├── AVAE.py
├── LICENSE
├── README.md
├── VAE_plus_plus.py
├── functions.py
└── structure of proposed Adversarial-Variational-Semi-supervised-Learning.PNG
/AVAE.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 | import scipy.io as sc
3 | import numpy as np
4 | import pickle
5 | import random
6 | import time
7 | from functions import *
8 | from VAE_plus_plus import *
9 | from sklearn.metrics import classification_report
10 | from scipy import stats
11 | from sklearn.metrics import accuracy_score
12 | import os
13 |
14 |
15 | """Dataset loading. All the datasets are public available.
16 | PAMAP2: http://archive.ics.uci.edu/ml/datasets/pamap2+physical+activity+monitoring
17 | TUH: https://www.isip.piconepress.com/projects/tuh_eeg/html/downloads.shtml
18 | MNIST: http://yann.lecun.com/exdb/mnist/
19 | Yelp: https://www.yelp.com/dataset
20 | Here we take the PAMAP2 dataset as an example.
21 | """
22 |
23 | """"PAMAP2: Here we use a subset of PAMAP2, totally contains 120,000 samples for 5 subjects (the more subjects,
24 | the better but more computational. We select 5 most commonly used activities (Cycling, standing, walking,
25 | lying, and running, labelled from 0 to4) as a subset for evaluation.12,000 samples for each sub, 51 features.
26 | The dataset is huge, using small subset for debugging is strongly recommended."""
27 |
28 | feature = sc.loadmat("/home/xiangzhang/scratch/AR_6p_8c.mat")
29 | data = feature['AR_6p_8c']
30 | data = np.concatenate((data[0:120000], data[200000*1:200000*1+120000], data[200000*2:200000*2+120000],
31 | data[200000*3:200000*3+120000], data[200000*4:200000*4+120000]
32 | , ), axis=0)
33 | print data.shape
34 | n_classes = 5 # number of classes
35 | keep_rate = 0.5 # kep rate for dropout
36 | len_block = 10 # the window size of each segment
37 | overlap = 0.5 # the overlapping
38 | # data_size_1 = data.shape[0]
39 | no_fea = data.shape[1] - 1 # the number of features
40 | # data segmentation
41 | new_x, new_y, data = extract(data, no_fea, len_block, int(overlap * len_block), n_classes)
42 | print new_x.shape, new_y.shape, data.shape
43 |
44 | # make the data size as a multiple of 5. This operation is prepare for batch splitting.
45 | data_size = data.shape[0] # data size
46 | dot = data_size % 5
47 | data = data[0:data_size - dot]
48 | data_size = data.shape[0] # update data_size
49 | no_fea_long = no_fea * len_block # the feature dimension after segmentation
50 |
51 | # split data into 80% and 20% for training and testing
52 | train_data = data[int(data_size*0.2):]
53 | test_data = data[:int(data_size*0.2)]
54 | np.random.shuffle(train_data)
55 | np.random.shuffle(test_data)
56 | supervision_rate = 0.6 # key parameter: supervision rate
57 | n_labeled = int(train_data.shape[0]*supervision_rate) # the number of labelled training samples
58 | # create training/testing features and labels
59 | train_x = train_data[:, :no_fea_long]
60 | train_y = one_hot(train_data[:, no_fea_long:no_fea_long + 1])
61 | test_x = test_data[:, :no_fea_long]
62 | test_y = one_hot(test_data[:, no_fea_long:no_fea_long + 1])
63 | print 'shape', train_x.shape, train_y.shape, test_x.shape, test_y.shape
64 |
65 | # split semi supervised dataset into labelled and unlabelled
66 | x_l = train_x[0:n_labeled]
67 | y_l = train_y[0:n_labeled]
68 | x_u = train_x[n_labeled:train_data.shape[0]]
69 | y_u = train_y[n_labeled:train_data.shape[0]]
70 | print 'shape_', train_data.shape, np.array(x_l).shape, np.array(x_u).shape
71 |
72 | # dimensions: n_fea_long, n_classes, 1
73 | label_data = np.hstack([x_l, y_l, np.ones([x_l.shape[0], 1])])
74 | unlabelled_data = np.hstack([x_u, y_u, np.zeros([x_u.shape[0], 1])])
75 | joint_data = np.vstack([label_data, unlabelled_data])
76 | # joint_data = label_data # only use labelled data for GAN
77 |
78 | np.random.shuffle(joint_data) # shuffle the labeled data and unlabelled data
79 | train_x_semi = joint_data[:, 0: no_fea_long]
80 | train_y_semi = joint_data[:, no_fea_long: no_fea_long+n_classes]
81 | # flag_ is a mark for labelled or unlabelled, which decide
82 | # use which loss function (L_{label} or L_{unlabel}) to calculate the loss
83 | flag_ = joint_data[:, no_fea_long+n_classes: no_fea_long+n_classes+1]
84 | print 'flag_ shape', flag_.shape
85 | # for the test data, we hope to recognize the specific class from the K classes, thus, we set the flag_test as 1
86 | flag_test = np.ones([test_x.shape[0], 1])
87 |
88 | dim_img = no_fea_long
89 | dim_z = int(no_fea_long)
90 | n_hidden = int(no_fea_long/4)
91 | global n_classes, dim_z
92 | print 'code neurons, hidden neurons', dim_z, n_hidden
93 | learn_rate = 0.0005
94 |
95 | lr_gan = 0.0001
96 | # train
97 | batch_size = test_data.shape[0]
98 | n_samples = train_data.shape[0]
99 | total_batch = int(n_samples / batch_size)
100 | # total_batch = 4 # n_groups
101 | n_epochs = 2000
102 | ADD_NOISE = False
103 |
104 | # input placeholders
105 | # In denoising-autoencoder, x_hat == x + noise, otherwise x_hat == x
106 | x_hat = tf.placeholder(tf.float32, shape=[None, dim_img], name='input_img')
107 | x = tf.placeholder(tf.float32, shape=[None, dim_img], name='target_img')
108 | # dropout
109 | keep_prob = tf.placeholder(tf.float32, name='keep_prob')
110 | """VAE++"""
111 | y, z_s, loss, neg_marginal_likelihood, KL_divergence, z_I, mu, sigma = autoencoder(x_hat, x, dim_img, dim_z, n_hidden,
112 | keep_prob)
113 |
114 | """ Prepare for the semi-supervised GAN, i.e., the discriminator"""
115 | # data preparation for discriminator
116 | n_classes_d = n_classes + 1 # the number of classes in semi-supervised GAN
117 | label = tf.placeholder(tf.float32, shape=[None, n_classes], name='inputlabel') # feed the label in, (n_classes)
118 | flag_in = tf.placeholder(tf.float32, shape=[None, 1], name='flag') # labeled: 1, unlabelled : 0
119 |
120 | final_z = tf.placeholder(tf.float32, shape=[None, dim_z])
121 | label_z = tf.concat([label, tf.zeros(shape=(batch_size, 1))], axis=1)
122 | label_z_s = tf.concat([label, tf.ones(shape=(batch_size, 1))], axis=1)
123 |
124 |
125 | un_label_z = tf.concat([tf.ones(shape=(batch_size, 1)), tf.zeros(shape=(batch_size, 1))], axis=1)
126 | un_label_z_s = tf.concat([tf.zeros(shape=(batch_size, 1)), tf.ones(shape=(batch_size, 1))], axis=1)
127 |
128 | # following with dim: dim_z, n_clsses_d, 2, 1, 1
129 | """Create False/True data for the discriminator"""
130 | z_data = tf.concat([z_I, label_z, un_label_z, flag_in], axis=1)
131 | z_s_data = tf.concat([z_s, label_z_s, un_label_z_s, flag_in], axis=1)
132 | Data = tf.concat([z_data, z_s_data], axis=0)
133 | # np.random.shuffle(Data)
134 | dim_d = dim_z + n_classes_d + 2 + 1 + 1
135 | print 'dim_z', dim_z
136 | # dim_z = 210
137 | d_data = Data[:, 0:dim_z]
138 | super_label = Data[:, dim_z:dim_z+n_classes_d]
139 | unsuper_label = Data[:, dim_d-4: dim_d-2]
140 | flag = Data[:, dim_d-2: dim_d-1]
141 |
142 |
143 | # just a very simple CNN classifier, used to compare the performance with KNN.
144 | # Please set your own parameters based on your situation
145 | def CNN(input):
146 | z_image = tf.reshape(input, [-1, 1, dim_z, 1])
147 | depth_1 = 10
148 | h_conv1 = tf.contrib.layers.convolution2d(z_image, depth_1, [3, 3], activation_fn=tf.nn.relu, padding='SAME')
149 | # fc1 layer
150 | input_size = dim_z*depth_1
151 | size3 = dim_z
152 | fc2_p = fc_relu(data=h_conv1, input_shape=input_size, output_size=size3)
153 | prediction = fc_nosig(data=fc2_p, input_shape=size3, output_size=n_classes_d)
154 |
155 | # change the unlabelled prediction to [1, 0] or [0, 1]
156 | aa = tf.reduce_max(prediction[:, 0:n_classes_d-1], axis=1, keep_dims=True)
157 | un_prediction = tf.concat([aa, prediction[:, n_classes_d-1:n_classes_d]], axis=1)
158 | return prediction, un_prediction, fc2_p
159 |
160 | super_prediction, unsuper_prediction, middle_feature = CNN(d_data)
161 | super_prediction_, _, __ = CNN(final_z)
162 |
163 | """ loss calculation"""
164 | # unsupervised loss
165 | unsuper_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(labels=unsuper_label*0.9,
166 | logits=unsuper_prediction))
167 | # supervised loss
168 | super_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(labels=super_label*0.9, logits=super_prediction))
169 | loss_featurematch = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(labels=d_data, logits=middle_feature))
170 | d_loss = 0.9*flag * super_loss + 0.1*(1-flag)*unsuper_loss # this weights of two loss: 0.9, 0.1
171 |
172 | loss_gan_supervised = super_loss
173 | loss_gan_unsupervised = unsuper_loss
174 | d_loss_plot = loss_gan_supervised + loss_gan_unsupervised
175 | # acc
176 | acc_gan_unsuper = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(unsuper_prediction, 1),
177 | tf.argmax(unsuper_label, 1)), tf.float32))
178 | acc_gan_super = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(super_prediction, 1), tf.argmax(super_label, 1)), tf.float32))
179 |
180 | """Build a CNN classifier"""
181 | acc_final = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(super_prediction_[:, :n_classes], 1),
182 | tf.argmax(label, 1)), tf.float32))
183 | final_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(labels=label * 0.9,
184 | logits=super_prediction_[:, :n_classes]))
185 | train_cnn = tf.train.AdamOptimizer(0.0004).minimize(final_loss)
186 | train_d = tf.train.AdamOptimizer(lr_gan).minimize(d_loss)
187 | # train VAE, add feature matching loss, but it seems the feature matching is not effective
188 | train_op = tf.train.AdamOptimizer(learn_rate).minimize(loss + loss_featurematch)
189 |
190 | """ training """
191 | os.environ['CUDA_VISIBLE_DEVICES'] = '0' # used to control the GPU device
192 | config = tf.ConfigProto()
193 | sess = tf.Session()
194 | init = tf.global_variables_initializer()
195 | sess.run(init)
196 | print 'Start training'
197 | time_s = time.clock()
198 | for epoch in range(n_epochs):
199 | # Loop over data batches
200 | for i in range(total_batch):
201 | # Compute the offset of the current minibatch in the data.
202 | offset = (i * batch_size) % (n_samples)
203 | batch_xs_input = train_x_semi[offset:(offset + batch_size), :] # semi supervised as input
204 | batch_y_input = train_y_semi[offset:(offset + batch_size), :]
205 | flag_input = flag_[offset:(offset + batch_size), :]
206 |
207 | batch_xs_target = batch_xs_input
208 | test_x = test_x
209 |
210 | train_feed = {x_hat: batch_xs_input, x: batch_xs_target, label: batch_y_input,
211 | flag_in: flag_input, keep_prob: keep_rate}
212 | test_feed = {x_hat: test_x, x: test_x, label: test_y, flag_in: flag_test, keep_prob: keep_rate}
213 | # train VAE++
214 | _, tot_loss, loss_likelihood, loss_divergence = sess.run(
215 | (train_op, loss, neg_marginal_likelihood, KL_divergence),
216 | feed_dict={x_hat: batch_xs_input,
217 | x: batch_xs_target,
218 | label: batch_y_input,
219 | flag_in:flag_input,
220 | keep_prob: keep_rate})
221 | # train GAN
222 | sess.run(train_d, feed_dict=train_feed)
223 |
224 | # print cost every epoch
225 | if epoch% 40 == 0:
226 | test_feed = {x_hat: test_x, x: test_x, label: test_y, flag_in: flag_test, keep_prob: keep_rate}
227 | tot_loss_t, loss_likelihood_t, loss_divergence_t, d_loss_t, acc_super, acc_unsuper = sess.run(
228 | (loss, neg_marginal_likelihood, KL_divergence, d_loss, acc_gan_super, acc_gan_unsuper),
229 | feed_dict=test_feed)
230 | print 'epoch', epoch, 'testing loss:', tot_loss_t, np.mean(loss_likelihood_t), np.mean(loss_divergence_t)
231 | print 'epoch', epoch, 'GAN loss, super acc, unsuper_acc', np.mean(d_loss_t), acc_super, acc_unsuper
232 | time_e = time.clock()
233 | test_z = sess.run(mu, feed_dict={x_hat: test_x, x: test_x, keep_prob: keep_rate})
234 | # creat semi_labeled_z based on x_l, the corresponding label is y_l
235 | # the semi_labeled_z is the exclusive code produced by our AVAE.
236 | semi_labeled_z = sess.run(z_I, feed_dict={x_hat: x_l, x: x_l, keep_prob: keep_rate})
237 |
238 | """Classification"""
239 | from sklearn.metrics import classification_report
240 | time1 = time.clock()
241 | print "This is the results:----------------------"
242 |
243 | # Prepare training and testing data for classifier
244 | feature_train = semi_labeled_z
245 | label_train = y_l
246 | feature_test = test_z
247 | label_test = test_y
248 | # KNN classifier. Personally, I prefer to KNN for the lightweight
249 | from sklearn.neighbors import KNeighborsClassifier
250 | time1 = time.clock()
251 | neigh = KNeighborsClassifier(n_neighbors=3)
252 | neigh.fit(feature_train, np.argmax(label_train, 1))
253 | time2 = time.clock()
254 | knn_acc = neigh.score(feature_test, np.argmax(label_test, 1))
255 | knn_prob = neigh.predict_proba(feature_test)
256 | time3 = time.clock()
257 | print classification_report(np.argmax(label_test, 1), np.argmax(knn_prob,1))
258 | print "KNN Accuracy, epoch:", knn_acc, epoch
259 | print "training time", time2 - time1, 'testing time', time3 - time2
260 |
261 | # CNN classifier, in some situations, CNN outperforms KNN
262 | # for i in range(1000):
263 | # train_acc, _, cnn_cost, pred = sess.run([acc_final, train_cnn, final_loss, super_prediction_],
264 | # feed_dict={final_z: feature_train, label: label_train, keep_prob: keep_rate})
265 | # if i % 100 == 0: # test
266 | # test_acc = sess.run(acc_final,
267 | # feed_dict={final_z: feature_test, label: label_test,keep_prob: keep_rate})
268 | # print 'iteration, CNN final acc', i, train_acc, test_acc, cnn_cost
269 |
270 |
--------------------------------------------------------------------------------
/LICENSE:
--------------------------------------------------------------------------------
1 | MIT License
2 |
3 | Copyright (c) 2022 Xiang Zhang
4 |
5 | Permission is hereby granted, free of charge, to any person obtaining a copy
6 | of this software and associated documentation files (the "Software"), to deal
7 | in the Software without restriction, including without limitation the rights
8 | to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
9 | copies of the Software, and to permit persons to whom the Software is
10 | furnished to do so, subject to the following conditions:
11 |
12 | The above copyright notice and this permission notice shall be included in all
13 | copies or substantial portions of the Software.
14 |
15 | THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
16 | IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
17 | FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
18 | AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
19 | LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
20 | OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
21 | SOFTWARE.
22 |
--------------------------------------------------------------------------------
/README.md:
--------------------------------------------------------------------------------
1 | # Adversarial-Variational-Semi-supervised-Learning
2 | ## Title: Adversarial variational embedding for robust semi-supervised learning
3 |
4 | **PDF: [KDD2019](https://dl.acm.org/doi/abs/10.1145/3292500.3330966), [arXiv](https://arxiv.org/abs/1905.02361)**
5 |
6 | **Authors: [Xiang Zhang](http://xiangzhang.info/) (xiang_zhang@hms.harvard.edu), [Lina Yao](https://www.linayao.com/) (lina.yao@unsw.edu.au), Feng Yuan**
7 |
8 | ## Overview
9 | This repository contains reproducible codes for the proposed AVAE model.
10 | In this paper, we present an effective and robust semi-supervised latent representation framework, AVAE, by proposing a modified VAE model and integration with generative adversarial networks. The VAE++ and GAN share the same generator. In order to automatically learn the exclusive latent code, in the VAE++, we explore the latent code’s posterior distribution and then stochastically generate a latent representation based on the posterior distribution. The discrepancy between the learned exclusive latent code and the generated latent representation is constrained by semi-supervised GAN. The latent code of AVAE is finally served as the learned feature for classification.
11 |
12 | 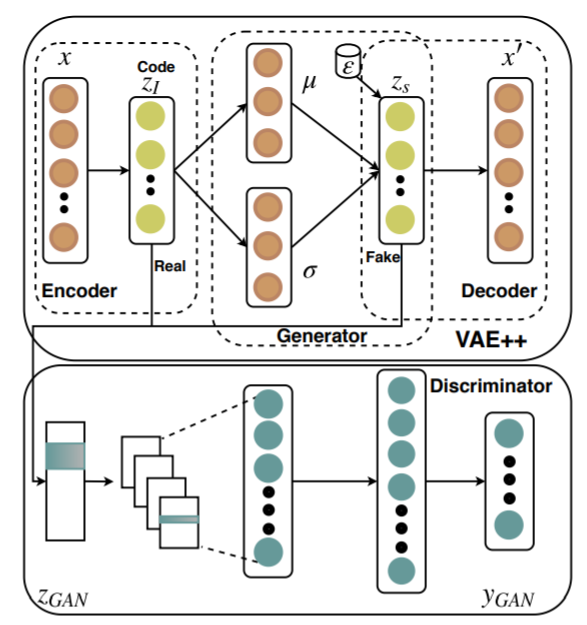 13 |
13 |
14 |
15 | Structure of the proposed AVAE model framewor
16 |
17 |
18 | ## Code
19 | [AVAE.py](https://github.com/xiangzhang1015/Adversarial-Variational-Semi-supervised-Learning/blob/master/AVAE.py) is the main file and other .py files are the related functions.
20 |
21 |
22 | ## Citing
23 | If you find our work useful for your research, please consider citing this paper:
24 |
25 | @inproceedings{zhang2019adversarial,
26 | title={Adversarial variational embedding for robust semi-supervised learning},
27 | author={Zhang, Xiang and Yao, Lina and Yuan, Feng},
28 | booktitle={Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery \& Data Mining},
29 | pages={139--147},
30 | year={2019}
31 | }
32 |
33 | ## Requirements
34 | > Python == 2.7
35 | Numpy == 1.11.2
36 | TensorFlow == 1.3.0
37 |
38 | ## Datasets
39 | All the datasets used in this paper are pretty large which are diffilut to upload to github. Fortunately, the datasets are public available and our paper basically used the raw data, so please access the original data in the following links.
40 |
41 | PAMAP2: http://archive.ics.uci.edu/ml/datasets/pamap2+physical+activity+monitoring
42 |
43 | TUH: https://www.isip.piconepress.com/projects/tuh_eeg/html/downloads.shtml
44 |
45 | MNIST: http://yann.lecun.com/exdb/mnist/
46 |
47 | Yelp: https://www.yelp.com/dataset
48 |
49 | The datasets are huge, using small subset for debugging is strongly recommended. There are very detail comments in the code in order to help understanding.
50 |
51 |
52 | ## Miscellaneous
53 |
54 | Please send any questions you might have about the code and/or the algorithm to .
55 |
56 |
57 | ## License
58 |
59 | This repository is licensed under the MIT License.
60 |
--------------------------------------------------------------------------------
/VAE_plus_plus.py:
--------------------------------------------------------------------------------
1 | import numpy as np
2 | import tensorflow as tf
3 |
4 |
5 | def mean_tile(input_, n_output):
6 | input_ = tf.reduce_mean(input_, axis=1, keep_dims=True)
7 | input_data = tf.tile(input_, [1, n_output])
8 | return input_data
9 |
10 |
11 | # Gaussian MLP as encoder
12 | def gaussian_MLP_encoder(x, n_output):
13 | with tf.variable_scope("gaussian_MLP_encoder"):
14 | # initializer
15 | w_init = tf.contrib.layers.variance_scaling_initializer()
16 | b_init = tf.constant_initializer(0.)
17 |
18 | # only one encoder layer
19 | w0 = tf.get_variable('w0', [x.get_shape()[1], n_output], initializer=w_init)
20 | b0 = tf.get_variable('b0', [n_output], initializer=b_init)
21 | h0 = tf.matmul(x, w0) + b0
22 | z_I = tf.nn.elu(h0)
23 |
24 | # Two methods to calculate mean and std, the results may depends on the dataset, try yourself.
25 | # method 1: the calculated mean and std
26 | # mean, std = tf.nn.moments(z, axes=[1], keep_dims=True)
27 | # mean = tf.tile(mean, [1, n_output])
28 | # stddev = 1e-6 + tf.nn.softplus(std)
29 | # stddev = tf.tile(stddev, [1, n_output])
30 |
31 | # method 2: mean, std under a number of distributions
32 | mean = tf.layers.dense(z_I, units=n_output) # presentation the z_I
33 | stddev = tf.layers.dense(z_I, units=n_output) # presentation the z_I
34 |
35 | return mean, stddev, z_I
36 |
37 |
38 | # Bernoulli MLP as decoder
39 | def bernoulli_MLP_decoder(z, n_hidden, n_output, keep_prob, reuse=False):
40 | with tf.variable_scope("bernoulli_MLP_decoder", reuse=reuse):
41 | # initializers
42 | w_init = tf.contrib.layers.variance_scaling_initializer()
43 | b_init = tf.constant_initializer(0.)
44 | # only one layer decoder
45 | w0 = tf.get_variable('w0', [z.get_shape()[1], n_output], initializer=w_init)
46 | b0 = tf.get_variable('b0', [n_output], initializer=b_init)
47 | y = tf.matmul(z, w0) + b0
48 | return y
49 |
50 |
51 | # Gateway
52 | def autoencoder(x_hat, x, dim_img, dim_z, n_hidden, keep_prob):
53 | # encoding
54 | mu, sigma, z_I = gaussian_MLP_encoder(x_hat, dim_z)
55 | print 'mu shape', mu.shape, sigma.shape
56 |
57 | # sampling by re-parameterization technique
58 | z_s = mu + sigma * tf.random_normal(tf.shape(mu), 0, 1, dtype=tf.float32)
59 | print 'mu, sigma, z_s, mu[0], sigma[0], z_s[0]', mu, sigma, z_s, mu[0], sigma[0], z_s[0]
60 | # decoding
61 | y = bernoulli_MLP_decoder(z_s, n_hidden, dim_img, keep_prob)
62 |
63 | # second loss
64 | logvar_encoder = tf.log(1e-8 + tf.square(sigma))
65 | KL_divergence = -0.5 * tf.reduce_sum(1 + logvar_encoder - tf.pow(mu, 2) - tf.exp(logvar_encoder),
66 | reduction_indices=1)
67 |
68 | loss_recog = tf.reduce_sum(tf.pow(tf.nn.sigmoid_cross_entropy_with_logits(logits=y, labels=x_hat), 2),
69 | reduction_indices=1)
70 |
71 | loss = tf.reduce_mean(loss_recog + KL_divergence)
72 | # loss = tf.reduce_mean(loss_recog) # No KL
73 | return y, z_s, loss, loss_recog, KL_divergence, z_I, mu, sigma
74 |
75 |
76 | def decoder(z, dim_img, n_hidden):
77 | y = bernoulli_MLP_decoder(z, n_hidden, dim_img, 1.0, reuse=True)
78 | return y
79 |
80 |
81 | def maxminnorm(data):
82 | data_norm = (data - np.min(data)) / (np.max(data) - np.min(data))
83 | return data_norm
--------------------------------------------------------------------------------
/functions.py:
--------------------------------------------------------------------------------
1 | import tensorflow as tf
2 | import numpy as np
3 | from sklearn import preprocessing
4 |
5 | def one_hot(y_):
6 | # Function to encode output labels from number indexes
7 | # e.g.: [[5], [0], [3]] --> [[0, 0, 0, 0, 0, 1], [1, 0, 0, 0, 0, 0], [0, 0, 0, 1, 0, 0]]
8 | y_ = y_.reshape(len(y_))
9 | n_values = np.max(y_) + 1
10 | return np.eye(n_values)[np.array(y_, dtype=np.int32)]
11 |
12 | def extract(input, n_fea, time_window, moving, n_classes):
13 | xx = input[:, :n_fea]
14 | xx = preprocessing.scale(xx) # z-score normalization
15 | yy = input[:, n_fea:n_fea+1]
16 | new_x = []
17 | new_y = []
18 | number = int((xx.shape[0]/moving)-1)
19 | for i in range(number):
20 | ave_y = np.average(yy[(i * moving):(i * moving + time_window)])
21 | if ave_y in range(n_classes+1):
22 | new_x.append(xx[(i * moving):(i * moving + time_window), :])
23 | new_y.append(ave_y)
24 | else:
25 | new_x.append(xx[(i * moving):(i * moving + time_window), :])
26 | new_y.append(0)
27 |
28 | new_x = np.array(new_x)
29 | new_x = new_x.reshape([-1, n_fea * time_window])
30 | new_y = np.array(new_y)
31 | new_y.shape =[new_y.shape[0], 1]
32 | data = np.hstack((new_x, new_y))
33 | # data = np.vstack((data[0], data))
34 | # print new_y.shape
35 | return new_x, new_y, data
36 |
37 | # def batch_split(feature, label, n_group, batch_size):
38 | # a = feature
39 | # train_fea = []
40 | # train_label = []
41 | #
42 | # for i in range(n_group):
43 | # f = a[(0 + batch_size * i):(batch_size + batch_size * i)]
44 | # train_fea.append(f)
45 | #
46 | # for i in range(n_group):
47 | # f = label[(0 + batch_size * i):(batch_size + batch_size * i), :]
48 | # train_label.append(f)
49 | # return train_fea, train_label
50 |
51 | # the CNN code
52 | def weight_variable(shape):
53 | initial = tf.truncated_normal(shape, stddev=0.1)
54 | return tf.Variable(initial)
55 |
56 | def bias_variable(shape):
57 | initial = tf.constant(0.1, shape=shape)
58 | return tf.Variable(initial)
59 |
60 | def conv2d(x, W):
61 | # stride [1, x_movement, y_movement, 1]
62 | # Must have strides[0] = strides[3] = 1
63 | print type(x), type(W)
64 | return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
65 |
66 | def max_pool_1x2(x):
67 | # stride [1, x_movement, y_movement, 1]
68 | return tf.nn.max_pool(x, ksize=[1,1,2,1], strides=[1,1, 2,1], padding='SAME')
69 |
70 | def conv(x_image, depth_1):
71 | W_conv1 = weight_variable([2, 2, 1, depth_1]) # patch 5x5, in size is 1, out size is 8
72 | b_conv1 = bias_variable([depth_1])
73 | h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1) + b_conv1) # output size 1*64*2
74 | # h_pool1 = max_pool_1x2(h_conv1) # output size 1*32x2
75 | return h_conv1
76 |
77 | def fc(data, input_shape, output_size):
78 | size2 = output_size
79 | W_fc1 = weight_variable([input_shape, size2])
80 | b_fc1 = bias_variable([size2])
81 | h_pool2_flat = tf.reshape(data, [-1, input_shape])
82 | h_fc1 = tf.nn.sigmoid(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
83 | # h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
84 | return h_fc1
85 | def fc_nosig(data, input_shape, output_size):
86 | size2 = output_size
87 | W_fc1 = weight_variable([input_shape, size2])
88 | b_fc1 = bias_variable([size2])
89 | h_pool2_flat = tf.reshape(data, [-1, input_shape])
90 | h_fc1 = tf.matmul(h_pool2_flat, W_fc1) + b_fc1
91 | # h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
92 | return h_fc1
93 |
94 | def fc_relu(data, input_shape, output_size):
95 | size2 = output_size
96 | W_fc1 = weight_variable([input_shape, size2])
97 | b_fc1 = bias_variable([size2])
98 | h_pool2_flat = tf.reshape(data, [-1, input_shape])
99 | h_fc1 = tf.nn.relu(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
100 | # h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
101 | return h_fc1
102 | def fc_tanh(data, input_shape, output_size):
103 | size2 = output_size
104 | W_fc1 = weight_variable([input_shape, size2])
105 | b_fc1 = bias_variable([size2])
106 | h_pool2_flat = tf.reshape(data, [-1, input_shape])
107 | h_fc1 = tf.tanh(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
108 | # h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
109 | return h_fc1
110 |
111 |
112 |
--------------------------------------------------------------------------------
/structure of proposed Adversarial-Variational-Semi-supervised-Learning.PNG:
--------------------------------------------------------------------------------
https://raw.githubusercontent.com/xiangzhang1015/Adversarial-Variational-Semi-supervised-Learning/6c327fb37085086a6129233170e28b429d1a4c70/structure of proposed Adversarial-Variational-Semi-supervised-Learning.PNG
--------------------------------------------------------------------------------
13 |