├── README.md
├── datasets
├── IMAGES.mat
├── IMAGES_RAW.mat
└── Sparse net.url
├── ica.py
├── network.py
├── results
├── ICA.png
├── PCA.png
├── RF.png
├── RF_cauchy_thresholding.png
└── error.png
├── sparse-coding.ipynb
├── sparse_coding.ipynb
└── train.py
/README.md:
--------------------------------------------------------------------------------
1 | # Sparse Coding (Olshausen & Field, 1996) Model
2 | Sparse Coding (Olshausen & Field, 1996) Model with **LCA** (locally competitive algorithm) ([Rozell et al., *Neural Comput*. 2008](https://www.ece.rice.edu/~eld1/papers/Rozell08.pdf)).
3 |
4 | Data is from .
5 |
6 | ## Requirement
7 | - `Python >= 3.5`
8 | - `numpy`, `matplotlib`, `scipy`, `tqdm`, `sklearn`
9 |
10 | ## Usage
11 | - Run `train.py` or `predictive-coding.ipynb` (written in Japanese).
12 | - `ica.py` is implementation of ICA and PCA for Natural images.
13 |
14 | ## Results
15 | ### Loss function
16 | 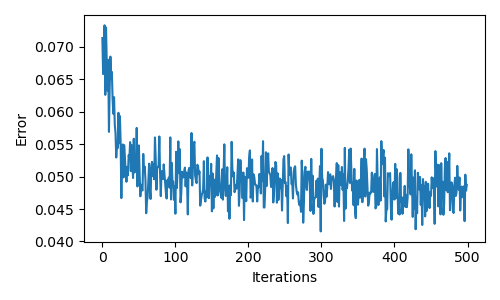 17 |
18 | ### Receptive fields (using soft threshold function)
19 |
17 |
18 | ### Receptive fields (using soft threshold function)
19 | 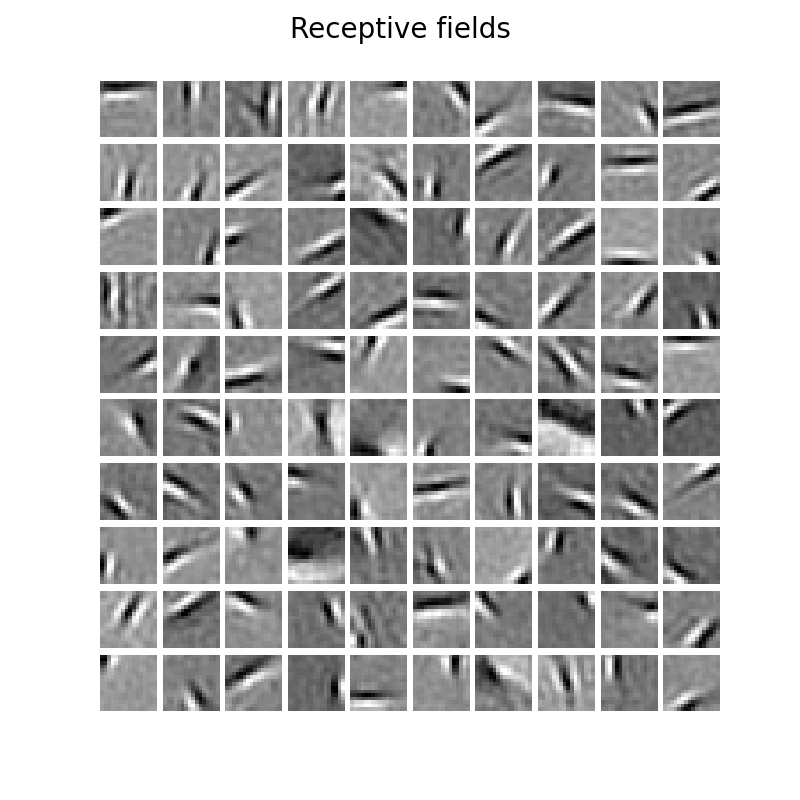 20 |
21 | ### Receptive fields (using Cauchy threshold function (Mayo et al., 2020))
22 |
20 |
21 | ### Receptive fields (using Cauchy threshold function (Mayo et al., 2020))
22 | 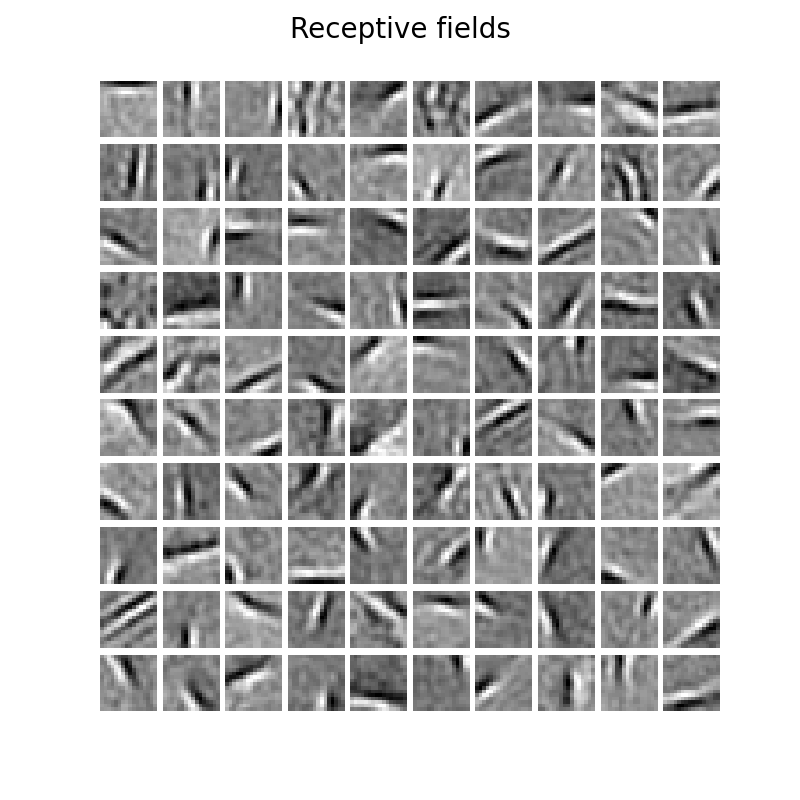 23 |
24 | ### ICA
25 |
23 |
24 | ### ICA
25 | 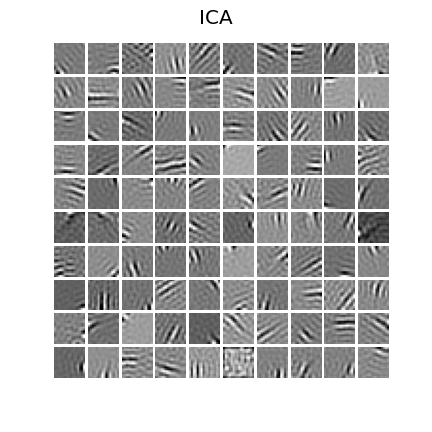 26 |
27 | ### PCA
28 |
26 |
27 | ### PCA
28 | 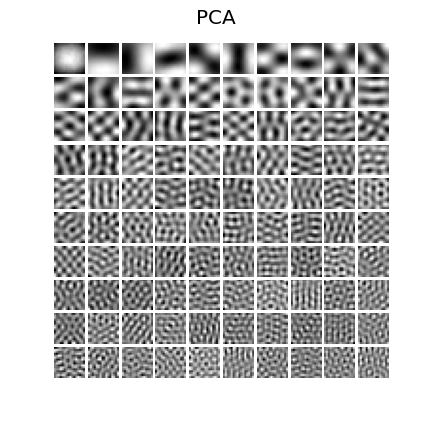 29 |
30 | ## Reference
31 | - Olshausen BA, Field DJ. [Emergence of simple-cell receptive field properties by learning a sparse code for natural images](https://www.nature.com/articles/381607a0). *Nature*. 1996;381(6583):607–609. [Data and Code](http://www.rctn.org/bruno/sparsenet/), [pdf](https://courses.cs.washington.edu/courses/cse528/11sp/Olshausen-nature-paper.pdf)
32 | - Rozell CJ, Johnson DH, Baraniuk RG, Olshausen BA. [Sparse coding via thresholding and local competition in neural circuits](http://www.mit.edu/~9.54/fall14/Classes/class07/Palm.pdf). *Neural Comput*. 2008;20(10):2526‐2563.
33 | - Mayo P, Holmes R, Achim A. [Iterative Cauchy Thresholding: Regularisation with a heavy-tailed prior](https://arxiv.org/abs/2003.12507). arXiv. 2020.
--------------------------------------------------------------------------------
/datasets/IMAGES.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES.mat
--------------------------------------------------------------------------------
/datasets/IMAGES_RAW.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES_RAW.mat
--------------------------------------------------------------------------------
/datasets/Sparse net.url:
--------------------------------------------------------------------------------
1 | [InternetShortcut]
2 | URL=http://www.rctn.org/bruno/sparsenet/
3 |
--------------------------------------------------------------------------------
/ica.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | from sklearn.decomposition import FastICA, PCA
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | # datasets from http://www.rctn.org/bruno/sparsenet/
10 | # mat_images = sio.loadmat('datasets/IMAGES.mat')
11 | # imgs = mat_images['IMAGES']
12 | mat_images_raw = sio.loadmat('datasets/IMAGES_RAW.mat')
13 | imgs_raw = mat_images_raw['IMAGESr']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs_raw.shape
17 |
18 | num_patches = 15000
19 | patchs_list = []
20 | w, h = 16, 16 # patch size
21 |
22 | # generate patches
23 | for i in tqdm(range(num_patches)):
24 | i = np.random.randint(0, num_images)
25 | # Get the coordinates of the upper left corner of clopping image randomly.
26 | beginx = np.random.randint(0, W-w-1)
27 | beginy = np.random.randint(0, H-h-1)
28 | img_clopped = imgs_raw[beginy:beginy+h, beginx:beginx+w, i]
29 | patchs_list.append(img_clopped.flatten())
30 |
31 | patches = np.array(patchs_list)
32 |
33 | # perform ICA
34 | print("perform ICA")
35 | n_comp = 100
36 | ica = FastICA(n_components=n_comp)
37 | ica.fit(patches)
38 | ica_filters = ica.components_
39 |
40 | # plot filters
41 | plt.figure(figsize=(6,6))
42 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
43 | for i in tqdm(range(n_comp)):
44 | plt.subplot(10, 10, i+1)
45 | plt.imshow(np.reshape(ica_filters[i], (w, h)), cmap="gray")
46 | plt.axis("off")
47 | plt.suptitle("ICA", fontsize=20)
48 | plt.subplots_adjust(top=0.9)
49 | plt.savefig("ICA.png")
50 | plt.show()
51 |
52 | # perform PCA
53 | print("perform PCA")
54 | pca = PCA(n_components=n_comp)
55 | pca.fit(patches)
56 | pca_filters = pca.components_
57 |
58 | # plot filters
59 | plt.figure(figsize=(6,6))
60 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
61 | for i in tqdm(range(n_comp)):
62 | plt.subplot(10, 10, i+1)
63 | plt.imshow(np.reshape(pca_filters[i], (w, h)), cmap="gray")
64 | plt.axis("off")
65 | plt.suptitle("PCA", fontsize=20)
66 | plt.subplots_adjust(top=0.9)
67 | plt.savefig("PCA.png")
68 | plt.show()
69 |
70 |
--------------------------------------------------------------------------------
/network.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 |
5 | class OlshausenField1996Model:
6 | def __init__(self, num_inputs, num_units, batch_size,
7 | lr_r=1e-2, lr_Phi=1e-2, lmda=5e-3):
8 | self.lr_r = lr_r # learning rate of r
9 | self.lr_Phi = lr_Phi # learning rate of Phi
10 | self.lmda = lmda # regularization parameter
11 |
12 | self.num_inputs = num_inputs
13 | self.num_units = num_units
14 | self.batch_size = batch_size
15 |
16 | # Weights
17 | Phi = np.random.randn(self.num_inputs, self.num_units).astype(np.float32)
18 | self.Phi = Phi * np.sqrt(1/self.num_units)
19 |
20 | # activity of neurons
21 | self.r = np.zeros((self.batch_size, self.num_units))
22 |
23 | def initialize_states(self):
24 | self.r = np.zeros((self.batch_size, self.num_units))
25 |
26 | def normalize_rows(self):
27 | self.Phi = self.Phi / np.maximum(np.linalg.norm(self.Phi, ord=2, axis=0, keepdims=True), 1e-8)
28 |
29 | # thresholding function of S(x)=|x|
30 | def soft_thresholding_func(self, x, lmda):
31 | return np.maximum(x - lmda, 0) - np.maximum(-x - lmda, 0)
32 |
33 | # thresholding function of S(x)=ln(1+x^2)
34 | def ln_thresholding_func(self, x, lmda):
35 | f = 9*lmda*x - 2*np.power(x, 3) - 18*x
36 | g = 3*lmda - np.square(x) + 3
37 | h = np.cbrt(np.sqrt(np.square(f) + 4*np.power(g, 3)) + f)
38 | two_croot = np.cbrt(2) # cubic root of two
39 | return (1/3)*(x - h / two_croot + two_croot*g / (1e-8+h))
40 |
41 | # thresholding function https://arxiv.org/abs/2003.12507
42 | def cauchy_thresholding_func(self, x, lmda):
43 | f = 0.5*(x + np.sqrt(np.maximum(x**2 - lmda,0)))
44 | g = 0.5*(x - np.sqrt(np.maximum(x**2 - lmda,0)))
45 | return f*(x>=lmda) + g*(x<=-lmda)
46 |
47 | def calculate_total_error(self, error):
48 | recon_error = np.mean(error**2)
49 | sparsity_r = self.lmda*np.mean(np.abs(self.r))
50 | return recon_error + sparsity_r
51 |
52 | def __call__(self, inputs, training=True):
53 | # Updates
54 | error = inputs - self.r @ self.Phi.T
55 |
56 | r = self.r + self.lr_r * error @ self.Phi
57 | self.r = self.soft_thresholding_func(r, self.lmda)
58 | #self.r = self.cauchy_thresholding_func(r, self.lmda)
59 |
60 | if training:
61 | error = inputs - self.r @ self.Phi.T
62 | dPhi = error.T @ self.r
63 | self.Phi += self.lr_Phi * dPhi
64 |
65 | return error, self.r
66 |
--------------------------------------------------------------------------------
/results/ICA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/ICA.png
--------------------------------------------------------------------------------
/results/PCA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/PCA.png
--------------------------------------------------------------------------------
/results/RF.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF.png
--------------------------------------------------------------------------------
/results/RF_cauchy_thresholding.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF_cauchy_thresholding.png
--------------------------------------------------------------------------------
/results/error.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/error.png
--------------------------------------------------------------------------------
/train.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | import network
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | np.random.seed(0)
10 |
11 | # datasets from http://www.rctn.org/bruno/sparsenet/
12 | mat_images = sio.loadmat('datasets/IMAGES.mat')
13 | imgs = mat_images['IMAGES']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs.shape
17 | num_iter = 500 # number of iterations
18 | nt_max = 1000 # Maximum number of simulation time
19 | batch_size = 250 # Batch size
20 |
21 | sz = 16 # image patch size
22 | num_units = 100 # number of neurons (units)
23 |
24 | eps = 1e-2 # small value which determines convergence
25 | error_list = [] # List to save errors
26 |
27 | # Define model
28 | model = network.OlshausenField1996Model(num_inputs=sz**2, num_units=num_units,
29 | batch_size=batch_size)
30 |
31 | # Run simulation
32 | for iter_ in tqdm(range(num_iter)):
33 | # Get the coordinates of the upper left corner of clopping image randomly.
34 | beginx = np.random.randint(0, W-sz, batch_size)
35 | beginy = np.random.randint(0, H-sz, batch_size)
36 |
37 | inputs_list = []
38 |
39 | # Get images randomly
40 | for i in range(batch_size):
41 | idx = np.random.randint(0, num_images)
42 | img = imgs[:, :, idx]
43 | clop = img[beginy[i]:beginy[i]+sz, beginx[i]:beginx[i]+sz].flatten()
44 | inputs_list.append(clop - np.mean(clop))
45 |
46 | inputs = np.array(inputs_list) # Input image patches
47 |
48 | model.initialize_states() # Reset states
49 | model.normalize_rows() # Normalize weights
50 |
51 | # Input an image patch until latent variables are converged
52 | r_tm1 = model.r # set previous r (t minus 1)
53 |
54 | for t in range(nt_max):
55 | # Update r without update weights
56 | error, r = model(inputs, training=False)
57 | dr = r - r_tm1
58 |
59 | # Compute norm of r
60 | dr_norm = np.linalg.norm(dr, ord=2) / (eps + np.linalg.norm(r_tm1, ord=2))
61 | r_tm1 = r # update r_tm1
62 |
63 | # Check convergence of r, then update weights
64 | if dr_norm < eps:
65 | error, r = model(inputs, training=True)
66 | break
67 |
68 | # If failure to convergence, break and print error
69 | if t >= nt_max-2:
70 | print("Error at patch:", iter_)
71 | print(dr_norm)
72 | break
73 |
74 | error_list.append(model.calculate_total_error(error)) # Append errors
75 |
76 | # Print moving average error
77 | if iter_ % 100 == 99:
78 | print("\n iter: "+str(iter_+1)+"/"+str(num_iter)+", Moving error:",
79 | np.mean(error_list[iter_-99:iter_]))
80 |
81 | # Plot error
82 | plt.figure(figsize=(5, 3))
83 | plt.ylabel("Error")
84 | plt.xlabel("Iterations")
85 | plt.plot(np.arange(len(error_list)), np.array(error_list))
86 | plt.tight_layout()
87 | plt.savefig("error.png")
88 | plt.show()
89 |
90 | # Plot Receptive fields
91 | fig = plt.figure(figsize=(8, 8))
92 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
93 | for i in tqdm(range(num_units)):
94 | plt.subplot(10, 10, i+1)
95 | plt.imshow(np.reshape(model.Phi[:, i], (sz, sz)), cmap="gray")
96 | plt.axis("off")
97 |
98 | fig.suptitle("Receptive fields", fontsize=20)
99 | plt.subplots_adjust(top=0.9)
100 | plt.savefig("RF.png")
101 | plt.show()
--------------------------------------------------------------------------------
29 |
30 | ## Reference
31 | - Olshausen BA, Field DJ. [Emergence of simple-cell receptive field properties by learning a sparse code for natural images](https://www.nature.com/articles/381607a0). *Nature*. 1996;381(6583):607–609. [Data and Code](http://www.rctn.org/bruno/sparsenet/), [pdf](https://courses.cs.washington.edu/courses/cse528/11sp/Olshausen-nature-paper.pdf)
32 | - Rozell CJ, Johnson DH, Baraniuk RG, Olshausen BA. [Sparse coding via thresholding and local competition in neural circuits](http://www.mit.edu/~9.54/fall14/Classes/class07/Palm.pdf). *Neural Comput*. 2008;20(10):2526‐2563.
33 | - Mayo P, Holmes R, Achim A. [Iterative Cauchy Thresholding: Regularisation with a heavy-tailed prior](https://arxiv.org/abs/2003.12507). arXiv. 2020.
--------------------------------------------------------------------------------
/datasets/IMAGES.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES.mat
--------------------------------------------------------------------------------
/datasets/IMAGES_RAW.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES_RAW.mat
--------------------------------------------------------------------------------
/datasets/Sparse net.url:
--------------------------------------------------------------------------------
1 | [InternetShortcut]
2 | URL=http://www.rctn.org/bruno/sparsenet/
3 |
--------------------------------------------------------------------------------
/ica.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | from sklearn.decomposition import FastICA, PCA
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | # datasets from http://www.rctn.org/bruno/sparsenet/
10 | # mat_images = sio.loadmat('datasets/IMAGES.mat')
11 | # imgs = mat_images['IMAGES']
12 | mat_images_raw = sio.loadmat('datasets/IMAGES_RAW.mat')
13 | imgs_raw = mat_images_raw['IMAGESr']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs_raw.shape
17 |
18 | num_patches = 15000
19 | patchs_list = []
20 | w, h = 16, 16 # patch size
21 |
22 | # generate patches
23 | for i in tqdm(range(num_patches)):
24 | i = np.random.randint(0, num_images)
25 | # Get the coordinates of the upper left corner of clopping image randomly.
26 | beginx = np.random.randint(0, W-w-1)
27 | beginy = np.random.randint(0, H-h-1)
28 | img_clopped = imgs_raw[beginy:beginy+h, beginx:beginx+w, i]
29 | patchs_list.append(img_clopped.flatten())
30 |
31 | patches = np.array(patchs_list)
32 |
33 | # perform ICA
34 | print("perform ICA")
35 | n_comp = 100
36 | ica = FastICA(n_components=n_comp)
37 | ica.fit(patches)
38 | ica_filters = ica.components_
39 |
40 | # plot filters
41 | plt.figure(figsize=(6,6))
42 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
43 | for i in tqdm(range(n_comp)):
44 | plt.subplot(10, 10, i+1)
45 | plt.imshow(np.reshape(ica_filters[i], (w, h)), cmap="gray")
46 | plt.axis("off")
47 | plt.suptitle("ICA", fontsize=20)
48 | plt.subplots_adjust(top=0.9)
49 | plt.savefig("ICA.png")
50 | plt.show()
51 |
52 | # perform PCA
53 | print("perform PCA")
54 | pca = PCA(n_components=n_comp)
55 | pca.fit(patches)
56 | pca_filters = pca.components_
57 |
58 | # plot filters
59 | plt.figure(figsize=(6,6))
60 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
61 | for i in tqdm(range(n_comp)):
62 | plt.subplot(10, 10, i+1)
63 | plt.imshow(np.reshape(pca_filters[i], (w, h)), cmap="gray")
64 | plt.axis("off")
65 | plt.suptitle("PCA", fontsize=20)
66 | plt.subplots_adjust(top=0.9)
67 | plt.savefig("PCA.png")
68 | plt.show()
69 |
70 |
--------------------------------------------------------------------------------
/network.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 |
5 | class OlshausenField1996Model:
6 | def __init__(self, num_inputs, num_units, batch_size,
7 | lr_r=1e-2, lr_Phi=1e-2, lmda=5e-3):
8 | self.lr_r = lr_r # learning rate of r
9 | self.lr_Phi = lr_Phi # learning rate of Phi
10 | self.lmda = lmda # regularization parameter
11 |
12 | self.num_inputs = num_inputs
13 | self.num_units = num_units
14 | self.batch_size = batch_size
15 |
16 | # Weights
17 | Phi = np.random.randn(self.num_inputs, self.num_units).astype(np.float32)
18 | self.Phi = Phi * np.sqrt(1/self.num_units)
19 |
20 | # activity of neurons
21 | self.r = np.zeros((self.batch_size, self.num_units))
22 |
23 | def initialize_states(self):
24 | self.r = np.zeros((self.batch_size, self.num_units))
25 |
26 | def normalize_rows(self):
27 | self.Phi = self.Phi / np.maximum(np.linalg.norm(self.Phi, ord=2, axis=0, keepdims=True), 1e-8)
28 |
29 | # thresholding function of S(x)=|x|
30 | def soft_thresholding_func(self, x, lmda):
31 | return np.maximum(x - lmda, 0) - np.maximum(-x - lmda, 0)
32 |
33 | # thresholding function of S(x)=ln(1+x^2)
34 | def ln_thresholding_func(self, x, lmda):
35 | f = 9*lmda*x - 2*np.power(x, 3) - 18*x
36 | g = 3*lmda - np.square(x) + 3
37 | h = np.cbrt(np.sqrt(np.square(f) + 4*np.power(g, 3)) + f)
38 | two_croot = np.cbrt(2) # cubic root of two
39 | return (1/3)*(x - h / two_croot + two_croot*g / (1e-8+h))
40 |
41 | # thresholding function https://arxiv.org/abs/2003.12507
42 | def cauchy_thresholding_func(self, x, lmda):
43 | f = 0.5*(x + np.sqrt(np.maximum(x**2 - lmda,0)))
44 | g = 0.5*(x - np.sqrt(np.maximum(x**2 - lmda,0)))
45 | return f*(x>=lmda) + g*(x<=-lmda)
46 |
47 | def calculate_total_error(self, error):
48 | recon_error = np.mean(error**2)
49 | sparsity_r = self.lmda*np.mean(np.abs(self.r))
50 | return recon_error + sparsity_r
51 |
52 | def __call__(self, inputs, training=True):
53 | # Updates
54 | error = inputs - self.r @ self.Phi.T
55 |
56 | r = self.r + self.lr_r * error @ self.Phi
57 | self.r = self.soft_thresholding_func(r, self.lmda)
58 | #self.r = self.cauchy_thresholding_func(r, self.lmda)
59 |
60 | if training:
61 | error = inputs - self.r @ self.Phi.T
62 | dPhi = error.T @ self.r
63 | self.Phi += self.lr_Phi * dPhi
64 |
65 | return error, self.r
66 |
--------------------------------------------------------------------------------
/results/ICA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/ICA.png
--------------------------------------------------------------------------------
/results/PCA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/PCA.png
--------------------------------------------------------------------------------
/results/RF.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF.png
--------------------------------------------------------------------------------
/results/RF_cauchy_thresholding.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF_cauchy_thresholding.png
--------------------------------------------------------------------------------
/results/error.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/error.png
--------------------------------------------------------------------------------
/train.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | import network
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | np.random.seed(0)
10 |
11 | # datasets from http://www.rctn.org/bruno/sparsenet/
12 | mat_images = sio.loadmat('datasets/IMAGES.mat')
13 | imgs = mat_images['IMAGES']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs.shape
17 | num_iter = 500 # number of iterations
18 | nt_max = 1000 # Maximum number of simulation time
19 | batch_size = 250 # Batch size
20 |
21 | sz = 16 # image patch size
22 | num_units = 100 # number of neurons (units)
23 |
24 | eps = 1e-2 # small value which determines convergence
25 | error_list = [] # List to save errors
26 |
27 | # Define model
28 | model = network.OlshausenField1996Model(num_inputs=sz**2, num_units=num_units,
29 | batch_size=batch_size)
30 |
31 | # Run simulation
32 | for iter_ in tqdm(range(num_iter)):
33 | # Get the coordinates of the upper left corner of clopping image randomly.
34 | beginx = np.random.randint(0, W-sz, batch_size)
35 | beginy = np.random.randint(0, H-sz, batch_size)
36 |
37 | inputs_list = []
38 |
39 | # Get images randomly
40 | for i in range(batch_size):
41 | idx = np.random.randint(0, num_images)
42 | img = imgs[:, :, idx]
43 | clop = img[beginy[i]:beginy[i]+sz, beginx[i]:beginx[i]+sz].flatten()
44 | inputs_list.append(clop - np.mean(clop))
45 |
46 | inputs = np.array(inputs_list) # Input image patches
47 |
48 | model.initialize_states() # Reset states
49 | model.normalize_rows() # Normalize weights
50 |
51 | # Input an image patch until latent variables are converged
52 | r_tm1 = model.r # set previous r (t minus 1)
53 |
54 | for t in range(nt_max):
55 | # Update r without update weights
56 | error, r = model(inputs, training=False)
57 | dr = r - r_tm1
58 |
59 | # Compute norm of r
60 | dr_norm = np.linalg.norm(dr, ord=2) / (eps + np.linalg.norm(r_tm1, ord=2))
61 | r_tm1 = r # update r_tm1
62 |
63 | # Check convergence of r, then update weights
64 | if dr_norm < eps:
65 | error, r = model(inputs, training=True)
66 | break
67 |
68 | # If failure to convergence, break and print error
69 | if t >= nt_max-2:
70 | print("Error at patch:", iter_)
71 | print(dr_norm)
72 | break
73 |
74 | error_list.append(model.calculate_total_error(error)) # Append errors
75 |
76 | # Print moving average error
77 | if iter_ % 100 == 99:
78 | print("\n iter: "+str(iter_+1)+"/"+str(num_iter)+", Moving error:",
79 | np.mean(error_list[iter_-99:iter_]))
80 |
81 | # Plot error
82 | plt.figure(figsize=(5, 3))
83 | plt.ylabel("Error")
84 | plt.xlabel("Iterations")
85 | plt.plot(np.arange(len(error_list)), np.array(error_list))
86 | plt.tight_layout()
87 | plt.savefig("error.png")
88 | plt.show()
89 |
90 | # Plot Receptive fields
91 | fig = plt.figure(figsize=(8, 8))
92 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
93 | for i in tqdm(range(num_units)):
94 | plt.subplot(10, 10, i+1)
95 | plt.imshow(np.reshape(model.Phi[:, i], (sz, sz)), cmap="gray")
96 | plt.axis("off")
97 |
98 | fig.suptitle("Receptive fields", fontsize=20)
99 | plt.subplots_adjust(top=0.9)
100 | plt.savefig("RF.png")
101 | plt.show()
--------------------------------------------------------------------------------
 17 |
18 | ### Receptive fields (using soft threshold function)
19 |
17 |
18 | ### Receptive fields (using soft threshold function)
19 |  20 |
21 | ### Receptive fields (using Cauchy threshold function (Mayo et al., 2020))
22 |
20 |
21 | ### Receptive fields (using Cauchy threshold function (Mayo et al., 2020))
22 |  23 |
24 | ### ICA
25 |
23 |
24 | ### ICA
25 |  26 |
27 | ### PCA
28 |
26 |
27 | ### PCA
28 |  29 |
30 | ## Reference
31 | - Olshausen BA, Field DJ. [Emergence of simple-cell receptive field properties by learning a sparse code for natural images](https://www.nature.com/articles/381607a0). *Nature*. 1996;381(6583):607–609. [Data and Code](http://www.rctn.org/bruno/sparsenet/), [pdf](https://courses.cs.washington.edu/courses/cse528/11sp/Olshausen-nature-paper.pdf)
32 | - Rozell CJ, Johnson DH, Baraniuk RG, Olshausen BA. [Sparse coding via thresholding and local competition in neural circuits](http://www.mit.edu/~9.54/fall14/Classes/class07/Palm.pdf). *Neural Comput*. 2008;20(10):2526‐2563.
33 | - Mayo P, Holmes R, Achim A. [Iterative Cauchy Thresholding: Regularisation with a heavy-tailed prior](https://arxiv.org/abs/2003.12507). arXiv. 2020.
--------------------------------------------------------------------------------
/datasets/IMAGES.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES.mat
--------------------------------------------------------------------------------
/datasets/IMAGES_RAW.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES_RAW.mat
--------------------------------------------------------------------------------
/datasets/Sparse net.url:
--------------------------------------------------------------------------------
1 | [InternetShortcut]
2 | URL=http://www.rctn.org/bruno/sparsenet/
3 |
--------------------------------------------------------------------------------
/ica.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | from sklearn.decomposition import FastICA, PCA
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | # datasets from http://www.rctn.org/bruno/sparsenet/
10 | # mat_images = sio.loadmat('datasets/IMAGES.mat')
11 | # imgs = mat_images['IMAGES']
12 | mat_images_raw = sio.loadmat('datasets/IMAGES_RAW.mat')
13 | imgs_raw = mat_images_raw['IMAGESr']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs_raw.shape
17 |
18 | num_patches = 15000
19 | patchs_list = []
20 | w, h = 16, 16 # patch size
21 |
22 | # generate patches
23 | for i in tqdm(range(num_patches)):
24 | i = np.random.randint(0, num_images)
25 | # Get the coordinates of the upper left corner of clopping image randomly.
26 | beginx = np.random.randint(0, W-w-1)
27 | beginy = np.random.randint(0, H-h-1)
28 | img_clopped = imgs_raw[beginy:beginy+h, beginx:beginx+w, i]
29 | patchs_list.append(img_clopped.flatten())
30 |
31 | patches = np.array(patchs_list)
32 |
33 | # perform ICA
34 | print("perform ICA")
35 | n_comp = 100
36 | ica = FastICA(n_components=n_comp)
37 | ica.fit(patches)
38 | ica_filters = ica.components_
39 |
40 | # plot filters
41 | plt.figure(figsize=(6,6))
42 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
43 | for i in tqdm(range(n_comp)):
44 | plt.subplot(10, 10, i+1)
45 | plt.imshow(np.reshape(ica_filters[i], (w, h)), cmap="gray")
46 | plt.axis("off")
47 | plt.suptitle("ICA", fontsize=20)
48 | plt.subplots_adjust(top=0.9)
49 | plt.savefig("ICA.png")
50 | plt.show()
51 |
52 | # perform PCA
53 | print("perform PCA")
54 | pca = PCA(n_components=n_comp)
55 | pca.fit(patches)
56 | pca_filters = pca.components_
57 |
58 | # plot filters
59 | plt.figure(figsize=(6,6))
60 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
61 | for i in tqdm(range(n_comp)):
62 | plt.subplot(10, 10, i+1)
63 | plt.imshow(np.reshape(pca_filters[i], (w, h)), cmap="gray")
64 | plt.axis("off")
65 | plt.suptitle("PCA", fontsize=20)
66 | plt.subplots_adjust(top=0.9)
67 | plt.savefig("PCA.png")
68 | plt.show()
69 |
70 |
--------------------------------------------------------------------------------
/network.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 |
5 | class OlshausenField1996Model:
6 | def __init__(self, num_inputs, num_units, batch_size,
7 | lr_r=1e-2, lr_Phi=1e-2, lmda=5e-3):
8 | self.lr_r = lr_r # learning rate of r
9 | self.lr_Phi = lr_Phi # learning rate of Phi
10 | self.lmda = lmda # regularization parameter
11 |
12 | self.num_inputs = num_inputs
13 | self.num_units = num_units
14 | self.batch_size = batch_size
15 |
16 | # Weights
17 | Phi = np.random.randn(self.num_inputs, self.num_units).astype(np.float32)
18 | self.Phi = Phi * np.sqrt(1/self.num_units)
19 |
20 | # activity of neurons
21 | self.r = np.zeros((self.batch_size, self.num_units))
22 |
23 | def initialize_states(self):
24 | self.r = np.zeros((self.batch_size, self.num_units))
25 |
26 | def normalize_rows(self):
27 | self.Phi = self.Phi / np.maximum(np.linalg.norm(self.Phi, ord=2, axis=0, keepdims=True), 1e-8)
28 |
29 | # thresholding function of S(x)=|x|
30 | def soft_thresholding_func(self, x, lmda):
31 | return np.maximum(x - lmda, 0) - np.maximum(-x - lmda, 0)
32 |
33 | # thresholding function of S(x)=ln(1+x^2)
34 | def ln_thresholding_func(self, x, lmda):
35 | f = 9*lmda*x - 2*np.power(x, 3) - 18*x
36 | g = 3*lmda - np.square(x) + 3
37 | h = np.cbrt(np.sqrt(np.square(f) + 4*np.power(g, 3)) + f)
38 | two_croot = np.cbrt(2) # cubic root of two
39 | return (1/3)*(x - h / two_croot + two_croot*g / (1e-8+h))
40 |
41 | # thresholding function https://arxiv.org/abs/2003.12507
42 | def cauchy_thresholding_func(self, x, lmda):
43 | f = 0.5*(x + np.sqrt(np.maximum(x**2 - lmda,0)))
44 | g = 0.5*(x - np.sqrt(np.maximum(x**2 - lmda,0)))
45 | return f*(x>=lmda) + g*(x<=-lmda)
46 |
47 | def calculate_total_error(self, error):
48 | recon_error = np.mean(error**2)
49 | sparsity_r = self.lmda*np.mean(np.abs(self.r))
50 | return recon_error + sparsity_r
51 |
52 | def __call__(self, inputs, training=True):
53 | # Updates
54 | error = inputs - self.r @ self.Phi.T
55 |
56 | r = self.r + self.lr_r * error @ self.Phi
57 | self.r = self.soft_thresholding_func(r, self.lmda)
58 | #self.r = self.cauchy_thresholding_func(r, self.lmda)
59 |
60 | if training:
61 | error = inputs - self.r @ self.Phi.T
62 | dPhi = error.T @ self.r
63 | self.Phi += self.lr_Phi * dPhi
64 |
65 | return error, self.r
66 |
--------------------------------------------------------------------------------
/results/ICA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/ICA.png
--------------------------------------------------------------------------------
/results/PCA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/PCA.png
--------------------------------------------------------------------------------
/results/RF.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF.png
--------------------------------------------------------------------------------
/results/RF_cauchy_thresholding.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF_cauchy_thresholding.png
--------------------------------------------------------------------------------
/results/error.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/error.png
--------------------------------------------------------------------------------
/train.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | import network
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | np.random.seed(0)
10 |
11 | # datasets from http://www.rctn.org/bruno/sparsenet/
12 | mat_images = sio.loadmat('datasets/IMAGES.mat')
13 | imgs = mat_images['IMAGES']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs.shape
17 | num_iter = 500 # number of iterations
18 | nt_max = 1000 # Maximum number of simulation time
19 | batch_size = 250 # Batch size
20 |
21 | sz = 16 # image patch size
22 | num_units = 100 # number of neurons (units)
23 |
24 | eps = 1e-2 # small value which determines convergence
25 | error_list = [] # List to save errors
26 |
27 | # Define model
28 | model = network.OlshausenField1996Model(num_inputs=sz**2, num_units=num_units,
29 | batch_size=batch_size)
30 |
31 | # Run simulation
32 | for iter_ in tqdm(range(num_iter)):
33 | # Get the coordinates of the upper left corner of clopping image randomly.
34 | beginx = np.random.randint(0, W-sz, batch_size)
35 | beginy = np.random.randint(0, H-sz, batch_size)
36 |
37 | inputs_list = []
38 |
39 | # Get images randomly
40 | for i in range(batch_size):
41 | idx = np.random.randint(0, num_images)
42 | img = imgs[:, :, idx]
43 | clop = img[beginy[i]:beginy[i]+sz, beginx[i]:beginx[i]+sz].flatten()
44 | inputs_list.append(clop - np.mean(clop))
45 |
46 | inputs = np.array(inputs_list) # Input image patches
47 |
48 | model.initialize_states() # Reset states
49 | model.normalize_rows() # Normalize weights
50 |
51 | # Input an image patch until latent variables are converged
52 | r_tm1 = model.r # set previous r (t minus 1)
53 |
54 | for t in range(nt_max):
55 | # Update r without update weights
56 | error, r = model(inputs, training=False)
57 | dr = r - r_tm1
58 |
59 | # Compute norm of r
60 | dr_norm = np.linalg.norm(dr, ord=2) / (eps + np.linalg.norm(r_tm1, ord=2))
61 | r_tm1 = r # update r_tm1
62 |
63 | # Check convergence of r, then update weights
64 | if dr_norm < eps:
65 | error, r = model(inputs, training=True)
66 | break
67 |
68 | # If failure to convergence, break and print error
69 | if t >= nt_max-2:
70 | print("Error at patch:", iter_)
71 | print(dr_norm)
72 | break
73 |
74 | error_list.append(model.calculate_total_error(error)) # Append errors
75 |
76 | # Print moving average error
77 | if iter_ % 100 == 99:
78 | print("\n iter: "+str(iter_+1)+"/"+str(num_iter)+", Moving error:",
79 | np.mean(error_list[iter_-99:iter_]))
80 |
81 | # Plot error
82 | plt.figure(figsize=(5, 3))
83 | plt.ylabel("Error")
84 | plt.xlabel("Iterations")
85 | plt.plot(np.arange(len(error_list)), np.array(error_list))
86 | plt.tight_layout()
87 | plt.savefig("error.png")
88 | plt.show()
89 |
90 | # Plot Receptive fields
91 | fig = plt.figure(figsize=(8, 8))
92 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
93 | for i in tqdm(range(num_units)):
94 | plt.subplot(10, 10, i+1)
95 | plt.imshow(np.reshape(model.Phi[:, i], (sz, sz)), cmap="gray")
96 | plt.axis("off")
97 |
98 | fig.suptitle("Receptive fields", fontsize=20)
99 | plt.subplots_adjust(top=0.9)
100 | plt.savefig("RF.png")
101 | plt.show()
--------------------------------------------------------------------------------
29 |
30 | ## Reference
31 | - Olshausen BA, Field DJ. [Emergence of simple-cell receptive field properties by learning a sparse code for natural images](https://www.nature.com/articles/381607a0). *Nature*. 1996;381(6583):607–609. [Data and Code](http://www.rctn.org/bruno/sparsenet/), [pdf](https://courses.cs.washington.edu/courses/cse528/11sp/Olshausen-nature-paper.pdf)
32 | - Rozell CJ, Johnson DH, Baraniuk RG, Olshausen BA. [Sparse coding via thresholding and local competition in neural circuits](http://www.mit.edu/~9.54/fall14/Classes/class07/Palm.pdf). *Neural Comput*. 2008;20(10):2526‐2563.
33 | - Mayo P, Holmes R, Achim A. [Iterative Cauchy Thresholding: Regularisation with a heavy-tailed prior](https://arxiv.org/abs/2003.12507). arXiv. 2020.
--------------------------------------------------------------------------------
/datasets/IMAGES.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES.mat
--------------------------------------------------------------------------------
/datasets/IMAGES_RAW.mat:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/datasets/IMAGES_RAW.mat
--------------------------------------------------------------------------------
/datasets/Sparse net.url:
--------------------------------------------------------------------------------
1 | [InternetShortcut]
2 | URL=http://www.rctn.org/bruno/sparsenet/
3 |
--------------------------------------------------------------------------------
/ica.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | from sklearn.decomposition import FastICA, PCA
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | # datasets from http://www.rctn.org/bruno/sparsenet/
10 | # mat_images = sio.loadmat('datasets/IMAGES.mat')
11 | # imgs = mat_images['IMAGES']
12 | mat_images_raw = sio.loadmat('datasets/IMAGES_RAW.mat')
13 | imgs_raw = mat_images_raw['IMAGESr']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs_raw.shape
17 |
18 | num_patches = 15000
19 | patchs_list = []
20 | w, h = 16, 16 # patch size
21 |
22 | # generate patches
23 | for i in tqdm(range(num_patches)):
24 | i = np.random.randint(0, num_images)
25 | # Get the coordinates of the upper left corner of clopping image randomly.
26 | beginx = np.random.randint(0, W-w-1)
27 | beginy = np.random.randint(0, H-h-1)
28 | img_clopped = imgs_raw[beginy:beginy+h, beginx:beginx+w, i]
29 | patchs_list.append(img_clopped.flatten())
30 |
31 | patches = np.array(patchs_list)
32 |
33 | # perform ICA
34 | print("perform ICA")
35 | n_comp = 100
36 | ica = FastICA(n_components=n_comp)
37 | ica.fit(patches)
38 | ica_filters = ica.components_
39 |
40 | # plot filters
41 | plt.figure(figsize=(6,6))
42 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
43 | for i in tqdm(range(n_comp)):
44 | plt.subplot(10, 10, i+1)
45 | plt.imshow(np.reshape(ica_filters[i], (w, h)), cmap="gray")
46 | plt.axis("off")
47 | plt.suptitle("ICA", fontsize=20)
48 | plt.subplots_adjust(top=0.9)
49 | plt.savefig("ICA.png")
50 | plt.show()
51 |
52 | # perform PCA
53 | print("perform PCA")
54 | pca = PCA(n_components=n_comp)
55 | pca.fit(patches)
56 | pca_filters = pca.components_
57 |
58 | # plot filters
59 | plt.figure(figsize=(6,6))
60 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
61 | for i in tqdm(range(n_comp)):
62 | plt.subplot(10, 10, i+1)
63 | plt.imshow(np.reshape(pca_filters[i], (w, h)), cmap="gray")
64 | plt.axis("off")
65 | plt.suptitle("PCA", fontsize=20)
66 | plt.subplots_adjust(top=0.9)
67 | plt.savefig("PCA.png")
68 | plt.show()
69 |
70 |
--------------------------------------------------------------------------------
/network.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 |
5 | class OlshausenField1996Model:
6 | def __init__(self, num_inputs, num_units, batch_size,
7 | lr_r=1e-2, lr_Phi=1e-2, lmda=5e-3):
8 | self.lr_r = lr_r # learning rate of r
9 | self.lr_Phi = lr_Phi # learning rate of Phi
10 | self.lmda = lmda # regularization parameter
11 |
12 | self.num_inputs = num_inputs
13 | self.num_units = num_units
14 | self.batch_size = batch_size
15 |
16 | # Weights
17 | Phi = np.random.randn(self.num_inputs, self.num_units).astype(np.float32)
18 | self.Phi = Phi * np.sqrt(1/self.num_units)
19 |
20 | # activity of neurons
21 | self.r = np.zeros((self.batch_size, self.num_units))
22 |
23 | def initialize_states(self):
24 | self.r = np.zeros((self.batch_size, self.num_units))
25 |
26 | def normalize_rows(self):
27 | self.Phi = self.Phi / np.maximum(np.linalg.norm(self.Phi, ord=2, axis=0, keepdims=True), 1e-8)
28 |
29 | # thresholding function of S(x)=|x|
30 | def soft_thresholding_func(self, x, lmda):
31 | return np.maximum(x - lmda, 0) - np.maximum(-x - lmda, 0)
32 |
33 | # thresholding function of S(x)=ln(1+x^2)
34 | def ln_thresholding_func(self, x, lmda):
35 | f = 9*lmda*x - 2*np.power(x, 3) - 18*x
36 | g = 3*lmda - np.square(x) + 3
37 | h = np.cbrt(np.sqrt(np.square(f) + 4*np.power(g, 3)) + f)
38 | two_croot = np.cbrt(2) # cubic root of two
39 | return (1/3)*(x - h / two_croot + two_croot*g / (1e-8+h))
40 |
41 | # thresholding function https://arxiv.org/abs/2003.12507
42 | def cauchy_thresholding_func(self, x, lmda):
43 | f = 0.5*(x + np.sqrt(np.maximum(x**2 - lmda,0)))
44 | g = 0.5*(x - np.sqrt(np.maximum(x**2 - lmda,0)))
45 | return f*(x>=lmda) + g*(x<=-lmda)
46 |
47 | def calculate_total_error(self, error):
48 | recon_error = np.mean(error**2)
49 | sparsity_r = self.lmda*np.mean(np.abs(self.r))
50 | return recon_error + sparsity_r
51 |
52 | def __call__(self, inputs, training=True):
53 | # Updates
54 | error = inputs - self.r @ self.Phi.T
55 |
56 | r = self.r + self.lr_r * error @ self.Phi
57 | self.r = self.soft_thresholding_func(r, self.lmda)
58 | #self.r = self.cauchy_thresholding_func(r, self.lmda)
59 |
60 | if training:
61 | error = inputs - self.r @ self.Phi.T
62 | dPhi = error.T @ self.r
63 | self.Phi += self.lr_Phi * dPhi
64 |
65 | return error, self.r
66 |
--------------------------------------------------------------------------------
/results/ICA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/ICA.png
--------------------------------------------------------------------------------
/results/PCA.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/PCA.png
--------------------------------------------------------------------------------
/results/RF.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF.png
--------------------------------------------------------------------------------
/results/RF_cauchy_thresholding.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/RF_cauchy_thresholding.png
--------------------------------------------------------------------------------
/results/error.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/takyamamoto/SparseCoding-OlshausenField-Model/6a73f2ca81b690b0d0fddbcfb14f508f597afd2c/results/error.png
--------------------------------------------------------------------------------
/train.py:
--------------------------------------------------------------------------------
1 | # -*- coding: utf-8 -*-
2 |
3 | import numpy as np
4 | import matplotlib.pyplot as plt
5 | import network
6 | from tqdm import tqdm
7 | import scipy.io as sio
8 |
9 | np.random.seed(0)
10 |
11 | # datasets from http://www.rctn.org/bruno/sparsenet/
12 | mat_images = sio.loadmat('datasets/IMAGES.mat')
13 | imgs = mat_images['IMAGES']
14 |
15 | # Simulation constants
16 | H, W, num_images = imgs.shape
17 | num_iter = 500 # number of iterations
18 | nt_max = 1000 # Maximum number of simulation time
19 | batch_size = 250 # Batch size
20 |
21 | sz = 16 # image patch size
22 | num_units = 100 # number of neurons (units)
23 |
24 | eps = 1e-2 # small value which determines convergence
25 | error_list = [] # List to save errors
26 |
27 | # Define model
28 | model = network.OlshausenField1996Model(num_inputs=sz**2, num_units=num_units,
29 | batch_size=batch_size)
30 |
31 | # Run simulation
32 | for iter_ in tqdm(range(num_iter)):
33 | # Get the coordinates of the upper left corner of clopping image randomly.
34 | beginx = np.random.randint(0, W-sz, batch_size)
35 | beginy = np.random.randint(0, H-sz, batch_size)
36 |
37 | inputs_list = []
38 |
39 | # Get images randomly
40 | for i in range(batch_size):
41 | idx = np.random.randint(0, num_images)
42 | img = imgs[:, :, idx]
43 | clop = img[beginy[i]:beginy[i]+sz, beginx[i]:beginx[i]+sz].flatten()
44 | inputs_list.append(clop - np.mean(clop))
45 |
46 | inputs = np.array(inputs_list) # Input image patches
47 |
48 | model.initialize_states() # Reset states
49 | model.normalize_rows() # Normalize weights
50 |
51 | # Input an image patch until latent variables are converged
52 | r_tm1 = model.r # set previous r (t minus 1)
53 |
54 | for t in range(nt_max):
55 | # Update r without update weights
56 | error, r = model(inputs, training=False)
57 | dr = r - r_tm1
58 |
59 | # Compute norm of r
60 | dr_norm = np.linalg.norm(dr, ord=2) / (eps + np.linalg.norm(r_tm1, ord=2))
61 | r_tm1 = r # update r_tm1
62 |
63 | # Check convergence of r, then update weights
64 | if dr_norm < eps:
65 | error, r = model(inputs, training=True)
66 | break
67 |
68 | # If failure to convergence, break and print error
69 | if t >= nt_max-2:
70 | print("Error at patch:", iter_)
71 | print(dr_norm)
72 | break
73 |
74 | error_list.append(model.calculate_total_error(error)) # Append errors
75 |
76 | # Print moving average error
77 | if iter_ % 100 == 99:
78 | print("\n iter: "+str(iter_+1)+"/"+str(num_iter)+", Moving error:",
79 | np.mean(error_list[iter_-99:iter_]))
80 |
81 | # Plot error
82 | plt.figure(figsize=(5, 3))
83 | plt.ylabel("Error")
84 | plt.xlabel("Iterations")
85 | plt.plot(np.arange(len(error_list)), np.array(error_list))
86 | plt.tight_layout()
87 | plt.savefig("error.png")
88 | plt.show()
89 |
90 | # Plot Receptive fields
91 | fig = plt.figure(figsize=(8, 8))
92 | plt.subplots_adjust(hspace=0.1, wspace=0.1)
93 | for i in tqdm(range(num_units)):
94 | plt.subplot(10, 10, i+1)
95 | plt.imshow(np.reshape(model.Phi[:, i], (sz, sz)), cmap="gray")
96 | plt.axis("off")
97 |
98 | fig.suptitle("Receptive fields", fontsize=20)
99 | plt.subplots_adjust(top=0.9)
100 | plt.savefig("RF.png")
101 | plt.show()
--------------------------------------------------------------------------------