├── requirements.txt
├── readme_src
├── notebook.png
├── sql_logo.jpg
├── sql_logo2.jpg
├── sql_logo_smaller.png
├── underConstruction.jpg
└── notebook_results
│ ├── 1.png
│ ├── 10.png
│ ├── 11.png
│ ├── 12.png
│ ├── 2.png
│ ├── 3.png
│ ├── 4.png
│ ├── 5.png
│ ├── 6.png
│ ├── 7.png
│ ├── 8.png
│ ├── 9.png
│ ├── data_sample.png
│ └── db_connected.png
├── .readthedocs.yaml
├── docs
├── source
│ ├── api.rst
│ ├── index.rst
│ ├── usage.rst
│ └── conf.py
├── publish_cmds.txt
├── Makefile
└── make.bat
├── sampleData
└── DataBaseBackup
│ └── INX.bak
├── pyproject.toml
├── README.rst
├── LICENSE
├── setup.py
├── .gitignore
├── README.md
├── example_notebook
└── SampleNoteBook_edaSQL.ipynb
└── edaSQL
└── src
└── edaSQL.py
/requirements.txt:
--------------------------------------------------------------------------------
1 | numpy
2 | pandas
3 | seaborn
4 | missingno
--------------------------------------------------------------------------------
/readme_src/notebook.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook.png
--------------------------------------------------------------------------------
/readme_src/sql_logo.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/sql_logo.jpg
--------------------------------------------------------------------------------
/readme_src/sql_logo2.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/sql_logo2.jpg
--------------------------------------------------------------------------------
/.readthedocs.yaml:
--------------------------------------------------------------------------------
1 | version: 2
2 |
3 | build:
4 | os: "ubuntu-20.04"
5 | tools:
6 | python: "3.8"
7 |
--------------------------------------------------------------------------------
/docs/source/api.rst:
--------------------------------------------------------------------------------
1 | API
2 | ===
3 |
4 | .. autosummary::
5 | :toctree: generated
6 |
7 | lumache
8 |
--------------------------------------------------------------------------------
/readme_src/sql_logo_smaller.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/sql_logo_smaller.png
--------------------------------------------------------------------------------
/readme_src/underConstruction.jpg:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/underConstruction.jpg
--------------------------------------------------------------------------------
/readme_src/notebook_results/1.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/1.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/10.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/10.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/11.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/11.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/12.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/12.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/2.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/2.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/3.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/3.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/4.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/4.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/5.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/5.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/6.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/6.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/7.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/7.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/8.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/8.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/9.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/9.png
--------------------------------------------------------------------------------
/sampleData/DataBaseBackup/INX.bak:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/sampleData/DataBaseBackup/INX.bak
--------------------------------------------------------------------------------
/readme_src/notebook_results/data_sample.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/data_sample.png
--------------------------------------------------------------------------------
/readme_src/notebook_results/db_connected.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/selva221724/edaSQL/HEAD/readme_src/notebook_results/db_connected.png
--------------------------------------------------------------------------------
/docs/publish_cmds.txt:
--------------------------------------------------------------------------------
1 | python setup.py sdist bdist_wheel
2 |
3 | python -m twine upload --repository testpypi dist/*
4 |
5 | python -m twine upload dist/*
--------------------------------------------------------------------------------
/pyproject.toml:
--------------------------------------------------------------------------------
1 | [build-system]
2 | requires = ["flit_core >=3.2,<4"]
3 | build-backend = "flit_core.buildapi"
4 |
5 | [project]
6 | name = "edaSQL"
7 | authors = [{name = "TamilSelvan", email = "selva221724@gmail.com"}]
8 | dynamic = ["version", "description"]
9 |
--------------------------------------------------------------------------------
/README.rst:
--------------------------------------------------------------------------------
1 | Template for the Read the Docs tutorial
2 | =======================================
3 |
4 | This GitHub template includes fictional Python library
5 | with some basic Sphinx docs.
6 |
7 | Read the tutorial here:
8 |
9 | https://docs.readthedocs.io/en/stable/tutorial/
10 |
--------------------------------------------------------------------------------
/docs/source/index.rst:
--------------------------------------------------------------------------------
1 | Welcome to edaSQL's documentation!
2 | ===================================
3 |
4 | **edaSQL** is a library to link SQL to Exploratory Data Analysis and further more in the Data Engineering. This will solve many limitations in the SQL studios available in the market. Use the SQL Query language to get your Table Results.
5 |
6 |
7 | Check out the :doc:`usage` section for further information, including
8 | how to :ref:`installation` the project.
9 |
10 | .. note::
11 |
12 | This project is under active development.
13 |
14 | Contents
15 | --------
16 |
17 | .. toctree::
18 |
19 | usage
20 | api
21 |
22 | edaSQL has its documentation hosted on Read the Docs.
23 |
--------------------------------------------------------------------------------
/docs/Makefile:
--------------------------------------------------------------------------------
1 | # Minimal makefile for Sphinx documentation
2 | #
3 |
4 | # You can set these variables from the command line, and also
5 | # from the environment for the first two.
6 | SPHINXOPTS ?=
7 | SPHINXBUILD ?= sphinx-build
8 | SOURCEDIR = source
9 | BUILDDIR = build
10 |
11 | # Put it first so that "make" without argument is like "make help".
12 | help:
13 | @$(SPHINXBUILD) -M help "$(SOURCEDIR)" "$(BUILDDIR)" $(SPHINXOPTS) $(O)
14 |
15 | .PHONY: help Makefile
16 |
17 | # Catch-all target: route all unknown targets to Sphinx using the new
18 | # "make mode" option. $(O) is meant as a shortcut for $(SPHINXOPTS).
19 | %: Makefile
20 | @$(SPHINXBUILD) -M $@ "$(SOURCEDIR)" "$(BUILDDIR)" $(SPHINXOPTS) $(O)
21 |
--------------------------------------------------------------------------------
/docs/source/usage.rst:
--------------------------------------------------------------------------------
1 | Usage
2 | =====
3 |
4 | .. _installation:
5 |
6 | Installation
7 | ------------
8 |
9 | To use edaSQL, first install it using pip:
10 |
11 | .. code-block:: console
12 |
13 | (.venv) $ pip install edaSQL
14 |
15 | Importing the Package and Iniate the eda object
16 | ----------------
17 | .. code-block:: python
18 |
19 | import edaSQL
20 | edasql = edaSQL.SQL()
21 |
22 | To retrieve a list of random ingredients,
23 | you can use the ``edaSQL.SQL()`` function:
24 |
25 | .. autofunction:: lumache.get_random_ingredients
26 |
27 | The ``kind`` parameter should be either ``"meat"``, ``"fish"``,
28 | or ``"veggies"``. Otherwise, :py:func:`lumache.get_random_ingredients`
29 | will raise an exception.
30 |
31 | .. autoexception:: lumache.InvalidKindError
32 |
33 |
--------------------------------------------------------------------------------
/docs/source/conf.py:
--------------------------------------------------------------------------------
1 | # Configuration file for the Sphinx documentation builder.
2 |
3 | # -- Project information
4 |
5 | project = 'edaSQL'
6 | copyright = '2021, TamilSelvan'
7 | author = 'TamilSelvan'
8 |
9 | release = '0.1'
10 | version = '0.1.0'
11 |
12 | # -- General configuration
13 |
14 | extensions = [
15 | 'sphinx.ext.duration',
16 | 'sphinx.ext.doctest',
17 | 'sphinx.ext.autodoc',
18 | 'sphinx.ext.autosummary',
19 | 'sphinx.ext.intersphinx',
20 | ]
21 |
22 | intersphinx_mapping = {
23 | 'python': ('https://docs.python.org/3/', None),
24 | 'sphinx': ('https://www.sphinx-doc.org/en/master/', None),
25 | }
26 | intersphinx_disabled_domains = ['std']
27 |
28 | templates_path = ['_templates']
29 |
30 | # -- Options for HTML output
31 |
32 | html_theme = 'sphinx_rtd_theme'
33 |
34 | # -- Options for EPUB output
35 | epub_show_urls = 'footnote'
36 |
--------------------------------------------------------------------------------
/docs/make.bat:
--------------------------------------------------------------------------------
1 | @ECHO OFF
2 |
3 | pushd %~dp0
4 |
5 | REM Command file for Sphinx documentation
6 |
7 | if "%SPHINXBUILD%" == "" (
8 | set SPHINXBUILD=sphinx-build
9 | )
10 | set SOURCEDIR=source
11 | set BUILDDIR=build
12 |
13 | if "%1" == "" goto help
14 |
15 | %SPHINXBUILD% >NUL 2>NUL
16 | if errorlevel 9009 (
17 | echo.
18 | echo.The 'sphinx-build' command was not found. Make sure you have Sphinx
19 | echo.installed, then set the SPHINXBUILD environment variable to point
20 | echo.to the full path of the 'sphinx-build' executable. Alternatively you
21 | echo.may add the Sphinx directory to PATH.
22 | echo.
23 | echo.If you don't have Sphinx installed, grab it from
24 | echo.http://sphinx-doc.org/
25 | exit /b 1
26 | )
27 |
28 | %SPHINXBUILD% -M %1 %SOURCEDIR% %BUILDDIR% %SPHINXOPTS% %O%
29 | goto end
30 |
31 | :help
32 | %SPHINXBUILD% -M help %SOURCEDIR% %BUILDDIR% %SPHINXOPTS% %O%
33 |
34 | :end
35 | popd
36 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2021 Tamil Selvan
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/setup.py:
--------------------------------------------------------------------------------
1 | import setuptools
2 |
3 | with open("README.md", "r") as fh:
4 | long_description = fh.read()
5 |
6 | # Read the requirements
7 | with open("requirements.txt",encoding="utf8") as f:
8 | requirements = f.read().splitlines()
9 |
10 | setuptools.setup(
11 | name="edaSQL", # This is the name of the package
12 | version="0.0.1.5", # The initial release version
13 | author="Tamil Selvan A V", # Full name of the author
14 | description="Exploratory Data Analytics tool for SQL",
15 | url="https://github.com/selva221724/edaSQL",
16 | license="MIT",
17 | include_package_data=True,
18 | long_description=long_description, # Long description read from the the readme file
19 | long_description_content_type="text/markdown",
20 | packages=setuptools.find_packages(), # List of all python modules to be installed
21 | classifiers=[
22 | "Programming Language :: Python :: 3",
23 | "Programming Language :: Python :: 3.6",
24 | "Programming Language :: Python :: 3.7",
25 | "Programming Language :: Python :: 3.8",
26 | "Programming Language :: Python :: 3.9",
27 | "Programming Language :: Python :: 3.10",
28 | "License :: OSI Approved :: MIT License",
29 | "Framework :: IPython",
30 | "Operating System :: OS Independent",
31 | "Intended Audience :: Science/Research",
32 | "Intended Audience :: Developers",
33 | "Intended Audience :: Financial and Insurance Industry",
34 | "Intended Audience :: Healthcare Industry",
35 | "Topic :: Scientific/Engineering",
36 | "Environment :: Console"
37 | ], # Information to filter the project on PyPi website
38 | python_requires='>=3.6', # Minimum version requirement of the package
39 | py_modules=["edaSQL"], # Name of the python package
40 | package_dir={'': 'edaSQL/src'}, # Directory of the source code of the package
41 | install_requires=requirements # Install other dependencies if any
42 |
43 | )
44 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 |
2 | # Created by https://www.toptal.com/developers/gitignore/api/python,jupyternotebooks
3 | # Edit at https://www.toptal.com/developers/gitignore?templates=python,jupyternotebooks
4 |
5 | ### JupyterNotebooks ###
6 | # gitignore template for Jupyter Notebooks
7 | # website: http://jupyter.org/
8 |
9 | .ipynb_checkpoints
10 | */.ipynb_checkpoints/*
11 |
12 | # IPython
13 | profile_default/
14 | ipython_config.py
15 |
16 | # Remove previous ipynb_checkpoints
17 | # git rm -r .ipynb_checkpoints/
18 |
19 | ### Python ###
20 | # Byte-compiled / optimized / DLL files
21 | __pycache__/
22 | *.py[cod]
23 | *$py.class
24 |
25 | # C extensions
26 | *.so
27 |
28 | # Distribution / packaging
29 | .Python
30 | build/
31 | develop-eggs/
32 | dist/
33 | downloads/
34 | eggs/
35 | .eggs/
36 | lib/
37 | lib64/
38 | parts/
39 | sdist/
40 | var/
41 | wheels/
42 | share/python-wheels/
43 | *.egg-info/
44 | .installed.cfg
45 | *.egg
46 | MANIFEST
47 |
48 | # PyInstaller
49 | # Usually these files are written by a python script from a template
50 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
51 | *.manifest
52 | *.spec
53 |
54 | # Installer logs
55 | pip-log.txt
56 | pip-delete-this-directory.txt
57 |
58 | # Unit test / coverage reports
59 | htmlcov/
60 | .tox/
61 | .nox/
62 | .coverage
63 | .coverage.*
64 | .cache
65 | nosetests.xml
66 | coverage.xml
67 | *.cover
68 | *.py,cover
69 | .hypothesis/
70 | .pytest_cache/
71 | cover/
72 |
73 | # Translations
74 | *.mo
75 | *.pot
76 |
77 | # Django stuff:
78 | *.log
79 | local_settings.py
80 | db.sqlite3
81 | db.sqlite3-journal
82 |
83 | # Flask stuff:
84 | instance/
85 | .webassets-cache
86 |
87 | # Scrapy stuff:
88 | .scrapy
89 |
90 | # Sphinx documentation
91 | docs/_build/
92 |
93 | # PyBuilder
94 | .pybuilder/
95 | target/
96 |
97 | # Jupyter Notebook
98 |
99 | # IPython
100 |
101 | # pyenv
102 | # For a library or package, you might want to ignore these files since the code is
103 | # intended to run in multiple environments; otherwise, check them in:
104 | # .python-version
105 |
106 | # pipenv
107 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
108 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
109 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
110 | # install all needed dependencies.
111 | #Pipfile.lock
112 |
113 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow
114 | __pypackages__/
115 |
116 | # Celery stuff
117 | celerybeat-schedule
118 | celerybeat.pid
119 |
120 | # SageMath parsed files
121 | *.sage.py
122 |
123 | # Environments
124 | .env
125 | .venv
126 | env/
127 | venv/
128 | ENV/
129 | env.bak/
130 | venv.bak/
131 |
132 | # Spyder project settings
133 | .spyderproject
134 | .spyproject
135 |
136 | # Rope project settings
137 | .ropeproject
138 |
139 | # mkdocs documentation
140 | /site
141 |
142 | # mypy
143 | .mypy_cache/
144 | .dmypy.json
145 | dmypy.json
146 |
147 | # Pyre type checker
148 | .pyre/
149 |
150 | # pytype static type analyzer
151 | .pytype/
152 |
153 | # Cython debug symbols
154 | cython_debug/
155 |
156 | # End of https://www.toptal.com/developers/gitignore/api/python,jupyternotebooks
157 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 |
2 |  3 |
3 |
4 |
5 |
6 | [ ](https://pypi.org/project/edaSQL/)
7 | [
](https://pypi.org/project/edaSQL/)
7 | [ ](https://edasql.readthedocs.io/en/latest/)
8 | [
](https://edasql.readthedocs.io/en/latest/)
8 | [ ](https://opensource.org/licenses/MIT)
9 |
](https://opensource.org/licenses/MIT)
9 |  10 |
10 |  11 |
11 |  12 |
12 |  13 |
14 | ## SQL Bridge Tool to Exploratory Data Analysis
15 |
16 |
17 | **edaSQL** is a library to link SQL to **Exploratory Data Analysis** and further more in the Data Engineering. This will solve many limitations in the SQL studios available in the market. Use the SQL Query language to get your Table Results.
18 |
19 | ## Installation
20 | Install dependency Packages before installing edaSQL
21 | ```shell
22 | pip install pyodbc
23 | pip install ipython
24 | ```
25 | Optional dependency for better visualization - [Jupyter Notebook](https://jupyter.org/install)
26 | ```shell
27 | pip install notebook
28 | ```
29 |
30 | **Now Install using pip** . [Offical Python Package Here!!](https://pypi.org/project/edaSQL/)
31 | ```shell
32 | pip install edaSQL
33 | ```
34 |
35 | (OR)
36 |
37 | Clone this Repository. Run this from the root directory to install
38 |
39 | ```shell
40 | python setup.py install
41 | ```
42 |
43 | ## Documentation
44 |
45 |
13 |
14 | ## SQL Bridge Tool to Exploratory Data Analysis
15 |
16 |
17 | **edaSQL** is a library to link SQL to **Exploratory Data Analysis** and further more in the Data Engineering. This will solve many limitations in the SQL studios available in the market. Use the SQL Query language to get your Table Results.
18 |
19 | ## Installation
20 | Install dependency Packages before installing edaSQL
21 | ```shell
22 | pip install pyodbc
23 | pip install ipython
24 | ```
25 | Optional dependency for better visualization - [Jupyter Notebook](https://jupyter.org/install)
26 | ```shell
27 | pip install notebook
28 | ```
29 |
30 | **Now Install using pip** . [Offical Python Package Here!!](https://pypi.org/project/edaSQL/)
31 | ```shell
32 | pip install edaSQL
33 | ```
34 |
35 | (OR)
36 |
37 | Clone this Repository. Run this from the root directory to install
38 |
39 | ```shell
40 | python setup.py install
41 | ```
42 |
43 | ## Documentation
44 |
45 |  46 |
47 | [Read the detailed documentation in readthedocs.io](https://edasql.readthedocs.io/en/latest/) (still under the development)
48 |
49 | ## License
50 | The license for edaSQL is MIT license
51 |
52 | ## Need help?
53 | Stuck on your edaSQL code or problem? Any other questions? Don't
54 | hestitate to send me an email (selva221724@gmail.com).
55 |
56 | ## edaSQL Jupyter NoteBook Tutorial
57 |
58 | Access the sample Jupyter Notebook [here!!](https://github.com/selva221724/edaSQL/blob/main/example_notebook/SampleNoteBook_edaSQL.ipynb)
59 |
60 | Access the Sample Data Used in this Repo
61 | - [CSV](https://github.com/selva221724/edaSQL/blob/main/sampleData/CSV/INX.csv)
62 | - [DataBase Backup](https://github.com/selva221724/edaSQL/blob/main/sampleData/DataBaseBackup/INX.bak) ( you can restore the DB in SQL Studio )
63 |
64 | **edaSQL for DataFrame:** If you are using the CSV or Excel as a source , Read using the Pandas & start from the [**3. Data Overview**](#Chapter1)
65 |
66 | ### Import Packages
67 | ```python
68 | import edaSQL
69 | import pandas as pd
70 | ```
71 |
72 | ### 1. Connect to the DataBase
73 | ```python
74 | edasql = edaSQL.SQL()
75 | edasql.connectToDataBase(server='your server name',
76 | database='your database',
77 | user='username',
78 | password='password',
79 | sqlDriver='ODBC Driver 17 for SQL Server')
80 | ```
81 |
82 |
46 |
47 | [Read the detailed documentation in readthedocs.io](https://edasql.readthedocs.io/en/latest/) (still under the development)
48 |
49 | ## License
50 | The license for edaSQL is MIT license
51 |
52 | ## Need help?
53 | Stuck on your edaSQL code or problem? Any other questions? Don't
54 | hestitate to send me an email (selva221724@gmail.com).
55 |
56 | ## edaSQL Jupyter NoteBook Tutorial
57 |
58 | Access the sample Jupyter Notebook [here!!](https://github.com/selva221724/edaSQL/blob/main/example_notebook/SampleNoteBook_edaSQL.ipynb)
59 |
60 | Access the Sample Data Used in this Repo
61 | - [CSV](https://github.com/selva221724/edaSQL/blob/main/sampleData/CSV/INX.csv)
62 | - [DataBase Backup](https://github.com/selva221724/edaSQL/blob/main/sampleData/DataBaseBackup/INX.bak) ( you can restore the DB in SQL Studio )
63 |
64 | **edaSQL for DataFrame:** If you are using the CSV or Excel as a source , Read using the Pandas & start from the [**3. Data Overview**](#Chapter1)
65 |
66 | ### Import Packages
67 | ```python
68 | import edaSQL
69 | import pandas as pd
70 | ```
71 |
72 | ### 1. Connect to the DataBase
73 | ```python
74 | edasql = edaSQL.SQL()
75 | edasql.connectToDataBase(server='your server name',
76 | database='your database',
77 | user='username',
78 | password='password',
79 | sqlDriver='ODBC Driver 17 for SQL Server')
80 | ```
81 |
82 |  83 |
84 | ### 2. Query Data
85 | ```python
86 | sampleQuery = "select * from INX"
87 | data = pd.read_sql(sampleQuery, edasql.dbConnection)
88 | ```
89 |
83 |
84 | ### 2. Query Data
85 | ```python
86 | sampleQuery = "select * from INX"
87 | data = pd.read_sql(sampleQuery, edasql.dbConnection)
88 | ```
89 | 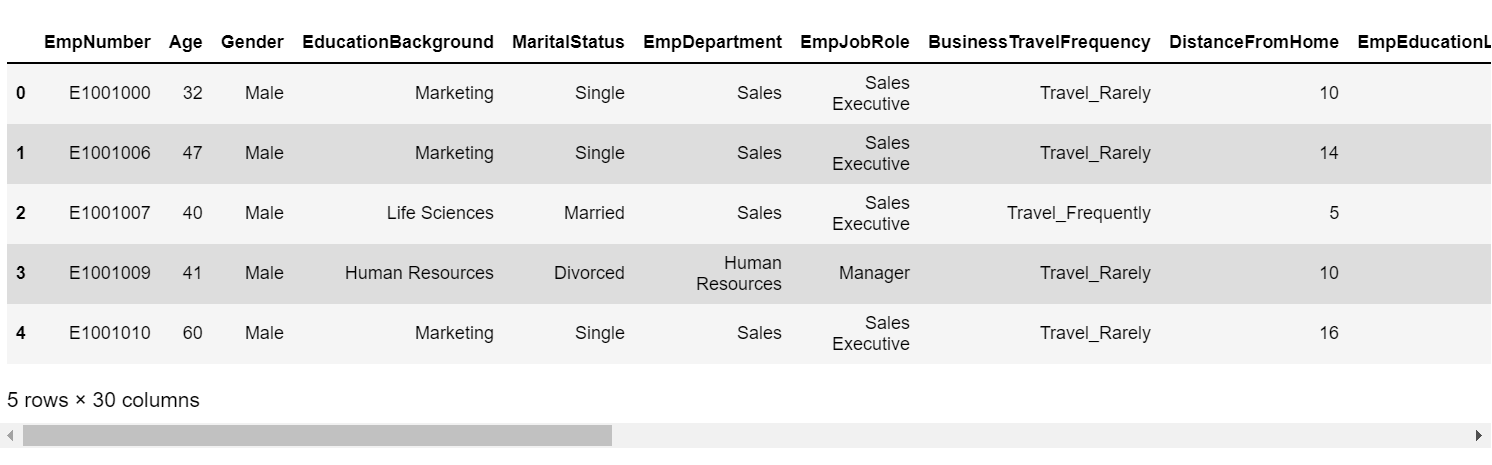 90 |
91 |
92 |
93 | ### 3. Data Overview
94 | ```python
95 | insights = edaSQL.EDA(dataFrame=data,HTMLDisplay=True)
96 | dataInsights =insights.dataInsights()
97 | ```
98 |
99 |
90 |
91 |
92 |
93 | ### 3. Data Overview
94 | ```python
95 | insights = edaSQL.EDA(dataFrame=data,HTMLDisplay=True)
96 | dataInsights =insights.dataInsights()
97 | ```
98 |
99 |  100 |
101 | ```python
102 | deepInsights = insights.deepInsights()
103 | ```
104 |
100 |
101 | ```python
102 | deepInsights = insights.deepInsights()
103 | ```
104 | 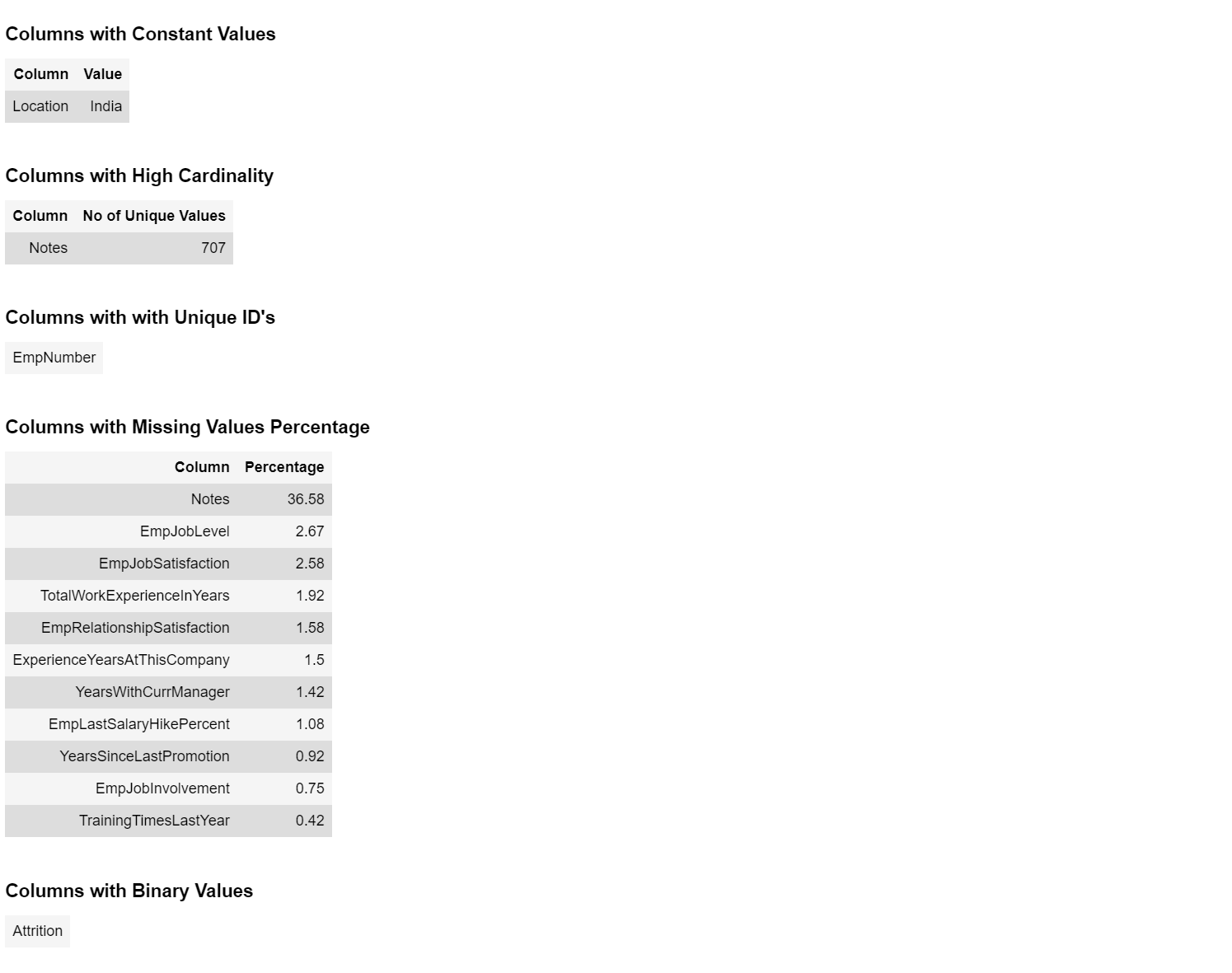 105 |
106 | ### 4. Correlation
107 | ```python
108 | eda = edaSQL.EDA(dataFrame=data)
109 | eda.pearsonCorrelation()
110 | ```
111 |
105 |
106 | ### 4. Correlation
107 | ```python
108 | eda = edaSQL.EDA(dataFrame=data)
109 | eda.pearsonCorrelation()
110 | ```
111 | 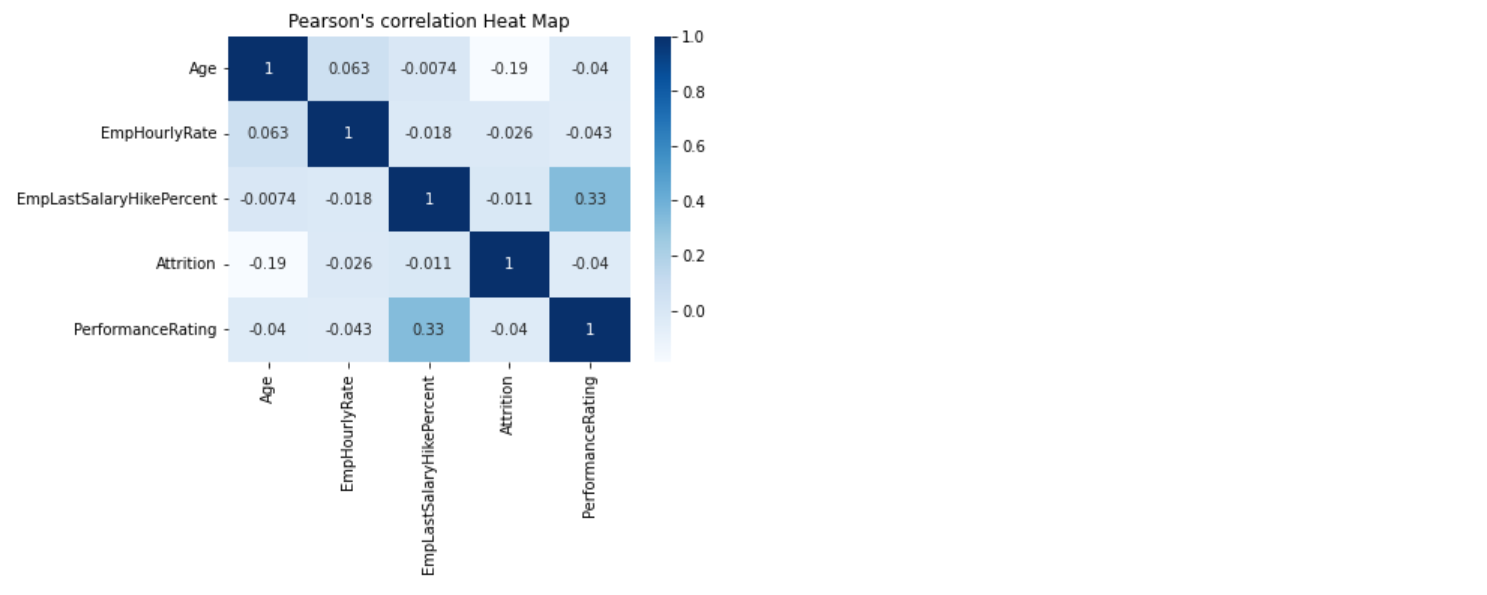 112 |
113 | ```python
114 | eda.spearmanCorrelation()
115 | ```
116 |
112 |
113 | ```python
114 | eda.spearmanCorrelation()
115 | ```
116 | 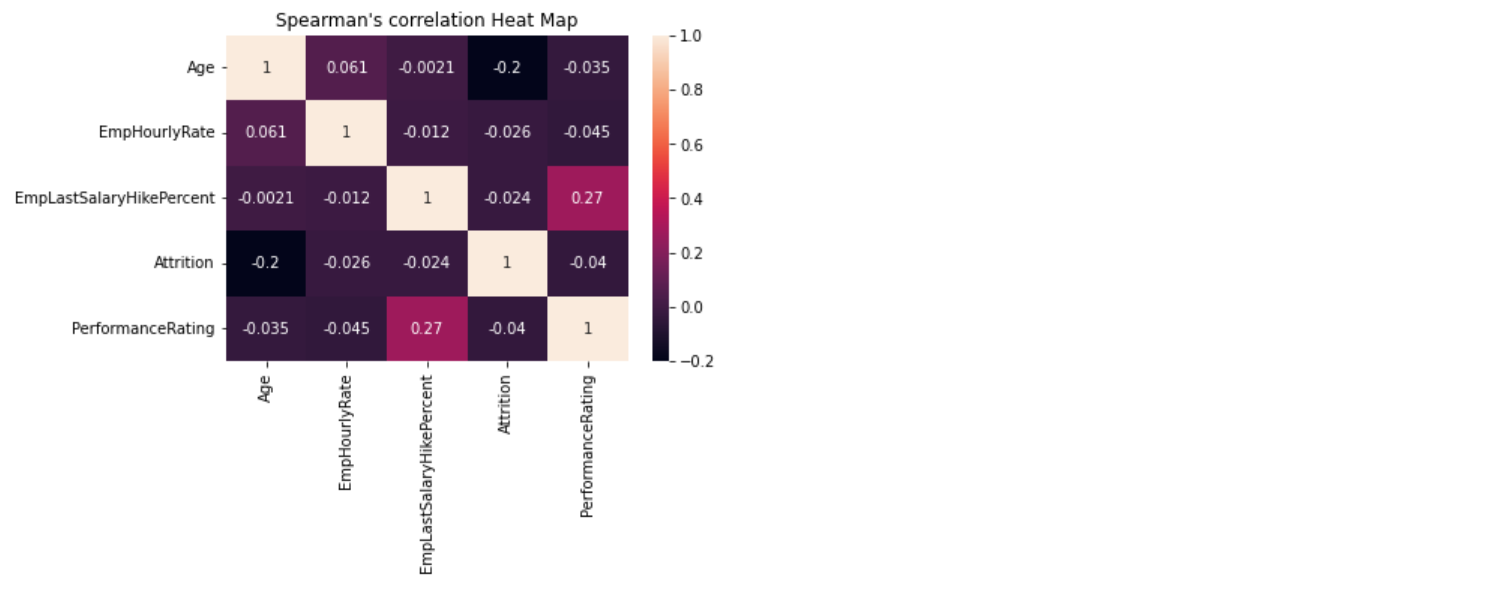 117 |
118 | ```python
119 | eda.kendallCorrelation()
120 | ```
121 |
117 |
118 | ```python
119 | eda.kendallCorrelation()
120 | ```
121 | 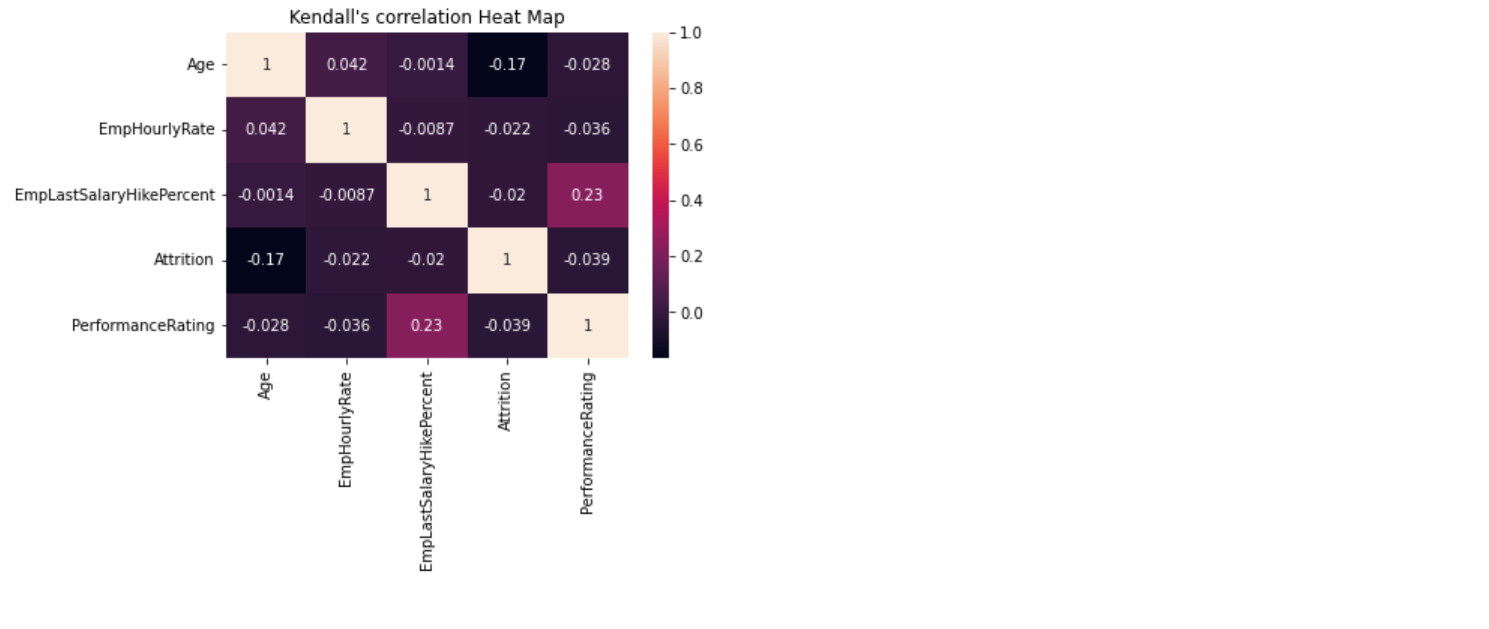 122 |
123 | ### 5. Missing Values
124 |
125 | ```python
126 | eda.missingValuesPlot(plot ='matrix')
127 | ```
128 |
122 |
123 | ### 5. Missing Values
124 |
125 | ```python
126 | eda.missingValuesPlot(plot ='matrix')
127 | ```
128 | 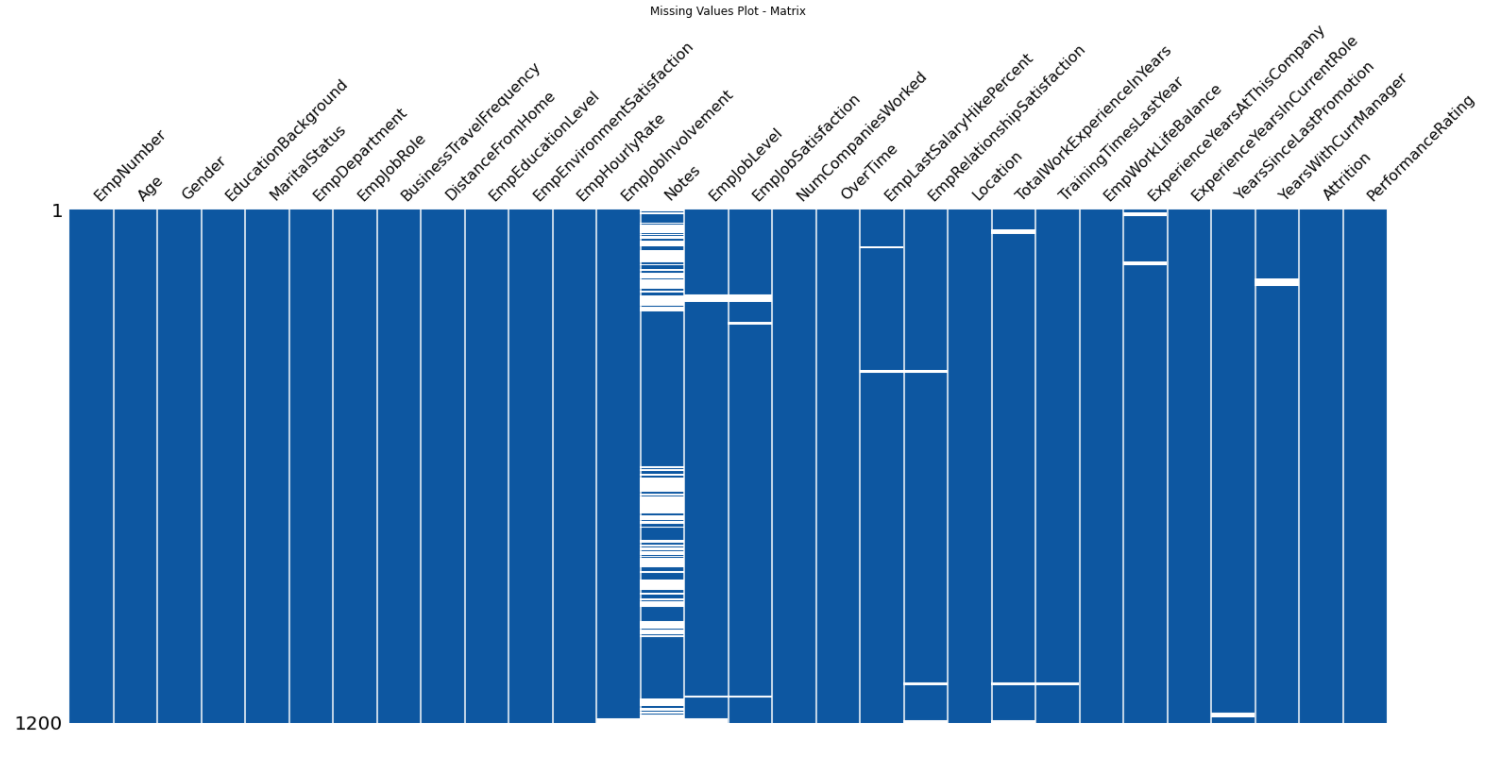 129 |
130 | ```python
131 | eda.missingValuesPlot(plot ='bar')
132 | ```
133 |
129 |
130 | ```python
131 | eda.missingValuesPlot(plot ='bar')
132 | ```
133 | 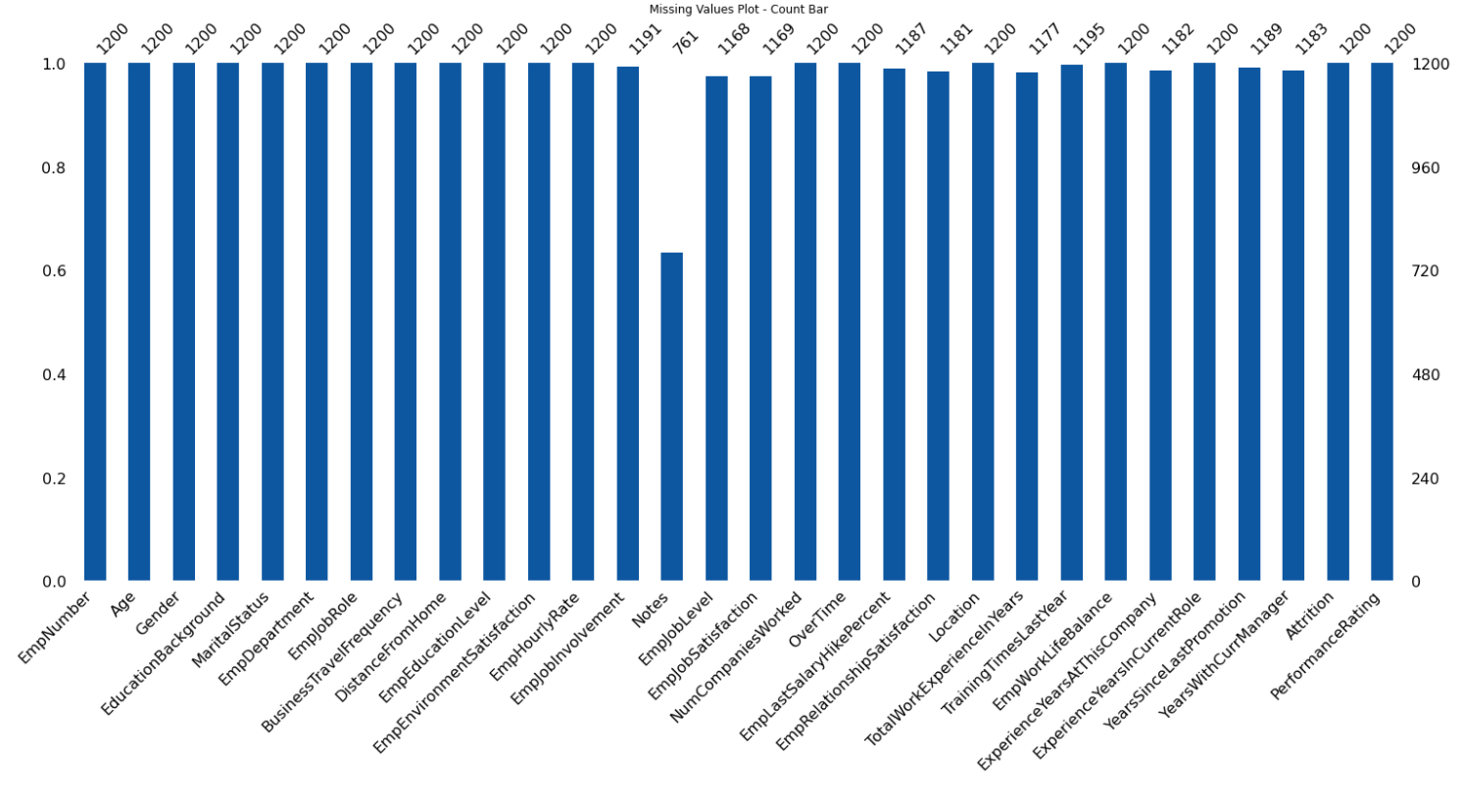 134 |
135 | ```python
136 | eda.missingValuesPlot(plot ='heatmap')
137 | ```
138 |
134 |
135 | ```python
136 | eda.missingValuesPlot(plot ='heatmap')
137 | ```
138 |  139 |
140 | ```python
141 | eda.missingValuesPlot(plot ='dendrogram')
142 | ```
143 |
139 |
140 | ```python
141 | eda.missingValuesPlot(plot ='dendrogram')
142 | ```
143 |  144 |
145 | ### 6. Outliers
146 |
147 | ```python
148 | eda.outliersVisualization(plot = 'box')
149 | ```
150 |
144 |
145 | ### 6. Outliers
146 |
147 | ```python
148 | eda.outliersVisualization(plot = 'box')
149 | ```
150 | 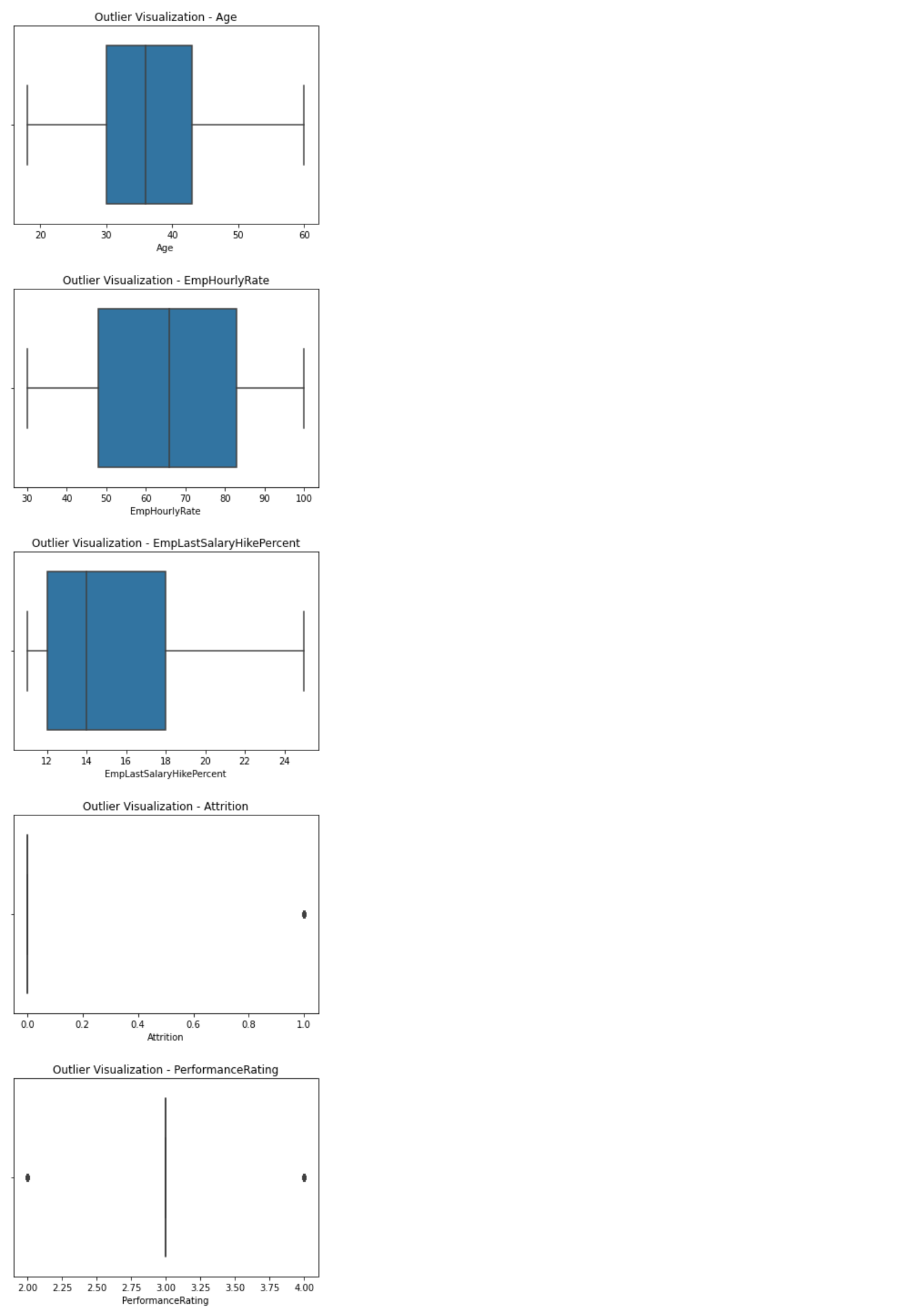 151 |
152 | ```python
153 | eda.outliersVisualization(plot = 'scatter')
154 | ```
155 |
151 |
152 | ```python
153 | eda.outliersVisualization(plot = 'scatter')
154 | ```
155 |  156 |
157 | ```python
158 | outliers = eda.getOutliers()
159 | ```
160 |
156 |
157 | ```python
158 | outliers = eda.getOutliers()
159 | ```
160 |  161 |
162 |
163 |
--------------------------------------------------------------------------------
/example_notebook/SampleNoteBook_edaSQL.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "ad11b57b",
6 | "metadata": {},
7 | "source": [
8 | "## Import Packages"
9 | ]
10 | },
11 | {
12 | "cell_type": "code",
13 | "execution_count": null,
14 | "id": "83529769",
15 | "metadata": {},
16 | "outputs": [],
17 | "source": [
18 | "import edaSQL\n",
19 | "import pandas as pd"
20 | ]
21 | },
22 | {
23 | "cell_type": "markdown",
24 | "id": "342b6675",
25 | "metadata": {},
26 | "source": [
27 | "
161 |
162 |
163 |
--------------------------------------------------------------------------------
/example_notebook/SampleNoteBook_edaSQL.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "ad11b57b",
6 | "metadata": {},
7 | "source": [
8 | "## Import Packages"
9 | ]
10 | },
11 | {
12 | "cell_type": "code",
13 | "execution_count": null,
14 | "id": "83529769",
15 | "metadata": {},
16 | "outputs": [],
17 | "source": [
18 | "import edaSQL\n",
19 | "import pandas as pd"
20 | ]
21 | },
22 | {
23 | "cell_type": "markdown",
24 | "id": "342b6675",
25 | "metadata": {},
26 | "source": [
27 | "
"
28 | ]
29 | },
30 | {
31 | "cell_type": "markdown",
32 | "id": "7842ea58",
33 | "metadata": {},
34 | "source": [
35 | "# 1. Connect to the DataBase"
36 | ]
37 | },
38 | {
39 | "cell_type": "code",
40 | "execution_count": null,
41 | "id": "368f6847",
42 | "metadata": {
43 | "scrolled": false
44 | },
45 | "outputs": [],
46 | "source": [
47 | "edasql = edaSQL.SQL(printAll=True)\n",
48 | "edasql.connectToDataBase(server='your server name', \n",
49 | " database='your database', \n",
50 | " user='username', \n",
51 | " password='password',\n",
52 | " sqlDriver='ODBC Driver 17 for SQL Server')\n"
53 | ]
54 | },
55 | {
56 | "cell_type": "markdown",
57 | "id": "01de9094",
58 | "metadata": {},
59 | "source": [
60 | "
"
61 | ]
62 | },
63 | {
64 | "cell_type": "markdown",
65 | "id": "e88ce7d1",
66 | "metadata": {},
67 | "source": [
68 | "# 2. Query Data "
69 | ]
70 | },
71 | {
72 | "cell_type": "code",
73 | "execution_count": null,
74 | "id": "01676089",
75 | "metadata": {},
76 | "outputs": [],
77 | "source": [
78 | "data = pd.read_sql(\"select * from INX\", edasql.dbConnection)"
79 | ]
80 | },
81 | {
82 | "cell_type": "code",
83 | "execution_count": null,
84 | "id": "f490c4f8",
85 | "metadata": {
86 | "scrolled": false
87 | },
88 | "outputs": [],
89 | "source": [
90 | "display(data.head())"
91 | ]
92 | },
93 | {
94 | "cell_type": "markdown",
95 | "id": "199c1796",
96 | "metadata": {},
97 | "source": [
98 | "
"
99 | ]
100 | },
101 | {
102 | "cell_type": "markdown",
103 | "id": "34836e46",
104 | "metadata": {},
105 | "source": [
106 | "# 3. Data Overview"
107 | ]
108 | },
109 | {
110 | "cell_type": "code",
111 | "execution_count": null,
112 | "id": "140fa6b0",
113 | "metadata": {},
114 | "outputs": [],
115 | "source": [
116 | "ins = edaSQL.EDA(dataFrame=data,HTMLDisplay=True)"
117 | ]
118 | },
119 | {
120 | "cell_type": "code",
121 | "execution_count": null,
122 | "id": "b5ac7e4d",
123 | "metadata": {},
124 | "outputs": [],
125 | "source": [
126 | "dataInsights =ins.dataInsights()\n",
127 | "# print(dataInsights)"
128 | ]
129 | },
130 | {
131 | "cell_type": "code",
132 | "execution_count": null,
133 | "id": "311bd16d",
134 | "metadata": {
135 | "scrolled": false
136 | },
137 | "outputs": [],
138 | "source": [
139 | "deepInsights = ins.deepInsights()"
140 | ]
141 | },
142 | {
143 | "cell_type": "markdown",
144 | "id": "bc8c9bd9",
145 | "metadata": {},
146 | "source": [
147 | "
"
148 | ]
149 | },
150 | {
151 | "cell_type": "markdown",
152 | "id": "c4e09807",
153 | "metadata": {},
154 | "source": [
155 | "# 4.Correlation"
156 | ]
157 | },
158 | {
159 | "cell_type": "code",
160 | "execution_count": null,
161 | "id": "7526aa90",
162 | "metadata": {},
163 | "outputs": [],
164 | "source": [
165 | "eda = edaSQL.EDA(dataFrame=data)"
166 | ]
167 | },
168 | {
169 | "cell_type": "markdown",
170 | "id": "a20ea377",
171 | "metadata": {},
172 | "source": [
173 | "## 4.1 Pearson Correlation"
174 | ]
175 | },
176 | {
177 | "cell_type": "code",
178 | "execution_count": null,
179 | "id": "8a4e29e3",
180 | "metadata": {},
181 | "outputs": [],
182 | "source": [
183 | "eda.pearsonCorrelation()"
184 | ]
185 | },
186 | {
187 | "cell_type": "markdown",
188 | "id": "93fe3213",
189 | "metadata": {},
190 | "source": [
191 | "## 4.2 Spearman Correlation"
192 | ]
193 | },
194 | {
195 | "cell_type": "code",
196 | "execution_count": null,
197 | "id": "1c17720f",
198 | "metadata": {},
199 | "outputs": [],
200 | "source": [
201 | "eda.spearmanCorrelation()"
202 | ]
203 | },
204 | {
205 | "cell_type": "markdown",
206 | "id": "65409fee",
207 | "metadata": {},
208 | "source": [
209 | "## 4.3 Kendall Correlation"
210 | ]
211 | },
212 | {
213 | "cell_type": "code",
214 | "execution_count": null,
215 | "id": "4037bf7b",
216 | "metadata": {},
217 | "outputs": [],
218 | "source": [
219 | "eda.kendallCorrelation()"
220 | ]
221 | },
222 | {
223 | "cell_type": "markdown",
224 | "id": "6deb6646",
225 | "metadata": {},
226 | "source": [
227 | "
"
228 | ]
229 | },
230 | {
231 | "cell_type": "markdown",
232 | "id": "46d21d8a",
233 | "metadata": {},
234 | "source": [
235 | "# 5. Missing Values"
236 | ]
237 | },
238 | {
239 | "cell_type": "markdown",
240 | "id": "55205766",
241 | "metadata": {},
242 | "source": [
243 | "## 5.1 Matrix Plot"
244 | ]
245 | },
246 | {
247 | "cell_type": "code",
248 | "execution_count": null,
249 | "id": "edf9ad2e",
250 | "metadata": {},
251 | "outputs": [],
252 | "source": [
253 | "eda.missingValuesPlot(plot ='matrix')"
254 | ]
255 | },
256 | {
257 | "cell_type": "markdown",
258 | "id": "e8e1d0b3",
259 | "metadata": {},
260 | "source": [
261 | "## 5.2 Count Bar Plot"
262 | ]
263 | },
264 | {
265 | "cell_type": "code",
266 | "execution_count": null,
267 | "id": "97e58073",
268 | "metadata": {},
269 | "outputs": [],
270 | "source": [
271 | "eda.missingValuesPlot(plot ='bar')"
272 | ]
273 | },
274 | {
275 | "cell_type": "markdown",
276 | "id": "e6175f2c",
277 | "metadata": {},
278 | "source": [
279 | "## 5.3 Heatmap Plot"
280 | ]
281 | },
282 | {
283 | "cell_type": "code",
284 | "execution_count": null,
285 | "id": "614e191c",
286 | "metadata": {},
287 | "outputs": [],
288 | "source": [
289 | "eda.missingValuesPlot(plot ='heatmap')"
290 | ]
291 | },
292 | {
293 | "cell_type": "markdown",
294 | "id": "84950b4a",
295 | "metadata": {},
296 | "source": [
297 | "## 5.4 Dendrogram Plot"
298 | ]
299 | },
300 | {
301 | "cell_type": "code",
302 | "execution_count": null,
303 | "id": "b746d285",
304 | "metadata": {},
305 | "outputs": [],
306 | "source": [
307 | "eda.missingValuesPlot(plot ='dendrogram')"
308 | ]
309 | },
310 | {
311 | "cell_type": "markdown",
312 | "id": "febfc343",
313 | "metadata": {},
314 | "source": [
315 | "
"
316 | ]
317 | },
318 | {
319 | "cell_type": "markdown",
320 | "id": "86e900f4",
321 | "metadata": {},
322 | "source": [
323 | "# 6. Outliers "
324 | ]
325 | },
326 | {
327 | "cell_type": "markdown",
328 | "id": "50770e59",
329 | "metadata": {},
330 | "source": [
331 | "## 6.1 Visualization"
332 | ]
333 | },
334 | {
335 | "cell_type": "markdown",
336 | "id": "759c458c",
337 | "metadata": {},
338 | "source": [
339 | "## 6.1.1 Box Plot"
340 | ]
341 | },
342 | {
343 | "cell_type": "code",
344 | "execution_count": null,
345 | "id": "a2df2234",
346 | "metadata": {},
347 | "outputs": [],
348 | "source": [
349 | "eda.outliersVisualization(plot = 'box')"
350 | ]
351 | },
352 | {
353 | "cell_type": "markdown",
354 | "id": "f8a5348c",
355 | "metadata": {},
356 | "source": [
357 | "## 6.1.2 Scatter Plot"
358 | ]
359 | },
360 | {
361 | "cell_type": "code",
362 | "execution_count": null,
363 | "id": "a6344961",
364 | "metadata": {
365 | "scrolled": false
366 | },
367 | "outputs": [],
368 | "source": [
369 | "eda.outliersVisualization(plot = 'scatter')"
370 | ]
371 | },
372 | {
373 | "cell_type": "markdown",
374 | "id": "74a25dbe",
375 | "metadata": {},
376 | "source": [
377 | "## 6.2 Get Outliers "
378 | ]
379 | },
380 | {
381 | "cell_type": "code",
382 | "execution_count": null,
383 | "id": "b9ef12de",
384 | "metadata": {
385 | "scrolled": false
386 | },
387 | "outputs": [],
388 | "source": [
389 | "outliers = eda.getOutliers()"
390 | ]
391 | },
392 | {
393 | "cell_type": "code",
394 | "execution_count": null,
395 | "id": "bcae93d9",
396 | "metadata": {},
397 | "outputs": [],
398 | "source": []
399 | }
400 | ],
401 | "metadata": {
402 | "kernelspec": {
403 | "display_name": "Python 3 (ipykernel)",
404 | "language": "python",
405 | "name": "python3"
406 | },
407 | "language_info": {
408 | "codemirror_mode": {
409 | "name": "ipython",
410 | "version": 3
411 | },
412 | "file_extension": ".py",
413 | "mimetype": "text/x-python",
414 | "name": "python",
415 | "nbconvert_exporter": "python",
416 | "pygments_lexer": "ipython3",
417 | "version": "3.8.12"
418 | }
419 | },
420 | "nbformat": 4,
421 | "nbformat_minor": 5
422 | }
423 |
--------------------------------------------------------------------------------
/edaSQL/src/edaSQL.py:
--------------------------------------------------------------------------------
1 | from IPython.core.display import display, HTML
2 | import pyodbc
3 | import numpy as np
4 | import seaborn as sb
5 | from matplotlib import pyplot as plt
6 | import missingno
7 | import warnings
8 | import pandas as pd
9 |
10 | warnings.filterwarnings('ignore')
11 |
12 |
13 | def isNaN(num):
14 | return num != num
15 |
16 |
17 | def findIfColumnsWithConstantValues(dataFrame: pd.core.frame.DataFrame) -> dict:
18 | """ This will return if there is any column with constant value present in all rows"""
19 |
20 | constantValues = {}
21 | for i in dataFrame.columns:
22 | uniqueValues = list(dataFrame[i].unique())
23 | if len(uniqueValues) == 1 and uniqueValues[0] not in [np.NAN, None, ''] and not isNaN(uniqueValues[0]):
24 | constantValues.update({i: uniqueValues[0]})
25 |
26 | return constantValues
27 |
28 |
29 | def findIfColumnsWithHighCardinality(dataFrame: pd.core.frame.DataFrame) -> dict:
30 | """ This will return if there is any column with High Cardinality.
31 | High-cardinality refers to columns with values that are very uncommon or unique"""
32 |
33 | cardinalityValues = {}
34 | for i in dataFrame.columns:
35 | uniqueValues = list(dataFrame[i].unique())
36 | if len(uniqueValues) > 100 and not len(uniqueValues) == len(dataFrame):
37 | cardinalityValues.update({i: str(len(uniqueValues))})
38 |
39 | return cardinalityValues
40 |
41 |
42 | def findIfColumnsWithUniqueIDs(dataFrame: pd.core.frame.DataFrame) -> list:
43 | """ This will return if there is any column with UniqueIDs"""
44 |
45 | IdColumns = []
46 | for i in dataFrame.columns:
47 | uniqueValues = list(dataFrame[i].unique())
48 | if len(uniqueValues) == len(dataFrame):
49 | IdColumns.append(i)
50 |
51 | return IdColumns
52 |
53 |
54 | def findMissingValuesPercentage(dataFrame: pd.core.frame.DataFrame) -> dict:
55 | """ This will return the percentage of missing values in each columns"""
56 |
57 | all_data_na = (dataFrame.isnull().sum() / len(dataFrame)) * 100
58 | all_data_na = all_data_na.drop(all_data_na[all_data_na == 0].index).sort_values(ascending=False)
59 | missing_data = pd.DataFrame({'Missing Ratio': all_data_na.apply(lambda x: str(round(x, 2)))}).to_dict()
60 | return missing_data['Missing Ratio']
61 |
62 |
63 | def findBinaryColumns(dataFrame: pd.core.frame.DataFrame) -> list:
64 | """ This will return the list of columns with Binary Values"""
65 |
66 | bool_cols = [col for col in dataFrame
67 | if np.isin(dataFrame[col].unique(), [0, 1]).all()]
68 | return bool_cols
69 |
70 |
71 | class SQL:
72 |
73 | def __init__(self, printAll=True):
74 | self.printAll = printAll
75 | self.server = self.database = self.user = self.password = self.sqlDriver = None
76 | self.cursor = self.dbConnection = None, None
77 | pass
78 |
79 | def connectToDataBase(self, server: str, user: str, password: str, database: str,
80 | sqlDriver='ODBC Driver 17 for SQL Server'):
81 | self.server = server
82 | self.database = database
83 | self.user = user
84 | self.password = password
85 | self.sqlDriver = sqlDriver
86 |
87 | self.dbConnection = pyodbc.connect(
88 | 'DRIVER={' + self.sqlDriver + '};SERVER='
89 | + self.server +
90 | ';DATABASE=' + self.database +
91 | ';UID=' + self.user +
92 | ';PWD=' + self.password)
93 |
94 | display(HTML(' '))
96 |
97 | if self.printAll:
98 | print('========== Connected to DataBase Successfully ===========')
99 | print('Server: ', self.server)
100 | print('DataBase: ', self.database)
101 | print('User : ', self.user)
102 | print('Password : ', self.password)
103 |
104 | self.cursor = self.dbConnection.cursor()
105 |
106 |
107 | class EDA:
108 |
109 | def __init__(self, dataFrame, HTMLDisplay=True):
110 | self.dataFrame = dataFrame
111 | self.HTMLDisplay = HTMLDisplay
112 | pass
113 |
114 | def getInfo(self):
115 | pass
116 |
117 | def dataInsights(self, printAll=True) -> dict:
118 | dataOverview = {
119 | 'Dataset Overview':
120 | {
121 | 'Number of Columns': str(len(self.dataFrame.columns)),
122 | 'Number of Rows': str(len(self.dataFrame)),
123 | 'Overall Missing cells': str(self.dataFrame.isnull().sum().sum()),
124 | 'Overall Missing cells (%)': str(
125 | round((self.dataFrame.isnull().sum().sum() / self.dataFrame.notnull().sum().sum()) * 100, 2)),
126 | 'Duplicate rows': str(self.dataFrame.duplicated().sum()),
127 | 'Duplicate rows (%)': str(round(self.dataFrame.duplicated().sum() / len(self.dataFrame) * 100, 2)),
128 | },
129 | 'Types of Columns':
130 | {
131 | 'Numeric': str(len(self.dataFrame.select_dtypes("number").columns)),
132 | 'Categorical': str(len(self.dataFrame.select_dtypes("object").columns)),
133 | 'Date and Time': str(len(self.dataFrame.select_dtypes("datetime64").columns))
134 | }
135 | }
136 |
137 | if self.HTMLDisplay and printAll:
138 | displayContent = """"""
139 | for parent, child in dataOverview.items():
140 | tempDisplay = ''
141 | tempDisplay += '\n\n\n\n\n\n\n
'))
96 |
97 | if self.printAll:
98 | print('========== Connected to DataBase Successfully ===========')
99 | print('Server: ', self.server)
100 | print('DataBase: ', self.database)
101 | print('User : ', self.user)
102 | print('Password : ', self.password)
103 |
104 | self.cursor = self.dbConnection.cursor()
105 |
106 |
107 | class EDA:
108 |
109 | def __init__(self, dataFrame, HTMLDisplay=True):
110 | self.dataFrame = dataFrame
111 | self.HTMLDisplay = HTMLDisplay
112 | pass
113 |
114 | def getInfo(self):
115 | pass
116 |
117 | def dataInsights(self, printAll=True) -> dict:
118 | dataOverview = {
119 | 'Dataset Overview':
120 | {
121 | 'Number of Columns': str(len(self.dataFrame.columns)),
122 | 'Number of Rows': str(len(self.dataFrame)),
123 | 'Overall Missing cells': str(self.dataFrame.isnull().sum().sum()),
124 | 'Overall Missing cells (%)': str(
125 | round((self.dataFrame.isnull().sum().sum() / self.dataFrame.notnull().sum().sum()) * 100, 2)),
126 | 'Duplicate rows': str(self.dataFrame.duplicated().sum()),
127 | 'Duplicate rows (%)': str(round(self.dataFrame.duplicated().sum() / len(self.dataFrame) * 100, 2)),
128 | },
129 | 'Types of Columns':
130 | {
131 | 'Numeric': str(len(self.dataFrame.select_dtypes("number").columns)),
132 | 'Categorical': str(len(self.dataFrame.select_dtypes("object").columns)),
133 | 'Date and Time': str(len(self.dataFrame.select_dtypes("datetime64").columns))
134 | }
135 | }
136 |
137 | if self.HTMLDisplay and printAll:
138 | displayContent = """"""
139 | for parent, child in dataOverview.items():
140 | tempDisplay = ''
141 | tempDisplay += '\n\n\n\n\n\n\n' + parent + '
'
146 | for cat, value in child.items():
147 | tempDisplay += '| ' + cat + ' : | ' + value + ' |
|---|
'
148 |
149 | tempDisplay += '
'
150 | displayContent += tempDisplay
151 |
152 | display(HTML(displayContent))
153 |
154 | else:
155 | printer = []
156 | for parent, child in dataOverview.items():
157 | printer.append(parent + '\n')
158 | for cat, value in child.items():
159 | printer.append(' ' + cat + ': ' + value + '\n')
160 |
161 | for pr in printer:
162 | print(pr)
163 |
164 | return dataOverview
165 |
166 | def deepInsights(self, printAll=True) -> dict:
167 | deepInsights = {
168 | "Columns with Constant Values": findIfColumnsWithConstantValues(self.dataFrame),
169 | "Columns with High Cardinality": findIfColumnsWithHighCardinality(self.dataFrame),
170 | "Columns with with Unique ID's": findIfColumnsWithUniqueIDs(self.dataFrame),
171 | "Columns with Missing Values Percentage": findMissingValuesPercentage(self.dataFrame),
172 | "Columns with Binary Values": findBinaryColumns(self.dataFrame)
173 | }
174 |
175 | mapper = {
176 | "Columns with Constant Values": ['Column', 'Value'],
177 | "Columns with High Cardinality": ['Column', 'No of Unique Values'],
178 | "Columns with Missing Values Percentage": ['Column', 'Percentage']
179 | }
180 |
181 | if self.HTMLDisplay and printAll:
182 | displayContent = """"""
183 | for parent, child in deepInsights.items():
184 | tempDisplay = ''
185 | tempDisplay += '\n\n\n\n\n\n\n' + parent + '
'
190 | if type(child) == dict:
191 | tempDisplay += '| ' + mapper[parent][0] + ' | ' + mapper[parent][1] + ' | '
192 | for cat, value in child.items():
193 | tempDisplay += '
|---|
| ' + cat + ' | ' + value + ' |
'
194 | else:
195 | for cols in child:
196 | tempDisplay += '| ' + cols + ' |
'
197 |
198 | tempDisplay += '
'
199 | displayContent += tempDisplay
200 |
201 | display(HTML(displayContent))
202 |
203 | else:
204 | printer = []
205 | for parent, child in deepInsights.items():
206 | printer.append(parent + '\n')
207 | if type(child) == dict:
208 | for cat, value in child.items():
209 | printer.append(' ' + cat + ': ' + value + '\n')
210 | else:
211 | for cols in child:
212 | printer.append(' ' + cols + '\n')
213 | for pr in printer:
214 | print(pr)
215 |
216 | return deepInsights
217 |
218 | def pearsonCorrelation(self, columns: list = None):
219 | """The Pearson correlation is also known as the “product moment correlation coefficient” (PMCC) or
220 | simply “correlation”. This will generate the Heat Map of Correlation between the Numerical Columns"""
221 |
222 | pearsonDataFrame = self.dataFrame.select_dtypes("number")
223 |
224 | corr = pearsonDataFrame.corr()
225 | sb.heatmap(corr, cmap="Blues", annot=True)
226 | plt.title("Pearson's correlation Heat Map")
227 |
228 | def spearmanCorrelation(self, columns: list = None):
229 | """In statistics, Spearman's rank correlation rank correlation coefficient or Spearman's ρ, named after Charles Spearman and
230 | often denoted by the Greek letter \rho or as r_{s}, is a nonparametric measure of rank correlation. It
231 | assesses how well the relationship between two variables can be described using a monotonic function. This
232 | will generate the Heat Map of Correlation between the Numerical Columns """
233 |
234 | spearmanDataFrame = self.dataFrame.select_dtypes("number")
235 |
236 | corr = spearmanDataFrame.corr(method='spearman')
237 | sb.heatmap(corr, annot=True)
238 | plt.title("Spearman's correlation Heat Map")
239 |
240 | def kendallCorrelation(self, columns: list = None):
241 | """Kendall's rank correlation provides a distribution free test of independence and a measure of the
242 | strength of dependence between two variables. This
243 | will generate the Heat Map of Correlation between the Numerical Columns """
244 |
245 | kendallDataFrame = self.dataFrame.select_dtypes("number")
246 |
247 | corr = kendallDataFrame.corr(method='kendall')
248 | sb.heatmap(corr, annot=True)
249 | plt.title("Kendall's correlation Heat Map")
250 |
251 | def missingValuesPlot(self, plot='matrix', color=(13, 87, 161)):
252 | color = (color[0] / 255, color[1] / 255, color[2] / 255)
253 | if plot == 'matrix':
254 | missingno.matrix(df=self.dataFrame, color=color, labels=True, sparkline=False)
255 | plt.title('Missing Values Plot - Matrix')
256 | if plot == 'bar':
257 | missingno.bar(df=self.dataFrame, color=color, labels=True)
258 | plt.title('Missing Values Plot - Count Bar')
259 | if plot == 'heatmap':
260 | missingno.heatmap(df=self.dataFrame, labels=True)
261 | plt.title('Missing Values Plot - HeatMap')
262 | if plot == 'dendrogram':
263 | missingno.dendrogram(df=self.dataFrame)
264 | plt.title('Missing Values Plot - Dendrogram')
265 |
266 | def outliersVisualization(self, plot='box'):
267 | if plot == 'box':
268 | for i in list(self.dataFrame.select_dtypes("number").columns):
269 | plt.figure()
270 | sb.boxplot(self.dataFrame[i])
271 | plt.title('Outlier Visualization - ' + str(i))
272 |
273 | if plot == 'scatter':
274 | for i in list(self.dataFrame.select_dtypes("number").columns):
275 | plt.figure()
276 | plt.scatter(range(len(self.dataFrame)), self.dataFrame[i])
277 | plt.title('Outlier Visualization - ' + str(i))
278 | plt.xlabel('Index')
279 | plt.ylabel('Values')
280 |
281 | def getOutliers(self, printAll=True) -> dict:
282 | outliersDict = {}
283 | for i in list(self.dataFrame.select_dtypes("number").columns):
284 | q_low = self.dataFrame[i].quantile(0.01)
285 | q_hi = self.dataFrame[i].quantile(0.99)
286 | filteredDf = self.dataFrame[(self.dataFrame[i] < q_hi) & (self.dataFrame[i] > q_low)]
287 | filteredDf1 = self.dataFrame.merge(filteredDf, indicator=True, how='outer').query(
288 | '_merge != "both"').drop('_merge', 1)
289 | outliers = filteredDf1[[i]]
290 | outliersDict.update({i: outliers})
291 | if printAll:
292 | display(HTML('' + i + '
'))
293 | display(outliers)
294 |
295 | return outliersDict
296 |
--------------------------------------------------------------------------------
 3 |
3 |  3 |
3 |  ](https://pypi.org/project/edaSQL/)
7 | [
](https://pypi.org/project/edaSQL/)
7 | [ ](https://edasql.readthedocs.io/en/latest/)
8 | [
](https://edasql.readthedocs.io/en/latest/)
8 | [ ](https://opensource.org/licenses/MIT)
9 |
](https://opensource.org/licenses/MIT)
9 |  10 |
10 |  11 |
11 |  12 |
12 |  13 |
14 | ## SQL Bridge Tool to Exploratory Data Analysis
15 |
16 |
17 | **edaSQL** is a library to link SQL to **Exploratory Data Analysis** and further more in the Data Engineering. This will solve many limitations in the SQL studios available in the market. Use the SQL Query language to get your Table Results.
18 |
19 | ## Installation
20 | Install dependency Packages before installing edaSQL
21 | ```shell
22 | pip install pyodbc
23 | pip install ipython
24 | ```
25 | Optional dependency for better visualization - [Jupyter Notebook](https://jupyter.org/install)
26 | ```shell
27 | pip install notebook
28 | ```
29 |
30 | **Now Install using pip** . [Offical Python Package Here!!](https://pypi.org/project/edaSQL/)
31 | ```shell
32 | pip install edaSQL
33 | ```
34 |
35 | (OR)
36 |
37 | Clone this Repository. Run this from the root directory to install
38 |
39 | ```shell
40 | python setup.py install
41 | ```
42 |
43 | ## Documentation
44 |
45 |
13 |
14 | ## SQL Bridge Tool to Exploratory Data Analysis
15 |
16 |
17 | **edaSQL** is a library to link SQL to **Exploratory Data Analysis** and further more in the Data Engineering. This will solve many limitations in the SQL studios available in the market. Use the SQL Query language to get your Table Results.
18 |
19 | ## Installation
20 | Install dependency Packages before installing edaSQL
21 | ```shell
22 | pip install pyodbc
23 | pip install ipython
24 | ```
25 | Optional dependency for better visualization - [Jupyter Notebook](https://jupyter.org/install)
26 | ```shell
27 | pip install notebook
28 | ```
29 |
30 | **Now Install using pip** . [Offical Python Package Here!!](https://pypi.org/project/edaSQL/)
31 | ```shell
32 | pip install edaSQL
33 | ```
34 |
35 | (OR)
36 |
37 | Clone this Repository. Run this from the root directory to install
38 |
39 | ```shell
40 | python setup.py install
41 | ```
42 |
43 | ## Documentation
44 |
45 |  46 |
47 | [Read the detailed documentation in readthedocs.io](https://edasql.readthedocs.io/en/latest/) (still under the development)
48 |
49 | ## License
50 | The license for edaSQL is MIT license
51 |
52 | ## Need help?
53 | Stuck on your edaSQL code or problem? Any other questions? Don't
54 | hestitate to send me an email (selva221724@gmail.com).
55 |
56 | ## edaSQL Jupyter NoteBook Tutorial
57 |
58 | Access the sample Jupyter Notebook [here!!](https://github.com/selva221724/edaSQL/blob/main/example_notebook/SampleNoteBook_edaSQL.ipynb)
59 |
60 | Access the Sample Data Used in this Repo
61 | - [CSV](https://github.com/selva221724/edaSQL/blob/main/sampleData/CSV/INX.csv)
62 | - [DataBase Backup](https://github.com/selva221724/edaSQL/blob/main/sampleData/DataBaseBackup/INX.bak) ( you can restore the DB in SQL Studio )
63 |
64 | **edaSQL for DataFrame:** If you are using the CSV or Excel as a source , Read using the Pandas & start from the [**3. Data Overview**](#Chapter1)
65 |
66 | ### Import Packages
67 | ```python
68 | import edaSQL
69 | import pandas as pd
70 | ```
71 |
72 | ### 1. Connect to the DataBase
73 | ```python
74 | edasql = edaSQL.SQL()
75 | edasql.connectToDataBase(server='your server name',
76 | database='your database',
77 | user='username',
78 | password='password',
79 | sqlDriver='ODBC Driver 17 for SQL Server')
80 | ```
81 |
82 |
46 |
47 | [Read the detailed documentation in readthedocs.io](https://edasql.readthedocs.io/en/latest/) (still under the development)
48 |
49 | ## License
50 | The license for edaSQL is MIT license
51 |
52 | ## Need help?
53 | Stuck on your edaSQL code or problem? Any other questions? Don't
54 | hestitate to send me an email (selva221724@gmail.com).
55 |
56 | ## edaSQL Jupyter NoteBook Tutorial
57 |
58 | Access the sample Jupyter Notebook [here!!](https://github.com/selva221724/edaSQL/blob/main/example_notebook/SampleNoteBook_edaSQL.ipynb)
59 |
60 | Access the Sample Data Used in this Repo
61 | - [CSV](https://github.com/selva221724/edaSQL/blob/main/sampleData/CSV/INX.csv)
62 | - [DataBase Backup](https://github.com/selva221724/edaSQL/blob/main/sampleData/DataBaseBackup/INX.bak) ( you can restore the DB in SQL Studio )
63 |
64 | **edaSQL for DataFrame:** If you are using the CSV or Excel as a source , Read using the Pandas & start from the [**3. Data Overview**](#Chapter1)
65 |
66 | ### Import Packages
67 | ```python
68 | import edaSQL
69 | import pandas as pd
70 | ```
71 |
72 | ### 1. Connect to the DataBase
73 | ```python
74 | edasql = edaSQL.SQL()
75 | edasql.connectToDataBase(server='your server name',
76 | database='your database',
77 | user='username',
78 | password='password',
79 | sqlDriver='ODBC Driver 17 for SQL Server')
80 | ```
81 |
82 |  83 |
84 | ### 2. Query Data
85 | ```python
86 | sampleQuery = "select * from INX"
87 | data = pd.read_sql(sampleQuery, edasql.dbConnection)
88 | ```
89 |
83 |
84 | ### 2. Query Data
85 | ```python
86 | sampleQuery = "select * from INX"
87 | data = pd.read_sql(sampleQuery, edasql.dbConnection)
88 | ```
89 |  90 |
91 |
92 |
93 | ### 3. Data Overview
94 | ```python
95 | insights = edaSQL.EDA(dataFrame=data,HTMLDisplay=True)
96 | dataInsights =insights.dataInsights()
97 | ```
98 |
99 |
90 |
91 |
92 |
93 | ### 3. Data Overview
94 | ```python
95 | insights = edaSQL.EDA(dataFrame=data,HTMLDisplay=True)
96 | dataInsights =insights.dataInsights()
97 | ```
98 |
99 |  100 |
101 | ```python
102 | deepInsights = insights.deepInsights()
103 | ```
104 |
100 |
101 | ```python
102 | deepInsights = insights.deepInsights()
103 | ```
104 |  105 |
106 | ### 4. Correlation
107 | ```python
108 | eda = edaSQL.EDA(dataFrame=data)
109 | eda.pearsonCorrelation()
110 | ```
111 |
105 |
106 | ### 4. Correlation
107 | ```python
108 | eda = edaSQL.EDA(dataFrame=data)
109 | eda.pearsonCorrelation()
110 | ```
111 |  112 |
113 | ```python
114 | eda.spearmanCorrelation()
115 | ```
116 |
112 |
113 | ```python
114 | eda.spearmanCorrelation()
115 | ```
116 |  117 |
118 | ```python
119 | eda.kendallCorrelation()
120 | ```
121 |
117 |
118 | ```python
119 | eda.kendallCorrelation()
120 | ```
121 |  122 |
123 | ### 5. Missing Values
124 |
125 | ```python
126 | eda.missingValuesPlot(plot ='matrix')
127 | ```
128 |
122 |
123 | ### 5. Missing Values
124 |
125 | ```python
126 | eda.missingValuesPlot(plot ='matrix')
127 | ```
128 |  129 |
130 | ```python
131 | eda.missingValuesPlot(plot ='bar')
132 | ```
133 |
129 |
130 | ```python
131 | eda.missingValuesPlot(plot ='bar')
132 | ```
133 |  134 |
135 | ```python
136 | eda.missingValuesPlot(plot ='heatmap')
137 | ```
138 |
134 |
135 | ```python
136 | eda.missingValuesPlot(plot ='heatmap')
137 | ```
138 |  139 |
140 | ```python
141 | eda.missingValuesPlot(plot ='dendrogram')
142 | ```
143 |
139 |
140 | ```python
141 | eda.missingValuesPlot(plot ='dendrogram')
142 | ```
143 |  144 |
145 | ### 6. Outliers
146 |
147 | ```python
148 | eda.outliersVisualization(plot = 'box')
149 | ```
150 |
144 |
145 | ### 6. Outliers
146 |
147 | ```python
148 | eda.outliersVisualization(plot = 'box')
149 | ```
150 |  151 |
152 | ```python
153 | eda.outliersVisualization(plot = 'scatter')
154 | ```
155 |
151 |
152 | ```python
153 | eda.outliersVisualization(plot = 'scatter')
154 | ```
155 |  156 |
157 | ```python
158 | outliers = eda.getOutliers()
159 | ```
160 |
156 |
157 | ```python
158 | outliers = eda.getOutliers()
159 | ```
160 |  161 |
162 |
163 |
--------------------------------------------------------------------------------
/example_notebook/SampleNoteBook_edaSQL.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "ad11b57b",
6 | "metadata": {},
7 | "source": [
8 | "## Import Packages"
9 | ]
10 | },
11 | {
12 | "cell_type": "code",
13 | "execution_count": null,
14 | "id": "83529769",
15 | "metadata": {},
16 | "outputs": [],
17 | "source": [
18 | "import edaSQL\n",
19 | "import pandas as pd"
20 | ]
21 | },
22 | {
23 | "cell_type": "markdown",
24 | "id": "342b6675",
25 | "metadata": {},
26 | "source": [
27 | "
161 |
162 |
163 |
--------------------------------------------------------------------------------
/example_notebook/SampleNoteBook_edaSQL.ipynb:
--------------------------------------------------------------------------------
1 | {
2 | "cells": [
3 | {
4 | "cell_type": "markdown",
5 | "id": "ad11b57b",
6 | "metadata": {},
7 | "source": [
8 | "## Import Packages"
9 | ]
10 | },
11 | {
12 | "cell_type": "code",
13 | "execution_count": null,
14 | "id": "83529769",
15 | "metadata": {},
16 | "outputs": [],
17 | "source": [
18 | "import edaSQL\n",
19 | "import pandas as pd"
20 | ]
21 | },
22 | {
23 | "cell_type": "markdown",
24 | "id": "342b6675",
25 | "metadata": {},
26 | "source": [
27 | " '))
96 |
97 | if self.printAll:
98 | print('========== Connected to DataBase Successfully ===========')
99 | print('Server: ', self.server)
100 | print('DataBase: ', self.database)
101 | print('User : ', self.user)
102 | print('Password : ', self.password)
103 |
104 | self.cursor = self.dbConnection.cursor()
105 |
106 |
107 | class EDA:
108 |
109 | def __init__(self, dataFrame, HTMLDisplay=True):
110 | self.dataFrame = dataFrame
111 | self.HTMLDisplay = HTMLDisplay
112 | pass
113 |
114 | def getInfo(self):
115 | pass
116 |
117 | def dataInsights(self, printAll=True) -> dict:
118 | dataOverview = {
119 | 'Dataset Overview':
120 | {
121 | 'Number of Columns': str(len(self.dataFrame.columns)),
122 | 'Number of Rows': str(len(self.dataFrame)),
123 | 'Overall Missing cells': str(self.dataFrame.isnull().sum().sum()),
124 | 'Overall Missing cells (%)': str(
125 | round((self.dataFrame.isnull().sum().sum() / self.dataFrame.notnull().sum().sum()) * 100, 2)),
126 | 'Duplicate rows': str(self.dataFrame.duplicated().sum()),
127 | 'Duplicate rows (%)': str(round(self.dataFrame.duplicated().sum() / len(self.dataFrame) * 100, 2)),
128 | },
129 | 'Types of Columns':
130 | {
131 | 'Numeric': str(len(self.dataFrame.select_dtypes("number").columns)),
132 | 'Categorical': str(len(self.dataFrame.select_dtypes("object").columns)),
133 | 'Date and Time': str(len(self.dataFrame.select_dtypes("datetime64").columns))
134 | }
135 | }
136 |
137 | if self.HTMLDisplay and printAll:
138 | displayContent = """"""
139 | for parent, child in dataOverview.items():
140 | tempDisplay = ''
141 | tempDisplay += '\n\n\n\n\n\n\n
'))
96 |
97 | if self.printAll:
98 | print('========== Connected to DataBase Successfully ===========')
99 | print('Server: ', self.server)
100 | print('DataBase: ', self.database)
101 | print('User : ', self.user)
102 | print('Password : ', self.password)
103 |
104 | self.cursor = self.dbConnection.cursor()
105 |
106 |
107 | class EDA:
108 |
109 | def __init__(self, dataFrame, HTMLDisplay=True):
110 | self.dataFrame = dataFrame
111 | self.HTMLDisplay = HTMLDisplay
112 | pass
113 |
114 | def getInfo(self):
115 | pass
116 |
117 | def dataInsights(self, printAll=True) -> dict:
118 | dataOverview = {
119 | 'Dataset Overview':
120 | {
121 | 'Number of Columns': str(len(self.dataFrame.columns)),
122 | 'Number of Rows': str(len(self.dataFrame)),
123 | 'Overall Missing cells': str(self.dataFrame.isnull().sum().sum()),
124 | 'Overall Missing cells (%)': str(
125 | round((self.dataFrame.isnull().sum().sum() / self.dataFrame.notnull().sum().sum()) * 100, 2)),
126 | 'Duplicate rows': str(self.dataFrame.duplicated().sum()),
127 | 'Duplicate rows (%)': str(round(self.dataFrame.duplicated().sum() / len(self.dataFrame) * 100, 2)),
128 | },
129 | 'Types of Columns':
130 | {
131 | 'Numeric': str(len(self.dataFrame.select_dtypes("number").columns)),
132 | 'Categorical': str(len(self.dataFrame.select_dtypes("object").columns)),
133 | 'Date and Time': str(len(self.dataFrame.select_dtypes("datetime64").columns))
134 | }
135 | }
136 |
137 | if self.HTMLDisplay and printAll:
138 | displayContent = """"""
139 | for parent, child in dataOverview.items():
140 | tempDisplay = ''
141 | tempDisplay += '\n\n\n\n\n\n\n