├── tests
├── __init__.py
├── scripts
│ ├── __init__.py
│ └── test_cli.py
├── conftest.py
├── testdata
│ └── example.faa
└── test_msaviz.py
├── src
└── pymsaviz
│ ├── scripts
│ ├── __init__.py
│ └── cli.py
│ ├── config
│ ├── testdata
│ │ ├── HIGD2A.fa
│ │ └── MRGPRG.fa
│ ├── __init__.py
│ └── color_schemes.tsv
│ ├── __init__.py
│ └── msaviz.py
├── .gitattributes
├── example
├── example.zip
├── cli_example_run.sh
├── HIGD2A.fa
└── MRGPRG.fa
├── docs
├── images
│ ├── api_example01.png
│ ├── api_example02.png
│ ├── api_example03.png
│ ├── cli_example01.png
│ ├── cli_example02.png
│ └── cli_example03.png
├── api-docs
│ └── msaviz.md
├── index.md
└── cli-docs
│ └── pymsaviz.md
├── CITATION.cff
├── .pre-commit-config.yaml
├── .github
└── workflows
│ ├── publish_mkdocs.yml
│ ├── publish_to_pypi.yml
│ └── ci.yml
├── requirements.lock
├── LICENSE
├── mkdocs.yml
├── pyproject.toml
├── .gitignore
├── README.md
└── requirements-dev.lock
/tests/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/tests/scripts/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/src/pymsaviz/scripts/__init__.py:
--------------------------------------------------------------------------------
1 |
--------------------------------------------------------------------------------
/.gitattributes:
--------------------------------------------------------------------------------
1 | *.ipynb linguist-documentation
2 |
--------------------------------------------------------------------------------
/example/example.zip:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/example/example.zip
--------------------------------------------------------------------------------
/docs/images/api_example01.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/api_example01.png
--------------------------------------------------------------------------------
/docs/images/api_example02.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/api_example02.png
--------------------------------------------------------------------------------
/docs/images/api_example03.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/api_example03.png
--------------------------------------------------------------------------------
/docs/images/cli_example01.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/cli_example01.png
--------------------------------------------------------------------------------
/docs/images/cli_example02.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/cli_example02.png
--------------------------------------------------------------------------------
/docs/images/cli_example03.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/moshi4/pyMSAviz/HEAD/docs/images/cli_example03.png
--------------------------------------------------------------------------------
/CITATION.cff:
--------------------------------------------------------------------------------
1 | cff-version: 1.2.0
2 | message: If you use this software, please cite it as below.
3 | authors:
4 | - family-names: Shimoyama
5 | given-names: Yuki
6 | title: "pyMSAviz: MSA visualization python package for sequence analysis"
7 | date-released: 2022-11-13
8 | url: https://github.com/moshi4/pyMSAviz
9 |
--------------------------------------------------------------------------------
/docs/api-docs/msaviz.md:
--------------------------------------------------------------------------------
1 | # MsaViz Class
2 |
3 | ::: pymsaviz.msaviz.MsaViz
4 | options:
5 | members:
6 | - available_color_schemes
7 | - set_plot_params
8 | - set_custom_color_scheme
9 | - set_custom_color_func
10 | - set_highlight_pos
11 | - set_highlight_pos_by_ident_thr
12 | - add_markers
13 | - add_text_annotation
14 | - plotfig
15 | - savefig
16 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | # See https://pre-commit.com for more information
2 | # See https://pre-commit.com/hooks.html for more hooks
3 | repos:
4 | - repo: https://github.com/astral-sh/ruff-pre-commit
5 | rev: v0.8.1
6 | hooks:
7 | - id: ruff
8 | name: ruff lint check
9 | types_or: [python, pyi]
10 | args: [--fix]
11 | - id: ruff-format
12 | name: ruff format check
13 | types_or: [python, pyi]
14 |
--------------------------------------------------------------------------------
/.github/workflows/publish_mkdocs.yml:
--------------------------------------------------------------------------------
1 | name: Publish MkDocs
2 |
3 | on:
4 | release:
5 | types: [released]
6 | workflow_dispatch:
7 |
8 | jobs:
9 | publish_mkdocs:
10 | runs-on: ubuntu-latest
11 | steps:
12 | - name: Checkout
13 | uses: actions/checkout@v4
14 |
15 | - name: Install Rye
16 | run: |
17 | curl -sSf https://rye.astral.sh/get | RYE_INSTALL_OPTION="--yes" bash
18 | echo "$HOME/.rye/shims" >> $GITHUB_PATH
19 |
20 | - name: Install Python & MkDocs & Plugins
21 | run: rye sync
22 |
23 | - name: Publish document
24 | run: rye run mkdocs gh-deploy --force
25 |
--------------------------------------------------------------------------------
/example/cli_example_run.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/bash
2 |
3 | # Example 01
4 | echo "Run pyMSAviz CLI example 01..."
5 | pymsaviz -i HIGD2A.fa -o cli_example01.png --color_scheme Identity --dpi 100
6 |
7 | # Example 02
8 | echo "Run pyMSAviz CLI example 02..."
9 | pymsaviz -i MRGPRG.fa -o cli_example02.png --wrap_length 80 --dpi 100 \
10 | --color_scheme Taylor --show_consensus --show_count
11 |
12 | # Example 03
13 | echo "Run pyMSAviz CLI example 03..."
14 | pymsaviz -i MRGPRG.fa -o cli_example03.png --start 100 --end 160 --dpi 100 \
15 | --color_scheme Flower --show_grid --show_consensus --consensus_color tomato
16 |
17 | echo -e "\nFinished all example CLI run."
18 |
--------------------------------------------------------------------------------
/tests/conftest.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import pytest
4 | from Bio.Align import MultipleSeqAlignment
5 | from Bio.Seq import Seq
6 | from Bio.SeqRecord import SeqRecord
7 |
8 |
9 | @pytest.fixture
10 | def testdata_dir() -> Path:
11 | """Test data directory"""
12 | return Path(__file__).parent / "testdata"
13 |
14 |
15 | @pytest.fixture
16 | def msa_fasta_file(testdata_dir: Path) -> Path:
17 | """MSA fasta file"""
18 | return testdata_dir / "example.faa"
19 |
20 |
21 | @pytest.fixture
22 | def dummy_msa() -> MultipleSeqAlignment:
23 | """Dummy MSA object"""
24 | return MultipleSeqAlignment([SeqRecord(Seq("ATGC")), SeqRecord(Seq("ATGC"))])
25 |
--------------------------------------------------------------------------------
/.github/workflows/publish_to_pypi.yml:
--------------------------------------------------------------------------------
1 | name: Publish to PyPI
2 | on:
3 | release:
4 | types: [released]
5 | workflow_dispatch:
6 |
7 | jobs:
8 | publish_to_pypi:
9 | name: Publish to PyPI
10 | runs-on: ubuntu-latest
11 | env:
12 | PYPI_USERNAME: ${{ secrets.PYPI_USERNAME }}
13 | PYPI_PASSWORD: ${{ secrets.PYPI_PASSWORD }}
14 | steps:
15 | - name: Checkout
16 | uses: actions/checkout@v4
17 |
18 | - name: Install Rye
19 | run: |
20 | curl -sSf https://rye.astral.sh/get | RYE_INSTALL_OPTION="--yes" bash
21 | echo "$HOME/.rye/shims" >> $GITHUB_PATH
22 |
23 | - name: Build
24 | run: rye build
25 |
26 | - name: Publish
27 | run: rye publish -u $PYPI_USERNAME --token $PYPI_PASSWORD -y
28 |
--------------------------------------------------------------------------------
/example/HIGD2A.fa:
--------------------------------------------------------------------------------
1 | >GorillaGorilla
2 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFLRKTRENPVVPIGCLATAAAL
3 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
4 | >HomoSapiens
5 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
6 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
7 | >NomascusLeucogenys
8 | MATPGPVIPEVPFEPSKPPVIEGFSPTVYRNPESFKGKFLRKTRENPVVPIGCLATAAAL
9 | TYGLYSFHRGDSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRPSAQGLASKAPQK
10 | >PanPaniscus
11 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
12 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
13 | >PanTroglodytes
14 | MATPGPVIQEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
15 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
16 | >PongoAbelii
17 | MATPGPVIPKVPFEPSKPPVIEGLSPTVYRNPESFKEKFLRKTRENPVVPIGCLATATAL
18 | SYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
19 |
--------------------------------------------------------------------------------
/src/pymsaviz/config/testdata/HIGD2A.fa:
--------------------------------------------------------------------------------
1 | >GorillaGorilla
2 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFLRKTRENPVVPIGCLATAAAL
3 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
4 | >HomoSapiens
5 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
6 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
7 | >NomascusLeucogenys
8 | MATPGPVIPEVPFEPSKPPVIEGFSPTVYRNPESFKGKFLRKTRENPVVPIGCLATAAAL
9 | TYGLYSFHRGDSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRPSAQGLASKAPQK
10 | >PanPaniscus
11 | MATPGPVIPEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
12 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
13 | >PanTroglodytes
14 | MATPGPVIQEVPFEPSKPPVIEGLSPTVYRNPESFKEKFVRKTRENPVVPIGCLATAAAL
15 | TYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
16 | >PongoAbelii
17 | MATPGPVIPKVPFEPSKPPVIEGLSPTVYRNPESFKEKFLRKTRENPVVPIGCLATATAL
18 | SYGLYSFHRGNSQRSQLMMRTRIAAQGFTVAAILLGLAVTAMKSRP------------
19 |
--------------------------------------------------------------------------------
/src/pymsaviz/__init__.py:
--------------------------------------------------------------------------------
1 | import warnings
2 |
3 | import matplotlib as mpl
4 |
5 | from pymsaviz.config import get_msa_testdata
6 | from pymsaviz.msaviz import MsaViz
7 |
8 | warnings.filterwarnings("ignore")
9 |
10 | __all__ = [
11 | "MsaViz",
12 | "get_msa_testdata",
13 | ]
14 |
15 | __version__ = "0.5.0"
16 |
17 | # Setting matplotlib rc(runtime configuration) parameters
18 | # https://matplotlib.org/stable/tutorials/introductory/customizing.html
19 | mpl_rc_params = {
20 | # Legend
21 | "legend.loc": "upper left", # Default: best
22 | "legend.frameon": False, # Default: True
23 | "legend.handlelength": 1, # Default: 2.0
24 | "legend.handleheight": 1, # Default: 0.7
25 | # Savefig
26 | "savefig.bbox": "tight", # Default: None
27 | "savefig.pad_inches": 0.5, # Default: 0.1
28 | # SVG
29 | "svg.fonttype": "none",
30 | }
31 | mpl.rcParams.update(mpl_rc_params)
32 |
--------------------------------------------------------------------------------
/tests/scripts/test_cli.py:
--------------------------------------------------------------------------------
1 | import subprocess
2 | from pathlib import Path
3 |
4 |
5 | def test_cli_default_run(msa_fasta_file: Path, tmp_path: Path):
6 | """Test CLI with default option"""
7 | fig_outfile = tmp_path / "test.png"
8 |
9 | cmd = f"pymsaviz -i {msa_fasta_file} -o {fig_outfile}"
10 | subprocess.run(cmd, shell=True)
11 |
12 | assert fig_outfile.exists()
13 |
14 |
15 | def test_cli_full_option_run(msa_fasta_file: Path, tmp_path: Path):
16 | """Test CLI with full option"""
17 | fig_outfile = tmp_path / "test.png"
18 |
19 | cmd = f"pymsaviz -i {msa_fasta_file} -o {fig_outfile} --format fasta "
20 | cmd += "--color_scheme Taylor --start 50 --end 250 --wrap_length 100 "
21 | cmd += "--wrap_space_size 3.0 --show_grid --show_count --show_consensus "
22 | cmd += "--consensus_color green --consensus_size 2.0 --sort --dpi 100"
23 | subprocess.run(cmd, shell=True)
24 |
25 | assert fig_outfile.exists()
26 |
--------------------------------------------------------------------------------
/requirements.lock:
--------------------------------------------------------------------------------
1 | # generated by rye
2 | # use `rye lock` or `rye sync` to update this lockfile
3 | #
4 | # last locked with the following flags:
5 | # pre: false
6 | # features: []
7 | # all-features: true
8 | # with-sources: false
9 | # generate-hashes: false
10 | # universal: false
11 |
12 | -e file:.
13 | biopython==1.84
14 | # via pymsaviz
15 | contourpy==1.3.0
16 | # via matplotlib
17 | cycler==0.12.1
18 | # via matplotlib
19 | fonttools==4.55.0
20 | # via matplotlib
21 | importlib-resources==6.4.5
22 | # via matplotlib
23 | kiwisolver==1.4.7
24 | # via matplotlib
25 | matplotlib==3.9.3
26 | # via pymsaviz
27 | numpy==2.0.2
28 | # via biopython
29 | # via contourpy
30 | # via matplotlib
31 | packaging==24.2
32 | # via matplotlib
33 | pillow==11.0.0
34 | # via matplotlib
35 | pyparsing==3.2.0
36 | # via matplotlib
37 | python-dateutil==2.9.0.post0
38 | # via matplotlib
39 | six==1.16.0
40 | # via python-dateutil
41 | zipp==3.21.0
42 | # via importlib-resources
43 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2022 moshi
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/.github/workflows/ci.yml:
--------------------------------------------------------------------------------
1 | name: CI
2 | on:
3 | push:
4 | branches: [main, develop]
5 | paths: ["src/**", "tests/**", ".github/workflows/ci.yml"]
6 | pull_request:
7 | branches: [main, develop]
8 | paths: ["src/**", "tests/**", ".github/workflows/ci.yml"]
9 | workflow_dispatch:
10 |
11 | jobs:
12 | CI:
13 | runs-on: ${{ matrix.os }}
14 | strategy:
15 | matrix:
16 | os: [ubuntu-latest, macos-latest]
17 | python-version: ["3.9", "3.10", "3.11", "3.12"]
18 | steps:
19 | - name: Checkout
20 | uses: actions/checkout@v4
21 |
22 | - name: Install Rye
23 | run: |
24 | curl -sSf https://rye.astral.sh/get | RYE_INSTALL_OPTION="--yes" bash
25 | echo "$HOME/.rye/shims" >> $GITHUB_PATH
26 |
27 | - name: Setup Python ${{matrix.python-version}} & Dependencies
28 | run: |
29 | rye pin ${{ matrix.python-version }}
30 | rye sync --update-all --all-features
31 |
32 | - name: Run ruff lint check
33 | run: rye run ruff check --diff
34 |
35 | - name: Run ruff format check
36 | run: rye run ruff format --check --diff
37 |
38 | - name: Run pytest
39 | run: rye run pytest
40 |
--------------------------------------------------------------------------------
/docs/index.md:
--------------------------------------------------------------------------------
1 | # pyMSAviz
2 |
3 | 
4 | 
5 | 
6 | [](https://pypi.python.org/pypi/pymsaviz)
7 | [](https://anaconda.org/bioconda/pymsaviz)
8 |
9 | ## Overview
10 |
11 | pyMSAviz is a MSA(Multiple Sequence Alignment) visualization python package for sequence analysis implemented based on matplotlib.
12 | This package is developed for the purpose of easily and beautifully plotting MSA in Python.
13 | It also implements the functionality to add markers, text annotations, highlights to specific positions and ranges in MSA.
14 | pyMSAviz was developed inspired by [Jalview](https://www.jalview.org/) and [ggmsa](https://github.com/YuLab-SMU/ggmsa).
15 |
16 |
17 | 
18 | Fig.1 Simple visualization result
19 |
20 |
21 |
22 | 
23 | Fig.2 Customized visualization result
24 |
25 |
26 | ## Installation
27 |
28 | `Python 3.9 or later` is required for installation.
29 |
30 | **Install PyPI package:**
31 |
32 | pip install pymsaviz
33 |
34 | **Install bioconda package:**
35 |
36 | conda install -c conda-forge -c bioconda pymsaviz
37 |
--------------------------------------------------------------------------------
/mkdocs.yml:
--------------------------------------------------------------------------------
1 | site_name: pyMSAviz
2 | site_description: MSA visualization python package for sequence analysis
3 | site_author: moshi4
4 | repo_name: moshi4/pyMSAviz
5 | repo_url: https://github.com/moshi4/pyMSAviz
6 | edit_uri: ""
7 | use_directory_urls: true
8 |
9 | nav:
10 | - Home: index.md
11 | - Getting Started: getting_started.ipynb
12 | - Color Schemes: color_schemes.ipynb
13 | - API Docs: api-docs/msaviz.md
14 | - CLI Docs: cli-docs/pymsaviz.md
15 |
16 | theme:
17 | name: material # material, readthedocs, mkdocs
18 | features:
19 | - navigation.top
20 | - navigation.expand
21 | # - navigation.tabs
22 | - navigation.tabs.sticky
23 | - navigation.sections
24 |
25 | markdown_extensions:
26 | - pymdownx.highlight:
27 | anchor_linenums: true
28 | - pymdownx.inlinehilite
29 | - pymdownx.snippets
30 | - pymdownx.superfences
31 | - pymdownx.details
32 | - admonition
33 | - attr_list
34 | - md_in_html
35 |
36 | plugins:
37 | - search
38 | - mkdocs-jupyter:
39 | execute: False
40 | - mkdocstrings:
41 | handlers:
42 | python:
43 | # Reference: https://mkdocstrings.github.io/python/usage/
44 | options:
45 | # Heading options
46 | heading_level: 2
47 | show_root_full_path: False
48 | show_root_heading: True
49 | # Member options
50 | members_order: source # alphabetical, source
51 | # Docstrings options
52 | docstring_style: numpy
53 | docstring_section_style: spacy # table, list, spacy
54 | line_length: 89
55 | merge_init_into_class: True
56 | # Signatures/annotations options
57 | show_signature_annotations: True
58 | separate_signature: True
59 | # Additional options
60 | show_source: False
61 |
--------------------------------------------------------------------------------
/example/MRGPRG.fa:
--------------------------------------------------------------------------------
1 | >GorillaGorilla
2 | MFGLFGLWRTFHSVVFYLTLIVGLGGPVGNGLVLWNLSFHVKKGPFSINLLHLAAADFLF
3 | LSCRVGFSVAQAALG---------------------------RCLSDLFPA---------
4 | ---VLCTLVWAPTLPAVLLPANACGLLCISARPLVCLRYHVASVTWFLVLARVAWTAGVV
5 | LFVWVTCCSTRLQPRLYGIVLGALLLLFLCGLPLVFYWSLQPLLNFLLPMFSPLATLLAC
6 | VNSSSKPLIYSGLGRQPGKRESLRSVLRRALGEGAKLGARGQSLPMGLL
7 | >HomoSapiens
8 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFRIKKGPFSIYLLHLAAADFLF
9 | LSCRVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
10 | ASAVLCALVWTPTLPAVPLPANACGLLRNSACPLVCPRYHVASVTWFLVLARVAWTAGVV
11 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPVFSPLATLLAC
12 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
13 | >NomascusLeucogenys
14 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
15 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLVVAFSVERCLSDLFPACYQGCRPRH
16 | TSVILCALVWALTLPAVLLPANACGLLHNSARPLVCLRYHVASVTWFLVLACVAWTAGVV
17 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPLVLYWSLQPLLNFLLPMFSPLATLLAC
18 | VNSSSKPLIYXXXXRQPGKREPLRVVLWRALGEGAELSARGQSLPMGLL
19 | >PanPaniscus
20 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFHIKKGTFSIYLLHLAAADFLF
21 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERSLSDLFPACYQGCRPRH
22 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
23 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWRLQPLLNFLLPMFSPLATLLAC
24 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
25 | >PanTroglodytes
26 | MFGLFGLWRTFDSVVFYLTPIVGLGGPVGNGLVLWNLGFHIKKGPFSIYLLHLAAADFLF
27 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
28 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
29 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPMFSPLATLLAC
30 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
31 | >PongoAbelii
32 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
33 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYHGCRPRH
34 | ASAVLCALVWALTLPAVLLPANACGLLRNSARPLVCLRYHVPASPGSCAGLR-AWTAGVV
35 | LFVLVTCCSMRARPR-YGIVLGALLLLF-CGLPLVFYWSLQPLLNFLLPMFSPLAMLLAC
36 | VNSSSKPLIYLGLGRQPGKREPLRVVLQRALGEGAELGARGQSLPMGLL
37 |
--------------------------------------------------------------------------------
/src/pymsaviz/config/testdata/MRGPRG.fa:
--------------------------------------------------------------------------------
1 | >GorillaGorilla
2 | MFGLFGLWRTFHSVVFYLTLIVGLGGPVGNGLVLWNLSFHVKKGPFSINLLHLAAADFLF
3 | LSCRVGFSVAQAALG---------------------------RCLSDLFPA---------
4 | ---VLCTLVWAPTLPAVLLPANACGLLCISARPLVCLRYHVASVTWFLVLARVAWTAGVV
5 | LFVWVTCCSTRLQPRLYGIVLGALLLLFLCGLPLVFYWSLQPLLNFLLPMFSPLATLLAC
6 | VNSSSKPLIYSGLGRQPGKRESLRSVLRRALGEGAKLGARGQSLPMGLL
7 | >HomoSapiens
8 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFRIKKGPFSIYLLHLAAADFLF
9 | LSCRVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
10 | ASAVLCALVWTPTLPAVPLPANACGLLRNSACPLVCPRYHVASVTWFLVLARVAWTAGVV
11 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPVFSPLATLLAC
12 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
13 | >NomascusLeucogenys
14 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
15 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLVVAFSVERCLSDLFPACYQGCRPRH
16 | TSVILCALVWALTLPAVLLPANACGLLHNSARPLVCLRYHVASVTWFLVLACVAWTAGVV
17 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPLVLYWSLQPLLNFLLPMFSPLATLLAC
18 | VNSSSKPLIYXXXXRQPGKREPLRVVLWRALGEGAELSARGQSLPMGLL
19 | >PanPaniscus
20 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFHIKKGTFSIYLLHLAAADFLF
21 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERSLSDLFPACYQGCRPRH
22 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
23 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWRLQPLLNFLLPMFSPLATLLAC
24 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
25 | >PanTroglodytes
26 | MFGLFGLWRTFDSVVFYLTPIVGLGGPVGNGLVLWNLGFHIKKGPFSIYLLHLAAADFLF
27 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
28 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
29 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPMFSPLATLLAC
30 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
31 | >PongoAbelii
32 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
33 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYHGCRPRH
34 | ASAVLCALVWALTLPAVLLPANACGLLRNSARPLVCLRYHVPASPGSCAGLR-AWTAGVV
35 | LFVLVTCCSMRARPR-YGIVLGALLLLF-CGLPLVFYWSLQPLLNFLLPMFSPLAMLLAC
36 | VNSSSKPLIYLGLGRQPGKREPLRVVLQRALGEGAELGARGQSLPMGLL
37 |
--------------------------------------------------------------------------------
/tests/testdata/example.faa:
--------------------------------------------------------------------------------
1 | >GorillaGorilla_ENSGGOP00000051206.1
2 | MFGLFGLWRTFHSVVFYLTLIVGLGGPVGNGLVLWNLSFHVKKGPFSINLLHLAAADFLF
3 | LSCRVGFSVAQAALG---------------------------RCLSDLFPA---------
4 | ---VLCTLVWAPTLPAVLLPANACGLLCISARPLVCLRYHVASVTWFLVLARVAWTAGVV
5 | LFVWVTCCSTRLQPRLYGIVLGALLLLFLCGLPLVFYWSLQPLLNFLLPMFSPLATLLAC
6 | VNSSSKPLIYSGLGRQPGKRESLRSVLRRALGEGAKLGARGQSLPMGLL
7 | >HomoSapiens_ENSP00000330612.3
8 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFRIKKGPFSIYLLHLAAADFLF
9 | LSCRVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
10 | ASAVLCALVWTPTLPAVPLPANACGLLRNSACPLVCPRYHVASVTWFLVLARVAWTAGVV
11 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPVFSPLATLLAC
12 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
13 | >NomascusLeucogenys_ENSNLEP00000013130.1
14 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
15 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLVVAFSVERCLSDLFPACYQGCRPRH
16 | TSVILCALVWALTLPAVLLPANACGLLHNSARPLVCLRYHVASVTWFLVLACVAWTAGVV
17 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPLVLYWSLQPLLNFLLPMFSPLATLLAC
18 | VNSSSKPLIYXXXXRQPGKREPLRVVLWRALGEGAELSARGQSLPMGLL
19 | >PanPaniscus_ENSPPAP00000002623.1
20 | MFGLFGLWRTFDSVVFYLTLIVGLGGPVGNGLVLWNLGFHIKKGTFSIYLLHLAAADFLF
21 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERSLSDLFPACYQGCRPRH
22 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
23 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWRLQPLLNFLLPMFSPLATLLAC
24 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
25 | >PanTroglodytes_ENSPTRP00000048296.3
26 | MFGLFGLWRTFDSVVFYLTPIVGLGGPVGNGLVLWNLGFHIKKGPFSIYLLHLAAADFLF
27 | LSCPVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYQGCRPRH
28 | ASAVLCALVWTPTLPALPLPANACGLLRNSACPLVCLRYHVASVTWFLVLARVAWTAGVV
29 | LFVWVTCCSTRPRPRLYGIVLGALLLLFFCGLPSVFYWSLQPLLNFLLPMFSPLATLLAC
30 | VNSSSKPLIYSGLGRQPGKREPLRSVLRRALGEGAELGARGQSLPMGLL
31 | >PongoAbelii_ENSPPYP00000003417.1
32 | MFGLFGLWRTFDSVVFYLTLIVGLGGLVGNGLVLWNLGFHIKKGPFSVYLLHLAAADFLF
33 | LSCHVGFSVAQAALGAQDTLYFVLTFLWFAVGLWLLAAFSVERCLSDLFPACYHGCRPRH
34 | ASAVLCALVWALTLPAVLLPANACGLLRNSARPLVCLRYHVPASPGSCAGLR-AWTAGVV
35 | LFVLVTCCSMRARPR-YGIVLGALLLLF-CGLPLVFYWSLQPLLNFLLPMFSPLAMLLAC
36 | VNSSSKPLIYLGLGRQPGKREPLRVVLQRALGEGAELGARGQSLPMGLL
37 |
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [project]
2 | name = "pyMSAviz"
3 | dynamic = ["version"]
4 | description = "MSA visualization python package for sequence analysis"
5 | authors = [{ name = "moshi4", email = "" }]
6 | license = "MIT"
7 | readme = "README.md"
8 | keywords = [
9 | "bioinformatics",
10 | "matplotlib",
11 | "visualization",
12 | "sequence-alignment",
13 | "sequence-analysis",
14 | ]
15 | classifiers = [
16 | "Intended Audience :: Science/Research",
17 | "Topic :: Scientific/Engineering :: Bio-Informatics",
18 | "Framework :: Matplotlib",
19 | ]
20 | requires-python = ">=3.9"
21 | dependencies = ["matplotlib>=3.5.2", "biopython>=1.79"]
22 |
23 | [project.urls]
24 | homepage = "https://moshi4.github.io/pyMSAviz/"
25 | repository = "https://github.com/moshi4/pyMSAviz/"
26 |
27 | [project.scripts]
28 | pymsaviz = "pymsaviz.scripts.cli:main"
29 |
30 | [tool.hatch.version]
31 | path = "src/pymsaviz/__init__.py"
32 |
33 | [tool.rye]
34 | managed = true
35 | dev-dependencies = [
36 | "ruff>=0.4.0",
37 | "pre-commit>=3.5.0",
38 | "pytest>=8.0.0",
39 | "pytest-cov>=4.0.0",
40 | "ipykernel>=6.13.0",
41 | # docs
42 | "mkdocs>=1.2",
43 | "mkdocstrings[python]>=0.19.0",
44 | "mkdocs-jupyter>=0.21.0",
45 | "mkdocs-material>=8.2",
46 | "black>=22.3.0",
47 | ]
48 |
49 | [tool.pytest.ini_options]
50 | minversion = "6.0"
51 | addopts = "--cov=src --tb=line --cov-report=xml --cov-report=term"
52 | testpaths = ["tests"]
53 |
54 | [tool.ruff]
55 | include = ["src/**.py", "tests/**.py"]
56 | line-length = 88
57 |
58 | # Lint Rules: https://docs.astral.sh/ruff/rules/
59 | [tool.ruff.lint]
60 | select = [
61 | "F", # pyflakes
62 | "E", # pycodestyle (Error)
63 | "W", # pycodestyle (Warning)
64 | "I", # isort
65 | "D", # pydocstyle
66 | ]
67 | ignore = [
68 | "D100", # Missing docstring in public module
69 | "D101", # Missing docstring in public class
70 | "D104", # Missing docstring in public package
71 | "D105", # Missing docstring in magic method
72 | "D205", # 1 blank line required between summary line and description
73 | "D400", # First line should end with a period
74 | "D401", # First line should be in imperative mood
75 | "D403", # First word of the first line should be properly capitalized
76 | "D415", # First line should end with a period, question mark, or exclamation point
77 | ]
78 |
79 | [tool.ruff.lint.pydocstyle]
80 | convention = "numpy"
81 |
82 | [build-system]
83 | requires = ["hatchling"]
84 | build-backend = "hatchling.build"
85 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | example
2 | .vscode/
3 | notebooks/
4 |
5 | # Byte-compiled / optimized / DLL files
6 | __pycache__/
7 | *.py[cod]
8 | *$py.class
9 |
10 | # C extensions
11 | *.so
12 |
13 | # Distribution / packaging
14 | .Python
15 | build/
16 | develop-eggs/
17 | dist/
18 | downloads/
19 | eggs/
20 | .eggs/
21 | lib/
22 | lib64/
23 | parts/
24 | sdist/
25 | var/
26 | wheels/
27 | pip-wheel-metadata/
28 | share/python-wheels/
29 | *.egg-info/
30 | .installed.cfg
31 | *.egg
32 | MANIFEST
33 |
34 | # PyInstaller
35 | # Usually these files are written by a python script from a template
36 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
37 | *.manifest
38 | *.spec
39 |
40 | # Installer logs
41 | pip-log.txt
42 | pip-delete-this-directory.txt
43 |
44 | # Unit test / coverage reports
45 | htmlcov/
46 | .tox/
47 | .nox/
48 | .coverage
49 | .coverage.*
50 | .cache
51 | nosetests.xml
52 | coverage.xml
53 | *.cover
54 | *.py,cover
55 | .hypothesis/
56 | .pytest_cache/
57 |
58 | # Translations
59 | *.mo
60 | *.pot
61 |
62 | # Django stuff:
63 | *.log
64 | local_settings.py
65 | db.sqlite3
66 | db.sqlite3-journal
67 |
68 | # Flask stuff:
69 | instance/
70 | .webassets-cache
71 |
72 | # Scrapy stuff:
73 | .scrapy
74 |
75 | # Sphinx documentation
76 | docs/_build/
77 |
78 | # PyBuilder
79 | target/
80 |
81 | # Jupyter Notebook
82 | .ipynb_checkpoints
83 |
84 | # IPython
85 | profile_default/
86 | ipython_config.py
87 |
88 | # pyenv
89 | .python-version
90 |
91 | # pipenv
92 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
93 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
94 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
95 | # install all needed dependencies.
96 | #Pipfile.lock
97 |
98 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
99 | __pypackages__/
100 |
101 | # Celery stuff
102 | celerybeat-schedule
103 | celerybeat.pid
104 |
105 | # SageMath parsed files
106 | *.sage.py
107 |
108 | # Environments
109 | .env
110 | .venv
111 | env/
112 | venv/
113 | ENV/
114 | env.bak/
115 | venv.bak/
116 |
117 | # Spyder project settings
118 | .spyderproject
119 | .spyproject
120 |
121 | # Rope project settings
122 | .ropeproject
123 |
124 | # mkdocs documentation
125 | /site
126 |
127 | # mypy
128 | .mypy_cache/
129 | .dmypy.json
130 | dmypy.json
131 |
132 | # Pyre type checker
133 | .pyre/
134 |
--------------------------------------------------------------------------------
/src/pymsaviz/config/__init__.py:
--------------------------------------------------------------------------------
1 | from __future__ import annotations
2 |
3 | import csv
4 | from enum import IntEnum, auto
5 | from pathlib import Path
6 |
7 | ###########################################################
8 | # Color Schemes Config
9 | ###########################################################
10 |
11 |
12 | def get_color_schemes() -> dict[str, dict[str, str]]:
13 | """Get color schemes
14 |
15 | Returns
16 | -------
17 | name2color_scheme : dict[str, dict[str, str]]
18 | Color schemes dict
19 | """
20 | COLOR_SCHEMES_FILE = Path(__file__).parent / "color_schemes.tsv"
21 | name2color_scheme = {}
22 | with open(COLOR_SCHEMES_FILE) as f:
23 | reader = csv.reader(f, delimiter="\t")
24 | header = next(reader)
25 | letters = header[1:]
26 | for row in reader:
27 | name, colors = row[0], row[1:]
28 | color_scheme = {}
29 | for letter, color in zip(letters, colors):
30 | color_scheme[letter] = color
31 | name2color_scheme[name] = color_scheme
32 | return name2color_scheme
33 |
34 |
35 | COLOR_SCHEMES = get_color_schemes()
36 |
37 | ###########################################################
38 | # Plot Config
39 | ###########################################################
40 |
41 |
42 | class AxesType(IntEnum):
43 | """Plot axes type enum"""
44 |
45 | MSA = auto()
46 | CONSENSUS = auto()
47 | SPACE = auto()
48 | WRAP_SPACE = auto()
49 |
50 |
51 | ###########################################################
52 | # Example MSA Dataset
53 | ###########################################################
54 |
55 |
56 | def get_msa_testdata(name: str = "MRGPRG.fa") -> Path:
57 | """Get MSA testdata file
58 |
59 | List of MSA testdata filename
60 | - `HIGD2A.fa` (6 species genes, 118 alignment length)
61 | - `MRGPRG.fa` (6 species genes, 289 alignment length)
62 |
63 | Parameters
64 | ----------
65 | name : str, optional
66 | Testdata name
67 |
68 | Returns
69 | -------
70 | msa_testdata_file : Path
71 | MSA testdata file
72 | """
73 | testdata_dir = Path(__file__).parent / "testdata"
74 | dataset_files = testdata_dir.glob("*")
75 | name2dataset_file = {f.name: f for f in dataset_files}
76 | if name not in name2dataset_file:
77 | err_msg = f"Dataset name = '{name}' not found. "
78 | err_msg += f"Available testdata name = {list(name2dataset_file.keys())}"
79 | raise ValueError(err_msg)
80 | return name2dataset_file[name]
81 |
--------------------------------------------------------------------------------
/docs/cli-docs/pymsaviz.md:
--------------------------------------------------------------------------------
1 | # pymsaviz CLI Document
2 |
3 | ## Usage
4 |
5 | ### Basic Command
6 |
7 | pymsaviz -i [MSA file] -o [MSA visualization file]
8 |
9 | ### Options
10 |
11 | $ pymsaviz --help
12 | usage: pymsaviz [options] -i msa.fa -o msa_viz.png
13 |
14 | MSA(Multiple Sequence Alignment) visualization CLI tool
15 |
16 | optional arguments:
17 | -i I, --infile I Input MSA file
18 | -o O, --outfile O Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)
19 | --format MSA file format (Default: 'fasta')
20 | --color_scheme Color scheme (Default: 'Zappo')
21 | --start Start position of MSA visualization (Default: 1)
22 | --end End position of MSA visualization (Default: 'MSA Length')
23 | --wrap_length Wrap length (Default: None)
24 | --wrap_space_size Space size between wrap MSA plot area (Default: 3.0)
25 | --label_type Label type ('id'[default]|'description')
26 | --show_grid Show grid (Default: OFF)
27 | --show_count Show seq char count without gap on right side (Default: OFF)
28 | --show_consensus Show consensus sequence (Default: OFF)
29 | --consensus_color Consensus identity bar color (Default: '#1f77b4')
30 | --consensus_size Consensus identity bar height size (Default: 2.0)
31 | --sort Sort MSA order by NJ tree constructed from MSA distance matrix (Default: OFF)
32 | --dpi Figure DPI (Default: 300)
33 | -v, --version Print version information
34 | -h, --help Show this help message and exit
35 |
36 | Available Color Schemes:
37 | ['Clustal', 'Zappo', 'Taylor', 'Flower', 'Blossom', 'Sunset', 'Ocean', 'Hydrophobicity', 'HelixPropensity', 'StrandPropensity', 'TurnPropensity', 'BuriedIndex', 'Nucleotide', 'Purine/Pyrimidine', 'Identity', 'None']
38 |
39 | ### Example Command
40 |
41 | Click [here](https://github.com/moshi4/pyMSAviz/raw/main/example/example.zip) to download example MSA files.
42 |
43 | #### Example 1
44 |
45 | pymsaviz -i ./example/HIGD2A.fa -o cli_example01.png --color_scheme Identity
46 |
47 | 
48 |
49 | #### Example 2
50 |
51 | pymsaviz -i ./example/MRGPRG.fa -o cli_example02.png --wrap_length 80 \
52 | --color_scheme Taylor --show_consensus --show_count
53 |
54 | 
55 |
56 | #### Example 3
57 |
58 | pymsaviz -i ./example/MRGPRG.fa -o cli_example03.png --start 100 --end 160 \
59 | --color_scheme Flower --show_grid --show_consensus --consensus_color tomato
60 |
61 | 
62 |
--------------------------------------------------------------------------------
/src/pymsaviz/config/color_schemes.tsv:
--------------------------------------------------------------------------------
1 | ColorScheme A R N D C Q E G H I L K M F P S T W Y V B X Z J O U -
2 | Clustal #80A0F0 #F01505 #00FF00 #C048C0 #F08080 #00FF00 #C048C0 #F09048 #15A4A4 #80A0F0 #80A0F0 #F01505 #80A0F0 #80A0F0 #FFFF00 #00FF00 #00FF00 #80A0F0 #15A4A4 #80A0F0 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
3 | Zappo #FFAFAF #6464FF #00FF00 #FF0000 #FFFF00 #00FF00 #FF0000 #FF00FF #6464FF #FFAFAF #FFAFAF #6464FF #FFAFAF #FFC800 #FF00FF #00FF00 #00FF00 #FFC800 #FFC800 #FFAFAF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
4 | Taylor #CCFF00 #0000FF #CC00FF #FF0000 #FFFF00 #FF00CC #FF0066 #FF9900 #0066FF #66FF00 #33FF00 #6600FF #00FF00 #00FF66 #FFCC00 #FF3300 #FF6600 #00CCFF #00FFCC #99FF00 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
5 | Flower #B18A51 #83BFF1 #38CEC6 #29A578 #F85604 #7295AE #2DA0A1 #B1C23C #2E94F9 #F27663 #DF6E75 #7FC3D7 #FA9DB0 #F9559D #4FA32A #B4BD9B #D2B576 #F92CED #C96ECF #FA997B #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
6 | Blossom #8BC4B4 #F99504 #B5C207 #5FA504 #2E93FE #BF8526 #DBB501 #36D382 #F85604 #9ABAF3 #CDA5DC #FAA527 #F5A1B8 #F74FA8 #35D631 #7E9D59 #2AA39B #F907FB #F84E7A #87C0E4 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
7 | Sunset #FBA0FD #85746A #ABC8F4 #2E7BBE #F90BFE #8C6E81 #677892 #3099FF #DBC58E #F821A1 #E01E82 #DEBECC #D13E7B #F8375D #5766F9 #E7B4FD #A658B7 #F83704 #CB5339 #F951B8 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
8 | Ocean #C6CA9B #2BA0A8 #3DDFC3 #4CDFA1 #C68136 #8BD3D1 #60DAC9 #33A551 #3CCFFE #F2BAAA #BB8A83 #40A090 #A48B88 #AB88AF #AFD364 #6D9B74 #8D9566 #758AEE #BAC3FC #E9BEA4 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
9 | Hydrophobicity #AD0052 #0000FF #0C00F3 #0C00F3 #C2003D #0C00F3 #0C00F3 #6A0095 #1500EA #FF0000 #EA0015 #0000FF #B0004F #CB0034 #4600B9 #5E00A1 #61009E #5B00A4 #4F00B0 #F60009 #0C00F3 #680097 #0C00F3 #FFFFFF #FFFFFF #FFFFFF #FFFFFF
10 | HelixPropensity #E718E7 #6F906F #39E41A #778877 #37DC23 #926D92 #F905FF #41FF00 #758A75 #8A758A #AE51AE #A05FA0 #EF10EF #986798 #41FF00 #36C936 #47B847 #8A758A #38DE20 #857A85 #49B649 #758A75 #C936C9 #FFFFFF #FFFFFF #FFFFFF #FFFFFF
11 | StrandPropensity #5858A7 #6B6B94 #64649C #2121DE #9D9D62 #8C8C73 #1D00FF #4949B6 #60609F #ECEC14 #B2B24D #4747B8 #82827D #C2C23C #2323DB #4949B6 #9D9D62 #C0C03E #D3D32C #FEFF03 #4343BC #797986 #4747B8 #FFFFFF #FFFFFF #FFFFFF #FFFFFF
12 | TurnPropensity #3AD3D3 #708F8F #F81502 #E81717 #A85757 #3FC0C0 #778888 #F81502 #708F8F #48FEFF #40E3E3 #7E8181 #3FE1E1 #3FE1E1 #F61507 #E11E1E #738C8C #738C8C #9D6262 #46F8F8 #F3140B #7C8383 #5BA4A4 #FFFFFF #FFFFFF #FFFFFF #FFFFFF
13 | BuriedIndex #28A35C #40FC03 #3BEB13 #3BEB13 #1D00FF #3DF10D #3DF10D #269D62 #35D529 #1954AB #1F7C84 #41FF00 #259768 #218778 #38E01F #35D529 #37DB23 #29A857 #3AE619 #1A5FA0 #3BEB13 #2DB649 #3DF10D #FFFFFF #FFFFFF #FFFFFF #FFFFFF
14 | Nucleotide #64F740 #FFFFFF #FFFFFF #FFFFFF #FAB340 #FFFFFF #FFFFFF #EB413B #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #3C88EE #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #3C88EE #FFFFFF
15 | Purine/Pyrimidine #FA82FA #FA82FA #FFFFFF #FFFFFF #40E0D0 #FFFFFF #FFFFFF #FA82FA #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #40E0D0 #FFFFFF #40E0D0 #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #40E0D0 #FFFFFF
16 | Identity #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
17 | None #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF #FFFFFF
18 |
--------------------------------------------------------------------------------
/tests/test_msaviz.py:
--------------------------------------------------------------------------------
1 | from pathlib import Path
2 |
3 | import pytest
4 | from Bio.Align import MultipleSeqAlignment
5 | from Bio.Seq import Seq
6 | from Bio.SeqRecord import SeqRecord
7 |
8 | from pymsaviz import MsaViz, get_msa_testdata
9 |
10 |
11 | def test_simple_all_run(msa_fasta_file: Path, tmp_path: Path):
12 | """Test simple all run (Only check if no error occurs)"""
13 | mv = MsaViz(msa_fasta_file)
14 |
15 | fig_outfile = tmp_path / "test.png"
16 | mv.savefig(fig_outfile)
17 |
18 | assert fig_outfile.exists()
19 |
20 |
21 | def test_all_run_with_options(msa_fasta_file: Path, tmp_path: Path):
22 | """Test all run with options (Only check if no error occurs)"""
23 | mv = MsaViz(
24 | msa_fasta_file,
25 | color_scheme="Identity",
26 | wrap_length=50,
27 | show_label=False,
28 | show_seq_char=False,
29 | sort=True,
30 | )
31 | mv.set_highlight_pos([1, 5, (10, 13), 18])
32 | mv.set_highlight_pos_by_ident_thr(min_thr=80, max_thr=100)
33 | mv.add_markers([50, 51, 52, (60, 70), 80], marker="x", color="blue", size=6)
34 | mv.add_text_annotation(
35 | (100, 120), text="test", text_color="blue", text_size=10, range_color="blue"
36 | )
37 |
38 | fig_outfile = tmp_path / "test.png"
39 | mv.savefig(fig_outfile)
40 |
41 | assert fig_outfile.exists()
42 |
43 |
44 | def test_basic_property():
45 | """Test basic property"""
46 | msa = MultipleSeqAlignment([])
47 | id_list = ["first", "second", "third", "fourth"]
48 | seq_list = [
49 | "CDNIPGFED",
50 | "ADNIPGFED",
51 | "BDNIPGFED",
52 | "DDNIPGFED",
53 | ]
54 | for id, seq in zip(id_list, seq_list):
55 | msa.append(SeqRecord(Seq(seq), id=id))
56 |
57 | mv = MsaViz(msa)
58 | assert mv.msa_count == 4
59 | assert mv.alignment_length == 9
60 | assert mv.id_list == id_list
61 | assert mv.seq_list == seq_list
62 | assert mv.wrap_num == 0
63 | assert mv.consensus_seq == "XDNIPGFED"
64 |
65 |
66 | def test_set_custom_color_scheme(dummy_msa: MultipleSeqAlignment):
67 | """Test set_custom_color_scheme"""
68 | mv = MsaViz(dummy_msa)
69 | # Case1: Set correct custom color scheme
70 | custom_color_scheme = {"A": "red", "T": "blue", "G": "green", "C": "orange"}
71 | mv.set_custom_color_scheme(custom_color_scheme)
72 | assert mv.color_scheme == custom_color_scheme
73 |
74 | # Case2: Set invalid custom color scheme
75 | invalid_color_scheme = {"A": "invalid", "T": "blue", "G": "green", "C": "orange"}
76 | with pytest.raises(ValueError):
77 | mv.set_custom_color_scheme(invalid_color_scheme)
78 |
79 |
80 | def test_set_custom_color_func(msa_fasta_file: Path, tmp_path: Path):
81 | """Test set_custom_color_func"""
82 | mv = MsaViz(msa_fasta_file)
83 |
84 | def custom_color_func(
85 | row_pos: int, col_pos: int, seq_char: str, msa: MultipleSeqAlignment
86 | ) -> str:
87 | if col_pos < 60 and seq_char != "-":
88 | return "salmon"
89 | if col_pos >= 60 and 1 <= row_pos <= 4:
90 | return "lime"
91 | return "white"
92 |
93 | mv.set_custom_color_func(custom_color_func)
94 |
95 | fig_outfile = tmp_path / "test.png"
96 | mv.savefig(fig_outfile)
97 |
98 | assert fig_outfile.exists()
99 |
100 |
101 | def test_consensus_identity():
102 | """Test consensus identity calculation"""
103 | msa = MultipleSeqAlignment([])
104 | # Test MSA summary
105 | # 1: 'ABCDE'(All different char) => 'X' [20 %]
106 | # 2: 'GGGGG'(All 'G') => 'G' [100 %]
107 | # 3: '-----'(All gaps) => 'X' [0 %]
108 | # 4: '--V--'(one char & gaps) => 'V' [20 %]
109 | # 5: '-AAAC'('A' is most common) => 'A' [60 %]
110 | # 6: 'RRTTI'('R' & 'T' is most common) => 'X' [40 %]
111 | # 7: 'XXAX-'('X' is most common) => 'X' [60 %]
112 | seq_list = [
113 | "AG---RX",
114 | "BG--ARX",
115 | "CG-VATA",
116 | "DG--ATX",
117 | "EG--CI-",
118 | ]
119 | for seq in seq_list:
120 | msa.append(SeqRecord(Seq(seq)))
121 |
122 | # Test consensus seq & identity
123 | mv = MsaViz(msa)
124 | assert mv.consensus_seq == "XGXVAXX"
125 | consensus_ident_list = mv._get_consensus_identity_list()
126 | assert consensus_ident_list == [20, 100, 0, 20, 60, 40, 60]
127 |
128 |
129 | def test_is_aa_msa():
130 | """Test `aa` or `nt` MSA check"""
131 | # Case1: AA MSA
132 | aa_msa = MultipleSeqAlignment(
133 | [
134 | SeqRecord(Seq("MFLTALLCRGRI")),
135 | SeqRecord(Seq("MFLT---TRGVI")),
136 | ]
137 | )
138 | assert MsaViz(aa_msa)._is_aa_msa() is True
139 |
140 | # Case2: NT MSA
141 | nt_msa = MultipleSeqAlignment(
142 | [
143 | SeqRecord(Seq("ATGC--TGCA")),
144 | SeqRecord(Seq("AAGCTCTGCA")),
145 | ]
146 | )
147 | assert MsaViz(nt_msa)._is_aa_msa() is False
148 |
149 |
150 | def test_parse_positions(dummy_msa: MultipleSeqAlignment):
151 | """Test parse_positions"""
152 | mv = MsaViz(dummy_msa)
153 | # Case1: int value
154 | assert mv._parse_positions([1]) == [0]
155 | # Case2: int values
156 | assert mv._parse_positions([1, 5, 10, 20]) == [0, 4, 9, 19]
157 | # Case3: tuple range
158 | assert mv._parse_positions([(5, 9)]) == [4, 5, 6, 7, 8]
159 | # Case4: int values & tuple range
160 | assert mv._parse_positions([1, 5, (10, 13), 18]) == [0, 4, 9, 10, 11, 12, 17]
161 |
162 |
163 | def test_get_msa_testdata():
164 | """Test get_msa_testdata"""
165 | assert get_msa_testdata().exists()
166 | assert get_msa_testdata("HIGD2A.fa").exists()
167 | with pytest.raises(ValueError):
168 | get_msa_testdata("invalid_name")
169 |
--------------------------------------------------------------------------------
/src/pymsaviz/scripts/cli.py:
--------------------------------------------------------------------------------
1 | from __future__ import annotations
2 |
3 | import argparse
4 | from pathlib import Path
5 |
6 | from pymsaviz import MsaViz, __version__
7 |
8 |
9 | def main():
10 | """Main function called from CLI"""
11 | args = get_args()
12 | run(**args.__dict__)
13 |
14 |
15 | def run(
16 | infile: str | Path,
17 | outfile: str | Path,
18 | format: str = "fasta",
19 | color_scheme: str = "Zappo",
20 | start: int = 1,
21 | end: int | None = None,
22 | wrap_length: int | None = None,
23 | wrap_space_size: float = 3.0,
24 | label_type: str = "id",
25 | show_grid: bool = False,
26 | show_count: bool = False,
27 | show_consensus: bool = False,
28 | consensus_color: str = "#1f77b4",
29 | consensus_size: float = 2.0,
30 | sort: bool = False,
31 | dpi: int = 300,
32 | ):
33 | """Run MSA visualization"""
34 | mv = MsaViz(
35 | msa=infile,
36 | format=format,
37 | start=start,
38 | end=end,
39 | wrap_length=wrap_length,
40 | wrap_space_size=wrap_space_size,
41 | label_type=label_type,
42 | color_scheme=color_scheme,
43 | show_grid=show_grid,
44 | show_count=show_count,
45 | show_consensus=show_consensus,
46 | consensus_color=consensus_color,
47 | consensus_size=consensus_size,
48 | sort=sort,
49 | )
50 | mv.savefig(outfile, dpi=dpi)
51 |
52 |
53 | def get_args() -> argparse.Namespace:
54 | """Get arguments
55 |
56 | Returns
57 | -------
58 | args : argparse.Namespace
59 | Argument parameters
60 | """
61 | description = "MSA(Multiple Sequence Alignment) visualization CLI tool"
62 | parser = argparse.ArgumentParser(
63 | description=description,
64 | usage="pymsaviz [options] -i msa.fa -o msa_viz.png",
65 | add_help=False,
66 | epilog=f"Available Color Schemes:\n{MsaViz.available_color_schemes()}",
67 | formatter_class=argparse.RawDescriptionHelpFormatter,
68 | )
69 |
70 | parser.add_argument(
71 | "-i",
72 | "--infile",
73 | type=Path,
74 | help="Input MSA file",
75 | metavar="I",
76 | )
77 | parser.add_argument(
78 | "-o",
79 | "--outfile",
80 | type=Path,
81 | help="Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)",
82 | required=True,
83 | metavar="O",
84 | )
85 | default_msa_format = "fasta"
86 | parser.add_argument(
87 | "--format",
88 | type=str,
89 | help=f"MSA file format (Default: '{default_msa_format}')",
90 | default=default_msa_format,
91 | metavar="",
92 | )
93 | default_color_scheme = "Zappo"
94 | parser.add_argument(
95 | "--color_scheme",
96 | type=str,
97 | help=f"Color scheme (Default: '{default_color_scheme}')",
98 | default=default_color_scheme,
99 | choices=MsaViz.available_color_schemes(),
100 | metavar="",
101 | )
102 | default_start = 1
103 | parser.add_argument(
104 | "--start",
105 | type=int,
106 | help=f"Start position of MSA visualization (Default: {default_start})",
107 | default=default_start,
108 | metavar="",
109 | )
110 | default_end = None
111 | parser.add_argument(
112 | "--end",
113 | type=int,
114 | help="End position of MSA visualization (Default: 'MSA Length')",
115 | default=default_end,
116 | metavar="",
117 | )
118 | default_wrap_length = None
119 | parser.add_argument(
120 | "--wrap_length",
121 | type=int,
122 | help=f"Wrap length (Default: {default_wrap_length})",
123 | default=default_wrap_length,
124 | metavar="",
125 | )
126 | default_wrap_space_size = 3.0
127 | parser.add_argument(
128 | "--wrap_space_size",
129 | type=float,
130 | help="Space size between wrap MSA plot area "
131 | f"(Default: {default_wrap_space_size})",
132 | default=default_wrap_space_size,
133 | metavar="",
134 | )
135 | default_label_type = "id"
136 | parser.add_argument(
137 | "--label_type",
138 | type=str,
139 | help="Label type ('id'[default]|'description')",
140 | default=default_label_type,
141 | choices=("id", "description"),
142 | metavar="",
143 | )
144 | parser.add_argument(
145 | "--show_grid",
146 | help="Show grid (Default: OFF)",
147 | action="store_true",
148 | )
149 | parser.add_argument(
150 | "--show_count",
151 | help="Show seq char count without gap on right side (Default: OFF)",
152 | action="store_true",
153 | )
154 | parser.add_argument(
155 | "--show_consensus",
156 | help="Show consensus sequence (Default: OFF)",
157 | action="store_true",
158 | )
159 | default_consensus_color = "#1f77b4"
160 | parser.add_argument(
161 | "--consensus_color",
162 | type=str,
163 | help=f"Consensus identity bar color (Default: '{default_consensus_color}')",
164 | default=default_consensus_color,
165 | metavar="",
166 | )
167 | default_consensus_size = 2.0
168 | parser.add_argument(
169 | "--consensus_size",
170 | type=float,
171 | help=f"Consensus identity bar height size (Default: {default_consensus_size})",
172 | default=default_consensus_size,
173 | metavar="",
174 | )
175 | parser.add_argument(
176 | "--sort",
177 | help="Sort MSA order by NJ tree constructed from MSA distance matrix "
178 | "(Default: OFF)",
179 | action="store_true",
180 | )
181 | default_dpi = 300
182 | parser.add_argument(
183 | "--dpi",
184 | type=int,
185 | help=f"Figure DPI (Default: {default_dpi})",
186 | default=default_dpi,
187 | metavar="",
188 | )
189 | parser.add_argument(
190 | "-v",
191 | "--version",
192 | version=f"v{__version__}",
193 | help="Print version information",
194 | action="version",
195 | )
196 | parser.add_argument(
197 | "-h",
198 | "--help",
199 | help="Show this help message and exit",
200 | action="help",
201 | )
202 | return parser.parse_args()
203 |
204 |
205 | if __name__ == "__main__":
206 | main()
207 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # pyMSAviz
2 |
3 | 
4 | 
5 | 
6 | [](https://pypi.python.org/pypi/pymsaviz)
7 | [](https://anaconda.org/bioconda/pymsaviz)
8 | [](https://github.com/moshi4/pyMSAviz/actions/workflows/ci.yml)
9 |
10 | ## Table of contents
11 |
12 | - [Overview](#overview)
13 | - [Installation](#installation)
14 | - [API Usage](#api-usage)
15 | - [CLI Usage](#cli-usage)
16 |
17 | ## Overview
18 |
19 | pyMSAviz is a MSA(Multiple Sequence Alignment) visualization python package for sequence analysis implemented based on matplotlib.
20 | This package is developed for the purpose of easily and beautifully plotting MSA in Python.
21 | It also implements the functionality to add markers, text annotations, highlights to specific positions and ranges in MSA.
22 | pyMSAviz was developed inspired by [Jalview](https://www.jalview.org/) and [ggmsa](https://github.com/YuLab-SMU/ggmsa).
23 | More detailed documentation is available [here](https://moshi4.github.io/pyMSAviz/).
24 |
25 | 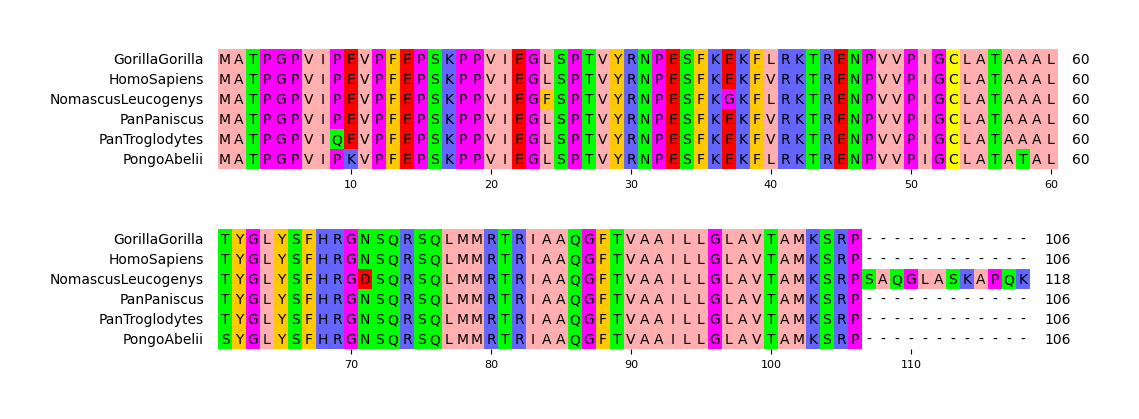
26 | **Fig.1 Simple visualization result**
27 |
28 | 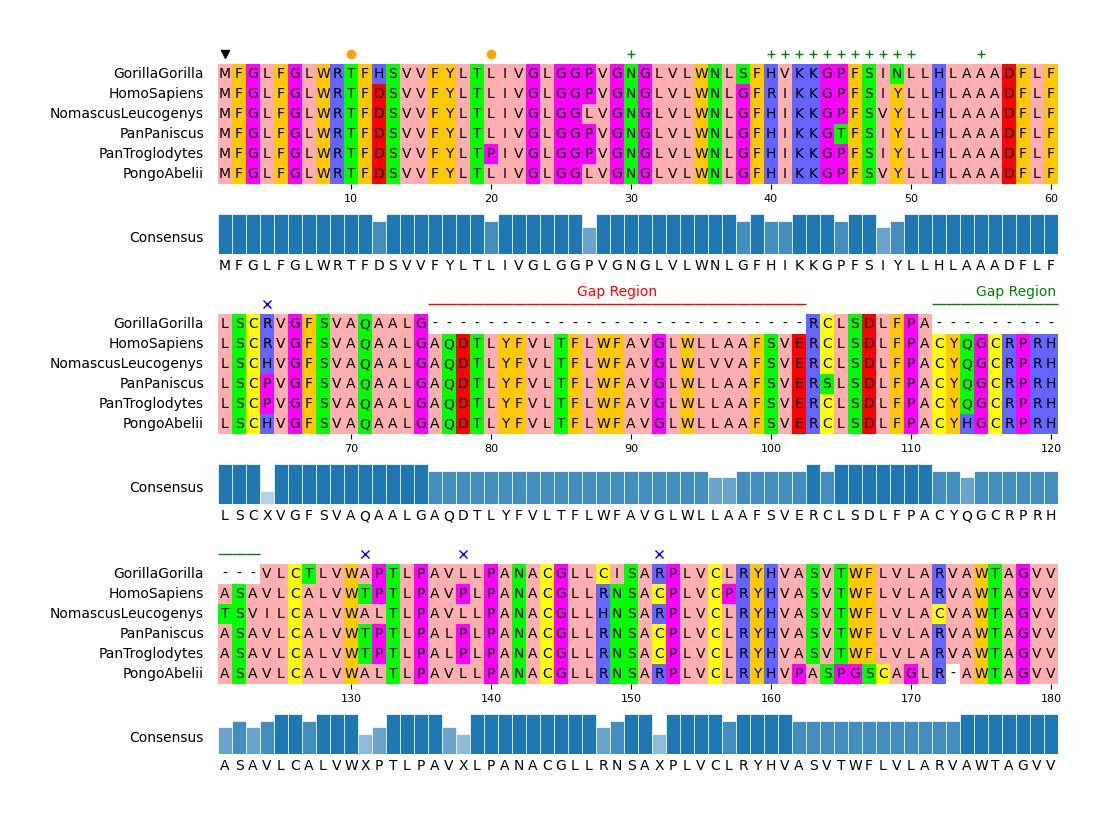
29 | **Fig.2 Customized visualization result**
30 |
31 | ## Installation
32 |
33 | `Python 3.9 or later` is required for installation.
34 |
35 | **Install PyPI package:**
36 |
37 | pip install pymsaviz
38 |
39 | **Install bioconda package:**
40 |
41 | conda install -c conda-forge -c bioconda pymsaviz
42 |
43 | ## API Usage
44 |
45 | Only simple example usage is described in this section.
46 | For more details, please see [Getting Started](https://moshi4.github.io/pyMSAviz/getting_started/) and [API Docs](https://moshi4.github.io/pyMSAviz/api-docs/msaviz/).
47 |
48 | ### API Example
49 |
50 | #### API Example 1
51 |
52 | ```python
53 | from pymsaviz import MsaViz, get_msa_testdata
54 |
55 | msa_file = get_msa_testdata("HIGD2A.fa")
56 | mv = MsaViz(msa_file, wrap_length=60, show_count=True)
57 | mv.savefig("api_example01.png")
58 | ```
59 |
60 | 
61 |
62 | #### API Example 2
63 |
64 | ```python
65 | from pymsaviz import MsaViz, get_msa_testdata
66 |
67 | msa_file = get_msa_testdata("MRGPRG.fa")
68 | mv = MsaViz(msa_file, color_scheme="Taylor", wrap_length=80, show_grid=True, show_consensus=True)
69 | mv.savefig("api_example02.png")
70 | ```
71 |
72 | 
73 |
74 | #### API Example 3
75 |
76 | ```python
77 | from pymsaviz import MsaViz, get_msa_testdata
78 |
79 | msa_file = get_msa_testdata("MRGPRG.fa")
80 | mv = MsaViz(msa_file, end=180, wrap_length=60, show_consensus=True)
81 |
82 | # Extract MSA positions less than 50% consensus identity
83 | pos_ident_less_than_50 = []
84 | ident_list = mv._get_consensus_identity_list()
85 | for pos, ident in enumerate(ident_list, 1):
86 | if ident <= 50:
87 | pos_ident_less_than_50.append(pos)
88 |
89 | # Add markers
90 | mv.add_markers([1])
91 | mv.add_markers([10, 20], color="orange", marker="o")
92 | mv.add_markers([30, (40, 50), 55], color="green", marker="+")
93 | mv.add_markers(pos_ident_less_than_50, marker="x", color="blue")
94 | # Add text annotations
95 | mv.add_text_annotation((76, 102), "Gap Region", text_color="red", range_color="red")
96 | mv.add_text_annotation((112, 123), "Gap Region", text_color="green", range_color="green")
97 |
98 | mv.savefig("api_example03.png")
99 | ```
100 |
101 | 
102 |
103 | ## CLI Usage
104 |
105 | pyMSAviz provides simple MSA visualization CLI.
106 |
107 | ### Basic Command
108 |
109 | pymsaviz -i [MSA file] -o [MSA visualization file]
110 |
111 | ### Options
112 |
113 | $ pymsaviz --help
114 | usage: pymsaviz [options] -i msa.fa -o msa_viz.png
115 |
116 | MSA(Multiple Sequence Alignment) visualization CLI tool
117 |
118 | optional arguments:
119 | -i I, --infile I Input MSA file
120 | -o O, --outfile O Output MSA visualization file (*.png|*.jpg|*.svg|*.pdf)
121 | --format MSA file format (Default: 'fasta')
122 | --color_scheme Color scheme (Default: 'Zappo')
123 | --start Start position of MSA visualization (Default: 1)
124 | --end End position of MSA visualization (Default: 'MSA Length')
125 | --wrap_length Wrap length (Default: None)
126 | --wrap_space_size Space size between wrap MSA plot area (Default: 3.0)
127 | --label_type Label type ('id'[default]|'description')
128 | --show_grid Show grid (Default: OFF)
129 | --show_count Show seq char count without gap on right side (Default: OFF)
130 | --show_consensus Show consensus sequence (Default: OFF)
131 | --consensus_color Consensus identity bar color (Default: '#1f77b4')
132 | --consensus_size Consensus identity bar height size (Default: 2.0)
133 | --sort Sort MSA order by NJ tree constructed from MSA distance matrix (Default: OFF)
134 | --dpi Figure DPI (Default: 300)
135 | -v, --version Print version information
136 | -h, --help Show this help message and exit
137 |
138 | Available Color Schemes:
139 | ['Clustal', 'Zappo', 'Taylor', 'Flower', 'Blossom', 'Sunset', 'Ocean', 'Hydrophobicity', 'HelixPropensity', 'StrandPropensity', 'TurnPropensity', 'BuriedIndex', 'Nucleotide', 'Purine/Pyrimidine', 'Identity', 'None']
140 |
141 | ### CLI Example
142 |
143 | Click [here](https://github.com/moshi4/pyMSAviz/raw/main/example/example.zip) to download example MSA files.
144 |
145 | #### CLI Example 1
146 |
147 | pymsaviz -i ./example/HIGD2A.fa -o cli_example01.png --color_scheme Identity
148 |
149 | 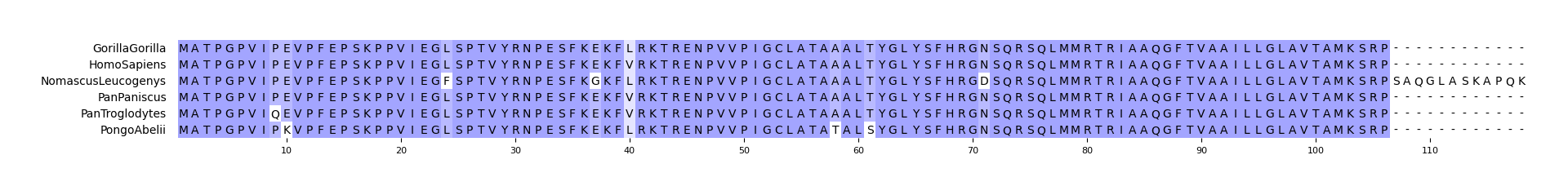
150 |
151 | #### CLI Example 2
152 |
153 | pymsaviz -i ./example/MRGPRG.fa -o cli_example02.png --wrap_length 80 \
154 | --color_scheme Taylor --show_consensus --show_count
155 |
156 | 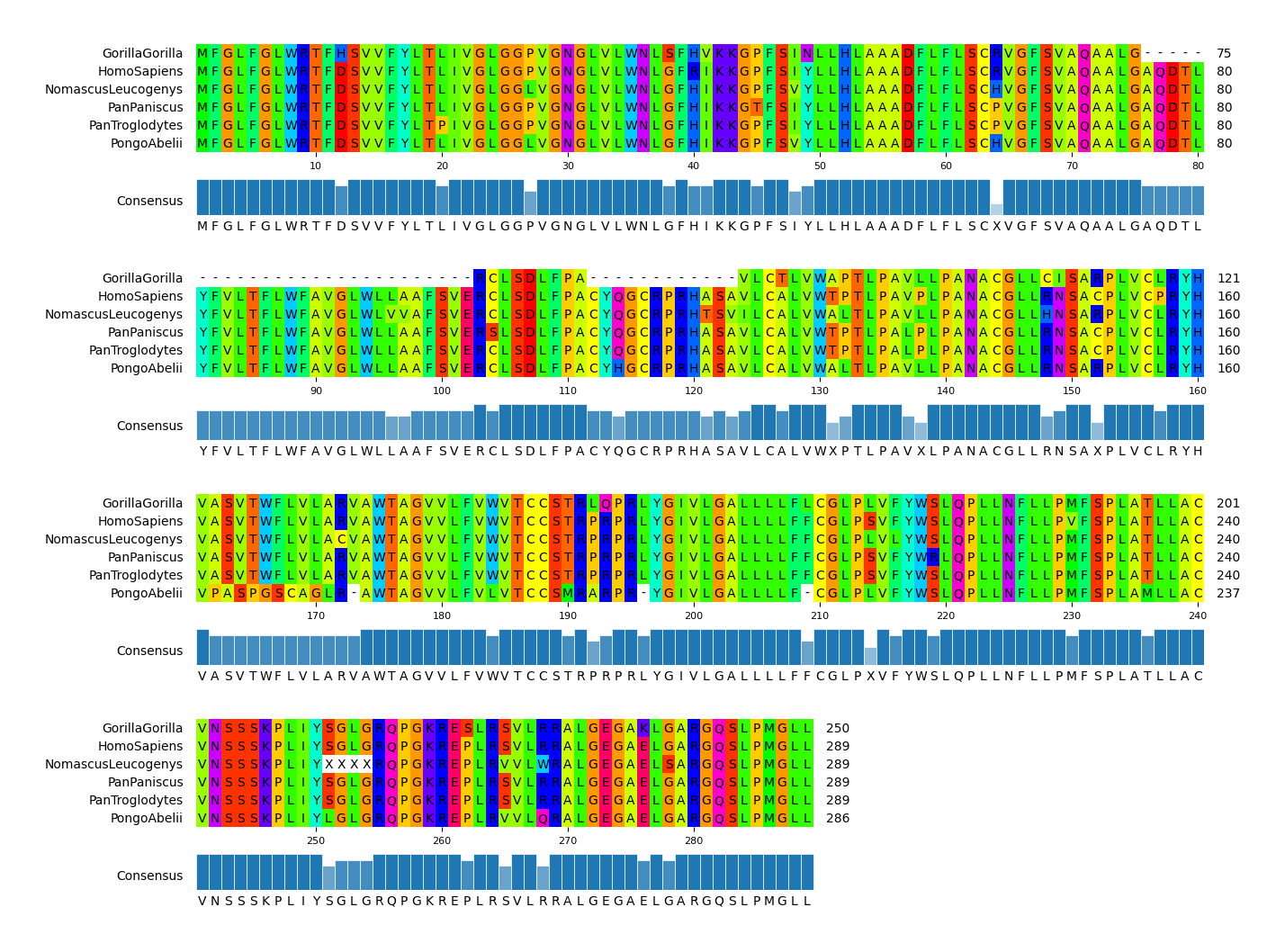
157 |
158 | #### CLI Example 3
159 |
160 | pymsaviz -i ./example/MRGPRG.fa -o cli_example03.png --start 100 --end 160 \
161 | --color_scheme Flower --show_grid --show_consensus --consensus_color tomato
162 |
163 | 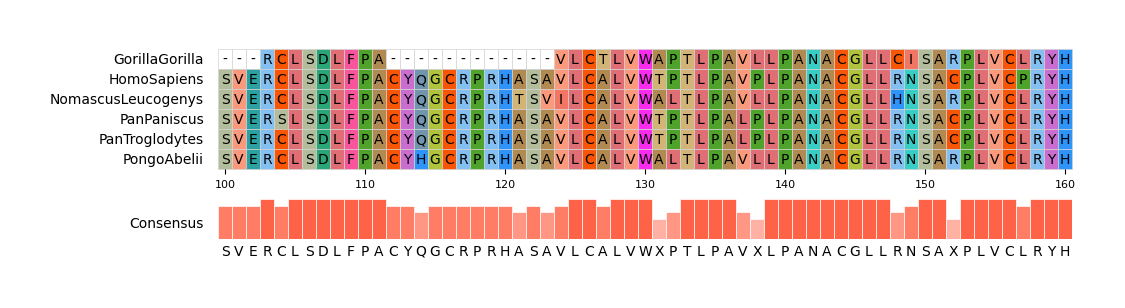
164 |
165 | ## Star History
166 |
167 | [](https://star-history.com/#moshi4/pyMSAviz&Date)
168 |
--------------------------------------------------------------------------------
/requirements-dev.lock:
--------------------------------------------------------------------------------
1 | # generated by rye
2 | # use `rye lock` or `rye sync` to update this lockfile
3 | #
4 | # last locked with the following flags:
5 | # pre: false
6 | # features: []
7 | # all-features: true
8 | # with-sources: false
9 | # generate-hashes: false
10 | # universal: false
11 |
12 | -e file:.
13 | asttokens==3.0.0
14 | # via stack-data
15 | attrs==24.2.0
16 | # via jsonschema
17 | # via referencing
18 | babel==2.16.0
19 | # via mkdocs-material
20 | beautifulsoup4==4.12.3
21 | # via nbconvert
22 | biopython==1.84
23 | # via pymsaviz
24 | black==24.10.0
25 | bleach==6.2.0
26 | # via nbconvert
27 | certifi==2024.8.30
28 | # via requests
29 | cfgv==3.4.0
30 | # via pre-commit
31 | charset-normalizer==3.4.0
32 | # via requests
33 | click==8.1.7
34 | # via black
35 | # via mkdocs
36 | # via mkdocstrings
37 | colorama==0.4.6
38 | # via griffe
39 | # via mkdocs-material
40 | comm==0.2.2
41 | # via ipykernel
42 | contourpy==1.3.0
43 | # via matplotlib
44 | coverage==7.6.8
45 | # via pytest-cov
46 | cycler==0.12.1
47 | # via matplotlib
48 | debugpy==1.8.9
49 | # via ipykernel
50 | decorator==5.1.1
51 | # via ipython
52 | defusedxml==0.7.1
53 | # via nbconvert
54 | distlib==0.3.9
55 | # via virtualenv
56 | exceptiongroup==1.2.2
57 | # via ipython
58 | # via pytest
59 | executing==2.1.0
60 | # via stack-data
61 | fastjsonschema==2.21.0
62 | # via nbformat
63 | filelock==3.16.1
64 | # via virtualenv
65 | fonttools==4.55.0
66 | # via matplotlib
67 | ghp-import==2.1.0
68 | # via mkdocs
69 | griffe==1.5.1
70 | # via mkdocstrings-python
71 | identify==2.6.3
72 | # via pre-commit
73 | idna==3.10
74 | # via requests
75 | importlib-metadata==8.5.0
76 | # via jupyter-client
77 | # via markdown
78 | # via mkdocs
79 | # via mkdocs-get-deps
80 | # via mkdocstrings
81 | # via nbconvert
82 | importlib-resources==6.4.5
83 | # via matplotlib
84 | iniconfig==2.0.0
85 | # via pytest
86 | ipykernel==6.29.5
87 | # via mkdocs-jupyter
88 | ipython==8.18.1
89 | # via ipykernel
90 | jedi==0.19.2
91 | # via ipython
92 | jinja2==3.1.4
93 | # via mkdocs
94 | # via mkdocs-material
95 | # via mkdocstrings
96 | # via nbconvert
97 | jsonschema==4.23.0
98 | # via nbformat
99 | jsonschema-specifications==2024.10.1
100 | # via jsonschema
101 | jupyter-client==8.6.3
102 | # via ipykernel
103 | # via nbclient
104 | jupyter-core==5.7.2
105 | # via ipykernel
106 | # via jupyter-client
107 | # via nbclient
108 | # via nbconvert
109 | # via nbformat
110 | jupyterlab-pygments==0.3.0
111 | # via nbconvert
112 | jupytext==1.16.4
113 | # via mkdocs-jupyter
114 | kiwisolver==1.4.7
115 | # via matplotlib
116 | markdown==3.7
117 | # via mkdocs
118 | # via mkdocs-autorefs
119 | # via mkdocs-material
120 | # via mkdocstrings
121 | # via pymdown-extensions
122 | markdown-it-py==3.0.0
123 | # via jupytext
124 | # via mdit-py-plugins
125 | markupsafe==3.0.2
126 | # via jinja2
127 | # via mkdocs
128 | # via mkdocs-autorefs
129 | # via mkdocstrings
130 | # via nbconvert
131 | matplotlib==3.9.3
132 | # via pymsaviz
133 | matplotlib-inline==0.1.7
134 | # via ipykernel
135 | # via ipython
136 | mdit-py-plugins==0.4.2
137 | # via jupytext
138 | mdurl==0.1.2
139 | # via markdown-it-py
140 | mergedeep==1.3.4
141 | # via mkdocs
142 | # via mkdocs-get-deps
143 | mistune==3.0.2
144 | # via nbconvert
145 | mkdocs==1.6.1
146 | # via mkdocs-autorefs
147 | # via mkdocs-jupyter
148 | # via mkdocs-material
149 | # via mkdocstrings
150 | mkdocs-autorefs==1.2.0
151 | # via mkdocstrings
152 | # via mkdocstrings-python
153 | mkdocs-get-deps==0.2.0
154 | # via mkdocs
155 | mkdocs-jupyter==0.25.1
156 | mkdocs-material==9.5.46
157 | # via mkdocs-jupyter
158 | mkdocs-material-extensions==1.3.1
159 | # via mkdocs-material
160 | mkdocstrings==0.27.0
161 | # via mkdocstrings-python
162 | mkdocstrings-python==1.12.2
163 | # via mkdocstrings
164 | mypy-extensions==1.0.0
165 | # via black
166 | nbclient==0.10.1
167 | # via nbconvert
168 | nbconvert==7.16.4
169 | # via mkdocs-jupyter
170 | nbformat==5.10.4
171 | # via jupytext
172 | # via nbclient
173 | # via nbconvert
174 | nest-asyncio==1.6.0
175 | # via ipykernel
176 | nodeenv==1.9.1
177 | # via pre-commit
178 | numpy==2.0.2
179 | # via biopython
180 | # via contourpy
181 | # via matplotlib
182 | packaging==24.2
183 | # via black
184 | # via ipykernel
185 | # via jupytext
186 | # via matplotlib
187 | # via mkdocs
188 | # via nbconvert
189 | # via pytest
190 | paginate==0.5.7
191 | # via mkdocs-material
192 | pandocfilters==1.5.1
193 | # via nbconvert
194 | parso==0.8.4
195 | # via jedi
196 | pathspec==0.12.1
197 | # via black
198 | # via mkdocs
199 | pexpect==4.9.0
200 | # via ipython
201 | pillow==11.0.0
202 | # via matplotlib
203 | platformdirs==4.3.6
204 | # via black
205 | # via jupyter-core
206 | # via mkdocs-get-deps
207 | # via mkdocstrings
208 | # via virtualenv
209 | pluggy==1.5.0
210 | # via pytest
211 | pre-commit==4.0.1
212 | prompt-toolkit==3.0.48
213 | # via ipython
214 | psutil==6.1.0
215 | # via ipykernel
216 | ptyprocess==0.7.0
217 | # via pexpect
218 | pure-eval==0.2.3
219 | # via stack-data
220 | pygments==2.18.0
221 | # via ipython
222 | # via mkdocs-jupyter
223 | # via mkdocs-material
224 | # via nbconvert
225 | pymdown-extensions==10.12
226 | # via mkdocs-material
227 | # via mkdocstrings
228 | pyparsing==3.2.0
229 | # via matplotlib
230 | pytest==8.3.3
231 | # via pytest-cov
232 | pytest-cov==6.0.0
233 | python-dateutil==2.9.0.post0
234 | # via ghp-import
235 | # via jupyter-client
236 | # via matplotlib
237 | pyyaml==6.0.2
238 | # via jupytext
239 | # via mkdocs
240 | # via mkdocs-get-deps
241 | # via pre-commit
242 | # via pymdown-extensions
243 | # via pyyaml-env-tag
244 | pyyaml-env-tag==0.1
245 | # via mkdocs
246 | pyzmq==26.2.0
247 | # via ipykernel
248 | # via jupyter-client
249 | referencing==0.35.1
250 | # via jsonschema
251 | # via jsonschema-specifications
252 | regex==2024.11.6
253 | # via mkdocs-material
254 | requests==2.32.3
255 | # via mkdocs-material
256 | rpds-py==0.21.0
257 | # via jsonschema
258 | # via referencing
259 | ruff==0.8.1
260 | six==1.16.0
261 | # via python-dateutil
262 | soupsieve==2.6
263 | # via beautifulsoup4
264 | stack-data==0.6.3
265 | # via ipython
266 | tinycss2==1.4.0
267 | # via nbconvert
268 | tomli==2.2.1
269 | # via black
270 | # via coverage
271 | # via jupytext
272 | # via pytest
273 | tornado==6.4.2
274 | # via ipykernel
275 | # via jupyter-client
276 | traitlets==5.14.3
277 | # via comm
278 | # via ipykernel
279 | # via ipython
280 | # via jupyter-client
281 | # via jupyter-core
282 | # via matplotlib-inline
283 | # via nbclient
284 | # via nbconvert
285 | # via nbformat
286 | typing-extensions==4.12.2
287 | # via black
288 | # via ipython
289 | # via mkdocstrings
290 | urllib3==2.2.3
291 | # via requests
292 | virtualenv==20.28.0
293 | # via pre-commit
294 | watchdog==6.0.0
295 | # via mkdocs
296 | wcwidth==0.2.13
297 | # via prompt-toolkit
298 | webencodings==0.5.1
299 | # via bleach
300 | # via tinycss2
301 | zipp==3.21.0
302 | # via importlib-metadata
303 | # via importlib-resources
304 |
--------------------------------------------------------------------------------
/src/pymsaviz/msaviz.py:
--------------------------------------------------------------------------------
1 | from __future__ import annotations
2 |
3 | import math
4 | from collections import Counter
5 | from io import StringIO
6 | from pathlib import Path
7 | from typing import Any, Callable
8 | from urllib.parse import urlparse
9 | from urllib.request import urlopen
10 |
11 | import matplotlib.pyplot as plt
12 | from Bio import AlignIO
13 | from Bio.AlignIO import MultipleSeqAlignment as MSA
14 | from Bio.Phylo.BaseTree import Tree
15 | from Bio.Phylo.TreeConstruction import DistanceCalculator, DistanceTreeConstructor
16 | from Bio.SeqRecord import SeqRecord
17 | from matplotlib import colors
18 | from matplotlib.axes import Axes
19 | from matplotlib.collections import PatchCollection
20 | from matplotlib.colors import is_color_like
21 | from matplotlib.figure import Figure
22 | from matplotlib.gridspec import GridSpec
23 | from matplotlib.patches import Rectangle
24 |
25 | from pymsaviz.config import COLOR_SCHEMES, AxesType
26 |
27 |
28 | class MsaViz:
29 | """Multiple Sequence Alignment Visualization"""
30 |
31 | def __init__(
32 | self,
33 | msa: str | Path | MSA,
34 | *,
35 | format: str = "fasta",
36 | color_scheme: str | None = None,
37 | start: int = 1,
38 | end: int | None = None,
39 | wrap_length: int | None = None,
40 | wrap_space_size: float = 3.0,
41 | show_label: bool = True,
42 | label_type: str = "id",
43 | show_seq_char: bool = True,
44 | show_grid: bool = False,
45 | show_count: bool = False,
46 | show_consensus: bool = False,
47 | consensus_color: str = "#1f77b4",
48 | consensus_size: float = 2.0,
49 | sort: bool = False,
50 | ):
51 | """
52 | Parameters
53 | ----------
54 | msa : str | Path | MultipleSeqAlignment
55 | MSA file, URL MSA file, MSA object
56 | format : str, optional
57 | Alignment file format (e.g. `fasta`, `phylip`, `clustal`, `emboss`, etc...)
58 | color_scheme : str | None, optional
59 | Color scheme. If None, `Zappo`(AA) or `Nucleotide`(NT) is set.
60 | [`Clustal`|`Zappo`|`Taylor`|`Flower`|`Blossom`|`Sunset`|`Ocean`|
61 | `Hydrophobicity`|`HelixPropensity`|`StrandPropensity`|`TurnPropensity`|
62 | `BuriedIndex`|`Nucleotide`|`Purine/Pyrimidine`|`Identity`|`None`]
63 | start : int, optional
64 | Start position of visualization (one-based coordinates)
65 | end : int | None, optional
66 | End position of visualization (one-based coordinates)
67 | wrap_length : int | None, optional
68 | Wrap sequence length. If None, no wrapping sequence.
69 | wrap_space_size: float, optional
70 | Space size between wrap MSA plot area
71 | show_label : bool, optional
72 | If True, show label
73 | label_type : str, optional
74 | Label type (`id`|`description`) to be shown when show_label=True.
75 | If `label_type="id"`, show omitted id label.
76 | If `label_type="description"`, show full description label.
77 | show_seq_char : bool, optional
78 | If True, show sequence character
79 | show_grid : bool, optional
80 | If True, show grid
81 | show_count : bool, optional
82 | If True, show seq char count without gap on right side
83 | show_consensus : bool, optional
84 | If True, show consensus sequence
85 | consensus_color : str, optional
86 | Consensus identity bar color
87 | consensus_size : float, optional
88 | Consensus identity bar height size
89 | sort : bool, optional
90 | Sort MSA order by NJ tree constructed from MSA distance matrix
91 | """
92 | # Load MSA
93 | if isinstance(msa, MSA):
94 | self._msa = msa
95 | elif isinstance(msa, str) and urlparse(msa).scheme in ("http", "https"):
96 | content = urlopen(msa).read().decode("utf-8")
97 | self._msa = AlignIO.read(StringIO(content), format)
98 | else:
99 | self._msa: MSA = AlignIO.read(msa, format)
100 | if sort:
101 | self._msa = self._sorted_msa_by_njtree(self._msa)
102 | self._consensus_seq = self._get_consensus_seq(self._msa)
103 | self._color_scheme_name = color_scheme
104 |
105 | # Check & Set start, end position
106 | end = self.alignment_length if end is None else end
107 | if not 1 <= start <= end <= self.alignment_length:
108 | err_msg = f"{start=}, {end=} is invalid MSA range "
109 | err_msg += f"(1 <= start <= end <= {self.alignment_length})"
110 | raise ValueError(err_msg)
111 | self._start, self._end = start - 1, end
112 | self._length = self._end - self._start
113 |
114 | # Set user-specified plot configs
115 | if wrap_length in (0, None) or wrap_length > self._length:
116 | self._wrap_length = self._length
117 | else:
118 | self._wrap_length = wrap_length
119 | self._wrap_space_size = wrap_space_size
120 | self._show_seq_char = show_seq_char
121 | self._show_label = show_label
122 | self._label_type = label_type
123 | self._show_grid = show_grid
124 | self._show_count = show_count
125 | self._show_consensus = show_consensus
126 | self._consensus_color = consensus_color
127 | self._consensus_size = consensus_size
128 | self._highlight_positions = None
129 | self._custom_color_func: Callable[[int, int, str, MSA], str | None] | None = (
130 | None
131 | )
132 | self._pos2marker_kws: dict[int, dict[str, Any]] = {}
133 | self._pos2text_kws: dict[int, dict[str, Any]] = {}
134 | self.set_plot_params()
135 |
136 | # Set color scheme
137 | if color_scheme is None:
138 | color_scheme = "Zappo" if self._is_aa_msa() else "Nucleotide"
139 | if color_scheme not in self.available_color_schemes():
140 | err_msg = f"{color_scheme=} is invalid.\n"
141 | err_msg += f"Available color scheme = {self.available_color_schemes()}"
142 | raise ValueError(err_msg)
143 | self._color_scheme = COLOR_SCHEMES[color_scheme]
144 |
145 | ############################################################
146 | # Property
147 | ############################################################
148 |
149 | @property
150 | def msa(self) -> MSA:
151 | """Multiple Sequence Alignment object (BioPython)"""
152 | return self._msa
153 |
154 | @property
155 | def msa_count(self) -> int:
156 | """MSA count"""
157 | return len(self._msa)
158 |
159 | @property
160 | def alignment_length(self) -> int:
161 | """Alignment length"""
162 | return self._msa.get_alignment_length()
163 |
164 | @property
165 | def id_list(self) -> list[str]:
166 | """MSA ID list"""
167 | return [rec.id for rec in self._msa]
168 |
169 | @property
170 | def desc_list(self) -> list[str]:
171 | """MSA description list"""
172 | return [rec.description for rec in self._msa]
173 |
174 | @property
175 | def seq_list(self) -> list[str]:
176 | """MSA sequence list"""
177 | return [str(rec.seq) for rec in self._msa]
178 |
179 | @property
180 | def wrap_num(self) -> int:
181 | """Wrap number"""
182 | if self._wrap_length is None:
183 | return 0
184 | else:
185 | return math.ceil(self._length / self._wrap_length) - 1
186 |
187 | @property
188 | def consensus_seq(self) -> str:
189 | """Consensus sequence"""
190 | return self._consensus_seq
191 |
192 | @property
193 | def color_scheme(self) -> dict[str, str]:
194 | """Color scheme"""
195 | return self._color_scheme
196 |
197 | ############################################################
198 | # Public Method
199 | ############################################################
200 |
201 | @staticmethod
202 | def available_color_schemes() -> list[str]:
203 | """Get available color schemes
204 |
205 | Returns
206 | -------
207 | color_scheme_names : list[str]
208 | Available color schemes
209 | """
210 | return list(COLOR_SCHEMES.keys())
211 |
212 | def set_plot_params(
213 | self,
214 | *,
215 | ticks_interval: int | None = 10,
216 | x_unit_size: float = 0.14,
217 | y_unit_size: float = 0.20,
218 | grid_color: str = "lightgrey",
219 | show_consensus_char: bool = True,
220 | identity_color: str = "#A3A5FF",
221 | identity_color_min_thr: float = 30,
222 | ) -> None:
223 | """Set plot parameters to adjust figure appearence in detail
224 |
225 | Parameters
226 | ----------
227 | ticks_interval : int | None, optional
228 | Ticks interval. If None, ticks interval is not displayed.
229 | x_unit_size : float, optional
230 | X-axis unit size of seq char rectangle
231 | y_unit_size : float, optional
232 | Y-axis unit size of seq char rectangle
233 | grid_color : str, optional
234 | Grid color
235 | show_consensus_char : bool, optional
236 | If True, show consensus character
237 | identity_color : str, optional
238 | Base color for `Identity` color scheme
239 | identity_color_min_thr : float, optional
240 | Min identity color threshold for `Identity` color scheme
241 | """
242 | self._ticks_interval = ticks_interval
243 | self._x_unit_size = x_unit_size
244 | self._y_unit_size = y_unit_size
245 | self._grid_color = grid_color
246 | self._show_consensus_char = show_consensus_char
247 | self._identity_color = identity_color
248 | self._identity_color_min_thr = identity_color_min_thr
249 |
250 | def set_custom_color_scheme(self, color_scheme: dict[str, str]) -> None:
251 | """Set user-defined custom color scheme (Overwrite color scheme setting)
252 |
253 | Parameters
254 | ----------
255 | color_scheme : dict[str, str]
256 | Custom color scheme dict (e.g. `{"A": "red", "R": "#F01505", ...}`)
257 | """
258 | if isinstance(color_scheme, dict):
259 | if not all(map(is_color_like, color_scheme.values())):
260 | raise ValueError(f"{color_scheme=} contains invalid color code.")
261 | self._color_scheme = color_scheme
262 | else:
263 | raise ValueError(f"{color_scheme=} is not dict type.")
264 |
265 | def set_custom_color_func(
266 | self,
267 | custom_color_func: Callable[[int, int, str, MSA], str | None],
268 | ):
269 | """Set user-defined custom color func (Overwrite all other color setting)
270 |
271 | User can change the color of each residue specified
272 | by the row and column position of the MSA.
273 |
274 | Parameters
275 | ----------
276 | custom_color_func : Callable[[int, int, str, MSA], str | None]
277 | Custom color function.
278 | `Callable[[int, int, str, MSA], str | None]` means
279 | `Callable[[row_pos, col_pos, seq_char, msa], hexcolor | None]`
280 | """
281 | self._custom_color_func = custom_color_func
282 |
283 | def set_highlight_pos(self, positions: list[tuple[int, int] | int]) -> None:
284 | """Set user-defined highlight MSA positions
285 |
286 | Parameters
287 | ----------
288 | positions : list[tuple[int, int] | int]
289 | Highlight positions. int and tuple range mixture positions can be specified.
290 | (e.g. If `[1, 5, (10, 13), 18]` is set, `1, 5, 10, 11, 12, 13, 18`
291 | positions are highlighted)
292 | """
293 | self._highlight_positions = self._parse_positions(positions)
294 |
295 | def set_highlight_pos_by_ident_thr(
296 | self, min_thr: float = 0, max_thr: float = 100
297 | ) -> None:
298 | """Set highlight MSA positions by consensus identity threshold
299 |
300 | Parameters
301 | ----------
302 | min_thr : float, optional
303 | Min identity threshold for highlight position selection
304 | max_thr : float, optional
305 | Max identity threshold for highlight position selection
306 | """
307 | ident_list = self._get_consensus_identity_list()
308 | highlight_positions: list[int] = []
309 | for pos, ident in enumerate(ident_list):
310 | if min_thr <= ident <= max_thr:
311 | highlight_positions.append(pos)
312 | self._highlight_positions = highlight_positions
313 |
314 | def add_markers(
315 | self,
316 | positions: list[tuple[int, int] | int],
317 | marker: str = "v",
318 | color: str = "black",
319 | size: float = 6,
320 | ) -> None:
321 | """Add markers on specified positions
322 |
323 | Parameters
324 | ----------
325 | positions : list[tuple[int, int] | int]
326 | Marker positions. int and tuple range mixture positions can be specified.
327 | (e.g. If `[1, 5, (10, 13), 18]` is set, markers are plotted on
328 | `1, 5, 10, 11, 12, 13, 18` positions)
329 | marker : str, optional
330 | Marker type of matplotlib.
331 | See for details.
332 | color : str, optional
333 | Marker color
334 | size : float, optional
335 | Marker size
336 | """
337 | for pos in self._parse_positions(positions):
338 | self._pos2marker_kws[pos] = dict(
339 | marker=marker,
340 | color=color,
341 | markersize=size,
342 | clip_on=False,
343 | )
344 |
345 | def add_text_annotation(
346 | self,
347 | range: tuple[int, int],
348 | text: str,
349 | *,
350 | text_color: str = "black",
351 | text_size: float = 10,
352 | range_color: str = "black",
353 | ) -> None:
354 | """Add text annotation in specified range
355 |
356 | Parameters
357 | ----------
358 | range : tuple[int, int]
359 | Annotation start-end range tuple
360 | text : str
361 | Annotation text
362 | text_color : str, optional
363 | Text color
364 | text_size : float, optional
365 | Text size
366 | range_color : str, optional
367 | Annotation range line color
368 | """
369 | # Add annotation text
370 | start, end = range[0] - 1, range[1]
371 | x = (start + end) / 2
372 | pos = int(x)
373 | self._pos2text_kws[pos] = dict(
374 | x=x,
375 | y=self.msa_count + 0.75,
376 | s=text,
377 | color=text_color,
378 | size=text_size,
379 | ha="center",
380 | va="bottom",
381 | )
382 | # Add annotation range line markers
383 | marker_size = 10 * (self._x_unit_size / 0.14)
384 | self.add_markers([range], marker="_", color=range_color, size=marker_size)
385 |
386 | def plotfig(self, dpi: int = 100) -> Figure:
387 | """Plot figure
388 |
389 | Parameters
390 | ----------
391 | dpi : int, optional
392 | Figure DPI
393 |
394 | Returns
395 | -------
396 | fig : Figure
397 | Figure
398 | """
399 | # Setup plot figure configs
400 | ax_type2y_size = {
401 | AxesType.MSA: self.msa_count * self._y_unit_size,
402 | AxesType.SPACE: self._y_unit_size * 1.5,
403 | AxesType.CONSENSUS: self._y_unit_size * self._consensus_size,
404 | AxesType.WRAP_SPACE: self._y_unit_size * self._wrap_space_size,
405 | }
406 |
407 | plot_ax_types = []
408 | for wrap_idx in range(self.wrap_num + 1):
409 | plot_ax_types.append(AxesType.MSA)
410 | if self._show_consensus:
411 | plot_ax_types.append(AxesType.SPACE)
412 | plot_ax_types.append(AxesType.CONSENSUS)
413 | if wrap_idx != self.wrap_num:
414 | plot_ax_types.append(AxesType.WRAP_SPACE)
415 |
416 | y_size_list = [ax_type2y_size[t] for t in plot_ax_types]
417 | figsize = (self._wrap_length * self._x_unit_size, sum(y_size_list))
418 | fig: Figure = plt.figure(figsize=figsize, dpi=dpi) # type: ignore
419 | fig.tight_layout()
420 | gs = GridSpec(nrows=len(plot_ax_types), ncols=1, height_ratios=y_size_list)
421 | gs.update(left=0, right=1, bottom=0, top=1, hspace=0, wspace=0)
422 |
423 | # Plot figure
424 | wrap_cnt = 0
425 | for idx, plot_ax_type in enumerate(plot_ax_types):
426 | ax: Axes = fig.add_subplot(gs[idx])
427 | if not isinstance(ax, Axes):
428 | raise TypeError("Error: Not matplotlib Axes class instance.")
429 |
430 | start = self._start + self._wrap_length * wrap_cnt
431 | end = self._start + self._wrap_length * (wrap_cnt + 1)
432 | end = self._end if end > self._end else end
433 |
434 | if plot_ax_type == AxesType.MSA:
435 | self._plot_msa(ax, start, end)
436 | elif plot_ax_type == AxesType.CONSENSUS:
437 | self._plot_consensus(ax, start, end)
438 | elif plot_ax_type == AxesType.SPACE:
439 | ax.axis("off")
440 | elif plot_ax_type == AxesType.WRAP_SPACE:
441 | ax.axis("off")
442 | wrap_cnt += 1
443 | else:
444 | raise NotImplementedError(f"{plot_ax_type=} is invalid.")
445 |
446 | return fig
447 |
448 | def savefig(
449 | self,
450 | savefile: str | Path,
451 | dpi: int = 100,
452 | pad_inches: float = 0.5,
453 | ) -> None:
454 | """Save figure to file
455 |
456 | Parameters
457 | ----------
458 | savefile : str | Path
459 | Save file
460 | dpi : int, optional
461 | DPI
462 | pad_inches : float, optional
463 | Padding inches
464 | """
465 | fig = self.plotfig(dpi=dpi)
466 | fig.savefig(
467 | fname=str(savefile),
468 | dpi=dpi,
469 | pad_inches=pad_inches,
470 | )
471 | # Clear & close figure to suppress memory leak
472 | fig.clear()
473 | plt.close(fig)
474 |
475 | ############################################################
476 | # Private Method
477 | ############################################################
478 |
479 | def _plot_msa(
480 | self, ax: Axes, start: int | None = None, end: int | None = None
481 | ) -> None:
482 | """Plot MSA
483 |
484 | Parameters
485 | ----------
486 | ax : Axes
487 | Matplotlib axes to be plotted
488 | start : int | None, optional
489 | Start position. If None, `0` is set.
490 | end : int | None, optional
491 | End position. If None, `alignment_length` is set.
492 | """
493 | # Set xlim, ylim
494 | start = 0 if start is None else start
495 | end = self.alignment_length if end is None else end
496 | ax.set_xlim(start, start + self._wrap_length)
497 | ax.set_ylim(0, self.msa_count)
498 |
499 | # Set spines & tick params (Only show bottom ticklables)
500 | for pos in ("left", "right", "top", "bottom"):
501 | ax.spines[pos].set_visible(False)
502 | ax.tick_params(left=False, labelleft=False)
503 |
504 | # Plot alignment position every 10 chars on xticks
505 | ticks_interval = self._ticks_interval
506 | if ticks_interval is None:
507 | ax.tick_params(bottom=False, labelbottom=False)
508 | else:
509 | tick_ranges = range(start + 1, end + 1)
510 | xticklabels = list(filter(lambda n: n % ticks_interval == 0, tick_ranges))
511 | xticks = [n - 0.5 for n in xticklabels]
512 | ax.set_xticks(xticks, xticklabels, size=8) # type: ignore
513 |
514 | plot_patches = []
515 | for cnt in range(self.msa_count):
516 | msa_seq = self.seq_list[cnt]
517 | y_lower = self.msa_count - (cnt + 1)
518 | y_center = y_lower + 0.5
519 | # Plot label text
520 | if self._show_label:
521 | if self._label_type == "id":
522 | label = self.id_list[cnt]
523 | elif self._label_type == "description":
524 | label = self.desc_list[cnt]
525 | else:
526 | err_msg = f"{self._label_type=} is invalid (`id`|`description`)"
527 | raise ValueError(err_msg)
528 | ax.text(start - 1, y_center, label, ha="right", va="center", size=10)
529 | # Plot count text
530 | if self._show_count:
531 | scale = end - self._start - msa_seq[self._start : end].count("-")
532 | ax.text(end + 1, y_center, str(scale), ha="left", va="center", size=10)
533 | for x_left in range(start, end):
534 | # Add colored rectangle patch
535 | seq_char = msa_seq[x_left]
536 | rect_prop: dict = dict(
537 | xy=(x_left, y_lower), width=1, height=1, color="none", lw=0
538 | )
539 | highlight_positions = self._highlight_positions
540 | if highlight_positions is None or x_left in highlight_positions:
541 | color = self.color_scheme.get(seq_char, "#FFFFFF")
542 | if self._color_scheme_name == "Identity":
543 | color = self._get_identity_color(seq_char, x_left)
544 | if self._custom_color_func is not None:
545 | custom_color = self._custom_color_func(
546 | cnt, x_left, seq_char, self.msa
547 | )
548 | color = color if custom_color is None else custom_color
549 | rect_prop.update(**dict(color=color, lw=0, fill=True))
550 | if self._show_grid:
551 | rect_prop.update(**dict(ec=self._grid_color, lw=0.5))
552 | plot_patches.append(Rectangle(**rect_prop))
553 |
554 | # Plot seq char text
555 | x_center = x_left + 0.5
556 | if self._show_seq_char:
557 | ax.text(
558 | x_center, y_center, seq_char, ha="center", va="center", size=10
559 | )
560 | # Plot marker
561 | if cnt == 0 and x_left in self._pos2marker_kws:
562 | marker_kws = self._pos2marker_kws[x_left]
563 | ax.plot(x_center, y_center + 1, **marker_kws)
564 | # Plot text annotation
565 | if cnt == 0 and x_left in self._pos2text_kws:
566 | text_kws = self._pos2text_kws[x_left]
567 | ax.text(**text_kws)
568 |
569 | # Plot colored rectangle patch collection (Use collection for speedup)
570 | collection = PatchCollection(plot_patches, match_original=True, clip_on=False)
571 | ax.add_collection(collection) # type: ignore
572 |
573 | def _plot_consensus(
574 | self, ax: Axes, start: int | None = None, end: int | None = None
575 | ) -> None:
576 | """Plot consensus seq char & identity bar
577 |
578 | Parameters
579 | ----------
580 | ax : Axes
581 | Matplotlib axes to be plotted
582 | start : int | None, optional
583 | Start position. If None, `0` is set.
584 | end : int | None, optional
585 | End position. If None, `alignment_length` is set.
586 | """
587 | # Set xlim, ylim
588 | start = 0 if start is None else start
589 | end = self.alignment_length if end is None else end
590 | ax.set_xlim(start, start + self._wrap_length)
591 | ax.set_ylim(0, 100) # 0 - 100 [%]
592 |
593 | # Plot label text
594 | if self._show_label and self._consensus_size != 0:
595 | ax.text(start - 1, 40, "Consensus", ha="right", va="center", size=10)
596 |
597 | # Set spines & tick params
598 | for pos in ("left", "right", "top", "bottom"):
599 | ax.spines[pos].set_visible(False)
600 | ax.tick_params(bottom=False, left=False, labelleft=False, pad=0)
601 |
602 | # Plot consensus seq chars on xticks
603 | xticks = list(map(lambda n: n + 0.5, range(start, end)))
604 | if self._show_consensus_char:
605 | xticklabels = list(self.consensus_seq[start:end])
606 | ax.set_xticks(xticks, xticklabels, size=10) # type: ignore
607 | else:
608 | ax.axis("off")
609 |
610 | # Plot consensus identity bar
611 | ident_list = self._get_consensus_identity_list(start, end)
612 | color_list = self._get_interpolate_colors(self._consensus_color, ident_list)
613 | ax.bar(xticks, ident_list, width=1, color=color_list, ec="white", lw=0.5)
614 |
615 | def _get_consensus_seq(self, msa: MSA) -> str:
616 | """Get consensus sequence
617 |
618 | Parameters
619 | ----------
620 | msa : MSA
621 | Multiple sequence alignment
622 |
623 | Returns
624 | -------
625 | consensus_seq : str
626 | Consensus suquence
627 | """
628 | consensus_seq = ""
629 | ambiguous_char = "X"
630 | aln_len = msa.get_alignment_length()
631 |
632 | for idx in range(aln_len):
633 | chars = ""
634 | for record in self._msa:
635 | char = str(record.seq)[idx]
636 | if char != "-" and char != ".":
637 | chars += str(record.seq)[idx]
638 | if len(chars) == 0:
639 | consensus_seq += ambiguous_char
640 | continue
641 |
642 | char2count = Counter(chars)
643 | most_freq_chars = []
644 | most_freq_count = char2count.most_common()[0][1]
645 | for char, count in char2count.most_common():
646 | if count == most_freq_count:
647 | most_freq_chars.append(char)
648 |
649 | if len(most_freq_chars) == 1:
650 | consensus_seq += most_freq_chars[0]
651 | else:
652 | consensus_seq += ambiguous_char
653 |
654 | return consensus_seq
655 |
656 | def _get_consensus_identity_list(
657 | self, start: int | None = None, end: int | None = None
658 | ) -> list[float]:
659 | """Get consensus identity list
660 |
661 | Parameters
662 | ----------
663 | start : int | None, optional
664 | Start position. If None, `0` is set.
665 | end : int | None, optional
666 | End position. If None, `alignment_length` is set.
667 |
668 | Returns
669 | -------
670 | consensus_identity_list : list[float]
671 | Consensus identity list (0 - 100 [%])
672 | """
673 | start = 0 if start is None else start
674 | end = self.alignment_length if end is None else end
675 | consensus_identity_list = []
676 | for idx, _ in enumerate(self.consensus_seq[start:end], start):
677 | column_chars = str(self.msa[:, idx])
678 | counter = Counter(filter(lambda c: c not in ("-", "*"), column_chars))
679 | count = counter.most_common()[0][1] if len(counter) != 0 else 0
680 | consensus_identity = (count / self.msa_count) * 100
681 | consensus_identity_list.append(consensus_identity)
682 | return consensus_identity_list
683 |
684 | def _get_interpolate_colors(
685 | self,
686 | color: str,
687 | values: list[float],
688 | vmin: float = 0,
689 | vmax: float = 100,
690 | ) -> list[str]:
691 | """Interpolate colors by size of values
692 |
693 | Parameters
694 | ----------
695 | color : str
696 | Base color for interpolation
697 | values : list[float]
698 | Values for interpolation
699 | vmin : float, optional
700 | Min value
701 | vmax : float, optional
702 | Max value
703 |
704 | Returns
705 | -------
706 | interpolated_colors : list[str]
707 | Interpolated colors based on values
708 | """
709 | cmap = colors.LinearSegmentedColormap.from_list("m", ["white", color])
710 | norm = colors.Normalize(vmin=vmin, vmax=vmax)
711 | return [colors.to_hex(cmap(norm(v))) for v in values] # type: ignore
712 |
713 | def _get_identity_color(self, seq_char: str, pos: int) -> str:
714 | """Get identity color for `Identity` color scheme
715 |
716 | Parameters
717 | ----------
718 | seq_char : str
719 | Seq character
720 | pos : int
721 | Seq character position
722 |
723 | Returns
724 | -------
725 | identity_color : str
726 | Identity color
727 | """
728 | # Exclude characters color
729 | exclude_chars = ("-", "*", "X")
730 | if seq_char in exclude_chars:
731 | return "#FFFFFF"
732 | # Get most common characters in target MSA position
733 | column_chars = str(self.msa[:, pos])
734 | counter = Counter(filter(lambda c: c not in exclude_chars, column_chars))
735 | most_common_count = counter.most_common()[0][1]
736 | most_common_chars = []
737 | for char, count in counter.most_common():
738 | if count == most_common_count:
739 | most_common_chars.append(char)
740 | # Calculate identity & color if target seq char is most common
741 | identity = (most_common_count / len(column_chars)) * 100
742 | if seq_char in most_common_chars and identity >= self._identity_color_min_thr:
743 | color, color_thr = self._identity_color, self._identity_color_min_thr
744 | return self._get_interpolate_colors(color, [identity], vmin=color_thr)[0]
745 | else:
746 | return "#FFFFFF"
747 |
748 | def _is_aa_msa(self) -> bool:
749 | """Check MSA is `aa` or `nt`
750 |
751 | If the ratio of `ATGCUN` char is less than 90%, return True.
752 |
753 | Returns
754 | -------
755 | check_result : bool
756 | Check result
757 | """
758 | nt_count, all_count = 0, 0
759 | for seq in self.seq_list:
760 | for seq_char in seq:
761 | if seq_char == "-":
762 | continue
763 | all_count += 1
764 | if seq_char in "ATGCUN":
765 | nt_count += 1
766 | return nt_count / all_count < 0.9
767 |
768 | def _parse_positions(self, positions: list[tuple[int, int] | int]) -> list[int]:
769 | """Parse int and tuple range mixture positions
770 |
771 | e.g. `[1, 5, (10, 13), 18]` means `1, 5, 10, 11, 12, 13, 18` positions
772 |
773 | Parameters
774 | ----------
775 | positions : list[tuple[int, int] | int]
776 | int and tuple range mixture positions (one-based coordinates)
777 |
778 | Returns
779 | -------
780 | result_positions : list[int]
781 | Parse result int positions (zero-based coordinates)
782 | """
783 | result_positions: list[int] = []
784 | for pos in positions:
785 | if isinstance(pos, (tuple, list)):
786 | result_positions.extend(list(range(pos[0] - 1, pos[1])))
787 | elif isinstance(pos, int):
788 | result_positions.append(pos - 1)
789 | else:
790 | raise ValueError(f"{positions=} is invalid.")
791 | return sorted(set(result_positions))
792 |
793 | def _sorted_msa_by_njtree(self, msa: MSA) -> MSA:
794 | """Sort MSA order by NJ tree constructed from MSA distance matrix
795 |
796 | Parameters
797 | ----------
798 | msa : MultipleSeqAlignment
799 | MSA
800 |
801 | Returns
802 | -------
803 | sorted_msa : MultipleSeqAlignment
804 | Sorted MSA
805 | """
806 | # Set unique id for MSA records to avoid duplicate name error
807 | uid2id = {}
808 | for idx, rec in enumerate(msa):
809 | uid = f"seq{idx}"
810 | uid2id[uid] = rec.id
811 | rec.id = uid
812 | uid2seq = {rec.id: rec.seq for rec in msa}
813 | uid2desc = {rec.id: rec.description for rec in msa}
814 | # Sort MSA order by NJ tree
815 | njtree = self._construct_njtree(msa)
816 | sorted_msa = MSA([])
817 | for leaf in njtree.get_terminals():
818 | uid = str(leaf.name)
819 | id, seq, desc = uid2id[uid], uid2seq[uid], uid2desc[uid]
820 | sorted_msa.append(SeqRecord(seq, id=id, description=desc))
821 | return sorted_msa
822 |
823 | def _construct_njtree(self, msa: MSA) -> Tree:
824 | """Construct NJ tree from MSA distance matrix
825 |
826 | Parameters
827 | ----------
828 | msa : MultipleSeqAlignment
829 | MSA
830 |
831 | Returns
832 | -------
833 | njtree : Tree
834 | NJ tree
835 | """
836 | # Calculate MSA distance matrix & construct NJ tree
837 | model = "blosum62" if self._is_aa_msa() else "identity"
838 | distance_matrix = DistanceCalculator(model).get_distance(msa)
839 | njtree = DistanceTreeConstructor().nj(distance_matrix)
840 | njtree.root_at_midpoint()
841 | return njtree
842 |
--------------------------------------------------------------------------------