #' x <- factor(c("apple", "bear", "banana", "dear"))
11 | #' fct_recode(x, fruit = "apple", fruit = "banana")
12 | #'
13 | #' # If you make a mistake you'll get a warning
14 | #' fct_recode(x, fruit = "apple", fruit = "bananana")
15 | #'
16 | #' # If you name the level NULL it will be removed
17 | #' fct_recode(x, NULL = "apple", fruit = "banana")
18 | #'
19 | #' # When passing a named vector to rename levels use !!! to splice
20 | #' x <- factor(c("apple", "bear", "banana", "dear"))
21 | #' levels <- c(fruit = "apple", fruit = "banana")
22 | #' fct_recode(x, !!!levels)
23 | fct_recode <- function(.f, ...) {
24 | f <- check_factor(.f)
25 |
26 | new_levels <- check_recode_levels(...)
27 |
28 | # Remove any named NULL and finish if all NULLs
29 | nulls <- names(new_levels) == "NULL"
30 | if (any(nulls)) {
31 | f <- factor(f, levels = setdiff(levels(f), new_levels[nulls]))
32 | new_levels <- new_levels[!nulls]
33 | }
34 |
35 | # Match old levels with new levels
36 | old_levels <- levels(f)
37 | idx <- match(new_levels, old_levels)
38 |
39 | # Handle levels that don't exist
40 | if (any(is.na(idx))) {
41 | bad <- new_levels[is.na(idx)]
42 | warning("Unknown levels in `f`: ", paste(bad, collapse = ", "), call. = FALSE)
43 |
44 | new_levels <- new_levels[!is.na(idx)]
45 | idx <- idx[!is.na(idx)]
46 | }
47 |
48 | old_levels[idx] <- names(new_levels)
49 |
50 | lvls_revalue(f, old_levels)
51 | }
52 |
53 | check_recode_levels <- function(...) {
54 | levels <- rlang::dots_list(...)
55 |
56 | is_ok <- function(x) is.character(x) && length(x) == 1
57 | ok <- vapply(levels, is_ok, logical(1))
58 |
59 | if (!all(ok)) {
60 | stop(
61 | "Each input to fct_recode must be a single named string. ",
62 | "Problems at positions: ", paste0(which(!ok), collapse = ", "),
63 | call. = FALSE
64 | )
65 | }
66 |
67 | unlist(levels)
68 | }
69 |
--------------------------------------------------------------------------------
/man/fct_reorder.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/reorder.R
3 | \name{fct_reorder}
4 | \alias{fct_reorder}
5 | \alias{fct_reorder2}
6 | \alias{last2}
7 | \alias{first2}
8 | \title{Reorder factor levels by sorting along another variable}
9 | \usage{
10 | fct_reorder(.f, .x, .fun = median, ..., .desc = FALSE)
11 |
12 | fct_reorder2(.f, .x, .y, .fun = last2, ..., .desc = TRUE)
13 |
14 | last2(.x, .y)
15 |
16 | first2(.x, .y)
17 | }
18 | \arguments{
19 | \item{.f}{A factor (or character vector).}
20 |
21 | \item{.x, .y}{The levels of \code{f} are reordered so that the values
22 | of \code{.fun(.x)} (for \code{fct_reorder()}) and \code{fun(.x, .y)} (for \code{fct_reorder2()})
23 | are in ascending order.}
24 |
25 | \item{.fun}{n summary function. It should take one vector for
26 | \code{fct_reorder}, and two vectors for \code{fct_reorder2}, and return a single
27 | value.}
28 |
29 | \item{...}{Other arguments passed on to \code{.fun}. A common argument is
30 | \code{na.rm = TRUE}.}
31 |
32 | \item{.desc}{Order in descending order? Note the default is different
33 | between \code{fct_reorder} and \code{fct_reorder2}, in order to
34 | match the default ordering of factors in the legend.}
35 | }
36 | \description{
37 | \code{fct_reorder()} is useful for 1d displays where the factor is mapped to
38 | position; \code{fct_reorder2()} for 2d displays where the factor is mapped to
39 | a non-position aesthetic. \code{last2()} and \code{first2()} are helpers for \code{fct_reorder2()};
40 | \code{last2()} finds the last value of \code{y} when sorted by \code{x}; \code{first2()} finds the first value.
41 | }
42 | \examples{
43 | boxplot(Sepal.Width ~ Species, data = iris)
44 | boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width), data = iris)
45 | boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width, .desc = TRUE), data = iris)
46 |
47 | chks <- subset(ChickWeight, as.integer(Chick) < 10)
48 | chks <- transform(chks, Chick = fct_shuffle(Chick))
49 |
50 | if (require("ggplot2")) {
51 | ggplot(chks, aes(Time, weight, colour = Chick)) +
52 | geom_point() +
53 | geom_line()
54 |
55 | # Note that lines match order in legend
56 | ggplot(chks, aes(Time, weight, colour = fct_reorder2(Chick, Time, weight))) +
57 | geom_point() +
58 | geom_line() +
59 | labs(colour = "Chick")
60 | }
61 | }
62 |
--------------------------------------------------------------------------------
/R/relevel.R:
--------------------------------------------------------------------------------
1 | #' Reorder factor levels by hand
2 | #'
3 | #' This is a generalisaton of [stats::relevel()] that allows you to move any
4 | #' number of levels to any location.
5 | #'
6 | #' @param .f A factor (or character vector).
7 | #' @param ... Either a function (or formula), or character levels.
8 | #'
9 | #' A function will be called with the current levels, and the return
10 | #' value (which must be a character vector) will be used to relevel the
11 | #' function.
12 | #'
13 | #' Any levels not mentioned will be left in existing order, after the
14 | #' explicitly mentioned levels. Supports tidy dots.
15 | #' @param after Where should the new values be placed?
16 | #' @export
17 | #' @examples
18 | #' f <- factor(c("a", "b", "c", "d"), levels = c("b", "c", "d", "a"))
19 | #' fct_relevel(f)
20 | #' fct_relevel(f, "a")
21 | #' fct_relevel(f, "b", "a")
22 | #'
23 | #' # Move to the third position
24 | #' fct_relevel(f, "a", after = 2)
25 | #'
26 | #' # Relevel to the end
27 | #' fct_relevel(f, "a", after = Inf)
28 | #' fct_relevel(f, "a", after = 3)
29 | #'
30 | #' # Relevel with a function
31 | #' fct_relevel(f, sort)
32 | #' fct_relevel(f, sample)
33 | #' fct_relevel(f, rev)
34 | #'
35 | #' # Using 'Inf' allows you to relevel to the end when the number
36 | #' # of levels is unknown or variable (e.g. vectorised operations)
37 | #' df <- forcats::gss_cat[, c("rincome", "denom")]
38 | #' lapply(df, levels)
39 | #'
40 | #' df2 <- lapply(df, fct_relevel, "Don't know", after = Inf)
41 | #' lapply(df2, levels)
42 | #'
43 | #' # You'll get a warning if the levels don't exist
44 | #' fct_relevel(f, "e")
45 | fct_relevel <- function(.f, ..., after = 0L) {
46 | f <- check_factor(.f)
47 |
48 | old_levels <- levels(f)
49 | if (rlang::dots_n(...) == 1L && (is.function(..1) || rlang::is_formula(..1))) {
50 | fun <- rlang::as_function(..1)
51 | first_levels <- fun(old_levels)
52 | if (!is.character(first_levels)) {
53 | stop("Re-leveling function must return character vector", call. = FALSE)
54 | }
55 | } else {
56 | first_levels <- rlang::chr(...)
57 | }
58 |
59 | unknown <- setdiff(first_levels, old_levels)
60 | if (length(unknown) > 0) {
61 | warning("Unknown levels in `f`: ", paste(unknown, collapse = ", "), call. = FALSE)
62 | first_levels <- intersect(first_levels, old_levels)
63 | }
64 |
65 | new_levels <- append(setdiff(old_levels, first_levels), first_levels, after = after)
66 |
67 | lvls_reorder(f, match(new_levels, old_levels))
68 | }
69 |
--------------------------------------------------------------------------------
/.github/SUPPORT.md:
--------------------------------------------------------------------------------
1 | # Getting help with forcats
2 |
3 | Thanks for using forcats!
4 | Before filing an issue, there are a few places to explore and pieces to put together to make the process as smooth as possible.
5 |
6 | ## Make a reprex
7 |
8 | Start by making a minimal **repr**oducible **ex**ample using the [reprex](https://reprex.tidyverse.org/) package.
9 | If you haven't heard of or used reprex before, you're in for a treat!
10 | Seriously, reprex will make all of your R-question-asking endeavors easier (which is a pretty insane ROI for the five to ten minutes it'll take you to learn what it's all about).
11 | For additional reprex pointers, check out the [Get help!](https://www.tidyverse.org/help/) section of the tidyverse site.
12 |
13 | ## Where to ask?
14 |
15 | Armed with your reprex, the next step is to figure out [where to ask](https://www.tidyverse.org/help/#where-to-ask).
16 |
17 | * If it's a question: start with [community.rstudio.com](https://community.rstudio.com/), and/or StackOverflow. There are more people there to answer questions.
18 |
19 | * If it's a bug: you're in the right place, [file an issue](https://github.com/batpigandme/forcats/issues/new).
20 |
21 | * If you're not sure: let the community help you figure it out!
22 | If your problem _is_ a bug or a feature request, you can easily return here and report it.
23 |
24 | Before opening a new issue, be sure to [search issues and pull requests](https://github.com/batpigandme/forcats/issues) to make sure the bug hasn't been reported and/or already fixed in the development version.
25 | By default, the search will be pre-populated with `is:issue is:open`.
26 | You can [edit the qualifiers](https://help.github.com/articles/searching-issues-and-pull-requests/) (e.g. `is:pr`, `is:closed`) as needed.
27 | For example, you'd simply remove `is:open` to search _all_ issues in the repo, open or closed.
28 |
29 | ## What happens next?
30 |
31 | To be as efficient as possible, development of tidyverse packages tends to be very bursty, so you shouldn't worry if you don't get an immediate response.
32 | Typically we don't look at a repo until a sufficient quantity of issues accumulates, then there’s a burst of intense activity as we focus our efforts.
33 | That makes development more efficient because it avoids expensive context switching between problems, at the cost of taking longer to get back to you.
34 | This process makes a good reprex particularly important because it might be multiple months between your initial report and when we start working on it.
35 | If we can’t reproduce the bug, we can’t fix it!
36 |
--------------------------------------------------------------------------------
/man/fct_lump.Rd:
--------------------------------------------------------------------------------
1 | % Generated by roxygen2: do not edit by hand
2 | % Please edit documentation in R/lump.R
3 | \name{fct_lump}
4 | \alias{fct_lump}
5 | \alias{fct_lump_min}
6 | \title{Lump together factor levels into "other"}
7 | \usage{
8 | fct_lump(
9 | f,
10 | n,
11 | prop,
12 | w = NULL,
13 | other_level = "Other",
14 | ties.method = c("min", "average", "first", "last", "random", "max")
15 | )
16 |
17 | fct_lump_min(f, min, w = NULL, other_level = "Other")
18 | }
19 | \arguments{
20 | \item{f}{A factor (or character vector).}
21 |
22 | \item{n, prop}{If both \code{n} and \code{prop} are missing, \code{fct_lump()} lumps
23 | together the least frequent levels into "other", while ensuring that
24 | "other" is still the smallest level. It's particularly useful in
25 | conjunction with \code{\link{fct_inorder}()}.
26 |
27 | Positive \code{n} preserves the most common \code{n} values.
28 | Negative \code{n} preserves the least common \code{-n} values.
29 | It there are ties, you will get at least \code{abs(n)} values.
30 |
31 | Positive \code{prop} preserves values that appear at least
32 | \code{prop} of the time. Negative \code{prop} preserves values that
33 | appear at most \code{-prop} of the time.}
34 |
35 | \item{w}{An optional numeric vector giving weights for frequency of
36 | each value (not level) in f.}
37 |
38 | \item{other_level}{Value of level used for "other" values. Always
39 | placed at end of levels.}
40 |
41 | \item{ties.method}{A character string specifying how ties are
42 | treated. See \code{\link[=rank]{rank()}} for details.}
43 |
44 | \item{min}{Preserves values that appear at least \code{min} number of times.}
45 | }
46 | \description{
47 | \code{fct_lump()} lumps together least/most common levels (as defined by \code{n} and
48 | \code{prop}) into "other".
49 | \code{fct_lump_min()} lumps together all levels which don't appear at least
50 | \code{min} number of times.
51 | }
52 | \examples{
53 | x <- factor(rep(LETTERS[1:9], times = c(40, 10, 5, 27, 1, 1, 1, 1, 1)))
54 | x \%>\% table()
55 | x \%>\% fct_lump() \%>\% table()
56 | x \%>\% fct_lump() \%>\% fct_inorder() \%>\% table()
57 |
58 | x <- factor(letters[rpois(100, 5)])
59 | x
60 | table(x)
61 | table(fct_lump(x))
62 |
63 | # Use positive values to collapse the rarest
64 | fct_lump(x, n = 3)
65 | fct_lump(x, prop = 0.1)
66 |
67 | # Use negative values to collapse the most common
68 | fct_lump(x, n = -3)
69 | fct_lump(x, prop = -0.1)
70 |
71 | # Use weighted frequencies
72 | w <- c(rep(2, 50), rep(1, 50))
73 | fct_lump(x, n = 5, w = w)
74 |
75 | # Use ties.method to control how tied factors are collapsed
76 | fct_lump(x, n = 6)
77 | fct_lump(x, n = 6, ties.method = "max")
78 |
79 | x <- factor(letters[rpois(100, 5)])
80 | fct_lump_min(x, min = 10)
81 | }
82 | \seealso{
83 | \code{\link[=fct_other]{fct_other()}} to convert specified levels to other.
84 | }
85 |
--------------------------------------------------------------------------------
/.github/CONTRIBUTING.md:
--------------------------------------------------------------------------------
1 | # Contributing to forcats
2 |
3 | This outlines how to propose a change to forcats.
4 | For more detailed info about contributing to this, and other tidyverse packages, please see the
5 | [**development contributing guide**](https://rstd.io/tidy-contrib).
6 |
7 | ## Fixing typos
8 |
9 | You can fix typos, spelling mistakes, or grammatical errors in the documentation directly using the GitHub web interface, as long as the changes are made in the _source_ file.
10 | This generally means you'll need to edit [roxygen2 comments](https://roxygen2.r-lib.org/articles/roxygen2.html) in an `.R`, not a `.Rd` file.
11 | You can find the `.R` file that generates the `.Rd` by reading the comment in the first line.
12 |
13 | ## Bigger changes
14 |
15 | If you want to make a bigger change, it's a good idea to first file an issue and make sure someone from the team agrees that it’s needed.
16 | If you’ve found a bug, please file an issue that illustrates the bug with a minimal
17 | [reprex](https://www.tidyverse.org/help/#reprex) (this will also help you write a unit test, if needed).
18 |
19 | ### Pull request process
20 |
21 | * Fork the package and clone onto your computer. If you haven't done this before, we recommend using `usethis::create_from_github("batpigandme/forcats", fork = TRUE)`.
22 |
23 | * Install all development dependences with `devtools::install_dev_deps()`, and then make sure the package passes R CMD check by running `devtools::check()`.
24 | If R CMD check doesn't pass cleanly, it's a good idea to ask for help before continuing.
25 | * Create a Git branch for your pull request (PR). We recommend using `usethis::pr_init("brief-description-of-change")`.
26 |
27 | * Make your changes, commit to git, and then create a PR by running `usethis::pr_push()`, and following the prompts in your browser.
28 | The title of your PR should briefly describe the change.
29 | The body of your PR should contain `Fixes #issue-number`.
30 |
31 | * For user-facing changes, add a bullet to the top of `NEWS.md` (i.e. just below the first header). Follow the style described in .
32 |
33 | ### Code style
34 |
35 | * New code should follow the tidyverse [style guide](https://style.tidyverse.org).
36 | You can use the [styler](https://CRAN.R-project.org/package=styler) package to apply these styles, but please don't restyle code that has nothing to do with your PR.
37 |
38 | * We use [roxygen2](https://cran.r-project.org/package=roxygen2), with [Markdown syntax](https://cran.r-project.org/web/packages/roxygen2/vignettes/markdown.html), for documentation.
39 |
40 | * We use [testthat](https://cran.r-project.org/package=testthat) for unit tests.
41 | Contributions with test cases included are easier to accept.
42 |
43 | ## Code of Conduct
44 |
45 | Please note that the forcats project is released with a
46 | [Contributor Code of Conduct](CODE_OF_CONDUCT.md). By contributing to this
47 | project you agree to abide by its terms.
48 |
--------------------------------------------------------------------------------
/data-raw/gss-cat.R:
--------------------------------------------------------------------------------
1 | # Extracted from GSS data explorer
2 | # Variables: year, id, marital, race, rincome, partyid, relig, denom
3 | # Years: 2000-2014
4 | # Export: xls and stata
5 | library(tibble)
6 | library(dplyr)
7 |
8 | # Warning because last two rows contains source info
9 | gss <- readxl::read_excel("data-raw/GSS.xls")
10 |
11 | vars <- readxl::read_excel("data-raw/GSS.xls", sheet = 2)
12 | names(gss) <- vars$Name
13 |
14 | na <- c("Not applicable", "No answer", "Don't know")
15 |
16 | gss_cat <- gss %>%
17 | filter(!is.na(id_)) %>%
18 | select(-id_) %>%
19 | mutate(
20 | year = as.integer(year),
21 | age = as.integer(readr::parse_number(age, na = na)), # ignore 148 "or older"
22 | tvhours = as.integer(readr::parse_double(tvhours, na = na))
23 | )
24 |
25 | # From .do file, copy factor definitions (to get levels)
26 | levels <- list(
27 | marital = c(
28 | "No answer",
29 | "Never married",
30 | "Separated",
31 | "Divorced",
32 | "Widowed",
33 | "Married"
34 | ),
35 | race = c(
36 | "Other",
37 | "Black",
38 | "White",
39 | "Not applicable"
40 | ),
41 | rincome = c(
42 | "No answer",
43 | "Don't know",
44 | "Refused",
45 | "$25000 or more",
46 | "$20000 - 24999",
47 | "$15000 - 19999",
48 | "$10000 - 14999",
49 | "$8000 to 9999",
50 | "$7000 to 7999",
51 | "$6000 to 6999",
52 | "$5000 to 5999",
53 | "$4000 to 4999",
54 | "$3000 to 3999",
55 | "$1000 to 2999",
56 | "Lt $1000",
57 | "Not applicable"
58 | ),
59 | partyid = c(
60 | "No answer",
61 | "Don't know",
62 | "Other party",
63 | "Strong republican",
64 | "Not str republican",
65 | "Ind,near rep",
66 | "Independent",
67 | "Ind,near dem",
68 | "Not str democrat",
69 | "Strong democrat"
70 | ),
71 | relig = c(
72 | "No answer",
73 | "Don't know",
74 | "Inter-nondenominational",
75 | "Native american",
76 | "Christian",
77 | "Orthodox-christian",
78 | "Moslem/islam",
79 | "Other eastern",
80 | "Hinduism",
81 | "Buddhism",
82 | "Other",
83 | "None",
84 | "Jewish",
85 | "Catholic",
86 | "Protestant",
87 | "Not applicable"
88 | ),
89 | denom = c(
90 | "No answer",
91 | "Don't know",
92 | "No denomination",
93 | "Other",
94 | "Episcopal",
95 | "Presbyterian-dk wh",
96 | "Presbyterian, merged",
97 | "Other presbyterian",
98 | "United pres ch in us",
99 | "Presbyterian c in us",
100 | "Lutheran-dk which",
101 | "Evangelical luth",
102 | "Other lutheran",
103 | "Wi evan luth synod",
104 | "Lutheran-mo synod",
105 | "Luth ch in america",
106 | "Am lutheran",
107 | "Methodist-dk which",
108 | "Other methodist",
109 | "United methodist",
110 | "Afr meth ep zion",
111 | "Afr meth episcopal",
112 | "Baptist-dk which",
113 | "Other baptists",
114 | "Southern baptist",
115 | "Nat bapt conv usa",
116 | "Nat bapt conv of am",
117 | "Am bapt ch in usa",
118 | "Am baptist asso",
119 | "Not applicable"
120 | )
121 | )
122 |
123 | for(name in names(levels)) {
124 | gss_cat[[name]] <- factor(gss_cat[[name]], level = levels[[name]])
125 | }
126 |
127 | use_data(gss_cat, overwrite = TRUE)
128 |
--------------------------------------------------------------------------------
/R/lvls.R:
--------------------------------------------------------------------------------
1 | #' Low-level functions for manipulating levels
2 | #'
3 | #' `lvls_reorder` leaves values as they are, but changes the order.
4 | #' `lvls_revalue` changes the values of existing levels; there must
5 | #' be one new level for each old level.
6 | #' `lvls_expand` expands the set of levels; the new levels must
7 | #' include the old levels.
8 | #'
9 | #' These functions are less helpful than the higher-level `fct_` functions,
10 | #' but are safer than the very low-level manipulation of levels directly,
11 | #' because they are more specific, and hence can more carefully check their
12 | #' arguments.
13 | #'
14 | #' @param f A factor (or character vector).
15 | #' @param idx A integer index, with one integer for each existing level.
16 | #' @param new_levels A character vector of new levels.
17 | #' @param ordered A logical which determines the "ordered" status of the
18 | #' output factor. `NA` preserves the existing status of the factor.
19 | #' @name lvls
20 | #' @examples
21 | #' f <- factor(c("a", "b", "c"))
22 | #' lvls_reorder(f, 3:1)
23 | #' lvls_revalue(f, c("apple", "banana", "carrot"))
24 | #' lvls_expand(f, c("a", "b", "c", "d"))
25 | NULL
26 |
27 | #' @export
28 | #' @rdname lvls
29 | lvls_reorder <- function(f, idx, ordered = NA) {
30 | f <- check_factor(f)
31 | if (!is.numeric(idx)) {
32 | stop("`idx` must be numeric", call. = FALSE)
33 | }
34 | if (!setequal(idx, lvls_seq(f)) || length(idx) != nlevels(f)) {
35 | stop("`idx` must contain one integer for each level of `f`", call. = FALSE)
36 | }
37 |
38 | refactor(f, levels(f)[idx], ordered = ordered)
39 | }
40 |
41 | #' @export
42 | #' @rdname lvls
43 | lvls_revalue <- function(f, new_levels) {
44 | f <- check_factor(f)
45 |
46 | if (!is.character(new_levels)) {
47 | stop("`new_levels` must be a character vector", call. = FALSE)
48 | }
49 |
50 | if (length(new_levels) != nlevels(f)) {
51 | stop(
52 | "`new_levels` must be the same length as `levels(f)`: expected ",
53 | nlevels(f), " new levels, got ", length(new_levels), ".",
54 | call. = FALSE
55 | )
56 | }
57 |
58 | if (anyDuplicated(new_levels)) {
59 | # Collapse levels, creating a new factor
60 | u_levels <- unique(new_levels)
61 | index <- match(new_levels, u_levels)
62 |
63 | out <- index[f]
64 | attributes(out) <- attributes(f)
65 | attr(out, "levels") <- u_levels
66 | out
67 | } else {
68 | attr(f, "levels") <- new_levels
69 | f
70 | }
71 | }
72 |
73 | #' @export
74 | #' @rdname lvls
75 | lvls_expand <- function(f, new_levels) {
76 | f <- check_factor(f)
77 |
78 | missing <- setdiff(levels(f), new_levels)
79 | if (length(missing) > 0) {
80 | stop(
81 | "Must include all existing levels. Missing: ", paste0(missing, collapse = ", "),
82 | call. = FALSE)
83 | }
84 |
85 | refactor(f, new_levels)
86 | }
87 |
88 | lvls_seq <- function(f) {
89 | seq_along(levels(f))
90 | }
91 |
92 | refactor <- function(f, new_levels, ordered = NA) {
93 | if (is.na(ordered)) {
94 | ordered <- is.ordered(f)
95 | }
96 |
97 | new_f <- factor(f, levels = new_levels, exclude = NULL, ordered = ordered)
98 | attributes(new_f) <- utils::modifyList(attributes(f), attributes(new_f))
99 | new_f
100 | }
101 |
102 |
103 | #' Find all levels in a list of factors
104 | #'
105 | #' @param fs A list of factors.
106 | #' @export
107 | #' @examples
108 | #' fs <- list(factor("a"), factor("b"), factor(c("a", "b")))
109 | #' lvls_union(fs)
110 | lvls_union <- function(fs) {
111 | fs <- check_factor_list(fs)
112 | Reduce(function(x, y) union(x, levels(y)), fs, init = character())
113 | }
114 |
--------------------------------------------------------------------------------
/README.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | output: github_document
3 | ---
4 |
5 |
6 |
7 | ```{r, include = FALSE}
8 | knitr::opts_chunk$set(
9 | collapse = TRUE,
10 | comment = "#>",
11 | fig.path = "man/figures/README-"
12 | )
13 | ```
14 |
15 | # forcats  16 |
17 |
18 | [](https://cran.r-project.org/package=forcats)
19 | [](https://travis-ci.org/tidyverse/forcats)
20 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
21 |
22 |
23 | ## Overview
24 |
25 | R uses __factors__ to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the __forcats__ package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. Some examples include:
26 |
27 | * `fct_reorder()`: Reordering a factor by another variable.
28 | * `fct_infreq()`: Reordering a factor by the frequency of values.
29 | * `fct_relevel()`: Changing the order of a factor by hand.
30 | * `fct_lump()`: Collapsing the least/most frequent values of a factor into "other".
31 |
32 | You can learn more about each of these in `vignette("forcats")`. If you're new to factors, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
33 |
34 | ## Installation
35 |
36 | ```
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 | ```
47 |
48 | ## Cheatsheet
49 |
50 |
16 |
17 |
18 | [](https://cran.r-project.org/package=forcats)
19 | [](https://travis-ci.org/tidyverse/forcats)
20 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
21 |
22 |
23 | ## Overview
24 |
25 | R uses __factors__ to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the __forcats__ package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. Some examples include:
26 |
27 | * `fct_reorder()`: Reordering a factor by another variable.
28 | * `fct_infreq()`: Reordering a factor by the frequency of values.
29 | * `fct_relevel()`: Changing the order of a factor by hand.
30 | * `fct_lump()`: Collapsing the least/most frequent values of a factor into "other".
31 |
32 | You can learn more about each of these in `vignette("forcats")`. If you're new to factors, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
33 |
34 | ## Installation
35 |
36 | ```
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 | ```
47 |
48 | ## Cheatsheet
49 |
50 | 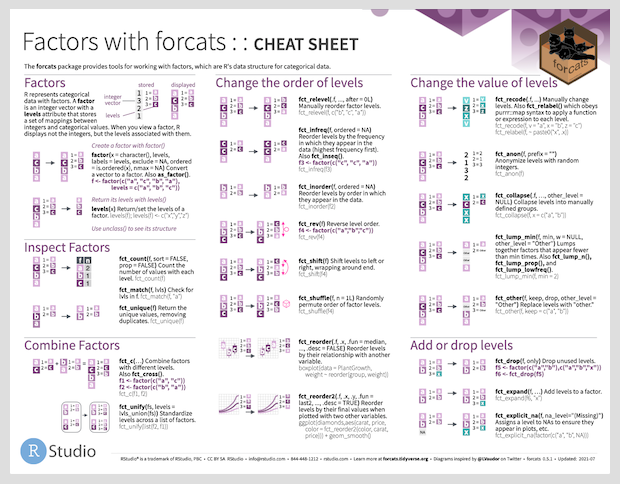 51 |
52 | ## Getting started
53 |
54 | forcats is part of the core tidyverse, so you can load it with `library(tidyverse)` or `library(forcats)`.
55 |
56 | ```{r setup, message = FALSE}
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ```{r}
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | ```
67 |
68 | ```{r}
69 | starwars %>%
70 | filter(!is.na(species)) %>%
71 | mutate(species = fct_lump(species, n = 3)) %>%
72 | count(species)
73 | ```
74 |
75 | ```{r unordered-plot}
76 | ggplot(starwars, aes(x = eye_color)) +
77 | geom_bar() +
78 | coord_flip()
79 | ```
80 |
81 | ```{r ordered-plot}
82 | starwars %>%
83 | mutate(eye_color = fct_infreq(eye_color)) %>%
84 | ggplot(aes(x = eye_color)) +
85 | geom_bar() +
86 | coord_flip()
87 | ```
88 |
89 | ## More resources
90 |
91 | For a history of factors, I recommend [_stringsAsFactors: An unauthorized biography_](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/) by Roger Peng and [_stringsAsFactors = \_](http://notstatschat.tumblr.com/post/124987394001/stringsasfactors-sigh) by Thomas Lumley. If you want to learn more about other approaches to working with factors and categorical data, I recommend [_Wrangling categorical data in R_](https://peerj.com/preprints/3163/), by Amelia McNamara and Nicholas Horton.
92 |
93 | ## Getting help
94 |
95 | If you encounter a clear bug, please file a minimal reproducible example on [github](https://github.com/tidyverse/forcats/issues). For questions and other discussion, please use [community.rstudio.com](https://community.rstudio.com/).
96 |
97 | ## Code of Conduct
98 |
99 | Please note that the 'forcats' project is released with a [Contributor Code of Conduct](.github/CODE_OF_CONDUCT.md). By contributing to this project, you agree to abide by its terms.
100 |
--------------------------------------------------------------------------------
/R/reorder.R:
--------------------------------------------------------------------------------
1 | #' Reorder factor levels by sorting along another variable
2 | #'
3 | #' `fct_reorder()` is useful for 1d displays where the factor is mapped to
4 | #' position; `fct_reorder2()` for 2d displays where the factor is mapped to
5 | #' a non-position aesthetic. `last2()` and `first2()` are helpers for `fct_reorder2()`;
6 | #' `last2()` finds the last value of `y` when sorted by `x`; `first2()` finds the first value.
7 | #'
8 | #' @param .f A factor (or character vector).
9 | #' @param .x,.y The levels of `f` are reordered so that the values
10 | #' of `.fun(.x)` (for `fct_reorder()`) and `fun(.x, .y)` (for `fct_reorder2()`)
11 | #' are in ascending order.
12 | #' @param .fun n summary function. It should take one vector for
13 | #' `fct_reorder`, and two vectors for `fct_reorder2`, and return a single
14 | #' value.
15 | #' @param ... Other arguments passed on to `.fun`. A common argument is
16 | #' `na.rm = TRUE`.
17 | #' @param .desc Order in descending order? Note the default is different
18 | #' between `fct_reorder` and `fct_reorder2`, in order to
19 | #' match the default ordering of factors in the legend.
20 | #' @importFrom stats median
21 | #' @export
22 | #' @examples
23 | #' boxplot(Sepal.Width ~ Species, data = iris)
24 | #' boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width), data = iris)
25 | #' boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width, .desc = TRUE), data = iris)
26 | #'
27 | #' chks <- subset(ChickWeight, as.integer(Chick) < 10)
28 | #' chks <- transform(chks, Chick = fct_shuffle(Chick))
29 | #'
30 | #' if (require("ggplot2")) {
31 | #' ggplot(chks, aes(Time, weight, colour = Chick)) +
32 | #' geom_point() +
33 | #' geom_line()
34 | #'
35 | #' # Note that lines match order in legend
36 | #' ggplot(chks, aes(Time, weight, colour = fct_reorder2(Chick, Time, weight))) +
37 | #' geom_point() +
38 | #' geom_line() +
39 | #' labs(colour = "Chick")
40 | #' }
41 | fct_reorder <- function(.f, .x, .fun = median, ..., .desc = FALSE) {
42 | f <- check_factor(.f)
43 | stopifnot(length(f) == length(.x))
44 | ellipsis::check_dots_used()
45 |
46 | summary <- tapply(.x, .f, .fun, ...)
47 | # This is a bit of a weak test, but should detect the most common case
48 | # where `.fun` returns multiple values.
49 | if (is.list(summary)) {
50 | stop("`fun` must return a single value per group", call. = FALSE)
51 | }
52 |
53 | lvls_reorder(f, order(summary, decreasing = .desc))
54 | }
55 |
56 | #' @export
57 | #' @rdname fct_reorder
58 | fct_reorder2 <- function(.f, .x, .y, .fun = last2, ..., .desc = TRUE) {
59 | f <- check_factor(.f)
60 | stopifnot(length(f) == length(.x), length(.x) == length(.y))
61 | ellipsis::check_dots_used()
62 |
63 | summary <- tapply(seq_along(.x), f, function(i) .fun(.x[i], .y[i], ...))
64 | if (is.list(summary)) {
65 | stop("`fun` must return a single value per group", call. = FALSE)
66 | }
67 |
68 | lvls_reorder(.f, order(summary, decreasing = .desc))
69 | }
70 |
71 |
72 | #' @export
73 | #' @rdname fct_reorder
74 | last2 <- function(.x, .y) {
75 | .y[order(.x, na.last = FALSE)][length(.y)]
76 | }
77 |
78 | #' @export
79 | #' @rdname fct_reorder
80 | first2 <- function(.x, .y) {
81 | .y[order(.x)][1]
82 | }
83 |
84 |
85 |
86 | #' Reorder factors levels by first appearance, frequency, or numeric order.
87 | #'
88 | #' @inheritParams lvls_reorder
89 | #' @param f A factor
90 | #' @export
91 | #' @examples
92 | #' f <- factor(c("b", "b", "a", "c", "c", "c"))
93 | #' f
94 | #' fct_inorder(f)
95 | #' fct_infreq(f)
96 | #'
97 | #' fct_inorder(f, ordered = TRUE)
98 | #'
99 | #' f <- factor(sample(1:10))
100 | #' fct_inseq(f)
101 | fct_inorder <- function(f, ordered = NA) {
102 | f <- check_factor(f)
103 |
104 | idx <- as.integer(f)[!duplicated(f)]
105 | idx <- idx[!is.na(idx)]
106 | lvls_reorder(f, idx, ordered = ordered)
107 | }
108 |
109 | #' @export
110 | #' @rdname fct_inorder
111 | fct_infreq <- function(f, ordered = NA) {
112 | f <- check_factor(f)
113 |
114 | lvls_reorder(f, order(table(f), decreasing = TRUE), ordered = ordered)
115 | }
116 |
117 | #' @export
118 | #' @rdname fct_inorder
119 | fct_inseq <- function(f, ordered = NA) {
120 | f <- check_factor(f)
121 |
122 | num_levels <- suppressWarnings(as.numeric(levels(f)))
123 | new_levels <- sort(num_levels)
124 |
125 | if (length(new_levels) == 0) {
126 | stop("At least one existing level must be coercible to numeric.", call. = FALSE)
127 | }
128 |
129 | refactor(f, new_levels, ordered = ordered)
130 | }
131 |
132 |
--------------------------------------------------------------------------------
/NEWS.md:
--------------------------------------------------------------------------------
1 | # forcats (development version)
2 |
3 | * `first2()`, a `fct_reorder2()` helper function, sorts `.y` by the first value of `.x` (@jtr13).
4 |
5 | * fixed bug in `fct_collapse()` so it now correctly collapses factors when `group_other = TRUE` (#172), and makes `"Other"` the last level (#202) (@gtm19, #172 & #202)
6 |
7 | # forcats 0.4.0
8 |
9 | ## New features
10 |
11 | * `fct_collapse()` gains a `group_other` argument to allow you to group all
12 | un-named levels into `"Other"`. (#100, @AmeliaMN)
13 |
14 | * `fct_cross()` creates a new factor containing the combined levels from two
15 | or more input factors, similar to `base::interaction` (@tslumley, #136)
16 |
17 | * `fct_inseq()` reorders labels in numeric order, if possible (#145, @kbodwin).
18 |
19 | * `fct_lump_min()` preserves levels that appear at least `min` times (can also
20 | be used with the `w` weighted argument) (@robinsones, #142).
21 |
22 | * `fct_match()` performs validated matching, providing a safer alternative to
23 | `f %in% c("x", "y")` which silently returns `FALSE` if `"x"` or `"y"`
24 | are not levels of `f` (e.g. because of a typo) (#126, @jonocarroll).
25 |
26 | * `fct_relevel()` can now level factors using a function that is passed the
27 | current levels (#117).
28 |
29 | * `as_factor()` now has a numeric method. By default, orders factors in numeric
30 | order, unlike the other methods which default to order of appearance.
31 | (#145, @kbodwin)
32 |
33 | ## Minor bug fixes and improvements
34 |
35 | * `fct_count()` gains a parameter to also compute the proportion
36 | (@zhiiiyang, #146).
37 |
38 | * `fct_lump()` now does not change the label if no lumping occurs
39 | (@zhiiiyang, #130).
40 |
41 | * `fct_relabel()` now accepts character input.
42 |
43 | * `fct_reorder()` and `fct_reorder2()` no longer require that the summary
44 | function return a numeric vector of length 1; instead it can return any
45 | orderable vector of length 1 (#147).
46 |
47 | * `fct_reorder()`, `fct_reorder2()` and `as_factor()` now use the ellipsis
48 | package to warn if you pass in named components to `...` (#174).
49 |
50 | # forcats 0.3.0
51 |
52 | ## API changes
53 |
54 | * `fct_c()` now requires explicit splicing with `!!!` if you have a
55 | list of factors that you want to combine. This is consistent with an emerging
56 | standards for handling `...` throughout the tidyverse.
57 |
58 | * `fct_reorder()` and `fct_reorder2()` have renamed `fun` to `.fun` to
59 | avoid spurious matching of named arguments.
60 |
61 | ## New features
62 |
63 | * All functions that take `...` use "tidy" dots: this means that you use can
64 | `!!!` to splice in a list of values, and trailing empty arguments are
65 | automatically removed. Additionally, all other arguments gain a `.` prefix
66 | in order to avoid unhelpful matching of named arguments (#110).
67 |
68 | * `fct_lump()` gains `w` argument (#70, @wilkox) to weight value
69 | frequencies before lumping them together (#68).
70 |
71 | ## Improvements to NA handling
72 |
73 | * `as_factor()` and `fct_inorder()` accept NA levels (#98).
74 |
75 | * `fct_explicit_na()` also replaces NAs encoded in levels.
76 |
77 | * `fct_lump()` correctly acccounts for `NA` values in input (#41)
78 |
79 | * `lvls_revalue()` preserves NA levels.
80 |
81 | ## Minor improvements and bug fixes
82 |
83 | * Test coverage increased from 80% to 99%.
84 |
85 | * `fct_drop()` now preserves attributes (#83).
86 |
87 | * `fct_expand()` and `lvls_expand()` now also take character vectors (#99).
88 |
89 | * `fct_relabel()` now accepts objects coercible to functions
90 | by `rlang::as_function` (#91, @alistaire47)
91 |
92 | # forcats 0.2.0
93 |
94 | ## New functions
95 |
96 | * `as_factor()` which works like `as.factor()` but orders levels by
97 | appearance to avoid differences between locales (#39).
98 |
99 | * `fct_other()` makes it easier to convert selected levels to "other" (#40)
100 |
101 | * `fct_relabel()` allows programmatic relabeling of levels (#50, @krlmlr).
102 |
103 | ## Minor improvements and bug fixes
104 |
105 | * `fct_c()` can take either a list of factors or individual factors (#42).
106 |
107 | * `fct_drop()` gains `only` argument to restrict which levels are dropped (#69)
108 | and no longer adds `NA` level if not present (#52).
109 |
110 | * `fct_recode()` is now checks that each new value is of length 1 (#56).
111 |
112 | * `fct_relevel()` gains `after` argument so you can also move levels
113 | to the end (or any other position you like) (#29).
114 |
115 | * `lvls_reorder()`, `fct_inorder()`, and `fct_infreq()` gain an `ordered`
116 | argument, allowing you to override the existing "ordered" status (#54).
117 |
118 | # forcats 0.1.1
119 |
120 | * Minor fixes for R CMD check
121 |
122 | * Add package docs
123 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | # forcats
51 |
52 | ## Getting started
53 |
54 | forcats is part of the core tidyverse, so you can load it with `library(tidyverse)` or `library(forcats)`.
55 |
56 | ```{r setup, message = FALSE}
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ```{r}
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | ```
67 |
68 | ```{r}
69 | starwars %>%
70 | filter(!is.na(species)) %>%
71 | mutate(species = fct_lump(species, n = 3)) %>%
72 | count(species)
73 | ```
74 |
75 | ```{r unordered-plot}
76 | ggplot(starwars, aes(x = eye_color)) +
77 | geom_bar() +
78 | coord_flip()
79 | ```
80 |
81 | ```{r ordered-plot}
82 | starwars %>%
83 | mutate(eye_color = fct_infreq(eye_color)) %>%
84 | ggplot(aes(x = eye_color)) +
85 | geom_bar() +
86 | coord_flip()
87 | ```
88 |
89 | ## More resources
90 |
91 | For a history of factors, I recommend [_stringsAsFactors: An unauthorized biography_](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/) by Roger Peng and [_stringsAsFactors = \_](http://notstatschat.tumblr.com/post/124987394001/stringsasfactors-sigh) by Thomas Lumley. If you want to learn more about other approaches to working with factors and categorical data, I recommend [_Wrangling categorical data in R_](https://peerj.com/preprints/3163/), by Amelia McNamara and Nicholas Horton.
92 |
93 | ## Getting help
94 |
95 | If you encounter a clear bug, please file a minimal reproducible example on [github](https://github.com/tidyverse/forcats/issues). For questions and other discussion, please use [community.rstudio.com](https://community.rstudio.com/).
96 |
97 | ## Code of Conduct
98 |
99 | Please note that the 'forcats' project is released with a [Contributor Code of Conduct](.github/CODE_OF_CONDUCT.md). By contributing to this project, you agree to abide by its terms.
100 |
--------------------------------------------------------------------------------
/R/reorder.R:
--------------------------------------------------------------------------------
1 | #' Reorder factor levels by sorting along another variable
2 | #'
3 | #' `fct_reorder()` is useful for 1d displays where the factor is mapped to
4 | #' position; `fct_reorder2()` for 2d displays where the factor is mapped to
5 | #' a non-position aesthetic. `last2()` and `first2()` are helpers for `fct_reorder2()`;
6 | #' `last2()` finds the last value of `y` when sorted by `x`; `first2()` finds the first value.
7 | #'
8 | #' @param .f A factor (or character vector).
9 | #' @param .x,.y The levels of `f` are reordered so that the values
10 | #' of `.fun(.x)` (for `fct_reorder()`) and `fun(.x, .y)` (for `fct_reorder2()`)
11 | #' are in ascending order.
12 | #' @param .fun n summary function. It should take one vector for
13 | #' `fct_reorder`, and two vectors for `fct_reorder2`, and return a single
14 | #' value.
15 | #' @param ... Other arguments passed on to `.fun`. A common argument is
16 | #' `na.rm = TRUE`.
17 | #' @param .desc Order in descending order? Note the default is different
18 | #' between `fct_reorder` and `fct_reorder2`, in order to
19 | #' match the default ordering of factors in the legend.
20 | #' @importFrom stats median
21 | #' @export
22 | #' @examples
23 | #' boxplot(Sepal.Width ~ Species, data = iris)
24 | #' boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width), data = iris)
25 | #' boxplot(Sepal.Width ~ fct_reorder(Species, Sepal.Width, .desc = TRUE), data = iris)
26 | #'
27 | #' chks <- subset(ChickWeight, as.integer(Chick) < 10)
28 | #' chks <- transform(chks, Chick = fct_shuffle(Chick))
29 | #'
30 | #' if (require("ggplot2")) {
31 | #' ggplot(chks, aes(Time, weight, colour = Chick)) +
32 | #' geom_point() +
33 | #' geom_line()
34 | #'
35 | #' # Note that lines match order in legend
36 | #' ggplot(chks, aes(Time, weight, colour = fct_reorder2(Chick, Time, weight))) +
37 | #' geom_point() +
38 | #' geom_line() +
39 | #' labs(colour = "Chick")
40 | #' }
41 | fct_reorder <- function(.f, .x, .fun = median, ..., .desc = FALSE) {
42 | f <- check_factor(.f)
43 | stopifnot(length(f) == length(.x))
44 | ellipsis::check_dots_used()
45 |
46 | summary <- tapply(.x, .f, .fun, ...)
47 | # This is a bit of a weak test, but should detect the most common case
48 | # where `.fun` returns multiple values.
49 | if (is.list(summary)) {
50 | stop("`fun` must return a single value per group", call. = FALSE)
51 | }
52 |
53 | lvls_reorder(f, order(summary, decreasing = .desc))
54 | }
55 |
56 | #' @export
57 | #' @rdname fct_reorder
58 | fct_reorder2 <- function(.f, .x, .y, .fun = last2, ..., .desc = TRUE) {
59 | f <- check_factor(.f)
60 | stopifnot(length(f) == length(.x), length(.x) == length(.y))
61 | ellipsis::check_dots_used()
62 |
63 | summary <- tapply(seq_along(.x), f, function(i) .fun(.x[i], .y[i], ...))
64 | if (is.list(summary)) {
65 | stop("`fun` must return a single value per group", call. = FALSE)
66 | }
67 |
68 | lvls_reorder(.f, order(summary, decreasing = .desc))
69 | }
70 |
71 |
72 | #' @export
73 | #' @rdname fct_reorder
74 | last2 <- function(.x, .y) {
75 | .y[order(.x, na.last = FALSE)][length(.y)]
76 | }
77 |
78 | #' @export
79 | #' @rdname fct_reorder
80 | first2 <- function(.x, .y) {
81 | .y[order(.x)][1]
82 | }
83 |
84 |
85 |
86 | #' Reorder factors levels by first appearance, frequency, or numeric order.
87 | #'
88 | #' @inheritParams lvls_reorder
89 | #' @param f A factor
90 | #' @export
91 | #' @examples
92 | #' f <- factor(c("b", "b", "a", "c", "c", "c"))
93 | #' f
94 | #' fct_inorder(f)
95 | #' fct_infreq(f)
96 | #'
97 | #' fct_inorder(f, ordered = TRUE)
98 | #'
99 | #' f <- factor(sample(1:10))
100 | #' fct_inseq(f)
101 | fct_inorder <- function(f, ordered = NA) {
102 | f <- check_factor(f)
103 |
104 | idx <- as.integer(f)[!duplicated(f)]
105 | idx <- idx[!is.na(idx)]
106 | lvls_reorder(f, idx, ordered = ordered)
107 | }
108 |
109 | #' @export
110 | #' @rdname fct_inorder
111 | fct_infreq <- function(f, ordered = NA) {
112 | f <- check_factor(f)
113 |
114 | lvls_reorder(f, order(table(f), decreasing = TRUE), ordered = ordered)
115 | }
116 |
117 | #' @export
118 | #' @rdname fct_inorder
119 | fct_inseq <- function(f, ordered = NA) {
120 | f <- check_factor(f)
121 |
122 | num_levels <- suppressWarnings(as.numeric(levels(f)))
123 | new_levels <- sort(num_levels)
124 |
125 | if (length(new_levels) == 0) {
126 | stop("At least one existing level must be coercible to numeric.", call. = FALSE)
127 | }
128 |
129 | refactor(f, new_levels, ordered = ordered)
130 | }
131 |
132 |
--------------------------------------------------------------------------------
/NEWS.md:
--------------------------------------------------------------------------------
1 | # forcats (development version)
2 |
3 | * `first2()`, a `fct_reorder2()` helper function, sorts `.y` by the first value of `.x` (@jtr13).
4 |
5 | * fixed bug in `fct_collapse()` so it now correctly collapses factors when `group_other = TRUE` (#172), and makes `"Other"` the last level (#202) (@gtm19, #172 & #202)
6 |
7 | # forcats 0.4.0
8 |
9 | ## New features
10 |
11 | * `fct_collapse()` gains a `group_other` argument to allow you to group all
12 | un-named levels into `"Other"`. (#100, @AmeliaMN)
13 |
14 | * `fct_cross()` creates a new factor containing the combined levels from two
15 | or more input factors, similar to `base::interaction` (@tslumley, #136)
16 |
17 | * `fct_inseq()` reorders labels in numeric order, if possible (#145, @kbodwin).
18 |
19 | * `fct_lump_min()` preserves levels that appear at least `min` times (can also
20 | be used with the `w` weighted argument) (@robinsones, #142).
21 |
22 | * `fct_match()` performs validated matching, providing a safer alternative to
23 | `f %in% c("x", "y")` which silently returns `FALSE` if `"x"` or `"y"`
24 | are not levels of `f` (e.g. because of a typo) (#126, @jonocarroll).
25 |
26 | * `fct_relevel()` can now level factors using a function that is passed the
27 | current levels (#117).
28 |
29 | * `as_factor()` now has a numeric method. By default, orders factors in numeric
30 | order, unlike the other methods which default to order of appearance.
31 | (#145, @kbodwin)
32 |
33 | ## Minor bug fixes and improvements
34 |
35 | * `fct_count()` gains a parameter to also compute the proportion
36 | (@zhiiiyang, #146).
37 |
38 | * `fct_lump()` now does not change the label if no lumping occurs
39 | (@zhiiiyang, #130).
40 |
41 | * `fct_relabel()` now accepts character input.
42 |
43 | * `fct_reorder()` and `fct_reorder2()` no longer require that the summary
44 | function return a numeric vector of length 1; instead it can return any
45 | orderable vector of length 1 (#147).
46 |
47 | * `fct_reorder()`, `fct_reorder2()` and `as_factor()` now use the ellipsis
48 | package to warn if you pass in named components to `...` (#174).
49 |
50 | # forcats 0.3.0
51 |
52 | ## API changes
53 |
54 | * `fct_c()` now requires explicit splicing with `!!!` if you have a
55 | list of factors that you want to combine. This is consistent with an emerging
56 | standards for handling `...` throughout the tidyverse.
57 |
58 | * `fct_reorder()` and `fct_reorder2()` have renamed `fun` to `.fun` to
59 | avoid spurious matching of named arguments.
60 |
61 | ## New features
62 |
63 | * All functions that take `...` use "tidy" dots: this means that you use can
64 | `!!!` to splice in a list of values, and trailing empty arguments are
65 | automatically removed. Additionally, all other arguments gain a `.` prefix
66 | in order to avoid unhelpful matching of named arguments (#110).
67 |
68 | * `fct_lump()` gains `w` argument (#70, @wilkox) to weight value
69 | frequencies before lumping them together (#68).
70 |
71 | ## Improvements to NA handling
72 |
73 | * `as_factor()` and `fct_inorder()` accept NA levels (#98).
74 |
75 | * `fct_explicit_na()` also replaces NAs encoded in levels.
76 |
77 | * `fct_lump()` correctly acccounts for `NA` values in input (#41)
78 |
79 | * `lvls_revalue()` preserves NA levels.
80 |
81 | ## Minor improvements and bug fixes
82 |
83 | * Test coverage increased from 80% to 99%.
84 |
85 | * `fct_drop()` now preserves attributes (#83).
86 |
87 | * `fct_expand()` and `lvls_expand()` now also take character vectors (#99).
88 |
89 | * `fct_relabel()` now accepts objects coercible to functions
90 | by `rlang::as_function` (#91, @alistaire47)
91 |
92 | # forcats 0.2.0
93 |
94 | ## New functions
95 |
96 | * `as_factor()` which works like `as.factor()` but orders levels by
97 | appearance to avoid differences between locales (#39).
98 |
99 | * `fct_other()` makes it easier to convert selected levels to "other" (#40)
100 |
101 | * `fct_relabel()` allows programmatic relabeling of levels (#50, @krlmlr).
102 |
103 | ## Minor improvements and bug fixes
104 |
105 | * `fct_c()` can take either a list of factors or individual factors (#42).
106 |
107 | * `fct_drop()` gains `only` argument to restrict which levels are dropped (#69)
108 | and no longer adds `NA` level if not present (#52).
109 |
110 | * `fct_recode()` is now checks that each new value is of length 1 (#56).
111 |
112 | * `fct_relevel()` gains `after` argument so you can also move levels
113 | to the end (or any other position you like) (#29).
114 |
115 | * `lvls_reorder()`, `fct_inorder()`, and `fct_infreq()` gain an `ordered`
116 | argument, allowing you to override the existing "ordered" status (#54).
117 |
118 | # forcats 0.1.1
119 |
120 | * Minor fixes for R CMD check
121 |
122 | * Add package docs
123 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | # forcats  5 |
6 |
7 |
8 | [](https://cran.r-project.org/package=forcats)
10 | [](https://travis-ci.org/tidyverse/forcats)
12 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
14 |
15 |
16 | ## Overview
17 |
18 | R uses **factors** to handle categorical variables, variables that have
19 | a fixed and known set of possible values. Factors are also helpful for
20 | reordering character vectors to improve display. The goal of the
21 | **forcats** package is to provide a suite of tools that solve common
22 | problems with factors, including changing the order of levels or the
23 | values. Some examples include:
24 |
25 | - `fct_reorder()`: Reordering a factor by another variable.
26 | - `fct_infreq()`: Reordering a factor by the frequency of values.
27 | - `fct_relevel()`: Changing the order of a factor by hand.
28 | - `fct_lump()`: Collapsing the least/most frequent values of a factor
29 | into “other”.
30 |
31 | You can learn more about each of these in `vignette("forcats")`. If
32 | you’re new to factors, the best place to start is the [chapter on
33 | factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
34 |
35 | ## Installation
36 |
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 |
47 | ## Cheatsheet
48 |
49 |
5 |
6 |
7 |
8 | [](https://cran.r-project.org/package=forcats)
10 | [](https://travis-ci.org/tidyverse/forcats)
12 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
14 |
15 |
16 | ## Overview
17 |
18 | R uses **factors** to handle categorical variables, variables that have
19 | a fixed and known set of possible values. Factors are also helpful for
20 | reordering character vectors to improve display. The goal of the
21 | **forcats** package is to provide a suite of tools that solve common
22 | problems with factors, including changing the order of levels or the
23 | values. Some examples include:
24 |
25 | - `fct_reorder()`: Reordering a factor by another variable.
26 | - `fct_infreq()`: Reordering a factor by the frequency of values.
27 | - `fct_relevel()`: Changing the order of a factor by hand.
28 | - `fct_lump()`: Collapsing the least/most frequent values of a factor
29 | into “other”.
30 |
31 | You can learn more about each of these in `vignette("forcats")`. If
32 | you’re new to factors, the best place to start is the [chapter on
33 | factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
34 |
35 | ## Installation
36 |
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 |
47 | ## Cheatsheet
48 |
49 |  50 |
51 | ## Getting started
52 |
53 | forcats is part of the core tidyverse, so you can load it with
54 | `library(tidyverse)` or `library(forcats)`.
55 |
56 | ``` r
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ``` r
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | #> # A tibble: 37 x 2
67 | #> species n
68 | #>
69 | #> 1 Human 35
70 | #> 2 Droid 5
71 | #> 3 Gungan 3
72 | #> 4 Kaminoan 2
73 | #> 5 Mirialan 2
74 | #> 6 Twi'lek 2
75 | #> 7 Wookiee 2
76 | #> 8 Zabrak 2
77 | #> 9 Aleena 1
78 | #> 10 Besalisk 1
79 | #> # … with 27 more rows
80 | ```
81 |
82 | ``` r
83 | starwars %>%
84 | filter(!is.na(species)) %>%
85 | mutate(species = fct_lump(species, n = 3)) %>%
86 | count(species)
87 | #> # A tibble: 4 x 2
88 | #> species n
89 | #>
90 | #> 1 Droid 5
91 | #> 2 Gungan 3
92 | #> 3 Human 35
93 | #> 4 Other 39
94 | ```
95 |
96 | ``` r
97 | ggplot(starwars, aes(x = eye_color)) +
98 | geom_bar() +
99 | coord_flip()
100 | ```
101 |
102 | 
103 |
104 | ``` r
105 | starwars %>%
106 | mutate(eye_color = fct_infreq(eye_color)) %>%
107 | ggplot(aes(x = eye_color)) +
108 | geom_bar() +
109 | coord_flip()
110 | ```
111 |
112 | 
113 |
114 | ## More resources
115 |
116 | For a history of factors, I recommend [*stringsAsFactors: An
117 | unauthorized
118 | biography*](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/)
119 | by Roger Peng and [*stringsAsFactors =
120 | \*](http://notstatschat.tumblr.com/post/124987394001/stringsasfactors-sigh)
121 | by Thomas Lumley. If you want to learn more about other approaches to
122 | working with factors and categorical data, I recommend [*Wrangling
123 | categorical data in R*](https://peerj.com/preprints/3163/), by Amelia
124 | McNamara and Nicholas Horton.
125 |
126 | ## Getting help
127 |
128 | If you encounter a clear bug, please file a minimal reproducible example
129 | on [github](https://github.com/tidyverse/forcats/issues). For questions
130 | and other discussion, please use
131 | [community.rstudio.com](https://community.rstudio.com/).
132 |
133 | ## Code of Conduct
134 |
135 | Please note that the ‘forcats’ project is released with a [Contributor
136 | Code of Conduct](.github/CODE_OF_CONDUCT.md). By contributing to this
137 | project, you agree to abide by its terms.
138 |
--------------------------------------------------------------------------------
/revdep/README.md:
--------------------------------------------------------------------------------
1 | # Platform
2 |
3 | |field |value |
4 | |:--------|:----------------------------|
5 | |version |R version 3.5.2 (2018-12-20) |
6 | |os |macOS Mojave 10.14.2 |
7 | |system |x86_64, darwin15.6.0 |
8 | |ui |RStudio |
9 | |language |(EN) |
10 | |collate |en_US.UTF-8 |
11 | |ctype |en_US.UTF-8 |
12 | |tz |America/Chicago |
13 | |date |2019-02-16 |
14 |
15 | # Dependencies
16 |
17 | |package |old |new |Δ |
18 | |:---------|:-----|:----------|:--|
19 | |forcats |0.3.0 |0.3.0.9000 |* |
20 | |ellipsis |NA |0.0.2 |* |
21 | |fansi |0.4.0 |0.4.0 | |
22 | |pillar |1.3.1 |1.3.1 | |
23 | |pkgconfig |2.0.2 |2.0.2 | |
24 | |rlang |0.3.1 |0.3.1 | |
25 | |tibble |2.0.1 |2.0.1 | |
26 | |utf8 |1.1.4 |1.1.4 | |
27 |
28 | # Revdeps
29 |
30 | ## Couldn't check (1)
31 |
32 | |package |version |error |warning |note |
33 | |:------------------------------------|:-------|:-----|:-------|:----|
34 | |[circumplex](problems.md#circumplex) |0.2.1 |1 | | |
35 |
36 | ## All (52)
37 |
38 | |package |version |error |warning |note |
39 | |:----------------------------------------------|:-------|:-----|:-------|:----|
40 | |apaTables |2.0.5 | | | |
41 | |bench |1.0.1 | | | |
42 | |[bupaR](problems.md#bupar) |0.4.1 | |1 | |
43 | |[circumplex](problems.md#circumplex) |0.2.1 |1 | | |
44 | |coalitions |0.6.5 | | | |
45 | |[cocktailApp](problems.md#cocktailapp) |0.2.0 | | |1 |
46 | |[dabestr](problems.md#dabestr) |0.2.0 | |1 |1 |
47 | |descriptr |0.5.0 | | | |
48 | |easyalluvial |0.1.8 | | | |
49 | |evaluator |0.3.2 | | | |
50 | |[ezplot](problems.md#ezplot) |0.2.2 | | |1 |
51 | |[factorMerger](problems.md#factormerger) |0.3.6 | | |1 |
52 | |[finalfit](problems.md#finalfit) |0.9.0 | | |1 |
53 | |forwards |0.1.1 | | | |
54 | |[furniture](problems.md#furniture) |1.8.7 | | |1 |
55 | |genogeographer |0.1.8 | | | |
56 | |[ggdag](problems.md#ggdag) |0.1.0 | | |1 |
57 | |[ggridges](problems.md#ggridges) |0.5.1 | | |1 |
58 | |[glmmfields](problems.md#glmmfields) |0.1.1 | | |1 |
59 | |[glmSparseNet](problems.md#glmsparsenet) |1.0.0 | |1 |2 |
60 | |glue |1.3.0 | | | |
61 | |[haven](problems.md#haven) |2.0.0 | | |1 |
62 | |healthcareai |2.3.0 | | | |
63 | |[idealstan](problems.md#idealstan) |0.5.1 | | |2 |
64 | |[iotables](problems.md#iotables) |0.4.2 | | |1 |
65 | |[konfound](problems.md#konfound) |0.1.1 | |1 |1 |
66 | |[MetamapsDB](problems.md#metamapsdb) |0.0.2 | | |1 |
67 | |naniar |0.4.2 | | | |
68 | |[noaastormevents](problems.md#noaastormevents) |0.1.0 | | |2 |
69 | |[nzelect](problems.md#nzelect) |0.4.0 | | |2 |
70 | |outcomerate |1.0.1 | | | |
71 | |OutliersO3 |0.6 | | | |
72 | |[phenopath](problems.md#phenopath) |1.6.0 | |1 | |
73 | |[plotly](problems.md#plotly) |4.8.0 | | |2 |
74 | |[processmapR](problems.md#processmapr) |0.3.2 | | |1 |
75 | |[questionr](problems.md#questionr) |0.7.0 | | |2 |
76 | |[railtrails](problems.md#railtrails) |0.1.1 | | |1 |
77 | |[rbin](problems.md#rbin) |0.1.1 | | |1 |
78 | |rfm |0.2.0 | | | |
79 | |[SCnorm](problems.md#scnorm) |1.4.5 | | |1 |
80 | |SEERaBomb |2018.1 | | | |
81 | |simstandard |0.3.0 | | | |
82 | |sjPlot |2.6.2 | | | |
83 | |survivalAnalysis |0.1.1 | | | |
84 | |[sweep](problems.md#sweep) |0.2.1.1 |1 | | |
85 | |[TextForecast](problems.md#textforecast) |0.1.0 | | |2 |
86 | |[tidybayes](problems.md#tidybayes) |1.0.3 |1 | | |
87 | |[tidyLPA](problems.md#tidylpa) |0.2.4 | | |1 |
88 | |tidytidbits |0.2.0 | | | |
89 | |[tidyverse](problems.md#tidyverse) |1.2.1 | | |1 |
90 | |[timetk](problems.md#timetk) |0.1.1.1 |1 | | |
91 | |unpivotr |0.5.0 | | | |
92 |
93 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_lump.R:
--------------------------------------------------------------------------------
1 | context("fct_lump")
2 |
3 | test_that("positive values keeps most commmon", {
4 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
5 |
6 | expect_equal(levels(fct_lump(f, n = 1)), c("a", "Other"))
7 | expect_equal(levels(fct_lump(f, n = 2)), c("a", "b", "Other"))
8 |
9 | expect_equal(levels(fct_lump(f, prop = 0.25)), c("a", "Other"))
10 | expect_equal(levels(fct_lump(f, prop = 0.15)), c("a", "b", "Other"))
11 | })
12 |
13 | test_that("ties are respected", {
14 | f <- c("a", "a", "a", "b", "b", "b", "c", "d")
15 | expect_equal(levels(fct_lump(f, 1)), c("a", "b", "Other"))
16 | })
17 |
18 | test_that("negative values drop most common" ,{
19 | f <- c("a", "a", "a", "a", "b", "b", "b", "b", "c", "d")
20 | expect_equal(levels(fct_lump(f, n = -1)), c("c", "d", "Other"))
21 | expect_equal(levels(fct_lump(f, prop = -0.2)), c("c", "d", "Other"))
22 | })
23 |
24 | test_that("return original factor when all element satisfy n / p condition", {
25 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
26 |
27 | expect_equal(levels(fct_lump(f, n = 4)), c("a", "b", "c", "d", "e", "f", "g"))
28 | expect_equal(levels(fct_lump(f, n = 10)), c("a", "b", "c", "d", "e", "f", "g"))
29 | expect_equal(levels(fct_lump(f, n = -10)), c("a", "b", "c", "d", "e", "f", "g"))
30 |
31 | expect_equal(levels(fct_lump(f, prop = 0.01)), c("a", "b", "c", "d", "e", "f", "g"))
32 | expect_equal(levels(fct_lump(f, prop = -1)), c("a", "b", "c", "d", "e", "f", "g"))
33 | })
34 |
35 | test_that("different behaviour when apply tie function", {
36 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
37 |
38 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "min")),
39 | c("a", "b", "c", "d", "e", "f", "g"))
40 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "max")),
41 | c("a", "b", "Other" ))
42 |
43 | # Rank of c, d, e, f, g is (3+4+5+6+7)/5 = 5
44 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "average")),
45 | c("a", "b", "Other" ))
46 | expect_equal(levels(fct_lump(f, n = 5, ties.method = "average")),
47 | c("a", "b", "c", "d", "e", "f", "g"))
48 |
49 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "first")),
50 | c("a", "b", "c", "d", "Other"))
51 |
52 | if (getRversion() >= "3.3.0") {
53 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "last")),
54 | c("a", "b", "f", "g", "Other"))
55 | }
56 | })
57 |
58 | test_that("NAs included in total", {
59 | f <- factor(c("a", "a", "b", "c", rep(NA, 7)))
60 |
61 | o1 <- fct_lump(f, prop = 0.10)
62 | expect_equal(levels(o1), c("a", "Other"))
63 |
64 | o2 <- fct_lump(f, w = rep(1, 11), prop = 0.10)
65 | expect_equal(levels(o2), c("a", "Other"))
66 | })
67 |
68 | test_that("bad weights generate error messages", {
69 | expect_error(fct_lump(letters, w = letters), "must be a numeric vector")
70 | expect_error(fct_lump(letters, w = 1:10), "must be the same length")
71 | expect_error(fct_lump(letters, w = rep(-1, 26)), "must be non-negative")
72 | })
73 |
74 | test_that("values are correctly weighted", {
75 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
76 | w <- c( 0.2, 0.2, 0.6, 2, 2, 6, 4, 2, 2, 1)
77 | f2 <- c(
78 | "a",
79 | rep("b", 4),
80 | rep("c", 6),
81 | rep("d", 4),
82 | rep("e", 2),
83 | rep("f", 2),
84 | "g"
85 | )
86 |
87 | expect_equal(levels(fct_lump(f, w = w)), levels(fct_lump(f2)))
88 | expect_equal(
89 | levels(fct_lump(f, n = 1, w = w)),
90 | levels(fct_lump(f2, n = 1))
91 | )
92 | expect_equal(
93 | levels(fct_lump(f, n = -2, w = w, ties.method = "first")),
94 | levels(fct_lump(f2, n = -2, ties.method = "first"))
95 | )
96 | expect_equal(
97 | levels(fct_lump(f, n = 99, w = w)),
98 | levels(fct_lump(f2, n = 99))
99 | )
100 | expect_equal(
101 | levels(fct_lump(f, prop = 0.01, w = w)),
102 | levels(fct_lump(f2, prop = 0.01))
103 | )
104 | expect_equal(
105 | levels(fct_lump(f, prop = -0.25, w = w, ties.method = "max")),
106 | levels(fct_lump(f2, prop = -0.25, ties.method = "max"))
107 | )
108 | })
109 |
110 | test_that("do not change the label when no lumping occurs", {
111 | f <- c("a", "a", "a", "a", "b", "b", "b", "c", "c", "d")
112 | expect_equal(levels(fct_lump(f, n = 3)), c("a", "b", "c", "d"))
113 | expect_equal(levels(fct_lump(f, prop = 0.1)), c("a", "b", "c", "d"))
114 | })
115 |
116 | test_that("fct_lump_min works when not weighted", {
117 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
118 |
119 | expect_equal(levels(fct_lump_min(f, min = 3)), c("a", "Other"))
120 | expect_equal(levels(fct_lump_min(f, min = 2)), c("a", "b", "Other"))
121 | })
122 |
123 | test_that("fct_lump_min works when weighted", {
124 | f <- c("a", "b", "c", "d", "e")
125 | w <- c( 0.2, 2, 6, 4, 1)
126 |
127 | expect_equal(levels(fct_lump_min(f, min = 6, w = w)), c("c", "Other"))
128 | expect_equal(levels(fct_lump_min(f, min = 1.5, w = w)), c("b", "c", "d", "Other"))
129 | })
130 |
131 | # Default -----------------------------------------------------------------
132 |
133 | test_that("lumps smallest", {
134 | expect_equal(lump_test(c(1, 2, 3, 6)), "Xbcd")
135 | expect_equal(lump_test(c(1, 2, 3, 7)), "XXXd")
136 |
137 | expect_equal(lump_test(c(1, 2, 3, 7, 13)), "XXXde")
138 | expect_equal(lump_test(c(1, 2, 3, 7, 14)), "XXXXe")
139 | })
140 |
141 | test_that("doesn't lump if none small enough", {
142 | expect_equal(lump_test(c(2, 2, 4)), "abc")
143 | })
144 |
145 | test_that("order doesn't matter", {
146 | expect_equal(lump_test(c(2, 2, 5)), "XXc")
147 | expect_equal(lump_test(c(2, 5, 2)), "XbX")
148 | expect_equal(lump_test(c(5, 2, 2)), "aXX")
149 | })
150 |
--------------------------------------------------------------------------------
/vignettes/forcats.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | title: "Introduction to forcats"
3 | author: "Emily Robinson"
4 | output: rmarkdown::html_vignette
5 | vignette: >
6 | %\VignetteIndexEntry{Introduction to forcats}
7 | %\VignetteEngine{knitr::rmarkdown}
8 | %\VignetteEncoding{UTF-8}
9 | ---
10 |
11 | ```{r setup, include = FALSE}
12 | knitr::opts_chunk$set(
13 | collapse = TRUE,
14 | comment = "#>"
15 | )
16 | ```
17 |
18 | The goal of the __forcats__ package is to provide a suite of useful tools that solve common problems with factors. Factors are useful when you have categorical data, variables that have a fixed and known set of values, and when you want to display character vectors in non-alphabetical order. If you want to learn more, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
19 |
20 | ## Ordering by frequency
21 |
22 | ```{r message = FALSE}
23 | library(dplyr)
24 | library(ggplot2)
25 | library(forcats)
26 | ```
27 |
28 | Let's try answering the question, "what are the most common hair colors of star wars characters?" Let's start off by making a bar plot:
29 |
30 | ```{r initial-plot}
31 | ggplot(starwars, aes(x = hair_color)) +
32 | geom_bar() +
33 | coord_flip()
34 | ```
35 |
36 | That's okay, but it would be more helpful the graph was ordered by count. This is a case of an **unordered** categorical variable where we want it ordered by its frequency. To do so, we can use the function `fct_infreq()`:
37 |
38 | ```{r fct-infreq-hair}
39 | ggplot(starwars, aes(x = fct_infreq(hair_color))) +
40 | geom_bar() +
41 | coord_flip()
42 | ```
43 |
44 | Note that `fct_infreq()` it automatically puts NA at the top, even though that doesn't have the smallest number of entries.
45 |
46 | ## Combining levels
47 |
48 | Let's take a look at skin color now:
49 |
50 | ```{r}
51 | starwars %>%
52 | count(skin_color, sort = TRUE)
53 | ```
54 |
55 | We see that there's 31 different skin colors - if we want to make a plot this would be way too many to display! Let's reduce it to only be the top 5. We can use `fct_lump()` to "lump" all the infrequent colors into one factor, "other." The argument `n` is the number of levels we want to keep.
56 |
57 | ```{r}
58 | starwars %>%
59 | mutate(skin_color = fct_lump(skin_color, n = 5)) %>%
60 | count(skin_color, sort = TRUE)
61 | ```
62 |
63 | We could also have used `prop` instead, which keeps all the levels that appear at least `prop` of the time. For example, let's keep skin colors that at least 10% of the characters have:
64 |
65 | ```{r}
66 | starwars %>%
67 | mutate(skin_color = fct_lump(skin_color, prop = .1)) %>%
68 | count(skin_color, sort = TRUE)
69 | ```
70 |
71 | Only light and fair remain; everything else is other.
72 |
73 | If you wanted to call it something than "other", you can change it with the argument `other_level`:
74 |
75 | ```{r}
76 | starwars %>%
77 | mutate(skin_color = fct_lump(skin_color, prop = .1, other_level = "extra")) %>%
78 | count(skin_color, sort = TRUE)
79 | ```
80 |
81 | What if we wanted to see if the average mass differed by eye color? We'll only look at the 6 most popular eye colors and remove `NA`s.
82 |
83 | ```{r fct-lump-mean}
84 | avg_mass_eye_color <- starwars %>%
85 | mutate(eye_color = fct_lump(eye_color, n = 6)) %>%
86 | group_by(eye_color) %>%
87 | summarise(mean_mass = mean(mass, na.rm = TRUE))
88 |
89 | avg_mass_eye_color
90 | ```
91 |
92 | ## Ordering by another variable
93 |
94 | It looks like people (or at least one person) with orange eyes are definitely heavier! If we wanted to make a graph, it would be nice if it was ordered by `mean_mass`. We can do this with `fct_reorder()`, which reorders one variable by another.
95 |
96 | ```{r fct-reorder}
97 | avg_mass_eye_color %>%
98 | mutate(eye_color = fct_reorder(eye_color, mean_mass)) %>%
99 | ggplot(aes(x = eye_color, y = mean_mass)) +
100 | geom_col()
101 | ```
102 |
103 | ## Manually reordering
104 |
105 | Let's switch to using another dataset, `gss_cat`, the general social survey. What is the income distribution among the respondents?
106 |
107 | ```{r}
108 | gss_cat %>%
109 | count(rincome)
110 | ```
111 |
112 | Notice that the income levels are in the correct order - they start with the non-answers and then go from highest to lowest. This is the same order you'd see if you plotted it as a bar chart. This is not a coincidence. When you're working with ordinal data, where there is an order, you can have an ordered factor. You can examine them with the base function `levels()`, which prints them in order:
113 |
114 | ```{r}
115 | levels(gss_cat$rincome)
116 | ```
117 |
118 | But what if your factor came in the wrong order? Let's simulate that by reordering the levels of `rincome` randomly with `fct_shuffle()`:
119 |
120 | ```{r}
121 | reshuffled_income <- gss_cat$rincome %>%
122 | fct_shuffle()
123 |
124 | levels(reshuffled_income)
125 | ```
126 |
127 | Now if we plotted it, it would show in this order, which is all over the place! How can we fix this and put it in the right order?
128 |
129 | We can use the function `fct_relevel()` when we need to manually reorder our factor levels. In addition to the factor, you give it a character vector of level names, and specify where you want to move them. It defaults to moving them to the front, but you can move them after another level with the argument `after`. If you want to move it to the end, you set `after` equal to `Inf`.
130 |
131 | For example, let's say we wanted to move `Lt $1000` and `$1000 to 2999` to the front. We would write:
132 |
133 | ```{r}
134 | fct_relevel(reshuffled_income, c("Lt $1000", "$1000 to 2999")) %>%
135 | levels()
136 | ```
137 |
138 | What if we want to move them to the second and third place?
139 |
140 | ```{r}
141 | fct_relevel(reshuffled_income, c("Lt $1000", "$1000 to 2999"), after = 1) %>%
142 | levels()
143 | ```
144 |
--------------------------------------------------------------------------------
/R/lump.R:
--------------------------------------------------------------------------------
1 | #' Lump together factor levels into "other"
2 | #'
3 | #' `fct_lump()` lumps together least/most common levels (as defined by `n` and
4 | #' `prop`) into "other".
5 | #' `fct_lump_min()` lumps together all levels which don't appear at least
6 | #' `min` number of times.
7 | #'
8 | #' @param f A factor (or character vector).
9 | #' @param n,prop
10 | #' If both `n` and `prop` are missing, `fct_lump()` lumps
11 | #' together the least frequent levels into "other", while ensuring that
12 | #' "other" is still the smallest level. It's particularly useful in
13 | #' conjunction with \code{\link{fct_inorder}()}.
14 | #'
15 | #' Positive `n` preserves the most common `n` values.
16 | #' Negative `n` preserves the least common `-n` values.
17 | #' It there are ties, you will get at least `abs(n)` values.
18 | #'
19 | #' Positive `prop` preserves values that appear at least
20 | #' `prop` of the time. Negative `prop` preserves values that
21 | #' appear at most `-prop` of the time.
22 | #' @param w An optional numeric vector giving weights for frequency of
23 | #' each value (not level) in f.
24 | #' @param other_level Value of level used for "other" values. Always
25 | #' placed at end of levels.
26 | #' @param ties.method A character string specifying how ties are
27 | #' treated. See [rank()] for details.
28 | #' @export
29 | #' @seealso [fct_other()] to convert specified levels to other.
30 | #' @examples
31 | #' x <- factor(rep(LETTERS[1:9], times = c(40, 10, 5, 27, 1, 1, 1, 1, 1)))
32 | #' x %>% table()
33 | #' x %>% fct_lump() %>% table()
34 | #' x %>% fct_lump() %>% fct_inorder() %>% table()
35 | #'
36 | #' x <- factor(letters[rpois(100, 5)])
37 | #' x
38 | #' table(x)
39 | #' table(fct_lump(x))
40 | #'
41 | #' # Use positive values to collapse the rarest
42 | #' fct_lump(x, n = 3)
43 | #' fct_lump(x, prop = 0.1)
44 | #'
45 | #' # Use negative values to collapse the most common
46 | #' fct_lump(x, n = -3)
47 | #' fct_lump(x, prop = -0.1)
48 | #'
49 | #' # Use weighted frequencies
50 | #' w <- c(rep(2, 50), rep(1, 50))
51 | #' fct_lump(x, n = 5, w = w)
52 | #'
53 | #' # Use ties.method to control how tied factors are collapsed
54 | #' fct_lump(x, n = 6)
55 | #' fct_lump(x, n = 6, ties.method = "max")
56 | #'
57 | fct_lump <- function(f, n, prop, w = NULL, other_level = "Other",
58 | ties.method = c("min", "average", "first", "last", "random", "max")) {
59 | f <- check_factor(f)
60 | w <- check_weights(w, length(f))
61 | ties.method <- match.arg(ties.method)
62 |
63 | levels <- levels(f)
64 | if (is.null(w)) {
65 | count <- as.vector(table(f))
66 | total <- length(f)
67 | } else {
68 | count <- as.vector(tapply(w, f, FUN = sum))
69 | total <- sum(w)
70 | }
71 |
72 | if (!xor(missing(n), missing(prop))) {
73 | new_levels <- ifelse(!in_smallest(count), levels, other_level)
74 | } else if (!missing(n)) {
75 | if (n < 0) {
76 | rank <- rank(count, ties = ties.method)
77 | n <- -n
78 | } else {
79 | rank <- rank(-count, ties = ties.method)
80 | }
81 |
82 | if (sum(rank > n) <= 1) {
83 | # No lumping needed
84 | return(f)
85 | }
86 |
87 | new_levels <- ifelse(rank <= n, levels, other_level)
88 | } else if (!missing(prop)) {
89 | prop_n <- count / total
90 | if (prop < 0) {
91 | new_levels <- ifelse(prop_n <= -prop, levels, other_level)
92 | } else {
93 | if (sum(prop_n <= prop) <= 1) {
94 | # No lumping needed
95 | return(f)
96 | }
97 |
98 | new_levels <- ifelse(prop_n > prop, levels, other_level)
99 | }
100 | }
101 |
102 | if (other_level %in% new_levels) {
103 | f <- lvls_revalue(f, new_levels)
104 | fct_relevel(f, other_level, after = Inf)
105 | } else {
106 | f
107 | }
108 | }
109 |
110 | #' @param min Preserves values that appear at least `min` number of times.
111 | #' @export
112 | #' @rdname fct_lump
113 | #' @examples
114 | #' x <- factor(letters[rpois(100, 5)])
115 | #' fct_lump_min(x, min = 10)
116 | fct_lump_min <- function(f, min, w = NULL, other_level = "Other") {
117 | f <- check_factor(f)
118 | w <- check_weights(w, length(f))
119 |
120 | levels <- levels(f)
121 | if (is.null(w)) {
122 | count <- as.vector(table(f))
123 | total <- length(f)
124 | } else {
125 | count <- as.vector(tapply(w, f, FUN = sum))
126 | total <- sum(w)
127 | }
128 |
129 | new_levels <- ifelse(count >= min, levels, other_level)
130 |
131 | if (other_level %in% new_levels) {
132 | f <- lvls_revalue(f, new_levels)
133 | fct_relevel(f, other_level, after = Inf)
134 | } else {
135 | f
136 | }
137 |
138 | }
139 |
140 | # Lump together smallest groups, ensuring that the collective
141 | # "other" is still the smallest group. Assumes x is vector
142 | # of counts in descending order
143 | lump_cutoff <- function(x) {
144 | left <- sum(x)
145 |
146 | for (i in seq_along(x)) {
147 | # After group, there are this many left
148 | left <- left - x[i]
149 |
150 | if (x[i] > left)
151 | return(i + 1)
152 | }
153 |
154 | length(x) + 1
155 | }
156 |
157 | # Given vector of counts, returns logical vector if in
158 | # smallest groups
159 | in_smallest <- function(x) {
160 | ord_x <- order(x, decreasing = TRUE)
161 | idx <- lump_cutoff(x[ord_x])
162 |
163 | to_lump <- seq_along(x) >= idx
164 | # Undo initial ordering
165 | to_lump[order(ord_x)]
166 | }
167 |
168 | check_weights <- function(w, n = length(w)) {
169 | if (is.null(w)) {
170 | return(w)
171 | }
172 |
173 | if (!is.numeric(w)) {
174 | stop("`w` must be a numeric vector", call. = FALSE)
175 | }
176 |
177 | if (length(w) != n) {
178 | stop(
179 | "`w` must be the same length as `f` (", n, "), not length ", length(w),
180 | call. = FALSE
181 | )
182 | }
183 |
184 | bad <- w < 0 | is.na(w)
185 | if (any(bad)) {
186 | stop(
187 | "All `w` must be non-negative and non-missing. Problems at positions: ",
188 | paste0(which(bad), collapse = ", "),
189 | call. = FALSE
190 | )
191 | }
192 |
193 | w
194 | }
195 |
--------------------------------------------------------------------------------
/revdep/problems.md:
--------------------------------------------------------------------------------
1 | # bupaR
2 |
3 | Version: 0.4.1
4 |
5 | ## In both
6 |
7 | * checking S3 generic/method consistency ... WARNING
8 | ```
9 | sample_n:
10 | function(tbl, size, replace, weight, .env, ...)
11 | sample_n.eventlog:
12 | function(tbl, size, replace, weight, .env)
13 |
14 | sample_n:
15 | function(tbl, size, replace, weight, .env, ...)
16 | sample_n.grouped_eventlog:
17 | function(tbl, size, replace, weight, .env)
18 |
19 | See section ‘Generic functions and methods’ in the ‘Writing R
20 | Extensions’ manual.
21 | ```
22 |
23 | # circumplex
24 |
25 | Version: 0.2.1
26 |
27 | ## In both
28 |
29 | * checking whether package ‘circumplex’ can be installed ... ERROR
30 | ```
31 | Installation failed.
32 | See ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/new/circumplex.Rcheck/00install.out’ for details.
33 | ```
34 |
35 | ## Installation
36 |
37 | ### Devel
38 |
39 | ```
40 | * installing *source* package ‘circumplex’ ...
41 | ** package ‘circumplex’ successfully unpacked and MD5 sums checked
42 | ** libs
43 | clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/Rcpp/include" -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/RcppArmadillo/include" -I/usr/local/include -fopenmp -fPIC -Wall -g -O2 -c RcppExports.cpp -o RcppExports.o
44 | clang: error: unsupported option '-fopenmp'
45 | make: *** [RcppExports.o] Error 1
46 | ERROR: compilation failed for package ‘circumplex’
47 | * removing ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/new/circumplex.Rcheck/circumplex’
48 |
49 | ```
50 | ### CRAN
51 |
52 | ```
53 | * installing *source* package ‘circumplex’ ...
54 | ** package ‘circumplex’ successfully unpacked and MD5 sums checked
55 | ** libs
56 | clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/Rcpp/include" -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/RcppArmadillo/include" -I/usr/local/include -fopenmp -fPIC -Wall -g -O2 -c RcppExports.cpp -o RcppExports.o
57 | clang: error: unsupported option '-fopenmp'
58 | make: *** [RcppExports.o] Error 1
59 | ERROR: compilation failed for package ‘circumplex’
60 | * removing ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/old/circumplex.Rcheck/circumplex’
61 |
62 | ```
63 | # cocktailApp
64 |
65 | Version: 0.2.0
66 |
67 | ## In both
68 |
69 | * checking data for non-ASCII characters ... NOTE
70 | ```
71 | Note: found 14661 marked UTF-8 strings
72 | ```
73 |
74 | # dabestr
75 |
76 | Version: 0.2.0
77 |
78 | ## In both
79 |
80 | * checking re-building of vignette outputs ... WARNING
81 | ```
82 | ...
83 |
84 | The following objects are masked from 'package:base':
85 |
86 | intersect, setdiff, setequal, union
87 |
88 |
89 | Attaching package: 'cowplot'
90 |
91 | The following object is masked from 'package:ggplot2':
92 |
93 | ggsave
94 |
95 | Loading required package: boot
96 | Warning: `data_frame()` is deprecated, use `tibble()`.
97 | This warning is displayed once per session.
98 | Loading required package: magrittr
99 | Warning: Some components of ... were not used: ..1
100 | Quitting from lines 110-166 (robust-statistical-visualization.Rmd)
101 | Error: processing vignette 'robust-statistical-visualization.Rmd' failed with diagnostics:
102 | polygon edge not found
103 | Execution halted
104 | ```
105 |
106 | * checking installed package size ... NOTE
107 | ```
108 | installed size is 6.6Mb
109 | sub-directories of 1Mb or more:
110 | doc 5.8Mb
111 | ```
112 |
113 | # ezplot
114 |
115 | Version: 0.2.2
116 |
117 | ## In both
118 |
119 | * checking dependencies in R code ... NOTE
120 | ```
121 | Namespace in Imports field not imported from: ‘magrittr’
122 | All declared Imports should be used.
123 | ```
124 |
125 | # factorMerger
126 |
127 | Version: 0.3.6
128 |
129 | ## In both
130 |
131 | * checking dependencies in R code ... NOTE
132 | ```
133 | Namespaces in Imports field not imported from:
134 | ‘forcats’ ‘formula.tools’
135 | All declared Imports should be used.
136 | ```
137 |
138 | # finalfit
139 |
140 | Version: 0.9.0
141 |
142 | ## In both
143 |
144 | * checking dependencies in R code ... NOTE
145 | ```
146 | Namespace in Imports field not imported from: ‘readr’
147 | All declared Imports should be used.
148 | ```
149 |
150 | # furniture
151 |
152 | Version: 1.8.7
153 |
154 | ## In both
155 |
156 | * checking dependencies in R code ... NOTE
157 | ```
158 | Namespace in Imports field not imported from: ‘magrittr’
159 | All declared Imports should be used.
160 | ```
161 |
162 | # ggdag
163 |
164 | Version: 0.1.0
165 |
166 | ## In both
167 |
168 | * checking dependencies in R code ... NOTE
169 | ```
170 | Namespaces in Imports field not imported from:
171 | ‘ggforce’ ‘plyr’
172 | All declared Imports should be used.
173 | ```
174 |
175 | # ggridges
176 |
177 | Version: 0.5.1
178 |
179 | ## In both
180 |
181 | * checking data for non-ASCII characters ... NOTE
182 | ```
183 | Note: found 6242 marked UTF-8 strings
184 | ```
185 |
186 | # glmmfields
187 |
188 | Version: 0.1.1
189 |
190 | ## In both
191 |
192 | * checking for GNU extensions in Makefiles ... NOTE
193 | ```
194 | GNU make is a SystemRequirements.
195 | ```
196 |
197 | # glmSparseNet
198 |
199 | Version: 1.0.0
200 |
201 | ## In both
202 |
203 | * checking re-building of vignette outputs ... WARNING
204 | ```
205 | Error in re-building vignettes:
206 | ...
207 |
208 | Attaching package: 'dplyr'
209 |
210 | The following objects are masked from 'package:stats':
211 |
212 | filter, lag
213 |
214 | The following objects are masked from 'package:base':
215 |
216 | intersect, setdiff, setequal, union
217 |
218 | Quitting from lines 28-43 (example_brca_logistic.Rmd)
219 | Error: processing vignette 'example_brca_logistic.Rmd' failed with diagnostics:
220 | there is no package called 'curatedTCGAData'
221 | Execution halted
222 | ```
223 |
224 | * checking package dependencies ... NOTE
225 | ```
226 | Package suggested but not available for checking: ‘curatedTCGAData’
227 | ```
228 |
229 | * checking installed package size ... NOTE
230 | ```
231 | installed size is 6.0Mb
232 | sub-directories of 1Mb or more:
233 | doc 5.2Mb
234 | ```
235 |
236 | # haven
237 |
238 | Version: 2.0.0
239 |
240 | ## In both
241 |
242 | * checking for GNU extensions in Makefiles ... NOTE
243 | ```
244 | GNU make is a SystemRequirements.
245 | ```
246 |
247 | # idealstan
248 |
249 | Version: 0.5.1
250 |
251 | ## In both
252 |
253 | * checking installed package size ... NOTE
254 | ```
255 | installed size is 6.2Mb
256 | sub-directories of 1Mb or more:
257 | libs 4.1Mb
258 | ```
259 |
260 | * checking for GNU extensions in Makefiles ... NOTE
261 | ```
262 | GNU make is a SystemRequirements.
263 | ```
264 |
265 | # iotables
266 |
267 | Version: 0.4.2
268 |
269 | ## In both
270 |
271 | * checking data for non-ASCII characters ... NOTE
272 | ```
273 | Note: found 53206 marked UTF-8 strings
274 | ```
275 |
276 | # konfound
277 |
278 | Version: 0.1.1
279 |
280 | ## In both
281 |
282 | * checking re-building of vignette outputs ... WARNING
283 | ```
284 | Error in re-building vignettes:
285 | ...
286 | Quitting from lines 50-51 (introduction-to-konfound.Rmd)
287 | Error: processing vignette 'introduction-to-konfound.Rmd' failed with diagnostics:
288 | there is no package called 'devtools'
289 | Execution halted
290 | ```
291 |
292 | * checking package dependencies ... NOTE
293 | ```
294 | Package suggested but not available for checking: ‘devtools’
295 | ```
296 |
297 | # MetamapsDB
298 |
299 | Version: 0.0.2
300 |
301 | ## In both
302 |
303 | * checking dependencies in R code ... NOTE
304 | ```
305 | Namespaces in Imports field not imported from:
306 | ‘Matrix’ ‘shiny’
307 | All declared Imports should be used.
308 | ```
309 |
310 | # noaastormevents
311 |
312 | Version: 0.1.0
313 |
314 | ## In both
315 |
316 | * checking package dependencies ... NOTE
317 | ```
318 | Package suggested but not available for checking: ‘hurricaneexposuredata’
319 | ```
320 |
321 | * checking dependencies in R code ... NOTE
322 | ```
323 | Namespaces in Imports field not imported from:
324 | ‘RColorBrewer’ ‘XML’ ‘choroplethr’ ‘choroplethrMaps’ ‘data.table’
325 | ‘forcats’ ‘hurricaneexposure’ ‘plyr’
326 | All declared Imports should be used.
327 | ```
328 |
329 | # nzelect
330 |
331 | Version: 0.4.0
332 |
333 | ## In both
334 |
335 | * checking installed package size ... NOTE
336 | ```
337 | installed size is 5.4Mb
338 | sub-directories of 1Mb or more:
339 | data 5.0Mb
340 | ```
341 |
342 | * checking data for non-ASCII characters ... NOTE
343 | ```
344 | Note: found 6409 marked UTF-8 strings
345 | ```
346 |
347 | # phenopath
348 |
349 | Version: 1.6.0
350 |
351 | ## In both
352 |
353 | * checking re-building of vignette outputs ... WARNING
354 | ```
355 | Error in re-building vignettes:
356 | ...
357 | Quitting from lines 64-72 (introduction_to_phenopath.Rmd)
358 | Error: processing vignette 'introduction_to_phenopath.Rmd' failed with diagnostics:
359 | Columns 1, 2, 3, 4, 5, … (and 3 more) must be named.
360 | Use .name_repair to specify repair.
361 | Execution halted
362 | ```
363 |
364 | # plotly
365 |
366 | Version: 4.8.0
367 |
368 | ## In both
369 |
370 | * checking package dependencies ... NOTE
371 | ```
372 | Package suggested but not available for checking: ‘devtools’
373 | ```
374 |
375 | * checking installed package size ... NOTE
376 | ```
377 | installed size is 7.0Mb
378 | sub-directories of 1Mb or more:
379 | R 2.3Mb
380 | htmlwidgets 3.1Mb
381 | ```
382 |
383 | # processmapR
384 |
385 | Version: 0.3.2
386 |
387 | ## In both
388 |
389 | * checking dependencies in R code ... NOTE
390 | ```
391 | Namespace in Imports field not imported from: ‘viridis’
392 | All declared Imports should be used.
393 | ```
394 |
395 | # questionr
396 |
397 | Version: 0.7.0
398 |
399 | ## In both
400 |
401 | * checking Rd cross-references ... NOTE
402 | ```
403 | Package unavailable to check Rd xrefs: ‘Hmisc’
404 | ```
405 |
406 | * checking data for non-ASCII characters ... NOTE
407 | ```
408 | Note: found 4145 marked UTF-8 strings
409 | ```
410 |
411 | # railtrails
412 |
413 | Version: 0.1.1
414 |
415 | ## In both
416 |
417 | * checking data for non-ASCII characters ... NOTE
418 | ```
419 | Note: found 1557 marked UTF-8 strings
420 | ```
421 |
422 | # rbin
423 |
424 | Version: 0.1.1
425 |
426 | ## In both
427 |
428 | * checking dependencies in R code ... NOTE
429 | ```
430 | Namespace in Imports field not imported from: ‘utils’
431 | All declared Imports should be used.
432 | ```
433 |
434 | # SCnorm
435 |
436 | Version: 1.4.5
437 |
438 | ## In both
439 |
440 | * checking package dependencies ... NOTE
441 | ```
442 | Package suggested but not available for checking: ‘devtools’
443 | ```
444 |
445 | # sweep
446 |
447 | Version: 0.2.1.1
448 |
449 | ## In both
450 |
451 | * checking package dependencies ... ERROR
452 | ```
453 | Package required but not available: ‘devtools’
454 |
455 | See section ‘The DESCRIPTION file’ in the ‘Writing R Extensions’
456 | manual.
457 | ```
458 |
459 | # TextForecast
460 |
461 | Version: 0.1.0
462 |
463 | ## In both
464 |
465 | * checking dependencies in R code ... NOTE
466 | ```
467 | Namespaces in Imports field not imported from:
468 | ‘doParallel’ ‘forecast’ ‘lars’ ‘parallel’ ‘tau’ ‘tsDyn’
469 | All declared Imports should be used.
470 | ```
471 |
472 | * checking data for non-ASCII characters ... NOTE
473 | ```
474 | Note: found 40 marked UTF-8 strings
475 | ```
476 |
477 | # tidybayes
478 |
479 | Version: 1.0.3
480 |
481 | ## In both
482 |
483 | * checking tests ...
484 | ```
485 | ERROR
486 | Running the tests in ‘tests/testthat.R’ failed.
487 | Last 13 lines of output:
488 | 9: setup.jagsfile(model = model, n.chains = n.chains, data = data, inits = inits, monitor = monitor, modules = modules,
489 | factories = factories, jags = jags, call.setup = TRUE, method = method, mutate = mutate)
490 | 10: setup.jags(model = outmodel, monitor = outmonitor, data = outdata, n.chains = n.chains, inits = outinits, modules = modules,
491 | factories = factories, response = response, fitted = fitted, residual = residual, jags = jags, method = method,
492 | mutate = mutate)
493 | 11: loadandcheckrjags()
494 | 12: stop("Loading the rjags package failed (diagnostics are given above this error message)", call. = FALSE)
495 |
496 | ══ testthat results ═════════════════════════════════════════════════════════════════════════════════════
497 | OK: 218 SKIPPED: 43 FAILED: 2
498 | 1. Failure: groups from spread_draws retain factor level names (@test.spread_draws.R#251)
499 | 2. Error: tidy_draws works with runjags (@test.tidy_draws.R#87)
500 |
501 | Error: testthat unit tests failed
502 | Execution halted
503 | ```
504 |
505 | # tidyLPA
506 |
507 | Version: 0.2.4
508 |
509 | ## In both
510 |
511 | * checking package dependencies ... NOTE
512 | ```
513 | Package suggested but not available for checking: ‘devtools’
514 | ```
515 |
516 | # tidyverse
517 |
518 | Version: 1.2.1
519 |
520 | ## In both
521 |
522 | * checking dependencies in R code ... NOTE
523 | ```
524 | Namespaces in Imports field not imported from:
525 | ‘dbplyr’ ‘reprex’ ‘rlang’
526 | All declared Imports should be used.
527 | ```
528 |
529 | # timetk
530 |
531 | Version: 0.1.1.1
532 |
533 | ## In both
534 |
535 | * checking package dependencies ... ERROR
536 | ```
537 | Package required but not available: ‘devtools’
538 |
539 | See section ‘The DESCRIPTION file’ in the ‘Writing R Extensions’
540 | manual.
541 | ```
542 |
543 |
--------------------------------------------------------------------------------
/LICENSE.md:
--------------------------------------------------------------------------------
1 | GNU General Public License
2 | ==========================
3 |
4 | _Version 3, 29 June 2007_
5 | _Copyright © 2007 Free Software Foundation, Inc. <>_
6 |
7 | Everyone is permitted to copy and distribute verbatim copies of this license
8 | document, but changing it is not allowed.
9 |
10 | ## Preamble
11 |
12 | The GNU General Public License is a free, copyleft license for software and other
13 | kinds of works.
14 |
15 | The licenses for most software and other practical works are designed to take away
16 | your freedom to share and change the works. By contrast, the GNU General Public
17 | License is intended to guarantee your freedom to share and change all versions of a
18 | program--to make sure it remains free software for all its users. We, the Free
19 | Software Foundation, use the GNU General Public License for most of our software; it
20 | applies also to any other work released this way by its authors. You can apply it to
21 | your programs, too.
22 |
23 | When we speak of free software, we are referring to freedom, not price. Our General
24 | Public Licenses are designed to make sure that you have the freedom to distribute
25 | copies of free software (and charge for them if you wish), that you receive source
26 | code or can get it if you want it, that you can change the software or use pieces of
27 | it in new free programs, and that you know you can do these things.
28 |
29 | To protect your rights, we need to prevent others from denying you these rights or
30 | asking you to surrender the rights. Therefore, you have certain responsibilities if
31 | you distribute copies of the software, or if you modify it: responsibilities to
32 | respect the freedom of others.
33 |
34 | For example, if you distribute copies of such a program, whether gratis or for a fee,
35 | you must pass on to the recipients the same freedoms that you received. You must make
36 | sure that they, too, receive or can get the source code. And you must show them these
37 | terms so they know their rights.
38 |
39 | Developers that use the GNU GPL protect your rights with two steps: **(1)** assert
40 | copyright on the software, and **(2)** offer you this License giving you legal permission
41 | to copy, distribute and/or modify it.
42 |
43 | For the developers' and authors' protection, the GPL clearly explains that there is
44 | no warranty for this free software. For both users' and authors' sake, the GPL
45 | requires that modified versions be marked as changed, so that their problems will not
46 | be attributed erroneously to authors of previous versions.
47 |
48 | Some devices are designed to deny users access to install or run modified versions of
49 | the software inside them, although the manufacturer can do so. This is fundamentally
50 | incompatible with the aim of protecting users' freedom to change the software. The
51 | systematic pattern of such abuse occurs in the area of products for individuals to
52 | use, which is precisely where it is most unacceptable. Therefore, we have designed
53 | this version of the GPL to prohibit the practice for those products. If such problems
54 | arise substantially in other domains, we stand ready to extend this provision to
55 | those domains in future versions of the GPL, as needed to protect the freedom of
56 | users.
57 |
58 | Finally, every program is threatened constantly by software patents. States should
59 | not allow patents to restrict development and use of software on general-purpose
60 | computers, but in those that do, we wish to avoid the special danger that patents
61 | applied to a free program could make it effectively proprietary. To prevent this, the
62 | GPL assures that patents cannot be used to render the program non-free.
63 |
64 | The precise terms and conditions for copying, distribution and modification follow.
65 |
66 | ## TERMS AND CONDITIONS
67 |
68 | ### 0. Definitions
69 |
70 | “This License” refers to version 3 of the GNU General Public License.
71 |
72 | “Copyright” also means copyright-like laws that apply to other kinds of
73 | works, such as semiconductor masks.
74 |
75 | “The Program” refers to any copyrightable work licensed under this
76 | License. Each licensee is addressed as “you”. “Licensees” and
77 | “recipients” may be individuals or organizations.
78 |
79 | To “modify” a work means to copy from or adapt all or part of the work in
80 | a fashion requiring copyright permission, other than the making of an exact copy. The
81 | resulting work is called a “modified version” of the earlier work or a
82 | work “based on” the earlier work.
83 |
84 | A “covered work” means either the unmodified Program or a work based on
85 | the Program.
86 |
87 | To “propagate” a work means to do anything with it that, without
88 | permission, would make you directly or secondarily liable for infringement under
89 | applicable copyright law, except executing it on a computer or modifying a private
90 | copy. Propagation includes copying, distribution (with or without modification),
91 | making available to the public, and in some countries other activities as well.
92 |
93 | To “convey” a work means any kind of propagation that enables other
94 | parties to make or receive copies. Mere interaction with a user through a computer
95 | network, with no transfer of a copy, is not conveying.
96 |
97 | An interactive user interface displays “Appropriate Legal Notices” to the
98 | extent that it includes a convenient and prominently visible feature that **(1)**
99 | displays an appropriate copyright notice, and **(2)** tells the user that there is no
100 | warranty for the work (except to the extent that warranties are provided), that
101 | licensees may convey the work under this License, and how to view a copy of this
102 | License. If the interface presents a list of user commands or options, such as a
103 | menu, a prominent item in the list meets this criterion.
104 |
105 | ### 1. Source Code
106 |
107 | The “source code” for a work means the preferred form of the work for
108 | making modifications to it. “Object code” means any non-source form of a
109 | work.
110 |
111 | A “Standard Interface” means an interface that either is an official
112 | standard defined by a recognized standards body, or, in the case of interfaces
113 | specified for a particular programming language, one that is widely used among
114 | developers working in that language.
115 |
116 | The “System Libraries” of an executable work include anything, other than
117 | the work as a whole, that **(a)** is included in the normal form of packaging a Major
118 | Component, but which is not part of that Major Component, and **(b)** serves only to
119 | enable use of the work with that Major Component, or to implement a Standard
120 | Interface for which an implementation is available to the public in source code form.
121 | A “Major Component”, in this context, means a major essential component
122 | (kernel, window system, and so on) of the specific operating system (if any) on which
123 | the executable work runs, or a compiler used to produce the work, or an object code
124 | interpreter used to run it.
125 |
126 | The “Corresponding Source” for a work in object code form means all the
127 | source code needed to generate, install, and (for an executable work) run the object
128 | code and to modify the work, including scripts to control those activities. However,
129 | it does not include the work's System Libraries, or general-purpose tools or
130 | generally available free programs which are used unmodified in performing those
131 | activities but which are not part of the work. For example, Corresponding Source
132 | includes interface definition files associated with source files for the work, and
133 | the source code for shared libraries and dynamically linked subprograms that the work
134 | is specifically designed to require, such as by intimate data communication or
135 | control flow between those subprograms and other parts of the work.
136 |
137 | The Corresponding Source need not include anything that users can regenerate
138 | automatically from other parts of the Corresponding Source.
139 |
140 | The Corresponding Source for a work in source code form is that same work.
141 |
142 | ### 2. Basic Permissions
143 |
144 | All rights granted under this License are granted for the term of copyright on the
145 | Program, and are irrevocable provided the stated conditions are met. This License
146 | explicitly affirms your unlimited permission to run the unmodified Program. The
147 | output from running a covered work is covered by this License only if the output,
148 | given its content, constitutes a covered work. This License acknowledges your rights
149 | of fair use or other equivalent, as provided by copyright law.
150 |
151 | You may make, run and propagate covered works that you do not convey, without
152 | conditions so long as your license otherwise remains in force. You may convey covered
153 | works to others for the sole purpose of having them make modifications exclusively
154 | for you, or provide you with facilities for running those works, provided that you
155 | comply with the terms of this License in conveying all material for which you do not
156 | control copyright. Those thus making or running the covered works for you must do so
157 | exclusively on your behalf, under your direction and control, on terms that prohibit
158 | them from making any copies of your copyrighted material outside their relationship
159 | with you.
160 |
161 | Conveying under any other circumstances is permitted solely under the conditions
162 | stated below. Sublicensing is not allowed; section 10 makes it unnecessary.
163 |
164 | ### 3. Protecting Users' Legal Rights From Anti-Circumvention Law
165 |
166 | No covered work shall be deemed part of an effective technological measure under any
167 | applicable law fulfilling obligations under article 11 of the WIPO copyright treaty
168 | adopted on 20 December 1996, or similar laws prohibiting or restricting circumvention
169 | of such measures.
170 |
171 | When you convey a covered work, you waive any legal power to forbid circumvention of
172 | technological measures to the extent such circumvention is effected by exercising
173 | rights under this License with respect to the covered work, and you disclaim any
174 | intention to limit operation or modification of the work as a means of enforcing,
175 | against the work's users, your or third parties' legal rights to forbid circumvention
176 | of technological measures.
177 |
178 | ### 4. Conveying Verbatim Copies
179 |
180 | You may convey verbatim copies of the Program's source code as you receive it, in any
181 | medium, provided that you conspicuously and appropriately publish on each copy an
182 | appropriate copyright notice; keep intact all notices stating that this License and
183 | any non-permissive terms added in accord with section 7 apply to the code; keep
184 | intact all notices of the absence of any warranty; and give all recipients a copy of
185 | this License along with the Program.
186 |
187 | You may charge any price or no price for each copy that you convey, and you may offer
188 | support or warranty protection for a fee.
189 |
190 | ### 5. Conveying Modified Source Versions
191 |
192 | You may convey a work based on the Program, or the modifications to produce it from
193 | the Program, in the form of source code under the terms of section 4, provided that
194 | you also meet all of these conditions:
195 |
196 | * **a)** The work must carry prominent notices stating that you modified it, and giving a
197 | relevant date.
198 | * **b)** The work must carry prominent notices stating that it is released under this
199 | License and any conditions added under section 7. This requirement modifies the
200 | requirement in section 4 to “keep intact all notices”.
201 | * **c)** You must license the entire work, as a whole, under this License to anyone who
202 | comes into possession of a copy. This License will therefore apply, along with any
203 | applicable section 7 additional terms, to the whole of the work, and all its parts,
204 | regardless of how they are packaged. This License gives no permission to license the
205 | work in any other way, but it does not invalidate such permission if you have
206 | separately received it.
207 | * **d)** If the work has interactive user interfaces, each must display Appropriate Legal
208 | Notices; however, if the Program has interactive interfaces that do not display
209 | Appropriate Legal Notices, your work need not make them do so.
210 |
211 | A compilation of a covered work with other separate and independent works, which are
212 | not by their nature extensions of the covered work, and which are not combined with
213 | it such as to form a larger program, in or on a volume of a storage or distribution
214 | medium, is called an “aggregate” if the compilation and its resulting
215 | copyright are not used to limit the access or legal rights of the compilation's users
216 | beyond what the individual works permit. Inclusion of a covered work in an aggregate
217 | does not cause this License to apply to the other parts of the aggregate.
218 |
219 | ### 6. Conveying Non-Source Forms
220 |
221 | You may convey a covered work in object code form under the terms of sections 4 and
222 | 5, provided that you also convey the machine-readable Corresponding Source under the
223 | terms of this License, in one of these ways:
224 |
225 | * **a)** Convey the object code in, or embodied in, a physical product (including a
226 | physical distribution medium), accompanied by the Corresponding Source fixed on a
227 | durable physical medium customarily used for software interchange.
228 | * **b)** Convey the object code in, or embodied in, a physical product (including a
229 | physical distribution medium), accompanied by a written offer, valid for at least
230 | three years and valid for as long as you offer spare parts or customer support for
231 | that product model, to give anyone who possesses the object code either **(1)** a copy of
232 | the Corresponding Source for all the software in the product that is covered by this
233 | License, on a durable physical medium customarily used for software interchange, for
234 | a price no more than your reasonable cost of physically performing this conveying of
235 | source, or **(2)** access to copy the Corresponding Source from a network server at no
236 | charge.
237 | * **c)** Convey individual copies of the object code with a copy of the written offer to
238 | provide the Corresponding Source. This alternative is allowed only occasionally and
239 | noncommercially, and only if you received the object code with such an offer, in
240 | accord with subsection 6b.
241 | * **d)** Convey the object code by offering access from a designated place (gratis or for
242 | a charge), and offer equivalent access to the Corresponding Source in the same way
243 | through the same place at no further charge. You need not require recipients to copy
244 | the Corresponding Source along with the object code. If the place to copy the object
245 | code is a network server, the Corresponding Source may be on a different server
246 | (operated by you or a third party) that supports equivalent copying facilities,
247 | provided you maintain clear directions next to the object code saying where to find
248 | the Corresponding Source. Regardless of what server hosts the Corresponding Source,
249 | you remain obligated to ensure that it is available for as long as needed to satisfy
250 | these requirements.
251 | * **e)** Convey the object code using peer-to-peer transmission, provided you inform
252 | other peers where the object code and Corresponding Source of the work are being
253 | offered to the general public at no charge under subsection 6d.
254 |
255 | A separable portion of the object code, whose source code is excluded from the
256 | Corresponding Source as a System Library, need not be included in conveying the
257 | object code work.
258 |
259 | A “User Product” is either **(1)** a “consumer product”, which
260 | means any tangible personal property which is normally used for personal, family, or
261 | household purposes, or **(2)** anything designed or sold for incorporation into a
262 | dwelling. In determining whether a product is a consumer product, doubtful cases
263 | shall be resolved in favor of coverage. For a particular product received by a
264 | particular user, “normally used” refers to a typical or common use of
265 | that class of product, regardless of the status of the particular user or of the way
266 | in which the particular user actually uses, or expects or is expected to use, the
267 | product. A product is a consumer product regardless of whether the product has
268 | substantial commercial, industrial or non-consumer uses, unless such uses represent
269 | the only significant mode of use of the product.
270 |
271 | “Installation Information” for a User Product means any methods,
272 | procedures, authorization keys, or other information required to install and execute
273 | modified versions of a covered work in that User Product from a modified version of
274 | its Corresponding Source. The information must suffice to ensure that the continued
275 | functioning of the modified object code is in no case prevented or interfered with
276 | solely because modification has been made.
277 |
278 | If you convey an object code work under this section in, or with, or specifically for
279 | use in, a User Product, and the conveying occurs as part of a transaction in which
280 | the right of possession and use of the User Product is transferred to the recipient
281 | in perpetuity or for a fixed term (regardless of how the transaction is
282 | characterized), the Corresponding Source conveyed under this section must be
283 | accompanied by the Installation Information. But this requirement does not apply if
284 | neither you nor any third party retains the ability to install modified object code
285 | on the User Product (for example, the work has been installed in ROM).
286 |
287 | The requirement to provide Installation Information does not include a requirement to
288 | continue to provide support service, warranty, or updates for a work that has been
289 | modified or installed by the recipient, or for the User Product in which it has been
290 | modified or installed. Access to a network may be denied when the modification itself
291 | materially and adversely affects the operation of the network or violates the rules
292 | and protocols for communication across the network.
293 |
294 | Corresponding Source conveyed, and Installation Information provided, in accord with
295 | this section must be in a format that is publicly documented (and with an
296 | implementation available to the public in source code form), and must require no
297 | special password or key for unpacking, reading or copying.
298 |
299 | ### 7. Additional Terms
300 |
301 | “Additional permissions” are terms that supplement the terms of this
302 | License by making exceptions from one or more of its conditions. Additional
303 | permissions that are applicable to the entire Program shall be treated as though they
304 | were included in this License, to the extent that they are valid under applicable
305 | law. If additional permissions apply only to part of the Program, that part may be
306 | used separately under those permissions, but the entire Program remains governed by
307 | this License without regard to the additional permissions.
308 |
309 | When you convey a copy of a covered work, you may at your option remove any
310 | additional permissions from that copy, or from any part of it. (Additional
311 | permissions may be written to require their own removal in certain cases when you
312 | modify the work.) You may place additional permissions on material, added by you to a
313 | covered work, for which you have or can give appropriate copyright permission.
314 |
315 | Notwithstanding any other provision of this License, for material you add to a
316 | covered work, you may (if authorized by the copyright holders of that material)
317 | supplement the terms of this License with terms:
318 |
319 | * **a)** Disclaiming warranty or limiting liability differently from the terms of
320 | sections 15 and 16 of this License; or
321 | * **b)** Requiring preservation of specified reasonable legal notices or author
322 | attributions in that material or in the Appropriate Legal Notices displayed by works
323 | containing it; or
324 | * **c)** Prohibiting misrepresentation of the origin of that material, or requiring that
325 | modified versions of such material be marked in reasonable ways as different from the
326 | original version; or
327 | * **d)** Limiting the use for publicity purposes of names of licensors or authors of the
328 | material; or
329 | * **e)** Declining to grant rights under trademark law for use of some trade names,
330 | trademarks, or service marks; or
331 | * **f)** Requiring indemnification of licensors and authors of that material by anyone
332 | who conveys the material (or modified versions of it) with contractual assumptions of
333 | liability to the recipient, for any liability that these contractual assumptions
334 | directly impose on those licensors and authors.
335 |
336 | All other non-permissive additional terms are considered “further
337 | restrictions” within the meaning of section 10. If the Program as you received
338 | it, or any part of it, contains a notice stating that it is governed by this License
339 | along with a term that is a further restriction, you may remove that term. If a
340 | license document contains a further restriction but permits relicensing or conveying
341 | under this License, you may add to a covered work material governed by the terms of
342 | that license document, provided that the further restriction does not survive such
343 | relicensing or conveying.
344 |
345 | If you add terms to a covered work in accord with this section, you must place, in
346 | the relevant source files, a statement of the additional terms that apply to those
347 | files, or a notice indicating where to find the applicable terms.
348 |
349 | Additional terms, permissive or non-permissive, may be stated in the form of a
350 | separately written license, or stated as exceptions; the above requirements apply
351 | either way.
352 |
353 | ### 8. Termination
354 |
355 | You may not propagate or modify a covered work except as expressly provided under
356 | this License. Any attempt otherwise to propagate or modify it is void, and will
357 | automatically terminate your rights under this License (including any patent licenses
358 | granted under the third paragraph of section 11).
359 |
360 | However, if you cease all violation of this License, then your license from a
361 | particular copyright holder is reinstated **(a)** provisionally, unless and until the
362 | copyright holder explicitly and finally terminates your license, and **(b)** permanently,
363 | if the copyright holder fails to notify you of the violation by some reasonable means
364 | prior to 60 days after the cessation.
365 |
366 | Moreover, your license from a particular copyright holder is reinstated permanently
367 | if the copyright holder notifies you of the violation by some reasonable means, this
368 | is the first time you have received notice of violation of this License (for any
369 | work) from that copyright holder, and you cure the violation prior to 30 days after
370 | your receipt of the notice.
371 |
372 | Termination of your rights under this section does not terminate the licenses of
373 | parties who have received copies or rights from you under this License. If your
374 | rights have been terminated and not permanently reinstated, you do not qualify to
375 | receive new licenses for the same material under section 10.
376 |
377 | ### 9. Acceptance Not Required for Having Copies
378 |
379 | You are not required to accept this License in order to receive or run a copy of the
380 | Program. Ancillary propagation of a covered work occurring solely as a consequence of
381 | using peer-to-peer transmission to receive a copy likewise does not require
382 | acceptance. However, nothing other than this License grants you permission to
383 | propagate or modify any covered work. These actions infringe copyright if you do not
384 | accept this License. Therefore, by modifying or propagating a covered work, you
385 | indicate your acceptance of this License to do so.
386 |
387 | ### 10. Automatic Licensing of Downstream Recipients
388 |
389 | Each time you convey a covered work, the recipient automatically receives a license
390 | from the original licensors, to run, modify and propagate that work, subject to this
391 | License. You are not responsible for enforcing compliance by third parties with this
392 | License.
393 |
394 | An “entity transaction” is a transaction transferring control of an
395 | organization, or substantially all assets of one, or subdividing an organization, or
396 | merging organizations. If propagation of a covered work results from an entity
397 | transaction, each party to that transaction who receives a copy of the work also
398 | receives whatever licenses to the work the party's predecessor in interest had or
399 | could give under the previous paragraph, plus a right to possession of the
400 | Corresponding Source of the work from the predecessor in interest, if the predecessor
401 | has it or can get it with reasonable efforts.
402 |
403 | You may not impose any further restrictions on the exercise of the rights granted or
404 | affirmed under this License. For example, you may not impose a license fee, royalty,
405 | or other charge for exercise of rights granted under this License, and you may not
406 | initiate litigation (including a cross-claim or counterclaim in a lawsuit) alleging
407 | that any patent claim is infringed by making, using, selling, offering for sale, or
408 | importing the Program or any portion of it.
409 |
410 | ### 11. Patents
411 |
412 | A “contributor” is a copyright holder who authorizes use under this
413 | License of the Program or a work on which the Program is based. The work thus
414 | licensed is called the contributor's “contributor version”.
415 |
416 | A contributor's “essential patent claims” are all patent claims owned or
417 | controlled by the contributor, whether already acquired or hereafter acquired, that
418 | would be infringed by some manner, permitted by this License, of making, using, or
419 | selling its contributor version, but do not include claims that would be infringed
420 | only as a consequence of further modification of the contributor version. For
421 | purposes of this definition, “control” includes the right to grant patent
422 | sublicenses in a manner consistent with the requirements of this License.
423 |
424 | Each contributor grants you a non-exclusive, worldwide, royalty-free patent license
425 | under the contributor's essential patent claims, to make, use, sell, offer for sale,
426 | import and otherwise run, modify and propagate the contents of its contributor
427 | version.
428 |
429 | In the following three paragraphs, a “patent license” is any express
430 | agreement or commitment, however denominated, not to enforce a patent (such as an
431 | express permission to practice a patent or covenant not to sue for patent
432 | infringement). To “grant” such a patent license to a party means to make
433 | such an agreement or commitment not to enforce a patent against the party.
434 |
435 | If you convey a covered work, knowingly relying on a patent license, and the
436 | Corresponding Source of the work is not available for anyone to copy, free of charge
437 | and under the terms of this License, through a publicly available network server or
438 | other readily accessible means, then you must either **(1)** cause the Corresponding
439 | Source to be so available, or **(2)** arrange to deprive yourself of the benefit of the
440 | patent license for this particular work, or **(3)** arrange, in a manner consistent with
441 | the requirements of this License, to extend the patent license to downstream
442 | recipients. “Knowingly relying” means you have actual knowledge that, but
443 | for the patent license, your conveying the covered work in a country, or your
444 | recipient's use of the covered work in a country, would infringe one or more
445 | identifiable patents in that country that you have reason to believe are valid.
446 |
447 | If, pursuant to or in connection with a single transaction or arrangement, you
448 | convey, or propagate by procuring conveyance of, a covered work, and grant a patent
449 | license to some of the parties receiving the covered work authorizing them to use,
450 | propagate, modify or convey a specific copy of the covered work, then the patent
451 | license you grant is automatically extended to all recipients of the covered work and
452 | works based on it.
453 |
454 | A patent license is “discriminatory” if it does not include within the
455 | scope of its coverage, prohibits the exercise of, or is conditioned on the
456 | non-exercise of one or more of the rights that are specifically granted under this
457 | License. You may not convey a covered work if you are a party to an arrangement with
458 | a third party that is in the business of distributing software, under which you make
459 | payment to the third party based on the extent of your activity of conveying the
460 | work, and under which the third party grants, to any of the parties who would receive
461 | the covered work from you, a discriminatory patent license **(a)** in connection with
462 | copies of the covered work conveyed by you (or copies made from those copies), or **(b)**
463 | primarily for and in connection with specific products or compilations that contain
464 | the covered work, unless you entered into that arrangement, or that patent license
465 | was granted, prior to 28 March 2007.
466 |
467 | Nothing in this License shall be construed as excluding or limiting any implied
468 | license or other defenses to infringement that may otherwise be available to you
469 | under applicable patent law.
470 |
471 | ### 12. No Surrender of Others' Freedom
472 |
473 | If conditions are imposed on you (whether by court order, agreement or otherwise)
474 | that contradict the conditions of this License, they do not excuse you from the
475 | conditions of this License. If you cannot convey a covered work so as to satisfy
476 | simultaneously your obligations under this License and any other pertinent
477 | obligations, then as a consequence you may not convey it at all. For example, if you
478 | agree to terms that obligate you to collect a royalty for further conveying from
479 | those to whom you convey the Program, the only way you could satisfy both those terms
480 | and this License would be to refrain entirely from conveying the Program.
481 |
482 | ### 13. Use with the GNU Affero General Public License
483 |
484 | Notwithstanding any other provision of this License, you have permission to link or
485 | combine any covered work with a work licensed under version 3 of the GNU Affero
486 | General Public License into a single combined work, and to convey the resulting work.
487 | The terms of this License will continue to apply to the part which is the covered
488 | work, but the special requirements of the GNU Affero General Public License, section
489 | 13, concerning interaction through a network will apply to the combination as such.
490 |
491 | ### 14. Revised Versions of this License
492 |
493 | The Free Software Foundation may publish revised and/or new versions of the GNU
494 | General Public License from time to time. Such new versions will be similar in spirit
495 | to the present version, but may differ in detail to address new problems or concerns.
496 |
497 | Each version is given a distinguishing version number. If the Program specifies that
498 | a certain numbered version of the GNU General Public License “or any later
499 | version” applies to it, you have the option of following the terms and
500 | conditions either of that numbered version or of any later version published by the
501 | Free Software Foundation. If the Program does not specify a version number of the GNU
502 | General Public License, you may choose any version ever published by the Free
503 | Software Foundation.
504 |
505 | If the Program specifies that a proxy can decide which future versions of the GNU
506 | General Public License can be used, that proxy's public statement of acceptance of a
507 | version permanently authorizes you to choose that version for the Program.
508 |
509 | Later license versions may give you additional or different permissions. However, no
510 | additional obligations are imposed on any author or copyright holder as a result of
511 | your choosing to follow a later version.
512 |
513 | ### 15. Disclaimer of Warranty
514 |
515 | THERE IS NO WARRANTY FOR THE PROGRAM, TO THE EXTENT PERMITTED BY APPLICABLE LAW.
516 | EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT HOLDERS AND/OR OTHER PARTIES
517 | PROVIDE THE PROGRAM “AS IS” WITHOUT WARRANTY OF ANY KIND, EITHER
518 | EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF
519 | MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE ENTIRE RISK AS TO THE
520 | QUALITY AND PERFORMANCE OF THE PROGRAM IS WITH YOU. SHOULD THE PROGRAM PROVE
521 | DEFECTIVE, YOU ASSUME THE COST OF ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
522 |
523 | ### 16. Limitation of Liability
524 |
525 | IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING WILL ANY
526 | COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MODIFIES AND/OR CONVEYS THE PROGRAM AS
527 | PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY GENERAL, SPECIAL,
528 | INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE USE OR INABILITY TO USE THE
529 | PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF DATA OR DATA BEING RENDERED INACCURATE
530 | OR LOSSES SUSTAINED BY YOU OR THIRD PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE
531 | WITH ANY OTHER PROGRAMS), EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE
532 | POSSIBILITY OF SUCH DAMAGES.
533 |
534 | ### 17. Interpretation of Sections 15 and 16
535 |
536 | If the disclaimer of warranty and limitation of liability provided above cannot be
537 | given local legal effect according to their terms, reviewing courts shall apply local
538 | law that most closely approximates an absolute waiver of all civil liability in
539 | connection with the Program, unless a warranty or assumption of liability accompanies
540 | a copy of the Program in return for a fee.
541 |
542 | _END OF TERMS AND CONDITIONS_
543 |
544 | ## How to Apply These Terms to Your New Programs
545 |
546 | If you develop a new program, and you want it to be of the greatest possible use to
547 | the public, the best way to achieve this is to make it free software which everyone
548 | can redistribute and change under these terms.
549 |
550 | To do so, attach the following notices to the program. It is safest to attach them
551 | to the start of each source file to most effectively state the exclusion of warranty;
552 | and each file should have at least the “copyright” line and a pointer to
553 | where the full notice is found.
554 |
555 |
556 | Copyright (C) 2019 Mara Averick
557 |
558 | This program is free software: you can redistribute it and/or modify
559 | it under the terms of the GNU General Public License as published by
560 | the Free Software Foundation, either version 3 of the License, or
561 | (at your option) any later version.
562 |
563 | This program is distributed in the hope that it will be useful,
564 | but WITHOUT ANY WARRANTY; without even the implied warranty of
565 | MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
566 | GNU General Public License for more details.
567 |
568 | You should have received a copy of the GNU General Public License
569 | along with this program. If not, see .
570 |
571 | Also add information on how to contact you by electronic and paper mail.
572 |
573 | If the program does terminal interaction, make it output a short notice like this
574 | when it starts in an interactive mode:
575 |
576 | forcats Copyright (C) 2019 Mara Averick
577 | This program comes with ABSOLUTELY NO WARRANTY; for details type 'show w'.
578 | This is free software, and you are welcome to redistribute it
579 | under certain conditions; type 'show c' for details.
580 |
581 | The hypothetical commands `show w` and `show c` should show the appropriate parts of
582 | the General Public License. Of course, your program's commands might be different;
583 | for a GUI interface, you would use an “about box”.
584 |
585 | You should also get your employer (if you work as a programmer) or school, if any, to
586 | sign a “copyright disclaimer” for the program, if necessary. For more
587 | information on this, and how to apply and follow the GNU GPL, see
588 | <>.
589 |
590 | The GNU General Public License does not permit incorporating your program into
591 | proprietary programs. If your program is a subroutine library, you may consider it
592 | more useful to permit linking proprietary applications with the library. If this is
593 | what you want to do, use the GNU Lesser General Public License instead of this
594 | License. But first, please read
595 | <>.
596 |
--------------------------------------------------------------------------------
50 |
51 | ## Getting started
52 |
53 | forcats is part of the core tidyverse, so you can load it with
54 | `library(tidyverse)` or `library(forcats)`.
55 |
56 | ``` r
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ``` r
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | #> # A tibble: 37 x 2
67 | #> species n
68 | #>
69 | #> 1 Human 35
70 | #> 2 Droid 5
71 | #> 3 Gungan 3
72 | #> 4 Kaminoan 2
73 | #> 5 Mirialan 2
74 | #> 6 Twi'lek 2
75 | #> 7 Wookiee 2
76 | #> 8 Zabrak 2
77 | #> 9 Aleena 1
78 | #> 10 Besalisk 1
79 | #> # … with 27 more rows
80 | ```
81 |
82 | ``` r
83 | starwars %>%
84 | filter(!is.na(species)) %>%
85 | mutate(species = fct_lump(species, n = 3)) %>%
86 | count(species)
87 | #> # A tibble: 4 x 2
88 | #> species n
89 | #>
90 | #> 1 Droid 5
91 | #> 2 Gungan 3
92 | #> 3 Human 35
93 | #> 4 Other 39
94 | ```
95 |
96 | ``` r
97 | ggplot(starwars, aes(x = eye_color)) +
98 | geom_bar() +
99 | coord_flip()
100 | ```
101 |
102 | 
103 |
104 | ``` r
105 | starwars %>%
106 | mutate(eye_color = fct_infreq(eye_color)) %>%
107 | ggplot(aes(x = eye_color)) +
108 | geom_bar() +
109 | coord_flip()
110 | ```
111 |
112 | 
113 |
114 | ## More resources
115 |
116 | For a history of factors, I recommend [*stringsAsFactors: An
117 | unauthorized
118 | biography*](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/)
119 | by Roger Peng and [*stringsAsFactors =
120 | \*](http://notstatschat.tumblr.com/post/124987394001/stringsasfactors-sigh)
121 | by Thomas Lumley. If you want to learn more about other approaches to
122 | working with factors and categorical data, I recommend [*Wrangling
123 | categorical data in R*](https://peerj.com/preprints/3163/), by Amelia
124 | McNamara and Nicholas Horton.
125 |
126 | ## Getting help
127 |
128 | If you encounter a clear bug, please file a minimal reproducible example
129 | on [github](https://github.com/tidyverse/forcats/issues). For questions
130 | and other discussion, please use
131 | [community.rstudio.com](https://community.rstudio.com/).
132 |
133 | ## Code of Conduct
134 |
135 | Please note that the ‘forcats’ project is released with a [Contributor
136 | Code of Conduct](.github/CODE_OF_CONDUCT.md). By contributing to this
137 | project, you agree to abide by its terms.
138 |
--------------------------------------------------------------------------------
/revdep/README.md:
--------------------------------------------------------------------------------
1 | # Platform
2 |
3 | |field |value |
4 | |:--------|:----------------------------|
5 | |version |R version 3.5.2 (2018-12-20) |
6 | |os |macOS Mojave 10.14.2 |
7 | |system |x86_64, darwin15.6.0 |
8 | |ui |RStudio |
9 | |language |(EN) |
10 | |collate |en_US.UTF-8 |
11 | |ctype |en_US.UTF-8 |
12 | |tz |America/Chicago |
13 | |date |2019-02-16 |
14 |
15 | # Dependencies
16 |
17 | |package |old |new |Δ |
18 | |:---------|:-----|:----------|:--|
19 | |forcats |0.3.0 |0.3.0.9000 |* |
20 | |ellipsis |NA |0.0.2 |* |
21 | |fansi |0.4.0 |0.4.0 | |
22 | |pillar |1.3.1 |1.3.1 | |
23 | |pkgconfig |2.0.2 |2.0.2 | |
24 | |rlang |0.3.1 |0.3.1 | |
25 | |tibble |2.0.1 |2.0.1 | |
26 | |utf8 |1.1.4 |1.1.4 | |
27 |
28 | # Revdeps
29 |
30 | ## Couldn't check (1)
31 |
32 | |package |version |error |warning |note |
33 | |:------------------------------------|:-------|:-----|:-------|:----|
34 | |[circumplex](problems.md#circumplex) |0.2.1 |1 | | |
35 |
36 | ## All (52)
37 |
38 | |package |version |error |warning |note |
39 | |:----------------------------------------------|:-------|:-----|:-------|:----|
40 | |apaTables |2.0.5 | | | |
41 | |bench |1.0.1 | | | |
42 | |[bupaR](problems.md#bupar) |0.4.1 | |1 | |
43 | |[circumplex](problems.md#circumplex) |0.2.1 |1 | | |
44 | |coalitions |0.6.5 | | | |
45 | |[cocktailApp](problems.md#cocktailapp) |0.2.0 | | |1 |
46 | |[dabestr](problems.md#dabestr) |0.2.0 | |1 |1 |
47 | |descriptr |0.5.0 | | | |
48 | |easyalluvial |0.1.8 | | | |
49 | |evaluator |0.3.2 | | | |
50 | |[ezplot](problems.md#ezplot) |0.2.2 | | |1 |
51 | |[factorMerger](problems.md#factormerger) |0.3.6 | | |1 |
52 | |[finalfit](problems.md#finalfit) |0.9.0 | | |1 |
53 | |forwards |0.1.1 | | | |
54 | |[furniture](problems.md#furniture) |1.8.7 | | |1 |
55 | |genogeographer |0.1.8 | | | |
56 | |[ggdag](problems.md#ggdag) |0.1.0 | | |1 |
57 | |[ggridges](problems.md#ggridges) |0.5.1 | | |1 |
58 | |[glmmfields](problems.md#glmmfields) |0.1.1 | | |1 |
59 | |[glmSparseNet](problems.md#glmsparsenet) |1.0.0 | |1 |2 |
60 | |glue |1.3.0 | | | |
61 | |[haven](problems.md#haven) |2.0.0 | | |1 |
62 | |healthcareai |2.3.0 | | | |
63 | |[idealstan](problems.md#idealstan) |0.5.1 | | |2 |
64 | |[iotables](problems.md#iotables) |0.4.2 | | |1 |
65 | |[konfound](problems.md#konfound) |0.1.1 | |1 |1 |
66 | |[MetamapsDB](problems.md#metamapsdb) |0.0.2 | | |1 |
67 | |naniar |0.4.2 | | | |
68 | |[noaastormevents](problems.md#noaastormevents) |0.1.0 | | |2 |
69 | |[nzelect](problems.md#nzelect) |0.4.0 | | |2 |
70 | |outcomerate |1.0.1 | | | |
71 | |OutliersO3 |0.6 | | | |
72 | |[phenopath](problems.md#phenopath) |1.6.0 | |1 | |
73 | |[plotly](problems.md#plotly) |4.8.0 | | |2 |
74 | |[processmapR](problems.md#processmapr) |0.3.2 | | |1 |
75 | |[questionr](problems.md#questionr) |0.7.0 | | |2 |
76 | |[railtrails](problems.md#railtrails) |0.1.1 | | |1 |
77 | |[rbin](problems.md#rbin) |0.1.1 | | |1 |
78 | |rfm |0.2.0 | | | |
79 | |[SCnorm](problems.md#scnorm) |1.4.5 | | |1 |
80 | |SEERaBomb |2018.1 | | | |
81 | |simstandard |0.3.0 | | | |
82 | |sjPlot |2.6.2 | | | |
83 | |survivalAnalysis |0.1.1 | | | |
84 | |[sweep](problems.md#sweep) |0.2.1.1 |1 | | |
85 | |[TextForecast](problems.md#textforecast) |0.1.0 | | |2 |
86 | |[tidybayes](problems.md#tidybayes) |1.0.3 |1 | | |
87 | |[tidyLPA](problems.md#tidylpa) |0.2.4 | | |1 |
88 | |tidytidbits |0.2.0 | | | |
89 | |[tidyverse](problems.md#tidyverse) |1.2.1 | | |1 |
90 | |[timetk](problems.md#timetk) |0.1.1.1 |1 | | |
91 | |unpivotr |0.5.0 | | | |
92 |
93 |
--------------------------------------------------------------------------------
/tests/testthat/test-fct_lump.R:
--------------------------------------------------------------------------------
1 | context("fct_lump")
2 |
3 | test_that("positive values keeps most commmon", {
4 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
5 |
6 | expect_equal(levels(fct_lump(f, n = 1)), c("a", "Other"))
7 | expect_equal(levels(fct_lump(f, n = 2)), c("a", "b", "Other"))
8 |
9 | expect_equal(levels(fct_lump(f, prop = 0.25)), c("a", "Other"))
10 | expect_equal(levels(fct_lump(f, prop = 0.15)), c("a", "b", "Other"))
11 | })
12 |
13 | test_that("ties are respected", {
14 | f <- c("a", "a", "a", "b", "b", "b", "c", "d")
15 | expect_equal(levels(fct_lump(f, 1)), c("a", "b", "Other"))
16 | })
17 |
18 | test_that("negative values drop most common" ,{
19 | f <- c("a", "a", "a", "a", "b", "b", "b", "b", "c", "d")
20 | expect_equal(levels(fct_lump(f, n = -1)), c("c", "d", "Other"))
21 | expect_equal(levels(fct_lump(f, prop = -0.2)), c("c", "d", "Other"))
22 | })
23 |
24 | test_that("return original factor when all element satisfy n / p condition", {
25 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
26 |
27 | expect_equal(levels(fct_lump(f, n = 4)), c("a", "b", "c", "d", "e", "f", "g"))
28 | expect_equal(levels(fct_lump(f, n = 10)), c("a", "b", "c", "d", "e", "f", "g"))
29 | expect_equal(levels(fct_lump(f, n = -10)), c("a", "b", "c", "d", "e", "f", "g"))
30 |
31 | expect_equal(levels(fct_lump(f, prop = 0.01)), c("a", "b", "c", "d", "e", "f", "g"))
32 | expect_equal(levels(fct_lump(f, prop = -1)), c("a", "b", "c", "d", "e", "f", "g"))
33 | })
34 |
35 | test_that("different behaviour when apply tie function", {
36 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
37 |
38 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "min")),
39 | c("a", "b", "c", "d", "e", "f", "g"))
40 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "max")),
41 | c("a", "b", "Other" ))
42 |
43 | # Rank of c, d, e, f, g is (3+4+5+6+7)/5 = 5
44 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "average")),
45 | c("a", "b", "Other" ))
46 | expect_equal(levels(fct_lump(f, n = 5, ties.method = "average")),
47 | c("a", "b", "c", "d", "e", "f", "g"))
48 |
49 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "first")),
50 | c("a", "b", "c", "d", "Other"))
51 |
52 | if (getRversion() >= "3.3.0") {
53 | expect_equal(levels(fct_lump(f, n = 4, ties.method = "last")),
54 | c("a", "b", "f", "g", "Other"))
55 | }
56 | })
57 |
58 | test_that("NAs included in total", {
59 | f <- factor(c("a", "a", "b", "c", rep(NA, 7)))
60 |
61 | o1 <- fct_lump(f, prop = 0.10)
62 | expect_equal(levels(o1), c("a", "Other"))
63 |
64 | o2 <- fct_lump(f, w = rep(1, 11), prop = 0.10)
65 | expect_equal(levels(o2), c("a", "Other"))
66 | })
67 |
68 | test_that("bad weights generate error messages", {
69 | expect_error(fct_lump(letters, w = letters), "must be a numeric vector")
70 | expect_error(fct_lump(letters, w = 1:10), "must be the same length")
71 | expect_error(fct_lump(letters, w = rep(-1, 26)), "must be non-negative")
72 | })
73 |
74 | test_that("values are correctly weighted", {
75 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
76 | w <- c( 0.2, 0.2, 0.6, 2, 2, 6, 4, 2, 2, 1)
77 | f2 <- c(
78 | "a",
79 | rep("b", 4),
80 | rep("c", 6),
81 | rep("d", 4),
82 | rep("e", 2),
83 | rep("f", 2),
84 | "g"
85 | )
86 |
87 | expect_equal(levels(fct_lump(f, w = w)), levels(fct_lump(f2)))
88 | expect_equal(
89 | levels(fct_lump(f, n = 1, w = w)),
90 | levels(fct_lump(f2, n = 1))
91 | )
92 | expect_equal(
93 | levels(fct_lump(f, n = -2, w = w, ties.method = "first")),
94 | levels(fct_lump(f2, n = -2, ties.method = "first"))
95 | )
96 | expect_equal(
97 | levels(fct_lump(f, n = 99, w = w)),
98 | levels(fct_lump(f2, n = 99))
99 | )
100 | expect_equal(
101 | levels(fct_lump(f, prop = 0.01, w = w)),
102 | levels(fct_lump(f2, prop = 0.01))
103 | )
104 | expect_equal(
105 | levels(fct_lump(f, prop = -0.25, w = w, ties.method = "max")),
106 | levels(fct_lump(f2, prop = -0.25, ties.method = "max"))
107 | )
108 | })
109 |
110 | test_that("do not change the label when no lumping occurs", {
111 | f <- c("a", "a", "a", "a", "b", "b", "b", "c", "c", "d")
112 | expect_equal(levels(fct_lump(f, n = 3)), c("a", "b", "c", "d"))
113 | expect_equal(levels(fct_lump(f, prop = 0.1)), c("a", "b", "c", "d"))
114 | })
115 |
116 | test_that("fct_lump_min works when not weighted", {
117 | f <- c("a", "a", "a", "b", "b", "c", "d", "e", "f", "g")
118 |
119 | expect_equal(levels(fct_lump_min(f, min = 3)), c("a", "Other"))
120 | expect_equal(levels(fct_lump_min(f, min = 2)), c("a", "b", "Other"))
121 | })
122 |
123 | test_that("fct_lump_min works when weighted", {
124 | f <- c("a", "b", "c", "d", "e")
125 | w <- c( 0.2, 2, 6, 4, 1)
126 |
127 | expect_equal(levels(fct_lump_min(f, min = 6, w = w)), c("c", "Other"))
128 | expect_equal(levels(fct_lump_min(f, min = 1.5, w = w)), c("b", "c", "d", "Other"))
129 | })
130 |
131 | # Default -----------------------------------------------------------------
132 |
133 | test_that("lumps smallest", {
134 | expect_equal(lump_test(c(1, 2, 3, 6)), "Xbcd")
135 | expect_equal(lump_test(c(1, 2, 3, 7)), "XXXd")
136 |
137 | expect_equal(lump_test(c(1, 2, 3, 7, 13)), "XXXde")
138 | expect_equal(lump_test(c(1, 2, 3, 7, 14)), "XXXXe")
139 | })
140 |
141 | test_that("doesn't lump if none small enough", {
142 | expect_equal(lump_test(c(2, 2, 4)), "abc")
143 | })
144 |
145 | test_that("order doesn't matter", {
146 | expect_equal(lump_test(c(2, 2, 5)), "XXc")
147 | expect_equal(lump_test(c(2, 5, 2)), "XbX")
148 | expect_equal(lump_test(c(5, 2, 2)), "aXX")
149 | })
150 |
--------------------------------------------------------------------------------
/vignettes/forcats.Rmd:
--------------------------------------------------------------------------------
1 | ---
2 | title: "Introduction to forcats"
3 | author: "Emily Robinson"
4 | output: rmarkdown::html_vignette
5 | vignette: >
6 | %\VignetteIndexEntry{Introduction to forcats}
7 | %\VignetteEngine{knitr::rmarkdown}
8 | %\VignetteEncoding{UTF-8}
9 | ---
10 |
11 | ```{r setup, include = FALSE}
12 | knitr::opts_chunk$set(
13 | collapse = TRUE,
14 | comment = "#>"
15 | )
16 | ```
17 |
18 | The goal of the __forcats__ package is to provide a suite of useful tools that solve common problems with factors. Factors are useful when you have categorical data, variables that have a fixed and known set of values, and when you want to display character vectors in non-alphabetical order. If you want to learn more, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
19 |
20 | ## Ordering by frequency
21 |
22 | ```{r message = FALSE}
23 | library(dplyr)
24 | library(ggplot2)
25 | library(forcats)
26 | ```
27 |
28 | Let's try answering the question, "what are the most common hair colors of star wars characters?" Let's start off by making a bar plot:
29 |
30 | ```{r initial-plot}
31 | ggplot(starwars, aes(x = hair_color)) +
32 | geom_bar() +
33 | coord_flip()
34 | ```
35 |
36 | That's okay, but it would be more helpful the graph was ordered by count. This is a case of an **unordered** categorical variable where we want it ordered by its frequency. To do so, we can use the function `fct_infreq()`:
37 |
38 | ```{r fct-infreq-hair}
39 | ggplot(starwars, aes(x = fct_infreq(hair_color))) +
40 | geom_bar() +
41 | coord_flip()
42 | ```
43 |
44 | Note that `fct_infreq()` it automatically puts NA at the top, even though that doesn't have the smallest number of entries.
45 |
46 | ## Combining levels
47 |
48 | Let's take a look at skin color now:
49 |
50 | ```{r}
51 | starwars %>%
52 | count(skin_color, sort = TRUE)
53 | ```
54 |
55 | We see that there's 31 different skin colors - if we want to make a plot this would be way too many to display! Let's reduce it to only be the top 5. We can use `fct_lump()` to "lump" all the infrequent colors into one factor, "other." The argument `n` is the number of levels we want to keep.
56 |
57 | ```{r}
58 | starwars %>%
59 | mutate(skin_color = fct_lump(skin_color, n = 5)) %>%
60 | count(skin_color, sort = TRUE)
61 | ```
62 |
63 | We could also have used `prop` instead, which keeps all the levels that appear at least `prop` of the time. For example, let's keep skin colors that at least 10% of the characters have:
64 |
65 | ```{r}
66 | starwars %>%
67 | mutate(skin_color = fct_lump(skin_color, prop = .1)) %>%
68 | count(skin_color, sort = TRUE)
69 | ```
70 |
71 | Only light and fair remain; everything else is other.
72 |
73 | If you wanted to call it something than "other", you can change it with the argument `other_level`:
74 |
75 | ```{r}
76 | starwars %>%
77 | mutate(skin_color = fct_lump(skin_color, prop = .1, other_level = "extra")) %>%
78 | count(skin_color, sort = TRUE)
79 | ```
80 |
81 | What if we wanted to see if the average mass differed by eye color? We'll only look at the 6 most popular eye colors and remove `NA`s.
82 |
83 | ```{r fct-lump-mean}
84 | avg_mass_eye_color <- starwars %>%
85 | mutate(eye_color = fct_lump(eye_color, n = 6)) %>%
86 | group_by(eye_color) %>%
87 | summarise(mean_mass = mean(mass, na.rm = TRUE))
88 |
89 | avg_mass_eye_color
90 | ```
91 |
92 | ## Ordering by another variable
93 |
94 | It looks like people (or at least one person) with orange eyes are definitely heavier! If we wanted to make a graph, it would be nice if it was ordered by `mean_mass`. We can do this with `fct_reorder()`, which reorders one variable by another.
95 |
96 | ```{r fct-reorder}
97 | avg_mass_eye_color %>%
98 | mutate(eye_color = fct_reorder(eye_color, mean_mass)) %>%
99 | ggplot(aes(x = eye_color, y = mean_mass)) +
100 | geom_col()
101 | ```
102 |
103 | ## Manually reordering
104 |
105 | Let's switch to using another dataset, `gss_cat`, the general social survey. What is the income distribution among the respondents?
106 |
107 | ```{r}
108 | gss_cat %>%
109 | count(rincome)
110 | ```
111 |
112 | Notice that the income levels are in the correct order - they start with the non-answers and then go from highest to lowest. This is the same order you'd see if you plotted it as a bar chart. This is not a coincidence. When you're working with ordinal data, where there is an order, you can have an ordered factor. You can examine them with the base function `levels()`, which prints them in order:
113 |
114 | ```{r}
115 | levels(gss_cat$rincome)
116 | ```
117 |
118 | But what if your factor came in the wrong order? Let's simulate that by reordering the levels of `rincome` randomly with `fct_shuffle()`:
119 |
120 | ```{r}
121 | reshuffled_income <- gss_cat$rincome %>%
122 | fct_shuffle()
123 |
124 | levels(reshuffled_income)
125 | ```
126 |
127 | Now if we plotted it, it would show in this order, which is all over the place! How can we fix this and put it in the right order?
128 |
129 | We can use the function `fct_relevel()` when we need to manually reorder our factor levels. In addition to the factor, you give it a character vector of level names, and specify where you want to move them. It defaults to moving them to the front, but you can move them after another level with the argument `after`. If you want to move it to the end, you set `after` equal to `Inf`.
130 |
131 | For example, let's say we wanted to move `Lt $1000` and `$1000 to 2999` to the front. We would write:
132 |
133 | ```{r}
134 | fct_relevel(reshuffled_income, c("Lt $1000", "$1000 to 2999")) %>%
135 | levels()
136 | ```
137 |
138 | What if we want to move them to the second and third place?
139 |
140 | ```{r}
141 | fct_relevel(reshuffled_income, c("Lt $1000", "$1000 to 2999"), after = 1) %>%
142 | levels()
143 | ```
144 |
--------------------------------------------------------------------------------
/R/lump.R:
--------------------------------------------------------------------------------
1 | #' Lump together factor levels into "other"
2 | #'
3 | #' `fct_lump()` lumps together least/most common levels (as defined by `n` and
4 | #' `prop`) into "other".
5 | #' `fct_lump_min()` lumps together all levels which don't appear at least
6 | #' `min` number of times.
7 | #'
8 | #' @param f A factor (or character vector).
9 | #' @param n,prop
10 | #' If both `n` and `prop` are missing, `fct_lump()` lumps
11 | #' together the least frequent levels into "other", while ensuring that
12 | #' "other" is still the smallest level. It's particularly useful in
13 | #' conjunction with \code{\link{fct_inorder}()}.
14 | #'
15 | #' Positive `n` preserves the most common `n` values.
16 | #' Negative `n` preserves the least common `-n` values.
17 | #' It there are ties, you will get at least `abs(n)` values.
18 | #'
19 | #' Positive `prop` preserves values that appear at least
20 | #' `prop` of the time. Negative `prop` preserves values that
21 | #' appear at most `-prop` of the time.
22 | #' @param w An optional numeric vector giving weights for frequency of
23 | #' each value (not level) in f.
24 | #' @param other_level Value of level used for "other" values. Always
25 | #' placed at end of levels.
26 | #' @param ties.method A character string specifying how ties are
27 | #' treated. See [rank()] for details.
28 | #' @export
29 | #' @seealso [fct_other()] to convert specified levels to other.
30 | #' @examples
31 | #' x <- factor(rep(LETTERS[1:9], times = c(40, 10, 5, 27, 1, 1, 1, 1, 1)))
32 | #' x %>% table()
33 | #' x %>% fct_lump() %>% table()
34 | #' x %>% fct_lump() %>% fct_inorder() %>% table()
35 | #'
36 | #' x <- factor(letters[rpois(100, 5)])
37 | #' x
38 | #' table(x)
39 | #' table(fct_lump(x))
40 | #'
41 | #' # Use positive values to collapse the rarest
42 | #' fct_lump(x, n = 3)
43 | #' fct_lump(x, prop = 0.1)
44 | #'
45 | #' # Use negative values to collapse the most common
46 | #' fct_lump(x, n = -3)
47 | #' fct_lump(x, prop = -0.1)
48 | #'
49 | #' # Use weighted frequencies
50 | #' w <- c(rep(2, 50), rep(1, 50))
51 | #' fct_lump(x, n = 5, w = w)
52 | #'
53 | #' # Use ties.method to control how tied factors are collapsed
54 | #' fct_lump(x, n = 6)
55 | #' fct_lump(x, n = 6, ties.method = "max")
56 | #'
57 | fct_lump <- function(f, n, prop, w = NULL, other_level = "Other",
58 | ties.method = c("min", "average", "first", "last", "random", "max")) {
59 | f <- check_factor(f)
60 | w <- check_weights(w, length(f))
61 | ties.method <- match.arg(ties.method)
62 |
63 | levels <- levels(f)
64 | if (is.null(w)) {
65 | count <- as.vector(table(f))
66 | total <- length(f)
67 | } else {
68 | count <- as.vector(tapply(w, f, FUN = sum))
69 | total <- sum(w)
70 | }
71 |
72 | if (!xor(missing(n), missing(prop))) {
73 | new_levels <- ifelse(!in_smallest(count), levels, other_level)
74 | } else if (!missing(n)) {
75 | if (n < 0) {
76 | rank <- rank(count, ties = ties.method)
77 | n <- -n
78 | } else {
79 | rank <- rank(-count, ties = ties.method)
80 | }
81 |
82 | if (sum(rank > n) <= 1) {
83 | # No lumping needed
84 | return(f)
85 | }
86 |
87 | new_levels <- ifelse(rank <= n, levels, other_level)
88 | } else if (!missing(prop)) {
89 | prop_n <- count / total
90 | if (prop < 0) {
91 | new_levels <- ifelse(prop_n <= -prop, levels, other_level)
92 | } else {
93 | if (sum(prop_n <= prop) <= 1) {
94 | # No lumping needed
95 | return(f)
96 | }
97 |
98 | new_levels <- ifelse(prop_n > prop, levels, other_level)
99 | }
100 | }
101 |
102 | if (other_level %in% new_levels) {
103 | f <- lvls_revalue(f, new_levels)
104 | fct_relevel(f, other_level, after = Inf)
105 | } else {
106 | f
107 | }
108 | }
109 |
110 | #' @param min Preserves values that appear at least `min` number of times.
111 | #' @export
112 | #' @rdname fct_lump
113 | #' @examples
114 | #' x <- factor(letters[rpois(100, 5)])
115 | #' fct_lump_min(x, min = 10)
116 | fct_lump_min <- function(f, min, w = NULL, other_level = "Other") {
117 | f <- check_factor(f)
118 | w <- check_weights(w, length(f))
119 |
120 | levels <- levels(f)
121 | if (is.null(w)) {
122 | count <- as.vector(table(f))
123 | total <- length(f)
124 | } else {
125 | count <- as.vector(tapply(w, f, FUN = sum))
126 | total <- sum(w)
127 | }
128 |
129 | new_levels <- ifelse(count >= min, levels, other_level)
130 |
131 | if (other_level %in% new_levels) {
132 | f <- lvls_revalue(f, new_levels)
133 | fct_relevel(f, other_level, after = Inf)
134 | } else {
135 | f
136 | }
137 |
138 | }
139 |
140 | # Lump together smallest groups, ensuring that the collective
141 | # "other" is still the smallest group. Assumes x is vector
142 | # of counts in descending order
143 | lump_cutoff <- function(x) {
144 | left <- sum(x)
145 |
146 | for (i in seq_along(x)) {
147 | # After group, there are this many left
148 | left <- left - x[i]
149 |
150 | if (x[i] > left)
151 | return(i + 1)
152 | }
153 |
154 | length(x) + 1
155 | }
156 |
157 | # Given vector of counts, returns logical vector if in
158 | # smallest groups
159 | in_smallest <- function(x) {
160 | ord_x <- order(x, decreasing = TRUE)
161 | idx <- lump_cutoff(x[ord_x])
162 |
163 | to_lump <- seq_along(x) >= idx
164 | # Undo initial ordering
165 | to_lump[order(ord_x)]
166 | }
167 |
168 | check_weights <- function(w, n = length(w)) {
169 | if (is.null(w)) {
170 | return(w)
171 | }
172 |
173 | if (!is.numeric(w)) {
174 | stop("`w` must be a numeric vector", call. = FALSE)
175 | }
176 |
177 | if (length(w) != n) {
178 | stop(
179 | "`w` must be the same length as `f` (", n, "), not length ", length(w),
180 | call. = FALSE
181 | )
182 | }
183 |
184 | bad <- w < 0 | is.na(w)
185 | if (any(bad)) {
186 | stop(
187 | "All `w` must be non-negative and non-missing. Problems at positions: ",
188 | paste0(which(bad), collapse = ", "),
189 | call. = FALSE
190 | )
191 | }
192 |
193 | w
194 | }
195 |
--------------------------------------------------------------------------------
/revdep/problems.md:
--------------------------------------------------------------------------------
1 | # bupaR
2 |
3 | Version: 0.4.1
4 |
5 | ## In both
6 |
7 | * checking S3 generic/method consistency ... WARNING
8 | ```
9 | sample_n:
10 | function(tbl, size, replace, weight, .env, ...)
11 | sample_n.eventlog:
12 | function(tbl, size, replace, weight, .env)
13 |
14 | sample_n:
15 | function(tbl, size, replace, weight, .env, ...)
16 | sample_n.grouped_eventlog:
17 | function(tbl, size, replace, weight, .env)
18 |
19 | See section ‘Generic functions and methods’ in the ‘Writing R
20 | Extensions’ manual.
21 | ```
22 |
23 | # circumplex
24 |
25 | Version: 0.2.1
26 |
27 | ## In both
28 |
29 | * checking whether package ‘circumplex’ can be installed ... ERROR
30 | ```
31 | Installation failed.
32 | See ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/new/circumplex.Rcheck/00install.out’ for details.
33 | ```
34 |
35 | ## Installation
36 |
37 | ### Devel
38 |
39 | ```
40 | * installing *source* package ‘circumplex’ ...
41 | ** package ‘circumplex’ successfully unpacked and MD5 sums checked
42 | ** libs
43 | clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/Rcpp/include" -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/RcppArmadillo/include" -I/usr/local/include -fopenmp -fPIC -Wall -g -O2 -c RcppExports.cpp -o RcppExports.o
44 | clang: error: unsupported option '-fopenmp'
45 | make: *** [RcppExports.o] Error 1
46 | ERROR: compilation failed for package ‘circumplex’
47 | * removing ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/new/circumplex.Rcheck/circumplex’
48 |
49 | ```
50 | ### CRAN
51 |
52 | ```
53 | * installing *source* package ‘circumplex’ ...
54 | ** package ‘circumplex’ successfully unpacked and MD5 sums checked
55 | ** libs
56 | clang++ -std=gnu++11 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/Rcpp/include" -I"/Users/hadley/Documents/tidyverse/forcats/revdep/library.noindex/circumplex/RcppArmadillo/include" -I/usr/local/include -fopenmp -fPIC -Wall -g -O2 -c RcppExports.cpp -o RcppExports.o
57 | clang: error: unsupported option '-fopenmp'
58 | make: *** [RcppExports.o] Error 1
59 | ERROR: compilation failed for package ‘circumplex’
60 | * removing ‘/Users/hadley/Documents/tidyverse/forcats/revdep/checks.noindex/circumplex/old/circumplex.Rcheck/circumplex’
61 |
62 | ```
63 | # cocktailApp
64 |
65 | Version: 0.2.0
66 |
67 | ## In both
68 |
69 | * checking data for non-ASCII characters ... NOTE
70 | ```
71 | Note: found 14661 marked UTF-8 strings
72 | ```
73 |
74 | # dabestr
75 |
76 | Version: 0.2.0
77 |
78 | ## In both
79 |
80 | * checking re-building of vignette outputs ... WARNING
81 | ```
82 | ...
83 |
84 | The following objects are masked from 'package:base':
85 |
86 | intersect, setdiff, setequal, union
87 |
88 |
89 | Attaching package: 'cowplot'
90 |
91 | The following object is masked from 'package:ggplot2':
92 |
93 | ggsave
94 |
95 | Loading required package: boot
96 | Warning: `data_frame()` is deprecated, use `tibble()`.
97 | This warning is displayed once per session.
98 | Loading required package: magrittr
99 | Warning: Some components of ... were not used: ..1
100 | Quitting from lines 110-166 (robust-statistical-visualization.Rmd)
101 | Error: processing vignette 'robust-statistical-visualization.Rmd' failed with diagnostics:
102 | polygon edge not found
103 | Execution halted
104 | ```
105 |
106 | * checking installed package size ... NOTE
107 | ```
108 | installed size is 6.6Mb
109 | sub-directories of 1Mb or more:
110 | doc 5.8Mb
111 | ```
112 |
113 | # ezplot
114 |
115 | Version: 0.2.2
116 |
117 | ## In both
118 |
119 | * checking dependencies in R code ... NOTE
120 | ```
121 | Namespace in Imports field not imported from: ‘magrittr’
122 | All declared Imports should be used.
123 | ```
124 |
125 | # factorMerger
126 |
127 | Version: 0.3.6
128 |
129 | ## In both
130 |
131 | * checking dependencies in R code ... NOTE
132 | ```
133 | Namespaces in Imports field not imported from:
134 | ‘forcats’ ‘formula.tools’
135 | All declared Imports should be used.
136 | ```
137 |
138 | # finalfit
139 |
140 | Version: 0.9.0
141 |
142 | ## In both
143 |
144 | * checking dependencies in R code ... NOTE
145 | ```
146 | Namespace in Imports field not imported from: ‘readr’
147 | All declared Imports should be used.
148 | ```
149 |
150 | # furniture
151 |
152 | Version: 1.8.7
153 |
154 | ## In both
155 |
156 | * checking dependencies in R code ... NOTE
157 | ```
158 | Namespace in Imports field not imported from: ‘magrittr’
159 | All declared Imports should be used.
160 | ```
161 |
162 | # ggdag
163 |
164 | Version: 0.1.0
165 |
166 | ## In both
167 |
168 | * checking dependencies in R code ... NOTE
169 | ```
170 | Namespaces in Imports field not imported from:
171 | ‘ggforce’ ‘plyr’
172 | All declared Imports should be used.
173 | ```
174 |
175 | # ggridges
176 |
177 | Version: 0.5.1
178 |
179 | ## In both
180 |
181 | * checking data for non-ASCII characters ... NOTE
182 | ```
183 | Note: found 6242 marked UTF-8 strings
184 | ```
185 |
186 | # glmmfields
187 |
188 | Version: 0.1.1
189 |
190 | ## In both
191 |
192 | * checking for GNU extensions in Makefiles ... NOTE
193 | ```
194 | GNU make is a SystemRequirements.
195 | ```
196 |
197 | # glmSparseNet
198 |
199 | Version: 1.0.0
200 |
201 | ## In both
202 |
203 | * checking re-building of vignette outputs ... WARNING
204 | ```
205 | Error in re-building vignettes:
206 | ...
207 |
208 | Attaching package: 'dplyr'
209 |
210 | The following objects are masked from 'package:stats':
211 |
212 | filter, lag
213 |
214 | The following objects are masked from 'package:base':
215 |
216 | intersect, setdiff, setequal, union
217 |
218 | Quitting from lines 28-43 (example_brca_logistic.Rmd)
219 | Error: processing vignette 'example_brca_logistic.Rmd' failed with diagnostics:
220 | there is no package called 'curatedTCGAData'
221 | Execution halted
222 | ```
223 |
224 | * checking package dependencies ... NOTE
225 | ```
226 | Package suggested but not available for checking: ‘curatedTCGAData’
227 | ```
228 |
229 | * checking installed package size ... NOTE
230 | ```
231 | installed size is 6.0Mb
232 | sub-directories of 1Mb or more:
233 | doc 5.2Mb
234 | ```
235 |
236 | # haven
237 |
238 | Version: 2.0.0
239 |
240 | ## In both
241 |
242 | * checking for GNU extensions in Makefiles ... NOTE
243 | ```
244 | GNU make is a SystemRequirements.
245 | ```
246 |
247 | # idealstan
248 |
249 | Version: 0.5.1
250 |
251 | ## In both
252 |
253 | * checking installed package size ... NOTE
254 | ```
255 | installed size is 6.2Mb
256 | sub-directories of 1Mb or more:
257 | libs 4.1Mb
258 | ```
259 |
260 | * checking for GNU extensions in Makefiles ... NOTE
261 | ```
262 | GNU make is a SystemRequirements.
263 | ```
264 |
265 | # iotables
266 |
267 | Version: 0.4.2
268 |
269 | ## In both
270 |
271 | * checking data for non-ASCII characters ... NOTE
272 | ```
273 | Note: found 53206 marked UTF-8 strings
274 | ```
275 |
276 | # konfound
277 |
278 | Version: 0.1.1
279 |
280 | ## In both
281 |
282 | * checking re-building of vignette outputs ... WARNING
283 | ```
284 | Error in re-building vignettes:
285 | ...
286 | Quitting from lines 50-51 (introduction-to-konfound.Rmd)
287 | Error: processing vignette 'introduction-to-konfound.Rmd' failed with diagnostics:
288 | there is no package called 'devtools'
289 | Execution halted
290 | ```
291 |
292 | * checking package dependencies ... NOTE
293 | ```
294 | Package suggested but not available for checking: ‘devtools’
295 | ```
296 |
297 | # MetamapsDB
298 |
299 | Version: 0.0.2
300 |
301 | ## In both
302 |
303 | * checking dependencies in R code ... NOTE
304 | ```
305 | Namespaces in Imports field not imported from:
306 | ‘Matrix’ ‘shiny’
307 | All declared Imports should be used.
308 | ```
309 |
310 | # noaastormevents
311 |
312 | Version: 0.1.0
313 |
314 | ## In both
315 |
316 | * checking package dependencies ... NOTE
317 | ```
318 | Package suggested but not available for checking: ‘hurricaneexposuredata’
319 | ```
320 |
321 | * checking dependencies in R code ... NOTE
322 | ```
323 | Namespaces in Imports field not imported from:
324 | ‘RColorBrewer’ ‘XML’ ‘choroplethr’ ‘choroplethrMaps’ ‘data.table’
325 | ‘forcats’ ‘hurricaneexposure’ ‘plyr’
326 | All declared Imports should be used.
327 | ```
328 |
329 | # nzelect
330 |
331 | Version: 0.4.0
332 |
333 | ## In both
334 |
335 | * checking installed package size ... NOTE
336 | ```
337 | installed size is 5.4Mb
338 | sub-directories of 1Mb or more:
339 | data 5.0Mb
340 | ```
341 |
342 | * checking data for non-ASCII characters ... NOTE
343 | ```
344 | Note: found 6409 marked UTF-8 strings
345 | ```
346 |
347 | # phenopath
348 |
349 | Version: 1.6.0
350 |
351 | ## In both
352 |
353 | * checking re-building of vignette outputs ... WARNING
354 | ```
355 | Error in re-building vignettes:
356 | ...
357 | Quitting from lines 64-72 (introduction_to_phenopath.Rmd)
358 | Error: processing vignette 'introduction_to_phenopath.Rmd' failed with diagnostics:
359 | Columns 1, 2, 3, 4, 5, … (and 3 more) must be named.
360 | Use .name_repair to specify repair.
361 | Execution halted
362 | ```
363 |
364 | # plotly
365 |
366 | Version: 4.8.0
367 |
368 | ## In both
369 |
370 | * checking package dependencies ... NOTE
371 | ```
372 | Package suggested but not available for checking: ‘devtools’
373 | ```
374 |
375 | * checking installed package size ... NOTE
376 | ```
377 | installed size is 7.0Mb
378 | sub-directories of 1Mb or more:
379 | R 2.3Mb
380 | htmlwidgets 3.1Mb
381 | ```
382 |
383 | # processmapR
384 |
385 | Version: 0.3.2
386 |
387 | ## In both
388 |
389 | * checking dependencies in R code ... NOTE
390 | ```
391 | Namespace in Imports field not imported from: ‘viridis’
392 | All declared Imports should be used.
393 | ```
394 |
395 | # questionr
396 |
397 | Version: 0.7.0
398 |
399 | ## In both
400 |
401 | * checking Rd cross-references ... NOTE
402 | ```
403 | Package unavailable to check Rd xrefs: ‘Hmisc’
404 | ```
405 |
406 | * checking data for non-ASCII characters ... NOTE
407 | ```
408 | Note: found 4145 marked UTF-8 strings
409 | ```
410 |
411 | # railtrails
412 |
413 | Version: 0.1.1
414 |
415 | ## In both
416 |
417 | * checking data for non-ASCII characters ... NOTE
418 | ```
419 | Note: found 1557 marked UTF-8 strings
420 | ```
421 |
422 | # rbin
423 |
424 | Version: 0.1.1
425 |
426 | ## In both
427 |
428 | * checking dependencies in R code ... NOTE
429 | ```
430 | Namespace in Imports field not imported from: ‘utils’
431 | All declared Imports should be used.
432 | ```
433 |
434 | # SCnorm
435 |
436 | Version: 1.4.5
437 |
438 | ## In both
439 |
440 | * checking package dependencies ... NOTE
441 | ```
442 | Package suggested but not available for checking: ‘devtools’
443 | ```
444 |
445 | # sweep
446 |
447 | Version: 0.2.1.1
448 |
449 | ## In both
450 |
451 | * checking package dependencies ... ERROR
452 | ```
453 | Package required but not available: ‘devtools’
454 |
455 | See section ‘The DESCRIPTION file’ in the ‘Writing R Extensions’
456 | manual.
457 | ```
458 |
459 | # TextForecast
460 |
461 | Version: 0.1.0
462 |
463 | ## In both
464 |
465 | * checking dependencies in R code ... NOTE
466 | ```
467 | Namespaces in Imports field not imported from:
468 | ‘doParallel’ ‘forecast’ ‘lars’ ‘parallel’ ‘tau’ ‘tsDyn’
469 | All declared Imports should be used.
470 | ```
471 |
472 | * checking data for non-ASCII characters ... NOTE
473 | ```
474 | Note: found 40 marked UTF-8 strings
475 | ```
476 |
477 | # tidybayes
478 |
479 | Version: 1.0.3
480 |
481 | ## In both
482 |
483 | * checking tests ...
484 | ```
485 | ERROR
486 | Running the tests in ‘tests/testthat.R’ failed.
487 | Last 13 lines of output:
488 | 9: setup.jagsfile(model = model, n.chains = n.chains, data = data, inits = inits, monitor = monitor, modules = modules,
489 | factories = factories, jags = jags, call.setup = TRUE, method = method, mutate = mutate)
490 | 10: setup.jags(model = outmodel, monitor = outmonitor, data = outdata, n.chains = n.chains, inits = outinits, modules = modules,
491 | factories = factories, response = response, fitted = fitted, residual = residual, jags = jags, method = method,
492 | mutate = mutate)
493 | 11: loadandcheckrjags()
494 | 12: stop("Loading the rjags package failed (diagnostics are given above this error message)", call. = FALSE)
495 |
496 | ══ testthat results ═════════════════════════════════════════════════════════════════════════════════════
497 | OK: 218 SKIPPED: 43 FAILED: 2
498 | 1. Failure: groups from spread_draws retain factor level names (@test.spread_draws.R#251)
499 | 2. Error: tidy_draws works with runjags (@test.tidy_draws.R#87)
500 |
501 | Error: testthat unit tests failed
502 | Execution halted
503 | ```
504 |
505 | # tidyLPA
506 |
507 | Version: 0.2.4
508 |
509 | ## In both
510 |
511 | * checking package dependencies ... NOTE
512 | ```
513 | Package suggested but not available for checking: ‘devtools’
514 | ```
515 |
516 | # tidyverse
517 |
518 | Version: 1.2.1
519 |
520 | ## In both
521 |
522 | * checking dependencies in R code ... NOTE
523 | ```
524 | Namespaces in Imports field not imported from:
525 | ‘dbplyr’ ‘reprex’ ‘rlang’
526 | All declared Imports should be used.
527 | ```
528 |
529 | # timetk
530 |
531 | Version: 0.1.1.1
532 |
533 | ## In both
534 |
535 | * checking package dependencies ... ERROR
536 | ```
537 | Package required but not available: ‘devtools’
538 |
539 | See section ‘The DESCRIPTION file’ in the ‘Writing R Extensions’
540 | manual.
541 | ```
542 |
543 |
--------------------------------------------------------------------------------
/LICENSE.md:
--------------------------------------------------------------------------------
1 | GNU General Public License
2 | ==========================
3 |
4 | _Version 3, 29 June 2007_
5 | _Copyright © 2007 Free Software Foundation, Inc. <>_
6 |
7 | Everyone is permitted to copy and distribute verbatim copies of this license
8 | document, but changing it is not allowed.
9 |
10 | ## Preamble
11 |
12 | The GNU General Public License is a free, copyleft license for software and other
13 | kinds of works.
14 |
15 | The licenses for most software and other practical works are designed to take away
16 | your freedom to share and change the works. By contrast, the GNU General Public
17 | License is intended to guarantee your freedom to share and change all versions of a
18 | program--to make sure it remains free software for all its users. We, the Free
19 | Software Foundation, use the GNU General Public License for most of our software; it
20 | applies also to any other work released this way by its authors. You can apply it to
21 | your programs, too.
22 |
23 | When we speak of free software, we are referring to freedom, not price. Our General
24 | Public Licenses are designed to make sure that you have the freedom to distribute
25 | copies of free software (and charge for them if you wish), that you receive source
26 | code or can get it if you want it, that you can change the software or use pieces of
27 | it in new free programs, and that you know you can do these things.
28 |
29 | To protect your rights, we need to prevent others from denying you these rights or
30 | asking you to surrender the rights. Therefore, you have certain responsibilities if
31 | you distribute copies of the software, or if you modify it: responsibilities to
32 | respect the freedom of others.
33 |
34 | For example, if you distribute copies of such a program, whether gratis or for a fee,
35 | you must pass on to the recipients the same freedoms that you received. You must make
36 | sure that they, too, receive or can get the source code. And you must show them these
37 | terms so they know their rights.
38 |
39 | Developers that use the GNU GPL protect your rights with two steps: **(1)** assert
40 | copyright on the software, and **(2)** offer you this License giving you legal permission
41 | to copy, distribute and/or modify it.
42 |
43 | For the developers' and authors' protection, the GPL clearly explains that there is
44 | no warranty for this free software. For both users' and authors' sake, the GPL
45 | requires that modified versions be marked as changed, so that their problems will not
46 | be attributed erroneously to authors of previous versions.
47 |
48 | Some devices are designed to deny users access to install or run modified versions of
49 | the software inside them, although the manufacturer can do so. This is fundamentally
50 | incompatible with the aim of protecting users' freedom to change the software. The
51 | systematic pattern of such abuse occurs in the area of products for individuals to
52 | use, which is precisely where it is most unacceptable. Therefore, we have designed
53 | this version of the GPL to prohibit the practice for those products. If such problems
54 | arise substantially in other domains, we stand ready to extend this provision to
55 | those domains in future versions of the GPL, as needed to protect the freedom of
56 | users.
57 |
58 | Finally, every program is threatened constantly by software patents. States should
59 | not allow patents to restrict development and use of software on general-purpose
60 | computers, but in those that do, we wish to avoid the special danger that patents
61 | applied to a free program could make it effectively proprietary. To prevent this, the
62 | GPL assures that patents cannot be used to render the program non-free.
63 |
64 | The precise terms and conditions for copying, distribution and modification follow.
65 |
66 | ## TERMS AND CONDITIONS
67 |
68 | ### 0. Definitions
69 |
70 | “This License” refers to version 3 of the GNU General Public License.
71 |
72 | “Copyright” also means copyright-like laws that apply to other kinds of
73 | works, such as semiconductor masks.
74 |
75 | “The Program” refers to any copyrightable work licensed under this
76 | License. Each licensee is addressed as “you”. “Licensees” and
77 | “recipients” may be individuals or organizations.
78 |
79 | To “modify” a work means to copy from or adapt all or part of the work in
80 | a fashion requiring copyright permission, other than the making of an exact copy. The
81 | resulting work is called a “modified version” of the earlier work or a
82 | work “based on” the earlier work.
83 |
84 | A “covered work” means either the unmodified Program or a work based on
85 | the Program.
86 |
87 | To “propagate” a work means to do anything with it that, without
88 | permission, would make you directly or secondarily liable for infringement under
89 | applicable copyright law, except executing it on a computer or modifying a private
90 | copy. Propagation includes copying, distribution (with or without modification),
91 | making available to the public, and in some countries other activities as well.
92 |
93 | To “convey” a work means any kind of propagation that enables other
94 | parties to make or receive copies. Mere interaction with a user through a computer
95 | network, with no transfer of a copy, is not conveying.
96 |
97 | An interactive user interface displays “Appropriate Legal Notices” to the
98 | extent that it includes a convenient and prominently visible feature that **(1)**
99 | displays an appropriate copyright notice, and **(2)** tells the user that there is no
100 | warranty for the work (except to the extent that warranties are provided), that
101 | licensees may convey the work under this License, and how to view a copy of this
102 | License. If the interface presents a list of user commands or options, such as a
103 | menu, a prominent item in the list meets this criterion.
104 |
105 | ### 1. Source Code
106 |
107 | The “source code” for a work means the preferred form of the work for
108 | making modifications to it. “Object code” means any non-source form of a
109 | work.
110 |
111 | A “Standard Interface” means an interface that either is an official
112 | standard defined by a recognized standards body, or, in the case of interfaces
113 | specified for a particular programming language, one that is widely used among
114 | developers working in that language.
115 |
116 | The “System Libraries” of an executable work include anything, other than
117 | the work as a whole, that **(a)** is included in the normal form of packaging a Major
118 | Component, but which is not part of that Major Component, and **(b)** serves only to
119 | enable use of the work with that Major Component, or to implement a Standard
120 | Interface for which an implementation is available to the public in source code form.
121 | A “Major Component”, in this context, means a major essential component
122 | (kernel, window system, and so on) of the specific operating system (if any) on which
123 | the executable work runs, or a compiler used to produce the work, or an object code
124 | interpreter used to run it.
125 |
126 | The “Corresponding Source” for a work in object code form means all the
127 | source code needed to generate, install, and (for an executable work) run the object
128 | code and to modify the work, including scripts to control those activities. However,
129 | it does not include the work's System Libraries, or general-purpose tools or
130 | generally available free programs which are used unmodified in performing those
131 | activities but which are not part of the work. For example, Corresponding Source
132 | includes interface definition files associated with source files for the work, and
133 | the source code for shared libraries and dynamically linked subprograms that the work
134 | is specifically designed to require, such as by intimate data communication or
135 | control flow between those subprograms and other parts of the work.
136 |
137 | The Corresponding Source need not include anything that users can regenerate
138 | automatically from other parts of the Corresponding Source.
139 |
140 | The Corresponding Source for a work in source code form is that same work.
141 |
142 | ### 2. Basic Permissions
143 |
144 | All rights granted under this License are granted for the term of copyright on the
145 | Program, and are irrevocable provided the stated conditions are met. This License
146 | explicitly affirms your unlimited permission to run the unmodified Program. The
147 | output from running a covered work is covered by this License only if the output,
148 | given its content, constitutes a covered work. This License acknowledges your rights
149 | of fair use or other equivalent, as provided by copyright law.
150 |
151 | You may make, run and propagate covered works that you do not convey, without
152 | conditions so long as your license otherwise remains in force. You may convey covered
153 | works to others for the sole purpose of having them make modifications exclusively
154 | for you, or provide you with facilities for running those works, provided that you
155 | comply with the terms of this License in conveying all material for which you do not
156 | control copyright. Those thus making or running the covered works for you must do so
157 | exclusively on your behalf, under your direction and control, on terms that prohibit
158 | them from making any copies of your copyrighted material outside their relationship
159 | with you.
160 |
161 | Conveying under any other circumstances is permitted solely under the conditions
162 | stated below. Sublicensing is not allowed; section 10 makes it unnecessary.
163 |
164 | ### 3. Protecting Users' Legal Rights From Anti-Circumvention Law
165 |
166 | No covered work shall be deemed part of an effective technological measure under any
167 | applicable law fulfilling obligations under article 11 of the WIPO copyright treaty
168 | adopted on 20 December 1996, or similar laws prohibiting or restricting circumvention
169 | of such measures.
170 |
171 | When you convey a covered work, you waive any legal power to forbid circumvention of
172 | technological measures to the extent such circumvention is effected by exercising
173 | rights under this License with respect to the covered work, and you disclaim any
174 | intention to limit operation or modification of the work as a means of enforcing,
175 | against the work's users, your or third parties' legal rights to forbid circumvention
176 | of technological measures.
177 |
178 | ### 4. Conveying Verbatim Copies
179 |
180 | You may convey verbatim copies of the Program's source code as you receive it, in any
181 | medium, provided that you conspicuously and appropriately publish on each copy an
182 | appropriate copyright notice; keep intact all notices stating that this License and
183 | any non-permissive terms added in accord with section 7 apply to the code; keep
184 | intact all notices of the absence of any warranty; and give all recipients a copy of
185 | this License along with the Program.
186 |
187 | You may charge any price or no price for each copy that you convey, and you may offer
188 | support or warranty protection for a fee.
189 |
190 | ### 5. Conveying Modified Source Versions
191 |
192 | You may convey a work based on the Program, or the modifications to produce it from
193 | the Program, in the form of source code under the terms of section 4, provided that
194 | you also meet all of these conditions:
195 |
196 | * **a)** The work must carry prominent notices stating that you modified it, and giving a
197 | relevant date.
198 | * **b)** The work must carry prominent notices stating that it is released under this
199 | License and any conditions added under section 7. This requirement modifies the
200 | requirement in section 4 to “keep intact all notices”.
201 | * **c)** You must license the entire work, as a whole, under this License to anyone who
202 | comes into possession of a copy. This License will therefore apply, along with any
203 | applicable section 7 additional terms, to the whole of the work, and all its parts,
204 | regardless of how they are packaged. This License gives no permission to license the
205 | work in any other way, but it does not invalidate such permission if you have
206 | separately received it.
207 | * **d)** If the work has interactive user interfaces, each must display Appropriate Legal
208 | Notices; however, if the Program has interactive interfaces that do not display
209 | Appropriate Legal Notices, your work need not make them do so.
210 |
211 | A compilation of a covered work with other separate and independent works, which are
212 | not by their nature extensions of the covered work, and which are not combined with
213 | it such as to form a larger program, in or on a volume of a storage or distribution
214 | medium, is called an “aggregate” if the compilation and its resulting
215 | copyright are not used to limit the access or legal rights of the compilation's users
216 | beyond what the individual works permit. Inclusion of a covered work in an aggregate
217 | does not cause this License to apply to the other parts of the aggregate.
218 |
219 | ### 6. Conveying Non-Source Forms
220 |
221 | You may convey a covered work in object code form under the terms of sections 4 and
222 | 5, provided that you also convey the machine-readable Corresponding Source under the
223 | terms of this License, in one of these ways:
224 |
225 | * **a)** Convey the object code in, or embodied in, a physical product (including a
226 | physical distribution medium), accompanied by the Corresponding Source fixed on a
227 | durable physical medium customarily used for software interchange.
228 | * **b)** Convey the object code in, or embodied in, a physical product (including a
229 | physical distribution medium), accompanied by a written offer, valid for at least
230 | three years and valid for as long as you offer spare parts or customer support for
231 | that product model, to give anyone who possesses the object code either **(1)** a copy of
232 | the Corresponding Source for all the software in the product that is covered by this
233 | License, on a durable physical medium customarily used for software interchange, for
234 | a price no more than your reasonable cost of physically performing this conveying of
235 | source, or **(2)** access to copy the Corresponding Source from a network server at no
236 | charge.
237 | * **c)** Convey individual copies of the object code with a copy of the written offer to
238 | provide the Corresponding Source. This alternative is allowed only occasionally and
239 | noncommercially, and only if you received the object code with such an offer, in
240 | accord with subsection 6b.
241 | * **d)** Convey the object code by offering access from a designated place (gratis or for
242 | a charge), and offer equivalent access to the Corresponding Source in the same way
243 | through the same place at no further charge. You need not require recipients to copy
244 | the Corresponding Source along with the object code. If the place to copy the object
245 | code is a network server, the Corresponding Source may be on a different server
246 | (operated by you or a third party) that supports equivalent copying facilities,
247 | provided you maintain clear directions next to the object code saying where to find
248 | the Corresponding Source. Regardless of what server hosts the Corresponding Source,
249 | you remain obligated to ensure that it is available for as long as needed to satisfy
250 | these requirements.
251 | * **e)** Convey the object code using peer-to-peer transmission, provided you inform
252 | other peers where the object code and Corresponding Source of the work are being
253 | offered to the general public at no charge under subsection 6d.
254 |
255 | A separable portion of the object code, whose source code is excluded from the
256 | Corresponding Source as a System Library, need not be included in conveying the
257 | object code work.
258 |
259 | A “User Product” is either **(1)** a “consumer product”, which
260 | means any tangible personal property which is normally used for personal, family, or
261 | household purposes, or **(2)** anything designed or sold for incorporation into a
262 | dwelling. In determining whether a product is a consumer product, doubtful cases
263 | shall be resolved in favor of coverage. For a particular product received by a
264 | particular user, “normally used” refers to a typical or common use of
265 | that class of product, regardless of the status of the particular user or of the way
266 | in which the particular user actually uses, or expects or is expected to use, the
267 | product. A product is a consumer product regardless of whether the product has
268 | substantial commercial, industrial or non-consumer uses, unless such uses represent
269 | the only significant mode of use of the product.
270 |
271 | “Installation Information” for a User Product means any methods,
272 | procedures, authorization keys, or other information required to install and execute
273 | modified versions of a covered work in that User Product from a modified version of
274 | its Corresponding Source. The information must suffice to ensure that the continued
275 | functioning of the modified object code is in no case prevented or interfered with
276 | solely because modification has been made.
277 |
278 | If you convey an object code work under this section in, or with, or specifically for
279 | use in, a User Product, and the conveying occurs as part of a transaction in which
280 | the right of possession and use of the User Product is transferred to the recipient
281 | in perpetuity or for a fixed term (regardless of how the transaction is
282 | characterized), the Corresponding Source conveyed under this section must be
283 | accompanied by the Installation Information. But this requirement does not apply if
284 | neither you nor any third party retains the ability to install modified object code
285 | on the User Product (for example, the work has been installed in ROM).
286 |
287 | The requirement to provide Installation Information does not include a requirement to
288 | continue to provide support service, warranty, or updates for a work that has been
289 | modified or installed by the recipient, or for the User Product in which it has been
290 | modified or installed. Access to a network may be denied when the modification itself
291 | materially and adversely affects the operation of the network or violates the rules
292 | and protocols for communication across the network.
293 |
294 | Corresponding Source conveyed, and Installation Information provided, in accord with
295 | this section must be in a format that is publicly documented (and with an
296 | implementation available to the public in source code form), and must require no
297 | special password or key for unpacking, reading or copying.
298 |
299 | ### 7. Additional Terms
300 |
301 | “Additional permissions” are terms that supplement the terms of this
302 | License by making exceptions from one or more of its conditions. Additional
303 | permissions that are applicable to the entire Program shall be treated as though they
304 | were included in this License, to the extent that they are valid under applicable
305 | law. If additional permissions apply only to part of the Program, that part may be
306 | used separately under those permissions, but the entire Program remains governed by
307 | this License without regard to the additional permissions.
308 |
309 | When you convey a copy of a covered work, you may at your option remove any
310 | additional permissions from that copy, or from any part of it. (Additional
311 | permissions may be written to require their own removal in certain cases when you
312 | modify the work.) You may place additional permissions on material, added by you to a
313 | covered work, for which you have or can give appropriate copyright permission.
314 |
315 | Notwithstanding any other provision of this License, for material you add to a
316 | covered work, you may (if authorized by the copyright holders of that material)
317 | supplement the terms of this License with terms:
318 |
319 | * **a)** Disclaiming warranty or limiting liability differently from the terms of
320 | sections 15 and 16 of this License; or
321 | * **b)** Requiring preservation of specified reasonable legal notices or author
322 | attributions in that material or in the Appropriate Legal Notices displayed by works
323 | containing it; or
324 | * **c)** Prohibiting misrepresentation of the origin of that material, or requiring that
325 | modified versions of such material be marked in reasonable ways as different from the
326 | original version; or
327 | * **d)** Limiting the use for publicity purposes of names of licensors or authors of the
328 | material; or
329 | * **e)** Declining to grant rights under trademark law for use of some trade names,
330 | trademarks, or service marks; or
331 | * **f)** Requiring indemnification of licensors and authors of that material by anyone
332 | who conveys the material (or modified versions of it) with contractual assumptions of
333 | liability to the recipient, for any liability that these contractual assumptions
334 | directly impose on those licensors and authors.
335 |
336 | All other non-permissive additional terms are considered “further
337 | restrictions” within the meaning of section 10. If the Program as you received
338 | it, or any part of it, contains a notice stating that it is governed by this License
339 | along with a term that is a further restriction, you may remove that term. If a
340 | license document contains a further restriction but permits relicensing or conveying
341 | under this License, you may add to a covered work material governed by the terms of
342 | that license document, provided that the further restriction does not survive such
343 | relicensing or conveying.
344 |
345 | If you add terms to a covered work in accord with this section, you must place, in
346 | the relevant source files, a statement of the additional terms that apply to those
347 | files, or a notice indicating where to find the applicable terms.
348 |
349 | Additional terms, permissive or non-permissive, may be stated in the form of a
350 | separately written license, or stated as exceptions; the above requirements apply
351 | either way.
352 |
353 | ### 8. Termination
354 |
355 | You may not propagate or modify a covered work except as expressly provided under
356 | this License. Any attempt otherwise to propagate or modify it is void, and will
357 | automatically terminate your rights under this License (including any patent licenses
358 | granted under the third paragraph of section 11).
359 |
360 | However, if you cease all violation of this License, then your license from a
361 | particular copyright holder is reinstated **(a)** provisionally, unless and until the
362 | copyright holder explicitly and finally terminates your license, and **(b)** permanently,
363 | if the copyright holder fails to notify you of the violation by some reasonable means
364 | prior to 60 days after the cessation.
365 |
366 | Moreover, your license from a particular copyright holder is reinstated permanently
367 | if the copyright holder notifies you of the violation by some reasonable means, this
368 | is the first time you have received notice of violation of this License (for any
369 | work) from that copyright holder, and you cure the violation prior to 30 days after
370 | your receipt of the notice.
371 |
372 | Termination of your rights under this section does not terminate the licenses of
373 | parties who have received copies or rights from you under this License. If your
374 | rights have been terminated and not permanently reinstated, you do not qualify to
375 | receive new licenses for the same material under section 10.
376 |
377 | ### 9. Acceptance Not Required for Having Copies
378 |
379 | You are not required to accept this License in order to receive or run a copy of the
380 | Program. Ancillary propagation of a covered work occurring solely as a consequence of
381 | using peer-to-peer transmission to receive a copy likewise does not require
382 | acceptance. However, nothing other than this License grants you permission to
383 | propagate or modify any covered work. These actions infringe copyright if you do not
384 | accept this License. Therefore, by modifying or propagating a covered work, you
385 | indicate your acceptance of this License to do so.
386 |
387 | ### 10. Automatic Licensing of Downstream Recipients
388 |
389 | Each time you convey a covered work, the recipient automatically receives a license
390 | from the original licensors, to run, modify and propagate that work, subject to this
391 | License. You are not responsible for enforcing compliance by third parties with this
392 | License.
393 |
394 | An “entity transaction” is a transaction transferring control of an
395 | organization, or substantially all assets of one, or subdividing an organization, or
396 | merging organizations. If propagation of a covered work results from an entity
397 | transaction, each party to that transaction who receives a copy of the work also
398 | receives whatever licenses to the work the party's predecessor in interest had or
399 | could give under the previous paragraph, plus a right to possession of the
400 | Corresponding Source of the work from the predecessor in interest, if the predecessor
401 | has it or can get it with reasonable efforts.
402 |
403 | You may not impose any further restrictions on the exercise of the rights granted or
404 | affirmed under this License. For example, you may not impose a license fee, royalty,
405 | or other charge for exercise of rights granted under this License, and you may not
406 | initiate litigation (including a cross-claim or counterclaim in a lawsuit) alleging
407 | that any patent claim is infringed by making, using, selling, offering for sale, or
408 | importing the Program or any portion of it.
409 |
410 | ### 11. Patents
411 |
412 | A “contributor” is a copyright holder who authorizes use under this
413 | License of the Program or a work on which the Program is based. The work thus
414 | licensed is called the contributor's “contributor version”.
415 |
416 | A contributor's “essential patent claims” are all patent claims owned or
417 | controlled by the contributor, whether already acquired or hereafter acquired, that
418 | would be infringed by some manner, permitted by this License, of making, using, or
419 | selling its contributor version, but do not include claims that would be infringed
420 | only as a consequence of further modification of the contributor version. For
421 | purposes of this definition, “control” includes the right to grant patent
422 | sublicenses in a manner consistent with the requirements of this License.
423 |
424 | Each contributor grants you a non-exclusive, worldwide, royalty-free patent license
425 | under the contributor's essential patent claims, to make, use, sell, offer for sale,
426 | import and otherwise run, modify and propagate the contents of its contributor
427 | version.
428 |
429 | In the following three paragraphs, a “patent license” is any express
430 | agreement or commitment, however denominated, not to enforce a patent (such as an
431 | express permission to practice a patent or covenant not to sue for patent
432 | infringement). To “grant” such a patent license to a party means to make
433 | such an agreement or commitment not to enforce a patent against the party.
434 |
435 | If you convey a covered work, knowingly relying on a patent license, and the
436 | Corresponding Source of the work is not available for anyone to copy, free of charge
437 | and under the terms of this License, through a publicly available network server or
438 | other readily accessible means, then you must either **(1)** cause the Corresponding
439 | Source to be so available, or **(2)** arrange to deprive yourself of the benefit of the
440 | patent license for this particular work, or **(3)** arrange, in a manner consistent with
441 | the requirements of this License, to extend the patent license to downstream
442 | recipients. “Knowingly relying” means you have actual knowledge that, but
443 | for the patent license, your conveying the covered work in a country, or your
444 | recipient's use of the covered work in a country, would infringe one or more
445 | identifiable patents in that country that you have reason to believe are valid.
446 |
447 | If, pursuant to or in connection with a single transaction or arrangement, you
448 | convey, or propagate by procuring conveyance of, a covered work, and grant a patent
449 | license to some of the parties receiving the covered work authorizing them to use,
450 | propagate, modify or convey a specific copy of the covered work, then the patent
451 | license you grant is automatically extended to all recipients of the covered work and
452 | works based on it.
453 |
454 | A patent license is “discriminatory” if it does not include within the
455 | scope of its coverage, prohibits the exercise of, or is conditioned on the
456 | non-exercise of one or more of the rights that are specifically granted under this
457 | License. You may not convey a covered work if you are a party to an arrangement with
458 | a third party that is in the business of distributing software, under which you make
459 | payment to the third party based on the extent of your activity of conveying the
460 | work, and under which the third party grants, to any of the parties who would receive
461 | the covered work from you, a discriminatory patent license **(a)** in connection with
462 | copies of the covered work conveyed by you (or copies made from those copies), or **(b)**
463 | primarily for and in connection with specific products or compilations that contain
464 | the covered work, unless you entered into that arrangement, or that patent license
465 | was granted, prior to 28 March 2007.
466 |
467 | Nothing in this License shall be construed as excluding or limiting any implied
468 | license or other defenses to infringement that may otherwise be available to you
469 | under applicable patent law.
470 |
471 | ### 12. No Surrender of Others' Freedom
472 |
473 | If conditions are imposed on you (whether by court order, agreement or otherwise)
474 | that contradict the conditions of this License, they do not excuse you from the
475 | conditions of this License. If you cannot convey a covered work so as to satisfy
476 | simultaneously your obligations under this License and any other pertinent
477 | obligations, then as a consequence you may not convey it at all. For example, if you
478 | agree to terms that obligate you to collect a royalty for further conveying from
479 | those to whom you convey the Program, the only way you could satisfy both those terms
480 | and this License would be to refrain entirely from conveying the Program.
481 |
482 | ### 13. Use with the GNU Affero General Public License
483 |
484 | Notwithstanding any other provision of this License, you have permission to link or
485 | combine any covered work with a work licensed under version 3 of the GNU Affero
486 | General Public License into a single combined work, and to convey the resulting work.
487 | The terms of this License will continue to apply to the part which is the covered
488 | work, but the special requirements of the GNU Affero General Public License, section
489 | 13, concerning interaction through a network will apply to the combination as such.
490 |
491 | ### 14. Revised Versions of this License
492 |
493 | The Free Software Foundation may publish revised and/or new versions of the GNU
494 | General Public License from time to time. Such new versions will be similar in spirit
495 | to the present version, but may differ in detail to address new problems or concerns.
496 |
497 | Each version is given a distinguishing version number. If the Program specifies that
498 | a certain numbered version of the GNU General Public License “or any later
499 | version” applies to it, you have the option of following the terms and
500 | conditions either of that numbered version or of any later version published by the
501 | Free Software Foundation. If the Program does not specify a version number of the GNU
502 | General Public License, you may choose any version ever published by the Free
503 | Software Foundation.
504 |
505 | If the Program specifies that a proxy can decide which future versions of the GNU
506 | General Public License can be used, that proxy's public statement of acceptance of a
507 | version permanently authorizes you to choose that version for the Program.
508 |
509 | Later license versions may give you additional or different permissions. However, no
510 | additional obligations are imposed on any author or copyright holder as a result of
511 | your choosing to follow a later version.
512 |
513 | ### 15. Disclaimer of Warranty
514 |
515 | THERE IS NO WARRANTY FOR THE PROGRAM, TO THE EXTENT PERMITTED BY APPLICABLE LAW.
516 | EXCEPT WHEN OTHERWISE STATED IN WRITING THE COPYRIGHT HOLDERS AND/OR OTHER PARTIES
517 | PROVIDE THE PROGRAM “AS IS” WITHOUT WARRANTY OF ANY KIND, EITHER
518 | EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF
519 | MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE ENTIRE RISK AS TO THE
520 | QUALITY AND PERFORMANCE OF THE PROGRAM IS WITH YOU. SHOULD THE PROGRAM PROVE
521 | DEFECTIVE, YOU ASSUME THE COST OF ALL NECESSARY SERVICING, REPAIR OR CORRECTION.
522 |
523 | ### 16. Limitation of Liability
524 |
525 | IN NO EVENT UNLESS REQUIRED BY APPLICABLE LAW OR AGREED TO IN WRITING WILL ANY
526 | COPYRIGHT HOLDER, OR ANY OTHER PARTY WHO MODIFIES AND/OR CONVEYS THE PROGRAM AS
527 | PERMITTED ABOVE, BE LIABLE TO YOU FOR DAMAGES, INCLUDING ANY GENERAL, SPECIAL,
528 | INCIDENTAL OR CONSEQUENTIAL DAMAGES ARISING OUT OF THE USE OR INABILITY TO USE THE
529 | PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF DATA OR DATA BEING RENDERED INACCURATE
530 | OR LOSSES SUSTAINED BY YOU OR THIRD PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE
531 | WITH ANY OTHER PROGRAMS), EVEN IF SUCH HOLDER OR OTHER PARTY HAS BEEN ADVISED OF THE
532 | POSSIBILITY OF SUCH DAMAGES.
533 |
534 | ### 17. Interpretation of Sections 15 and 16
535 |
536 | If the disclaimer of warranty and limitation of liability provided above cannot be
537 | given local legal effect according to their terms, reviewing courts shall apply local
538 | law that most closely approximates an absolute waiver of all civil liability in
539 | connection with the Program, unless a warranty or assumption of liability accompanies
540 | a copy of the Program in return for a fee.
541 |
542 | _END OF TERMS AND CONDITIONS_
543 |
544 | ## How to Apply These Terms to Your New Programs
545 |
546 | If you develop a new program, and you want it to be of the greatest possible use to
547 | the public, the best way to achieve this is to make it free software which everyone
548 | can redistribute and change under these terms.
549 |
550 | To do so, attach the following notices to the program. It is safest to attach them
551 | to the start of each source file to most effectively state the exclusion of warranty;
552 | and each file should have at least the “copyright” line and a pointer to
553 | where the full notice is found.
554 |
555 |
556 | Copyright (C) 2019 Mara Averick
557 |
558 | This program is free software: you can redistribute it and/or modify
559 | it under the terms of the GNU General Public License as published by
560 | the Free Software Foundation, either version 3 of the License, or
561 | (at your option) any later version.
562 |
563 | This program is distributed in the hope that it will be useful,
564 | but WITHOUT ANY WARRANTY; without even the implied warranty of
565 | MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
566 | GNU General Public License for more details.
567 |
568 | You should have received a copy of the GNU General Public License
569 | along with this program. If not, see .
570 |
571 | Also add information on how to contact you by electronic and paper mail.
572 |
573 | If the program does terminal interaction, make it output a short notice like this
574 | when it starts in an interactive mode:
575 |
576 | forcats Copyright (C) 2019 Mara Averick
577 | This program comes with ABSOLUTELY NO WARRANTY; for details type 'show w'.
578 | This is free software, and you are welcome to redistribute it
579 | under certain conditions; type 'show c' for details.
580 |
581 | The hypothetical commands `show w` and `show c` should show the appropriate parts of
582 | the General Public License. Of course, your program's commands might be different;
583 | for a GUI interface, you would use an “about box”.
584 |
585 | You should also get your employer (if you work as a programmer) or school, if any, to
586 | sign a “copyright disclaimer” for the program, if necessary. For more
587 | information on this, and how to apply and follow the GNU GPL, see
588 | <>.
589 |
590 | The GNU General Public License does not permit incorporating your program into
591 | proprietary programs. If your program is a subroutine library, you may consider it
592 | more useful to permit linking proprietary applications with the library. If this is
593 | what you want to do, use the GNU Lesser General Public License instead of this
594 | License. But first, please read
595 | <>.
596 |
--------------------------------------------------------------------------------
 16 |
17 |
18 | [](https://cran.r-project.org/package=forcats)
19 | [](https://travis-ci.org/tidyverse/forcats)
20 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
21 |
22 |
23 | ## Overview
24 |
25 | R uses __factors__ to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the __forcats__ package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. Some examples include:
26 |
27 | * `fct_reorder()`: Reordering a factor by another variable.
28 | * `fct_infreq()`: Reordering a factor by the frequency of values.
29 | * `fct_relevel()`: Changing the order of a factor by hand.
30 | * `fct_lump()`: Collapsing the least/most frequent values of a factor into "other".
31 |
32 | You can learn more about each of these in `vignette("forcats")`. If you're new to factors, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
33 |
34 | ## Installation
35 |
36 | ```
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 | ```
47 |
48 | ## Cheatsheet
49 |
50 |
16 |
17 |
18 | [](https://cran.r-project.org/package=forcats)
19 | [](https://travis-ci.org/tidyverse/forcats)
20 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
21 |
22 |
23 | ## Overview
24 |
25 | R uses __factors__ to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the __forcats__ package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. Some examples include:
26 |
27 | * `fct_reorder()`: Reordering a factor by another variable.
28 | * `fct_infreq()`: Reordering a factor by the frequency of values.
29 | * `fct_relevel()`: Changing the order of a factor by hand.
30 | * `fct_lump()`: Collapsing the least/most frequent values of a factor into "other".
31 |
32 | You can learn more about each of these in `vignette("forcats")`. If you're new to factors, the best place to start is the [chapter on factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
33 |
34 | ## Installation
35 |
36 | ```
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 | ```
47 |
48 | ## Cheatsheet
49 |
50 |  51 |
52 | ## Getting started
53 |
54 | forcats is part of the core tidyverse, so you can load it with `library(tidyverse)` or `library(forcats)`.
55 |
56 | ```{r setup, message = FALSE}
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ```{r}
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | ```
67 |
68 | ```{r}
69 | starwars %>%
70 | filter(!is.na(species)) %>%
71 | mutate(species = fct_lump(species, n = 3)) %>%
72 | count(species)
73 | ```
74 |
75 | ```{r unordered-plot}
76 | ggplot(starwars, aes(x = eye_color)) +
77 | geom_bar() +
78 | coord_flip()
79 | ```
80 |
81 | ```{r ordered-plot}
82 | starwars %>%
83 | mutate(eye_color = fct_infreq(eye_color)) %>%
84 | ggplot(aes(x = eye_color)) +
85 | geom_bar() +
86 | coord_flip()
87 | ```
88 |
89 | ## More resources
90 |
91 | For a history of factors, I recommend [_stringsAsFactors: An unauthorized biography_](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/) by Roger Peng and [_stringsAsFactors = \
51 |
52 | ## Getting started
53 |
54 | forcats is part of the core tidyverse, so you can load it with `library(tidyverse)` or `library(forcats)`.
55 |
56 | ```{r setup, message = FALSE}
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ```{r}
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | ```
67 |
68 | ```{r}
69 | starwars %>%
70 | filter(!is.na(species)) %>%
71 | mutate(species = fct_lump(species, n = 3)) %>%
72 | count(species)
73 | ```
74 |
75 | ```{r unordered-plot}
76 | ggplot(starwars, aes(x = eye_color)) +
77 | geom_bar() +
78 | coord_flip()
79 | ```
80 |
81 | ```{r ordered-plot}
82 | starwars %>%
83 | mutate(eye_color = fct_infreq(eye_color)) %>%
84 | ggplot(aes(x = eye_color)) +
85 | geom_bar() +
86 | coord_flip()
87 | ```
88 |
89 | ## More resources
90 |
91 | For a history of factors, I recommend [_stringsAsFactors: An unauthorized biography_](http://simplystatistics.org/2015/07/24/stringsasfactors-an-unauthorized-biography/) by Roger Peng and [_stringsAsFactors = \ 5 |
6 |
7 |
8 | [](https://cran.r-project.org/package=forcats)
10 | [](https://travis-ci.org/tidyverse/forcats)
12 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
14 |
15 |
16 | ## Overview
17 |
18 | R uses **factors** to handle categorical variables, variables that have
19 | a fixed and known set of possible values. Factors are also helpful for
20 | reordering character vectors to improve display. The goal of the
21 | **forcats** package is to provide a suite of tools that solve common
22 | problems with factors, including changing the order of levels or the
23 | values. Some examples include:
24 |
25 | - `fct_reorder()`: Reordering a factor by another variable.
26 | - `fct_infreq()`: Reordering a factor by the frequency of values.
27 | - `fct_relevel()`: Changing the order of a factor by hand.
28 | - `fct_lump()`: Collapsing the least/most frequent values of a factor
29 | into “other”.
30 |
31 | You can learn more about each of these in `vignette("forcats")`. If
32 | you’re new to factors, the best place to start is the [chapter on
33 | factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
34 |
35 | ## Installation
36 |
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 |
47 | ## Cheatsheet
48 |
49 |
5 |
6 |
7 |
8 | [](https://cran.r-project.org/package=forcats)
10 | [](https://travis-ci.org/tidyverse/forcats)
12 | [](https://codecov.io/gh/tidyverse/forcats?branch=master)
14 |
15 |
16 | ## Overview
17 |
18 | R uses **factors** to handle categorical variables, variables that have
19 | a fixed and known set of possible values. Factors are also helpful for
20 | reordering character vectors to improve display. The goal of the
21 | **forcats** package is to provide a suite of tools that solve common
22 | problems with factors, including changing the order of levels or the
23 | values. Some examples include:
24 |
25 | - `fct_reorder()`: Reordering a factor by another variable.
26 | - `fct_infreq()`: Reordering a factor by the frequency of values.
27 | - `fct_relevel()`: Changing the order of a factor by hand.
28 | - `fct_lump()`: Collapsing the least/most frequent values of a factor
29 | into “other”.
30 |
31 | You can learn more about each of these in `vignette("forcats")`. If
32 | you’re new to factors, the best place to start is the [chapter on
33 | factors](http://r4ds.had.co.nz/factors.html) in R for Data Science.
34 |
35 | ## Installation
36 |
37 | # The easiest way to get forcats is to install the whole tidyverse:
38 | install.packages("tidyverse")
39 |
40 | # Alternatively, install just forcats:
41 | install.packages("forcats")
42 |
43 | # Or the the development version from GitHub:
44 | # install.packages("devtools")
45 | devtools::install_github("tidyverse/forcats")
46 |
47 | ## Cheatsheet
48 |
49 |  50 |
51 | ## Getting started
52 |
53 | forcats is part of the core tidyverse, so you can load it with
54 | `library(tidyverse)` or `library(forcats)`.
55 |
56 | ``` r
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ``` r
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | #> # A tibble: 37 x 2
67 | #> species n
68 | #>
50 |
51 | ## Getting started
52 |
53 | forcats is part of the core tidyverse, so you can load it with
54 | `library(tidyverse)` or `library(forcats)`.
55 |
56 | ``` r
57 | library(forcats)
58 | library(dplyr)
59 | library(ggplot2)
60 | ```
61 |
62 | ``` r
63 | starwars %>%
64 | filter(!is.na(species)) %>%
65 | count(species, sort = TRUE)
66 | #> # A tibble: 37 x 2
67 | #> species n
68 | #>