├── LICENSE
├── PCA
├── Principal Component Methods in R.ipynb
└── Principal Component Methods in R.r
├── README.md
├── Visualization
├── 基于R语言实现树形图的绘制
│ ├── 基于R语言实现树形图的绘制.ipynb
│ └── 基于R语言实现树形图的绘制.r
├── 基于R语言实现环状条形图的绘制
│ ├── 基于R语言实现环状条形图的绘制.ipynb
│ └── 基于R语言实现环状条形图的绘制.r
└── 数据绘图要点
│ ├── [数据分析与可视化] 数据绘图要点1-注重数据排序.ipynb
│ ├── [数据分析与可视化] 数据绘图要点10-图例的构建.ipynb
│ ├── [数据分析与可视化] 数据绘图要点11-雷达图的注意事项.ipynb
│ ├── [数据分析与可视化] 数据绘图要点12-图表注释的重要性.ipynb
│ ├── [数据分析与可视化] 数据绘图要点2-Y轴的开始与结束.ipynb
│ ├── [数据分析与可视化] 数据绘图要点3-意大利面条图.ipynb
│ ├── [数据分析与可视化] 数据绘图要点4-饼图的问题.ipynb
│ ├── [数据分析与可视化] 数据绘图要点5-误差线的问题.ipynb
│ ├── [数据分析与可视化] 数据绘图要点6-数据组过多.ipynb
│ ├── [数据分析与可视化] 数据绘图要点7-过度绘图.ipynb
│ ├── [数据分析与可视化] 数据绘图要点8-环状条形图的使用.ipynb
│ ├── [数据分析与可视化] 数据绘图要点9-颜色的选择.ipynb
│ └── image
│ ├── img5_1.png
│ ├── img8_1.png
│ ├── img9_1.png
│ ├── img9_2.png
│ ├── img9_3.png
│ ├── img9_4.png
│ ├── img9_5.png
│ ├── img9_6.png
│ ├── img9_7.png
│ ├── img9_8.png
│ └── img9_9.png
├── WGCNA
├── wgcna_tutorial.ipynb
└── wgcna_tutorial.r
└── ggplot2入门笔记
├── [R语言] ggplot2入门笔记1—ggplot2简要教程.ipynb
├── [R语言] ggplot2入门笔记1—ggplot2简要教程.r
├── [R语言] ggplot2入门笔记2—通用教程ggplot2简介.ipynb
├── [R语言] ggplot2入门笔记2—通用教程ggplot2简介.r
├── [R语言] ggplot2入门笔记3—通用教程如何自定义ggplot2.ipynb
├── [R语言] ggplot2入门笔记3—通用教程如何自定义ggplot2.r
├── [R语言] ggplot2入门笔记4—前50个ggplot2可视化效果.ipynb
└── [R语言] ggplot2入门笔记4—前50个ggplot2可视化效果.r
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2020 luohenyueji
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/PCA/Principal Component Methods in R.r:
--------------------------------------------------------------------------------
1 |
2 | # 调用R包
3 | library("FactoMineR");
4 | library("factoextra");
5 |
6 | data(decathlon2)
7 | head(decathlon2)
8 |
9 | decathlon2.active <- decathlon2[1:23, 1:10]
10 | head(decathlon2.active[, 1:6], 4)
11 |
12 | # 获得数据
13 | library("FactoMineR")
14 | library("factoextra")
15 | data(decathlon2)
16 | decathlon2.active <- decathlon2[1:23, 1:10]
17 |
18 | # PCA计算
19 | res.pca <- PCA(decathlon2.active, graph = FALSE)
20 | # 提取变量的分析结果
21 | var <- get_pca_var(res.pca)
22 | var
23 |
24 | # Coordinates of variables
25 | head(var$coord, 4)
26 | # col.var设定线条颜色
27 | fviz_pca_var(res.pca, col.var = "black")
28 |
29 | head(var$cos2)
30 | library("corrplot")
31 | # is.corr表示输入的矩阵不是相关系数矩阵
32 | corrplot(var$cos2, is.corr=FALSE)
33 |
34 | # Total cos2 of variables on Dim.1 and Dim.2
35 | # 在第一第二主成分是显示结果(通过值的叠加显示)
36 | fviz_cos2(res.pca, choice = "var", axes = 1:2)
37 |
38 | # Color by cos2 values: quality on the factor map
39 | fviz_pca_var(res.pca, col.var = "cos2",
40 | gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07"),
41 | repel = TRUE # Avoid text overlapping
42 | )
43 |
44 | # Change the transparency by cos2 values
45 | fviz_pca_var(res.pca, alpha.var = "cos2")
46 |
47 | head(var$contrib, 4)
48 | library("corrplot")
49 | corrplot(var$contrib, is.corr=FALSE)
50 |
51 | # Contributions of variables to PC1
52 | # 各变量对第一主成分的贡献
53 | fviz_contrib(res.pca, choice = "var", axes = 1, top = 10)
54 | # Contributions of variables to PC2
55 | # 各变量对第二主成分的贡献
56 | fviz_contrib(res.pca, choice = "var", axes = 2, top = 10)
57 |
58 | fviz_contrib(res.pca, choice = "var", axes = 1:2, top = 10)
59 |
60 | fviz_pca_var(res.pca, col.var = "contrib",
61 | gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07")
62 | )
63 |
64 | # Change the transparency by contrib values
65 | fviz_pca_var(res.pca, alpha.var = "contrib")
66 |
67 | # Create a random continuous variable of length 10
68 | # 生成随机数
69 | set.seed(123)
70 | my.cont.var <- rnorm(10)
71 | # Color variables by the continuous variable
72 | # col.var设置颜色
73 | # gradient.cols设置颜色渐变范围
74 | fviz_pca_var(res.pca, col.var = my.cont.var,
75 | gradient.cols = c("blue", "yellow", "red"),
76 | legend.title = "Cont.Var")
77 |
78 | # Create a grouping variable using kmeans
79 | # Create 3 groups of variables (centers = 3)
80 | set.seed(123)
81 | # 进行聚类
82 | # center聚类数量
83 | res.km <- kmeans(var$coord, centers = 3, nstart = 25)
84 | # 将向量编码为因子
85 | grp <- as.factor(res.km$cluster)
86 | # Color variables by groups
87 | fviz_pca_var(res.pca, col.var = grp,
88 | palette = c("#0073C2FF", "#EFC000FF", "#868686FF"),
89 | legend.title = "Cluster")
90 |
91 | #proba用于表征维度的显着性阈值,
92 | res.desc <- dimdesc(res.pca, axes = c(1,2), proba = 0.05)
93 | # Description of dimension 1 第一主成分
94 | res.desc$Dim.1
95 |
96 | # 第二主成分
97 | res.desc$Dim.2
98 |
99 | ind <- get_pca_ind(res.pca)
100 | ind
101 |
102 | fviz_pca_ind(res.pca)
103 |
104 | # Quality of individuals
105 | head(ind$cos2)
106 | # repel=TRUE能够避免部分重合的点重叠
107 | fviz_pca_ind(res.pca, col.ind = "cos2",
108 | gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07"),
109 | repel = TRUE # Avoid text overlapping (slow if many points)
110 | )
111 |
112 | fviz_pca_ind(res.pca, col.ind = "cos2", pointsize = "cos2",
113 | gradient.cols = c("#00AFBB", "#E7B800", "#FC4E07"),
114 | repel = TRUE # Avoid text overlapping (slow if many points)
115 | )
116 |
117 | fviz_cos2(res.pca, choice = "ind")
118 |
119 | # Total contribution on PC1 and PC2
120 | fviz_contrib(res.pca, choice = "ind", axes = 1:2)
121 |
122 | head(iris, 3)
123 |
124 | # The variable Species (index = 5) is removed before PCA analysis
125 | # 第5列不进行PCA运算
126 | iris.pca <- PCA(iris[,-5], graph = FALSE)
127 |
128 | fviz_pca_ind(iris.pca,
129 | # show points only (nbut not "text") 只显示点而不显示文本,默认都显示
130 | geom.ind = "point",
131 | # 设定分类种类
132 | col.ind = iris$Species,
133 | # 设定颜色
134 | palette = c("#00AFBB", "#E7B800", "#FC4E07"),
135 | # 添加椭圆 Concentration ellipses
136 | addEllipses = TRUE,
137 | legend.title = "Groups",
138 | )
139 |
140 | fviz_pca_ind(iris.pca,
141 | label = "none", # hide individual labels
142 | habillage = iris$Species, # color by groups
143 | addEllipses = TRUE, # Concentration ellipses
144 | palette = "jco"
145 | )
146 |
147 | # Variables on dimensions 2 and 3
148 | fviz_pca_var(res.pca, axes = c(2, 3))
149 | # Individuals on dimensions 2 and 3
150 | fviz_pca_ind(res.pca, axes = c(2, 3))
151 |
152 | # Show variable points and text labels
153 | fviz_pca_var(res.pca, geom.var = c("point", "text"))
154 |
155 | # Show individuals text labels only
156 | fviz_pca_ind(res.pca, geom.ind = "text")
157 |
158 | # Change the size of arrows an labels
159 | fviz_pca_var(res.pca, arrowsize = 1, labelsize = 5,
160 | repel = TRUE)
161 | # Change points size, shape and fill color
162 | # Change labelsize
163 | fviz_pca_ind(res.pca,
164 | pointsize = 3, pointshape = 21, fill = "lightblue",
165 | labelsize = 5, repel = TRUE)
166 |
167 | # Add confidence ellipses
168 | fviz_pca_ind(iris.pca, geom.ind = "point",
169 | # 使用iris数据集
170 | col.ind = iris$Species, # color by groups
171 | palette = c("#00AFBB", "#E7B800", "#FC4E07"),
172 | addEllipses = TRUE, ellipse.type = "confidence",
173 | legend.title = "Groups"
174 | )
175 | # Convex hull
176 | fviz_pca_ind(iris.pca, geom.ind = "point",
177 | col.ind = iris$Species, # color by groups
178 | palette = c("#00AFBB", "#E7B800", "#FC4E07"),
179 | # 用凸包多边形代替椭圆
180 | addEllipses = TRUE, ellipse.type = "convex",
181 | legend.title = "Groups"
182 | )
183 |
184 |
185 | fviz_pca_var(res.pca, axes.linetype = "dotted")
186 |
187 | ind.p <- fviz_pca_ind(iris.pca, geom = "point", col.ind = iris$Species)
188 | ggpubr::ggpar(ind.p,
189 | title = "Principal Component Analysis",
190 | # 下标题
191 | subtitle = "Iris data set",
192 | # 说明
193 | caption = "Source: factoextra",
194 | # x,y轴标题
195 | xlab = "PC1", ylab = "PC2",
196 | # 标题名字位置
197 | legend.title = "Species", legend.position = "top",
198 | # 主题和配设
199 | ggtheme = theme_gray(), palette = "jco"
200 | )
201 |
202 | fviz_pca_biplot(res.pca, repel = TRUE,
203 | col.var = "#2E9FDF", # Variables color
204 | col.ind = "#696969" # Individuals color
205 | )
206 |
207 | fviz_pca_biplot(iris.pca,
208 | # 观测量颜色
209 | col.ind = iris$Species, palette = "jco",
210 | # 添加椭圆
211 | addEllipses = TRUE, label = "var",
212 | # 线条颜色
213 | col.var = "black", repel = TRUE,
214 | legend.title = "Species")
215 |
216 | fviz_pca_biplot(iris.pca,
217 | # Fill individuals by groups
218 | geom.ind = "point",
219 | # 点的形状
220 | pointshape = 21,
221 | # 点的大小
222 | pointsize = 2.5,
223 | # 按照组类特定形状
224 | fill.ind = iris$Species,
225 | col.ind = "black",
226 | # Color variable by groups
227 | # 颜色
228 | col.var = factor(c("sepal", "sepal", "petal", "petal")),
229 | # 标题
230 | legend.title = list(fill = "Species", color = "Clusters"),
231 | repel = TRUE # Avoid label overplotting
232 | )+
233 | ggpubr::fill_palette("jco")+ # Indiviual fill color
234 | ggpubr::color_palette("npg") # Variable colors
235 |
236 | fviz_pca_biplot(iris.pca,
237 | # Individuals
238 | geom.ind = "point",
239 | fill.ind = iris$Species, col.ind = "black",
240 | pointshape = 21, pointsize = 2,
241 | palette = "jco",
242 | addEllipses = TRUE,
243 | # Variables
244 | alpha.var ="contrib", col.var = "contrib",
245 | gradient.cols = "RdYlBu",
246 |
247 | legend.title = list(fill = "Species", color = "Contrib",
248 | alpha = "Contrib")
249 | )
250 |
251 | # Visualize variable with cos2 >= 0.6
252 | # 可视化cos2>0.6
253 | fviz_pca_var(res.pca, select.var = list(cos2 = 0.6))
254 |
255 | # Select by names
256 | # 根据名字显示
257 | name <- list(name = c("Long.jump", "High.jump", "X100m"))
258 | fviz_pca_var(res.pca, select.var = name)
259 |
260 | # 根据前五贡献
261 | # top 5 contributing individuals and variable
262 | fviz_pca_biplot(res.pca, select.ind = list(contrib = 5),
263 | select.var = list(contrib = 5),
264 | ggtheme = theme_minimal())
265 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # R-Study-Notes
2 | 我的R语言代码笔记和代码仓库
3 |
4 | ---
5 |
6 | R语言使用平台为jupyter notebook,提供.ipynb和.r文件源码,推荐使用jupyter notebook,.r文件只提供代码没有相关笔记。如果没有相关环境,查看我的个人博客有对应的笔记和代码。
7 | **个人博客:[LuohenYJ](https://blog.csdn.net/luohenyj/category_9152388.html)**
--------------------------------------------------------------------------------
/Visualization/基于R语言实现树形图的绘制/基于R语言实现树形图的绘制.r:
--------------------------------------------------------------------------------
1 |
2 | # libraries
3 | # 包
4 | library(ggraph)
5 | library(igraph)
6 | library(tidyverse)
7 |
8 | # create an edge list data frame giving the hierarchical structure of your individuals
9 | # 创建层级数据
10 | d1 <- data.frame(from="origin", to=paste("group", seq(1,3), sep=""))
11 | d1
12 | d2 <- data.frame(from=rep(d1$to, each=3), to=paste("subgroup", seq(1,9), sep="_"))
13 | d2
14 | # 汇总
15 | edges <- rbind(d1, d2)
16 | edges
17 |
18 | # Create a graph object
19 | mygraph <- graph_from_data_frame( edges )
20 | mygraph
21 |
22 | # Basic tree
23 | # 基础树形图
24 | # layout表示布局方式,circular表示是否为环状树形图
25 | ggraph(mygraph, layout = 'dendrogram', circular = FALSE) +
26 | # 画边
27 | geom_edge_diagonal() +
28 | # 画节点
29 | geom_node_point() +
30 | # 设置主题
31 | theme_void()
32 |
33 | # libraries
34 | library(ggraph)
35 | library(igraph)
36 | library(tidyverse)
37 |
38 | # create a data frame
39 | data <- data.frame(
40 | level1="CEO",
41 | level2=c( rep("boss1",4), rep("boss2",4)),
42 | level3=paste0("mister_", letters[1:8])
43 | )
44 | data
45 |
46 | # transform it to a edge list!
47 | edges_level1_2 <- data %>% select(level1, level2) %>% unique %>% rename(from=level1, to=level2)
48 | edges_level2_3 <- data %>% select(level2, level3) %>% unique %>% rename(from=level2, to=level3)
49 | edges_level1_2
50 | edges_level2_3
51 | edge_list=rbind(edges_level1_2, edges_level2_3)
52 | edge_list
53 |
54 | # Now we can plot that
55 | mygraph<- graph_from_data_frame( edge_list )
56 | ggraph(mygraph, layout = 'dendrogram', circular = FALSE) +
57 | geom_edge_diagonal() +
58 | geom_node_point() +
59 | theme_void()
60 |
61 | # Libraries
62 | library(ggraph)
63 | library(igraph)
64 | library(tidyverse)
65 | theme_set(theme_void())
66 |
67 | # data: edge list
68 | # 边数据
69 | d1 <- data.frame(from="origin", to=paste("group", seq(1,7), sep=""))
70 | d2 <- data.frame(from=rep(d1$to, each=7), to=paste("subgroup", seq(1,49), sep="_"))
71 | edges <- rbind(d1, d2)
72 |
73 | # We can add a second data frame with information for each node!
74 | # 为每个节点设置信息
75 | name <- unique(c(as.character(edges$from), as.character(edges$to)))
76 | # 设置每个节点对应的聚类信息和值
77 | vertices <- data.frame(
78 | name=name,
79 | group=c( rep(NA,8) , rep( paste("group", seq(1,7), sep=""), each=7)),
80 | cluster=sample(letters[1:4], length(name), replace=T),
81 | value=sample(seq(10,30), length(name), replace=T)
82 | )
83 | vertices[0:10,]
84 | # Create a graph object
85 | mygraph <- graph_from_data_frame( edges, vertices=vertices)
86 |
87 | # 线形布局
88 | ggraph(mygraph, layout = 'dendrogram', circular = FALSE) +
89 | geom_edge_diagonal()
90 | # 环形布局
91 | ggraph(mygraph, layout = 'dendrogram', circular = TRUE) +
92 | geom_edge_diagonal()
93 |
94 | # 折线
95 | ggraph(mygraph, layout = 'dendrogram') +
96 | geom_edge_link()
97 | # 弧线
98 | ggraph(mygraph, layout = 'dendrogram') +
99 | geom_edge_diagonal()
100 |
101 | ggraph(mygraph, layout = 'dendrogram') +
102 | # 设置边
103 | geom_edge_diagonal() +
104 | # 设置节点名,label表示节点名,filter=leaf表示跳过叶子节点,angle标签方向,hjust和nudge_y标签和节点距离
105 | geom_node_text(aes( label=name, filter=leaf) , angle=90 , hjust=1, nudge_y = -0.01) +
106 | # 设置y轴范围
107 | ylim(-.4, NA)
108 |
109 | ggraph(mygraph, layout = 'dendrogram') +
110 | geom_edge_diagonal() +
111 | geom_node_text(aes( label=name, filter=leaf) , angle=90 , hjust=1, nudge_y = -0.04) +
112 | # 为每个节点添加点
113 | geom_node_point(aes(filter=leaf) , alpha=0.6) +
114 | ylim(-.5, NA)
115 |
116 | ggraph(mygraph, layout = 'dendrogram') +
117 | geom_edge_diagonal() +
118 | geom_node_text(aes( label=name, filter=leaf, color=group) , angle=90 , hjust=1, nudge_y=-0.1) +
119 | geom_node_point(aes(filter=leaf, size=value, color=group) , alpha=0.6) +
120 | ylim(-.6, NA) +
121 | theme(legend.position="none")
122 |
123 | # Libraries
124 | library(ggraph)
125 | library(igraph)
126 | library(tidyverse)
127 | library(RColorBrewer)
128 |
129 | # 创建数据,类似前面的步骤
130 | # create a data frame giving the hierarchical structure of your individuals
131 | d1=data.frame(from="origin", to=paste("group", seq(1,5), sep=""))
132 | d2=data.frame(from=rep(d1$to, each=5), to=paste("subgroup", seq(1,25), sep="_"))
133 | edges=rbind(d1, d2)
134 |
135 | # create a vertices data.frame. One line per object of our hierarchy
136 | # 为每个节点添加值
137 | vertices = data.frame(
138 | name = unique(c(as.character(edges$from), as.character(edges$to))),
139 | # 正态分布随机取值,共获得31个值。如果是其他数据,去掉value = runif(31),查看运行后的dim(vertices)就知道该填多少了
140 | value = runif(31)
141 | )
142 | # Let's add a column with the group of each name. It will be useful later to color points
143 | # 为每个节点添加分组信息
144 | vertices$group = edges$from[ match( vertices$name, edges$to ) ]
145 | dim(vertices)
146 | head(vertices)
147 |
148 | # Let's add information concerning the label we are going to add: angle, horizontal adjustement and potential flip calculate the ANGLE of the labels
149 | # 让我们添加有关我们将要添加的标签的信息:角度、水平调整和翻转,计算标签的角度
150 | # 添加id值
151 | vertices$id=NA
152 | myleaves=which(is.na( match(vertices$name, edges$from) ))
153 | nleaves=length(myleaves)
154 | vertices$id[ myleaves ] = seq(1:nleaves)
155 | # 添加角度
156 | vertices$angle= -360 * vertices$id / nleaves
157 | vertices
158 |
159 | # calculate the alignment of labels: right or left
160 | # 判断标签是偏向左边还是右边
161 | # hjust表示是否水平翻转
162 | vertices$hjust<-ifelse(vertices$angle < -90 & vertices$angle > -270, 1, 0)
163 |
164 | # flip angle BY to make them readable
165 | # 是否翻转标签
166 | vertices$angle<-ifelse(vertices$angle < -90 & vertices$angle > -270, vertices$angle+180, vertices$angle)
167 | vertices[12:20,]
168 | # Create a graph object
169 | # 创建图
170 | mygraph <- graph_from_data_frame( edges, vertices=vertices )
171 |
172 | vertices
173 |
174 | # Make the plot
175 | p<-ggraph(mygraph, layout = 'dendrogram', circular = TRUE) +
176 | # 设置边
177 | geom_edge_diagonal(colour="grey") +
178 | # 设置边的颜色
179 | scale_edge_colour_distiller(palette = "RdPu") +
180 | # 设置点的标签
181 | geom_node_text(aes(x = x*1.15, y=y*1.15, filter = leaf, label=name, angle = angle, hjust=hjust, colour=group), size=2.7, alpha=1) +
182 | # 设置点的形状
183 | geom_node_point(aes(filter = leaf, x = x*1.07, y=y*1.07, colour=group, size=value, alpha=0.2)) +
184 | # 控制颜色
185 | scale_colour_manual(values= rep( brewer.pal(9,"Paired") , 30)) +
186 | scale_size_continuous( range = c(0.1,10) ) +
187 | theme_void() +
188 | theme(
189 | # 不显示图例
190 | legend.position="none",
191 | plot.margin=unit(c(0,0,0,0),"cm"),
192 | ) +
193 | expand_limits(x = c(-1.3, 1.3), y = c(-1.3, 1.3))
194 | p
195 | # 保存数据 Save at png
196 | ggsave(p, file="output.png", width=10, height=10,dpi=300)
197 |
198 | # Dataset
199 | data <- matrix( sample(seq(1,2000),200), ncol = 10 )
200 | rownames(data) <- paste0("sample_" , seq(1,20))
201 | colnames(data) <- paste0("variable",seq(1,10))
202 | data
203 | dim(data)
204 |
205 | # Euclidean distance
206 | # 计算欧式距离
207 | dist <- dist(data[ , c(4:8)] , diag=TRUE)
208 |
209 | # Hierarchical Clustering with hclust
210 | # 分层聚类
211 | hc <- hclust(dist)
212 |
213 | # Plot the result
214 | plot(hc)
215 |
216 | # store the dedrogram in an object
217 | # 保存聚类结果为dhc变量
218 | dhc <- as.dendrogram(hc)
219 |
220 | # set the margin
221 | par(mar=c(4,4,2,2))
222 | # 打印会告诉你分支情况
223 | print(dhc[[2]])
224 | # Plot the Second group
225 | # 绘图
226 | plot(dhc[[2]] , main= "zoom on a part of the dendrogram")
227 |
228 | # store the dedrogram in an object
229 | dhc <- as.dendrogram(hc)
230 |
231 | # set the margin
232 | par(mar=c(4,4,2,2))
233 |
234 | print(dhc[[2]][[1]])
235 | # Plot the Second group
236 | plot(dhc[[2]][[1]] , main= "zoom on a part of the dendrogram")
237 |
238 | # Build dataset (just copy and paste, this is NOT interesting)
239 | # 生成数据,可以跳过
240 | sample <- paste(rep("sample_",24) , seq(1,24) , sep="")
241 | specie <- c(rep("dicoccoides" , 8) , rep("dicoccum" , 8) , rep("durum" , 8))
242 | treatment <- rep(c(rep("High",4 ) , rep("Low",4)),3)
243 | data <- data.frame(sample,specie,treatment)
244 | for (i in seq(1:5)){
245 | gene=sample(c(1:40) , 24 )
246 | data=cbind(data , gene)
247 | colnames(data)[ncol(data)]=paste("gene_",i,sep="")
248 | }
249 | data[data$treatment=="High" , c(4:8)]=data[data$treatment=="High" , c(4:8)]+100
250 | data[data$specie=="durum" , c(4:8)]=data[data$specie=="durum" , c(4:8)]-30
251 | rownames(data) <- data[,1]
252 | head(data)
253 |
254 | # Compute Euclidean distance between samples
255 | dist=dist(data[ , c(4:8)] , diag=TRUE)
256 |
257 | # Perfor clustering with hclust
258 | # 聚类并保存结果
259 | hc <- hclust(dist)
260 | dhc <- as.dendrogram(hc)
261 | dhc
262 |
263 | # Actually, each leaf of the tree has several attributes, like the color, the shape.. Have a look to it:
264 | # 选择特别的节点
265 | specific_leaf <- dhc[[1]][[1]][[1]]
266 | specific_leaf
267 | attributes(specific_leaf)
268 |
269 | i=0
270 | colLab<-function(n)
271 | {
272 | # 判断是否为节点
273 | if(is.leaf(n))
274 | {
275 | # 获得节点的属性
276 | a=attributes(n)
277 |
278 | # I deduce the line in the original data, and so the treatment and the specie.
279 | # 获得该点的信息
280 | ligne=match(attributes(n)$label,data[,1])
281 | # 根据自己的结果设置
282 | treatment=data[ligne,3];
283 | if(treatment=="Low"){col_treatment="blue"};if(treatment=="High"){col_treatment="red"}
284 | # 根据种类设置颜色,根据自己的结果设置
285 | specie=data[ligne,2];
286 | if(specie=="dicoccoides"){col_specie="red"};if(specie=="dicoccum"){col_specie="Darkgreen"};if(specie=="durum"){col_specie="blue"}
287 |
288 | # M odification of leaf attribute
289 | # 修改节点的属性
290 | attr(n,"nodePar")<-c(a$nodePar,list(cex=1.5,lab.cex=1,pch=20,col=col_treatment,lab.col=col_specie,lab.font=1,lab.cex=1))
291 | }
292 | return(n)
293 | }
294 |
295 | # 应用函数
296 | dL <- dendrapply(dhc, colLab)
297 |
298 | # And the plot
299 | plot(dL , main="structure of the population")
300 | # 图例
301 | legend("topright",
302 | # 文字
303 | legend = c("High Nitrogen" , "Low Nitrogen" , "Durum" , "Dicoccoides" , "Dicoccum"),
304 | # 颜色
305 | col = c("red", "blue" , "blue" , "red" , "Darkgreen"),
306 | pch = c(20,20,4,4,4), bty = "n", pt.cex = 1.5, cex = 0.8 ,
307 | text.col = "black", horiz = FALSE, inset = c(0, 0.1))
308 |
309 | # Library
310 | library(tidyverse)
311 |
312 | # Data
313 | head(mtcars)
314 |
315 | # Clusterisation using 3 variables
316 | # 聚类,使用管道
317 | mtcars %>%
318 | select(mpg, cyl, disp) %>%
319 | dist() %>%
320 | hclust() %>%
321 | as.dendrogram() -> dend
322 |
323 | # Plot
324 | # 绘图
325 | par(mar=c(7,3,1,1)) # Increase bottom margin to have the complete label
326 | plot(dend)
327 |
328 | # library
329 | library(dendextend)
330 |
331 | # 绘图dend是设置函数
332 | dend %>%
333 | # Custom branches
334 | # 自定义树枝的颜色
335 | set("branches_col", "red") %>%
336 | # 自定义树枝宽度
337 | set("branches_lwd", 3) %>%
338 | # Custom labels
339 | # 自定义标签颜色
340 | set("labels_col", "blue") %>%
341 | # 自定义标签字体大小
342 | set("labels_cex", 0.8) %>%
343 | plot()
344 |
345 | dend %>%
346 | # 自定义树枝节点形状
347 | set("nodes_pch", 20) %>%
348 | # 自定义树枝节点大小
349 | set("nodes_cex", 1.5) %>%
350 | # 自定义树枝节点颜色
351 | set("nodes_col", "red") %>%
352 | plot()
353 |
354 | dend %>%
355 | # 最后一层节点形状
356 | set("leaves_pch", 22) %>%
357 | # 最后一层节点宽度
358 | set("leaves_cex", 1) %>%
359 | # 最后一层节点颜色
360 | set("leaves_col", "red") %>%
361 | plot()
362 |
363 |
364 | par(mar=c(1,1,1,7))
365 | dend %>%
366 | # 根据第一层分支结果自定义标签颜色
367 | set("labels_col", value = c("skyblue", "orange", "grey"), k=3) %>%
368 | # 根据第一层分支结果自定义分支颜色
369 | set("branches_k_color", value = c("skyblue", "orange", "grey"), k = 3) %>%
370 | # horize是否水平放置,axes是否显示旁边的距离尺
371 | plot(horiz=TRUE, axes=FALSE)
372 | # 画线条,v高度,lty线条类型
373 | abline(v = 350, lty = 2)
374 |
375 | # 使用
376 | par(mar=c(9,1,1,1))
377 | dend %>%
378 | set("labels_col", value = c("skyblue", "orange", "grey"), k=3) %>%
379 | set("branches_k_color", value = c("skyblue", "orange", "grey"), k = 3) %>%
380 | plot(axes=FALSE)
381 | # 画矩形框
382 | # k表示将类切割为k个簇,lty矩形框线条类型,lwd矩形框线条宽度,col填充颜色,x表示从第几个类开始画簇
383 | rect.dendrogram( dend, k=3, lty = 2, lwd = 5, x=17, col=rgb(0.1, 0.2, 0.4, 0.1) )
384 |
385 |
386 | # Create a vector of colors, darkgreen if am is 0, green if 1.
387 | # 获得数据,如果am为0就是forestgreen颜色
388 | my_colors <- ifelse(mtcars$am==0, "forestgreen", "green")
389 |
390 | # Make the dendrogram
391 | # 设置图像空白区域
392 | par(mar=c(10,1,1,1))
393 | dend %>%
394 | set("labels_col", value = c("skyblue", "orange", "grey"), k=3) %>%
395 | set("branches_k_color", value = c("skyblue", "orange", "grey"), k = 3) %>%
396 | set("leaves_pch", 19) %>%
397 | set("nodes_cex", 0.7) %>%
398 | plot(axes=FALSE,horiz =FALSE)
399 |
400 | # Add the colored bar
401 | # 添加颜色bar
402 | # colors颜色,dend聚类图,rowLabels名字
403 | colored_bars(colors = my_colors, dend = dend, rowLabels = "am",horiz =FALSE)
404 |
405 | # Make 2 dendrograms, using 2 different clustering methods
406 | # 使用两种完全不同的聚类方法
407 | d1 <- USArrests %>% dist() %>% hclust( method="average" ) %>% as.dendrogram()
408 | d2 <- USArrests %>% dist() %>% hclust( method="complete" ) %>% as.dendrogram()
409 |
410 | # Custom these kendo, and place them in a list
411 | # 定制树列表
412 | dl <- dendlist(
413 | d1 %>%
414 | set("labels_col", value = c("skyblue", "orange", "grey"), k=3) %>%
415 | set("branches_lty", 1) %>%
416 | set("branches_k_color", value = c("skyblue", "orange", "grey"), k = 3),
417 | d2 %>%
418 | set("labels_col", value = c("skyblue", "orange", "grey"), k=3) %>%
419 | set("branches_lty", 1) %>%
420 | set("branches_k_color", value = c("skyblue", "orange", "grey"), k = 3)
421 | )
422 |
423 | # Plot them together
424 | tanglegram(dl,
425 | # 子树是否带颜色
426 | common_subtrees_color_lines = FALSE,
427 | # 是否突出显示边
428 | highlight_distinct_edges = TRUE,
429 | # 是否突出分支
430 | highlight_branches_lwd=FALSE,
431 | # 两个树的距离
432 | margin_inner=7,
433 | # 两个树之间线条宽度
434 | lwd=2
435 | )
436 |
--------------------------------------------------------------------------------
/Visualization/基于R语言实现环状条形图的绘制/基于R语言实现环状条形图的绘制.r:
--------------------------------------------------------------------------------
1 |
2 | # Libraries
3 | # 导入包
4 | library(tidyverse)
5 |
6 | # Create dataset
7 | # 创建数据
8 | data <- data.frame(
9 | id=seq(1,60),
10 | individual=paste( "Mister ", seq(1,60), sep=""),
11 | value=sample( seq(10,100), 60, replace=T)
12 | )
13 | head(data)
14 |
15 | # Make the plot
16 | # 画图
17 | p <- ggplot(data, aes(x=as.factor(id), y=value)) +
18 | # This add the bars with a blue color
19 | # 添加蓝色条形,stat表示数据统计方式,也就是说identity提取横坐标x对应的y值
20 | geom_bar(stat="identity", fill=alpha("blue", 0.3)) +
21 | # The negative value controls the size of the inner circle, the positive one is useful to add size over each bar
22 | # 设置y的范围,负值设定内圆的大小,正值设定各个条柱的最高高度
23 | ylim(-100,120)+
24 | # theme_minimal简约主题
25 | theme_minimal() +
26 | # Custom the theme: no axis title and no cartesian grid
27 | # 自定义主题
28 | theme(
29 | # 移除标题坐标文字
30 | axis.text = element_blank(),
31 | axis.title = element_blank(),
32 | # 移除网格
33 | panel.grid = element_blank(),
34 | # This remove unnecessary margin around plot
35 | # 移除不必要空白
36 | plot.margin = unit(rep(-2,4), "cm"))+
37 | # This makes the coordinate polar instead of cartesian.
38 | # 使用极坐标系
39 | coord_polar(start = 0)
40 | p
41 |
42 | # Libraries
43 | library(tidyverse)
44 |
45 | # Create dataset
46 | # 创建数据
47 | data <- data.frame(
48 | id=seq(1,60),
49 | individual=paste( "Mister ", seq(1,60), sep=""),
50 | value=sample( seq(10,100), 60, replace=T)
51 | )
52 | # ----- This section prepare a dataframe for labels ---- #
53 | # 准备数据标签
54 | # Get the name and the y position of each label
55 | label_data <- data
56 | # calculate the ANGLE of the labels
57 | # 计算标签角度
58 | number_of_bar <- nrow(label_data)
59 | number_of_bar
60 |
61 | # I substract 0.5 because the letter must have the angle of the center of the bars. Not extreme right(1) or extreme left (0)
62 | # 减去0.5是为了让标签位于条柱中心
63 | # angle是标签角度
64 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
65 |
66 | # calculate the alignment of labels: right or left
67 | # If I am on the left part of the plot, my labels have currently an angle < -90
68 | # 判断标签左对齐还是右对齐,也就是标签是朝向左边还是右边
69 | label_data$hjust<-ifelse( angle < -90, 1, 0)
70 |
71 | # flip angle BY to make them readable
72 | # 翻转标签
73 | label_data$angle<-ifelse(angle < -90, angle+180, angle)
74 | # ----- ------------------------------------------- ---- #
75 | head(label_data)
76 |
77 |
78 | # Start the plot
79 | # 开始绘图
80 | p <- ggplot(data, aes(x=as.factor(id), y=value)) +
81 | # This add the bars with a bskyblue color
82 | # 添加蓝色条形,stat表示数据统计方式,也就是说identity提取横坐标x对应的y值
83 | geom_bar(stat="identity", fill=alpha("skyblue", 0.7)) +
84 |
85 | # The negative value controls the size of the inner circle, the positive one is useful to add size over each bar
86 | # 设置y的范围,负值设定内圆的大小,正值设定各个条柱的最高高度
87 | ylim(-100,120)+
88 |
89 | # theme_minimal简约主题
90 | theme_minimal() +
91 | # Custom the theme: no axis title and no cartesian grid
92 | # 自定义主题
93 | theme(
94 | # 移除标题坐标文字
95 | axis.text = element_blank(),
96 | axis.title = element_blank(),
97 | # 移除网格
98 | panel.grid = element_blank(),
99 | # This remove unnecessary margin around plot
100 | # 移除不必要空白
101 | plot.margin = unit(rep(-2,4), "cm"))+
102 |
103 | # This makes the coordinate polar instead of
104 | # 设置极坐标系
105 | coord_polar(start = 0) +
106 |
107 | # Add the labels, using the label_data dataframe that we have created before
108 | # 添加标签
109 | geom_text(data=label_data, aes(x=id, y=value+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=2.5, angle= label_data$angle, inherit.aes = FALSE )
110 |
111 | p
112 |
113 | # library
114 | library(tidyverse)
115 |
116 | # Create dataset
117 | # 添加数据
118 | data <- data.frame(
119 | individual=paste( "Mister ", seq(1,60), sep=""),
120 | value=sample( seq(10,100), 60, replace=T)
121 | )
122 |
123 | # Set a number of 'empty bar'
124 | # 设置空白柱的个数
125 | empty_bar <- 10
126 |
127 | # 在原始数据中添加空白数据
128 | # Add lines to the initial dataset
129 | to_add <- matrix(NA, empty_bar, ncol(data))
130 | colnames(to_add) <- colnames(data)
131 | data <- rbind(data, to_add)
132 | data$id <- seq(1, nrow(data))
133 |
134 | # Get the name and the y position of each label
135 | # 和上一步一样,获得标签角度信息
136 | label_data <- data
137 | number_of_bar <- nrow(label_data)
138 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
139 | label_data$hjust <- ifelse( angle < -90, 1, 0)
140 | label_data$angle <- ifelse(angle < -90, angle+180, angle)
141 | head(label_data)
142 |
143 | # Make the plot
144 | # 绘图
145 | p <- ggplot(data, aes(x=as.factor(id), y=value)) + # Note that id is a factor. If x is numeric, there is some space between the first bar
146 | geom_bar(stat="identity", fill=alpha("green", 0.3)) +
147 | ylim(-100,120) +
148 | theme_minimal() +
149 | theme(
150 | axis.text = element_blank(),
151 | axis.title = element_blank(),
152 | panel.grid = element_blank(),
153 | plot.margin = unit(rep(-1,4), "cm")
154 | ) +
155 | coord_polar(start = 0) +
156 | geom_text(data=label_data, aes(x=id, y=value+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=2.5, angle= label_data$angle, inherit.aes = FALSE )
157 |
158 | p;
159 |
160 | # library
161 | library(tidyverse)
162 |
163 | # Create dataset
164 | # 创建数据集
165 | data <- data.frame(
166 | individual=paste( "Mister ", seq(1,60), sep=""),
167 | group=c( rep('A', 10), rep('B', 30), rep('C', 14), rep('D', 6)) ,
168 | value=sample( seq(10,100), 60, replace=T)
169 | )
170 |
171 | # Set a number of 'empty bar' to add at the end of each group
172 | # 在原始数据中添加空白数据
173 | # empty_bar 表示组之间的空白距离
174 | empty_bar <- 4
175 | # 每一组之间4个空白
176 | to_add <- data.frame( matrix(NA, empty_bar*nlevels(data$group), ncol(data)) )
177 | colnames(to_add) <- colnames(data)
178 | # 为每个空白值提供组信息,rep函数的意思就是复制值,levels(data$group)为复制的对象,each为复制的次数
179 | to_add$group <- rep(levels(data$group), each=empty_bar)
180 | head(to_add)
181 |
182 | colnames(to_add) <- colnames(data)
183 | to_add$group <- rep(levels(data$group), each=empty_bar)
184 | data <- rbind(data, to_add)
185 | # 管道操作类似 data<-arrange(data,data$group)
186 | data <- data %>% arrange(group)
187 | # 设置id
188 | data$id <- seq(1, nrow(data))
189 | head(data)
190 |
191 | # Get the name and the y position of each label
192 | # 设定角度值
193 | label_data <- data
194 | number_of_bar <- nrow(label_data)
195 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
196 | label_data$hjust <- ifelse( angle < -90, 1, 0)
197 | label_data$angle <- ifelse(angle < -90, angle+180, angle)
198 |
199 | # Make the plot
200 | # fill 按组填充颜色
201 | p <- ggplot(data, aes(x=as.factor(id), y=value, fill=group)) +
202 | geom_bar(stat="identity", alpha=0.5) +
203 | ylim(-100,120) +
204 | theme_minimal() +
205 | theme(
206 | legend.position = "none",
207 | axis.text = element_blank(),
208 | axis.title = element_blank(),
209 | panel.grid = element_blank(),
210 | plot.margin = unit(rep(-1,4), "cm")
211 | ) +
212 | coord_polar() +
213 | geom_text(data=label_data, aes(x=id, y=value+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=2.5, angle= label_data$angle, inherit.aes = FALSE )
214 | p
215 |
216 | # library
217 | library(tidyverse)
218 |
219 | # Create dataset
220 | # 创建数据集
221 | data <- data.frame(
222 | individual=paste( "Mister ", seq(1,60), sep=""),
223 | group=c( rep('A', 10), rep('B', 30), rep('C', 14), rep('D', 6)) ,
224 | value=sample( seq(10,100), 60, replace=T)
225 | )
226 |
227 | # Set a number of 'empty bar' to add at the end of each group
228 | # 在原始数据中添加空白数据
229 | # empty_bar 表示组之间的空白距离
230 | empty_bar <- 4
231 | # 每一组之间4个空白
232 | to_add <- data.frame( matrix(NA, empty_bar*nlevels(data$group), ncol(data)) )
233 | colnames(to_add) <- colnames(data)
234 | # 为每个空白值提供组信息,rep函数的意思就是复制值,levels(data$group)为复制的对象,each为复制的次数
235 | to_add$group <- rep(levels(data$group), each=empty_bar)
236 | head(to_add)
237 |
238 | colnames(to_add) <- colnames(data)
239 | to_add$group <- rep(levels(data$group), each=empty_bar)
240 | data <- rbind(data, to_add)
241 | # 管道操作类似 data<-arrange(data,data$group)
242 | data <- data %>% arrange(group, value)
243 | # 设置id
244 | data$id <- seq(1, nrow(data))
245 | head(data)
246 |
247 | # Get the name and the y position of each label
248 | # 设定角度值
249 | label_data <- data
250 | number_of_bar <- nrow(label_data)
251 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
252 | label_data$hjust <- ifelse( angle < -90, 1, 0)
253 | label_data$angle <- ifelse(angle < -90, angle+180, angle)
254 |
255 | # Make the plot
256 | # fill 按组填充颜色
257 | p <- ggplot(data, aes(x=as.factor(id), y=value, fill=group)) +
258 | geom_bar(stat="identity", alpha=0.5) +
259 | ylim(-100,120) +
260 | theme_minimal() +
261 | theme(
262 | legend.position = "none",
263 | axis.text = element_blank(),
264 | axis.title = element_blank(),
265 | panel.grid = element_blank(),
266 | plot.margin = unit(rep(-1,4), "cm")
267 | ) +
268 | coord_polar() +
269 | geom_text(data=label_data, aes(x=id, y=value+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=2.5, angle= label_data$angle, inherit.aes = FALSE )
270 | p
271 |
272 | # library
273 | library(tidyverse)
274 |
275 | # Create dataset
276 | data <- data.frame(
277 | individual=paste( "Mister ", seq(1,60), sep=""),
278 | group=c( rep('A', 10), rep('B', 30), rep('C', 14), rep('D', 6)) ,
279 | value=sample( seq(10,100), 60, replace=T)
280 | )
281 |
282 | # Set a number of 'empty bar' to add at the end of each group
283 | empty_bar <- 3
284 | to_add <- data.frame( matrix(NA, empty_bar*nlevels(data$group), ncol(data)) )
285 | colnames(to_add) <- colnames(data)
286 | to_add$group <- rep(levels(data$group), each=empty_bar)
287 | data <- rbind(data, to_add)
288 | data <- data %>% arrange(group)
289 | data$id <- seq(1, nrow(data))
290 |
291 | # Get the name and the y position of each label
292 | label_data <- data
293 | number_of_bar <- nrow(label_data)
294 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
295 | label_data$hjust <- ifelse( angle < -90, 1, 0)
296 | label_data$angle <- ifelse(angle < -90, angle+180, angle)
297 | head(label_data)
298 |
299 | # prepare a data frame for base lines
300 | base_data <- data %>%
301 | group_by(group) %>%
302 | summarize(start=min(id), end=max(id) - empty_bar) %>%
303 | rowwise() %>%
304 | mutate(title=mean(c(start, end)))
305 | head(base_data)
306 |
307 | # prepare a data frame for grid (scales)
308 | grid_data <- base_data
309 | grid_data$end <- grid_data$end[ c( nrow(grid_data), 1:nrow(grid_data)-1)] + 1
310 | grid_data$start <- grid_data$start - 1
311 | grid_data <- grid_data[-1,]
312 | grid_data
313 |
314 | # Make the plot
315 | p <- ggplot(data, aes(x=as.factor(id), y=value, fill=group)) +
316 | # 添加条形图
317 | geom_bar(aes(x=as.factor(id), y=value, fill=group), stat="identity", alpha=0.5) +
318 |
319 | # 添加各组之间的线条,可以注释
320 | geom_segment(data=grid_data, aes(x = end, y = 80, xend = start, yend = 80), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
321 | geom_segment(data=grid_data, aes(x = end, y = 60, xend = start, yend = 60), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
322 | geom_segment(data=grid_data, aes(x = end, y = 40, xend = start, yend = 40), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
323 | geom_segment(data=grid_data, aes(x = end, y = 20, xend = start, yend = 20), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
324 |
325 | # Add text showing the value of each 100/75/50/25 lines,设置值坐标,可以注释
326 | annotate("text", x = rep(max(data$id),4), y = c(20, 40, 60, 80), label = c("20", "40", "60", "80") , color="grey", size=3 , angle=0, fontface="bold", hjust=1) +
327 |
328 | # 和前面一样
329 | ylim(-100,120) +
330 | theme_minimal() +
331 | theme(
332 | legend.position = "none",
333 | axis.text = element_blank(),
334 | axis.title = element_blank(),
335 | panel.grid = element_blank(),
336 | plot.margin = unit(rep(-1,4), "cm")
337 | ) +
338 | coord_polar() +
339 | geom_text(data=label_data, aes(x=id, y=value+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=2.5, angle= label_data$angle, inherit.aes = FALSE ) +
340 |
341 | # Add base line information
342 | # 添加下划线
343 | geom_segment(data=base_data, aes(x = start, y = -5, xend = end, yend = -5), colour = "black", alpha=0.8, size=0.6 , inherit.aes = FALSE ) +
344 | # 添加各组的名字
345 | geom_text(data=base_data, aes(x = title, y = -18, label=group), hjust=c(1,1,0,0), colour = "black", alpha=0.8, size=4, fontface="bold", inherit.aes = FALSE)
346 | p
347 |
348 | # library
349 | library(tidyverse)
350 | library(viridis)
351 |
352 | # Create dataset
353 | # 创建数据集
354 | data <- data.frame(
355 | individual=paste( "Mister ", seq(1,60), sep=""),
356 | group=c( rep('A', 10), rep('B', 30), rep('C', 14), rep('D', 6)) ,
357 | value1=sample( seq(10,100), 60, replace=T),
358 | value2=sample( seq(10,100), 60, replace=T),
359 | value3=sample( seq(10,100), 60, replace=T)
360 | )
361 | head(data)
362 |
363 | # Transform data in a tidy format (long format)
364 | # key表示观察的变量就是value1,value2,value3;value代表值,-c(1,2)表示不对第一列和第二列进行转换
365 | data <- data %>% gather(key = "observation", value="value", -c(1,2))
366 | head(data)
367 | dim(data)
368 |
369 | # Set a number of 'empty bar' to add at the end of each group
370 | empty_bar <- 2
371 | nObsType <- nlevels(as.factor(data$observation))
372 | to_add <- data.frame( matrix(NA, empty_bar*nlevels(data$group)*nObsType, ncol(data)) )
373 | colnames(to_add) <- colnames(data)
374 | to_add$group <- rep(levels(data$group), each=empty_bar*nObsType )
375 | data <- rbind(data, to_add)
376 | data <- data %>% arrange(group, individual)
377 | data$id <- rep( seq(1, nrow(data)/nObsType) , each=nObsType)
378 |
379 | # Get the name and the y position of each label
380 | label_data <- data %>% group_by(id, individual) %>% summarize(tot=sum(value))
381 | number_of_bar <- nrow(label_data)
382 | angle <- 90 - 360 * (label_data$id-0.5) /number_of_bar
383 | label_data$hjust <- ifelse( angle < -90, 1, 0)
384 | label_data$angle <- ifelse(angle < -90, angle+180, angle)
385 |
386 | # prepare a data frame for base lines
387 | base_data <- data %>%
388 | group_by(group) %>%
389 | summarize(start=min(id), end=max(id) - empty_bar) %>%
390 | rowwise() %>%
391 | mutate(title=mean(c(start, end)))
392 |
393 | # prepare a data frame for grid (scales)
394 | grid_data <- base_data
395 | grid_data$end <- grid_data$end[ c( nrow(grid_data), 1:nrow(grid_data)-1)] + 1
396 | grid_data$start <- grid_data$start - 1
397 | grid_data <- grid_data[-1,]
398 |
399 |
400 | # Make the plot
401 | p <- ggplot(data) +
402 |
403 | # Add the stacked bar
404 | geom_bar(aes(x=as.factor(id), y=value, fill=observation), stat="identity", alpha=0.5) +
405 | scale_fill_viridis(discrete=TRUE) +
406 |
407 | # Add a val=100/75/50/25 lines. I do it at the beginning to make sur barplots are OVER it.
408 | geom_segment(data=grid_data, aes(x = end, y = 0, xend = start, yend = 0), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
409 | geom_segment(data=grid_data, aes(x = end, y = 50, xend = start, yend = 50), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
410 | geom_segment(data=grid_data, aes(x = end, y = 100, xend = start, yend = 100), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
411 | geom_segment(data=grid_data, aes(x = end, y = 150, xend = start, yend = 150), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
412 | geom_segment(data=grid_data, aes(x = end, y = 200, xend = start, yend = 200), colour = "grey", alpha=1, size=0.3 , inherit.aes = FALSE ) +
413 |
414 | # Add text showing the value of each 100/75/50/25 lines
415 | ggplot2::annotate("text", x = rep(max(data$id),5), y = c(0, 50, 100, 150, 200), label = c("0", "50", "100", "150", "200") , color="grey", size=6 , angle=0, fontface="bold", hjust=1) +
416 |
417 | ylim(-150,max(label_data$tot, na.rm=T)) +
418 | theme_minimal() +
419 | theme(
420 | legend.position = "none",
421 | axis.text = element_blank(),
422 | axis.title = element_blank(),
423 | panel.grid = element_blank(),
424 | plot.margin = unit(rep(-1,4), "cm")
425 | ) +
426 | coord_polar() +
427 |
428 | # Add labels on top of each bar
429 | geom_text(data=label_data, aes(x=id, y=tot+10, label=individual, hjust=hjust), color="black", fontface="bold",alpha=0.6, size=5, angle= label_data$angle, inherit.aes = FALSE ) +
430 |
431 | # Add base line information
432 | geom_segment(data=base_data, aes(x = start, y = -5, xend = end, yend = -5), colour = "black", alpha=0.8, size=0.6 , inherit.aes = FALSE ) +
433 | geom_text(data=base_data, aes(x = title, y = -18, label=group), hjust=c(1,1,0,0), colour = "black", alpha=0.8, size=4, fontface="bold", inherit.aes = FALSE)
434 |
435 | p
436 | # 保存数据 Save at png
437 | ggsave(p, file="output.png", width=10, height=10)
438 |
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/[数据分析与可视化] 数据绘图要点1-注重数据排序.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "8bafdde3",
6 | "metadata": {},
7 | "source": [
8 | "# 数据绘图要点1-注重数据排序\n",
9 | "默认情况下,大多数数据可视化工具将使用字母顺序或使用输入表中的出现顺序对分类变量组进行排序。当显示多个实体项的值时,对它们进行排序会使得图表更具洞察力。"
10 | ]
11 | },
12 | {
13 | "cell_type": "markdown",
14 | "id": "979ae494",
15 | "metadata": {},
16 | "source": [
17 | "## 实例\n",
18 | "\n",
19 | "### 无序棒棒糖图\n",
20 | "\n",
21 | "让我们从一个无序棒棒糖图开始,下面代码展示了一些国家出口的某一重要物品的数量。这里每一行代表一个国家,X 轴显示2017 年销售的重要物品数量。默认情况下,国家按字母顺序排列。"
22 | ]
23 | },
24 | {
25 | "cell_type": "code",
26 | "execution_count": 7,
27 | "id": "3743784d",
28 | "metadata": {},
29 | "outputs": [
30 | {
31 | "data": {
32 | "text/html": [
33 | "\n",
34 | "A data.frame: 10 × 2\n",
35 | "\n",
36 | "\t | Country | Value |
|---|
\n",
37 | "\t | <fct> | <int> |
|---|
\n",
38 | "\n",
39 | "\n",
40 | "\t| 1 | United States | 12394 |
|---|
\n",
41 | "\t| 2 | Russia | 6148 |
|---|
\n",
42 | "\t| 3 | Germany (FRG) | 1653 |
|---|
\n",
43 | "\t| 4 | France | 2162 |
|---|
\n",
44 | "\t| 5 | United Kingdom | 1214 |
|---|
\n",
45 | "\t| 6 | China | 1131 |
|---|
\n",
46 | "\t| 7 | Soviet Union | NA |
|---|
\n",
47 | "\t| 8 | Netherlands | 1167 |
|---|
\n",
48 | "\t| 9 | Italy | 660 |
|---|
\n",
49 | "\t| 10 | Israel | 1263 |
|---|
\n",
50 | "\n",
51 | "
\n"
52 | ],
53 | "text/latex": [
54 | "A data.frame: 10 × 2\n",
55 | "\\begin{tabular}{r|ll}\n",
56 | " & Country & Value\\\\\n",
57 | " & & \\\\\n",
58 | "\\hline\n",
59 | "\t1 & United States & 12394\\\\\n",
60 | "\t2 & Russia & 6148\\\\\n",
61 | "\t3 & Germany (FRG) & 1653\\\\\n",
62 | "\t4 & France & 2162\\\\\n",
63 | "\t5 & United Kingdom & 1214\\\\\n",
64 | "\t6 & China & 1131\\\\\n",
65 | "\t7 & Soviet Union & NA\\\\\n",
66 | "\t8 & Netherlands & 1167\\\\\n",
67 | "\t9 & Italy & 660\\\\\n",
68 | "\t10 & Israel & 1263\\\\\n",
69 | "\\end{tabular}\n"
70 | ],
71 | "text/markdown": [
72 | "\n",
73 | "A data.frame: 10 × 2\n",
74 | "\n",
75 | "| | Country <fct> | Value <int> |\n",
76 | "|---|---|---|\n",
77 | "| 1 | United States | 12394 |\n",
78 | "| 2 | Russia | 6148 |\n",
79 | "| 3 | Germany (FRG) | 1653 |\n",
80 | "| 4 | France | 2162 |\n",
81 | "| 5 | United Kingdom | 1214 |\n",
82 | "| 6 | China | 1131 |\n",
83 | "| 7 | Soviet Union | NA |\n",
84 | "| 8 | Netherlands | 1167 |\n",
85 | "| 9 | Italy | 660 |\n",
86 | "| 10 | Israel | 1263 |\n",
87 | "\n"
88 | ],
89 | "text/plain": [
90 | " Country Value\n",
91 | "1 United States 12394\n",
92 | "2 Russia 6148\n",
93 | "3 Germany (FRG) 1653\n",
94 | "4 France 2162\n",
95 | "5 United Kingdom 1214\n",
96 | "6 China 1131\n",
97 | "7 Soviet Union NA\n",
98 | "8 Netherlands 1167\n",
99 | "9 Italy 660\n",
100 | "10 Israel 1263"
101 | ]
102 | },
103 | "metadata": {},
104 | "output_type": "display_data"
105 | },
106 | {
107 | "data": {
108 | "text/html": [
109 | "51"
110 | ],
111 | "text/latex": [
112 | "51"
113 | ],
114 | "text/markdown": [
115 | "51"
116 | ],
117 | "text/plain": [
118 | "[1] 51"
119 | ]
120 | },
121 | "metadata": {},
122 | "output_type": "display_data"
123 | }
124 | ],
125 | "source": [
126 | "# Libraries\n",
127 | "library(tidyverse)\n",
128 | "library(hrbrthemes)\n",
129 | "library(kableExtra)\n",
130 | "options(knitr.table.format = \"html\")\n",
131 | "\n",
132 | "# 从github加载数据\n",
133 | "data <- read.table(\"https://raw.githubusercontent.com/holtzy/data_to_viz/master/Example_dataset/7_OneCatOneNum.csv\", header=TRUE, sep=\",\")\n",
134 | "# 展示数据\n",
135 | "head(data,10)\n",
136 | "# 长度\n",
137 | "nrow(data)"
138 | ]
139 | },
140 | {
141 | "cell_type": "code",
142 | "execution_count": 8,
143 | "id": "0a651dc6",
144 | "metadata": {},
145 | "outputs": [
146 | {
147 | "data": {
148 | "text/html": [
149 | "38"
150 | ],
151 | "text/latex": [
152 | "38"
153 | ],
154 | "text/markdown": [
155 | "38"
156 | ],
157 | "text/plain": [
158 | "[1] 38"
159 | ]
160 | },
161 | "metadata": {},
162 | "output_type": "display_data"
163 | }
164 | ],
165 | "source": [
166 | "# 清除空值数据\n",
167 | "data <- filter(data,!is.na(Value))\n",
168 | "nrow(data)"
169 | ]
170 | },
171 | {
172 | "cell_type": "code",
173 | "execution_count": 9,
174 | "id": "f9d6f837",
175 | "metadata": {
176 | "scrolled": true
177 | },
178 | "outputs": [
179 | {
180 | "data": {
181 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAA0gAAANICAMAAADKOT/pAAAAPFBMVEUAAAAzMzNNTU1oaGhp\ns6J8fHyMjIyampqnp6eysrK9vb2+vr7Hx8fQ0NDZ2dnh4eHp6enr6+vw8PD///9rsEd/AAAA\nCXBIWXMAABJ0AAASdAHeZh94AAAgAElEQVR4nO2di3rrxpGEtbATx+s4Gxnv/65r8YIaXNlD\nojHdU9Xf7qFE8XSdatcfUuDM8GtUqVQf11frf4BK1UMJJJXqhBJIKtUJJZBUqhNKIKlUJ5RA\nUqlOKIGkUp1QAkmlOqFSgfR9WC9+/HF5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA\n2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2ewGpF//rvcnaBqab/sOBPgM\nIJudgPTroz6Z4suheTbvQoDPALL5EqRhdrP48oQathruqO36+fXXC0hKH5N4OYwmkByk4/uK\nb3b9CKQQAnwGkM0uQPr11ytISh+TeDmMJnANSMM4DMPznmH69udmmO6d/wg/nn6OLtP9Q/mo\nYXhIDs8eIGnPjkCKIcBn4D2QytzPvx2Hx70TDPOnsgK0Yf3XN366+uN/fmrPjkCKIcBn4D2Q\ncDt/iinunmNR3Gw9fAukOT7j/Ks9OwIphgCfgQ9BmsX9BUhj+QJtH6Rh+QJxnF4cvgZJFxti\nCPAZ+Ayk5yu4Lb6GFUiL12sHz0jj4m+af0cSSDEE+Ax8BtIeEXsg7QH0CqTVt/uG9IZsBAE+\nAxUgIcwvQHq+Ilvkf/a3hvHVxYY5PmaQtEQoggCfgRqQpuvQi7ADoPLnK5Dw4+HxbHV8+Xt8\nvqTD3zOBFHDKdAJ8BqpAOrfWlxD2H7P85uQp1A7NuX9+AT4DyKZAsg/NuX9+AT4DyGZIkMof\nlo87eQq1Q3Pun1+AzwCy2ck2iremUDs05/75BfgMIJsCyT405/75BfgMIJsCyT405/75BfgM\nIJsCyT405/75BfgMIJsCyT405/75Bfoz8OptfmRTIJkrfUw0ocr+rxeeIZsngzTMbhZfru8b\nig2AhqqawumVPiaaUF1/w1JoZNMdpNV34+K9pGHxiAOyaqZwfqWPiSZU1z8NSIuF5XuPRtVM\n4fxKHxNNqKq/ZbsosukG0mxx6rBauzpDpnzE6tFaIpRGoC8DUUCabZNY7aaYU7KxS2Oxq+Lw\nzIZ7/aFSnVhRQMLtNhoTTOUdW49GVfzPiUOl/99bTaiqf1CQ1ocyLOCZXtUVjx7nF/UqpuBQ\n6WOiCdX1j3GxYesZaTwEqdzmV/xUvyOlEejMQBqQ5g9e/Y606CmQwgv0ZqDhG7KLt4ZeXWwo\nH/z8ycFvVFVTOL3Sx0QTqu7fcInQxgkPe5e/53dOhzro8ndWAT4DjiB51slTqB2ac//8AnwG\nkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+a\nc//8AnwGkE2BZB+ac//8AnwGkE2BZD5+P31M4uUwmoBAemsKt7J/IEz6mMTLYTSBdiAtt+it\ndj5sPGq9PeLwLxd18hR+quIjytLHJF4OowlEAmkLh9mjjmARSKkF+Awgm5eDdMjK5SBZNm99\n0r+q0gvwGUA2TwPJeEbDsHz89IDidurz3EErkFII8BnwAAn78na3Tew8Hrsnyn1M09+83eN0\nZoNAytQ/ngEPkHC7t6NoGJ9PTsP6AXu3vvuRBFKm/vEMXADSxhkNwxZhbUHSxYZM/eMZuACk\n8gGriw0CqUsBPgMNQXr+NAhIekM2Uf94Bk4DCVE/+B1pmIFUPOjwYsNFIGmJUJ7+8QycB1LN\nGQ14lipB2b38fRVI5qE5988vwGfgRJBOqeH1Q37q5CnUDs25f34BPgPIZnOQFk86h3XyFGqH\n5tw/vwCfAWSzOUg1HzZ28hRqh+bcP78AnwFksz1IFXXyFGqH5tw/vwCfAWRTINmH5tw/vwCf\nAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWSzR5CsSxVqh+bQsy8BPgPI\nZn8g2RfP1Q7t9I69CfAZQDabgLRY6rr9042yTKFiOXft0M5u2J0AnwFkMyBI+2WZgkBqJ8Bn\nANnsDaSaLa+1Qzu5X38CfAaQzQgg3bagD+W67/JklII1wxQEUkMBPgPIZgCQhvUm9PJklPsf\n1sNPBFJDAT4DyHR7kMr9tFu7/ur2IwmkhgJ8BpDp5iDN9qI/z0uZQBpmr+0sU9DFhnYCfAaQ\n6dYgDcUzEl7PzZ6RUJYpCKR2AnwGkM3WII2Ll3Afg6Q3ZNsJ8BlANtusbJgdmTI/wOHD35F+\nSkuEGgnwGUCkGy0RWhyZMjzvXDwj1V/+dqz0MdGETu+PbPa31s6t0sdEEzq9P7IpkOxDc+6f\nX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIp\nkOxDc+6fX4DPALLZE0hOb8RO/R179yHAZwDZ7Ackt6VB09DcOvciwGcA2QwD0uyjX6aVDvbV\n336LVaeheTXuRoDPAPIbEqRhucjuUUeWBFJ7AT4DyGYvIDlu6JuG5tS3HwE+A8imQLIPzalv\nPwJ8BpDNiCCV//+sV2c2CKQAAnwGENDQIFVcbBBIAQT4DCC/AUEa5l8UdWRJFxvaC/AZQDYj\ngvR8HhJIyQT4DCCbEUGaf1HUoSe9IdtcgM8AshkGpNWBJ0MlSFoi1FyAzwCyGQek4sTi+7d1\nKxsCTplOgM8A0hsIpNd18hRqh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZ\nh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFks0eQdIh+IwE+\nA8hmfyDpY12aCfAZQDabgTSs1i3c7z76O5Yp6IPG2gnwGUA2mz4jbVAjkBIL8BlANnsDyXGD\nX/qYxMthNIHcIGHZ94DDGsodSRNchikIpIYCfAaQ5UggrT8LEw94dWbD3/XHTwmkhgJ8BpDl\nSCAVt6uvb2WYgkBqKMBnAFmOB9L9pthtXrcfSRcb2gnwGUCWw4E0zOGZ7ZO1TEEgtRPgM4Bs\nhgOp+Pr+fe2nmusN2WYCfAaQzSAgDVsg3b+uBUlLhJoJ8BlANgOA9PML0QweXP4ey9+QtGg1\nugCfAWQz/Fo7gZRHgM8AsimQ7ENz7p9fgM8AshkcpPma1pOnUDs05/75BfgMIJvBQZrXyVOo\nHZpz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wC\nfAaQzf5AcvtMivQxiZfDaAICaZqC46ckpY9JvBxGEwgK0vpok8Od5Hs/q9lG4fm5feljEi+H\n0QRigoS1cztcGH8gkLII8BlASv1AGmY3W1wYf1ABkuP+2A5iEi+H0QQigzRf1X3/GL5pAyw+\npe+5oQ/fP3fKCqQ0AnwGLgGp3Agx4E98tOX63JOt7+9dbIefCKSmAnwGrgDpAdOTiGHE/z05\nWm+OXXy/+DXr5RQEUlMBPgNXgTSCnm2QijNOcM/7IOliQ1MBPgNXg1S+ris3vpYv86bXegIp\nqwCfAeT8iqt2dpD2bs0g6Q3ZlgJ8Bi4BafZb0Lg+12QHoOVLu6EKJC0RaijAZ+AKkFYHmDxB\n2rz8/fz54hyUysvfrpU+JprQ6f2vAen0OnkKtUNz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz\n7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9fgM8A\nstkVSG7vxT76O1d6AT4DyGZHIDmuDroPzatxNwJ8BpDNBiBtHOUw//nuTw49ea5XvQ/NqW8/\nAnwGkM3rQdo5ymHxgM069CSQmgvwGUA2Lwdp7yiH1SM26siS656++9B82nYkwGcA2WwF0nKL\n7OKMhuXZDfc6siSQ2gvwGUA2rwdp5yiH7W0U09Lwl2c2CKT2AnwGkOsmV+22jnL4dGOfQGov\nwGcAmW51+XtxlMPi7Ib1WQ63OvSkiw3NBfgMINBNQSp3oD/vHbe+f9ShJ4HUXIDPAALd8Krd\nySDpDdnmAnwGkOsGFxsmOGY70HfPbKjYaq4lQm0F+Awg1y1XNhR/7l7+HmtA8q30MdGETu+P\nVHe01i7clOkE+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn\n/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+Awgm2wgffCebfqYxMthNAGBZJzCR6uI0sck\nXg6jCeQGae8Mh/Wdn07hs3Wt6WMSL4fRBFKD9PIMB9SnUxBIufvHM4BsNgfp9RkOqA+n8OHe\nv/QxiZfDaAIdgDQWBzQM5Q5a65kNAqm9AJ8BZLM9SBMqoGe6rTiz4fv7j1clkJL3j2cAOW4O\n0k9t7JmYn+XwqA+nIJCS949nABkOAdK4oGY6sOHk/Ui62JC7fzwDCHBEkPASTyClEuAzgAA3\nB6k8w2F1e/YOWb0hm7p/PAPIcXuQ8ERUXGzweWn3rSVCqfvHM4AcNwepOK9hnB/c4ALSB5U+\nJprQ6f2R4gAgPev1e7InT6F2aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2A4H0\nuk6eQu3QnPvnF+AzgGwKJPvQnPvnF+AzgGwKJPvQnPvnF+AzgGwKJPvQnPvnF+AzgGwKJPvQ\nnPvnF+AzgGxSgHTOp1Skj0m8HEYTEEhHUzjrc5PSxyReDqMJxANp70CT28/WX9uObHgPpNM+\nyS99TOLlMJpAOJB2DzQZxi2QjBwJpOACfAaQTReQ9p9nNkGycvQWSB9ui33Z/8RKL8BnANn0\nBOn25Wwb+XBf2I3XfQVY0yNvWyjm56AIpBQCfAa8QcKBJo8/lh8Su9zPVzxi+ZmyHx5+IpCu\nE+Az4AzSA6Y9kCY6bs8/s4+SnZ3c8Ow09XxnCgLpOgE+AxeAtAZoE6T5HdMjh83Xdm9NQRcb\nLhPgMxAHpNUf88sP84sTb01BIF0mwGcAYXe+amcCaXbv+lXdp1vN9YbsVQJ8BrxBwoEmJUC7\nIG0hN7828QlIWiJ0lQCfAWeQil9thsWBJsOwBdKIK96bf+1DkM6p9DHRhE7v7w6ST508hdqh\nOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8Bn\nANkUSPahOffPL8BnANkUSPahOffPL8BnANnsCaRz3nfd7+/Yuw8BPgPIZj8gnbUSaH9obp17\nEeAzgGx6bqPY3fk6bH47DJs/LerI0mlrU/eH5tW4GwE+A8imF0i7e8iH1f3D/oMXdWRJILUX\n4DOAbHqttZvdLH/iANJ5+/f2h+bUtx8BPgPIpi9Ity+PT214gjT9AN+NyxeIB44EUgABPgP+\nIJlPbZieosofrPdUvDqzQSAFEOAz4A7SA6Y9kCY+liCVDx6LB9zrwJFACiDAZwAp9b38XbNH\ndhsk85kNutjQXoDPAKLeHqTnQSebIM0vTRxZEkjtBfgMuIO0v4W86hlpcTnv0JPekG0uwGfA\nH6QJgeVFhgVIQGb3FyrzVnMtEWoswGfAHaTRemrDNkg6syGlAJ+BC0DyqJOnUDs05/75BfgM\nIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT40\n5/75BfgMIJsCyT405/75BfgMIJv9gOT8dmwHMYmXw2gCAsl/gVAHMYmXw2gCqUCaFi3sP2J2\nU9S+If8lqx3EJF4OowlkAmm+CG//IVu1b0ggRRDgM4BsXg3SYln40WPWtevngm19HcQkXg6j\nCeQDaSzXpmInxfx4h3G0ntkgkEII8BlANi8HqTzMoSBmusVOi4ozGwRSCAE+Awh2i6t2w9YZ\nJ8VevvU9j9r1I5BCCPAZQKgbXf4etkF6vtTDM5HxzAZdbIggwGcAiQ4BUonO7VelCS3rmQ0C\nKYIAnwEkuulVuwVIR78j3erAkd6QDSDAZwDBvv5iw5ONGSfFHvMlSNYzG7REqLkAnwEEu+3K\nBhzs8PzR4mKezmzIJMBnALHuZa3dO1OoHZpz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wC\nfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFk\nH5pz//wCfAaQzW5Acl/YkD8m8XIYTUAgXbDULn9M4uUwmkBakOYbYJ93Lm5Ru36uWPydPybx\nchhNICtI26cJ7R/msOtHIIUQ4DOAbLYEaefYrXqQLtkgmz8m8XIYTSA3SLcvn9vPb6u9cQaK\n7fATgRRDgM8AstkUpPIglOfe2OcfNYefCKQYAnwGEObWV+1mB3DNN/nZDz8RSDEE+AwgyK1B\nGqcD7YZNkGyHn+hiQwgBPgNIcRCQ8OX6bCGBlEWAzwBSHOBiwwFI5sNP9IZsBAE+Awhz24sN\ntz/H1au5sQTJePiJlgi1F+AzgDAHWdmwOPW7vPytw0/SCPAZQJQD/I5kr5OnUDs05/75BfgM\nIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT405/75BfgMIJsCyT40\n5/75BfgMIJsCyT405/75BfgMIJsCyT405/4XCTi+c51+QgLprSnUDs25/yUCrmup0k8oIUjD\n8Fzxvf+Q1T0nT6F2aM79rxDwXd2bfkIZQVrcmurkKdQOzbm/QGouIJDemkLt0Jz7XyDgvAMy\n/YRyg7T6/L6h9syGN6dQOzTn/gKpuUAfIG3cGs9suNcfqlclkE7uj0AHuNiwd0aD/cyGN6dQ\nOzTn/npGai6QEaTpj/knmo+4rTiz4b0p1A7Nub8uNjQXyArSfGv5uECp4syG96ZQOzTn/gKp\nuUA3IC1/dzKf2fDWFGqH5txfb8g2F0gLEp519i826KXduQJaInRifwQ6wFW76WCG7cvfekZK\nI8BnAIHWWjv70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4Bs\nCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsdgSSPo2itQCfAWSzG5D0iX3t\nBfgMIJtXg/T6qIb1X5m+OnB0wUeNpY9JvBxGE8gE0hKO7e+3f3bgSCAFEOAzgJR2ApLzjrX7\n0LwadyPAZwApbQjStDb18WqvPLth/oBn7RsSSBEE+Awg2O1AmnZLzDbwFXsnZtsqXpzZIJAi\nCPAZQLDbXWxYbj9a31F+f6t9QwIpggCfAQS70TPSaALpjpwuNmQR4DOAYEcGaRjNz0gCKYIA\nnwEEOzBIy2O69IZsdAE+Awh2MJCeL+WWt/c6NqUlQq0F+Awg2O1Aml3dvv8yhMve5a3ObMgi\nwGcAaY661m7zHdqTp1A7NOf++QX4DCCbAUFaHGZX1MlTqB2ac//8AnwGkM2AII27q1pPnkLt\n0Jz75xfgM4BsRgRpt06eQu3QnPvnF+AzgGwKJPvQnPvnF+AzgGwKJPvQnPvnF+AzgGwKJPvQ\nnPvnF+AzgGwKJPvQnPvnF+AzgGx2A5L3uoYOYhIvh9EEBJL/SrsOYhIvh9EEMoK0PgVlb7u5\nYYnQBWu/O4hJvBxGE0gJ0uxm8eXWI0eBFF2AzwBS2gVIV+yP7SAm8XIYTSA5SPNP6JsORBlH\n6yf2CaQYAnwGkOcAIBUbk5YHojy3JL04/EQgxRDgM4A8B7jYsNwJu7FJ9lF7dgRSDAE+A8hz\n62ekcUnNsPUB54/a9aOLDSEE+Awgz8FAKs87EUjZBPgMIM+xQNo7/+RR+4b0hmwEAT4DyHM8\nkJYv7QYLSFoiFEGAzwDyHACkxeXv+clCpsvfb06hdmjO/fML8BlAiDtZa/fWFGqH5tw/vwCf\nAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH\n5tw/vwCfAWRTINmH5tw/vwCfAWSzJ5C0RKixAJ8BZLMfkPRBY80F+Awgmy1Bmp3ZsLc/tqwj\nS/oM2fYCfAaQzXYgPbfAjsXn9b2oI0sCqb0AnwFksyFIuDkBpAv29qWPSbwcRhPICNKAL4b7\n0lQc0/A8tmH5CS8HjgRSAAE+A8hmBJCwW2Lj2IbHH4dnNnwLpBACfAaQ5zggFV9v7Om71YEj\ngRRAgM8AshkPpKE4tmGcf3TfkSVdbGgvwGcA2YwH0uLnxt+RBFIAAT4DyGasq3YbINnObPjW\nG7IBBPgMIM6x3kfa/B3JvNVcS4QaC/AZQJxDrGyYTmYYcO8wf8itTp5C7dCc++cX4DOAbPaz\n1i7elOkE+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF\n+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+Awgm52BpCVCLQX4DCCbXYHku0oofUzi5TCaQDaQ\nhtmNvV6Ycl63mj4m8XIYTSAdSI/1P7V/74UpgdRYgM8AstnoGWlr297rOjblvbcvfUzi5TCa\nQGKQihMaZuu9Z7v7nn/v2JRAai3AZwCZbvU70sYJDQ+gxgI0gPTqzIZvgdRegM8AMh0FpOl2\nvrdvflDXsSmB1FqAzwAy3eyq3fACJLy2q9rYp4sNDQX4DCDT7S5/Dxsgla/uytd6j3phSiA1\nFuAzgGwGBQkXIypA0huyjQX4DCCbDd+QHZYgFX8O43ScQ9VWcy0RainAZwDZbL2yYX5CA/6c\nHePwrJOnUDs05/75BfgMIJvh19oJpDwCfAaQTYFkH5pz//wCfAaQzeAg2Y8sDjhlOgE+A8hm\ncJDmdfIUaofm3D+/AJ8BZFMg2Yfm3D+/AJ8BZFMg2Yfm3D+/AJ8BZFMg2Yfm3D+/AJ8BZFMg\n2Yfm3D+/AJ8BZJMeJPtaiPQxiZfDaAIC6a0pfNd9qFL6mMTLYTSBgCBtn2+yubl8b8f5+v6T\np1D5MX/pYxIvh9EEIoK0Ot9kvrVonP/Adv/JUxBIyfrHM4Bs+j0jLZecBgSpbk9t+pjEy2E0\ngdggPfa63tdzP884wZkn22egzO8XSCkE+AxcAdLWsQzD8r6D78vtSqbDT/6oLoGUq388A81A\nGtf3Lc86Wd2iTp6CnpGS9Y9n4BKQnuebDItzTBb3Lc86me809139rYsNqfrHM3ANSOPycK3V\nM81Q/mD7mcr1U80FUqr+8QxEA+nVSz4vkPSGbKr+8QxcBNLqfJMjcFaPXQKlJULRBfgMXAXS\n7NL2sHXkN75f/G50xeXvyqE5988vwGfAHySXOnkKtUNz7p9fgM8AsimQ7ENz7p9fgM8AsimQ\n7ENz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9fgM8AsimQ7ENz7p9f\ngM8AstkTSJ4fRfHdQUzi5TCagED6rlvt81alj0m8HEYTSAvSYL7zVkeWnD+u77uDmMTLYTQB\ngSSQIgjwGUA2ewGpbo/eW5U+JvFyGE0gNUjl8Q3LxarTGQ73OnAkkAII8BlANpuDtDyuYb6d\nYloi/urMBoEUQIDPAKIcB6TH7eYZDo86cCSQAgjwGUCUo4K0PNPhVkeWdLGhvQCfAUQ5KEjD\n8s5bHVkSSO0F+Awgys1A2j+mYe/sBr0hG12AzwDyHA+k6eiu6jMbtESosQCfAeS53Uu7AacT\nT9fmysvftc9I8aZMJ8BnAHHuZ61dvCnTCfAZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYF\nkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDa7Acn53djv\nDmISL4fRBASS+/qg7w5iEi+H0QRyg7S/I3ZZu378V6x+dxCTeDmMJiCQBFIEAT4DyGYXIF2w\nq++7g5jEy2E0gfwgLc9oqDuzQSDFEOAzgGzGAGl5RkPlmQ0CKYYAnwGkOAZIz9s3z2wQSDEE\n+AwgxVFAWp7RUHdmgy42hBDgM4AUBwFpeUZD5ZkNAimEAJ8BpLj54Seb+82rz2zQG7IRBPgM\nIMpxQBoWFxveOI7r/QmahubbvgMBPgOIcuOXdsXBDXeAlt+POrMhjwCfASQ5wO9IRb14b/bk\nKdQOzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8hmLJBe1MlTqB2ac//8AnwGkE2B\nZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE0akD5f+ZA+\nJvFyGE1AIL2awhlr8dLHJF4Oown0AtJw+O0HIJ2yOjx9TOLlMJpARyAN82/n9fYUBNIlAnwG\nkE0KkM7ZQZs+JvFyGE2gH5AeG2Ono08edz/q3SkIpGsE+Awgu8FAmrbHPjZVPO8+PvzkXn/s\nlkC6RoDPALIbEaTia3y67E+9OwWBdI0AnwFkNyZI09En54Ckiw3XCPAZQHZDglRuOC8vObw9\nBYF0iQCfAWQzIkizkxxOAUlvyF4iwGcA2YwK0nAuSFoidIUAnwFkMxxIj8vfG0efaNFqdAE+\nA8hmKJC2SiDlEeAzgGwKJPvQnPvnF+AzgGwGB6n8UBeBFF2AzwCyGRykeZ08hdqhOffPL8Bn\nANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSD9lerc2fUzi5TCagEB6\nawrPMq4fSh+TeDmMJpAbpBcHfrt/rIt1RWv6mMTLYTQBgfTWFB4lkLL0j2cAKRVI5l1/6WMS\nL4fRBPKDhFMacGQD7nzWyVO4l0BK0z+eAaQ4BkjTDqTldqTp9qMzGw5LIKXpH88AUhwIpLF8\n8lmCdKuTp3AvgZSmfzwDSHEMkJ6LUx/M4FCuwR8kXWxI0z+eAaQ4CEjlq7ryyAb8/KdOnsKj\nBFKW/vEMIMVhQFm0pboAABdpSURBVJoffXIlSHpDNkv/eAaQ4hggLa8vTEc2XPA70k9piVCK\n/vEMIMUxQJpf6X4c2XDR5W/70Jz75xfgM4AUBwDJXidPoXZozv3zC/AZQDYFkn1ozv3zC/AZ\nQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1o\nzv3zC/AZQDa7AenzQ/JfDs23fQcCfAaQzU5AOuNjW14OzbN5FwJ8BpDNWCC92Cy76+eUDxJ7\nOTTH3n0I8BlANgWSfWiOvfsQ4DOAbHYBknlv3keVPibxchhNoCeQZsc2DONQfh7Fnh2BFEOA\nzwCyGQ+k2bEN0zeHZzYIpBgCfAYQ3Xggrb55vY1CIMUQ4DOA6EYFqTiuwbAfSRcbQgjwGUB0\ng4JUHtsgkLII8BlAdGOCtNp6fq99Q3pDNoIAnwFENy5INS/tvrVEKIIAnwFENyZIz2MbKkAK\nOGU6AT4DiG4skF7UyVOoHZpz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz\n//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQTYFkH5pz//wCfAaQ\nzb5Acl3ekD4m8XIYTUAg3Uw5L7hLH5N4OYwmkACk5SbyF5vKtx9ybMp7CXj6mMTLYTSBPkFa\n17EpgdRagM8AstkPSO7bZNPHJF4OownkAGk6yWT1iXy4v7wtHiKQUgjwGWgDUrHtdfYZsYvt\nsMUttlEcHn5yK4HUXIDPQBuQam7H9a2ekaIL8BlAwCOBNDxf0+Hchvlru2NTutjQWoDPAAIe\nCaTi0dPvSKN9h6xAai3AZwCRjQnS+nekW70wpTdkGwvwGUBgG4E02MAqXugZQNISocYCfAaa\ngrS+/I37Ny5/6/CTNAJ8Bq4H6ZQ6eQq1Q3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+A\nzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCy2RFI\n+liX1gJ8BpDNbkDSR1+2F+AzgGxGAWljB23VmQ0XfGZf+pjEy2E0gT5BWteBI4EUQIDPALLZ\nCUjuu/q+O4hJvBxGE+gDpOWZDjVnNgikCAJ8BpDNSCCtz3Qwn9kgkCII8BlAgCOBtHU72jb2\nCaQIAnwGEODoIFnPbNDFhgACfAYQ4OAgmc9sEEgBBPgMIMCxQao4s0FvyLYX4DOAAAcFqbjY\nUHUc17sTNA3Ns3kXAnwGEOCAIC0uf+vMhjQCfAYQ4CggmerkKdQOzbl/fgE+A8imQLIPzbl/\nfgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8im\nQLIPzbl/fgE+A8imQNqp9du76WMSL4fRBATSW1M4qK0FR+ljEi+H0QTagTTMd97V//3nB/Xh\ni2HeuOx+8hT2a3MJbPqYxMthNIFmIBWfUfn23x/Kb+Z3Lkk6eQr7JZBC9o9nANn8CKT11rs3\nGmyBtGbqVidPYbe2twmmj0m8HEYTaA3S7evnp4Pdlpk+X6stfjLMn8TmzzgbIC1UTp7Cbgmk\nmP3jGTgfpGkPxLTzoTyCoTiS4U2QXpzZ8FN/nFYCKWb/eAbOBGkYtj4QdkHJxsbX8u8Pw/RF\n8WmYq8sNJ09htwRSzP7xDJwJ0lhce9sGaY3aDKRZt/nli0Yg6WJDzP7xDDiAtIo+9ukVjyhf\n3m2/tFs+kQmkPAJ8Bq4EafWiLwFIekM2ZP94Bk4Cqcj+/u9IzwsPRpDWr/BagKQlQhH7xzNw\nFkjFyobZZYLZH8URDEt0hnENUnEJAg+6HqTr++cX4DNwGkhv1bDxleXRAim4AJ8BZFMg2Yfm\n3D+/AJ8BZPN6kGaLXF+TVD7i5CnUDs25f34BPgPIprZR2Ifm3D+/AJ8BZFMg2Yfm3D+/AJ8B\nZFMg2Yfm3D+/AJ8BZFMg2Yfm3D+/AJ8BZFMg2Yfm3D+/AJ8BZFMg2Yd2+9PxMy/4chhNQCC9\nNYXaoX07fwoTXw6jCSQEqTzoxFwnT6F2aN6fC8iXw2gCGUF65y+dPIXaoQmkxv3jGUA2BZJ9\naN6fnc6Xw2gCmUEaxueZdo9v5tvMZy//Tp5C7dAEUuP+8Qwgm+1Bmu1mWuymxa6LSw8/qTsU\n5bTiy2E0gYwgzc45ud+1AGgcF9uX9IwUXIDPAALd/hnpcbM+i2gscLvXyVOoHZouNjTuH88A\nAh0EpK2XdOWj7nXyFGqHJpAa949nANmMAdLmQSljPJD0hmzT/vEMIJtxQNo6ZjLY70g/pSVC\n7frHM4BAxwBpdUBKzMvfEmjbP54BZFNr7exDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6f\nX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALLZ\nEUiOb5Xeh+bZvAsBPgPIZjcguS7euQ/Nr3UnAnwGkM1GIE1HNhTrFvY2zZqWCPkuJ70Pza1z\nLwJ8BpDSViAZ75vff+BIIAUQ4DOAlHYCkvOWu/vQvBp3I8BnACltDdKAgxqKxarl2Q0CKY0A\nnwEkOgJI2NW3dXbDYDmzQSBFEOAzgEQ3vdgwzul5/uyN/UgCKYIAnwEkOsIz0ux2fXaDLjZk\nEeAzgEQHA2n3OK5bHTgSSAEE+Awg0bFA+mCrud6QbS/AZwCJjgfS8qWd+TguLRFqLcBnAImO\nBdLW2Q3BDj+RQLv+8Qwg0d2stQs4ZToBPgPIpkCyD825f34BPgPIpkCyD825f34BPgPIpkCy\nD825f34BPgPIpkCyD825f34BPgPIpkCyD825f34BPgPIpkCyD825f34BPgPIZjcgeb8d20FM\n4uUwmoBA8l8g1EFM4uUwmkAKkIbFx+/Z/tLsu10/FyxZ7SAm8XIYTSAHSFtoGP/So3b9CKQQ\nAnwGkM3LQaojyQbSFdv6OohJvBxGE0gH0uJohsft/MPFfu5c7KPYsyOQYgjwGUC8m4C0Opph\n2i6x8cPx9ZkNAimGAJ+BKCAtbvcfdK89OwIphgCfAWSzDUjD3tEM63Mbyt+Sdv3oYkMIAT4D\nyGa7Z6RxD6TZS7zB8owkkGII8BlANuOBtLrTskNWb8hGEOAzgHg3eR/p8Hek5bkNFYefvD9B\n09B823cgwGcA8W6zsgGnQ269tFuc2/D68vebU6gdmnP//AJ8BhDvTtbavTWF2qE5988vwGcA\n2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5\n988vwGcA2RRI9qE5988vwGcA2ewLJH2sS1MBPgPIZk8gOa8TSh+TeDmMJtASpFfnMJQ/HF6f\n27CxIg91bMp75Wr6mMTLYTSBdiAN8zU8B2QsKHn18M2HHJsSSK0F+Awgm5+CZCbjJSWGhxx6\nct/dlz4m8XIYTaAZSEXc8Tnl66MXlnRsHtswPl7PzT7tfP5C8NCTQGouwGfAAaQJkImIcX70\nwhyk5Y83vp+tAD88s+FWAqm5AJ8BH5DW+/Nmd84uNuxtSSpu19cbDj0JpOYCfAY8QSrPY1iC\nhK/3jm14HyRdbGguwGfABaRhdtcuSPPNegKpIwE+A2eBtPVU0wwkvSHbWoDPwHkgzX4vOiJj\ncbN1sWE4uNhgAUlLhBoL8Bk4DaRxdrV7+/L3CqSty984qeHdy9/xpkwnwGfgRJA+rOH1Q1An\nT6F2aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2W4NUVSdPoXZozv3zC/AZQDYF\nkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDbJQapZCpE+\nJvFyGE1AIL01hcpPVUofk3g5jCaQG6RpTdHz+91HnjyFyuXi6WMSL4fRBDKDBIhevzd78hQE\nUrL+8Qwgm+1Bmm6uBql2S236mMTLYTSBxCAN5ZfFwu/5EvJHnTsFgZStfzwDyGYkkIqtSMVO\nCuvhJ99/VJZAytY/ngGkNxJIjz+WG/oqNvZVlUDK1j+eAaQ3Okiz13YnT0EXG5L1j2cA6Q0O\n0vxq3slTEEjJ+sczgPS2Bml51W5YfukIkt6QTdY/ngHkuD1I8/eR1iD5vbT71hKhXP3jGUCO\nm4O0WNmwfGl3PxLlUSdPoXZozv3zC/AZQIoDgGSvk6dQOzTn/vkF+AwgmwLJPjTn/vkF+Awg\nmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn\n/vkF+Awgm12B5PphFPljEi+H0QQE0k85fzxS/pjEy2E0gXQgzffr7T1odc+hJ+8P7Msfk3g5\njCaQDKRhsRZ172Grew49CaTmAnwGkM0mIM1uXj2sqCNLtdv06it9TOLlMJpALpBKQDY+t6/8\n/L6KMxsEUnsBPgPIZluQtj5JdvaJstODXp7ZIJDaC/AZQJQbPyM9vl+e0fDGmQ0Cqb0AnwGk\nuDVI99duyzMa3jmzQRcbmgvwGUCQW/+ONG6e0fDOmQ0CqbkAnwEkue1Vu+XZDHu3jzo2pTdk\nWwvwGUCo276PtLyoUH5f3j7q1RS0RKitAJ8BhLrxyobHZe7VGQ3L23udPIXaoTn3zy/AZwCR\n7mitXbgp0wnwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM79\n8wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2OwLJ9+3YDmISL4fRBASS/wKhDmISL4fR\nBDKDNNxr98fGtXbuS1Y7iEm8HEYTSA3S7ObgEaNAii7AZwAp7QQk/219HcQkXg6jCfQB0nBf\n8P04xWGYtvgJpCwCfAaQ4yAgTfslyh1I5S6/F2c2CKQIAnwGkOP2IE0XG/DMA7j0jJRJgM8A\nctwepOnm+dW0V2l+/6iLDdEF+Awgx+FAWry+E0iJBPgMIMfRQFr8YQZJb8gGEOAzgBxHBKn8\njckMkpYItRfgM4ActwdpcbHheYpD3eXvt6ZQOzTn/vkF+Awgx81BqqmTp1A7NOf++QX4DCCb\nAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf+\n+QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCb3YDkva6hg5jEy2E0AYHkv9Kug5jEy2E0\ngUwgrQ87GTa/3KpdPxes/e4gJvFyGE0gFUizm8WXL2rXj0AKIcBnANnsAqQr9sd2EJN4OYwm\nkBSkYtU3PrkP+2KHx1ko+Lt7dgRSDAE+A8hmI5DKfbDzz4yd7p3vTDo8/EQgxRDgM4BcN7zY\nsABp/eX8TKFRz0jRBfgMINcNX9ot9+69D5IuNsQQ4DOAXLcEaZx/KZCyC/AZQK77AElvyIYQ\n4DOAXLcGafNiw1ANkpYIRRDgM4Bct1zZsHn5u/yyBqSAU6YT4DOAXAdaa/f6jdmTp1A7NOf+\n+QX4DCCbMUBaPPPs1clTqB2ac//8AnwGkM0YII2Ldaw7dfIUaofm3D+/AJ8BZDMISLY6eQq1\nQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCy2Q9I7u/Ipo9J\nvBxGExBIV6wRSh+TeDmMJpAMpMNjG45q35C2UUQQ4DOAbDYBaXaz+PKo9g0JpAgCfAaQzT5A\numRnX/qYxMthNIG8IM3Xrd6/vC9and35qF0/AimEAJ8BZLMdSHu7zRf3W85sEEgxBPgMINRt\nLzbMgBkX7Ayrl3y7fgRSCAE+A8hm25d2xbENz5dx831I86Ws+4Z0sSGCAJ8BZLMxSMU9w7h+\nRhrnKO0bEkgRBPgMIJthQNo/q8G0Q1ZvyAYQ4DOAUAcAaeeshvnrvFsdetISoeYCfAYQ6sYr\nG7aPbZi+sV7+fm8KtUNz7p9fgM8AstnLWrt3plA7NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+\nNOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4\nDCCb/YCkN2SbC/AZQDZ7AUlLhAII8BlANluC9GJf7PrH+4a0aDWCAJ8BZLMhSMM2SQd47RsS\nSBEE+Awgm32ApI19IQT4DCCb7UFanM8w3Ner1i1aFUghBPgMIJvtQCo28602JD2XhOvMhlQC\nfAYQ52AgLe4by1s9I0UX4DOAODcDaTpKaHk+wxyk2Wu7fUO62BBBgM8A8twOpPnBJ4/7xgVI\n0ze32jckkCII8BlAngM9IxXfrw9wuNWBI70hG0CAzwDy3AokkLM8n2EJkuml3beWCAUQ4DOA\nQLcHaXk+w7A4sth6+Em8KdMJ8BlAoHtZa/fOFGqH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH\n5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCf\nAWRTINmH5tw/vwCfAWSzK5C01q6tAJ8BZLMjkLyX26WPSbwcRhPIBNKLE0+Kh9UcfvJ9wQLw\n9DGJl8NoAolAsh7UsPmoQ08CqbkAnwFksxuQ/DfJpo9JvBxGE8gH0u6JJ8Vn+NV9Yp9Aai/A\nZwDZvBgky4knxafKWg8/+RZIEQT4DCDZEUAq7xvxg4rDT74FUgQBPgNI9rUgvT7xZHqxV3f4\nybcuNgQQ4DOAaF8M0ssTT55fDYtnr1sdehJIzQX4DCDarZ+Riu9nVyGWvyPd6tiU3pBtLcBn\nANG+FCTbiSfjMAPJ+tJOS4SaC/AZQLYbgbR74snzK1z+1uEnaQT4DCDbHa21CzdlOgE+A8im\nQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8hmKpCO639a\n/wM+LRloXR8YEEhxSgZal0D6Keb/jDGK2YBAilMy0LoEkkrVtgSSSnVCCSSV6oQSSCrVCSWQ\nVKoTSiCpVCdUNyDND+7KU7OTKZa34WvvH57EwLQF7nMTvYC02JSeppZnkG3tsI9be//wJAaG\n5ekGH5gQSE1r2D3lL4WZ3CAtDisVSD+V4z/dshanVeTK4ZgdpPKfKJAeleU/3bwEUtsSSKvK\n8p9uVsOYHKTipPaUBobyC4H0U1n+05U1/Zuz5rCfZySB9Kws/+nKWp07my2H3YA0lH8IpAz/\n6ValZ6SGNZQ3AumnsvynW5VAalgFQQLpUUneS1/V/T9XzoUB+//wJAamf+vzuN/3TXQDkkrV\nsgSSSnVCCSSV6oQSSCrVCSWQVKoTSiCpVCeUQFKpTiiBpFKdUAJJpTqhBFLP9dfXL4+vfvn6\nL+7+0n/100sj7br+8eDnv1//KO4VSOeXRtp1/fvr99vt71//Lu4VSOeXRtp1PV/b/fL11zj+\n559fX8O/xjtId5huf/7129fXb381/Ff2UAKp7/rn7bXd7ZXdn1+3+tcKpOHn7l+O+6helEDq\nu/68vba7vbL75eeP/3tCBJB+/2HrX1//2/hfmrwEUud1e6q5vbL7+4npz9//sQbpl/tX/2z4\nj+ygBFLn9dvfr+3+e6fkH/fXdkuQvr6e96veL42v8/p5bff715/jD1K//O+f/xVIPqXx9V5/\nv7b7pbhCNwPpv3hpp/qsNMTe67evP79++/ni6+s/41/T70jD178f3/3r52LDv2fv2KqqSyD1\nXj9XvX9e2f0NTPk70u2733+++ut2+fvr/1r/Q3OXQOq+huevP799ff3jP9PLun8Nf//2dH+B\nd/tBy39iByWQVKoTSiCpVCeUQFKpTiiBpFKdUAJJpTqhBJJKdUIJJJXqhBJIKtUJJZBUqhNK\nIKlUJ5RAUqlOKIGkUp1Q/w8i+oh3Y2WLtgAAAABJRU5ErkJggg==",
182 | "text/plain": [
183 | "plot without title"
184 | ]
185 | },
186 | "metadata": {
187 | "image/png": {
188 | "height": 420,
189 | "width": 420
190 | }
191 | },
192 | "output_type": "display_data"
193 | }
194 | ],
195 | "source": [
196 | "# 绘图\n",
197 | "ggplot(data,aes(x=Country, y=Value) ) +\n",
198 | "# 定义数据轴\n",
199 | "geom_segment( aes(x=Country ,xend=Country, y=0, yend=Value), color=\"grey\") +\n",
200 | "# 绘制点\n",
201 | "geom_point(size=3, color=\"#69b3a2\") +\n",
202 | "# x,y轴调换\n",
203 | "coord_flip() +\n",
204 | "# 设置主题\n",
205 | "theme(\n",
206 | " # 将内部线条设置为空\n",
207 | " panel.grid.minor.y = element_blank(),\n",
208 | " panel.grid.major.y = element_blank(),\n",
209 | " legend.position=\"none\"\n",
210 | ") +\n",
211 | "# 原来x轴也就是现在图像中y轴的轴标题设置为空\n",
212 | "xlab(\"\")"
213 | ]
214 | },
215 | {
216 | "cell_type": "markdown",
217 | "id": "1cddd2a3",
218 | "metadata": {},
219 | "source": [
220 | "很明显,美国和俄罗斯出售物品数量比其他国家多得多。然而,很难看出任何其他国家之间的差异,读者必须从一个国家到另一个国家进行比较。这是很多工作,肯定会放弃对您的图形的关注。"
221 | ]
222 | },
223 | {
224 | "cell_type": "markdown",
225 | "id": "976c6bda",
226 | "metadata": {},
227 | "source": [
228 | "### 有序棒棒糖图\n",
229 | "相反,让我们制作完全相同的图表,但使用它们的值重新排序每个组。\n"
230 | ]
231 | },
232 | {
233 | "cell_type": "code",
234 | "execution_count": 10,
235 | "id": "73cf38bc",
236 | "metadata": {},
237 | "outputs": [
238 | {
239 | "data": {
240 | "text/html": [
241 | "38"
242 | ],
243 | "text/latex": [
244 | "38"
245 | ],
246 | "text/markdown": [
247 | "38"
248 | ],
249 | "text/plain": [
250 | "[1] 38"
251 | ]
252 | },
253 | "metadata": {},
254 | "output_type": "display_data"
255 | },
256 | {
257 | "data": {
258 | "text/html": [
259 | "\n",
260 | "A data.frame: 6 × 2\n",
261 | "\n",
262 | "\t | Country | Value |
|---|
\n",
263 | "\t | <fct> | <int> |
|---|
\n",
264 | "\n",
265 | "\n",
266 | "\t| 1 | United States | 12394 |
|---|
\n",
267 | "\t| 2 | Russia | 6148 |
|---|
\n",
268 | "\t| 3 | Germany (FRG) | 1653 |
|---|
\n",
269 | "\t| 4 | France | 2162 |
|---|
\n",
270 | "\t| 5 | United Kingdom | 1214 |
|---|
\n",
271 | "\t| 6 | China | 1131 |
|---|
\n",
272 | "\n",
273 | "
\n"
274 | ],
275 | "text/latex": [
276 | "A data.frame: 6 × 2\n",
277 | "\\begin{tabular}{r|ll}\n",
278 | " & Country & Value\\\\\n",
279 | " & & \\\\\n",
280 | "\\hline\n",
281 | "\t1 & United States & 12394\\\\\n",
282 | "\t2 & Russia & 6148\\\\\n",
283 | "\t3 & Germany (FRG) & 1653\\\\\n",
284 | "\t4 & France & 2162\\\\\n",
285 | "\t5 & United Kingdom & 1214\\\\\n",
286 | "\t6 & China & 1131\\\\\n",
287 | "\\end{tabular}\n"
288 | ],
289 | "text/markdown": [
290 | "\n",
291 | "A data.frame: 6 × 2\n",
292 | "\n",
293 | "| | Country <fct> | Value <int> |\n",
294 | "|---|---|---|\n",
295 | "| 1 | United States | 12394 |\n",
296 | "| 2 | Russia | 6148 |\n",
297 | "| 3 | Germany (FRG) | 1653 |\n",
298 | "| 4 | France | 2162 |\n",
299 | "| 5 | United Kingdom | 1214 |\n",
300 | "| 6 | China | 1131 |\n",
301 | "\n"
302 | ],
303 | "text/plain": [
304 | " Country Value\n",
305 | "1 United States 12394\n",
306 | "2 Russia 6148\n",
307 | "3 Germany (FRG) 1653\n",
308 | "4 France 2162\n",
309 | "5 United Kingdom 1214\n",
310 | "6 China 1131"
311 | ]
312 | },
313 | "metadata": {},
314 | "output_type": "display_data"
315 | }
316 | ],
317 | "source": [
318 | "# Libraries\n",
319 | "library(tidyverse)\n",
320 | "library(hrbrthemes)\n",
321 | "library(kableExtra)\n",
322 | "options(knitr.table.format = \"html\")\n",
323 | "\n",
324 | "# 从github加载数据\n",
325 | "data <- read.table(\"https://raw.githubusercontent.com/holtzy/data_to_viz/master/Example_dataset/7_OneCatOneNum.csv\", header=TRUE, sep=\",\")\n",
326 | "# 清除空值数据\n",
327 | "data <- filter(data,!is.na(Value))\n",
328 | "nrow(data)\n",
329 | "head(data)"
330 | ]
331 | },
332 | {
333 | "cell_type": "code",
334 | "execution_count": 11,
335 | "id": "efe98df7",
336 | "metadata": {},
337 | "outputs": [],
338 | "source": [
339 | "# 排列数据\n",
340 | "data<- arrange(data,Value)\n",
341 | "# 将Contry转换为factor项,来表示分类数据\n",
342 | "data<- mutate(data,Country=factor(Country, Country)) "
343 | ]
344 | },
345 | {
346 | "cell_type": "code",
347 | "execution_count": 12,
348 | "id": "b26de11a",
349 | "metadata": {},
350 | "outputs": [
351 | {
352 | "data": {
353 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAA0gAAANICAMAAADKOT/pAAAAPFBMVEUAAAAzMzNNTU1oaGhp\ns6J8fHyMjIyampqnp6eysrK9vb2+vr7Hx8fQ0NDZ2dnh4eHp6enr6+vw8PD///9rsEd/AAAA\nCXBIWXMAABJ0AAASdAHeZh94AAAgAElEQVR4nO2di3brRq5EPUwySeYmMznh///rjfVgdfMl\nUGqwAVZhrbFkWUalcGqPaKrZ+hpVKtXH9dX7P0ClukIJJJWqQQkklapBCSSVqkEJJJWqQQkk\nlapBCSSVqkEJJJWqQaUC6cduvfjxx+XdP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFk\nUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP7/A9Qz8/E/t/sJU\nAslc6WOiCR3s//Ojdn5hqsYgDdXN7O7yseGfOtD90BSaV/qYaELH+v/880uSkE13kBbfFQ8M\nzy/D3rNRR6bQvtLHRBM61j8NSLgRSJcQuJaBn39+TRKy6QbSMN4P24bH8dvzKO77Zo5b+YzF\nswuwDkzBodLHRBM61D8KSMMTKBy8Pe8/nzVRsnhGcf/+5V/ftT+F/1OpGlYUkHC7jsYEU/nA\n2rNRB/7vxKHS//+tJnSof1CQhscx2wykEhg84/nssT6pd2AKDpU+JprQsf4xTjasvSKNuyDh\nWLB+ycL9I1NoX+ljogkd658GpPrJi7+RZj0FUniBqxno+Ibs7K2hVycbyic/f7LzF9WhKTSv\n9DHRhA7377hEaDpxXYC0cfq7fvD5E53+TivAZ8ARJM9qPIWjQ3Pun1+AzwCyKZDsQ3Pun1+A\nzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDs\nQ3Pun1+AzwCyKZDsQ3Puf5LAq/cYP+3vWeH+iZFNgWQfmnP/UwRer3r5rL9vhfsnRjZ7gTQM\n0wrvzacsHmk8haNDc+5/hoBhHeZH/Z0r3D8xstkNpNmtqRpP4ejQnPsLpO4CAumtKRwdmnP/\nEwQs16p90t+h56kCuUHCGvB6pWr50L0aT+Ho0Jz7C6TuAtcAaeVWeza0LYHUuD8CHeBkw3yD\nhq1bvSJ9LCCQGvdHoLu+Ig313fqCv3F5QVLjKRwdmnN/nWzoLpAVpAKn5/YnI27L/92r8RSO\nDs25v0DqLnAZkOZ/O+lS89YCekO2aX8EuvPJhvlmDasbeU2/1HgKR4fm3F9LhLoL5AXpeY57\n5/S3XpHSCPAZQKC11s4+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf+\n+QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+tNtXvZ/Zr388\nA8imQLIP7YdW2PTtH88Astn5Mopjv9R4CkeHZvroqc8EfCtcDqMJZATpnV9qPIWjQxNInfvH\nM4BsCiT70Ewfz/uRgHOFy2E0gcwgDWPxwWLDOPuAsVh7Ngikvv3jGUA2+4NUXW3++Gb50Zch\n9mwQSH37xzOAQPc92TCWx3hzgMYSpFs1nsLRoekVqXP/eAYQ6P6vSI+bYXFt31jgdq/GUzg6\nNJ1s6Nw/ngEEOghIa4d05bPu1XgKR4cmkDr3j2cA2YwB0urWdmM8kPSGbNf+8Qwgm3FA2ti2\nIdLfSN+lJUL9+sczgEDHAOm5N0NBVcTT3xLo2z+eAWRTa+3sQ3Pun1+AzwCyKZDsQ3Pun1+A\nzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDs\nQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZA2armCIX1M4uUwmoBAemsKO7W2pi59TOLlMJpAP5De\n2MGk/v1iG5TiY2WLxmX3xlPYrtVV3uljEi+H0QS6gVR87t7bvz+/Mql4cE5S4ylsl0AK2T+e\nAWTzI5A2rnY41mANpCVTt2o8hc1avxI2fUzi5TCaQG+QbvexfclzD5P5B/DhItjVV5wVkGYq\njaewWQIpZv94BtqDVG9fgi+LjU3eBOnUzU8EUsz+8Qy0BGllt4XZlxlQeHj6/WGY7hS7Ci1O\nNzSewmYJpJj94xloCdJYnHtbB2mJWgVS1a0+fdHrClmdbAjZP54BB5AW0cfVrsUzysO79UO7\n+QuZQMojwGfgTJAWB30JQNIbsiH7xzPQCKQi+9t/Iz1PPBhBWh7h9QBJS4Qi9o9noBVIxcqG\n6jRB9WW+sUmBzjAuQSpOQeBJ54N0fv/8AnwGmoH0Vg0r9yzPFkjBBfgMIJsCyT405/75BfgM\nIJvng1Qtcn1NUvmMxlM4OjTn/vkF+Awgm7qMwj405/75BfgMIJsCyT405/75BfgMIJsCyT40\n5/75BfgMIJsCyT405/75BfgMIJsC6btMHzGRPibxchhNQCC9NYVnGT/0KH1M4uUwmkBukF6d\nAvde2WD9GL70MYmXw2gCAumtKTxKIGXpH88AUiqQ1q/ia9ffXukF+AwgpUFAKnd8mH9cn0DK\nIsBnACmOAdJ0ycXaFRm3W789GwRSmv7xDCDFgUAaZ8tZh9kP9YoUXYDPAFIcA6TnStbntX3F\nzqs62ZBHgM8AUhwEpOoiv3KTh1Eg5RHgM4AUhwFpeaH6WSDpDdks/eMZQIpjgLTcIOW8v5G+\nS0uEUvSPZwApjgFSfab7scnDSae/7UNz7p9fgM8AUhwAJHs1nsLRoTn3zy/AZwDZFEj2oTn3\nzy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZ\nFEj2oTn3zy/AZwDZFEi2d2M/6G+u9AJ8BpBNepCM64Pe7n+g0gvwGUA2P/00iupmdnfjWcP6\nk7Z+uajGU/gu64rVd/sfqfQCfAaQzeYgreFQPWsPFoGUWoDPALJ5Oki7rJwOkvmqvjf7H6r0\nAnwGkM1mIA0jtl2YbbxQgjTMn18uUZ1v33BvI5CyCPAZ8AAJl+PNN14YStxWnj9dNFFdPDH9\npueeDQIpU/94BjxAwu3aDib3m+eL08olSFu3vpdRCKRM/eMZOAGkcsOFaSuGFcL6gqSTDZn6\nxzNwAkjlExYnGwTSJQX4DHQE6fnTICDpDdlE/eMZaAYSor7zN9JQgVQ8afdkw0kgaYlQnv7x\nDLQDCae5C0Z2Tn/fb0tQNk9/nwWSeWjO/fML8BloCFKTGl4/5bsaT+Ho0Jz75xfgM4Bsdgdp\n9qKzW42ncHRozv3zC/AZQDa7gzQdAxqq8RSODs25f34BPgPIZn+QDlTjKRwdmnP//AJ8BpBN\ngWQfmnP//AJ8BpBNgWQfmnP//AJ8BpBNgWQfmnP//AJ8BpBNgWQfmnP//AJ8BpBNepCs6xou\nEJN4OYwmIJDemsKPIyvtLhCTeDmMJhAQpNmqoHH1250HVx9vPIVDa78vEJN4OYwmEBEkLPUu\nuBBIlxbgM4Bs+r0izdf+BATpyPWxF4hJvBxGE4gNUrEZSrnjyR2s1R1PFkvBBVIKAT4DZ4C0\ndgXfMH9s5/v6QnWXzU8EUq7+8Qx0A2lcPlZeTLv6c1TjKegVKVn/eAZOAQmX+U3boOCVZ741\nSv19cVjoCZJONuTqH8/AOSAVl8KOK69IYwnMuPqKNNYoNZ6CQErWP56BaCC9OuTzAklvyKbq\nH8/ASSAtdjPZA2fx3DlQWiIUXYDPwFkgVae2h8Up8er72d9GZ5z+Pjg05/75BfgM+IPkUo2n\ncHRozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3z\nC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDbJQbK/HXuBmMTLYTQBgfTWFA4tELpATOLl\nMJpAbpCqD4YZyyVB82o8hUNLVi8Qk3g5jCaQGSRAtA3QsxpPQSAl6x/PALLZH6Tp5myQjl3W\nd4GYxMthNIHEIA3l3QGXmtd7Ozyq7RQEUrb+8Qwgm5FAKq+qHcdqNbjHng0CKVv/eAaQ3kgg\nPb6cdWGfQMrWP54BpDc6SNWxXeMp6GRDsv7xDCC9wUGqz+Y1noJAStY/ngGktzdI87N2w/yu\n56XmekM2V/94BpDj/iDV7yMtQfI7tPuhJUK5+sczgBx3B2m2smF+aPfc6OFWjadwdGjO/fML\n8BlAigOAZK/GUzg6NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4\nDCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf++QX4DCCbbCAdeQf2nf6fVHoBPgPI\nJhdIx9YEHe//WaUX4DOAbAYAqb54r3h88cinUzi4SvVw/w8rvQCfAWSzP0izhal79ekUBFLu\n/vEMIJvdQRqqm/36cApHr+Q72v/TSi/AZwDZjALSuNyoody74V4fTkEgJe8fzwCy2R+kCZX5\nRg3F3g1t9mwQSMn7xzOAHHcH6btmn4P5uG1+YZ9ASt4/ngFkOARI4+pGDe2vkNXJhtz94xlA\ngCOCVGzL9XzoXp9OQSDl7h/PAALcHaTVPRucQNIbsrn7xzOAHPcHqbisfKw2anDZ/ERLhBL3\nj2cAOe4OUrlnA05/O70ifVTpY6IJNe+PFAcA6Vlnb6J/eGjO/fML8BlANgWSfWjO/fML8BlA\nNgWSfWjO/fML8BlANgOB9LoaT+Ho0Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4Bs\nCiT70Jz75xfgM4BsCiT70Jz75xfgM4Bs0oD0wZIGU/8GlV6AzwCySQLSR4vsDP2bVHoBPgPI\nZiiQht1vPwDps2Xfr/u3qfQCfAaQzWggDfW3db09BYF0igCfAWSTAqQPL4192b9RpRfgM4Bs\nBgPpcW3s89rz4fHwo96dgkA6R4DPALIbDKTZh8kCpI82PxFI5wjwGUB2I4JU3J+4utW7UxBI\n5wjwGUB2Y4JU7X7S4MI+nWw4RYDPALIbEqTymvPylMPbUxBIpwjwGUA2I4I0VPeb7LSqN2TP\nEOAzgGxGBWloC5KWCJ0hwGcA2QwH0uP098rWJ1q0Gl2AzwCyGQqktRJIeQT4DCCbAsk+NOf+\n+QX4DCCbwUGqP8uv8RSODs25f34BPgPIZnCQ6mo8haNDc+6fX4DPALIpkOxDc+6fX4DPALIp\nkOxDc+6fX4DPALIpkOxDc+6fX4DPALJJAdLnb8bu929U6QX4DCCbBCC1WB60179ZpRfgM4Bs\nOoE0DPWJ6+pny/uvt/2+1VtTaLJgdad/u0ovwGcA2fQBabG4p/zBEiQjRwIpuACfAWTTBaTt\n15lVkKwcvQVSm4v6tvs3rPQCfAaQTU+Qbnef2y+M05YM0yfzjRVY2KgBuzbMDhHfmYJAOk+A\nz4A3SFP8Vz5geRxKeuqXr6H8abX+++09GwTSeQJ8BpxBesC0BdJEx/1yo+LvpIqd8lXrXu9M\nQSCdJ8Bn4ASQlgCtglQ/MD1zWD22e2sKOtlwmgCfgTggLb7Upx8abH4ikE4T4DOAsDuftTOB\ntNiooT6q+xAkvSF7mgCfAW+QJgDmJxnWQVpDrj438QlIWiJ0lgCfAWeQij9tir91nlsyrIA0\n4oz36q99CFKbSh8TTah5f3eQfKrxFI4Ozbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+\nA8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8jmFUFq\n8/7rdn+vSi/AZwDZvB5IrVYEbfX3q/QCfAaQzW4gDc+aPbz3O5YpNFujutHfsdIL8BlANru+\nIm1ci75ZlikIpH4CfAaQzauB1O46vvX+npVegM8AshkAJCz1Lla0Th/HPBZwGaYgkDoK8BlA\nliOB9Fz8jYsp8ATrng0CqaMAnwFkORJIxe3i/q0MUxBIHQX4DCDL8UCqNm04fqm5Tjb0E+Az\ngCyHA2mo4Tn8qeYCqZ8AnwFkMxxIxf379wdB0huy/QT4DCCbQUAa1kC63z8KkpYIdRPgM4Bs\nBgBp2s9hegg7ONRvLDWewtGhOffPL8BnANkMv9ZOIOUR4DOAbAok+9Cc++cX4DOAbAYHqV7T\n2ngKR4fm3D+/AJ8BZDM4SHU1nsLRoTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZ\nFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZvCJIWtnQSYDPALJ5PZC01q6bAJ8BZLMLSLNPRVr/\n6UpZpqDV3/0E+AwgmwFB2i7LFARSPwE+A8jm1UDSFbIdBfgMIJsRQBru+54Un9k3jGsfai6Q\nggvwGUA2A4BU7nvyeAxAPR/X5icJBPgMINP9Qaq3ZRgqgMqPmx31ihRdgM8AMt0dpOqjmZ8b\nnpSX9x38VHOdbOgnwGcAme4NUrnXCY7nqlcklGUKAqmfAJ8BZLM3SOPsEO5jkPSGbD8BPgPI\nZp+VDeU+J/VeJ5/+jfRdWiLUSYDPACLdaYnQ80+fclPIYhOUAqyDp78dK31MNKHm/ZHN6621\nc6v0MdGEmvdHNgWSfWjO/fML8BlANgWSfWjO/fML8BlANgWSfWjO/fML8BlANgWSfWjO/fML\n8BlANgWSfWjO/fML8BlANgWSfWjO/fML8BlANq8HktPbsReISbwcRhMQSNMU3BYIXSAm8XIY\nTSAoSPWyhPtDe09/+fjrKfgtWb1ATOLlMJpATJBmC+WWXBh/IJCyCPAZQEr9QBqqmzUujD84\nAJLjZX0XiEm8HEYTiAxSfY3EcD/emy7Ym7ZpeF6YhO+fezYIpDQCfAZOAan88MoBX59cVNs0\nzC6nKL8fDuzZIJC6CvAZOAOkB0xPIqbrI8pDvtXrjj64HkkgdRXgM3AWSCPoWQdpmLZpKB75\n4MI+nWzoKcBn4GyQyuM6HO/Vh3lrV8wKpFQCfAaQ8zPO2tlB2ro1g6Q3ZHsK8Bk4BaTqr6Bx\nuT/DBkDzQ7uD23FpiVA/AT4DZ4A0O28HkFZPfz9/PgwVbAdPf7tW+phoQs37nwNS82o8haND\nc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DP\nALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALJ5MZDc3o39cYGYxMthNAGBdJ+C4/qgHxeISbwc\nRhPIBtLWxbOv6oUpzxWrPy4Qk3g5jCaQDqR6yaq5XpgSSJ0F+Awgm51ekWbLuo21b8r1qr4f\nF4hJvBxGE0gM0mKThseX4gI/ALdvSiD1FuAzgEz3+htpZZOG2d4O083tq2HPBoHUW4DPADId\nBaTpFhejF0971L4pgdRbgM8AMt3trN3wAiQc25mvR9LJhs4CfAaQ6X6nv4cVkMqju/JY71Ev\nTAmkzgJ8BpDNoCAVu6HYQdIbsp0F+Awgmx3fkF3su1Vfkv7cJfIASFoi1FeAzwCy2XtlQ7lp\nQ/l12qyhfLOp8RSODs25f34BPgPIZvi1dgIpjwCfAWRTINmH5tw/vwCfAWQzOEj1J5U1nsLR\noTn3zy/AZwDZDA5SXY2ncHRozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1ozv3zC/AZQDYFkn1o\nzv3zC/AZQDYFkn1ozv3zC/AZQDavBZLnwob8MYmXw2gCAulmynepXf6YxMthNIGeIE2f3bL1\n89lzXzx/di1SXfumnBd/549JvBxGE+gH0rBYD7dFxoySV09ffcq+KYHUW4DPALL5KUhmMl5S\nYnjKrifvC2TzxyReDqMJdAOpiPv8Q/gW25rMKJn9uLhE9vl92UwgZRDgM+AA0gTIRMQ4bWsy\nf+Fa+fHK9+X1fYbNTwRSdwE+Az4gDeWXYfFgdbJhuVnD4nZ5vmHXk0DqLsBnwBOkodySbgYS\n7ldPawKSTjZ0F+Az4ALSUD20CVJ94atAupAAn4FWIK291HQDSW/I9hbgM9AOpOrvoj0yZjdr\nJxuGnZMNFpC0RKizAJ+BZiCN1dnu9dPfC5DWTn8/dzt5//R3vCnTCfAZaAjShzW8fgqq8RSO\nDs25f34BPgPIpkCyD825f34BPgPIpkCyD825f34BPgPIZm+QDlXjKRwdmnP//AJ8BpBNgWQf\nmnP//AJ8BpBNgWQfmnP//AJ8BpBNgWQfmnP//AJ8BpDNa4GkN2S7CvAZQDavBJKWCHUW4DOA\nbJ4F0vw0t+G09/Ip+6a0aLW3AJ8BZDMwSMvaNyWQegvwGUA2rwOSLuzrLsBnANk8EaT5+tS1\ndavz5a7asyGTAJ+BPiDNr5hYeXx5SdKoPRvSCPAZ6APSkdtxeatXpOgCfAYQ8Egg4YomAFQd\n2+2b0smG3gJ8BhDwSCAVz57+RhrtV8gKpN4CfAYQ2ZggvXepud6Q7SzAZwCB7QTSi524SpDM\nh3ZaItRbgM9AV5CWp7/x+Mrp7yObn7hW+phoQs37nw9Sk2o8haNDc+6fX4DPALIpkOxDc+6f\nX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIpkOxDc+6fX4DPALIp\nkOxDc+6fX4DPALJ5KZBc34/NH5N4OYwmIJC+y3mFUP6YxMthNIFMIBkvjZ2vvbvVrifvNav5\nYxIvh9EEEoE0rJO0eHD1WbueBFJ3AT4DyOZlQHK/ri9/TOLlMJpAPpBmC1WH++LU6W6x9Nu6\nZ4NA6i/AZwDZPBmk6mK9lUsrZtdQHNizQSD1F+AzgGRHAKl8bMQPju3ZIJD6C/AZQLLPBWna\njKE4tJt+8ADmcbB3eM8GnWzoLsBnANE+GaTnBifFNeTV/gzTvWH26nWrXU8CqbsAnwFEu/cr\nUvF9dRbijT0b9IZsbwE+A4j2qSCBnLV9G6rDvbf2bNASob4CfAaQ7U4gLfZtmPZmeNzD6W/t\n2ZBGgM8Asn2htXbhpkwnwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA\n2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2bwUSFrZ\n0FeAzwCyeSGQtNautwCfAWSzD0j1ha9bT1o8sutJq7+7C/AZQDZ7gDTMFnVvPW3xyK4ngdRd\ngM8AstkFpOrm1dOK2rOkK2T7C/AZQDY7gFQCMm11Un9Y33xx+KP2LAmk/gJ8BpDNviBNlyMV\nGwetfcysNj9JIcBnAFHu/Ir0+H7tglltfpJOgM8AUtwbpPkWDuvfP2rXk042dBfgM4Ag9/4b\naZxtKrT+/aN2PQmk7gJ8BpDkvmftBuPto/ZN6Q3Z3gJ8BhDqvu8jre6BsnL7qFdT0BKhvgJ8\nBhDqzisbsNVJvdnJ/PZejadwdGjO/fML8BlApC+01i7clOkE+AwgmwLJPjTn/vkF+AwgmwLJ\nPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF\n+Awgm5cCSW/I9hXgM4BsXggkLRHqLcBnANnssWj1xYYN2z/c9aRFq90F+Awgm+eD9HLDBoGU\nVYDPALJ5OkivN2x4DyRd2NdfgM8AstkLpPlHxq7v1TDt5XCvPUsCqb8AnwFk83yQpoXf4/Rh\nspt7NUyfeK49GzII8BlArructRuw2Qk+LvbDC/sEUn8BPgPIdK/T30MN0lDv1TD//lG7nnSy\nobsAnwEEuitI03FdtWHQyveP2vUkkLoL8BlAoDuetWsMkt6Q7S7AZwC57nCyYYKjPOkwrp9s\nwNO+69UUtESorwCfAeS658qG4uvm6e/xCEi+lT4mmlDz/kj1hdbahZsynQCfAWRTINmH5tw/\nvwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWRT\nINmH5tw/vwCfAWTzSiD5vh+bPybxchhNQCD98F8hlD8m8XIYTSApSNXmDbvbODxqz5L7mtX8\nMYmXw2gCKUF6rgka55/Lt117lgRSfwE+A8hmR5Bw0wAk/+v68sckXg6jCWQEqdi7YbgvVcW6\n1ed2DfOtu3YcCaQAAnwGkM0IIOETLle3azDt2SCQAgjwGUCe44BU3F+5FOlWO44EUgABPgPI\nZjyQyu0axnpP1j1LOtnQX4DPALIZD6TZz4/va/f+HF8MzavxZQT4DCCbsc7arYB0ePOTt6f4\ncmhuna8iwGcAcY71PtLq30jmS821RKizAJ8BxDnEyobnTg2L7Rrsp7/fmcLRoTn3zy/AZwDZ\nvM5au3hTphPgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz7\n5xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsCiT70Jz75xfgM4BsXgkkrWzoLMBnANm8\nDkhaa9ddgM8AstkXpNXry7cvOt+zpNXf/QX4DCCbAsk+NK/GlxHgM4BsXgUkXSEbQIDPALLZ\nH6T5ZifzB7X5SRoBPgPIZneQFpudDGsPavOTDAJ8BhDlOCA9bqtrZO0X9gmkAAJ8BhDlqCAV\nO6DYrpDVyYb+AnwGEOWgIA3zB2+1Z0kg9RfgM4Ao991FaAukjc3tdj3pDdnuAnwGkOd4ID0P\n6Q4e2mmJUH8BPgPIc89dhGZ7fT++eZ7+PvqKFG/KdAJ8BhDn66y1izdlOgE+A8imQLIPzbl/\nfgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8im\nQLIPzbl/fgE+A8jmlUDSG7KdBfgMIJvXAUlLhLoL8BlANt1Aqj+QZfaz1W+fz9el5lkF+Awg\nm14grX2W8vSDYfbI5pNntWdJIPUX4DOAbDqBtP3i4gSSLuwLIMBnANn0Bel2t1iUOty/q7di\nKHZpuG3cUOzZYP/EPoEUQIDPgD9IQ/UXz+xyiaF8WSo/RPb5g+X1SNqzIYEAnwF3kB4wbYE0\n8TEHqXzyWDzhXi4rfiIAABbOSURBVDuOBFIAAT4DSKnv6e+VF5c5SMO48oPiuK88ttuzpJMN\n/QX4DCDq/UG61wZI9amJPUsCqb8AnwF3kPBy89ErkvZsSCXAZ8AfJGzBUHKyBAnIbP5BpT0b\nsgjwGXAHqfjjpjiTfWNjGF6DNP8lG0jvjs86NOf++QX4DJwAkkc1nsLRoTn3zy/AZwDZFEj2\noTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/A\nZwDZFEj2oTn3zy/AZwDZvBJIekO2swCfAWTzOiBpiVB3AT4DyGYYkOZL78ZpPasWrWYR4DOA\n/IYEaevi8z1LAqm/AJ8BZPMqIOnCvgACfAaQTYFkH5pT3+sI8BlANiOCVF14/ijt2ZBAgM8A\nAhoapAMnGwRSAAE+A8hvQJCG+k5Re5Z0sqG/AJ8BZDMiSOUuDlXtWRJI/QX4DCCbEUGq7xS1\n60lvyHYX4DOAbIYBaWXjk2MgaYlQdwE+A8hmHJCqvY1vt4dWNgScMp0AnwGkNxBIr6vxFI4O\nzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+\nA8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8jmhUByXtiQPybxchhNQCCd\nsNQuf0zi5TCaQD6Q1lZ4L5bWLR7fceS/+Dt/TOLlMJpAQpCMj9WP7zgSSAEE+AwgpRcB6YQL\nZPPHJF4OowkkBml4LPMeq0/qez5k/sQ+gRRBgM8AEh0BpAdF5WfH4qHHZ8i+2PxEIEUQ4DOA\nRHc92TDW9Dx/tvxQ5kdtGxJIEQT4DCDREV6RqlucyzsCkk42RBDgM4BEBwNp7UVKIGUR4DOA\nRMcCaQ6QHSS9IRtAgM8AEh0PpPmhnXnPBi0R6i3AZwCJjgXS83T39EpkPf391hSODs25f34B\nPgNI9GXW2gWcMp0AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZ\nh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFk80Ig6Q3Z3gJ8BpDNy4CkJUL9BfgM\nIJtRQJpfHrv2OWNatBpcgM8AshkYpGXtOBJIAQT4DCCbFwFJF/ZFEOAzgGwGAmm+UUOxh8Oj\ntg0JpAgCfAaQzUggzTZqqK5H0p4NCQT4DCDAkUBaux21Z0MeAT4DCHB0kKwfxqyTDQEE+Awg\nwMFBev7vXjuOBFIAAT4DCHBskLRnQyoBPgMIcFCQipMN2rMhjQCfAQQ4IEiz09/asyGNAJ8B\nBDgKSKZqPIWjQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCy\nKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyKZDsQ3Pun1+AzwCyeSGQ9IZsbwE+A8jmZUDSEqH+\nAnwGkM2zQRqmD720/8p0b8eRFq0GEOAzgJSeDtIcjvXv13+240ggBRDgM4CUXgQkXdgXQYDP\nAFLaEaTpc80fR3uLTxfDB58/atuQQIogwGcAwe4H0nTVRHUB38bnX2rPhgwCfAYQ7H4nGxYf\nuLz6CcyDXpHyCPAZQLA7vSKNJpDuyOlkQxYBPgMIdmSQiu257rXjSCAFEOAzgGAHBmm+LZfe\nkI0uwGcAwQ4G0vNQbn57r31TWiLUW4DPAILdD6Tq7Pb9jyGc9p7v3XCvxlM4OjTn/vkF+Awg\nzVHX2q2+Q9t4CkeH5tw/vwCfAWQzIEizzeyKajyFo0Nz7p9fgM8AshkQpHFzVWvjKRwdmnP/\n/AJ8BpDNiCBtVuMpHB2ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8\nAnwGkE2BZB+ac//8AnwGkM0LgaSVDb0F+Awgm5cBSWvt+gvwGUA2u4P0YjcUrf5OJMBnAEHt\nD1J1s/OMUSBFF+AzgJReBCRdIRtBgM8AUhoIpGF8LAF/bnxSfP7lvbYNCaQIAnwGkOMgIE0f\nFluuWC2vkNXmJwkE+Awgx/1BqndDuT+Gr3pFSiTAZwA57g/SdPO89zyHd+hSc51sCCDAZwA5\nDgfS7PhOICUS4DOAHEcDafZFm59kEuAzgBxHBKn8i0mbnyQS4DOAHPcHaXay4bnjybHT329N\n4ejQnPvnF+AzgBx3B+lINZ7C0aE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI\n9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2RRI9qE5988vwGcA2bwOSN7vx+aP\nSbwcRhMQSCesEMofk3g5jCaQDKTlNg0718eWtW3ohDWr+WMSL4fRBLKBVN3M7u7VtiGBFEGA\nzwCyeQ2QzriuL39M4uUwmkBekIr1qtPdx+f1lQ8+atOPQAohwGcA2ewH0uzzl6e7a5/L/GrP\nBoEUQoDPAELd92TD/Gry+eV9s0O+TT8CKYQAnwFks++hXXHV0fMwrr5Ett6CdduQTjZEEOAz\ngGx2Bql4pNx8q7zW3PI3kkAKIcBnANkMA9Lyz6KhfvqoN2SjC/AZQKgDgISTDdWhXX2cd6td\nT1oi1F2AzwBC3XllQ336u9iU63EO3Hj6+70pHB2ac//8AnwGkM2rrLV7ZwpHh+bcP78AnwFk\nUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bcP78AnwFkUyDZh+bc\nP78AnwFkUyDZh+bcP78AnwFk8zog6Q3Z7gJ8BpDNq4CkJUIBBPgMIJs9QXpxXezyx9uGtGg1\nggCfAWSzI0jDOkk7eG0bEkgRBPgMIJvXAEkX9oUQ4DOAbPYHabY/w3Bfr3ps0apACiHAZwDZ\n7AdScTHf4oKk55Jw7dmQSoDPAOIcDKTZY2N5q1ek6AJ8BhDnbiBNWwnN92eoQaqO7bYN6WRD\nBAE+A8hzP5DqjU8ej40zkKZvbrVtSCBFEOAzgDwHekUqvl9u4HCrHUd6QzaAAJ8B5LkXSCBn\nvj/DHCTTod0PLREKIMBnAIHuD9J8f4ZhtmWxdfOTeFOmE+AzgEBfZa3dO1M4OjTn/vkF+Awg\nmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn/vkF+AwgmwLJPjTn\n/vkF+AwgmwLJPjTn/vkF+Awgm9cBSW/IdhfgM4BsXgUkLREKIMBnANk8H6Tpgyi2n1HdFLVt\nSItWIwjwGUA2TwepWti985S12jYkkCII8BlANs8GaflpfZvPWdamH13YF0KAzwCy2Qmksdqm\n4fl9/Vnn2rMhmwCfAWTzdJAmMoaKmHIvFGzYoD0bUgnwGUCwO3305VhxUv7JNKw98qhNPwIp\nhACfAYS60+nvYR2k56Ge9mxIKcBnAIkOAVKJzu1PpWq/BoGURYDPABLd9azdDKS9v5FuteNI\nb8gGEOAzgGCff7LhyUbFSbUnVw2S9mxII8BnAMHuu7Jh+gvouR3XMDuZpz0bMgnwGUCsr7LW\n7p0pHB2ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+a\nc//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkE2BZB+ac//8AnwGkM3LgOS+sCF/TOLl\nMJqAQDphqV3+mMTLYTSBFCBNH9F36Jeq7zb9nLH4O39M4uUwmkAOkNbQMP7Sozb9CKQQAnwG\nkM3TQTpGkg2kUy6QzR+TeDmMJpAOpGmXk+r2uSwcD84uSNqyI5BiCPAZQLy7gDRdwTeUt/ON\nHKorZHc3PxFIMQT4DEQBaXa7/aR7bdkRSDEE+Awgm31AGooN7VaulC1/WP6VtOlHJxtCCPAZ\nQDb7vSKNWyBVh3iD5RVJIMUQ4DOAbMYDafHgkV2EPpniy6F5Nr+EAJ8BxLvL+0i7fyOVW6CY\nQdISoQgCfAYQ7z4rG+pNvhcbCQ3LB2/VeApHh+bcP78AnwHE+yJr7d6awtGhOffPL8BnANkU\nSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffPL8BnANkUSPahOffP\nL8BnANkUSPahOffPL8BnANm8DEh6Q7a/AJ8BZPMiIGmJUAQBPgPI5ukgLbduGFbvrtWmHy1a\nDSHAZwDZPB+k6mZ290Vt+hFIIQT4DCCblwBJF/bFEOAzgGz2BKlYuoqFqljIWu7jcK8tOwIp\nhgCfAWSzE0iztd/FpRP1lg3asyGTAJ8B5LrjyYYZSMu786so9IoUXIDPAHLd8dCu2LnhQ5B0\nsiGGAJ8B5LonSGN9VyBlF+AzgFxfAyS9IRtCgM8Act0bpNWTDcNhkLREKIIAnwHkuufKhtXT\n3ztbNmjRanABPgPIdaC1dq/fmG08haNDc+6fX4DPALIZA6TZK89WNZ7C0aE5988vwGcA2YwB\n0jhbx7pRjadwdGjO/fML8BlANoOAZKvGUzg6NOf++QX4DCCbAsk+NOf++QX4DCCbAsk+NOf+\n+QX4DCCbAsk+NOf++QX4DCCblwFJb8j2F+AzgGxeBCQtEYogwGcA2YwFkvZsSC3AZwDZFEj2\noTn2voYAnwFk8xIg6cK+GAJ8BpDNcCAN42Orhvt6Vu3ZkEiAzwCyGQ+k6ip07dmQSYDPAKIb\nD6TFN9qzIYsAnwFENypI03YOutQ8jwCfAUQ3KEjDeOQVSSDFEOAzgOjGBGm5ccOttg3pDdkI\nAnwGEN24IB05tPuhJUIRBPgMILoxQVpu3HCrxlM4OjTn/vkF+AwgurFAelGNp3B0aM798wvw\nGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2BZJ9\naM798wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2LwOSVjb0F+AzgGxeBCSttYsgwGcA\n2XwJ0lDdzO42qGGt4Ybaph+t/g4hwGcA2YwA0v5jhkvNBVIMAT4DyOYlQNIVsjEE+AwgmwdA\neu5E8liSPW1MMr+YtfgRflxcYlTva4LP6Xs+igXgQ/lfIJDCC/AZeA+kMvf1t+PweHSCoX4p\nKz8xdvnrKz9dfNHmJwkE+Ay8BxJu65eY4uEai+Jm7elrINX4jPW9LTsCKYYAn4EPQVq9HHwD\npLE8QNsGaVjsdjIdHBou7NPJhhACfAY+A+l5BLfG19qFrcP60yuQSrU1BAVSAgE+A5+BtEXE\nFkhbAL0CafHttiG9IRtBgM/AAZAQ5hcgPY/IZvmvfmsYX51sqPExg6QlQhEE+AwcAWk6Dz0L\nOwAqf74ACT9+7meyf/p7fB7S4fdMIAWcMp0An4FDILWt5SmE7efMv2k8haNDc+6fX4DPALIp\nkOxDc+6fX4DPALIZEqTyh+XzGk/h6NCc++cX4DOAbF7kMoq3pnB0aM798wvwGUA2BZJ9aM79\n8wvwGUA2BZJ9aM798wvwGUA2BZJ9aM798wvwGUA2LwOS3pDtL8BnANm8CEhaIhRBgM8AstkN\npOG54BuPbD1zurfpR4tWQwjwGUBK+4FU3czurj1zFEjRBfgMIKWXAEkX9sUQ4DOAlEYAqViw\nOtQrWp8rXR+1ZUcgxRDgM4A8BwCpvjJpfo3FY/W39mxIIMBnAHkOcLKhujxjnLOlQ7s8AnwG\nkOfer0jjnJpq6wbrhX062RBCgM8A8hwMpK2rb++16UcghRDgM4A8xwJpDpD5UnO9IRtBgM8A\n8hwPpPmhnfFScy0R6i/AZwB5DgDS7PR3vaGK6fT3m1M4OjTn/vkF+AwgxBdZa/fWFI4Ozbl/\nfgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8imQLIPzbl/fgE+A8im\nQLIPzbl/fgE+A8imQLIPzbl/fgE+A8jmZUDSG7L9BfgMIJsXAUlLhCII8BlANvuCNNu14fHg\n7Ba16UeLVkMI8BlANruCNFuTWj66Wpt+BFIIAT4DyGZPkJbbNqx+j9qyowv7YgjwGUA2A4B0\nu4u9GsqPIJsd+23ZEUgxBPgMIJtdQZooqfdqwNV9OPbTng0JBPgMIMy9z9qtbNuwsofDo7bs\nCKQYAnwGEOTeII2PV6J6r4b5Hg6P2vSjkw0hBPgMIMVBQMLd1T0cHrXpRyCFEOAzgBQHONmw\nA5L2bEglwGcAYe57suH2dVwczY0lSNqzIY0AnwGEOcjKhnqvyMUeDo9qPIWjQ3Pun1+AzwCi\nHOBvJHs1nsLRoTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZ\nFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZFEj2oTn3zy/AZwDZvAxIWtnQ\nX4DPALJ5EZC01i6CAJ8BZDMASNuXls9r049Wf4cQ4DOAbAok+9Ace19DgM8AsnkJkHSFbAwB\nPgPIZhCQis1Pygtjp+/vtWVHIMUQ4DOAbMYAqd78ZPm9Nj9JIcBnACmOAdLzdtj7Xq9I0QX4\nDCDFUUBabH4y+/5Wm350siGEAJ8BpDgISLNDusX399r0I5BCCPAZQIo7X2r++PLq9lHbhvSG\nbAQBPgOIchyQhtnJhiOHdj+0RCiCAJ8BRLnzod2A0913gObfj1aQAk6ZToDPAJIc4G+kol68\nN9t4CkeH5tw/vwCfAWRTINmH5tw/vwCfAWRTINmH5tw/vwCfAWQzFkgvqvEUjg7NuX9+AT4D\nyKZAsg/NuX9+AT4DyGYqkPbrX73/Az4tGehdHxgQSHFKBnqXQPou5n/GGMVsQCDFKRnoXQJJ\npepbAkmlalACSaVqUAJJpWpQAkmlalACSaVqUJcBaag27spT5e5ji9vwtfUfnsRA9Ungn5m4\nCkizSwDTVPlh7ltX2Mct4xYBUWsoNtv51IRA6lrDKJC61TAKpEXl+Keb13N7iuJ+nhyO2UEq\n/xMF0qOy/NPVJZD6lkBaVJZ/uqqGMTlI2KN9TGlgKO8IpO/K8k9X1vTfnDWH13lFEkjPyvJP\nV9Zwr7w5vAxIQ/lFIGX4p1uUXpE61lDeCKTvyvJPtyiB1LEKggTSo5K8l76o+z9XzoUB2//h\nSQxM/633UyZa2aBSdS6BpFI1KIGkUjUogaRSNSiBpFI1KIGkUjUogaRSNSiBpFI1KIGkUjUo\ngXTl+vvrp8e9n77+wsNf+ldvXhrppeuXBz9/ff1SPCqQ2pdGeun64+v32+3vX38Ujwqk9qWR\nXrqex3Y/ff09jv/999fX8Nt4B+kO0+3r379+ff36d8f/yiuUQLp2/ft2bHc7svvz61a/LUAa\nvh/+ab+P6kUJpGvXn7dju9uR3U/fX/73hAgg/f7N1m9f/+n8X5q8BNLF6/ZSczuy++eF6c/f\nf1mC9NP93r87/kdeoATSxevXf47t/rpT8sv92G4O0tfX83HV+6XxXby+j+1+//pz/Ebqp//8\n+ZdA8imN7+r1z7HdT8UZugqkv3Bop/qsNMSr169ff379+n3n6+u/49/T30jD1x+P7377Ptnw\nR/WOrepwCaSr1/dZ7+8ju3+AKf9Gun33+/e9v2+nv7/+1/s/NHcJpMvX8Pzz59evr1/+Ox3W\n/Tb889fT/QDv9oOe/4kXKIGkUjUogaRSNSiBpFI1KIGkUjUogaRSNSiBpFI1KIGkUjUogaRS\nNSiBpFI1KIGkUjUogaRSNSiBpFI1qP8HvLyId2hUfqsAAAAASUVORK5CYII=",
354 | "text/plain": [
355 | "plot without title"
356 | ]
357 | },
358 | "metadata": {
359 | "image/png": {
360 | "height": 420,
361 | "width": 420
362 | }
363 | },
364 | "output_type": "display_data"
365 | }
366 | ],
367 | "source": [
368 | "# 绘图\n",
369 | "ggplot(data,aes(x=Country, y=Value) ) +\n",
370 | "# 定义数据轴\n",
371 | "geom_segment( aes(x=Country ,xend=Country, y=0, yend=Value), color=\"grey\") +\n",
372 | "# 绘制点\n",
373 | "geom_point(size=3, color=\"#69b3a2\") +\n",
374 | "# x,y轴调换\n",
375 | "coord_flip() +\n",
376 | "# 设置主题\n",
377 | "theme(\n",
378 | " # 将内部线条设置为空\n",
379 | " panel.grid.minor.y = element_blank(),\n",
380 | " panel.grid.major.y = element_blank(),\n",
381 | " legend.position=\"none\"\n",
382 | ") +\n",
383 | "# 原来x轴也就是现在图像中y轴的轴标题设置为空\n",
384 | "xlab(\"\")"
385 | ]
386 | },
387 | {
388 | "cell_type": "markdown",
389 | "id": "2ea94485",
390 | "metadata": {},
391 | "source": [
392 | "这个数字现在更具洞察力,法国是第三大出口国,其次是德国、以色列和英国。当然,请注意,将每个国家的人口归一化该图形以获得更多可比数据是有意义的。重新排序数据是构建图表时应始终考虑的简单步骤。当然,有时组的顺序必须由它们的特征而不是它们的值来设置,例如一年中的几个月,这是值得考虑。"
393 | ]
394 | },
395 | {
396 | "cell_type": "markdown",
397 | "id": "e6a47f94",
398 | "metadata": {},
399 | "source": [
400 | "## 参考\n",
401 | "+ [WHY YOU SHOULD ORDER YOUR DATA](https://www.data-to-viz.com/caveat/order_data.html)\n",
402 | "+ [如何理解R中因子(factor)的概念](https://www.zhihu.com/question/48472404)"
403 | ]
404 | }
405 | ],
406 | "metadata": {
407 | "kernelspec": {
408 | "display_name": "R",
409 | "language": "R",
410 | "name": "ir"
411 | },
412 | "language_info": {
413 | "codemirror_mode": "r",
414 | "file_extension": ".r",
415 | "mimetype": "text/x-r-source",
416 | "name": "R",
417 | "pygments_lexer": "r",
418 | "version": "3.6.0"
419 | }
420 | },
421 | "nbformat": 4,
422 | "nbformat_minor": 5
423 | }
424 |
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/[数据分析与可视化] 数据绘图要点2-Y轴的开始与结束.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "7890093b",
6 | "metadata": {},
7 | "source": [
8 | "# 数据绘图要点2-Y轴的开始与结束\n",
9 | "\n",
10 | "\n",
11 | "切割或不切割Y轴可能是数据可视化中最具争议的话题之一。基本上,主要问题在于 Y 轴是否应始终从零开始。数据可视化的目的是讲述一个故事,图形表达方式会对可视化讲述的故事产生重大影响。好的可视化可以带出数据的重要方面,但可视化也可以用来隐藏或误导。我们将探讨这一看似简单的问题的一些微妙之处。\n",
12 | "\n"
13 | ]
14 | },
15 | {
16 | "cell_type": "markdown",
17 | "id": "a112b6e2",
18 | "metadata": {},
19 | "source": [
20 | "## Y轴的开始\n",
21 | "### 不好的实例\n",
22 | "\n",
23 | "让我们从显示问题的条形图开始。我们从一个众所周知的问题开始:绘制Y轴(因变量)不从0开始的条形图。条形图由德国经济发展机构 GTAI 创建,来自一个有关[德国劳动力市场的网页](https://www.gtai.de/gtai-en/invest/business-location-germany/business-climate/motivated-and-dependable-employees-81844)。在随附的文本中,该机构吹嘘德国工人比其他欧盟国家的工人更有动力,工作时间更长。\n",
24 | "\n",
25 | "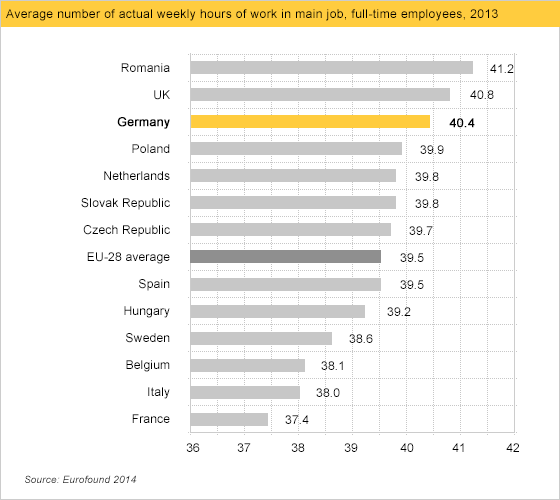"
26 | ]
27 | },
28 | {
29 | "cell_type": "markdown",
30 | "id": "f36d8688",
31 | "metadata": {},
32 | "source": [
33 | "看起来德国比瑞典等其他国家有很大的优势,更不用说法国了,对吧?不,这个差距的大小是一种错觉。该图具有误导性,因为表示工作时间的横轴并未归零,而是在 36 处截断。下面,我们重新绘制了该图,其中因变量轴一直归零。现在国家之间的差异似乎可以忽略不计。您可能会注意到,在重新绘制的图表中,我们删除了分隔国家/地区的水平网格线。这些并不是特别具有误导性,但它们增加了视觉混乱,没有任何作用。\n",
34 | "\n",
35 | "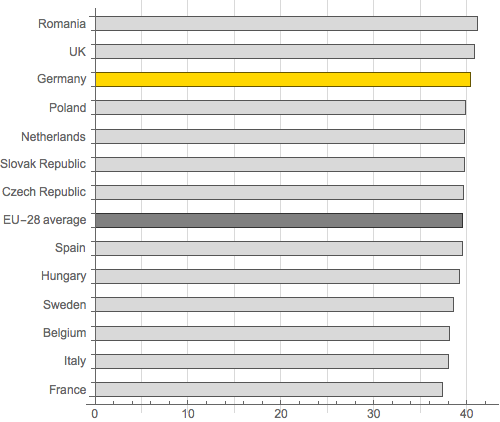"
36 | ]
37 | },
38 | {
39 | "cell_type": "markdown",
40 | "id": "5d322d43",
41 | "metadata": {},
42 | "source": [
43 | "### 好的实例\n",
44 | "\n",
45 | "虽然条形图中的条形应该(几乎)总是延伸到零,但折线图不需要在因变量轴上包含零。例如,下面来自加利福尼亚预算和政策中心的折线图非常好,尽管Y轴不从0开始。\n",
46 | "\n",
47 | "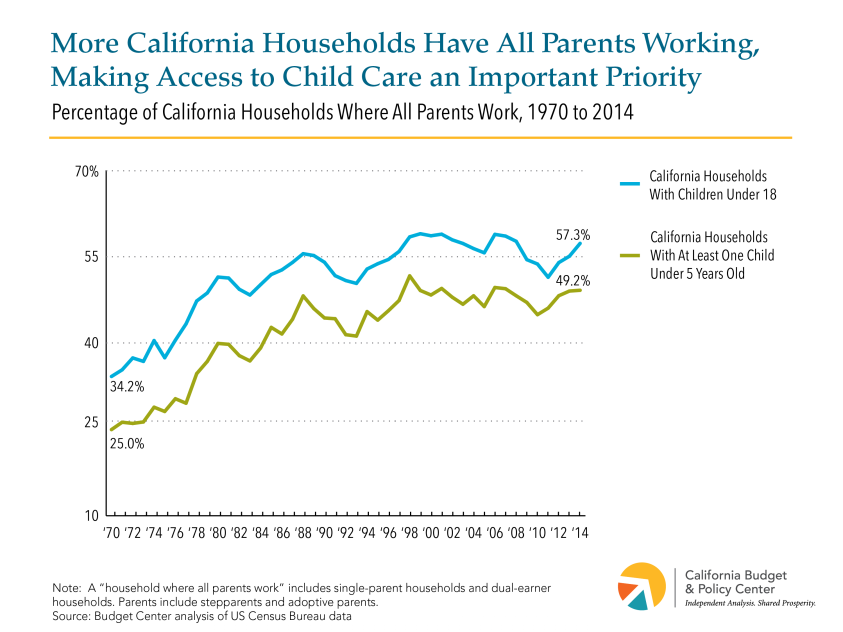\n",
48 | "\n",
49 | "有什么区别?为什么条形图需要在Y轴上包含0,而折线图不需要这样做?一种观点是,这两种类型的图表讲述了不同的故事。通过其设计,条形图强调与每个类别相关的值的绝对大小,而折线图强调因变量(通常为Y值)随自变量(通常为X值)的变化而变化。"
50 | ]
51 | },
52 | {
53 | "cell_type": "markdown",
54 | "id": "6bc404b8",
55 | "metadata": {},
56 | "source": [
57 | "那么在某些情况下,如果折线图的Y轴从0开始,会出现什么样的情况? 下图是一张被广泛分享用于表现气候变化的折线图,该折线图的Y轴从0开始。可以得出结论,气候根本没什么变化。视觉效果看起来像一条扁平线。我们不会看到任何变化,这样这张图对我们来说将变得毫无意义。\n",
58 | "\n",
59 | "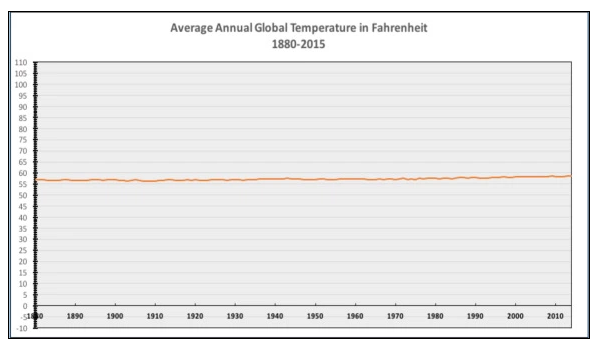\n",
60 | "\n",
61 | "但是如果我们通过适当的选择尺度显示,该折线图可能变成下面这样。很显然,这一变化与我们平时所看到的,感受到的是一致的。\n",
62 | "\n",
63 | ""
64 | ]
65 | },
66 | {
67 | "cell_type": "markdown",
68 | "id": "ac521238",
69 | "metadata": {},
70 | "source": [
71 | "因此除了条形图,一般提倡使用基于数据合理的Y轴。也许轴的最小值是您历史上的最低点,也许是您的团队决定需要采取不同行动的触发点。随便设置,只要让它有意义,而不是软件自动为您决定,这是您需要思考的地方。对于那些0不在可能的数据点范围内,则更加不能够将其包含在Y轴中。\n",
72 | "\n"
73 | ]
74 | },
75 | {
76 | "cell_type": "markdown",
77 | "id": "d0f9b384",
78 | "metadata": {},
79 | "source": [
80 | "## Y轴的结束"
81 | ]
82 | },
83 | {
84 | "cell_type": "markdown",
85 | "id": "4b07af9d",
86 | "metadata": {},
87 | "source": [
88 | "有些人认为Y轴必须以值的理论最大值为结束。下图表示的是一项调查情况,这会压缩数据并形成一个尴尬的图表,我们无法完全看到发生了什么。这些项中的任何一条实际中都不可能达到100%,因此,Y轴可以不以100%为结束。\n",
89 | "\n",
90 | "\n",
91 | "\n",
92 | "\n",
93 | "\n",
94 | "如果我们选择一个更接近真实数据结束位置的轴,我们实际上可以更清楚地看到数据。\n",
95 | "\n",
96 | ""
97 | ]
98 | },
99 | {
100 | "cell_type": "markdown",
101 | "id": "c70017ac",
102 | "metadata": {},
103 | "source": [
104 | "虽然这确实让我们的数据全面可见,但它可能会遗漏部分故事。如果我们定了一个目标,设置这些项离我们的目标有多远。比如我们有将报告为盟友ally的百分比提高到 75% 的目标。那么75%可以成为该条形图Y轴的结束点。最好让我们这样标记目标,以便我们的目标是显而易见的。\n",
105 | "\n",
106 | "\n",
107 | "\n",
108 | "\n"
109 | ]
110 | },
111 | {
112 | "cell_type": "markdown",
113 | "id": "b487707a",
114 | "metadata": {},
115 | "source": [
116 | "## 总结\n",
117 | "总之,数据可视化会讲故事。相对微妙的选择,例如条形图或折线图中轴的范围,会对图形讲述的故事产生重大影响。当您查看数据图形时,您需要问问自己图形是否旨在讲述一个准确反映基础数据的故事,或者它是否旨在讲述一个希望大家能看到的故事。\n",
118 | "\n",
119 | "条形图强调与每个类别相关的值的绝对值,而折线图强调因变量通常是Y值随着自变量(通常是X值)的变化而发生的变化,这是重点。因此:\n",
120 | "\n",
121 | "+ 条形图:对于这种图表有共识:Y轴应该从0开始。\n",
122 | "+ 折线图:对于这种图表没有达成共识,Y轴通常不必从0开始。\n",
123 | "\n",
124 | "至于Y轴的结束点,关键问题是你应该为你的Y轴选择一个有意义的最大值。也许最大值是您的目标,这样轴本身就成为您需要讲述数据的故事的一部分。"
125 | ]
126 | },
127 | {
128 | "cell_type": "markdown",
129 | "id": "7cdda6ac",
130 | "metadata": {},
131 | "source": [
132 | "## 参考\n",
133 | "\n",
134 | "+ [TO CUT OR NOT TO CUT](https://www.data-to-viz.com/caveat/cut_y_axis.html)\n",
135 | "+ [Misleading axes on graphs](https://www.callingbullshit.org/tools/tools_misleading_axes.html)\n",
136 | "+ [Where to Start and End Your Y-Axis Scale](http://stephanieevergreen.com/y-axis/)\n"
137 | ]
138 | }
139 | ],
140 | "metadata": {
141 | "kernelspec": {
142 | "display_name": "R",
143 | "language": "R",
144 | "name": "ir"
145 | },
146 | "language_info": {
147 | "codemirror_mode": "r",
148 | "file_extension": ".r",
149 | "mimetype": "text/x-r-source",
150 | "name": "R",
151 | "pygments_lexer": "r",
152 | "version": "3.6.0"
153 | }
154 | },
155 | "nbformat": 4,
156 | "nbformat_minor": 5
157 | }
158 |
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/[数据分析与可视化] 数据绘图要点5-误差线的问题.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "9f498e92",
6 | "metadata": {},
7 | "source": [
8 | "# 数据绘图要点5-误差线的问题\n",
9 | "\n",
10 | "误差线给出了测量精确度的一般概念,真实(无误差)值可能与报告值相差多远。如果条形图上显示的值是聚合的结果(如多个数据点的平均值),您可能需要显示误差线。但我们必须要谨慎使用误差线,具体原因将在后续给出。"
11 | ]
12 | },
13 | {
14 | "cell_type": "markdown",
15 | "id": "82931290",
16 | "metadata": {},
17 | "source": [
18 | "## 误差线的绘制\n",
19 | "\n",
20 | "在下图中,报告了5个group。条形高度代表它们的平均值。黑色误差线提供有关单个观测值如何分散在平均值周围的信息。例如,似乎groupB中的测量结果比groupE中的更精确。"
21 | ]
22 | },
23 | {
24 | "cell_type": "code",
25 | "execution_count": 7,
26 | "id": "edc404bf",
27 | "metadata": {},
28 | "outputs": [
29 | {
30 | "data": {
31 | "text/html": [
32 | "\n",
33 | "A data.frame: 5 × 3\n",
34 | "\n",
35 | "\t | name | value | sd |
|---|
\n",
36 | "\t | <fct> | <int> | <dbl> |
|---|
\n",
37 | "\n",
38 | "\n",
39 | "\t| 1 | a | 10 | 1.0 |
|---|
\n",
40 | "\t| 2 | b | 5 | 0.2 |
|---|
\n",
41 | "\t| 3 | c | 12 | 3.0 |
|---|
\n",
42 | "\t| 4 | d | 9 | 2.0 |
|---|
\n",
43 | "\t| 5 | e | 7 | 4.0 |
|---|
\n",
44 | "\n",
45 | "
\n"
46 | ],
47 | "text/latex": [
48 | "A data.frame: 5 × 3\n",
49 | "\\begin{tabular}{r|lll}\n",
50 | " & name & value & sd\\\\\n",
51 | " & & & \\\\\n",
52 | "\\hline\n",
53 | "\t1 & a & 10 & 1.0\\\\\n",
54 | "\t2 & b & 5 & 0.2\\\\\n",
55 | "\t3 & c & 12 & 3.0\\\\\n",
56 | "\t4 & d & 9 & 2.0\\\\\n",
57 | "\t5 & e & 7 & 4.0\\\\\n",
58 | "\\end{tabular}\n"
59 | ],
60 | "text/markdown": [
61 | "\n",
62 | "A data.frame: 5 × 3\n",
63 | "\n",
64 | "| | name <fct> | value <int> | sd <dbl> |\n",
65 | "|---|---|---|---|\n",
66 | "| 1 | a | 10 | 1.0 |\n",
67 | "| 2 | b | 5 | 0.2 |\n",
68 | "| 3 | c | 12 | 3.0 |\n",
69 | "| 4 | d | 9 | 2.0 |\n",
70 | "| 5 | e | 7 | 4.0 |\n",
71 | "\n"
72 | ],
73 | "text/plain": [
74 | " name value sd \n",
75 | "1 a 10 1.0\n",
76 | "2 b 5 0.2\n",
77 | "3 c 12 3.0\n",
78 | "4 d 9 2.0\n",
79 | "5 e 7 4.0"
80 | ]
81 | },
82 | "metadata": {},
83 | "output_type": "display_data"
84 | }
85 | ],
86 | "source": [
87 | "# 加载库\n",
88 | "library(tidyverse)\n",
89 | "library(hrbrthemes)\n",
90 | "library(viridis)\n",
91 | "library(patchwork)\n",
92 | "\n",
93 | "# 创建数据\n",
94 | "data <- data.frame(\n",
95 | " # 创建小写数字\n",
96 | " name=letters[1:5],\n",
97 | " value=sample(seq(4,15),5),\n",
98 | " sd=c(1,0.2,3,2,4)\n",
99 | ")\n",
100 | "\n",
101 | "# 展示数据\n",
102 | "head(data)"
103 | ]
104 | },
105 | {
106 | "cell_type": "code",
107 | "execution_count": 8,
108 | "id": "0c41d90a",
109 | "metadata": {},
110 | "outputs": [
111 | {
112 | "data": {
113 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAA0gAAANICAMAAADKOT/pAAAATlBMVEUAAAABAgICAgIOExIP\nFBMXFxcZGRkzMzNNTU1oaGh8fHyMjIyQxLiWyr6ampqnp6eysrK9vb3Hx8fQ0NDZ2dnh4eHp\n6enr6+vw8PD///+8zlEtAAAACXBIWXMAABJ0AAASdAHeZh94AAAfrklEQVR4nO2dgVYayRZF\n2yTjoL7RJGZs+f8ffSCoTSZVnqIucLy191rjUhHct6t2ukEnmdYA0M10aQGADBASQACEBBAA\nIQEEQEgAARASQACEBBAAIQEEQEgAARASQAA9IU3TtPxAucPyox+r4he93KQ8ovJ9AE5Px5Z7\n3IT0+P5AzSEV7rH99MtNhASfh44tdz/dTffvDxQV0ttNhASfh44tN03Pix27efduun3avvvr\nbppWDy+f+3d1u7jl5cuf7qfp/unlwnB378eXHH+9nN3ups15bn/T5r+H3ePseN7e8XnxsNu3\nbw/3+nHdBeA0HB/Sdv/fv1/bTZvz02bPPu8u+TY8bD93u/mizUbf37LN43k17T56C2k9bZ8R\nPUy77b5ehHS3f5wdL3e8Wb8/7Pbt28PtP/7ABeA0HB/SNqLH97252ajP69vtlr2Zfq7X/+5S\neDi4Zfu5h2lzXnj7aP9I/24zWW3vdb98jrS53/fp9SWJ79v7PEw/3h92+3b5cA8fuwCchuND\n+u15zDRtLqWeXs4Y66fH77e7zbu/5trfsv3czdvXvd33cfq+ubJ7mH6tf2ziXIT0tPwONy/v\nTHfvNzytDx/u6WMXgNNwdEj7i6a3a7vdhn95e7u75eBzb30cfrS/7+3mzPK8OWXcLb9oee/1\n7jnVnx/294eruACchqO31/1+X79e271v3vvp5sfjU0tI99Pz6m59t9qfbwJD+t0F4DQcvb1W\n0/YFtOe3pzDT4SXb83LHv9xyW7y0217bbZ7L/Nxc3f0sh3SzuIp8f/unh6u4AJyGY7fXr/2p\n6H7zzGb3QJtSnm83z3Y27/3avrcI6f2WP77Y8HK2edru9+m5HNLLy3o/X3t8fVt4uJILwGk4\ndns97AN6fH0x7P0l54f/XINtn6i8/tLP8vXqt98Run95YeBmG8U+pNV/QtrdcfvCxfKG5cO9\nD1V0ATgNx26v1eq3d15+CHr38srY5unT7a+DS7vb15+Zrhc/Qf3xHtIux+313e6LfvwhpJc7\n3v5a/3bD4gey70MVXQBOwxm2F1sY8kNIAAEQEkAAhAQQAJscIABCAgiAkAACICSAAAgJIABC\nAgiAkAACODak+VSc7pE7sJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTy\ntBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTy\ntBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTy\ntBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTy\ntBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTytBpLipDasJTy\ntBpLipDasJTytBpLqi2k1e7tFkIywtJqLKmmkPb5rA4+eSqzwRaiC0ursaRaQlqtCclSytNq\nLKmmM9IuocOOCOks/FXlunbjhYwHW79jQnp7ivT3FuFu0M11jS9fvlRuvbT6aLSdkXix4bxU\nT0ibkDgjaTidkX5771Rmgy1ED5tLu0sr/AHLQ0VIJlhKEZKMU0hc2rlBSDJuIS1euTuV2WAL\n0QMhyTiFtD74xQZCujyEJGMS0p84ldlgC9EDIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSC\npRQhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlgKUVI\nMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCSCZZShCRDSCZYShGSDCGZ\nYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoR\nkgwhmWApRUgyhGSCpRQhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENI\nJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCSCZZS\nhCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQ\nkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKl\nFCHJEJIJllKEJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgy\nhGSCpRQhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlg\nKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCSCZZShCRDSCZYShGS\nDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gm\nWEoRkgwhmWApRUgyhGSCpRQhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKE\nJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCS\nCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZKMcUhwca6vry+tAG9w\nRlKwlOKMJGN8RjqV2WAL0QMhyRCSCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKE\nJENIJlhKEZIMIZlgKUVIMoRkgqUUIckQkgmWUoQkQ0gmWEoRkgwhmWApRUgyhGSCpRQhyRCS\nCZZShCRDSCZYShGSDCGZYClFSDKEZIKlFCHJEJIJllKEJENIJlhKEZIMIZlgKUVIMoRkgqUU\nIckQkgnnlvqfwtdvX4WvOrP5YOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dI\nbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjr\nR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLM\nWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0Q\nksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dI\nbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY60dIbRCSzFjr\nR0htEJLMWOtHSG0QksxY60dIbRCSzFjrR0htEJLMWOtHSG0QksxY63fRkP6qcl29NewItEFI\nCyzX70JSviF9+fLlIgtRh5AWWK7fhaQIqQ1CWmC5fheSumhIVTZn4VM9dAeEpGK5fqeTIqQ2\nCEnFcv0IyQVCUrFcP0JygZBULNePkFwgJBXL9SMkFwhJxXL9CMkFQlKxXD9CcoGQVCzXj5Bc\nICQVy/UjJBcIScVy/QjJBUJSsVw/QnKBkFQs14+QXCAkFcv1IyQXCEnFcv0IyQVCUrFcP0Jy\ngZBULNePkFwgJBXL9SMkFwhJxXL9CMkFQlKxXD9CcoGQVCzXj5BcICQVy/UjJBcIScVy/QjJ\nBUJSsVw/QnKBkFQs14+QXCAkFcv1IyQXCEnFcv0IyQVCUrFcP0JygZBULNePkFwgJBXL9SMk\nFwhJxXL9CMkFQlKxXD9CcoGQVCzXj5BcICQVy/UjJBcIScVy/QjJBUJSsVw/QnKBkFQs14+Q\nXCAkFcv1IyQXCEnFcv0IyQVCUrFcP5eQVru3GwjpPBBSKCYh7fp5f0NIJ4eQQvEIabUmJEJS\nsVw/j5DWhERIMpbrZxrS31uEu/2HfwS+ffumfNkx3/4ToRwC8VCd2fz6+vrM31HgHFLnPCPF\n/TF77j9nOSOpcEYipAqEpEJIhFSBkFQIiZAqEJIKIRFSBUJSIaSPQgr4zYa43UFI8qE6szkh\nNXLMd43bHYQkH6ozmxMSIVUgJBVCIqQKhKRCSIRUgZBUCImQKhCSCiERUgVCUiEkQqpASCqE\nREgVCEmFkAipAiGpEBIhVSAkFUIipAqEpEJIhFSBkFQIiZAqEJIKIRFSBUJSISRCqkBIKoRE\nSBUISYWQCKkCIakQEiFVICQVQiKkCoSkQkiEVIGQVAiJkCoQkgohEVIFQlIhJEKqQEgqhERI\nFQhJhZAIqQIhqRASIVUgJBVCIqQKhKRCSIRUgZBUCImQKhCSCiERUgVCUiEkQqpASCqEREgV\nCEmFkAipAiGpEBIhVSAkFUIipAqEpEJIhFSBkFQIiZAqEJIKIRFSBUJSIaTzhfS1xtXVVe3m\nwULqOFSE9M6IIW3+mCUkLaT6oYo/Upwm/wshtUFIMyH9iYuGVOcf6auiD8gHWD5H0g7VmaUI\niZAqEJIqRUiEVIGQVClCIqQKhKRKERIhVSAkVYqQCKkCIalShERIFQhJlSIkQqpASKoUIRFS\nBUJSpQiJkCoQkipFSIRUgZBUKUIipAqEpEoREiFVICRVipAIqQIhqVKEREgVCEmVIiRCqkBI\nqhQhEVIFQlKlCImQKhCSKkVIhFSBkFQpQiKkCoSkShESIVUgJFWKkAipAiGpUoRESBUISZUi\nJEKqQEiqFCERUgVCUqUIiZAqEJIqRUiEVIGQVClCIqQKhKRKERIhVSAkVYqQCKkCIalShERI\nFQhJlSIkQqpASKoUIRFSBUJSpQiJkCoQkipFSIRUgZBUKUIipAqEpEoREiFVICRVipAIqQIh\nqVKEREgVCEmVIiRCqkBIqhQhEVIFQlKlCImQKhCSKkVIhFSBkFQpQiKkCoSkShESIVUgJFWK\nkAipAiGpUoT0yo+7aVrf/ktISwhJlSKkHc8304b1NP0ipAWEpEoR0o776WFT0frndEtICwhJ\nlSKk/YfT+3+E9AYhqVKEREgVCEmVIqQd+0u7h+mekBYQkipFSDueV9MLqydCWkBIqhQhvfL9\nZppuHp4/6IiQTop0DAhJhR/IuqwEIalShERIFQhJlSKk/YdvENICQlKlCImQKhCSKkVIS55u\nv3/QESGdFOkYEJLK5Z4jPU8flXTMd5VWgpBmQgrmgi82cGl3ACGpUoR0wM9pRUgLCEmVIqTX\nM9GeB0JaQEiqFCEdhLT6qCNCOinSMSAkFX4g67IShKRKERIhVSAkVYqQ1sufxn78A9lj+CeO\nE9g5YXmkpG/37du3s0pJXF9fn/x7HBvSMflKf6RxRpo5IwXDpZ3LShCSKkVIhFSBkFQpQtrz\nwKXdHyAkVYqQfuuIkJYQkipFSDtW07+309PzLX9B5AGEpEoR0v7Daf19elw/8xdEHkBIqhQh\nvYX0OP3gt79/g5BUKULacTf9fJpu1r8I6QBCUqUIace2oNvtaw38BZFLCEmVIqQ9jzfbv271\nw/+LgpBOinQMCEnlAiF9+O8iEdI5kI4BIalc4sWGm0dC+i+EpEoR0o6baVp9//DvKyakEyMd\nA0JSucRzpKeH1TTdffTjWEI6LdIxICSVC/3S6q+Habr5SUgLCEmVIqSD0xK/a3cIIalShLQ4\nI91vzkg/CGkBIalShPR6Mto+R7rnOdIhhKRKEdKO7b8y9oNX7X6HkFQpQtp/eMfPkf4AIalS\nhLRDORkR0smRjsGlQvpa4+rqqnbzZZaPv7PhDNtDgpBmMaTNGem8ISnH4HSnSUJqg5BmQvoT\nhNQGIblKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk2XTPWkoRUglCmk33rKUU\nIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk2XTPWkoRUglCmk33\nrKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk2XTPWkoRUglC\nmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk2XTPWkoR\nUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk2XTP\nWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQhlSCk\n2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfespRQh\nlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKaTfes\npRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFSCUKa\nTfespRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9aShFS\nCUKaTfespRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZdM9a\nShFSCUKaTfespRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGVIKTZ\ndM9aShFSCUKaTfespRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVIJQppN96ylFCGV\nIKTZdM9aShFSCUKaTfespRQhlSCk2XTPWkoRUglCmk33rKUUIZUgpNl0z1pKEVKJS4X0V43r\n6q3xMtIxICTVipBCh65TK+XLly+E5ClFSCUIaTbds5ZShFTC8TnS5tLuvN9QOgaEpFoRUujQ\nx0NItlKEVIKQZtM9ayn1CUNabSGksyAdA0JSrcxC4ox0NqRjQEiqFSGFDn08hGQr9flCOuyI\nkE6KdAwISbXyCuntKdLfW9S7LfknjmO+/fFS375985MSSS4VuH59Hm1npIQvNijfTfsTLfsf\n/pZSn++M9F4TIVlIEZJsRUihQx8vRUi2Up8vJC7tzKQISbayC2nxyt2pZiYkVYqQZCurkNYH\nv9hASBeXIiTZyiukQ041MyGpUoQkWxFS6NB1qa8Vrq6uajefTirgUCWXIqQSjiFtFoKQPKUI\nqQQhzaZ71lKKkEo4PkfylCIk1YqQQofOJkVIqhUhhQ6dTYqQVCtCCh06mxQhqVaEFDp0NilC\nUq0IKXTobFKEpFoRUujQ2aQISbUipNChs0kRkmpFSKFDZ5MiJNWKkEKHziZFSKoVIYUOnU2K\nkFQrQgodOpsUIalWhBQ6dDYpQlKtCCl06GxShKRaEVLo0NmkCEm1IqTQobNJEZJqRUihQ2eT\nIiTVipBCh84mRUiqFSGFDp1NipBUK0IKHTqbFCGpVoQUOnQ2KUJSrQgpdOhsUoSkWhFS6NDZ\npAhJtSKk0KGzSRGSakVIoUNnkyIk1YqQQofOJkVIqhUhhQ6dTYqQVCtCCh06mxQhqVaEFDp0\nNilCUq0IKXTobFKEpFoRUujQ2aQISbUipNChs0kRkmpFSKFDZ5MiJNWKkEKHziZFSKoVIYUO\nnU2KkFQrQgodOpsUIalWhBQ6dDYpQlKtCCl06GxShKRaEVLo0NmkCEm1IqTQobNJEZJqRUih\nQ2eTIiTVipBCh84mRUiqFSGFDp1NipBUK0IKHTqbFCGpVoQUOnQ2KUJSrQgpdOhsUoSkWhFS\n6NDZpAhJtSKk0KGzSRGSakVIoUNnkyIk1YqQQofOJkVIqhUhhQ6dTYqQVCtCCh06mxQhqVaE\nFDp0NilCUq0IKXTobFKEpFoRUujQ2aQISbUipNChs0kRkmpFSKFDZ5MiJNWKkEKHziZFSKoV\nIYUOnU2KkFQrQgodOpsUIalWhBQ6dDYpQlKtCCl06GxShKRaEVLo0NmkCEm1IqTQobNJEZJq\nRUihQ2eTIiTVipBCh84mRUiqFSGFDp1NipBUK0IKHTqbFCGpVoQUOnQ2KUJSrQgpdOhsUoSk\nWhFS6NDZpAhJtSKk0KGzSRGSakVIoUNnkyIk1YqQQofOJkVIqhUhhQ6dTYqQVCtCCh06mxQh\nqVaEFDp0NilCUq0IKXTobFKEpFoRUujQ2aQISbUipNChs0kR0sLqa42rq6vazR1ShJRAipAW\nVtWQNmckQjr1SnxeKUJaWBHSUQtBSLJVcqnA9TvmuxNSAilCkq0IKXLobFKEJFsRUuTQ2aQI\nSbYipMihs0kRkmxFSJFDZ5MiJNmKkCKHziZFSLIVIUUOnU2KkGQrQoocOpsUIclWhBQ5dDYp\nQpKtCCly6GxShCRbEVLk0NmkCEm2IqTIobNJEZJsRUiRQ2eTIiTZipAih84mRUiyFSFFDp1N\nipBkK0KKHDqbFCHJVoQUOXQ2KUKSrQgpcuhsUoQkWxFS5NDZpAhJtjIO6Rj+iQMppKKt+jw4\nI31aKc5IspXxGemyMxOSbJVcipC6ZiYk2Sq5FCF1zUxIslVyKULqmpmQZKvkUoTUNTMhyVbJ\npQipa2ZCkq2SSxFS18yEJFsllyKkrpkJSbZKLkVIXTMTkmyVXIqQumYmJNkquRQhdc1MSLJV\ncilC6pqZkGSr5FKE1DUzIclWyaUIqWtmQpKtkksRUtfMhCRbJZcipK6ZCUm2Si5FSF0zE5Js\nlVyKkLpmJiTZKrkUIXXNTEiyVXIpQuqamZBkq+RShNQ1MyHJVsmlCKlrZkKSrZJLEVLXzIQk\nWyWXIqSumQlJtkouRUhdMxOSbJVcipC6ZiYk2Sq5FCF1zUxIslVyKULqmpmQZKvkUoTUNTMh\nyVbJpQipa2ZCkq2SSxFS18yEJFsllyKkrpkJSbZKLkVIXTMTkmyVXIqQumYmJNkquRQhdc1M\nSLJVcilC6pqZkGSr5FKE1DUzIclWyaUIqWtmQpKtkksRUtfMhCRbJZcipK6ZCUm2Si5FSF0z\nE5JslVyKkLpmJiTZKrkUIXXNTEiyVXIpQuqamZBkq+RShNQ1MyHJVsmlCKlrZkKSrZJLEVLX\nzIQkWyWXIqSumQlJtkouRUhdMxOSbJVcipC6ZiYk2Sq5FCF1zUxIslVyKULqmpmQZKvkUoTU\nNTMhyVbJpQipa2ZCkq2SSxFS18yEJFsllyKkrpkJSbZKLkVIXTMTkmyVXIqQumYmJNkquRQh\ndc1MSLJVcilC6pqZkGSr5FKE1DUzIclWyaUIqWtmQpKtkksRUtfMhCRbJZcipK6ZCUm2Si5F\nSF0zE5JslVyKkLpmJiTZKrkUIXXNTEiyVXIpQuqamZBkq+RShNQ1MyHJVsmlCKlrZkKSrZJL\nEVLXzIQkWyWXIqSumQlJtkouRUhdMxOSbJVcipC6ZiYk2Sq5FCF1zUxIslVyKULqmpmQZKvk\nUoTUNTMhyVbJpQipa2ZCkq2SSxFS18yEJFsllyKkrpkJSbZKLkVIXTMTkmyVXIqQumYmJNkq\nuRQhdc1MSLJVcilC6pqZkGSr5FKE1DUzIclWyaUIqWtmQpKtkksRUtfMhCRbJZcipK6ZCUm2\nSi5FSF0zE5JslVyKkLpmJiTZKrkUIXXNTEiyVXIpQuqamZBkq+RShNQ1MyHJVsmlCKlrZkKS\nrZJLEVLXzIQkWyWXIqSumQlJtkouRUhdMxOSbJVcipC6ZiYk2Sq5FCF1zUxIslVyKULqmpmQ\nZKvkUoTUNTMhyVbJpQipa2ZCkq2SSxFS18yEJFsllyKkrpkJSbZKLkVIXTMTkmyVXIqQumYm\nJNkquRQhdc1MSLJVcilC6pqZkGSr5FKfMKTVBkIykiIk2coppNXbG0LykCIk2YqQIofOJkVI\nshUhRQ6dTYqQZCvPkP7eot4NYAjOeUbSGj/ZI3dgKeVpNZYUIbVhKeVpNZYUIbVhKeVpNZYU\nIbVhKeVpNZYUIbVhKeVpNZbUESEF/GaDpnayR+7AUsrTaiypY0I65FRmgy1EF5ZWY0kRUhuW\nUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuW\nUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuW\nUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuW\nUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuWUp5WY0kRUhuW\nUp5WY0kRUhuWUp5WY0n1h3QyLP8tQEspT6tRpQhJwVLK02pUKUJSsJTytBpVipAULKU8rUaV\nsgsJ4DNCSAABEBJAAIQEEAAhAQRASAABENKHrD7+Elgw5vEipA8Zc2Mcz5jHi5A+ZMyNcTxj\nHi+zkFYbLu3wO6u1oZTnoXo5UnZWZzlSXiGt3t4Y8bIKblKmh2q9P1xOnOdIeYX0wpgL0Yqd\n0BbLQzVmSIYXLJa7w09oi+WhWq3OsanMQnK8irLcHX5CWywP1Xl8vELyXQg3KT+hLZaHatSQ\n/C7tHE+TvnvWb/3e3pwSr5AsXz7l5W8dx/Ub8eVvgE8KIQEEQEgAARASQACEBBAAIQEEQEgA\nARASQACEBBAAIQEEQEgAARASQACEBBAAIQEEQEgAARASQACEBBAAIQEEQEgAARASQACEBBAA\nIQEEQEgAARASQACEBBAAIQEEQEgAARASQACEBBAAIQEEQEgAARASQACEBBAAIQEE8H9TIMhh\ndZxcTwAAAABJRU5ErkJggg==",
114 | "text/plain": [
115 | "plot without title"
116 | ]
117 | },
118 | "metadata": {
119 | "image/png": {
120 | "height": 420,
121 | "width": 420
122 | }
123 | },
124 | "output_type": "display_data"
125 | }
126 | ],
127 | "source": [
128 | "# 绘图\n",
129 | "ggplot(data) +\n",
130 | "# 绘制条形\n",
131 | "geom_bar( aes(x=name, y=value), stat=\"identity\", fill=\"#69b3a2\", alpha=0.7, width=0.5) +\n",
132 | "# 绘制误差线\n",
133 | "geom_errorbar( aes(x=name, ymin=value-sd, ymax=value+sd), width=0.4, colour=\"black\", alpha=0.9, size=1) +\n",
134 | "theme(\n",
135 | " legend.position=\"none\",\n",
136 | " plot.title = element_text(size=11)\n",
137 | ") +\n",
138 | "ggtitle(\"A barplot with error bar\") +\n",
139 | "xlab(\"\")"
140 | ]
141 | },
142 | {
143 | "cell_type": "markdown",
144 | "id": "9504c6d1",
145 | "metadata": {},
146 | "source": [
147 | "## 误差线中的问题\n",
148 | "### 误差线隐藏信息\n",
149 | "\n",
150 | "误差线可能会隐藏很多信息。如下图所示,这是PLOS Biology期刊一篇论文[Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm](http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002128)中的图。它说明完整的数据可能暗示与汇总统计数据不同的结论。其中A图是一张用于汇总数据带有误差线的条形图。但是从A图中,我们并不能得到A图中两个数据组明确的数据分布信息,因为A图可能对应不同数据组分布信息。A可能会对应B,C,D,E四张图,而这四张图表示了完全不同的数据分布信息。B图表示两个数据组具有相同类型的数据分布,C图表示第二个数据组有异常值,D图表示两组数据分布不同,E图表示两组数据样本数不同。\n",
151 | "\n",
152 | "\n",
153 | "\n",
154 | "\n",
155 | "因此,带有误差线的相同条形图实际上可以讲述非常不同的故事,对读者来说这些数据是隐藏的。所以尽可能显示个人数据信息。"
156 | ]
157 | },
158 | {
159 | "cell_type": "markdown",
160 | "id": "7f9d067a",
161 | "metadata": {},
162 | "source": [
163 | "### 误差线的计算方式\n",
164 | "\n",
165 | "误差线的第二个问题是误差线有多种计算方式,并且并不总是清楚显示的是哪一个。误差线通常使用三种不同的计算方式,选择不同的计算方式有时给出非常不同的结果。下面是它们的定义以及如何在R上计算。\n",
166 | "\n",
167 | "**标准偏差(SD)**\n",
168 | "\n",
169 | "表示变量的分散量。计算公式为方差的平方根\n",
170 | "\n",
171 | "```\n",
172 | "# 计算方差\n",
173 | "sd <- sd(vec)\n",
174 | "# 计算平方根\n",
175 | "sd <- sqrt(var(vec))\n",
176 | "```\n",
177 | "\n",
178 | "**标准误差(SE)** \n",
179 | "\n",
180 | "表示变量均值的标准偏差,计算方法为SD除以样本大小的平方根。通过计算方法,SE小于SD。对于非常大的样本量,SE趋向于0。\n",
181 | "\n",
182 | "```\n",
183 | "se = sd(vec) / sqrt(length(vec))\n",
184 | "```\n",
185 | "\n",
186 | "**置信区间(CI)**\n",
187 | "\n",
188 | "表示使某个值存在于其内的特定概率。它计算为t*SE,其中t值是t检验在特定显著水平alpha下的统计量值。其值在具有较大的样本量时通常四舍五入到1.96。但是,如果样本量很大或分布不正态,则最好使用bootstrap方法计算CI。\n",
189 | "\n",
190 | "```\n",
191 | "alpha=0.05\n",
192 | "t=qt((1-alpha)/2 + .5, length(vec)-1) \n",
193 | "# 数据量很大是取为1.96\n",
194 | "# t = 1.96\n",
195 | "CI=t*se\n",
196 | "```"
197 | ]
198 | },
199 | {
200 | "cell_type": "markdown",
201 | "id": "41ad1c69",
202 | "metadata": {},
203 | "source": [
204 | "以上3个指标在著名的Iris数据集上应用时。三种鸢尾花的平均萼片长度和平均长度用误差线表示的结果完全不同。\n",
205 | "\n"
206 | ]
207 | },
208 | {
209 | "cell_type": "code",
210 | "execution_count": 9,
211 | "id": "61a55f3b",
212 | "metadata": {},
213 | "outputs": [
214 | {
215 | "data": {
216 | "text/html": [
217 | "\n",
218 | "A data.frame: 6 × 2\n",
219 | "\n",
220 | "\t | Species | Sepal.Length |
|---|
\n",
221 | "\t | <fct> | <dbl> |
|---|
\n",
222 | "\n",
223 | "\n",
224 | "\t| 1 | setosa | 5.1 |
|---|
\n",
225 | "\t| 2 | setosa | 4.9 |
|---|
\n",
226 | "\t| 3 | setosa | 4.7 |
|---|
\n",
227 | "\t| 4 | setosa | 4.6 |
|---|
\n",
228 | "\t| 5 | setosa | 5.0 |
|---|
\n",
229 | "\t| 6 | setosa | 5.4 |
|---|
\n",
230 | "\n",
231 | "
\n"
232 | ],
233 | "text/latex": [
234 | "A data.frame: 6 × 2\n",
235 | "\\begin{tabular}{r|ll}\n",
236 | " & Species & Sepal.Length\\\\\n",
237 | " & & \\\\\n",
238 | "\\hline\n",
239 | "\t1 & setosa & 5.1\\\\\n",
240 | "\t2 & setosa & 4.9\\\\\n",
241 | "\t3 & setosa & 4.7\\\\\n",
242 | "\t4 & setosa & 4.6\\\\\n",
243 | "\t5 & setosa & 5.0\\\\\n",
244 | "\t6 & setosa & 5.4\\\\\n",
245 | "\\end{tabular}\n"
246 | ],
247 | "text/markdown": [
248 | "\n",
249 | "A data.frame: 6 × 2\n",
250 | "\n",
251 | "| | Species <fct> | Sepal.Length <dbl> |\n",
252 | "|---|---|---|\n",
253 | "| 1 | setosa | 5.1 |\n",
254 | "| 2 | setosa | 4.9 |\n",
255 | "| 3 | setosa | 4.7 |\n",
256 | "| 4 | setosa | 4.6 |\n",
257 | "| 5 | setosa | 5.0 |\n",
258 | "| 6 | setosa | 5.4 |\n",
259 | "\n"
260 | ],
261 | "text/plain": [
262 | " Species Sepal.Length\n",
263 | "1 setosa 5.1 \n",
264 | "2 setosa 4.9 \n",
265 | "3 setosa 4.7 \n",
266 | "4 setosa 4.6 \n",
267 | "5 setosa 5.0 \n",
268 | "6 setosa 5.4 "
269 | ]
270 | },
271 | "metadata": {},
272 | "output_type": "display_data"
273 | }
274 | ],
275 | "source": [
276 | "# 读取数据\n",
277 | "data <- iris %>% select(Species, Sepal.Length) \n",
278 | "head(data)"
279 | ]
280 | },
281 | {
282 | "cell_type": "code",
283 | "execution_count": 10,
284 | "id": "ec5fdd62",
285 | "metadata": {},
286 | "outputs": [],
287 | "source": [
288 | "# 分别计算标准偏差,标准误差,置信区间\n",
289 | "my_sum <- data %>%\n",
290 | " group_by(Species) %>%\n",
291 | " summarise( \n",
292 | " n=n(),\n",
293 | " mean=mean(Sepal.Length),\n",
294 | " sd=sd(Sepal.Length)\n",
295 | " ) %>%\n",
296 | " mutate( se=sd/sqrt(n)) %>%\n",
297 | " mutate( ic=se * qt((1-0.05)/2 + .5, n-1))"
298 | ]
299 | },
300 | {
301 | "cell_type": "code",
302 | "execution_count": 11,
303 | "id": "a8766d67",
304 | "metadata": {},
305 | "outputs": [
306 | {
307 | "data": {
308 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAA0gAAANICAMAAADKOT/pAAAATlBMVEUAAAABAgICAgIOExIP\nFBMXFxcZGRkzMzNNTU1oaGh8fHyMjIyQxLiWyr6ampqnp6eysrK9vb3Hx8fQ0NDZ2dnh4eHp\n6enr6+vw8PD///+8zlEtAAAACXBIWXMAABJ0AAASdAHeZh94AAAgAElEQVR4nO2dC1sbyZJE\n256HBbMznqev+f9/dMEG06qKTmW3Qq1K6cR3d2bNlEOZhzoIA4bpiRBydqZrD0DILQSRCDEE\nkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEI\nMQSRCDEEkQgxBJEIMQSRCDHkTJH63z4t/HJafPFtuZwncp+ZXgGEGBZuTf/fx8mVRHI9/nhB\npDCn7sPS8dSLr5kzRpq+5du/X//x9q/Xl/x46fvLvz/g97dKLy/79n+v/+X9dNWsI/L9H2//\n4an68jrT8Wv3/fXdvtaPGBwxnJ368aJrrBJn+0jT+2+fjv/3FP5yWnpx+XfzthMpv/pSZq/v\nxP9iOtPRfxwtZz0jPb2/dTm6D8cvefrx/7++fOn3jUloRdYQUdvfXt7etuj/tVSmt+frhVNz\nkqPlrJFyb2ye2v//5PG6WUPk6bZWl5lOIHk6+uV09Nv6UyOz2j7S2/uv87crP96znf/yx+H3\nd3Rnb2nmf0YaklA+64n8eOFT8dUXMzWv3SMMPYPuGWmaPQe9nhjzmgw4EiH1gkiEGIJIhBiC\nSIQYgkiEGIJIhBiCSIQYgkiEGIJIhBiCSIQYgkiEGLJdpP9lkz95yYp0x3aW9lEuWwGRzR1i\nVUTqzhFb7K+cy1YgkrODa9Odg0h7rg8idecg0p6DSHuuDyJ15yDSnoNIe64PInXnINKeg0h7\nrg8idecg0p6DSHuuDyJ15yDSnoNIe64PInXnINKeg0h7rg8idecg0p6DSHuuDyJ15yDSniOn\ng0jdOYi05yDSnuuDSN05iLTnINKe64NI3TmItOcg0p7rg0jdOYi05yDSnuuDSN05iLTnINKe\n64NI3TmItOcg0p7rg0jdOYi05yDSnuuDSN05iLTnINKe64NI3TmItOcg0p7rg0jdOYi05yDS\nnuuDSN05iLTnINKe64NI3TmItOcg0p7rg0jdOYi05yDSnuuDSN05iLTnINKe64NI3TmItOcg\n0p7rg0jdOYi05yDSnuuDSN05iLTnINKe64NI3TmItOcg0p7rg0jdOYi05yDSnuuDSN05iLTn\nINKe64NI3TmItOcg0p7rc8si/XKUX+e/iB4LIu1jQaR9rD53I9LHjx+5NhBpYyOCSN1jQaR9\nLIi0j9XnlkU6yvPTdvKxINI+FkTax+qDSN1jQaR9LIi0j9UHkbrHgkj7WBBpH6sPInWPBZH2\nsSDSPlYfROoeCyLtY0Gkfaw+iNQ9FkTax4JI+1h9EKl7LIi0jwWR9rH6IFL3WDdJ5P/6/PTz\nT/0L5WPdJBERREoEkRDpVBApEURCpFNBpEQQCZFOBZESQSREOhVESgSREOlUECkRREKkU0Gk\nRBAJkU4FkRJBpBd35vnw4cPsV4iESKkgUivS8zPSHYsknqLPeo5GpO6xbpIIIjVBpI1BpC6/\ni5fJx7pFIuOIVCy//vrrpR/iUq9zRwUiNRlHpEtteKEKnpEQaR5E2hhEQqR5EGljEAmR5kGk\njUEkRJoHkTYGkRBpHkTaGERCpHkQaWMQCZHmQaSNQSREmgeRNgaREGkeRNoYREKkeRBpYxAJ\nkeZBpI1BJESaB5E2BpEQaR5E2hhEQqR53txx/Z1hROoe6yaJIFITJdI5f9URkbrHukkiiNQE\nkTYGkRBpHgXkHCKI1D3WTRJBpCaItDGIhEjzINLGIBIizYNIG4NIiDQPIm0MIiHSPIi0MYiE\nSPMg0sYgEiLNg0gbg0iINA8ibQwiIdI8iLQxiIRI8yDSxiASIs2DSBuDSIg0DyJtDCIh0jyI\ntDGIhEjzINLGIBIizYNIG4NIiDQPIm0MIiHSPIi0MYiESPMg0sYgEiLNg0gbg0iINA8iZSJ4\nnPPN/26TCCIh0qkgUhtEaoJImSBSG0RqgkiZIFIbRGqCSJkgUhtEaoJImewl0uE5ZYkgEiKd\nyk4iHX78oyIRREKkU7lJkX6Z59ejX53+zYjUBJEy2VOk3YjMzfn48SMinfWmBZEy2U2kH39G\n+vSczShz+XWWZ5Hmvzz9m39PxjvyxV7B33POmxZEymQvkV4susqfkdJf8PQW87UZ444gUiLu\nPxHU/zPSPIjUZi2RuxTJ8ScCREKkeRAJkf43sEhX+8waIiXiuDaItIdIdYggUiY7ibTT299z\ntgk6EMlJBJG6cYcjcs42QcfFRCpxRxQQRGpzztXbDASR5p9Z+7TDZ9bmSX02bZbsJ9ayn1lD\npG7c4YhUEqnMZ9YUEJ6R2pxz9TYDQST+jDTStRFBpM3bBB2I5CSCSN24wxF522CWDx8+zH+J\nSIiUCSK1Ij1vg0hHQaREEKmKSHU+s6aAIFKbc0BvBrLntUm/yh0dJYggUiKItPlV7ugoQQSR\nEkGkza9yR0cJIoiUCCJtfpU7OkoQQaREEGnzq9zRUYIIIiWCSJtf5Y6OEkQQKRFE2vwqd3SU\nIIJIiSDS5le5o6MEEURKBJE2v8odHSWIIFIiiLT5Ve7oKEEEkRJBpM2vckdHCSKIlAgibX6V\nOzpKEEGkRBBp86vc0VGCCCIlgkibX+WOjmGJnPM3tBQQRGojeCCS89qMQeScv1iigCBSG8ED\nkZzXZgwiiLQyiLT5Ve7ouEkiqgKR2ggeiOS8NvWJqApEaiN4IJLz2tQnoioQqY3ggUjOa1Of\niKpApDaCByI5r019IqoCkdoIHojkvDb1iagKRGojeCCS89rUJ6IqEKmN4IFIzmtTn4iqQKQ2\nggciOa9NfSKqApHaCB6I5Lw29Ymoir1EKvODdhHJ2nGTRFTFTiLV+QbpiGTtuEkiqgKR2gge\niOS8NvWJqIo9Rbo8JBFEOvfaIFKmYjeRfvwZafAftKt+0u7PP/+8+QftJjPwtUGkTMVeIpX5\nQbs8I1k7bpKIquDPSG0ED0RyXpv6RFQFIrURPBDJeW3qE1EViNTmO4Kjv3t8/M0x7u3aIFKm\nApHafEdwJNLx3+m/t2uDSJmK2/rKBrGLfLfsNCVE8nTcJBFVcVtfayd22SiSDdJ4RBDpXCKq\nApEylO762kAkU4FIGUp3fW0gkqlApAylu742EMlUIFKG0l1fG4hkKhApQ+murw1EMhWIlKF0\n19cGIpkKRMpQuutrA5FMBSJlKN31tYFIpgKRMpTu+tpAJFOBSBlKd31tIJKpQKQMpbu+NhDJ\nVCBShtJdXxuIZCoQKUPprq8NRDIViJShdNfXBiKZCkTKULrrawORTAUiZSjd9bWBSKYCkTKU\n7vraQCRTgUgZSnd9bSCSqUCkDKW7vjYQyVQgUobSXV8biGQqEClD6a6vDUQyFYiUoXTX1wYi\nmQpEylC662sDkUwFImUo3fW1gUimApEylO762kAkU4FIGUp3fW0gkqlApAylu742EMlUIFKG\n0l1fG4hkKhApQ+murw1EMhWIlKF019cGIpkKRMpQuutrA5FMBSJlKN31tYFIpgKRMpTu+tpA\nJFOBSBlKd31tIJKpQKQMpbu+NhDJVCBShtJdXxuIZCoQKUPprq8NRDIViJShdNfXBiKZCkTK\nULrrawORTAUiZSjd9bWBSKYCkTKUrnptfjnKr/NfbCKCSOcSURWIlKE0jkgfP35EpOYh0idd\nRFQFImUoIdK4RK7wHK0qEClDaZy3v8/X5lwiNybSFd60qApEylBCpKGIINLaa7NRpJ9m+fDh\nw/yX2yghEkROVNy8SM/PSFVFUpPIJ9gLbTMeERVEyuRuRFL5XeTnn3/uX7iyQ2XtHGs71if7\nOkekTC70oc3xRMpOwjNSG0TKBJGaIFIbRFrgssfnCEqJdPSu6fHHTi67zXhEVBBpgcseH9qs\nK9LxH/kuu814RFQQaYELIjWTIFIYRLoipFIibRvFsE0NIlf4U6OqQKStHUlI5mtzPyKpDPIJ\ngWxF9hMCiOS7NtlJ7kek7CQ8IyWCSJtHMWxTgwgiJYJIm0cxbFODCCIlgkibRzFsU4MIIiWC\nSJtHMWxTgwgiJYJIm0cxbFODCCIlgkibRzFsU4MIIiWCSJtHMWwzLJFrf/WhqkCkrR1JSOdf\nm22jGLYZlsi1v2hKVSDS1o4kpPOvzbZRDNsMSwSRIkgqiLR5FMM2EFlRgUhbO5KQ6l8biFjH\nEKsiUgJS/WsDEesYYlVESkCqf20gYh1DrIpICUj1rw1ErGOIVREpAan+tYGIdQyxKiIlINW/\nNhCxjiFWRaQEpPrXBiLWMcSqiJSAVP/aQMQ6hlgVkRKQ6l8biFjHEKsiUgJS/WsDEesYYlVE\nSkCqf20gYh1DrIpICUj1rw1ErGOIVREpAan+tYGIdQyxKiIlINW/NhCxjiFWRaQEpPrXBiLW\nMcSqiJSAVP/aQMQ6hlgVkRKQ6l8biFjHEKsiUgJS/WsDEesYYlWvSGq4S32rJURaUQER6xhi\n1e0iqez5Izsu/XPA12fga4NI1jHEqjwjJSDVvzYQsY4hVkWkBKT61wYi1jHEqoiUgFT/2kDE\nOoZYFZESkOpfG4hYxxCrIlICUv1rAxHrGGJVREpAqn9tIGIdQ6yKSAlI9a8NRKxjiFURKQGp\n/rWBiHUMsSoiJSDVvzYQsY4hVkWkBKT61wYi1jHEqoiUgFT/2kDEOoZYFZESkOpfG4hYxxCr\nIlICUv1rAxHrGGJVREpAqn9tIGIdQ6yKSAlI9a8NRKxjiFURKQGp/rWBiHUMsSoiJSDVvzYQ\nsY4hVkWkBKT61wYi1jHEqoiUgFT/2kDEOoZYFZESkOpfG4hYxxCrIlICUv1rAxHrGGJVREpA\nqn9tIGIdQ6yKSAlI9a8NRKxjiFURKQGp/rWBiHUMsSoiJSDVvzYQsY4hVkWkBKT61wYi1jHE\nqoiUgFT/2kDEOoZYFZESkOpfG4hYxxCrIlICUv1rAxHrGGJVREpAqn9tIGIdQ6x6EZF+mufD\nhw+zX/kgIdKKCohYxxCrXl6k52ckRNo2imEbiKyoQCQn6Ju8NhCxjiFWvfyfkS4FCZFWVEDE\nOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOI\nVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWR\nEpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGp\nCJGgAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJ\nKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi\n1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgRTmMQySoQCTrGGJVREpACnJApIHv\nyMoKRHKCXgXhwDPSyHdkZQUiOUGvYXDgXbuh78jKCkRygl7D4F2kT89RJ37PJngUQ4WlY32u\nfUdWViCSE/QKBIcnnpE0lfGJnDWG2BqREpCiG1Pj2vDhF+sYYm1ESkBavDLfMgyRoIIPv1jH\nEHsjUgJSfHGGIRJU8OEX6xhicURKQDpxc0YhElTsL9KnwT/84hhjHkQ659o8IVJPowaRs8YQ\nmyNSAlIRIkEFH36xjiFWR6QEpCJEggo+/GIdQ6yOSAlIRYgEFbyzax1DbI1ICUhFiAQViGQd\nQ2yNSAlIRYgEFYhkHUNsjUgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsuivT5MH3PqJB2F2l4\nIkEFRKxjiFWXGHyepsEh7S3S+ESCCohYxxCrLjE4TH8u0RkE0t4ijU8kqICIdQyx6pJIy29l\nRoG0t0jjEwkqIGIdQ8FYQPB5+jo4pP3ftRudSFABEesYYtXFNyq/PX4ZG9LuH2wYnkhQARHr\nGGJVKdI0z6iQdhWpBJGgAiLWMRSPspAQaUUFRKxjKB5LDE7m2pD4hOyKCohYxxCrIlICUhEi\nQQVErGOIVU99+Hv+PWHGgnStD3+PSySogIh1DAVD7X+o8P7vriKVIBJUQMQ6hlhVMvhrxuiv\nY3zjQNpVpGUiNa4NRKxjiFXXfWXDSN/8j69sWFEBEesYCsYpFvMM9c3/+GDDigqIWMcQqy4/\nI73l8Mfbp6/H+uZ/+z8j9UTKXBuIWMdQME5Ces4rpbF+9sKlf35DhshxBr42FxapIJGzxlAw\nFiD9M718HdWXx+nvp8/TH99eNNg3/9v7GUkQqXNtIGIdQ6y6JNLD61f2Tg9vf6gc7Zv/7S1S\nT6TQtYGIdQyx6qmP2r38+1Wkwb7537U+avdOpNC1gYh1DAVjAdLj29P249O/L29xXnPHz0gL\nRGpcG4hYxxCrLon05fUz14cvT/NPuN2xSAtEalwbiFjHEKsufh7p658P0/Tw+fm94Onz+0vv\nWKQFIjWuDUSsY4hV+ervBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hV+QaRCUhFiAQVELGOIVbl\nG0QmIBUhElRAxDqGWHX5G0Qufm38IJD2Fml8IkEFRKxjiFX5BpEJSEWIBBUQsY6hYCwg+G34\nb/63t0jjEwkqIGIdQ6y6/AnZ0b/53/6fkB2dSFABEesYYtXTf41iVEj7v2s3OpGgAiLWMRSM\nspAQaUUFRKxjKBhLDE7m2pD4hOyKCohYxxCrIlICUhEiQQVErGOIVZdF+uu356fsx/+GhbS/\nSKMTCSogYh1DrLok0teHb+/7TtO/o0LaW6TxiQQVELGOIVZdEumP6fPLJ9z+nh5HhbS3SOMT\nCSogYh1DrBp9ZcPyXyEeAdI1vrJhbCJBBUSsYygYZSEh0ooKiFjHUDAWELw+bS98m6URIF3p\nXbuBiQQVELGOIVZd/GDD+9/HHxTS7h9sGJ5IUAER6xhi1eUPf//4+/iDQtr/w9+jEwkqIGId\nQ6zKJ2QTkIoQCSogYh1DrIpICUhFiAQVELGOIVY9JdK4H5G5lkjjEgkqIGIdQ0EoCwmRVlRA\nxDqGglAWEiKtqICIdQwFoSwkRFpRARHrGApCWUiItKICItYxFISykBBpRQVErGMoCJrMNP5f\nI95VpBJEggqIWMdQPMpCQqQVFRCxjqF4LDE4mWtD4hOyKyogYh1DrIpICUhFiAQVELGOIVZF\npASkIkSCCohYxxCrIlICUhEiQQVErGOIVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpA\nKkIkqICIdQyxKp9HSkAqQiSogIh1DMWjLCREWlEBEesYiscSg5O5NiTetVtRARHrGGJVREpA\nKkIkqICIdQyx6qJIn0d/2t5dpOGJBBUQsY4hVl1iMP6Pft9bpPGJBBUQsY4hVl1icJj+e5y+\nfH0c9ycN7C3S+ESCCohYxxCrBt/7+8/pn6ev4/6kgb1FGp9IUAER6xgKxjKkf6a/Rv7bj1cQ\naXAiQQVErGMoGAsIfpv+/jI9PP07LqS9RRqfSFABEesYYtUlBi90Hl/+HDnsTxrYW6TxiQQV\nELGOIVZdfGPyz8PLz+2YPi/996tD2v3D38MTCSogYh1DrMonZBOQihAJKiBiHUOsikgJSEWI\nBBUQsY4hVl0U6evnh2l6/HNcSLuLNDyRoAIi1jHEqksifRn+p7HtLdL4RIIKiFjHEKsuifQ4\nPT7j+fI47kdk9hZpfCJBBUSsY4hVo59q/pyv436O4AqfkB2cSFABEesYCsYCgt+m7z8ZdNwv\n/9j/E7KjEwkqIGIdQ6y6+Mbkj8f/Xp62H4d9/3f3DzYMTySogIh1DLHq8rt2p/4q8bUh7f+u\n3ehEggqIWMdQMMpCQqQVFRCxjqFgLFE6mWtD4hOyKyogYh1DrIpICUhFiAQVELGOIVZdFumv\n316+uPe/YSHtL9LoRIIKiFjHEKsuifT14dv7vdO4f414b5HGJxJUQMQ6hlh1SaQ/ps8vn3D7\ne9zPEewt0mkiKr9nY+hwzLFmt/HvyMqKS31lw9v/jQnpGl/ZMDaRoAIi1jEUjLKQEGlFBUSs\nYygYCwhen7Y/j/sFiVd6125gIkEFRKxjiFUXP9gw/JfI7/7BhuGJBBVXIqIyyJ8aHWPMs/zh\n7z8fpunh89fF/37ta7P/h79HJxJUQMQ6hliVT8gmIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yq\nRfr6+eXlfx+m35bf/b02pH1FqkAkqICIdQyxqhbp8PIRzX+//UFy8R3ga0PaV6QKRIIKiFjH\nEKtKkf6aHp/ZPDy+/OSOxe/+d21Iu4pUgkhQARHrGGJVKdLj9PJNLV4+PfB1OowKaVeRShAJ\nKiBiHUOsuvDDmJ//8fe3NzTjftZ6V5FKEAkqIGIdQ/FQ+x9eXvp5+m9oSLuKVIJIUAER6xhi\nVcng27eHeXh4evnD5LBf2burSCWIBBUQsY4hVl34YMMfT/9Mfz6/+/v48oOkxoS08wcbChAJ\nKiBiHUOsKkX69kVULx/UnKaHJUZXh7SrSCWIBBUQsY4hVtXv3v738P3TbMGPvrk6pH0/j1SB\nSFABEesYYlW+RCgBqQiRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSES\nVEDEOoZYFZESkIoQCSogYh1DrLpdJJU9/xrxpb+B1foMfG0QyTqGWJVnpASkIkSCCohYxxCr\nIlICUhEiQQVErGOIVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIl\nIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIR\nIkEFRKxjiFURKQGpCJGgAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJU\nQMQ6hlgVkRKQihAJKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSs\nY4hVESkBqQiRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZY\nFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOIVREp\nAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWREpCK\nEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGpCJGg\nAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJKiBi\nHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi1jHE\nqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkIoQCSogYh1DrIpI\nCUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOIVREpAakIkaACItYxxKqIlIBU\nhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yKSAlIRYgE\nFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGpCJGgAiLWMcSqiJSAVIRIUAER\n6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJKiBiHUOsikgJSEWIBBUQsY4h\nVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi1jHEqoiUgFSESFABEesYYlVE\nSkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASk\nxRyeMw6RoAKRrGOIVREpAWkphx//GIJIUIFI1jHEqoiUgLQURBr8jqysQCQn6LUcEOmIRpV3\nds8aQyyOSAlI8dX59s9Pz1H/9fdsgkcwVFg6EqnzHH3WGGJzREpAOnl1xiASVPDOrnUMsTki\nJSCdvDpjEAkqeGfXOoZYGpESkFKX5vpEgor9Rfo0+Du7jjHmQaSzrs3co6sTCSp4jraOIbZG\npASkzJ0ZgEhQgUjWMcTWiJSAtHhlDkcf7b02kaCCd3atY4i1ESkBqQiRoIJ3dq1jiL0RKQGp\nCJGggnd2rWOIxREpAakIkaCCd3atY4jNESkBqQiRoAIi1jHEqmtEGuvrqBBpRQVErGOIVVeI\nNNiXfyDSigqIWMcQqyJSAlL9awMR6xhi1bV/RkKkktcGItYxxKrbRBrh66gu/RcP1mfga4NI\n1jHEqitF4oMNNa8NRKxjiFURKQGp/rWBiHUMseo6kUb6OipEWlEBEesYYtVVIg31dVSItKIC\nItYxxKqrPiE7FCREWlEBEesYYtU1n0ca6+uoEGlFBUSsY4hV+Vq7BKQiRIIKiFjHEKsiUgJS\nESJBBUSsY4hVESkBqQiRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSES\nVEDEOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVE\nrGOIVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqG\nWBWREpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFUR\nKQGpCJGgAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQ\nihAJKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiR\noAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogPvPWWMAAAt9SURBVIh1DLEqIiUgFSESVEDE\nOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOI\nVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWR\nEpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGp\nCJGgAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJ\nKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi\n1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkIoQCSogYh1D\nrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOIVREpAakIkaACItYxxKqI\nlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yKSAlI\nRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGpCJGgAiLWMcSqiJSAVIRI\nUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJKiBiHUOsikgJSEWIBBUQ\nsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi1jHEqoiUgFSESFABEesY\nYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkM7I79kYOhxzeLe/9h1ZWYFITtA3eW14\n03KJMeZBJN+1uTaRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDE\nOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVbdLpLKnh/avPRnXtZn4GuDSNYxxKo8IyUg\nFSESVEDEOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEi\nQQVErGOIVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRA\nxDqGWBWREpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxj\niFURKQGpCJGgAiLWMcSqiJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgV\nkRKQihAJKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkB\nqQiRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkIoQ\nCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOIVREpAakIkaAC\nItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWREpCKEAkqIGId\nQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGpCJGgAiLWMcSq\niJSAVIRIUAER6xhiVURKQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJKiBiHUOsikgJ\nSEWIBBUQsY4hVkWkBKQiRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi1jHEqoiUgFSE\nSFABEesYYlVESkAqQiSogIh1DLEqIiUgFSESVEDEOoZYFZESkIoQCSogYh1DrIpICUhFiAQV\nELGOIVZFpASkIkSCCohYxxCrIlICUhEiQQVErGOIVREpAakIkaACItYxxKqIlIBUhEhQARHr\nGGJVREpAKkIkqICIdQyxKiIlIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFW\nRaQEpCJEggqIWMcQqyJSAlIRIkEFRKxjiFURKQGpCJGgAiLWMcSqiJSAVIRIUAER6xhiVURK\nQCpCJKiAiHUMsSoiJSAVIRJUQMQ6hlgVkRKQihAJKiBiHUOsikgJSEWIBBUQsY4hVkWkBKQi\nRIIKiFjHEKsiUgJSESJBBUSsY4hVESkBqQiRoAIi1jHEqoiUgFSESFABEesYYlVESkAqQiSo\ngIh1DLEqIiUgFSESVEDEOoZYFZESkIoQCSogYh1DrIpICUhFiAQVELGOIVZFpASkIkSCCohY\nxxCrIlICUhEiQQVErGOIVREpAakIkaACItYxxKqIlIBUhEhQARHrGGJVREpAKkIkqICIdQyx\nKiIlIBUhElRAxDqGWBWREpCKEAkqIGIdQ6yKSAlIRYgEFRCxjiFWRaQEpCJEggqIWMcQqyJS\nAlIRIkEFRKxjiFURKQGpCJGgAiLWMcSqiJSAVIRIUAER6xhi1TUiHZ4zDqQRRBqLSFABEesY\nYvEVIh1+/GMISAOINBiRoAIi1jHE5oiUgFSESFABEesYYnNESkAqQiSogIh1DLH5NpE+PWcF\n3lsNRNq8E/l0Z0S8z0gy+ZOXrEh3rIAHEYi8BZG6cxBpz0GkPdcHkbpzEGnPQaQ91weRunMQ\nac9BpD3XB5G6cxBpz0GkPdfH+5UNZ0132YpLXBuIQOQt3q+1O2u6y1Zc5NpABCKvQaTuHETa\ncxBpz/VBpO4cRNpzEGnP9UGk7hxE2nMQac/1QaTuHETacxBpz/VBpO4cRNpzEGnP9UGk7hxE\n2nMQac/1QaTuHETacxBpz/VBpO4cRNpzEGnP9UGk7hxE2nMQac/1QaTuHETacxBpz/VBpO4c\nRNpzEGnP9UGk7hxE2nMQac/1QaTuHETacxBpz/VBpO4cRNpzEGnP9UGk7hxE2nMQac/1QaTu\nHETacxBpz/VBpO4cRNpzEGnP9UGk7hxE2nMQac/1QaTuHETacxBpz/VBpO4cRNpzEGnP9UGk\n7hxE2nMQac/1QaTuHETacxBpz/VBpO4cRNpzEGnP9UGk7hxE2nMQac/1QaTuHETacxBpz/XZ\nLlI6hh835fiJVQP91CuItKlPBJGuEIi0qU8Eka4QiLSpTwSRrhCItKlPBJGuEIi0qU9kB5EI\nuf0gEiGGIBIhhiASIYYgEiGGIBIhhhQQ6ZB6UXzgsPyfCgYiba5P5LIiXepVVPdVD5E2N0IE\nkfYNRNrcCBGzSIfnvP97/s/+3/Pf9vbP1/92eDrMf9H93sNTc7QtOsw6vv/i268X6i4ZiLS5\nTSJekQ6v/5j/W72sfXMx2+2V1fFvFB1HR1XRjMhR54lRzIFImxslcgGRno6nOJqoPTj/9dL0\nB9kra+dFh+XfEI7iDUTa3CgR+7t2r/96fcaezfn+dmHheftw/Pveyy4I6fLvyUCkzW0SsX+w\n4f0Z96mB9Lb1/MD7b3tqf9+8LA/pvSgDSY7iDkTa3CKRC3zU7rAIqRn++DcpSO1WZkh6FH8g\n0ub2iFzngw3iubLdaZlIDKnvOHS/4cQozkDk1DQ3QuSiH/5+WvrQplD8MP/93S+iD22qosOs\nQ5KJRrEGIt04N0mkwJcInZ0LX4yCgUibs4ncuEjvz//keyDSxkPkxkXa4zP1xQKRNhYity4S\nIbsEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgx\nBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSR\nCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgxBJEIMQSRCDEEkQgx\nBJEIMeT/AVdHgtX3QaLjAAAAAElFTkSuQmCC",
309 | "text/plain": [
310 | "plot without title"
311 | ]
312 | },
313 | "metadata": {
314 | "image/png": {
315 | "height": 420,
316 | "width": 420
317 | }
318 | },
319 | "output_type": "display_data"
320 | }
321 | ],
322 | "source": [
323 | "\n",
324 | "# 标准偏差\n",
325 | "p1 <- ggplot(my_sum) +\n",
326 | " geom_bar( aes(x=Species, y=mean), stat=\"identity\", fill=\"#69b3a2\", alpha=0.7, width=0.6) + \n",
327 | " geom_errorbar( aes(x=Species, ymin=mean-sd, ymax=mean+sd), width=0.4, colour=\"black\", alpha=0.9, size=1) +\n",
328 | " ggtitle(\"standard deviation\") +\n",
329 | " theme(\n",
330 | " plot.title = element_text(size=6)\n",
331 | " ) +\n",
332 | " xlab(\"\") +\n",
333 | " ylab(\"Sepal Length\")\n",
334 | " \n",
335 | "# 标准误差\n",
336 | "p2 <- ggplot(my_sum) +\n",
337 | " geom_bar( aes(x=Species, y=mean), stat=\"identity\", fill=\"#69b3a2\", alpha=0.7, width=0.6) + \n",
338 | " geom_errorbar( aes(x=Species, ymin=mean-se, ymax=mean+se),width=0.4, colour=\"black\", alpha=0.9, size=1) +\n",
339 | " ggtitle(\"standard error\") +\n",
340 | " theme(\n",
341 | " plot.title = element_text(size=6)\n",
342 | " ) +\n",
343 | " xlab(\"\") +\n",
344 | " ylab(\"Sepal Length\")\n",
345 | " \n",
346 | "# 置信区间\n",
347 | "p3 <- ggplot(my_sum) +\n",
348 | " geom_bar( aes(x=Species, y=mean), stat=\"identity\", fill=\"#69b3a2\", alpha=0.7, width=0.6) + \n",
349 | " geom_errorbar( aes(x=Species, ymin=mean-ic, ymax=mean+ic), width=0.4, colour=\"black\", alpha=0.9, size=1) +\n",
350 | " ggtitle(\"confidence interval\") +\n",
351 | " theme(\n",
352 | " plot.title = element_text(size=6)\n",
353 | " ) +\n",
354 | " xlab(\"\") +\n",
355 | " ylab(\"Sepal Length\")\n",
356 | "\n",
357 | "p1 + p2 + p3"
358 | ]
359 | },
360 | {
361 | "cell_type": "markdown",
362 | "id": "e3765f3f",
363 | "metadata": {},
364 | "source": [
365 | "很明显,这 3 个指标报告了非常不同的可视化和结论。所以应该始终指定用于误差线的指标。"
366 | ]
367 | },
368 | {
369 | "cell_type": "markdown",
370 | "id": "c64cb91a",
371 | "metadata": {},
372 | "source": [
373 | "### 解决方法\n",
374 | "\n",
375 | "最好尽可能避免误差线。当然,如果您只有汇总统计数据,这是不可能的。但是,如果您知道各个数据点,请显示它们。有几种解决方法是可能的。带有散点信息的箱形图适用于相对少量的数据。当数据量较多时,使用小提琴数据图是另一种办法。\n",
376 | "\n"
377 | ]
378 | },
379 | {
380 | "cell_type": "code",
381 | "execution_count": 12,
382 | "id": "ea46d3f0",
383 | "metadata": {},
384 | "outputs": [
385 | {
386 | "data": {
387 | "image/png": "iVBORw0KGgoAAAANSUhEUgAAA0gAAANICAMAAADKOT/pAAAAPFBMVEUAAAAzMzNNTU1oaGhp\ns6J8fHyMjIyampqnp6eysrK9vb3Hx8fQ0NDZ2dnh4eHp6enr6+vw8PD/pQD///+lTq0HAAAA\nCXBIWXMAABJ0AAASdAHeZh94AAAgAElEQVR4nO2dC3vaSBZEAY8zTmZ2Qsz//69rAcYCJPPQ\nrVZd3XO+3WTsMJ4qqU+6EXqsdgAwmdXcAQCWACIBBIBIAAEgEkAAiAQQACIBBIBIAAEgEkAA\niAQQACIBBBAg0ur4Q779UaurfwjPATAf0wfwLUXu+w8iEqTm8QG8Wn39sjr8cvqi9/3+Cw//\nndPL+686fSukDcBMPDyAV8e13H3/Oyky/uenPwTIyzMifWPO5+zyqcbqODuNveprYossBdCa\n8Blpd/bl6uxfu37VxfcAkjLpPVLfglXvbdPnd07fPpuOenPQ8RWIBNlhAAMEgEgAASASQACI\nBBAAIgEEgEgAASASQACIBBAAIgEE8KhIWz8cM8mpWNqwMyJlp2Jpw86IlJ2KpQ07I1J2KpY2\n7IxI2alY2rAzImWnYmnDzoiUnYqlDTsjUnYqljbsjEjZqVjasDMiZadiacPOiJSdiqUNOyNS\ndiqWNuz8iEibDxDJjYqlDTs/INLm9AsiGVGxtGFnRMpOxdKGnREpOxVLG3Z+SqR1x62XA5SE\ngw05qVjasPMDIrG0s6RiacPOiJSdiqUNOyNSdiqWNuyMSNmpWNqw8wMicbDBkoqlDTs/ItI5\ncycfwDGTnIqlDTsjUnYqljbsjEjZqVjasDMiZadiacPOiJSdiqUNOyNSdiqWNuyMSNmpWNqw\nMyJlp2Jpw86IlJ2KpQ07I1J2KpY27IxI2alY2rAzImWnYmnDzoiUnYql13MHuAaRslOxNCJp\nccwkp2JpRNLimElOxdKIpMUxk5xipd/ft4ikxjGTnFql3987kxBJi2MmObVKI1ILHDPJKVaa\npV0DHDPJqVgakbQ4ZpJTsTQiaXHMJKdiaUTS4phJTsXSiKTFMZOciqURSYtjJjkVSyOSFsdM\nciqWRiQtjpnkVCyNSFocM8mpWBqRtDhmklOxNCJpccwkp2JpRNLimElOxdKIpMUxk5yKpRFJ\ni2MmORVLI5IWx0xyKpZGJC2OmeRULI1IWhwzyalYGpG0OGaSU7E0ImlxzCSnYmlE0uKYSU7F\n0oikxTGTnIqlEUmLYyY5yy69v2nQFYikxTGTnEWXPtzG7gpE0uKYSc6iSyPSLDhmkrPs0izt\n5sAxk5yKpRFJi2MmORVLI5IWx0xyKpZGJC2OmeRULI1IWhwzyalYei/S8HGIuUCk7FQs3Yk0\ncmR8LhApOxVLI5IWx0xyKpZmaafFMZOcJZcec4WDDVocM8lZcOnR1RsiaXHMJGfBpY8iXduE\nSFocM8lZcumjR1cmIZIWx0xyFl8akZrjmEnO8kuztGuNYyY5FUsjkhbHTHIqlkYkLY6Z5FQs\njUhaHDPJqVgakbQ4ZpJTsTQiaXHMJKdi6Z5ILmfcIVJ2Kpb+EsnmHHBEyk7F0oikxTGTnIql\nWdppccwkp2JpDjZoccwkp2JpRNLimElOxdKIpMUxk5yKpRFJi2MmORVLI5IWx0xyKpZGJC2O\nmeRULI1IWhwzyalYGpG0OGaSU7E0ImlxzCSnYmlE0uKYSU7F0oikxTGTnIqlEUmLYyY5FUsj\nkhbHTHIqlkYkLY6Z5FQsjUhaHDPJqVgakbQ4ZpJTsTQiaXHMJKdiaUTS4phJTsXSiKTFMZOc\niqUXJRLAXKznDvAdzEg5qVh6UTPS3MkHcMwkp2JpRNLimElOxdKIpMUxk5yKpRFJi2MmORVL\nI5IWx0xyKpZGJC2OmeRULI1IWhwzyalYGpG0OGaSU7E0ImlxzCSnYmlE0uKYSU7F0oikxTGT\nnIqlEUmLYyY5FUsjkhbHTHIqlkYkLY6Z5FQsjUhaHDPJqVgakbQ4ZpJTsTQiaXHMJKdiaUTS\n4phJTsXSiKTFMZOciqURSYtjJjkVSyOSFsdMciqWRiQtjpnkVCyNSFocM8mpWHov0vv73DH6\nIFJ2KpbuRHp/tzIJkbJTsTQiaXHMJKdiaZZ2WhwzyalYmoMNWhwzyalYGpG0OGaSU7E0Imlx\nzCSnYmlE0uKYSU7F0oikxTGTnIqlEUmLYyY5FUsjkhbHTHIqlkYkLY6Z5FQsjUhaHDPJqVga\nkbQ4ZpJTsTQiaXHMJKdiaUTS4phJTsXSiKTFMZOciqURSYtjJjkVSyOSFsdMciqWHhBp7quT\nECk7FUtfizT79bKIlJ2KpRFJi2MmORVLs7TT4phJTsXSHGzQ4phJTsXSiKTFMZOciqURSYtj\nJjkVSyOSFsdMciqWRiQtjpnkVCyNSFocM8mpV/r9HZG0OGaSU6707J+9DoJI2SlXGpHkOGaS\nU680Szs1jpnkLL/09QQ0KNK88xQiZWfxpQeWckMizbziQ6TsLL40IjXHMZOc5Zdmadcax0xy\nKpbmYIMWx0xyKpZGJC2OmeRULI1IWhwzyalYGpG0OGaSU7E0ImlxzCSnYmlE0uKYSU7F0oik\nxTGTnIqlEUmLYyY5FUsjkhbHTHIqlkYkLY6Z5FQsjUhaHDPJqVgakbQ4ZpJTsTQiaXHMJKdi\naUTS4phJTsXSiKTFMZOciqURSYtjJjkVS1+KZHBbIUTKTsXSFyI53KALkbJTsTQiaXHMJKdi\naZZ2WhwzyalYmoMNWhwzyalYGpG0OGaSU7E0ImlxzCSnYmlE0uKYSU7F0oikxTGTnIqlEUmL\nYyY5FUsjkhbHTPFcfGhSo/Q5iKTFMVM4lx/jlyh9ASJpccwUDiIhkhrHTPGwtEsu0mYPIplR\nsXRukQ42IZIZFUvnF+nLI8f955hJTsXSiKTFMZOciqXTi3T0aN1xj3egp+KOsO78gEh75v4r\nYADHTHIM/3aWY9gZkbJjOKjkGHZ+SKS+R46D1jGTHMNBJcewMyJlx3BQyTHsjEjZMRxUcgw7\nI1J2DAeVHMPOD4l0xtzJB3DMJMdwUMkx7IxI2TEcVHIMOyNSdgwHlRzDzoiUHcNBJcewMyJl\nx3BQSekuxzLsjEjZMRxUSvYXCBt2RqTsGA6qYM4uCUakBjhmkmM4qGK5uEkFSzs9jpnkGA6q\nWAaefmTYGZGyYziogrl++pFhZ0TKTn9QGTxwqwmIpMUxk5zeoHJ4BGQTEEmLYyY5tiKtk/Jc\nW0TKju3Sbv1XShAJkbxoK9L7e9APQiRE8qKpSN2qNuYnIRIieYFIiJQJRNrD0i4Qx0xyECkW\nREIkLxAJkTKBSLEgEiJ5gUiIlAlEigWREMkLREKkTCBSLIiESF4gEiJlApFiQSRE8gKRECkT\niBQLIiGSF4iESJkoK1LYaarnIBIieREm0rAxcRdOnINIiNTD4KLzKJFGjEEkHY6Z5AzueIfb\noIhFYmmnwzGTnMWLpDJmBERCpB7ze8RRO0RKxfIPNrQFkRDJC0RCpEwgUiyIhEheIBIiZQKR\nYkEkRPICkRApE4gUCyIVEenyEyJEigWRaoh0dc5CVZFU5zsgEiJ5oRVJdc4qIm1riMTS7gAi\nCXHMJKeoSCzthDhmCqfsjNToJHBEqiFS2fdIsrXcBYiESF40EylWMESqIRJLu+vvh85ViFRE\npEvKiDTGe6xJiIRIXrQR6UMhRIrGMZOc2iIdHGJpF4tjJiX7t0uIFPsTEamcSIcDeLVFiv90\nCZEQyQvO/kakJLC0E4BI9UTag0ixIBIieYFIiJQJW5G264QqrZ/dnIiUHV+REk5Kz29MRMqO\ns0jJJqWnp6MtIuXHWqRMKk3RCJFScnYCuLlIadZ3E7cjIuXj/JIke5FSTErTpqMtImUkm0gJ\nJqXpGxGREpJradfhPSlNno62iJSfFCI5qxShESLlJ4lItuu7oO2HSMkxeFjsvThOSjHT0RaR\nsuPw+PL7sTMpbjpHpNzkEslsUgqbjraIlJ5UHm2dJqVIjRApP2kONhxxmZSCtxsiZSebSB4q\nxU5HW0TKTz6RDNZ38RsNkbKTUaSZJ6Xw6WiLSPlJKdITk1LcfbckWwyRspNUpEcnpbA7QSqm\noy0i5SerSA+qFCSSSCNEyk9ekR5b39326A7TdBsLkbKTWaTQgw635yzZdLRFpPykFinySPhN\nkaRbCpGyk1ykwEnpe4+U09EWkfKTXaQ2HyqJNUKk/OQX6a713bRjdvqNhEjZWYJIH5PSDbo3\nQDdfNIJ+OtpOEQlm5f39+A/rWWOEcdujp0VqvIWYkTLxdUFfhRlpmkfmM5I+28M4ZhKxLJFu\nvUeafFoD75EewjGTitOVsflFuuOo3eTTgzhq9wiOmeRkF6nVFRV8jnQ/jpnkJBep4YVJnNlw\nL46Z5KQWqe0FfpxrdyeOmeRkFqn5dbKc/X0Xjpnk5BVpjuvNuR7pHhwzyckq0ly3beAK2ds4\nZpKTVKQZ737CPRtu4ZhJTkqRuIsQIrmRUaQnNIq7idBepfBKiJSdyzHhfzPwZ6ajsJsIfZrE\nnVa/wTGTnIsRYf94iudWddEice/v73DMJGdAJGeTnn1zNO7Rs4bxNIpRHDPJuV7aGYv06HQ0\nKsnXH0yYq3g+0giOmeRcDQZnjx4c6aOS9P5g0qKPJ/YN4phJTp6jdo+/OzpJculK355Jb554\nhuwQjpnkpBHpmXdHJ4+uTZpgzxk81fwax0xykog06SPY+GN2PWImJUTKTgqRpp7JIPTorxiV\nECk7GUSa/Ql9t5i+EREpO/4iOTwz9haTJ6Vxkd42qwOIZI29SAk06pi4HUdFelutECkD5iJl\nmI4OTJuURkXarH5+s65DJBusRcqjUccUlUZFGp2JEMkLZ5FSadTx/MYcFelt9QeRMuArUq7p\n6MDTk9KoSLsfr78RKQG2IiXUqOPJ7Tks0qoPIjnif8tiREIkfxLcRB+R7iN2q4fgmEkEIqlA\npFIilV/avb+LTsCLFulzSbfZIJI1NUV6PyD4yaEibXiPlIblizTkSxKRfvU8+oVI1ixepGFh\n0i3tRond6iE4ZpJTVCQZHGwwFunp5wjPzLTSUUO7qUeCGemTzd+DpzhM2sgaHDPtqXUkuGTp\nO0T6YMikSRtZg2OmPbXGVMnSoyL9u+rOtfv9uvpn97b6G5EmUWtMzVM6agEYLdLL8ezv1cvI\ngYdJG1mDY6Y9jcaU1+2xG5TuFQ47JKE6atf9jkgTEY+p0bu/TcRVpM+a57eJNBXp9XNp97r7\nXzcrIdIE1GfLHMZQFZF69189m5Jifnq0SL+PZzdsfu+GP5SdtJE1OGba00akKku7L38UB8bD\nP0f68/NltXp5+3intHob8Mhx0Dpm2tNmaReOqUjaD5b4QLauSCpcRTrDZBpGpBYgkgyXN4bj\nInGDyDgQSYa9SNwgMpDQMdXu1LMMItkv7Taj108g0sNEjqmGJ0OnECkaLqNApGgmipSU59qO\nivSDG0TGUXJpJ8Qw2KhIvzfcIDKM6FVOI5cMx+uBtd8Dp0dF4p4NgQSL1Gp1ZyvS183IbECk\nFiBSLJlEusncyQdwzLSHpV0smZZ2iBRIrSPBegyDfSPSrx8fy7rX/xBpOogUi2GwUZH+vOzf\nH61W/0OkySBSLIbBRkX6e/XWfSj7z+oVkSaDSLEYBhsV6fMSc47aBYBIsRgGQ6QWIFIshsFu\nLe2Gb8WFSI+BSLEYBhsV6c/XPRsQaSqIFIthsFGRdrvTPRsQaSqIFIthsG9EusHcyQdwzLQH\nkWIxDIZILUCkWAyD3RSJo3YBIFIshsEQqQWctBqLYTBEakGISDfvcx2ul+F4PWAYDJFaECFS\nz56xp6qa3FBHj2EwRGpBsEjDcw8izQkitSB4affsCx7Fb7weL+jzCzYi0tlzLxFpMhy1m8rB\noM9LzI2CfYJILUCkiRwNSifSPcydfADHTHsQaSKfBmVb2iFSLHyONJXzu50YBfsEkVrA7bhi\nMQyGSC1ApFgMgyFSC1jaxWIYDJFawMGGWAyDIVILECkWw2DDIvE5UiyIFIthMERqASLFYhhs\nWKR7mDv5AI6ZDqwTqvTss+saYJgMkRqRziTDwXrCMNu4SG8s7ULJNSkZT0fbXCK98R4pmjwq\neWuUS6TN6r/X1e8/rzyNIpAkJhmO03MMA46K9DET/Vz9u/vTexrF5gNEmkSGScl9OtqmE+nf\n1a/+FbKb0y+I9Dz2JhkO0isMM46K9GP1z+/Vy+5/iBSM96SUYDra5hKpM+i1O9ZwehrF5vwF\ncycfwDHTAAEqic5azaFRLpF2/750z3ZZvZ2+sdl9vkdad+zgaZ4yqSeP6DoKv336/j74bb+g\nPW5+ILu3iKVdCE9MSue34xoQ6cbdum5r5Pf3/OcV5hffzDUjXYt0+gWRpvOwSWfyDHr0+c3n\n5ivDwTkoUvc9w6zjIv15e1mtXn8ikoiHJ6UbdkwTyXA66hickHKJ9PvqiX2IFEz4BejX/5Rb\no2GSLe1eV68fCv1+vThqh0iBuBwJNxyX32MYeFSk4+dHf3rn2nFmQzgOKqWajg4YJh4V6cfq\n8PTY3ilC58ydfADHTLeY3STDQXkTw8yjIu3+fv2vW9q98lRzLfNOSgmno20ukVa3LjefO/kA\njpnuYEaTDEfkPRjGRiQD5pqUck5H21wi3WTu5AM4ZrqPOVRKqxEiqXHMdC/NTTIcjHdjmP0b\nkX796E4A/w+R2tB2Uko8HW1zifTnZf/eaMWl5s1oaJLhSHwEw/ijIv29eus+lP2Hz5Ha0WpS\nyj0dbXOJ1B2p+/w/IrWihUrpNUIkNY6ZHkVukuEgfBjDDreWdm9fJ60iUhPWSpYwHX2wHri8\nYmZGRfpzdRkFIjVCadLc3WIYvHJ2XkZF2u1+vqxWL29/RjxyHLSOmR7mhgnv71M8WsiMlEuk\nG8ydfADHTI9y4z3S9NueLMGkTEs7RJqDm0ftAu4ftIBJybDBiEh/3rov/9msfoy9RXIctI6Z\nHuKOQ3bDHj1ml+E4fAzDAiMibbqj3v/bH2wYe5M0d/IBHDM9wPMfIj06T2WflAzjD4v0a/X6\n4c/La/d0l7crhRBJwoRPkB5f8BkOxQcwTD8s0uuqu/FJ9xHSn9XmSiFEEjDtnIbF3H/rPgyz\nD4u0P53hn/1kxJkNLeB6pIcwTD4s0qb74m31HyK1Ya4rZOfu/SyGwYdF2t9C6OVl1x1w4Oxv\nNfPd/iTHpHT9oZFh7GGRfn28Pfp39fPjLdJr97AxRFIy712E5m5/m4HTGAxTD4u0P9GuO/C9\nWr2MeOQ4aB0z3WLuW0T6T0qZRdr993L4KHb04DcixTD7/SETTEqjSzunE4VGRLqDuZMP4Jjp\nW+aejg74T0qXHAJbnbqaW6SLDWmR6QEsNOrIZhIixXK5JR0y3Y/HdHQg2aTE0i6W1CJN1Cj6\nIbKpTDIMm1qkxEu7qdPR6Nl1TwuWaVIyjJpbpAscMw0zeVU3JtKU65UMh+cIhkkXJZLh9h0k\n4t3R6IQ0Yc2XZlIyzIlI7ZEeZJj23inLFpw7wDWI1Bqng3XX5JiU1l5H7DoQqS3eGnVkUGnt\n9RlSByI1xV6jjgSbEZGkuI8A4UGGUOwnJZZ2Wsz3f8R0FHA7rntw35JzB7gGkVoR8+6okUjm\nk5JhOERqRNS7ozYe/eU9KRlmQ6Qm+B+su8Z4UjJMhkgtSKhRh+/2nDvANYjUAkSKxTAYIrUA\nkWIxDIZILVCKJDz64Ls95w5wTUqRxj6MW9/3svbEfII08m2dSYbj9YBhsIwijZ4esr7vZe0J\nOaMh/gKkWxiO1wO9YC77GJFaIBTpr9oi2ezkjCKxtDv/vswkRLqflCKN4bvjRSO9o7ZINn9b\nIlILjK+J/Q7f7Tl3gGsQqQXxItU+B9wwGCK1IFykNieB+27PuQNcg0gtQKRYDIMhUgsiRToY\nxNLODERqQaBIra7s6/DdnnMHuAaRWoBIsRgGQ6QWxC/tmuC7PecOcA0itYDLKGIxDIZILUCk\nWAyDIVILECkWw2CI1AJEisUwGCK1YLpIDQ8xfOG7PecOcA0itUD2XDEtvttz7gDXIFIL9CJJ\nPPPdnnMHuAaRWiBf2mlmLN/tOXeAaxCpBfKDDYg0N4jUAv1RO4lJvttz7gDXIFILECkWw2CI\n1IK1nvd3wQ+de7uNYTj4ECk7p9IutwFpwGnw+XRGpOx8lra5MVUDPgefUWdEyg4iWYBISTmN\nIJZ2FiBSTr7+Li5U+oTh4EOknCCSGYiUlOulXSEMBx8iZadiacPBh0jZqVjacPClFunymM3S\nx9TgMaqllx7CYfBdkFmkq08RFj6mhj81uSrtc0hYhsHguwSR8nCfSMdXLVong8F3SWaRWNp1\nrC++fRDJ6DN/AQ6D74LUIl2ydJEGWV86c5yQEKkpiJSXgypXIvX/cKEYDj5ESstRoMulXQUM\nB9+SRCo2oE4i1cNw8C1IpGW/LRjguLSbO8YM+A2+CSLZ0Yk0d4YZWM8dAM5JPyNVW9odYUay\nYEkilRxTJUsbDj5Eyk7F0oaDD5GyU7G04eBDpOxULG04+BApOxVLGw4+RMpOxdKGgw+RslOx\ntOHgQ6TsVCxtOPgQKSP9j57LlO5hOPgQKSFnZxVWKd3HcPAhUkIQae4A1yBSRlja2YFI2alY\n2nDwLVqkEqeDI5IFyxTpYFCNK/0QyYIFifT+fvHMLURaKn6Db0Ei9bT5/McKHiGSB8sS6cuk\nWaM0BZEsWI5IZyYVApEsWJBIRd4TXYJIFixJpJomIZIFiJQdRLJgUSKtC3qESB4sS6S5A8xB\nxdKGgw+RslOxtOHgQ6TsVCxtOPgQKTsVSxsOPkTKTsXShoMPkbJTsbTh4EOk7FQsbTj4ECk7\nFUsbDr4liHT6GLbimCpZ2mjwfbIAkb5ODKo4pkqW9hl8JxApOxVL+wy+EwsQiaVdOYwG3ydL\nEOlExTFVsrTh4EOk7FQsbTj4ECk7FUsbDj5Eyk7F0oaDD5GyU7G04eBDpOxULG04+BApOxVL\nGw4+RMpOxdKGgw+RslOxtOHgQ6TsVCxtOPgQKTsVSxsOPkTKTr90lfv6GQ6+ZYpUZUB19EQq\nc6dZw8G3SJHKDKgORLIAkbLD0s6CRYpUZkB1VHxjaDj4lilSJSqWNhx8iJSdiqUNB19ukS6W\ncDzWpQgWg++c1CJdHlQodZDhE0SyoLlI60A6cb77OpTQzR6IbTAhiPSx3/8K5P39+68DsR2v\ntsGEIFKwSA2xHa+2wYQgEiKFYxtMCCIhUji2wYQgEiKFYxtMCCIhUji2wYQgEiKFYxtMCCIh\nUji2wYQgEiKFYxtMCCK1FSny81nb8WobTAgiBYl0nyHdGUMR/7U9tuPVNpgQRIoR6Q5Duhcg\n0lJBpFYiHV6x0KXd+SnuRsGagUhNlnYffxo6Ge3xGa8XF4v4BGsHIrU42LCXKPw8cJ/xikiI\ntNWIdG5N/GzUYTReWdohkkSkS3O+vuJgwzJBpDiReo6MTkGRc5PteLUNJgSRwkQ6c2RMF0Ra\nKIikEembV8X81/5CJCsQqbv5SczAHnJEecsG2/FqG0wIInVEqXSF5nBdh69GiGTCDCLtb8ml\nGO4ikYxvxdVhHU4EIp2QqCT5/Mh9oLrnU4BIPUTTUijmk9Ee/4TxINI53i5lsGiLSCbMKtJW\neORhKjks2iKSCXOL5DktJZmM9uRJGgciDWOmUiKLtohkgoVITtOS92Q08Nga57gqEOkbLFSy\ntmj4AVA8jNkCH5FE09IDny15T0YdN0Qq86A1RLpFuEr3n+1gb1HH90s7RJoPM5HCp6U7RfKf\njEZhaWfBIyJtOtQibYNdusOjxBZtOdhgwkMitZiRDtyn0g1J7lvUpbZoi0gmmIp017R0Y9l2\nz6ou92S0J32BJ8gt0rlHDbp8qPTtc8ZvPMT81jPO809Ge5bQ4VGSi3R6i7Qfh7deHsG4I3eI\nsr7xx20qyFlGiwVx34zU4mDDkfXojHRQaEykm/4cWcTCjhnJhAdE+rKphUjfvUU6vvsZfgv0\n2HWy+VVKX+AJEOlO1jeONAyocvrWoxecZ5+Wcqd/jtwitVra3bJoRK2eSY/+y6ldShz9afKL\n1DtyJwr0mEVPz0PXLon6yEkbfAK5Rdqdndgg6fLoZDRpHrpSKeeQzJl6GslFOic8yhNLuuAb\ncKV0KWHkySDSKE+9MRLcgCufSukCB4BII1hc1Hck27SUK20MiDTEk5ORkFQuJYoaBiJdIbbo\n6aVfHpey5IwEkS4IsGjyCeCjJFEpR8opXF+uiEh9Qiaj71WZeFQvxbSUIOI0Bi6gR6QTUUu6\nm9ckTfz5/i6555sMIo0T+MZI+HCxI+YqDaZb1M0bWNoN43eU7hbW09JQtIXfTshxfzQXKZ1F\nBwx33ZF6Iu2vJps7xCXtRZKP+f5ir+pTzRfvkZ1JyxOpf/gh8Fw8tx13wvD9ghZE2oNIwSCS\nBcsTqdrSrpxIvEfak/NYAyI5YegRIt2L3647UlAkx86IdCeIZIRhZ0S6E0QywrAzIt0JIhlh\n2BmR7gSRjDDsnFok/QmrXyCSEYadM4sUfA+h70EkIww7I9KdIJIRhp0zi8TSrsNwUMkx7Jxa\npJYgkhGGnRcvUtSshUhGGHZeukhh76MQyQjDzslFumkJIi0Rw865RbpDE5Z2C8Swc36RGh26\nQyQjDDu3FymUg0mxP3OE0M0eiOGgkmPYublIsRxE+vzKdrAr8dgRbTHsnFukc48QqQqGnfOL\n1PsSkYpg2Dm3SBf3b0OkIhh2Ti7SOYhUBMPOiJQdxx2hxrAzImXHcUeoMeyMSNlx3BFqDDsj\nUnYcd4Qaw86IlB3HHaHGsHNWkQafW4JIRTDsnFSk4SdpIVIRDDsjUnYMB5Ucw85JRWJpd8Jw\nUMkx7JxVpEEQqQiGnREpO447Qo1hZ0TKjuOOUGPYGZGy47gj1Bh2RqTsOO4INYadESk7jjtC\njWHn7CJxhazJjmiKYefkIp1/MItIRTDsjEjZ8dgRbTHsnFwklnaOg0qOYefsIp2BSEUw7IxI\n2XHcEWoMOyNSdhx3hBrDzoiUHccdocawMyJlx3FHqDHsvGCRBi9ZWh6OO0KNYeflijR8Ee3y\ncNwRagw7I1J2HIMJevsAAAVFSURBVHeEGsPOyxWJpd1yMey8NJGK2NPDcUeoMey8MJGqrOd6\nOO4INYad84o0YAwiFcGwc1qRhpRhaVcEw85LE6kec++IOTDsnFak4aVdQWbfETNg2DmvSAMg\nUhEMOyNSdhx3hBrDzoiUHccdocaw87JFqnAMz3FHqDHsvGiRSnyq5Lgj1Bh2RqTsOO4INYad\nFy0SS7uFYth52SJVwHFHqDHsjEjZcdwRagw7I1J2HHeEGsPOiJQdxx2hxrAzImXHcUeoMeyM\nSNlx3BFqDDsjUnYcd4Qaw86IlB3HHaHGsDMiZcdxR6gx7IxI2XHcEWoMOz8vkiHruQMAMCMl\nxXFHqDHsnF+k3ompiFQEw87pRepfKoFIRTDsjEjZcdkRLTHsnF4klnZzB5gBw875ReqBSEUw\n7IxI2XHcEWoMOyNSdhx3hBrDzoiUHccdocawMyJlx3FHqDHsjEjZcdwRagw7I1J2HHeEGsPO\niJQdxx2hxrDzUkTafyyLSEUw7LwQkQ4nCiFSEQw7I1J2vHZEGww7L0QklnalMOy8FJH2IFIR\nDDsjUnYcd4Qaw86IlB3HHaHGsDMiZcdxR6gx7IxI2XHcEWoMOyNSdhx3hBrDzqlFunyyJSIV\nwbBzZpEOn8Jyz4Z6GHZOLxJ3EZo7wAwYds4s0vYwISFSOQw7pxZpD0u7ehh2zi9SD0QqgmFn\nRMqO445QY9g5uUjnB8ARqQiGnXOL1D/SsEWkMhh2RqTsWOyIxhh2zi0SSzvLQSXHsHNykc5B\npCIYdkak7DjuCDWGnREpO447Qo1h50WJ5JhJTsXShp0RKTsVSxt2RqTsVCxt2BmRslOxtGFn\nRMpOxdKGnREpOxVLG3ZGpOxULG3YGZGyU7G0YWdEyk7F0oadESk7FUsbdkak7FQsbdgZkbJT\nsbRhZ0TKTsXShp0RKTsVSxt2RqTsVCxt2BmRslOxtGFnRMpOxdKGnREpOxVLG3ZGpOxULG3Y\nGZGyU7G0YWdEyk7F0oadESk7FUsbdkak7FQsbdgZkbJTsbRhZ0TKTsXShp0RKTsVSxt2RqTs\nVCxt2BmRslOxtGFnRMpOxdKGnREpOxVLG3ZGpOxULG3YGZGyU7G0YWdEyk7F0oadESk7FUsb\ndkak7FQsbdj5eZEMWc8dYA4qlrbujEg5qVjaujMi5aRiaevOiJSTiqWtOy9AJID5QSSAABAJ\nIABEAggAkQACQCSAABApIZu7vpWby0Kb8T+yILdIlpt0FtgSM4NIy4AtMTPJRNp88PV7/9fr\n31Oz+fz12Gaz2/S/uGq72V28NCufxTe9zocv9l+P1J+fXCJtjr/0fx/6Xv6/oHvj6ejT7qzi\nQOuzl6alL9KXNWfbwHJnJxRpd74dz7bp5Qvz8jVorkfMZnBLDG6IfFzbMlDQb2fnEunzb9vN\n5riq2/U29+lvYpvpfgqf46nX9Ks+In293GRnJxOpv8bZXYj0Oc76L0jMZnfZdP97f6baLVGk\nr+L3iGSzs9OJdDWMBhc9Ftt2GsMiXY6k4iL57OxcIl2+Y9iMfc9i207kchyNW7Mska47b64K\n+u3sXCJdHfQdOfydfzDtvparvcXsnYe/k3P+N0L/8PeuZ4/Zzk4mEpTEQpXvQSSwxudd0Pcg\nEnjjcnz7BogEEAAiAQSASAABIBJAAIgEEAAiAQSASAABIBJAAIgEEAAiAQSASAABIBJAAIgE\nEAAiAQSASAABIBJAAIgEEAAiAQSASAABIBJAAIgEEAAiAQSASAABIBJAAIgEEAAiAQSASAAB\nIBJAAIgEEAAiAQSASAAB/B/WwgBK3OdvKgAAAABJRU5ErkJggg==",
388 | "text/plain": [
389 | "plot without title"
390 | ]
391 | },
392 | "metadata": {
393 | "image/png": {
394 | "height": 420,
395 | "width": 420
396 | }
397 | },
398 | "output_type": "display_data"
399 | }
400 | ],
401 | "source": [
402 | "ggplot(data,aes(x=Species, y=Sepal.Length)) +\n",
403 | "# 绘图,notch为TRUE表示绘制小提琴图,否则为绘制箱形图\n",
404 | "geom_boxplot( fill=\"#69b3a2\", notch=T) +\n",
405 | "# 绘制数据点信息\n",
406 | "geom_jitter( size=0.9, color=\"orange\", width=0.1) +\n",
407 | "ggtitle(\"confidence interval\") +\n",
408 | "theme(\n",
409 | " plot.title = element_text(size=6)\n",
410 | ") +\n",
411 | "xlab(\"\") +\n",
412 | "ylab(\"Sepal Length\")"
413 | ]
414 | },
415 | {
416 | "cell_type": "markdown",
417 | "id": "e591c82e",
418 | "metadata": {},
419 | "source": [
420 | "## 参考\n",
421 | "\n",
422 | "+ [Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm](http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002128)\n",
423 | "+ [THE ISSUE WITH ERROR BARS](https://www.data-to-viz.com/caveat/error_bar.html)"
424 | ]
425 | }
426 | ],
427 | "metadata": {
428 | "kernelspec": {
429 | "display_name": "R",
430 | "language": "R",
431 | "name": "ir"
432 | },
433 | "language_info": {
434 | "codemirror_mode": "r",
435 | "file_extension": ".r",
436 | "mimetype": "text/x-r-source",
437 | "name": "R",
438 | "pygments_lexer": "r",
439 | "version": "3.6.0"
440 | }
441 | },
442 | "nbformat": 4,
443 | "nbformat_minor": 5
444 | }
445 |
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img5_1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img5_1.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img8_1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img8_1.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_1.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_2.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_3.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_4.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_4.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_5.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_5.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_6.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_6.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_7.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_7.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_8.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_8.png
--------------------------------------------------------------------------------
/Visualization/数据绘图要点/image/img9_9.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/luohenyueji/R-Study-Notes/e6021d86a15f46a981c09c5e2c1b6766c6470462/Visualization/数据绘图要点/image/img9_9.png
--------------------------------------------------------------------------------
/WGCNA/wgcna_tutorial.r:
--------------------------------------------------------------------------------
1 |
2 | # 加载库
3 | library(WGCNA);
4 |
5 | # 读取文件
6 | # The following setting is important, do not omit.
7 | # 如果没有显式地指定“stringsAsFactors=FALSE”,默认会将所有的字符串转换为因子,导致数据处理速度较慢
8 | options(stringsAsFactors = FALSE)
9 | # Read in the female liver data set 读取135个雌性小鼠的数据
10 | femData = read.csv("./data/LiverFemale3600.csv")
11 | # Take a quick look at what is in the data set:
12 | # 查看数据的维度
13 | dim(femData)
14 | # 预览数据
15 | head(femData)
16 |
17 | # 删除冗余数据-c(1:8)删除前8列数据,t()转置数据
18 | datExpr0 = as.data.frame(t(femData[, -c(1:8)]));
19 | head(datExpr0)
20 |
21 | # 将原数据的行列名复制过来
22 | names(datExpr0) = femData$substanceBXH;
23 | rownames(datExpr0) = names(femData)[-c(1:8)];
24 | head(datExpr0)
25 |
26 | gsg = goodSamplesGenes(datExpr0, verbose = 3)
27 | gsg$allOK;
28 |
29 | if (!gsg$allOK)
30 | {
31 | # Optionally, print the gene and sample names that were removed:
32 | # 打印删除的基因和样本名称
33 | if (sum(!gsg$goodGenes)>0)
34 | printFlush(paste("Removing genes:", paste(names(datExpr0)[!gsg$goodGenes], collapse = ", ")));
35 | if (sum(!gsg$goodSamples)>0)
36 | printFlush(paste("Removing samples:", paste(rownames(datExpr0)[!gsg$goodSamples], collapse = ", ")));
37 | # Remove the offending genes and samples from the data:
38 | # 从数据中删除有问题的基因和样本
39 | datExpr0 = datExpr0[gsg$goodSamples, gsg$goodGenes]
40 | }
41 |
42 | # hclusts聚类算法, dist计算基因之间的距离
43 | sampleTree = hclust(dist(datExpr0), method = "average");
44 | # Plot the sample tree: Open a graphic output window of size 12 by 9 inches
45 | # The user should change the dimensions if the window is too large or too small.
46 | # 绘制聚类树,sizeGrWindow设置绘图窗口大小
47 | # sizeGrWindow(16,9)
48 | pdf(file = "./plot/sampleClustering.pdf", width = 12, height = 9);
49 | # 设置文字大小
50 | par(cex = 0.5);
51 | # 设置图像边距c(bottom, left, top, right)
52 | # par(mar = c(0,4,2,0))
53 | # 画图 main标题,sub子标题,xlab x轴标题,cex.lab标题字体大小,cex.axis坐标轴刻度大小,cex.main主标题字体
54 | plot(sampleTree, main = "Sample clustering to detect outliers", sub="", xlab="", cex.lab = 1.5, cex.axis = 1.5, cex.main = 2)
55 | dev.off()
56 |
57 | # Plot a line to show the cut
58 | # 设置文字大小
59 | par(cex = 0.5);
60 | plot(sampleTree, main = "Sample clustering to detect outliers", sub="", xlab="", cex.lab = 1.5, cex.axis = 1.5, cex.main = 2)
61 | # 在上图上画红线
62 | abline(h = 15, col = "red");
63 | # Determine cluster under the line
64 | # 剪枝算法,cutHeight 修剪树枝的高度 minSize集群最小数
65 | clust = cutreeStatic(sampleTree, cutHeight = 15, minSize = 10)
66 | # 剪枝结果
67 | table(clust)
68 | # clust 1 contains the samples we want to keep
69 | keepSamples = (clust==1)
70 | # 符合要求的数据
71 | datExpr = datExpr0[keepSamples, ]
72 | # 提取行
73 | nSamples = nrow(datExpr)
74 | # 提取列
75 | nGenes = ncol(datExpr)
76 |
77 | traitData = read.csv("./data/ClinicalTraits.csv");
78 | dim(traitData)
79 | #names(traitData)
80 | # remove columns that hold information we do not need.
81 | # 删除不需要的列
82 | allTraits = traitData[, -c(31, 16)];
83 | allTraits = allTraits[, c(2, 11:36) ];
84 | dim(allTraits)
85 | head(allTraits)
86 | # names(allTraits)
87 |
88 | # 形成一个类似于表达数据的数据框架,以保存临床特征
89 | # 提取行名
90 | femaleSamples = rownames(datExpr)
91 | # 数据匹配 返回匹配行
92 | traitRows = match(femaleSamples, allTraits$Mice);
93 | # 提取指定要求行
94 | datTraits = allTraits[traitRows, -1];
95 | # 提取行名
96 | rownames(datTraits) = allTraits[traitRows, 1];
97 | # 垃圾回收
98 | collectGarbage();
99 |
100 | # Re-cluster samples
101 | # 画聚类图
102 | sampleTree2 = hclust(dist(datExpr), method = "average")
103 | # Convert traits to a color representation: white means low, red means high, grey means missing entry
104 | # 画表型的热图
105 | # 将特征转换为颜色表示:白色表示低,红色表示高,灰色表示缺少条目
106 | # 如果signed为true 以绿色开头代表最大负值,以白色开头代表零附近的值,然后变为红色代表正值
107 | traitColors = numbers2colors(datTraits, signed =FALSE);
108 | # Plot the sample dendrogram and the colors underneath.
109 | # 绘制出树状图和下面的颜色
110 | plotDendroAndColors(sampleTree2, traitColors,groupLabels = names(datTraits),main = "Sample dendrogram and trait heatmap")
111 |
112 | # Choose a set of soft-thresholding powers
113 | # 给出候选的β值,c(1:10)表示1到10;seq(from = 12, to=20, by=2)表示从12开始间隔两个数到20
114 | powers = c(c(1:10), seq(from = 12, to=20, by=2))
115 | powers
116 | # Call the network topology analysis function 调用网络拓扑分析函数
117 | # verbose表示输出结果详细程度
118 | sft = pickSoftThreshold(datExpr, powerVector = powers, verbose = 0);
119 |
120 | # sft这中保存了每个powers值计算出来的网络特征,其中powerEstimate就是最佳power值,fitIndices保存了每个power对应的网络的特征。
121 | str(sft)
122 |
123 | # Plot the results 结果绘图
124 | # 设置窗格大小
125 | #sizeGrWindow(9, 5)
126 | # 设置图的显示一行两列
127 | # par(mfrow = c(1,2));
128 | cex1 = 0.9;
129 | # Scale-free topology fit index as a function of the soft-thresholding power
130 | # 生成阈值和网络的特征之间的关系函数
131 | plot(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
132 | xlab="Soft Threshold (power)",ylab="Scale Free Topology Model Fit,signed R^2",type="n",
133 | main = paste("Scale independence"))
134 | text(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
135 | labels=powers,cex=cex1,col="red");
136 | # this line corresponds to using an R^2 cut-off of h
137 | abline(h=0.90,col="red")
138 |
139 | # sft$fitIndices 保存了每个power构建的相关性网络中的连接度的统计值,k就是连接度值,每个power值提供了max, median, max3种连接度的统计量
140 | # 对连接度的均值进行可视化
141 | # Mean connectivity as a function of the soft-thresholding power
142 | plot(sft$fitIndices[,1], sft$fitIndices[,5],
143 | xlab="Soft Threshold (power)",ylab="Mean Connectivity", type="n",
144 | main = paste("Mean connectivity"))
145 | text(sft$fitIndices[,1], sft$fitIndices[,5], labels=powers, cex=cex1,col="red")
146 |
147 | # datExpr表达数据,TOMType拓扑重叠矩阵计算方式,minModuleSize用于模块检测的最小模块尺寸,
148 | # reassignThreshold 是否在模块之间重新分配基因的p值比率阈值,mergeCutHeight 树状图切割高度
149 | # numericLabels 返回的模块应该用颜色(FALSE)还是数字(TRUE)标记,pamRespectsDendro树状图相关参数
150 | # saveTOMs 字符串的向量,saveTOMFileBase 包含包含共识拓扑重叠文件的文件名库的字符串
151 | net = blockwiseModules(datExpr, power = sft$powerEstimate,TOMType = "unsigned", minModuleSize = 30,reassignThreshold = 0,
152 | mergeCutHeight = 0.25,numericLabels = TRUE, pamRespectsDendro = FALSE,saveTOMs = TRUE,
153 | saveTOMFileBase = "femaleMouseTOM",verbose = 3)
154 |
155 | table(net$colors)
156 |
157 | # open a graphics window
158 | # sizeGrWindow(12, 9)
159 | # Convert labels to colors for plotting
160 | # 将标签转化为绘图颜色
161 | mergedColors = labels2colors(net$colors)
162 | # Plot the dendrogram and the module colors underneath
163 | # 绘制树状图和下面的模块颜色
164 | # dendroLabels树状图标签。设置为FALSE完全禁用树状图标签;设置为NULL使用的行标签datExpr
165 | # addGuide是否应在树状图中添加垂直的“指导线”?线条使识别单个样本的颜色代码更加容易。
166 | plotDendroAndColors(net$dendrograms[[1]], mergedColors[net$blockGenes[[1]]],"Module colors",
167 | dendroLabels = FALSE, hang = 0.03,addGuide = TRUE, guideHang = 0.05)
168 |
169 | moduleLabels = net$colors
170 | moduleColors = labels2colors(net$colors)
171 | MEs = net$MEs;
172 | geneTree = net$dendrograms[[1]];
173 | save(MEs, moduleLabels, moduleColors, geneTree,
174 | file = "FemaleLiver-02-networkConstruction-auto.RData")
175 |
176 | # Define numbers of genes and samples
177 | # 获得基因数和样本数
178 | nGenes = ncol(datExpr);
179 | nSamples = nrow(datExpr);
180 |
181 | # Recalculate MEs with color labels
182 | # 用彩色标签重新计算MEs
183 | # 在给定的单个数据集中计算模块的模块本征基因
184 | MEs0 = moduleEigengenes(datExpr, moduleColors)$eigengenes
185 | # 对给定的(特征)向量进行重新排序,以使相似的向量(通过相关性度量)彼此相邻
186 | MEs = orderMEs(MEs0)
187 |
188 | # 计算module的ME值与表型的相关系数
189 | moduleTraitCor = cor(MEs, datTraits, use = "p");
190 | moduleTraitPvalue = corPvalueStudent(moduleTraitCor, nSamples);
191 |
192 | names(MEs)
193 |
194 | # sizeGrWindow(10,6)
195 | # 显示相关性及其p值
196 | textMatrix = paste(signif(moduleTraitCor, 2), "\n(",signif(moduleTraitPvalue, 1), ")", sep = "");
197 | dim(textMatrix) = dim(moduleTraitCor)
198 | par(mar = c(6, 8.5, 3, 3));
199 | # Display the correlation values within a heatmap plot\
200 | # ySymbols 当ylabels使用时所使用的其他标签; colorLabels 应该使用颜色标签吗
201 | # colors 颜色; textMatrix 单元格名字
202 | labeledHeatmap(Matrix = moduleTraitCor,xLabels = names(datTraits),yLabels = names(MEs),ySymbols = names(MEs),
203 | colorLabels = FALSE,colors = greenWhiteRed(50),textMatrix = textMatrix,setStdMargins = FALSE,
204 | cex.text = 0.4,zlim = c(-1,1),
205 | main = paste("Module-trait relationships"))
206 |
207 | sizeGrWindow(10,6)
208 | # Will display correlations and their p-values
209 |
210 | dim(textMatrix) = dim(moduleTraitCor)
211 | par(mar = c(6, 8.5, 3, 3));
212 | # Display the correlation values within a heatmap plot
213 | labeledHeatmap(Matrix = moduleTraitCor,
214 | xLabels = names(datTraits),
215 | yLabels = names(MEs),
216 | ySymbols = names(MEs),
217 | colorLabels = FALSE,
218 | colors = greenWhiteRed(50),
219 | textMatrix = textMatrix,
220 | setStdMargins = FALSE,
221 | cex.text = 0.5,
222 | zlim = c(-1,1),
223 | main = paste("Module-trait relationships"))
224 |
225 | # Define variable weight containing the weight column of datTrait
226 | # 定义包含数据特征权重列的变量权重
227 | weight = as.data.frame(datTraits$weight_g);
228 | names(weight) = "weight"
229 | geneModuleMembership = as.data.frame(cor(datExpr, MEs, use = "p"));
230 | # 模块的名称(颜色) substring提取文本从第3个字母开始
231 | modNames = substring(names(MEs), 3)
232 | # 基因和模块的相关系数
233 | geneModuleMembership = as.data.frame(cor(datExpr, MEs, use = "p"));
234 | MMPvalue = as.data.frame(corPvalueStudent(as.matrix(geneModuleMembership), nSamples));
235 | names(geneModuleMembership) = paste("MM", modNames, sep="");
236 | names(MMPvalue) = paste("p.MM", modNames, sep="");
237 |
238 | #gene和性状的关系
239 | geneTraitSignificance = as.data.frame(cor(datExpr, weight, use = "p"));
240 | GSPvalue = as.data.frame(corPvalueStudent(as.matrix(geneTraitSignificance), nSamples));
241 | names(geneTraitSignificance) = paste("GS.", names(weight), sep="");
242 | names(GSPvalue) = paste("p.GS.", names(weight), sep="");
243 |
244 | # 模型颜色
245 | module = "brown"
246 | # 匹配列
247 | column = match(module, modNames);
248 | moduleGenes = moduleColors==module;
249 | #sizeGrWindow(7, 7);
250 | par(mfrow = c(1,1));
251 | # 画散点图
252 | verboseScatterplot(abs(geneModuleMembership[moduleGenes, column]),
253 | abs(geneTraitSignificance[moduleGenes, 1]),
254 | xlab = paste("Module Membership in", module, "module"),
255 | ylab = "Gene significance for body weight",
256 | main = paste("Module membership vs. gene significance\n"),
257 | cex.main = 1.2, cex.lab = 1.2, cex.axis = 1.2, col = module)
258 |
259 | # 提取表带数据样本名称
260 | # names(datExpr);
261 | # 指定颜色数据名称
262 | # names(datExpr)[moduleColors=="brown"]
263 |
264 | # 基因注释数据
265 | annot = read.csv(file = "./data/GeneAnnotation.csv");
266 | dim(annot)
267 | names(annot)
268 | probes = names(datExpr)
269 | probes2annot = match(probes, annot$substanceBXH)
270 | # The following is the number or probes without annotation:
271 | sum(is.na(probes2annot))
272 |
273 | # Create the starting data frame
274 | geneInfo0 = data.frame(substanceBXH = probes,
275 | geneSymbol = annot$gene_symbol[probes2annot],
276 | LocusLinkID = annot$LocusLinkID[probes2annot],
277 | moduleColor = moduleColors,
278 | geneTraitSignificance,
279 | GSPvalue)
280 | # Order modules by their significance for weight
281 | modOrder = order(-abs(cor(MEs, weight, use = "p")));
282 | # Add module membership information in the chosen order
283 | for (mod in 1:ncol(geneModuleMembership))
284 | {
285 | oldNames = names(geneInfo0)
286 | geneInfo0 = data.frame(geneInfo0, geneModuleMembership[, modOrder[mod]],
287 | MMPvalue[, modOrder[mod]]);
288 | names(geneInfo0) = c(oldNames, paste("MM.", modNames[modOrder[mod]], sep=""),
289 | paste("p.MM.", modNames[modOrder[mod]], sep=""))
290 | }
291 | # Order the genes in the geneInfo variable first by module color, then by geneTraitSignificance
292 | geneOrder = order(geneInfo0$moduleColor, -abs(geneInfo0$GS.weight));
293 | geneInfo = geneInfo0[geneOrder, ]
294 | write.csv(geneInfo, file = "geneInfo.csv")
295 |
296 | # Calculate topological overlap anew: this could be done more efficiently by saving the TOM
297 | # calculated during module detection, but let us do it again here.
298 | # 重新计算拓扑重叠:通过保存TOM可以更有效地完成此操作
299 | # 是在模块检测期间计算的,但让我们在这里再次进行。
300 | dissTOM = 1-TOMsimilarityFromExpr(datExpr, power = 6);
301 | # Transform dissTOM with a power to make moderately strong connections more visible in the heatmap
302 | # 变换dissTOM
303 | plotTOM = dissTOM^7;
304 | # Set diagonal to NA for a nicer plot
305 | diag(plotTOM) = NA;
306 | # Call the plot function
307 | # sizeGrWindow(9,9)
308 | # 基因的聚类树聚类时的距离为1-TOM值结合基因间的距离,即1-TOM值,用热图展示
309 | # TOMplot(plotTOM, geneTree, moduleColors, main = "Network heatmap plot, all genes")
310 |
311 | nSelect = 400
312 | # For reproducibility, we set the random seed
313 | set.seed(10);
314 | select = sample(nGenes, size = nSelect);
315 | selectTOM = dissTOM[select, select];
316 | # There’s no simple way of restricting a clustering tree to a subset of genes, so we must re-cluster.
317 | # 重新画聚类图
318 | selectTree = hclust(as.dist(selectTOM), method = "average")
319 | selectColors = moduleColors[select];
320 | # Open a graphical window
321 | # sizeGrWindow(9,9)
322 | # Taking the dissimilarity to a power, say 10, makes the plot more informative by effectively changing
323 | # the color palette; setting the diagonal to NA also improves the clarity of the plot
324 | plotDiss = selectTOM^7;
325 | diag(plotDiss) = NA;
326 | TOMplot(plotDiss, selectTree, selectColors, main = "Network heatmap plot, selected genes")
327 |
328 | # Recalculate module eigengenes
329 | # 重新计算基因特征值
330 | MEs = moduleEigengenes(datExpr, moduleColors)$eigengenes
331 | # Isolate weight from the clinical traits
332 | weight = as.data.frame(datTraits$weight_g);
333 | names(weight) = "weight"
334 | # Add the weight to existing module eigengenes
335 | MET = orderMEs(cbind(MEs, weight))
336 | # Plot the relationships among the eigengenes and the trait
337 | #sizeGrWindow(5,7.5);
338 | par(cex = 0.9)
339 | # 画树形图
340 | # marDendro给出树状图的边距设置,marHeatmap热图边距设置
341 | plotEigengeneNetworks(MET, "", marDendro = c(0,4,1,2), marHeatmap = c(3,4,1,2), cex.lab = 0.8, xLabelsAngle= 90)
342 |
343 | # Plot the dendrogram
344 | # sizeGrWindow(6,6);
345 | par(cex = 1.0)
346 | plotEigengeneNetworks(MET, "Eigengene dendrogram", marDendro = c(0,4,2,0),
347 | plotHeatmaps = FALSE)
348 | # Plot the heatmap matrix (note: this plot will overwrite the dendrogram plot)
349 | par(cex = 1.0)
350 | plotEigengeneNetworks(MET, "Eigengene adjacency heatmap", marHeatmap = c(3,4,2,2),plotDendrograms = FALSE, xLabelsAngle = 90)
351 |
--------------------------------------------------------------------------------
/ggplot2入门笔记/[R语言] ggplot2入门笔记1—ggplot2简要教程.r:
--------------------------------------------------------------------------------
1 |
2 | # 调用ggplot2库
3 | library(ggplot2)
4 | # 展示金刚石数据集
5 | head(diamonds)
6 |
7 | # if only the dataset is known. 只显示数据
8 | ggplot(diamonds)
9 |
10 | # if only X-axis is known. The Y-axis can be specified in respective geoms.
11 | # 只设定x轴,y轴数据可以在geoms中指定
12 | ggplot(diamonds, aes(x=carat))
13 |
14 | # if both X and Y axes are fixed for all layers.
15 | # 指定x轴和y轴
16 | ggplot(diamonds, aes(x=carat, y=price))
17 |
18 | # Each category of the 'cut' variable will now have a distinct color, once a geom is added.
19 | # 指定颜色类别cut
20 | ggplot(diamonds, aes(x=carat, color=cut))
21 |
22 | ggplot(diamonds, aes(x=carat), color="steelblue")
23 |
24 | ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
25 | # Adding scatterplot geom (layer1) 添加散点图
26 | geom_point() +
27 | # Adding moothing geom (layer2) 在散点图的基础上添加一条平滑的趋势曲线
28 | geom_smooth()
29 |
30 | # Same as above but specifying the aesthetics inside the geoms. 类似上面的结果
31 | ggplot(diamonds) +
32 | geom_point(aes(x=carat, y=price, color=cut)) +
33 | geom_smooth(aes(x=carat, y=price, color=cut))
34 |
35 | library(ggplot2)
36 | ggplot(diamonds) +
37 | geom_point(aes(x=carat, y=price, color=cut)) +
38 | # Remove color from geom_smooth 只画一条拟合平滑线
39 | geom_smooth(aes(x=carat, y=price))
40 |
41 | # same but simpler 类似上图同样的功能
42 | ggplot(diamonds, aes(x=carat, y=price)) +
43 | geom_point(aes(color=cut)) +
44 | geom_smooth()
45 |
46 | # Answer to the challenge 设置形状点
47 | ggplot(diamonds, aes(x=carat, y=price, color=cut, shape=color)) +
48 | geom_point()
49 |
50 | gg <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
51 | geom_point() +
52 | # add axis lables and plot title 添加标签
53 | labs(title="Scatterplot", x="Carat", y="Price")
54 | print(gg)
55 |
56 | gg1 <- gg +
57 | theme(
58 | # 设置标题大小,face="bold"字体加粗
59 | plot.title=element_text(size=30, face="bold"),
60 | axis.text.x=element_text(size=15),
61 | axis.text.y=element_text(size=15),
62 | axis.title.x=element_text(size=25),
63 | axis.title.y=element_text(size=25)) +
64 | # add title and axis text, change legend title.
65 | # 添加渐变色,并设置颜色条图例标题
66 | scale_color_discrete(name="Cut of diamonds")
67 | print(gg1) # print the plot
68 |
69 | gg1 + facet_wrap( ~cut , ncol=3)
70 |
71 | # row: color, column: cut
72 | gg1 + facet_wrap(color ~ cut)
73 |
74 | # row: color, column: cut
75 | # gg1 + facet_wrap(color ~ cut, scales="free")
76 |
77 | gg1 + facet_grid(color ~ cut)
78 |
79 | # 载入库
80 | library(ggfortify)
81 | # 查看数据
82 | AirPassengers
83 |
84 | autoplot(AirPassengers) +
85 | labs(title="AirPassengers")
86 |
87 | # Approach 1:
88 | data(economics, package="ggplot2") # init data
89 | economics <- data.frame(economics) # convert to dataframe
90 | # 展示数据
91 | head(economics)
92 |
93 | # 画图
94 | ggplot(economics) +
95 | # 画线条
96 | geom_line(aes(x=date, y=pce, color="pcs")) +
97 | geom_line(aes(x=date, y=unemploy, col="unemploy")) +
98 | # 设定颜色
99 | scale_color_discrete(name="Legend") +
100 | labs(title="Economics")
101 |
102 | # Approach 2:
103 | library(reshape2)
104 | # 融合数据
105 | df <- melt(economics[, c("date", "pce", "unemploy")], id="date")
106 | head(df)
107 |
108 | # 绘图
109 | ggplot(df) +
110 | geom_line(aes(x=date, y=value, color=variable)) +
111 | labs(title="Economics")
112 |
113 | df <- melt(economics[, c("date", "pce", "unemploy", "psavert")], id="date")
114 | ggplot(df) +
115 | geom_line(aes(x=date, y=value, color=variable)) +
116 | facet_wrap( ~ variable, scales="free")
117 |
118 | # 显示数据
119 | head(mtcars)
120 | plot1 <- ggplot(mtcars, aes(x=cyl)) +
121 | # 画柱状图
122 | geom_bar() +
123 | # Y axis derived from counts of X item
124 | labs(title="Frequency bar chart")
125 | print(plot1)
126 |
127 | df <- data.frame(var=c("a", "b", "c"), nums=c(1:3))
128 | # 显示数据
129 | df
130 | # Y axis is explicit. 'stat=identit
131 | # 显示y
132 | plot2 <- ggplot(df, aes(x=var, y=nums)) +
133 | geom_bar(stat = "identity")
134 | print(plot2)
135 |
136 | library(gridExtra)
137 | # 分配图像
138 | grid.arrange(plot1, plot2, ncol=2)
139 |
140 | df <- data.frame(var=c("a", "b", "c"), nums=c(1:3))
141 | ggplot(df, aes(x=var, y=nums)) +
142 | geom_bar(stat = "identity") +
143 | # 翻转坐标轴
144 | coord_flip() +
145 | labs(title="Coordinates are flipped")
146 |
147 | ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
148 | geom_point() +
149 | geom_smooth() +
150 | # 设置y轴范围
151 | coord_cartesian(ylim=c(0, 10000)) +
152 | labs(title="Coord_cartesian zoomed in!")
153 |
154 | ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
155 | geom_point() +
156 | geom_smooth() +
157 | # 设定范围
158 | ylim(c(0, 10000)) +
159 | labs(title="Datapoints deleted: Note the change in smoothing lines!")
160 |
161 | ggplot(diamonds, aes(x=price, y=price+runif(nrow(diamonds), 100, 10000), color=cut)) +
162 | geom_point() +
163 | geom_smooth() +
164 | coord_equal()
165 |
166 | ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
167 | geom_point() +
168 | geom_smooth() +
169 | # 更改主题
170 | theme_bw() +
171 | labs(title="bw Theme")
172 |
173 | # 无图例
174 | p1 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
175 | geom_point() +
176 | geom_smooth() +
177 | # 无图例
178 | theme(legend.position="none") +
179 | labs(title="legend.position='none'")
180 |
181 | p2 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
182 | geom_point() +
183 | geom_smooth() +
184 | # legend at top 设置图例在图形顶部
185 | theme(legend.position="top") +
186 | labs(title="legend.position='top'")
187 |
188 | p3 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) +
189 | geom_point() +
190 | geom_smooth() +
191 | labs(title="legend.position='coords inside plot'") +
192 | # legend inside the plot 设置图形位置
193 | theme(legend.justification=c(1,0), legend.position=c(1,0))
194 |
195 | # arrange统一显示图像
196 | grid.arrange(p1, p2, p3, ncol=3)
197 |
198 | ggplot(mtcars, aes(x=cyl)) +
199 | geom_bar(fill='darkgoldenrod2') +
200 | theme(panel.background = element_rect(fill = 'steelblue'),
201 | # 设置主网格线
202 | panel.grid.major = element_line(colour = "firebrick", size=3),
203 | panel.grid.minor = element_line(colour = "blue", size=1))
204 |
205 | ggplot(mtcars, aes(x=cyl)) +
206 | geom_bar(fill="firebrick") +
207 | # top, right, bottom, left
208 | # plot.background设置背景,plot.margain设置边距
209 | theme(plot.background=element_rect(fill="steelblue"), plot.margin = unit(c(2, 4, 1, 3), "cm"))
210 |
211 | library(grid)
212 | # 添加注释
213 | my_grob = grobTree(textGrob("This text is at x=0.1 and y=0.9, relative!\n Anchor point is at 0,0", x=0.1, y=0.9, hjust=0,
214 | gp=gpar(col="firebrick", fontsize=25, fontface="bold")))
215 | ggplot(mtcars, aes(x=cyl)) +
216 | geom_bar() +
217 | annotation_custom(my_grob) +
218 | labs(title="Annotation Example")
219 |
220 | plot1 <- ggplot(mtcars, aes(x=cyl)) +
221 | geom_bar()
222 | # 保存图像
223 | ggsave("myggplot.png") # saves the last plot.
224 | ggsave("myggplot.png", plot=plot1) # save a stored ggplot
225 |
--------------------------------------------------------------------------------
/ggplot2入门笔记/[R语言] ggplot2入门笔记2—通用教程ggplot2简介.r:
--------------------------------------------------------------------------------
1 |
2 | # Setup
3 | # #关闭科学记数法,如1e+06
4 | # turn off scientific notation like 1e+06
5 | options(scipen=999)
6 | library(ggplot2)
7 | # load the data 载入数据
8 | data("midwest", package = "ggplot2")
9 | # 显示数据
10 | head(midwest)
11 | # Init Ggplot 初始化图像
12 | # area and poptotal are columns in 'midwest'
13 | ggplot(midwest, aes(x=area, y=poptotal))
14 |
15 | library(ggplot2)
16 | ggplot(midwest, aes(x=area, y=poptotal)) +
17 | geom_point()
18 |
19 | g <- ggplot(midwest, aes(x=area, y=poptotal)) +
20 | geom_point() +
21 | # set se=FALSE to turnoff confidence bands
22 | # 设置se=FALSE来关闭置信区间
23 | geom_smooth(method="lm", se=TRUE)
24 | plot(g)
25 |
26 | library(ggplot2)
27 | # set se=FALSE to turnoff confidence bands
28 | # 设置se=FALSE来关闭置信区间
29 | g <- ggplot(midwest, aes(x=area, y=poptotal)) +
30 | geom_point() +
31 | geom_smooth(method="lm")
32 |
33 | # Delete the points outside the limits
34 | # deletes points 删除点
35 | g + xlim(c(0, 0.1)) + ylim(c(0, 1000000))
36 | # g + xlim(0, 0.1) + ylim(0, 1000000)
37 |
38 | library(ggplot2)
39 | g <- ggplot(midwest, aes(x=area, y=poptotal)) +
40 | geom_point() +
41 | # set se=FALSE to turnoff confidence bands
42 | geom_smooth(method="lm")
43 |
44 | # Zoom in without deleting the points outside the limits.
45 | # As a result, the line of best fit is the same as the original plot.
46 | # 放大而不删除超出限制的点。因此,最佳拟合线与原始图相同。
47 | g1 <- g + coord_cartesian(xlim=c(0,0.1), ylim=c(0, 1000000))
48 | plot(g1)
49 |
50 | library(ggplot2)
51 | # 画图
52 | # set se=FALSE to turnoff confidence bands
53 | g <- ggplot(midwest, aes(x=area, y=poptotal)) + geom_point() + geom_smooth(method="lm")
54 | # 限制范围
55 | g1 <- g + coord_cartesian(xlim=c(0,0.1), ylim=c(0, 1000000)) # zooms in
56 | # Add Title and Labels
57 | # 添加标签,标题名,小标题名,说明文字
58 | g1 + labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
59 |
60 | # 另外一种方法
61 | g1 + ggtitle("Area Vs Population", subtitle="From midwest dataset") + xlab("Area") + ylab("Population")
62 |
63 | # Full Plot call
64 | library(ggplot2)
65 | ggplot(midwest, aes(x=area, y=poptotal)) +
66 | geom_point() +
67 | geom_smooth(method="lm") +
68 | coord_cartesian(xlim=c(0,0.1), ylim=c(0, 1000000)) +
69 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
70 |
71 | library(ggplot2)
72 | # 画图
73 | ggplot(midwest, aes(x=area, y=poptotal)) +
74 | # Set static color and size for points
75 | # 设置固定颜色和尺寸
76 | geom_point(col="steelblue", size=3) +
77 | # change the color of line
78 | # 更改拟合直线颜色
79 | geom_smooth(method="lm", col="firebrick") +
80 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
81 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
82 |
83 | library(ggplot2)
84 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
85 | # Set color to vary based on state categories.
86 | # 根据状态类别将颜色设置为不同。
87 | geom_point(aes(col=state), size=3) +
88 | geom_smooth(method="lm", col="firebrick", size=2) +
89 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
90 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
91 | plot(gg)
92 |
93 | # remove legend 移除图例
94 | gg + theme(legend.position="None")
95 |
96 | # change color palette 更改调色板
97 | gg + scale_colour_brewer(palette = "Set1")
98 |
99 | library(RColorBrewer)
100 | head(brewer.pal.info, 10)
101 |
102 | library(ggplot2)
103 |
104 | # Base plot
105 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
106 | # Set color to vary based on state categories
107 | # 设置颜色
108 | geom_point(aes(col=state), size=3) +
109 | geom_smooth(method="lm", col="firebrick", size=2) +
110 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
111 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
112 |
113 | # Change breaks
114 | # 改变间距
115 | gg + scale_x_continuous(breaks=seq(0, 0.1, 0.01))
116 |
117 | library(ggplot2)
118 |
119 | # Base plot
120 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
121 | # Set color to vary based on state categories
122 | # 设置颜色
123 | geom_point(aes(col=state), size=3) +
124 | geom_smooth(method="lm", col="firebrick", size=2) +
125 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
126 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
127 |
128 | # Change breaks + label
129 | # letters字母表
130 | gg + scale_x_continuous(breaks=seq(0, 0.1, 0.01), labels = letters[1:11])
131 |
132 | library(ggplot2)
133 |
134 | # Base plot
135 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
136 | # Set color to vary based on state categories
137 | # 设置颜色
138 | geom_point(aes(col=state), size=3) +
139 | geom_smooth(method="lm", col="firebrick", size=2) +
140 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
141 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
142 |
143 | # Reverse X Axis Scale
144 | # 反转x轴
145 | gg + scale_x_reverse()
146 |
147 | library(ggplot2)
148 |
149 | # Base plot
150 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
151 | # Set color to vary based on state categories
152 | # 设置颜色
153 | geom_point(aes(col=state), size=3) +
154 | geom_smooth(method="lm", col="firebrick", size=2) +
155 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
156 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
157 |
158 | # Change Axis Texts
159 | gg +
160 | # 更改x轴
161 | scale_x_continuous(breaks=seq(0, 0.1, 0.01), labels = sprintf("%1.2f%%", seq(0, 0.1, 0.01))) +
162 | # 更改y轴
163 | scale_y_continuous(breaks=seq(0, 1000000, 200000), labels = function(x){paste0(x/1000, 'K')})
164 |
165 | library(ggplot2)
166 |
167 | # Base plot
168 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
169 | # Set color to vary based on state categories.
170 | geom_point(aes(col=state), size=3) +
171 | geom_smooth(method="lm", col="firebrick", size=2) +
172 | coord_cartesian(xlim=c(0, 0.1), ylim=c(0, 1000000)) +
173 | labs(title="Area Vs Population", subtitle="From midwest dataset", y="Population", x="Area", caption="Midwest Demographics")
174 |
175 | gg <- gg + scale_x_continuous(breaks=seq(0, 0.1, 0.01))
176 |
177 | # method 1: Using theme_set()
178 | theme_set(theme_classic())
179 | gg
180 |
181 | # method 2: Adding theme Layer itself.
182 | # 添加主题层
183 | gg + theme_bw() + labs(subtitle="BW Theme")
184 |
185 | gg + theme_classic() + labs(subtitle="Classic Theme")
186 |
--------------------------------------------------------------------------------
/ggplot2入门笔记/[R语言] ggplot2入门笔记3—通用教程如何自定义ggplot2.r:
--------------------------------------------------------------------------------
1 |
2 | # Setup
3 | options(scipen=999)
4 | library(ggplot2)
5 | data("midwest", package = "ggplot2")
6 | theme_set(theme_bw())
7 |
8 | # Add plot components --------------------------------
9 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
10 | geom_point(aes(col=state, size=popdensity)) +
11 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
12 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
13 |
14 | # Call plot ------------------------------------------
15 | plot(gg)
16 |
17 | library(ggplot2)
18 |
19 | # Base Plot 基础绘图
20 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
21 | geom_point(aes(col=state, size=popdensity)) +
22 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
23 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
24 |
25 | library(showtext)
26 | showtext.auto(enable = TRUE)
27 | # 添加字体
28 | font.add('SimSun', 'simsun.ttc')
29 |
30 | # Modify theme components
31 | # 修改主题
32 | gg + theme(
33 | # 设置标题
34 | plot.title=element_text(size=20, # 字体大小
35 | face="bold", # 字体加粗
36 | family="SimSun", # 字体类型
37 | color="tomato", # 字体颜色
38 | hjust=0.5, # 标题离左边距距离
39 | lineheight=1.2), # 线条高度
40 | # 设置子标题
41 | plot.subtitle=element_text(size=15, # 字体大小
42 | family="SimSun", # 字体类型
43 | face="bold", # 字体加粗
44 | hjust=0.5), # 标题离左边距距离
45 | # caption 注释
46 | plot.caption=element_text(size=15),
47 | # X axis title X轴标题
48 | axis.title.x=element_text(vjust=0,
49 | size=15),
50 | # Y axis title Y轴标题
51 | axis.title.y=element_text(size=15),
52 | # X axis text X轴文字
53 | axis.text.x=element_text(size=10,
54 | angle = 30,
55 | vjust=.5),
56 | # Y axis text Y轴文字
57 | axis.text.y=element_text(size=10))
58 |
59 | library(ggplot2)
60 |
61 | # Base Plot
62 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
63 | geom_point(aes(col=state, size=popdensity)) +
64 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
65 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
66 |
67 | # modify legend title
68 | # 单独调用labs修改颜色和字体
69 | gg + labs(color="State", size="Density")
70 |
71 | library(ggplot2)
72 |
73 | # Base Plot
74 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
75 | geom_point(aes(col=state, size=popdensity)) +
76 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
77 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
78 |
79 | # modify legend title
80 | # 修改legend
81 | gg <- gg + guides(color=guide_legend("State"), size=guide_legend("Density"))
82 | plot(gg)
83 |
84 | library(ggplot2)
85 |
86 | # Base Plot
87 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
88 | geom_point(aes(col=state, size=popdensity)) +
89 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
90 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
91 |
92 | # Modify Legend 修改图例
93 | # guide = FALSE turn off legend for size 关闭size的图例
94 | # scale_color_discrete(name="States") 设置离散颜色变量的图例
95 | gg + scale_color_discrete(name="States") + scale_size_continuous(name = "Density", guide = FALSE)
96 |
97 | library(ggplot2)
98 |
99 | # Base Plot
100 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
101 | geom_point(aes(col=state, size=popdensity)) +
102 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
103 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
104 |
105 | gg + scale_color_manual(name="State",
106 | # 设置标签
107 | labels = c("Illinois",
108 | "Indiana",
109 | "Michigan",
110 | "Ohio",
111 | "Wisconsin"),
112 | # 设置标签对应的颜色
113 | values = c("IL"="blue",
114 | "IN"="red",
115 | "MI"="green",
116 | "OH"="brown",
117 | "WI"="orange"))
118 |
119 | library(ggplot2)
120 |
121 | # Base Plot
122 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
123 | geom_point(aes(col=state, size=popdensity)) +
124 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
125 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
126 |
127 | # order设置位置顺序
128 | gg + guides(colour = guide_legend(order = 2), size = guide_legend(order = 1))
129 |
130 | library(ggplot2)
131 |
132 | # Base Plot
133 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
134 | geom_point(aes(col=state, size=popdensity)) +
135 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
136 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
137 |
138 | gg + theme(
139 | # 设置图例标题字体颜色和大小
140 | legend.title = element_text(size=12, color = "firebrick"),
141 | # 设置图例内容文字大小
142 | legend.text = element_text(size=10),
143 | # 设置背景色
144 | legend.key=element_rect(fill='springgreen')) +
145 | # 设置内部图例圆圈大小和间距
146 | guides(colour = guide_legend(override.aes = list(size=2, stroke=1.5)))
147 |
148 | library(ggplot2)
149 |
150 | # Base Plot
151 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
152 | geom_point(aes(col=state, size=popdensity)) +
153 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
154 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
155 |
156 | # No legend
157 | # 无图例
158 | gg + theme(legend.position="None") + labs(subtitle="No Legend")
159 |
160 | # Legend to the left 图例位置在左边
161 | gg + theme(legend.position="left") + labs(subtitle="Legend on the Left")
162 |
163 | # legend at the bottom and horizontal
164 | # 图例位于图像底部,图例水平摆放
165 | gg + theme(legend.position="bottom", legend.box = "horizontal") + labs(subtitle="Legend at Bottom")
166 |
167 | # legend at bottom-right, inside the plot
168 | # 图例位于图像内部右下角
169 | gg + theme(
170 | # 设置图像标题
171 | legend.title = element_text(size=12, color = "salmon", face="bold"),
172 | # 设置图像铰点为图内左下角
173 | legend.justification=c(1,0),
174 | # 图例位置
175 | legend.position=c(0.95, 0.05),
176 | # 图例背景
177 | legend.background = element_blank(),
178 | # 图例填充颜色
179 | legend.key = element_blank()) +
180 | labs(subtitle="Legend: Bottom-Right Inside the Plot")
181 |
182 | # legend at top-left, inside the plot
183 | # 图例位于图像内部左上角
184 | gg + theme(
185 | # 设置标题名
186 | legend.title = element_text(size=12, color = "salmon", face="bold"),
187 | # 设置图像铰点为图内右上角
188 | legend.justification=c(0,1),
189 | legend.position=c(0.05, 0.95),
190 | legend.background = element_blank(),
191 | legend.key = element_blank()) +
192 | labs(subtitle="Legend: Top-Left Inside the Plot")
193 |
194 | library(ggplot2)
195 |
196 | # Filter required rows.
197 | # 获取数据
198 | midwest_sub <- midwest[midwest$poptotal > 300000, ]
199 | midwest_sub$large_county <- ifelse(midwest_sub$poptotal > 300000, midwest_sub$county, "")
200 |
201 | # Base Plot
202 | # 基础绘图
203 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
204 | geom_point(aes(col=state, size=popdensity)) +
205 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
206 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
207 |
208 | # Plot text and label
209 | # 添加标签
210 | gg + geom_text(aes(label=large_county), size=2, data=midwest_sub) +
211 | # 小标题
212 | labs(subtitle="With ggplot2::geom_text") +
213 | # 无图例
214 | theme(legend.position = "None")
215 |
216 | # 添加标签和透明度
217 | gg + geom_label(aes(label=large_county), size=2, data=midwest_sub, alpha=0.25) +
218 | labs(subtitle="With ggplot2::geom_label") +
219 | theme(legend.position = "None")
220 |
221 | library(ggrepel)
222 | # 调用ggrepel库添加标签
223 | gg + geom_text_repel(aes(label=large_county), size=2, data=midwest_sub) +
224 | labs(subtitle="With ggrepel::geom_text_repel") + theme(legend.position = "None")
225 |
226 | gg + geom_label_repel(aes(label=large_county), size=2, data=midwest_sub) +
227 | labs(subtitle="With ggrepel::geom_label_repel") + theme(legend.position = "None") # label
228 |
229 | library(ggplot2)
230 |
231 | # Base Plot
232 | # 基础绘图
233 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
234 | geom_point(aes(col=state, size=popdensity)) +
235 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
236 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest")
237 |
238 | # Define and add annotation
239 | library(grid)
240 | #文本
241 | my_text <- "This text is at x=0.7 and y=0.8!"
242 | #my_grob = grid.text(my_text, x=0.7 and y=0.8, gp=gpar(col="firebrick", fontsize=14, fontface="bold"))
243 | #gg + annotation_custom(my_grob)
244 |
245 | library(ggplot2)
246 |
247 | # Base Plot
248 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
249 | geom_point(aes(col=state, size=popdensity)) +
250 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
251 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest", subtitle="X and Y axis Flipped") +
252 | theme(legend.position = "None")
253 |
254 | # Flip the X and Y axis -------------------------------------------------
255 | # 翻转X和Y轴
256 | gg + coord_flip()
257 |
258 | library(ggplot2)
259 |
260 | # Base Plot
261 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
262 | geom_point(aes(col=state, size=popdensity)) +
263 | geom_smooth(method="loess", se=F) + xlim(c(0, 0.1)) + ylim(c(0, 500000)) +
264 | labs(title="Area Vs Population", y="Population", x="Area", caption="Source: midwest", subtitle="Axis Scales Reversed") +
265 | theme(legend.position = "None")
266 |
267 | # Reverse the X and Y Axis ---------------------------
268 | # 反转X轴和Y轴
269 | gg + scale_x_reverse() + scale_y_reverse()
270 |
271 | library(ggplot2)
272 | # 载入数据
273 | data(mpg, package="ggplot2")
274 | # 展示数据
275 | head(mpg)
276 | # 画图
277 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
278 | geom_point() +
279 | labs(title="hwy vs displ", caption = "Source: mpg") +
280 | geom_smooth(method="lm", se=FALSE) +
281 | theme_bw()
282 | plot(g)
283 |
284 | library(ggplot2)
285 |
286 | # Base Plot
287 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
288 | geom_point() +
289 | geom_smooth(method="lm", se=FALSE) +
290 | theme_bw()
291 |
292 | # Facet wrap with common scales
293 | # 分面
294 | # 以为class为列,分为3行
295 | g + facet_wrap( ~ class, nrow=3) +
296 | # 共享标尺
297 | labs(title="hwy vs displ", caption = "Source: mpg", subtitle="Ggplot2 - Faceting - Multiple plots in one figure")
298 |
299 | # Facet wrap with free scales
300 | # 以列作为分块
301 | g + facet_wrap( ~ class, scales = "free") +
302 | labs(title="hwy vs displ", caption = "Source: mpg", subtitle="Ggplot2 - Faceting - Multiple plots in one figure with free scales")
303 |
304 | library(ggplot2)
305 |
306 | # Base Plot
307 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
308 | geom_point() +
309 | labs(title="hwy vs displ", caption = "Source: mpg", subtitle="Ggplot2 - Faceting - Multiple plots in one figure") +
310 | geom_smooth(method="lm", se=FALSE) +
311 | theme_bw()
312 | # Add Facet Grid
313 | # manufacturer in rows and class in columns
314 | # 添加分面,列为class,行为manufacturer
315 | g1 <- g + facet_grid(manufacturer ~ class)
316 | plot(g1)
317 |
318 | library(ggplot2)
319 |
320 | # Base Plot
321 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
322 | geom_point() +
323 | geom_smooth(method="lm", se=FALSE) +
324 | labs(title="hwy vs displ", caption = "Source: mpg", subtitle="Ggplot2 - Facet Grid - Multiple plots in one figure") +
325 | theme_bw()
326 |
327 | # Add Facet Grid
328 | # cyl in rows and class in columns.
329 | g2 <- g + facet_grid(cyl ~ class)
330 | plot(g2)
331 |
332 | # Draw Multiple plots in same figure.
333 | library(gridExtra)
334 | gridExtra::grid.arrange(g1, g2, ncol=2)
335 |
336 | library(ggplot2)
337 |
338 | # Base Plot
339 | # 基础绘图
340 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
341 | geom_point() +
342 | geom_smooth(method="lm", se=FALSE) +
343 | theme_bw()
344 | g
345 |
346 | # Change Plot Background elements
347 | # 改变图像背景
348 | g + theme(
349 | # 设置背景色
350 | panel.background = element_rect(fill = 'khaki'),
351 | # 设置图像网格主间隔
352 | panel.grid.major = element_line(colour = "burlywood", size=1.5),
353 | # 设置图像网格次间隔
354 | panel.grid.minor = element_line(colour = "tomato", size=.25, linetype = "dashed"),
355 | # 设置图像边缘
356 | panel.border = element_blank(),
357 | # x轴颜色宽度
358 | axis.line.x = element_line(colour = "darkorange", size=1.5, lineend = "butt"),
359 | # y轴颜色宽度
360 | axis.line.y = element_line(colour = "darkorange", size=1.5)) +
361 | labs(title="Modified Background", subtitle="How to Change Major and Minor grid, Axis Lines, No Border")
362 |
363 | # Change Plot Margins
364 | g + theme(plot.background=element_rect(fill="salmon"),
365 | # top, right, bottom, left
366 | # 设置图像边缘
367 | plot.margin = unit(c(2, 2, 1, 1), "cm")) +
368 | labs(title="Modified Background", subtitle="How to Change Plot Margin")
369 |
370 | library(ggplot2)
371 |
372 | # Base Plot
373 | # 基础绘图
374 | g <- ggplot(mpg, aes(x=displ, y=hwy)) +
375 | geom_point() +
376 | geom_smooth(method="lm", se=FALSE) +
377 | theme_bw()
378 |
379 | g + theme(
380 | # 主网格空白
381 | panel.grid.major = element_blank(),
382 | # 次网格空白
383 | panel.grid.minor = element_blank(),
384 | # 边缘空白
385 | panel.border = element_blank(),
386 | # 标题空白
387 | axis.title = element_blank(),
388 | # 轴文字空白
389 | axis.text = element_blank(),
390 | axis.ticks = element_blank()) +
391 |
392 | labs(title="Modified Background", subtitle="How to remove major and minor axis grid, border, axis title, text and ticks")
393 |
--------------------------------------------------------------------------------
/ggplot2入门笔记/[R语言] ggplot2入门笔记4—前50个ggplot2可视化效果.r:
--------------------------------------------------------------------------------
1 |
2 | # install.packages("ggplot2")
3 | # load package and data
4 |
5 | # turn-off scientific notation like 1e+48 关闭科学计数法
6 | options(scipen=999)
7 | library(ggplot2)
8 | # pre-set the bw theme. 设置主题
9 | theme_set(theme_bw())
10 | # 调用数据集
11 | data("midwest", package = "ggplot2")
12 | # midwest <- read.csv("http://goo.gl/G1K41K") # bkup data source
13 | head(midwest)
14 |
15 | # Scatterplot
16 | # 画散点图
17 | gg <- ggplot(midwest, aes(x=area, y=poptotal)) +
18 | geom_point(aes(col=state, size=popdensity)) +
19 | # 画平滑曲线
20 | geom_smooth(method="loess", se=F) +
21 | xlim(c(0, 0.1)) +
22 | ylim(c(0, 500000)) +
23 | labs(subtitle="Area Vs Population",
24 | y="Population",
25 | x="Area",
26 | title="Scatterplot",
27 | caption = "Source: midwest")
28 |
29 | plot(gg)
30 |
31 | # install 'ggalt' pkg
32 | # devtools::install_github("hrbrmstr/ggalt")
33 | options(scipen = 999)
34 | library(ggplot2)
35 | library(ggalt)
36 |
37 | # 筛选符合要求的点
38 | midwest_select <- midwest[midwest$poptotal > 350000 &
39 | midwest$poptotal <= 500000 &
40 | midwest$area > 0.01 &
41 | midwest$area < 0.1, ]
42 |
43 | head(midwest_select)
44 |
45 | # Plot
46 | ggplot(midwest, aes(x=area, y=poptotal)) +
47 | geom_point(aes(col=state, size=popdensity)) + # draw points
48 | geom_smooth(method="loess", se=F) +
49 | xlim(c(0, 0.1)) +
50 | # draw smoothing line
51 | ylim(c(0, 500000)) +
52 | # encircle 画边界
53 | geom_encircle(aes(x=area, y=poptotal),
54 | data=midwest_select,
55 | color="red",
56 | size=2,
57 | expand=0.08) + #
58 | labs(subtitle="Area Vs Population",
59 | y="Population",
60 | x="Area",
61 | title="Scatterplot + Encircle",
62 | caption="Source: midwest")
63 |
64 | # load package and data
65 | library(ggplot2)
66 | data(mpg, package="ggplot2")
67 | # pre-set the bw theme. 提前设置主题
68 | theme_set(theme_bw())
69 |
70 | g <- ggplot(mpg, aes(cty, hwy))
71 |
72 | # Scatterplot
73 | g + geom_point() +
74 | geom_smooth(method="lm", se=F) +
75 | labs(subtitle="mpg: city vs highway mileage",
76 | y="hwy",
77 | x="cty",
78 | title="Scatterplot with overlapping points",
79 | caption="Source: midwest")
80 |
81 | dim(mpg)
82 |
83 | # load package and data
84 | library(ggplot2)
85 | data(mpg, package="ggplot2")
86 | # mpg <- read.csv("http://goo.gl/uEeRGu")
87 |
88 | # Scatterplot
89 | # pre-set the bw theme.
90 | theme_set(theme_bw())
91 | g <- ggplot(mpg, aes(cty, hwy))
92 | # 画抖动图
93 | g + geom_jitter(width = 0.5, size=1) +
94 | labs(subtitle="mpg: city vs highway mileage",
95 | y="hwy",
96 | x="cty",
97 | title="Jittered Points")
98 |
99 | # load package and data
100 | library(ggplot2)
101 | data(mpg, package="ggplot2")
102 | # mpg <- read.csv("http://goo.gl/uEeRGu")
103 |
104 | # Scatterplot
105 | # pre-set the bw theme.
106 | theme_set(theme_bw())
107 | g <- ggplot(mpg, aes(cty, hwy))
108 | # 画计数图,show.legent设置图例
109 | g + geom_count(col="tomato3", show.legend=F) +
110 | labs(subtitle="mpg: city vs highway mileage",
111 | y="hwy",
112 | x="cty",
113 | title="Counts Plot")
114 |
115 |
116 | # load package and data
117 | library(ggplot2)
118 | data(mpg, package="ggplot2")
119 | # mpg <- read.csv("http://goo.gl/uEeRGu")
120 |
121 | mpg_select <- mpg[mpg$manufacturer %in% c("audi", "ford", "honda", "hyundai"), ]
122 |
123 | # Scatterplot
124 | theme_set(theme_bw()) # pre-set the bw theme.
125 | g <- ggplot(mpg_select, aes(displ, cty)) +
126 | labs(subtitle="mpg: Displacement vs City Mileage",
127 | title="Bubble chart")
128 |
129 | g + geom_jitter(aes(col=manufacturer, size=hwy)) +
130 | # 画平滑曲线
131 | geom_smooth(aes(col=manufacturer), method="lm", se=F)
132 |
133 | # load package and data
134 | library(ggplot2)
135 | library(ggExtra)
136 | data(mpg, package="ggplot2")
137 | # mpg <- read.csv("http://goo.gl/uEeRGu")
138 |
139 | # Scatterplot
140 | theme_set(theme_bw()) # pre-set the bw theme.
141 | mpg_select <- mpg[mpg$hwy >= 35 & mpg$cty > 27, ]
142 | g <- ggplot(mpg, aes(cty, hwy)) +
143 | # 绘制计数图
144 | geom_count() +
145 | # se是否绘制置信区间
146 | geom_smooth(method="lm", se=FALSE)
147 |
148 | plot(g)
149 |
150 | # 绘制边际直方图
151 | ggMarginal(g, type = "histogram", fill="transparent")
152 |
153 | # 绘制边际箱形图
154 | ggMarginal(g, type = "boxplot", fill="transparent")
155 |
156 | # 绘制边际核密度图
157 | ggMarginal(g, type = "density", fill="transparent")
158 |
159 | # devtools::install_github("kassambara/ggcorrplot")
160 | library(ggplot2)
161 | library(ggcorrplot)
162 |
163 | # Correlation matrix
164 | data(mtcars)
165 | # 计算相关性结果
166 | corr <- round(cor(mtcars), 1)
167 | corr
168 |
169 | # Plot
170 | # 画相关系图
171 | ggcorrplot(corr,
172 | # hc.order是否对相关性矩阵排序
173 | hc.order = FALSE,
174 | # 下三角形显示
175 | type = "lower",
176 | # 是否显示图中数字
177 | lab = TRUE,
178 | # 图中点的大小
179 | lab_size = 3,
180 | # 点的形状square or circle
181 | method="circle",
182 | # 颜色
183 | colors = c("tomato2", "white", "springgreen3"),
184 | title="Correlogram of mtcars",
185 | ggtheme=theme_bw)
186 |
187 | library(ggplot2)
188 | theme_set(theme_bw())
189 |
190 | # Data Prep
191 | # load data 加载数据
192 | data("mtcars")
193 | mtcars$`car name` <- rownames(mtcars) # create new column for car names
194 | mtcars$mpg_z <- round((mtcars$mpg - mean(mtcars$mpg))/sd(mtcars$mpg), 2) # compute normalized mpg
195 | # # above / below avg flag
196 | mtcars$mpg_type <- ifelse(mtcars$mpg_z < 0, "below", "above")
197 | mtcars <- mtcars[order(mtcars$mpg_z), ] # sort
198 | mtcars$`car name` <- factor(mtcars$`car name`, levels = mtcars$`car name`) # convert to factor to retain sorted order in plot.
199 |
200 |
201 | # Diverging Barcharts
202 | ggplot(mtcars, aes(x=`car name`, y=mpg_z, label=mpg_z)) +
203 | #条形图
204 | geom_bar(stat='identity', aes(fill=mpg_type), width=.5) +
205 | # 自定义颜色
206 | scale_fill_manual(name="Mileage",
207 | labels = c("Above Average", "Below Average"),
208 | values = c("above"="#00ba38", "below"="#f8766d")) +
209 | labs(subtitle="Normalised mileage from 'mtcars'",
210 | title= "Diverging Bars") +
211 | # 翻转坐标轴
212 | coord_flip()
213 |
214 | library(ggplot2)
215 | theme_set(theme_bw())
216 |
217 | ggplot(mtcars, aes(x=`car name`, y=mpg_z, label=mpg_z)) +
218 | geom_point(stat='identity', fill="black", size=6) +
219 | # 绘制点x,y到xend,yend的直线
220 | geom_segment(aes(y = 0,
221 | x = `car name`,
222 | yend = mpg_z,
223 | xend = `car name`),
224 | color = "black") +
225 | geom_text(color="white", size=2) +
226 | labs(title="Diverging Lollipop Chart",
227 | subtitle="Normalized mileage from 'mtcars': Lollipop") +
228 | ylim(-2.5, 2.5) +
229 | coord_flip()
230 |
231 | library(ggplot2)
232 | theme_set(theme_bw())
233 |
234 | # Plot
235 | ggplot(mtcars, aes(x=`car name`, y=mpg_z, label=mpg_z)) +
236 | geom_point(stat='identity', aes(col=mpg_type), size=6) +
237 | scale_color_manual(name="Mileage",
238 | labels = c("Above Average", "Below Average"),
239 | values = c("above"="#00ba38", "below"="#f8766d")) +
240 | geom_text(color="white", size=2) +
241 | labs(title="Diverging Dot Plot",
242 | subtitle="Normalized mileage from 'mtcars': Dotplot") +
243 | ylim(-2.5, 2.5) +
244 | coord_flip()
245 |
246 | library(ggplot2)
247 | library(quantmod)
248 | data("economics", package = "ggplot2")
249 |
250 | # Compute % Returns
251 | economics$returns_perc <- c(0, diff(economics$psavert)/economics$psavert[-length(economics$psavert)])
252 |
253 | # Create break points and labels for axis ticks
254 | brks <- economics$date[seq(1, length(economics$date), 12)]
255 | lbls <- lubridate::year(economics$date[seq(1, length(economics$date), 12)])
256 |
257 | # Plot
258 | ggplot(economics[1:100, ], aes(date, returns_perc)) +
259 | # 画面积图
260 | geom_area() +
261 | scale_x_date(breaks=brks, labels=lbls) +
262 | theme(axis.text.x = element_text(angle=90)) +
263 | labs(title="Area Chart",
264 | subtitle = "Perc Returns for Personal Savings",
265 | y="% Returns for Personal savings",
266 | caption="Source: economics")
267 |
268 | # Prepare data: group mean city mileage by manufacturer.
269 | cty_mpg <- aggregate(mpg$cty, by=list(mpg$manufacturer), FUN=mean) # aggregate
270 | colnames(cty_mpg) <- c("make", "mileage") # change column names
271 | cty_mpg <- cty_mpg[order(cty_mpg$mileage), ] # sort
272 | cty_mpg$make <- factor(cty_mpg$make, levels = cty_mpg$make) # to retain the order in plot.
273 | head(cty_mpg, 4)
274 |
275 | library(ggplot2)
276 | theme_set(theme_bw())
277 |
278 | # Draw plot
279 | ggplot(cty_mpg, aes(x=make, y=mileage)) +
280 | geom_bar(stat="identity", width=.5, fill="tomato3") +
281 | labs(title="Ordered Bar Chart",
282 | subtitle="Make Vs Avg. Mileage",
283 | caption="source: mpg") +
284 | theme(axis.text.x = element_text(angle=65, vjust=0.6))
285 |
286 | library(ggplot2)
287 | theme_set(theme_bw())
288 |
289 | # Plot
290 | ggplot(cty_mpg, aes(x=make, y=mileage)) +
291 | geom_point(size=3) +
292 | geom_segment(aes(x=make,
293 | xend=make,
294 | y=0,
295 | yend=mileage)) +
296 | labs(title="Lollipop Chart",
297 | subtitle="Make Vs Avg. Mileage",
298 | caption="source: mpg") +
299 | theme(axis.text.x = element_text(angle=65, vjust=0.6))
300 |
301 | library(ggplot2)
302 | library(scales)
303 | theme_set(theme_classic())
304 |
305 | # Plot
306 | ggplot(cty_mpg, aes(x=make, y=mileage)) +
307 | geom_point(col="tomato2", size=3) + # Draw points
308 | geom_segment(aes(x=make,
309 | xend=make,
310 | y=min(mileage),
311 | yend=max(mileage)),
312 | linetype="dashed",
313 | size=0.1) + # Draw dashed lines
314 | labs(title="Dot Plot",
315 | subtitle="Make Vs Avg. Mileage",
316 | caption="source: mpg") +
317 | coord_flip()
318 |
319 | library(ggplot2)
320 | library(scales)
321 | theme_set(theme_classic())
322 |
323 | # prep data
324 | df <- read.csv("https://raw.githubusercontent.com/selva86/datasets/master/gdppercap.csv")
325 | colnames(df) <- c("continent", "1952", "1957")
326 | left_label <- paste(df$continent, round(df$`1952`),sep=", ")
327 | right_label <- paste(df$continent, round(df$`1957`),sep=", ")
328 | df$class <- ifelse((df$`1957` - df$`1952`) < 0, "red", "green")
329 |
330 | # Plot
331 | p <- ggplot(df) + geom_segment(aes(x=1, xend=2, y=`1952`, yend=`1957`, col=class), size=.75, show.legend=F) +
332 | geom_vline(xintercept=1, linetype="dashed", size=.1) +
333 | geom_vline(xintercept=2, linetype="dashed", size=.1) +
334 | scale_color_manual(labels = c("Up", "Down"),
335 | values = c("green"="#00ba38", "red"="#f8766d")) + # color of lines
336 | labs(x="", y="Mean GdpPerCap") + # Axis labels
337 | xlim(.5, 2.5) + ylim(0,(1.1*(max(df$`1952`, df$`1957`)))) # X and Y axis limits
338 |
339 | # Add texts
340 | p <- p + geom_text(label=left_label, y=df$`1952`, x=rep(1, NROW(df)), hjust=1.1, size=3.5)
341 | p <- p + geom_text(label=right_label, y=df$`1957`, x=rep(2, NROW(df)), hjust=-0.1, size=3.5)
342 | p <- p + geom_text(label="Time 1", x=1, y=1.1*(max(df$`1952`, df$`1957`)), hjust=1.2, size=5) # title
343 | p <- p + geom_text(label="Time 2", x=2, y=1.1*(max(df$`1952`, df$`1957`)), hjust=-0.1, size=5) # title
344 |
345 | # Minify theme
346 | p + theme(panel.background = element_blank(),
347 | panel.grid = element_blank(),
348 | axis.ticks = element_blank(),
349 | axis.text.x = element_blank(),
350 | panel.border = element_blank(),
351 | plot.margin = unit(c(1,2,1,2), "cm"))
352 |
353 | # devtools::install_github("hrbrmstr/ggalt")
354 | library(ggplot2)
355 | library(ggalt)
356 | theme_set(theme_classic())
357 |
358 | health <- read.csv("https://raw.githubusercontent.com/selva86/datasets/master/health.csv")
359 | health$Area <- factor(health$Area, levels=as.character(health$Area)) # for right ordering of the dumbells
360 |
361 | # health$Area <- factor(health$Area)
362 | gg <- ggplot(health, aes(x=pct_2013, xend=pct_2014, y=Area, group=Area)) +
363 | # 画哑铃图
364 | geom_dumbbell(color="#a3c4dc",
365 | size=0.75,
366 | point.colour.l="#0e668b") +
367 | scale_x_continuous(label=percent) +
368 | labs(x=NULL,
369 | y=NULL,
370 | title="Dumbbell Chart",
371 | subtitle="Pct Change: 2013 vs 2014",
372 | caption="Source: https://github.com/hrbrmstr/ggalt") +
373 | theme(plot.title = element_text(hjust=0.5, face="bold"),
374 | plot.background=element_rect(fill="#f7f7f7"),
375 | panel.background=element_rect(fill="#f7f7f7"),
376 | panel.grid.minor=element_blank(),
377 | panel.grid.major.y=element_blank(),
378 | panel.grid.major.x=element_line(),
379 | axis.ticks=element_blank(),
380 | legend.position="top",
381 | panel.border=element_blank())
382 | plot(gg)
383 |
384 | library(ggplot2)
385 | theme_set(theme_classic())
386 |
387 | # Histogram on a Continuous (Numeric) Variable
388 | g <- ggplot(mpg, aes(displ)) +
389 | # 设置颜色
390 | scale_fill_brewer(palette = "Spectral")
391 |
392 | g + geom_histogram(aes(fill=class),
393 | binwidth = .1,
394 | col="black",
395 | size=.1) + # change binwidth
396 | labs(title="Histogram with Auto Binning",
397 | subtitle="Engine Displacement across Vehicle Classes")
398 |
399 | g + geom_histogram(aes(fill=class),
400 | bins=5,
401 | col="black",
402 | size=.1) + # change number of bins
403 | labs(title="Histogram with Fixed Bins",
404 | subtitle="Engine Displacement across Vehicle Classes")
405 |
406 | library(ggplot2)
407 | theme_set(theme_classic())
408 |
409 | # Histogram on a Categorical variable
410 | g <- ggplot(mpg, aes(manufacturer))
411 | g + geom_bar(aes(fill=class), width = 0.5) +
412 | theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
413 | labs(title="Histogram on Categorical Variable",
414 | subtitle="Manufacturer across Vehicle Classes")
415 |
416 | library(ggplot2)
417 | theme_set(theme_classic())
418 |
419 | # Plot
420 | g <- ggplot(mpg, aes(cty))
421 | # 密度图
422 | g + geom_density(aes(fill=factor(cyl)), alpha=0.8) +
423 | labs(title="Density plot",
424 | subtitle="City Mileage Grouped by Number of cylinders",
425 | caption="Source: mpg",
426 | x="City Mileage",
427 | fill="# Cylinders")
428 |
429 | head(mpg)
430 |
431 | library(ggplot2)
432 | theme_set(theme_classic())
433 |
434 | # Plot
435 | g <- ggplot(mpg, aes(class, cty))
436 | g + geom_boxplot(varwidth=T, fill="plum") +
437 | labs(title="Box plot",
438 | subtitle="City Mileage grouped by Class of vehicle",
439 | caption="Source: mpg",
440 | x="Class of Vehicle",
441 | y="City Mileage")
442 |
443 | library(ggthemes)
444 | g <- ggplot(mpg, aes(class, cty))
445 | g + geom_boxplot(aes(fill=factor(cyl))) +
446 | theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
447 | labs(title="Box plot",
448 | subtitle="City Mileage grouped by Class of vehicle",
449 | caption="Source: mpg",
450 | x="Class of Vehicle",
451 | y="City Mileage")
452 |
453 | library(ggplot2)
454 | theme_set(theme_bw())
455 |
456 | # plot
457 | g <- ggplot(mpg, aes(manufacturer, cty))
458 | g + geom_boxplot() +
459 | # binaxis bin x or y
460 | # stackdir 点在箱形图的位置
461 | geom_dotplot(binaxis='y',
462 | stackdir='center',
463 | dotsize = .5,
464 | fill="red") +
465 | theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
466 | labs(title="Box plot + Dot plot",
467 | subtitle="City Mileage vs Class: Each dot represents 1 row in source data",
468 | caption="Source: mpg",
469 | x="Class of Vehicle",
470 | y="City Mileage")
471 |
472 | library(ggthemes)
473 | library(ggplot2)
474 | theme_set(theme_tufte()) # from ggthemes
475 |
476 | # plot
477 | g <- ggplot(mpg, aes(manufacturer, cty))
478 | g + geom_tufteboxplot() +
479 | theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
480 | labs(title="Tufte Styled Boxplot",
481 | subtitle="City Mileage grouped by Class of vehicle",
482 | caption="Source: mpg",
483 | x="Class of Vehicle",
484 | y="City Mileage")
485 |
486 | library(ggplot2)
487 | theme_set(theme_bw())
488 |
489 | # plot
490 | g <- ggplot(mpg, aes(class, cty))
491 | g + geom_violin() +
492 | labs(title="Violin plot",
493 | subtitle="City Mileage vs Class of vehicle",
494 | caption="Source: mpg",
495 | x="Class of Vehicle",
496 | y="City Mileage")
497 |
498 | library(ggplot2)
499 | library(ggthemes)
500 | # turns of scientific notations like 1e+40 不使用科学计数
501 | options(scipen = 999)
502 |
503 | # Read data
504 | email_campaign_funnel <- read.csv("https://raw.githubusercontent.com/selva86/datasets/master/email_campaign_funnel.csv")
505 |
506 | # X Axis Breaks and Labels
507 | brks <- seq(-15000000, 15000000, 5000000)
508 | lbls = paste0(as.character(c(seq(15, 0, -5), seq(5, 15, 5))), "m")
509 |
510 | # Plot
511 | ggplot(email_campaign_funnel, aes(x = Stage, y = Users, fill = Gender)) + # Fill column
512 | # draw the bars 绘柱状图
513 | geom_bar(stat = "identity", width = .6) +
514 | scale_y_continuous(breaks = brks, # Breaks
515 | labels = lbls) + # Labels
516 | coord_flip() + # Flip axes
517 | labs(title="Email Campaign Funnel") +
518 | theme_tufte() + # Tufte theme from ggfortify
519 | theme(plot.title = element_text(hjust = .5),
520 | axis.ticks = element_blank()) + # Centre plot title
521 | scale_fill_brewer(palette = "Dark2") # Color palette
522 |
523 | var <- mpg$class # the categorical data
524 |
525 | ## Prep data (nothing to change here)
526 | nrows <- 10
527 | df <- expand.grid(y = 1:nrows, x = 1:nrows)
528 | categ_table <- round(table(var) * ((nrows*nrows)/(length(var))))
529 | categ_table
530 | #> 2seater compact midsize minivan pickup subcompact suv
531 | #> 2 20 18 5 14 15 26
532 |
533 | # 打印数据
534 | df$category <- factor(rep(names(categ_table), categ_table))
535 | # NOTE: if sum(categ_table) is not 100 (i.e. nrows^2), it will need adjustment to make the sum to 100.
536 |
537 |
538 |
539 | ## Plot
540 | ggplot(df, aes(x = x, y = y, fill = category)) +
541 | # 画热图
542 | geom_tile(color = "black", size = 0.5) +
543 | scale_x_continuous(expand = c(0, 0)) +
544 | scale_y_continuous(expand = c(0, 0), trans = 'reverse') +
545 | scale_fill_brewer(palette = "Set3") +
546 | labs(title="Waffle Chart", subtitle="'Class' of vehicles",
547 | caption="Source: mpg") +
548 | theme(panel.border = element_rect(size = 2),
549 | plot.title = element_text(size = rel(1.2)),
550 | axis.text = element_blank(),
551 | axis.title = element_blank(),
552 | axis.ticks = element_blank(),
553 | legend.title = element_blank(),
554 | legend.position = "right")
555 |
556 | library(ggplot2)
557 | theme_set(theme_classic())
558 |
559 | # Source: Frequency table
560 | df <- as.data.frame(table(mpg$class))
561 | colnames(df) <- c("class", "freq")
562 | pie <- ggplot(df, aes(x = "", y=freq, fill = factor(class))) +
563 | geom_bar(width = 1, stat = "identity") +
564 | theme(axis.line = element_blank(),
565 | plot.title = element_text(hjust=0.5)) +
566 | labs(fill="class",
567 | x=NULL,
568 | y=NULL,
569 | title="Pie Chart of class",
570 | caption="Source: mpg")
571 |
572 | pie + coord_polar(theta = "y", start=0)
573 |
574 |
575 |
576 | # Source: Categorical variable.
577 | # mpg$class
578 | pie <- ggplot(mpg, aes(x = "", fill = factor(class))) +
579 | geom_bar(width = 1) +
580 | theme(axis.line = element_blank(),
581 | plot.title = element_text(hjust=0.5)) +
582 | labs(fill="class",
583 | x=NULL,
584 | y=NULL,
585 | title="Pie Chart of class",
586 | caption="Source: mpg")
587 |
588 | pie + coord_polar(theta = "y", start=0)
589 |
590 | library(ggplot2)
591 | library(treemapify)
592 | head(G20)
593 |
594 | ggplot(G20, aes(area = gdp_mil_usd, fill = hdi)) +
595 | geom_treemap()
596 |
597 | ggplot(G20, aes(area = gdp_mil_usd, fill = hdi, label = country)) +
598 | geom_treemap() +
599 | geom_treemap_text(fontface = "italic", colour = "white", place = "centre",
600 | grow = TRUE)
601 |
602 | ggplot(G20, aes(area = gdp_mil_usd, fill = hdi, label = country,
603 | subgroup = region)) +
604 | geom_treemap() +
605 | geom_treemap_subgroup_border() +
606 | geom_treemap_subgroup_text(place = "centre", grow = T, alpha = 0.5, colour =
607 | "black", fontface = "italic", min.size = 0) +
608 | geom_treemap_text(colour = "white", place = "topleft", reflow = T)
609 |
610 | ggplot(G20, aes(area = 1, label = country, subgroup = hemisphere,
611 | subgroup2 = region, subgroup3 = econ_classification)) +
612 | geom_treemap() +
613 | geom_treemap_subgroup3_border(colour = "blue", size = 1) +
614 | geom_treemap_subgroup2_border(colour = "white", size = 3) +
615 | geom_treemap_subgroup_border(colour = "red", size = 5) +
616 | geom_treemap_subgroup_text(place = "middle", colour = "red", alpha = 0.5, grow = T) +
617 | geom_treemap_subgroup2_text(colour = "white", alpha = 0.5, fontface = "italic") +
618 | geom_treemap_subgroup3_text(place = "top", colour = "blue", alpha = 0.5) +
619 | geom_treemap_text(colour = "white", place = "middle", reflow = T)
620 |
621 | ggplot(G20, aes(area = gdp_mil_usd, fill = region, label = country, subgroup = region)) +
622 | geom_treemap() +
623 | geom_treemap_text(grow = T, reflow = T, colour = "black") +
624 | facet_wrap( ~ hemisphere) +
625 | scale_fill_brewer(palette = "Set1") +
626 | theme(legend.position = "bottom") +
627 | labs(
628 | title = "The G-20 major economies by hemisphere",
629 | caption = "The area of each tile represents the country's GDP as a
630 | proportion of all countries in that hemisphere",
631 | fill = "Region"
632 | )
633 |
634 | freqtable <- table(mpg$manufacturer)
635 | df <- as.data.frame.table(freqtable)
636 | head(df)
637 |
638 | # plot
639 | library(ggplot2)
640 | theme_set(theme_classic())
641 |
642 | # Plot
643 | g <- ggplot(df, aes(Var1, Freq))
644 | g + geom_bar(stat="identity", width = 0.5, fill="tomato2") +
645 | labs(title="Bar Chart",
646 | subtitle="Manufacturer of vehicles",
647 | caption="Source: Frequency of Manufacturers from 'mpg' dataset") +
648 | theme(axis.text.x = element_text(angle=65, vjust=0.6))
649 |
650 | # From on a categorical column variable
651 | g <- ggplot(mpg, aes(manufacturer))
652 | g + geom_bar(aes(fill=class), width = 0.5) +
653 | theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
654 | labs(title="Categorywise Bar Chart",
655 | subtitle="Manufacturer of vehicles",
656 | caption="Source: Manufacturers from 'mpg' dataset")
657 |
658 | ## From Timeseries object (ts)
659 | library(ggplot2)
660 | library(ggfortify)
661 | theme_set(theme_classic())
662 |
663 | economics$returns_perc <- c(0, diff(economics$psavert)/economics$psavert[-length(economics$psavert)])
664 |
665 | # Plot
666 | autoplot(AirPassengers) +
667 | labs(title="AirPassengers") +
668 | theme(plot.title = element_text(hjust=0.5))
669 |
670 | library(ggplot2)
671 | theme_set(theme_classic())
672 |
673 | # Allow Default X Axis Labels
674 | ggplot(economics, aes(x=date)) +
675 | geom_line(aes(y=returns_perc)) +
676 | labs(title="Time Series Chart",
677 | subtitle="Returns Percentage from 'Economics' Dataset",
678 | caption="Source: Economics",
679 | y="Returns %")
680 |
681 | library(ggplot2)
682 | library(lubridate)
683 | theme_set(theme_bw())
684 |
685 | economics_m <- economics[1:24, ]
686 |
687 | # labels and breaks for X axis text
688 | lbls <- paste0(month.abb[month(economics_m$date)], " ", lubridate::year(economics_m$date))
689 | brks <- economics_m$date
690 |
691 | # plot
692 | ggplot(economics_m, aes(x=date)) +
693 | geom_line(aes(y=returns_perc)) +
694 | labs(title="Monthly Time Series",
695 | subtitle="Returns Percentage from Economics Dataset",
696 | caption="Source: Economics",
697 | y="Returns %") + # title and caption
698 | scale_x_date(labels = lbls,
699 | breaks = brks) + # change to monthly ticks and labels
700 | theme(axis.text.x = element_text(angle = 90, vjust=0.5), # rotate x axis text
701 | panel.grid.minor = element_blank()) # turn off minor grid
702 |
703 | library(ggplot2)
704 | library(lubridate)
705 | theme_set(theme_bw())
706 |
707 | economics_y <- economics[1:90, ]
708 |
709 | # labels and breaks for X axis text
710 | brks <- economics_y$date[seq(1, length(economics_y$date), 12)]
711 | lbls <- lubridate::year(brks)
712 |
713 | # plot
714 | ggplot(economics_y, aes(x=date)) +
715 | geom_line(aes(y=returns_perc)) +
716 | labs(title="Yearly Time Series",
717 | subtitle="Returns Percentage from Economics Dataset",
718 | caption="Source: Economics",
719 | y="Returns %") + # title and caption
720 | scale_x_date(labels = lbls,
721 | breaks = brks) + # change to monthly ticks and labels
722 | theme(axis.text.x = element_text(angle = 90, vjust=0.5), # rotate x axis text
723 | panel.grid.minor = element_blank()) # turn off minor grid
724 |
725 | data(economics_long, package = "ggplot2")
726 | head(economics_long)
727 |
728 | library(ggplot2)
729 | library(lubridate)
730 | theme_set(theme_bw())
731 |
732 | df <- economics_long[economics_long$variable %in% c("psavert", "uempmed"), ]
733 | df <- df[lubridate::year(df$date) %in% c(1967:1981), ]
734 |
735 | # labels and breaks for X axis text
736 | brks <- df$date[seq(1, length(df$date), 12)]
737 | lbls <- lubridate::year(brks)
738 |
739 | # plot
740 | ggplot(df, aes(x=date)) +
741 | geom_line(aes(y=value, col=variable)) +
742 | labs(title="Time Series of Returns Percentage",
743 | subtitle="Drawn from Long Data format",
744 | caption="Source: Economics",
745 | y="Returns %",
746 | color=NULL) + # title and caption
747 | scale_x_date(labels = lbls, breaks = brks) + # change to monthly ticks and labels
748 | scale_color_manual(labels = c("psavert", "uempmed"),
749 | values = c("psavert"="#00ba38", "uempmed"="#f8766d")) + # line color
750 | theme(axis.text.x = element_text(angle = 90, vjust=0.5, size = 8), # rotate x axis text
751 | panel.grid.minor = element_blank()) # turn off minor grid
752 |
753 | library(ggplot2)
754 | library(lubridate)
755 | theme_set(theme_bw())
756 |
757 | df <- economics[, c("date", "psavert", "uempmed")]
758 | df <- df[lubridate::year(df$date) %in% c(1967:1981), ]
759 |
760 | # labels and breaks for X axis text
761 | brks <- df$date[seq(1, length(df$date), 12)]
762 | lbls <- lubridate::year(brks)
763 |
764 | # plot
765 | ggplot(df, aes(x=date)) +
766 | geom_line(aes(y=psavert, col="psavert")) +
767 | geom_line(aes(y=uempmed, col="uempmed")) +
768 | labs(title="Time Series of Returns Percentage",
769 | subtitle="Drawn From Wide Data format",
770 | caption="Source: Economics", y="Returns %") + # title and caption
771 | scale_x_date(labels = lbls, breaks = brks) + # change to monthly ticks and labels
772 | scale_color_manual(name="",
773 | values = c("psavert"="#00ba38", "uempmed"="#f8766d")) + # line color
774 | theme(panel.grid.minor = element_blank()) # turn off minor grid
775 |
776 | library(ggplot2)
777 | library(lubridate)
778 | theme_set(theme_bw())
779 |
780 | df <- economics[, c("date", "psavert", "uempmed")]
781 | df <- df[lubridate::year(df$date) %in% c(1967:1981), ]
782 |
783 | # labels and breaks for X axis text
784 | brks <- df$date[seq(1, length(df$date), 12)]
785 | lbls <- lubridate::year(brks)
786 |
787 | # plot
788 | ggplot(df, aes(x=date)) +
789 | geom_area(aes(y=psavert+uempmed, fill="psavert")) +
790 | geom_area(aes(y=uempmed, fill="uempmed")) +
791 | labs(title="Area Chart of Returns Percentage",
792 | subtitle="From Wide Data format",
793 | caption="Source: Economics",
794 | y="Returns %") + # title and caption
795 | scale_x_date(labels = lbls, breaks = brks) + # change to monthly ticks and labels
796 | scale_fill_manual(name="",
797 | values = c("psavert"="#00ba38", "uempmed"="#f8766d")) + # line color
798 | theme(panel.grid.minor = element_blank()) # turn off minor grid
799 |
800 | # http://margintale.blogspot.in/2012/04/ggplot2-time-series-heatmaps.html
801 | library(ggplot2)
802 | library(plyr)
803 | library(scales)
804 | library(zoo)
805 |
806 | df <- read.csv("https://raw.githubusercontent.com/selva86/datasets/master/yahoo.csv")
807 | df$date <- as.Date(df$date) # format date
808 | df <- df[df$year >= 2012, ] # filter reqd years
809 |
810 | # Create Month Week
811 | df$yearmonth <- as.yearmon(df$date)
812 | df$yearmonthf <- factor(df$yearmonth)
813 | df <- ddply(df,.(yearmonthf), transform, monthweek=1+week-min(week)) # compute week number of month
814 | df <- df[, c("year", "yearmonthf", "monthf", "week", "monthweek", "weekdayf", "VIX.Close")]
815 | head(df)
816 | #> year yearmonthf monthf week monthweek weekdayf VIX.Close
817 | #> 1 2012 Jan 2012 Jan 1 1 Tue 22.97
818 | #> 2 2012 Jan 2012 Jan 1 1 Wed 22.22
819 | #> 3 2012 Jan 2012 Jan 1 1 Thu 21.48
820 | #> 4 2012 Jan 2012 Jan 1 1 Fri 20.63
821 | #> 5 2012 Jan 2012 Jan 2 2 Mon 21.07
822 | #> 6 2012 Jan 2012 Jan 2 2 Tue 20.69
823 |
824 |
825 | # Plot
826 | ggplot(df, aes(monthweek, weekdayf, fill = VIX.Close)) +
827 | geom_tile(colour = "white") +
828 | facet_grid(year~monthf) +
829 | scale_fill_gradient(low="red", high="green") +
830 | labs(x="Week of Month",
831 | y="",
832 | title = "Time-Series Calendar Heatmap",
833 | subtitle="Yahoo Closing Price",
834 | fill="Close")
835 |
836 | library(ggplot2)
837 | library(forecast)
838 | theme_set(theme_classic())
839 |
840 | # Subset data
841 | nottem_small <- window(nottem, start=c(1920, 1), end=c(1925, 12)) # subset a smaller timewindow
842 |
843 | # Plot
844 | ggseasonplot(AirPassengers) + labs(title="Seasonal plot: International Airline Passengers");
845 |
846 | ggseasonplot(nottem_small) + labs(title="Seasonal plot: Air temperatures at Nottingham Castle");
847 |
848 | # install.packages("ggdendro")
849 | library(ggplot2)
850 | library(ggdendro)
851 | theme_set(theme_bw())
852 |
853 | hc <- hclust(dist(USArrests), "ave") # hierarchical clustering
854 |
855 | # plot
856 | ggdendrogram(hc, rotate = TRUE, size = 2)
857 |
858 | # devtools::install_github("hrbrmstr/ggalt")
859 | library(ggplot2)
860 | library(ggalt)
861 | library(ggfortify)
862 | theme_set(theme_classic())
863 |
864 | # Compute data with principal components ------------------
865 | df <- iris[c(1, 2, 3, 4)]
866 | pca_mod <- prcomp(df) # compute principal components
867 |
868 | # Data frame of principal components ----------------------
869 | df_pc <- data.frame(pca_mod$x, Species=iris$Species) # dataframe of principal components
870 | df_pc_vir <- df_pc[df_pc$Species == "virginica", ] # df for 'virginica'
871 | df_pc_set <- df_pc[df_pc$Species == "setosa", ] # df for 'setosa'
872 | df_pc_ver <- df_pc[df_pc$Species == "versicolor", ] # df for 'versicolor'
873 |
874 | # Plot ----------------------------------------------------
875 | ggplot(df_pc, aes(PC1, PC2, col=Species)) +
876 | geom_point(aes(shape=Species), size=2) + # draw points
877 | labs(title="Iris Clustering",
878 | subtitle="With principal components PC1 and PC2 as X and Y axis",
879 | caption="Source: Iris") +
880 | coord_cartesian(xlim = 1.2 * c(min(df_pc$PC1), max(df_pc$PC1)),
881 | ylim = 1.2 * c(min(df_pc$PC2), max(df_pc$PC2))) + # change axis limits
882 | geom_encircle(data = df_pc_vir, aes(x=PC1, y=PC2)) + # draw circles
883 | geom_encircle(data = df_pc_set, aes(x=PC1, y=PC2)) +
884 | geom_encircle(data = df_pc_ver, aes(x=PC1, y=PC2))
885 |
--------------------------------------------------------------------------------