├── .git-blame-ignore-revs
├── .github
├── ISSUE_TEMPLATE
│ ├── bug_report.md
│ ├── documentation.md
│ └── feature_request.md
└── workflows
│ ├── pytests.yml
│ └── test_and_deploy.yml
├── .gitignore
├── .pre-commit-config.yaml
├── LICENSE
├── README.md
├── docs
├── README.md
├── __init__.py
├── buyers-guide.md
├── calibration-guide.md
├── data-schema.md
├── development-guide.md
├── images
│ ├── HSV_legend.png
│ ├── JCh_Color_legend.png
│ ├── JCh_legend.png
│ ├── acq_finished.png
│ ├── acquire_buttons.png
│ ├── acquisition_settings.png
│ ├── advanced.png
│ ├── cap_bg.png
│ ├── cli_structure.png
│ ├── comms_video_screenshot.png
│ ├── connect_to_mm.png
│ ├── create_group.png
│ ├── create_group_voltage.png
│ ├── create_preset.png
│ ├── create_preset_voltage.png
│ ├── general_reconstruction_settings.png
│ ├── ideal_plot.png
│ ├── modulation.png
│ ├── no-overlay.png
│ ├── overlay-demo.png
│ ├── overlay.png
│ ├── phase_reconstruction_settings.png
│ ├── poincare_swing.svg
│ ├── recOrder_Fig1_Overview.png
│ ├── recOrder_plugin_logo.png
│ ├── reconstruction_birefriengence.png
│ ├── reconstruction_data.png
│ ├── reconstruction_data_info.png
│ ├── reconstruction_models.png
│ ├── reconstruction_queue.png
│ ├── run_calib.png
│ └── run_port.png
├── microscope-installation-guide.md
├── napari-plugin-guide.md
├── reconstruction-guide.md
└── software-installation-guide.md
├── examples
├── README.md
├── birefringence-and-phase.yml
├── birefringence.yml
├── fluorescence.yml
└── phase.yml
├── pyproject.toml
├── recOrder
├── __init__.py
├── acq
│ ├── __init__.py
│ ├── acq_functions.py
│ └── acquisition_workers.py
├── calib
│ ├── Calibration.py
│ ├── Optimization.py
│ ├── __init__.py
│ └── calibration_workers.py
├── cli
│ ├── apply_inverse_models.py
│ ├── apply_inverse_transfer_function.py
│ ├── compute_transfer_function.py
│ ├── gui_widget.py
│ ├── jobs_mgmt.py
│ ├── main.py
│ ├── monitor.py
│ ├── option_eat_all.py
│ ├── parsing.py
│ ├── printing.py
│ ├── reconstruct.py
│ ├── settings.py
│ └── utils.py
├── io
│ ├── __init__.py
│ ├── _reader.py
│ ├── core_functions.py
│ ├── metadata_reader.py
│ ├── utils.py
│ └── visualization.py
├── napari.yaml
├── plugin
│ ├── __init__.py
│ ├── gui.py
│ ├── gui.ui
│ ├── main_widget.py
│ └── tab_recon.py
├── scripts
│ ├── __init__.py
│ ├── launch_napari.py
│ ├── repeat-cal-acq-rec.py
│ ├── repeat-calibration.py
│ ├── samples.py
│ └── simulate_zarr_acq.py
└── tests
│ ├── acq_tests
│ └── test_acq.py
│ ├── calibration_tests
│ └── test_calibration.py
│ ├── cli_tests
│ ├── test_cli.py
│ ├── test_compute_tf.py
│ ├── test_reconstruct.py
│ └── test_settings.py
│ ├── conftest.py
│ ├── mmcore_tests
│ └── test_core_func.py
│ ├── util_tests
│ ├── test_create_empty.py
│ ├── test_io.py
│ └── test_overlays.py
│ └── widget_tests
│ ├── test_dock_widget.py
│ └── test_sample_contributions.py
├── setup.cfg
├── setup.py
└── tox.ini
/.git-blame-ignore-revs:
--------------------------------------------------------------------------------
1 | # .git-blame-ignore-revs
2 | # created as described in: https://docs.github.com/en/repositories/working-with-files/using-files/viewing-a-file#ignore-commits-in-the-blame-view
3 | # black-format all `.py` files except recorder_ui.py
4 | 82f6df5ed34460374ce7c0fdca089d8caa570b9f
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/bug_report.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Bug report
3 | about: Create a report to help us improve
4 | title: "[BUG]"
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Summary and expected behavior**
11 |
12 | **Code for reproduction (using python script or command line interface)**
13 | ```
14 | # paste your code here
15 |
16 | ```
17 |
18 | **Screenshots or steps for reproduction (using napari GUI)**
19 |
20 | **Include relevant logs which are created next to the output dir, name of the dataset, yaml file(s) if encountering reconstruction errors.**
21 |
22 | **Expected behavior**
23 | A clear and concise description of what you expected to happen.
24 |
25 | **Environment:**
26 |
27 | Operating system:
28 | Python version:
29 | Python environment (command line, IDE, Jupyter notebook, etc):
30 | Micro-Manager/pycromanager version:
31 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/documentation.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Documentation
3 | about: Help us improve documentation
4 | title: "[DOC]"
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Suggested improvement**
11 |
12 | **Optional: Pull request with better documentation**
13 |
--------------------------------------------------------------------------------
/.github/ISSUE_TEMPLATE/feature_request.md:
--------------------------------------------------------------------------------
1 | ---
2 | name: Feature request
3 | about: Suggest an idea for this project
4 | title: "[FEATURE]"
5 | labels: ''
6 | assignees: ''
7 |
8 | ---
9 |
10 | **Problem**
11 |
12 | **Proposed solution**
13 |
14 |
15 | **Alternatives you have considered, if any**
16 |
17 | **Additional context**
18 | Note relevant experimental conditions or datasets
19 |
--------------------------------------------------------------------------------
/.github/workflows/pytests.yml:
--------------------------------------------------------------------------------
1 | # This workflow will install Python dependencies, run tests and lint with a single version of Python
2 | # For more information see: https://help.github.com/actions/language-and-framework-guides/using-python-with-github-actions
3 |
4 | name: pytests
5 |
6 | on: [push]
7 |
8 | jobs:
9 | build:
10 | runs-on: ubuntu-latest
11 | strategy:
12 | matrix:
13 | python-version: ["3.10", "3.11"]
14 |
15 | steps:
16 | - name: Checkout repo
17 | uses: actions/checkout@v2
18 |
19 | - name: Set up Python ${{ matrix.python-version }}
20 | uses: actions/setup-python@v1
21 | with:
22 | python-version: ${{ matrix.python-version }}
23 |
24 | - name: Install dependencies
25 | run: |
26 | python -m pip install --upgrade pip
27 | pip install ".[all,dev]"
28 |
29 | # - name: Lint with flake8
30 | # run: |
31 | # pip install flake8
32 | # # stop the build if there are Python syntax errors or undefined names
33 | # flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
34 | # # exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
35 | # flake8 . --count --exit-zero --max-complexity=10 --max-line-length=127 --statistics
36 |
37 | - name: Test with pytest
38 | run: |
39 | pytest -v
40 | pytest --cov=./ --cov-report=xml
41 |
42 | # - name: Upload coverage to Codecov
43 | # uses: codecov/codecov-action@v1
44 | # with:
45 | # flags: unittest
46 | # name: codecov-umbrella
47 | # fail_ci_if_error: true
48 |
--------------------------------------------------------------------------------
/.github/workflows/test_and_deploy.yml:
--------------------------------------------------------------------------------
1 | # Modified from cookiecutter-napari-plugin
2 | # This workflows will upload a Python Package using Twine when a release is created
3 | # For more information see: https://help.github.com/en/actions/language-and-framework-guides/using-python-with-github-actions#publishing-to-package-registries
4 | name: test-and-deploy
5 |
6 | on:
7 | push:
8 | branches:
9 | - main
10 | tags:
11 | - "*"
12 | pull_request:
13 | branches:
14 | - "*"
15 | workflow_dispatch:
16 |
17 | jobs:
18 | test:
19 | name: ${{ matrix.platform }} py${{ matrix.python-version }}
20 | runs-on: ${{ matrix.platform }}
21 | strategy:
22 | matrix:
23 | platform: [ubuntu-latest, windows-latest, macos-latest]
24 | python-version: ["3.10", "3.11"]
25 |
26 | steps:
27 | - uses: actions/checkout@v3
28 |
29 | - name: Set up Python ${{ matrix.python-version }}
30 | uses: actions/setup-python@v4
31 | with:
32 | python-version: ${{ matrix.python-version }}
33 |
34 | # these libraries enable testing on Qt on linux
35 | - uses: tlambert03/setup-qt-libs@v1
36 |
37 | # strategy borrowed from vispy for installing opengl libs on windows
38 | - name: Install Windows OpenGL
39 | if: runner.os == 'Windows'
40 | run: |
41 | git clone --depth 1 https://github.com/pyvista/gl-ci-helpers.git
42 | powershell gl-ci-helpers/appveyor/install_opengl.ps1
43 |
44 | # note: if you need dependencies from conda, considering using
45 | # setup-miniconda: https://github.com/conda-incubator/setup-miniconda
46 | # and
47 | # tox-conda: https://github.com/tox-dev/tox-conda
48 | - name: Install dependencies
49 | run: |
50 | python -m pip install --upgrade pip
51 | python -m pip install setuptools tox tox-gh-actions
52 |
53 | # https://github.com/napari/cookiecutter-napari-plugin/commit/cb9a8c152b68473e8beabf44e7ab11fc46483b5d
54 | - name: Test
55 | uses: aganders3/headless-gui@v1
56 | with:
57 | run: python -m tox
58 |
59 | - name: Coverage
60 | uses: codecov/codecov-action@v3
61 |
62 | deploy:
63 | # this will run when you have tagged a commit with a version number
64 | # and requires that you have put your twine API key in your
65 | # github secrets (see readme for details)
66 | needs: [test]
67 | runs-on: ubuntu-latest
68 | if: contains(github.ref, 'tags')
69 | steps:
70 | - uses: actions/checkout@v2
71 | - name: Set up Python
72 | uses: actions/setup-python@v2
73 | with:
74 | python-version: "3.x"
75 | - name: Install dependencies
76 | run: |
77 | python -m pip install --upgrade pip
78 | pip install -U setuptools setuptools_scm wheel twine build
79 |
80 | # skip build and publish for now
81 | # - name: Build and publish
82 | # env:
83 | # TWINE_USERNAME: __token__
84 | # TWINE_PASSWORD: ${{ secrets.TWINE_API_KEY }}
85 | # run: |
86 | # git tag
87 | # python -m build .
88 | # twine upload --repository testpypi dist/* # Commented until API key is on github
89 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Binary image files.
2 | *.tif[f]
3 | *.jp[e]g
4 | *.zar[r]
5 | *.json
6 |
7 | # pycharm IDE
8 | .idea
9 | .DS_Store
10 |
11 | # Byte-compiled / optimized / DLL files
12 | __pycache__/
13 | *.py[cod]
14 | *$py.class
15 |
16 | # C extensions
17 | *.so
18 |
19 | # Distribution / packaging
20 | .Python
21 | build/
22 | develop-eggs/
23 | dist/
24 | downloads/
25 | eggs/
26 | .eggs/
27 | lib/
28 | lib64/
29 | parts/
30 | sdist/
31 | var/
32 | wheels/
33 | pip-wheel-metadata/
34 | share/python-wheels/
35 | *.egg-info/
36 | .installed.cfg
37 | *.egg
38 | MANIFEST
39 |
40 | # PyInstaller
41 | # Usually these files are written by a python script from a template
42 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
43 | *.manifest
44 | *.spec
45 |
46 | # Installer logs
47 | pip-log.txt
48 | pip-delete-this-directory.txt
49 |
50 | # Unit test / coverage reports
51 | htmlcov/
52 | .tox/
53 | .nox/
54 | .coverage
55 | .coverage.*
56 | .cache
57 | nosetests.xml
58 | coverage.xml
59 | *.cover
60 | *.py,cover
61 | .hypothesis/
62 | .pytest_cache/
63 | pytest_temp/
64 |
65 | # Translations
66 | *.mo
67 | *.pot
68 |
69 | # Django stuff:
70 | *.log
71 | local_settings.py
72 | db.sqlite3
73 | db.sqlite3-journal
74 |
75 | # Flask stuff:
76 | instance/
77 | .webassets-cache
78 |
79 | # Scrapy stuff:
80 | .scrapy
81 |
82 | # Sphinx documentation
83 | docs/_build/
84 |
85 | # PyBuilder
86 | target/
87 |
88 | # Jupyter Notebook
89 | .ipynb_checkpoints
90 |
91 | # IPython
92 | profile_default/
93 | ipython_config.py
94 |
95 | # pyenv
96 | .python-version
97 |
98 | # pipenv
99 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
100 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
101 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
102 | # install all needed dependencies.
103 | #Pipfile.lock
104 |
105 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
106 | __pypackages__/
107 |
108 | # Celery stuff
109 | celerybeat-schedule
110 | celerybeat.pid
111 |
112 | # SageMath parsed files
113 | *.sage.py
114 |
115 | # Environments
116 | .env
117 | .venv
118 | env/
119 | venv/
120 | ENV/
121 | env.bak/
122 | venv.bak/
123 |
124 | # Spyder project settings
125 | .spyderproject
126 | .spyproject
127 |
128 | # Rope project settings
129 | .ropeproject
130 |
131 | # mkdocs documentation
132 | /site
133 |
134 | # mypy
135 | .mypy_cache/
136 | .dmypy.json
137 | dmypy.json
138 |
139 | # Pyre type checker
140 | .pyre/
141 |
142 | .DS_Store
143 |
144 | # written by setuptools_scm
145 | */_version.py
146 | recOrder/_version.py
147 | *.autosave
148 |
149 | # example data
150 | /examples/data_temp/*
151 | /logs/*
152 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | repos:
2 | - repo: https://github.com/python/black
3 | rev: 21.6b0
4 | hooks:
5 | - id: black
6 | language_version: python3
7 | pass_filenames: false
8 | args: [.]
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | BSD 3-Clause License
2 |
3 | Copyright (c) 2020, Chan Zuckerberg Biohub
4 |
5 | Redistribution and use in source and binary forms, with or without
6 | modification, are permitted provided that the following conditions are met:
7 |
8 | 1. Redistributions of source code must retain the above copyright notice, this
9 | list of conditions and the following disclaimer.
10 |
11 | 2. Redistributions in binary form must reproduce the above copyright notice,
12 | this list of conditions and the following disclaimer in the documentation
13 | and/or other materials provided with the distribution.
14 |
15 | 3. Neither the name of the copyright holder nor the names of its
16 | contributors may be used to endorse or promote products derived from
17 | this software without specific prior written permission.
18 |
19 | THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
20 | AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
21 | IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
22 | DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
23 | FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
24 | DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
25 | SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
26 | CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
27 | OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
28 | OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
29 |
30 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # recOrder
2 | [](https://pypi.org/project/recOrder-napari)
3 | [](https://pypistats.org/packages/recOrder-napari)
4 | [](https://pepy.tech/project/recOrder-napari)
5 | [](https://github.com/mehta-lab/recOrder/graphs/contributors)
6 | 
7 | 
8 | 
9 |
10 |
11 |
12 | `recOrder` is a collection of computational imaging methods. It currently provides QLIPP (quantitative label-free imaging with phase and polarization), phase from defocus, and fluorescence deconvolution.
13 |
14 | [](https://www.youtube.com/watch?v=JEZAaPeZhck)
15 |
16 | Acquisition, calibration, background correction, reconstruction, and applications of QLIPP are described in the following [E-Life Paper](https://elifesciences.org/articles/55502):
17 |
18 | ```bibtex
19 | Syuan-Ming Guo, Li-Hao Yeh, Jenny Folkesson, Ivan E Ivanov, Anitha P Krishnan, Matthew G Keefe, Ezzat Hashemi, David Shin, Bryant B Chhun, Nathan H Cho, Manuel D Leonetti, May H Han, Tomasz J Nowakowski, Shalin B Mehta, "Revealing architectural order with quantitative label-free imaging and deep learning," eLife 2020;9:e55502 DOI: 10.7554/eLife.55502 (2020).
20 | ```
21 |
22 | These are the kinds of data you can acquire with `recOrder` and QLIPP:
23 |
24 | https://user-images.githubusercontent.com/9554101/271128301-cc71da57-df6f-401b-a955-796750a96d88.mov
25 |
26 | https://user-images.githubusercontent.com/9554101/271128510-aa2180af-607f-4c0c-912c-c18dc4f29432.mp4
27 |
28 | ## What do I need to use `recOrder`
29 | `recOrder` is to be used alongside a conventional widefield microscope. For QLIPP, the microscope must be fitted with an analyzer and a universal polarizer:
30 |
31 | https://user-images.githubusercontent.com/9554101/273073475-70afb05a-1eb7-4019-9c42-af3e07bef723.mp4
32 |
33 | For phase-from-defocus or fluorescence deconvolution methods, the universal polarizer is optional.
34 |
35 | The overall structure of `recOrder` is shown in Panel B, highlighting the structure of the graphical user interface (GUI) through a napari plugin and the command-line interface (CLI) that allows users to perform reconstructions.
36 |
37 | 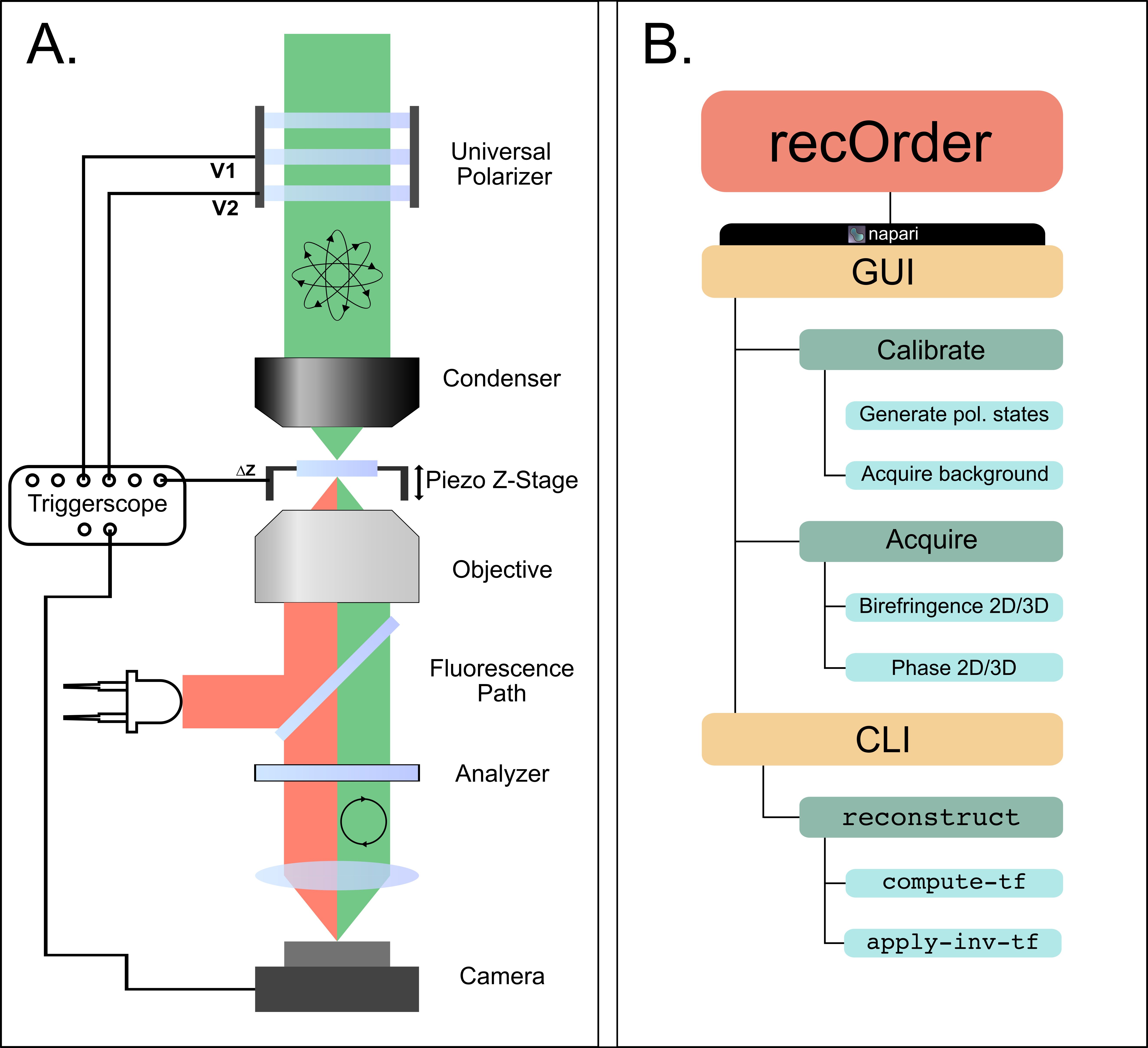
38 |
39 |
40 |
41 | ## Software Quick Start
42 |
43 | (Optional but recommended) install [anaconda](https://www.anaconda.com/products/distribution) and create a virtual environment:
44 |
45 | ```sh
46 | conda create -y -n recOrder python=3.10
47 | conda activate recOrder
48 | ```
49 |
50 | Install `recOrder-napari` with acquisition dependencies
51 | (napari with PyQt6 and pycro-manager):
52 |
53 | ```sh
54 | pip install recOrder-napari[all]
55 | ```
56 |
57 | Install `recOrder-napari` without napari, QtBindings (PyQt/PySide) and pycro-manager dependencies:
58 |
59 | ```sh
60 | pip install recOrder-napari
61 | ```
62 |
63 | Open `napari` with `recOrder-napari`:
64 |
65 | ```sh
66 | napari -w recOrder-napari
67 | ```
68 |
69 | For more help, see [`recOrder`'s documentation](https://github.com/mehta-lab/recOrder/tree/main/docs). To install `recOrder`
70 | on a microscope, see the [microscope installation guide](https://github.com/mehta-lab/recOrder/blob/main/docs/microscope-installation-guide.md).
71 |
72 | ## Dataset
73 |
74 | [Slides](https://doi.org/10.5281/zenodo.5135889) and a [dataset](https://doi.org/10.5281/zenodo.5178487) shared during a workshop on QLIPP and recOrder can be found on Zenodo, and the napari plugin's sample contributions (`File > Open Sample > recOrder-napari` in napari).
75 |
76 | [](https://doi.org/10.5281/zenodo.5178487)
77 | [](https://doi.org/10.5281/zenodo.5135889)
--------------------------------------------------------------------------------
/docs/README.md:
--------------------------------------------------------------------------------
1 | # Welcome to `recOrder`'s documentation
2 |
3 | **I want to buy hardware for a polarized-light installation:** start with the [buyers guide](./buyers-guide.md).
4 |
5 | **I would like to install `recOrder` on my microscope:** start with the [microscope installation guide](./microscope-installation-guide.md).
6 |
7 | **I would like to use the `napari plugin`:** start with the [plugin guide](./napari-plugin-guide.md).
8 |
9 | **I would like to reconstruct existing data:** start with the [reconstruction guide](./reconstruction-guide.md) and consult the [data schema](./data-schema.md) for `recOrder`'s format.
10 |
11 | **I would like to set up a development environment and test `recOrder`**: start with the [development guide](./development-guide.md).
12 |
13 | **I would like understand `recOrder`'s calibration routine**: read the [calibration guide](./calibration-guide.md).
14 |
15 | **I noticed an error in the documentation or code:** [open an issue](https://github.com/mehta-lab/recOrder/issues/new/choose) or [send us an email](mailto:shalin.mehta@czbiohub.org,talon.chandler@czbiohub.org). We appreciate your help!
--------------------------------------------------------------------------------
/docs/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/__init__.py
--------------------------------------------------------------------------------
/docs/buyers-guide.md:
--------------------------------------------------------------------------------
1 | # Buyer's Guide
2 |
3 | ## Quantitative phase imaging:
4 |
5 | You can use a transmitted light source (LED or a lamp) and a condenser commonly available on almost all microscopes. In addition to the transmitted light imaging path, you will need a motorized stage for acquiring through-focus image stacks.
6 |
7 | ## Quantitative polarization imaging (PolScope):
8 |
9 | The following list of components assumes that you already have a transmitted light source (LED or a lamp) and a condenser.
10 |
11 | Buyers have two options:
12 | 1. buy a complete hardware kit from the OpenPolScope project, or
13 | 2. assemble your own kit piece by piece.
14 |
15 | ### Buy a kit from the OpenPolScope project
16 |
17 | - Read about the [OpenPolScope Hardware Kit](https://openpolscope.org/pages/OPS_Hardware.htm).
18 | - Complete the [OpenPolScope information request form](https://openpolscope.org/pages/Info_Request_Form.htm).
19 |
20 | ### Buy individual components
21 |
22 | The components are listed in the order in which they process light. See the build video here to see how to assemble these components on your microscope.
23 |
24 | https://github.com/user-attachments/assets/a0a8bffb-bf81-4401-9ace-3b4955436b57
25 |
26 | | Part | Approximate Price | Notes |
27 | |--------------------------|-------------------|-----------------------------|
28 | | Illumination filter | $200 | We suggest [a Thorlabs CWL = 530 nm, FWHM = 10 nm notch filter](https://www.thorlabs.com/thorproduct.cfm?partnumber=FBH530-10).|
29 | | Circular polarizer | $350 | We suggest [a Thorlabs 532 nm, left-hand circular polarizer](https://www.thorlabs.com/thorproduct.cfm?partnumber=CP1L532).|
30 | | Liquid crystal compensator | $6,000 | Meadowlark optics LVR-42x52mm-VIS-ASSY or LVR-50x60mm-VIS-POL-ASSY. Although near-variants are listed in the [Meadowlowlark catalog](https://www.meadowlark.com/product/liquid-crystal-variable-retarder/), this is a custom part with two liquid crystals in a custom housing. [Contact Meadowlark](https://www.meadowlark.com/contact-us/) for a quote.|
31 | | Liquid crystal control electronics | $2,000 | [Meadowlark optics D5020-20V](https://www.meadowlark.com/product/liquid-crystal-digital-interface-controller/). Choose the high-voltage 20V version.
32 | | Liquid crystal adapter | $25-$500 | A 3D printed part that aligns the liquid crystal compensator in a microscope stand's illumination path. Check for your stand among the [OpenPolScope `.stl` files](https://github.com/amitabhverma/Microscope-LC-adapters/tree/main/stl_files) or [contact us](compmicro@czbiohub.org) for more options.|
33 | | Circular analyzer (opposite handedness) | $350 | We suggest [a Thorlabs 532 nm, right-hand circular polarizer](https://www.thorlabs.com/thorproduct.cfm?partnumber=CP1R532).|
34 |
35 | If you need help selecting or assembling the components, please start an issue on this GitHub repository or contact us at compmicro@czbiohub.org.
36 |
37 | ## Quantitative phase and polarization imaging (QLIPP):
38 |
39 | Combining the Z-stage and the PolScope components listed above enables joint phase and polarization imaging with `recOrder`.
--------------------------------------------------------------------------------
/docs/calibration-guide.md:
--------------------------------------------------------------------------------
1 | # Calibration guide
2 | This guide describes `recOrder`'s calibration routine with details about its goals, parameters, and evaluation metrics.
3 |
4 | ## Why calibrate?
5 |
6 | `recOrder` sends commands via Micro-Manager (or a TriggerScope) to apply voltages to the liquid crystals which modify the polarization of the light that illuminates the sample. `recOrder` could apply a fixed set of voltages so the user would never have to worry about these details, but this approach leads to extremely poor performance because
7 |
8 | - the sample, the sample holder, lenses, dichroics, and other optical elements introduce small changes in polarization, and
9 | - the liquid crystals' voltage response drifts over time.
10 |
11 | Therefore, recalibrating the liquid crystals regularly (definitely between imaging sessions, often between different samples) is essential for acquiring optimal images.
12 |
13 | ## Finding the extinction state
14 |
15 | Every calibration starts with a routine that finds the **extinction state**: the polarization state (and corresponding voltages) that minimizes the intensity that reaches the camera. If the analyzer is a right-hand-circular polarizer, then the extinction state is the set of voltages that correspond to left-hand-circular light in the sample.
16 |
17 | ## Setting a goal for the remaining states: swing
18 |
19 | After finding the circular extinction state, the calibration routine finds the remaining states. The **swing** parameter sets the target ellipticity of the remaining states and is best understood using [the Poincare sphere](https://en.wikipedia.org/wiki/Unpolarized_light#Poincar%C3%A9_sphere), a diagram that organizes all pure polarization states onto the surface of a sphere.
20 |
21 |  22 |
23 | On the Poincare sphere, the extinction state corresponds to the north pole, and the swing value corresponds to the targeted line of [colatitude](https://en.m.wikipedia.org/wiki/File:Spherical_Coordinates_%28Colatitude,_Longitude%29.svg) for the remaining states. For example, a swing value of 0.25 (above left) sets the targeted polarization states to the states on the equator: a set of linear polarization states. Similarly, a swing value of 0.125 (above right) sets the targeted polarization states to the states on the line of colatitude 45 degrees ( $\pi$/4 radians) away from the north pole: a set of elliptical polarization states.
24 |
25 | The Poincare sphere is also useful for calculating the ratio of intensities measured before and after an analyzer illuminated with a polarized beam. First, find the point on the Poincare sphere that corresponds to the analyzer; in our case we have a right-circular analyzer corresponding to the south pole. Next, find the point that corresponds to the polarization state of the light incident on the analyzer; this could be any arbitrary point on the Poincare sphere. To find the ratio of intensities before and after the analyzer $I/I_0$, find the great-circle angle between the two points on the Poincare sphere, $\alpha$, and calculate $I/I_0 = \cos^2(\alpha/2)$. As expected, points that are close together transmit perfectly ( $\alpha = 0$ implies $I/I_0 = 1$), while antipodal points lead to extinction ( $\alpha = \pi$ implies $I/I_0 = 0$).
26 |
27 | This geometric construction illustrates that all non-extinction polarization states have the same intensity after the analyzer because they live on the same line of colatitude and have the same great-circle angle to the south pole (the analyzer). We use this fact to help us find our non-extinction states.
28 |
29 | Practically, we find our first non-extinction state immediately using the liquid crystal manufacturer's measurements from the factory. In other words, we apply a fixed voltage offset to the extinction-state voltages to find the first non-extinction state, and this requires no iteration or optimization. To find the remaining non-extinction states, we keep the polarization orientation fixed and search through neighboring states with different ellipticity to find states that transmit the same intensity as the first non-extinction state.
30 |
31 | ## Evaluating a calibration: extinction ratio
32 |
33 | At the end of a calibration we report the **extinction ratio**, the ratio of the largest and smallest intensities that the imaging system can transmit above background. This metric measures the quality of the entire optical path including the liquid crystals and their calibrated states, and all depolarization, scattering, or absorption caused by optical elements in the light path will reduce the extinction ratio.
34 |
35 | ## Calculating extinction ratio from measured intensities (advanced topic)
36 |
37 | To calculate the extinction ratio, we could optimize the liquid crystal voltages to maximize measured intensity then calculate the ratio of that result with the earlier extinction intensity, but this approach requires a time-consuming optimization and it does not characterize the quality of the calibrated states of the liquid crystals.
38 |
39 | Instead, we estimate the extinction ratio from the intensities we measure during the calibration process. Specifically, we measure the black-level intensity $I_{\text{bl}}$, the extinction intensity $I_{\text{ext}}$, and the intensity under the first elliptical state $I_{\text{ellip}}(S)$ where $S$ is the swing. We proceed to algebraically express the extinction ratio in terms of these three quantities.
40 |
41 | We can decompose $I_{\text{ellip}}(S)$ into a constant term $I_{\text{ellip}}(0) = I_{\text{ext}}$, and a modulation term given by
42 |
43 | $$I_{\text{ellip}}(S) = I_{\text{mod}}\sin^2(\pi S) + I_{\text{ext}},\qquad\qquad (1)$$
44 | where $I_{\text{mod}}$ is the modulation depth, and the $\sin^2(\pi S)$ term can be understood using the Poincare sphere (the intensity behind the circular analyzer is proportional to $\cos^2(\alpha/2)$ and for a given swing we have $\alpha = \pi - 2\pi S$ so $\cos^2(\frac{\pi - 2\pi S}{2}) = \sin^2(\pi S)$ ).
45 |
46 | Next, we decompose $I_{\text{ext}}$ into the sum of two terms, the black level intensity and a leakage intensity $I_{\text{leak}}$
47 | $$I_{\text{ext}} = I_{\text{bl}} + I_{\text{leak}}.\qquad\qquad (2)$$
48 |
49 | The following diagram clarifies our definitions and shows how the measured $I_{\text{ellip}}(S)$ depends on the swing (green line).
50 |
51 |
22 |
23 | On the Poincare sphere, the extinction state corresponds to the north pole, and the swing value corresponds to the targeted line of [colatitude](https://en.m.wikipedia.org/wiki/File:Spherical_Coordinates_%28Colatitude,_Longitude%29.svg) for the remaining states. For example, a swing value of 0.25 (above left) sets the targeted polarization states to the states on the equator: a set of linear polarization states. Similarly, a swing value of 0.125 (above right) sets the targeted polarization states to the states on the line of colatitude 45 degrees ( $\pi$/4 radians) away from the north pole: a set of elliptical polarization states.
24 |
25 | The Poincare sphere is also useful for calculating the ratio of intensities measured before and after an analyzer illuminated with a polarized beam. First, find the point on the Poincare sphere that corresponds to the analyzer; in our case we have a right-circular analyzer corresponding to the south pole. Next, find the point that corresponds to the polarization state of the light incident on the analyzer; this could be any arbitrary point on the Poincare sphere. To find the ratio of intensities before and after the analyzer $I/I_0$, find the great-circle angle between the two points on the Poincare sphere, $\alpha$, and calculate $I/I_0 = \cos^2(\alpha/2)$. As expected, points that are close together transmit perfectly ( $\alpha = 0$ implies $I/I_0 = 1$), while antipodal points lead to extinction ( $\alpha = \pi$ implies $I/I_0 = 0$).
26 |
27 | This geometric construction illustrates that all non-extinction polarization states have the same intensity after the analyzer because they live on the same line of colatitude and have the same great-circle angle to the south pole (the analyzer). We use this fact to help us find our non-extinction states.
28 |
29 | Practically, we find our first non-extinction state immediately using the liquid crystal manufacturer's measurements from the factory. In other words, we apply a fixed voltage offset to the extinction-state voltages to find the first non-extinction state, and this requires no iteration or optimization. To find the remaining non-extinction states, we keep the polarization orientation fixed and search through neighboring states with different ellipticity to find states that transmit the same intensity as the first non-extinction state.
30 |
31 | ## Evaluating a calibration: extinction ratio
32 |
33 | At the end of a calibration we report the **extinction ratio**, the ratio of the largest and smallest intensities that the imaging system can transmit above background. This metric measures the quality of the entire optical path including the liquid crystals and their calibrated states, and all depolarization, scattering, or absorption caused by optical elements in the light path will reduce the extinction ratio.
34 |
35 | ## Calculating extinction ratio from measured intensities (advanced topic)
36 |
37 | To calculate the extinction ratio, we could optimize the liquid crystal voltages to maximize measured intensity then calculate the ratio of that result with the earlier extinction intensity, but this approach requires a time-consuming optimization and it does not characterize the quality of the calibrated states of the liquid crystals.
38 |
39 | Instead, we estimate the extinction ratio from the intensities we measure during the calibration process. Specifically, we measure the black-level intensity $I_{\text{bl}}$, the extinction intensity $I_{\text{ext}}$, and the intensity under the first elliptical state $I_{\text{ellip}}(S)$ where $S$ is the swing. We proceed to algebraically express the extinction ratio in terms of these three quantities.
40 |
41 | We can decompose $I_{\text{ellip}}(S)$ into a constant term $I_{\text{ellip}}(0) = I_{\text{ext}}$, and a modulation term given by
42 |
43 | $$I_{\text{ellip}}(S) = I_{\text{mod}}\sin^2(\pi S) + I_{\text{ext}},\qquad\qquad (1)$$
44 | where $I_{\text{mod}}$ is the modulation depth, and the $\sin^2(\pi S)$ term can be understood using the Poincare sphere (the intensity behind the circular analyzer is proportional to $\cos^2(\alpha/2)$ and for a given swing we have $\alpha = \pi - 2\pi S$ so $\cos^2(\frac{\pi - 2\pi S}{2}) = \sin^2(\pi S)$ ).
45 |
46 | Next, we decompose $I_{\text{ext}}$ into the sum of two terms, the black level intensity and a leakage intensity $I_{\text{leak}}$
47 | $$I_{\text{ext}} = I_{\text{bl}} + I_{\text{leak}}.\qquad\qquad (2)$$
48 |
49 | The following diagram clarifies our definitions and shows how the measured $I_{\text{ellip}}(S)$ depends on the swing (green line).
50 |
51 |  52 |
53 | The extinction ratio is the ratio of the largest and smallest intensities that the imaging system can transmit above background, which is most easily expressed in terms of $I_{\text{mod}}$ and $I_{\text{leak}}$
54 | $$\text{Extinction Ratio} = \frac{I_{\text{mod}} + I_{\text{leak}}}{I_{\text{leak}}}.\qquad\qquad (3)$$
55 |
56 | Substituting Eqs. (1) and (2) into Eq. (3) gives the extinction ratio in terms of the measured intensities
57 | $$\text{Extinction Ratio} = \frac{1}{\sin^2(\pi S)}\frac{I_{\text{ellip}}(S) - I_{\text{ext}}}{I_{\text{ext}} - I_{\text{bl}}} + 1.$$
58 |
59 | ## Summary: `recOrder`'s step-by-step calibration procedure
60 | 1. Close the shutter, measure the black level, then reopen the shutter.
61 | 2. Find the extinction state by finding voltages that minimize the intensity that reaches the camera.
62 | 3. Use the swing value to immediately find the first elliptical state, and record the intensity on the camera.
63 | 4. For each remaining elliptical state, keep the polarization orientation fixed and optimize the voltages to match the intensity of the first elliptical state.
64 | 5. Store the voltages and calculate the extinction ratio.
--------------------------------------------------------------------------------
/docs/data-schema.md:
--------------------------------------------------------------------------------
1 | # Data schema
2 |
3 | This document defines the standard for data acquired with `recOrder`.
4 |
5 | ## Raw directory organization
6 |
7 | Currently, we structure raw data in the following hierarchy:
8 |
9 | ```text
10 | working_directory/ # commonly YYYY_MM_DD_exp_name, but not enforced
11 | ├── polarization_calibration_0.txt
12 | │ ...
13 | ├── polarization_calibration_.txt # i calibration repeats
14 | │
15 | ├── bg_0
16 | │ ...
17 | ├── bg_ # j background repeats
18 | │ ├── background.zarr
19 | │ ├── polarization_calibration.txt # copied into each bg folder
20 | │ ├── reconstruction.zarr
21 | │ ├── reconstruction_settings.yml # for use with `recorder reconstruct`
22 | │ └── transfer_function.zarr # for use with `recorder apply-inv-tf`
23 | │
24 | ├── _snap_0
25 | ├── _snap_1
26 | │ ├── raw_data.zarr
27 | │ ├── reconstruction.zarr
28 | │ ├── reconstruction_settings.yml
29 | │ └── transfer_function.zarr
30 | │ ...
31 | ├── _snap_ # k repeats with the first acquisition name
32 | │ ├── raw_data.zarr
33 | │ ├── reconstruction.zarr
34 | │ ├── reconstruction_settings.yml
35 | │ └── transfer_function.zarr
36 | │ ...
37 | │
38 | ├── _snap_0 # l different acquisition names
39 | │ ...
40 | ├── _snap_ # m repeats for this acquisition name
41 | ├── raw_data.zarr
42 | ├── reconstruction.zarr
43 | ├── reconstruction_settings.yml
44 | └── transfer_function.zarr
45 | ```
46 |
47 | Each `.zarr` contains an [OME-NGFF v0.4](https://ngff.openmicroscopy.org/0.4/) in HCS format with a single field of view.
--------------------------------------------------------------------------------
/docs/development-guide.md:
--------------------------------------------------------------------------------
1 | # `recOrder` development guide
2 |
3 | ## Install `recOrder` for development
4 |
5 | 1. Install [conda](https://github.com/conda-forge/miniforge) and create a virtual environment:
6 |
7 | ```sh
8 | conda create -y -n recOrder python=3.10
9 | conda activate recOrder
10 | ```
11 |
12 | 2. Clone the `recOrder` directory:
13 |
14 | ```sh
15 | git clone https://github.com/mehta-lab/recOrder.git
16 | ```
17 |
18 | 3. Install `recOrder` in editable mode with development dependencies
19 |
20 | ```sh
21 | cd recOrder
22 | pip install -e ".[all,dev]"
23 | ```
24 |
25 | 4. Optionally, for the co-development of [`waveorder`](https://github.com/mehta-lab/waveorder) and `recOrder`:
26 |
27 | > Note that `pip` will raise an 'error' complaining that the dependency of `recOrder-napari` has been broken if you do the following.
28 | > This does not affect the installation, but can be suppressed by removing [this line](https://github.com/mehta-lab/recOrder/blob/5bc9314a9bacf6f4e235eaffb06c297cf20e4b65/setup.cfg#L40) before installing `waveorder`.
29 | > (Just remember to revert the change afterwards!)
30 | > We expect a nicer behavior to be possible once we release a stable version of `waveorder`.
31 |
32 | ```sh
33 | cd # where you want to clone the repo
34 | git clone https://github.com/mehta-lab/waveorder.git
35 | pip install ./waveorder -e ".[dev]"
36 | ```
37 |
38 | > Importing from a local and/or editable package can cause issues with static code analyzers such due to the absence of the source code from the paths they expect.
39 | > VS Code users can refer to [this guide](https://github.com/microsoft/pylance-release/blob/main/TROUBLESHOOTING.md#common-questions-and-issues) to resolve typing warnings.
40 |
41 | ## Set up a development environment
42 |
43 | ### Code linting
44 |
45 | We are not currently specifying a code linter as most modern Python code editors already have their own. If not, add a plugin to your editor to help catch bugs pre-commit!
46 |
47 | ### Code formatting
48 |
49 | We use `black` to format Python code, and a specific version is installed as a development dependency. Use the `black` in the `recOrder` virtual environment, either from commandline or the editor of your choice.
50 |
51 | > *VS Code users*: Install the [Black Formatter](https://marketplace.visualstudio.com/items?itemName=ms-python.black-formatter) plugin. Press `^/⌘ ⇧ P` and type 'format document with...', choose the Black Formatter and start formatting!
52 |
53 | ### Docstring style
54 |
55 | The [NumPy style](https://numpydoc.readthedocs.io/en/latest/format.html) docstrings are used in `recOrder`.
56 |
57 | > *VS Code users*: [this popular plugin](https://marketplace.visualstudio.com/items?itemName=njpwerner.autodocstring) helps auto-generate most popular docstring styles (including `numpydoc`).
58 |
59 | ## Run automated tests
60 |
61 | From within the `recOrder` directory run:
62 |
63 | ```sh
64 | pytest
65 | ```
66 |
67 | Running `pytest` for the first time will download ~50 MB of test data from Zenodo, and subsequent runs will reuse the downloaded data.
68 |
69 | ## Run manual tests
70 |
71 | Although many of `recOrder`'s tests are automated, many features require manual testing. The following is a summary of features that need to be tested manually before release:
72 |
73 | * Install a compatible version of Micro-Manager and check that `recOrder` can connect.
74 | * Perform calibrations with and without an ROI; with and without a shutter configured in Micro-Manager, in 4- and 5-state modes; and in MM-Voltage, MM-Retardance, and DAC modes (if the TriggerScope is available).
75 | * Test "Load Calibration" and "Calculate Extinction" buttons.
76 | * Test "Capture Background" button.
77 | * Test the "Acquire Birefringence" button on a background FOV. Does a background-corrected background acquisition give random orientations?
78 | * Test the four "Acquire" buttons with varied combinations of 2D/3D, background correction settings, "Phase from BF" checkbox, and regularization parameters.
79 | * Use the data you collected to test "Offline" mode reconstructions with varied combinations of parameters.
80 |
81 | ## GUI development
82 |
83 | We use `QT Creator` for large parts of `recOrder`'s GUI. To modify the GUI, install `QT Creator` from [its website](https://www.qt.io/product/development-tools) or with `brew install --cask qt-creator`
84 |
85 | Open `./recOrder/plugin/gui.ui` in `QT Creator` and make your changes.
86 |
87 | Next, convert the `.ui` to a `.py` file with:
88 |
89 | ```sh
90 | pyuic5 -x gui.ui -o gui.py
91 | ```
92 |
93 | Note: `pyuic5` is installed alongside `PyQt5`, so you can expect to find it installed in your `recOrder` conda environement.
94 |
95 | Finally, change the `gui.py` file's to import `qtpy` instead of `PyQt5` to adhere to [napari plugin best practices](https://napari.org/stable/plugins/best_practices.html#don-t-include-pyside2-or-pyqt5-in-your-plugin-s-dependencies).
96 | On macOS, you can modify the file in place with:
97 |
98 | ```sh
99 | sed -i '' 's/from PyQt5/from qtpy/g' gui.py
100 | ```
101 |
102 | > This is specific for BSD `sed`, omit `''` with GNU.
103 |
104 | Note: although much of the GUI is specified in the generated `recOrder_ui.py` file, the `main_widget.py` file makes extensive modifications to the GUI.
105 |
106 | ## Make `git blame` ignore formatting commits
107 |
108 | **Note:** `git --version` must be `>=2.23` to use this feature.
109 |
110 | If you would like `git blame` to ignore formatting commits, run this line:
111 |
112 | ```sh
113 | git config --global blame.ignoreRevsFile .git-blame-ignore-revs
114 | ```
115 |
116 | The `\.git-blame-ignore-revs` file contains a list of commit hashes corresponding to formatting commits.
117 | If you make a formatting commit, please add the commit's hash to this file.
118 |
119 | ## Pre-release checklist
120 | - merge `README.md` figures to `main`, then update the links to point to these uploaded figures. We do not upload figures to PyPI, so without this step the README figure will not appear on PyPI or napari-hub.
121 | - update version numbers and links in [the microscope dependency guide](./microscope-installation-guide.md).
122 |
--------------------------------------------------------------------------------
/docs/images/HSV_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/HSV_legend.png
--------------------------------------------------------------------------------
/docs/images/JCh_Color_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/JCh_Color_legend.png
--------------------------------------------------------------------------------
/docs/images/JCh_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/JCh_legend.png
--------------------------------------------------------------------------------
/docs/images/acq_finished.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acq_finished.png
--------------------------------------------------------------------------------
/docs/images/acquire_buttons.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acquire_buttons.png
--------------------------------------------------------------------------------
/docs/images/acquisition_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acquisition_settings.png
--------------------------------------------------------------------------------
/docs/images/advanced.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/advanced.png
--------------------------------------------------------------------------------
/docs/images/cap_bg.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/cap_bg.png
--------------------------------------------------------------------------------
/docs/images/cli_structure.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/cli_structure.png
--------------------------------------------------------------------------------
/docs/images/comms_video_screenshot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/comms_video_screenshot.png
--------------------------------------------------------------------------------
/docs/images/connect_to_mm.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/connect_to_mm.png
--------------------------------------------------------------------------------
/docs/images/create_group.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_group.png
--------------------------------------------------------------------------------
/docs/images/create_group_voltage.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_group_voltage.png
--------------------------------------------------------------------------------
/docs/images/create_preset.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_preset.png
--------------------------------------------------------------------------------
/docs/images/create_preset_voltage.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_preset_voltage.png
--------------------------------------------------------------------------------
/docs/images/general_reconstruction_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/general_reconstruction_settings.png
--------------------------------------------------------------------------------
/docs/images/ideal_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/ideal_plot.png
--------------------------------------------------------------------------------
/docs/images/modulation.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/modulation.png
--------------------------------------------------------------------------------
/docs/images/no-overlay.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/no-overlay.png
--------------------------------------------------------------------------------
/docs/images/overlay-demo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/overlay-demo.png
--------------------------------------------------------------------------------
/docs/images/overlay.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/overlay.png
--------------------------------------------------------------------------------
/docs/images/phase_reconstruction_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/phase_reconstruction_settings.png
--------------------------------------------------------------------------------
/docs/images/recOrder_Fig1_Overview.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/recOrder_Fig1_Overview.png
--------------------------------------------------------------------------------
/docs/images/recOrder_plugin_logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/recOrder_plugin_logo.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_birefriengence.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_birefriengence.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_data.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_data.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_data_info.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_data_info.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_models.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_models.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_queue.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_queue.png
--------------------------------------------------------------------------------
/docs/images/run_calib.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/run_calib.png
--------------------------------------------------------------------------------
/docs/images/run_port.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/run_port.png
--------------------------------------------------------------------------------
/docs/microscope-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Microscope Installation Guide
2 |
3 | This guide will walk through a complete recOrder installation consisting of:
4 | 1. Checking pre-requisites for compatibility.
5 | 2. Installing Meadowlark DS5020 and liquid crystals.
6 | 3. Installing and launching the latest stable version of `recOrder` via `pip`.
7 | 4. Installing a compatible version of Micro-Manager and LC device drivers.
8 | 5. Connecting `recOrder` to Micro-Manager via a `pycromanager` connection.
9 |
10 | ## Compatibility Summary
11 | Before you start you will need to confirm that your system is compatible with the following software:
12 |
13 | | Software | Version |
14 | | :--- | :--- |
15 | | `recOrder` | 0.4.0 |
16 | | OS | Windows 10 |
17 | | Micro-Manager version | [2023-04-26 (160 MB)](https://download.micro-manager.org/nightly/2.0/Windows/MMSetup_64bit_2.0.1_20230426.exe) |
18 | | Meadowlark drivers | [USB driver (70 kB)](https://github.com/mehta-lab/recOrder/releases/download/0.4.0/usbdrvd.dll) |
19 | | Meadowlark PC software version | 1.08 |
20 | | Meadowlark controller firmware version | >=1.04 |
21 |
22 | ## Install Meadowlark DS5020 and liquid crystals
23 |
24 | Start by installing the Meadowlark DS5020 and liquid crystals using the software on the USB stick provided by Meadowlark. You will need to install the USB drivers and CellDrive5000.
25 |
26 | **Check your installation versions** by opening CellDrive5000 and double clicking the Meadowlark Optics logo. Confirm that **"PC software version = 1.08" and "Controller firmware version >= 1.04".**
27 |
28 | If you need to change your PC software version, follow these steps:
29 | - From "Add and remove programs", remove CellDrive5000 and "National Instruments Software".
30 | - From "Device manager", open the "Meadowlark Optics" group, right click `mlousb`, click "Uninstall device", check "Delete the driver software for this device", and click "Uninstall". Uninstall `Meadowlark Optics D5020 LC Driver` following the same steps.
31 | - Using the USB stick provided by Meadowlark, reinstall the USB drivers and CellDrive5000.
32 |
33 | ## Install recOrder software
34 |
35 | (Optional but recommended) install [anaconda](https://www.anaconda.com/products/distribution) and create a virtual environment
36 | ```
37 | conda create -y -n recOrder python=3.10
38 | conda activate recOrder
39 | ```
40 |
41 | Install `recOrder` with acquisition dependencies (napari and pycro-manager):
42 | ```
43 | pip install recOrder-napari[all]
44 | ```
45 | Check your installation:
46 | ```
47 | napari -w recOrder-napari
48 | ```
49 | should launch napari with the recOrder plugin (may take 15 seconds on a fresh installation).
50 |
51 | ## Install and configure Micro-Manager
52 |
53 | Download and install [`Micro-Manager 2.0` nightly build `20230426` (~150 MB link).](https://download.micro-manager.org/nightly/2.0/Windows/MMSetup_64bit_2.0.1_20230426.exe)
54 |
55 | **Note:** We have tested recOrder with `20230426`, but most features will work with newer builds. We recommend testing a minimal installation with `20230426` before testing with a different nightly build or additional device drivers.
56 |
57 | Before launching Micro-Manager, download the [USB driver](https://github.com/mehta-lab/recOrder/releases/download/0.4.0rc0/usbdrvd.dll) and place this file into your Micro-Manager folder (likely `C:\Program Files\Micro-Manager` or similar).
58 |
59 | Launch Micro-Manager, open `Devices > Hardware Configuration Wizard...`, and add the `MeadowlarkLC` device to your configuration. Confirm your installation by opening `Devices > Device Property Browser...` and confirming that `MeadowlarkLC` properties appear.
60 |
61 | **Upgrading users:** you will need to reinstall the Meadowlark device to your Micro-Manager configuration file, because the device driver's name has changed to from `MeadowlarkLcOpenSource` to `MeadowlarkLC`.
62 |
63 | ### Option 1 (recommended): Voltage-mode calibration installation
64 | Create a new channel group and add the `MeadowlarkLC-Voltage (V) LC-A` and `MeadowlarkLC-Voltage (V) LC-B` properties.
65 |
66 | 
67 |
68 | Add 5 presets to this group named `State0`, `State1`, `State2`, `State3`, and `State4`. You can set random voltages to add these presets, and `recOrder` will calibrate and set these voltages later.
69 |
70 | 
71 |
72 | ### Option 2 (soon deprecated): retardance mode calibration installation
73 |
74 | Create a new channel group and add the property `MeadowlarkLC-String send to -`.
75 |
76 | 
77 |
78 | Add 5 presets to this group named `State0`, `State1`, `State2`, `State3`, and `State4` and set the corresponding preset values to `state0`, `state1`, `state2`, `state3`, `state4` in the `MeadowlarkLC-String send to –`* property.
79 |
80 | 
81 |
82 | ### (Optional) Enable "Phase From BF" acquisition
83 |
84 | If you would like to reconstruct phase from brightfield, add a Micro-Manager preset with brightfield properties (e.g. moving the polarization analyzer out the light path) and give the preset a name that contains one of the following case-insensitive keywords:
85 |
86 | `["bf", "brightfield", "bright", "labelfree", "label-free", "lf", "label", "phase, "ph"]`
87 |
88 | In `recOrder` you can select this preset using the `Acquisition Settings > BF Channel` dropdown menu.
89 |
90 | ### Enable port access
91 |
92 | Finally, enable port access so that Micro-Manager can communicate with recOrder through the `pycromanager` bridge. To do so open Micro-Manager and navigate to `Tools > Options` and check the box that says `Run server on port 4827`
93 |
94 | 
95 |
96 | ## Connect `recOrder` to Micro-Manager
97 |
98 | From the `recOrder` window, click `Switch to Online`. If you see `Success`, your installation is complete and you can [proceed to the napari plugin guide](./napari-plugin-guide.md).
99 |
100 | If you you see `Failed`, check that Micro-Manager is open, check that you've enabled `Run server on port 4827`. If the connection continues to fail, report an issue with your stack trace for support.
101 |
--------------------------------------------------------------------------------
/docs/napari-plugin-guide.md:
--------------------------------------------------------------------------------
1 | # Napari Plugin Guide
2 | This guide summarizes a complete `recOrder` workflow.
3 |
4 | ## Launch `recOrder`
5 | Activate the `recOrder` environment
6 | ```
7 | conda activate recOrder
8 | ```
9 |
10 | Launch `napari` with `recOrder`
11 | ```
12 | napari -w recOrder-napari
13 | ```
14 | ## Connect to Micro-Manager
15 | Click “Connect to MM”. If the connection succeeds, proceed to calibration. If not, revisit the [microscope installation guide](./microscope-installation-guide.md).
16 |
17 | 
18 |
19 | For polarization imaging, start with the **Calibration** tab. For phase-from-brightfield imaging, you can skip the calibration and go to the **Aquisition / Reconstruction** tab.
20 |
21 | ## Calibration tab
22 | The first step in the acquisition process is to calibrate the liquid crystals and measure a background. In the `recOrder` plugin you will see the following options for controlling the calibration:
23 |
24 | 
25 |
26 |
27 | ### Prepare for a calibration
28 | Place your sample on the stage, focus on the surface of the coverslip/well, navigate to **an empty FOV**, then align the light source into **Kohler illumination** [following these steps](https://www.microscopyu.com/tutorials/kohler).
29 |
30 | ### Choose calibration parameters
31 | Browse for and choose a **Directory** where you calibration and background images will be saved.
32 |
33 | Choose a **Swing** based on the anisotropy of your sample. We recommend
34 |
35 | * Tissue Imaging: `swing = 0.1 - 0.05`
36 | * Live or fixed Cells: `swing = 0.05 – 0.03`
37 |
38 | We recommend starting with a swing of **0.1** for tissue samples and **0.05** for cells then reducing the swing to measure smaller structures. See the [calibration guide](./calibration-guide.md) for more information about this parameter and the calibration process.
39 |
40 | Choose an **Illumination Scheme** to decides how many polarization states you will calibrate and use. We recommend starting with the *4-State (Ext, 0, 60, 120)* scheme as it requires one less illumination state than the *5-State* scheme.
41 |
42 | **Calibration Mode** is set automatically, so the default value is a good place to start. Different modes allow calibrations with voltages, retardances, or hardware sequencing.
43 |
44 | The **Config Group** is set automatically to the Micro-Manager configuration group that contains the `State*` presets. You can modify this option if you have multple configuration groups with these presets.
45 |
46 | ### Run the calibration
47 | Start a calibration with **Run Calibration**.
48 |
49 | The progress bar will show the progress of calibration, and it should take less than 2 minutes on most systems.
50 |
51 | The plot shows the intensities over time during calibration. One way to diagnose an in-progress calibration is to watch the intensity plot. An ideal plot will look similar to the following:
52 |
53 | 
54 |
55 | Once finished, you will get a calibration assessment and an extinction value. The extinction value gives you a metric for calibration quality: the higher the extinction, the cleaner the light path and the greater the sensitivity of QLIPP.
56 |
57 | * **Extinction 0 – 50**: Very poor. The alignment of the universal compensator may be off or the sample chamber may be highly birefringent.
58 |
59 | * **Extinction 50 - 100**: Okay extinction, could be okay for tissue imaging and strong anisotropic structures. Most likely not suitable for cell imaging
60 |
61 | * **Extinction 100 - 200**: Good Extinction. These are the typical values we get on our microscopes.
62 |
63 | * **Extinction 200+**: Excellent. Indicates a very well-aligned and clean light path and high sensitivity of the system.
64 |
65 | For a deeper discussion of the calibration procedure, swing, and the extinction ratio, see the [calibration guide](./calibration-guide.md).
66 |
67 | ### Optional: Load Calibration
68 | The **Load Calibration** button allows earlier calibrations to be reused. Select a *polarization_calibration.txt* file and Micro-Manager's presets will be updated with these settings. `recOrder` will also collect a few images to update the extinction ratio to reflect the current condition of the light path. Once this short acquisition has finished, the user can acquire data as normal.
69 |
70 | This feature is useful if Micro-Manager and/or `recOrder` crashes. If the sample and imaging setup haven't changed, it is safe to reuse a calibration. Otherwise, if the sample or the microscope changes, we recommend performing a new calibration.
71 |
72 | ### Optional: Calculate Extinction
73 | The **Calculate Extinction** button acquires a few images and recalculates the extinction value.
74 |
75 | This feature is useful for checking if a new region of your sample requires a recalibration. If the sample or background varies as you move around the sample, the extinction will drop and you should recalibrate and acquire background images as close to the area you will be imaging as possible.
76 |
77 | ### Capture Background
78 |
79 | The **Capture Background** button will acquire several images under each of the calibrated polarization states, average them (we recommend 5), save them to specified **Background Folder Name** within the main **Directory**, then display the result in napari layers.
80 |
81 | 
82 |
83 | It is normal to see background retardance and orientation. We will use these background images to correct the data we collect our acquisitions of the sample.
84 |
85 | ### Advanced Tab
86 | The advanced tab gives the user a log output which can be useful for debugging purposes. There is a log level “debugging” which serves as a verbose output. Look here for any hints as to what may have gone wrong during calibration or acquisition.
87 |
88 | ## Acquisition / Reconstruction Tab
89 | This acquisition tab is designed to acquire and reconstruct single volumes of both phase and birefringence measurements to allow the user to test their calibration and background. We recommend this tab for quick testing and the Micro-Manager MDA acquisition for high-throughput data collection.
90 |
91 | ### Acquire Buttons
92 | 
93 |
94 | The **Retardance + Orientation**, **Phase From BF**, and **Retardance + Orientation + Phase** buttons set off Micro-Manager acquisitions that use the upcoming acquisition settings. After the acquisition is complete, these routines will set off `recOrder` reconstructions that estimate the named parameters.
95 |
96 | The **STOP** button will end the acquisition as soon as possible, though Micro-Manager acquisitions cannot always be interrupted.
97 |
98 | ### Acquisition Settings
99 | 
100 |
101 | The **Acquisition Mode** sets the target dimensions for the reconstruction. Perhaps surprisingly, all 2D reconstructions require 3D data except for **Retardance + Orientation** in **2D Acquisition Mode**. The following table summarizes the data that will be acquired when an acquisition button is pressed in **2D** and **3D** acquisition modes:
102 |
103 | | **Acquisition** \ Acquisition Mode | 2D mode | 3D mode |
104 | | :--- | :--- | :--- |
105 | | **Retardance + Orientation** | CYX data | CZYX data |
106 | | **Phase From BF** | ZYX data | ZYX data |
107 | | **Retardance + Orientation + Phase** | CZYX data | CZYX data |
108 |
109 | Unless a **Retardance + Orientation** reconstruction in **2D Acquisition Mode** is requested, `recOrder` uses Micro-Manager's z-stage to acquire 3D data. **Z Start**, **Z End**, and **Z Step** are stage settings for acquiring an image volume, relative to the current position of the stage. Values are in the stage's default units, typically in micrometers.
110 |
111 | For example, to image a 20 um thick cell the user would focus in the middle of the cell then choose
112 |
113 | * **Z Start** = -12
114 | * **Z End** = 12
115 | * **Z Step** = 0.25
116 |
117 | For phase reconstruction, the stack should have about two depths-of-focus above and below the edges of the sample because the reconstruction algorithm uses defocus information to more accurately reconstruct phase.
118 |
119 | ### General Reconstruction Settings
120 | 
121 |
122 | The **Save Directory** and **Save Name** are where the acquired data (`/_snap_/raw_data.zarr`) and reconstructions (`/_snap_/reconstruction.zarr`) will be saved.
123 |
124 | The **Background Correction** menu has several options (each with mouseover explanations):
125 | * **None**: No background correction is performed.
126 | * **Measured**: Corrects sample images with a background image acquired at an empty field of view, loaded from **Background Path**, by default the most recent background acquisition.
127 | * **Estimated**: Estimates the sample background by fitting a 2D surface to the sample images. Works well when structures are spatially distributed across the field of view and a clear background is unavailable.
128 | * **Measured + Estimated**: Applies a **Measured** background correction then an **Estimated** background correction. Use to remove residual background after the sample retardance is corrected with measured background.
129 |
130 | The remaining parameters are used by the reconstructions:
131 |
132 | * **GPU ID**: Not implemented
133 | * **Wavelength (nm)**: illumination wavelength
134 | * **Objective NA**: numerical aperture of the objective, typically found next to magnification
135 | * **Condenser NA**: numerical aperture of the condenser
136 | * **Camera Pixel Size (um)**: pixel size of the camera in micrometers (e.g. 6.5 μm)
137 | * **RI of Obj. Media**: refractive index of the objective media, typical values are 1.0 (air), 1.3 (water), 1.473 (glycerol), or 1.512 (oil)
138 | * **Magnification**: magnification of the objective
139 | * **Rotate Orientation (90 deg)**: rotates "Orientation" reconstructions by +90 degrees clockwise and saves the result, most useful when a known-orientation sample is available
140 | * **Flip Orientation**: flips "Orientation" reconstructions about napari's horizontal axis before saving the result
141 | * **Invert Phase Contrast**: inverts the phase reconstruction's contrast by flipping the positive and negative directions of the stage during the reconstruction, and saves the result
142 |
143 | ### Phase Reconstruction Settings
144 | 
145 |
146 | These parameters are used only by phase reconstructions
147 |
148 | * **Z Padding**: The number of slices to pad on either end of the stack, necessary if the sample is not fully out of focus on either end of the stack
149 | * **Regularizer**: Choose "Tikhonov", the "TV" regularizer is not implemented
150 | * **Strength**: The Tikhonov regularization strength, too small/large will result in reconstructions that are too noisy/smooth
151 |
152 | The acquired data will then be displayed in napari layers. Note that phase reconstruction is more computationally expensive and may take several minutes depending on your system.
153 |
154 | Examples of acquiring 2D birefringence data (kidney tissue) with this snap method are below:
155 |
156 | 
157 |
158 | ### Recreating reconstructions
159 | `recOrder`'s GUI acquires data from Micro-Manager, reads the GUI to generate a configuration file, then uses a CLI to reconstruct the acquired data with the configuration file, which makes all reconstructions exactly reproducible via a CLI. See the terminal that started napari for a log of the exact CLI commands that will reproduce the results in the napari window.
160 |
161 | See the [reconstruction guide](./reconstruction-guide.md) for CLI usage instructions.
162 |
163 | ## Reconstruction Tab
164 | The **Reconstruction** tab is designed to reconstruct `birefriengence, phase, birefrignence with phase, and flurescenece` datasets that have been either acquired or coverted to `.zarr` store as well as acquisitions that are in progress.
165 |
166 | 
167 |
168 | The **Input Store** and **Output Directory** point to the input and output `.zarr` data locations. Once an Input Store is selected some metadata parameters can be viewed by hovering the cursor over the `info label` ⓘ.
169 |
170 | 
171 |
172 | A `Model` defines the reconstruction parameters. Multiple models can be run against a dataset with varying parameters. The model generates a configuration file `.yml`, then uses the CLI to reconstruct the data with the configuration file, which makes all reconstructions exactly reproducible via a CLI.
173 | * **New**: Builds a model based on the `Checkbox` selection.
174 | * **Load**: Allows a model to be imported using a previous reconstruction `.yml` file.
175 | * **Clear**: This will clear all defined models.
176 |
177 | 
178 |
179 | Once a `New` model is built, it is pre-populated with default values that can be accessed by clicking on the ► icon and the parameters can be changed as required.
180 | See the [reconstruction guide](./reconstruction-guide.md) for further information on the parameters.
181 |
182 | 
183 |
184 | Once the **RUN** button is triggered, the reconstruction will proceed based on the defined model(s) concurrently.
185 |
186 | > [!CAUTION]
187 | > Since the models run concurrently, it is the users responsibility to manage compute resources accordingly on a local or SLURM system.
188 |
189 | The `Reconstruction Queue` section will display the progress of the reconstruction in the form of text output. Once a reconstruction finishes the queue will self clear. Only in the case of any issues or error that are encountered the entry will remain.
190 |
191 | Once the reconstruction processing finishes, based on the option `Show after Reconstruction` the reconstructed images will show up in the napari viewer.
192 |
193 | ## Visualizations
194 | When an **Orientation*** layer appears at the top of the layers list, `recOrder` will automatically color it with an HSV color map that indicates the orientation.
195 |
196 | If the **Orientation*** layer has a matching **Retardance*** layer in the layer list, a **BirefringenceOverlay*** layer that only shows orientation colors in regions with large retardance is generated. This overlay is computed lazily (when the slider moves), and this computation can be turned off by hiding the layer (eyeball in the layer list).
197 |
198 | 
199 |
200 | If the **BirefringenceOverlay*** needs to be regenerated, an **Orientation*** layer can be dragged to the top of the layer list:
201 | 
202 |
203 | The **Visualization** tab shows the mapping between HSV colors and the orientation, and the **Overlay Retardance Maximum** slider controls the mapping between retardance values and saturated colors in the overlay.
204 | 
--------------------------------------------------------------------------------
/docs/reconstruction-guide.md:
--------------------------------------------------------------------------------
1 | # Automating reconstructions
2 |
3 | `recOrder` uses a configuration-file-based command-line interface (CLI) to perform all reconstructions.
4 |
5 | ## Preparing your data
6 |
7 | `recOrder` is compatible with OME-Zarr, a chunked next generation file format with an [open specification](https://ngff.openmicroscopy.org/0.4/). All acquisitions completed with the `recOrder` plugin will be automatically converted to `.zarr` format, and existing data can be converted using `iohub`'s `convert` utility.
8 |
9 | Inside a `recOrder` environment, convert a Micro-Manager TIFF sequence, OME-TIFF, or pycromanager NDTiff dataset with
10 | ```
11 | iohub convert `
12 | -i ./dataset/ `
13 | -o ./dataset.zarr
14 | ```
15 |
16 | ## How can I use `recOrder`'s CLI to perform reconstructions?
17 | `recOrder`'s CLI is summarized in the following figure:
18 |
52 |
53 | The extinction ratio is the ratio of the largest and smallest intensities that the imaging system can transmit above background, which is most easily expressed in terms of $I_{\text{mod}}$ and $I_{\text{leak}}$
54 | $$\text{Extinction Ratio} = \frac{I_{\text{mod}} + I_{\text{leak}}}{I_{\text{leak}}}.\qquad\qquad (3)$$
55 |
56 | Substituting Eqs. (1) and (2) into Eq. (3) gives the extinction ratio in terms of the measured intensities
57 | $$\text{Extinction Ratio} = \frac{1}{\sin^2(\pi S)}\frac{I_{\text{ellip}}(S) - I_{\text{ext}}}{I_{\text{ext}} - I_{\text{bl}}} + 1.$$
58 |
59 | ## Summary: `recOrder`'s step-by-step calibration procedure
60 | 1. Close the shutter, measure the black level, then reopen the shutter.
61 | 2. Find the extinction state by finding voltages that minimize the intensity that reaches the camera.
62 | 3. Use the swing value to immediately find the first elliptical state, and record the intensity on the camera.
63 | 4. For each remaining elliptical state, keep the polarization orientation fixed and optimize the voltages to match the intensity of the first elliptical state.
64 | 5. Store the voltages and calculate the extinction ratio.
--------------------------------------------------------------------------------
/docs/data-schema.md:
--------------------------------------------------------------------------------
1 | # Data schema
2 |
3 | This document defines the standard for data acquired with `recOrder`.
4 |
5 | ## Raw directory organization
6 |
7 | Currently, we structure raw data in the following hierarchy:
8 |
9 | ```text
10 | working_directory/ # commonly YYYY_MM_DD_exp_name, but not enforced
11 | ├── polarization_calibration_0.txt
12 | │ ...
13 | ├── polarization_calibration_.txt # i calibration repeats
14 | │
15 | ├── bg_0
16 | │ ...
17 | ├── bg_ # j background repeats
18 | │ ├── background.zarr
19 | │ ├── polarization_calibration.txt # copied into each bg folder
20 | │ ├── reconstruction.zarr
21 | │ ├── reconstruction_settings.yml # for use with `recorder reconstruct`
22 | │ └── transfer_function.zarr # for use with `recorder apply-inv-tf`
23 | │
24 | ├── _snap_0
25 | ├── _snap_1
26 | │ ├── raw_data.zarr
27 | │ ├── reconstruction.zarr
28 | │ ├── reconstruction_settings.yml
29 | │ └── transfer_function.zarr
30 | │ ...
31 | ├── _snap_ # k repeats with the first acquisition name
32 | │ ├── raw_data.zarr
33 | │ ├── reconstruction.zarr
34 | │ ├── reconstruction_settings.yml
35 | │ └── transfer_function.zarr
36 | │ ...
37 | │
38 | ├── _snap_0 # l different acquisition names
39 | │ ...
40 | ├── _snap_ # m repeats for this acquisition name
41 | ├── raw_data.zarr
42 | ├── reconstruction.zarr
43 | ├── reconstruction_settings.yml
44 | └── transfer_function.zarr
45 | ```
46 |
47 | Each `.zarr` contains an [OME-NGFF v0.4](https://ngff.openmicroscopy.org/0.4/) in HCS format with a single field of view.
--------------------------------------------------------------------------------
/docs/development-guide.md:
--------------------------------------------------------------------------------
1 | # `recOrder` development guide
2 |
3 | ## Install `recOrder` for development
4 |
5 | 1. Install [conda](https://github.com/conda-forge/miniforge) and create a virtual environment:
6 |
7 | ```sh
8 | conda create -y -n recOrder python=3.10
9 | conda activate recOrder
10 | ```
11 |
12 | 2. Clone the `recOrder` directory:
13 |
14 | ```sh
15 | git clone https://github.com/mehta-lab/recOrder.git
16 | ```
17 |
18 | 3. Install `recOrder` in editable mode with development dependencies
19 |
20 | ```sh
21 | cd recOrder
22 | pip install -e ".[all,dev]"
23 | ```
24 |
25 | 4. Optionally, for the co-development of [`waveorder`](https://github.com/mehta-lab/waveorder) and `recOrder`:
26 |
27 | > Note that `pip` will raise an 'error' complaining that the dependency of `recOrder-napari` has been broken if you do the following.
28 | > This does not affect the installation, but can be suppressed by removing [this line](https://github.com/mehta-lab/recOrder/blob/5bc9314a9bacf6f4e235eaffb06c297cf20e4b65/setup.cfg#L40) before installing `waveorder`.
29 | > (Just remember to revert the change afterwards!)
30 | > We expect a nicer behavior to be possible once we release a stable version of `waveorder`.
31 |
32 | ```sh
33 | cd # where you want to clone the repo
34 | git clone https://github.com/mehta-lab/waveorder.git
35 | pip install ./waveorder -e ".[dev]"
36 | ```
37 |
38 | > Importing from a local and/or editable package can cause issues with static code analyzers such due to the absence of the source code from the paths they expect.
39 | > VS Code users can refer to [this guide](https://github.com/microsoft/pylance-release/blob/main/TROUBLESHOOTING.md#common-questions-and-issues) to resolve typing warnings.
40 |
41 | ## Set up a development environment
42 |
43 | ### Code linting
44 |
45 | We are not currently specifying a code linter as most modern Python code editors already have their own. If not, add a plugin to your editor to help catch bugs pre-commit!
46 |
47 | ### Code formatting
48 |
49 | We use `black` to format Python code, and a specific version is installed as a development dependency. Use the `black` in the `recOrder` virtual environment, either from commandline or the editor of your choice.
50 |
51 | > *VS Code users*: Install the [Black Formatter](https://marketplace.visualstudio.com/items?itemName=ms-python.black-formatter) plugin. Press `^/⌘ ⇧ P` and type 'format document with...', choose the Black Formatter and start formatting!
52 |
53 | ### Docstring style
54 |
55 | The [NumPy style](https://numpydoc.readthedocs.io/en/latest/format.html) docstrings are used in `recOrder`.
56 |
57 | > *VS Code users*: [this popular plugin](https://marketplace.visualstudio.com/items?itemName=njpwerner.autodocstring) helps auto-generate most popular docstring styles (including `numpydoc`).
58 |
59 | ## Run automated tests
60 |
61 | From within the `recOrder` directory run:
62 |
63 | ```sh
64 | pytest
65 | ```
66 |
67 | Running `pytest` for the first time will download ~50 MB of test data from Zenodo, and subsequent runs will reuse the downloaded data.
68 |
69 | ## Run manual tests
70 |
71 | Although many of `recOrder`'s tests are automated, many features require manual testing. The following is a summary of features that need to be tested manually before release:
72 |
73 | * Install a compatible version of Micro-Manager and check that `recOrder` can connect.
74 | * Perform calibrations with and without an ROI; with and without a shutter configured in Micro-Manager, in 4- and 5-state modes; and in MM-Voltage, MM-Retardance, and DAC modes (if the TriggerScope is available).
75 | * Test "Load Calibration" and "Calculate Extinction" buttons.
76 | * Test "Capture Background" button.
77 | * Test the "Acquire Birefringence" button on a background FOV. Does a background-corrected background acquisition give random orientations?
78 | * Test the four "Acquire" buttons with varied combinations of 2D/3D, background correction settings, "Phase from BF" checkbox, and regularization parameters.
79 | * Use the data you collected to test "Offline" mode reconstructions with varied combinations of parameters.
80 |
81 | ## GUI development
82 |
83 | We use `QT Creator` for large parts of `recOrder`'s GUI. To modify the GUI, install `QT Creator` from [its website](https://www.qt.io/product/development-tools) or with `brew install --cask qt-creator`
84 |
85 | Open `./recOrder/plugin/gui.ui` in `QT Creator` and make your changes.
86 |
87 | Next, convert the `.ui` to a `.py` file with:
88 |
89 | ```sh
90 | pyuic5 -x gui.ui -o gui.py
91 | ```
92 |
93 | Note: `pyuic5` is installed alongside `PyQt5`, so you can expect to find it installed in your `recOrder` conda environement.
94 |
95 | Finally, change the `gui.py` file's to import `qtpy` instead of `PyQt5` to adhere to [napari plugin best practices](https://napari.org/stable/plugins/best_practices.html#don-t-include-pyside2-or-pyqt5-in-your-plugin-s-dependencies).
96 | On macOS, you can modify the file in place with:
97 |
98 | ```sh
99 | sed -i '' 's/from PyQt5/from qtpy/g' gui.py
100 | ```
101 |
102 | > This is specific for BSD `sed`, omit `''` with GNU.
103 |
104 | Note: although much of the GUI is specified in the generated `recOrder_ui.py` file, the `main_widget.py` file makes extensive modifications to the GUI.
105 |
106 | ## Make `git blame` ignore formatting commits
107 |
108 | **Note:** `git --version` must be `>=2.23` to use this feature.
109 |
110 | If you would like `git blame` to ignore formatting commits, run this line:
111 |
112 | ```sh
113 | git config --global blame.ignoreRevsFile .git-blame-ignore-revs
114 | ```
115 |
116 | The `\.git-blame-ignore-revs` file contains a list of commit hashes corresponding to formatting commits.
117 | If you make a formatting commit, please add the commit's hash to this file.
118 |
119 | ## Pre-release checklist
120 | - merge `README.md` figures to `main`, then update the links to point to these uploaded figures. We do not upload figures to PyPI, so without this step the README figure will not appear on PyPI or napari-hub.
121 | - update version numbers and links in [the microscope dependency guide](./microscope-installation-guide.md).
122 |
--------------------------------------------------------------------------------
/docs/images/HSV_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/HSV_legend.png
--------------------------------------------------------------------------------
/docs/images/JCh_Color_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/JCh_Color_legend.png
--------------------------------------------------------------------------------
/docs/images/JCh_legend.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/JCh_legend.png
--------------------------------------------------------------------------------
/docs/images/acq_finished.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acq_finished.png
--------------------------------------------------------------------------------
/docs/images/acquire_buttons.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acquire_buttons.png
--------------------------------------------------------------------------------
/docs/images/acquisition_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/acquisition_settings.png
--------------------------------------------------------------------------------
/docs/images/advanced.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/advanced.png
--------------------------------------------------------------------------------
/docs/images/cap_bg.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/cap_bg.png
--------------------------------------------------------------------------------
/docs/images/cli_structure.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/cli_structure.png
--------------------------------------------------------------------------------
/docs/images/comms_video_screenshot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/comms_video_screenshot.png
--------------------------------------------------------------------------------
/docs/images/connect_to_mm.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/connect_to_mm.png
--------------------------------------------------------------------------------
/docs/images/create_group.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_group.png
--------------------------------------------------------------------------------
/docs/images/create_group_voltage.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_group_voltage.png
--------------------------------------------------------------------------------
/docs/images/create_preset.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_preset.png
--------------------------------------------------------------------------------
/docs/images/create_preset_voltage.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/create_preset_voltage.png
--------------------------------------------------------------------------------
/docs/images/general_reconstruction_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/general_reconstruction_settings.png
--------------------------------------------------------------------------------
/docs/images/ideal_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/ideal_plot.png
--------------------------------------------------------------------------------
/docs/images/modulation.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/modulation.png
--------------------------------------------------------------------------------
/docs/images/no-overlay.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/no-overlay.png
--------------------------------------------------------------------------------
/docs/images/overlay-demo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/overlay-demo.png
--------------------------------------------------------------------------------
/docs/images/overlay.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/overlay.png
--------------------------------------------------------------------------------
/docs/images/phase_reconstruction_settings.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/phase_reconstruction_settings.png
--------------------------------------------------------------------------------
/docs/images/recOrder_Fig1_Overview.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/recOrder_Fig1_Overview.png
--------------------------------------------------------------------------------
/docs/images/recOrder_plugin_logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/recOrder_plugin_logo.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_birefriengence.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_birefriengence.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_data.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_data.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_data_info.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_data_info.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_models.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_models.png
--------------------------------------------------------------------------------
/docs/images/reconstruction_queue.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/reconstruction_queue.png
--------------------------------------------------------------------------------
/docs/images/run_calib.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/run_calib.png
--------------------------------------------------------------------------------
/docs/images/run_port.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/docs/images/run_port.png
--------------------------------------------------------------------------------
/docs/microscope-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Microscope Installation Guide
2 |
3 | This guide will walk through a complete recOrder installation consisting of:
4 | 1. Checking pre-requisites for compatibility.
5 | 2. Installing Meadowlark DS5020 and liquid crystals.
6 | 3. Installing and launching the latest stable version of `recOrder` via `pip`.
7 | 4. Installing a compatible version of Micro-Manager and LC device drivers.
8 | 5. Connecting `recOrder` to Micro-Manager via a `pycromanager` connection.
9 |
10 | ## Compatibility Summary
11 | Before you start you will need to confirm that your system is compatible with the following software:
12 |
13 | | Software | Version |
14 | | :--- | :--- |
15 | | `recOrder` | 0.4.0 |
16 | | OS | Windows 10 |
17 | | Micro-Manager version | [2023-04-26 (160 MB)](https://download.micro-manager.org/nightly/2.0/Windows/MMSetup_64bit_2.0.1_20230426.exe) |
18 | | Meadowlark drivers | [USB driver (70 kB)](https://github.com/mehta-lab/recOrder/releases/download/0.4.0/usbdrvd.dll) |
19 | | Meadowlark PC software version | 1.08 |
20 | | Meadowlark controller firmware version | >=1.04 |
21 |
22 | ## Install Meadowlark DS5020 and liquid crystals
23 |
24 | Start by installing the Meadowlark DS5020 and liquid crystals using the software on the USB stick provided by Meadowlark. You will need to install the USB drivers and CellDrive5000.
25 |
26 | **Check your installation versions** by opening CellDrive5000 and double clicking the Meadowlark Optics logo. Confirm that **"PC software version = 1.08" and "Controller firmware version >= 1.04".**
27 |
28 | If you need to change your PC software version, follow these steps:
29 | - From "Add and remove programs", remove CellDrive5000 and "National Instruments Software".
30 | - From "Device manager", open the "Meadowlark Optics" group, right click `mlousb`, click "Uninstall device", check "Delete the driver software for this device", and click "Uninstall". Uninstall `Meadowlark Optics D5020 LC Driver` following the same steps.
31 | - Using the USB stick provided by Meadowlark, reinstall the USB drivers and CellDrive5000.
32 |
33 | ## Install recOrder software
34 |
35 | (Optional but recommended) install [anaconda](https://www.anaconda.com/products/distribution) and create a virtual environment
36 | ```
37 | conda create -y -n recOrder python=3.10
38 | conda activate recOrder
39 | ```
40 |
41 | Install `recOrder` with acquisition dependencies (napari and pycro-manager):
42 | ```
43 | pip install recOrder-napari[all]
44 | ```
45 | Check your installation:
46 | ```
47 | napari -w recOrder-napari
48 | ```
49 | should launch napari with the recOrder plugin (may take 15 seconds on a fresh installation).
50 |
51 | ## Install and configure Micro-Manager
52 |
53 | Download and install [`Micro-Manager 2.0` nightly build `20230426` (~150 MB link).](https://download.micro-manager.org/nightly/2.0/Windows/MMSetup_64bit_2.0.1_20230426.exe)
54 |
55 | **Note:** We have tested recOrder with `20230426`, but most features will work with newer builds. We recommend testing a minimal installation with `20230426` before testing with a different nightly build or additional device drivers.
56 |
57 | Before launching Micro-Manager, download the [USB driver](https://github.com/mehta-lab/recOrder/releases/download/0.4.0rc0/usbdrvd.dll) and place this file into your Micro-Manager folder (likely `C:\Program Files\Micro-Manager` or similar).
58 |
59 | Launch Micro-Manager, open `Devices > Hardware Configuration Wizard...`, and add the `MeadowlarkLC` device to your configuration. Confirm your installation by opening `Devices > Device Property Browser...` and confirming that `MeadowlarkLC` properties appear.
60 |
61 | **Upgrading users:** you will need to reinstall the Meadowlark device to your Micro-Manager configuration file, because the device driver's name has changed to from `MeadowlarkLcOpenSource` to `MeadowlarkLC`.
62 |
63 | ### Option 1 (recommended): Voltage-mode calibration installation
64 | Create a new channel group and add the `MeadowlarkLC-Voltage (V) LC-A` and `MeadowlarkLC-Voltage (V) LC-B` properties.
65 |
66 | 
67 |
68 | Add 5 presets to this group named `State0`, `State1`, `State2`, `State3`, and `State4`. You can set random voltages to add these presets, and `recOrder` will calibrate and set these voltages later.
69 |
70 | 
71 |
72 | ### Option 2 (soon deprecated): retardance mode calibration installation
73 |
74 | Create a new channel group and add the property `MeadowlarkLC-String send to -`.
75 |
76 | 
77 |
78 | Add 5 presets to this group named `State0`, `State1`, `State2`, `State3`, and `State4` and set the corresponding preset values to `state0`, `state1`, `state2`, `state3`, `state4` in the `MeadowlarkLC-String send to –`* property.
79 |
80 | 
81 |
82 | ### (Optional) Enable "Phase From BF" acquisition
83 |
84 | If you would like to reconstruct phase from brightfield, add a Micro-Manager preset with brightfield properties (e.g. moving the polarization analyzer out the light path) and give the preset a name that contains one of the following case-insensitive keywords:
85 |
86 | `["bf", "brightfield", "bright", "labelfree", "label-free", "lf", "label", "phase, "ph"]`
87 |
88 | In `recOrder` you can select this preset using the `Acquisition Settings > BF Channel` dropdown menu.
89 |
90 | ### Enable port access
91 |
92 | Finally, enable port access so that Micro-Manager can communicate with recOrder through the `pycromanager` bridge. To do so open Micro-Manager and navigate to `Tools > Options` and check the box that says `Run server on port 4827`
93 |
94 | 
95 |
96 | ## Connect `recOrder` to Micro-Manager
97 |
98 | From the `recOrder` window, click `Switch to Online`. If you see `Success`, your installation is complete and you can [proceed to the napari plugin guide](./napari-plugin-guide.md).
99 |
100 | If you you see `Failed`, check that Micro-Manager is open, check that you've enabled `Run server on port 4827`. If the connection continues to fail, report an issue with your stack trace for support.
101 |
--------------------------------------------------------------------------------
/docs/napari-plugin-guide.md:
--------------------------------------------------------------------------------
1 | # Napari Plugin Guide
2 | This guide summarizes a complete `recOrder` workflow.
3 |
4 | ## Launch `recOrder`
5 | Activate the `recOrder` environment
6 | ```
7 | conda activate recOrder
8 | ```
9 |
10 | Launch `napari` with `recOrder`
11 | ```
12 | napari -w recOrder-napari
13 | ```
14 | ## Connect to Micro-Manager
15 | Click “Connect to MM”. If the connection succeeds, proceed to calibration. If not, revisit the [microscope installation guide](./microscope-installation-guide.md).
16 |
17 | 
18 |
19 | For polarization imaging, start with the **Calibration** tab. For phase-from-brightfield imaging, you can skip the calibration and go to the **Aquisition / Reconstruction** tab.
20 |
21 | ## Calibration tab
22 | The first step in the acquisition process is to calibrate the liquid crystals and measure a background. In the `recOrder` plugin you will see the following options for controlling the calibration:
23 |
24 | 
25 |
26 |
27 | ### Prepare for a calibration
28 | Place your sample on the stage, focus on the surface of the coverslip/well, navigate to **an empty FOV**, then align the light source into **Kohler illumination** [following these steps](https://www.microscopyu.com/tutorials/kohler).
29 |
30 | ### Choose calibration parameters
31 | Browse for and choose a **Directory** where you calibration and background images will be saved.
32 |
33 | Choose a **Swing** based on the anisotropy of your sample. We recommend
34 |
35 | * Tissue Imaging: `swing = 0.1 - 0.05`
36 | * Live or fixed Cells: `swing = 0.05 – 0.03`
37 |
38 | We recommend starting with a swing of **0.1** for tissue samples and **0.05** for cells then reducing the swing to measure smaller structures. See the [calibration guide](./calibration-guide.md) for more information about this parameter and the calibration process.
39 |
40 | Choose an **Illumination Scheme** to decides how many polarization states you will calibrate and use. We recommend starting with the *4-State (Ext, 0, 60, 120)* scheme as it requires one less illumination state than the *5-State* scheme.
41 |
42 | **Calibration Mode** is set automatically, so the default value is a good place to start. Different modes allow calibrations with voltages, retardances, or hardware sequencing.
43 |
44 | The **Config Group** is set automatically to the Micro-Manager configuration group that contains the `State*` presets. You can modify this option if you have multple configuration groups with these presets.
45 |
46 | ### Run the calibration
47 | Start a calibration with **Run Calibration**.
48 |
49 | The progress bar will show the progress of calibration, and it should take less than 2 minutes on most systems.
50 |
51 | The plot shows the intensities over time during calibration. One way to diagnose an in-progress calibration is to watch the intensity plot. An ideal plot will look similar to the following:
52 |
53 | 
54 |
55 | Once finished, you will get a calibration assessment and an extinction value. The extinction value gives you a metric for calibration quality: the higher the extinction, the cleaner the light path and the greater the sensitivity of QLIPP.
56 |
57 | * **Extinction 0 – 50**: Very poor. The alignment of the universal compensator may be off or the sample chamber may be highly birefringent.
58 |
59 | * **Extinction 50 - 100**: Okay extinction, could be okay for tissue imaging and strong anisotropic structures. Most likely not suitable for cell imaging
60 |
61 | * **Extinction 100 - 200**: Good Extinction. These are the typical values we get on our microscopes.
62 |
63 | * **Extinction 200+**: Excellent. Indicates a very well-aligned and clean light path and high sensitivity of the system.
64 |
65 | For a deeper discussion of the calibration procedure, swing, and the extinction ratio, see the [calibration guide](./calibration-guide.md).
66 |
67 | ### Optional: Load Calibration
68 | The **Load Calibration** button allows earlier calibrations to be reused. Select a *polarization_calibration.txt* file and Micro-Manager's presets will be updated with these settings. `recOrder` will also collect a few images to update the extinction ratio to reflect the current condition of the light path. Once this short acquisition has finished, the user can acquire data as normal.
69 |
70 | This feature is useful if Micro-Manager and/or `recOrder` crashes. If the sample and imaging setup haven't changed, it is safe to reuse a calibration. Otherwise, if the sample or the microscope changes, we recommend performing a new calibration.
71 |
72 | ### Optional: Calculate Extinction
73 | The **Calculate Extinction** button acquires a few images and recalculates the extinction value.
74 |
75 | This feature is useful for checking if a new region of your sample requires a recalibration. If the sample or background varies as you move around the sample, the extinction will drop and you should recalibrate and acquire background images as close to the area you will be imaging as possible.
76 |
77 | ### Capture Background
78 |
79 | The **Capture Background** button will acquire several images under each of the calibrated polarization states, average them (we recommend 5), save them to specified **Background Folder Name** within the main **Directory**, then display the result in napari layers.
80 |
81 | 
82 |
83 | It is normal to see background retardance and orientation. We will use these background images to correct the data we collect our acquisitions of the sample.
84 |
85 | ### Advanced Tab
86 | The advanced tab gives the user a log output which can be useful for debugging purposes. There is a log level “debugging” which serves as a verbose output. Look here for any hints as to what may have gone wrong during calibration or acquisition.
87 |
88 | ## Acquisition / Reconstruction Tab
89 | This acquisition tab is designed to acquire and reconstruct single volumes of both phase and birefringence measurements to allow the user to test their calibration and background. We recommend this tab for quick testing and the Micro-Manager MDA acquisition for high-throughput data collection.
90 |
91 | ### Acquire Buttons
92 | 
93 |
94 | The **Retardance + Orientation**, **Phase From BF**, and **Retardance + Orientation + Phase** buttons set off Micro-Manager acquisitions that use the upcoming acquisition settings. After the acquisition is complete, these routines will set off `recOrder` reconstructions that estimate the named parameters.
95 |
96 | The **STOP** button will end the acquisition as soon as possible, though Micro-Manager acquisitions cannot always be interrupted.
97 |
98 | ### Acquisition Settings
99 | 
100 |
101 | The **Acquisition Mode** sets the target dimensions for the reconstruction. Perhaps surprisingly, all 2D reconstructions require 3D data except for **Retardance + Orientation** in **2D Acquisition Mode**. The following table summarizes the data that will be acquired when an acquisition button is pressed in **2D** and **3D** acquisition modes:
102 |
103 | | **Acquisition** \ Acquisition Mode | 2D mode | 3D mode |
104 | | :--- | :--- | :--- |
105 | | **Retardance + Orientation** | CYX data | CZYX data |
106 | | **Phase From BF** | ZYX data | ZYX data |
107 | | **Retardance + Orientation + Phase** | CZYX data | CZYX data |
108 |
109 | Unless a **Retardance + Orientation** reconstruction in **2D Acquisition Mode** is requested, `recOrder` uses Micro-Manager's z-stage to acquire 3D data. **Z Start**, **Z End**, and **Z Step** are stage settings for acquiring an image volume, relative to the current position of the stage. Values are in the stage's default units, typically in micrometers.
110 |
111 | For example, to image a 20 um thick cell the user would focus in the middle of the cell then choose
112 |
113 | * **Z Start** = -12
114 | * **Z End** = 12
115 | * **Z Step** = 0.25
116 |
117 | For phase reconstruction, the stack should have about two depths-of-focus above and below the edges of the sample because the reconstruction algorithm uses defocus information to more accurately reconstruct phase.
118 |
119 | ### General Reconstruction Settings
120 | 
121 |
122 | The **Save Directory** and **Save Name** are where the acquired data (`/_snap_/raw_data.zarr`) and reconstructions (`/_snap_/reconstruction.zarr`) will be saved.
123 |
124 | The **Background Correction** menu has several options (each with mouseover explanations):
125 | * **None**: No background correction is performed.
126 | * **Measured**: Corrects sample images with a background image acquired at an empty field of view, loaded from **Background Path**, by default the most recent background acquisition.
127 | * **Estimated**: Estimates the sample background by fitting a 2D surface to the sample images. Works well when structures are spatially distributed across the field of view and a clear background is unavailable.
128 | * **Measured + Estimated**: Applies a **Measured** background correction then an **Estimated** background correction. Use to remove residual background after the sample retardance is corrected with measured background.
129 |
130 | The remaining parameters are used by the reconstructions:
131 |
132 | * **GPU ID**: Not implemented
133 | * **Wavelength (nm)**: illumination wavelength
134 | * **Objective NA**: numerical aperture of the objective, typically found next to magnification
135 | * **Condenser NA**: numerical aperture of the condenser
136 | * **Camera Pixel Size (um)**: pixel size of the camera in micrometers (e.g. 6.5 μm)
137 | * **RI of Obj. Media**: refractive index of the objective media, typical values are 1.0 (air), 1.3 (water), 1.473 (glycerol), or 1.512 (oil)
138 | * **Magnification**: magnification of the objective
139 | * **Rotate Orientation (90 deg)**: rotates "Orientation" reconstructions by +90 degrees clockwise and saves the result, most useful when a known-orientation sample is available
140 | * **Flip Orientation**: flips "Orientation" reconstructions about napari's horizontal axis before saving the result
141 | * **Invert Phase Contrast**: inverts the phase reconstruction's contrast by flipping the positive and negative directions of the stage during the reconstruction, and saves the result
142 |
143 | ### Phase Reconstruction Settings
144 | 
145 |
146 | These parameters are used only by phase reconstructions
147 |
148 | * **Z Padding**: The number of slices to pad on either end of the stack, necessary if the sample is not fully out of focus on either end of the stack
149 | * **Regularizer**: Choose "Tikhonov", the "TV" regularizer is not implemented
150 | * **Strength**: The Tikhonov regularization strength, too small/large will result in reconstructions that are too noisy/smooth
151 |
152 | The acquired data will then be displayed in napari layers. Note that phase reconstruction is more computationally expensive and may take several minutes depending on your system.
153 |
154 | Examples of acquiring 2D birefringence data (kidney tissue) with this snap method are below:
155 |
156 | 
157 |
158 | ### Recreating reconstructions
159 | `recOrder`'s GUI acquires data from Micro-Manager, reads the GUI to generate a configuration file, then uses a CLI to reconstruct the acquired data with the configuration file, which makes all reconstructions exactly reproducible via a CLI. See the terminal that started napari for a log of the exact CLI commands that will reproduce the results in the napari window.
160 |
161 | See the [reconstruction guide](./reconstruction-guide.md) for CLI usage instructions.
162 |
163 | ## Reconstruction Tab
164 | The **Reconstruction** tab is designed to reconstruct `birefriengence, phase, birefrignence with phase, and flurescenece` datasets that have been either acquired or coverted to `.zarr` store as well as acquisitions that are in progress.
165 |
166 | 
167 |
168 | The **Input Store** and **Output Directory** point to the input and output `.zarr` data locations. Once an Input Store is selected some metadata parameters can be viewed by hovering the cursor over the `info label` ⓘ.
169 |
170 | 
171 |
172 | A `Model` defines the reconstruction parameters. Multiple models can be run against a dataset with varying parameters. The model generates a configuration file `.yml`, then uses the CLI to reconstruct the data with the configuration file, which makes all reconstructions exactly reproducible via a CLI.
173 | * **New**: Builds a model based on the `Checkbox` selection.
174 | * **Load**: Allows a model to be imported using a previous reconstruction `.yml` file.
175 | * **Clear**: This will clear all defined models.
176 |
177 | 
178 |
179 | Once a `New` model is built, it is pre-populated with default values that can be accessed by clicking on the ► icon and the parameters can be changed as required.
180 | See the [reconstruction guide](./reconstruction-guide.md) for further information on the parameters.
181 |
182 | 
183 |
184 | Once the **RUN** button is triggered, the reconstruction will proceed based on the defined model(s) concurrently.
185 |
186 | > [!CAUTION]
187 | > Since the models run concurrently, it is the users responsibility to manage compute resources accordingly on a local or SLURM system.
188 |
189 | The `Reconstruction Queue` section will display the progress of the reconstruction in the form of text output. Once a reconstruction finishes the queue will self clear. Only in the case of any issues or error that are encountered the entry will remain.
190 |
191 | Once the reconstruction processing finishes, based on the option `Show after Reconstruction` the reconstructed images will show up in the napari viewer.
192 |
193 | ## Visualizations
194 | When an **Orientation*** layer appears at the top of the layers list, `recOrder` will automatically color it with an HSV color map that indicates the orientation.
195 |
196 | If the **Orientation*** layer has a matching **Retardance*** layer in the layer list, a **BirefringenceOverlay*** layer that only shows orientation colors in regions with large retardance is generated. This overlay is computed lazily (when the slider moves), and this computation can be turned off by hiding the layer (eyeball in the layer list).
197 |
198 | 
199 |
200 | If the **BirefringenceOverlay*** needs to be regenerated, an **Orientation*** layer can be dragged to the top of the layer list:
201 | 
202 |
203 | The **Visualization** tab shows the mapping between HSV colors and the orientation, and the **Overlay Retardance Maximum** slider controls the mapping between retardance values and saturated colors in the overlay.
204 | 
--------------------------------------------------------------------------------
/docs/reconstruction-guide.md:
--------------------------------------------------------------------------------
1 | # Automating reconstructions
2 |
3 | `recOrder` uses a configuration-file-based command-line interface (CLI) to perform all reconstructions.
4 |
5 | ## Preparing your data
6 |
7 | `recOrder` is compatible with OME-Zarr, a chunked next generation file format with an [open specification](https://ngff.openmicroscopy.org/0.4/). All acquisitions completed with the `recOrder` plugin will be automatically converted to `.zarr` format, and existing data can be converted using `iohub`'s `convert` utility.
8 |
9 | Inside a `recOrder` environment, convert a Micro-Manager TIFF sequence, OME-TIFF, or pycromanager NDTiff dataset with
10 | ```
11 | iohub convert `
12 | -i ./dataset/ `
13 | -o ./dataset.zarr
14 | ```
15 |
16 | ## How can I use `recOrder`'s CLI to perform reconstructions?
17 | `recOrder`'s CLI is summarized in the following figure:
18 |  19 |
20 | The main command `reconstruct` command is composed of two subcommands: `compute-tf` and `apply-inv-tf`.
21 |
22 | A reconstruction can be performed with a single `reconstruct` call. For example:
23 | ```
24 | recorder reconstruct `
25 | -i ./data.zarr/*/*/* `
26 | -c ./config.yml `
27 | -o ./reconstruction.zarr
28 | ```
29 | Equivalently, a reconstruction can be performed with a `compute-tf` call followed by an `apply-inv-tf` call. For example:
30 | ```
31 | recorder compute-tf `
32 | -i ./data.zarr/0/0/0 `
33 | -c ./config.yml `
34 | -o ./tf.zarr
35 |
36 | recorder apply-inv-tf
37 | -i ./data.zarr/*/*/* `
38 | -t ./tf.zarr `
39 | -c ./config.yml `
40 | -o ./reconstruction.zarr
41 | ```
42 | Computing the transfer function is typically the most expensive part of the reconstruction, so saving a transfer function then applying it to many datasets can save time.
43 |
44 | ## Input options
45 |
46 | The input `-i` flag always accepts a list of inputs, either explicitly e.g. `-i ./data.zarr/A/1/0 ./data.zarr/A/2/0` or through wildcards `-i ./data.zarr/*/*/*`. The positions in a high-content screening `.zarr` store are organized into `/row/col/fov` folders, so `./input.zarr/*/*/*` creates a list of all positions in a dataset.
47 |
48 | The `recorder compute-tf` command accepts a list of inputs, but it only computes the transfer function for the first position in the list. The `apply-inv-tf` command accepts a list of inputs and applies the same transfer function to all of the inputs, which requires that all positions contain arrays with matching TCZYX shapes.
49 |
50 | ## What types of reconstructions are supported?
51 | See `/recOrder/examples/` for a list of example configuration files.
52 |
53 | WIP: This documentation will be expanded for each reconstruction type and parameter.
54 |
--------------------------------------------------------------------------------
/docs/software-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Software Installation Guide
2 |
3 | 1. (Optional but recommended) install [`conda`](https://github.com/conda-forge/miniforge) and create a virtual environment
4 |
5 | ```sh
6 | conda create -y -n recOrder python=3.10
7 | conda activate recOrder
8 | ```
9 |
10 | 2. Install `recOrder-napari`:
11 |
12 | ```sh
13 | pip install recOrder-napari
14 | ```
15 |
16 | 3. To use the GUI: open `napari` with `recOrder-napari`:
17 |
18 | ```sh
19 | napari -w recOrder-napari
20 | ```
21 |
22 | 4. View command-line help by running
23 |
24 | ```sh
25 | recOrder
26 | ```
27 |
28 | 5. To acquire data via Micro-Manager`, follow the [microscope installation guide](./microscope-installation-guide.md).
29 |
--------------------------------------------------------------------------------
/examples/README.md:
--------------------------------------------------------------------------------
1 | # `recOrder` CLI examples
2 |
3 | `recOrder` uses a configuration-file-based command-line inferface (CLI) to
4 | calculate transfer functions and apply these transfer functions to datasets.
5 |
6 | This page demonstrates `recOrder`'s CLI.
7 |
8 | ## Getting started
9 |
10 | ### 1. Check your installation
11 | First, [install `recOrder`](../docs/software-installation-guide.md) and run
12 | ```bash
13 | recOrder
14 | ```
15 | in a shell. If `recOrder` is installed correctly, you will see a usage string and
16 | ```
17 | recOrder: Computational Toolkit for Label-Free Imaging
18 | ```
19 |
20 | ### 2. Download and convert a test dataset
21 | Next, [download the test data from zenodo (47 MB)](https://zenodo.org/record/6983916/files/recOrder_test_data.zip?download=1), and convert a dataset to the latest version of `.zarr` with
22 | ```
23 | cd /path/to/
24 | iohub convert -i /path/to/recOrder_test_data/2022_08_04_recOrder_pytest_20x_04NA/2T_3P_16Z_128Y_256X_Kazansky_1/
25 | -o ./dataset.zarr
26 | ```
27 |
28 | You can view the test dataset with
29 | ```
30 | napari ./dataset.zarr --plugin recOrder-napari
31 | ```
32 |
33 | ### 3. Run a reconstruction
34 | Run an example reconstruction with
35 | ```
36 | recOrder reconstruct ./dataset.zarr/0/0/0 -c /path/to/recOrder/examples/settings/birefringence-and-phase.yml -o ./reconstruction.zarr
37 | ```
38 | then view the reconstruction with
39 | ```
40 | napari ./reconstruction.zarr --plugin recOrder-napari
41 | ```
42 |
43 | Try modifying the configuration file to see how the regularization parameter changes the results.
44 |
45 | ### 4. Parallelize over positions or time points
46 |
47 | TODO: @Ed I think we'll use the same strategy as ```mantis deskew```
48 |
49 | ## FAQ
50 | 1. **Q: Which configuration file should I use?**
51 |
52 | If you are acquiring:
53 |
54 | **3D data with calibrated liquid-crystal polarizers via `recOrder`** use `birefringence.yml`.
55 |
56 | **3D fluorescence data** use `fluorescence.yml`.
57 |
58 | **3D brightfield data** use `phase.yml`.
59 |
60 | **Multi-modal data**, start by reconstructing the individual modaliities, each with a single config file and CLI call. Then combine the reconstructions by ***TODO: @Ziwen do can you help me append to the zarrs to help me fix this? ***
61 |
62 | 2. **Q: Should I use `reconstruction_dimension` = 2 or 3?
63 |
64 | If your downstream processing requires 3D information or if you're unsure, then you should use `reconstruction_dimension = 3`. If your sample is very thin compared to the depth of field of the microscope, if you're in a noise-limited regime, or if your downstream processing requires 2D information, then you should use `reconstruction_dimension = 2`. Empirically, we have found that 2D reconstructions reduce the noise in our reconstructions because it uses 3D information to make a single estimate for each pixel.
65 |
66 | 3. **Q: What regularization parameter should I use?**
67 |
68 | We recommend starting with the defaults then testing over a few orders of magnitude and choosing a result that isn't too noisy or too smooth.
69 |
70 | ### Developers note
71 |
72 | These configuration files are automatically generated when the tests run. See `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
73 |
74 | To keep these settings up to date, run `pytest` locally when `cli/settings.py` changes.
75 |
--------------------------------------------------------------------------------
/examples/birefringence-and-phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 | phase:
18 | transfer_function:
19 | wavelength_illumination: 0.532
20 | yx_pixel_size: 0.325
21 | z_pixel_size: 2.0
22 | z_padding: 0
23 | index_of_refraction_media: 1.3
24 | numerical_aperture_detection: 1.2
25 | numerical_aperture_illumination: 0.5
26 | invert_phase_contrast: false
27 | apply_inverse:
28 | reconstruction_algorithm: Tikhonov
29 | regularization_strength: 0.001
30 | TV_rho_strength: 0.001
31 | TV_iterations: 1
32 |
--------------------------------------------------------------------------------
/examples/birefringence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 |
--------------------------------------------------------------------------------
/examples/fluorescence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - GFP
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | fluorescence:
6 | transfer_function:
7 | yx_pixel_size: 0.325
8 | z_pixel_size: 2.0
9 | z_padding: 0
10 | index_of_refraction_media: 1.3
11 | numerical_aperture_detection: 1.2
12 | wavelength_emission: 0.507
13 | apply_inverse:
14 | reconstruction_algorithm: Tikhonov
15 | regularization_strength: 0.001
16 | TV_rho_strength: 0.001
17 | TV_iterations: 1
18 |
--------------------------------------------------------------------------------
/examples/phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - BF
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | phase:
6 | transfer_function:
7 | wavelength_illumination: 0.532
8 | yx_pixel_size: 0.325
9 | z_pixel_size: 2.0
10 | z_padding: 0
11 | index_of_refraction_media: 1.3
12 | numerical_aperture_detection: 1.2
13 | numerical_aperture_illumination: 0.5
14 | invert_phase_contrast: false
15 | apply_inverse:
16 | reconstruction_algorithm: Tikhonov
17 | regularization_strength: 0.001
18 | TV_rho_strength: 0.001
19 | TV_iterations: 1
20 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = ["setuptools", "wheel", "setuptools_scm"]
3 | build-backend = "setuptools.build_meta"
4 |
5 | [tool.setuptools_scm]
6 | write_to = "recOrder/_version.py"
7 |
8 | [tool.black]
9 | line-length = 79
10 |
11 | [tool.isort]
12 | profile = "black"
13 | line_length = 79
--------------------------------------------------------------------------------
/recOrder/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/acq/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/acq_functions.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import json
3 | import os
4 | import time
5 |
6 | import numpy as np
7 | from iohub import read_micromanager
8 | try:
9 | from pycromanager import Studio
10 | except:pass
11 |
12 | def generate_acq_settings(
13 | mm,
14 | channel_group,
15 | channels=None,
16 | zstart=None,
17 | zend=None,
18 | zstep=None,
19 | save_dir=None,
20 | prefix=None,
21 | keep_shutter_open_channels=False,
22 | keep_shutter_open_slices=False,
23 | ):
24 | """
25 | This function generates a json file specific to the Micro-Manager SequenceSettings.
26 | It has default parameters for a multi-channels z-stack acquisition but does not yet
27 | support multi-position or multi-frame acquisitions.
28 |
29 | This also has default values for QLIPP Acquisition. Can be used as a framework for other types
30 | of acquisitions
31 |
32 | Parameters
33 | ----------
34 | mm: (object) MM Studio API object
35 | scheme: (str) '4-State' or '5-State'
36 | zstart: (float) relative starting position for the z-stack
37 | zend: (float) relative ending position for the z-stack
38 | zstep: (float) step size for the z-stack
39 | save_dir: (str) path to save directory
40 | prefix: (str) name to save the data under
41 |
42 | Returns
43 | -------
44 | settings: (json) json dictionary conforming to MM SequenceSettings
45 | """
46 |
47 | # Get API Objects
48 | am = mm.getAcquisitionManager()
49 | ss = am.getAcquisitionSettings()
50 | app = mm.app()
51 |
52 | # Get current SequenceSettings to modify
53 | original_ss = ss.toJSONStream(ss)
54 | original_json = json.loads(original_ss).copy()
55 |
56 | if zstart:
57 | do_z = True
58 | else:
59 | do_z = False
60 |
61 | # Structure of the channel properties
62 | channel_dict = {
63 | "channelGroup": channel_group,
64 | "config": None,
65 | "exposure": None,
66 | "zOffset": 0,
67 | "doZStack": do_z,
68 | "color": {"value": -16747854, "falpha": 0.0},
69 | "skipFactorFrame": 0,
70 | "useChannel": True if channels else False,
71 | }
72 |
73 | channel_list = None
74 | if channels:
75 | # Append all the channels with their current exposure settings
76 | channel_list = []
77 | for chan in channels:
78 | # todo: think about how to deal with missing exposure
79 | exposure = app.getChannelExposureTime(
80 | channel_group, chan, 10
81 | ) # sets exposure to 10 if not found
82 | channel = channel_dict.copy()
83 | channel["config"] = chan
84 | channel["exposure"] = exposure
85 |
86 | channel_list.append(channel)
87 |
88 | # set other parameters
89 | original_json["numFrames"] = 1
90 | original_json["intervalMs"] = 0
91 | original_json["relativeZSlice"] = True
92 | original_json["slicesFirst"] = True
93 | original_json["timeFirst"] = False
94 | original_json["keepShutterOpenSlices"] = keep_shutter_open_slices
95 | original_json["keepShutterOpenChannels"] = keep_shutter_open_channels
96 | original_json["useAutofocus"] = False
97 | original_json["saveMode"] = "MULTIPAGE_TIFF"
98 | original_json["save"] = True if save_dir else False
99 | original_json["root"] = save_dir if save_dir else ""
100 | original_json["prefix"] = prefix if prefix else "Untitled"
101 | original_json["channels"] = channel_list
102 | original_json["zReference"] = 0.0
103 | original_json["channelGroup"] = channel_group
104 | original_json["usePositionList"] = False
105 | original_json["shouldDisplayImages"] = True
106 | original_json["useSlices"] = do_z
107 | original_json["useFrames"] = False
108 | original_json["useChannels"] = True if channels else False
109 | original_json["slices"] = (
110 | list(np.arange(float(zstart), float(zend + zstep), float(zstep)))
111 | if zstart
112 | else []
113 | )
114 | original_json["sliceZStepUm"] = zstep
115 | original_json["sliceZBottomUm"] = zstart
116 | original_json["sliceZTopUm"] = zend
117 | original_json["acqOrderMode"] = 1
118 |
119 | return original_json
120 |
121 |
122 | def acquire_from_settings(
123 | mm: Studio,
124 | settings: dict,
125 | grab_images: bool = True,
126 | restore_settings: bool = True,
127 | ):
128 | """Function to acquire an MDA acquisition with the native MM MDA Engine.
129 | Assumes single position acquisition.
130 |
131 | Parameters
132 | ----------
133 | mm : Studio
134 | settings : dict
135 | JSON dictionary conforming to MM SequenceSettings
136 | grab_images : bool, optional
137 | return the acquired array, by default True
138 | restore_settings : bool, optional
139 | restore MDA settings before acquisition, by default True
140 |
141 | Returns
142 | -------
143 | NDArray
144 | acquired images
145 | """
146 | am = mm.getAcquisitionManager()
147 | ss = am.getAcquisitionSettings()
148 |
149 | ss_new = ss.fromJSONStream(json.dumps(settings))
150 | am.runAcquisitionWithSettings(ss_new, True)
151 |
152 | time.sleep(3)
153 |
154 | if restore_settings:
155 | am.setAcquisitionSettings(ss)
156 |
157 | # TODO: speed improvements in reading the data with pycromanager acquisition?
158 | if grab_images:
159 | # get the most recent acquisition if multiple
160 | path = os.path.join(settings["root"], settings["prefix"])
161 | files = glob.glob(path + "*")
162 | index = max([int(x.split(path + "_")[1]) for x in files])
163 |
164 | reader = read_micromanager(
165 | path + f"_{index}", data_type="ometiff", extract_data=True
166 | )
167 |
168 | return reader.get_array(0)
169 |
--------------------------------------------------------------------------------
/recOrder/calib/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/calib/__init__.py
--------------------------------------------------------------------------------
/recOrder/cli/apply_inverse_models.py:
--------------------------------------------------------------------------------
1 | """
2 | This module converts recOrder's reconstructions into waveorder calls
3 | """
4 |
5 | import numpy as np
6 | import torch
7 | from waveorder.models import (
8 | inplane_oriented_thick_pol3d,
9 | isotropic_fluorescent_thick_3d,
10 | isotropic_thin_3d,
11 | phase_thick_3d,
12 | )
13 |
14 |
15 | def radians_to_nanometers(retardance_rad, wavelength_illumination_um):
16 | """
17 | waveorder returns retardance in radians, while recorder displays and saves
18 | retardance in nanometers. This function converts from radians to nanometers
19 | using the illumination wavelength (which is internally handled in um
20 | in recOrder).
21 | """
22 | return retardance_rad * wavelength_illumination_um * 1e3 / (2 * np.pi)
23 |
24 |
25 | def birefringence(
26 | czyx_data,
27 | cyx_no_sample_data,
28 | wavelength_illumination,
29 | recon_dim,
30 | biref_inverse_dict,
31 | transfer_function_dataset,
32 | ):
33 | # Load transfer function
34 | intensity_to_stokes_matrix = torch.tensor(
35 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
36 | )
37 |
38 | # Apply reconstruction
39 | # (retardance, orientation, transmittance, depolarization)
40 | reconstructed_parameters = (
41 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
42 | czyx_data,
43 | intensity_to_stokes_matrix,
44 | cyx_no_sample_data=cyx_no_sample_data,
45 | project_stokes_to_2d=(recon_dim == 2),
46 | **biref_inverse_dict,
47 | )
48 | )
49 |
50 | # Convert retardance
51 | retardance = radians_to_nanometers(
52 | reconstructed_parameters[0], wavelength_illumination
53 | )
54 |
55 | return torch.stack((retardance,) + reconstructed_parameters[1:])
56 |
57 |
58 | def phase(
59 | czyx_data,
60 | recon_dim,
61 | settings_phase,
62 | transfer_function_dataset,
63 | ):
64 | # [phase only, 2]
65 | if recon_dim == 2:
66 | # Load transfer functions
67 | absorption_transfer_function = torch.tensor(
68 | transfer_function_dataset["absorption_transfer_function"][0, 0]
69 | )
70 | phase_transfer_function = torch.tensor(
71 | transfer_function_dataset["phase_transfer_function"][0, 0]
72 | )
73 |
74 | # Apply
75 | (
76 | _,
77 | output,
78 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
79 | czyx_data[0],
80 | absorption_transfer_function,
81 | phase_transfer_function,
82 | **settings_phase.apply_inverse.dict(),
83 | )
84 |
85 | # [phase only, 3]
86 | elif recon_dim == 3:
87 | # Load transfer functions
88 | real_potential_transfer_function = torch.tensor(
89 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
90 | )
91 | imaginary_potential_transfer_function = torch.tensor(
92 | transfer_function_dataset["imaginary_potential_transfer_function"][
93 | 0, 0
94 | ]
95 | )
96 |
97 | # Apply
98 | output = phase_thick_3d.apply_inverse_transfer_function(
99 | czyx_data[0],
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | z_padding=settings_phase.transfer_function.z_padding,
103 | **settings_phase.apply_inverse.dict(),
104 | )
105 |
106 | # Pad to CZYX
107 | while output.ndim != 4:
108 | output = torch.unsqueeze(output, 0)
109 |
110 | return output
111 |
112 |
113 | def birefringence_and_phase(
114 | czyx_data,
115 | cyx_no_sample_data,
116 | wavelength_illumination,
117 | recon_dim,
118 | biref_inverse_dict,
119 | settings_phase,
120 | transfer_function_dataset,
121 | ):
122 | # Load birefringence transfer function
123 | intensity_to_stokes_matrix = torch.tensor(
124 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
125 | )
126 |
127 | # [biref and phase, 2]
128 | if recon_dim == 2:
129 | # Load phase transfer functions
130 | absorption_transfer_function = torch.tensor(
131 | transfer_function_dataset["absorption_transfer_function"][0, 0]
132 | )
133 | phase_transfer_function = torch.tensor(
134 | transfer_function_dataset["phase_transfer_function"][0, 0]
135 | )
136 |
137 | # Apply
138 | reconstructed_parameters_2d = (
139 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
140 | czyx_data,
141 | intensity_to_stokes_matrix,
142 | cyx_no_sample_data=cyx_no_sample_data,
143 | project_stokes_to_2d=True,
144 | **biref_inverse_dict,
145 | )
146 | )
147 |
148 | reconstructed_parameters_3d = (
149 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
150 | czyx_data,
151 | intensity_to_stokes_matrix,
152 | cyx_no_sample_data=cyx_no_sample_data,
153 | project_stokes_to_2d=False,
154 | **biref_inverse_dict,
155 | )
156 | )
157 |
158 | brightfield_3d = reconstructed_parameters_3d[2]
159 |
160 | (
161 | _,
162 | yx_phase,
163 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
164 | brightfield_3d,
165 | absorption_transfer_function,
166 | phase_transfer_function,
167 | **settings_phase.apply_inverse.dict(),
168 | )
169 |
170 | # Convert retardance
171 | retardance = radians_to_nanometers(
172 | reconstructed_parameters_2d[0], wavelength_illumination
173 | )
174 |
175 | output = torch.stack(
176 | (retardance,)
177 | + reconstructed_parameters_2d[1:]
178 | + (torch.unsqueeze(yx_phase, 0),)
179 | ) # CZYX
180 |
181 | # [biref and phase, 3]
182 | elif recon_dim == 3:
183 | # Load phase transfer functions
184 | intensity_to_stokes_matrix = torch.tensor(
185 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
186 | )
187 | # Load transfer functions
188 | real_potential_transfer_function = torch.tensor(
189 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
190 | )
191 | imaginary_potential_transfer_function = torch.tensor(
192 | transfer_function_dataset["imaginary_potential_transfer_function"][
193 | 0, 0
194 | ]
195 | )
196 |
197 | # Apply
198 | reconstructed_parameters_3d = (
199 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
200 | czyx_data,
201 | intensity_to_stokes_matrix,
202 | cyx_no_sample_data=cyx_no_sample_data,
203 | project_stokes_to_2d=False,
204 | **biref_inverse_dict,
205 | )
206 | )
207 |
208 | brightfield_3d = reconstructed_parameters_3d[2]
209 |
210 | zyx_phase = phase_thick_3d.apply_inverse_transfer_function(
211 | brightfield_3d,
212 | real_potential_transfer_function,
213 | imaginary_potential_transfer_function,
214 | z_padding=settings_phase.transfer_function.z_padding,
215 | **settings_phase.apply_inverse.dict(),
216 | )
217 |
218 | # Convert retardance
219 | retardance = radians_to_nanometers(
220 | reconstructed_parameters_3d[0], wavelength_illumination
221 | )
222 |
223 | # Save
224 | output = torch.stack(
225 | (retardance,) + reconstructed_parameters_3d[1:] + (zyx_phase,)
226 | )
227 | return output
228 |
229 |
230 | def fluorescence(
231 | czyx_data, recon_dim, settings_fluorescence, transfer_function_dataset
232 | ):
233 | # [fluo, 2]
234 | if recon_dim == 2:
235 | raise NotImplementedError
236 | # [fluo, 3]
237 | elif recon_dim == 3:
238 | # Load transfer functions
239 | optical_transfer_function = torch.tensor(
240 | transfer_function_dataset["optical_transfer_function"][0, 0]
241 | )
242 |
243 | # Apply
244 | output = (
245 | isotropic_fluorescent_thick_3d.apply_inverse_transfer_function(
246 | czyx_data[0],
247 | optical_transfer_function,
248 | settings_fluorescence.transfer_function.z_padding,
249 | **settings_fluorescence.apply_inverse.dict(),
250 | )
251 | )
252 | # Pad to CZYX
253 | while output.ndim != 4:
254 | output = torch.unsqueeze(output, 0)
255 |
256 | return output
257 |
--------------------------------------------------------------------------------
/recOrder/cli/compute_transfer_function.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 | import numpy as np
5 | from iohub.ngff import open_ome_zarr, Position
6 | from waveorder.models import (
7 | inplane_oriented_thick_pol3d,
8 | isotropic_fluorescent_thick_3d,
9 | isotropic_thin_3d,
10 | phase_thick_3d,
11 | )
12 |
13 | from recOrder.cli.parsing import (
14 | config_filepath,
15 | input_position_dirpaths,
16 | output_dirpath,
17 | )
18 | from recOrder.cli.printing import echo_headline, echo_settings
19 | from recOrder.cli.settings import ReconstructionSettings
20 | from recOrder.io import utils
21 |
22 |
23 | def generate_and_save_birefringence_transfer_function(settings, dataset):
24 | """Generates and saves the birefringence transfer function to the dataset, based on the settings.
25 |
26 | Parameters
27 | ----------
28 | settings: ReconstructionSettings
29 | dataset: NGFF Node
30 | The dataset that will be updated.
31 | """
32 | echo_headline("Generating birefringence transfer function with settings:")
33 | echo_settings(settings.birefringence.transfer_function)
34 |
35 | # Calculate transfer functions
36 | intensity_to_stokes_matrix = (
37 | inplane_oriented_thick_pol3d.calculate_transfer_function(
38 | scheme=str(len(settings.input_channel_names)) + "-State",
39 | **settings.birefringence.transfer_function.dict(),
40 | )
41 | )
42 | # Save
43 | dataset[
44 | "intensity_to_stokes_matrix"
45 | ] = intensity_to_stokes_matrix.cpu().numpy()[None, None, None, ...]
46 |
47 |
48 | def generate_and_save_phase_transfer_function(
49 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
50 | ):
51 | """Generates and saves the phase transfer function to the dataset, based on the settings.
52 |
53 | Parameters
54 | ----------
55 | settings: ReconstructionSettings
56 | dataset: Position
57 | The dataset that will be updated.
58 | zyx_shape : tuple
59 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
60 | """
61 | echo_headline("Generating phase transfer function with settings:")
62 | echo_settings(settings.phase.transfer_function)
63 |
64 | if settings.reconstruction_dimension == 2:
65 | # Convert zyx_shape and z_pixel_size into yx_shape and z_position_list
66 | settings_dict = settings.phase.transfer_function.dict()
67 | settings_dict["yx_shape"] = [zyx_shape[1], zyx_shape[2]]
68 | settings_dict["z_position_list"] = list(
69 | -(np.arange(zyx_shape[0]) - zyx_shape[0] // 2)
70 | * settings_dict["z_pixel_size"]
71 | )
72 |
73 | # Remove unused parameters
74 | settings_dict.pop("z_pixel_size")
75 | settings_dict.pop("z_padding")
76 |
77 | # Calculate transfer functions
78 | (
79 | absorption_transfer_function,
80 | phase_transfer_function,
81 | ) = isotropic_thin_3d.calculate_transfer_function(
82 | **settings_dict,
83 | )
84 |
85 | # Save
86 | dataset.create_image(
87 | "absorption_transfer_function",
88 | absorption_transfer_function.cpu().numpy()[None, None, ...],
89 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
90 | )
91 | dataset.create_image(
92 | "phase_transfer_function",
93 | phase_transfer_function.cpu().numpy()[None, None, ...],
94 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
95 | )
96 |
97 | elif settings.reconstruction_dimension == 3:
98 | # Calculate transfer functions
99 | (
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | ) = phase_thick_3d.calculate_transfer_function(
103 | zyx_shape=zyx_shape,
104 | **settings.phase.transfer_function.dict(),
105 | )
106 | # Save
107 | dataset.create_image(
108 | "real_potential_transfer_function",
109 | real_potential_transfer_function.cpu().numpy()[None, None, ...],
110 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
111 | )
112 | dataset.create_image(
113 | "imaginary_potential_transfer_function",
114 | imaginary_potential_transfer_function.cpu().numpy()[
115 | None, None, ...
116 | ],
117 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
118 | )
119 |
120 |
121 | def generate_and_save_fluorescence_transfer_function(
122 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
123 | ):
124 | """Generates and saves the fluorescence transfer function to the dataset, based on the settings.
125 |
126 | Parameters
127 | ----------
128 | settings: ReconstructionSettings
129 | dataset: Position
130 | The dataset that will be updated.
131 | zyx_shape : tuple
132 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
133 | """
134 | echo_headline("Generating fluorescence transfer function with settings:")
135 | echo_settings(settings.fluorescence.transfer_function)
136 |
137 | if settings.reconstruction_dimension == 2:
138 | raise NotImplementedError
139 | elif settings.reconstruction_dimension == 3:
140 | # Calculate transfer functions
141 | optical_transfer_function = (
142 | isotropic_fluorescent_thick_3d.calculate_transfer_function(
143 | zyx_shape=zyx_shape,
144 | **settings.fluorescence.transfer_function.dict(),
145 | )
146 | )
147 | # Save
148 | dataset.create_image(
149 | "optical_transfer_function",
150 | optical_transfer_function.cpu().numpy()[None, None, ...],
151 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
152 | )
153 |
154 |
155 | def compute_transfer_function_cli(
156 | input_position_dirpath: Path, config_filepath: Path, output_dirpath: Path
157 | ) -> None:
158 | """CLI command to compute the transfer function given a configuration file path
159 | and a desired output path.
160 | """
161 |
162 | # Load config file
163 | settings = utils.yaml_to_model(config_filepath, ReconstructionSettings)
164 |
165 | echo_headline(
166 | f"Generating transfer functions and storing in {output_dirpath}\n"
167 | )
168 |
169 | # Read shape from input dataset
170 | input_dataset = open_ome_zarr(
171 | input_position_dirpath, layout="fov", mode="r"
172 | )
173 | zyx_shape = input_dataset.data.shape[
174 | 2:
175 | ] # only loads a single position "0"
176 |
177 | # Check input channel names
178 | if not set(settings.input_channel_names).issubset(
179 | input_dataset.channel_names
180 | ):
181 | raise ValueError(
182 | f"Each of the input_channel_names = {settings.input_channel_names} in {config_filepath} must appear in the dataset {input_position_dirpaths[0]} which currently contains channel_names = {input_dataset.channel_names}."
183 | )
184 |
185 | # Prepare output dataset
186 | output_dataset = open_ome_zarr(
187 | output_dirpath, layout="fov", mode="w", channel_names=["None"]
188 | )
189 |
190 | # Pass settings to appropriate calculate_transfer_function and save
191 | if settings.birefringence is not None:
192 | generate_and_save_birefringence_transfer_function(
193 | settings, output_dataset
194 | )
195 | if settings.phase is not None:
196 | generate_and_save_phase_transfer_function(
197 | settings, output_dataset, zyx_shape
198 | )
199 | if settings.fluorescence is not None:
200 | generate_and_save_fluorescence_transfer_function(
201 | settings, output_dataset, zyx_shape
202 | )
203 |
204 | # Write settings to metadata
205 | output_dataset.zattrs["settings"] = settings.dict()
206 |
207 | echo_headline(f"Closing {output_dirpath}\n")
208 | output_dataset.close()
209 |
210 | echo_headline(
211 | f"Recreate this transfer function with:\n$ recorder compute-tf {input_position_dirpaths} -c {config_filepath} -o {output_dirpath}"
212 | )
213 |
214 |
215 | @click.command()
216 | @input_position_dirpaths()
217 | @config_filepath()
218 | @output_dirpath()
219 | def compute_tf(
220 | input_position_dirpaths: list[Path],
221 | config_filepath: Path,
222 | output_dirpath: Path,
223 | ) -> None:
224 | """
225 | Compute a transfer function using a dataset and configuration file.
226 |
227 | Calculates the transfer function based on the shape of the first position
228 | in the list `input-position-dirpaths`.
229 |
230 | See /examples for example configuration files.
231 |
232 | >> recorder compute-tf -i ./input.zarr/0/0/0 -c ./examples/birefringence.yml -o ./transfer_function.zarr
233 | """
234 | compute_transfer_function_cli(
235 | input_position_dirpaths[0], config_filepath, output_dirpath
236 | )
237 |
--------------------------------------------------------------------------------
/recOrder/cli/gui_widget.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import click
3 |
4 | try:
5 | from recOrder.plugin import tab_recon
6 | except:pass
7 |
8 | try:
9 | from qtpy.QtWidgets import QApplication, QWidget, QVBoxLayout, QStyle
10 | except:pass
11 |
12 | try:
13 | import qdarktheme
14 | except:pass
15 |
16 | PLUGIN_NAME = "recOrder: Computational Toolkit for Label-Free Imaging"
17 | PLUGIN_ICON = "🔬"
18 |
19 | @click.command()
20 | def gui():
21 | """GUI for recOrder: Computational Toolkit for Label-Free Imaging"""
22 |
23 | app = QApplication(sys.argv)
24 | app.setStyle("Fusion") # Other options: "Fusion", "Windows", "macOS", "WindowsVista"
25 | try:
26 | qdarktheme.setup_theme("dark")
27 | except:pass
28 | window = MainWindow()
29 | window.setWindowTitle(PLUGIN_ICON + " " + PLUGIN_NAME + " " + PLUGIN_ICON)
30 |
31 | pixmapi = getattr(QStyle.StandardPixmap, "SP_TitleBarMenuButton")
32 | icon = app.style().standardIcon(pixmapi)
33 | window.setWindowIcon(icon)
34 |
35 | window.show()
36 | sys.exit(app.exec())
37 |
38 | class MainWindow(QWidget):
39 | def __init__(self):
40 | super().__init__()
41 | recon_tab = tab_recon.Ui_ReconTab_Form(stand_alone=True)
42 | layout = QVBoxLayout()
43 | self.setLayout(layout)
44 | layout.addWidget(recon_tab.recon_tab_mainScrollArea)
45 |
46 | if __name__ == "__main__":
47 | gui()
48 |
--------------------------------------------------------------------------------
/recOrder/cli/jobs_mgmt.py:
--------------------------------------------------------------------------------
1 | import os, json
2 | from pathlib import Path
3 | import socket
4 | import submitit
5 | import threading, time
6 |
7 | DIR_PATH = os.path.dirname(os.path.realpath(__file__))
8 | FILE_PATH = os.path.join(DIR_PATH, "main.py")
9 |
10 | SERVER_PORT = 8089 # Choose an available port
11 | JOBS_TIMEOUT = 5 # 5 mins
12 | SERVER_uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
13 |
14 | class JobsManagement():
15 |
16 | def __init__(self, *args, **kwargs):
17 | self.clientsocket = None
18 | self.uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
19 | self.DATA_QUEUE = []
20 |
21 | def check_for_jobID_File(self, jobID, logs_path, extension="out"):
22 |
23 | if Path(logs_path).exists():
24 | files = os.listdir(logs_path)
25 | try:
26 | for file in files:
27 | if file.endswith(extension):
28 | if jobID in file:
29 | file_path = os.path.join(logs_path, file)
30 | f = open(file_path, "r")
31 | txt = f.read()
32 | f.close()
33 | return txt
34 | except Exception as exc:
35 | print(exc.args)
36 | return ""
37 |
38 | def set_shorter_timeout(self):
39 | self.clientsocket.settimeout(30)
40 |
41 | def start_client(self):
42 | try:
43 | self.clientsocket = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
44 | self.clientsocket.settimeout(300)
45 | self.clientsocket.connect(('localhost', SERVER_PORT))
46 | self.clientsocket.settimeout(None)
47 |
48 | thread = threading.Thread(target=self.stop_client)
49 | thread.start()
50 | except Exception as exc:
51 | print(exc.args)
52 |
53 | # The stopClient() is called right with the startClient() but does not stop
54 | # and essentially is a wait thread listening and is triggered by either a
55 | # connection or timeout. Based on condition triggered by user, reconstruction
56 | # completion or errors the end goal is to close the socket connection which
57 | # would let the CLI exit. I could break it down to 2 parts but the idea was to
58 | # keep the clientsocket.close() call within one method to make it easier to follow.
59 | def stop_client(self):
60 | try:
61 | time.sleep(2)
62 | while True:
63 | time.sleep(1)
64 | buf = ""
65 | try:
66 | buf = self.clientsocket.recv(1024)
67 | except:
68 | pass
69 | if len(buf) > 0:
70 | if b"\n" in buf:
71 | dataList = buf.split(b"\n")
72 | for data in dataList:
73 | if len(data)>0:
74 | decoded_string = data.decode()
75 | json_str = str(decoded_string)
76 | json_obj = json.loads(json_str)
77 | u_idx = json_obj["uID"]
78 | job_idx = str(json_obj["jID"])
79 | cmd = json_obj["command"]
80 | if cmd == "clientRelease":
81 | if self.has_submitted_job(u_idx, job_idx):
82 | self.clientsocket.close()
83 | break
84 | if cmd == "cancel":
85 | if self.has_submitted_job(u_idx, job_idx):
86 | try:
87 | job = self.uIDsjobIDs[u_idx][job_idx]
88 | job.cancel()
89 | except Exception as exc:
90 | pass # possibility of throwing an exception based on diff. OS
91 | forDeletions = []
92 | for uID in self.uIDsjobIDs.keys():
93 | for jID in self.uIDsjobIDs[uID].keys():
94 | job = self.uIDsjobIDs[uID][jID]
95 | if job.done():
96 | forDeletions.append((uID, jID))
97 | for idx in range(len(forDeletions)):
98 | del self.uIDsjobIDs[forDeletions[idx][0]][forDeletions[idx][1]]
99 | forDeletions = []
100 | for uID in self.uIDsjobIDs.keys():
101 | if len(self.uIDsjobIDs[uID].keys()) == 0:
102 | forDeletions.append(uID)
103 | for idx in range(len(forDeletions)):
104 | del self.uIDsjobIDs[forDeletions[idx]]
105 | if len(self.uIDsjobIDs.keys()) == 0:
106 | self.clientsocket.close()
107 | break
108 | except Exception as exc:
109 | self.clientsocket.close()
110 | print(exc.args)
111 |
112 | def check_all_ExpJobs_completion(self, uID):
113 | if uID in SERVER_uIDsjobIDs.keys():
114 | for jobEntry in SERVER_uIDsjobIDs[uID].keys():
115 | job:submitit.Job = SERVER_uIDsjobIDs[uID][jobEntry]["job"]
116 | jobBool = SERVER_uIDsjobIDs[uID][jobEntry]["bool"]
117 | if job is not None and job.done() == False:
118 | return False
119 | if jobBool == False:
120 | return False
121 | return True
122 |
123 | def put_Job_completion_in_list(self, job_bool, uID: str, jID: str, mode="client"):
124 | if uID in SERVER_uIDsjobIDs.keys():
125 | if jID in SERVER_uIDsjobIDs[uID].keys():
126 | SERVER_uIDsjobIDs[uID][jID]["bool"] = job_bool
127 |
128 | def add_data(self, data):
129 | self.DATA_QUEUE.append(data)
130 |
131 | def send_data_thread(self):
132 | thread = threading.Thread(target=self.send_data)
133 | thread.start()
134 |
135 | def send_data(self):
136 | data = "".join(self.DATA_QUEUE)
137 | self.clientsocket.send(data.encode())
138 | self.DATA_QUEUE = []
139 |
140 | def put_Job_in_list(self, job, uID: str, jID: str, well:str, log_folder_path:str="", mode="client"):
141 | try:
142 | well = str(well)
143 | jID = str(jID)
144 | if ".zarr" in well:
145 | wells = well.split(".zarr")
146 | well = wells[1].replace("\\","-").replace("/","-")[1:]
147 | if mode == "client":
148 | if uID not in self.uIDsjobIDs.keys():
149 | self.uIDsjobIDs[uID] = {}
150 | self.uIDsjobIDs[uID][jID] = job
151 | else:
152 | if jID not in self.uIDsjobIDs[uID].keys():
153 | self.uIDsjobIDs[uID][jID] = job
154 | json_obj = {uID:{"jID": str(jID), "pos": well, "log": log_folder_path}}
155 | json_str = json.dumps(json_obj)+"\n"
156 | self.add_data(json_str)
157 | else:
158 | # from server side jobs object entry is a None object
159 | # this will be later checked as completion boolean for a ExpID which might
160 | # have several Jobs associated with it
161 | if uID not in SERVER_uIDsjobIDs.keys():
162 | SERVER_uIDsjobIDs[uID] = {}
163 | SERVER_uIDsjobIDs[uID][jID] = {}
164 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
165 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
166 | else:
167 | SERVER_uIDsjobIDs[uID][jID] = {}
168 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
169 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
170 | except Exception as exc:
171 | print(exc.args)
172 |
173 | def has_submitted_job(self, uID: str, jID: str, mode="client")->bool:
174 | jID = str(jID)
175 | if mode == "client":
176 | if uID in self.uIDsjobIDs.keys():
177 | if jID in self.uIDsjobIDs[uID].keys():
178 | return True
179 | return False
180 | else:
181 | if uID in SERVER_uIDsjobIDs.keys():
182 | if jID in SERVER_uIDsjobIDs[uID].keys():
183 | return True
184 | return False
185 |
--------------------------------------------------------------------------------
/recOrder/cli/main.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 | from recOrder.cli.apply_inverse_transfer_function import apply_inv_tf
4 | from recOrder.cli.compute_transfer_function import compute_tf
5 | from recOrder.cli.reconstruct import reconstruct
6 | try:
7 | from recOrder.cli.gui_widget import gui
8 | except:pass
9 |
10 |
11 | CONTEXT = {"help_option_names": ["-h", "--help"]}
12 |
13 |
14 | # `recorder -h` will show subcommands in the order they are added
15 | class NaturalOrderGroup(click.Group):
16 | def list_commands(self, ctx):
17 | return self.commands.keys()
18 |
19 |
20 | @click.group(context_settings=CONTEXT, cls=NaturalOrderGroup)
21 | def cli():
22 | """\033[92mrecOrder: Computational Toolkit for Label-Free Imaging\033[0m\n"""
23 |
24 |
25 | cli.add_command(reconstruct)
26 | cli.add_command(compute_tf)
27 | cli.add_command(apply_inv_tf)
28 | try:
29 | cli.add_command(gui)
30 | except:pass
31 |
32 | if __name__ == "__main__":
33 | cli()
34 |
--------------------------------------------------------------------------------
/recOrder/cli/monitor.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import time
4 | import numpy as np
5 | import shutil
6 | import submitit
7 | import sys
8 |
9 |
10 | def _move_cursor_up(n_lines, do_print=True):
11 | if do_print:
12 | sys.stdout.write("\033[F" * n_lines)
13 |
14 |
15 | def _print_status(jobs, position_dirpaths, elapsed_list, print_indices=None, do_print=True):
16 |
17 | columns = [15, 30, 40, 50]
18 |
19 | # header
20 | if do_print:
21 | sys.stdout.write(
22 | "\033[K" # clear line
23 | "\033[96mID" # cyan
24 | f"\033[{columns[0]}G WELL "
25 | f"\033[{columns[1]}G STATUS "
26 | f"\033[{columns[2]}G NODE "
27 | f"\033[{columns[2]}G ELAPSED\n"
28 | )
29 |
30 | if print_indices is None:
31 | print_indices = range(len(jobs))
32 |
33 | complete_count = 0
34 |
35 | for i, (job, position_dirpath) in enumerate(zip(jobs, position_dirpaths)):
36 | try:
37 | node_name = job.get_info()["NodeList"] # slowest, so do this first
38 | except:
39 | node_name = "SUBMITTED"
40 |
41 | if job.state == "COMPLETED":
42 | color = "\033[32m" # green
43 | complete_count += 1
44 | elif job.state == "RUNNING":
45 | color = "\033[93m" # yellow
46 | elapsed_list[i] += 1 # inexact timing
47 | else:
48 | color = "\033[91m" # red
49 |
50 | if i in print_indices:
51 | if do_print:
52 | sys.stdout.write(
53 | f"\033[K" # clear line
54 | f"{color}{job.job_id}"
55 | f"\033[{columns[0]}G {'/'.join(position_dirpath.parts[-3:])}"
56 | f"\033[{columns[1]}G {job.state}"
57 | f"\033[{columns[2]}G {node_name}"
58 | f"\033[{columns[3]}G {elapsed_list[i]} s\n"
59 | )
60 | sys.stdout.flush()
61 | if do_print:
62 | print(
63 | f"\033[32m{complete_count}/{len(jobs)} jobs complete. "
64 | " to move monitor to background. "
65 | " twice to cancel jobs."
66 | )
67 |
68 | return elapsed_list
69 |
70 |
71 | def _get_jobs_to_print(jobs, num_to_print):
72 | job_indices_to_print = []
73 |
74 | # if number of jobs is smaller than termanal size, print all
75 | if len(jobs) <= num_to_print:
76 | return list(range(len(jobs)))
77 |
78 | # prioritize incomplete jobs
79 | for i, job in enumerate(jobs):
80 | if not job.done():
81 | job_indices_to_print.append(i)
82 | if len(job_indices_to_print) == num_to_print:
83 | return job_indices_to_print
84 |
85 | # fill in the rest with complete jobs
86 | for i, job in enumerate(jobs):
87 | job_indices_to_print.append(i)
88 | if len(job_indices_to_print) == num_to_print:
89 | return job_indices_to_print
90 |
91 | # shouldn't reach here
92 | return job_indices_to_print
93 |
94 |
95 | def monitor_jobs(jobs: list[submitit.Job], position_dirpaths: list[Path], do_print=True):
96 | """Displays the status of a list of submitit jobs with corresponding paths.
97 |
98 | Parameters
99 | ----------
100 | jobs : list[submitit.Job]
101 | List of submitit jobs
102 | position_dirpaths : list[Path]
103 | List of corresponding position paths
104 | """
105 | NON_JOB_LINES = 3
106 |
107 | if not len(jobs) == len(position_dirpaths):
108 | raise ValueError(
109 | "The number of jobs and position_dirpaths should be the same."
110 | )
111 |
112 | elapsed_list = [0] * len(jobs) # timer for each job
113 |
114 | # print all jobs once if terminal is too small
115 | if shutil.get_terminal_size().lines - NON_JOB_LINES < len(jobs):
116 | _print_status(jobs, position_dirpaths, elapsed_list, do_print)
117 |
118 | # main monitor loop
119 | try:
120 | while not all(job.done() for job in jobs):
121 | terminal_lines = shutil.get_terminal_size().lines
122 | num_jobs_to_print = np.min(
123 | [terminal_lines - NON_JOB_LINES, len(jobs)]
124 | )
125 |
126 | job_indices_to_print = _get_jobs_to_print(jobs, num_jobs_to_print)
127 |
128 | elapsed_list = _print_status(
129 | jobs,
130 | position_dirpaths,
131 | elapsed_list,
132 | job_indices_to_print,
133 | do_print,
134 | )
135 |
136 | time.sleep(1)
137 | _move_cursor_up(num_jobs_to_print + 2, do_print)
138 |
139 | # Print final status
140 | time.sleep(1)
141 | _print_status(jobs, position_dirpaths, elapsed_list, do_print=do_print)
142 |
143 | # cancel jobs if ctrl+c
144 | except KeyboardInterrupt:
145 | for job in jobs:
146 | job.cancel()
147 | print("All jobs cancelled.\033[97m")
148 |
149 | # Print STDOUT and STDERR for first incomplete job
150 | incomplete_count = 0
151 | for job in jobs:
152 | if not job.done():
153 | if incomplete_count == 0:
154 | print("\033[32mSTDOUT")
155 | print(job.stdout())
156 | print("\033[91mSTDERR")
157 | print(job.stderr())
158 |
159 | print("\033[97m") # print white
160 |
--------------------------------------------------------------------------------
/recOrder/cli/option_eat_all.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 |
4 | # Copied directly from https://stackoverflow.com/a/48394004
5 | # Enables `-i ./input.zarr/*/*/*`
6 | class OptionEatAll(click.Option):
7 | def __init__(self, *args, **kwargs):
8 | self.save_other_options = kwargs.pop("save_other_options", True)
9 | nargs = kwargs.pop("nargs", -1)
10 | assert nargs == -1, "nargs, if set, must be -1 not {}".format(nargs)
11 | super(OptionEatAll, self).__init__(*args, **kwargs)
12 | self._previous_parser_process = None
13 | self._eat_all_parser = None
14 |
15 | def add_to_parser(self, parser, ctx):
16 | def parser_process(value, state):
17 | # method to hook to the parser.process
18 | done = False
19 | value = [value]
20 | if self.save_other_options:
21 | # grab everything up to the next option
22 | while state.rargs and not done:
23 | for prefix in self._eat_all_parser.prefixes:
24 | if state.rargs[0].startswith(prefix):
25 | done = True
26 | if not done:
27 | value.append(state.rargs.pop(0))
28 | else:
29 | # grab everything remaining
30 | value += state.rargs
31 | state.rargs[:] = []
32 | value = tuple(value)
33 |
34 | # call the actual process
35 | self._previous_parser_process(value, state)
36 |
37 | retval = super(OptionEatAll, self).add_to_parser(parser, ctx)
38 | for name in self.opts:
39 | our_parser = parser._long_opt.get(name) or parser._short_opt.get(
40 | name
41 | )
42 | if our_parser:
43 | self._eat_all_parser = our_parser
44 | self._previous_parser_process = our_parser.process
45 | our_parser.process = parser_process
46 | break
47 | return retval
48 |
--------------------------------------------------------------------------------
/recOrder/cli/parsing.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from typing import Callable
3 |
4 | import click
5 | import torch.multiprocessing as mp
6 | from iohub.ngff import Plate, open_ome_zarr
7 | from natsort import natsorted
8 |
9 | from recOrder.cli.option_eat_all import OptionEatAll
10 |
11 |
12 | def _validate_and_process_paths(
13 | ctx: click.Context, opt: click.Option, value: str
14 | ) -> list[Path]:
15 | # Sort and validate the input paths, expanding plates into lists of positions
16 | input_paths = [Path(path) for path in natsorted(value)]
17 | for path in input_paths:
18 | with open_ome_zarr(path, mode="r") as dataset:
19 | if isinstance(dataset, Plate):
20 | plate_path = input_paths.pop()
21 | for position in dataset.positions():
22 | input_paths.append(plate_path / position[0])
23 |
24 | return input_paths
25 |
26 |
27 | def _str_to_path(ctx: click.Context, opt: click.Option, value: str) -> Path:
28 | return Path(value)
29 |
30 |
31 | def input_position_dirpaths() -> Callable:
32 | def decorator(f: Callable) -> Callable:
33 | return click.option(
34 | "--input-position-dirpaths",
35 | "-i",
36 | cls=OptionEatAll,
37 | type=tuple,
38 | required=True,
39 | callback=_validate_and_process_paths,
40 | help="List of paths to input positions, each with the same TCZYX shape. Supports wildcards e.g. 'input.zarr/*/*/*'.",
41 | )(f)
42 |
43 | return decorator
44 |
45 |
46 | def config_filepath() -> Callable:
47 | def decorator(f: Callable) -> Callable:
48 | return click.option(
49 | "--config-filepath",
50 | "-c",
51 | required=True,
52 | type=click.Path(exists=True, file_okay=True, dir_okay=False),
53 | callback=_str_to_path,
54 | help="Path to YAML configuration file.",
55 | )(f)
56 |
57 | return decorator
58 |
59 |
60 | def transfer_function_dirpath() -> Callable:
61 | def decorator(f: Callable) -> Callable:
62 | return click.option(
63 | "--transfer-function-dirpath",
64 | "-t",
65 | required=True,
66 | type=click.Path(exists=False),
67 | callback=_str_to_path,
68 | help="Path to transfer function .zarr.",
69 | )(f)
70 |
71 | return decorator
72 |
73 |
74 | def output_dirpath() -> Callable:
75 | def decorator(f: Callable) -> Callable:
76 | return click.option(
77 | "--output-dirpath",
78 | "-o",
79 | required=True,
80 | type=click.Path(exists=False),
81 | callback=_str_to_path,
82 | help="Path to output directory.",
83 | )(f)
84 |
85 | return decorator
86 |
87 |
88 | # TODO: this setting will have to be collected from SLURM?
89 | def processes_option(default: int = None) -> Callable:
90 | def check_processes_option(ctx, param, value):
91 | max_processes = mp.cpu_count()
92 | if value > max_processes:
93 | raise click.BadParameter(
94 | f"Maximum number of processes is {max_processes}"
95 | )
96 | return value
97 |
98 | def decorator(f: Callable) -> Callable:
99 | return click.option(
100 | "--num_processes",
101 | "-j",
102 | default=default or mp.cpu_count(),
103 | type=int,
104 | help="Number of processes to run in parallel.",
105 | callback=check_processes_option,
106 | )(f)
107 |
108 | return decorator
109 |
110 |
111 | def ram_multiplier() -> Callable:
112 | def decorator(f: Callable) -> Callable:
113 | return click.option(
114 | "--ram-multiplier",
115 | "-rx",
116 | default=1.0,
117 | type=float,
118 | help="SLURM RAM multiplier.",
119 | )(f)
120 |
121 | return decorator
122 |
123 | def unique_id() -> Callable:
124 | def decorator(f: Callable) -> Callable:
125 | return click.option(
126 | "--unique-id",
127 | "-uid",
128 | default="",
129 | required=False,
130 | type=str,
131 | help="Unique ID.",
132 | )(f)
133 |
134 | return decorator
--------------------------------------------------------------------------------
/recOrder/cli/printing.py:
--------------------------------------------------------------------------------
1 | import click
2 | import yaml
3 |
4 |
5 | def echo_settings(settings):
6 | click.echo(
7 | yaml.dump(settings.dict(), default_flow_style=False, sort_keys=False)
8 | )
9 |
10 |
11 | def echo_headline(headline):
12 | click.echo(click.style(headline, fg="green"))
13 |
--------------------------------------------------------------------------------
/recOrder/cli/reconstruct.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 |
5 | from recOrder.cli.apply_inverse_transfer_function import (
6 | apply_inverse_transfer_function_cli,
7 | )
8 | from recOrder.cli.compute_transfer_function import (
9 | compute_transfer_function_cli,
10 | )
11 | from recOrder.cli.parsing import (
12 | config_filepath,
13 | input_position_dirpaths,
14 | output_dirpath,
15 | processes_option,
16 | ram_multiplier,
17 | unique_id,
18 | )
19 |
20 |
21 | @click.command()
22 | @input_position_dirpaths()
23 | @config_filepath()
24 | @output_dirpath()
25 | @processes_option(default=1)
26 | @ram_multiplier()
27 | @unique_id()
28 | def reconstruct(
29 | input_position_dirpaths,
30 | config_filepath,
31 | output_dirpath,
32 | num_processes,
33 | ram_multiplier,
34 | unique_id,
35 | ):
36 | """
37 | Reconstruct a dataset using a configuration file. This is a
38 | convenience function for a `compute-tf` call followed by a `apply-inv-tf`
39 | call.
40 |
41 | Calculates the transfer function based on the shape of the first position
42 | in the list `input-position-dirpaths`, then applies that transfer function
43 | to all positions in the list `input-position-dirpaths`, so all positions
44 | must have the same TCZYX shape.
45 |
46 | See /examples for example configuration files.
47 |
48 | >> recorder reconstruct -i ./input.zarr/*/*/* -c ./examples/birefringence.yml -o ./output.zarr
49 | """
50 |
51 | # Handle transfer function path
52 | transfer_function_path = output_dirpath.parent / Path(
53 | "transfer_function_" + config_filepath.stem + ".zarr"
54 | )
55 |
56 | # Compute transfer function
57 | compute_transfer_function_cli(
58 | input_position_dirpaths[0],
59 | config_filepath,
60 | transfer_function_path,
61 | )
62 |
63 | # Apply inverse transfer function
64 | apply_inverse_transfer_function_cli(
65 | input_position_dirpaths,
66 | transfer_function_path,
67 | config_filepath,
68 | output_dirpath,
69 | num_processes,
70 | ram_multiplier,
71 | unique_id,
72 | )
73 |
--------------------------------------------------------------------------------
/recOrder/cli/settings.py:
--------------------------------------------------------------------------------
1 | import os
2 | from typing import List, Literal, Optional, Union
3 | from pathlib import Path
4 |
5 | from pydantic.v1 import (
6 | BaseModel,
7 | Extra,
8 | NonNegativeFloat,
9 | NonNegativeInt,
10 | PositiveFloat,
11 | root_validator,
12 | validator,
13 | )
14 |
15 | # This file defines the configuration settings for the CLI.
16 |
17 | # Example settings files in `/examples/settings/` are autmatically generated
18 | # by the tests in `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
19 |
20 | # To keep the example settings up to date, run `pytest` locally when this file changes.
21 |
22 |
23 | # All settings classes inherit from MyBaseModel, which forbids extra parameters to guard against typos
24 | class MyBaseModel(BaseModel, extra=Extra.forbid):
25 | pass
26 |
27 |
28 | # Bottom level settings
29 | class WavelengthIllumination(MyBaseModel):
30 | wavelength_illumination: PositiveFloat = 0.532
31 |
32 |
33 | class BirefringenceTransferFunctionSettings(MyBaseModel):
34 | swing: float = 0.1
35 |
36 | @validator("swing")
37 | def swing_range(cls, v):
38 | if v <= 0 or v >= 1.0:
39 | raise ValueError(f"swing = {v} should be between 0 and 1.")
40 | return v
41 |
42 |
43 | class BirefringenceApplyInverseSettings(WavelengthIllumination):
44 | background_path: Union[str, Path] = ""
45 | remove_estimated_background: bool = False
46 | flip_orientation: bool = False
47 | rotate_orientation: bool = False
48 |
49 | @validator("background_path")
50 | def check_background_path(cls, v):
51 | if v == "":
52 | return v
53 |
54 | raw_dir = r"{}".format(v)
55 | if not os.path.isdir(raw_dir):
56 | raise ValueError(f"{v} is not a existing directory")
57 | return raw_dir
58 |
59 |
60 | class FourierTransferFunctionSettings(MyBaseModel):
61 | yx_pixel_size: PositiveFloat = 6.5 / 20
62 | z_pixel_size: PositiveFloat = 2.0
63 | z_padding: NonNegativeInt = 0
64 | index_of_refraction_media: PositiveFloat = 1.3
65 | numerical_aperture_detection: PositiveFloat = 1.2
66 |
67 | @validator("numerical_aperture_detection")
68 | def na_det(cls, v, values):
69 | n = values["index_of_refraction_media"]
70 | if v > n:
71 | raise ValueError(
72 | f"numerical_aperture_detection = {v} must be less than or equal to index_of_refraction_media = {n}"
73 | )
74 | return v
75 |
76 | @validator("z_pixel_size")
77 | def warn_unit_consistency(cls, v, values):
78 | yx_pixel_size = values["yx_pixel_size"]
79 | ratio = yx_pixel_size / v
80 | if ratio < 1.0 / 20 or ratio > 20:
81 | raise Warning(
82 | f"yx_pixel_size ({yx_pixel_size}) / z_pixel_size ({v}) = {ratio}. Did you use consistent units?"
83 | )
84 | return v
85 |
86 |
87 | class FourierApplyInverseSettings(MyBaseModel):
88 | reconstruction_algorithm: Literal["Tikhonov", "TV"] = "Tikhonov"
89 | regularization_strength: NonNegativeFloat = 1e-3
90 | TV_rho_strength: PositiveFloat = 1e-3
91 | TV_iterations: NonNegativeInt = 1
92 |
93 |
94 | class PhaseTransferFunctionSettings(
95 | FourierTransferFunctionSettings,
96 | WavelengthIllumination,
97 | ):
98 | numerical_aperture_illumination: NonNegativeFloat = 0.5
99 | invert_phase_contrast: bool = False
100 |
101 | @validator("numerical_aperture_illumination")
102 | def na_ill(cls, v, values):
103 | n = values.get("index_of_refraction_media")
104 | if v > n:
105 | raise ValueError(
106 | f"numerical_aperture_illumination = {v} must be less than or equal to index_of_refraction_media = {n}"
107 | )
108 | return v
109 |
110 |
111 | class FluorescenceTransferFunctionSettings(FourierTransferFunctionSettings):
112 | wavelength_emission: PositiveFloat = 0.507
113 |
114 | @validator("wavelength_emission")
115 | def warn_unit_consistency(cls, v, values):
116 | yx_pixel_size = values.get("yx_pixel_size")
117 | ratio = yx_pixel_size / v
118 | if ratio < 1.0 / 20 or ratio > 20:
119 | raise Warning(
120 | f"yx_pixel_size ({yx_pixel_size}) / wavelength_illumination ({v}) = {ratio}. Did you use consistent units?"
121 | )
122 | return v
123 |
124 |
125 | # Second level settings
126 | class BirefringenceSettings(MyBaseModel):
127 | transfer_function: BirefringenceTransferFunctionSettings = (
128 | BirefringenceTransferFunctionSettings()
129 | )
130 | apply_inverse: BirefringenceApplyInverseSettings = (
131 | BirefringenceApplyInverseSettings()
132 | )
133 |
134 |
135 | class PhaseSettings(MyBaseModel):
136 | transfer_function: PhaseTransferFunctionSettings = (
137 | PhaseTransferFunctionSettings()
138 | )
139 | apply_inverse: FourierApplyInverseSettings = FourierApplyInverseSettings()
140 |
141 |
142 | class FluorescenceSettings(MyBaseModel):
143 | transfer_function: FluorescenceTransferFunctionSettings = (
144 | FluorescenceTransferFunctionSettings()
145 | )
146 | apply_inverse: FourierApplyInverseSettings = FourierApplyInverseSettings()
147 |
148 |
149 | # Top level settings
150 | class ReconstructionSettings(MyBaseModel):
151 | input_channel_names: List[str] = [f"State{i}" for i in range(4)]
152 | time_indices: Union[
153 | NonNegativeInt, List[NonNegativeInt], Literal["all"]
154 | ] = "all"
155 | reconstruction_dimension: Literal[2, 3] = 3
156 | birefringence: Optional[BirefringenceSettings]
157 | phase: Optional[PhaseSettings]
158 | fluorescence: Optional[FluorescenceSettings]
159 |
160 | @root_validator(pre=False)
161 | def validate_reconstruction_types(cls, values):
162 | if (values.get("birefringence") or values.get("phase")) and values.get(
163 | "fluorescence"
164 | ) is not None:
165 | raise ValueError(
166 | '"fluorescence" cannot be present alongside "birefringence" or "phase". Please use one configuration file for a "fluorescence" reconstruction and another configuration file for a "birefringence" and/or "phase" reconstructions.'
167 | )
168 | num_channel_names = len(values.get("input_channel_names"))
169 | if values.get("birefringence") is None:
170 | if (
171 | values.get("phase") is None
172 | and values.get("fluorescence") is None
173 | ):
174 | raise ValueError(
175 | "Provide settings for either birefringence, phase, birefringence + phase, or fluorescence."
176 | )
177 | if num_channel_names != 1:
178 | raise ValueError(
179 | f"{num_channel_names} channels names provided. Please provide a single channel for fluorescence/phase reconstructions."
180 | )
181 |
182 | return values
183 |
--------------------------------------------------------------------------------
/recOrder/cli/utils.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from typing import Tuple
3 |
4 | import click
5 | import numpy as np
6 | import torch
7 | from iohub.ngff import Position, open_ome_zarr
8 | from iohub.ngff_meta import TransformationMeta
9 | from numpy.typing import DTypeLike
10 |
11 |
12 | def create_empty_hcs_zarr(
13 | store_path: Path,
14 | position_keys: list[Tuple[str]],
15 | shape: Tuple[int],
16 | chunks: Tuple[int],
17 | scale: Tuple[float],
18 | channel_names: list[str],
19 | dtype: DTypeLike,
20 | plate_metadata: dict = {},
21 | ) -> None:
22 | """If the plate does not exist, create an empty zarr plate.
23 |
24 | If the plate exists, append positions and channels if they are not
25 | already in the plate.
26 |
27 | Parameters
28 | ----------
29 | store_path : Path
30 | hcs plate path
31 | position_keys : list[Tuple[str]]
32 | Position keys, will append if not present in the plate.
33 | e.g. [("A", "1", "0"), ("A", "1", "1")]
34 | shape : Tuple[int]
35 | chunks : Tuple[int]
36 | scale : Tuple[float]
37 | channel_names : list[str]
38 | Channel names, will append if not present in metadata.
39 | dtype : DTypeLike
40 | plate_metadata : dict
41 | """
42 |
43 | # Create plate
44 | output_plate = open_ome_zarr(
45 | str(store_path), layout="hcs", mode="a", channel_names=channel_names
46 | )
47 |
48 | # Pass metadata
49 | output_plate.zattrs.update(plate_metadata)
50 |
51 | # Create positions
52 | for position_key in position_keys:
53 | position_key_string = "/".join(position_key)
54 | # Check if position is already in the store, if not create it
55 | if position_key_string not in output_plate.zgroup:
56 | position = output_plate.create_position(*position_key)
57 |
58 | _ = position.create_zeros(
59 | name="0",
60 | shape=shape,
61 | chunks=chunks,
62 | dtype=dtype,
63 | transform=[TransformationMeta(type="scale", scale=scale)],
64 | )
65 | else:
66 | position = output_plate[position_key_string]
67 |
68 | # Check if channel_names are already in the store, if not append them
69 | for channel_name in channel_names:
70 | # Read channel names directly from metadata to avoid race conditions
71 | metadata_channel_names = [
72 | channel.label for channel in position.metadata.omero.channels
73 | ]
74 | if channel_name not in metadata_channel_names:
75 | position.append_channel(channel_name, resize_arrays=True)
76 |
77 |
78 | def apply_inverse_to_zyx_and_save(
79 | func,
80 | position: Position,
81 | output_path: Path,
82 | input_channel_indices: list[int],
83 | output_channel_indices: list[int],
84 | t_idx: int = 0,
85 | **kwargs,
86 | ) -> None:
87 | """Load a zyx array from a Position object, apply a transformation and save the result to file"""
88 | click.echo(f"Reconstructing t={t_idx}")
89 |
90 | # Load data
91 | czyx_uint16_numpy = position.data.oindex[t_idx, input_channel_indices]
92 |
93 | # convert to np.int32 (torch doesn't accept np.uint16), then convert to tensor float32
94 | czyx_data = torch.tensor(np.int32(czyx_uint16_numpy), dtype=torch.float32)
95 |
96 | # Apply transformation
97 | reconstruction_czyx = func(czyx_data, **kwargs)
98 |
99 | # Write to file
100 | # for c, recon_zyx in enumerate(reconstruction_zyx):
101 | with open_ome_zarr(output_path, mode="r+") as output_dataset:
102 | output_dataset[0].oindex[
103 | t_idx, output_channel_indices
104 | ] = reconstruction_czyx
105 | click.echo(f"Finished Writing.. t={t_idx}")
106 |
107 |
--------------------------------------------------------------------------------
/recOrder/io/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/io/__init__.py
--------------------------------------------------------------------------------
/recOrder/io/_reader.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Tuple, Union
2 |
3 | import zarr
4 | from iohub import read_micromanager
5 | from napari_ome_zarr._reader import napari_get_reader as fallback_reader
6 |
7 |
8 | def napari_get_reader(path):
9 | if isinstance(path, str):
10 | if ".zarr" in path:
11 | with zarr.open(path) as root:
12 | if "plate" in root.attrs:
13 | return hcs_zarr_reader
14 | else:
15 | return fallback_reader(path)
16 | else:
17 | return ome_tif_reader
18 | else:
19 | return None
20 |
21 |
22 | def hcs_zarr_reader(
23 | path: Union[str, List[str]]
24 | ) -> List[Tuple[zarr.Array, Dict]]:
25 | reader = read_micromanager(path)
26 | results = list()
27 |
28 | zs = zarr.open(path, "r")
29 | names = []
30 |
31 | dict_ = zs.attrs.asdict()
32 | wells = dict_["plate"]["wells"]
33 | for well in wells:
34 | path = well["path"]
35 | well_dict = zs[path].attrs.asdict()
36 | for name in well_dict["well"]["images"]:
37 | names.append(name["path"])
38 | for pos in range(reader.get_num_positions()):
39 | meta = dict()
40 | name = names[pos]
41 | meta["name"] = name
42 | results.append((reader.get_zarr(pos), meta))
43 | return results
44 |
45 |
46 | def ome_tif_reader(
47 | path: Union[str, List[str]]
48 | ) -> List[Tuple[zarr.Array, Dict]]:
49 | reader = read_micromanager(path)
50 | results = list()
51 |
52 | npos = reader.get_num_positions()
53 | for pos in range(npos):
54 | meta = dict()
55 | if npos == 1:
56 | meta["name"] = "Pos000_000"
57 | else:
58 | meta["name"] = reader.stage_positions[pos]["Label"][2:]
59 | results.append((reader.get_zarr(pos), meta))
60 |
61 | return results
62 |
--------------------------------------------------------------------------------
/recOrder/io/core_functions.py:
--------------------------------------------------------------------------------
1 | import time
2 | from contextlib import contextmanager
3 |
4 | import numpy as np
5 |
6 |
7 | @contextmanager
8 | def suspend_live_sm(snap_manager):
9 | """Context manager that suspends/unsuspends MM live mode for `SnapLiveManager`.
10 |
11 | Parameters
12 | ----------
13 | snap_manager : object
14 | `org.micromanager.internal.SnapLiveManager` object via pycromanager
15 |
16 | Yields
17 | ------
18 | object
19 | `org.micromanager.internal.SnapLiveManager` object via pycromanager
20 |
21 | Usage
22 | -----
23 | ```py
24 | with suspend_live_sm(snap_manager) as sm:
25 | pass # do something with MM that can't be done in live mode
26 | ```
27 | """
28 | snap_manager.setSuspended(True)
29 | try:
30 | yield snap_manager
31 | finally:
32 | snap_manager.setSuspended(False)
33 |
34 |
35 | def snap_and_get_image(snap_manager):
36 | """
37 | Snap and get image using Snap Live Window Manager + transfer of ZMQ
38 |
39 | Parameters
40 | ----------
41 | snap_manager: (object) MM Snap Live Window object
42 |
43 | Returns
44 | -------
45 | image: (array) 2D array of size (Y, X)
46 |
47 | """
48 | snap_manager.snap(True)
49 | time.sleep(
50 | 0.3
51 | ) # sleep after snap to make sure the image we grab is the correct one
52 |
53 | # get pixels + dimensions
54 | height = snap_manager.getDisplay().getDisplayedImages().get(0).getHeight()
55 | width = snap_manager.getDisplay().getDisplayedImages().get(0).getWidth()
56 | array = (
57 | snap_manager.getDisplay().getDisplayedImages().get(0).getRawPixels()
58 | )
59 |
60 | return np.reshape(array, (height, width))
61 |
62 |
63 | def snap_and_average(snap_manager, display=True):
64 | """
65 | Snap an image with Snap Live manager + grab only the mean (computed in java)
66 |
67 | Parameters
68 | ----------
69 | snap_manager: (object) MM Snap Live Window object
70 | display: (bool) Whether to show the snap on the Snap Live Window in MM
71 |
72 | Returns
73 | -------
74 | mean: (float) mean of snapped image
75 |
76 | """
77 |
78 | snap_manager.snap(display)
79 | time.sleep(
80 | 0.3
81 | ) # sleep after snap to make sure the image we grab is the correct one
82 |
83 | return snap_manager.getDisplay().getImagePlus().getStatistics().umean
84 |
85 |
86 | def set_lc_waves(mmc, device_property: tuple, value: float):
87 | """
88 | Set retardance in waves for LC in device_property
89 |
90 | Parameters
91 | ----------
92 | mmc : object
93 | MM Core object
94 | device_property : tuple
95 | (device_name, property_name) set
96 | value : float
97 | Retardance to set as fraction of a wavelength
98 |
99 | Returns
100 | -------
101 |
102 | """
103 | device_name = device_property[0]
104 | prop_name = device_property[1]
105 |
106 | if value > 1.6 or value < 0.001:
107 | raise ValueError(
108 | f"Requested retardance value is {value} waves. "
109 | f"Retardance must be greater than 0.001 and less than 1.6 waves."
110 | )
111 |
112 | mmc.setProperty(device_name, prop_name, str(value))
113 | time.sleep(20 / 1000)
114 |
115 |
116 | def set_lc_voltage(mmc, device_property: tuple, value: float):
117 | """
118 | Set LC retardance by specifying LC voltage
119 |
120 | Parameters
121 | ----------
122 | mmc : object
123 | MM Core object
124 | device_property : tuple
125 | (device_name, property_name) set
126 | value : float
127 | LC voltage in volts. Applied voltage is limited to 20V
128 |
129 | Returns
130 | -------
131 |
132 | """
133 | device_name = device_property[0]
134 | prop_name = device_property[1]
135 |
136 | if value > 20.0 or value < 0.0:
137 | raise ValueError(
138 | f"Requested LC voltage is {value} V. "
139 | f"LC voltage must be greater than 0.0 and less than 20.0 V."
140 | )
141 |
142 | mmc.setProperty(device_name, prop_name, str(value))
143 | time.sleep(20 / 1000)
144 |
145 |
146 | def set_lc_daq(mmc, device_property: tuple, value: float):
147 | """
148 | Set LC retardance based on DAQ output
149 |
150 | Parameters
151 | ----------
152 | mmc : object
153 | MM Core object
154 | device_property : tuple
155 | (device_name, property_name) set
156 | value : float
157 | DAQ output voltage in volts. DAQ output must be in 0-5V range
158 |
159 | Returns
160 | -------
161 |
162 | """
163 | device_name = device_property[0]
164 | prop_name = device_property[1]
165 |
166 | if value > 5.0 or value < 0.0:
167 | raise ValueError(
168 | "DAC voltage must be greater than 0.0 and less than 5.0"
169 | )
170 |
171 | mmc.setProperty(device_name, prop_name, str(value))
172 | time.sleep(20 / 1000)
173 |
174 |
175 | def get_lc(mmc, device_property: tuple):
176 | """
177 | Get LC state in the native units of the device property
178 |
179 | Parameters
180 | ----------
181 | mmc : object
182 | MM Core object
183 | device_property : tuple
184 | (device_name, property_name) set
185 |
186 | Returns
187 | -------
188 |
189 | """
190 |
191 | device_name = device_property[0]
192 | prop_name = device_property[1]
193 |
194 | val = float(mmc.getProperty(device_name, prop_name))
195 | return val
196 |
197 |
198 | def define_meadowlark_state(mmc, device_property: tuple):
199 | """
200 | Defines pallet element in the Meadowlark device adapter for the given state.
201 | Make sure LC values for this state are set before calling this function
202 |
203 | Parameters
204 | ----------
205 | mmc : object
206 | MM Core object
207 | device_property : tuple
208 | (device_name, property_name) set, e.g.
209 | ('MeadowlarkLC', 'Pal. elem. 00; enter 0 to define; 1 to activate')
210 |
211 | Returns
212 | -------
213 |
214 | """
215 |
216 | device_name = device_property[0]
217 | prop_name = device_property[1]
218 |
219 | # define LC state
220 | # setting pallet elements to 0 defines LC state
221 | mmc.setProperty(device_name, prop_name, 0)
222 | mmc.waitForDevice(device_name)
223 |
224 |
225 | def define_config_state(
226 | mmc, group: str, config: str, device_properties: list, values: list
227 | ):
228 | """
229 | Define config state by specifying the values for all device properties in this config
230 |
231 | Parameters
232 | ----------
233 | mmc : object
234 | MM Core object
235 | group : str
236 | Name of config group
237 | config : str
238 | Name of config, e.g. State0
239 | device_properties: list
240 | List of (device_name, property_name) tuples in config
241 | values: list
242 | List of matching device property values
243 |
244 | Returns
245 | -------
246 |
247 | """
248 |
249 | for device_property, value in zip(device_properties, values):
250 | device_name = device_property[0]
251 | prop_name = device_property[1]
252 | mmc.defineConfig(group, config, device_name, prop_name, str(value))
253 | mmc.waitForConfig(group, config)
254 |
255 |
256 | def set_lc_state(mmc, group: str, config: str):
257 | """
258 | Change to the specific LC State
259 |
260 | Parameters
261 | ----------
262 | mmc : object
263 | MM Core object
264 | group : str
265 | Name of config group
266 | config : str
267 | Name of config, e.g. State0
268 |
269 | """
270 |
271 | mmc.setConfig(group, config)
272 | time.sleep(20 / 1000) # delay for LC settle time
273 |
--------------------------------------------------------------------------------
/recOrder/io/metadata_reader.py:
--------------------------------------------------------------------------------
1 | import json
2 | import os
3 |
4 | from natsort import natsorted
5 |
6 |
7 | def load_json(path):
8 | with open(path, "r") as f:
9 | data = json.load(f)
10 |
11 | return data
12 |
13 |
14 | def get_last_metadata_file(path):
15 | last_metadata_file = natsorted(
16 | [
17 | file
18 | for file in os.listdir(path)
19 | if file.startswith("calibration_metadata")
20 | ]
21 | )[-1]
22 | return os.path.join(path, last_metadata_file)
23 |
24 |
25 | class MetadataReader:
26 | """
27 | Calibration metadata reader class. Helps load metadata from different metadata formats and naming conventions
28 | """
29 |
30 | def __init__(self, path: str):
31 | """
32 |
33 | Parameters
34 | ----------
35 | path: full path to calibration metadata
36 | """
37 | self.metadata_path = path
38 | self.json_metadata = load_json(self.metadata_path)
39 |
40 | self.Timestamp = self.get_summary_calibration_attr("Timestamp")
41 | self.recOrder_napari_verion = self.get_summary_calibration_attr(
42 | "recOrder-napari version"
43 | )
44 | self.waveorder_version = self.get_summary_calibration_attr(
45 | "waveorder version"
46 | )
47 | self.Calibration_scheme = self.get_calibration_scheme()
48 | self.Swing = self.get_swing()
49 | self.Wavelength = self.get_summary_calibration_attr("Wavelength (nm)")
50 | self.Black_level = self.get_black_level()
51 | self.Extinction_ratio = self.get_extinction_ratio()
52 | self.Channel_names = self.get_channel_names()
53 | self.LCA_retardance = self.get_lc_retardance("LCA")
54 | self.LCB_retardance = self.get_lc_retardance("LCB")
55 | self.LCA_voltage = self.get_lc_voltage("LCA")
56 | self.LCB_voltage = self.get_lc_voltage("LCB")
57 | self.Swing_measured = self.get_swing_measured()
58 | self.Notes = self.get_notes()
59 |
60 | def get_summary_calibration_attr(self, attr):
61 | try:
62 | val = self.json_metadata["Summary"][attr]
63 | except KeyError:
64 | try:
65 | val = self.json_metadata["Calibration"][attr]
66 | except KeyError:
67 | val = None
68 | return val
69 |
70 | def get_cal_states(self):
71 | if self.Calibration_scheme == "4-State":
72 | states = ["ext", "0", "60", "120"]
73 | elif self.Calibration_scheme == "5-State":
74 | states = ["ext", "0", "45", "90", "135"]
75 | return states
76 |
77 | def get_lc_retardance(self, lc):
78 | """
79 |
80 | Parameters
81 | ----------
82 | lc: 'LCA' or 'LCB'
83 |
84 | Returns
85 | -------
86 |
87 | """
88 | states = self.get_cal_states()
89 |
90 | val = None
91 | try:
92 | val = [
93 | self.json_metadata["Calibration"]["LC retardance"][
94 | f"{lc}_{state}"
95 | ]
96 | for state in states
97 | ]
98 | except KeyError:
99 | states[0] = "Ext"
100 | if lc == "LCA":
101 | val = [
102 | self.json_metadata["Summary"][
103 | f"[LCA_{state}, LCB_{state}]"
104 | ][0]
105 | for state in states
106 | ]
107 | elif lc == "LCB":
108 | val = [
109 | self.json_metadata["Summary"][
110 | f"[LCA_{state}, LCB_{state}]"
111 | ][1]
112 | for state in states
113 | ]
114 |
115 | return val
116 |
117 | def get_lc_voltage(self, lc):
118 | """
119 |
120 | Parameters
121 | ----------
122 | lc: 'LCA' or 'LCB'
123 |
124 | Returns
125 | -------
126 |
127 | """
128 | states = self.get_cal_states()
129 |
130 | val = None
131 | if "Calibration" in self.json_metadata:
132 | lc_voltage = self.json_metadata["Calibration"]["LC voltage"]
133 | if lc_voltage:
134 | val = [
135 | self.json_metadata["Calibration"]["LC voltage"][
136 | f"{lc}_{state}"
137 | ]
138 | for state in states
139 | ]
140 |
141 | return val
142 |
143 | def get_swing(self):
144 | try:
145 | val = self.json_metadata["Calibration"]["Swing (waves)"]
146 | except KeyError:
147 | val = self.json_metadata["Summary"]["Swing (fraction)"]
148 | return val
149 |
150 | def get_swing_measured(self):
151 | states = self.get_cal_states()
152 | try:
153 | val = [
154 | self.json_metadata["Calibration"][f"Swing_{state}"]
155 | for state in states[1:]
156 | ]
157 | except KeyError:
158 | val = [
159 | self.json_metadata["Summary"][f"Swing{state}"]
160 | for state in states[1:]
161 | ]
162 |
163 | return val
164 |
165 | def get_calibration_scheme(self):
166 | try:
167 | val = self.json_metadata["Calibration"]["Calibration scheme"]
168 | except KeyError:
169 | val = self.json_metadata["Summary"]["Acquired Using"]
170 | return val
171 |
172 | def get_black_level(self):
173 | try:
174 | val = self.json_metadata["Calibration"]["Black level"]
175 | except KeyError:
176 | val = self.json_metadata["Summary"]["BlackLevel"]
177 | return val
178 |
179 | def get_extinction_ratio(self):
180 | try:
181 | val = self.json_metadata["Calibration"]["Extinction ratio"]
182 | except KeyError:
183 | val = self.json_metadata["Summary"]["Extinction Ratio"]
184 | return val
185 |
186 | def get_channel_names(self):
187 | try:
188 | val = self.json_metadata["Calibration"]["Channel names"]
189 | except KeyError:
190 | val = self.json_metadata["Summary"]["ChNames"]
191 | return val
192 |
193 | def get_notes(self):
194 | try:
195 | val = self.json_metadata["Notes"]
196 | except KeyError:
197 | val = None
198 | return val
199 |
--------------------------------------------------------------------------------
/recOrder/io/utils.py:
--------------------------------------------------------------------------------
1 | import os
2 | import textwrap
3 | from pathlib import Path
4 |
5 | import psutil
6 | import torch
7 | import yaml
8 | from iohub import open_ome_zarr
9 |
10 |
11 |
12 | def add_index_to_path(path: Path):
13 | """Takes a path to a file or folder and appends the smallest index that does
14 | not already exist in that folder.
15 |
16 | For example:
17 | './output.txt' -> './output_0.txt' if no other files named './output*.txt' exist.
18 | './output.txt' -> './output_2.txt' if './output_0.txt' and './output_1.txt' already exist.
19 |
20 | Parameters
21 | ----------
22 | path: Path
23 | Base path to add index to
24 |
25 | Returns
26 | -------
27 | Path
28 | """
29 | index = 0
30 | new_stem = f"{path.stem}_{index}"
31 |
32 | while (path.parent / (new_stem + path.suffix)).exists():
33 | index += 1
34 | new_stem = f"{path.stem}_{index}"
35 |
36 | return path.parent / (new_stem + path.suffix)
37 |
38 |
39 | def load_background(background_path):
40 | with open_ome_zarr(
41 | os.path.join(background_path, "background.zarr", "0", "0", "0")
42 | ) as dataset:
43 | cyx_data = dataset["0"][0, :, 0]
44 | return torch.tensor(cyx_data, dtype=torch.float32)
45 |
46 |
47 | class MockEmitter:
48 | def emit(self, value):
49 | pass
50 |
51 |
52 | def ram_message():
53 | """
54 | Determine if the system's RAM capacity is sufficient for running reconstruction.
55 | The message should be treated as a warning if the RAM detected is less than 32 GB.
56 |
57 | Returns
58 | -------
59 | ram_report (is_warning, message)
60 | """
61 | BYTES_PER_GB = 2**30
62 | gb_available = psutil.virtual_memory().total / BYTES_PER_GB
63 | is_warning = gb_available < 32
64 |
65 | if is_warning:
66 | message = " \n".join(
67 | textwrap.wrap(
68 | f"recOrder reconstructions often require more than the {gb_available:.1f} "
69 | f"GB of RAM that this computer is equipped with. We recommend starting with reconstructions of small "
70 | f"volumes ~1000 x 1000 x 10 and working up to larger volumes while monitoring your RAM usage with "
71 | f"Task Manager or htop.",
72 | )
73 | )
74 | else:
75 | message = f"{gb_available:.1f} GB of RAM is available."
76 |
77 | return (is_warning, message)
78 |
79 |
80 | def model_to_yaml(model, yaml_path: Path) -> None:
81 | """
82 | Save a model's dictionary representation to a YAML file.
83 |
84 | Parameters
85 | ----------
86 | model : object
87 | The model object to convert to YAML.
88 | yaml_path : Path
89 | The path to the output YAML file.
90 |
91 | Raises
92 | ------
93 | TypeError
94 | If the `model` object does not have a `dict()` method.
95 |
96 | Notes
97 | -----
98 | This function converts a model object into a dictionary representation
99 | using the `dict()` method. It removes any fields with None values before
100 | writing the dictionary to a YAML file.
101 |
102 | Examples
103 | --------
104 | >>> from my_model import MyModel
105 | >>> model = MyModel()
106 | >>> model_to_yaml(model, 'model.yaml')
107 |
108 | """
109 | yaml_path = Path(yaml_path)
110 |
111 | if not hasattr(model, "dict"):
112 | raise TypeError("The 'model' object does not have a 'dict()' method.")
113 |
114 | model_dict = model.dict()
115 |
116 | # Remove None-valued fields
117 | clean_model_dict = {
118 | key: value for key, value in model_dict.items() if value is not None
119 | }
120 |
121 | with open(yaml_path, "w+") as f:
122 | yaml.dump(
123 | clean_model_dict, f, default_flow_style=False, sort_keys=False
124 | )
125 |
126 |
127 | def yaml_to_model(yaml_path: Path, model):
128 | """
129 | Load model settings from a YAML file and create a model instance.
130 |
131 | Parameters
132 | ----------
133 | yaml_path : Path
134 | The path to the YAML file containing the model settings.
135 | model : class
136 | The model class used to create an instance with the loaded settings.
137 |
138 | Returns
139 | -------
140 | object
141 | An instance of the model class with the loaded settings.
142 |
143 | Raises
144 | ------
145 | TypeError
146 | If the provided model is not a class or does not have a callable constructor.
147 | FileNotFoundError
148 | If the YAML file specified by `yaml_path` does not exist.
149 |
150 | Notes
151 | -----
152 | This function loads model settings from a YAML file using `yaml.safe_load()`.
153 | It then creates an instance of the provided `model` class using the loaded settings.
154 |
155 | Examples
156 | --------
157 | # >>> from my_model import MyModel
158 | # >>> model = yaml_to_model('model.yaml', MyModel)
159 |
160 | """
161 | yaml_path = Path(yaml_path)
162 |
163 | if not callable(getattr(model, "__init__", None)):
164 | raise TypeError(
165 | "The provided model must be a class with a callable constructor."
166 | )
167 |
168 | try:
169 | with open(yaml_path, "r") as file:
170 | raw_settings = yaml.safe_load(file)
171 | except FileNotFoundError:
172 | raise FileNotFoundError(f"The YAML file '{yaml_path}' does not exist.")
173 |
174 | return model(**raw_settings)
175 |
--------------------------------------------------------------------------------
/recOrder/io/visualization.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 | from typing import Literal, Union
4 | from colorspacious import cspace_convert
5 | from matplotlib.colors import hsv_to_rgb
6 | from skimage.color import hsv2rgb
7 | from skimage.exposure import rescale_intensity
8 |
9 |
10 | def ret_ori_overlay(
11 | czyx,
12 | ret_min: float = 1,
13 | ret_max: Union[float, Literal["auto"]] = 10,
14 | cmap: Literal["JCh", "HSV"] = "HSV",
15 | ):
16 | """
17 | Creates an overlay of retardance and orientation with two different colormap options.
18 | "HSV" maps orientation to hue and retardance to value with maximum saturation.
19 | "JCh" is a similar colormap but is perceptually uniform.
20 |
21 | Parameters
22 | ----------
23 | czyx: (nd-array) czyx[0] is retardance in nanometers, czyx[1] is orientation in radians [0, pi],
24 | czyx.shape = (2, ...)
25 |
26 | ret_min: (float) minimum displayed retardance. Typically a noise floor.
27 | ret_max: (float) maximum displayed retardance. Typically used to adjust contrast limits.
28 |
29 | cmap: (str) 'JCh' or 'HSV'
30 |
31 | Returns
32 | -------
33 | overlay (nd-array) RGB image with shape (3, ...)
34 |
35 | """
36 | if czyx.shape[0] != 2:
37 | raise ValueError(
38 | f"Input must have shape (2, ...) instead of ({czyx.shape[0]}, ...)"
39 | )
40 |
41 | retardance = czyx[0]
42 | orientation = czyx[1]
43 |
44 | if ret_max == "auto":
45 | ret_max = np.percentile(np.ravel(retardance), 99.99)
46 |
47 | # Prepare input and output arrays
48 | ret_ = np.clip(retardance, 0, ret_max) # clip and copy
49 | # Convert 180 degree range into 360 to match periodicity of hue.
50 | ori_ = orientation * 360 / np.pi
51 | overlay_final = np.zeros_like(retardance)
52 |
53 | if cmap == "JCh":
54 | J = ret_
55 | C = np.ones_like(J) * 60
56 | C[ret_ < ret_min] = 0
57 | h = ori_
58 |
59 | JCh = np.stack((J, C, h), axis=-1)
60 | JCh_rgb = cspace_convert(JCh, "JCh", "sRGB1")
61 |
62 | JCh_rgb[JCh_rgb < 0] = 0
63 | JCh_rgb[JCh_rgb > 1] = 1
64 |
65 | overlay_final = JCh_rgb
66 | elif cmap == "HSV":
67 | I_hsv = np.moveaxis(
68 | np.stack(
69 | [
70 | ori_ / 360,
71 | np.ones_like(ori_),
72 | ret_ / np.max(ret_),
73 | ]
74 | ),
75 | source=0,
76 | destination=-1,
77 | )
78 | overlay_final = hsv_to_rgb(I_hsv)
79 | else:
80 | raise ValueError(f"Colormap {cmap} not understood")
81 |
82 | return np.moveaxis(
83 | overlay_final, source=-1, destination=0
84 | ) # .shape = (3, ...)
85 |
86 |
87 | def ret_ori_phase_overlay(
88 | czyx, max_val_V: float = 1.0, max_val_S: float = 1.0
89 | ):
90 | """
91 | Creates an overlay of retardance, orientation, and phase.
92 | Maps orientation to hue, retardance to saturation, and phase to value.
93 |
94 | HSV encoding of retardance + orientation + phase image with hsv colormap
95 | (orientation in h, retardance in s, phase in v)
96 | Parameters
97 | ----------
98 | czyx : numpy.ndarray
99 | czyx[0] corresponds to the retardance image
100 | czyx[1]is the orientation image (range from 0 to pi)
101 | czyx[2] is the the phase image
102 |
103 | max_val_V : float
104 | raise the brightness of the phase channel by 1/max_val_V
105 |
106 | max_val_S : float
107 | raise the brightness of the retardance channel by 1/max_val_S
108 |
109 | Returns
110 | -------
111 | overlay (nd-array) RGB image with shape (3, ...)
112 |
113 | Returns:
114 | RGB with HSV
115 | """
116 |
117 | if czyx.shape[0] != 3:
118 | raise ValueError(
119 | f"Input must have shape (3, ...) instead of ({czyx.shape[0]}, ...)"
120 | )
121 |
122 | czyx_out = np.zeros_like(czyx, dtype=np.float32)
123 |
124 | retardance = czyx[0]
125 | orientation = czyx[1]
126 | phase = czyx[2]
127 |
128 | # Normalize the stack
129 | ordered_stack = np.stack(
130 | (
131 | # Normalize the first channel by dividing by pi
132 | orientation / np.pi,
133 | # Normalize the second channel and rescale intensity
134 | rescale_intensity(
135 | retardance,
136 | in_range=(
137 | np.min(retardance),
138 | np.max(retardance),
139 | ),
140 | out_range=(0, 1),
141 | )
142 | / max_val_S,
143 | # Normalize the third channel and rescale intensity
144 | rescale_intensity(
145 | phase,
146 | in_range=(
147 | np.min(phase),
148 | np.max(phase),
149 | ),
150 | out_range=(0, 1),
151 | )
152 | / max_val_V,
153 | ),
154 | axis=0,
155 | )
156 | czyx_out = hsv2rgb(ordered_stack, channel_axis=0)
157 | return czyx_out
158 |
--------------------------------------------------------------------------------
/recOrder/napari.yaml:
--------------------------------------------------------------------------------
1 | name: recOrder-napari
2 | display_name: recOrder-napari
3 | contributions:
4 | commands:
5 | - id: recOrder-napari.MainWidget
6 | title: Create Main Widget
7 | python_name: recOrder.plugin.main_widget:MainWidget
8 | - id: recOrder-napari.get_reader
9 | title: Read ome-zarr and ome-tif files
10 | python_name: recOrder.io._reader:napari_get_reader
11 | - id: recOrder-napari.polarization_target_data
12 | title: Polarization Target Data
13 | python_name: recOrder.scripts.samples:read_polarization_target_data
14 | - id: recOrder-napari.polarization_target_reconstruction
15 | title: Polarization Target Data
16 | python_name: recOrder.scripts.samples:read_polarization_target_reconstruction
17 | - id: recOrder-napari.zebrafish_embryo_reconstruction
18 | title: Zebrafish Embryo Reconstruction
19 | python_name: recOrder.scripts.samples:read_zebrafish_embryo_reconstruction
20 | readers:
21 | - command: recOrder-napari.get_reader
22 | accepts_directories: true
23 | filename_patterns: ['*.zarr', '*.tif']
24 | widgets:
25 | - command: recOrder-napari.MainWidget
26 | display_name: Main Menu
27 | sample_data:
28 | - command: recOrder-napari.polarization_target_data
29 | key: polarization-target-data
30 | display_name: Polarization Target Data (10 MB)
31 | - command: recOrder-napari.polarization_target_reconstruction
32 | key: polarization-target-reconstruction
33 | display_name: Polarization Target Reconstruction (10 MB)
34 | - command: recOrder-napari.zebrafish_embryo_reconstruction
35 | key: zebrafish-embryo-reconstruction
36 | display_name: Zebrafish Embryo Reconstruction (92 MB)
37 |
38 |
--------------------------------------------------------------------------------
/recOrder/plugin/__init__.py:
--------------------------------------------------------------------------------
1 | # qtpy defaults to PyQt5/PySide2 which can be present in upgraded environments
2 | try:
3 | import qtpy
4 |
5 | qtpy.API_NAME # check qtpy API name - one is required for GUI
6 |
7 | except RuntimeError as error:
8 | if type(error).__name__ == "QtBindingsNotFoundError":
9 | print("WARNING: QtBindings (PyQT or PySide) was not found for GUI")
10 |
--------------------------------------------------------------------------------
/recOrder/scripts/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/scripts/__init__.py
--------------------------------------------------------------------------------
/recOrder/scripts/launch_napari.py:
--------------------------------------------------------------------------------
1 | import napari
2 | from recOrder.plugin.main_widget import MainWidget
3 |
4 |
5 | def main():
6 | viewer = napari.Viewer()
7 | viewer.window.add_dock_widget(MainWidget(viewer))
8 | napari.run()
9 |
10 |
11 | if __name__ == "__main__":
12 | main()
13 |
--------------------------------------------------------------------------------
/recOrder/scripts/repeat-cal-acq-rec.py:
--------------------------------------------------------------------------------
1 | # This script can be modified to debug and test calibrations
2 |
3 | import random
4 | import time
5 | from contextlib import contextmanager
6 |
7 | import napari
8 | from pycromanager import Core
9 |
10 | from recOrder.plugin.main_widget import MainWidget
11 |
12 | SAVE_DIR = "."
13 | SWING = 0.05
14 | CAL_REPEATS = 3
15 | BKG_REPEATS = 3
16 |
17 |
18 | @contextmanager
19 | def stage_detour(app: MainWidget, dx: float, dy: float, wait=5):
20 | """Context manager to temporarily move the stage to a new XY-position.
21 |
22 | Parameters
23 | ----------
24 | app : MainWidget
25 | recOrder main widget instance
26 | dx : float
27 | relative x to translate
28 | dy : float
29 | relative y to translate
30 | wait : int, optional
31 | time to wait for the stage to complete movement, by default 5
32 |
33 | Yields
34 | ------
35 | MainWidget
36 | recOrder main widget instance
37 |
38 | Usage
39 | -----
40 | ```py
41 | with stage_detour(app) as app:
42 | pass # do something at the new location
43 | ```
44 | """
45 | xy_stage = app.mmc.getXYStageDevice()
46 | # get the original position
47 | ox = app.mmc.getXPosition(xy_stage)
48 | oy = app.mmc.getYPosition(xy_stage)
49 | # go to a translated position
50 | # TODO: args are floored due to a pycromanager bug: https://github.com/micro-manager/pycro-manager/issues/67
51 | app.mmc.setRelativeXYPosition(int(dx), int(dy))
52 | time.sleep(wait)
53 | try:
54 | yield app
55 | finally:
56 | # go back to the original position
57 | # TODO: args are floored due to a pycromanager bug: https://github.com/micro-manager/pycro-manager/issues/67
58 | app.mmc.setXYPosition(int(ox), int(oy))
59 | time.sleep(wait)
60 |
61 |
62 | def measure_fov(mmc: Core):
63 | """Calculate the MM FOV in micrometers.
64 |
65 | Parameters
66 | ----------
67 | mmc : Core
68 | MMCore object via pycromanager (with CamelCase set to `True`)
69 |
70 | Returns
71 | -------
72 | tuple[float, float]
73 | FOV size (x, y)
74 | """
75 | pixel_size = float(mmc.getPixelSizeUm())
76 | if pixel_size == 0:
77 | float(
78 | input(
79 | "Pixel size is not calibrated. Please provide an estimate (in micrometers):"
80 | )
81 | )
82 | fov_x = pixel_size * float(mmc.getImageWidth())
83 | fov_y = pixel_size * float(mmc.getImageHeight())
84 | return fov_x, fov_y
85 |
86 |
87 | def rand_shift(length: float):
88 | """Randomly signed shift of a certain length.
89 |
90 | Parameters
91 | ----------
92 | length : float
93 | absolote length in micrometers
94 |
95 | Returns
96 | -------
97 | float
98 | +length or -length
99 | """
100 | sign = random.randint(0, 1) * 2 - 1
101 | return sign * length
102 |
103 |
104 | def main():
105 | viewer = napari.Viewer()
106 | app = MainWidget(viewer)
107 | viewer.window.add_dock_widget(app)
108 | app.ui.qbutton_gui_mode.click()
109 | app.calib_scheme = "5-State"
110 | app.directory = SAVE_DIR
111 | app.save_directory = SAVE_DIR
112 |
113 | fov_x, fov_y = measure_fov(app.mmc)
114 |
115 | input("Please center the target in the FOV and hit ")
116 |
117 | for cal_repeat in range(CAL_REPEATS):
118 | dx = rand_shift(fov_x)
119 | dy = rand_shift(fov_y)
120 | # run calibration
121 | with stage_detour(app, dx, dy) as app:
122 | print(f"Calibration repeat # {cal_repeat}")
123 | app.swing = SWING
124 |
125 | print(f"Calibrating with swing = {SWING}")
126 | app.run_calibration()
127 | time.sleep(90)
128 |
129 | for bkg_repeat in range(BKG_REPEATS):
130 | # capture background
131 | with stage_detour(app, dx, dy) as app:
132 | print(f">>> Background repeat # {bkg_repeat}")
133 | app.last_calib_meta_file = app.calib.meta_file
134 | app.capture_bg()
135 | time.sleep(20)
136 | app.ui.cb_bg_method.setCurrentIndex(
137 | 1
138 | ) # Set to "Measured" bg correction
139 | app.enter_bg_correction()
140 | app.save_name = f"cal-{cal_repeat}-bkg-{bkg_repeat}"
141 | app.enter_acq_bg_path()
142 | app.acq_ret_ori()
143 | time.sleep(15)
144 |
145 |
146 | if __name__ == "__main__":
147 | main()
148 |
--------------------------------------------------------------------------------
/recOrder/scripts/repeat-calibration.py:
--------------------------------------------------------------------------------
1 | # This script can be modified to debug and test calibrations
2 |
3 | import napari
4 | import time
5 | from recOrder.plugin.main_widget import MainWidget
6 |
7 | SAVE_DIR = "./"
8 | SWINGS = [0.1, 0.03, 0.01, 0.005]
9 | REPEATS = 5
10 |
11 |
12 | def main():
13 | viewer = napari.Viewer()
14 | recorder = MainWidget(viewer)
15 | viewer.window.add_dock_widget(recorder)
16 | recorder.ui.qbutton_connect_to_mm.click()
17 | recorder.calib_scheme = "5-State"
18 |
19 | for repeat in range(REPEATS):
20 | for swing in SWINGS:
21 | print("Calibrating with swing = " + str(swing))
22 | recorder.swing = swing
23 | recorder.directory = SAVE_DIR
24 | recorder.run_calibration()
25 | time.sleep(100)
26 |
27 |
28 | if __name__ == "__main__":
29 | main()
30 |
--------------------------------------------------------------------------------
/recOrder/scripts/samples.py:
--------------------------------------------------------------------------------
1 | import shutil
2 | from pathlib import Path
3 |
4 | from typing import Literal
5 | from iohub import open_ome_zarr
6 | from iohub.ngff import Plate
7 | from napari.utils.notifications import show_warning
8 | from platformdirs import user_data_dir
9 | from wget import download
10 |
11 |
12 | def _build_layer_list(dataset: Plate, layer_names: list[str]):
13 | layer_list = []
14 | for channel_name in layer_names:
15 | channel_index = dataset.channel_names.index(channel_name)
16 | position = dataset["0/0/0"]
17 | data = (position["0"][:, channel_index],)
18 | layer_dict = {"name": channel_name, "scale": position.scale[3:]}
19 | layer_list.append((data, layer_dict))
20 |
21 | return layer_list
22 |
23 |
24 | def download_and_unzip(data_type: Literal["target", "embryo"]) -> tuple[Path]:
25 | """Downloads sample data .zip from zenodo, unzips, and returns Paths to the .zarr datasets.
26 |
27 | Skips the download if the files already exist.
28 |
29 | Uses platformdirs.user_data_dir to store data.
30 | """
31 |
32 | # Delete old data
33 | old_data_dirs = ["recOrder-sample-v1.4"]
34 | for old_data_dir in old_data_dirs:
35 | old_data_path = Path(user_data_dir(old_data_dir))
36 | if old_data_path.exists():
37 | shutil.rmtree(str(old_data_path))
38 |
39 | temp_dirpath = Path(user_data_dir("recOrder-sample-v1.5"))
40 | temp_dirpath.mkdir(exist_ok=True, parents=True)
41 |
42 | if data_type == "target":
43 | data_dirpath = temp_dirpath / "sample_contribution"
44 | data_size = "10 MB"
45 | data_url = "https://zenodo.org/record/8386856/files/sample_contribution.zip?download=1"
46 | elif data_type == "embryo":
47 | data_dirpath = temp_dirpath / "sample_contribution_embryo"
48 | data_size = "92 MB"
49 | data_url = "https://zenodo.org/record/8386856/files/sample_contribution_embryo.zip?download=1"
50 |
51 | if not data_dirpath.with_suffix(".zip").exists():
52 | show_warning(
53 | f"Downloading {data_size} sample contribution. This might take a moment..."
54 | )
55 | download(data_url, out=str(temp_dirpath))
56 |
57 | if not data_dirpath.exists():
58 | shutil.unpack_archive(
59 | data_dirpath.with_suffix(".zip"), extract_dir=temp_dirpath
60 | )
61 |
62 | data_path = data_dirpath / "raw_data.zarr"
63 | recon_path = data_dirpath / "reconstruction.zarr"
64 | return data_path, recon_path
65 |
66 |
67 | def read_polarization_target_data():
68 | """Returns the polarization data sample contribution"""
69 | data_path, _ = download_and_unzip("target")
70 | dataset = open_ome_zarr(data_path)
71 | return _build_layer_list(dataset, dataset.channel_names)
72 |
73 |
74 | def read_polarization_target_reconstruction():
75 | """Returns the polarization target reconstruction sample contribution"""
76 | _, recon_path = download_and_unzip("target")
77 | dataset = open_ome_zarr(recon_path)
78 | return _build_layer_list(dataset, ["Phase3D", "Retardance", "Orientation"])
79 |
80 |
81 | def read_zebrafish_embryo_reconstruction():
82 | """Returns the embryo reconstruction sample contribution"""
83 | _, recon_path = download_and_unzip("embryo")
84 | dataset = open_ome_zarr(recon_path)
85 | return _build_layer_list(dataset, ["Retardance", "Orientation"])
86 |
--------------------------------------------------------------------------------
/recOrder/scripts/simulate_zarr_acq.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from iohub.convert import TIFFConverter
3 | from iohub.ngff import open_ome_zarr
4 | from recOrder.cli.utils import create_empty_hcs_zarr
5 | from recOrder.cli import jobs_mgmt
6 |
7 | import time, threading, os, shutil, subprocess
8 |
9 | # This script is a demo .zarr acquisition simulation from an acquired .zarr store
10 | # The script copies and writes additional metadata to .zattrs inserting two keys
11 | # The two keys are "FinalDimensions" and "CurrentDimensions".
12 | # The "FinalDimensions" key with (t,p,z,c) needs to be inserted when the dataset is created
13 | # and then should be updated at close to ensure aborted acquisitions represent correct dimensions.
14 | # The "CurrentDimensions" key should have the same (t,p,z,c) information and should be written out
15 | # either with every new image, end of dimension OR at frequent intervals.
16 | # Refer further notes below in the example regarding encountered issues.
17 | #
18 | # Refer to steps at the end of the file on steps to run this file
19 |
20 | #%% #############################################
21 | def convert_data(tif_path, latest_out_path, prefix="", data_type_str="ometiff"):
22 | converter = TIFFConverter(
23 | os.path.join(tif_path , prefix),
24 | latest_out_path,
25 | data_type=data_type_str,
26 | grid_layout=False,
27 | )
28 | converter.run()
29 |
30 | def run_convert(ome_tif_path):
31 | out_path = os.path.join(Path(ome_tif_path).parent.absolute(), ("raw_" + Path(ome_tif_path).name + ".zarr"))
32 | convert_data(ome_tif_path, out_path)
33 |
34 | #%% #############################################
35 |
36 | def run_acq(input_path="", waitBetweenT=30):
37 |
38 | output_store_path = os.path.join(Path(input_path).parent.absolute(), ("acq_sim_" + Path(input_path).name))
39 |
40 | if Path(output_store_path).exists():
41 | shutil.rmtree(output_store_path)
42 | time.sleep(1)
43 |
44 | input_data = open_ome_zarr(input_path, mode="r")
45 | channel_names = input_data.channel_names
46 |
47 | position_keys: list[tuple[str]] = []
48 |
49 | for path, pos in input_data.positions():
50 | shape = pos["0"].shape
51 | dtype = pos["0"].dtype
52 | chunks = pos["0"].chunks
53 | scale = (1, 1, 1, 1, 1)
54 | position_keys.append(path.split("/"))
55 |
56 | create_empty_hcs_zarr(
57 | output_store_path,
58 | position_keys,
59 | shape,
60 | chunks,
61 | scale,
62 | channel_names,
63 | dtype,
64 | {},
65 | )
66 | output_dataset = open_ome_zarr(output_store_path, mode="r+")
67 |
68 | if "Summary" in input_data.zattrs.keys():
69 | output_dataset.zattrs["Summary"] = input_data.zattrs["Summary"]
70 |
71 | output_dataset.zattrs.update({"FinalDimensions": {
72 | "channel": shape[1],
73 | "position": len(position_keys),
74 | "time": shape[0],
75 | "z": shape[2]
76 | }
77 | })
78 |
79 | total_time = shape[0]
80 | total_pos = len(position_keys)
81 | total_z = shape[2]
82 | total_c = shape[1]
83 | for t in range(total_time):
84 | for p in range(total_pos):
85 | for z in range(total_z):
86 | for c in range(total_c):
87 | position_key_string = "/".join(position_keys[p])
88 | img_src = input_data[position_key_string][0][t, c, z]
89 |
90 | img_data = output_dataset[position_key_string][0]
91 | img_data[t, c, z] = img_src
92 |
93 | # Note: On-The-Fly dataset reconstruction will throw Permission Denied when being written
94 | # Maybe we can read the zaatrs directly in that case as a file which is less blocking
95 | # If this write/read is a constant issue then the zattrs 'CurrentDimensions' key
96 | # should be updated less frequently, instead of current design of updating with
97 | # each image
98 | output_dataset.zattrs.update({"CurrentDimensions": {
99 | "channel": total_c,
100 | "position": p+1,
101 | "time": t+1,
102 | "z": z+1
103 | }
104 | })
105 |
106 | required_order = ['time', 'position', 'z', 'channel']
107 | my_dict = output_dataset.zattrs["CurrentDimensions"]
108 | sorted_dict_acq = {k: my_dict[k] for k in sorted(my_dict, key=lambda x: required_order.index(x))}
109 | print("Writer thread - Acquisition Dim:", sorted_dict_acq)
110 |
111 |
112 | # reconThread = threading.Thread(target=doReconstruct, args=(output_store_path, t))
113 | # reconThread.start()
114 |
115 | time.sleep(waitBetweenT) # sleep after every t
116 |
117 | output_dataset.close
118 |
119 | def do_reconstruct(input_path, time_point):
120 |
121 | config_path = os.path.join(Path(input_path).parent.absolute(), "Bire-"+str(time_point)+".yml")
122 | output_path = os.path.join(Path(input_path).parent.absolute(), "Recon_"+Path(input_path).name)
123 | mainfp = str(jobs_mgmt.FILE_PATH)
124 |
125 | print("Processing {input} time_point={tp}".format(input=input_path, tp=time_point))
126 |
127 | try:
128 | proc = subprocess.run(
129 | [
130 | "python",
131 | mainfp,
132 | "reconstruct",
133 | "-i",
134 | input_path,
135 | "-c",
136 | config_path,
137 | "-o",
138 | output_path,
139 | "-rx",
140 | str(20)
141 | ]
142 | )
143 | if proc.returncode != 0:
144 | raise Exception("An error occurred in processing ! Check terminal output.")
145 | except Exception as exc:
146 | print(exc.args)
147 |
148 | #%% #############################################
149 | def run_acquire(input_path, waitBetweenT):
150 | runThread1Acq = threading.Thread(target=run_acq, args=(input_path, waitBetweenT))
151 | runThread1Acq.start()
152 |
153 | #%% #############################################
154 | # Step 1:
155 | # Convert an existing ome-tif recOrder acquisition, preferably with all dims (t, p, z, c)
156 | # This will convert an existing ome-tif to a .zarr storage
157 |
158 | # ome_tif_path = "/ome-zarr_data/recOrderAcq/test/snap_6D_ometiff_1"
159 | # runConvert(ome_tif_path)
160 |
161 | #%% #############################################
162 | # Step 2:
163 | # run the test to simulate Acquiring a recOrder .zarr store
164 |
165 | input_path = "/ome-zarr_data/recOrderAcq/test/raw_snap_6D_ometiff_1.zarr"
166 | waitBetweenT = 60
167 | run_acquire(input_path, waitBetweenT)
168 |
169 |
170 |
171 |
172 |
--------------------------------------------------------------------------------
/recOrder/tests/acq_tests/test_acq.py:
--------------------------------------------------------------------------------
1 | from unittest.mock import patch
2 |
3 | import numpy as np
4 | from recOrder.acq.acquisition_workers import _check_scale_mismatch
5 |
6 |
7 | def test_check_scale_mismatch():
8 | warn_fn_path = "recOrder.acq.acquisition_workers.show_warning"
9 | identity = np.array((1.0, 1.0, 1.0))
10 | with patch(warn_fn_path) as mock:
11 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1))
12 | mock.assert_not_called()
13 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1.001))
14 | mock.assert_not_called()
15 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1.1))
16 | mock.assert_called_once()
17 |
--------------------------------------------------------------------------------
/recOrder/tests/calibration_tests/test_calibration.py:
--------------------------------------------------------------------------------
1 | def test_calib_imports():
2 | from recOrder.calib import Calibration, Optimization
3 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_cli.py:
--------------------------------------------------------------------------------
1 | from recOrder.cli.main import cli
2 | from click.testing import CliRunner
3 |
4 |
5 | def test_main():
6 | runner = CliRunner()
7 | result = runner.invoke(cli)
8 |
9 | assert result.exit_code == 0
10 | assert "Toolkit" in result.output
11 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_compute_tf.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 | from click.testing import CliRunner
4 |
5 | from recOrder.cli import settings
6 | from recOrder.cli.compute_transfer_function import (
7 | generate_and_save_birefringence_transfer_function,
8 | generate_and_save_fluorescence_transfer_function,
9 | generate_and_save_phase_transfer_function,
10 | )
11 | from recOrder.cli.main import cli
12 | from recOrder.io import utils
13 |
14 |
15 | def test_compute_transfer(tmp_path, example_plate):
16 | recon_settings = settings.ReconstructionSettings(
17 | input_channel_names=[f"State{i}" for i in range(4)],
18 | reconstruction_dimension=3,
19 | birefringence=settings.BirefringenceSettings(),
20 | phase=settings.PhaseSettings(),
21 | )
22 | config_path = tmp_path / "test.yml"
23 | utils.model_to_yaml(recon_settings, config_path)
24 |
25 | output_path = tmp_path / "output.zarr"
26 |
27 | plate_path, _ = example_plate
28 | runner = CliRunner()
29 | result = runner.invoke(
30 | cli,
31 | [
32 | "compute-tf",
33 | "-i",
34 | str(plate_path / "A" / "1" / "0"),
35 | "-c",
36 | str(config_path),
37 | "-o",
38 | str(output_path),
39 | ],
40 | )
41 | assert result.exit_code == 0
42 |
43 |
44 | def test_compute_transfer_blank_config():
45 | runner = CliRunner()
46 | for option in ("-c ", "--config-path "):
47 | cmd = "compute-tf " + option
48 | result = runner.invoke(cli, cmd)
49 | assert result.exit_code == 2
50 | assert "Error" in result.output
51 |
52 |
53 | def test_compute_transfer_blank_output():
54 | runner = CliRunner()
55 | for option in ("-o ", "--output-path "):
56 | cmd = "compute-tf " + option
57 | result = runner.invoke(cli, cmd)
58 | assert result.exit_code == 2
59 | assert "Error" in result.output
60 |
61 |
62 | def test_compute_transfer_output_file(tmp_path, example_plate):
63 | recon_settings = settings.ReconstructionSettings(
64 | input_channel_names=["BF"],
65 | reconstruction_dimension=3,
66 | phase=settings.PhaseSettings(),
67 | )
68 | config_path = tmp_path / "test.yml"
69 | utils.model_to_yaml(recon_settings, config_path)
70 |
71 | plate_path, _ = example_plate
72 | runner = CliRunner()
73 | for option in ("-o", "--output-dirpath"):
74 | for output_folder in ["test1.zarr", "test2/test.zarr"]:
75 | output_path = tmp_path.joinpath(output_folder)
76 | result = runner.invoke(
77 | cli,
78 | [
79 | "compute-tf",
80 | "-i",
81 | str(plate_path / "A" / "1" / "0"),
82 | "-c",
83 | str(config_path),
84 | str(option),
85 | str(output_path),
86 | ],
87 | )
88 | assert result.exit_code == 0
89 | assert str(output_path) in result.output
90 | assert output_path.exists()
91 |
92 |
93 | def test_stokes_matrix_write(birefringence_phase_recon_settings_function):
94 | settings, dataset = birefringence_phase_recon_settings_function
95 | generate_and_save_birefringence_transfer_function(settings, dataset)
96 | assert dataset["intensity_to_stokes_matrix"]
97 |
98 |
99 | def test_absorption_and_phase_write(
100 | birefringence_phase_recon_settings_function,
101 | ):
102 | settings, dataset = birefringence_phase_recon_settings_function
103 | generate_and_save_phase_transfer_function(settings, dataset, (3, 4, 5))
104 | assert dataset["real_potential_transfer_function"]
105 | assert dataset["imaginary_potential_transfer_function"]
106 | assert dataset["imaginary_potential_transfer_function"].shape == (
107 | 1,
108 | 1,
109 | 3,
110 | 4,
111 | 5,
112 | )
113 | assert "absorption_transfer_function" not in dataset
114 | assert "phase_transfer_function" not in dataset
115 |
116 |
117 | def test_phase_3dim_write(birefringence_phase_recon_settings_function):
118 | settings, dataset = birefringence_phase_recon_settings_function
119 | settings.reconstruction_dimension = 2
120 | generate_and_save_phase_transfer_function(settings, dataset, (3, 4, 5))
121 | assert dataset["absorption_transfer_function"]
122 | assert dataset["phase_transfer_function"]
123 | assert dataset["phase_transfer_function"].shape == (1, 1, 3, 4, 5)
124 | assert "real_potential_transfer_function" not in dataset
125 | assert "imaginary_potential_transfer_function" not in dataset
126 |
127 |
128 | def test_fluorescence_write(fluorescence_recon_settings_function):
129 | settings, dataset = fluorescence_recon_settings_function
130 | generate_and_save_fluorescence_transfer_function(
131 | settings, dataset, (3, 4, 5)
132 | )
133 | assert dataset["optical_transfer_function"]
134 | assert dataset["optical_transfer_function"].shape == (1, 1, 3, 4, 5)
135 | assert "real_potential_transfer_function" not in dataset
136 | assert "imaginary_potential_transfer_function" not in dataset
137 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_reconstruct.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 | import numpy as np
4 | from click.testing import CliRunner
5 | from iohub.ngff import open_ome_zarr
6 | from iohub.ngff_meta import TransformationMeta
7 |
8 | from recOrder.cli import settings
9 | from recOrder.cli.main import cli
10 | from recOrder.io import utils
11 | from recOrder.cli.apply_inverse_transfer_function import (
12 | apply_inverse_transfer_function_cli,
13 | )
14 | from unittest.mock import patch
15 | import pytest
16 | from pathlib import Path
17 | from tempfile import TemporaryDirectory
18 |
19 |
20 | input_scale = [1, 2, 3, 4, 5]
21 | # Setup options
22 | birefringence_settings = settings.BirefringenceSettings(
23 | transfer_function=settings.BirefringenceTransferFunctionSettings()
24 | )
25 |
26 | # birefringence_option, time_indices, phase_option, dimension_option, time_length_target
27 | all_options = [
28 | (birefringence_settings, [0, 3, 4], None, 2, 5),
29 | (birefringence_settings, 0, settings.PhaseSettings(), 2, 5),
30 | (birefringence_settings, [0, 1], None, 3, 5),
31 | (birefringence_settings, "all", settings.PhaseSettings(), 3, 5),
32 | ]
33 |
34 |
35 | @pytest.fixture(scope="session")
36 | def tmp_input_path_zarr():
37 | tmp_path = TemporaryDirectory()
38 | yield Path(tmp_path.name) / "input.zarr", Path(tmp_path.name) / "test.yml"
39 | tmp_path.cleanup()
40 |
41 |
42 | def test_reconstruct(tmp_input_path_zarr):
43 | input_path, tmp_config_yml = tmp_input_path_zarr
44 | # Generate input "dataset"
45 | channel_names = [f"State{x}" for x in range(4)]
46 | dataset = open_ome_zarr(
47 | input_path,
48 | layout="hcs",
49 | mode="w",
50 | channel_names=channel_names,
51 | )
52 |
53 | position = dataset.create_position("0", "0", "0")
54 | position.create_zeros(
55 | "0",
56 | (5, 4, 4, 5, 6),
57 | dtype=np.uint16,
58 | transform=[TransformationMeta(type="scale", scale=input_scale)],
59 | )
60 |
61 | for i, (
62 | birefringence_option,
63 | time_indices,
64 | phase_option,
65 | dimension_option,
66 | time_length_target,
67 | ) in enumerate(all_options):
68 | if (birefringence_option is None) and (phase_option is None):
69 | continue
70 |
71 | # Generate recon settings
72 | recon_settings = settings.ReconstructionSettings(
73 | input_channel_names=channel_names,
74 | time_indices=time_indices,
75 | reconstruction_dimension=dimension_option,
76 | birefringence=birefringence_option,
77 | phase=phase_option,
78 | )
79 | config_path = tmp_config_yml.with_name(f"{i}.yml")
80 | utils.model_to_yaml(recon_settings, config_path)
81 |
82 | # Run CLI
83 | runner = CliRunner()
84 | tf_path = input_path.with_name(f"tf_{i}.zarr")
85 | runner.invoke(
86 | cli,
87 | [
88 | "compute-tf",
89 | "-i",
90 | str(input_path / "0" / "0" / "0"),
91 | "-c",
92 | str(config_path),
93 | "-o",
94 | str(tf_path),
95 | ],
96 | catch_exceptions=False,
97 | )
98 | assert tf_path.exists()
99 |
100 |
101 | def test_append_channel_reconstruction(tmp_input_path_zarr):
102 | input_path, tmp_config_yml = tmp_input_path_zarr
103 | output_path = input_path.with_name(f"output.zarr")
104 |
105 | # Generate input "dataset"
106 | channel_names = [f"State{x}" for x in range(4)] + ["GFP"]
107 | dataset = open_ome_zarr(
108 | input_path,
109 | layout="hcs",
110 | mode="w",
111 | channel_names=channel_names,
112 | )
113 | position = dataset.create_position("0", "0", "0")
114 | position.create_zeros(
115 | "0",
116 | (5, 5, 4, 5, 6),
117 | dtype=np.uint16,
118 | transform=[TransformationMeta(type="scale", scale=input_scale)],
119 | )
120 |
121 | # Generate recon settings
122 | biref_settings = settings.ReconstructionSettings(
123 | input_channel_names=[f"State{x}" for x in range(4)],
124 | time_indices="all",
125 | reconstruction_dimension=3,
126 | birefringence=settings.BirefringenceSettings(),
127 | phase=None,
128 | fluorescence=None,
129 | )
130 | fluor_settings = settings.ReconstructionSettings(
131 | input_channel_names=["GFP"],
132 | time_indices="all",
133 | reconstruction_dimension=3,
134 | birefringence=None,
135 | phase=None,
136 | fluorescence=settings.FluorescenceSettings(),
137 | )
138 | biref_config_path = tmp_config_yml.with_name(f"biref.yml")
139 | fluor_config_path = tmp_config_yml.with_name(f"fluor.yml")
140 |
141 | utils.model_to_yaml(biref_settings, biref_config_path)
142 | utils.model_to_yaml(fluor_settings, fluor_config_path)
143 |
144 | # Apply birefringence reconstruction
145 | runner = CliRunner()
146 | runner.invoke(
147 | cli,
148 | [
149 | "reconstruct",
150 | "-i",
151 | str(input_path / "0" / "0" / "0"),
152 | "-c",
153 | str(biref_config_path),
154 | "-o",
155 | str(output_path),
156 | ],
157 | catch_exceptions=False,
158 | )
159 | assert output_path.exists()
160 | with open_ome_zarr(output_path) as dataset:
161 | assert dataset["0/0/0"]["0"].shape[1] == 4
162 |
163 | # Append fluoresncence reconstruction
164 | runner.invoke(
165 | cli,
166 | [
167 | "reconstruct",
168 | "-i",
169 | str(input_path / "0" / "0" / "0"),
170 | "-c",
171 | str(fluor_config_path),
172 | "-o",
173 | str(output_path),
174 | ],
175 | catch_exceptions=False,
176 | )
177 | assert output_path.exists()
178 | with open_ome_zarr(output_path) as dataset:
179 | assert dataset["0/0/0"]["0"].shape[1] == 5
180 | assert dataset.channel_names[-1] == "GFP_Density3D"
181 | assert dataset.channel_names[-2] == "Pol"
182 |
183 |
184 | def test_cli_apply_inv_tf_mock(tmp_input_path_zarr):
185 | tmp_input_zarr, tmp_config_yml = tmp_input_path_zarr
186 | tmp_config_yml = tmp_config_yml.with_name("0.yml").resolve()
187 | tf_path = tmp_input_zarr.with_name("tf_0.zarr").resolve()
188 | input_path = (tmp_input_zarr / "0" / "0" / "0").resolve()
189 | result_path = tmp_input_zarr.with_name("result.zarr").resolve()
190 |
191 | assert tmp_config_yml.exists()
192 | assert tf_path.exists()
193 | assert input_path.exists()
194 | assert not result_path.exists()
195 |
196 | runner = CliRunner()
197 | with patch(
198 | "recOrder.cli.apply_inverse_transfer_function.apply_inverse_transfer_function_cli"
199 | ) as mock:
200 | cmd = [
201 | "apply-inv-tf",
202 | "-i",
203 | str(input_path),
204 | "-t",
205 | str(tf_path),
206 | "-c",
207 | str(tmp_config_yml),
208 | "-o",
209 | str(result_path),
210 | "-j",
211 | str(1),
212 | ]
213 | result_inv = runner.invoke(
214 | cli,
215 | cmd,
216 | catch_exceptions=False,
217 | )
218 | mock.assert_called_with(
219 | [input_path],
220 | Path(tf_path),
221 | Path(tmp_config_yml),
222 | Path(result_path),
223 | 1,
224 | 1,
225 | )
226 | assert result_inv.exit_code == 0
227 |
228 |
229 | def test_cli_apply_inv_tf_output(tmp_input_path_zarr, capsys):
230 | tmp_input_zarr, tmp_config_yml = tmp_input_path_zarr
231 | input_path = tmp_input_zarr / "0" / "0" / "0"
232 |
233 | for i, (
234 | birefringence_option,
235 | time_indices,
236 | phase_option,
237 | dimension_option,
238 | time_length_target,
239 | ) in enumerate(all_options):
240 | if (birefringence_option is None) and (phase_option is None):
241 | continue
242 |
243 | result_path = tmp_input_zarr.with_name(f"result{i}.zarr").resolve()
244 |
245 | tf_path = tmp_input_zarr.with_name(f"tf_{i}.zarr")
246 | tmp_config_yml = tmp_config_yml.with_name(f"{i}.yml")
247 |
248 | # # Check output
249 | apply_inverse_transfer_function_cli(
250 | [input_path], tf_path, tmp_config_yml, result_path, 1
251 | )
252 |
253 | result_dataset = open_ome_zarr(str(result_path / "0" / "0" / "0"))
254 | assert result_dataset["0"].shape[0] == time_length_target
255 | assert result_dataset["0"].shape[3:] == (5, 6)
256 |
257 | assert result_path.exists()
258 | captured = capsys.readouterr()
259 | assert "submitted" in captured.out
260 |
261 | # Check scale transformations pass through
262 | assert input_scale == result_dataset.scale
263 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_settings.py:
--------------------------------------------------------------------------------
1 | import pytest
2 | from recOrder.cli import settings
3 | from recOrder.io import utils
4 | from pydantic.v1 import ValidationError
5 |
6 |
7 | def test_reconstruction_settings():
8 | # Test defaults
9 | s = settings.ReconstructionSettings(
10 | birefringence=settings.BirefringenceSettings()
11 | )
12 | assert len(s.input_channel_names) == 4
13 | assert s.birefringence.apply_inverse.background_path == ""
14 | assert s.phase == None
15 | assert s.fluorescence == None
16 |
17 | # Test logic that "fluorescence" or ("phase" and/or "birefringence")
18 | s = settings.ReconstructionSettings(
19 | input_channel_names=["GFP"],
20 | birefringence=None,
21 | phase=None,
22 | fluorescence=settings.FluorescenceSettings(),
23 | )
24 |
25 | assert s.fluorescence.apply_inverse.reconstruction_algorithm == "Tikhonov"
26 |
27 | # Not allowed to supply both phase/biref and fluorescence
28 | with pytest.raises(ValidationError):
29 | settings.ReconstructionSettings(
30 | phase=settings.PhaseSettings(),
31 | fluorescence=settings.FluorescenceSettings(),
32 | )
33 |
34 | # Test incorrect settings
35 | with pytest.raises(ValidationError):
36 | settings.ReconstructionSettings(input_channel_names=3)
37 |
38 | with pytest.raises(ValidationError):
39 | settings.ReconstructionSettings(reconstruction_dimension=1)
40 |
41 | # Test typo
42 | with pytest.raises(ValidationError):
43 | settings.ReconstructionSettings(
44 | flurescence=settings.FluorescenceSettings()
45 | )
46 |
47 |
48 | def test_biref_tf_settings():
49 | settings.BirefringenceTransferFunctionSettings(swing=0.1)
50 |
51 | with pytest.raises(ValidationError):
52 | settings.BirefringenceTransferFunctionSettings(swing=1.1)
53 |
54 | with pytest.raises(ValidationError):
55 | settings.BirefringenceTransferFunctionSettings(scheme="Test")
56 |
57 |
58 | def test_phase_tf_settings():
59 | settings.PhaseTransferFunctionSettings(
60 | index_of_refraction_media=1.0, numerical_aperture_detection=0.8
61 | )
62 |
63 | with pytest.raises(ValidationError):
64 | settings.PhaseTransferFunctionSettings(
65 | index_of_refraction_media=1.0, numerical_aperture_detection=1.1
66 | )
67 |
68 | # Inconsistent units
69 | with pytest.raises(Warning):
70 | settings.PhaseTransferFunctionSettings(
71 | yx_pixel_size=650, z_pixel_size=0.3
72 | )
73 |
74 | # Extra parameter
75 | with pytest.raises(ValidationError):
76 | settings.PhaseTransferFunctionSettings(zyx_pixel_size=650)
77 |
78 |
79 | def test_fluor_tf_settings():
80 | settings.FluorescenceTransferFunctionSettings(
81 | wavelength_emission=0.500, yx_pixel_size=0.2
82 | )
83 |
84 | with pytest.raises(Warning):
85 | settings.FluorescenceTransferFunctionSettings(
86 | wavelength_emission=0.500, yx_pixel_size=2000
87 | )
88 |
89 |
90 | def test_generate_example_settings():
91 | example_path = "./examples/"
92 |
93 | s0 = settings.ReconstructionSettings(
94 | birefringence=settings.BirefringenceSettings(),
95 | phase=settings.PhaseSettings(),
96 | )
97 | s1 = settings.ReconstructionSettings(
98 | input_channel_names=["BF"],
99 | phase=settings.PhaseSettings(),
100 | )
101 | s2 = settings.ReconstructionSettings(
102 | birefringence=settings.BirefringenceSettings(),
103 | )
104 | s3 = settings.ReconstructionSettings(
105 | input_channel_names=["GFP"],
106 | fluorescence=settings.FluorescenceSettings(),
107 | )
108 | file_names = [
109 | "birefringence-and-phase.yml",
110 | "phase.yml",
111 | "birefringence.yml",
112 | "fluorescence.yml",
113 | ]
114 | settings_list = [s0, s1, s2, s3]
115 |
116 | # Save to examples folder and test roundtrip
117 | for file_name, settings_obj in zip(file_names, settings_list):
118 | utils.model_to_yaml(settings_obj, example_path + file_name)
119 | settings_roundtrip = utils.yaml_to_model(
120 | example_path + file_name, settings.ReconstructionSettings
121 | )
122 | assert settings_obj.dict() == settings_roundtrip.dict()
123 |
--------------------------------------------------------------------------------
/recOrder/tests/conftest.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pytest
3 | from iohub.ngff import open_ome_zarr
4 |
5 | from recOrder.cli import settings
6 |
7 |
8 | @pytest.fixture(scope="function")
9 | def example_plate(tmp_path):
10 | plate_path = tmp_path / "input.zarr"
11 |
12 | position_list = (
13 | ("A", "1", "0"),
14 | ("B", "1", "0"),
15 | ("B", "2", "0"),
16 | )
17 |
18 | plate_dataset = open_ome_zarr(
19 | plate_path,
20 | layout="hcs",
21 | mode="w",
22 | channel_names=[f"State{i}" for i in range(4)] + ["BF"],
23 | )
24 |
25 | for row, col, fov in position_list:
26 | position = plate_dataset.create_position(row, col, fov)
27 | position.create_zeros("0", (2, 5, 4, 5, 6), dtype=np.uint16)
28 |
29 | yield plate_path, plate_dataset
30 |
31 |
32 | @pytest.fixture(scope="function")
33 | def birefringence_phase_recon_settings_function(tmp_path):
34 | recon_settings = settings.ReconstructionSettings(
35 | birefringence=settings.BirefringenceSettings(),
36 | phase=settings.PhaseSettings(),
37 | )
38 | dataset = open_ome_zarr(
39 | tmp_path,
40 | layout="fov",
41 | mode="w",
42 | channel_names=[f"State{i}" for i in range(4)],
43 | )
44 | yield recon_settings, dataset
45 |
46 |
47 | @pytest.fixture(scope="function")
48 | def fluorescence_recon_settings_function(tmp_path):
49 | recon_settings = settings.ReconstructionSettings(
50 | input_channel_names=["GFP"],
51 | fluorescence=settings.FluorescenceSettings(),
52 | )
53 | dataset = open_ome_zarr(
54 | tmp_path,
55 | layout="fov",
56 | mode="w",
57 | channel_names=[f"State{i}" for i in range(4)],
58 | )
59 | yield recon_settings, dataset
60 |
--------------------------------------------------------------------------------
/recOrder/tests/mmcore_tests/test_core_func.py:
--------------------------------------------------------------------------------
1 | from unittest.mock import MagicMock, Mock, call
2 | import pytest
3 | import numpy as np
4 | from numpy import ndarray
5 | from typing import Callable, Tuple
6 |
7 | # tested components
8 | from recOrder.io.core_functions import *
9 |
10 |
11 | # TODO: move these to fixture or generate with Hypothesis
12 | # dynamic range
13 | TIFF_I_MAX = 2**16
14 | # image size
15 | IMAGE_WIDTH = np.random.randint(1, 2**12)
16 | IMAGE_HEIGHT = np.random.randint(1, 2**12)
17 | PIXEL_COUNT = IMAGE_HEIGHT * IMAGE_WIDTH
18 | # serialized image from the pycromanager bridge
19 | SERIAL_IMAGE = np.random.randint(0, TIFF_I_MAX, size=(PIXEL_COUNT,))
20 | # LC device parameters
21 | # TODO: parameterize this example
22 | DEVICE_PROPERTY = ("deviceName", "propertyName")
23 | CONFIG_GROUP = "configGroup"
24 | CONFIG_NAME = "State0"
25 | # LC state in native units
26 | LC_STATE = np.random.rand(1)[0] * 10
27 |
28 |
29 | def _get_mmcore_mock():
30 | """Creates a mock for the `pycromanager.Core` object.
31 |
32 | Returns
33 | -------
34 | MagicMock
35 | MMCore mock object
36 | """
37 | mmcore_mock_config = {
38 | "getImage": Mock(return_value=SERIAL_IMAGE),
39 | "getProperty": Mock(return_value=str(LC_STATE)),
40 | }
41 | return MagicMock(**mmcore_mock_config)
42 |
43 |
44 | def _get_snap_manager_mock():
45 | """Creates a mock for the pycromanager remote Snap Live Window Manager object.
46 |
47 | Returns
48 | -------
49 | MagicMock
50 | Mock object for `org.micromanager.internal.SnapLiveManager` via pycromanager
51 | """
52 | sm = MagicMock()
53 | get_snap_mocks = {
54 | "getHeight": Mock(return_value=IMAGE_HEIGHT),
55 | "getWidth": Mock(return_value=IMAGE_WIDTH),
56 | "getRawPixels": Mock(return_value=SERIAL_IMAGE),
57 | }
58 | # TODO: break down these JAVA call stack chains for maintainability
59 | sm.getDisplay.return_value.getDisplayedImages.return_value.get = Mock(
60 | # return image object mock with H, W, and pixel values
61 | return_value=Mock(**get_snap_mocks)
62 | )
63 | sm.getDisplay.return_value.getImagePlus.return_value.getStatistics = Mock(
64 | # return statistics object mock with the attribute "umean"
65 | return_value=Mock(umean=SERIAL_IMAGE.mean())
66 | )
67 | return sm
68 |
69 |
70 | def _is_int(data: ndarray):
71 | """Check if the data type is integer.
72 |
73 | Parameters
74 | ----------
75 | data
76 |
77 | Returns
78 | -------
79 | bool
80 | True if the data type is any integer type.
81 | """
82 | return np.issubdtype(data.dtype, np.integer)
83 |
84 |
85 | def _get_examples(low: float, high: float):
86 | """Generate 4 valid and 4 invalid floating numbers for closed interval [low, high].
87 |
88 | Parameters
89 | ----------
90 | low : float
91 | high : float
92 |
93 | Returns
94 | -------
95 | tuple(1d-array, 1d-array)
96 | valid and invalid values
97 | """
98 | epsilon = np.finfo(float).eps
99 | samples = np.random.rand(4)
100 | valid_values = samples * (high - low) + low + epsilon
101 | invalid_values = np.array(
102 | [
103 | low - samples[0],
104 | low - samples[1],
105 | high + samples[2],
106 | high + samples[3],
107 | ]
108 | )
109 | return valid_values, invalid_values
110 |
111 |
112 | def test_suspend_live_sm():
113 | """Test `recOrder.io.core_functions.suspend_live_sm`."""
114 | snap_manager = _get_snap_manager_mock()
115 | with suspend_live_sm(snap_manager) as sm:
116 | sm.setSuspended.assert_called_once_with(True)
117 | snap_manager.setSuspended.assert_called_with(False)
118 |
119 |
120 | def test_snap_and_get_image():
121 | """Test `recOrder.io.core_functions.snap_and_get_image`."""
122 | sm = _get_snap_manager_mock()
123 | image = snap_and_get_image(sm)
124 | assert _is_int(image), image.dtype
125 | assert image.shape == (IMAGE_HEIGHT, IMAGE_WIDTH), image.shape
126 |
127 |
128 | def test_snap_and_average():
129 | """Test `recOrder.io.core_functions.snap_and_average`."""
130 | sm = _get_snap_manager_mock()
131 | mean = snap_and_average(sm)
132 | np.testing.assert_almost_equal(mean, SERIAL_IMAGE.mean())
133 |
134 |
135 | def _set_lc_test(
136 | tested_func: Callable[[object, Tuple[str, str], float], None],
137 | value_range: Tuple[float, float],
138 | ):
139 | mmc = _get_mmcore_mock()
140 | valid_values, invalid_values = _get_examples(*value_range)
141 | for value in valid_values:
142 | tested_func(mmc, DEVICE_PROPERTY, value)
143 | mmc.setProperty.assert_called_with(
144 | DEVICE_PROPERTY[0], DEVICE_PROPERTY[1], str(value)
145 | )
146 | for value in invalid_values:
147 | with pytest.raises(ValueError):
148 | tested_func(mmc, DEVICE_PROPERTY, value)
149 |
150 |
151 | def test_set_lc_waves():
152 | """Test `recOrder.io.core_functions.set_lc_waves`."""
153 | _set_lc_test(set_lc_waves, (0.001, 1.6))
154 |
155 |
156 | def test_set_lc_voltage():
157 | """Test `recOrder.io.core_functions.set_lc_voltage`."""
158 | _set_lc_test(set_lc_voltage, (0.0, 20.0))
159 |
160 |
161 | def test_set_lc_daq():
162 | """Test `recOrder.io.core_functions.set_lc_daq`."""
163 | _set_lc_test(set_lc_daq, (0.0, 5.0))
164 |
165 |
166 | def test_get_lc():
167 | """Test `recOrder.io.core_functions.get_lc`."""
168 | mmc = _get_mmcore_mock()
169 | state = get_lc(mmc, DEVICE_PROPERTY)
170 | mmc.getProperty.assert_called_once_with(*DEVICE_PROPERTY)
171 | np.testing.assert_almost_equal(state, LC_STATE)
172 |
173 |
174 | def test_define_meadowlark_state():
175 | """Test `recOrder.io.core_functions.define_meadowlark_state`."""
176 | mmc = _get_mmcore_mock()
177 | define_meadowlark_state(mmc, DEVICE_PROPERTY)
178 | mmc.setProperty.assert_called_once_with(*DEVICE_PROPERTY, 0)
179 | mmc.waitForDevice.assert_called_once_with(DEVICE_PROPERTY[0])
180 |
181 |
182 | def test_define_config_state():
183 | """Test `recOrder.io.core_functions.define_config_state`."""
184 | mmc = _get_mmcore_mock()
185 | device_properties = [DEVICE_PROPERTY] * 4
186 | values = _get_examples(0, 10)[0].tolist()
187 | define_config_state(
188 | mmc, CONFIG_GROUP, CONFIG_NAME, device_properties, values
189 | )
190 | expected_calls = [
191 | call(CONFIG_GROUP, CONFIG_NAME, *d, str(v))
192 | for d, v in zip(device_properties, values)
193 | ]
194 | got_calls = mmc.defineConfig.call_args_list
195 | assert got_calls == expected_calls, got_calls
196 |
197 |
198 | def test_set_lc_state():
199 | """Test `recOrder.io.core_functions.set_lc_state`."""
200 | mmc = _get_mmcore_mock()
201 | set_lc_state(mmc, CONFIG_GROUP, CONFIG_NAME)
202 | mmc.setConfig.assert_called_once_with(CONFIG_GROUP, CONFIG_NAME)
203 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_create_empty.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import numpy as np
4 | from iohub.ngff import Position, open_ome_zarr
5 |
6 | from recOrder.cli.utils import create_empty_hcs_zarr
7 |
8 |
9 | def test_create_empty_hcs_zarr(tmp_path):
10 | store_path = tmp_path / Path("test_store.zarr")
11 | position_keys: list[tuple[str]] = [
12 | ("A", "0", "3"),
13 | ("B", "10", "4"),
14 | ]
15 | shape = (1, 2, 1, 1024, 1024)
16 | chunks = (1, 1, 1, 256, 256)
17 | scale = (1, 1, 1, 0.5, 0.5)
18 | channel_names = ["Channel1", "Channel2"]
19 | dtype = np.uint16
20 | plate_metadata = {"test": 2}
21 |
22 | create_empty_hcs_zarr(
23 | store_path,
24 | position_keys,
25 | shape,
26 | chunks,
27 | scale,
28 | channel_names,
29 | dtype,
30 | plate_metadata,
31 | )
32 |
33 | # Verify existence of positions and channels
34 | with open_ome_zarr(store_path, mode="r") as plate:
35 | assert plate.zattrs["test"] == 2
36 | for position_key in position_keys:
37 | position = plate["/".join(position_key)]
38 | assert isinstance(position, Position)
39 | assert position[0].shape == shape
40 |
41 | # Repeat creation should not fail
42 | more_channel_names = ["Channel3"]
43 | create_empty_hcs_zarr(
44 | store_path,
45 | position_keys,
46 | shape,
47 | chunks,
48 | scale,
49 | more_channel_names,
50 | dtype,
51 | )
52 |
53 | # Verify existence of appended channel names
54 | channel_names += more_channel_names
55 | for position_key in position_keys:
56 | position_path = store_path
57 | for element in position_key:
58 | position_path /= element
59 | with open_ome_zarr(position_path, mode="r") as position:
60 | assert position.channel_names == channel_names
61 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_io.py:
--------------------------------------------------------------------------------
1 | import os
2 | from pathlib import Path
3 |
4 | import pytest
5 | import yaml
6 |
7 | from recOrder.cli import settings
8 | from recOrder.io.utils import add_index_to_path, model_to_yaml
9 |
10 |
11 | @pytest.fixture
12 | def model():
13 | # Create a sample model object
14 | return settings.ReconstructionSettings(
15 | birefringence=settings.BirefringenceSettings()
16 | )
17 |
18 |
19 | @pytest.fixture
20 | def yaml_path(tmpdir):
21 | # Create a temporary YAML file path
22 | return os.path.join(tmpdir, "model.yaml")
23 |
24 |
25 | def test_model_to_yaml(model, yaml_path):
26 | # Call the function under test
27 | model_to_yaml(model, yaml_path)
28 |
29 | # Check if the YAML file is created
30 | assert os.path.exists(yaml_path)
31 |
32 | # Load the YAML file and verify its contents
33 | with open(yaml_path, "r") as f:
34 | yaml_data = yaml.safe_load(f)
35 |
36 | # Check if the YAML data is a dictionary
37 | assert isinstance(yaml_data, dict)
38 |
39 | # Check YAML data
40 | assert "input_channel_names" in yaml_data
41 |
42 |
43 | def test_model_to_yaml_invalid_model():
44 | # Create an object that does not have a 'dict()' method
45 | invalid_model = "not a model object"
46 |
47 | # Call the function and expect a TypeError
48 | with pytest.raises(TypeError):
49 | model_to_yaml(invalid_model, "model.yaml")
50 |
51 |
52 | def test_add_index_to_path(tmp_path):
53 | test_cases = [
54 | ("output.txt", "output_0.txt"),
55 | ("output.txt", "output_1.txt"),
56 | ("output.txt", "output_2.txt"),
57 | ("output.png", "output_0.png"),
58 | ("output.png", "output_1.png"),
59 | ("output.png", "output_2.png"),
60 | ("folder", "folder_0"),
61 | ("folder", "folder_1"),
62 | ("folder", "folder_2"),
63 | ]
64 |
65 | for input_path_str, expected_output_str in test_cases:
66 | input_path = tmp_path / Path(input_path_str)
67 | expected_output = tmp_path / Path(expected_output_str)
68 |
69 | output_path = add_index_to_path(input_path)
70 | assert output_path == expected_output
71 |
72 | output_path.touch() # Create a file/folder at the expected output path for testing
73 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_overlays.py:
--------------------------------------------------------------------------------
1 | import hypothesis.extra.numpy as npst
2 | import hypothesis.strategies as st
3 | import numpy as np
4 | from hypothesis import given
5 | from numpy.typing import NDArray
6 | from numpy.testing import assert_equal
7 |
8 | from recOrder.io.visualization import ret_ori_overlay, ret_ori_phase_overlay
9 |
10 |
11 | @st.composite
12 | def _birefringence(draw):
13 | shape = (2,) + tuple(
14 | draw(st.lists(st.integers(1, 16), min_size=2, max_size=4))
15 | )
16 | dtype = draw(npst.floating_dtypes(sizes=(32, 64)))
17 | bit_width = dtype.itemsize * 8
18 | retardance = draw(
19 | npst.arrays(
20 | dtype,
21 | shape=shape,
22 | elements=st.floats(
23 | min_value=1.0000000168623835e-16,
24 | max_value=50,
25 | exclude_min=True,
26 | width=bit_width,
27 | ),
28 | )
29 | )
30 | orientation = draw(
31 | npst.arrays(
32 | dtype,
33 | shape=shape,
34 | elements=st.floats(
35 | min_value=0,
36 | max_value=dtype.type(np.pi),
37 | exclude_min=True,
38 | exclude_max=True,
39 | width=bit_width,
40 | ),
41 | )
42 | )
43 |
44 | return retardance, orientation
45 |
46 |
47 | @given(briefringence=_birefringence(), jch=st.booleans())
48 | def test_ret_ori_overlay(briefringence: tuple[NDArray, NDArray], jch: bool):
49 | """Test recOrder.io.utils.ret_ori_overlay()"""
50 | retardance, orientation = briefringence
51 | retardance_copy = retardance.copy()
52 | orientation_copy = orientation.copy()
53 | cmap = "JCh" if jch else "HSV"
54 | overlay = ret_ori_overlay(
55 | np.stack((retardance, orientation)),
56 | ret_max=np.percentile(retardance, 99),
57 | cmap=cmap,
58 | )
59 |
60 | overlay2 = ret_ori_phase_overlay(
61 | np.stack((retardance, orientation, retardance)), # dummy phase
62 | )
63 |
64 | # check that the function did not mutate input data
65 | assert_equal(retardance, retardance_copy)
66 | assert_equal(orientation, orientation_copy)
67 | # check output properties
68 | # output contains NaN, pending further investigation
69 | # assert overlay.min() >= 0
70 | # assert overlay.max() <= 1
71 | assert overlay.shape == (3,) + retardance.shape
72 | assert overlay2.shape == (3,) + retardance.shape
73 |
--------------------------------------------------------------------------------
/recOrder/tests/widget_tests/test_dock_widget.py:
--------------------------------------------------------------------------------
1 | from napari.viewer import ViewerModel
2 |
3 | from recOrder.plugin.main_widget import MainWidget
4 |
5 |
6 | def test_dock_widget(make_napari_viewer):
7 | viewer: ViewerModel = make_napari_viewer()
8 | viewer.window.add_dock_widget(MainWidget(viewer))
9 | assert "recOrder" in list(viewer._window._dock_widgets.keys())[0]
10 |
--------------------------------------------------------------------------------
/recOrder/tests/widget_tests/test_sample_contributions.py:
--------------------------------------------------------------------------------
1 | from recOrder.scripts.samples import download_and_unzip
2 |
3 |
4 | def test_download_and_unzip():
5 | p1, p2 = download_and_unzip("target")
6 |
7 | assert p1.exists()
8 | assert p2.exists()
9 |
--------------------------------------------------------------------------------
/setup.cfg:
--------------------------------------------------------------------------------
1 | [metadata]
2 | name = recOrder-napari
3 | author = Computational Microscopy Platform, CZ Biohub
4 | author_email = shalin.mehta@czbiohub.org
5 | url = https://github.com/mehta-lab/recOrder
6 | license = BSD 3-Clause License
7 | description = Computational microscopy toolkit for label-free imaging
8 | long_description = file: README.md
9 | long_description_content_type = text/markdown
10 | classifiers =
11 | License :: OSI Approved :: BSD License
12 | Programming Language :: Python
13 | Programming Language :: Python :: 3 :: Only
14 | Programming Language :: Python :: 3.10
15 | Programming Language :: Python :: 3.11
16 | Topic :: Scientific/Engineering
17 | Topic :: Scientific/Engineering :: Visualization
18 | Topic :: Scientific/Engineering :: Information Analysis
19 | Topic :: Scientific/Engineering :: Bio-Informatics
20 | Topic :: Utilities
21 | Framework :: napari
22 | Operating System :: Microsoft :: Windows
23 | Operating System :: POSIX
24 | Operating System :: Unix
25 | Operating System :: MacOS
26 | project_urls =
27 | Bug Tracker = https://github.com/mehta-lab/recOrder/issues
28 | Documentation = https://github.com/mehta-lab/recOrder/wiki
29 | Source Code = https://github.com/mehta-lab/recOrder/tree/main/recOrder
30 | User Support = https://github.com/mehta-lab/recOrder/issues
31 |
32 | [options]
33 | packages = find:
34 | include_package_data = True
35 | python_requires = >=3.10
36 | setup_requires = setuptools_scm
37 | install_requires =
38 | waveorder==2.2.0rc0
39 | click>=8.0.1
40 | natsort>=7.1.1
41 | colorspacious>=1.1.2
42 | importlib-metadata
43 | iohub==0.1.0
44 | wget>=3.2
45 | psutil
46 | submitit
47 | pydantic==1.10.19
48 | ome-zarr==0.8.3 # unpin when resolved: https://github.com/ome/napari-ome-zarr/issues/111
49 | qtpy
50 | pyqtgraph>=0.12.3
51 |
52 | [options.extras_require]
53 | all =
54 | napari[pyqt6]
55 | napari-ome-zarr>=0.3.2 # drag and drop convenience
56 | pycromanager==0.27.2
57 |
58 | dev =
59 | pytest>=5.0.0
60 | pytest-cov
61 | pytest-qt
62 | tox
63 | pre-commit
64 | black
65 | hypothesis
66 |
67 | [options.package_data]
68 | * = *.yaml
69 |
70 | [options.entry_points]
71 | console_scripts =
72 | recorder = recOrder.cli.main:cli
73 | recOrder = recOrder.cli.main:cli
74 |
75 | napari.manifest =
76 | recOrder = recOrder:napari.yaml
77 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | from setuptools import setup
2 |
3 | setup()
4 |
--------------------------------------------------------------------------------
/tox.ini:
--------------------------------------------------------------------------------
1 | # Modified from from cookiecutter-napari-plugin
2 | # For more information about tox, see https://tox.readthedocs.io/en/latest/
3 | [tox]
4 | envlist = py{310,311}-{linux,macos,windows}
5 | isolated_build=true
6 |
7 | [gh-actions]
8 | python =
9 | 3.10: py310
10 | 3.11: py311
11 |
12 | [gh-actions:env]
13 | PLATFORM =
14 | ubuntu-latest: linux
15 | macos-latest: macos
16 | windows-latest: windows
17 |
18 | [testenv]
19 | platform =
20 | macos: darwin
21 | linux: linux
22 | windows: win32
23 | passenv =
24 | CI
25 | GITHUB_ACTIONS
26 | DISPLAY
27 | XAUTHORITY
28 | NUMPY_EXPERIMENTAL_ARRAY_FUNCTION
29 | PYVISTA_OFF_SCREEN
30 | extras =
31 | dev
32 | all
33 | commands = pytest -v --color=yes --cov=recOrder --cov-report=xml
--------------------------------------------------------------------------------
19 |
20 | The main command `reconstruct` command is composed of two subcommands: `compute-tf` and `apply-inv-tf`.
21 |
22 | A reconstruction can be performed with a single `reconstruct` call. For example:
23 | ```
24 | recorder reconstruct `
25 | -i ./data.zarr/*/*/* `
26 | -c ./config.yml `
27 | -o ./reconstruction.zarr
28 | ```
29 | Equivalently, a reconstruction can be performed with a `compute-tf` call followed by an `apply-inv-tf` call. For example:
30 | ```
31 | recorder compute-tf `
32 | -i ./data.zarr/0/0/0 `
33 | -c ./config.yml `
34 | -o ./tf.zarr
35 |
36 | recorder apply-inv-tf
37 | -i ./data.zarr/*/*/* `
38 | -t ./tf.zarr `
39 | -c ./config.yml `
40 | -o ./reconstruction.zarr
41 | ```
42 | Computing the transfer function is typically the most expensive part of the reconstruction, so saving a transfer function then applying it to many datasets can save time.
43 |
44 | ## Input options
45 |
46 | The input `-i` flag always accepts a list of inputs, either explicitly e.g. `-i ./data.zarr/A/1/0 ./data.zarr/A/2/0` or through wildcards `-i ./data.zarr/*/*/*`. The positions in a high-content screening `.zarr` store are organized into `/row/col/fov` folders, so `./input.zarr/*/*/*` creates a list of all positions in a dataset.
47 |
48 | The `recorder compute-tf` command accepts a list of inputs, but it only computes the transfer function for the first position in the list. The `apply-inv-tf` command accepts a list of inputs and applies the same transfer function to all of the inputs, which requires that all positions contain arrays with matching TCZYX shapes.
49 |
50 | ## What types of reconstructions are supported?
51 | See `/recOrder/examples/` for a list of example configuration files.
52 |
53 | WIP: This documentation will be expanded for each reconstruction type and parameter.
54 |
--------------------------------------------------------------------------------
/docs/software-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Software Installation Guide
2 |
3 | 1. (Optional but recommended) install [`conda`](https://github.com/conda-forge/miniforge) and create a virtual environment
4 |
5 | ```sh
6 | conda create -y -n recOrder python=3.10
7 | conda activate recOrder
8 | ```
9 |
10 | 2. Install `recOrder-napari`:
11 |
12 | ```sh
13 | pip install recOrder-napari
14 | ```
15 |
16 | 3. To use the GUI: open `napari` with `recOrder-napari`:
17 |
18 | ```sh
19 | napari -w recOrder-napari
20 | ```
21 |
22 | 4. View command-line help by running
23 |
24 | ```sh
25 | recOrder
26 | ```
27 |
28 | 5. To acquire data via Micro-Manager`, follow the [microscope installation guide](./microscope-installation-guide.md).
29 |
--------------------------------------------------------------------------------
/examples/README.md:
--------------------------------------------------------------------------------
1 | # `recOrder` CLI examples
2 |
3 | `recOrder` uses a configuration-file-based command-line inferface (CLI) to
4 | calculate transfer functions and apply these transfer functions to datasets.
5 |
6 | This page demonstrates `recOrder`'s CLI.
7 |
8 | ## Getting started
9 |
10 | ### 1. Check your installation
11 | First, [install `recOrder`](../docs/software-installation-guide.md) and run
12 | ```bash
13 | recOrder
14 | ```
15 | in a shell. If `recOrder` is installed correctly, you will see a usage string and
16 | ```
17 | recOrder: Computational Toolkit for Label-Free Imaging
18 | ```
19 |
20 | ### 2. Download and convert a test dataset
21 | Next, [download the test data from zenodo (47 MB)](https://zenodo.org/record/6983916/files/recOrder_test_data.zip?download=1), and convert a dataset to the latest version of `.zarr` with
22 | ```
23 | cd /path/to/
24 | iohub convert -i /path/to/recOrder_test_data/2022_08_04_recOrder_pytest_20x_04NA/2T_3P_16Z_128Y_256X_Kazansky_1/
25 | -o ./dataset.zarr
26 | ```
27 |
28 | You can view the test dataset with
29 | ```
30 | napari ./dataset.zarr --plugin recOrder-napari
31 | ```
32 |
33 | ### 3. Run a reconstruction
34 | Run an example reconstruction with
35 | ```
36 | recOrder reconstruct ./dataset.zarr/0/0/0 -c /path/to/recOrder/examples/settings/birefringence-and-phase.yml -o ./reconstruction.zarr
37 | ```
38 | then view the reconstruction with
39 | ```
40 | napari ./reconstruction.zarr --plugin recOrder-napari
41 | ```
42 |
43 | Try modifying the configuration file to see how the regularization parameter changes the results.
44 |
45 | ### 4. Parallelize over positions or time points
46 |
47 | TODO: @Ed I think we'll use the same strategy as ```mantis deskew```
48 |
49 | ## FAQ
50 | 1. **Q: Which configuration file should I use?**
51 |
52 | If you are acquiring:
53 |
54 | **3D data with calibrated liquid-crystal polarizers via `recOrder`** use `birefringence.yml`.
55 |
56 | **3D fluorescence data** use `fluorescence.yml`.
57 |
58 | **3D brightfield data** use `phase.yml`.
59 |
60 | **Multi-modal data**, start by reconstructing the individual modaliities, each with a single config file and CLI call. Then combine the reconstructions by ***TODO: @Ziwen do can you help me append to the zarrs to help me fix this? ***
61 |
62 | 2. **Q: Should I use `reconstruction_dimension` = 2 or 3?
63 |
64 | If your downstream processing requires 3D information or if you're unsure, then you should use `reconstruction_dimension = 3`. If your sample is very thin compared to the depth of field of the microscope, if you're in a noise-limited regime, or if your downstream processing requires 2D information, then you should use `reconstruction_dimension = 2`. Empirically, we have found that 2D reconstructions reduce the noise in our reconstructions because it uses 3D information to make a single estimate for each pixel.
65 |
66 | 3. **Q: What regularization parameter should I use?**
67 |
68 | We recommend starting with the defaults then testing over a few orders of magnitude and choosing a result that isn't too noisy or too smooth.
69 |
70 | ### Developers note
71 |
72 | These configuration files are automatically generated when the tests run. See `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
73 |
74 | To keep these settings up to date, run `pytest` locally when `cli/settings.py` changes.
75 |
--------------------------------------------------------------------------------
/examples/birefringence-and-phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 | phase:
18 | transfer_function:
19 | wavelength_illumination: 0.532
20 | yx_pixel_size: 0.325
21 | z_pixel_size: 2.0
22 | z_padding: 0
23 | index_of_refraction_media: 1.3
24 | numerical_aperture_detection: 1.2
25 | numerical_aperture_illumination: 0.5
26 | invert_phase_contrast: false
27 | apply_inverse:
28 | reconstruction_algorithm: Tikhonov
29 | regularization_strength: 0.001
30 | TV_rho_strength: 0.001
31 | TV_iterations: 1
32 |
--------------------------------------------------------------------------------
/examples/birefringence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 |
--------------------------------------------------------------------------------
/examples/fluorescence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - GFP
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | fluorescence:
6 | transfer_function:
7 | yx_pixel_size: 0.325
8 | z_pixel_size: 2.0
9 | z_padding: 0
10 | index_of_refraction_media: 1.3
11 | numerical_aperture_detection: 1.2
12 | wavelength_emission: 0.507
13 | apply_inverse:
14 | reconstruction_algorithm: Tikhonov
15 | regularization_strength: 0.001
16 | TV_rho_strength: 0.001
17 | TV_iterations: 1
18 |
--------------------------------------------------------------------------------
/examples/phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - BF
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | phase:
6 | transfer_function:
7 | wavelength_illumination: 0.532
8 | yx_pixel_size: 0.325
9 | z_pixel_size: 2.0
10 | z_padding: 0
11 | index_of_refraction_media: 1.3
12 | numerical_aperture_detection: 1.2
13 | numerical_aperture_illumination: 0.5
14 | invert_phase_contrast: false
15 | apply_inverse:
16 | reconstruction_algorithm: Tikhonov
17 | regularization_strength: 0.001
18 | TV_rho_strength: 0.001
19 | TV_iterations: 1
20 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = ["setuptools", "wheel", "setuptools_scm"]
3 | build-backend = "setuptools.build_meta"
4 |
5 | [tool.setuptools_scm]
6 | write_to = "recOrder/_version.py"
7 |
8 | [tool.black]
9 | line-length = 79
10 |
11 | [tool.isort]
12 | profile = "black"
13 | line_length = 79
--------------------------------------------------------------------------------
/recOrder/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/acq/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/acq_functions.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import json
3 | import os
4 | import time
5 |
6 | import numpy as np
7 | from iohub import read_micromanager
8 | try:
9 | from pycromanager import Studio
10 | except:pass
11 |
12 | def generate_acq_settings(
13 | mm,
14 | channel_group,
15 | channels=None,
16 | zstart=None,
17 | zend=None,
18 | zstep=None,
19 | save_dir=None,
20 | prefix=None,
21 | keep_shutter_open_channels=False,
22 | keep_shutter_open_slices=False,
23 | ):
24 | """
25 | This function generates a json file specific to the Micro-Manager SequenceSettings.
26 | It has default parameters for a multi-channels z-stack acquisition but does not yet
27 | support multi-position or multi-frame acquisitions.
28 |
29 | This also has default values for QLIPP Acquisition. Can be used as a framework for other types
30 | of acquisitions
31 |
32 | Parameters
33 | ----------
34 | mm: (object) MM Studio API object
35 | scheme: (str) '4-State' or '5-State'
36 | zstart: (float) relative starting position for the z-stack
37 | zend: (float) relative ending position for the z-stack
38 | zstep: (float) step size for the z-stack
39 | save_dir: (str) path to save directory
40 | prefix: (str) name to save the data under
41 |
42 | Returns
43 | -------
44 | settings: (json) json dictionary conforming to MM SequenceSettings
45 | """
46 |
47 | # Get API Objects
48 | am = mm.getAcquisitionManager()
49 | ss = am.getAcquisitionSettings()
50 | app = mm.app()
51 |
52 | # Get current SequenceSettings to modify
53 | original_ss = ss.toJSONStream(ss)
54 | original_json = json.loads(original_ss).copy()
55 |
56 | if zstart:
57 | do_z = True
58 | else:
59 | do_z = False
60 |
61 | # Structure of the channel properties
62 | channel_dict = {
63 | "channelGroup": channel_group,
64 | "config": None,
65 | "exposure": None,
66 | "zOffset": 0,
67 | "doZStack": do_z,
68 | "color": {"value": -16747854, "falpha": 0.0},
69 | "skipFactorFrame": 0,
70 | "useChannel": True if channels else False,
71 | }
72 |
73 | channel_list = None
74 | if channels:
75 | # Append all the channels with their current exposure settings
76 | channel_list = []
77 | for chan in channels:
78 | # todo: think about how to deal with missing exposure
79 | exposure = app.getChannelExposureTime(
80 | channel_group, chan, 10
81 | ) # sets exposure to 10 if not found
82 | channel = channel_dict.copy()
83 | channel["config"] = chan
84 | channel["exposure"] = exposure
85 |
86 | channel_list.append(channel)
87 |
88 | # set other parameters
89 | original_json["numFrames"] = 1
90 | original_json["intervalMs"] = 0
91 | original_json["relativeZSlice"] = True
92 | original_json["slicesFirst"] = True
93 | original_json["timeFirst"] = False
94 | original_json["keepShutterOpenSlices"] = keep_shutter_open_slices
95 | original_json["keepShutterOpenChannels"] = keep_shutter_open_channels
96 | original_json["useAutofocus"] = False
97 | original_json["saveMode"] = "MULTIPAGE_TIFF"
98 | original_json["save"] = True if save_dir else False
99 | original_json["root"] = save_dir if save_dir else ""
100 | original_json["prefix"] = prefix if prefix else "Untitled"
101 | original_json["channels"] = channel_list
102 | original_json["zReference"] = 0.0
103 | original_json["channelGroup"] = channel_group
104 | original_json["usePositionList"] = False
105 | original_json["shouldDisplayImages"] = True
106 | original_json["useSlices"] = do_z
107 | original_json["useFrames"] = False
108 | original_json["useChannels"] = True if channels else False
109 | original_json["slices"] = (
110 | list(np.arange(float(zstart), float(zend + zstep), float(zstep)))
111 | if zstart
112 | else []
113 | )
114 | original_json["sliceZStepUm"] = zstep
115 | original_json["sliceZBottomUm"] = zstart
116 | original_json["sliceZTopUm"] = zend
117 | original_json["acqOrderMode"] = 1
118 |
119 | return original_json
120 |
121 |
122 | def acquire_from_settings(
123 | mm: Studio,
124 | settings: dict,
125 | grab_images: bool = True,
126 | restore_settings: bool = True,
127 | ):
128 | """Function to acquire an MDA acquisition with the native MM MDA Engine.
129 | Assumes single position acquisition.
130 |
131 | Parameters
132 | ----------
133 | mm : Studio
134 | settings : dict
135 | JSON dictionary conforming to MM SequenceSettings
136 | grab_images : bool, optional
137 | return the acquired array, by default True
138 | restore_settings : bool, optional
139 | restore MDA settings before acquisition, by default True
140 |
141 | Returns
142 | -------
143 | NDArray
144 | acquired images
145 | """
146 | am = mm.getAcquisitionManager()
147 | ss = am.getAcquisitionSettings()
148 |
149 | ss_new = ss.fromJSONStream(json.dumps(settings))
150 | am.runAcquisitionWithSettings(ss_new, True)
151 |
152 | time.sleep(3)
153 |
154 | if restore_settings:
155 | am.setAcquisitionSettings(ss)
156 |
157 | # TODO: speed improvements in reading the data with pycromanager acquisition?
158 | if grab_images:
159 | # get the most recent acquisition if multiple
160 | path = os.path.join(settings["root"], settings["prefix"])
161 | files = glob.glob(path + "*")
162 | index = max([int(x.split(path + "_")[1]) for x in files])
163 |
164 | reader = read_micromanager(
165 | path + f"_{index}", data_type="ometiff", extract_data=True
166 | )
167 |
168 | return reader.get_array(0)
169 |
--------------------------------------------------------------------------------
/recOrder/calib/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/calib/__init__.py
--------------------------------------------------------------------------------
/recOrder/cli/apply_inverse_models.py:
--------------------------------------------------------------------------------
1 | """
2 | This module converts recOrder's reconstructions into waveorder calls
3 | """
4 |
5 | import numpy as np
6 | import torch
7 | from waveorder.models import (
8 | inplane_oriented_thick_pol3d,
9 | isotropic_fluorescent_thick_3d,
10 | isotropic_thin_3d,
11 | phase_thick_3d,
12 | )
13 |
14 |
15 | def radians_to_nanometers(retardance_rad, wavelength_illumination_um):
16 | """
17 | waveorder returns retardance in radians, while recorder displays and saves
18 | retardance in nanometers. This function converts from radians to nanometers
19 | using the illumination wavelength (which is internally handled in um
20 | in recOrder).
21 | """
22 | return retardance_rad * wavelength_illumination_um * 1e3 / (2 * np.pi)
23 |
24 |
25 | def birefringence(
26 | czyx_data,
27 | cyx_no_sample_data,
28 | wavelength_illumination,
29 | recon_dim,
30 | biref_inverse_dict,
31 | transfer_function_dataset,
32 | ):
33 | # Load transfer function
34 | intensity_to_stokes_matrix = torch.tensor(
35 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
36 | )
37 |
38 | # Apply reconstruction
39 | # (retardance, orientation, transmittance, depolarization)
40 | reconstructed_parameters = (
41 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
42 | czyx_data,
43 | intensity_to_stokes_matrix,
44 | cyx_no_sample_data=cyx_no_sample_data,
45 | project_stokes_to_2d=(recon_dim == 2),
46 | **biref_inverse_dict,
47 | )
48 | )
49 |
50 | # Convert retardance
51 | retardance = radians_to_nanometers(
52 | reconstructed_parameters[0], wavelength_illumination
53 | )
54 |
55 | return torch.stack((retardance,) + reconstructed_parameters[1:])
56 |
57 |
58 | def phase(
59 | czyx_data,
60 | recon_dim,
61 | settings_phase,
62 | transfer_function_dataset,
63 | ):
64 | # [phase only, 2]
65 | if recon_dim == 2:
66 | # Load transfer functions
67 | absorption_transfer_function = torch.tensor(
68 | transfer_function_dataset["absorption_transfer_function"][0, 0]
69 | )
70 | phase_transfer_function = torch.tensor(
71 | transfer_function_dataset["phase_transfer_function"][0, 0]
72 | )
73 |
74 | # Apply
75 | (
76 | _,
77 | output,
78 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
79 | czyx_data[0],
80 | absorption_transfer_function,
81 | phase_transfer_function,
82 | **settings_phase.apply_inverse.dict(),
83 | )
84 |
85 | # [phase only, 3]
86 | elif recon_dim == 3:
87 | # Load transfer functions
88 | real_potential_transfer_function = torch.tensor(
89 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
90 | )
91 | imaginary_potential_transfer_function = torch.tensor(
92 | transfer_function_dataset["imaginary_potential_transfer_function"][
93 | 0, 0
94 | ]
95 | )
96 |
97 | # Apply
98 | output = phase_thick_3d.apply_inverse_transfer_function(
99 | czyx_data[0],
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | z_padding=settings_phase.transfer_function.z_padding,
103 | **settings_phase.apply_inverse.dict(),
104 | )
105 |
106 | # Pad to CZYX
107 | while output.ndim != 4:
108 | output = torch.unsqueeze(output, 0)
109 |
110 | return output
111 |
112 |
113 | def birefringence_and_phase(
114 | czyx_data,
115 | cyx_no_sample_data,
116 | wavelength_illumination,
117 | recon_dim,
118 | biref_inverse_dict,
119 | settings_phase,
120 | transfer_function_dataset,
121 | ):
122 | # Load birefringence transfer function
123 | intensity_to_stokes_matrix = torch.tensor(
124 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
125 | )
126 |
127 | # [biref and phase, 2]
128 | if recon_dim == 2:
129 | # Load phase transfer functions
130 | absorption_transfer_function = torch.tensor(
131 | transfer_function_dataset["absorption_transfer_function"][0, 0]
132 | )
133 | phase_transfer_function = torch.tensor(
134 | transfer_function_dataset["phase_transfer_function"][0, 0]
135 | )
136 |
137 | # Apply
138 | reconstructed_parameters_2d = (
139 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
140 | czyx_data,
141 | intensity_to_stokes_matrix,
142 | cyx_no_sample_data=cyx_no_sample_data,
143 | project_stokes_to_2d=True,
144 | **biref_inverse_dict,
145 | )
146 | )
147 |
148 | reconstructed_parameters_3d = (
149 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
150 | czyx_data,
151 | intensity_to_stokes_matrix,
152 | cyx_no_sample_data=cyx_no_sample_data,
153 | project_stokes_to_2d=False,
154 | **biref_inverse_dict,
155 | )
156 | )
157 |
158 | brightfield_3d = reconstructed_parameters_3d[2]
159 |
160 | (
161 | _,
162 | yx_phase,
163 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
164 | brightfield_3d,
165 | absorption_transfer_function,
166 | phase_transfer_function,
167 | **settings_phase.apply_inverse.dict(),
168 | )
169 |
170 | # Convert retardance
171 | retardance = radians_to_nanometers(
172 | reconstructed_parameters_2d[0], wavelength_illumination
173 | )
174 |
175 | output = torch.stack(
176 | (retardance,)
177 | + reconstructed_parameters_2d[1:]
178 | + (torch.unsqueeze(yx_phase, 0),)
179 | ) # CZYX
180 |
181 | # [biref and phase, 3]
182 | elif recon_dim == 3:
183 | # Load phase transfer functions
184 | intensity_to_stokes_matrix = torch.tensor(
185 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
186 | )
187 | # Load transfer functions
188 | real_potential_transfer_function = torch.tensor(
189 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
190 | )
191 | imaginary_potential_transfer_function = torch.tensor(
192 | transfer_function_dataset["imaginary_potential_transfer_function"][
193 | 0, 0
194 | ]
195 | )
196 |
197 | # Apply
198 | reconstructed_parameters_3d = (
199 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
200 | czyx_data,
201 | intensity_to_stokes_matrix,
202 | cyx_no_sample_data=cyx_no_sample_data,
203 | project_stokes_to_2d=False,
204 | **biref_inverse_dict,
205 | )
206 | )
207 |
208 | brightfield_3d = reconstructed_parameters_3d[2]
209 |
210 | zyx_phase = phase_thick_3d.apply_inverse_transfer_function(
211 | brightfield_3d,
212 | real_potential_transfer_function,
213 | imaginary_potential_transfer_function,
214 | z_padding=settings_phase.transfer_function.z_padding,
215 | **settings_phase.apply_inverse.dict(),
216 | )
217 |
218 | # Convert retardance
219 | retardance = radians_to_nanometers(
220 | reconstructed_parameters_3d[0], wavelength_illumination
221 | )
222 |
223 | # Save
224 | output = torch.stack(
225 | (retardance,) + reconstructed_parameters_3d[1:] + (zyx_phase,)
226 | )
227 | return output
228 |
229 |
230 | def fluorescence(
231 | czyx_data, recon_dim, settings_fluorescence, transfer_function_dataset
232 | ):
233 | # [fluo, 2]
234 | if recon_dim == 2:
235 | raise NotImplementedError
236 | # [fluo, 3]
237 | elif recon_dim == 3:
238 | # Load transfer functions
239 | optical_transfer_function = torch.tensor(
240 | transfer_function_dataset["optical_transfer_function"][0, 0]
241 | )
242 |
243 | # Apply
244 | output = (
245 | isotropic_fluorescent_thick_3d.apply_inverse_transfer_function(
246 | czyx_data[0],
247 | optical_transfer_function,
248 | settings_fluorescence.transfer_function.z_padding,
249 | **settings_fluorescence.apply_inverse.dict(),
250 | )
251 | )
252 | # Pad to CZYX
253 | while output.ndim != 4:
254 | output = torch.unsqueeze(output, 0)
255 |
256 | return output
257 |
--------------------------------------------------------------------------------
/recOrder/cli/compute_transfer_function.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 | import numpy as np
5 | from iohub.ngff import open_ome_zarr, Position
6 | from waveorder.models import (
7 | inplane_oriented_thick_pol3d,
8 | isotropic_fluorescent_thick_3d,
9 | isotropic_thin_3d,
10 | phase_thick_3d,
11 | )
12 |
13 | from recOrder.cli.parsing import (
14 | config_filepath,
15 | input_position_dirpaths,
16 | output_dirpath,
17 | )
18 | from recOrder.cli.printing import echo_headline, echo_settings
19 | from recOrder.cli.settings import ReconstructionSettings
20 | from recOrder.io import utils
21 |
22 |
23 | def generate_and_save_birefringence_transfer_function(settings, dataset):
24 | """Generates and saves the birefringence transfer function to the dataset, based on the settings.
25 |
26 | Parameters
27 | ----------
28 | settings: ReconstructionSettings
29 | dataset: NGFF Node
30 | The dataset that will be updated.
31 | """
32 | echo_headline("Generating birefringence transfer function with settings:")
33 | echo_settings(settings.birefringence.transfer_function)
34 |
35 | # Calculate transfer functions
36 | intensity_to_stokes_matrix = (
37 | inplane_oriented_thick_pol3d.calculate_transfer_function(
38 | scheme=str(len(settings.input_channel_names)) + "-State",
39 | **settings.birefringence.transfer_function.dict(),
40 | )
41 | )
42 | # Save
43 | dataset[
44 | "intensity_to_stokes_matrix"
45 | ] = intensity_to_stokes_matrix.cpu().numpy()[None, None, None, ...]
46 |
47 |
48 | def generate_and_save_phase_transfer_function(
49 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
50 | ):
51 | """Generates and saves the phase transfer function to the dataset, based on the settings.
52 |
53 | Parameters
54 | ----------
55 | settings: ReconstructionSettings
56 | dataset: Position
57 | The dataset that will be updated.
58 | zyx_shape : tuple
59 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
60 | """
61 | echo_headline("Generating phase transfer function with settings:")
62 | echo_settings(settings.phase.transfer_function)
63 |
64 | if settings.reconstruction_dimension == 2:
65 | # Convert zyx_shape and z_pixel_size into yx_shape and z_position_list
66 | settings_dict = settings.phase.transfer_function.dict()
67 | settings_dict["yx_shape"] = [zyx_shape[1], zyx_shape[2]]
68 | settings_dict["z_position_list"] = list(
69 | -(np.arange(zyx_shape[0]) - zyx_shape[0] // 2)
70 | * settings_dict["z_pixel_size"]
71 | )
72 |
73 | # Remove unused parameters
74 | settings_dict.pop("z_pixel_size")
75 | settings_dict.pop("z_padding")
76 |
77 | # Calculate transfer functions
78 | (
79 | absorption_transfer_function,
80 | phase_transfer_function,
81 | ) = isotropic_thin_3d.calculate_transfer_function(
82 | **settings_dict,
83 | )
84 |
85 | # Save
86 | dataset.create_image(
87 | "absorption_transfer_function",
88 | absorption_transfer_function.cpu().numpy()[None, None, ...],
89 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
90 | )
91 | dataset.create_image(
92 | "phase_transfer_function",
93 | phase_transfer_function.cpu().numpy()[None, None, ...],
94 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
95 | )
96 |
97 | elif settings.reconstruction_dimension == 3:
98 | # Calculate transfer functions
99 | (
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | ) = phase_thick_3d.calculate_transfer_function(
103 | zyx_shape=zyx_shape,
104 | **settings.phase.transfer_function.dict(),
105 | )
106 | # Save
107 | dataset.create_image(
108 | "real_potential_transfer_function",
109 | real_potential_transfer_function.cpu().numpy()[None, None, ...],
110 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
111 | )
112 | dataset.create_image(
113 | "imaginary_potential_transfer_function",
114 | imaginary_potential_transfer_function.cpu().numpy()[
115 | None, None, ...
116 | ],
117 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
118 | )
119 |
120 |
121 | def generate_and_save_fluorescence_transfer_function(
122 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
123 | ):
124 | """Generates and saves the fluorescence transfer function to the dataset, based on the settings.
125 |
126 | Parameters
127 | ----------
128 | settings: ReconstructionSettings
129 | dataset: Position
130 | The dataset that will be updated.
131 | zyx_shape : tuple
132 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
133 | """
134 | echo_headline("Generating fluorescence transfer function with settings:")
135 | echo_settings(settings.fluorescence.transfer_function)
136 |
137 | if settings.reconstruction_dimension == 2:
138 | raise NotImplementedError
139 | elif settings.reconstruction_dimension == 3:
140 | # Calculate transfer functions
141 | optical_transfer_function = (
142 | isotropic_fluorescent_thick_3d.calculate_transfer_function(
143 | zyx_shape=zyx_shape,
144 | **settings.fluorescence.transfer_function.dict(),
145 | )
146 | )
147 | # Save
148 | dataset.create_image(
149 | "optical_transfer_function",
150 | optical_transfer_function.cpu().numpy()[None, None, ...],
151 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
152 | )
153 |
154 |
155 | def compute_transfer_function_cli(
156 | input_position_dirpath: Path, config_filepath: Path, output_dirpath: Path
157 | ) -> None:
158 | """CLI command to compute the transfer function given a configuration file path
159 | and a desired output path.
160 | """
161 |
162 | # Load config file
163 | settings = utils.yaml_to_model(config_filepath, ReconstructionSettings)
164 |
165 | echo_headline(
166 | f"Generating transfer functions and storing in {output_dirpath}\n"
167 | )
168 |
169 | # Read shape from input dataset
170 | input_dataset = open_ome_zarr(
171 | input_position_dirpath, layout="fov", mode="r"
172 | )
173 | zyx_shape = input_dataset.data.shape[
174 | 2:
175 | ] # only loads a single position "0"
176 |
177 | # Check input channel names
178 | if not set(settings.input_channel_names).issubset(
179 | input_dataset.channel_names
180 | ):
181 | raise ValueError(
182 | f"Each of the input_channel_names = {settings.input_channel_names} in {config_filepath} must appear in the dataset {input_position_dirpaths[0]} which currently contains channel_names = {input_dataset.channel_names}."
183 | )
184 |
185 | # Prepare output dataset
186 | output_dataset = open_ome_zarr(
187 | output_dirpath, layout="fov", mode="w", channel_names=["None"]
188 | )
189 |
190 | # Pass settings to appropriate calculate_transfer_function and save
191 | if settings.birefringence is not None:
192 | generate_and_save_birefringence_transfer_function(
193 | settings, output_dataset
194 | )
195 | if settings.phase is not None:
196 | generate_and_save_phase_transfer_function(
197 | settings, output_dataset, zyx_shape
198 | )
199 | if settings.fluorescence is not None:
200 | generate_and_save_fluorescence_transfer_function(
201 | settings, output_dataset, zyx_shape
202 | )
203 |
204 | # Write settings to metadata
205 | output_dataset.zattrs["settings"] = settings.dict()
206 |
207 | echo_headline(f"Closing {output_dirpath}\n")
208 | output_dataset.close()
209 |
210 | echo_headline(
211 | f"Recreate this transfer function with:\n$ recorder compute-tf {input_position_dirpaths} -c {config_filepath} -o {output_dirpath}"
212 | )
213 |
214 |
215 | @click.command()
216 | @input_position_dirpaths()
217 | @config_filepath()
218 | @output_dirpath()
219 | def compute_tf(
220 | input_position_dirpaths: list[Path],
221 | config_filepath: Path,
222 | output_dirpath: Path,
223 | ) -> None:
224 | """
225 | Compute a transfer function using a dataset and configuration file.
226 |
227 | Calculates the transfer function based on the shape of the first position
228 | in the list `input-position-dirpaths`.
229 |
230 | See /examples for example configuration files.
231 |
232 | >> recorder compute-tf -i ./input.zarr/0/0/0 -c ./examples/birefringence.yml -o ./transfer_function.zarr
233 | """
234 | compute_transfer_function_cli(
235 | input_position_dirpaths[0], config_filepath, output_dirpath
236 | )
237 |
--------------------------------------------------------------------------------
/recOrder/cli/gui_widget.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import click
3 |
4 | try:
5 | from recOrder.plugin import tab_recon
6 | except:pass
7 |
8 | try:
9 | from qtpy.QtWidgets import QApplication, QWidget, QVBoxLayout, QStyle
10 | except:pass
11 |
12 | try:
13 | import qdarktheme
14 | except:pass
15 |
16 | PLUGIN_NAME = "recOrder: Computational Toolkit for Label-Free Imaging"
17 | PLUGIN_ICON = "🔬"
18 |
19 | @click.command()
20 | def gui():
21 | """GUI for recOrder: Computational Toolkit for Label-Free Imaging"""
22 |
23 | app = QApplication(sys.argv)
24 | app.setStyle("Fusion") # Other options: "Fusion", "Windows", "macOS", "WindowsVista"
25 | try:
26 | qdarktheme.setup_theme("dark")
27 | except:pass
28 | window = MainWindow()
29 | window.setWindowTitle(PLUGIN_ICON + " " + PLUGIN_NAME + " " + PLUGIN_ICON)
30 |
31 | pixmapi = getattr(QStyle.StandardPixmap, "SP_TitleBarMenuButton")
32 | icon = app.style().standardIcon(pixmapi)
33 | window.setWindowIcon(icon)
34 |
35 | window.show()
36 | sys.exit(app.exec())
37 |
38 | class MainWindow(QWidget):
39 | def __init__(self):
40 | super().__init__()
41 | recon_tab = tab_recon.Ui_ReconTab_Form(stand_alone=True)
42 | layout = QVBoxLayout()
43 | self.setLayout(layout)
44 | layout.addWidget(recon_tab.recon_tab_mainScrollArea)
45 |
46 | if __name__ == "__main__":
47 | gui()
48 |
--------------------------------------------------------------------------------
/recOrder/cli/jobs_mgmt.py:
--------------------------------------------------------------------------------
1 | import os, json
2 | from pathlib import Path
3 | import socket
4 | import submitit
5 | import threading, time
6 |
7 | DIR_PATH = os.path.dirname(os.path.realpath(__file__))
8 | FILE_PATH = os.path.join(DIR_PATH, "main.py")
9 |
10 | SERVER_PORT = 8089 # Choose an available port
11 | JOBS_TIMEOUT = 5 # 5 mins
12 | SERVER_uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
13 |
14 | class JobsManagement():
15 |
16 | def __init__(self, *args, **kwargs):
17 | self.clientsocket = None
18 | self.uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
19 | self.DATA_QUEUE = []
20 |
21 | def check_for_jobID_File(self, jobID, logs_path, extension="out"):
22 |
23 | if Path(logs_path).exists():
24 | files = os.listdir(logs_path)
25 | try:
26 | for file in files:
27 | if file.endswith(extension):
28 | if jobID in file:
29 | file_path = os.path.join(logs_path, file)
30 | f = open(file_path, "r")
31 | txt = f.read()
32 | f.close()
33 | return txt
34 | except Exception as exc:
35 | print(exc.args)
36 | return ""
37 |
38 | def set_shorter_timeout(self):
39 | self.clientsocket.settimeout(30)
40 |
41 | def start_client(self):
42 | try:
43 | self.clientsocket = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
44 | self.clientsocket.settimeout(300)
45 | self.clientsocket.connect(('localhost', SERVER_PORT))
46 | self.clientsocket.settimeout(None)
47 |
48 | thread = threading.Thread(target=self.stop_client)
49 | thread.start()
50 | except Exception as exc:
51 | print(exc.args)
52 |
53 | # The stopClient() is called right with the startClient() but does not stop
54 | # and essentially is a wait thread listening and is triggered by either a
55 | # connection or timeout. Based on condition triggered by user, reconstruction
56 | # completion or errors the end goal is to close the socket connection which
57 | # would let the CLI exit. I could break it down to 2 parts but the idea was to
58 | # keep the clientsocket.close() call within one method to make it easier to follow.
59 | def stop_client(self):
60 | try:
61 | time.sleep(2)
62 | while True:
63 | time.sleep(1)
64 | buf = ""
65 | try:
66 | buf = self.clientsocket.recv(1024)
67 | except:
68 | pass
69 | if len(buf) > 0:
70 | if b"\n" in buf:
71 | dataList = buf.split(b"\n")
72 | for data in dataList:
73 | if len(data)>0:
74 | decoded_string = data.decode()
75 | json_str = str(decoded_string)
76 | json_obj = json.loads(json_str)
77 | u_idx = json_obj["uID"]
78 | job_idx = str(json_obj["jID"])
79 | cmd = json_obj["command"]
80 | if cmd == "clientRelease":
81 | if self.has_submitted_job(u_idx, job_idx):
82 | self.clientsocket.close()
83 | break
84 | if cmd == "cancel":
85 | if self.has_submitted_job(u_idx, job_idx):
86 | try:
87 | job = self.uIDsjobIDs[u_idx][job_idx]
88 | job.cancel()
89 | except Exception as exc:
90 | pass # possibility of throwing an exception based on diff. OS
91 | forDeletions = []
92 | for uID in self.uIDsjobIDs.keys():
93 | for jID in self.uIDsjobIDs[uID].keys():
94 | job = self.uIDsjobIDs[uID][jID]
95 | if job.done():
96 | forDeletions.append((uID, jID))
97 | for idx in range(len(forDeletions)):
98 | del self.uIDsjobIDs[forDeletions[idx][0]][forDeletions[idx][1]]
99 | forDeletions = []
100 | for uID in self.uIDsjobIDs.keys():
101 | if len(self.uIDsjobIDs[uID].keys()) == 0:
102 | forDeletions.append(uID)
103 | for idx in range(len(forDeletions)):
104 | del self.uIDsjobIDs[forDeletions[idx]]
105 | if len(self.uIDsjobIDs.keys()) == 0:
106 | self.clientsocket.close()
107 | break
108 | except Exception as exc:
109 | self.clientsocket.close()
110 | print(exc.args)
111 |
112 | def check_all_ExpJobs_completion(self, uID):
113 | if uID in SERVER_uIDsjobIDs.keys():
114 | for jobEntry in SERVER_uIDsjobIDs[uID].keys():
115 | job:submitit.Job = SERVER_uIDsjobIDs[uID][jobEntry]["job"]
116 | jobBool = SERVER_uIDsjobIDs[uID][jobEntry]["bool"]
117 | if job is not None and job.done() == False:
118 | return False
119 | if jobBool == False:
120 | return False
121 | return True
122 |
123 | def put_Job_completion_in_list(self, job_bool, uID: str, jID: str, mode="client"):
124 | if uID in SERVER_uIDsjobIDs.keys():
125 | if jID in SERVER_uIDsjobIDs[uID].keys():
126 | SERVER_uIDsjobIDs[uID][jID]["bool"] = job_bool
127 |
128 | def add_data(self, data):
129 | self.DATA_QUEUE.append(data)
130 |
131 | def send_data_thread(self):
132 | thread = threading.Thread(target=self.send_data)
133 | thread.start()
134 |
135 | def send_data(self):
136 | data = "".join(self.DATA_QUEUE)
137 | self.clientsocket.send(data.encode())
138 | self.DATA_QUEUE = []
139 |
140 | def put_Job_in_list(self, job, uID: str, jID: str, well:str, log_folder_path:str="", mode="client"):
141 | try:
142 | well = str(well)
143 | jID = str(jID)
144 | if ".zarr" in well:
145 | wells = well.split(".zarr")
146 | well = wells[1].replace("\\","-").replace("/","-")[1:]
147 | if mode == "client":
148 | if uID not in self.uIDsjobIDs.keys():
149 | self.uIDsjobIDs[uID] = {}
150 | self.uIDsjobIDs[uID][jID] = job
151 | else:
152 | if jID not in self.uIDsjobIDs[uID].keys():
153 | self.uIDsjobIDs[uID][jID] = job
154 | json_obj = {uID:{"jID": str(jID), "pos": well, "log": log_folder_path}}
155 | json_str = json.dumps(json_obj)+"\n"
156 | self.add_data(json_str)
157 | else:
158 | # from server side jobs object entry is a None object
159 | # this will be later checked as completion boolean for a ExpID which might
160 | # have several Jobs associated with it
161 | if uID not in SERVER_uIDsjobIDs.keys():
162 | SERVER_uIDsjobIDs[uID] = {}
163 | SERVER_uIDsjobIDs[uID][jID] = {}
164 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
165 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
166 | else:
167 | SERVER_uIDsjobIDs[uID][jID] = {}
168 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
169 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
170 | except Exception as exc:
171 | print(exc.args)
172 |
173 | def has_submitted_job(self, uID: str, jID: str, mode="client")->bool:
174 | jID = str(jID)
175 | if mode == "client":
176 | if uID in self.uIDsjobIDs.keys():
177 | if jID in self.uIDsjobIDs[uID].keys():
178 | return True
179 | return False
180 | else:
181 | if uID in SERVER_uIDsjobIDs.keys():
182 | if jID in SERVER_uIDsjobIDs[uID].keys():
183 | return True
184 | return False
185 |
--------------------------------------------------------------------------------
/recOrder/cli/main.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 | from recOrder.cli.apply_inverse_transfer_function import apply_inv_tf
4 | from recOrder.cli.compute_transfer_function import compute_tf
5 | from recOrder.cli.reconstruct import reconstruct
6 | try:
7 | from recOrder.cli.gui_widget import gui
8 | except:pass
9 |
10 |
11 | CONTEXT = {"help_option_names": ["-h", "--help"]}
12 |
13 |
14 | # `recorder -h` will show subcommands in the order they are added
15 | class NaturalOrderGroup(click.Group):
16 | def list_commands(self, ctx):
17 | return self.commands.keys()
18 |
19 |
20 | @click.group(context_settings=CONTEXT, cls=NaturalOrderGroup)
21 | def cli():
22 | """\033[92mrecOrder: Computational Toolkit for Label-Free Imaging\033[0m\n"""
23 |
24 |
25 | cli.add_command(reconstruct)
26 | cli.add_command(compute_tf)
27 | cli.add_command(apply_inv_tf)
28 | try:
29 | cli.add_command(gui)
30 | except:pass
31 |
32 | if __name__ == "__main__":
33 | cli()
34 |
--------------------------------------------------------------------------------
/recOrder/cli/monitor.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import time
4 | import numpy as np
5 | import shutil
6 | import submitit
7 | import sys
8 |
9 |
10 | def _move_cursor_up(n_lines, do_print=True):
11 | if do_print:
12 | sys.stdout.write("\033[F" * n_lines)
13 |
14 |
15 | def _print_status(jobs, position_dirpaths, elapsed_list, print_indices=None, do_print=True):
16 |
17 | columns = [15, 30, 40, 50]
18 |
19 | # header
20 | if do_print:
21 | sys.stdout.write(
22 | "\033[K" # clear line
23 | "\033[96mID" # cyan
24 | f"\033[{columns[0]}G WELL "
25 | f"\033[{columns[1]}G STATUS "
26 | f"\033[{columns[2]}G NODE "
27 | f"\033[{columns[2]}G ELAPSED\n"
28 | )
29 |
30 | if print_indices is None:
31 | print_indices = range(len(jobs))
32 |
33 | complete_count = 0
34 |
35 | for i, (job, position_dirpath) in enumerate(zip(jobs, position_dirpaths)):
36 | try:
37 | node_name = job.get_info()["NodeList"] # slowest, so do this first
38 | except:
39 | node_name = "SUBMITTED"
40 |
41 | if job.state == "COMPLETED":
42 | color = "\033[32m" # green
43 | complete_count += 1
44 | elif job.state == "RUNNING":
45 | color = "\033[93m" # yellow
46 | elapsed_list[i] += 1 # inexact timing
47 | else:
48 | color = "\033[91m" # red
49 |
50 | if i in print_indices:
51 | if do_print:
52 | sys.stdout.write(
53 | f"\033[K" # clear line
54 | f"{color}{job.job_id}"
55 | f"\033[{columns[0]}G {'/'.join(position_dirpath.parts[-3:])}"
56 | f"\033[{columns[1]}G {job.state}"
57 | f"\033[{columns[2]}G {node_name}"
58 | f"\033[{columns[3]}G {elapsed_list[i]} s\n"
59 | )
60 | sys.stdout.flush()
61 | if do_print:
62 | print(
63 | f"\033[32m{complete_count}/{len(jobs)} jobs complete. "
64 | " to move monitor to background. "
65 | " twice to cancel jobs."
66 | )
67 |
68 | return elapsed_list
69 |
70 |
71 | def _get_jobs_to_print(jobs, num_to_print):
72 | job_indices_to_print = []
73 |
74 | # if number of jobs is smaller than termanal size, print all
75 | if len(jobs) <= num_to_print:
76 | return list(range(len(jobs)))
77 |
78 | # prioritize incomplete jobs
79 | for i, job in enumerate(jobs):
80 | if not job.done():
81 | job_indices_to_print.append(i)
82 | if len(job_indices_to_print) == num_to_print:
83 | return job_indices_to_print
84 |
85 | # fill in the rest with complete jobs
86 | for i, job in enumerate(jobs):
87 | job_indices_to_print.append(i)
88 | if len(job_indices_to_print) == num_to_print:
89 | return job_indices_to_print
90 |
91 | # shouldn't reach here
92 | return job_indices_to_print
93 |
94 |
95 | def monitor_jobs(jobs: list[submitit.Job], position_dirpaths: list[Path], do_print=True):
96 | """Displays the status of a list of submitit jobs with corresponding paths.
97 |
98 | Parameters
99 | ----------
100 | jobs : list[submitit.Job]
101 | List of submitit jobs
102 | position_dirpaths : list[Path]
103 | List of corresponding position paths
104 | """
105 | NON_JOB_LINES = 3
106 |
107 | if not len(jobs) == len(position_dirpaths):
108 | raise ValueError(
109 | "The number of jobs and position_dirpaths should be the same."
110 | )
111 |
112 | elapsed_list = [0] * len(jobs) # timer for each job
113 |
114 | # print all jobs once if terminal is too small
115 | if shutil.get_terminal_size().lines - NON_JOB_LINES < len(jobs):
116 | _print_status(jobs, position_dirpaths, elapsed_list, do_print)
117 |
118 | # main monitor loop
119 | try:
120 | while not all(job.done() for job in jobs):
121 | terminal_lines = shutil.get_terminal_size().lines
122 | num_jobs_to_print = np.min(
123 | [terminal_lines - NON_JOB_LINES, len(jobs)]
124 | )
125 |
126 | job_indices_to_print = _get_jobs_to_print(jobs, num_jobs_to_print)
127 |
128 | elapsed_list = _print_status(
129 | jobs,
130 | position_dirpaths,
131 | elapsed_list,
132 | job_indices_to_print,
133 | do_print,
134 | )
135 |
136 | time.sleep(1)
137 | _move_cursor_up(num_jobs_to_print + 2, do_print)
138 |
139 | # Print final status
140 | time.sleep(1)
141 | _print_status(jobs, position_dirpaths, elapsed_list, do_print=do_print)
142 |
143 | # cancel jobs if ctrl+c
144 | except KeyboardInterrupt:
145 | for job in jobs:
146 | job.cancel()
147 | print("All jobs cancelled.\033[97m")
148 |
149 | # Print STDOUT and STDERR for first incomplete job
150 | incomplete_count = 0
151 | for job in jobs:
152 | if not job.done():
153 | if incomplete_count == 0:
154 | print("\033[32mSTDOUT")
155 | print(job.stdout())
156 | print("\033[91mSTDERR")
157 | print(job.stderr())
158 |
159 | print("\033[97m") # print white
160 |
--------------------------------------------------------------------------------
/recOrder/cli/option_eat_all.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 |
4 | # Copied directly from https://stackoverflow.com/a/48394004
5 | # Enables `-i ./input.zarr/*/*/*`
6 | class OptionEatAll(click.Option):
7 | def __init__(self, *args, **kwargs):
8 | self.save_other_options = kwargs.pop("save_other_options", True)
9 | nargs = kwargs.pop("nargs", -1)
10 | assert nargs == -1, "nargs, if set, must be -1 not {}".format(nargs)
11 | super(OptionEatAll, self).__init__(*args, **kwargs)
12 | self._previous_parser_process = None
13 | self._eat_all_parser = None
14 |
15 | def add_to_parser(self, parser, ctx):
16 | def parser_process(value, state):
17 | # method to hook to the parser.process
18 | done = False
19 | value = [value]
20 | if self.save_other_options:
21 | # grab everything up to the next option
22 | while state.rargs and not done:
23 | for prefix in self._eat_all_parser.prefixes:
24 | if state.rargs[0].startswith(prefix):
25 | done = True
26 | if not done:
27 | value.append(state.rargs.pop(0))
28 | else:
29 | # grab everything remaining
30 | value += state.rargs
31 | state.rargs[:] = []
32 | value = tuple(value)
33 |
34 | # call the actual process
35 | self._previous_parser_process(value, state)
36 |
37 | retval = super(OptionEatAll, self).add_to_parser(parser, ctx)
38 | for name in self.opts:
39 | our_parser = parser._long_opt.get(name) or parser._short_opt.get(
40 | name
41 | )
42 | if our_parser:
43 | self._eat_all_parser = our_parser
44 | self._previous_parser_process = our_parser.process
45 | our_parser.process = parser_process
46 | break
47 | return retval
48 |
--------------------------------------------------------------------------------
/recOrder/cli/parsing.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from typing import Callable
3 |
4 | import click
5 | import torch.multiprocessing as mp
6 | from iohub.ngff import Plate, open_ome_zarr
7 | from natsort import natsorted
8 |
9 | from recOrder.cli.option_eat_all import OptionEatAll
10 |
11 |
12 | def _validate_and_process_paths(
13 | ctx: click.Context, opt: click.Option, value: str
14 | ) -> list[Path]:
15 | # Sort and validate the input paths, expanding plates into lists of positions
16 | input_paths = [Path(path) for path in natsorted(value)]
17 | for path in input_paths:
18 | with open_ome_zarr(path, mode="r") as dataset:
19 | if isinstance(dataset, Plate):
20 | plate_path = input_paths.pop()
21 | for position in dataset.positions():
22 | input_paths.append(plate_path / position[0])
23 |
24 | return input_paths
25 |
26 |
27 | def _str_to_path(ctx: click.Context, opt: click.Option, value: str) -> Path:
28 | return Path(value)
29 |
30 |
31 | def input_position_dirpaths() -> Callable:
32 | def decorator(f: Callable) -> Callable:
33 | return click.option(
34 | "--input-position-dirpaths",
35 | "-i",
36 | cls=OptionEatAll,
37 | type=tuple,
38 | required=True,
39 | callback=_validate_and_process_paths,
40 | help="List of paths to input positions, each with the same TCZYX shape. Supports wildcards e.g. 'input.zarr/*/*/*'.",
41 | )(f)
42 |
43 | return decorator
44 |
45 |
46 | def config_filepath() -> Callable:
47 | def decorator(f: Callable) -> Callable:
48 | return click.option(
49 | "--config-filepath",
50 | "-c",
51 | required=True,
52 | type=click.Path(exists=True, file_okay=True, dir_okay=False),
53 | callback=_str_to_path,
54 | help="Path to YAML configuration file.",
55 | )(f)
56 |
57 | return decorator
58 |
59 |
60 | def transfer_function_dirpath() -> Callable:
61 | def decorator(f: Callable) -> Callable:
62 | return click.option(
63 | "--transfer-function-dirpath",
64 | "-t",
65 | required=True,
66 | type=click.Path(exists=False),
67 | callback=_str_to_path,
68 | help="Path to transfer function .zarr.",
69 | )(f)
70 |
71 | return decorator
72 |
73 |
74 | def output_dirpath() -> Callable:
75 | def decorator(f: Callable) -> Callable:
76 | return click.option(
77 | "--output-dirpath",
78 | "-o",
79 | required=True,
80 | type=click.Path(exists=False),
81 | callback=_str_to_path,
82 | help="Path to output directory.",
83 | )(f)
84 |
85 | return decorator
86 |
87 |
88 | # TODO: this setting will have to be collected from SLURM?
89 | def processes_option(default: int = None) -> Callable:
90 | def check_processes_option(ctx, param, value):
91 | max_processes = mp.cpu_count()
92 | if value > max_processes:
93 | raise click.BadParameter(
94 | f"Maximum number of processes is {max_processes}"
95 | )
96 | return value
97 |
98 | def decorator(f: Callable) -> Callable:
99 | return click.option(
100 | "--num_processes",
101 | "-j",
102 | default=default or mp.cpu_count(),
103 | type=int,
104 | help="Number of processes to run in parallel.",
105 | callback=check_processes_option,
106 | )(f)
107 |
108 | return decorator
109 |
110 |
111 | def ram_multiplier() -> Callable:
112 | def decorator(f: Callable) -> Callable:
113 | return click.option(
114 | "--ram-multiplier",
115 | "-rx",
116 | default=1.0,
117 | type=float,
118 | help="SLURM RAM multiplier.",
119 | )(f)
120 |
121 | return decorator
122 |
123 | def unique_id() -> Callable:
124 | def decorator(f: Callable) -> Callable:
125 | return click.option(
126 | "--unique-id",
127 | "-uid",
128 | default="",
129 | required=False,
130 | type=str,
131 | help="Unique ID.",
132 | )(f)
133 |
134 | return decorator
--------------------------------------------------------------------------------
/recOrder/cli/printing.py:
--------------------------------------------------------------------------------
1 | import click
2 | import yaml
3 |
4 |
5 | def echo_settings(settings):
6 | click.echo(
7 | yaml.dump(settings.dict(), default_flow_style=False, sort_keys=False)
8 | )
9 |
10 |
11 | def echo_headline(headline):
12 | click.echo(click.style(headline, fg="green"))
13 |
--------------------------------------------------------------------------------
/recOrder/cli/reconstruct.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 |
5 | from recOrder.cli.apply_inverse_transfer_function import (
6 | apply_inverse_transfer_function_cli,
7 | )
8 | from recOrder.cli.compute_transfer_function import (
9 | compute_transfer_function_cli,
10 | )
11 | from recOrder.cli.parsing import (
12 | config_filepath,
13 | input_position_dirpaths,
14 | output_dirpath,
15 | processes_option,
16 | ram_multiplier,
17 | unique_id,
18 | )
19 |
20 |
21 | @click.command()
22 | @input_position_dirpaths()
23 | @config_filepath()
24 | @output_dirpath()
25 | @processes_option(default=1)
26 | @ram_multiplier()
27 | @unique_id()
28 | def reconstruct(
29 | input_position_dirpaths,
30 | config_filepath,
31 | output_dirpath,
32 | num_processes,
33 | ram_multiplier,
34 | unique_id,
35 | ):
36 | """
37 | Reconstruct a dataset using a configuration file. This is a
38 | convenience function for a `compute-tf` call followed by a `apply-inv-tf`
39 | call.
40 |
41 | Calculates the transfer function based on the shape of the first position
42 | in the list `input-position-dirpaths`, then applies that transfer function
43 | to all positions in the list `input-position-dirpaths`, so all positions
44 | must have the same TCZYX shape.
45 |
46 | See /examples for example configuration files.
47 |
48 | >> recorder reconstruct -i ./input.zarr/*/*/* -c ./examples/birefringence.yml -o ./output.zarr
49 | """
50 |
51 | # Handle transfer function path
52 | transfer_function_path = output_dirpath.parent / Path(
53 | "transfer_function_" + config_filepath.stem + ".zarr"
54 | )
55 |
56 | # Compute transfer function
57 | compute_transfer_function_cli(
58 | input_position_dirpaths[0],
59 | config_filepath,
60 | transfer_function_path,
61 | )
62 |
63 | # Apply inverse transfer function
64 | apply_inverse_transfer_function_cli(
65 | input_position_dirpaths,
66 | transfer_function_path,
67 | config_filepath,
68 | output_dirpath,
69 | num_processes,
70 | ram_multiplier,
71 | unique_id,
72 | )
73 |
--------------------------------------------------------------------------------
/recOrder/cli/settings.py:
--------------------------------------------------------------------------------
1 | import os
2 | from typing import List, Literal, Optional, Union
3 | from pathlib import Path
4 |
5 | from pydantic.v1 import (
6 | BaseModel,
7 | Extra,
8 | NonNegativeFloat,
9 | NonNegativeInt,
10 | PositiveFloat,
11 | root_validator,
12 | validator,
13 | )
14 |
15 | # This file defines the configuration settings for the CLI.
16 |
17 | # Example settings files in `/examples/settings/` are autmatically generated
18 | # by the tests in `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
19 |
20 | # To keep the example settings up to date, run `pytest` locally when this file changes.
21 |
22 |
23 | # All settings classes inherit from MyBaseModel, which forbids extra parameters to guard against typos
24 | class MyBaseModel(BaseModel, extra=Extra.forbid):
25 | pass
26 |
27 |
28 | # Bottom level settings
29 | class WavelengthIllumination(MyBaseModel):
30 | wavelength_illumination: PositiveFloat = 0.532
31 |
32 |
33 | class BirefringenceTransferFunctionSettings(MyBaseModel):
34 | swing: float = 0.1
35 |
36 | @validator("swing")
37 | def swing_range(cls, v):
38 | if v <= 0 or v >= 1.0:
39 | raise ValueError(f"swing = {v} should be between 0 and 1.")
40 | return v
41 |
42 |
43 | class BirefringenceApplyInverseSettings(WavelengthIllumination):
44 | background_path: Union[str, Path] = ""
45 | remove_estimated_background: bool = False
46 | flip_orientation: bool = False
47 | rotate_orientation: bool = False
48 |
49 | @validator("background_path")
50 | def check_background_path(cls, v):
51 | if v == "":
52 | return v
53 |
54 | raw_dir = r"{}".format(v)
55 | if not os.path.isdir(raw_dir):
56 | raise ValueError(f"{v} is not a existing directory")
57 | return raw_dir
58 |
59 |
60 | class FourierTransferFunctionSettings(MyBaseModel):
61 | yx_pixel_size: PositiveFloat = 6.5 / 20
62 | z_pixel_size: PositiveFloat = 2.0
63 | z_padding: NonNegativeInt = 0
64 | index_of_refraction_media: PositiveFloat = 1.3
65 | numerical_aperture_detection: PositiveFloat = 1.2
66 |
67 | @validator("numerical_aperture_detection")
68 | def na_det(cls, v, values):
69 | n = values["index_of_refraction_media"]
70 | if v > n:
71 | raise ValueError(
72 | f"numerical_aperture_detection = {v} must be less than or equal to index_of_refraction_media = {n}"
73 | )
74 | return v
75 |
76 | @validator("z_pixel_size")
77 | def warn_unit_consistency(cls, v, values):
78 | yx_pixel_size = values["yx_pixel_size"]
79 | ratio = yx_pixel_size / v
80 | if ratio < 1.0 / 20 or ratio > 20:
81 | raise Warning(
82 | f"yx_pixel_size ({yx_pixel_size}) / z_pixel_size ({v}) = {ratio}. Did you use consistent units?"
83 | )
84 | return v
85 |
86 |
87 | class FourierApplyInverseSettings(MyBaseModel):
88 | reconstruction_algorithm: Literal["Tikhonov", "TV"] = "Tikhonov"
89 | regularization_strength: NonNegativeFloat = 1e-3
90 | TV_rho_strength: PositiveFloat = 1e-3
91 | TV_iterations: NonNegativeInt = 1
92 |
93 |
94 | class PhaseTransferFunctionSettings(
95 | FourierTransferFunctionSettings,
96 | WavelengthIllumination,
97 | ):
98 | numerical_aperture_illumination: NonNegativeFloat = 0.5
99 | invert_phase_contrast: bool = False
100 |
101 | @validator("numerical_aperture_illumination")
102 | def na_ill(cls, v, values):
103 | n = values.get("index_of_refraction_media")
104 | if v > n:
105 | raise ValueError(
106 | f"numerical_aperture_illumination = {v} must be less than or equal to index_of_refraction_media = {n}"
107 | )
108 | return v
109 |
110 |
111 | class FluorescenceTransferFunctionSettings(FourierTransferFunctionSettings):
112 | wavelength_emission: PositiveFloat = 0.507
113 |
114 | @validator("wavelength_emission")
115 | def warn_unit_consistency(cls, v, values):
116 | yx_pixel_size = values.get("yx_pixel_size")
117 | ratio = yx_pixel_size / v
118 | if ratio < 1.0 / 20 or ratio > 20:
119 | raise Warning(
120 | f"yx_pixel_size ({yx_pixel_size}) / wavelength_illumination ({v}) = {ratio}. Did you use consistent units?"
121 | )
122 | return v
123 |
124 |
125 | # Second level settings
126 | class BirefringenceSettings(MyBaseModel):
127 | transfer_function: BirefringenceTransferFunctionSettings = (
128 | BirefringenceTransferFunctionSettings()
129 | )
130 | apply_inverse: BirefringenceApplyInverseSettings = (
131 | BirefringenceApplyInverseSettings()
132 | )
133 |
134 |
135 | class PhaseSettings(MyBaseModel):
136 | transfer_function: PhaseTransferFunctionSettings = (
137 | PhaseTransferFunctionSettings()
138 | )
139 | apply_inverse: FourierApplyInverseSettings = FourierApplyInverseSettings()
140 |
141 |
142 | class FluorescenceSettings(MyBaseModel):
143 | transfer_function: FluorescenceTransferFunctionSettings = (
144 | FluorescenceTransferFunctionSettings()
145 | )
146 | apply_inverse: FourierApplyInverseSettings = FourierApplyInverseSettings()
147 |
148 |
149 | # Top level settings
150 | class ReconstructionSettings(MyBaseModel):
151 | input_channel_names: List[str] = [f"State{i}" for i in range(4)]
152 | time_indices: Union[
153 | NonNegativeInt, List[NonNegativeInt], Literal["all"]
154 | ] = "all"
155 | reconstruction_dimension: Literal[2, 3] = 3
156 | birefringence: Optional[BirefringenceSettings]
157 | phase: Optional[PhaseSettings]
158 | fluorescence: Optional[FluorescenceSettings]
159 |
160 | @root_validator(pre=False)
161 | def validate_reconstruction_types(cls, values):
162 | if (values.get("birefringence") or values.get("phase")) and values.get(
163 | "fluorescence"
164 | ) is not None:
165 | raise ValueError(
166 | '"fluorescence" cannot be present alongside "birefringence" or "phase". Please use one configuration file for a "fluorescence" reconstruction and another configuration file for a "birefringence" and/or "phase" reconstructions.'
167 | )
168 | num_channel_names = len(values.get("input_channel_names"))
169 | if values.get("birefringence") is None:
170 | if (
171 | values.get("phase") is None
172 | and values.get("fluorescence") is None
173 | ):
174 | raise ValueError(
175 | "Provide settings for either birefringence, phase, birefringence + phase, or fluorescence."
176 | )
177 | if num_channel_names != 1:
178 | raise ValueError(
179 | f"{num_channel_names} channels names provided. Please provide a single channel for fluorescence/phase reconstructions."
180 | )
181 |
182 | return values
183 |
--------------------------------------------------------------------------------
/recOrder/cli/utils.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from typing import Tuple
3 |
4 | import click
5 | import numpy as np
6 | import torch
7 | from iohub.ngff import Position, open_ome_zarr
8 | from iohub.ngff_meta import TransformationMeta
9 | from numpy.typing import DTypeLike
10 |
11 |
12 | def create_empty_hcs_zarr(
13 | store_path: Path,
14 | position_keys: list[Tuple[str]],
15 | shape: Tuple[int],
16 | chunks: Tuple[int],
17 | scale: Tuple[float],
18 | channel_names: list[str],
19 | dtype: DTypeLike,
20 | plate_metadata: dict = {},
21 | ) -> None:
22 | """If the plate does not exist, create an empty zarr plate.
23 |
24 | If the plate exists, append positions and channels if they are not
25 | already in the plate.
26 |
27 | Parameters
28 | ----------
29 | store_path : Path
30 | hcs plate path
31 | position_keys : list[Tuple[str]]
32 | Position keys, will append if not present in the plate.
33 | e.g. [("A", "1", "0"), ("A", "1", "1")]
34 | shape : Tuple[int]
35 | chunks : Tuple[int]
36 | scale : Tuple[float]
37 | channel_names : list[str]
38 | Channel names, will append if not present in metadata.
39 | dtype : DTypeLike
40 | plate_metadata : dict
41 | """
42 |
43 | # Create plate
44 | output_plate = open_ome_zarr(
45 | str(store_path), layout="hcs", mode="a", channel_names=channel_names
46 | )
47 |
48 | # Pass metadata
49 | output_plate.zattrs.update(plate_metadata)
50 |
51 | # Create positions
52 | for position_key in position_keys:
53 | position_key_string = "/".join(position_key)
54 | # Check if position is already in the store, if not create it
55 | if position_key_string not in output_plate.zgroup:
56 | position = output_plate.create_position(*position_key)
57 |
58 | _ = position.create_zeros(
59 | name="0",
60 | shape=shape,
61 | chunks=chunks,
62 | dtype=dtype,
63 | transform=[TransformationMeta(type="scale", scale=scale)],
64 | )
65 | else:
66 | position = output_plate[position_key_string]
67 |
68 | # Check if channel_names are already in the store, if not append them
69 | for channel_name in channel_names:
70 | # Read channel names directly from metadata to avoid race conditions
71 | metadata_channel_names = [
72 | channel.label for channel in position.metadata.omero.channels
73 | ]
74 | if channel_name not in metadata_channel_names:
75 | position.append_channel(channel_name, resize_arrays=True)
76 |
77 |
78 | def apply_inverse_to_zyx_and_save(
79 | func,
80 | position: Position,
81 | output_path: Path,
82 | input_channel_indices: list[int],
83 | output_channel_indices: list[int],
84 | t_idx: int = 0,
85 | **kwargs,
86 | ) -> None:
87 | """Load a zyx array from a Position object, apply a transformation and save the result to file"""
88 | click.echo(f"Reconstructing t={t_idx}")
89 |
90 | # Load data
91 | czyx_uint16_numpy = position.data.oindex[t_idx, input_channel_indices]
92 |
93 | # convert to np.int32 (torch doesn't accept np.uint16), then convert to tensor float32
94 | czyx_data = torch.tensor(np.int32(czyx_uint16_numpy), dtype=torch.float32)
95 |
96 | # Apply transformation
97 | reconstruction_czyx = func(czyx_data, **kwargs)
98 |
99 | # Write to file
100 | # for c, recon_zyx in enumerate(reconstruction_zyx):
101 | with open_ome_zarr(output_path, mode="r+") as output_dataset:
102 | output_dataset[0].oindex[
103 | t_idx, output_channel_indices
104 | ] = reconstruction_czyx
105 | click.echo(f"Finished Writing.. t={t_idx}")
106 |
107 |
--------------------------------------------------------------------------------
/recOrder/io/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/io/__init__.py
--------------------------------------------------------------------------------
/recOrder/io/_reader.py:
--------------------------------------------------------------------------------
1 | from typing import Dict, List, Tuple, Union
2 |
3 | import zarr
4 | from iohub import read_micromanager
5 | from napari_ome_zarr._reader import napari_get_reader as fallback_reader
6 |
7 |
8 | def napari_get_reader(path):
9 | if isinstance(path, str):
10 | if ".zarr" in path:
11 | with zarr.open(path) as root:
12 | if "plate" in root.attrs:
13 | return hcs_zarr_reader
14 | else:
15 | return fallback_reader(path)
16 | else:
17 | return ome_tif_reader
18 | else:
19 | return None
20 |
21 |
22 | def hcs_zarr_reader(
23 | path: Union[str, List[str]]
24 | ) -> List[Tuple[zarr.Array, Dict]]:
25 | reader = read_micromanager(path)
26 | results = list()
27 |
28 | zs = zarr.open(path, "r")
29 | names = []
30 |
31 | dict_ = zs.attrs.asdict()
32 | wells = dict_["plate"]["wells"]
33 | for well in wells:
34 | path = well["path"]
35 | well_dict = zs[path].attrs.asdict()
36 | for name in well_dict["well"]["images"]:
37 | names.append(name["path"])
38 | for pos in range(reader.get_num_positions()):
39 | meta = dict()
40 | name = names[pos]
41 | meta["name"] = name
42 | results.append((reader.get_zarr(pos), meta))
43 | return results
44 |
45 |
46 | def ome_tif_reader(
47 | path: Union[str, List[str]]
48 | ) -> List[Tuple[zarr.Array, Dict]]:
49 | reader = read_micromanager(path)
50 | results = list()
51 |
52 | npos = reader.get_num_positions()
53 | for pos in range(npos):
54 | meta = dict()
55 | if npos == 1:
56 | meta["name"] = "Pos000_000"
57 | else:
58 | meta["name"] = reader.stage_positions[pos]["Label"][2:]
59 | results.append((reader.get_zarr(pos), meta))
60 |
61 | return results
62 |
--------------------------------------------------------------------------------
/recOrder/io/core_functions.py:
--------------------------------------------------------------------------------
1 | import time
2 | from contextlib import contextmanager
3 |
4 | import numpy as np
5 |
6 |
7 | @contextmanager
8 | def suspend_live_sm(snap_manager):
9 | """Context manager that suspends/unsuspends MM live mode for `SnapLiveManager`.
10 |
11 | Parameters
12 | ----------
13 | snap_manager : object
14 | `org.micromanager.internal.SnapLiveManager` object via pycromanager
15 |
16 | Yields
17 | ------
18 | object
19 | `org.micromanager.internal.SnapLiveManager` object via pycromanager
20 |
21 | Usage
22 | -----
23 | ```py
24 | with suspend_live_sm(snap_manager) as sm:
25 | pass # do something with MM that can't be done in live mode
26 | ```
27 | """
28 | snap_manager.setSuspended(True)
29 | try:
30 | yield snap_manager
31 | finally:
32 | snap_manager.setSuspended(False)
33 |
34 |
35 | def snap_and_get_image(snap_manager):
36 | """
37 | Snap and get image using Snap Live Window Manager + transfer of ZMQ
38 |
39 | Parameters
40 | ----------
41 | snap_manager: (object) MM Snap Live Window object
42 |
43 | Returns
44 | -------
45 | image: (array) 2D array of size (Y, X)
46 |
47 | """
48 | snap_manager.snap(True)
49 | time.sleep(
50 | 0.3
51 | ) # sleep after snap to make sure the image we grab is the correct one
52 |
53 | # get pixels + dimensions
54 | height = snap_manager.getDisplay().getDisplayedImages().get(0).getHeight()
55 | width = snap_manager.getDisplay().getDisplayedImages().get(0).getWidth()
56 | array = (
57 | snap_manager.getDisplay().getDisplayedImages().get(0).getRawPixels()
58 | )
59 |
60 | return np.reshape(array, (height, width))
61 |
62 |
63 | def snap_and_average(snap_manager, display=True):
64 | """
65 | Snap an image with Snap Live manager + grab only the mean (computed in java)
66 |
67 | Parameters
68 | ----------
69 | snap_manager: (object) MM Snap Live Window object
70 | display: (bool) Whether to show the snap on the Snap Live Window in MM
71 |
72 | Returns
73 | -------
74 | mean: (float) mean of snapped image
75 |
76 | """
77 |
78 | snap_manager.snap(display)
79 | time.sleep(
80 | 0.3
81 | ) # sleep after snap to make sure the image we grab is the correct one
82 |
83 | return snap_manager.getDisplay().getImagePlus().getStatistics().umean
84 |
85 |
86 | def set_lc_waves(mmc, device_property: tuple, value: float):
87 | """
88 | Set retardance in waves for LC in device_property
89 |
90 | Parameters
91 | ----------
92 | mmc : object
93 | MM Core object
94 | device_property : tuple
95 | (device_name, property_name) set
96 | value : float
97 | Retardance to set as fraction of a wavelength
98 |
99 | Returns
100 | -------
101 |
102 | """
103 | device_name = device_property[0]
104 | prop_name = device_property[1]
105 |
106 | if value > 1.6 or value < 0.001:
107 | raise ValueError(
108 | f"Requested retardance value is {value} waves. "
109 | f"Retardance must be greater than 0.001 and less than 1.6 waves."
110 | )
111 |
112 | mmc.setProperty(device_name, prop_name, str(value))
113 | time.sleep(20 / 1000)
114 |
115 |
116 | def set_lc_voltage(mmc, device_property: tuple, value: float):
117 | """
118 | Set LC retardance by specifying LC voltage
119 |
120 | Parameters
121 | ----------
122 | mmc : object
123 | MM Core object
124 | device_property : tuple
125 | (device_name, property_name) set
126 | value : float
127 | LC voltage in volts. Applied voltage is limited to 20V
128 |
129 | Returns
130 | -------
131 |
132 | """
133 | device_name = device_property[0]
134 | prop_name = device_property[1]
135 |
136 | if value > 20.0 or value < 0.0:
137 | raise ValueError(
138 | f"Requested LC voltage is {value} V. "
139 | f"LC voltage must be greater than 0.0 and less than 20.0 V."
140 | )
141 |
142 | mmc.setProperty(device_name, prop_name, str(value))
143 | time.sleep(20 / 1000)
144 |
145 |
146 | def set_lc_daq(mmc, device_property: tuple, value: float):
147 | """
148 | Set LC retardance based on DAQ output
149 |
150 | Parameters
151 | ----------
152 | mmc : object
153 | MM Core object
154 | device_property : tuple
155 | (device_name, property_name) set
156 | value : float
157 | DAQ output voltage in volts. DAQ output must be in 0-5V range
158 |
159 | Returns
160 | -------
161 |
162 | """
163 | device_name = device_property[0]
164 | prop_name = device_property[1]
165 |
166 | if value > 5.0 or value < 0.0:
167 | raise ValueError(
168 | "DAC voltage must be greater than 0.0 and less than 5.0"
169 | )
170 |
171 | mmc.setProperty(device_name, prop_name, str(value))
172 | time.sleep(20 / 1000)
173 |
174 |
175 | def get_lc(mmc, device_property: tuple):
176 | """
177 | Get LC state in the native units of the device property
178 |
179 | Parameters
180 | ----------
181 | mmc : object
182 | MM Core object
183 | device_property : tuple
184 | (device_name, property_name) set
185 |
186 | Returns
187 | -------
188 |
189 | """
190 |
191 | device_name = device_property[0]
192 | prop_name = device_property[1]
193 |
194 | val = float(mmc.getProperty(device_name, prop_name))
195 | return val
196 |
197 |
198 | def define_meadowlark_state(mmc, device_property: tuple):
199 | """
200 | Defines pallet element in the Meadowlark device adapter for the given state.
201 | Make sure LC values for this state are set before calling this function
202 |
203 | Parameters
204 | ----------
205 | mmc : object
206 | MM Core object
207 | device_property : tuple
208 | (device_name, property_name) set, e.g.
209 | ('MeadowlarkLC', 'Pal. elem. 00; enter 0 to define; 1 to activate')
210 |
211 | Returns
212 | -------
213 |
214 | """
215 |
216 | device_name = device_property[0]
217 | prop_name = device_property[1]
218 |
219 | # define LC state
220 | # setting pallet elements to 0 defines LC state
221 | mmc.setProperty(device_name, prop_name, 0)
222 | mmc.waitForDevice(device_name)
223 |
224 |
225 | def define_config_state(
226 | mmc, group: str, config: str, device_properties: list, values: list
227 | ):
228 | """
229 | Define config state by specifying the values for all device properties in this config
230 |
231 | Parameters
232 | ----------
233 | mmc : object
234 | MM Core object
235 | group : str
236 | Name of config group
237 | config : str
238 | Name of config, e.g. State0
239 | device_properties: list
240 | List of (device_name, property_name) tuples in config
241 | values: list
242 | List of matching device property values
243 |
244 | Returns
245 | -------
246 |
247 | """
248 |
249 | for device_property, value in zip(device_properties, values):
250 | device_name = device_property[0]
251 | prop_name = device_property[1]
252 | mmc.defineConfig(group, config, device_name, prop_name, str(value))
253 | mmc.waitForConfig(group, config)
254 |
255 |
256 | def set_lc_state(mmc, group: str, config: str):
257 | """
258 | Change to the specific LC State
259 |
260 | Parameters
261 | ----------
262 | mmc : object
263 | MM Core object
264 | group : str
265 | Name of config group
266 | config : str
267 | Name of config, e.g. State0
268 |
269 | """
270 |
271 | mmc.setConfig(group, config)
272 | time.sleep(20 / 1000) # delay for LC settle time
273 |
--------------------------------------------------------------------------------
/recOrder/io/metadata_reader.py:
--------------------------------------------------------------------------------
1 | import json
2 | import os
3 |
4 | from natsort import natsorted
5 |
6 |
7 | def load_json(path):
8 | with open(path, "r") as f:
9 | data = json.load(f)
10 |
11 | return data
12 |
13 |
14 | def get_last_metadata_file(path):
15 | last_metadata_file = natsorted(
16 | [
17 | file
18 | for file in os.listdir(path)
19 | if file.startswith("calibration_metadata")
20 | ]
21 | )[-1]
22 | return os.path.join(path, last_metadata_file)
23 |
24 |
25 | class MetadataReader:
26 | """
27 | Calibration metadata reader class. Helps load metadata from different metadata formats and naming conventions
28 | """
29 |
30 | def __init__(self, path: str):
31 | """
32 |
33 | Parameters
34 | ----------
35 | path: full path to calibration metadata
36 | """
37 | self.metadata_path = path
38 | self.json_metadata = load_json(self.metadata_path)
39 |
40 | self.Timestamp = self.get_summary_calibration_attr("Timestamp")
41 | self.recOrder_napari_verion = self.get_summary_calibration_attr(
42 | "recOrder-napari version"
43 | )
44 | self.waveorder_version = self.get_summary_calibration_attr(
45 | "waveorder version"
46 | )
47 | self.Calibration_scheme = self.get_calibration_scheme()
48 | self.Swing = self.get_swing()
49 | self.Wavelength = self.get_summary_calibration_attr("Wavelength (nm)")
50 | self.Black_level = self.get_black_level()
51 | self.Extinction_ratio = self.get_extinction_ratio()
52 | self.Channel_names = self.get_channel_names()
53 | self.LCA_retardance = self.get_lc_retardance("LCA")
54 | self.LCB_retardance = self.get_lc_retardance("LCB")
55 | self.LCA_voltage = self.get_lc_voltage("LCA")
56 | self.LCB_voltage = self.get_lc_voltage("LCB")
57 | self.Swing_measured = self.get_swing_measured()
58 | self.Notes = self.get_notes()
59 |
60 | def get_summary_calibration_attr(self, attr):
61 | try:
62 | val = self.json_metadata["Summary"][attr]
63 | except KeyError:
64 | try:
65 | val = self.json_metadata["Calibration"][attr]
66 | except KeyError:
67 | val = None
68 | return val
69 |
70 | def get_cal_states(self):
71 | if self.Calibration_scheme == "4-State":
72 | states = ["ext", "0", "60", "120"]
73 | elif self.Calibration_scheme == "5-State":
74 | states = ["ext", "0", "45", "90", "135"]
75 | return states
76 |
77 | def get_lc_retardance(self, lc):
78 | """
79 |
80 | Parameters
81 | ----------
82 | lc: 'LCA' or 'LCB'
83 |
84 | Returns
85 | -------
86 |
87 | """
88 | states = self.get_cal_states()
89 |
90 | val = None
91 | try:
92 | val = [
93 | self.json_metadata["Calibration"]["LC retardance"][
94 | f"{lc}_{state}"
95 | ]
96 | for state in states
97 | ]
98 | except KeyError:
99 | states[0] = "Ext"
100 | if lc == "LCA":
101 | val = [
102 | self.json_metadata["Summary"][
103 | f"[LCA_{state}, LCB_{state}]"
104 | ][0]
105 | for state in states
106 | ]
107 | elif lc == "LCB":
108 | val = [
109 | self.json_metadata["Summary"][
110 | f"[LCA_{state}, LCB_{state}]"
111 | ][1]
112 | for state in states
113 | ]
114 |
115 | return val
116 |
117 | def get_lc_voltage(self, lc):
118 | """
119 |
120 | Parameters
121 | ----------
122 | lc: 'LCA' or 'LCB'
123 |
124 | Returns
125 | -------
126 |
127 | """
128 | states = self.get_cal_states()
129 |
130 | val = None
131 | if "Calibration" in self.json_metadata:
132 | lc_voltage = self.json_metadata["Calibration"]["LC voltage"]
133 | if lc_voltage:
134 | val = [
135 | self.json_metadata["Calibration"]["LC voltage"][
136 | f"{lc}_{state}"
137 | ]
138 | for state in states
139 | ]
140 |
141 | return val
142 |
143 | def get_swing(self):
144 | try:
145 | val = self.json_metadata["Calibration"]["Swing (waves)"]
146 | except KeyError:
147 | val = self.json_metadata["Summary"]["Swing (fraction)"]
148 | return val
149 |
150 | def get_swing_measured(self):
151 | states = self.get_cal_states()
152 | try:
153 | val = [
154 | self.json_metadata["Calibration"][f"Swing_{state}"]
155 | for state in states[1:]
156 | ]
157 | except KeyError:
158 | val = [
159 | self.json_metadata["Summary"][f"Swing{state}"]
160 | for state in states[1:]
161 | ]
162 |
163 | return val
164 |
165 | def get_calibration_scheme(self):
166 | try:
167 | val = self.json_metadata["Calibration"]["Calibration scheme"]
168 | except KeyError:
169 | val = self.json_metadata["Summary"]["Acquired Using"]
170 | return val
171 |
172 | def get_black_level(self):
173 | try:
174 | val = self.json_metadata["Calibration"]["Black level"]
175 | except KeyError:
176 | val = self.json_metadata["Summary"]["BlackLevel"]
177 | return val
178 |
179 | def get_extinction_ratio(self):
180 | try:
181 | val = self.json_metadata["Calibration"]["Extinction ratio"]
182 | except KeyError:
183 | val = self.json_metadata["Summary"]["Extinction Ratio"]
184 | return val
185 |
186 | def get_channel_names(self):
187 | try:
188 | val = self.json_metadata["Calibration"]["Channel names"]
189 | except KeyError:
190 | val = self.json_metadata["Summary"]["ChNames"]
191 | return val
192 |
193 | def get_notes(self):
194 | try:
195 | val = self.json_metadata["Notes"]
196 | except KeyError:
197 | val = None
198 | return val
199 |
--------------------------------------------------------------------------------
/recOrder/io/utils.py:
--------------------------------------------------------------------------------
1 | import os
2 | import textwrap
3 | from pathlib import Path
4 |
5 | import psutil
6 | import torch
7 | import yaml
8 | from iohub import open_ome_zarr
9 |
10 |
11 |
12 | def add_index_to_path(path: Path):
13 | """Takes a path to a file or folder and appends the smallest index that does
14 | not already exist in that folder.
15 |
16 | For example:
17 | './output.txt' -> './output_0.txt' if no other files named './output*.txt' exist.
18 | './output.txt' -> './output_2.txt' if './output_0.txt' and './output_1.txt' already exist.
19 |
20 | Parameters
21 | ----------
22 | path: Path
23 | Base path to add index to
24 |
25 | Returns
26 | -------
27 | Path
28 | """
29 | index = 0
30 | new_stem = f"{path.stem}_{index}"
31 |
32 | while (path.parent / (new_stem + path.suffix)).exists():
33 | index += 1
34 | new_stem = f"{path.stem}_{index}"
35 |
36 | return path.parent / (new_stem + path.suffix)
37 |
38 |
39 | def load_background(background_path):
40 | with open_ome_zarr(
41 | os.path.join(background_path, "background.zarr", "0", "0", "0")
42 | ) as dataset:
43 | cyx_data = dataset["0"][0, :, 0]
44 | return torch.tensor(cyx_data, dtype=torch.float32)
45 |
46 |
47 | class MockEmitter:
48 | def emit(self, value):
49 | pass
50 |
51 |
52 | def ram_message():
53 | """
54 | Determine if the system's RAM capacity is sufficient for running reconstruction.
55 | The message should be treated as a warning if the RAM detected is less than 32 GB.
56 |
57 | Returns
58 | -------
59 | ram_report (is_warning, message)
60 | """
61 | BYTES_PER_GB = 2**30
62 | gb_available = psutil.virtual_memory().total / BYTES_PER_GB
63 | is_warning = gb_available < 32
64 |
65 | if is_warning:
66 | message = " \n".join(
67 | textwrap.wrap(
68 | f"recOrder reconstructions often require more than the {gb_available:.1f} "
69 | f"GB of RAM that this computer is equipped with. We recommend starting with reconstructions of small "
70 | f"volumes ~1000 x 1000 x 10 and working up to larger volumes while monitoring your RAM usage with "
71 | f"Task Manager or htop.",
72 | )
73 | )
74 | else:
75 | message = f"{gb_available:.1f} GB of RAM is available."
76 |
77 | return (is_warning, message)
78 |
79 |
80 | def model_to_yaml(model, yaml_path: Path) -> None:
81 | """
82 | Save a model's dictionary representation to a YAML file.
83 |
84 | Parameters
85 | ----------
86 | model : object
87 | The model object to convert to YAML.
88 | yaml_path : Path
89 | The path to the output YAML file.
90 |
91 | Raises
92 | ------
93 | TypeError
94 | If the `model` object does not have a `dict()` method.
95 |
96 | Notes
97 | -----
98 | This function converts a model object into a dictionary representation
99 | using the `dict()` method. It removes any fields with None values before
100 | writing the dictionary to a YAML file.
101 |
102 | Examples
103 | --------
104 | >>> from my_model import MyModel
105 | >>> model = MyModel()
106 | >>> model_to_yaml(model, 'model.yaml')
107 |
108 | """
109 | yaml_path = Path(yaml_path)
110 |
111 | if not hasattr(model, "dict"):
112 | raise TypeError("The 'model' object does not have a 'dict()' method.")
113 |
114 | model_dict = model.dict()
115 |
116 | # Remove None-valued fields
117 | clean_model_dict = {
118 | key: value for key, value in model_dict.items() if value is not None
119 | }
120 |
121 | with open(yaml_path, "w+") as f:
122 | yaml.dump(
123 | clean_model_dict, f, default_flow_style=False, sort_keys=False
124 | )
125 |
126 |
127 | def yaml_to_model(yaml_path: Path, model):
128 | """
129 | Load model settings from a YAML file and create a model instance.
130 |
131 | Parameters
132 | ----------
133 | yaml_path : Path
134 | The path to the YAML file containing the model settings.
135 | model : class
136 | The model class used to create an instance with the loaded settings.
137 |
138 | Returns
139 | -------
140 | object
141 | An instance of the model class with the loaded settings.
142 |
143 | Raises
144 | ------
145 | TypeError
146 | If the provided model is not a class or does not have a callable constructor.
147 | FileNotFoundError
148 | If the YAML file specified by `yaml_path` does not exist.
149 |
150 | Notes
151 | -----
152 | This function loads model settings from a YAML file using `yaml.safe_load()`.
153 | It then creates an instance of the provided `model` class using the loaded settings.
154 |
155 | Examples
156 | --------
157 | # >>> from my_model import MyModel
158 | # >>> model = yaml_to_model('model.yaml', MyModel)
159 |
160 | """
161 | yaml_path = Path(yaml_path)
162 |
163 | if not callable(getattr(model, "__init__", None)):
164 | raise TypeError(
165 | "The provided model must be a class with a callable constructor."
166 | )
167 |
168 | try:
169 | with open(yaml_path, "r") as file:
170 | raw_settings = yaml.safe_load(file)
171 | except FileNotFoundError:
172 | raise FileNotFoundError(f"The YAML file '{yaml_path}' does not exist.")
173 |
174 | return model(**raw_settings)
175 |
--------------------------------------------------------------------------------
/recOrder/io/visualization.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 |
3 | from typing import Literal, Union
4 | from colorspacious import cspace_convert
5 | from matplotlib.colors import hsv_to_rgb
6 | from skimage.color import hsv2rgb
7 | from skimage.exposure import rescale_intensity
8 |
9 |
10 | def ret_ori_overlay(
11 | czyx,
12 | ret_min: float = 1,
13 | ret_max: Union[float, Literal["auto"]] = 10,
14 | cmap: Literal["JCh", "HSV"] = "HSV",
15 | ):
16 | """
17 | Creates an overlay of retardance and orientation with two different colormap options.
18 | "HSV" maps orientation to hue and retardance to value with maximum saturation.
19 | "JCh" is a similar colormap but is perceptually uniform.
20 |
21 | Parameters
22 | ----------
23 | czyx: (nd-array) czyx[0] is retardance in nanometers, czyx[1] is orientation in radians [0, pi],
24 | czyx.shape = (2, ...)
25 |
26 | ret_min: (float) minimum displayed retardance. Typically a noise floor.
27 | ret_max: (float) maximum displayed retardance. Typically used to adjust contrast limits.
28 |
29 | cmap: (str) 'JCh' or 'HSV'
30 |
31 | Returns
32 | -------
33 | overlay (nd-array) RGB image with shape (3, ...)
34 |
35 | """
36 | if czyx.shape[0] != 2:
37 | raise ValueError(
38 | f"Input must have shape (2, ...) instead of ({czyx.shape[0]}, ...)"
39 | )
40 |
41 | retardance = czyx[0]
42 | orientation = czyx[1]
43 |
44 | if ret_max == "auto":
45 | ret_max = np.percentile(np.ravel(retardance), 99.99)
46 |
47 | # Prepare input and output arrays
48 | ret_ = np.clip(retardance, 0, ret_max) # clip and copy
49 | # Convert 180 degree range into 360 to match periodicity of hue.
50 | ori_ = orientation * 360 / np.pi
51 | overlay_final = np.zeros_like(retardance)
52 |
53 | if cmap == "JCh":
54 | J = ret_
55 | C = np.ones_like(J) * 60
56 | C[ret_ < ret_min] = 0
57 | h = ori_
58 |
59 | JCh = np.stack((J, C, h), axis=-1)
60 | JCh_rgb = cspace_convert(JCh, "JCh", "sRGB1")
61 |
62 | JCh_rgb[JCh_rgb < 0] = 0
63 | JCh_rgb[JCh_rgb > 1] = 1
64 |
65 | overlay_final = JCh_rgb
66 | elif cmap == "HSV":
67 | I_hsv = np.moveaxis(
68 | np.stack(
69 | [
70 | ori_ / 360,
71 | np.ones_like(ori_),
72 | ret_ / np.max(ret_),
73 | ]
74 | ),
75 | source=0,
76 | destination=-1,
77 | )
78 | overlay_final = hsv_to_rgb(I_hsv)
79 | else:
80 | raise ValueError(f"Colormap {cmap} not understood")
81 |
82 | return np.moveaxis(
83 | overlay_final, source=-1, destination=0
84 | ) # .shape = (3, ...)
85 |
86 |
87 | def ret_ori_phase_overlay(
88 | czyx, max_val_V: float = 1.0, max_val_S: float = 1.0
89 | ):
90 | """
91 | Creates an overlay of retardance, orientation, and phase.
92 | Maps orientation to hue, retardance to saturation, and phase to value.
93 |
94 | HSV encoding of retardance + orientation + phase image with hsv colormap
95 | (orientation in h, retardance in s, phase in v)
96 | Parameters
97 | ----------
98 | czyx : numpy.ndarray
99 | czyx[0] corresponds to the retardance image
100 | czyx[1]is the orientation image (range from 0 to pi)
101 | czyx[2] is the the phase image
102 |
103 | max_val_V : float
104 | raise the brightness of the phase channel by 1/max_val_V
105 |
106 | max_val_S : float
107 | raise the brightness of the retardance channel by 1/max_val_S
108 |
109 | Returns
110 | -------
111 | overlay (nd-array) RGB image with shape (3, ...)
112 |
113 | Returns:
114 | RGB with HSV
115 | """
116 |
117 | if czyx.shape[0] != 3:
118 | raise ValueError(
119 | f"Input must have shape (3, ...) instead of ({czyx.shape[0]}, ...)"
120 | )
121 |
122 | czyx_out = np.zeros_like(czyx, dtype=np.float32)
123 |
124 | retardance = czyx[0]
125 | orientation = czyx[1]
126 | phase = czyx[2]
127 |
128 | # Normalize the stack
129 | ordered_stack = np.stack(
130 | (
131 | # Normalize the first channel by dividing by pi
132 | orientation / np.pi,
133 | # Normalize the second channel and rescale intensity
134 | rescale_intensity(
135 | retardance,
136 | in_range=(
137 | np.min(retardance),
138 | np.max(retardance),
139 | ),
140 | out_range=(0, 1),
141 | )
142 | / max_val_S,
143 | # Normalize the third channel and rescale intensity
144 | rescale_intensity(
145 | phase,
146 | in_range=(
147 | np.min(phase),
148 | np.max(phase),
149 | ),
150 | out_range=(0, 1),
151 | )
152 | / max_val_V,
153 | ),
154 | axis=0,
155 | )
156 | czyx_out = hsv2rgb(ordered_stack, channel_axis=0)
157 | return czyx_out
158 |
--------------------------------------------------------------------------------
/recOrder/napari.yaml:
--------------------------------------------------------------------------------
1 | name: recOrder-napari
2 | display_name: recOrder-napari
3 | contributions:
4 | commands:
5 | - id: recOrder-napari.MainWidget
6 | title: Create Main Widget
7 | python_name: recOrder.plugin.main_widget:MainWidget
8 | - id: recOrder-napari.get_reader
9 | title: Read ome-zarr and ome-tif files
10 | python_name: recOrder.io._reader:napari_get_reader
11 | - id: recOrder-napari.polarization_target_data
12 | title: Polarization Target Data
13 | python_name: recOrder.scripts.samples:read_polarization_target_data
14 | - id: recOrder-napari.polarization_target_reconstruction
15 | title: Polarization Target Data
16 | python_name: recOrder.scripts.samples:read_polarization_target_reconstruction
17 | - id: recOrder-napari.zebrafish_embryo_reconstruction
18 | title: Zebrafish Embryo Reconstruction
19 | python_name: recOrder.scripts.samples:read_zebrafish_embryo_reconstruction
20 | readers:
21 | - command: recOrder-napari.get_reader
22 | accepts_directories: true
23 | filename_patterns: ['*.zarr', '*.tif']
24 | widgets:
25 | - command: recOrder-napari.MainWidget
26 | display_name: Main Menu
27 | sample_data:
28 | - command: recOrder-napari.polarization_target_data
29 | key: polarization-target-data
30 | display_name: Polarization Target Data (10 MB)
31 | - command: recOrder-napari.polarization_target_reconstruction
32 | key: polarization-target-reconstruction
33 | display_name: Polarization Target Reconstruction (10 MB)
34 | - command: recOrder-napari.zebrafish_embryo_reconstruction
35 | key: zebrafish-embryo-reconstruction
36 | display_name: Zebrafish Embryo Reconstruction (92 MB)
37 |
38 |
--------------------------------------------------------------------------------
/recOrder/plugin/__init__.py:
--------------------------------------------------------------------------------
1 | # qtpy defaults to PyQt5/PySide2 which can be present in upgraded environments
2 | try:
3 | import qtpy
4 |
5 | qtpy.API_NAME # check qtpy API name - one is required for GUI
6 |
7 | except RuntimeError as error:
8 | if type(error).__name__ == "QtBindingsNotFoundError":
9 | print("WARNING: QtBindings (PyQT or PySide) was not found for GUI")
10 |
--------------------------------------------------------------------------------
/recOrder/scripts/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/scripts/__init__.py
--------------------------------------------------------------------------------
/recOrder/scripts/launch_napari.py:
--------------------------------------------------------------------------------
1 | import napari
2 | from recOrder.plugin.main_widget import MainWidget
3 |
4 |
5 | def main():
6 | viewer = napari.Viewer()
7 | viewer.window.add_dock_widget(MainWidget(viewer))
8 | napari.run()
9 |
10 |
11 | if __name__ == "__main__":
12 | main()
13 |
--------------------------------------------------------------------------------
/recOrder/scripts/repeat-cal-acq-rec.py:
--------------------------------------------------------------------------------
1 | # This script can be modified to debug and test calibrations
2 |
3 | import random
4 | import time
5 | from contextlib import contextmanager
6 |
7 | import napari
8 | from pycromanager import Core
9 |
10 | from recOrder.plugin.main_widget import MainWidget
11 |
12 | SAVE_DIR = "."
13 | SWING = 0.05
14 | CAL_REPEATS = 3
15 | BKG_REPEATS = 3
16 |
17 |
18 | @contextmanager
19 | def stage_detour(app: MainWidget, dx: float, dy: float, wait=5):
20 | """Context manager to temporarily move the stage to a new XY-position.
21 |
22 | Parameters
23 | ----------
24 | app : MainWidget
25 | recOrder main widget instance
26 | dx : float
27 | relative x to translate
28 | dy : float
29 | relative y to translate
30 | wait : int, optional
31 | time to wait for the stage to complete movement, by default 5
32 |
33 | Yields
34 | ------
35 | MainWidget
36 | recOrder main widget instance
37 |
38 | Usage
39 | -----
40 | ```py
41 | with stage_detour(app) as app:
42 | pass # do something at the new location
43 | ```
44 | """
45 | xy_stage = app.mmc.getXYStageDevice()
46 | # get the original position
47 | ox = app.mmc.getXPosition(xy_stage)
48 | oy = app.mmc.getYPosition(xy_stage)
49 | # go to a translated position
50 | # TODO: args are floored due to a pycromanager bug: https://github.com/micro-manager/pycro-manager/issues/67
51 | app.mmc.setRelativeXYPosition(int(dx), int(dy))
52 | time.sleep(wait)
53 | try:
54 | yield app
55 | finally:
56 | # go back to the original position
57 | # TODO: args are floored due to a pycromanager bug: https://github.com/micro-manager/pycro-manager/issues/67
58 | app.mmc.setXYPosition(int(ox), int(oy))
59 | time.sleep(wait)
60 |
61 |
62 | def measure_fov(mmc: Core):
63 | """Calculate the MM FOV in micrometers.
64 |
65 | Parameters
66 | ----------
67 | mmc : Core
68 | MMCore object via pycromanager (with CamelCase set to `True`)
69 |
70 | Returns
71 | -------
72 | tuple[float, float]
73 | FOV size (x, y)
74 | """
75 | pixel_size = float(mmc.getPixelSizeUm())
76 | if pixel_size == 0:
77 | float(
78 | input(
79 | "Pixel size is not calibrated. Please provide an estimate (in micrometers):"
80 | )
81 | )
82 | fov_x = pixel_size * float(mmc.getImageWidth())
83 | fov_y = pixel_size * float(mmc.getImageHeight())
84 | return fov_x, fov_y
85 |
86 |
87 | def rand_shift(length: float):
88 | """Randomly signed shift of a certain length.
89 |
90 | Parameters
91 | ----------
92 | length : float
93 | absolote length in micrometers
94 |
95 | Returns
96 | -------
97 | float
98 | +length or -length
99 | """
100 | sign = random.randint(0, 1) * 2 - 1
101 | return sign * length
102 |
103 |
104 | def main():
105 | viewer = napari.Viewer()
106 | app = MainWidget(viewer)
107 | viewer.window.add_dock_widget(app)
108 | app.ui.qbutton_gui_mode.click()
109 | app.calib_scheme = "5-State"
110 | app.directory = SAVE_DIR
111 | app.save_directory = SAVE_DIR
112 |
113 | fov_x, fov_y = measure_fov(app.mmc)
114 |
115 | input("Please center the target in the FOV and hit ")
116 |
117 | for cal_repeat in range(CAL_REPEATS):
118 | dx = rand_shift(fov_x)
119 | dy = rand_shift(fov_y)
120 | # run calibration
121 | with stage_detour(app, dx, dy) as app:
122 | print(f"Calibration repeat # {cal_repeat}")
123 | app.swing = SWING
124 |
125 | print(f"Calibrating with swing = {SWING}")
126 | app.run_calibration()
127 | time.sleep(90)
128 |
129 | for bkg_repeat in range(BKG_REPEATS):
130 | # capture background
131 | with stage_detour(app, dx, dy) as app:
132 | print(f">>> Background repeat # {bkg_repeat}")
133 | app.last_calib_meta_file = app.calib.meta_file
134 | app.capture_bg()
135 | time.sleep(20)
136 | app.ui.cb_bg_method.setCurrentIndex(
137 | 1
138 | ) # Set to "Measured" bg correction
139 | app.enter_bg_correction()
140 | app.save_name = f"cal-{cal_repeat}-bkg-{bkg_repeat}"
141 | app.enter_acq_bg_path()
142 | app.acq_ret_ori()
143 | time.sleep(15)
144 |
145 |
146 | if __name__ == "__main__":
147 | main()
148 |
--------------------------------------------------------------------------------
/recOrder/scripts/repeat-calibration.py:
--------------------------------------------------------------------------------
1 | # This script can be modified to debug and test calibrations
2 |
3 | import napari
4 | import time
5 | from recOrder.plugin.main_widget import MainWidget
6 |
7 | SAVE_DIR = "./"
8 | SWINGS = [0.1, 0.03, 0.01, 0.005]
9 | REPEATS = 5
10 |
11 |
12 | def main():
13 | viewer = napari.Viewer()
14 | recorder = MainWidget(viewer)
15 | viewer.window.add_dock_widget(recorder)
16 | recorder.ui.qbutton_connect_to_mm.click()
17 | recorder.calib_scheme = "5-State"
18 |
19 | for repeat in range(REPEATS):
20 | for swing in SWINGS:
21 | print("Calibrating with swing = " + str(swing))
22 | recorder.swing = swing
23 | recorder.directory = SAVE_DIR
24 | recorder.run_calibration()
25 | time.sleep(100)
26 |
27 |
28 | if __name__ == "__main__":
29 | main()
30 |
--------------------------------------------------------------------------------
/recOrder/scripts/samples.py:
--------------------------------------------------------------------------------
1 | import shutil
2 | from pathlib import Path
3 |
4 | from typing import Literal
5 | from iohub import open_ome_zarr
6 | from iohub.ngff import Plate
7 | from napari.utils.notifications import show_warning
8 | from platformdirs import user_data_dir
9 | from wget import download
10 |
11 |
12 | def _build_layer_list(dataset: Plate, layer_names: list[str]):
13 | layer_list = []
14 | for channel_name in layer_names:
15 | channel_index = dataset.channel_names.index(channel_name)
16 | position = dataset["0/0/0"]
17 | data = (position["0"][:, channel_index],)
18 | layer_dict = {"name": channel_name, "scale": position.scale[3:]}
19 | layer_list.append((data, layer_dict))
20 |
21 | return layer_list
22 |
23 |
24 | def download_and_unzip(data_type: Literal["target", "embryo"]) -> tuple[Path]:
25 | """Downloads sample data .zip from zenodo, unzips, and returns Paths to the .zarr datasets.
26 |
27 | Skips the download if the files already exist.
28 |
29 | Uses platformdirs.user_data_dir to store data.
30 | """
31 |
32 | # Delete old data
33 | old_data_dirs = ["recOrder-sample-v1.4"]
34 | for old_data_dir in old_data_dirs:
35 | old_data_path = Path(user_data_dir(old_data_dir))
36 | if old_data_path.exists():
37 | shutil.rmtree(str(old_data_path))
38 |
39 | temp_dirpath = Path(user_data_dir("recOrder-sample-v1.5"))
40 | temp_dirpath.mkdir(exist_ok=True, parents=True)
41 |
42 | if data_type == "target":
43 | data_dirpath = temp_dirpath / "sample_contribution"
44 | data_size = "10 MB"
45 | data_url = "https://zenodo.org/record/8386856/files/sample_contribution.zip?download=1"
46 | elif data_type == "embryo":
47 | data_dirpath = temp_dirpath / "sample_contribution_embryo"
48 | data_size = "92 MB"
49 | data_url = "https://zenodo.org/record/8386856/files/sample_contribution_embryo.zip?download=1"
50 |
51 | if not data_dirpath.with_suffix(".zip").exists():
52 | show_warning(
53 | f"Downloading {data_size} sample contribution. This might take a moment..."
54 | )
55 | download(data_url, out=str(temp_dirpath))
56 |
57 | if not data_dirpath.exists():
58 | shutil.unpack_archive(
59 | data_dirpath.with_suffix(".zip"), extract_dir=temp_dirpath
60 | )
61 |
62 | data_path = data_dirpath / "raw_data.zarr"
63 | recon_path = data_dirpath / "reconstruction.zarr"
64 | return data_path, recon_path
65 |
66 |
67 | def read_polarization_target_data():
68 | """Returns the polarization data sample contribution"""
69 | data_path, _ = download_and_unzip("target")
70 | dataset = open_ome_zarr(data_path)
71 | return _build_layer_list(dataset, dataset.channel_names)
72 |
73 |
74 | def read_polarization_target_reconstruction():
75 | """Returns the polarization target reconstruction sample contribution"""
76 | _, recon_path = download_and_unzip("target")
77 | dataset = open_ome_zarr(recon_path)
78 | return _build_layer_list(dataset, ["Phase3D", "Retardance", "Orientation"])
79 |
80 |
81 | def read_zebrafish_embryo_reconstruction():
82 | """Returns the embryo reconstruction sample contribution"""
83 | _, recon_path = download_and_unzip("embryo")
84 | dataset = open_ome_zarr(recon_path)
85 | return _build_layer_list(dataset, ["Retardance", "Orientation"])
86 |
--------------------------------------------------------------------------------
/recOrder/scripts/simulate_zarr_acq.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 | from iohub.convert import TIFFConverter
3 | from iohub.ngff import open_ome_zarr
4 | from recOrder.cli.utils import create_empty_hcs_zarr
5 | from recOrder.cli import jobs_mgmt
6 |
7 | import time, threading, os, shutil, subprocess
8 |
9 | # This script is a demo .zarr acquisition simulation from an acquired .zarr store
10 | # The script copies and writes additional metadata to .zattrs inserting two keys
11 | # The two keys are "FinalDimensions" and "CurrentDimensions".
12 | # The "FinalDimensions" key with (t,p,z,c) needs to be inserted when the dataset is created
13 | # and then should be updated at close to ensure aborted acquisitions represent correct dimensions.
14 | # The "CurrentDimensions" key should have the same (t,p,z,c) information and should be written out
15 | # either with every new image, end of dimension OR at frequent intervals.
16 | # Refer further notes below in the example regarding encountered issues.
17 | #
18 | # Refer to steps at the end of the file on steps to run this file
19 |
20 | #%% #############################################
21 | def convert_data(tif_path, latest_out_path, prefix="", data_type_str="ometiff"):
22 | converter = TIFFConverter(
23 | os.path.join(tif_path , prefix),
24 | latest_out_path,
25 | data_type=data_type_str,
26 | grid_layout=False,
27 | )
28 | converter.run()
29 |
30 | def run_convert(ome_tif_path):
31 | out_path = os.path.join(Path(ome_tif_path).parent.absolute(), ("raw_" + Path(ome_tif_path).name + ".zarr"))
32 | convert_data(ome_tif_path, out_path)
33 |
34 | #%% #############################################
35 |
36 | def run_acq(input_path="", waitBetweenT=30):
37 |
38 | output_store_path = os.path.join(Path(input_path).parent.absolute(), ("acq_sim_" + Path(input_path).name))
39 |
40 | if Path(output_store_path).exists():
41 | shutil.rmtree(output_store_path)
42 | time.sleep(1)
43 |
44 | input_data = open_ome_zarr(input_path, mode="r")
45 | channel_names = input_data.channel_names
46 |
47 | position_keys: list[tuple[str]] = []
48 |
49 | for path, pos in input_data.positions():
50 | shape = pos["0"].shape
51 | dtype = pos["0"].dtype
52 | chunks = pos["0"].chunks
53 | scale = (1, 1, 1, 1, 1)
54 | position_keys.append(path.split("/"))
55 |
56 | create_empty_hcs_zarr(
57 | output_store_path,
58 | position_keys,
59 | shape,
60 | chunks,
61 | scale,
62 | channel_names,
63 | dtype,
64 | {},
65 | )
66 | output_dataset = open_ome_zarr(output_store_path, mode="r+")
67 |
68 | if "Summary" in input_data.zattrs.keys():
69 | output_dataset.zattrs["Summary"] = input_data.zattrs["Summary"]
70 |
71 | output_dataset.zattrs.update({"FinalDimensions": {
72 | "channel": shape[1],
73 | "position": len(position_keys),
74 | "time": shape[0],
75 | "z": shape[2]
76 | }
77 | })
78 |
79 | total_time = shape[0]
80 | total_pos = len(position_keys)
81 | total_z = shape[2]
82 | total_c = shape[1]
83 | for t in range(total_time):
84 | for p in range(total_pos):
85 | for z in range(total_z):
86 | for c in range(total_c):
87 | position_key_string = "/".join(position_keys[p])
88 | img_src = input_data[position_key_string][0][t, c, z]
89 |
90 | img_data = output_dataset[position_key_string][0]
91 | img_data[t, c, z] = img_src
92 |
93 | # Note: On-The-Fly dataset reconstruction will throw Permission Denied when being written
94 | # Maybe we can read the zaatrs directly in that case as a file which is less blocking
95 | # If this write/read is a constant issue then the zattrs 'CurrentDimensions' key
96 | # should be updated less frequently, instead of current design of updating with
97 | # each image
98 | output_dataset.zattrs.update({"CurrentDimensions": {
99 | "channel": total_c,
100 | "position": p+1,
101 | "time": t+1,
102 | "z": z+1
103 | }
104 | })
105 |
106 | required_order = ['time', 'position', 'z', 'channel']
107 | my_dict = output_dataset.zattrs["CurrentDimensions"]
108 | sorted_dict_acq = {k: my_dict[k] for k in sorted(my_dict, key=lambda x: required_order.index(x))}
109 | print("Writer thread - Acquisition Dim:", sorted_dict_acq)
110 |
111 |
112 | # reconThread = threading.Thread(target=doReconstruct, args=(output_store_path, t))
113 | # reconThread.start()
114 |
115 | time.sleep(waitBetweenT) # sleep after every t
116 |
117 | output_dataset.close
118 |
119 | def do_reconstruct(input_path, time_point):
120 |
121 | config_path = os.path.join(Path(input_path).parent.absolute(), "Bire-"+str(time_point)+".yml")
122 | output_path = os.path.join(Path(input_path).parent.absolute(), "Recon_"+Path(input_path).name)
123 | mainfp = str(jobs_mgmt.FILE_PATH)
124 |
125 | print("Processing {input} time_point={tp}".format(input=input_path, tp=time_point))
126 |
127 | try:
128 | proc = subprocess.run(
129 | [
130 | "python",
131 | mainfp,
132 | "reconstruct",
133 | "-i",
134 | input_path,
135 | "-c",
136 | config_path,
137 | "-o",
138 | output_path,
139 | "-rx",
140 | str(20)
141 | ]
142 | )
143 | if proc.returncode != 0:
144 | raise Exception("An error occurred in processing ! Check terminal output.")
145 | except Exception as exc:
146 | print(exc.args)
147 |
148 | #%% #############################################
149 | def run_acquire(input_path, waitBetweenT):
150 | runThread1Acq = threading.Thread(target=run_acq, args=(input_path, waitBetweenT))
151 | runThread1Acq.start()
152 |
153 | #%% #############################################
154 | # Step 1:
155 | # Convert an existing ome-tif recOrder acquisition, preferably with all dims (t, p, z, c)
156 | # This will convert an existing ome-tif to a .zarr storage
157 |
158 | # ome_tif_path = "/ome-zarr_data/recOrderAcq/test/snap_6D_ometiff_1"
159 | # runConvert(ome_tif_path)
160 |
161 | #%% #############################################
162 | # Step 2:
163 | # run the test to simulate Acquiring a recOrder .zarr store
164 |
165 | input_path = "/ome-zarr_data/recOrderAcq/test/raw_snap_6D_ometiff_1.zarr"
166 | waitBetweenT = 60
167 | run_acquire(input_path, waitBetweenT)
168 |
169 |
170 |
171 |
172 |
--------------------------------------------------------------------------------
/recOrder/tests/acq_tests/test_acq.py:
--------------------------------------------------------------------------------
1 | from unittest.mock import patch
2 |
3 | import numpy as np
4 | from recOrder.acq.acquisition_workers import _check_scale_mismatch
5 |
6 |
7 | def test_check_scale_mismatch():
8 | warn_fn_path = "recOrder.acq.acquisition_workers.show_warning"
9 | identity = np.array((1.0, 1.0, 1.0))
10 | with patch(warn_fn_path) as mock:
11 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1))
12 | mock.assert_not_called()
13 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1.001))
14 | mock.assert_not_called()
15 | _check_scale_mismatch(identity, (1, 1, 1, 1, 1.1))
16 | mock.assert_called_once()
17 |
--------------------------------------------------------------------------------
/recOrder/tests/calibration_tests/test_calibration.py:
--------------------------------------------------------------------------------
1 | def test_calib_imports():
2 | from recOrder.calib import Calibration, Optimization
3 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_cli.py:
--------------------------------------------------------------------------------
1 | from recOrder.cli.main import cli
2 | from click.testing import CliRunner
3 |
4 |
5 | def test_main():
6 | runner = CliRunner()
7 | result = runner.invoke(cli)
8 |
9 | assert result.exit_code == 0
10 | assert "Toolkit" in result.output
11 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_compute_tf.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 | from click.testing import CliRunner
4 |
5 | from recOrder.cli import settings
6 | from recOrder.cli.compute_transfer_function import (
7 | generate_and_save_birefringence_transfer_function,
8 | generate_and_save_fluorescence_transfer_function,
9 | generate_and_save_phase_transfer_function,
10 | )
11 | from recOrder.cli.main import cli
12 | from recOrder.io import utils
13 |
14 |
15 | def test_compute_transfer(tmp_path, example_plate):
16 | recon_settings = settings.ReconstructionSettings(
17 | input_channel_names=[f"State{i}" for i in range(4)],
18 | reconstruction_dimension=3,
19 | birefringence=settings.BirefringenceSettings(),
20 | phase=settings.PhaseSettings(),
21 | )
22 | config_path = tmp_path / "test.yml"
23 | utils.model_to_yaml(recon_settings, config_path)
24 |
25 | output_path = tmp_path / "output.zarr"
26 |
27 | plate_path, _ = example_plate
28 | runner = CliRunner()
29 | result = runner.invoke(
30 | cli,
31 | [
32 | "compute-tf",
33 | "-i",
34 | str(plate_path / "A" / "1" / "0"),
35 | "-c",
36 | str(config_path),
37 | "-o",
38 | str(output_path),
39 | ],
40 | )
41 | assert result.exit_code == 0
42 |
43 |
44 | def test_compute_transfer_blank_config():
45 | runner = CliRunner()
46 | for option in ("-c ", "--config-path "):
47 | cmd = "compute-tf " + option
48 | result = runner.invoke(cli, cmd)
49 | assert result.exit_code == 2
50 | assert "Error" in result.output
51 |
52 |
53 | def test_compute_transfer_blank_output():
54 | runner = CliRunner()
55 | for option in ("-o ", "--output-path "):
56 | cmd = "compute-tf " + option
57 | result = runner.invoke(cli, cmd)
58 | assert result.exit_code == 2
59 | assert "Error" in result.output
60 |
61 |
62 | def test_compute_transfer_output_file(tmp_path, example_plate):
63 | recon_settings = settings.ReconstructionSettings(
64 | input_channel_names=["BF"],
65 | reconstruction_dimension=3,
66 | phase=settings.PhaseSettings(),
67 | )
68 | config_path = tmp_path / "test.yml"
69 | utils.model_to_yaml(recon_settings, config_path)
70 |
71 | plate_path, _ = example_plate
72 | runner = CliRunner()
73 | for option in ("-o", "--output-dirpath"):
74 | for output_folder in ["test1.zarr", "test2/test.zarr"]:
75 | output_path = tmp_path.joinpath(output_folder)
76 | result = runner.invoke(
77 | cli,
78 | [
79 | "compute-tf",
80 | "-i",
81 | str(plate_path / "A" / "1" / "0"),
82 | "-c",
83 | str(config_path),
84 | str(option),
85 | str(output_path),
86 | ],
87 | )
88 | assert result.exit_code == 0
89 | assert str(output_path) in result.output
90 | assert output_path.exists()
91 |
92 |
93 | def test_stokes_matrix_write(birefringence_phase_recon_settings_function):
94 | settings, dataset = birefringence_phase_recon_settings_function
95 | generate_and_save_birefringence_transfer_function(settings, dataset)
96 | assert dataset["intensity_to_stokes_matrix"]
97 |
98 |
99 | def test_absorption_and_phase_write(
100 | birefringence_phase_recon_settings_function,
101 | ):
102 | settings, dataset = birefringence_phase_recon_settings_function
103 | generate_and_save_phase_transfer_function(settings, dataset, (3, 4, 5))
104 | assert dataset["real_potential_transfer_function"]
105 | assert dataset["imaginary_potential_transfer_function"]
106 | assert dataset["imaginary_potential_transfer_function"].shape == (
107 | 1,
108 | 1,
109 | 3,
110 | 4,
111 | 5,
112 | )
113 | assert "absorption_transfer_function" not in dataset
114 | assert "phase_transfer_function" not in dataset
115 |
116 |
117 | def test_phase_3dim_write(birefringence_phase_recon_settings_function):
118 | settings, dataset = birefringence_phase_recon_settings_function
119 | settings.reconstruction_dimension = 2
120 | generate_and_save_phase_transfer_function(settings, dataset, (3, 4, 5))
121 | assert dataset["absorption_transfer_function"]
122 | assert dataset["phase_transfer_function"]
123 | assert dataset["phase_transfer_function"].shape == (1, 1, 3, 4, 5)
124 | assert "real_potential_transfer_function" not in dataset
125 | assert "imaginary_potential_transfer_function" not in dataset
126 |
127 |
128 | def test_fluorescence_write(fluorescence_recon_settings_function):
129 | settings, dataset = fluorescence_recon_settings_function
130 | generate_and_save_fluorescence_transfer_function(
131 | settings, dataset, (3, 4, 5)
132 | )
133 | assert dataset["optical_transfer_function"]
134 | assert dataset["optical_transfer_function"].shape == (1, 1, 3, 4, 5)
135 | assert "real_potential_transfer_function" not in dataset
136 | assert "imaginary_potential_transfer_function" not in dataset
137 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_reconstruct.py:
--------------------------------------------------------------------------------
1 | import os
2 |
3 | import numpy as np
4 | from click.testing import CliRunner
5 | from iohub.ngff import open_ome_zarr
6 | from iohub.ngff_meta import TransformationMeta
7 |
8 | from recOrder.cli import settings
9 | from recOrder.cli.main import cli
10 | from recOrder.io import utils
11 | from recOrder.cli.apply_inverse_transfer_function import (
12 | apply_inverse_transfer_function_cli,
13 | )
14 | from unittest.mock import patch
15 | import pytest
16 | from pathlib import Path
17 | from tempfile import TemporaryDirectory
18 |
19 |
20 | input_scale = [1, 2, 3, 4, 5]
21 | # Setup options
22 | birefringence_settings = settings.BirefringenceSettings(
23 | transfer_function=settings.BirefringenceTransferFunctionSettings()
24 | )
25 |
26 | # birefringence_option, time_indices, phase_option, dimension_option, time_length_target
27 | all_options = [
28 | (birefringence_settings, [0, 3, 4], None, 2, 5),
29 | (birefringence_settings, 0, settings.PhaseSettings(), 2, 5),
30 | (birefringence_settings, [0, 1], None, 3, 5),
31 | (birefringence_settings, "all", settings.PhaseSettings(), 3, 5),
32 | ]
33 |
34 |
35 | @pytest.fixture(scope="session")
36 | def tmp_input_path_zarr():
37 | tmp_path = TemporaryDirectory()
38 | yield Path(tmp_path.name) / "input.zarr", Path(tmp_path.name) / "test.yml"
39 | tmp_path.cleanup()
40 |
41 |
42 | def test_reconstruct(tmp_input_path_zarr):
43 | input_path, tmp_config_yml = tmp_input_path_zarr
44 | # Generate input "dataset"
45 | channel_names = [f"State{x}" for x in range(4)]
46 | dataset = open_ome_zarr(
47 | input_path,
48 | layout="hcs",
49 | mode="w",
50 | channel_names=channel_names,
51 | )
52 |
53 | position = dataset.create_position("0", "0", "0")
54 | position.create_zeros(
55 | "0",
56 | (5, 4, 4, 5, 6),
57 | dtype=np.uint16,
58 | transform=[TransformationMeta(type="scale", scale=input_scale)],
59 | )
60 |
61 | for i, (
62 | birefringence_option,
63 | time_indices,
64 | phase_option,
65 | dimension_option,
66 | time_length_target,
67 | ) in enumerate(all_options):
68 | if (birefringence_option is None) and (phase_option is None):
69 | continue
70 |
71 | # Generate recon settings
72 | recon_settings = settings.ReconstructionSettings(
73 | input_channel_names=channel_names,
74 | time_indices=time_indices,
75 | reconstruction_dimension=dimension_option,
76 | birefringence=birefringence_option,
77 | phase=phase_option,
78 | )
79 | config_path = tmp_config_yml.with_name(f"{i}.yml")
80 | utils.model_to_yaml(recon_settings, config_path)
81 |
82 | # Run CLI
83 | runner = CliRunner()
84 | tf_path = input_path.with_name(f"tf_{i}.zarr")
85 | runner.invoke(
86 | cli,
87 | [
88 | "compute-tf",
89 | "-i",
90 | str(input_path / "0" / "0" / "0"),
91 | "-c",
92 | str(config_path),
93 | "-o",
94 | str(tf_path),
95 | ],
96 | catch_exceptions=False,
97 | )
98 | assert tf_path.exists()
99 |
100 |
101 | def test_append_channel_reconstruction(tmp_input_path_zarr):
102 | input_path, tmp_config_yml = tmp_input_path_zarr
103 | output_path = input_path.with_name(f"output.zarr")
104 |
105 | # Generate input "dataset"
106 | channel_names = [f"State{x}" for x in range(4)] + ["GFP"]
107 | dataset = open_ome_zarr(
108 | input_path,
109 | layout="hcs",
110 | mode="w",
111 | channel_names=channel_names,
112 | )
113 | position = dataset.create_position("0", "0", "0")
114 | position.create_zeros(
115 | "0",
116 | (5, 5, 4, 5, 6),
117 | dtype=np.uint16,
118 | transform=[TransformationMeta(type="scale", scale=input_scale)],
119 | )
120 |
121 | # Generate recon settings
122 | biref_settings = settings.ReconstructionSettings(
123 | input_channel_names=[f"State{x}" for x in range(4)],
124 | time_indices="all",
125 | reconstruction_dimension=3,
126 | birefringence=settings.BirefringenceSettings(),
127 | phase=None,
128 | fluorescence=None,
129 | )
130 | fluor_settings = settings.ReconstructionSettings(
131 | input_channel_names=["GFP"],
132 | time_indices="all",
133 | reconstruction_dimension=3,
134 | birefringence=None,
135 | phase=None,
136 | fluorescence=settings.FluorescenceSettings(),
137 | )
138 | biref_config_path = tmp_config_yml.with_name(f"biref.yml")
139 | fluor_config_path = tmp_config_yml.with_name(f"fluor.yml")
140 |
141 | utils.model_to_yaml(biref_settings, biref_config_path)
142 | utils.model_to_yaml(fluor_settings, fluor_config_path)
143 |
144 | # Apply birefringence reconstruction
145 | runner = CliRunner()
146 | runner.invoke(
147 | cli,
148 | [
149 | "reconstruct",
150 | "-i",
151 | str(input_path / "0" / "0" / "0"),
152 | "-c",
153 | str(biref_config_path),
154 | "-o",
155 | str(output_path),
156 | ],
157 | catch_exceptions=False,
158 | )
159 | assert output_path.exists()
160 | with open_ome_zarr(output_path) as dataset:
161 | assert dataset["0/0/0"]["0"].shape[1] == 4
162 |
163 | # Append fluoresncence reconstruction
164 | runner.invoke(
165 | cli,
166 | [
167 | "reconstruct",
168 | "-i",
169 | str(input_path / "0" / "0" / "0"),
170 | "-c",
171 | str(fluor_config_path),
172 | "-o",
173 | str(output_path),
174 | ],
175 | catch_exceptions=False,
176 | )
177 | assert output_path.exists()
178 | with open_ome_zarr(output_path) as dataset:
179 | assert dataset["0/0/0"]["0"].shape[1] == 5
180 | assert dataset.channel_names[-1] == "GFP_Density3D"
181 | assert dataset.channel_names[-2] == "Pol"
182 |
183 |
184 | def test_cli_apply_inv_tf_mock(tmp_input_path_zarr):
185 | tmp_input_zarr, tmp_config_yml = tmp_input_path_zarr
186 | tmp_config_yml = tmp_config_yml.with_name("0.yml").resolve()
187 | tf_path = tmp_input_zarr.with_name("tf_0.zarr").resolve()
188 | input_path = (tmp_input_zarr / "0" / "0" / "0").resolve()
189 | result_path = tmp_input_zarr.with_name("result.zarr").resolve()
190 |
191 | assert tmp_config_yml.exists()
192 | assert tf_path.exists()
193 | assert input_path.exists()
194 | assert not result_path.exists()
195 |
196 | runner = CliRunner()
197 | with patch(
198 | "recOrder.cli.apply_inverse_transfer_function.apply_inverse_transfer_function_cli"
199 | ) as mock:
200 | cmd = [
201 | "apply-inv-tf",
202 | "-i",
203 | str(input_path),
204 | "-t",
205 | str(tf_path),
206 | "-c",
207 | str(tmp_config_yml),
208 | "-o",
209 | str(result_path),
210 | "-j",
211 | str(1),
212 | ]
213 | result_inv = runner.invoke(
214 | cli,
215 | cmd,
216 | catch_exceptions=False,
217 | )
218 | mock.assert_called_with(
219 | [input_path],
220 | Path(tf_path),
221 | Path(tmp_config_yml),
222 | Path(result_path),
223 | 1,
224 | 1,
225 | )
226 | assert result_inv.exit_code == 0
227 |
228 |
229 | def test_cli_apply_inv_tf_output(tmp_input_path_zarr, capsys):
230 | tmp_input_zarr, tmp_config_yml = tmp_input_path_zarr
231 | input_path = tmp_input_zarr / "0" / "0" / "0"
232 |
233 | for i, (
234 | birefringence_option,
235 | time_indices,
236 | phase_option,
237 | dimension_option,
238 | time_length_target,
239 | ) in enumerate(all_options):
240 | if (birefringence_option is None) and (phase_option is None):
241 | continue
242 |
243 | result_path = tmp_input_zarr.with_name(f"result{i}.zarr").resolve()
244 |
245 | tf_path = tmp_input_zarr.with_name(f"tf_{i}.zarr")
246 | tmp_config_yml = tmp_config_yml.with_name(f"{i}.yml")
247 |
248 | # # Check output
249 | apply_inverse_transfer_function_cli(
250 | [input_path], tf_path, tmp_config_yml, result_path, 1
251 | )
252 |
253 | result_dataset = open_ome_zarr(str(result_path / "0" / "0" / "0"))
254 | assert result_dataset["0"].shape[0] == time_length_target
255 | assert result_dataset["0"].shape[3:] == (5, 6)
256 |
257 | assert result_path.exists()
258 | captured = capsys.readouterr()
259 | assert "submitted" in captured.out
260 |
261 | # Check scale transformations pass through
262 | assert input_scale == result_dataset.scale
263 |
--------------------------------------------------------------------------------
/recOrder/tests/cli_tests/test_settings.py:
--------------------------------------------------------------------------------
1 | import pytest
2 | from recOrder.cli import settings
3 | from recOrder.io import utils
4 | from pydantic.v1 import ValidationError
5 |
6 |
7 | def test_reconstruction_settings():
8 | # Test defaults
9 | s = settings.ReconstructionSettings(
10 | birefringence=settings.BirefringenceSettings()
11 | )
12 | assert len(s.input_channel_names) == 4
13 | assert s.birefringence.apply_inverse.background_path == ""
14 | assert s.phase == None
15 | assert s.fluorescence == None
16 |
17 | # Test logic that "fluorescence" or ("phase" and/or "birefringence")
18 | s = settings.ReconstructionSettings(
19 | input_channel_names=["GFP"],
20 | birefringence=None,
21 | phase=None,
22 | fluorescence=settings.FluorescenceSettings(),
23 | )
24 |
25 | assert s.fluorescence.apply_inverse.reconstruction_algorithm == "Tikhonov"
26 |
27 | # Not allowed to supply both phase/biref and fluorescence
28 | with pytest.raises(ValidationError):
29 | settings.ReconstructionSettings(
30 | phase=settings.PhaseSettings(),
31 | fluorescence=settings.FluorescenceSettings(),
32 | )
33 |
34 | # Test incorrect settings
35 | with pytest.raises(ValidationError):
36 | settings.ReconstructionSettings(input_channel_names=3)
37 |
38 | with pytest.raises(ValidationError):
39 | settings.ReconstructionSettings(reconstruction_dimension=1)
40 |
41 | # Test typo
42 | with pytest.raises(ValidationError):
43 | settings.ReconstructionSettings(
44 | flurescence=settings.FluorescenceSettings()
45 | )
46 |
47 |
48 | def test_biref_tf_settings():
49 | settings.BirefringenceTransferFunctionSettings(swing=0.1)
50 |
51 | with pytest.raises(ValidationError):
52 | settings.BirefringenceTransferFunctionSettings(swing=1.1)
53 |
54 | with pytest.raises(ValidationError):
55 | settings.BirefringenceTransferFunctionSettings(scheme="Test")
56 |
57 |
58 | def test_phase_tf_settings():
59 | settings.PhaseTransferFunctionSettings(
60 | index_of_refraction_media=1.0, numerical_aperture_detection=0.8
61 | )
62 |
63 | with pytest.raises(ValidationError):
64 | settings.PhaseTransferFunctionSettings(
65 | index_of_refraction_media=1.0, numerical_aperture_detection=1.1
66 | )
67 |
68 | # Inconsistent units
69 | with pytest.raises(Warning):
70 | settings.PhaseTransferFunctionSettings(
71 | yx_pixel_size=650, z_pixel_size=0.3
72 | )
73 |
74 | # Extra parameter
75 | with pytest.raises(ValidationError):
76 | settings.PhaseTransferFunctionSettings(zyx_pixel_size=650)
77 |
78 |
79 | def test_fluor_tf_settings():
80 | settings.FluorescenceTransferFunctionSettings(
81 | wavelength_emission=0.500, yx_pixel_size=0.2
82 | )
83 |
84 | with pytest.raises(Warning):
85 | settings.FluorescenceTransferFunctionSettings(
86 | wavelength_emission=0.500, yx_pixel_size=2000
87 | )
88 |
89 |
90 | def test_generate_example_settings():
91 | example_path = "./examples/"
92 |
93 | s0 = settings.ReconstructionSettings(
94 | birefringence=settings.BirefringenceSettings(),
95 | phase=settings.PhaseSettings(),
96 | )
97 | s1 = settings.ReconstructionSettings(
98 | input_channel_names=["BF"],
99 | phase=settings.PhaseSettings(),
100 | )
101 | s2 = settings.ReconstructionSettings(
102 | birefringence=settings.BirefringenceSettings(),
103 | )
104 | s3 = settings.ReconstructionSettings(
105 | input_channel_names=["GFP"],
106 | fluorescence=settings.FluorescenceSettings(),
107 | )
108 | file_names = [
109 | "birefringence-and-phase.yml",
110 | "phase.yml",
111 | "birefringence.yml",
112 | "fluorescence.yml",
113 | ]
114 | settings_list = [s0, s1, s2, s3]
115 |
116 | # Save to examples folder and test roundtrip
117 | for file_name, settings_obj in zip(file_names, settings_list):
118 | utils.model_to_yaml(settings_obj, example_path + file_name)
119 | settings_roundtrip = utils.yaml_to_model(
120 | example_path + file_name, settings.ReconstructionSettings
121 | )
122 | assert settings_obj.dict() == settings_roundtrip.dict()
123 |
--------------------------------------------------------------------------------
/recOrder/tests/conftest.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import pytest
3 | from iohub.ngff import open_ome_zarr
4 |
5 | from recOrder.cli import settings
6 |
7 |
8 | @pytest.fixture(scope="function")
9 | def example_plate(tmp_path):
10 | plate_path = tmp_path / "input.zarr"
11 |
12 | position_list = (
13 | ("A", "1", "0"),
14 | ("B", "1", "0"),
15 | ("B", "2", "0"),
16 | )
17 |
18 | plate_dataset = open_ome_zarr(
19 | plate_path,
20 | layout="hcs",
21 | mode="w",
22 | channel_names=[f"State{i}" for i in range(4)] + ["BF"],
23 | )
24 |
25 | for row, col, fov in position_list:
26 | position = plate_dataset.create_position(row, col, fov)
27 | position.create_zeros("0", (2, 5, 4, 5, 6), dtype=np.uint16)
28 |
29 | yield plate_path, plate_dataset
30 |
31 |
32 | @pytest.fixture(scope="function")
33 | def birefringence_phase_recon_settings_function(tmp_path):
34 | recon_settings = settings.ReconstructionSettings(
35 | birefringence=settings.BirefringenceSettings(),
36 | phase=settings.PhaseSettings(),
37 | )
38 | dataset = open_ome_zarr(
39 | tmp_path,
40 | layout="fov",
41 | mode="w",
42 | channel_names=[f"State{i}" for i in range(4)],
43 | )
44 | yield recon_settings, dataset
45 |
46 |
47 | @pytest.fixture(scope="function")
48 | def fluorescence_recon_settings_function(tmp_path):
49 | recon_settings = settings.ReconstructionSettings(
50 | input_channel_names=["GFP"],
51 | fluorescence=settings.FluorescenceSettings(),
52 | )
53 | dataset = open_ome_zarr(
54 | tmp_path,
55 | layout="fov",
56 | mode="w",
57 | channel_names=[f"State{i}" for i in range(4)],
58 | )
59 | yield recon_settings, dataset
60 |
--------------------------------------------------------------------------------
/recOrder/tests/mmcore_tests/test_core_func.py:
--------------------------------------------------------------------------------
1 | from unittest.mock import MagicMock, Mock, call
2 | import pytest
3 | import numpy as np
4 | from numpy import ndarray
5 | from typing import Callable, Tuple
6 |
7 | # tested components
8 | from recOrder.io.core_functions import *
9 |
10 |
11 | # TODO: move these to fixture or generate with Hypothesis
12 | # dynamic range
13 | TIFF_I_MAX = 2**16
14 | # image size
15 | IMAGE_WIDTH = np.random.randint(1, 2**12)
16 | IMAGE_HEIGHT = np.random.randint(1, 2**12)
17 | PIXEL_COUNT = IMAGE_HEIGHT * IMAGE_WIDTH
18 | # serialized image from the pycromanager bridge
19 | SERIAL_IMAGE = np.random.randint(0, TIFF_I_MAX, size=(PIXEL_COUNT,))
20 | # LC device parameters
21 | # TODO: parameterize this example
22 | DEVICE_PROPERTY = ("deviceName", "propertyName")
23 | CONFIG_GROUP = "configGroup"
24 | CONFIG_NAME = "State0"
25 | # LC state in native units
26 | LC_STATE = np.random.rand(1)[0] * 10
27 |
28 |
29 | def _get_mmcore_mock():
30 | """Creates a mock for the `pycromanager.Core` object.
31 |
32 | Returns
33 | -------
34 | MagicMock
35 | MMCore mock object
36 | """
37 | mmcore_mock_config = {
38 | "getImage": Mock(return_value=SERIAL_IMAGE),
39 | "getProperty": Mock(return_value=str(LC_STATE)),
40 | }
41 | return MagicMock(**mmcore_mock_config)
42 |
43 |
44 | def _get_snap_manager_mock():
45 | """Creates a mock for the pycromanager remote Snap Live Window Manager object.
46 |
47 | Returns
48 | -------
49 | MagicMock
50 | Mock object for `org.micromanager.internal.SnapLiveManager` via pycromanager
51 | """
52 | sm = MagicMock()
53 | get_snap_mocks = {
54 | "getHeight": Mock(return_value=IMAGE_HEIGHT),
55 | "getWidth": Mock(return_value=IMAGE_WIDTH),
56 | "getRawPixels": Mock(return_value=SERIAL_IMAGE),
57 | }
58 | # TODO: break down these JAVA call stack chains for maintainability
59 | sm.getDisplay.return_value.getDisplayedImages.return_value.get = Mock(
60 | # return image object mock with H, W, and pixel values
61 | return_value=Mock(**get_snap_mocks)
62 | )
63 | sm.getDisplay.return_value.getImagePlus.return_value.getStatistics = Mock(
64 | # return statistics object mock with the attribute "umean"
65 | return_value=Mock(umean=SERIAL_IMAGE.mean())
66 | )
67 | return sm
68 |
69 |
70 | def _is_int(data: ndarray):
71 | """Check if the data type is integer.
72 |
73 | Parameters
74 | ----------
75 | data
76 |
77 | Returns
78 | -------
79 | bool
80 | True if the data type is any integer type.
81 | """
82 | return np.issubdtype(data.dtype, np.integer)
83 |
84 |
85 | def _get_examples(low: float, high: float):
86 | """Generate 4 valid and 4 invalid floating numbers for closed interval [low, high].
87 |
88 | Parameters
89 | ----------
90 | low : float
91 | high : float
92 |
93 | Returns
94 | -------
95 | tuple(1d-array, 1d-array)
96 | valid and invalid values
97 | """
98 | epsilon = np.finfo(float).eps
99 | samples = np.random.rand(4)
100 | valid_values = samples * (high - low) + low + epsilon
101 | invalid_values = np.array(
102 | [
103 | low - samples[0],
104 | low - samples[1],
105 | high + samples[2],
106 | high + samples[3],
107 | ]
108 | )
109 | return valid_values, invalid_values
110 |
111 |
112 | def test_suspend_live_sm():
113 | """Test `recOrder.io.core_functions.suspend_live_sm`."""
114 | snap_manager = _get_snap_manager_mock()
115 | with suspend_live_sm(snap_manager) as sm:
116 | sm.setSuspended.assert_called_once_with(True)
117 | snap_manager.setSuspended.assert_called_with(False)
118 |
119 |
120 | def test_snap_and_get_image():
121 | """Test `recOrder.io.core_functions.snap_and_get_image`."""
122 | sm = _get_snap_manager_mock()
123 | image = snap_and_get_image(sm)
124 | assert _is_int(image), image.dtype
125 | assert image.shape == (IMAGE_HEIGHT, IMAGE_WIDTH), image.shape
126 |
127 |
128 | def test_snap_and_average():
129 | """Test `recOrder.io.core_functions.snap_and_average`."""
130 | sm = _get_snap_manager_mock()
131 | mean = snap_and_average(sm)
132 | np.testing.assert_almost_equal(mean, SERIAL_IMAGE.mean())
133 |
134 |
135 | def _set_lc_test(
136 | tested_func: Callable[[object, Tuple[str, str], float], None],
137 | value_range: Tuple[float, float],
138 | ):
139 | mmc = _get_mmcore_mock()
140 | valid_values, invalid_values = _get_examples(*value_range)
141 | for value in valid_values:
142 | tested_func(mmc, DEVICE_PROPERTY, value)
143 | mmc.setProperty.assert_called_with(
144 | DEVICE_PROPERTY[0], DEVICE_PROPERTY[1], str(value)
145 | )
146 | for value in invalid_values:
147 | with pytest.raises(ValueError):
148 | tested_func(mmc, DEVICE_PROPERTY, value)
149 |
150 |
151 | def test_set_lc_waves():
152 | """Test `recOrder.io.core_functions.set_lc_waves`."""
153 | _set_lc_test(set_lc_waves, (0.001, 1.6))
154 |
155 |
156 | def test_set_lc_voltage():
157 | """Test `recOrder.io.core_functions.set_lc_voltage`."""
158 | _set_lc_test(set_lc_voltage, (0.0, 20.0))
159 |
160 |
161 | def test_set_lc_daq():
162 | """Test `recOrder.io.core_functions.set_lc_daq`."""
163 | _set_lc_test(set_lc_daq, (0.0, 5.0))
164 |
165 |
166 | def test_get_lc():
167 | """Test `recOrder.io.core_functions.get_lc`."""
168 | mmc = _get_mmcore_mock()
169 | state = get_lc(mmc, DEVICE_PROPERTY)
170 | mmc.getProperty.assert_called_once_with(*DEVICE_PROPERTY)
171 | np.testing.assert_almost_equal(state, LC_STATE)
172 |
173 |
174 | def test_define_meadowlark_state():
175 | """Test `recOrder.io.core_functions.define_meadowlark_state`."""
176 | mmc = _get_mmcore_mock()
177 | define_meadowlark_state(mmc, DEVICE_PROPERTY)
178 | mmc.setProperty.assert_called_once_with(*DEVICE_PROPERTY, 0)
179 | mmc.waitForDevice.assert_called_once_with(DEVICE_PROPERTY[0])
180 |
181 |
182 | def test_define_config_state():
183 | """Test `recOrder.io.core_functions.define_config_state`."""
184 | mmc = _get_mmcore_mock()
185 | device_properties = [DEVICE_PROPERTY] * 4
186 | values = _get_examples(0, 10)[0].tolist()
187 | define_config_state(
188 | mmc, CONFIG_GROUP, CONFIG_NAME, device_properties, values
189 | )
190 | expected_calls = [
191 | call(CONFIG_GROUP, CONFIG_NAME, *d, str(v))
192 | for d, v in zip(device_properties, values)
193 | ]
194 | got_calls = mmc.defineConfig.call_args_list
195 | assert got_calls == expected_calls, got_calls
196 |
197 |
198 | def test_set_lc_state():
199 | """Test `recOrder.io.core_functions.set_lc_state`."""
200 | mmc = _get_mmcore_mock()
201 | set_lc_state(mmc, CONFIG_GROUP, CONFIG_NAME)
202 | mmc.setConfig.assert_called_once_with(CONFIG_GROUP, CONFIG_NAME)
203 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_create_empty.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import numpy as np
4 | from iohub.ngff import Position, open_ome_zarr
5 |
6 | from recOrder.cli.utils import create_empty_hcs_zarr
7 |
8 |
9 | def test_create_empty_hcs_zarr(tmp_path):
10 | store_path = tmp_path / Path("test_store.zarr")
11 | position_keys: list[tuple[str]] = [
12 | ("A", "0", "3"),
13 | ("B", "10", "4"),
14 | ]
15 | shape = (1, 2, 1, 1024, 1024)
16 | chunks = (1, 1, 1, 256, 256)
17 | scale = (1, 1, 1, 0.5, 0.5)
18 | channel_names = ["Channel1", "Channel2"]
19 | dtype = np.uint16
20 | plate_metadata = {"test": 2}
21 |
22 | create_empty_hcs_zarr(
23 | store_path,
24 | position_keys,
25 | shape,
26 | chunks,
27 | scale,
28 | channel_names,
29 | dtype,
30 | plate_metadata,
31 | )
32 |
33 | # Verify existence of positions and channels
34 | with open_ome_zarr(store_path, mode="r") as plate:
35 | assert plate.zattrs["test"] == 2
36 | for position_key in position_keys:
37 | position = plate["/".join(position_key)]
38 | assert isinstance(position, Position)
39 | assert position[0].shape == shape
40 |
41 | # Repeat creation should not fail
42 | more_channel_names = ["Channel3"]
43 | create_empty_hcs_zarr(
44 | store_path,
45 | position_keys,
46 | shape,
47 | chunks,
48 | scale,
49 | more_channel_names,
50 | dtype,
51 | )
52 |
53 | # Verify existence of appended channel names
54 | channel_names += more_channel_names
55 | for position_key in position_keys:
56 | position_path = store_path
57 | for element in position_key:
58 | position_path /= element
59 | with open_ome_zarr(position_path, mode="r") as position:
60 | assert position.channel_names == channel_names
61 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_io.py:
--------------------------------------------------------------------------------
1 | import os
2 | from pathlib import Path
3 |
4 | import pytest
5 | import yaml
6 |
7 | from recOrder.cli import settings
8 | from recOrder.io.utils import add_index_to_path, model_to_yaml
9 |
10 |
11 | @pytest.fixture
12 | def model():
13 | # Create a sample model object
14 | return settings.ReconstructionSettings(
15 | birefringence=settings.BirefringenceSettings()
16 | )
17 |
18 |
19 | @pytest.fixture
20 | def yaml_path(tmpdir):
21 | # Create a temporary YAML file path
22 | return os.path.join(tmpdir, "model.yaml")
23 |
24 |
25 | def test_model_to_yaml(model, yaml_path):
26 | # Call the function under test
27 | model_to_yaml(model, yaml_path)
28 |
29 | # Check if the YAML file is created
30 | assert os.path.exists(yaml_path)
31 |
32 | # Load the YAML file and verify its contents
33 | with open(yaml_path, "r") as f:
34 | yaml_data = yaml.safe_load(f)
35 |
36 | # Check if the YAML data is a dictionary
37 | assert isinstance(yaml_data, dict)
38 |
39 | # Check YAML data
40 | assert "input_channel_names" in yaml_data
41 |
42 |
43 | def test_model_to_yaml_invalid_model():
44 | # Create an object that does not have a 'dict()' method
45 | invalid_model = "not a model object"
46 |
47 | # Call the function and expect a TypeError
48 | with pytest.raises(TypeError):
49 | model_to_yaml(invalid_model, "model.yaml")
50 |
51 |
52 | def test_add_index_to_path(tmp_path):
53 | test_cases = [
54 | ("output.txt", "output_0.txt"),
55 | ("output.txt", "output_1.txt"),
56 | ("output.txt", "output_2.txt"),
57 | ("output.png", "output_0.png"),
58 | ("output.png", "output_1.png"),
59 | ("output.png", "output_2.png"),
60 | ("folder", "folder_0"),
61 | ("folder", "folder_1"),
62 | ("folder", "folder_2"),
63 | ]
64 |
65 | for input_path_str, expected_output_str in test_cases:
66 | input_path = tmp_path / Path(input_path_str)
67 | expected_output = tmp_path / Path(expected_output_str)
68 |
69 | output_path = add_index_to_path(input_path)
70 | assert output_path == expected_output
71 |
72 | output_path.touch() # Create a file/folder at the expected output path for testing
73 |
--------------------------------------------------------------------------------
/recOrder/tests/util_tests/test_overlays.py:
--------------------------------------------------------------------------------
1 | import hypothesis.extra.numpy as npst
2 | import hypothesis.strategies as st
3 | import numpy as np
4 | from hypothesis import given
5 | from numpy.typing import NDArray
6 | from numpy.testing import assert_equal
7 |
8 | from recOrder.io.visualization import ret_ori_overlay, ret_ori_phase_overlay
9 |
10 |
11 | @st.composite
12 | def _birefringence(draw):
13 | shape = (2,) + tuple(
14 | draw(st.lists(st.integers(1, 16), min_size=2, max_size=4))
15 | )
16 | dtype = draw(npst.floating_dtypes(sizes=(32, 64)))
17 | bit_width = dtype.itemsize * 8
18 | retardance = draw(
19 | npst.arrays(
20 | dtype,
21 | shape=shape,
22 | elements=st.floats(
23 | min_value=1.0000000168623835e-16,
24 | max_value=50,
25 | exclude_min=True,
26 | width=bit_width,
27 | ),
28 | )
29 | )
30 | orientation = draw(
31 | npst.arrays(
32 | dtype,
33 | shape=shape,
34 | elements=st.floats(
35 | min_value=0,
36 | max_value=dtype.type(np.pi),
37 | exclude_min=True,
38 | exclude_max=True,
39 | width=bit_width,
40 | ),
41 | )
42 | )
43 |
44 | return retardance, orientation
45 |
46 |
47 | @given(briefringence=_birefringence(), jch=st.booleans())
48 | def test_ret_ori_overlay(briefringence: tuple[NDArray, NDArray], jch: bool):
49 | """Test recOrder.io.utils.ret_ori_overlay()"""
50 | retardance, orientation = briefringence
51 | retardance_copy = retardance.copy()
52 | orientation_copy = orientation.copy()
53 | cmap = "JCh" if jch else "HSV"
54 | overlay = ret_ori_overlay(
55 | np.stack((retardance, orientation)),
56 | ret_max=np.percentile(retardance, 99),
57 | cmap=cmap,
58 | )
59 |
60 | overlay2 = ret_ori_phase_overlay(
61 | np.stack((retardance, orientation, retardance)), # dummy phase
62 | )
63 |
64 | # check that the function did not mutate input data
65 | assert_equal(retardance, retardance_copy)
66 | assert_equal(orientation, orientation_copy)
67 | # check output properties
68 | # output contains NaN, pending further investigation
69 | # assert overlay.min() >= 0
70 | # assert overlay.max() <= 1
71 | assert overlay.shape == (3,) + retardance.shape
72 | assert overlay2.shape == (3,) + retardance.shape
73 |
--------------------------------------------------------------------------------
/recOrder/tests/widget_tests/test_dock_widget.py:
--------------------------------------------------------------------------------
1 | from napari.viewer import ViewerModel
2 |
3 | from recOrder.plugin.main_widget import MainWidget
4 |
5 |
6 | def test_dock_widget(make_napari_viewer):
7 | viewer: ViewerModel = make_napari_viewer()
8 | viewer.window.add_dock_widget(MainWidget(viewer))
9 | assert "recOrder" in list(viewer._window._dock_widgets.keys())[0]
10 |
--------------------------------------------------------------------------------
/recOrder/tests/widget_tests/test_sample_contributions.py:
--------------------------------------------------------------------------------
1 | from recOrder.scripts.samples import download_and_unzip
2 |
3 |
4 | def test_download_and_unzip():
5 | p1, p2 = download_and_unzip("target")
6 |
7 | assert p1.exists()
8 | assert p2.exists()
9 |
--------------------------------------------------------------------------------
/setup.cfg:
--------------------------------------------------------------------------------
1 | [metadata]
2 | name = recOrder-napari
3 | author = Computational Microscopy Platform, CZ Biohub
4 | author_email = shalin.mehta@czbiohub.org
5 | url = https://github.com/mehta-lab/recOrder
6 | license = BSD 3-Clause License
7 | description = Computational microscopy toolkit for label-free imaging
8 | long_description = file: README.md
9 | long_description_content_type = text/markdown
10 | classifiers =
11 | License :: OSI Approved :: BSD License
12 | Programming Language :: Python
13 | Programming Language :: Python :: 3 :: Only
14 | Programming Language :: Python :: 3.10
15 | Programming Language :: Python :: 3.11
16 | Topic :: Scientific/Engineering
17 | Topic :: Scientific/Engineering :: Visualization
18 | Topic :: Scientific/Engineering :: Information Analysis
19 | Topic :: Scientific/Engineering :: Bio-Informatics
20 | Topic :: Utilities
21 | Framework :: napari
22 | Operating System :: Microsoft :: Windows
23 | Operating System :: POSIX
24 | Operating System :: Unix
25 | Operating System :: MacOS
26 | project_urls =
27 | Bug Tracker = https://github.com/mehta-lab/recOrder/issues
28 | Documentation = https://github.com/mehta-lab/recOrder/wiki
29 | Source Code = https://github.com/mehta-lab/recOrder/tree/main/recOrder
30 | User Support = https://github.com/mehta-lab/recOrder/issues
31 |
32 | [options]
33 | packages = find:
34 | include_package_data = True
35 | python_requires = >=3.10
36 | setup_requires = setuptools_scm
37 | install_requires =
38 | waveorder==2.2.0rc0
39 | click>=8.0.1
40 | natsort>=7.1.1
41 | colorspacious>=1.1.2
42 | importlib-metadata
43 | iohub==0.1.0
44 | wget>=3.2
45 | psutil
46 | submitit
47 | pydantic==1.10.19
48 | ome-zarr==0.8.3 # unpin when resolved: https://github.com/ome/napari-ome-zarr/issues/111
49 | qtpy
50 | pyqtgraph>=0.12.3
51 |
52 | [options.extras_require]
53 | all =
54 | napari[pyqt6]
55 | napari-ome-zarr>=0.3.2 # drag and drop convenience
56 | pycromanager==0.27.2
57 |
58 | dev =
59 | pytest>=5.0.0
60 | pytest-cov
61 | pytest-qt
62 | tox
63 | pre-commit
64 | black
65 | hypothesis
66 |
67 | [options.package_data]
68 | * = *.yaml
69 |
70 | [options.entry_points]
71 | console_scripts =
72 | recorder = recOrder.cli.main:cli
73 | recOrder = recOrder.cli.main:cli
74 |
75 | napari.manifest =
76 | recOrder = recOrder:napari.yaml
77 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | from setuptools import setup
2 |
3 | setup()
4 |
--------------------------------------------------------------------------------
/tox.ini:
--------------------------------------------------------------------------------
1 | # Modified from from cookiecutter-napari-plugin
2 | # For more information about tox, see https://tox.readthedocs.io/en/latest/
3 | [tox]
4 | envlist = py{310,311}-{linux,macos,windows}
5 | isolated_build=true
6 |
7 | [gh-actions]
8 | python =
9 | 3.10: py310
10 | 3.11: py311
11 |
12 | [gh-actions:env]
13 | PLATFORM =
14 | ubuntu-latest: linux
15 | macos-latest: macos
16 | windows-latest: windows
17 |
18 | [testenv]
19 | platform =
20 | macos: darwin
21 | linux: linux
22 | windows: win32
23 | passenv =
24 | CI
25 | GITHUB_ACTIONS
26 | DISPLAY
27 | XAUTHORITY
28 | NUMPY_EXPERIMENTAL_ARRAY_FUNCTION
29 | PYVISTA_OFF_SCREEN
30 | extras =
31 | dev
32 | all
33 | commands = pytest -v --color=yes --cov=recOrder --cov-report=xml
--------------------------------------------------------------------------------
 19 |
20 | The main command `reconstruct` command is composed of two subcommands: `compute-tf` and `apply-inv-tf`.
21 |
22 | A reconstruction can be performed with a single `reconstruct` call. For example:
23 | ```
24 | recorder reconstruct `
25 | -i ./data.zarr/*/*/* `
26 | -c ./config.yml `
27 | -o ./reconstruction.zarr
28 | ```
29 | Equivalently, a reconstruction can be performed with a `compute-tf` call followed by an `apply-inv-tf` call. For example:
30 | ```
31 | recorder compute-tf `
32 | -i ./data.zarr/0/0/0 `
33 | -c ./config.yml `
34 | -o ./tf.zarr
35 |
36 | recorder apply-inv-tf
37 | -i ./data.zarr/*/*/* `
38 | -t ./tf.zarr `
39 | -c ./config.yml `
40 | -o ./reconstruction.zarr
41 | ```
42 | Computing the transfer function is typically the most expensive part of the reconstruction, so saving a transfer function then applying it to many datasets can save time.
43 |
44 | ## Input options
45 |
46 | The input `-i` flag always accepts a list of inputs, either explicitly e.g. `-i ./data.zarr/A/1/0 ./data.zarr/A/2/0` or through wildcards `-i ./data.zarr/*/*/*`. The positions in a high-content screening `.zarr` store are organized into `/row/col/fov` folders, so `./input.zarr/*/*/*` creates a list of all positions in a dataset.
47 |
48 | The `recorder compute-tf` command accepts a list of inputs, but it only computes the transfer function for the first position in the list. The `apply-inv-tf` command accepts a list of inputs and applies the same transfer function to all of the inputs, which requires that all positions contain arrays with matching TCZYX shapes.
49 |
50 | ## What types of reconstructions are supported?
51 | See `/recOrder/examples/` for a list of example configuration files.
52 |
53 | WIP: This documentation will be expanded for each reconstruction type and parameter.
54 |
--------------------------------------------------------------------------------
/docs/software-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Software Installation Guide
2 |
3 | 1. (Optional but recommended) install [`conda`](https://github.com/conda-forge/miniforge) and create a virtual environment
4 |
5 | ```sh
6 | conda create -y -n recOrder python=3.10
7 | conda activate recOrder
8 | ```
9 |
10 | 2. Install `recOrder-napari`:
11 |
12 | ```sh
13 | pip install recOrder-napari
14 | ```
15 |
16 | 3. To use the GUI: open `napari` with `recOrder-napari`:
17 |
18 | ```sh
19 | napari -w recOrder-napari
20 | ```
21 |
22 | 4. View command-line help by running
23 |
24 | ```sh
25 | recOrder
26 | ```
27 |
28 | 5. To acquire data via Micro-Manager`, follow the [microscope installation guide](./microscope-installation-guide.md).
29 |
--------------------------------------------------------------------------------
/examples/README.md:
--------------------------------------------------------------------------------
1 | # `recOrder` CLI examples
2 |
3 | `recOrder` uses a configuration-file-based command-line inferface (CLI) to
4 | calculate transfer functions and apply these transfer functions to datasets.
5 |
6 | This page demonstrates `recOrder`'s CLI.
7 |
8 | ## Getting started
9 |
10 | ### 1. Check your installation
11 | First, [install `recOrder`](../docs/software-installation-guide.md) and run
12 | ```bash
13 | recOrder
14 | ```
15 | in a shell. If `recOrder` is installed correctly, you will see a usage string and
16 | ```
17 | recOrder: Computational Toolkit for Label-Free Imaging
18 | ```
19 |
20 | ### 2. Download and convert a test dataset
21 | Next, [download the test data from zenodo (47 MB)](https://zenodo.org/record/6983916/files/recOrder_test_data.zip?download=1), and convert a dataset to the latest version of `.zarr` with
22 | ```
23 | cd /path/to/
24 | iohub convert -i /path/to/recOrder_test_data/2022_08_04_recOrder_pytest_20x_04NA/2T_3P_16Z_128Y_256X_Kazansky_1/
25 | -o ./dataset.zarr
26 | ```
27 |
28 | You can view the test dataset with
29 | ```
30 | napari ./dataset.zarr --plugin recOrder-napari
31 | ```
32 |
33 | ### 3. Run a reconstruction
34 | Run an example reconstruction with
35 | ```
36 | recOrder reconstruct ./dataset.zarr/0/0/0 -c /path/to/recOrder/examples/settings/birefringence-and-phase.yml -o ./reconstruction.zarr
37 | ```
38 | then view the reconstruction with
39 | ```
40 | napari ./reconstruction.zarr --plugin recOrder-napari
41 | ```
42 |
43 | Try modifying the configuration file to see how the regularization parameter changes the results.
44 |
45 | ### 4. Parallelize over positions or time points
46 |
47 | TODO: @Ed I think we'll use the same strategy as ```mantis deskew```
48 |
49 | ## FAQ
50 | 1. **Q: Which configuration file should I use?**
51 |
52 | If you are acquiring:
53 |
54 | **3D data with calibrated liquid-crystal polarizers via `recOrder`** use `birefringence.yml`.
55 |
56 | **3D fluorescence data** use `fluorescence.yml`.
57 |
58 | **3D brightfield data** use `phase.yml`.
59 |
60 | **Multi-modal data**, start by reconstructing the individual modaliities, each with a single config file and CLI call. Then combine the reconstructions by ***TODO: @Ziwen do can you help me append to the zarrs to help me fix this? ***
61 |
62 | 2. **Q: Should I use `reconstruction_dimension` = 2 or 3?
63 |
64 | If your downstream processing requires 3D information or if you're unsure, then you should use `reconstruction_dimension = 3`. If your sample is very thin compared to the depth of field of the microscope, if you're in a noise-limited regime, or if your downstream processing requires 2D information, then you should use `reconstruction_dimension = 2`. Empirically, we have found that 2D reconstructions reduce the noise in our reconstructions because it uses 3D information to make a single estimate for each pixel.
65 |
66 | 3. **Q: What regularization parameter should I use?**
67 |
68 | We recommend starting with the defaults then testing over a few orders of magnitude and choosing a result that isn't too noisy or too smooth.
69 |
70 | ### Developers note
71 |
72 | These configuration files are automatically generated when the tests run. See `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
73 |
74 | To keep these settings up to date, run `pytest` locally when `cli/settings.py` changes.
75 |
--------------------------------------------------------------------------------
/examples/birefringence-and-phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 | phase:
18 | transfer_function:
19 | wavelength_illumination: 0.532
20 | yx_pixel_size: 0.325
21 | z_pixel_size: 2.0
22 | z_padding: 0
23 | index_of_refraction_media: 1.3
24 | numerical_aperture_detection: 1.2
25 | numerical_aperture_illumination: 0.5
26 | invert_phase_contrast: false
27 | apply_inverse:
28 | reconstruction_algorithm: Tikhonov
29 | regularization_strength: 0.001
30 | TV_rho_strength: 0.001
31 | TV_iterations: 1
32 |
--------------------------------------------------------------------------------
/examples/birefringence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 |
--------------------------------------------------------------------------------
/examples/fluorescence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - GFP
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | fluorescence:
6 | transfer_function:
7 | yx_pixel_size: 0.325
8 | z_pixel_size: 2.0
9 | z_padding: 0
10 | index_of_refraction_media: 1.3
11 | numerical_aperture_detection: 1.2
12 | wavelength_emission: 0.507
13 | apply_inverse:
14 | reconstruction_algorithm: Tikhonov
15 | regularization_strength: 0.001
16 | TV_rho_strength: 0.001
17 | TV_iterations: 1
18 |
--------------------------------------------------------------------------------
/examples/phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - BF
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | phase:
6 | transfer_function:
7 | wavelength_illumination: 0.532
8 | yx_pixel_size: 0.325
9 | z_pixel_size: 2.0
10 | z_padding: 0
11 | index_of_refraction_media: 1.3
12 | numerical_aperture_detection: 1.2
13 | numerical_aperture_illumination: 0.5
14 | invert_phase_contrast: false
15 | apply_inverse:
16 | reconstruction_algorithm: Tikhonov
17 | regularization_strength: 0.001
18 | TV_rho_strength: 0.001
19 | TV_iterations: 1
20 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = ["setuptools", "wheel", "setuptools_scm"]
3 | build-backend = "setuptools.build_meta"
4 |
5 | [tool.setuptools_scm]
6 | write_to = "recOrder/_version.py"
7 |
8 | [tool.black]
9 | line-length = 79
10 |
11 | [tool.isort]
12 | profile = "black"
13 | line_length = 79
--------------------------------------------------------------------------------
/recOrder/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/acq/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/acq_functions.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import json
3 | import os
4 | import time
5 |
6 | import numpy as np
7 | from iohub import read_micromanager
8 | try:
9 | from pycromanager import Studio
10 | except:pass
11 |
12 | def generate_acq_settings(
13 | mm,
14 | channel_group,
15 | channels=None,
16 | zstart=None,
17 | zend=None,
18 | zstep=None,
19 | save_dir=None,
20 | prefix=None,
21 | keep_shutter_open_channels=False,
22 | keep_shutter_open_slices=False,
23 | ):
24 | """
25 | This function generates a json file specific to the Micro-Manager SequenceSettings.
26 | It has default parameters for a multi-channels z-stack acquisition but does not yet
27 | support multi-position or multi-frame acquisitions.
28 |
29 | This also has default values for QLIPP Acquisition. Can be used as a framework for other types
30 | of acquisitions
31 |
32 | Parameters
33 | ----------
34 | mm: (object) MM Studio API object
35 | scheme: (str) '4-State' or '5-State'
36 | zstart: (float) relative starting position for the z-stack
37 | zend: (float) relative ending position for the z-stack
38 | zstep: (float) step size for the z-stack
39 | save_dir: (str) path to save directory
40 | prefix: (str) name to save the data under
41 |
42 | Returns
43 | -------
44 | settings: (json) json dictionary conforming to MM SequenceSettings
45 | """
46 |
47 | # Get API Objects
48 | am = mm.getAcquisitionManager()
49 | ss = am.getAcquisitionSettings()
50 | app = mm.app()
51 |
52 | # Get current SequenceSettings to modify
53 | original_ss = ss.toJSONStream(ss)
54 | original_json = json.loads(original_ss).copy()
55 |
56 | if zstart:
57 | do_z = True
58 | else:
59 | do_z = False
60 |
61 | # Structure of the channel properties
62 | channel_dict = {
63 | "channelGroup": channel_group,
64 | "config": None,
65 | "exposure": None,
66 | "zOffset": 0,
67 | "doZStack": do_z,
68 | "color": {"value": -16747854, "falpha": 0.0},
69 | "skipFactorFrame": 0,
70 | "useChannel": True if channels else False,
71 | }
72 |
73 | channel_list = None
74 | if channels:
75 | # Append all the channels with their current exposure settings
76 | channel_list = []
77 | for chan in channels:
78 | # todo: think about how to deal with missing exposure
79 | exposure = app.getChannelExposureTime(
80 | channel_group, chan, 10
81 | ) # sets exposure to 10 if not found
82 | channel = channel_dict.copy()
83 | channel["config"] = chan
84 | channel["exposure"] = exposure
85 |
86 | channel_list.append(channel)
87 |
88 | # set other parameters
89 | original_json["numFrames"] = 1
90 | original_json["intervalMs"] = 0
91 | original_json["relativeZSlice"] = True
92 | original_json["slicesFirst"] = True
93 | original_json["timeFirst"] = False
94 | original_json["keepShutterOpenSlices"] = keep_shutter_open_slices
95 | original_json["keepShutterOpenChannels"] = keep_shutter_open_channels
96 | original_json["useAutofocus"] = False
97 | original_json["saveMode"] = "MULTIPAGE_TIFF"
98 | original_json["save"] = True if save_dir else False
99 | original_json["root"] = save_dir if save_dir else ""
100 | original_json["prefix"] = prefix if prefix else "Untitled"
101 | original_json["channels"] = channel_list
102 | original_json["zReference"] = 0.0
103 | original_json["channelGroup"] = channel_group
104 | original_json["usePositionList"] = False
105 | original_json["shouldDisplayImages"] = True
106 | original_json["useSlices"] = do_z
107 | original_json["useFrames"] = False
108 | original_json["useChannels"] = True if channels else False
109 | original_json["slices"] = (
110 | list(np.arange(float(zstart), float(zend + zstep), float(zstep)))
111 | if zstart
112 | else []
113 | )
114 | original_json["sliceZStepUm"] = zstep
115 | original_json["sliceZBottomUm"] = zstart
116 | original_json["sliceZTopUm"] = zend
117 | original_json["acqOrderMode"] = 1
118 |
119 | return original_json
120 |
121 |
122 | def acquire_from_settings(
123 | mm: Studio,
124 | settings: dict,
125 | grab_images: bool = True,
126 | restore_settings: bool = True,
127 | ):
128 | """Function to acquire an MDA acquisition with the native MM MDA Engine.
129 | Assumes single position acquisition.
130 |
131 | Parameters
132 | ----------
133 | mm : Studio
134 | settings : dict
135 | JSON dictionary conforming to MM SequenceSettings
136 | grab_images : bool, optional
137 | return the acquired array, by default True
138 | restore_settings : bool, optional
139 | restore MDA settings before acquisition, by default True
140 |
141 | Returns
142 | -------
143 | NDArray
144 | acquired images
145 | """
146 | am = mm.getAcquisitionManager()
147 | ss = am.getAcquisitionSettings()
148 |
149 | ss_new = ss.fromJSONStream(json.dumps(settings))
150 | am.runAcquisitionWithSettings(ss_new, True)
151 |
152 | time.sleep(3)
153 |
154 | if restore_settings:
155 | am.setAcquisitionSettings(ss)
156 |
157 | # TODO: speed improvements in reading the data with pycromanager acquisition?
158 | if grab_images:
159 | # get the most recent acquisition if multiple
160 | path = os.path.join(settings["root"], settings["prefix"])
161 | files = glob.glob(path + "*")
162 | index = max([int(x.split(path + "_")[1]) for x in files])
163 |
164 | reader = read_micromanager(
165 | path + f"_{index}", data_type="ometiff", extract_data=True
166 | )
167 |
168 | return reader.get_array(0)
169 |
--------------------------------------------------------------------------------
/recOrder/calib/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/calib/__init__.py
--------------------------------------------------------------------------------
/recOrder/cli/apply_inverse_models.py:
--------------------------------------------------------------------------------
1 | """
2 | This module converts recOrder's reconstructions into waveorder calls
3 | """
4 |
5 | import numpy as np
6 | import torch
7 | from waveorder.models import (
8 | inplane_oriented_thick_pol3d,
9 | isotropic_fluorescent_thick_3d,
10 | isotropic_thin_3d,
11 | phase_thick_3d,
12 | )
13 |
14 |
15 | def radians_to_nanometers(retardance_rad, wavelength_illumination_um):
16 | """
17 | waveorder returns retardance in radians, while recorder displays and saves
18 | retardance in nanometers. This function converts from radians to nanometers
19 | using the illumination wavelength (which is internally handled in um
20 | in recOrder).
21 | """
22 | return retardance_rad * wavelength_illumination_um * 1e3 / (2 * np.pi)
23 |
24 |
25 | def birefringence(
26 | czyx_data,
27 | cyx_no_sample_data,
28 | wavelength_illumination,
29 | recon_dim,
30 | biref_inverse_dict,
31 | transfer_function_dataset,
32 | ):
33 | # Load transfer function
34 | intensity_to_stokes_matrix = torch.tensor(
35 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
36 | )
37 |
38 | # Apply reconstruction
39 | # (retardance, orientation, transmittance, depolarization)
40 | reconstructed_parameters = (
41 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
42 | czyx_data,
43 | intensity_to_stokes_matrix,
44 | cyx_no_sample_data=cyx_no_sample_data,
45 | project_stokes_to_2d=(recon_dim == 2),
46 | **biref_inverse_dict,
47 | )
48 | )
49 |
50 | # Convert retardance
51 | retardance = radians_to_nanometers(
52 | reconstructed_parameters[0], wavelength_illumination
53 | )
54 |
55 | return torch.stack((retardance,) + reconstructed_parameters[1:])
56 |
57 |
58 | def phase(
59 | czyx_data,
60 | recon_dim,
61 | settings_phase,
62 | transfer_function_dataset,
63 | ):
64 | # [phase only, 2]
65 | if recon_dim == 2:
66 | # Load transfer functions
67 | absorption_transfer_function = torch.tensor(
68 | transfer_function_dataset["absorption_transfer_function"][0, 0]
69 | )
70 | phase_transfer_function = torch.tensor(
71 | transfer_function_dataset["phase_transfer_function"][0, 0]
72 | )
73 |
74 | # Apply
75 | (
76 | _,
77 | output,
78 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
79 | czyx_data[0],
80 | absorption_transfer_function,
81 | phase_transfer_function,
82 | **settings_phase.apply_inverse.dict(),
83 | )
84 |
85 | # [phase only, 3]
86 | elif recon_dim == 3:
87 | # Load transfer functions
88 | real_potential_transfer_function = torch.tensor(
89 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
90 | )
91 | imaginary_potential_transfer_function = torch.tensor(
92 | transfer_function_dataset["imaginary_potential_transfer_function"][
93 | 0, 0
94 | ]
95 | )
96 |
97 | # Apply
98 | output = phase_thick_3d.apply_inverse_transfer_function(
99 | czyx_data[0],
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | z_padding=settings_phase.transfer_function.z_padding,
103 | **settings_phase.apply_inverse.dict(),
104 | )
105 |
106 | # Pad to CZYX
107 | while output.ndim != 4:
108 | output = torch.unsqueeze(output, 0)
109 |
110 | return output
111 |
112 |
113 | def birefringence_and_phase(
114 | czyx_data,
115 | cyx_no_sample_data,
116 | wavelength_illumination,
117 | recon_dim,
118 | biref_inverse_dict,
119 | settings_phase,
120 | transfer_function_dataset,
121 | ):
122 | # Load birefringence transfer function
123 | intensity_to_stokes_matrix = torch.tensor(
124 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
125 | )
126 |
127 | # [biref and phase, 2]
128 | if recon_dim == 2:
129 | # Load phase transfer functions
130 | absorption_transfer_function = torch.tensor(
131 | transfer_function_dataset["absorption_transfer_function"][0, 0]
132 | )
133 | phase_transfer_function = torch.tensor(
134 | transfer_function_dataset["phase_transfer_function"][0, 0]
135 | )
136 |
137 | # Apply
138 | reconstructed_parameters_2d = (
139 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
140 | czyx_data,
141 | intensity_to_stokes_matrix,
142 | cyx_no_sample_data=cyx_no_sample_data,
143 | project_stokes_to_2d=True,
144 | **biref_inverse_dict,
145 | )
146 | )
147 |
148 | reconstructed_parameters_3d = (
149 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
150 | czyx_data,
151 | intensity_to_stokes_matrix,
152 | cyx_no_sample_data=cyx_no_sample_data,
153 | project_stokes_to_2d=False,
154 | **biref_inverse_dict,
155 | )
156 | )
157 |
158 | brightfield_3d = reconstructed_parameters_3d[2]
159 |
160 | (
161 | _,
162 | yx_phase,
163 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
164 | brightfield_3d,
165 | absorption_transfer_function,
166 | phase_transfer_function,
167 | **settings_phase.apply_inverse.dict(),
168 | )
169 |
170 | # Convert retardance
171 | retardance = radians_to_nanometers(
172 | reconstructed_parameters_2d[0], wavelength_illumination
173 | )
174 |
175 | output = torch.stack(
176 | (retardance,)
177 | + reconstructed_parameters_2d[1:]
178 | + (torch.unsqueeze(yx_phase, 0),)
179 | ) # CZYX
180 |
181 | # [biref and phase, 3]
182 | elif recon_dim == 3:
183 | # Load phase transfer functions
184 | intensity_to_stokes_matrix = torch.tensor(
185 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
186 | )
187 | # Load transfer functions
188 | real_potential_transfer_function = torch.tensor(
189 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
190 | )
191 | imaginary_potential_transfer_function = torch.tensor(
192 | transfer_function_dataset["imaginary_potential_transfer_function"][
193 | 0, 0
194 | ]
195 | )
196 |
197 | # Apply
198 | reconstructed_parameters_3d = (
199 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
200 | czyx_data,
201 | intensity_to_stokes_matrix,
202 | cyx_no_sample_data=cyx_no_sample_data,
203 | project_stokes_to_2d=False,
204 | **biref_inverse_dict,
205 | )
206 | )
207 |
208 | brightfield_3d = reconstructed_parameters_3d[2]
209 |
210 | zyx_phase = phase_thick_3d.apply_inverse_transfer_function(
211 | brightfield_3d,
212 | real_potential_transfer_function,
213 | imaginary_potential_transfer_function,
214 | z_padding=settings_phase.transfer_function.z_padding,
215 | **settings_phase.apply_inverse.dict(),
216 | )
217 |
218 | # Convert retardance
219 | retardance = radians_to_nanometers(
220 | reconstructed_parameters_3d[0], wavelength_illumination
221 | )
222 |
223 | # Save
224 | output = torch.stack(
225 | (retardance,) + reconstructed_parameters_3d[1:] + (zyx_phase,)
226 | )
227 | return output
228 |
229 |
230 | def fluorescence(
231 | czyx_data, recon_dim, settings_fluorescence, transfer_function_dataset
232 | ):
233 | # [fluo, 2]
234 | if recon_dim == 2:
235 | raise NotImplementedError
236 | # [fluo, 3]
237 | elif recon_dim == 3:
238 | # Load transfer functions
239 | optical_transfer_function = torch.tensor(
240 | transfer_function_dataset["optical_transfer_function"][0, 0]
241 | )
242 |
243 | # Apply
244 | output = (
245 | isotropic_fluorescent_thick_3d.apply_inverse_transfer_function(
246 | czyx_data[0],
247 | optical_transfer_function,
248 | settings_fluorescence.transfer_function.z_padding,
249 | **settings_fluorescence.apply_inverse.dict(),
250 | )
251 | )
252 | # Pad to CZYX
253 | while output.ndim != 4:
254 | output = torch.unsqueeze(output, 0)
255 |
256 | return output
257 |
--------------------------------------------------------------------------------
/recOrder/cli/compute_transfer_function.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 | import numpy as np
5 | from iohub.ngff import open_ome_zarr, Position
6 | from waveorder.models import (
7 | inplane_oriented_thick_pol3d,
8 | isotropic_fluorescent_thick_3d,
9 | isotropic_thin_3d,
10 | phase_thick_3d,
11 | )
12 |
13 | from recOrder.cli.parsing import (
14 | config_filepath,
15 | input_position_dirpaths,
16 | output_dirpath,
17 | )
18 | from recOrder.cli.printing import echo_headline, echo_settings
19 | from recOrder.cli.settings import ReconstructionSettings
20 | from recOrder.io import utils
21 |
22 |
23 | def generate_and_save_birefringence_transfer_function(settings, dataset):
24 | """Generates and saves the birefringence transfer function to the dataset, based on the settings.
25 |
26 | Parameters
27 | ----------
28 | settings: ReconstructionSettings
29 | dataset: NGFF Node
30 | The dataset that will be updated.
31 | """
32 | echo_headline("Generating birefringence transfer function with settings:")
33 | echo_settings(settings.birefringence.transfer_function)
34 |
35 | # Calculate transfer functions
36 | intensity_to_stokes_matrix = (
37 | inplane_oriented_thick_pol3d.calculate_transfer_function(
38 | scheme=str(len(settings.input_channel_names)) + "-State",
39 | **settings.birefringence.transfer_function.dict(),
40 | )
41 | )
42 | # Save
43 | dataset[
44 | "intensity_to_stokes_matrix"
45 | ] = intensity_to_stokes_matrix.cpu().numpy()[None, None, None, ...]
46 |
47 |
48 | def generate_and_save_phase_transfer_function(
49 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
50 | ):
51 | """Generates and saves the phase transfer function to the dataset, based on the settings.
52 |
53 | Parameters
54 | ----------
55 | settings: ReconstructionSettings
56 | dataset: Position
57 | The dataset that will be updated.
58 | zyx_shape : tuple
59 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
60 | """
61 | echo_headline("Generating phase transfer function with settings:")
62 | echo_settings(settings.phase.transfer_function)
63 |
64 | if settings.reconstruction_dimension == 2:
65 | # Convert zyx_shape and z_pixel_size into yx_shape and z_position_list
66 | settings_dict = settings.phase.transfer_function.dict()
67 | settings_dict["yx_shape"] = [zyx_shape[1], zyx_shape[2]]
68 | settings_dict["z_position_list"] = list(
69 | -(np.arange(zyx_shape[0]) - zyx_shape[0] // 2)
70 | * settings_dict["z_pixel_size"]
71 | )
72 |
73 | # Remove unused parameters
74 | settings_dict.pop("z_pixel_size")
75 | settings_dict.pop("z_padding")
76 |
77 | # Calculate transfer functions
78 | (
79 | absorption_transfer_function,
80 | phase_transfer_function,
81 | ) = isotropic_thin_3d.calculate_transfer_function(
82 | **settings_dict,
83 | )
84 |
85 | # Save
86 | dataset.create_image(
87 | "absorption_transfer_function",
88 | absorption_transfer_function.cpu().numpy()[None, None, ...],
89 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
90 | )
91 | dataset.create_image(
92 | "phase_transfer_function",
93 | phase_transfer_function.cpu().numpy()[None, None, ...],
94 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
95 | )
96 |
97 | elif settings.reconstruction_dimension == 3:
98 | # Calculate transfer functions
99 | (
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | ) = phase_thick_3d.calculate_transfer_function(
103 | zyx_shape=zyx_shape,
104 | **settings.phase.transfer_function.dict(),
105 | )
106 | # Save
107 | dataset.create_image(
108 | "real_potential_transfer_function",
109 | real_potential_transfer_function.cpu().numpy()[None, None, ...],
110 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
111 | )
112 | dataset.create_image(
113 | "imaginary_potential_transfer_function",
114 | imaginary_potential_transfer_function.cpu().numpy()[
115 | None, None, ...
116 | ],
117 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
118 | )
119 |
120 |
121 | def generate_and_save_fluorescence_transfer_function(
122 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
123 | ):
124 | """Generates and saves the fluorescence transfer function to the dataset, based on the settings.
125 |
126 | Parameters
127 | ----------
128 | settings: ReconstructionSettings
129 | dataset: Position
130 | The dataset that will be updated.
131 | zyx_shape : tuple
132 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
133 | """
134 | echo_headline("Generating fluorescence transfer function with settings:")
135 | echo_settings(settings.fluorescence.transfer_function)
136 |
137 | if settings.reconstruction_dimension == 2:
138 | raise NotImplementedError
139 | elif settings.reconstruction_dimension == 3:
140 | # Calculate transfer functions
141 | optical_transfer_function = (
142 | isotropic_fluorescent_thick_3d.calculate_transfer_function(
143 | zyx_shape=zyx_shape,
144 | **settings.fluorescence.transfer_function.dict(),
145 | )
146 | )
147 | # Save
148 | dataset.create_image(
149 | "optical_transfer_function",
150 | optical_transfer_function.cpu().numpy()[None, None, ...],
151 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
152 | )
153 |
154 |
155 | def compute_transfer_function_cli(
156 | input_position_dirpath: Path, config_filepath: Path, output_dirpath: Path
157 | ) -> None:
158 | """CLI command to compute the transfer function given a configuration file path
159 | and a desired output path.
160 | """
161 |
162 | # Load config file
163 | settings = utils.yaml_to_model(config_filepath, ReconstructionSettings)
164 |
165 | echo_headline(
166 | f"Generating transfer functions and storing in {output_dirpath}\n"
167 | )
168 |
169 | # Read shape from input dataset
170 | input_dataset = open_ome_zarr(
171 | input_position_dirpath, layout="fov", mode="r"
172 | )
173 | zyx_shape = input_dataset.data.shape[
174 | 2:
175 | ] # only loads a single position "0"
176 |
177 | # Check input channel names
178 | if not set(settings.input_channel_names).issubset(
179 | input_dataset.channel_names
180 | ):
181 | raise ValueError(
182 | f"Each of the input_channel_names = {settings.input_channel_names} in {config_filepath} must appear in the dataset {input_position_dirpaths[0]} which currently contains channel_names = {input_dataset.channel_names}."
183 | )
184 |
185 | # Prepare output dataset
186 | output_dataset = open_ome_zarr(
187 | output_dirpath, layout="fov", mode="w", channel_names=["None"]
188 | )
189 |
190 | # Pass settings to appropriate calculate_transfer_function and save
191 | if settings.birefringence is not None:
192 | generate_and_save_birefringence_transfer_function(
193 | settings, output_dataset
194 | )
195 | if settings.phase is not None:
196 | generate_and_save_phase_transfer_function(
197 | settings, output_dataset, zyx_shape
198 | )
199 | if settings.fluorescence is not None:
200 | generate_and_save_fluorescence_transfer_function(
201 | settings, output_dataset, zyx_shape
202 | )
203 |
204 | # Write settings to metadata
205 | output_dataset.zattrs["settings"] = settings.dict()
206 |
207 | echo_headline(f"Closing {output_dirpath}\n")
208 | output_dataset.close()
209 |
210 | echo_headline(
211 | f"Recreate this transfer function with:\n$ recorder compute-tf {input_position_dirpaths} -c {config_filepath} -o {output_dirpath}"
212 | )
213 |
214 |
215 | @click.command()
216 | @input_position_dirpaths()
217 | @config_filepath()
218 | @output_dirpath()
219 | def compute_tf(
220 | input_position_dirpaths: list[Path],
221 | config_filepath: Path,
222 | output_dirpath: Path,
223 | ) -> None:
224 | """
225 | Compute a transfer function using a dataset and configuration file.
226 |
227 | Calculates the transfer function based on the shape of the first position
228 | in the list `input-position-dirpaths`.
229 |
230 | See /examples for example configuration files.
231 |
232 | >> recorder compute-tf -i ./input.zarr/0/0/0 -c ./examples/birefringence.yml -o ./transfer_function.zarr
233 | """
234 | compute_transfer_function_cli(
235 | input_position_dirpaths[0], config_filepath, output_dirpath
236 | )
237 |
--------------------------------------------------------------------------------
/recOrder/cli/gui_widget.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import click
3 |
4 | try:
5 | from recOrder.plugin import tab_recon
6 | except:pass
7 |
8 | try:
9 | from qtpy.QtWidgets import QApplication, QWidget, QVBoxLayout, QStyle
10 | except:pass
11 |
12 | try:
13 | import qdarktheme
14 | except:pass
15 |
16 | PLUGIN_NAME = "recOrder: Computational Toolkit for Label-Free Imaging"
17 | PLUGIN_ICON = "🔬"
18 |
19 | @click.command()
20 | def gui():
21 | """GUI for recOrder: Computational Toolkit for Label-Free Imaging"""
22 |
23 | app = QApplication(sys.argv)
24 | app.setStyle("Fusion") # Other options: "Fusion", "Windows", "macOS", "WindowsVista"
25 | try:
26 | qdarktheme.setup_theme("dark")
27 | except:pass
28 | window = MainWindow()
29 | window.setWindowTitle(PLUGIN_ICON + " " + PLUGIN_NAME + " " + PLUGIN_ICON)
30 |
31 | pixmapi = getattr(QStyle.StandardPixmap, "SP_TitleBarMenuButton")
32 | icon = app.style().standardIcon(pixmapi)
33 | window.setWindowIcon(icon)
34 |
35 | window.show()
36 | sys.exit(app.exec())
37 |
38 | class MainWindow(QWidget):
39 | def __init__(self):
40 | super().__init__()
41 | recon_tab = tab_recon.Ui_ReconTab_Form(stand_alone=True)
42 | layout = QVBoxLayout()
43 | self.setLayout(layout)
44 | layout.addWidget(recon_tab.recon_tab_mainScrollArea)
45 |
46 | if __name__ == "__main__":
47 | gui()
48 |
--------------------------------------------------------------------------------
/recOrder/cli/jobs_mgmt.py:
--------------------------------------------------------------------------------
1 | import os, json
2 | from pathlib import Path
3 | import socket
4 | import submitit
5 | import threading, time
6 |
7 | DIR_PATH = os.path.dirname(os.path.realpath(__file__))
8 | FILE_PATH = os.path.join(DIR_PATH, "main.py")
9 |
10 | SERVER_PORT = 8089 # Choose an available port
11 | JOBS_TIMEOUT = 5 # 5 mins
12 | SERVER_uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
13 |
14 | class JobsManagement():
15 |
16 | def __init__(self, *args, **kwargs):
17 | self.clientsocket = None
18 | self.uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
19 | self.DATA_QUEUE = []
20 |
21 | def check_for_jobID_File(self, jobID, logs_path, extension="out"):
22 |
23 | if Path(logs_path).exists():
24 | files = os.listdir(logs_path)
25 | try:
26 | for file in files:
27 | if file.endswith(extension):
28 | if jobID in file:
29 | file_path = os.path.join(logs_path, file)
30 | f = open(file_path, "r")
31 | txt = f.read()
32 | f.close()
33 | return txt
34 | except Exception as exc:
35 | print(exc.args)
36 | return ""
37 |
38 | def set_shorter_timeout(self):
39 | self.clientsocket.settimeout(30)
40 |
41 | def start_client(self):
42 | try:
43 | self.clientsocket = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
44 | self.clientsocket.settimeout(300)
45 | self.clientsocket.connect(('localhost', SERVER_PORT))
46 | self.clientsocket.settimeout(None)
47 |
48 | thread = threading.Thread(target=self.stop_client)
49 | thread.start()
50 | except Exception as exc:
51 | print(exc.args)
52 |
53 | # The stopClient() is called right with the startClient() but does not stop
54 | # and essentially is a wait thread listening and is triggered by either a
55 | # connection or timeout. Based on condition triggered by user, reconstruction
56 | # completion or errors the end goal is to close the socket connection which
57 | # would let the CLI exit. I could break it down to 2 parts but the idea was to
58 | # keep the clientsocket.close() call within one method to make it easier to follow.
59 | def stop_client(self):
60 | try:
61 | time.sleep(2)
62 | while True:
63 | time.sleep(1)
64 | buf = ""
65 | try:
66 | buf = self.clientsocket.recv(1024)
67 | except:
68 | pass
69 | if len(buf) > 0:
70 | if b"\n" in buf:
71 | dataList = buf.split(b"\n")
72 | for data in dataList:
73 | if len(data)>0:
74 | decoded_string = data.decode()
75 | json_str = str(decoded_string)
76 | json_obj = json.loads(json_str)
77 | u_idx = json_obj["uID"]
78 | job_idx = str(json_obj["jID"])
79 | cmd = json_obj["command"]
80 | if cmd == "clientRelease":
81 | if self.has_submitted_job(u_idx, job_idx):
82 | self.clientsocket.close()
83 | break
84 | if cmd == "cancel":
85 | if self.has_submitted_job(u_idx, job_idx):
86 | try:
87 | job = self.uIDsjobIDs[u_idx][job_idx]
88 | job.cancel()
89 | except Exception as exc:
90 | pass # possibility of throwing an exception based on diff. OS
91 | forDeletions = []
92 | for uID in self.uIDsjobIDs.keys():
93 | for jID in self.uIDsjobIDs[uID].keys():
94 | job = self.uIDsjobIDs[uID][jID]
95 | if job.done():
96 | forDeletions.append((uID, jID))
97 | for idx in range(len(forDeletions)):
98 | del self.uIDsjobIDs[forDeletions[idx][0]][forDeletions[idx][1]]
99 | forDeletions = []
100 | for uID in self.uIDsjobIDs.keys():
101 | if len(self.uIDsjobIDs[uID].keys()) == 0:
102 | forDeletions.append(uID)
103 | for idx in range(len(forDeletions)):
104 | del self.uIDsjobIDs[forDeletions[idx]]
105 | if len(self.uIDsjobIDs.keys()) == 0:
106 | self.clientsocket.close()
107 | break
108 | except Exception as exc:
109 | self.clientsocket.close()
110 | print(exc.args)
111 |
112 | def check_all_ExpJobs_completion(self, uID):
113 | if uID in SERVER_uIDsjobIDs.keys():
114 | for jobEntry in SERVER_uIDsjobIDs[uID].keys():
115 | job:submitit.Job = SERVER_uIDsjobIDs[uID][jobEntry]["job"]
116 | jobBool = SERVER_uIDsjobIDs[uID][jobEntry]["bool"]
117 | if job is not None and job.done() == False:
118 | return False
119 | if jobBool == False:
120 | return False
121 | return True
122 |
123 | def put_Job_completion_in_list(self, job_bool, uID: str, jID: str, mode="client"):
124 | if uID in SERVER_uIDsjobIDs.keys():
125 | if jID in SERVER_uIDsjobIDs[uID].keys():
126 | SERVER_uIDsjobIDs[uID][jID]["bool"] = job_bool
127 |
128 | def add_data(self, data):
129 | self.DATA_QUEUE.append(data)
130 |
131 | def send_data_thread(self):
132 | thread = threading.Thread(target=self.send_data)
133 | thread.start()
134 |
135 | def send_data(self):
136 | data = "".join(self.DATA_QUEUE)
137 | self.clientsocket.send(data.encode())
138 | self.DATA_QUEUE = []
139 |
140 | def put_Job_in_list(self, job, uID: str, jID: str, well:str, log_folder_path:str="", mode="client"):
141 | try:
142 | well = str(well)
143 | jID = str(jID)
144 | if ".zarr" in well:
145 | wells = well.split(".zarr")
146 | well = wells[1].replace("\\","-").replace("/","-")[1:]
147 | if mode == "client":
148 | if uID not in self.uIDsjobIDs.keys():
149 | self.uIDsjobIDs[uID] = {}
150 | self.uIDsjobIDs[uID][jID] = job
151 | else:
152 | if jID not in self.uIDsjobIDs[uID].keys():
153 | self.uIDsjobIDs[uID][jID] = job
154 | json_obj = {uID:{"jID": str(jID), "pos": well, "log": log_folder_path}}
155 | json_str = json.dumps(json_obj)+"\n"
156 | self.add_data(json_str)
157 | else:
158 | # from server side jobs object entry is a None object
159 | # this will be later checked as completion boolean for a ExpID which might
160 | # have several Jobs associated with it
161 | if uID not in SERVER_uIDsjobIDs.keys():
162 | SERVER_uIDsjobIDs[uID] = {}
163 | SERVER_uIDsjobIDs[uID][jID] = {}
164 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
165 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
166 | else:
167 | SERVER_uIDsjobIDs[uID][jID] = {}
168 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
169 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
170 | except Exception as exc:
171 | print(exc.args)
172 |
173 | def has_submitted_job(self, uID: str, jID: str, mode="client")->bool:
174 | jID = str(jID)
175 | if mode == "client":
176 | if uID in self.uIDsjobIDs.keys():
177 | if jID in self.uIDsjobIDs[uID].keys():
178 | return True
179 | return False
180 | else:
181 | if uID in SERVER_uIDsjobIDs.keys():
182 | if jID in SERVER_uIDsjobIDs[uID].keys():
183 | return True
184 | return False
185 |
--------------------------------------------------------------------------------
/recOrder/cli/main.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 | from recOrder.cli.apply_inverse_transfer_function import apply_inv_tf
4 | from recOrder.cli.compute_transfer_function import compute_tf
5 | from recOrder.cli.reconstruct import reconstruct
6 | try:
7 | from recOrder.cli.gui_widget import gui
8 | except:pass
9 |
10 |
11 | CONTEXT = {"help_option_names": ["-h", "--help"]}
12 |
13 |
14 | # `recorder -h` will show subcommands in the order they are added
15 | class NaturalOrderGroup(click.Group):
16 | def list_commands(self, ctx):
17 | return self.commands.keys()
18 |
19 |
20 | @click.group(context_settings=CONTEXT, cls=NaturalOrderGroup)
21 | def cli():
22 | """\033[92mrecOrder: Computational Toolkit for Label-Free Imaging\033[0m\n"""
23 |
24 |
25 | cli.add_command(reconstruct)
26 | cli.add_command(compute_tf)
27 | cli.add_command(apply_inv_tf)
28 | try:
29 | cli.add_command(gui)
30 | except:pass
31 |
32 | if __name__ == "__main__":
33 | cli()
34 |
--------------------------------------------------------------------------------
/recOrder/cli/monitor.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import time
4 | import numpy as np
5 | import shutil
6 | import submitit
7 | import sys
8 |
9 |
10 | def _move_cursor_up(n_lines, do_print=True):
11 | if do_print:
12 | sys.stdout.write("\033[F" * n_lines)
13 |
14 |
15 | def _print_status(jobs, position_dirpaths, elapsed_list, print_indices=None, do_print=True):
16 |
17 | columns = [15, 30, 40, 50]
18 |
19 | # header
20 | if do_print:
21 | sys.stdout.write(
22 | "\033[K" # clear line
23 | "\033[96mID" # cyan
24 | f"\033[{columns[0]}G WELL "
25 | f"\033[{columns[1]}G STATUS "
26 | f"\033[{columns[2]}G NODE "
27 | f"\033[{columns[2]}G ELAPSED\n"
28 | )
29 |
30 | if print_indices is None:
31 | print_indices = range(len(jobs))
32 |
33 | complete_count = 0
34 |
35 | for i, (job, position_dirpath) in enumerate(zip(jobs, position_dirpaths)):
36 | try:
37 | node_name = job.get_info()["NodeList"] # slowest, so do this first
38 | except:
39 | node_name = "SUBMITTED"
40 |
41 | if job.state == "COMPLETED":
42 | color = "\033[32m" # green
43 | complete_count += 1
44 | elif job.state == "RUNNING":
45 | color = "\033[93m" # yellow
46 | elapsed_list[i] += 1 # inexact timing
47 | else:
48 | color = "\033[91m" # red
49 |
50 | if i in print_indices:
51 | if do_print:
52 | sys.stdout.write(
53 | f"\033[K" # clear line
54 | f"{color}{job.job_id}"
55 | f"\033[{columns[0]}G {'/'.join(position_dirpath.parts[-3:])}"
56 | f"\033[{columns[1]}G {job.state}"
57 | f"\033[{columns[2]}G {node_name}"
58 | f"\033[{columns[3]}G {elapsed_list[i]} s\n"
59 | )
60 | sys.stdout.flush()
61 | if do_print:
62 | print(
63 | f"\033[32m{complete_count}/{len(jobs)} jobs complete. "
64 | "
19 |
20 | The main command `reconstruct` command is composed of two subcommands: `compute-tf` and `apply-inv-tf`.
21 |
22 | A reconstruction can be performed with a single `reconstruct` call. For example:
23 | ```
24 | recorder reconstruct `
25 | -i ./data.zarr/*/*/* `
26 | -c ./config.yml `
27 | -o ./reconstruction.zarr
28 | ```
29 | Equivalently, a reconstruction can be performed with a `compute-tf` call followed by an `apply-inv-tf` call. For example:
30 | ```
31 | recorder compute-tf `
32 | -i ./data.zarr/0/0/0 `
33 | -c ./config.yml `
34 | -o ./tf.zarr
35 |
36 | recorder apply-inv-tf
37 | -i ./data.zarr/*/*/* `
38 | -t ./tf.zarr `
39 | -c ./config.yml `
40 | -o ./reconstruction.zarr
41 | ```
42 | Computing the transfer function is typically the most expensive part of the reconstruction, so saving a transfer function then applying it to many datasets can save time.
43 |
44 | ## Input options
45 |
46 | The input `-i` flag always accepts a list of inputs, either explicitly e.g. `-i ./data.zarr/A/1/0 ./data.zarr/A/2/0` or through wildcards `-i ./data.zarr/*/*/*`. The positions in a high-content screening `.zarr` store are organized into `/row/col/fov` folders, so `./input.zarr/*/*/*` creates a list of all positions in a dataset.
47 |
48 | The `recorder compute-tf` command accepts a list of inputs, but it only computes the transfer function for the first position in the list. The `apply-inv-tf` command accepts a list of inputs and applies the same transfer function to all of the inputs, which requires that all positions contain arrays with matching TCZYX shapes.
49 |
50 | ## What types of reconstructions are supported?
51 | See `/recOrder/examples/` for a list of example configuration files.
52 |
53 | WIP: This documentation will be expanded for each reconstruction type and parameter.
54 |
--------------------------------------------------------------------------------
/docs/software-installation-guide.md:
--------------------------------------------------------------------------------
1 | # Software Installation Guide
2 |
3 | 1. (Optional but recommended) install [`conda`](https://github.com/conda-forge/miniforge) and create a virtual environment
4 |
5 | ```sh
6 | conda create -y -n recOrder python=3.10
7 | conda activate recOrder
8 | ```
9 |
10 | 2. Install `recOrder-napari`:
11 |
12 | ```sh
13 | pip install recOrder-napari
14 | ```
15 |
16 | 3. To use the GUI: open `napari` with `recOrder-napari`:
17 |
18 | ```sh
19 | napari -w recOrder-napari
20 | ```
21 |
22 | 4. View command-line help by running
23 |
24 | ```sh
25 | recOrder
26 | ```
27 |
28 | 5. To acquire data via Micro-Manager`, follow the [microscope installation guide](./microscope-installation-guide.md).
29 |
--------------------------------------------------------------------------------
/examples/README.md:
--------------------------------------------------------------------------------
1 | # `recOrder` CLI examples
2 |
3 | `recOrder` uses a configuration-file-based command-line inferface (CLI) to
4 | calculate transfer functions and apply these transfer functions to datasets.
5 |
6 | This page demonstrates `recOrder`'s CLI.
7 |
8 | ## Getting started
9 |
10 | ### 1. Check your installation
11 | First, [install `recOrder`](../docs/software-installation-guide.md) and run
12 | ```bash
13 | recOrder
14 | ```
15 | in a shell. If `recOrder` is installed correctly, you will see a usage string and
16 | ```
17 | recOrder: Computational Toolkit for Label-Free Imaging
18 | ```
19 |
20 | ### 2. Download and convert a test dataset
21 | Next, [download the test data from zenodo (47 MB)](https://zenodo.org/record/6983916/files/recOrder_test_data.zip?download=1), and convert a dataset to the latest version of `.zarr` with
22 | ```
23 | cd /path/to/
24 | iohub convert -i /path/to/recOrder_test_data/2022_08_04_recOrder_pytest_20x_04NA/2T_3P_16Z_128Y_256X_Kazansky_1/
25 | -o ./dataset.zarr
26 | ```
27 |
28 | You can view the test dataset with
29 | ```
30 | napari ./dataset.zarr --plugin recOrder-napari
31 | ```
32 |
33 | ### 3. Run a reconstruction
34 | Run an example reconstruction with
35 | ```
36 | recOrder reconstruct ./dataset.zarr/0/0/0 -c /path/to/recOrder/examples/settings/birefringence-and-phase.yml -o ./reconstruction.zarr
37 | ```
38 | then view the reconstruction with
39 | ```
40 | napari ./reconstruction.zarr --plugin recOrder-napari
41 | ```
42 |
43 | Try modifying the configuration file to see how the regularization parameter changes the results.
44 |
45 | ### 4. Parallelize over positions or time points
46 |
47 | TODO: @Ed I think we'll use the same strategy as ```mantis deskew```
48 |
49 | ## FAQ
50 | 1. **Q: Which configuration file should I use?**
51 |
52 | If you are acquiring:
53 |
54 | **3D data with calibrated liquid-crystal polarizers via `recOrder`** use `birefringence.yml`.
55 |
56 | **3D fluorescence data** use `fluorescence.yml`.
57 |
58 | **3D brightfield data** use `phase.yml`.
59 |
60 | **Multi-modal data**, start by reconstructing the individual modaliities, each with a single config file and CLI call. Then combine the reconstructions by ***TODO: @Ziwen do can you help me append to the zarrs to help me fix this? ***
61 |
62 | 2. **Q: Should I use `reconstruction_dimension` = 2 or 3?
63 |
64 | If your downstream processing requires 3D information or if you're unsure, then you should use `reconstruction_dimension = 3`. If your sample is very thin compared to the depth of field of the microscope, if you're in a noise-limited regime, or if your downstream processing requires 2D information, then you should use `reconstruction_dimension = 2`. Empirically, we have found that 2D reconstructions reduce the noise in our reconstructions because it uses 3D information to make a single estimate for each pixel.
65 |
66 | 3. **Q: What regularization parameter should I use?**
67 |
68 | We recommend starting with the defaults then testing over a few orders of magnitude and choosing a result that isn't too noisy or too smooth.
69 |
70 | ### Developers note
71 |
72 | These configuration files are automatically generated when the tests run. See `/tests/cli_tests/test_settings.py` - `test_generate_example_settings`.
73 |
74 | To keep these settings up to date, run `pytest` locally when `cli/settings.py` changes.
75 |
--------------------------------------------------------------------------------
/examples/birefringence-and-phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 | phase:
18 | transfer_function:
19 | wavelength_illumination: 0.532
20 | yx_pixel_size: 0.325
21 | z_pixel_size: 2.0
22 | z_padding: 0
23 | index_of_refraction_media: 1.3
24 | numerical_aperture_detection: 1.2
25 | numerical_aperture_illumination: 0.5
26 | invert_phase_contrast: false
27 | apply_inverse:
28 | reconstruction_algorithm: Tikhonov
29 | regularization_strength: 0.001
30 | TV_rho_strength: 0.001
31 | TV_iterations: 1
32 |
--------------------------------------------------------------------------------
/examples/birefringence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - State0
3 | - State1
4 | - State2
5 | - State3
6 | time_indices: all
7 | reconstruction_dimension: 3
8 | birefringence:
9 | transfer_function:
10 | swing: 0.1
11 | apply_inverse:
12 | wavelength_illumination: 0.532
13 | background_path: ''
14 | remove_estimated_background: false
15 | flip_orientation: false
16 | rotate_orientation: false
17 |
--------------------------------------------------------------------------------
/examples/fluorescence.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - GFP
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | fluorescence:
6 | transfer_function:
7 | yx_pixel_size: 0.325
8 | z_pixel_size: 2.0
9 | z_padding: 0
10 | index_of_refraction_media: 1.3
11 | numerical_aperture_detection: 1.2
12 | wavelength_emission: 0.507
13 | apply_inverse:
14 | reconstruction_algorithm: Tikhonov
15 | regularization_strength: 0.001
16 | TV_rho_strength: 0.001
17 | TV_iterations: 1
18 |
--------------------------------------------------------------------------------
/examples/phase.yml:
--------------------------------------------------------------------------------
1 | input_channel_names:
2 | - BF
3 | time_indices: all
4 | reconstruction_dimension: 3
5 | phase:
6 | transfer_function:
7 | wavelength_illumination: 0.532
8 | yx_pixel_size: 0.325
9 | z_pixel_size: 2.0
10 | z_padding: 0
11 | index_of_refraction_media: 1.3
12 | numerical_aperture_detection: 1.2
13 | numerical_aperture_illumination: 0.5
14 | invert_phase_contrast: false
15 | apply_inverse:
16 | reconstruction_algorithm: Tikhonov
17 | regularization_strength: 0.001
18 | TV_rho_strength: 0.001
19 | TV_iterations: 1
20 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = ["setuptools", "wheel", "setuptools_scm"]
3 | build-backend = "setuptools.build_meta"
4 |
5 | [tool.setuptools_scm]
6 | write_to = "recOrder/_version.py"
7 |
8 | [tool.black]
9 | line-length = 79
10 |
11 | [tool.isort]
12 | profile = "black"
13 | line_length = 79
--------------------------------------------------------------------------------
/recOrder/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/acq/__init__.py
--------------------------------------------------------------------------------
/recOrder/acq/acq_functions.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import json
3 | import os
4 | import time
5 |
6 | import numpy as np
7 | from iohub import read_micromanager
8 | try:
9 | from pycromanager import Studio
10 | except:pass
11 |
12 | def generate_acq_settings(
13 | mm,
14 | channel_group,
15 | channels=None,
16 | zstart=None,
17 | zend=None,
18 | zstep=None,
19 | save_dir=None,
20 | prefix=None,
21 | keep_shutter_open_channels=False,
22 | keep_shutter_open_slices=False,
23 | ):
24 | """
25 | This function generates a json file specific to the Micro-Manager SequenceSettings.
26 | It has default parameters for a multi-channels z-stack acquisition but does not yet
27 | support multi-position or multi-frame acquisitions.
28 |
29 | This also has default values for QLIPP Acquisition. Can be used as a framework for other types
30 | of acquisitions
31 |
32 | Parameters
33 | ----------
34 | mm: (object) MM Studio API object
35 | scheme: (str) '4-State' or '5-State'
36 | zstart: (float) relative starting position for the z-stack
37 | zend: (float) relative ending position for the z-stack
38 | zstep: (float) step size for the z-stack
39 | save_dir: (str) path to save directory
40 | prefix: (str) name to save the data under
41 |
42 | Returns
43 | -------
44 | settings: (json) json dictionary conforming to MM SequenceSettings
45 | """
46 |
47 | # Get API Objects
48 | am = mm.getAcquisitionManager()
49 | ss = am.getAcquisitionSettings()
50 | app = mm.app()
51 |
52 | # Get current SequenceSettings to modify
53 | original_ss = ss.toJSONStream(ss)
54 | original_json = json.loads(original_ss).copy()
55 |
56 | if zstart:
57 | do_z = True
58 | else:
59 | do_z = False
60 |
61 | # Structure of the channel properties
62 | channel_dict = {
63 | "channelGroup": channel_group,
64 | "config": None,
65 | "exposure": None,
66 | "zOffset": 0,
67 | "doZStack": do_z,
68 | "color": {"value": -16747854, "falpha": 0.0},
69 | "skipFactorFrame": 0,
70 | "useChannel": True if channels else False,
71 | }
72 |
73 | channel_list = None
74 | if channels:
75 | # Append all the channels with their current exposure settings
76 | channel_list = []
77 | for chan in channels:
78 | # todo: think about how to deal with missing exposure
79 | exposure = app.getChannelExposureTime(
80 | channel_group, chan, 10
81 | ) # sets exposure to 10 if not found
82 | channel = channel_dict.copy()
83 | channel["config"] = chan
84 | channel["exposure"] = exposure
85 |
86 | channel_list.append(channel)
87 |
88 | # set other parameters
89 | original_json["numFrames"] = 1
90 | original_json["intervalMs"] = 0
91 | original_json["relativeZSlice"] = True
92 | original_json["slicesFirst"] = True
93 | original_json["timeFirst"] = False
94 | original_json["keepShutterOpenSlices"] = keep_shutter_open_slices
95 | original_json["keepShutterOpenChannels"] = keep_shutter_open_channels
96 | original_json["useAutofocus"] = False
97 | original_json["saveMode"] = "MULTIPAGE_TIFF"
98 | original_json["save"] = True if save_dir else False
99 | original_json["root"] = save_dir if save_dir else ""
100 | original_json["prefix"] = prefix if prefix else "Untitled"
101 | original_json["channels"] = channel_list
102 | original_json["zReference"] = 0.0
103 | original_json["channelGroup"] = channel_group
104 | original_json["usePositionList"] = False
105 | original_json["shouldDisplayImages"] = True
106 | original_json["useSlices"] = do_z
107 | original_json["useFrames"] = False
108 | original_json["useChannels"] = True if channels else False
109 | original_json["slices"] = (
110 | list(np.arange(float(zstart), float(zend + zstep), float(zstep)))
111 | if zstart
112 | else []
113 | )
114 | original_json["sliceZStepUm"] = zstep
115 | original_json["sliceZBottomUm"] = zstart
116 | original_json["sliceZTopUm"] = zend
117 | original_json["acqOrderMode"] = 1
118 |
119 | return original_json
120 |
121 |
122 | def acquire_from_settings(
123 | mm: Studio,
124 | settings: dict,
125 | grab_images: bool = True,
126 | restore_settings: bool = True,
127 | ):
128 | """Function to acquire an MDA acquisition with the native MM MDA Engine.
129 | Assumes single position acquisition.
130 |
131 | Parameters
132 | ----------
133 | mm : Studio
134 | settings : dict
135 | JSON dictionary conforming to MM SequenceSettings
136 | grab_images : bool, optional
137 | return the acquired array, by default True
138 | restore_settings : bool, optional
139 | restore MDA settings before acquisition, by default True
140 |
141 | Returns
142 | -------
143 | NDArray
144 | acquired images
145 | """
146 | am = mm.getAcquisitionManager()
147 | ss = am.getAcquisitionSettings()
148 |
149 | ss_new = ss.fromJSONStream(json.dumps(settings))
150 | am.runAcquisitionWithSettings(ss_new, True)
151 |
152 | time.sleep(3)
153 |
154 | if restore_settings:
155 | am.setAcquisitionSettings(ss)
156 |
157 | # TODO: speed improvements in reading the data with pycromanager acquisition?
158 | if grab_images:
159 | # get the most recent acquisition if multiple
160 | path = os.path.join(settings["root"], settings["prefix"])
161 | files = glob.glob(path + "*")
162 | index = max([int(x.split(path + "_")[1]) for x in files])
163 |
164 | reader = read_micromanager(
165 | path + f"_{index}", data_type="ometiff", extract_data=True
166 | )
167 |
168 | return reader.get_array(0)
169 |
--------------------------------------------------------------------------------
/recOrder/calib/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/mehta-lab/recOrder/c4fed09c0ce0d0fa259951c5940d4ca4f5ef4f5d/recOrder/calib/__init__.py
--------------------------------------------------------------------------------
/recOrder/cli/apply_inverse_models.py:
--------------------------------------------------------------------------------
1 | """
2 | This module converts recOrder's reconstructions into waveorder calls
3 | """
4 |
5 | import numpy as np
6 | import torch
7 | from waveorder.models import (
8 | inplane_oriented_thick_pol3d,
9 | isotropic_fluorescent_thick_3d,
10 | isotropic_thin_3d,
11 | phase_thick_3d,
12 | )
13 |
14 |
15 | def radians_to_nanometers(retardance_rad, wavelength_illumination_um):
16 | """
17 | waveorder returns retardance in radians, while recorder displays and saves
18 | retardance in nanometers. This function converts from radians to nanometers
19 | using the illumination wavelength (which is internally handled in um
20 | in recOrder).
21 | """
22 | return retardance_rad * wavelength_illumination_um * 1e3 / (2 * np.pi)
23 |
24 |
25 | def birefringence(
26 | czyx_data,
27 | cyx_no_sample_data,
28 | wavelength_illumination,
29 | recon_dim,
30 | biref_inverse_dict,
31 | transfer_function_dataset,
32 | ):
33 | # Load transfer function
34 | intensity_to_stokes_matrix = torch.tensor(
35 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
36 | )
37 |
38 | # Apply reconstruction
39 | # (retardance, orientation, transmittance, depolarization)
40 | reconstructed_parameters = (
41 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
42 | czyx_data,
43 | intensity_to_stokes_matrix,
44 | cyx_no_sample_data=cyx_no_sample_data,
45 | project_stokes_to_2d=(recon_dim == 2),
46 | **biref_inverse_dict,
47 | )
48 | )
49 |
50 | # Convert retardance
51 | retardance = radians_to_nanometers(
52 | reconstructed_parameters[0], wavelength_illumination
53 | )
54 |
55 | return torch.stack((retardance,) + reconstructed_parameters[1:])
56 |
57 |
58 | def phase(
59 | czyx_data,
60 | recon_dim,
61 | settings_phase,
62 | transfer_function_dataset,
63 | ):
64 | # [phase only, 2]
65 | if recon_dim == 2:
66 | # Load transfer functions
67 | absorption_transfer_function = torch.tensor(
68 | transfer_function_dataset["absorption_transfer_function"][0, 0]
69 | )
70 | phase_transfer_function = torch.tensor(
71 | transfer_function_dataset["phase_transfer_function"][0, 0]
72 | )
73 |
74 | # Apply
75 | (
76 | _,
77 | output,
78 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
79 | czyx_data[0],
80 | absorption_transfer_function,
81 | phase_transfer_function,
82 | **settings_phase.apply_inverse.dict(),
83 | )
84 |
85 | # [phase only, 3]
86 | elif recon_dim == 3:
87 | # Load transfer functions
88 | real_potential_transfer_function = torch.tensor(
89 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
90 | )
91 | imaginary_potential_transfer_function = torch.tensor(
92 | transfer_function_dataset["imaginary_potential_transfer_function"][
93 | 0, 0
94 | ]
95 | )
96 |
97 | # Apply
98 | output = phase_thick_3d.apply_inverse_transfer_function(
99 | czyx_data[0],
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | z_padding=settings_phase.transfer_function.z_padding,
103 | **settings_phase.apply_inverse.dict(),
104 | )
105 |
106 | # Pad to CZYX
107 | while output.ndim != 4:
108 | output = torch.unsqueeze(output, 0)
109 |
110 | return output
111 |
112 |
113 | def birefringence_and_phase(
114 | czyx_data,
115 | cyx_no_sample_data,
116 | wavelength_illumination,
117 | recon_dim,
118 | biref_inverse_dict,
119 | settings_phase,
120 | transfer_function_dataset,
121 | ):
122 | # Load birefringence transfer function
123 | intensity_to_stokes_matrix = torch.tensor(
124 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
125 | )
126 |
127 | # [biref and phase, 2]
128 | if recon_dim == 2:
129 | # Load phase transfer functions
130 | absorption_transfer_function = torch.tensor(
131 | transfer_function_dataset["absorption_transfer_function"][0, 0]
132 | )
133 | phase_transfer_function = torch.tensor(
134 | transfer_function_dataset["phase_transfer_function"][0, 0]
135 | )
136 |
137 | # Apply
138 | reconstructed_parameters_2d = (
139 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
140 | czyx_data,
141 | intensity_to_stokes_matrix,
142 | cyx_no_sample_data=cyx_no_sample_data,
143 | project_stokes_to_2d=True,
144 | **biref_inverse_dict,
145 | )
146 | )
147 |
148 | reconstructed_parameters_3d = (
149 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
150 | czyx_data,
151 | intensity_to_stokes_matrix,
152 | cyx_no_sample_data=cyx_no_sample_data,
153 | project_stokes_to_2d=False,
154 | **biref_inverse_dict,
155 | )
156 | )
157 |
158 | brightfield_3d = reconstructed_parameters_3d[2]
159 |
160 | (
161 | _,
162 | yx_phase,
163 | ) = isotropic_thin_3d.apply_inverse_transfer_function(
164 | brightfield_3d,
165 | absorption_transfer_function,
166 | phase_transfer_function,
167 | **settings_phase.apply_inverse.dict(),
168 | )
169 |
170 | # Convert retardance
171 | retardance = radians_to_nanometers(
172 | reconstructed_parameters_2d[0], wavelength_illumination
173 | )
174 |
175 | output = torch.stack(
176 | (retardance,)
177 | + reconstructed_parameters_2d[1:]
178 | + (torch.unsqueeze(yx_phase, 0),)
179 | ) # CZYX
180 |
181 | # [biref and phase, 3]
182 | elif recon_dim == 3:
183 | # Load phase transfer functions
184 | intensity_to_stokes_matrix = torch.tensor(
185 | transfer_function_dataset["intensity_to_stokes_matrix"][0, 0, 0]
186 | )
187 | # Load transfer functions
188 | real_potential_transfer_function = torch.tensor(
189 | transfer_function_dataset["real_potential_transfer_function"][0, 0]
190 | )
191 | imaginary_potential_transfer_function = torch.tensor(
192 | transfer_function_dataset["imaginary_potential_transfer_function"][
193 | 0, 0
194 | ]
195 | )
196 |
197 | # Apply
198 | reconstructed_parameters_3d = (
199 | inplane_oriented_thick_pol3d.apply_inverse_transfer_function(
200 | czyx_data,
201 | intensity_to_stokes_matrix,
202 | cyx_no_sample_data=cyx_no_sample_data,
203 | project_stokes_to_2d=False,
204 | **biref_inverse_dict,
205 | )
206 | )
207 |
208 | brightfield_3d = reconstructed_parameters_3d[2]
209 |
210 | zyx_phase = phase_thick_3d.apply_inverse_transfer_function(
211 | brightfield_3d,
212 | real_potential_transfer_function,
213 | imaginary_potential_transfer_function,
214 | z_padding=settings_phase.transfer_function.z_padding,
215 | **settings_phase.apply_inverse.dict(),
216 | )
217 |
218 | # Convert retardance
219 | retardance = radians_to_nanometers(
220 | reconstructed_parameters_3d[0], wavelength_illumination
221 | )
222 |
223 | # Save
224 | output = torch.stack(
225 | (retardance,) + reconstructed_parameters_3d[1:] + (zyx_phase,)
226 | )
227 | return output
228 |
229 |
230 | def fluorescence(
231 | czyx_data, recon_dim, settings_fluorescence, transfer_function_dataset
232 | ):
233 | # [fluo, 2]
234 | if recon_dim == 2:
235 | raise NotImplementedError
236 | # [fluo, 3]
237 | elif recon_dim == 3:
238 | # Load transfer functions
239 | optical_transfer_function = torch.tensor(
240 | transfer_function_dataset["optical_transfer_function"][0, 0]
241 | )
242 |
243 | # Apply
244 | output = (
245 | isotropic_fluorescent_thick_3d.apply_inverse_transfer_function(
246 | czyx_data[0],
247 | optical_transfer_function,
248 | settings_fluorescence.transfer_function.z_padding,
249 | **settings_fluorescence.apply_inverse.dict(),
250 | )
251 | )
252 | # Pad to CZYX
253 | while output.ndim != 4:
254 | output = torch.unsqueeze(output, 0)
255 |
256 | return output
257 |
--------------------------------------------------------------------------------
/recOrder/cli/compute_transfer_function.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import click
4 | import numpy as np
5 | from iohub.ngff import open_ome_zarr, Position
6 | from waveorder.models import (
7 | inplane_oriented_thick_pol3d,
8 | isotropic_fluorescent_thick_3d,
9 | isotropic_thin_3d,
10 | phase_thick_3d,
11 | )
12 |
13 | from recOrder.cli.parsing import (
14 | config_filepath,
15 | input_position_dirpaths,
16 | output_dirpath,
17 | )
18 | from recOrder.cli.printing import echo_headline, echo_settings
19 | from recOrder.cli.settings import ReconstructionSettings
20 | from recOrder.io import utils
21 |
22 |
23 | def generate_and_save_birefringence_transfer_function(settings, dataset):
24 | """Generates and saves the birefringence transfer function to the dataset, based on the settings.
25 |
26 | Parameters
27 | ----------
28 | settings: ReconstructionSettings
29 | dataset: NGFF Node
30 | The dataset that will be updated.
31 | """
32 | echo_headline("Generating birefringence transfer function with settings:")
33 | echo_settings(settings.birefringence.transfer_function)
34 |
35 | # Calculate transfer functions
36 | intensity_to_stokes_matrix = (
37 | inplane_oriented_thick_pol3d.calculate_transfer_function(
38 | scheme=str(len(settings.input_channel_names)) + "-State",
39 | **settings.birefringence.transfer_function.dict(),
40 | )
41 | )
42 | # Save
43 | dataset[
44 | "intensity_to_stokes_matrix"
45 | ] = intensity_to_stokes_matrix.cpu().numpy()[None, None, None, ...]
46 |
47 |
48 | def generate_and_save_phase_transfer_function(
49 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
50 | ):
51 | """Generates and saves the phase transfer function to the dataset, based on the settings.
52 |
53 | Parameters
54 | ----------
55 | settings: ReconstructionSettings
56 | dataset: Position
57 | The dataset that will be updated.
58 | zyx_shape : tuple
59 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
60 | """
61 | echo_headline("Generating phase transfer function with settings:")
62 | echo_settings(settings.phase.transfer_function)
63 |
64 | if settings.reconstruction_dimension == 2:
65 | # Convert zyx_shape and z_pixel_size into yx_shape and z_position_list
66 | settings_dict = settings.phase.transfer_function.dict()
67 | settings_dict["yx_shape"] = [zyx_shape[1], zyx_shape[2]]
68 | settings_dict["z_position_list"] = list(
69 | -(np.arange(zyx_shape[0]) - zyx_shape[0] // 2)
70 | * settings_dict["z_pixel_size"]
71 | )
72 |
73 | # Remove unused parameters
74 | settings_dict.pop("z_pixel_size")
75 | settings_dict.pop("z_padding")
76 |
77 | # Calculate transfer functions
78 | (
79 | absorption_transfer_function,
80 | phase_transfer_function,
81 | ) = isotropic_thin_3d.calculate_transfer_function(
82 | **settings_dict,
83 | )
84 |
85 | # Save
86 | dataset.create_image(
87 | "absorption_transfer_function",
88 | absorption_transfer_function.cpu().numpy()[None, None, ...],
89 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
90 | )
91 | dataset.create_image(
92 | "phase_transfer_function",
93 | phase_transfer_function.cpu().numpy()[None, None, ...],
94 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
95 | )
96 |
97 | elif settings.reconstruction_dimension == 3:
98 | # Calculate transfer functions
99 | (
100 | real_potential_transfer_function,
101 | imaginary_potential_transfer_function,
102 | ) = phase_thick_3d.calculate_transfer_function(
103 | zyx_shape=zyx_shape,
104 | **settings.phase.transfer_function.dict(),
105 | )
106 | # Save
107 | dataset.create_image(
108 | "real_potential_transfer_function",
109 | real_potential_transfer_function.cpu().numpy()[None, None, ...],
110 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
111 | )
112 | dataset.create_image(
113 | "imaginary_potential_transfer_function",
114 | imaginary_potential_transfer_function.cpu().numpy()[
115 | None, None, ...
116 | ],
117 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
118 | )
119 |
120 |
121 | def generate_and_save_fluorescence_transfer_function(
122 | settings: ReconstructionSettings, dataset: Position, zyx_shape: tuple
123 | ):
124 | """Generates and saves the fluorescence transfer function to the dataset, based on the settings.
125 |
126 | Parameters
127 | ----------
128 | settings: ReconstructionSettings
129 | dataset: Position
130 | The dataset that will be updated.
131 | zyx_shape : tuple
132 | A tuple of integers specifying the input data's shape in (Z, Y, X) order
133 | """
134 | echo_headline("Generating fluorescence transfer function with settings:")
135 | echo_settings(settings.fluorescence.transfer_function)
136 |
137 | if settings.reconstruction_dimension == 2:
138 | raise NotImplementedError
139 | elif settings.reconstruction_dimension == 3:
140 | # Calculate transfer functions
141 | optical_transfer_function = (
142 | isotropic_fluorescent_thick_3d.calculate_transfer_function(
143 | zyx_shape=zyx_shape,
144 | **settings.fluorescence.transfer_function.dict(),
145 | )
146 | )
147 | # Save
148 | dataset.create_image(
149 | "optical_transfer_function",
150 | optical_transfer_function.cpu().numpy()[None, None, ...],
151 | chunks=(1, 1, 1, zyx_shape[1], zyx_shape[2]),
152 | )
153 |
154 |
155 | def compute_transfer_function_cli(
156 | input_position_dirpath: Path, config_filepath: Path, output_dirpath: Path
157 | ) -> None:
158 | """CLI command to compute the transfer function given a configuration file path
159 | and a desired output path.
160 | """
161 |
162 | # Load config file
163 | settings = utils.yaml_to_model(config_filepath, ReconstructionSettings)
164 |
165 | echo_headline(
166 | f"Generating transfer functions and storing in {output_dirpath}\n"
167 | )
168 |
169 | # Read shape from input dataset
170 | input_dataset = open_ome_zarr(
171 | input_position_dirpath, layout="fov", mode="r"
172 | )
173 | zyx_shape = input_dataset.data.shape[
174 | 2:
175 | ] # only loads a single position "0"
176 |
177 | # Check input channel names
178 | if not set(settings.input_channel_names).issubset(
179 | input_dataset.channel_names
180 | ):
181 | raise ValueError(
182 | f"Each of the input_channel_names = {settings.input_channel_names} in {config_filepath} must appear in the dataset {input_position_dirpaths[0]} which currently contains channel_names = {input_dataset.channel_names}."
183 | )
184 |
185 | # Prepare output dataset
186 | output_dataset = open_ome_zarr(
187 | output_dirpath, layout="fov", mode="w", channel_names=["None"]
188 | )
189 |
190 | # Pass settings to appropriate calculate_transfer_function and save
191 | if settings.birefringence is not None:
192 | generate_and_save_birefringence_transfer_function(
193 | settings, output_dataset
194 | )
195 | if settings.phase is not None:
196 | generate_and_save_phase_transfer_function(
197 | settings, output_dataset, zyx_shape
198 | )
199 | if settings.fluorescence is not None:
200 | generate_and_save_fluorescence_transfer_function(
201 | settings, output_dataset, zyx_shape
202 | )
203 |
204 | # Write settings to metadata
205 | output_dataset.zattrs["settings"] = settings.dict()
206 |
207 | echo_headline(f"Closing {output_dirpath}\n")
208 | output_dataset.close()
209 |
210 | echo_headline(
211 | f"Recreate this transfer function with:\n$ recorder compute-tf {input_position_dirpaths} -c {config_filepath} -o {output_dirpath}"
212 | )
213 |
214 |
215 | @click.command()
216 | @input_position_dirpaths()
217 | @config_filepath()
218 | @output_dirpath()
219 | def compute_tf(
220 | input_position_dirpaths: list[Path],
221 | config_filepath: Path,
222 | output_dirpath: Path,
223 | ) -> None:
224 | """
225 | Compute a transfer function using a dataset and configuration file.
226 |
227 | Calculates the transfer function based on the shape of the first position
228 | in the list `input-position-dirpaths`.
229 |
230 | See /examples for example configuration files.
231 |
232 | >> recorder compute-tf -i ./input.zarr/0/0/0 -c ./examples/birefringence.yml -o ./transfer_function.zarr
233 | """
234 | compute_transfer_function_cli(
235 | input_position_dirpaths[0], config_filepath, output_dirpath
236 | )
237 |
--------------------------------------------------------------------------------
/recOrder/cli/gui_widget.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import click
3 |
4 | try:
5 | from recOrder.plugin import tab_recon
6 | except:pass
7 |
8 | try:
9 | from qtpy.QtWidgets import QApplication, QWidget, QVBoxLayout, QStyle
10 | except:pass
11 |
12 | try:
13 | import qdarktheme
14 | except:pass
15 |
16 | PLUGIN_NAME = "recOrder: Computational Toolkit for Label-Free Imaging"
17 | PLUGIN_ICON = "🔬"
18 |
19 | @click.command()
20 | def gui():
21 | """GUI for recOrder: Computational Toolkit for Label-Free Imaging"""
22 |
23 | app = QApplication(sys.argv)
24 | app.setStyle("Fusion") # Other options: "Fusion", "Windows", "macOS", "WindowsVista"
25 | try:
26 | qdarktheme.setup_theme("dark")
27 | except:pass
28 | window = MainWindow()
29 | window.setWindowTitle(PLUGIN_ICON + " " + PLUGIN_NAME + " " + PLUGIN_ICON)
30 |
31 | pixmapi = getattr(QStyle.StandardPixmap, "SP_TitleBarMenuButton")
32 | icon = app.style().standardIcon(pixmapi)
33 | window.setWindowIcon(icon)
34 |
35 | window.show()
36 | sys.exit(app.exec())
37 |
38 | class MainWindow(QWidget):
39 | def __init__(self):
40 | super().__init__()
41 | recon_tab = tab_recon.Ui_ReconTab_Form(stand_alone=True)
42 | layout = QVBoxLayout()
43 | self.setLayout(layout)
44 | layout.addWidget(recon_tab.recon_tab_mainScrollArea)
45 |
46 | if __name__ == "__main__":
47 | gui()
48 |
--------------------------------------------------------------------------------
/recOrder/cli/jobs_mgmt.py:
--------------------------------------------------------------------------------
1 | import os, json
2 | from pathlib import Path
3 | import socket
4 | import submitit
5 | import threading, time
6 |
7 | DIR_PATH = os.path.dirname(os.path.realpath(__file__))
8 | FILE_PATH = os.path.join(DIR_PATH, "main.py")
9 |
10 | SERVER_PORT = 8089 # Choose an available port
11 | JOBS_TIMEOUT = 5 # 5 mins
12 | SERVER_uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
13 |
14 | class JobsManagement():
15 |
16 | def __init__(self, *args, **kwargs):
17 | self.clientsocket = None
18 | self.uIDsjobIDs = {} # uIDsjobIDs[uid][jid] = job
19 | self.DATA_QUEUE = []
20 |
21 | def check_for_jobID_File(self, jobID, logs_path, extension="out"):
22 |
23 | if Path(logs_path).exists():
24 | files = os.listdir(logs_path)
25 | try:
26 | for file in files:
27 | if file.endswith(extension):
28 | if jobID in file:
29 | file_path = os.path.join(logs_path, file)
30 | f = open(file_path, "r")
31 | txt = f.read()
32 | f.close()
33 | return txt
34 | except Exception as exc:
35 | print(exc.args)
36 | return ""
37 |
38 | def set_shorter_timeout(self):
39 | self.clientsocket.settimeout(30)
40 |
41 | def start_client(self):
42 | try:
43 | self.clientsocket = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
44 | self.clientsocket.settimeout(300)
45 | self.clientsocket.connect(('localhost', SERVER_PORT))
46 | self.clientsocket.settimeout(None)
47 |
48 | thread = threading.Thread(target=self.stop_client)
49 | thread.start()
50 | except Exception as exc:
51 | print(exc.args)
52 |
53 | # The stopClient() is called right with the startClient() but does not stop
54 | # and essentially is a wait thread listening and is triggered by either a
55 | # connection or timeout. Based on condition triggered by user, reconstruction
56 | # completion or errors the end goal is to close the socket connection which
57 | # would let the CLI exit. I could break it down to 2 parts but the idea was to
58 | # keep the clientsocket.close() call within one method to make it easier to follow.
59 | def stop_client(self):
60 | try:
61 | time.sleep(2)
62 | while True:
63 | time.sleep(1)
64 | buf = ""
65 | try:
66 | buf = self.clientsocket.recv(1024)
67 | except:
68 | pass
69 | if len(buf) > 0:
70 | if b"\n" in buf:
71 | dataList = buf.split(b"\n")
72 | for data in dataList:
73 | if len(data)>0:
74 | decoded_string = data.decode()
75 | json_str = str(decoded_string)
76 | json_obj = json.loads(json_str)
77 | u_idx = json_obj["uID"]
78 | job_idx = str(json_obj["jID"])
79 | cmd = json_obj["command"]
80 | if cmd == "clientRelease":
81 | if self.has_submitted_job(u_idx, job_idx):
82 | self.clientsocket.close()
83 | break
84 | if cmd == "cancel":
85 | if self.has_submitted_job(u_idx, job_idx):
86 | try:
87 | job = self.uIDsjobIDs[u_idx][job_idx]
88 | job.cancel()
89 | except Exception as exc:
90 | pass # possibility of throwing an exception based on diff. OS
91 | forDeletions = []
92 | for uID in self.uIDsjobIDs.keys():
93 | for jID in self.uIDsjobIDs[uID].keys():
94 | job = self.uIDsjobIDs[uID][jID]
95 | if job.done():
96 | forDeletions.append((uID, jID))
97 | for idx in range(len(forDeletions)):
98 | del self.uIDsjobIDs[forDeletions[idx][0]][forDeletions[idx][1]]
99 | forDeletions = []
100 | for uID in self.uIDsjobIDs.keys():
101 | if len(self.uIDsjobIDs[uID].keys()) == 0:
102 | forDeletions.append(uID)
103 | for idx in range(len(forDeletions)):
104 | del self.uIDsjobIDs[forDeletions[idx]]
105 | if len(self.uIDsjobIDs.keys()) == 0:
106 | self.clientsocket.close()
107 | break
108 | except Exception as exc:
109 | self.clientsocket.close()
110 | print(exc.args)
111 |
112 | def check_all_ExpJobs_completion(self, uID):
113 | if uID in SERVER_uIDsjobIDs.keys():
114 | for jobEntry in SERVER_uIDsjobIDs[uID].keys():
115 | job:submitit.Job = SERVER_uIDsjobIDs[uID][jobEntry]["job"]
116 | jobBool = SERVER_uIDsjobIDs[uID][jobEntry]["bool"]
117 | if job is not None and job.done() == False:
118 | return False
119 | if jobBool == False:
120 | return False
121 | return True
122 |
123 | def put_Job_completion_in_list(self, job_bool, uID: str, jID: str, mode="client"):
124 | if uID in SERVER_uIDsjobIDs.keys():
125 | if jID in SERVER_uIDsjobIDs[uID].keys():
126 | SERVER_uIDsjobIDs[uID][jID]["bool"] = job_bool
127 |
128 | def add_data(self, data):
129 | self.DATA_QUEUE.append(data)
130 |
131 | def send_data_thread(self):
132 | thread = threading.Thread(target=self.send_data)
133 | thread.start()
134 |
135 | def send_data(self):
136 | data = "".join(self.DATA_QUEUE)
137 | self.clientsocket.send(data.encode())
138 | self.DATA_QUEUE = []
139 |
140 | def put_Job_in_list(self, job, uID: str, jID: str, well:str, log_folder_path:str="", mode="client"):
141 | try:
142 | well = str(well)
143 | jID = str(jID)
144 | if ".zarr" in well:
145 | wells = well.split(".zarr")
146 | well = wells[1].replace("\\","-").replace("/","-")[1:]
147 | if mode == "client":
148 | if uID not in self.uIDsjobIDs.keys():
149 | self.uIDsjobIDs[uID] = {}
150 | self.uIDsjobIDs[uID][jID] = job
151 | else:
152 | if jID not in self.uIDsjobIDs[uID].keys():
153 | self.uIDsjobIDs[uID][jID] = job
154 | json_obj = {uID:{"jID": str(jID), "pos": well, "log": log_folder_path}}
155 | json_str = json.dumps(json_obj)+"\n"
156 | self.add_data(json_str)
157 | else:
158 | # from server side jobs object entry is a None object
159 | # this will be later checked as completion boolean for a ExpID which might
160 | # have several Jobs associated with it
161 | if uID not in SERVER_uIDsjobIDs.keys():
162 | SERVER_uIDsjobIDs[uID] = {}
163 | SERVER_uIDsjobIDs[uID][jID] = {}
164 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
165 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
166 | else:
167 | SERVER_uIDsjobIDs[uID][jID] = {}
168 | SERVER_uIDsjobIDs[uID][jID]["job"] = job
169 | SERVER_uIDsjobIDs[uID][jID]["bool"] = False
170 | except Exception as exc:
171 | print(exc.args)
172 |
173 | def has_submitted_job(self, uID: str, jID: str, mode="client")->bool:
174 | jID = str(jID)
175 | if mode == "client":
176 | if uID in self.uIDsjobIDs.keys():
177 | if jID in self.uIDsjobIDs[uID].keys():
178 | return True
179 | return False
180 | else:
181 | if uID in SERVER_uIDsjobIDs.keys():
182 | if jID in SERVER_uIDsjobIDs[uID].keys():
183 | return True
184 | return False
185 |
--------------------------------------------------------------------------------
/recOrder/cli/main.py:
--------------------------------------------------------------------------------
1 | import click
2 |
3 | from recOrder.cli.apply_inverse_transfer_function import apply_inv_tf
4 | from recOrder.cli.compute_transfer_function import compute_tf
5 | from recOrder.cli.reconstruct import reconstruct
6 | try:
7 | from recOrder.cli.gui_widget import gui
8 | except:pass
9 |
10 |
11 | CONTEXT = {"help_option_names": ["-h", "--help"]}
12 |
13 |
14 | # `recorder -h` will show subcommands in the order they are added
15 | class NaturalOrderGroup(click.Group):
16 | def list_commands(self, ctx):
17 | return self.commands.keys()
18 |
19 |
20 | @click.group(context_settings=CONTEXT, cls=NaturalOrderGroup)
21 | def cli():
22 | """\033[92mrecOrder: Computational Toolkit for Label-Free Imaging\033[0m\n"""
23 |
24 |
25 | cli.add_command(reconstruct)
26 | cli.add_command(compute_tf)
27 | cli.add_command(apply_inv_tf)
28 | try:
29 | cli.add_command(gui)
30 | except:pass
31 |
32 | if __name__ == "__main__":
33 | cli()
34 |
--------------------------------------------------------------------------------
/recOrder/cli/monitor.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import time
4 | import numpy as np
5 | import shutil
6 | import submitit
7 | import sys
8 |
9 |
10 | def _move_cursor_up(n_lines, do_print=True):
11 | if do_print:
12 | sys.stdout.write("\033[F" * n_lines)
13 |
14 |
15 | def _print_status(jobs, position_dirpaths, elapsed_list, print_indices=None, do_print=True):
16 |
17 | columns = [15, 30, 40, 50]
18 |
19 | # header
20 | if do_print:
21 | sys.stdout.write(
22 | "\033[K" # clear line
23 | "\033[96mID" # cyan
24 | f"\033[{columns[0]}G WELL "
25 | f"\033[{columns[1]}G STATUS "
26 | f"\033[{columns[2]}G NODE "
27 | f"\033[{columns[2]}G ELAPSED\n"
28 | )
29 |
30 | if print_indices is None:
31 | print_indices = range(len(jobs))
32 |
33 | complete_count = 0
34 |
35 | for i, (job, position_dirpath) in enumerate(zip(jobs, position_dirpaths)):
36 | try:
37 | node_name = job.get_info()["NodeList"] # slowest, so do this first
38 | except:
39 | node_name = "SUBMITTED"
40 |
41 | if job.state == "COMPLETED":
42 | color = "\033[32m" # green
43 | complete_count += 1
44 | elif job.state == "RUNNING":
45 | color = "\033[93m" # yellow
46 | elapsed_list[i] += 1 # inexact timing
47 | else:
48 | color = "\033[91m" # red
49 |
50 | if i in print_indices:
51 | if do_print:
52 | sys.stdout.write(
53 | f"\033[K" # clear line
54 | f"{color}{job.job_id}"
55 | f"\033[{columns[0]}G {'/'.join(position_dirpath.parts[-3:])}"
56 | f"\033[{columns[1]}G {job.state}"
57 | f"\033[{columns[2]}G {node_name}"
58 | f"\033[{columns[3]}G {elapsed_list[i]} s\n"
59 | )
60 | sys.stdout.flush()
61 | if do_print:
62 | print(
63 | f"\033[32m{complete_count}/{len(jobs)} jobs complete. "
64 | " 22 |
23 | On the Poincare sphere, the extinction state corresponds to the north pole, and the swing value corresponds to the targeted line of [colatitude](https://en.m.wikipedia.org/wiki/File:Spherical_Coordinates_%28Colatitude,_Longitude%29.svg) for the remaining states. For example, a swing value of 0.25 (above left) sets the targeted polarization states to the states on the equator: a set of linear polarization states. Similarly, a swing value of 0.125 (above right) sets the targeted polarization states to the states on the line of colatitude 45 degrees ( $\pi$/4 radians) away from the north pole: a set of elliptical polarization states.
24 |
25 | The Poincare sphere is also useful for calculating the ratio of intensities measured before and after an analyzer illuminated with a polarized beam. First, find the point on the Poincare sphere that corresponds to the analyzer; in our case we have a right-circular analyzer corresponding to the south pole. Next, find the point that corresponds to the polarization state of the light incident on the analyzer; this could be any arbitrary point on the Poincare sphere. To find the ratio of intensities before and after the analyzer $I/I_0$, find the great-circle angle between the two points on the Poincare sphere, $\alpha$, and calculate $I/I_0 = \cos^2(\alpha/2)$. As expected, points that are close together transmit perfectly ( $\alpha = 0$ implies $I/I_0 = 1$), while antipodal points lead to extinction ( $\alpha = \pi$ implies $I/I_0 = 0$).
26 |
27 | This geometric construction illustrates that all non-extinction polarization states have the same intensity after the analyzer because they live on the same line of colatitude and have the same great-circle angle to the south pole (the analyzer). We use this fact to help us find our non-extinction states.
28 |
29 | Practically, we find our first non-extinction state immediately using the liquid crystal manufacturer's measurements from the factory. In other words, we apply a fixed voltage offset to the extinction-state voltages to find the first non-extinction state, and this requires no iteration or optimization. To find the remaining non-extinction states, we keep the polarization orientation fixed and search through neighboring states with different ellipticity to find states that transmit the same intensity as the first non-extinction state.
30 |
31 | ## Evaluating a calibration: extinction ratio
32 |
33 | At the end of a calibration we report the **extinction ratio**, the ratio of the largest and smallest intensities that the imaging system can transmit above background. This metric measures the quality of the entire optical path including the liquid crystals and their calibrated states, and all depolarization, scattering, or absorption caused by optical elements in the light path will reduce the extinction ratio.
34 |
35 | ## Calculating extinction ratio from measured intensities (advanced topic)
36 |
37 | To calculate the extinction ratio, we could optimize the liquid crystal voltages to maximize measured intensity then calculate the ratio of that result with the earlier extinction intensity, but this approach requires a time-consuming optimization and it does not characterize the quality of the calibrated states of the liquid crystals.
38 |
39 | Instead, we estimate the extinction ratio from the intensities we measure during the calibration process. Specifically, we measure the black-level intensity $I_{\text{bl}}$, the extinction intensity $I_{\text{ext}}$, and the intensity under the first elliptical state $I_{\text{ellip}}(S)$ where $S$ is the swing. We proceed to algebraically express the extinction ratio in terms of these three quantities.
40 |
41 | We can decompose $I_{\text{ellip}}(S)$ into a constant term $I_{\text{ellip}}(0) = I_{\text{ext}}$, and a modulation term given by
42 |
43 | $$I_{\text{ellip}}(S) = I_{\text{mod}}\sin^2(\pi S) + I_{\text{ext}},\qquad\qquad (1)$$
44 | where $I_{\text{mod}}$ is the modulation depth, and the $\sin^2(\pi S)$ term can be understood using the Poincare sphere (the intensity behind the circular analyzer is proportional to $\cos^2(\alpha/2)$ and for a given swing we have $\alpha = \pi - 2\pi S$ so $\cos^2(\frac{\pi - 2\pi S}{2}) = \sin^2(\pi S)$ ).
45 |
46 | Next, we decompose $I_{\text{ext}}$ into the sum of two terms, the black level intensity and a leakage intensity $I_{\text{leak}}$
47 | $$I_{\text{ext}} = I_{\text{bl}} + I_{\text{leak}}.\qquad\qquad (2)$$
48 |
49 | The following diagram clarifies our definitions and shows how the measured $I_{\text{ellip}}(S)$ depends on the swing (green line).
50 |
51 |
22 |
23 | On the Poincare sphere, the extinction state corresponds to the north pole, and the swing value corresponds to the targeted line of [colatitude](https://en.m.wikipedia.org/wiki/File:Spherical_Coordinates_%28Colatitude,_Longitude%29.svg) for the remaining states. For example, a swing value of 0.25 (above left) sets the targeted polarization states to the states on the equator: a set of linear polarization states. Similarly, a swing value of 0.125 (above right) sets the targeted polarization states to the states on the line of colatitude 45 degrees ( $\pi$/4 radians) away from the north pole: a set of elliptical polarization states.
24 |
25 | The Poincare sphere is also useful for calculating the ratio of intensities measured before and after an analyzer illuminated with a polarized beam. First, find the point on the Poincare sphere that corresponds to the analyzer; in our case we have a right-circular analyzer corresponding to the south pole. Next, find the point that corresponds to the polarization state of the light incident on the analyzer; this could be any arbitrary point on the Poincare sphere. To find the ratio of intensities before and after the analyzer $I/I_0$, find the great-circle angle between the two points on the Poincare sphere, $\alpha$, and calculate $I/I_0 = \cos^2(\alpha/2)$. As expected, points that are close together transmit perfectly ( $\alpha = 0$ implies $I/I_0 = 1$), while antipodal points lead to extinction ( $\alpha = \pi$ implies $I/I_0 = 0$).
26 |
27 | This geometric construction illustrates that all non-extinction polarization states have the same intensity after the analyzer because they live on the same line of colatitude and have the same great-circle angle to the south pole (the analyzer). We use this fact to help us find our non-extinction states.
28 |
29 | Practically, we find our first non-extinction state immediately using the liquid crystal manufacturer's measurements from the factory. In other words, we apply a fixed voltage offset to the extinction-state voltages to find the first non-extinction state, and this requires no iteration or optimization. To find the remaining non-extinction states, we keep the polarization orientation fixed and search through neighboring states with different ellipticity to find states that transmit the same intensity as the first non-extinction state.
30 |
31 | ## Evaluating a calibration: extinction ratio
32 |
33 | At the end of a calibration we report the **extinction ratio**, the ratio of the largest and smallest intensities that the imaging system can transmit above background. This metric measures the quality of the entire optical path including the liquid crystals and their calibrated states, and all depolarization, scattering, or absorption caused by optical elements in the light path will reduce the extinction ratio.
34 |
35 | ## Calculating extinction ratio from measured intensities (advanced topic)
36 |
37 | To calculate the extinction ratio, we could optimize the liquid crystal voltages to maximize measured intensity then calculate the ratio of that result with the earlier extinction intensity, but this approach requires a time-consuming optimization and it does not characterize the quality of the calibrated states of the liquid crystals.
38 |
39 | Instead, we estimate the extinction ratio from the intensities we measure during the calibration process. Specifically, we measure the black-level intensity $I_{\text{bl}}$, the extinction intensity $I_{\text{ext}}$, and the intensity under the first elliptical state $I_{\text{ellip}}(S)$ where $S$ is the swing. We proceed to algebraically express the extinction ratio in terms of these three quantities.
40 |
41 | We can decompose $I_{\text{ellip}}(S)$ into a constant term $I_{\text{ellip}}(0) = I_{\text{ext}}$, and a modulation term given by
42 |
43 | $$I_{\text{ellip}}(S) = I_{\text{mod}}\sin^2(\pi S) + I_{\text{ext}},\qquad\qquad (1)$$
44 | where $I_{\text{mod}}$ is the modulation depth, and the $\sin^2(\pi S)$ term can be understood using the Poincare sphere (the intensity behind the circular analyzer is proportional to $\cos^2(\alpha/2)$ and for a given swing we have $\alpha = \pi - 2\pi S$ so $\cos^2(\frac{\pi - 2\pi S}{2}) = \sin^2(\pi S)$ ).
45 |
46 | Next, we decompose $I_{\text{ext}}$ into the sum of two terms, the black level intensity and a leakage intensity $I_{\text{leak}}$
47 | $$I_{\text{ext}} = I_{\text{bl}} + I_{\text{leak}}.\qquad\qquad (2)$$
48 |
49 | The following diagram clarifies our definitions and shows how the measured $I_{\text{ellip}}(S)$ depends on the swing (green line).
50 |
51 |  52 |
53 | The extinction ratio is the ratio of the largest and smallest intensities that the imaging system can transmit above background, which is most easily expressed in terms of $I_{\text{mod}}$ and $I_{\text{leak}}$
54 | $$\text{Extinction Ratio} = \frac{I_{\text{mod}} + I_{\text{leak}}}{I_{\text{leak}}}.\qquad\qquad (3)$$
55 |
56 | Substituting Eqs. (1) and (2) into Eq. (3) gives the extinction ratio in terms of the measured intensities
57 | $$\text{Extinction Ratio} = \frac{1}{\sin^2(\pi S)}\frac{I_{\text{ellip}}(S) - I_{\text{ext}}}{I_{\text{ext}} - I_{\text{bl}}} + 1.$$
58 |
59 | ## Summary: `recOrder`'s step-by-step calibration procedure
60 | 1. Close the shutter, measure the black level, then reopen the shutter.
61 | 2. Find the extinction state by finding voltages that minimize the intensity that reaches the camera.
62 | 3. Use the swing value to immediately find the first elliptical state, and record the intensity on the camera.
63 | 4. For each remaining elliptical state, keep the polarization orientation fixed and optimize the voltages to match the intensity of the first elliptical state.
64 | 5. Store the voltages and calculate the extinction ratio.
--------------------------------------------------------------------------------
/docs/data-schema.md:
--------------------------------------------------------------------------------
1 | # Data schema
2 |
3 | This document defines the standard for data acquired with `recOrder`.
4 |
5 | ## Raw directory organization
6 |
7 | Currently, we structure raw data in the following hierarchy:
8 |
9 | ```text
10 | working_directory/ # commonly YYYY_MM_DD_exp_name, but not enforced
11 | ├── polarization_calibration_0.txt
12 | │ ...
13 | ├── polarization_calibration_.txt # i calibration repeats
14 | │
15 | ├── bg_0
16 | │ ...
17 | ├── bg_
52 |
53 | The extinction ratio is the ratio of the largest and smallest intensities that the imaging system can transmit above background, which is most easily expressed in terms of $I_{\text{mod}}$ and $I_{\text{leak}}$
54 | $$\text{Extinction Ratio} = \frac{I_{\text{mod}} + I_{\text{leak}}}{I_{\text{leak}}}.\qquad\qquad (3)$$
55 |
56 | Substituting Eqs. (1) and (2) into Eq. (3) gives the extinction ratio in terms of the measured intensities
57 | $$\text{Extinction Ratio} = \frac{1}{\sin^2(\pi S)}\frac{I_{\text{ellip}}(S) - I_{\text{ext}}}{I_{\text{ext}} - I_{\text{bl}}} + 1.$$
58 |
59 | ## Summary: `recOrder`'s step-by-step calibration procedure
60 | 1. Close the shutter, measure the black level, then reopen the shutter.
61 | 2. Find the extinction state by finding voltages that minimize the intensity that reaches the camera.
62 | 3. Use the swing value to immediately find the first elliptical state, and record the intensity on the camera.
63 | 4. For each remaining elliptical state, keep the polarization orientation fixed and optimize the voltages to match the intensity of the first elliptical state.
64 | 5. Store the voltages and calculate the extinction ratio.
--------------------------------------------------------------------------------
/docs/data-schema.md:
--------------------------------------------------------------------------------
1 | # Data schema
2 |
3 | This document defines the standard for data acquired with `recOrder`.
4 |
5 | ## Raw directory organization
6 |
7 | Currently, we structure raw data in the following hierarchy:
8 |
9 | ```text
10 | working_directory/ # commonly YYYY_MM_DD_exp_name, but not enforced
11 | ├── polarization_calibration_0.txt
12 | │ ...
13 | ├── polarization_calibration_.txt # i calibration repeats
14 | │
15 | ├── bg_0
16 | │ ...
17 | ├── bg_