├── LICENSE
├── README.md
├── best-config
└── .readme.md
├── cmds
├── download-caltech-ucsd-birds-200-2011-dataset.sh
└── download-glas-dataset.sh
├── config_bash
└── .readme.md
├── config_yaml
└── .readme.md
├── constants.py
├── create_folds.py
├── data
└── .readme.md
├── deeplearning
├── __init__.py
├── aspp.py
├── backbone
│ ├── __init__.py
│ ├── resnet.py
│ ├── resnet_atrous.py
│ └── xception.py
├── criteria.py
├── decay.py
├── decision_pooling.py
├── deeplab.py
├── lr_scheduler.py
├── models_cl.py
├── models_seg.py
├── optimize_mask.py
├── pairwise_similarity.py
├── sampling.py
├── syncbn
│ ├── .gitignore
│ ├── Dockerfile
│ ├── LICENSE
│ ├── README.md
│ ├── __init__.py
│ ├── functional
│ │ ├── __init__.py
│ │ ├── _csrc.py
│ │ ├── csrc
│ │ │ ├── bn.h
│ │ │ ├── cuda
│ │ │ │ ├── bn_cuda.cu
│ │ │ │ ├── common.h
│ │ │ │ └── ext_lib.h
│ │ │ └── ext_lib.cpp
│ │ └── syncbn.py
│ ├── nn
│ │ ├── __init__.py
│ │ └── syncbn.py
│ ├── requirements.txt
│ └── test.py
├── train.py
├── utils.py
└── wildcat.py
├── dependencies
├── .readme.md
├── get_dependencies.sh
└── requirements.txt
├── doc
├── .readme.md
├── algo.png
├── arch.png
├── knn.png
├── perf.png
├── proposal.png
├── pseudo-labeling.png
├── results.png
└── wacv2021-active-learning-weak-annotator.pdf
├── exps
└── .readme.md
├── folds
├── .readme.md
├── Caltech-UCSD-Birds-200-2011
│ ├── encoding.yaml
│ ├── log-stats-ds-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.txt
│ ├── readme.md
│ ├── size-stats-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.png
│ └── split_0
│ │ ├── fold_0
│ │ ├── .~lock.train_s_0_f_0.csv#
│ │ ├── encoding.yaml
│ │ ├── readme.md
│ │ ├── seed.txt
│ │ ├── test_s_0_f_0.csv
│ │ ├── train_s_0_f_0.csv
│ │ └── valid_s_0_f_0.csv
│ │ ├── fold_1
│ │ ├── encoding.yaml
│ │ ├── readme.md
│ │ ├── seed.txt
│ │ ├── test_s_0_f_1.csv
│ │ ├── train_s_0_f_1.csv
│ │ └── valid_s_0_f_1.csv
│ │ ├── fold_2
│ │ ├── encoding.yaml
│ │ ├── readme.md
│ │ ├── seed.txt
│ │ ├── test_s_0_f_2.csv
│ │ ├── train_s_0_f_2.csv
│ │ └── valid_s_0_f_2.csv
│ │ ├── fold_3
│ │ ├── encoding.yaml
│ │ ├── readme.md
│ │ ├── seed.txt
│ │ ├── test_s_0_f_3.csv
│ │ ├── train_s_0_f_3.csv
│ │ └── valid_s_0_f_3.csv
│ │ └── fold_4
│ │ ├── encoding.yaml
│ │ ├── readme.md
│ │ ├── seed.txt
│ │ ├── test_s_0_f_4.csv
│ │ ├── train_s_0_f_4.csv
│ │ └── valid_s_0_f_4.csv
└── glas
│ ├── encoding.yaml
│ ├── log-stats-ds-ds-glas-s-0-f-0-subset-train.txt
│ ├── readme.md
│ ├── size-stats-ds-glas-s-0-f-0-subset-train.png
│ └── split_0
│ ├── fold_0
│ ├── .~lock.test_s_0_f_0.csv#
│ ├── encoding.yaml
│ ├── readme.md
│ ├── seed.txt
│ ├── test_s_0_f_0.csv
│ ├── train_s_0_f_0.csv
│ └── valid_s_0_f_0.csv
│ ├── fold_1
│ ├── encoding.yaml
│ ├── readme.md
│ ├── seed.txt
│ ├── test_s_0_f_1.csv
│ ├── train_s_0_f_1.csv
│ └── valid_s_0_f_1.csv
│ ├── fold_2
│ ├── encoding.yaml
│ ├── readme.md
│ ├── seed.txt
│ ├── test_s_0_f_2.csv
│ ├── train_s_0_f_2.csv
│ └── valid_s_0_f_2.csv
│ ├── fold_3
│ ├── encoding.yaml
│ ├── readme.md
│ ├── seed.txt
│ ├── test_s_0_f_3.csv
│ ├── train_s_0_f_3.csv
│ └── valid_s_0_f_3.csv
│ └── fold_4
│ ├── encoding.yaml
│ ├── readme.md
│ ├── seed.txt
│ ├── test_s_0_f_4.csv
│ ├── train_s_0_f_4.csv
│ └── valid_s_0_f_4.csv
├── fonts
├── .readme.md
├── Inconsolata.zip
└── Inconsolata
│ ├── Inconsolata-Bold.ttf
│ ├── Inconsolata-Regular.ttf
│ └── OFL.txt
├── gen_configs.py
├── instantiators.py
├── jobs
└── .readme.md
├── loader.py
├── main.py
├── outputjobs
└── .readme.md
├── pairwise_sims
└── .readme.md
├── paper
├── .readme.md
├── results-per-method-each-run-dataset-Caltech-UCSD-Birds-200-2011.pkl
└── results-per-method-each-run-dataset-glas.pkl
├── parseit.py
├── plot_active_learning_curves.py
├── pretrained-imgnet
└── .readme.md
├── pretrained
└── .readme.md
├── prologues.py
├── reproducibility.py
├── results
└── .readme.md
├── scheduler.sh
├── shared.py
├── tmp
└── .readme.md
├── tools.py
├── vision.py
└── yaml-gen.py
/best-config/.readme.md:
--------------------------------------------------------------------------------
1 | Contains the best config.
--------------------------------------------------------------------------------
/cmds/download-caltech-ucsd-birds-200-2011-dataset.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
2 | # Script to download and extract the dataset: Caltech-UCSD-Birds-200-2011
3 | # See: http://www.vision.caltech.edu/visipedia/CUB-200-2011.html
4 |
5 | # cd to your folder where you want to save the data.
6 | cd $1
7 | mkdir Caltech-UCSD-Birds-200-2011
8 | cd Caltech-UCSD-Birds-200-2011
9 |
10 | # Download the images.
11 | echo "Downloading images (1.1GB) ..."
12 | wget http://www.vision.caltech.edu/visipedia-data/CUB-200-2011/CUB_200_2011.tgz
13 |
14 | # Download masks (birds segmentations)
15 | echo "Downloading segmentation (37MB) ..."
16 | wget http://www.vision.caltech.edu/visipedia-data/CUB-200-2011/segmentations.tgz
17 |
18 | # Downlaod the readme
19 | echo "Downloading README.txt ..."
20 | wget http://www.vision.caltech.edu/visipedia-data/CUB-200-2011/README.txt
21 |
22 |
23 | echo "Finished downloading Caltech-UCSD-Birds-200-2011 dataset."
24 |
25 | echo "Extracting files ..."
26 |
27 | tar -zxvf CUB_200_2011.tgz
28 | tar -zxvf segmentations.tgz
29 |
30 |
31 | echo "Finished extracting Caltech-UCSD-Birds-200-2011 dataset."
--------------------------------------------------------------------------------
/cmds/download-glas-dataset.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
2 | # Script to download and extract the dataset: Glas. (GlaS-2015)
3 | # See: https://warwick.ac.uk/fac/sci/dcs/research/tia/glascontest/download/
4 |
5 | # cd to your folder where you want to save the data.
6 | cd $1
7 | mkdir GlaS-2015
8 | cd GlaS-2015
9 |
10 | # Download the images.
11 | echo "Downloading ..."
12 | wget https://warwick.ac.uk/fac/sci/dcs/research/tia/glascontest/download/warwick_qu_dataset_released_2016_07_08.zip
13 |
14 | echo "Finished downloading GlaS-2015."
15 |
16 | echo "Extracting files ..."
17 |
18 | unzip warwick_qu_dataset_released_2016_07_08.zip
19 |
20 | echo "========================================================================="
21 | echo " We are aware of the SPACES in the folder name. "
22 | echo " Not sure whose idea was to put space in the folder name. "
23 | echo " I let you to deal with it. "
24 | echo "========================================================================="
25 |
26 |
27 | echo "Finished extracting GlaS-2015."
--------------------------------------------------------------------------------
/config_bash/.readme.md:
--------------------------------------------------------------------------------
1 | Contains bash files.
--------------------------------------------------------------------------------
/config_yaml/.readme.md:
--------------------------------------------------------------------------------
1 | Contains yaml files.
--------------------------------------------------------------------------------
/constants.py:
--------------------------------------------------------------------------------
1 | # baseline
2 | AL_WSL = "WSL" # using only global annotation over the enter dataset.
3 | # similar to AL_FULL_SUP but no segmentation training is done. The predicted
4 | # mask is estimated from the CAMs.
5 |
6 | # type of active learning
7 | AL_RANDOM = 'Random' # random selection
8 | AL_LP = "Label_prop" # our method. (label-propagation)
9 | AL_FULL_SUP = "Full_sup" # no active learning. use entire dataset with full
10 | # supervision
11 | AL_ENTROPY = "Entropy" # uncertainty based on entropy of the classification
12 | # scores.
13 | AL_MCDROPOUT = "MC_Dropout" # uncertainty based on MC dropout.
14 |
15 | al_types = [AL_WSL, AL_RANDOM, AL_LP, AL_FULL_SUP, AL_ENTROPY, AL_MCDROPOUT]
16 |
17 | # loss
18 | CE = 'CE'
19 | KL = 'KL'
20 |
21 | # total segmentation loss: seg + cl.
22 | HYBRIDLOSS = "HybridLoss"
23 |
24 | losses = [CE, KL, HYBRIDLOSS]
25 |
26 | # how to cluster samples in our method for selection.
27 | CLUSTER_DENSITY_DIVERSITY = 'Density_and_diversity'
28 | CLUSTER_DENSITY_LABELLING = 'Density_and_labelling'
29 | CLUSTER_DENSITY_DIVERSITY_LABELLING = "Density,_diversity,_and_labelling"
30 | # clustering that is based on standard active learning selection criteria.
31 |
32 | CLUSTER_RANDOM = 'Random' # random sampling.
33 | CLUSTER_ENTROPY = 'Entropy' # entropy sampling.
34 | ours_clustering = [CLUSTER_DENSITY_DIVERSITY,
35 | CLUSTER_DENSITY_LABELLING,
36 | CLUSTER_DENSITY_DIVERSITY_LABELLING,
37 | CLUSTER_RANDOM,
38 | CLUSTER_ENTROPY]
39 |
40 | # models
41 | LENET5 = "lenet5" # lenet5
42 | SOTASSL = "sota_ssl" # sota_ssl
43 | HYBRIDMODEL = 'hybrid_model' # for SEG task.
44 |
45 | nets = [LENET5, SOTASSL, HYBRIDMODEL]
46 |
47 | # datasets
48 | # CL
49 | CIFAR_10 = "cifar-10"
50 | CIFAR_100 = "cifar-100"
51 | SVHN = "svhn"
52 | MNIST = "mnist"
53 |
54 | # SEG
55 | GLAS = "glas"
56 | CUB = "Caltech-UCSD-Birds-200-2011"
57 | OXF = "Oxford-flowers-102"
58 | CAM16 = "camelyon16"
59 |
60 | datasets = [CIFAR_10, CIFAR_100, SVHN, MNIST, GLAS]
61 | CL_DATASETS = [CIFAR_10, CIFAR_100, SVHN, MNIST]
62 | SEG_DATASETS = [GLAS, CUB, OXF]
63 |
64 |
65 | # task
66 | CL = 'CLASSIFICATION'
67 | SEG = 'SEGMENTATION'
68 | tasks = [CL, SEG]
69 |
70 | # subtasks
71 | SUBCL = CL
72 | SUBSEG = SEG
73 | SUBCLSEG = "Classification_Segmentation"
74 | subtasks = [SUBCL, SUBSEG, SUBCLSEG]
75 | # ==============================================================================
76 | # Types of attention
77 | NONE = 'NONE'
78 | LEARNABLE = 'LEARNABLE'
79 | STOCHASTICXXX = 'STOCHASTICXXX'
80 |
81 | attention_types = [NONE, LEARNABLE, STOCHASTICXXX]
82 | attentionz = [LEARNABLE, STOCHASTICXXX]
83 |
84 | # Types of similarity measure between scores
85 | JSD = "JSD" # "Jensen-Shannon divergence"
86 | MSE = "MSE" # "Mean-squarred error"
87 |
88 | sim_scores = [JSD, MSE]
89 |
90 |
91 | # Tags for samples
92 | L = 0 # Labeled samples
93 | U = 1 # Unlabeled sample
94 | PL = 2 # unlabeled sample that has been Pseudo-Labeled.
95 |
96 | samples_tags = [L, U, PL] # list of possible sample tags.
97 |
98 | # indicator on how to find the best when looking to labele unlabeled samples.
99 | LOW = "low"
100 | HIGH = "high"
101 |
102 | best = [LOW, HIGH] # list of possible choices for the best criterion.
103 |
104 | # Colours
105 | COLOR_WHITE = "white"
106 | COLOR_BLACK = "black"
107 |
108 | # backbones.
109 |
110 | RESNET18 = "resnet18"
111 | RESNET34 = "resnet34"
112 | RESNET50 = 'resnet50'
113 | RESNET101 = 'resnet101'
114 | RESNET152 = 'resnet152'
115 | RESNEXT50_32X4D = 'resnext50_32x4d'
116 | RESNEXT101_32X8D = 'resnext101_32x8d'
117 | WIDE_RESNET50_2 = 'wide_resnet50_2'
118 | WIDE_RESNET101_2 = 'wide_resnet101_2'
119 |
120 | backbones = [RESNET18,
121 | RESNET34,
122 | RESNET50,
123 | RESNET101,
124 | RESNET152,
125 | RESNEXT50_32X4D,
126 | RESNEXT101_32X8D,
127 | WIDE_RESNET50_2,
128 | WIDE_RESNET101_2
129 | ]
130 | resnet_backbones = [RESNET18,
131 | RESNET34,
132 | RESNET50,
133 | RESNET101,
134 | RESNET152,
135 | RESNEXT50_32X4D,
136 | RESNEXT101_32X8D,
137 | WIDE_RESNET50_2,
138 | WIDE_RESNET101_2
139 | ]
140 |
141 | # segmentation losses
142 | BinSoftInvDiceLoss = 'BinSoftInvDiceLoss'
143 | BinCrossEntropySegmLoss = 'BinCrossEntropySegmLoss'
144 | BCEAndSoftDiceLoss = 'BCEAndSoftDiceLoss'
145 | BinL1SegmLoss = 'BinL1SegmLoss'
146 | BinIOUSegmLoss = 'BinIOUSegmLoss'
147 | BinL1SegAndIOUSegmLoss = 'BinL1SegAndIOUSegmLoss'
148 |
149 | seglosses = [
150 | BinCrossEntropySegmLoss, BinSoftInvDiceLoss, BCEAndSoftDiceLoss,
151 | BinL1SegmLoss, BinIOUSegmLoss, BinL1SegAndIOUSegmLoss
152 | ]
153 |
154 | # scale decay
155 |
156 | ConstantWeight = 'ConstantWeight'
157 | LinearAnnealedWeight = 'LinearAnnealedWeight'
158 | ExponentialDecayWeight = 'ExponentialDecayWeight'
159 |
160 | scale_decay = [ConstantWeight, LinearAnnealedWeight,ExponentialDecayWeight]
161 |
162 |
--------------------------------------------------------------------------------
/data/.readme.md:
--------------------------------------------------------------------------------
1 | Contains some data.
--------------------------------------------------------------------------------
/deeplearning/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/deeplearning/__init__.py
--------------------------------------------------------------------------------
/deeplearning/aspp.py:
--------------------------------------------------------------------------------
1 | """
2 | Main refs:
3 | https://github.com/YudeWang/deeplabv3plus-pytorch
4 | https://github.com/VainF/DeepLabV3Plus-Pytorch
5 | """
6 | import sys
7 | import os
8 | import datetime as dt
9 |
10 | import torch
11 | from torch import nn
12 | from torch.nn import functional as F
13 |
14 |
15 | sys.path.append("..")
16 | sys.path.append("../..")
17 |
18 | from shared import check_if_allow_multgpu_mode, announce_msg
19 | from deeplearning.utils import initialize_weights
20 |
21 | ACTIVATE_SYNC_BN = False
22 | # Override ACTIVATE_SYNC_BN using variable environment in Bash:
23 | # $ export ACTIVATE_SYNC_BN="True" ----> Activate
24 | # $ export ACTIVATE_SYNC_BN="False" ----> Deactivate
25 |
26 | if "ACTIVATE_SYNC_BN" in os.environ.keys():
27 | ACTIVATE_SYNC_BN = (os.environ['ACTIVATE_SYNC_BN'] == "True")
28 |
29 | announce_msg("ACTIVATE_SYNC_BN was set to {}".format(ACTIVATE_SYNC_BN))
30 |
31 | if check_if_allow_multgpu_mode() and ACTIVATE_SYNC_BN: # Activate Synch-BN.
32 | from deeplearning.syncbn import nn as NN_Sync_BN
33 | BatchNorm2d = NN_Sync_BN.BatchNorm2d
34 | announce_msg("Synchronized BN has been activated. \n"
35 | "MultiGPU mode has been activated. "
36 | "{} GPUs".format(torch.cuda.device_count()))
37 | else:
38 | BatchNorm2d = nn.BatchNorm2d
39 | if check_if_allow_multgpu_mode():
40 | announce_msg("Synchronized BN has been deactivated.\n"
41 | "MultiGPU mode has been activated. "
42 | "{} GPUs".format(torch.cuda.device_count()))
43 | else:
44 | announce_msg("Synchronized BN has been deactivated.\n"
45 | "MultiGPU mode has been deactivated. "
46 | "{} GPUs".format(torch.cuda.device_count()))
47 |
48 | __all__ = ['aspp']
49 |
50 |
51 | # The Atrous Spatial Pyramid Pooling
52 |
53 |
54 | def assp_branch(in_channels, out_channles, kernel_size, dilation):
55 | padding = 0 if kernel_size == 1 else dilation
56 | return nn.Sequential(
57 | nn.Conv2d(in_channels, out_channles, kernel_size, padding=padding,
58 | dilation=dilation, bias=False),
59 | BatchNorm2d(out_channles),
60 | nn.ReLU(inplace=True))
61 |
62 |
63 | class ASPP(nn.Module):
64 | """
65 | Implements Atrous Spatial Pyramid Pooling (ASPP).
66 | """
67 | def __init__(self, in_channels, output_stride):
68 | super(ASPP, self).__init__()
69 |
70 | msg = 'Only output strides of 8 or 16 are supported.'
71 | assert output_stride in [8, 16], msg
72 | if output_stride == 16:
73 | dilations = [1, 6, 12, 18]
74 | elif output_stride == 8:
75 | dilations = [1, 12, 24, 36]
76 |
77 | self.aspp1 = assp_branch(in_channels, 256, 1, dilation=dilations[0])
78 | self.aspp2 = assp_branch(in_channels, 256, 3, dilation=dilations[1])
79 | self.aspp3 = assp_branch(in_channels, 256, 3, dilation=dilations[2])
80 | self.aspp4 = assp_branch(in_channels, 256, 3, dilation=dilations[3])
81 |
82 | self.avg_pool = nn.Sequential(
83 | nn.AdaptiveAvgPool2d((1, 1)),
84 | nn.Conv2d(in_channels, 256, 1, bias=False),
85 | BatchNorm2d(256),
86 | nn.ReLU(inplace=True))
87 |
88 | self.conv1 = nn.Conv2d(256 * 5, 256, 1, bias=False)
89 | self.bn1 = BatchNorm2d(256)
90 | self.relu = nn.ReLU(inplace=True)
91 | self.dropout = nn.Dropout(0.5)
92 |
93 | initialize_weights(self)
94 |
95 | def forward(self, x):
96 | x1 = self.aspp1(x)
97 | x2 = self.aspp2(x)
98 | x3 = self.aspp3(x)
99 | x4 = self.aspp4(x)
100 | x5 = F.interpolate(self.avg_pool(x), size=(x.size(2), x.size(3)),
101 | mode='bilinear', align_corners=True)
102 |
103 | x = self.conv1(torch.cat((x1, x2, x3, x4, x5), dim=1))
104 | x = self.bn1(x)
105 | x = self.dropout(self.relu(x))
106 |
107 | return x
108 |
109 | def get_nbr_params(self):
110 | """
111 | Compute the number of parameters of the model.
112 | :return:
113 | """
114 | return sum([p.numel() for p in self.parameters()])
115 |
116 |
117 | def aspp(in_channels, output_stride):
118 | return ASPP(in_channels=in_channels, output_stride=output_stride)
119 |
120 |
121 | if __name__ == "__main__":
122 | torch.manual_seed(0)
123 |
124 | cuda = "0"
125 | print("cuda:{}".format(cuda))
126 | print("DEVICE BEFORE: ", torch.cuda.current_device())
127 | DEVICE = torch.device(
128 | "cuda:{}".format(cuda) if torch.cuda.is_available() else "cpu")
129 | if torch.cuda.is_available():
130 | # torch.cuda.set_device(int(cuda))
131 | pass
132 |
133 | print("DEVICE AFTER: ", torch.cuda.current_device())
134 | batch, h, w = 4, 32, 32
135 | x = torch.randn(batch, 3, h, w)
136 | x = x.to(DEVICE)
137 |
138 | model = aspp(in_channels=3, output_stride=16)
139 | announce_msg('testing {}.'.format(ASPP))
140 | model.to(DEVICE)
141 | t0 = dt.datetime.now()
142 | out = model(x)
143 | print(
144 | "in-shape {} \t output-shape {} "
145 | ". Forward time {}.".format(x.shape, out.shape,
146 | dt.datetime.now() - t0))
147 | print("NBR-PARAMS: ", model.get_nbr_params())
148 |
--------------------------------------------------------------------------------
/deeplearning/backbone/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/deeplearning/backbone/__init__.py
--------------------------------------------------------------------------------
/deeplearning/backbone/xception.py:
--------------------------------------------------------------------------------
1 | """
2 | Main refs:
3 | https://github.com/YudeWang/deeplabv3plus-pytorch
4 | https://github.com/VainF/DeepLabV3Plus-Pytorch
5 | https://github.com/yassouali/pytorch_segmentation
6 | """
7 | import sys
8 | import os
9 | import datetime as dt
10 | import math
11 |
12 | import torch
13 | import torch.nn as nn
14 | from torchvision.models.utils import load_state_dict_from_url
15 | import torch.nn.functional as F
16 |
17 |
18 | sys.path.append("..")
19 | sys.path.append("../..")
20 |
21 | from shared import check_if_allow_multgpu_mode, announce_msg
22 | from deeplearning.utils import initialize_weights
23 |
24 | ACTIVATE_SYNC_BN = False
25 | # Override ACTIVATE_SYNC_BN using variable environment in Bash:
26 | # $ export ACTIVATE_SYNC_BN="True" ----> Activate
27 | # $ export ACTIVATE_SYNC_BN="False" ----> Deactivate
28 |
29 | if "ACTIVATE_SYNC_BN" in os.environ.keys():
30 | ACTIVATE_SYNC_BN = (os.environ['ACTIVATE_SYNC_BN'] == "True")
31 |
32 | announce_msg("ACTIVATE_SYNC_BN was set to {}".format(ACTIVATE_SYNC_BN))
33 |
34 | if check_if_allow_multgpu_mode() and ACTIVATE_SYNC_BN: # Activate Synch-BN.
35 | from deeplearning.syncbn import nn as NN_Sync_BN
36 | BatchNorm2d = NN_Sync_BN.BatchNorm2d
37 | announce_msg("Synchronized BN has been activated. \n"
38 | "MultiGPU mode has been activated. "
39 | "{} GPUs".format(torch.cuda.device_count()))
40 | else:
41 | BatchNorm2d = nn.BatchNorm2d
42 | if check_if_allow_multgpu_mode():
43 | announce_msg("Synchronized BN has been deactivated.\n"
44 | "MultiGPU mode has been activated. "
45 | "{} GPUs".format(torch.cuda.device_count()))
46 | else:
47 | announce_msg("Synchronized BN has been deactivated.\n"
48 | "MultiGPU mode has been deactivated. "
49 | "{} GPUs".format(torch.cuda.device_count()))
50 |
51 | bn_mom = 0.0003
52 |
53 | __all__ = ['xception']
54 |
55 | model_urls = {
56 | 'xception': 'http://data.lip6.fr/cadene/pretrainedmodels/xception-b5690688.pth'
57 | # 'https://www.dropbox.com/s/1hplpzet9d7dv29/xception-c0a72b38.pth.tar?dl=1'
58 | # 'http://data.lip6.fr/cadene/pretrainedmodels/xception-b5690688.pth'
59 | }
60 |
61 |
62 | class SeparableConv2d(nn.Module):
63 | def __init__(self, in_channels, out_channels, kernel_size=3, stride=1,
64 | dilation=1, bias=False):
65 | super(SeparableConv2d, self).__init__()
66 |
67 | if dilation > kernel_size // 2:
68 | padding = dilation
69 | else:
70 | padding = kernel_size // 2
71 |
72 | self.conv1 = nn.Conv2d(in_channels, in_channels, kernel_size, stride,
73 | padding=padding,
74 | dilation=dilation, groups=in_channels, bias=bias)
75 | self.bn = BatchNorm2d(in_channels)
76 | self.pointwise = nn.Conv2d(in_channels, out_channels, 1, 1, bias=bias)
77 |
78 | def forward(self, x):
79 | x = self.conv1(x)
80 | x = self.bn(x)
81 | x = self.pointwise(x)
82 | return x
83 |
84 |
85 | class Block(nn.Module):
86 | def __init__(self, in_channels, out_channels, stride=1, dilation=1,
87 | exit_flow=False, use_1st_relu=True):
88 | super(Block, self).__init__()
89 |
90 | if in_channels != out_channels or stride != 1:
91 | self.skip = nn.Conv2d(in_channels, out_channels, 1, stride=stride,
92 | bias=False)
93 | self.skipbn = BatchNorm2d(out_channels)
94 | else:
95 | self.skip = None
96 |
97 | rep = []
98 | self.relu = nn.ReLU(inplace=True)
99 |
100 | rep.append(self.relu)
101 | rep.append(SeparableConv2d(in_channels, out_channels, 3, stride=1,
102 | dilation=dilation))
103 | rep.append(BatchNorm2d(out_channels))
104 |
105 | rep.append(self.relu)
106 | rep.append(SeparableConv2d(out_channels, out_channels, 3, stride=1,
107 | dilation=dilation))
108 | rep.append(BatchNorm2d(out_channels))
109 |
110 | rep.append(self.relu)
111 | rep.append(SeparableConv2d(out_channels, out_channels, 3, stride=stride,
112 | dilation=dilation))
113 | rep.append(BatchNorm2d(out_channels))
114 |
115 | if exit_flow:
116 | rep[3:6] = rep[:3]
117 | rep[:3] = [
118 | self.relu,
119 | SeparableConv2d(in_channels, in_channels, 3, 1, dilation),
120 | BatchNorm2d(in_channels)]
121 |
122 | if not use_1st_relu: rep = rep[1:]
123 | self.rep = nn.Sequential(*rep)
124 |
125 | def forward(self, x):
126 | output = self.rep(x)
127 | if self.skip is not None:
128 | skip = self.skip(x)
129 | skip = self.skipbn(skip)
130 | else:
131 | skip = x
132 |
133 | x = output + skip
134 | return x
135 |
136 |
137 | class Xception(nn.Module):
138 | def __init__(self, output_stride=16, in_channels=3, pretrained=True):
139 | super(Xception, self).__init__()
140 |
141 | self.name = "xception"
142 |
143 | # Stride for block 3 (entry flow), and the dilation rates for middle
144 | # flow and exit flow
145 | if output_stride == 16:
146 | b3_s, mf_d, ef_d = 2, 1, (1, 2)
147 | if output_stride == 8:
148 | b3_s, mf_d, ef_d = 1, 2, (2, 4)
149 |
150 | # Entry Flow
151 | self.conv1 = nn.Conv2d(in_channels, 32, 3, 2, padding=1, bias=False)
152 | self.bn1 = BatchNorm2d(32)
153 | self.relu = nn.ReLU(inplace=True)
154 | self.conv2 = nn.Conv2d(32, 64, 3, 1, padding=1, bias=False)
155 | self.bn2 = BatchNorm2d(64)
156 |
157 | self.block1 = Block(64, 128, stride=2, dilation=1, use_1st_relu=False)
158 | self.block2 = Block(128, 256, stride=2, dilation=1)
159 | self.block3 = Block(256, 728, stride=b3_s, dilation=1)
160 |
161 | # Middle Flow
162 | for i in range(16):
163 | exec(
164 | f'self.block{i + 4} = Block(728, 728, stride=1, dilation=mf_d)')
165 |
166 | # Exit flow
167 | self.block20 = Block(728, 1024, stride=1, dilation=ef_d[0],
168 | exit_flow=True)
169 |
170 | self.conv3 = SeparableConv2d(1024, 1536, 3, stride=1, dilation=ef_d[1])
171 | self.bn3 = BatchNorm2d(1536)
172 | self.conv4 = SeparableConv2d(1536, 1536, 3, stride=1, dilation=ef_d[1])
173 | self.bn4 = BatchNorm2d(1536)
174 | self.conv5 = SeparableConv2d(1536, 2048, 3, stride=1, dilation=ef_d[1])
175 | self.bn5 = BatchNorm2d(2048)
176 |
177 | initialize_weights(self)
178 | if pretrained:
179 | self._load_pretrained_model()
180 |
181 | def _load_pretrained_model(self):
182 | """
183 | Load imagenet pretrained xception.
184 | :return:
185 | """
186 | pretrained_weights = load_state_dict_from_url(
187 | url=model_urls['xception'], model_dir='./pretrained-imgnet',
188 | map_location=torch.device('cpu'), progress=True)
189 | state_dict = self.state_dict()
190 | model_dict = {}
191 |
192 | for k, v in pretrained_weights.items():
193 | if k in state_dict:
194 | if 'pointwise' in k:
195 | v = v.unsqueeze(-1).unsqueeze(-1) # [C, C] -> [C, C, 1, 1]
196 | if k.startswith('block11'):
197 | # In Xception there is only 8 blocks in Middle flow
198 | model_dict[k] = v
199 | for i in range(8):
200 | model_dict[k.replace('block11', f'block{i + 12}')] = v
201 | elif k.startswith('block12'):
202 | model_dict[k.replace('block12', 'block20')] = v
203 | elif k.startswith('bn3'):
204 | model_dict[k] = v
205 | model_dict[k.replace('bn3', 'bn4')] = v
206 | elif k.startswith('conv4'):
207 | model_dict[k.replace('conv4', 'conv5')] = v
208 | elif k.startswith('bn4'):
209 | model_dict[k.replace('bn4', 'bn5')] = v
210 | else:
211 | model_dict[k] = v
212 |
213 | state_dict.update(model_dict)
214 | self.load_state_dict(state_dict)
215 |
216 | def forward(self, x):
217 | # Entry flow
218 | x = self.conv1(x)
219 | x = self.bn1(x)
220 | x = self.relu(x)
221 | x = self.conv2(x)
222 | x = self.bn2(x)
223 | x = self.block1(x)
224 | low_level_features = x
225 | x = F.relu(x)
226 | x = self.block2(x)
227 | x = self.block3(x)
228 |
229 | # Middle flow
230 | x = self.block4(x)
231 | x = self.block5(x)
232 | x = self.block6(x)
233 | x = self.block7(x)
234 | x = self.block8(x)
235 | x = self.block9(x)
236 | x = self.block10(x)
237 | x = self.block11(x)
238 | x = self.block12(x)
239 | x = self.block13(x)
240 | x = self.block14(x)
241 | x = self.block15(x)
242 | x = self.block16(x)
243 | x = self.block17(x)
244 | x = self.block18(x)
245 | x = self.block19(x)
246 |
247 | # Exit flow
248 | x = self.block20(x)
249 | x = self.relu(x)
250 |
251 | x = self.conv3(x)
252 | x = self.bn3(x)
253 | x = self.relu(x)
254 |
255 | x = self.conv4(x)
256 | x = self.bn4(x)
257 | x = self.relu(x)
258 |
259 | x = self.conv5(x)
260 | x = self.bn5(x)
261 | x = self.relu(x)
262 |
263 | return low_level_features, x
264 |
265 | def get_nbr_params(self):
266 | """
267 | Compute the number of parameters of the model.
268 | :return:
269 | """
270 | return sum([p.numel() for p in self.parameters()])
271 |
272 | def __str__(self):
273 | return "{}: MODIFIED XCEPTION .".format(self.name)
274 |
275 |
276 | def xception(pretrained=True, output_stride=16):
277 | return Xception(
278 | output_stride=output_stride, in_channels=3, pretrained=pretrained)
279 |
280 |
281 | if __name__ == "__main__":
282 | torch.manual_seed(0)
283 |
284 | cuda = "0"
285 | print("cuda:{}".format(cuda))
286 | print("DEVICE BEFORE: ", torch.cuda.current_device())
287 | DEVICE = torch.device(
288 | "cuda:{}".format(cuda) if torch.cuda.is_available() else "cpu")
289 | if torch.cuda.is_available():
290 | # torch.cuda.set_device(int(cuda))
291 | pass
292 |
293 | print("DEVICE AFTER: ", torch.cuda.current_device())
294 | batch, h, w = 4, 256, 256
295 | x = torch.randn(batch, 3, h, w)
296 | x = x.to(DEVICE)
297 | for pretrained in [False, True]:
298 | announce_msg('testing {}. pretrained: {}'.format('xception',
299 | pretrained))
300 | model = sys.modules["__main__"].__dict__['xception'](
301 | pretrained=pretrained)
302 | model.to(DEVICE)
303 | t0 = dt.datetime.now()
304 | out = model(x)
305 | print(
306 | "in-shape {} \t output-low-shape {} \t output-high-shape {}"
307 | ". Forward time {}.".format(x.shape, out[0].shape,
308 | out[1].shape,
309 | dt.datetime.now() - t0))
310 | print("NBR-PARAMS: ", model.get_nbr_params())
311 | print("Model: {}".format(model))
312 |
--------------------------------------------------------------------------------
/deeplearning/decay.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import matplotlib.pyplot as plt
3 |
4 |
5 | __all__ = ['ConstantWeight', 'LinearAnnealedWeight', 'ExponentialDecayWeight']
6 |
7 |
8 | class Decay(object):
9 | """
10 | Parent class for decay.
11 | """
12 | def __init__(self, init_val, end_val, max_epochs, sigma):
13 | """
14 | Init function.
15 | :param init_val: float. initial value of the weight.
16 | :param end_val: float. final value (or minimum value allowed).
17 | :param max_epochs: int. maximum number of epochs.
18 | :param sigma: float. scaling factor to change the rate of the curve.
19 | the higher the value, the slower the slope.
20 | """
21 | pass
22 |
23 | def __call__(self):
24 | """
25 | Update the weight according to the predefine schedule.
26 | """
27 | raise NotImplementedError
28 |
29 | def get_current_weight(self):
30 | """
31 | Return the current value of the weight.
32 | """
33 | raise NotImplementedError
34 | def __repr__(self):
35 | """
36 | rep of the class.
37 | """
38 | raise NotImplementedError
39 |
40 |
41 | class ConstantWeight(Decay):
42 | """
43 | A callback to adjust the weight.
44 | Schedule: keep the weight fixed to some specific value.
45 | """
46 | def __init__(self, init_val, end_val, max_epochs, sigma):
47 | """
48 | Init. function.
49 | :param init_val: float. initial value of the weight.
50 | :param end_val: float. final value (or minimum value allowed).
51 | :param max_epochs: int. maximum number of epochs.
52 | :param sigma: float. scaling factor to change the rate of the curve.
53 | the higher the value, the slower the slope.
54 | """
55 | super(ConstantWeight, self).__init__(
56 | init_val, end_val, max_epochs, sigma)
57 | self._init_val = init_val
58 |
59 | def __call__(self):
60 | """
61 | Update the weight according the annealing schedule.
62 | """
63 | return self.get_current_weight()
64 |
65 | def get_current_weight(self):
66 | """
67 | Calculate the current weight according to the annealing
68 | schedule.
69 | """
70 | return self._init_val
71 |
72 | def __repr__(self):
73 | return "{}(init_val={})".format(
74 | self.__class__.__name__, self._init_val
75 | )
76 |
77 |
78 |
79 | class LinearAnnealedWeight(Decay):
80 | """
81 | A callback to adjust a weight linearly.
82 |
83 | Linearly anneal a weight from init_value to end_value.
84 | w(t) := w(t-1) - rate.
85 | where:
86 | rate := (init_value - end_value) / max_epochs.
87 |
88 | the scale is computed based on the maximum epochs.
89 |
90 | ref:
91 | S. Belharbi, R. Hérault, C. Chatelain and S. Adam,
92 | “Deep multi-task learning with evolving weights”, in European Symposium
93 | on Artificial Neural Networks (ESANN), 2016.
94 | """
95 | def __init__(self, init_val, end_val, max_epochs, sigma):
96 | """
97 | Init. function.
98 | :param init_val: float. initial value of the weight.
99 | :param end_val: float. final value (or minimum value allowed).
100 | :param max_epochs: int. maximum number of epochs.
101 | :param sigma: float. scaling factor to change the rate of the curve.
102 | the higher the value, the slower the slope.
103 | """
104 | super(LinearAnnealedWeight, self).__init__(
105 | init_val, end_val, max_epochs, sigma)
106 |
107 | self._count = 0.
108 | self._anneal_start = init_val
109 | self._anneal_end = end_val
110 | msg = "'init_val' must be >= 'end_val'"
111 | assert init_val >= end_val, msg
112 | self._max_epochs = max_epochs
113 | self.anneal_rate = (init_val - end_val) / float(max_epochs)
114 |
115 | self.weight = init_val # holds the current value.
116 | self._count = 0
117 |
118 | def __call__(self):
119 | """
120 | Updates the weight according to the annealing schedule.
121 |
122 | return: float. the new updated value.
123 | """
124 | if self._count == 0:
125 | self._count += 1
126 | return self.weight # return the initial value.
127 | else:
128 | return self.get_current_weight()
129 |
130 | def get_current_weight(self):
131 | """
132 | Calculate the current weight according to the annealing
133 | schedule.
134 | """
135 | self.weight = self.weight - self.anneal_rate
136 | return max(self._anneal_end, self.weight)
137 |
138 | def __repr__(self):
139 | return "{}(init_val={}, end_val={}, max_epochs={})".format(
140 | self.__class__.__name__, self._anneal_start, self._anneal_end,
141 | self._max_epochs
142 | )
143 |
144 |
145 | class ExponentialDecayWeight(Decay):
146 | """
147 | This anneals the weight exponentially with respect to the current epoch.
148 |
149 | w(t) = exp(-t/sigma).
150 |
151 | where `t` is the current epoch number, and `sigma`, is a constant that
152 | affects the slope of the function.
153 |
154 | ref:
155 | S. Belharbi, R. Hérault, C. Chatelain and S. Adam,
156 | “Deep multi-task learning with evolving weights”, in European Symposium
157 | on Artificial Neural Networks (ESANN), 2016.
158 | """
159 | def __init__(self, init_val, end_val, max_epochs, sigma):
160 | """
161 | Init. function.
162 | :param end_val: float. minimal value allowed.
163 | :param sigma: float. scaling factor to change the rate of the curve.
164 | the higher the value, the slower the slope.
165 | """
166 | super(ExponentialDecayWeight, self).__init__(
167 | init_val, end_val, max_epochs, sigma)
168 |
169 | self._count = 0
170 | assert sigma != 0, "'sigma'=0. must be different than zero."
171 |
172 | self._sigma = float(sigma)
173 | self._end_val = end_val
174 |
175 | self.weight = self.get_current_weight()
176 |
177 | def __call__(self):
178 | """Update the learning rate according to the exponential decay
179 | schedule.
180 |
181 | """
182 | if self._count == 0:
183 | self._count += 1
184 | return self.weight # return the initial value.
185 | else:
186 | return self.get_current_weight()
187 |
188 | def get_current_weight(self):
189 | """
190 | Calculate the current weight according to the annealing
191 | schedule.
192 | """
193 | self.weight = np.exp(- self._count / float(self._sigma))
194 | self._count += 1
195 |
196 | return max(self.weight, self._end_val)
197 |
198 | def __repr__(self):
199 | return "{}(end_val={}, sigma={})".format(
200 | self.__class__.__name__, self._end_val, self._sigma
201 | )
202 |
203 | if __name__ == "__main__":
204 | init_val, end_val, max_epochs, sigma = 1., 0.0001, 1000, 150.
205 |
206 | instances = [ConstantWeight(init_val, end_val, max_epochs, sigma),
207 | LinearAnnealedWeight(init_val, end_val, max_epochs, sigma),
208 | ExponentialDecayWeight(init_val, end_val, max_epochs, sigma)
209 | ]
210 | fig = plt.figure()
211 | for inst in instances:
212 | plt.plot(
213 | [inst() for _ in range(max_epochs)],
214 | label=str(inst)
215 | )
216 | plt.legend(loc="lower right")
217 | fig.savefig('test-decay.png')
218 | plt.close('all')
219 |
220 |
--------------------------------------------------------------------------------
/deeplearning/decision_pooling.py:
--------------------------------------------------------------------------------
1 | import threading
2 | import sys
3 |
4 | import numpy as np
5 |
6 |
7 | import torch
8 | import torch.nn as nn
9 |
10 | # lock for threads to protect the instruction that cause randomness and make
11 | # them thread-safe.
12 | thread_lock = threading.Lock()
13 |
14 | import reproducibility
15 | from shared import check_tensor_inf_nan
16 |
17 |
18 | __all__ = ["WildCatPoolDecision", "ClassWisePooling"]
19 |
20 |
21 | class WildCatPoolDecision(nn.Module):
22 | """Compute the score of each class using wildcat pooling strategy.
23 | Reference to wildcat pooling:
24 | http://webia.lip6.fr/~cord/pdfs/publis/Durand_WILDCAT_CVPR_2017.pdf
25 | """
26 | def __init__(self, kmax=0.5, kmin=None, alpha=1, dropout=0.0):
27 | """
28 | Input:
29 | kmax: int or float scalar. The number of maximum features to
30 | consider.
31 | kmin: int or float scalar. If None, it takes the same value as
32 | kmax. The number of minimal features to consider.
33 | alpha: float scalar. A weight , used to compute the final score.
34 | dropout: float scalar. If not zero, a dropout is performed over the
35 | min and max selected features.
36 | """
37 | super(WildCatPoolDecision, self).__init__()
38 |
39 | msg = "kmax must be an integer or a float in ]0, 1]"
40 | assert isinstance(kmax, (int, float)) and kmax > 0, msg
41 | msg = "kmin must be None or the same type as kmax, and it must be >= " \
42 | "0 or None"
43 | assert kmin is None or (
44 | isinstance(kmin, (int, float)) and kmin >= 0), msg
45 | self.kmax = kmax
46 | self.kmin = kmax if kmin is None else kmin
47 | self.alpha = alpha
48 | self.dropout = dropout
49 |
50 | self.dropout_md = nn.Dropout(p=dropout, inplace=False)
51 |
52 | def get_k(self, k, n):
53 | if k <= 0:
54 | return 0
55 | elif k < 1:
56 | return round(k * n)
57 | elif k == 1 and isinstance(k, float):
58 | return int(n)
59 | elif k == 1 and isinstance(k, int):
60 | return 1
61 | elif k > n:
62 | return int(n)
63 | else:
64 | return int(k)
65 |
66 | def forward(self, x, seed=None, prngs_cuda=None):
67 | """
68 | Input:
69 | In the case of K classes:
70 | x: torch tensor of size (n, c, h, w), where n is the batch
71 | size, c is the number of classes,
72 | h is the height of the feature map, w is its width.
73 | seed: int, seed for the thread to guarantee reproducibility over a
74 | fixed number of gpus.
75 | Output:
76 | scores: torch vector of size (k). Contains the wildcat score of
77 | each class. A score is a linear combination
78 | of different features. The class with the highest features is the
79 | winner.
80 | """
81 | b, c, h, w = x.shape
82 | activations = x.view(b, c, h * w)
83 |
84 | n = h * w
85 |
86 | sorted_features = torch.sort(activations, dim=-1, descending=True)[0]
87 | kmax = self.get_k(self.kmax, n)
88 | kmin = self.get_k(self.kmin, n)
89 |
90 | # assert kmin != 0, "kmin=0"
91 | assert kmax != 0, "kmax=0"
92 |
93 | # dropout
94 | if self.dropout != 0.:
95 | if seed is not None:
96 | thread_lock.acquire()
97 | msg = "`prngs_cuda` is expected to not be None. Exiting " \
98 | ".... [NOT OK]"
99 | assert prngs_cuda is not None, msg
100 | prng_state = (torch.cuda.get_rng_state().cpu())
101 | reproducibility.reset_seed(seed)
102 | torch.cuda.set_rng_state(prngs_cuda.cpu())
103 |
104 | # instruction that causes randomness.
105 | sorted_features = self.dropout_md(sorted_features)
106 |
107 | reproducibility.reset_seed(seed)
108 | torch.cuda.set_rng_state(prng_state)
109 | thread_lock.release()
110 | else:
111 | sorted_features = self.dropout_md(sorted_features)
112 |
113 | scores = sorted_features.narrow(-1, 0, kmax).sum(-1).div_(kmax)
114 |
115 | if kmin > 0 and self.alpha != 0.:
116 | scores.add(
117 | sorted_features.narrow(
118 | -1, n - kmin, kmin).sum(-1).mul_(
119 | self.alpha / kmin)).div_(2.)
120 |
121 | return scores
122 |
123 | def __str__(self):
124 | return self.__class__.__name__ + "(kmax={}, kmin={}, alpha={}, " \
125 | "dropout={}".format(
126 | self.kmax, self.kmin, self.alpha,
127 | self.dropout)
128 |
129 | def __repr__(self):
130 | return super(WildCatPoolDecision, self).__repr__()
131 |
132 |
133 | class ClassWisePooling(nn.Module):
134 | """
135 | Pull a feature map per class.
136 | Reference to wildcat:
137 | http://webia.lip6.fr/~cord/pdfs/publis/Durand_WILDCAT_CVPR_2017.pdf
138 | """
139 | def __init__(self, classes, modalities):
140 | """
141 | Init. function.
142 | :param classes: int, number of classes.
143 | :param modalities: int, number of modalities per class.
144 | """
145 | super(ClassWisePooling, self).__init__()
146 |
147 | self.C = classes
148 | self.M = modalities
149 |
150 | def forward(self, inputs):
151 | N, C, H, W = inputs.size()
152 | msg = 'Wrong number of channels, expected {} ' \
153 | 'channels but got {}'.format(self.C * self.M, C)
154 | assert C == self.C * self.M, msg
155 | return torch.mean(
156 | inputs.view(N, self.C, self.M, -1), dim=2).view(N, self.C, H, W)
157 |
158 | def __str__(self):
159 | return self.__class__.__name__ +\
160 | '(classes={}, modalities={})'.format(self.C, self.M)
161 |
162 | def __repr__(self):
163 | return super(ClassWisePooling, self).__repr__()
164 |
165 |
166 | if __name__ == "__main__":
167 | b, c = 10, 2
168 | reproducibility.reset_seed(0)
169 | funcs = [WildCatPoolDecision(dropout=0.5)]
170 | x = torch.randn(b, c, 12, 12)
171 | for func in funcs:
172 | out = func(x)
173 | print(func.__class__.__name__, '->', out.size(), out)
174 |
175 |
--------------------------------------------------------------------------------
/deeplearning/deeplab.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import os
3 | import datetime as dt
4 |
5 | import torch
6 | from torch import nn
7 | from torch.nn import functional as F
8 |
9 | sys.path.append("..")
10 |
11 | from shared import check_if_allow_multgpu_mode, announce_msg
12 | from deeplearning.utils import initialize_weights
13 | from deeplearning.aspp import aspp
14 |
15 | import reproducibility
16 | import constants
17 |
18 | ACTIVATE_SYNC_BN = False
19 | # Override ACTIVATE_SYNC_BN using variable environment in Bash:
20 | # $ export ACTIVATE_SYNC_BN="True" ----> Activate
21 | # $ export ACTIVATE_SYNC_BN="False" ----> Deactivate
22 |
23 | if "ACTIVATE_SYNC_BN" in os.environ.keys():
24 | ACTIVATE_SYNC_BN = (os.environ['ACTIVATE_SYNC_BN'] == "True")
25 |
26 | announce_msg("ACTIVATE_SYNC_BN was set to {}".format(ACTIVATE_SYNC_BN))
27 |
28 | if check_if_allow_multgpu_mode() and ACTIVATE_SYNC_BN: # Activate Synch-BN.
29 | from deeplearning.syncbn import nn as NN_Sync_BN
30 | BatchNorm2d = NN_Sync_BN.BatchNorm2d
31 | announce_msg("Synchronized BN has been activated. \n"
32 | "MultiGPU mode has been activated. "
33 | "{} GPUs".format(torch.cuda.device_count()))
34 | else:
35 | BatchNorm2d = nn.BatchNorm2d
36 | if check_if_allow_multgpu_mode():
37 | announce_msg("Synchronized BN has been deactivated.\n"
38 | "MultiGPU mode has been activated. "
39 | "{} GPUs".format(torch.cuda.device_count()))
40 | else:
41 | announce_msg("Synchronized BN has been deactivated.\n"
42 | "MultiGPU mode has been deactivated. "
43 | "{} GPUs".format(torch.cuda.device_count()))
44 |
45 |
46 | __all__ = ["deeplab_v3_plus_head"]
47 |
48 |
49 | class Decoder(nn.Module):
50 | """
51 | Implement a decoder for deeplabv3+.
52 | """
53 | def __init__(self, low_level_channels, num_classes):
54 | super(Decoder, self).__init__()
55 | self.conv1 = nn.Conv2d(low_level_channels, 48, 1, bias=False)
56 | self.bn1 = nn.BatchNorm2d(48)
57 | self.relu = nn.ReLU(inplace=True)
58 |
59 | # Table 2, best performance with two 3x3 convs
60 | self.output = nn.Sequential(
61 | nn.Conv2d(48+256, 256, 3, stride=1, padding=1, bias=False),
62 | nn.BatchNorm2d(256),
63 | nn.ReLU(inplace=True),
64 | nn.Conv2d(256, 256, 3, stride=1, padding=1, bias=False),
65 | nn.BatchNorm2d(256),
66 | nn.ReLU(inplace=True),
67 | nn.Dropout(0.1),
68 | nn.Conv2d(256, num_classes, 1, stride=1),
69 | )
70 | initialize_weights(self)
71 |
72 | def forward(self, x, low_level_features):
73 | low_level_features = self.conv1(low_level_features)
74 | low_level_features = self.relu(self.bn1(low_level_features))
75 | h, w = low_level_features.size(2), low_level_features.size(3)
76 |
77 | x = F.interpolate(x, size=(h, w), mode='bilinear', align_corners=True)
78 | x = self.output(torch.cat((low_level_features, x), dim=1))
79 | return x
80 |
81 |
82 | class DeepLabV3PlusHead(nn.Module):
83 | """
84 | DeepLabV3+ head.
85 | """
86 | def __init__(self, num_classes=1, backbone='xception',
87 | output_stride=16, freeze_bn=False):
88 | """
89 | Init function.
90 |
91 | :param num_classes: int. number of segmentation masks to output.
92 | :param backbone: str. name of the backbone.
93 | :param output_stride: output stride. supported: 8, 16.
94 | :param freeze_bn: bool. if true, the batchnorm parameters are frozen.
95 | """
96 | super(DeepLabV3PlusHead, self).__init__()
97 |
98 | self.name = "deeblabv3plus"
99 | assert ('xception' or 'resnet' in backbone)
100 | if 'resnet' in backbone:
101 | low_level_channels = 256
102 | elif 'xception' in backbone:
103 | low_level_channels = 128
104 | else:
105 | raise ValueError('How did you get here?')
106 |
107 | self.ASSP = aspp(in_channels=2048, output_stride=output_stride)
108 | self.decoder = Decoder(low_level_channels, num_classes)
109 |
110 | if freeze_bn:
111 | self.freeze_bn()
112 |
113 | def forward(self, low_level_features, x, h, w):
114 | """
115 | The forward function.
116 | :param low_level_features: feature at low level.
117 | :param x: features at high level.
118 | :param h: int. original height of the image.
119 | :param w: int. original width of the image.
120 | :return: x: row output scores (unnormalized).
121 | """
122 | x = self.ASSP(x)
123 | x = self.decoder(x, low_level_features)
124 | x = F.interpolate(x, size=(h, w), mode='bilinear', align_corners=True)
125 | return x
126 |
127 | def freeze_bn(self):
128 | for module in self.modules():
129 | if isinstance(module, BatchNorm2d):
130 | module.eval()
131 |
132 | def get_nbr_params(self):
133 | """
134 | Compute the number of parameters of the model.
135 | :return:
136 | """
137 | return sum([p.numel() for p in self.parameters()])
138 |
139 | def __str__(self):
140 | return "{}: DEEPLABV3+.".format(self.name)
141 |
142 |
143 | def deeplab_v3_plus_head(
144 | num_classes=1, backbone='xception', output_stride=16, freeze_bn=False):
145 | return DeepLabV3PlusHead(
146 | num_classes=num_classes, backbone=backbone, output_stride=output_stride,
147 | freeze_bn=freeze_bn)
148 |
149 |
150 | if __name__ == "__main__":
151 | torch.manual_seed(0)
152 |
153 | cuda = "0"
154 | print("cuda:{}".format(cuda))
155 | print("DEVICE BEFORE: ", torch.cuda.current_device())
156 | DEVICE = torch.device(
157 | "cuda:{}".format(cuda) if torch.cuda.is_available() else "cpu")
158 | if torch.cuda.is_available():
159 | # torch.cuda.set_device(int(cuda))
160 | pass
161 |
162 | print("DEVICE AFTER: ", torch.cuda.current_device())
163 | batch, h, w = 4, 32, 32
164 | in_channels = 2048
165 | for backbone in constants.backbones:
166 | if 'resnet' in backbone:
167 | low_level_channels = 256
168 | elif 'xception' in backbone:
169 | low_level_channels = 128
170 |
171 | x = torch.randn(batch, in_channels, h // 4, w // 4)
172 | low_level_features = torch.randn(
173 | batch, low_level_channels, h // 2, w // 2)
174 | x = x.to(DEVICE)
175 | low_level_features = low_level_features.to(DEVICE)
176 | announce_msg('testing {}. backbone {}.'.format(
177 | DeepLabV3PlusHead, backbone))
178 |
179 | model = deeplab_v3_plus_head(num_classes=1, backbone=backbone,
180 | output_stride=16, freeze_bn=False)
181 | model.to(DEVICE)
182 | t0 = dt.datetime.now()
183 | out = model(low_level_features, x, h, w)
184 | print(
185 | "in-shape {} \t output-shape {} "
186 | ". Forward time {}.".format(x.shape, out.shape,
187 | dt.datetime.now() - t0))
188 | print("NBR-PARAMS: ", model.get_nbr_params())
189 | print("Model: {}".format(model))
190 | print("Min-max output: {}, {}".format(out.min(), out.max()))
191 |
--------------------------------------------------------------------------------
/deeplearning/lr_scheduler.py:
--------------------------------------------------------------------------------
1 | import warnings
2 | import math
3 |
4 | import torch.optim.lr_scheduler as lr_scheduler
5 |
6 |
7 | class MyStepLR(lr_scheduler.StepLR):
8 | """
9 | Override: https://pytorch.org/docs/1.0.0/_modules/torch/optim/lr_scheduler.html#StepLR
10 | Reason: we want to fix the learning rate to not get lower than some value:
11 | min_lr.
12 |
13 | Sets the learning rate of each parameter group to the initial lr
14 | decayed by gamma every step_size epochs. When last_epoch=-1, sets
15 | initial lr as lr.
16 |
17 | Args:
18 | optimizer (Optimizer): Wrapped optimizer.
19 | step_size (int): Period of learning rate decay.
20 | gamma (float): Multiplicative factor of learning rate decay.
21 | Default: 0.1.
22 | last_epoch (int): The index of last epoch. Default: -1.
23 | min_lr (float): The lowest allowed value for the learning rate.
24 | """
25 |
26 | def __init__(self, optimizer, step_size, gamma=0.1, last_epoch=-1,
27 | min_lr=1e-6):
28 | self.step_size = step_size
29 | self.gamma = gamma
30 | self.min_lr = min_lr
31 | super(lr_scheduler.StepLR, self).__init__(optimizer, last_epoch)
32 |
33 | def get_lr(self):
34 | return [
35 | max(base_lr * self.gamma ** (self.last_epoch // self.step_size),
36 | self.min_lr) for base_lr in self.base_lrs]
37 |
38 |
39 | class MyCosineLR(lr_scheduler.StepLR):
40 | """
41 | Override: https://pytorch.org/docs/1.0.0/_modules/torch/optim/
42 | lr_scheduler.html#StepLR
43 | Reason: use a cosine evolution of lr.
44 | paper:
45 | `S. Qiao, W. Shen, Z. Zhang, B. Wang, and A. Yuille. Deepco-training for
46 | semi-supervised image recognition. InECCV,2018`
47 |
48 |
49 | for the epoch T:
50 | lr = base_lr * coef × (1.0 + cos((T − 1) × π/max_epochs)).
51 |
52 | Args:
53 | optimizer (Optimizer): Wrapped optimizer.
54 | coef (float): float coefficient. e.g. 0.005
55 | max_epochs (int): maximum epochs.
56 | last_epoch (int): The index of last epoch. Default: -1.
57 | min_lr (float): The lowest allowed value for the learning rate.
58 | """
59 |
60 | def __init__(self, optimizer, coef, max_epochs, min_lr=1e-9,
61 | last_epoch=-1):
62 | assert isinstance(coef, float), "'coef' must be a float. found {}" \
63 | "...[not ok]".format(type(coef))
64 | assert coef > 0., "'coef' must be > 0. found {} ....[NOT OK]".format(
65 | coef
66 | )
67 | assert max_epochs > 0, "'max_epochs' must be > 0. found {}" \
68 | "...[NOT OK]".format(max_epochs)
69 | self.max_epochs = float(max_epochs)
70 | self.coef = coef
71 | self.min_lr = min_lr
72 | super(lr_scheduler.StepLR, self).__init__(optimizer, last_epoch)
73 |
74 | def get_lr(self):
75 | return [
76 | max(base_lr * self.coef * (
77 | 1. + math.cos((self.last_epoch - 1) * math.pi /
78 | self.max_epochs)),
79 | self.min_lr) for base_lr in self.base_lrs]
80 |
81 |
82 | if __name__ == "__main__":
83 | from torch.optim import SGD
84 | import torch
85 | import matplotlib.pyplot as plt
86 |
87 | optimizer = SGD(torch.nn.Linear(10, 20).parameters(), lr=0.001)
88 | lr_sch = MyCosineLR(optimizer, coef=0.5, max_epochs=600, min_lr=1e-9)
89 | vals = []
90 | for i in range(1000):
91 | optimizer.step()
92 | vals.append(lr_sch.get_lr())
93 | lr_sch.step()
94 | plt.plot(vals)
95 | plt.savefig("lr_evolve_{}.png".format(lr_sch.__class__.__name__))
96 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/.gitignore:
--------------------------------------------------------------------------------
1 | # Prerequisites
2 | *.d
3 |
4 | # Compiled Object files
5 | *.slo
6 | *.lo
7 | *.o
8 | *.obj
9 |
10 | # Precompiled Headers
11 | *.gch
12 | *.pch
13 |
14 | # Compiled Dynamic libraries
15 | *.so
16 | *.dylib
17 | *.dll
18 |
19 | # Fortran module files
20 | *.mod
21 | *.smod
22 |
23 | # Compiled Static libraries
24 | *.lai
25 | *.la
26 | *.a
27 | *.lib
28 |
29 | # Executables
30 | *.exe
31 | *.out
32 | *.app
33 |
34 | # CMake
35 | CMakeCache.txt
36 | CMakeFiles
37 | CMakeScripts
38 | Testing
39 | Makefile
40 | cmake_install.cmake
41 | install_manifest.txt
42 | compile_commands.json
43 | CTestTestfile.cmake
44 |
45 | # IDE
46 | .cproject
47 | .project
48 | .pydevproject
49 | .travis.yml
50 |
51 | # Compiled protocol buffers
52 | *.pb.h
53 | *.pb.cc
54 | *_pb2.py
55 |
56 | # IPython notebook checkpoints
57 | .ipynb_checkpoints
58 |
59 | # Editor temporaries
60 | *.swp
61 | *~
62 |
63 | # Sublime Text settings
64 | *.sublime-workspace
65 | *.sublime-project
66 |
67 | # Eclipse Project settings
68 | *.*project
69 | .settings
70 |
71 | # QtCreator files
72 | *.user
73 |
74 | # PyCharm files
75 | .idea
76 |
77 | # Visual Studio Code files
78 | .vscode
79 |
80 | # OSX dir files
81 | .DS_Store
82 |
83 | # Byte-compiled / optimized / DLL files
84 | __pycache__/
85 | *.py[cod]
86 | *$py.class
87 |
88 | # C extensions
89 | *.so
90 |
91 | # Distribution / packaging
92 | .Python
93 | build/
94 | develop-eggs/
95 | dist/
96 | downloads/
97 | eggs/
98 | .eggs/
99 | lib/
100 | lib64/
101 | parts/
102 | sdist/
103 | var/
104 | wheels/
105 | *.egg-info/
106 | .installed.cfg
107 | *.egg
108 |
109 | # PyInstaller
110 | # Usually these files are written by a python script from a template
111 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
112 | *.manifest
113 | *.spec
114 |
115 | # Installer logs
116 | pip-log.txt
117 | pip-delete-this-directory.txt
118 |
119 | # Unit test / coverage reports
120 | htmlcov/
121 | .tox/
122 | .coverage
123 | .coverage.*
124 | .cache
125 | nosetests.xml
126 | coverage.xml
127 | *.cover
128 | .hypothesis/
129 |
130 | # Translations
131 | *.mo
132 | *.pot
133 |
134 | # Django stuff:
135 | *.log
136 | local_settings.py
137 |
138 | # Flask stuff:
139 | instance/
140 | .webassets-cache
141 |
142 | # Scrapy stuff:
143 | .scrapy
144 |
145 | # Sphinx documentation
146 | docs/_build/
147 |
148 | # PyBuilder
149 | target/
150 |
151 | # Jupyter Notebook
152 | .ipynb_checkpoints
153 |
154 | # pyenv
155 | .python-version
156 |
157 | # celery beat schedule file
158 | celerybeat-schedule
159 |
160 | # SageMath parsed files
161 | *.sage.py
162 |
163 | # Environments
164 | .env
165 | .venv
166 | env/
167 | venv/
168 | ENV/
169 |
170 | # Spyder project settings
171 | .spyderproject
172 | .spyproject
173 |
174 | # Rope project settings
175 | .ropeproject
176 |
177 | # mkdocs documentation
178 | /site
179 |
180 | # mypy
181 | .mypy_cache/
182 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/Dockerfile:
--------------------------------------------------------------------------------
1 | # base container
2 | FROM nvidia/cuda::10.0-devel-ubuntu18.04
3 |
4 | # update
5 | ENV DEBIAN_FRONTEND "noninteractive"
6 | RUN apt-get update -y
7 | RUN apt-get -y \
8 | -o Dpkg::Options::="--force-confdef" \
9 | -o Dpkg::Options::="--force-confold" dist-upgrade
10 |
11 | # install basic
12 | RUN apt-get install -y --no-install-recommends \
13 | less sudo ssh \
14 | build-essential \
15 | unzip git curl wget vim tree htop \
16 | python3-dev python3-tk \
17 | ninja-build
18 |
19 | # python libs
20 | RUN curl https://bootstrap.pypa.io/get-pip.py | python3
21 | RUN pip3 install \
22 | future six cffi numpy pillow tqdm Cython awscli ninja
23 |
24 | # install pytorch
25 | RUN pip3 install https://download.pytorch.org/whl/cu100/torch-1.0.0-cp36-cp36m-linux_x86_64.whl
26 | RUN pip3 install torchvision
27 |
28 | # clean up
29 | RUN apt-get update -y && apt-get upgrade -y && apt-get autoremove -y
30 | RUN apt-get clean -y && apt-get autoclean -y
31 | RUN rm -rf /var/lib/apt/lists/* /var/cache/apt/archives/*
32 |
33 | # create mountpoint from host
34 | RUN mkdir -p /workspace
35 |
36 | # create non-root user
37 | ARG user_name=ubuntu
38 | ARG user_id=1000
39 | ARG group_name=ubuntu
40 | ARG group_id=1000
41 | RUN groupadd -g ${group_id} ${group_name}
42 | RUN useradd -u ${user_id} -g ${group_id} -d /home/${user_name} --create-home --shell /bin/bash ${user_name}
43 | RUN echo "${user_name} ALL=(ALL) NOPASSWD:ALL" >> /etc/sudoers
44 | RUN chown -R ${user_name}:${group_name} /home/${user_name}
45 | RUN chown -R ${user_name}:${group_name} /workspace
46 | RUN chsh -s /bin/bash ${user_name}
47 | USER ubuntu
48 | WORKDIR /home/ubuntu
49 | ENV HOME /home/ubuntu
50 |
51 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2018 Tamaki Kojima
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/README.md:
--------------------------------------------------------------------------------
1 | # All credits of Synchronized BN go to Tamaki Kojima(tamakoji@gmail.com) (https://github.com/tamakoji/pytorch-syncbn)
2 | # pytorch-syncbn
3 |
4 | Tamaki Kojima(tamakoji@gmail.com)
5 |
6 | ## Announcement
7 |
8 | **Pytorch 1.0 support**
9 |
10 | ## Overview
11 | This is alternative implementation of "Synchronized Multi-GPU Batch Normalization" which computes global stats across gpus instead of locally computed. SyncBN are getting important for those input image is large, and must use multi-gpu to increase the minibatch-size for the training.
12 |
13 | The code was inspired by [Pytorch-Encoding](https://github.com/zhanghang1989/PyTorch-Encoding) and [Inplace-ABN](https://github.com/mapillary/inplace_abn)
14 |
15 | ## Remarks
16 | - Unlike [Pytorch-Encoding](https://github.com/zhanghang1989/PyTorch-Encoding), you don't need custom `nn.DataParallel`
17 | - Unlike [Inplace-ABN](https://github.com/mapillary/inplace_abn), you can just replace your `nn.BatchNorm2d` to this module implementation, since it will not mark for inplace operation
18 | - You can plug into arbitrary module written in PyTorch to enable Synchronized BatchNorm
19 | - Backward computation is rewritten and tested against behavior of `nn.BatchNorm2d`
20 |
21 | ## Requirements
22 | For PyTorch, please refer to https://pytorch.org/
23 |
24 | NOTE : The code is tested only with PyTorch v1.0.0, CUDA10/CuDNN7.4.2 on ubuntu18.04
25 |
26 | It utilize Pytorch JIT mechanism to compile seamlessly, using ninja. Please install ninja-build before use.
27 |
28 | ```
29 | sudo apt-get install ninja-build
30 | ```
31 |
32 | Also install all dependencies for python. For pip, run:
33 |
34 |
35 | ```
36 | pip install -U -r requirements.txt
37 | ```
38 |
39 | ## Build
40 |
41 | There is no need to build. just run and JIT will take care.
42 | JIT and cpp extensions are supported after PyTorch0.4, however it is highly recommended to use PyTorch > 1.0 due to huge design changes.
43 |
44 | ## Usage
45 |
46 | Please refer to [`test.py`](test.py) for testing the difference between `nn.BatchNorm2d` and `modules.nn.BatchNorm2d`
47 |
48 | ```
49 | import torch
50 | from modules import nn as NN
51 | num_gpu = torch.cuda.device_count()
52 | model = nn.Sequential(
53 | nn.Conv2d(3, 3, 1, 1, bias=False),
54 | NN.BatchNorm2d(3),
55 | nn.ReLU(inplace=True),
56 | nn.Conv2d(3, 3, 1, 1, bias=False),
57 | NN.BatchNorm2d(3),
58 | ).cuda()
59 | model = nn.DataParallel(model, device_ids=range(num_gpu))
60 | x = torch.rand(num_gpu, 3, 2, 2).cuda()
61 | z = model(x)
62 | ```
63 |
64 | ## Math
65 |
66 | ### Forward
67 | 1. compute 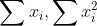 in each gpu
68 | 2. gather all
in each gpu
68 | 2. gather all 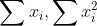 from workers to master and compute
from workers to master and compute 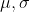 where
69 |
70 |
where
69 |
70 | 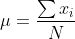 71 |
72 | and
73 |
74 |
71 |
72 | and
73 |
74 | 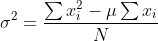 75 |
76 | and then above global stats to be shared to all gpus, update running_mean and running_var by moving average using global stats.
77 |
78 | 3. forward batchnorm using global stats by
79 |
80 |
75 |
76 | and then above global stats to be shared to all gpus, update running_mean and running_var by moving average using global stats.
77 |
78 | 3. forward batchnorm using global stats by
79 |
80 | 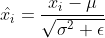 81 |
82 | and then
83 |
84 |
81 |
82 | and then
83 |
84 | 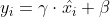 85 |
86 | where
85 |
86 | where 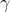 is weight parameter and
is weight parameter and 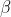 is bias parameter.
87 |
88 | 4. save
is bias parameter.
87 |
88 | 4. save 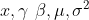 for backward
89 |
90 | ### Backward
91 |
92 | 1. Restore saved
for backward
89 |
90 | ### Backward
91 |
92 | 1. Restore saved 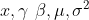 93 |
94 | 2. Compute below sums on each gpu
95 |
96 |
93 |
94 | 2. Compute below sums on each gpu
95 |
96 | ) 97 |
98 | and
99 |
100 |
97 |
98 | and
99 |
100 | ) 101 |
102 | where
101 |
102 | where 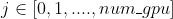 103 |
104 | then gather them at master node to sum up global, and normalize with N where N is total number of elements for each channels. Global sums are then shared among all gpus.
105 |
106 | 3. compute gradients using global stats
107 |
108 |
103 |
104 | then gather them at master node to sum up global, and normalize with N where N is total number of elements for each channels. Global sums are then shared among all gpus.
105 |
106 | 3. compute gradients using global stats
107 |
108 | 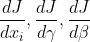 109 |
110 | where
111 |
112 |
109 |
110 | where
111 |
112 | ) 113 |
114 | and
115 |
116 |
113 |
114 | and
115 |
116 | ) 117 |
118 | and finally,
119 |
120 |
117 |
118 | and finally,
119 |
120 | 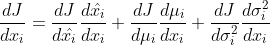 121 |
122 |
121 |
122 | }}(N\frac{dJ}{d\hat{x_i}}-\sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}})-\hat{x_i}\sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}}\hat{x_j}))) 123 |
124 |
123 |
124 | }}(N\frac{dJ}{dy_i}-\sum_{j=1}^{N}(\frac{dJ}{dy_j})-\hat{x_i}\sum_{j=1}^{N}(\frac{dJ}{dy_j}\hat{x_j}))) 125 |
126 | Note that in the implementation, normalization with N is performed at step (2) and above equation and implementation is not exactly the same, but mathematically is same.
127 |
128 | You can go deeper on above explanation at [Kevin Zakka's Blog](https://kevinzakka.github.io/2016/09/14/batch_normalization/)
--------------------------------------------------------------------------------
/deeplearning/syncbn/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/deeplearning/syncbn/__init__.py
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/__init__.py:
--------------------------------------------------------------------------------
1 | from .syncbn import batchnorm2d_sync
2 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/_csrc.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | Extension module loader
5 |
6 | code referenced from : https://github.com/facebookresearch/maskrcnn-benchmark
7 |
8 | /*****************************************************************************/
9 | """
10 | from __future__ import absolute_import
11 | from __future__ import division
12 | from __future__ import print_function
13 |
14 | import glob
15 | import os.path
16 |
17 | import torch
18 |
19 | try:
20 | from torch.utils.cpp_extension import load
21 | from torch.utils.cpp_extension import CUDA_HOME
22 | except ImportError:
23 | raise ImportError(
24 | "The cpp layer extensions requires PyTorch 0.4 or higher")
25 |
26 |
27 | def _load_C_extensions():
28 | this_dir = os.path.dirname(os.path.abspath(__file__))
29 | this_dir = os.path.join(this_dir, "csrc")
30 |
31 | main_file = glob.glob(os.path.join(this_dir, "*.cpp"))
32 | sources_cpu = glob.glob(os.path.join(this_dir, "cpu", "*.cpp"))
33 | sources_cuda = glob.glob(os.path.join(this_dir, "cuda", "*.cu"))
34 |
35 | sources = main_file + sources_cpu
36 |
37 | extra_cflags = []
38 | extra_cuda_cflags = []
39 | if torch.cuda.is_available() and CUDA_HOME is not None:
40 | sources.extend(sources_cuda)
41 | extra_cflags = ["-O3", "-DWITH_CUDA"]
42 | extra_cuda_cflags = ["--expt-extended-lambda"]
43 | sources = [os.path.join(this_dir, s) for s in sources]
44 | extra_include_paths = [this_dir]
45 | return load(
46 | name="ext_lib",
47 | sources=sources,
48 | extra_cflags=extra_cflags,
49 | extra_include_paths=extra_include_paths,
50 | extra_cuda_cflags=extra_cuda_cflags,
51 | )
52 |
53 |
54 | _backend = _load_C_extensions()
55 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/bn.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | SyncBN
4 |
5 | *****************************************************************************/
6 | #pragma once
7 |
8 | #ifdef WITH_CUDA
9 | #include "cuda/ext_lib.h"
10 | #endif
11 |
12 | /// SyncBN
13 |
14 | std::vector syncbn_sum_sqsum(const at::Tensor& x) {
15 | if (x.is_cuda()) {
16 | #ifdef WITH_CUDA
17 | return syncbn_sum_sqsum_cuda(x);
18 | #else
19 | AT_ERROR("Not compiled with GPU support");
20 | #endif
21 | } else {
22 | AT_ERROR("CPU implementation not supported");
23 | }
24 | }
25 |
26 | at::Tensor syncbn_forward(const at::Tensor& x, const at::Tensor& weight,
27 | const at::Tensor& bias, const at::Tensor& mean,
28 | const at::Tensor& var, bool affine, float eps) {
29 | if (x.is_cuda()) {
30 | #ifdef WITH_CUDA
31 | return syncbn_forward_cuda(x, weight, bias, mean, var, affine, eps);
32 | #else
33 | AT_ERROR("Not compiled with GPU support");
34 | #endif
35 | } else {
36 | AT_ERROR("CPU implementation not supported");
37 | }
38 | }
39 |
40 | std::vector syncbn_backward_xhat(const at::Tensor& dz,
41 | const at::Tensor& x,
42 | const at::Tensor& mean,
43 | const at::Tensor& var, float eps) {
44 | if (dz.is_cuda()) {
45 | #ifdef WITH_CUDA
46 | return syncbn_backward_xhat_cuda(dz, x, mean, var, eps);
47 | #else
48 | AT_ERROR("Not compiled with GPU support");
49 | #endif
50 | } else {
51 | AT_ERROR("CPU implementation not supported");
52 | }
53 | }
54 |
55 | std::vector syncbn_backward(
56 | const at::Tensor& dz, const at::Tensor& x, const at::Tensor& weight,
57 | const at::Tensor& bias, const at::Tensor& mean, const at::Tensor& var,
58 | const at::Tensor& sum_dz, const at::Tensor& sum_dz_xhat, bool affine,
59 | float eps) {
60 | if (dz.is_cuda()) {
61 | #ifdef WITH_CUDA
62 | return syncbn_backward_cuda(dz, x, weight, bias, mean, var, sum_dz,
63 | sum_dz_xhat, affine, eps);
64 | #else
65 | AT_ERROR("Not compiled with GPU support");
66 | #endif
67 | } else {
68 | AT_ERROR("CPU implementation not supported");
69 | }
70 | }
71 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/bn_cuda.cu:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA SyncBN code
4 |
5 | code referenced from : https://github.com/mapillary/inplace_abn
6 |
7 | *****************************************************************************/
8 | #include
9 | #include
10 | #include
11 | #include
12 | #include "cuda/common.h"
13 |
14 | // Utilities
15 | void get_dims(at::Tensor x, int64_t &num, int64_t &chn, int64_t &sp) {
16 | num = x.size(0);
17 | chn = x.size(1);
18 | sp = 1;
19 | for (int64_t i = 2; i < x.ndimension(); ++i) sp *= x.size(i);
20 | }
21 |
22 | /// SyncBN
23 |
24 | template
25 | struct SqSumOp {
26 | __device__ SqSumOp(const T *t, int c, int s) : tensor(t), chn(c), sp(s) {}
27 | __device__ __forceinline__ Pair operator()(int batch, int plane, int n) {
28 | T x = tensor[(batch * chn + plane) * sp + n];
29 | return Pair(x, x * x); // x, x^2
30 | }
31 | const T *tensor;
32 | const int chn;
33 | const int sp;
34 | };
35 |
36 | template
37 | __global__ void syncbn_sum_sqsum_kernel(const T *x, T *sum, T *sqsum,
38 | int num, int chn, int sp) {
39 | int plane = blockIdx.x;
40 | Pair res =
41 | reduce, SqSumOp>(SqSumOp(x, chn, sp), plane, num, chn, sp);
42 | __syncthreads();
43 | if (threadIdx.x == 0) {
44 | sum[plane] = res.v1;
45 | sqsum[plane] = res.v2;
46 | }
47 | }
48 |
49 | std::vector syncbn_sum_sqsum_cuda(const at::Tensor &x) {
50 | CHECK_INPUT(x);
51 |
52 | // Extract dimensions
53 | int64_t num, chn, sp;
54 | get_dims(x, num, chn, sp);
55 |
56 | // Prepare output tensors
57 | auto sum = at::empty({chn}, x.options());
58 | auto sqsum = at::empty({chn}, x.options());
59 |
60 | // Run kernel
61 | dim3 blocks(chn);
62 | dim3 threads(getNumThreads(sp));
63 | AT_DISPATCH_FLOATING_TYPES(
64 | x.type(), "syncbn_sum_sqsum_cuda", ([&] {
65 | syncbn_sum_sqsum_kernel<<>>(
66 | x.data(), sum.data(),
67 | sqsum.data(), num, chn, sp);
68 | }));
69 | return {sum, sqsum};
70 | }
71 |
72 | template
73 | __global__ void syncbn_forward_kernel(T *z, const T *x, const T *weight,
74 | const T *bias, const T *mean,
75 | const T *var, bool affine, float eps,
76 | int num, int chn, int sp) {
77 | int plane = blockIdx.x;
78 | T _mean = mean[plane];
79 | T _var = var[plane];
80 | T _weight = affine ? weight[plane] : T(1);
81 | T _bias = affine ? bias[plane] : T(0);
82 | float _invstd = T(0);
83 | if (_var || eps) {
84 | _invstd = rsqrt(_var + eps);
85 | }
86 | for (int batch = 0; batch < num; ++batch) {
87 | for (int n = threadIdx.x; n < sp; n += blockDim.x) {
88 | T _x = x[(batch * chn + plane) * sp + n];
89 | T _xhat = (_x - _mean) * _invstd;
90 | T _z = _xhat * _weight + _bias;
91 | z[(batch * chn + plane) * sp + n] = _z;
92 | }
93 | }
94 | }
95 |
96 | at::Tensor syncbn_forward_cuda(const at::Tensor &x, const at::Tensor &weight,
97 | const at::Tensor &bias, const at::Tensor &mean,
98 | const at::Tensor &var, bool affine, float eps) {
99 | CHECK_INPUT(x);
100 | CHECK_INPUT(weight);
101 | CHECK_INPUT(bias);
102 | CHECK_INPUT(mean);
103 | CHECK_INPUT(var);
104 |
105 | // Extract dimensions
106 | int64_t num, chn, sp;
107 | get_dims(x, num, chn, sp);
108 |
109 | auto z = at::zeros_like(x);
110 |
111 | // Run kernel
112 | dim3 blocks(chn);
113 | dim3 threads(getNumThreads(sp));
114 | AT_DISPATCH_FLOATING_TYPES(
115 | x.type(), "syncbn_forward_cuda", ([&] {
116 | syncbn_forward_kernel<<>>(
117 | z.data(), x.data(),
118 | weight.data(), bias.data(),
119 | mean.data(), var.data(),
120 | affine, eps, num, chn, sp);
121 | }));
122 | return z;
123 | }
124 |
125 | template
126 | struct XHatOp {
127 | __device__ XHatOp(T _weight, T _bias, const T *_dz, const T *_x, int c, int s)
128 | : weight(_weight), bias(_bias), x(_x), dz(_dz), chn(c), sp(s) {}
129 | __device__ __forceinline__ Pair operator()(int batch, int plane, int n) {
130 | // xhat = (x - bias) * weight
131 | T _xhat = (x[(batch * chn + plane) * sp + n] - bias) * weight;
132 | // dxhat * x_hat
133 | T _dz = dz[(batch * chn + plane) * sp + n];
134 | return Pair(_dz, _dz * _xhat);
135 | }
136 | const T weight;
137 | const T bias;
138 | const T *dz;
139 | const T *x;
140 | const int chn;
141 | const int sp;
142 | };
143 |
144 | template
145 | __global__ void syncbn_backward_xhat_kernel(const T *dz, const T *x,
146 | const T *mean, const T *var,
147 | T *sum_dz, T *sum_dz_xhat,
148 | float eps, int num, int chn,
149 | int sp) {
150 | int plane = blockIdx.x;
151 | T _mean = mean[plane];
152 | T _var = var[plane];
153 | T _invstd = T(0);
154 | if (_var || eps) {
155 | _invstd = rsqrt(_var + eps);
156 | }
157 | Pair res = reduce, XHatOp>(
158 | XHatOp(_invstd, _mean, dz, x, chn, sp), plane, num, chn, sp);
159 | __syncthreads();
160 | if (threadIdx.x == 0) {
161 | // \sum(\frac{dJ}{dy_i})
162 | sum_dz[plane] = res.v1;
163 | // \sum(\frac{dJ}{dy_i}*\hat{x_i})
164 | sum_dz_xhat[plane] = res.v2;
165 | }
166 | }
167 |
168 | std::vector syncbn_backward_xhat_cuda(const at::Tensor &dz,
169 | const at::Tensor &x,
170 | const at::Tensor &mean,
171 | const at::Tensor &var,
172 | float eps) {
173 | CHECK_INPUT(dz);
174 | CHECK_INPUT(x);
175 | CHECK_INPUT(mean);
176 | CHECK_INPUT(var);
177 | // Extract dimensions
178 | int64_t num, chn, sp;
179 | get_dims(x, num, chn, sp);
180 | // Prepare output tensors
181 | auto sum_dz = at::empty({chn}, x.options());
182 | auto sum_dz_xhat = at::empty({chn}, x.options());
183 | // Run kernel
184 | dim3 blocks(chn);
185 | dim3 threads(getNumThreads(sp));
186 | AT_DISPATCH_FLOATING_TYPES(
187 | x.type(), "syncbn_backward_xhat_cuda", ([&] {
188 | syncbn_backward_xhat_kernel<<>>(
189 | dz.data(), x.data(), mean.data(),
190 | var.data(), sum_dz.data(),
191 | sum_dz_xhat.data(), eps, num, chn, sp);
192 | }));
193 | return {sum_dz, sum_dz_xhat};
194 | }
195 |
196 | template
197 | __global__ void syncbn_backward_kernel(const T *dz, const T *x, const T *weight,
198 | const T *bias, const T *mean,
199 | const T *var, const T *sum_dz,
200 | const T *sum_dz_xhat, T *dx, T *dweight,
201 | T *dbias, bool affine, float eps,
202 | int num, int chn, int sp) {
203 | int plane = blockIdx.x;
204 | T _mean = mean[plane];
205 | T _var = var[plane];
206 | T _weight = affine ? weight[plane] : T(1);
207 | T _sum_dz = sum_dz[plane];
208 | T _sum_dz_xhat = sum_dz_xhat[plane];
209 | T _invstd = T(0);
210 | if (_var || eps) {
211 | _invstd = rsqrt(_var + eps);
212 | }
213 | /*
214 | \frac{dJ}{dx_i} = \frac{1}{N\sqrt{(\sigma^2+\epsilon)}} (
215 | N\frac{dJ}{d\hat{x_i}} -
216 | \sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}}) -
217 | \hat{x_i}\sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}}\hat{x_j})
218 | )
219 | Note : N is omitted here since it will be accumulated and
220 | _sum_dz and _sum_dz_xhat expected to be already normalized
221 | before the call.
222 | */
223 | if (dx) {
224 | T _mul = _weight * _invstd;
225 | for (int batch = 0; batch < num; ++batch) {
226 | for (int n = threadIdx.x; n < sp; n += blockDim.x) {
227 | T _dz = dz[(batch * chn + plane) * sp + n];
228 | T _xhat = (x[(batch * chn + plane) * sp + n] - _mean) * _invstd;
229 | T _dx = (_dz - _sum_dz - _xhat * _sum_dz_xhat) * _mul;
230 | dx[(batch * chn + plane) * sp + n] = _dx;
231 | }
232 | }
233 | }
234 | __syncthreads();
235 | if (threadIdx.x == 0) {
236 | if (affine) {
237 | T _norm = num * sp;
238 | dweight[plane] += _sum_dz_xhat * _norm;
239 | dbias[plane] += _sum_dz * _norm;
240 | }
241 | }
242 | }
243 |
244 | std::vector syncbn_backward_cuda(
245 | const at::Tensor &dz, const at::Tensor &x, const at::Tensor &weight,
246 | const at::Tensor &bias, const at::Tensor &mean, const at::Tensor &var,
247 | const at::Tensor &sum_dz, const at::Tensor &sum_dz_xhat, bool affine,
248 | float eps) {
249 | CHECK_INPUT(dz);
250 | CHECK_INPUT(x);
251 | CHECK_INPUT(weight);
252 | CHECK_INPUT(bias);
253 | CHECK_INPUT(mean);
254 | CHECK_INPUT(var);
255 | CHECK_INPUT(sum_dz);
256 | CHECK_INPUT(sum_dz_xhat);
257 |
258 | // Extract dimensions
259 | int64_t num, chn, sp;
260 | get_dims(x, num, chn, sp);
261 |
262 | // Prepare output tensors
263 | auto dx = at::zeros_like(dz);
264 | auto dweight = at::zeros_like(weight);
265 | auto dbias = at::zeros_like(bias);
266 |
267 | // Run kernel
268 | dim3 blocks(chn);
269 | dim3 threads(getNumThreads(sp));
270 | AT_DISPATCH_FLOATING_TYPES(
271 | x.type(), "syncbn_backward_cuda", ([&] {
272 | syncbn_backward_kernel<<>>(
273 | dz.data(), x.data(), weight.data(),
274 | bias.data(), mean.data(), var.data(),
275 | sum_dz.data(), sum_dz_xhat.data(),
276 | dx.data(), dweight.data(),
277 | dbias.data(), affine, eps, num, chn, sp);
278 | }));
279 | return {dx, dweight, dbias};
280 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/common.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA utility funcs
4 |
5 | code referenced from : https://github.com/mapillary/inplace_abn
6 |
7 | *****************************************************************************/
8 | #pragma once
9 |
10 | #include
11 |
12 | // Checks
13 | #ifndef AT_CHECK
14 | #define AT_CHECK AT_ASSERT
15 | #endif
16 | #define CHECK_CUDA(x) AT_CHECK(x.type().is_cuda(), #x " must be a CUDA tensor")
17 | #define CHECK_CONTIGUOUS(x) AT_CHECK(x.is_contiguous(), #x " must be contiguous")
18 | #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)
19 |
20 | /*
21 | * General settings
22 | */
23 | const int WARP_SIZE = 32;
24 | const int MAX_BLOCK_SIZE = 512;
25 |
26 | template
27 | struct Pair {
28 | T v1, v2;
29 | __device__ Pair() {}
30 | __device__ Pair(T _v1, T _v2) : v1(_v1), v2(_v2) {}
31 | __device__ Pair(T v) : v1(v), v2(v) {}
32 | __device__ Pair(int v) : v1(v), v2(v) {}
33 | __device__ Pair &operator+=(const Pair &a) {
34 | v1 += a.v1;
35 | v2 += a.v2;
36 | return *this;
37 | }
38 | };
39 |
40 | /*
41 | * Utility functions
42 | */
43 | template
44 | __device__ __forceinline__ T WARP_SHFL_XOR(T value, int laneMask,
45 | int width = warpSize,
46 | unsigned int mask = 0xffffffff) {
47 | #if CUDART_VERSION >= 9000
48 | return __shfl_xor_sync(mask, value, laneMask, width);
49 | #else
50 | return __shfl_xor(value, laneMask, width);
51 | #endif
52 | }
53 |
54 | __device__ __forceinline__ int getMSB(int val) { return 31 - __clz(val); }
55 |
56 | static int getNumThreads(int nElem) {
57 | int threadSizes[5] = {32, 64, 128, 256, MAX_BLOCK_SIZE};
58 | for (int i = 0; i != 5; ++i) {
59 | if (nElem <= threadSizes[i]) {
60 | return threadSizes[i];
61 | }
62 | }
63 | return MAX_BLOCK_SIZE;
64 | }
65 |
66 | template
67 | static __device__ __forceinline__ T warpSum(T val) {
68 | #if __CUDA_ARCH__ >= 300
69 | for (int i = 0; i < getMSB(WARP_SIZE); ++i) {
70 | val += WARP_SHFL_XOR(val, 1 << i, WARP_SIZE);
71 | }
72 | #else
73 | __shared__ T values[MAX_BLOCK_SIZE];
74 | values[threadIdx.x] = val;
75 | __threadfence_block();
76 | const int base = (threadIdx.x / WARP_SIZE) * WARP_SIZE;
77 | for (int i = 1; i < WARP_SIZE; i++) {
78 | val += values[base + ((i + threadIdx.x) % WARP_SIZE)];
79 | }
80 | #endif
81 | return val;
82 | }
83 |
84 | template
85 | static __device__ __forceinline__ Pair warpSum(Pair value) {
86 | value.v1 = warpSum(value.v1);
87 | value.v2 = warpSum(value.v2);
88 | return value;
89 | }
90 |
91 | template
92 | __device__ T reduce(Op op, int plane, int N, int C, int S) {
93 | T sum = (T)0;
94 | for (int batch = 0; batch < N; ++batch) {
95 | for (int x = threadIdx.x; x < S; x += blockDim.x) {
96 | sum += op(batch, plane, x);
97 | }
98 | }
99 |
100 | // sum over NumThreads within a warp
101 | sum = warpSum(sum);
102 |
103 | // 'transpose', and reduce within warp again
104 | __shared__ T shared[32];
105 | __syncthreads();
106 | if (threadIdx.x % WARP_SIZE == 0) {
107 | shared[threadIdx.x / WARP_SIZE] = sum;
108 | }
109 | if (threadIdx.x >= blockDim.x / WARP_SIZE && threadIdx.x < WARP_SIZE) {
110 | // zero out the other entries in shared
111 | shared[threadIdx.x] = (T)0;
112 | }
113 | __syncthreads();
114 | if (threadIdx.x / WARP_SIZE == 0) {
115 | sum = warpSum(shared[threadIdx.x]);
116 | if (threadIdx.x == 0) {
117 | shared[0] = sum;
118 | }
119 | }
120 | __syncthreads();
121 |
122 | // Everyone picks it up, should be broadcast into the whole gradInput
123 | return shared[0];
124 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/ext_lib.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA SyncBN code
4 |
5 | *****************************************************************************/
6 | #pragma once
7 | #include
8 | #include
9 |
10 | /// Sync-BN
11 | std::vector syncbn_sum_sqsum_cuda(const at::Tensor& x);
12 | at::Tensor syncbn_forward_cuda(const at::Tensor& x, const at::Tensor& weight,
13 | const at::Tensor& bias, const at::Tensor& mean,
14 | const at::Tensor& var, bool affine, float eps);
15 | std::vector syncbn_backward_xhat_cuda(const at::Tensor& dz,
16 | const at::Tensor& x,
17 | const at::Tensor& mean,
18 | const at::Tensor& var,
19 | float eps);
20 | std::vector syncbn_backward_cuda(
21 | const at::Tensor& dz, const at::Tensor& x, const at::Tensor& weight,
22 | const at::Tensor& bias, const at::Tensor& mean, const at::Tensor& var,
23 | const at::Tensor& sum_dz, const at::Tensor& sum_dz_xhat, bool affine,
24 | float eps);
25 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/ext_lib.cpp:
--------------------------------------------------------------------------------
1 | #include "bn.h"
2 |
3 | PYBIND11_MODULE(TORCH_EXTENSION_NAME, m) {
4 | m.def("syncbn_sum_sqsum", &syncbn_sum_sqsum, "Sum and Sum^2 computation");
5 | m.def("syncbn_forward", &syncbn_forward, "SyncBN forward computation");
6 | m.def("syncbn_backward_xhat", &syncbn_backward_xhat,
7 | "First part of SyncBN backward computation");
8 | m.def("syncbn_backward", &syncbn_backward,
9 | "Second part of SyncBN backward computation");
10 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/syncbn.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | BatchNorm2dSync with multi-gpu
5 |
6 | code referenced from : https://github.com/mapillary/inplace_abn
7 |
8 | /*****************************************************************************/
9 | """
10 | from __future__ import absolute_import
11 | from __future__ import division
12 | from __future__ import print_function
13 |
14 | import torch.cuda.comm as comm
15 | from torch.autograd import Function
16 | from torch.autograd.function import once_differentiable
17 | from ._csrc import _backend
18 |
19 |
20 | def _count_samples(x):
21 | count = 1
22 | for i, s in enumerate(x.size()):
23 | if i != 1:
24 | count *= s
25 | return count
26 |
27 |
28 | class BatchNorm2dSyncFunc(Function):
29 |
30 | @staticmethod
31 | def forward(ctx, x, weight, bias, running_mean, running_var,
32 | extra, compute_stats=True, momentum=0.1, eps=1e-05):
33 | def _parse_extra(ctx, extra):
34 | ctx.is_master = extra["is_master"]
35 | if ctx.is_master:
36 | ctx.master_queue = extra["master_queue"]
37 | ctx.worker_queues = extra["worker_queues"]
38 | ctx.worker_ids = extra["worker_ids"]
39 | else:
40 | ctx.master_queue = extra["master_queue"]
41 | ctx.worker_queue = extra["worker_queue"]
42 | # Save context
43 | if extra is not None:
44 | _parse_extra(ctx, extra)
45 | ctx.compute_stats = compute_stats
46 | ctx.momentum = momentum
47 | ctx.eps = eps
48 | ctx.affine = weight is not None and bias is not None
49 | if ctx.compute_stats:
50 | N = _count_samples(x) * (ctx.master_queue.maxsize + 1)
51 | assert N > 1
52 | # 1. compute sum(x) and sum(x^2)
53 | xsum, xsqsum = _backend.syncbn_sum_sqsum(x.detach())
54 | if ctx.is_master:
55 | xsums, xsqsums = [xsum], [xsqsum]
56 | # master : gatther all sum(x) and sum(x^2) from slaves
57 | for _ in range(ctx.master_queue.maxsize):
58 | xsum_w, xsqsum_w = ctx.master_queue.get()
59 | ctx.master_queue.task_done()
60 | xsums.append(xsum_w)
61 | xsqsums.append(xsqsum_w)
62 | xsum = comm.reduce_add(xsums)
63 | xsqsum = comm.reduce_add(xsqsums)
64 | mean = xsum / N
65 | sumvar = xsqsum - xsum * mean
66 | var = sumvar / N
67 | uvar = sumvar / (N - 1)
68 | # master : broadcast global mean, variance to all slaves
69 | tensors = comm.broadcast_coalesced(
70 | (mean, uvar, var), [mean.get_device()] + ctx.worker_ids)
71 | for ts, queue in zip(tensors[1:], ctx.worker_queues):
72 | queue.put(ts)
73 | else:
74 | # slave : send sum(x) and sum(x^2) to master
75 | ctx.master_queue.put((xsum, xsqsum))
76 | # slave : get global mean and variance
77 | mean, uvar, var = ctx.worker_queue.get()
78 | ctx.worker_queue.task_done()
79 |
80 | # Update running stats
81 | running_mean.mul_((1 - ctx.momentum)).add_(ctx.momentum * mean)
82 | running_var.mul_((1 - ctx.momentum)).add_(ctx.momentum * uvar)
83 | ctx.N = N
84 | ctx.save_for_backward(x, weight, bias, mean, var)
85 | else:
86 | mean, var = running_mean, running_var
87 |
88 | # do batch norm forward
89 | z = _backend.syncbn_forward(x, weight, bias, mean, var,
90 | ctx.affine, ctx.eps)
91 | return z
92 |
93 | @staticmethod
94 | @once_differentiable
95 | def backward(ctx, dz):
96 | x, weight, bias, mean, var = ctx.saved_tensors
97 | dz = dz.contiguous()
98 |
99 | # 1. compute \sum(\frac{dJ}{dy_i}) and \sum(\frac{dJ}{dy_i}*\hat{x_i})
100 | sum_dz, sum_dz_xhat = _backend.syncbn_backward_xhat(
101 | dz, x, mean, var, ctx.eps)
102 | if ctx.is_master:
103 | sum_dzs, sum_dz_xhats = [sum_dz], [sum_dz_xhat]
104 | # master : gatther from slaves
105 | for _ in range(ctx.master_queue.maxsize):
106 | sum_dz_w, sum_dz_xhat_w = ctx.master_queue.get()
107 | ctx.master_queue.task_done()
108 | sum_dzs.append(sum_dz_w)
109 | sum_dz_xhats.append(sum_dz_xhat_w)
110 | # master : compute global stats
111 | sum_dz = comm.reduce_add(sum_dzs)
112 | sum_dz_xhat = comm.reduce_add(sum_dz_xhats)
113 | sum_dz /= ctx.N
114 | sum_dz_xhat /= ctx.N
115 | # master : broadcast global stats
116 | tensors = comm.broadcast_coalesced(
117 | (sum_dz, sum_dz_xhat), [mean.get_device()] + ctx.worker_ids)
118 | for ts, queue in zip(tensors[1:], ctx.worker_queues):
119 | queue.put(ts)
120 | else:
121 | # slave : send to master
122 | ctx.master_queue.put((sum_dz, sum_dz_xhat))
123 | # slave : get global stats
124 | sum_dz, sum_dz_xhat = ctx.worker_queue.get()
125 | ctx.worker_queue.task_done()
126 |
127 | # do batch norm backward
128 | dx, dweight, dbias = _backend.syncbn_backward(
129 | dz, x, weight, bias, mean, var, sum_dz, sum_dz_xhat,
130 | ctx.affine, ctx.eps)
131 |
132 | return dx, dweight, dbias, \
133 | None, None, None, None, None, None
134 |
135 | batchnorm2d_sync = BatchNorm2dSyncFunc.apply
136 |
137 | __all__ = ["batchnorm2d_sync"]
138 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/nn/__init__.py:
--------------------------------------------------------------------------------
1 | from .syncbn import *
2 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/nn/syncbn.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | BatchNorm2dSync with multi-gpu
5 |
6 | /*****************************************************************************/
7 | """
8 | from __future__ import absolute_import
9 | from __future__ import division
10 | from __future__ import print_function
11 |
12 | try:
13 | # python 3
14 | from queue import Queue
15 | except ImportError:
16 | # python 2
17 | from Queue import Queue
18 |
19 | import torch
20 | import torch.nn as nn
21 | from torch.nn import functional as F
22 | from torch.nn.parameter import Parameter
23 | import sys
24 | sys.path.append("..")
25 | from deeplearning.syncbn.functional import batchnorm2d_sync
26 |
27 |
28 | class _BatchNorm(nn.Module):
29 | """
30 | Customized BatchNorm from nn.BatchNorm
31 | >> added freeze attribute to enable bn freeze.
32 | """
33 |
34 | def __init__(self, num_features, eps=1e-5, momentum=0.1, affine=True,
35 | track_running_stats=True):
36 | super(_BatchNorm, self).__init__()
37 | self.num_features = num_features
38 | self.eps = eps
39 | self.momentum = momentum
40 | self.affine = affine
41 | self.track_running_stats = track_running_stats
42 | self.freezed = False
43 | if self.affine:
44 | self.weight = Parameter(torch.Tensor(num_features))
45 | self.bias = Parameter(torch.Tensor(num_features))
46 | else:
47 | self.register_parameter('weight', None)
48 | self.register_parameter('bias', None)
49 | if self.track_running_stats:
50 | self.register_buffer('running_mean', torch.zeros(num_features))
51 | self.register_buffer('running_var', torch.ones(num_features))
52 | else:
53 | self.register_parameter('running_mean', None)
54 | self.register_parameter('running_var', None)
55 | self.reset_parameters()
56 |
57 | def reset_parameters(self):
58 | if self.track_running_stats:
59 | self.running_mean.zero_()
60 | self.running_var.fill_(1)
61 | if self.affine:
62 | self.weight.data.uniform_()
63 | self.bias.data.zero_()
64 |
65 | def _check_input_dim(self, input):
66 | return NotImplemented
67 |
68 | def forward(self, input):
69 | self._check_input_dim(input)
70 |
71 | compute_stats = not self.freezed and \
72 | self.training and self.track_running_stats
73 |
74 | ret = F.batch_norm(input, self.running_mean, self.running_var,
75 | self.weight, self.bias, compute_stats,
76 | self.momentum, self.eps)
77 | return ret
78 |
79 | def extra_repr(self):
80 | return '{num_features}, eps={eps}, momentum={momentum}, '\
81 | 'affine={affine}, ' \
82 | 'track_running_stats={track_running_stats}'.format(
83 | **self.__dict__)
84 |
85 |
86 | class BatchNorm2dNoSync(_BatchNorm):
87 | """
88 | Equivalent to nn.BatchNorm2d
89 | """
90 |
91 | def _check_input_dim(self, input):
92 | if input.dim() != 4:
93 | raise ValueError('expected 4D input (got {}D input)'
94 | .format(input.dim()))
95 |
96 |
97 | class BatchNorm2dSync(BatchNorm2dNoSync):
98 | """
99 | BatchNorm2d with automatic multi-GPU Sync
100 | """
101 |
102 | def __init__(self, num_features, eps=1e-5, momentum=0.1, affine=True,
103 | track_running_stats=True):
104 | super(BatchNorm2dSync, self).__init__(
105 | num_features, eps=eps, momentum=momentum, affine=affine,
106 | track_running_stats=track_running_stats)
107 | self.sync_enabled = True
108 | self.devices = list(range(torch.cuda.device_count()))
109 | if len(self.devices) > 1:
110 | # Initialize queues
111 | self.worker_ids = self.devices[1:]
112 | self.master_queue = Queue(len(self.worker_ids))
113 | self.worker_queues = [Queue(1) for _ in self.worker_ids]
114 |
115 | def forward(self, x):
116 | compute_stats = not self.freezed and \
117 | self.training and self.track_running_stats
118 | if self.sync_enabled and compute_stats and len(self.devices) > 1:
119 | if x.get_device() == self.devices[0]:
120 | # Master mode
121 | extra = {
122 | "is_master": True,
123 | "master_queue": self.master_queue,
124 | "worker_queues": self.worker_queues,
125 | "worker_ids": self.worker_ids
126 | }
127 | else:
128 | # Worker mode
129 | extra = {
130 | "is_master": False,
131 | "master_queue": self.master_queue,
132 | "worker_queue": self.worker_queues[
133 | self.worker_ids.index(x.get_device())]
134 | }
135 | return batchnorm2d_sync(x, self.weight, self.bias,
136 | self.running_mean, self.running_var,
137 | extra, compute_stats, self.momentum,
138 | self.eps)
139 | return super(BatchNorm2dSync, self).forward(x)
140 |
141 | def __repr__(self):
142 | """repr"""
143 | rep = '{name}({num_features}, eps={eps}, momentum={momentum},' \
144 | 'affine={affine}, ' \
145 | 'track_running_stats={track_running_stats},' \
146 | 'devices={devices})'
147 | return rep.format(name=self.__class__.__name__, **self.__dict__)
148 |
149 | #BatchNorm2d = BatchNorm2dNoSync
150 | BatchNorm2d = BatchNorm2dSync
151 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/requirements.txt:
--------------------------------------------------------------------------------
1 | future
2 | cffi
3 | ninja
--------------------------------------------------------------------------------
/deeplearning/syncbn/test.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | Test for BatchNorm2dSync with multi-gpu
5 |
6 | /*****************************************************************************/
7 | """
8 | from __future__ import absolute_import
9 | from __future__ import division
10 | from __future__ import print_function
11 |
12 | import sys
13 | import numpy as np
14 | import torch
15 | from torch import nn
16 | from torch.nn import functional as F
17 | sys.path.append("./")
18 | import nn as NN
19 |
20 | torch.backends.cudnn.deterministic = True
21 |
22 |
23 | def init_weight(model):
24 | for m in model.modules():
25 | if isinstance(m, nn.Conv2d):
26 | n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels
27 | m.weight.data.normal_(0, np.sqrt(2. / n))
28 | elif isinstance(m, NN.BatchNorm2d) or isinstance(m, nn.BatchNorm2d):
29 | m.weight.data.fill_(1)
30 | m.bias.data.zero_()
31 | elif isinstance(m, nn.Linear):
32 | m.bias.data.zero_()
33 |
34 | num_gpu = torch.cuda.device_count()
35 | print("num_gpu={}".format(num_gpu))
36 | if num_gpu < 2:
37 | print("No multi-gpu found. NN.BatchNorm2d will act as normal nn.BatchNorm2d")
38 |
39 | m1 = nn.Sequential(
40 | nn.Conv2d(3, 3, 1, 1, bias=False),
41 | nn.BatchNorm2d(3),
42 | nn.ReLU(inplace=True),
43 | nn.Conv2d(3, 3, 1, 1, bias=False),
44 | nn.BatchNorm2d(3),
45 | ).cuda()
46 | torch.manual_seed(123)
47 | init_weight(m1)
48 | m2 = nn.Sequential(
49 | nn.Conv2d(3, 3, 1, 1, bias=False),
50 | NN.BatchNorm2d(3),
51 | nn.ReLU(inplace=True),
52 | nn.Conv2d(3, 3, 1, 1, bias=False),
53 | NN.BatchNorm2d(3),
54 | ).cuda()
55 | torch.manual_seed(123)
56 | init_weight(m2)
57 | m2 = nn.DataParallel(m2, device_ids=range(num_gpu))
58 | o1 = torch.optim.SGD(m1.parameters(), 1e-3)
59 | o2 = torch.optim.SGD(m2.parameters(), 1e-3)
60 | y = torch.ones(num_gpu).float().cuda()
61 | torch.manual_seed(123)
62 | for _ in range(100):
63 | x = torch.rand(num_gpu, 3, 2, 2).cuda()
64 | o1.zero_grad()
65 | z1 = m1(x)
66 | l1 = F.mse_loss(z1.mean(-1).mean(-1).mean(-1), y)
67 | l1.backward()
68 | o1.step()
69 | o2.zero_grad()
70 | z2 = m2(x)
71 | l2 = F.mse_loss(z2.mean(-1).mean(-1).mean(-1), y)
72 | l2.backward()
73 | o2.step()
74 | print(m2.module[1].bias.grad - m1[1].bias.grad)
75 | print(m2.module[1].weight.grad - m1[1].weight.grad)
76 | print(m2.module[-1].bias.grad - m1[-1].bias.grad)
77 | print(m2.module[-1].weight.grad - m1[-1].weight.grad)
78 | m2 = m2.module
79 | print("===============================")
80 | print("m1(nn.BatchNorm2d) running_mean",
81 | m1[1].running_mean, m1[-1].running_mean)
82 | print("m2(NN.BatchNorm2d) running_mean",
83 | m2[1].running_mean, m2[-1].running_mean)
84 | print("m1(nn.BatchNorm2d) running_var", m1[1].running_var, m1[-1].running_var)
85 | print("m2(NN.BatchNorm2d) running_var", m2[1].running_var, m2[-1].running_var)
86 | print("m1(nn.BatchNorm2d) weight", m1[1].weight, m1[-1].weight)
87 | print("m2(NN.BatchNorm2d) weight", m2[1].weight, m2[-1].weight)
88 | print("m1(nn.BatchNorm2d) bias", m1[1].bias, m1[-1].bias)
89 | print("m2(NN.BatchNorm2d) bias", m2[1].bias, m2[-1].bias)
90 |
--------------------------------------------------------------------------------
/deeplearning/utils.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import os
3 |
4 |
5 | import torch
6 | import torch.nn as nn
7 | import numpy as np
8 | import torch.nn.functional as F
9 | from collections import OrderedDict
10 |
11 |
12 | sys.path.append("..")
13 |
14 | from shared import check_if_allow_multgpu_mode, announce_msg
15 |
16 | ACTIVATE_SYNC_BN = False
17 | # Override ACTIVATE_SYNC_BN using variable environment in Bash:
18 | # $ export ACTIVATE_SYNC_BN="True" ----> Activate
19 | # $ export ACTIVATE_SYNC_BN="False" ----> Deactivate
20 |
21 | if "ACTIVATE_SYNC_BN" in os.environ.keys():
22 | ACTIVATE_SYNC_BN = (os.environ['ACTIVATE_SYNC_BN'] == "True")
23 |
24 | announce_msg("ACTIVATE_SYNC_BN was set to {}".format(ACTIVATE_SYNC_BN))
25 |
26 | if check_if_allow_multgpu_mode() and ACTIVATE_SYNC_BN: # Activate Synch-BN.
27 | from deeplearning.syncbn import nn as NN_Sync_BN
28 | BatchNorm2d = NN_Sync_BN.BatchNorm2d
29 | announce_msg("Synchronized BN has been activated. \n"
30 | "MultiGPU mode has been activated. "
31 | "{} GPUs".format(torch.cuda.device_count()))
32 | else:
33 | BatchNorm2d = nn.BatchNorm2d
34 | if check_if_allow_multgpu_mode():
35 | announce_msg("Synchronized BN has been deactivated.\n"
36 | "MultiGPU mode has been activated. "
37 | "{} GPUs".format(torch.cuda.device_count()))
38 | else:
39 | announce_msg("Synchronized BN has been deactivated.\n"
40 | "MultiGPU mode has been deactivated. "
41 | "{} GPUs".format(torch.cuda.device_count()))
42 |
43 |
44 | def initialize_weights(*models):
45 | for model in models:
46 | for m in model.modules():
47 | if isinstance(m, nn.Conv2d):
48 | nn.init.kaiming_normal_(m.weight.data, nonlinearity='relu')
49 | elif isinstance(m, BatchNorm2d):
50 | m.weight.data.fill_(1.)
51 | m.bias.data.fill_(1e-4)
52 | elif isinstance(m, nn.Linear):
53 | m.weight.data.normal_(0.0, 0.0001)
54 | m.bias.data.zero_()
55 |
56 | class _SimpleSegmentationModel(nn.Module):
57 | def __init__(self, backbone, classifier):
58 | super(_SimpleSegmentationModel, self).__init__()
59 | self.backbone = backbone
60 | self.classifier = classifier
61 |
62 | def forward(self, x):
63 | input_shape = x.shape[-2:]
64 | features = self.backbone(x)
65 | x = self.classifier(features)
66 | x = F.interpolate(x, size=input_shape, mode='bilinear',
67 | align_corners=False)
68 | return x
69 |
70 |
71 | class IntermediateLayerGetter(nn.ModuleDict):
72 | """
73 | Module wrapper that returns intermediate layers from a model
74 |
75 | It has a strong assumption that the modules have been registered
76 | into the model in the same order as they are used.
77 | This means that one should **not** reuse the same nn.Module
78 | twice in the forward if you want this to work.
79 |
80 | Additionally, it is only able to query submodules that are directly
81 | assigned to the model. So if `model` is passed, `model.feature1` can
82 | be returned, but not `model.feature1.layer2`.
83 |
84 | Arguments:
85 | model (nn.Module): model on which we will extract the features
86 | return_layers (Dict[name, new_name]): a dict containing the names
87 | of the modules for which the activations will be returned as
88 | the key of the dict, and the value of the dict is the name
89 | of the returned activation (which the user can specify).
90 |
91 | Examples::
92 |
93 | >>> m = torchvision.models.resnet18(pretrained=True)
94 | >>> # extract layer1 and layer3, giving as names `feat1` and feat2`
95 | >>> new_m = torchvision.models._utils.IntermediateLayerGetter(m,
96 | >>> {'layer1': 'feat1', 'layer3': 'feat2'})
97 | >>> out = new_m(torch.rand(1, 3, 224, 224))
98 | >>> print([(k, v.shape) for k, v in out.items()])
99 | >>> [('feat1', torch.Size([1, 64, 56, 56])),
100 | >>> ('feat2', torch.Size([1, 256, 14, 14]))]
101 | """
102 |
103 | def __init__(self, model, return_layers):
104 | if not set(return_layers).issubset(
105 | [name for name, _ in model.named_children()]):
106 | raise ValueError("return_layers are not present in model")
107 |
108 | orig_return_layers = return_layers
109 | return_layers = {k: v for k, v in return_layers.items()}
110 | layers = OrderedDict()
111 | for name, module in model.named_children():
112 | layers[name] = module
113 | if name in return_layers:
114 | del return_layers[name]
115 | if not return_layers:
116 | break
117 |
118 | super(IntermediateLayerGetter, self).__init__(layers)
119 | self.return_layers = orig_return_layers
120 |
121 | def forward(self, x):
122 | out = OrderedDict()
123 | for name, module in self.named_children():
124 | x = module(x)
125 | if name in self.return_layers:
126 | out_name = self.return_layers[name]
127 | out[out_name] = x
128 | return out
129 |
--------------------------------------------------------------------------------
/deeplearning/wildcat.py:
--------------------------------------------------------------------------------
1 | import sys
2 |
3 |
4 | import torch.nn as nn
5 |
6 | sys.path.append("..")
7 |
8 | from deeplearning.decision_pooling import WildCatPoolDecision, ClassWisePooling
9 |
10 |
11 | class WildCatClassifierHead(nn.Module):
12 | """
13 | A WILDCAT type classifier head.
14 | `WILDCAT: Weakly Supervised Learning of Deep ConvNets for
15 | Image Classification, Pointwise Localization and Segmentation`,
16 | Thibaut Durand, Taylor Mordan, Nicolas Thome, Matthieu Cord.
17 | """
18 | def __init__(self, inplans, modalities, num_classes, kmax=0.5, kmin=None,
19 | alpha=0.6, dropout=0.0):
20 | super(WildCatClassifierHead, self).__init__()
21 | self.name = "wildcat"
22 |
23 | self.num_classes = num_classes
24 |
25 | self.to_modalities = nn.Conv2d(
26 | inplans, num_classes * modalities, kernel_size=1, bias=True)
27 | self.to_maps = ClassWisePooling(num_classes, modalities)
28 | self.wildcat = WildCatPoolDecision(
29 | kmax=kmax, kmin=kmin, alpha=alpha, dropout=dropout)
30 |
31 | def forward(self, x, seed=None, prngs_cuda=None):
32 | """
33 | The forward function.
34 | :param x: input tensor.
35 | :param seed:
36 | :param prngs_cuda:
37 | :return: scores, maps.
38 | """
39 | modalities = self.to_modalities(x)
40 | maps = self.to_maps(modalities)
41 | scores = self.wildcat(x=maps, seed=seed, prngs_cuda=prngs_cuda)
42 |
43 | return scores, maps
44 |
45 | def get_nbr_params(self):
46 | """
47 | Compute the number of parameters of the model.
48 | :return:
49 | """
50 | return sum([p.numel() for p in self.parameters()])
51 |
52 | def __str__(self):
53 | return "{}: WILDCAT.".format(self.name)
54 |
55 | def __repr__(self):
56 | return super(WildCatClassifierHead, self).__repr__()
57 |
58 |
59 | if __name__ == "__main__":
60 | inst = WildCatClassifierHead(inplans=10, modalities=5, num_classes=2)
61 | print(repr(inst))
62 | print(inst)
--------------------------------------------------------------------------------
/dependencies/.readme.md:
--------------------------------------------------------------------------------
1 | Contains files about the dependencies.
--------------------------------------------------------------------------------
/dependencies/get_dependencies.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
2 | # =============================================================================================
3 | # PURE PYTHON 3.7.0 VIRTUAL ENVIRONMENT
4 | # ==============================================================================================
5 |
6 | # virtualenvwrapper commands are not recognized WITHIN bash files. Not sure why.
7 | # Need to source virtualenvwrapper.sh in order to be recognized.
8 | # First: create a bash variable pointing to the path of virtualenvwrapper.sh
9 | # name it: export VIRTUALENVWRAPPER_SH=/usr/local/bin/virtualenvwrapper.sh
10 |
11 | # You can omit this line if you do not use virtual environment wrapper. You can create your virtual environment
12 | # using pip and install the requirements.
13 | source $VIRTUALENVWRAPPER_SH
14 |
15 | # You can ignore this line as well. (same reason)
16 | workon pytorch.1.4.0
17 | pip freeze > requirements.txt
18 |
19 | # Create a new V.E using virtualenvwrapper
20 | # mkvirtualenv -r requirements.txt pytorch.1.2.0-dev
21 | # mkvirtualenv -p $HOME/anaconda3/bin/python3 -r requirements.txt pytorch.1.2.0-dev
22 |
23 | # Create a new V.E using virtualenv
24 | # virtualenv ~/Venvs/pytorch.1.0.1
25 |
26 | # Install requirements. Up to you for --no-index
27 | # pip install --no-index -r requirements.txt
28 |
--------------------------------------------------------------------------------
/dependencies/requirements.txt:
--------------------------------------------------------------------------------
1 | attrs==19.1.0
2 | backcall==0.1.0
3 | bleach==2.1.3
4 | compress-pickle==1.1.0
5 | cycler==0.10.0
6 | Cython==0.29.2
7 | decorator==4.3.2
8 | entrypoints==0.2.3
9 | future==0.16.0
10 | html5lib==1.0.1
11 | imageio==2.4.1
12 | ipykernel==4.8.2
13 | ipython==6.5.0
14 | ipython-genutils==0.2.0
15 | ipywidgets==7.4.2
16 | jedi==0.12.1
17 | Jinja2==2.10
18 | jsonschema==2.6.0
19 | jupyter==1.0.0
20 | jupyter-client==5.2.4
21 | jupyter-console==5.2.0
22 | jupyter-core==4.4.0
23 | kiwisolver==1.0.1
24 | MarkupSafe==1.1.1
25 | matplotlib==3.0.2
26 | mistune==0.8.4
27 | nbconvert==5.3.1
28 | nbformat==4.4.0
29 | notebook==5.7.4
30 | numpy==1.16.2
31 | opencv-python==3.4.4.19
32 | pandocfilters==1.4.2
33 | parso==0.3.1
34 | pexpect==4.6.0
35 | pickleshare==0.7.5
36 | Pillow==7.1.1
37 | prometheus-client==0.3.1
38 | prompt-toolkit==1.0.15
39 | protobuf==3.7.1
40 | ptyprocess==0.6.0
41 | pygifsicle==1.0.1
42 | Pygments==2.3.1
43 | pyparsing==2.3.1
44 | python-dateutil==2.8.0
45 | PyYAML==3.13
46 | pyzmq==17.1.2
47 | qtconsole==4.3.1
48 | scikit-learn==0.20.2
49 | scipy==1.2.1
50 | Send2Trash==1.5.0
51 | simplegeneric==0.8.1
52 | six==1.12.0
53 | terminado==0.8.1
54 | testpath==0.3.1
55 | texttable==1.6.2
56 | torch==1.4.0+cu100
57 | torchvision==0.5.0+cu100
58 | tornado==5.1.1
59 | tqdm==4.31.1
60 | traitlets==4.3.2
61 | typing==3.6.6
62 | wcwidth==0.1.7
63 | webencodings==0.5.1
64 | widgetsnbextension==3.4.2
65 |
66 |
--------------------------------------------------------------------------------
/doc/.readme.md:
--------------------------------------------------------------------------------
1 | idea?
--------------------------------------------------------------------------------
/doc/algo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/algo.png
--------------------------------------------------------------------------------
/doc/arch.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/arch.png
--------------------------------------------------------------------------------
/doc/knn.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/knn.png
--------------------------------------------------------------------------------
/doc/perf.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/perf.png
--------------------------------------------------------------------------------
/doc/proposal.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/proposal.png
--------------------------------------------------------------------------------
/doc/pseudo-labeling.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/pseudo-labeling.png
--------------------------------------------------------------------------------
/doc/results.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/results.png
--------------------------------------------------------------------------------
/doc/wacv2021-active-learning-weak-annotator.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/wacv2021-active-learning-weak-annotator.pdf
--------------------------------------------------------------------------------
/exps/.readme.md:
--------------------------------------------------------------------------------
1 | Contains exps results.
--------------------------------------------------------------------------------
/folds/.readme.md:
--------------------------------------------------------------------------------
1 | contains folds.
2 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/log-stats-ds-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.txt:
--------------------------------------------------------------------------------
1 | min h 120, max h 500
2 | min w 121, max w 500
3 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/size-stats-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/folds/Caltech-UCSD-Birds-200-2011/size-stats-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.png
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/.~lock.train_s_0_f_0.csv#:
--------------------------------------------------------------------------------
1 | ,brian,dell,16.09.2020 19:52,file:///home/brian/.config/libreoffice/4;
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/log-stats-ds-ds-glas-s-0-f-0-subset-train.txt:
--------------------------------------------------------------------------------
1 | min h 430, max h 522
2 | min w 567, max w 775
3 |
--------------------------------------------------------------------------------
/folds/glas/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/size-stats-ds-glas-s-0-f-0-subset-train.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/folds/glas/size-stats-ds-glas-s-0-f-0-subset-train.png
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/.~lock.test_s_0_f_0.csv#:
--------------------------------------------------------------------------------
1 | ,brian,dell,24.07.2020 12:46,file:///home/brian/.config/libreoffice/4;
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/test_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/train_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
2 | 105,train_32.bmp,train_32_anno.bmp,benign,1
3 | 128,train_53.bmp,train_53_anno.bmp,benign,1
4 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
5 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
6 | 157,train_8.bmp,train_8_anno.bmp,benign,1
7 | 135,train_6.bmp,train_6_anno.bmp,benign,1
8 | 107,train_34.bmp,train_34_anno.bmp,benign,1
9 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
10 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
11 | 106,train_33.bmp,train_33_anno.bmp,benign,1
12 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
13 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
14 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
15 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
16 | 138,train_62.bmp,train_62_anno.bmp,benign,1
17 | 145,train_69.bmp,train_69_anno.bmp,benign,1
18 | 162,train_84.bmp,train_84_anno.bmp,benign,1
19 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
20 | 130,train_55.bmp,train_55_anno.bmp,benign,1
21 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
22 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
23 | 121,train_47.bmp,train_47_anno.bmp,benign,1
24 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
25 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
26 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
27 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
28 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
29 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
30 | 140,train_64.bmp,train_64_anno.bmp,benign,1
31 | 92,train_20.bmp,train_20_anno.bmp,benign,1
32 | 103,train_30.bmp,train_30_anno.bmp,benign,1
33 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
34 | 134,train_59.bmp,train_59_anno.bmp,benign,1
35 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
36 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
37 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
38 | 97,train_25.bmp,train_25_anno.bmp,benign,1
39 | 120,train_46.bmp,train_46_anno.bmp,benign,1
40 | 164,train_9.bmp,train_9_anno.bmp,benign,1
41 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
42 | 119,train_45.bmp,train_45_anno.bmp,benign,1
43 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
44 | 93,train_21.bmp,train_21_anno.bmp,benign,1
45 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
46 | 158,train_80.bmp,train_80_anno.bmp,benign,1
47 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
48 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
49 | 137,train_61.bmp,train_61_anno.bmp,benign,1
50 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
51 | 94,train_22.bmp,train_22_anno.bmp,benign,1
52 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
53 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
54 | 113,train_4.bmp,train_4_anno.bmp,benign,1
55 | 163,train_85.bmp,train_85_anno.bmp,benign,1
56 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
57 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
58 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
59 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
60 | 143,train_67.bmp,train_67_anno.bmp,benign,1
61 | 156,train_79.bmp,train_79_anno.bmp,benign,1
62 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
63 | 131,train_56.bmp,train_56_anno.bmp,benign,1
64 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
65 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
66 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
67 | 86,train_15.bmp,train_15_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/valid_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 96,train_24.bmp,train_24_anno.bmp,malignant,0
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,0
3 | 124,train_5.bmp,train_5_anno.bmp,malignant,0
4 | 109,train_36.bmp,train_36_anno.bmp,malignant,0
5 | 98,train_26.bmp,train_26_anno.bmp,malignant,0
6 | 154,train_77.bmp,train_77_anno.bmp,malignant,0
7 | 151,train_74.bmp,train_74_anno.bmp,malignant,0
8 | 89,train_18.bmp,train_18_anno.bmp,malignant,0
9 | 161,train_83.bmp,train_83_anno.bmp,malignant,0
10 | 144,train_68.bmp,train_68_anno.bmp,malignant,0
11 | 111,train_38.bmp,train_38_anno.bmp,benign,0
12 | 149,train_72.bmp,train_72_anno.bmp,benign,0
13 | 141,train_65.bmp,train_65_anno.bmp,benign,0
14 | 126,train_51.bmp,train_51_anno.bmp,benign,0
15 | 83,train_12.bmp,train_12_anno.bmp,benign,0
16 | 139,train_63.bmp,train_63_anno.bmp,benign,0
17 | 91,train_2.bmp,train_2_anno.bmp,benign,0
18 | 127,train_52.bmp,train_52_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/test_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/train_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
2 | 139,train_63.bmp,train_63_anno.bmp,benign,1
3 | 140,train_64.bmp,train_64_anno.bmp,benign,1
4 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
5 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
6 | 141,train_65.bmp,train_65_anno.bmp,benign,1
7 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
8 | 119,train_45.bmp,train_45_anno.bmp,benign,1
9 | 92,train_20.bmp,train_20_anno.bmp,benign,1
10 | 121,train_47.bmp,train_47_anno.bmp,benign,1
11 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
12 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
13 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
14 | 149,train_72.bmp,train_72_anno.bmp,benign,1
15 | 127,train_52.bmp,train_52_anno.bmp,benign,1
16 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
17 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
18 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
19 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
20 | 107,train_34.bmp,train_34_anno.bmp,benign,1
21 | 158,train_80.bmp,train_80_anno.bmp,benign,1
22 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
23 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
24 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
25 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
26 | 126,train_51.bmp,train_51_anno.bmp,benign,1
27 | 113,train_4.bmp,train_4_anno.bmp,benign,1
28 | 94,train_22.bmp,train_22_anno.bmp,benign,1
29 | 162,train_84.bmp,train_84_anno.bmp,benign,1
30 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
31 | 138,train_62.bmp,train_62_anno.bmp,benign,1
32 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
33 | 163,train_85.bmp,train_85_anno.bmp,benign,1
34 | 128,train_53.bmp,train_53_anno.bmp,benign,1
35 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
36 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
37 | 111,train_38.bmp,train_38_anno.bmp,benign,1
38 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
39 | 143,train_67.bmp,train_67_anno.bmp,benign,1
40 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
41 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
42 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
43 | 97,train_25.bmp,train_25_anno.bmp,benign,1
44 | 86,train_15.bmp,train_15_anno.bmp,benign,1
45 | 83,train_12.bmp,train_12_anno.bmp,benign,1
46 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
47 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
48 | 105,train_32.bmp,train_32_anno.bmp,benign,1
49 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
50 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
51 | 93,train_21.bmp,train_21_anno.bmp,benign,1
52 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
53 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
54 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
55 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
56 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
57 | 156,train_79.bmp,train_79_anno.bmp,benign,1
58 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
59 | 131,train_56.bmp,train_56_anno.bmp,benign,1
60 | 135,train_6.bmp,train_6_anno.bmp,benign,1
61 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
62 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
63 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
64 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
65 | 91,train_2.bmp,train_2_anno.bmp,benign,1
66 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
67 | 157,train_8.bmp,train_8_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/valid_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 125,train_50.bmp,train_50_anno.bmp,malignant,0
2 | 85,train_14.bmp,train_14_anno.bmp,malignant,0
3 | 146,train_7.bmp,train_7_anno.bmp,malignant,0
4 | 87,train_16.bmp,train_16_anno.bmp,malignant,0
5 | 152,train_75.bmp,train_75_anno.bmp,malignant,0
6 | 153,train_76.bmp,train_76_anno.bmp,malignant,0
7 | 160,train_82.bmp,train_82_anno.bmp,malignant,0
8 | 116,train_42.bmp,train_42_anno.bmp,malignant,0
9 | 150,train_73.bmp,train_73_anno.bmp,malignant,0
10 | 122,train_48.bmp,train_48_anno.bmp,malignant,0
11 | 103,train_30.bmp,train_30_anno.bmp,benign,0
12 | 130,train_55.bmp,train_55_anno.bmp,benign,0
13 | 137,train_61.bmp,train_61_anno.bmp,benign,0
14 | 164,train_9.bmp,train_9_anno.bmp,benign,0
15 | 145,train_69.bmp,train_69_anno.bmp,benign,0
16 | 106,train_33.bmp,train_33_anno.bmp,benign,0
17 | 120,train_46.bmp,train_46_anno.bmp,benign,0
18 | 134,train_59.bmp,train_59_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/test_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/train_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 135,train_6.bmp,train_6_anno.bmp,benign,1
2 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
3 | 131,train_56.bmp,train_56_anno.bmp,benign,1
4 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
5 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
6 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
7 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
8 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
9 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
10 | 127,train_52.bmp,train_52_anno.bmp,benign,1
11 | 164,train_9.bmp,train_9_anno.bmp,benign,1
12 | 93,train_21.bmp,train_21_anno.bmp,benign,1
13 | 141,train_65.bmp,train_65_anno.bmp,benign,1
14 | 86,train_15.bmp,train_15_anno.bmp,benign,1
15 | 106,train_33.bmp,train_33_anno.bmp,benign,1
16 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
17 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
18 | 134,train_59.bmp,train_59_anno.bmp,benign,1
19 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
20 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
21 | 130,train_55.bmp,train_55_anno.bmp,benign,1
22 | 119,train_45.bmp,train_45_anno.bmp,benign,1
23 | 113,train_4.bmp,train_4_anno.bmp,benign,1
24 | 120,train_46.bmp,train_46_anno.bmp,benign,1
25 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
26 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
27 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
28 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
29 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
30 | 156,train_79.bmp,train_79_anno.bmp,benign,1
31 | 94,train_22.bmp,train_22_anno.bmp,benign,1
32 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
33 | 162,train_84.bmp,train_84_anno.bmp,benign,1
34 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
35 | 137,train_61.bmp,train_61_anno.bmp,benign,1
36 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
37 | 163,train_85.bmp,train_85_anno.bmp,benign,1
38 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
39 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
40 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
41 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
42 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
43 | 149,train_72.bmp,train_72_anno.bmp,benign,1
44 | 128,train_53.bmp,train_53_anno.bmp,benign,1
45 | 91,train_2.bmp,train_2_anno.bmp,benign,1
46 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
47 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
48 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
49 | 145,train_69.bmp,train_69_anno.bmp,benign,1
50 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
51 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
52 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
53 | 92,train_20.bmp,train_20_anno.bmp,benign,1
54 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
55 | 126,train_51.bmp,train_51_anno.bmp,benign,1
56 | 83,train_12.bmp,train_12_anno.bmp,benign,1
57 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
58 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
59 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
60 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
61 | 139,train_63.bmp,train_63_anno.bmp,benign,1
62 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
63 | 111,train_38.bmp,train_38_anno.bmp,benign,1
64 | 143,train_67.bmp,train_67_anno.bmp,benign,1
65 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
66 | 103,train_30.bmp,train_30_anno.bmp,benign,1
67 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/valid_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 118,train_44.bmp,train_44_anno.bmp,malignant,0
2 | 90,train_19.bmp,train_19_anno.bmp,malignant,0
3 | 132,train_57.bmp,train_57_anno.bmp,malignant,0
4 | 117,train_43.bmp,train_43_anno.bmp,malignant,0
5 | 133,train_58.bmp,train_58_anno.bmp,malignant,0
6 | 147,train_70.bmp,train_70_anno.bmp,malignant,0
7 | 104,train_31.bmp,train_31_anno.bmp,malignant,0
8 | 110,train_37.bmp,train_37_anno.bmp,malignant,0
9 | 80,train_1.bmp,train_1_anno.bmp,malignant,0
10 | 82,train_11.bmp,train_11_anno.bmp,malignant,0
11 | 140,train_64.bmp,train_64_anno.bmp,benign,0
12 | 157,train_8.bmp,train_8_anno.bmp,benign,0
13 | 138,train_62.bmp,train_62_anno.bmp,benign,0
14 | 107,train_34.bmp,train_34_anno.bmp,benign,0
15 | 97,train_25.bmp,train_25_anno.bmp,benign,0
16 | 158,train_80.bmp,train_80_anno.bmp,benign,0
17 | 105,train_32.bmp,train_32_anno.bmp,benign,0
18 | 121,train_47.bmp,train_47_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/test_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/train_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 107,train_34.bmp,train_34_anno.bmp,benign,1
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
3 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
4 | 138,train_62.bmp,train_62_anno.bmp,benign,1
5 | 97,train_25.bmp,train_25_anno.bmp,benign,1
6 | 139,train_63.bmp,train_63_anno.bmp,benign,1
7 | 91,train_2.bmp,train_2_anno.bmp,benign,1
8 | 120,train_46.bmp,train_46_anno.bmp,benign,1
9 | 149,train_72.bmp,train_72_anno.bmp,benign,1
10 | 86,train_15.bmp,train_15_anno.bmp,benign,1
11 | 105,train_32.bmp,train_32_anno.bmp,benign,1
12 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
13 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
14 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
15 | 130,train_55.bmp,train_55_anno.bmp,benign,1
16 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
17 | 145,train_69.bmp,train_69_anno.bmp,benign,1
18 | 83,train_12.bmp,train_12_anno.bmp,benign,1
19 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
20 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
21 | 158,train_80.bmp,train_80_anno.bmp,benign,1
22 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
23 | 121,train_47.bmp,train_47_anno.bmp,benign,1
24 | 162,train_84.bmp,train_84_anno.bmp,benign,1
25 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
26 | 106,train_33.bmp,train_33_anno.bmp,benign,1
27 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
28 | 111,train_38.bmp,train_38_anno.bmp,benign,1
29 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
30 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
31 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
32 | 126,train_51.bmp,train_51_anno.bmp,benign,1
33 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
34 | 164,train_9.bmp,train_9_anno.bmp,benign,1
35 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
36 | 103,train_30.bmp,train_30_anno.bmp,benign,1
37 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
38 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
39 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
40 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
41 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
42 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
43 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
44 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
45 | 163,train_85.bmp,train_85_anno.bmp,benign,1
46 | 127,train_52.bmp,train_52_anno.bmp,benign,1
47 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
48 | 140,train_64.bmp,train_64_anno.bmp,benign,1
49 | 119,train_45.bmp,train_45_anno.bmp,benign,1
50 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
51 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
52 | 143,train_67.bmp,train_67_anno.bmp,benign,1
53 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
54 | 141,train_65.bmp,train_65_anno.bmp,benign,1
55 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
56 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
57 | 137,train_61.bmp,train_61_anno.bmp,benign,1
58 | 157,train_8.bmp,train_8_anno.bmp,benign,1
59 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
60 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
61 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
62 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
63 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
64 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
65 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
66 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
67 | 134,train_59.bmp,train_59_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/valid_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 159,train_81.bmp,train_81_anno.bmp,malignant,0
2 | 136,train_60.bmp,train_60_anno.bmp,malignant,0
3 | 99,train_27.bmp,train_27_anno.bmp,malignant,0
4 | 114,train_40.bmp,train_40_anno.bmp,malignant,0
5 | 148,train_71.bmp,train_71_anno.bmp,malignant,0
6 | 102,train_3.bmp,train_3_anno.bmp,malignant,0
7 | 88,train_17.bmp,train_17_anno.bmp,malignant,0
8 | 115,train_41.bmp,train_41_anno.bmp,malignant,0
9 | 100,train_28.bmp,train_28_anno.bmp,malignant,0
10 | 155,train_78.bmp,train_78_anno.bmp,malignant,0
11 | 93,train_21.bmp,train_21_anno.bmp,benign,0
12 | 156,train_79.bmp,train_79_anno.bmp,benign,0
13 | 128,train_53.bmp,train_53_anno.bmp,benign,0
14 | 135,train_6.bmp,train_6_anno.bmp,benign,0
15 | 94,train_22.bmp,train_22_anno.bmp,benign,0
16 | 92,train_20.bmp,train_20_anno.bmp,benign,0
17 | 131,train_56.bmp,train_56_anno.bmp,benign,0
18 | 113,train_4.bmp,train_4_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/test_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/train_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
3 | 83,train_12.bmp,train_12_anno.bmp,benign,1
4 | 94,train_22.bmp,train_22_anno.bmp,benign,1
5 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
6 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
7 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
8 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
9 | 145,train_69.bmp,train_69_anno.bmp,benign,1
10 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
11 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
12 | 158,train_80.bmp,train_80_anno.bmp,benign,1
13 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
14 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
15 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
16 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
17 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
18 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
19 | 107,train_34.bmp,train_34_anno.bmp,benign,1
20 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
21 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
22 | 149,train_72.bmp,train_72_anno.bmp,benign,1
23 | 93,train_21.bmp,train_21_anno.bmp,benign,1
24 | 91,train_2.bmp,train_2_anno.bmp,benign,1
25 | 127,train_52.bmp,train_52_anno.bmp,benign,1
26 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
27 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
28 | 126,train_51.bmp,train_51_anno.bmp,benign,1
29 | 106,train_33.bmp,train_33_anno.bmp,benign,1
30 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
31 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
32 | 157,train_8.bmp,train_8_anno.bmp,benign,1
33 | 164,train_9.bmp,train_9_anno.bmp,benign,1
34 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
35 | 105,train_32.bmp,train_32_anno.bmp,benign,1
36 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
37 | 156,train_79.bmp,train_79_anno.bmp,benign,1
38 | 128,train_53.bmp,train_53_anno.bmp,benign,1
39 | 97,train_25.bmp,train_25_anno.bmp,benign,1
40 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
41 | 130,train_55.bmp,train_55_anno.bmp,benign,1
42 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
43 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
44 | 92,train_20.bmp,train_20_anno.bmp,benign,1
45 | 103,train_30.bmp,train_30_anno.bmp,benign,1
46 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
47 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
48 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
49 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
50 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
51 | 141,train_65.bmp,train_65_anno.bmp,benign,1
52 | 134,train_59.bmp,train_59_anno.bmp,benign,1
53 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
54 | 121,train_47.bmp,train_47_anno.bmp,benign,1
55 | 137,train_61.bmp,train_61_anno.bmp,benign,1
56 | 120,train_46.bmp,train_46_anno.bmp,benign,1
57 | 140,train_64.bmp,train_64_anno.bmp,benign,1
58 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
59 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
60 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
61 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
62 | 135,train_6.bmp,train_6_anno.bmp,benign,1
63 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
64 | 138,train_62.bmp,train_62_anno.bmp,benign,1
65 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
66 | 111,train_38.bmp,train_38_anno.bmp,benign,1
67 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
68 | 139,train_63.bmp,train_63_anno.bmp,benign,1
69 | 113,train_4.bmp,train_4_anno.bmp,benign,1
70 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
71 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
72 | 131,train_56.bmp,train_56_anno.bmp,benign,1
73 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/valid_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 108,train_35.bmp,train_35_anno.bmp,malignant,0
2 | 142,train_66.bmp,train_66_anno.bmp,malignant,0
3 | 84,train_13.bmp,train_13_anno.bmp,malignant,0
4 | 81,train_10.bmp,train_10_anno.bmp,malignant,0
5 | 112,train_39.bmp,train_39_anno.bmp,malignant,0
6 | 123,train_49.bmp,train_49_anno.bmp,malignant,0
7 | 101,train_29.bmp,train_29_anno.bmp,malignant,0
8 | 95,train_23.bmp,train_23_anno.bmp,malignant,0
9 | 143,train_67.bmp,train_67_anno.bmp,benign,0
10 | 119,train_45.bmp,train_45_anno.bmp,benign,0
11 | 163,train_85.bmp,train_85_anno.bmp,benign,0
12 | 86,train_15.bmp,train_15_anno.bmp,benign,0
13 | 162,train_84.bmp,train_84_anno.bmp,benign,0
14 |
--------------------------------------------------------------------------------
/fonts/.readme.md:
--------------------------------------------------------------------------------
1 | Contains fonts used for drawing.
2 | Source: https://fonts.google.com/specimen/Inconsolata?selection.family=Inconsolata
3 |
--------------------------------------------------------------------------------
/fonts/Inconsolata.zip:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata.zip
--------------------------------------------------------------------------------
/fonts/Inconsolata/Inconsolata-Bold.ttf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata/Inconsolata-Bold.ttf
--------------------------------------------------------------------------------
/fonts/Inconsolata/Inconsolata-Regular.ttf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata/Inconsolata-Regular.ttf
--------------------------------------------------------------------------------
/fonts/Inconsolata/OFL.txt:
--------------------------------------------------------------------------------
1 | Copyright 2006 The Inconsolata Project Authors
2 |
3 | This Font Software is licensed under the SIL Open Font License, Version 1.1.
4 | This license is copied below, and is also available with a FAQ at:
5 | http://scripts.sil.org/OFL
6 |
7 |
8 | -----------------------------------------------------------
9 | SIL OPEN FONT LICENSE Version 1.1 - 26 February 2007
10 | -----------------------------------------------------------
11 |
12 | PREAMBLE
13 | The goals of the Open Font License (OFL) are to stimulate worldwide
14 | development of collaborative font projects, to support the font creation
15 | efforts of academic and linguistic communities, and to provide a free and
16 | open framework in which fonts may be shared and improved in partnership
17 | with others.
18 |
19 | The OFL allows the licensed fonts to be used, studied, modified and
20 | redistributed freely as long as they are not sold by themselves. The
21 | fonts, including any derivative works, can be bundled, embedded,
22 | redistributed and/or sold with any software provided that any reserved
23 | names are not used by derivative works. The fonts and derivatives,
24 | however, cannot be released under any other type of license. The
25 | requirement for fonts to remain under this license does not apply
26 | to any document created using the fonts or their derivatives.
27 |
28 | DEFINITIONS
29 | "Font Software" refers to the set of files released by the Copyright
30 | Holder(s) under this license and clearly marked as such. This may
31 | include source files, build scripts and documentation.
32 |
33 | "Reserved Font Name" refers to any names specified as such after the
34 | copyright statement(s).
35 |
36 | "Original Version" refers to the collection of Font Software components as
37 | distributed by the Copyright Holder(s).
38 |
39 | "Modified Version" refers to any derivative made by adding to, deleting,
40 | or substituting -- in part or in whole -- any of the components of the
41 | Original Version, by changing formats or by porting the Font Software to a
42 | new environment.
43 |
44 | "Author" refers to any designer, engineer, programmer, technical
45 | writer or other person who contributed to the Font Software.
46 |
47 | PERMISSION & CONDITIONS
48 | Permission is hereby granted, free of charge, to any person obtaining
49 | a copy of the Font Software, to use, study, copy, merge, embed, modify,
50 | redistribute, and sell modified and unmodified copies of the Font
51 | Software, subject to the following conditions:
52 |
53 | 1) Neither the Font Software nor any of its individual components,
54 | in Original or Modified Versions, may be sold by itself.
55 |
56 | 2) Original or Modified Versions of the Font Software may be bundled,
57 | redistributed and/or sold with any software, provided that each copy
58 | contains the above copyright notice and this license. These can be
59 | included either as stand-alone text files, human-readable headers or
60 | in the appropriate machine-readable metadata fields within text or
61 | binary files as long as those fields can be easily viewed by the user.
62 |
63 | 3) No Modified Version of the Font Software may use the Reserved Font
64 | Name(s) unless explicit written permission is granted by the corresponding
65 | Copyright Holder. This restriction only applies to the primary font name as
66 | presented to the users.
67 |
68 | 4) The name(s) of the Copyright Holder(s) or the Author(s) of the Font
69 | Software shall not be used to promote, endorse or advertise any
70 | Modified Version, except to acknowledge the contribution(s) of the
71 | Copyright Holder(s) and the Author(s) or with their explicit written
72 | permission.
73 |
74 | 5) The Font Software, modified or unmodified, in part or in whole,
75 | must be distributed entirely under this license, and must not be
76 | distributed under any other license. The requirement for fonts to
77 | remain under this license does not apply to any document created
78 | using the Font Software.
79 |
80 | TERMINATION
81 | This license becomes null and void if any of the above conditions are
82 | not met.
83 |
84 | DISCLAIMER
85 | THE FONT SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
86 | EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO ANY WARRANTIES OF
87 | MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT
88 | OF COPYRIGHT, PATENT, TRADEMARK, OR OTHER RIGHT. IN NO EVENT SHALL THE
89 | COPYRIGHT HOLDER BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY,
90 | INCLUDING ANY GENERAL, SPECIAL, INDIRECT, INCIDENTAL, OR CONSEQUENTIAL
91 | DAMAGES, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
92 | FROM, OUT OF THE USE OR INABILITY TO USE THE FONT SOFTWARE OR FROM
93 | OTHER DEALINGS IN THE FONT SOFTWARE.
94 |
--------------------------------------------------------------------------------
/jobs/.readme.md:
--------------------------------------------------------------------------------
1 | Contains slurm jobs.
--------------------------------------------------------------------------------
/outputjobs/.readme.md:
--------------------------------------------------------------------------------
1 | Contains the output of the jobs (errors, standard output redirected to files).
--------------------------------------------------------------------------------
/pairwise_sims/.readme.md:
--------------------------------------------------------------------------------
1 | folder contains pairwise similarities.
2 |
--------------------------------------------------------------------------------
/paper/.readme.md:
--------------------------------------------------------------------------------
1 | contains files produced for the results.
2 |
--------------------------------------------------------------------------------
/paper/results-per-method-each-run-dataset-Caltech-UCSD-Birds-200-2011.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/paper/results-per-method-each-run-dataset-Caltech-UCSD-Birds-200-2011.pkl
--------------------------------------------------------------------------------
/paper/results-per-method-each-run-dataset-glas.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/paper/results-per-method-each-run-dataset-glas.pkl
--------------------------------------------------------------------------------
/pretrained-imgnet/.readme.md:
--------------------------------------------------------------------------------
1 | contains imagenet pretrained weights.
2 |
--------------------------------------------------------------------------------
/pretrained/.readme.md:
--------------------------------------------------------------------------------
1 | contains pretrained models.
2 |
--------------------------------------------------------------------------------
/reproducibility.py:
--------------------------------------------------------------------------------
1 | # self-contained-as-possible module.
2 | # handles reproducibility procedures.
3 |
4 | import random
5 | import os
6 | import warnings
7 |
8 |
9 | import numpy as np
10 | import torch
11 | from torch._C import default_generator
12 |
13 |
14 | DEFAULT_SEED = 0 # the default seed.
15 |
16 |
17 | def check_if_allow_multgpu_mode():
18 | """
19 | Check if we can do multigpu.
20 | If yes, allow multigpu.
21 | :return: ALLOW_MULTIGPUS: bool. If True, we enter multigpu mode:
22 | 1. Computation will be dispatched over the AVAILABLE GPUs.
23 | 2. Synch-BN is activated.
24 | """
25 | if "CC_CLUSTER" in os.environ.keys():
26 | ALLOW_MULTIGPUS = True # CC.
27 | else:
28 | ALLOW_MULTIGPUS = False # others.
29 |
30 | # ALLOW_MULTIGPUS = True
31 | os.environ["ALLOW_MULTIGPUS"] = str(ALLOW_MULTIGPUS)
32 | NBRGPUS = torch.cuda.device_count()
33 | ALLOW_MULTIGPUS = ALLOW_MULTIGPUS and (NBRGPUS > 1)
34 |
35 | return ALLOW_MULTIGPUS
36 |
37 |
38 | def announce_msg(msg, upper=True):
39 | """
40 | Display sa message in the standard output. Something like this:
41 | =================================================================
42 | message
43 | =================================================================
44 |
45 | :param msg: str, text message to display.
46 | :param upper: True/False, if True, the entire message is converted into
47 | uppercase. Else, the message is displayed
48 | as it is.
49 | :return: str, what was printed in the standard output.
50 | """
51 | if upper:
52 | msg = msg.upper()
53 | n = min(120, max(80, len(msg)))
54 | top = "\n" + "=" * n
55 | middle = " " * (int(n / 2) - int(len(msg) / 2)) + " {}".format(msg)
56 | bottom = "=" * n + "\n"
57 |
58 | output_msg = "\n".join([top, middle, bottom])
59 |
60 | print(output_msg)
61 |
62 | return output_msg
63 |
64 |
65 | def get_seed():
66 | """
67 | Get the default seed from the environment variable.
68 | If not set, we use our default seed.
69 | :return: int, a seed.
70 | """
71 | try:
72 | msg = "REQUIRED SEED: {} ".format(os.environ["MYSEED"])
73 | announce_msg(msg)
74 |
75 | return int(os.environ["MYSEED"])
76 | except KeyError:
77 | print("`os.environ` does not have a key named `MYSEED`."
78 | "This key is supposed to hold the current seed. Please set it,"
79 | "and try again, if you want.")
80 |
81 | warnings.warn("MEANWHILE, .... WE ARE GOING TO USE OUR DEFAULT SEED: "
82 | "{}".format(DEFAULT_SEED))
83 | os.environ["MYSEED"] = str(DEFAULT_SEED)
84 | msg = "DEFAULT SEED: {} ".format(os.environ["MYSEED"])
85 | announce_msg(msg)
86 | return DEFAULT_SEED
87 |
88 |
89 | def init_seed(seed=None):
90 | """
91 | * initialize the seed.
92 | * Set a seed to some modules for reproducibility.
93 |
94 | Note:
95 |
96 | While this attempts to ensure reproducibility, it does not offer an
97 | absolute guarantee. The results may be similar to some precision.
98 | Also, they may be different due to an amplification to extremely
99 | small differences.
100 |
101 | See:
102 |
103 | https://pytorch.org/docs/stable/notes/randomness.html
104 | https://stackoverflow.com/questions/50744565/
105 | how-to-handle-non-determinism-when-training-on-a-gpu
106 |
107 | :param seed: int, a seed. Default is None: use the default seed (0).
108 | :return:
109 | """
110 | if seed is None:
111 | seed = get_seed()
112 | else:
113 | os.environ["MYSEED"] = str(seed)
114 | announce_msg("SEED: {} ".format(os.environ["MYSEED"]))
115 |
116 | check_if_allow_multgpu_mode()
117 | reset_seed(seed)
118 |
119 |
120 | def reset_seed(seed, check_cudnn=True):
121 | """
122 | Reset seed to some modules.
123 | :param seed: int. The current seed.
124 | :param check_cudnn: boo. if true, we check if we are in multi-gpu to
125 | disable the cudnn use. `ALLOW_MULTIGPUS` variable has to be already
126 | created in os.environ otherwise an error will be raise.
127 | :return:
128 | """
129 | torch.manual_seed(seed)
130 | np.random.seed(seed)
131 | random.seed(seed)
132 | torch.cuda.manual_seed(seed)
133 | torch.cuda.manual_seed_all(seed)
134 | torch.backends.cudnn.benchmark = False
135 | # Deterministic mode can have a performance impact, depending on your
136 | torch.backends.cudnn.deterministic = True
137 | # model: https://pytorch.org/docs/stable/notes/randomness.html#cudnn
138 | # If multigpu is on, deactivate cudnn since it has many random things
139 | # that we can not control.
140 | if check_cudnn:
141 | cond = torch.cuda.device_count() > 1
142 | cond = cond and (os.environ["ALLOW_MULTIGPUS"] == 'True')
143 | if cond:
144 | torch.backends.cudnn.enabled = False
145 |
146 |
147 | def set_default_seed():
148 | """
149 | Set the default seed.
150 | :return:
151 | """
152 | assert "MYSEED" in os.environ.keys(), "`MYSEED` key is not found in " \
153 | "os.environ.keys() ...." \
154 | "[NOT OK]"
155 | reset_seed(int(os.environ["MYSEED"]))
156 |
157 |
158 | def manual_seed(seed):
159 | r"""Sets the seed for generating random numbers. Returns a
160 | `torch._C.Generator` object.
161 |
162 | NOTE: WE REMOVE MANUAL RESEEDING ALL THE GPUS. At this point,
163 | it is not necessary; and there is not logic/reason
164 | to do it since we want only to reseed the current device.
165 |
166 | Args:
167 | seed (int): The desired seed.
168 | """
169 | return default_generator.manual_seed(int(seed))
170 |
171 |
172 | def force_seed_thread(seed):
173 | """
174 | For seed to some modules.
175 | :param seed:
176 | :return:
177 | """
178 | manual_seed(seed)
179 | np.random.seed(seed)
180 | random.seed(seed)
181 | torch.cuda.manual_seed(seed)
182 | torch.backends.cudnn.enabled = False
183 | # torch.backends.cudnn.enabled = True
184 | # torch.backends.cudnn.enabled = False
185 | # torch.backends.cudnn.benchmark = False
186 | # Deterministic mode can have a performance impact, depending on your
187 | # torch.backends.cudnn.deterministic = True
188 | # model: https://pytorch.org/docs/stable/notes/randomness.html#cudnn
189 |
--------------------------------------------------------------------------------
/results/.readme.md:
--------------------------------------------------------------------------------
1 | contains results.
2 |
--------------------------------------------------------------------------------
/scheduler.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
--------------------------------------------------------------------------------
/shared.py:
--------------------------------------------------------------------------------
1 | # This module shouldn't import any of our modules to avoid recursive importing.
2 | import os
3 | import argparse
4 | import textwrap
5 | import csv
6 | from os.path import join
7 | import fnmatch
8 |
9 |
10 | from sklearn.metrics import auc
11 | import torch
12 | import numpy as np
13 |
14 |
15 | CONST1 = 1000 # used to generate random numbers.
16 |
17 |
18 | def str2bool(v):
19 | """
20 | Read `v`: and returns a boolean value:
21 | True: if `v== "True"`
22 | False: if `v=="False"`
23 | :param v: str.
24 | :return: bool.
25 | """
26 | if isinstance(v, bool):
27 | return v
28 | if isinstance(v, str):
29 | if v == "True":
30 | return True
31 | elif v == "False":
32 | return False
33 | else:
34 | raise ValueError(
35 | "Expected value: 'True'/'False'. found {}.".format(v))
36 | else:
37 | raise argparse.ArgumentTypeError('String boolean value expected: '
38 | '"True"/"Flse"')
39 |
40 |
41 | def announce_msg(msg, upper=True):
42 | """
43 | Display sa message in the standard output. Something like this:
44 | =================================================================
45 | message
46 | =================================================================
47 |
48 | :param msg: str, text message to display.
49 | :param upper: True/False, if True, the entire message is converted into
50 | uppercase. Else, the message is displayed
51 | as it is.
52 | :return: str, what was printed in the standard output.
53 | """
54 | if upper:
55 | msg = msg.upper()
56 | n = min(120, max(80, len(msg)))
57 | top = "\n" + "=" * n
58 | middle = " " * (int(n / 2) - int(len(msg) / 2)) + " {}".format(msg)
59 | bottom = "=" * n + "\n"
60 |
61 | output_msg = "\n".join([top, middle, bottom])
62 |
63 | print(output_msg)
64 |
65 | return output_msg
66 |
67 |
68 | def check_if_allow_multgpu_mode():
69 | """
70 | Check if we can do multigpu.
71 | If yes, allow multigpu.
72 | :return: ALLOW_MULTIGPUS: bool. If True, we enter multigpu mode:
73 | 1. Computation will be dispatched over the AVAILABLE GPUs.
74 | 2. Synch-BN is activated.
75 | """
76 | if "CC_CLUSTER" in os.environ.keys():
77 | ALLOW_MULTIGPUS = True # CC.
78 | else:
79 | ALLOW_MULTIGPUS = False # others.
80 |
81 | # ALLOW_MULTIGPUS = True
82 | os.environ["ALLOW_MULTIGPUS"] = str(ALLOW_MULTIGPUS)
83 | NBRGPUS = torch.cuda.device_count()
84 | ALLOW_MULTIGPUS = ALLOW_MULTIGPUS and (NBRGPUS > 1)
85 |
86 | return ALLOW_MULTIGPUS
87 |
88 |
89 | def check_tensor_inf_nan(tn):
90 | """
91 | Check if a tensor has any inf or nan.
92 | """
93 | if any(torch.isinf(tn.view(-1))):
94 | raise ValueError("Found inf in projection.")
95 | if any(torch.isnan(tn.view(-1))):
96 | raise ValueError("Found nan in projection.")
97 |
98 |
99 | def wrap_command_line(cmd):
100 | """
101 | Wrap command line
102 | :param cmd: str. command line with space as a separator.
103 | :return:
104 | """
105 | return " \\\n".join(textwrap.wrap(
106 | cmd, width=77, break_long_words=False, break_on_hyphens=False))
107 |
108 |
109 | def drop_normal_samples(l_samples):
110 | """
111 | Remove normal samples from the list of samples.
112 |
113 | When to call this?
114 | # drop normal samples and keep metastatic if: 1. dataset=CAM16. 2.
115 | # al_type != AL_WSL.
116 |
117 | :param l_samples: list of samples resulting from csv_loader().
118 | :return: l_samples without any normal sample.
119 | """
120 | return [el for el in l_samples if el[3] == 'tumor']
121 |
122 |
123 | def csv_loader(fname, rootpath, drop_normal=False):
124 | """
125 | Read a *.csv file. Each line contains:
126 | 0. id_: str
127 | 1. img: str
128 | 2. mask: str or '' or None
129 | 3. label: str
130 | 4. tag: int in {0, 1}
131 |
132 | Example: 50162.0, test/img_50162_label_frog.jpeg, , frog, 0
133 |
134 | :param fname: Path to the *.csv file.
135 | :param rootpath: The root path to the folders of the images.
136 | :return: List of elements.
137 | :param drop_normal: bool. if true, normal samples are dropped.
138 | Each element is the path to an image: image path, mask path [optional],
139 | class name.
140 | """
141 | with open(fname, 'r') as f:
142 | out = [
143 | [row[0],
144 | join(rootpath, row[1]),
145 | join(rootpath, row[2]) if row[2] else None,
146 | row[3],
147 | int(row[4])
148 | ]
149 | for row in csv.reader(f)
150 | ]
151 |
152 | if drop_normal:
153 | out = drop_normal_samples(out)
154 |
155 | return out
156 |
157 |
158 | def csv_writer(data, fname):
159 | """
160 | Write a list of rows into a file.
161 | """
162 | msg = "'data' must be a list. found {}".format(type(data))
163 | assert isinstance(data, list), msg
164 |
165 | with open(fname, 'w') as fcsv:
166 | filewriter = csv.writer(
167 | fcsv, delimiter=',', quotechar='|', quoting=csv.QUOTE_MINIMAL)
168 | for row in data:
169 | filewriter.writerow(row)
170 |
171 |
172 | def find_files_pattern(fd_in_, pattern_):
173 | """
174 | Find paths to files with pattern within a folder recursively.
175 | :return:
176 | """
177 | assert os.path.exists(fd_in_), "Folder {} does not exist " \
178 | ".... [NOT OK]".format(fd_in_)
179 | files = []
180 | for r, d, f in os.walk(fd_in_):
181 | for file in f:
182 | if fnmatch.fnmatch(file, pattern_):
183 | files.append(os.path.join(r, file))
184 |
185 | return files
186 |
187 |
188 | def check_nans(tens, msg=''):
189 | """
190 | Check if the tensor 'tens' contains any 'nan' values, and how many.
191 |
192 | :param tens: torch tensor.
193 | :param msg: str. message to display if there is nan.
194 | :return:
195 | """
196 | nbr_nans = torch.isnan(tens).float().sum().item()
197 | if nbr_nans > 0:
198 | print("NAN-CHECK: {}. Found: {} NANs.".format(msg, nbr_nans))
199 |
200 |
201 | def compute_auc(vec, nbr_p):
202 | """
203 | Compute the area under a curve.
204 | :param vec: vector contains values in [0, 100.].
205 | :param nbr_p: int. number of points in the x-axis. it is expected to be
206 | the same as the number of values in `vec`.
207 | :return: float in [0, 100]. percentage of the area from the perfect area.
208 | """
209 | if vec.size == 1:
210 | return float(vec[0])
211 | else:
212 | area_under_c = auc(x=np.array(list(range(vec.size))), y=vec)
213 | area_under_c /= (100. * (nbr_p - 1))
214 | area_under_c *= 100. # (%)
215 | return area_under_c
216 |
217 | # ==============================================================================
218 | # TEST
219 | # ==============================================================================
220 |
221 |
222 | def test_announce_msg():
223 | """
224 | Test announce_msg()
225 | :return:
226 | """
227 | announce_msg("Hello world!!!")
--------------------------------------------------------------------------------
/tmp/.readme.md:
--------------------------------------------------------------------------------
1 | Contains temporary files.
--------------------------------------------------------------------------------
125 |
126 | Note that in the implementation, normalization with N is performed at step (2) and above equation and implementation is not exactly the same, but mathematically is same.
127 |
128 | You can go deeper on above explanation at [Kevin Zakka's Blog](https://kevinzakka.github.io/2016/09/14/batch_normalization/)
--------------------------------------------------------------------------------
/deeplearning/syncbn/__init__.py:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/deeplearning/syncbn/__init__.py
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/__init__.py:
--------------------------------------------------------------------------------
1 | from .syncbn import batchnorm2d_sync
2 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/_csrc.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | Extension module loader
5 |
6 | code referenced from : https://github.com/facebookresearch/maskrcnn-benchmark
7 |
8 | /*****************************************************************************/
9 | """
10 | from __future__ import absolute_import
11 | from __future__ import division
12 | from __future__ import print_function
13 |
14 | import glob
15 | import os.path
16 |
17 | import torch
18 |
19 | try:
20 | from torch.utils.cpp_extension import load
21 | from torch.utils.cpp_extension import CUDA_HOME
22 | except ImportError:
23 | raise ImportError(
24 | "The cpp layer extensions requires PyTorch 0.4 or higher")
25 |
26 |
27 | def _load_C_extensions():
28 | this_dir = os.path.dirname(os.path.abspath(__file__))
29 | this_dir = os.path.join(this_dir, "csrc")
30 |
31 | main_file = glob.glob(os.path.join(this_dir, "*.cpp"))
32 | sources_cpu = glob.glob(os.path.join(this_dir, "cpu", "*.cpp"))
33 | sources_cuda = glob.glob(os.path.join(this_dir, "cuda", "*.cu"))
34 |
35 | sources = main_file + sources_cpu
36 |
37 | extra_cflags = []
38 | extra_cuda_cflags = []
39 | if torch.cuda.is_available() and CUDA_HOME is not None:
40 | sources.extend(sources_cuda)
41 | extra_cflags = ["-O3", "-DWITH_CUDA"]
42 | extra_cuda_cflags = ["--expt-extended-lambda"]
43 | sources = [os.path.join(this_dir, s) for s in sources]
44 | extra_include_paths = [this_dir]
45 | return load(
46 | name="ext_lib",
47 | sources=sources,
48 | extra_cflags=extra_cflags,
49 | extra_include_paths=extra_include_paths,
50 | extra_cuda_cflags=extra_cuda_cflags,
51 | )
52 |
53 |
54 | _backend = _load_C_extensions()
55 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/bn.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | SyncBN
4 |
5 | *****************************************************************************/
6 | #pragma once
7 |
8 | #ifdef WITH_CUDA
9 | #include "cuda/ext_lib.h"
10 | #endif
11 |
12 | /// SyncBN
13 |
14 | std::vector syncbn_sum_sqsum(const at::Tensor& x) {
15 | if (x.is_cuda()) {
16 | #ifdef WITH_CUDA
17 | return syncbn_sum_sqsum_cuda(x);
18 | #else
19 | AT_ERROR("Not compiled with GPU support");
20 | #endif
21 | } else {
22 | AT_ERROR("CPU implementation not supported");
23 | }
24 | }
25 |
26 | at::Tensor syncbn_forward(const at::Tensor& x, const at::Tensor& weight,
27 | const at::Tensor& bias, const at::Tensor& mean,
28 | const at::Tensor& var, bool affine, float eps) {
29 | if (x.is_cuda()) {
30 | #ifdef WITH_CUDA
31 | return syncbn_forward_cuda(x, weight, bias, mean, var, affine, eps);
32 | #else
33 | AT_ERROR("Not compiled with GPU support");
34 | #endif
35 | } else {
36 | AT_ERROR("CPU implementation not supported");
37 | }
38 | }
39 |
40 | std::vector syncbn_backward_xhat(const at::Tensor& dz,
41 | const at::Tensor& x,
42 | const at::Tensor& mean,
43 | const at::Tensor& var, float eps) {
44 | if (dz.is_cuda()) {
45 | #ifdef WITH_CUDA
46 | return syncbn_backward_xhat_cuda(dz, x, mean, var, eps);
47 | #else
48 | AT_ERROR("Not compiled with GPU support");
49 | #endif
50 | } else {
51 | AT_ERROR("CPU implementation not supported");
52 | }
53 | }
54 |
55 | std::vector syncbn_backward(
56 | const at::Tensor& dz, const at::Tensor& x, const at::Tensor& weight,
57 | const at::Tensor& bias, const at::Tensor& mean, const at::Tensor& var,
58 | const at::Tensor& sum_dz, const at::Tensor& sum_dz_xhat, bool affine,
59 | float eps) {
60 | if (dz.is_cuda()) {
61 | #ifdef WITH_CUDA
62 | return syncbn_backward_cuda(dz, x, weight, bias, mean, var, sum_dz,
63 | sum_dz_xhat, affine, eps);
64 | #else
65 | AT_ERROR("Not compiled with GPU support");
66 | #endif
67 | } else {
68 | AT_ERROR("CPU implementation not supported");
69 | }
70 | }
71 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/bn_cuda.cu:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA SyncBN code
4 |
5 | code referenced from : https://github.com/mapillary/inplace_abn
6 |
7 | *****************************************************************************/
8 | #include
9 | #include
10 | #include
11 | #include
12 | #include "cuda/common.h"
13 |
14 | // Utilities
15 | void get_dims(at::Tensor x, int64_t &num, int64_t &chn, int64_t &sp) {
16 | num = x.size(0);
17 | chn = x.size(1);
18 | sp = 1;
19 | for (int64_t i = 2; i < x.ndimension(); ++i) sp *= x.size(i);
20 | }
21 |
22 | /// SyncBN
23 |
24 | template
25 | struct SqSumOp {
26 | __device__ SqSumOp(const T *t, int c, int s) : tensor(t), chn(c), sp(s) {}
27 | __device__ __forceinline__ Pair operator()(int batch, int plane, int n) {
28 | T x = tensor[(batch * chn + plane) * sp + n];
29 | return Pair(x, x * x); // x, x^2
30 | }
31 | const T *tensor;
32 | const int chn;
33 | const int sp;
34 | };
35 |
36 | template
37 | __global__ void syncbn_sum_sqsum_kernel(const T *x, T *sum, T *sqsum,
38 | int num, int chn, int sp) {
39 | int plane = blockIdx.x;
40 | Pair res =
41 | reduce, SqSumOp>(SqSumOp(x, chn, sp), plane, num, chn, sp);
42 | __syncthreads();
43 | if (threadIdx.x == 0) {
44 | sum[plane] = res.v1;
45 | sqsum[plane] = res.v2;
46 | }
47 | }
48 |
49 | std::vector syncbn_sum_sqsum_cuda(const at::Tensor &x) {
50 | CHECK_INPUT(x);
51 |
52 | // Extract dimensions
53 | int64_t num, chn, sp;
54 | get_dims(x, num, chn, sp);
55 |
56 | // Prepare output tensors
57 | auto sum = at::empty({chn}, x.options());
58 | auto sqsum = at::empty({chn}, x.options());
59 |
60 | // Run kernel
61 | dim3 blocks(chn);
62 | dim3 threads(getNumThreads(sp));
63 | AT_DISPATCH_FLOATING_TYPES(
64 | x.type(), "syncbn_sum_sqsum_cuda", ([&] {
65 | syncbn_sum_sqsum_kernel<<>>(
66 | x.data(), sum.data(),
67 | sqsum.data(), num, chn, sp);
68 | }));
69 | return {sum, sqsum};
70 | }
71 |
72 | template
73 | __global__ void syncbn_forward_kernel(T *z, const T *x, const T *weight,
74 | const T *bias, const T *mean,
75 | const T *var, bool affine, float eps,
76 | int num, int chn, int sp) {
77 | int plane = blockIdx.x;
78 | T _mean = mean[plane];
79 | T _var = var[plane];
80 | T _weight = affine ? weight[plane] : T(1);
81 | T _bias = affine ? bias[plane] : T(0);
82 | float _invstd = T(0);
83 | if (_var || eps) {
84 | _invstd = rsqrt(_var + eps);
85 | }
86 | for (int batch = 0; batch < num; ++batch) {
87 | for (int n = threadIdx.x; n < sp; n += blockDim.x) {
88 | T _x = x[(batch * chn + plane) * sp + n];
89 | T _xhat = (_x - _mean) * _invstd;
90 | T _z = _xhat * _weight + _bias;
91 | z[(batch * chn + plane) * sp + n] = _z;
92 | }
93 | }
94 | }
95 |
96 | at::Tensor syncbn_forward_cuda(const at::Tensor &x, const at::Tensor &weight,
97 | const at::Tensor &bias, const at::Tensor &mean,
98 | const at::Tensor &var, bool affine, float eps) {
99 | CHECK_INPUT(x);
100 | CHECK_INPUT(weight);
101 | CHECK_INPUT(bias);
102 | CHECK_INPUT(mean);
103 | CHECK_INPUT(var);
104 |
105 | // Extract dimensions
106 | int64_t num, chn, sp;
107 | get_dims(x, num, chn, sp);
108 |
109 | auto z = at::zeros_like(x);
110 |
111 | // Run kernel
112 | dim3 blocks(chn);
113 | dim3 threads(getNumThreads(sp));
114 | AT_DISPATCH_FLOATING_TYPES(
115 | x.type(), "syncbn_forward_cuda", ([&] {
116 | syncbn_forward_kernel<<>>(
117 | z.data(), x.data(),
118 | weight.data(), bias.data(),
119 | mean.data(), var.data(),
120 | affine, eps, num, chn, sp);
121 | }));
122 | return z;
123 | }
124 |
125 | template
126 | struct XHatOp {
127 | __device__ XHatOp(T _weight, T _bias, const T *_dz, const T *_x, int c, int s)
128 | : weight(_weight), bias(_bias), x(_x), dz(_dz), chn(c), sp(s) {}
129 | __device__ __forceinline__ Pair operator()(int batch, int plane, int n) {
130 | // xhat = (x - bias) * weight
131 | T _xhat = (x[(batch * chn + plane) * sp + n] - bias) * weight;
132 | // dxhat * x_hat
133 | T _dz = dz[(batch * chn + plane) * sp + n];
134 | return Pair(_dz, _dz * _xhat);
135 | }
136 | const T weight;
137 | const T bias;
138 | const T *dz;
139 | const T *x;
140 | const int chn;
141 | const int sp;
142 | };
143 |
144 | template
145 | __global__ void syncbn_backward_xhat_kernel(const T *dz, const T *x,
146 | const T *mean, const T *var,
147 | T *sum_dz, T *sum_dz_xhat,
148 | float eps, int num, int chn,
149 | int sp) {
150 | int plane = blockIdx.x;
151 | T _mean = mean[plane];
152 | T _var = var[plane];
153 | T _invstd = T(0);
154 | if (_var || eps) {
155 | _invstd = rsqrt(_var + eps);
156 | }
157 | Pair res = reduce, XHatOp>(
158 | XHatOp(_invstd, _mean, dz, x, chn, sp), plane, num, chn, sp);
159 | __syncthreads();
160 | if (threadIdx.x == 0) {
161 | // \sum(\frac{dJ}{dy_i})
162 | sum_dz[plane] = res.v1;
163 | // \sum(\frac{dJ}{dy_i}*\hat{x_i})
164 | sum_dz_xhat[plane] = res.v2;
165 | }
166 | }
167 |
168 | std::vector syncbn_backward_xhat_cuda(const at::Tensor &dz,
169 | const at::Tensor &x,
170 | const at::Tensor &mean,
171 | const at::Tensor &var,
172 | float eps) {
173 | CHECK_INPUT(dz);
174 | CHECK_INPUT(x);
175 | CHECK_INPUT(mean);
176 | CHECK_INPUT(var);
177 | // Extract dimensions
178 | int64_t num, chn, sp;
179 | get_dims(x, num, chn, sp);
180 | // Prepare output tensors
181 | auto sum_dz = at::empty({chn}, x.options());
182 | auto sum_dz_xhat = at::empty({chn}, x.options());
183 | // Run kernel
184 | dim3 blocks(chn);
185 | dim3 threads(getNumThreads(sp));
186 | AT_DISPATCH_FLOATING_TYPES(
187 | x.type(), "syncbn_backward_xhat_cuda", ([&] {
188 | syncbn_backward_xhat_kernel<<>>(
189 | dz.data(), x.data(), mean.data(),
190 | var.data(), sum_dz.data(),
191 | sum_dz_xhat.data(), eps, num, chn, sp);
192 | }));
193 | return {sum_dz, sum_dz_xhat};
194 | }
195 |
196 | template
197 | __global__ void syncbn_backward_kernel(const T *dz, const T *x, const T *weight,
198 | const T *bias, const T *mean,
199 | const T *var, const T *sum_dz,
200 | const T *sum_dz_xhat, T *dx, T *dweight,
201 | T *dbias, bool affine, float eps,
202 | int num, int chn, int sp) {
203 | int plane = blockIdx.x;
204 | T _mean = mean[plane];
205 | T _var = var[plane];
206 | T _weight = affine ? weight[plane] : T(1);
207 | T _sum_dz = sum_dz[plane];
208 | T _sum_dz_xhat = sum_dz_xhat[plane];
209 | T _invstd = T(0);
210 | if (_var || eps) {
211 | _invstd = rsqrt(_var + eps);
212 | }
213 | /*
214 | \frac{dJ}{dx_i} = \frac{1}{N\sqrt{(\sigma^2+\epsilon)}} (
215 | N\frac{dJ}{d\hat{x_i}} -
216 | \sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}}) -
217 | \hat{x_i}\sum_{j=1}^{N}(\frac{dJ}{d\hat{x_j}}\hat{x_j})
218 | )
219 | Note : N is omitted here since it will be accumulated and
220 | _sum_dz and _sum_dz_xhat expected to be already normalized
221 | before the call.
222 | */
223 | if (dx) {
224 | T _mul = _weight * _invstd;
225 | for (int batch = 0; batch < num; ++batch) {
226 | for (int n = threadIdx.x; n < sp; n += blockDim.x) {
227 | T _dz = dz[(batch * chn + plane) * sp + n];
228 | T _xhat = (x[(batch * chn + plane) * sp + n] - _mean) * _invstd;
229 | T _dx = (_dz - _sum_dz - _xhat * _sum_dz_xhat) * _mul;
230 | dx[(batch * chn + plane) * sp + n] = _dx;
231 | }
232 | }
233 | }
234 | __syncthreads();
235 | if (threadIdx.x == 0) {
236 | if (affine) {
237 | T _norm = num * sp;
238 | dweight[plane] += _sum_dz_xhat * _norm;
239 | dbias[plane] += _sum_dz * _norm;
240 | }
241 | }
242 | }
243 |
244 | std::vector syncbn_backward_cuda(
245 | const at::Tensor &dz, const at::Tensor &x, const at::Tensor &weight,
246 | const at::Tensor &bias, const at::Tensor &mean, const at::Tensor &var,
247 | const at::Tensor &sum_dz, const at::Tensor &sum_dz_xhat, bool affine,
248 | float eps) {
249 | CHECK_INPUT(dz);
250 | CHECK_INPUT(x);
251 | CHECK_INPUT(weight);
252 | CHECK_INPUT(bias);
253 | CHECK_INPUT(mean);
254 | CHECK_INPUT(var);
255 | CHECK_INPUT(sum_dz);
256 | CHECK_INPUT(sum_dz_xhat);
257 |
258 | // Extract dimensions
259 | int64_t num, chn, sp;
260 | get_dims(x, num, chn, sp);
261 |
262 | // Prepare output tensors
263 | auto dx = at::zeros_like(dz);
264 | auto dweight = at::zeros_like(weight);
265 | auto dbias = at::zeros_like(bias);
266 |
267 | // Run kernel
268 | dim3 blocks(chn);
269 | dim3 threads(getNumThreads(sp));
270 | AT_DISPATCH_FLOATING_TYPES(
271 | x.type(), "syncbn_backward_cuda", ([&] {
272 | syncbn_backward_kernel<<>>(
273 | dz.data(), x.data(), weight.data(),
274 | bias.data(), mean.data(), var.data(),
275 | sum_dz.data(), sum_dz_xhat.data(),
276 | dx.data(), dweight.data(),
277 | dbias.data(), affine, eps, num, chn, sp);
278 | }));
279 | return {dx, dweight, dbias};
280 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/common.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA utility funcs
4 |
5 | code referenced from : https://github.com/mapillary/inplace_abn
6 |
7 | *****************************************************************************/
8 | #pragma once
9 |
10 | #include
11 |
12 | // Checks
13 | #ifndef AT_CHECK
14 | #define AT_CHECK AT_ASSERT
15 | #endif
16 | #define CHECK_CUDA(x) AT_CHECK(x.type().is_cuda(), #x " must be a CUDA tensor")
17 | #define CHECK_CONTIGUOUS(x) AT_CHECK(x.is_contiguous(), #x " must be contiguous")
18 | #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)
19 |
20 | /*
21 | * General settings
22 | */
23 | const int WARP_SIZE = 32;
24 | const int MAX_BLOCK_SIZE = 512;
25 |
26 | template
27 | struct Pair {
28 | T v1, v2;
29 | __device__ Pair() {}
30 | __device__ Pair(T _v1, T _v2) : v1(_v1), v2(_v2) {}
31 | __device__ Pair(T v) : v1(v), v2(v) {}
32 | __device__ Pair(int v) : v1(v), v2(v) {}
33 | __device__ Pair &operator+=(const Pair &a) {
34 | v1 += a.v1;
35 | v2 += a.v2;
36 | return *this;
37 | }
38 | };
39 |
40 | /*
41 | * Utility functions
42 | */
43 | template
44 | __device__ __forceinline__ T WARP_SHFL_XOR(T value, int laneMask,
45 | int width = warpSize,
46 | unsigned int mask = 0xffffffff) {
47 | #if CUDART_VERSION >= 9000
48 | return __shfl_xor_sync(mask, value, laneMask, width);
49 | #else
50 | return __shfl_xor(value, laneMask, width);
51 | #endif
52 | }
53 |
54 | __device__ __forceinline__ int getMSB(int val) { return 31 - __clz(val); }
55 |
56 | static int getNumThreads(int nElem) {
57 | int threadSizes[5] = {32, 64, 128, 256, MAX_BLOCK_SIZE};
58 | for (int i = 0; i != 5; ++i) {
59 | if (nElem <= threadSizes[i]) {
60 | return threadSizes[i];
61 | }
62 | }
63 | return MAX_BLOCK_SIZE;
64 | }
65 |
66 | template
67 | static __device__ __forceinline__ T warpSum(T val) {
68 | #if __CUDA_ARCH__ >= 300
69 | for (int i = 0; i < getMSB(WARP_SIZE); ++i) {
70 | val += WARP_SHFL_XOR(val, 1 << i, WARP_SIZE);
71 | }
72 | #else
73 | __shared__ T values[MAX_BLOCK_SIZE];
74 | values[threadIdx.x] = val;
75 | __threadfence_block();
76 | const int base = (threadIdx.x / WARP_SIZE) * WARP_SIZE;
77 | for (int i = 1; i < WARP_SIZE; i++) {
78 | val += values[base + ((i + threadIdx.x) % WARP_SIZE)];
79 | }
80 | #endif
81 | return val;
82 | }
83 |
84 | template
85 | static __device__ __forceinline__ Pair warpSum(Pair value) {
86 | value.v1 = warpSum(value.v1);
87 | value.v2 = warpSum(value.v2);
88 | return value;
89 | }
90 |
91 | template
92 | __device__ T reduce(Op op, int plane, int N, int C, int S) {
93 | T sum = (T)0;
94 | for (int batch = 0; batch < N; ++batch) {
95 | for (int x = threadIdx.x; x < S; x += blockDim.x) {
96 | sum += op(batch, plane, x);
97 | }
98 | }
99 |
100 | // sum over NumThreads within a warp
101 | sum = warpSum(sum);
102 |
103 | // 'transpose', and reduce within warp again
104 | __shared__ T shared[32];
105 | __syncthreads();
106 | if (threadIdx.x % WARP_SIZE == 0) {
107 | shared[threadIdx.x / WARP_SIZE] = sum;
108 | }
109 | if (threadIdx.x >= blockDim.x / WARP_SIZE && threadIdx.x < WARP_SIZE) {
110 | // zero out the other entries in shared
111 | shared[threadIdx.x] = (T)0;
112 | }
113 | __syncthreads();
114 | if (threadIdx.x / WARP_SIZE == 0) {
115 | sum = warpSum(shared[threadIdx.x]);
116 | if (threadIdx.x == 0) {
117 | shared[0] = sum;
118 | }
119 | }
120 | __syncthreads();
121 |
122 | // Everyone picks it up, should be broadcast into the whole gradInput
123 | return shared[0];
124 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/cuda/ext_lib.h:
--------------------------------------------------------------------------------
1 | /*****************************************************************************
2 |
3 | CUDA SyncBN code
4 |
5 | *****************************************************************************/
6 | #pragma once
7 | #include
8 | #include
9 |
10 | /// Sync-BN
11 | std::vector syncbn_sum_sqsum_cuda(const at::Tensor& x);
12 | at::Tensor syncbn_forward_cuda(const at::Tensor& x, const at::Tensor& weight,
13 | const at::Tensor& bias, const at::Tensor& mean,
14 | const at::Tensor& var, bool affine, float eps);
15 | std::vector syncbn_backward_xhat_cuda(const at::Tensor& dz,
16 | const at::Tensor& x,
17 | const at::Tensor& mean,
18 | const at::Tensor& var,
19 | float eps);
20 | std::vector syncbn_backward_cuda(
21 | const at::Tensor& dz, const at::Tensor& x, const at::Tensor& weight,
22 | const at::Tensor& bias, const at::Tensor& mean, const at::Tensor& var,
23 | const at::Tensor& sum_dz, const at::Tensor& sum_dz_xhat, bool affine,
24 | float eps);
25 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/csrc/ext_lib.cpp:
--------------------------------------------------------------------------------
1 | #include "bn.h"
2 |
3 | PYBIND11_MODULE(TORCH_EXTENSION_NAME, m) {
4 | m.def("syncbn_sum_sqsum", &syncbn_sum_sqsum, "Sum and Sum^2 computation");
5 | m.def("syncbn_forward", &syncbn_forward, "SyncBN forward computation");
6 | m.def("syncbn_backward_xhat", &syncbn_backward_xhat,
7 | "First part of SyncBN backward computation");
8 | m.def("syncbn_backward", &syncbn_backward,
9 | "Second part of SyncBN backward computation");
10 | }
--------------------------------------------------------------------------------
/deeplearning/syncbn/functional/syncbn.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | BatchNorm2dSync with multi-gpu
5 |
6 | code referenced from : https://github.com/mapillary/inplace_abn
7 |
8 | /*****************************************************************************/
9 | """
10 | from __future__ import absolute_import
11 | from __future__ import division
12 | from __future__ import print_function
13 |
14 | import torch.cuda.comm as comm
15 | from torch.autograd import Function
16 | from torch.autograd.function import once_differentiable
17 | from ._csrc import _backend
18 |
19 |
20 | def _count_samples(x):
21 | count = 1
22 | for i, s in enumerate(x.size()):
23 | if i != 1:
24 | count *= s
25 | return count
26 |
27 |
28 | class BatchNorm2dSyncFunc(Function):
29 |
30 | @staticmethod
31 | def forward(ctx, x, weight, bias, running_mean, running_var,
32 | extra, compute_stats=True, momentum=0.1, eps=1e-05):
33 | def _parse_extra(ctx, extra):
34 | ctx.is_master = extra["is_master"]
35 | if ctx.is_master:
36 | ctx.master_queue = extra["master_queue"]
37 | ctx.worker_queues = extra["worker_queues"]
38 | ctx.worker_ids = extra["worker_ids"]
39 | else:
40 | ctx.master_queue = extra["master_queue"]
41 | ctx.worker_queue = extra["worker_queue"]
42 | # Save context
43 | if extra is not None:
44 | _parse_extra(ctx, extra)
45 | ctx.compute_stats = compute_stats
46 | ctx.momentum = momentum
47 | ctx.eps = eps
48 | ctx.affine = weight is not None and bias is not None
49 | if ctx.compute_stats:
50 | N = _count_samples(x) * (ctx.master_queue.maxsize + 1)
51 | assert N > 1
52 | # 1. compute sum(x) and sum(x^2)
53 | xsum, xsqsum = _backend.syncbn_sum_sqsum(x.detach())
54 | if ctx.is_master:
55 | xsums, xsqsums = [xsum], [xsqsum]
56 | # master : gatther all sum(x) and sum(x^2) from slaves
57 | for _ in range(ctx.master_queue.maxsize):
58 | xsum_w, xsqsum_w = ctx.master_queue.get()
59 | ctx.master_queue.task_done()
60 | xsums.append(xsum_w)
61 | xsqsums.append(xsqsum_w)
62 | xsum = comm.reduce_add(xsums)
63 | xsqsum = comm.reduce_add(xsqsums)
64 | mean = xsum / N
65 | sumvar = xsqsum - xsum * mean
66 | var = sumvar / N
67 | uvar = sumvar / (N - 1)
68 | # master : broadcast global mean, variance to all slaves
69 | tensors = comm.broadcast_coalesced(
70 | (mean, uvar, var), [mean.get_device()] + ctx.worker_ids)
71 | for ts, queue in zip(tensors[1:], ctx.worker_queues):
72 | queue.put(ts)
73 | else:
74 | # slave : send sum(x) and sum(x^2) to master
75 | ctx.master_queue.put((xsum, xsqsum))
76 | # slave : get global mean and variance
77 | mean, uvar, var = ctx.worker_queue.get()
78 | ctx.worker_queue.task_done()
79 |
80 | # Update running stats
81 | running_mean.mul_((1 - ctx.momentum)).add_(ctx.momentum * mean)
82 | running_var.mul_((1 - ctx.momentum)).add_(ctx.momentum * uvar)
83 | ctx.N = N
84 | ctx.save_for_backward(x, weight, bias, mean, var)
85 | else:
86 | mean, var = running_mean, running_var
87 |
88 | # do batch norm forward
89 | z = _backend.syncbn_forward(x, weight, bias, mean, var,
90 | ctx.affine, ctx.eps)
91 | return z
92 |
93 | @staticmethod
94 | @once_differentiable
95 | def backward(ctx, dz):
96 | x, weight, bias, mean, var = ctx.saved_tensors
97 | dz = dz.contiguous()
98 |
99 | # 1. compute \sum(\frac{dJ}{dy_i}) and \sum(\frac{dJ}{dy_i}*\hat{x_i})
100 | sum_dz, sum_dz_xhat = _backend.syncbn_backward_xhat(
101 | dz, x, mean, var, ctx.eps)
102 | if ctx.is_master:
103 | sum_dzs, sum_dz_xhats = [sum_dz], [sum_dz_xhat]
104 | # master : gatther from slaves
105 | for _ in range(ctx.master_queue.maxsize):
106 | sum_dz_w, sum_dz_xhat_w = ctx.master_queue.get()
107 | ctx.master_queue.task_done()
108 | sum_dzs.append(sum_dz_w)
109 | sum_dz_xhats.append(sum_dz_xhat_w)
110 | # master : compute global stats
111 | sum_dz = comm.reduce_add(sum_dzs)
112 | sum_dz_xhat = comm.reduce_add(sum_dz_xhats)
113 | sum_dz /= ctx.N
114 | sum_dz_xhat /= ctx.N
115 | # master : broadcast global stats
116 | tensors = comm.broadcast_coalesced(
117 | (sum_dz, sum_dz_xhat), [mean.get_device()] + ctx.worker_ids)
118 | for ts, queue in zip(tensors[1:], ctx.worker_queues):
119 | queue.put(ts)
120 | else:
121 | # slave : send to master
122 | ctx.master_queue.put((sum_dz, sum_dz_xhat))
123 | # slave : get global stats
124 | sum_dz, sum_dz_xhat = ctx.worker_queue.get()
125 | ctx.worker_queue.task_done()
126 |
127 | # do batch norm backward
128 | dx, dweight, dbias = _backend.syncbn_backward(
129 | dz, x, weight, bias, mean, var, sum_dz, sum_dz_xhat,
130 | ctx.affine, ctx.eps)
131 |
132 | return dx, dweight, dbias, \
133 | None, None, None, None, None, None
134 |
135 | batchnorm2d_sync = BatchNorm2dSyncFunc.apply
136 |
137 | __all__ = ["batchnorm2d_sync"]
138 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/nn/__init__.py:
--------------------------------------------------------------------------------
1 | from .syncbn import *
2 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/nn/syncbn.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | BatchNorm2dSync with multi-gpu
5 |
6 | /*****************************************************************************/
7 | """
8 | from __future__ import absolute_import
9 | from __future__ import division
10 | from __future__ import print_function
11 |
12 | try:
13 | # python 3
14 | from queue import Queue
15 | except ImportError:
16 | # python 2
17 | from Queue import Queue
18 |
19 | import torch
20 | import torch.nn as nn
21 | from torch.nn import functional as F
22 | from torch.nn.parameter import Parameter
23 | import sys
24 | sys.path.append("..")
25 | from deeplearning.syncbn.functional import batchnorm2d_sync
26 |
27 |
28 | class _BatchNorm(nn.Module):
29 | """
30 | Customized BatchNorm from nn.BatchNorm
31 | >> added freeze attribute to enable bn freeze.
32 | """
33 |
34 | def __init__(self, num_features, eps=1e-5, momentum=0.1, affine=True,
35 | track_running_stats=True):
36 | super(_BatchNorm, self).__init__()
37 | self.num_features = num_features
38 | self.eps = eps
39 | self.momentum = momentum
40 | self.affine = affine
41 | self.track_running_stats = track_running_stats
42 | self.freezed = False
43 | if self.affine:
44 | self.weight = Parameter(torch.Tensor(num_features))
45 | self.bias = Parameter(torch.Tensor(num_features))
46 | else:
47 | self.register_parameter('weight', None)
48 | self.register_parameter('bias', None)
49 | if self.track_running_stats:
50 | self.register_buffer('running_mean', torch.zeros(num_features))
51 | self.register_buffer('running_var', torch.ones(num_features))
52 | else:
53 | self.register_parameter('running_mean', None)
54 | self.register_parameter('running_var', None)
55 | self.reset_parameters()
56 |
57 | def reset_parameters(self):
58 | if self.track_running_stats:
59 | self.running_mean.zero_()
60 | self.running_var.fill_(1)
61 | if self.affine:
62 | self.weight.data.uniform_()
63 | self.bias.data.zero_()
64 |
65 | def _check_input_dim(self, input):
66 | return NotImplemented
67 |
68 | def forward(self, input):
69 | self._check_input_dim(input)
70 |
71 | compute_stats = not self.freezed and \
72 | self.training and self.track_running_stats
73 |
74 | ret = F.batch_norm(input, self.running_mean, self.running_var,
75 | self.weight, self.bias, compute_stats,
76 | self.momentum, self.eps)
77 | return ret
78 |
79 | def extra_repr(self):
80 | return '{num_features}, eps={eps}, momentum={momentum}, '\
81 | 'affine={affine}, ' \
82 | 'track_running_stats={track_running_stats}'.format(
83 | **self.__dict__)
84 |
85 |
86 | class BatchNorm2dNoSync(_BatchNorm):
87 | """
88 | Equivalent to nn.BatchNorm2d
89 | """
90 |
91 | def _check_input_dim(self, input):
92 | if input.dim() != 4:
93 | raise ValueError('expected 4D input (got {}D input)'
94 | .format(input.dim()))
95 |
96 |
97 | class BatchNorm2dSync(BatchNorm2dNoSync):
98 | """
99 | BatchNorm2d with automatic multi-GPU Sync
100 | """
101 |
102 | def __init__(self, num_features, eps=1e-5, momentum=0.1, affine=True,
103 | track_running_stats=True):
104 | super(BatchNorm2dSync, self).__init__(
105 | num_features, eps=eps, momentum=momentum, affine=affine,
106 | track_running_stats=track_running_stats)
107 | self.sync_enabled = True
108 | self.devices = list(range(torch.cuda.device_count()))
109 | if len(self.devices) > 1:
110 | # Initialize queues
111 | self.worker_ids = self.devices[1:]
112 | self.master_queue = Queue(len(self.worker_ids))
113 | self.worker_queues = [Queue(1) for _ in self.worker_ids]
114 |
115 | def forward(self, x):
116 | compute_stats = not self.freezed and \
117 | self.training and self.track_running_stats
118 | if self.sync_enabled and compute_stats and len(self.devices) > 1:
119 | if x.get_device() == self.devices[0]:
120 | # Master mode
121 | extra = {
122 | "is_master": True,
123 | "master_queue": self.master_queue,
124 | "worker_queues": self.worker_queues,
125 | "worker_ids": self.worker_ids
126 | }
127 | else:
128 | # Worker mode
129 | extra = {
130 | "is_master": False,
131 | "master_queue": self.master_queue,
132 | "worker_queue": self.worker_queues[
133 | self.worker_ids.index(x.get_device())]
134 | }
135 | return batchnorm2d_sync(x, self.weight, self.bias,
136 | self.running_mean, self.running_var,
137 | extra, compute_stats, self.momentum,
138 | self.eps)
139 | return super(BatchNorm2dSync, self).forward(x)
140 |
141 | def __repr__(self):
142 | """repr"""
143 | rep = '{name}({num_features}, eps={eps}, momentum={momentum},' \
144 | 'affine={affine}, ' \
145 | 'track_running_stats={track_running_stats},' \
146 | 'devices={devices})'
147 | return rep.format(name=self.__class__.__name__, **self.__dict__)
148 |
149 | #BatchNorm2d = BatchNorm2dNoSync
150 | BatchNorm2d = BatchNorm2dSync
151 |
--------------------------------------------------------------------------------
/deeplearning/syncbn/requirements.txt:
--------------------------------------------------------------------------------
1 | future
2 | cffi
3 | ninja
--------------------------------------------------------------------------------
/deeplearning/syncbn/test.py:
--------------------------------------------------------------------------------
1 | """
2 | /*****************************************************************************/
3 |
4 | Test for BatchNorm2dSync with multi-gpu
5 |
6 | /*****************************************************************************/
7 | """
8 | from __future__ import absolute_import
9 | from __future__ import division
10 | from __future__ import print_function
11 |
12 | import sys
13 | import numpy as np
14 | import torch
15 | from torch import nn
16 | from torch.nn import functional as F
17 | sys.path.append("./")
18 | import nn as NN
19 |
20 | torch.backends.cudnn.deterministic = True
21 |
22 |
23 | def init_weight(model):
24 | for m in model.modules():
25 | if isinstance(m, nn.Conv2d):
26 | n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels
27 | m.weight.data.normal_(0, np.sqrt(2. / n))
28 | elif isinstance(m, NN.BatchNorm2d) or isinstance(m, nn.BatchNorm2d):
29 | m.weight.data.fill_(1)
30 | m.bias.data.zero_()
31 | elif isinstance(m, nn.Linear):
32 | m.bias.data.zero_()
33 |
34 | num_gpu = torch.cuda.device_count()
35 | print("num_gpu={}".format(num_gpu))
36 | if num_gpu < 2:
37 | print("No multi-gpu found. NN.BatchNorm2d will act as normal nn.BatchNorm2d")
38 |
39 | m1 = nn.Sequential(
40 | nn.Conv2d(3, 3, 1, 1, bias=False),
41 | nn.BatchNorm2d(3),
42 | nn.ReLU(inplace=True),
43 | nn.Conv2d(3, 3, 1, 1, bias=False),
44 | nn.BatchNorm2d(3),
45 | ).cuda()
46 | torch.manual_seed(123)
47 | init_weight(m1)
48 | m2 = nn.Sequential(
49 | nn.Conv2d(3, 3, 1, 1, bias=False),
50 | NN.BatchNorm2d(3),
51 | nn.ReLU(inplace=True),
52 | nn.Conv2d(3, 3, 1, 1, bias=False),
53 | NN.BatchNorm2d(3),
54 | ).cuda()
55 | torch.manual_seed(123)
56 | init_weight(m2)
57 | m2 = nn.DataParallel(m2, device_ids=range(num_gpu))
58 | o1 = torch.optim.SGD(m1.parameters(), 1e-3)
59 | o2 = torch.optim.SGD(m2.parameters(), 1e-3)
60 | y = torch.ones(num_gpu).float().cuda()
61 | torch.manual_seed(123)
62 | for _ in range(100):
63 | x = torch.rand(num_gpu, 3, 2, 2).cuda()
64 | o1.zero_grad()
65 | z1 = m1(x)
66 | l1 = F.mse_loss(z1.mean(-1).mean(-1).mean(-1), y)
67 | l1.backward()
68 | o1.step()
69 | o2.zero_grad()
70 | z2 = m2(x)
71 | l2 = F.mse_loss(z2.mean(-1).mean(-1).mean(-1), y)
72 | l2.backward()
73 | o2.step()
74 | print(m2.module[1].bias.grad - m1[1].bias.grad)
75 | print(m2.module[1].weight.grad - m1[1].weight.grad)
76 | print(m2.module[-1].bias.grad - m1[-1].bias.grad)
77 | print(m2.module[-1].weight.grad - m1[-1].weight.grad)
78 | m2 = m2.module
79 | print("===============================")
80 | print("m1(nn.BatchNorm2d) running_mean",
81 | m1[1].running_mean, m1[-1].running_mean)
82 | print("m2(NN.BatchNorm2d) running_mean",
83 | m2[1].running_mean, m2[-1].running_mean)
84 | print("m1(nn.BatchNorm2d) running_var", m1[1].running_var, m1[-1].running_var)
85 | print("m2(NN.BatchNorm2d) running_var", m2[1].running_var, m2[-1].running_var)
86 | print("m1(nn.BatchNorm2d) weight", m1[1].weight, m1[-1].weight)
87 | print("m2(NN.BatchNorm2d) weight", m2[1].weight, m2[-1].weight)
88 | print("m1(nn.BatchNorm2d) bias", m1[1].bias, m1[-1].bias)
89 | print("m2(NN.BatchNorm2d) bias", m2[1].bias, m2[-1].bias)
90 |
--------------------------------------------------------------------------------
/deeplearning/utils.py:
--------------------------------------------------------------------------------
1 | import sys
2 | import os
3 |
4 |
5 | import torch
6 | import torch.nn as nn
7 | import numpy as np
8 | import torch.nn.functional as F
9 | from collections import OrderedDict
10 |
11 |
12 | sys.path.append("..")
13 |
14 | from shared import check_if_allow_multgpu_mode, announce_msg
15 |
16 | ACTIVATE_SYNC_BN = False
17 | # Override ACTIVATE_SYNC_BN using variable environment in Bash:
18 | # $ export ACTIVATE_SYNC_BN="True" ----> Activate
19 | # $ export ACTIVATE_SYNC_BN="False" ----> Deactivate
20 |
21 | if "ACTIVATE_SYNC_BN" in os.environ.keys():
22 | ACTIVATE_SYNC_BN = (os.environ['ACTIVATE_SYNC_BN'] == "True")
23 |
24 | announce_msg("ACTIVATE_SYNC_BN was set to {}".format(ACTIVATE_SYNC_BN))
25 |
26 | if check_if_allow_multgpu_mode() and ACTIVATE_SYNC_BN: # Activate Synch-BN.
27 | from deeplearning.syncbn import nn as NN_Sync_BN
28 | BatchNorm2d = NN_Sync_BN.BatchNorm2d
29 | announce_msg("Synchronized BN has been activated. \n"
30 | "MultiGPU mode has been activated. "
31 | "{} GPUs".format(torch.cuda.device_count()))
32 | else:
33 | BatchNorm2d = nn.BatchNorm2d
34 | if check_if_allow_multgpu_mode():
35 | announce_msg("Synchronized BN has been deactivated.\n"
36 | "MultiGPU mode has been activated. "
37 | "{} GPUs".format(torch.cuda.device_count()))
38 | else:
39 | announce_msg("Synchronized BN has been deactivated.\n"
40 | "MultiGPU mode has been deactivated. "
41 | "{} GPUs".format(torch.cuda.device_count()))
42 |
43 |
44 | def initialize_weights(*models):
45 | for model in models:
46 | for m in model.modules():
47 | if isinstance(m, nn.Conv2d):
48 | nn.init.kaiming_normal_(m.weight.data, nonlinearity='relu')
49 | elif isinstance(m, BatchNorm2d):
50 | m.weight.data.fill_(1.)
51 | m.bias.data.fill_(1e-4)
52 | elif isinstance(m, nn.Linear):
53 | m.weight.data.normal_(0.0, 0.0001)
54 | m.bias.data.zero_()
55 |
56 | class _SimpleSegmentationModel(nn.Module):
57 | def __init__(self, backbone, classifier):
58 | super(_SimpleSegmentationModel, self).__init__()
59 | self.backbone = backbone
60 | self.classifier = classifier
61 |
62 | def forward(self, x):
63 | input_shape = x.shape[-2:]
64 | features = self.backbone(x)
65 | x = self.classifier(features)
66 | x = F.interpolate(x, size=input_shape, mode='bilinear',
67 | align_corners=False)
68 | return x
69 |
70 |
71 | class IntermediateLayerGetter(nn.ModuleDict):
72 | """
73 | Module wrapper that returns intermediate layers from a model
74 |
75 | It has a strong assumption that the modules have been registered
76 | into the model in the same order as they are used.
77 | This means that one should **not** reuse the same nn.Module
78 | twice in the forward if you want this to work.
79 |
80 | Additionally, it is only able to query submodules that are directly
81 | assigned to the model. So if `model` is passed, `model.feature1` can
82 | be returned, but not `model.feature1.layer2`.
83 |
84 | Arguments:
85 | model (nn.Module): model on which we will extract the features
86 | return_layers (Dict[name, new_name]): a dict containing the names
87 | of the modules for which the activations will be returned as
88 | the key of the dict, and the value of the dict is the name
89 | of the returned activation (which the user can specify).
90 |
91 | Examples::
92 |
93 | >>> m = torchvision.models.resnet18(pretrained=True)
94 | >>> # extract layer1 and layer3, giving as names `feat1` and feat2`
95 | >>> new_m = torchvision.models._utils.IntermediateLayerGetter(m,
96 | >>> {'layer1': 'feat1', 'layer3': 'feat2'})
97 | >>> out = new_m(torch.rand(1, 3, 224, 224))
98 | >>> print([(k, v.shape) for k, v in out.items()])
99 | >>> [('feat1', torch.Size([1, 64, 56, 56])),
100 | >>> ('feat2', torch.Size([1, 256, 14, 14]))]
101 | """
102 |
103 | def __init__(self, model, return_layers):
104 | if not set(return_layers).issubset(
105 | [name for name, _ in model.named_children()]):
106 | raise ValueError("return_layers are not present in model")
107 |
108 | orig_return_layers = return_layers
109 | return_layers = {k: v for k, v in return_layers.items()}
110 | layers = OrderedDict()
111 | for name, module in model.named_children():
112 | layers[name] = module
113 | if name in return_layers:
114 | del return_layers[name]
115 | if not return_layers:
116 | break
117 |
118 | super(IntermediateLayerGetter, self).__init__(layers)
119 | self.return_layers = orig_return_layers
120 |
121 | def forward(self, x):
122 | out = OrderedDict()
123 | for name, module in self.named_children():
124 | x = module(x)
125 | if name in self.return_layers:
126 | out_name = self.return_layers[name]
127 | out[out_name] = x
128 | return out
129 |
--------------------------------------------------------------------------------
/deeplearning/wildcat.py:
--------------------------------------------------------------------------------
1 | import sys
2 |
3 |
4 | import torch.nn as nn
5 |
6 | sys.path.append("..")
7 |
8 | from deeplearning.decision_pooling import WildCatPoolDecision, ClassWisePooling
9 |
10 |
11 | class WildCatClassifierHead(nn.Module):
12 | """
13 | A WILDCAT type classifier head.
14 | `WILDCAT: Weakly Supervised Learning of Deep ConvNets for
15 | Image Classification, Pointwise Localization and Segmentation`,
16 | Thibaut Durand, Taylor Mordan, Nicolas Thome, Matthieu Cord.
17 | """
18 | def __init__(self, inplans, modalities, num_classes, kmax=0.5, kmin=None,
19 | alpha=0.6, dropout=0.0):
20 | super(WildCatClassifierHead, self).__init__()
21 | self.name = "wildcat"
22 |
23 | self.num_classes = num_classes
24 |
25 | self.to_modalities = nn.Conv2d(
26 | inplans, num_classes * modalities, kernel_size=1, bias=True)
27 | self.to_maps = ClassWisePooling(num_classes, modalities)
28 | self.wildcat = WildCatPoolDecision(
29 | kmax=kmax, kmin=kmin, alpha=alpha, dropout=dropout)
30 |
31 | def forward(self, x, seed=None, prngs_cuda=None):
32 | """
33 | The forward function.
34 | :param x: input tensor.
35 | :param seed:
36 | :param prngs_cuda:
37 | :return: scores, maps.
38 | """
39 | modalities = self.to_modalities(x)
40 | maps = self.to_maps(modalities)
41 | scores = self.wildcat(x=maps, seed=seed, prngs_cuda=prngs_cuda)
42 |
43 | return scores, maps
44 |
45 | def get_nbr_params(self):
46 | """
47 | Compute the number of parameters of the model.
48 | :return:
49 | """
50 | return sum([p.numel() for p in self.parameters()])
51 |
52 | def __str__(self):
53 | return "{}: WILDCAT.".format(self.name)
54 |
55 | def __repr__(self):
56 | return super(WildCatClassifierHead, self).__repr__()
57 |
58 |
59 | if __name__ == "__main__":
60 | inst = WildCatClassifierHead(inplans=10, modalities=5, num_classes=2)
61 | print(repr(inst))
62 | print(inst)
--------------------------------------------------------------------------------
/dependencies/.readme.md:
--------------------------------------------------------------------------------
1 | Contains files about the dependencies.
--------------------------------------------------------------------------------
/dependencies/get_dependencies.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
2 | # =============================================================================================
3 | # PURE PYTHON 3.7.0 VIRTUAL ENVIRONMENT
4 | # ==============================================================================================
5 |
6 | # virtualenvwrapper commands are not recognized WITHIN bash files. Not sure why.
7 | # Need to source virtualenvwrapper.sh in order to be recognized.
8 | # First: create a bash variable pointing to the path of virtualenvwrapper.sh
9 | # name it: export VIRTUALENVWRAPPER_SH=/usr/local/bin/virtualenvwrapper.sh
10 |
11 | # You can omit this line if you do not use virtual environment wrapper. You can create your virtual environment
12 | # using pip and install the requirements.
13 | source $VIRTUALENVWRAPPER_SH
14 |
15 | # You can ignore this line as well. (same reason)
16 | workon pytorch.1.4.0
17 | pip freeze > requirements.txt
18 |
19 | # Create a new V.E using virtualenvwrapper
20 | # mkvirtualenv -r requirements.txt pytorch.1.2.0-dev
21 | # mkvirtualenv -p $HOME/anaconda3/bin/python3 -r requirements.txt pytorch.1.2.0-dev
22 |
23 | # Create a new V.E using virtualenv
24 | # virtualenv ~/Venvs/pytorch.1.0.1
25 |
26 | # Install requirements. Up to you for --no-index
27 | # pip install --no-index -r requirements.txt
28 |
--------------------------------------------------------------------------------
/dependencies/requirements.txt:
--------------------------------------------------------------------------------
1 | attrs==19.1.0
2 | backcall==0.1.0
3 | bleach==2.1.3
4 | compress-pickle==1.1.0
5 | cycler==0.10.0
6 | Cython==0.29.2
7 | decorator==4.3.2
8 | entrypoints==0.2.3
9 | future==0.16.0
10 | html5lib==1.0.1
11 | imageio==2.4.1
12 | ipykernel==4.8.2
13 | ipython==6.5.0
14 | ipython-genutils==0.2.0
15 | ipywidgets==7.4.2
16 | jedi==0.12.1
17 | Jinja2==2.10
18 | jsonschema==2.6.0
19 | jupyter==1.0.0
20 | jupyter-client==5.2.4
21 | jupyter-console==5.2.0
22 | jupyter-core==4.4.0
23 | kiwisolver==1.0.1
24 | MarkupSafe==1.1.1
25 | matplotlib==3.0.2
26 | mistune==0.8.4
27 | nbconvert==5.3.1
28 | nbformat==4.4.0
29 | notebook==5.7.4
30 | numpy==1.16.2
31 | opencv-python==3.4.4.19
32 | pandocfilters==1.4.2
33 | parso==0.3.1
34 | pexpect==4.6.0
35 | pickleshare==0.7.5
36 | Pillow==7.1.1
37 | prometheus-client==0.3.1
38 | prompt-toolkit==1.0.15
39 | protobuf==3.7.1
40 | ptyprocess==0.6.0
41 | pygifsicle==1.0.1
42 | Pygments==2.3.1
43 | pyparsing==2.3.1
44 | python-dateutil==2.8.0
45 | PyYAML==3.13
46 | pyzmq==17.1.2
47 | qtconsole==4.3.1
48 | scikit-learn==0.20.2
49 | scipy==1.2.1
50 | Send2Trash==1.5.0
51 | simplegeneric==0.8.1
52 | six==1.12.0
53 | terminado==0.8.1
54 | testpath==0.3.1
55 | texttable==1.6.2
56 | torch==1.4.0+cu100
57 | torchvision==0.5.0+cu100
58 | tornado==5.1.1
59 | tqdm==4.31.1
60 | traitlets==4.3.2
61 | typing==3.6.6
62 | wcwidth==0.1.7
63 | webencodings==0.5.1
64 | widgetsnbextension==3.4.2
65 |
66 |
--------------------------------------------------------------------------------
/doc/.readme.md:
--------------------------------------------------------------------------------
1 | idea?
--------------------------------------------------------------------------------
/doc/algo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/algo.png
--------------------------------------------------------------------------------
/doc/arch.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/arch.png
--------------------------------------------------------------------------------
/doc/knn.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/knn.png
--------------------------------------------------------------------------------
/doc/perf.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/perf.png
--------------------------------------------------------------------------------
/doc/proposal.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/proposal.png
--------------------------------------------------------------------------------
/doc/pseudo-labeling.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/pseudo-labeling.png
--------------------------------------------------------------------------------
/doc/results.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/results.png
--------------------------------------------------------------------------------
/doc/wacv2021-active-learning-weak-annotator.pdf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/doc/wacv2021-active-learning-weak-annotator.pdf
--------------------------------------------------------------------------------
/exps/.readme.md:
--------------------------------------------------------------------------------
1 | Contains exps results.
--------------------------------------------------------------------------------
/folds/.readme.md:
--------------------------------------------------------------------------------
1 | contains folds.
2 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/log-stats-ds-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.txt:
--------------------------------------------------------------------------------
1 | min h 120, max h 500
2 | min w 121, max w 500
3 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/size-stats-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/folds/Caltech-UCSD-Birds-200-2011/size-stats-ds-Caltech-UCSD-Birds-200-2011-s-0-f-0-subset-train.png
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/.~lock.train_s_0_f_0.csv#:
--------------------------------------------------------------------------------
1 | ,brian,dell,16.09.2020 19:52,file:///home/brian/.config/libreoffice/4;
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_0/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_1/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_2/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_3/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/encoding.yaml:
--------------------------------------------------------------------------------
1 | {001.Black_footed_Albatross: 0, 002.Laysan_Albatross: 1, 003.Sooty_Albatross: 2, 004.Groove_billed_Ani: 3,
2 | 005.Crested_Auklet: 4, 006.Least_Auklet: 5, 007.Parakeet_Auklet: 6, 008.Rhinoceros_Auklet: 7,
3 | 009.Brewer_Blackbird: 8, 010.Red_winged_Blackbird: 9, 011.Rusty_Blackbird: 10, 012.Yellow_headed_Blackbird: 11,
4 | 013.Bobolink: 12, 014.Indigo_Bunting: 13, 015.Lazuli_Bunting: 14, 016.Painted_Bunting: 15,
5 | 017.Cardinal: 16, 018.Spotted_Catbird: 17, 019.Gray_Catbird: 18, 020.Yellow_breasted_Chat: 19,
6 | 021.Eastern_Towhee: 20, 022.Chuck_will_Widow: 21, 023.Brandt_Cormorant: 22, 024.Red_faced_Cormorant: 23,
7 | 025.Pelagic_Cormorant: 24, 026.Bronzed_Cowbird: 25, 027.Shiny_Cowbird: 26, 028.Brown_Creeper: 27,
8 | 029.American_Crow: 28, 030.Fish_Crow: 29, 031.Black_billed_Cuckoo: 30, 032.Mangrove_Cuckoo: 31,
9 | 033.Yellow_billed_Cuckoo: 32, 034.Gray_crowned_Rosy_Finch: 33, 035.Purple_Finch: 34,
10 | 036.Northern_Flicker: 35, 037.Acadian_Flycatcher: 36, 038.Great_Crested_Flycatcher: 37,
11 | 039.Least_Flycatcher: 38, 040.Olive_sided_Flycatcher: 39, 041.Scissor_tailed_Flycatcher: 40,
12 | 042.Vermilion_Flycatcher: 41, 043.Yellow_bellied_Flycatcher: 42, 044.Frigatebird: 43,
13 | 045.Northern_Fulmar: 44, 046.Gadwall: 45, 047.American_Goldfinch: 46, 048.European_Goldfinch: 47,
14 | 049.Boat_tailed_Grackle: 48, 050.Eared_Grebe: 49, 051.Horned_Grebe: 50, 052.Pied_billed_Grebe: 51,
15 | 053.Western_Grebe: 52, 054.Blue_Grosbeak: 53, 055.Evening_Grosbeak: 54, 056.Pine_Grosbeak: 55,
16 | 057.Rose_breasted_Grosbeak: 56, 058.Pigeon_Guillemot: 57, 059.California_Gull: 58,
17 | 060.Glaucous_winged_Gull: 59, 061.Heermann_Gull: 60, 062.Herring_Gull: 61, 063.Ivory_Gull: 62,
18 | 064.Ring_billed_Gull: 63, 065.Slaty_backed_Gull: 64, 066.Western_Gull: 65, 067.Anna_Hummingbird: 66,
19 | 068.Ruby_throated_Hummingbird: 67, 069.Rufous_Hummingbird: 68, 070.Green_Violetear: 69,
20 | 071.Long_tailed_Jaeger: 70, 072.Pomarine_Jaeger: 71, 073.Blue_Jay: 72, 074.Florida_Jay: 73,
21 | 075.Green_Jay: 74, 076.Dark_eyed_Junco: 75, 077.Tropical_Kingbird: 76, 078.Gray_Kingbird: 77,
22 | 079.Belted_Kingfisher: 78, 080.Green_Kingfisher: 79, 081.Pied_Kingfisher: 80, 082.Ringed_Kingfisher: 81,
23 | 083.White_breasted_Kingfisher: 82, 084.Red_legged_Kittiwake: 83, 085.Horned_Lark: 84,
24 | 086.Pacific_Loon: 85, 087.Mallard: 86, 088.Western_Meadowlark: 87, 089.Hooded_Merganser: 88,
25 | 090.Red_breasted_Merganser: 89, 091.Mockingbird: 90, 092.Nighthawk: 91, 093.Clark_Nutcracker: 92,
26 | 094.White_breasted_Nuthatch: 93, 095.Baltimore_Oriole: 94, 096.Hooded_Oriole: 95,
27 | 097.Orchard_Oriole: 96, 098.Scott_Oriole: 97, 099.Ovenbird: 98, 100.Brown_Pelican: 99,
28 | 101.White_Pelican: 100, 102.Western_Wood_Pewee: 101, 103.Sayornis: 102, 104.American_Pipit: 103,
29 | 105.Whip_poor_Will: 104, 106.Horned_Puffin: 105, 107.Common_Raven: 106, 108.White_necked_Raven: 107,
30 | 109.American_Redstart: 108, 110.Geococcyx: 109, 111.Loggerhead_Shrike: 110, 112.Great_Grey_Shrike: 111,
31 | 113.Baird_Sparrow: 112, 114.Black_throated_Sparrow: 113, 115.Brewer_Sparrow: 114,
32 | 116.Chipping_Sparrow: 115, 117.Clay_colored_Sparrow: 116, 118.House_Sparrow: 117,
33 | 119.Field_Sparrow: 118, 120.Fox_Sparrow: 119, 121.Grasshopper_Sparrow: 120, 122.Harris_Sparrow: 121,
34 | 123.Henslow_Sparrow: 122, 124.Le_Conte_Sparrow: 123, 125.Lincoln_Sparrow: 124, 126.Nelson_Sharp_tailed_Sparrow: 125,
35 | 127.Savannah_Sparrow: 126, 128.Seaside_Sparrow: 127, 129.Song_Sparrow: 128, 130.Tree_Sparrow: 129,
36 | 131.Vesper_Sparrow: 130, 132.White_crowned_Sparrow: 131, 133.White_throated_Sparrow: 132,
37 | 134.Cape_Glossy_Starling: 133, 135.Bank_Swallow: 134, 136.Barn_Swallow: 135, 137.Cliff_Swallow: 136,
38 | 138.Tree_Swallow: 137, 139.Scarlet_Tanager: 138, 140.Summer_Tanager: 139, 141.Artic_Tern: 140,
39 | 142.Black_Tern: 141, 143.Caspian_Tern: 142, 144.Common_Tern: 143, 145.Elegant_Tern: 144,
40 | 146.Forsters_Tern: 145, 147.Least_Tern: 146, 148.Green_tailed_Towhee: 147, 149.Brown_Thrasher: 148,
41 | 150.Sage_Thrasher: 149, 151.Black_capped_Vireo: 150, 152.Blue_headed_Vireo: 151,
42 | 153.Philadelphia_Vireo: 152, 154.Red_eyed_Vireo: 153, 155.Warbling_Vireo: 154, 156.White_eyed_Vireo: 155,
43 | 157.Yellow_throated_Vireo: 156, 158.Bay_breasted_Warbler: 157, 159.Black_and_white_Warbler: 158,
44 | 160.Black_throated_Blue_Warbler: 159, 161.Blue_winged_Warbler: 160, 162.Canada_Warbler: 161,
45 | 163.Cape_May_Warbler: 162, 164.Cerulean_Warbler: 163, 165.Chestnut_sided_Warbler: 164,
46 | 166.Golden_winged_Warbler: 165, 167.Hooded_Warbler: 166, 168.Kentucky_Warbler: 167,
47 | 169.Magnolia_Warbler: 168, 170.Mourning_Warbler: 169, 171.Myrtle_Warbler: 170, 172.Nashville_Warbler: 171,
48 | 173.Orange_crowned_Warbler: 172, 174.Palm_Warbler: 173, 175.Pine_Warbler: 174, 176.Prairie_Warbler: 175,
49 | 177.Prothonotary_Warbler: 176, 178.Swainson_Warbler: 177, 179.Tennessee_Warbler: 178,
50 | 180.Wilson_Warbler: 179, 181.Worm_eating_Warbler: 180, 182.Yellow_Warbler: 181,
51 | 183.Northern_Waterthrush: 182, 184.Louisiana_Waterthrush: 183, 185.Bohemian_Waxwing: 184,
52 | 186.Cedar_Waxwing: 185, 187.American_Three_toed_Woodpecker: 186, 188.Pileated_Woodpecker: 187,
53 | 189.Red_bellied_Woodpecker: 188, 190.Red_cockaded_Woodpecker: 189, 191.Red_headed_Woodpecker: 190,
54 | 192.Downy_Woodpecker: 191, 193.Bewick_Wren: 192, 194.Cactus_Wren: 193, 195.Carolina_Wren: 194,
55 | 196.House_Wren: 195, 197.Marsh_Wren: 196, 198.Rock_Wren: 197, 199.Winter_Wren: 198,
56 | 200.Common_Yellowthroat: 199}
57 |
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/Caltech-UCSD-Birds-200-2011/split_0/fold_4/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/log-stats-ds-ds-glas-s-0-f-0-subset-train.txt:
--------------------------------------------------------------------------------
1 | min h 430, max h 522
2 | min w 567, max w 775
3 |
--------------------------------------------------------------------------------
/folds/glas/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/size-stats-ds-glas-s-0-f-0-subset-train.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/folds/glas/size-stats-ds-glas-s-0-f-0-subset-train.png
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/.~lock.test_s_0_f_0.csv#:
--------------------------------------------------------------------------------
1 | ,brian,dell,24.07.2020 12:46,file:///home/brian/.config/libreoffice/4;
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/test_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/train_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
2 | 105,train_32.bmp,train_32_anno.bmp,benign,1
3 | 128,train_53.bmp,train_53_anno.bmp,benign,1
4 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
5 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
6 | 157,train_8.bmp,train_8_anno.bmp,benign,1
7 | 135,train_6.bmp,train_6_anno.bmp,benign,1
8 | 107,train_34.bmp,train_34_anno.bmp,benign,1
9 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
10 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
11 | 106,train_33.bmp,train_33_anno.bmp,benign,1
12 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
13 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
14 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
15 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
16 | 138,train_62.bmp,train_62_anno.bmp,benign,1
17 | 145,train_69.bmp,train_69_anno.bmp,benign,1
18 | 162,train_84.bmp,train_84_anno.bmp,benign,1
19 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
20 | 130,train_55.bmp,train_55_anno.bmp,benign,1
21 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
22 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
23 | 121,train_47.bmp,train_47_anno.bmp,benign,1
24 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
25 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
26 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
27 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
28 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
29 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
30 | 140,train_64.bmp,train_64_anno.bmp,benign,1
31 | 92,train_20.bmp,train_20_anno.bmp,benign,1
32 | 103,train_30.bmp,train_30_anno.bmp,benign,1
33 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
34 | 134,train_59.bmp,train_59_anno.bmp,benign,1
35 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
36 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
37 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
38 | 97,train_25.bmp,train_25_anno.bmp,benign,1
39 | 120,train_46.bmp,train_46_anno.bmp,benign,1
40 | 164,train_9.bmp,train_9_anno.bmp,benign,1
41 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
42 | 119,train_45.bmp,train_45_anno.bmp,benign,1
43 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
44 | 93,train_21.bmp,train_21_anno.bmp,benign,1
45 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
46 | 158,train_80.bmp,train_80_anno.bmp,benign,1
47 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
48 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
49 | 137,train_61.bmp,train_61_anno.bmp,benign,1
50 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
51 | 94,train_22.bmp,train_22_anno.bmp,benign,1
52 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
53 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
54 | 113,train_4.bmp,train_4_anno.bmp,benign,1
55 | 163,train_85.bmp,train_85_anno.bmp,benign,1
56 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
57 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
58 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
59 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
60 | 143,train_67.bmp,train_67_anno.bmp,benign,1
61 | 156,train_79.bmp,train_79_anno.bmp,benign,1
62 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
63 | 131,train_56.bmp,train_56_anno.bmp,benign,1
64 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
65 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
66 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
67 | 86,train_15.bmp,train_15_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_0/valid_s_0_f_0.csv:
--------------------------------------------------------------------------------
1 | 96,train_24.bmp,train_24_anno.bmp,malignant,0
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,0
3 | 124,train_5.bmp,train_5_anno.bmp,malignant,0
4 | 109,train_36.bmp,train_36_anno.bmp,malignant,0
5 | 98,train_26.bmp,train_26_anno.bmp,malignant,0
6 | 154,train_77.bmp,train_77_anno.bmp,malignant,0
7 | 151,train_74.bmp,train_74_anno.bmp,malignant,0
8 | 89,train_18.bmp,train_18_anno.bmp,malignant,0
9 | 161,train_83.bmp,train_83_anno.bmp,malignant,0
10 | 144,train_68.bmp,train_68_anno.bmp,malignant,0
11 | 111,train_38.bmp,train_38_anno.bmp,benign,0
12 | 149,train_72.bmp,train_72_anno.bmp,benign,0
13 | 141,train_65.bmp,train_65_anno.bmp,benign,0
14 | 126,train_51.bmp,train_51_anno.bmp,benign,0
15 | 83,train_12.bmp,train_12_anno.bmp,benign,0
16 | 139,train_63.bmp,train_63_anno.bmp,benign,0
17 | 91,train_2.bmp,train_2_anno.bmp,benign,0
18 | 127,train_52.bmp,train_52_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/test_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/train_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
2 | 139,train_63.bmp,train_63_anno.bmp,benign,1
3 | 140,train_64.bmp,train_64_anno.bmp,benign,1
4 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
5 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
6 | 141,train_65.bmp,train_65_anno.bmp,benign,1
7 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
8 | 119,train_45.bmp,train_45_anno.bmp,benign,1
9 | 92,train_20.bmp,train_20_anno.bmp,benign,1
10 | 121,train_47.bmp,train_47_anno.bmp,benign,1
11 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
12 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
13 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
14 | 149,train_72.bmp,train_72_anno.bmp,benign,1
15 | 127,train_52.bmp,train_52_anno.bmp,benign,1
16 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
17 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
18 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
19 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
20 | 107,train_34.bmp,train_34_anno.bmp,benign,1
21 | 158,train_80.bmp,train_80_anno.bmp,benign,1
22 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
23 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
24 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
25 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
26 | 126,train_51.bmp,train_51_anno.bmp,benign,1
27 | 113,train_4.bmp,train_4_anno.bmp,benign,1
28 | 94,train_22.bmp,train_22_anno.bmp,benign,1
29 | 162,train_84.bmp,train_84_anno.bmp,benign,1
30 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
31 | 138,train_62.bmp,train_62_anno.bmp,benign,1
32 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
33 | 163,train_85.bmp,train_85_anno.bmp,benign,1
34 | 128,train_53.bmp,train_53_anno.bmp,benign,1
35 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
36 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
37 | 111,train_38.bmp,train_38_anno.bmp,benign,1
38 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
39 | 143,train_67.bmp,train_67_anno.bmp,benign,1
40 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
41 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
42 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
43 | 97,train_25.bmp,train_25_anno.bmp,benign,1
44 | 86,train_15.bmp,train_15_anno.bmp,benign,1
45 | 83,train_12.bmp,train_12_anno.bmp,benign,1
46 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
47 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
48 | 105,train_32.bmp,train_32_anno.bmp,benign,1
49 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
50 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
51 | 93,train_21.bmp,train_21_anno.bmp,benign,1
52 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
53 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
54 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
55 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
56 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
57 | 156,train_79.bmp,train_79_anno.bmp,benign,1
58 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
59 | 131,train_56.bmp,train_56_anno.bmp,benign,1
60 | 135,train_6.bmp,train_6_anno.bmp,benign,1
61 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
62 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
63 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
64 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
65 | 91,train_2.bmp,train_2_anno.bmp,benign,1
66 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
67 | 157,train_8.bmp,train_8_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_1/valid_s_0_f_1.csv:
--------------------------------------------------------------------------------
1 | 125,train_50.bmp,train_50_anno.bmp,malignant,0
2 | 85,train_14.bmp,train_14_anno.bmp,malignant,0
3 | 146,train_7.bmp,train_7_anno.bmp,malignant,0
4 | 87,train_16.bmp,train_16_anno.bmp,malignant,0
5 | 152,train_75.bmp,train_75_anno.bmp,malignant,0
6 | 153,train_76.bmp,train_76_anno.bmp,malignant,0
7 | 160,train_82.bmp,train_82_anno.bmp,malignant,0
8 | 116,train_42.bmp,train_42_anno.bmp,malignant,0
9 | 150,train_73.bmp,train_73_anno.bmp,malignant,0
10 | 122,train_48.bmp,train_48_anno.bmp,malignant,0
11 | 103,train_30.bmp,train_30_anno.bmp,benign,0
12 | 130,train_55.bmp,train_55_anno.bmp,benign,0
13 | 137,train_61.bmp,train_61_anno.bmp,benign,0
14 | 164,train_9.bmp,train_9_anno.bmp,benign,0
15 | 145,train_69.bmp,train_69_anno.bmp,benign,0
16 | 106,train_33.bmp,train_33_anno.bmp,benign,0
17 | 120,train_46.bmp,train_46_anno.bmp,benign,0
18 | 134,train_59.bmp,train_59_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/test_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/train_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 135,train_6.bmp,train_6_anno.bmp,benign,1
2 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
3 | 131,train_56.bmp,train_56_anno.bmp,benign,1
4 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
5 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
6 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
7 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
8 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
9 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
10 | 127,train_52.bmp,train_52_anno.bmp,benign,1
11 | 164,train_9.bmp,train_9_anno.bmp,benign,1
12 | 93,train_21.bmp,train_21_anno.bmp,benign,1
13 | 141,train_65.bmp,train_65_anno.bmp,benign,1
14 | 86,train_15.bmp,train_15_anno.bmp,benign,1
15 | 106,train_33.bmp,train_33_anno.bmp,benign,1
16 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
17 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
18 | 134,train_59.bmp,train_59_anno.bmp,benign,1
19 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
20 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
21 | 130,train_55.bmp,train_55_anno.bmp,benign,1
22 | 119,train_45.bmp,train_45_anno.bmp,benign,1
23 | 113,train_4.bmp,train_4_anno.bmp,benign,1
24 | 120,train_46.bmp,train_46_anno.bmp,benign,1
25 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
26 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
27 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
28 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
29 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
30 | 156,train_79.bmp,train_79_anno.bmp,benign,1
31 | 94,train_22.bmp,train_22_anno.bmp,benign,1
32 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
33 | 162,train_84.bmp,train_84_anno.bmp,benign,1
34 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
35 | 137,train_61.bmp,train_61_anno.bmp,benign,1
36 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
37 | 163,train_85.bmp,train_85_anno.bmp,benign,1
38 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
39 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
40 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
41 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
42 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
43 | 149,train_72.bmp,train_72_anno.bmp,benign,1
44 | 128,train_53.bmp,train_53_anno.bmp,benign,1
45 | 91,train_2.bmp,train_2_anno.bmp,benign,1
46 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
47 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
48 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
49 | 145,train_69.bmp,train_69_anno.bmp,benign,1
50 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
51 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
52 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
53 | 92,train_20.bmp,train_20_anno.bmp,benign,1
54 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
55 | 126,train_51.bmp,train_51_anno.bmp,benign,1
56 | 83,train_12.bmp,train_12_anno.bmp,benign,1
57 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
58 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
59 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
60 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
61 | 139,train_63.bmp,train_63_anno.bmp,benign,1
62 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
63 | 111,train_38.bmp,train_38_anno.bmp,benign,1
64 | 143,train_67.bmp,train_67_anno.bmp,benign,1
65 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
66 | 103,train_30.bmp,train_30_anno.bmp,benign,1
67 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_2/valid_s_0_f_2.csv:
--------------------------------------------------------------------------------
1 | 118,train_44.bmp,train_44_anno.bmp,malignant,0
2 | 90,train_19.bmp,train_19_anno.bmp,malignant,0
3 | 132,train_57.bmp,train_57_anno.bmp,malignant,0
4 | 117,train_43.bmp,train_43_anno.bmp,malignant,0
5 | 133,train_58.bmp,train_58_anno.bmp,malignant,0
6 | 147,train_70.bmp,train_70_anno.bmp,malignant,0
7 | 104,train_31.bmp,train_31_anno.bmp,malignant,0
8 | 110,train_37.bmp,train_37_anno.bmp,malignant,0
9 | 80,train_1.bmp,train_1_anno.bmp,malignant,0
10 | 82,train_11.bmp,train_11_anno.bmp,malignant,0
11 | 140,train_64.bmp,train_64_anno.bmp,benign,0
12 | 157,train_8.bmp,train_8_anno.bmp,benign,0
13 | 138,train_62.bmp,train_62_anno.bmp,benign,0
14 | 107,train_34.bmp,train_34_anno.bmp,benign,0
15 | 97,train_25.bmp,train_25_anno.bmp,benign,0
16 | 158,train_80.bmp,train_80_anno.bmp,benign,0
17 | 105,train_32.bmp,train_32_anno.bmp,benign,0
18 | 121,train_47.bmp,train_47_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/test_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/train_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 107,train_34.bmp,train_34_anno.bmp,benign,1
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
3 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
4 | 138,train_62.bmp,train_62_anno.bmp,benign,1
5 | 97,train_25.bmp,train_25_anno.bmp,benign,1
6 | 139,train_63.bmp,train_63_anno.bmp,benign,1
7 | 91,train_2.bmp,train_2_anno.bmp,benign,1
8 | 120,train_46.bmp,train_46_anno.bmp,benign,1
9 | 149,train_72.bmp,train_72_anno.bmp,benign,1
10 | 86,train_15.bmp,train_15_anno.bmp,benign,1
11 | 105,train_32.bmp,train_32_anno.bmp,benign,1
12 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
13 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
14 | 95,train_23.bmp,train_23_anno.bmp,malignant,1
15 | 130,train_55.bmp,train_55_anno.bmp,benign,1
16 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
17 | 145,train_69.bmp,train_69_anno.bmp,benign,1
18 | 83,train_12.bmp,train_12_anno.bmp,benign,1
19 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
20 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
21 | 158,train_80.bmp,train_80_anno.bmp,benign,1
22 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
23 | 121,train_47.bmp,train_47_anno.bmp,benign,1
24 | 162,train_84.bmp,train_84_anno.bmp,benign,1
25 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
26 | 106,train_33.bmp,train_33_anno.bmp,benign,1
27 | 84,train_13.bmp,train_13_anno.bmp,malignant,1
28 | 111,train_38.bmp,train_38_anno.bmp,benign,1
29 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
30 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
31 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
32 | 126,train_51.bmp,train_51_anno.bmp,benign,1
33 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
34 | 164,train_9.bmp,train_9_anno.bmp,benign,1
35 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
36 | 103,train_30.bmp,train_30_anno.bmp,benign,1
37 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
38 | 123,train_49.bmp,train_49_anno.bmp,malignant,1
39 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
40 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
41 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
42 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
43 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
44 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
45 | 163,train_85.bmp,train_85_anno.bmp,benign,1
46 | 127,train_52.bmp,train_52_anno.bmp,benign,1
47 | 112,train_39.bmp,train_39_anno.bmp,malignant,1
48 | 140,train_64.bmp,train_64_anno.bmp,benign,1
49 | 119,train_45.bmp,train_45_anno.bmp,benign,1
50 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
51 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
52 | 143,train_67.bmp,train_67_anno.bmp,benign,1
53 | 108,train_35.bmp,train_35_anno.bmp,malignant,1
54 | 141,train_65.bmp,train_65_anno.bmp,benign,1
55 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
56 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
57 | 137,train_61.bmp,train_61_anno.bmp,benign,1
58 | 157,train_8.bmp,train_8_anno.bmp,benign,1
59 | 142,train_66.bmp,train_66_anno.bmp,malignant,1
60 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
61 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
62 | 101,train_29.bmp,train_29_anno.bmp,malignant,1
63 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
64 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
65 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
66 | 81,train_10.bmp,train_10_anno.bmp,malignant,1
67 | 134,train_59.bmp,train_59_anno.bmp,benign,1
68 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_3/valid_s_0_f_3.csv:
--------------------------------------------------------------------------------
1 | 159,train_81.bmp,train_81_anno.bmp,malignant,0
2 | 136,train_60.bmp,train_60_anno.bmp,malignant,0
3 | 99,train_27.bmp,train_27_anno.bmp,malignant,0
4 | 114,train_40.bmp,train_40_anno.bmp,malignant,0
5 | 148,train_71.bmp,train_71_anno.bmp,malignant,0
6 | 102,train_3.bmp,train_3_anno.bmp,malignant,0
7 | 88,train_17.bmp,train_17_anno.bmp,malignant,0
8 | 115,train_41.bmp,train_41_anno.bmp,malignant,0
9 | 100,train_28.bmp,train_28_anno.bmp,malignant,0
10 | 155,train_78.bmp,train_78_anno.bmp,malignant,0
11 | 93,train_21.bmp,train_21_anno.bmp,benign,0
12 | 156,train_79.bmp,train_79_anno.bmp,benign,0
13 | 128,train_53.bmp,train_53_anno.bmp,benign,0
14 | 135,train_6.bmp,train_6_anno.bmp,benign,0
15 | 94,train_22.bmp,train_22_anno.bmp,benign,0
16 | 92,train_20.bmp,train_20_anno.bmp,benign,0
17 | 131,train_56.bmp,train_56_anno.bmp,benign,0
18 | 113,train_4.bmp,train_4_anno.bmp,benign,0
19 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/encoding.yaml:
--------------------------------------------------------------------------------
1 | {benign: 0, malignant: 1}
2 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/readme.md:
--------------------------------------------------------------------------------
1 | Format: float `id`: 0, str `img`: 1, None `mask`: 2, str `label`: 3, int `tag`: 4
2 | Possible tags:
3 | 0: labeled
4 | 1: unlabeled
5 | 2: labeled but came from unlabeled set. [not possible at this level].
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/seed.txt:
--------------------------------------------------------------------------------
1 | MYSEED: 0
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/test_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 0,testA_1.bmp,testA_1_anno.bmp,benign,0
2 | 1,testA_10.bmp,testA_10_anno.bmp,benign,0
3 | 2,testA_11.bmp,testA_11_anno.bmp,benign,0
4 | 3,testA_12.bmp,testA_12_anno.bmp,malignant,0
5 | 4,testA_13.bmp,testA_13_anno.bmp,malignant,0
6 | 5,testA_14.bmp,testA_14_anno.bmp,malignant,0
7 | 6,testA_15.bmp,testA_15_anno.bmp,malignant,0
8 | 7,testA_16.bmp,testA_16_anno.bmp,malignant,0
9 | 8,testA_17.bmp,testA_17_anno.bmp,malignant,0
10 | 9,testA_18.bmp,testA_18_anno.bmp,benign,0
11 | 10,testA_19.bmp,testA_19_anno.bmp,benign,0
12 | 11,testA_2.bmp,testA_2_anno.bmp,benign,0
13 | 12,testA_20.bmp,testA_20_anno.bmp,benign,0
14 | 13,testA_21.bmp,testA_21_anno.bmp,benign,0
15 | 14,testA_22.bmp,testA_22_anno.bmp,malignant,0
16 | 15,testA_23.bmp,testA_23_anno.bmp,malignant,0
17 | 16,testA_24.bmp,testA_24_anno.bmp,malignant,0
18 | 17,testA_25.bmp,testA_25_anno.bmp,benign,0
19 | 18,testA_26.bmp,testA_26_anno.bmp,malignant,0
20 | 19,testA_27.bmp,testA_27_anno.bmp,benign,0
21 | 20,testA_28.bmp,testA_28_anno.bmp,benign,0
22 | 21,testA_29.bmp,testA_29_anno.bmp,malignant,0
23 | 22,testA_3.bmp,testA_3_anno.bmp,malignant,0
24 | 23,testA_30.bmp,testA_30_anno.bmp,benign,0
25 | 24,testA_31.bmp,testA_31_anno.bmp,benign,0
26 | 25,testA_32.bmp,testA_32_anno.bmp,malignant,0
27 | 26,testA_33.bmp,testA_33_anno.bmp,benign,0
28 | 27,testA_34.bmp,testA_34_anno.bmp,malignant,0
29 | 28,testA_35.bmp,testA_35_anno.bmp,benign,0
30 | 29,testA_36.bmp,testA_36_anno.bmp,benign,0
31 | 30,testA_37.bmp,testA_37_anno.bmp,benign,0
32 | 31,testA_38.bmp,testA_38_anno.bmp,malignant,0
33 | 32,testA_39.bmp,testA_39_anno.bmp,malignant,0
34 | 33,testA_4.bmp,testA_4_anno.bmp,benign,0
35 | 34,testA_40.bmp,testA_40_anno.bmp,benign,0
36 | 35,testA_41.bmp,testA_41_anno.bmp,malignant,0
37 | 36,testA_42.bmp,testA_42_anno.bmp,malignant,0
38 | 37,testA_43.bmp,testA_43_anno.bmp,benign,0
39 | 38,testA_44.bmp,testA_44_anno.bmp,benign,0
40 | 39,testA_45.bmp,testA_45_anno.bmp,malignant,0
41 | 40,testA_46.bmp,testA_46_anno.bmp,benign,0
42 | 41,testA_47.bmp,testA_47_anno.bmp,malignant,0
43 | 42,testA_48.bmp,testA_48_anno.bmp,malignant,0
44 | 43,testA_49.bmp,testA_49_anno.bmp,benign,0
45 | 44,testA_5.bmp,testA_5_anno.bmp,benign,0
46 | 45,testA_50.bmp,testA_50_anno.bmp,benign,0
47 | 46,testA_51.bmp,testA_51_anno.bmp,malignant,0
48 | 47,testA_52.bmp,testA_52_anno.bmp,benign,0
49 | 48,testA_53.bmp,testA_53_anno.bmp,malignant,0
50 | 49,testA_54.bmp,testA_54_anno.bmp,benign,0
51 | 50,testA_55.bmp,testA_55_anno.bmp,benign,0
52 | 51,testA_56.bmp,testA_56_anno.bmp,malignant,0
53 | 52,testA_57.bmp,testA_57_anno.bmp,malignant,0
54 | 53,testA_58.bmp,testA_58_anno.bmp,benign,0
55 | 54,testA_59.bmp,testA_59_anno.bmp,malignant,0
56 | 55,testA_6.bmp,testA_6_anno.bmp,benign,0
57 | 56,testA_60.bmp,testA_60_anno.bmp,benign,0
58 | 57,testA_7.bmp,testA_7_anno.bmp,benign,0
59 | 58,testA_8.bmp,testA_8_anno.bmp,malignant,0
60 | 59,testA_9.bmp,testA_9_anno.bmp,benign,0
61 | 60,testB_1.bmp,testB_1_anno.bmp,malignant,0
62 | 61,testB_10.bmp,testB_10_anno.bmp,malignant,0
63 | 62,testB_11.bmp,testB_11_anno.bmp,malignant,0
64 | 63,testB_12.bmp,testB_12_anno.bmp,malignant,0
65 | 64,testB_13.bmp,testB_13_anno.bmp,malignant,0
66 | 65,testB_14.bmp,testB_14_anno.bmp,malignant,0
67 | 66,testB_15.bmp,testB_15_anno.bmp,malignant,0
68 | 67,testB_16.bmp,testB_16_anno.bmp,malignant,0
69 | 68,testB_17.bmp,testB_17_anno.bmp,benign,0
70 | 69,testB_18.bmp,testB_18_anno.bmp,malignant,0
71 | 70,testB_19.bmp,testB_19_anno.bmp,malignant,0
72 | 71,testB_2.bmp,testB_2_anno.bmp,malignant,0
73 | 72,testB_20.bmp,testB_20_anno.bmp,malignant,0
74 | 73,testB_3.bmp,testB_3_anno.bmp,malignant,0
75 | 74,testB_4.bmp,testB_4_anno.bmp,benign,0
76 | 75,testB_5.bmp,testB_5_anno.bmp,benign,0
77 | 76,testB_6.bmp,testB_6_anno.bmp,malignant,0
78 | 77,testB_7.bmp,testB_7_anno.bmp,benign,0
79 | 78,testB_8.bmp,testB_8_anno.bmp,malignant,0
80 | 79,testB_9.bmp,testB_9_anno.bmp,malignant,0
81 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/train_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 116,train_42.bmp,train_42_anno.bmp,malignant,1
2 | 129,train_54.bmp,train_54_anno.bmp,malignant,1
3 | 83,train_12.bmp,train_12_anno.bmp,benign,1
4 | 94,train_22.bmp,train_22_anno.bmp,benign,1
5 | 159,train_81.bmp,train_81_anno.bmp,malignant,1
6 | 80,train_1.bmp,train_1_anno.bmp,malignant,1
7 | 87,train_16.bmp,train_16_anno.bmp,malignant,1
8 | 102,train_3.bmp,train_3_anno.bmp,malignant,1
9 | 145,train_69.bmp,train_69_anno.bmp,benign,1
10 | 161,train_83.bmp,train_83_anno.bmp,malignant,1
11 | 114,train_40.bmp,train_40_anno.bmp,malignant,1
12 | 158,train_80.bmp,train_80_anno.bmp,benign,1
13 | 154,train_77.bmp,train_77_anno.bmp,malignant,1
14 | 115,train_41.bmp,train_41_anno.bmp,malignant,1
15 | 125,train_50.bmp,train_50_anno.bmp,malignant,1
16 | 144,train_68.bmp,train_68_anno.bmp,malignant,1
17 | 146,train_7.bmp,train_7_anno.bmp,malignant,1
18 | 122,train_48.bmp,train_48_anno.bmp,malignant,1
19 | 107,train_34.bmp,train_34_anno.bmp,benign,1
20 | 98,train_26.bmp,train_26_anno.bmp,malignant,1
21 | 110,train_37.bmp,train_37_anno.bmp,malignant,1
22 | 149,train_72.bmp,train_72_anno.bmp,benign,1
23 | 93,train_21.bmp,train_21_anno.bmp,benign,1
24 | 91,train_2.bmp,train_2_anno.bmp,benign,1
25 | 127,train_52.bmp,train_52_anno.bmp,benign,1
26 | 152,train_75.bmp,train_75_anno.bmp,malignant,1
27 | 155,train_78.bmp,train_78_anno.bmp,malignant,1
28 | 126,train_51.bmp,train_51_anno.bmp,benign,1
29 | 106,train_33.bmp,train_33_anno.bmp,benign,1
30 | 117,train_43.bmp,train_43_anno.bmp,malignant,1
31 | 99,train_27.bmp,train_27_anno.bmp,malignant,1
32 | 157,train_8.bmp,train_8_anno.bmp,benign,1
33 | 164,train_9.bmp,train_9_anno.bmp,benign,1
34 | 82,train_11.bmp,train_11_anno.bmp,malignant,1
35 | 105,train_32.bmp,train_32_anno.bmp,benign,1
36 | 100,train_28.bmp,train_28_anno.bmp,malignant,1
37 | 156,train_79.bmp,train_79_anno.bmp,benign,1
38 | 128,train_53.bmp,train_53_anno.bmp,benign,1
39 | 97,train_25.bmp,train_25_anno.bmp,benign,1
40 | 136,train_60.bmp,train_60_anno.bmp,malignant,1
41 | 130,train_55.bmp,train_55_anno.bmp,benign,1
42 | 88,train_17.bmp,train_17_anno.bmp,malignant,1
43 | 148,train_71.bmp,train_71_anno.bmp,malignant,1
44 | 92,train_20.bmp,train_20_anno.bmp,benign,1
45 | 103,train_30.bmp,train_30_anno.bmp,benign,1
46 | 89,train_18.bmp,train_18_anno.bmp,malignant,1
47 | 133,train_58.bmp,train_58_anno.bmp,malignant,1
48 | 132,train_57.bmp,train_57_anno.bmp,malignant,1
49 | 118,train_44.bmp,train_44_anno.bmp,malignant,1
50 | 124,train_5.bmp,train_5_anno.bmp,malignant,1
51 | 141,train_65.bmp,train_65_anno.bmp,benign,1
52 | 134,train_59.bmp,train_59_anno.bmp,benign,1
53 | 85,train_14.bmp,train_14_anno.bmp,malignant,1
54 | 121,train_47.bmp,train_47_anno.bmp,benign,1
55 | 137,train_61.bmp,train_61_anno.bmp,benign,1
56 | 120,train_46.bmp,train_46_anno.bmp,benign,1
57 | 140,train_64.bmp,train_64_anno.bmp,benign,1
58 | 109,train_36.bmp,train_36_anno.bmp,malignant,1
59 | 147,train_70.bmp,train_70_anno.bmp,malignant,1
60 | 150,train_73.bmp,train_73_anno.bmp,malignant,1
61 | 90,train_19.bmp,train_19_anno.bmp,malignant,1
62 | 135,train_6.bmp,train_6_anno.bmp,benign,1
63 | 153,train_76.bmp,train_76_anno.bmp,malignant,1
64 | 138,train_62.bmp,train_62_anno.bmp,benign,1
65 | 151,train_74.bmp,train_74_anno.bmp,malignant,1
66 | 111,train_38.bmp,train_38_anno.bmp,benign,1
67 | 160,train_82.bmp,train_82_anno.bmp,malignant,1
68 | 139,train_63.bmp,train_63_anno.bmp,benign,1
69 | 113,train_4.bmp,train_4_anno.bmp,benign,1
70 | 96,train_24.bmp,train_24_anno.bmp,malignant,1
71 | 104,train_31.bmp,train_31_anno.bmp,malignant,1
72 | 131,train_56.bmp,train_56_anno.bmp,benign,1
73 |
--------------------------------------------------------------------------------
/folds/glas/split_0/fold_4/valid_s_0_f_4.csv:
--------------------------------------------------------------------------------
1 | 108,train_35.bmp,train_35_anno.bmp,malignant,0
2 | 142,train_66.bmp,train_66_anno.bmp,malignant,0
3 | 84,train_13.bmp,train_13_anno.bmp,malignant,0
4 | 81,train_10.bmp,train_10_anno.bmp,malignant,0
5 | 112,train_39.bmp,train_39_anno.bmp,malignant,0
6 | 123,train_49.bmp,train_49_anno.bmp,malignant,0
7 | 101,train_29.bmp,train_29_anno.bmp,malignant,0
8 | 95,train_23.bmp,train_23_anno.bmp,malignant,0
9 | 143,train_67.bmp,train_67_anno.bmp,benign,0
10 | 119,train_45.bmp,train_45_anno.bmp,benign,0
11 | 163,train_85.bmp,train_85_anno.bmp,benign,0
12 | 86,train_15.bmp,train_15_anno.bmp,benign,0
13 | 162,train_84.bmp,train_84_anno.bmp,benign,0
14 |
--------------------------------------------------------------------------------
/fonts/.readme.md:
--------------------------------------------------------------------------------
1 | Contains fonts used for drawing.
2 | Source: https://fonts.google.com/specimen/Inconsolata?selection.family=Inconsolata
3 |
--------------------------------------------------------------------------------
/fonts/Inconsolata.zip:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata.zip
--------------------------------------------------------------------------------
/fonts/Inconsolata/Inconsolata-Bold.ttf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata/Inconsolata-Bold.ttf
--------------------------------------------------------------------------------
/fonts/Inconsolata/Inconsolata-Regular.ttf:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/fonts/Inconsolata/Inconsolata-Regular.ttf
--------------------------------------------------------------------------------
/fonts/Inconsolata/OFL.txt:
--------------------------------------------------------------------------------
1 | Copyright 2006 The Inconsolata Project Authors
2 |
3 | This Font Software is licensed under the SIL Open Font License, Version 1.1.
4 | This license is copied below, and is also available with a FAQ at:
5 | http://scripts.sil.org/OFL
6 |
7 |
8 | -----------------------------------------------------------
9 | SIL OPEN FONT LICENSE Version 1.1 - 26 February 2007
10 | -----------------------------------------------------------
11 |
12 | PREAMBLE
13 | The goals of the Open Font License (OFL) are to stimulate worldwide
14 | development of collaborative font projects, to support the font creation
15 | efforts of academic and linguistic communities, and to provide a free and
16 | open framework in which fonts may be shared and improved in partnership
17 | with others.
18 |
19 | The OFL allows the licensed fonts to be used, studied, modified and
20 | redistributed freely as long as they are not sold by themselves. The
21 | fonts, including any derivative works, can be bundled, embedded,
22 | redistributed and/or sold with any software provided that any reserved
23 | names are not used by derivative works. The fonts and derivatives,
24 | however, cannot be released under any other type of license. The
25 | requirement for fonts to remain under this license does not apply
26 | to any document created using the fonts or their derivatives.
27 |
28 | DEFINITIONS
29 | "Font Software" refers to the set of files released by the Copyright
30 | Holder(s) under this license and clearly marked as such. This may
31 | include source files, build scripts and documentation.
32 |
33 | "Reserved Font Name" refers to any names specified as such after the
34 | copyright statement(s).
35 |
36 | "Original Version" refers to the collection of Font Software components as
37 | distributed by the Copyright Holder(s).
38 |
39 | "Modified Version" refers to any derivative made by adding to, deleting,
40 | or substituting -- in part or in whole -- any of the components of the
41 | Original Version, by changing formats or by porting the Font Software to a
42 | new environment.
43 |
44 | "Author" refers to any designer, engineer, programmer, technical
45 | writer or other person who contributed to the Font Software.
46 |
47 | PERMISSION & CONDITIONS
48 | Permission is hereby granted, free of charge, to any person obtaining
49 | a copy of the Font Software, to use, study, copy, merge, embed, modify,
50 | redistribute, and sell modified and unmodified copies of the Font
51 | Software, subject to the following conditions:
52 |
53 | 1) Neither the Font Software nor any of its individual components,
54 | in Original or Modified Versions, may be sold by itself.
55 |
56 | 2) Original or Modified Versions of the Font Software may be bundled,
57 | redistributed and/or sold with any software, provided that each copy
58 | contains the above copyright notice and this license. These can be
59 | included either as stand-alone text files, human-readable headers or
60 | in the appropriate machine-readable metadata fields within text or
61 | binary files as long as those fields can be easily viewed by the user.
62 |
63 | 3) No Modified Version of the Font Software may use the Reserved Font
64 | Name(s) unless explicit written permission is granted by the corresponding
65 | Copyright Holder. This restriction only applies to the primary font name as
66 | presented to the users.
67 |
68 | 4) The name(s) of the Copyright Holder(s) or the Author(s) of the Font
69 | Software shall not be used to promote, endorse or advertise any
70 | Modified Version, except to acknowledge the contribution(s) of the
71 | Copyright Holder(s) and the Author(s) or with their explicit written
72 | permission.
73 |
74 | 5) The Font Software, modified or unmodified, in part or in whole,
75 | must be distributed entirely under this license, and must not be
76 | distributed under any other license. The requirement for fonts to
77 | remain under this license does not apply to any document created
78 | using the Font Software.
79 |
80 | TERMINATION
81 | This license becomes null and void if any of the above conditions are
82 | not met.
83 |
84 | DISCLAIMER
85 | THE FONT SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND,
86 | EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO ANY WARRANTIES OF
87 | MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT
88 | OF COPYRIGHT, PATENT, TRADEMARK, OR OTHER RIGHT. IN NO EVENT SHALL THE
89 | COPYRIGHT HOLDER BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY,
90 | INCLUDING ANY GENERAL, SPECIAL, INDIRECT, INCIDENTAL, OR CONSEQUENTIAL
91 | DAMAGES, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
92 | FROM, OUT OF THE USE OR INABILITY TO USE THE FONT SOFTWARE OR FROM
93 | OTHER DEALINGS IN THE FONT SOFTWARE.
94 |
--------------------------------------------------------------------------------
/jobs/.readme.md:
--------------------------------------------------------------------------------
1 | Contains slurm jobs.
--------------------------------------------------------------------------------
/outputjobs/.readme.md:
--------------------------------------------------------------------------------
1 | Contains the output of the jobs (errors, standard output redirected to files).
--------------------------------------------------------------------------------
/pairwise_sims/.readme.md:
--------------------------------------------------------------------------------
1 | folder contains pairwise similarities.
2 |
--------------------------------------------------------------------------------
/paper/.readme.md:
--------------------------------------------------------------------------------
1 | contains files produced for the results.
2 |
--------------------------------------------------------------------------------
/paper/results-per-method-each-run-dataset-Caltech-UCSD-Birds-200-2011.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/paper/results-per-method-each-run-dataset-Caltech-UCSD-Birds-200-2011.pkl
--------------------------------------------------------------------------------
/paper/results-per-method-each-run-dataset-glas.pkl:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/sbelharbi/deep-active-learning-for-joint-classification-and-segmentation-with-weak-annotator/f9eeb5f4901f4fb192d4cdc341abad7da6735944/paper/results-per-method-each-run-dataset-glas.pkl
--------------------------------------------------------------------------------
/pretrained-imgnet/.readme.md:
--------------------------------------------------------------------------------
1 | contains imagenet pretrained weights.
2 |
--------------------------------------------------------------------------------
/pretrained/.readme.md:
--------------------------------------------------------------------------------
1 | contains pretrained models.
2 |
--------------------------------------------------------------------------------
/reproducibility.py:
--------------------------------------------------------------------------------
1 | # self-contained-as-possible module.
2 | # handles reproducibility procedures.
3 |
4 | import random
5 | import os
6 | import warnings
7 |
8 |
9 | import numpy as np
10 | import torch
11 | from torch._C import default_generator
12 |
13 |
14 | DEFAULT_SEED = 0 # the default seed.
15 |
16 |
17 | def check_if_allow_multgpu_mode():
18 | """
19 | Check if we can do multigpu.
20 | If yes, allow multigpu.
21 | :return: ALLOW_MULTIGPUS: bool. If True, we enter multigpu mode:
22 | 1. Computation will be dispatched over the AVAILABLE GPUs.
23 | 2. Synch-BN is activated.
24 | """
25 | if "CC_CLUSTER" in os.environ.keys():
26 | ALLOW_MULTIGPUS = True # CC.
27 | else:
28 | ALLOW_MULTIGPUS = False # others.
29 |
30 | # ALLOW_MULTIGPUS = True
31 | os.environ["ALLOW_MULTIGPUS"] = str(ALLOW_MULTIGPUS)
32 | NBRGPUS = torch.cuda.device_count()
33 | ALLOW_MULTIGPUS = ALLOW_MULTIGPUS and (NBRGPUS > 1)
34 |
35 | return ALLOW_MULTIGPUS
36 |
37 |
38 | def announce_msg(msg, upper=True):
39 | """
40 | Display sa message in the standard output. Something like this:
41 | =================================================================
42 | message
43 | =================================================================
44 |
45 | :param msg: str, text message to display.
46 | :param upper: True/False, if True, the entire message is converted into
47 | uppercase. Else, the message is displayed
48 | as it is.
49 | :return: str, what was printed in the standard output.
50 | """
51 | if upper:
52 | msg = msg.upper()
53 | n = min(120, max(80, len(msg)))
54 | top = "\n" + "=" * n
55 | middle = " " * (int(n / 2) - int(len(msg) / 2)) + " {}".format(msg)
56 | bottom = "=" * n + "\n"
57 |
58 | output_msg = "\n".join([top, middle, bottom])
59 |
60 | print(output_msg)
61 |
62 | return output_msg
63 |
64 |
65 | def get_seed():
66 | """
67 | Get the default seed from the environment variable.
68 | If not set, we use our default seed.
69 | :return: int, a seed.
70 | """
71 | try:
72 | msg = "REQUIRED SEED: {} ".format(os.environ["MYSEED"])
73 | announce_msg(msg)
74 |
75 | return int(os.environ["MYSEED"])
76 | except KeyError:
77 | print("`os.environ` does not have a key named `MYSEED`."
78 | "This key is supposed to hold the current seed. Please set it,"
79 | "and try again, if you want.")
80 |
81 | warnings.warn("MEANWHILE, .... WE ARE GOING TO USE OUR DEFAULT SEED: "
82 | "{}".format(DEFAULT_SEED))
83 | os.environ["MYSEED"] = str(DEFAULT_SEED)
84 | msg = "DEFAULT SEED: {} ".format(os.environ["MYSEED"])
85 | announce_msg(msg)
86 | return DEFAULT_SEED
87 |
88 |
89 | def init_seed(seed=None):
90 | """
91 | * initialize the seed.
92 | * Set a seed to some modules for reproducibility.
93 |
94 | Note:
95 |
96 | While this attempts to ensure reproducibility, it does not offer an
97 | absolute guarantee. The results may be similar to some precision.
98 | Also, they may be different due to an amplification to extremely
99 | small differences.
100 |
101 | See:
102 |
103 | https://pytorch.org/docs/stable/notes/randomness.html
104 | https://stackoverflow.com/questions/50744565/
105 | how-to-handle-non-determinism-when-training-on-a-gpu
106 |
107 | :param seed: int, a seed. Default is None: use the default seed (0).
108 | :return:
109 | """
110 | if seed is None:
111 | seed = get_seed()
112 | else:
113 | os.environ["MYSEED"] = str(seed)
114 | announce_msg("SEED: {} ".format(os.environ["MYSEED"]))
115 |
116 | check_if_allow_multgpu_mode()
117 | reset_seed(seed)
118 |
119 |
120 | def reset_seed(seed, check_cudnn=True):
121 | """
122 | Reset seed to some modules.
123 | :param seed: int. The current seed.
124 | :param check_cudnn: boo. if true, we check if we are in multi-gpu to
125 | disable the cudnn use. `ALLOW_MULTIGPUS` variable has to be already
126 | created in os.environ otherwise an error will be raise.
127 | :return:
128 | """
129 | torch.manual_seed(seed)
130 | np.random.seed(seed)
131 | random.seed(seed)
132 | torch.cuda.manual_seed(seed)
133 | torch.cuda.manual_seed_all(seed)
134 | torch.backends.cudnn.benchmark = False
135 | # Deterministic mode can have a performance impact, depending on your
136 | torch.backends.cudnn.deterministic = True
137 | # model: https://pytorch.org/docs/stable/notes/randomness.html#cudnn
138 | # If multigpu is on, deactivate cudnn since it has many random things
139 | # that we can not control.
140 | if check_cudnn:
141 | cond = torch.cuda.device_count() > 1
142 | cond = cond and (os.environ["ALLOW_MULTIGPUS"] == 'True')
143 | if cond:
144 | torch.backends.cudnn.enabled = False
145 |
146 |
147 | def set_default_seed():
148 | """
149 | Set the default seed.
150 | :return:
151 | """
152 | assert "MYSEED" in os.environ.keys(), "`MYSEED` key is not found in " \
153 | "os.environ.keys() ...." \
154 | "[NOT OK]"
155 | reset_seed(int(os.environ["MYSEED"]))
156 |
157 |
158 | def manual_seed(seed):
159 | r"""Sets the seed for generating random numbers. Returns a
160 | `torch._C.Generator` object.
161 |
162 | NOTE: WE REMOVE MANUAL RESEEDING ALL THE GPUS. At this point,
163 | it is not necessary; and there is not logic/reason
164 | to do it since we want only to reseed the current device.
165 |
166 | Args:
167 | seed (int): The desired seed.
168 | """
169 | return default_generator.manual_seed(int(seed))
170 |
171 |
172 | def force_seed_thread(seed):
173 | """
174 | For seed to some modules.
175 | :param seed:
176 | :return:
177 | """
178 | manual_seed(seed)
179 | np.random.seed(seed)
180 | random.seed(seed)
181 | torch.cuda.manual_seed(seed)
182 | torch.backends.cudnn.enabled = False
183 | # torch.backends.cudnn.enabled = True
184 | # torch.backends.cudnn.enabled = False
185 | # torch.backends.cudnn.benchmark = False
186 | # Deterministic mode can have a performance impact, depending on your
187 | # torch.backends.cudnn.deterministic = True
188 | # model: https://pytorch.org/docs/stable/notes/randomness.html#cudnn
189 |
--------------------------------------------------------------------------------
/results/.readme.md:
--------------------------------------------------------------------------------
1 | contains results.
2 |
--------------------------------------------------------------------------------
/scheduler.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
--------------------------------------------------------------------------------
/shared.py:
--------------------------------------------------------------------------------
1 | # This module shouldn't import any of our modules to avoid recursive importing.
2 | import os
3 | import argparse
4 | import textwrap
5 | import csv
6 | from os.path import join
7 | import fnmatch
8 |
9 |
10 | from sklearn.metrics import auc
11 | import torch
12 | import numpy as np
13 |
14 |
15 | CONST1 = 1000 # used to generate random numbers.
16 |
17 |
18 | def str2bool(v):
19 | """
20 | Read `v`: and returns a boolean value:
21 | True: if `v== "True"`
22 | False: if `v=="False"`
23 | :param v: str.
24 | :return: bool.
25 | """
26 | if isinstance(v, bool):
27 | return v
28 | if isinstance(v, str):
29 | if v == "True":
30 | return True
31 | elif v == "False":
32 | return False
33 | else:
34 | raise ValueError(
35 | "Expected value: 'True'/'False'. found {}.".format(v))
36 | else:
37 | raise argparse.ArgumentTypeError('String boolean value expected: '
38 | '"True"/"Flse"')
39 |
40 |
41 | def announce_msg(msg, upper=True):
42 | """
43 | Display sa message in the standard output. Something like this:
44 | =================================================================
45 | message
46 | =================================================================
47 |
48 | :param msg: str, text message to display.
49 | :param upper: True/False, if True, the entire message is converted into
50 | uppercase. Else, the message is displayed
51 | as it is.
52 | :return: str, what was printed in the standard output.
53 | """
54 | if upper:
55 | msg = msg.upper()
56 | n = min(120, max(80, len(msg)))
57 | top = "\n" + "=" * n
58 | middle = " " * (int(n / 2) - int(len(msg) / 2)) + " {}".format(msg)
59 | bottom = "=" * n + "\n"
60 |
61 | output_msg = "\n".join([top, middle, bottom])
62 |
63 | print(output_msg)
64 |
65 | return output_msg
66 |
67 |
68 | def check_if_allow_multgpu_mode():
69 | """
70 | Check if we can do multigpu.
71 | If yes, allow multigpu.
72 | :return: ALLOW_MULTIGPUS: bool. If True, we enter multigpu mode:
73 | 1. Computation will be dispatched over the AVAILABLE GPUs.
74 | 2. Synch-BN is activated.
75 | """
76 | if "CC_CLUSTER" in os.environ.keys():
77 | ALLOW_MULTIGPUS = True # CC.
78 | else:
79 | ALLOW_MULTIGPUS = False # others.
80 |
81 | # ALLOW_MULTIGPUS = True
82 | os.environ["ALLOW_MULTIGPUS"] = str(ALLOW_MULTIGPUS)
83 | NBRGPUS = torch.cuda.device_count()
84 | ALLOW_MULTIGPUS = ALLOW_MULTIGPUS and (NBRGPUS > 1)
85 |
86 | return ALLOW_MULTIGPUS
87 |
88 |
89 | def check_tensor_inf_nan(tn):
90 | """
91 | Check if a tensor has any inf or nan.
92 | """
93 | if any(torch.isinf(tn.view(-1))):
94 | raise ValueError("Found inf in projection.")
95 | if any(torch.isnan(tn.view(-1))):
96 | raise ValueError("Found nan in projection.")
97 |
98 |
99 | def wrap_command_line(cmd):
100 | """
101 | Wrap command line
102 | :param cmd: str. command line with space as a separator.
103 | :return:
104 | """
105 | return " \\\n".join(textwrap.wrap(
106 | cmd, width=77, break_long_words=False, break_on_hyphens=False))
107 |
108 |
109 | def drop_normal_samples(l_samples):
110 | """
111 | Remove normal samples from the list of samples.
112 |
113 | When to call this?
114 | # drop normal samples and keep metastatic if: 1. dataset=CAM16. 2.
115 | # al_type != AL_WSL.
116 |
117 | :param l_samples: list of samples resulting from csv_loader().
118 | :return: l_samples without any normal sample.
119 | """
120 | return [el for el in l_samples if el[3] == 'tumor']
121 |
122 |
123 | def csv_loader(fname, rootpath, drop_normal=False):
124 | """
125 | Read a *.csv file. Each line contains:
126 | 0. id_: str
127 | 1. img: str
128 | 2. mask: str or '' or None
129 | 3. label: str
130 | 4. tag: int in {0, 1}
131 |
132 | Example: 50162.0, test/img_50162_label_frog.jpeg, , frog, 0
133 |
134 | :param fname: Path to the *.csv file.
135 | :param rootpath: The root path to the folders of the images.
136 | :return: List of elements.
137 | :param drop_normal: bool. if true, normal samples are dropped.
138 | Each element is the path to an image: image path, mask path [optional],
139 | class name.
140 | """
141 | with open(fname, 'r') as f:
142 | out = [
143 | [row[0],
144 | join(rootpath, row[1]),
145 | join(rootpath, row[2]) if row[2] else None,
146 | row[3],
147 | int(row[4])
148 | ]
149 | for row in csv.reader(f)
150 | ]
151 |
152 | if drop_normal:
153 | out = drop_normal_samples(out)
154 |
155 | return out
156 |
157 |
158 | def csv_writer(data, fname):
159 | """
160 | Write a list of rows into a file.
161 | """
162 | msg = "'data' must be a list. found {}".format(type(data))
163 | assert isinstance(data, list), msg
164 |
165 | with open(fname, 'w') as fcsv:
166 | filewriter = csv.writer(
167 | fcsv, delimiter=',', quotechar='|', quoting=csv.QUOTE_MINIMAL)
168 | for row in data:
169 | filewriter.writerow(row)
170 |
171 |
172 | def find_files_pattern(fd_in_, pattern_):
173 | """
174 | Find paths to files with pattern within a folder recursively.
175 | :return:
176 | """
177 | assert os.path.exists(fd_in_), "Folder {} does not exist " \
178 | ".... [NOT OK]".format(fd_in_)
179 | files = []
180 | for r, d, f in os.walk(fd_in_):
181 | for file in f:
182 | if fnmatch.fnmatch(file, pattern_):
183 | files.append(os.path.join(r, file))
184 |
185 | return files
186 |
187 |
188 | def check_nans(tens, msg=''):
189 | """
190 | Check if the tensor 'tens' contains any 'nan' values, and how many.
191 |
192 | :param tens: torch tensor.
193 | :param msg: str. message to display if there is nan.
194 | :return:
195 | """
196 | nbr_nans = torch.isnan(tens).float().sum().item()
197 | if nbr_nans > 0:
198 | print("NAN-CHECK: {}. Found: {} NANs.".format(msg, nbr_nans))
199 |
200 |
201 | def compute_auc(vec, nbr_p):
202 | """
203 | Compute the area under a curve.
204 | :param vec: vector contains values in [0, 100.].
205 | :param nbr_p: int. number of points in the x-axis. it is expected to be
206 | the same as the number of values in `vec`.
207 | :return: float in [0, 100]. percentage of the area from the perfect area.
208 | """
209 | if vec.size == 1:
210 | return float(vec[0])
211 | else:
212 | area_under_c = auc(x=np.array(list(range(vec.size))), y=vec)
213 | area_under_c /= (100. * (nbr_p - 1))
214 | area_under_c *= 100. # (%)
215 | return area_under_c
216 |
217 | # ==============================================================================
218 | # TEST
219 | # ==============================================================================
220 |
221 |
222 | def test_announce_msg():
223 | """
224 | Test announce_msg()
225 | :return:
226 | """
227 | announce_msg("Hello world!!!")
--------------------------------------------------------------------------------
/tmp/.readme.md:
--------------------------------------------------------------------------------
1 | Contains temporary files.
--------------------------------------------------------------------------------