├── .gitignore
├── Code
├── network
│ ├── deeplab
│ │ ├── README.md
│ │ └── deeplab.py
│ ├── segnet
│ │ ├── README.md
│ │ ├── custom_layers.py
│ │ ├── segnet.py
│ │ └── segnet_plot.png
│ ├── unet
│ │ ├── README.md
│ │ ├── u_net.py
│ │ └── unet_plot.png
│ └── unetmod
│ │ ├── README.md
│ │ ├── u_net_mod.py
│ │ └── unet_plot.png
└── utils
│ ├── lossfunctions.py
│ └── metricfunctions.py
├── Datasets
├── MonuSeg-20200319T073151Z-001.zip
├── README.md
└── Samples
│ ├── TCGA-RD-A8N9-01A-01-TS1.png
│ ├── TCGA-RD-A8N9-01A-01-TS1_bin_mask.png
│ └── Test
│ ├── TCGA-HT-8564-01Z-00-DX1.png
│ └── TCGA-HT-8564-01Z-00-DX1_bin_mask.png
├── README.md
├── Results
├── outputs
│ └── TCGA-HT-8564-01Z-00-DX1.jpg
└── plots
│ ├── DEEPLAB
│ ├── train_accuracy.png
│ ├── train_dice.png
│ ├── train_f1.png
│ └── train_loss.png
│ ├── README.md
│ ├── SEGNET
│ ├── train_accuracy.png
│ ├── train_dice.png
│ ├── train_f1.png
│ └── train_loss.png
│ ├── UNET
│ ├── train_accuracy.png
│ ├── train_dice.png
│ ├── train_f1.png
│ └── train_loss.png
│ └── UNETMOD
│ ├── train_accuracy.png
│ ├── train_dice.png
│ ├── train_f1.png
│ └── train_loss.png
├── Test.py
├── Train.py
├── Train_Bak.py
├── config.json
├── requirements.txt
└── temp.py
/.gitignore:
--------------------------------------------------------------------------------
1 | Datasets/MonuSeg/
2 | Results/weights/
3 | __pycache__/
4 | Results/weights.zip
5 |
--------------------------------------------------------------------------------
/Code/network/deeplab/README.md:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Code/network/deeplab/README.md
--------------------------------------------------------------------------------
/Code/network/deeplab/deeplab.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 |
3 | from tensorflow.keras.models import Model, load_model
4 | from tensorflow.keras.layers import Input, BatchNormalization, Activation, Dense, Dropout
5 | from tensorflow.keras.layers import Lambda, RepeatVector, Reshape, Add, Concatenate, ZeroPadding2D
6 | from tensorflow.keras.layers import Conv2D, Conv2DTranspose, DepthwiseConv2D
7 | from tensorflow.keras.layers import MaxPooling2D, GlobalMaxPool2D, GlobalAveragePooling2D
8 | from tensorflow.keras.layers import concatenate, add
9 | from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
10 | from tensorflow.keras.optimizers import Adam
11 | from tensorflow.keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
12 | from tensorflow.python.keras.activations import relu
13 |
14 | from tensorflow.keras import backend as K
15 |
16 | WEIGHTS_PATH_X = "https://github.com/bonlime/keras-deeplab-v3-plus/releases/download/1.1/deeplabv3_xception_tf_dim_ordering_tf_kernels.h5"
17 | WEIGHTS_PATH_MOBILE = "https://github.com/bonlime/keras-deeplab-v3-plus/releases/download/1.1/deeplabv3_mobilenetv2_tf_dim_ordering_tf_kernels.h5"
18 | WEIGHTS_PATH_X_CS = "https://github.com/bonlime/keras-deeplab-v3-plus/releases/download/1.2/deeplabv3_xception_tf_dim_ordering_tf_kernels_cityscapes.h5"
19 | WEIGHTS_PATH_MOBILE_CS = "https://github.com/bonlime/keras-deeplab-v3-plus/releases/download/1.2/deeplabv3_mobilenetv2_tf_dim_ordering_tf_kernels_cityscapes.h5"
20 |
21 |
22 | def SepConv_BN(x, filters, prefix, stride=1, kernel_size=3, rate=1, depth_activation=False, epsilon=1e-3):
23 | """ SepConv with BN between depthwise & pointwise. Optionally add activation after BN
24 | Implements right "same" padding for even kernel sizes
25 | Args:
26 | x: input tensor
27 | filters: num of filters in pointwise convolution
28 | prefix: prefix before name
29 | stride: stride at depthwise conv
30 | kernel_size: kernel size for depthwise convolution
31 | rate: atrous rate for depthwise convolution
32 | depth_activation: flag to use activation between depthwise & poinwise convs
33 | epsilon: epsilon to use in BN layer
34 | """
35 |
36 | if stride == 1:
37 | depth_padding = 'same'
38 | else:
39 | kernel_size_effective = kernel_size + (kernel_size - 1) * (rate - 1)
40 | pad_total = kernel_size_effective - 1
41 | pad_beg = pad_total // 2
42 | pad_end = pad_total - pad_beg
43 | x = ZeroPadding2D((pad_beg, pad_end))(x)
44 | depth_padding = 'valid'
45 |
46 | if not depth_activation:
47 | x = Activation('relu')(x)
48 | x = DepthwiseConv2D((kernel_size, kernel_size), strides=(stride, stride), dilation_rate=(rate, rate),
49 | padding=depth_padding, use_bias=False, name=prefix + '_depthwise')(x)
50 | x = BatchNormalization(name=prefix + '_depthwise_BN', epsilon=epsilon)(x)
51 | if depth_activation:
52 | x = Activation('relu')(x)
53 | x = Conv2D(filters, (1, 1), padding='same',
54 | use_bias=False, name=prefix + '_pointwise')(x)
55 | x = BatchNormalization(name=prefix + '_pointwise_BN', epsilon=epsilon)(x)
56 | if depth_activation:
57 | x = Activation('relu')(x)
58 |
59 | return x
60 |
61 |
62 | def _conv2d_same(x, filters, prefix, stride=1, kernel_size=3, rate=1):
63 | """Implements right 'same' padding for even kernel sizes
64 | Without this there is a 1 pixel drift when stride = 2
65 | Args:
66 | x: input tensor
67 | filters: num of filters in pointwise convolution
68 | prefix: prefix before name
69 | stride: stride at depthwise conv
70 | kernel_size: kernel size for depthwise convolution
71 | rate: atrous rate for depthwise convolution
72 | """
73 | if stride == 1:
74 | return Conv2D(filters,
75 | (kernel_size, kernel_size),
76 | strides=(stride, stride),
77 | padding='same', use_bias=False,
78 | dilation_rate=(rate, rate),
79 | name=prefix)(x)
80 | else:
81 | kernel_size_effective = kernel_size + (kernel_size - 1) * (rate - 1)
82 | pad_total = kernel_size_effective - 1
83 | pad_beg = pad_total // 2
84 | pad_end = pad_total - pad_beg
85 | x = ZeroPadding2D((pad_beg, pad_end))(x)
86 | return Conv2D(filters,
87 | (kernel_size, kernel_size),
88 | strides=(stride, stride),
89 | padding='valid', use_bias=False,
90 | dilation_rate=(rate, rate),
91 | name=prefix)(x)
92 |
93 |
94 | def _xception_block(inputs, depth_list, prefix, skip_connection_type, stride,
95 | rate=1, depth_activation=False, return_skip=False):
96 | """ Basic building block of modified Xception network
97 | Args:
98 | inputs: input tensor
99 | depth_list: number of filters in each SepConv layer. len(depth_list) == 3
100 | prefix: prefix before name
101 | skip_connection_type: one of {'conv','sum','none'}
102 | stride: stride at last depthwise conv
103 | rate: atrous rate for depthwise convolution
104 | depth_activation: flag to use activation between depthwise & pointwise convs
105 | return_skip: flag to return additional tensor after 2 SepConvs for decoder
106 | """
107 | residual = inputs
108 | for i in range(3):
109 | residual = SepConv_BN(residual,
110 | depth_list[i],
111 | prefix + '_separable_conv{}'.format(i + 1),
112 | stride=stride if i == 2 else 1,
113 | rate=rate,
114 | depth_activation=depth_activation)
115 | if i == 1:
116 | skip = residual
117 | if skip_connection_type == 'conv':
118 | shortcut = _conv2d_same(inputs, depth_list[-1], prefix + '_shortcut',
119 | kernel_size=1,
120 | stride=stride)
121 | shortcut = BatchNormalization(name=prefix + '_shortcut_BN')(shortcut)
122 | outputs = tf.keras.layers.add([residual, shortcut])
123 | elif skip_connection_type == 'sum':
124 | outputs = tf.keras.layers.add([residual, inputs])
125 | elif skip_connection_type == 'none':
126 | outputs = residual
127 | if return_skip:
128 | return outputs, skip
129 | else:
130 | return outputs
131 |
132 |
133 | def relu6(x):
134 | return relu(x, max_value=6)
135 |
136 |
137 | def _make_divisible(v, divisor, min_value=None):
138 | if min_value is None:
139 | min_value = divisor
140 | new_v = max(min_value, int(v + divisor / 2) // divisor * divisor)
141 | # Make sure that round down does not go down by more than 10%.
142 | if new_v < 0.9 * v:
143 | new_v += divisor
144 | return new_v
145 |
146 |
147 | def _inverted_res_block(inputs, expansion, stride, alpha, filters, block_id, skip_connection, rate=1):
148 | in_channels = inputs.shape[-1] # inputs._keras_shape[-1]
149 | pointwise_conv_filters = int(filters * alpha)
150 | pointwise_filters = _make_divisible(pointwise_conv_filters, 8)
151 | x = inputs
152 | prefix = 'expanded_conv_{}_'.format(block_id)
153 | if block_id:
154 | # Expand
155 |
156 | x = Conv2D(expansion * in_channels, kernel_size=1, padding='same',
157 | use_bias=False, activation=None,
158 | name=prefix + 'expand')(x)
159 | x = BatchNormalization(epsilon=1e-3, momentum=0.999,
160 | name=prefix + 'expand_BN')(x)
161 | x = Activation(relu6, name=prefix + 'expand_relu')(x)
162 | else:
163 | prefix = 'expanded_conv_'
164 | # Depthwise
165 | x = DepthwiseConv2D(kernel_size=3, strides=stride, activation=None,
166 | use_bias=False, padding='same', dilation_rate=(rate, rate),
167 | name=prefix + 'depthwise')(x)
168 | x = BatchNormalization(epsilon=1e-3, momentum=0.999,
169 | name=prefix + 'depthwise_BN')(x)
170 |
171 | x = Activation(relu6, name=prefix + 'depthwise_relu')(x)
172 |
173 | # Project
174 | x = Conv2D(pointwise_filters,
175 | kernel_size=1, padding='same', use_bias=False, activation=None,
176 | name=prefix + 'project')(x)
177 | x = BatchNormalization(epsilon=1e-3, momentum=0.999,
178 | name=prefix + 'project_BN')(x)
179 |
180 | if skip_connection:
181 | return Add(name=prefix + 'add')([inputs, x])
182 |
183 | # if in_channels == pointwise_filters and stride == 1:
184 | # return Add(name='res_connect_' + str(block_id))([inputs, x])
185 |
186 | return x
187 |
188 |
189 | def Deeplabv3(weights='pascal_voc', input_tensor=None, input_shape=(512, 512, 3), classes=21, backbone='mobilenetv2',
190 | OS=16, alpha=1., activation=None):
191 | """ Instantiates the Deeplabv3+ architecture
192 | Optionally loads weights pre-trained
193 | on PASCAL VOC or Cityscapes. This model is available for TensorFlow only.
194 | # Arguments
195 | weights: one of 'pascal_voc' (pre-trained on pascal voc),
196 | 'cityscapes' (pre-trained on cityscape) or None (random initialization)
197 | input_tensor: optional Keras tensor (i.e. output of `layers.Input()`)
198 | to use as image input for the model.
199 | input_shape: shape of input image. format HxWxC
200 | PASCAL VOC model was trained on (512,512,3) images. None is allowed as shape/width
201 | classes: number of desired classes. PASCAL VOC has 21 classes, Cityscapes has 19 classes.
202 | If number of classes not aligned with the weights used, last layer is initialized randomly
203 | backbone: backbone to use. one of {'xception','mobilenetv2'}
204 | activation: optional activation to add to the top of the network.
205 | One of 'softmax', 'sigmoid' or None
206 | OS: determines input_shape/feature_extractor_output ratio. One of {8,16}.
207 | Used only for xception backbone.
208 | alpha: controls the width of the MobileNetV2 network. This is known as the
209 | width multiplier in the MobileNetV2 paper.
210 | - If `alpha` < 1.0, proportionally decreases the number
211 | of filters in each layer.
212 | - If `alpha` > 1.0, proportionally increases the number
213 | of filters in each layer.
214 | - If `alpha` = 1, default number of filters from the paper
215 | are used at each layer.
216 | Used only for mobilenetv2 backbone. Pretrained is only available for alpha=1.
217 | # Returns
218 | A Keras model instance.
219 | # Raises
220 | RuntimeError: If attempting to run this model with a
221 | backend that does not support separable convolutions.
222 | ValueError: in case of invalid argument for `weights` or `backbone`
223 | """
224 |
225 | if not (weights in {'pascal_voc', 'cityscapes', None}):
226 | raise ValueError('The `weights` argument should be either '
227 | '`None` (random initialization), `pascal_voc`, or `cityscapes` '
228 | '(pre-trained on PASCAL VOC)')
229 |
230 | if not (backbone in {'xception', 'mobilenetv2'}):
231 | raise ValueError('The `backbone` argument should be either '
232 | '`xception` or `mobilenetv2` ')
233 |
234 | if input_tensor is None:
235 | img_input = Input(shape=input_shape)
236 | else:
237 | img_input = input_tensor
238 |
239 | if backbone == 'xception':
240 | if OS == 8:
241 | entry_block3_stride = 1
242 | middle_block_rate = 2 # ! Not mentioned in paper, but required
243 | exit_block_rates = (2, 4)
244 | atrous_rates = (12, 24, 36)
245 | else:
246 | entry_block3_stride = 2
247 | middle_block_rate = 1
248 | exit_block_rates = (1, 2)
249 | atrous_rates = (6, 12, 18)
250 |
251 | x = Conv2D(32, (3, 3), strides=(2, 2),
252 | name='entry_flow_conv1_1', use_bias=False, padding='same')(img_input)
253 | x = BatchNormalization(name='entry_flow_conv1_1_BN')(x)
254 | x = Activation('relu')(x)
255 |
256 | x = _conv2d_same(x, 64, 'entry_flow_conv1_2', kernel_size=3, stride=1)
257 | x = BatchNormalization(name='entry_flow_conv1_2_BN')(x)
258 | x = Activation('relu')(x)
259 |
260 | x = _xception_block(x, [128, 128, 128], 'entry_flow_block1',
261 | skip_connection_type='conv', stride=2,
262 | depth_activation=False)
263 | x, skip1 = _xception_block(x, [256, 256, 256], 'entry_flow_block2',
264 | skip_connection_type='conv', stride=2,
265 | depth_activation=False, return_skip=True)
266 |
267 | x = _xception_block(x, [728, 728, 728], 'entry_flow_block3',
268 | skip_connection_type='conv', stride=entry_block3_stride,
269 | depth_activation=False)
270 | for i in range(16):
271 | x = _xception_block(x, [728, 728, 728], 'middle_flow_unit_{}'.format(i + 1),
272 | skip_connection_type='sum', stride=1, rate=middle_block_rate,
273 | depth_activation=False)
274 |
275 | x = _xception_block(x, [728, 1024, 1024], 'exit_flow_block1',
276 | skip_connection_type='conv', stride=1, rate=exit_block_rates[0],
277 | depth_activation=False)
278 | x = _xception_block(x, [1536, 1536, 2048], 'exit_flow_block2',
279 | skip_connection_type='none', stride=1, rate=exit_block_rates[1],
280 | depth_activation=True)
281 |
282 | else:
283 | OS = 8
284 | first_block_filters = _make_divisible(32 * alpha, 8)
285 | x = Conv2D(first_block_filters,
286 | kernel_size=3,
287 | strides=(2, 2), padding='same',
288 | use_bias=False, name='Conv')(img_input)
289 | x = BatchNormalization(
290 | epsilon=1e-3, momentum=0.999, name='Conv_BN')(x)
291 | x = Activation(relu6, name='Conv_Relu6')(x)
292 |
293 | x = _inverted_res_block(x, filters=16, alpha=alpha, stride=1,
294 | expansion=1, block_id=0, skip_connection=False)
295 |
296 | x = _inverted_res_block(x, filters=24, alpha=alpha, stride=2,
297 | expansion=6, block_id=1, skip_connection=False)
298 | x = _inverted_res_block(x, filters=24, alpha=alpha, stride=1,
299 | expansion=6, block_id=2, skip_connection=True)
300 |
301 | x = _inverted_res_block(x, filters=32, alpha=alpha, stride=2,
302 | expansion=6, block_id=3, skip_connection=False)
303 | x = _inverted_res_block(x, filters=32, alpha=alpha, stride=1,
304 | expansion=6, block_id=4, skip_connection=True)
305 | x = _inverted_res_block(x, filters=32, alpha=alpha, stride=1,
306 | expansion=6, block_id=5, skip_connection=True)
307 |
308 | # stride in block 6 changed from 2 -> 1, so we need to use rate = 2
309 | x = _inverted_res_block(x, filters=64, alpha=alpha, stride=1, # 1!
310 | expansion=6, block_id=6, skip_connection=False)
311 | x = _inverted_res_block(x, filters=64, alpha=alpha, stride=1, rate=2,
312 | expansion=6, block_id=7, skip_connection=True)
313 | x = _inverted_res_block(x, filters=64, alpha=alpha, stride=1, rate=2,
314 | expansion=6, block_id=8, skip_connection=True)
315 | x = _inverted_res_block(x, filters=64, alpha=alpha, stride=1, rate=2,
316 | expansion=6, block_id=9, skip_connection=True)
317 |
318 | x = _inverted_res_block(x, filters=96, alpha=alpha, stride=1, rate=2,
319 | expansion=6, block_id=10, skip_connection=False)
320 | x = _inverted_res_block(x, filters=96, alpha=alpha, stride=1, rate=2,
321 | expansion=6, block_id=11, skip_connection=True)
322 | x = _inverted_res_block(x, filters=96, alpha=alpha, stride=1, rate=2,
323 | expansion=6, block_id=12, skip_connection=True)
324 |
325 | x = _inverted_res_block(x, filters=160, alpha=alpha, stride=1, rate=2, # 1!
326 | expansion=6, block_id=13, skip_connection=False)
327 | x = _inverted_res_block(x, filters=160, alpha=alpha, stride=1, rate=4,
328 | expansion=6, block_id=14, skip_connection=True)
329 | x = _inverted_res_block(x, filters=160, alpha=alpha, stride=1, rate=4,
330 | expansion=6, block_id=15, skip_connection=True)
331 |
332 | x = _inverted_res_block(x, filters=320, alpha=alpha, stride=1, rate=4,
333 | expansion=6, block_id=16, skip_connection=False)
334 |
335 | # end of feature extractor
336 |
337 | # branching for Atrous Spatial Pyramid Pooling
338 |
339 | # Image Feature branch
340 | shape_before = tf.shape(x)
341 | b4 = GlobalAveragePooling2D()(x)
342 | # from (b_size, channels)->(b_size, 1, 1, channels)
343 | b4 = Lambda(lambda x: K.expand_dims(x, 1))(b4)
344 | b4 = Lambda(lambda x: K.expand_dims(x, 1))(b4)
345 | b4 = Conv2D(256, (1, 1), padding='same',
346 | use_bias=False, name='image_pooling')(b4)
347 | b4 = BatchNormalization(name='image_pooling_BN', epsilon=1e-5)(b4)
348 | b4 = Activation('relu')(b4)
349 | # upsample. have to use compat because of the option align_corners
350 | size_before = tf.keras.backend.int_shape(x)
351 | b4 = Lambda(lambda x: tf.compat.v1.image.resize(x, size_before[1:3],
352 | method='bilinear', align_corners=True))(b4)
353 | # simple 1x1
354 | b0 = Conv2D(256, (1, 1), padding='same', use_bias=False, name='aspp0')(x)
355 | b0 = BatchNormalization(name='aspp0_BN', epsilon=1e-5)(b0)
356 | b0 = Activation('relu', name='aspp0_activation')(b0)
357 |
358 | # there are only 2 branches in mobilenetV2. not sure why
359 | if backbone == 'xception':
360 | # rate = 6 (12)
361 | b1 = SepConv_BN(x, 256, 'aspp1',
362 | rate=atrous_rates[0], depth_activation=True, epsilon=1e-5)
363 | # rate = 12 (24)
364 | b2 = SepConv_BN(x, 256, 'aspp2',
365 | rate=atrous_rates[1], depth_activation=True, epsilon=1e-5)
366 | # rate = 18 (36)
367 | b3 = SepConv_BN(x, 256, 'aspp3',
368 | rate=atrous_rates[2], depth_activation=True, epsilon=1e-5)
369 |
370 | # concatenate ASPP branches & project
371 | x = Concatenate()([b4, b0, b1, b2, b3])
372 | else:
373 | x = Concatenate()([b4, b0])
374 |
375 | x = Conv2D(256, (1, 1), padding='same',
376 | use_bias=False, name='concat_projection')(x)
377 | x = BatchNormalization(name='concat_projection_BN', epsilon=1e-5)(x)
378 | x = Activation('relu')(x)
379 | x = Dropout(0.1)(x)
380 | # DeepLab v.3+ decoder
381 |
382 | if backbone == 'xception':
383 | # Feature projection
384 | # x4 (x2) block
385 | size_before2 = tf.keras.backend.int_shape(x)
386 | x = Lambda(lambda xx: tf.compat.v1.image.resize(xx,
387 | size_before2[1:3] * tf.constant(OS // 4),
388 | method='bilinear', align_corners=True))(x)
389 |
390 | dec_skip1 = Conv2D(48, (1, 1), padding='same',
391 | use_bias=False, name='feature_projection0')(skip1)
392 | dec_skip1 = BatchNormalization(
393 | name='feature_projection0_BN', epsilon=1e-5)(dec_skip1)
394 | dec_skip1 = Activation('relu')(dec_skip1)
395 | x = Concatenate()([x, dec_skip1])
396 | x = SepConv_BN(x, 256, 'decoder_conv0',
397 | depth_activation=True, epsilon=1e-5)
398 | x = SepConv_BN(x, 256, 'decoder_conv1',
399 | depth_activation=True, epsilon=1e-5)
400 |
401 | # you can use it with arbitary number of classes
402 | if (weights == 'pascal_voc' and classes == 21) or (weights == 'cityscapes' and classes == 19):
403 | last_layer_name = 'logits_semantic'
404 | else:
405 | last_layer_name = 'custom_logits_semantic'
406 |
407 | x = Conv2D(classes, (1, 1), padding='same', name=last_layer_name)(x)
408 | size_before3 = tf.keras.backend.int_shape(img_input)
409 | x = Lambda(lambda xx: tf.compat.v1.image.resize(xx,
410 | size_before3[1:3],
411 | method='bilinear', align_corners=True))(x)
412 |
413 | # Ensure that the model takes into account
414 | # any potential predecessors of `input_tensor`.
415 | if input_tensor is not None:
416 | inputs = get_source_inputs(input_tensor)
417 | else:
418 | inputs = img_input
419 |
420 | if activation in {'softmax', 'sigmoid'}:
421 | x = tf.keras.layers.Activation(activation)(x)
422 |
423 | model = Model(inputs, x, name='deeplabv3plus')
424 |
425 | # load weights
426 |
427 | if weights == 'pascal_voc':
428 | if backbone == 'xception':
429 | weights_path = get_file('deeplabv3_xception_tf_dim_ordering_tf_kernels.h5',
430 | WEIGHTS_PATH_X,
431 | cache_subdir='models')
432 | else:
433 | weights_path = get_file('deeplabv3_mobilenetv2_tf_dim_ordering_tf_kernels.h5',
434 | WEIGHTS_PATH_MOBILE,
435 | cache_subdir='models')
436 | model.load_weights(weights_path, by_name=True)
437 | elif weights == 'cityscapes':
438 | if backbone == 'xception':
439 | weights_path = get_file('deeplabv3_xception_tf_dim_ordering_tf_kernels_cityscapes.h5',
440 | WEIGHTS_PATH_X_CS,
441 | cache_subdir='models')
442 | else:
443 | weights_path = get_file('deeplabv3_mobilenetv2_tf_dim_ordering_tf_kernels_cityscapes.h5',
444 | WEIGHTS_PATH_MOBILE_CS,
445 | cache_subdir='models')
446 | model.load_weights(weights_path, by_name=True)
447 | return model
448 |
449 | def preprocess_input(x):
450 | """Preprocesses a numpy array encoding a batch of images.
451 | # Arguments
452 | x: a 4D numpy array consists of RGB values within [0, 255].
453 | # Returns
454 | Input array scaled to [-1.,1.]
455 | """
456 | return preprocess_input(x, mode='tf')
457 |
--------------------------------------------------------------------------------
/Code/network/segnet/README.md:
--------------------------------------------------------------------------------
1 | 
2 |
--------------------------------------------------------------------------------
/Code/network/segnet/custom_layers.py:
--------------------------------------------------------------------------------
1 | from keras.layers import Activation,BatchNormalization,Conv2D
2 | from keras.engine.topology import Layer
3 | import keras.backend as K
4 | import tensorflow as tf
5 |
6 | class MaxPoolingWithIndices(Layer):
7 | def __init__(self, pool_size=2,strides=2,padding='SAME',**kwargs):
8 | super(MaxPoolingWithIndices, self).__init__(**kwargs)
9 | self.pool_size=pool_size
10 | self.strides=strides
11 | self.padding=padding
12 | return
13 | def call(self,x):
14 | pool_size=self.pool_size
15 | strides=self.strides
16 | if isinstance(pool_size,int):
17 | ps=[1,pool_size,pool_size,1]
18 | else:
19 | ps=[1,pool_size[0],pool_size[1],1]
20 | if isinstance(strides,int):
21 | st=[1,strides,strides,1]
22 | else:
23 | st=[1,strides[0],strides[1],1]
24 | output1,output2=tf.nn.max_pool_with_argmax(x,ps,st,self.padding)
25 | return [output1,output2]
26 | def compute_output_shape(self, input_shape):
27 | if isinstance(self.pool_size,int):
28 | output_shape=(input_shape[0],input_shape[1]//self.pool_size,input_shape[2]//self.pool_size,input_shape[3])
29 | else:
30 | output_shape=(input_shape[0],input_shape[1]//self.pool_size[0],input_shape[2]//self.pool_size[1],input_shape[3])
31 | return [output_shape,output_shape]
32 |
33 |
34 | class UpSamplingWithIndices(Layer):
35 | def __init__(self, **kwargs):
36 | super(UpSamplingWithIndices, self).__init__(**kwargs)
37 | return
38 | def call(self,x):
39 | argmax=K.cast(K.flatten(x[1]),'int32')
40 | max_value=K.flatten(x[0])
41 | with tf.compat.v1.variable_scope(self.name):
42 | input_shape=K.shape(x[0])
43 | batch_size=input_shape[0]
44 | image_size=input_shape[1]*input_shape[2]*input_shape[3]
45 | output_shape=[input_shape[0],input_shape[1]*2,input_shape[2]*2,input_shape[3]]
46 | indices_0=K.flatten(tf.matmul(K.reshape(tf.range(batch_size),(batch_size,1)),K.ones((1,image_size),dtype='int32')))
47 | indices_1=argmax%(image_size*4)//(output_shape[2]*output_shape[3])
48 | indices_2=argmax%(output_shape[2]*output_shape[3])//output_shape[3]
49 | indices_3=argmax%output_shape[3]
50 | indices=tf.stack([indices_0,indices_1,indices_2,indices_3])
51 | output=tf.scatter_nd(K.transpose(indices),max_value,output_shape)
52 | return output
53 | def compute_output_shape(self, input_shape):
54 | return input_shape[0][0],input_shape[0][1]*2,input_shape[0][2]*2,input_shape[0][3]
55 |
56 | def CompositeConv(inputs,num_layers,num_features):
57 | output=inputs
58 | if isinstance(num_features,int):
59 | for i in range(num_layers):
60 | output=Conv2D(num_features,(7,7),padding='same')(output)
61 | output=BatchNormalization(axis=3)(output)
62 | output=Activation('relu')(output)
63 | return output
64 | for i in range(num_layers):
65 | output=Conv2D(num_features[i],(7,7),padding='same')(output)
66 | output=BatchNormalization(axis=3)(output)
67 | output=Activation('relu')(output)
68 | return output
69 |
--------------------------------------------------------------------------------
/Code/network/segnet/segnet.py:
--------------------------------------------------------------------------------
1 | from keras.models import Model
2 | from keras.layers import Activation,Input,ZeroPadding2D,Cropping2D
3 | from .custom_layers import MaxPoolingWithIndices,UpSamplingWithIndices,CompositeConv
4 | #import config as cf
5 |
6 | def get_segnet(image_shape):
7 | padding=((0,0),(0,0))
8 | #image_shape=(1024,1024,3)
9 | num_classes = 1
10 | inputs=Input(shape=image_shape)
11 |
12 | x = ZeroPadding2D(padding)(inputs)
13 |
14 | x=CompositeConv(x,2,64)
15 | x,argmax1=MaxPoolingWithIndices(pool_size=2,strides=2)(x)
16 |
17 | x=CompositeConv(x,2,64)
18 | x,argmax2=MaxPoolingWithIndices(pool_size=2,strides=2)(x)
19 |
20 | x=CompositeConv(x,3,64)
21 | x,argmax3=MaxPoolingWithIndices(pool_size=2,strides=2)(x)
22 |
23 | x=CompositeConv(x,3,64)
24 | x,argmax4=MaxPoolingWithIndices(pool_size=2,strides=2)(x)
25 |
26 | x=CompositeConv(x,3,64)
27 | x,argmax5=MaxPoolingWithIndices(pool_size=2,strides=2)(x)
28 |
29 | x=UpSamplingWithIndices()([x,argmax5])
30 | x=CompositeConv(x,3,64)
31 |
32 | x=UpSamplingWithIndices()([x,argmax4])
33 | x=CompositeConv(x,3,64)
34 |
35 | x=UpSamplingWithIndices()([x,argmax3])
36 | x=CompositeConv(x,3,64)
37 |
38 | x=UpSamplingWithIndices()([x,argmax2])
39 | x=CompositeConv(x,2,64)
40 |
41 | x=UpSamplingWithIndices()([x,argmax1])
42 | x=CompositeConv(x,2,[64,num_classes])
43 |
44 | x=Activation('sigmoid')(x)
45 |
46 | y=Cropping2D(padding)(x)
47 | my_model=Model(inputs=inputs,outputs=y)
48 |
49 | return my_model
50 |
51 |
--------------------------------------------------------------------------------
/Code/network/segnet/segnet_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Code/network/segnet/segnet_plot.png
--------------------------------------------------------------------------------
/Code/network/unet/README.md:
--------------------------------------------------------------------------------
1 | 
2 |
--------------------------------------------------------------------------------
/Code/network/unet/u_net.py:
--------------------------------------------------------------------------------
1 | import os
2 | import random
3 |
4 | import tensorflow as tf
5 | from keras import backend as K
6 | from keras.models import Model, load_model
7 | from keras.layers import Input, BatchNormalization, Activation, Dense, Dropout
8 | from keras.layers.core import Lambda, RepeatVector, Reshape
9 | from keras.layers.convolutional import Conv2D, Conv2DTranspose
10 | from keras.layers.pooling import MaxPooling2D, GlobalMaxPool2D
11 | from keras.layers.merge import concatenate, add
12 |
13 | def conv2d_block(input_tensor, n_filters, kernel_size = 3, batchnorm = True):
14 | """Function to add 2 convolutional layers with the parameters passed to it"""
15 | # first layer
16 | x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
17 | kernel_initializer = 'he_normal', padding = 'same')(input_tensor)
18 | if batchnorm:
19 | x = BatchNormalization()(x)
20 | x = Activation('relu')(x)
21 |
22 | # second layer
23 | x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
24 | kernel_initializer = 'he_normal', padding = 'same')(x)
25 | if batchnorm:
26 | x = BatchNormalization()(x)
27 | x = Activation('relu')(x)
28 |
29 | return x
30 |
31 | def get_unet(input_img, n_filters = 16, dropout = 0.1, batchnorm = True):
32 | """Function to define the UNET Model"""
33 | # Contracting Path
34 | c1 = conv2d_block(input_img, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
35 | p1 = MaxPooling2D((2, 2))(c1)
36 | p1 = Dropout(dropout)(p1)
37 |
38 | c2 = conv2d_block(p1, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
39 | p2 = MaxPooling2D((2, 2))(c2)
40 | p2 = Dropout(dropout)(p2)
41 |
42 | c3 = conv2d_block(p2, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
43 | p3 = MaxPooling2D((2, 2))(c3)
44 | p3 = Dropout(dropout)(p3)
45 |
46 | c4 = conv2d_block(p3, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
47 | p4 = MaxPooling2D((2, 2))(c4)
48 | p4 = Dropout(dropout)(p4)
49 |
50 | c5 = conv2d_block(p4, n_filters = n_filters * 16, kernel_size = 3, batchnorm = batchnorm)
51 |
52 | # Expansive Path
53 | u6 = Conv2DTranspose(n_filters * 8, (3, 3), strides = (2, 2), padding = 'same')(c5)
54 | u6 = concatenate([u6, c4])
55 | u6 = Dropout(dropout)(u6)
56 | c6 = conv2d_block(u6, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
57 |
58 | u7 = Conv2DTranspose(n_filters * 4, (3, 3), strides = (2, 2), padding = 'same')(c6)
59 | u7 = concatenate([u7, c3])
60 | u7 = Dropout(dropout)(u7)
61 | c7 = conv2d_block(u7, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
62 |
63 | u8 = Conv2DTranspose(n_filters * 2, (3, 3), strides = (2, 2), padding = 'same')(c7)
64 | u8 = concatenate([u8, c2])
65 | u8 = Dropout(dropout)(u8)
66 | c8 = conv2d_block(u8, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
67 |
68 | u9 = Conv2DTranspose(n_filters * 1, (3, 3), strides = (2, 2), padding = 'same')(c8)
69 | u9 = concatenate([u9, c1])
70 | u9 = Dropout(dropout)(u9)
71 | c9 = conv2d_block(u9, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
72 |
73 | outputs = Conv2D(1, (1, 1), activation='sigmoid')(c9)
74 | model = Model(inputs=[input_img], outputs=[outputs])
75 | return model
76 |
77 |
--------------------------------------------------------------------------------
/Code/network/unet/unet_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Code/network/unet/unet_plot.png
--------------------------------------------------------------------------------
/Code/network/unetmod/README.md:
--------------------------------------------------------------------------------
1 | 
2 |
--------------------------------------------------------------------------------
/Code/network/unetmod/u_net_mod.py:
--------------------------------------------------------------------------------
1 | import os
2 | import random
3 |
4 | import tensorflow as tf
5 | from keras import backend as K
6 | from keras.models import Model, load_model
7 | from keras.layers import Input, BatchNormalization, Activation, Dense, Dropout, ZeroPadding2D, AveragePooling2D, GlobalAveragePooling2D, multiply
8 | from keras.layers.core import Lambda, RepeatVector, Reshape
9 | from keras.layers.convolutional import Conv2D, Conv2DTranspose
10 | from keras.layers.pooling import MaxPooling2D, GlobalMaxPool2D
11 | from keras.layers.merge import concatenate, add
12 | from keras.engine.topology import Layer
13 | from keras.engine import InputSpec
14 | from keras.layers import merge
15 | from keras import layers
16 |
17 | def squeeze_excite_block(tensor, ratio=16):
18 | init = tensor
19 | channel_axis = 1 if K.image_data_format() == "channels_first" else -1
20 | filters = init._keras_shape[channel_axis]
21 | se_shape = (1, 1, filters)
22 |
23 | se = GlobalAveragePooling2D()(init)
24 | se = Reshape(se_shape)(se)
25 | se = Dense(filters // ratio, activation='relu', kernel_initializer='he_normal', use_bias=False)(se)
26 | se = Dense(filters, activation='sigmoid', kernel_initializer='he_normal', use_bias=False)(se)
27 |
28 | if K.image_data_format() == 'channels_first':
29 | se = Permute((3, 1, 2))(se)
30 |
31 | x = multiply([init, se])
32 | return x
33 |
34 | def conv2d_block(input_tensor, n_filters, kernel_size = 3, batchnorm = True):
35 | """Function to add 2 convolutional layers with the parameters passed to it"""
36 | # first layer
37 | se = squeeze_excite_block(input_tensor)
38 | x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
39 | kernel_initializer = 'he_normal', padding = 'same')(input_tensor)
40 | if batchnorm:
41 | x = BatchNormalization()(x)
42 | x = Activation('relu')(x)
43 |
44 | # second layer
45 | x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
46 | kernel_initializer = 'he_normal', padding = 'same')(x)
47 | if batchnorm:
48 | x = BatchNormalization()(x)
49 | x = Activation('relu')(x)
50 | c1 = layers.add([x, se])
51 |
52 | return x
53 |
54 | def get_unet_mod(input_img, n_filters = 16, dropout = 0.1, batchnorm = True):
55 | """Function to define the UNET Model"""
56 | # Contracting Path
57 | skip = Conv2D(filters = n_filters * 1, kernel_size = (3, 3),\
58 | kernel_initializer = 'he_normal', padding = 'same')(input_img)
59 | c1 = conv2d_block(skip, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
60 | c1 = layers.add([c1, skip])
61 | p1 = MaxPooling2D((2, 2))(c1)
62 | p1 = Dropout(dropout)(p1)
63 |
64 | skip = Conv2D(filters = n_filters * 2, kernel_size = (3, 3),\
65 | kernel_initializer = 'he_normal', padding = 'same')(p1)
66 | c2 = conv2d_block(skip, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
67 | c2 = layers.add([c2, skip])
68 | p2 = MaxPooling2D((2, 2))(c2)
69 | p2 = Dropout(dropout)(p2)
70 |
71 | skip = Conv2D(filters = n_filters * 4, kernel_size = (3, 3),\
72 | kernel_initializer = 'he_normal', padding = 'same')(p2)
73 | c3 = conv2d_block(skip, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
74 | c3 = layers.add([c3, skip])
75 | p3 = MaxPooling2D((2, 2))(c3)
76 | p3 = Dropout(dropout)(p3)
77 |
78 | skip = Conv2D(filters = n_filters * 8, kernel_size = (3, 3),\
79 | kernel_initializer = 'he_normal', padding = 'same')(p3)
80 | c4 = conv2d_block(skip, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
81 | c4 = layers.add([c4, skip])
82 | p4 = MaxPooling2D((2, 2))(c4)
83 | p4 = Dropout(dropout)(p4)
84 |
85 | skip = Conv2D(filters = n_filters * 16, kernel_size = (3, 3),\
86 | kernel_initializer = 'he_normal', padding = 'same')(p4)
87 | c5 = conv2d_block(skip, n_filters = n_filters * 16, kernel_size = 3, batchnorm = batchnorm)
88 |
89 | c5 = aspp_block(c5,num_filters=256,rate_scale=1,output_stride=16,input_shape=(256,256,3))
90 |
91 | # Expansive Path
92 |
93 | u6 = Conv2DTranspose(n_filters * 8, (3, 3), strides = (2, 2), padding = 'same')(c5)
94 | u6 = concatenate([u6, c4])
95 | u6 = Dropout(dropout)(u6)
96 | skip = Conv2D(filters = n_filters * 8, kernel_size = (3, 3),\
97 | kernel_initializer = 'he_normal', padding = 'same')(u6)
98 | c6 = conv2d_block(skip, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
99 | c6 = layers.add([c6, skip])
100 |
101 |
102 | u7 = Conv2DTranspose(n_filters * 4, (3, 3), strides = (2, 2), padding = 'same')(c6)
103 | u7 = concatenate([u7, c3])
104 | u7 = Dropout(dropout)(u7)
105 | skip = Conv2D(filters = n_filters * 4, kernel_size = (3, 3),\
106 | kernel_initializer = 'he_normal', padding = 'same')(u7)
107 | c7 = conv2d_block(skip, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
108 | c6 = layers.add([c7, skip])
109 |
110 |
111 | u8 = Conv2DTranspose(n_filters * 2, (3, 3), strides = (2, 2), padding = 'same')(c7)
112 | u8 = concatenate([u8, c2])
113 | u8 = Dropout(dropout)(u8)
114 | skip = Conv2D(filters = n_filters * 2, kernel_size = (3, 3),\
115 | kernel_initializer = 'he_normal', padding = 'same')(u8)
116 | c8 = conv2d_block(skip, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
117 | c6 = layers.add([c8, skip])
118 |
119 |
120 | u9 = Conv2DTranspose(n_filters * 1, (3, 3), strides = (2, 2), padding = 'same')(c8)
121 | u9 = concatenate([u9, c1])
122 | u9 = Dropout(dropout)(u9)
123 | skip = Conv2D(filters = n_filters * 1, kernel_size = (3, 3),\
124 | kernel_initializer = 'he_normal', padding = 'same')(u9)
125 | c9 = conv2d_block(skip, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
126 | c6 = layers.add([c9, skip])
127 |
128 | #outputs = Conv2D(1, (1, 1), activation='sigmoid')(c5)
129 | #c9 = aspp_block(c5,num_filters=256,rate_scale=1,output_stride=16,input_shape=(256,256,3))
130 | outputs = Conv2D(1, (1, 1), activation='sigmoid')(c9)
131 | model = Model(inputs=[input_img], outputs=[outputs])
132 | return model
133 |
134 |
135 | def aspp_block(x,num_filters=256,rate_scale=1,output_stride=16,input_shape=(256,256,3)):
136 | if K.image_data_format() == 'channels_last':
137 | bn_axis = 3
138 | else:
139 | bn_axis = 1
140 | conv3_3_1 = ZeroPadding2D(padding=(6*rate_scale, 6*rate_scale))(x)
141 | conv3_3_1 = _conv(filters=num_filters, kernel_size=(3, 3),dilation_rate=(6*rate_scale, 6*rate_scale),padding='valid')(conv3_3_1)
142 | conv3_3_1 = BatchNormalization(axis=bn_axis)(conv3_3_1)
143 |
144 | conv3_3_2 = ZeroPadding2D(padding=(12*rate_scale, 12*rate_scale))(x)
145 | conv3_3_2 = _conv(filters=num_filters, kernel_size=(3, 3),dilation_rate=(12*rate_scale, 12*rate_scale),padding='valid')(conv3_3_2)
146 | conv3_3_2 = BatchNormalization(axis=bn_axis)(conv3_3_2)
147 |
148 | conv3_3_3 = ZeroPadding2D(padding=(18*rate_scale, 18*rate_scale))(x)
149 | conv3_3_3 = _conv(filters=num_filters, kernel_size=(3, 3),dilation_rate=(18*rate_scale, 18*rate_scale),padding='valid')(conv3_3_3)
150 | conv3_3_3 = BatchNormalization(axis=bn_axis)(conv3_3_3)
151 |
152 |
153 | # conv3_3_4 = ZeroPadding2D(padding=(24*rate_scale, 24*rate_scale))(x)
154 | # conv3_3_4 = _conv(filters=num_filters, kernel_size=(3, 3),dilation_rate=(24*rate_scale, 24*rate_scale),padding='valid')(conv3_3_4)
155 | # conv3_3_4 = BatchNormalization()(conv3_3_4)
156 |
157 | conv1_1 = _conv(filters=num_filters, kernel_size=(1, 1),padding='same')(x)
158 | conv1_1 = BatchNormalization(axis=bn_axis)(conv1_1)
159 |
160 | global_feat = AveragePooling2D((input_shape[0]/output_stride,input_shape[1]/output_stride))(x)
161 | global_feat = _conv(filters=num_filters, kernel_size=(1, 1),padding='same')(global_feat)
162 | global_feat = BatchNormalization()(global_feat)
163 | global_feat = BilinearUpSampling2D((256,input_shape[0]/output_stride,input_shape[1]/output_stride),factor=input_shape[1]/output_stride)(global_feat)
164 |
165 | # y = merge([conv3_3_1,conv3_3_2,conv3_3_3,conv1_1,global_feat,], mode='concat', concat_axis=3)
166 | y = concatenate([conv3_3_1,conv3_3_2,conv3_3_3,conv1_1,global_feat])
167 | """
168 | y = merge([
169 | conv3_3_1,
170 | conv3_3_2,
171 | conv3_3_3,
172 | # conv3_3_4,
173 | conv1_1,
174 | global_feat,
175 | ], mode='concat', concat_axis=3)
176 | """
177 |
178 | # y = _conv_bn_relu(filters=1, kernel_size=(1, 1),padding='same')(y)
179 | y = _conv(filters=256, kernel_size=(1, 1),padding='same')(y)
180 | y = BatchNormalization()(y)
181 | return y
182 |
183 | def _conv(**conv_params):
184 | """Helper to build a conv -> BN -> relu block
185 | """
186 | filters = conv_params["filters"]
187 | kernel_size = conv_params["kernel_size"]
188 | strides = conv_params.setdefault("strides", (1, 1))
189 | dilation_rate = conv_params.setdefault('dilation_rate',(1,1))
190 | # dilation_rate = conv_params["dilation_rate"]
191 | kernel_initializer = conv_params.setdefault("kernel_initializer", "he_normal")
192 | padding = conv_params.setdefault("padding", "same")
193 |
194 | def f(input):
195 | conv = Conv2D(filters=filters, kernel_size=kernel_size,
196 | strides=strides, padding=padding,
197 | dilation_rate=dilation_rate,
198 | kernel_initializer=kernel_initializer,activation='linear')(input)
199 | return conv
200 | return f
201 |
202 | class BilinearUpSampling2D(Layer):
203 | """Upsampling2D with bilinear interpolation."""
204 |
205 | def __init__(self, target_shape=None,factor=16, data_format=None, **kwargs):
206 | if data_format is None:
207 | data_format = K.image_data_format()
208 | assert data_format in {
209 | 'channels_last', 'channels_first'}
210 | self.data_format = data_format
211 | self.input_spec = [InputSpec(ndim=4)]
212 | self.target_shape = target_shape
213 | self.factor = factor
214 | if self.data_format == 'channels_first':
215 | self.target_size = (target_shape[2], target_shape[3])
216 | elif self.data_format == 'channels_last':
217 | self.target_size = (target_shape[1], target_shape[2])
218 | super(BilinearUpSampling2D, self).__init__(**kwargs)
219 |
220 | def compute_output_shape(self, input_shape):
221 | if self.data_format == 'channels_last':
222 | return (input_shape[0], self.target_size[0],
223 | self.target_size[1], input_shape[3])
224 | else:
225 | return (input_shape[0], input_shape[1],
226 | self.target_size[0], self.target_size[1])
227 |
228 | def call(self, inputs):
229 | return K.resize_images(inputs, self.factor, self.factor, self.data_format)
230 |
231 | def get_config(self):
232 | config = {'target_shape': self.target_shape,
233 | 'data_format': self.data_format}
234 | base_config = super(BilinearUpSampling2D, self).get_config()
235 | return dict(list(base_config.items()) + list(config.items()))
236 |
--------------------------------------------------------------------------------
/Code/network/unetmod/unet_plot.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Code/network/unetmod/unet_plot.png
--------------------------------------------------------------------------------
/Code/utils/lossfunctions.py:
--------------------------------------------------------------------------------
1 | from keras import backend as K
2 | import tensorflow as tf
3 |
4 |
5 | def jaccard_distance_loss(y_true, y_pred, smooth=100):
6 | """
7 | Jaccard = (|X & Y|)/ (|X|+ |Y| - |X & Y|)
8 | = sum(|A*B|)/(sum(|A|)+sum(|B|)-sum(|A*B|))
9 |
10 | The jaccard distance loss is usefull for unbalanced datasets. This has been
11 | shifted so it converges on 0 and is smoothed to avoid exploding or disapearing
12 | gradient.
13 |
14 | Ref: https://en.wikipedia.org/wiki/Jaccard_index
15 |
16 | @url: https://gist.github.com/wassname/f1452b748efcbeb4cb9b1d059dce6f96
17 | @author: wassname
18 | """
19 | intersection = K.sum(K.abs(y_true * y_pred), axis=-1)
20 | sum_ = K.sum(K.abs(y_true) + K.abs(y_pred), axis=-1)
21 | jac = (intersection + smooth) / (sum_ - intersection + smooth)
22 | return (1 - jac) * smooth
23 |
24 | def dice_loss(y_true, y_pred):
25 | numerator = 2 * tf.reduce_sum(y_true * y_pred, axis=(1,2,3))
26 | denominator = tf.reduce_sum(y_true + y_pred, axis=(1,2,3))
27 |
28 | return 1 - numerator / denominator
29 |
30 | def focal_loss(alpha=0.25, gamma=2):
31 | def focal_loss_with_logits(logits, targets, alpha, gamma, y_pred):
32 | weight_a = alpha * (1 - y_pred) ** gamma * targets
33 | weight_b = (1 - alpha) * y_pred ** gamma * (1 - targets)
34 |
35 | return (tf.math.log1p(tf.exp(-tf.abs(logits))) + tf.nn.relu(-logits)) * (weight_a + weight_b) + logits * weight_b
36 |
37 | def loss(y_true, y_pred):
38 | y_pred = tf.clip_by_value(y_pred, tf.keras.backend.epsilon(), 1 - tf.keras.backend.epsilon())
39 | logits = tf.math.log(y_pred / (1 - y_pred))
40 |
41 | loss = focal_loss_with_logits(logits=logits, targets=y_true, alpha=alpha, gamma=gamma, y_pred=y_pred)
42 |
43 | # or reduce_sum and/or axis=-1
44 | return tf.reduce_mean(loss)
45 |
46 | return loss
--------------------------------------------------------------------------------
/Code/utils/metricfunctions.py:
--------------------------------------------------------------------------------
1 | from keras import backend as K
2 |
3 | def dice_coef(y_true, y_pred):
4 | y_true_f = K.flatten(y_true)
5 | y_pred_f = K.flatten(y_pred)
6 | intersection = K.sum(y_true_f * y_pred_f)
7 | return (2. * intersection + K.epsilon()) / (K.sum(y_true_f) + K.sum(y_pred_f) + K.epsilon())
8 |
9 | """
10 | def dice_coef(y_true, y_pred, smooth=1.):
11 | y_true_f = K.flatten(y_true)
12 | y_pred_f = K.flatten(y_pred)
13 | intersection = K.sum(y_true_f * y_pred_f)
14 | return (2. * intersection + smooth) / (
15 | K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
16 | """
17 | def f1(y_true, y_pred):
18 | def recall(y_true, y_pred):
19 | """Recall metric.
20 |
21 | Only computes a batch-wise average of recall.
22 |
23 | Computes the recall, a metric for multi-label classification of
24 | how many relevant items are selected.
25 | """
26 | true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
27 | possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
28 | recall = true_positives / (possible_positives + K.epsilon())
29 | return recall
30 |
31 | def precision(y_true, y_pred):
32 | """Precision metric.
33 |
34 | Only computes a batch-wise average of precision.
35 |
36 | Computes the precision, a metric for multi-label classification of
37 | how many selected items are relevant.
38 | """
39 | true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
40 | predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
41 | precision = true_positives / (predicted_positives + K.epsilon())
42 | return precision
43 | precision = precision(y_true, y_pred)
44 | recall = recall(y_true, y_pred)
45 | return 2*((precision*recall)/(precision+recall+K.epsilon()))
--------------------------------------------------------------------------------
/Datasets/MonuSeg-20200319T073151Z-001.zip:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Datasets/MonuSeg-20200319T073151Z-001.zip
--------------------------------------------------------------------------------
/Datasets/README.md:
--------------------------------------------------------------------------------
1 | # 01_Datasets README File
2 |
--------------------------------------------------------------------------------
/Datasets/Samples/TCGA-RD-A8N9-01A-01-TS1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Datasets/Samples/TCGA-RD-A8N9-01A-01-TS1.png
--------------------------------------------------------------------------------
/Datasets/Samples/TCGA-RD-A8N9-01A-01-TS1_bin_mask.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Datasets/Samples/TCGA-RD-A8N9-01A-01-TS1_bin_mask.png
--------------------------------------------------------------------------------
/Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1.png
--------------------------------------------------------------------------------
/Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1_bin_mask.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1_bin_mask.png
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | ## Repository for [A Multi-organ Nucleus Segmentation Challenge (MoNuSeg)](https://ieeexplore.ieee.org/document/8880654).
2 |
3 | Note: If you're interested in using it, feel free to ⭐️ the repo so we know!
4 |
5 | ### Current Features
6 | - [x] Config File
7 | - [x] Training Graphs
8 | - [x] Patch-Wise Input
9 | - [x] Updation of README Files
10 | - [x] Inference Files
11 | - [x] Quantitative Results
12 | - [x] Visualization of Results
13 | - [x] Train File
14 | - [x] Directory Structure
15 | - [x] Weights Save With Model
16 |
17 | Legend
18 | - [x] Resolved
19 | - [ ] Work In-Progess
20 |
21 | ## Dataset
22 | The dataset for this challenge was obtained by carefully annotating tissue images of several patients with tumors of different organs and who were diagnosed at multiple hospitals. This dataset was created by downloading H&E stained tissue images captured at 40x magnification from TCGA archive. H&E staining is a routine protocol to enhance the contrast of a tissue section and is commonly used for tumor assessment (grading, staging, etc.). Given the diversity of nuclei appearances across multiple organs and patients, and the richness of staining protocols adopted at multiple hospitals, the training datatset will enable the development of robust and generalizable nuclei segmentation techniques that will work right out of the box.
23 |
24 | ### Training Data
25 | Training data containing 30 images and around 22,000 nuclear boundary annotations has been released to the public previously as a dataset article in IEEE Transactions on Medical imaging in 2017.
26 |
27 | ### Testing Data
28 | Test set images with additional 7000 nuclear boundary annotations are available here MoNuSeg 2018 Testing data.
29 |
30 | Dataset can be downloaded from [Grand Challenge Webiste](https://monuseg.grand-challenge.org/)
31 |
32 | A training sample with segmentation mask from training set can be seen below:
33 | | Tissue | Segmentation Mask (Ground Truth) |
34 | :-------------------------:|:-------------------------:
35 |  | 
36 |
37 | ### Patch Generation
38 | Since the size of data set is small and was eaisly loaded into the memmory so we have created patches in online mode. . All the images of training set and test set were reshape to 1024x1024 and then patches were extracted from them. The patch dimensions comprised of **256x256** with 50% overlap among them.
39 |
40 | ## Models Diagrams
41 |
42 | ### U-Net Modified
43 | Blocks used to modify U-Net are:
44 | #### Atrous Spatial Pyramid Pooling(ASPP)
45 |  46 |
47 | #### Squeeze-and-Excitation Network
48 |
46 |
47 | #### Squeeze-and-Excitation Network
48 |  49 |
50 |
51 | ### SegNet
52 | 
53 |
54 |
55 | ### U-Net
56 | 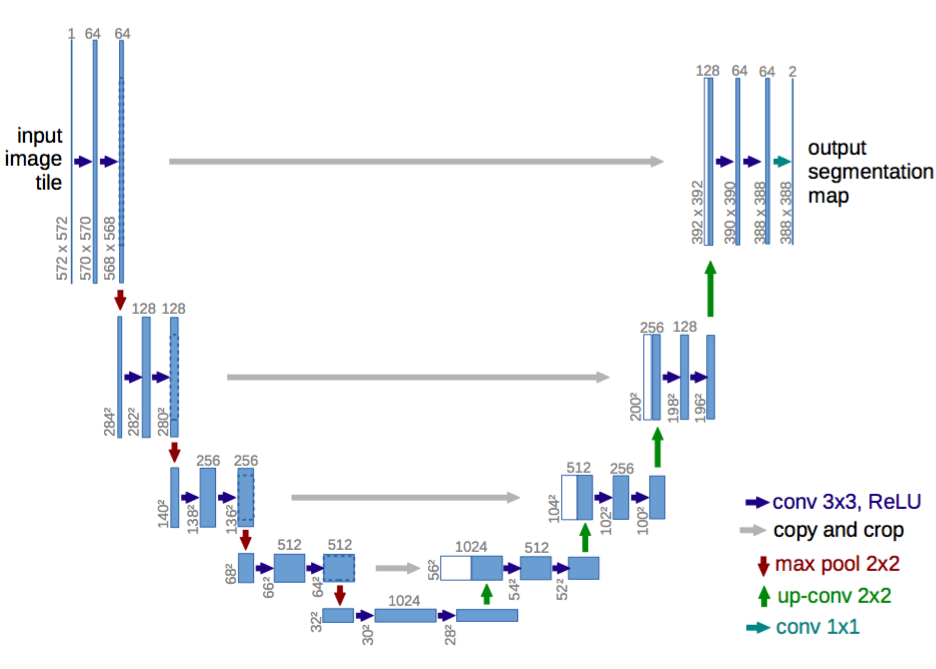
57 |
58 | ### Deep Lab v3+
59 | 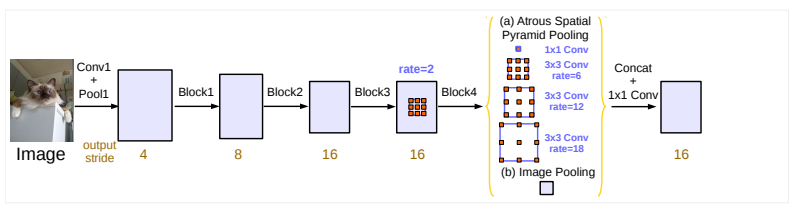
60 |
61 |
62 | ## Pre-Trained Models
63 | The Pre-Trained models can be downloaded from [google drive](https://drive.google.com/drive/folders/1g5SdbW8q1Z0e9dk6cW431JO01BDq4g0H).
64 |
65 | ## Installation
66 | To get this repo work please install all the dependencies using the command below:
67 | ```
68 | pip install -r requirments.txt
69 | ```
70 |
71 | ## Training
72 | To start training run the Train.py script from the command below. For training configurations refer to the [config.json](./config.json) file. You can update the file according to your training settings. Model avaible for training are U-NET,SegNet, DeepLabv3+.
73 | ```
74 | python Train.py
75 | ```
76 |
77 | ## Testing
78 | To test the trained models on Test Images you first have to download the weights and place them in the [results](./Results/). After downliading the weights you unzip them and then run the Inference by using the command below. For testing configurations please refer to the [config.json](./config.json) file.
79 | ```
80 | python Test.py
81 | ```
82 |
83 | ## Visualization of Results
84 | | Tissue | Mask | Predicted Mask |
85 | :-------------------------:|:-------------------------:|:-------------------------:
86 |  |  | 
87 |
88 | ## Quantitatvie Results
89 |
90 | | Model | Loss | Accuracy | F1 Score | Dice Score |
91 | | ----- | ---- | ---- | ---- | ---- |
92 | | Unet | 0.0835 | 0.9150 | 0.7910 | 0.7906
93 | | Segnet | 0.4820 | 0.8077 | 0.5798 | 0.3684
94 | | DeeplabV3+ | 0.0783 | 0.9120 | 0.7750 | 0.7743
95 | | Unet + Skip Connections + ASPP + SE Block | 0.0770 | 0.9210 | 0.801 | 0.8005
96 |
97 | ## Results
98 | Three Segmentation models have been trained and the model is evaluated on three metrics namely:
99 | * Accuracy
100 | * F1-Score
101 | * Dice Score
102 | ### U-Net + Skip Connections + ASPP
103 |
49 |
50 |
51 | ### SegNet
52 | 
53 |
54 |
55 | ### U-Net
56 | 
57 |
58 | ### Deep Lab v3+
59 | 
60 |
61 |
62 | ## Pre-Trained Models
63 | The Pre-Trained models can be downloaded from [google drive](https://drive.google.com/drive/folders/1g5SdbW8q1Z0e9dk6cW431JO01BDq4g0H).
64 |
65 | ## Installation
66 | To get this repo work please install all the dependencies using the command below:
67 | ```
68 | pip install -r requirments.txt
69 | ```
70 |
71 | ## Training
72 | To start training run the Train.py script from the command below. For training configurations refer to the [config.json](./config.json) file. You can update the file according to your training settings. Model avaible for training are U-NET,SegNet, DeepLabv3+.
73 | ```
74 | python Train.py
75 | ```
76 |
77 | ## Testing
78 | To test the trained models on Test Images you first have to download the weights and place them in the [results](./Results/). After downliading the weights you unzip them and then run the Inference by using the command below. For testing configurations please refer to the [config.json](./config.json) file.
79 | ```
80 | python Test.py
81 | ```
82 |
83 | ## Visualization of Results
84 | | Tissue | Mask | Predicted Mask |
85 | :-------------------------:|:-------------------------:|:-------------------------:
86 |  |  | 
87 |
88 | ## Quantitatvie Results
89 |
90 | | Model | Loss | Accuracy | F1 Score | Dice Score |
91 | | ----- | ---- | ---- | ---- | ---- |
92 | | Unet | 0.0835 | 0.9150 | 0.7910 | 0.7906
93 | | Segnet | 0.4820 | 0.8077 | 0.5798 | 0.3684
94 | | DeeplabV3+ | 0.0783 | 0.9120 | 0.7750 | 0.7743
95 | | Unet + Skip Connections + ASPP + SE Block | 0.0770 | 0.9210 | 0.801 | 0.8005
96 |
97 | ## Results
98 | Three Segmentation models have been trained and the model is evaluated on three metrics namely:
99 | * Accuracy
100 | * F1-Score
101 | * Dice Score
102 | ### U-Net + Skip Connections + ASPP
103 |
104 |  105 |
105 |  106 |
106 |  107 |
107 |  108 |
108 |
109 |
110 | ### U-Net
111 |
112 |  113 |
113 |  114 |
114 |  115 |
115 |  116 |
116 |
117 |
118 | ### SegNet
119 |
120 |  121 |
121 |  122 |
122 |  123 |
123 |  124 |
124 |
125 |
126 | ### DeepLab v3
127 |
128 |  129 |
129 |  130 |
130 |  131 |
131 |  132 |
132 |
133 |
134 | ## Author
135 | `Maintainer` [Syed Nauyan Rashid](https://https://github.com/nauyan) (nauyan@hotmail.com)
136 |
137 |
138 |
139 |
--------------------------------------------------------------------------------
/Results/outputs/TCGA-HT-8564-01Z-00-DX1.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/outputs/TCGA-HT-8564-01Z-00-DX1.jpg
--------------------------------------------------------------------------------
/Results/plots/DEEPLAB/train_accuracy.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/DEEPLAB/train_accuracy.png
--------------------------------------------------------------------------------
/Results/plots/DEEPLAB/train_dice.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/DEEPLAB/train_dice.png
--------------------------------------------------------------------------------
/Results/plots/DEEPLAB/train_f1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/DEEPLAB/train_f1.png
--------------------------------------------------------------------------------
/Results/plots/DEEPLAB/train_loss.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/DEEPLAB/train_loss.png
--------------------------------------------------------------------------------
/Results/plots/README.md:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/README.md
--------------------------------------------------------------------------------
/Results/plots/SEGNET/train_accuracy.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/SEGNET/train_accuracy.png
--------------------------------------------------------------------------------
/Results/plots/SEGNET/train_dice.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/SEGNET/train_dice.png
--------------------------------------------------------------------------------
/Results/plots/SEGNET/train_f1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/SEGNET/train_f1.png
--------------------------------------------------------------------------------
/Results/plots/SEGNET/train_loss.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/SEGNET/train_loss.png
--------------------------------------------------------------------------------
/Results/plots/UNET/train_accuracy.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNET/train_accuracy.png
--------------------------------------------------------------------------------
/Results/plots/UNET/train_dice.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNET/train_dice.png
--------------------------------------------------------------------------------
/Results/plots/UNET/train_f1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNET/train_f1.png
--------------------------------------------------------------------------------
/Results/plots/UNET/train_loss.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNET/train_loss.png
--------------------------------------------------------------------------------
/Results/plots/UNETMOD/train_accuracy.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNETMOD/train_accuracy.png
--------------------------------------------------------------------------------
/Results/plots/UNETMOD/train_dice.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNETMOD/train_dice.png
--------------------------------------------------------------------------------
/Results/plots/UNETMOD/train_f1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNETMOD/train_f1.png
--------------------------------------------------------------------------------
/Results/plots/UNETMOD/train_loss.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/nauyan/Segmentation/fce3667286ef9c4a7f3e7053a260280ed948eb04/Results/plots/UNETMOD/train_loss.png
--------------------------------------------------------------------------------
/Test.py:
--------------------------------------------------------------------------------
1 | import keras
2 | from keras import backend as K
3 | import json
4 | from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img, save_img
5 | import os
6 | from Code.network.segnet.custom_layers import MaxPoolingWithIndices,UpSamplingWithIndices,CompositeConv
7 | from Code.network.unetmod.u_net_mod import BilinearUpSampling2D
8 | import tensorflow as tf
9 | import cv2
10 | import numpy as np
11 | from skimage.util.shape import view_as_windows
12 | import math
13 |
14 |
15 |
16 | def createTiles(img):
17 | size = img.shape[0]
18 | sizeNew = 0
19 | for x in range(0,100):
20 | if size<2**x:
21 | sizeNew = 2**x
22 | break
23 |
24 | pad = sizeNew - size
25 | imgNew = np.zeros((sizeNew,sizeNew,3))
26 | imgNew[pad//2:sizeNew-(pad//2),pad//2:sizeNew-(pad//2),:] = img
27 |
28 | #save_img("Check.png", imgNew)
29 |
30 | new_imgs = view_as_windows(imgNew, (patch_width, patch_height, 3), (patch_width, patch_height, 3))
31 | new_imgs = new_imgs.reshape(-1,patch_width, patch_height, 3)
32 | #print(new_imgs.shape)
33 |
34 | return new_imgs
35 |
36 | def mergeTiles(tiles):
37 | #print(int(math.sqrt(tiles.shape[0])))
38 | num = int(math.sqrt(tiles.shape[0]))
39 | img = np.zeros((patch_width*num,patch_height*num,1))
40 | count = 0
41 | for x in range(0,num):
42 | for y in range(0,num):
43 | startX = (x)*(patch_width)
44 | endX = (x+1)*(patch_width)
45 | startY = (y)*(patch_width)
46 | endY = (y+1)*(patch_width)
47 |
48 | img[startX:endX,startY:endY,:]= tiles[count]
49 | count = count + 1
50 |
51 | pad = img.shape[0]-im_width
52 | img = img[pad//2:im_width+(pad//2),pad//2:im_width+(pad//2),:]
53 | return img
54 |
55 |
56 | os.environ["CUDA_VISIBLE_DEVICES"] = "1"
57 |
58 | with open('./config.json') as config_file:
59 | config = json.load(config_file)
60 |
61 |
62 | im_width = config['im_width']
63 | im_height = config['im_height']
64 | patch_width = config['patch_width']
65 | patch_height = config['patch_height']
66 |
67 | if config['Model'] == "UNETMOD":
68 | model = keras.models.load_model("./Results/weights/" + config['Model'] + "/" +config['Model']+"-Best.h5", compile=False,
69 | custom_objects={'BilinearUpSampling2D':BilinearUpSampling2D(target_shape=(256,16,16),data_format=K.image_data_format())})
70 |
71 | if config['Model']=="UNET":
72 | model = keras.models.load_model("./Results/weights/" + config['Model'] + "/" +config['Model']+"-Best.h5", compile=False)
73 | if config['Model']=="SEGNET":
74 | model = keras.models.load_model("./Results/weights/" + config['Model'] + "/" +config['Model']+"-Best.h5", compile=False,
75 | custom_objects={'MaxPoolingWithIndices':MaxPoolingWithIndices,'UpSamplingWithIndices':UpSamplingWithIndices})
76 | if config['Model']=="DEEPLAB":
77 | model = tf.keras.models.load_model("./Results/weights/" + config['Model'] + "/" +config['Model']+"-Best.h5", compile=False, custom_objects={'tf': tf})
78 |
79 | #print(model.summary())
80 | #config['sample_test_image']
81 | #config['sample_test_mask']
82 | image = img_to_array(load_img(config['sample_test_image'], color_mode='rgb', target_size=[config['im_width'],config['im_height']]))/255.0
83 | mask = img_to_array(load_img(config['sample_test_mask'], color_mode='grayscale', target_size=[config['im_width'],config['im_height']]))/255.0
84 |
85 | image = createTiles(image)
86 |
87 |
88 | #print(image.shape)
89 | pred = model.predict(image)
90 | #print(pred.shape)
91 |
92 | pred = mergeTiles(pred)
93 | #print(pred.shape)
94 | img_array = pred
95 | #img_array = img_to_array(pred)
96 | # save the image with a new filename
97 |
98 | base=os.path.basename(config['sample_test_image'])
99 | fn = os.path.splitext(base)[0]
100 | filename = './Results/outputs/'+fn+'.jpg'
101 | save_img(filename, img_array*255.0)
102 | print("The Output mask is stored at "+ filename)
103 |
104 |
--------------------------------------------------------------------------------
/Train.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import numpy as np
3 | import matplotlib.pyplot as plt
4 | import os
5 |
6 | from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
7 | from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
8 | from keras.optimizers import Adam, nadam,SGD
9 | from keras.layers import Input
10 | # from Code.utils.lossfunctions import jaccard_distance_loss,dice_coef_loss
11 | from Code.utils.metricfunctions import dice_coef,f1
12 | from Code.utils.lossfunctions import *
13 |
14 | #from Code.network.unetmod.u_net_mod import get_unet_mod
15 | from Code.network.unetmod.u_net_mod import *
16 | from Code.network.unet.u_net import get_unet
17 | from Code.network.segnet.segnet import get_segnet
18 | from Code.network.deeplab.deeplab import Deeplabv3
19 | import argparse
20 | import tensorflow as tf
21 |
22 | from skimage.util.shape import view_as_windows
23 | import json
24 |
25 | os.environ["CUDA_VISIBLE_DEVICES"] = "1"
26 |
27 |
28 | with open('./config.json') as config_file:
29 | config = json.load(config_file)
30 | # print (config)
31 | im_width = config['im_width']
32 | im_height = config['im_height']
33 | patch_width = config['patch_width']
34 | patch_height = config['patch_height']
35 | Epochs = config['Epochs']
36 |
37 |
38 | TRAIN_PATH_IMAGES = config['TRAIN_PATH_IMAGES']
39 | TRAIN_PATH_GT = config['TRAIN_PATH_GT']
40 | TEST_PATH_IMAGES = config['TEST_PATH_IMAGES']
41 | TEST_PATH_GT = config['TEST_PATH_GT']
42 |
43 |
44 | ids_train_x = glob.glob(TRAIN_PATH_IMAGES)
45 | ids_train_y = glob.glob(TRAIN_PATH_GT)
46 | print("No. of training images = ", len(ids_train_x))
47 | ids_test_x = glob.glob(TEST_PATH_IMAGES)
48 | ids_test_y = glob.glob(TEST_PATH_GT)
49 | print("No. of testing images = ", len(ids_test_x))
50 |
51 | #X_train = np.zeros((len(ids_train_x), im_height, im_width, 3), dtype=np.float32)
52 | #y_train = np.zeros((len(ids_train_y), im_height, im_width, 1), dtype=np.float32)
53 |
54 | #X_test = np.zeros((len(ids_test_x), im_height, im_width, 3), dtype=np.float32)
55 | #y_test = np.zeros((len(ids_test_y), im_height, im_width, 1), dtype=np.float32)
56 |
57 | X_train = []
58 | y_train = []
59 | X_test = []
60 | y_test = []
61 |
62 | print("Loading Training Data")
63 | count =0
64 | for x in (ids_train_x):
65 | base=os.path.basename(x)
66 | fn = os.path.splitext(base)[0]
67 | y = glob.glob(config['TRAIN_PATH_GT']+fn+'*')[0]
68 | x_img = img_to_array(load_img(x, color_mode='rgb', target_size=[im_width,im_height]))
69 | x_img = x_img/255.0
70 | # Load masks
71 | mask = img_to_array(load_img(y, color_mode='grayscale', target_size=[im_width,im_height]))
72 | mask = mask/255.0
73 | #X_train[count] = x_img/255.0
74 | #y_train[count] = mask/255.0
75 | new_imgs = view_as_windows(x_img, (patch_width, patch_height, 3), (patch_width//2, patch_height//2, 3))
76 | #print("Number of Patches")
77 | #print(new_imgs.shape)
78 | for patch in new_imgs:
79 | X_train.append(patch)
80 | new_masks = view_as_windows(mask, (patch_width, patch_height, 1), (patch_width//2, patch_height//2, 1))
81 | for patch in new_masks:
82 | y_train.append(patch)
83 | count = count+1
84 |
85 |
86 |
87 | print("Loading Testing Data")
88 | count =0
89 | for x in (ids_test_x):
90 | base=os.path.basename(x)
91 | fn = os.path.splitext(base)[0]
92 | y = glob.glob(config['TEST_PATH_GT']+fn+'*')[0]
93 | x_img = img_to_array(load_img(x, color_mode='rgb', target_size=[im_width,im_height]))
94 | x_img = x_img/255.0
95 | # Load masks
96 | mask = img_to_array(load_img(y, color_mode='grayscale', target_size=[im_width,im_height]))
97 | mask = mask/255.0
98 | #X_test[count] = x_img/255.0
99 | #y_test[count] = mask/255.0
100 | new_imgs = view_as_windows(x_img, (patch_width, patch_height, 3), (patch_width//2, patch_height//2, 3))
101 | for patch in new_imgs:
102 | X_test.append(patch)
103 | new_masks = view_as_windows(mask, (patch_width, patch_height, 1), (patch_width//2, patch_height//2, 1))
104 | for patch in new_masks:
105 | y_test.append(patch)
106 | count = count+1
107 |
108 |
109 | #print(len(X_train),len(y_train))
110 | #print(len(X_test),len(y_test))
111 | X_train = np.array(X_train)
112 | y_train = np.array(y_train)
113 | X_test = np.array(X_test)
114 | y_test = np.array(y_test)
115 |
116 | input_img = Input((256, 256, 3), name='img')
117 | #from tensorflow.keras.utils.vis_utils import plot_model
118 |
119 | if config['Model'] == "UNETMOD":
120 | print("Loading UNETMOD Model")
121 | model = get_unet_mod(input_img, n_filters=16, dropout=0.1, batchnorm=True) #32
122 | # model.compile(optimizer=Adam(1e-5), loss=jaccard_distance_loss, metrics=[iou,dice_coef])
123 | model.compile(optimizer=Adam(amsgrad=True), loss=jaccard_distance_loss, metrics=["accuracy", dice_coef, f1])
124 | print("Printing Model Summary")
125 | print (model.summary())
126 | tf.keras.utils.plot_model(model, './Code/network/unetmod/unet_plot.png')
127 |
128 | if config['Model'] == "UNET":
129 | print("Loading UNET Model")
130 | model = get_unet(input_img, n_filters=16, dropout=0.1, batchnorm=True)
131 | # model.compile(optimizer=Adam(1e-5), loss=jaccard_distance_loss, metrics=[iou,dice_coef])
132 | model.compile(optimizer=Adam(amsgrad=True), loss=jaccard_distance_loss, metrics=["accuracy", dice_coef, f1])
133 | print("Printing Model Summary")
134 | print (model.summary())
135 | tf.keras.utils.plot_model(model, './Code/network/unet/unet_plot.png')
136 |
137 | if config['Model'] == "SEGNET":
138 | print("Loading SEGNET Model")
139 | model = get_segnet((patch_height, patch_width, 3))
140 | #n_labels=3,
141 | #kernel=3,
142 | #pool_size=(2, 2),
143 | #output_mode="softmax")
144 | model.compile(optimizer=Adam(amsgrad=True), loss=jaccard_distance_loss, metrics=["accuracy", dice_coef, f1])
145 | print("Printing Model Summary")
146 | print (model.summary())
147 | tf.keras.utils.plot_model(model, './Code/network/segnet/segnet_plot.png')
148 |
149 | if config['Model'] == "DEEPLAB":
150 | print("Loading DEEPLAB Model")
151 | model = Deeplabv3(weights=None, input_tensor=None, input_shape=(patch_height, patch_width, 3), classes=1, backbone='xception',
152 | OS=16, alpha=1., activation='sigmoid')
153 | model.compile(optimizer=tf.keras.optimizers.Adam(amsgrad=True), loss=jaccard_distance_loss, metrics=["accuracy", dice_coef, f1])
154 | #plot_model(model, to_file='./Code/network/deeplab/deeplab_plot.png', show_shapes=True, show_layer_names=True)
155 |
156 | print("Compiling Model")
157 | #model.compile(optimizer=sgd(), loss="binary_crossentropy"dice_coef_loss,jaccard_distance_loss metrics=["accuracy"]) # ,f1_m,iou_coef,dice_coef
158 | #
159 |
160 |
161 | #nadam(lr=1e-5)
162 | #Adam(1e-5, amsgrad=True, clipnorm=5.0)
163 | #Adam()
164 | #SGD(lr=1e-5, momentum=0.95)
165 | callbacks = [
166 | EarlyStopping(patience=10, verbose=1),
167 | ReduceLROnPlateau(factor=0.1, patience=10, min_lr=0.00001, verbose=1),
168 | ModelCheckpoint('./Results/weights/'+str(config['Model'])+'/'+str(config['Model'])+'-Best.h5', monitor='val_dice_coef',mode = 'max' , verbose=1, save_best_only=True, save_weights_only=False)
169 | ]
170 | X_train = X_train.reshape(-1,patch_height,patch_width,3)
171 | y_train = y_train.reshape(-1,patch_height,patch_width,1)
172 | X_test = X_test.reshape(-1,patch_height,patch_width,3)
173 | y_test = y_test.reshape(-1,patch_height,patch_width,1)
174 |
175 | print(X_train.shape, y_train.shape)
176 | print(X_test.shape, y_test.shape)
177 |
178 |
179 | results = model.fit(X_train, y_train, batch_size=config['Batch'], verbose=1, epochs=Epochs, callbacks=callbacks,\
180 | validation_data=(X_test, y_test))
181 |
182 | print(model.evaluate(X_test, y_test, verbose=1))
183 |
184 | plt.figure(figsize=(8, 8))
185 | plt.title("Learning curve")
186 | plt.plot(results.history["loss"], label="loss")
187 | plt.plot(results.history["val_loss"], label="val_loss")
188 | plt.plot( np.argmin(results.history["val_loss"]), np.min(results.history["val_loss"]), marker="x", color="r", label="best model")
189 | plt.xlabel("Epochs")
190 | plt.ylabel("log_loss")
191 | plt.legend();
192 | plt.savefig('./Results/plots/'+str(config['Model'])+'/train_loss.png')
193 |
194 | plt.figure(figsize=(8, 8))
195 | plt.title("Learning curve")
196 | plt.plot(results.history["dice_coef"], label="dice_coef")
197 | plt.plot(results.history["val_dice_coef"], label="val_dice_coef")
198 | plt.plot( np.argmax(results.history["val_dice_coef"]), np.max(results.history["val_dice_coef"]), marker="x", color="r", label="best model")
199 | plt.xlabel("Epochs")
200 | plt.ylabel("Dice Coeff")
201 | plt.legend();
202 | plt.savefig('./Results/plots/'+str(config['Model'])+'/train_dice.png')
203 |
204 | plt.figure(figsize=(8, 8))
205 | plt.title("Learning curve")
206 | plt.plot(results.history["f1"], label="f1")
207 | plt.plot(results.history["val_f1"], label="val_f1")
208 | plt.plot( np.argmax(results.history["val_f1"]), np.max(results.history["val_f1"]), marker="x", color="r", label="best model")
209 | plt.xlabel("Epochs")
210 | plt.ylabel("f1")
211 | plt.legend();
212 | plt.savefig('./Results/plots/'+str(config['Model'])+'/train_f1.png')
213 |
214 | plt.figure(figsize=(8, 8))
215 | plt.title("Learning curve")
216 | plt.plot(results.history["accuracy"], label="accuracy")
217 | plt.plot(results.history["val_accuracy"], label="val_accuracy")
218 | plt.plot( np.argmax(results.history["val_accuracy"]), np.max(results.history["val_accuracy"]), marker="x", color="r", label="best model")
219 | plt.xlabel("Epochs")
220 | plt.ylabel("accuracy")
221 | plt.legend();
222 | plt.savefig('./Results/plots/'+str(config['Model'])+'/train_accuracy.png')
223 |
224 |
225 |
--------------------------------------------------------------------------------
/Train_Bak.py:
--------------------------------------------------------------------------------
1 | import glob
2 | import numpy as np
3 | import matplotlib.pyplot as plt
4 |
5 |
6 | from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
7 | from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
8 | from keras.optimizers import Adam, nadam,SGD
9 | from keras.layers import Input
10 | from Code.utils.lossfunctions import jaccard_distance_loss,dice_coef_loss
11 | from Code.utils.metricfunctions import iou,dice_coef
12 |
13 | from Code.network.unet.u_net import get_unet
14 | from Code.network.segnet.segnet import get_segnet
15 | from Code.network.deeplab.deeplab import Deeplabv3
16 | import argparse
17 | import tensorflow as tf
18 |
19 | from skimage.util.shape import view_as_windows
20 |
21 |
22 | parser = argparse.ArgumentParser(description='Simple training script for training a U-Net network.')
23 | parser.add_argument('--TRAIN_PATH_IMAGES', help='Path to Training Images', type=str, default='./Datasets/MonuSeg/Training/TissueImages/*')
24 | parser.add_argument('--TRAIN_PATH_GT', help='Path to Training Ground Truth Images', type=str, default='./Datasets/MonuSeg/Training/GroundTruth/*')
25 | parser.add_argument('--TEST_PATH_IMAGES', help='Path to Test Images', type=str, default='./Datasets/MonuSeg/Test/TissueImages/*')
26 | parser.add_argument('--TEST_PATH_GT', help='Path to Test Ground Truth Images', type=str, default='./Datasets/MonuSeg/Test/GroundTruth/*')
27 | parser.add_argument('--im_width', help='Width of Image', type=int, default=1024)
28 | parser.add_argument('--im_height', help='Height of Image', type=int, default=1024)
29 | parser.add_argument('--Epochs', help='Epochs to Train', type=int, default=1)
30 | parser.add_argument('--Model', help='Model to Train (UNET,SEGNET,DEEPLAB)', type=str, default="UNET")
31 |
32 | args = parser.parse_args()
33 |
34 | im_width = args.im_width
35 | im_height = args.im_height
36 | Epochs = args.Epochs
37 |
38 |
39 | TRAIN_PATH_IMAGES = args.TRAIN_PATH_IMAGES
40 | TRAIN_PATH_GT = args.TRAIN_PATH_GT
41 | TEST_PATH_IMAGES = args.TEST_PATH_IMAGES
42 | TEST_PATH_GT = args.TEST_PATH_GT
43 |
44 |
45 | ids_train_x = glob.glob(TRAIN_PATH_IMAGES)
46 | ids_train_y = glob.glob(TRAIN_PATH_GT)
47 | print("No. of training images = ", len(ids_train_x))
48 | ids_test_x = glob.glob(TEST_PATH_IMAGES)
49 | ids_test_y = glob.glob(TEST_PATH_GT)
50 | print("No. of testing images = ", len(ids_test_x))
51 |
52 | X_train = np.zeros((len(ids_train_x), im_height, im_width, 1), dtype=np.float32)
53 | y_train = np.zeros((len(ids_train_y), im_height, im_width, 1), dtype=np.float32)
54 |
55 | X_test = np.zeros((len(ids_test_x), im_height, im_width, 1), dtype=np.float32)
56 | y_test = np.zeros((len(ids_test_y), im_height, im_width, 1), dtype=np.float32)
57 |
58 |
59 | print("Loading Training Data")
60 | count =0
61 | for x in (ids_train_x):
62 | y = glob.glob(x[:-4]+'*')[0]
63 | x_img = img_to_array(load_img(x, color_mode='grayscale', target_size=[im_width,im_height]))
64 | # Load masks
65 | mask = img_to_array(load_img(y, color_mode='grayscale', target_size=[im_width,im_height]))
66 | X_train[count] = x_img/255.0
67 | y_train[count] = mask/255.0
68 | count = count+1
69 |
70 |
71 | print("Loading Testing Data")
72 | count =0
73 | for x in (ids_test_x):
74 | y = glob.glob(x[:-4]+'*')[0]
75 | x_img = img_to_array(load_img(x, color_mode='grayscale', target_size=[im_width,im_height]))
76 | # Load masks
77 | mask = img_to_array(load_img(y, color_mode='grayscale', target_size=[im_width,im_height]))
78 | X_test[count] = x_img/255.0
79 | y_test[count] = mask/255.0
80 | count = count+1
81 |
82 |
83 | input_img = Input((im_width, im_height, 1), name='img')
84 | #from tensorflow.keras.utils.vis_utils import plot_model
85 |
86 | if args.Model == "UNET":
87 | print("Loading UNET Model")
88 | model = get_unet(input_img, n_filters=32, dropout=0.05, batchnorm=True)
89 | model.compile(optimizer=Adam(1e-5), loss=jaccard_distance_loss, metrics=[iou,dice_coef])
90 | print("Printing Model Summary")
91 | print (model.summary())
92 | tf.keras.utils.plot_model(model, './Code/network/unet/unet_plot.png')
93 |
94 | if args.Model == "SEGNET":
95 | print("Loading SEGNET Model")
96 | model = get_segnet((im_width, im_height, 3))
97 | #n_labels=3,
98 | #kernel=3,
99 | #pool_size=(2, 2),
100 | #output_mode="softmax")
101 | model.compile(optimizer=SGD(lr=1e-5, momentum=0.95), loss=dice_coef_loss, metrics=[iou,dice_coef])
102 | print("Printing Model Summary")
103 | print (model.summary())
104 | tf.keras.utils.plot_model(model, './Code/network/segnet/segnet_plot.png')
105 |
106 | if args.Model == "DEEPLAB":
107 | print("Loading DEEPLAB Model")
108 | model = Deeplabv3(weights=None, input_tensor=None, input_shape=(im_width, im_height, 3), classes=1, backbone='mobilenetv2',
109 | OS=16, alpha=1., activation=None)
110 | model.compile(optimizer=tf.keras.optimizers.Adam(), loss="binary_crossentropy", metrics=[iou,dice_coef])
111 | #plot_model(model, to_file='./Code/network/deeplab/deeplab_plot.png', show_shapes=True, show_layer_names=True)
112 |
113 | print("Compiling Model")
114 | #model.compile(optimizer=sgd(), loss="binary_crossentropy"dice_coef_loss,jaccard_distance_loss metrics=["accuracy"]) # ,f1_m,iou_coef,dice_coef
115 | #
116 |
117 |
118 | #nadam(lr=1e-5)
119 | #Adam(1e-5, amsgrad=True, clipnorm=5.0)
120 | #Adam()
121 | #SGD(lr=1e-5, momentum=0.95)
122 | callbacks = [
123 | EarlyStopping(patience=10, verbose=1),
124 | ReduceLROnPlateau(factor=0.1, patience=5, min_lr=0.00001, verbose=1),
125 | ModelCheckpoint('./Results/weights/'+str(args.Model)+'/'+str(args.Model)+'-Best.h5', verbose=1, save_best_only=True, save_weights_only=False)
126 | ]
127 |
128 | print(X_train.shape, y_train.shape)
129 | print(X_test.shape, y_test.shape)
130 |
131 |
132 | results = model.fit(X_train, y_train, batch_size=1, verbose=1, epochs=Epochs, callbacks=callbacks,\
133 | validation_data=(X_test, y_test))
134 |
135 |
136 | plt.figure(figsize=(8, 8))
137 | plt.title("Learning curve")
138 | plt.plot(results.history["loss"], label="loss")
139 | plt.plot(results.history["val_loss"], label="val_loss")
140 | plt.plot( np.argmin(results.history["val_loss"]), np.min(results.history["val_loss"]), marker="x", color="r", label="best model")
141 | plt.xlabel("Epochs")
142 | plt.ylabel("log_loss")
143 | plt.legend();
144 | plt.savefig('./Results/plots/'+str(args.Model)+'/train_loss.png')
145 |
146 | plt.figure(figsize=(8, 8))
147 | plt.title("Learning curve")
148 | plt.plot(results.history["dice_coef"], label="dice_coef")
149 | plt.plot(results.history["val_dice_coef"], label="val_dice_coef")
150 | plt.plot( np.argmax(results.history["val_dice_coef"]), np.max(results.history["val_dice_coef"]), marker="x", color="r", label="best model")
151 | plt.xlabel("Epochs")
152 | plt.ylabel("Dice Coeff")
153 | plt.legend();
154 | plt.savefig('./Results/plots/'+str(args.Model)+'/train_dice.png')
155 |
156 | plt.figure(figsize=(8, 8))
157 | plt.title("Learning curve")
158 | plt.plot(results.history["iou"], label="iou")
159 | plt.plot(results.history["val_iou"], label="val_iou")
160 | plt.plot( np.argmax(results.history["val_iou"]), np.max(results.history["val_iou"]), marker="x", color="r", label="best model")
161 | plt.xlabel("Epochs")
162 | plt.ylabel("iou")
163 | plt.legend();
164 | plt.savefig('./Results/plots/'+str(args.Model)+'/train_iou.png')
165 |
166 | """
167 | plt.figure(figsize=(8, 8))
168 | plt.title("Learning curve")
169 | plt.plot(results.history["acc"], label="acc")
170 | plt.plot(results.history["val_acc"], label="val_acc")
171 | plt.plot( np.argmax(results.history["val_acc"]), np.max(results.history["val_acc"]), marker="x", color="r", label="best model")
172 | plt.xlabel("Epochs")
173 | plt.ylabel("Accuracy")
174 | plt.legend();
175 | plt.savefig('./train_acc.png')
176 |
177 | plt.figure(figsize=(8, 8))
178 | plt.title("Learning curve")
179 | plt.plot(results.history["f1_m"], label="f1_m")
180 | plt.plot(results.history["val_f1_m"], label="val_f1_m")
181 | plt.plot( np.argmax(results.history["val_f1_m"]), np.max(results.history["val_f1_m"]), marker="x", color="r", label="best model")
182 | plt.xlabel("Epochs")
183 | plt.ylabel("F1-Score")
184 | plt.legend();
185 | plt.savefig('./train_F1.png')
186 | """
187 |
--------------------------------------------------------------------------------
/config.json:
--------------------------------------------------------------------------------
1 | {
2 | "im_width" : 1000,
3 | "im_height" : 1000,
4 | "TRAIN_PATH_IMAGES" : "./Datasets/MonuSeg/Training/TissueImages/*",
5 | "TRAIN_PATH_GT" : "./Datasets/MonuSeg/Training/GroundTruth/",
6 | "TEST_PATH_IMAGES" : "./Datasets/MonuSeg/Test/TissueImages/*",
7 | "TEST_PATH_GT" : "./Datasets/MonuSeg/Test/GroundTruth/",
8 | "Epochs" : 30,
9 | "Batch" : 8,
10 | "Model" : "UNETMOD",
11 | "patch_width" : 256 ,
12 | "patch_height" : 256,
13 | "sample_test_image" : "./Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1.png",
14 | "sample_test_mask" : "./Datasets/Samples/Test/TCGA-HT-8564-01Z-00-DX1_bin_mask.png"
15 | }
16 |
--------------------------------------------------------------------------------
/requirements.txt:
--------------------------------------------------------------------------------
1 | absl-py==0.9.0
2 | asn1crypto==1.3.0
3 | astor==0.8.0
4 | blinker==1.4
5 | cachetools==3.1.1
6 | certifi==2019.11.28
7 | cffi==1.14.0
8 | chardet==3.0.4
9 | click==7.1.1
10 | cryptography==2.8
11 | cycler==0.10.0
12 | Cython==0.29.15
13 | decorator==4.4.2
14 | gast==0.2.2
15 | google-auth==1.11.2
16 | google-auth-oauthlib==0.4.1
17 | google-pasta==0.1.8
18 | grpcio==1.27.2
19 | h5py==2.10.0
20 | idna==2.9
21 | imageio==2.8.0
22 | joblib==0.14.1
23 | Keras==2.3.1
24 | Keras-Applications==1.0.8
25 | Keras-Preprocessing==1.1.0
26 | keras-resnet==0.1.0
27 | keras-retinanet==0.5.1
28 | kiwisolver==1.1.0
29 | Markdown==3.1.1
30 | matplotlib==3.2.1
31 | mkl-fft==1.0.15
32 | mkl-random==1.1.0
33 | mkl-service==2.3.0
34 | networkx==2.4
35 | numpy==1.18.1
36 | oauthlib==3.1.0
37 | opencv-python==4.2.0.32
38 | opt-einsum==3.1.0
39 | Pillow==7.0.0
40 | progressbar2==3.50.0
41 | protobuf==3.11.4
42 | pyasn1==0.4.8
43 | pyasn1-modules==0.2.7
44 | pycparser==2.20
45 | pydot==1.4.1
46 | PyJWT==1.7.1
47 | pyOpenSSL==19.1.0
48 | pyparsing==2.4.6
49 | PySocks==1.7.1
50 | python-dateutil==2.8.1
51 | python-utils==2.4.0
52 | PyWavelets==1.1.1
53 | PyYAML==5.3
54 | requests==2.23.0
55 | requests-oauthlib==1.3.0
56 | rsa==4.0
57 | scikit-image==0.16.2
58 | scikit-learn==0.22.2.post1
59 | scipy==1.4.1
60 | six==1.14.0
61 | tensorboard==2.1.0
62 | tensorflow==2.0.0
63 | tensorflow-estimator==2.0.0
64 | termcolor==1.1.0
65 | urllib3==1.25.8
66 | Werkzeug==0.16.1
67 | wrapt==1.12.1
68 |
--------------------------------------------------------------------------------
/temp.py:
--------------------------------------------------------------------------------
1 | from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
2 | import glob
3 | import cv2
4 | import sys
5 | import numpy
6 | import os
7 | numpy.set_printoptions(threshold=sys.maxsize)
8 | ids_train_x = glob.glob('./Datasets/MonuSeg/Training/TissueImages/*')
9 | ids_train_y = glob.glob('./Datasets/MonuSeg/Training/GroundTruth/*')
10 | #print(ids_train_x)
11 | for x in (ids_train_x):
12 | base=os.path.basename(x)
13 | fn = os.path.splitext(base)[0]
14 | y = glob.glob('./Datasets/MonuSeg/Training/GroundTruth/'+fn+'*')[0]
15 | print(y)
16 | #print(x[:-4])
17 |
18 | #x_img = img_to_array(load_img(x, color_mode='rgb', target_size=[1024,1024]))
19 | mask = img_to_array(load_img(y, color_mode='grayscale', target_size=[1024,1024]))
20 | #mask = img_to_array(load_img(y, color_mode='rgb'))
21 |
22 |
23 | #mask = cv2.imread(y)
24 | #print(mask)
25 | print(numpy.unique(mask, return_counts=True))
26 |
27 |
28 |
29 |
30 |
31 |
32 |
33 |
34 |
35 |
--------------------------------------------------------------------------------
 105 |
105 |  106 |
106 |  107 |
107 |  108 |
108 |  46 |
47 | #### Squeeze-and-Excitation Network
48 |
46 |
47 | #### Squeeze-and-Excitation Network
48 |  49 |
50 |
51 | ### SegNet
52 | 
53 |
54 |
55 | ### U-Net
56 | 
57 |
58 | ### Deep Lab v3+
59 | 
60 |
61 |
62 | ## Pre-Trained Models
63 | The Pre-Trained models can be downloaded from [google drive](https://drive.google.com/drive/folders/1g5SdbW8q1Z0e9dk6cW431JO01BDq4g0H).
64 |
65 | ## Installation
66 | To get this repo work please install all the dependencies using the command below:
67 | ```
68 | pip install -r requirments.txt
69 | ```
70 |
71 | ## Training
72 | To start training run the Train.py script from the command below. For training configurations refer to the [config.json](./config.json) file. You can update the file according to your training settings. Model avaible for training are U-NET,SegNet, DeepLabv3+.
73 | ```
74 | python Train.py
75 | ```
76 |
77 | ## Testing
78 | To test the trained models on Test Images you first have to download the weights and place them in the [results](./Results/). After downliading the weights you unzip them and then run the Inference by using the command below. For testing configurations please refer to the [config.json](./config.json) file.
79 | ```
80 | python Test.py
81 | ```
82 |
83 | ## Visualization of Results
84 | | Tissue | Mask | Predicted Mask |
85 | :-------------------------:|:-------------------------:|:-------------------------:
86 |  |  | 
87 |
88 | ## Quantitatvie Results
89 |
90 | | Model | Loss | Accuracy | F1 Score | Dice Score |
91 | | ----- | ---- | ---- | ---- | ---- |
92 | | Unet | 0.0835 | 0.9150 | 0.7910 | 0.7906
93 | | Segnet | 0.4820 | 0.8077 | 0.5798 | 0.3684
94 | | DeeplabV3+ | 0.0783 | 0.9120 | 0.7750 | 0.7743
95 | | Unet + Skip Connections + ASPP + SE Block | 0.0770 | 0.9210 | 0.801 | 0.8005
96 |
97 | ## Results
98 | Three Segmentation models have been trained and the model is evaluated on three metrics namely:
99 | * Accuracy
100 | * F1-Score
101 | * Dice Score
102 | ### U-Net + Skip Connections + ASPP
103 |
49 |
50 |
51 | ### SegNet
52 | 
53 |
54 |
55 | ### U-Net
56 | 
57 |
58 | ### Deep Lab v3+
59 | 
60 |
61 |
62 | ## Pre-Trained Models
63 | The Pre-Trained models can be downloaded from [google drive](https://drive.google.com/drive/folders/1g5SdbW8q1Z0e9dk6cW431JO01BDq4g0H).
64 |
65 | ## Installation
66 | To get this repo work please install all the dependencies using the command below:
67 | ```
68 | pip install -r requirments.txt
69 | ```
70 |
71 | ## Training
72 | To start training run the Train.py script from the command below. For training configurations refer to the [config.json](./config.json) file. You can update the file according to your training settings. Model avaible for training are U-NET,SegNet, DeepLabv3+.
73 | ```
74 | python Train.py
75 | ```
76 |
77 | ## Testing
78 | To test the trained models on Test Images you first have to download the weights and place them in the [results](./Results/). After downliading the weights you unzip them and then run the Inference by using the command below. For testing configurations please refer to the [config.json](./config.json) file.
79 | ```
80 | python Test.py
81 | ```
82 |
83 | ## Visualization of Results
84 | | Tissue | Mask | Predicted Mask |
85 | :-------------------------:|:-------------------------:|:-------------------------:
86 |  |  | 
87 |
88 | ## Quantitatvie Results
89 |
90 | | Model | Loss | Accuracy | F1 Score | Dice Score |
91 | | ----- | ---- | ---- | ---- | ---- |
92 | | Unet | 0.0835 | 0.9150 | 0.7910 | 0.7906
93 | | Segnet | 0.4820 | 0.8077 | 0.5798 | 0.3684
94 | | DeeplabV3+ | 0.0783 | 0.9120 | 0.7750 | 0.7743
95 | | Unet + Skip Connections + ASPP + SE Block | 0.0770 | 0.9210 | 0.801 | 0.8005
96 |
97 | ## Results
98 | Three Segmentation models have been trained and the model is evaluated on three metrics namely:
99 | * Accuracy
100 | * F1-Score
101 | * Dice Score
102 | ### U-Net + Skip Connections + ASPP
103 |  105 |
105 |  106 |
106 |  107 |
107 |  108 |
108 |  113 |
113 |  114 |
114 |  115 |
115 |  116 |
116 |  121 |
121 |  122 |
122 |  123 |
123 |  124 |
124 |  129 |
129 |  130 |
130 |  131 |
131 |  132 |
132 |