├── .github
├── PULL_REQUEST_TEMPLATE.md
├── dependabot.yml
├── release.yml
└── workflows
│ ├── ci.yml
│ ├── nightly.yml
│ └── pypipublish.yml
├── .gitignore
├── .pre-commit-config.yaml
├── .readthedocs.yaml
├── CITATION.cff
├── LICENSE
├── README.md
├── ci
├── docs.yml
├── environment.yml
└── install-upstream-wheels.sh
├── design_doc.md
├── docs
├── _static
│ └── logos
│ │ ├── xdggs_logo.png
│ │ └── xdggs_logo.svg
├── api-hidden.rst
├── api.rst
├── changelog.md
├── conf.py
├── contributor_guide
│ └── docs.md
├── getting_started
│ └── installation.md
├── index.md
├── reference_guide
│ ├── publications.bib
│ └── publications.md
└── tutorials
│ ├── h3.ipynb
│ └── healpix.ipynb
├── environment.yml
├── examples
├── example_h3.ipynb
├── example_healpy.ipynb
└── prepare_dataset_h3.ipynb
├── pyproject.toml
├── xdggs-cropped.gif
└── xdggs
├── __init__.py
├── accessor.py
├── grid.py
├── h3.py
├── healpix.py
├── index.py
├── itertools.py
├── plotting.py
├── tests
├── __init__.py
├── matchers.py
├── test_accessor.py
├── test_h3.py
├── test_healpix.py
├── test_index.py
├── test_matchers.py
├── test_plotting.py
├── test_tutorial.py
└── test_utils.py
├── tutorial.py
└── utils.py
/.github/PULL_REQUEST_TEMPLATE.md:
--------------------------------------------------------------------------------
1 | - [ ] closes #xxxx
2 | - [ ] Tests added
3 | - [ ] User visible changes (including notable bug fixes) are documented in `changelog.md`
4 | - [ ] New functions/methods are listed in `api.rst`
5 |
--------------------------------------------------------------------------------
/.github/dependabot.yml:
--------------------------------------------------------------------------------
1 | version: 2
2 | updates:
3 | - package-ecosystem: "github-actions"
4 | directory: "/"
5 | schedule:
6 | # Check for updates once a week
7 | interval: "weekly"
8 |
--------------------------------------------------------------------------------
/.github/release.yml:
--------------------------------------------------------------------------------
1 | changelog:
2 | exclude:
3 | authors:
4 | - dependabot
5 | - pre-commit-ci
6 |
--------------------------------------------------------------------------------
/.github/workflows/ci.yml:
--------------------------------------------------------------------------------
1 | name: CI
2 |
3 | on:
4 | push:
5 | branches: ["main"]
6 | pull_request:
7 | branches: ["main"]
8 | workflow_dispatch:
9 |
10 | concurrency:
11 | group: ${{ github.workflow }}-${{ github.ref }}
12 | cancel-in-progress: true
13 |
14 | env:
15 | FORCE_COLOR: 3
16 |
17 | jobs:
18 | detect-ci-trigger:
19 | name: detect ci trigger
20 | runs-on: ubuntu-latest
21 | if: |
22 | github.repository_owner == 'xarray-contrib'
23 | && (github.event_name == 'pull_request' || github.event_name == 'push')
24 | outputs:
25 | triggered: ${{ steps.detect-trigger.outputs.trigger-found }}
26 | steps:
27 | - uses: actions/checkout@v4
28 | with:
29 | fetch-depth: 2
30 | - uses: xarray-contrib/ci-trigger@v1
31 | id: detect-trigger
32 | with:
33 | keyword: "[skip-ci]"
34 |
35 | tests:
36 | name: ${{ matrix.os }} py${{ matrix.python-version }}
37 | runs-on: ${{ matrix.os }}
38 | needs: detect-ci-trigger
39 | if: needs.detect-ci-trigger.outputs.triggered == 'false'
40 | defaults:
41 | run:
42 | shell: bash -leo pipefail {0} {0}
43 |
44 | strategy:
45 | fail-fast: false
46 |
47 | matrix:
48 | os: ["ubuntu-latest", "macos-latest", "windows-latest"]
49 | python-version: ["3.10", "3.11", "3.12"]
50 |
51 | steps:
52 | - name: Clone repository
53 | uses: actions/checkout@v4
54 | with:
55 | fetch-depth: 0 # Fetch all history for all branches and tags
56 |
57 | - name: Setup environment
58 | run: >-
59 | echo "CONDA_ENV_FILE=ci/environment.yml" >> $GITHUB_ENV
60 |
61 | - name: Setup micromamba
62 | uses: mamba-org/setup-micromamba@v2
63 | with:
64 | micromamba-version: "1.5.10-0"
65 | environment-file: ${{ env.CONDA_ENV_FILE }}
66 | environment-name: xdggs-tests
67 | cache-environment: true
68 | cache-environment-key: "${{runner.os}}-${{runner.arch}}-py${{matrix.python-version}}-${{env.TODAY}}-${{hashFiles(env.CONDA_ENV_FILE)}}"
69 | create-args: >-
70 | python=${{matrix.python-version}}
71 |
72 | - name: Install xdggs
73 | run: |
74 | python -m pip install --no-deps -e .
75 |
76 | - name: Import xdggs
77 | run: |
78 | python -c "import xdggs"
79 |
80 | - name: Run tests
81 | run: |
82 | python -m pytest
83 |
--------------------------------------------------------------------------------
/.github/workflows/nightly.yml:
--------------------------------------------------------------------------------
1 | name: Upstream-dev CI
2 |
3 | on:
4 | push:
5 | branches: ["main"]
6 | pull_request:
7 | branches: ["main"]
8 | types: [opened, reopened, synchronize, labeled]
9 | schedule:
10 | - cron: "0 0 * * 1" # Mondays “At 00:00” UTC
11 | workflow_dispatch: # allows you to trigger the workflow run manually
12 |
13 | concurrency:
14 | group: ${{ github.workflow }}-${{ github.ref }}
15 | cancel-in-progress: true
16 |
17 | jobs:
18 | detect-ci-trigger:
19 | name: detect ci trigger
20 | runs-on: ubuntu-latest
21 | if: |
22 | github.repository_owner == 'xarray-contrib' && github.event_name == 'pull_request'
23 | outputs:
24 | triggered: ${{ steps.detect-trigger.outputs.trigger-found }}
25 | steps:
26 | - uses: actions/checkout@v4
27 | with:
28 | fetch-depth: 2
29 | - uses: xarray-contrib/ci-trigger@v1
30 | id: detect-trigger
31 | with:
32 | keyword: "[test-upstream]"
33 |
34 | tests:
35 | name: upstream-dev
36 | runs-on: ubuntu-latest
37 | needs: detect-ci-trigger
38 | if: |
39 | always()
40 | && (
41 | (github.event_name == 'schedule' || github.event_name == 'workflow_dispatch')
42 | || needs.detect-ci-trigger.outputs.triggered == 'true'
43 | || contains( github.event.pull_request.labels.*.name, 'run-upstream')
44 | )
45 |
46 | defaults:
47 | run:
48 | shell: bash -leo pipefail {0} {0}
49 |
50 | strategy:
51 | fail-fast: false
52 |
53 | matrix:
54 | python-version: ["3.12"]
55 |
56 | permissions:
57 | issues: write
58 |
59 | steps:
60 | - name: Clone repository
61 | uses: actions/checkout@v4
62 | with:
63 | fetch-depth: 0 # Fetch all history for all branches and tags
64 |
65 | - name: Setup micromamba
66 | uses: mamba-org/setup-micromamba@v2
67 | with:
68 | environment-file: ci/environment.yml
69 | environment-name: xdggs-tests
70 | cache-environment: false
71 | create-args: >-
72 | python=${{matrix.python-version}}
73 | pytest-reportlog
74 | conda
75 |
76 | - name: Install upstream versions
77 | run: |
78 | bash ci/install-upstream-wheels.sh
79 |
80 | - name: Install xdggs
81 | run: |
82 | python -m pip install --no-deps -e .
83 |
84 | - name: Import xdggs
85 | run: |
86 | python -c "import xdggs"
87 |
88 | - name: Run tests
89 | if: success()
90 | id: run-tests
91 | run: |
92 | python -m pytest --report-log output-${{ matrix.python-version }}-log.jsonl
93 |
94 | - name: Generate and publish a failure report
95 | if: |
96 | failure()
97 | && steps.run-tests.outcome == 'failure'
98 | && github.event_name == 'schedule'
99 | && github.repository_owner == 'xarray-contrib'
100 | uses: xarray-contrib/issue-from-pytest-log@v1
101 | with:
102 | log-path: output-${{ matrix.python-version }}-log.jsonl
103 |
--------------------------------------------------------------------------------
/.github/workflows/pypipublish.yml:
--------------------------------------------------------------------------------
1 | name: Upload Python Package on PyPI
2 |
3 | on:
4 | release:
5 | types: [published]
6 |
7 | jobs:
8 | deploy:
9 | runs-on: ubuntu-latest
10 | environment:
11 | name: pypi
12 | url: https://pypi.org/p/xdggs
13 | permissions:
14 | id-token: write
15 | steps:
16 | - name: Checkout
17 | uses: actions/checkout@v4
18 | - name: Set up Python
19 | uses: actions/setup-python@v5
20 | with:
21 | python-version: "3.x"
22 | - name: Install publish dependencies
23 | run: python -m pip install build
24 | - name: Build package
25 | run: python -m build . -o py_dist
26 | - name: Publish package to PyPI
27 | uses: pypa/gh-action-pypi-publish@release/v1
28 | with:
29 | packages-dir: py_dist/
30 |
--------------------------------------------------------------------------------
/.gitignore:
--------------------------------------------------------------------------------
1 | # Byte-compiled / optimized / DLL files
2 | __pycache__/
3 | *.py[cod]
4 | *$py.class
5 |
6 | # C extensions
7 | *.so
8 |
9 | # Distribution / packaging
10 | .Python
11 | build/
12 | develop-eggs/
13 | dist/
14 | downloads/
15 | eggs/
16 | .eggs/
17 | lib/
18 | lib64/
19 | parts/
20 | sdist/
21 | var/
22 | wheels/
23 | share/python-wheels/
24 | *.egg-info/
25 | .installed.cfg
26 | *.egg
27 | MANIFEST
28 |
29 | # PyInstaller
30 | # Usually these files are written by a python script from a template
31 | # before PyInstaller builds the exe, so as to inject date/other infos into it.
32 | *.manifest
33 | *.spec
34 |
35 | # Installer logs
36 | pip-log.txt
37 | pip-delete-this-directory.txt
38 |
39 | # Unit test / coverage reports
40 | htmlcov/
41 | .tox/
42 | .nox/
43 | .coverage
44 | .coverage.*
45 | .cache
46 | nosetests.xml
47 | coverage.xml
48 | *.cover
49 | *.py,cover
50 | .hypothesis/
51 | .pytest_cache/
52 | cover/
53 |
54 | # Translations
55 | *.mo
56 | *.pot
57 |
58 | # Django stuff:
59 | *.log
60 | local_settings.py
61 | db.sqlite3
62 | db.sqlite3-journal
63 |
64 | # Flask stuff:
65 | instance/
66 | .webassets-cache
67 |
68 | # Scrapy stuff:
69 | .scrapy
70 |
71 | # Sphinx documentation
72 | docs/_build/
73 | docs/**/generated/
74 |
75 | # PyBuilder
76 | .pybuilder/
77 | target/
78 |
79 | # Jupyter Notebook
80 | .ipynb_checkpoints/
81 | .virtual_documents/
82 |
83 | # IPython
84 | profile_default/
85 | ipython_config.py
86 |
87 | # pyenv
88 | # For a library or package, you might want to ignore these files since the code is
89 | # intended to run in multiple environments; otherwise, check them in:
90 | # .python-version
91 |
92 | # pipenv
93 | # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
94 | # However, in case of collaboration, if having platform-specific dependencies or dependencies
95 | # having no cross-platform support, pipenv may install dependencies that don't work, or not
96 | # install all needed dependencies.
97 | #Pipfile.lock
98 |
99 | # poetry
100 | # Similar to Pipfile.lock, it is generally recommended to include poetry.lock in version control.
101 | # This is especially recommended for binary packages to ensure reproducibility, and is more
102 | # commonly ignored for libraries.
103 | # https://python-poetry.org/docs/basic-usage/#commit-your-poetrylock-file-to-version-control
104 | #poetry.lock
105 |
106 | # pdm
107 | # Similar to Pipfile.lock, it is generally recommended to include pdm.lock in version control.
108 | #pdm.lock
109 | # pdm stores project-wide configurations in .pdm.toml, but it is recommended to not include it
110 | # in version control.
111 | # https://pdm.fming.dev/#use-with-ide
112 | .pdm.toml
113 |
114 | # PEP 582; used by e.g. github.com/David-OConnor/pyflow and github.com/pdm-project/pdm
115 | __pypackages__/
116 |
117 | # Celery stuff
118 | celerybeat-schedule
119 | celerybeat.pid
120 |
121 | # SageMath parsed files
122 | *.sage.py
123 |
124 | # Environments
125 | .env
126 | .venv
127 | env/
128 | venv/
129 | ENV/
130 | env.bak/
131 | venv.bak/
132 |
133 | # Spyder project settings
134 | .spyderproject
135 | .spyproject
136 |
137 | # Rope project settings
138 | .ropeproject
139 |
140 | # mkdocs documentation
141 | /site
142 |
143 | # code formatter cache
144 | .prettier_cache/
145 |
146 | # mypy
147 | .mypy_cache/
148 | .dmypy.json
149 | dmypy.json
150 |
151 | # Pyre type checker
152 | .pyre/
153 |
154 | # pytype static type analyzer
155 | .pytype/
156 |
157 | # Cython debug symbols
158 | cython_debug/
159 |
160 | # PyCharm
161 | # JetBrains specific template is maintained in a separate JetBrains.gitignore that can
162 | # be found at https://github.com/github/gitignore/blob/main/Global/JetBrains.gitignore
163 | # and can be added to the global gitignore or merged into this file. For a more nuclear
164 | # option (not recommended) you can uncomment the following to ignore the entire idea folder.
165 | #.idea/
166 |
167 | # bibtex
168 | .auctex-auto
169 | *.bib.untidy
170 |
--------------------------------------------------------------------------------
/.pre-commit-config.yaml:
--------------------------------------------------------------------------------
1 | repos:
2 | - repo: https://github.com/pre-commit/pre-commit-hooks
3 | rev: v5.0.0
4 | hooks:
5 | - id: trailing-whitespace
6 | - id: end-of-file-fixer
7 | - id: check-docstring-first
8 | - repo: https://github.com/rbubley/mirrors-prettier
9 | rev: v3.5.3
10 | hooks:
11 | - id: prettier
12 | args: [--cache-location=.prettier_cache/cache]
13 | - repo: https://github.com/ComPWA/taplo-pre-commit
14 | rev: v0.9.3
15 | hooks:
16 | - id: taplo-format
17 | args: [--option, array_auto_collapse=false]
18 | - id: taplo-lint
19 | args: [--no-schema]
20 | - repo: https://github.com/abravalheri/validate-pyproject
21 | rev: v0.24.1
22 | hooks:
23 | - id: validate-pyproject
24 | - repo: https://github.com/astral-sh/ruff-pre-commit
25 | rev: v0.11.12
26 | hooks:
27 | - id: ruff
28 | args: [--fix]
29 | - repo: https://github.com/psf/black-pre-commit-mirror
30 | rev: 25.1.0
31 | hooks:
32 | - id: black-jupyter
33 | - repo: https://github.com/keewis/blackdoc
34 | rev: v0.3.9
35 | hooks:
36 | - id: blackdoc
37 | additional_dependencies: ["black==25.1.0"]
38 | - id: blackdoc-autoupdate-black
39 | - repo: https://github.com/citation-file-format/cffconvert
40 | rev: b6045d78aac9e02b039703b030588d54d53262ac
41 | hooks:
42 | - id: validate-cff
43 | - repo: https://github.com/kynan/nbstripout
44 | rev: 0.8.1
45 | hooks:
46 | - id: nbstripout
47 | args: [--extra-keys=metadata.kernelspec metadata.langauge_info.version]
48 | - repo: https://github.com/FlamingTempura/bibtex-tidy
49 | rev: v1.14.0
50 | hooks:
51 | - id: bibtex-tidy
52 | stages: [manual]
53 | args:
54 | - "--modify"
55 | - "--blank-lines"
56 | - "--sort=-year,name"
57 | - "--duplicates"
58 | - "--escape"
59 | - "--trailing-commas"
60 |
61 | ci:
62 | autofix_prs: true
63 | autoupdate_schedule: monthly
64 |
--------------------------------------------------------------------------------
/.readthedocs.yaml:
--------------------------------------------------------------------------------
1 | version: 2
2 |
3 | build:
4 | os: ubuntu-lts-latest
5 | tools:
6 | python: mambaforge-latest
7 | jobs:
8 | post_checkout:
9 | - (git --no-pager log --pretty="tformat:%s" -1 | grep -vqF "[skip-rtd]") || exit 183
10 | - git fetch --unshallow || true
11 | pre_install:
12 | - git update-index --assume-unchanged docs/conf.py ci/docs.yml
13 |
14 | conda:
15 | environment: ci/docs.yml
16 |
17 | sphinx:
18 | fail_on_warning: true
19 | configuration: docs/conf.py
20 |
21 | formats: []
22 |
--------------------------------------------------------------------------------
/CITATION.cff:
--------------------------------------------------------------------------------
1 | cff-version: 1.2.0
2 | title: xdggs
3 | message: "If you use this software, please cite it as below."
4 | type: software
5 | authors:
6 | - family-names: "Magin"
7 | given-names: "Justus"
8 | orcid: "https://orcid.org/0000-0002-4254-8002"
9 | - family-names: "Bovy"
10 | given-names: "Benoît"
11 | orcid: "https://orcid.org/0009-0001-4011-3574"
12 | - family-names: "Kmoch"
13 | given-names: "Alexander"
14 | orcid: "https://orcid.org/0000-0003-4386-4450"
15 | - family-names: "Abernathey"
16 | given-names: "Ryan"
17 | orcid: "https://orcid.org/0000-0001-5999-4917"
18 | - family-names: "Coca-Castro"
19 | given-names: "Alejandro"
20 | orcid: "https://orcid.org/0000-0002-9264-1539"
21 | - family-names: "Strobl"
22 | given-names: "Peter"
23 | orcid: "https://orcid.org/0000-0003-2733-1822"
24 | - family-names: "Fouilloux"
25 | given-names: "Anne"
26 | orcid: "https://orcid.org/0000-0002-1784-2920"
27 | - family-names: "Loos"
28 | given-names: "Daniel"
29 | orcid: "https://orcid.org/0000-0002-4024-4443"
30 | - family-names: "Chan"
31 | given-names: "Wai Tik"
32 | orcid: "https://orcid.org/0009-0005-3779-139X"
33 | - family-names: "Delouis"
34 | given-names: "Jean-Marc"
35 | orcid: "https://orcid.org/0000-0002-0713-1658"
36 | - family-names: "Odaka"
37 | given-names: "Tina"
38 | orcid: "https://orcid.org/0000-0002-1500-0156"
39 | repository-code: "https://github.com/xarray-contrib/xdggs"
40 | url: "https://xdggs.readthedocs.io"
41 | keywords:
42 | - xarray
43 | - discrete global grid systems
44 | - dggs
45 | license: Apache-2.0
46 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | Apache License
2 | Version 2.0, January 2004
3 | http://www.apache.org/licenses/
4 |
5 | TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
6 |
7 | 1. Definitions.
8 |
9 | "License" shall mean the terms and conditions for use, reproduction,
10 | and distribution as defined by Sections 1 through 9 of this document.
11 |
12 | "Licensor" shall mean the copyright owner or entity authorized by
13 | the copyright owner that is granting the License.
14 |
15 | "Legal Entity" shall mean the union of the acting entity and all
16 | other entities that control, are controlled by, or are under common
17 | control with that entity. For the purposes of this definition,
18 | "control" means (i) the power, direct or indirect, to cause the
19 | direction or management of such entity, whether by contract or
20 | otherwise, or (ii) ownership of fifty percent (50%) or more of the
21 | outstanding shares, or (iii) beneficial ownership of such entity.
22 |
23 | "You" (or "Your") shall mean an individual or Legal Entity
24 | exercising permissions granted by this License.
25 |
26 | "Source" form shall mean the preferred form for making modifications,
27 | including but not limited to software source code, documentation
28 | source, and configuration files.
29 |
30 | "Object" form shall mean any form resulting from mechanical
31 | transformation or translation of a Source form, including but
32 | not limited to compiled object code, generated documentation,
33 | and conversions to other media types.
34 |
35 | "Work" shall mean the work of authorship, whether in Source or
36 | Object form, made available under the License, as indicated by a
37 | copyright notice that is included in or attached to the work
38 | (an example is provided in the Appendix below).
39 |
40 | "Derivative Works" shall mean any work, whether in Source or Object
41 | form, that is based on (or derived from) the Work and for which the
42 | editorial revisions, annotations, elaborations, or other modifications
43 | represent, as a whole, an original work of authorship. For the purposes
44 | of this License, Derivative Works shall not include works that remain
45 | separable from, or merely link (or bind by name) to the interfaces of,
46 | the Work and Derivative Works thereof.
47 |
48 | "Contribution" shall mean any work of authorship, including
49 | the original version of the Work and any modifications or additions
50 | to that Work or Derivative Works thereof, that is intentionally
51 | submitted to Licensor for inclusion in the Work by the copyright owner
52 | or by an individual or Legal Entity authorized to submit on behalf of
53 | the copyright owner. For the purposes of this definition, "submitted"
54 | means any form of electronic, verbal, or written communication sent
55 | to the Licensor or its representatives, including but not limited to
56 | communication on electronic mailing lists, source code control systems,
57 | and issue tracking systems that are managed by, or on behalf of, the
58 | Licensor for the purpose of discussing and improving the Work, but
59 | excluding communication that is conspicuously marked or otherwise
60 | designated in writing by the copyright owner as "Not a Contribution."
61 |
62 | "Contributor" shall mean Licensor and any individual or Legal Entity
63 | on behalf of whom a Contribution has been received by Licensor and
64 | subsequently incorporated within the Work.
65 |
66 | 2. Grant of Copyright License. Subject to the terms and conditions of
67 | this License, each Contributor hereby grants to You a perpetual,
68 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
69 | copyright license to reproduce, prepare Derivative Works of,
70 | publicly display, publicly perform, sublicense, and distribute the
71 | Work and such Derivative Works in Source or Object form.
72 |
73 | 3. Grant of Patent License. Subject to the terms and conditions of
74 | this License, each Contributor hereby grants to You a perpetual,
75 | worldwide, non-exclusive, no-charge, royalty-free, irrevocable
76 | (except as stated in this section) patent license to make, have made,
77 | use, offer to sell, sell, import, and otherwise transfer the Work,
78 | where such license applies only to those patent claims licensable
79 | by such Contributor that are necessarily infringed by their
80 | Contribution(s) alone or by combination of their Contribution(s)
81 | with the Work to which such Contribution(s) was submitted. If You
82 | institute patent litigation against any entity (including a
83 | cross-claim or counterclaim in a lawsuit) alleging that the Work
84 | or a Contribution incorporated within the Work constitutes direct
85 | or contributory patent infringement, then any patent licenses
86 | granted to You under this License for that Work shall terminate
87 | as of the date such litigation is filed.
88 |
89 | 4. Redistribution. You may reproduce and distribute copies of the
90 | Work or Derivative Works thereof in any medium, with or without
91 | modifications, and in Source or Object form, provided that You
92 | meet the following conditions:
93 |
94 | (a) You must give any other recipients of the Work or
95 | Derivative Works a copy of this License; and
96 |
97 | (b) You must cause any modified files to carry prominent notices
98 | stating that You changed the files; and
99 |
100 | (c) You must retain, in the Source form of any Derivative Works
101 | that You distribute, all copyright, patent, trademark, and
102 | attribution notices from the Source form of the Work,
103 | excluding those notices that do not pertain to any part of
104 | the Derivative Works; and
105 |
106 | (d) If the Work includes a "NOTICE" text file as part of its
107 | distribution, then any Derivative Works that You distribute must
108 | include a readable copy of the attribution notices contained
109 | within such NOTICE file, excluding those notices that do not
110 | pertain to any part of the Derivative Works, in at least one
111 | of the following places: within a NOTICE text file distributed
112 | as part of the Derivative Works; within the Source form or
113 | documentation, if provided along with the Derivative Works; or,
114 | within a display generated by the Derivative Works, if and
115 | wherever such third-party notices normally appear. The contents
116 | of the NOTICE file are for informational purposes only and
117 | do not modify the License. You may add Your own attribution
118 | notices within Derivative Works that You distribute, alongside
119 | or as an addendum to the NOTICE text from the Work, provided
120 | that such additional attribution notices cannot be construed
121 | as modifying the License.
122 |
123 | You may add Your own copyright statement to Your modifications and
124 | may provide additional or different license terms and conditions
125 | for use, reproduction, or distribution of Your modifications, or
126 | for any such Derivative Works as a whole, provided Your use,
127 | reproduction, and distribution of the Work otherwise complies with
128 | the conditions stated in this License.
129 |
130 | 5. Submission of Contributions. Unless You explicitly state otherwise,
131 | any Contribution intentionally submitted for inclusion in the Work
132 | by You to the Licensor shall be under the terms and conditions of
133 | this License, without any additional terms or conditions.
134 | Notwithstanding the above, nothing herein shall supersede or modify
135 | the terms of any separate license agreement you may have executed
136 | with Licensor regarding such Contributions.
137 |
138 | 6. Trademarks. This License does not grant permission to use the trade
139 | names, trademarks, service marks, or product names of the Licensor,
140 | except as required for reasonable and customary use in describing the
141 | origin of the Work and reproducing the content of the NOTICE file.
142 |
143 | 7. Disclaimer of Warranty. Unless required by applicable law or
144 | agreed to in writing, Licensor provides the Work (and each
145 | Contributor provides its Contributions) on an "AS IS" BASIS,
146 | WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
147 | implied, including, without limitation, any warranties or conditions
148 | of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
149 | PARTICULAR PURPOSE. You are solely responsible for determining the
150 | appropriateness of using or redistributing the Work and assume any
151 | risks associated with Your exercise of permissions under this License.
152 |
153 | 8. Limitation of Liability. In no event and under no legal theory,
154 | whether in tort (including negligence), contract, or otherwise,
155 | unless required by applicable law (such as deliberate and grossly
156 | negligent acts) or agreed to in writing, shall any Contributor be
157 | liable to You for damages, including any direct, indirect, special,
158 | incidental, or consequential damages of any character arising as a
159 | result of this License or out of the use or inability to use the
160 | Work (including but not limited to damages for loss of goodwill,
161 | work stoppage, computer failure or malfunction, or any and all
162 | other commercial damages or losses), even if such Contributor
163 | has been advised of the possibility of such damages.
164 |
165 | 9. Accepting Warranty or Additional Liability. While redistributing
166 | the Work or Derivative Works thereof, You may choose to offer,
167 | and charge a fee for, acceptance of support, warranty, indemnity,
168 | or other liability obligations and/or rights consistent with this
169 | License. However, in accepting such obligations, You may act only
170 | on Your own behalf and on Your sole responsibility, not on behalf
171 | of any other Contributor, and only if You agree to indemnify,
172 | defend, and hold each Contributor harmless for any liability
173 | incurred by, or claims asserted against, such Contributor by reason
174 | of your accepting any such warranty or additional liability.
175 |
176 | END OF TERMS AND CONDITIONS
177 |
178 | APPENDIX: How to apply the Apache License to your work.
179 |
180 | To apply the Apache License to your work, attach the following
181 | boilerplate notice, with the fields enclosed by brackets "[]"
182 | replaced with your own identifying information. (Don't include
183 | the brackets!) The text should be enclosed in the appropriate
184 | comment syntax for the file format. We also recommend that a
185 | file or class name and description of purpose be included on the
186 | same "printed page" as the copyright notice for easier
187 | identification within third-party archives.
188 |
189 | Copyright [yyyy] [name of copyright owner]
190 |
191 | Licensed under the Apache License, Version 2.0 (the "License");
192 | you may not use this file except in compliance with the License.
193 | You may obtain a copy of the License at
194 |
195 | http://www.apache.org/licenses/LICENSE-2.0
196 |
197 | Unless required by applicable law or agreed to in writing, software
198 | distributed under the License is distributed on an "AS IS" BASIS,
199 | WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
200 | See the License for the specific language governing permissions and
201 | limitations under the License.
202 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | [](https://github.com/xarray-contrib/xdggs/actions/ci.yml?query=branch%3Amain+event%3Apush)
2 | [](https://xdggs.readthedocs.io)
3 | [](https://pypi.org/project/xdggs)
4 | [](https://github.com/python/black)
5 | [](https://github.com/conda-forge/xdggs-feedstock)
6 |
7 | ---
8 |
9 |
10 |
11 | # xdggs: discrete global grid systems with xarray
12 |
13 | `xdggs` is an open-source Python package that provides tools for handling geospatial data using Discrete Global Grid Systems (DGGS).
14 |
15 | It enables efficient manipulation and analysis of multi-dimensional gridded data within a DGGS framework, supporting spatial data processing, resampling, and aggregation on both global and regional scales.
16 |
17 | Inspired by the growing need for scalable geospatial data analysis with DGGS, `xdggs` is built upon the robust [Xarray](https://xarray.pydata.org/) ecosystem, which simplifies working with labeled multi-dimensional arrays.
18 |
19 | As an extension of Xarray, `xdggs` leverages Xarray's capabilities, including seamless access to formats like [NetCDF](https://www.unidata.ucar.edu/software/netcdf/), [Zarr](https://zarr.readthedocs.io/), and parallelization through [Dask](https://www.dask.org/), to provide a powerful and flexible toolkit for geospatial analysis.
20 |
21 | ## Key Features
22 |
23 | - **Seamless Integration with Xarray**: Use `xdggs` alongside Xarray's powerful tools for managing labeled, multi-dimensional data.
24 | - **Support for DGGS**: Convert geospatial data into DGGS representations, allowing for uniform spatial partitioning of the Earth's surface.
25 | - **Spatial Resampling**: Resample data on DGGS grids, enabling downscaling or upscaling across multiple resolutions.
26 | - **DGGS Aggregation**: Perform spatial aggregation of data on DGGS cells.
27 | - **Efficient Data Management**: Manage large datasets with Xarray's lazy loading, Dask integration, and chunking to optimize performance.
28 |
29 | ## Documentation
30 |
31 | You can find the documentation in [https://xdggs.readthedocs.io/en/latest/](https://xdggs.readthedocs.io/en/latest/).
32 |
33 | ## Demo
34 |
35 | 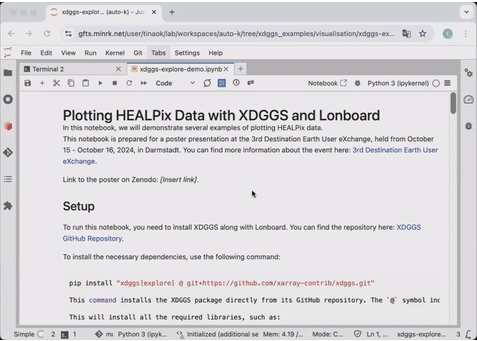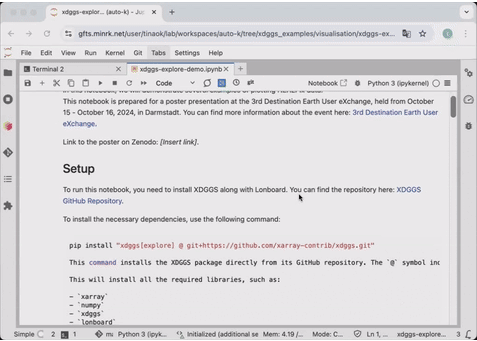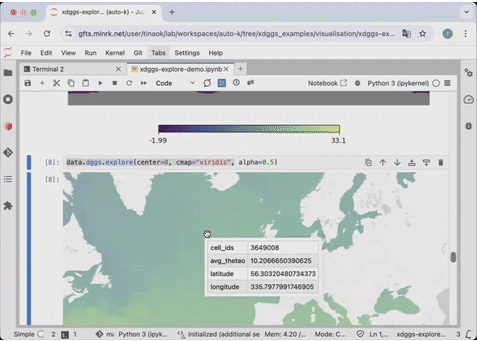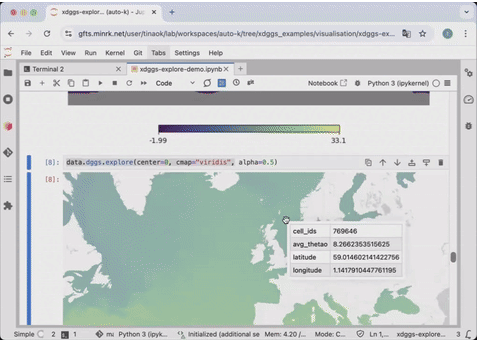
36 |
37 | ## Getting Started
38 |
39 | As an example, this is how you would use `xdggs` to reconstruct geographical coordinates from the cell ids then create an interactive plot indicating cell ids, data values and the associated geographical coordinates:
40 |
41 | ```python
42 | import xarray as xr
43 | import xdggs
44 |
45 | ds = xdggs.tutorial.open_dataset("air_temperature", "h3")
46 |
47 | # Decode DGGS coordinates
48 | ds_idx = ds.pipe(xdggs.decode)
49 |
50 | # Assign geographical coordinates
51 | ds_idx = ds_idx.dggs.assign_latlon_coords()
52 |
53 | # Interactive visualization
54 | ds_idx['air'].isel(time=0).compute().dggs.explore(center=0, cmap="viridis", alpha=0.5)
55 |
56 | ```
57 |
58 | ## Roadmap
59 |
60 | We have exciting plans to expand xdggs with new features and improvements. You can check out our roadmap in the [design_doc.md](https://github.com/xarray-contrib/xdggs/blob/main/design_doc.md) file for details on the design of xdggs, upcoming features, and future enhancements.
61 |
62 | ## Contributing
63 |
64 | We welcome contributions to `xdggs`! Please follow these steps to get involved:
65 |
66 | 1. Fork the repository.
67 | 2. Create a new branch (`git checkout -b feature-branch`).
68 | 3. Make your changes and write tests.
69 | 4. Ensure all tests pass (`pytest`).
70 | 5. Submit a pull request!
71 |
72 | ## License
73 |
74 | `xdggs` is licensed under the Apache License License. See [LICENSE](https://github.com/xarray-contrib/xdggs/blob/main/LICENSE) for more details.
75 |
76 | ## Acknowledgments
77 |
78 | This project was initiated using funding from CNES (PANGEO IAOCEA, contract R&T R-S23/DU-0002-025-01) and the European Union (ERC, WaterSmartLand, 101125476, Interreg-BSR, HyTruck, #C031).
79 |
--------------------------------------------------------------------------------
/ci/docs.yml:
--------------------------------------------------------------------------------
1 | name: xdggs-docs
2 | channels:
3 | - conda-forge
4 | dependencies:
5 | - python=3.12
6 | - sphinx
7 | - myst-nb
8 | - sphinx-autobuild
9 | - sphinx-book-theme
10 | - sphinx-autosummary-accessors
11 | - sphinx-copybutton
12 | - sphinx-design
13 | - sphinx-inline-tabs
14 | - sphinxcontrib-bibtex
15 | - xarray
16 | - h5netcdf
17 | - netcdf4
18 | - pooch
19 | - matplotlib-base
20 | - shapely
21 | - pytest
22 | - hypothesis
23 | - ruff
24 | - typing-extensions
25 | - geoarrow-pyarrow
26 | - lonboard
27 | - arro3-core
28 | - h3ronpy
29 | - cdshealpix
30 | - pip
31 | - pip:
32 | - -e ..
33 |
--------------------------------------------------------------------------------
/ci/environment.yml:
--------------------------------------------------------------------------------
1 | channels:
2 | - conda-forge
3 | dependencies:
4 | - xarray
5 | - h5netcdf
6 | - netcdf4
7 | - pooch
8 | - matplotlib-base
9 | - shapely
10 | - pytest
11 | - hypothesis
12 | - ruff
13 | - typing-extensions
14 | - geoarrow-pyarrow
15 | - lonboard
16 | - arro3-core
17 | - cdshealpix
18 | - h3ronpy
19 |
--------------------------------------------------------------------------------
/ci/install-upstream-wheels.sh:
--------------------------------------------------------------------------------
1 | #!/usr/bin/env bash
2 |
3 | if which micromamba; then
4 | conda=micromamba
5 | else
6 | conda=mamba
7 | fi
8 |
9 | # force-remove re-installed versions
10 | $conda remove -y --force \
11 | xarray \
12 | pandas \
13 | cdshealpix
14 | python -m pip uninstall -y h3ronpy
15 |
16 | # install from scientific-python wheels
17 | python -m pip install \
18 | -i https://pypi.anaconda.org/scientific-python-nightly-wheels/simple \
19 | --no-deps \
20 | --pre \
21 | --upgrade \
22 | pandas \
23 | numpy \
24 | xarray
25 | # pyarrow nightly builds
26 | python -m pip install \
27 | -i https://pypi.fury.io/arrow-nightlies/ \
28 | --prefer-binary \
29 | --no-deps \
30 | --pre \
31 | --upgrade \
32 | pyarrow
33 |
34 |
35 | # install from github
36 | python -m pip install --no-deps --upgrade \
37 | git+https://github.com/Unidata/cftime \
38 | git+https://github.com/astropy/astropy \
39 | "git+https://github.com/nmandery/h3ronpy#subdirectory=h3ronpy" \
40 | git+https://github.com/cds-astro/cds-healpix-python
41 |
--------------------------------------------------------------------------------
/design_doc.md:
--------------------------------------------------------------------------------
1 | # XDGGS - Design document
2 |
3 | Xarrays extension for DGGS. Technical specifications.
4 |
5 | ## Goals
6 |
7 | The goal of the `xdggs` library is to facilitate working with multiple Discrete Global Grid Systems (DGGSs) via a unified, high-level and user-friendly API that is deeply integrated with [Xarray](https://xarray.dev).
8 | This document describes the in-memory representation of DGGS data in Python environments.
9 |
10 | Examples of common DGGS features that `xdggs` should provide or facilitate:
11 |
12 | - convert a DGGS from/to another grid (e.g., a DGGS, a latitude/longitude rectilinear grid, a raster grid, an unstructured mesh)
13 | - convert a DGGS from/to vector data (points, lines, polygons, envelopes)
14 | - convert between different cell id representations of a same DGGS (e.g., uint64 vs. string)
15 | - select data on a DGGS by cell ids or by geometries (spatial indexing)
16 | - expand and reduce the available resolutions of a DGGS using down and upsampling, respectively.
17 | - operations between similar DGGS (with auto-alignment)
18 | - re-organize cell ids (e.g., spatial shuffling / partitioning)
19 | - plotting
20 |
21 | Conversion between DGGS and other grids or vector features may require specific interpolation or regridding methods.
22 |
23 | `xdggs` should leverage the current recommended Xarray extension mechanisms ([apply_ufunc](https://docs.xarray.dev/en/stable/examples/apply_ufunc_vectorize_1d.html), [accessors](https://docs.xarray.dev/en/stable/internals/extending-xarray.html), [custom indexes](https://docs.xarray.dev/en/stable/internals/how-to-create-custom-index.html)) and possibly the future ones (e.g., variable encoders/decoders) to provide DGGS-specific functionality on top of Xarray core features.
24 |
25 | `xdggs` should facilitate interoperability with other existing Xarray extensions (e.g., [xvec](https://github.com/xarray-contrib/xvec) for vector data or [uxarray](https://github.com/UXARRAY/uxarray) for unstructured grids).
26 |
27 | `xdggs` should also leverage the existing implementation of various DGGSs exposed to Python via 3rd-party libraries, here below referred as "backends". Preferrably, those backends would expose DGGS functionality in an efficient way as vectorized functions (e.g., working with NumPy arrays).
28 |
29 | `xdggs` should try to follow standards and/or conventions defined for DGGS (see below). However, we may need to depart from them for practical reasons (e.g., common practices in popular DGGS libraries that do not fit well with the proposed standards). Strict adherence to a standard is welcome but shouldn't be enforced by all means.
30 |
31 | `xdggs` should also try to support applications in both GIS and Earth-System communities, which may each use DGGS in slightly different ways (see examples below).
32 |
33 | When possible, `xdggs` operations should scale to fine DGGS resolutions (billions of cells). This can be done vertically using backends with vectorized bindings of DGGS implementations written in low-level languages and/or horizontally leveraging Xarray interoperability with Dask. Some operations like spatial indexing may be hard to scale horizontally, though. For the latter, we should probably focus `xdggs` development first towards good vertical scaling before figuring out how they can be scaled horizontally (for reference, see [dask-geopandas](https://github.com/geopandas/dask-geopandas) and [spatialpandas](https://github.com/holoviz/spatialpandas)).
34 |
35 | ## Non-Goals
36 |
37 | `xdggs` should focus on providing the core DGGS functionality and operations that are listed above. Higher-level operations that can be implemented by combining together those core operations are out-of-scope and should be implemented in downstream libraries. Likewise, there may be many ways of resampling a grid to a DGGS ; `xdggs` should support the most common methods but not try to support _all of them_.
38 |
39 | `xdggs` should try not re-inventing the wheel and delegate to Xarray API when possible.
40 |
41 | `xdggs` does not implement any particular DGGS from scratch. `xdggs` does not aim at providing _all the functionality provided by each grid_ (e.g., some functionality may be very specific to one DGGS and not supported by other DGGSs, or some functionality may not be available yet in one DGGS Python backend).
42 |
43 | Although some DGGS may handle both the spatial and temporal domains in a joint fashion, `xdggs` focuses primarily on the spatial domain. The temporal domain is considered as orthogonal and already benefits from many core features provided by Xarray.
44 |
45 | ## Discrete Global Grid Systems
46 |
47 | A Discrete Global Grid System (DGGS) can be roughly defined as a partitioning or tessellation of the entire Earth's surface into a finite number of "cells" or "zones". The shape and the properties of these cells generally vary from one DGGS to another. Most DGGSs are also hierarchical, i.e., the cells are arranged recursively on multiple levels or resolutions. Follow the links in the subsection below for a more strict and detailed definition of a DGGS.
48 |
49 | DGGSs may be used in various ways, e.g.,
50 |
51 | - Applications in Earth-system modelling seem to use DGGS as grids of contiguous, fixed-resolution cells covering the entire Earth's surface or a region of interest (figure 1). This makes the analysis of simulation outputs on large extents of the Earth's surface easier. DGGS may also be used as pyramid data (multiple stacked datasets at different resolutions)
52 | - Applications in GIS often consist of using DGGS to display aggregated (vector) data as a collection of cells with a more complex spatial distribution (sparse) and sometimes with mixed resolutions (figures 2 and 3).
53 |
54 | 
55 | Figure 1: DGGS data as contiguous cells of fixed resolution ([source](https://danlooo.github.io/DGGS.jl/))
56 |
57 | 
58 |
59 | Figure 2: Data aggreated on DGGS (H3) sparsely distributed cells of fixed resolution ([source](https://medium.com/@jesse.b.nestler/how-to-convert-h3-cell-boundaries-to-shapely-polygons-in-python-f7558add2f63)).
60 |
61 | 
62 |
63 | Figure 3: Raster data converted as DGGS (H3) cells of mixed resolutions ([source](https://github.com/nmandery/h3ronpy)).
64 |
65 | ### Standards and Conventions
66 |
67 | The [OGC abstract specification topic 21](http://www.opengis.net/doc/AS/dggs/2.0) defines properties of a DGGS including the reference systems of its grids.
68 |
69 | However, there is no consensus yet about the actual specification on how to work with DGGS-indexed data.
70 | [OGC API draft](https://github.com/opengeospatial/ogcapi-discrete-global-grid-systems) defines ways of how to access DGGS data via web-based API.
71 | A [DGGS data specification draft](https://github.com/danlooo/dggs-data-spec) aims to specify the storage format of DGGS data.
72 |
73 | There are discrepancies between the proposed standards and popular DGGS libraries (H3, S2, HealPIX). For example regarding the term used to define a grid unit: The two specifications above use "zone", S2/H3 use "cell" and HealPIX uses "pixel".
74 | OGC abstract specification topic 21 defines the region as a zone and its boundary geometry as a cell.
75 | Although in this document we use "cell", the term to choose for `xdggs` is still open for discussion.
76 |
77 | Furthermore, several libraries allow for customised instantiation of a DGGS. This makes it crucial to be able to specify the particular DGGS type and potentially additional parameters. The OGC DGGS working group is discussing how to define a DGGS reference system, analogously to the spatial/coordinate reference systems registries (PROJ, EPSG, ..).
78 |
79 | The OGC is [currently discussing](https://github.com/opengeospatial/ogcapi-discrete-global-grid-systems/issues/41) ways to describe and identify unique DGGS types. This is slightly different from storing DGGS data. But the important info here is to be able to store the metadata about the specifics of the used DGGS in form e.g. an identifier, label, link to a detailed definition. This would likely need to include the main type e.g. H3, S2, HEALPIX, RHEALPIX, DGGGRID_ISEAxxx, but also required parameters like HEALPIX (indexing: nested or ring), RHEALPIX (ellipsoid, orientation/rotation ..), DGGRID-icosahedron-based DGGS types (orientation, potentially mixed apertures..), GeoSOT. It would be good to have synergies here and not reinvent the wheel.
80 |
81 | ### Backends (Python)
82 |
83 | Several Python packages are currently available for handling certain DGGSs. They mostly consist of Python bindings of DGGS implementations written in C/C++/Rust. Here is a list (probably incomplete):
84 |
85 | - [healpy](https://github.com/healpy/healpy): Python bindings of [HealPix](https://healpix.sourceforge.io/)
86 | - mostly vectorized
87 | - [rhealpixdggs-py](https://github.com/manaakiwhenua/rhealpixdggs-py): Python/Numpy implementation of rHEALPix
88 | - [h3-py](https://github.com/uber/h3-py): "official" Python bindings of [H3](https://h3geo.org/)
89 | - experimental and incomplete vectorized version of H3's API (removed in the forthcoming v4 release?)
90 | - [h3pandas](https://github.com/DahnJ/H3-Pandas): integration of h3-py (non-vectorized) with pandas and geopandas
91 | - [h3ronpy](https://github.com/nmandery/h3ronpy): Python bindings of [h3o](https://github.com/HydroniumLabs/h3o) (Rust implementation of H3)
92 | - provides high-level features (conversion, etc.) working with arrow, numpy (?), pandas/geopandas and polars
93 | - [s2geometry](https://github.com/google/s2geometry): Python bindings generated with SWIG
94 | - not vectorized nor very "pythonic"
95 | - plans to switch to pybind11 (no time frame given)
96 | - [spherely](https://github.com/benbovy/spherely): Python bindings of S2, mostly copying shapely's API
97 | - provides numpy-like universal functions
98 | - not yet ready for use
99 | - [dggrid4py](https://github.com/allixender/dggrid4py): Python wrapper for [DGGRID](https://github.com/sahrk/DGGRID)
100 | - DGGRID implements many DGGS variants!
101 | - DGGRID current design makes it hardly reusable from within Python in an optimal way (the dggrid wrapper communicates with DGGRID through OS processes and I/O generated files)
102 |
103 | ## Representation of DGGS Data in Xdggs
104 |
105 | `xdggs` represents a DGGS as an Xarray Dataset or DataArray containing a 1-dimensional coordinate with cell ids as labels and with grid name, resolution & parameters (optional) as attributes. This coordinate is indexed using a custom, Xarray-compatible `DGGSIndex`. Multiple dimensions may be used if the coordinate consists of multiple parts, e.g., polyhedron face, x, and y on that face in DGGRID PROJTRI.
106 |
107 | `xdggs` does not support a Dataset or DataArray with multiple coordinates indexed with a `DGGSIndex` (only one DGGS per object is supported).
108 |
109 | The cell ids in the 1-dimensional coordinate are all relative to the _exact same_ grid, i.e., same grid system, same grid parameter values and same grid resolution! For simplicity, `xdggs` does not support cell ids of mixed-resolutions in the same coordinate.
110 |
111 | ### DGGSIndex
112 |
113 | `xdggs.DGGSIndex` is the base class for all Xarray DGGS-aware indexes. It inherits from `xarray.indexes.Index` and has the following specifications:
114 |
115 | - It encapsulates an `xarray.indexes.PandasIndex` built from cell ids so that selection and alignment by cell id is possible
116 | - It might also eventually encapsulate a spatial index (RTree, KDTree) to enable data selection by geometries, e.g., find nearest cell centroids, extract all cells intersecting a polygon, etc.
117 | - Alternatively, spatial indexing might be enabled by explicit conversion of cells to vector geometries and then by reusing the Xarray spatial indexes available in [xvec](https://github.com/xarray-contrib/xvec)
118 | - It partially implements the Xarray Index API to enable DGGS-aware alignment and selection
119 | - Calls are most often redirected to the encapsulated `PandasIndex`
120 | - Some pre/post checks or processing may be done, e.g., to prevent the alignment of two indexes that are not on the exact same grid.
121 | - The `DGGSIndex.__init__()` constructor only requires cell ids and the name of the cell (array) dimension
122 | - The `DGGSIndex.from_variables()` factory method parses the attributes of the given cell ids coordinates and creates the right index object (subclass) accordingly
123 | - It declares a few abstract methods for grid-aware operations (e.g., convert between cell id and lat/lon point or geometry, etc.)
124 | - They can be implemented in subclasses (see below)
125 | - They are either called from within the DGGSIndex or from the `.dggs` Dataset/DataArray accessors
126 |
127 | Each DGGS supported in `xdggs` has its own subclass of `DGGSIndex`, e.g.,
128 |
129 | - `HealpixIndex` for Healpix
130 | - `H3Index` for H3
131 | - ...
132 |
133 | A DGGSIndex can be set directly from a cell ids coordinate using the Xarray API:
134 |
135 | ```python
136 | import xarray as xr
137 | import xdggs
138 |
139 | ds = xr.Dataset(
140 | coords={"cell": ("cell", [...], {"grid_name": "h3", "resolution": 3})}
141 | )
142 |
143 | # auto-detect grid system and parameters
144 | ds.set_xindex("cell", xdggs.DGGSIndex)
145 |
146 | # set the grid system and parameters manually
147 | ds.set_xindex("cell", xdggs.H3Index, resolution=3)
148 | ```
149 |
150 | The DGGSIndex is set automatically when converting a gridded or vector dataset to a DGGS dataset (see below).
151 |
152 | ## Conversion from/to DGGS
153 |
154 | DGGS data may be created from various sources, e.g.,
155 |
156 | - regridded from a latitude/longitude rectilinear grid
157 | - regridded from an unstructured grid
158 | - regridded and reprojected from a raster having a local projection
159 | - aggregated from vector point data
160 | - filled from polygon data
161 |
162 | Conversely, DGGS data may be converted to various forms, e.g.,
163 |
164 | - regridded on a latitude/longitude rectilinear grid
165 | - rasterized (resampling / projection)
166 | - conversion to vector point data (cell centroids)
167 | - conversion to vector polygon data (cell boundaries)
168 |
169 | Here is a tentative API based on Dataset/DataArray `.dggs` accessors (note: other options are discussed in [this issue](https://github.com/xarray-contrib/xdggs/issues/13)):
170 |
171 | ```python
172 | # "convert" directly from existing cell ids coordinate to DGGS
173 | # basically an alias to ds.set_xindex(..., DGGSIndex)
174 | ds.dggs.from_cell_ids(...)
175 |
176 | # convert from lat/lon grid
177 | ds.dggs.from_latlon_grid(...)
178 |
179 | # convert from raster
180 | ds.dggs.from_raster(...)
181 |
182 | # convert from point data
183 | ds.dggs.from_points(...)
184 |
185 | # convert from point data (with aggregation)
186 | ds.dggs.from_points_aggregate(...)
187 |
188 | # convert from point data (with aggregation using Xarray API)
189 | ds.dggs.from_points(...).groupby(...).mean()
190 |
191 | # convert to lat/lon grid
192 | ds.dggs.to_latlon_grid(...)
193 |
194 | # convert to raster
195 | ds.dggs.to_raster(...)
196 |
197 | # convert to points (cell centroids)
198 | ds.dggs.to_points(...)
199 |
200 | # convert to polygons (cell boundaries)
201 | ds.dggs.to_polygons(...)
202 | ```
203 |
204 | In the API methods above, the "dggs" accessor name serves as a prefix.
205 |
206 | Those methods are all called from an existing xarray Dataset (DataArray) and should all return another Dataset (DataArray):
207 |
208 | - Xarray has built-in support for regular grids
209 | - for rasters, we could return objects that are [rioxarray](https://github.com/corteva/rioxarray)-friendly
210 | - for vector data, we could return objects that are [xvec](https://github.com/xarray-contrib/xvec)-friendly (coordinate of shapely objects)
211 | - etc.
212 |
213 | ## Extracting DGGS Cell Geometries
214 |
215 | DGGS cell geometries could be extracted using the conversion methods proposed above. Alternatively, it would be convenient to get them directly as xarray DataArrays so that we can for example manually assign them as coordinates.
216 |
217 | The API may look like:
218 |
219 | ```python
220 | # return a DataArray of DGGS cell centroids as shapely.POINT objects
221 | ds.dggs.cell_centroids()
222 |
223 | # return two DataArrays of DGGS cell centroids as lat/lon coordinates
224 | ds.dggs.cell_centroids_coords()
225 |
226 | # return a DataArray of DGGS cell boundaries as shapely.POLYGON objects
227 | ds.dggs.cell_boundaries()
228 |
229 | # return a DataArray of DGGS cell envelopes as shapely.POLYGON objects
230 | ds.dggs.cell_envelopes()
231 | ```
232 |
233 | ## Indexing and Selecting DGGS Data
234 |
235 | ### Selection by Cell IDs
236 |
237 | The simplest way to select DGGS data is by cell ids. This can be done directly using Xarray's API (`.sel()`):
238 |
239 | ```python
240 | ds.sel(cell=value)
241 | ```
242 |
243 | where `value` can be a single cell id (integer or string/token?) or an array-like of cell ids. This is easily supported by the DGGSIndex encapsulating a PandasIndex. We might also want to support other `value` types, e.g.,
244 |
245 | - assuming that DGGS cell ids are defined such that contiguous cells in space have contiguous id values, we could provide a `slice` to define a range of cell ids.
246 | - DGGSIndex might implement some DGGS-aware logic such that it auto-detects if the given input cells are parent cells (lower DGGS resolution) and then selects all child cells accordingly.
247 |
248 | We might want to select cell neighbors (i.e., return a new Dataset/DataArray with a new neighbor dimension), probably via a specific API (`.dggs` accessors).
249 |
250 | ### Selection by Geometries (Spatial Indexing)
251 |
252 | Another useful way of selecting DGGS data is from input geometries (spatial queries), e.g.,
253 |
254 | - Select all cells that are the closest to a collection of data points
255 | - Select all cells that intersects with or are fully contained in a polygon
256 |
257 | This kind of selection requires spatial indexes as this can not be done with a pandas index (see [this issue](https://github.com/xarray-contrib/xdggs/issues/16)).
258 |
259 | If we support spatial indexing directly in `xdggs`, we can hardly reuse Xarray's `.sel()` for spatial queries as `ds.sel(cell=shapely.Polygon(...))` would look quite confusing. Perhaps better would be to align with [xvec](https://github.com/xarray-contrib/xvec) and have a separate `.dggs.query()` method.
260 |
261 | Alternatively, we could just get away with the conversion and cell geometry extraction methods proposed above and leave spatial indexes/queries to [xvec](https://github.com/xarray-contrib/xvec).
262 |
263 | ## Handling hierarchical DGGS
264 |
265 | DGGS are grid systems with grids of the same topology but different spatial resolution.
266 | There is a hierarchical relationship between grids of different resolutions.
267 | Even though the coordinate of one grid in the DGGS of a Dataset (DataArray) is limited to cell ids of same resolution (no mixed-resolutions), `xdggs` can still provide functionality to deal with the hierarchical aspect of DGGSs.
268 |
269 | Selection by parent cell ids may be in example (see section above). Another example would be to have utility methods to explicitly change the grid resolution (see [issue #18](https://github.com/xarray-contrib/xdggs/issues/18) for more details and discussion).
270 | One can also store DGGS data at all resolutions as a list of datasets.
271 |
272 | However, like in hexagonal grids of aperture 3 or 4 (e.g. DGGRID ISEA4H), the parent child relationship can be also ambiguous.
273 | The actual spatial aggregation functions in the subclasses might be implemented differently depending on the selected DGGS.
274 |
275 | ## Operations between similar DGGS (alignment)
276 |
277 | Computation involving multiple DGGS datasets (or dataarrays) often requires to align them together. Sometimes this can be trivial (same DGGS with same resolution and parameter values) but in other cases this can be complex (requires regridding or a change of DGGS resolution).
278 |
279 | In Xarray, alignment of datasets (dataarrays) is done primarily via their indexes. Since a DGGSIndex wraps a PandasIndex, it is easy to support alignment by cells ids (trivial case). At the very least, a DGGSIndex should raise an error when trying to align cell ids that do not refer to the exact same DGGS (i.e., same system, resolution and parameter values). For the complex cases, it may be preferable to handle them manually instead of trying to make the DGGSIndex perform the alignment automatically. Regridding and/or changing the resolution of a DGGS (+ data aggregation) often highly depend on the use-case so it might be hard to find a default behavior. Also performing those operations automatically and implicitly would probably feel too magical. That being said, in order to help alignment `xdggs` may provide some utility methods to change the grid resolution (see section above) and/or to convert from one DGGS to another.
280 |
281 | ## Plotting
282 |
283 | Three approaches are possible (non-mutually exclusive):
284 |
285 | 1. convert cell data into gridded or raster data (choose grid/raster resolution depending on the resolution of the rendered figure) and then reuse existing python plotting libraries (matplotlib, cartopy) maybe through xarray plotting API
286 | 2. convert cell data into vector data and plot the latter via, e.g., [xvec](https://github.com/xarray-contrib/xvec) or [geopandas](https://github.com/geopandas/geopandas) API
287 | 3. leverage libraries that support plotting DGGS data, e.g., [lonboard](https://github.com/developmentseed/lonboard) enables interactive plotting in Jupyter via deck.gl, which has support of H3 and S2 cell data.
288 |
289 | The 3rd approach (lonboard) is efficient for plotting large DGGS data: we would only need to transfer cell ids (tokens) and cell data and then let deck.gl render the cells efficiently in the web browser using the GPU. For approach 1, we might want to investigate using [datashader](https://github.com/holoviz/datashader) to set both the resolution and raster extent dynamically. Likewise for approach 2, we could dynamically downgrade the DGGS resolution and aggregate the data before converting it into vector data in order to allow (interactive) plotting of large DGGS data.
290 |
--------------------------------------------------------------------------------
/docs/_static/logos/xdggs_logo.png:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/xarray-contrib/xdggs/0a37d34c601f8521dec9228115ecba248e0b751e/docs/_static/logos/xdggs_logo.png
--------------------------------------------------------------------------------
/docs/api-hidden.rst:
--------------------------------------------------------------------------------
1 | :orphan:
2 |
3 | .. currentmodule:: xdggs
4 |
5 | .. autosummary::
6 | :toctree: generated
7 |
8 | DGGSInfo.level
9 |
10 | DGGSInfo.from_dict
11 | DGGSInfo.to_dict
12 | DGGSInfo.cell_boundaries
13 | DGGSInfo.cell_ids2geographic
14 | DGGSInfo.geographic2cell_ids
15 |
16 | HealpixInfo.level
17 | HealpixInfo.indexing_scheme
18 | HealpixInfo.valid_parameters
19 | HealpixInfo.nside
20 | HealpixInfo.nest
21 |
22 | HealpixInfo.from_dict
23 | HealpixInfo.to_dict
24 | HealpixInfo.cell_boundaries
25 | HealpixInfo.cell_ids2geographic
26 | HealpixInfo.geographic2cell_ids
27 |
28 | H3Info.level
29 | H3Info.valid_parameters
30 |
31 | H3Info.from_dict
32 | H3Info.to_dict
33 | H3Info.cell_boundaries
34 | H3Info.cell_ids2geographic
35 | H3Info.geographic2cell_ids
36 |
--------------------------------------------------------------------------------
/docs/api.rst:
--------------------------------------------------------------------------------
1 | .. _api:
2 |
3 | #############
4 | API reference
5 | #############
6 |
7 | Top-level functions
8 | ===================
9 |
10 | .. currentmodule:: xdggs
11 |
12 | .. autosummary::
13 | :toctree: generated
14 |
15 | decode
16 |
17 | Grid parameter objects
18 | ======================
19 |
20 | .. autosummary::
21 | :toctree: generated
22 |
23 | DGGSInfo
24 |
25 | HealpixInfo
26 | H3Info
27 |
28 | .. currentmodule:: xarray
29 |
30 | Dataset
31 | =======
32 |
33 | Parameters
34 | ----------
35 | .. autosummary::
36 | :toctree: generated
37 | :template: autosummary/accessor_attribute.rst
38 |
39 | Dataset.dggs.grid_info
40 | Dataset.dggs.params
41 | Dataset.dggs.decode
42 |
43 |
44 | Data inference
45 | --------------
46 |
47 | .. autosummary::
48 | :toctree: generated
49 | :template: autosummary/accessor_method.rst
50 |
51 | Dataset.dggs.cell_centers
52 | Dataset.dggs.cell_boundaries
53 |
54 | DataArray
55 | =========
56 |
57 | Parameters
58 | ----------
59 | .. autosummary::
60 | :toctree: generated
61 | :template: autosummary/accessor_attribute.rst
62 |
63 | DataArray.dggs.grid_info
64 | DataArray.dggs.params
65 | DataArray.dggs.decode
66 |
67 |
68 | Data inference
69 | --------------
70 |
71 | .. autosummary::
72 | :toctree: generated
73 | :template: autosummary/accessor_method.rst
74 |
75 | DataArray.dggs.cell_centers
76 | DataArray.dggs.cell_boundaries
77 |
78 | Plotting
79 | --------
80 | .. autosummary::
81 | :toctree: generated
82 | :template: autosummary/accessor_method.rst
83 |
84 | DataArray.dggs.explore
85 |
86 | Tutorial
87 | ========
88 |
89 | .. currentmodule:: xdggs
90 |
91 | .. autosummary::
92 | :toctree: generated
93 |
94 | tutorial.open_dataset
95 |
--------------------------------------------------------------------------------
/docs/changelog.md:
--------------------------------------------------------------------------------
1 | # Changelog
2 |
3 | ## 0.2.1 (_unreleased_)
4 |
5 | ### New features
6 |
7 | ### Bug fixes
8 |
9 | ### Documentation
10 |
11 | - Documentation Contributer Guide + Github Button ({pull}`137`)
12 |
13 | ### Internal changes
14 |
15 | ## 0.2.0 (2025-02-12)
16 |
17 | ## New features
18 |
19 | - allow adding additional coords to the cell inspection table in the map ({pull}`122`)
20 | - allow passing `matplotlib` colormap objects to `explore` ({pull}`120`)
21 | - support plotting multi-dimensional data ({pull}`124`)
22 | - allow overriding the grid info data using the function or a new accessor ({pull}`63`, {pull}`121`)
23 |
24 | ## Bug fixes
25 |
26 | - use explicit `arrow` API to extract cell coordinates ({issue}`113`, {pull}`114`)
27 | - correct the `HealpixIndex` `repr` ({pull}`119`)
28 |
29 | ## Internal changes
30 |
31 | - add initial set of tests for `explore` ({pull}`127`)
32 | - adapt to recent changes on RTD ({pull}`122`)
33 |
34 | ## 0.1.1 (2024-11-25)
35 |
36 | ### Bug fixes
37 |
38 | - properly reference the readme in the package metadata ({pull}`106`)
39 |
40 | ## 0.1.0 (2024-11-25)
41 |
42 | ### Enhancements
43 |
44 | - derive cell boundaries ({pull}`30`)
45 | - add grid objects ({pull}`39`, {pull}`57`)
46 | - decoder function ({pull}`47`, {pull}`48`)
47 | - rename the primary grid parameter to `level` ({pull}`65`)
48 | - interactive plotting with `lonboard` ({pull}`67`)
49 | - expose example datasets through `xdggs.tutorial` ({pull}`84`)
50 | - add a preliminary logo ({pull}`101`, {pull}`103`)
51 |
52 | ### Bug fixes
53 |
54 | - fix the cell centers computation ({pull}`61`)
55 | - work around blocked HTTP requests from RTD to github ({pull}`93`)
56 |
57 | ### Documentation
58 |
59 | - create a readme ({pull}`70`)
60 | - create the documentation ({pull}`79`, {pull}`80`, {pull}`81`, {pull}`89`)
61 | - fix headings in tutorials ({pull}`90`)
62 | - rewrite the readme ({pull}`97`)
63 |
64 | ### Internal changes
65 |
66 | - replace `h3` with `h3ronpy` ({pull}`28`)
67 | - setup CI ({pull}`31`)
68 | - tests for the healpix index ({pull}`36`)
69 | - testing utils for exception groups ({pull}`55`)
70 |
71 | ## 0.0.1 (2023-11-28)
72 |
73 | Preliminary version of `xdggs` python package.
74 |
--------------------------------------------------------------------------------
/docs/conf.py:
--------------------------------------------------------------------------------
1 | # -- Project information -----------------------------------------------------
2 | import datetime as dt
3 |
4 | import sphinx_autosummary_accessors
5 |
6 | import xdggs # noqa: F401
7 |
8 | project = "xdggs"
9 | author = f"{project} developers"
10 | initial_year = "2023"
11 | year = dt.datetime.now().year
12 | copyright = f"{initial_year}-{year}, {author}"
13 |
14 | # The root toctree document.
15 | root_doc = "index"
16 |
17 | # -- General configuration ---------------------------------------------------
18 |
19 | # Add any Sphinx extension module names here, as strings. They can be
20 | # extensions coming with Sphinx (named 'sphinx.ext.*') or your custom
21 | # ones.
22 |
23 | source_suffix = {
24 | ".rst": "restructuredtext",
25 | ".md": "myst-nb", # enables myst-nb support for plain md files
26 | }
27 |
28 | extensions = [
29 | "sphinx.ext.extlinks",
30 | "sphinx.ext.intersphinx",
31 | "sphinx.ext.autodoc",
32 | "sphinx.ext.autosummary",
33 | "sphinx.ext.napoleon",
34 | "IPython.sphinxext.ipython_directive",
35 | "IPython.sphinxext.ipython_console_highlighting",
36 | "sphinx_autosummary_accessors",
37 | "myst_nb",
38 | "sphinx_design",
39 | "sphinx_copybutton",
40 | "sphinxcontrib.bibtex",
41 | ]
42 |
43 | extlinks = {
44 | "issue": ("https://github.com/xarray-contrib/xdggs/issues/%s", "GH%s"),
45 | "pull": ("https://github.com/xarray-contrib/xdggs/pull/%s", "PR%s"),
46 | }
47 |
48 | # Add any paths that contain templates here, relative to this directory.
49 | templates_path = ["_templates", sphinx_autosummary_accessors.templates_path]
50 |
51 | # List of patterns, relative to source directory, that match files and
52 | # directories to ignore when looking for source files.
53 | # This pattern also affects html_static_path and html_extra_path.
54 | exclude_patterns = ["_build", "directory"]
55 |
56 | ## Github Buttons
57 | html_theme_options = {
58 | "repository_url": "https://github.com/xarray-contrib/xdggs",
59 | "use_repository_button": True,
60 | "use_issues_button": True,
61 | }
62 |
63 | # -- autosummary / autodoc ---------------------------------------------------
64 |
65 | autosummary_generate = True
66 | autodoc_typehints = "none"
67 |
68 | # -- napoleon ----------------------------------------------------------------
69 |
70 | napoleon_numpy_docstring = True
71 | napoleon_use_param = False
72 | napoleon_use_rtype = False
73 | napoleon_preprocess_types = True
74 | napoleon_type_aliases = {
75 | # general terms
76 | "sequence": ":term:`sequence`",
77 | "iterable": ":term:`iterable`",
78 | "callable": ":py:func:`callable`",
79 | "dict_like": ":term:`dict-like `",
80 | "dict-like": ":term:`dict-like `",
81 | "path-like": ":term:`path-like `",
82 | "mapping": ":term:`mapping`",
83 | "file-like": ":term:`file-like `",
84 | "any": ":py:class:`any