├── .DS_Store
├── .gitattributes
├── LICENSE
├── MedMNIST_tutorial
├── .DS_Store
├── mednist_CE.ipynb
├── mednist_CM.ipynb
├── mednist_HC.ipynb
└── pretty_confusion_matrix_local.py
├── NIFTI
├── affine.m
├── bipolar.m
├── bresenham_line3d.m
├── clip_nii.m
├── collapse_nii_scan.m
├── examples.txt
├── expand_nii_scan.m
├── extra_nii_hdr.m
├── flip_lr.m
├── get_nii_frame.m
├── license.txt
├── load_nii.m
├── load_nii_ext.m
├── load_nii_hdr.m
├── load_nii_img.m
├── load_untouch0_nii_hdr.m
├── load_untouch_header_only.m
├── load_untouch_nii.m
├── load_untouch_nii_hdr.m
├── load_untouch_nii_img.m
├── make_ana.m
├── make_nii.m
├── mat_into_hdr.m
├── reslice_nii.m
├── rri_file_menu.m
├── rri_orient.m
├── rri_orient_ui.m
├── rri_select_file.m
├── rri_xhair.m
├── rri_zoom_menu.m
├── save_nii.m
├── save_nii_ext.m
├── save_nii_hdr.m
├── save_untouch0_nii_hdr.m
├── save_untouch_nii.m
├── save_untouch_nii_hdr.m
├── save_untouch_slice.m
├── unxform_nii.m
├── verify_nii_ext.m
├── view_nii.m
├── view_nii_menu.m
└── xform_nii.m
├── README.md
├── UNETR_matrixpenalty_v5_2d.csv
├── build_container_v08.sh
├── combine_masks.m
├── doc
├── DOMINO_MICCAI22_PPT.pdf
├── ReadMeDoc.md
└── poster1693.pdf
├── make_datalist_json.m
├── test.sh
├── test_domino.py
├── tissue_cond_11.xlsx
├── train.sh
└── train_domino.py
/.DS_Store:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/lab-smile/DOMINO/3d261da4dec8ba338818babfa471c47c51d0866d/.DS_Store
--------------------------------------------------------------------------------
/.gitattributes:
--------------------------------------------------------------------------------
1 | .pth filter=lfs diff=lfs merge=lfs -text
2 | *.pth filter=lfs diff=lfs merge=lfs -text
3 |
--------------------------------------------------------------------------------
/MedMNIST_tutorial/.DS_Store:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/lab-smile/DOMINO/3d261da4dec8ba338818babfa471c47c51d0866d/MedMNIST_tutorial/.DS_Store
--------------------------------------------------------------------------------
/MedMNIST_tutorial/pretty_confusion_matrix_local.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 | """
3 | plot a pretty confusion matrix with seaborn
4 | Created on Mon Jun 25 14:17:37 2018
5 | @author: Wagner Cipriano - wagnerbhbr - gmail - CEFETMG / MMC
6 | REFerences:
7 | https://www.mathworks.com/help/nnet/ref/plotconfusion.html
8 | https://stackoverflow.com/questions/28200786/how-to-plot-scikit-learn-classification-report
9 | https://stackoverflow.com/questions/5821125/how-to-plot-confusion-matrix-with-string-axis-rather-than-integer-in-python
10 | https://www.programcreek.com/python/example/96197/seaborn.heatmap
11 | https://stackoverflow.com/questions/19233771/sklearn-plot-confusion-matrix-with-labels/31720054
12 | http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html#sphx-glr-auto-examples-model-selection-plot-confusion-matrix-py
13 | """
14 |
15 | import matplotlib.font_manager as fm
16 | import matplotlib.pyplot as plt
17 | import numpy as np
18 | import seaborn as sn
19 | from matplotlib.collections import QuadMesh
20 |
21 |
22 | def get_new_fig(fn, figsize=[9, 9]):
23 | """Init graphics"""
24 | fig1 = plt.figure(fn, figsize)
25 | ax1 = fig1.gca() # Get Current Axis
26 | ax1.cla() # clear existing plot
27 | return fig1, ax1
28 |
29 |
30 | def configcell_text_and_colors(
31 | array_df, lin, col, oText, facecolors, posi, fz, fmt, show_null_values=0
32 | ):
33 | """
34 | config cell text and colors
35 | and return text elements to add and to dell
36 | @TODO: use fmt
37 | """
38 | text_add = []
39 | text_del = []
40 | cell_val = array_df[lin][col]
41 | tot_all = array_df[-1][-1]
42 | per = float(cell_val) / array_df[lin][-1] * 100#(float(cell_val) / tot_all) * 100
43 | curr_column = array_df[:, col]
44 | ccl = len(curr_column)
45 |
46 | # last line and/or last column
47 | if (col == (ccl - 1)) or (lin == (ccl - 1)):
48 | # tots and percents
49 | if cell_val != 0:
50 | if (col == ccl - 1) and (lin == ccl - 1):
51 | tot_rig = 0
52 | for i in range(array_df.shape[0] - 1):
53 | tot_rig += array_df[i][i]
54 | per_ok = (float(tot_rig) / cell_val) * 100
55 | elif col == ccl - 1:
56 | tot_rig = array_df[lin][lin]
57 | per_ok = (float(tot_rig) / cell_val) * 100
58 | elif lin == ccl - 1:
59 | tot_rig = array_df[col][col]
60 | per_ok = (float(tot_rig) / cell_val) * 100
61 | per_err = 100 - per_ok
62 | else:
63 | per_ok = per_err = 0
64 |
65 | per_ok_s = ["%.2f%%" % (per_ok), "100%"][per_ok == 100]
66 |
67 | # text to DEL
68 | text_del.append(oText)
69 |
70 | # text to ADD
71 | font_prop = fm.FontProperties(weight="bold", size=fz)
72 | text_kwargs = dict(
73 | color="w",
74 | ha="center",
75 | va="center",
76 | gid="sum",
77 | fontproperties=font_prop,
78 | )
79 | lis_txt = ["%d" % (cell_val), per_ok_s, "%.2f%%" % (per_err)]
80 | lis_kwa = [text_kwargs]
81 | dic = text_kwargs.copy()
82 | dic["color"] = "g"
83 | lis_kwa.append(dic)
84 | dic = text_kwargs.copy()
85 | dic["color"] = "r"
86 | lis_kwa.append(dic)
87 | lis_pos = [
88 | (oText._x, oText._y - 0.3),
89 | (oText._x, oText._y),
90 | (oText._x, oText._y + 0.3),
91 | ]
92 | for i in range(len(lis_txt)):
93 | newText = dict(
94 | x=lis_pos[i][0],
95 | y=lis_pos[i][1],
96 | text=lis_txt[i],
97 | kw=lis_kwa[i],
98 | )
99 | text_add.append(newText)
100 |
101 | # set background color for sum cells (last line and last column)
102 | carr = [0.27, 0.30, 0.27, 1.0]
103 | if (col == ccl - 1) and (lin == ccl - 1):
104 | carr = [0.17, 0.20, 0.17, 1.0]
105 | facecolors[posi] = carr
106 |
107 | else:
108 | if per > 0:
109 | txt = "%s\n%.2f%%" % (cell_val, per)
110 | else:
111 | if show_null_values == 0:
112 | txt = ""
113 | elif show_null_values == 1:
114 | txt = "0\n0.00%"
115 | else:
116 | txt = "0\n0.00%"
117 | oText.set_text(txt)
118 |
119 | # main diagonal
120 | if col == lin:
121 | # set color of the textin the diagonal to white
122 | oText.set_color("k")
123 | # set background color in the diagonal to blue
124 | facecolors[posi] = [0.35, 0.8, 0.55, 1.0]
125 | else:
126 | oText.set_color("r")
127 |
128 | return text_add, text_del
129 |
130 |
131 | def insert_totals(df_cm):
132 | """insert total column and line (the last ones)"""
133 | sum_col = []
134 | for c in df_cm.columns:
135 | sum_col.append(df_cm[c].sum())
136 | sum_lin = []
137 | for item_line in df_cm.iterrows():
138 | sum_lin.append(item_line[1].sum())
139 | df_cm["sum_lin"] = sum_lin
140 | sum_col.append(np.sum(sum_lin))
141 | df_cm.loc["sum_col"] = sum_col
142 |
143 |
144 | def pp_matrix(
145 | df_cm,
146 | annot=True,
147 | cmap="Oranges",
148 | fmt=".2f",
149 | fz=11,
150 | lw=0.5,
151 | cbar=False,
152 | figsize=[8, 8],
153 | show_null_values=0,
154 | pred_val_axis="y",
155 | ):
156 | """
157 | print conf matrix with default layout (like matlab)
158 | params:
159 | df_cm dataframe (pandas) without totals
160 | annot print text in each cell
161 | cmap Oranges,Oranges_r,YlGnBu,Blues,RdBu, ... see:
162 | fz fontsize
163 | lw linewidth
164 | pred_val_axis where to show the prediction values (x or y axis)
165 | 'col' or 'x': show predicted values in columns (x axis) instead lines
166 | 'lin' or 'y': show predicted values in lines (y axis)
167 | """
168 | if pred_val_axis in ("col", "x"):
169 | xlbl = "Predicted"
170 | ylbl = "Actual"
171 | else:
172 | xlbl = "Actual"
173 | ylbl = "Predicted"
174 | df_cm = df_cm.T
175 |
176 | # create "Total" column
177 | insert_totals(df_cm)
178 |
179 | # this is for print allways in the same window
180 | fig, ax1 = get_new_fig("Conf matrix default", figsize)
181 |
182 | ax = sn.heatmap(
183 | df_cm,
184 | annot=annot,
185 | annot_kws={"size": fz},
186 | linewidths=lw,
187 | ax=ax1,
188 | cbar=cbar,
189 | cmap=cmap,
190 | linecolor="w",
191 | fmt=fmt,
192 | )
193 |
194 | # set ticklabels rotation
195 | ax.set_xticklabels(ax.get_xticklabels(), rotation=45, fontsize=16)
196 | ax.set_yticklabels(ax.get_yticklabels(), rotation=25, fontsize=16)
197 |
198 | # Turn off all the ticks
199 | for t in ax.xaxis.get_major_ticks():
200 | t.tick1On = False
201 | t.tick2On = False
202 | for t in ax.yaxis.get_major_ticks():
203 | t.tick1On = False
204 | t.tick2On = False
205 |
206 | # face colors list
207 | quadmesh = ax.findobj(QuadMesh)[0]
208 | facecolors = quadmesh.get_facecolors()
209 |
210 | # iter in text elements
211 | array_df = np.array(df_cm.to_records(index=False).tolist())
212 | text_add = []
213 | text_del = []
214 | posi = -1 # from left to right, bottom to top.
215 | for t in ax.collections[0].axes.texts: # ax.texts:
216 | pos = np.array(t.get_position()) - [0.5, 0.5]
217 | lin = int(pos[1])
218 | col = int(pos[0])
219 | posi += 1
220 |
221 | # set text
222 | txt_res = configcell_text_and_colors(
223 | array_df, lin, col, t, facecolors, posi, fz, fmt, show_null_values
224 | )
225 |
226 | text_add.extend(txt_res[0])
227 | text_del.extend(txt_res[1])
228 |
229 | # remove the old ones
230 | for item in text_del:
231 | item.remove()
232 | # append the new ones
233 | for item in text_add:
234 | ax.text(item["x"], item["y"], item["text"], **item["kw"])
235 |
236 | # titles and legends
237 | ax.set_title("Confusion matrix")
238 | ax.set_xlabel(xlbl)
239 | ax.set_ylabel(ylbl)
240 | plt.tight_layout() # set layout slim
241 | plt.show()
242 |
243 |
244 | def pp_matrix_from_data(
245 | y_test,
246 | predictions,
247 | columns=None,
248 | annot=True,
249 | cmap="Oranges",
250 | fmt=".2f",
251 | fz=11,
252 | lw=0.5,
253 | cbar=False,
254 | figsize=[8, 8],
255 | show_null_values=0,
256 | pred_val_axis="lin",
257 | ):
258 | """
259 | plot confusion matrix function with y_test (actual values) and predictions (predic),
260 | whitout a confusion matrix yet
261 | """
262 | from pandas import DataFrame
263 | from sklearn.metrics import confusion_matrix
264 |
265 | # data
266 | if not columns:
267 | from string import ascii_uppercase
268 |
269 | columns = [
270 | "class %s" % (i)
271 | for i in list(ascii_uppercase)[0 : len(np.unique(y_test))]
272 | ]
273 |
274 | confm = confusion_matrix(y_test, predictions)
275 | df_cm = DataFrame(confm, index=columns, columns=columns)
276 | pp_matrix(
277 | df_cm,
278 | fz=fz,
279 | cmap=cmap,
280 | figsize=figsize,
281 | show_null_values=show_null_values,
282 | pred_val_axis=pred_val_axis,
283 | )
--------------------------------------------------------------------------------
/NIFTI/bipolar.m:
--------------------------------------------------------------------------------
1 | %BIPOLAR returns an M-by-3 matrix containing a blue-red colormap, in
2 | % in which red stands for positive, blue stands for negative,
3 | % and white stands for 0.
4 | %
5 | % Usage: cmap = bipolar(M, lo, hi, contrast); or cmap = bipolar;
6 | %
7 | % cmap: output M-by-3 matrix for BIPOLAR colormap.
8 | % M: number of shades in the colormap. By default, it is the
9 | % same length as the current colormap.
10 | % lo: the lowest value to represent.
11 | % hi: the highest value to represent.
12 | %
13 | % Inspired from the LORETA PASCAL program:
14 | % http://www.unizh.ch/keyinst/NewLORETA
15 | %
16 | % jimmy@rotman-baycrest.on.ca
17 | %
18 | %----------------------------------------------------------------
19 | function cmap = bipolar(M, lo, hi, contrast)
20 |
21 | if ~exist('contrast','var')
22 | contrast = 128;
23 | end
24 |
25 | if ~exist('lo','var')
26 | lo = -1;
27 | end
28 |
29 | if ~exist('hi','var')
30 | hi = 1;
31 | end
32 |
33 | if ~exist('M','var')
34 | cmap = colormap;
35 | M = size(cmap,1);
36 | end
37 |
38 | steepness = 10 ^ (1 - (contrast-1)/127);
39 | pos_infs = 1e-99;
40 | neg_infs = -1e-99;
41 |
42 | doubleredc = [];
43 | doublebluec = [];
44 |
45 | if lo >= 0 % all positive
46 |
47 | if lo == 0
48 | lo = pos_infs;
49 | end
50 |
51 | for i=linspace(hi/M, hi, M)
52 | t = exp(log(i/hi)*steepness);
53 | doubleredc = [doubleredc; [(1-t)+t,(1-t)+0,(1-t)+0]];
54 | end
55 |
56 | cmap = doubleredc;
57 |
58 | elseif hi <= 0 % all negative
59 |
60 | if hi == 0
61 | hi = neg_infs;
62 | end
63 |
64 | for i=linspace(abs(lo)/M, abs(lo), M)
65 | t = exp(log(i/abs(lo))*steepness);
66 | doublebluec = [doublebluec; [(1-t)+0,(1-t)+0,(1-t)+t]];
67 | end
68 |
69 | cmap = flipud(doublebluec);
70 |
71 | else
72 |
73 | if hi > abs(lo)
74 | maxc = hi;
75 | else
76 | maxc = abs(lo);
77 | end
78 |
79 | for i=linspace(maxc/M, hi, round(M*hi/(hi-lo)))
80 | t = exp(log(i/maxc)*steepness);
81 | doubleredc = [doubleredc; [(1-t)+t,(1-t)+0,(1-t)+0]];
82 | end
83 |
84 | for i=linspace(maxc/M, abs(lo), round(M*abs(lo)/(hi-lo)))
85 | t = exp(log(i/maxc)*steepness);
86 | doublebluec = [doublebluec; [(1-t)+0,(1-t)+0,(1-t)+t]];
87 | end
88 |
89 | cmap = [flipud(doublebluec); doubleredc];
90 |
91 | end
92 |
93 | return; % bipolar
94 |
95 |
--------------------------------------------------------------------------------
/NIFTI/bresenham_line3d.m:
--------------------------------------------------------------------------------
1 | % Generate X Y Z coordinates of a 3D Bresenham's line between
2 | % two given points.
3 | %
4 | % A very useful application of this algorithm can be found in the
5 | % implementation of Fischer's Bresenham interpolation method in my
6 | % another program that can rotate three dimensional image volume
7 | % with an affine matrix:
8 | % http://www.mathworks.com/matlabcentral/fileexchange/loadFile.do?objectId=21080
9 | %
10 | % Usage: [X Y Z] = bresenham_line3d(P1, P2, [precision]);
11 | %
12 | % P1 - vector for Point1, where P1 = [x1 y1 z1]
13 | %
14 | % P2 - vector for Point2, where P2 = [x2 y2 z2]
15 | %
16 | % precision (optional) - Although according to Bresenham's line
17 | % algorithm, point coordinates x1 y1 z1 and x2 y2 z2 should
18 | % be integer numbers, this program extends its limit to all
19 | % real numbers. If any of them are floating numbers, you
20 | % should specify how many digits of decimal that you would
21 | % like to preserve. Be aware that the length of output X Y
22 | % Z coordinates will increase in 10 times for each decimal
23 | % digit that you want to preserve. By default, the precision
24 | % is 0, which means that they will be rounded to the nearest
25 | % integer.
26 | %
27 | % X - a set of x coordinates on Bresenham's line
28 | %
29 | % Y - a set of y coordinates on Bresenham's line
30 | %
31 | % Z - a set of z coordinates on Bresenham's line
32 | %
33 | % Therefore, all points in XYZ set (i.e. P(i) = [X(i) Y(i) Z(i)])
34 | % will constitute the Bresenham's line between P1 and P1.
35 | %
36 | % Example:
37 | % P1 = [12 37 6]; P2 = [46 3 35];
38 | % [X Y Z] = bresenham_line3d(P1, P2);
39 | % figure; plot3(X,Y,Z,'s','markerface','b');
40 | %
41 | % This program is ported to MATLAB from:
42 | %
43 | % B.Pendleton. line3d - 3D Bresenham's (a 3D line drawing algorithm)
44 | % ftp://ftp.isc.org/pub/usenet/comp.sources.unix/volume26/line3d, 1992
45 | %
46 | % Which is also referenced by:
47 | %
48 | % Fischer, J., A. del Rio (2004). A Fast Method for Applying Rigid
49 | % Transformations to Volume Data, WSCG2004 Conference.
50 | % http://wscg.zcu.cz/wscg2004/Papers_2004_Short/M19.pdf
51 | %
52 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
53 | %
54 | function [X,Y,Z] = bresenham_line3d(P1, P2, precision)
55 |
56 | if ~exist('precision','var') | isempty(precision) | round(precision) == 0
57 | precision = 0;
58 | P1 = round(P1);
59 | P2 = round(P2);
60 | else
61 | precision = round(precision);
62 | P1 = round(P1*(10^precision));
63 | P2 = round(P2*(10^precision));
64 | end
65 |
66 | d = max(abs(P2-P1)+1);
67 | X = zeros(1, d);
68 | Y = zeros(1, d);
69 | Z = zeros(1, d);
70 |
71 | x1 = P1(1);
72 | y1 = P1(2);
73 | z1 = P1(3);
74 |
75 | x2 = P2(1);
76 | y2 = P2(2);
77 | z2 = P2(3);
78 |
79 | dx = x2 - x1;
80 | dy = y2 - y1;
81 | dz = z2 - z1;
82 |
83 | ax = abs(dx)*2;
84 | ay = abs(dy)*2;
85 | az = abs(dz)*2;
86 |
87 | sx = sign(dx);
88 | sy = sign(dy);
89 | sz = sign(dz);
90 |
91 | x = x1;
92 | y = y1;
93 | z = z1;
94 | idx = 1;
95 |
96 | if(ax>=max(ay,az)) % x dominant
97 | yd = ay - ax/2;
98 | zd = az - ax/2;
99 |
100 | while(1)
101 | X(idx) = x;
102 | Y(idx) = y;
103 | Z(idx) = z;
104 | idx = idx + 1;
105 |
106 | if(x == x2) % end
107 | break;
108 | end
109 |
110 | if(yd >= 0) % move along y
111 | y = y + sy;
112 | yd = yd - ax;
113 | end
114 |
115 | if(zd >= 0) % move along z
116 | z = z + sz;
117 | zd = zd - ax;
118 | end
119 |
120 | x = x + sx; % move along x
121 | yd = yd + ay;
122 | zd = zd + az;
123 | end
124 | elseif(ay>=max(ax,az)) % y dominant
125 | xd = ax - ay/2;
126 | zd = az - ay/2;
127 |
128 | while(1)
129 | X(idx) = x;
130 | Y(idx) = y;
131 | Z(idx) = z;
132 | idx = idx + 1;
133 |
134 | if(y == y2) % end
135 | break;
136 | end
137 |
138 | if(xd >= 0) % move along x
139 | x = x + sx;
140 | xd = xd - ay;
141 | end
142 |
143 | if(zd >= 0) % move along z
144 | z = z + sz;
145 | zd = zd - ay;
146 | end
147 |
148 | y = y + sy; % move along y
149 | xd = xd + ax;
150 | zd = zd + az;
151 | end
152 | elseif(az>=max(ax,ay)) % z dominant
153 | xd = ax - az/2;
154 | yd = ay - az/2;
155 |
156 | while(1)
157 | X(idx) = x;
158 | Y(idx) = y;
159 | Z(idx) = z;
160 | idx = idx + 1;
161 |
162 | if(z == z2) % end
163 | break;
164 | end
165 |
166 | if(xd >= 0) % move along x
167 | x = x + sx;
168 | xd = xd - az;
169 | end
170 |
171 | if(yd >= 0) % move along y
172 | y = y + sy;
173 | yd = yd - az;
174 | end
175 |
176 | z = z + sz; % move along z
177 | xd = xd + ax;
178 | yd = yd + ay;
179 | end
180 | end
181 |
182 | if precision ~= 0
183 | X = X/(10^precision);
184 | Y = Y/(10^precision);
185 | Z = Z/(10^precision);
186 | end
187 |
188 | return; % bresenham_line3d
189 |
190 |
--------------------------------------------------------------------------------
/NIFTI/clip_nii.m:
--------------------------------------------------------------------------------
1 | % CLIP_NII: Clip the NIfTI volume from any of the 6 sides
2 | %
3 | % Usage: nii = clip_nii(nii, [option])
4 | %
5 | % Inputs:
6 | %
7 | % nii - NIfTI volume.
8 | %
9 | % option - struct instructing how many voxel to be cut from which side.
10 | %
11 | % option.cut_from_L = ( number of voxel )
12 | % option.cut_from_R = ( number of voxel )
13 | % option.cut_from_P = ( number of voxel )
14 | % option.cut_from_A = ( number of voxel )
15 | % option.cut_from_I = ( number of voxel )
16 | % option.cut_from_S = ( number of voxel )
17 | %
18 | % Options description in detail:
19 | % ==============================
20 | %
21 | % cut_from_L: Number of voxels from Left side will be clipped.
22 | %
23 | % cut_from_R: Number of voxels from Right side will be clipped.
24 | %
25 | % cut_from_P: Number of voxels from Posterior side will be clipped.
26 | %
27 | % cut_from_A: Number of voxels from Anterior side will be clipped.
28 | %
29 | % cut_from_I: Number of voxels from Inferior side will be clipped.
30 | %
31 | % cut_from_S: Number of voxels from Superior side will be clipped.
32 | %
33 | % NIfTI data format can be found on: http://nifti.nimh.nih.gov
34 | %
35 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
36 | %
37 | function nii = clip_nii(nii, opt)
38 |
39 | dims = abs(nii.hdr.dime.dim(2:4));
40 | origin = abs(nii.hdr.hist.originator(1:3));
41 |
42 | if isempty(origin) | all(origin == 0) % according to SPM
43 | origin = round((dims+1)/2);
44 | end
45 |
46 | cut_from_L = 0;
47 | cut_from_R = 0;
48 | cut_from_P = 0;
49 | cut_from_A = 0;
50 | cut_from_I = 0;
51 | cut_from_S = 0;
52 |
53 | if nargin > 1 & ~isempty(opt)
54 | if ~isstruct(opt)
55 | error('option argument should be a struct');
56 | end
57 |
58 | if isfield(opt,'cut_from_L')
59 | cut_from_L = round(opt.cut_from_L);
60 |
61 | if cut_from_L >= origin(1) | cut_from_L < 0
62 | error('cut_from_L cannot be negative or cut beyond originator');
63 | end
64 | end
65 |
66 | if isfield(opt,'cut_from_P')
67 | cut_from_P = round(opt.cut_from_P);
68 |
69 | if cut_from_P >= origin(2) | cut_from_P < 0
70 | error('cut_from_P cannot be negative or cut beyond originator');
71 | end
72 | end
73 |
74 | if isfield(opt,'cut_from_I')
75 | cut_from_I = round(opt.cut_from_I);
76 |
77 | if cut_from_I >= origin(3) | cut_from_I < 0
78 | error('cut_from_I cannot be negative or cut beyond originator');
79 | end
80 | end
81 |
82 | if isfield(opt,'cut_from_R')
83 | cut_from_R = round(opt.cut_from_R);
84 |
85 | if cut_from_R > dims(1)-origin(1) | cut_from_R < 0
86 | error('cut_from_R cannot be negative or cut beyond originator');
87 | end
88 | end
89 |
90 | if isfield(opt,'cut_from_A')
91 | cut_from_A = round(opt.cut_from_A);
92 |
93 | if cut_from_A > dims(2)-origin(2) | cut_from_A < 0
94 | error('cut_from_A cannot be negative or cut beyond originator');
95 | end
96 | end
97 |
98 | if isfield(opt,'cut_from_S')
99 | cut_from_S = round(opt.cut_from_S);
100 |
101 | if cut_from_S > dims(3)-origin(3) | cut_from_S < 0
102 | error('cut_from_S cannot be negative or cut beyond originator');
103 | end
104 | end
105 | end
106 |
107 | nii = make_nii(nii.img( (cut_from_L+1) : (dims(1)-cut_from_R), ...
108 | (cut_from_P+1) : (dims(2)-cut_from_A), ...
109 | (cut_from_I+1) : (dims(3)-cut_from_S), ...
110 | :,:,:,:,:), nii.hdr.dime.pixdim(2:4), ...
111 | [origin(1)-cut_from_L origin(2)-cut_from_P origin(3)-cut_from_I], ...
112 | nii.hdr.dime.datatype, nii.hdr.hist.descrip);
113 |
114 | return;
115 |
116 |
--------------------------------------------------------------------------------
/NIFTI/collapse_nii_scan.m:
--------------------------------------------------------------------------------
1 | % Collapse multiple single-scan NIFTI files into a multiple-scan NIFTI file
2 | %
3 | % Usage: collapse_nii_scan(scan_file_pattern, [collapsed_fileprefix], [scan_file_folder])
4 | %
5 | % Here, scan_file_pattern should look like: 'myscan_0*.img'
6 | % If collapsed_fileprefix is omit, 'multi_scan' will be used
7 | % If scan_file_folder is omit, current file folder will be used
8 | %
9 | % The order of volumes in the collapsed file will be the order of
10 | % corresponding filenames for those selected scan files.
11 | %
12 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
13 | %
14 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
15 | %
16 | function collapse_nii_scan(scan_pattern, fileprefix, scan_path)
17 |
18 | if ~exist('fileprefix','var')
19 | fileprefix = 'multi_scan';
20 | else
21 | [tmp fileprefix] = fileparts(fileprefix);
22 | end
23 |
24 | if ~exist('scan_path','var'), scan_path = pwd; end
25 | pnfn = fullfile(scan_path, scan_pattern);
26 |
27 | file_lst = dir(pnfn);
28 | flist = {file_lst.name};
29 | flist = flist(:);
30 | filename = flist{1};
31 |
32 | v = version;
33 |
34 | % Check file extension. If .gz, unpack it into temp folder
35 | %
36 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
37 |
38 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
39 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
40 | ~strcmp(filename(end-6:end), '.nii.gz')
41 |
42 | error('Please check filename.');
43 | end
44 |
45 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
46 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
47 | else
48 | gzFile = 1;
49 | end
50 | else
51 | if ~strcmp(filename(end-3:end), '.img') & ...
52 | ~strcmp(filename(end-3:end), '.hdr') & ...
53 | ~strcmp(filename(end-3:end), '.nii')
54 |
55 | error('Please check filename.');
56 | end
57 | end
58 |
59 | nii = load_untouch_nii(fullfile(scan_path,filename));
60 | nii.hdr.dime.dim(5) = length(flist);
61 |
62 | if nii.hdr.dime.dim(1) < 4
63 | nii.hdr.dime.dim(1) = 4;
64 | end
65 |
66 | hdr = nii.hdr;
67 | filetype = nii.filetype;
68 |
69 | if isfield(nii,'ext') & ~isempty(nii.ext)
70 | ext = nii.ext;
71 | [ext, esize_total] = verify_nii_ext(ext);
72 | else
73 | ext = [];
74 | end
75 |
76 | switch double(hdr.dime.datatype),
77 | case 1,
78 | hdr.dime.bitpix = int16(1 ); precision = 'ubit1';
79 | case 2,
80 | hdr.dime.bitpix = int16(8 ); precision = 'uint8';
81 | case 4,
82 | hdr.dime.bitpix = int16(16); precision = 'int16';
83 | case 8,
84 | hdr.dime.bitpix = int16(32); precision = 'int32';
85 | case 16,
86 | hdr.dime.bitpix = int16(32); precision = 'float32';
87 | case 32,
88 | hdr.dime.bitpix = int16(64); precision = 'float32';
89 | case 64,
90 | hdr.dime.bitpix = int16(64); precision = 'float64';
91 | case 128,
92 | hdr.dime.bitpix = int16(24); precision = 'uint8';
93 | case 256
94 | hdr.dime.bitpix = int16(8 ); precision = 'int8';
95 | case 512
96 | hdr.dime.bitpix = int16(16); precision = 'uint16';
97 | case 768
98 | hdr.dime.bitpix = int16(32); precision = 'uint32';
99 | case 1024
100 | hdr.dime.bitpix = int16(64); precision = 'int64';
101 | case 1280
102 | hdr.dime.bitpix = int16(64); precision = 'uint64';
103 | case 1792,
104 | hdr.dime.bitpix = int16(128); precision = 'float64';
105 | otherwise
106 | error('This datatype is not supported');
107 | end
108 |
109 | if filetype == 2
110 | fid = fopen(sprintf('%s.nii',fileprefix),'w');
111 |
112 | if fid < 0,
113 | msg = sprintf('Cannot open file %s.nii.',fileprefix);
114 | error(msg);
115 | end
116 |
117 | hdr.dime.vox_offset = 352;
118 |
119 | if ~isempty(ext)
120 | hdr.dime.vox_offset = hdr.dime.vox_offset + esize_total;
121 | end
122 |
123 | hdr.hist.magic = 'n+1';
124 | save_untouch_nii_hdr(hdr, fid);
125 |
126 | if ~isempty(ext)
127 | save_nii_ext(ext, fid);
128 | end
129 | elseif filetype == 1
130 | fid = fopen(sprintf('%s.hdr',fileprefix),'w');
131 |
132 | if fid < 0,
133 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
134 | error(msg);
135 | end
136 |
137 | hdr.dime.vox_offset = 0;
138 | hdr.hist.magic = 'ni1';
139 | save_untouch_nii_hdr(hdr, fid);
140 |
141 | if ~isempty(ext)
142 | save_nii_ext(ext, fid);
143 | end
144 |
145 | fclose(fid);
146 | fid = fopen(sprintf('%s.img',fileprefix),'w');
147 | else
148 | fid = fopen(sprintf('%s.hdr',fileprefix),'w');
149 |
150 | if fid < 0,
151 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
152 | error(msg);
153 | end
154 |
155 | save_untouch0_nii_hdr(hdr, fid);
156 |
157 | fclose(fid);
158 | fid = fopen(sprintf('%s.img',fileprefix),'w');
159 | end
160 |
161 | if filetype == 2 & isempty(ext)
162 | skip_bytes = double(hdr.dime.vox_offset) - 348;
163 | else
164 | skip_bytes = 0;
165 | end
166 |
167 | if skip_bytes
168 | fwrite(fid, zeros(1,skip_bytes), 'uint8');
169 | end
170 |
171 | glmax = -inf;
172 | glmin = inf;
173 |
174 | for i = 1:length(flist)
175 | nii = load_untouch_nii(fullfile(scan_path,flist{i}));
176 |
177 | if double(hdr.dime.datatype) == 128
178 |
179 | % RGB planes are expected to be in the 4th dimension of nii.img
180 | %
181 | if(size(nii.img,4)~=3)
182 | error(['The NII structure does not appear to have 3 RGB color planes in the 4th dimension']);
183 | end
184 |
185 | nii.img = permute(nii.img, [4 1 2 3 5 6 7 8]);
186 | end
187 |
188 | % For complex float32 or complex float64, voxel values
189 | % include [real, imag]
190 | %
191 | if hdr.dime.datatype == 32 | hdr.dime.datatype == 1792
192 | real_img = real(nii.img(:))';
193 | nii.img = imag(nii.img(:))';
194 | nii.img = [real_img; nii.img];
195 | end
196 |
197 | if nii.hdr.dime.glmax > glmax
198 | glmax = nii.hdr.dime.glmax;
199 | end
200 |
201 | if nii.hdr.dime.glmin < glmin
202 | glmin = nii.hdr.dime.glmin;
203 | end
204 |
205 | fwrite(fid, nii.img, precision);
206 | end
207 |
208 | hdr.dime.glmax = round(glmax);
209 | hdr.dime.glmin = round(glmin);
210 |
211 | if filetype == 2
212 | fseek(fid, 140, 'bof');
213 | fwrite(fid, hdr.dime.glmax, 'int32');
214 | fwrite(fid, hdr.dime.glmin, 'int32');
215 | elseif filetype == 1
216 | fid2 = fopen(sprintf('%s.hdr',fileprefix),'w');
217 |
218 | if fid2 < 0,
219 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
220 | error(msg);
221 | end
222 |
223 | save_untouch_nii_hdr(hdr, fid2);
224 |
225 | if ~isempty(ext)
226 | save_nii_ext(ext, fid2);
227 | end
228 |
229 | fclose(fid2);
230 | else
231 | fid2 = fopen(sprintf('%s.hdr',fileprefix),'w');
232 |
233 | if fid2 < 0,
234 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
235 | error(msg);
236 | end
237 |
238 | save_untouch0_nii_hdr(hdr, fid2);
239 |
240 | fclose(fid2);
241 | end
242 |
243 | fclose(fid);
244 |
245 | % gzip output file if requested
246 | %
247 | if exist('gzFile', 'var')
248 | if filetype == 1
249 | gzip([fileprefix, '.img']);
250 | delete([fileprefix, '.img']);
251 | gzip([fileprefix, '.hdr']);
252 | delete([fileprefix, '.hdr']);
253 | elseif filetype == 2
254 | gzip([fileprefix, '.nii']);
255 | delete([fileprefix, '.nii']);
256 | end;
257 | end;
258 |
259 | return; % collapse_nii_scan
260 |
261 |
--------------------------------------------------------------------------------
/NIFTI/examples.txt:
--------------------------------------------------------------------------------
1 |
2 | - Examples to load, make and save a nii struct:
3 |
4 | To load Analyze data or NIFTI data to a structure:
5 |
6 | nii = load_nii(NIFTI_file_name, [img_idx], [old_RGB24]);
7 |
8 | img_idx is a numerical array of image indices along the temporal
9 | axis, which is only available in NIFTI data. After you specify
10 | img_idx, only those images indexed by img_idx will be loaded. If
11 | there is no img_idx or img_idx is empty, all available images
12 | will be loaded.
13 |
14 | For RGB image, most people use RGB triple sequentially for each
15 | voxel, like [R1 G1 B1 R2 G2 B2 ...]. However, some program like
16 | Analyze 6.0 developed by AnalyzeDirect uses old RGB24, in a way
17 | like [R1 R2 ... G1 G2 ... B1 B2 ...] for each slices. In this
18 | case, you can set old_RGB24 flag to 1 and load data correctly:
19 |
20 | nii = load_nii(NIFTI_file_name, [], 1);
21 |
22 | To get a total number of images along the temporal axis:
23 |
24 | num_scan = get_nii_frame(NIFTI_file_name);
25 |
26 | You can also load the header extension if it exists:
27 |
28 | nii.ext = load_nii_ext(NIFTI_file_name);
29 |
30 | You can just load the Analyze or NIFTI header:
31 | (header contains: hk, dime, and hist)
32 |
33 | hdr = load_nii_hdr(NIFTI_file_name);
34 |
35 | You can also save the structure to a new file:
36 | (header extension will be saved if there is nii.ext structure)
37 |
38 | save_nii(nii, NIFTI_file_name);
39 |
40 | To make the structure from any 3D (or 4D) data:
41 |
42 | img = rand(91,109,91); or
43 | img = rand(64,64,21,18);
44 | nii = make_nii(img [, voxel_size, origin, datatype] );
45 |
46 | Use "help load_nii", "help save_nii", "help make_nii" etc.
47 | to get more detail information.
48 |
49 |
50 | - Examples to plot a nii struct:
51 | (More detail descriptions are available on top of "view_nii.m")
52 |
53 | Simple way to plot a nii struct:
54 |
55 | view_nii(nii);
56 |
57 | The default colormap will use the Gray if all data values are
58 | non-negative; otherwise, the default colormap will use BiPolar.
59 | You can choose other colormap, including customized colormap
60 | from panel.
61 |
62 | To imbed the plot into your existing figure:
63 |
64 | h = gcf;
65 | opt.command = 'init';
66 | opt.setarea = [0.3 0.1 0.6 0.8];
67 | view_nii(h, nii, opt);
68 |
69 | To add a colorbar:
70 |
71 | opt.usecolorbar = 1;
72 | view_nii(gcf, opt);
73 |
74 | Here, opt.command is implicitly set to 'update'.
75 |
76 | To display in real aspect ratio:
77 |
78 | opt.usestretch = 0;
79 | view_nii(gcf, opt);
80 |
81 | If you want the data value to be directly used as the index
82 | of colormap, instead of scale to the whole colormap:
83 |
84 | opt.useimagesc = 0;
85 | view_nii(gcf, opt);
86 |
87 | If you modified the data value without changing the dimension,
88 | voxel_size, and origin, you can update the display by:
89 |
90 | opt.command = 'updateimg';
91 | view_nii(gcf, nii.img, opt);
92 |
93 | If the data is completely different, display can be updated by:
94 |
95 | opt.command = 'updatenii';
96 | view_nii(gcf, nii, opt);
97 |
98 | This is an example to plot EEG source imaging on top of T1 background:
99 | 1. download overlay.zip and unzip it from:
100 | http://www.rotman-baycrest.on.ca/~jimmy/NIFTI/overlay.zip
101 | 2. T1 = load_nii('T1.nii');
102 | 3. EEG = load_nii('EEG.nii');
103 | 4. option.setvalue.idx = find(EEG.img);
104 | 5. option.setvalue.val = EEG.img(option.setvalue.idx);
105 | 6. option.useinterp = 1;
106 | 7. option.setviewpoint = [62 48 46];
107 | 8. view_nii(T1, option);
108 |
109 |
110 | - Contrast and Brightness are available under Gray and Bipolar colormap:
111 |
112 | Increase contrast in Gray colormap will make high end values
113 | more distinguishable by sacrificing the low end values; The
114 | minimum contrast (default) will display the whole range.
115 |
116 | Increase or decrease contrast in BiPolar colormap will shift
117 | the distinguishable position for both positive and negative
118 | values.
119 |
120 | Increase or decrease brightness in Gray colormap will shift
121 | the distinguishable position.
122 |
123 | Increase or decrease brightness in BiPolar colormap will make
124 | both positive and negative values more distinguishable.
125 |
126 |
127 | - Required files:
128 |
129 | All files in this package.
130 |
131 |
--------------------------------------------------------------------------------
/NIFTI/expand_nii_scan.m:
--------------------------------------------------------------------------------
1 | % Expand a multiple-scan NIFTI file into multiple single-scan NIFTI files
2 | %
3 | % Usage: expand_nii_scan(multi_scan_filename, [img_idx], [path_to_save])
4 | %
5 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
6 | %
7 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
8 | %
9 | function expand_nii_scan(filename, img_idx, newpath)
10 |
11 | v = version;
12 |
13 | % Check file extension. If .gz, unpack it into temp folder
14 | %
15 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
16 |
17 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
18 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
19 | ~strcmp(filename(end-6:end), '.nii.gz')
20 |
21 | error('Please check filename.');
22 | end
23 |
24 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
25 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
26 | else
27 | gzFile = 1;
28 | end
29 | end
30 |
31 | if ~exist('newpath','var') | isempty(newpath), newpath = pwd; end
32 | if ~exist('img_idx','var') | isempty(img_idx), img_idx = 1:get_nii_frame(filename); end
33 |
34 | for i=img_idx
35 | nii_i = load_untouch_nii(filename, i);
36 |
37 | fn = [nii_i.fileprefix '_' sprintf('%04d',i)];
38 | pnfn = fullfile(newpath, fn);
39 |
40 | if exist('gzFile', 'var')
41 | pnfn = [pnfn '.nii.gz'];
42 | end
43 |
44 | save_untouch_nii(nii_i, pnfn);
45 | end
46 |
47 | return; % expand_nii_scan
48 |
49 |
--------------------------------------------------------------------------------
/NIFTI/extra_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % Decode extra NIFTI header information into hdr.extra
2 | %
3 | % Usage: hdr = extra_nii_hdr(hdr)
4 | %
5 | % hdr can be obtained from load_nii_hdr
6 | %
7 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
8 | %
9 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
10 | %
11 | function hdr = extra_nii_hdr(hdr)
12 |
13 | switch hdr.dime.datatype
14 | case 1
15 | extra.NIFTI_DATATYPES = 'DT_BINARY';
16 | case 2
17 | extra.NIFTI_DATATYPES = 'DT_UINT8';
18 | case 4
19 | extra.NIFTI_DATATYPES = 'DT_INT16';
20 | case 8
21 | extra.NIFTI_DATATYPES = 'DT_INT32';
22 | case 16
23 | extra.NIFTI_DATATYPES = 'DT_FLOAT32';
24 | case 32
25 | extra.NIFTI_DATATYPES = 'DT_COMPLEX64';
26 | case 64
27 | extra.NIFTI_DATATYPES = 'DT_FLOAT64';
28 | case 128
29 | extra.NIFTI_DATATYPES = 'DT_RGB24';
30 | case 256
31 | extra.NIFTI_DATATYPES = 'DT_INT8';
32 | case 512
33 | extra.NIFTI_DATATYPES = 'DT_UINT16';
34 | case 768
35 | extra.NIFTI_DATATYPES = 'DT_UINT32';
36 | case 1024
37 | extra.NIFTI_DATATYPES = 'DT_INT64';
38 | case 1280
39 | extra.NIFTI_DATATYPES = 'DT_UINT64';
40 | case 1536
41 | extra.NIFTI_DATATYPES = 'DT_FLOAT128';
42 | case 1792
43 | extra.NIFTI_DATATYPES = 'DT_COMPLEX128';
44 | case 2048
45 | extra.NIFTI_DATATYPES = 'DT_COMPLEX256';
46 | otherwise
47 | extra.NIFTI_DATATYPES = 'DT_UNKNOWN';
48 | end
49 |
50 | switch hdr.dime.intent_code
51 | case 2
52 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_CORREL';
53 | case 3
54 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_TTEST';
55 | case 4
56 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_FTEST';
57 | case 5
58 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_ZSCORE';
59 | case 6
60 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_CHISQ';
61 | case 7
62 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_BETA';
63 | case 8
64 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_BINOM';
65 | case 9
66 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_GAMMA';
67 | case 10
68 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_POISSON';
69 | case 11
70 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_NORMAL';
71 | case 12
72 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_FTEST_NONC';
73 | case 13
74 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_CHISQ_NONC';
75 | case 14
76 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_LOGISTIC';
77 | case 15
78 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_LAPLACE';
79 | case 16
80 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_UNIFORM';
81 | case 17
82 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_TTEST_NONC';
83 | case 18
84 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_WEIBULL';
85 | case 19
86 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_CHI';
87 | case 20
88 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_INVGAUSS';
89 | case 21
90 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_EXTVAL';
91 | case 22
92 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_PVAL';
93 | case 23
94 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_LOGPVAL';
95 | case 24
96 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_LOG10PVAL';
97 | case 1001
98 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_ESTIMATE';
99 | case 1002
100 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_LABEL';
101 | case 1003
102 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_NEURONAME';
103 | case 1004
104 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_GENMATRIX';
105 | case 1005
106 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_SYMMATRIX';
107 | case 1006
108 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_DISPVECT';
109 | case 1007
110 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_VECTOR';
111 | case 1008
112 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_POINTSET';
113 | case 1009
114 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_TRIANGLE';
115 | case 1010
116 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_QUATERNION';
117 | case 1011
118 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_DIMLESS';
119 | otherwise
120 | extra.NIFTI_INTENT_CODES = 'NIFTI_INTENT_NONE';
121 | end
122 |
123 | extra.NIFTI_INTENT_NAMES = hdr.hist.intent_name;
124 |
125 | if hdr.hist.sform_code > 0
126 | switch hdr.hist.sform_code
127 | case 1
128 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_SCANNER_ANAT';
129 | case 2
130 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_ALIGNED_ANAT';
131 | case 3
132 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_TALAIRACH';
133 | case 4
134 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_MNI_152';
135 | otherwise
136 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
137 | end
138 |
139 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
140 | elseif hdr.hist.qform_code > 0
141 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
142 |
143 | switch hdr.hist.qform_code
144 | case 1
145 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_SCANNER_ANAT';
146 | case 2
147 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_ALIGNED_ANAT';
148 | case 3
149 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_TALAIRACH';
150 | case 4
151 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_MNI_152';
152 | otherwise
153 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
154 | end
155 | else
156 | extra.NIFTI_SFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
157 | extra.NIFTI_QFORM_CODES = 'NIFTI_XFORM_UNKNOWN';
158 | end
159 |
160 | switch bitand(hdr.dime.xyzt_units, 7) % mask with 0x07
161 | case 1
162 | extra.NIFTI_SPACE_UNIT = 'NIFTI_UNITS_METER';
163 | case 2
164 | extra.NIFTI_SPACE_UNIT = 'NIFTI_UNITS_MM'; % millimeter
165 | case 3
166 | extra.NIFTI_SPACE_UNIT = 'NIFTI_UNITS_MICRO';
167 | otherwise
168 | extra.NIFTI_SPACE_UNIT = 'NIFTI_UNITS_UNKNOWN';
169 | end
170 |
171 | switch bitand(hdr.dime.xyzt_units, 56) % mask with 0x38
172 | case 8
173 | extra.NIFTI_TIME_UNIT = 'NIFTI_UNITS_SEC';
174 | case 16
175 | extra.NIFTI_TIME_UNIT = 'NIFTI_UNITS_MSEC';
176 | case 24
177 | extra.NIFTI_TIME_UNIT = 'NIFTI_UNITS_USEC'; % microsecond
178 | otherwise
179 | extra.NIFTI_TIME_UNIT = 'NIFTI_UNITS_UNKNOWN';
180 | end
181 |
182 | switch hdr.dime.xyzt_units
183 | case 32
184 | extra.NIFTI_SPECTRAL_UNIT = 'NIFTI_UNITS_HZ';
185 | case 40
186 | extra.NIFTI_SPECTRAL_UNIT = 'NIFTI_UNITS_PPM'; % part per million
187 | case 48
188 | extra.NIFTI_SPECTRAL_UNIT = 'NIFTI_UNITS_RADS'; % radians per second
189 | otherwise
190 | extra.NIFTI_SPECTRAL_UNIT = 'NIFTI_UNITS_UNKNOWN';
191 | end
192 |
193 | % MRI-specific spatial and temporal information
194 | %
195 | dim_info = hdr.hk.dim_info;
196 | extra.NIFTI_FREQ_DIM = bitand(dim_info, 3);
197 | extra.NIFTI_PHASE_DIM = bitand(bitshift(dim_info, -2), 3);
198 | extra.NIFTI_SLICE_DIM = bitand(bitshift(dim_info, -4), 3);
199 |
200 | % Check slice code
201 | %
202 | switch hdr.dime.slice_code
203 | case 1
204 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_SEQ_INC'; % sequential increasing
205 | case 2
206 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_SEQ_DEC'; % sequential decreasing
207 | case 3
208 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_ALT_INC'; % alternating increasing
209 | case 4
210 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_ALT_DEC'; % alternating decreasing
211 | case 5
212 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_ALT_INC2'; % ALT_INC # 2
213 | case 6
214 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_ALT_DEC2'; % ALT_DEC # 2
215 | otherwise
216 | extra.NIFTI_SLICE_ORDER = 'NIFTI_SLICE_UNKNOWN';
217 | end
218 |
219 | % Check NIFTI version
220 | %
221 | if ~isempty(hdr.hist.magic) & strcmp(hdr.hist.magic(1),'n') & ...
222 | ( strcmp(hdr.hist.magic(2),'i') | strcmp(hdr.hist.magic(2),'+') ) & ...
223 | str2num(hdr.hist.magic(3)) >= 1 & str2num(hdr.hist.magic(3)) <= 9
224 |
225 | extra.NIFTI_VERSION = str2num(hdr.hist.magic(3));
226 | else
227 | extra.NIFTI_VERSION = 0;
228 | end
229 |
230 | % Check if data stored in the same file (*.nii) or separate
231 | % files (*.hdr/*.img)
232 | %
233 | if isempty(hdr.hist.magic)
234 | extra.NIFTI_ONEFILE = 0;
235 | else
236 | extra.NIFTI_ONEFILE = strcmp(hdr.hist.magic(2), '+');
237 | end
238 |
239 | % Swap has been taken care of by checking whether sizeof_hdr is
240 | % 348 (machine is 'ieee-le' or 'ieee-be' etc)
241 | %
242 | % extra.NIFTI_NEEDS_SWAP = (hdr.dime.dim(1) < 0 | hdr.dime.dim(1) > 7);
243 |
244 | % Check NIFTI header struct contains a 5th (vector) dimension
245 | %
246 | if hdr.dime.dim(1) > 4 & hdr.dime.dim(6) > 1
247 | extra.NIFTI_5TH_DIM = hdr.dime.dim(6);
248 | else
249 | extra.NIFTI_5TH_DIM = 0;
250 | end
251 |
252 | hdr.extra = extra;
253 |
254 | return; % extra_nii_hdr
255 |
256 |
--------------------------------------------------------------------------------
/NIFTI/flip_lr.m:
--------------------------------------------------------------------------------
1 | % When you load any ANALYZE or NIfTI file with 'load_nii.m', and view
2 | % it with 'view_nii.m', you may find that the image is L-R flipped.
3 | % This is because of the confusion of radiological and neurological
4 | % convention in the medical image before NIfTI format is adopted. You
5 | % can find more details from:
6 | %
7 | % http://www.rotman-baycrest.on.ca/~jimmy/UseANALYZE.htm
8 | %
9 | % Sometime, people even want to convert RAS (standard orientation) back
10 | % to LAS orientation to satisfy the legend programs or processes. This
11 | % program is only written for those purpose. So PLEASE BE VERY CAUTIOUS
12 | % WHEN USING THIS 'FLIP_LR.M' PROGRAM.
13 | %
14 | % With 'flip_lr.m', you can convert any ANALYZE or NIfTI (no matter
15 | % 3D or 4D) file to a flipped NIfTI file. This is implemented simply
16 | % by flipping the affine matrix in the NIfTI header. Since the L-R
17 | % orientation is determined there, so the image will be flipped.
18 | %
19 | % Usage: flip_lr(original_fn, flipped_fn, [old_RGB],[tolerance],[preferredForm])
20 | %
21 | % original_fn - filename of the original ANALYZE or NIfTI (3D or 4D) file
22 | %
23 | % flipped_fn - filename of the L-R flipped NIfTI file
24 | %
25 | % old_RGB (optional) - a scale number to tell difference of new RGB24
26 | % from old RGB24. New RGB24 uses RGB triple sequentially for each
27 | % voxel, like [R1 G1 B1 R2 G2 B2 ...]. Analyze 6.0 from AnalyzeDirect
28 | % uses old RGB24, in a way like [R1 R2 ... G1 G2 ... B1 B2 ...] for
29 | % each slices. If the image that you view is garbled, try to set
30 | % old_RGB variable to 1 and try again, because it could be in
31 | % old RGB24. It will be set to 0, if it is default or empty.
32 | %
33 | % tolerance (optional) - distortion allowed for non-orthogonal rotation
34 | % or shearing in NIfTI affine matrix. It will be set to 0.1 (10%),
35 | % if it is default or empty.
36 | %
37 | % preferredForm (optional) - selects which transformation from voxels
38 | % to RAS coordinates; values are s,q,S,Q. Lower case s,q indicate
39 | % "prefer sform or qform, but use others if preferred not present".
40 | % Upper case indicate the program is forced to use the specificied
41 | % tranform or fail loading. 'preferredForm' will be 's', if it is
42 | % default or empty. - Jeff Gunter
43 | %
44 | % Example: flip_lr('avg152T1_LR_nifti.nii', 'flipped_lr.nii');
45 | % flip_lr('avg152T1_RL_nifti.nii', 'flipped_rl.nii');
46 | %
47 | % You will find that 'avg152T1_LR_nifti.nii' and 'avg152T1_RL_nifti.nii'
48 | % are the same, and 'flipped_lr.nii' and 'flipped_rl.nii' are also the
49 | % the same, but they are L-R flipped from 'avg152T1_*'.
50 | %
51 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
52 | %
53 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
54 | %
55 | function flip_lr(original_fn, flipped_fn, old_RGB, tolerance, preferredForm)
56 |
57 | if ~exist('original_fn','var') | ~exist('flipped_fn','var')
58 | error('Usage: flip_lr(original_fn, flipped_fn, [old_RGB],[tolerance])');
59 | end
60 |
61 | if ~exist('old_RGB','var') | isempty(old_RGB)
62 | old_RGB = 0;
63 | end
64 |
65 | if ~exist('tolerance','var') | isempty(tolerance)
66 | tolerance = 0.1;
67 | end
68 |
69 | if ~exist('preferredForm','var') | isempty(preferredForm)
70 | preferredForm= 's'; % Jeff
71 | end
72 |
73 | nii = load_nii(original_fn, [], [], [], [], old_RGB, tolerance, preferredForm);
74 | M = diag(nii.hdr.dime.pixdim(2:5));
75 | M(1:3,4) = -M(1:3,1:3)*(nii.hdr.hist.originator(1:3)-1)';
76 | M(1,:) = -1*M(1,:);
77 | nii.hdr.hist.sform_code = 1;
78 | nii.hdr.hist.srow_x = M(1,:);
79 | nii.hdr.hist.srow_y = M(2,:);
80 | nii.hdr.hist.srow_z = M(3,:);
81 | save_nii(nii, flipped_fn);
82 |
83 | return; % flip_lr
84 |
85 |
--------------------------------------------------------------------------------
/NIFTI/get_nii_frame.m:
--------------------------------------------------------------------------------
1 | % Return time frame of a NIFTI dataset. Support both *.nii and
2 | % *.hdr/*.img file extension. If file extension is not provided,
3 | % *.hdr/*.img will be used as default.
4 | %
5 | % It is a lightweighted "load_nii_hdr", and is equivalent to
6 | % hdr.dime.dim(5)
7 | %
8 | % Usage: [ total_scan ] = get_nii_frame(filename)
9 | %
10 | % filename - NIFTI file name.
11 | %
12 | % Returned values:
13 | %
14 | % total_scan - total number of image scans for the time frame
15 | %
16 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
17 | %
18 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
19 | %
20 | function [ total_scan ] = get_nii_frame(filename)
21 |
22 | if ~exist('filename','var'),
23 | error('Usage: [ total_scan ] = get_nii_frame(filename)');
24 | end
25 |
26 | v = version;
27 |
28 | % Check file extension. If .gz, unpack it into temp folder
29 | %
30 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
31 |
32 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
33 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
34 | ~strcmp(filename(end-6:end), '.nii.gz')
35 |
36 | error('Please check filename.');
37 | end
38 |
39 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
40 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
41 | elseif strcmp(filename(end-6:end), '.img.gz')
42 | filename1 = filename;
43 | filename2 = filename;
44 | filename2(end-6:end) = '';
45 | filename2 = [filename2, '.hdr.gz'];
46 |
47 | tmpDir = tempname;
48 | mkdir(tmpDir);
49 | gzFileName = filename;
50 |
51 | filename1 = gunzip(filename1, tmpDir);
52 | filename2 = gunzip(filename2, tmpDir);
53 | filename = char(filename1); % convert from cell to string
54 | elseif strcmp(filename(end-6:end), '.hdr.gz')
55 | filename1 = filename;
56 | filename2 = filename;
57 | filename2(end-6:end) = '';
58 | filename2 = [filename2, '.img.gz'];
59 |
60 | tmpDir = tempname;
61 | mkdir(tmpDir);
62 | gzFileName = filename;
63 |
64 | filename1 = gunzip(filename1, tmpDir);
65 | filename2 = gunzip(filename2, tmpDir);
66 | filename = char(filename1); % convert from cell to string
67 | elseif strcmp(filename(end-6:end), '.nii.gz')
68 | tmpDir = tempname;
69 | mkdir(tmpDir);
70 | gzFileName = filename;
71 | filename = gunzip(filename, tmpDir);
72 | filename = char(filename); % convert from cell to string
73 | end
74 | end
75 |
76 | fileprefix = filename;
77 | machine = 'ieee-le';
78 | new_ext = 0;
79 |
80 | if findstr('.nii',fileprefix) & strcmp(fileprefix(end-3:end), '.nii')
81 | new_ext = 1;
82 | fileprefix(end-3:end)='';
83 | end
84 |
85 | if findstr('.hdr',fileprefix) & strcmp(fileprefix(end-3:end), '.hdr')
86 | fileprefix(end-3:end)='';
87 | end
88 |
89 | if findstr('.img',fileprefix) & strcmp(fileprefix(end-3:end), '.img')
90 | fileprefix(end-3:end)='';
91 | end
92 |
93 | if new_ext
94 | fn = sprintf('%s.nii',fileprefix);
95 |
96 | if ~exist(fn)
97 | msg = sprintf('Cannot find file "%s.nii".', fileprefix);
98 | error(msg);
99 | end
100 | else
101 | fn = sprintf('%s.hdr',fileprefix);
102 |
103 | if ~exist(fn)
104 | msg = sprintf('Cannot find file "%s.hdr".', fileprefix);
105 | error(msg);
106 | end
107 | end
108 |
109 | fid = fopen(fn,'r',machine);
110 |
111 | if fid < 0,
112 | msg = sprintf('Cannot open file %s.',fn);
113 | error(msg);
114 | else
115 | hdr = read_header(fid);
116 | fclose(fid);
117 | end
118 |
119 | if hdr.sizeof_hdr ~= 348
120 | % first try reading the opposite endian to 'machine'
121 | switch machine,
122 | case 'ieee-le', machine = 'ieee-be';
123 | case 'ieee-be', machine = 'ieee-le';
124 | end
125 |
126 | fid = fopen(fn,'r',machine);
127 |
128 | if fid < 0,
129 | msg = sprintf('Cannot open file %s.',fn);
130 | error(msg);
131 | else
132 | hdr = read_header(fid);
133 | fclose(fid);
134 | end
135 | end
136 |
137 | if hdr.sizeof_hdr ~= 348
138 | % Now throw an error
139 | msg = sprintf('File "%s" is corrupted.',fn);
140 | error(msg);
141 | end
142 |
143 | total_scan = hdr.dim(5);
144 |
145 | % Clean up after gunzip
146 | %
147 | if exist('gzFileName', 'var')
148 | rmdir(tmpDir,'s');
149 | end
150 |
151 | return; % get_nii_frame
152 |
153 |
154 | %---------------------------------------------------------------------
155 | function [ dsr ] = read_header(fid)
156 |
157 | fseek(fid,0,'bof');
158 | dsr.sizeof_hdr = fread(fid,1,'int32')'; % should be 348!
159 |
160 | fseek(fid,40,'bof');
161 | dsr.dim = fread(fid,8,'int16')';
162 |
163 | return; % read_header
164 |
165 |
--------------------------------------------------------------------------------
/NIFTI/license.txt:
--------------------------------------------------------------------------------
1 | Copyright (c) 2009, Jimmy Shen
2 | All rights reserved.
3 |
4 | Redistribution and use in source and binary forms, with or without
5 | modification, are permitted provided that the following conditions are
6 | met:
7 |
8 | * Redistributions of source code must retain the above copyright
9 | notice, this list of conditions and the following disclaimer.
10 | * Redistributions in binary form must reproduce the above copyright
11 | notice, this list of conditions and the following disclaimer in
12 | the documentation and/or other materials provided with the distribution
13 |
14 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
15 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
16 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE
17 | ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE
18 | LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR
19 | CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF
20 | SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS
21 | INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN
22 | CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE)
23 | ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE
24 | POSSIBILITY OF SUCH DAMAGE.
25 |
--------------------------------------------------------------------------------
/NIFTI/load_nii.m:
--------------------------------------------------------------------------------
1 | % Load NIFTI or ANALYZE dataset. Support both *.nii and *.hdr/*.img
2 | % file extension. If file extension is not provided, *.hdr/*.img will
3 | % be used as default.
4 | %
5 | % A subset of NIFTI transform is included. For non-orthogonal rotation,
6 | % shearing etc., please use 'reslice_nii.m' to reslice the NIFTI file.
7 | % It will not cause negative effect, as long as you remember not to do
8 | % slice time correction after reslicing the NIFTI file. Output variable
9 | % nii will be in RAS orientation, i.e. X axis from Left to Right,
10 | % Y axis from Posterior to Anterior, and Z axis from Inferior to
11 | % Superior.
12 | %
13 | % Usage: nii = load_nii(filename, [img_idx], [dim5_idx], [dim6_idx], ...
14 | % [dim7_idx], [old_RGB], [tolerance], [preferredForm])
15 | %
16 | % filename - NIFTI or ANALYZE file name.
17 | %
18 | % img_idx (optional) - a numerical array of 4th dimension indices,
19 | % which is the indices of image scan volume. The number of images
20 | % scan volumes can be obtained from get_nii_frame.m, or simply
21 | % hdr.dime.dim(5). Only the specified volumes will be loaded.
22 | % All available image volumes will be loaded, if it is default or
23 | % empty.
24 | %
25 | % dim5_idx (optional) - a numerical array of 5th dimension indices.
26 | % Only the specified range will be loaded. All available range
27 | % will be loaded, if it is default or empty.
28 | %
29 | % dim6_idx (optional) - a numerical array of 6th dimension indices.
30 | % Only the specified range will be loaded. All available range
31 | % will be loaded, if it is default or empty.
32 | %
33 | % dim7_idx (optional) - a numerical array of 7th dimension indices.
34 | % Only the specified range will be loaded. All available range
35 | % will be loaded, if it is default or empty.
36 | %
37 | % old_RGB (optional) - a scale number to tell difference of new RGB24
38 | % from old RGB24. New RGB24 uses RGB triple sequentially for each
39 | % voxel, like [R1 G1 B1 R2 G2 B2 ...]. Analyze 6.0 from AnalyzeDirect
40 | % uses old RGB24, in a way like [R1 R2 ... G1 G2 ... B1 B2 ...] for
41 | % each slices. If the image that you view is garbled, try to set
42 | % old_RGB variable to 1 and try again, because it could be in
43 | % old RGB24. It will be set to 0, if it is default or empty.
44 | %
45 | % tolerance (optional) - distortion allowed in the loaded image for any
46 | % non-orthogonal rotation or shearing of NIfTI affine matrix. If
47 | % you set 'tolerance' to 0, it means that you do not allow any
48 | % distortion. If you set 'tolerance' to 1, it means that you do
49 | % not care any distortion. The image will fail to be loaded if it
50 | % can not be tolerated. The tolerance will be set to 0.1 (10%), if
51 | % it is default or empty.
52 | %
53 | % preferredForm (optional) - selects which transformation from voxels
54 | % to RAS coordinates; values are s,q,S,Q. Lower case s,q indicate
55 | % "prefer sform or qform, but use others if preferred not present".
56 | % Upper case indicate the program is forced to use the specificied
57 | % tranform or fail loading. 'preferredForm' will be 's', if it is
58 | % default or empty. - Jeff Gunter
59 | %
60 | % Returned values:
61 | %
62 | % nii structure:
63 | %
64 | % hdr - struct with NIFTI header fields.
65 | %
66 | % filetype - Analyze format .hdr/.img (0);
67 | % NIFTI .hdr/.img (1);
68 | % NIFTI .nii (2)
69 | %
70 | % fileprefix - NIFTI filename without extension.
71 | %
72 | % machine - machine string variable.
73 | %
74 | % img - 3D (or 4D) matrix of NIFTI data.
75 | %
76 | % original - the original header before any affine transform.
77 | %
78 | % Part of this file is copied and modified from:
79 | % http://www.mathworks.com/matlabcentral/fileexchange/1878-mri-analyze-tools
80 | %
81 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
82 | %
83 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
84 | %
85 | function nii = load_nii(filename, img_idx, dim5_idx, dim6_idx, dim7_idx, ...
86 | old_RGB, tolerance, preferredForm)
87 |
88 | if ~exist('filename','var')
89 | error('Usage: nii = load_nii(filename, [img_idx], [dim5_idx], [dim6_idx], [dim7_idx], [old_RGB], [tolerance], [preferredForm])');
90 | end
91 |
92 | if ~exist('img_idx','var') | isempty(img_idx)
93 | img_idx = [];
94 | end
95 |
96 | if ~exist('dim5_idx','var') | isempty(dim5_idx)
97 | dim5_idx = [];
98 | end
99 |
100 | if ~exist('dim6_idx','var') | isempty(dim6_idx)
101 | dim6_idx = [];
102 | end
103 |
104 | if ~exist('dim7_idx','var') | isempty(dim7_idx)
105 | dim7_idx = [];
106 | end
107 |
108 | if ~exist('old_RGB','var') | isempty(old_RGB)
109 | old_RGB = 0;
110 | end
111 |
112 | if ~exist('tolerance','var') | isempty(tolerance)
113 | tolerance = 0.1; % 10 percent
114 | end
115 |
116 | if ~exist('preferredForm','var') | isempty(preferredForm)
117 | preferredForm= 's'; % Jeff
118 | end
119 |
120 | v = version;
121 |
122 | % Check file extension. If .gz, unpack it into temp folder
123 | %
124 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
125 |
126 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
127 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
128 | ~strcmp(filename(end-6:end), '.nii.gz')
129 |
130 | error('Please check filename.');
131 | end
132 |
133 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
134 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
135 | elseif strcmp(filename(end-6:end), '.img.gz')
136 | filename1 = filename;
137 | filename2 = filename;
138 | filename2(end-6:end) = '';
139 | filename2 = [filename2, '.hdr.gz'];
140 |
141 | tmpDir = tempname;
142 | mkdir(tmpDir);
143 | gzFileName = filename;
144 |
145 | filename1 = gunzip(filename1, tmpDir);
146 | filename2 = gunzip(filename2, tmpDir);
147 | filename = char(filename1); % convert from cell to string
148 | elseif strcmp(filename(end-6:end), '.hdr.gz')
149 | filename1 = filename;
150 | filename2 = filename;
151 | filename2(end-6:end) = '';
152 | filename2 = [filename2, '.img.gz'];
153 |
154 | tmpDir = tempname;
155 | mkdir(tmpDir);
156 | gzFileName = filename;
157 |
158 | filename1 = gunzip(filename1, tmpDir);
159 | filename2 = gunzip(filename2, tmpDir);
160 | filename = char(filename1); % convert from cell to string
161 | elseif strcmp(filename(end-6:end), '.nii.gz')
162 | tmpDir = tempname;
163 | mkdir(tmpDir);
164 | gzFileName = filename;
165 | filename = gunzip(filename, tmpDir);
166 | filename = char(filename); % convert from cell to string
167 | end
168 | end
169 |
170 | % Read the dataset header
171 | %
172 | [nii.hdr,nii.filetype,nii.fileprefix,nii.machine] = load_nii_hdr(filename);
173 |
174 | % Read the header extension
175 | %
176 | % nii.ext = load_nii_ext(filename);
177 |
178 | % Read the dataset body
179 | %
180 | [nii.img,nii.hdr] = load_nii_img(nii.hdr,nii.filetype,nii.fileprefix, ...
181 | nii.machine,img_idx,dim5_idx,dim6_idx,dim7_idx,old_RGB);
182 |
183 | % Perform some of sform/qform transform
184 | %
185 | nii = xform_nii(nii, tolerance, preferredForm);
186 |
187 | % Clean up after gunzip
188 | %
189 | if exist('gzFileName', 'var')
190 |
191 | % fix fileprefix so it doesn't point to temp location
192 | %
193 | nii.fileprefix = gzFileName(1:end-7);
194 | rmdir(tmpDir,'s');
195 | end
196 |
197 | return % load_nii
198 |
199 |
--------------------------------------------------------------------------------
/NIFTI/load_nii_ext.m:
--------------------------------------------------------------------------------
1 | % Load NIFTI header extension after its header is loaded using load_nii_hdr.
2 | %
3 | % Usage: ext = load_nii_ext(filename)
4 | %
5 | % filename - NIFTI file name.
6 | %
7 | % Returned values:
8 | %
9 | % ext - Structure of NIFTI header extension, which includes num_ext,
10 | % and all the extended header sections in the header extension.
11 | % Each extended header section will have its esize, ecode, and
12 | % edata, where edata can be plain text, xml, or any raw data
13 | % that was saved in the extended header section.
14 | %
15 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
16 | %
17 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
18 | %
19 | function ext = load_nii_ext(filename)
20 |
21 | if ~exist('filename','var'),
22 | error('Usage: ext = load_nii_ext(filename)');
23 | end
24 |
25 |
26 | v = version;
27 |
28 | % Check file extension. If .gz, unpack it into temp folder
29 | %
30 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
31 |
32 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
33 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
34 | ~strcmp(filename(end-6:end), '.nii.gz')
35 |
36 | error('Please check filename.');
37 | end
38 |
39 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
40 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
41 | elseif strcmp(filename(end-6:end), '.img.gz')
42 | filename1 = filename;
43 | filename2 = filename;
44 | filename2(end-6:end) = '';
45 | filename2 = [filename2, '.hdr.gz'];
46 |
47 | tmpDir = tempname;
48 | mkdir(tmpDir);
49 | gzFileName = filename;
50 |
51 | filename1 = gunzip(filename1, tmpDir);
52 | filename2 = gunzip(filename2, tmpDir);
53 | filename = char(filename1); % convert from cell to string
54 | elseif strcmp(filename(end-6:end), '.hdr.gz')
55 | filename1 = filename;
56 | filename2 = filename;
57 | filename2(end-6:end) = '';
58 | filename2 = [filename2, '.img.gz'];
59 |
60 | tmpDir = tempname;
61 | mkdir(tmpDir);
62 | gzFileName = filename;
63 |
64 | filename1 = gunzip(filename1, tmpDir);
65 | filename2 = gunzip(filename2, tmpDir);
66 | filename = char(filename1); % convert from cell to string
67 | elseif strcmp(filename(end-6:end), '.nii.gz')
68 | tmpDir = tempname;
69 | mkdir(tmpDir);
70 | gzFileName = filename;
71 | filename = gunzip(filename, tmpDir);
72 | filename = char(filename); % convert from cell to string
73 | end
74 | end
75 |
76 | machine = 'ieee-le';
77 | new_ext = 0;
78 |
79 | if findstr('.nii',filename) & strcmp(filename(end-3:end), '.nii')
80 | new_ext = 1;
81 | filename(end-3:end)='';
82 | end
83 |
84 | if findstr('.hdr',filename) & strcmp(filename(end-3:end), '.hdr')
85 | filename(end-3:end)='';
86 | end
87 |
88 | if findstr('.img',filename) & strcmp(filename(end-3:end), '.img')
89 | filename(end-3:end)='';
90 | end
91 |
92 | if new_ext
93 | fn = sprintf('%s.nii',filename);
94 |
95 | if ~exist(fn)
96 | msg = sprintf('Cannot find file "%s.nii".', filename);
97 | error(msg);
98 | end
99 | else
100 | fn = sprintf('%s.hdr',filename);
101 |
102 | if ~exist(fn)
103 | msg = sprintf('Cannot find file "%s.hdr".', filename);
104 | error(msg);
105 | end
106 | end
107 |
108 | fid = fopen(fn,'r',machine);
109 | vox_offset = 0;
110 |
111 | if fid < 0,
112 | msg = sprintf('Cannot open file %s.',fn);
113 | error(msg);
114 | else
115 | fseek(fid,0,'bof');
116 |
117 | if fread(fid,1,'int32') == 348
118 | if new_ext
119 | fseek(fid,108,'bof');
120 | vox_offset = fread(fid,1,'float32');
121 | end
122 |
123 | ext = read_extension(fid, vox_offset);
124 | fclose(fid);
125 | else
126 | fclose(fid);
127 |
128 | % first try reading the opposite endian to 'machine'
129 | %
130 | switch machine,
131 | case 'ieee-le', machine = 'ieee-be';
132 | case 'ieee-be', machine = 'ieee-le';
133 | end
134 |
135 | fid = fopen(fn,'r',machine);
136 |
137 | if fid < 0,
138 | msg = sprintf('Cannot open file %s.',fn);
139 | error(msg);
140 | else
141 | fseek(fid,0,'bof');
142 |
143 | if fread(fid,1,'int32') ~= 348
144 |
145 | % Now throw an error
146 | %
147 | msg = sprintf('File "%s" is corrupted.',fn);
148 | error(msg);
149 | end
150 |
151 | if new_ext
152 | fseek(fid,108,'bof');
153 | vox_offset = fread(fid,1,'float32');
154 | end
155 |
156 | ext = read_extension(fid, vox_offset);
157 | fclose(fid);
158 | end
159 | end
160 | end
161 |
162 |
163 | % Clean up after gunzip
164 | %
165 | if exist('gzFileName', 'var')
166 | rmdir(tmpDir,'s');
167 | end
168 |
169 |
170 | return % load_nii_ext
171 |
172 |
173 | %---------------------------------------------------------------------
174 | function ext = read_extension(fid, vox_offset)
175 |
176 | ext = [];
177 |
178 | if vox_offset
179 | end_of_ext = vox_offset;
180 | else
181 | fseek(fid, 0, 'eof');
182 | end_of_ext = ftell(fid);
183 | end
184 |
185 | if end_of_ext > 352
186 | fseek(fid, 348, 'bof');
187 | ext.extension = fread(fid,4)';
188 | end
189 |
190 | if isempty(ext) | ext.extension(1) == 0
191 | ext = [];
192 | return;
193 | end
194 |
195 | i = 1;
196 |
197 | while(ftell(fid) < end_of_ext)
198 | ext.section(i).esize = fread(fid,1,'int32');
199 | ext.section(i).ecode = fread(fid,1,'int32');
200 | ext.section(i).edata = char(fread(fid,ext.section(i).esize-8)');
201 | i = i + 1;
202 | end
203 |
204 | ext.num_ext = length(ext.section);
205 |
206 | return % read_extension
207 |
208 |
--------------------------------------------------------------------------------
/NIFTI/load_untouch0_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % internal function
2 |
3 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
4 |
5 | function hdr = load_nii_hdr(fileprefix, machine)
6 |
7 | fn = sprintf('%s.hdr',fileprefix);
8 | fid = fopen(fn,'r',machine);
9 |
10 | if fid < 0,
11 | msg = sprintf('Cannot open file %s.',fn);
12 | error(msg);
13 | else
14 | fseek(fid,0,'bof');
15 | hdr = read_header(fid);
16 | fclose(fid);
17 | end
18 |
19 | return % load_nii_hdr
20 |
21 |
22 | %---------------------------------------------------------------------
23 | function [ dsr ] = read_header(fid)
24 |
25 | % Original header structures
26 | % struct dsr
27 | % {
28 | % struct header_key hk; /* 0 + 40 */
29 | % struct image_dimension dime; /* 40 + 108 */
30 | % struct data_history hist; /* 148 + 200 */
31 | % }; /* total= 348 bytes*/
32 |
33 | dsr.hk = header_key(fid);
34 | dsr.dime = image_dimension(fid);
35 | dsr.hist = data_history(fid);
36 |
37 | return % read_header
38 |
39 |
40 | %---------------------------------------------------------------------
41 | function [ hk ] = header_key(fid)
42 |
43 | fseek(fid,0,'bof');
44 |
45 | % Original header structures
46 | % struct header_key /* header key */
47 | % { /* off + size */
48 | % int sizeof_hdr /* 0 + 4 */
49 | % char data_type[10]; /* 4 + 10 */
50 | % char db_name[18]; /* 14 + 18 */
51 | % int extents; /* 32 + 4 */
52 | % short int session_error; /* 36 + 2 */
53 | % char regular; /* 38 + 1 */
54 | % char hkey_un0; /* 39 + 1 */
55 | % }; /* total=40 bytes */
56 | %
57 | % int sizeof_header Should be 348.
58 | % char regular Must be 'r' to indicate that all images and
59 | % volumes are the same size.
60 |
61 | v6 = version;
62 | if str2num(v6(1))<6
63 | directchar = '*char';
64 | else
65 | directchar = 'uchar=>char';
66 | end
67 |
68 | hk.sizeof_hdr = fread(fid, 1,'int32')'; % should be 348!
69 | hk.data_type = deblank(fread(fid,10,directchar)');
70 | hk.db_name = deblank(fread(fid,18,directchar)');
71 | hk.extents = fread(fid, 1,'int32')';
72 | hk.session_error = fread(fid, 1,'int16')';
73 | hk.regular = fread(fid, 1,directchar)';
74 | hk.hkey_un0 = fread(fid, 1,directchar)';
75 |
76 | return % header_key
77 |
78 |
79 | %---------------------------------------------------------------------
80 | function [ dime ] = image_dimension(fid)
81 |
82 | %struct image_dimension
83 | % { /* off + size */
84 | % short int dim[8]; /* 0 + 16 */
85 | % /*
86 | % dim[0] Number of dimensions in database; usually 4.
87 | % dim[1] Image X dimension; number of *pixels* in an image row.
88 | % dim[2] Image Y dimension; number of *pixel rows* in slice.
89 | % dim[3] Volume Z dimension; number of *slices* in a volume.
90 | % dim[4] Time points; number of volumes in database
91 | % */
92 | % char vox_units[4]; /* 16 + 4 */
93 | % char cal_units[8]; /* 20 + 8 */
94 | % short int unused1; /* 28 + 2 */
95 | % short int datatype; /* 30 + 2 */
96 | % short int bitpix; /* 32 + 2 */
97 | % short int dim_un0; /* 34 + 2 */
98 | % float pixdim[8]; /* 36 + 32 */
99 | % /*
100 | % pixdim[] specifies the voxel dimensions:
101 | % pixdim[1] - voxel width, mm
102 | % pixdim[2] - voxel height, mm
103 | % pixdim[3] - slice thickness, mm
104 | % pixdim[4] - volume timing, in msec
105 | % ..etc
106 | % */
107 | % float vox_offset; /* 68 + 4 */
108 | % float roi_scale; /* 72 + 4 */
109 | % float funused1; /* 76 + 4 */
110 | % float funused2; /* 80 + 4 */
111 | % float cal_max; /* 84 + 4 */
112 | % float cal_min; /* 88 + 4 */
113 | % int compressed; /* 92 + 4 */
114 | % int verified; /* 96 + 4 */
115 | % int glmax; /* 100 + 4 */
116 | % int glmin; /* 104 + 4 */
117 | % }; /* total=108 bytes */
118 |
119 | v6 = version;
120 | if str2num(v6(1))<6
121 | directchar = '*char';
122 | else
123 | directchar = 'uchar=>char';
124 | end
125 |
126 | dime.dim = fread(fid,8,'int16')';
127 | dime.vox_units = deblank(fread(fid,4,directchar)');
128 | dime.cal_units = deblank(fread(fid,8,directchar)');
129 | dime.unused1 = fread(fid,1,'int16')';
130 | dime.datatype = fread(fid,1,'int16')';
131 | dime.bitpix = fread(fid,1,'int16')';
132 | dime.dim_un0 = fread(fid,1,'int16')';
133 | dime.pixdim = fread(fid,8,'float32')';
134 | dime.vox_offset = fread(fid,1,'float32')';
135 | dime.roi_scale = fread(fid,1,'float32')';
136 | dime.funused1 = fread(fid,1,'float32')';
137 | dime.funused2 = fread(fid,1,'float32')';
138 | dime.cal_max = fread(fid,1,'float32')';
139 | dime.cal_min = fread(fid,1,'float32')';
140 | dime.compressed = fread(fid,1,'int32')';
141 | dime.verified = fread(fid,1,'int32')';
142 | dime.glmax = fread(fid,1,'int32')';
143 | dime.glmin = fread(fid,1,'int32')';
144 |
145 | return % image_dimension
146 |
147 |
148 | %---------------------------------------------------------------------
149 | function [ hist ] = data_history(fid)
150 |
151 | %struct data_history
152 | % { /* off + size */
153 | % char descrip[80]; /* 0 + 80 */
154 | % char aux_file[24]; /* 80 + 24 */
155 | % char orient; /* 104 + 1 */
156 | % char originator[10]; /* 105 + 10 */
157 | % char generated[10]; /* 115 + 10 */
158 | % char scannum[10]; /* 125 + 10 */
159 | % char patient_id[10]; /* 135 + 10 */
160 | % char exp_date[10]; /* 145 + 10 */
161 | % char exp_time[10]; /* 155 + 10 */
162 | % char hist_un0[3]; /* 165 + 3 */
163 | % int views /* 168 + 4 */
164 | % int vols_added; /* 172 + 4 */
165 | % int start_field; /* 176 + 4 */
166 | % int field_skip; /* 180 + 4 */
167 | % int omax; /* 184 + 4 */
168 | % int omin; /* 188 + 4 */
169 | % int smax; /* 192 + 4 */
170 | % int smin; /* 196 + 4 */
171 | % }; /* total=200 bytes */
172 |
173 | v6 = version;
174 | if str2num(v6(1))<6
175 | directchar = '*char';
176 | else
177 | directchar = 'uchar=>char';
178 | end
179 |
180 | hist.descrip = deblank(fread(fid,80,directchar)');
181 | hist.aux_file = deblank(fread(fid,24,directchar)');
182 | hist.orient = fread(fid, 1,'char')';
183 | hist.originator = fread(fid, 5,'int16')';
184 | hist.generated = deblank(fread(fid,10,directchar)');
185 | hist.scannum = deblank(fread(fid,10,directchar)');
186 | hist.patient_id = deblank(fread(fid,10,directchar)');
187 | hist.exp_date = deblank(fread(fid,10,directchar)');
188 | hist.exp_time = deblank(fread(fid,10,directchar)');
189 | hist.hist_un0 = deblank(fread(fid, 3,directchar)');
190 | hist.views = fread(fid, 1,'int32')';
191 | hist.vols_added = fread(fid, 1,'int32')';
192 | hist.start_field = fread(fid, 1,'int32')';
193 | hist.field_skip = fread(fid, 1,'int32')';
194 | hist.omax = fread(fid, 1,'int32')';
195 | hist.omin = fread(fid, 1,'int32')';
196 | hist.smax = fread(fid, 1,'int32')';
197 | hist.smin = fread(fid, 1,'int32')';
198 |
199 | return % data_history
200 |
201 |
--------------------------------------------------------------------------------
/NIFTI/load_untouch_header_only.m:
--------------------------------------------------------------------------------
1 | % Load NIfTI / Analyze header without applying any appropriate affine
2 | % geometric transform or voxel intensity scaling. It is equivalent to
3 | % hdr field when using load_untouch_nii to load dataset. Support both
4 | % *.nii and *.hdr file extension. If file extension is not provided,
5 | % *.hdr will be used as default.
6 | %
7 | % Usage: [header, ext, filetype, machine] = load_untouch_header_only(filename)
8 | %
9 | % filename - NIfTI / Analyze file name.
10 | %
11 | % Returned values:
12 | %
13 | % header - struct with NIfTI / Analyze header fields.
14 | %
15 | % ext - NIfTI extension if it is not empty.
16 | %
17 | % filetype - 0 for Analyze format (*.hdr/*.img);
18 | % 1 for NIFTI format in 2 files (*.hdr/*.img);
19 | % 2 for NIFTI format in 1 file (*.nii).
20 | %
21 | % machine - a string, see below for details. The default here is 'ieee-le'.
22 | %
23 | % 'native' or 'n' - local machine format - the default
24 | % 'ieee-le' or 'l' - IEEE floating point with little-endian
25 | % byte ordering
26 | % 'ieee-be' or 'b' - IEEE floating point with big-endian
27 | % byte ordering
28 | % 'vaxd' or 'd' - VAX D floating point and VAX ordering
29 | % 'vaxg' or 'g' - VAX G floating point and VAX ordering
30 | % 'cray' or 'c' - Cray floating point with big-endian
31 | % byte ordering
32 | % 'ieee-le.l64' or 'a' - IEEE floating point with little-endian
33 | % byte ordering and 64 bit long data type
34 | % 'ieee-be.l64' or 's' - IEEE floating point with big-endian byte
35 | % ordering and 64 bit long data type.
36 | %
37 | % Part of this file is copied and modified from:
38 | % http://www.mathworks.com/matlabcentral/fileexchange/1878-mri-analyze-tools
39 | %
40 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
41 | %
42 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
43 | %
44 | function [hdr, ext, filetype, machine] = load_untouch_header_only(filename)
45 |
46 | if ~exist('filename','var')
47 | error('Usage: [header, ext, filetype, machine] = load_untouch_header_only(filename)');
48 | end
49 |
50 |
51 | v = version;
52 |
53 | % Check file extension. If .gz, unpack it into temp folder
54 | %
55 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
56 |
57 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
58 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
59 | ~strcmp(filename(end-6:end), '.nii.gz')
60 |
61 | error('Please check filename.');
62 | end
63 |
64 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
65 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
66 | elseif strcmp(filename(end-6:end), '.img.gz')
67 | filename1 = filename;
68 | filename2 = filename;
69 | filename2(end-6:end) = '';
70 | filename2 = [filename2, '.hdr.gz'];
71 |
72 | tmpDir = tempname;
73 | mkdir(tmpDir);
74 | gzFileName = filename;
75 |
76 | filename1 = gunzip(filename1, tmpDir);
77 | filename2 = gunzip(filename2, tmpDir);
78 | filename = char(filename1); % convert from cell to string
79 | elseif strcmp(filename(end-6:end), '.hdr.gz')
80 | filename1 = filename;
81 | filename2 = filename;
82 | filename2(end-6:end) = '';
83 | filename2 = [filename2, '.img.gz'];
84 |
85 | tmpDir = tempname;
86 | mkdir(tmpDir);
87 | gzFileName = filename;
88 |
89 | filename1 = gunzip(filename1, tmpDir);
90 | filename2 = gunzip(filename2, tmpDir);
91 | filename = char(filename1); % convert from cell to string
92 | elseif strcmp(filename(end-6:end), '.nii.gz')
93 | tmpDir = tempname;
94 | mkdir(tmpDir);
95 | gzFileName = filename;
96 | filename = gunzip(filename, tmpDir);
97 | filename = char(filename); % convert from cell to string
98 | end
99 | end
100 |
101 | % Read the dataset header
102 | %
103 | [hdr, filetype, fileprefix, machine] = load_nii_hdr(filename);
104 |
105 | if filetype == 0
106 | hdr = load_untouch0_nii_hdr(fileprefix, machine);
107 | ext = [];
108 | else
109 | hdr = load_untouch_nii_hdr(fileprefix, machine, filetype);
110 |
111 | % Read the header extension
112 | %
113 | ext = load_nii_ext(filename);
114 | end
115 |
116 | % Set bitpix according to datatype
117 | %
118 | % /*Acceptable values for datatype are*/
119 | %
120 | % 0 None (Unknown bit per voxel) % DT_NONE, DT_UNKNOWN
121 | % 1 Binary (ubit1, bitpix=1) % DT_BINARY
122 | % 2 Unsigned char (uchar or uint8, bitpix=8) % DT_UINT8, NIFTI_TYPE_UINT8
123 | % 4 Signed short (int16, bitpix=16) % DT_INT16, NIFTI_TYPE_INT16
124 | % 8 Signed integer (int32, bitpix=32) % DT_INT32, NIFTI_TYPE_INT32
125 | % 16 Floating point (single or float32, bitpix=32) % DT_FLOAT32, NIFTI_TYPE_FLOAT32
126 | % 32 Complex, 2 float32 (Use float32, bitpix=64) % DT_COMPLEX64, NIFTI_TYPE_COMPLEX64
127 | % 64 Double precision (double or float64, bitpix=64) % DT_FLOAT64, NIFTI_TYPE_FLOAT64
128 | % 128 uint8 RGB (Use uint8, bitpix=24) % DT_RGB24, NIFTI_TYPE_RGB24
129 | % 256 Signed char (schar or int8, bitpix=8) % DT_INT8, NIFTI_TYPE_INT8
130 | % 511 Single RGB (Use float32, bitpix=96) % DT_RGB96, NIFTI_TYPE_RGB96
131 | % 512 Unsigned short (uint16, bitpix=16) % DT_UNINT16, NIFTI_TYPE_UNINT16
132 | % 768 Unsigned integer (uint32, bitpix=32) % DT_UNINT32, NIFTI_TYPE_UNINT32
133 | % 1024 Signed long long (int64, bitpix=64) % DT_INT64, NIFTI_TYPE_INT64
134 | % 1280 Unsigned long long (uint64, bitpix=64) % DT_UINT64, NIFTI_TYPE_UINT64
135 | % 1536 Long double, float128 (Unsupported, bitpix=128) % DT_FLOAT128, NIFTI_TYPE_FLOAT128
136 | % 1792 Complex128, 2 float64 (Use float64, bitpix=128) % DT_COMPLEX128, NIFTI_TYPE_COMPLEX128
137 | % 2048 Complex256, 2 float128 (Unsupported, bitpix=256) % DT_COMPLEX128, NIFTI_TYPE_COMPLEX128
138 | %
139 | switch hdr.dime.datatype
140 | case 1,
141 | hdr.dime.bitpix = 1; precision = 'ubit1';
142 | case 2,
143 | hdr.dime.bitpix = 8; precision = 'uint8';
144 | case 4,
145 | hdr.dime.bitpix = 16; precision = 'int16';
146 | case 8,
147 | hdr.dime.bitpix = 32; precision = 'int32';
148 | case 16,
149 | hdr.dime.bitpix = 32; precision = 'float32';
150 | case 32,
151 | hdr.dime.bitpix = 64; precision = 'float32';
152 | case 64,
153 | hdr.dime.bitpix = 64; precision = 'float64';
154 | case 128,
155 | hdr.dime.bitpix = 24; precision = 'uint8';
156 | case 256
157 | hdr.dime.bitpix = 8; precision = 'int8';
158 | case 511
159 | hdr.dime.bitpix = 96; precision = 'float32';
160 | case 512
161 | hdr.dime.bitpix = 16; precision = 'uint16';
162 | case 768
163 | hdr.dime.bitpix = 32; precision = 'uint32';
164 | case 1024
165 | hdr.dime.bitpix = 64; precision = 'int64';
166 | case 1280
167 | hdr.dime.bitpix = 64; precision = 'uint64';
168 | case 1792,
169 | hdr.dime.bitpix = 128; precision = 'float64';
170 | otherwise
171 | error('This datatype is not supported');
172 | end
173 |
174 | tmp = hdr.dime.dim(2:end);
175 | tmp(find(tmp < 1)) = 1;
176 | hdr.dime.dim(2:end) = tmp;
177 |
178 |
179 | % Clean up after gunzip
180 | %
181 | if exist('gzFileName', 'var')
182 | rmdir(tmpDir,'s');
183 | end
184 |
185 |
186 | return % load_untouch_header_only
187 |
188 |
--------------------------------------------------------------------------------
/NIFTI/load_untouch_nii.m:
--------------------------------------------------------------------------------
1 | % Load NIFTI or ANALYZE dataset, but not applying any appropriate affine
2 | % geometric transform or voxel intensity scaling.
3 | %
4 | % Although according to NIFTI website, all those header information are
5 | % supposed to be applied to the loaded NIFTI image, there are some
6 | % situations that people do want to leave the original NIFTI header and
7 | % data untouched. They will probably just use MATLAB to do certain image
8 | % processing regardless of image orientation, and to save data back with
9 | % the same NIfTI header.
10 | %
11 | % Since this program is only served for those situations, please use it
12 | % together with "save_untouch_nii.m", and do not use "save_nii.m" or
13 | % "view_nii.m" for the data that is loaded by "load_untouch_nii.m". For
14 | % normal situation, you should use "load_nii.m" instead.
15 | %
16 | % Usage: nii = load_untouch_nii(filename, [img_idx], [dim5_idx], [dim6_idx], ...
17 | % [dim7_idx], [old_RGB], [slice_idx])
18 | %
19 | % filename - NIFTI or ANALYZE file name.
20 | %
21 | % img_idx (optional) - a numerical array of image volume indices.
22 | % Only the specified volumes will be loaded. All available image
23 | % volumes will be loaded, if it is default or empty.
24 | %
25 | % The number of images scans can be obtained from get_nii_frame.m,
26 | % or simply: hdr.dime.dim(5).
27 | %
28 | % dim5_idx (optional) - a numerical array of 5th dimension indices.
29 | % Only the specified range will be loaded. All available range
30 | % will be loaded, if it is default or empty.
31 | %
32 | % dim6_idx (optional) - a numerical array of 6th dimension indices.
33 | % Only the specified range will be loaded. All available range
34 | % will be loaded, if it is default or empty.
35 | %
36 | % dim7_idx (optional) - a numerical array of 7th dimension indices.
37 | % Only the specified range will be loaded. All available range
38 | % will be loaded, if it is default or empty.
39 | %
40 | % old_RGB (optional) - a scale number to tell difference of new RGB24
41 | % from old RGB24. New RGB24 uses RGB triple sequentially for each
42 | % voxel, like [R1 G1 B1 R2 G2 B2 ...]. Analyze 6.0 from AnalyzeDirect
43 | % uses old RGB24, in a way like [R1 R2 ... G1 G2 ... B1 B2 ...] for
44 | % each slices. If the image that you view is garbled, try to set
45 | % old_RGB variable to 1 and try again, because it could be in
46 | % old RGB24. It will be set to 0, if it is default or empty.
47 | %
48 | % slice_idx (optional) - a numerical array of image slice indices.
49 | % Only the specified slices will be loaded. All available image
50 | % slices will be loaded, if it is default or empty.
51 | %
52 | % Returned values:
53 | %
54 | % nii structure:
55 | %
56 | % hdr - struct with NIFTI header fields.
57 | %
58 | % filetype - Analyze format .hdr/.img (0);
59 | % NIFTI .hdr/.img (1);
60 | % NIFTI .nii (2)
61 | %
62 | % fileprefix - NIFTI filename without extension.
63 | %
64 | % machine - machine string variable.
65 | %
66 | % img - 3D (or 4D) matrix of NIFTI data.
67 | %
68 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
69 | %
70 | function nii = load_untouch_nii(filename, img_idx, dim5_idx, dim6_idx, dim7_idx, ...
71 | old_RGB, slice_idx)
72 |
73 | if ~exist('filename','var')
74 | error('Usage: nii = load_untouch_nii(filename, [img_idx], [dim5_idx], [dim6_idx], [dim7_idx], [old_RGB], [slice_idx])');
75 | end
76 |
77 | if ~exist('img_idx','var') | isempty(img_idx)
78 | img_idx = [];
79 | end
80 |
81 | if ~exist('dim5_idx','var') | isempty(dim5_idx)
82 | dim5_idx = [];
83 | end
84 |
85 | if ~exist('dim6_idx','var') | isempty(dim6_idx)
86 | dim6_idx = [];
87 | end
88 |
89 | if ~exist('dim7_idx','var') | isempty(dim7_idx)
90 | dim7_idx = [];
91 | end
92 |
93 | if ~exist('old_RGB','var') | isempty(old_RGB)
94 | old_RGB = 0;
95 | end
96 |
97 | if ~exist('slice_idx','var') | isempty(slice_idx)
98 | slice_idx = [];
99 | end
100 |
101 |
102 | v = version;
103 |
104 | % Check file extension. If .gz, unpack it into temp folder
105 | %
106 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
107 |
108 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
109 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
110 | ~strcmp(filename(end-6:end), '.nii.gz')
111 |
112 | error('Please check filename.');
113 | end
114 |
115 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
116 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
117 | elseif strcmp(filename(end-6:end), '.img.gz')
118 | filename1 = filename;
119 | filename2 = filename;
120 | filename2(end-6:end) = '';
121 | filename2 = [filename2, '.hdr.gz'];
122 |
123 | tmpDir = tempname;
124 | mkdir(tmpDir);
125 | gzFileName = filename;
126 |
127 | filename1 = gunzip(filename1, tmpDir);
128 | filename2 = gunzip(filename2, tmpDir);
129 | filename = char(filename1); % convert from cell to string
130 | elseif strcmp(filename(end-6:end), '.hdr.gz')
131 | filename1 = filename;
132 | filename2 = filename;
133 | filename2(end-6:end) = '';
134 | filename2 = [filename2, '.img.gz'];
135 |
136 | tmpDir = tempname;
137 | mkdir(tmpDir);
138 | gzFileName = filename;
139 |
140 | filename1 = gunzip(filename1, tmpDir);
141 | filename2 = gunzip(filename2, tmpDir);
142 | filename = char(filename1); % convert from cell to string
143 | elseif strcmp(filename(end-6:end), '.nii.gz')
144 | tmpDir = tempname;
145 | mkdir(tmpDir);
146 | gzFileName = filename;
147 | filename = gunzip(filename, tmpDir);
148 | filename = char(filename); % convert from cell to string
149 | end
150 | end
151 |
152 | % Read the dataset header
153 | %
154 | [nii.hdr,nii.filetype,nii.fileprefix,nii.machine] = load_nii_hdr(filename);
155 |
156 | if nii.filetype == 0
157 | nii.hdr = load_untouch0_nii_hdr(nii.fileprefix,nii.machine);
158 | nii.ext = [];
159 | else
160 | nii.hdr = load_untouch_nii_hdr(nii.fileprefix,nii.machine,nii.filetype);
161 |
162 | % Read the header extension
163 | %
164 | nii.ext = load_nii_ext(filename);
165 | end
166 |
167 | % Read the dataset body
168 | %
169 | [nii.img,nii.hdr] = load_untouch_nii_img(nii.hdr,nii.filetype,nii.fileprefix, ...
170 | nii.machine,img_idx,dim5_idx,dim6_idx,dim7_idx,old_RGB,slice_idx);
171 |
172 | % Perform some of sform/qform transform
173 | %
174 | % nii = xform_nii(nii, tolerance, preferredForm);
175 |
176 | nii.untouch = 1;
177 |
178 |
179 | % Clean up after gunzip
180 | %
181 | if exist('gzFileName', 'var')
182 |
183 | % fix fileprefix so it doesn't point to temp location
184 | %
185 | nii.fileprefix = gzFileName(1:end-7);
186 | rmdir(tmpDir,'s');
187 | end
188 |
189 |
190 | return % load_untouch_nii
191 |
192 |
--------------------------------------------------------------------------------
/NIFTI/load_untouch_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % internal function
2 |
3 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
4 |
5 | function hdr = load_nii_hdr(fileprefix, machine, filetype)
6 |
7 | if filetype == 2
8 | fn = sprintf('%s.nii',fileprefix);
9 |
10 | if ~exist(fn)
11 | msg = sprintf('Cannot find file "%s.nii".', fileprefix);
12 | error(msg);
13 | end
14 | else
15 | fn = sprintf('%s.hdr',fileprefix);
16 |
17 | if ~exist(fn)
18 | msg = sprintf('Cannot find file "%s.hdr".', fileprefix);
19 | error(msg);
20 | end
21 | end
22 |
23 | fid = fopen(fn,'r',machine);
24 |
25 | if fid < 0,
26 | msg = sprintf('Cannot open file %s.',fn);
27 | error(msg);
28 | else
29 | fseek(fid,0,'bof');
30 | hdr = read_header(fid);
31 | fclose(fid);
32 | end
33 |
34 | return % load_nii_hdr
35 |
36 |
37 | %---------------------------------------------------------------------
38 | function [ dsr ] = read_header(fid)
39 |
40 | % Original header structures
41 | % struct dsr
42 | % {
43 | % struct header_key hk; /* 0 + 40 */
44 | % struct image_dimension dime; /* 40 + 108 */
45 | % struct data_history hist; /* 148 + 200 */
46 | % }; /* total= 348 bytes*/
47 |
48 | dsr.hk = header_key(fid);
49 | dsr.dime = image_dimension(fid);
50 | dsr.hist = data_history(fid);
51 |
52 | % For Analyze data format
53 | %
54 | if ~strcmp(dsr.hist.magic, 'n+1') & ~strcmp(dsr.hist.magic, 'ni1')
55 | dsr.hist.qform_code = 0;
56 | dsr.hist.sform_code = 0;
57 | end
58 |
59 | return % read_header
60 |
61 |
62 | %---------------------------------------------------------------------
63 | function [ hk ] = header_key(fid)
64 |
65 | fseek(fid,0,'bof');
66 |
67 | % Original header structures
68 | % struct header_key /* header key */
69 | % { /* off + size */
70 | % int sizeof_hdr /* 0 + 4 */
71 | % char data_type[10]; /* 4 + 10 */
72 | % char db_name[18]; /* 14 + 18 */
73 | % int extents; /* 32 + 4 */
74 | % short int session_error; /* 36 + 2 */
75 | % char regular; /* 38 + 1 */
76 | % char dim_info; % char hkey_un0; /* 39 + 1 */
77 | % }; /* total=40 bytes */
78 | %
79 | % int sizeof_header Should be 348.
80 | % char regular Must be 'r' to indicate that all images and
81 | % volumes are the same size.
82 |
83 | v6 = version;
84 | if str2num(v6(1))<6
85 | directchar = '*char';
86 | else
87 | directchar = 'uchar=>char';
88 | end
89 |
90 | hk.sizeof_hdr = fread(fid, 1,'int32')'; % should be 348!
91 | hk.data_type = deblank(fread(fid,10,directchar)');
92 | hk.db_name = deblank(fread(fid,18,directchar)');
93 | hk.extents = fread(fid, 1,'int32')';
94 | hk.session_error = fread(fid, 1,'int16')';
95 | hk.regular = fread(fid, 1,directchar)';
96 | hk.dim_info = fread(fid, 1,'uchar')';
97 |
98 | return % header_key
99 |
100 |
101 | %---------------------------------------------------------------------
102 | function [ dime ] = image_dimension(fid)
103 |
104 | % Original header structures
105 | % struct image_dimension
106 | % { /* off + size */
107 | % short int dim[8]; /* 0 + 16 */
108 | % /*
109 | % dim[0] Number of dimensions in database; usually 4.
110 | % dim[1] Image X dimension; number of *pixels* in an image row.
111 | % dim[2] Image Y dimension; number of *pixel rows* in slice.
112 | % dim[3] Volume Z dimension; number of *slices* in a volume.
113 | % dim[4] Time points; number of volumes in database
114 | % */
115 | % float intent_p1; % char vox_units[4]; /* 16 + 4 */
116 | % float intent_p2; % char cal_units[8]; /* 20 + 4 */
117 | % float intent_p3; % char cal_units[8]; /* 24 + 4 */
118 | % short int intent_code; % short int unused1; /* 28 + 2 */
119 | % short int datatype; /* 30 + 2 */
120 | % short int bitpix; /* 32 + 2 */
121 | % short int slice_start; % short int dim_un0; /* 34 + 2 */
122 | % float pixdim[8]; /* 36 + 32 */
123 | % /*
124 | % pixdim[] specifies the voxel dimensions:
125 | % pixdim[1] - voxel width, mm
126 | % pixdim[2] - voxel height, mm
127 | % pixdim[3] - slice thickness, mm
128 | % pixdim[4] - volume timing, in msec
129 | % ..etc
130 | % */
131 | % float vox_offset; /* 68 + 4 */
132 | % float scl_slope; % float roi_scale; /* 72 + 4 */
133 | % float scl_inter; % float funused1; /* 76 + 4 */
134 | % short slice_end; % float funused2; /* 80 + 2 */
135 | % char slice_code; % float funused2; /* 82 + 1 */
136 | % char xyzt_units; % float funused2; /* 83 + 1 */
137 | % float cal_max; /* 84 + 4 */
138 | % float cal_min; /* 88 + 4 */
139 | % float slice_duration; % int compressed; /* 92 + 4 */
140 | % float toffset; % int verified; /* 96 + 4 */
141 | % int glmax; /* 100 + 4 */
142 | % int glmin; /* 104 + 4 */
143 | % }; /* total=108 bytes */
144 |
145 | dime.dim = fread(fid,8,'int16')';
146 | dime.intent_p1 = fread(fid,1,'float32')';
147 | dime.intent_p2 = fread(fid,1,'float32')';
148 | dime.intent_p3 = fread(fid,1,'float32')';
149 | dime.intent_code = fread(fid,1,'int16')';
150 | dime.datatype = fread(fid,1,'int16')';
151 | dime.bitpix = fread(fid,1,'int16')';
152 | dime.slice_start = fread(fid,1,'int16')';
153 | dime.pixdim = fread(fid,8,'float32')';
154 | dime.vox_offset = fread(fid,1,'float32')';
155 | dime.scl_slope = fread(fid,1,'float32')';
156 | dime.scl_inter = fread(fid,1,'float32')';
157 | dime.slice_end = fread(fid,1,'int16')';
158 | dime.slice_code = fread(fid,1,'uchar')';
159 | dime.xyzt_units = fread(fid,1,'uchar')';
160 | dime.cal_max = fread(fid,1,'float32')';
161 | dime.cal_min = fread(fid,1,'float32')';
162 | dime.slice_duration = fread(fid,1,'float32')';
163 | dime.toffset = fread(fid,1,'float32')';
164 | dime.glmax = fread(fid,1,'int32')';
165 | dime.glmin = fread(fid,1,'int32')';
166 |
167 | return % image_dimension
168 |
169 |
170 | %---------------------------------------------------------------------
171 | function [ hist ] = data_history(fid)
172 |
173 | % Original header structures
174 | % struct data_history
175 | % { /* off + size */
176 | % char descrip[80]; /* 0 + 80 */
177 | % char aux_file[24]; /* 80 + 24 */
178 | % short int qform_code; /* 104 + 2 */
179 | % short int sform_code; /* 106 + 2 */
180 | % float quatern_b; /* 108 + 4 */
181 | % float quatern_c; /* 112 + 4 */
182 | % float quatern_d; /* 116 + 4 */
183 | % float qoffset_x; /* 120 + 4 */
184 | % float qoffset_y; /* 124 + 4 */
185 | % float qoffset_z; /* 128 + 4 */
186 | % float srow_x[4]; /* 132 + 16 */
187 | % float srow_y[4]; /* 148 + 16 */
188 | % float srow_z[4]; /* 164 + 16 */
189 | % char intent_name[16]; /* 180 + 16 */
190 | % char magic[4]; % int smin; /* 196 + 4 */
191 | % }; /* total=200 bytes */
192 |
193 | v6 = version;
194 | if str2num(v6(1))<6
195 | directchar = '*char';
196 | else
197 | directchar = 'uchar=>char';
198 | end
199 |
200 | hist.descrip = deblank(fread(fid,80,directchar)');
201 | hist.aux_file = deblank(fread(fid,24,directchar)');
202 | hist.qform_code = fread(fid,1,'int16')';
203 | hist.sform_code = fread(fid,1,'int16')';
204 | hist.quatern_b = fread(fid,1,'float32')';
205 | hist.quatern_c = fread(fid,1,'float32')';
206 | hist.quatern_d = fread(fid,1,'float32')';
207 | hist.qoffset_x = fread(fid,1,'float32')';
208 | hist.qoffset_y = fread(fid,1,'float32')';
209 | hist.qoffset_z = fread(fid,1,'float32')';
210 | hist.srow_x = fread(fid,4,'float32')';
211 | hist.srow_y = fread(fid,4,'float32')';
212 | hist.srow_z = fread(fid,4,'float32')';
213 | hist.intent_name = deblank(fread(fid,16,directchar)');
214 | hist.magic = deblank(fread(fid,4,directchar)');
215 |
216 | return % data_history
217 |
218 |
--------------------------------------------------------------------------------
/NIFTI/make_ana.m:
--------------------------------------------------------------------------------
1 | % Make ANALYZE 7.5 data structure specified by a 3D or 4D matrix.
2 | % Optional parameters can also be included, such as: voxel_size,
3 | % origin, datatype, and description.
4 | %
5 | % Once the ANALYZE structure is made, it can be saved into ANALYZE 7.5
6 | % format data file using "save_untouch_nii" command (for more detail,
7 | % type: help save_untouch_nii).
8 | %
9 | % Usage: ana = make_ana(img, [voxel_size], [origin], [datatype], [description])
10 | %
11 | % Where:
12 | %
13 | % img: a 3D matrix [x y z], or a 4D matrix with time
14 | % series [x y z t]. When image is in RGB format,
15 | % make sure that the size of 4th dimension is
16 | % always 3 (i.e. [R G B]). In that case, make
17 | % sure that you must specify RGB datatype to 128.
18 | %
19 | % voxel_size (optional): Voxel size in millimeter for each
20 | % dimension. Default is [1 1 1].
21 | %
22 | % origin (optional): The AC origin. Default is [0 0 0].
23 | %
24 | % datatype (optional): Storage data type:
25 | % 2 - uint8, 4 - int16, 8 - int32, 16 - float32,
26 | % 64 - float64, 128 - RGB24
27 | % Default will use the data type of 'img' matrix
28 | % For RGB image, you must specify it to 128.
29 | %
30 | % description (optional): Description of data. Default is ''.
31 | %
32 | % e.g.:
33 | % origin = [33 44 13]; datatype = 64;
34 | % ana = make_ana(img, [], origin, datatype); % default voxel_size
35 | %
36 | % ANALYZE 7.5 format: http://www.rotman-baycrest.on.ca/~jimmy/ANALYZE75.pdf
37 | %
38 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
39 | %
40 | function ana = make_ana(varargin)
41 |
42 | ana.img = varargin{1};
43 | dims = size(ana.img);
44 | dims = [4 dims ones(1,8)];
45 | dims = dims(1:8);
46 |
47 | voxel_size = [0 ones(1,3) zeros(1,4)];
48 | origin = zeros(1,5);
49 | descrip = '';
50 |
51 | switch class(ana.img)
52 | case 'uint8'
53 | datatype = 2;
54 | case 'int16'
55 | datatype = 4;
56 | case 'int32'
57 | datatype = 8;
58 | case 'single'
59 | datatype = 16;
60 | case 'double'
61 | datatype = 64;
62 | otherwise
63 | error('Datatype is not supported by make_ana.');
64 | end
65 |
66 | if nargin > 1 & ~isempty(varargin{2})
67 | voxel_size(2:4) = double(varargin{2});
68 | end
69 |
70 | if nargin > 2 & ~isempty(varargin{3})
71 | origin(1:3) = double(varargin{3});
72 | end
73 |
74 | if nargin > 3 & ~isempty(varargin{4})
75 | datatype = double(varargin{4});
76 |
77 | if datatype == 128 | datatype == 511
78 | dims(5) = [];
79 | dims = [dims 1];
80 | end
81 | end
82 |
83 | if nargin > 4 & ~isempty(varargin{5})
84 | descrip = varargin{5};
85 | end
86 |
87 | if ndims(ana.img) > 4
88 | error('NIfTI only allows a maximum of 4 Dimension matrix.');
89 | end
90 |
91 | maxval = round(double(max(ana.img(:))));
92 | minval = round(double(min(ana.img(:))));
93 |
94 | ana.hdr = make_header(dims, voxel_size, origin, datatype, ...
95 | descrip, maxval, minval);
96 | ana.filetype = 0;
97 | ana.ext = [];
98 | ana.untouch = 1;
99 |
100 | switch ana.hdr.dime.datatype
101 | case 2
102 | ana.img = uint8(ana.img);
103 | case 4
104 | ana.img = int16(ana.img);
105 | case 8
106 | ana.img = int32(ana.img);
107 | case 16
108 | ana.img = single(ana.img);
109 | case 64
110 | ana.img = double(ana.img);
111 | case 128
112 | ana.img = uint8(ana.img);
113 | otherwise
114 | error('Datatype is not supported by make_ana.');
115 | end
116 |
117 | return; % make_ana
118 |
119 |
120 | %---------------------------------------------------------------------
121 | function hdr = make_header(dims, voxel_size, origin, datatype, ...
122 | descrip, maxval, minval)
123 |
124 | hdr.hk = header_key;
125 | hdr.dime = image_dimension(dims, voxel_size, datatype, maxval, minval);
126 | hdr.hist = data_history(origin, descrip);
127 |
128 | return; % make_header
129 |
130 |
131 | %---------------------------------------------------------------------
132 | function hk = header_key
133 |
134 | hk.sizeof_hdr = 348; % must be 348!
135 | hk.data_type = '';
136 | hk.db_name = '';
137 | hk.extents = 0;
138 | hk.session_error = 0;
139 | hk.regular = 'r';
140 | hk.hkey_un0 = '0';
141 |
142 | return; % header_key
143 |
144 |
145 | %---------------------------------------------------------------------
146 | function dime = image_dimension(dims, voxel_size, datatype, maxval, minval)
147 |
148 | dime.dim = dims;
149 | dime.vox_units = 'mm';
150 | dime.cal_units = '';
151 | dime.unused1 = 0;
152 | dime.datatype = datatype;
153 |
154 | switch dime.datatype
155 | case 2,
156 | dime.bitpix = 8; precision = 'uint8';
157 | case 4,
158 | dime.bitpix = 16; precision = 'int16';
159 | case 8,
160 | dime.bitpix = 32; precision = 'int32';
161 | case 16,
162 | dime.bitpix = 32; precision = 'float32';
163 | case 64,
164 | dime.bitpix = 64; precision = 'float64';
165 | case 128
166 | dime.bitpix = 24; precision = 'uint8';
167 | otherwise
168 | error('Datatype is not supported by make_ana.');
169 | end
170 |
171 | dime.dim_un0 = 0;
172 | dime.pixdim = voxel_size;
173 | dime.vox_offset = 0;
174 | dime.roi_scale = 1;
175 | dime.funused1 = 0;
176 | dime.funused2 = 0;
177 | dime.cal_max = 0;
178 | dime.cal_min = 0;
179 | dime.compressed = 0;
180 | dime.verified = 0;
181 | dime.glmax = maxval;
182 | dime.glmin = minval;
183 |
184 | return; % image_dimension
185 |
186 |
187 | %---------------------------------------------------------------------
188 | function hist = data_history(origin, descrip)
189 |
190 | hist.descrip = descrip;
191 | hist.aux_file = 'none';
192 | hist.orient = 0;
193 | hist.originator = origin;
194 | hist.generated = '';
195 | hist.scannum = '';
196 | hist.patient_id = '';
197 | hist.exp_date = '';
198 | hist.exp_time = '';
199 | hist.hist_un0 = '';
200 | hist.views = 0;
201 | hist.vols_added = 0;
202 | hist.start_field = 0;
203 | hist.field_skip = 0;
204 | hist.omax = 0;
205 | hist.omin = 0;
206 | hist.smax = 0;
207 | hist.smin = 0;
208 |
209 | return; % data_history
210 |
211 |
--------------------------------------------------------------------------------
/NIFTI/make_nii.m:
--------------------------------------------------------------------------------
1 | % Make NIfTI structure specified by an N-D matrix. Usually, N is 3 for
2 | % 3D matrix [x y z], or 4 for 4D matrix with time series [x y z t].
3 | % Optional parameters can also be included, such as: voxel_size,
4 | % origin, datatype, and description.
5 | %
6 | % Once the NIfTI structure is made, it can be saved into NIfTI file
7 | % using "save_nii" command (for more detail, type: help save_nii).
8 | %
9 | % Usage: nii = make_nii(img, [voxel_size], [origin], [datatype], [description])
10 | %
11 | % Where:
12 | %
13 | % img: Usually, img is a 3D matrix [x y z], or a 4D
14 | % matrix with time series [x y z t]. However,
15 | % NIfTI allows a maximum of 7D matrix. When the
16 | % image is in RGB format, make sure that the size
17 | % of 4th dimension is always 3 (i.e. [R G B]). In
18 | % that case, make sure that you must specify RGB
19 | % datatype, which is either 128 or 511.
20 | %
21 | % voxel_size (optional): Voxel size in millimeter for each
22 | % dimension. Default is [1 1 1].
23 | %
24 | % origin (optional): The AC origin. Default is [0 0 0].
25 | %
26 | % datatype (optional): Storage data type:
27 | % 2 - uint8, 4 - int16, 8 - int32, 16 - float32,
28 | % 32 - complex64, 64 - float64, 128 - RGB24,
29 | % 256 - int8, 511 - RGB96, 512 - uint16,
30 | % 768 - uint32, 1792 - complex128
31 | % Default will use the data type of 'img' matrix
32 | % For RGB image, you must specify it to either 128
33 | % or 511.
34 | %
35 | % description (optional): Description of data. Default is ''.
36 | %
37 | % e.g.:
38 | % origin = [33 44 13]; datatype = 64;
39 | % nii = make_nii(img, [], origin, datatype); % default voxel_size
40 | %
41 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
42 | %
43 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
44 | %
45 | function nii = make_nii(varargin)
46 |
47 | nii.img = varargin{1};
48 | dims = size(nii.img);
49 | dims = [length(dims) dims ones(1,8)];
50 | dims = dims(1:8);
51 |
52 | voxel_size = [0 ones(1,7)];
53 | origin = zeros(1,5);
54 | descrip = '';
55 |
56 | switch class(nii.img)
57 | case 'uint8'
58 | datatype = 2;
59 | case 'int16'
60 | datatype = 4;
61 | case 'int32'
62 | datatype = 8;
63 | case 'single'
64 | if isreal(nii.img)
65 | datatype = 16;
66 | else
67 | datatype = 32;
68 | end
69 | case 'double'
70 | if isreal(nii.img)
71 | datatype = 64;
72 | else
73 | datatype = 1792;
74 | end
75 | case 'int8'
76 | datatype = 256;
77 | case 'uint16'
78 | datatype = 512;
79 | case 'uint32'
80 | datatype = 768;
81 | otherwise
82 | error('Datatype is not supported by make_nii.');
83 | end
84 |
85 | if nargin > 1 & ~isempty(varargin{2})
86 | voxel_size(2:4) = double(varargin{2});
87 | end
88 |
89 | if nargin > 2 & ~isempty(varargin{3})
90 | origin(1:3) = double(varargin{3});
91 | end
92 |
93 | if nargin > 3 & ~isempty(varargin{4})
94 | datatype = double(varargin{4});
95 |
96 | if datatype == 128 | datatype == 511
97 | dims(5) = [];
98 | dims(1) = dims(1) - 1;
99 | dims = [dims 1];
100 | end
101 | end
102 |
103 | if nargin > 4 & ~isempty(varargin{5})
104 | descrip = varargin{5};
105 | end
106 |
107 | if ndims(nii.img) > 7
108 | error('NIfTI only allows a maximum of 7 Dimension matrix.');
109 | end
110 |
111 | maxval = round(double(max(nii.img(:))));
112 | minval = round(double(min(nii.img(:))));
113 |
114 | nii.hdr = make_header(dims, voxel_size, origin, datatype, ...
115 | descrip, maxval, minval);
116 |
117 | switch nii.hdr.dime.datatype
118 | case 2
119 | nii.img = uint8(nii.img);
120 | case 4
121 | nii.img = int16(nii.img);
122 | case 8
123 | nii.img = int32(nii.img);
124 | case 16
125 | nii.img = single(nii.img);
126 | case 32
127 | nii.img = single(nii.img);
128 | case 64
129 | nii.img = double(nii.img);
130 | case 128
131 | nii.img = uint8(nii.img);
132 | case 256
133 | nii.img = int8(nii.img);
134 | case 511
135 | img = double(nii.img(:));
136 | img = single((img - min(img))/(max(img) - min(img)));
137 | nii.img = reshape(img, size(nii.img));
138 | nii.hdr.dime.glmax = double(max(img));
139 | nii.hdr.dime.glmin = double(min(img));
140 | case 512
141 | nii.img = uint16(nii.img);
142 | case 768
143 | nii.img = uint32(nii.img);
144 | case 1792
145 | nii.img = double(nii.img);
146 | otherwise
147 | error('Datatype is not supported by make_nii.');
148 | end
149 |
150 | return; % make_nii
151 |
152 |
153 | %---------------------------------------------------------------------
154 | function hdr = make_header(dims, voxel_size, origin, datatype, ...
155 | descrip, maxval, minval)
156 |
157 | hdr.hk = header_key;
158 | hdr.dime = image_dimension(dims, voxel_size, datatype, maxval, minval);
159 | hdr.hist = data_history(origin, descrip);
160 |
161 | return; % make_header
162 |
163 |
164 | %---------------------------------------------------------------------
165 | function hk = header_key
166 |
167 | hk.sizeof_hdr = 348; % must be 348!
168 | hk.data_type = '';
169 | hk.db_name = '';
170 | hk.extents = 0;
171 | hk.session_error = 0;
172 | hk.regular = 'r';
173 | hk.dim_info = 0;
174 |
175 | return; % header_key
176 |
177 |
178 | %---------------------------------------------------------------------
179 | function dime = image_dimension(dims, voxel_size, datatype, maxval, minval)
180 |
181 | dime.dim = dims;
182 | dime.intent_p1 = 0;
183 | dime.intent_p2 = 0;
184 | dime.intent_p3 = 0;
185 | dime.intent_code = 0;
186 | dime.datatype = datatype;
187 |
188 | switch dime.datatype
189 | case 2,
190 | dime.bitpix = 8; precision = 'uint8';
191 | case 4,
192 | dime.bitpix = 16; precision = 'int16';
193 | case 8,
194 | dime.bitpix = 32; precision = 'int32';
195 | case 16,
196 | dime.bitpix = 32; precision = 'float32';
197 | case 32,
198 | dime.bitpix = 64; precision = 'float32';
199 | case 64,
200 | dime.bitpix = 64; precision = 'float64';
201 | case 128
202 | dime.bitpix = 24; precision = 'uint8';
203 | case 256
204 | dime.bitpix = 8; precision = 'int8';
205 | case 511
206 | dime.bitpix = 96; precision = 'float32';
207 | case 512
208 | dime.bitpix = 16; precision = 'uint16';
209 | case 768
210 | dime.bitpix = 32; precision = 'uint32';
211 | case 1792,
212 | dime.bitpix = 128; precision = 'float64';
213 | otherwise
214 | error('Datatype is not supported by make_nii.');
215 | end
216 |
217 | dime.slice_start = 0;
218 | dime.pixdim = voxel_size;
219 | dime.vox_offset = 0;
220 | dime.scl_slope = 0;
221 | dime.scl_inter = 0;
222 | dime.slice_end = 0;
223 | dime.slice_code = 0;
224 | dime.xyzt_units = 0;
225 | dime.cal_max = 0;
226 | dime.cal_min = 0;
227 | dime.slice_duration = 0;
228 | dime.toffset = 0;
229 | dime.glmax = maxval;
230 | dime.glmin = minval;
231 |
232 | return; % image_dimension
233 |
234 |
235 | %---------------------------------------------------------------------
236 | function hist = data_history(origin, descrip)
237 |
238 | hist.descrip = descrip;
239 | hist.aux_file = 'none';
240 | hist.qform_code = 0;
241 | hist.sform_code = 0;
242 | hist.quatern_b = 0;
243 | hist.quatern_c = 0;
244 | hist.quatern_d = 0;
245 | hist.qoffset_x = 0;
246 | hist.qoffset_y = 0;
247 | hist.qoffset_z = 0;
248 | hist.srow_x = zeros(1,4);
249 | hist.srow_y = zeros(1,4);
250 | hist.srow_z = zeros(1,4);
251 | hist.intent_name = '';
252 | hist.magic = '';
253 | hist.originator = origin;
254 |
255 | return; % data_history
256 |
257 |
--------------------------------------------------------------------------------
/NIFTI/mat_into_hdr.m:
--------------------------------------------------------------------------------
1 | %MAT_INTO_HDR The old versions of SPM (any version before SPM5) store
2 | % an affine matrix of the SPM Reoriented image into a matlab file
3 | % (.mat extension). The file name of this SPM matlab file is the
4 | % same as the SPM Reoriented image file (.img/.hdr extension).
5 | %
6 | % This program will convert the ANALYZE 7.5 SPM Reoriented image
7 | % file into NIfTI format, and integrate the affine matrix in the
8 | % SPM matlab file into its header file (.hdr extension).
9 | %
10 | % WARNING: Before you run this program, please save the header
11 | % file (.hdr extension) into another file name or into another
12 | % folder location, because all header files (.hdr extension)
13 | % will be overwritten after they are converted into NIfTI
14 | % format.
15 | %
16 | % Usage: mat_into_hdr(filename);
17 | %
18 | % filename: file name(s) with .hdr or .mat file extension, like:
19 | % '*.hdr', or '*.mat', or a single .hdr or .mat file.
20 | % e.g. mat_into_hdr('T1.hdr')
21 | % mat_into_hdr('*.mat')
22 | %
23 |
24 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
25 | %
26 | %-------------------------------------------------------------------------
27 | function mat_into_hdr(files)
28 |
29 | pn = fileparts(files);
30 | file_lst = dir(files);
31 | file_lst = {file_lst.name};
32 | file1 = file_lst{1};
33 | [p n e]= fileparts(file1);
34 |
35 | for i=1:length(file_lst)
36 | [p n e]= fileparts(file_lst{i});
37 | disp(['working on file ', num2str(i) ,' of ', num2str(length(file_lst)), ': ', n,e]);
38 | process=1;
39 |
40 | if isequal(e,'.hdr')
41 | mat=fullfile(pn, [n,'.mat']);
42 | hdr=fullfile(pn, file_lst{i});
43 |
44 | if ~exist(mat,'file')
45 | warning(['Cannot find file "',mat , '". File "', n, e, '" will not be processed.']);
46 | process=0;
47 | end
48 | elseif isequal(e,'.mat')
49 | hdr=fullfile(pn, [n,'.hdr']);
50 | mat=fullfile(pn, file_lst{i});
51 |
52 | if ~exist(hdr,'file')
53 | warning(['Can not find file "',hdr , '". File "', n, e, '" will not be processed.']);
54 | process=0;
55 | end
56 | else
57 | warning(['Input file must have .mat or .hdr extension. File "', n, e, '" will not be processed.']);
58 | process=0;
59 | end
60 |
61 | if process
62 | load(mat);
63 | R=M(1:3,1:3);
64 | T=M(1:3,4);
65 | T=R*ones(3,1)+T;
66 | M(1:3,4)=T;
67 |
68 | [h filetype fileprefix machine]=load_nii_hdr(hdr);
69 | h.hist.qform_code=0;
70 | h.hist.sform_code=1;
71 | h.hist.srow_x=M(1,:);
72 | h.hist.srow_y=M(2,:);
73 | h.hist.srow_z=M(3,:);
74 | h.hist.magic='ni1';
75 |
76 | fid = fopen(hdr,'w',machine);
77 | save_nii_hdr(h,fid);
78 | fclose(fid);
79 | end
80 | end
81 |
82 | return; % mat_into_hdr
83 |
84 |
--------------------------------------------------------------------------------
/NIFTI/rri_file_menu.m:
--------------------------------------------------------------------------------
1 | % Imbed a file menu to any figure. If file menu exist, it will append
2 | % to the existing file menu. This file menu includes: Copy to clipboard,
3 | % print, save, close etc.
4 | %
5 | % Usage: rri_file_menu(fig);
6 | %
7 | % rri_file_menu(fig,0) means no 'Close' menu.
8 | %
9 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
10 | %
11 | %--------------------------------------------------------------------
12 |
13 | function rri_file_menu(action, varargin)
14 |
15 | if isnumeric(action)
16 | fig = action;
17 | action = 'init';
18 | end
19 |
20 | % clear the message line,
21 | %
22 | h = findobj(gcf,'Tag','MessageLine');
23 | set(h,'String','');
24 |
25 | if ~strcmp(action, 'init')
26 | set(gcbf, 'InvertHardcopy','off');
27 | % set(gcbf, 'PaperPositionMode','auto');
28 | end
29 |
30 | switch action
31 | case {'init'}
32 | if nargin > 1
33 | init(fig, 1); % no 'close' menu
34 | else
35 | init(fig, 0);
36 | end
37 | case {'print_fig'}
38 | printdlg(gcbf);
39 | case {'copy_fig'}
40 | copy_fig;
41 | case {'export_fig'}
42 | export_fig;
43 | end
44 |
45 | return % rri_file_menu
46 |
47 |
48 | %------------------------------------------------
49 | %
50 | % Create (or append) File menu
51 | %
52 | function init(fig, no_close)
53 |
54 | % search for file menu
55 | %
56 | h_file = [];
57 | menuitems = findobj(fig, 'type', 'uimenu');
58 |
59 | for i=1:length(menuitems)
60 | filelabel = get(menuitems(i),'label');
61 |
62 | if strcmpi(strrep(filelabel, '&', ''), 'file')
63 | h_file = menuitems(i);
64 | break;
65 | end
66 | end
67 |
68 | set(fig, 'menubar', 'none');
69 |
70 | if isempty(h_file)
71 | if isempty(menuitems)
72 | h_file = uimenu('parent', fig, 'label', 'File');
73 | else
74 | h_file = uimenu('parent', fig, 'label', 'Copy Figure');
75 | end
76 |
77 | h1 = uimenu('parent', h_file, ...
78 | 'callback','rri_file_menu(''copy_fig'');', ...
79 | 'label','Copy to Clipboard');
80 | else
81 | h1 = uimenu('parent', h_file, ...
82 | 'callback','rri_file_menu(''copy_fig'');', ...
83 | 'separator','on', ...
84 | 'label','Copy to Clipboard');
85 | end
86 |
87 | h2 = uimenu(h_file, ...
88 | 'callback','pagesetupdlg(gcbf);', ...
89 | 'label','Page Setup...');
90 |
91 | h2 = uimenu(h_file, ...
92 | 'callback','printpreview(gcbf);', ...
93 | 'label','Print Preview...');
94 |
95 | h2 = uimenu('parent', h_file, ...
96 | 'callback','printdlg(gcbf);', ...
97 | 'label','Print Figure ...');
98 |
99 | h2 = uimenu('parent', h_file, ...
100 | 'callback','rri_file_menu(''export_fig'');', ...
101 | 'label','Save Figure ...');
102 |
103 | arch = computer;
104 | if ~strcmpi(arch(1:2),'PC')
105 | set(h1, 'enable', 'off');
106 | end
107 |
108 | if ~no_close
109 | h1 = uimenu('parent', h_file, ...
110 | 'callback','close(gcbf);', ...
111 | 'separator','on', ...
112 | 'label','Close');
113 | end
114 |
115 | return; % init
116 |
117 |
118 | %------------------------------------------------
119 | %
120 | % Copy to clipboard
121 | %
122 | function copy_fig
123 |
124 | arch = computer;
125 | if(~strcmpi(arch(1:2),'PC'))
126 | error('copy to clipboard can only be used under MS Windows');
127 | return;
128 | end
129 |
130 | print -noui -dbitmap;
131 |
132 | return % copy_fig
133 |
134 |
135 | %------------------------------------------------
136 | %
137 | % Save as an image file
138 | %
139 | function export_fig
140 |
141 | curr = pwd;
142 | if isempty(curr)
143 | curr = filesep;
144 | end
145 |
146 | [selected_file, selected_path] = rri_select_file(curr,'Save As');
147 |
148 | if isempty(selected_file) | isempty(selected_path)
149 | return;
150 | end
151 |
152 | filename = [selected_path selected_file];
153 |
154 | if(exist(filename,'file')==2) % file exist
155 |
156 | dlg_title = 'Confirm File Overwrite';
157 | msg = ['File ',filename,' exist. Are you sure you want to overwrite it?'];
158 | response = questdlg(msg,dlg_title,'Yes','No','Yes');
159 |
160 | if(strcmp(response,'No'))
161 | return;
162 | end
163 |
164 | end
165 |
166 | old_pointer = get(gcbf,'pointer');
167 | set(gcbf,'pointer','watch');
168 |
169 | try
170 | saveas(gcbf,filename);
171 | catch
172 | msg = 'ERROR: Cannot save file';
173 | set(findobj(gcf,'Tag','MessageLine'),'String',msg);
174 | end
175 |
176 | set(gcbf,'pointer',old_pointer);
177 |
178 | return; % export_fig
179 |
180 |
--------------------------------------------------------------------------------
/NIFTI/rri_orient.m:
--------------------------------------------------------------------------------
1 | % Convert image of different orientations to standard Analyze orientation
2 | %
3 | % Usage: nii = rri_orient(nii);
4 |
5 | % Jimmy Shen (jimmy@rotman-baycrest.on.ca), 26-APR-04
6 | %___________________________________________________________________
7 |
8 | function [nii, orient, pattern] = rri_orient(nii, varargin)
9 |
10 | if nargin > 1
11 | pattern = varargin{1};
12 | else
13 | pattern = [];
14 | end
15 |
16 | orient = [1 2 3];
17 | dim = double(nii.hdr.dime.dim([2:4]));
18 |
19 | if ~isempty(pattern) & ~isequal(length(pattern), prod(dim))
20 | return;
21 | end
22 |
23 | % get orient of the current image
24 | %
25 | orient = rri_orient_ui;
26 | pause(.1);
27 |
28 | % no need for conversion
29 | %
30 | if isequal(orient, [1 2 3])
31 | return;
32 | end
33 |
34 | if isempty(pattern)
35 | pattern = 1:prod(dim);
36 | end
37 |
38 | pattern = reshape(pattern, dim);
39 | img = nii.img;
40 |
41 | % calculate after flip orient
42 | %
43 | rot_orient = mod(orient + 2, 3) + 1;
44 |
45 | % do flip:
46 | %
47 | flip_orient = orient - rot_orient;
48 |
49 | for i = 1:3
50 | if flip_orient(i)
51 | pattern = flipdim(pattern, i);
52 | img = flipdim(img, i);
53 | end
54 | end
55 |
56 | % get index of orient (do inverse)
57 | %
58 | [tmp rot_orient] = sort(rot_orient);
59 |
60 | % do rotation:
61 | %
62 | pattern = permute(pattern, rot_orient);
63 | img = permute(img, [rot_orient 4 5 6]);
64 |
65 | % rotate resolution, or 'dim'
66 | %
67 | new_dim = nii.hdr.dime.dim([2:4]);
68 | new_dim = new_dim(rot_orient);
69 | nii.hdr.dime.dim([2:4]) = new_dim;
70 |
71 | % rotate voxel_size, or 'pixdim'
72 | %
73 | tmp = nii.hdr.dime.pixdim([2:4]);
74 | tmp = tmp(rot_orient);
75 | nii.hdr.dime.pixdim([2:4]) = tmp;
76 |
77 | % re-calculate originator
78 | %
79 | tmp = nii.hdr.hist.originator([1:3]);
80 | tmp = tmp(rot_orient);
81 | flip_orient = flip_orient(rot_orient);
82 |
83 | for i = 1:3
84 | if flip_orient(i) & ~isequal(double(tmp(i)), 0)
85 | tmp(i) = int16(double(new_dim(i)) - double(tmp(i)) + 1);
86 | end
87 | end
88 |
89 | nii.hdr.hist.originator([1:3]) = tmp;
90 |
91 | nii.img = img;
92 | pattern = pattern(:);
93 |
94 | return; % rri_orient
95 |
96 |
--------------------------------------------------------------------------------
/NIFTI/rri_orient_ui.m:
--------------------------------------------------------------------------------
1 | % Return orientation of the current image:
2 | % orient is orientation 1x3 matrix, in that:
3 | % Three elements represent: [x y z]
4 | % Element value: 1 - Left to Right; 2 - Posterior to Anterior;
5 | % 3 - Inferior to Superior; 4 - Right to Left;
6 | % 5 - Anterior to Posterior; 6 - Superior to Inferior;
7 | % e.g.:
8 | % Standard RAS Orientation: [1 2 3]
9 | % Standard RHOS Orientation: [2 4 3]
10 |
11 | % Jimmy Shen (jimmy@rotman-baycrest.on.ca), 26-APR-04
12 | %
13 | function orient = rri_orient_ui(varargin)
14 |
15 | if nargin == 0
16 | init;
17 | orient_ui_fig = gcf;
18 | uiwait; % wait for user finish
19 |
20 | orient = getappdata(gcf, 'orient');
21 |
22 | if isempty(orient)
23 | orient = [1 2 3];
24 | end
25 |
26 | if ishandle(orient_ui_fig)
27 | close(gcf);
28 | end
29 |
30 | return;
31 | end
32 |
33 | action = varargin{1};
34 |
35 | if strcmp(action, 'done')
36 | click_done;
37 | elseif strcmp(action, 'cancel')
38 | uiresume;
39 | end
40 |
41 | return; % rri_orient_ui
42 |
43 |
44 | %----------------------------------------------------------------------
45 | function init
46 |
47 | save_setting_status = 'on';
48 | rri_orient_pos = [];
49 |
50 | try
51 | load('pls_profile');
52 | catch
53 | end
54 |
55 | try
56 | load('rri_pos_profile');
57 | catch
58 | end
59 |
60 | if ~isempty(rri_orient_pos) & strcmp(save_setting_status,'on')
61 |
62 | pos = rri_orient_pos;
63 |
64 | else
65 |
66 | w = 0.35;
67 | h = 0.4;
68 | x = (1-w)/2;
69 | y = (1-h)/2;
70 |
71 | pos = [x y w h];
72 |
73 | end
74 |
75 | handles.figure = figure('Color',[0.8 0.8 0.8], ...
76 | 'Units','normal', ...
77 | 'Name', 'Convert to standard RAS orientation', ...
78 | 'NumberTitle','off', ...
79 | 'MenuBar','none', ...
80 | 'Position',pos, ...
81 | 'WindowStyle', 'normal', ...
82 | 'ToolBar','none');
83 |
84 | h0 = handles.figure;
85 | Font.FontUnits = 'point';
86 | Font.FontSize = 12;
87 |
88 | margin = .1;
89 | line_num = 6;
90 | line_ht = (1 - margin*2) / line_num;
91 |
92 | x = margin;
93 | y = 1 - margin - line_ht;
94 | w = 1 - margin * 2;
95 | h = line_ht * .7;
96 |
97 | pos = [x y w h];
98 |
99 | handles.Ttit = uicontrol('parent', h0, ...
100 | 'style','text', ...
101 | 'unit', 'normal', ...
102 | Font, ...
103 | 'Position',pos, ...
104 | 'HorizontalAlignment','left',...
105 | 'background', [0.8 0.8 0.8], ...
106 | 'string', 'Please input orientation of the current image:');
107 |
108 | y = y - line_ht;
109 | w = .2;
110 |
111 | pos = [x y w h];
112 |
113 | handles.Tx_orient = uicontrol('parent', h0, ...
114 | 'style','text', ...
115 | 'unit', 'normal', ...
116 | Font, ...
117 | 'Position',pos, ...
118 | 'HorizontalAlignment','left',...
119 | 'background', [0.8 0.8 0.8], ...
120 | 'string', 'X Axes:');
121 |
122 | y = y - line_ht;
123 |
124 | pos = [x y w h];
125 |
126 | handles.Ty_orient = uicontrol('parent', h0, ...
127 | 'style','text', ...
128 | 'unit', 'normal', ...
129 | Font, ...
130 | 'Position',pos, ...
131 | 'HorizontalAlignment','left',...
132 | 'background', [0.8 0.8 0.8], ...
133 | 'string', 'Y Axes:');
134 |
135 | y = y - line_ht;
136 |
137 | pos = [x y w h];
138 |
139 | handles.Tz_orient = uicontrol('parent', h0, ...
140 | 'style','text', ...
141 | 'unit', 'normal', ...
142 | Font, ...
143 | 'Position',pos, ...
144 | 'HorizontalAlignment','left',...
145 | 'background', [0.8 0.8 0.8], ...
146 | 'string', 'Z Axes:');
147 |
148 | choice = { 'From Left to Right', 'From Posterior to Anterior', ...
149 | 'From Inferior to Superior', 'From Right to Left', ...
150 | 'From Anterior to Posterior', 'From Superior to Inferior' };
151 |
152 | y = 1 - margin - line_ht;
153 | y = y - line_ht;
154 | w = 1 - margin - x - w;
155 | x = 1 - margin - w;
156 |
157 | pos = [x y w h];
158 |
159 | handles.x_orient = uicontrol('parent', h0, ...
160 | 'style','popupmenu', ...
161 | 'unit', 'normal', ...
162 | Font, ...
163 | 'Position',pos, ...
164 | 'HorizontalAlignment','left',...
165 | 'string', choice, ...
166 | 'value', 1, ...
167 | 'background', [1 1 1]);
168 |
169 | y = y - line_ht;
170 |
171 | pos = [x y w h];
172 |
173 | handles.y_orient = uicontrol('parent', h0, ...
174 | 'style','popupmenu', ...
175 | 'unit', 'normal', ...
176 | Font, ...
177 | 'Position',pos, ...
178 | 'HorizontalAlignment','left',...
179 | 'string', choice, ...
180 | 'value', 2, ...
181 | 'background', [1 1 1]);
182 |
183 | y = y - line_ht;
184 |
185 | pos = [x y w h];
186 |
187 | handles.z_orient = uicontrol('parent', h0, ...
188 | 'style','popupmenu', ...
189 | 'unit', 'normal', ...
190 | Font, ...
191 | 'Position',pos, ...
192 | 'HorizontalAlignment','left',...
193 | 'string', choice, ...
194 | 'value', 3, ...
195 | 'background', [1 1 1]);
196 |
197 | x = margin;
198 | y = y - line_ht * 1.5;
199 | w = .3;
200 |
201 | pos = [x y w h];
202 |
203 | handles.done = uicontrol('parent', h0, ...
204 | 'unit', 'normal', ...
205 | Font, ...
206 | 'Position',pos, ...
207 | 'HorizontalAlignment','center',...
208 | 'callback', 'rri_orient_ui(''done'');', ...
209 | 'string', 'Done');
210 |
211 | x = 1 - margin - w;
212 |
213 | pos = [x y w h];
214 |
215 | handles.cancel = uicontrol('parent', h0, ...
216 | 'unit', 'normal', ...
217 | Font, ...
218 | 'Position',pos, ...
219 | 'HorizontalAlignment','center',...
220 | 'callback', 'rri_orient_ui(''cancel'');', ...
221 | 'string', 'Cancel');
222 |
223 | setappdata(h0, 'handles', handles);
224 | setappdata(h0, 'orient', [1 2 3]);
225 |
226 | return; % init
227 |
228 |
229 | %----------------------------------------------------------------------
230 | function click_done
231 |
232 | handles = getappdata(gcf, 'handles');
233 |
234 | x_orient = get(handles.x_orient, 'value');
235 | y_orient = get(handles.y_orient, 'value');
236 | z_orient = get(handles.z_orient, 'value');
237 |
238 | orient = [x_orient y_orient z_orient];
239 | test_orient = [orient, orient + 3];
240 | test_orient = mod(test_orient, 3);
241 |
242 | if length(unique(test_orient)) ~= 3
243 | msgbox('Please don''t choose same or opposite direction','Error','modal');
244 | return;
245 | end
246 |
247 | setappdata(gcf, 'orient', [x_orient y_orient z_orient]);
248 | uiresume;
249 |
250 | return; % click_done
251 |
252 |
--------------------------------------------------------------------------------
/NIFTI/rri_xhair.m:
--------------------------------------------------------------------------------
1 | % rri_xhair: create a pair of full_cross_hair at point [x y] in
2 | % axes h_ax, and return xhair struct
3 | %
4 | % Usage: xhair = rri_xhair([x y], xhair, h_ax);
5 | %
6 | % If omit xhair, rri_xhair will create a pair of xhair; otherwise,
7 | % rri_xhair will update the xhair. If omit h_ax, current axes will
8 | % be used.
9 | %
10 |
11 | % 24-nov-2003 jimmy (jimmy@rotman-baycrest.on.ca)
12 | %
13 | %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
14 |
15 | function xhair = rri_xhair(varargin)
16 |
17 | if nargin == 0
18 | error('Please enter a point position as first argument');
19 | return;
20 | end
21 |
22 | if nargin > 0
23 | p = varargin{1};
24 |
25 | if ~isnumeric(p) | length(p) ~= 2
26 | error('Invalid point position');
27 | return;
28 | else
29 | xhair = [];
30 | end
31 | end
32 |
33 | if nargin > 1
34 | xhair = varargin{2};
35 |
36 | if ~isempty(xhair)
37 | if ~isstruct(xhair)
38 | error('Invalid xhair struct');
39 | return;
40 | elseif ~isfield(xhair,'lx') | ~isfield(xhair,'ly')
41 | error('Invalid xhair struct');
42 | return;
43 | elseif ~ishandle(xhair.lx) | ~ishandle(xhair.ly)
44 | error('Invalid xhair struct');
45 | return;
46 | end
47 |
48 | lx = xhair.lx;
49 | ly = xhair.ly;
50 | else
51 | lx = [];
52 | ly = [];
53 | end
54 | end
55 |
56 | if nargin > 2
57 | h_ax = varargin{3};
58 |
59 | if ~ishandle(h_ax)
60 | error('Invalid axes handle');
61 | return;
62 | elseif ~strcmp(lower(get(h_ax,'type')), 'axes')
63 | error('Invalid axes handle');
64 | return;
65 | end
66 | else
67 | h_ax = gca;
68 | end
69 |
70 | x_range = get(h_ax,'xlim');
71 | y_range = get(h_ax,'ylim');
72 |
73 | if ~isempty(xhair)
74 | set(lx, 'ydata', [p(2) p(2)]);
75 | set(ly, 'xdata', [p(1) p(1)]);
76 | set(h_ax, 'selected', 'on');
77 | set(h_ax, 'selected', 'off');
78 | else
79 | figure(get(h_ax,'parent'));
80 | axes(h_ax);
81 |
82 | xhair.lx = line('xdata', x_range, 'ydata', [p(2) p(2)], ...
83 | 'zdata', [11 11], 'color', [1 0 0], 'hittest', 'off');
84 | xhair.ly = line('xdata', [p(1) p(1)], 'ydata', y_range, ...
85 | 'zdata', [11 11], 'color', [1 0 0], 'hittest', 'off');
86 | end
87 |
88 | set(h_ax,'xlim',x_range);
89 | set(h_ax,'ylim',y_range);
90 |
91 | return;
92 |
93 |
--------------------------------------------------------------------------------
/NIFTI/rri_zoom_menu.m:
--------------------------------------------------------------------------------
1 | % Imbed a zoom menu to any figure.
2 | %
3 | % Usage: rri_zoom_menu(fig);
4 | %
5 |
6 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
7 | %
8 | %--------------------------------------------------------------------
9 | function menu_hdl = rri_zoom_menu(fig)
10 |
11 | if isnumeric(fig)
12 | menu_hdl = uimenu('Parent',fig, ...
13 | 'Label','Zoom on', ...
14 | 'Userdata', 1, ...
15 | 'Callback','rri_zoom_menu(''zoom'');');
16 |
17 | return;
18 | end
19 |
20 | zoom_on_state = get(gcbo,'Userdata');
21 |
22 | if (zoom_on_state == 1)
23 | zoom on;
24 | set(gcbo,'Userdata',0,'Label','Zoom off');
25 | set(gcbf,'pointer','crosshair');
26 | else
27 | zoom off;
28 | set(gcbo,'Userdata',1,'Label','Zoom on');
29 | set(gcbf,'pointer','arrow');
30 | end
31 |
32 | return % rri_zoom_menu
33 |
34 |
--------------------------------------------------------------------------------
/NIFTI/save_nii.m:
--------------------------------------------------------------------------------
1 | % Save NIFTI dataset. Support both *.nii and *.hdr/*.img file extension.
2 | % If file extension is not provided, *.hdr/*.img will be used as default.
3 | %
4 | % Usage: save_nii(nii, filename, [old_RGB])
5 | %
6 | % nii.hdr - struct with NIFTI header fields (from load_nii.m or make_nii.m)
7 | %
8 | % nii.img - 3D (or 4D) matrix of NIFTI data.
9 | %
10 | % filename - NIFTI file name.
11 | %
12 | % old_RGB - an optional boolean variable to handle special RGB data
13 | % sequence [R1 R2 ... G1 G2 ... B1 B2 ...] that is used only by
14 | % AnalyzeDirect (Analyze Software). Since both NIfTI and Analyze
15 | % file format use RGB triple [R1 G1 B1 R2 G2 B2 ...] sequentially
16 | % for each voxel, this variable is set to FALSE by default. If you
17 | % would like the saved image only to be opened by AnalyzeDirect
18 | % Software, set old_RGB to TRUE (or 1). It will be set to 0, if it
19 | % is default or empty.
20 | %
21 | % Tip: to change the data type, set nii.hdr.dime.datatype,
22 | % and nii.hdr.dime.bitpix to:
23 | %
24 | % 0 None (Unknown bit per voxel) % DT_NONE, DT_UNKNOWN
25 | % 1 Binary (ubit1, bitpix=1) % DT_BINARY
26 | % 2 Unsigned char (uchar or uint8, bitpix=8) % DT_UINT8, NIFTI_TYPE_UINT8
27 | % 4 Signed short (int16, bitpix=16) % DT_INT16, NIFTI_TYPE_INT16
28 | % 8 Signed integer (int32, bitpix=32) % DT_INT32, NIFTI_TYPE_INT32
29 | % 16 Floating point (single or float32, bitpix=32) % DT_FLOAT32, NIFTI_TYPE_FLOAT32
30 | % 32 Complex, 2 float32 (Use float32, bitpix=64) % DT_COMPLEX64, NIFTI_TYPE_COMPLEX64
31 | % 64 Double precision (double or float64, bitpix=64) % DT_FLOAT64, NIFTI_TYPE_FLOAT64
32 | % 128 uint RGB (Use uint8, bitpix=24) % DT_RGB24, NIFTI_TYPE_RGB24
33 | % 256 Signed char (schar or int8, bitpix=8) % DT_INT8, NIFTI_TYPE_INT8
34 | % 511 Single RGB (Use float32, bitpix=96) % DT_RGB96, NIFTI_TYPE_RGB96
35 | % 512 Unsigned short (uint16, bitpix=16) % DT_UNINT16, NIFTI_TYPE_UNINT16
36 | % 768 Unsigned integer (uint32, bitpix=32) % DT_UNINT32, NIFTI_TYPE_UNINT32
37 | % 1024 Signed long long (int64, bitpix=64) % DT_INT64, NIFTI_TYPE_INT64
38 | % 1280 Unsigned long long (uint64, bitpix=64) % DT_UINT64, NIFTI_TYPE_UINT64
39 | % 1536 Long double, float128 (Unsupported, bitpix=128) % DT_FLOAT128, NIFTI_TYPE_FLOAT128
40 | % 1792 Complex128, 2 float64 (Use float64, bitpix=128) % DT_COMPLEX128, NIFTI_TYPE_COMPLEX128
41 | % 2048 Complex256, 2 float128 (Unsupported, bitpix=256) % DT_COMPLEX128, NIFTI_TYPE_COMPLEX128
42 | %
43 | % Part of this file is copied and modified from:
44 | % http://www.mathworks.com/matlabcentral/fileexchange/1878-mri-analyze-tools
45 | %
46 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
47 | %
48 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
49 | % - "old_RGB" related codes in "save_nii.m" are added by Mike Harms (2006.06.28)
50 | %
51 | function save_nii(nii, fileprefix, old_RGB)
52 |
53 | if ~exist('nii','var') | isempty(nii) | ~isfield(nii,'hdr') | ...
54 | ~isfield(nii,'img') | ~exist('fileprefix','var') | isempty(fileprefix)
55 |
56 | error('Usage: save_nii(nii, filename, [old_RGB])');
57 | end
58 |
59 | if isfield(nii,'untouch') & nii.untouch == 1
60 | error('Usage: please use ''save_untouch_nii.m'' for the untouched structure.');
61 | end
62 |
63 | if ~exist('old_RGB','var') | isempty(old_RGB)
64 | old_RGB = 0;
65 | end
66 |
67 | v = version;
68 |
69 | % Check file extension. If .gz, unpack it into temp folder
70 | %
71 | if length(fileprefix) > 2 & strcmp(fileprefix(end-2:end), '.gz')
72 |
73 | if ~strcmp(fileprefix(end-6:end), '.img.gz') & ...
74 | ~strcmp(fileprefix(end-6:end), '.hdr.gz') & ...
75 | ~strcmp(fileprefix(end-6:end), '.nii.gz')
76 |
77 | error('Please check filename.');
78 | end
79 |
80 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
81 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
82 | else
83 | gzFile = 1;
84 | fileprefix = fileprefix(1:end-3);
85 | end
86 | end
87 |
88 | filetype = 1;
89 |
90 | % Note: fileprefix is actually the filename you want to save
91 | %
92 | if findstr('.nii',fileprefix) & strcmp(fileprefix(end-3:end), '.nii')
93 | filetype = 2;
94 | fileprefix(end-3:end)='';
95 | end
96 |
97 | if findstr('.hdr',fileprefix) & strcmp(fileprefix(end-3:end), '.hdr')
98 | fileprefix(end-3:end)='';
99 | end
100 |
101 | if findstr('.img',fileprefix) & strcmp(fileprefix(end-3:end), '.img')
102 | fileprefix(end-3:end)='';
103 | end
104 |
105 | write_nii(nii, filetype, fileprefix, old_RGB);
106 |
107 | % gzip output file if requested
108 | %
109 | if exist('gzFile', 'var')
110 | if filetype == 1
111 | gzip([fileprefix, '.img']);
112 | delete([fileprefix, '.img']);
113 | gzip([fileprefix, '.hdr']);
114 | delete([fileprefix, '.hdr']);
115 | elseif filetype == 2
116 | gzip([fileprefix, '.nii']);
117 | delete([fileprefix, '.nii']);
118 | end;
119 | end;
120 |
121 | if filetype == 1
122 |
123 | % So earlier versions of SPM can also open it with correct originator
124 | %
125 | M=[[diag(nii.hdr.dime.pixdim(2:4)) -[nii.hdr.hist.originator(1:3).*nii.hdr.dime.pixdim(2:4)]'];[0 0 0 1]];
126 | save([fileprefix '.mat'], 'M');
127 | end
128 |
129 | return % save_nii
130 |
131 |
132 | %-----------------------------------------------------------------------------------
133 | function write_nii(nii, filetype, fileprefix, old_RGB)
134 |
135 | hdr = nii.hdr;
136 |
137 | if isfield(nii,'ext') & ~isempty(nii.ext)
138 | ext = nii.ext;
139 | [ext, esize_total] = verify_nii_ext(ext);
140 | else
141 | ext = [];
142 | end
143 |
144 | switch double(hdr.dime.datatype),
145 | case 1,
146 | hdr.dime.bitpix = int16(1 ); precision = 'ubit1';
147 | case 2,
148 | hdr.dime.bitpix = int16(8 ); precision = 'uint8';
149 | case 4,
150 | hdr.dime.bitpix = int16(16); precision = 'int16';
151 | case 8,
152 | hdr.dime.bitpix = int16(32); precision = 'int32';
153 | case 16,
154 | hdr.dime.bitpix = int16(32); precision = 'float32';
155 | case 32,
156 | hdr.dime.bitpix = int16(64); precision = 'float32';
157 | case 64,
158 | hdr.dime.bitpix = int16(64); precision = 'float64';
159 | case 128,
160 | hdr.dime.bitpix = int16(24); precision = 'uint8';
161 | case 256
162 | hdr.dime.bitpix = int16(8 ); precision = 'int8';
163 | case 511,
164 | hdr.dime.bitpix = int16(96); precision = 'float32';
165 | case 512
166 | hdr.dime.bitpix = int16(16); precision = 'uint16';
167 | case 768
168 | hdr.dime.bitpix = int16(32); precision = 'uint32';
169 | case 1024
170 | hdr.dime.bitpix = int16(64); precision = 'int64';
171 | case 1280
172 | hdr.dime.bitpix = int16(64); precision = 'uint64';
173 | case 1792,
174 | hdr.dime.bitpix = int16(128); precision = 'float64';
175 | otherwise
176 | error('This datatype is not supported');
177 | end
178 |
179 | hdr.dime.glmax = round(double(max(nii.img(:))));
180 | hdr.dime.glmin = round(double(min(nii.img(:))));
181 |
182 | if filetype == 2
183 | fid = fopen(sprintf('%s.nii',fileprefix),'w');
184 |
185 | if fid < 0,
186 | msg = sprintf('Cannot open file %s.nii.',fileprefix);
187 | error(msg);
188 | end
189 |
190 | hdr.dime.vox_offset = 352;
191 |
192 | if ~isempty(ext)
193 | hdr.dime.vox_offset = hdr.dime.vox_offset + esize_total;
194 | end
195 |
196 | hdr.hist.magic = 'n+1';
197 | save_nii_hdr(hdr, fid);
198 |
199 | if ~isempty(ext)
200 | save_nii_ext(ext, fid);
201 | end
202 | else
203 | fid = fopen(sprintf('%s.hdr',fileprefix),'w');
204 |
205 | if fid < 0,
206 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
207 | error(msg);
208 | end
209 |
210 | hdr.dime.vox_offset = 0;
211 | hdr.hist.magic = 'ni1';
212 | save_nii_hdr(hdr, fid);
213 |
214 | if ~isempty(ext)
215 | save_nii_ext(ext, fid);
216 | end
217 |
218 | fclose(fid);
219 | fid = fopen(sprintf('%s.img',fileprefix),'w');
220 | end
221 |
222 | ScanDim = double(hdr.dime.dim(5)); % t
223 | SliceDim = double(hdr.dime.dim(4)); % z
224 | RowDim = double(hdr.dime.dim(3)); % y
225 | PixelDim = double(hdr.dime.dim(2)); % x
226 | SliceSz = double(hdr.dime.pixdim(4));
227 | RowSz = double(hdr.dime.pixdim(3));
228 | PixelSz = double(hdr.dime.pixdim(2));
229 |
230 | x = 1:PixelDim;
231 |
232 | if filetype == 2 & isempty(ext)
233 | skip_bytes = double(hdr.dime.vox_offset) - 348;
234 | else
235 | skip_bytes = 0;
236 | end
237 |
238 | if double(hdr.dime.datatype) == 128
239 |
240 | % RGB planes are expected to be in the 4th dimension of nii.img
241 | %
242 | if(size(nii.img,4)~=3)
243 | error(['The NII structure does not appear to have 3 RGB color planes in the 4th dimension']);
244 | end
245 |
246 | if old_RGB
247 | nii.img = permute(nii.img, [1 2 4 3 5 6 7 8]);
248 | else
249 | nii.img = permute(nii.img, [4 1 2 3 5 6 7 8]);
250 | end
251 | end
252 |
253 | if double(hdr.dime.datatype) == 511
254 |
255 | % RGB planes are expected to be in the 4th dimension of nii.img
256 | %
257 | if(size(nii.img,4)~=3)
258 | error(['The NII structure does not appear to have 3 RGB color planes in the 4th dimension']);
259 | end
260 |
261 | if old_RGB
262 | nii.img = permute(nii.img, [1 2 4 3 5 6 7 8]);
263 | else
264 | nii.img = permute(nii.img, [4 1 2 3 5 6 7 8]);
265 | end
266 | end
267 |
268 | % For complex float32 or complex float64, voxel values
269 | % include [real, imag]
270 | %
271 | if hdr.dime.datatype == 32 | hdr.dime.datatype == 1792
272 | real_img = real(nii.img(:))';
273 | nii.img = imag(nii.img(:))';
274 | nii.img = [real_img; nii.img];
275 | end
276 |
277 | if skip_bytes
278 | fwrite(fid, zeros(1,skip_bytes), 'uint8');
279 | end
280 |
281 | fwrite(fid, nii.img, precision);
282 | % fwrite(fid, nii.img, precision, skip_bytes); % error using skip
283 | fclose(fid);
284 |
285 | return; % write_nii
286 |
287 |
--------------------------------------------------------------------------------
/NIFTI/save_nii_ext.m:
--------------------------------------------------------------------------------
1 | % Save NIFTI header extension.
2 | %
3 | % Usage: save_nii_ext(ext, fid)
4 | %
5 | % ext - struct with NIFTI header extension fields.
6 | %
7 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
8 | %
9 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
10 | %
11 | function save_nii_ext(ext, fid)
12 |
13 | if ~exist('ext','var') | ~exist('fid','var')
14 | error('Usage: save_nii_ext(ext, fid)');
15 | end
16 |
17 | if ~isfield(ext,'extension') | ~isfield(ext,'section') | ~isfield(ext,'num_ext')
18 | error('Wrong header extension');
19 | end
20 |
21 | write_ext(ext, fid);
22 |
23 | return; % save_nii_ext
24 |
25 |
26 | %---------------------------------------------------------------------
27 | function write_ext(ext, fid)
28 |
29 | fwrite(fid, ext.extension, 'uchar');
30 |
31 | for i=1:ext.num_ext
32 | fwrite(fid, ext.section(i).esize, 'int32');
33 | fwrite(fid, ext.section(i).ecode, 'int32');
34 | fwrite(fid, ext.section(i).edata, 'uchar');
35 | end
36 |
37 | return; % write_ext
38 |
39 |
--------------------------------------------------------------------------------
/NIFTI/save_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % internal function
2 |
3 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
4 |
5 | function save_nii_hdr(hdr, fid)
6 |
7 | if ~exist('hdr','var') | ~exist('fid','var')
8 | error('Usage: save_nii_hdr(hdr, fid)');
9 | end
10 |
11 | if ~isequal(hdr.hk.sizeof_hdr,348),
12 | error('hdr.hk.sizeof_hdr must be 348.');
13 | end
14 |
15 | if hdr.hist.qform_code == 0 & hdr.hist.sform_code == 0
16 | hdr.hist.sform_code = 1;
17 | hdr.hist.srow_x(1) = hdr.dime.pixdim(2);
18 | hdr.hist.srow_x(2) = 0;
19 | hdr.hist.srow_x(3) = 0;
20 | hdr.hist.srow_y(1) = 0;
21 | hdr.hist.srow_y(2) = hdr.dime.pixdim(3);
22 | hdr.hist.srow_y(3) = 0;
23 | hdr.hist.srow_z(1) = 0;
24 | hdr.hist.srow_z(2) = 0;
25 | hdr.hist.srow_z(3) = hdr.dime.pixdim(4);
26 | hdr.hist.srow_x(4) = (1-hdr.hist.originator(1))*hdr.dime.pixdim(2);
27 | hdr.hist.srow_y(4) = (1-hdr.hist.originator(2))*hdr.dime.pixdim(3);
28 | hdr.hist.srow_z(4) = (1-hdr.hist.originator(3))*hdr.dime.pixdim(4);
29 | end
30 |
31 | write_header(hdr, fid);
32 |
33 | return; % save_nii_hdr
34 |
35 |
36 | %---------------------------------------------------------------------
37 | function write_header(hdr, fid)
38 |
39 | % Original header structures
40 | % struct dsr /* dsr = hdr */
41 | % {
42 | % struct header_key hk; /* 0 + 40 */
43 | % struct image_dimension dime; /* 40 + 108 */
44 | % struct data_history hist; /* 148 + 200 */
45 | % }; /* total= 348 bytes*/
46 |
47 | header_key(fid, hdr.hk);
48 | image_dimension(fid, hdr.dime);
49 | data_history(fid, hdr.hist);
50 |
51 | % check the file size is 348 bytes
52 | %
53 | fbytes = ftell(fid);
54 |
55 | if ~isequal(fbytes,348),
56 | msg = sprintf('Header size is not 348 bytes.');
57 | warning(msg);
58 | end
59 |
60 | return; % write_header
61 |
62 |
63 | %---------------------------------------------------------------------
64 | function header_key(fid, hk)
65 |
66 | fseek(fid,0,'bof');
67 |
68 | % Original header structures
69 | % struct header_key /* header key */
70 | % { /* off + size */
71 | % int sizeof_hdr /* 0 + 4 */
72 | % char data_type[10]; /* 4 + 10 */
73 | % char db_name[18]; /* 14 + 18 */
74 | % int extents; /* 32 + 4 */
75 | % short int session_error; /* 36 + 2 */
76 | % char regular; /* 38 + 1 */
77 | % char dim_info; % char hkey_un0; /* 39 + 1 */
78 | % }; /* total=40 bytes */
79 |
80 | fwrite(fid, hk.sizeof_hdr(1), 'int32'); % must be 348.
81 |
82 | % data_type = sprintf('%-10s',hk.data_type); % ensure it is 10 chars from left
83 | % fwrite(fid, data_type(1:10), 'uchar');
84 | pad = zeros(1, 10-length(hk.data_type));
85 | hk.data_type = [hk.data_type char(pad)];
86 | fwrite(fid, hk.data_type(1:10), 'uchar');
87 |
88 | % db_name = sprintf('%-18s', hk.db_name); % ensure it is 18 chars from left
89 | % fwrite(fid, db_name(1:18), 'uchar');
90 | pad = zeros(1, 18-length(hk.db_name));
91 | hk.db_name = [hk.db_name char(pad)];
92 | fwrite(fid, hk.db_name(1:18), 'uchar');
93 |
94 | fwrite(fid, hk.extents(1), 'int32');
95 | fwrite(fid, hk.session_error(1), 'int16');
96 | fwrite(fid, hk.regular(1), 'uchar'); % might be uint8
97 |
98 | % fwrite(fid, hk.hkey_un0(1), 'uchar');
99 | % fwrite(fid, hk.hkey_un0(1), 'uint8');
100 | fwrite(fid, hk.dim_info(1), 'uchar');
101 |
102 | return; % header_key
103 |
104 |
105 | %---------------------------------------------------------------------
106 | function image_dimension(fid, dime)
107 |
108 | % Original header structures
109 | % struct image_dimension
110 | % { /* off + size */
111 | % short int dim[8]; /* 0 + 16 */
112 | % float intent_p1; % char vox_units[4]; /* 16 + 4 */
113 | % float intent_p2; % char cal_units[8]; /* 20 + 4 */
114 | % float intent_p3; % char cal_units[8]; /* 24 + 4 */
115 | % short int intent_code; % short int unused1; /* 28 + 2 */
116 | % short int datatype; /* 30 + 2 */
117 | % short int bitpix; /* 32 + 2 */
118 | % short int slice_start; % short int dim_un0; /* 34 + 2 */
119 | % float pixdim[8]; /* 36 + 32 */
120 | % /*
121 | % pixdim[] specifies the voxel dimensions:
122 | % pixdim[1] - voxel width
123 | % pixdim[2] - voxel height
124 | % pixdim[3] - interslice distance
125 | % pixdim[4] - volume timing, in msec
126 | % ..etc
127 | % */
128 | % float vox_offset; /* 68 + 4 */
129 | % float scl_slope; % float roi_scale; /* 72 + 4 */
130 | % float scl_inter; % float funused1; /* 76 + 4 */
131 | % short slice_end; % float funused2; /* 80 + 2 */
132 | % char slice_code; % float funused2; /* 82 + 1 */
133 | % char xyzt_units; % float funused2; /* 83 + 1 */
134 | % float cal_max; /* 84 + 4 */
135 | % float cal_min; /* 88 + 4 */
136 | % float slice_duration; % int compressed; /* 92 + 4 */
137 | % float toffset; % int verified; /* 96 + 4 */
138 | % int glmax; /* 100 + 4 */
139 | % int glmin; /* 104 + 4 */
140 | % }; /* total=108 bytes */
141 |
142 | fwrite(fid, dime.dim(1:8), 'int16');
143 | fwrite(fid, dime.intent_p1(1), 'float32');
144 | fwrite(fid, dime.intent_p2(1), 'float32');
145 | fwrite(fid, dime.intent_p3(1), 'float32');
146 | fwrite(fid, dime.intent_code(1), 'int16');
147 | fwrite(fid, dime.datatype(1), 'int16');
148 | fwrite(fid, dime.bitpix(1), 'int16');
149 | fwrite(fid, dime.slice_start(1), 'int16');
150 | fwrite(fid, dime.pixdim(1:8), 'float32');
151 | fwrite(fid, dime.vox_offset(1), 'float32');
152 | fwrite(fid, dime.scl_slope(1), 'float32');
153 | fwrite(fid, dime.scl_inter(1), 'float32');
154 | fwrite(fid, dime.slice_end(1), 'int16');
155 | fwrite(fid, dime.slice_code(1), 'uchar');
156 | fwrite(fid, dime.xyzt_units(1), 'uchar');
157 | fwrite(fid, dime.cal_max(1), 'float32');
158 | fwrite(fid, dime.cal_min(1), 'float32');
159 | fwrite(fid, dime.slice_duration(1), 'float32');
160 | fwrite(fid, dime.toffset(1), 'float32');
161 | fwrite(fid, dime.glmax(1), 'int32');
162 | fwrite(fid, dime.glmin(1), 'int32');
163 |
164 | return; % image_dimension
165 |
166 |
167 | %---------------------------------------------------------------------
168 | function data_history(fid, hist)
169 |
170 | % Original header structures
171 | %struct data_history
172 | % { /* off + size */
173 | % char descrip[80]; /* 0 + 80 */
174 | % char aux_file[24]; /* 80 + 24 */
175 | % short int qform_code; /* 104 + 2 */
176 | % short int sform_code; /* 106 + 2 */
177 | % float quatern_b; /* 108 + 4 */
178 | % float quatern_c; /* 112 + 4 */
179 | % float quatern_d; /* 116 + 4 */
180 | % float qoffset_x; /* 120 + 4 */
181 | % float qoffset_y; /* 124 + 4 */
182 | % float qoffset_z; /* 128 + 4 */
183 | % float srow_x[4]; /* 132 + 16 */
184 | % float srow_y[4]; /* 148 + 16 */
185 | % float srow_z[4]; /* 164 + 16 */
186 | % char intent_name[16]; /* 180 + 16 */
187 | % char magic[4]; % int smin; /* 196 + 4 */
188 | % }; /* total=200 bytes */
189 |
190 | % descrip = sprintf('%-80s', hist.descrip); % 80 chars from left
191 | % fwrite(fid, descrip(1:80), 'uchar');
192 | pad = zeros(1, 80-length(hist.descrip));
193 | hist.descrip = [hist.descrip char(pad)];
194 | fwrite(fid, hist.descrip(1:80), 'uchar');
195 |
196 | % aux_file = sprintf('%-24s', hist.aux_file); % 24 chars from left
197 | % fwrite(fid, aux_file(1:24), 'uchar');
198 | pad = zeros(1, 24-length(hist.aux_file));
199 | hist.aux_file = [hist.aux_file char(pad)];
200 | fwrite(fid, hist.aux_file(1:24), 'uchar');
201 |
202 | fwrite(fid, hist.qform_code, 'int16');
203 | fwrite(fid, hist.sform_code, 'int16');
204 | fwrite(fid, hist.quatern_b, 'float32');
205 | fwrite(fid, hist.quatern_c, 'float32');
206 | fwrite(fid, hist.quatern_d, 'float32');
207 | fwrite(fid, hist.qoffset_x, 'float32');
208 | fwrite(fid, hist.qoffset_y, 'float32');
209 | fwrite(fid, hist.qoffset_z, 'float32');
210 | fwrite(fid, hist.srow_x(1:4), 'float32');
211 | fwrite(fid, hist.srow_y(1:4), 'float32');
212 | fwrite(fid, hist.srow_z(1:4), 'float32');
213 |
214 | % intent_name = sprintf('%-16s', hist.intent_name); % 16 chars from left
215 | % fwrite(fid, intent_name(1:16), 'uchar');
216 | pad = zeros(1, 16-length(hist.intent_name));
217 | hist.intent_name = [hist.intent_name char(pad)];
218 | fwrite(fid, hist.intent_name(1:16), 'uchar');
219 |
220 | % magic = sprintf('%-4s', hist.magic); % 4 chars from left
221 | % fwrite(fid, magic(1:4), 'uchar');
222 | pad = zeros(1, 4-length(hist.magic));
223 | hist.magic = [hist.magic char(pad)];
224 | fwrite(fid, hist.magic(1:4), 'uchar');
225 |
226 | return; % data_history
227 |
228 |
--------------------------------------------------------------------------------
/NIFTI/save_untouch0_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % internal function
2 |
3 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
4 |
5 | function save_nii_hdr(hdr, fid)
6 |
7 | if ~isequal(hdr.hk.sizeof_hdr,348),
8 | error('hdr.hk.sizeof_hdr must be 348.');
9 | end
10 |
11 | write_header(hdr, fid);
12 |
13 | return; % save_nii_hdr
14 |
15 |
16 | %---------------------------------------------------------------------
17 | function write_header(hdr, fid)
18 |
19 | % Original header structures
20 | % struct dsr /* dsr = hdr */
21 | % {
22 | % struct header_key hk; /* 0 + 40 */
23 | % struct image_dimension dime; /* 40 + 108 */
24 | % struct data_history hist; /* 148 + 200 */
25 | % }; /* total= 348 bytes*/
26 |
27 | header_key(fid, hdr.hk);
28 | image_dimension(fid, hdr.dime);
29 | data_history(fid, hdr.hist);
30 |
31 | % check the file size is 348 bytes
32 | %
33 | fbytes = ftell(fid);
34 |
35 | if ~isequal(fbytes,348),
36 | msg = sprintf('Header size is not 348 bytes.');
37 | warning(msg);
38 | end
39 |
40 | return; % write_header
41 |
42 |
43 | %---------------------------------------------------------------------
44 | function header_key(fid, hk)
45 |
46 | fseek(fid,0,'bof');
47 |
48 | % Original header structures
49 | % struct header_key /* header key */
50 | % { /* off + size */
51 | % int sizeof_hdr /* 0 + 4 */
52 | % char data_type[10]; /* 4 + 10 */
53 | % char db_name[18]; /* 14 + 18 */
54 | % int extents; /* 32 + 4 */
55 | % short int session_error; /* 36 + 2 */
56 | % char regular; /* 38 + 1 */
57 | % char hkey_un0; /* 39 + 1 */
58 | % }; /* total=40 bytes */
59 |
60 | fwrite(fid, hk.sizeof_hdr(1), 'int32'); % must be 348.
61 |
62 | % data_type = sprintf('%-10s',hk.data_type); % ensure it is 10 chars from left
63 | % fwrite(fid, data_type(1:10), 'uchar');
64 | pad = zeros(1, 10-length(hk.data_type));
65 | hk.data_type = [hk.data_type char(pad)];
66 | fwrite(fid, hk.data_type(1:10), 'uchar');
67 |
68 | % db_name = sprintf('%-18s', hk.db_name); % ensure it is 18 chars from left
69 | % fwrite(fid, db_name(1:18), 'uchar');
70 | pad = zeros(1, 18-length(hk.db_name));
71 | hk.db_name = [hk.db_name char(pad)];
72 | fwrite(fid, hk.db_name(1:18), 'uchar');
73 |
74 | fwrite(fid, hk.extents(1), 'int32');
75 | fwrite(fid, hk.session_error(1), 'int16');
76 | fwrite(fid, hk.regular(1), 'uchar');
77 |
78 | fwrite(fid, hk.hkey_un0(1), 'uchar');
79 |
80 | return; % header_key
81 |
82 |
83 | %---------------------------------------------------------------------
84 | function image_dimension(fid, dime)
85 |
86 | %struct image_dimension
87 | % { /* off + size */
88 | % short int dim[8]; /* 0 + 16 */
89 | % char vox_units[4]; /* 16 + 4 */
90 | % char cal_units[8]; /* 20 + 8 */
91 | % short int unused1; /* 28 + 2 */
92 | % short int datatype; /* 30 + 2 */
93 | % short int bitpix; /* 32 + 2 */
94 | % short int dim_un0; /* 34 + 2 */
95 | % float pixdim[8]; /* 36 + 32 */
96 | % /*
97 | % pixdim[] specifies the voxel dimensions:
98 | % pixdim[1] - voxel width
99 | % pixdim[2] - voxel height
100 | % pixdim[3] - interslice distance
101 | % ..etc
102 | % */
103 | % float vox_offset; /* 68 + 4 */
104 | % float roi_scale; /* 72 + 4 */
105 | % float funused1; /* 76 + 4 */
106 | % float funused2; /* 80 + 4 */
107 | % float cal_max; /* 84 + 4 */
108 | % float cal_min; /* 88 + 4 */
109 | % int compressed; /* 92 + 4 */
110 | % int verified; /* 96 + 4 */
111 | % int glmax; /* 100 + 4 */
112 | % int glmin; /* 104 + 4 */
113 | % }; /* total=108 bytes */
114 |
115 | fwrite(fid, dime.dim(1:8), 'int16');
116 |
117 | pad = zeros(1, 4-length(dime.vox_units));
118 | dime.vox_units = [dime.vox_units char(pad)];
119 | fwrite(fid, dime.vox_units(1:4), 'uchar');
120 |
121 | pad = zeros(1, 8-length(dime.cal_units));
122 | dime.cal_units = [dime.cal_units char(pad)];
123 | fwrite(fid, dime.cal_units(1:8), 'uchar');
124 |
125 | fwrite(fid, dime.unused1(1), 'int16');
126 | fwrite(fid, dime.datatype(1), 'int16');
127 | fwrite(fid, dime.bitpix(1), 'int16');
128 | fwrite(fid, dime.dim_un0(1), 'int16');
129 | fwrite(fid, dime.pixdim(1:8), 'float32');
130 | fwrite(fid, dime.vox_offset(1), 'float32');
131 | fwrite(fid, dime.roi_scale(1), 'float32');
132 | fwrite(fid, dime.funused1(1), 'float32');
133 | fwrite(fid, dime.funused2(1), 'float32');
134 | fwrite(fid, dime.cal_max(1), 'float32');
135 | fwrite(fid, dime.cal_min(1), 'float32');
136 | fwrite(fid, dime.compressed(1), 'int32');
137 | fwrite(fid, dime.verified(1), 'int32');
138 | fwrite(fid, dime.glmax(1), 'int32');
139 | fwrite(fid, dime.glmin(1), 'int32');
140 |
141 | return; % image_dimension
142 |
143 |
144 | %---------------------------------------------------------------------
145 | function data_history(fid, hist)
146 |
147 | % Original header structures - ANALYZE 7.5
148 | %struct data_history
149 | % { /* off + size */
150 | % char descrip[80]; /* 0 + 80 */
151 | % char aux_file[24]; /* 80 + 24 */
152 | % char orient; /* 104 + 1 */
153 | % char originator[10]; /* 105 + 10 */
154 | % char generated[10]; /* 115 + 10 */
155 | % char scannum[10]; /* 125 + 10 */
156 | % char patient_id[10]; /* 135 + 10 */
157 | % char exp_date[10]; /* 145 + 10 */
158 | % char exp_time[10]; /* 155 + 10 */
159 | % char hist_un0[3]; /* 165 + 3 */
160 | % int views /* 168 + 4 */
161 | % int vols_added; /* 172 + 4 */
162 | % int start_field; /* 176 + 4 */
163 | % int field_skip; /* 180 + 4 */
164 | % int omax; /* 184 + 4 */
165 | % int omin; /* 188 + 4 */
166 | % int smax; /* 192 + 4 */
167 | % int smin; /* 196 + 4 */
168 | % }; /* total=200 bytes */
169 |
170 | % descrip = sprintf('%-80s', hist.descrip); % 80 chars from left
171 | % fwrite(fid, descrip(1:80), 'uchar');
172 | pad = zeros(1, 80-length(hist.descrip));
173 | hist.descrip = [hist.descrip char(pad)];
174 | fwrite(fid, hist.descrip(1:80), 'uchar');
175 |
176 | % aux_file = sprintf('%-24s', hist.aux_file); % 24 chars from left
177 | % fwrite(fid, aux_file(1:24), 'uchar');
178 | pad = zeros(1, 24-length(hist.aux_file));
179 | hist.aux_file = [hist.aux_file char(pad)];
180 | fwrite(fid, hist.aux_file(1:24), 'uchar');
181 |
182 | fwrite(fid, hist.orient(1), 'uchar');
183 | fwrite(fid, hist.originator(1:5), 'int16');
184 |
185 | pad = zeros(1, 10-length(hist.generated));
186 | hist.generated = [hist.generated char(pad)];
187 | fwrite(fid, hist.generated(1:10), 'uchar');
188 |
189 | pad = zeros(1, 10-length(hist.scannum));
190 | hist.scannum = [hist.scannum char(pad)];
191 | fwrite(fid, hist.scannum(1:10), 'uchar');
192 |
193 | pad = zeros(1, 10-length(hist.patient_id));
194 | hist.patient_id = [hist.patient_id char(pad)];
195 | fwrite(fid, hist.patient_id(1:10), 'uchar');
196 |
197 | pad = zeros(1, 10-length(hist.exp_date));
198 | hist.exp_date = [hist.exp_date char(pad)];
199 | fwrite(fid, hist.exp_date(1:10), 'uchar');
200 |
201 | pad = zeros(1, 10-length(hist.exp_time));
202 | hist.exp_time = [hist.exp_time char(pad)];
203 | fwrite(fid, hist.exp_time(1:10), 'uchar');
204 |
205 | pad = zeros(1, 3-length(hist.hist_un0));
206 | hist.hist_un0 = [hist.hist_un0 char(pad)];
207 | fwrite(fid, hist.hist_un0(1:3), 'uchar');
208 |
209 | fwrite(fid, hist.views(1), 'int32');
210 | fwrite(fid, hist.vols_added(1), 'int32');
211 | fwrite(fid, hist.start_field(1),'int32');
212 | fwrite(fid, hist.field_skip(1), 'int32');
213 | fwrite(fid, hist.omax(1), 'int32');
214 | fwrite(fid, hist.omin(1), 'int32');
215 | fwrite(fid, hist.smax(1), 'int32');
216 | fwrite(fid, hist.smin(1), 'int32');
217 |
218 | return; % data_history
219 |
220 |
--------------------------------------------------------------------------------
/NIFTI/save_untouch_nii.m:
--------------------------------------------------------------------------------
1 | % Save NIFTI or ANALYZE dataset that is loaded by "load_untouch_nii.m".
2 | % The output image format and file extension will be the same as the
3 | % input one (NIFTI.nii, NIFTI.img or ANALYZE.img). Therefore, any file
4 | % extension that you specified will be ignored.
5 | %
6 | % Usage: save_untouch_nii(nii, filename)
7 | %
8 | % nii - nii structure that is loaded by "load_untouch_nii.m"
9 | %
10 | % filename - NIFTI or ANALYZE file name.
11 | %
12 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
13 | %
14 | function save_untouch_nii(nii, filename)
15 |
16 | if ~exist('nii','var') | isempty(nii) | ~isfield(nii,'hdr') | ...
17 | ~isfield(nii,'img') | ~exist('filename','var') | isempty(filename)
18 |
19 | error('Usage: save_untouch_nii(nii, filename)');
20 | end
21 |
22 | if ~isfield(nii,'untouch') | nii.untouch == 0
23 | error('Usage: please use ''save_nii.m'' for the modified structure.');
24 | end
25 |
26 | if isfield(nii.hdr.hist,'magic') & strcmp(nii.hdr.hist.magic(1:3),'ni1')
27 | filetype = 1;
28 | elseif isfield(nii.hdr.hist,'magic') & strcmp(nii.hdr.hist.magic(1:3),'n+1')
29 | filetype = 2;
30 | else
31 | filetype = 0;
32 | end
33 |
34 | v = version;
35 |
36 | % Check file extension. If .gz, unpack it into temp folder
37 | %
38 | if length(filename) > 2 & strcmp(filename(end-2:end), '.gz')
39 |
40 | if ~strcmp(filename(end-6:end), '.img.gz') & ...
41 | ~strcmp(filename(end-6:end), '.hdr.gz') & ...
42 | ~strcmp(filename(end-6:end), '.nii.gz')
43 |
44 | error('Please check filename.');
45 | end
46 |
47 | if str2num(v(1:3)) < 7.1 | ~usejava('jvm')
48 | error('Please use MATLAB 7.1 (with java) and above, or run gunzip outside MATLAB.');
49 | else
50 | gzFile = 1;
51 | filename = filename(1:end-3);
52 | end
53 | end
54 |
55 | [p,f] = fileparts(filename);
56 | fileprefix = fullfile(p, f);
57 |
58 | write_nii(nii, filetype, fileprefix);
59 |
60 | % gzip output file if requested
61 | %
62 | if exist('gzFile', 'var')
63 | if filetype == 1
64 | gzip([fileprefix, '.img']);
65 | delete([fileprefix, '.img']);
66 | gzip([fileprefix, '.hdr']);
67 | delete([fileprefix, '.hdr']);
68 | elseif filetype == 2
69 | gzip([fileprefix, '.nii']);
70 | delete([fileprefix, '.nii']);
71 | end;
72 | end;
73 |
74 | % % So earlier versions of SPM can also open it with correct originator
75 | % %
76 | % if filetype == 0
77 | % M=[[diag(nii.hdr.dime.pixdim(2:4)) -[nii.hdr.hist.originator(1:3).*nii.hdr.dime.pixdim(2:4)]'];[0 0 0 1]];
78 | % save(fileprefix, 'M');
79 | % elseif filetype == 1
80 | % M=[];
81 | % save(fileprefix, 'M');
82 | %end
83 |
84 | return % save_untouch_nii
85 |
86 |
87 | %-----------------------------------------------------------------------------------
88 | function write_nii(nii, filetype, fileprefix)
89 |
90 | hdr = nii.hdr;
91 |
92 | if isfield(nii,'ext') & ~isempty(nii.ext)
93 | ext = nii.ext;
94 | [ext, esize_total] = verify_nii_ext(ext);
95 | else
96 | ext = [];
97 | end
98 |
99 | switch double(hdr.dime.datatype),
100 | case 1,
101 | hdr.dime.bitpix = int16(1 ); precision = 'ubit1';
102 | case 2,
103 | hdr.dime.bitpix = int16(8 ); precision = 'uint8';
104 | case 4,
105 | hdr.dime.bitpix = int16(16); precision = 'int16';
106 | case 8,
107 | hdr.dime.bitpix = int16(32); precision = 'int32';
108 | case 16,
109 | hdr.dime.bitpix = int16(32); precision = 'float32';
110 | case 32,
111 | hdr.dime.bitpix = int16(64); precision = 'float32';
112 | case 64,
113 | hdr.dime.bitpix = int16(64); precision = 'float64';
114 | case 128,
115 | hdr.dime.bitpix = int16(24); precision = 'uint8';
116 | case 256
117 | hdr.dime.bitpix = int16(8 ); precision = 'int8';
118 | case 512
119 | hdr.dime.bitpix = int16(16); precision = 'uint16';
120 | case 768
121 | hdr.dime.bitpix = int16(32); precision = 'uint32';
122 | case 1024
123 | hdr.dime.bitpix = int16(64); precision = 'int64';
124 | case 1280
125 | hdr.dime.bitpix = int16(64); precision = 'uint64';
126 | case 1792,
127 | hdr.dime.bitpix = int16(128); precision = 'float64';
128 | otherwise
129 | error('This datatype is not supported');
130 | end
131 |
132 | % hdr.dime.glmax = round(double(max(nii.img(:))));
133 | % hdr.dime.glmin = round(double(min(nii.img(:))));
134 |
135 | if filetype == 2
136 | fid = fopen(sprintf('%s.nii',fileprefix),'w');
137 |
138 | if fid < 0,
139 | msg = sprintf('Cannot open file %s.nii.',fileprefix);
140 | error(msg);
141 | end
142 |
143 | hdr.dime.vox_offset = 352;
144 |
145 | if ~isempty(ext)
146 | hdr.dime.vox_offset = hdr.dime.vox_offset + esize_total;
147 | end
148 |
149 | hdr.hist.magic = 'n+1';
150 | save_untouch_nii_hdr(hdr, fid);
151 |
152 | if ~isempty(ext)
153 | save_nii_ext(ext, fid);
154 | end
155 | elseif filetype == 1
156 | fid = fopen(sprintf('%s.hdr',fileprefix),'w');
157 |
158 | if fid < 0,
159 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
160 | error(msg);
161 | end
162 |
163 | hdr.dime.vox_offset = 0;
164 | hdr.hist.magic = 'ni1';
165 | save_untouch_nii_hdr(hdr, fid);
166 |
167 | if ~isempty(ext)
168 | save_nii_ext(ext, fid);
169 | end
170 |
171 | fclose(fid);
172 | fid = fopen(sprintf('%s.img',fileprefix),'w');
173 | else
174 | fid = fopen(sprintf('%s.hdr',fileprefix),'w');
175 |
176 | if fid < 0,
177 | msg = sprintf('Cannot open file %s.hdr.',fileprefix);
178 | error(msg);
179 | end
180 |
181 | save_untouch0_nii_hdr(hdr, fid);
182 |
183 | fclose(fid);
184 | fid = fopen(sprintf('%s.img',fileprefix),'w');
185 | end
186 |
187 | ScanDim = double(hdr.dime.dim(5)); % t
188 | SliceDim = double(hdr.dime.dim(4)); % z
189 | RowDim = double(hdr.dime.dim(3)); % y
190 | PixelDim = double(hdr.dime.dim(2)); % x
191 | SliceSz = double(hdr.dime.pixdim(4));
192 | RowSz = double(hdr.dime.pixdim(3));
193 | PixelSz = double(hdr.dime.pixdim(2));
194 |
195 | x = 1:PixelDim;
196 |
197 | if filetype == 2 & isempty(ext)
198 | skip_bytes = double(hdr.dime.vox_offset) - 348;
199 | else
200 | skip_bytes = 0;
201 | end
202 |
203 | if double(hdr.dime.datatype) == 128
204 |
205 | % RGB planes are expected to be in the 4th dimension of nii.img
206 | %
207 | if(size(nii.img,4)~=3)
208 | error(['The NII structure does not appear to have 3 RGB color planes in the 4th dimension']);
209 | end
210 |
211 | nii.img = permute(nii.img, [4 1 2 3 5 6 7 8]);
212 | end
213 |
214 | % For complex float32 or complex float64, voxel values
215 | % include [real, imag]
216 | %
217 | if hdr.dime.datatype == 32 | hdr.dime.datatype == 1792
218 | real_img = real(nii.img(:))';
219 | nii.img = imag(nii.img(:))';
220 | nii.img = [real_img; nii.img];
221 | end
222 |
223 | if skip_bytes
224 | fwrite(fid, zeros(1,skip_bytes), 'uint8');
225 | end
226 |
227 | fwrite(fid, nii.img, precision);
228 | % fwrite(fid, nii.img, precision, skip_bytes); % error using skip
229 | fclose(fid);
230 |
231 | return; % write_nii
232 |
233 |
--------------------------------------------------------------------------------
/NIFTI/save_untouch_nii_hdr.m:
--------------------------------------------------------------------------------
1 | % internal function
2 |
3 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
4 |
5 | function save_nii_hdr(hdr, fid)
6 |
7 | if ~isequal(hdr.hk.sizeof_hdr,348),
8 | error('hdr.hk.sizeof_hdr must be 348.');
9 | end
10 |
11 | write_header(hdr, fid);
12 |
13 | return; % save_nii_hdr
14 |
15 |
16 | %---------------------------------------------------------------------
17 | function write_header(hdr, fid)
18 |
19 | % Original header structures
20 | % struct dsr /* dsr = hdr */
21 | % {
22 | % struct header_key hk; /* 0 + 40 */
23 | % struct image_dimension dime; /* 40 + 108 */
24 | % struct data_history hist; /* 148 + 200 */
25 | % }; /* total= 348 bytes*/
26 |
27 | header_key(fid, hdr.hk);
28 | image_dimension(fid, hdr.dime);
29 | data_history(fid, hdr.hist);
30 |
31 | % check the file size is 348 bytes
32 | %
33 | fbytes = ftell(fid);
34 |
35 | if ~isequal(fbytes,348),
36 | msg = sprintf('Header size is not 348 bytes.');
37 | warning(msg);
38 | end
39 |
40 | return; % write_header
41 |
42 |
43 | %---------------------------------------------------------------------
44 | function header_key(fid, hk)
45 |
46 | fseek(fid,0,'bof');
47 |
48 | % Original header structures
49 | % struct header_key /* header key */
50 | % { /* off + size */
51 | % int sizeof_hdr /* 0 + 4 */
52 | % char data_type[10]; /* 4 + 10 */
53 | % char db_name[18]; /* 14 + 18 */
54 | % int extents; /* 32 + 4 */
55 | % short int session_error; /* 36 + 2 */
56 | % char regular; /* 38 + 1 */
57 | % char dim_info; % char hkey_un0; /* 39 + 1 */
58 | % }; /* total=40 bytes */
59 |
60 | fwrite(fid, hk.sizeof_hdr(1), 'int32'); % must be 348.
61 |
62 | % data_type = sprintf('%-10s',hk.data_type); % ensure it is 10 chars from left
63 | % fwrite(fid, data_type(1:10), 'uchar');
64 | pad = zeros(1, 10-length(hk.data_type));
65 | hk.data_type = [hk.data_type char(pad)];
66 | fwrite(fid, hk.data_type(1:10), 'uchar');
67 |
68 | % db_name = sprintf('%-18s', hk.db_name); % ensure it is 18 chars from left
69 | % fwrite(fid, db_name(1:18), 'uchar');
70 | pad = zeros(1, 18-length(hk.db_name));
71 | hk.db_name = [hk.db_name char(pad)];
72 | fwrite(fid, hk.db_name(1:18), 'uchar');
73 |
74 | fwrite(fid, hk.extents(1), 'int32');
75 | fwrite(fid, hk.session_error(1), 'int16');
76 | fwrite(fid, hk.regular(1), 'uchar'); % might be uint8
77 |
78 | % fwrite(fid, hk.hkey_un0(1), 'uchar');
79 | % fwrite(fid, hk.hkey_un0(1), 'uint8');
80 | fwrite(fid, hk.dim_info(1), 'uchar');
81 |
82 | return; % header_key
83 |
84 |
85 | %---------------------------------------------------------------------
86 | function image_dimension(fid, dime)
87 |
88 | % Original header structures
89 | % struct image_dimension
90 | % { /* off + size */
91 | % short int dim[8]; /* 0 + 16 */
92 | % float intent_p1; % char vox_units[4]; /* 16 + 4 */
93 | % float intent_p2; % char cal_units[8]; /* 20 + 4 */
94 | % float intent_p3; % char cal_units[8]; /* 24 + 4 */
95 | % short int intent_code; % short int unused1; /* 28 + 2 */
96 | % short int datatype; /* 30 + 2 */
97 | % short int bitpix; /* 32 + 2 */
98 | % short int slice_start; % short int dim_un0; /* 34 + 2 */
99 | % float pixdim[8]; /* 36 + 32 */
100 | % /*
101 | % pixdim[] specifies the voxel dimensions:
102 | % pixdim[1] - voxel width
103 | % pixdim[2] - voxel height
104 | % pixdim[3] - interslice distance
105 | % pixdim[4] - volume timing, in msec
106 | % ..etc
107 | % */
108 | % float vox_offset; /* 68 + 4 */
109 | % float scl_slope; % float roi_scale; /* 72 + 4 */
110 | % float scl_inter; % float funused1; /* 76 + 4 */
111 | % short slice_end; % float funused2; /* 80 + 2 */
112 | % char slice_code; % float funused2; /* 82 + 1 */
113 | % char xyzt_units; % float funused2; /* 83 + 1 */
114 | % float cal_max; /* 84 + 4 */
115 | % float cal_min; /* 88 + 4 */
116 | % float slice_duration; % int compressed; /* 92 + 4 */
117 | % float toffset; % int verified; /* 96 + 4 */
118 | % int glmax; /* 100 + 4 */
119 | % int glmin; /* 104 + 4 */
120 | % }; /* total=108 bytes */
121 |
122 | fwrite(fid, dime.dim(1:8), 'int16');
123 | fwrite(fid, dime.intent_p1(1), 'float32');
124 | fwrite(fid, dime.intent_p2(1), 'float32');
125 | fwrite(fid, dime.intent_p3(1), 'float32');
126 | fwrite(fid, dime.intent_code(1), 'int16');
127 | fwrite(fid, dime.datatype(1), 'int16');
128 | fwrite(fid, dime.bitpix(1), 'int16');
129 | fwrite(fid, dime.slice_start(1), 'int16');
130 | fwrite(fid, dime.pixdim(1:8), 'float32');
131 | fwrite(fid, dime.vox_offset(1), 'float32');
132 | fwrite(fid, dime.scl_slope(1), 'float32');
133 | fwrite(fid, dime.scl_inter(1), 'float32');
134 | fwrite(fid, dime.slice_end(1), 'int16');
135 | fwrite(fid, dime.slice_code(1), 'uchar');
136 | fwrite(fid, dime.xyzt_units(1), 'uchar');
137 | fwrite(fid, dime.cal_max(1), 'float32');
138 | fwrite(fid, dime.cal_min(1), 'float32');
139 | fwrite(fid, dime.slice_duration(1), 'float32');
140 | fwrite(fid, dime.toffset(1), 'float32');
141 | fwrite(fid, dime.glmax(1), 'int32');
142 | fwrite(fid, dime.glmin(1), 'int32');
143 |
144 | return; % image_dimension
145 |
146 |
147 | %---------------------------------------------------------------------
148 | function data_history(fid, hist)
149 |
150 | % Original header structures
151 | %struct data_history
152 | % { /* off + size */
153 | % char descrip[80]; /* 0 + 80 */
154 | % char aux_file[24]; /* 80 + 24 */
155 | % short int qform_code; /* 104 + 2 */
156 | % short int sform_code; /* 106 + 2 */
157 | % float quatern_b; /* 108 + 4 */
158 | % float quatern_c; /* 112 + 4 */
159 | % float quatern_d; /* 116 + 4 */
160 | % float qoffset_x; /* 120 + 4 */
161 | % float qoffset_y; /* 124 + 4 */
162 | % float qoffset_z; /* 128 + 4 */
163 | % float srow_x[4]; /* 132 + 16 */
164 | % float srow_y[4]; /* 148 + 16 */
165 | % float srow_z[4]; /* 164 + 16 */
166 | % char intent_name[16]; /* 180 + 16 */
167 | % char magic[4]; % int smin; /* 196 + 4 */
168 | % }; /* total=200 bytes */
169 |
170 | % descrip = sprintf('%-80s', hist.descrip); % 80 chars from left
171 | % fwrite(fid, descrip(1:80), 'uchar');
172 | pad = zeros(1, 80-length(hist.descrip));
173 | hist.descrip = [hist.descrip char(pad)];
174 | fwrite(fid, hist.descrip(1:80), 'uchar');
175 |
176 | % aux_file = sprintf('%-24s', hist.aux_file); % 24 chars from left
177 | % fwrite(fid, aux_file(1:24), 'uchar');
178 | pad = zeros(1, 24-length(hist.aux_file));
179 | hist.aux_file = [hist.aux_file char(pad)];
180 | fwrite(fid, hist.aux_file(1:24), 'uchar');
181 |
182 | fwrite(fid, hist.qform_code, 'int16');
183 | fwrite(fid, hist.sform_code, 'int16');
184 | fwrite(fid, hist.quatern_b, 'float32');
185 | fwrite(fid, hist.quatern_c, 'float32');
186 | fwrite(fid, hist.quatern_d, 'float32');
187 | fwrite(fid, hist.qoffset_x, 'float32');
188 | fwrite(fid, hist.qoffset_y, 'float32');
189 | fwrite(fid, hist.qoffset_z, 'float32');
190 | fwrite(fid, hist.srow_x(1:4), 'float32');
191 | fwrite(fid, hist.srow_y(1:4), 'float32');
192 | fwrite(fid, hist.srow_z(1:4), 'float32');
193 |
194 | % intent_name = sprintf('%-16s', hist.intent_name); % 16 chars from left
195 | % fwrite(fid, intent_name(1:16), 'uchar');
196 | pad = zeros(1, 16-length(hist.intent_name));
197 | hist.intent_name = [hist.intent_name char(pad)];
198 | fwrite(fid, hist.intent_name(1:16), 'uchar');
199 |
200 | % magic = sprintf('%-4s', hist.magic); % 4 chars from left
201 | % fwrite(fid, magic(1:4), 'uchar');
202 | pad = zeros(1, 4-length(hist.magic));
203 | hist.magic = [hist.magic char(pad)];
204 | fwrite(fid, hist.magic(1:4), 'uchar');
205 |
206 | return; % data_history
207 |
208 |
--------------------------------------------------------------------------------
/NIFTI/unxform_nii.m:
--------------------------------------------------------------------------------
1 | % Undo the flipping and rotations performed by xform_nii; spit back only
2 | % the raw img data block. Initial cut will only deal with 3D volumes
3 | % strongly assume we have called xform_nii to write down the steps used
4 | % in xform_nii.
5 | %
6 | % Usage: a = load_nii('original_name');
7 | % manipulate a.img to make array b;
8 | %
9 | % if you use unxform_nii to un-tranform the image (img) data
10 | % block, then nii.original.hdr is the corresponding header.
11 | %
12 | % nii.original.img = unxform_nii(a, b);
13 | % save_nii(nii.original,'newname');
14 | %
15 | % Where, 'newname' is created with data in the same space as the
16 | % original_name data
17 | %
18 | % - Jeff Gunter, 26-JUN-06
19 | %
20 | function outblock = unxform_nii(nii, inblock)
21 |
22 | if isempty(nii.hdr.hist.rot_orient)
23 | outblock=inblock;
24 | else
25 | [dummy unrotate_orient] = sort(nii.hdr.hist.rot_orient);
26 | outblock = permute(inblock, unrotate_orient);
27 | end
28 |
29 | if ~isempty(nii.hdr.hist.flip_orient)
30 | flip_orient = nii.hdr.hist.flip_orient(unrotate_orient);
31 |
32 | for i = 1:3
33 | if flip_orient(i)
34 | outblock = flipdim(outblock, i);
35 | end
36 | end
37 | end;
38 |
39 | return;
40 |
41 |
--------------------------------------------------------------------------------
/NIFTI/verify_nii_ext.m:
--------------------------------------------------------------------------------
1 | % Verify NIFTI header extension to make sure that each extension section
2 | % must be an integer multiple of 16 byte long that includes the first 8
3 | % bytes of esize and ecode. If the length of extension section is not the
4 | % above mentioned case, edata should be padded with all 0.
5 | %
6 | % Usage: [ext, esize_total] = verify_nii_ext(ext)
7 | %
8 | % ext - Structure of NIFTI header extension, which includes num_ext,
9 | % and all the extended header sections in the header extension.
10 | % Each extended header section will have its esize, ecode, and
11 | % edata, where edata can be plain text, xml, or any raw data
12 | % that was saved in the extended header section.
13 | %
14 | % esize_total - Sum of all esize variable in all header sections.
15 | %
16 | % NIFTI data format can be found on: http://nifti.nimh.nih.gov
17 | %
18 | % - Jimmy Shen (jimmy@rotman-baycrest.on.ca)
19 | %
20 | function [ext, esize_total] = verify_nii_ext(ext)
21 |
22 | if ~isfield(ext, 'section')
23 | error('Incorrect NIFTI header extension structure.');
24 | elseif ~isfield(ext, 'num_ext')
25 | ext.num_ext = length(ext.section);
26 | elseif ~isfield(ext, 'extension')
27 | ext.extension = [1 0 0 0];
28 | end
29 |
30 | esize_total = 0;
31 |
32 | for i=1:ext.num_ext
33 | if ~isfield(ext.section(i), 'ecode') | ~isfield(ext.section(i), 'edata')
34 | error('Incorrect NIFTI header extension structure.');
35 | end
36 |
37 | ext.section(i).esize = ceil((length(ext.section(i).edata)+8)/16)*16;
38 | ext.section(i).edata = ...
39 | [ext.section(i).edata ...
40 | zeros(1,ext.section(i).esize-length(ext.section(i).edata)-8)];
41 | esize_total = esize_total + ext.section(i).esize;
42 | end
43 |
44 | return % verify_nii_ext
45 |
46 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # DOMINO: Domain-aware Model Calibration in Medical Image Segmentation
2 | We create and provide open-source code of a model called a **domain-aware model calibration method**, nicknamed **DOMINO** that leverages the semantic confusability and hierarchical similarity between class labels. Our experiments demonstrate that our DOMINO-calibrated deep neural networks outperform non-calibrated models and state-of-the-art morphometric methods in head image segmentation. Our results show that our method can consistently achieve better calibration, higher accuracy, and faster inference times than these methods, especially on rarer classes. This performance is attributed to our domain-aware regularization to inform semantic model calibration. These findings show the importance of semantic ties between class labels in building confidence in deep learning models. The framework has the potential to improve the trustworthiness and reliability of generic medical image segmentation models.
3 |
4 | ## Paper
5 | This repository provides the official implemantation of training DOMINO as well as the usage the model DOMINO in the following paper:
6 |
7 | **DOMINO: Domain-aware Model Calibration in Medical Image Segmentation**
8 |
9 | Skylar E. Stolte1, Kyle Volle2, Aprinda Indahlastari3,4, Alejandro Albizu3,5, Adam J. Woods3,4,5, Kevin Brink6, Matthew Hale2, and Ruogu Fang1,3,7*
10 |
11 | 1 J. Crayton Pruitt Family Department of Biomedical Engineering, HerbertWertheim College of Engineering, University of Florida (UF), USA
12 | 2 Department of Mechanical and Aerospace Engineering, Herbert Wertheim Collegeof Engineering, UF, USA
13 | 3 Center for Cognitive Aging and Memory, McKnight Brain Institute, UF, USA
14 | 4 Department of Clinical and Health Psychology, College of Public Health andHealth Professions, UF, USA
15 | 5 Department of Neuroscience, College of Medicine, UF, USA
16 | 6 United States Air Force Research Laboratory, Eglin Air Force Base, Florida, USA
17 | 7 Department of Electrical and Computer Engineering, Herbert Wertheim College ofEngineering, UF, USA
18 |
19 | International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022
20 | [paper](https://arxiv.org/abs/2209.06077) | [code](https://github.com/lab-smile/DOMINO) | [slides](https://github.com/lab-smile/DOMINO/blob/5378ad4ad099efe2700ad75c23ecbc5ce585d266/doc/DOMINO_MICCAI22_PPT.pdf) | [poster](https://github.com/lab-smile/DOMINO/blob/5378ad4ad099efe2700ad75c23ecbc5ce585d266/doc/poster1693.pdf) | [talk](https://www.youtube.com/watch?v=tzdPTrUWlHk)
21 | 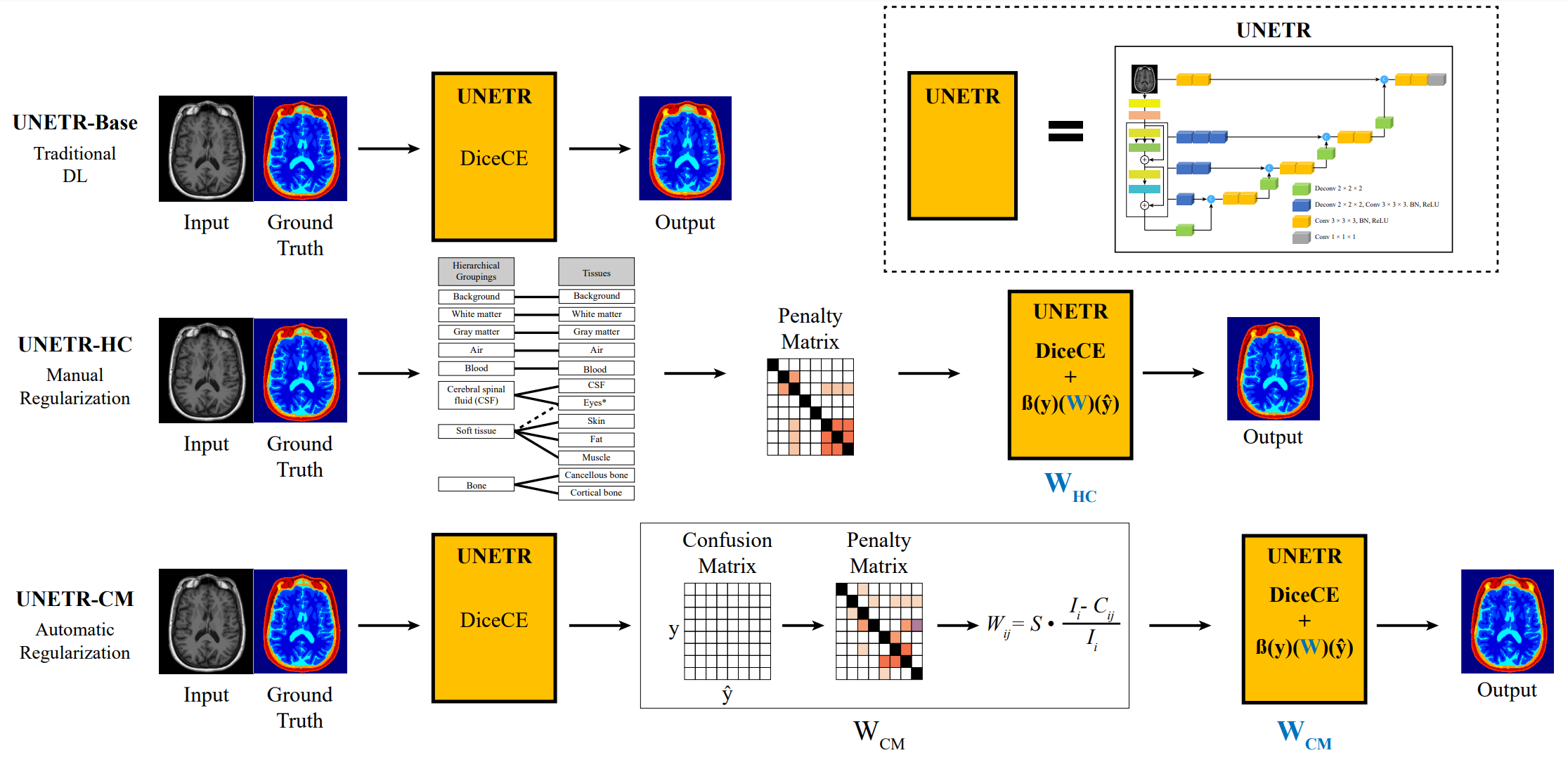
22 |
23 | ## Major results from our work
24 |
25 | - Our DOMINO methods improve calibration and accuracy in head segmentation problems from T1 MRIs
26 | - DOMINO-CM achieves higher Top-1, Top-2, and Top-3 accuracy than DOMINO-HC or an uncalibrated model. This indicates superior regional performance and higher relevance to the non-selected classes (important in calibration).
27 | - DOMINO-HC achieves more precise boundary detection when compared to DOMINO-CM or an uncalibrated model. This is important to calibration problems because boundaries are the most uncertain areas in segmentation challenges.
28 |
29 |
30 |
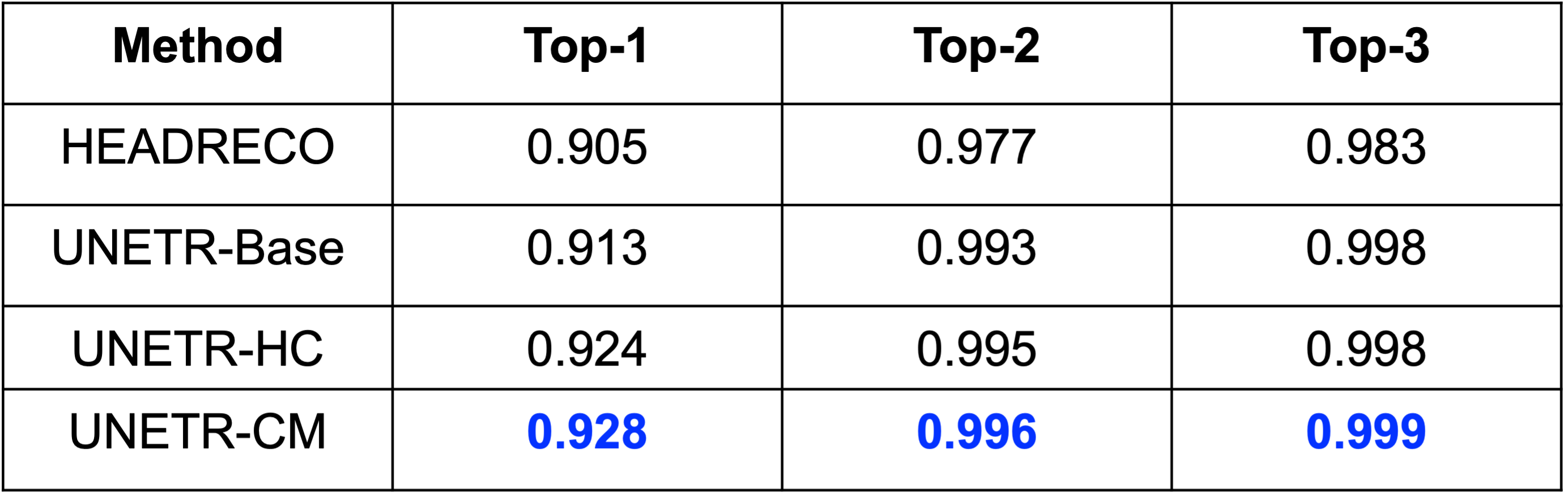
31 |
34 | fig. 1: Top-N Accuracy on 6 classes
35 |
36 |
37 |
38 |
39 | 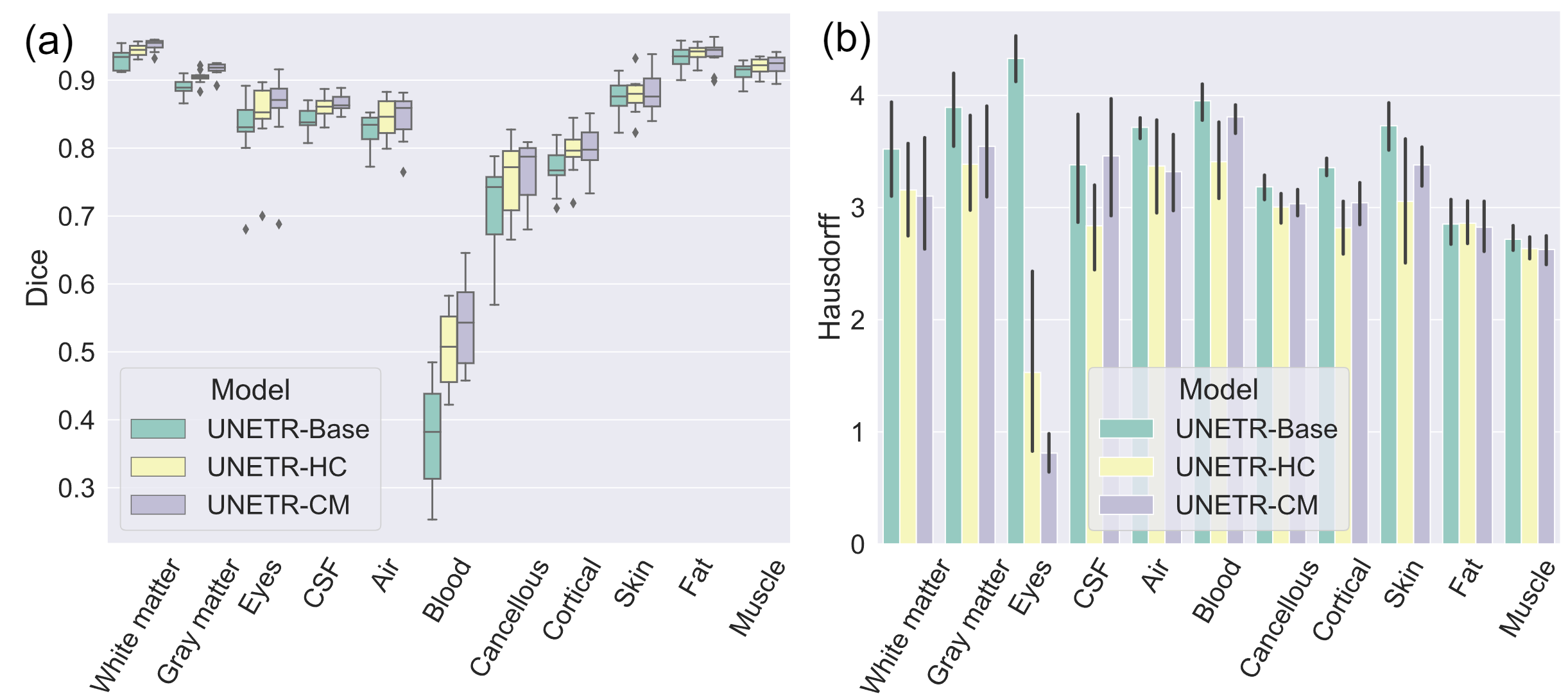
40 |
41 | fig. 2: (a) Dice scores and (b) Hausdorff distances in 11-class segmentation
42 |
43 |
44 |
45 |
46 |
47 | 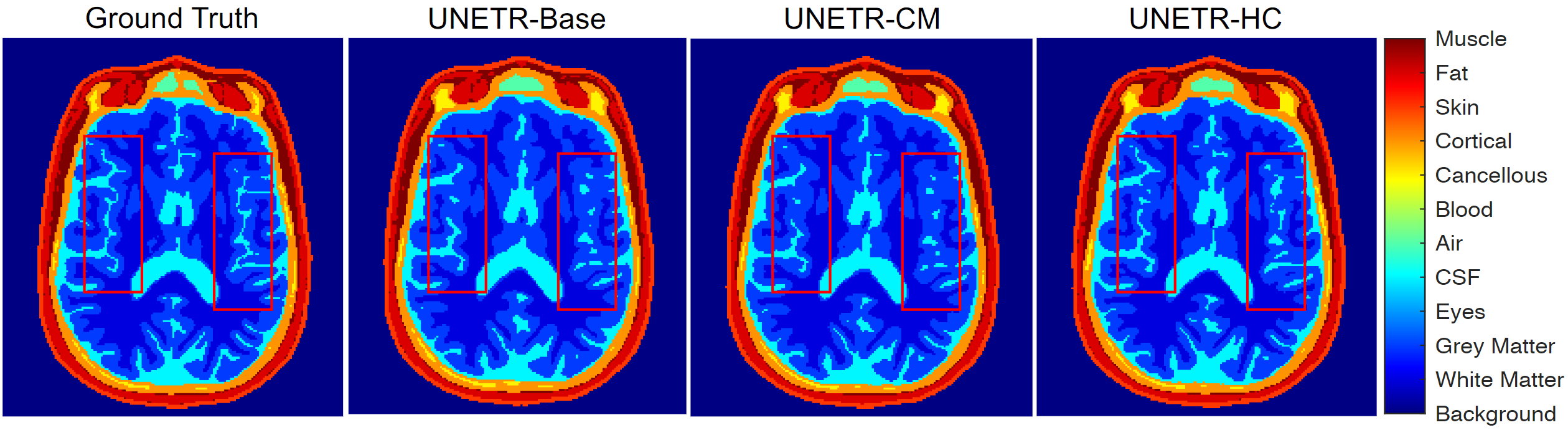
48 |
49 | fig. 3: Sample image slice for 11-tissue segmentation
50 |
51 |
52 |
53 | 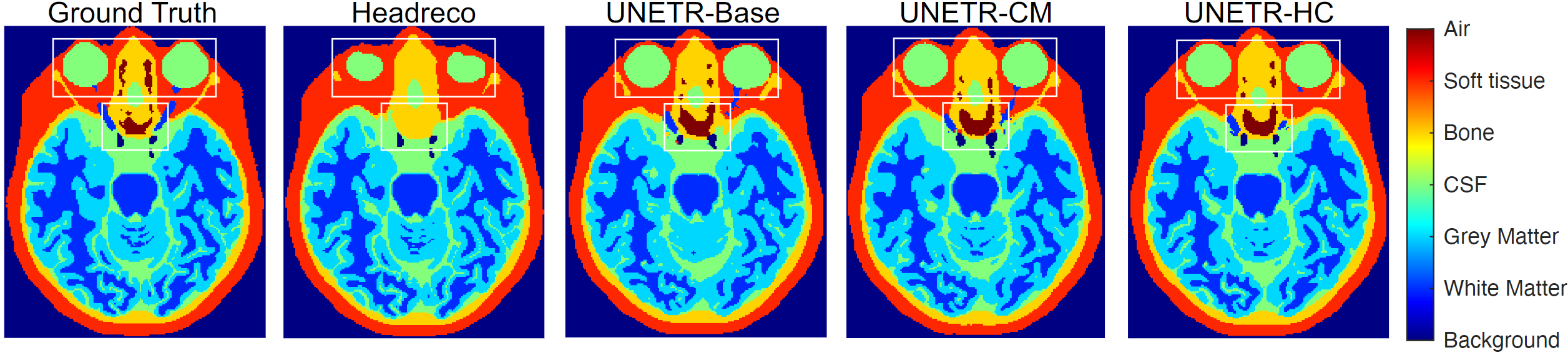
54 |
55 | fig. 4: Sample image slice for 6-tissue segmentation
56 |
57 |
58 |
59 | ## Usage
60 | You can find there are two MATLAB codes, you can directly change the directory to your own data. You need to select the DOMINO working folder and add to path before you running these two MATLAB codes.
61 |
62 | To run the combineIn case of you are using different version of MATLAB, if you are using MATLAB 2020b, you need to change line 56 to :
63 | ```
64 | image(index) = tissue_cond_updated.Labels(k)
65 | ```
66 | Then you can run the combine_mask.m. The output should be a Data folder with the following structure:
67 | ```
68 | Data ImagesTr sub-TrX_T1.nii sub-TrXX_T1.nii ...
69 | ImagesTs sub-TsX_T1.nii sub-TsXX_T1.nii ...
70 | LabelsTr sub-TrX_seg.nii sub-TrXX_seg.nii ...
71 | LabelsTs sub-TsX_seg.nii sub-TsX_seg.nii ...
72 | ```
73 | Maneuver to the /your_data/Data/. Run make_datalist_json.m
74 |
75 | After this code is done, you may exit MATLAB and open the terminal to run the other codes.
76 |
77 | ### Build container
78 | The DOMINO code uses the MONAI, an open-source foundation. We provide a .sh script to help you to build your own container for running your code.
79 |
80 | Run the following code in the terminal, you need to change the line after --sandbox to your desired writable directory and change the line after --nv to your own directory.
81 | ```
82 | sbatch building_container_v08.sh
83 | ```
84 |
85 | The output should be a folder named monaicore08 under your desired directory.
86 |
87 | ### Training
88 | Once the data and the container are ready, you can train the model by using the following command:
89 | ```
90 | sbatch train.sh
91 | ```
92 | Before you training the model, you need to make sure change the following directory:
93 | - change the first singularity exec -nv to the directory includes monaicore08, for example: /user/DOMINO/monaicore08
94 | - change the line after --bind to the directory includes monaicore08
95 | - change the data_dir to your data directory
96 | - change the model name to your desired model name
97 | You can also specify the max iteration number for training. For the iterations = 100, the training progress might take about one hours, and for the iterations = 25,000, the training progress might take about 24 hours.
98 |
99 | ### Testing
100 | The test progress is very similar to the training progress. You need to change all paths and make sure the model_save_name matches your model name in runMONAI.sh. Then running the runMONAI_test.sh with the following command:
101 | ```
102 | sbatch test.sh
103 | ```
104 | The outputs for each test subject is saved as a mat file.
105 |
106 | ### Pre-trained models
107 | You can also use the pre-trained models we provide for testing, please fill out the following request form before accessing to DOMINO models.
108 | Download pre-trained models [here](https://forms.gle/3GPnXXvWgaM6RZvr5)
109 |
110 | ### Code Ocean
111 | You can alternatively find the link to our reproducible capsule for our code on Code Ocean. The link to this capsule is as follows: https://codeocean.com/capsule/6022409/tree/v2.
112 |
113 | ## Citation
114 | If you use this code, please cite our papers:
115 | ```
116 | @InProceedings{stolte2022DOMINO,
117 | author="Stolte, Skylar E. and Volle, Kyle and Indahlastari, Aprinda and Albizu, Alejandro and Woods, Adam J. and Brink, Kevin and Hale, Matthew and Fang, Ruogu",
118 | title="DOMINO: Domain-aware Model Calibration in Medical Image Segmentation",
119 | booktitle="Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022",
120 | year="2022",
121 | url="https://arxiv.org/abs/2209.06077"
122 | }
123 | ```
124 | ## Acknowledgement
125 |
126 | This work was supported by the National Institutes ofHealth/National Institute on Aging (NIA RF1AG071469, NIA R01AG054077),the National Science Foundation (1908299), and the NSF-AFRL INTERN Sup-plement (2130885).
127 |
128 | We acknowledge NVIDIA AI Technology Center (NVAITC)for their suggestions. We also thank Jiaqing Zhang for formatting assistance.
129 |
130 | We employ UNETR as our base model from:
131 | https://github.com/Project-MONAI/research-contributions/tree/main/UNETR
132 | ## Contact
133 | Any discussion, suggestions and questions please contact: [Skylar Stolte](mailto:skylastolte4444@ufl.edu), [Dr. Ruogu Fang](mailto:ruogu.fang@bme.ufl.edu).
134 |
135 | *Smart Medical Informatics Learning & Evaluation Laboratory, Dept. of Biomedical Engineering, University of Florida*
136 |
--------------------------------------------------------------------------------
/UNETR_matrixpenalty_v5_2d.csv:
--------------------------------------------------------------------------------
1 | 0,0.999999992,1,1,0.999999984,0.99999029,1,1,0.999998278,0.999623564,0.999999516,0.99998146
2 | 0.999990477,0,0.930067945,0.999950797,0.998069989,1,0.999956239,0.999999773,0.999571688,0.999948303,0.999738569,0.999414331
3 | 1,0.980327666,0,1,0.947401465,1,1,0.999991598,0.999437618,1,1,0.99999825
4 | 0.999885009,0.999012994,1,0,0.968214573,1,1,1,0.999597532,0.962321285,0.966432213,0.94579133
5 | 0.999947667,0.999447344,0.903345211,0.998282222,0,0.999998275,0.999411401,0.998227301,0.937511861,0.999994824,0.999995974,0.999762202
6 | 0.997312003,0.999998553,1,1,0.999998553,0,1,0.999817616,0.886309454,0.999675761,1,0.97068245
7 | 0.994282344,0.99907697,0.999794882,1,0.831700938,0.999615404,0,0.975514076,0.940438952,0.9982565,0.800292293,0.304445926
8 | 1,0.999977785,0.999861772,1,0.992594951,0.999997532,0.999039812,0,0.803091526,0.99961576,0.995874565,0.925463248
9 | 0.999753484,0.999956423,0.999329366,1,0.95887749,0.98052425,0.999110466,0.951492661,0,0.999548012,0.995745135,0.906914118
10 | 0.859514759,1,1,0.999682116,1,0.998863924,1,0.999996119,0.999639809,0,0.988288756,0.978038459
11 | 0.999994015,0.999967898,0.999999637,0.999758603,0.999971888,1,0.999892088,0.999546587,0.99878975,0.9799031,0,0.979812962
12 | 0.999525959,0.999920496,0.999977167,0.999553626,0.999880898,0.998755089,0.999362528,0.9985066,0.977906838,0.973810891,0.957159484,0
--------------------------------------------------------------------------------
/build_container_v08.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=build_container
3 | #SBATCH --mail-type=END,FAIL
4 | #SBATCH --mail-user=skylastolte4444@ufl.edu
5 | #SBATCH --nodes=1
6 | #SBATCH --cpus-per-task=1
7 | #SBATCH --mem-per-cpu=30gb
8 | #SBATCH --time=08:00:00
9 | #SBATCH --output=%x.%j.out
10 | date;hostname;pwd
11 |
12 | module load singularity
13 |
14 | # build a Singularity sandbox container (container in a writable directory) from MONAI Core docker image
15 | singularity build --sandbox /red/nvidia-ai/SkylarStolte/monaicore08/ docker://projectmonai/monai:0.8.1
16 | # check nsys environment
17 | singularity exec --nv /red/nvidia-ai/SkylarStolte/monaicore08 nsys status -e
--------------------------------------------------------------------------------
/combine_masks.m:
--------------------------------------------------------------------------------
1 | clc; clear;
2 | ACT_dir = dir('.');
3 | for i = length(ACT_dir) : -1 : 1
4 | if ~isdir(ACT_dir(i).name)
5 | ACT_dir(i)=[];
6 | elseif strcmp(ACT_dir(i).name,'.') || strcmp(ACT_dir(i).name,'..')
7 | ACT_dir(i)=[];
8 | end
9 | end
10 | names = [];
11 | for i = 1 : length(ACT_dir)
12 | names(i) = str2num(ACT_dir(i).name(11:16));
13 | end
14 | x = 256; y = 256; z = 256;
15 | ccnt=x*y*z;
16 |
17 | tissues_cond = readtable('tissue_cond_11.xlsx');
18 | tissue_cond_updated = tissues_cond;
19 | tissue_cond_updated.TissueType(2) = {'wm'};
20 | tissue_cond_updated.TissueType(3) = {'gm'};
21 | tissue_cond_updated.TissueType(8) = {'cancellous'};
22 | tissue_cond_updated.TissueType(9) = {'cortical'};
23 | tissue_cond_updated(13:14,:)=[];
24 |
25 | T = length(ACT_dir); Te = round(T*0.10);
26 |
27 | mkdir('data')
28 | imagesTr = fullfile('data', 'imagesTr'); mkdir(imagesTr);
29 | imagesTs = fullfile('data', 'imagesTs'); mkdir(imagesTs);
30 | labelsTr = fullfile('data', 'labelsTr'); mkdir(labelsTr);
31 | labelsTs = fullfile('data', 'labelsTs'); mkdir(labelsTs);
32 |
33 | for i = 1 : length(ACT_dir)
34 | path_raw = strcat('./',ACT_dir(i).name,'/idv_mask/');
35 | path_save = strcat('./',ACT_dir(i).name,'/comb_mask/');
36 |
37 | if ~exist(path_save, 'dir')
38 | mkdir(path_save)
39 | end
40 |
41 | path_dir = dir(fullfile(path_raw, '*.raw'));
42 | image = zeros(x*y*z,1);
43 |
44 | for k = height(tissue_cond_updated):-1:1
45 |
46 | for j = 1 : length(path_dir)
47 | fileID = fopen(strcat('/', path_dir(j).folder,'//',path_dir(j).name),'r');
48 | A=fread(fileID,ccnt,'uint8=>uint8');
49 | A=reshape(A,x,y,z);
50 | fclose(fileID);
51 |
52 | index = find(A==255);
53 |
54 | if contains(path_dir(j).name, string(tissue_cond_updated.TissueType(k)), 'IgnoreCase', true)
55 | %labels = tissue_cond_updated.Labels{k} * ones(length(index),1);
56 | image(index)=str2num(tissue_cond_updated.Labels{k});
57 |
58 | end
59 |
60 | end
61 | end
62 |
63 | image = reshape(image,x,y,z);
64 | figure; imshow(image(:,:,floor(z/2)),[0 13]);
65 | save(strcat(path_save,ACT_dir(i).name(7:16),'_seg.mat'),'image');
66 |
67 | seg_file = make_nii(image);
68 | seg_file.filetype = 2;
69 | seg_file.fileprefix = strcat(path_save,ACT_dir(i).name(7:16),'_seg');
70 | seg_file.machine = 'ieee-le';
71 | seg_file.original.hdr = seg_file.hdr;
72 | save_nii(seg_file, char(strcat(path_save,ACT_dir(i).name(7:16),'_seg.nii')));
73 |
74 | copyfile(fullfile(ACT_dir(i).name, 'T1.nii'), fullfile(ACT_dir(i).name,strcat(ACT_dir(i).name(7:16), '_T1.nii')));
75 |
76 | if (i>(T-Te-1)) && (i<=T)
77 | copyfile(fullfile(ACT_dir(i).name,'comb_mask', strcat(ACT_dir(i).name(7:16), '_seg.nii')), labelsTs);
78 | copyfile(fullfile(ACT_dir(i).name,strcat(ACT_dir(i).name(7:16), '_T1.nii')), imagesTs);
79 | else
80 | copyfile(fullfile(ACT_dir(i).name,'comb_mask', strcat(ACT_dir(i).name(7:16), '_seg.nii')), labelsTr);
81 | copyfile(fullfile(ACT_dir(i).name,strcat(ACT_dir(i).name(7:16), '_T1.nii')), imagesTr);
82 | end
83 |
84 | end
--------------------------------------------------------------------------------
/doc/DOMINO_MICCAI22_PPT.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/lab-smile/DOMINO/3d261da4dec8ba338818babfa471c47c51d0866d/doc/DOMINO_MICCAI22_PPT.pdf
--------------------------------------------------------------------------------
/doc/ReadMeDoc.md:
--------------------------------------------------------------------------------
1 | This is the ReadMe file for the MICCAI presentation documents.
2 |
--------------------------------------------------------------------------------
/doc/poster1693.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/lab-smile/DOMINO/3d261da4dec8ba338818babfa471c47c51d0866d/doc/poster1693.pdf
--------------------------------------------------------------------------------
/make_datalist_json.m:
--------------------------------------------------------------------------------
1 | % fname = 'dataset_2.json';
2 | % fid = fopen(fname);
3 | % raw = fread(fid,inf);
4 | % str = char(raw');
5 | % fclose(fid);
6 | % val = jsondecode(str);
7 |
8 | description = 'AISEG V5 - Code Validation';
9 | labels = struct('xO', 'background', ...
10 | 'x1', 'wm', ...
11 | 'x2', 'gm', ...
12 | 'x3', 'eyes', ...
13 | 'x4', 'csf', ...
14 | 'x5', 'air', ...
15 | 'x6', 'blood', ...
16 | 'x7', 'cancellous', ...
17 | 'x8', 'cortical', ...
18 | 'x9', 'skin', ...
19 | 'x10', 'fat', ...
20 | 'x11', 'muscle');
21 | license = 'UF';
22 | modality = struct('x0', 'T1');
23 | numTest = length(dir(fullfile('imagesTs', '*.nii')));
24 | numTraining = length(dir(fullfile('imagesTr', '*.nii')));
25 |
26 | testdir = dir(fullfile('imagesTs', '*.nii'));
27 | for i = 1 : length(testdir)
28 | test(i,1) = {fullfile('imagesTs',testdir(i).name)};
29 | end
30 |
31 | traindir = dir(fullfile('imagesTr', '*.nii'));
32 | trainlabeldir = dir(fullfile('labelsTr', '*.nii'));
33 |
34 | T = length(traindir); Te = round(T*0.10);
35 | traindir_fin = traindir(1:T-Te-1);
36 | validdir_fin = traindir(T-Te:T);
37 | traindir_label_fin = trainlabeldir(1:T-Te-1);
38 | validdir_label_fin = trainlabeldir(T-Te:T);
39 |
40 | %training = struct('image', fullfile('imagesTr',traindir_fin(:).name), 'label', fullfile('labelsTr',traindir_label_fin(:).name));
41 | % training = struct();
42 | % training.image = {fullfile('imagesTr',traindir_fin(:).name)};
43 | % training.label = {fullfile('labelsTr',traindir_label_fin(:).name)};
44 | % training = struct('image', fullfile('imagesTr',traindir_fin(:).name), 'label', fullfile('labelsTr',traindir_label_fin(:).name));
45 | % training = [image, label];
46 | for i = 1 : length(traindir_fin)
47 | newtrain = struct('image', fullfile('imagesTr',traindir_fin(i).name), 'label', fullfile('labelsTr',traindir_label_fin(i).name));
48 | if i == 1
49 | training = newtrain;
50 | else
51 | training = [training; newtrain];
52 | end
53 | end
54 | for i = 1 : length(validdir_fin)
55 | newtrain = struct('image', fullfile('imagesTr',validdir_fin(i).name), 'label', fullfile('labelsTr',validdir_label_fin(i).name));
56 | if i == 1
57 | validation = newtrain;
58 | else
59 | validation = [validation; newtrain];
60 | end
61 | end
62 |
63 | % s = struct('description', description, ...
64 | % 'license', license, ...
65 | % 'labels', labels, ...
66 | % 'modality', modality, ...
67 | % 'name', 'ACT', ...
68 | % 'numTest', numTest, ...
69 | % 'numTraining', numTraining, ...
70 | % 'reference', 'NA', ...
71 | % 'release', 'NA', ...
72 | % 'tensorImageSize', '3D', ...
73 | % 'test', test, ...
74 | % 'training', training, ...
75 | % 'validation', validation);
76 | s = struct();
77 | s.description = description;
78 | s.labels = labels;
79 | s.license = license;
80 | s.modality = modality;
81 | s.name = 'ACT';
82 | s.numTest = numTest;
83 | s.numTraining = numTraining;
84 | s.reference = 'NA';
85 | s.release = 'NA';
86 | s.tensorImageSize = '3D';
87 | s.test = test;
88 | s.training = training;
89 | s.validation = validation;
90 |
91 | s = jsonencode(s);
92 | fid = fopen('dataset_1.json','w');
93 | fprintf(fid,s);
94 | fclose(fid);
95 |
96 |
97 |
--------------------------------------------------------------------------------
/test.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=monai_test
3 | #SBATCH --mail-type=END,FAIL
4 | #SBATCH --mail-user=
5 | #SBATCH --ntasks=1
6 | #SBATCH --cpus-per-task=4
7 | #SBATCH --distribution=block:block
8 | #SBATCH --partition=hpg-ai
9 | #SBATCH --gres=gpu:a100:2
10 | #SBATCH --mem=30gb
11 | #SBATCH --time=72:00:00
12 | #SBATCH --output=%x_%j.log
13 |
14 | #module load pytorch
15 | module load singularity
16 | #module load monai
17 |
18 | # Check if cuda enabled
19 | #python -c "import torch; print(torch.cuda.is_available())"
20 | singularity exec --nv /monaicore081 python3 -c "import torch; print(torch.cuda.is_available())"
21 |
22 | #run code
23 | #python test.py --num_gpu 2 --data_dir "" --N_classes 12 --model_load_name "unetr_v5_bfc.pth" --dataparallel "True"
24 |
25 | singularity exec --nv --bind :/mnt /monaicore081 python3 /mnt/test.py --num_gpu 2 --data_dir '/mnt//' --model_load_name "unetr_v5_06-19-22.pth" --N_classes 12 --dataparallel "True" --a_max_value 255 --spatial_size 64
--------------------------------------------------------------------------------
/test_domino.py:
--------------------------------------------------------------------------------
1 | # coding=utf-8
2 |
3 | #load packages:
4 |
5 | #standard packages -
6 |
7 | import os
8 | import shutil
9 | import tempfile
10 | import matplotlib.pyplot as plt
11 | import numpy as np
12 | from tqdm import tqdm
13 | import torch
14 | import torch.nn as nn
15 | import time
16 | import argparse
17 | from scipy.io import savemat
18 |
19 | #load monai functions -
20 |
21 | from monai.losses import DiceCELoss
22 | from monai.inferers import sliding_window_inference
23 | from monai.transforms import (
24 | AsDiscrete,
25 | AddChanneld,
26 | Compose,
27 | CropForegroundd,
28 | LoadImaged,
29 | Orientationd,
30 | RandFlipd,
31 | RandCropByPosNegLabeld,
32 | RandShiftIntensityd,
33 | ScaleIntensityRanged,
34 | Spacingd,
35 | RandRotate90d,
36 | ToTensord,
37 | SpatialPadd,
38 | RandGaussianNoised,
39 | ToDeviced,
40 | )
41 |
42 | from monai.config import print_config
43 | from monai.metrics import DiceMetric
44 | from monai.networks.nets import UNETR, BasicUNet
45 | #from monai.networks.nets import UNet
46 |
47 | from monai.data import (
48 | DataLoader,
49 | load_decathlon_datalist,
50 | decollate_batch,
51 | Dataset,
52 | pad_list_data_collate,
53 | )
54 |
55 | #-----------------------------------
56 |
57 | #set up starting conditions:
58 |
59 | #start_time = time.time()
60 | print_config()
61 |
62 | # our CLI parser
63 | parser = argparse.ArgumentParser()
64 | parser.add_argument("--data_dir", type=str, default="/red/nvidia-ai/SkylarStolte/training_pairs_v5_bfc/", help="directory the dataset is in")
65 | parser.add_argument("--batch_size_test", type=int, default=1, help="batch size testing data")
66 | parser.add_argument("--num_gpu", type=int, default=1, help="number of gpus")
67 | parser.add_argument("--N_classes", type=int, default=12, help="number of tissues classes")
68 | parser.add_argument("--spatial_size", type=int, default=256, help="one patch dimension")
69 | parser.add_argument("--model_load_name", type=str, default="unetr_v5_bfc.pth", help="model to load")
70 | parser.add_argument("--dataparallel", type=str, default="True", help="did your model use multi-gpu")
71 | parser.add_argument("--a_max_value", type=int, default=255, help="maximum image intensity")
72 | parser.add_argument("--a_min_value", type=int, default=0, help="minimum image intensity")
73 | args = parser.parse_args()
74 |
75 | split_JSON = "dataset_1.json"
76 | datasets = args.data_dir + split_JSON
77 |
78 | #os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
79 | #device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
80 |
81 | #-----------------------------------
82 |
83 | #data transformations:
84 |
85 | test_transforms = Compose(
86 | [
87 | LoadImaged(keys=["image"]),
88 | AddChanneld(keys=["image"]),
89 | Spacingd(
90 | keys=["image"],
91 | pixdim=(1.0, 1.0, 1.0),
92 | mode=("bilinear"),
93 | ),
94 | Orientationd(keys=["image"], axcodes="RAS"),
95 | ScaleIntensityRanged(keys=["image"], a_min=args.a_min_value, a_max=args.a_max_value, b_min=0.0, b_max=1.0, clip=True),
96 | #CropForegroundd(keys=["image", "label"], source_key="image"),
97 | ToTensord(keys=["image"]),
98 | #ToDeviced(keys=["image", "label"], device=device),
99 | ]
100 | )
101 |
102 | #-----------------------------------
103 |
104 | #set up data loaders
105 |
106 | test_files = load_decathlon_datalist(datasets, True, "test")
107 |
108 | test_ds = Dataset(
109 | data=test_files, transform=test_transforms,
110 | )
111 | test_loader = DataLoader(
112 | test_ds, batch_size=args.batch_size_test, shuffle=False, num_workers=4, pin_memory=True, collate_fn=pad_list_data_collate,
113 | )
114 |
115 | #-----------------------------------
116 |
117 | #set up gpu device and unetr model
118 |
119 | #os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
120 | #device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
121 |
122 | if args.dataparallel == "True":
123 |
124 | model = nn.DataParallel(
125 | UNETR(
126 | in_channels=1,
127 | out_channels=args.N_classes, #12 for all tissues
128 | img_size=(args.spatial_size, args.spatial_size, args.spatial_size),
129 | feature_size=16,
130 | hidden_size=768,
131 | mlp_dim=3072,
132 | num_heads=12,
133 | pos_embed="perceptron",
134 | norm_name="instance",
135 | res_block=True,
136 | dropout_rate=0.0,
137 | ), device_ids=[i for i in range(args.num_gpu)]).cuda()
138 |
139 | elif args.dataparallel == "False":
140 |
141 | #device = torch.device(f"cuda:{args.local_rank}")
142 | #torch.cuda.set_device(device)
143 | os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
144 | device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
145 |
146 | model = UNETR(
147 | in_channels=1,
148 | out_channels=args.N_classes, #12 for all tissues
149 | img_size=(args.spatial_size, args.spatial_size, args.spatial_size),
150 | feature_size=16,
151 | hidden_size=768,
152 | mlp_dim=3072,
153 | num_heads=12,
154 | pos_embed="perceptron",
155 | norm_name="instance",
156 | res_block=True,
157 | dropout_rate=0.0,
158 | ).to(device)
159 |
160 | loss_function = DiceCELoss(to_onehot_y=True, softmax=True)
161 | torch.backends.cudnn.benchmark = True
162 | optimizer = torch.optim.AdamW(model.parameters(), lr=1e-4, weight_decay=1e-5)
163 |
164 | #-----------------------------------
165 |
166 | model.load_state_dict(torch.load(os.path.join(args.data_dir, args.model_load_name)))
167 | model.eval()
168 |
169 | case_num = len(test_ds)
170 | for i in range(case_num):
171 | #start_time = time.time()
172 | with torch.no_grad():
173 | img_name = os.path.split(test_ds[i]["image_meta_dict"]["filename_or_obj"])[1]
174 | img = test_ds[i]["image"]
175 | test_inputs = torch.unsqueeze(img, 1).cuda()
176 | #start_time = time.time()
177 | test_outputs = sliding_window_inference(test_inputs, (args.spatial_size, args.spatial_size, args.spatial_size), 4, model, overlap=0.8)
178 | #print("--- %s seconds ---" % (time.time() - start_time))
179 | #logits = test_outputs.detach().cpu().numpy()
180 | testimage = torch.argmax(test_outputs, dim=1).detach().cpu().numpy()
181 | #savepath = 'testimage' + str(i) + '.mat'
182 | filename, file_extension = os.path.splitext(img_name)
183 | savepath = filename + '.mat'
184 | savemat(os.path.join(args.data_dir,savepath), {'testimage':testimage})#, 'logits':logits})
185 | #print("--- %s seconds ---" % (time.time() - start_time))
186 |
187 | #------------------------------------
188 |
189 | #time since start
190 |
191 | #print("--- %s seconds ---" % (time.time() - start_time))
192 |
--------------------------------------------------------------------------------
/tissue_cond_11.xlsx:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/lab-smile/DOMINO/3d261da4dec8ba338818babfa471c47c51d0866d/tissue_cond_11.xlsx
--------------------------------------------------------------------------------
/train.sh:
--------------------------------------------------------------------------------
1 | #!/bin/bash
2 | #SBATCH --job-name=train
3 | #SBATCH --mail-type=END,FAIL
4 | #SBATCH --mail-user=
5 | #SBATCH --ntasks=1
6 | #SBATCH --cpus-per-task=4
7 | #SBATCH --partition=hpg-ai
8 | #SBATCH --gres=gpu:a100:1
9 | #SBATCH --mem=90gb
10 | #SBATCH --time=72:00:00
11 | #SBATCH --output=%x_%j.log
12 |
13 | #module load pytorch
14 | module load singularity
15 | #module load monai
16 |
17 | # Check if cuda enabled
18 | singularity exec --nv /monaicore081 python3 -c "import torch; print(torch.cuda.is_available())"
19 |
20 | #run code
21 | #singularity exec --nv /monaicore08 python3 /train_domino.py --num_gpu 3
22 | #python3 /train_domino.py --num_gpu 3 --a_min_value 0 --a_max_value 255
23 |
24 | singularity exec --nv --bind :/mnt /monaicore081 python3 /mnt/train_domino.py --num_gpu 1 --data_dir '/mnt//' --model_save_name "unetr_v5_domino_06-20-22" --N_classes 12 --max_iteration 100
--------------------------------------------------------------------------------
 31 |
31 |  31 |
31 |