55 |
56 |
90 |
91 |
92 |
93 |
94 |
145 |
146 |
147 |
148 |
149 |
150 |
151 |
--------------------------------------------------------------------------------
/docs/reference/drjacoby_cols.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
95 |
134 |
135 |
144 |
96 |
113 |
123 |
124 |
125 |
126 |
133 |
97 |
100 |
101 |
102 | Replace with default values
98 | 99 |define_default.Rd
103 |
104 |
108 |
109 | If NULL then replace with chosen value, otherwise keep original 105 | value.
106 | 107 |define_default(x, default)110 | 111 |
Arguments
112 || x | 116 |object to define default. |
117 |
|---|---|
| default | 120 |default value. |
121 |
64 |
65 |
119 |
120 |
121 |
122 |
123 |
124 |
161 |
162 |
163 |
164 |
165 |
166 |
167 |
168 |
169 |
--------------------------------------------------------------------------------
/docs/reference/ess.html:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
125 |
148 |
149 |
150 |
160 |
126 |
142 |
147 |
127 |
130 |
131 |
132 | Series of standard colours used within the drjacoby package
128 | Source:R/plot.R
129 | drjacoby_cols.Rd
133 |
136 |
137 | Returns a series of standard colours used within the drjacoby 134 | package.
135 |drjacoby_cols()
138 |
139 |
140 |
141 |
57 |
58 |
113 |
114 |
115 |
116 |
117 |
172 |
173 |
174 |
175 |
176 |
177 |
178 |
--------------------------------------------------------------------------------
/docs/sitemap.xml:
--------------------------------------------------------------------------------
1 |
2 |
118 |
161 |
162 |
171 |
119 |
138 |
144 |
145 |
151 |
160 |
120 |
123 |
124 |
125 | Effective sample size
121 | 122 |ess.Rd
126 |
127 |
133 |
134 | Estimates effective sample size. 128 | Note 1: Lag is trunctaed at when autocorrelation < 0.05, 129 | if -ve autocorrelation occurs it is possible to get ess > N. 130 | Note 2: ESS estimates will be too optimistic for chains that haven’t mixed.
131 | 132 |ess(x)135 | 136 |
Arguments
137 || x | 141 |chain |
142 |
|---|
Value
146 | 147 |The effective sample size of a chain
148 | 149 | 150 |Dr. Jacoby 
9 | 10 | *drjacoby* is a package for running flexible Markov chain Monte Carlo (MCMC) with minimal fiddling required by the user. The likelihood and the priors that go into the model can be written as either R or C++ functions, with the latter typically being much faster to run. Outputs are produced in a standardised format, and can be explored using a range of built in diagnostic plots and statistics. 11 | 12 |
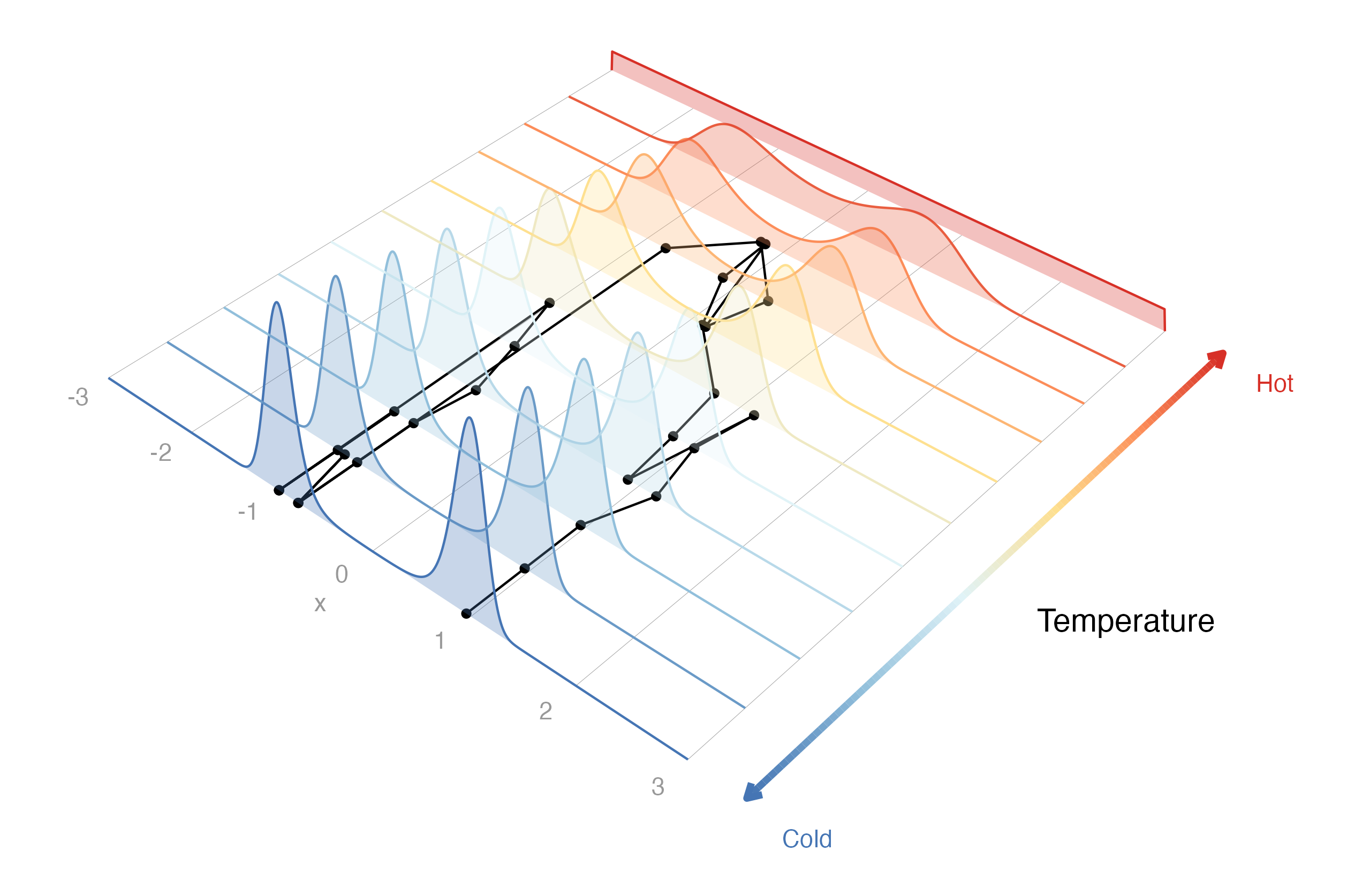
*drjacoby* uses parallel tempering to "melt" the target distribution,
thereby allowing information to pass between well-separated peaks.
 17 | ===================================================
18 |
19 |
20 | [](https://github.com/mrc-ide/drjacoby/actions)
21 | [](https://github.com/mrc-ide/drjacoby/actions)
22 | [](https://codecov.io/github/mrc-ide/drjacoby?branch=master)
23 |
24 |
25 | *drjacoby* is an R package for performing Bayesian inference via Markov chain monte carlo (MCMC). In addition to being highly flexible it implements some advanced techniques that can improve mixing in tricky situations. Full details can be found on the drjacoby [website](https://mrc-ide.github.io/drjacoby/).
26 |
27 | So far, *drjacoby* has been used in the following projects:
28 |
29 | * The paper [Estimates of the severity of coronavirus disease 2019: a model-based analysis](https://doi.org/10.1016/S1473-3099(20)30243-7) used *drjacoby* **version 1.0.1**
30 | * The report [COVID-19 Infection Fatality Ratio: Estimates from Seroprevalence](https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-34-ifr/) used *drjacoby* **version 1.2.0**
31 |
32 |
33 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | # DrJacoby
17 | ===================================================
18 |
19 |
20 | [](https://github.com/mrc-ide/drjacoby/actions)
21 | [](https://github.com/mrc-ide/drjacoby/actions)
22 | [](https://codecov.io/github/mrc-ide/drjacoby?branch=master)
23 |
24 |
25 | *drjacoby* is an R package for performing Bayesian inference via Markov chain monte carlo (MCMC). In addition to being highly flexible it implements some advanced techniques that can improve mixing in tricky situations. Full details can be found on the drjacoby [website](https://mrc-ide.github.io/drjacoby/).
26 |
27 | So far, *drjacoby* has been used in the following projects:
28 |
29 | * The paper [Estimates of the severity of coronavirus disease 2019: a model-based analysis](https://doi.org/10.1016/S1473-3099(20)30243-7) used *drjacoby* **version 1.0.1**
30 | * The report [COVID-19 Infection Fatality Ratio: Estimates from Seroprevalence](https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-34-ifr/) used *drjacoby* **version 1.2.0**
31 |
32 |
33 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |
3 |
4 | # DrJacoby